ABSTRACT
In 2008, we published the first set of guidelines for standardizing research in autophagy. Since then, this topic has received increasing attention, and many scientists have entered the field. Our knowledge base and relevant new technologies have also been expanding. Thus, it is important to formulate on a regular basis updated guidelines for monitoring autophagy in different organisms. Despite numerous reviews, there continues to be confusion regarding acceptable methods to evaluate autophagy, especially in multicellular eukaryotes. Here, we present a set of guidelines for investigators to select and interpret methods to examine autophagy and related processes, and for reviewers to provide realistic and reasonable critiques of reports that are focused on these processes. These guidelines are not meant to be a dogmatic set of rules, because the appropriateness of any assay largely depends on the question being asked and the system being used. Moreover, no individual assay is perfect for every situation, calling for the use of multiple techniques to properly monitor autophagy in each experimental setting. Finally, several core components of the autophagy machinery have been implicated in distinct autophagic processes (canonical and noncanonical autophagy), implying that genetic approaches to block autophagy should rely on targeting two or more autophagy-related genes that ideally participate in distinct steps of the pathway. Along similar lines, because multiple proteins involved in autophagy also regulate other cellular pathways including apoptosis, not all of them can be used as a specific marker for bona fide autophagic responses. Here, we critically discuss current methods of assessing autophagy and the information they can, or cannot, provide. Our ultimate goal is to encourage intellectual and technical innovation in the field.
KEYWORDS: Autophagosome, cancer, flux, LC3, lysosome, macroautophagy, neurodegeneration, phagophore, stress, vacuole
Table of Contents
A. NomenclatureXX
B. Methods for monitoring autophagyXX
- Additional autophagy-related protein markersXX
- Atg9/ATG9A....XX
- ATG12–ATG5.XX
- ATG14....XX
- ATG16L1....XX
- Atg18/WIPI family....XX
- BECN1/Vps30/Atg6....XX
- STX17XX
- TECPR1XX
- Selective types of autophagy...XX
- Aggrephagy....XX
- Allophagy......XX
- Chlorophagy..XX
- Chromatophagy......XX
- Granulophagy...XX
- Lipophagy....XX
- Lysophagy...XX
- Myelinophagy...XX
- Oxiapoptophagy..XX
- Proteaphagy.XX
- Reticulophagy...XX
- Ribophagy....XX
- Virophagy....XX
- Xenophagy....XX
- Zymophagy....XX
Autophagosome-lysosome colocalization and dequenching assay XX
Tissue fractionation....XX
Analyses in vivo....XX
Clinical setting..XX
Cell death and autophagy......XX
Microautophagy......XX
C. Comments on additional topics.....XX
1. Acidotropic dyes......XX
2. Autophagy inhibitors and inducers.....XX
3. Basal autophagy......XX
4. Experimental systems....XX
5. Bimolecular fluorescence complementation.XX
6. Nanoparticles.XX
7. Nomenclature....XX
8. General considerations for experimental manipulations...XX
D. Methods and challenges of specialized topics/model systems....XX
Chlamydomonas....XX
Filamentous fungi...XX
Honeybee....XX
Human...XX
Hydra..XX
Lepidoptera....XX
Tooth..XX
Parasitic helminths....XX
Planarians...XX
Plants...XX
Protists...XX
Rainbow trout....XX
Sea urchin..XX
Ticks....XX
E. Noncanonical use of autophagy-related proteins....XX
F. Interpretation of in silico assays for monitoring autophagy..XX
1. Sequence comparison and comparative genomics approaches..XX
2. Web-based resources related to autophagyXX
a. The THANATOS database.XX
b. The human autophagy database (HADb)XX
d. The Autophagy Regulatory Network (ARN).XX
e. Prediction of Atg8-family interacting proteins..XX
f. The iLIR server..XX
g. The Eukaryotic Linear Motif resource (ELM).XX
h. Molecular modeling of interactions between Atg8-family proteins and LIR-containing proteinsXX
i. The ncRNA-associated cell death database (ncRDeathDB).XX
j. Predicting impact for autophagy-related gene copy number alterations in cancerXX
k. KFERQ finder....XX
l. Autophagy to Disease (ATD)......XX
m. LysoQuant.XX
3. Mathematical models of autophagy dynamics....XX
Conclusions and Future Perspectives...XX
Table 1. Genetic and pharmacological regulation of autophagy..XX
Table 1.
Genetic and pharmacological regulation of autophagy.2
| Method | Comments |
|---|---|
| 1. 3-methyladenine | A PtdIns3K inhibitor that effectively blocks an early stage of autophagy by inhibiting the class III PtdIns3K, but it is important to note that it is not a specific autophagy inhibitor. 3-MA also inhibits the class I PI3K and can thus, at suboptimal concentrations in long-term experiments, promote autophagy in some systems [436,437], as well as affect cell survival through AKT and other kinases. 3-MA does not inhibit BECN1-independent autophagy. |
| 2. 10-NCP | 10-(4′-N-diethylamino)butyl)-2-chlorophenoxazine; an AKT inhibitor that induces autophagy in neurons [1950]. |
| 3. 17-AAG | An inhibitor of the HSP90-CDC37 chaperone complex; induces autophagy in certain systems (e.g., neurons), but impairs starvation-induced autophagy and mitophagy in others by promoting the turnover of ULK1 [647]. |
| 4. ABG33 | ABG33 (7-aminobenzo[cd]indol-2-(1 H)-one 33 is a small molecule inhibitor of ATG4B enzymatic activity in vitro. In cells, ABG33 results in a dose-dependent increase in LC3B-II levels [2576]. |
| 5. AC220/quizartinib | An FLT3 inhibitor that enhances the inhibitory activity of spautin-1. A70 is an improved derivative of AC220. Treatment sensitizes cancer cells to autophagy inhibition [5]. |
| 6. ACY-1215/ricolinostat | ACY-1215 is a selective HDAC6 inhibitor that inhibits the fusion of lysosomes with autophagosomes and abrogates the clearance of autophagosomes [2577]. |
| 7. AZD8055 | A catalytic MTOR inhibitor that acts as a potent autophagy inducer [2578]. |
| 8. Akti-1/2 | An allosteric inhibitor of AKT1 and AKT2 that promotes autophagy in B-cell lymphoma [2579]. |
| 9. AR-12 (OSU-03012) | A broad-specificity anti-viral celecoxib-derivative that stimulates autophagosome formation and viral protein degradation [2580]. |
| 10. AR7 | AR7 was developed as a highly potent and selective enhancer of CMA through antagonizing RARA/RARα; AR7 is the first small molecule developed to selectively stimulate CMA without affecting autophagy [2581]. |
| 11. ARN5187 | Lysosomotropic compound with a dual inhibitory activity against the circadian regulator NR1D2/REV-ERBβ and autophagy [2582]. |
| 12. AS-605,240 | A selective PIK3CG/PI3Kγ inhibitor that activates autophagy in the heart [2583]. |
| 13. ATG4C74A | An active site mutant of ATG4 that is defective for autophagy [2584]. |
| 14. Autophinib | An autophagy inhibitor that targets the lipid kinase PIK3C3/VPS34 [2585]. |
| 15. Bafilomycin A1 | A V-ATPase inhibitor that causes an increase in lysosomal/vacuolar pH, and, ultimately, blocks fusion of autophagosomes with the vacuole; the latter may result from inhibition of ATP2A/SERCA [303]. |
| 16. Benzothiadiazole | A chemical analog of salicylic acid, which can be used to induce autophagy and autophagosome formation in plant cells including A. thaliana [128,2586]. |
| 17. Betulinic acid | A pentacyclic triterpenoid that promotes parallel damage in mitochondrial and lysosomal compartments, and, ultimately, jeopardizes lysosomal degradative capacity, which results in autophagy-associated cell death [314] or aging [315]. |
| 18. Butein | A plant-derived natural molecule that induces autophagy through the activation of AMPK [2587]. |
| 19. C12TPP | Dodecyltriphenylphosphonium is a penetrating cation that selectively accumulates in mitochondria, uncouples oxidative phosphorylation and stimulates autophagy and mitophagy without inhibition of autophagosome-lysosome fusion, in contrast to protonophores [2588]. |
| 20. Calcium | An intracellular signal that can promote autophagy at different steps. Calcium can be released from the ER upon physiological stimulation or from lysosomal stores under stress conditions, or can enter from the extracellular space [2001]. However, calcium has a complex effect as it can also inhibit autophagy, and the abrogation of calcium signaling can trigger autophagy [216,1990,1994,2589]. |
| 21. Carbamazepine | Induces autophagy by reducing inositol levels, and inhibits autophagy via neuronal voltage-gated sodium channels [1976,2590]. |
| 22. CB-5083 | A selective inhibitor of VCP/p97-mediated protein degradation that activates autophagy in human cancer cells [2591,2592]. |
| 23. CCCP | Carbonyl cyanide m-chlorophenylhydrazone is a prototype protonophore, uncoupler of oxidative phosphorylation that stimulates autophagy via the AMPK-ULK1 pathway [671,672] or alternative pathways [2593] and mitophagy [339], but inhibits autophagosome-lysosome fusion due to the increase of intralysosomal pH [215]. |
| 24. Chloroquine, NH4Cl | Lysosomotropic compounds that elevate/neutralize the lysosomal/vacuolar pH [225]. |
| 25. Cinacalcet HCl | A calcimimetic that increases the sensitivity of CASR (calcium sensing receptor) to extracellular calcium. In some models, cinacalcet induces the formation of GFP-LC3 puncta [494] during starvation, whereas in others it causes an increase in LC3-II accumulation in basal [1563,2594,2595] and CQ conditions [2594]. In a diabetic nephropathy model, the proposed pathway through cinacalcet-induced autophagy is CAMKK2/CaMKKβ-STK11/LKB1-AMPK-PPARGC1A/PGC1α to decrease oxidative stress, which results in a decrease of apoptosis (increased BCL2:BAX ratio) and increased autophagy (increase of BECN1 and LC3-I to LC3-II conversion) [2595]. Cinacalcet may have a dual effect inducing autophagosome formation and inhibiting the late steps of autophagy. |
| 26. Clonidine | Activates the imidazoline receptor, which decrease cAMP in cells. An MTOR-independent inducer of autophagy [1951] |
| 27. Concanamycin A | A specific inhibitor of V-ATPases that reduces acidification of the lysosome or vacuole, and will block the degradation of autophagic bodies within the vacuole [128,2586]. |
| 28. DFMO | α-difluoromethylornithine is an irreversible inhibitor of ODC1 (ornithine decarboxylase 1) that blocks spermidine synthesis and ATG gene expression [2596]. |
| 29. DMMB | A photosensitizer derivative of methylene blue that promotes parallel damage in lysosomes and mitochondria after photoactivation with red light, leading to accumulation of non-functional autolysosomes and autophagy-associated cell death [316]. |
| 30. Docosahexaenoic | An omega-3 polyunsaturated fatty acid, that has been described as acid (DHA) an activator of autophagy, which could potentially be used in cancer therapy either alone or in combinatorial strategies, as well as in neurodegenerative, cardiovascular or infectious diseases [2597-2599]. |
| 31. E-64c | A derivative of E-64, a cysteine protease inhibitor. |
| 32. E-64d | A membrane-permeable cysteine protease inhibitor that can block the activity of a subset of lysosomal hydrolases; should be used in combination with pepstatin A to inhibit lysosomal protein degradation. The ethyl ester of E-64c. |
| 33. Eriocalyxin B | An autophagy inducer that exerts anti-tumor activity in breast cancer by inhibition of the AKT-MTOR-RPS6KB signaling pathway [2600]. |
| 34. ESC8 | A cationic estradiol derivative that induces autophagy and apoptosis simultaneously by downregulating the MTOR kinase pathway in breast cancer cells. |
| 35. Everolimus | An inhibitor of MTORC1 that induces both autophagy and apoptosis in B-cell lymphoma primary cultures [2579]. |
| 36. Ezetimibe | A cholesterol absorption inhibitor that acts by binding to NPC1L1, which induces autophagy via MTORC1-dependent [2601] and -independent [2602] pathways. Ezetimibe also activates TFEB and could potentially exert therapeutic effects on steatohepatitis and fibrosis [2601,2602]. |
| 37. Fasudil | An inhibitor of ROCK (Rho associated coiled-coil containing protein kinase) enhancing autophagy via phosphorylation of MAPK8/JNK1 and BCL2, and promoting BECN1-PIK3C3/VPS34 complex formation; shRNA-mediated approaches to inhibiting ROCK have similar results [2603-2605]. |
| 38. Flavonoids | A large class of polyphenols that have been described as autophagy modulators, which could potentially constitute useful adjuvant agents of conventional therapies for different human pathologies such as cancer, neurodegenerative, cardiovascular, hepatic or infectious diseases [2606]. |
| 39. Fumonisin B1 | An inhibitor of ceramide synthesis that interferes with autophagy. |
| 40. Gene deletion | This method provides the most direct evidence for the role of an autophagic component; however, more than one gene involved in autophagy should be targeted to avoid indirect effects. |
| 41. HBHA | Heparin-binding hemagglutinin of M. tuberculosis that inhibits autophagy. HBHA treatment inhibits LC3 expression and the maturation of autophagosomes, eventually inducing apoptosis [2607]. |
| 42. HMOX1 induction | Mitophagy and the formation of iron-containing cytoplasmic inclusions and corpora amylacea are accelerated in HMOX1-transfected rat astroglia and astrocytes of GFAP HMOX1 transgenic mice. Heme-derived ferrous iron and carbon monoxide, products of the HMOX1 reaction, promote autophagy in these cells [2608,2609]. |
| 43. Knockdown | This method (including miRNA, RNAi, shRNA and siRNA) can be used to inhibit gene expression and provides relatively direct evidence for the role of an autophagic component. However, the efficiency of knockdown varies, as does the stability of the targeted protein. In addition, more than one gene involved in autophagy should be targeted to avoid misinterpreting indirect effects. |
| 44. KU-0063794 | An MTOR inhibitor that binds the catalytic site and activates autophagy [453,2610]. |
| 45. Leupeptin | An inhibitor of cysteine, serine and threonine proteases that can be used in combination with pepstatin A and/or E-64d to block lysosomal protein degradation. Leupeptin is not membrane permeable, so its effect on cathepsins may depend on endocytic activity. |
| 46. LV-320 | A small molecule inhibitor of ATG4A and ATG4B enzymatic activity in vitro. In cells, LV-320 results in a dose-dependent increase in LC3B-II levels, reduces GABARAP levels and reduces autophagic flux [2611]. |
| 47. MB | A phenothiazine photosensitizer that promotes specific photodamage in lysosomes when used at low doses and photoactivated with red light. By targeting lysosomes to photodamage, MB can promptly switch autophagy to favor cell demise when parallel mitochondrial membrane damage by hydrogen peroxide or rotenone occurs [316]. |
| 48. Melatonin | N-acetyl-5-methoxy tryptamine is a sleep–wake cycle regulating and antioxidant hormone that inhibits autophagy in animal models of fibrosis [2612], cancer [2613] and acute organ failure [2614]. |
| 49. Metformin | Activates both AMPK-dependent and -independent autophagy [2615-2617]. |
| 50. microRNA | Can be used to reduce the levels of target mRNA(s) or block translation. |
| 51. MK2206 | A small molecule inhibitor of AKT that is able to induce Autophagy independently of MTORC1 activity [973,2618]. |
| 52. MLN4924 | A small molecule inhibitor of NAE (NEDD8 activating enzyme) [2619]; induces autophagy by blockage of MTOR activity via both DEPTOR and the HIF1A-DDIT4/REDD1-TSC1/2 axis as a result of inactivation of cullin-RING ligases [2620]. |
| 53. Mycolactone | A polyketide lactone and virulence exotoxin of Mycobacterium ulcerans that functions by blocking SEC61-dependent translocation of proteins into the ER [2022]. Mycolactone induces the integrated stress response [2021] and autophagy [2020,2021]. |
| 54. NAADP-AM | Activates the lysosomal TPCN/two-pore channel and induces autophagy [1996]. |
| 55. NED-19 | Inhibits the lysosomal TPCN and NAADP-induced autophagy [1996]. |
| 56. NeuroHeal | A combination of acamprosate and ribavirin that activates SIRT1 and autophagy, promoting neuroprotection [2015,2016]. |
| 57. NSC611216 | A small molecule inhibitor of ATG4B enzymatic activity in vitro [2576]. |
| 58. NVP-BEZ235 | A dual inhibitor of PIK3CA/p110 and the MTOR catalytic site that activates autophagy [2621,2622]. |
| 59. p140/Lupuzor™ | Small peptide that inhibits LAMP2A overexpression in lupus B cells and binds to the NBD domain of HSPA8 [2623,2624]. Furthermore, this drug has been described as a potent CMA inhibitor [2625]. |
| 60. Pathogen-derived factors | Virally-encoded autophagy inhibitors including HSV-1 ICP34.5, Kaposi sarcoma-associated herpesvirus vBCL2, γ-herpesvirus 68 M11, ASFV vBCL2, HIV-1 Nef and influenza A virus M2 [848,1418,1423,1424,1966]. |
| 61. Pepstatin A | An aspartyl protease inhibitor that can be used to partially block lysosomal degradation; should be used in combination with other inhibitors such as E-64d. Pepstatin A is not membrane permeable. |
| 62. PMI | SQSTM1/p62-mediated mitophagy inducer is a pharmacological activator of autophagic selection of mitochondria that operates without collapsing the mitochondrial membrane potential (ΔΨm) and hence by exploiting the autophagic component of the process [714]. |
| 63. Propolis | An inducer of autophagy that may be related with the classical autophagy pathway [2626]. |
| 64. Protease inhibitors | These chemicals inhibit the degradation of autophagic substrates within the lysosome/vacuole lumen. A combination of inhibitors (e.g., leupeptin, pepstatin A and E-64d) is needed for complete blockage of degradation. |
| 65. Rapamycin | Binds to FKBP1A/FKBP12 and inhibits MTORC1; the complex binds to the FRB domain of MTOR and limits its interaction with RPTOR, thus inducing autophagy, but only providing partial MTORC1 inhibition. Rapamycin also inhibits yeast TOR. |
| 66. Resveratrol | A natural polyphenol that affects many proteins [2627] and induces autophagy via activation of AMPK [2628, 2629]. |
| 67. RNAi | Can be used to inhibit gene expression. |
| 68. RSVAs | Synthetic small-molecule analogs of resveratrol that potently activate AMPK and induce autophagy [2630]. |
| 69. Saikosaponin-d | A natural small-molecule inhibitor of ATP2A/SERCA that induces autophagy and autophagy-dependent cell death in apoptosis-resistant cells [1909]. |
| 70. SAR405 | A low-molecular-mass kinase inhibitor of PIK3C3/VPS34 that interacts within the ATP binding cleft of human PIK3C3 and inhibits autophagy [1883]. |
| 71. SB02024 | Potent and selective PIK3C3/VPS34 inhibitor that binds in the active site of PIK3C3, thus inhibiting its catalytic function [2631]. |
| 72. SBI-0206965 | A highly selective ULK1 kinase inhibitor in vitro that suppresses ULK1-mediated phosphorylation events in cells, regulating autophagy and cell survival [2632]. This compound is also an inhibitor of AMPK, competitively inhibiting ATP binding, and also inhibiting the binding of AMPK to its substrates [2633]. |
| 73. Sorafenib | An antitumoral inhibitor of tyrosine kinase receptors whose sustained administration induces a shift from early induction of survival autophagy to apoptosis [2634]. |
| 74. SMER28 | An MTOR-independent inducer of autophagy [2013]. |
| 75. Spautin-1 | An autophagy inhibitor that acts via suppression of USP10 and USP13, and degradation of the PIK3C3/VSP34-BECN1 complex [2635]. |
| 76. Spermidine | A chemical originally isolated from semen and enriched in many food products; it promotes autophagy flux by depleting cytosolic HDAC4 to enhance MAP1S-mediated autophagy [956]. Spermidine maintains basal autophagy in NIH 3T3 cells and B cells of mice or humans via hypusination of EIF5A and subsequent upregulation of TFEB [958]. |
| 77. Sulforaphane | A natural isothiocyanate, alone and in combination with cytostatics induces cell death via autophagy, and elevates the level of LC3-II [1905,2636,2637]. |
| 78. Tat-beclin 1 | A cell penetrating peptide that potently induces autophagy [2004,2638]. |
| 79. Thapsigargin | An inhibitor of ATP2A/SERCA that inhibits autophagic sequestration through the depletion of intracellular calcium stores [216,2639]; however, thapsigargin may also block fusion of autophagosomes with endosomes by interfering with recruitment of RAB7, resulting in autophagosome accumulation [2640]. |
| 80. TMS | Trans-3,5,4-trimethoxystilbene upregulates the expression of TRPC4, resulting in MTOR inhibition [2641]. |
| 81. Torin1 | A catalytic MTOR inhibitor that induces autophagy and provides more complete inhibition than rapamycin (it inhibits all forms of MTOR) [802]. |
| 82. TPCK | An inducer of autophagic cell death. |
| 83. TPPS2a | TPPS2a photoexcitation promotes mainly lysosomal damage leading to autophagy-associated cell death [317]. |
| 84. Trehalose | A membrane-protective agent [2642] and inducer of autophagy that may be relevant for the treatment of different neurodegenerative diseases [973, 2036, 2643-2645]. |
| 85. Tunicamycin | A glycosylation inhibitor that induces autophagy due to ER stress [473,2646]. |
| 86. Vacuolin-1 | A RAB5A activator that reversibly blocks autophagosome-lysosome fusion [2647]. |
| 87. Verteporfin | An FDA-approved drug; used in photodynamic therapy, but it inhibits the formation of autophagosomes in vivo without light activation [2648]. |
| 88. Vinblastine | A depolymerizer of both normal and acetylated microtubules that interferes with autophagosome-lysosome fusion [304]. |
| 89. VP2.51 | A small molecule, ATP-competitive inhibitor of GSK3B enzymatic activity in vitro. In vivo, VP2.51 modulates autophagy and ameliorates motor neuron disease [2649]. |
| 90. VPS34-IN1 | A low-molecular-mass kinase inhibitor of PIK3C3/VPS34 similar to SAR-405 that interacts within the ATP binding cleft of human PIK3C3 and inhibits autophagy [2650]. |
| 91. Wortmannin | An inhibitor of PI3K and PtdIns3K that blocks autophagy, but is not a specific inhibitor (see 3-MA above). |
| 92. Yessotoxin (YTX) | A small molecule marine compound that can potentially induce autophagic-associated cell death [1515]. YTX can induce various cell death modalites [2651]; its molecular target and mode of action are not yet clarified. |
Table 2. Phosphorylation targets of AKT, AMPK, GSK3B, MTORC1, PKA and Atg1/ULK1......XX
Table 2.
Phosphorylation targets of AKT, AMPK, GSK3B, MTORC1, PKA and Atg1/ULK1
| Protein and phosphorylation site | Main kinase | Function | Ref |
|---|---|---|---|
| Acc1 (S1157 in yeast) | Snf1 | Inhibits de novo lipogenesis required for stationary phase autophagy | [655] |
| AMBRA1 S52 | MTORC1 | Inhibits AMBRA1-dependent activation of ULK1 | [741] |
| Atg1 | TORC1 | Inhibits Atg1 kinase activity | [744] |
| Atg1 | PKA | Regulation of kinase activity | [2652] |
| ATG4B S316 | ULK1 | Inhibits ATG4B activity and LC3 processing | [708] |
| Atg9 | Atg1 | Recruitment of Atg protein to the PAS | [2653] |
| ATG9 S14 | ULK1 | Promotes ATG9 trafficking in response to starvation | [707] |
| ATG9 S761 | AMPK | Participates in the recruitment of lipids to the phagophore | [2654] |
| Atg13 | TORC1 | Interaction with Atg1, assembly of Atg1 kinase complex | [744, 2655] |
| Atg13 | PKA | Regulates localization to the PAS | [2656] |
| ATG13 S318 | ULK1 | Required for clearance of depolarized mitochondria | [647] |
| ATG14 S29 | ULK1 | Promotes autophagy by increasing PtdIns3K complex activity | [702, 2657] |
| BECN1 S14 | ULK1 | Increases the activity of the PtdIns3K | [705] |
| BECN1 S30 | ULK1 | Activates the ATG14-containing PtdIns3K complex and stimulates autophagosome formation in response to amino acid starvation, hypoxia, and MTORC1 inhibition. | [706] |
| BECN1 S90 | MAPKAPK2-MAPKAPK3 | Stimulates autophagy | [2658] |
| BECN1 S91, S94 (S93, S96 in human) | AMPK | Required for glucose starvation-induced autophagy | [868] |
| BECN1 Y229, Y233 | EGFR | Inhibits autophagy | [775] |
| BECN1 S234, S295 | AKT | Suppresses autophagy | [774] |
| BECN1 unknown site | ERBB2/HER2 | Inhibits autophagy | [2659, 2660] |
| CCNY (cyclin Y) S326 | AMPK | Stimulates interaction with CDK16 and promotes autophagy | [762] |
| FUNDC1 S17 | ULK1 | Promotes mitophagy by enhancing FUNDC1 binding to LC3 | [735] |
| HTT S421 | AKT | Activates HTT clearance | [2661] |
| LC3 S12 | PKA | Inhibits autophagy by reducing recruitment to phagophores | [293] |
| MTOR S2448 | AKT | Correlates with the activity of MTORC1 | [2662] |
| MTOR S2481 | Autophosphorylation | Necessary for MTORC1 formation and kinase activity | [2663] |
| NBR1 T586 | GSK3A/B | Modulates protein aggregation | [2664] |
| RPS6KB T389 | MTORC1 (apparently indirect, through reduction of dephosphorylation) | Necessary for protein activity | [2665] |
| RPS6KB S371 | GSK3B | Necessary for T389 phosphorylation and the activity of RPS6KB | [2666] |
| RPTOR S792 | AMPK | Suppresses MTORC1 | [689] |
| RUBCNL/Pacer S157 | MTORC1 | Repress RUBCNL interaction with STX17 and HOPS complex | [2667] |
| SQSTM1 S293 | AMPK (S293/S294 in rat and human sequence, respectively) | Promotes autophagic cell death | [657] |
| SQSTM1 S403 | ULK1 (also TBK1, CSNK, CDK1) | Promotes autophagic degradation of SQSTM1 and its substrates | [2668] |
| TFEB S122, S142, S211 | MTORC1 | Inhibits TFEB nuclear translocation | [970–972, 2669] |
| TFEB S467 | AKT1 | Inhibits TFEB nuclear translocation | [973] |
| TSC2 T1227, S1345 |
AMPK | Negative regulator of MTORC1 | [670] |
| ULK1 S317, S555, S574, S673 | AMPK | Required for mitophagy, mitochondrial homeostasis, and cell survival | [672] |
| ULK1 S467, S777 (mouse) | AMPK | Increase the kinase activity of ULK1 and promote autophagy | [672, 673] |
| ULK1 S757/S758 (mouse/human) | MTORC1 | Facilitates ULK1 interaction with AMPK | [688] |
| ULK1 S757 | MTORC1 | Prevents ULK1 interaction with AMPK | [673] |
| ULK1 S637 | MTORC1, AMPK | Facilitates ULK1 interaction with AMPK | [688] |
| ULK1 (uncertain site between 278 and 351) | Autophosphorylation | Modulates the conformation of the C-terminal tail and prevents its interaction with ATG13 | [646, 699] |
| USP14 S432 | AKT | Overcomes negative regulation of DNA repair | [2670] |
| UVRAG S498 | MTORC1 | Negatively regulates autophagosome and endosome maturation | [2671] |
| UVRAG S550, S571 | MTORC1 | Activates the PtdIns3K-UVRAG complex to regulate autolysosomal tubulation | [2672] |
Table 3. Eukaryotic linear motif entries related to the LIR-motif....XX
Table 3.
Eukaryotic linear motif entries related to the LIR motif.4
| ELM identifier | ELM | Description | Status |
|---|---|---|---|
| LIG_LIR_Gen_1 | [EDST].{0,2}[WFY]..[ILV] | Canonical LIR motif that binds to Atg8-family protein members to mediate processes involved in autophagy. | ELM |
| LIG_LIR_Apic_2 | [EDST].{0,2}[WFY]..P | Apicomplexa-specific variant of the canonical LIR motif that binds to Atg8-family protein members to mediate processes involved in autophagy. | ELM |
| LIG_LIR_Nem_3 | [EDST].{0,2}[WFY]..[ILVFY] | Nematode-specific variant of the canonical LIR motif that binds to Atg8-family protein members to mediate processes involved in autophagy. | ELM |
| LIG_LIR_LC3C_4 | [EDST].{0,2}LVV | Noncanonical variant of the LIR motif that binds to Atg8-family protein members to mediate processes involved in autophagy. | ELM |
| LIG_AIM | [WY]..[ILV] | Atg8-family protein interacting motif found in Atg19, SQSTM1, ATG4B and CALR (calreticulin), involved in autophagy-related processes. | Candidate |
| LIG_LIR | WxxL or [WYF]xx[LIV] | The LIR might link ubiquitinated substrates that should be degraded to the autophagy-related proteins in the phagophore membrane. | Candidate |
| LIG_GABARAP | W.FL | GABAA receptor binding to clathrin and CALR; possibly linked to trafficking. | Candidate |
Table 4. Recommended methods for monitoring autophagy....XX
Table 4.
Recommended methods for monitoring autophagy.5
| Method | Description | |
|---|---|---|
| 1 | Atg8-family protein western blotting | Western blot. The analysis is carried out in the absence and presence of lysosomal protease or fusion inhibitors to monitor flux; an increase in the LC3-II amount in the presence of the inhibitor is usually indicative of flux. |
| 2 | Atg18 oligomerization | FRET stopped-flow assay, chemical cross-linking, mass spectrometry. |
| 3 | Autophagic protein degradation | Turnover of long-lived proteins to monitor flux. |
| 4 | Autophagic sequestration assays | Accumulation of cargo in autophagic compartments in the presence of lysosomal protease or fusion inhibitors by biochemical or multilabel fluorescence techniques. |
| 5 | Autophagosome quantification | FACS/flow cytometry. |
| 6 | Autophagosome-lysosome colocalization and dequenching assay | Fluorescence microscopy. |
| 7 | Bimolecular fluorescence complementation | Can be used to monitor protein-proteim interaction in vivo. |
| 8 | Degradation of endogenous lipofuscin | Fluorescence microscopy. |
| 9 | Electron microscopy | Quantitative electron microscopy, immuno-TEM; monitor autophagosome number, volume, and content/cargo. |
| 10 | FRET | Interaction of LC3 with gangliosides to monitor autophagosome formation. |
| 11 | GFP-Atg8-family protein fluorescence microscopy | Fluorescence microscopy, flow cytometry to monitor vacuolar/lysosomal localization. Also, increase in punctate GFP-Atg8-family protein or Atg18/WIPI, and live time-lapse fluorescence microscopy to track the dynamics of GFP-Atg8-family protein-positive structures. |
| 12 | GFP-Atg8-family protein lysosomal delivery and proteolysis | Western blot +/- lysosomal fusion or degradation inhibitors; the generation of free GFP indicates lysosomal/vacuolar delivery. |
| 13 | Immunofluorescence for endogenous LC3 puncta | Can be used to identify autophagosomes in cells difficult to transfect with a GFP-LC3 chimera. |
| 14 | Keima | Confocal microscopy, flow cytometry, western blotting to monitor transfer of Keima or various Keima fusion variants to acidic and proteolytically active environments. |
| 15 | MTOR, AMPK and Atg1/ULK1 kinase activity | Western blot, immunoprecipitation or kinase assays. |
| 16 | Pex14-GFP, GFP-Atg8, Om45-GFP, mitoPho8Δ60 | A range of assays can be used to monitor selective types of autophagy. These typically involve proteolytic maturation of a resident enzyme or degradation of a chimera, which can be followed enzymatically or by western blot. |
| 17 | Sequestration and processing assays in plants | Chimeric RFP fluorescence and processing, and light and electron microscopy. |
| 18 | SQSTM1- and related LC3-binding protein turnover | The amount of SQSTM1 increases when autophagy is inhibited and decreases when autophagy is induced, but the potential impact of transcriptional and/or translational regulation or the formation of insoluble aggregates should be addressed in individual experimental systems. |
| 19 | Tandem mRFP/mCherry-GFP fluorescence microscopy, Rosella | Flux can be monitored as a decrease in green/red (yellow) fluorescence (phagophores, autophagosomes) and an increase in red fluorescence (autolysosomes). |
| 20 | Tissue fractionation | Centrifugation, western blot and electron microscopy. |
| 21 | Transcriptional and translational regulation | Northern blot, or RT-PCR, autophagy-dedicated microarray. |
| 22 | Turnover of autophagic compartments | Electron microscopy with morphometry/stereology at different time points. |
| 23 | WIPI fluorescence microscopy | Quantitative fluorescence analysis using endogenous WIPI proteins, or GFP- or MYC-tagged versions. Suitable for high-throughput imaging procedures. |
References......XX
Glossary..XX
Quick guide..XX
Index..XX
Abbreviations: 3-MA, 3-methyladenine; ABC, avidin-biotin peroxidase complex; AD, Alzheimer disease; ADCD, autophagy-dependent cell death; AIM, Atg8-family interacting motif; ALIS, aggresome-like induced structures; ALS, amyotrophic lateral sclerosis; AMA, antimycin A; AMCD, autophagy-mediated cell death; AML, acute myeloid leukemia; ARN, Autophagy Regulatory Network; ASFV, African swine fever virus; Atg, autophagy-related; AV, autophagic vacuole; BDI, bright detail intensity; CASA, chaperone-assisted selective autophagy; CF, cystic fibrosis; CHX, cycloheximide; CLEAR, coordinated lysosomal enhancement and regulation; CLEM, correlative light and electron microscopy; CMA, chaperone-mediated autophagy; CQ, chloroquine; cryo-SXT, cryo-soft X-ray tomography; CS, centriolar satellite; Cvt, cytoplasm-to-vacuole targeting; DAMP, danger/damage-associated molecular pattern; DN, diabetic nephropathy; DQ-BSA, dequenched bovine serum albumin; e-MI, endosomal microautophagy; EBSS, Earle’s balanced salt solution; EM, electron microscopy; ER, endoplasmic reticulum; ERAD, endoplasmic reticulum associated degradation; ERLAD, ER-to-lysosome-associated degradation; ESCRT, endosomal sorting complex required for transport; EST, expressed sequence tag; FACS, fluorescence-activated cell sorter; FIB-SEM, focused ion beam-scanning electron microscopy; FRET, Förster resonance energy transfer; FTD, frontotemporal dementia; GAP, GTPase activating protein; GBP, guanylate binding protein; GFP, green fluorescent protein; GPCR, G protein-coupled receptor; GSCs, glioma stem-like cells; HCQ, hydroxychloroquine; HD, Huntington disease; HIV-1, human immunodeficiency virus type 1; HKP, housekeeping protein; HOPS, homotypic fusion and vacuole protein sorting; HSV-1, herpes simplex virus type 1; Hyp-PDT, hypericin-based photodynamic therapy; ICD, immunogenic cell death; IHC, immunohistochemistry; ILVs, intralumenal vesicles; IMP, intramembrane particle; LAP, LC3-associated phagocytosis; LDS, LIR/AIM docking site; LIR, LC3-interacting region; LIRCPs, LIR-containing proteins; LN, late nucleophagy; LPS, lipopolysaccharide; LRP1-ICD, LRP1 intracellular domain; MAMs, mitochondria-associated ER membranes; MAP1LC3/LC3, microtubule associated protein 1 light chain 3; MDC, monodansylcadaverine; MDH, monodansylpentane; MEC, mammary epithelial cells; MEFs, mouse embryonic fibroblasts; MELAS, mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; mRFP, monomeric red fluorescent protein; MS, mass spectrometry; mtDNA, mitochondrial DNA; MTOR, mechanistic target of rapamycin kinase; MTORC1, mechanistic target of rapamycin kinase complex 1; MVB, multivesicular body; NAADP, nicotinic acid adenine dinucleotide phosphate; NASH, nonalcoholic steatohepatitis; ncRNA, noncoding RNA; NETs, neutrophil extracellular traps; NMR, nuclear magnetic resonance; NPs, nanoparticles; NTs, nanotubes; NVJ, nucleus-vacuole junction; OMM, outer mitochondrial membrane; OS, outer segments; PAMP, pathogen-associated molecular pattern; paNPs, photo-activable NPs; PAS, phagophore assembly site; PD, Parkinson disease; PDT, photodynamic therapy; PE, phosphatidylethanolamine; PI, phosphoinositide; PI3K, phosphoinositide 3-kinase; PMN, piecemeal microautophagy of the nucleus; PMSF, phenylmethylsulphonylfluoride; POFs, postovulatory follicles; POS, photoreceptor outer segments; PPI, protein-protein interaction; PS, phosphatidylserine; PSSM, position-specific scoring matrix; PtdIns3K, phosphatidylinositol 3-kinase; PtdIns3P, phosphatidylinositol-3-phosphate; PTECs, proximal tubular epithelial cells; PTM, posttranslational modification; PVM, parasitophorus vacuole membrane; qPCR/qRT-PCR, quantitative polymerase chain reaction; RBC, red blood cell; RCBs, Rubisco-containing bodies; RCD, regulated cell death; Rluc, Renilla reniformis luciferase; ROS, reactive oxygen species; RPE, retinal pigment epithelium; RT-PCR, reverse transcription polymerase chain reaction; S1P, sphingosine-1-phosphate; SD, standard deviation; SEM, scanning electron microscopy; SINGD, stress-induced nascent granule degradation; SKL, serine-lysine-leucine (a peroxisome targeting signal); SLE, systemic lupus erythematosus; SOD, superoxide dismutase; SVCV, spring viremia of carp virus; TEM, transmission electron microscopy; tfLC3, tandem fluorescent LC3; TORC1, TOR complex I; TR-FRET, time-resolved Förster resonance energy transfer; TVA, tubulovesicular autophagosome; UDS, UIM docking site; UIM, ubiquitin-interacting motif; UPR, unfolded protein response; UPS, ubiquitin-proteasome system; V-ATPase, vacuolar-type H+-translocating ATPase; VHSV, viral hemorraghic septicemia virus; WSM, water stress medium; xLIR, extended LIR-motif; ZIKV, Zika virus
Introduction
Many researchers, especially those new to the field, need to determine which criteria are essential for demonstrating autophagy, either for the purposes of their own research, or in the capacity of a manuscript or grant review [1,2]. Acceptable standards are an important issue, particularly considering that each of us may have her/his own opinion regarding the answer. Furthermore, as science progresses and the field evolves, the answer is in part a “moving target” [3]. This can be extremely frustrating for researchers who may think they have met those criteria, only to find out that the reviewers of their work disagree. Conversely, as a reviewer, it is tiresome to raise the same objections repeatedly, wondering why researchers have not fulfilled some of the basic requirements for establishing the occurrence of an autophagic process. In addition, drugs that potentially modulate autophagy are increasingly being used in clinical trials, and screens are being carried out for new drugs that can modulate autophagy for therapeutic purposes. Clearly, it is important to determine whether these drugs are truly affecting autophagy, and which step(s) of the process/es is/are affected, based on a set of accepted criteria. To this aim, we describe here a basic set of updated guidelines that can be used by researchers to plan and interpret their experiments, by clinicians to evaluate the literature with regard to autophagy-modulating therapies, and by both authors and reviewers to justify or criticize an experimental approach.
Several fundamental points must be kept in mind as we establish guidelines for the selection of appropriate methods to monitor autophagy [2]. Importantly, there are no absolute criteria for determining autophagic status that are applicable in every single biological or experimental context. This is because some assays are unsuitable, problematic or may not work at all in particular cells, tissues or organisms [1–4]. For example, autophagic responses to drugs may be different in transformed versus nontransformed cells, in confluent versus nonconfluent cells, or in cells grown with or without glucose [5]. These guidelines are likely to evolve as new methodologies are developed and current assays are superseded. Nonetheless, it is useful to establish a reference for acceptable assays that can reliably monitor autophagy in many experimental systems. It is important to note that in this set of guidelines the term “autophagy” generally refers to macroautophagy; other autophagy-related processes are specifically designated when appropriate.
For the purposes of this review, the autophagic compartments (Figure 1) are referred to as the sequestering (pre-autophagosomal) phagophore (PG; previously called the isolation or sequestration membrane [6, 7]) [8], the double-membrane autophagosome (AP; generated by scission of the phagophore membrane [9, 10]), the single-membrane amphisome (AM; generated by the fusion of the outer autophagosomal membrane with endosomes) [11], the lysosome (LY), the autolysosome (AL; generated by fusion of the outer autophagosomal membrane or amphisome with a lysosome), and the autophagic body (AB; generated by fusion of the outer autophagosomal membrane with, typically, the vacuole in fungi and plants followed by the release of the internal autophagosomal compartment into the vacuole lumen). Except for cases of highly stimulated autophagic sequestration (Figure 2), autophagic bodies are not seen in animal cells, because lysosomes/autolysosomes are typically smaller than autophagosomes [11]. One critical point is that autophagy is a highly dynamic, multi-step process. Like other cellular pathways, it can be modulated at several steps, both positively and negatively. An accumulation of autophagosomes measured by transmission electron microscopy (TEM) image analysis [12], identified as green fluorescent protein (GFP)-MAP1LC3 (GFP-LC3) puncta under fluorescence microscopy, or as changes in the amount of lipidated LC3 (LC3-II) on a western blot, could reflect a reduction in autophagosome turnover [13–15], or the inability of turnover to keep pace with increased autophagosome formation (Figure 1B) [16]. For example, inefficient fusion with endosomes and/or lysosomes, or perturbation of the transport machinery [17], would inhibit autophagosome maturation to amphisomes or autolysosomes (Figure 1C), whereas decreased flux could also be due to inefficient degradation of the cargo once fusion has occurred [18]. Moreover, GFP-LC3 puncta and LC3 lipidation can reflect the induction of a different/modified pathway such as LC3-associated phagocytosis (LAP) [19], or the noncanonical destruction pathway of paternal mitochondria after egg fertilization [20, 21].
Figure 1.
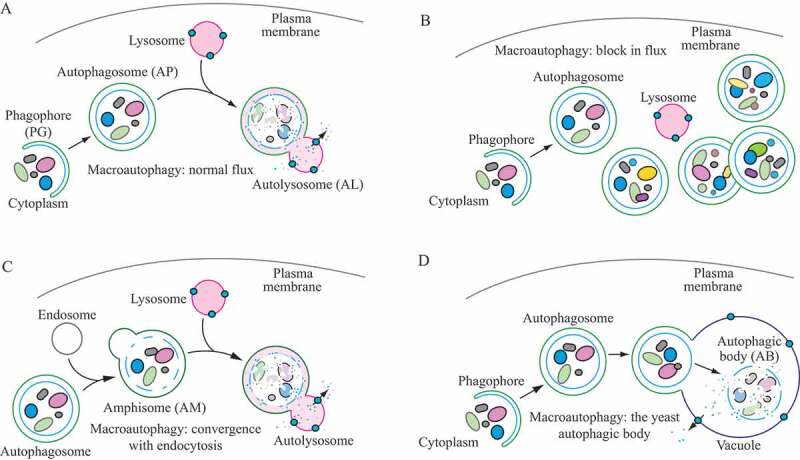
Schematic model demonstrating the induction of autophagosome formation when turnover is blocked versus normal autophagic flux, and illustrating the morphological intermediates of autophagy. (A) The initiation of autophagy includes the formation and expansion of the phagophore, the initial sequestering compartment, which expands into an autophagosome. Completion of the autophagosome requires an intraphagophore membrane scission step and is followed by fusion of the outer autophagosomal membrane with lysosomes and degradation of the contents, allowing complete flux, or flow, through the entire pathway. This is a different outcome than the situation shown in (B) where induction results in the initiation of autophagy, but a defect in autophagosome turnover due, for example, to a block in fusion with lysosomes or disruption of lysosomal functions will result in an increased number of autophagosomes. In this scenario, autophagy has been induced, but there is no or limited autophagic flux. (C) An autophagosome can fuse with an endosome to generate an amphisome, prior to fusion with the lysosome. (D) Schematic drawing showing the formation of an autophagic body in fungi. The large size of the fungal vacuole relative to autophagosomes allows the release of the single-membrane autophagic body within the vacuole lumen. In cells that lack vacuolar hydrolase activity, or in the presence of inhibitors that block hydrolase activity, intact autophagic bodies accumulate within the vacuole lumen and can be detected by light microscopy. The lysosome of most more complex eukaryotes is too small to accommodate an autophagic body
Figure 2.
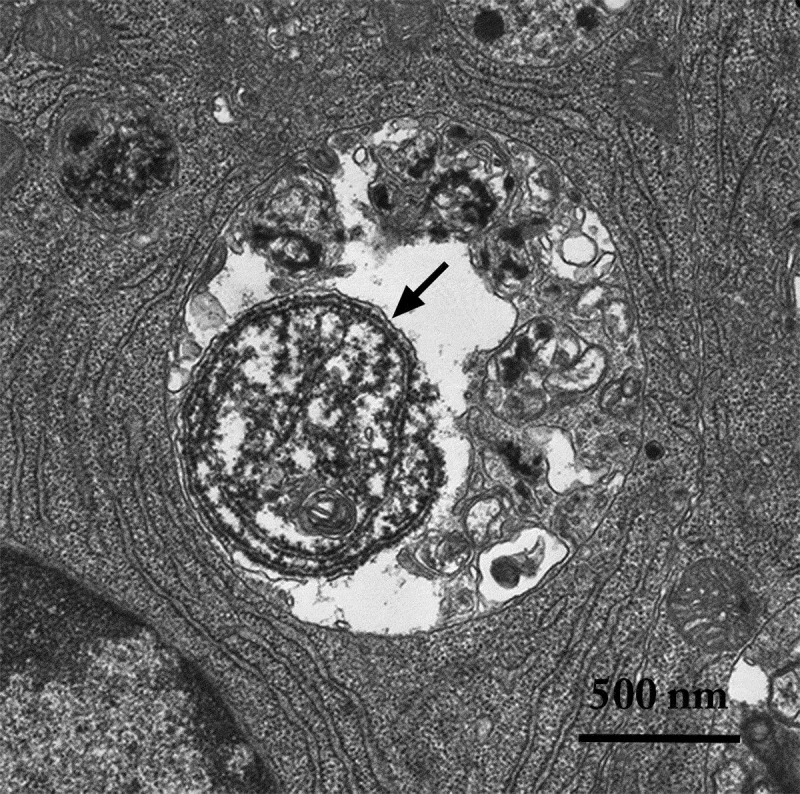
An autophagic body in a large lysosome of a mammalian epithelial cell in a mouse seminal vesicle in vitro. The arrow shows the single limiting membrane covering the sequestered rough ER. Image provided by A.L. Kovács
Thus, the use of autophagy markers such as LC3-II must be complemented by assays to estimate overall autophagic flux, or flow, to permit a correct interpretation of the results. That is, autophagic activity includes not just the increased synthesis or lipidation of Atg8/LC3/GABARAP (LC3 and GABARAP subfamilies constitute the mammalian homologs of yeast Atg8), or an increase in the formation of autophagosomes, but, most importantly, flux through the entire system, including lysosomes or the vacuole, and the subsequent release of the breakdown products. Therefore, autophagic substrates need to be monitored dynamically over time to verify that they have reached the lysosome/vacuole, and whether or not they are degraded. By responding to perturbations in the extracellular environment, cells tune the autophagic flux to meet intracellular metabolic demands and support repair mechanisms. The impact of autophagic flux on cell death and human pathologies, therefore, demands accurate tools to measure not only the current flux of the system, but also its capacity [22], and its response time, when exposed to a defined stress [23].
One approach to evaluate autophagic flux is to measure the rate of general protein breakdown by autophagy [6, 24, 25]. It is possible to arrest the autophagic flux at a given point, and then record the time-dependent accumulation of an organelle, an organelle marker, a cargo marker, or the entire cargo at the point of blockage; however, this approach assumes there is no feedback of the accumulating structure on its own rate of formation [26]. Thus, the chase period should be kept short, ideally with more than one time point. In an alternative approach, one can follow the time-dependent decrease of an autophagy-degradable marker following inhibition of protein synthesis (with the caveat that the potential contribution of other proteolytic systems needs to be experimentally addressed). A potential complication here is that inhibition of protein synthesis, for example, by cycloheximide (CHX), can activate MTORC1 signaling, which in turn impairs autophagy [27]. In theory, these nonautophagic processes can be assessed if degradation persists after blocking autophagic sequestration [13, 15, 28]. The key issue is to differentiate between the often transient accumulation of autophagosomes due to increased induction, and their accumulation due to inefficient clearance of sequestered cargos. This can be done by both measuring the levels of autophagosomes at static time points, and by measuring changes in the rates of autophagic degradation of cellular components, or, in neurons, by assaying autophagosome transport [18, 29]. Multiple strategies have been used to estimate “autophagy,” but unless the experiments can relate changes in autophagosome quantity to a direct or indirect measurement for autophagic flux, the results may be difficult to interpret [30]. A general caution regarding the use of the term “steady state” is warranted at this point. It should not be assumed that an autophagic system is at steady state in the strict biochemical meaning of this term, as this implies that the level of autophagosomes does not change with time, and the flux through the system is constant. In these guidelines, we use “steady state” to refer to the baseline range of autophagic flux in a system that is not subjected to specific perturbations that increase or decrease that flux.
Autophagic flux refers to the entire process of autophagy over a period of time, which encompasses the selection of cargo and its inclusion within the autophagosome, the delivery of cargo to lysosomes (via fusion of the latter with autophagosomes or amphisomes) and its subsequent breakdown and release of the resulting macromolecules back into the cytosol, which may be referred to as productive or complete autophagy. Thus, increases in the level of phosphatidylethanolamine (PE)-modified Atg8-family proteins (Atg8–PE, or LC3/GABARAP-II), or even the appearance of autophagosomes, are not measures of autophagic flux per se, but can reflect the induction of autophagic sequestration and/or inhibition of autophagosome or amphisome clearance. Also, it is important to realize that while formation of Atg8–PE (or LC3/GABARAP-II) appears to correlate with the induction of autophagy, we do not know, at present, the actual mechanistic relationship between Atg8–PE (or LC3/GABARAP-II) formation and the rest of the autophagic process; indeed, some variants of autophagy proceed in the absence of LC3-II [31–35].
In addition, as the metabolic control of autophagy is becoming increasingly clear, highlighting a tight network between the autophagy machinery, energy sensing pathways and the cell’s metabolic circuits [36, 37], mitochondrial parameters such as fission and fusion rate and the cell’s ATP demand should be monitored and correlated with autophagic flux data. In this regard, the use of mitochondria-localized mCherry-GFP tandem reporters (such as the mito-QC mouse [38]), may be important in understanding how deregulated mitophagy affects the progression of metabolic disorders, including diabetes [39]. These types of studies will provide a better understanding on the variability of autophagy and cell death susceptibility.
As a final note, we also recommend that researchers refrain from the use of the expression “percent autophagy” when describing experimental results, as in “The cells displayed a 25% increase in autophagy.” Instead, it is appropriate to indicate that the average number of GFP-Atg8-family protein puncta per cell is increased or a certain percentage of cells displayed punctate GFP-Atg8-family proteins that exceeds a particular threshold (and this threshold should be clearly defined in the Methods section), or that there is a specific increase or decrease in the rate of cargo sequestration or the degradation of long-lived proteins, when these are the actual measurements being quantified.
In previous versions of these guidelines [1, 3], the methods were separated into two main sections—steady state and flux. In some instances, a lack of clear distinction between the actual methodologies and their potential uses made such a separation somewhat artificial. For example, fluorescence microscopy was initially listed as a steady-state method, although this approach can clearly be used to monitor flux as described in this article, especially when considering the increasing availability of new technologies such as microfluidic chambers. Furthermore, the use of multiple time points and/or lysosomal fusion/degradation inhibitors can turn even a typically static method such as TEM into one that monitors flux. Therefore, although we maintain the importance of monitoring autophagic flux and not just induction, this revised set of guidelines does not separate the methods based on this criterion. Readers should be aware that this article is not meant to present protocols, but rather guidelines, including information that is typically not presented in protocol papers. For detailed information on experimental procedures we refer readers to various protocols that have been published elsewhere [25, 40–56]. Finally, throughout the guidelines we provide specific cautionary notes, and these are important to consider when planning experiments and interpreting data; however, these cautions are not meant to be a deterrent to undertaking any of these experiments or a hindrance to data interpretation.
Collectively, we propose the following guidelines for measuring various aspects of selective and nonselective autophagy in eukaryotes.
A. Nomenclature
To minimize confusion regarding nomenclature, we make the following notes: In general, we follow the conventions established by the nomenclature committees for each model organism whenever appropriate guidelines are available, and briefly summarize the information here using “ATG1” as an example for yeast and “ULK1” for mammals. The standard nomenclature of autophagy-related wild-type genes, mutants and proteins for yeast is ATG1, atg1 (or atg1∆ in the case of deletions) and Atg1, respectively, according to the guidelines adopted by the Saccharomyces Genome Database (https://www.yeastgenome.org/). For mammals we follow the recommendations of the International Committee on Standardized Genetic Nomenclature for Mice (http://www.informatics.jax.org/mgihome/nomen/), which dictates the designations Ulk1, ulk1 and ULK1 (for all rodents), respectively, and the guidelines for human genes established by the HUGO Nomenclature Committee (http://www.genenames.org/guidelines.html), which states that human gene symbols are in the form ULK1 and recommends that proteins use the same designation without italics, as with ULK1; mutants are written for example as ULK1−/− [57]. For simplicity unless referring to a specific species, the human gene/protein symbols and definitions will be used throughout the guidelines.
B. Methods for monitoring autophagy
1. Transmission electron microscopy
Autophagy was first detected by TEM in the 1950s (reviewed in ref. [6]). This process was originally observed as focal degradation of cytoplasmic areas performed by lysosomes. Later analysis revealed that autophagy starts with the sequestration of portions of the cytoplasm by a special double-membrane structure (termed the phagophore), which matures into the autophagosome, also delimited by a double membrane. Subsequent fusion events expose the cargo to the lysosome (or the vacuole in fungi or plants) for enzymatic breakdown.
The importance of TEM in autophagy research lies in several qualities. It is the only tool that reveals the morphology of autophagic structures at a resolution in the nm range; shows these structures in their natural environment and position among all other cellular components; allows their exact identification; and, in addition, can support quantitative studies if the rules of proper sampling are followed [12].
Autophagy can be both selective and nonselective, and TEM can be used to monitor both. In the case of selective autophagy, the cargo is the specific substrate being targeted for sequestration—bulk cytoplasm is essentially excluded. In contrast, during nonselective autophagy, disposable cytoplasmic constituents are sequestered. Sequestration of larger structures (such as big lipid droplets, extremely elongated or branching mitochondria or the entire Golgi complex) is rare, indicating an apparent upper size limit for individual autophagosomes. However, it has been observed that under special circumstances the potential exists for the formation of huge autophagosomes, which can even engulf a complete nucleus [28]. Cellular components that form large confluent areas excluding bulk cytoplasm, such as organized, functional myofibrillar structures, do not seem to be sequestered by autophagy. The situation is less clear with regard to glycogen [58–60].
Plant cell-specific structures called provacuoles have a striking similarity to a phagophore, but form in an autophagy-independent manner [61]. These structures have been detected in cells undergoing major changes in vacuolar morphology, such as meristematic cells [62]. Thus, using TEM to detect autophagosomes in plant cells must be done while comparing with an appropriate autophagy-deficient control sample.
After sequestration, the content of the autophagosome and its bordering double membrane remain morphologically unchanged, and recognizable for at least several minutes. During this period, the membranes of the sequestered organelles (for example the ER or mitochondria) remain intact, and the electron density of ribosomes is conserved at normal levels. Degradation of the sequestered material and the corresponding deterioration of ultrastructure commences and runs to completion within the amphisome and the autolysosome after fusion with a late endosome and lysosome (the vacuole in fungi and plants), respectively (Figure 1) [63]. The sequential morphological changes during the autophagic process can be followed by TEM [64]. The maturation from the phagophore through the autolysosome is a dynamic and continuous process [65], and, thus, the classification of compartments into discrete morphological subsets can be problematic; therefore, some basic guidelines for such classifications are offered below.
In the preceding sections the “autophagosome”, the “amphisome” and the “autolysosome” were terms used to describe or indicate three basic stages and compartments of autophagy. It is important to make it clear that for instances (which may be many) when we cannot or do not want to differentiate among the autophagosomal, amphisomal and autolysosomal stage we use the general term “autophagic vacuole”. In the yeast autophagy field, the term “autophagic vesicle” is used to avoid confusion with the primary vacuole, and by now the two terms are used in parallel and can be considered synonyms. It is strongly recommended, however, to use only the term “autophagic vacuole” when referring to autophagy in more complex eukaryotic cells. Autophagosomes, also referred to as initial autophagic vacuoles (AVi), typically have a double membrane. This structure is usually distinctly visible by TEM as two parallel membrane layers (bilayers) separated by a relatively narrower or wider electron-translucent cleft, even when applying the simplest routine TEM fixation procedure (Figure 3A) [66, 67]. This electron-translucent cleft, however, is less visible in freeze-fixed samples, suggesting it may be an artefact of sample preparation (see Fig. S3 in ref. [68]). Amphisomes [69] can sometimes be identified by the presence of small intralumenal vesicles [70]. These intralumenal vesicles are delivered into the lumen by fusion of the autophagosome/autophagic vacuole (AV) limiting membrane with multivesicular endosomes, and care should, therefore, be taken in the identification of the organelles, especially in cells that produce large numbers of multivesicular body (MVB)-derived exosomes (such as tumor or stem cells) [71]. Late/degradative autophagic vacuoles/autolysosomes (AVd or AVl) typically have only one limiting membrane; frequently they contain electron-dense cytoplasmic material and/or organelles at various stages of degradation (Figure 3A and B) [63, 72]; however, late in the digestion process they may contain only a few membrane fragments and be difficult to distinguish from lysosomes, endosomes, or tubular smooth ER cut in cross-section. It is not always easy to morphologically distinguish amphisomes, autolysosomes and lysosomes, even for an expert [6]. A simple solution to assess autophagy progression is to group all of these structures, which are typically stained dark in TEM samples, and define them as degradative compartments/vacuoles. As autophagy induction leads to an increase of autophagosomes, amphisomes and autolysosomes, an increase of degradative compartments per cell area provides a simple measurement to determine whether this degradative pathway is enhanced [73–75]. Unequivocal identification of these structures and of lysosomes devoid of visible content requires immuno-EM detection of a cathepsin or other lysosomal hydrolase (e.g., ACP2 [acid phosphatase 2, lysosomal] [76, 77]) that is detected on the limiting membrane of the lysosome [78]. Smaller, often electron dense, lysosomes may predominate in some cells and exhibit hydrolase immunoreactivity within the lumen and on the limiting membrane [79].
Figure 3.
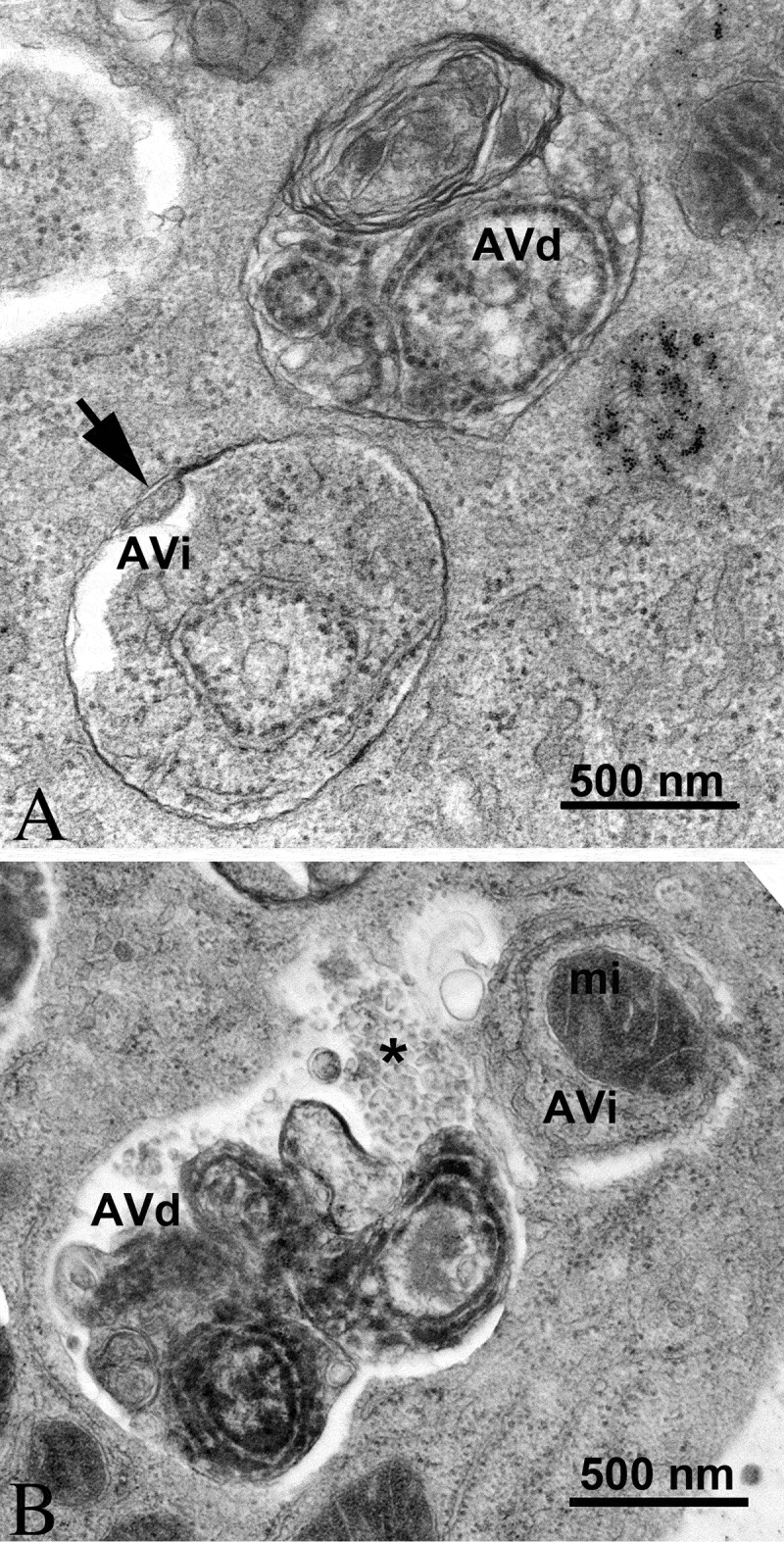
TEM images of autophagic vacuoles in isolated mouse hepatocytes. (A) One autophagosome or early autophagic vacuole (AVi) and one degradative autophagic vacuole (AVd) are shown. The AVi can be identified by its contents (morphologically intact cytoplasm, including ribosomes, and rough ER), and the limiting membrane that is partially visible as two bilayers separated by a narrow electron-lucent cleft, i. e., as a double membrane (arrow). The AVd can be identified by its contents, partially degraded, electron-dense rough ER. The vesicle next to the AVd is an endosomal/lysosomal structure containing 5-nm gold particles that were added to the culture medium to trace the endocytic pathway. (B) One AVi, containing rough ER and a mitochondrion, and one AVd, containing partially degraded rough ER, are shown. Note that the limiting membrane of the AVi is not clearly visible, possibly because it is tangentially sectioned. However, the electron-lucent cleft between the two limiting membranes is visible and helps in the identification of the AVi. The AVd contains a region filled by small internal vesicles (asterisk), indicating that the AVd has fused with a multivesicular endosome. mi, mitochondrion. Image provided by E.-L. Eskelinen
In addition, structural proteins of the lysosome/late endosomes, such as LAMP1 and LAMP2 or SCARB2/LIMP-2, can be used for confirmation. No single protein marker, however, has been effective in discriminating autolysosomes from the compartments mentioned above, in part due to the dynamic fusion and “kiss-and-run” events that promote interchange of components that can occur between these organelle subtypes. Rigorous further discrimination of these compartments from each other and other vesicles ultimately requires demonstrating the colocalization of a second marker indicating the presence of an autophagic substrate (e.g., LC3 and CTSD [cathepsin D] colocalization) or the acidification of the compartment (e.g., mRFP/mCherry-GFP-LC3 probes or LysoTracker™ dyes; see Tandem mRFP/mCherry-GFP fluorescence microscopy), Keima probes, or BODIPY-pepstatin A that allows detection of CTSD in an activated form within an acidic compartment), and, when appropriate, by excluding markers of other vesicular components [76, 80, 81].
The sequential deterioration of cytoplasmic structures being digested can be used for identifying autolysosomes by TEM. Even when the partially digested and destroyed structure of the cytoplasmic cargo cannot be recognized in itself, it can be traced back to earlier forms by identifying preceding stages of sequential morphological deterioration. Degradation usually leads first to the increased electron density of still recognizable organelles, then to vacuoles with heterogeneous density, which become more homogeneous and amorphous, mostly electron dense, but sometimes light (i.e., electron translucent). It should be noted that, in pathological states, it is not uncommon that active autophagy of autolysosomes and damaged lysosomes (“lysophagy”) may yield populations of double-membrane limited autophagosomes containing partially digested amorphous substrate in the lumen. These structures, which are enriched in hydrolases, are seen in swollen dystrophic neurites in some neurodegenerative diseases, and in cerebellar slices cultured in vitro and infected with prions. Alternatively, it is possible to inhibit the fusion of autophagosomes and lysosomes using bafilomycin A1 (a vacuolar-type H+-translocating ATPase [V-ATPase] inhibitor). It is then possible to both visualize the cargo(s) that are being actively sequestered within AVi structures during the chase period, as well as quantify their rates of formation provided the chase period is kept short [82] (Figure 4).
Figure 4.
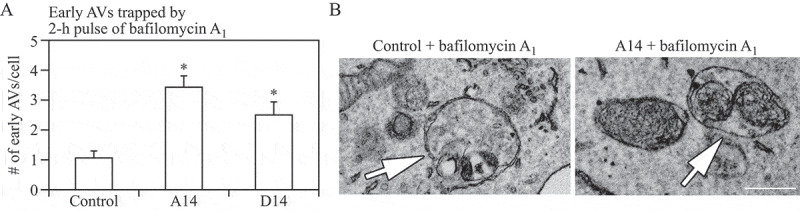
Autophagosomes with recognizable cargo are rare in cells. (A) To assess relative rates of autophagosome formation, the lysosomal inhibitor bafilomycin A1 (10 nM) was applied for 2 h prior to fixation with 2% glutaraldehyde in order to trap newly formed autophagosomes (note that whereas short-term treatment with bafilomycin A1 in most cases primarily blocks autolysosomal degradation, it can also inhibit autophagosome-lysosome fusion). Two different PINK1 shRNA lines (A14 and D14) exhibit increased AV formation over 2 h compared to the control shRNA line. *, p < 0.05 vs. Control. (B) Autophagosomes in bafilomycin A1-treated control cells contain a variety of cytoplasmic structures (left, arrow), whereas mitochondria comprise a prominent component of autophagosomes in bafilomycin A1-treated (PINK1 shRNA) cells (right, arrow). Scale bar: 500 nm. These data indicate induction of selective mitophagy in PINK1-deficient cells. This figure was modified from Figure 2 published in Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Human Molecular Genetics 2010; 19:R28-R37
It must be emphasized that in addition to the autophagic input, other processes (e.g., endosomal, phagosomal, chaperone-mediated) also carry cargo to the lysosomes [64,65], in some cases through the intermediate step of direct endosome fusion with an autophagosome to form an amphisome. This process is exceptionally common in the axons of neurons [83, 84]. Therefore, strictly speaking, we can only have a lytic compartment containing cargos arriving from several possible sources; however, we still may use the term “autolysosome” if the content appears to be overwhelmingly autophagic. Note that the engulfment of dying cells via phagocytosis also produces lysosomes that contain cytoplasmic structures, but in this case, it originates from the dying cell [85]; hence the possibility of an extracellular origin for such content must be considered when monitoring autophagy in settings where apoptotic cell death may be reasonably expected or anticipated.
For many physiological and pathological situations, the examination of both early and late autophagic vacuoles yields valuable data regarding the overall autophagy status in the cells [16, 86]. Along these lines, it is possible to use immunocytochemistry to follow particular cytosolic proteins such as SOD1/Cu,Zn-superoxide dismutase and CA (carbonic anhydrase) to determine the stage of autophagy; the former is much more resistant to lysosomal degradation [87].
In some autophagy-inducing conditions it is possible to observe multi-lamellar membrane structures in addition to the conventional double-membrane autophagosomes, although the nature of these structures is not fully understood. These multi-lamellar structures may indeed be multiple double layers of phagophores [88] and positive for LC3 [89], they could be autolysosomes [90], or they may form as an artefact of fixation. Depending on the cell type, it may be necessary to distinguish these from myelin or surfactant, both of which are also multilamellar. These multi-lamellar bodies are typical in lysosomal storage diseases, such as Niemann-Pick disease type I [91] and Parkinson disease (PD) [92–94]. In addition, cells treated with U18666A, an inhibitor of cholesterol transport [95, 96], or chloroquine (CQ) that induces phospholipidosis [97], produce numerous large multi-lamellar bodies with concentric membrane stacks that represent dysfunctional lysosomes, containing undegraded phospholipids and cholesterol. Multi-lamellar bodies are formed through cellular autophagy, and the implication of various lysosomal enzymes in their formation suggests a lysosomal nature. Initially, single or multiple foci of lamella appear within an autophagic vacuole and then progress into multi-lamellar structures [90, 93] as they are getting filled with lipids; these lipids are cholesterol-containing rafts in late endocytic/lysosomes organelles [94].
Special features of the autophagic process may be clarified by immuno-TEM with gold-labeling [98, 99], using antibodies, for example, to cargo proteins of cytoplasmic origin and to LC3 to verify the autophagic nature of the compartment. LC3 immunogold labeling also enables the detection of novel degradative organelles within autophagy compartments. This is the case with the autophagoproteasome [100] that consists of single-, double-, or multiple-membrane LC3-positive autophagosomes costaining for specific components of the ubiquitin-proteasome system (UPS). It may be that a rich multi-enzymatic (both autophagic and UPS) activity takes place within these organelles instead of being segregated within different domains of the cell. Also in plants, TEM immunogold labelling for ATG8 ultrastructural detection can be performed. This can be approached using either anti-GFP antibodies for GFP-ATG8 fusion proteins, or anti-ATG8 antibodies for direct labeling [101, 102]. Freeze-substitution followed by cryo embedding in acrylic resins is the most convenient and feasible processing method for ATG8 immunogold labelling in plant cells.
Although labeling of LC3 can be difficult, an increasing number of commercial antibodies are becoming available, including reagents that enable visualization of the GFP moiety of GFP-LC3 reporter constructs [103]. It is important to keep in mind that LC3 can be associated with nonautophagic structures (see Xenophagy, and Noncanonical use of autophagy-related proteins), and that LC3 puncta can be observed in autophagy-deficient cells [104]. LC3 is involved in specialized forms of endocytosis such as LC3-associated phagocytosis. In addition, LC3 can decorate vesicles dedicated to exocytosis in nonconventional secretion systems (reviewed in ref. [105, 106]). Antibodies against an abundant cytosolic protein will result in high labeling all over the cytoplasm; however, organelle markers work well. Because there are very few characterized proteins that remain associated with the closed autophagosomes, the choices for confirmation of their autophagic nature are limited. Furthermore, autophagosome-associated proteins may be cell-, age-, sex- and/or condition-specific. Sex-specific expression of autophagic markers are observed both in humans and in rats [107–111]. At any rate, the success of this methodology depends on the quality of the antibodies and also on the TEM preparation and fixation procedures utilized. With immuno-TEM, authors should provide controls showing that labeling is specific. This may require a quantitative comparison of labeling over different cellular compartments not expected to contain antigen and those containing the antigen of interest.
It is difficult to clearly monitor autophagy in tissues of formalin-fixed and paraffin-embedded biopsy samples retrospectively, because (a) tissues fixed in formalin have low or no LC3 detectable by routine immunostaining, (b) because phospholipids melt together with paraffin during the sample preparation, and (c) immuno-EM of many tissues not optimally fixed for this purpose (e.g., using rapid fixation) produces low-quality images. Combining antigen retrieval with the avidin-biotin peroxidase complex (ABC) method may be quite useful for these situations. For example, immunohistochemistry can be performed using an antigen retrieval method, and then tissues are stained by the ABC technique using a labeled anti-human LC3 antibody. After imaging by light microscopy, the same prepared slides can be remade into sections for TEM examination, which can reveal peroxidase reaction deposits in vacuoles within the region that is LC3-immunopositive by light microscopy [112].
In addition, statistical information should be provided due to the necessity of showing only a selective number of sections in publications. Again, we note that for quantitative data it is necessary to use proper volumetric analysis rather than just counting numbers of sectioned objects. On the one hand, it must be kept in mind that even volumetric morphometry/stereology only shows either steady-state levels, or a snapshot in a changing dynamic process. Such data by themselves are not informative regarding autophagic flux, unless carried out over multiple time points. Alternatively, investigation in the presence and absence of flux inhibitors can reveal the dynamic changes in various stages of the autophagic process [13, 22, 55, 113, 114]. On the other hand, if the turnover of autolysosomes is very rapid, a low number/volume in the experimental compared to the basal condition, will not necessarily be an accurate reflection of low autophagic activity; as with autophagosomes, a smaller number of autolysosomes can reflect increased degradation or decreased formation. However, quantitative analyses indicate that autophagosome volume in many cases does correlate with the rates of protein degradation [115–117]. One potential compromise is to perform whole cell quantification of autophagosomes using fluorescence methods, with qualitative verification by TEM [118], to show that the changes in fluorescent puncta reflect corresponding changes in autophagic structures.
One additional caveat with TEM, and to some extent with confocal fluorescence microscopy, is that the analysis of a single plane within a cell can be misleading and may make the identification of autophagic structures difficult. Confocal microscopy and fluorescence microscopy with deconvolution software (or with much more work, 3-dimensional TEM) can be used to generate multiple/serial sections of the same cell to reduce this concern; however, in many cases where there is sufficient structural resolution, analysis of a single plane in a relatively large cell population can suffice given practical limitations. EM technologies, such as focused ion beam scanning electron microscopy (SEM), Serial Block Face-SEM, and Automatic Tape-collecting Ultramicrotomy for SEM, should make it much easier to apply 3-dimensional analyses. An additional methodology to assess autophagosome accumulation is correlative light and electron microscopy (CLEM), which is helpful in confirming that fluorescent structures are autophagosomes [119–122]. Along these lines, it is important to note that even though GFP fluorescence will be quenched in the acidic environment of the autolysosome, some of the GFP puncta detected by fluorescence microscopy may correspond to early autolysosomes prior to GFP quenching. These numbers may increase substantially in pathological conditions where lysosomal/autolysosomal acidification is impaired. The mini Singlet Oxygen Generator (miniSOG) fluorescent flavoprotein, which is less than half the size of GFP, provides an additional means to genetically tag proteins for CLEM analysis under conditions that are particularly suited to subsequent TEM analysis [123], with the caveat that single oxygen targets aromatic amino acids, promoting artefactual protein damage as well as double bonds in lipids, promoting lipid peroxidation [124]. Combinatorial assays using tandem monomeric red fluorescent protein (mRFP)-GFP-LC3 (see Tandem mRFP/mCherry-GFP fluorescence microscopy) or other markers for acidic autophagic vacuoles (e.g., Keima) along with static TEM images should help in the analysis of flux and the visualization of cargo structures [125].
Another technique that has proven quite useful for analyzing the complex membrane structures that participate in autophagy is 3-dimensional electron tomography [126–128], and cryoelectron microscopy (cryo-EM; Figure 5) [129]. More sophisticated, cryo-soft X-ray tomography (cryo-SXT) is an emerging imaging technique used to visualize autophagosomes [130]. Cryo-SXT extracts ultrastructural information from whole, unstained mammalian cells as close to the “near-native” fully-hydrated (living) state as possible. Correlative studies combining cryo-fluorescence and cryo-SXT workflow (cryo-CLXM) have been applied to capture early autophagosomes. In order to study the structural biology of purified autophagy components and complexes, high-resolution cryo-EM combined with 3-dimensional structure determination is also increasingly being used as an alternative to X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy [131, 132].
Finally, although only as an indirect measurement, the comparison of the ratio of autophagosomes to autolysosomes by TEM can support alterations in autophagy identified by other procedures [133]. In this case, it is important to always compare samples to the control of the same cell type and in the same growth phase, and to acquire data at different time points, as the autophagosome:autolysosome ratio varies in time in a cell context-dependent fashion, depending on their clearance activity. An additional category of lysosomal compartments, especially common in disease states and aged postmitotic cells such as neurons, muscle cells and retinal pigment epithelium, is represented by residual bodies. This category includes ceroid and lipofuscin, lobulated vesicular compartments of varying size composed of highly indigestible complexes of protein and lipid, and abundant, mostly inactive, acid hydrolases. Reflecting end-stage unsuccessful incomplete autolysosomal digestion, lipofuscin is fairly easily distinguished from AVs and lysosomes by TEM but can be easily confused with autolysosomes in immunocytochemistry studies at the light microscopy level [76, 134]; lipofuscin has broad spectral emission, and is the main cause of autofluorescence in tissues.
TEM observations of platinum-carbon replicas obtained by the freeze fracture technique can also supply useful ultrastructural information on the autophagic process. In quickly frozen and fractured cells the fracture runs preferentially along the hydrophobic plane of the membranes, allowing characterization of the limiting membranes of the different types of autophagic vacuoles, and visualization of their limited protein intramembrane particles/integral membrane proteins (IMPs). Several studies have been carried out using this technique on yeast [135], as well as on mammalian cells or tissues including the mouse exocrine pancreas [136], the mouse and rat liver [137, 138], mouse seminal vesicle epithelium [28, 88], rat tumor and heart [139], and cancer cell lines (e.g., breast cancer MDA-MB-231) [140] to investigate the various phases of autophagosome maturation, and to reveal useful details about the origin and evolution of their limiting membranes [6, 141–144].
The phagophore and the limiting membranes of autophagosomes contain few, or no detectable, IMPs (Figure 6A,B), when compared to other cellular membranes and to the membranes of lysosomes. In subsequent stages of the autophagic process the fusion of the autophagosome with an endosome and a lysosome results in increased density of IMPs in the membrane of the formed autophagic compartments (amphisomes, autolysosomes; Figure 6C) [6, 28, 135–138, 145, 146]. Autolysosomes are delimited by a single membrane because, in addition to the engulfed material, the inner membrane is also degraded by the lytic enzymes. Similarly, the limiting membrane of autophagic bodies in yeast (and presumably plants) is also quickly broken down under normal conditions. Autophagic bodies can be stabilized, however, by the addition of phenylmethylsulphonylfluoride (PMSF) or genetically by the deletion of the yeast PEP4 gene (see The Cvt pathway, mitophagy, pexophagy, piecemeal microautophagy of the nucleus and late nucleophagy in yeast and filamentous fungi). Thus, another method to consider for monitoring autophagy in yeast (and potentially in plants) is to count autophagic bodies by TEM using at least two time points [147]. The advantage of this approach is that it can provide accurate information on flux even when the autophagosomes are abnormally small [148, 149]. Thus, although a high frequency of “abnormal” structures presents a challenge, TEM is still very helpful in analyzing autophagy.
Figure 6.
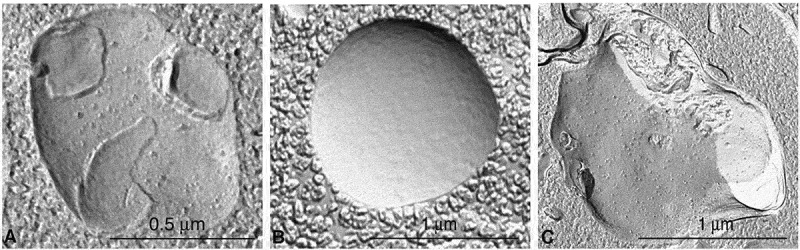
Different autophagic vacuoles observed after freeze fracturing in cultured osteosarcoma cells after treatment with the autophagy inducer voacamine [143]. (A) Early autophagosome delimited by a double membrane. (B) Inner monolayer of an autophagosome membrane deprived of protein particles. (C) Autolysosome delimited by a single membrane rich in protein particles. In the cross-fractured portion (on the right) the profile of the single membrane and the inner digested material are easily visible. Images provided by S. Meschini, M. Condello and A. Giuseppe
Cautionary notes: Despite the introduction of many new methods, TEM maintains its special role in autophagy research. There are, however, difficulties in utilizing TEM. It is relatively time consuming and needs technical expertise to ensure proper handling of samples in all stages of preparation from fixation to sectioning and staining. It should be noted that some of the hurdles linked to ultrathin section preparation can be overcome by using focused ion beam scanning electron microscopy (FIB-SEM) technology, which enables the operator to selectively ablate in a nanometer scale a previously marked region of the sample by using a focused ion current from a gallium source. The milling process can be interrupted every few nanometers to take high-resolution images of cross sections by the SEM column [150]. Moreover, the prospects for application of cryopreparation techniques have been improved; the notoriously slow process of freeze substitution of frozen samples can be accelerated tremendously by sample agitation using either an experimental setup or agitation modules within automated freeze-substitution units [151, 152].
After the criteria for sample preparation are met, an important problem is the proper identification of autophagic structures. This is crucial for both qualitative and quantitative characterization, and needs considerable experience, even in the case of one cell type. The difficulty lies in the fact that many subcellular components may be mistaken for autophagic structures. For example, some authors (or reviewers of manuscripts) assume that almost all cytoplasmic structures that, in the section plane, are surrounded by two (more or less) parallel membranes are autophagosomes. Structures appearing to be limited by a double membrane, however, may include swollen mitochondria, plastids in plant cells, cellular interdigitations, endocytosed apoptotic bodies, circular structures of lamellar smooth endoplasmic reticulum (ER), and even areas surrounded by rough ER. Endosomes, phagosomes and secretory vacuoles may have heterogeneous content that makes it possible to confuse them with autolysosomes. Additional identification problems may arise from damage caused by improper sample collection or fixation artefacts [66, 67, 153, 154].
Whereas fixation of in vitro samples is relatively straightforward, fixation of excised tissues requires care to avoid sampling a nonrepresentative, uninformative, or damaged part of the tissue. For instance, if 95% of a tumor is necrotic, TEM analysis of the necrotic core may not be informative, and if the sampling is from the viable rim, this needs to be specified when reported. Clearly, this introduces the potential for subjectivity because reviewers of a paper cannot request multiple images with a careful statistical analysis with these types of samples. In addition, ex vivo samples are not typically randomized during processing, further complicating the possibility of valid statistical analyses. Ex vivo tissue should be fixed immediately and systematically across samples to avoid changes in autophagy that may occur simply due to the elapsed time ex vivo. It is recommended that for tissue samples, perfusion fixation should be used when possible. Rapid freezing techniques such as high-pressure freezing followed by freeze substitution (i.e., dehydration and chemical fixation at low temperature) have a widely accepted potential for improved sample preparation. Consequently, cryopreparation protocols have been established for many molecular biological model organisms and tissue culture [155]. Such cryopreparation techniques have already proven especially useful for elucidation of autophagy in yeast [156, 157].
Quantification of autophagy by TEM morphometry can be very useful and accurate, but, unfortunately, unreliable procedures still continue to be used. For the principles of reliable quantification and to avoid misleading results, excellent reviews are available [12, 158–160]. In line with the basic principles of morphometry we find it necessary to emphasize here some common problems with regard to quantification. Counting autophagic vacuole profiles in sections of cells (i.e., number of autophagic profiles per cell profile) may give unreliable results, partly because both cell areas and profile areas are variable and also because the frequency of section profiles depends on the size of the vacuoles. However, estimation of the number of autophagic profiles per cell area is more reliable and correlates well with the volume fraction mentioned below [161]. There are morphometric procedures to measure or estimate the size range and the number of spherical objects by profiles in sections [160]; however, such methods have been used in autophagy research only a few times [42, 149, 162, 163].
Proper morphometric procedures return data as µm3 autophagic vacuole/µm3 cytoplasm for relative volume (also called volume fraction or volume density), or µm2 autophagic vacuole surface/µm3 cytoplasm for relative surface (surface density). Examples of actual morphometric measurements for the characterization of autophagic processes can be found in several articles [22, 154, 160, 164, 165]. It is appropriate to note here that a change in the volume fraction of the autophagic compartment may come from two sources; from the real growth of its size in a given cytoplasmic volume, or from the decrease of the cytoplasmic volume itself. To avoid this so-called “reference trap,” the reference space volume can be determined by different methods [158, 166]. If different magnifications are used for measuring the autophagic vacuoles and the cytoplasm (which may be practical when autophagy is less intense) correction factors should always be used.
In some cases, it may be prudent to employ tomographic reconstructions of TEM images to confirm that the autophagic compartments are spherical and are not being confused with interdigitations observed between neighboring cells, endomembrane cisternae or damaged mitochondria with similar appearance in thin-sections (e.g., see ref. [167]), but this is obviously a time-consuming approach requiring sophisticated equipment. In addition, interpretation of tomographic images can be problematic. For example, starvation-induced autophagosomes should contain cytoplasm (i.e., cytosol and possibly organelles), but autophagosome-related structures involved in specific types of autophagy should show the selective cytoplasmic target, but may be relatively devoid of bulk cytoplasm. Such processes include selective peroxisome or mitochondria degradation (pexophagy or mitophagy, respectively) [168, 169], targeted degradation of pathogenic microbes (xenophagy) [170–175], a combination of xenophagy and stress-induced mitophagy [176], as well as the yeast biosynthetic cytoplasm-to-vacuole targeting (Cvt) pathway [177]. Furthermore, some pathogenic microbes express membrane-disrupting factors during infection (e.g., phospholipases) that disrupt the normal double-membrane architecture of autophagosomes [178]. It is not even clear if the sequestering compartments used for specific organelle degradation or xenophagy should be termed autophagosomes or if alternate terms such as pexophagosome [179], mitophagosome and xenophagosome should be used, even though the membrane and mechanisms involved in their formation may be identical to those for starvation-induced autophagosomes. Indeed, the double-membrane vesicle of the Cvt pathway is referred to as a Cvt vesicle [180].
The confusion of heterophagic structures with autophagic ones is a major source of misinterpretation. A prominent example of this is related to cell death. Apoptotic bodies from neighboring cells can be readily phagocytosed by surviving cells of the same tissue [181, 182]. Immediately after phagocytic uptake of apoptotic bodies, phagosomes may appear as double membraned. The inner one is the plasma membrane of the apoptotic body and the outer one is that of the phagocytizing cell. The early heterophagic vacuole formed in this way may appear similar to an autophagosome or, in a later stage, an early autolysosome in that it contains recognizable or identifiable cytoplasmic material. A major difference, however, is that the surrounding membranes are the thicker plasma membrane type, rather than the thinner sequestration membrane type [153]. A good feature to distinguish between autophagosomes and double plasma membrane-bound structures is the lack of the distended empty space (characteristic for the sequestration membranes of autophagosomes) between the two membranes of the phagocytic vacuoles. In addition, engulfed apoptotic bodies usually have a larger average size than autophagosomes [183, 184]. The problem of heterophagic elements interfering with the identification of autophagic ones is most prominent in cell types with particularly intense heterophagic activity (such as macrophages, and amoeboid or ciliate protists). Special attention has to be paid to this problem in cell cultures or in vivo treatments (e.g., with toxic or chemotherapeutic agents) causing extensive cell death.
The most common organelles confused with autophagic vacuoles are mitochondria, ER, endosomes, and also (depending on their structure) plastids in plants. Due to the cisternal structure of the ER, double membrane-like structures surrounding mitochondria or other organelles are often observed after sectioning [185], but these can also correspond to cisternae of the ER coming into and out of the section plane [66]. If there are ribosomes associated with these membranes they can help in distinguishing them from the ribosome-free double-membrane of the phagophore and autophagosome. Observation of a mixture of early and late autophagic vacuoles that is modulated by the time point of collection and/or brief pulses of bafilomycin A1 to trap the cargo in a recognizable early state [55] increases the confidence that an autophagic process is being observed. In these cases, however, the possibility that feedback activation of sequestration gets involved in the autophagic process has to be carefully considered. To minimize the impact of errors, exact categorization of autophagic elements should be applied. Efforts should be made to clarify the nature of questionable structures by extensive preliminary comparison in many test areas. Elements that still remain questionable should be categorized into special groups and measured separately. Should their later identification become possible, they can be added to the proper category or, if not, kept separate.
For nonspecialists it can be particularly difficult to distinguish among amphisomes, autolysosomes and lysosomes, which are all single-membrane compartments containing material that has been more or less degraded. Therefore, we suggest in general to measure autophagosomes as a separate category for a start, and to compile another category of degradative compartments (including amphisomes, autolysosomes and lysosomes). All of the autophagic compartments increase in quantity upon true autophagy induction; however, in pathological states, it may be informative to discriminate among these different forms of degradative compartments, which may be differentially affected by disease factors. By applying both immuno-TEM and Airyscan confocal imaging, it is possible to obtain a comprehensive and quantitative analysis of LAMP1 distribution in various autophagic organelles in neurons [186, 187]. A significant portion of LAMP1-labeled organelles lack major lysosomal hydrolases, and LAMP1 intensity is not a sensitive readout to assess autophagic deficits in familial amyotrophic lateral sclerosis-linked motor neurons in vivo [188, 189]. Thus, caution is warranted when interpreting LAMP1-labeled autolysosomes and labeling a set of active lysosomal hydrolases combined with various autophagic markers would be necessary to assess degradative autolysosomes under physiological and pathological conditions.
A new and fast developing technique is combining the temporal resolution of time-lapse fluorescence microscopy with the spatial resolution of super-resolution microscopy. HEK293 cells that express recombinant proteins of interest fused to fluorescent tags are imaged live to capture the formation of autophagosomes, fixed on stage to “snap-freeze” these structures, stained with appropriate antibodies, relocated, and imaged at super resolution by direct stochastic optical reconstruction microscopy [190].
Super-resolution microscopy techniques at ∼20 nm spatial resolution via 3-color, 3-dimensional super-resolution fluorescence microscopy, makes it possible to image the structural organization of the ULK1 complex that scaffolds the formation of cup-like structures located at SEC12-enriched remodeled ER-exit sites prior to LC3 lipidation. This cup scaffold provides a structural asymmetry to enforce the directional recruitment of downstream components, including the ATG12–ATG5-ATG16L1 complex, WIPI2, and LC3, to the convex side of the cup [191].
In yeast, it is convenient to identify autophagic bodies that reside within the vacuole lumen, and to quantify them as an alternative to the direct examination of autophagosomes. However, it is important to keep in mind that it may not be possible to distinguish between autophagic bodies that are derived from the fusion of autophagosomes with the vacuole, and the single-membrane vesicles that are generated during microautophagy-like processes such as micropexophagy and micromitophagy.
Conclusion: EM is an extremely informative and powerful method for monitoring autophagy and is one of the few techniques that shows autophagy in its complex cellular environment with subcellular resolution. The cornerstone of successfully using TEM is the proper identification of autophagic structures, which is also the prerequisite to get reliable quantitative results by TEM morphometry. EM is best used in combination with other methods to ensure the complex and holistic approach that is becoming increasingly necessary for further progress in autophagy research.
2. Atg8-family protein detection and quantification
Atg8 and the Atg8-family proteins are the most widely monitored autophagy-related proteins. In this section we describe multiple assays that utilize these proteins.
a. Western blotting and ubiquitin-like protein conjugation systems
Atg8 is a ubiquitin-like protein that can be conjugated to PE (and possibly to phosphatidylserine [PS] [192]). In yeast and several other organisms, the conjugated form is referred to as Atg8–PE. The mammalian homologs of Atg8 constitute a family of proteins subdivided in two major subfamilies: MAP1LC3/LC3 and GABARAP. The former consists of LC3A (two splice variants), LC3B, LC3B2 and LC3C, whereas the latter family includes GABARAP, GABARAPL1, and GABARAPL2/GATE-16 [193]. After cleavage of the precursor protein mostly by the cysteine protease ATG4B [194, 195], the nonlipidated and lipidated forms are usually referred to respectively as LC3-I and LC3-II, or GABARAP and GABARAP–PE, etc. The PE-conjugated form of Atg8-family proteins, although larger in mass, shows faster electrophoretic mobility in SDS-PAGE gels, probably as a consequence of increased hydrophobicity. The positions of both the unconjugated (approximately 16-18 kDa) and lipid conjugated (approximately 14-16 kDa) forms of the Atg8-family proteins should be indicated on western blots whenever both are detectable. The differences among the LC3/GABARAP proteins with regard to function and tissue-specific expression are not well defined; however, new evidence suggests that LC3 proteins have distinct subcellular distributions and mediate different types of selective autophagy [196, 197]. Therefore, it is important to indicate the isoform being analyzed just as it is for the GABARAP subfamily, and to specify which antibody is being used.
The mammalian Atg8 homologs share from 29% to 94% sequence identity with the yeast protein and have all been demonstrated to be involved in autophagosome biogenesis [198]. LC3 proteins are involved in autophagosome formation, with participation of GABARAP subfamily members in later stages of autophagosome formation [199]. Some evidence, however, suggests that, at least in certain cell types, the LC3 subfamily may be dispensable for bulk autophagic sequestration of cytosolic proteins, whereas the GABARAP subfamily is absolutely required [32]. Also, PINK1-PRKN-dependent mitophagy strongly requires the GABARAP subfamily, with little or no requirement for the LC3 subfamily [34, 35]. Due to unique features in their molecular surface charge distribution [200], emerging evidence indicates that LC3 and GABARAP proteins may be involved in recognizing distinct sets of cargoes for selective autophagy [201–203]. Nevertheless, in most published studies, LC3 has been the primary Atg8-family homolog examined in mammalian cells and the one that is typically characterized as an autophagosome marker per se. Note that although this protein is referred to as “Atg8” in many other systems, we primarily refer to it in this section as LC3 to distinguish it from the yeast protein and from the GABARAP subfamily, whereas we generally refer to the “Atg8-family proteins” throughout the rest of these guidelines. LC3, like the other Atg8 homologs, is initially synthesized in an unprocessed form, proLC3, which is converted into a proteolytically processed form lacking amino acids from the C terminus, LC3-I, and is finally modified into the PE-conjugated form, LC3-II (Figure 7). Atg8–PE/LC3-II is the only protein marker that is reliably associated with completed autophagosomes, but is also localized to phagophores. In yeast, Atg8 protein levels increase at least 10-fold when autophagy is induced [204]. In mammalian cells, however, the total levels of LC3 do not necessarily change in a predictable manner, as there may be an increase in the conversion of LC3-I to LC3-II, or a decrease in LC3-II relative to LC3-I if degradation of LC3-II via lysosomal turnover is particularly rapid (this can also be a concern in yeast with regard to vacuolar turnover of Atg8–PE). Both of these events can be seen sequentially in several cell types as a response to total nutrient and serum starvation. It is also possible that following the induction of autophagy there is a decrease in both LC3-I and LC3-II due to rapid LC3-I conversion together with rapid LC3-II degradation [205]. In cells of neural lineage, a high ratio of LC3-I to LC3-II is a common finding [206]. For instance, SH-SY5Y neuroblastoma cell lines display only a slight increase of LC3-II after nutrient deprivation, whereas LC3-I is clearly reduced. This is likely related to a high basal autophagic flux, as suggested by the higher increase in LC3-II when cells are treated with NH4Cl [207, 208], although cell-specific differences in transcriptional regulation of LC3 may also play a role. In fact, stimuli or stress that inhibit transcription or translation of LC3 might actually be misinterpreted as inhibition of autophagy, and vice versa—stimuli or stress that increase transcription or translation of LC3 might be misinterpreted as activation of autophagy. The LC3-I:LC3-II ratio can vary across brain cancer cells depending on the basal level of autophagy, a phenomenon that can influence further analysis of autophagy activation upon stressful conditions such as hypoxia [209]. Importantly, in brain spinal cord and dorsal root ganglia tissue, LC3-I is much more abundant than LC3-II [210, 211] and the latter form is most easily discernible in enriched fractions of autophagosomes, autolysosomes and ER, and may be more difficult to detect in crude homogenate or cytosol [212]. It is possible to readily detect both LC3-I and LC3-II in brain and spinal cord lysates with the use of a gel that allows sufficient separation of the LC3-I/LC3-II bands so the strong LC3-I band does not interfere with detection of the much weaker LC3-II band (e.g., a 4-20% gradient gel or a 4-12% Bis-Tris gel using MES buffer) [213, 214]. In studies of the brain, immunoblot analysis of the membrane and cytosol fraction from a cell lysate, upon appropriate loading of samples to achieve quantifiable and comparative signals, can be useful to measure LC3 forms. For more accurate quantification of LC3-I and LC3-II levels, a correction factor for differential immunoreactivity of the two forms can be obtained through analyses of LC3-I and LC3-II protein levels upon ATG4-mediated delipidation [32].
Figure 7.
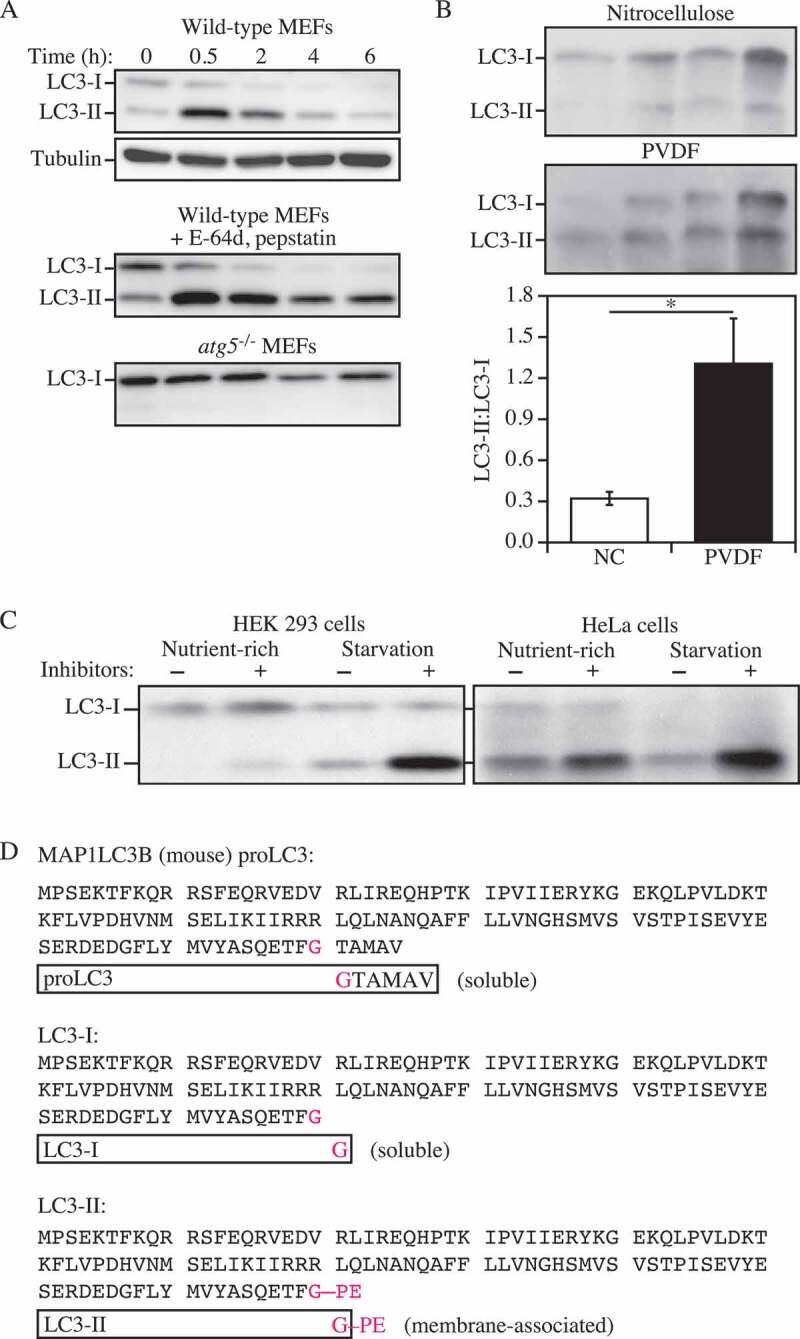
LC3-I conversion and LC3-II turnover. (A) Expression levels of LC3-I and LC3-II during starvation. Atg5+/+ (wild-type) and atg5−/− MEFs were cultured in DMEM without amino acids and serum for the indicated times, and then subjected to immunoblot analysis using anti-LC3 antibody and anti-tubulin antibody. E-64d (10 µg/ml) and pepstatin A (10 µg/ml) were added to the medium where indicated. Positions of LC3-I and LC3-II are marked. The inclusion of lysosomal protease inhibitors reveals that the apparent decrease in LC3-II is due to lysosomal degradation as easily seen by comparing samples with and without inhibitors at the same time points (the overall decrease seen in the presence of inhibitors may reflect decreasing effectiveness of the inhibitors over time). Monitoring autophagy by following steady-state amounts of LC3-II without including inhibitors in the analysis can result in an incorrect interpretation that autophagy is not taking place (due to the apparent absence of LC3-II). Conversely, if there are high levels of LC3-II but there is no change in the presence of inhibitors this may indicate that induction has occurred but that the final steps of autophagy are blocked, resulting in stabilization of this protein. This figure was modified from data previously published in ref. [30], and is reproduced by permission of Landes Bioscience, copyright 2007. (B) Lysates of four human adipose tissue biopsies were resolved on two 12% polyacrylamide gels, as described previously [292]. Proteins were transferred in parallel to either a PVDF or a nitrocellulose membrane, and blotted with anti-LC3 antibody, and then identified by reacting the membranes with an HRP-conjugated anti-rabbit IgG antibody, followed by ECL. The LC3-II:LC3-I ratio was calculated based on densitometry analysis of both bands. *, P< 0.05. (C) HEK 293 and HeLa cells were cultured in nutrient-rich medium (DMEM containing 10% fetal calf serum) or incubated for 4 h in starvation conditions (Krebs-Ringer medium) in the absence (-) or presence (+) of E-64d and pepstatin at 10 µg/ml each (Inhibitors). Cells were then lysed and the proteins resolved by SDS-PAGE. Endogenous LC3 was detected by immunoblotting. Positions of LC3-I and LC3-II are indicated. In the absence of lysosomal protease inhibitors, starvation results in a modest increase (HEK 293 cells) or even a decrease (HeLa cells) in the amount of LC3-II. The use of inhibitors reveals that this apparent decrease is due to lysosome-dependent degradation. This figure was modified from data previously published in ref. [240], and is reproduced by permission of Landes Bioscience, copyright 2005. (D) Sequence and schematic representation of the different forms of LC3B. The sequence for the nascent (proLC3) from mouse is shown. The glycine at position 120 indicates the cleavage site for ATG4. After this cleavage, the truncated LC3 is referred to as LC3-I, which is still a soluble form of the protein. Conjugation to PE generates the membrane-associated LC3-II form (equivalent to Atg8–PE)
The pattern of LC3-I to LC3-II conversion seems to be not only cell specific, but also related to the kind of stress to which cells are subjected. For example, SH-SY5Y cells display a strong increase of LC3-II when treated with the proton gradient uncoupler CCCP, a well-known disruptor of the mitochondrial membrane potential and inducer of mitophagy (although it has also been reported that CCCP may actually inhibit mitophagy [215]). Thus, neither assessment of LC3-I consumption nor the evaluation of LC3-II levels would necessarily reveal a slight induction of autophagy (e.g., by rapamycin). Also, there is not always a clear precursor/product relationship between LC3-I and LC3-II, because the conversion of the former to the latter is cell type-specific and dependent on the treatment used to induce autophagy. Accumulation of LC3-II, which is generally proportional with time, can be obtained through the following: i) By interrupting the autophagosome-lysosome fusion step (e.g., by depolymerizing acetylated microtubules with vinblastine); ii) by inhibiting the ATP2A/SERCA calcium pump with thapsigargin [216]; iii) by specifically inhibiting the V-ATPase with bafilomycin A1 [217–219]; iv) or by raising the lysosomal pH by the addition of CQ [220, 221]. It should be noted that some of these treatments may increase autophagosome numbers by: i) Disrupting the lysosome-dependent activation of MTOR (mechanistic target of rapamycin kinase) complex 1 (MTORC1), a major suppressor of autophagy induction [222, 223] (note that the original term “mTOR” was named to distinguish the “mammalian” target of rapamycin from the yeast proteins [224]); ii) by inhibiting lysosome-mediated proteolysis (e.g., with a cysteine protease inhibitor such as E-64d, the aspartic protease inhibitor pepstatin A, the cysteine, serine and threonine protease inhibitor leupeptin or treatment with bafilomycin A1, NH4Cl or CQ [220, 225–227]); iii) by inhibiting autophagosome-lysosome fusion (by treatment with bafilomycin A1 [218]). It should also be noted that low concentration treatment with lysosomal inhibitors increases lysosomal activity [228]. Western blotting can be used to monitor changes in LC3 amounts (Figure 7) [30, 229]; however, even if the total amount of LC3 does increase, the magnitude of the response is generally less than that documented in yeast. It is worth noting that because the conjugated forms of the GABARAP subfamily members are usually undetectable without induction of autophagy in mammalian and other vertebrate cells [230, 231], these proteins might be more suitable than LC3 to study and quantify subtle changes in autophagy induction.
As Atg8-family proteins are often synthesized with a C-terminal extension that is removed by Atg4, this processing event can be used to monitor Atg4 activity. For example, when GFP or tags such as HA, MYC or FLAG are fused at the C terminus of Atg8 (Atg8-GFP, etc.), the epitope is removed in the cytosol to generate free Atg8 and the corresponding tag. This processing can be easily monitored by western blot [232, 233]. It is also possible to use assays with an artificial fluorogenic substrate, or a fusion of LC3B to PLA2 (phospholipase A2) that allows the release of the active phospholipase for a subsequent fluorogenic assay [234], and there is a Förster resonance energy transfer (FRET)-based assay utilizing CFP- and YFP-tagged versions of LC3B and GABARAPL2 that can be used for high-throughput screening [235]. Another method to monitor ATG4 activity in vivo uses the release of Gaussia luciferase from the C terminus of LC3 that is tethered to actin [236]. Note that there are 4 homologs of yeast Atg4 in mammals, and they have different activities with regard to the Atg8-family proteins [237]. ATG4A is able to cleave the GABARAP subfamily, but has very limited activity toward the LC3 subfamily, whereas ATG4B is apparently active against most or all of these proteins [194, 195]. The ATG4C and ATG4D isoforms have minimal activity for any of the Atg8-family protein homologs. In particular, because Atg4/ATG4 will cleave a C-terminal fusion immediately, researchers should be careful to specify whether they are using GFP-Atg8 (or GFP-LC3/GABARAP; an N-terminal fusion, which can be used to monitor various steps of autophagy) or Atg8-GFP (or LC3/GBARAP-GFP; a C-terminal fusion, which can only be used to monitor Atg4/ATG4 activity) [238].
Cautionary notes: There are several important caveats to using Atg8/LC3-II/GABARAP-II to visualize fluctuations in autophagy. First, changes in LC3-II amounts are tissue- and cell context-dependent [239, 240]. Indeed, in some cases, autophagosome accumulation detected by TEM does not correlate well with the amount of LC3-II (Tallóczy Z, de Vries RLA, and Sulzer D, unpublished results; Eskelinen E-L, unpublished results). This is particularly evident in those cells that show low levels of LC3-II (based on western blotting) because of an intense autophagic flux that consumes this protein, or in cell lines having high levels of LC3-II that are tumor-derived, such as HeLa cells [240]. Conversely, the detectable formation of LC3-II is not sufficient evidence for autophagy. For example, homozygous deletion of Becn1 does not prevent the formation of LC3-II in embryonic stem cells even though autophagy is substantially reduced, whereas deletion of Atg5 results in the complete absence of LC3-II (see Fig. 6A and supplemental data in ref. [241]). The same is true for the generation of Atg8–PE in yeast in the absence of VPS30/ATG6 (see Fig. 7 in ref. [242]). Thus, it is important to remember that not all of the autophagy-related proteins are required for Atg8-family protein processing, including lipidation [242]. Fluctuations in the detection and amounts of LC3-I versus LC3-II present technical problems. For example, LC3-I is very abundant in brain tissue, and the intensity of the LC3-I band may obscure detection of LC3-II, unless the polyacrylamide crosslinking density is optimized, or the membrane fraction of LC3 is first separated from the cytosolic fraction [41]. Conversely, some cell lines have much less visible LC3-I compared to LC3-II. In addition, tissues may have asynchronous and heterogeneous cell populations, and this variability may present challenges when analyzing LC3 by western blotting.
Second, LC3-II also associates with the membranes of nonautophagic structures. For example, some members of the γ-protocadherin family undergo clustering to form intracellular tubules that emanate from lysosomes [243]. LC3-II is recruited to these tubules, where it appears to promote or stabilize membrane expansion. Furthermore, LC3 can be recruited directly to apoptotic cell-containing phagosome membranes [244, 245], macropinosomes [244], the parasitophorous vacuole of Toxoplasma gondii [246], and single-membrane entotic vacuoles [244], as well as to bacteria-containing phagosome membranes under certain immune activating conditions, for example, toll-like receptor (TLR)-mediated stimulation in LC3-associated phagocytosis [247, 248]. Importantly, LC3 is involved in secretory trafficking as it has been associated with secretory granules in mast cells [249] and PC12 hormone-secreting cells [250]. LC3 is also detected on secretory lysosomes in osteoclasts [251] and in amphisome-like structures involved in mucin secretion by goblet cells [252]. Therefore, in studies of infection of mammalian cells by bacterial pathogens, the identity of the LC3-II labelled compartment as an autophagosome should be confirmed by a second method, such as TEM. It is also worth noting that autophagy induced in response to bacterial infection is not directed solely against the bacteria but can also be a response to remnants of the phagocytic membrane [253]. Similar cautions apply with regard to viral infection. For example, coronaviruses induce autophagosomes during infection through the expression of nsp6; however, coronaviruses also induce the formation of double-membrane vesicles that are coated with LC3-I, and this plays an autophagy-independent role in viral replication [254, 255]. Similarly, nonlipidated LC3 marks replication complexes in flavivirus (Japanese encephalitis virus)-infected cells and is essential for viral replication [256]. Along these lines, during herpes simplex virus type 1 (HSV-1) infection, an LC3+ autophagosome-like organelle that is derived from nuclear membranes and that contains viral proteins is observed [257], whereas influenza A virus directs LC3 to the plasma membrane via an LC3-interacting region (LIR) motif in its M2 protein [258]. In addition, shedding microvesicles isolated from HSV-1-infected cells are positive for LC3-II, suggesting a role for the autophagic pathway in microvesicle-mediated HSV-1 spread [259]. Moreover, in vivo studies have shown that coxsackievirus (an enterovirus) induces formation of autophagy-like vesicles in pancreatic acinar cells, together with extremely large autophagy-related compartments that have been termed megaphagosomes [260]; the absence of ATG5 disrupts viral replication and prevents the formation of these structures [261]. Of note, LC3 not only attaches to membrane lipids, but can also be covalently linked to other proteins [262], thus complicating interpretation of its distribution in cells.
Third, caution must be exercised in general when evaluating LC3 by western blotting, and appropriate standardization controls are necessary. For example, LC3-I may be less sensitive to detection by certain anti-LC3 antibodies; antibodies targeting the N-terminal region show lower binding efficiency of LC3-I compared to polyclonal antibodies against the entire protein, leading to a different interpretation of LC3 turnover (Figure 8) (C. Leschczyk, P. Cebollada Rica, U.E. Schaible, unpublished results) [263]. Moreover, LC3-I is more labile than LC3-II, being more sensitive to freezing-thawing and to degradation in SDS sample buffer. Therefore, fresh samples should be boiled and assessed as soon as possible and should not be subjected to repeated freeze-thaw cycles. Alternatively, trichloroacetic acid precipitation of protein from fresh cell homogenates can be used to protect against degradation of LC3 by proteases that may be present in the sample. A general point to consider when examining transfected cells concerns the efficiency of transfection. A western blot will detect LC3 in the entire cell population, including those that are not transfected. Thus, if transfection efficiency is too low, it may be necessary to use methods, such as fluorescence microscopy, that allow autophagy to be monitored in single cells. In summary, the analysis of the gel shift of transfected LC3 or GFP-LC3 can be employed to follow LC3 lipidation only in highly transfectable cells [264].
Figure 8.
Different LC3B-I:LC3B-II ratios indicating turnover were assessed using a mono- as well as polyclonal anti-LC3B antibody. Monocytes were isolated from human whole blood and differentiated into monocyte-derived macrophages (MDMs) by incubation in human CSF1/M-CSF for 1 week. To induce autophagy, cells were starved by reducing the FCS concentration to 1% for one day. Monocytes, and resting and starved MDMs were lysed with Laemmli buffer; the proteins were separated by SDS-PAGE and analyzed by western blot. Membranes were labeled using a monoclonal antibody to the N terminus of LC3B (Novus, clone 1251D, NBP2-59800) or polyclonal antibodies (Sigma, L7543). Relative intensity of LC3B-I and LC3B-II was quantified with Image Lab™ to calculate LC3B-II:LC3B-I ratios
When dealing with animal tissues, western blotting of LC3 should be performed on frozen biopsy samples homogenized in the presence of general protease inhibitors (C. Isidoro, personal communication; see also Human) [265]. Caveats regarding detection of LC3 by western blotting have been covered in a dedicated review [30]. For example, PVDF membranes may result in a stronger LC3-II retention than nitrocellulose membranes, possibly due to a higher affinity for hydrophobic proteins (Figure 7B; J. Kovsan and A. Rudich, personal communication), and Triton X-100 may not efficiently solubilize LC3-II in some systems [266]. Heating in the presence of 1% SDS, or analysis of membrane fractions [41], may assist in the detection of the lipidated form of this protein. This observation is particularly relevant for cells with a high nucleocytoplasmic ratio, such as lymphocytes. Under these constraints, direct lysis in Laemmli loading buffer, containing SDS, just before heating, greatly improves LC3 detection on PVDF membranes, especially when working with a small number of cells (F. Gros, unpublished observations) [267]. Analysis of a membrane fraction is particularly useful for brain, where levels of soluble LC3-I greatly exceed the level of LC3-II.
One of the most important issues is the quantification of changes in LC3-II, because this assay is one of the most widely used in the field and is rather prone to misinterpretation. Levels of LC3-II should be compared not to LC3-I (see the caveat in the next paragraph), but ideally to more than one “housekeeping” protein (HKP) such as ACTB/β-actin. Actin and other HKPs, however, are usually abundant and can easily be overloaded on the gel [268] such that their density is saturated and, as such, they are not detected within a linear range. Moreover, actin levels may decrease when autophagy is induced in many organisms from yeast to mammals. Similar considerations apply to GAPDH, at least in some cell types (L. Galluzzi, personal communication) [269]. For any proteins used as “loading controls” (including actin, tubulin, MAPK1 [270–272] and GAPDH) multiple exposures of the western blot are generally necessary to ensure that the signals are detected in the linear range when using film. Alternatively, the western blot signals can be detected using a gel imaging system compatible with secondary antibodies with infrared fluorescence, or an instrument that takes multiple chemiluminescence exposures and automatically selects the optimal exposure times. Another alternative approach is to stain for total cellular proteins with Coomassie Brilliant Blue or Ponceau Red [273] instead of using HKPs, but that approach is generally less sensitive and may not reveal small differences in protein loading. Stain-Free gels, which also stain for total cellular proteins, have been shown to be an excellent alternative to HKPs [274].
It is important to realize that ignoring the level of LC3-I in favor of LC3-II normalized to HKPs may not provide the full picture of the cellular autophagic response [239, 275]. For example, in aging rat skeletal muscle, the increase in LC3-I is at least as important as that for LC3-II [276, 277]. Yet in other settings, autophagy induction triggers a significant decrease in LC3-I levels, along with an increase in LC3-II levels, presumably due to its increased conversion into LC3-II [278]. Quantification of both isoforms is therefore informative, but requires adequate conditions of electrophoretic separation. This is particularly important for samples where the amount of LC3-I is high relative to LC3-II (as in brain tissues, where the LC3-I signal can be overwhelming). Under such a scenario, it may be helpful to use 15% or 16% polyacrylamide gels or gradient gels to increase the separation of LC3-I from LC3-II. Furthermore, because the dynamic range of LC3 immunoblots is generally quite limited, it is imperative that other assays be used in parallel in order to draw valid conclusions about changes in autophagy activity.
Fourth, in mammalian cells LC3 is expressed as multiple isoforms (LC3A, LC3B, LC3B2 and LC3C [279, 280]), which exhibit different tissue distributions and whose functions are still poorly understood. A point of caution along these lines is that the increase in LC3A-II versus LC3B-II levels may not display equivalent changes in all organisms under autophagy-inducing conditions, and it should not be assumed that LC3B is the optimal protein to monitor [281]. A key technical consideration is that the isoforms may exhibit different specificities for antisera or antibodies. Thus, it is highly recommended that investigators report exactly the source and catalog number of the antibodies used to detect LC3 as this might help avoid discrepancies between studies (reporting company and catalog number is a requirement for publishing in the journal Autophagy [282]). The current commercialized anti-LC3B antibodies also recognize LC3A, but do not recognize LC3C, which shares less sequence homology. It is important to note that LC3C possesses in its primary amino acid sequence the DYKD motif that is recognized with a high affinity by anti-FLAG antibodies. Thus, the standard anti-FLAG M2 antibody can detect and immunoprecipitate overexpressed LC3C, and caution has to be taken in experiments using FLAG-tagged proteins (M. Biard-Piechaczyk and L. Espert, personal communication). Note that according to Ensembl there is no LC3C in mouse or rat.
In addition, it is important to keep in mind the other subfamily of Atg8 proteins, the GABARAP subfamily (see above) [198, 283]. Both starvation-induced autophagy and PINK1-PRKN-dependent mitophagy, as noted above, predominantly require the GABARAP subfamily over the LC3 subfamily [32, 34, 35]. Moreover, certain types of mitophagy induced by BNIP3L/NIX are highly dependent on GABARAP and less dependent on LC3 proteins [284, 285]. Furthermore, commercial antibodies for GABARAPL1 also recognize GABARAP [32, 193], which might lead to misinterpretation of experiments, in particular those using immunohistochemical techniques. Sometimes the problem with cross-reactivity of the anti-GABARAPL1 antibody can be overcome when analyzing these proteins by western blot because the isoforms can be resolved during SDS-PAGE using high concentration (15%) gels, as GABARAP migrates faster than GABARAPL1 (M. Boyer-Guittaut, personal communication; also see Fig. S4 in ref. [32]). Because GABARAP and GABARAPL1 can both be proteolytically processed and lipidated, generating GABARAP-I or GABARAPL1-I and GABARAP-II or GABARAPL1-II, respectively, this may lead to a misassignment of the different bands. As soon as highly specific antibodies that are able to discriminate between GABARAP and GABARAPL1 become available, we strongly advise their use; until then, we recommend caution in interpreting results based on the detection of these proteins by western blot. Antibody specificity can be assessed after complete inhibition of GABARAP (or any other Atg8-family protein) expression by RNA interference [32, 231]. In general, we advise caution in choosing antibodies for western blotting and immunofluorescence experiments and in interpreting results based on stated affinities of antibodies unless these have been clearly determined.
As with any western blot, proper methods of quantification must be used, which are, unfortunately, often not well disseminated; readers are referred to an excellent paper on this subject (see ref. [286]). Unlike the other members of the GABARAP family, almost no information is available on GABARAPL3, perhaps because it is not yet possible to differentiate between GABARAPL1 and GABARAPL3 proteins, which have 94% identity. As stated by the laboratory that described the cloning of the human GABARAPL1 and GABARAPL3 genes [283], their expression patterns are apparently identical. It is worth noting that GABARAPL3 is the only gene of the GABARAP subfamily that seems to lack an ortholog in mice [283]. GABARAPL3 might therefore be considered as a pseudogene without an intron that is derived from GABARAPL1. Hence, until new data are published, GABARAPL3 should not be considered as the fourth member of the GABARAP family. Another important consideration is that lipidated LC3/GABARAP isoforms (particularly GABARAP and GABARAPL1) can be unstable in non-denatured cell lysates due to ATG4B delipidation activity, even in the presence of a protease inhibitor cocktail. This can result in an underestimation of the true physiological levels of lipidated LC3/GABARAP detected by western blotting. To avoid this artefact, N-ethylmaleimide can be included in lysis buffer to irreversibly inhibit ATG4B, or lysis can be performed under reducing and denaturing conditions [287].
Fifth, in non-mammalian species, the discrimination of Atg8–PE from the nonlipidated form can be complicated by their nearly identical SDS-PAGE mobilities and the presence of multiple isoforms (e.g., there are nine in Arabidopsis). In yeast, it is possible to resolve Atg8 (the nonlipidated form) from Atg8–PE by including 6 M urea in the SDS-PAGE separating gel [288], or by using a 15% resolving gel without urea (F. Reggiori, personal communication). Similarly, urea combined with prior treatment of the samples with (or without) PLD (phospholipase D; that will remove the PE moiety) can often resolve the ATG8 species in plants [289, 290]. It is also possible to label cells with radioactive ethanolamine, followed by autoradiography to identify Atg8–PE, and a C-terminal peptide can be analyzed by mass spectrometry (MS) to identify the lipid modification at the terminal glycine residue. Special treatments are not needed for the separation of mammalian LC3-I from LC3-II. However, in human cells, pro-LC3B and LC3B-II are indistinguishable by western blotting [291], and a PLD cleavage assay may be required to discriminate between the two isoforms [287], which is particularly important under conditions where ATG4 activity is reduced.
Sixth, it is important to keep in mind that ATG8, and to a lesser extent LC3, undergoes substantial transcriptional and posttranscriptional regulation. Accordingly, to obtain an accurate interpretation of Atg8-family protein levels it is also necessary to monitor the mRNA levels. Without analyzing the corresponding mRNA, it is not possible to discriminate between changes that are strictly reflected in altered amounts of protein versus those that are due to changes in transcription (e.g., the rate of transcription, or mRNA stability). For example, in cells treated with the calcium ionophore A23187 or the ER calcium pump blocker thapsigargin, an obvious correlation is found between the time-dependent increases in LC3B-I and LC3B-II protein levels, as well as with the observed increase in LC3B mRNA levels [216]. Clinically, in human adipose tissue, protein and mRNA levels of LC3 in omental fat are similarly elevated in obese compared to lean individuals [292]. Post-translational modifications, such as phosphorylation of LC3, may also affect its migration and/or the avidity of certain antibodies [293].
Seventh, LC3-I can be fully degraded by the 20S proteasome or, more problematically, processed to a form (LC3-T) appearing equal in size to LC3-II on a western blot; LC3-T was identified in HeLa cells and is devoid of the ubiquitin conjugation domain, thus lacking its adaptor function for autophagy [294].
Eighth, although it is usually possible to distinguish the nonlipidated (LC3-I) and lipidated (LC3-II) forms of LC3 using standard SDS-PAGE and western blotting (see above), some other protein separation systems fail to differentiate between them. For example, the widely used WES system, based on capillary electrophoresis and Simple Western™ technology (in which all assay steps, from protein separation, immunoprobing, detection and analysis of data are fully automated), can solve many problems found in traditional western blotting [295]; however, using this system it is not possible to distinguish LC3-I and LC3-II forms (see Figure 9 for comparison of separation of LC3 forms in traditional western blotting and WES). Most likely, this is due to unusual (i.e., inconsistent with the actual molecular mass) migration of LC3-II in SDS-PAGE which does not take place during gel-free capillary electrophoresis [296]. Therefore, although the WES system is excellent for rapid and accurate detection of the vast majority of proteins, and makes it possible to avoid various technical problems met in traditional western blotting, including those met in studies on subjects related to autophagy (see for example [297, 298]), it is not recommended for experiments where it is important to resolve LC3-I and LC3-II. This problem has been widely discussed with representatives of the WES system manufacturer who confirmed that it is technically not possible to separate these two forms of LC3 using Simple Western™ technology (K. Pierzynowska and G. Wegrzyn, personal communication). Nonetheless, the WES system can still be used to monitor changes in the total amount of LC3, and can thus provide useful information, especially in conjunction with other assays.
Figure 9.
Detection of nonlipidated (LC3-I) and lipidated (LC3-II) forms of the LC3 protein using (A) traditional SDS-PAGE and western blotting or (B) the WES System (WES - Automated Western Blots with Simple Western; ProteinSimple, San Jose, CA, USA). HEK 293 cells were cultured in DMEM medium, containing 10% fetal bovine serum and a penicillin-streptomycin mixture, at 37°C in a humidified atmosphere with 5% CO2. Cell cultures were treated with medium devoid of fetal bovine serum (-FBS) or with chloroquine (CQ; final concentration 10 µM) for 2 h. The forms of the LC3 protein were detected using anti-LC3 antibodies (MBL International, PM036). Materials from the same samples were used in experiments presented in panels A and B. The LC3-I and LC3-II forms can be effectively separated using traditional SDS-PAGE and western blotting, whereas these two forms of LC3 cannot be distinguished by using the WES system. Results provided by K. Pierzynowska, L. Gaffke and G. Wegrzyn
Conclusion: Atg8-family proteins are often excellent markers for autophagic structures; however, it must be kept in mind that there are multiple LC3 isoforms, there is a second family of mammalian Atg8-like proteins (GABARAPs), and antibody affinity (for LC3-I versus LC3-II) and specificity (for example, for LC3A versus LC3B) must be considered and/or determined. Moreover, LC3/GABARAP levels on their own do not address issues of autophagic flux. Finally, even when flux assays are carried out, there is a problem with the limited dynamic range of LC3/GABARAP immunoblots; accordingly, this method should not be used by itself to analyze changes in autophagy.
b. Turnover of LC3-II/Atg8–PE: Autophagic flux
Autophagic flux is often inferred on the basis of LC3-II turnover, measured by western blot (Figure 7C) [240] in both the presence and absence of lysosomal, or vacuolar degradation. However, it should be cautioned that such LC3 assays are merely indicative of autophagic “carrier flux”, not of actual autophagic cargo/substrate flux. It has, in fact, been observed that in rat hepatocytes, an autophagic-lysosomal flux of LC3-II can take place in the absence of an accompanying flux of cytosolic bulk cargo [299]. The relevant parameter in LC3 assays is the difference in the amount of LC3-II in both the presence and absence of saturating levels of inhibitors, which can be used to examine the transit of LC3-II through the autophagic pathway; if flux is occurring, the amount of LC3-II will be higher in the presence of the inhibitor [240]. Lysosomal degradation can be prevented through the use of protease inhibitors (e.g., pepstatin A, leupeptin and E-64d), compounds that neutralize the lysosomal pH such as bafilomycin A1, CQ or NH4Cl [17, 206, 220, 226, 300, 301], or by treatment with agents that block the fusion of autophagosomes with lysosomes (note that CQ blocks autophagy predominantly by inhibiting autophagosome-lysosome fusion [302] and that bafilomycin A1 will ultimately cause a fusion block as well as neutralize the pH [218], but the inhibition of fusion may be due to a block in ATP2A/SERCA activity [303]) [217–219, 304]. Alternatively, knocking down or knocking out LAMP2 (lysosomal associated membrane protein 2) represents a genetic approach to block the fusion of autophagosomes and lysosomes (for example, inhibiting LAMP2 in leukemic cells results in a marked increase of GFP-LC3 puncta and endogenous LC3-II protein compared to control cells upon autophagy induction during myeloid differentiation [M.P. Tschan, unpublished data], whereas in prostate cancer LNCaP cells, knocking down LAMP2 prevents autophagy [305]) [306]. This approach, however, is only valid when the knockdown of LAMP2 is directed against the mRNA region specific for the LAMP2B spliced variant, as targeting the region common to the three variants would also inhibit chaperone-mediated autophagy (CMA), which may result in the compensatory upregulation of autophagy [133, 307, 308].
Increased levels of LC3-II in the presence of lysosomal inhibition or interfering with autophagosome-lysosome fusion alone (e.g., with bafilomycin A1), may be indicative of greater induction and cargo sequestration, but to assess whether a particular treatment alters complete autophagic flux through substrate digestion, the treatment plus bafilomycin A1 must be compared with results obtained with treatment alone as well as with bafilomycin A1 alone. An additive or supra-additive effect in LC3-II levels may indicate that the treatment enhances autophagic flux (Figure 7C). Moreover, higher LC3-II levels with treatment plus bafilomycin A1 compared to bafilomycin A1 alone may indicate that the treatment increases the synthesis of autophagy-related membranes. If the treatment by itself increases LC3-II levels, but the treatment plus bafilomycin A1 does not increase LC3-II levels compared to bafilomycin A1 alone, this may indicate that the treatment induced a partial block in autophagic flux. Thus, a treatment condition increasing LC3-II on its own that has no difference in LC3-II in the presence of bafilomycin A1 compared to treatment alone may suggest a complete block in autophagy at the terminal stages [309]. This procedure has been validated with several autophagy modulators [310]. With each of these techniques, it is essential to avoid assay saturation. The duration of the bafilomycin A1 treatment (or any other inhibitor of autophagic flux such as CQ) needs to be relatively short (1-4 h) [311] to allow comparisons of the amount of LC3 that is lysosomally degraded over a given time frame under one treatment condition to another treatment condition. A concentration-curve and time-course standardization for the use of autophagic flux inhibitors is required for the initial optimization of the conditions to detect LC3-II accumulation and avoid nonspecific or secondary effects, and to exclude the possibility of a remaining residual flux, if inhibition is incomplete [312]. By using a rapid screening approach, such as a colorimetric based-platform method [313], it is possible to monitor a long time frame for autolysosome accumulation, which closely associates with autophagy activation [314–317]. Positive control experiments using treatment with known autophagy inducers, along with bafilomycin A1 versus vehicle, are important to demonstrate the utility of this approach in each experimental context.
In some circumstances it may be important to evaluate alterations in autophagy flux once autophagy is induced by a particular agent or genetic manipulation. In that case, steady-state measurements are not adequate. This can be useful for example to evaluate if a gene modification by itself enhances or impairs autophagosome synthesis or degradation quantitatively (e.g., Clec16a [318–320]). With this aim, cells should be treated with an autophagy inducer in the presence and absence of a degradation inhibitor. As the LC3-II basal levels in the steady state may be different, it is necessary to establish a ratio to evaluate LC3-II synthesis and degradation flux. Therefore, the synthesis ratio can be considered as the rate of LC3-II levels in the presence of the inducer and the inhibitor divided by the LC3-II level in the presence of the inhibitor alone. Similarly, the degradation ratio would be the ratio of LC3-II levels in the cells treated with the inducer and the inhibitor divided by the LC3-II levels in the presence of the inducer alone. By comparing LC3-II synthesis and degradation ratios among different conditions, such as a gene modification, we can evaluate whether autophagy flux is modified by increasing or decreasing LC3-II synthesis or degradation phases [321, 322]. Alternatively, the degradation can be determined by calculating LC3-II levels in the presence of inducer and the inhibitor minus the levels in the presence of inducer alone [323].
The same type of assay monitoring the turnover of Atg8–PE can be used to monitor flux in yeast, by comparing the amount of Atg8 present in a wild-type versus a pep4∆ strain following autophagy induction [324]; however, it is important to be aware that the PEP4 knockout can influence yeast cell physiology (e.g., the inability to degrade and hence recycle autophagic cargo may trigger a starvation response). PMSF, which inhibits the activity of Prb1, can also be used to block Atg8–PE turnover.
Due to the advances in time-lapse fluorescence microscopy and the development of photoswitchable fluorescent proteins, autophagic flux can also be monitored by assessing the half-life of the LC3 protein [325, 326] post-photoactivation, by quantitatively measuring the autophagosomal pool size and its transition time [327], or by quantifying the rate of autophagosome formation [328]. Here, single-cell fluorescence live-cell imaging-based approaches, in combination with micropatterning, have shown accurate quantitative monitoring of autophagic flux that allows standardization of basal and induced flux in key cell types and model systems [312, 329] (Figure 10). These approaches deliver invaluable information on the kinetics of the system and the time required to clear a complete autophagosomal pool. Nonetheless, care must be taken for this type of analysis as changes in transcriptional/translational regulation of LC3 might also affect the readout.
Figure 10.
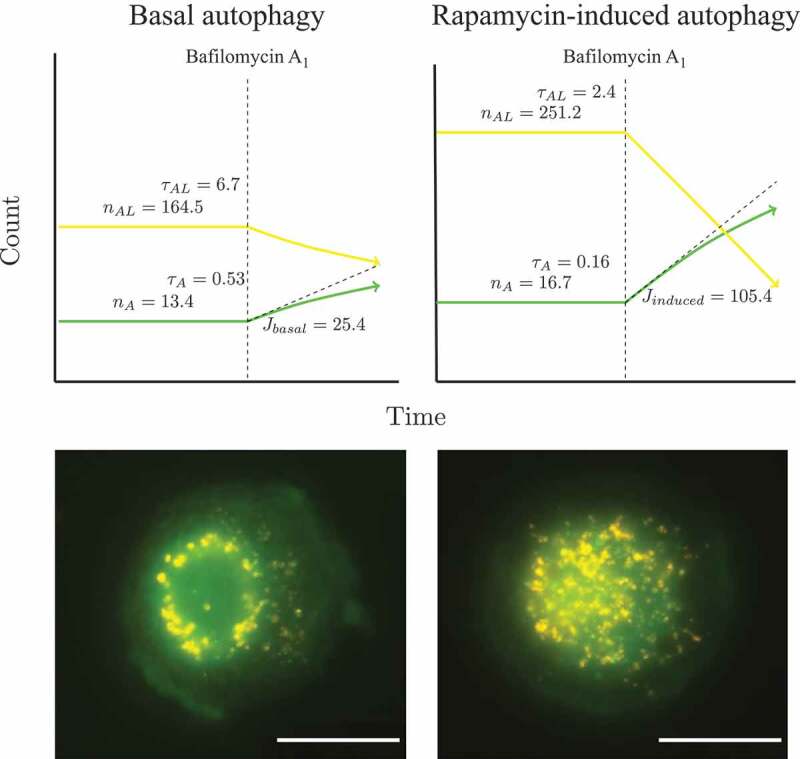
Measuring autophagic flux and pool size of pathway intermediates at the single-cell level: autophagosome, autolysosome and lysosome pool size and flux data, characterizing MEF cells with a basal flux of 25 autophagosomes/h/cell, which increases upon rapamycin treatment to 105 autophagosomes/h/cell. Scale bar: 20 µm. This figure was previously published in ref. [312]
Finally, autophagic flux can be monitored based on the turnover of LC3-II, by utilizing a luminescence-based assay. For example, a reporter assay based on the degradation of Renilla reniformis luciferase (Rluc)-LC3 fusion proteins is well suited for screening compounds affecting autophagic flux [330]. In this assay, Rluc is fused N-terminally to either wild-type LC3 or a lipidation-deficient mutant of LC3 (G120A). Because WT Rluc-LC3, in contrast to Rluc-LC3G120A, specifically associates with autophagosomal membranes, WT Rluc-LC3 is more sensitive to autophagic degradation. A change in autophagy-dependent LC3 turnover can thus be estimated by monitoring the change in the ratio of luciferase activities between the two cell populations expressing either WT Rluc-LC3 or Rluc-LC3G120A. In its simplest form, the Rluc-LC3-assay can be used to estimate autophagic flux at a single time point by defining the luciferase activities in cell extracts. Moreover, the use of a live cell luciferase substrate makes it possible to monitor changes in autophagic activity in live cells in real time. This method has been successfully used to identify positive and negative regulators of autophagy from cells treated with microRNA, siRNA and small molecule libraries [330–336].
Cautionary notes: The use of a radioactive pulse-chase analysis, which assesses complete autophagic flux, provides an alternative to lysosomal protease inhibitors [204]. Although such inhibitors should still be used to verify that degradation is lysosome dependent. In addition, drugs must be used at concentrations and for time spans that are effective in inhibiting fusion or degradation, but that do not provoke cell death. Thus, these techniques may not be practical in all cell types or in tissues from whole organisms where the use of protease inhibitors is problematic, and where pulse labeling requires artificial short-term culture conditions that may induce autophagy. Another concern when monitoring flux via LC3-II turnover may be seen in the case of a partial autophagy block; in this situation, agents that disrupt autophagy (e.g., bafilomycin A1) will still result in an increase in LC3-II. Thus, care is needed in interpretation. For characterizing new autophagy modulators, it is ideal to test autophagic flux at early (e.g., 4 h) and late (e.g., 24 h) time points, because in certain instances, such as with calcium phosphate precipitates, a compound may increase or decrease flux at these two time points, respectively [204]. Moreover, it is important to consider assaying autophagy modulators in a long-term response in order to further understand their effects. Finally, many of the chemicals used to inhibit autophagy, such as bafilomycin A1, NH4Cl or CQ (see Autophagy inhibitors and inducers), also directly inhibit the endocytosis/uncoating of viruses (D.R. Smith, personal communication), and other endocytic events requiring low pH, as well as exit from the Golgi (S. Tooze, personal communication). As such, agents that neutralize endosomal compartments should be used only with extreme caution in studies investigating autophagy-virus interactions. One means to address this is to carefully titrate the amounts of inhibitors to use, because, for example, low nanomolar amounts of bafilomycin A1 can affect autophagy without apparently affecting acidification during influenza virus infections [337].
One additional consideration is that it may not be absolutely necessary to follow LC3-II turnover if other substrates are being monitored simultaneously. For example, an increase in LC3-II levels in combination with the lysosomal (or ideally autophagy-specific) removal of an autophagic substrate (such as an organelle [338, 339]) that is not a good proteasomal substrate provides an independent assessment of autophagic flux. However, it is probably prudent to monitor both turnover of LC3-II and an autophagosome substrate in parallel, due to the fact that LC3 might be coupled to endosomal membranes and not just autophagosomes, and the levels of well-characterized autophagosome substrates such as SQSTM1/p62 can also be affected by proteasome inhibitors [340].
Another issue relates to the use of protease inhibitors (see Autophagy inhibitors and inducers). When using lysosomal protease inhibitors, it is of fundamental importance to assess proper conditions of inhibitor concentration and time of pre-incubation to ensure full inhibition of lysosomal cathepsins. In this respect, 1 h of pre-incubation with 10-20 µM E-64d is sufficient in most cases, because this inhibitor is membrane permeable and rapidly accumulates within lysosomes, but another frequently used inhibitor, leupeptin, requires at least 6 h pre-incubation [78, 341]. Moreover, pepstatin A is membrane impermeable (ethanol or preferably DMSO must be employed as a vehicle) and requires a prolonged incubation (> 8 h) and a relatively high concentration (>50-100 µM) to fully inhibit lysosomal CTSD (Figure 11). An incubation of this duration, however, can be problematic due to indirect effects (see GFP-Atg8-family protein lysosomal delivery and partial proteolysis). At least in neurons, pepstatin A alone is a less effective lysosomal proteolytic block, and combining a lysosomal cysteine protease (i.e., cathepsin) inhibitor with it is most effective [78]. Also, note that the relative amount of lysosomal CTSB (cathepsin B) and CTSD is cell-specific and changes with culture conditions. A possible alternative to pepstatin A is the pepstatin A BODIPY® FL conjugate [342, 343], which is transported to lysosomes via endocytosis. In contrast to the protease inhibitors, CQ (10-40 µM) or bafilomycin A1 (1-100 nM) can be added to cells immediately prior to autophagy induction, although in some cases a pre-incubation with bafilomycin A1 should be considered. bafilomycin A1 requires ~30 min to increase lysosomal pH [226, 344]; therefore, a pre-incubation of 30 min is required in case of short autophagy induction times. Because cysteine protease inhibitors may upregulate CTSD and some such as E-64d and its derivatives have potential inhibitory activity toward calpains, whereas bafilomycin A1 can have potential significant cytotoxicity, especially in cultured neurons and pathological states, the use of both methods may be important in some experiments to exclude off-target effects of a single method.
Figure 11.
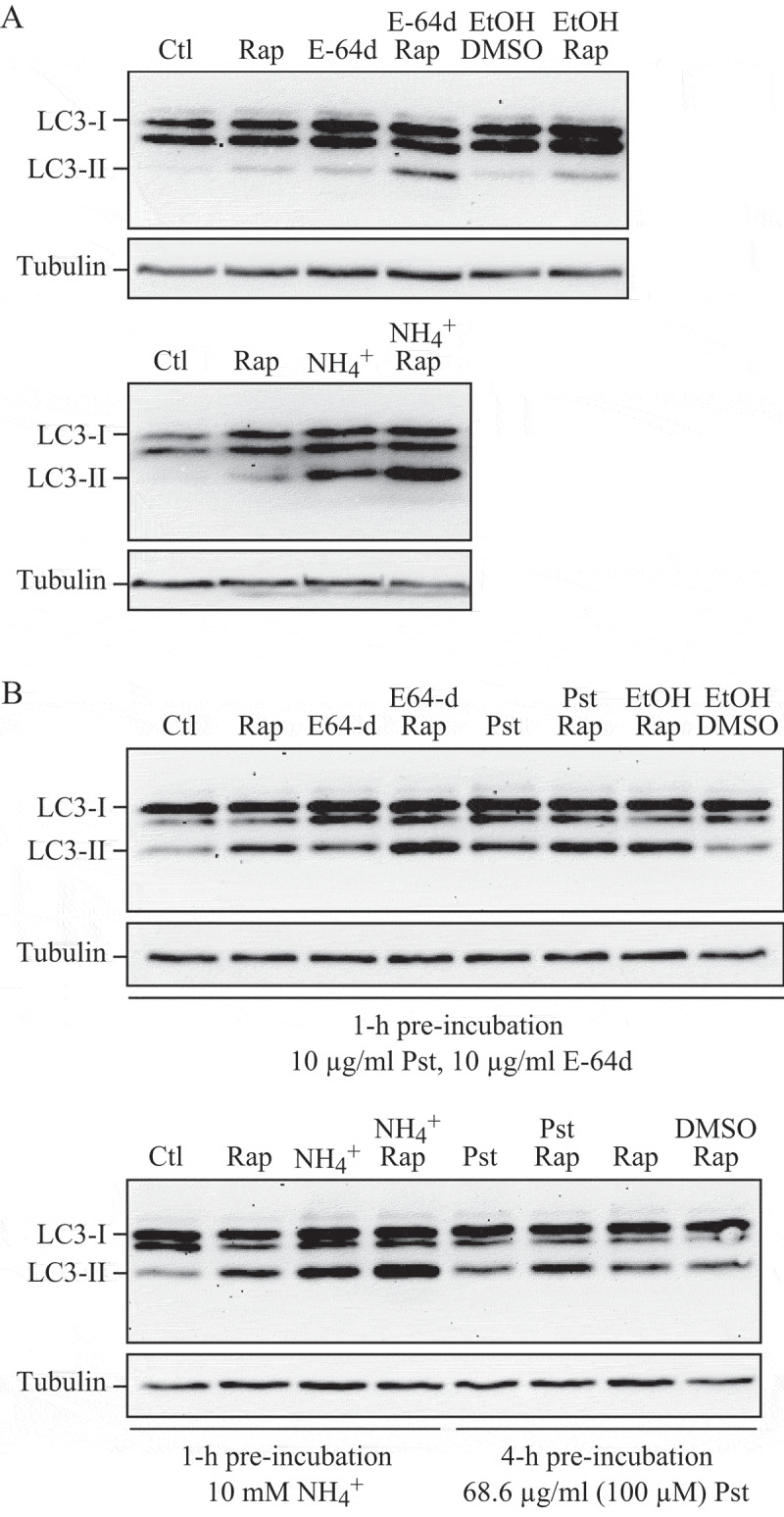
Effect of different inhibitors on LC3-II accumulation. SH-SY5Y human neuroblastoma cells were plated and allowed to adhere for a minimum of 24 h, then treated in fresh medium. Treatments were as follows: rapamycin (Rap), (A) 1 µM, 4 h or (B) 10 µM, 4 h; E-64d, final concentration 10 µg/ml from a 1 mg/ml stock in ethanol (EtOH); NH4Cl (NH4+), final concentration 10 mM from a 1 M stock in water; pepstatin A (Pst), final concentration 10 µg/ml from a 1 mg/ml stock in ethanol, or 68.6 µg/ml from a 6.86 mg/ml stock in DMSO; ethanol or DMSO, final concentration 1%. Pre-incubations in (B) were for 1 or 4 h as indicated. 10 mM NH4Cl (or 30 µM CQ, not shown) were the most effective compounds for demonstrating the accumulation of LC3-II. E-64d was also effective in preventing the degradation of LC3-II, with or without a preincubation, but ammonium chloride (or CQ) may be more effective. Pepstatin A at 10 µg/ml with a 1-h pre-incubation was not effective at blocking degradation, whereas a 100 µM concentration with 4-h pre-incubation had a partial effect. Thus, alkalinizing compounds are more effective in blocking LC3-II degradation, and pepstatin A must be used at saturating conditions to have any noticeable effect. Images provided by C. Isidoro. Note that the band running just below LC3-I at approximately 17.5 kDa may be a processing intermediate of LC3-I; it is detectable in freshly prepared homogenates, but is less visible after the sample is subjected to a freeze-thaw cycle
Conclusion: It is important to be aware of the difference between monitoring the steady-state level of Atg8-family proteins and autophagic flux. The latter may be assessed by following Atg8-family proteins in the absence and presence of autophagy flux inhibitors (such as lysosomal degradation inhibitors), and by examining the autophagy-dependent degradation of appropriate substrates. In particular, if there is any evidence of an increase in LC3-II (or autophagosomes), it is essential to determine whether this represents an induction of autophagy and increased synthesis of LC3, or decreased flux and the subsequent accumulation of LC3 due to a block in fusion or degradation, through the use of inhibitors such as CQ, bafilomycin A1 or lysosomal protease inhibitors. In the case of a suspected impaired degradation, assessment of lysosomal function (i.e., pH or activity of lysosomal enzymes) is then required to validate the conclusion and to establish the basis.
c. GFP-Atg8-family protein lysosomal delivery and partial proteolysis
GFP-LC3B (hereafter referred to as GFP-LC3) has also been used to follow flux. It should be cautioned that, as with endogenous LC3, an assessment of autophagic GFP-LC3 flux is a carrier flux that cannot be equated with, and is not necessarily representative of, an autophagic cargo flux. When GFP-Atg8 or GFP-LC3 is delivered to a lysosome/vacuole, the Atg8-family protein part of the chimera is sensitive to degradation, whereas the GFP protein is relatively resistant to hydrolysis (note, however, that GFP fluorescence is quenched by low pH; see GFP-Atg8-family protein fluorescence microscopy and Tandem mRFP/mCherry-GFP fluorescence microscopy). Therefore, the appearance of free GFP on western blots can be used to monitor lysis of the inner autophagosome membrane and breakdown of the cargo in metazoans (Figure 12A) [324, 345, 346], or the delivery of autophagosomes to, and the breakdown of autophagic bodies within, the fungal [347–349] and plant vacuole [289, 290, 324, 350]. Reports on Dictyostelium discoideum and mammalian cells highlight the importance of lysosomal pH as a critical factor in the detection of free GFP that results from the degradation of fused proteins. In these cell types, free GFP fragments are only detectable in the presence of nonsaturating levels of lysosomotropic compounds (NH4Cl or CQ) or under conditions that attenuate lysosomal acidity; otherwise, the autophagic/degradative machinery appears to be too efficient to allow the accumulation of the proteolytic fragment (Figure 12B,C) [40, 64, 351]. Hence, a reduction in the intensity of the free GFP band may indicate reduced flux, but it may also be due to efficient turnover. Using a range of concentrations and treatment times of compounds that inhibit autophagy can be useful in distinguishing between these possibilities [352]. Because the pH in the yeast vacuole is higher than that in mammalian or D. discoideum lysosomes, the levels of free GFP fragments are detectable in yeast even in the absence of lysosomotropic compounds [52]. Additionally, in yeast the diffuse fluorescent haze from the released GFP moiety within the vacuole lumen can be observed by fluorescence microscopy.
Figure 12.
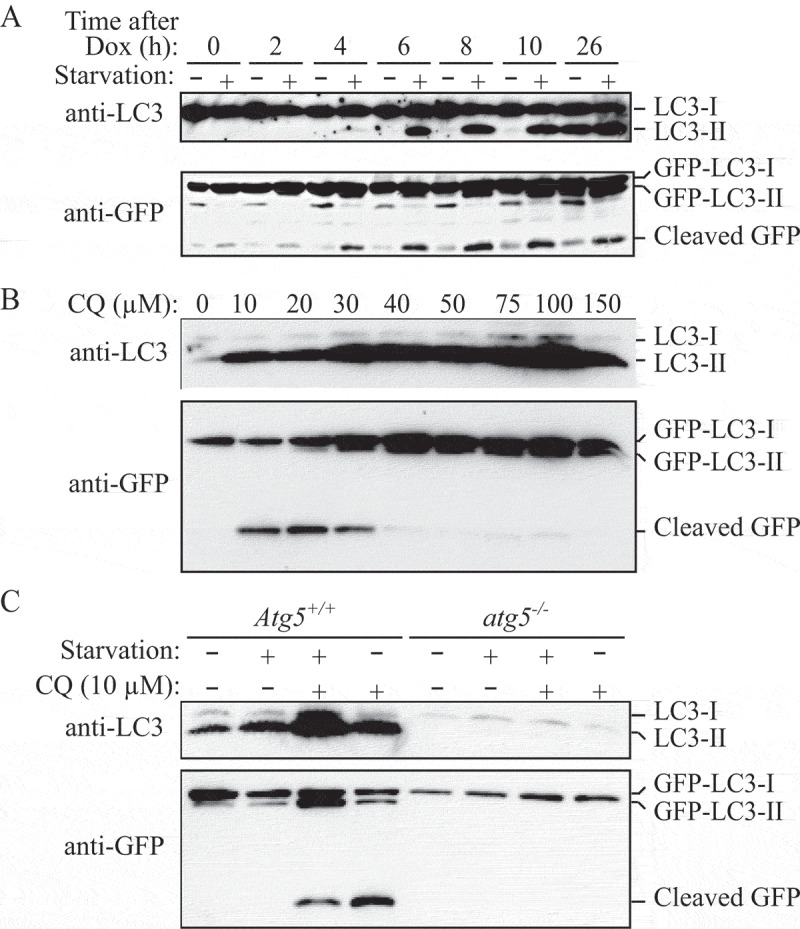
GFP-LC3 processing can be used to monitor delivery of autophagosomal membranes. (A) atg5−/− MEFs engineered to express Atg5 under the control of the Tet-off promoter were grown in the presence of doxycycline (Dox; 10 ng/ml) for one week to suppress autophagy. Cells were then cultured in the absence of drug for the indicated times, with or without a final 2-h starvation. Protein lysates were analyzed by western blot using anti-LC3 and anti-GFP antibodies. The positions of untagged and GFP-tagged LC3-I and LC3-II, and free GFP are indicated. This figure was modified from data previously published in ref. [346], FEBS Letters, 580, Hosokawa N, Hara Y, Mizushima N, Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size, pp. 2623-2629, copyright 2006, with permission from Elsevier. (B) Differential role of unsaturating and saturating concentrations of lysosomal inhibitors on GFP-LC3 cleavage. HeLa cells stably transfected with GFP-LC3 were treated with various concentrations of CQ for 6 h. Total lysates were prepared and subjected to immunoblot analysis. (C) CQ-induced free GFP fragments require classical autophagy machinery. Wild-type and atg5−/− MEFs were first infected with adenovirus GFP-LC3 (100 viral particles per cell) for 24 h. The cells were then either cultured in regular culture medium with or without CQ (10 µM), or subjected to starvation in EBSS buffer in the absence or presence of CQ for 6 h. Total lysates were prepared and subjected to immunoblot analysis. Panel B and C are modified from the data previously published in ref. [351]
The dynamic movement to lysosomes of GFP-LC3, or of its associated cargo, also can be monitored by time-lapse fluorescence microscopy, although, as mentioned above, the GFP fluorescent signal is more sensitive to acidic pH than other fluorophores (see GFP-Atg8-family protein fluorescence microscopy). A time-course evaluation of the cell population showing GFP-LC3 puncta can serve to monitor the autophagic flux, because a constant increase in the number of cells accumulating GFP-LC3 puncta is suggestive of defective fusion of autophagosomes with lysosomes. Conversely, a decline implies that GFP-LC3 is delivered to properly acidified lysosomes and may, in addition, reflect proteolytic elimination within them, although the latter needs to be independently established. In either case, it can be problematic to use GFP fluorescence to follow flux, as new GFP-LC3 is continuously being synthesized. A potential solution to this problem is to follow the fluorescence of a photoactivatable version of the fluorescent protein [353], which allows this assay to be performed essentially as a pulse/chase analysis. Another alternative to follow flux is to monitor GFP-LC3 fluorescence by adding lysosomal protease or fusion inhibitors to cells expressing GFP-LC3 and monitoring changes in the number of puncta. In this case, the presence of lysosomal inhibitors should increase the number of GFP-LC3-positive structures, and the absence of an effect on the total number of GFP-LC3 puncta or on the percentage of cells displaying numerous puncta is indicative of a defect(s) in autophagic flux [64, 354]. The combination of protease inhibitors (to prevent the degradation of GFP) or compounds that modify lysosomal pH and/or block fusion of autophagosomes such as NH4Cl, bafilomycin A1 or CQ, or compounds that block fusion of autophagosomes with lysosomes such as bafilomycin A1 or others (e.g., vinblastine) may be most effective in preventing lysosome-dependent decreases in GFP-LC3 puncta. However, because the stability of GFP is affected by lysosomal pH, researchers may also consider the use of protease inhibitors whether or not lysosomotropic compounds or fusion inhibitors are included.
Cautionary notes: The GFP-Atg8 processing assay is used routinely to monitor autophagy in yeast. One caveat, however, is that this assay is not always carried out in a quantitative manner. For example, western blot exposures need to be in the linear range. Accordingly, an enzymatic assay such as the Pho8∆60 assay may be preferred (see Autophagic protein degradation) [355, 356], especially when the differences in autophagic activity need to be determined precisely (note that an equivalent assay has not been developed for more complex eukaryotic cells); however, as with any enzyme assay, appropriate caution must be used regarding, for example, substrate concentrations and linearity. The Pho8∆60 assay also requires a control to verify equal Pho8∆60 expression in the different genetic backgrounds or conditions to be tested [356]; differences in Pho8∆60 expression potentially affect its activity and may thus cause misinterpretation of results. Another issue to keep in mind is that GFP-Atg8 processing correlates with the surface area of the inner sphere of the autophagosome, and thus provides a smaller signal than assays that measure the volume of the autophagosome. Pgk1 (3-phosphoglycerate kinase)-GFP processing [52] is another assay that can be used to monitor autophagy.
A thorough analysis of GFP proteolysis in plant roots reveals the importance of normalizing to tissue-specific reporter expression and autophagic activity range [102, 357]. For instance, GFP-ATG8 expression in Arabidopsis thaliana is typically highest in the root apical meristem, but the response to the autophagy-inducing conditions in this root zone is much lower compared to the rest of the root. Thus, excluding this root zone from the samples for western blot provides a much more reliable readout of the GFP-ATG8 proteolysis.
As a note of caution, GFP-LC3 has been demonstrated to be present in protein aggregates in an autophagy-unrelated manner and this association is dependent on its interaction with SQSTM1. This interaction poses potential difficulties to distinguish LC3 bound to aggregates from those on autophagosomes [358]. The main limitation of the GFP-LC3 processing assay in mammalian cells is that its usefulness seems to depend on cell type and culture conditions (N. Hosokawa and N. Mizushima, unpublished data). Apparently, GFP is more sensitive to mammalian lysosomal hydrolases than to the degradative milieu of the yeast vacuole or the lysosomes in Drosophila. Alternatively, the lower pH of mammalian lysosomes relative to that of the yeast vacuole may contribute to differences in detecting free GFP. Under certain conditions (such as Earle’s balanced salt solution [EBSS]-induced starvation) in some cell lines, when the lysosomal pH becomes particularly low, free GFP is undetectable because both the LC3-II and free GFP fragments are quickly degraded [273]. Therefore, if this method is used it should be accompanied by immunoblotting and include controls to address the stability of nonlysosomal GFP such as GFP-LC3-I. It should also be noted that free GFP can be detected when cells are treated with nonsaturating doses of inhibitors such as CQ, E-64d and bafilomycin A1. The saturating concentrations of these lysosomal inhibitors vary in different cell lines, and it would be better to use a saturating concentration of lysosomal inhibitors when performing an autophagic flux assay [273]. Therefore, caution must be exercised in interpreting the data using this assay; it would be helpful to combine an analysis of GFP-LC3 processing with other assays, such as the monitoring of endogenous LC3-II by western blot.
Along these lines, a caution concerning the use of the EGFP fluorescent protein for microscopy is that this fluorophore has a relatively neutral pH optimum for fluorescence [263], and its signal diminishes quickly during live cell imaging due to the acidic environment of the lysosome. It is possible to circumvent this latter problem by imaging paraformaldehyde-fixed cultures that are maintained in a neutral pH buffer, which retains EGFP fluorescence (M. Kleinman and J.J. Reiners, personal communication). Alternatively, it may be preferable to use a different fluorophore such as mRFP or mCherry, which retain fluorescence even at acidic pH [341]. On the one hand, a putative advantage of mCherry over mRFP is its enhanced photostability and intensity, which are an order of magnitude higher (and comparable to GFP), enabling acquisition of images at similar exposure settings as are used for GFP, thus minimizing potential bias in interpretation [342]. On the other hand, caution is required when evaluating the localization of mCherry fusion proteins during autophagy due to the persistence of the mCherry signal in acidic environments; all tagged proteins are prone to show enrichment in lysosomes during nonselective autophagy of the cytoplasm, especially at higher expression levels. In addition, red fluorescent proteins (even the monomeric forms) can be toxic due to oligomer formation [343]; the tendency to form abnormal accumulations may be a general feature of coral- and anemone-derived fluorescent proteins. Dendra2 is an improved version of the green-to-red photoswitchable fluorescent protein Dendra, which is derived from the octocoral Dendronephthya sp [344]. Dendra2 is capable of irreversible photoconversion from a green to a red fluorescent form, but can be used also as normal GFP or RFP vector. This modified version of the fluorophore has certain properties including a monomeric state, low phototoxic activation and efficient chromophore maturation, which make it suitable for real-time tracking of LC3 and SQSTM1 (Figure 13; [359]). A newer generation of photoswitchable proteins, EOS, are now available that are brighter than Dendra2 and display more efficient photoswitching (N.A. Castello and S. Finkbeiner, in press). Another alternative to mRFP or mCherry is to use the Venus variant of YFP, which is brighter than mRFP and less sensitive to pH than GFP [345].
Figure 13.
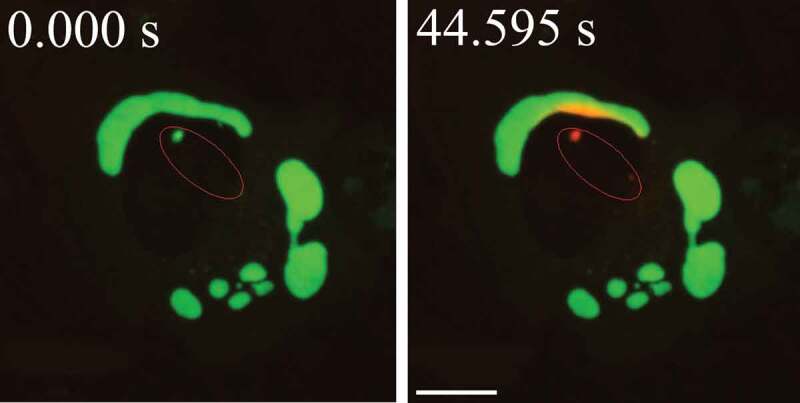
Movement of activated pDendra2-hp62 (SQSTM1; orange) from the nucleus (middle) to an aggregate in ARPE-19 cells, revealed by confocal microscopy. Cells were exposed to 5 µM MG132 for 24 h to induce the formation of perinuclear aggregates [4083]. The cells were then exposed to a UV pulse (the UV-induced area is shown by red lines that are inside of the nucleus) that converts Dendra2 from green to red, and the time shown after the pulse is indicated. SQSTM1 is present in a small nuclear aggregate, and is shuttled from the nucleus to a perinuclear large protein aggregate (detected as red). Scale bar: 5 µm. Image provided by K. Kaarniranta
The pH optimum of EGFP is important to consider when using GFP-LC3 constructs, as the original GFP-LC3 marker [346] uses the EGFP variant, which may result in a reduced signal upon the formation of amphisomes or autolysosomes. An additional caveat when using the photoactivatable construct PA-GFP [301] is that the process of activation by photons may induce DNA damage, which could, in turn, induce autophagy. Also, GFP is relatively resistant to denaturation, and boiling for 5 min may be needed to prevent the folded protein from being trapped in the stacking gel during SDS-PAGE.
As noted above (see Western blotting and ubiquitin-like protein conjugation systems), Atg4/ATG4 cleaves the residue(s) that follow the C-terminal glycine of Atg8-family proteins that will be conjugated to PE. Accordingly, it is critical that any chimeras should be constructed with the fluorescent tag at the amino terminus of Atg8-family proteins (unless the goal is to monitor Atg4/ATG4 activity).
Finally, lysosomal inhibition needs to be carefully controlled. Prolonged inhibition of lysosomal hydrolases (>6 h) is likely to induce a secondary autophagic response triggered by the accumulated undigested autophagy cargo. This secondary autophagic response can complicate the analysis of the autophagic flux, making it appear more vigorous than it would in the absence of the lysosomal inhibitors.
Conclusion: The GFP-Atg8 (or GFP-LC3/GABARAP) processing assay, which monitors free GFP generated within the vacuole/lysosome, is a convenient way to follow autophagy, but it does not work in all cell types, and is not as easy to quantify as enzyme-based assays. Furthermore, the assay measures the flux of an autophagic carrier, which may not necessarily be equivalent to autophagic cargo flux.
d. HaloTag-LC3 autophagosome completion assay
Upon phagophore closure, LC3-II on the convex side of the membrane is delipidated and recycled back into the cytosol, while that on the concave side is sequestered within the vacuole and delivered into the lysosome for degradation [240]. Exploiting the topological property of LC3, the HaloTag-LC3 (HT-LC3) assay is designed to analyze the process of phagophore closure (Figure 14A) [360]. The HaloTag is a modified haloalkane dehalogenase that covalently binds to synthetic HaloTag ligands [361]. The HT-LC3 assay employs the HaloTag-conjugated LC3 reporter in combination with membrane-permeable and -impermeable HaloTag ligands labelled with two different fluorescent dyes to distinguish membrane-unenclosed and -enclosed HT-LC3-II. By sequentially incubating plasma membrane-permeabilized HT-LC3-expressing cells with a saturating dose of membrane-impermeable ligands (MILs) followed by membrane-permeable ligands (MPLs), phagophores, nascent autophagosomes, and mature autophagosomes or autolysosomes are visualized as MIL+ MPL−, MIL+ MPL+, and MIL− MPL+ structures, respectively (Figure 14B). Because the cytosolic HT-LC3-I is released upon plasma membrane permeabilization, the assay provides a superior signal-to-noise ratio and the data can be semi-quantitatively analyzed by confocal or fluorescence microscopy. As MPL fluorescent signals are not retained in functional lysosomes, autophagic flux can also be measured by monitoring MPL signal accumulation upon exposure to a lysosomal inhibitor. Moreover, the assay has been successfully adapted to a fluorescence-activated cell sorting (FACS)-based high-throughput platform to screen genes required for phagophore closure [362].
Figure 14.
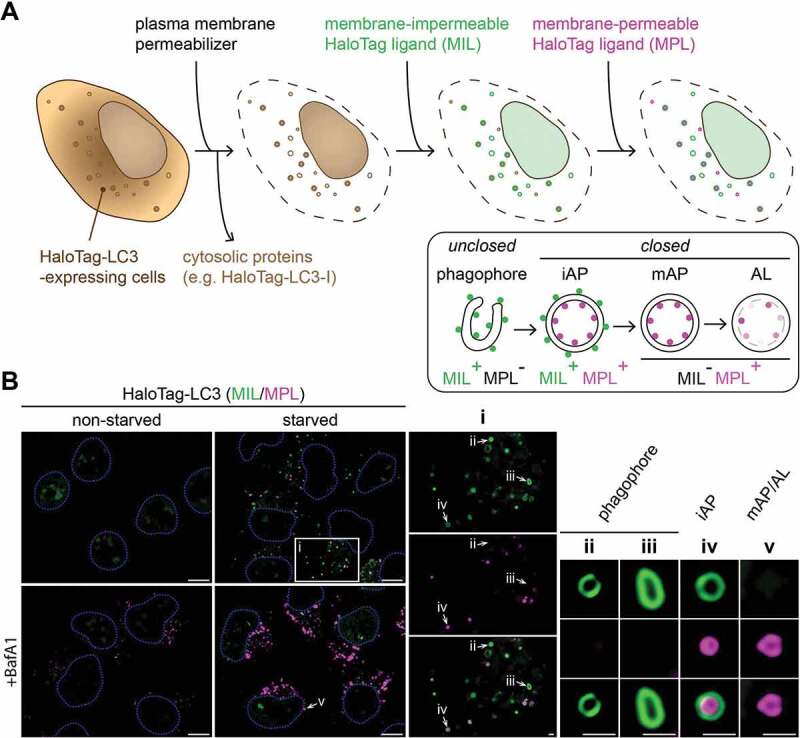
The HaloTag-LC3 assay distinguishes phagophores, immature autophagosomes, and mature autophagosomes and autolysosomes. (A) Schematic diagram of the HaloTag-LC3 (HT-LC3) assay. Cells expressing HT-LC3 are treated with a cholesterol-dependent plasma membrane permeabilizer to release cytosolic proteins including HT-LC3-I and sequentially labeled with a saturated dose of membrane-impermeable HaloTag ligands (MILs) conjugated with Alexa Fluor 488 (or 660) followed by membrane-permeable HaloTag ligands (MPLs) conjugated with tetramethylrhodamine to detect phagophores (MIL+ MPL−), immature autophagosomes (iAP; MIL+ MPL+), and mature autophagosomes and autolysosomes (AP and AL; MIL− MPL+). (B) U-2 OS cells were stably transduced with HT-LC3-encoding lentiviruses, incubated in starvation medium or control complete medium in the presence or absence of 100 nM bafilomycin A1 (BafA1) for 4 h, and subjected to the HT-LC3 assay followed by confocal microscopy. Magnified images of the boxed (i) and arrow-indicated (ii-v) areas are shown in the right panels. Scale bars: 10 μm (1 μm in the magnified images)
Cautionary notes: Similar to fluorescent protein(s)-tagged LC3 assays, the HT-LC3 assay requires a system amenable to exogenous introduction. In addition, the assay requires plasma membrane permeabilization. Therefore, it would be challenging to use the assay in 3-dimensional-cultured cells, tissue samples, or live-cell imaging. Moreover, the current method employs cholesterol-dependent pore-forming agents such as recombinant perfringolysin O [363] and digitonin to permeabilize the plasma membrane. Therefore, it would also be challenging when a treatment or gene manipulation perturbs plasma membrane cholesterol distribution (e.g., prolonged treatment with a lysosomal inhibitor [Y. Takahashi and H.G. Wang, personal communication]). Along this line, because the plasma membrane cholesterol concentration is different among cell types, it is important to find an optimal permeabilization condition. If plasma membrane permeabilization fails or is incomplete, diffuse MPL signals, which represent cytosolic HT-LC3-I, will be detected in addition to cytoplasmic HT-LC3-II foci. In addition, it is critical to ensure the saturation of all available binding sites with each ligand. A secondary incubation with the same type of ligands conjugated with a different fluorophore (e.g., primary incubation with Alexa Fluor 488-conjugated MILs followed by secondary incubation with Alexa Fluor 600-conjugated MILs) make it possible to determine an appropriate staining condition. Another concern for the assay is that the detection of membrane-unenclosed HT-LC3-II relies on the accessibility of MILs. Therefore, if the pore size of the closure site is too small to pass through MILs, HT-LC3-II on the concave side of phagophores will be falsely negative for MILs; the structure will be detected as MIL+ MPL+ in this case.
Conclusion: Using two HaloTag ligands with different membrane permeability and fluorophores, the HT-LC3 assay can determine each step of autophagy by distinguishing membrane-unenclosed and -enclosed HT-LC3-II. However, unlike a fluorescent protein-tagged LC3 assay, the HT-LC3 assay requires several optimization steps to ensure the staining specificity. Once optimized, this assay provides a superior signal-to-noise ratio and is compatible with high-throughput screening platforms.
e. GFP-Atg8-family protein fluorescence microscopy
LC3B, or the protein tagged at its N terminus with a fluorescent protein such as GFP (GFP-LC3), has been used to monitor autophagy through indirect immunofluorescence or direct fluorescence microscopy (Figure 15), measured as an increase in punctate LC3 or GFP-LC3 [364, 365]. The detection of GFP-Atg8 (or GFP-LC3/GABARAP/LGG-1/2) is also useful for in vivo studies using transgenic organisms such as Saccharomyces cerevisiae [366], Aspergillus nidulans [348], Caenorhabditis elegans [367], D. discoideum [368], filamentous ascomycetes [369–373], Ciona intestinalis [374], Drosophila melanogaster [375–377], A. thaliana [378], Zea mays [379], Trypanosoma brucei [380–382], Leishmania major [383–385], Trypanosoma cruzi [386, 387], zebrafish [328, 388] and mice [239]. “Super-resolution” fluorescence images of GFP-LC3-positive phagophores have been shown in platelets prepared from GFP-LC3 mice by “super-resolution” microscopy (specifically, 3-dimensional structured illumination microscopy/3D-SIM) to be similar to what was observed by TEM [389, 390]. It is also possible to use anti-Atg8-family protein antibodies for immunocytochemistry or immunohistochemistry (IHC) [265, 391–397], procedures that have the advantages of detecting the endogenous protein, obviating the need for transfection and/or the generation of a transgenic organism, as well as avoiding potential artefacts resulting from overexpression. For example, high levels of overexpressed GFP-LC3 can result in its nuclear localization, although the protein can still relocate to the cytosol upon starvation. The use of imaging cytometry allows rapid and quantitative measures of the number of LC3 puncta and their relative number in individual or mixed cell types, using computerized assessment, enumeration, and data display (e.g., see refs. [41, 398]). In this respect, the alternative use of an automated counting system may be helpful for obtaining an objective number of puncta per cell. For this purpose, the WatershedCounting3D plug-in for ImageJ may be useful [399, 400]. Changes in the number of GFP-Atg8 puncta can also be monitored using flow cytometry (see Autophagic flux determination using flow and multispectral imaging cytometry) [382]. An alternative way to quantify LC3 immunofluorescence staining is to estimate the percentage of LC3 signals originating from puncta over total LC3 signals in the same cell [401]. This approach is useful if it is difficult to define the number of puncta per cell due to widely varying size or clustering of the puncta. A key control to perform when using these approaches is the use of a non-lipidatable mutant version of the Atg8-family protein that does not associate with autophagosomes.
Figure 15.
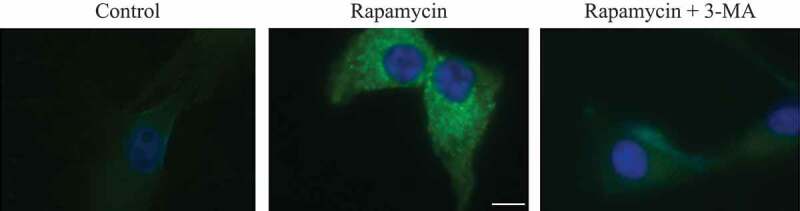
Changes in the detection and localization of GFP-LC3 upon the induction of autophagy. U87 cells stably expressing GFP-LC3 were treated with PBS (Control), rapamycin (200 nM), or rapamycin in combination with 3-MA (2 mM) for 24 h. Representative fluorescence images of cells counterstained with DAPI (nuclei) are shown. Scale bar: 10 µm. This figure was modified from Figure 6 published in ref. [364], Badr et al. Lanatoside C sensitizes glioblastoma cells to tumor necrosis factor–related apoptosis-inducing ligand and induces an alternative cell death pathway. Neuro-Oncology, 13(11):1213-24, 2011, by permission of Oxford University Press
LC3-positive autophagosomes can be quantified by confocal microscopy using a software program called Imaris (Oxford Instruments). Confocal Z-stacks of samples immunolabeled with an antibody to LC3 are reconstructed into 3-dimensional animations with the aid of Imaris software. The Spot function in Imaris automatically locates and enumerates autophagosomes within individual cells based on size and intensity thresholds [402, 403].
Monitoring the endogenous Atg8-family proteins obviously depends on the ability to detect these proteins in the system of interest, which is not always possible. If the endogenous amount is below the level of detection, the use of an exogenous construct is warranted. In this case, it is important to consider the use of stable transformants versus transient transfections. On the one hand, stable transformants may have reduced background resulting from the lower gene expression, and artefacts resulting from recent exposure to transfection reagents (see below) are eliminated. Furthermore, with stable transformants more cells can be easily analyzed because nearly 100% of the population will express tagged LC3. On the other hand, a disadvantage of stable transfectants is that the integration sites cannot always be predicted, and expression levels may not be optimal. Therefore, it is worth considering the use of stable episomal plasmids that avoid the problem of unsuitable integration [344]. An important advantage of transient transfection is that this approach is better for examining the immediate effects of the transfected protein on autophagy; however, the transient transfection approach restricts the length of time that the analysis can be performed, and consideration must be given to the induction of autophagy resulting from exposure to the transfection reagents (see below). One word of caution is that optimizing the time of transient expression of GFP-LC3 is necessary, as some cell types (e.g., HeLa cells) may require 1 day for achieving optimal expression to visualize GFP-LC3 puncta, whereas neuronal cell lines such as SH-SY5Y cells typically need at least 48 h of expression prior to performing GFP-LC3 puncta analyses. In addition, a double transfection can be used (e.g., with GFP-LC3 and the protein of interest) to visually tag the cells that express the protein being examined.
A disadvantage of transfecting GFP-LC3 with liposomes is that frequently it leads to an unstable efficiency of transfection, causing a reduction in the number of cells effectively expressing GFP-LC3, and degradation of the plasmid, thus decreasing the numbers of GFP-LC3 puncta. Stable cell lines expressing GFP-LC3 can be generated using lentiviral systems and efficiently selected through antibiotic resistance leading to uniform and prolonged expression levels. These stable cell lines are sensitive to autophagy inducers as measured by the LC3-II:LC3-I ratio by western blot, and also show increased numbers of cytoplasmic GFP-LC3 puncta upon autophagic stimuli (unpublished results R. Muñoz-Moreno, R.I. Galindo, L. Barrado-Gil and C. Alonso).
In conclusion, there is no simple rule for the use of stable versus transient transfections. When stable transfections are utilized through a nonlentiviral system it is worthwhile screening for stable clones that give the best signal to noise ratio; when transient transfections are used, it is worthwhile optimizing the GFP-LC3 DNA concentration to give the best signal-to-noise ratio (note potential problems with transfections under Western blotting and ubiquitin-like protein conjugation systems). In clones, the uniformity of expression of GFP-LC3 facilitates “thresholding” when scoring puncta-positive cells (see below). However, there is also a need to be aware that a single cell clone may not be representative of the overall pool. Using a pool of multiple selected clones may reduce artefacts that can arise from the selection and propagation of individual clones from a single transfected cell (although the use of a pool is also problematic as its composition will change over time). Another possibility is to select a mixed stable population with uniform GFP-LC3 expression levels by the use of a fluorescence-activated cell sorter (FACS) [404]. Optimization, together with including the appropriate controls (e.g., transfecting GFP-LC3G120A as a negative control), will help to overcome the effects of the inherent variability in these analyses. For accurate interpretations, it is also important to assess the level of overexpression of the GFP-LC3 constructs relative to endogenous LC3 by western blot. Finally, a recent advent of CRISPR-Cas9 gene-editing technologies provides a promising alternative to overcome potential pitfalls of GFP-LC3 overexpression—the generation of knockin cell lines, in which the coding sequence of GFP is added in frame with the 5ʹ sequence (encoding the N-terminal part) of endogenous LC3 [120, 405].
An additional use of GFP-LC3 is to monitor colocalization with a target during autophagy-related processes such as organelle degradation or the sequestration of pathogenic microbes [299-302]. Preincubation of cells stably expressing GFP-LC3 with leupeptin can help stabilize the GFP-LC3 signal during fluorescence microscopy, especially under conditions of induced autophagic flux. Leupeptin is an inhibitor of lysosomal cysteine and serine proteases and will therefore inhibit degradation of membrane-conjugated GFP-LC3 that is present within autolysosomes.
Cautionary notes: Quantification of autophagy by measuring GFP-LC3 puncta (or LC3 by immunofluorescence) can, depending on the method used, be more tedious than monitoring LC3-II by western blot; however, the former may be more sensitive and quantitative. Ideally, it is preferable to include both assays and to compare the two sets of results. In addition, if GFP-LC3 is being quantified, it is better to determine the number of puncta corresponding to GFP-LC3 on a per cell basis (or per cell area basis) rather than simply the total number (or percentage) of cells displaying puncta. This latter point is critical because, even in nutrient-rich conditions, cells display some basal level of GFP-LC3 puncta. There are, however, practical issues with counting puncta manually and reliably, especially if there are large numbers per cell. Nevertheless, manual scoring may be more accurate than relying on a software program, in which case it is important to ensure that only appropriate puncta are being counted (applicable programs include ImageJ, Imaris, and the open-source software CellProfiler [406]). Moreover, when autophagosome-lysosome fusion is blocked, larger autophagosomes are detected, possibly due to autophagosome-autophagosome fusion, or to an inability to resolve individual autophagosomes when they are present in large numbers. Although it is possible to detect changes in the size of GFP-Atg8-family protein puncta by fluorescence microscopy, it is not possible to correlate size with autophagy activity without additional assay methods. Size determinations can be problematic by fluorescence microscopy unless careful standardization is carried out [407], and size estimation on its own without considering puncta number per cell is not recommended as a method for monitoring autophagy; however, it is possible to quantify the fluorescence intensity of GFP-Atg8-family proteins at specific puncta, which does provide a valid measure of protein recruitment [408].
In addition to autophagosome size, the number of puncta visible to the eye will also be influenced by both the level of expression of GFP-LC3 in a given cell (an issue that can be avoided by analyzing endogenous LC3 by immunofluorescence) and by the exposure time of the microscope, if using widefield microscopy. Another way to account for differential GFP-LC3 expression levels and/or exposure is to normalize the intensity of GFP-LC3 present in the puncta to the total GFP-LC3 intensity in the cell. This can be done either on the population level [306] or individual cell level [404]. The approach to measuring the proportion of total LC3 signals originating from puncta is also suitable for quantification of immunofluorescence staining of endogenous LC3. In many cell types it may be possible to establish a threshold value for the number of puncta per cell in conditions of “low” and “high” autophagy [409]. This can be tested empirically by exposing cells to autophagy-inducing and -blocking agents. Thus, cell populations showing significantly greater proportions of cells with autophagosome numbers higher than the threshold in perturbation conditions compared to the control cells could provide quantitative evidence of altered autophagy. It is then possible to score the population as the percentage of cells displaying numerous autophagosomes. This approach will only be feasible if the background number of puncta is relatively low. For this method, it is particularly important to count a large number of cells and multiple representative sections of the sample. Typically, it is appropriate to score on the order of 50 or more cells, preferably in at least three different fields, depending on the particular system and experiment, but the critical point is that this determination should be based on statistical power analysis. Accordingly, high-content imaging analysis methods enable quantification of GFP-LC3 puncta (or overall fluorescence intensity) in thousands of cells per sample (e.g., see refs. [334, 352, 410]). When using automated analysis methods, care must be taken to manually evaluate parameters used to establish background threshold values for different treatment conditions and cell types, particularly as many systems image at lower magnifications that may be insufficient to resolve individual puncta. Another note of caution is that treatments affecting cell morphology, leading to the “rounding-up” of cells for example, can result in apparent changes in the number of GFP-LC3 puncta per cell. To avoid misinterpretation of results due to such potential artefacts, manual review of cell images is highly recommended. If cells are rounding up due to apoptosis or mitosis, it is easy to automatically remove them from analysis based on nuclear morphology (using DAPI or Hoechst staining) or cell roundness. If levels of autophagy in the rounded-up cells are of particular interest, images can be acquired as z-stacks and either analyzed as a z-series or processed to generate maximum projection or extended depth-of-field images and then analyzed [411].
To allow comparisons by other researchers attempting to repeat these experiments, it is critical that the authors also specify the baseline number of puncta that are used to define “normal” or “low” autophagy. Furthermore, the cells should be counted using unbiased procedures (e.g., using a random start point followed by inclusion of all cells at regular intervals), and statistical information should be provided for both baseline and altered conditions, as these assays can be highly variable. One possible method to obtain unbiased counting of GFP-LC3 puncta in a large number of cells is to perform multispectral imaging flow cytometry (see Autophagic flux determination using flow and multispectral imaging cytometry) [412, 413]. Multispectral imaging flow cytometry allows characterization of single cells within a population by assessing a combination of morphology and immunofluorescence patterns, thereby providing statistically meaningful data [414]. This method can also be used for endogenous LC3, and, therefore, is useful for nontransfected primary cells [415]. For adherent cell cultures, one caution for flow cytometry is that the techniques necessary to produce single cell suspensions can cause significant injury to the cells, leading to secondary changes in autophagy. Therefore, staining for plasma membrane permeabilization (e.g., cell death) before versus after isolation is an important control, and allowing a period of recovery between harvesting the culture and staining is also advisable [416].
An important caveat in the use of GFP-LC3 is that this chimera can associate with aggregates, especially when expressed at high levels in the presence of aggregate-prone proteins, which can lead to a misinterpretation of the results [417]. Of note, GFP-LC3 can associate with ubiquitinated protein aggregates [418]; however, this does not occur if the GFP-LC3 is expressed at low levels (D.C. Rubinsztein, unpublished observations). These aggregates have been described in many systems and are also referred to as aggresome-like induced structures (ALIS) [418–420], dendritic cell ALIS/DCALIS [421], SQSTM1 bodies/sequestosomes [422, 423] and inclusions. Indeed, many microbe-associated molecular patterns (MAMPs) described to induce the formation of autophagosomes in fact trigger massive formation of SQSTM1 bodies (L.H. Travassos, unpublished observations). Inhibition of autophagy in vitro and in vivo leads to the accumulation of these aggregates, suggesting a role for autophagy in mediating their clearance [418, 420, 422, 424, 425]. One way to control for background levels of puncta is to determine fluorescence from untagged GFP.
The receptor protein SQSTM1 is required for the formation of ubiquitinated protein aggregates in vitro (see SQSTM1 and related LC3 binding protein turnover assays) [423]. In this case, the interaction of SQSTM1 with both ubiquitinated proteins and LC3 is thought to mediate delivery of these aggregates to the autophagy system [426, 427]. Many cellular stresses can induce the formation of aggregates, including transfection reagents [418], or foreign DNA (especially if the DNA is not extracted free of endotoxin). SQSTM1-positive aggregates are also formed by proteasome inhibition or puromycin treatment, and can be found in cells exposed to rapamycin for extended periods where the rates of autophagy are elevated [428]. Calcium phosphate transfection of COS7 cells or lipofectamine transfection of MEFs (R. Pinkas-Kramarski, personal communication), primary neurons (A.R. La Spada, personal communication) or neuronal cells (C.T. Chu, personal communication; [429]) transiently increases basal levels of GFP-LC3 puncta and/or the amount of LC3-II. One solution to this artefact is to examine GFP-LC3 puncta in cells stably expressing GFP-LC3; however, as transfection-induced increases in GFP-LC3 puncta and LC3-II are often transient, another approach is to use cells transfected with GFP, with cells subjected to a mock time-matched transfection as the background (negative) control. A lipidation-defective LC3 mutant where glycine 120 is mutated to alanine is targeted to these aggregates independently of autophagy (likely via its interaction with SQSTM1, see above); as a result, this mutant can serve as another specificity control [418]. When carrying out transfections it may be necessary to alter the protocol depending on the level of background fluorescence. For example, changing the medium and waiting 24 to 48 h after the transfection can help to reduce the background level of GFP-LC3 puncta that is due to the transfection reagent (M. I. Colombo, personal communication). Similarly, when using an mCherry-GFP-SQSTM1 double tag (see Tandem mRFP/mCherry-GFP fluorescence microscopy) in transient transfections it is best to wait 48 h after transfection to reduce the level of aggregate formation and potential inhibition of autophagy (T. Johansen, personal communication). An additional consideration is that, in addition to transfection, viral infection can activate stress pathways in some cells and possibly induce autophagy. Influenza virus induces autophagy and autophagy is required for subsequent viral-induced apoptosis [337]. Proteomic screens show that several viruses, including influenza virus [430] and Zika virus [431], can significantly alter the expression of numerous proteins involved in autophagy and other cell stress pathways. This again emphasizes the importance of appropriate controls, such as control viruses expressing GFP [432].
The formation and clearance of ubiquitinated protein aggregates appear to represent a cellular recycling process. Aggregate formation can occur when autophagy is either inhibited or when its capacity for degradation is exceeded by the formation of proteins delivered to the aggregates. In principle, formation of GFP-LC3-positive aggregates represents a component of the autophagy process. However, the formation of GFP-LC3-positive ubiquitinated protein aggregates does not directly reflect either the induction of autophagy (or autophagosome formation) or flux through the system. Indeed, formation of ubiquitinated protein aggregates that are GFP-LC3 positive can occur in autophagy-deficient cells [418]. Therefore, it should be remembered that GFP-LC3 puncta likely represent a mix of ubiquitinated protein aggregates in the cytosol, ubiquitinated protein aggregates within autophagosomes and/or more “conventional” phagophores and autophagosomes bearing other cytoplasmic cargo (this is one example where CLEM could help in resolving this question [119]). In D. discoideum, inhibition of autophagy leads to large ubiquitinated protein aggregates containing SQSTM1 and GFP-Atg8, when the latter is co-expressed [422]; the large size of the aggregates makes them easily distinguishable from autophagosomes. Saponin treatment has been used to reduce background fluorescence under conditions where no aggregation of GFP-LC3 is detected in hepatocytes, GFP-LC3 stably-transfected HEK 293 [432] and human osteosarcoma cells, and in nontransfected cells [433]; however, because treatment with saponin and other detergents can provoke artefactual GFP-LC3 puncta formation [434], specificity controls need to be included in such experiments. In general, it is preferable to include additional assays that measure autophagy rather than relying solely on monitoring GFP-LC3. In addition, we recommend that researchers validate their assays by demonstrating the absence or reversal of GFP-LC3 puncta formation in cells treated with pharmacological or RNA interference-based autophagy inhibitors (Table 1). For example, 3-methyladenine (3-MA) is commonly used to inhibit starvation- or rapamycin-induced autophagy [435], but it has no effect on BECN1-independent forms of autophagy [118, 208], and some data indicate that this compound can also have stimulatory effects on autophagy (see Autophagy inhibitors and inducers) [436, 437], as well as induce cell death at progressively higher concentrations [438].
Another general limitation of the GFP-LC3 assay is that it requires a system amenable to the introduction of an exogenous gene. Accordingly, the use of GFP-LC3 in primary non-transgenic cells is more challenging. Here again, controls need to be included to verify that the transfection protocol itself does not artefactually induce GFP-LC3 puncta or cause LC3 aggregation. Furthermore, transfection should be performed with low levels of constructs, and the transfected cells should be followed to determine: i) when sufficient expression for detection is achieved, and ii) that, during the time frame of the assay, basal GFP-LC3 puncta remain appropriately low. In addition, the demonstration of a reduction in the number of induced GFP-LC3 puncta under conditions of autophagy inhibition is helpful. For some primary cells, delivering GFP-LC3 to precursor cells by infection with recombinant lentivirus, retrovirus or adenovirus [439], and subsequent differentiation into the cell type of interest, is a powerful alternative to transfection of the already differentiated cell type [103].
To implement the scoring of autophagy via fluorescence microscopy, one option is to measure pixel intensity. Because the expression of GFP-LC3 may not be the same in all cells—as discussed above—it is possible to use specific imaging software to calculate the standard deviation (SD) of pixel intensity within the fluorescence image and divide this by the mean intensity of the pixels within the area of analysis. This will provide a ratio useful for establishing differences in the degree of autophagy between cells. Cells with increased levels of autophagic activity, and hence a greater number of autophagosomes in their cytosol, are associated with a greater variability in pixel intensity (i.e., a high SD). Conversely, in cells where autophagy is not occurring, GFP-LC3 is uniformly distributed throughout the cytosol, and a variation in pixel intensity is not observed (i.e., a low SD; M. Campanella, personal communication).
Although LC3-II is primarily membrane-associated, it is not necessarily associated with autophagosomes as is often assumed; the protein is also found on phagophores, the precursors to autophagosomes, as well as on amphisomes and phagosomes (see Western blotting and ubiquitin-like protein conjugation systems) [247, 440, 441]. Along these lines, yeast Atg8 can associate with the vacuole membrane independent of lipidation, so that a punctate pattern does not necessarily correspond to autophagic compartments [442]. Thus, the use of additional markers is necessary to specify the identity of an LC3-positive structure; for example, ATG12–ATG5-ATG16L1 would be present on a phagophore, but not on an autophagosome, and thus colocalization of LC3 with any of these proteins would indicate the former structure. In addition, the site(s) of LC3 conjugation to PE is not definitively known, and levels of Atg8–PE/LC3-II can increase even in autophagy mutants that cannot form autophagosomes [443]. One method that can be used to examine LC3-II membrane association is differential extraction in Triton X-114, which can be used with mammalian cells [439], or western blot analysis of total membrane fractions following solubilization with Triton X-100, which is helpful in plants [289, 290]. Importantly, we stress again that numbers of GFP-LC3 puncta, similar to steady state LC3-II levels, reflect only a snapshot of the numbers of autophagy-related structures (e.g., autophagosomes) in a cell at one time, not autophagic flux. A potential solution to determine the effect of a given perturbation on flux, is to count GFP-LC3 puncta at various time points following the addition of 3-MA (to prevent formation of new puncta), with the rate of puncta disappearance essentially indicting the flux of disposal [444].
GFP-LC3 expression can perturb autophagy and cellular function, both in the basal state and disease models, as found in the exocrine pancreas of GFP-LC3 mice [445]. Compared to the wild type, the pancreatic ATG4B level is markedly decreased in GFP-LC3 mice, resulting in an increase of the endogenous LC3-II. These effects are organ specific (e.g., there are no effects on ATG4B and LC3 levels in lung and spleen). Autophagic flux analysis (using the lysosomal protease inhibitors E64d plus pepstatin A) indicate that in GFP-LC3 pancreatic acinar cells the basal autophagosome formation is enhanced several-fold but is not fully counterbalanced by increased autophagic degradation. As a result, the exocrine pancreas of GFP-LC3 mice displays accumulation of enlarged autophagic vacuoles. GFP-LC3 expression affects functional parameters of acinar cells and worsens key pathological responses in mouse models of acute pancreatitis. The study referenced above demonstrates organ-specific effects of GFP-LC3 expression and indicates that application of GFP-LC3 mice in disease models should be done cautiously.
Finally, we offer a general note of caution with regard to GFP. First, the GFP tag is large, in particular relative to the size of LC3; therefore, it is possible that a chimera may behave differently from the native protein in some respects. Second, GFP is not native to most systems, and as such (i) it may be recognized as an aberrant protein and targeted for degradation, which has obvious implications when studying autophagy, and (ii) it may elicit immune responses targeting GFP-expressing cells in vivo. Third, some forms of GFP tend to oligomerize, which may interfere with protein function and/or localization. Fourth, EGFP inhibits polyubiquitination [446], and may cause defects in other cellular processes. Fifth, not all LC3 puncta represent LC3-II and correspond to autophagosomes [255, 256, 447, 448]. Accordingly, it would be prudent to complement any assays that rely on GFP fusions (to Atg8-family proteins or any protein) with additional methods that avoid the use of this fluorophore. Similarly, with the emergence of “super-resolution” microscopy methods such as photoactivated localization microscopy (PALM), new tags are being used (e.g., the EosFP green-to-red photoconvertible fluorescent protein, or the Dronpa GFP-like protein) that will need to be tested and validated [449].
Conclusion: GFP-LC3 provides a marker that is relatively easy to use for monitoring autophagy induction (based on the appearance of puncta), or colocalization with cargo; however, monitoring this chimera does not determine flux unless utilized in conjunction with inhibitors of lysosomal fusion and/or degradation. In addition, it is recommended that results obtained by GFP-LC3 fluorescence microscopy are verified by additional assays.
f. Tandem mRFP/mCherry-GFP fluorescence microscopy
A fluorescence assay that is designed to monitor flux relies on the use of a tandem monomeric RFP-GFP-tagged LC3 (tfLC3; Figure 16) [344]. The GFP signal is sensitive to the acidic and/or proteolytic conditions of the lysosome lumen, whereas mRFP is more stable. Therefore, colocalization of both GFP and mRFP fluorescence indicates a compartment that has not fused with a lysosome, such as a phagophore, an amphisome or an autophagosome [450]. In a pathological state where acidification mechanisms are impaired, fusion may occur without GFP becoming quenched, and additional markers of fusion must be applied [451]. Although inhibiting lysosomal acidification may impede fusion in some cell types, fusion may proceed in other cell types under these conditions [218]. In contrast, an mRFP signal without GFP corresponds to an autolysosome. Other fluorophores such as mCherry are also suitable instead of mRFP [423], and an image-recognition algorithm has been developed to quantify flux of the reporter to acidified compartments [452–454]. One of the major advantages of the tandem mRFP/mCherry-GFP reporter method is that it enables simultaneous estimation of both the induction of autophagy and flux through autophagic compartments. However, determining the efficiency of the actual degradation of the substrate or carrier in the lysosome still requires the use of lysosomal protease inhibitors such as E64d and pepstatin. The competence of lysosomal digestion of the substrate requires additional analysis using methods described above. The use of more than one time point allows visualization of increased early autophagosomes followed by increases in late autophagosomes as an additional assurance that flux has been maintained [293]. In addition, this method can be used to monitor autophagy in high-throughput drug screening studies [453]. The quantification of “yellow” (where the yellow signal results from merging the red and green channels) and “red only” puncta in a stable tandem-fluorescent LC3-reporter cell line can be automated by a Cellomics microscope that can be used to assess a huge population of cells (1,000 or more) over a large number of random fields of view [311, 455]. In the presence of a lysosomal acidification defect, additional markers of autophagosome-lysosome fusion need to be applied to assess autophagy flux alterations [451]. The use of late inhibitors of autophagy such as CQ or bafilomycin A1, which prevent the formation of autolysosomes, is recommended as a useful experimental control for the visualization of “yellow” puncta. Note that “green-only” dots may occur under certain conditions due to more rapid maturation of the GFP chromophore, allowing similar fusions to be used as timers [456, 457]. Notably, organelle-specific variations of the tandem mRFP/mCherry-GFP reporter system have successfully been used to analyze selective types of autophagy, such as pexophagy [458, 459], mitophagy [460–463] and reticulophagy [464, 465] in mammalian cells. This tandem reporter is technically less challenging in plant cells due to accumulation of red fluorescent signal in the large relatively static plant vacuoles instead of small mobile dot-like lysosomes. Optimization of the tandem-tag assay for monitoring autophagic activity in plant roots has been described, providing a pipeline for automated high-throughput image analysis [357]. Importantly, in vivo systems to detect mitophagy have been generated employing Drosophila [466] and mouse models [38].
Figure 16.
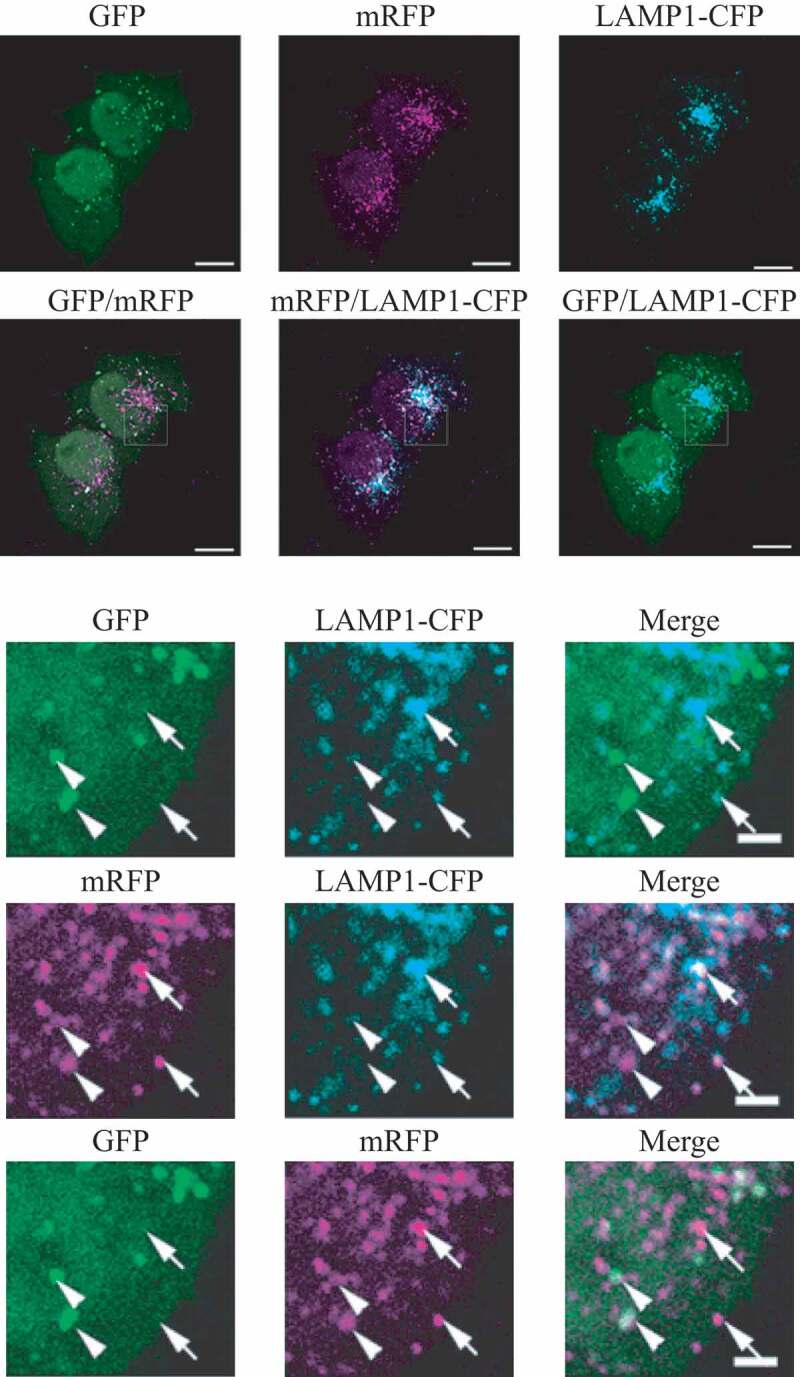
The GFP and mRFP signals of tandem fluorescent LC3 (tfLC3, mRFP-GFP-LC3) show different localization patterns. HeLa cells were cotransfected with plasmids expressing either tfLC3 or LAMP1-CFP. Twenty-four h after the transfection, the cells were starved in Hanks balanced salt solution for 2 h, fixed and analyzed by microscopy. The lower panels are a higher magnification of the upper panels. Bar: 10 µm in the upper panels and 2 µm in the lower panels. Arrows in the lower panels point to (or mark the location of) typical examples of colocalized signals of mRFP and LAMP1. Arrowheads point to (or mark the location of) typical examples of colocalized particles of GFP and mRFP signals. This figure was previously published in ref. [344], and is reproduced by permission of Landes Bioscience, copyright 2007
An alternative dual fluorescence assay involves the Rosella pH biosensor. This assay monitors the uptake of material to the lysosome/vacuole and complements the use of the tandem mRFP/mCherry-GFP reporter. The assay is based upon the genetically encoded dual color-emission biosensor Rosella, a fusion between a relatively pH-stable fast-maturing RFP variant, and a pH-sensitive GFP variant. When targeted to specific cellular compartments or fused to an individual protein, the Rosella biosensor provides information about the identity of the cellular component being delivered to the lysosome/vacuole for degradation. Importantly, the pH-sensitive dual color fluorescence emission provides information about the environment of the biosensor during autophagy of various cellular components. In yeast, Rosella has been successfully used to monitor autophagy of cytosol, mitochondria (mitophagy) and the nucleus (nucleophagy) [156, 467, 468]. Furthermore, the Rosella biosensor can be used as a reporter under various conditions including nitrogen depletion-dependent induction of autophagy [467, 468]. The Rosella biosensor can also be expressed in mammalian cells to follow either nonselective autophagy (cytoplasmic turnover), or mitophagy [467, 469]. A Rosella-based mitophagy reporter mouse line has been created to assess mitophagy activity in the heart [470].
Cautionary notes: The motion of puncta corresponding to Atg8-family proteins can complicate the use of tandem mRFP/mCherry-GFP-Atg-family protein reporters in live imaging experiments. As a consequence, conventional confocal microscopy may not allow visualization of colocalized mRFP/mCherry-GFP puncta. In this case, mRFP/mCherry-GFP colocalized puncta represent newly formed autophagic structures whereas mRFP/mCherry-only puncta are ambiguous. Spinning disk confocal microscopy or rapid acquisition times may be required for imaging tandem mRFP/mCherry-GFP proteins, although these techniques require a brighter fluorescent signal potentially associated with undesirably higher levels of transgene expression. Overexpression of these sensors can target the proteins to acidified lysosomes, which also results in mRFP/mCherry-only puncta. A good control is the non-lipidatable form of the sensor expressed at the same levels as the wild-type experimental sensor, to assess baseline targeting of the tandem proteins to lysosomes [471]. Another optimization is to use the mTagRFP-mWasabi-LC3 chimera [472, 473], as mTagRFP is brighter than mRFP and mCherry, and mWasabi is brighter than EGFP [474]. An improved version of tfLC3 is pHluorin-mKate2-hLC3 reporter (PK-hLC3), because pHluorin is more sensitive to acidic pH (pKa 7.6, quenched at pH 6.5) than EGFP and mWasabi [450, 475]. In the latter case, however, organelles that only achieve a lower level of acidification, such as amphisomes, may not be differentiated from fully acidified (i.e., mature) lysosomes [476]. A good quantitative technique for cells in suspension, which also require identification by surface markers (such as immune cells), is the detection of LC3-II by flow cytometry. Here LC3-I is washed out after treatment with a mild detergent, and only membrane-bound LC3-II is retained for staining with an anti-LC3 antibody. Early fixation avoids the induction of artefacts due to centrifugation or mixing. This approach has been established for both cell lines [433] and primary cells [477].
Another possibility is to use fixed cells; however, this presents an additional concern: The use of tandem mRFP/mCherry-GFP relies on the quenching of the GFP signal in the acidic autolysosome; however, fixation solutions are often neutral or weak bases, which will increase the pH of the entire cell. Accordingly, the GFP signal may be restored after fixation (Figure 17), which would cause an underestimation of the amount of signal that corresponds only to RFP (i.e., in the autolysosome). Thus, the tissue or cell samples must be properly processed to avoid losing the acidic environment of the autolysosomes. In addition, there may be weak fluorescence of EGFP even in an acidic environment (pH between 4 and 5) [439, 478]. Therefore, it may be desirable to choose a monomeric green fluorescent protein that is more acid sensitive than EGFP for assaying autophagic flux. For example, the pHluorin-based probe (PK-hLC3) referred to above can solve these problems [450]; in a PK-hLC3 transgenic mouse autophagic responses in the neurons are easily detectable, whereas such responses in the neurons of a GFP-LC3 transgenic mouse are hardly recognized [479]. pHluorin-LC3-mCherry is also an improved autophagic flux probe variant of GFP-LC3-RFP-LC3ΔG [480]. Finally, photobleaching, light-induced degradation of fluorophores, is a significant problem in live-cell imaging [481]. When examining live tissue, it is important to remember that marker fluorescence, particularly GFP fluorescence, can diminish rapidly with the decay of cell physiology. The use of the minimal possible exposure and light power level is therefore recommended. If sequential acquisition of fluorescence emissions is needed, they should be acquired in the order GFP then RFP, as RFP exhibits higher photostability. The experimenter can also take advantage of anti-fading media developed for live imaging [482]. In some tissues (e.g., Drosophila brain) it may be necessary to image each sample no more than 30-40 min after dissection of the individual sample.
Figure 17.
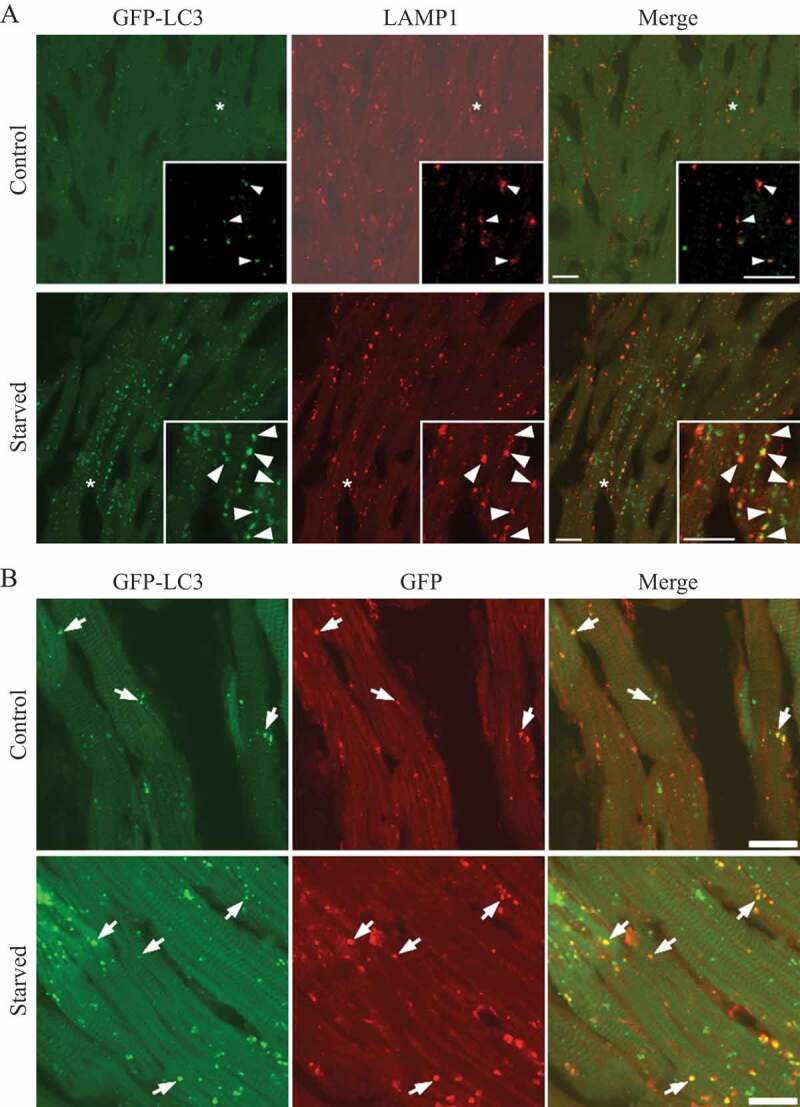
GFP fluorescence in the autolysosome can be recovered upon neutralization of the pH. (A) GFP-LC3 emits green fluorescence in the autolysosomes of post-mortem processed heart sections. Cryosections of 3.8% paraformaldehyde fixed ventricular myocardium from 3-week-old GFP-LC3 transgenic mice at the baseline (Control) or starved for 24 h (Starved) were processed for immunostaining using a standard protocol (buffered at pH 7.4). Most of the GFP-LC3 puncta are positive for LAMP1, suggesting that the autolysosomes had recovered GFP fluorescence. (B) Colocalization between GFP-LC3 direct fluorescence (green) and indirect immunostaining for GFP (red). Sections processed as in (A) were immunostained for GFP using a red fluorescence-tagged secondary antibody, and the colocalization with GFP fluorescence was examined by confocal microscopy. Almost all of the red puncta emit green fluorescence. Scale bar: 10 µm. Image provided by Xuejun Wang
Another caution in the interpretation of the tandem fluorescent marker is that an enhanced degree of colocalization of GFP and mRFP/mCherry might also be seen in the case of impaired proteolytic degradation within autolysosomes or altered lysosomal pH. This limitation may be overcome by incorporating two strategies in the experimental design: i) direct measurement of lysosomal pH in the in vitro model of interest, to discount lysosomal alkanization as a cause for increased GFP+ RFP+ puncta [483–485], and ii) Immunohistochemical analysis against lysosomal markers such as CTSD and LAMP2. Whereas measuring lysosomal pH or proteolytic activity in vivo or in fixed tissue is not possible, colocalization of GFP+ RFP+ puncta, or target autophagic cargo with CTSD or LAMP2 may be indicative of lysosomal dysfunction [485].
RFP-GFP-LC3 and GFP-LC3 (or other Atg8-family proteins) methodology, which involves in vitro or in vivo overexpression of the fluorescent construct, requires careful microscopy by including a GFP-expressing control. A comparable in vitro or in vivo system overexpressing GFP is necessary to “titer” the minimum laser intensity, minimum gain, and offset parameters on the confocal microscope that are required to detect true GFP. Excessive laser intensity/gain often leads to an undesirable increase in signal:background ratio, may also contribute to increased false-positive counts, and often leads to rapid photobleaching as well. The same laser intensity/gain/offset setting to detect true GFP should strictly be applied to the RFP-GFP-LC3-expressing in vitro or in vivo system of interest. The other experimental positive control is CQ or bafilomycin A1 treatment to induce lysosomal alkalinization. The intensity/gain settings (sufficient to detect true GFP) should be minimally sufficient to detect GFP+ RFP+ puncta in CQ-treated samples. Autophagosomes, typically, are 900 nm to 1.5 µm in diameter. Low magnification images (using 20X/40X objectives) do not provide enough resolution to obtain quantifiable data. Only higher magnification images should be used to monitor autophagy. Once the microscope “parameters” to detect GFP have been determined (using a GFP-overexpressing control), it is important that the user captures the fluorescence images in the green channel in experimental conditions, and the CQ treatment control, in a “blinded” manner; ImageJ, or other equivalent software, should be used for objective quantification of GFP/RFP puncta (S. Ramachandra Rao and S.J. Fliesler, upublished results).
Finally, expression of tandem mRFP-GFP-LC3 is toxic to some cancer cell lines relative to GFP-LC3 or RFP-LC3 (K.S. Choi, personal communication). By contrast, transgenic expression of mRFP-GFP-LC3 in neurons, which generates strong fluorescence signals at low levels of expression of the reporter construct, exhibit no evident toxicity or effects on baseline autophagy or lifespan [451]. The cytotoxicity of DsRed and its variants such as mRFP is associated with downregulation of BCL2L1/Bcl-XL [486]. In contrast to mRFP-GFP-LC3, overexpression of mTagRFP-mWasabi-LC3 does not appear to be toxic to HeLa cells (J. Lin, personal communication) or LNCaP cells (N. Engedal, personal communication).
The Rosella assay has not been tested in a wide range of mammalian cell types. Accordingly, the sensitivity and the specificity of the assay must be verified independently until this method has been tested more extensively and used more widely.
Finally, it may be desirable to capture the dynamic behavior of autophagy in real time, to generate data revealing the rate of formation and clearance of autophagosomes over time, rather than single data points. For example, by acquiring signals from two fluorescent constructs in real time, the rate of change in colocalization signal as a measure of the fusion rate and recycling rate between autophagosomes and lysosomes can be assessed [487]. Importantly, due to the integral dynamic relationship of autophagic flux with the onset of apoptosis and necrosis, it is advantageous to monitor cell death and autophagic flux parameters concomitantly over time, which FRET-based reporter constructs make possible [488].
Tandem fluorescent markers show real-time changes in autophagosome fusion with lysosomes, due to entry into an acidic environment; however, fusion is not definitive evidence of substrate or carrier degradation. Lysosomes may be able to fuse, but be unable to degrade newly delivered cargo, as occurs in some lysosomal storage diseases and aging-related neurodegenerative diseases. Best practice would be to perform an autophagic flux assay in parallel with quantification of tandem fluorescent markers to confirm completion of carrier flux.
Conclusion: The use of tandem fluorescent constructs, which display different emission signals depending on the environment (in particular, GFP fluorescence is sensitive to an acidic pH), provides a convenient way to monitor autophagic flux in many cell types.
g. Autophagic flux determination using flow and multispectral imaging cytometry
Whereas fluorescence microscopy, in combination with novel autophagy probes, has permitted single-cell analysis of autophagic flux, automation for allowing medium- to high-throughput analysis has been challenging. A number of methods have been developed that allow the determination of autophagic flux using flow cytometry [300, 414, 433, 489–492], and commercial kits are now available for monitoring autophagy by flow cytometry. These approaches make it possible to capture data or, in specialized instruments, high-content, multiparametric images of cells in flow (at rates of up to 1,000 cells/s for imaging, and higher in nonimaging flow cytometers), and are particularly useful for cells that grow in suspension. This quantitative method is simple and can be used for high-content studies with simultaneous analysis of multiple parameters. This is especially useful for the study of complex mixtures of cell types, for example in the analysis of immune cells where it might require discrimination of the autophagic state of each cell type or even subsets. The employment of a vital nuclear dye in combination with other markers makes it possible not only to exclude dead cells by detection of nuclear fragmentation, but also to analyze a cell population in a specific cell cycle phase. Notably, as living cells expressing fluorescence proteins are amenable to analysis by flow cytometry, this method may also be utilized to sort specific subpopulations for further characterization.
Optimization of image analysis permits the study of cells with heterogeneous LC3 puncta, thus making it possible to quantify autophagic flux accurately in situations that might perturb normal processes (e.g., microbial infection or drug treatment) [489, 493]. Because EGFP-LC3 is a substrate for autophagic degradation, total fluorescence intensity of EGFP-LC3 can be used to indicate levels of autophagy in living mammalian cells [492]. When autophagy is induced, the decrease in total cellular fluorescence can be precisely quantified in large numbers of cells to obtain robust data; flux can also be directly associated with an increase of detectable puncta [413]. Moreover, current technology makes it possible to investigate the colocalization of EGFP-LC3 puncta and other specific proteins, identifying novel molecules degraded during autophagic flux. In another approach, soluble EGFP-LC3-I can be depleted from the cell by a brief saponin (or digitonin) extraction so that the total fluorescence of EGFP-LC3 then represents that of EGFP-LC3-II alone (Figure 18A) [432, 433]. Because EGFP-LC3 transfection typically results in high relative levels of EGFP-LC3-I, this treatment significantly reduces the background fluorescence due to non-phagophore and non-autophagosome-associated reporter protein. By comparing treatments in the presence or absence of lysosomal degradation inhibitors, subtle changes in the flux rate of the GFP-LC3 reporter construct can be detected. If it is not desirable to treat cells with lysosomal inhibitors to determine rates of autophagic flux, a tandem mRFP/mCherry-EGFP-LC3 (or similar) construct can also be used for autophagic flux measurements in flow cytometry experiments (see Tandem mRFP/mCherry-GFP fluorescence microscopy) [473, 491].
Figure 18.
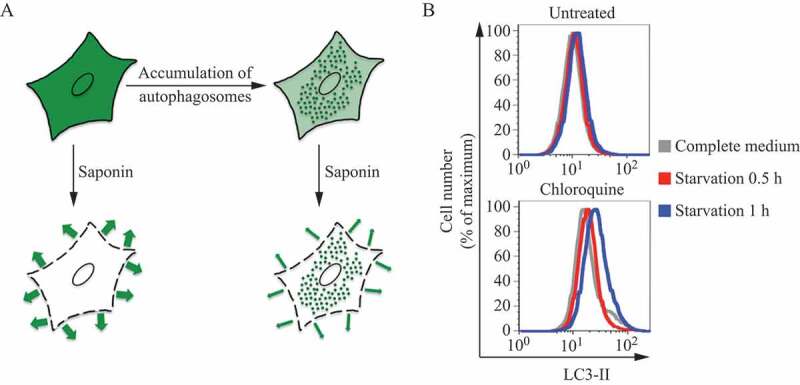
Saponin extraction allows quantification of LC3-II fluorescence by FACS. (A) Schematic diagram of the effects of the saponin wash. Due to the reorganization of the EGFP-LC3 reporter protein, induction of autophagosome formation does not change the total levels of fluorescence in EGFP-LC3-transfected cells. However, extraction of EGFP-LC3-I with saponin results in a higher level of fluorescence in cells with proportionally higher levels of EGFP-LC3-II-containing autophagosomes. This figure was previously published in ref. [433]. (B) Saponin extraction can also be used to measure flux of endogenous LC3 protein. Human osteosarcoma cells were starved of amino acids and serum by incubation in EBSS, for the indicated times in the presence or absence of a 1-h CQ (50 μM) treatment. Cells were then washed with PBS containing 0.05% saponin and processed for FACS analysis for endogenous LC3. Image provided by K.E. Eng and G.M. McInerney
These methods, however, require the cells of interest to be transfected with reporter constructs. Because the saponin extraction method can also be combined with intracellular staining for endogenous LC3 protein, subtle changes in autophagic flux can be measured without the need for reporter transfections (Figure 18B).
In addition to GFP-LC3, a novel probe has emerged in recent years: GFP-LC3-RFP-LC3∆G and GFP-LC3-RFP (without LC3∆G) that bypass the weaknesses of GFP-LC3 [494]. This probe is cleaved by endogenous ATG4 and releases an equal amount of GFP-LC3 and RFP-LC3∆G (or RFP) in the cells. While GFP-LC3 is lipidated and localizes to phagophores and autophagosomes (as described previously), the RFP-LC3∆G (or RFP) cannot be conjugated with PE and remains in the cytoplasm, acting as an internal control. The GFP-LC3:RFP-LC3∆G (or RFP) ratio (or GFP:RFP ratio) indicates the autophagic flux. The advantage of this probe compared to the “traditional” GFP-LC3 probe is that the release of the internal control makes it possible to discriminate the changes of overall GFP-LC3 levels caused by the autophagic flux to the ones resulting from variation of gene expression. The measurement of both GFP-LC3 and RFP-LC3∆G (or RFP) can be performed in a high-throughput manner using FACS for single-cell analysis or some plate readers. The most precise way to monitor autophagy with this probe is to use a single-cell derived colony of stable cell lines, in order to have the most homogeneous population. This is also because the DNA sequence corresponding to GFP-LC3-RFP-LC3ΔG sometimes undergoes homologous recombination between the two LC3-encoding fragments during retrovirus infection (this does not occur with GFP-LC3-RFP).
Cautionary notes: Care must be taken when applying flow cytometry measurements to adherent cells, particularly neurons and other cells with interdigitated processes, as the preparation of single cell suspensions entails significant levels of plasma membrane disruption and injury that can secondarily induce autophagy.
Users of the saponin or digitonin extraction method should carefully titrate detergent concentrations and times of treatment to ensure specific extraction of LC3-I in their systems. Also, it has been observed in some cell types that saponin treatment can lead to nonautophagic aggregation of LC3 [434], which should be controlled for in these assays (see GFP-Atg8-family protein fluorescence microscopy). Similarly, for treatment with other detergents, such as Triton X-100, it is also important to carefully titrate the concentrations and times of treatment.
Cell membrane permeabilization with digitonin and extraction of the nonmembrane-bound form of LC3 allows combined staining of membrane-associated LC3-II protein and any markers for detection of autophagy in relation to other cellular events/processes. Based on this approach, a method for monitoring autophagy in different stages of the cell cycle was developed [495]. Thus, the presence of basal or starvation-induced autophagy is detected in G1, S, and G2/M phases of the cell cycle in MEFs with doxycycline-regulated ATG5 expression. In these experiments, cells were gated based on their DNA content and the relative intensity of GFP-LC3-II and LC3-II expression. This approach might also be used for the detection of autophagic flux in different stages of the cell cycle, or the subG1 apoptotic cell population by measuring accumulation of LC3-II in the presence or absence of lysosomal inhibitors.
Although GFP-LC3 can be used as a reporter for flow cytometry, it is more stable (which is not necessarily ideal for flux measurements) than GFP-SQSTM1 or GFP-NBR1 (NBR1 is an autophagy receptor with structural similarity to SQSTM1 [496]). GFP-SQSTM1 displays the largest magnitude change following the induction of autophagy by amino acid deprivation or rapamycin treatment, and may thus be a better marker for following autophagic flux by this method (confirmed in SH-SY5Y neuronal cell lines stably expressing GFP-SQSTM1; E.M. Valente, personal communication) [497]. In addition, to reduce/eliminate potential effects on transcription or translation of the reporter, a doxycycline-inducible version of GFP-SQSTM1 can be used [497]. Flow cytometry for LC3, SQSTM1 or using commercial autophagy kits can also be used to measure autophagy in a specific cell sub-population isolated from tissue. For example, this approach can measure autophagy levels specifically in microglia and infiltrating macrophages in the mouse brain after traumatic brain injury (M. Lipinski, unpublished data).
Using purification of intracellular vesicles, flow cytometry can be adapted for a deeper understanding and better characterization of individual autophagosomes. Single organelle fluorescence analysis can be applied for the analysis of endosomes [498], mitochondria [499], phagosomes [500], autophagosomes and lysosomes [501], using various fluorescent probes.
Finally, probes measuring the autophagic flux without requiring transfection or permeabilization have also been developed. Such methods are based on dyes that selectively label autophagic vesicles (autophagosomes and autolysosomes) but not lysosomes, and are used for both primary cells [502, 503] and cell lines [504, 505] from different species (including non-mammals). See also: https://bio-protocol.org/e1090.
Conclusion: Medium- to high-throughput analysis of autophagy is possible using flow and multispectral imaging cytometry (Figure 19). The advantage of this approach is that larger numbers of cells can be analyzed with regard to GFP-LC3 puncta, cell morphology and/or autophagic flux, and concomitant detection of surface markers can be included, potentially providing more robust data than is achieved with other methods. A major disadvantage, however, is that flow cytometry only measures changes in total GFP-LC3 levels, which can be subject to modification by changes in transcription or translation, or by pH, and this approach cannot accurately evaluate localization (e.g., to autophagosomes) or lipidation (generation of LC3-II) without further permeabilization of the cell.
Figure 19.
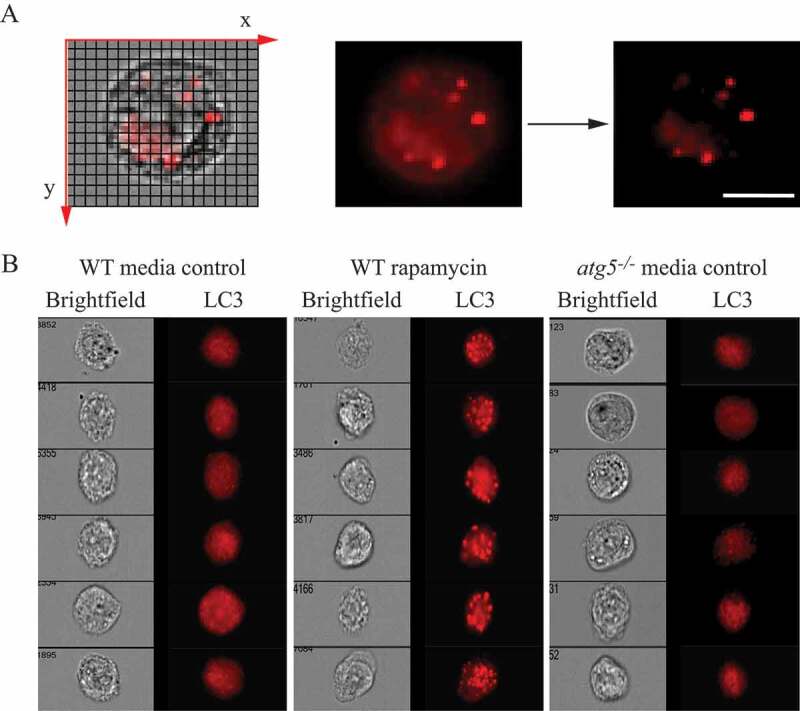
Assessing autophagy with multispectral imaging cytometry. (A) Bright Detail Intensity (BDI) measures the foreground intensity of bright puncta (that are 3 pixels or less) within the cell image. For each cell, the local background around the spots is removed before intensity calculation. Thus, autophagic cells with puncta have higher BDI values. (B) Media control (untreated wild type), rapamycin-treated wild-type and atg5−/− MEFs were gated based on BDI. Representative images of cells with high or low BDI values. Scale bar: 10 µm. Images provided by M.L. Albert
h. Autophagosome-lytic compartment fusion
Technical limitations have prevented insight into the mechanism of autophagosome-lytic compartment fusion. Disrupting genes encoding components that play a role in membrane fusion in intact cells may not only affect autophagy directly but will affect general vesicular trafficking, which can cause indirect effects on autophagy. In addition, if a fusion component is involved in early steps of autophagosome formation, this requirement will mask its function in late stages of autophagy such as fusion with the lytic compartment. As a result, it is difficult to analyze the molecular mechanisms of autophagosome-lytic compartment fusion in intact cells.
In vitro reconstitutions of autophagosome-vacuole and autophagosome-lysosome fusion have partially overcome this problem and made it possible to identify relevant proteins and their functions in this specific step of autophagy. Both autophagosome-vacuole fusion in yeast and autophagosome-lysosome fusion in mammals has been recently reconstituted, using partially purified fractions of autophagosomes, vacuoles/lysosomes and cytosol [506–508].
i. Immunohistochemistry and immunofluorescent staining
Immunodetection of ATG and related proteins (particularly LC3 and BECN1) has been reported as a prognostic factor in various human carcinomas, including lymphoma [265, 509], breast carcinoma [510, 511], endometrial adenocarcinoma [512, 513], head and neck squamous cell carcinoma [514–516], hepatocellular carcinoma [517, 518], gliomas [519], non-small cell lung carcinomas [520], pancreatic [521] and colon adenocarcinomas [522–524], as well as in cutaneous and uveal melanomas [525, 526]. Unfortunately, the reported changes often reflect overall diffuse staining intensity rather than appropriately compartmentalized puncta. Therefore, the observation of increased levels of diffuse LC3 staining (which may reflect a decrease in autophagy) should not be used to draw conclusions that autophagy is increased in cancer or other tissue samples [527]. Assessing LC3 puncta fails to show prognostic significance in non-small cell lung cancer [528, 529]. Importantly, this kind of assay should be performed as recommended by the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) [530]. As we identify new drugs for modulating autophagy in clinical applications, this type of information may prove useful in the identification of subgroups of patients for targeted therapy [531–533].
In the brain of hypoxic-ischemic encephalopathy dead human newborns, LC3 immunostaining on paraffin sections has been used to quantify increased autophagosome presence in dying neurons as shown by increased LC3-positive dots [534, 535]. In mouse and rat tissues, endogenous LC3, ATG4B, and ATG9A have been detected by immunohistochemical analyses using both paraffin sections and cryosections [85, 391, 536–539]. When autophagosomes are absent, the localization pattern of LC3 in the cells of various tissues is diffuse and cytosolic. Moreover, intense fibrillary staining of LC3 is detectable along dendrites of intact neurons, whereas granular staining for LC3 appears mainly in the perinuclear area of neurons in CTSD- or CTSB- and CTSL (cathepsin L)-deficient mouse brains [393]. LC3 puncta are also observed in mice in the peripheral nerves, specifically in Schwann cells after neurodegeneration [540], and Paneth cells of the small intestine from human Crohn disease patients and mouse models of intestinal inflammation driven by ER-stress and acute radiation injury exhibit strong LC3 puncta staining [541–543]. In various neurodegenerative states, LC3 puncta may be numerous in neurites, especially within dystrophic swellings and, in many cases, these vacuoles are amphisomes or autolysosomes, reflecting the delayed or inhibited degradation of LC3 despite the presence of abundant hydrolase activity [76, 83]. In developing inner ear and retinal tissue in chicken, BECN1 is detected by immunofluorescence; in chick retina AMBRA1 is also detected [393-395]. IHC using ABC and 3,3′-diamino-benzidine (DAB) as chromogen has also been used to detect AMBRA1, thus accomplishing a complete map of AMBRA1 protein distribution in the mouse brain, and highlighting differential expression in neuronal/glial cell populations. Differences in AMBRA1 content have been related to specific neuronal features and properties, particularly concerning susceptibility to neurodegeneration, during aging and amyloid pathology [544]. AMBRA1 and BECN1 IHC distribution have also been studied in rat brain, after anti-NGF administration, which results in increased levels of autophagic proteins in specific brain regions (olfactory bulb, neocortex and hippocampus), suggesting NGF-modulated autophagic pathways [545]. In mouse platelets, endogenous PtdIns3P, the product of the BECN1-PIK3C3/VPS34 protein complex-mediated enzymatic reaction, can be detected using recombinant GST-2×FYVE followed by anti-GST immunofluorescence [389]. Finally, in non-mammalian vertebrates, BECN1 is detected during follicular atresia in the ovary of three fish species using paraffin sections; a punctate immunofluorescent staining for BECN1 is scattered throughout the cytoplasm of the follicular cells when they are in intense phagocytic activity for yolk removal [546].
Cautionary notes: One problem with LC3 IHC is that in some tissues this protein can be localized in structures other than autophagosomes. For example, in murine hepatocytes and cardiomyocytes under starved conditions, endogenous LC3 is detected not only in autophagosomes but also on lipid droplets [547]. In neurons in ATG7-deficient mice, LC3 accumulates in ubiquitin- and SQSTM1-positive aggregates [548]. In neurons in aging or neurodegenerative disease states, LC3 is commonly present in autolysosomes and may be abundant in lipofuscin and other lysosomal residual bodies [76]. Similarly, accumulation of large LC3-positive puncta occurring during methamphetamine intoxication does not derive from stagnant autophagic vacuoles. In fact, the polarization of LC3 within granules is greatly reduced and LC3 IHC even monitored by confocal microscopy demonstrates cytosolic accumulation of the protein, which, despite being increased, loses its polarization within autophagic granules [549]. This is clearly demonstrated by counting stoichiometrically immunogold-stained LC3 particles within the cytosol compared with granules. Thus, immunodetection of LC3 in cytoplasmic granules is not sufficient to monitor autophagy in vivo. To evaluate autophagy by the methods of IHC, it is necessary to identify the autophagosomes directly using the ABC technique for TEM observation (see Transmission electron microscopy) [77]. Peroxidase depositions in the vacuoles indicate LC3 expression, detected by IHC, and therefore identify those structures as autophagic vacuoles [550].
Conclusion: It has not been clearly demonstrated that IHC of ATG proteins in tissues corresponds to autophagy activity, and this area of research needs to be further explored before we can make specific recommendations.
j. LC3-HiBiT reporter assay
The Autophagy LC3-HiBiT reporter assay system is a method that measures autophagic flux by monitoring total LC3-reporter levels [551]. A plasmid coding for a human LC3B is tagged to a HiBiT peptide through a linker. The approach is based on the high affinity of the HiBiT peptide to the inactive luciferase subunit LgBiT, that, upon binding, produces an active NanoBiT luciferase that generates luminescence proportional to the amount of autophagy (Figure 20). It is recommended to generate stable cell lines using G418 selection to avoid the variability due to transfection efficiencies among different cell lines and experiments. The amount of LC3-reporter within the cell can be measured by the addition of lysis buffer mixed with the LgBiT protein and the substrate. After incubation at room temperature for 10 min, luminescence can be measured in a microplate reader (integration time of 0.5–2 s) and it is stable for up to three h. Induction of autophagy, such as through starvation, or MTORC1 inhibition via PP242 treatment or siRNA knockdown of RPTOR, decrease luminescence readings. Conversely, blockade of autophagy flux, for example by treatment with CQ or bafilomycin A1, increases the luminescence readings (Figure 20). A major advantage of this reporter system is that it allows for determination of autophagic flux upon exposure to a large number of conditions at the same time and, therefore, is suitable for large-scale screens using 96- or 386-well plate formats.
Figure 20.
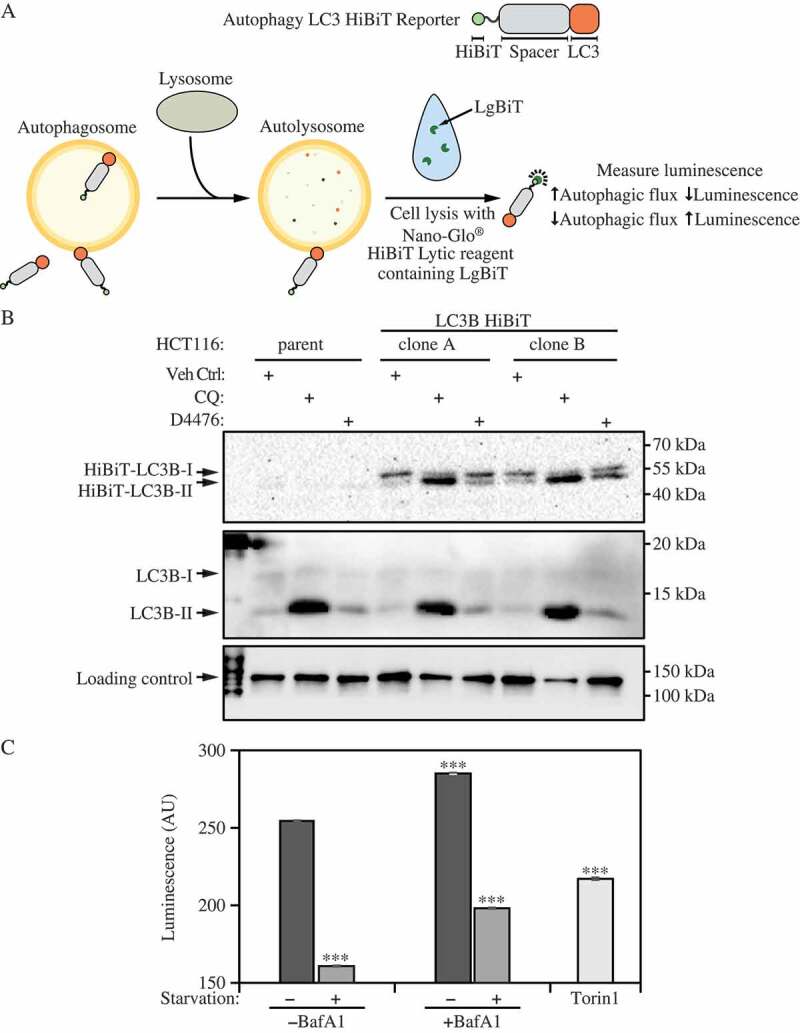
HiBiT-LC3B imitates endogenous LC3B response upon autophagy perturbation. (A) Principle of the Autophagy LC3-HiBiT reporter assay system. (B) HCT116 (parent) and stable HCT116-LC3 HiBiT cell lines were exposed to vehicle control (Veh Ctrl), chloroquine (CQ; 25 μM) or the CSNK1A1/CK1α (casein kinase 1 alpha 1) inhibitor D4476 (10 μM) for 24 h, which increases LC3 abundance [4084]. Cell lysates (30 μg) were resolved in 12% SDS-PAGE gels and transblotted to nitrocellulose membranes. Nano-Glo HiBiT Blotting System (Promega, N2410) was used to detect HiBiT-LC3B (expected molecular size of 55 kDa) using Tris-buffered saline supplemented with 0.1% Tween 20. The HiBiT-LC3B blot was imaged after 20 min incubation with the substrate. Loading control (KIF11/Eg5; Cell Signaling Technology, 4203) and endogenous LC3B (Cell Signaling Technology, 2775) were detected via standard immunoblotting (leftmost lane in immunoblots: protein ladder). Image provided by John J.E. Chua and Jit K. Cheong. (C) U2OS LC3 HiBiT cells treated with autophagy inducers or inhibitors. Effect on autophagic flux was measured in cells incubated in normal or starvation medium (HBSS) alone or containing bafilomycin A1 (50 nM) or torin1 (25 nM) for 3 h using the HiBiT luminescence assay. Bars are mean ± s.e.m. of triplicate samples. ***P < 0.001 vs. untreated normal media control; t-test. AU, arbitrary units. Image provided by Silvia Vega-Rubin-de-Celis
Cautionary notes: One caveat to the immunostaining validation of cells that stably express LC3-HiBiT is the inability of widely-used LC3 antibodies (such as the LC3B antibody from Cell Signaling Technology [2775] that targets the N terminus of LC3B) to recognize the amino terminal HiBiT-tagged LC3 resolved on protein blots. This limitation can be overcome by the use of the Nano-Glo® HiBiT blotting system (Promega, N2410) that detects the amino terminal HiBiT-tagged LC3B-I/II as proteins of approximately 55 kDa (Figure 20). For conditions that might affect cell number/viability, it is recommended to prepare a separate culture plate(s) with an identical treatment condition(s) in parallel for cell number/viability measurement (e.g., using Hoechst staining of the nucleus followed by quantification). Alternatively, the Autophagy LC3-HiBiT reporter assay system could also be multiplexed with the CellToxTM Green Cytotoxicity Assay (Promega, G8741) for cell viability assessment within the same sample well.
k. In vitro enzymatic lipidation of human Atg8-family proteins: Preparation of fluorescent Atg8–PE conjugates
After activation by ATG4B, covalent attachment of an Atg8-family protein to PE is mediated by a ubiquitin-like chain of enzymatic steps involving the E1-like ATG7 and the E2-like ATG3. These reactions can be reconstituted in vitro, using recombinant purified proteins, liposomes and ATP. To study the role of these protein-lipid complexes in membrane tethering and fusion processes, the enzymatically driven lipidation reaction of the human Atg8-family proteins can be reconstituted. Reaction systems including ATG7, ATG3, ATP, and liposomes lead to the formation of a more rapidly migrating band that is readily visualized by Coomassie Brilliant Blue staining. To confirm the lipidation reaction, conjugation mixtures are prepared with liposomes containing 10% of the fluorescent phospholipid derivative NBD-PE. In each case, reactions lead to the formation of fluorescent, faster-migrating bands representing the lipidated products of Atg8-family proteins [552].
3. SQSTM1 and related LC3-binding protein turnover assays
In addition to LC3, SQSTM1, or other receptors such as NBR1, can also be used as protein markers, at least in certain settings [30, 553]. For example, SQSTM1 can be detected as puncta by IHC in cancer cells or primary neurons, similar to LC3 [515]; however, note that it is critical to freshly cut formalin fixed paraffin embedded/FFPE tissue before IHC for LC3 [554, 555]. The SQSTM1 protein serves as a link between LC3 and ubiquitinated substrates [119]. SQSTM1 and SQSTM1-bound polyubiquitinated proteins become incorporated into the completed autophagosome and are degraded in autolysosomes, thus serving as an index of autophagic degradation (Figure 21). In addition, SQSTM1 can also bind RNA substrates, which controls RNA turnover via autolysosomes [556]. Inhibition of autophagy can correlate with increased levels of SQSTM1 in mammals, C. elegans and Drosophila, suggesting that steady state levels of this protein reflect the autophagic status [81, 536, 557–562]. Deficiency of the LIR domain-containing, SQSTM1-interacting SPRED2 results in an accumulation of SQSTM1 in the heart in vivo, accompanied by an altered LC3 turnover and reduced autophagy [563]. Similarly, decreased SQSTM1 levels are associated with autophagy activation; however, similar to LC3-II, lysosomal inhibitors (such as CQ) can be used to assess increased autophagy flux based on an accumulation of SQSTM1 [86, 564]. The phosphorylation of SQSTM1 at Ser403 appears to regulate its role in the autophagic clearance of ubiquitinated proteins, and anti-phospho-SQSTM1 antibodies can be used to detect the modified form of the protein [427].
Figure 21.
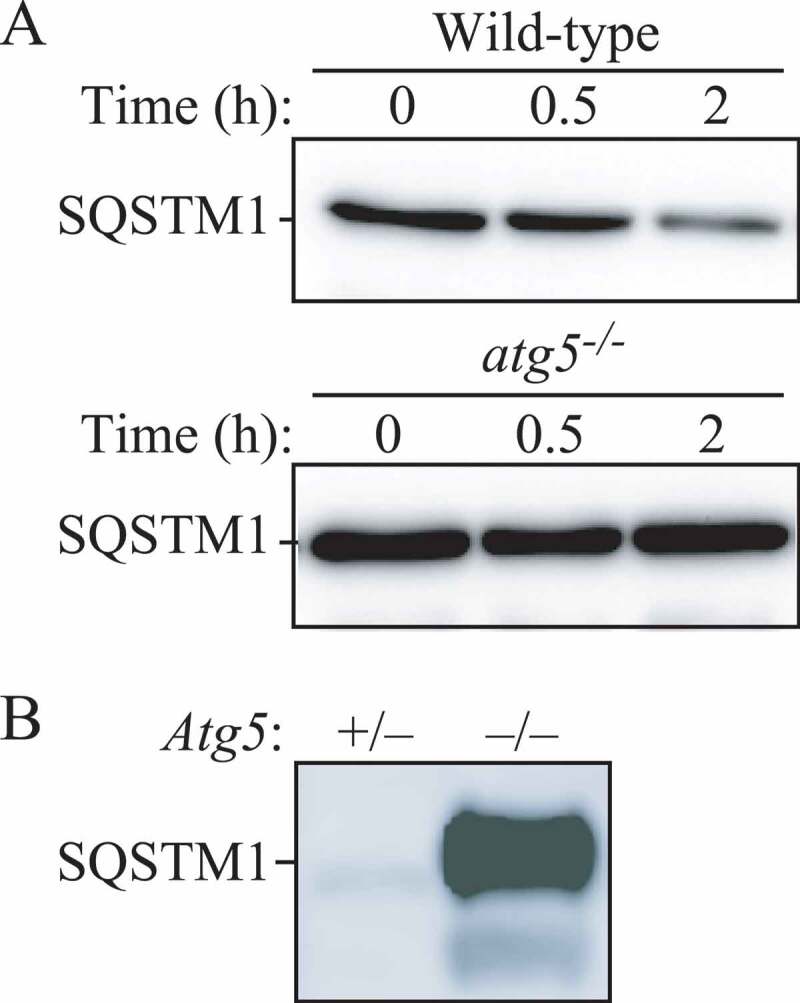
Regulation of the SQSTM1 protein during autophagy. (A) The level of SQSTM1 during starvation. Atg5+/+ and atg5−/− MEFs were cultured in DMEM without amino acids and serum for the indicated times, and then subjected to immunoblot analysis using anti-SQSTM1antibody (Progen Biotechnik, GP62). This figure was previously published in ref. [30], and is reproduced by permission of Landes Bioscience, copyright 2007. (B) The level of SQSTM1 in the brain of neural-cell specific atg5 knockout mice. Image provided by T. Hara
Cautionary notes: SQSTM1 changes can be cell-type and context specific. In some cell types, there is no change in the overall amount of SQSTM1 despite strong levels of autophagy induction, verified by the tandem mRFP/mCherry-GFP-LC3 reporter as well as ATG7- and lysosome-dependent turnover of cargo proteins (C.T. Chu, personal observation). In other contexts, a robust loss of SQSTM1 does not correlate with increased autophagic flux as assessed by a luciferase-based measure of flux [336]; a decrease of SQSTM1 can even relate to a blockage of autophagy due to cleavage of the protein, together with other autophagy proteins, by caspases or calpains [565].
In some systems, even transgenic constructs may not allow reliable detection of SQSTM1. For instance, although very informative to monitor autophagy levels in the C. elegans embryo, SQST-1::GFP (a tagged version of the C. elegans SQSTM1 homolog) is not detectable in most adult tissues unless it is stabilized with background mutations such as rpl-43 (encoding the ribosomal protein RPL-43). Stabilization of SQST-1::GFP via rpl-43 mutation does not affect the degradation of the autophagy substrates so far tested, and reduced SQST-1::GFP signal is observed in conditions of increased autophagic flux such as starvation [566]; however, animals show signs of generalized sickness, and altered lifespan, and RNAi against some autophagy genes (e.g., vps-34) leads to increased, instead of reduced, SQST-1::GFP signal (E.J. O’Rourke, personal communication). SQSTM1 changes can be treatment specific such that chemotherapy-induced autophagy increases LC3-II without changing SQSTM1, whereas radiation-induced autophagy increases LC3-II and decreases SQSTM1 in ERBB2/HER2-overexpressing mouse mammary carcinoma cells [567].
SQSTM1 may be transcriptionally upregulated under some conditions [419, 568–571], as observed in several C. elegans longevity models [572, 573], further complicating the interpretation of results. For example, SQSTM1 upregulation, and at least transient increases in the amount of SQSTM1, is seen in some situations where there is an increase in autophagic flux [574–576]. One such case is seen during retinoic acid-induced differentiation of acute myeloid leukemia (AML) cells where SQSTM1 is upregulated [569] with concomitant increased autophagic flux [577]. Synovial fibroblasts obtained from patients with rheumatoid arthritis also exhibit a significant upregulation of SQSTM1 with concomitant increased autophagy flux [578]. Activation of a signaling pathway, e.g., RAF1/Raf-MAP2K/MEK-MAPK/ERK, can also upregulate SQSTM1 transcription [579]. SQSTM1 mRNA is also upregulated following prolonged starvation, which can restore the SQSTM1 protein level to that before starvation [580, 581]. In the same way, physical exercise, especially when performed during starvation, increases the SQSTM1 mRNA level in skeletal muscle, and can lead to an incorrect interpretation of autophagic flux if only the protein level is measured [582, 583]. Another instance when both mRNA and protein levels of SQSTM1 are elevated, even though autophagic flux is not impaired, is observed in aneuploid human and murine cells that are generated by introduction of one or two extra chromosomes [584, 585].
The SQSTM1 protein level also increases when autophagy needs to be triggered. SQSTM1 expression can be positively regulated post-transcriptionally by the ELAVL1/HuR protein which binds to the SQSTM1 transcript in ARPE19 cells exposed to 24-h MG132 treatment [359]. Two-h AICAR + MG132 pro-autophagic cotreatment similarly induces the binding of ELAVL1/HuR protein to SQSTM1 mRNA, its loading on polysomes and its translation into de novo protein, an effect that is required to trigger autophagy and is prevented by the protein synthesis inhibitor puromycin [586]. Moreover, SQSTM1 can be regulated by the integrated stress response, which can promote accumulation of protein in cells that is dependent on the EIF2S1/eIF2α pathway of translational control (R.E. Simmonds, personal communication). Thus, appropriate positive and negative controls are needed prior to the use of SQSTM1 as a flux indicator in a particular cellular context, and we recommend monitoring the SQSTM1 mRNA level as part of a complete analysis, or determining the SQSTM1 protein level in the presence of the transcription inhibitor actinomycin D or the p-EIF2S1 antagonist ISRIB.
Of interest, SQSTM1 overexpression at both the gene and protein levels can be observed in muscle atrophy induced by cancer, though not by glucocorticoids, suggesting that the stimulus inducing autophagy may also be relevant to the differential regulation of autophagy-related proteins [587]. One solution to problems relating to variations in SQSTM1 expression levels is to use a HaloTag®-p62 (SQSTM1) chimera [588]. The chimeric protein can be covalently labeled with HaloTag® ligands, and the loss of signal can then be monitored without interference by subsequent changes in protein synthesis. Similarly, a stable cell line expressing EGFP-tagged SQSTM1 under the control of an inducible promoter can be used to assess the rates of SQSTM1 degradation, taking into account the limitations outlined above (see Autophagic flux determination using flow and multispectral imaging cytometry) [497]. A similar system exists in Drosophila in which a GFP-tagged ref(2)P/SQSTM1 can be expressed using the UAS-GAL4 system [589]. It is worth noting that tetracycline can reduce autophagy levels; therefore, the appropriate control of only tetracycline addition has to be included if using an inducible promoter that responds to this drug [590]. Furthermore, the toxicity of tetracycline antibiotics toward mitochondria is well known (these drugs induce a mitochondrial unfolded protein response) [591], such that their use may trigger mitophagy, or other mitochondrial signaling events that interface with the autophagic machinery, thus complicating the interpretation of any results. Yet another solution is to employ a radioactive pulse-chase assay to measure the rates of SQSTM1 degradation [592].
SQSTM1 contains a LIR as well as a ubiquitin binding domain and appears to act by linking ubiquitinated substrates with the autophagic machinery. Nonetheless, it would be prudent to keep in mind that SQSTM1 contains domains that interact with several signaling molecules [593], and SQSTM1 may be part of MTORC1 [594]. Thus, SQSTM1 may have additional functions that need to be considered with regard to its role in autophagy. In the context of autophagy as a stress response, the complexity of using SQSTM1 as an autophagy marker protein is underscored by its capacity to modulate the NFE2L2/NRF2 anti-oxidant response pathway through a KEAP1 binding domain [595, 596]. In fact, SQSTM1 may, itself, be transcriptionally induced by NFE2L2 [597]. Furthermore, it is preferable to examine endogenous SQSTM1 because overexpression of this protein leads to the formation of protein inclusions. In fact, even endogenous SQSTM1 becomes Triton X-100-insoluble in the presence of protein aggregates and when autophagic degradation is inhibited; thus, results with this protein are often context-dependent.
Indeed, there is a reciprocal crosstalk between the UPS and autophagy, with SQSTM1 being a key link between them [598, 599]. First, SQSTM1 participates in proteasomal degradation, and its level may also increase when the proteasome is inhibited [600]. Accordingly, the SQSTM1 degradation rate should be analyzed in the presence of a proteasomal inhibitor such as epoxomicin or lactacystin to determine the contribution from the proteasome (see Autophagy inhibitors and inducers for potential problems with MG132) [601]. Second, the accumulation of SQSTM1 due to autophagy inhibition can impair UPS function by competitively binding ubiquitinated proteins, preventing their delivery to, and degradation by, the proteasome [602]. Inhibition of autophagy by treatment with 3-MA (5 mM, 4 h) increases the accumulation of MAPT/tau oligomers within neurites of primary transgenic (prepared from PS19 mouse embryos, expressing the frontotemporal dementia P301S mutant MAPT [603]) cultured neurons, reducing their access to the soma and lysosomes for degradation [604]. Furthermore, USP14, a major proteasomal deubiquitinase that regulates degradation through the proteasome, interacts with the UBA domain of SQSTM1 as well as LC3. In addition, levels as well as chromatin recruitment of USP14 are upregulated in autophagy-deficient cells upon DNA damage, and knockdown of SQSTM1 in autophagy-deficient cells decreases USP14 levels [605, 606]. These data clearly indicate that autophagy regulates USP14 degradation in an SQSTM1-dependent manner. Accordingly, it may be advisable to measure the UPS flux by using UbG76V-GFP, a ubiquitin-proteasome activity reporter [607], when SQSTM1 accumulation is observed. Thus, it is very important to determine whether autophagy alone or in conjunction with the UPS accounts for substrate degradation induced by a particular biological change. A number of stressors that impair the UPS induce the aggregation/dimerization of SQSTM1, and this can be seen by the detection of a high molecular mass (~150 kDa) protein complex by western blot, which is recognized by SQSTM1 antibodies [564, 608, 609] Although the accumulation of this protein complex can be related to the accumulation of ubiquitinated SQSTM1-bound proteins, or the dimerization/inactivation of SQSTM1 [564, 610], evaluation of the ratio between SQSTM1 aggregates/dimers and SQSTM1 monomers is likely a better measurement of changes in SQSTM1 dynamics linked to autophagy or the UPS.
SQSTM1 is also a substrate for CASP6 (caspase 6) and CASP8 (as well as CAPN1 [calpain 1]), which may confound its use in examining cell death and autophagy [611]. This is one reason why SQSTM1 degradation should also be analyzed in the presence of a pan-caspase inhibitor such as Q-VD-OPh before concluding that autophagy is activated based on a decrease of this protein [565]. Another issue is that some phosphatidylinositol 3-kinase (PtdIns3K) inhibitors such as LY294002, and to a lesser extent wortmannin (but apparently not 3-MA) [435], can inhibit protein synthesis [612]; this might in turn affect the turnover of SQSTM1 and LC3, which could influence conclusions that are drawn from the status of these proteins regarding autophagic flux or ALIS formation. Accordingly, it may be advisable to measure protein synthesis and proteasome activity along with autophagy under inhibitory or activating conditions [613]. With regard to protein synthesis, it is worth noting that this can be monitored through a nonradioactive method [614].
Western blot analysis of cell lysates prepared using NP40- or Triton X-100-containing lysis buffers in autophagic conditions typically shows a reduction in SQSTM1 levels. However, this does not necessarily indicate that SQSTM1 is degraded, because SQSTM1 aggregates are insoluble in these detergent lysis conditions [419, 615]. Moreover, in some instances SQSTM1 levels do not change in the soluble fractions despite autophagic degradation, a finding that might be explained by simultaneous transcriptional and translational induction of the gene encoding SQSTM1, because the soluble fraction accounts only for the diffuse or free form of SQSTM1. Accumulation of SQSTM1 in the Triton X-100-insoluble fraction can be observed when autophagy-mediated degradation is inhibited. Under conditions of higher autophagic flux, accumulation of SQSTM1 in Triton X-100-insoluble fractions may not be observed, and SQSTM1 levels may be reduced or maintained. The simplest approach to circumvent many of these problems is using lysis buffer that allows identification of the entire cellular pool of SQSTM1 (e.g., containing 1% SDS); however, additional assessment of both Triton X-100-soluble and -insoluble fractions will provide further information regarding the extent of SQSTM1 oligomerization [548]. Note, when performing a western blot using an SQSTM1 antibody, it is always a good idea to include a positive control in which SQSTM1 accumulates, such as an atg8a mutant (e.g., see Fig. S3 in ref. [616]).
To conclusively establish SQSTM1 degradation by autophagy, SQSTM1 levels in both Triton X-100-soluble and -insoluble fractions need to be determined upon treatment with autophagy inducers in combination with autophagy inhibitors, such as those that inhibit the autolysosomal degradation steps (e.g., protease inhibitors, CQ or bafilomycin A1). Additionally, an alteration in the level of SQSTM1 may not be immediately evident with changes observed in autophagic flux upon certain chemical perturbations (S. Sarkar, personal communication). Whereas LC3 changes may be rapid, clearance of autophagy substrates may require a longer time. Therefore, if LC3 changes are assessed at six h or 24 h after a drug treatment, SQSTM1 levels can be tested not only at the same time points, but also at later time points (24 h or 48 h) to determine the maximal impact on substrate clearance. An alternative method is immunostaining, with and without autophagy inhibitors, for SQSTM1, which will appear as either a diffuse or punctate pattern. Experiments with autophagy inducers and inhibitors, in combination with western blot and immunostaining analyses, best establish autophagic degradation based on SQSTM1 turnover. A final point, however, is that empirical evidence suggests that the species-specificity of antibodies for detecting SQSTM1 must be taken into account. For example, some commercial antibodies recognize both human and mouse SQSTM1, whereas others detect the human, but not the mouse protein [617]. Another issue with detecting SQSTM1 in the context of human diseases is that it can be mutated (e.g., in Paget disease of bone) [618]. Thus, care should be taken to ensure that potential mutations are not affecting the epitopes that are recognized by anti-SQSTM1 antibodies when using western blotting to detect this protein.
As an alternative, the SQSTM1:BECN1 protein level ratio can be used as a readout of autophagy [619]. Because both decreased SQSTM1 levels and increased BECN1 levels correlate with enhanced autophagy, a decreased SQSTM1:BECN1 protein level ratio (when derived from the same protein extract) may, cautiously, be interpreted as augmented autophagy, keeping in mind that SQSTM1 gene expression varies significantly under different conditions and may obscure the meaning of a change in the amount of SQSTM1 protein. Another substantial alternate is analysis of neomycinephosphotransferase II (NeoR) degradation. NeoR is an exclusive autophagic substrate [620, 621]. NeoR-GFP degradation is completely blocked by autophagic inhibitors such as 3-MA, but does not respond to inhibitors of proteasomal degradation. Inhibition of autophagy leads to accumulation of NeoR-GFP, resulting in enhanced GFP fluorescence [621]. NeoR-GFP gene expression is not affected by most autophagy inducers including H2O2 (that transcriptionally upregulate SQSTM1), however, degradation can be evaluated by accumulation of NeoR-GFP puncta under confocal microscopy or by analyzing total protein level by western blot of GFP.
As a general note, using ratios of the levels of proteins changing in opposite directions, rather than the protein levels themselves, could be beneficial because it overcomes the loading normalization issue. The often-used alternative approach of housekeeping proteins to normalize for loading biases among samples is sometimes problematic as levels of the HKPs change under various physiological, pathological and pharmacological conditions [269, 622–626].
Finally, a novel protein family of autophagy receptors, named CUET (from Cue5/TOLLIP), was identified, which in contrast to SQSTM1 and NBR1 has members that are present in all eukaryotes [627, 628]. The CUET proteins also possess a ubiquitin-binding CUE-domain and an Atg8-family interacting motif (AIM)/LIR sequence that interacts with Atg8-family proteins. In their absence, cells are more vulnerable to the toxicity resulting from aggregation-prone proteins, showing that CUET proteins, and more generally autophagy, play a critical evolutionarily conserved role in the clearance of cytotoxic protein aggregates [627]. Experiments in yeast have shown that Cue5 and the cytoplasmic proteins that require this autophagy receptor for rapid degradation under starvation conditions could be potentially good marker proteins for measuring autophagic flux [629]. Studies with mammalian immune cells indicate that TOLLIP is primarily responsible for the final step of autophagy, and facilitates the fusion of lysosomes with autophagosomes, lipid droplets, or peroxisomes [630]. TOLLIP may fulfill its critical function of lysosome fusion through its interaction with phospholipid [631, 632]. TOLLIP-deficient monocytes are defective in lysosome fusion and are programmed into an inflamed state with elevated CCR5 and enhanced expression of chemokines such as CCL2/MCP1 [631, 633]. Pathologically, TOLLIP-deficient mice tend to develop more severe atherosclerosis as well as neurological defects [633, 634].
Another recent study demonstrated a functional link between CLEC16A and disrupted mitophagy in murine splenic immune cells and showed that incomplete mitophagy predisposes clec16a knockout mice to a cascade of altered immune signaling functions resulting in pathogenic inflammation [319, 320].
Special caution must be taken when evaluating SQSTM1 levels in models of protein aggregation. Small protoaggregates often stain positively for SQSTM1 and may be similar in size to autophagic puncta. Similarly, GFP-u/GFP-degron reporters (designed as an unstable variant that undergoes proteasome-dependent degradation) will mark SQSTM1-positive protein inclusions [635]. Finally, some types of aggregates and inclusions will release soluble SQSTM1 or GFP-u/GFP-degron under cell lysis or denaturing conditions, which can skew the interpretation of soluble SQSTM1 and/or proteasomal function, accordingly.
Conclusion: There is not always a clear correlation between increases in LC3-II and decreases in SQSTM1. Thus, although analysis of SQSTM1 can assist in assessing the impairment of autophagy or autophagic flux, we recommend using SQSTM1 only in combination with other methods detailed in these guidelines to monitor flux. See also the discussion in Autophagic flux determination using flow and multispectral imaging cytometry.
4. TOR/MTOR, AMPK and Atg1/ULK1
Atg1/ULK1 are central components in autophagy that likely act at more than one stage of the process. There are multiple ULK isoforms in mammalian cells including ULK1, ULK2, ULK3, ULK4 and STK36 [636]. ULK3 is a positive regulator of the Hedgehog signaling pathway [637], and its overexpression induces both autophagy and senescence [638]. Along these lines, ectopic ULK3 displays a punctate pattern upon starvation-induced autophagy induction [638]. ULK3, ULK4 and STK36, however, lack the domains present on ULK1 and ULK2 that bind ATG13 and RB1CC1/FIP200 [639]. Thus, ULK3 may play a role that is restricted to senescence and that is independent of the core autophagy machinery. ULK2 has a higher degree of identity with ULK1 than any of the other homologs, but they may have both similar and distinct functions that are tissue- or cell-type specific [640–644]. Specifically in relation to autophagy, pharmacological inhibition of ULK1 and ULK2, with the compound MRT68921, blocks the process, and expression of a drug-resistant ULK1 mutant is sufficient to rescue this block [457]. However, at least in some cell types, ULK2 can likely compensate for loss of ULK1. For instance, in LNCaP cells, combined knockdown of ULK1 and ULK2 provides a substantially stronger inhibition of basal and starvation-induced autophagic sequestration and degradation activity than knockdown of ULK1 alone (N. Engedal, personal communication). ULK1 activity can also be inhibited by the expression of a dominant-negative ULK1 mutant [645, 646]. The stability and activation of ULK1, but not ULK2, is dependent on its interaction with the HSP90-CDC37 chaperone complex. Pharmacological or genetic inhibition of the chaperone complex increases proteasome-mediated turnover of ULK1, impairing its kinase activity and ability to promote both starvation-induced autophagy and mitophagy [647]. In addition, ULK1 is ubiquitinated for its activation through TRAF6-dependent K63-linked ubiquitination [501], or for degradation through CUL3-KLHL20-dependent K48-linked ubiquitination [648]. GCA (grancalcin) inhibits K48-linked ubiquitination and activates TRAF6-dependent K63-linked ubiquitination of ULK1 to induce autophagy [649].
AMPK (AMP-activated protein kinase) is a multimeric serine/threonine protein kinase comprised of PRKAA1/AMPKα1 or PRKAA2/AMPKα2 (α, catalytic), the PRKAB1/AMPKβ1 or PRKAB2/AMPKβ2 (β, scaffold), and the PRKAG1/AMPKγ1, PRKAG2/AMPKγ2 or PRKAG3/AMPKγ3 (γ, regulatory) subunits. The enzyme activity of AMPK is dependent on phosphorylation of the PRKAA2/α2-subunit on Thr172 (corresponds to Thr183 in α1) [459,460], and, therefore, can be conveniently monitored by western blotting with a phosphospecific antibody against this site. Depending on the stimulus and cell type, Thr172 is phosphorylated either by CAMKK2/CaMKKβ, STK11/LKB1 or MAP3K7/TAK1. Inhibition of AMPK activity is mediated primarily by Thr172-dephosphorylating protein phosphatases such as PPP1/PP1 (protein phosphatase 1) and PPP2/PP2A (protein phosphatase 2) [650]. Thr172 dephosphorylation is modulated by adenine nucleotides that bind competitively to regulatory sites in the PRKAG/γ-subunit. AMP and ADP promote phosphorylation and AMPK activity, whereas Mg2+-ATP has the opposite effect [651]. Moreover, Thr172 phosphorylation and AMPK activation can be enhanced by PRKDC (protein kinase, DNA-activated, catalytic subunit)-mediated phosphorylation of PRKAG1/AMPKγ1, which promotes the lysosomal localization of the AMPK complex [652]. Thus, AMPK acts as a fine-tuned sensor of the overall cellular energy charge that regulates cellular metabolism to maintain energy homeostasis. Overexpression of a dominant negative mutant (R531G) of PRKAG2, the γ−subunit isoform 2 of AMPK that is unable to bind AMP, makes it possible to analyze the relationship between AMP modulation (or alteration of energetic metabolism) and AMPK activity [653, 654]. Activation of AMPK is also associated with the phosphorylation of downstream enzymes involved in ATP-consuming processes, such as fatty acid (ACAC [acetyl-CoA carboxylase]) and cholesterol (HMGCR [3-hydroxy-3-methylglutaryl-CoA reductase]) biosynthesis.
The role of AMPK in autophagy is complex and highly dependent on both cell type and metabolic conditions. In yeast, the AMPK ortholog Snf1 shows autophagy inhibitory functions dependent on its ability to inhibit cytosolic Acc1 (acetyl-CoA carboxylase)-mediated lipogenesis, which is required for autophagy in stationary phase cells [655]. AMPK also exerts autophagy inhibitory effects through distinct ULK1-dependent effects on autophagosome formation and lysosomal acidification in cancer cell lines [656]. Furthermore, as noted above, there are two isoforms of the catalytic subunit, PRKAA1/AMPKα1 and PRKAA2/AMPKα2, and these may have distinct effects with regard to autophagy (C. Koumenis, personal communication) [657]. In liver cells, AMPK suppresses autophagy at the level of cargo sequestration, as indicated by the rapid sequestration-inhibitory effects of a variety of AMPK activators, whereas it appears to stimulate autophagy in many other cell types, including fibroblasts, colon carcinoma cells and skeletal muscle [658–667], and there appears to be a completely AMPK-dependent type of autophagy [668]. Autophagy-promoting effects of AMPK are most evident in cells cultured in a complete medium with serum and amino acids, where cargo sequestration is otherwise largely suppressed [664]. Amino acids acutely activate AMPK, which sustains autophagy under nutrient sufficiency [669]. Presumably, AMPK antagonizes the autophagy-inhibitory effect of amino acids (at the level of phagophore assembly) by phosphorylating proteins involved in MTORC1 signaling, such as TSC2 [670] and RPTOR/raptor [670] as well as the MTORC1 target ULK1 (see below) [671–673].
Compound C is an effective and widely used inhibitor of activated (phosphorylated) AMPK [674, 675]. However, being a nonspecific inhibitor of oxidative phosphorylation [676, 677], this drug has been observed to inhibit autophagy under conditions where AMPK is already inactive or knocked out [678, 679], and it has even been shown to stimulate autophagy by an AMP-independent mechanism [677, 680]. Compound C thus cannot be used as a stand-alone indicator of AMPK involvement, but can be used along with shRNA-mediated inhibition of AMPK.
TORC1 is an autophagy-suppressive regulator that integrates growth factor, nutrient and energy signals. In most systems, inhibition of MTOR leads to induction of autophagy, and AMPK activity is generally antagonistic toward MTOR function. MTORC1 mediates the autophagy-inhibitory effect of amino acids, which stimulate the MTOR protein kinase through a RRAG GTPase heterodimer. INS (insulin) and growth factors activate MTORC1 through upstream kinases including AKT/protein kinase B and MAPK1/ERK2-MAPK3/ERK1 when the energy supply is sufficient, whereas energy depletion may induce AMPK-mediated MTORC1 inhibition and autophagy stimulation, for example, during glucose starvation. In contrast, amino acid starvation can strongly induce autophagy even in cells completely lacking AMPK catalytic activity [681]. The impact of MTORC1 on autophagy is furthermore underlined in the pathological setting of a lysosomal storage disease based on insufficient MTORC1 activation and subsequent increased autophagosome formation due to hereditary TBCK (TBC1 domain containing kinase) deficiency [682, 683].
MTORC1-mediated autophagy is negatively regulated by SHOC2, a scaffold protein that activates the RAS-RAF-MAPK signaling pathway [684, 685]. Specifically, SHOC2 binds to RPTOR and dislodges it from MTORC1, leading to MTORC1 inactivation and autophagy induction [686, 687]. Thus, MTORC1 signaling can be negatively regulated by MAPK signaling.
AMPK and MTORC1 regulate autophagy through coordinated phosphorylation of ULK1. Under glucose starvation, AMPK apparently promotes autophagy by directly activating ULK1 through phosphorylation, although the exact AMPK-mediated ULK1 phosphorylation site(s) remains controversial (Table 2) [667, 671–673]. Under conditions of nutrient sufficiency, high MTORC1 activity prevents ULK1 activation by phosphorylating alternate ULK1 residues and disrupting the interaction between ULK1 and AMPK. There are commercially available phospho-specific antibodies that recognize different forms of ULK1. For example, phosphorylation at Ser556 in human (corresponds to Ser555 in mouse), an AMPK site, is indicative of increased autophagy in response to nutrient stress, whereas Ser758 in human (corresponds to Ser757 in mouse) is targeted by MTOR to inhibit autophagy. Even the autophagy-suppressive effects of AMPK could, conceivably, be mediated through ULK1 phosphorylation, for example, at the inhibitory site Ser638 [688]. AMPK inhibits MTORC1 by phosphorylating and activating TSC2 [670], as well as by phosphorylating the MTOR binding partner RPTOR [689]. Therefore, AMPK is involved in processes that synergize to activate autophagy, by directly activating ULK1, and indirectly impairing MTOR-dependent inhibition of ULK1. In addition, IPMK (inositol polyphosphate multikinase) can act as a scaffold protein to influence AMPK-dependent ULK phosphorylation [690]. The identification of ULK1 as a direct target of MTORC1 and AMPK represents a significant step toward the definition of new tools to monitor the induction of autophagy. ULK1 and ATG13 are also phosphorylated by CCNB/cyclin B in mitosis to activate autophagy [691].
In addition to ULK1 regulation by AMPK and MTORC1 under conditions of glucose starvation, in skeletal muscle ULK1 is activated by MAPK11/p38β in response to a tumor burden through phosphorylation of Ser555 in mice. Despite AMPK activation (phosphorylation on Thr172) by factors released from tumor cells, inhibition of AMPK with compound C does not alter ULK1 phosphorylation on Ser555 and activation of autophagy in these conditions. Conversely, MAPK11 gain- and loss-of-function assays indicate that MAPK11 is a key activator of ULK1 and autophagy in the cancer milieu [692].
Further studies directed at identifying physiological substrates of ULK1 will be essential to understand how ULK1 activation results in initiation of the autophagy program. So far, several ULK1 substrates have been reported, and these can be classified into 4 subgroups: 1) components of the ULK1 complex; 2) components of the class III PtdIns3K complex I; 3) other autophagy-related proteins; or 4) non-autophagy-related proteins. Numerous groups have shown that ULK1 autophosphorylates and transphosphorylates its binding partners ATG13, RB1CC1, and ATG101 [646, 647, 693–701]. So far, only the ULK1 autophosphorylation at Thr180 and Ser1047, and the phosphorylation of ATG13 at Ser318 (human isoform 2) have been shown to be functionally relevant. ATG13 phosphorylation at Ser318 by ULK1 is required for efficient clearance of damaged mitochondria [647]. The functional relevance of ULK1-dependent phosphorylation of RB1CC1 and ATG101 awaits further clarification. With regard to the components of the class III PtdIns3K complex I, ULK1-dependent phospho-acceptor sites have been identified in PIK3C3/VPS34 (phosphatidylinositol 3-kinase catalytic subunit type 3), BECN1, ATG14, and AMBRA1 [695, 702–706]. Following amino acid starvation or MTOR inhibition, the activated ULK1 phosphorylates ATG14 on Ser29 and BECN1 on Ser14 and Ser30, enhancing the activity of the complexes containing ATG14 and PIK3C3/VPS34. These ATG14 and BECN1 phosphorylations by ULK1 are required for full autophagic induction in response to amino acid starvation or MTOR inhibition [702, 705]. ULK1-dependent phosphorylation of BECN1 at Ser30 also stimulates autophagosome formation in response to hypoxia [706]. Next to the two autophagy-initiating complexes, other ATG proteins have been identified as ULK1 substrates, notably ATG4B, ATG9A, and ATG16L1 [707–709]. Finally, there are several ULK1 substrates that are not specifically ATG proteins. These proteins are involved in the execution of autophagy, or fulfill additional cellular functions. These substrates include RPTOR, AMPK, SQSTM1, FUNDC1, DAPK3, MAPK14/p38alpha, FLCN, enzymes involved in glucose metabolic flux, DENND3, SMCR8, TBK1, PDPK1, SEC16A, SEC23A, SEC23B, EXOC7, SDCBP, STING1/TMEM173, CDC37, MAD1L1, VCP/p97, DVL1, NR3C2, YAP1, WWTR1, and RIPK1 [710–735]. The ULK1-dependent phosphorylation of RPTOR leads to inhibition of MTORC1 [711, 718], and the ULK1-dependent inhibitory phosphorylation of AMPK subunits appears to generate a negative feedback loop [710]. Note that caution should be taken to use appropriate inhibitors of phosphatases (e.g., sodium fluoride, and β-glycerophosphate) in cell lysis buffer before analyzing the phosphorylation of AMPK and ULK1 at serine and threonine sites.
MTORC1 activity can be monitored by following the phosphorylation of its substrates, such as EIF4EBP1/4E-BP1/PHAS-I and RPS6KB/p70S6 kinase or the latter’s downstream target, RPS6/S6, for which good commercial antibodies are available [736–738]. In mammalian cells, the analysis should focus on the phosphorylation of S6K1 at Thr389, and EIF4EBP1 at Ser65, a serum-responsive and rapamycin-sensitive site; phosphorylation of EIF4EBP1 at Thr37 and Thr46 primes the protein for phosphorylation at Ser65, and although directly phosphorylated by MTORC1, the modifications at Thr37 and Thr46 are only partially sensitive to serum and rapamycin [739]. The MTORC1-dependent phosphorylation of EIF4EBP1 can be detected as a molecular mass shift by western blot [736]. Examining the phosphorylation status of RPS6KB and EIF4EBP1 may be a better method for monitoring MTORC1 activity than following the phosphorylation of proteins such as RPS6, because the latter is not a direct substrate of MTORC1 (although RPS6 phosphorylation is a good readout for RPS6KB1/2 activities, which are directly dependent on MTOR), and it can also be phosphorylated by other kinases such as RPS6KA/RSK. Whereas RPS6KB1/2 phosphorylates RPS6 at Ser235, Ser236, Ser240, and Ser244, RPS6KA/RSK exclusively phosphorylates RPS6 at Ser235 and Ser236 in vitro and in vivo in a manner independent of MTORC1 [740]. Thus, the use of RPS6 phospho-Ser240/244 antibody is necessary for monitoring cellular MTORC1-RPS6KB1/2 activity specifically in western blot or immunocytochemistry.
Furthermore, the mechanisms that determine the selectivity as well as the sensitivity of MTORC1 for its substrates seem to be dependent on the integrity and configuration of MTORC1. For example, rapamycin strongly reduces RPS6KB1 phosphorylation, whereas its effect on EIF4EBP1 is more variable. In the case of rapamycin treatment, EIF4EBP1 can be phosphorylated by MTORC1 until rapamycin disrupts MTORC1 dimerization and its integrity, whereas RPS6KB1 phosphorylation is quickly reduced when rapamycin simply interacts with MTOR in MTORC1 (see Autophagy inhibitors and inducers for information on catalytic MTOR inhibitors such as torin1) [739]. Because it is likely that other inhibitors, stress, and stimuli may also affect the integrity of MTORC1, a decrease or increase in the phosphorylation status of one MTORC1 substrate does not necessarily correlate with changes in others, including ULK1. Therefore, reliable anti-phospho-ULK1 antibodies should be used to directly examine the phosphorylation state of ULK1, along with additional experimental approaches to analyze the role of the MTOR complex in regulating autophagy. The MTORC1-mediated phosphorylation of AMBRA1 on Ser52 has also been described as relevant to ULK1 regulation and autophagy induction [703, 741]. In line with what is described for ULK1, the anti-phospho-AMBRA1 antibody, which is commercially available, could be used to indirectly measure MTORC1 activity [741].
Activation/assembly of the Atg1 complex in yeast (composed of at least Atg1-Atg13-Atg17-Atg31-Atg29) or the ULK1 complex in mammals (ULK1-RB1CC1-ATG13-ATG101) is one of the first steps of autophagy induction. Therefore, activation of this complex can be assessed to monitor autophagy induction. In yeast, dephosphorylation of Atg13 is associated with activation/assembly of the core complex that reflects the reduction of TORC1 and PKA activities. Therefore, assessing the phosphorylation levels of this protein by immunoprecipitation or western blotting [742–745] can be used not only to follow the early steps of autophagy but also to monitor the activity of some of the upstream nutrient-sensing kinases. Because this protein is not easily detected when cells are lysed using conventional procedures, a detailed protocol has been described [746]. In addition, the autophosphorylation of Atg1 at Thr226 is required for its kinase activity and for autophagy induction; this can be detected using phospho-specific antibodies, by immunoprecipitation or western blotting (Figure 22) [747, 748]. In Drosophila, TORC1-dependent phosphorylation of Atg1 and Atg1-dependent phosphorylation of Atg13 can be indirectly determined by monitoring phosphorylation-induced electromobility retardation (gel shift) of protein bands in immunoblot images [423,47,509,510]. Nutritional starvation suppresses TORC1-mediated Atg1 phosphorylation [423,509] while stimulating Atg1-mediated Atg13 phosphorylation [589, 749, 750]. In mammalian cells, the phosphorylation status of ULK1 at the activating sites (Ser317, 777 [position in the murine sequence, not conserved in human], 467, 556, 638, or Thr575 in the human sequence) or dephosphorylation at inactivating sites (Ser638, 758 in the human sequence) can be determined by western blot using phospho-specific antibodies [672–674, 688, 751, 752]. In general, the core complex is stable in mammalian cells, although, as noted above, upstream inhibitors (MTOR) or activators (AMPK) may interact dynamically with it, thereby determining the status of autophagy.
Figure 22.
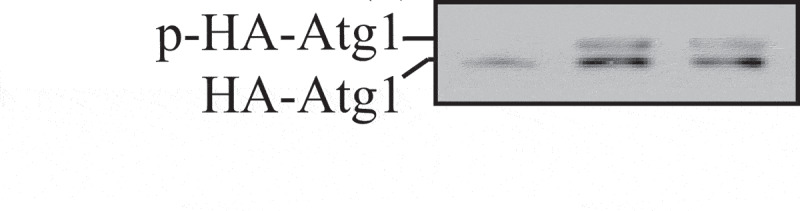
S. cerevisiae cells transformed with a plasmid encoding HA-Atg1 were cultured to mid-log phase and shifted to SD-N (minimal medium lacking nitrogen that induces a starvation response). Immunoblotting was done with anti-HA antibody. The upper band corresponds to autophosphorylation of Atg1. This figure was modified from data previously published in ref. [747], and is reproduced by permission of the American Society for Cell Biology, copyright 2011
Alternatively, the activation of the ULK1 complex can be monitored by assessing the localization pattern of ATG13 by immunofluorescence. In fact, following ULK1 complex activation, ATG13 relocates to the omegasome, which results in a punctate pattern [753, 754]. In mesothelioma ex vivo 3-dimensional models, the percentage of tumor cells with ATG13 puncta correlates with the level of autophagy, and the analysis of ATG13 puncta has been proposed as an assay to monitor autophagy in mesothelioma formalin-fixed, paraffin-embedded and 3-dimensional models [755]. In the same model, the ULK1/2 inhibitor MRT68921 blocks both autophagy and the formation of ATG13 puncta [756]. As a cautionary note, ATG13 puncta do not reflect autophagy in traditional monolayer cultures of mesothelioma cells. Therefore, studies in 3-dimensional models of other tumors are needed to confirm the validity of ATG13 as a marker of autophagy.
One additional topic that bears on ULK1 concerns the process of LC3-associated phagocytosis (see Noncanonical use of autophagy-related proteins). LAP is a type of phagocytosis in macrophages that involves the conjugation of LC3 to single-membrane pathogen-containing phagosomes, a process that promotes phagosome maturation [247]. Although ULK1 is not required for the clearance of cell corpses by LAP, in mammals [245], and UNC-51 (the Atg1/ULK1 homolog in C. elegans) is not required for the clearance of neuroblast corpses in larval worms or released cell fragments in worm embryos [757, 758], it is important to note that an increased number of apoptotic cell corpses persist during embryonic development in unc-51 mutant worms [759], suggesting that UNC-51 could have a role in cell death or cell corpse clearance. A recent study shows that pancreatic acinar cells also have the ability to process post-exocytic organelles via LAP [760]. LAP-deficient tumor-associated macrophages also aid in promoting an anti-tumor response in T cells in a tumor microenvironment [761].
An additional substrate that is required for efficient AMPK-induced autophagy is CCNY (cyclin Y)-CDK16 [762]. AMPK phosphorylates CCNY, which promotes its interaction with CDK16, a PCTAIRE kinase family member. The loss of CCNY-CDK16 impairs AMPK-stimulated autophagy, whereas overexpression of CCNY-CDK16 is sufficient to induce autophagy. This outcome is dependent on the catalytic activity of CCNY-CDK16, albeit the substrates of this kinase have not been identified yet.
Cautionary notes: A decrease in TORC1 activity is a good measure for autophagy induction; however, TORC1 activity does not necessarily preclude autophagy induction because there are TOR-independent mechanisms that induce autophagy both in mammals and yeast [763–767] Along these lines, the disassociation of the AMPK-MTORC1 axis is observed in some AML cells such as the KG-1 cell line, as well as in primary AML cells treated with the specific AMPK agonist GSK621 [768, 769]. The co-activation of AMPK and MTORC1 in these cancer cells is associated with increased autophagy flux, with AMPK as the major regulator of autophagy in these conditions [768, 769]. Whereas in most systems inhibition of MTOR leads to the induction of autophagy, there are instances in commonly used cancer cell lines and influenza A virus-infected cells in which MTOR appears to be a positive effector [770, 771]. Also, MTOR suppression does not always induce autophagy, such as when BECN1 undergoes inhibitory phosphorylation by the growth factor signaling molecules EGFR and AKT, when microglia are activated by lipopolysaccharide (LPS), a TLR4 ligand [772], or during Salmonella infection [773–775]. Note that the effect of everolimus in EGFR-transgenic mice is not mainly attributable to autophagy although it suppresses MTOR and induces autophagy in EGFR-driven lung cancer cell lines [776]. In adult skeletal muscle, active MTORC1 phosphorylates ULK1 at Ser757 to inhibit the induction of autophagosome formation. Thus, induction of autophagy requires inhibition of MTORC1 and not of MTORC2 [777, 778]. There is also evidence that inhibition of MTORC1 is not sufficient to maintain autophagic flux, but requires additional activation of FOXO transcription factors for the upregulation of autophagy gene expression [662]. In addition, MTORC1 is downstream of AKT; however, oxidative stress inhibits MTOR, thus allowing autophagy induction, despite the concomitant activation of AKT [207]. For neural cells, following administration of the class I phosphoinositide 3-kinase (PI3K) inhibitor LY294002, the phosphorylation levels of AKT and MTOR decrease, and the ratio of LC3-II:LC3-I is higher in the inhibitor-treated injury group than in the simple-injury group [779]. Also, persistent MTORC1 inhibition can cause downregulation of negative feedback loops on IRS-MTORC2-AKT that results in the reactivation of MTORC2 under conditions of ongoing starvation [581, 780, 781]. Along these lines, both TORC1 and autophagy can be active in specific cell subpopulations of yeast colonies [763]. Similarly, mature autophagosomes and MTOR accumulate in the TOR-autophagy spatial coupling compartment (TASCC) during RAS-induced senescence [782]. Thus, it is necessary to be cautious in deciding how to monitor the TOR/MTOR pathway, and to verify that the pathway being analyzed displays TOR/MTOR-dependent inhibition.
Another point is that the regulation of autophagy by MTOR can be ULK1-independent. During mycobacterial infection of macrophages, MTOR induces the expression of MIR155 and MIR31 to sustain the activation of the WNT5A and SHH/sonic hedgehog pathways. Together, these pathways contribute to the expression of lipoxygenases and downregulation of IFNG-induced autophagy [783]. Signaling pathways can be monitored by western blotting, and TaqMan miRNA assays are available to detect these miRNAs.
One problem in monitoring assembly of the ULK1 complex is the low abundance of endogenous ULK1 in many systems, which makes it difficult to detect phospho-ULK1 by western blot analysis. In addition, Atg1/ULK1 is phosphorylated by multiple kinases, and the amount of phosphorylation at different sites can increase or decrease during autophagy induction. Thus, although there is an increase in phosphorylation at the activating sites upon induction, the overall phosphorylation states of ULK1 and ATG13 are decreased under conditions that lead to induction of autophagy; therefore, monitoring changes in phosphorylation by following molecular mass shifts upon SDS-PAGE may not be informative. In addition, such phosphorylation/dephosphorylation events are expected to occur relatively early (1-2 h) in the signaling cascade of autophagy. Therefore, it is necessary to optimize treatment time conditions. Finally, in Arabidopsis and possibly other eukaryotes, the ATG1 and ATG13 proteins are targets of autophagy, which means that their levels may drop substantially under conditions that induce autophagic turnover [350].
At present, the use of Atg1/ULK1 kinase activity as a tool to monitor autophagy is limited because only a few physiological substrates have been identified, and the importance of Atg1/ULK1-dependent phosphorylation has not always been determined. Nonetheless, Atg1/ULK1 kinase activity appears to increase when autophagy is induced, irrespective of the pathway leading to induction. As additional physiological substrates of Atg1/ULK1 are identified, it will be possible to follow their phosphorylation in vivo as is done with analyses for MTOR. Nonetheless, it must be kept in mind that monitoring changes in the activity of Atg1/ULK1 is not a direct assay for autophagy, although such changes may correlate with autophagy activity. Furthermore, the ULK1 substrates described above and additional ULK1-interacting proteins (e.g., PARP1) [784] already indicate that ULK1—next to its essential role for the induction of autophagy—participates in several additional physiological processes including axon guidance during brain development, type I interferon production, ER-Golgi trafficking, regulation of chaperone function, mitosis, stress granule dynamics, WNT-CTNNB1/β-catenin signaling, NR3C2/mineralocorticoid receptor signaling, and non-autophagic regulation of cell death. In a C. elegans Parkinson disease model, RNAi knockdown of UNC-51/ULK1 results in the accumulation of a human SNCA-GFP fusion [785]. Accordingly, the ULK activity state may thus reflect its role in these processes [786–792]. Therefore, other methods as described throughout these guidelines should also be used to follow autophagy directly.
Finally, there is not a complete consensus on the specific residues of ULK1 that are targeted by AMPK or MTOR. Similarly, apparently contradictory data have been published regarding the association of AMPK and MTOR with the ULK1 kinase complex under different conditions. Therefore, caution should be used in monitoring ULK1 phosphorylation or the status of ULK1 association with AMPK until these issues are resolved.
Conclusion: Assays for Atg1/ULK1 can provide detailed insight into the induction of autophagy, but they are not a direct measurement of the process. Similarly, because MTOR substrates such as RPS6KB1 and EIF4EBP1 are not recommended readouts for autophagy, their analysis needs to be combined with other assays that directly monitor autophagy activity.
5. Estimation of PtdIns3K (PIK3C3/VPS34) activity
PIK3C3/VPS34 is highly conserved through evolution, and belongs to the class III PtdIns3K that phosphorylates the 3ʹ-OH position of phosphatidylinositol (PtdIns) to synthesize PtdIns3P [793, 794]. PtdIns3P is essential for the regulation of endocytic pathways and for the generation of various types of autophagosomes and phagosomes. However, PIK3C3/VPS34 cannot be found alone in the cell but mainly is present in two types of mutually exclusive complexes, complexes I and II. Complex I is composed of PIK3C3/VPS34, PIK3R4/VPS15/p150, BECN1, ATG14 and NRBF2 for mammals, or Vps34, Vps15, Vps30/Atg6, Atg14 and Atg38 for yeast. Complex II replaces ATG14/Atg14 with UVRAG, or Vps38 for mammals and yeast, respectively [795–799]. Complex I regulates autophagy, whereas complex II regulates endocytic pathways, LAP, and cytokinesis [800–802]. Both in yeast and mammals, PIK3C3/VPS34 shows higher activity in complexes than on its own. For example, yeast complexes I and II show higher activity than a Vps34-Vps15 heterodimer [800], and human PIK3C3 activity is increased by coexpressing it with PIK3R4/VPS15 [803]. Also, various post-translational modifications of the subunits of complexes I and II affect the kinase activity [804].
The most commonly used method to estimate PIK3C3/VPS34 activity is to immunoprecipitate the protein complex from cells, immobilize it on beads, mix with the substrate (PtdIns) and radioactive ATP, then measure the PtdIns3P production by autoradiography. Furthermore, two commercial kits are available; an ELISA-based kit from Echelon (K-3300), and a kit for measuring ADP generation from ATP from Promega (V6930). The PIK3C3/VPS34 activity is affected by enzyme concentration and substrate structure. First, for all methods it is important to estimate the concentration and purity of immobilized PIK3C3/VPS34 complex by Coomassie Brilliant Blue or silver staining. Second, the PtdIns structure and the environment where PtdIns is surrounded such as the length of acyl chain, and the size and composition of liposomes largely affect the activity. The PtdIns provided with the Echelon kit is water-soluble diC8-PtdIns. There are also PtdIns:phosphatidylserine mixture substrates at a 1:9 molar ratio (Thermo Fisher, PV5122), or at a 1:3 molar ratio (Promega, V1711). Although they are good substrates for drug screening, they are not physiological. If researchers are examining the kinase activity to reflect the intracellular conditions, it is recommended to make liposomes that mimic the lipid compositions of the organelle of interest. Therefore, it is necessary to describe the lipid compositions of liposomes, the catalog number for each lipid species, and procedures for making liposomes (just sonication, or whether the liposome size was adjusted by extruding) for publication.
For ADP-Glo assays, because it measures the ATP-ADP conversion, if the purified enzyme is contaminated with chaperones, the ATPase activities of the latter dominate the values. Therefore, it is important to check the purity of the purified enzyme in advance and additionally measure the luminescence values of the enzyme without substrate. Also, the measured luminescence values need to be subtracted by the background values, which can be the intercept value of the standard curve or the measured luminescence of a mixture of 0% ADP and 100% ATP (this should contain ATP in case of impurities). This means that the enzyme concentration needs to be adjusted high enough so that the luminescence values of enzyme plus substrate are higher than the background values (i.e., the measured luminescence values of enzyme without substrate or the mixture of 0% ADP and 100% ATP). The above points need to be considered not only for the PIK3C3/VPS34 assays, but also for all lipid kinase and phosphatase activity assays.
6. Additional autophagy-related protein markers
Although Atg8-family proteins have been the most extensively used proteins for monitoring autophagy, other proteins can also be used for this purpose. Here, we discuss some of the more commonly used or better-characterized possibilities.
a. Atg9/ATG9A
Atg9/ATG9A is the only integral membrane Atg protein that is essential for autophagosome formation in all eukaryotes. Mammalian ATG9A displays partial colocalization with GFP-LC3 [805], and ATG9A deficiency in the mouse brain causes axon-specific lesions including neuronal circuit dysgenesis [806]. Perhaps the most unique feature of Atg9, however, is that it localizes to multiple discrete puncta, whereas most Atg proteins are detected primarily in a single punctum or diffusely within the cytosol. Yeast Atg9 may cycle between the phagophore assembly site (PAS) and peripheral reservoirs [807]; the latter correspond to tubulovesicular clusters that are precursors to the phagophore [808]. Anterograde movement to the PAS is dependent on Atg11, Atg23, Atg27 and actin. Retrograde movement requires Atg1-Atg13, Atg2-Atg18 and the PtdIns3K complex I [809]. Mutants such as atg1∆ accumulate Atg9 primarily at the PAS, and this phenotype forms the basis of the “transport of Atg9 after knocking out ATG1” (TAKA) assay [148]. In brief, this is an epistasis analysis in which a double-mutant strain is constructed (one of the mutations being atg1∆) that expresses Atg9-GFP. If the second mutated gene encodes a protein that is needed for Atg9 anterograde transport, the double mutant will display multiple Atg9-GFP puncta. In contrast, if the protein acts along with or after Atg1, all of the Atg9-GFP will be confined to the PAS. One such example is a septin complex that regulates Atg9 retrograde transport. The temperature-sensitive point mutations in Cdc10 (P3S and G44D) show accumulation of Atg9 at the PAS at non-permissive temperatures [810]. Monitoring the localization of ATG9A has not been used as extensively in more complex eukaryotes, but this protein displays the same type of dependence on Atg1/ULK1 and PtdIns3P for cycling as seen in yeast [805, 809], suggesting that it is possible to follow this ATG9A as an indication of ULK1 and ATG13 function [646, 805, 809].
There are two conserved classical adaptor protein sorting signals within the cytosolic N terminus of ATG9, which mediate trafficking of ATG9 from the plasma membrane and trans-Golgi network (TGN) via interaction with AP-1/2 [707, 811]. SRC phosphorylates ATG9 at Tyr8 to maintain its endocytic and constitutive trafficking in unstressed conditions. In response to starvation, phosphorylation of ATG9 at Tyr8 by SRC, and at Ser14 by ULK1, functionally cooperate to promote interactions between ATG9 and the AP-1/2 complex, leading to redistribution of ATG9 from the plasma membrane and juxta-nuclear region to the peripheral pool for autophagy initiation [707]. Furthermore, the localization of mammalian ATG9A is regulated by cellular sphingomyelin levels. In cells with excess sphingomyelin, ATG9A is trapped in juxtanuclear recycling endosomes, and its failure to be recruited to autophagic membranes results in defective phagophore closure [812]. In neurons ATG9 localizes to axons and presynaptic sites, and requires active transport by the kinesin motor KIF1A to direct its localization into distal neurites [29, 537].
ATG9 is also conserved in plants including the model plant A. thaliana. A protease protection assay with microsomes isolated from A. thaliana cells shows that ATG9 has a similar membrane topology, with its N- and C-termini facing the cytosol [128]. Subcellular analysis indicates that A. thaliana ATG9 displays similar discrete puncta within the cytosol in close proximity to the trans-Golgi network and late endosomes, whereas ATG9-GFP fusion proteins show a transient association with the autophagosomal marker ATG8. However, in contrast to the yeast and mammalian atg9 mutants, Arabidopsis atg9 mutants accumulate numerous abnormal tubular autophagosomal structures, which are dynamically associated with the ER membranes. Using 3-dimensional electron tomography analysis, direct connections between these ATG8-positive tubular structures and the ER have been observed, implying that plant ATG9 plays an essential role in autophagosome progression from the ER, particularly under stress conditions [128]. Recently, the homotrimeric structure of A. thaliana ATG9 was resolved by cryo-EM at subnanometer resolution, which provides a structural basis for future studies of ATG9 function in eukayotes [129].
b. Atg12–Atg5
ATG5, ATG12 and ATG16L1 associate with the phagophore and have been detected by fluorescence and immunofluorescence (Figure 23) [813, 814]. The endogenous proteins form puncta that can be followed to monitor autophagy upregulation. Under non-stressed, nutrient-rich conditions, these proteins are predominantly diffusely distributed throughout the cytoplasm. Upon induction of autophagy, for example during starvation, there is a marked increase in the proportion of cells with punctate ATG5, ATG12 and ATG16L1. Furthermore, upstream inhibitors of autophagosome formation result in a block in this starvation-induced puncta formation, and this assay is very robust in some mammalian cells. Conversely, downstream inhibition of autophagy at the level of phagophore expansion, such as with inhibition of LC3/GABARAP expression, results in an accumulation of the phagophore-associated ATG5, ATG12 and ATG16L1 immunofluorescent puncta [815]. Moreover, PLSCR1 (phospholipid scramblase 1) may play an inhibitory role in the autophagic process interfering with ATG12–ATG5-ATG16L1 complex formation and phagophore elongation as shown through co-immunoprecipitation experiments. Indeed, PLSCR1 binds the ATG12–ATG5 complex preventing ATG16L1 association [413]; therefore, the evaluation of active complexes by co-immunoprecipitation and subsequent immunoblotting analysis can be a further indirect way to evaluate autophagy activation.
Figure 23.
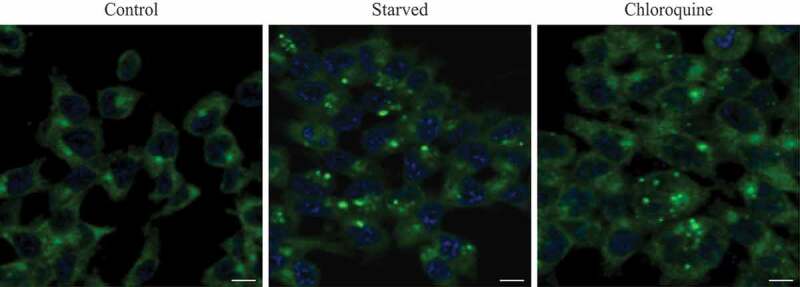
Confocal microscopy image of HCT116 cells immunostained with antibody specific to human ATG12. Cells were starved for 8 h or treated with CQ (50 µM) for 3 h. Scale bar: 10 µm. Image provided by M. Llanos Valero, M.A de la Cruz and R. Sanchez-Prieto
ATG12–ATG5 conjugation has been used in some studies to measure autophagy. In Arabidopsis and some mammalian cells it appears that essentially all of the ATG5 and ATG12 proteins exist in the conjugated form, and the expression levels do not change, at least during short-term starvation [289, 813, 814, 816]. Therefore, monitoring ATG12–ATG5 conjugation per se may not be a useful method for following the induction of autophagy. It is worth noting, however, that in some cell lines free ATG5 can be detected [817], suggesting that the amount of free ATG5 may be cell line-dependent; free ATG5 levels also vary in response to stress such as DNA damage [818]. Furthermore, free ATG12 can be detected in some cell lines and tissues and has ATG5-independent roles in cell signaling [819–821].One final parameter that may be considered is that the total amount of the ATG12–ATG5 conjugate may increase following prolonged starvation as has been observed in hepatocytes and both mouse and human fibroblasts (A.M. Cuervo, personal communication; S. Sarkar, personal communication), even though in these conditions part of the ATG12–ATG5 population is secreted in association with exosomes [822].
c. ATG14
Yeast Atg14 is the autophagy-specific subunit of the Vps34 complex I [796], and a human homolog, named ATG14/ATG14L/BARKOR, has been identified [795, 798, 799, 823]. ATG14 localizes primarily to phagophores. The C-terminal fragment of the protein, named the BATS domain, is able to direct GFP and BECN1 to autophagosomes in the context of a chimeric protein [824]. ATG14-GFP or BATS-GFP detected by fluorescence microscopy or TEM can be used as a phagophore marker protein; however, ATG14 is not localized exclusively to phagophores, as it can also be detected on mature autophagosomes as well as the ER [824, 825]. Accordingly, detection of ATG14 should be carried out in combination with other phagophore and autophagosome markers. A good antibody that can be used to detect endogenous ATG14 by immunostaining has been described [702].
d. ATG16L1
ATG16L1 has been used to monitor the movement of plasma membrane as a donor for autophagy, and thus an early step in the process. Indeed, ATG16L1 is located on phagophores, but not on completed autophagosomes [455, 826]. ATG16L1 can be detected by immuno-TEM, by immunostaining of Flag epitope-tagged ATG16L1, and/or by the use of GFP-tagged ATG16L1. ATG16L1 is phosphorylated on a serine residue at amino acid position 278 by ULK1 under autophagy-inducing conditions. Detection of endogenous phospho-ATG16L1 [827] has been demonstrated as a novel method to monitor autophagy induction. Because ATG16L1 is specifically located on phagophores but not complete autophagosomes, phospho-ATG16L1-based autophagy assays are predicted to be unaffected by a late stage autophagy block, and thus able to circumvent a major caveat of LC3-based assays while serving as an alternative tool with unique advantages to monitor autophagy.
ATG16L1 is ubiquitinated by the GAN (gigaxonin) E3 ligase, through interaction with the WD40 domain [828]. GAN causes the clearance of ATG16L1 in cell lines, whereas its repression in primary neurons derived from the gan−/− mouse induces an abnormal bundling of ATG16L1 within the soma. Action of GAN is dynamic, as restoration of its expression using lentiviral vector clears the aggregate and the endogenous ATG16L1, respectively, in GAN mutant and wild-type neurons. GAN mutant neurons exhibit a failure in producing autophagosomes over time upon autophagy induction, hence leading to a defective autophagic flux in subsequent steps. Thus, GAN is the first E3 ligase fine-tuning autophagosome production through ATG16L1.
Finally, the coding polymorphism of ATG16L1 (T300A, rs2241880), which is associated with Crohn disease, renders the protein sensitive to CASP3- and CASP7-mediated cleavage in the WD40 domain; this leads to decreased ATG16L1 function and can be detected by western blot [829, 830].
Cautionary notes: The expression level of ATG16L1 does not always correlate with other components of the autophagic machinery, and in some cases may be altered in a manner that is independent of autophagy; for example, this can be seen in samples from patients with acute myeloid leukemia (P. Ludovico, unpublished results).
e. Atg18/WIPI family
Yeast Atg18 [831, 832] and Atg21 [443] (or the mammalian WIPI homologs [833]) are required for both autophagy (i.e., nonselective sequestration of cytoplasm) and autophagy-related processes (e.g., the Cvt pathway [834, 835], specific organelle degradation [168], and autophagic elimination of invasive microbes [171, 172, 174, 175, 836]). These proteins bind PtdIns3P that is present at the phagophore and autophagosome [837, 838] and also PtdIns(3,5)P2. Furthermore, fluorescence stopped-flow [839] and chemical cross‐linking assays [840] show that Atg18 oligomerizes upon membrane binding, whereas it is mainly monomeric when unbound. Human WIPI1 and WIPI2 function downstream of the class III phosphatidylinositol 3-kinase complex I (PIK3C3/VPS34, BECN1, PIK3R4/VPS15, ATG14, NRBF2) and upstream of both the ATG12 and LC3 ubiquitin-like conjugation systems [837, 841, 842]. Upon the initiation of the autophagic pathway, WIPI1 and WIPI2 bind PtdIns3P and accumulate at limiting membranes, such as those of the ER, where they participate in the formation of omegasomes and/or autophagosomes [843, 844]. On the basis of quantitative fluorescence microscopy, this specific WIPI protein localization has been used as an assay to monitor autophagy in human cells [838].
Using either endogenous WIPI1 or WIPI2, detected by indirect fluorescence microscopy or EM, or transiently or stably expressed tagged fusions of GFP to WIPI1 or WIPI2, basal autophagy can be detected in cells that display WIPI puncta at autophagosomal membranes. In circumstances of increased autophagic activity, such as nutrient starvation or rapamycin administration, the induction of autophagy is reflected by the elevated number of cells that display WIPI puncta when compared to the control setting. Also, in circumstances of reduced autophagic activity such as upon wortmannin treatment, the reduced number of WIPI puncta-positive cells reflects the inhibition of autophagy. Basal, induced and inhibited formation of WIPI puncta closely correlates with both the protein level of LC3-II and the formation of GFP-LC3 puncta [838, 842]. Accordingly, WIPI puncta can be assessed as an alternative to LC3. Automated imaging and analysis of fluorescent WIPI1 (Figure 24) or WIPI2 puncta represent an efficient and reliable opportunity to combine the detection of WIPI proteins with other parameters. It should be noted that there are two isoforms of WIPI2 (2B and 2D) [842], and in C. elegans EPG-6/WDR45/WIPI4 has been identified as the WIPI homolog required for autophagy [845]. Thus, these proteins, along with the currently uncharacterized WDR45B/WIPI3, provide additional possibilities for monitoring phagophore and autophagosome formation.
Figure 24.
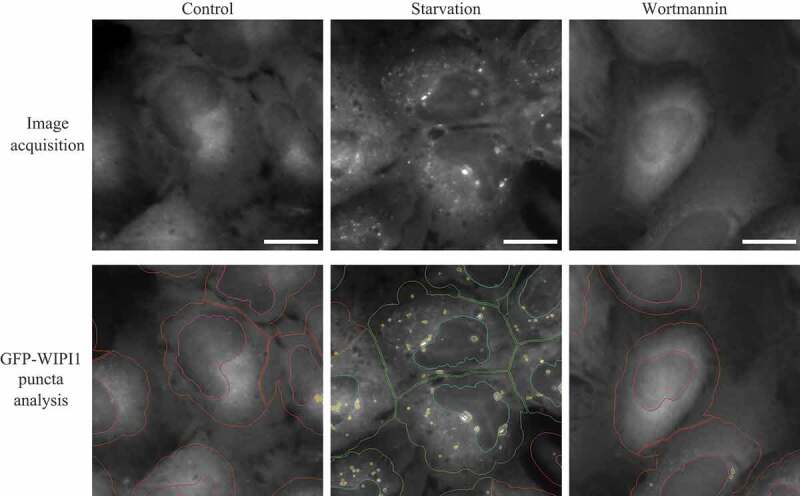
Automated WIPI1 puncta image acquisition and analysis monitors the induction and inhibition of autophagy. Stable U2OS clones expressing GFP-WIPI1 were selected using 0.6 μg/ml G418 and then cultured in 96-well plates. Cells were treated for 3 h with nutrient-rich medium (Control), nutrient-free medium (EBSS), or with 233 nM wortmannin. Cells were fixed in 3.7% paraformaldehyde and stained with DAPI (5 μg/ml in PBS). An automated imaging and analysis platform was used to determine the number of both GFP-WIPI1 puncta-positive cells and the number of GFP-WIPI1 puncta per individual cell [664]. Cells without GFP-WIPI1 puncta are highlighted in red (cell detection) and purple (nuclei detection), whereas GFP-WIPI1 puncta-positive cells are highlighted in yellow (GFP-WIPI1 puncta detection), green (cell detection) and blue (nuclei detection). Bars: 20 µm. Images provided by S. Pfisterer and T. Proikas-Cezanne
Cautionary notes: With regard to detection of the WIPI proteins, endogenous WIPI1 puncta cannot be detected in many cell types [837], and the level of transiently expressed GFP-WIPI1 puncta is cell context-dependent [837, 838]. However, this approach has been used in human and mouse cell systems [664, 838] and mCherry-Atg18 also works well for monitoring autophagy in transgenic Drosophila [183], although one caution with regard to the latter is that GFP-Atg18 expression enhances Atg8 lipidation in the fat body of fed larvae. GFP-WIPI1 and GFP-WIPI2 have been detected on the completed (mature) autophagosome by freeze-fracture analysis [144], but endogenous WIPI2 has not been detected on mRFP-LC3- or LAMP2-positive autophagosomes or autolysosomes using immunolabeling [837]. Accordingly, it may be possible to follow the formation and subsequent disappearance of WIPI puncta to monitor autophagy induction and flux using specific techniques. As with GFP-LC3, overexpression of WIPI1 or WIPI2 can lead to the formation of aggregates, which are stable in the presence of PtdIns3K inhibitors.
f. BECN1/Vps30/Atg6
BECN1 (yeast Vps30/Atg6) and PIK3C3/VPS34 are essential partners in the autophagy interactome that signals the onset of autophagy [796, 846, 847], and many researchers use this protein as a way to monitor autophagy. Binding to the anti-apoptotic protein BCL2 inhibits BECN1 [848]. BECN1 also binds other anti-apoptotic BCL2-family members via its putative BH3 domain [849, 850]. Autophagy is induced by the release of BECN1 from BCL2 by pro-apoptotic BH3 proteins, phosphorylation of BECN1 by DAPK1 and DAPK2 (at Thr119, located in the BH3 domain) [851, 852], or phosphorylation of BCL2 by MAPK8/JNK1 (at Thr69, Ser70 and Ser87) [853, 854]. Release of BECN1 can also be achieved by the expression of the F121A mutant, which leads to enhanced basal autophagy in vivo [855]. The relationship between BECN1 and BCL2 is more complex in developing cerebellar neurons, as it appears that the cellular levels of BCL2 are, in turn, post-translationally regulated by an autophagic mechanism linked to a switch from immaturity to maturity [856, 857]. It is important to be aware, however, that certain forms of autophagy are induced in a BECN1-independent manner and are not blocked by PtdIns3K inhibitors [118, 858, 859]. Interestingly, caspase-mediated cleavage of BECN1 inactivates BECN1-induced autophagy and enhances apoptosis in several cell types [860], emphasizing that the crosstalk between apoptosis and autophagy is complex.
Although a population of BECN1 may localize in proximity to the trans-Golgi network [861], it is also present at the ER and mitochondria [848]. In keeping with these observations, in cerebellar organotypic cultures BECN1 co-immunoprecipitates with BCL2 that is primarily localized at the mitochondria and ER; and in a mouse model of neurodegeneration, autophagic vacuoles in Purkinje neurons contain partially digested organelles that are immunoreactive for BCL2 [856, 862]. In addition, as BECN1-PIK3C3/VPS34 are the major source of cellular PtdIns3P lipids and can be present in multiple complexes that act during endosome maturation in addition to autophagy [863], caution must be exercised when monitoring localization. On induction of autophagy by various stimuli, the presence of BECN1- and PIK3C3/VPS34-positive macroaggregates can be detected in the region of the Golgi complex by immunofluorescence [207, 864]. Thus, BECN1-GFP puncta detected by fluorescence microscopy or TEM may serve as an additional marker for autophagy induction [865]; however, it should be noted that caspase cleavage of BECN1 can be detected in normal culture conditions (S. Luo, personal communication), and cleaved BECN1 is translocated into the nucleus [866]. Thus, care needs to be taken with these assays under stress conditions in which more pronounced BECN1 cleavage occurs. In addition, as with any GFP chimeras there is a concern that the GFP moiety interferes with correct localization of BECN1.
To demonstrate that BECN1 or PtdIns3K macroaggregates are an indirect indication of ongoing autophagy, it is mandatory to show their specific association with the process by including appropriate controls with inhibitors or preferably by autophagy gene silencing. When a BECN1-independent autophagy pathway is induced, such aggregates are not formed regardless of the fact that the cell expresses BECN1 (e.g., as assessed by western blotting; C. Isidoro, personal communication). As BECN1-associated PtdIns3K activity is crucial in autophagosome formation in BECN1-dependent autophagy, the measurement of PtdInsk3K in vitro lipid kinase activity in BECN1 immunoprecipitates can be a useful technique to monitor the functional activity of this complex during autophagy modulation [774, 775, 867]. It is important to note that an in vitro lipid kinase assay with BECN1 immunoprecipitates represents the total PtdIns3K activity and does not make it possible to distinguish between the production of PtdIns3P by PIK3C3/VPS34 in complex I versus that in complex II. Therefore, the most accurate measure of complex-specific activity of the class 3 PtdIns3K would be an in vitro lipid kinase assay using ATG14 and UVRAG immunoprecipitates [868, 869].
g. STX17
STX17 is a SNARE protein implicated in autophagosome-endolysosome fusion in cooperation with SNAP29 and VAMP8 [795, 870]. STX17 was initially reported to be recruited to completely sealed autophagosomes, but not to phagophores [871–875]. STX17 as a competence factor may be recruited just prior to fusion of autophagosomes with lysosomes, and not all autophagosomes are positive for this protein. However, later studies demonstrate that upon starvation STX17 colocalizes with the omegasome marker ZFYVE1/DFCP1 [876, 877], consistent with the view that STX17 is also implicated in autophagosome formation in starvation-induced autophagy [872, 873, 877] and mitophagy [878, 879]. In fed cells, STX17 principally localizes to the ER, mitochondria-associated ER membranes (MAMs), and mitochondria [871–873]. Some STX17 is phosphorylated by TBK1 at Ser202, and the phosphorylated form localizes to the Golgi apparatus [877]. STX17 also has a critical role in mediating the retrograde transport of autophagosomes upon their fusion with late endosome (LEs) in distal neuronal axons [880, 881]. Neurons are highly polarized cells with long axons, and thus face special challenges to transport AVs toward the soma where mature lysosomes are relatively enriched. LE-loaded dynein-SNAPIN motor-adaptor complexes are recruited to AVs upon STX17-mediated LE-AV fusion. This motor sharing ride-on service enables neurons to maintain effective autophagic clearance in the soma, thus reducing autophagic stress in axons.
h. TECPR1
TECPR1 binds ATG5 through an AFIM (ATG5 [five] interacting motif). TECPR1 competes with ATG16L1 for binding to ATG5, suggesting that there is a transition from the ATG5-ATG16L1 complex that is involved in phagophore expansion to an ATG5-TECPR1 complex that plays a role in autophagosome-lysosome fusion [882]. TECPR1 thus marks lysosomes and autolysosomes [883].
i. ZFYVE1/DFCP1
ZFYVE1 binds PtdIns3P that localizes to the ER and Golgi. Starvation induces the translocation of ZFYVE1 to punctate structures on the ER; the ER population of ZFYVE1 marks the site of omegasome formation [884]. ZFYVE1 partially colocalizes with WIPI1 upon nutrient starvation [842] and also with WIPI2 [837].
Conclusion: Components of the autophagic machinery other than Atg8-family proteins can be monitored to follow autophagy, and these can be important tools to define specific steps of the process. For example, WIPI puncta formation can be used to monitor autophagy, but, similar to Atg8-family proteins, should be examined in the presence and absence of lysosomal inhibitors. Analysis of WIPI puncta should be combined with other assays because individual members of the WIPI family might also participate in additional, uncharacterized functions apart from their role in autophagy. At present, we caution against the use of changes in BECN1 localization as a marker of autophagy induction, given its other cellular roles. It is also worth considering the use of different markers depending on the specific autophagic stimuli.
7. Sphingolipids
Sphingolipids are ubiquitous membrane lipids that can be produced in a de novo manner in the ER and Golgi apparatus or by cleavage involving phosphodiesterases (sphingomyelinases), hydrolases (glycosphingolipid glycosidases), sphingolipid ceramide N-deacylase (SCDase), phosphatases (acting on sphingosine-1-phosphate [S1P] and ceramide-1-phosphate) or lyases (e.g., SGPL1 [sphingosine-1-phosphate lyase 1]) [885–887]. For instance, SGPL1 is a ubiquitiously expressed enzyme having a wide-range of functions in different cellular processes, including proliferation, motility and death. Moreover, SGPL1 is a critical determinant for the degredation of the sphingolipid S1P. The S1P pathway is a crucial mechanism for neuronal autophagy by providing PE for LC3 conjugation [888, 889]. Ablation and deletion of Sgpl1 result in reduced autophagic activity in mouse brain [888]. Likewise, mutations in SGPL1 and alterations in neuronal autophagy lead to severe neurodevelopmental phenotypes ranging from fetal hydrops to congenital brain malformations and neuropathies in humans [890]. The multiple different metabolites of the sphingolipid pathway, which are distinct by even a single double bond, carbon chain length of the fatty acid, or presence of a phosphate group, can have quite varied cellular functions. Sphingolipids were first recognized for their role in the architecture of membrane bilayers affecting parameters such as bilayer stiffness, neighboring lipid order parameter and microdomain/raft formation. They also act as second messengers in vital cellular signaling pathways and as key determinants of cellular homeostasis in what is called a sphingolipid rheostat [891]. Sphingolipids participate in the formation of different membrane structures and subcellular organelles, such as mitochondria and ER, and are also involved in the fusion and biophysical properties of cell membranes [892]. Moreover, they are constitutive components of MAMs, subdomains of the ER that interact with mitochondria [893].
Ceramides, positioned at the core of sphingolipid metabolism, play several roles that affect multiple steps of autophagy, by inhibition of nutrient transporters [894], by modulation of BCL2-BECN1 association at the level of AKT signaling [895], and by regulation of mitophagy [896]. The latter function is regulated by a particular ceramide species, stearoyl (C18:0)-ceramide, a sphingolipid generated by CERS1 (ceramide synthase 1). C18-ceramide, in association with LC3-II, targets damaged mitochondria for phagophore sequestration in response to ceramide stress, leading to tumor suppression [896–900]. The binding of ceramide to LC3-II can be detected using anti-ceramide and anti-LC3 antibodies by immunofluorescence and confocal microscopy, co-immunoprecipitation using anti-LC3 antibody followed by liquid chromatography-tandem MS, using appropriate standards (targeted lipidomics), or labeling cells with biotin-sphingosine to generate biotin-ceramide, and immunoprecipitation using avidin columns followed by western blotting to detect LC3-II. It should be noted that inhibitors of ceramide generation, mutants of LC3 with altered ceramide binding (F52A or I35A), and/or that are conjugation defective (e.g., G120A), should be used as negative controls [901]. The generation of C18-ceramide in the outer mitochondrial membrane, which recruits LC3-containing phagophores for mitophagy induction, is regulated through the trafficking of the metabolic enzyme CERS1 by RPL29P31/p17/PERMIT [901]. shRNA-mediated knockdown or deletion of this gene prevent mitophagy in response to cellular stress both in cultured cells and in knockout mice [901]. Thus, colocalization of CERS1 or RPL29P31/p17/PERMIT with TOMM20 using immunofluorescence can also be used to detect mitophagy signals in response to acute or chronic stress in situ and in vivo.
Other sphingolipids are also involved in autophagy. For example, accumulation of endogenous sphingosine-1-phosphate, a pro-survival downstream metabolite from ceramide triggers ER-stress associated autophagy, by activation of AKT [902], excess sphingomyelin inhibits phagophore closure by disturbing the trafficking of ATG9A [812], and SMPD1/acid sphingomyelinase inhibits autophagy through the activation of the MTOR pathway [903], whereas it is required for LC3-associated phagocytosis [904]. Likewise, dihydroceramides, the penultimate metabolite of ceramide biosynthesis have been implicated in the regulation of autophagy [905]. Specifically, changes in the levels of C16:0 and C18:0 dihydroceramides cause the destabilization of autolyososomal membranes thereby leading to the induction of autophagy-associated cell death [906]. In addition, gangliosides, have been implicated in autolysosome morphogenesis [907]. Moreover, a molecular interaction of the ganglioside GD3 with core-initiator proteins of autophagy, such as AMBRA1 and WIPI1, is revealed within lipid microdomains in MAMs, indicating that MAM raft-like microdomains can play a role in the initial organelle scrambling activity that finally leads to the formation of the autophagosome [908].
To analyze the role of gangliosides in autophagy, two main technical approaches can be used: co-immunoprecipitation and Förster resonance energy transfer. For the first method, lysates from untreated or autophagy-induced cells have to be immunoprecipitated with an anti-LC3 polyclonal antibody (a rabbit IgG isotypic antibody should be used as a negative control). The obtained immunoprecipitates are subjected to ganglioside extraction, and the extracts run on an HPTLC aluminum-backed silica gel and analyzed for the presence of specific gangliosides by using monoclonal antibodies. Alternatively, the use of FRET by flow cytometry appears to be highly sensitive to small changes in distance between two molecules, and is thus suitable to study molecular interactions, for example, between ganglioside and LC3. Furthermore, FRET requires ~10 times less biological material than immunoprecipitation.
Conclusion: Sphingolipids are bioactive molecules that play key roles in the regulation of autophagy at various stages, including upstream signal transduction pathways to regulate autophagy via transcriptional and/or translational mechanisms, autolysosome morphogenesis, and/or targeting phagophores to mitochondria for degradation via sphingolipid-LC3 association [276, 897, 899–901, 909].
8. Transcriptional and translational regulation
The induction of autophagy in certain scenarios is accompanied by an increase in the mRNA levels of certain autophagy-related genes, such as ATG1 [910], ATG6 [911], ATG7 [912, 913], ATG8/Lc3 [64, 473, 914–916], GABARAPL1 [473, 916], ATG9 [917], Atg12 [918], ATG13 [473, 916], Atg14 [919], ATG29 [910], WIPI1 [473, 916], and SQSTM1 [64], and an autophagy-dedicated microarray was developed as a high-throughput tool to simultaneously monitor the transcriptional regulation of all genes involved in, and related to, autophagy [920]. The mammalian gene that shows the greatest transcriptional regulation in the liver (in response to starvation and circadian signals) is Ulk1, but others also show more limited changes in mRNA levels including Gabarapl1, Bnip3 and, to a minor extent, Lc3b [921]. In skin cancer and HeLa cells ULK1 and ULK2 expression is negatively regulated at the transcriptional level by the chromatin non-histone protein HMGA1 (high mobility group AT-hook 1) [922]. In several mouse and human cancer cell lines, ER stress and hypoxia increase the transcription of Lc3/LC3, GABARAPL1, Atg5/ATG5, Atg12/ATG12, ATG13, and WIPI1 by a mechanism involving the unfolded protein response (UPR). Similarly, a stimulus-dependent increase in LC3B expression is detected in neural stem cells undergoing autophagy induction [923]. The ATG9A promoter, similar to those of BNIP3 and BNIP3L, but in contrast to other ATG family members such as ATG5 and ATG7, contains HIF1A-responsive elements and is transcriptionally activated in hypoxic glioblastoma cells [209]. Increased expression of Atg5 in vivo after optic nerve axotomy in mice [924] and increased expression of Atg7, Becn1 and Lc3a during neurogenesis at different embryonic stages in the mouse olfactory bulb are also seen [925]. LC3 and ATG5 are not required for the initiation of autophagy, but mediate phagophore expansion and autophagosome formation. In this regard, the transcriptional induction of LC3 may be necessary to replenish the LC3 protein that is turned over during extensive ER stress- and hypoxia-induced autophagy [918, 926]. Of note, however, a recent study showed that although tunicamycin-induced ER stress activates autophagy and triggers a strong transcriptional increase in LC3 mRNA and protein levels (via ATF4), depletion of LC3 does not reduce ER stress-induced autophagy [473].
In the clinical setting, tissue expression of ATG5, LC3A and LC3B and their respective proteins accompanies elevated autophagy flux in human adipose tissue in obesity [292, 927]. Thus, assessing the mRNA levels of LC3 and other autophagy-related genes by northern blot or reverse transcriptase polymerase chain reaction (RT-PCR) may provide correlative data relating to the induction of autophagy; in addition, proteomic profiling of de novo protein synthesis in starvation-induced autophagy using bioorthogonal noncanonical amino acid tagging can further validate the function of the corresponding proteins in autophagy induction [928]. However, a time course may be necessary to obtain accurate information because mRNA levels are likely to change substantially over time. In addition, mRNA may be sequestered in P-bodies, resulting in suppression of protein translation and time-dependent loss of autophagy-related proteins, as was shown for AMBRA1 and BECN1 in cells exposed to the hypoxia mimetic CoCl2 [929]. Downregulation of autophagy-related mRNAs has been observed in human islets under conditions of lipotoxicity [571] that impair autophagic flux [930]. It is not clear if these changes are sufficient to regulate autophagy, however, and therefore these are not direct measurements.
Several transcription factors of the nuclear receptor superfamily modulate expression of genes related to autophagy. For instance, NR1D1/Rev-erbα modulates autophagy-associated genes in a tissue-specific manner. Whereas NR1D1 represses Ulk1, Bnip3, Atg5, Becn1 and Prkn/park2/parkin gene expression in mouse skeletal muscle [931] as well as ulk1a and atp6v1d in zebrafish larvae [932] by directly binding to regulatory regions in their DNA sequences, STRA8 suppresses autophagy and at the same time transcriptionally represses Nr1d1 and, thereby, inhibits the expression of Ulk1 in mouse testis [933]. NR1D1 upregulates Ulk1 by direct engagement of distal RAR-related orphan receptor DNA elements as evaluated in stra8−/− nr1d1−/− double-knockout mice. Moreover, in human macrophages, NR1D1 promotes lysosome biogenesis and autophagy, contributing to its antimicrobial properties against M. tuberculosis [934]. Whereas NR1D1 represses autophagic flux in skeletal muscles, it upregulates the expression of autophagy- and lysosome-associated genes in mouse testis and human macrophages. Furthermore, NR1D1 induces mitochondrial biogenesis in skeletal muscles, leads to improved oxidative capacity of cells, and induces lysosome biogenesis in human macrophages, augmenting antimicrobial properties.
The nuclear receptors PPARA and NR1H4/FXR also regulate hepatic autophagy in mice. Indeed, PPARA and NR1H4 compete for the control of hepatic lipophagy in response to fasting and feeding nutritional cues, respectively [921]. In addition, activation of PPARA-mediated autophagy and the lysosomal pathway in the nervous system, contributes to Aβ clearance, and thus reduces Alzheimer disease (AD)-like pathology and cognitive decline in a mouse model [935]. NR1H4 may also inhibit autophagy via inhibition of CREB-CRTC2 complex assembly [936]. Consistent with in vitro studies utilizing human cancer cell lines [937, 938], in human adipose tissue explants, E2F1 binds the LC3B promoter, in association with increased expression of several autophagy genes and elevated adipose tissue autophagic flux [292, 927]. In this instance, classical promoter analysis studies, including chromatin immunoprecipitation and ATG promoter-luciferase constructs, provide insights into the putative transcriptional regulation of autophagy genes by demonstrating promoter binding in situ, and promoter activity in vitro [927].
Of note, large changes in Atg gene transcription just prior to Drosophila salivary gland cell death (that is accompanied by an increase in autophagy) are detected for Atg2, Atg4, Atg5 and Atg7, whereas there is no significant change in Atg8a or Atg8b mRNA [939, 940]. Autophagy is critical for Drosophila midgut cell death, which is accompanied by transcriptional upregulation of all of the Atg genes tested, including Atg8a (Figure 25) [375, 941]. Similarly, in the silkworm (Bombyx mori) larval midgut [942], fat body [943] and silk gland [944] the occurrence of autophagy is accompanied by an upregulation of the mRNA levels of several Atg genes. Transcriptional upregulation of Drosophila Atg8a and Atg8b is also observed in the fat body following induction of autophagy at the end of larval development [945], and these genes as well as Atg2, Atg9 and Atg18 show a more than 10-fold induction during starvation [946]. Atg5, Atg6, Atg8a and Atg18 are upregulated in the ovary of starved flies [947], and an increase in Drosophila Atg8b is observed in cultured Drosophila l(2)mbn cells following starvation (S. Gorski, personal communication). An upregulation of plant ATG8 may be needed during the adaptation to reproductive growth; a T-DNA inserted mutation of rice ATG8b blocked the change from vegetative growth to reproductive growth in both homozygous and heterozygous plant lines (M.-Y. Zhang, H. Budak, unpublished results).
Figure 25.
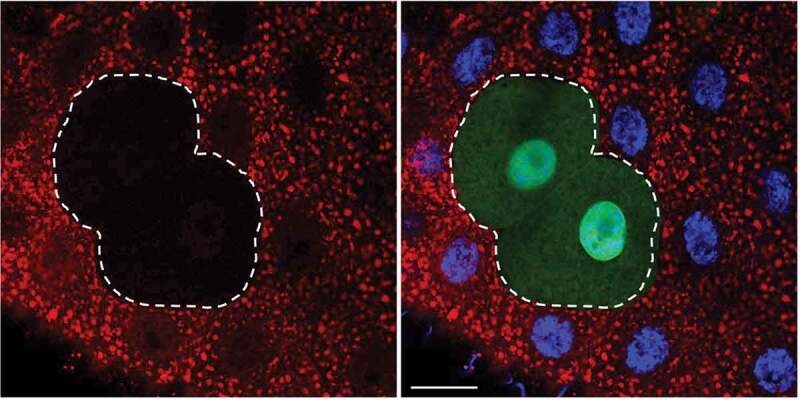
Clonal analysis of autophagy in the Drosophila larval midgut. Inhibition of autophagy in somatic clone cells marked by GFP (green, outlined) have decreased levels of mCherry-Atg8a puncta (red) compared to the control wild-type cells (non-GFP) with nuclei in blue (merged image, right panel). Bar: 20 µm. Image provided by D. Denton and S. Kumar
Similarly, the upregulation of autophagy-related and -associated genes (Atg4b, Atg12, Atg13, Bnip3, Gabarapl1, Lc3, WIPI1) has been documented at the transcriptional and translational level in several other species (e.g., C. elegans [948], mouse, rat, human [949], trout, Arabidopsis and maize) under conditions of ER stress [473, 918], and diverse types of prolonged (several days) catabolic situations including cancer cachexia, diabetes mellitus, uremia and fasting [290, 662, 950–953]. Along these lines, the mRNA levels of atg1, atg8a/b and sqstm1 increase in D. discoideum upon infection with the fish and frog pathogen Mycobacterium marinum [64], a close relative of M. tuberculosis. Similarly, ATG9 and ATG16L1 are transcriptionally upregulated upon influenza virus infection (H. Khalil, personal communication), and in C. elegans, the FOXA transcription factor PHA-4 and the TFEB (transcription factor EB) ortholog HLH-30 regulate the expression of several autophagy-related genes (see Methods and challenges of specialized topics/model systems. C. elegans) [573, 948]. Such prolonged induction of the expression of ATG genes has been thought to allow the replenishment of critical proteins (e.g., LC3 and GABARAP) that are destroyed during autophagosome fusion with the lysosome [954].
The polyamine spermidine increases life span and induces autophagy in cultured yeast and mammalian cells, as well as in nematodes, flies and mice. In aging yeast, spermidine treatment triggers epigenetic deacetylation of histone H3 through inhibition of histone acetyltransferases, leading to significant upregulation of various autophagy-related transcripts [955]. In mammalian cells, spermidine promotes autophagy flux by depleting cytosolic HDAC4 to enhance the acetylation and stability of MAP1S (microtubule-associated protein 1S) to prolong mouse lifespan and prevent liver fibrosis and hepatocellular carcinomas [956]; however, the functional relevance of autophagy for liver fibrosis and cancer is highly dependent on the cell type. Whereas autophagy maintains cellular homeostasis in hepatocytes, Kupffer cells (macrophages), and liver sinusoidal endothelial cells, thereby counteracting fibrogenesis in the liver, it is the prime process of providing energy for the activation of hepatic stellate cells, which leads to collagen production and fibrogenesis [957]. Spermidine also drives the hypusination of the translation factor EIF5A, which in turn controls the translation of TFEB to rejuvenate B cell immunity [958]. In addition, spermidine stimulates mitophagy in cardiomyocytes and prevents typical age-related cardiac deterioration in an autophagy-dependent manner [959]. IPMK, can alter histone H4 acetylation and influence gene expression of LC3B, BNIP3, BNIP3L, SQSTM1, GABARAPL1 and ATG12; loss of IPMK in liver prevents lipophagy and liver regeneration [621, 690].
In addition to spermidine, melatonin, a hormone present in both mammals and plants, plays a critical role in inducing the expression of ATG genes under heat stress in tomato [960]. Both foliar application of an optimal dose of melatonin and the overexpression of the ASMT (N-acetylserotonin O-methyltransferase) gene results in an upregulation of the expression of ATG genes and the formation of autophagosomes leading to the degradation of denatured proteins resulting from heat stress in tomato [960]. Under cadmium stress, HSFA1A (heat shock factor 1A) promotes the accumulation of melatonin through directly activating the transcription of COMT1 (caffeate O-methyltransferase 1), a key gene in melatonin biosynthesis [961], indicating that HSFA1A might mediate autophagy in response to stress in plants. Indeed, silencing of HSFA1A completely blocks drought stress-induced expression of ATG10 and ATG18F, whereas the expression of these genes is increased in HSFA1A-overexpressing plants [962]. An electrophoretic mobility shift assay and ChIP-qPCR analysis show that HSFA1A binds to the promoters of these two ATG genes and directly regulates their expression to trigger autophagy under drought stress in tomato plants [962]. Furthermore, BZR1 (brassinazole-resistant 1), a phytohormone brassinosteroid-activated transcription factor, induces the expression of ATG2 and ATG6 to form autophagosomes, which mediate the response to nitrogen starvation in tomato [963].
In addition to the ATG genes, transcriptional upregulation of VMP1 can be detected in mammalian cells subjected to rapamycin treatment or starvation, and in tissues undergoing disease-induced autophagy such as cancer [964]. VMP1 is an essential autophagy gene that is conserved from D. discoideum to mammals [422, 965], and the VMP1 protein regulates early steps of the autophagic pathway and is essential for correct functioning of membrane contact sites between the ER and other organelles including autophagosomes [841, 966]. VMP1 is poorly expressed in mammalian cells under nutrient-normal conditions, but is highly upregulated in cells undergoing autophagy, and the expression of VMP1 induces autophagosome formation. The GLI3 transcription factor is an effector of KRAS that regulates the expression and promoter activity of VMP1, using the histone acetyltransferase EP300/p300 as a co-activator [967].
A gene regulatory network, named CLEAR (coordinated lysosomal expression and regulation) that controls both lysosome and autophagosome biogenesis was identified using a systems-biology approach [949, 968, 969],635,636]. The basic helix-loop-helix transcription factor TFEB acts as a master gene of the CLEAR network and positively regulates the expression of both lysosomal and autophagy genes, thus linking the biogenesis of two distinct types of cellular compartments (i.e., autophagosomes and lysosomes) that cooperate in the autophagic pathway. TFEB activity is regulated by starvation and is controlled by both MAPK1/ERK2-, MTOR-, and AKT-mediated phosphorylation at specific serine residues [949, 970–973]; thus, it can serve as a new tool for monitoring transcriptional regulation connected with autophagy. TFEB is phosphorylated by MTORC1 on the lysosomal surface, preventing its nuclear translocation. A lysosome-to-nucleus signaling mechanism transcriptionally regulates autophagy and lysosomal biogenesis via MTOR and TFEB [971]. TFEB phosphorylation on specific residues also occurs in the nuclear compartment and enables TFEB nuclear export [974–976]. Thus, TFEB activity is tightly regulated by different phosphorylation events that control TFEB nuclear import and export rates. Therefore, a very useful readout of endogenous TFEB activity is the evaluation of TFEB subcellular localization, as activation of TFEB correlates with its relocalization from the cytoplasm to the nucleus. This shift can be monitored by immunofluorescence using antibodies against TFEB. TFEB localization may also be studied to monitor MTOR activity, as in most cases TFEB nuclear localization correlates with inhibition of MTOR. However, due to the low expression levels of TFEB in most cells and tissues, it may be difficult to visualize the endogenous protein. Thus, a TFEB nuclear translocation assay was developed in a HeLa cell line stably transfected with TFEB-GFP. This fluorescence assay can be used to identify the conditions and factors that promote TFEB activation [971]. TFE3 and MITF, two other members of the MiT/TFE family of transcription factors, in some cases can compensate for TFEB and are regulated in a similar manner [973, 977, 978]. In response to histone deacetylase inhibitors, TFEB acetylation exerts an important function in control of its transcriptional activity and lysosomal function [979]. Finally, an AMPK-SKP2-CARM1 signaling cascade has also been reported to play a role in transcriptional regulation of autophagy [980]; CARM1 exerts a transcriptional coactivator function on autophagy and lysosomal genes through TFEB.
Similar to TFEB, the erythroid transcription factor GATA1 and its coregulator ZFPM1/FOG1 as well as the myeloid master regulator SPI1/PU.1 induce the transcription of multiple genes encoding autophagy components. This developmentally regulated transcriptional response is coupled to increases in autophagosome number as well as the percent of cells that contain autophagosomes [981–983]. FOXO transcription factors, especially FOXO1 and FOXO3, also play critical roles in the regulation of autophagy gene expression [662, 919, 984]. A zinc finger family DNA-binding protein, ZKSCAN3, is a master transcriptional repressor of autophagy and lysosome biogenesis; starvation and MTOR inhibition with torin1 induce nucleus-to-cytoplasm translocation of ZKSCAN3 [985]. The expression of the transcription factor EGR1 (early growth response 1) is rapidly increased upon nutrient deprivation and can directly increase transcription of multiple components of the autophagy machinery. The EGR1 DNA-binding motif is significantly enriched in the promoters/enhancers of autophagy-associated genes; EGR1 positively regulates the transcription of these genes (including ATG2A, ATG14, ATG3, ATG13, ATG101, LC3B, PIK3C3, PPM1D, ULK1, and ZFYVE1), and thereby increases the autophagic flux [986]. Transcription factor NFE2L2/NRF2, considered as the master regulator of cellular homeostasis, modulates the expression of autophagy-related genes, including the already mentioned Sqstm1 but also Atg2b, Atg4d, Atg5, Atg7, Calcoco2/Ndp52, Gabarapl1 and Ulk1 [987]. Moreover, NFE2L2/NRF2 is a regulator of Lamp2a transcription, and therefore, it controls CMA [988]. This transcription factor may have a relevant role upon stressful conditions, including proteotoxic or oxidative insults. Finally, CEBPB/C/EBPβ is a transcription factor that regulates autophagy in response to the circadian cycle in mice [989] and zebrafish [932].
Although less work has been done on post-transcriptional regulation, several studies implicate microRNAs in controlling the expression of proteins associated with autophagy [332–334, 990–993]. In this context, an important player is represented by MIR27A. Autophagy implementation is linked to ATP and HMGB1 release and ecto-CALR (calreticulin) exposure in HCT116 colon cancer cells with knockdown of MIR27A. This pathway is active in basal conditions, as indicated by the presence of the mature LC3-II form and acquisition of autophagic morphological features (large bodies, multiple or multilobated nuclei, cytosolic vacuoles and granules) when compared to control and MIR27A-overexpressing HCT116 cells. Methotrexate treatment triggers autophagy in time-course experiments, as the mature LC3-II form rapidly increases following MIR27A knockdown, whereas the change is limited in control and MIR27A-overexpressing HCT116 cells. Treatment with the lysosomotropic agent CQ confirms that the higher LC3-II levels reveal an augmented autophagic flux leading to autophagosome development. The mature LC3-II form shows a remarkable dose-dependent increase upon MIR27A knockdown with respect to control and especially MIR27A-overexpressing HCT116 cells [994].
Cautionary notes: Most of the ATG genes do not show significant changes in mRNA levels when autophagy is induced. Even increases in LC3 mRNA can be quite modest and are cell type- and organism-dependent [995]. In addition, it is generally better to follow protein levels, which, ultimately, are the significant parameter with regard to the initiation and completion of autophagy. However, ATG protein amounts do not always change significantly, and the extent of increase is again cell type- and tissue-dependent. Finally, changes in autophagy protein levels are not sufficient evidence of autophagy induction and must be accompanied by additional assays as described herein. Thus, monitoring changes in mRNA levels for either ATG genes or autophagy regulators may provide some evidence supporting upregulation of the potential to undergo autophagy, but should be used along with other methods.
Another general caution pertains to the fact that in any cell culture system mixed populations of cells (for example, those undergoing autophagy or not) exist simultaneously. Therefore, only an average level of protein or mRNA expression can be evaluated with most methods. This means that the results regarding specific changes in autophagic cells could be hidden due to the background of the average data. Along these lines, experiments using single-cell qPCR to examine gene expression in individual cardiomyocytes with and without signs of autophagy reveal that the transcription of MTOR markedly and significantly increases in autophagic cells in intact cultures (spontaneously undergoing autophagy) as well as in cultures treated with proteasome inhibitors to induce autophagy (V. Dosenko, personal communication). Finally, researchers need to realize that mammalian cell lines may have mutations that alter autophagy signaling or execution; this problem can be avoided by using primary cells.
Conclusion: Although there are changes in ATG gene expression that coincide with, and may be needed for, autophagy, in most cases this has not been carefully studied experimentally. Therefore, at the present time we do not recommend the monitoring of ATG gene transcription as a general readout for autophagy unless there is clear documentation that the change(s) correlates with autophagy activity.
9. Posttranslational modifications
Autophagy is controlled by posttranslational modification (PTM) of ATG proteins such as phosphorylation, ubiquitination, acetylation, O-GlcNAcylation, N6-methyladenosine modification, oxidation and cleavage, which can be monitored to analyze the status of the process [293, 611, 767, 775, 996–1000]. The global deacetylation of proteins, which often accompanies autophagy, can be conveniently measured by quantitative immunofluorescence and western blotting with antibodies specifically recognizing acetylated lysine residues [1001]. Indeed, depletion of the nutrient supply causes autophagy in yeast or mammalian cells by reducing the nucleo-cytosolic pool of acetyl-coenzyme A, which provides acetyl groups to acetyltransferases, thus reducing the acetylation level of hundreds of cytoplasmic and nuclear proteins [1002]. A global deacetylation of cellular proteins is also observed in response to so-called “caloric restriction mimetics”, that is, a class of pharmacological agents that deplete the nucleo-cytosolic pool of acetyl-coenzyme A, inhibit acetyltransferases (such as EP300) or activate deacetylases (such as SIRT1). All these agents reduce protein acetylation levels in cells as they induce autophagy [1003]. One prominent ATG protein that is subjected to pro-autophagic deacetylation is LC3 [1004, 1005]. Moreover, SIRT1 inhibition by EX-527 decreases the lipidation of LC3 [1006]. Recently, ULK1 O-GlcNAcylation was shown to be crucial for autophagy initiation [1007, 1008]; this modification potentiatiates AMPK-dependent phosphorylation of ULK1 and allows binding to and phosphorylation of ATG14, and subsequent activation of PIK3C3/VPS34.
Another mechanism through which autophagy-related proteins are regulated is by means of S-nitrosylation, the covalent binding of nitric oxide (NO) to specific cysteine residues [1009]. High levels of free NO have been linked to an overall inhibitory effect on autophagic machinery [1010]. Conversely, the modulation of the amount of S-nitrosylated proteins triggered by changes in the activity or expression of the denitrosylase ADH5/GSNOR (alcohol dehydrogenase 5 [class III], chi polypeptide), seems to have no major effects on nonselective autophagy, whereas there is an effect on the recognition of damaged mitochondria to be targeted for selective mitophagy [1011, 1012]. Persulfidation (S-sulfhydration) plays an important role in mitophagy-related proteins such as PRKN, whose catalytic activity is stimulated by persulfidation, whereas nitrosylation inactivates it [1013]. Mitophagy is also promoted by persulfidation of USP8 (ubiquitin specific peptidase 8), which enhances deubiquitination of PRKN [1014]. Other important autophagy-related proteins such as ATG3, ATG5, ATG7 and ATG18A in plants are also targets for persulfidation, but the role of this modification needs further clarification [1015].
Phosphorylation of other autophagic proteins plays a critical role in the regulation of autophagy activity. For example, CSNK2 (casein kinase 2) and ULK1 induce phosphorylation of SQSTM1 at serine 403 and serine 409, respectively, increasing the binding affinity of SQSTM1 for ubiquitin, and enhancing the autophagic degradation of ubiquitinated proteins [427, 714]. Also, EGFR signaling induces multi-site tyrosine phosphorylation of BECN1 to inhibit core autophagy machinery activation [775].
Finally, N6-methyladenosine (m6A) mRNA modification plays an important role in regulating autophagy. ULK1 mRNA undergoes m6A modification in the 3ʹ UTR, and the m6A-marked ULK1 transcripts can further be targeted for degradation by YTHDF2 (YTH N6-methyladenosine RNA binding protein 2). Moreover, FTO (FTO alpha-ketoglutarate dependent dioxygenase) reverses the m6A mRNA modification of ULK1 transcripts, thereby promoting the initiation of autophagy [1016].
10. Autophagic protein degradation
Protein degradation assays represent a well-established methodology for measuring autophagic flux, and they allow good quantification. The general strategy is first to label cellular proteins by incorporation of a radioactive amino acid (e.g., [14C]- or [3H]-leucine, [14C]-valine or [35S]-methionine; although valine may be preferred over leucine due to the strong inhibitory effects of the latter on autophagy), preferably for a period sufficient to achieve labeling of the long-lived proteins that best represent autophagic substrates, and then to follow this with a long cold-chase so that the assay starts well after labeled short-lived proteins are degraded (which occurs predominantly via the proteasome). Next, the time-dependent release of acid-soluble radioactivity from the labeled protein in intact cells or perfused organs is measured [4, 25, 1017]. Note that the inclusion of the appropriate unlabeled amino acid (i.e., valine, leucine or methionine) in the starvation medium at a concentration equivalent to that of other amino acids in the chase medium is necessary; otherwise, the released [14C]-amino acid is effectively re-incorporated into cellular proteins, which results in a significant underestimation of protein degradation. A newer method of quantifying autophagic protein degradation is based on L-azidohomoalanine (AHA) labeling [1018, 1019]. When added to cultured cells, L-azidohomoalanine is incorporated into proteins during active protein synthesis. After a click reaction between an azide and an alkyne, the azide-containing proteins can be detected with an alkyne-tagged fluorescent dye, coupled with flow cytometry. The turnover of specific proteins can also be measured in a pulse-chase regimen using the Tet-ON/OFF or GeneSwitch systems and subsequent western blot analysis [1020–1022].
In this type of assay a considerable fraction of the measured degradation will be nonautophagic, and thus it is important to also measure, in parallel, cell samples treated with autophagy-suppressive concentrations of 3-MA, SAR-405, bafilomycin A1, CQ, ammonia, or amino acids, or generated under conditions of amino acid depletion, or in samples obtained from mutants missing central ATG components; these values are then subtracted from the total readouts. The complementary approach of using compounds that block other degradative pathways, such as proteasome and ER-associated degradation (ERAD) inhibitors, can also provide valuable information [216]. However, these inhibitors may sometimes cause unexpected results and should be interpreted with caution due to potential nonspecific effects and crosstalk among the degradative systems. For example, blocking proteasome function may activate autophagy [613, 1023–1026], although those studies did not assess long-lived protein degradation. Studies that have directly compared the effects of proteasomal and lysosomal degradation inhibitors—alone and in combination—on long-lived protein degradation have demonstrated that proteasomal and lysosomal inhibitors have near perfectly additive effects [216, 1027], thus suggesting that the crosstalk between the proteasomal and autophagic systems does not appreciably affect the results obtained in the long-lived protein degradation assay (although this does not exclude the possibility that this may occur under other conditions, so this needs to be tested from case to case). Conversely, interference with the CMA pathway does seem to activate a compensatory form of autophagy that increases the overall degradation of long-lived proteins [133, 307]. In general, when using inhibitors, it is critical to know whether the inhibitors being used alter autophagy in the particular cell type and context being examined. In addition, because 3-MA could have some autophagy-independent effects in particular settings it is advisable to verify that the 3-MA-sensitive degradation is also sensitive to specific class III PtdIns3K inhibitors such as SAR-405, and to general lysosomal inhibitors (such as NH4Cl or leupeptin) [25, 216].
The use of stable isotopes, such as 13C and 15N, in quantitative MS-based proteomics allows the recording of degradation rates of thousands of proteins simultaneously. These assays may be applied to autophagy-related questions enabling researchers to investigate differential effects in global protein or even organelle degradation studies [1028, 1029]. Stable isotope labeling with amino acids in cell culture (SILAC) can also provide comparative information between different treatment conditions, or between a wild type and mutant.
Another assay that could be considered relies on the limited proteolysis of a BHMT (betaine–homocysteine S-methyltransferase) fusion protein. The 44-kDa full-length BHMT protein is cleaved in hepatocyte amphisomes in the presence of leupeptin to generate 32-kDa and 10-kDa fragments [1030–1033]. Accumulation of these fragments is time dependent and is blocked by treatment with autophagy inhibitors. A modified version of this marker, GST-BHMT, can be expressed in other cell lines where it behaves similar to the wild-type protein [1034]. Additional substrates may be considered for similar types of assays. For example, the neomycin phosphotransferase II-GFP (NeoR-GFP) fusion protein is a target of autophagy [620]. Transfection of lymphoblastoid cells with a plasmid encoding NeoR-GFP followed by incubation in the presence of 3-MA leads to an accumulation of the NeoR-GFP protein as measured by flow cytometry [1035].
A similar western blot assay is based on the degradation of a cytosolic protein fused to GFP. This method has been used in yeast and D. discoideum cells using GFP-Pgk1 and GFP-Tkt-1 (phosphoglycerate kinase and transketolase, respectively). In this case the relative amount of free GFP and the complete fusion protein is the relevant parameter for quantification; although it may not be possible to detect clear changes in the amount of the full-length chimera, especially under conditions of limited flux [40, 52]. As described above for the marker GFP-Atg8-family proteins, nonsaturating levels of lysosomal inhibitors are also needed in D. discoideum cells to slow down the autophagic degradation, allowing the accumulation and detection of free GFP. It should be noted that this method monitors bulk autophagy because it relies on the passive transit of a cytoplasmic marker to the lysosome. Consequently, it is important to determine that the marker is distributed homogeneously in the cytoplasm.
Recently, the fluorescent coral protein Keima, which is resistant to lysosomal degradation, and which can be used to measure autophagic cargo flux to acidic environments [1036] has been fused (through genetic engineering) to a variety of cellular proteins, for example ribosomal, proteasomal, mitochondrial, or cytosolic proteins [1037]. These fusion proteins are proteolytically cleaved off from Keima and degraded (whereas Keima is stable). The cleavage can be detected by western blotting for Keima, where an increase in non-fused Keima reflects delivery of the fusion proteins to lysosomes. Thus, this approach represents a very versatile method to determine delivery of various cargo for lysosomal proteolysis and thereby monitor both nonselective and selective autophagy [1037]. Generation of stable cell lines with inducible expression of the Keima fusion proteins may provide a more reliable result under certain conditions. For example, during oxidative stress the expression of the Keima fusion proteins themselves seem to be increased, possibly due to stress-induced activation of the CMV promoter. More reliable data are produced, especially for the high-turnover probe Keima-LC3, by inducing expression of the Keima-probe prior to the stimulus of interest, and then following the generation of cleaved Keima during a chase period (M. Torgersen, unpublished results).
Of note, however, the assay only assesses proteolytic activity, and cannot be used to tell whether the cargo has reached fully active autolysosomes or whether the degraded cargo is recycled to the cytosol. This is as opposed to the long-lived protein degradation assay, which is a true end-point measurement of autophagy, because it (with the inclusion of proper controls) can measure the amount of degraded, free amino acids (and short peptides) that have been released from the autolysosomes.
One of the most useful methods for monitoring autophagy in S. cerevisiae is the Pho8∆60 assay. PHO8 encodes a vacuolar phosphatase, which is synthesized as a zymogen before finally being transported to and activated in the vacuole [1038]. A molecular genetic modification that eliminates the first 60 amino acids prevents the mutant (Pho8∆60) from entering the ER, leaving the zymogen in the cytosol. When autophagy is induced, the mutant zymogen is delivered to the vacuole nonselectively inside autophagosomes along with other cytoplasmic material. The resulting activation of the zymogen can be easily measured by enzymatic assays for phosphatase activity [356]. To minimize background activity, it is preferable to have the gene encoding the cytosolic Pho13 phosphatase additionally deleted (although this is not necessary when assaying certain substrates).
Cautionary notes: Measuring the degradation of long-lived proteins requires prior radiolabeling of the cells, and subsequent separation of acid-soluble from acid-insoluble radioactivity. The labeling can be done with relative ease both in cultured cells and in live animals [4], and has recently been scaled down to minimize the amount of radioactivity needed in cell culture experiments [25]. In cells, it is also possible to measure the release of an unlabeled amino acid by chromatographic methods, thereby obviating the need for prelabeling [1039]; however, it is important to keep in mind that amino acid release is also regulated by protein synthesis, which in turn is modulated by many different factors. In either case, one potential problem is that the released amino acid may be further metabolized. For example, branched chain amino acids are good indicators of proteolysis in hepatocytes, but not in muscle cells where they are further oxidized (A.J. Meijer, personal communication). In addition, the amino acid can be reincorporated into protein; for this reason, such experiments can be carried out in the presence of CHX, but this raises additional concerns (see Turnover of autophagic compartments). In the case of labeled amino acids, a nonlabeled chase is added where the tracer amino acid is present in excess (being cautious to avoid using an amino acid that inhibits autophagy), or by use of single-pass perfused organs or superfused cells [1040, 1041]. The perfused organ system also allows for testing the reversibility of effects on proteolysis and the use of autophagy-specific inhibitors in the same experimental preparation, which are crucial controls for proper assessment.
If the autophagic protein degradation is low (as it will be in cells in replete medium), it may be difficult to measure it reliably above the relatively high background of nonautophagic degradation. It should also be noted that the usual practice of incubating the cells under “degradation conditions,” that is, in a saline buffer, indicates the potential autophagic capacity (maximal attainable activity) of the cells rather than the autophagic activity that prevails in vivo or under rich-culture conditions. Finally, inhibition of a particular degradative pathway is typically accompanied by an increase in a separate pathway as the cell attempts to compensate for the loss of degradative capacity [1025]. This compensation might interfere with control measurements under conditions that attempt to inhibit autophagy; however, as the latter is the major degradative pathway, the contributions of other types of degradation over the course of this type of experiment are most often negligible. Another issue of concern, however, is that most pharmacological protease inhibitors have “off target” effects that complicate the interpretation of the data.
The Pho8∆60 assay requires standard positive and negative (such as an atg1∆ strain) controls, and care must be taken to ensure the efficiency of cell lysis. Glass beads lysis works well in general, provided that the agitation speed of the instrument is adequate. Instruments designed for liquid mixing with lower speeds should be avoided. We also recommend against holding individual sample tubes on a vortex, as it is difficult to maintain reproducibility; devices or attachments are available to allow multiple tubes to be agitated simultaneously. Finally, it is also important to realize that the deletion of PHO8 can affect yeast cell physiology, especially depending on the growth conditions, and this may in turn have consequences for the cell wall; cells under starvation stress generate thicker cell walls that can be difficult to degrade enzymatically.
Conclusion: Measuring the turnover of long-lived proteins is a standard method for determining autophagic flux. Newer proteomic techniques that compare protein levels in autophagy-deficient animals relative to wild-type animals are promising [1042, 1043], but the current ratiometric methods are affected by both protein synthesis and degradation, and thus analyze protein turnover, rather than degradation.
11. Selective types of autophagy
Although autophagy can be nonselective, in particular during starvation, there are many examples of selective types of autophagy.
a. The Cvt pathway, mitophagy, pexophagy, piecemeal microautophagy of the nucleus and late nucleophagy in yeast and filamentous fungi
The precursor form of aminopeptidase I (prApe1) is the major cargo of the Cvt pathway in yeast, a biosynthetic autophagy-related pathway [177]. The propeptide of prApe1 is proteolytically cleaved upon vacuolar delivery, and the resulting shift in molecular mass can be monitored by western blot. Under starvation conditions, prApe1 can enter the vacuole through nonselective autophagy, and thus has been used as a marker for both the Cvt pathway and autophagy.
The yeast Cvt pathway is unique in that it is a biosynthetic route that utilizes the autophagy-related protein machinery, whereas other types of selective autophagy are degradative. The latter include pexophagy, mitophagy, reticulophagy and xenophagy, and each process has its own marker proteins, although these are typically variations of other assays used to monitor the Cvt pathway or autophagy. One common type of assay involves the processing of a GFP chimera similar to the GFP-Atg8 processing assay (see GFP-Atg8-family protein lysosomal delivery and partial proteolysis). For example, yeast pexophagy utilizes the processing of Pex14-GFP and Pot1/Fox3/thiolase-GFP [1044, 1045], whereas mitophagy can be monitored by the generation of free GFP from Om45-GFP, Idh1-GFP, Idp1-GFP or mito-DHFR-GFP [1046–1050]. Important differences, however, can be observed between GFP chimera of endogenous mitochondrial proteins and an artificial construct such as mito-DHFR-GFP [1051]. In filamentous fungi, NBR1-dependent pexophagy can be monitored by inducing peroxisome proliferation through growth in fatty acid-containing medium and shifting the mycelium back to complete medium to visualize DsRED-labeled peroxisome degradation in the vacuole [1052]. Localization of mitochondrially-targeted proteins (or specific MitoTracker® dyes) or similar organelle markers such as those for the peroxisome (e.g., GFP-SKL with Ser-Lys-Leu at the C terminus that acts as a peroxisomal targeting signal, Aox3 [acyl-CoA oxidase 3]-EYFP that allows simultaneous observation of peroxisome-vacuole dynamics with the single FITC filter set, or GFP-Cta1 [catalase A]) can also be followed by fluorescence microscopy [831, 1045, 1053–1055]. In addition, yeast mitophagy requires both the Slt2 and Hog1 signaling pathways; the activation and phosphorylation of Slt2 and Hog1 can be monitored with commercially available phospho-specific antibodies (Figure 26) [747]. It is also possible to monitor pexophagy in yeasts by the disappearance of activities of specific peroxisome markers such as catalase, alcohol oxidase or amine oxidase in cell-free extracts [1056], or permeabilized cell suspensions. Catalase activity, however, is a useful marker only when peroxisomal catalases are the only such enzymes present or when activities of different catalases can be distinguished. In S. cerevisiae there are two genes, CTT1 and CTA1, encoding catalase activity, and only one of these gene products, Cta1, is localized in peroxisomes. Activities of both catalases can be distinguished using an in-gel activity assay after PAGE under nondenaturing conditions by staining with diaminobenzidine [1057, 1058]. Plate assays for monitoring the activity of peroxisomal oxidases in yeast colonies are also available [1054]. The decrease in the level of endogenous proteins such as alcohol oxidase, Pex14 or Pot1 can be followed by western blotting [831, 1059–1062], TEM [1063], fluorescence microscopy [831, 1064, 1065] or laser confocal scanning microscopy of GFP-labeled peroxisomes [1066, 1067].
Figure 26.
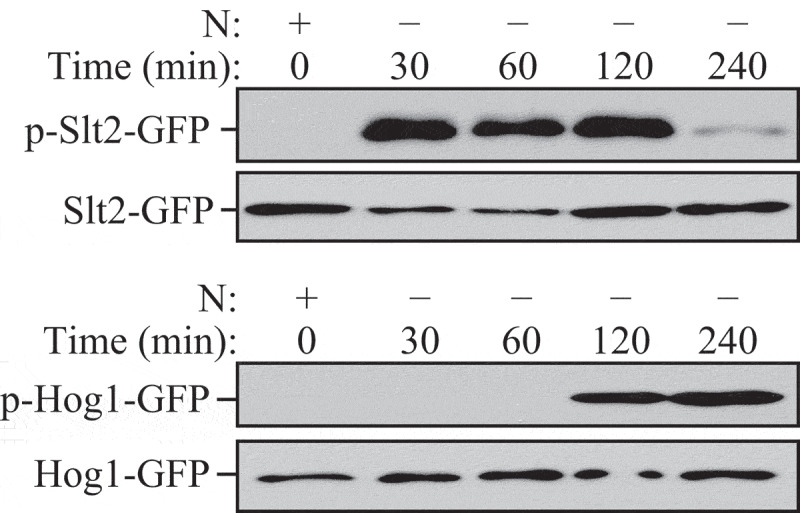
S. cerevisiae cells were cultured to mid-log phase and shifted to SD-N for the indicated times. Samples were taken before (+) and at the indicated times after (–) nitrogen starvation. Immunoblotting was done with anti-phospho-Slt2 and anti-phospho-Hog1 antibody. This figure was modified from data previously published in ref. [747], and is reproduced by permission of the American Society for Cell Biology, copyright 2011
In yeast, nonselective autophagy can be induced by nitrogen-starvation conditions, whereas degradative types of selective autophagy generally require a carbon source change or ER stress for efficient induction. For example, in S. cerevisiae, to induce a substantial level of mitophagy, cells need to be precultured in a nonfermentable carbon source such as lactate or glycerol to stimulate the proliferation of mitochondria (although this is not the case in Komagataella phaffii/Pichia pastoris). After sufficient mitochondria proliferation, shifting the cells back to a fermentable carbon source such as glucose will cause the autophagic degradation of superfluous mitochondria [1047]. It should be noted that in addition to carbon source change, simultaneous nitrogen starvation is also required for efficient mitophagy induction. This is possibly because excessive mitochondria can be segregated into daughter cells by cell division if growth continues [1047]. A similar carbon source change from oleic acid or methanol to ethanol or glucose (with or without nitrogen starvation) can be used to assay for pexophagy [1068]; whereas a shift to glucose induces micropexophagy, a shift to ethanol induces macropexophagy [1061]. Mitophagy can apparently be induced in Magnaporthe oryzae by treatment with ROS to induce mitochondrial damage [1069]; however, ROS or mitochondrial oxidative phosphorylation uncouplers such as CCCP do not induce mitophagy in S. cerevisiae [1048, 1070]. Mitophagy can be induced by culturing yeast cells in a nonfermentable carbon source to post-log phase or before nitrogen starvation [1070, 1071]. In this case, mitophagy may be induced because the energy demand is lower at post-log phase and the mitochondrial mass exceeds the cell’s needs [169, 1072, 1073]. It has been suggested that this type of mitophagy, also known as “stationary phase mitophagy,” reflects a quality-control function that culls defective mitochondria that accumulate in nondividing, respiring cells [1074]. Furthermore, there is some evidence that mitophagy can be induced in cells cultured with a fermentable carbon source such as glucose by a shift from nutrient-rich to nitrogen-starvation conditions, which makes it possible to examine mitophagy even in respiratory-deficient cells, although the amount of mitochondrial turnover may be quite low [1075]. Similarly, pexophagy can be induced by culturing the cells in a peroxisome proliferation medium to post-log phase (J.-C. Farré, unpublished results). Along these lines, it should also be realized that some types of selective autophagy continuously occur at a low level under noninducing conditions. Thus, organelles such as peroxisomes have a finite life span and are turned over at a slow rate by autophagy-related pathways [1076].
Piecemeal microautophagy of the nucleus (PMN, also termed micronucleophagy) is another selective autophagic subtype, which targets portions of the nucleus for degradation [157, 1077, 1078]. In S. cerevisiae, the nuclear outer membrane, which is continuous with the nuclear ER, forms contact sites with the vacuolar membrane. These nucleus-vacuole junctions (NVJs) are generated by interaction of the outer nuclear membrane protein Nvj1 with the vacuolar protein Vac8 [1079]. Nvj1 further recruits the ER-membrane protein Tsc13, which is involved in the synthesis of very-long-chain fatty acids (VLCFAs) and Swh1/Osh1, a member of a family of oxysterol-binding proteins. Upon starvation the NVJs bulge into the vacuole and subsequently a PMN-vesicle pinches off into the vacuole. PMN vesicles thus contain nuclear material and are limited by three membranes with the outermost derived from the vacuole, and the two inner ones from the nuclear ER. It is not clear which nuclear components are removed by PMN, but because PMN is not a cell death mechanism per se, it seems most likely that superfluous material is recycled. During PMN the NVJs are selectively incorporated into the PMN vesicles and degraded. Accordingly, PMN can be monitored using the proteins that are associated with the NVJs as markers. To quantitatively follow PMN, an assay analogous to the above-described GFP-Atg8 processing assay has been established using either GFP-Swh1/Osh1 or Nvj1-GFP. These GFP chimeras are, together with the PMN-vesicles, degraded in the vacuole. Thus, the formation of the relatively proteolysis-resistant GFP detected in western blots correlates with the PMN rate. In fluorescence microscopy, PMN can be visualized with the same constructs, and a chimera of mCherry fused to a nuclear localization signal (NLS-mCherry) can also be used. To assure that the measured PMN rate is indeed due to selective PMN/micronucleophagy, appropriate controls such as cells lacking Nvj1 or Vac8 should be included. Detailed protocols for the described assays are provided in ref. [1080].
Late nucleophagy (LN) is another type of selective degradation of the nucleus, which specifically targets bulk nucleoplasm for degradation after prolonged periods (20-24 h) of nitrogen starvation [721]. LN induction occurs in the absence of the essential PMN proteins Nvj1 and Vac8 and, therefore, the formation of NVJs. Although, some components of the core Atg machinery are required for LN, Atg11 and the Vps34-containing PtdIns3K complex I are not needed. LN can be monitored by employing a nuclear-targeted version of the Rosella biosensor (n-Rosella) and following either its accumulation (by confocal microscopy), or degradation (by immunoblotting), within the vacuole [1081]. Dual labeling of cells with Nvj1-EYFP, a nuclear membrane reporter of PMN, and the nucleoplasm-targeted NAB35-DsRed.T3 (NAB35 is a target sequence for the Nab2 RNA-binding protein, and DsRed.T3 is the pH-stable, red fluorescent component of n-Rosella) allows detection of PMN soon after the commencement of nitrogen starvation, whereas delivery to the vacuole of the nucleoplasm reporter, indicative of LN, is observed only after prolonged periods of nitrogen starvation. Few cells show simultaneous accumulation of both reporters in the vacuole, indicating that PMN and LN are temporally and spatially separated [1081].
In contrast to unicellular yeasts, filamentous fungi form an interconnected mycelium of multinucleate hyphae containing up to 100 nuclei in a single hyphal compartment. A mycelial colony grows by tip extension with actively growing hyphae at the colony margin surrounded by an older, inner hyphal network that recycles nutrients to fuel the hyphal tips. By labeling organelle markers with GFP it is possible to show in Aspergillus oryzae that autophagy mediates degradation of basal hyphal organelles such as peroxisomes, mitochondria and entire nuclei [1082]. In contrast to yeast, PMN has not been observed in filamentous ascomycetes. In M. oryzae, germination of the condiospore and formation of the appressorium is accompanied by nuclear degeneration in the spore [373]. The degradation of nuclei in spores requires the nonselective autophagy machinery, whereas conserved components of the PMN pathway such as Vac8 and Tsc13 are dispensable for nuclear breakdown during plant infection [1083]. Nuclei are proposed to function in storage of growth-limiting nutrients such as phosphate and nitrogen [1084, 1085]. Similar to nuclei, mitochondria and peroxisomes are also preferentially degraded in the basal hyphae of filamentous ascomycetes [373, 1082–1086].
Cautionary notes: The Cvt pathway has been demonstrated to occur only in yeast. In addition, the sequestration of prApe1 is specific, even under starvation conditions, as it involves the recognition of the propeptide by a receptor, Atg19, which in turn interacts with the scaffold protein Atg11 [1087, 1088]. Thus, unless the propeptide is removed or the genes encoding Atg11 or Atg19 are deleted, prApe1 is recognized as a selective substrate. Overexpression of prApe1 saturates import by the Cvt pathway, and the precursor form accumulates, but is rapidly matured upon autophagy induction [408]. In addition, mutants such as vac8∆ and tlg2∆ accumulate prApe1 under nutrient-rich conditions, but not during autophagy [745, 1089]. Accordingly, it is possible to monitor the processing of prApe1 when overexpressed, or in certain mutant strains to follow autophagy induction. However, under the latter conditions it must be kept in mind that the sequestering vesicles are substantially smaller than typical autophagosomes generated during nonselective autophagy; the Cvt complex (prApe1 bound to Atg19) is smaller than typical peroxisomes or mitochondrial fragments that are subject to autophagic degradation. Accordingly, particular mutants may display complete maturation of prApe1 under autophagy-inducing conditions, but may still have a defect in other types of selective autophagy, as well as being unable to induce a normal level of nonselective autophagy [148]. For this reason, it is good practice to evaluate autophagosome size and number by TEM. Actually, it is much simpler to monitor autophagic bodies (rather than autophagosomes) in yeast. First, the vacuole is easily identified, making the identification of autophagic bodies much simpler. Second, autophagic bodies can be accumulated within the vacuole, allowing for an increased sample size. It is best to use a strain background that is pep4∆ vps4∆ to prevent the breakdown of the autophagic bodies, and to eliminate confounding vesicles from the multivesicular body pathway. One caveat to the detection of autophagic bodies, however, is that they may coalesce in the vacuole lumen, making it difficult to obtain an accurate quantification. Finally, it is important to account for biases in sample sectioning to obtain an accurate estimate of autophagic body number or size [147].
In general, when working with yeast it is preferable to use strains that have the marker proteins integrated into the chromosome rather than relying on plasmid-based expression, because plasmid numbers can vary from cell to cell. The GFP-Atg8, or a similar, processing assay is easy to perform and is suitable for analysis by microscopy as well as western blotting; however, particular care is needed to obtain quantitative data for GFP-Atg8, Pex14-GFP or Om45-GFP, etc. processing assays (see cautionary notes for GFP-Atg8-family protein lysosomal delivery and partial proteolysis).
A pHluorin-Atg8 chimera can be used to determine the breakdown of autophagic bodies in budding yeast by live cell fluorescence microscopy. In WT cells, fluorescence of pHluorin-Atg8 is detectable at neutral pH in the cytosol and at the PAS or on autophagosomes, but not at the lower pH within the vacuole upon starvation. In mutants that are either deficient in vacuolar peptidases (atg42∆, pep4∆, prb1∆, prc1∆) or vacuolar acidification (vma4∆) pHluorin-Atg8 is not quenched and pHluorin-Atg8-positive vesicular structures are detected inside their vacuoles, suggesting that autophagic bodies are not efficiently lysed. Hence, pHluorin-Atg8 is a useful tool to detect defects in the breakdown of autophagic bodies inside vacuoles [1090].
An alternative method to monitor selective autophagy is to use an organelle-targeted Pho8∆60 assay. For example, mitoPho8∆60 can be used to quantitatively measure mitophagy [1048]. In addition, for the GFP-Atg8 processing assay, 2 h of starvation is generally sufficient to detect a significant level of free (i.e., vacuolar) GFP by western blotting as a measure of nonselective autophagy. For selective types of autophagy, the length of induction needed for a clearly detectable free GFP band will vary depending on the rate of cargo delivery/degradation. Usually 6 h of mitophagy induction is needed to be able to detect free GFP (e.g., from Om45-GFP) by western blot under starvation conditions, whereas stationary phase mitophagy typically requires 2 days before a free GFP band is observed. However, as with animal systems (see Animal mitophagy and pexophagy), it would be prudent to follow more than one GFP-tagged protein, as the kinetics, and even the occurrence of mitophagic trafficking, seems to be protein species-dependent, even within the mitochondrial matrix [1091]. The use of an artificial, non-mitochondrial protein as a chimeric mitophagy reporter (such as mtDHFR-GFP) can apparently be used as a reporter for “general” mitophagy as it does not appear to have any endogenous “selectivity” cues [1051].
b. Aggrephagy
Aggrephagy is the selective removal of aggregates by autophagy [1092]. This process can be followed in vitro (in cell culture) and in vivo (in mice) by monitoring the levels of an aggregate-prone protein such as an expanded polyglutamine (polyQ)-containing protein or mutant MAPT/tau or SNCA/α-synuclein (synuclein alpha). Levels are quantified by immunofluorescence, immunogold labeling, filter-trap assay or traditional immunoblot. In yeast, degradation of SNCA aggregates can be followed by promoter shut-off assays. Expression of the inducible GAL1 promoter of GFP-tagged SNCA is stopped by glucose repression. The removal of aggregates is thus monitored with fluorescence microscopy.
The relationship between SNCA clearnace and autophagy has also been exploited in yeast studies during chronological aging with SNCA expressed under the control of a constitutive promoter [347, 349, 366, 629]. In this model, SNCA toxicity is dependent on Atg11 [366] and promotes cell cycle re‐entry, S‐phase arrest, and DNA damage response activation, which is responsible for a dramatic increase in autophagy [349]. This selective pathway of autophagy has been termed genotoxin‐induced targeted autophagy (GTA) and, in addition to Atg11, requires the involvement of the Mec1 and Rad53 kinases [1093].
The contribution of autophagy to SNCA aggregate clearance can be studied by the use of different autophagy mutants or by pharmacological treatment with the proteinase B inhibitor PMSF [1094–1096]. Similarly, fluorescently tagged aggregated proteins such as polyQ80-CFP can be monitored via immunoblot and immunofluorescence. In addition to fluorescence methods, aggregates formed by a splice variant of CCND2 (cyclin D2) can also be monitored in electron-dense lysosomes and autophagosomes by immunogold labeling and TEM techniques [1097]. A polyQ80-luciferase reporter, which forms aggregates, can also be used to follow aggrephagy [1098]. A nonaggregating polyQ19-luciferase or untagged full-length luciferase serves as a control. The ratio of luciferase activity from these two constructs can be calculated to determine autophagic flux.
Autophagic clearance of mutated human HTT (huntingtin) protein with a polyQ expansion (HTT103Q) can also be observed in budding yeast. After overnight induction from a galactose inducible promotor, HTT103Q proteins form inclusion bodies in yeast cells. When glucose is added into the cell culture to shut off HTT103Q expression, obvious vacuolar localization of the protein is detected within 1 h, and this localization depends on the core autophagy machinery. Moreover, the absence of the ubiquilin protein Dsk2 and some heat-shock proteins compromises the vacuolar localization of HTT103Q [1099, 1100]. Therefore, mutated HTT protein can be used as a model substrate to study aggrephagy.
Autophagic degradation of endogenous aggregates such as lipofuscin can be monitored in some cell types by fluorescence microscopy, utilizing the autofluorescence of lipofuscin particles. Although under normal conditions almost 99% of the lipofuscin particles are located in autophagosomes or lysosomes, an impairment of autophagy leads to free lipofuscin in the cytosol [1101, 1102]. The amount of lipofuscin in primary human adipocytes can be reduced by activation of autophagy, and the amount of lipofuscin is dramatically reduced in adipocytes from patients with type 2 diabetes and chronically enhanced autophagy [396]. Monitoring autophagy in tissues with lipofuscin accumulation is not possible using a mouse reporter model expressing GFP-LC3, because cytosolic lipofuscin appears as a hyperfluorescent punctum in the green channel [485]. A tandem tagged LC3 reporter model (CAG-mRFP-EGFP-LC3 [1103]) will be better suited to study pathologies involving lipofuscin accumulation. ImageJ, or other equivalent software, should be utilized to detect GFP-positive puncta that colocalize with RFP-positive structures. Cytosolic lipofuscin will appear as an RFP-independent GFP (green) punctum.
Similarly, TFEB overexpression either in neurons or oligodendrocytes reduces neurodegeneration and the pathological burden of SNCA in many experimental models of synucleinopathies reported by independent investigators [1104–1106].
Cautionary notes: Caution must be used when performing immunoblots of aggregated proteins, as many protein aggregates fail to enter the resolving gel and are retained in the stacking gel. This drawback can be bypassed by performing a filter-trap assay in which protein extracts are forced by mild suction through a nitrocellulose membrane, and protein aggregates larger than the nitrocellulose pores are stuck on the membrane and can then be detected by traditional immunoblot [1107]. In addition, the polyQ80-luciferase in the aggregated state lacks luciferase activity, whereas soluble polyQ80-luciferase retains activity. Therefore, caution must be used when interpreting results with these vectors, as treatments that increase aggrephagy or enhance protein aggregation can lead to a decrease in luciferase activity [1108]. Finally, soluble polyQ reporters can be degraded by the proteasome; thus, changes in the ratio of polyQ19-luciferase:polyQ80-luciferase may also reflect proteasomal effects and not just changes in autophagic flux.
c. Allophagy
In C. elegans, mitochondria, and hence paternal mitochondrial DNA, from sperm are eliminated by an autophagic process. This process of allogeneic (nonself) organelle autophagy is termed “allophagy” [1109, 1110]. During allophagy in C. elegans, both paternal mitochondria and membranous organelles (a sperm-specific membrane compartment) are eliminated by the 16-cell stage (100-120 min post-fertilization) [1111, 1112]. The degradation process can be monitored in living embryos with GFP::ubiquitin, which appears in the vicinity of the sperm chromatin (labeled for example with mCherry-histone H2B) on the membranous organelles within 3 min after fertilization. GFP fusions and antibodies specific for LGG-1 and LGG-2 (Atg8-family protein homologs), which appear next to the sperm DNA, membranous organelles and mitochondria (labeled with CMXRos or mitochondria-targeted GFP) within 15 to 30 min post-fertilization, can be used to verify the autophagic nature of the degradation. TEM [1113–1115] can also be utilized to demonstrate the presence of mitochondria within autophagosomes in the early embryo. The respective functions of LGG-1 and LGG-2 have been addressed by RNAi depletion or through the use of genetic loss-of-function mutants lgg-1(tm3489) and lgg-2(tm5755). LGG-1 is essential for allophagosome formation, whereas LGG-2 contributes to their efficient maturation [1114]. Ubiquitination of the substrates was first described for the membranous organelles and not for sperm-inherited mitochondria [1111, 1112], but studies suggest that ubiquitination of sperm-mitochondria could be required for the initial step of allophagy [1116, 1117]. The autophagy receptor ALLO-1 and its kinase IKKE-1 are required for the recruitment of LGG-1 around sperm-inherited organelles [1116]. This autophagy targeting requires both the ubiquitination of substrates and the loss of sperm mitochondrial membrane potential [1116, 1118, 1119].
Conclusion: There are many assays that can be used to monitor selective types of autophagy, but caution must be used in choosing an appropriate marker(s). The potential role of other degradative pathways for any individual organelle or cargo marker should be considered, and it is advisable to use more than one marker or technique.
d. Animal mitophagy and pexophagy
There is no consensus at the present time with regard to the best method for monitoring mitophagy in animal cells. As with any organelle-specific form of autophagy, it is necessary to demonstrate: i) increased levels of phagophores interacting with, or autophagosomes containing, mitochondria; ii) maturation of these autophagosomes that culminates with mitochondrial degradation, which can be blocked by specific inhibitors of autophagy or of lysosomal degradation; and iii) whether the changes are due to selective mitophagy or increased mitochondrial degradation during nonselective autophagy. Techniques to address each of these points have been reviewed [55, 1120]. Note that a common misconception is that mitophagy can be monitored via RT-qPCR of mRNA transcripts encoding mitophagy-associated factors (e.g., PINK1, PRKN, etc.); in fact, changes in mRNA levels of these factors do not necessarily reflect mitophagic activity and should not be used to infer changes in mitophagy in the absence of other assays.
The following methods can be used to follow all forms of mitophagy: Ultrastructural analysis by TEM at early time points can be used to establish selective mitophagy. It should be noted that a detailed handbook on how to specifically dissect the several phases of the mitophagic process by TEM is not available. This should ideally include an initial phase of mitochondrial fragmentation, followed by formation of a double-layered membrane that expands around the selected organelle to form a double-membrane mitophagosome that contains mitochondria-like structure. TEM can be used to demonstrate the presence of mitochondria within these vesicles, and this can be coupled with bafilomycin A1 or CQ treatment to prevent fusion with the lysosome to trap early autophagosomes with recognizable cargo [55] (Figure 4). In the later phase, and in the absence of maturation inhibitors, it might become difficult to clearly identify mitochondria-like structures inside the mitophagosomes; however, these should be appropriate in size, retain a double-membrane structure, and contain remnants of mitochondrial cristae. Depending on the use of specific imaging techniques, dyes for living cells or antibodies for fixed cells have to be chosen. In any case, transfection of the phagophore and autophagosome marker GFP-LC3 to monitor the initiation of mitophagy, or RFP-LC3 to assess mitophagy progression, and visualization of mitochondria (independent of their mitochondrial membrane potential) makes it possible to determine the association of these two cellular compartments. Qualitatively, this may appear as fluorescence colocalization or as rings of GFP-LC3 surrounding mitochondria in higher-resolution images [201, 1113, 1121].
Care must be taken in interpreting these results, as some data indicate that autophagosomes form at ER-mitochondria contact sites [872]; hence, there will be some degree of colocalization between forming (non-mitophagic) autophagosomes and mitochondria. Fluorescence microscopy-based approaches for monitoring autophagosome or lysosome colocalization with mitochondria in cells in which cytoplasm is almost fully occupied by mitochondria, such as brown adipocytes, may be particularly challenging. Background thresholds should be accurately set to avoid false positive results.
For live-cell imaging microscopy, mitochondria should be labeled by a matrix-targeted fluorescent protein through transfection or by the use of mitochondria-specific dyes. When using matrix-targeted fluorophores for certain cell lines (e.g., SH-SY5Y), it is important to allow at least 48 h of transient expression for sufficient targeting/import of mitochondrial GFP/RFP prior to analyzing mitophagy. Among the MitoTracker® probes are lipophilic cations that include a chloromethyl group and a fluorescent moiety. These probes concentrate in mitochondria due to their negative charge and react with the reduced thiols present in mitochondrial matrix proteins [1122–1124]. After this reaction, the probe can be fixed and remains in the mitochondria independent of subsequent alterations in mitochondrial function or mitochondrial membrane potential [1123, 1125, 1126]. This method can thus be used when cells remain healthy when the dye is applied, as the dye will remain in the mitochondria and is retained after fixation, although, as stated above, accumulation is dependent on the membrane potential. In addition, it is important to note that the various mitochondrial dyes are not identical in terms of their properties, and not all are suitable for use following fixation. For example, MitoTracker® Green FM is not retained well after aldehyde fixation, whereas MitoTracker® Red CMXRos works under these conditions. Although in some cases it is convenient to utilize the fixation step, it is possible to evaluate fresh, unfixed cells, and, consequently, with less manipulated mitochondria, obtain good results with both flow cytometry and confocal microscopy [1127]. Transfection with mitochondrially targeted fluorescent proteins can also be used with similar results to MitoTracker® Green FM [201]. Antibodies that specifically recognize mitochondrial proteins such as VDAC, TOMM20/TOM20 (translocase of outer mitochondrial membrane 20), SOD2 (superoxide dismutase 2), HSPD1/HSP60 (heat shock protein family D (Hsp60) member 1), HSPA9/mtHSP70 or COX4I1 (cytochrome c oxidase subunit 4I1) may be used to visualize mitochondria in immunohistochemical experimental procedures [1128–1132] or in human patient samples [1133].
Colocalization analyses of mitochondria and autophagosomes provide an indication of the degree of autophagic sequestration. To quantify early mitophagy, the percentage of LC3 puncta (endogenous, RFP- or GFP-LC3 puncta) that colocalize with mitochondria and the number of colocalizing LC3 puncta per cell—as assessed by confocal microscopy—in response to mitophagic stimuli can be employed as well [1134]. Of note, PINK1-PRKN-dependent mitophagy is independent of the LC3 subfamily, but strongly requires the GABARAPs [34, 35]. Conversely, LC3 is involved in cardiolipin-mediated mitophagy [201]. Thus, monitoring of more than on Atg8 subfamily may be necessary. In addition, the percentage of lysosomes that colocalize with mitochondria can be used to quantify autophagy-mediated delivery of mitochondria. Furthermore, induction of mitophagy also promotes the formation of ring-shaped/spheroid mitochondria interacting with structures positive for LC3 and lysosomal proteins (based on immuno-EM). It is not clear whether these structures represent forming autophagomes dedicated to the degradation of mitochondria, or whether they represent a distinct process of mitochondrial dynamics [1135, 1136]. Overall, it is important to quantify mitophagy at various stages (initiation, progression, and late mitophagy) to identify stimuli that elicit this process [1137, 1138].
The fusion process of mitophagosomes with hydrolase-containing lysosomes represents the next step in the degradation process. To monitor the amount of fused organelles via live cell imaging microscopy, MitoTracker® Green FM and LysoTracker™ Red DND-99 may be used to visualize the fusion process (Figure 27). Independent of the cell-type specific concentration used for both dyes, we recommend exchanging MitoTracker® Green FM medium with normal medium (preferably phenol red-free and CO2 independent to reduce unwanted autofluorescence) after incubation with the dye, whereas it is best to maintain the LysoTracker™ Red stain in the incubation medium during the acquisition of images. Given that these fluorescent dyes are extremely sensitive to photobleaching, it is critical to perform live cell mitophagy experiments via confocal microscopy, preferably by using a spinning disc confocal microscope for long-term imaging experiments. For immunocytochemical experiments, antibodies specific for mitochondrial proteins and an antibody against LAMP1 (lysosomal associated membrane protein 1) can be used. Overlapping signals appear as a merged color and can be used as indicators for successful fusion of autophagosomes that contain mitochondria with lysosomal structures [1139]. To measure the correlation between two variables by imaging techniques, such as the colocalization of two different fluorescent signals, we recommend some form of correlation analysis to assess the value correlating with the strength of the association. This may use, for example, ImageJ software or other colocalization scores that can be derived from consideration not only of pixel colocalization, but also from a determination that the structures have the appropriate shape. During live-cell imaging, the two structures (autophagosomes and mitochondria) should move together in more than one frame. Mitophagy can also be quantitatively monitored using a mitochondria-targeted version of the pH-dependent Keima protein [1036]. The peak of the excitation spectrum of the protein shifts from 440 nm to 586 nm when mitochondria are delivered to acidic lysosomes, which can provide a quantitative readout of mitophagy (Figure 28). However, it should be noted that long exposure time of the specimen to intense laser light leads to a similar spectral change. mt-Keima in combination with flow cytometry has been used to quantitatively monitor mitophagy flux [1130, 1140, 1141].
Figure 27.
Human fibroblasts showing colocalization of mitochondria with lysosomes. The degree of colocalization of mitochondria with lysosomes in human fibroblasts was measured via live cell imaging microscopy at 37°C and 5% CO2 atmosphere using the ApoTome® technique. LysoTracker® Red DND-99 staining was applied to mark lysosomal structures (red), and MitoTracker® Green FM to visualize mitochondria (green). Hoechst 33342 dye was used to stain nuclei (blue). A positive colocalization is indicated by yellow signals (Merge) due to the overlap of LysoTracker® Red and MitoTracker® Green staining (white arrows). Scale bar: 10 μm. Statistical evaluation is performed by calculating the Pearson’s coefficient for colocalizing pixels. Image provided by L. Burbulla and R. Krüger
Figure 28.
Detection of mitophagy in primary cortical neurons using mitochondria-targeted Keima. Neurons transfected with mito-Keima were visualized using 458-nm (green, mitochondria at neutral pH) and 561-nm (red, mitochondria in acidic pH) laser lines and 575-nm band pass filter. Compared with the control (A) wild-type PINK1 overexpression (B) increases the number of the mitochondria exposed to acidic conditions. Scale bar: 2 µm. (C) Quantification of red puncta suggests increased mitophagy in wild-type PINK1 but not in the kinase dead (kd) PINK1K219M-overexpressing neurons. Image provided by V. Choubey and A. Kaasik
It is important to note that in a process distinct from mitophagy, mitochondria and lysosomes can also become dynamically tethered to one another in a RAB7A-GTP hydrolysis-dependent manner at inter-organelle mitochondria-lysosome contact sites, which are important for regulating mitochondrial dynamics [1142–1144]. Thus, high-resolution microscopy and preferably live cell imaging are strongly recommended to differentiate mitophagy (which results in mitochondria engulfed within the lysosomal membrane) from stably tethered mitochondria-lysosome contacts (mitochondria that are in contact [<10 nm] from a lysosome and can subsequently untether from one another without undergoing bulk mitochondrial degradation).
Finally, a mitochondria-targeted version of the tandem mCherry-GFP fluorescent reporter (see Tandem mRFP/mCherry-GFP fluorescence microscopy) using a targeting sequence from the mitochondrial membrane protein FIS1 [460, 461] can be used to monitor mitophagic flux [460]. In addition, transgenic mice and Drosophila expressing mt-Keima, mito-QC or mt-mCherry-GFP provide useful tools for analysis of mitophagy in vivo in many physiological and pathological conditions [38, 466, 1145–1148]. The tandem fluorescent and the mitochondrially-targeted Keima fluorescence microscopy approaches both assess delivery of mitochondria to acidic (endo-lysosomal) environments. To evaluate whether these acidic environments are proteolytically active, the cleavage of ectopically expressed TOMM20-Keima (or other mitochondria-targeted Keima fusion proteins) can be followed by western blotting [1037]. Whereas TOMM20 is sensitive to proteolytic enzymes, Keima is resistant, and thus the appearance of free Keima in the western blot indicates arrival of the mitochondria-targeted fusion protein to a proteolytic environment (lysosomes). The fold-change in TOMM20-Keima cleavage upon treatment with an autophagic stimulus can be compared with the fold change in the cleavage of a cytosolic Keima fusion protein (e.g., LDHB-Keima), to thereby assess the degree of selectivity of the autophagic response towards mitochondria over cytosolic proteins.
The third and last step of monitoring the degradation process is to examine the amount of remaining mitochondria by analyzing the mitochondrial mass. This final step provides the opportunity to determine the efficiency of degradation of dysfunctional, aged or impaired mitochondria. Mitochondrial mass can either be measured by a flow cytometry technique using MitoTracker® Green FM (or MitoTracker® Deep Red FM to monitor mitochondria with a polarized membrane) [1123] on a single-cell basis, by either live cell imaging or immunocytochemistry (using antibodies specifically raised against different mitochondrial proteins or, less specifically, by staining with acridine orange 10-nonyl bromide applied after chemical fixation [1149, 1150]). Alternatively, mitochondrial content in response to mitophagic stimuli (in the presence and absence of autophagy inhibitors to assess the contribution of mitophagy) in live or fixed cells can be quantified at the single-cell level as the percentage of cytosol occupied by mitochondrial-specific fluorescent pixels using NIH ImageJ [1138, 1151], specifically by using the MiNA plugin [1152]. One caveat of the latter is that mitochondrial mass may be overestimated when organelle swelling has occurred. Immunoblot analysis of the levels of mitochondrial proteins from different mitochondrial subcompartments is valuable for validating the data from flow cytometry or microscopy studies, and it should be noted that OMM proteins, such as MFNs (mitofusins), TOMM complex proteins, and VDACs, but also PRKN, can be degraded by the proteasome, especially in the context of mitochondrial depolarization [1153–1155]. EM can also be used to verify loss of entire mitochondria, and qPCR (or fluorescence microscopy) to quantify mitochondrial DNA. A reliable estimation of mtDNA copy number per cell can be performed by qPCR of the MT-ND1 (mitochondrially encoded NADH dehydrogenase 1) or MT-ND2 gene expressed as a ratio of mtDNA:nuclear DNA by normalizing to that of the single nuclear-encoded PKM (pyruvate kinase M1/2) or TERT (telomerase reverse transcriptase) genomic DNA [764]. The spectrophotometric measurement of the activity of CS (citrate synthase) [1156], a mitochondrial matrix enzyme of the tricarboxylic acid cycle, which remains highly constant in these organelles and is considered a reliable marker of their intracellular content, has been used as a marker of mitochondrial mass in a variety of systems [1156–1159]. Mitophagy induction can also be examined by using mitochondrial fractionation followed by immunoblot to detect the levels of mitophagic or autophagosome-associated proteins (e.g., PRKN, LC3-II and SQSTM1) in the mitochondrial fraction. The levels of mitochondria-localized DNM1L/Drp1 (dynamin 1 like), which is involved in mitochondrial fission, could also be used to detect early events of mitophagy induction, because mitochondrial fission is required for mitophagy [1160], although mitochondrial DNM1L levels do not necessarily reflect a change in mitophagy.
Each of these techniques to monitor structures associated with the different steps of mitophagy—whether by single-cell analyses of Atg8-family protein mitochondrial colocalization or by immunoblotting for mitochondrial markers—can be combined with strategic use of inhibitors to determine whether mitophagy is impaired or activated in response to stimuli, and at which steps. Therefore, appropriate treatment (pharmacological inhibition and/or siRNA-mediated knockdown of ATG genes) may be applied to prevent mitochondrial degradation at distinct steps of the process. A recent method using flow cytometry in combination with autophagy and mitophagy inhibitors has been developed to determine mitophagic flux using MitoTracker® probes [1123]. Alternatively, mitophagic flux can be monitored by flow cytometry in cells from mito-Keima mice. In this case, it is important to remove dead cells on the basis of SYTOX Blue staining. As a positive control of the assay, carbonyl cyanide p-trifluoromethoxyphenyl-hydrazone (FCCP) is a potent mitochondrial uncoupler that stimulates mitophagic activity [1161].
Certain cellular models require stress conditions to measure the mitochondrial degradation capacity, as basal levels are too low to reliably assess organelle clearance. Exceptions include developmental clearance of large amounts of mitochondria as observed in erythrocyte maturation [1162], and during neuronal development where massive mitophagy is essential to promote a metabolic change towards glycolysis that is required for neurogenesis [1151]. Hence, it may be useful to treat cells with uncoupling agents, such as CCCP, that stimulate mitochondrial degradation and allow measurements of mitophagic activity. In this scenario, it has recently been proposed that assessing the amount of mitochondrial proteins through western blot at basal level and after CCCP administration in human cells may be useful to assess the mitophagic flux [1163, 1164]; however, it should be kept in mind that this treatment is not physiological and promotes the rapid degradation of outer membrane-localized mitochondrial proteins in addition to the loss of mitochondrially-derived ATP used for cellular work. In part for this reason a milder mitophagy stimulus has been developed that relies on a combination of antimycin A (AMA) and oligomycin, inhibitors of the electron transport chain and ATP synthase, respectively [1165]; this treatment is less toxic, and the resulting damage is time dependent. However, this treatment not only blocks ATP production by mitochondria but also substantially enhances mitochondrial ROS production inducing mitochondrial damage. The pharmacological compound PMI that pharmacologically induces mitophagy without disrupting mitochondrial respiration [1166] should provide further insight as it circumvents the acute, chemically induced, blockade of mitochondrial respiration. In addition, the molecule cloxyquin (not to be confused with chloroquine) also induces mitophagy via a mild uncoupling mechanism [444]. In certain conditions/cell types, mitophagy can be induced by NAD-boosting strategies [1167, 1168]. Another method to induce mitophagy is by the treatment of cells with hypoxia-inducing and iron-deprivation agents. Mitochondria are the major site for oxygen consumption, and deprivation of oxygen induces receptor (FUNDC1, BNIP3, BNIP3L)-dependent mitophagy [1169–1171] Treatment of animals including mice, Drosophila and C. elegans [1145, 1148, 1172] under hypoxic conditions or by exposure to iron-deprivation agents (deferiprone/DFP) induces mitochondrial degradation in different tissues, although the degrees of mitophagic activation are not the same in different organs. More specific induction of mitophagy can be achieved by expressing and activating a mitochondrially-localized fluorescent protein photosensitizer such as Killer Red [1173]. The excitation of Killer Red results in an acute increase of superoxide, due to phototoxicity, that causes mitochondrial damage resulting in mitophagy [462]. The advantage of using a genetically encoded photosensitizer is that it allows for both spatial and temporal control in inducing mitophagy. The forced targeting of AMBRA1 to the external mitochondrial membrane is sufficient to induce mitophagy [1174], and expression of constitutively active MAPK1 is sufficient to drive mitophagy in otherwise uninjured tumor cells [1138]. Finally, mitophagy can also be induced in vitro in different cell types by inhibiting the proteasome with the specific inhibitor IU1 [1175]. This type of mitophagy is induced following proteasome recruitment to mitochondria to expose the inner mitochondrial membrane mitophagy receptor PHB2 [1176], and is PINK1- and PRKN-independent [1175].
Mitochondrial turnover, mitochondrial oxidative stress and mitophagy can also be monitored through the use of MitoTimer, a time-sensitive fluorescent protein that targets to the mitochondrial matrix; the emission of MitoTimer shifts from green to red over time [1177–1179]. A lentiviral inducible system encoding MitoTimer is available allowing the controlled expression of this transgene in a wide range of cells [1180]. A constitutively active plasmid DNA encoding MitoTimer as well as inducible transgenic flies and mice allow quantification of mitochondrial structure (fluorescent labeling of mitochondria), oxidative tension (red:green ratio) and mitophagy (pure red puncta that are positive for the mitochondrial protein COX4I1/Cox4 and the lysosomal marker LAMP1) in a variety of tissues, organs and whole animals [1177, 1179, 1181–1187]. Mitophagy can be monitored in mouse primary cells by exploiting the mitoQC mouse model, which ubiquitously expresses a GFP-mCherry tandem protein targeting the mitochondrial outer membrane [38], and the mt-Keima mouse model, which expresses a pH-sensitive protein targeting the mitochondrial matrix [1145].
It is important to keep in mind that there are multiple distinct or partially overlapping pathways of cargo recognition for selective mitophagy [1188]. These include PINK1-PRKN-dependent pathways utilizing p-S65-Ub, receptor-mediated mitophagy involving LIR-domain proteins, and the recognition of mitochondrial phospholipids such as cardiolipin by the LC3 phagophore system [201, 1189, 1190]; among others. Thus, it would be inappropriate to conclude that selective mitophagy is not occurring if markers of only one cargo recognition system are considered.
Antibodies against phosphorylated ubiquitin (p-S65-Ub) have been described as novel tools to detect PINK1-PRKN-mediated mitophagy [1191–1193]. p-S65-Ub is formed by the kinase PINK1 specifically upon mitochondrial stress, and is amplified in the presence of the E3 Ub ligase PRKN (reviewed in [1194]) [1195]. p-S65-Ub antibodies have been used to demonstrate stress-induced activation of PINK1 in various cells including primary human fibroblasts (Figure 29) and dopaminergic neurons differentiated from iPS cells [1193]. Phosphorylated poly-ubiquitin chains specifically accumulate on damaged mitochondria, and staining with p-S65-Ub antibodies can be used, in addition to translocation of PRKN, to monitor the initiation of mitophagy. Given the complete conservation of the epitopes across species, mitochondrial p-S65-Ub can also be detected in mouse primary neurons upon mitochondrial depolarization and park/PRKN-deficient Drosophila. Furthermore, the p-S65-Ub signal partially colocalizes with mitochondrial, lysosomal, and total ubiquitin markers in cytoplasmic granules that appear to increase with age and disease in human postmortem brain samples [1191, 1193]. Examination of the phosphorylation status of outer mitochondrial membrane (OMM) autophagy receptors such as FUNDC1 and BNIP3L is also useful for measuring mitophagy activity [1196, 1197]. Note that care should be taken when choosing antibodies to assess the degree of mitochondrial protein removal by autophagy; the quality and clarity of the result may vary depending on the specifics of the antibody. In testing the efficiency of mitophagy, clearer results may be obtained by using antibodies against mitochondrial DNA (mtDNA)-encoded proteins. This experimental precaution may prove critical to uncover subtle differences that could be missed when evaluating the process with antibodies against nuclear encoded, mitochondrially imported proteins (M. Campanella, personal communication).
Figure 29.
PINK1-dependent phosphorylation of ubiquitin (p-S65-Ub) upon mitophagic stress. (A) Human dermal fibroblasts from healthy controls or PD patients carrying a PINK1 loss-of-function mutation (Q456X) were treated with valinomycin for the indicated times, and lysates were analyzed by western blot. The p-S65-Ub signal is almost undetectable under nonstress conditions in controls, but is strongly induced in a PINK1 kinase-dependent manner during its stabilization on the outer mitochondrial membrane. MFN2 serves as a control substrate and VCL (vinculin) as a loading control. (B) HeLa cells stably expressing GFP-PRKN (wild type) were treated with CCCP for the indicated times, fixed and stained with p-S65-Ub (red) and GFP-PRKN (green) as well as mitochondrial (TOMM20, cyan) and nuclear (Hoechst, blue) markers. The p-S65-Ub staining is almost undetectable in nonstressed cells, but rapidly accumulates on damaged mitochondria where it functions to activate PRKN. On mitochondria, PINK1 and PRKN together amplify the p-S65-Ub signal. Scale bar: 10 µm. Image provided by F.C. Fiesel and W. Springer
Stabilized, unprocessed PINK1 that accumulates on the mitochondrial outer membrane in response to certain forms of acute mitochondrial damage can be used to differentiate between healthy mitochondria and those that have lost their membrane potential. However, caution should be taken with this approach in cells where mitochondria exhibit physiological uncoupling and lowered membrane potential, such as thermogenic brown adipocytes. Similarly, redistribution of cardiolipin to the OMM acts as an elimination signal to trigger mitophagy induction in mammalian cells, including primary neurons [201]. In addition, during CCCP-induced mitophagy, the hexameric protein NME4/NDPKD/NM23-H4 localizes to the mitochondrial intermembrane space, binds cardiolipin and facilitates its redistribution to the OMM [1189], and the ANXA5 (annexin A5) binding assay for externalized cardiolipin can be used as a marker for damaged mitochondria and early mitophagy [201]. The charge of multiple anionic phospholipids present on the OMM can change in response to mild alterations in mitochondrial function. These signals are important to the regulation of protein signaling between mitochondria and the cytosol. Changes in surface charge can be estimated by detecting the binding of ANXA5. Mild metabolic insults (e.g., a 50% inhibition of the mitochondrial enzyme OGDH/ketoglutarate dehydrogenase) increase ANXA5 binding nearly three-fold, while stimulating translocation of DNM1L and LC3 to mitochondria without altering cardiolipin translocation, ATP or the mitochondrial membrane potential [1198]; DNM1L is a fundamental component of mitochondrial fission, which helps facilitate mitophagy. Finally, many of the LIR domain-containing mitophagy receptors undergo transcriptional upregulation during developmental stages when mitochondria are eliminated, or during hypoxia [1151, 1188]. Changes in their expression can be used to gauge the potential for undergoing mitophagy, rather than as an estimate of mitophagy activity.
Previously, it was suggested that mitophagy can be divided into three types [1199]; however, this was based largely upon in vitro data. In vivo data from reporter animals suggests a simpler classification that has reached consensus in the field. In terms of mitophagy classifications in vitro: Type 1 mitophagy, involves the formation of a phagophore, and typically also requires mitochondrial fission; the PtdIns3K complex containing BECN1 mediates this process. In contrast, type 2 mitophagy is independent of BECN1 and takes place when mitochondria have been damaged [118], resulting in depolarization; sequestration involves the coalescence of GFP-LC3 membranes around the mitochondria rather than through fission and engulfment within a phagophore. Receptor-dependent mitophagy is found in the BECN1-independent pathway. In type 3 mitophagy, mitochondrial fragments or vesicles from damaged organelles are sequestered through a microautophagy-like process named micromitophagy that is independent of ATG5 and LC3, but requires PINK1 and PRKN; in mammals, this process occurs through the formation of mitochondria-derived vesicles/MDVs, small vesicles delivering damaged mitochondrial components to lysosomes for degradation.
Although the process of pexophagy is prominent and well described in yeast cells [1059, 1200], relatively little work has been done in the area of selective mammalian peroxisome degradation by autophagy (for a review see ref. [1201]). Typically, peroxisomes are induced by treatment with hypolipidemic drugs such as clofibrate, ciprofibrate or dioctyl phthalate, which bind to a subfamily of nuclear receptors, referred to as PPARs (peroxisome proliferator activated receptors) [1202]. Of note, while inducing peroxisomal proliferation, PPARA/PPARα may regulate neuronal autophagy, in physiological or pathological settings, such as AD models [921, 1203]. Degradation of excess organelles is induced by drug withdrawal, although starvation without prior proliferation can also be used. EPAS1 activation in liver-specific vhl−/− and vhl−/− hif1a−/− mice reduces peroxisome abundance by pexophagy, whereas ER and mitochondrial protein levels are not affected [774]. Pexophagy can also be induced by amino acid starvation, which induces the stabilization of the peroxisomal E3 ubiquitin ligase PEX2 [1204]. PEX2 is destabilized by MTORC1 such that the overexpression of PEX2 can induce pexophagy. PEX2 ubiquitinates PEX5 and ABCD3/PMP70 (ATP binding cassette subfamily D member 3), which then recruit NBR1 to target the peroxisome for pexophagy [1204]. The action of PEX2 is counteracted by the deubiquitinating enzyme USP30 [1205, 1206]. Pexophagy can also be induced by the expression of a nondegradable active EPAS1 variant [1207]. Induction of pexophagy in response to endogenous and exogenous reactive oxygen species (ROS) and reactive nitrogen species has been observed in mammalian cells. In this setting, pexophagy is induced via ROS- or reactive nitrogen species-mediated activation of ATM/ataxia telangiectasia mutated (ATM serine/threonine kinase) [1208, 1209], repression of MTORC1 and phosphorylation of PEX5 by ATM [1210, 1211]; ATM phosphorylation of PEX5 at S141 triggers PEX5 ubiquitination and binding of SQSTM1 to peroxisomes targeted for pexophagy [1211].
Loss of peroxisomes can be followed enzymatically or by immunoblot, monitoring enzymes such as ACOX/fatty acyl-CoA oxidase (note that this enzyme is sometimes abbreviated “AOX,” but should not be confused with the enzyme alcohol oxidase that is frequently used in assays for yeast pexophagy) or CAT (catalase), and also by EM, cytochemistry or immunocytochemistry [1212–1215]. Finally, a HaloTag®-PTS1 marker that is targeted to peroxisomes has been used to fluorescently label the organelle [1216]. An alternative approach uses a peroxisome-specific tandem fluorochrome assay (RFP-EGFP localizing to peroxisomes by the C-terminal addition of the tripeptide SKL, or a peroxisomal membrane protein tagged with mCherry-mGFP), which has been used to demonstrate the involvement of ACBD5/Atg37, NBR1 and SQSTM1 in mammalian and fungal pexophagy [458, 459, 1052]. By showing that PEX14 directly interacts with LC3-II, which is competitively inhibited by PEX5, PEX14 is demonstrated to function in the dual processes of biogenesis and degradation of peroxisomes with the coordination of PEX5 in response to environmental changes [1217, 1218]. Peroxisomal proteins are degraded preferentially over cytosolic proteins in CHO-K1 cells when starved and then cultured in a normal culture medium. Degradation of peroxisomes is dependent on LC3 and PEX14 [1219]. By making use of autophagy inhibitors or siRNA against NBR1, ubiquitin- and NBR1-mediated pexophagy is shown to be induced by increased expression of PEX3 in mammalian cells, where ubiquitination of PEX3 is dispensable for pexophagy [1220, 1221]. Another autophagic receptor protein, SQSTM1, is required only for the clustering of peroxisomes.
Cautionary notes: There are many assays that can be used to monitor specific types of autophagy, but caution must be used in choosing an appropriate marker(s). To follow mitophagy it is required to monitor more than one protein and to include an inner membrane and a matrix component (and preferably encoded by the mitochondrial DNA) in the analysis to evaluate mass, and not be biased by selective clearance of proteins located in different submitochondrial compartments. In this regard, it is not sufficient to follow a single mitochondrial outer membrane protein because it can be degraded independently of mitophagy through the UPS. Although the localization of PRKN to mitochondria as monitored by fluorescence microscopy is associated with the early stages of CCCP-driven mitochondria degradation [339], this by itself cannot be used as a marker for mitophagy, as these events can be dissociated [1222]. Even with PRKN translocation and ubiquitination, FCCP-induced donut mitochondria resist autophagy, by failing to recruit autophagy receptors CALCOCO2/NDP52 and OPTN [1223]. Moreover, mitophagy elicited in a number of disease models and by pharmacological means [1224]) does not involve mitochondrial PRKN translocation [201, 460, 1225]. Along these lines, recent studies implicate an essential role for TRAF2, an E3 ubiquitin ligase, as a mitophagy effector in concert with PRKN in cardiac myocytes; whereby mitochondrial proteins accumulate differentially with deficiency of either, indicating nonredundant roles for these E3 ubiquitin ligases in mitophagy [1226]. This finding necessitates an integrated approach to assess mitophagy based on a broad evaluation of multiple mitochondrial effectors and proteins. Because PINK1-PRKN-dependent mitophagy can only be detected under certain non-physiological conditions, it is a controversial matter of debate as to the role of PINK1-PRKN during mitophagy, and whether basal and stimulus (e.g., age)-induced mitophagy are regulated through the same pathways or employ distinct machineries [1227].
During canonical PRKN-mediated mitophagy, PRKN translocates to damaged mitochondria and ubiquitinates a wide range of outer membrane proteins including VDAC1, MFN1/2 and TOMM20 [1128, 1153, 1154, 1228, 1229]. This results in the preferential degradation of OMM proteins by the proteasome, while inner membrane proteins and mitochondrial DNA [1230] remain intact. Monitoring loss of a single protein such as TOMM20 by western blot or fluorescence microscopy to follow mitophagy may thus be misleading, as noted above [1228]. Similarly, following the level of DNM1L may provide some information with regard to mitophagy, but it must be kept in mind that alterations in mitochondrial dynamics and DNM1L recruitment to mitochondria mostly occur in response to conditions other than mitophagy, such as changing nutrient concentrations. MitoTracker® dyes are widely used to stain mitochondria and, when colocalized with GFP-LC3, they can function as markers for mitophagy. However, staining with MitoTracker® dyes depends on mitochondrial membrane potential (although MitoTracker® Green FM is less sensitive to loss of membrane potential), so that damaged, or sequestered nonfunctional mitochondria may not be stained. In vitro this can be avoided by labeling the cells with MitoTracker® before the induction by the mitophagic stimuli [1123]. One additional point is that MitoTracker® dyes might influence mitochondrial motility in axons (D. Ebrahimi-Fakhari, personal communication).
Although it is widely assumed that autophagy is the major mechanism for degradation of entire organelles, there are multiple mitochondrial quality control mechanisms that may account for the disappearance of mitochondrial markers. These include proteasomal degradation of outer membrane proteins and/or proteins that fail to correctly translocate into the mitochondria, degradation due to proteases within the mitochondria, and reduced biosynthesis or import of mitochondrial proteins. PINK1 and PRKN are not essential for all types of mitophagy in vitro or in vivo [466, 1188, 1231]. Moreover, these two proteins also participate in an ATG gene-independent pathway for lysosomal degradation of small mitochondria-derived vesicles [791]. An unbiased proteomic study in vivo shows that PRKN ubiquitinates not only OMM proteins during mitophagy but also several proteins that a priori are unrelated to mitophagy [1229]. Furthermore, the PINK1-PRKN mitophagy pathway is also transcriptionally upregulated in response to starvation-triggered generalized autophagy, and is intertwined with the lipogenesis pathway [1232–1235]. In addition to mitophagy, mitochondria can be eliminated by extrusion from the cell (mitoptosis) [1112, 1128, 1139, 1236]. Transcellular degradation of mitochondria, or transmitophagy, also occurs in the nervous system when astrocytes degrade axon-derived mitochondria [1237]. Thus, it is advisable to use a variety of complementary methods to monitor mitochondria loss including TEM, single-cell analysis of Atg-family protein fluorescent puncta that colocalize with mitochondria, and western blot, in conjunction with flux inhibitors and specific inhibitors of autophagy induction compared with inhibitors of the other major degradation systems (see cautions in Autophagy inhibitors and inducers).
To monitor and/or rule out changes in cellular capacity to undergo mitochondrial biogenesis, a process that is tightly coordinated with mitophagy and can dictate the outcome following mitophagy-inducing insults especially in primary neurons and other mitochondria-dependent cells, colocalization analysis after double staining for the mitochondrial marker TOMM20 and BrdU (for visualization of newly synthesized mtDNA) can be performed (Figure 30). Alternatively, direct assay for translation of mtDNA-encoded proteins is a straightforward assay for mitochondrial biogenesis, which can be combined with analysis of transcripts driven by mtDNA promoters [1238, 1239].
Figure 30.
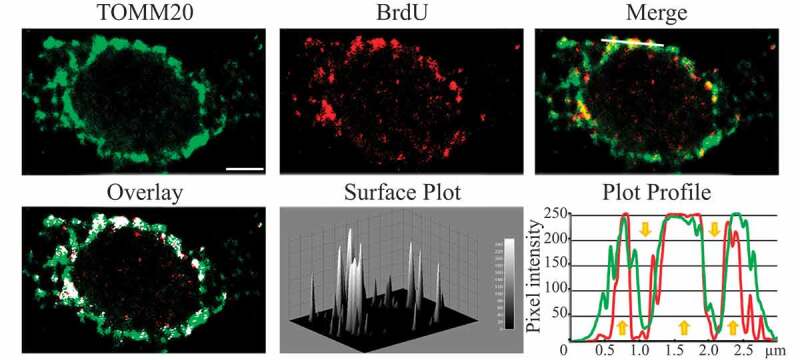
Confocal microscopy deconvolved (AutoQuant X3) images and colocalization image analysis (ImageJ 1.47; Imaris 7.6) through a local approach showing perinuclear mitochondrial biogenesis in hippocampal neuronal cultures. The upper channels show TOMM20 (green channel), BrdU (for visualization of newly synthesized mitochondrial DNA, red channel), and merged fluorescence channels. Overlay, corresponds to the spatial pattern of software thresholded colocalized structures (white spots) layered on the merged fluorescence channels. Surface Plot, or luminance intensity height, is proportional to the colocalization strength of the colocalized structures (white spots). Plot Profile, corresponds to the spatial intensity profiles of the fluorescence channels of the white line positioned in the Merge image. Yellow arrows indicate a qualitative evaluation of the spatial association trends for the fluorescence intensities. Arrows pointing up indicate an increase in the colocalization, whereas arrows pointing down show a decrease. Scale bar: 2 μm. This figure was modified from previously published data [4085] and is provided by F. Florenzano
Likewise, although the mechanism(s) of peroxisomal protein degradation in mammals awaits further elucidation, it can occur by both autophagic and proteasome-dependent mechanisms [1219]. Thus, controls are needed to determine the extent of degradation that is due to the proteasome. Moreover, two additional degradation mechanisms have been suggested: the action of the peroxisome-specific LONP2/Lon (lon peptidase 2, peroxisomal) protease and the membrane disruption effect of 15-lipoxygenase [1240].
e. Chlorophagy
Besides functioning as the primary energy suppliers for plants, chloroplasts represent a major source of fixed carbon and nitrogen to be remobilized from senescing leaves to storage organs and newly developing tissues. As such, the turnover of these organelles has long been considered to occur via an autophagy-type mechanism. However, while the detection of chloroplasts within autophagic body-like vesicles or within vacuole-like compartments has been observed for decades, only recently has a direct connection between chloroplast turnover and autophagy been made through the analysis of atg mutants combined with the use of fluorescent ATG8 reporters [1241–1244]. In fact, it is now clear that chlorophagy, the selective degradation of chloroplasts by autophagy, can occur via several routes, including the encapsulation of whole chloroplasts by the tonoplast via a microautophagy-type process [1244], or the budding of chloroplast material into small distinct autophagic vesicles called Rubisco-containing bodies (RCBs) and ATI1 (ATG8-interacting protein 1) plastid-associated (ATI-PS) bodies, which then transport chloroplast cargo to the vacuole [1241, 1245]. Chloroplasts produce long tubes called stromules that project out from the organelle outer membrane. Recent studies suggest that stromules are part of the chlorophagy process, by which the stromule tips, presumably containing unwanted or damaged chloroplast material, are engulfed by autophagic membranes using ESCRT-II endocytic machinery that depends on ATG8 [1246]. Chloroplast morphology can easily be monitored by TEM, whereas chloroplast abundance and association with autophagic membranes can be studied by confocal fluorescence microscopy using chlorophyll autofluorescence in combination with appropriate fluorescent protein markers (e.g., stromally-targeted GFP, GFP-ATG8, or tonoplast markers such as GFP-TIP2/δTIP or VHP1-GFP). The appearance of RCBs is tightly linked with leaf carbon status, indicating that chlorophagy through RCBs represents an important route for recycling plant nutrients provided in plastid stores. As such, it is critical to maintain consistent plant growth conditions, particularly with respect to light quality and intensity, and to take into account that different responses may be observed depending on the time of day experiments are performed.
f. Chromatophagy
Autophagy is best known for its pro-survival role in cells under metabolic stress and other conditions. However, excessively induced autophagy may be cytotoxic and may lead to cell death (Figure 31) [1247]. Chromatophagy (chromatin-specific autophagy) comes into view as one of the autophagic responses that can contribute to cell death [1248]. Chromatophagy can be seen in cells during nutrient depletion, such as arginine starvation, and its phenotype consists of giant-autophagosome formation, nucleus membrane rupture and histone-associated-chromatin/DNA leakage that is captured by phagophores. Arginine starvation can be achieved by adding purified arginine deiminase to remove arginine from the culture medium, or by using arginine-dropout medium. The degradation of leaked nuclear DNA/chromatin can be observed by fluorescence microscopy; with GFP-LC3 or anti-LC3 antibody, and LysoTracker™ Red or anti-LAMP1, multiple giant autophagosomes or autolysosomes containing leaked nuclear DNA can be detected. In addition, the chromatophagy-related autophagosomes also contain parts of the nuclear outer-membrane, including NUP98 (nucleoporin 98 and 96 precursor), indicating that the process involves a fusion event [1248].
Figure 31.
Pathways that follow DNA damage may result in PCD or cell survival through autophagy, or checkpoint deficiency. (A) DNA damage inhibits TOR signaling, which promotes the formation of an ATG protein complex, thereby bringing about autophagy. Autophagy may contribute to cell survival as well as to cell death by PCD. (B) DNA damage may cause mutations in genes encoding checkpoint RAD proteins and/or proteins involved in DNA replication initiation, as well as the simultaneous deletion of genes encoding checkpoint kinases. Such protein defects lead to deficient checkpoints, thereby causing cells to enter M-phase prematurely with damaged DNA or incompletely replicated DNA, resulting in ROS generation, mitotic catastrophe, and subsequently PCD. This figure was previously published in ref. [1247]
g. Clockophagy
Clockophagy is the process of selective autophagic degradation of the key circadian clock protein ARNTL/BMAL1 (aryl hydrocarbon receptor nuclear translocator-like) during RSL3-induced ferroptosis in Calu-1 and HT1080 cells [1249]. SQSTM1 is a cargo receptor responsible for clockophagy-dependent ARNTL degradation during ferroptosis. Clockophagy-dependent ARNTL degradation dramatically promotes ferroptotic cancer cell death through EGLN2/PHD1 (egl-9 family hypoxia-inducible factor 2)-mediated oxidative injury in vitro and in vivo. The interactome map of the arntl/bmal1−/− mouse, an arrhythmic circadian rhythms model, reveals significant loss of genes encoding proteins such as COL6A/collagen VI and autophagy-related genes such as SQSTM1 [1250]. Given the importance of the accumulated data from both mice and human studies, deregulation of Clock genes might lead to enhanced autophagy through ATG14, whereas downregulated autophagy through the AKT pathway may be involved in the pathogenesis of COL6A myopathy and potentially contribute to other muscle-wasting diseases.
h. Crinophagy and the SINGD pathway
Distinct from cargo disposal that involves autophagosomes, crinophagy, the degradation of secretory granules via direct fusion with lysosomes, was discovered in the 1960s as a pituitary gland response to the inhibition of exocytosis [1251]. Crinophagy has been observed in different types of secretory cells including cells of the anterior pituitary gland, pancreatic α cells and β cells [1251–1254]. Traditionally, crinophagy was monitored using electron microscopy and immunoelectron microscopy. Newer molecular biology techniques have been employed to study crinophagy in the salivary gland of Drosophila [1255] and in mammalian pancreatic β cells [1256]. In Drosophila, reporter lines expressing granule and lysosomal markers with fluorescent tags have been used to assess crinophagic degradation of glue granules at different time points and elucidate the molecular mechanisms of this pathway [1255]. In β cells, short-term nutrient deprivation evokes rapid autophagy-independent lysosomal degradation of nascent INS (insulin) secretory granules, the pathway termed “stress-induced nascent granule degradation” (SINGD; pronounced ˈsindi). SINGD occurs via crinophagy and counters autophagy through localized activation of MTORC1; the depletion of secretory granules together with the inhibition of autophagy protect against unwanted INS release during fasting [120]. The major regulator of secretory granule biogenesis at the trans-Golgi network, PRKD (protein kinase D), controls SINGD, thus routing secretory granules to secretion or degradation depending on the nutrient availability. Furthermore, erroneous activation of the SINGD pathway contributes to β cell failure in type 2 diabetes [1256]. To further characterize the dynamics of the crinophagic SINGD pathway in β cells, the sequences coding fluorescent tags have been inserted directly into the endogenous loci of the secretory granule marker PTPRN2/Phogrin and the lysosomal protein CD63 using CRISPR-Cas9 gene-editing. This tool makes it possible to follow crinophagy in real time using several imaging techniques, including live-cell imaging combined with CLEM (live-CLEM). In addition, 3-dimensional reconstruction of large cellular volumes achieved by FIB-SEM is particularly helpful to detect crinophagic events in primary islets.
i. Doryphagy
Centriolar satellites (CSs) are protein complexes associated with microtubules and clustering around the centrosome. Whereas CSs have long been described as the structures regulating centrosome composition, the mechanisms controlling CS homeostasis and function are not yet understood in detail [1257]. A process targeting CSs for selective autophagy has been identified and termed “doryphagy”, from the Greek word “doryphoros” for satellites [1258]. Of note, the selective degradation of CSs is achieved by a LIR-mediated interaction between PCM1, a component of CSs, and GABARAPs. As a consequence of CS function in regulating centrosomes, disruption of doryphagy results in centrosome abnormalities and aberrant mitosis.
j. Ferritinophagy
Ferritinophagy is a selective form of autophagy that functions in intracellular iron processing [805]. Iron is recruited to ferritin for storage and to prevent the generation of oxygen free radicals through the Fenton and Haber-Weiss reactions [806,807]. Because ferritin is largely degraded by autophagy [1259, 1260] the ferritin status can be used as a marker of the autophagic flux in a given cell. To release iron from ferritin, the iron-bound form is sequestered within an autophagosome [1261]. Fusion with a lysosome leads to breakdown of ferritin and release of iron. Furthermore, iron can be acidified in the lysosome, converting it from an inactive state of Fe3+ to Fe2+ [809,810]. Iron can be detected in the autolysosome via TEM [809]. Colocalization of iron with autolysosomes may also be determined utilizing calcein AM to tag iron [1262, 1263]. NCOA4 is a cargo receptor that recruits ferritin to the autophagosome [1264]. NCOA4-dependent ferritinophagy promotes ferroptosis, an iron-dependent form of regulated cell death (RCD) [1265, 1266], by the degradation of ferritin in multiple cells [1267] as discussed below. Note that ferritinophagy can be co-opted by pathogens for their own survival. For example, uropathogenic E. coli persist in host cells by taking advantage of ferritinophagy. Iron overload in urothelial cells induces ferritinophagy in a NCOA4-dependent manner causing increased iron availability for uropathogenic E. coli to overgrow, which can be reversed by inhibition of autophagy [1268].
Ferroptosis is currently defined as a form of programmed cell death initiated by oxidative perturbations of the intracellular microenvironment, that is under constitutive control by GPX4. This form of programmed cell death may be accompanied by excessive autophagy initiated after administration of erastin and glutamate, which results in inactivation of SLC7A11/cystine transporter/xCT. Uptake of cystine is essential for glutathione synthesis, and, therefore, a deficient SLC7A11 transporter will promote lipid peroxidation due to depletion of GPX4 (glutathione peroxidase 4) protein and activity [1269]. Cysteine deprivation also causes endoplasmic reticulum stress resulting in induction of DDIT4/REDD1 [1270]. DDIT4 acts as an inducer of autophagy by binding and inhibiting YWHA/14-3-3, which otherwise inhibits the TSC1-TSC2 complex, and ultimately leads to inhibition of MTOR. Increased autophagy induced by DDIT4 causes ferritin to be degraded and iron released to promote ferroptosis, whereas inhibition of autophagy protects against ferroptosis [1267]. Recent reports indicate that GPX4 depletion is facilitated by CMA involving HSP90. Inhibition of HSP90 using 2-amino-5-chloro-N,3-dimethylbenzamide (CDDO) can spare GPX4 depletion and rescue erastin-mediated cell death [1271]. Moreover, upregulation of the RNA-binding protein ELAVL1/HuR promotes BECN1 production via binding to the AU-rich elements (AREs) in the 3ʹ UTR of BECN1 mRNA, thus triggering autophagy activation, promoting autophagic ferritin degradation, and eventually leading to iron-dependent ferroptosis [1272]. Conversely, upregulation of the RNA-binding protein ZFP36 (ZFP36 ring finger protein) can result in ATG16L1 mRNA decay via binding to the AREs in the 3ʹ UTR, thus triggering autophagy inactivation, blocking autophagic ferritin degradation, and eventually conferring resistance to ferroptosis [1273].
k. Granulophagy
Granulophagy is a term generally applicable to the autophagic clearance of mRNA-protein granules in eukaryotic cells. First termed to describe the clearance of stress granules in S. cerevisiae and human cell lines [1274], other mRNP granules subject to autophagic clearance include P-bodies in mammalian cells [1275] and P-granules in C. elegans [1276]. Evidence that granulophagy is a selective autophagic process includes the identification of granule-specific autophagic receptor proteins, including SQSTM1 for stress granules in human cells, CALCOCO2 for P-bodies in human cells and SEPA-1 for P-granules in C. elegans [1275, 1276]. In all cases, the receptor proteins colocalize in their respective mRNP granules while autophagic clearance occurs, and the absence of said receptor proteins leads to accumulation of the mRNP granule. SQSTM1- and LC3-adorned bodies resembling stress granules also localize in autophagosomes as revealed by electron microscopy [1277].
Granulophagy studies with stress granules suggest induction varies depending on cellular context. For example, yeast stress granules induced by transient nutrient deprivation or oxidative stress are not targeted by granulophagy, whereas diauxic shift and inhibition of mRNA decay do induce granulopahgy [1274]. Additionally, studies involving various stress stimuli (e.g., heat shock, proteasome inhibition, or arsenite stress) in human cell lines reveal differing degrees of importance of autophagic versus chaperone-based mechanisms in the disassembly or degradation of stress granules [1274, 1275, 1277–1279]. Stress granule clearance following heat shock may involve migration via microtubules of stress granules to aggresomes, based on colocalization studies in the presence and absence of autophagic inhibitors [1279]. Thus, granulophagy and aggrephagy mechanisms may overlap in at least some cases. Moreover, stalled 48S translation pre-initiation complexes, forming stress granules upon accumulation and condensation, are found within exosomes secreted by cells submitted to prolonged serum starvation, a process enhanced by ATG5 depletion [822].
Granulophagy may affect the pathology of amyotrophic lateral sclerosis (ALS). Aberrant persistence or formation of stress granules has been hypothesized to facilitate formation of toxic cytoplasmic aggregates containing TARDBP or FUS RNA-binding proteins [1280]. Mutations in VCP that are associated with ALS onset also impair granulophagy and lead to persistence of TARDBP-containing stress granules in human cell models [1274]. ALS-mutant forms of FUS also induce aberrant stress granule assembly in neuronal cells, and lead to increased stress granule association with autophagosomes versus stress granules formed in control cells, suggesting granulophagy exerts selective clearance of potentially pathological stress granules [1281]. Finally, the most commonly mutated gene in ALS patients, C9orf72, may also function with SQSTM1 in autophagic clearance of FUS-containing stress granules. Supporting this, C9orf72 physically interacts with SQSTM1 and localizes in stress granules, and its depletion impairs stress granule clearance following arsenite stress [1277].
l. Intraplastidial autophagy
Intraplastidial autophagy is a process whereby plastids of some cell types adopt autophagic functions, engulfing and digesting portions of the cytoplasm. These plastids are characterized by formation of invaginations in their double-membrane envelopes that eventually generate a cytoplasmic compartment within the plastidial stroma, isolated from the outer cytoplasm. W. Nagl coined the term plastolysome to define this special plastid type [1282]. Initially, the engulfed cytoplasm is identical to the outer cytoplasm, containing ribosomes, vesicles and even larger organelles. Lytic activity was demonstrated in these plastids, in both the cytoplasmic compartment and the stroma. Therefore, it was suggested that plastolysomes digest themselves together with their cytoplasmic cargo, and transform into lytic vacuoles. Intraplastidial autophagy has been reported in plastids of suspensor cells of Phaseolus coccineus [1282] and Phaseolus vulgaris [1283], where plastids transformed into autophagic vacuoles during the senescence of the suspensor. This process was also demonstrated in petal cells of Dendrobium [1284], and in Brassica napus microspores experimentally induced towards embryogenesis [1285]. All these reports established a clear link between these plastid transformations and their engagement in autophagy. At present, descriptions of this process are limited to a few, specialized plant cell types. However, pictures of cytoplasm-containing plastids in other plant cell types have been occasionally published, although the authors did not make any mention of this special plastid type. For example, this has been seen in pictures of fertile and Ogu-INRA male sterile tetrads of Brassica napus [1286], and Phaseolus vulgaris root cells [1287]. Possibly, this process is not as rare as initially thought, but authors have only paid attention to it in those cell types where it is particularly frequent.
m. Lipophagy
The specific autophagic degradation of lipid droplets represents another type of selective autophagy [1288]. Lipophagy requires the core autophagic machinery and can be monitored by following triglyceride content, or total lipid levels using BODIPY 493/503 or HCS LipidTOX neutral lipid stains with fluorescence microscopy, cell staining with Oil Red O, the cholesterol dye filipin III [1289], or ideally label-free techniques such as coherent anti-stokes Raman scattering/CARS or spontaneous Raman scattering/SRS microscopy. BODIPY 493/503 should be used with caution, however, when performing costains (especially in the green and red spectra) because this commonly used fluorescent marker of neutral lipids is highly susceptible to bleed-through into the other fluorescence channels (hence often yielding false positives), unlike the LipidTOX stain that has a narrow emission spectrum [1290]. In addition, BODIPY 493/503 cannot be used to monitor lipophagy in C. elegans because it stains both lipid droplets and the lysosome [1291]. TEM can also be used to monitor lipid droplet size and number, as well as lipid droplet-associated double-membrane structures, which correspond to autophagosomes [1288, 1292, 1293].
The transcription factor TFEB positively regulates lipophagy [948], and promotes fatty acid β-oxidation [1294], thus providing a regulatory link between different lipid degradation pathways [1295]. Accordingly, TFEB overexpression rescues fat accumulation and metabolic syndrome in a diet-induced model of obesity [1294, 1296] and in alcohol-induced fatty liver in mice [1297]. As a coactivator for TFEB and PPARG, CARM1 regulates lysosome biogenesis and lipid metabolism through processes that are partially dependent on lipophagy [980, 1298]. Under conditions of nutrient starvation, CARM1-TFEB-mediated lipophagy is regulated by C9orf72 [1299, 1300]. Genetic mutations in C9orf72 are linked to neurodegenerative diseases including ALS and frontotemporal dementia (FTD) [1301, 1302]. Spermidine can also stimulate autophagy in adipose tissue, reducing visceral fat and obesity-associated alterations upon hypercaloric regimens [1303]. Expression of the C. elegans lysosomal lipases lipl-1, lipl-3, and lipl-4 tightly correlates with activation of autophagy in the conditions so far tested [948, 1304, 1305], and this transcriptional activation is necessary for optimal lipid mobilization in conditions of autophagy activation such as fasting [948, 1304].
The antioxidant enzyme PRDX1 (peroxiredoxin 1) is expressed most highly in macrophages, and plays an essential role in regulation of lipophagic flux and maintenance of cholesterol homeostasis against oxidative stress within atherosclerotic macrophages [1306]. The regulation of expression of lipid droplet regulators (such as the PLIN/perilipin family) and of autophagy adaptors (such as the TBC1D1 family) during starvation and disease deserves further exploration [1307–1309]. Members of the PNPLA (patatin like phospholipase domain containing) protein family, PNPLA1 [1310], PNPLA2 [1311, 1312] and PNPLA3 [1313], as lipid droplet residents, play essential roles in lipophagy by regulating lipid droplet size and autophagic flux. Although a physiological receptor protein and specific induction signal for lipophagy are poorly understood, expression of a fusion protein of SQSTM1 and a lipid droplet-binding domain can induce forced lipophagy to promote the breakdown of lipid droplets [1314]. Coating PLINs (perilipins) can also be degraded through CMA, facilitating access of cytosolic lipases to the esterified lipids stored in the droplet [1315]. Lipophagy is often monitored in vitro using cell culture media supplemented with fatty acids to promote the formation of intracellular lipid droplets. Caution should be taken with the assessment and interpretation of lipophagy data in adipocytes. This cell type shows spontaneous physiological accumulation of lipid droplets, in contrast with cells in which lipid droplet accumulation is experimentally forced and is associated with lipotoxicity.
Cautionary notes: With regard to changes in the cellular neutral lipid content, the presence and potential activation of cytoplasmic lipases that are unrelated to lysosomal degradation must be considered. Caution should also be taken when interpreting lipophagy data using autophagy-related gene knockout mice. In response to fasting or diet-induced obesity, liver-specific rb1cc1, atg7 or atg5 knockout mice have decreased hepatic lipid accumulation, which is likely due to an adaptive response that includes increased FGF21 production and NFE2L2 activation in these knockout mice as a result of chronic impaired hepatic autophagy [1316–1318].
n. Lysophagy
Lysophagy is a selective autophagy process that participates in cellular quality control through lysosome turnover. By eliminating ruptured lysosomes, lysophagy prevents the subsequent activation of the inflammasome complex and innate response [1319–1321]. The conserved autophagy machinery of D. discoideum also localizes at lysosomes damaged by lysosomotropic agents such as LLOMe (polymers of Leu-Leu-OMe). It has been proposed that autophagy, which also occurs at damaged compartments containing the bacterial pathogen M. marinum, plays a role in both the repair of the damaged compartment and its total engulfment for degradation [1322].
o. Myelinophagy
Myelinophagy or Schwann cell autophagy refers to selective autophagic degradation of myelin from Schwann cells in order to avoid or to reduce myelin debris and aggregates following peripheral nerve injury [1323]. An efficient Schwann cells myelin clearance, an early event in Wallerian degeneration, counteracts inflammatory processes facilitating recovery and nerve regeneration [1324, 1325].
Schwann cells form autophagosomes in response to nerve injury. Inhibition of autophagy using both pharmacological inhibitors or genetic manipulation of autophagic genes (such as Ambra1 and Atg7) leads to a severe neuropathy in response to injury, in vitro and in vivo [540, 1326, 1327]. A fundamental role of myelinophagy in peripheral neurodegeneration (i.e., demyelinating diseases) has been recognized [1328].
p. Nucleophagy
Nuclear autophagy is a mechanism by which cells maintain cellular homeostasis and ensure nuclear integrity, stability and correctness of gene expression. Targeted removal of nuclear material, part of or the entire nucleus, from a cell by autophagy (i.e., nucleophagy) has been reported as a selective mode occurring by autophagy as well as microautophagy [4]. The nuclear membrane may contribute to the phagophore membrane in addition to being an autophagic target. In autophagy, phagophores can sequester the nucleus-derived cargo, and autophagosomes subsequently merge with the vacuole or lysosomes, leading to the degradation of their contents [1329–1331]. In micronucleophagy, satellite nuclei are formed due to stress or genome instability and then engulfed directly [1077, 1332, 1333]. An alternative mechanism of nucleophagy has been reported in Saccharomyces cerevisiae, which is mediated by Atg39, a nuclear envelope receptor inducing autophagic sequestration of localized parts of the nucleus [1334].
The autophagy marker LC3 is expressed in the nucleus of human primary fibroblasts where it can directly interact with the nuclear lamina protein LMNB1 (lamin B1) [1335]; this process is associated with extensive DNA damage, and is triggered by oncogenic insult and senescence. The interaction of LC3 with LMNB1 does not downregulate LMNB1 during starvation, but can mediate its degradation upon oncogenic stress, providing a general mechanism to protect the cells from oncogene-induced senescence and tumorigenesis. Nucleophagy can thus be monitored through a quantification of the colocalization between LMNB1 and (GFP)-LC3 in puncta in the cytoplasm and in the nucleus, or through a dual fluorescent RFP-GFP-LMNB1 [1335].
q. Oxiapoptophagy
There are now several lines of evidence indicating that autophagy is an essential process in vascular and neurological functions. Autophagy can be considered as atheroprotective in the early stages of atherosclerosis, and dysfunctional in advanced atherosclerotic plaques [1336]. A deregulated, amplified or attenuated autophagy process at different levels of the activation pathway appears to be associated with several neurodegenerative diseases [1337]. Currently, little is known about the molecules that promote autophagy on the cells of the vascular wall and on neural cells (glial and microglial cells, neurons). As increased levels of cholesterol oxidation products (named oxysterols) are found in atherosclerotic lesions [1338], and in the brain, cerebrospinal fluid and/or plasma of patients with neurodegenerative diseases [1339], the part taken by these molecules has been investigated, and several studies support the idea that some of them could contribute to the induction of autophagy [1339–1342]. There are several lines of evidence that oxysterols, especially 7-ketocholesterol and 7β-hydroxycholesterol, which can be increased under various stress conditions in several age-related diseases including vascular and neurodegenerative diseases [1339], could trigger a particular type of autophagy termed oxiapoptophagy (OXIdation + APOPTOsis + autoPHAGY) [1343] characterized by the simultaneous induction of oxidative stress associated with apoptosis, and autophagic criteria in different cell types from different species [1344–1346]. As oxiapoptophagy has also been observed with 7β-hydroxycholesterol and 24(S)-hydroxycholesterol, which are potent inducers of cell death, it is suggested that oxiapoptophagy could characterize the effect of cytotoxic oxysterols [1344]. In addition, following treatment with 7β-hydroxycholesterol, in 158N murine oligodendrocytes, there is evidence of a link between 7β-hydroxycholesterol-induced oxiapoptophagy and inflammation [1346, 1347].
In any case, care must be taken in assigning an autophagy activating role to cholesterol-related compounds. Most of these studies usually consider such compounds as autophagy inducers because of their ability to convert LC3-I to LC3-II. However, the conversion of LC3 and/or the accumulation of LC3-labeled autophagosomes might be due to the blockade of this pathway at a later stage, as happens for some autophagy blockers such as CQ [302, 1348, 1349]. Furthermore, an increase in ROS generation is also commonly reported in these studies, which other authors have associated with lysosomal pH increases that ultimately prevent the fusion of lysosomes with autophagosomes [1348]. In this context, it is notable that the imbalance of membrane cholesterol has already been described to induce the generation of ROS [1350, 1351].
r. Proteaphagy
The autophagic degradation of 26S proteasome complexes has been reported in plants [1352–1354], yeast [1355, 1356], and humans [1357]. Two pathways for degradation have been reported: an ATG1-dependent pathway triggered by nutrient starvation, and an ATG1-independent pathway stimulated by chemical or genetic inhibition [1352, 1353]. Starvation-induced proteaphagy occurs in response to nitrogen but not carbon starvation in Arabidopsis and yeast [1355], as carbon starvation instead triggers relocalization of proteasomes into cytoplasmic proteasome storage granules that offer protection against autophagy [1358]. However, if proteasome storage granule formation is blocked, proteaphagy also becomes the default response to carbon starvation. While little is currently known about the selectivity of starvation-induced proteaphagy in plants and yeast, in humans it appears to involve subunit ubiquitination and the autophagy receptor SQSTM1 [1357].
Inhibitor-induced proteaphagy also involves extensive ubiquitination of proteasome subunits to facilitate binding of autophagy receptors. In yeast, proteasomes first aggregate in the cytosol in an Hsp42-dependent manner, before the receptor Cue5 tethers the ubiquitinated, aggregated proteasomes to the expanding phagophore [1353]. In Arabidopsis, RPN10 instead acts as the receptor [1352]. RPN10 is a ubiquitin receptor within the proteasome regulatory particle, but is an unusual proteasome subunit as it also exists as a free form in the cytosol. The free form can bind ubiquitinated proteasome subunits via a standard ubiquitin-interacting motif (UIM), and also binds ATG8 via a related UIM-like sequence, rather than a canonical AIM/LIR [1359]. This casts RPN10 as the founding member of a new class of UIM-containing autophagy adaptors and receptors that are conserved across kingdoms. The exact subunits and residues to be ubiquitinated during proteaphagy, and the E3 ligases involved, are currently unknown.
As with other types of selective autophagy, proteaphagy can easily be monitored using fluorescently tagged proteasome subunits. Numerous core protease and regulatory particle subunits have been successfully tagged [1360], although care should be taken to ensure that the tag does not interfere with incorporation of the subunit into the proteasome particle. Once tagged, proteasome delivery to the vacuole can be studied by both confocal fluorescence microscopy, and by monitoring the release of free fluorescent protein by immunoblot. It is important to note that proteasome subunit levels do not necessarily correlate with levels of proteaphagy, particularly when studying the inhibitor-induced pathway. This is because synthesis of proteasome subunits is strongly induced upon proteasome inhibition by transcriptional feedback loops involving Rpn4 in yeast, NRF1 in humans and AT5G04410/NAC78 and AT3G10500/NAC53 in Arabidopsis [1360].
When yeast are grown under very low levels of glucose, proteasomes are also taken up directly into vacuoles by microautophagy [1361]. Microautophagy appears biased toward aberrant or inactive proteasomes, with functional proteasomes accumulating in proteasome storage granules. AMPK and ESCRT factors are required for proteasome microautophagy and also affect proteasome storage granule dissipation and nuclear reimport of proteasomes upon glucose refeeding.
s. Reticulophagy
Starvation in yeast induces a type of selective autophagy of the ER [1362], which depends on the autophagy receptors Atg39 and Atg40 [1334]. ER stress also triggers an autophagic response [1363], which includes the formation of multi-lamellar ER whorls and their degradation by a microautophagic mechanism [1364]. ER-selective autophagy has been termed reticulophagy/ER-phagy [1365]. Selective autophagy of the ER has also been observed in mammalian cells [1366], where multiple receptors have been recently characterized [1367–1369]. Reticulophagy receptors are selective not only for the ER itself, but they can also lead to the degradation of specific ER subdomains [1370]. RETREG1/FAM134B was the first ER protein identified as an ER-specific autophagy receptor specific for ER sheets [74]. RTN3 and ATL3 have been described as reticulophagy receptors committed to the degradation of ER tubules ([1371, 1372]; whereas TEX264 is mainly located in the ER 3-way junctions [464, 1373]. SEC62 and CCPG1 are two other reticulophagy receptors with a broader ER distribution. SEC62 is involved in a particular form of reticulophagy (recovER reticulophagy), which reduces the ER size to a normal level after an ER stress is resolved via ESCRT-III driven microreticulophagy [1374, 1375]. In contrast, CCPG1 is activated directly under ER stress conditions [1376]. Because reticulophagy is selective, it is able to act in ER quality control [1370, 1377, 1378], and eliminate protein aggregates that cannot be removed in other ways. In the clearance of specific protein aggregates, the reticulophagy receptors cooperate with other ER proteins such as specific chaperones or elements of the COPII complex [1379–1382]. Moreover, reticulophagy functions to sequester parts of the ER that are damaged by the presence of pathogens such as viruses and bacteria [1383, 1384]. The acetylation of ATG9A within the ER lumen seems to regulate its ability to interact with RETREG1/FAM134B and SEC62, and induce reticulophagy [1385, 1386], a process that might be involved in the maintenance of proteostasis within the ER [1386, 1387]. Reticulophagy can be monitored using reticulophagy reporters such as eGFP-mCherry-SERP1/RAMP4 [465], mCherry-GFP-REEP5 [1372], and ssRFP-GFP-KDEL [464]. These tandem fluorescent protein reporters are detected as yellow signals in the ER, but when they are delivered to lysosomes by autophagy, they become red, as the GFP signal is quenched. Cleavage of these reporters in lysosomes can also be monitored by immunoblotting.
The COPII complex has also been associated with an additional, less understood pathway involving noncanonical, microautophagy-like degradation of ER exit sites (ERES) containing misfolded procollagen [1388]. This pathway is characterized by cargo colocalization with COPII proteins and lysosomal markers without ER membrane or lumen markers; the colocalization is further enhanced by lysosomal hydrolase inhibitors. Cargo selectivity and activation mechanisms for this recently identified pathway have not yet been established.
t. Ribophagy
Autophagy has been reported for the selective removal of ribosomes in yeast, particularly upon nitrogen starvation [1389]; however, it remains unclear whether yeast has a dedicated ribophagy pathway that is activated under conditions of nitrogen starvation. Published papers monitor this process by western blot, following the generation of free GFP from Rpl5-GFP or Rpl25-GFP [1390], or the disappearance of ribosomal subunits such as Rps3. Vacuolar localization of Rpl5-GFP or Rpl25-GFP can also be seen by fluorescence microscopy. The Rkr1/Ltn1 ubiquitin ligase is reported to act as an inhibitor of 60S ribosomal subunit ribophagy via, at least, Rpl25 as a target, and is antagonized by the deubiquitinating Ubp3-Bre5 complex [1389, 1390]. Rkr1/Ltn1 and Ubp3-Bre5 are proposed to contribute to adapt ribophagy activity to both nutrient supply and protein translation. Ribophagy has also been observed in animal cells, for instance in arsenite-treated mammalian cells, as was demonstrated with Ribo-Keima flux assays alongside a variety of other Keima-based flux assays [1037].
u. RNA-silencing components
Several components of the RNA-silencing machinery are selectively degraded by autophagy in different organisms. This was first shown for the plant AGO1/ARGONAUTE1 protein, a key component of the Arabidopsis RNA-induced silencing complex (RISC) that, after ubiquitination by a virus encoded F-box protein, is targeted to the vacuole [1391]. AGO1 colocalizes with Arabidopsis ATG8a-positive bodies, and its degradation is impaired by various drugs such as 3-MA and E64d, or in Arabidopsis mutants in which autophagy is compromised such as the TOR-overexpressing mutant line G548 or the atg7-2 mutant allele [1391]. Moreover, this pathway also degrades AGO1 in a nonviral context, especially when the production of miRNAs is impaired. Defects in miRNA biogenesis also cause autophagic degradation of Drosophila AGO1 [1392]. In mammalian cells, not only the main miRNA effector AGO2, but also the miRNA-processing enzyme DICER1, is degraded as a miRNA-free entity by selective autophagy [1393]. Chemical inhibitors of autophagy (bafilomycin A1 and CQ) and, in HeLa cells, depletion of key autophagy components ATG5, BECN1 or ATG7 using short interfering RNAs, blocks the degradation of both proteins. Electron microscopy shows that DICER1 is associated with membrane-bound structures having the hallmarks of autophagosomes. Moreover, the selectivity of DICER1 and AGO2 degradation might depend on the autophagy receptor CALCOCO2, at least in these cell types. Finally, in C. elegans, AIN-1, a homolog of mammalian TNRC6A/GW182 that interacts with AGO and mediates silencing, is also degraded by autophagy [1394]. AIN-1 colocalizes with the C. elegans SQSTM1 homolog SQST-1 that acts as a receptor for autophagic degradation of ubiquitinated protein aggregates, and also directly interacts with Atg8-family proteins contributing to cargo specificity.
v. RNautophagy and DNautophagy
RNautophagy and DNautophagy are non-macroautophagic pathways, where RNA and DNA, respectively, are taken up by lysosomes directly [1395–1398]. LAMP2C, one of the LAMP2 isoforms, can function as an RNA/DNA receptor in RNautophagy and DNautophagy. SIDT2 is another molecule that has been identified to mediate nucleic acid transport during RNautophagy and DNautophagy [1399–1401]. SIDT2 is a lysosomal multipass transmembrane protein, and a vertebrate ortholog of the C. elegans RNA transporter SID-1.
RNautophagy and DNautophagy were first discovered using in vitro assays with isolated lysosomes derived from mouse brains [1395, 1397], and are also confirmed in isolated lysosomes from HeLa, Neuro2a cells, and MEFs. In vitro assays can be used to detect the activity of RNautophagy or DNautophagy in isolated lysosomes. The activity of RNautophagy at the cellular level can be detected in mammalian cells using a pulse-chase assay [1400]. For example, overexpression of SIDT2 in Neuro2a cells remarkably promotes lysosomal degradation of RNA at the cellular level [1401]. Knockdown of SIDT2 significantly inhibits lysosomal degradation of cellular RNA in MEFs [1400]. Currently, it remains unclear whether there is a SIDT2-independent pathway in RNautophagy and DNautophagy. The activity of DNautophagy at the cellular level has not been reported to date. G/dG sequences in nucleic acids could be motifs that are recognized by RNautophagy and DNautophagy, because poly-G/dG are a substrate of RNautophagy and DNautophagy in vitro, but poly-C/dC, poly-A/dA, poly-U or poly-T are not [1398].
Conclusion: Currently, RNautophagy and DNautophagy activities can be significantly manipulated only by knockdown or overexpression of SIDT2. Identification of nucleic acid sequences that are recognized by RNautophagy or DNautophagy, or specific inhibitors of these pathways, would contribute to the development of novel methods that can monitor RNautophagy and DNautophagy more accurately.
w. Vacuole import and degradation pathway
In yeast, gluconeogenic enzymes such as Fbp1/FBPase (fructose-1,6-bisphosphatase), Mdh2 (malate dehydrogenase), Icl1 (isocitrate lyase) and Pck1 (phosphoenolpyruvate carboxykinase) constitute the cargo of the vacuole import and degradation (Vid) pathway [1402]. These enzymes are induced when yeast cells are glucose starved (grown in a medium containing 0.5% glucose and potassium acetate). Upon replenishing these cells with fresh glucose (a medium containing 2% glucose), these enzymes are degraded in either the proteasome [1403–1405] or the vacuole [1402, 1406] depending on the duration of starvation. Following glucose replenishment after 3 days of glucose starvation, the gluconeogenic enzymes are delivered to the vacuole for degradation [1407]. These enzymes are sequestered in specialized 30- to 50-nm Vid vesicles [1408]. Vid vesicles can be purified by fractionation and gradient centrifugation; western blotting analysis using antibodies against organelle markers and Fbp1, and the subsequent verification of fractions by EM facilitate their identification [1408]. Furthermore, the amount of marker proteins in the cytosol compared to the Vid vesicles can be examined by differential centrifugation. In this case, yeast cells are lysed and subjected to differential centrifugation. The Vid vesicle-enriched pellet fraction and the cytosolic supernatant fraction are examined with antibodies against Vid24, Vid30, Sec28 and Fbp1 [1409–1411].
x. Virophagy
Virophagy is a type of xenophagy, and refers to the autophagic clearance of viruses. An important point when considering the convergence of autophagy and viral infection is that some viruses have evolved mechanisms to block autophagy or to subvert the process to promote viral replication. For example, infection of a cell by influenza and dengue viruses [1415, 1416] or enforced expression of the hepatitis B virus X protein [1417] have profound consequences for autophagy, as viral proteins such as NS4A stimulate autophagy and protect the infected cell against apoptosis, thus extending the time in which the virus can replicate. Conversely, the HSV-1 ICP34.5 protein inhibits autophagy by targeting BECN1 [1418]. Whereas the impact of ICP34.5’s targeting of BECN1 on viral replication in cultured permissive cells is minimal, it has a significant impact upon pathogenesis in vivo, most likely through interfering with activation of CD4+ T cells [1419, 1420], and through cell-intrinsic antiviral effects in neurons [1421]. In addition, the ICP0 protein of HSV-1 downregulates major autophagy receptors such as SQSTM1 and OPTN during the early stages of HSV-1 infection. This could be a mechanism of HSV-1 to counteract the pleiotropic functions of these autophagy receptors, because in SQSTM1-overexpressing cells HSV-1 yields decrease [1422]. Also, viral BCL2 proteins, encoded by large DNA viruses, are able to inhibit autophagy by interacting with BECN1 [848] through their BH3 homology domain. Examples of these include γ-herpesvirus 68 [1423], Kaposi sarcoma-associated herpesvirus [848] and African swine fever virus (ASFV) vBCL2 homologs [1424]. ASFV encodes a protein homologous to HSV-1 ICP34.5, which, similar to its herpesvirus counterpart, inhibits the ER stress response activating PPP1/protein phosphatase 1; however, in contrast to HSV-1 ICP34.5 it does not interact with BECN1. ASFV vBCL2 strongly inhibits both autophagy (reviewed in ref. [1425]) and apoptosis [1426]. The polyQ repeats in some viral proteins could also affect BECN1-mediated autophagy and play a role in virus survival [1427].
HIV has evolved to employ different strategies to finely regulate autophagy to favor its replication and dissemination. In particular, the HIV proteins TAT, NEF and ENV are involved in this regulation by either blocking or stimulating autophagy through direct interaction with autophagy proteins and/or modulation of the MTOR pathway [1428, 1429].
Autophagy contributes to limiting viral pathogenesis in HIV-1 nonprogressor-infected patients by targeting viral components for degradation [1430]. Innate immune stimulation induces antiviral autophagy against Rift Valley fever virus from insects to humans [1431]. One of the Fanconi anemia (FA) genes, Fancc, is required for virophagy of two genetically distinct viruses, Sindbis virus and HSV-1ΔICP34.5BBD, but not for starvation-induced autophagy. Knockout of Fancc in mice increases susceptibility to lethal viral encephalitis [1432]
In the case of Epstein-Barr virus (EBV), several EBV proteins including EBNA1, EBNA3C, LMP1, LMP2A and Rta/Zta interact with the autophagy machinery in B cells. Autophagy is involved in the processing and MHC-II presentation of EBNA1 [1433]. Conversely, EBNA3C, LMP1, LMP2A and Rta initiate and accelerate autophagy progression [1434–1437]. Moreover, autophagy inhibition by 3-MA or ATG5 knockdown diminishes EBV lytic protein expression and viral particle production in B cells [1438]. Autophagy also plays a key role in B-cell proliferation and survival early after infection [1439].
Adenoviruses rupture the endosomal membrane upon entry, thereby triggering antiviral autophagy mediated by LGALS8/galectin-8 (lectin, galactoside-binding, soluble, 8). Adenovirus subsequently limit the autophagic response by recruiting the cellular ubiquitin ligase NEDD4L/NEDD4.2 and escape from the endosome into the cytosol [1440]. In addition, autophagosomes may fuse with intermediate endosomes in response to certain specific viral infections, thus forming amphisomes [1441–1443].
Care must be taken in determining the role of autophagy in viral replication, as some viruses such as vaccinia virus use double-membrane structures that form independently of the autophagy machinery [1444]. Similarly, dengue virus replication, which appears to involve a double-membrane compartment, requires the ER rather than autophagosomes [1445, 1446], whereas coronaviruses and Japanese encephalitis virus use a nonlipidated version of LC3 (see Atg8-family protein detection and quantification) [255, 256]. Yet another type of variation is seen with hepatitis C virus, which requires BECN1, ATG4B, ATG5 and ATG12 for initiating replication, but does not require these proteins once an infection is established [1447].
Autophagy has been highlighted as a critical player in the process of Zika virus (ZIKV) infection and pathogenesis, particularly during pregnancy [1448]. In mammals, autophagy activation is triggered by ZIKV infection likely due to inhibition of the AKT-MTOR pathway, which is co-opted to facilitate viral entry, replication, and release [1448–1450]. Pharmacological blockade of autophagy activity, for example, treatment with lysosomotropic agents (especially hydroxychloroquine [HCQ]), is proposed as a promising therapeutic to counteract ZIKV infection and limit vertical transmission [1448].
After viral hemorraghic septicemia virus (VHSV) entry into rainbow trout red blood cells, autophagy is induced as a mechanism for viral protein degradation. VHSV triggers an increase of LC3A/B protein levels and upregulation of autophagy-related genes such as ULK1, BECN1, and ATG9A, whereas SQSTM1 undergoes degradation early after VHSV exposure. Inhibition of autophagosome degradation with niclosamide results in intracellular VHSV and SQSTM1 accumulation [1451].
y. Xenophagy
Xenophagy refers to the autophagic pathway for the capture and lysosomal degradation of cytosolic pathogens, and pathogens in damaged intracellular vacuoles. Many in vitro and in vivo studies have demonstrated that genes encoding autophagy components are required for host defense against infection by bacteria, parasites and viruses. In a quest for survival, microbial pathogens have evolved strategies to overcome xenophagic clearance. The interactions of these pathogens with the host autophagy system are complex and have been the subject of several excellent reviews [170–175, 628, 1452–1460]. There are a few key considerations when studying interactions of microbial pathogens with the autophagy system [1461]. Importantly, autophagy should no longer be considered as strictly antibacterial, and several studies have described the fact that autophagy may serve to either restrict or promote bacterial replication both in vivo [1462] and in vitro (reviewed in refs. [1463, 1464]). Moreover, special care should be taken when evaluating bacterial-induced specific autophagy and autophagic flux, because an increased basal autophagy and flux perceived by western blot may be unlinked to the cellular compartment of the bacterial vacuole, which can be revealed by careful examination of the bacterial compartment using IHC and colocalization studies [1465]. For example, autophagy has been proposed to both support the survival of intraphagosomal M. marinum, by providing cytosolic material and/or membranes to the bacteria-containing compartment, and to restrict the proliferation of the cytosolic mycobacteria in D. discoideum [64, 1322]. In addition to pathogenic bacteria, autophagy can be induced by beneficial bacteria, contributing to alleviation of the hepatotoxicity induced by acetaminophen, in vitro [1466].
LC3 is commonly used as a marker of autophagy. However, studies have established that LC3 can promote phagosome maturation independently of autophagy through LC3-associated phagocytosis (see cautionary notes in Atg8-family protein detection and quantification, and Noncanonical use of autophagy-related proteins). Other studies show that autophagy of Salmonella enterica serovar Typhimurium (S. Typhimurium) is dependent on ATG9, an essential autophagy protein, whereas LC3 recruitment to a bacteria-containing phagosome does not require ATG9 [1467]. In contrast, autophagy of these bacteria requires either glycan-dependent binding of LGALS8 to damaged membranes and subsequent recruitment of the cargo receptor CALCOCO2 [1468], or ubiquitination of target proteins (not yet identified) and recruitment of at least four different ubiquitin-binding receptor proteins, SQSTM1 [1469], CALCOCO2 [1470], TAX1BP1/CALCOCO3 [1471] and OPTN [1472]. In fibroblasts, S. Typhimurium triggers the formation of host endomembrane-containing aggresomes that are further captured together with intravacuolar bacteria by phagophores harboring LC3 and SQSTM1, but devoid of CALCOCO2 and ubiquitin [1473]. Therefore, the available criteria to differentiate LAP from autophagy include: i) LAP involves LC3 recruitment to a bacteria-containing phagosome in a manner that requires ROS production by an NADPH oxidase. It should be noted that most cells express at least one member of the NADPH oxidase family. Targeting expression of the common CYBA/p22phox subunit is an effective way to disrupt the NADPH oxidases. Scavenging of ROS by antioxidants such as NAC, resveratrol and alpha-tocopherol is also an effective way to inhibit LAP. ii) Autophagy of bacteria requires ATG9, whereas LAP apparently does not [1467]. iii) LAP involves single-membrane structures surrounding the bacterial cargo. CLEM is expected to show single-membrane structures that are LC3+ with LAP [247]. In contrast, autophagy is expected to generate double-membrane structures surrounding cargo (which may include single-membrane phagosomes, giving rise to triple-membrane structures around the bacterial membrane(s), corresponding to an autophagolysosome [1467]). It is anticipated that more specific markers of LAP will be identified as these phagosomes are further characterized. In vivo xenophagy studies in mice show that S. Typhimurium reduces the level of basal autophagy in tissues such as intestine as seen by LC3-II levels at later times of infection [1474]. This suggests that pathogens have the ability to decrease host autophagy for their survival. Recently identified xenophagy-enhancing compounds show enhanced capture and degradation of S. Typhimurium in both cellular and in vivo models with enhanced LC3-II levels in tissues [1475].
Elegant mechanisms that differentiate autophagy from LAP have emerged that demonstrate that there are mechanistic differences between these processes. For example, ATG16L1 recruitment to the phagosome in Salmonellae-infected cells occurs through a carboxy-terminal WD40 domain that binds to the V-ATPase on the phagosome, which is dispensable for canonical autophagy [1476, 1477]. This domain is also required in influenza infection [1478]. These studies illustrate that while LC3 targeting of a pathogen-containing vacuole uses components shared with canonical autophagy, it utilizes a distinct mechanism.
Nonmotile Listeria monocytogenes can be targeted to phagophores upon antibiotic treatment [883], which indicates that autophagy serves as a cellular defense against microbes in the cytosol. However, subsequent studies have revealed that autophagy can also target pathogens within phagosomes, damaged phagosomes or the cytosol, as illustrated by the various phases of infection of M. marinum in D. discoideum [1322, 1459]. Therefore, when studying microbial interactions by EM, many structures can be visualized, with any number of membranes encompassing microbes, all of which may be LC3+ [1479, 1480]. As discussed above, single-membrane structures that are LC3+ may arise through LAP, and we cannot rule out the possibility that both LAP and autophagy may operate at the same time to target the same phagosome. Indeed, autophagy may facilitate phagocytosis and subsequent bacterial clearance [1481]. Autophagy is not only induced by intracellular bacteria, but also can be activated by extracellular bacteria such as Pseudomonas aeruginosa and Klebsiella pneumoniae, which may involve complex mechanisms [1482–1484]. Furthermore, autophagy can be induced by Gram-negative bacteria via a common mechanism involving naturally-produced bacterial outer membrane vesicles [1485, 1486]; these vesicles enter human epithelial cells, resulting in autophagosome formation and inflammatory responses mediated via the host pathogen recognition receptor NOD1 [1485, 1487]. In addition, highly purified outer membrane proteins from bacteria and mitochondria can trigger autophagy [1488]. Upon specific stimulation, NOD1 binds to LC3 inducing an increased autophagy flux and autolysosome formation, and LC3-NLRP3 inflammasome interaction, in epithelial Sertoli cells [1489]. The ability of NOD1 to sense ER stress and cell damage and induce pro-inflammatory signaling is regulated by ATG16L1 [1490], implicating autophagy and inflammasomes in environmental stress responses. NOD2 also regulates autophagy upon stimulation by danger/damage-associated molecular patterns (DAMPs) such as the bacterial NOD2 ligand sulfatide. NOD2 connects inflammation hypoxia and autophagy, as NOD2 is a direct transcriptional target of HIF1A, the main oxygen sensor in mammalian cells induced by reduced oxygen. Hypoxia-induced NOD2 functions upstream of CQ and directly binds to the V-ATPAse complex, regulating vesicular pH [1491].
Viruses can also be targeted by autophagy, and in turn can act to inhibit autophagy (see Virophagy). Xenophagy has also been observed with intracellular parasites. Mice deficient in autophagy develop a more severe Trypanosoma cruzi infection, characterized by higher peaks of parasitemia, higher cardiac amastigote nests and premature death, compared to controls. Peritoneal macrophages from these mice display higher levels of infection that correlate with the minor recruitment of LC3 and other proteins, such as CALCOCO2 and SQSTM1, to amastigotes, observed in the cytoplasm of RAW cells in the presence of inhibitors of autophagy [1492].
Finally, it is important to realize that there may be other autophagy-like pathways that have yet to be characterized. For example, in response to cytotoxic stress (treatment with etoposide), autophagosomes are formed in an ATG5- and ATG7-independent manner (see Noncanonical use of autophagy-related proteins) [31]. While this does not rule out involvement of other autophagy regulators/components in the formation of these autophagosomes, it does establish that the canonical autophagy pathway involving LC3 conjugation is not involved. In contrast, RAB9 is required for this alternative pathway, potentially providing a useful marker for analysis of these structures. Returning to xenophagy, M. marinum can be targeted to phagophores in an ATG5-independent manner [1493]. Furthermore, up to 25% of intracellular S. typhimurium are observed in multi-lamellar membrane structures resembling autophagosomes in atg5−/− MEFs [1469]. These findings indicate that an alternate autophagy pathway is relevant to host-pathogen interactions. Moreover, differences are observed that depend on the cell type being studied. Yersinia pseudotuberculosis is targeted to autophagosomes where it can replicate in bone marrow-derived macrophages [1494], whereas in RAW 264.7 and J774 cells, bacteria are targeted both to autophagosomes, and LC3-negative, single-membrane vacuoles (F. Lafont, personal communication).
One key consideration has recently emerged in studying xenophagy. Whereas the basal autophagic flux in most cells is essential for their survival, infecting pathogens can selectively modulate antibacterial autophagy (i.e., xenophagy) without influencing basal autophagy. This may help pathogens ensure prolonged cellular (i.e., host) survival. Thus, in the case of xenophagy it would be prudent to monitor substrate (pathogen)-specific autophagic flux to understand the true nature of the perturbation of infecting pathogens on autophagy [1495, 1496]. Furthermore, this consideration particularly limits the sensitivity of LC3 western blots for use in monitoring autophagy regulation, and stresses that other techniques such as those enabling subcellular analysis of the pathogen-specific compartment/vacuole are additionally used. For instance, to verify that the effect of a total reduction in LC3-II during autophagy induction by western blot also extends to the subcellular compartment of the pathogen/bacterial vacuole by using LC3-based microscopy [1465].
z. Zymophagy
Zymophagy was originally defined as a specific mechanism that eliminates zymogen granules in the pancreatic acinar cells and, thus, prevents deleterious effects of prematurely activated and intracellularly released proteolytic enzymes, when impairment of secretory function occurs [1497]. Therefore, zymophagy is primarily considered to be a protective mechanism implemented to sustain secretory homeostasis and to mitigate pancreatitis. The presence of zymogen granules, however, is not only attributed to pancreatic acinar cells. Thus, zymophagy was also reported in activated secretory Paneth cells of the crypts of Lieberkühn in the small intestine [542]. Note that one of the major functions of Paneth cells is to prevent translocation of intestinal bacteria by secreting hydrolytic enzymes and antibacterial peptides to the crypt lumens. The similarity in mechanisms of degradation of secretory granules in these two different types of secretory cells sustains the concept of the protective role of autophagy when “self-inflicted” damage may occur due to overreaction and/or secretory malfunction in specialized cells.
Zymophagy can be monitored by TEM, identifying autophagosomes containing secretory granules, by following SQSTM1 degradation by western blot, and by examining the subcellular localization of VMP1-EGFP, which relocates to granular areas of the cell upon zymophagy induction. Colocalization of PRSS1/trypsinogen (which is packaged within zymogen granules) and LC3, or of GFP-ubiquitin (which is recruited to the activated granules) with RFP-LC3 can also be observed by indirect or direct immunofluorescence microscopy, respectively. Active trypsin is also detectable in zymophagosomes and participates in the early onset of acute pancreatitis (F. Fortunato et al., unpublished data). In addition, isolated zymogen granules from alcohol-fed mouse pancreas also contain LC3-II based on western blot analysis, which may also serve as another indirect quantitative marker for zymophagy [1498].
Of note, studies from the past decade have shown an essential role of autophagy in maintaining pancreatic acinar cell homeostasis and function, and strongly implicate impaired autophagy in initiation and development of pancreatitis (see Large animals and rodents). In particular, immunofluorescence data [1499] indicate autolysosomes as one compartment in which trypsinogen activation occurs in pancreatitis, as evidenced by colocalization of LC3-II and LAMP2 with trypsinogen activation peptide (an oligopeptide cleaved off trypsinogen in the process of its conversion to active trypsin). Impaired TFEB-mediated lysosomal biogenesis has also been shown to promote cerulein or alcohol-induced pancreatitis in mice. In addition to experimental pancreatitis, acinar cell nuclear TFEB staining markedly decreased in both human alcoholic and non-alcoholic pancreatitis, supporting a critical role of autophagy and lysosomal biogenesis in the pathogenesis of pancreatitis [1498, 1500].
12. Autophagic sequestration assays
Although it is useful to employ autophagic markers such as LC3 in studies of autophagy, LC3-II levels or LC3 puncta cannot quantify actual autophagic activity, because LC3-II is not involved in all cargo sequestration events, and LC3-II can be found on phagophores and nonautophagosomal membranes in addition to autophagosomes. Thus, quantification of autophagic markers such as LC3 does not tell how much cargo material has actually been sequestered inside autophagosomes. Moreover, LC3 and several other autophagic markers cannot be used to monitor noncanonical autophagy. Autophagic sequestration assays constitute marker-independent methods to measure the sequestration of autophagic cargo into autophagosomal compartments, and are among the few functional autophagy assays described to date.
Autophagic cargo sequestration activity can be monitored using either an (electro)injected, inert cytosolic marker such as [3H]-raffinose [1501] or an endogenous cytosolic protein such as LDH (lactate dehydrogenase) [1502], in the latter case along with treatment with a protease inhibitor (e.g., leupeptin) or other inhibitors of lysosomal activity or autophagosome-lysosome fusion (e.g., bafilomycin A1, concanamycin A, or CQ) [216, 302, 1503] to prevent intralysosomal degradation of the protein marker. The assay simply measures the transfer of cargo from the soluble (cytosol) to the insoluble (sedimentable) cell fraction (which includes autophagic compartments), with no need for a sophisticated subcellular fractionation. Electrodisruption of the plasma membrane followed by centrifugation through a density cushion was originally used to separate cytosol from sedimentable cell fractions in primary hepatocytes [1504]. This method has also been used in various human cancer cell lines and mouse embryonic fibroblasts, where the LDH sequestration assay has been validated with pharmacological agents as well as genetic silencing or knockout of key factors of the autophagic machinery (N. Engedal, unpublished results) [32, 56, 216, 473, 1503, 1505]. Moreover, a downscaling and simplification of the method that avoids the density cushion has been introduced and validated [56, 473, 1503, 1506]. Homogenization and sonication techniques have also been successfully used for the LDH sequestration assay [1017, 1507]. The endogenous LDH cargo marker can be quantified by an enzymatic assay, or by western blotting. In principle, any intracellular component can be used as a cargo marker, but cytosolic enzymes having low sedimentable backgrounds are preferable. Membrane-associated markers are less suitable, and proteins such as LC3, which are part of the sequestering system itself, will have a much more complex relationship to the autophagic flux than a pure cargo marker such as LDH.
In yeast, sequestration assays are typically done by monitoring protease protection of an autophagosome marker or a cargo protein. For example, prApe1, and GFP-Atg8 have been used to follow completion of the autophagosome [1508]. The relative resistance or sensitivity to an exogenous protease in the absence of detergent is an indication of whether the autophagosome (or other sequestering vesicle) is complete or incomplete, respectively. Thus, this method also distinguishes between a block in autophagosome formation versus fusion with the vacuole. The critical issues to keep in mind involve the use of appropriate control strains and/or proteins, and deciding on the correct reporter protein. In addition to protease protection assays, sequestration can be monitored by fluorescence microscopy during pexophagy of methanol-induced peroxisomes, using GFP-Atg8 as a pexophagosome marker and BFP-SKL to label the peroxisomes. The vacuolar sequestration process during micropexophagy can also be monitored by formation of the vacuolar sequestering membrane stained with FM 4-64 [1053].
Sequestration assays can be designed to measure flux through individual steps of the autophagy pathway. For example, whereas electroinjected [3H]-raffinose or endogenous LDH can be used to measure the sequestration step, electroinjected [14C]-lactose can be used to monitor cargo flux to amphisomes and proteolytically active autolysosomes (as explained below). Whereas [3H]-raffinose is completely resistant to (auto)lysosomal degradation, the [14C]-lactose that reaches active autolysosomes is rapidly hydrolyzed into [14C]-glucose and galactose (by GLB1/beta-galactosidase), measurable by chromatography. [14C]-lactose thus marks prelysosomal compartments (autophagosomes and amphisomes), whereas [14C]-glucose marks the autolysosomal compartment. Experimental conditions or treatments that block autophagosome-lysosome fusion (e.g., asparagine or the microtubule inhibitor vinblastine) lead to an accumulation of lactose in prelysosomal compartments [11, 1509]. By adding exogenous beta-galactosidase (that is endocytosed by the cells) in the presence of asparagine (which blocks autophagosome-lysosome fusion), the fusion of autophagosomes with endosomes (thus producing amphisomes) can be studied. In fact, this was the experimental approach that first identified the amphisome [11].
One caveat with using lysosome or autophagosome-lysosome inhibitors is that they may affect sequestration indirectly, for example, by modifying the uptake and metabolism (including protein synthesis) of autophagy-suppressive amino acids (see Autophagy inhibitors and inducers). Therefore, the time period of treatment with the inhibitor should be as short as possible (typically 2-3 h). Note that for measuring autophagic sequestration and degradation activity with electroinjected [3H]-raffinose or [14C]-lactose, respectively, no inhibitors are needed. Also note that the LDH sequestration assay, when used without addition of lysosomal degradation inhibitors, can be used to identify treatments or conditions that block autophagic flux at a post-sequestration step. For instance, autophagically sequestered LDH accumulates in cells depleted of RAB7A (but not RAB7B) [1505], thus confirming the role of RAB7A in autophagosome-lysosome fusion [344, 1510, 1511].
A variation of this approach applicable to mammalian cells includes live cell imaging. Autophagy induction is monitored as the movement of cargo, such as mitochondria, to GFP-LC3-colocalizing compartments, and then fusion/flux is measured by delivery of cargo to lysosomal compartments [439, 1512]. In addition, sequestration of fluorescently tagged cytosolic proteins into membranous compartments can be measured, as fluorescent puncta become resistant to the detergent digitonin [1513]. Use of multiple time points and monitoring colocalization of a particular cargo with GFP-LC3 and lysosomes can also be used to assess sequestration of cargo with autophagosomes as well as delivery to lysosomes [1138]. Moreover, colocalization of cargo with endogeneous LC3 puncta using immunofluorescent staining can be used [606].
Time-lapse microscopy allows direct visualization of vacuole transfer from mother cells to their daughters as seen for A549 lung cancer cells exposed to yessotoxin (YTX) [1514]. Such effects on downstream lineages may be significant for the interpretation of observations related to autophagy signaling especially for cells in environments where the stress varies. Autophagic activity caused by this toxin results in the sequestration and degradation, by an autophagic-like process, of ribosomes and lipid droplets associated with autophagic compartments and lamellar bodies in BC3H1 cells [1515].
In the Drosophila fat body, the localization of free cytosolic RFP-family proteins changes from a diffuse to a punctate pattern in an Atg gene-dependent manner, and these mCherry puncta colocalize with the lysosomal marker Lamp1-GFP during starvation [1516]. Thus, the redistribution of free cytosolic mCherry may be used to follow bulk, nonselective autophagy due to its stability and accumulation in autolysosomes.
Cautionary notes: The electro-injection of radiolabeled probes is technically demanding, but the use of an endogenous cytosolic protein probe is very simple and requires no pretreatment of the cells other than with a protease inhibitor. Another concern with electro-injection is that it can affect cellular physiology, so it is necessary to verify that the cells behave properly under control situations such as amino acid deprivation. An alternate approach for incorporating exogenous proteins into mammalian cell cytosol is to use “scrape-loading,” a method that works for cells that are adherent to tissue culture plates [1517]. Finally, these assays work well with hepatocytes but may be problematic with other cell types, and it can be difficult to load the cell while retaining the integrity of the compartments in the post-nuclear supernatant (S. Tooze, unpublished results). General points of caution to be addressed with regard to live cell imaging relate to photobleaching of the fluorophore, cell injury due to repetitive imaging, autofluorescence in tissues containing lipofuscin, and the pH sensitivity of the fluorophore.
There are several issues to keep in mind when monitoring sequestration by the protease protection assay in yeast [1508]. First, as discussed in Selective types of autophagy, prApe1 is not an accurate marker for nonselective autophagy; import of prApe1 utilizes a receptor (Atg19) and a scaffold (Atg11) that make the process specific. In addition, vesicles that are substantially smaller than autophagosomes can effectively sequester the Cvt complex. Another problem is that prApe1 cannot be used as an autophagy reporter for mutants that are not defective in the Cvt pathway, although this can be bypassed by using a vac8∆ background [1518]. At present, the prApe1 assay cannot be used in any system other than yeast. The GFP-Atg8 protease protection assay avoids these problems, but the signal-to-noise ratio is typically substantially lower. In theory, it should be possible to use this assay in other cell types, and protease protection of GFP-LC3 and GFP-SQSTM1 has been analyzed in HeLa cells [1519]. Finally, tendencies of GFP-LC3 and particularly GFP-SQSTM1 to aggregate may make LC3 and SQSTM1 inaccessible to proteases.
Conclusion: Sequestration assays represent the most direct method for monitoring autophagy, and in particular for discriminating between conditions where the autophagosome is complete (but not fused with the lysosome/vacuole) or open (i.e., a phagophore). These assays can also be modified to measure autophagic flux.
13. Turnover of autophagic compartments
Inhibitors of autophagic sequestration (e.g., amino acids, 3-MA, wortmannin, SAR-405, BAPTA-AM, MRT67307, or thapsigargin) [32, 56, 216, 299, 1032, 1503, 1520] can be used to monitor the disappearance of autophagic elements (phagophores, autophagosomes, autolysosomes) to estimate their half-life by TEM morphometry/stereology. The turnover of the autophagosome or the autolysosome will be differentially affected if fusion or intralysosomal degradation is inhibited [13, 15, 28, 1521]. The duration of such experiments is usually only a few hours; therefore, long-term side effects or declining effectiveness of the inhibitors can be avoided. It should be noted that fluorescence microscopy has also been used to monitor the half-life of autophagosomes, monitoring GFP-LC3 in the presence and absence of bafilomycin A1 or following GFP-LC3 after starvation and recovery in amino acid-rich medium (see Atg8-family protein detection and quantification) [17, 1522].
Cautionary notes: The inhibitory effect must be strong, and the efficiency of the inhibitor needs to be tested under the experimental conditions to be employed. Cycloheximide is sometimes used as an autophagy inhibitor, but its use in long-term experiments is problematic because of the many potential indirect effects. CHX inhibits translational elongation, and therefore protein synthesis. In addition, CHX decreases the efficiency of protein degradation in several cell types (A.M. Cuervo, personal communication) including hematopoietic cells (A. Edinger, personal communication). Treatment with CHX causes a potent increase in MTORC1 activity, which can decrease autophagy in part as a result of the increase in the amino acid pool resulting from suppressed protein synthesis (H.-M. Shen, personal communication; I. Topisirovic, personal communication) [27, 1523]. In addition, at high concentrations (in the millimolar range) CHX inhibits complex I of the mitochondrial respiratory chain [1524, 1525], but this is not a problem, at least in hepatocytes, at low concentrations (10 -20 µM) that are sufficient to prevent protein synthesis (A.J. Meijer, personal communication).
Conclusion: The turnover of autophagic compartments is a valid method for monitoring autophagic-lysosomal flux, but CHX must be used with caution in long-term experiments.
14. Autophagosome-lysosome colocalization and dequenching assay
Another method to demonstrate the convergence of the autophagic pathway with a functional degradative compartment is to incubate cells with the bovine serum albumin derivative dequenched (DQ)-BSA that is labeled with the red-fluorescent BODIPY TR-X dye; this conjugate will accumulate in lysosomes. The labeling of DQ-BSA is so extensive that the fluorophore is self-quenched. Proteolysis of this compound results in dequenching and the release of brightly fluorescent fragments. Thus, DQ-BSA is useful for detecting intracellular proteolytic activity as a measure of a functional lysosome [1526].
Furthermore, DQ-BSA labeling can be combined with GFP-LC3 to monitor colocalization, and thus visualize the convergence, of amphisomes with a functional degradative compartment (DQ-BSA is internalized by endocytosis). This method can also be used to visualize fusion events in real-time experiments by confocal microscopy (live cell imaging). Along similar lines, other approaches for monitoring convergence are to follow the colocalization of RFP-LC3 and LysoSensor Green (M. Bains and K.A. Heidenreich, personal communication), mCherry-LC3 and LysoSensor Blue [441], or tagged versions of LC3 and LAMP1 (K. Macleod, personal communication) or CD63 [439] as a measure of the fusion of autophagosomes with lysosomes. It is also possible to trace autophagic events by visualizing the pH-dependent excitation changes of the coral protein Keima [1036]. This quantitative technique is capable of monitoring the fusion of autophagosomes with lysosomes, that is, the formation of an autolysosome, and the assay does not depend on the analysis of LC3.
Cautionary notes: Some experiments require the use of inhibitors (e.g., 3-MA or wortmannin) or overexpression of proteins (e.g., RAB7 dominant negative mutants) that may also affect the endocytic pathway or the delivery of DQ-BSA to lysosomes (e.g., wortmannin causes the swelling of late endosomes [1527]). In this case, the lysosomal compartment can be labeled with DQ-BSA overnight before treating the cells with the drugs, or prior to the transfection.
Conclusion: DQ-BSA provides a relatively convenient means for monitoring lysosomal protease function and can also be used to follow the fusion of amphisomes with the lysosome. Colocalization of autophagosomes (fluorescently tagged LC3) with lysosomal proteins or dyes can also be monitored.
15. Tissue fractionation
The study of autophagy in the organs of larger animals, in large numbers of organisms with very similar characteristics, or in tissue culture cells provides an opportunity to use tissue fractionation techniques as has been possible with autophagy in rat liver [50, 69, 1528–1533]. Because of their sizes (smaller than nuclei but larger than membrane fragments [microsomes]), differential centrifugation can be used to obtain a subcellular fraction enriched in mitochondria and organelles of the autophagy-lysosomal system, which can then be subjected to density gradient centrifugation to enrich autophagosomes, amphisomes, autolysosomes and lysosomes [50, 69, 1533–1537]. Please see previous versions of the guidelines [1, 2] for a discussion of the uses and limitations of tissue fractionation.
16. In vitro determination of autophagosome formation
Mobilization of membranes from intracellular resources is required for autophagosome biogenesis. A cell-free assay was established to identify organelle membranes that form a precursor for autophagosome formation. The membrane from ATG5 mutant cells is defective in autophagosome formation in vivo during starvation [814]. In the cell-free assay, membranes from atg5 knockout MEFs are mixed with cytosolic fractions from starved or untreated wild-type cells. These cytosolic fractions include a high amount of LC3-I and lack the lipidated form, LC3-II, which is sedimented with the membrane. The reaction is performed in the presence of GTP and an ATP regeneration system. The assay measures cell-free LC3 lipidation by the formation of LC3-II [1538]. The reaction thus identifies membranes responsible for LC3-II generation. A three-step membrane fractionation is performed along with monitoring of lipidation enrichment with respect to different membrane markers. First, differential centrifugation is performed to obtain four membrane pellets with different markers. The 25K fraction reveals the highest lipidation activity and includes peroxisomes (ABCD3/PMP70), late endosomes (LAMP2), cis-Golgi (GOLGA2/GM130) ER-Golgi intermediate compartment (ERGIC; SEC22B and LMAN1/ERGIC53), plasma membrane/early endosomes (TFRC), ER (RPN1), ER exit sites (ERES, active sites on the ER that generate COPII-coated vesicles; PREB/SEC12), lysosomes (CTSD), and ATG9 vesicles. The 25K membrane is further fractionated using step-gradient ultracentrifugation, where the fraction with higher lipidation activity is determined to include ERGIC, cis-Golgi, ATG9 vesicles and plasma membrane/early endosomes.
This assay recapitulates the early cellular steps of autophagosome formation in different aspects. The cells are stimulated by starvation, and rapamycin or torin1 treatment and are inhibited in the absence of ULK1, which reflects the involvement of the MTORC1 pathway. PtdIns3K inhibitors abolish the LC3 lipidation, and LC3 lipidation is prohibited in the absence of ATG proteins such as ATG3, ATG5 or ATG7 [1539].
The contribution of different organelles to autophagosome biogenesis was tested using different fractionation and purification steps to obtain the ERGIC, which represents a primary membrane determinant that triggers LC3 lipidation. The ERGIC is a recycling compartment located in the ER and cis-Golgi compartments. PtdIns3K is activated upon starvation, and this enzyme facilitates the recruitment of COPII proteins to the ERGIC membrane. Subsequently, the ERGIC-derived COPII vesicles form a potential membrane source of the autophagosome and LC3 lipidation vesicles [1540].
A COPII vesicle-labelling system using the transmembrane cargo protein Axl2 was investigated by immuno-EM in yeast, showing that COPII acts as precursor for the formation of the autophagosome membrane [1541]. Another study employing super-resolution microscopy showed that starvation results in ER-exit site enlargement. COPII production served as positive control, and demonstrated contribution to autophagosome formation [1542].
Conclusion: The cell-free assay implicates the ERGIC as one of the primary cellular membrane determinants that facilitates LC3 lipidation. Further application of this method may reveal more with regard to functional forms of the cytosol and the triggering factors for autophagosome membrane formation.
17. Analyses in vivo
Monitoring autophagic flux in vivo or in organs is one of the least developed areas at present, and ideal methods relative to the techniques possible with cell culture may not exist. Importantly, the level of basal autophagy, time course of autophagic induction, and the bioavailability of autophagy-stimulating and -inhibiting drugs is likely tissue specific. Moreover, basal autophagy or sensitivity to autophagic induction may vary with animal age, sex or strain background. Therefore, methods may need to be optimized for the tissue of interest. One method for in vivo studies is the analysis of GFP-Atg8-family proteins (see GFP-Atg8-family protein fluorescence microscopy). Autophagy can be monitored in tissue (e.g., skeletal muscle, heart, kidney, liver, brain, spinal cord, dorsal root ganglia, peripheral nerve, retina and platelets) in vivo in transgenic mice and zebrafish systemically expressing GFP-LC3 [109, 210, 214, 239, 388, 390, 540, 561, 924, 1543, 1544], or in other models by transfection with GFP-LC3-encoding plasmids or in transgenic strains that possess either mCherry- or GFP-Atg8-family proteins under the control of either inducible or Atg8-family protein gene promoter sequences [375, 662]. All of these in vivo approaches require appropriate negative controls for Atg8-family protein localization to autophagosomes, through the use of point mutants that cannot be lipidated or associated with the autophagosomes [1545] or, in genetically tractable systems, mutations that predictably disrupt their association with autophagosomes [562].
It should be noted that tissues such as white adipose tissue, ovary, and testes, and some brain regions such as the hypothalamus, do not appear to express the Actb promoter-driven GFP-Lc3 transgene strongly enough to allow detection of the fluorescent protein [239]. In addition, tissue-specific GFP-LC3 mice have been generated for monitoring cardiac myocytes [1546, 1547]. In these settings, GFP fluorescent puncta are indicative of autophagic structures; however, the use of a lysosomal fusion or protease inhibitor would be needed to assess flux. Cleavage of GFP-LC3 to generate free GFP can be evaluated as one method to monitor the completion of autophagy. This has been successfully performed in mouse liver [351], suggesting the GFP-LC3 cleavage assay may also be applied to in vivo studies. Note that the accumulation of free GFP in the mouse brain is minimal after autophagy is induced with rapamycin (autophagy induction based on GFP-LC3 imaging and SQSTM1 IHC; M. Lipinski, personal communication), but significant when autophagic flux is partially blocked after traumatic brain injury [214]. Thus, caution needs to be taken when interpreting results of these assays in different tissues. We also recommend including a control under conditions known to induce autophagic flux such as starvation.
A simple methodology to measure autophagic flux in the brain was described [1548]. This strategy combines the generation of adeno-associated virus and the use of the dynamic fluorescent reporter mCherry-GFP-LC3 that allows an extended transduction and stable expression of mCherry-GFP-LC3 after intracerebroventricular injection in newborn animals. With this approach, a widespread transduction level is achieved along neurons at the central nervous system when newborn pups are injected, including pyramidal cortical and hippocampal neurons, Purkinje cells, and motor neurons in the spinal cord and also, to a lesser extent, in oligodendrocytes [1548]. The different serotypes of adeno-associated virus can be used to transduce other cell types at the CNS [1548–1550]. This methodology allows a reproducible and sensitive mCherry-GFP-LC3 detection, and a strong LC3 flux when animals are treated with autophagy inducers including rapamycin and trehalose [1550, 1551]. Therefore, using these combined strategies can be applied to follow autophagy activity in mice or rats and can be particularly useful to evaluate it in animal models of diseases affecting the nervous system [1548–1550]. A transgenic mouse with a low level neuron-specific expression of mCherry-RFP-GFP-LC3 was generated that has possible advantages over viral-expression models in achieving a relatively uniform expression reproducibly in a given mouse throughout its life or among different experimental groups of mice [451]. Alternatively, confocal laser scanning microscopy, which makes it possible to obtain numerous sections and substantial data about spatial localization features, can be a suitable system for studying autophagic structures (especially for whole mount embryo in vivo analysis) [1552]. In addition, this method can be used to obtain quantitative data through densitometric analysis of fluorescent signals [1553].
A number of transgenic autophagy mouse and Drosophila models have now been generated that rely on the expression of pH-sensitive fluorophores as mentioned above. In terms of monitoring general autophagy, mice stably expressing mRFP/mCherry-GFP-LC3, from the ubiquitous ROSA26 locus, allow monitoring of autophagic flux in multiple organs [1103, 1231]. When combined with immunohistochemical staining using cell-specific markers, autophagy can be quantified in distinct cell types within tissues. As with utilization of this marker in cell lines (see above), the same caveats apply, and care must be taken to maintain pH during fixation [1554].
Similar fluorescence methodology has been used to measure mitophagy in mouse and Drosophila tissue, either using mitochondrial matrix-localized mt-Keima [1145, 1147] or OMM-localized mCherry-GFP in the case of the mito-QC mouse [38]. mito-QC is very similar to the mCherry-GFP-LC3 mouse (only differing in the fluorophore-targeting peptide), and thus allows an in vivo comparison between autophagy and mitophagy, which do not necessarily occur under the same conditions [1146, 1231]. The mito-QC mouse has been used to monitor mitophagy in disease models, as shown with diabetes through the generation of mito-QC Ins2Akita mice [39]. Analyses of tissues from both mito-QC and mt-Keima demonstrate the basal nature of mammalian mitophagy in vivo and its conservation to Drosophila. An important distinction between these mitophagy reporter mouse models is that tissues from the mito-QC mouse are compatible with fixation, whereas fluorescence in cells and tissues from the mt-Keima mouse is lost upon fixation [1554]. This difference has implications for applications where high throughput analyses of mitophagy in tissues and cells are required. Furthermore, because mito-QC is compatible with fixation, it is also possible to confirm the lysosomal localization of mCherry puncta using the mito-QC approach [38, 1554]. Similarly, Drosophila harboring GAL4/UAS responsive transgenes for mt-mCherry-GFP (mito-QC) or mt-Keima have been developed, which allows spatiotemporal restricted expression analysis [466]. Utilizing such mitophagy reporters in Drosophila is particularly useful for rapidly and economically screening putative genetic or pharmacological regulators of mitophagy in vivo.
Another possibility is immunohistochemical staining, an important procedure that may be applicable to human studies as well, considering the role of autophagy in neurodegeneration, myopathies and cardiac disease where samples may be limited to biopsy/autopsy tissue. In this sense, special attention should be taken in the sample extraction and preservation, as LC3B-II could undergo degradation. Immunodetection of LC3 as definite puncta is possible in paraffin-embedded tissue sections and fresh frozen tissue, by either IHC or immunofluorescence [265, 1555–1562]. Immunostaining of LC3 puncta in peripheral nerve has been initially evaluated and compared to that obtained in GFP-LC3 mice (measured by means ImageJ RGB pixels analysis, which automatically converts pixels in brightness values) [540]. This method is, therefore, widely utilized in this kind of tissue [1327, 1563, 1564]; however, this methodology has not received extensive evaluation, and does not lend itself well to dynamic assays.
Other autophagic substrates can be evaluated via IHC and include SQSTM1, NBR1, ubiquitinated inclusions and protein aggregates [1562]. Similarly, autophagy can be evaluated by measuring levels of these autophagic substrates via traditional immunoblot; however, their presence or absence needs to be cautiously interpreted as some of these substrates can accumulate with either an increase or a decrease in autophagic flux (see SQSTM1 and related LC3 binding protein turnover assays). Bone marrow transfer has been used to document in vivo the role of autophagy in the reverse cholesterol transport pathway from peripheral tissues or cells (e.g., macrophages) to the liver for secretion in bile and for excretion [966], and a study shows that TGM2 (transglutaminase 2) protein levels decrease in mouse liver in vivo upon starvation in an autophagy-dependent manner (and in human cell lines in vitro in response to various stimuli; M. Piacentini, personal communication), presenting additional possible methods for following autophagy activity. In that respect, it is noteworthy to mention that TGM2 can also inhibit autophagic flux at the level of autophagosome-lysosome fusion by modifying ITPR1 (inositol 1,4,5-trisphosphate receptor, type 1) and suppressing its calcium-release activity [1565].
It is also possible to analyze tissues ex vivo, and these studies can be particularly helpful in assessing autophagic flux as they avoid the risks of toxicity and bioavailability of compounds such as bafilomycin A1 or other autophagy inhibitors. Along these lines, autophagic flux can be determined by western blot in retinas placed in culture for 4 h with protease inhibitors [968,969]. This method could be used in tissues that can remain “alive” for several hours in culture such as the retina [1566–1568], brain slices [214, 1569] (particularly organotypic brain slices that can be cultured in vitro for weeks, allowing for treatments with autophagy stimulators or inhibitors for long periods [1570]), and spinal cord slices [1571]. Ex vivo tumors are relevant models of autophagy in mesothelioma. In these models, basal autophagy and its modulation can be measured by immunofluorescence to assess the presence of LC3 puncta when combined with lysosomal inhibitor treatment, or of ATG13 puncta without lysosomal inhibition [755, 756, 1572].
Several studies have demonstrated the feasibility of monitoring autophagic flux in vivo in skeletal muscle. Starvation is one of the easiest and most rapid methods for stimulating the autophagic machinery in skeletal muscles. Twelve h of fasting in mice may be sufficient to trigger autophagy in muscle [1573–1575], but the appropriate time should be determined empirically. It is also important to consider that the expression of autophagy-related factors, as well as the autophagic response to various stimuli and disease states, can differ between muscles of different fiber type, metabolic, and contractile properties [239, 1576–1579]. Thus, which muscle(s) or portion of muscle(s) used for analysis should be carefully considered and clearly outlined. Moreover, given that skeletal muscle properties can change during stress, exercise, and disease, attention should be given to the potential influence of these changes on the observed autophagic expression/signaling (J. Quadrilatero, personal communication). Although food deprivation does not induce detectable autophagy in the brain, it induces autophagy in the retina, and by the use of in vivo injection of leupeptin autophagic flux can be evaluated with LC3 lipidation by western blot [1567]. Although difficult to standardize and multifactorial, exercise may be a particularly appropriate stimulus to use for assessing autophagy in skeletal muscle [1543, 1580]. Data about the autophagic flux can be obtained by treating mice or rats with, for example, CQ [86, 1574], leupeptin [1567, 1581] or colchicine [301] and then monitoring the change in accumulation of LC3 (see cautionary notes). It should be noted, however, that surgery itself profoundly affects intracellular signaling pathways such as those involving MTOR, MAPK/ERK, and autophagic flux itself (C.N. Brown and C.L. Edelstein, personal communication). Thus, proper validation of such models should be carefully conducted before their use can be accepted. This type of flux analysis can also be done with liver, by comparing the LC3-II level in untreated liver (obtained by a partial hepatectomy) to that following subsequent exposure to CQ (V. Skop, Z. Papackova and M. Cahová, personal communication). Moreover, after peripheral nerve degeneration, to verify whether the increase in rapamycin-induced Schwann cell autophagy, can be attributed to increased autophagosome formation, the lysosomal inhibitor CQ can be injected both in vehicle- and rapamycin-treated mice, and 3 h after the injection, LC3 conversion is measured in sciatic nerves by western blot. [540].
Additional reporter assays to monitor autophagic flux in vivo need to be developed, including tandem fluorescent-LC3 transgenic mice, expressing the construct in specific cell types beyond the existing neuron-specific model [451], or viral vectors to express this construct in vivo in localized areas. Moreover, LC3-independent approaches are also needed. The LDH sequestration assay is an LC3-independent method that may be useful to study autophagic sequestration activity in vivo, and which does not require any genetic modification of the experimental animals. Indeed, injection of leupeptin in rats results in accumulation of LDH within autophagic vacuoles in hepatocytes [1582]. One of the challenges of studying autophagic flux in intact animals is the demonstration of cargo clearance, but studies of fly intestines that combine sophisticated mosaic mutant cell genetics with imaging of mitochondrial clearance reveal that such analyses are possible [1162].
Another organ particularly amenable to ex vivo analysis is the heart, with rodent hearts easily subjected to perfusion by the methods of Langendorff established in 1895 (for review see [1583]). Autophagy has been monitored in perfused hearts [1584], where it is thought to be an important process in several modes of cardioprotection against ischemic injury [1585]. It should be noted that baseline autophagy levels (as indicated by LC3-II) appear relatively high in the perfused heart, although this may be due to perceived starvation by the ex vivo organ (e.g., the lack of protein in the perfusion medium may result in osmotic stress and edema, which could trigger a starvation-like stress that accelerates autophagy), highlighting the need to ensure adequate delivery of metabolic substrates in perfusion media, which may include the addition of INS (insulin). Another concern may be that the high partial pressure of oxygen of the perfusate (e.g., buffer perfused with 95%:5% [O2:CO2]) used in the Langendorff method makes this preparation problematic for the study of autophagy because of the high levels of oxidation (redox disturbances) that could result from the preparation. However, the absence of hemoglobin means that even at a high partial pressure of oxygen these hearts may be at the limit of oxygen availability, and perfused hearts have normal levels of glutathione, NADH and other measures of redox. Due to these potential effects, great caution should be exercised in interpretation of these results. As a guide to correct interpretation of these data, we recommend a review that covers the diverse array of “state of the art” methods to analyze autophagy in cardiac physiopathology [1586].
The role of autophagy in pregnancy has been extensively reviewed [1587, 1588], and human placenta represents an organ suitable for ex vivo studies, such as to investigate pregnancy outcome abnormalities. Autophagy has been evaluated in placentae from normal pregnancies [1589–1591] identifying a baseline autophagy level (as indicated by LC3-II) in uneventful gestation. In cases with abnormal pregnancy outcome, LC3-II is increased in placentae complicated by intrauterine growth restriction in cases both from singleton pregnancies [1592] and from monochorionic twins pregnancies [1593]. Moreover, placentae from pregnancies complicated by preeclampsia show a higher level of LC3-II than normal pregnancies [1594]. Finally, placentae from acidotic newborns developing neonatal encephalopathy exhibit a higher IHC LC3 expression than placentae from newborn without neonatal encephalopathy [1595]. For this reported association, further investigations are needed to assess if autophagy protein expression in placentae with severe neonatal acidosis could be a potential marker for poor neurological outcome.
The retina is a very suitable organ for ex vivo as well as in vivo autophagy determination. The retina is a part of the central nervous system, is readily accessible and can be maintained in organotypic cultures for some time, allowing treatment with protease and autophagy inhibitors. This allows determination of autophagic flux ex vivo in adult and embryonic retinas by western blot [1566, 1596, 1597] as well as by flow cytometry and microscopy analysis [1567, 1597]. Moreover, only 4 h of leupeptin injection in fasted mice allows for autophagic flux assessment in the retina [1567] indicating two things: first, food deprivation induces autophagy in selected areas of the central nervous system; and second, leupeptin can cross the blood-retinal barrier.Accordingly, the intravitreal injection of beta-adrenergic receptor blockers in a mouse model of oxygen-induced retinopathy stimulates autophagic turnover of retinal neurons [1598].
In vivo analysis of the autophagic flux in the brain tissue of neonatal rats can also be performed. These studies use the intraperitoneal administration of the acidotropic dye monodansylcadaverine (MDC) to pup rats 1 h before sacrifice, followed by the analysis of tissue labeling through fluorescence or confocal laser scanning microscopy (365/525-nm excitation/emission filter). This method was adapted to study autophagy in the central nervous system after its validation in cardiac tissue [1599]. MDC labels acidic endosomes, lysosomes, and late-stage autophagosomes, and its labeling is upregulated under conditions that increase autophagy [1600]. In a neonatal model of hypoxic-ischemic brain injury, where autophagy activation is a direct consequence of the insult [1601], MDC labeling is detectable only in the ischemic tissue, and colocalizes with LC3-II [1602]. The number of MDC- and LC3-II-positive structures changes when autophagy is pharmacologically up- or downregulated [1601, 1602]. Whether this method can also be used in adult animals needs to be determined. Furthermore, it should be kept in mind that staining with MDC is not, by itself, a sufficient method for monitoring autophagy in live cells (see Acidotropic dyes). A better alternative approach in live cells is the MDC derivative monodansylpentane (MDH) which stains lipid-containing vacuoles such as late autophagic vacuoles [1603]. In formaldehyde-fixed cells MDC and MDH both stain lipid-containing vacuoles/late autophagosomes.
Cell-type specific observation of autophagy flux in vivo in adult brain and spinal cord is possible. Adult mice can be stereotaxically injected with lentivirus expressing mRFP-GFP-LC3 under the control of the Nes promoter in hippocampus. Using this approach, it was demonstrated that restraint stress increases autophagy flux in adult hippocampal neural stem cells, and induces autophagic death of neural stem cells without signs of apoptosis [1604]. Intrathecal injection of adenoassociated viral vector AAV9rh10, that infects spinal motoneurons, and expressing the mCherry-GFP-LC3 reporter, can be used to demonstrate autophagy flux blockage in the neurodegenerative process after proximal axotomy or nerve root avulsion [1550].
Another approach that can be used in vivo in brain tissue is to stain for lysosomal enzymes. In situations where an increase in autophagosomes has been shown (e.g., by immunostaining for LC3 and immunoblotting for LC3-II), it is important to show whether this is due to a shutdown of the lysosomal system, causing an accumulation of autophagosomes and/or incompletely acidified autolysosomes, or whether this is due to a true increase in autophagic flux. The standard methods described above for in vitro research, such as the study of clearance of a substrate, are difficult to use in vivo, but if it can be demonstrated that the increase in autophagosomes is accompanied by an increase in lysosomes, this makes it very likely that there has been a true increase in autophagic flux [1605]. Conversely, a decrease in lysosomal enzyme levels and activity can indicate that accumulation of autophagosomes is caused by lysosomal damage and a consequent decrease in flux [1606, 1607]. Lysosomal enzymes can be detected by IHC (e.g., for LAMP1 or CTSD) or by classical histochemistry to reveal their activity (e.g., ACP/acid phosphatase or HEX/β-hexosaminidase) [1608, 1609]. It should be noted, however, that this combination of measures will not exclude a defect in lysosomal acidification, increasingly reported in several major neurodegenerative diseases [1610]. In this situation, flux is blocked, and incompletely acidified autolysosomes accumulate, which cannot be discriminated from autophagosomes using the mCherry/RFP-GFP-LC3 probe (or other measures of LC3) because both vesicle types will fluoresce yellow. Only by applying a third fluorescent marker for lysosomes (e.g., CTSD, CTSB) by IHC can the deacidified autolysosomes be identified [451]. Lysosomal enzyme activity can be also separately assessed in lysosomes and cytosol following tissue fractionation. In this case, a decrease in enzyme activity in the lysosomal fraction accompanied by an increase in the cytosol can indicate lysosomal membrane permeabilization (LMP) as a potential cause for lysosomal dysfunction. LMP may also be detected in vivo in the brain by comparing the pattern of IHC staining for lysosomal membrane proteins (such as LAMP1/2) to soluble lysosomal enzymes (such as CTSB, CTSD or CTSL) [1606, 1611].
Some biochemical assays may be used to at least provide indirect correlative data relating to autophagy, in particular when examining the role of autophagy in cell death. For example, cellular viability is related to high CTSB activity and low CTSD activities [1612]. Therefore, the appearance of the opposite levels of activities may be one indication of the initiation of autophagy (lysosome)-dependent cell death. The question of “high” versus “low” activities can be determined by comparison to the same tissue under control conditions, or to a different tissue in the same organism, depending on the specific question.
Cautionary notes: The major hurdle with most in vivo analyses is the identification of autophagy-specific substrates and the ability to “block” autophagosome degradation with a compound such as bafilomycin A1. Regardless, it is still essential to adapt the same rigors for measuring autophagic flux in vitro to measurements made with in vivo systems. Moreover, as with cell culture, to substantiate a change in autophagic flux it is not adequate to rely solely on the analysis of static levels or changes in LC3-II protein levels on western blot using tissue samples. To truly measure in vivo autophagic flux using LC3-II as a biomarker, it is necessary to block lysosomal degradation of the protein. Several studies have successfully done this in selected tissues in vivo. Certain general principles need to be kept in mind: (a) Any autophagic blocker, whether leupeptin, bafilomycin A1, CQ or microtubule depolarizing agents such as colchicine or vinblastine, must significantly increase basal LC3-II levels in control cells or tissues. The turnover of LC3-II or rate of basal autophagic flux is not known for tissues in vivo, and therefore short treatments (e.g., 4 h) may not be as effective as blocking for longer times (e.g., 12 to 24 h). (b) The toxicity of the blocking agent needs to be considered (e.g., treating animals with doses higher than 2 mg/kg bafilomycin A1 for 2 h can be quite toxic), and food intake must be monitored. If long-term treatment is needed to see a change in LC3-II levels, then confirmation that the animals have not lost weight may be needed. Mice may lose a substantial portion of their body weight when deprived of food for 24 h, and starvation is a potent stimulus for the activation of autophagy. (c) The bioavailability of the agent needs to be considered. For example, many inhibitors such as bafilomycin A1 or CQ have relatively poor bioavailability to the central nervous system. To overcome this problem, intracerebroventricular injection can be performed.
A dramatic increase of intracellular free poly-unsaturated fatty acid levels can be observed by proton nuclear magnetic resonance spectroscopy in living pancreatic cancer cells within 4 h of autophagy inhibition by omeprazole, which interacts with the V-ATPase and probably inhibits autophagosome-lysosome fusion [1613]. Omeprazole is one of the most frequently prescribed drugs worldwide and shows only minor side effects even in higher doses. Proton nuclear magnetic resonance spectroscopy is a noninvasive method that can also be applied as localized spectroscopy in magnetic resonance tomography, and therefore opens the possibility of a noninvasive, clinically applicable autophagy monitoring method, although technical issues still have to be solved [1614].
In terms of measuring mitophagy in tissues, recently developed reporter systems represent a more rigorous choice than monitoring any particular pathway. This is especially true for stress-induced PINK1-dependent PRKN phosphorylation, where KO-validated reagents to monitor this signaling pathway in mice have only just become available. It is important to note that despite a plethora of publications, many commercially available anti-PINK1 and anti-PRKN antibodies are not specific; that is, although it is possible to run a western blot with these reagents and detect a band at the predicted size, it is highly likely that this band will also be present in KO tissue (especially for endogenous mouse PINK1). The first endogenous detection of mouse PINK1 from tissues verified using KO controls and mass spectrometry has been published [1231]. Readers should be aware that the detection of bona fide PINK1 is technically challenging, and the current state of the art necessitates an immunoprecipitation-immunoblot approach to ensure optimal results. This approach has been successfully replicated in other mouse cell types. In cells, researchers also use the PINK1-dependent phosphorylation of PRKN or ubiquitin at Ser65 to monitor pathway activation. Monitoring PRKN substrate ubiquitination is another useful approach. While these methods are tractable for in vitro paradigms, the activation of this pathway requires substantial levels of stress (often treatment with harsh mitochondrial uncouplers). Thus, PINK1-mediated generation of phospho-ubiquitin, phospho-PRKN or substrate ubiquitination can be difficult to detect without mitochondrial depolarization. Nonetheless, the activation of this endogenous pathway has been performed in mature primary neurons using a combination of ubiquitin-enrichment and highly specific antibodies [1615]. Researchers should also be mindful that while detection of Pink1 or Prkn mRNA may seem like a useful approach, changes in the levels of these genes do not infer any reliable alterations in mitophagy.
When analyzing autophagic flux in vivo, one major limitation is the variability between animals. Different animals do not always activate autophagy at the same time. To improve the statistical relevance and avoid unclear results, these experiments should be repeated more than once, with each experiment including several animals; it may also be important to consider age and gender [1616] as additional variables. Induction of autophagy in a time-dependent manner by fasting mice for different times requires appropriate caution. Mice are nocturnal animals, so they preferentially move and eat during the night, while they mostly rest during daylight. Therefore, in such experiments it is better to start food deprivation early in the morning, to avoid the possibility that the animals have already been fasting for several hours. The use of CQ is technically easier, because it only needs one intraperitoneal injection per day, but the main concern is that CQ has some toxicity (mouse intraperitoneal LD50: 68 mg/kg). CQ suppresses the immunological response in a manner that is not due to its pH-dependent lysosomotropic accumulation (CQ interferes with LPS-induced Tnf/Tnf-α gene expression by a nonlysosmotropic mechanism) [1617], as well as through its pH-dependent inhibition of antigen presentation [1433]. Therefore, CQ treatment should be used for short times and at doses that do not induce severe collateral effects, which may invalidate the measurement of the autophagic flux, and care must be exercised in using CQ for studies on autophagy that involve immunological aspects.
It is also important to have time-matched controls for in vivo analyses. That is, having only a zero-hour time point control is not sufficient because there may be substantial diurnal changes in basal autophagy [989]. For example, variations in basal flux in the liver associated with circadian rhythm may be several fold [989], which can equal or exceed the changes due to starvation. Along these lines, to allow comparisons of a single time-point it is important to specify what time of day the measurement is taken and the lighting conditions under which the animals are housed. It is also important that the replicate experiments are conducted at the same time of day. Controlling for circadian effects can greatly reduce the mouse-to-mouse variability in autophagy markers and flux [1618]. Note, when handling litters, autophagy flux should be analyzed within a restricted range of weight; nursing mothers have a limited production of nutrients, and therefore an increased variability is detected between groups of big and small litter number.
When analyzing the basal autophagic level in vivo using GFP-LC3 transgenic mice [239], one pitfall is that GFP-LC3 expression is driven by the Cmv/cytomegalovirus enhancer and Actb/β-actin (CAG) promoter, so that the intensity of the GFP signal may not always represent the actual autophagic activity, but rather the CAG promoter activity in individual cells. For example, GFP-LC3 transgenic mice exhibit prominent fluorescence in podocytes, but rarely in tubular epithelial cells in the kidney [239], but a similar GFP pattern is observed in transgenic mice carrying CAG promoter-driven non-tagged GFP [1619]. Furthermore, proximal tubule-specific ATG5-deficient mice [1620] display a degeneration phenotype earlier than podocyte-specific ATG5-deficient mice [1621], suggesting that autophagy, and hence LC3 levels, might actually be more prominent in the former.
One caution in using approaches that monitor ubiquitinated aggregates is that the accumulation of ubiquitin may indicate a block in autophagy or inhibition of proteasomal degradation, or it may correspond to structural changes in the substrate proteins that hinder their degradation. In addition, only cytosolic and not nuclear ubiquitin is subject to autophagic degradation. It is helpful to analyze aggregate degradation in an autophagy-deficient control strain, such as an autophagy mutant mouse, whenever possible to determine whether an aggregate is being degraded by an autophagic mechanism. This type of control will be impractical for some tissues such as those of the central nervous system because the absence of autophagy leads to rapid degeneration. Accordingly, the use of Atg16l1 hypomorphs, Becn1 heterozygotes or Atg4b homozygotes, with systemic autophagy impairment, may help circumvent this problem.
Conclusion: Although the techniques for analyzing autophagy in vivo are not as advanced as those for cell culture, it is still possible to follow this process (including flux) by monitoring, for example, GFP-LC3 or mCherry/RFP-GFP-LC3 by fluorescence microscopy, and SQSTM1 and NBR1 by IHC and/or western blotting.
18. Proteomic readouts of autophagy
An alternate approach for evaluating autophagy is with proteomics, which enables the identification of hundreds to thousands of protein species in a sample. The main advantage of proteomics is that it provides a direct, holistic readout of how autophagic activity affects the protein composition of a cell. Proteomics also avoids an assumption of common “marker-based” autophagy assays (LC3B-based or otherwise)—that dynamic changes to either the abundance or localization of a marker protein is generally reflective of total autophagic activity. Although proteomics requires specialized equipment and data processing, gradual improvements in technology, declining cost, and availability through core facilities and companies are making proteomics increasingly accessible.
Over the last decade, dozens of studies employing proteomics to examine autophagic activity have been published, and the pace of novel publications is accelerating [1622, 1623]. While these studies differ significantly in their technical execution (on-label versus label free, instrumentation, sample processing, and quantification), conceptually they can be subdivided into three general experimental approaches. In the first approach, proteomics is used to examine changes to total cellular protein composition in the setting of autophagy inhibition or stimulation. As an example, an on-label proteomic approach known as stable isotope labeling by amino acids in cell culture (SILAC) has been used to analyze cells subjected to autophagy activation by amino acid starvation [1029]. The results indicate that autophagy activation is accompanied by an orderly progression of substrates that are targeted for disposal, starting with cytosolic proteins and followed later by mitochondrial and other organellar proteins. This kind of whole cell proteomics analysis provides a holistic picture of how autophagy affects cellular proteostasis, but it does not distinguish between proteins that are directly degraded by autophagy and proteins whose steady-state levels change through indirect effects (regulation of transcription, translation, or export) or through off-pathway functions of ATG proteins.
Another example is seen from experiments conducted in maize, where the protein composition in autophagy mutants was determined using a label-free MS analysis of the total protein extract [1624]. One remarkable observation was the ~2-fold increase in protein content/fresh weight in the absence of autophagy, which was at least partially due to a retention of various organelles. Global comparisons between affected transcript and protein abundances, made it possible to pinpoint putative autophagic cargo (solely elevated protein levels) and proteins that are actively engaged (elevated transcript and protein levels). Although protein-transcript comparisons are potentially flawed due to misassigned protein-coding mRNAs (due to homology), or due to differences in translation efficiencies, consistent trends were observed for several protein groups. For example, strong increases of peroxisomal, endoplasmic reticulum, Golgi, ribosomal and proteasomal proteins are evident without any associated transcripts being affected, indicating that these organelles and protein complexes are autophagic targets. In contrast, proteins involved in secondary, amino acid, glutathione and lipid metabolism are elevated in both protein and corresponding mRNA abundances, which strongly correlate with alterations in associated metabolites, indicative of an active response to restore cellular homeostasis.
In the second approach, proteomics is used to catalog the composition of autophagosomes or autolysosomes that are isolated using biochemical fractionation or affinity purification. This approach can identify specific autophagy substrates, and through these substrates it can suggest cellular functions that autophagy is affecting. To cite some examples, label-free proteomics of biochemically fractionated autolysosomes was used to identify the cargo receptor NCOA4 that regulates iron homeostasis by recruiting ferritin to phagophores [1264] (see Ferritinophagy). Another study [1625] used label-free proteomics to compare the substrates of CMA-competent versus CMA-incompetent lysosomes in mouse liver, thereby inferring unique substrate specificity of CMA compared to autophagy. A novel chemical labeling approach [1626] transfected APEX-Atg8-family fusion proteins into cells, which enables the biotinylation and subsequent purification of autophagosome contents using streptavidin resin. Combined with a SILAC-based proteomics analysis, this technique identified a novel PRKN-independent mitophagy mechanism that is dependent on LC3C.
In the third approach, proteomics is used in a quantitative or semi-quantitative manner to measure autophagic flux. This approach enables simultaneous examination of how a stimulus affects the rate of autophagic activity and the composition of the autophagy substrate proteome. For example, SILAC was used to conduct a pulse-chase experiment in human fibroblasts, enabling the proteome-wide calculation of protein half-lives under basal conditions [1627]. By comparing cells with atg5 or atg7 deletion to wild-type cells, they were able to infer degradation rates via autophagy in many hundreds of proteins simultaneously. In another example, a label-free approach was used to examine circadian variations in autophagic flux in mouse liver [1618].
Cautionary notes: Current proteomic platforms identify on the order of 104 to 105 protein spectra (similar in concept to RNA sequencing reads) per sample. By comparison, RNA sequencing provides on the order of 107 reads per sample, although it does not specifically address the issue of RNA turnover. The limited sensitivity of proteomics means that the technique favors detection of abundant proteins and is less reliable for reproducibly detecting rarer protein species. To some extent this can be overcome by reducing the complexity of the sample being analyzed (for example, by analyzing purified autolysosomes rather than whole cell homogenates), but it is routine for non-abundant proteins to be detected in some biological replicates but not in others.
Because cellular material must be homogenized, proteomic readouts do not preserve subcellular localization information precisely, even when samples are carefully biochemically fractionated. Particularly with human biological samples, care must be taken to avoid contamination with exogenous human proteins, especially with samples that have small quantities of protein to begin with [1628].
In proteomics, proteins are identified by matching peptide sequences against a database (akin to RNA sequencing). In some instances, peptides can be misassigned to a protein because the peptide sequence in question maps to a conserved region shared by multiple different protein species. Finally, the sensitivity of proteomic detection depends on the ionizability of different oligopeptides which varies from protein to protein. As a result, the linear relationship between a proteomic metric such as spectral counts, and absolute protein abundance varies in slope from protein species to protein species. What this means is that while shotgun proteomics can distinguish between the relative amounts of a given protein in different samples, it cannot reliably compare the abundances of two different protein species without the addition of reference protein standards of known quantity.
Conclusion: Even with all the technical caveats, proteomics is unique in allowing the application of “omics” approaches to autophagy measurement and can be used to validate the conclusions of marker-based autophagy assays. As the technology continues to improve and as the costs of experiments decline, proteomics is likely to become an increasingly standard approach to examining the role of autophagy in cellular physiology and pathophysiology.
19. Metabolic markers of autophagy
Metabolites play an essential role in autophagy regulation and therefore constitute key targets for the understanding of biological processes that are involved in autophagy and are misregulated in autophagy-related diseases. Recent metabolomics approaches have been developed in order to identify the key metabolites involved in the regulation of autophagy [1629]. These approaches rely on two main and complementary methods, which are MS and NMR spectroscopy. On the one hand, NMR provides access to unique structural information, is quantitative and highly reproducible. On the other hand, MS is more sensitive than NMR, but suffers from the ambiguity of spectral signatures.
The regulation of autophagy is mediated by various conditions including (a) starvation and (b) protein acetylation status. Under normal growth conditions, associated with abundant nutrients, autophagy is kept at a basal level making it possible to maintain essential cellular processes such as the turnover of damaged cellular organelles and the degradation of proteins. Under conditions of nutrient starvation, autophagy is further induced to provide cells with additional internal nutrient supplies and is associated with a dramatic change in the cellular metabolome profile. Indeed, low glucose levels result in decreased cellular capacity to convert ATP to cAMP and are therefore linked to a decreased activation of autophagy-related proteins via the PRKA/cAMP-dependent protein kinase A pathway [36, 1629]. Therefore, monitoring the levels of glucose and cAMP as well as the AMP:ATP ratio are efficient readouts associated with autophagic capacity, and these can be quantitatively detected using both NMR spectroscopy and MS approaches.
Several studies underlined the role of protein acetylation in the regulation of autophagy, and show that a decreased cellular acetylation level is associated with increased autophagy [996, 1630]. For instance, Atg proteins mediate autophagy via formation of autophagosomes only in their de-acetylated state [1631, 1632]. Protein acetylation status is regulated by the cellular balance between acetyltransferases and deacetylases, which use acetyl-CoA and NAD+ as cofactors, respectively. Therefore, monitoring acetyl-CoA and NAD+ metabolites are efficient readouts of protein acetylation marks and associated autophagic flux. Several studies also underline the role of polyamines, spermidine and spermine in the regulation of autophagy via inhibition of histone-acetyltransferases [1633–1636]. Nevertheless, the exact cellular mechanisms linking histone deacetylation and autophagy regulation are still unclear but likely involve a transcription-dependent activation/repression of autophagy-related genes. The cellular NAD+ and spermidine levels can be detected by both MS and NMR spectroscopy, whereas, due to its low cellular abundance, acetyl-CoA can only be detected using MS.
Other metabolites also reflect the autophagic capacity of the cell. As previously mentioned, autophagy allows protein turnover via activation of proteolysis. Therefore, levels of free amino acids, which are building blocks of proteins are suitable markers for (in)activation of autophagy and can be quantitatively detected using NMR spectroscopy and MS [1637, 1638]. Finally, elevated levels of free fatty acids or triglycerides as well as production of PtdIns3P are linked to induction of autophagy [36]. Detection and quantification of this complex class of lipids is usually performed using MS [1639], as NMR spectroscopy provides mainly information regarding the chemical nature of apolar metabolites.
In conclusion, metabolomics studies provide essential information in the field of autophagy and contribute to the deep-understanding of its complex regulatory mechanisms in living cells and organisms. Given the recent advances in method development using NMR and MS metabolomics approaches, it is to be expected that more metabolites involved in autophagy regulation [1640–1642] will be identified in the coming years.
20. Clinical setting
Altered autophagy is clearly relevant in neurodegenerative diseases, as demonstrated by the accumulation of protein aggregates and gene dysregulation, for example in AD [1643, 1644], adult brain ischemia [1645, 1646], PD [1647], Huntington disease (HD) and other polyglutamine repeat expansion diseases [1648, 1649], muscle diseases [1650, 1651], and ALS [1652]. Elevated levels of autophagosomes or mitophagosomes have been identified ultrastructurally in aging, brain ischemia, vacuolar myopathies, PD, AD and Lewy body dementia [79, 1133, 1188, 1653]. Of note, depending on the disease being considered, autophagy is not necessarily impaired but could be, in particular conditions, excessively activated (i.e., an increase in the autophagic flux) such as in neonatal models of cerebral ischemia [1608, 1654, 1655]. Autophagy defects with autophagosome accumulation are also associated with different forms of hereditary spastic paraplegia/HSP [1656]. Of note, the expression levels of ATG5 and the ratio between LC3A and LC3B significantly increase in 3xTgAD mouse brain, following treatment with near infrared light, thus emphasizing the involvement of autophagic machinery in the degradation of dysfunctional MAPT protein [1657]. Further evidence comes from the observations that the stress-inducible mitophagy regulators PINK1 and PRKN show loss-of-function mutations in autosomal recessive juvenile parkinsonism [1658]. Along these lines, it is important to dissociate the clinical significance of these PD-associated loci in patients from the depolarization-induced “PINK1-PRKN signaling pathway” as it is traditionally studied in cultured cells.
A very useful nonspecific indicator of deficient aggrephagy in autopsy brain or biopsy tissue is SQSTM1 IHC [1659, 1660]. For clinical attempts to monitor autophagy alterations in peripheral tissues such as blood, it is important to know that eating behavior may be altered as a consequence of the disease [1661], resulting in a need to control feeding-fasting conditions during the analyses. Recently, altered autophagy was also implicated in schizophrenia, with BECN1 transcript levels decreasing in the postmortem hippocampus in comparison to appropriate controls [1662]. In the same hippocampal postmortem samples, the correlation between the RNA transcript content for ADNP (activity-dependent neuroprotective homeobox) and its sister protein ADNP2 is deregulated [1663], and ADNP as well as ADNP2 RNA levels increase in peripheral lymphocytes from schizophrenia patients compared to matched healthy controls, suggesting a potential biomarker [1662].
Over the past decade, our depth of knowledge and understanding on therapeutic potentials of autophagy inhibition for treating cancer has been vastly improved. Particularly, after tumors have been formed, cancer cells actively undergo autophagy to survive and grow under conditions of nutrient limitation and hypoxia. Therefore, autophagy inhibitors are becoming emerging therapeutics to combat cancer [1664, 1665]. To this end, more pharmacological molecules that are designed to suppress autophagy have been examined for clinical use such as 3-MA, wortmannin, LY294002, CQ, and HCQ [1665]. For example, class III PtdIns3K inhibitors including 3-MA, wortmannin and LY294002 prevent autophagosome formation, and thus inhibit autophagy. However, these inhibitors are not specific for inhibiting autophagy and can activate autophagy at higher doses. Thus, the PtdIns3K inhibitors are not suitable for clinical settings. Other commonly used autophagy inhibitors such as CQ and its derivative HCQ that are FDA approved anti-malaria drugs, have been extensively studied and tested in clinical trials. Although CQ and HCQ show moderate anti-neoplastic effects, these largely come from the modulation of pathways other than autophagy inhibition per se [1666, 1667]. Moreover, the mechanism by which CQ and HCQ inhibit autophagy is still not fully understood. Therefore, developing molecules that specifically regulate autophagy will surely broaden clinical utility in combating cancer.
In addition to neurodegenerative diseases, alterations in autophagy have also been implicated in other neurological diseases including some epilepsies, neurometabolic and neurodevelopmental disorders [1569, 1668–1670], and inherited autophagic vacuolar myopathies (including Danon disease, acid maltase deficiency/Pompe disease, X-linked myopathy with excessive autophagy/XMEA, etc.), which are characterized by lysosomal defects and an accumulation of autophagic vacuoles [1671]. Autophagic vacuolar myopathies and cardiomyopathies can also be secondary to treatment with autophagy-inhibiting drugs (CQ, HCQ and colchicine), which are used experimentally to interrogate autophagic flux and clinically to treat malaria, rheumatological diseases, and gout [1561]. Autophagy impairment has also been implicated in the pathogenesis of inclusion body myositis, an age-associated inflammatory myopathy that is currently refractory to any form of treatment [1672–1675], along with some muscular dystrophies such as tibial muscular dystrophy [1676]. In all these striated muscle disorders, accumulated autophagic vacuoles can be seen by electron microscopy, or, alternatively, LC3 and/or SQSTM1 can be detected by IHC [1560, 1561, 1651, 1677]. In addition, autophagy defects can also lead to the formation of an eosinophilic cytoplasmic inclusion, which is a round to oval homogeneous cytoplasmic eosinophilic globule composed of protein aggregates and/or organelles; SQSTM1, BECN1, NBR1, LC3 and/or peroxisomes are deposited in the inclusion, and both the proteins and organelles can be detected by IHC, immunofluorescence, or TEM [1678].
Whereas autophagosomes and autolysosomes are not always distinguishable using only morphological methods to confirm whether or not an autophagic structure has fused with a lysosome, “autophagic vacuoles” are easily recognized by electron microscopy in the cardiomyocytes of patients with dilated cardiomyopathy [550]. Autophagic vacuoles are easily observed not only in secondary cardiomyopathy but also in failing cardiomyocytes of dilated cardiomyopathy [550]. These vacuoles display LC3 expression by using the ABC technique for TEM observation (see Transmission electron microscopy) [112]. Dilated cardiomyopathy with autophagic vacuoles indicates a good prognosis, confirming that autophagy resists cardiomyocyte degeneration. In dilated cardiomyopathy, it is suggested that autophagy is not always the cause of the disease but also a process that occurs to prevent the disease.
In addition, altered basal autophagy levels are seen in rheumatoid arthritis [1028,1029], systemic lupus erythematosus (SLE) [1679–1681], and osteoarthritis [1682]. Other aspects of the immune response associated with dysfunctional autophagy are seen in neutrophils from patients with familial Mediterranean fever [1683] and in monocytes from patients with TNF receptor-associated periodic syndrome [1684], two autoinflammatory disorders. Aberrant elevation of IL17A plays a critical role in the pathogenesis of pulmonary fibrosis through suppressing the autophagic degradation of collagen in fibrotic lung tissue [1685]. In lung epithelial cells, IL17A-activated PIK3CA inhibits the kinase activity of GSK3B by stimulating its phosphorylation at Ser9, which consequently attenuates activation of an autophagic core complex via inhibiting the ubiquitination-dependent degradation of BCL2 and its interaction with BECN1 [1686]. ANXA2 is identified as a specific bleomycin target linked to interstitial pulmonary fibrosis as bleomycin binding to ANXA2 impedes TFEB-induced autophagic flux to cause pulmonary fibrosis proliferation [1687].
Moreover, autophagy regulates an important neutrophil function, the generation of neutrophil extracellular traps (NETs) [1674, 1688]. The important role of autophagy in the induction of NET formation has been studied in several neutrophil-associated disorders such as gout [1689] and other IL1B autoinflammatory disorders [1690–1692], ulcerative colitis [1693], sepsis [1694], thromboinflammation [1695, 1696] and lung fibrosis [1697], including the inflammatory remodeling associated with systemic sclerosis [1698]. The prototypical DAMP and autophagy inducer, HMGB1, released by activated platelets appears to play a role in neutrophil autophagic flux induction [1674, 1698, 1699], and studies of patients with systemic sclerosis have shown that platelet-derived, microparticle-associated HMGB1 promotes neutrophil autophagy, as evidenced by Cyto-ID labeling, leading to the production of NETs [1698].
Furthermore, there is an intersection between autophagy and the secretory pathway in mammalian macrophages for the release of IL1B [1700], demonstrating a possible alternative role of autophagy for protein trafficking. This role has also been implied in neutrophils through exposure of protein epitopes on NETs by acidified LC3-positive vacuoles in sepsis [1694] and anti-neutrophil cytoplasmic antibody associated vasculitis [1701]. Patients with chronic kidney disease also have impaired autophagy, which results in NLRP3 activation, IL1B release and leukocyte influx. However, autophagy was also shown to play an important role in the development in vitro of giant phagocytes, a long-lived neutrophil subpopulation, derived from neutrophils of healthy individuals [1702, 1703]. Recently, evidence from genetic, cell biology and animal models suggests that autophagy plays a pivotal role in the occurrence and development of SLE. For example, altered basal autophagy levels are seen in immune cells, such as B cells, T cells, and neutrophils in SLE [1679]. There is also evidence for altered autophagy in pancreatic beta cells [1704, 1705], and in adipocytes [292, 396, 1706] of patients with type 2 diabetes [1707].
Photodynamic therapy (PDT), an FDA-approved anticancer therapy, is based on electromagnetic radiation and has applications in the selective eradication of delineated tumor lesions and infection sites. It is a two-step process whereby cells are first incubated with photosensitizers and then exposed to light, usually in the red spectral region. Although these components (i.e., photosensitizers and light) are harmless alone, when combined they provide a localized therapeutic archetype avoiding attack to healthy cells and preventing side effects [1708, 1709]. This combination results in the generation of singlet oxygen (1O2) and other ROS that can cause cancer cell death [1710]. PDT can prompt AKT-MTOR pathway downregulation and stimulate autophagy in eukaryotic cells [1711]. The mechanism of PDT that modulates autophagy depends on several factors, such as photosensitizer molecular properties and concentrations, light dose and the preferential intracellular target of the photosensitizers. Particularly, photosensitizers that target lysosomes (e.g., chlorophyllin e4, chlorophyllin f, NPe6, WST11, TPPS2a, MB, and DMMB) can modulate autophagy [316, 317, 1712–1715]. PDT fulfills the need to merge a direct cytotoxic action on tumor cells with potent immunostimulatory effects (i.e., immunogenic cell death, ICD) [1716]. A few photosensitizers, such as Photofrin, hypericin, Foscan, 5-ALA and Rose Bengal acetate, are associated with DAMP exposure and/or release that is a requisite to elicit ICD. Rose Bengal acetate PDT is the first treatment to induce autophagic HeLa cells to express and release DAMPs, thus suggesting a possible role of the autophagic cells in ICD induction [1717]. Similarly, the photosensitizer hypocrellin B-acetate is able to induce autophagy at very low concentrations [1718].
A crucial role for therapy-induced autophagy in cancer cells has recently emerged, in modulating the interface of cancer cells and the immune system [1719]; primarily, by affecting the nature of danger signaling (i.e., the signaling cascade that facilitates the exposure and/or release of danger signals) associated with ICD [1716, 1719–1722]. This is an important point considering the recent clinical surge in the success of cancer immunotherapy in patients, and the emerging clinical relevance of ICD for positive patient prognosis. Several notorious autophagy-inducing anticancer therapies induce ICD including mitoxantrone, doxorubicin, oxaliplatin, radiotherapy, certain oncolytic viruses and hypericin-based photodynamic therapy (Hyp-PDT) [1709, 1722–1724]. In fact, in the setting of Hyp-PDT, ER stress-induced autophagy in human cancer cells suppresses CALR (calreticulin) surface exposure (a danger signal crucial for ICD) thereby leading to suppression of human dendritic cell maturation and human CD4+ and CD8+ T cell stimulation [1724]. Similarly, ATG5- and ATG7-dependent autophagic responses limit the secretion of type I interferon by cancer cells undergoing radiotherapy-driven ICD, largely as a consequence of decreased cytosolic accumulation of mitochondrial DNA and consequent inhibition of CGAS-STING1 signaling [1725].
Conversely, chemotherapy (mitoxantrone or oxaliplatin)-induced autophagy facilitates ATP secretion (another crucial ICD-associated danger signal) thereby facilitating ICD and anti-tumor immunity in the murine system, the first documented instance of autophagy-based ICD modulation [1726]. The role of ATP as a DAMP becomes clear when the extracellular concentration of ATP becomes high and elicits activation of the purinergic receptor P2RX7. P2RX7 is involved in several pathways, including the sterile immune response, and its activation induces cancer cell death through PI3K, AKT and MTOR [1727, 1728]. In addition, cells lacking the essential CMA gene LAMP2A fail to expose surface CALR after treatment with both Hyp-PDT and mitoxantrone [1729].
Although autophagy has been linked to fibrosis in many tissues, not much is known about it with regard to respiratory diseases per se. Initial observations have demonstrated that there is an increased formation of autophagosomes in mesenchymal cells from asthmatic donors with an increase in ATG5 in the lung [1730, 1731]. Basal autophagy markers can be measured using IHC in the lung tissue, and with this approach it is possible to measure expression of BECN1, ATG5, LC3B and SQSTM1 in the airway epithelium and mesenchymal layer (airway wall) of asthmatic and non-asthmatic human tissues in both small and large airways [1732]. The actual expression of these markers may vary in the airway wall and is largely dependent upon cell type as observed in the lung tissue; however, these observations provide a tool to monitor basal autophagy in health vs disease and can provide useful information on how it varies from one cell type to another in a clinical setting.
Finally, it is important to note that disease-associated autophagy defects are not restricted to macroautophagy but also concern other forms of autophagy. CMA impairment, for instance, is associated with several disease conditions, including neurodegenerative disorders [307, 1733], lysosomal storage diseases [1734, 1735], nephropathies [1736] and diabetes [1737]. In addition, it is very important to keep in mind that although human disease is mostly associated with inhibited autophagy, enhanced autophagy has also been proposed to participate in, and even contribute to, the pathogenesis of human diseases, such as chronic obstructive pulmonary disease [1738], adipocyte/adipose tissue dysfunction in obesity [292, 396] and bilirubin-induced neurotoxicity [1739]. Along these lines, CQ was reported to decrease diabetes risk in patients treated with the drug for rheumatoid arthritis [1740].
A set of recommendations regarding the design of clinical trials modulating autophagy can be found in ref. [1741].
Cautionary notes: Although the protein products of several genes mutated in different neurodegenerative diseases are involved in regulating selective autophagy [1188], several of these gene products also act together to regulate other important aspects of neuronal structure and function. For example, PINK1 (implicated in mitophagy) interacts with VCP/p97 (implicated in ribophagy and granulophagy) to promote the growth and extension of neuronal processes through activation of PRKA/PKA signaling, and not via degradative mechanisms [1742]. To establish a role for autophagy in disease states, whether neurodegenerative or immunological, specific tests need to be performed where genes encoding autophagy-relevant components (e.g., ATG5, ATG7 or BECN1) have been knocked down through RNA silencing or other protein- or gene-specific targeting technologies [1724, 1726, 1729]. Usage of chemical inhibitors such as bafilomycin A1, 3-MA or CQ can create problems owing to their off-target effects, especially on immune cells, and thus their use should be subjected to due caution, and relevant controls are critical to account for any off-target effects. In the context of ICD, consideration should be given to the observations that autophagy can play a context-dependent role in modulating danger signaling [1724, 1726, 1729]; and thus, all the relevant danger signals (e.g., surface exposed CALR or secreted ATP) should be (re-)tested for new agents/therapies in the presence of targeted ablation of autophagy-relevant proteins/genes, accompanied by relevant immunological assays (e.g., in vivo rodent vaccination/anti-tumor immunity studies or ex vivo immune cell stimulation assays), in order to imply a role for autophagy in regulating ICD or general immune responses.
21. Cell death and autophagy
Autophagy is often seen in tumor tissue accompanying cell death; however, the function of autophagy mediating cell death is more limiting, and mostly confined to specific settings [1743–1745]. It is important to carefully establish the contribution of autophagy to the execution of cell death before making claims that autophagy is involved in the cell death process. Published literature often suffer from ambiguous use of the term “autophagic cell death,” which was coined in the 1970s [1746] in a purely morphological context to refer to cell death with autophagic features (especially the presence of numerous secondary lysosomes); this was sometimes taken to suggest a role of autophagy in the cell death mechanism, but death-mediation was not part of the definition [1747]. Recent nomenclature guidelines suggest that autophagy-dependent cell death (ADCD) is a distinct mechanism of cell death, independent of apoptosis or necrosis [1266]. Additional contributions of autophagy to cell death can be: (a) autophagy-associated cell death, where autophagy accompanies other cell death modalities and (b) autophagy-mediated cell death (AMCD), which could involve a standard mechanism of cell death such as apoptosis, but triggered by autophagy. The contribution of autophagy to cell death needs to be established by genetic and pharmacological means where autophagy inhibition blocks or reduces cell death, especially when distinct pathways of cell death appear to be simultaneously triggered by certain events [1748, 1749]. However, while evidence for the need of autophagy in the context of cell death alone may support the definition of autophagy-mediated cell death, it is important when establishing ADCD that further proof is required that other established modes of programmed (or regulated) cell death do not contribute to cellular demise. It is preferable to use the term AMCD when it is proven that autophagy is a pre-requisite for the occurrence of cell death, but it is not proven that autophagy mechanistically mediates the switch to cell death [1750].
Inhibition of the full autophagy degradation cycle has also been proposed to lead to specific forms of autophagy-associated cell death, such as karyoptosis, involving the nucleophagy machinery and clearance by expulsion into the extracellular space [1750–1752]. Induction of the autophagy degradation cycle also promotes other cell death pathways, such as apoptosis, and cell cycle arrest [1753, 1754]. It is important to note that a stress stimulus can in many circumstances induce different cell death pathways at the same time, which might lead to a “type” of cell death with mixed phenotypes [678, 1755–1757]. Here, autophagy can be one of a range of adaptive mechanisms induced in the face of cellular stress, which precedes cell death if the stress cannot be overcome. Furthermore, inhibition of one cell death pathway (e.g., apoptosis) can either induce the compensatory activation of a secondary mechanism (e.g., necrosis) [1758, 1759], or attenuate a primary mechanism (e.g., liponecrosis) [1755].
The role of autophagy in the death of plant cells is well established, because plants are devoid of the apoptotic machinery and use lytic vacuoles to disassemble dying cells from inside [1760]. This mode of cell death governs many plant developmental processes, as well as stress-induced cell death in some plant systems [911] and was named “vacuolar cell death” [1761]. Recent studies have revealed a key role of autophagy in the execution of vacuolar cell death, where autophagy sustains the growth of lytic vacuoles [1762, 1763]. Besides being an executioner of vacuolar cell death, autophagy can also play an upstream, initiator role in immunity-associated cell death related to the pathogen-triggered hypersensitive response [1760, 1764].
Upon induction by starvation of multicellular development in the protist D. discoideum, autophagy (or at least Atg1) is required to protect against starvation-induced cell death, allowing vacuolar developmental cell death to take place instead [1765, 1766]. Autophagy may be involved not only in allowing this death to occur, but also, as during vacuolar cell death in plants, in the vacuolization process itself [1767]. D. discoideum provides the ability to rapidly identify and characterize defects in lysosomal activity and autophagic degradation in relation to model diseases, such as a non-proteolytic activity for the gamma secretase complex [1768, 1769].
The best known physiologically relevant demonstration of cell death that involves autophagy, and not apoptosis, is during Drosophila development. Drosophila is a powerful genetically amenable model system to study ADCD, as the process of autophagy and the function of Atg genes are highly conserved, enabling genetic analysis of the autophagy machinery components and interactions with other pathways (see Drosophila melanogaster). During Drosophila metamorphosis temporal increases in the steroid hormone ecdysone trigger the degradation of obsolete larval tissues including the midgut and salivary gland. Larval midgut degradation is dependent on autophagy and not apoptosis, as the inhibition of autophagy significantly delays midgut degradation whereas in the absence of apoptosis degradation occurs normally [375]. Many Atg genes are transcriptionally upregulated immediately prior to larval midgut degradation in an ecdysone receptor-dependent manner [375, 1770]. Yet only a subset of the multi-subunit complexes that are required for autophagy induced during cell survival are essential for ADCD [1162, 1771]. In contrast to the midgut, destruction of the salivary gland requires both caspase-dependent apoptosis and autophagy in parallel [1772–1774]. Inhibition of either autophagy or apoptosis alone results in a partial block in degradation, whereas combined inhibition completely blocks salivary gland degradation [1772]. As in the midgut, in response to ecdysone the expression of several Atg and apoptosis genes increase during salivary gland degradation [939, 940, 1775]. Although larval midgut and salivary gland degradation utilize autophagy for cell death, there are clear differences in the requirement of other cell death pathways between these tissues.
While there are numerous examples where autophagy promotes cell death in cultured cells, evidence for the physiological roles of ADCD in mammals have been more difficult to establish. The first description of autophagic cell death under physiological conditions in mammals is the terminal cell death in keratinocyte lineage cells of the skin [1776]. Under pathological conditions, an authentic case of autophagic cell death is the death of adult hippocampal neural stem cells following chronic restraint stress or injection of corticosterone, a stress-mediating hormone in mice [1777].
Along these lines, recent evidence suggests that ferroptosis is a type of autophagy-dependent cell death with increased autophagic flux [1778]. Mechanistically, NCOA4-facilitated ferritinophagy [1267, 1272], RAB7A-dependent lipophagy [1779], BECN1-mediated SLC7A11/system xc- inhibition [1780, 1781], STAT3-induced lysosomal membrane permeabilization [1782], HSP90-associated CMA [1271], and SQSTM1-dependent clockophagy [1249] can trigger ferroptosis through increasing iron accumulation or lipid peroxidation.
Another programmed death pathway, paraptosis [1783], is non-apoptotic in nature and has been linked to autophagy. There are several reports showing continuous increase in the autophagy marker protein LC3 and in SQSTM1 in ER stress-induced paraptosis [1784–1791]; in particular LC3 is indispensable for paraptosis as its knockdown significantly abrogates the cell death process [1784]. Pretreatment with autophagy inhibitors cannot interrupt, but rather enhances, the induction of cytoplasmic vacuolization and cell death during paraptosis. Increased SQSTM1 levels clearly indicate that the autophagy is impaired or inhibited during paraptosis-mediated cell death [1792]. Wheat germ agglutinin- and 8-p-hydroxybenzoyl tovarol-induced autophagy can antagonize paraptosis in cancer cells [1793, 1794]. In contrast, a mitophagy-dependent pathway plays a crucial role in paraptosis induction by activating PINK1 [1795]. TEM analysis would be the best way to characterize the big empty vacuoles observed during paraptosis.
Cautionary notes: In brief, rigorous criteria must be met in order to establish a death-mediating role of autophagy (AMCD or ADCD), as this process typically promotes cell survival. These include a clear demonstration of autophagic flux as described in this article, as well as verification that inhibition of autophagy prevents cell death (if using a knockdown approach, multiple ATG genes should be targeted), and that, in the case of ADCD, other mechanisms of cell death are not responsible. It is imperative to assess the genetic inhibition of autophagy using multiple ATG gene ablation, especially given the emerging non-autophagy role of ATG proteins [1796, 1797]. Another caution concerns the stability of ATG proteins; for some proteins the half-life may exceed several days, making a 24- to 48-h knockdown experiment problematic. In addition, depending on the experimental model system, appropriate protocols are needed to determine cellular viability or cell death. For example, long-term clonogenic assays should be employed when possible to measure the effective functional survival of cells. Together, care is needed to establish that the cell death is primarily dependent on autophagy rather than contributions from other modes of cell death.
Conclusion: In most systems, ascribing death to autophagy based solely on morphological criteria is insufficient; ADCD can only be demonstrated as death that is suppressed by the inhibition of autophagy, through either genetic and/or chemical means, noting that there a very few pharmacological inhibitors of autophagy induction [1798].
22. Chaperone-mediated autophagy
The primary characteristic that makes CMA different from the other autophagic variants described in these guidelines is that it does not require formation of intermediate vesicular compartments (autophagosomes or microvesicles) for the import of cargo into lysosomes [1799, 1800]. Instead, the CMA substrates are translocated across the lysosomal membrane through the action of HSPA8/HSC70 (heat shock protein family A (Hsp70) member 8) located in the cytosol and lysosome lumen, and the lysosome membrane protein LAMP2A. This machinery makes CMA unique and distinct from the other two major types of autophagy [1801, 1802]. CMA was originally identified in mammalian cells, and this section refers only to studies in mammals; however, this process has now been investigated in birds [1803], fish [1804, 1805], Drosophila [1806] and C. elegans [1807]; in C. elegans, the process may actually be ESCRT-mediated sorting at the endosome, also referred to as endosomal microautophagy (e-MI), because the lmp-1 and lmp-2 genes are more closely related to mammalian LAMP1. In fact, in a large variety of fish species there exist expressed sequences displaying high homology with mammalian LAMP2A, suggesting that a functional CMA activity might not be solely restricted to mammals and birds, and therefore likely appeared much earlier during evolution than initially thought [1805]. Along these lines, a CMA activity is indeed present in fish [1808]. These data provide new information on the evolution of CMA, and also bring new perspectives on the possible use of complementary genetic models, such as zebrafish or medaka for studying CMA function from a comparative angle.
Furthermore, a complete in silico analysis has shed further light on the definition of CMA-competent and -incompetent species. In this case, the authors used two essential features that differentiate the LAMP2A splice variant from the other two LAMP2 variants, which are the presence of (a) three to four basic amino acids in the C-terminal proximal region of the cytosolic tail [1806] and (b) the sequence GYEQF at the C terminus of the LAMP2A protein [1809]. Following these systematic approaches, CMA-competent species include mammals, some types of birds [1810], reptiles [1809] and fish [1808]. Interestingly, not all the mammalian species can perform CMA, such as the Methateria, which could indicate a diversified evolution for autophagy in this branch of the mammalian kingdom [1809]. It should also be noted that although most teleost fish display the consensus sequence GYXXF, the divergence of that motif in zebrafish, encoding an additional C-terminal amino acid, raises questions about the ability of this species to specifically perform CMA, and deserves special attention. Therefore, CMA-competent species should at present be restricted to mammals, some birds, fish and reptiles, until convincing data are provided regarding the presence of LAMP2A homologs in other species.
The following section discusses methods commonly utilized to determine if a protein is a CMA substrate (see ref. [1811] for experimental details):
a. Analysis of the amino acid sequence of the protein to identify the presence of a KFERQ-related motif, which is recognized by HSPA8, and is an absolute requirement for all CMA substrates [1812]. A free web-based resource is available to perform searches for KFERQ-like motifs in proteins [1809]. Modifications by signaling or stress may generate a novel CMA motif in proteins without such a motif and then make them suitable to be degraded via CMA. For example, acetylation can make the lysine (K) mimic a glutamine (Q), leading to a new CMA substrate motif in the protein, which is accessible for recognition by the CMA chaperone protein [1813]. In experimental CMA activity assays, mutation of the KFERQ-related motif in a protein substrate of interest to alter its physical properties for CMA recognition is one of the strategies to elucidate the specificity of CMA-mediated protein degradation [1814].
b. Colocalization studies with lysosomal markers (typically LAMP2A and/or LysoTracker™) to identify a fraction of the protein associated with lysosomes. The increase in association of the putative substrate under conditions that upregulate CMA (such as prolonged starvation) or upon blockage of lysosomal proteases (to prevent the degradation of the protein) helps support the hypothesis that the protein of interest is a CMA substrate. However, association with lysosomes is necessary, but not sufficient, to consider a protein an authentic CMA substrate, because proteins delivered by other pathways to lysosomes will also behave in a similar manner. A higher degree of confidence can be attained if the association is preferentially with the subset of lysosomes active for CMA (i.e., those containing HSPA8 in their lumen), which can be separated from other lysosomes following published procedures [1815].
c. Co-immunoprecipitation of the protein of interest with cytosolic HSPA8. Due to the large number of proteins that interact with this chaperone, it is usually better to perform affinity isolation with the protein of interest and then analyze the isolated proteins for the presence of HSPA8 rather than vice versa.
d. Co-immunoprecipitation of the protein of interest with LAMP2A [1816]. Due to the fact that the only antibodies specific for the LAMP2A variant (the only one of the three LAMP2 variants involved in CMA [133, 1817]) are generated against the cytosolic tail of LAMP2A, where the substrate also binds, it is necessary to affinity isolate the protein of interest and then analyze for the presence of LAMP2A. Immunoblot for LAMP2A in the precipitate can only be done with the antibodies specific for LAMP2A and not just those that recognize the lumenal portion of the protein that is identical in the other LAMP2 variants. If the protein of interest is abundant inside cells, co-immunoprecipitations with LAMP2A can be done in total cellular lysates, but for low-abundance cellular proteins, preparation of a membrane fraction (enriched in lysosomes) by differential centrifugation may facilitate the detection of the population of the protein bound to LAMP2A.
e. Selective upregulation and blockage of CMA to demonstrate that degradation of the protein of interest changes with these manipulations. Selective chemical inhibitors for CMA are not currently available. Note that general inhibitors of lysosomal proteases (e.g., bafilomycin A1, NH4Cl, leupeptin) also block the degradation of proteins delivered to lysosomes by other autophagic and endosomal pathways. The most selective way to block CMA is by knockdown of LAMP2A, which causes this protein to become a limiting factor [133]. The other components involved in CMA, including HSPA8, HSP90AA1, GFAP, and EEF1A/eF1α, are all multifunctional cellular proteins, making it difficult to interpret the effects of knockdowns. Overexpression of LAMP2A [1816] is also a better approach to upregulate CMA than the use of chemical modulators. The two compounds demonstrated to affect degradation of long-lived proteins in lysosomes [1818], 6-aminonicotinamide and geldanamycin, lack selectivity, as they affect many other cellular processes. In addition, in the case of geldanamycin, the effect on CMA can be the opposite (inhibition rather than stimulation) depending on the cell type (this is due to the fact that the observed stimulation of CMA is actually a compensatory response to the blockage of HSP90AA1 in lysosomes, and different cells activate different compensatory responses) [1819].
f. The most conclusive way to prove that a protein is a CMA substrate is by reconstituting its direct translocation into lysosomes using a cell-free system [1811]. This method is only possible when the protein of interest can be purified, and it requires the isolation of the population of lysosomes active for CMA. Internalization of the protein of interest inside lysosomes upon incubation with the isolated organelle can be monitored using protease protection assays (in which addition of an exogenous protease removes the protein bound to the cytosolic side of lysosomes, whereas it is inaccessible to the protein that has reached the lysosomal lumen; note that pre-incubation of lysosomes with lysosomal protease inhibitors before adding the substrate is required to prevent the degradation of the translocated substrate inside lysosomes) [1820]. The use of exogenous protease requires numerous controls (see ref. [1811]) to guarantee that the amount of protease is sufficient to remove all the substrate outside lysosomes, but will not penetrate inside the lysosomal lumen upon breaking the lysosomal membrane.
The difficulties in the adjustment of the amount of protease have led to the development of a second method that is more suitable for laboratories that have no previous experience with these procedures. In this case, the substrate is incubated with lysosomes untreated or previously incubated with inhibitors of lysosomal proteases, and then uptake is determined as the difference of protein associated with lysosomes not incubated with inhibitors (in which the only remaining protein will be the one associated with the cytosolic side of the lysosomal membrane) and those incubated with the protease inhibitors (which contain both the protein bound to the membrane and that translocated into the lumen) [1821].
Confidence that the lysosomal internalization is by CMA increases if the uptake of the substrate can be competed with proteins previously identified as substrates for CMA (e.g., GAPDH [glyceraldehyde-3-phosphate dehydrogenase] or RNASE1 [ribonuclease A family member 1, pancreatic], both commercially available as purified proteins), but is not affected by the presence of similar amounts of nonsubstrate proteins (such as SERPINB/ovalbumin or PPIA/cyclophilin A). Blockage of uptake by pre-incubation of the lysosomes with antibodies against the cytosolic tail of LAMP2A also reinforces the hypothesis that the protein is a CMA substrate. It should be noted that several commercially available kits for lysosome isolation separate a mixture of lysosomal populations and do not enrich in the subgroup of lysosomes active for CMA, which limits their use for CMA uptake assays.
Further to the limitations in purifying CMA-active lysosomes for cell-free assays, CMA activity of these lysosomes may be blocked by proteins that bind abnormally to LAMP2A on the lysosomal surface, e.g., mutant LRRK2 [1733]. Such protein binding, which happens in vivo, may be inadvertently removed during stringent washes and centrifugation during the lysosome isolation and purification processes. Hence, assaying the degradation of an artificial CMA substrate in isolated lysosomes may not necessarily reflect the in vivo CMA activity [1814].
In other instances, rather than determining if a particular protein is a CMA substrate, the interest may be to analyze possible changes in CMA activity under different conditions or in response to different modifications. We enumerate here the methods, from lower to higher complexity, that can be utilized to measure CMA in cultured cells and in tissues (see ref. [1811] for detailed experimental procedures).
a. Measurement of changes in the intracellular rates of degradation of long-lived proteins, when combined with inhibitors of other autophagic pathways, can provide a first demonstration in support of changes that are due to CMA. For example, CMA is defined in part as lysosomal degradation upregulated in response to serum removal but insensitive to PtdIns3K inhibitors.
b. Measurement of levels of CMA components is insufficient to conclude changes in CMA because this does not provide functional information, and changes in CMA components can also occur under other conditions. However, analysis of the levels of LAMP2A can be used to support changes in CMA detected by other procedures. Cytosolic levels of HSPA8 remain constant and are not limiting for CMA, thus providing no information about this pathway. Likewise, changes in total cellular levels of LAMP2A do not have an impact on this pathway unless they also affect their lysosomal levels (i.e., conditions in which LAMP2A is massively overexpressed lead to its targeting to the plasma membrane where it cannot function in CMA). It is advisable that changes in the levels of these two CMA components are confirmed to occur in lysosomes, either by colocalization with lysosomal markers when using image-based procedures or by performing immunoblot of a lysosomal enriched fraction (purification of this fraction does not require the large amounts of cells/tissue necessary for the isolation of the subset of lysosomes active for CMA).
Given that specific LAMP2A antibody is available for immunohistochemistry, comparison of puncta sizes from LAMP2A-positive lysosomes under different conditions provides useful information on the availability of LAMP2A at lysosomal levels. Although LAMP2A plays a key role in CMA activity, an increased LAMP2A level does not necessarily reflect increased CMA activity. Blockage of substrate translocation and disassembly of LAMP2A-binding complexes can result in accumulation of lysosomal LAMP2A, thus impairing LAMP2A turnover and CMA activity [1814].
c. Tracking changes in the subset of lysosomes active for CMA. This group of lysosomes is defined as those containing HSPA8 in their lumen (note that LAMP2A is present both in lysosomes that are active or inactive for CMA, and it is the presence of HSPA8 that confers CMA capability). Immunogold or immunofluorescence against these two proteins (LAMP2A and HSPA8) makes it possible to quantify changes in the levels of these lysosomes present at a given time, which correlates well with CMA activity [1815].
d. Analysis of lysosomal association of fluorescent artificial CMA substrates. Two different fluorescent probes have been generated to track changes in CMA activity in cultured cells using immunofluorescence or flow cytometry analysis [1815]. These probes contain the KFERQ and context sequences in frame with photoswitchable or photoactivated fluorescent proteins. Activation of CMA results in the mobilization of a fraction of the cytosolic probe to lysosomes and the subsequent change from a diffuse to a punctate pattern. CMA activity can be quantified as the number of fluorescent puncta per cell or as the decay in fluorescence activity over time because of degradation of the artificial substrate. Because the assay does not allow measuring accumulation of the substrate (which must unfold for translocation), it is advisable to perform a time-course analysis to determine gradual changes in CMA activity. Antibodies against the fluorescent protein in combination with inhibitors of lysosomal proteases can be used to monitor accumulation of the probe in lysosomes over a period of time, but both the photoswitchable and the unmodified probe will be detected by this procedure [1822]. As for any other fluorescence probe based on analysis of intracellular “puncta” it is essential to include controls to confirm that the puncta are indeed lysosomes (colocalization with LysoTracker™ or LAMPs and lack of colocalization with markers of cytosolic aggregation such as ubiquitin) and do not reach the lysosomes through other autophagic pathways (insensitivity to PtdIns3K inhibitors and sensitivity to LAMP2A knockdown are good controls in this respect).
e. Direct measurement of CMA using in vitro cell-free assays. Although the introduction of the fluorescent probes should facilitate measurement of CMA in many instances, they are not applicable for tissue samples. In addition, because the probes measure binding of substrate to lysosomal membranes it is important to confirm that enhanced binding does not result from defective translocation. Last, the in vitro uptake assays are also the most efficient way to determine primary changes in CMA independently of changes in other proteolytic systems in the cells. These in vitro assays are the same ones described in the previous section on the identification of proteins as substrates of CMA, but are performed in this case with purified proteins previously characterized to be substrates for CMA. In this case the substrate protein is always the same, and what changes is the source of lysosomes (from the different tissues or cells that are to be compared). As described in the previous section, binding and uptake can be analyzed separately using lysosomes previously treated or not with protease inhibitors. The analysis of the purity of the lysosomal fractions prior to performing functional analysis is essential to conclude that changes in the efficiency to take up the substrates results from changes in CMA rather than from different levels of lysosomes in the isolated fractions. Control of the integrity of the lysosomal membrane and sufficiency of the proteases are also essential to discard the possibility that degradation is occurring outside lysosomes because of leakage, or that accumulation of substrates inside lysosomes is due to enhanced uptake rather than to decreased degradation.
f. Time-course analysis to determine CMA activity in live cells. Cells of interest can be developed to express a photoactivatable fluorescent reporter protein (e.g., PA-mCherry) conjugated to a KFERQ-like recognition motif. Photoactivation induces emission of fluorescence of reporter substrates already expressed in the cells. Any decline in substrate fluorescence levels after photoactivation can be monitored and quantified at different time points using flow cytometry. It is crucial that the culture medium be refreshed prior to photoactivation (typically 2 to 4 h) to minimize the confounding effects of basal autophagy due to depletion of nutrients in the old culture medium. Furthermore, CMA reference substrates may be degraded via non-CMA pathways. To confirm whether the degradation of the substrate is CMA-specific, it is important to include a control assay after lamp2a knockdown [1814].
Cautionary notes: The discovery of a new selective form of protein degradation in mammals named endosomal microautophagy [1823] has made it necessary to reconsider some of the criteria that applied in the past for the definition of a protein as a CMA substrate. The KFERQ-like motif, previously considered to be exclusive for CMA, is also used to mediate selective targeting of cytosolic proteins to the surface of late endosomes. For example, MAPT containing the KFERQ-like motifs has been found to be a CMA substrate [1824]; however, it was also revealed to be a substrate of e-MI [1825]. Once there, substrates can be internalized in microvesicles that form from the inward invagination of the limiting membrane of these organelles in an ESCRT-dependent manner. HSPA8 has been identified as the chaperone that binds this subset of substrates and directly interacts with lipids in the late endosomal membrane, thus acting as a receptor for cytosolic substrates in this compartment [1826]; accordingly, e-MI is a variation of the MVB pathway, and as such can be referred to as ESCRT-mediated sorting at the endosome. At a practical level, to determine if a KFERQ-containing protein is being degraded by CMA or e-MI the following criteria can be applied: (a) Inhibition of lysosomal proteolysis (for example with NH4Cl and leupeptin) blocks degradation by both pathways. (b) Knockdown of LAMP2A inhibits CMA but not e-MI. (c) Knockdown of components of ESCRT-I and ESCRT-II (e.g., VPS4 and TSG101) inhibits e-MI but not CMA. (d) Interfering with the capability to unfold the substrate protein blocks its degradation by CMA, but does not affect e-MI of the protein. In this respect, soluble proteins, oligomers and protein aggregates can undergo e-MI, but only soluble proteins can be CMA substrates. (e) In vitro uptake of e-MI substrates can be reconstituted using isolated late endosomes whereas in vitro uptake of CMA substrates can only be reconstituted using lysosomes. e-MI has also been described in Drosophila neuromuscular junctions and fat body. Using photoactivatable PA-mCherry or split-GFP sensors it was shown to genetically depend on Hsc70-4, a homolog of HSPA8, and components of the ESCRT machinery [1806, 1827].
Another pathway that needs to be considered relative to CMA is chaperone-assisted selective autophagy (CASA) [1828]. CASA is dependent on HSPA8 and LAMP2 (although it is not yet known if it is dependent solely on the LAMP2A isoform). Thus, a requirement for these two proteins is not sufficient to conclude that a protein is degraded by CMA.
It should also be noted that LAMP1 and LAMP2 share common function as revealed by the embryonic lethal phenotype of lamp1−/− lamp2y/− double-deficient mice [1829]. LAMP2 is involved in the fusion of late endosomes and autophagosomes or phagosomes [1830, 1831]. LAMP2C, one of the LAMP2 isoforms, can also function as an RNA/DNA receptor in RNautophagy and DNautophagy pathways, where RNA or DNA is taken up directly by lysosomes in an ATP-dependent manner [1395–1398]. Whereas LAMP2A and LAMP2B are expressed in most mammalian cells, LAMP2C is selectively expressed in different tissues and cell types. Increased expression of LAMP2C is induced in human lymphocytes upon cellular exposure to inflammatory stimuli, with ectopic LAMP2C expression disrupting CMA [1832]. In human melanoma tumors, increased cellular LAMP2C reduces the expression of LAMP2A and LAMP2B, disrupting CMA and autophagy, as well as cell cycle progression and tumor growth in vivo [1833]. Finally, LAMP1 and LAMP2 deficiency does not necessarily affect protein degradation under conditions when CMA is active [1829], and the expression levels of neuronal CMA substrates does not change upon loss of LAMP2 [1397, 1834, 1835].
Conclusion: One of the key issues with the analysis of CMA is verifying that the protein of interest is an authentic substrate. Methods for monitoring CMA that utilize fluorescent probes are available that eliminate the need for the isolation of CMA-competent lysosomes, one of the most difficult aspects of assaying this process.
23. Chaperone-assisted selective autophagy
CASA is a specialized form of macroautophagy whereby substrate proteins are ubiquitinated and targeted for lysosomal degradation by chaperone and co-chaperone proteins [1828]. The substrate protein does not require a KFERQ motif, which differentiates CASA from CMA. In addition, in CASA the cargo protein destined for degradation is delivered to the phagophore instead of directly to the lysosome as occurs in CMA. In CASA the substrate protein is recognized by the CASA complex that is formed by the assembly of the co-chaperone BAG3, which forms a multidomain complex with HSPA8, the small heat shock proteins HSPB6 and HSPB8, the ubiquitin ligase STUB1/CHIP, and the receptor proteins SYNPO2/myopodin (synaptopodin 2) and SQSTM1. The co-chaperone DNAJB6 also interacts with the core CASA machinery [1836], although its precise role in the pathway awaits confirmation. Following ubiquitination, the substrate protein is loaded onto the CASA machinery. SYNPO2 and SQSTM1 then bind to core components of the phagophore (VPS18 and LC3, respectively) resulting in sequestration of the substrate protein and associated multidomain complex within the autophagosome, and subsequent lysosomal degradation [1828, 1837]. Note that association of BAG1 with STUB1/CHIP and HSPA8, displacing or preventing the association of HSPA8 with BAG3, causes the cargoes to be re-routed from CASA to the proteasome [1838–1841]. Along these lines, BAG3 Pro209 mutants, associated with neuromuscular diseases and peripheral neuropathies, relocate chaperones of the CASA complex to aggresomes; the mutant BAG3 protein traps ubiquitinated client proteins at the aggresome preventing their efficient clearance [1842]. Finally, CASA has been observed primarily in mammalian cells, but is also found in Aspergillus [1843]. See also Filamentous fungi.
Conclusion: Given that the autophagy machinery involved in CASA is very similar to that in other forms of autophagy there are currently no specific markers or inhibitors available to study this process specifically, but the involvement of HSPA8, BAG3 and ubiquitination of client proteins is highly suggestive of CASA activity.
24. Microautophagy
Microautophagy is a category of autophagic pathway that is driven by morphological changes of the lysosomal (vacuolar) or endosomal membrane. Protrusion (type 1) or invagination (type 2) of the lysosomal membrane leads to an uptake of the cytoplasmic components, while invagination of the endosomal membrane producing multivesicular bodies is also known to transport cytoplasmic components into the organelle lumen (type 3), and eventually to the lysosomal lumen [1844]. This category encompasses both bulk and selective autophagic pathways; several of the latter, termed micropexophagy (selective microautophagy of peroxisomes) in the yeast Komagataella phaffii/Pichia pastoris, and PMN in Saccharomyces cerevisiae, have been extensively studied [157, 1061]. Whereas both micropexophagy and PMN are dependent on “core” Atg proteins [1845] responsible for the biogenesis of autophagic membrane structures [1846, 1847], recent studies identified several microautophagic pathways independent of such Atg proteins, that function in selective degradation of lipid droplets (microlipophagy) in the yeast S. cerevisiae [1848, 1849] or in the formation of anthocyanin vacuole inclusions in A. thaliana [1850]. Endosomal microautophagy (type 3) requires ESCRT and HSPA8-family proteins, and similar pathways have been found in Schizosaccharomyces pombe [1851] and D. melanogaster [1827].
Because microautophagy accompanies incorporation of part of the lysosomal membrane into the organelle lumen together with the cargo (target) components, the most authentic method to monitor microautophagy is detection of transport of lysosomal transmembrane proteins into the lumen. In yeast studies, the vacuolar transmembrane protein Vph1 can be expressed with a C-terminal GFP, and subjected to immunoblot analysis for the detection of the cleaved GFP moiety produced in the vacuolar lumen through microautophagic activity [1849, 1852, 1853]. For specific monitoring of selective microautophagy, similar detection of free GFP from GFP-tagged, organelle-specific proteins have been utilized for monitoring their activity. For example, Nvj1 or Swh1/Osh1 can be used for monitoring PMN [1847]; and Erg6 [1854, 1855], Faa4 [1856], or Ldo16/Osw5 [1849] are useful for the detection of microlipophagy. For in vitro detection of bulk microautophagic activity in S. cerevisiae, luciferase incorporation into the purified vacuole fraction can be monitored [1857]. In mammalian cells, exosomes correspond to intralumenal vesicles (ILVs) of multivesicular endosomes (MVEs) that are secreted in the extracellular space upon fusion of MVEs with the plasma membrane instead of lysosomes. Exosome analysis can be used as a readout of ILV biogenesis [1858]. Some elements of commonality are achieved between exosome/ILV biogenesis (type 3) and vacuolar microautophagy (type 2), such as lipid domain and ESCRT involvement [1859]. Moreover, a crosstalk exists between the exosomal and autophagic pathways allowing cell clearance of unwanted components [822].
Cautionary notes: It should be noted that these assay systems monitoring the dynamics of organelle-specific proteins other than lysosomal transmembrane proteins, do not discriminate between autophagic and microautophagic activities, and thus the data should be interpreted in combination with other morphological analyses or immunoblot data from the lysosomal membrane protein(s). In monitoring microlipophagy, care should be taken for the choice of the lipid droplet marker proteins for the immunoblot analysis, as several of the marker proteins, e.g., Erg6, exhibit dual localization to the ER and lipid droplets, depending on culture conditions [1849]; the use of such dual-localized marker proteins makes it difficult to discriminate reticulophagy and (micro)lipophagy. It also should be noted that overexpression of lipid marker proteins easily leads to release of the proteins into the cytosol, which renders the degradation dependent on bulk autophagy and “core” Atg factors, because all of the lipid droplet proteins are peripherally associated with the organelle surface.
C. Comments on additional topics
1. Acidotropic dyes
Among the older methods for following autophagy is staining with acidotropic dyes such as MDC [1603], acridine orange [1860], neutral red [1552], LysoSensor Blue [1861] and LysoTracker™ Red [377, 1862]. It should be emphasized that, whereas these dyes are useful to identify acidified vesicular compartments, they should not be relied upon to compare differences in endosomal or lysosomal pH between cells due to variables that can alter the intensity of the signal. For example, excessive incubation time and/or concentrations of LysoTracker™ Red can oversaturate labeling of the cell and mask differences in signal intensity that reflect different degrees of acidification within populations of compartments [1863]. Use of these dyes to detect, size, and quantify numbers of acidic compartments must involve careful standardization of the conditions of labeling and ideally should be confirmed by ancillary TEM and/or immunoblot analysis. Reliable measurements of vesicle pH require ratiometric measurements of two dyes with different peaks of optimal fluorescence (e.g., LysoSensor Blue and LysoSensor Yellow), or the use of a molecule with two emission wavelengths that are differentially affected by pH to exclude variables related to uptake and cell size [80, 1863, 1864]. Another method to validate the fluorescent signal of an acidotropic dye is by defining a cut-off level for the net fluorescence intensity resulting from each dye’s incorporation in acidic lysosomes. Coupling acidotropic dye staining with flow cytometric analysis is highly recommended for a numerically validated and objective determination of autophagy, and the use of a control such as CQ or HCQ will further validate this inexpensive and convenient technique [1865].
Finally, degradation of lysosomal cargo depends on acidification which leads to enzyme activation. Most cathepsins are abundant and activated at the low pH of the lysosomal lumen. For validation of cathepsin activation, and thus validation of functional lysosomes, co-staining with Magic Red dye and a lysosomal marker such as LAMP2 is recommended. Selective inhibitors of cathepsin, such as E64d for cathepsin B, must be included as an experimental control for Magic Red specificity.
Cautionary notes: Although MDC was first described as a specific marker of autophagic vacuoles [1866] subsequent studies have suggested that this, and other acidotropic dyes, are not specific markers for early autophagosomes [439], but rather label later stages in the degradation process. For example, autophagosomes are not acidic, and MDC staining can be seen in autophagy-defective mutants [814] and in the absence of autophagy activation [1867]. MDC may also show confounding levels of background labeling unless narrow bandpass filters are used. However, in the presence of vinblastine, which blocks fusion with lysosomes, MDC labeling increases, suggesting that under these conditions MDC can label late-stage autophagosomes [1600]. Along these lines, cells that overexpress a dominant negative version of RAB7 (the T22N mutant) show colocalization of this protein with MDC; in this case fusion with lysosomes is also blocked [1510] indicating that MDC does not just label lysosomes. Nevertheless, MDC labeling could be considered as an indicator of autophagy when the increased labeling of cellular compartments by this dye is prevented by treatment with specific autophagy inhibitors.
Overall, staining with MDC or its derivative monodansylpentane [1603] is not, by itself, a sufficient method for monitoring autophagy. Similarly, LysoTracker™ Red, neutral red and acridine orange are not ideal markers for autophagy because they primarily detect lysosomes, and an increase in lysosome size or number could reflect an increase in nonprofessional phagocytosis (often seen in embryonic tissues [1868]) rather than autophagy. These markers are, however, useful for monitoring selective autophagy when used in conjunction with protein markers or other dyes. For example, increased colocalization of mitochondria with both GFP-LC3 and LysoTracker™ Red can be used as evidence of autophagic cargo delivery to lysosomes. Moreover, LysoTracker™ Red has been used to provide correlative data on autophagy in D. melanogaster fat body cells (Figure 32) [376, 377]. However, additional assays, such as GFP-Atg8-family protein fluorescence and EM, should be used to substantiate results obtained with acidotropic dyes whenever possible to rule out the possibility that LAP is involved (see Noncanonical use of autophagy-related proteins). Finally, one important caution when co-imaging with LysoTracker™ Red and a green-fluorescing marker (e.g., GFP-LC3 or MitoTracker™ Green) is that it is necessary to control for rapid red-to-green photoconversion of the LysoTracker™, which can otherwise result in an incorrect interpretation of colocalization [1869].
Figure 32.
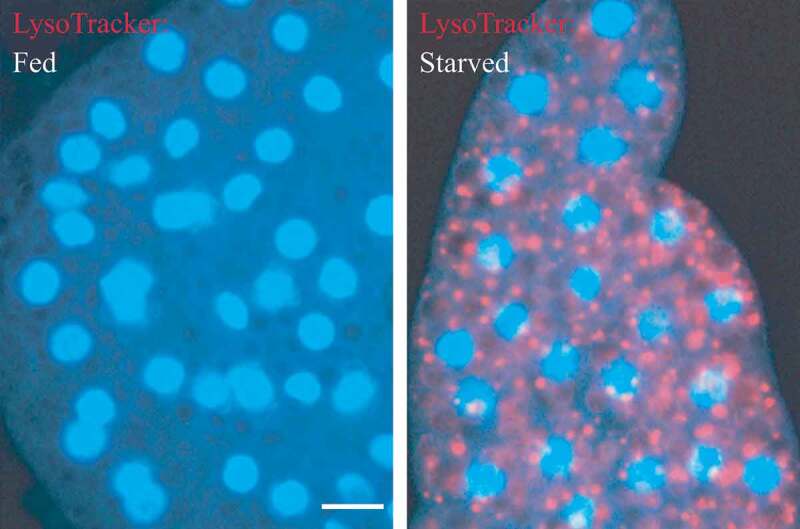
LysoTracker™ Red stains lysosomes and can be used to monitor autophagy in Drosophila. Live fat body tissues from Drosophila were stained with LysoTracker™ Red (red) and Hoechst 33342 (blue) to stain the nucleus. Tissues were isolated from fed (left) or 3-h starved (right) animals. Bar: 25 µm. This figure was modified from data presented in ref. [377], Developmental Cell, 7, Scott RC, Schuldiner O, Neufeld TP, Role and regulation of starvation-induced autophagy in the Drosophila fat body, pp. 167-78, copyright 2004, with permission from Elsevier
Some of the confusion regarding the interpretation of results with these dyes stems in part from the nomenclature in this field. Indeed, the discussion of acidotropic dyes points out why it is advisable to differentiate between the terms “autophagosome” and “autophagic vacuole,” although they are occasionally, and incorrectly, used interchangeably. The autophagosome is the sequestering compartment generated by the phagophore. The fusion of an autophagosome with an endosome or a lysosome generates an amphisome or an autolysosome, respectively [1480]. The early autophagosome is not an acidic compartment, whereas amphisomes and autolysosomes are acidic. As noted in the section Transmission electron microscopy, earlier names for these compartments are “initial autophagic vacuole (AVi),” “intermediate or intermediate/degradative autophagic vacuole (AVi/d)” and “degradative autophagic vacuole (AVd),” respectively. Thus, acidotropic dyes can stain late autophagic vacuoles (in particular autolysosomes), but not the initial autophagic vacuole, the early autophagosome.
A recently developed dye for monitoring autophagy, Cyto-ID (Enzo Life Sciences), stains vesicular structures shortly after amino acid deprivation, which extensively colocalize with RFP-LC3-positive structures, while colocalizing partially with lysosomal probes [1870]. Moreover, unlike MDC, Cyto-ID does not show background fluorescence under control conditions and the two dyes colocalize only marginally. Furthermore, the Cyto-ID signal responds to well-known autophagy modulators. Therefore, this amphiphilic dye, which partitions in hydrophobic environments, may prove more selective for autophagic vacuoles than the previously discussed lysosomotropic dyes.
With the above caveats in mind, the combined use of early and late markers of autophagy is highly encouraged, and, when quantifying mammalian lysosomes, it is important to note that increases in both lysosome size and number are frequently observed. Finally, to avoid confusion with the plant and fungal vacuole, the equivalent organelle to the lysosome, we recommend the use of the term “autophagosome” instead of “autophagic vacuole” when possible, that is, when the specific nature of the structure is known.
Conclusion: Given the development of better techniques that are indicators of autophagy, relying entirely on the use of acidotropic dyes to study this process is not acceptable.
2. Autophagy inhibitors and inducers
In many situations it is important to demonstrate an effect resulting from inhibition or stimulation of autophagy (see ref. [1871] for a partial listing of regulatory compounds), and a few words of caution are worthwhile in this regard. Most chemical inhibitors of autophagy are not entirely specific, and it is important to consider possible dose- and time-dependent effects. Accordingly, it is generally preferable to analyze specific loss-of-function Atg mutants. However, it must be kept in mind that some apparently specific Atg gene products may have autophagy-independent roles (e.g., ATG5 in cell death, and the PIK3C3/VPS34-containing complexes—including BECN1—in apoptosis, endosomal function and protein trafficking), or may be dispensable for autophagy (see Noncanonical use of autophagy-related proteins) [31, 817, 860, 1550, 1872–1875]. Therefore, the experimental conditions of inhibitor application and their side effects must be carefully considered.
In addition, it must be emphasized once again that autophagy, as a multistep process, can be inhibited at different stages. Sequestration inhibitors, including 3-MA, LY294002 and wortmannin, inhibit class I PI3Ks as well as class III PtdIns3Ks [181, 437, 1876]. The class I enzymes generate products such as PtdIns(3,4,5)P3 that inhibit autophagic sequestration, whereas the class III product (PtdIns3P) generally stimulates autophagic sequestration. The overall effect of these inhibitors is typically to block autophagy because the class III enzymes that are required to activate autophagy act downstream of the negative regulatory class I enzymes, although cell death may ensue in cell types that are dependent upon high levels of AKT for survival. The effect of 3-MA (but not that of wortmannin) is further complicated by the fact that it has different temporal patterns of inhibition, causing a long-term suppression of the class I PI3K, but only a transient inhibition of the class III enzyme. In cells incubated in a complete medium for extended periods of time, 3-MA may, therefore (particularly at suboptimal concentrations), promote autophagy by inhibition of the class I enzyme [436, 437]. Thus, wortmannin may be considered as an alternative to 3-MA for autophagy inhibition [437]. However, wortmannin can induce the formation of vacuoles that may have the appearance of autophagosomes, although they are swollen late endocytic compartments [1527]. In addition, treatment of human alveolar macrophages with wortmannin or 3-MA in complete medium or HBSS results in increased levels of LC3-I and LC3-II as detected by western blotting. Neither wortmannin nor 3-MA blocks rapamycin-induced conversion of LC3-I to LC3-II in these cells; rather there seems to be an additive effect. Consequently, these inhibitors should be used with caution when investigating autophagy in macrophages (M. O’Sullivan and S. O’Leary, unpublished observation). Furthermore, studies have demonstrated that inhibition of autophagy with 3-MA or wortmannin can have effects on cytokine transcription, processing and secretion, particularly of IL1 family members [1877–1879], but 3-MA and wortmannin also inhibit the secretion of some cytokines and chemokines (e.g., TNF, IL6, CCL2/MCP-1) in an autophagy-independent manner (J. Harris, unpublished observations) [1877, 1880]. Moreover, 3-MA inhibits the production of nitric oxide in IFNG-activated bone-marrow derived macrophages [1881]. Thus, in studies where the effect of autophagy inhibition on specific cellular processes is being investigated, it is important to confirm results using other methods, such as RNA silencing. Due to these issues, it is of great interest that inhibitors with specificity for the class III PtdIns3Ks, and their consequent effects on autophagy, have been described [331, 1882, 1883]. For instance, the selective class III PtdIns3K inhibitor SAR405 is an efficient blocker of autophagic (LDH) sequestration and (long-lived protein) degradation activity [25, 56, 302]. Finally, it is important to stress that the efficacy of wortmannin as an inhibitor of PI3Ks and PtdIns3Ks may be decreased by its non-enzymatic covalent binding to free amino acids [1884, 1885].
A mutant mouse line carrying a floxed allele of Pik3c3 has been created [1886]. This provides a useful genetic tool that will help in defining the physiological role of the class III PtdIns3K with bona fide specificity by deleting the class III kinase in a cell type-specific manner in a whole animal using the Cre-LoxP strategy. For example, the phenotype resulting from a knockout of Pik3c3 specifically in the kidney glomerular podocytes (pik3c3 [pdKO]) indicates that there is no compensation by other classes of PtdIns3Ks or related Atg genes, thus highlighting the functional specificity and physiological importance of the class III PtdIns3K in these cells.
CHX, a commonly used protein synthesis inhibitor in mammals, is also an inhibitor of sequestration in vivo [13–15, 113, 1522, 1887–1890], and in various cell types in vitro [216, 660, 1891], and it has been utilized to investigate the dynamic nature of the regression of various autophagic elements [13–15, 28, 113, 1887, 1888]. The mechanism of action of CHX in short-term experiments is not clear, but it has no direct relation to the inhibition of protein synthesis [660]. This latter activity, however, may complicate certain types of analysis when using this drug.
A significant challenge for a more detailed analysis of the dynamic role of autophagy in physiological and pathophysiological processes, for instance with regard to cancer and cancer therapy, is to find more specific inhibitors of autophagy signaling which do not affect other signaling cascades (reviewed in ref. [527]). For example, in the context of cellular radiation responses it is well known that PI3Ks, in addition to signaling through the PI3K-AKT pathway, have a major role in the regulation of DNA-damage repair [1891]. However, 3-MA, which is a nonspecific inhibitor of these lipid kinases, can alter the function of other classes of this enzyme, which are involved in the DNA-damage repair response. This is of particular importance for investigations into the role of radiation-induced autophagy in cellular radiation sensitivity or resistance [1892, 1893]. CQ, through the induction of ROS, increases DNA damage and can be used to synergistically enhance the therapeutic effect of otherwise toxic NOTCH and gamma secretase inhibitors that target the oncogenic NOTCH signaling pathway [1894].
Most other inhibitory chemicals act at post-sequestration steps. These types of agents have been used in many experiments to both inhibit endogenous protein degradation and to increase the number of autophagic compartments. These chemicals cause the accumulation of sequestered material in either autophagosomes or autolysosomes, or both, because they allow autophagic sequestration to proceed. The main categories of these types of inhibitors include the vinca alkaloids (e.g., vinblastine) and other microtubule poisons that inhibit fusion, inhibitors of lysosomal enzymes (e.g., leupeptin, pepstatin A and E-64d), and compounds that elevate lysosomal pH (e.g., inhibitors of V-ATPases such as bafilomycin A1, concanamycin A and concanamycin B [64], and weak base amines including methyl- or propylamine, CQ, and neutral red, some of which slow down fusion). Ammonia is a very useful agent for the elevation of lysosomal pH in short-term experiments, but it has been reported to cause a stimulation of autophagy during long-term incubation of cells in a full medium [1895], under which conditions a good alternative might be methylamine or propylamine [1896]. Along these lines, it should be noted that the half-life of glutamine in cell culture media is approximately two weeks due to chemical decomposition, which results in media with lowered glutamine and elevated ammonia concentrations that can affect the autophagic flux (either inhibiting or stimulating autophagy, depending on the concentration [1897]). Thus, to help reduce experimental variation, the use of freshly prepared cell culture media with glutamine is advised. Alternatively, GlutaMAX is recommended for culture media without glutamine [1898].
A special note of caution is also warranted in regard to CQ. Although this chemical is commonly used as an autophagy inhibitor, CQ may initially stimulate autophagy (F.C. Dorsey, personal communication; R. Franco, personal communication). In addition, culture conditions requiring acidic media preclude the use of CQ because intracellular accumulation of the chemical is dramatically reduced by low pH [1899]. To overcome this issue, it is possible to use acid compounds that modulate autophagy, such as betulinic acid and its derivatives [314, 1900–1902]. Betulinic acid damages lysosomal function differing from traditional inhibitors (e.g., CQ, NH4Cl or bafilomycin A1) that raise the lysosomal pH; betulinic acid interacts with pure phospholipid membranes [314,1170], and is capable of changing membrane permeability [314, 1903, 1904]. The lysosomal damage mediated by betulinic acid is capable of compromising autophagy without any incremental damage when lysosomal function is altered by lysosomal inhibitors (e.g., CQ or bafilomycin A1) [314]; however, betulinic acid is not lysosome specific, and will affect other organelles such as mitochondria.
Other natural compounds, such as sulforaphane (in breast cancer MDA-MB-231 cell line), the Phellinus linteus fungus extract (in the breast cancer MDA-MB-231 cell line) and neferine (in the lung adenocarcinoma A549 cell line) combined with anticancer drugs synergistically induce autophagic cell death [1905]. Dehydroandrographolide and polyphyllin G trigger activation of MAPK8/9 and an inhibition of AKT and MAPK/p38, inducing oral cancer autophagic cell death [1906, 1907]. Notably, a significant number of natural compounds have been identified to overcome drug-resistant or apoptosis-resistant cancer via induction of autophagic cell death: For example, an ATP2A/SERCA inhibitor, saikosaponin-d, N-desmethyldauricine and celastrol [1908–1911]; a group of AMPK activators (liensinine, isoliensinine, dauricine, cepharanthine, hernandezine and thalidezine) [1912–1915]; and the PRKCA/PKC-α inhibitor, tetrandrine [1916]. Other natural small-molecules, such as thonningianin A from Penthorum chinense Pursh, steroidal saponin and polyphyllin VI from Trillium tschonoskii Maxi were reported to show their anti-oxidative effect via autophagy induction [1917].
Some data suggest that particular nanomaterials may also be novel modulators of autophagy, by as yet unidentified mechanisms [1918, 1919] (See Nanoparticles).
It is worth noting that lysosomal proteases fall into three general groups, cysteine, aspartic acid and serine proteases. Therefore, the fact that leupeptin, a serine and cysteine protease inhibitor, has little or no effect does not necessarily indicate that lysosomal degradation is not taking place; a combination of leupeptin, pepstatin A and E-64d may be a more effective treatment. However, it should also be pointed out that these protease inhibitors can exert inhibitory effects not only on lysosomal proteases, but also on cytosolic proteases; that is, degradation of proteins might be blocked through inhibition of cytosolic instead of lysosomal proteases. Conversely, it should be noted that MG132 (Z-leu-leu-leu-al) and its related peptide aldehydes are commonly used as proteasomal inhibitors, but they can also inhibit certain lysosomal hydrolases such as cathepsins and calpains [1920]. Thus, any positive results using MG132 do not rule out the possibility of involvement of the autophagy-lysosomal system. Therefore, even if MG132 is effective in inhibiting autophagy, it is important to confirm the result using more specific proteasomal inhibitors such as lactacystin or epoxomicin. Finally, there are significant differences in cell permeability among protease inhibitors. For example, E-64d is membrane permeable, whereas leupeptin and pepstatin A are not (although there are derivatives that display greater permeability such as pepstatin A methyl ester) [1921]. Thus, when analyzing whether a protein is an autophagy substrate, caution should be taken in utilizing these protease inhibitors to block autophagy.
As with the PtdIns3K inhibitors, many autophagy-suppressive compounds are not specific. For example, okadaic acid [1922] is a powerful general inhibitor of both type 1 (PPP1) and type 2A (PPP2) protein phosphatases [1923]. Bafilomycin A1 and other compounds that raise the lysosomal pH may have indirect effects on any acidified compartments. Moreover, treatment with bafilomycin A1 for extended periods (18 h) can cause significant disruption of the mitochondrial network in cultured cells (M.E. Gegg, personal communication), and either bafilomycin A1 or concanamycin A cause swelling of the Golgi in plants [1924], and increase cell death by apoptosis in cancer cells (V.A. Rao, personal communication) [216]. Furthermore, bafilomycin A1 may have off-target effects on the cell, particularly on MTORC1 [781, 1925, 1926]. Bafilomycin A1 is often used at a final concentration of 100 nM, but much lower concentrations such as 1 nM may be sufficient to inhibit autophagic-lysosomal degradation and are less likely to cause indirect effects [300, 1927]. For example, in pulmonary A549 epithelial cells bafilomycin A1 exhibits concentration-dependent effects on cellular morphology and on protein expression; at concentrations of 10 and 100 nM the cells become more rounded accompanied by increased expression of VIM (vimentin) and a decrease in CDH1/E-cadherin (B. Yeganeh, M. Post and S. Ghavami, unpublished observations). Thus, appropriate inhibitory concentrations should be empirically determined for each cell type [309]. As elaborated earlier in these guidelines, there is a lacuna in the field due to lack of specific autophagy inhibitors. Nonetheless, a small molecule inhibitor of autophagosome-lysosome fusion, EACC (ethyl [2-{5-nitrothiophene-2-carboxamido} thiophene-3-carbonyl] carbamate), has been identified [1928]. This molecule selectively blocks autophagic flux by inhibiting STX17 translocation onto autophagosomes.
Although these various agents can inhibit different steps of the autophagic pathway, their potential side effects must be considered in interpretation of the secondary consequences of autophagy inhibition, especially in long-term studies. For example, lysosomotropic compounds can increase the rate of autophagosome formation by inhibiting MTORC1, as activation of lysosomally localized MTORC1 depends on an active V-ATPase (as well as RRAG GTPases [223]) [1925, 1929]. Along these lines, CQ treatment may cause an apparent increase in the formation of autophagosomes possibly by blocking fusion with the lysosome (F.C. Dorsey and J.L. Cleveland, personal communication). This conclusion is supported by the finding that CQ reduces the colocalization of LC3 and LysoTracker™ despite the presence of autophagosomes and lysosomes (A.K. Simon, personal communication). In addition, CQ, but not bafilomycin A1, blocks autophagosome-lysosome fusion in U2OS, HeLa and MEFs [302]. This mechanism might be cell-type specific, as other studies report that CQ prevents autolysosome clearance and degradation of cargo content, but not autophagosome-lysosome fusion [1930–1933]. Concanamycin A blocks sorting of vacuolar proteins and diverts the route of autophagy in plant cells along with inhibiting vacuolar acidification [1934, 1935]. Furthermore, in addition to causing the accumulation of autophagic compartments, many of these drugs seem to stimulate sequestration in many cell types, especially in vivo [114, 432, 1521, 1582, 1887, 1890, 1936–1939]. Although it is clear why these drugs cause the accumulation of autophagic compartments, it is not known why they stimulate sequestration. One possibility, at least for hepatocytes, is that the inhibition of protein degradation reduces the intracellular amino acid pool, which in turn upregulates sequestration. A time-course study of the changes in both the intra- and extracellular fractions may provide accurate information regarding amino acid metabolism. For these various reasons, it is important to include appropriate controls; along these lines, MTOR inhibitors such as rapamycin or amino acid deprivation can be utilized as positive controls for inducing autophagy. In many cell types, as well as in D. discoideum [1940], however, the induction of autophagy by rapamycin is relatively slow, or transient, allowing more time for indirect effects.
Amino acid starvation induces autophagy through deactivation of MTOR. Whereas autophagy is induced equally well by pharmacological inhibition of MTOR and amino acid starvation, amino acid starvation additionally causes depletion of intracellular amino acids, which strongly repress protein synthesis. For that reason, the protein expression levels of classical substrates of autophagy (e.g., SQSTM1-like receptors [SLRs]) decrease more rapidly during amino acid starvation than during pharmacological inhibition of MTOR. Additionally, endosomal microautophagy, which also targets SLRs and certain Atg8-family protein homologs, is also active during amino acid starvation contributing to the overall decreased expression of many classical substrates of autophagy [1941].
Several small molecule inhibitors, including torin1, PP242, KU-0063794, PI-103 and NVP-BEZ235, have been developed that target the catalytic domain of MTOR in an ATP-competitive manner [300, 802, 1942–1945]. In comparison to rapamycin, these catalytic MTOR inhibitors are more potent, and hence are stronger autophagy agonists in most cell lines [453, 802, 1946]. The use of these second-generation MTOR inhibitors may reveal that some reports of MTOR-independent autophagy may actually reflect the use of the relatively weak inhibitor rapamycin. Furthermore, the use of these compounds has revealed a role for MTORC1 and MTORC2 as independent regulators of autophagy [1947].
Neurons, however, seem to be a particular case in regard to their response to MTOR inhibitors. Rapamycin may fail to activate autophagy in cultured primary neurons, despite its potent stimulation of autophagy in some cancer cell lines [106, 818, 1948]. Interestingly, both rapamycin and catalytic MTOR inhibitors do not induce a robust autophagy in either cultured primary mouse neurons or human neuroblastoma SH-SY5Y cells, which can differentiate into neuron-like cells, whereas the drugs do elicit a potent autophagic response in cultured astrocytes (J. Diaz-Nido and R. Gargini, personal communication). This observation suggests a differential regulation of autophagy in neurons. It has been suggested that control of neuronal autophagy may reflect the particular physiological adaptations and metabolic requirements of neurons, which are very different from most peripheral cell types [1949]. For example, acute starvation in transgenic mice expressing GFP-LC3 leads to a potent induction of autophagy in the liver, muscle and heart but not in the brain [239]. Along these lines, glucose depletion may be much more efficient at inducing autophagy than rapamycin or amino acid starvation in neurons in culture (M. Germain and R. Slack, personal communication). Indeed, treatment of cultured primary mouse neurons and human neuroblastoma SH-SY5Y cells with 2-deoxy-glucose, which hampers glucose metabolism and leads to activation of AMPK, results in robust autophagy induction (J. Diaz-Nido and R. Gargini, personal communication). A number of compounds can also be quite efficient autophagy inducers in neurons including the CAPN (calpain) inhibitor calpeptin [1950–1952]. Thus, it has been suggested that autophagy induction in neurons may be achieved by molecular mechanisms relying on AMPK or increases in intracellular calcium concentration [1949]. An example where changes in cytosolic calcium levels, due to the incapacity of the mitochondria to buffer calcium release, result in an increase in autophagy is seen in a cellular model of the neurodegenerative disease Friedreich ataxia, based on FXN (frataxin) silencing in SH-SY5Y human neuroblastoma cells [1953].
Finally, a specialized class of compounds with α,β-unsaturated ketone structure tends to induce autophagic cell death, accompanied by changes in mitochondrial morphology. Because the cytotoxic action of these compounds is efficiently blocked by N-acetyl-L-cysteine, the β-position in the structure may interact with an SH group of the targeted molecules [1954]. Due to the potential pleiotropic effects of various drug treatments, it is incumbent upon the researcher to demonstrate that autophagy is indeed inhibited, by using the methodologies described herein. Accordingly, it is critical to verify the effect of a particular biochemical treatment with regard to its effects on autophagy induction or inhibition when using a cell line that was previously uncharacterized for the chemical being used. Similarly, cytotoxicity of the relevant chemical should be assessed.
The use of gene deletions/inactivations (e.g., in primary or immortalized atg−/− MEFs [814], plant T-DNA or transposon insertion mutants [378, 1955], or in vivo using transgenic knockout models [1956, 1957] including Cre-lox based “conditional” knockouts [424, 425]) or functional knockdowns (e.g., with RNAi against ATG genes) is the preferred approach when possible because these methods allow a more direct assessment of the resulting phenotype; however, different floxed genes are deleted with varying efficiency, and the proportion deleted must be carefully quantified [1958]. Studies also suggest that microRNAs may be used for blocking gene expression [332–335, 990, 991, 1959]. In most contexts, it is advisable when using a knockout or knockdown approach to examine multiple autophagy-related genes to exclude the possibility that the phenotype observed is due to effects on a nonautophagic function(s) of the corresponding protein, especially when examining the possibility of autophagic cell death. This is particularly the case in evaluating BECN1, which interacts with anti-apoptotic BCL2 family proteins [848], or when low levels of a target protein are sufficient for maintaining autophagy as is the case with ATG5 [346]. With regard to ATG5, a better approach may be to use a dominant negative (K130R) version [1875, 1948, 1960]. Also noteworthy is the role of ATG5 in mitotic catastrophe [818] and several other nonautophagic roles of ATG proteins (see Noncanonical use of autophagy-related proteins) [106]. Along these lines, and as stated above for the use of inhibitors, when employing a knockout or especially a knockdown approach, it is again incumbent upon the researcher to demonstrate that autophagy is actually inhibited, by using the methodologies described herein.
Finally, we note that the long-term secondary consequences of gene knockouts or knockdowns are likely much more complex than the immediate effects of the actual autophagy inhibition. To overcome this concern, inducible knockout systems might be useful [346, 559]. One additional caveat to knockdown experiments is that PAMP recognition pathways can be triggered by double-stranded RNAs (dsRNA), such as siRNA probes, or the viral vector systems that deliver shRNA [1961]. Some of these, including TLR-mediated RNA recognition [1962], can influence autophagy by either masking any inhibitory effect or compromising autophagy independent of the knockdown probe. Therefore, nontargeting (scrambled) siRNA or shRNA controls should be used with the respective transfection or transduction methods in the experiments that employ ATG knockdown. Another strategy to specifically interfere with autophagy is to use dominant negative inhibitors. Delivery of these agents by transient transfection, adenovirus, or TAT-mediated protein transduction offers the possibility of their use in cell culture or in vivo [1960]. However, because autophagy is an essential metabolic process for many cell types and tissues, loss of viability due to autophagy inhibition always has to be a concern when analyzing cell death-unrelated questions. In this respect it is noteworthy that some cell-types of the immune system such as dendritic cells [440] seem to tolerate loss of autophagy fairly well, whereas others such as T and B cells are compromised in their development and function after autophagy inhibition [1963, 1964].
In addition to pharmacological inhibition, RNA silencing, gene knockout and dominant negative RAB and ATG protein expression, pathogen-derived autophagy inhibitors can also be considered for use in manipulating autophagy. Along these lines ICP34.5, viral BCL2 homologs and viral FLIP of herpesviruses block autophagosome formation [848, 1418, 1965], whereas M2 of influenza virus and HIV-1 Nef block autophagosome degradation [493, 1966]. However, as with other tools discussed in this section, transfection or transduction of viral autophagy inhibitors should be used in parallel with other means of autophagy manipulation, because these proteins are used for the regulation of usually more than one cellular pathway by the respective pathogens. Finally, RavZ is an example of a bacterial protein that blocks autophagy. RavZ is a Legionella effector that inhibits host autophagy by irreversible deconjugation of LC3 [1967]. RavZ has 3 LIR motifs in its N- and C-terminal regions for interacting with LC3 [1968] and a catalytic cysteine protease domain that cleaves the peptide bond between the PE-modified C-terminal glycine residue and the adjacent aromatic residue in Atg8-family proteins [1967]. In addition, all Legionella pneumophila strains sequenced to date [1969] encode a homolog of the eukaryotic enzyme SGPL1 (sphingosine-1-phosphate lyase 1) that was named LpSPL for Legionella pneumophila SPL. This gene was most likely acquired from a protist host [1970]. The translocated LpSPL effector protein targets host sphingosine biosynthesis to curtail autophagy. LpSPL activity alone is sufficient to prevent an increase in sphingosine levels in infected host cells and to inhibit autophagy during macrophage infection [1971].
There are fewer compounds that act as inducers of autophagy, but the initial characterization of this process was due in large part to the inducing effects of glucagon, which appears to act through indirect inhibition of MTOR via the activation of STK11/LKB1-AMPK [1531, 1532, 1972]. Recent studies demonstrate that glucagon directly induces the CAMK2-OGT-ULK cascade that potentiates AMPK-dependent ULK phosphorylation [1007]. Currently, the most commonly used inducer of autophagy is rapamycin, an allosteric inhibitor of MTORC1 (although as mentioned above, catalytic inhibitors such as torin1 are increasingly being used). Nevertheless, one caution is that MTOR is a major regulatory protein that is part of several signaling pathways, including for example those that respond to INS (insulin), EGF (epidermal growth factor) and amino acids, and it thus controls processes other than autophagy, so rapamycin will ultimately affect many metabolic pathways [744, 1973–1975]. In particular, the strong effects of MTOR on protein synthesis may be a confounding factor when analyzing the effects of rapamycin. MTOR-independent regulation can be achieved through lithium, sodium valproate and carbamazepine, compounds that lower the myo-inositol 1,4,5-triphosphate levels [1976], as well as FDA-approved compounds such as verapamil, trifluoperazine and clonidine [1977, 1978]. Regarding trifluoperazine, studies have shown that other structurally related antipsychotic phenothiazine derivatives, such as chlorpromazine and thioridazine, induce autophagy in tumor cells in vitro through the inhibition of AKT-MTOR [1979, 1980] and by modulating the WNT-CTNNB1/β-catenin signaling pathway [1981] in glioma cells. The antihistamine phenothiazine derivative promethazine also induces autophagy-associated cell death in a Philadelphia chromosome-positive chronic myeloid leukemia model (K562) mediated by activation of AMPK [1982].
In vivo treatment of embryos with cadmium results in an increase in autophagy, probably to counter the stress, allowing cell survival through the elimination/recycling of damaged structures [1552]. Autophagy may also be regulated by the release of calcium from the ER under stress conditions [216, 1922, 1983–1987]. Studies have demonstrated that a natural compound, celastrol, inhibits ATP2A/SERCA, a sarcoplasmic/endoplasmic reticulum calcium-ATPase pump to induce autophagy-dependent cytotoxicity in rheumatoid arthritis synovial fibroblasts and rheumatoid arthritis fibroblast-like synoviocytes via the CAMK2B (calcium/calmodulin dependent kinase kinase II beta)-AMPK-MTOR pathway [1988]. Conversely, some compounds can achieve their biological effect by inhibition of calcium-regulated autophagy. For instance, 2-aminoethoxydiphenylborane sensitizes the anti-tumor effect of bortezomib via suppression of calcium-mediated autophagy [1989].
ITPRs as ER-resident intracellular calcium-release channels, which also localize at MAMs, have a dual role in autophagy [1990]. In non-starved conditions, ITPRs appear to suppress basal autophagy by funneling calcium into the mitochondria, thereby promoting mitochondrial bio-energetics [668]. Upon starvation, ITPRs are involved in the augmented autophagic flux through the CAMK2-OGT-ULK cascade as well as a process that involves ITPR sensitization through the recruitment of BECN1 [1007, 1983]. The essential role of ITPRs and/or intracellular calcium signaling to drive autophagic flux has also been observed after treatment with rapamycin [1985], resveratrol [1991] or some chemical inducers of ER stress [1992]. Confluency-induced differentiation of Caco-2 cells increases the expression of the master transcriptional regulator HNF4A/HNF4α, which in turn induces ER stress via the increased the expression of XBP1 and ATF6, accompanied by an increase in the intracellular calcium levels and autophagy [1993]. However, additional calcium signals from other stores such as lysosomes could also play an important role in autophagy induction [1994]. The activation of the lysosomal TPCN/two-pore channel (two pore segment channel), by nicotinic acid adenine dinucleotide phosphate (NAADP) induces autophagy through an AMPK-ACAC pathway independently of MTORC1 in neural cells [1995]. Autophagosome formation mediated by NAADP can be selectively inhibited by the TPCN blocker NED-19, by pre-incubation with a cell-permeable acetoxymethyl ester version of BAPTA (BAPTA-AM), or in cells overexpressing TPCN2 mutated within the putative pore region (TPCN2L265P), indicating that lysosomal calcium selectively induces autophagy [1996]. Furthermore, a possible NAADP-agonist, glutamate, is able to induce autophagy via a TPC1/2-AMPK-ACAC pathway [1995].
Lysosomal cation-permeable channels such as TPCN2 associate with MTORC1, a key nutrient sensor and upstream control mechanism of autophagy, to regulate autophagy flux [1997]. Lysosomal calcium release via TPCN2 occurs upon inhibition of MTOR in response to starvation or rapamycin treatment and is an essential component to drive autophagic flux in these conditions. In addition to MTORC1 control of TPCN2 activity, calcium release via TPCN2 also supports MTORC1 activity [1998]. Furthermore, upon starvation, MCOLN1 (mucolipin 1) also contributes to lysosomal calcium release. This release results in the calcium-dependent activation of PPP3/calcineurin, which dephosphorylates TFEB, triggering its nuclear translocation. In the nucleus, TFEB upregulates several autophagy and lysosomal biogenesis genes. MCOLN1-mediated calcium release also contributes to the reactivation of MTORC1 in conditions of prolonged starvation via a mechanism that requires the calcium-binding protein CALM (calmodulin) [1999]. As such, the dynamic nature of calcium signaling involving ER, mitochondria and lysosomes contribute to the fine-tuned control of autophagic flux [2000, 2001]. The use of BAPTA-AM to implicate calcium signaling in autophagy comes with a caution, as intracellular BAPTA can also exert calcium-independent effects, such as inhibition of the Na+/K+ ATPase [2002, 2003] as well as autophagy [1994]. Along these lines, it is becoming increasingly clear that BAPTA, and related molecules such as calcium indicator dyes, have cellular effects that are not related to calcium buffering [2002, 2003]. Low-affinity analogs of BAPTA (e.g., dibromo- and difluoro-BAPTA) can inhibit autophagy triggered by PP242 (M.D. Bootman, personal communication).
Cell-penetrating autophagy-inducing peptides, such as Tat-vFLIP or Tat-beclin 1, are also potent inducers of autophagy in cultured cells as well as in mice [1965, 2004]. Other cell-penetrating peptides, such as Tat-wtBH3D or Tat-dsBH3D, designed to disrupt very specific regulatory interactions such as the BCL2-BECN1 interaction, are potent, yet very specific, inducers of autophagy in cultured cells [2005].
In contrast to other PtdIns3K inhibitors, caffeine induces autophagy in the food spoilage yeast Zygosaccharomyces bailii [2006], mouse embryonic fibroblasts [2007], and S. cerevisiae [2008] at millimolar concentrations. In more complex eukaryotes, this is accompanied by inhibition of the MTOR pathway. Similarly, in budding yeast caffeine is a potent TORC1 inhibitor suggesting that this drug induces autophagy via inhibition of the TORC1 signaling pathway; however, as with other PtdIns3K inhibitors caffeine targets other proteins, notably Mec1/ATR and Tel1/ATM, and affects the cellular response to DNA damage.
Another autophagy inducer is the histone deacetylase inhibitor valproic acid [2009, 2010]. The mechanism by which valproic acid stimulates autophagy is not entirely clear but may occur due to inhibition of the histone deacetylase Rpd3, which negatively regulates the transcription of ATG genes (most notably ATG8 [2011]) and, via deacetylation of Atg3, controls Atg8 lipidation [2012]. SMER28 is an MTOR-independent inducer of autophagy that acts through largely unknown mechanisms [2013]. Dasatinib, a dual SRC and BCR-ABL kinase inhibitor, also stimulates autophagy through unknown mechanisms to further induce myeloid differentiation of AML cells [2014].
A new promising drug, NeuroHeal, has emerged that activates autophagy through a SIRT1-dependent mechanism [2015, 2016]. NeuroHeal treatment protects from ER-stress and promotes in vivo neuroprotection in several models where neurons remain isolated and disconnected from their targets, a common characteristic in any neurodegenerative process [2017, 2018]. An efficient high-throughput method for screening of autophagy modulators has been carried out employing a dual-luciferase assay. In this case, degradation of the individual luciferases indicates the degradation of general cytoplasmic contents and the selective degradation of specific cargoes. The levels of cytosolic Renilla luciferase, and targeted firefly luciferase, which can be delivered to a specific cargo (such as peroxisomes), is measured and interpreted as rates of general and selective autophagy flux, respectively [2019].
Induction of autophagy can also be involved in virulence mechanisms for infection; the Buruli ulcer causative agent Mycobacterium ulcerans produces an exotoxin, mycolactone, that induces autophagy [2020, 2021]. The mechanism is most likely a protective response to mycolatone’s inhibition of SEC61A1, the major subunit of the SEC61 translocon (R.E. Simmonds, personal communication), which causes the accumulation of mislocalized proteins in the cytosol [2022] and a consequent integrated stress response [2021]. Notably, polymorphisms in autophagy-related genes may be involved in the risk of acquiring M. ulcerans infection from the environment [2023].
It is also possible, depending on the organism or cell system, to modulate autophagy through transcriptional control. For example, this can be achieved either through overexpression or post-translational activation of TFEB (see Transcriptional and translational regulation), a transcriptional regulator of the biogenesis of both lysosomes and autophagosomes [947, 949]. Along these lines, inhibition or genetic deletion of CTSB downregulates MTOR, causing TFEB to activate autophagy [2024]. Similarly, adenoviral-mediated expression of the transcription factor CEBPB induces autophagy in hepatocytes [989]. Either the genetic ablation or the knockdown of the nucleolar transcription factor RRN3/TIF-IA, a crucial regulator of the recruitment of POLR1 (RNA polymerase I) to ribosomal DNA promoters, induces autophagy in neurons and in MCF-7 cancer cells, respectively, linking ribosomal DNA transcription to autophagy [2025, 2026]. Likewise, inhibition of POLR1 by the small molecule inhibitor CX-5461 induces autophagy. A growing body of evidence suggests the involvement of nucleolar ribosome biogenesis factors and the so-called nucleolar stress response in autophagy [2027, 2028]. A class of diseases connected to impaired ribosome biogenesis, termed ribosomopathies, reveal activation of autophagy (see Erythroid cells). Nucleolar-stress induced autophagy seems to engage both TP53-dependent as well as -independent mechanisms. Also, induction of autophagy is commonly connected to MTOR signaling in this context. However, the underlying mechanisms connecting nucleolar stress and autophagy have to be better elucidated in future studies.
Relatively little is known about direct regulation via the ATG proteins, but there is some indication that tamoxifen acts to induce autophagy by increasing the expression of BECN1 in MCF7 cells [2029]. However, BECN1 does not appear to be upregulated in U87MG cells treated with tamoxifen, whereas the levels of LC3-II and SQSTM1 are increased, while LAMP2B is downregulated and CTSD and CTSL activities are almost completely blocked (K.S. Choi, personal communication). Thus, the effect of tamoxifen may differ depending on the cell type. Other data suggest that tamoxifen acts by blocking cholesterol biosynthesis, and that the sterol balance may determine whether autophagy acts in a protective versus cytotoxic manner [2030, 2031]. Finally, screens have identified small molecules that induce autophagy independently of rapamycin and allow the removal of misfolded or aggregate-prone proteins [1978, 2032], suggesting that they may prove useful in therapeutic applications.
One novel autophagy inducer that does not target MTOR, is KYP-2047, a small-molecule inhibitor for PREP (prolyl endopeptidase), a serine protease belonging to the prolyl oligopeptidase family (clan SC) [2033]. Although the exact mechanism as to how PREP regulates autophagy is not clear, PREP inhibition by KYP-2047 elevates BECN1 mRNA and protein levels in HEK 293 cells after a 24-h incubation. This inhibition results in decreased aggregation-prone protein levels in several cellular and animal models [2034]. Moreover, removal of PREP from HEK 293 cells induces autophagic flux, and also decreases proteasomal activity [2035]. However, caution should be taken because of the crosstalk between autophagy and the proteasomal system. For example, trehalose, an MTOR-independent autophagy inducer [2036], can compromise proteasomal activity in cultured primary neurons [2037]. Trehalose activates autophagy by inhibiting AKT-mediated phosphorylation of TFEB, thereby promoting TFEB nuclear translocation and subsequent activation of CLEAR-regulated autophagy and lysosomal genes in cells and in vivo [973]. For experiments in cells, it must be considered that at the concentration usually tested (>mM) trehalose effects can be potentially ascribed to hyperosmotic signaling (R. Franco, personal communication). Trehalose treatment also results in subtle lysosomal damage (possibly because of an osmotic shock to this organelle), which causes activation of PPP3/calcineurin, a calcium-dependent phosphatase capable of dephosphorylating and activating TFEB [2038]. This activation permits enhanced lysophagy to degrade damaged lysosomes, but at the same time enhances the overall autophagic capacity of trehalose-treated cells. Lysosomal impairment by trehalose might underlie observations that cell treatment with this disaccharide can decrease degradation of APP in lysosomes [2039], increase GFP:RFP ratios using the GFP-LC3-RFP-LC3∆G fluorescent probe [494], or result in the accumulation of autophagosomes [2040]. Finally, several disease-causing and aggregation-prone proteins in the secretory pathway that are targeted for autophagy are also targeted for ERAD, which requires proteasome function [2041].
Another autophagy inducer, genistein (trihydroxyisoflavone or 5, 7-dihydroxy-3-[4-hydroxyphenyl]-4H-1-benzopyran-4-one), has been suggested previously to stimulate autophagy in various cancer cell lines, including ovarian cancer [2042], colon cancer [2043], breast cancer [2044], pancreatic cancer [2045], and uterine leiomyoma cells [2046]. Other studies demonstrate that this isoflavone effectively induces autophagy in cellular and animal models of HD and AD, respectively [2047, 2048]. Such genistein-mediated autophagy stimulation is responsible for correction of phenotypes of these diseases through degradation of pathological protein aggregates, which otherwise accumulate in cells and organs. In fact, induction of autophagy by this isoflavone has been proposed as a therapeutic approach in various genetic and neurodegenerative disorders caused by accumulation of undegraded proteins or other macromolecules [2049, 2050]. The molecular mechanism of genistein-mediated induction of autophagy is not clear, however, it appears that inhibition of MTOR and subsequent activation of TFEB contributes significantly to this process [2051–2053].
While likely to be nonspecific, inhibition of NFKB activation—through either an NKFBIA/IκBα kinase inhibitor or SERPINA1/alpha-1-antitrypsin—may augment autophagy in macrophages infected with mycobacteria [2054, 2055]. One possible mechanism is the inhibition of NFKB-mediated induction of TNFAIP3/A20, a deubiquitinating enzyme that normally deactivates BECN1; hence, by sequentially inhibiting NFαB activation and TNFAIP3/A20 expression, a pathway that inhibits BECN1 is mitigated [2054].
Because gangliosides are implicated in autophagosome morphogenesis, pharmacological or genetic impairment of gangliosidic compartment integrity and function can provide useful information in the analysis of autophagy. To deplete cells of gangliosides, an inhibitor of CERS (ceramide synthase), such as a fungal metabolite produced by Fusarium moniliforme (fumonisin B1), or, alternatively, siRNA to CERS or ST8SIA1, can be used [907].
Antimicrobial peptides (AMPs) were originally described for their activity in killing microbes; however, they also have a role in immune system modulation [2056]. Autophagy induction produces AMPs by proteolysis of cytosolic proteins of infected cells [2057, 2058]). Furthermore, three AMPs (indolicidin, and two peptides derived from PYY2/Seminalplasmin) induce autophagy in Leishmania cells [2059], and the anti-microbial peptide LL-37 induces autophagy in human cells as a way to eliminate tuberculosis infection [2060].
Finally, in addition to genetic and chemical compounds, it was reported that electromagnetic fields can induce autophagy in mammalian cells. Studies of biological effects of novel therapeutic approaches for cancer therapy based on the use of noninvasive radiofrequency fields reveal that autophagy, but not apoptosis, is induced in cancer cells in response to this treatment, which leads to cell death [2061]. This effect is tumor specific and different from traditional ionizing radiation therapy that induces apoptosis in cells.
Conclusion: Considering that pharmacological inhibitors or activators of autophagy have an impact on many other cellular pathways, the use of more than one methodology, including molecular methods, is desirable. Rapamycin is less effective at inhibiting MTOR and inducing autophagy than catalytic inhibitors; however, it must be kept in mind that catalytic inhibitors also affect MTORC2. The main concern with pharmacological manipulations is pleiotropic effects of the compound being used. Accordingly, genetic confirmation is preferred whenever possible. Alternatively, pharmacological compounds that do not target cell survival and maintenance pathways such as MTOR, can be used for temporal regulation of autophagy [2062].
3. Basal autophagy
Basal levels of LC3-II or GFP-LC3 puncta may change according to the time after addition of fresh medium to cells, and this can lead to misinterpretations of what basal autophagy means. This is particularly important when comparing the levels of basal autophagy between different cell populations (such as knockout versus wild-type clones). If cells are very sensitive to nutrient supply and display a high variability of basal autophagy, the best experimental condition is to monitor the levels of basal autophagy at different times after the addition of fresh medium. One example is the chicken lymphoma DT40 cells (see Chicken B-lymphoid DT40 cells) and their knockout variant for all three ITPR isoforms [668, 1990, 2063]. In these cells, no differences in basal levels of LC3-II can be observed up to 4 h after addition of fresh medium, but differences can be observed after longer times (J.M. Vicencio and G. Szabadkai, personal communication). This concept should also be applied to experiments in which the effect of a drug upon autophagy is the subject of study. If the drugs are added after a time in which basal autophagy is already high, then the effects of the drug can be masked by the cell’s basal autophagy, and wrong conclusions may be drawn. To avoid this, fresh medium should be added first (followed by incubation for 2-4 h) in order to reduce and equilibrate basal autophagy in cells under all conditions, and then the drugs can be added. The basal autophagy levels of the cell under study must be identified beforehand to know the time needed to reduce basal autophagy.
A similar caution must be exercised with regard to cell culture density and hypoxia. When cells are grown in normoxic conditions at high cell density, HIF1A/HIF-1α is stabilized at levels similar to that obtained with low-density cultures under hypoxic conditions [2064]. This results in the induction of BNIP3 and BNIP3L and “hypoxia”-induced autophagy, even though the conditions are theoretically normoxic [1170]. Therefore, researchers need to be careful about cell density to avoid accidental induction of autophagy.
It should be realized that in yeast species, medium changes can trigger a higher “basal” level of autophagy in the cells. In the methylotrophic yeast species K. phaffii/P. pastoris and Hansenula polymorpha, a shift of cells grown in batch from glucose to methanol results in stimulation of autophagy [2065, 2066]. A shift to a new medium can be considered a stress situation. Thus, it appears to be essential to cultivate the yeast cells for a number of hours to stabilize the level of basal autophagy before performing experiments intended to study levels of (selective) autophagy (e.g., pexophagy). Finally, plant root tips cultured in nutrient-sufficient medium display constitutive autophagic flux (i.e., a basal level), which is enhanced in nutrient-deprived medium [2067–2069].
Conclusion: The levels of basal autophagy can vary substantially and can mask the effects of the experimental parameters being tested. Changes in media and growth conditions need to be examined empirically to determine effects on basal autophagy and the appropriate times for subsequent manipulations.
4. Experimental systems
Throughout these guidelines we have noted that it is not possible to state explicit rules that can be applied to all experimental systems. For example, some techniques may not work in particular cell types or organisms. In each case, efficacy of autophagy promotors, inhibitors and measurement techniques must be empirically determined, which is why it is important to include appropriate controls. Differences may also be seen between in vivo or perfused organ studies and cell culture analyses. For example, INS (insulin) has no effect on proteolysis in suspended rat hepatocytes, in contrast to the result with perfused rat liver. The INS effect reappears, however, when isolated hepatocytes are incubated in stationary dishes [2070, 2071] or are allowed to settle down on the matrix (D. Häussinger, personal communication). The reason for this might be that autophagy regulation by INS and some amino acids requires volume sensing via integrin-matrix interactions and also intact microtubules [2072–2074]. Along these lines, the use of whole embryos makes it possible to investigate autophagy in multipotent cells, which interact among themselves in their natural environment, bypassing the disadvantages of isolated cells that are deprived of their normal network of interactions [1552]. In general, it is important to keep in mind that results from one particular system may not be generally applicable to others.
Conclusion: Although autophagy is conserved from yeast to human, there may be tremendous differences in the specific details among systems. Thus, results based on one system should not be assumed to be applicable to another.
5. Bimolecular fluorescence complementation
Bimolecular fluorescence complementation (BiFC) may be useful to study protein-protein interactions in the autophagic pathway [2075] In this assay, a protein of interest is cloned into a vector containing one half of a fluorescent reporter (e.g., YFP), while a second protein is cloned into a different vector containing the other half of the reporter. Constructs are cotransfected into cells. If the two proteins of interest interact, the two halves of the reporter are brought into close proximity and a fluorescent signal is reconstituted, which can be monitored by confocal microscopy. This assay can be used to determine protein interactions without prior knowledge of the location or structural nature of the interaction interface. Moreover, this approach is applicable to living cells, and relatively low concentrations of recombinant protein are required to generate a detectable signal. One issue with BiFC is that once the two halves of the fluorophore interact with each other the binding is extremely stable, which can result in the amplification of weak signals. For the same reason, the localization of the BiFC interaction may not represent the normal physiological site. Conversely, the stable nature of this interaction may be utilized as an alternative to chemical cross-linking for studying protein-protein interactions.
6. Nanoparticles
Nanoparticles (NPs) are tiny particulate materials, ranging in size from 1 to 100 nm in diameter. Due to their physicochemical properties and small size, some NPs may cross biological membranes and are often used to deliver a cytotoxic agent or as a tool to modulate cellular processes [2076]. NPs may act both as inhibitors and inducers of autophagy [1919]. Indeed, most endocytic routes of NP uptake converge on the lysosome, making this organelle a common site of NP sequestration and degradation [2077]. Autophagy induction is of relevance for NPs due to the similarities in sizes and shapes between NPs and pathogens [2078]. The exact mechanism of autophagy induction by NPs is not well understood, although studies have shown that the surface properties including surface charge, may play a decisive role, as shown in studies using ammonium-functionalized gold NPs [2079]. For example, the positively charged surface of cationic NPs might facilitate their interaction with the negatively charged plasma or endolysosomal membranes harboring the members of the MTOR signaling pathway. Moreover, lysosomal alkalinization by the “proton sponge” effect of cationic NPs could cause lysosomal dysfunction and subsequent defects in lysosomal recruitment and activation of MTORC1 [2080].
NPs also promote autophagy through specific modulation of lysosomal pH. Biodegradable nanoparticles such as photo-activable NPs (paNPs) and poly (lactide-co-glycotide) (PLGA) NPs induce autophagy through acidification of the lysosomal environment in cellular models of type II diabetes [2081, 2082], AD [80] and PD [2083, 2084]. In pancreatic beta cells under lipotoxicity or PC-12 cells under MPP+ neurotoxin treatment, lysosomal pH is elevated and autophagy is inhibited due to impaired fusion between lysosomes and autophagosomes. PLGA NPs and paNPs localize to lysosomes, and lower lysosomal pH in both types of cellular models, thereby restoring autophagy and cellular functions. Of note, the paNPs are stimuli-responsive NPs that can acutely release acids to lower lysosomal pH only upon application of UV light. This allows for paNPs to be a useful tool that can temporally control the outcomes of autophagy by an external stimulus [2081].
ROS production by NPs may also play a role in autophagy induction [2085, 2086]. Furthermore, NPs with ROS-quenching capacity (e.g., non-photo-excited graphene quantum dots) induce subsequent tolerogenic effects in human dendritic cells in an autophagy-dependent manner, and this is reversed by ATG5 silencing [2087]. Conversely, LC3 silencing reduces the oxidative stress-dependent cytotoxicity of photoexcited graphene quantum dots, indicating a role of autophagy in their photodynamic anticancer activity [2088]. Nitrogen-doped TiO2 NPs can induce autophagy-dependent differentiation or autophagy-associated cell death in leukemia cells, depending on the dose of the NPs, and pre-incubation of leukemic cells with ROS scavengers diminishes the effect of the NPs [2089]. Expansile nanoparticles/eNPs can also induce autophagy-associated cell death through disruption of autophagosomal trafficking, and offer a new opportunity to develop autophagy modulators for cancer therapy when used in conjugation with a chemotherapeutic [2090]. Using a combinatorial library of multi-walled carbon nanotubes (CNTs), it is possible to show that autophagy induction can be “tuned” by varying surface ligands on the CNTs [2091]. Furthermore, the autophagy-inducing activity of certain metallic NPs can be modulated through surface coating with specific peptides [2092]. Size-dependent autophagy induction has also been observed [2093], and it is suggested that quantum dots (i.e., semiconductor crystals) may serve as potentially useful probes for autophagy studies, in light of their unique optical properties [2094].
It should be noted that NPs may also block autophagy, and shape-related targeting of lysosomes may explain NP-mediated inhibition of autophagic flux [2095]. It is important to distinguish between autophagosome accumulation resulting from blockade of autophagic flux as opposed to the induction of autophagy, and, as with other areas of study, the failure to distinguish one from the other may result in the misinterpretation of NP effects on cells [2096]. Silica (SiO2) NPs are among the most widely produced and most intensively studied nanomaterials, and the persistent presence of enlarged autolysosomes is seen in hepatocytes after exposure to SiO2 NPs [2097]. This accumulation is due to a defect in the autophagy termination process known as autophagic lysosome reformation (ALR). Similarly, the blockade of autophagic flux by SiO2 NPs was reported in lung epithelial cells, and evidence was provided for a suppressive effect of the NPs on lysosomal acidification, thereby contributing to the decreased autophagic degradation in these cells [2097]. Others have shown that SiO2 NPs induce autophagosome accumulation in hepatocytes via the activation of the EIF2AK3- and ATF6-dependent UPR pathways [2098]. To further compound the situation, autophagic cell death induction was shown, in a study of single-walled CNTs, to occur through the AKT-TSC2-MTOR pathway. Inhibition of autophagy using both pharmacological and genetic approaches significantly reduces the CNT-induced autophagic cell death as well as the acute lung injury evidenced in mice [2099].
Graphene oxide combined with cisplatin (GO-CDDP) not only elicits autophagy, but induces the nuclear import of cisplatin as well as LC3 [2100]. The nuclear LC3 does not colocalize with SQSTM1 or LAMP2, and blocking autolysosome formation does not significantly hinder the nuclear import of LC3-CDDP, indicating that autophagosome and autolysosome formation is dispensable. Furthermore, direct binding between silica nanoparticles and LC3 and SQSTM1 was demonstrated in osteoblast cells [2101].
ATG4 proteases are essential enzymes in the autophagic process, and ATG4B appears to be most relevant for autophagy. Several peptide-conjugated polymeric nanoprobes have been developed for real-time monitoring of ATG4B activity in vitro and in vivo. There is an “in vivo self-assembly”-based nanoprobe that consists of a TFGF peptide (a peptide from LC3, which specifically responds to ATG4B), an aggregation-induced emission molecule and a hydrophilic carrier, that in situ self-assembles into new nanostructures that “turn on” signals in the presence of ATG4B [2102, 2103]. These nanoprobes do not induce autophagy at the used dose and can be applied for real-time and quantitative evaluation of ATG4B in living tumor cells, as well as the zebrafish and mouse models. Another probe is a FRET-based nanoparticle that uses the fluorescent dye FITC and the quencher BHQ1 attached to the TFGF peptide, which is nonfluorescent, but fluoresces in autophagy-inducing cells [2104].
To summarize, careful, case-by-case evaluation of the role of autophagy for each NP is required, and a direct interaction between NPs and the cellular autophagic machinery seems possible. In addition, it is important to determine whether each type of NP alters the net autophagic degradation capacity by employing one of the functional assays described in these guidelines. This will provide a more comprehensive picture than merely determining the levels of autophagic markers and the number of autophagosomes.
NPs may provide useful tools with which to study autophagy, as suggested in early work on quantum dots and using photo-activated nanoparticles [2081, 2093]. NPs can either induce or inhibit autophagy; thus, the autophagy-inducing/inhibiting efficacy of nanoprobes should be carefully explored before using them in vivo or in clinical research.
Another example of the use of NPs to study autophagy is seen with nanotubes (NTs) in the study of ATG3-membrane interaction. As shown in flotation studies with sonicated unilamellar vesicles, ATG3 increases its binding to neutral membranes when vesicle size decreases. To visualize this effect under the microscope, lipid NTs can be generated, starting from a compositionally well-defined unilamellar membrane system, such as SUPER templates [2105]. The formed NTs are thin tubules with high membrane curvature, reported to be a powerful tool to analyze curvature-dependent binding of proteins. ATG3-Alexa Fluor 488 interacts with electrically neutral (PC:DOPE) NTs, whereas GABARAP does not exhibit any binding to NTs with similar curvature [2106].
7. Nomenclature
8. General considerations for experimental manipulations
One general issue with regard to any assay is that experimental manipulation could introduce some type of stress—for example, mechanical stress due to lysis, temperature stress due to heating or cooling a sample, or oxidative stress on a microscope slide, which could lead to potential artefacts including the induction of autophagy—even maintaining cells in higher than physiologically normal oxygen levels can be a stress condition [2107, 2108]. Special care should be taken with cells in suspension, as the stress resulting from mixing and/or centrifugation can induce autophagy. This point is not intended to limit the use of any specific methodology, but rather to note that there are no perfect assays. Therefore, it is important to verify that the positive (e.g., treatment with rapamycin, torin1 or other inducers) and negative (e.g., inhibitor treatment) controls behave as expected in any assays being utilized.
Similarly, plasmid transfection or nucleofection can result in the potent induction of autophagy (based on increases in LC3-II or degradation of SQSTM1), and certain transfection agents promote selective autophagy [429]. In some cell types, the amount of autophagy induced by transfection of a control empty vector may be so high that it is virtually impossible to examine the effect of enforced gene expression on autophagy (B. Levine, personal communication). It is thus advisable to perform time-course experiments to determine when the transfection effect returns to acceptably low levels and to use appropriate time-matched transfection controls (see also the discussion in GFP-Atg8-family protein fluorescence microscopy). This effect is generally not observed with siRNA transfection; however, it is an issue for plasmid expression constructs including those for shRNA and for viral delivery systems. The use of endotoxin-free DNA reduces, but does not eliminate, this problem. In many cells the cationic polymers used for DNA transfection, such as liposomes and polyplex, induce large tubulovesicular autophagosomes (TVAs) in the absence of DNA [2109]. These structures accumulate SQSTM1 and fuse slowly with lysosomes. In addition, these TVAs appear to reduce gene delivery, which increases 8-10 fold in cells that are unable to make TVAs due to the absence of ATG5.
Finally, the precise composition of media components and the density of cells in culture can have profound effects on basal autophagy levels and may need to be modified empirically depending on the cell lines being used. Along these lines various types of media, in particular those with different serum levels (ranging from 0-15%), may have profound effects with regard to how cells (or organs) perceive a fed versus starved state. For example, normal serum contains significant levels of cytokines and hormones that likely regulate the basal levels of autophagy and/or have an impact upon its modulation by additional stress or stimuli; thus, the use of dialyzed serum might be an alternative for these studies. In addition, the amino acid composition of the medium/assay buffer may have profound effects on initiation or progression of autophagy. For example, in the protist parasite Trypanosoma brucei starvation-induced autophagy can be prevented by addition of histidine to the incubation buffer [382]. For these reasons, the cell culture conditions should be fully described. It is also important to specify duration of autophagy stimulation, as long-term autophagy can modify signal transduction pathways of importance in cell survival [780].
D. Methods and challenges of specialized topics/model systems
There are now a large number of model systems being used to study autophagy. These guidelines cannot cover every detail, and as stated in the Introduction, this article is not meant to provide detailed protocols. Nonetheless, we think it is useful to briefly discuss what techniques can be used in these systems and to highlight some of the specific concerns and/or challenges. We also refer readers to the three volumes of Methods in Enzymology that provide additional information for “nonstandard” model systems [46–48].
1. Caenorhabditis elegans
C. elegans has a single ortholog of most yeast Atg proteins; however, two nematode homologs exist for Atg4, Atg8 and Atg16 [1477, 2110, 2111]. Multiple studies have established C. elegans as a useful multicellular genetic model to delineate the autophagy pathway and associated functions (see for example refs. [367, 965, 1109, 1112, 1276]). The LGG-1/Atg8 reporter is the most commonly used tool to detect autophagy in C. elegans. Similar to Atg8, which is incorporated into the double membrane of autophagic vacuoles during autophagy [204, 365, 914], the C. elegans LGG-1 localizes into cytoplasmic puncta under conditions known to induce autophagy. Fluorescent reporter fusions of LGG-1/Atg8 with GFP, DsRED or mCherry have been used to monitor autophagosome formation in vivo, in the nematode. These reporters can be expressed either in specific cells and tissues or throughout the animal [367, 1112, 2112, 2113]. Caution should be taken, however, when using protein markers fused to mCherry in worms. mCherry can accumulate in lysosomes [471] and might aggregate in autophagy-inducing conditions, such as fasting, even if not fused to LGG-1 or other autophagy markers (E. O’Rourke, personal communication); therefore, caution should be employed when using mCherry puncta as a readout to monitor autophagy in C. elegans. LGG-2 is the second LC3 homolog and is also a convenient marker for autophagy either using specific antibodies [1111] or fused to GFP [2114], especially when expressed from an integrated transgene to prevent its germline silencing [1111]. The exact function of LGG-1 versus LGG-2 remains to be addressed [1115].
For observing autophagy by GFP-LC3 fluorescence in C. elegans, it is best to use integrated versions of the marker [1111, 1112, 2115] (GFP::LGG-1 and GFP::LGG-2; Figure 33) rather than extrachromosomal transgenic strains [367, 2114] because the latter show variable expression among different animals or mosaic expression (C. Kang, personal communication; V. Galy, personal communication; [2116]). Integration of the markers requires mutagenesis, and care should be taken to outcross the strains to ensure that any remaining background mutations do not affect the examined phenotypes by, for example, examining the phenotypes in independent integrants. However, it is important to note that some integrated strains overexpress these chimeras because they are driven by heterologous promoters. One approach to overcome this problem is to monitor cleavage of a dual fluorescent protein marker consisting of tandem monomeric RFP (mRFP) joined by a flexible linker and attached to LGG-1 [2117]. Autophagic flux can be monitored as the ratio of free mRFP (mFP) to the uncleaved full-length protein (dFP) normalized to a loading control (i.e., actin or tubulin). However, this readout needs to be used with caution in the adult worm. Although relative mFP abundance is reported to change in L3-L4 larvae treated with RNAi against essential autophagy genes (i.e., bec-1) or CQ, and in 5-days starved L1 larvae [2117], no changes are observed in 6- to 12-h fasted adults, even when increased autophagic flux can be detected in aliquots of the same samples when using anti-LGG-1 antibodies (V.K. Mony, personal communication). Furthermore, the original studies characterizing this readout reported dFP-to-mFP cleavage in animals incubated for 18 h in a concentrated suspension of E. coli in M9 with or without CQ, an incubation condition that activates caloric restriction responses including autophagy (V. K. Mony, personal communication). Therefore, further validation of the dFP-mFP readout may be necessary to confidently use it in adult C. elegans.
Figure 33.
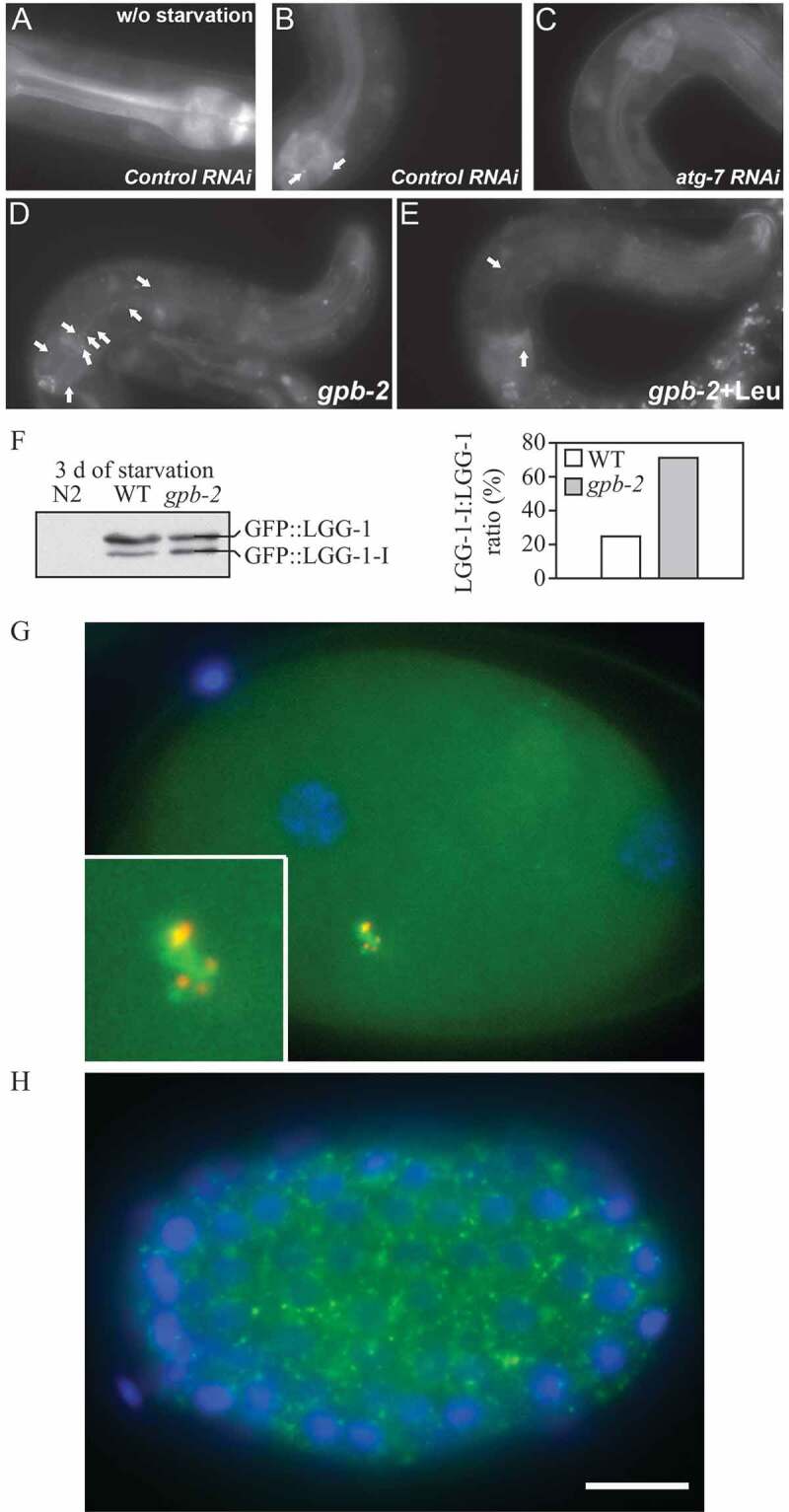
GFP::LGG-1 and GFP::LGG-2 are autophagy markers in C. elegans. (A-F) Animals were generated that carry an integrated transgene expressing a GFP-tagged version of lgg-1, the C. elegans ortholog of mammalian MAP1LC3. Representative green fluorescence images in the pharyngeal muscles of (A) control RNAi animals without starvation, (B) control RNAi animals after 9 d of starvation, (C) atg-7 RNAi animals after 9 d of starvation, (D) starvation-hypersensitive gpb-2 mutants without leucine after 3 d of starvation, and (E) gpb-2 mutants with leucine after 3 d of starvation. The arrows show representative GFP::LGG-1-positive punctate areas that label pre-autophagosomal and autophagosomal structures. (F) The relative levels of PE-conjugated and unconjugated GFP::LGG-1 were determined by western blotting. These figures were modified from data previously published in ref. [2115], Kang, C., Y.J. You, and L. Avery. 2007. Dual roles of autophagy in the survival of C. elegans during starvation. Genes & Development. 21:2161-2171, Copyright © 2007, Genes & Development by Cold Spring Harbor Laboratory Press, and ref. [4086], Kang, C., and L. Avery. 2009. Systemic regulation of starvation response in C. elegans. Genes & development. 23:12-17, Copyright © 2011, Genes & Development by Cold Spring Harbor Laboratory Press, www.genesdev.org. (G-H) GFP:LGG-2 serves as a marker for autophagosomes in early C. elegans embryos. (G) GFP::LGG-2 expressed in the germline from an integrated transgene reveals the formation of autophagosomes (green) around sperm-inherited membranous organelles (red). DNA of the two pronuclei is stained (blue). (H) Later during development, GFP::LGG-2-positive structures are present in all cells of the embryo. Scale bar: 10 µm. Images provided by V. Galy
To increase signal to noise, it is also possible to carry out indirect immunofluorescence microscopy using antibodies against endogenous LGG-1 [965, 1112], or LGG-2 [1111]; however, anti-LGG-1 and anti-LGG-2 antibodies are not commercially available. In addition, with the integrated version, or with antibodies directed against endogenous LGG-1, it is possible to perform a western blot analysis for lipidation, at least in embryos (LGG-1-I is the nonlipidated soluble form and LGG-1-II/LGG-1–PE is the lipidated form) [965, 1112, 2115]. In contrast to the yeast and mammalian autophagosomal membrane proteins Atg8 and LC3, lipidation of the C. elegans ortholog LGG-1 with phosphatidylethanolamine has rarely been investigated by western blotting; this is likely due to technical problems with separating the nonlipidated from the lipidated LGG-1 protein by gel electrophoresis. A new protocol for western blot analysis, taking advantage of improved antibodies to LGG-1 and SQST-1/SQSTM1, is applicable for both the detection of transgenic and endogenous proteins and provides a quantifiable method to assess autophagic flux [2118].
The LGG-1 precursor accumulates in the atg-4.1 mutant, but is undetectable in wild-type embryos [1261]. In fact, LGG-1 phenotypes vary in atg-4.1 and atg-4.2 mutants, indicative of distinct functions for these two genes [471]. Moreover, the banding pattern of LGG-1 or LGG-1 fused to fluorescent proteins in western blots may not be easy to interpret in larvae or the adult C. elegans because enrichment for a fast running band (the lipidated form) is not observed in some autophagy-inducing conditions including fasting (E.J. O’Rourke, personal communication). In the embryos of some autophagy mutants, including epg-3, epg-4, epg-5, and epg-6 mutants, levels of LGG-1-I and LGG-1-II are elevated [845, 965]. In an immunostaining assay, endogenous LGG-1 forms distinct punctate structures, mostly at the ~64- to 100-cell embryonic stage. LGG-1 puncta are absent in atg-3, atg-7, atg-5 and atg-10 mutant embryos [965], but dramatically accumulate in other autophagy mutants [845, 965]. The widely used GFP::LGG-1 reporter forms aggregates in atg-3 and atg-7 mutant embryos, in which endogenous LGG-1 puncta are absent, indicating that GFP::LGG-1 could be incorporated into protein aggregates during embryogenesis. Immunostaining for endogenous VPS-34 is also a useful marker of autophagy induction in C. elegans embryos [1271].
A variety of protein aggregates, including PGL granules (PGL-1-PGL-3-SEPA-1) and the C. elegans SQSTM1 homolog SQST-1, are selectively degraded by autophagy during embryogenesis; impaired autophagy activity results in their accumulation and the generation of numerous aggregates [965]. Thus, degradation of these autophagy substrates can also be used to monitor autophagy activity, with similar cautionary notes to those described in section A3 (see SQSTM1 and related LC3 binding protein turnover assays) for the SQST-1 turnover assay. Similar to mammalian cells, the total amount of GFP::LGG-1 along with SQST-1::GFP transcriptional expression coupled with its posttranscriptional accumulation can be informative with regard to autophagic flux in the embryo (again with the same cautionary notes described in section A3) [573].
As with its mammalian counterpart, loss of the C. elegans TP53 ortholog, cep-1, increases autophagosome accumulation [2119] and extends the animal’s life span [2120]. bec-1- and cep-1-regulated autophagy is also required for optimal life-span extension. However, non-autophagic roles for bec-1 and cep-1 have been reported. Hence, bec-1 or cep-1 inactivation is insufficient to define a longevity mechanism as autophagy dependent. The TFEB ortholog HLH-30 transcriptionally regulates autophagy including the expression of bec-1 [948, 1294], and life-span analyses uncovered an anti-aging role for HLH-30/TFEB in C. elegans, and possibly in mammals [573, 948, 1294]. However, it remains to be definitively demonstrated whether HLH-30/TFEB longevity is exclusively, or even mostly, mediated by activation of autophagy. bec-1- and cep-1 are also required to reduce lipid accumulation in response to silencing FRH-1/FXN, a protein involved in mitochondrial respiratory chain functionality [2121]. FRH-1 silencing also induces mitophagy in an evolutionarily conserved manner [1172]. Moreover, the products of C. elegans mitophagy regulatory gene homologs (PDR-1/PRKN, PINK-1/PINK1, DCT-1/BNIP3, and SQST-1/SQSTM1) are required for induction of mitophagy (monitored through the Rosella biosensor [2122]) and life-span extension following FRH-1 silencing and iron deprivation [1172]. HLH-30/TFEB transcriptionally regulates autophagy and promotes lipid degradation [948, 1294], and life-span analyses uncovered a direct role for HLH-30/TFEB in aging in C. elegans, and possibly in mammals [573, 948, 1294].
C. elegans body wall muscle is a useful tissue for studying autophagy. In addition to the methods discussed above, in this tissue transgenic reporter proteins can be used to monitor rates of protein degradation in specific subcellular compartments [2123], mutations in at least two signaling pathways can be used to modulate autophagy [2124], drugs can be used to inhibit autophagy [2125], and mutations and drugs can be used to inhibit the proteasome [2126], calpains [2125], and caspases [2127] thus allowing both positive and negative controls. Finally, knockdown of a substantial number of kinases [2128] and phosphatases [2129] appears to induce autophagy, potentially enabling further study of the upstream signals that modulate this process.
For a more complete review of methods for monitoring autophagy in C. elegans see ref. [562]. These approaches can be used to monitor autophagy in embryos, early larval stages, and adult C. elegans, including during aging [2130].
2. Chicken B-lymphoid DT40 cells, retina and inner ear
The chicken B-lymphoid DT40 cell line represents a suitable tool for the analysis of autophagic processes in a nonmammalian vertebrate system. In DT40 cells, foreign DNA integrates with a very high frequency by homologous recombination compared to random integration. This feature was—prior to the CRISPR-Cas9 era—employed in order to generate cellular gene knockouts. Different Atg-deficient DT40 cell lines already exist, including atg13−/−, ulk1−/−, ulk2−/−, and ulk1−/− ulk2−/− [693]. Many additional non-autophagy-related gene knockout DT40 cell lines have been generated and are commercially available [2131].
DT40 cells mount an autophagic response upon starvation in EBSS [693], and autophagy can be analyzed by a variety of assays in this cell line. Steady state methods that can be used include TEM, LC3 western blotting and fluorescence microscopy; flux measurements include monitoring LC3-II turnover and tandem mRFP/mCherry-GFP-LC3 fluorescence microscopy. Using atg13−/− and ulk1−/− ulk2−/− DT40 cells, it was shown that ATG13 and its binding capacity for RB1CC1 are mandatory for both basal and starvation-induced autophagy, whereas ULK1/2 and in vitro-mapped ULK1-dependent phosphorylation sites of ATG13 appear to be dispensable for these processes [693].
Another useful system is chick retina, which can be used for monitoring autophagy at different stages of development. For example, lipidation of LC3 is observed during starvation, and can be blocked with a short-term incubation with 3-MA [1596, 2132]. LEP-100 antibody is commercially available for the detection of this lysosomal protein. In the developing chicken inner ear, LC3 flux can be detected in otic vesicles cultured in a serum-free medium exposed to either 3-MA or CQ [2133].
One of the salient features of chicken cells, including primary cells such as chicken embryo fibroblasts, is the capacity of obtaining rapid, efficient and sustained transcript/protein downregulation with replication-competent retrovirus for shRNA expression [2134]. In chicken embryo fibroblasts, nearly complete and general (i.e., in nearly all cells) protein downregulation can be observed within a few days after transfection of the shRNA retroviral vector [231].
Cautionary notes: It is possible that there is some divergence within the signaling pathways between mammalian and nonmammalian model systems. One example might be the role of ULK1/2 in starvation-induced autophagy described above. Additionally, DT40 cells represent a transformed cell line, being derived from an avian leukosis virus-induced bursal lymphoma. Thus, DT40 cells release avian leukosis virus into the medium, and the 3ʹ-long terminal repeat has integrated upstream of the MYC gene, leading to increased MYC expression [2135]. Both circumstances might influence basal and starvation-induced autophagy.
3. Chlamydomonas
The unicellular green alga Chlamydomonas reinhardtii is an excellent model system to investigate autophagy in photosynthetic eukaryotes. Most of the ATG genes that constitute the autophagy core machinery including the ATG8 and ATG12 ubiquitin-like systems are conserved as single-copy genes in the nuclear genome of this model alga. Autophagy can be monitored in Chlamydomonas by western blotting through the detection of Atg8 lipidation as well as an increase in the abundance of this protein in response to autophagy activation [397]. Localization of Atg8 by immunofluorescence microscopy can also be used to study autophagy in Chlamydomonas because the cellular distribution of this protein changes drastically upon autophagy induction. The Atg8 signal is weak and usually detected as a single spot in nonstressed cells, whereas autophagy activation results in the localization of Atg8 in multiple spots with a very intense signal [397, 2136, 2137]. A red fluorescent protein (mCherry) tagged-Atg8 has been developed that allows the observation of autophagosomes in living microalgal cells [2138]. Autophagic flux can also be monitored in Chlamydomonas by analyzing the abundance and lipidation of Atg8 protein in cells treated with the vacuolar-type ATPase inhibitor concanamycin A. Inhibition of autophagic flux results in the accumulation of total Atg8 and detection of the Atg8 lipidated form [2139]. Finally, enhanced expression of ATG8 and other ATG genes has also been reported in stressed Chlamydomonas cells [2136, 2140]. These methodological approaches have been used to investigate the activation of autophagy in Chlamydomonas under different stress conditions including nutrient (nitrogen or carbon) limitation, rapamycin treatment, ER stress, oxidative stress, photo-oxidative damage or high light stress [397, 2136, 2137].
4. Drosophila melanogaster
Drosophila provides an excellent and highly amenable system for in vivo analysis of autophagy as the machinery is highly conserved with well-characterized functions in several tissues including oocyte, embryo, larval/pupal fat body, midgut, salivary gland and imaginal disc, larval motor neurons and adult neurons [183, 1744, 2141–2144]. The advantage of using Drosophila as a model is the ability to undertake genetic analysis of individual components of the autophagy machinery [1771, 2141]. Another major advantage of Drosophila is that the problem of animal-to-animal variability can be circumvented by the use of clonal mutant cell analysis [183, 2141, 2143, 2145]. In this scenario, somatic clones of cells are induced that either overexpress the gene of interest, or silence the gene through expression of a transgenic RNA interference construct, or gene mutation/deletion. These gain- or loss-of-function clones are surrounded by wild-type cells, which serve as an internal control for autophagy induction. In such an analysis, autophagy in these genetically distinct cells is always compared to neighboring cells of the same tissue, thus eliminating most of the variability and also ruling out potential non-cell-autonomous effects that may arise in mutant animals (Figure 25). Along these lines, clonal analysis should be an integral part of in vivo Drosophila studies when possible.
Multiple steps of the autophagic pathway can be monitored in Drosophila due to the development of useful markers, corresponding to every step of the process. Interested readers may find further information in several reviews with a detailed discussion of the currently available assays and reagents for the study of autophagy in Drosophila [183, 2141, 2146]. For example, the level of autophagy can be examined live in vivo using transgenic lines that express fluorescently-tagged specific components of the autophagy pathway. Moreover, fluorescent reporters for components of the autophagy pathway can be used in genetic screens for new regulators of autophagy [2147]. The expression of fluorescently tagged Atg8a from the endogenous Atg8a promoter is a useful reporter, that does not require a driver line [375, 2148]. In addition, autophagy has been successfully monitored in Drosophila expressing various components of the pathway including (but not limited to) human UAS-GFP-LC3 [123, 376, 2149], UAS-GFP-Atg8a [2150], UAS-mCherry-Atg8a [589], mCherry-Atg18 [2151], UAS-GFP-Atg5 [376], UAS-RFP-Atg5 [947], UAS-GFP-Atg6 [2152], UAS-GFP-DFCP1 [2153], UAS-GFP-ref(2)P (corresponding to the Drosophila SQSTM1 homolog) [589], and the tandem fluorescent reporter UASp-GFP-mCherry-Atg8a [2154, 2155], with an increasing list of addition transgenic fluorescence reagents, including protein traps, that are being made available through the Drosophila stock centers.
There are also a limited number of commercially available antibodies, including a rabbit monoclonal anti-GABARAP antibody and a rabbit polyclonal anti-ref(2)P antibody that can be used to detect endogenous levels of Drosophila Atg8a and ref(2)P, respectively, in both immunostaining and immunoblotting experiments [560, 2141, 2156, 2157]. The advantage of UAS-ref(2)P-GFP over the antibody against endogenous ref(2)P is that its accumulation is independent of ref(2)P promoter regulation and unambiguously reflects autophagy impairment [557, 2155]. Of note, immunoblot analysis of ref(2)P levels should include both soluble and insoluble fractions [557, 2157]. Several laboratories have also generated antibodies, including those against Atg8a and ref(2)P [560, 2149, 2158]. Finally, it is worth noting that a commercial Atg5 antibody can also be used for Drosophila [750, 2159].
Cultured Drosophila (S2) cells can also be stably transfected with GFP fused to Drosophila Atg8a, which generates easily resolvable GFP-Atg8a and GFP-Atg8a–PE forms that respond to autophagic stimuli (S. Wilkinson, personal communication); stable S2 cells with GFP-Atg8a under the control of a 2-kb Atg8a 5ʹ UTR are also available [2160]. Similarly, cultured Drosophila cells (l[2]mbn or S2) stably transfected with EGFP-HsLC3B respond to autophagy stimuli (nutrient deprivation) and inhibitors (3-MA, bafilomycin A1) as expected, and can be used to quantify GFP-LC3 puncta, which works best using fixed cells with the aid of an anti-GFP antibody [2161].
The selective degradation of cargo can also be used to assay for autophagy in this system. The Drosophila components of the IKK complex, key/kenny and IKKβ/ird5, are selectively degraded by autophagy, and transgenic lines are available (UAS-GFP-key/kenny, UAS-mCherry-IKKβ/ird5 and UAS-mCherry-GFP-IKKβ/ird5) to follow key and ird5 expression and localization [2162].
With the distinct morphology of autophagy, TEM is also an indispensable and reliable method for monitoring autophagy in Drosophila. Finally, in addition to genetic analysis, pharmacological modulation of autophagy can be examined in Drosophila. For example, rapamycin can be fed to larvae or adults to induce autophagy, and CQ can be used to block lysosomal degradation [589, 1770, 2141].
Cautionary notes: In the Drosophila eye, overexpression of GFP-Atg8 results in a significant increase in Atg8–PE based on immunoblot, and this occurs even in control flies in which punctate GFP-Atg8 is not detected by immunofluorescence (M. Fanto, personal communication/unpublished results), and in transfected Drosophila Kc167 cells, uninducible but persistent GFP-Atg8 puncta are detected (A. Kiger, personal communication/unpublished results). In contrast, expression of GFP-LC3 under the control of the ninaE/rh1 promoter in wild-type flies does not result in the formation of LC3-II detectable by immunoblot, nor the formation of punctate staining; however, increased GFP-LC3 puncta by immunofluorescence or LC3-II by immunoblot are observed upon activation of autophagy [616]. Finally, most Drosophila food contains the anti-fungal nipagin (methylparaben), which has certain redox and anti-oxidant effects; these could interfere with particular experiments.
5. Erythroid cells
The unique morphology of red blood cells (RBCs) is instrumental to their function. The bi-concave shape provided by a highly flexible membrane and the absence of organelles is critical to their long lifespan in the peripheral circulation (120 days), allowing unimpeded circulation of the RBC even through the thinnest blood vessels, thereby delivering O2 to all the tissues of the body. Erythroid cells acquire this unique morphology upon terminal erythroid maturation, which commences in the bone marrow with the release of reticulocytes that become mature RBCs in the peripheral circulation. This process involves extrusion of the pyknotic nucleus through a specialized form of asymmetric division, and degradation of the ribosome and mitochondria machinery along with a reduction in cell volume via a specialized form of autophagy (Figure 34). In the context of RBC biogenesis, autophagy exerts a unique function to sculpt the cytoplasm, with the mature autophagic vacuoles engulfing and degrading organelles, such as mitochondria and ribosomes, whose presence would impair the flexibility of the cells.
Figure 34.
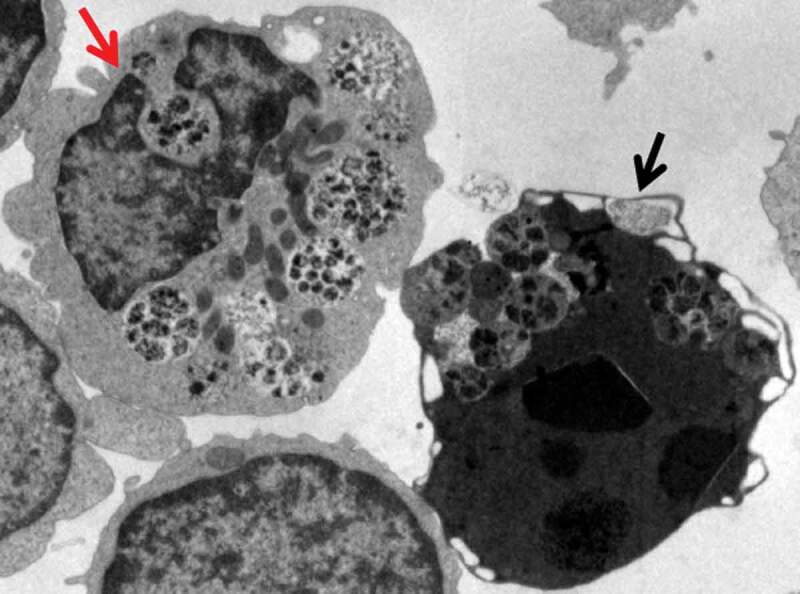
Transmission electron micrograph of erythroblasts obtained from the blood of regular donors after ten days of culture in the presence of KITLG/SCF, IL3, EPO and dexamethasone. Original magnification 3000X. This figure shows two erythroblasts containing autophagic vacuoles. One erythroblast (red arrow) has the morphology of a live cell with several autophagic vacuoles that have engulfed cytoplasmic organelles. The other erythroblast (black arrow) has the electron-dense cytoplasm characteristic of a dead cell and is in the process of shedding its autolysosomes from the cytoplasm to the extracellular space. Image provided by A.R. Migliaccio and M. Zingariello
Another unique feature of erythropoiesis is that expression of genes required for autophagosome assembly/function, such as LC3B, does not appear to be regulated by nutrient deprivation, but rather is upregulated by the erythroid-specific transcription factor GATA1 [981]. FOXO3, a transcription factor that modulates RBC production based on the levels of O2 present in the tissues [2163], amplifies GATA1-mediated activation of autophagy genes [981] and additional genes required for erythroid maturation [2164]. Furthermore, lipidation of the cytosolic form of LC3B into the lipidated LC3-II form is controlled by EPO (erythropoietin), the erythroid-specific growth factor that ensures survival of the maturing erythroid cells. The fact that the genes encoding the autophagic machinery are controlled by the same factors that regulate expression of genes encoding important red cell constituents (such as red blood cell antigens and cytoskeletal components, globin, and proteins mediating heme biosynthesis) [2165–2167], ensures that the process of terminal maturation progresses in a highly ordered fashion.
The importance of autophagy for RBC production has been established through the use of mutant mouse strains lacking genes encoding proteins of the autophagy machinery (BNIP3L, ULK1, ATG7) [2168–2171]. These mutant mice exhibit ineffective erythropoiesis with erythroid cells blocked at various stages of terminal erythroid maturation and anemia. Abnormalities of the autophagic machinery are also linked to erythroid disorders such as Diamond-Blackfan anemia or myelodysplastic syndrome, which are characterized by either congenic or acquired loss-of-function mutations of genes encoding ribosomal proteins (ribosomopathies), and involve erythroid progenitors. As in other cell types, in erythroid cells TP53 activation may influence the functional consequences of autophagy—to determine cell death rather than maturation. TP53, through MDM2, is the gatekeeper to ensure normal ribosome biosynthesis by inducing death of cells lacking sufficient levels of ribosomal proteins. In these disorders, activated TP53 and abnormally high levels of autophagic death of erythroid progenitors, promote anemia and bone marrow failure. Glucocorticoids might improve anemia in some Diamond-Blackfan anemia patients by inhibiting TP53 activity. Of note, impairment of autophagy in the late phase of erythropoiesis, involving erythroid precursors, has been reported in hereditary red cell disorders such as β-thalassemic syndromes, characterized by ineffective erythropoiesis [2172, 2173], or in chorea-acanthocytosis, a neurodegenerative disorder linked to VPS13 mutations and characterized by circulating acanthocytes containing multivesicular bodies and double-membrane remnants [2174]. Recent evidence also links the abnormal regulation of the redox sensitive transcription factor NFE2L2 to ineffective erythropoiesis and impairment of autophagy, with accumulation in erythroid precursors of non-functional proteins, further amplifying oxidation and promoting cell apoptosis [2175].
6. Filamentous fungi
As in yeast, autophagy is involved in nutrient recycling during starvation [369, 373, 2176–2182]. In addition, autophagy seems to be involved in many normal developmental processes such as sexual and asexual reproduction, where there is a need for reallocation of nutrients from one part of the mycelium to another to supply the developing spores and spore-bearing structures [369, 1086, 2176, 2177, 2179, 2183–2186]. Similarly, autophagy also affects conidial germination under nitrogen-limiting conditions [369]. In Podospora anserina, autophagy has been studied in relation to incompatibility reactions between mating strains where it seems to play a prosurvival role [372, 2183]. During aging of this long-standing aging model, autophagy is increased (based on numbers of GFP-Atg8 puncta and increased autophagy-dependent degradation of a GFP reporter protein) and acts as a prosurvival pathway [2187]. Moreover, mitophagy has been demonstrated to exert pro-survival effects under mild stress conditions, while displaying Atg1-dependent pro-death features under elevated stress.
In Sordaria macrospora, the pexophagy receptor Nbr1 is involved in fruiting-body development and maturation of sexual ascospores [1052]. Of special interest to many researchers of autophagy in filamentous fungi has been the possible involvement of autophagy in plant and insect pathogen infection and growth inside the host [373, 1069, 1078, 2176, 2177, 2188–2193]. For example, treatment with amiodarone promotes movement of the blast fungus M. oryzae between living rice cells during the early biotrophic stage of infection when the fungus inhabits epidermal cells but before symptoms develop, whereas inhibiting autophagy with 3-MA attenuates cell-to-cell movement and disrupts the biotrophic interface between the fungus and living host rice cells. In conjunction with the analysis of a mutant strain impaired in autophagy induction, these results suggest a fundamental role for autophagy in mediating intracellular host-microbe interactions [2193]. Autophagy also appears to be necessary for the development of aerial hyphae [369, 2176, 2182, 2183, 2189], and for appresorium function in M. oryzae, Colletotrichum orbiculare and Metarhizium robertsii [373, 2188, 2189, 2191, 2194]. In particular, invasion-associated ER stress can promote autophagy to enhance the cell wall integrity-associated MAPK pathway in order to help with infection by M. oryzae [2195]. Some of these effects could be caused by the absence of autophagic processing of storage lipids (lipophagy) to generate glycerol for increasing turgor and recycling the contents of spores into the incipient appressorium, as a prerequisite to infection [2176, 2189, 2190].
Methods for functional analysis of autophagy have been covered in a review article (see ref. [2196]). Most studies on autophagy in filamentous fungi have involved deleting some of the key genes necessary for autophagy, followed by an investigation of what effects this has on the biology of the fungus. Most commonly, ATG1, ATG4, ATG8 and/or ATG9 have been deleted [373, 1843, 2176, 2177, 2179, 2180, 2183, 2186, 2189, 2191, 2197, 2198]. To confirm that the deletion(s) affects autophagy, the formation of autophagic bodies in the wild type and the mutant can be compared. In filamentous fungi the presence of autophagic bodies can be detected using MDC staining [373, 2176], TEM [373, 2177] or fluorescence microscopy to monitor Atg8 tagged with a fluorescent protein [369, 2179, 2183, 2186]. This type of analysis is most effective after increasing the number of autophagic bodies by starvation or alternatively by adding the autophagy-inducing drug rapamycin [369, 2176], in combination with decreasing the degradation of the autophagic bodies through the use of the protease inhibitor PMSF [373, 2177, 2179, 2183]. In filamentous fungi it might also be possible to detect the accumulation of autophagic bodies in the vacuoles using differential interference contrast microscopy, especially following PMSF treatment [2179, 2183]. Additional information regarding the timing of autophagy induction can be gained by monitoring transcript accumulation of ATG1 and/or ATG8 using qPCR [2177].
Autophagy has been investigated intensively in Aspergilli, and in particular in the genetically amenable species Aspergillus nidulans, which is well suited to investigate intracellular traffic [1843, 2199, 2200]. In A. oryzae, autophagy has been monitored by the rapamycin-induced and Atg8-dependent delivery of DsRed2, which is normally cytosolic, to the vacuoles [369]. In A. nidulans, the more “canonical” GFP-Atg8 proteolysis assays have been used, by monitoring the delivery of GFP-Atg8 to the vacuole (by time-lapse microscopy), and by directly following the biogenesis of GFP-Atg8-labeled phagophores and autophagosomes [1843], which can be tracked in large numbers using kymographs traced across the hyphal axis. In these kymographs, the autophagosome cycle starting from a PAS “draws” a cone whose apex and base correspond to the “parental” PAS punctum and to the diameter of the “final” autophagosome, respectively [348]. Genetic analyses revealed that autophagosomes normally fuse with the vacuole in a Rab7-dependent manner. However, should Rab7 fusogenic activity be mutationally inactivated, autophagosomes can traffic to the endosomes in a RabB/Rab5- and CORVET-dependent manner [348]. An important finding was that RabO/Rab1 plays a key role in A. nidulans autophagy (and actually can be observed on the phagophore membranes). This finding agrees with previous work in S. cerevisiae demonstrating that Ypt1 (the homolog of RAB1) is activated by the Trs85-containing version of TRAPP, TRAPPIII, for autophagy [2201, 2202]. This crucial involvement of RabO/Ypt1 points at the ER as one source of membrane for autophagosomes.
In A. nidulans, specific misfolded transporters, which are retained in the ER, are degraded by chaperone-assisted selective autophagy. The chaperone involved was identified as BsdA, which is an ER transmembrane protein acting as an adaptor for the recruitment of the HECT-type ubiquitin ligase HulA (NEDD4/Rsp5 type), which ubiquitinates the misfolded transporter and elicits its recognition by maturing autophagosomes. The process involves Atg8 and Atg9. Epifluorescence microscopy has shown that the misfolded transporter tagged with GFP colocalizes with Atg8-RFP and vacuoles stained with CMAC, revealing a direct translocation from the ER to the vacuole via autophagosomes. Knockout of the gene encoding the BsdA chaperone allows the misfolded transporter to escape autophagy and be sorted to the plasma membrane. Distinct homologs of BsdA might be present in metazoa. Based on the present guidelines the Aspergillus example classifies as CASA rather than CMA.
The suitability of A. nidulans for in vivo microscopy has been exploited to demonstrate that nascent phagophores are cradled by ER-associated structures resembling mammalian omegasomes [348]. The autophagic degradation of whole nuclei that has been observed in A. oryzae [1082] might be considered as a specialized version of reticulophagy. Finally, autophagosome biogenesis has also been observed using a PtdIns3P-binding GFP-tagged FYVE domain probe in mutant cells lacking RabB/Rab5. Under these genetic conditions Vps34 cannot be recruited to endosomes and is entirely at the disposition of autophagy [348], such that PtdIns3P is only present in autophagic membranes.
Mitophagy has been studied in M. oryzae, by detecting the endogenous level of porin (a mitochondrial outer membrane protein) by western blot, and by microscopy observation of vacuolar accumulation of mito-GFP [1069]. Mitophagy is involved in regulating the dynamics of mitochondrial morphology and/or mitochondrial quality control, during asexual development and invasive growth in M. oryzae. Pexophagy has also been studied in rice-blast fungus and it serves no obvious biological function, but is naturally induced during appressorial development, likely for clearance of excessive peroxisomes prior to cell death [2203]. In turn, normal mitochondrial and peroxisomal fission is also essential for mitophagy and pexophagy [2204]. Methods to monitor pexophagy in M. oryzae include microscopy observation of the vacuolar accumulation of GFP-SRL (peroxisome-localized GFP), and detection of the endogenous thiolase [2203], or Pex14 levels.
The existence of crosstalk between autophagy and endocytosis has been explored in M. oryzae, by analyzing the biological functions of Vps9-domain containing proteins [2205, 2206]. Pyricularia oryzae (the asexual stage of, and hence essentially a synonym of, M. oryzae) Vps9 recruits PoVps34 and targets it to endosomes by activating PoVps21; PoAtg6 is then recruited by PoVps34 under the action of PoVps38 to target endosomes in endocytosis. Additionally, PoAtg6 is recruited to the PAS by PoVps34 to participate in autophagy by activating PoVps21. Methods to monitor the crosstalk include microscopy observation of the endosomes and autophagosomes, affinity isolation and co-IP.
7. Food biotechnology
Required for yeast cell survival under a variety of stress conditions, autophagy has the potential to contribute to the outcome of many food fermentation processes. For example, autophagy induction is observed during the primary fermentation of synthetic grape must [2207] and during sparkling wine production (secondary fermentation) [2208]. A number of genome-wide studies have identified vacuolar functions and autophagy as relevant processes during primary wine fermentation or for ethanol tolerance, based on gene expression data or cell viability of knockout yeast strains [2207, 2209–2213]. However, determining the relevance of autophagy to yeast-driven food fermentation processes requires experimentation using some of the methods available for S. cerevisiae as described in these guidelines.
Autophagy is a target for some widespread food preservatives used to prevent yeast-dependent spoilage. For example, the effect of benzoic acid is exacerbated when concurrent with nitrogen starvation [2214]. This observation opened the way to devise strategies to improve the usefulness of sorbic and benzoic acid, taking advantage of their combination with stress conditions that would require functional autophagy for yeast cell survival [2006]. Practical application of these findings would also require extending this research to other relevant food spoilage yeast species, which would be of obvious practical interest.
In the food/health interface, the effect of some food bioactive compounds on autophagy in different human cell types has already attracted some attention [2215, 2216]. Interpreting the results of this type of research, however, warrants two cautionary notes [2217]. First, the relationship between health status and autophagic activity is obviously far from being direct. Second, experimental design in this field must take into account the actual levels of these molecules in the target organs after ingestion, as well as exposure time and their transformations in the human body. In addition, attention must be paid to the fact that several mechanisms might contribute to the observed biological effects. Thus, relevant conclusions about the actual involvement of autophagy on the health-related effect of food bioactive compounds would only be possible by assaying the correct molecules in the appropriate concentrations.
8. Honeybee
The reproductive system of bees, or insects whose ovaries exhibit a meroistic polytrophic developmental cycle can be a useful tool to analyze and monitor physiological autophagy. Both queen and worker ovaries of Africanized A. mellifera display time-regulated features of cell death that are linked to external stimuli [2218]. Features of apoptosis and autophagy are frequently associated with the degeneration process in bee organs, but only more recently has the role of autophagy been highlighted in degenerating bee tissues. The primary method currently being used to monitor autophagy is to follow the formation of autophagosomes and autolysosomes by TEM. This technique can be combined with cytochemical and immunohistochemical detection of acid phosphatase as a marker for autolysosomes [2219, 2220]. Acidotropic dyes can also be used to follow autophagy in bee organs, as long as the cautions noted in this article are followed. The honeybee genome has been sequenced, and differential gene expression has been used to monitor Atg18 in bees parasitized by Varroa destructor [2221].
9. Human
Considering that much of the research conducted today is directed at understanding the functioning of the human body, in both normal and disease states, it is pertinent to include humans and primary human tissues and cells as important models for the investigation of autophagy. Although clinical studies are not readily amenable to these types of analyses, it should be kept in mind that the MTORC1 inhibitor rapamycin, the lysosomal inhibitors CQ and HCQ, and the microtubule depolymerizing agent colchicine are all available as clinically approved drugs. However, these drugs are not highly selective, having numerous off-targets, and have serious side effects, which often impede their clinical use to study autophagy (e.g., severe immunosuppressive effects of rapamycin; gastrointestinal complaints, bone marrow depression, neuropathy and rhabdomyolysis induced by colchicine; gastrointestinal complaints, neuropathy and convulsions, retinopathy and heart disease induced by HCQ). These side effects may in part be exacerbated by potential inhibition of autophagy tself by these drugs [2222]. In cancer treatment, for example, autophagy-inhibiting drugs are used in combination with other anticancer drugs to increase their potency. Conversely, normal tissues such as kidney induce autophagy in response to anticancer drugs to resist against their toxicity [2223]; additional blockade of autophagy could worsen normal tissue toxicity and cause serious side effects. Therefore, the potential for serious adverse effects and toxicity of these drugs warrants caution, especially when studying a role of autophagy in high-risk patients, such as the critically ill.
Fortunately, it is possible to obtain fresh biopsies of some human tissues. Blood, in particular, as well as samples of adipose and muscle tissues, can be obtained from needle biopsies or from elective surgery. For example, in a large study, adipocytes were isolated from pieces of adipose tissue (obtained during surgery) and examined for INS (insulin) signaling and autophagy. It was demonstrated that autophagy was strongly upregulated (based on LC3 flux, EM, and lipofuscin degradation) in adipocytes obtained from obese patients with type 2 diabetes compared with nondiabetic subjects [396]. In another study utilizing human adipose tissue biopsies and explants, elevated autophagic flux in obesity was associated with increased expression of several autophagy genes [292, 927, 2224]. Conversely, by using fibroblasts from a patient with X-linked myopathy with excessive autophagy, it was shown that deficiency of VMA21 blocks vacuolar ATPase assembly and causes autophagic vacuolar myopathy due to increased pH of lysosomes, reduced lysosomal protein degradation and enhanced macroautophagy [2225].
The study of autophagy in the blood has revealed that SNCA may represent a further marker to evaluate the autophagy level in T lymphocytes isolated from peripheral blood [2226]. In these cells it has been shown that (a) knocking down the SNCA gene results in increased autophagy, (b) autophagy induction by energy deprivation is associated with a significant decrease of SNCA levels, (c) autophagy inhibition (e.g., with 3-MA or knocking down ATG5) leads to a significant increase of SNCA levels, and d) SNCA levels negatively correlate with LC3-II levels. Thus, SNCA, and in particular the 14-kDa monomeric form, can be detected by western blot as a useful tool for the evaluation of autophagy in primary T lymphocytes. In contrast, the analysis of SQSTM1 or NBR1 in freshly isolated T lymphocytes fails to reveal any correlation with either LC3-II or SNCA, suggesting that these markers cannot be used to evaluate basal autophagy in these primary cells. Conversely, LC3-II upregulation is correlated with SQSTM1 degradation in neutrophils, as demonstrated in a human sepsis model [1694].
A major caveat of the work concerning autophagy in human tissue is the problem of tissue heterogeneity, postmortem times, agonal state, genetic heterogeneity, premortem clinical history (medication, diet, etc.) and tissue fixation. Time to fixation is typically longer in autopsy material than when biopsies are obtained. For tumors, careful sampling to avoid necrosis, hemorrhagic areas and non-neoplastic tissue is required. The problem of fixation is that it can diminish the antibody binding capability; in addition, especially in autopsies, material is not obtained immediately after death [2227, 2228]. The possibilities of postmortem autolysis and fixation artefacts must always be taken into consideration when interpreting changes attributed to autophagy [2229]. Analyses of these types of samples require not only special antigen retrieval techniques, but also histopathological experience to interpret autophagy studies by IHC, immunofluorescence or TEM. Nonetheless, at least one recent study demonstrated that LC3 and SQSTM1 accumulation can be readily detected in autopsy-derived cardiac tissue from patients with CQ- and HCQ-induced autophagic vacuolar cardiomyopathy [1559]. Despite significant postmortem intervals, sections of a few millimeters thickness cut from fresh autopsy brain and fixed in appropriate glutaraldehyde-formalin fixative for EM, can yield TEM images of sufficient ultrastructural morphology to discriminate different autophagic vacuole subtypes and their relative regional abundance in some cases (R. Nixon, personal communication).
The situation is even worse with TEM, where postmortem delays can cause vacuolization. Researchers experienced in the analysis of TEM images corresponding to autophagy should be able to identify these potential artefacts because autophagic vacuoles should contain cytoplasm. While brain biopsies may be usable for high quality TEM (Figures 35, 36), this depends upon proper handling at the intraoperative consultation stage, and such biopsies are performed infrequently except for brain tumor diagnostic studies. Conversely, biopsies of organs such as the digestive tract, the liver, muscle and the skin are routinely performed and thus nearly always yield high-quality TEM images. When possible, nonsurgical biopsies are preferable because surgery is usually performed in anesthetized and fasting patients, two conditions possibly affecting autophagy. Moreover, certain surgical procedures require tissue ischemia-reperfusion strategies that can also affect autophagy level [2230]. An analysis that examined liver and skeletal muscle from critically ill patients utilized tissue biopsies that were taken within 30 ± 20 min after death and were flash-frozen in liquid nitrogen followed by storage at -80°C [2231]. Samples could subsequently be used for EM and western blot analysis.
Figure 35.
A large dystrophic neurite from a brain biopsy of a patient with Gerstmann-Sträussler-Scheinker disease not unlike those reported for AD [79]. This structure is filled with innumerable autophagic vacuoles, some of which are covered by a double membrane. Electron dense lysosomal-like structures are also visible. The red arrow points to a double-membrane autophagic vacuole. Scale bar: 200 nm. Image provided by P. Liberski
Figure 36.
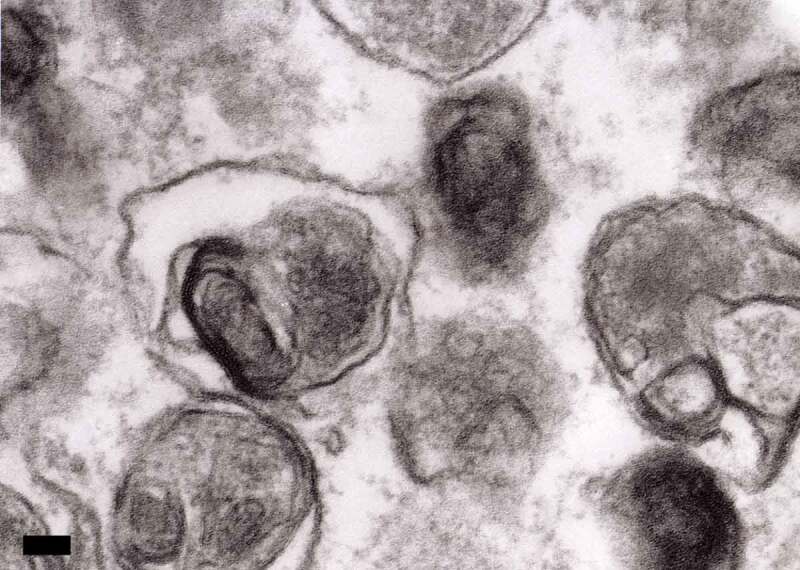
A high-power electron micrograph from a brain biopsy showing autophagic vacuoles in a case of ganglioglioma. Scale bar: 200 nm. Image provided by P. Liberski
A major limitation of studying patient biopsies is that only static measurements can be performed. This limitation does not apply, however, for dynamic experiments on tissue biopsies or cells derived from biopsies, as described above [396]. Multiple measurements over time, especially when deep (vital) organs are involved, are impossible and ethically not justifiable. Hence, quantitative flux measurements are virtually impossible in patients. To overcome these problems to the extent possible and to gain a more robust picture of the autophagic status, observational studies need to include two different aspects. First, a static marker for phagophore or autophagosome formation needs to be measured. This can be done by assessing ultrastructural changes with TEM and/or on the molecular level by measuring LC3-II protein levels. Second, accumulation of autophagy substrates, such as SQSTM1 and (poly)ubiquitinated proteins, can provide information on the overall efficacy of the pathway and can be a surrogate marker of the consequences of altered autophagic flux, especially when autophagy is insufficient, although these changes can also be affected by the ubiquitin-proteasome system as mentioned above.
In addition, and even more so when problems with selective pathways are suspected (e.g., mitophagy), specific substrates of these pathways should be determined. Again, none of these measurements on its own provides enough information on (the efficacy of) autophagy, because other processes may confound every single parameter. However, the combination of multiple analyses should be informative. Of note, there has been interest in assessing markers of autophagy and autophagic flux in right atrial biopsy samples obtained from patients undergoing cardiac surgery [2232, 2233]. Evidence to date suggests that cardiac surgery may be associated with an increase in autophagic flux, and that this response may protect the heart from perioperative cardiac ischemia-reperfusion injury [2232]. The autophagy deficiency also correlates with the decline of serum testosterone in some hypogonadism patients, as the LC3 expression and puncta number per square micrometer are significantly decreased in the Leydig cells from the patients compared with those of the control group [2234]. In the brain of hypoxic-ischemic encephalopathy human neonates, punctate LC3 labelling combined with increased number and size in CTSD- and LAMP1-positive dots (presumably autolysosomes) and decreased SQSTM1 expression is detected in dying neurons, suggesting that the autophagy flux is enhanced and associated with neuronal death occurring in neonatal brain injury [534, 535]. Although still in its infancy with regard to autophagy, it is worth pointing out that mathematical modeling has the power to bridge whole body in vivo data with in vitro data from tissues and cells. The usefulness of so-called hierarchical or multilevel modeling has been demonstrated when examining the relevance of INS (insulin) signaling to glucose uptake in primary human adipocytes compared with whole-body glucose homeostasis [2235].
In contrast to tissue samples, blood samples for autophagy study can be more easily obtained from living donors, from non-diseased donors, and from a wide age range including infants up to adults. However, current medication history is especially important in blood samples, as high concentrations of medications that can alter autophagy may be present. For example, in patients with cystic fibrosis (CF) repurposed medications such as cysteamine are undergoing clinical trial evaluation. Cysteamine improves autophagy in CF [2236–2238], whereas other chronic medications such as azithromycin may suppress autophagy. Therefore a careful record of daily and study medications should be accounted for when examining human blood cells. Further, many autophagy regulators are differentially expressed across human sample and cell types. This was shown for CF sputum, where high expression of a microRNA cluster that regulates autophagy was found, in contrast to low expression in the blood [2239]. It is recommended that autophagy studies in humans account for multiple biological sources (including cells from the affected tissues or organs) when making definitive conclusions about the state of overall autophagy under- or overexpression. Likewise, therapeutic testing of new compounds in human samples ex vivo should be validated in multiple sample types. In the case of CF, this is often done in local tissues such as airway epithelial brushings, and validated in blood cells that are recruited to the local site of action [2240]. Autophagy studies in CF models can be easily adopted for other disease states. Alternatively, several pathologies with an aberrant autophagy process have been identified in humans through genome-wide studies. Cells derived from the affected tissues of such patients could be used for testing the therapeutic potency of new molecules.
A stepwise process can be proposed for linking changes in the autophagic pathway to changes in disease outcome. First, in an observational study, the changes in the autophagic pathway should be quantified and linked to changes in disease outcome. To prove causality, a subsequent autophagy-modifying intervention should be tested in a randomized study. Before an intervention study is performed in human patients, the phenotype of (in)active autophagy contributing to poor outcome should be established in a validated animal model of the disease. For the validation of the hypothesis in an animal model, a similar two-step process is suggested, with the assessment of the phenotype in a first stage, followed by a proof-of-concept intervention study (see Large animals).
10. Hydra
Hydra is a freshwater cnidarian animal that provides a unique model system to test autophagy. The process can be analyzed either in the context of nutrient deprivation, as these animals easily survive several weeks of starvation [2241], or in the context of regeneration, because in the absence of protease inhibitors, bisection of the animals leads to an uncontrolled wave of autophagy. In the latter case, an excess of autophagy in the regenerating tip immediately after amputation is deleterious [2242, 2243]. Most components of the autophagy and MTOR pathways are evolutionarily conserved in Hydra [2244]. For steady-state measurements, autophagy can be monitored by western blot for Atg8-family proteins, by immunofluorescence (using antibodies to Atg8-family proteins, lysobisphosphatidic acid or RPS6KA/RSK), or with dyes such as MitoFluor™ Red 589 and LysoTracker™ Red. Flux measurements can be made by following Atg8-family protein turnover using lysosomal protease inhibitors (leupeptin and pepstatin A) or in vivo labeling using LysoTracker™ Red. It is also possible to monitor MTOR activity with phosphospecific antibodies to RPS6KB and EIF4EBP1 or to examine gene expression by semiquantitative RT-PCR, using primers that are designed for Hydra. Autophagy can be induced by RNAi-mediated knockdown of Kazal1 [2242, 2243], or with rapamycin treatment, and can be inhibited with wortmannin or bafilomycin A1 [2241, 2244].
In situ hybridization shows high expression of ATG12 transcripts specifically in nematoblasts, and of ATG5 in the budding region and growing buds. The utilization of both knockdown and RNAi approaches indicates a crucial role of autophagy in various developmental and physiological processes, including the regeneration processes in adults [2245].
11. Induced pluripotent stem cells
Previous studies typically used patient biopsies and post-mortem tissues to investigate the role of autophagy in the pathogenesis of human disease. Nonetheless, the availability, preparation, and fixation of human biopsied tissues and organs, as well as the quantity and quality of biopsies, limit the dynamic measurement of autophagic flux. Furthermore, insufficiency of tissue biopsies from healthy controls for comparison challenges the snapshot results obtained from patient biopsies. To overcome the limitation of sample sources for investigating autophagy in human disease, various animal models and immortalized cell lines have been used to represent these diseases. Valuable results from these model systems have provided a fundamental pathomechanism for the role of autophagy in various human diseases; however, the species discrepancy between animal and human, and the tumorous genetic background of cell lines elicit concerns for the implications of the results as they pertain to humans.
Recently, the development of induced pluripotent stem (iPS) cells provides a valuable experimental system to uncover disease mechanisms and novel therapeutic strategies in human disease [2246, 2247]. Diverse tissue-specific cells differentiated from iPS cells offer great potential to model different systemic diseases. Furthermore, the unique genetic and molecular signature of the affected individuals allows researchers to address disorder-relevant phenotypes at a cellular level. Multiple somatic cell sources such as skin, adipose tissues and peripheral blood for reprogramming to iPS cells can be obtained in non-invasive procedures. Thus, both disease and control iPS cells can be made available for comparison.
iPS cells modeling mitochondrial disease have been used for investigating the impact of mtDNA mutation on autophagy [2252]. Both isogenic iPS cell clones with high mutant mtDNA burden and without mtDNA mutation can be isolated simultaneously during cell passages. Thus, the impact of mtDNA heteroplasmy on autophagy involving pathogenesis of mitochondrial diseases can be observed directly. iPS cells with high mutant mtDNA burden modeling mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes (MELAS) syndrome show elevated levels of autophagy, superoxide, intracellular calcium and mitochondrial depolarization at basal conditions in comparison with control iPS cells [2252]. It is noted that oxidative stress exacerbates the accumulation of autophagosomes and autolysosomes, increases levels of superoxide and enhances calcium flux into the cytoplasm, leading to robust depolarization of mitochondrial membrane potential and enhanced mitophagy in MELAS-iPS cells. Mitophagy is very scarce in MELAS-iPS cells at the basal condition, consistent with previous observations that selective elimination of mitochondria containing pathogenic mtDNA is spared in mitochondrial diseases under physiological conditions [2252, 2253].
Moreover, work describing the changes occurring in long-term cultures of iPS cells, suggest this as a suitable model to study aging processes. In this context, autophagy increases in senescent cells (Figure 37), so that identifying autophagic mechanisms triggered by cellular senescence could suggest potential therapeutic strategies against premature aging [150, 2254].
Figure 37.
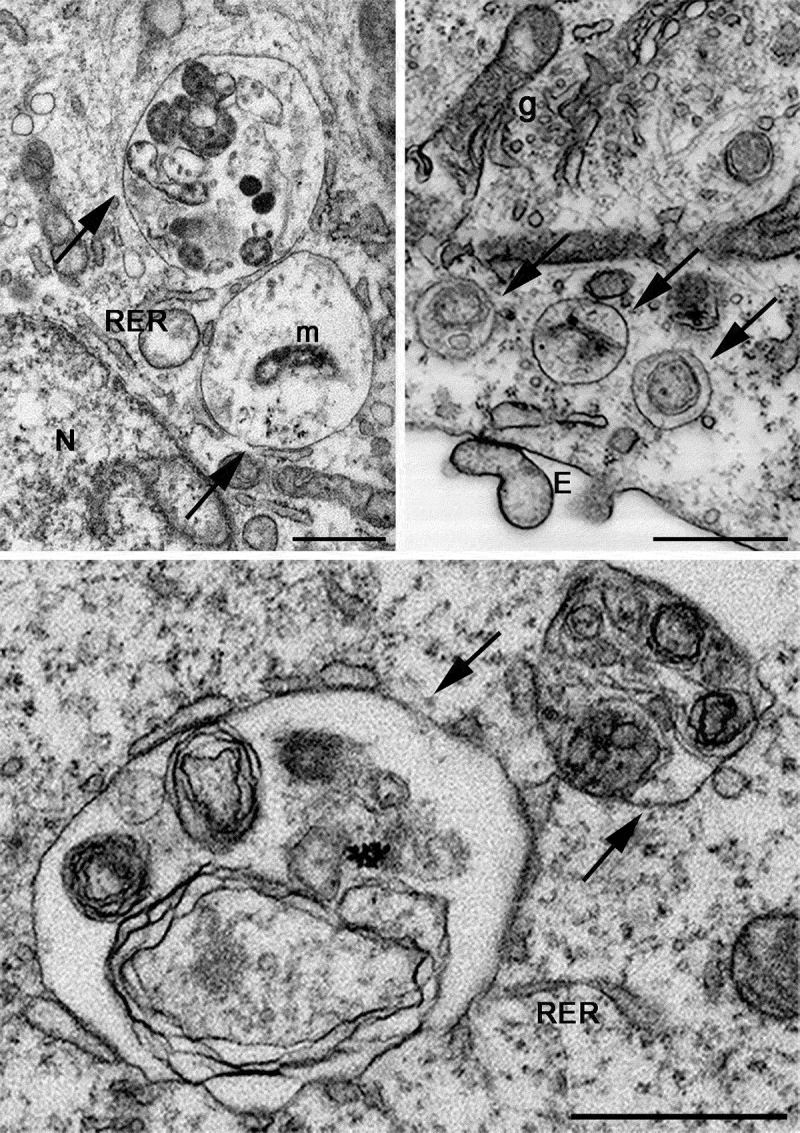
FIB-SEM images showing ultrastructural details of aging iPS cells. Arrows indicate autophagosomes containing mitochondria or other partially digested cytoplasmic material. E, exosomes; g, Golgi apparatus; m, mitochondrion; N, nucleus; RER, rough endoplasmic reticulum. Scale bars: 1 µm. Image provided by F. Colasuonno, modified from ref. [150]
Cumulative evidence from emerging research indicates that the iPS cellular model is a useful and promising tool to recapitulate the pathogenesis of human diseases, allowing better understanding of the mechanism, and facilitating development of potential therapeutic targets.
Large animals and rodents. This section refers in particular to mammals other than humans. Assessment of autophagy (and, in particular, autophagic flux) in clinically relevant large animal models is critical in establishing its (patho)physiological role in multiple disease states. For example, evidence obtained in swine suggests that upregulation of autophagy may protect the heart against damage caused by acute myocardial infarction/heart attack [2255]. Ovine models of placental insufficiency leading to intrauterine growth restriction have shown that there is no change in the expression of markers of autophagy in the fetus in late gestation [2256] or in the lamb at 21 days after birth [2257]. Furthermore, there is an increase in markers of autophagy in the placenta of human intrauterine growth restriction pregnancies [2258]. Studies in rabbits suggest a protective role of upregulated autophagy against critical illness-induced multiple organ failure and muscle weakness [2259, 2260], which is corroborated by human studies [2231, 2261]. Conversely, autophagy may contribute to the pathogenesis of some types of tissue injury, at least in the lung [2262, 2263] and different regions of the CNS [39, 86, 539]. Similarly, autophagy may play different roles in ischemic stroke as ischemia progresses [2264], or during subsequent reperfusion [2265]. For example, studies in rats demonstrate that activation of autophagy [539] and disruption of autophagosome-lysosome fusion [86] may induce ischemic neuronal damage in the hippocampal CA1 region after transient global cerebral ischemia. The autophagic flux was also demonstrated to be activated, and an autophagic mechanism may contribute to ischemic neuronal injury in rats subjected to focal ischemia [2266] and neonatal cerebral hypoxic ischemia [535, 2267]. Dysregulation of autophagy and mitophagy genes with parallel Casp3 gene expression is observed in rats after complete cerebral ischemia with survival of 2-30 days after ischemia, and post-ischemic studies in rats suggest a lack of a protective role of the dysregulated autophagy in the brain as assessed by the expression of the Bace1 gene [1645, 1646]. In the mouse retina, mitophagy is dramatically impaired during prolonged diabetes, suggesting a pathogenic role in the development of neurovascular complications and premature senescence [39]. Finally, there is an increase in LC3-II in the kidney in normal wild-type mice treated with bafilomycin A1, but no increase in LC3-II in mice with polycystic kidney disease, suggesting suppressed autophagic flux in cys1/cpk mouse kidneys [2268].
Studies in rodent and cellular models have shown a critical role of dysregulated autophagy in pancreatitis [1499, 2269]. Experimental pancreatitis stimulates autophagosome formation, but at the same time inhibits autophagic degradation, resulting in impaired autophagic flux evidenced by accumulation of enlarged autolysosomes, decreased rate of long-lived protein degradation in pancreatic acinar cells, and increases in both LC3-II and SQSTM1. Mice with pancreas-specific ablation of Atg5 or Atg7, double knockout of Tfeb and Tfe3, or Lamp2 knockout, all develop spontaneous pancreatitis. Further, manifestations of impaired autophagy are prominent in human pancreatitis, such as acinar cell vacuolization (a long-noted, but poorly understood hallmark response of this disease), increases in pancreatic LC3-II and SQSTM1, and decreases in LAMP2 and TFEB.
Autophagy also plays an important role in the development and remodeling of the bovine mammary gland. In vitro studies with the use of a 3-dimensional culture model of bovine mammary epithelial cells (MECs) have shown that this process is involved in the formation of fully developed alveoli-like structures [2270]. Earlier studies show that intensified autophagy is observed in bovine MECs at the end of lactation and during the dry period, when there is a decrease in the levels of lactogenic hormones, increased expression of auto/paracrine apoptogenic peptides, increased influence of sex steroids and enhanced competition between the intensively developing fetus and the mother organism for nutritional and bioactive compounds [2271, 2272]. These studies were based on some of the methods described elsewhere in these guidelines, including GFP-Atg8-family protein fluorescence microscopy, TEM, and western blotting of LC3 and BECN1. Creation of a specific GFP-LC3 construct by insertion of cDNA encoding bovine LC3 into the pEGFP-C1 vector makes it possible to observe induction of autophagy in bovine MECs in a more specific manner than can be achieved by immunofluorescence techniques, in which the antibodies do not show specific reactivity to bovine cells and tissues [2270, 2272]. However, it is important to remember that definitive confirmation of cause-and-effect is challenging for studies on large animals, given the lack or poor availability of specific antibodies and other molecular tools, the frequent inability to utilize genetic approaches, and the often prohibitive costs of administering pharmacological inhibitors in these translational preparations.
In contrast with cell culture experiments, precise monitoring of autophagic flux is practically impossible in vivo in large animals. Theoretically, repetitive analyses of small tissue biopsies should be performed to study ultrastructural and molecular alterations over time in the presence or absence of an autophagy inhibitor (e.g., CQ). However, several practical problems impede applicability of this approach. First, repetitive sampling of small needle biopsies in the same animal (a major challenge by itself) could be assumed to induce artefacts following repetitive tissue destruction, especially when deep (vital) organs are involved. In addition, chemical inhibitors of autophagy have considerable side effects and toxicity, hampering their usage. Also, the general physical condition of an animal may confound results obtained with administration of a certain compound, for instance altered uptake of the compound when perfusion is worse.
Therefore, in contrast to cells, where it is more practical to accurately document autophagic flux, we suggest the use of a stepwise approach in animal models to provide a proof of concept with an initial evaluation of sequelae of (in)active autophagy and the relation to the outcome of interest.
First, prior to an intervention, the static ultrastructural and molecular changes in the autophagic pathway should be documented and linked to the outcome of interest (organ function, muscle mass or strength, survival, etc.). These changes can be evaluated by light microscopy, EM and/or by molecular markers such as LC3-II. In addition, the cellular content of specific substrates normally cleared by autophagy should be quantified, as, despite its static nature, such measurement could provide a clue about the results of altered autophagic flux in vivo. These autophagic substrates can include SQSTM1 and (poly)ubiquitinated substrates or aggregates, but also specific substrates such as damaged mitochondria. As noted above, measurement of these autophagic substrates is mainly informative when autophagic flux is prohibited/insufficient, and, individually, all have specific limitations for interpretation. As mentioned several times in these guidelines, no single measurement provides enough information on its own to reliably assess autophagy, and all measurements should be interpreted in view of the whole picture. In every case, both static measurements reflecting the number of autophagosomes (ultrastructural and/or molecular) and measurements of autophagic substrates as surrogate markers of autophagic flux need to be combined. Depending on the study hypothesis, essential molecular markers can further be studied to pinpoint at which stage of the process autophagy may be disrupted.
Second, after having identified a potential role of autophagy in mediating an outcome in a clinically relevant large animal model, an autophagy-modifying intervention should be tested. For this purpose, an adequately designed, randomized controlled study of sufficient size on the effect of a certain intervention on the phenotype and outcome can be performed in a large animal model. Alternatively, the effect of a genetic intervention can be studied in a small animal model with clinical relevance to the studied disease.
As mentioned above, exact assessment of autophagic flux requires multiple time points, which cannot be done in the same animal. Alternatively, different animals can be studied for different periods of time. Due to the high variability between animals, however, it is important to include an appropriate control group, and a sufficiently high number of animals per time point as corroborated by statistical power analyses. This requirement limits feasibility and the number of time points that can be investigated. The right approach to studying autophagy in large animals likely differs depending on the question that is being addressed. Several shortcomings regarding the methodology, inherent to working with large animals, can be overcome by an adequate study design. As for every study question, the use of an appropriate control group with a sufficient number of animals is crucial in this regard.
12. Lepidoptera
Some of the earliest work in the autophagy field was carried out in the area of insect metamorphosis. Microscopy and biochemical research revealed autophagy during the metamorphosis of American silkmoths and the tobacco hornworm, Manduca sexta, and included studies of the intersegmental muscles, but they did not include molecular analysis of autophagy [2273]. Overall, these tissues cannot be easily maintained in culture, and antibodies against mammalian proteins do not often work. Accordingly, these studies were confined to biochemical measurements and electron micrographs. During metamorphosis, the bulk of the larval tissue is removed by autophagy and other forms of proteolysis [2274]. Bombyx mori is now used as a representative model among Lepidoptera, for studying not only the regulation of autophagy in a developmental setting, but also the relations between autophagy and apoptosis. The advantages of this model are the large amount of information gathered on its developmental biology, physiology and endocrinology, the availability of numerous genetic and molecular biology tools, and a completely sequenced genome [2275]. The basic studies of B. mori autophagy have been mainly carried out in four larval organs: the silk gland, the fat body, the midgut and the ovary.
The techniques used for these studies are comparatively similar, starting from EM, which is the most widely used method to follow the changes of various autophagic structures and other features of the cytosol and organelles that are degraded during autophagy [942, 2276–2279]. Immuno-TEM also can be used, when specific antibodies for autophagic markers are available. As in other model systems the use of Atg8 antibodies has been reported in Lepidoptera. In B. mori midgut [942, 2280], fat body [943] and silk gland [944] as well as in various larval tissues of Galleria mellonella [2281] and Helicoverpa armigera [2282], the use of both custom and commercial antibodies makes it possible to monitor Atg8 conversion to Atg8–PE by western blotting. Moreover, transfection of GFP-Atg8 or mCherry-GFP-Atg8 has been used to study autophagy in several lepidopteran cell lines [2282]. In addition, an antibody against Sqstm1 was generated, and it is efficient in detecting its autophagic degradation during autophagic processes in B. mori [2283, 2284]. Activation of MTOR can be monitored with an antibody against p-EIF4EBP1 [943, 2280, 2284]. Acidotropic dyes such as MDC and LysoTracker™ Red staining have been used as markers for autophagy in silkmoth egg chambers, always combined with additional assays [2276, 2277]. Acid phosphatase also can be used as a marker for autolysosomal participation in these tissues [942, 2278, 2285]. Systematic cloning and analysis revealed that homologs of most of the Atg genes identified in other insect species such as Drosophila are present in B. mori, and 15 Atg genes have now been identified in the silkworm genome, as well as other genes involved in the TOR signal transduction pathway [2286–2288]. Variations in the expression of several of these genes have been monitored not only in silkworm larval organs, where autophagy is associated with development [942, 944, 2286, 2287, 2289], but also in the fat body of larvae undergoing starvation [2286, 2290].
In the IPLB-LdFB cell line, derived from the fat body of the caterpillar of the gypsy moth Lymantria dispar, indirect immunofluorescence experiments have demonstrated an increased number of Atg8-positive dots in cells with increased autophagic activity; however, in contrast to larval tissues, western blotting did not reveal the conversion of Atg8 into Atg8–PE. In fact, a single band with an approximate molecular mass of 42 kDa was observed that was independent of the percentage of cells displaying punctate Atg8 (D. Malagoli, unpublished results). Thus, the utility of monitoring Atg8 in insects may depend on the species and antibody.
13. Marine invertebrates
The invaluable diversity of biological properties in marine invertebrates offers a unique opportunity to explore the different facets of autophagy at various levels from cell to tissue, and throughout development and evolution. For example, work on the tunicate Ciona intestinalis has highlighted the key role of autophagy during the late phases of development in lecithotrophic organisms (those in which the larvae during metamorphosis feed exclusively from the egg yolk resources) [374, 2291]. This work has also helped in pinpointing the coexistence of autophagy and apoptosis in cells, as well as the beneficial value of combining complementary experimental data such as LC3 immunolabeling and TUNEL detection. This type of approach could shed a new light on the close relationship between autophagy and apoptosis, and provide valuable information about how molecular mechanisms control the existing continuum between these two forms of programmed cell death. Autophagy also appears to play a role in the cell renewal process observed during the regeneration of the carnivorous sponge Asbestopluma hypogea [2292].
The identification of a growing number of autophagy-related sequences in different species has opened a much wider scenario for investigating the molecular mechanisms of autophagy and its role in a variety of processes. For example, in the “living fossil”, the sponge Astrosclera willeyana, molecular, histochemical, and morphological evidence indicate that specialized cells involved in the formation of a highly calcified skeleton actively degrade their intracellular microbial community using the autophagy pathway (namely ATG8) [2293]. This is the first observation suggesting an association between the process of autophagy and biomineralization in a metazoan. Analysis of the expression patterns of 13 genes involved in autophagy and apoptosis in the sea urchin Paracentrotus lividus highlights the simultaneous involvement of both processes in early embryo development [2294].
Bivalve molluscs provide useful models for studying autophagic function [2295]. Autophagy plays a key role in the resistance to nutritional stress as is known to be the case in many Mediterranean bivalve molluscs in the winter. For example, the European clam Ruditapes decussatus is able to withstand strict fasting for two months, and this resistant characteristic is accompanied by massive autophagy in the digestive gland (Figure 38). This phenomenon, observed by TEM, demonstrates once again the advantage of using this classical ultrastructural method to study autophagy in unconventional biological models for which molecular tools may not be operational. Autophagy has been also demonstrated by different types of lysosomal reactions in digestive gland cells in response to a variety of environmental stressors (starvation, salinity change, hyperthermia, hypoxia, pollutant-induced stress). In the Mediterranean mussel Mytilus galloprovincialis, dephosphorylation of MTOR, evaluated by immunohistochemistry with antibodies generated to the mammalian protein, contributes to increased lysosomal membrane permeability and autophagy induced by contaminant exposure [2296].
Figure 38.
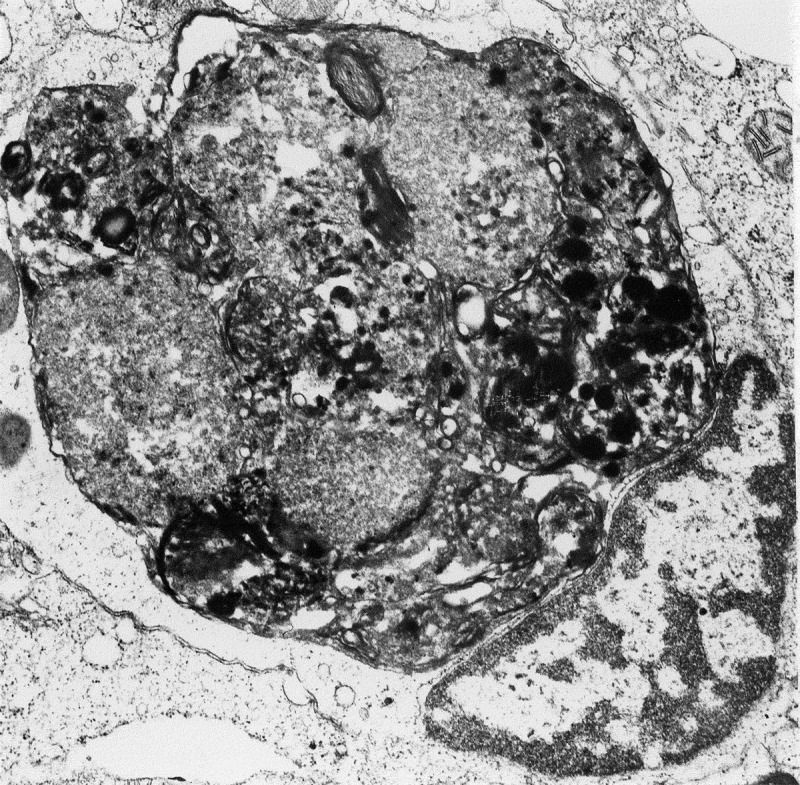
Autophagy in the digestive gland of Ruditapes decussatus (Mollusca, Bivalvia) subjected to a strict starvation of 2 months. Image provided by S. Baghdiguian
Autophagy also plays a role during pathogen infections, as has been observed in the Pacific oyster, Crassostrea gigas. In the mantle of this bivalve mollusc, autophagy is modulated in response to a viral (Ostreid herpesvirus 1 [OsHV-1]) and a bacterial (Vibrio aestuarianus) infection [2297]. Autophagy may therefore play a protective role in oysters against infections as suggested by a survival assay when autophagy is inhibited by NH4Cl treatment or induced by carbamazepine or starvation. Furthermore, autophagy occurs in the hemocytes of the Pacific oyster [502], which are the main effectors of its immune system, and thus play a key role in the defense against pathogens. Hemocyte autophagy activity characterized by flow cytometry, fluorescence microscopy and TEM analysis shows the importance of combining different approaches to investigate autophagy in marine invertebrate models.
Although the different facets of autophagy are increasingly studied, the molecular mechanism of autophagy is still poorly understood in these models. For the first time, an identification of the ATG proteins that constitute the core molecular machinery of autophagy in a bivalve mollusc, C. gigas, has been established [502]. The autophagy machinery in this organism is conserved with other eukaryotic organisms. These results will provide new possibilities to better understand the autophagy processes and mechanism in marine invertebrates.
At present, the use of TEM still represents a unique tool to confirm the presence of autophagic structures in bivalves at the subcellular level [502, 2297, 2298]. In M. galloprovincialis hemocytes, rapid autophagosome formation is observed within 5-15 min of in vitro challenge with Vibrio tapetis [2298]. This observation, together with increased LC3-II expression, decreased levels of phosphorylated MTOR and of SQSTM1, represents the first direct evidence for modulation of autophagic processes induced in bivalve immune cells by bacterial challenge.
Genome sequencing and transcriptomic data in different bivalve species are revealing a growing number of autophagy-related genes that are involved in the immune response [2299, 2300]. Overall, available data in bivalves underlines the point that autophagy is not involved in pathogen degradation, but in protection against viral and bacterial infection.
A relationship between autophagy and resistance to disease has also been described in corals. Comparison of transcriptomics data on the immune response of four coral species, with a range of disease susceptibility, shows activation of apoptosis and autophagic pathways prevailing, respectively, in susceptible species (Orbicella faveolata) and disease-tolerant species (Porites porites and P. astreoides), indicating that apoptotic and autophagic pathways might have a significant impact on the susceptibility of corals to disease [2301].
In crustaceans, gene expression, miRNA silencing and proteome analysis are revealing the role of autophagy-related mechanisms in immune and stress responses. Different miRNAs play key roles in immunity and host autophagy after infection by white spot syndrome virus, one of the main causes of disease in aquacultured species [2302, 2303]. In the crab Eirocheir sinensis, EsBECN1 (Vps30/Atg6) is involved in regulating the expression of antimicrobial peptides in the immune responses to bacterial infection [2304]. In copepods, ocean acidification enhances lysosome-autophagy pathway proteomes that are responsible for repairing and removing proteins and enzymes damaged under stress, possibly mitigating mercury-induced toxicity [2305].
The intralysosomal subcellular distribution of C60 fullerene nanoparticles in digestive gland cells of the marine mussel (M. galloprovincialis), following experimental exposure to C60 nanoparticles in seawater results in lysosomal membrane permeabilization and inhibition of MTOR, and provokes an excessive induction of autophagy [2296, 2306]. The effects of C60 fullerene nanoparticles indicate that moderate to severe ROS production and oxidative damage are not necessary under these conditions to inhibit the MTOR pathways, although lysosomal membrane permeabilization, probably caused by lysosomal overload of foreign material (i.e., C60 fullerene), will result in release of intralysosomal iron that will produce ROS [2077]. Consequently, autophagic induction by C60 (as for other nanoparticles [2077, 2094]), may represent a protective degradation in autolysosomes of material that is recognized by the cell as foreign or aberrant, such as pathogens or damaged intracellular proteins and membranes.
The cytoskeletal alterations induced by C60 fullerene nanoparticles may impair the growth of the cells and their organization in the digestive tubules of the digestive gland. Overall, dysregulation of MTORC1 and MTORC2 may reduce the capacity of the cells, and organisms, to properly grow and reproduce. Consequently, MTOR dephosphorylation should be considered a diagnostic biomarker for the toxic effects of the C60 nanoparticles and polycyclic aromatic hydrocarbons as previously demonstrated [2296]; and, under chronic stressful conditions, prognostic for potential harmful effects at the whole animal and population level.
Overall, knowledge on the autophagic machinery in marine invertebrates will help not only in elucidating the molecular networks that regulate autophagy within an evolutionary framework; this information will also contribute to understanding the response to infection of those species that are affected by pathogen-induced mass mortalities or other environmental stressors, also in the context of rapid global changes that affect their survival and distribution in oceans.
14. Neotropical teleosts
In tropical environments, fish have developed different reproductive strategies, and many species have the potential for use as a biological model in cell and molecular biology, especially for studying the mechanisms that regulate gametogenesis and embryo development. In these fish, the ovary is a suitable experimental model system for studying autophagy and its interplay with cell death programs due to the presence of postovulatory follicles (POFs) and atretic follicles, which follow different routes during ovarian remodeling after spawning [2307]. In fish reproductive biology, POFs are excellent morphological indicators of spawning, whereas atretic follicles are relevant biomarkers of environmental stress. In addition, many freshwater teleosts of commercial value do not spawn spontaneously in captivity, providing a suitable model for studying the mechanisms of follicular atresia under controlled conditions [2308]. When these species are subjected to induced spawning, the final oocyte maturation (resumption of meiosis) occurs, and POFs are formed and quickly reabsorbed in ovaries after spawning [2309]. Assessment of autophagy in fish has been primarily made using TEM at different times of ovarian regression [2310]. Due to the difficulty of obtaining antibodies specific for each fish species, immunodetection of ATG proteins (mainly LC3 and BECN1) by IHC associated with analyses by western blotting can be performed using antibodies that are commercially available for other vertebrates [546]. Such studies suggest dual roles for autophagy in follicular cells [2307]; however, evaluation of the autophagic flux in different conditions is critical for establishing its physiological role during follicular regression and ovarian remodeling after spawning. Given the ease of obtaining samples and monitoring them during development, embryos of these fish are also suitable models for studying autophagy that is activated in response to different environmental stressors, particularly in studies in vivo.
15. Odontoblasts
Odontoblasts are long-lived dentin-forming postmitotic cells, which evolved from neural crest cells early during vertebrate evolution. These cells are aligned at the periphery of the dental pulp and are maintained during the entire healthy life of a tooth. As opposed to other permanent postmitotic cells such as cardiac myocytes or central nervous system neurons, odontoblasts are significantly less protected from environmental insults such as dental caries and trauma. Mature odontoblasts develop a well-characterized autophagy-lysosomal system, including a conspicuous autophagic vacuole that ensures turnover and degradation of cell components. Immunocytochemical and TEM studies make it possible to monitor age-related changes in autophagic activity in human odontoblasts [2311]. Tooth pulp cells, in contrast, process minor autophagic activities; however, the autophagy level in those cells can be highly induced in stress conditions [2312]. Furthermore, in the periodontal ligament mesenchymal cells, increased autophagy has a protective role in apoptosis prevention [2313], and it plays a role in healing of the oral mucosa [2314].
16. Parasitic helminths
Parasitic helminths comprise parasitic flatworms (Monogenea, Trematoda [flukes], and Cestoda [tapeworms] of the class Neodermatans [2315]) that infect vertebrates and cause certain human neglected tropical diseases such as neurocysticercosis and taeniasis (Taenia sp.), echinococcosis (Echinococcus sp.), schistosomiasis (Schistosoma sp.), fascioliasis (Fasciola hepatica), clonorchiasis (Clonorchis sinensis) and opisthorchiasis (Opisthorchis viverrini) among others [2316]. Although autophagy is a fundamental catabolic pathway conserved from yeast to mammals, it remains understudied in these parasites. Since the 1960s, autophagy and particularly glycophagy have been described via TEM for these parasitic helminths through ultrastructural changes in the syncytial tegument of larval stages and adult worms during in vitro and in vivo drug chemotherapy [2317–2320]. In addition, data obtained by TEM analysis led to the proposal that specialized biomineralized cells, termed calcareous corpuscles, are the result of continuous cytoplasmic autophagy in tapeworms [2320, 2321]. These cells show multi-lamellar structures coincident with the typical ultrastructure of autophagy activation induced by endoplasmic reticulum stress, and different from that seen in cells deprived of nutrients (Figure 39A) [2322]. The calcareous corpuscles play key roles in the physiology of tapeworms; they are involved in bioaccumulation of ions (calcium, magnesium, carbonate and phosphate, and traces of aluminum, boron, copper and iron), metamorphosis of parasitic tissues (the corpuscles are formed, reorganized and resorbed in different hosts) and they correlate with previous or ongoing active metabolic activity (high content of carbohydrate metabolism enzymes and glycogen) [2323]. Studies carried out with confocal IHC using a commercial polyclonal antibody directed against the N terminus of human LC3, make it possible to verify the autophagy activity of calcareous corpuscles in Echinococcus granulosus larval stages exposed to arsenic trioxide, metformin and rapamycin (Figure 39B) [2324].
Figure 39.
Detection of autophagy in Echinococcus granulosus larval stage. (A) Scanning electron micrographs of a sectioned larva (or protoscolex) (i) showing big oval-shaped cells named calcareous corpuscles (red arrowheads) developed by cytoplasmic autophagy. Ultrastructural details of different developmental stages of these parenchymatic cells showing a central vacuole (ii-iii) at the initial development phase and concentric membranes that marginalize a thin layer of cytoplasm in mature corpuscles at the end of the autophagic process (iv-v). Energy-dispersive X-ray elemental microanalysis of the calcareous corpuscles in a sectioned protoscolex demonstrates the colocalization of accumulated ions into corpuscles: calcium (vi), phosphorus (vii). Scale bar: 10 µm. (B) Optical transmission (i) and confocal (ii-iv) microscopy images of a protoscolex treated with metformin (10 mM) for 48 h (i-iii) and an untreated microcyst (or metacestode) (iv) incubated with an anti-LC3 antibody and revealed with an antibody conjugated with Alexa Fluor 488 (green fluorescence) and counterstained with propidium iodide (red fluorescence) to observe cell nuclei under optimal contrast conditions. Fluorescent punctate images are often detected in the tegument of rapamycin-treated protoscoleces (ii-iii) and microcysts originated by vesicular de-differentiation from protoscoleces (iv) with high Atg8 polypeptide levels within the free cytoplasmic matrix of these cells, demonstrating pharmacological autophagy induction in corpuscles (ii-iii) and basal autophagy in small cysts in development even under nutrient-rich conditions (iv). Scale bar: 100 µm. Inset images correspond to TEM. bo, body; gl, germinal layer; su, sucker; tg, tegument. Images provided by A. C. Cumino and J. A. Loos. Only images in panel B were previously published in ref. [327]
Currently, the availability of genome sequences together with the extensive transcriptomic and/or expressed sequence tag (EST) data allow in silico confirmation of the occurrence of the autophagy-related genes for the parasitic flatworms that cause the most serious problems among 50 helminth genome draft assemblies [2325–2328]. Most components of the autophagy core machinery and related key signaling pathways such as those involving AKT, PI3K, TOR, AMPK, FOXO and TFEB are evolutionarily preserved in these parasitic flatworms; however, only in some parasites such as Echinococcus sp. has the autophagy pathway been formally analyzed [2324, 2329]. Basic studies performed in metacestodes and protoscoleces, larval forms of the cestode Echinococcus that can develop in humans, allow the detection of active basal autophagy both in cellular systems and during the vesicular de-differentiation of protoscolex to metacestode [2324]. All Atg homologs (encoded by fourteen genes including two paralogs for Atg8) involved in induction, vesicle nucleation, autophagosome expansion and membrane recycling (except Atg10, which was also not identified in D. melanogaster nor in Apis mellifera [2]) have been found in Echinococcus sp. [2324]. These autophagy-related proteins conserve all domains corresponding to specific functions, including the key amino acids involved in protein-protein or protein-membrane interactions.
Autophagy in Echinococcus can be regulated by transcription-dependent upregulation via FOXO and non-transcriptional inhibition through TOR [2330] (J. Loos and V. Dávila, personal communication). As in other invertebrates, a single FOXO transcription factor is identified in the cestode. Likewise, the consensus core recognition motif for FOXO binding (TTGTTTAC) is conserved in autophagy genes (atg8 and atg12). Furthermore, it has been demonstrated that rapamycin, metformin and bortezomib are able to induce autophagy, dose-dependent pharmacological effects and death in these parasites even under nutrient-rich conditions. These results were verified by detection of diverse autophagic structures through TEM (including the phagophore, autophagosomes, autolysosomes with lamellar stacks, and glycogen surrounded by double-membrane vesicles), Atg8 punctate images detected by confocal microscopy, conversion of Atg8 to the Atg8–PE conjugate by western blotting, and an increase in the mRNA levels of autophagy genes (atg5, atg6, atg8, atg12, atg16 and atg18) by RT-PCR, proportional to the drug concentration employed [2324, 2329–2331]). Although autophagy is predominantly a homeostatic mechanism, drug-induced excessive autophagy might also play a role in cell death. Therefore, from a therapeutic perspective, it will be of great importance to understand how autophagy can be pharmacologically manipulated to favor pro-death signaling in these parasites. The establishment of new molecular tools and studies involving specific related atg mutants would be of great value in order to get insights into the role of autophagy in parasitic flatworms.
17. Planarians
Because planarians are one of the favorite model systems in which to study regeneration and stem cell biology, these flatworms represent a unique model where it is possible to investigate autophagy in the context of regeneration, stem cells and growth. Currently the method used to detect autophagy is TEM. A detailed protocol adapted to planarians has been described [2332, 2333]. However, complementary methods to detect autophagy are also needed, because TEM cannot easily distinguish between activation and blockage of autophagy, which would both be observed as an accumulation of autophagosomes. Other methods to detect autophagy are being developed (C. González-Estévez, personal communication), including IHC and western blotting approaches for the planarian homolog of LC3. Several commercial antibodies against human LC3 have been tried for cross-reactivity without success, and three planarian-specific antibodies have been generated. Some preliminary results show that LysoTracker™ Red can be a useful reagent to analyze whole-mount planarians. Most of the components of the autophagy and MTOR signaling machinery are evolutionarily conserved in planarians. Whether autophagy genes vary at the mRNA level during starvation and after depletion of MTOR signaling components is still to be determined.
18. Plants
As stated above with regard to other organisms, staining with MDC or derivatives (such as monodansylamylamine) is not sufficient for detection of autophagy, as these stains also detect vacuoles. The same is the case with the use of LysoTracker™ Red, neutral red or acridine orange. The fluorophore of the red fluorescent protein shows a relatively high stability under acidic pH conditions. Thus, chimeric RFP fusion proteins that are sequestered within autophagosomes and delivered to the plant vacuole can be easily detected by fluorescence microscopy. Furthermore, fusion proteins with some versions of RFP tend to form intracellular aggregates, allowing the development of a visible autophagic assay for plant cells [2334]. For example, fusion of cytochrome b5 and the original (tetrameric) RFP generate an aggregated cargo protein that displays cytosolic puncta of red fluorescence and, following vacuolar delivery, diffuse staining throughout the vacuolar lumen. However, it is not certain whether these puncta represent autophagosomes or small vacuoles, and therefore these data should be combined with immuno-TEM or with conventional TEM using high-pressure frozen and freeze-substituted samples [2335].
In plant studies, GFP-Atg8 fluorescence is typically assumed to correspond to autophagosomes; however, as with other systems, caution needs to be exercised because it cannot be ruled out that Atg8 is involved in processes other than autophagy. Immunolabeled GFP-Atg8 can be detected both on the inner and outer membrane of an autophagosome in an Arabidopsis root cell, using chemical fixation (see Fig. 6b in ref. [2336]), suggesting that it will be a useful marker to monitor autophagy. Arabidopsis cells can be stably transfected with GFP fused to plant ATG8, and the lipidated and nonlipidated forms can be separated by SDS-PAGE [289]. Furthermore, the GFP-ATG8 processing assay is particularly robust in Arabidopsis and can be observed by western blotting [290, 350]. Two kinds of GFP-ATG8 transgenic seeds are currently available from the Arabidopsis Biological Resource Center, each expressing similar GFP-ATG8A transgenes but having different promoter strength. One transgene is under the control of the stronger Cauliflower mosaic virus 35S promoter [816], whereas the other uses a promoter of the Arabidopsis AT4G05320.2/ubiquitin10 gene [2337]. In the GFP-ATG8 processing assay, the former has a higher ratio of GFP-ATG8A band intensity to that of free GFP than does the latter [2337]. Because free GFP level reflects vacuolar delivery of GFP-ATG8, the ubiquitin promoter line may be useful when studying an inhibitory effect of a drug/mutation on autophagic delivery. Likewise, the 35S promoter line may be used for testing potential autophagy inducers. GFP-ATG8CL Nicotiana benthamiana seeds are also available upon request3 (unpublished). The transgene is under a 35S promoter, and the plants can be used for both confocal microscopy and western blotting to monitor autophagic flux and image autophagosomes in vivo. Immunofluorescence with anti-ATG8 antibodies followed by confocal microscopy imaging has been also used to visualize autophagic structures in plant cells, during developmental events, from tissue differentiation [2338] to senescence [2339], as well as in stress-treated barley microspores [911].
Thus, as with other systems, autophagosome formation in plants can be monitored through the combined use of fluorescent protein fusions to ATG8, immunolabeling and TEM (Figure 40). A tandem fluorescence reporter system is also available in Arabidopsis [2340]. The number of fluorescent ATG8-labeled vesicles can be increased by pretreatment with concanamycin A, which inhibits vacuolar acidification [1763, 2336]; however, this may interfere with the detection of MDC and LysoTracker™ Red. It is also possible to use plant and fungal homologs of SQSTM1 and NBR1 in Arabidopsis [1052, 2340] (the NBR1 homolog is called JOKA2 in tobacco [2341]) as markers for selective autophagy when constructed as fluorescent chimeras. In addition, detection of the NBR1 protein level by western blot, preferably accompanied by qPCR analysis of its transcript level, provides reliable semi-quantitative data about autophagic flux in plant cells [2342].
Figure 40.
Detection of autophagy in tobacco BY-2 cells. (A) Induction of autophagosomes in tobacco BY-2 cells expressing YFP-NtAtg8 (shown in green for ease of visualization) under conditions of nitrogen limitation (Induced). Arrowheads indicate autophagosomes that can be seen as a bright green dot. No such structure was found in cells grown in normal culture medium (Control). Bar: 10 µm. N, nucleus; V, vacuole. (B) Ultrastructure of an autophagosome in a tobacco BY-2 cell cultured for 24 h without a nitrogen source. Bar: 200 µm. AP, autophagosome; CW, cell wall; ER, endoplasmic reticulum; P, plastid. Image provided by K. Toyooka
Another approach for assessing autophagic flux is based on the observation that autophagy mutants in Arabidopsis exhibit peroxisomal abnormalities [2343, 2344]. Consequently, peroxisome abundance can provide information on autophagic flux. Peroxisome abundance can be measured in total tissue extracts by spectrofluorometry using the small fluorescent probe Nitro-BODIPY [2345, 2346]. This approach demonstrates that knockout of Arabidopsis ATG5 correlates with both a greater number of peroxisomes per cell and higher Nitro-BODIPY fluorescence in the total extracts from leaves [2345]. Although, low cost and ease of the procedure makes the Nitro-BODIPY assay applicable for the identification of autophagy mutants in large populations, direct markers should be used to examine autophagic flux in the identified genotypes.
Hydrotropism determines the degree of root bending towards the water source, which consequently compensates for the effects of drought. Hydrotropism modulates the development of the root system, and it has an effect on plant support, as well as water and nutrient intake. A water potential gradient system (using a water stress medium [WSM]) [2347] can be used to demonstrate that autophagy is required for the hydrotropic response. Looking for autophagosome accumulation in the root bending zone, 4-days-post germination 35S-ATG8A seedlings [378] are transferred to the WSM, and accumulation of autophagosomes is followed from 0 to 6 h by confocal microscopy using a 40X dry objective in order to avoid manipulation that may affect the root bending. During this time the root bending is achieved and autophagosome accumulation can be observed in the bending zone. Autophagosomes accumulate in the epidermal cells of the root bending zone 2 h after the transfer of seedlings to WSM. WSM supplemented with CQ can be used to monitor the requirement of autophagy flux. Several ATG mutants do not show hydrotropic curvature in WSM [2348]. Thus, the WSM system also allows the observation of autophagosomes in situ using confocal microscopy without seedling manipulation.
It has been assumed that, just as in yeast, autophagic bodies are found in the vacuoles of plant cells, because both microautophagy and autophagy are detected in these cells [2349]. The data supporting this conclusion are mainly based on EM studies showing vesicles filled with material in the vacuole of the epidermis cells of Arabidopsis roots; these vesicles are absent in ATG4A and ATG4B mutant plants [378]. However, it cannot be excluded that these vacuolar vesicles are in fact cytoplasmic/protoplasmic strands, or that they arrived at the vacuole independent of autophagy; although the amount of such strands would not be expected to increase following treatment with concanamycin. Immunolabeling with an antibody to detect ATG8 could clarify this issue.
The Phytophthora infestans RXLR effector PexRD54 has been published as an inducer of ATG8CL autophagosome formation and can be used in N. benthamiana as a tool to transiently activate autophagy [2350]. ATG4 and ATG9 RNAi constructs can also be used to knock down gene expression of the core autophagy components and transiently suppress autophagy in N. benthamiana [2351].
Other methods described throughout these guidelines can also be used in plants [2352]. For example, in tobacco cells cultured in sucrose starvation medium, the net degradation of cellular proteins can be measured by a standard protein assay; this degradation is inhibited by 3-MA and E-64c (an analog of E-64d), and is thus presumed to be due to autophagy [1862, 2353].
Cautionary notes: Although the detection of vacuolar RFP can be applied to both plant cell lines and to intact plants, it is not practical to measure RFP fluorescence in intact plant leaves, due to the very high red autofluorescence of chlorophyll in the chloroplasts. Furthermore, different autophagic induction conditions cause differences in protein synthesis rates; thus, special care should be taken to monitor the efficiency of autophagy by quantifying the intact and processed cargo proteins.
19. Protists
An essential role of autophagy during the differentiation of some parasitic protists (formerly called protozoa) is clearly emerging. Only a few of the known ATG genes are present in these organisms, which raises the question about the minimal system that is necessary for the normal functioning of autophagy. The reduced complexity of the autophagic machinery in many protists provides a simplified model to investigate the core mechanisms of autophagosome formation necessary for selective proteolysis; accordingly, protist models have the potential to open a completely new area in autophagy research. Some of the standard techniques used in other systems can be applied to protists including indirect immunofluorescence using antibodies generated against ATG8 and the generation of stable lines expressing mCherry- or GFP-fused ATG8 for live microscopy and immuno-TEM analyses. Extrachromosomal constructs of GFP-ATG8 also work well with less complex eukaryotes [384, 385, 2354], as do other fluorescently-tagged ATG proteins including ATG5 and ATG12.
The unicellular amoeba D. discoideum provides another useful system for monitoring autophagy [1940, 2355]. The primary advantage of D. discoideum is that it has a unique life cycle that involves a transition from a unicellular to a multicellular form. Upon starvation, up to 100,000 single cells aggregate by chemotaxis and form a multicellular structure that undergoes morphogenesis and cell-type differentiation. Development proceeds via the mound stage, the tipped aggregate and a motile slug, and culminates with the formation of a fruiting body that is composed of a ball of spores supported by a thin, long stalk made of vacuolized dead cells. Development is dependent on autophagy and, at present, all of the generated mutants in D. discoideum autophagy genes display developmental phenotypes of varying severity [628, 2356]. D. discoideum is also a versatile model to study infection with human pathogens and the role of autophagy in the infection process. The susceptibility of D. discoideum to microbial infection and its strategies to counteract pathogens are similar to those in more complex eukaryotes [1460, 2357]. Along these lines, D. discoideum utilizes some of the proteins involved in autophagy that are not present in S. cerevisiae including ATG101 and VMP1, in addition to the core Atg proteins. The classical markers GFP-ATG8 and GFP-ATG18 can be used to detect autophagosomes by fluorescence microscopy [64]. Flux assays based on the proteolytic cleavage of cytoplasmic substrates are also available [40, 422].
One cautionary note with regard to the use of GFP-ATG8 in protists is that these organisms display some “nonclassical” variations in their ATG proteins (see LC3-associated apicoplast) and possibly a wide phylogenetic variation because they constitute a paraphyletic taxon [2358]. For example, Leishmania contains many apparent ATG8-like proteins (the number varying per species; e.g., up to 25 in L. major) grouped in four families, but only one labels true autophagosomes even though the others form puncta [384], and ATG12 requires truncation to provide the C-terminal glycine before it functions in the canonical manner. Unusual variants in protein structures also exist in other protists, including apicomplexan parasites, for example, the malaria parasite Plasmodium spp. or T. gondii, which express ATG8 with a terminal glycine not requiring cleavage to be membrane associated [2359]. Thus, in each case care needs to be applied and the use of the protein to monitor autophagy validated. In addition, due to possible divergence in the upstream signaling kinases, classical inhibitors such as 3-MA or wortmannin, or inducers such as rapamycin, must be used with caution. Although they are not as potent for T. brucei [2360] or apicomplexan parasites as in mammalian cells or yeast (I. Coppens, personal communication) [2354]; they are efficient for T. cruzi [2361]. Likewise, RNAi knockdown of TORC1 (e.g., TOR1 or RPTOR) is effective in inducing autophagy in trypanosomes. Conversely, the inhibitory effect of bafilomycin A1 on trypanosome autophagy seems to occur during formation, resulting in a low number of ATG8-positive compartments, in contrast to what occurs in mammalian cells [2361, 2362]. In addition, small molecule inhibitors of the protein-protein interaction of ATG8 and ATG3 in Plasmodium falciparum have been discovered that are potent in cell-based assays and useable at 1-10 µM final concentration [2363, 2364]. Note that although the lysosomal protease inhibitors E64 and pepstatin block lysosomal degradative activity in Plasmodium, these inhibitors do not affect ATG8 levels and associated structures, suggesting a need for alternate methodologies to investigate autophagy in this model system [2365].
In conventional autophagy, the final destination of autophagosomes is their fusion with lysosomes for intracellular degradation. However, certain stages of Plasmodium (insect and hepatic) lack degradative lysosomes, which makes questionable the presence of canonical autophagosomes and a process of autophagy in this parasite. Nevertheless, if protists employ their autophagic machineries in unconventional manners, studies of their core machinery of autophagy will provide information as to how autophagy has changed and adapted through evolution. For example, although lysosome-like structures were not observed initially in the apicomplexa T. gondii, it is now clear that this protist harbors an organelle, named the vacuolar compartment/VAC or plant-like vacuole/PLV, with the characteristics of an acidic degradative compartment similar to lysosomes [2366, 2367]. Autophagic markers, such as the T. gondii ortholog of ATG8 and ATG9, colocalize with the vacuolar compartment markers CPL and CRT, indicating that in T. gondii autophagosomes fuse with this lysosome-like organelle [2368, 2369]. The ability of T. gondii to sustain prolonged extracellular stress relies on a functional autophagic machinery, although autophagy is dispensable for tachyzoite intracellular growth in normal in vitro culture conditions [2368]. The chronic form of this parasite, the bradyzoite stage, requires a basal autophagy flux for survival also when intracellular, perhaps because of reduced access to host cell nutrient due to the thick wall surrounding the vacuole containing the bradyzoites [2369, 2370].
The scuticociliate Philasterides dicentrarchi has proven to be a good experimental organism for identifying autophagy-inducing drugs or for autophagy initiation by starvation-like conditions, because this process can be easily induced and visualized in this ciliate [2371]. In scuticociliates, the presence of autophagic vacuoles can be detected by TEM, fluorescence microscopy or confocal laser scanning microscopy by using dyes such as MitoTracker® Deep Red FM and MDC.
Finally, a novel autophagy event has been found in Tetrahymena thermophila, which is a free-living ciliated protist. A remarkable, virtually unique feature of the ciliates is that they maintain spatially differentiated germline and somatic nuclear genomes within a single cell. The germline genome is housed in the micronucleus, while the somatic genome is housed in the macronucleus. These nuclei are produced during sexual reproduction (conjugation), which involves not only meiosis and mitosis of the micronucleus and its products, but also degradation of some of these nuclei as well as the parental old macronucleus. Hence, there should be a mechanism governing the degradation of these nuclei. The inhibition of PtdIns3Ks with wortmannin or LY294002 results in the accumulation of additional nuclei during conjugation [2372]. During degradation of the parental old macronucleus, the envelope of the nucleus becomes MDC- and LysoTracker™ Red-stainable without sequestration of the nucleus by a double membrane, and with the exposure of certain sugars and PS on the envelope [2373]. Subsequently, lysosomes fuse only to the old parental macronucleus, but other co-existing nuclei such as developing new macro- and micronuclei are unaffected [2373]. Using gene technology, it has been shown that ATG8 and VPS34 play critical roles in nuclear degradation [1330, 2373]. Knockout mutations of the corresponding genes result in a block in nuclear acidification, suggesting that these proteins function in lysosome-nucleus fusion. In addition, the envelope of the nucleus in the VPS34 knockout mutant does not become stainable with MDC. This evidence suggests that selective autophagy may be involved in the degradation of the parental macronucleus and implies a link between VPS34 and ATG8 in controlling this event. In Trypanosoma cruzi, there is a complex consisting of the PtdIns3K TcVPS34 and the serine-threonine kinase TcVPS15, which participates in autophagy. It has also been observed that TcVPS34 participates in fundamental processes for T. cruzi such as endocytosis, osmoregulation and acidification [387, 2374].
20. Rainbow trout
Salmonids (e.g., salmon, rainbow trout) experience long periods of fasting often associated with seasonal reductions in water temperature and prey availability or spawning migrations. As such, they represent an interesting model system for studying and monitoring the long-term induction of autophagy. Moreover, the rainbow trout (Oncorhynchus mykiss) displays unusual metabolic features that may allow us to gain a better understanding of the nutritional regulation of this degradative system (i.e., a high dietary protein requirement, an important use of amino acids as energy sources, and an apparent inability to metabolize dietary carbohydrates). It is also probably one of the most deeply studied fish species with a long history of research carried out in physiology, nutrition, ecology, genetics, pathology, carcinogenesis and toxicology [2375]. Its relatively large size compared to model fish, such as zebrafish or medaka, makes rainbow trout a particularly well-suited alternative model to carry out biochemical and molecular studies on specific tissues or cells that are impossible to decipher in small fish models. The genomic resources in rainbow trout are now being extensively developed; a high-throughput DNA sequencing program of ESTs has been initiated associated with numerous transcriptomics studies [2376–2379], and the full genome sequence is now available.
Most components of the autophagy and associated signaling pathways (AKT, TOR, AMPK, FOXO) are evolutionarily conserved in rainbow trout [952, 2380–2382]; however, not all ATG proteins and autophagy-regulatory proteins are detected by the commercially available antibodies produced against their mammalian orthologs. Nonetheless, the EST databases facilitate the design of targeting constructs. For steady-state measurement, autophagy can be monitored by western blot or by immunofluorescence using antibodies to Atg8-family proteins [2380]. Flux measurements can be made in a trout cell culture model (e.g., in primary culture of trout myocytes) by following Atg8-family protein turnover in the absence and presence of bafilomycin A1. It is also possible to monitor the mRNA levels of ATG genes by qPCR using primer sequences chosen from trout sequences available in the above-mentioned EST database. A major challenge in the near future for this model will be to develop the use of RNAi-mediated gene silencing to analyze the role of some signaling proteins in the control of autophagy, and also the function of autophagy-related proteins in this species.
21. Retinal pigment epithelium
The retinal pigment epithelium (RPE) is a single polarized layer of cells that form the outer blood retinal barrier and play a central role in maintaining metabolic homeostasis in the outer retina through transport of nutrients and waste products. These terminally differentiated cells also phagocytose lipid and protein-rich photoreceptor outer segments (POS) derived from the underlying photoreceptor cells on a daily basis. RPE develops a well-characterized autophagy-lysosomal system as well as a LAP pathway that ensures the turnover and degradation of cell content, and the daily degradation of ingested POS lipids, respectively [2383]. The RPE may be the only example in which autophagy and LAP are regulated in a light- and circadian-dependent manner that is postulated to occur through RUBCN [2384]. Immuno-histochemical, biochemical and TEM studies make it possible to monitor both circadian and age-related changes in autophagy activity in mouse models [2385–2389]. Moreover, lipidomic and metabolism studies have highlighted the critical role played by LC3-associated processes in RPE health and photoreceptor function [2390]. Lowering of lysosomal pH in diseased cells through pharmacological means or using acidic nanoparticles can enhance autophagic turnover [2391–2393]. The P2Y12 antagonist ticagrelor can reduce loss of photoreceptors and visual function when added to food; the decreased lysosomal pH and autofluorescent lipofuscin waste are consistent with enhanced lysosomal function [2394, 2395].
Autophagy plays an important role in maintaining retinal functions. Excessive upregulation of autophagy or depletion of key proteins for autophagy will disrupt functions of photoreceptor cells. Haploinsufficiency of TUBGCP4 (tubulin, gamma complex associated protein 4) impairs assembly of TUBG/γ-tubulin ring complexes and disturbs autophagy homeostasis of the retina. TUBGCP4 can inhibit autophagy by competing with ATG3 to interact with ATG7, thus interfering with lipidation of LC3B. Both cytoplasmic and nuclear autophagy have been observed in photoreceptor cell segments [2396, 2397].
POS phagocytosis shares functional similarity with efferocytosis, the ingestion and degradation of dead cell corpses (or apoptotic cells). On a molecular level, both processes rely on PS as an “eat me” signal [245, 2398]. Upon ingestion, both dead cells and POS stimulate the recruitment of LC3B via LAP [245, 2389, 2399, 2400]. The extent of LC3B association with phagosomes in the RPE remains unclear, and the percent of LAPosomes is an open question; in vitro [2389] and in vivo studies [2390, 2401] suggest that ~ 30-45% of ingested phagosomes are LC3B positive. In those studies, the levels of endogenous LC3B associated with OPN (opsin)-positive phagosomes were analyzed. Higher percentages are observed when GFP-LC3B is expressed in vitro in ARPE19 cells (between 80-90%) or in GFP-LC3B mice overexpressing this tag [2400] where almost 90% of OPN-containing structures are also GFP-LC3B positive. Further studies using DQ-BSA quantified the extent of LAPosome-lysosome association in vitro [2388]. An assessment of LAPosome levels in models of age-related retinal disease would provide valuable insight into the balance between two LC3-requiring processes—stress-mediated autophagosome formation and OS degradation.
Aberrant MTORC1 signaling has been implicated in aging and age-related degeneration of the human RPE [2387]. The phagocytosed POS serve as a physiological stimulus of MTORC1 activation through lysosome-independent mechanisms in the RPE [2402]. Whereas synchronized photoreceptor disk shedding and RPE phagocytosis activate MTORC1 during the morning burst, this is subsequently followed by MTORC1 inactivation and maintenance of retinal homeostasis. Reports suggest that excessive and sustained activation of MTORC1 in response to stress and independent of nutrient stimulation, leads to RPE cell death and senescence. Furthermore, genetic ablation of RPE mitochondrial oxidative phosphorylation in mice activates the AKT-MTOR pathway leading to dedifferentiation and hypertrophy of the RPE [2386], suggesting that inhibition of MTORC1 could protect the RPE against chronic metabolic stress and acute oxidative stress. It is well known that proteins of the autophagic machinery in the RPE participate in POS trafficking through a non-canonical autophagy pathway independent of the ULK1 complex, namely LAP [2400]. These studies showed that the MTORC1-independent interplay between autophagy and phagocytosis in the RPE is critical for POS degradation.
The cancer stem cell biomarker PROM1/CD133 (prominin 1), was demonstrated to play a critical role in maintaining RPE homeostasis through regulation of autophagy flux [2403]. Whereas overexpression of PROM1 increases autophagy flux, genomic deletion of PROM1 (using CRISPR-Cas9) in the RPE blocks autophagy through both upstream activation of MTORC1/2 and downstream disruption of a macromolecular complex involving PROM1, SQSTM1, and HDAC6 in the forming autophagosome. These findings have important implications because defective autophagosomal-lysosomal-phagocytic pathways can lead to ineffective clearance of POS and damaged organelles, all of which have been linked to the pathogenesis of retinal diseases, including age-related macular degeneration/AMD. Therefore, PROM1-mediated targeting of MTORC1/2 signaling in the RPE, could provide a therapeutic strategy for retinal degenerative diseases.
22. Sea urchin
Sea urchin embryo is an appropriate model system for studying and monitoring autophagy and other defense mechanisms activated during physiological development and in response to stress [1552]. This experimental model offers the possibility of detecting LC3 through both western blot and immunofluorescence in situ analysis. Furthermore, in vivo staining of autolysosomes with acidotropic dyes can also be carried out. Studies on whole embryos make it possible to obtain qualitative and quantitative data for autophagy and also to get information about spatial localization aspects in cells that interact among themselves in their natural environment. Furthermore, because embryogenesis of this model system occurs simply in a culture of sea water, it is very easy to study the effects of inducers or inhibitors of autophagy by adding these substances directly into the culture. Exploiting this potential, it has recently been possible to understand the functional relationship between autophagy and apoptosis induced by cadmium stress during sea urchin development. In fact, inhibition of autophagy by 3-MA results in a concurrent reduction of apoptosis; however, using a substrate for ATP production, methyl pyruvate, apoptosis (assessed by TUNEL assay and cleaved CASP3 immunocytochemistry) is substantially induced in cadmium-treated embryos where autophagy is inhibited. Therefore, autophagy could play a crucial role in the stress response of this organism because it could energetically contribute to apoptotic execution through its catabolic role [2404]. Cautionary notes include the standard recommendation that it is always preferable to combine molecular and morphological parameters to validate the data.
23. Ticks
In the hard tick Haemaphysalis longicornis, endogenous autophagy-related proteins (Atg6 and Atg12) can be detected by western blotting and/or by immunohistochemical analysis of midgut sections [2405, 2406]. It is also possible to detect endogenous Atg3 and Atg8 by western blotting using antibodies produced against the H. longicornis proteins (R. Umemiya-Shirafuji, unpublished results). Commercial antibodies against mammalian ATG orthologs (ATG3, ATG5, and BECN1) can also be used for western blotting. However, when the tick samples include blood of a host animal, the animal species immunized with autophagy-related proteins should be checked before use to avoid nonspecific background cross-reactivity.
In addition to these methods, TEM is recommended to detect autophagosomes and autolysosomes. Although acidotropic dyes can be useful as a marker for autolysosomes in some animals, careful attention should be taken when using the dyes in ticks. Because the midgut epithelial cells contain acidic organelles (e.g., lysosomes) that are related to blood digestion during blood feeding, this method may cause confusion. It is difficult to distinguish between autophagy (autolysosomes) and blood digestion (lysosomes) with acidotropic dyes.
Another available monitoring method is to assess the mRNA levels of tick ATG genes by qPCR [2407, 2408]. However, this method should be used along with other approaches such as western blotting, immunostaining, and TEM as described in this article. Unlike model insects, such as Drosophila, powerful genetic tools to assess autophagy are still not established in ticks. However, RNAi-mediated gene silencing is now well established in ticks [2409], and is being developed to analyze the function of autophagy-related genes in ticks during nonfeeding periods (R. Umemiya-Shirafuji, unpublished results) and in response to pathogen infection. Recently, “omics” technologies such as transcriptomics and proteomics have been applied to the study of apoptosis pathways in Ixodes scapularis ticks in response to infection with Anaplasma phagocytophilum [2410]. I. scapularis, the vector of Lyme disease and human granulocytic anaplasmosis, is the only tick species for which genome sequence information is available (assembly JCVI_ISG_i3_1.0; http://www.ncbi.nlm.nih.gov/nuccore/NZ_ABJB000000000). For related tick species such as I. ricinus, mapping to the I. scapularis genome sequence is possible [2411], but for other tick species more sequence information is needed for these analyses.
24. Zebrafish (Danio rerio)
Zebrafish have many characteristics that make them a valuable vertebrate model organism for the analysis of autophagy. For example, taking advantage of the transparency of embryos, autophagosome formation can be visualized in vivo during development using transgenic GFP-Lc3 and GFP-Gabarap fish [44, 2412, 2413] and in specific cell types such as neurons [328]. It has been reported that conventional anti-pigmentation strategies including 1-phenyl-2-thiourea/PTU treatment and genetic targeting of TYR (tyrosinase) induce autophagy in various tissues; however, in vivo visualization of later-stage embryos can still be performed using light-sheet fluorescence microscopy, and image quality is only minimally affected by developed pigments (X.K. Chen, J.S. Kwan, R.C. Chang and A.C. Ma, in press). Lysosomes can also be readily detected in vivo by the addition of LysoTracker™ Red to fish media prior to visualization. Additionally, protocols have been developed to monitor the rate of autophagosome accumulation in vivo [328], and Lc3 protein levels and conjugation to PE by western blot analysis using commercially available Lc3 antibodies [44, 388]. It should be noted that in addition to Lc3-I and Lc3-II, a third, lower-sized protein product is frequently evident following western blot analysis in zebrafish [2414].
Because of their translucent character and external fertilization and development, zebrafish have proven to be an exceptional choice for developmental research. In situ hybridization of whole embryos can be performed to determine expression patterns. Knockdown of gene function is performed by treatment with morpholinos; the core autophagy machinery proteins Gabarap [2415], Atg5 [2416, 2417] and Atg13 [2418], and regulatory proteins such as the phosphoinositide phosphatase Mtmr14 [2419], Rubcn [2418], Raptor and Mtor [2420], have all been successfully knocked down by morpholino treatment. However, a number of papers have raised concerns about the cellular stress pathway inducing, off-target effects of this approach [2421–2423], therefore, validation of these phenotypes in bona fide mutants is necessary. The CRISPR-Cas9 system has been used for efficient targeted gene deletions of Epg5, Sqstm1, Optn and Snap29 [2424–2426] and should continue to be of great help in future analyses [2427].
It is well known that the aquatic environment is frequently compromised by the action of chemical substances and/or their metabolites. According to a study that applied a computational model for investigating biocidal compounds, approximately 50–60% of those substances are highly toxic for different aquatic compartments and organisms [2428]. For this reason, zebrafish are ideal organisms for in vivo drug discovery and/or verification because of their relatively small size allowing easy handling, and several chemicals have been identified that modulate zebrafish autophagy activity [388]. Many chemicals can be added to the media and are absorbed directly through the skin. Because of simple drug delivery and rapid embryonic development, zebrafish are a promising organism for the study of autophagy’s role in disease including HD [1951], AD [2429], PD [2430] and myofibrillar myopathy [2431–2433]. In the case of infection, studies in zebrafish have made important contributions to understanding the role of bacterially- [2418, 2425, 2434, 2435] and virally [2436–2438]-induced autophagy. In vivo zebrafish studies have also contributed to understanding the role of autophagy in different aspects of development, including cardiac morphogenesis, caudal fin regeneration [2439], and muscle and brain development [2412, 2440, 2441].
In vitro studies in the zebrafish cell line ZF4 (zebrafish embryonic fibroblast) [2442] show that autophagy is required for fish rhabdovirus (spring viremia of carp virus, SVCV) replication [1351, 2443]. In fact, several standardized autophagy blockers (also including cholesterol-related molecules such as C-reactive protein, 25-hydroxycholesterol, methyl-beta-cyclodextrin and cholesterol itself) inhibit SVCV infectivity in this cell line [1351, 2444]. Moreover, the glycoprotein G of viral hemorrhagic septicemia (rhabdo)virus/VHSV and SVCV induce a cell’s antiviral autophagic program in ZF4 cells [2438, 2443]. In this regard, autophagy is also induced in GFP-LC3 transgenic zebrafish that are experimentally infected with SVCV [2436].
E. Noncanonical use of autophagy-related proteins
Multiple components of the autophagy machinery mediate non-autophagic functions [1797], as described here below.
1. LC3-associated phagocytosis (LAP)
Although the lipidation of LC3 to form LC3-II is a commonly used marker of autophagy, studies have established that LC3-II can also be present on phagosomes, acting to promote maturation independently of traditional autophagy, in a noncanonical autophagic process termed LC3-associated phagocytosis [2, 30, 2445, 2446]. LAP requires RUBCN and occurs upon engulfment of particles (such as dead cells, and pathogens including Aspergillus fumigatus, Burkholderia pseudomallei, Bacteroides fragilis, and Yersinia pestis) that engage a receptor-mediated signaling pathway, resulting in the recruitment of some but not all of the autophagic machinery to the phagosome. LAP requires the association of RUBCN with the UVRAG-containing class III PtdIns3K complex, and it facilitates generation and localization of PtdIns3P. This PtdIns3P then binds and stabilizes the CYBB/NOX2/gp91phox complex resulting in ROS production for processing the engulfed cargo [245, 801, 2447]. These autophagic components facilitate rapid phagosome maturation and degradation of engulfed cargo, and play roles in the generation of signaling molecules and regulation of immune responses [244, 245, 904, 2448, 2449]. LAP thus represents a unique process that marries the ancient pathways of phagocytosis and autophagy.
Despite overlap in molecular machinery, there currently exist several criteria by which to differentiate LAP from autophagy: (a) Whereas LC3-decorated autophagosomes can take hours to form, LC3 can be detected on LAP-engaged phagosomes as early as 10 min after phagocytosis, and PtdIns3P can also be seen at LAP-engaged phagosomes minutes after phagocytosis [245, 247, 2448]. (b) EM analysis reveals that LAP involves single-membrane structures [247]. In contrast, autophagy is expected to generate double-membrane structures surrounding cargo. However, this can be confusing if the engulfed structure already possesses a membrane before engulfment, as in the case of cell corpses [244, 2450, 2451]. (c) Whereas most of the core autophagy components are required for LAP, the two processes can be distinguished by the involvement of the pre-initiation complex. RB1CC1, ATG13, ULK1 and ULK2 are dispensable for LAP, which provides a convenient means for distinguishing between the two processes [245, 2448]. (d) LAP requires the WD repeats of ATG16L1, whereas autophagy does not have this requirement [1477, 1478]. (e) LAP involves LC3 recruitment in a manner that requires ROS production by the NADPH oxidase family, notably CYBB/NOX2/gp91phox. It should be noted that most cells express at least one member of the NADPH oxidase family. Silencing of the common subunits, CYBB or CYBA/p22phox, is an effective way to disrupt NADPH oxidase activity and therefore LAP. It is anticipated that more specific markers of LAP will be identified as this process is further characterized. (f) In human macrophages infected with Mycobacterium tuberculosis, MORN2 (MORN repeat containing 2) is recruited at the phagosome membrane containing M. tuberculosis to induce the recruitment of LC3, and subsequent maturation into phagolysosomes. In addition, MORN2 drives trafficking of M. tuberculosis to a single-membrane compartment. Thus, in certain conditions, MORN2 can be used to help to make the distinction between autophagy and LAP [2452].
Of note, an ATG5- and CTSL-dependent cell death process has been reported that can be activated by the small molecule NID-1; this process depends on PtdIns3K signaling, generates LC3B puncta and single-membrane vacuoles, and results in the clearance of SQSTM1. Thus, LAP and/or related processes can be co-opted to cause cell death in some cases [2453].
A very similar process to LAP occurs during the cell cannibalism process of entosis. After engulfment of an epithelial cell by a neighboring cell, LC3 is recruited on the single membrane entotic vacuole before lysosome fusion and death of the inner cell [244]. It is worth noting that many lysosomotropic compounds, including CQ, activate a LAP-like noncanonical autophagy pathway that drives LC3 lipidation on endolysosomal membranes and potentially interferes with the interpretation of LC3 lipidation data [2454, 2455]. In a zebrafish model, LAP in macrophages is important in clearing intracellular bacteria such as Salmonella [2418]. LAP-like non-canonical autophagy is also observed in pancreatic acinar cells and involves LC3-conjugation to the membrane of endocytic vacuoles (organelles formed as a consequence of compound exocytosis followed by compensatory membrane retrieval) [760].
Mouse models have also been developed to study LAP in vivo. RUBCN stabilizes the CYBA/p22PHOX-CYBB/NOX2/gp91phox complex during LAP [2456] allowing ROS to induce binding of ATG16L1 to endo-lysosome membranes [801]. rubcn−/− mice [2457] have systemic loss of LAP and have been useful as a source of LAP-deficient cells for “in vitro” studies, and for “in vivo” studies of autoimmunity and β-amyloid trafficking [2457, 2458]. RUBCN is a multidomain adaptor protein that suppresses NFKB signaling and pro-inflammatory responses [2456]. Exaggerated proinflammatory responses mean that rubcn−/− mice are difficult to use in infection studies. The mice also fail to gain weight and have defects in the clearance of dying and apoptotic cells, leading to autoimmune disease that resembles systemic lupus erythematosus [2457]. An alternative approach to the study of LAP “in vivo” has targeted pathways downstream of RUBCN [2459]. LAP and autophagy require the E3-ligase like activity of the ATG12–ATG5-ATG16L1 complex, but conjugation of LC3 to endo-lysosome membranes during LAP requires the WD domain of ATG16L1 [1478]. This has led to the generation of mice lacking the WD and linker domain of ATG16L1, which have been developed to study the role played by non-canonical autophagy in vivo. These mice have systemic loss of LAP and LC3-associated endocytosis (termed LANDO) [2459], but retain the N-terminal CCD and ATG5-binding domains of ATG16L1 required for conventional autophagy. This allows the mice to activate autophagy, grow normally and maintain tissue homeostasis. These mice also maintain inflammatory and immunological homeostasis and can be used to study the role played by LAP and LANDO during infection in vivo.
2. LC3-associated apicoplast
In several important parasitic protists of the phylum Apicomplexa (e.g., T. gondii and Plasmodium spp.), the single ATG8 homolog localizes to an endosymbiotic nonphotosynthetic plastid, called the apicoplast [2359, 2460–2463]. This organelle is the product of a secondary endosymbiotic event, by which a red alga was endocytosed by an auxotrophic eukaryote (the ancestor Apicomplexa); the apicoplast is the main remnant of this red alga. This organelle is approximately 300 nm in diameter, and is composed of four membranes that trace their ancestry to three different organisms. The successive endosymbiotic events that led to its incorporation into an ancestor of the Apicomplexa imply that its outermost membrane could be of phagosomal origin, although it might also have incorporated elements of the host ER. It is possible that ATG8-containing vesicles are generated from apicoplastic membranes to form phagophores, as evidenced in Plasmodium liver forms. Interestingly, it has been shown that in a parasite strain of Plasmodium overexpressing ATG8, the apicoplast forms an abnormally large, reticulate network that ultimately collapses, leading to poorly infectious parasites [2464]. This finding suggests that ATG8 may supply the apicoplast with lipids, controlling the maintenance and homeostasis of this organelle. On the apicoplast of T. gondii, ATG8 plays a role in the centrosome-mediated inheritance of the organelle in daughter cells during parasite division, which highlights unconventional functions of ATG8 in protists [2465]. Interestingly, both ATG8 and PtdIns3P-binding PROPPINs of the WIPI/Atg18 family are essential for apicoplast function [2466, 2467]. Because of this peculiar ATG8 localization and potential morphological similarities between the multi-membrane apicoplast and stress-induced autophagosomes, caution must be taken when identifying these structures by electron microscopy or by fluorescence microscopy with ATG8 labeling in these parasites.
3. LC3 conjugation system for IFNG-mediated pathogen control
Similar to LAP, LC3 localizes on the parasitophorus vacuole membrane (PVM) of T. gondii [246]. The parasitophorus vacuole is a vesicle-like structure formed from host plasma membrane during the invasion of T. gondii, and it sequesters and protects the invasive T. gondii from the hostile host cytoplasm. The cell-autonomous immune system uses IFNG-induced effectors, such as immunity-related GTPases and guanylate binding proteins (GBPs), to attack and disrupt this type of membrane structure; consequently, naked T. gondii in the cytoplasm are killed by a currently unknown mechanism. Intriguingly, proper targeting of these effectors onto the PVM of T. gondii requires the autophagic ubiquitin-like conjugation system, including ATG7, ATG3, and the ATG12–ATG5-ATG16L1 complex [2468], although the necessity of LC3-conjugation itself for the targeting is not yet clear [2469]. In contrast, up- or downregulation of canonical autophagy using rapamycin, wortmannin, or starvation do not significantly affect the IFNG-mediated control of T. gondii. Furthermore, the degradative function or other components of the autophagy pathway, such as ULK1/2 and ATG14, are dispensable. Many groups have confirmed the essential nature of the LC3-conjugation system for the control of T. gondii [2470–2472], and the same or a similar mechanism also functions against other pathogens such as murine norovirus and Chlamydia trachomatis [1958, 2470]. Although topologically and mechanistically similar to LAP, the one notable difference is that the parasitophorous vacuole of T. gondii is actively made by the pathogen itself using host membrane, and the LC3-conjugation system-dependent targeting happens even in nonphagocytic cells. GBP-mediated lysis of pathogen-containing vacuoles is important for the activation of noncanonical inflammasomes [2473], but the targeting mechanism of GBPs to the vacuoles is unknown. Considering the necessity of the LC3-conjugation system to target GBPs to the PVM of T. gondii, this system may play crucial roles in the general guidance of various effector molecules to target membranes, as well as in selective phagophore-dependent sequestration, phagophore membrane expansion and autophagosome maturation.
4. Intracellular trafficking of bacterial pathogens
Some ATG proteins are involved in the intracellular trafficking and cell-to-cell spread of bacterial pathogens by noncanonical autophagic pathways. For example, ATG9 and WIPI1, but not ULK1, BECN1, ATG5, ATG7 or LC3B are required for the establishment of an endoplasmic reticulum-derived replicative niche after cell invasion with Brucella abortus [2474]. In addition, the cell-to-cell transmission of B. abortus seems to be dependent on ULK1, ATG14 and PIK3C3/VPS34, but independent of ATG5, ATG7, ATG4B and ATG16L1 [2475].
5. Exocytosis with LC3-associated membranes
The Atg8-family protein lipidation machinery is also involved in non-canonical secretion and exocytosis of extracellular vesicles [105, 106]. This role has been initially described for yeast Acb1 [2476, 2477] and IL1B and CFTR in more complex eukaryotes [1700, 2478, 2479]. In addition to Atg8-family protein lipidation, this pathway seems to require Golgi reassembly-stacking proteins (GORASPs) and components of ESCRT complexes [2480, 2481]. The associated release of extracellular vesicles with Atg8-family protein-conjugated membranes is also hijacked by viruses for their efficient exocytosis [2482–2484]. In the filamentous fungus Aspergillus nidulans, a protein denoted AN4171/BapH (BAR- and PH domain-containing) is an effector of RAB11 that binds PtdIns(4,5)P2 and localizes to exocytic membranes. In mutants lacking BapH, basal autophagy under nitrogen-replete conditions is increased, suggesting that it acts as a liaison between exocytosis/endocytic recycling and autophagy [2200].
6. Other processes
ATG proteins are involved in various other nonautophagic processes, particularly apoptosis, membraneless organelle dynamics, COPII-mediated ER export, and noncanonical protein secretion, as discussed in various papers [31, 105, 106, 818, 858, 2399, 2448, 2485–2489]. For example, ATG5 and RUBCN, but not RB1CC1, are required for LC3-associated endocytosis (LANDO), identified in microglial cells and the macrophage RAW264.7 cell line [2458], whereas the requirement of ATG5, RUBCN, and the lack of a requirement for RB1CC1 are well-established for the non-canonical function of autophagy proteins in the LAP pathway. LANDO is also required for the recycling of putative beta-amyloid receptors (CD36, TREM2, and TLR4) from internalized endosomes to the plasma membrane.
F. Interpretation of in silico assays for monitoring autophagy
The increasing availability of complete (or near-complete) genomes for key species spanning the eukaryotic domain provides a unique opportunity for delineating the spread of autophagic machinery components in the eukaryotic world [2490, 2491]. Fast and sensitive sequence similarity search procedures are already available; an increasing number of experimental biologists are now comfortable “BLASTing” their favorite sequences against the ever-increasing sequence databases for identifying putative homologs in different species [2492]. Nevertheless, several limiting factors and potential pitfalls need to be taken into account.
In addition to sequence comparison approaches, a number of computational tools and resources related to autophagy have become available online. All the aforementioned methods and approaches may be collectively considered as “in silico assays” for monitoring autophagy, in the sense that they can be used to identify the presence of autophagy components in different species and provide information on their known or predicted associations.
In the following sections we briefly present relevant in silico approaches, highlighting their strengths while underscoring some inherent limitations, with the hope that this information will provide guidelines for the most appropriate usage of these resources.
1. Sequence comparison and comparative genomic approaches
Apart from the generic shortcomings when performing sequence comparisons (discussed in ref. [2493]), there are some important issues that need to be taken into account, especially for autophagy-related proteins. Because autophagy components seem to be conserved throughout the eukaryotic domain of life, the deep divergent relations of key subunits may reside in the so called “midnight zone” of sequence similarity: i.e., genuine orthologs may share even less than 10% sequence identity at the amino acid sequence level [2494]. This is the case with autophagy subunits in protists [2495, 2496] and with other universally conserved eukaryotic systems, as for example the nuclear pore complex [2497]. This low sequence identity is especially pronounced in proteins that contain large intrinsically disordered regions [2498]. In such cases, sophisticated (manual) iterative database search protocols, including proper handling of compositionally biased subsequences and considering domain architecture may assist in eliminating spurious similarities or in the identification of homologs that share low sequence identity with the search molecule [2496–2498].
Genome-aware comparative genomics methods [2499] can also provide invaluable information on yet unidentified components of autophagy. However, care should be taken to avoid possible Next Generation Sequencing artefacts (usually incorrect genome assemblies): these may directly (via a similarity to a protein encoded in an incorrectly assembled genomic region) or indirectly (via propagating erroneous annotations in databases) give misleading homolog assignments [2500]. In addition, taking into account other types of high-throughput data available in publicly accessible repositories (e.g., EST/RNAseq data, expression data) can provide orthogonal evidence for validation purposes when sequence similarities are marginal [2497].
2. Web-based resources related to autophagy
A number of autophagy-related resources are now available online, providing access to diverse data types ranging from gene lists and sequences to comprehensive catalogs of physical and indirect interactions. In the following we do not attempt to review all functionalities offered by the different servers, but to highlight those that (a) offer possibilities for identifying novel autophagy-related proteins, or (b) characterize features that may link specific proteins to autophagic processes. Two comments regarding biological databases in general also apply to autophagy-related resources as well: (a) the need for regular updates, and (b) data and annotation quality. Nevertheless, these issues are not discussed further herein.
a. The THANATOS database
THANATOS (THe Apoptosis, Necrosis, AuTophagy OrchestratorS) is a comprehensive data resource developed by the CUCKOO Workgroup, which contains 191,543 proteins potentially associated with autophagy and cell death pathways in 164 eukaryotes [2501]. THANATOS was started from the manual collection of 4,237 experimentally identified proteins regulated in autophagy and cell death pathways from the literature, whereas potential orthologs of these known proteins were computationally detected. Besides sequence data, known PTMs, protein-protein interactions (PPIs) and functional annotations are also integrated. A simple web interface assists in data retrieval, using keyword searches, browsing by species and cell death type, performing BLAST searches with user-defined sequences, and by requesting the display of orthologs among predefined species. Using the data in THANATOS, an evolutionary analysis demonstrates that the machinery of the autophagy pathway is highly conserved across eukaryotes, whereas statistical analyses suggest human autophagy proteins are enriched among cancer gene products and drug targets. A reconstruction of a kinase-substrate phosphorylation network for ATG proteins supports a critical role of phosphorylation in regulating autophagy. The THANATOS database is publicly available online at the URL http://thanatos.biocuckoo.org/.
With the help of THANATOS, a network-based algorithm of in silico Kinome Activity Profiling/iKAP was designed to computationally infer protein kinases differentially regulated by two natural neuroprotective autophagy enhancers, corynoxine (Cory) and corynoxine B (Cory B) [2502]. This algorithm predicted and verified that two kinases, MAP2K2/MEK2 (mitogen-activated protein kinase kinase 2) and PLK1 (polo like kinase 1), are essential for Cory-induced autophagy to promote the clearance of AD-associated APP (amyloid beta precursor protein) and PD-associated SNCA/α-synuclein (synuclein alpha). The CUCKOO workgroup is mainly focused on PTM bioinformatics, and has developed fourteen PTM site predictors, five tools for biological data analysis, and twelve PTM-related databases at the URL http://www.biocuckoo.org/, including DeepPhagy (deep learning for autophagy) for quantitatively analyzing four types of autophagic phenotypes in Saccharomyces cerevisiae, including the vacuolar targeting of GFP-Atg8, the targeting of Atg1-GFP to the vacuole, the vacuolar delivery of GFP-Atg19, and the disintegration of autophagic bodies indicated by GFP-Atg8 [2503]. DeepPhagy was implemented in a 5-layer convolutional neural network framework, containing three connected convolutional blocks and two fully connected layers, and is freely available online at the URL http://deepphagy.biocuckoo.org/. This workgroup also developed CGDB, the Circadian Gene DataBase at URL http://cgdb.biocuckoo.org/ [2504] (see Clockophagy).
b. The human autophagy database (HADb)
The human autophagy database, developed in the Tumor Immunotherapy and Microenvironment (TIME) group at the Luxembourg Institute of Health, lists over 200 human genes/proteins related to autophagy [920]. These entries have been manually collected from the biomedical literature and other online resources. An update of the initially published list is currently underway. For each gene there exists information on its sequence, transcripts and isoforms (including exon boundaries) as well as links to external resources. HADb provides basic search and browsing functionalities and is publicly available online at the URL http://autophagy.lu/.
c. The autophagy database
The Autophagy Database is a multifaceted online resource providing information for proteins related to autophagy and their homologs across several eukaryotic species, with a focus on functional and structural data [2505]. It is developed by the National Institute of Genetics (Japan) under the Targeted Proteins Research Program of the Ministry of Education, Culture, Sports, Science and Technology (http://www.tanpaku.org/). This resource is regularly updated and as of August 2014 contained information regarding 312 reviewed protein entries; when additional data regarding orthologous/homologous proteins from more than 50 eukaryotes is considered, the total number of entries reaches approximately 9,000. In addition to the browse functionalities offered under the “Protein List” and the “Homologs” menus, an instance of the NCBI-BLAST software facilitates sequence-based queries against the database entries. Moreover, interested users may download the gene list or the autophagy dump files licensed under a Creative Commons Attribution-ShareAlike 2.1 Japan License. The Autophagy Database is publicly available online at the URL http://www.tanpaku.org/autophagy/index.html.
d. The Autophagy Regulatory Network (ARN)
Another addition to the web-based resources relevant to autophagy research is the Autophagy Regulatory Network (ARN), originally developed at the Eötvös Loránd University and Semmelweis University (Budapest, Hungary) in collaboration with the Quadram Institute and the Earlham Institute (Norfolk, UK). Maintenance and hosting the ARN resource is secured at The Genome Analysis Centre until at least 2022. ARN is an integrated systems-level resource aiming to collect and provide an interactive user interface enabling access to validated or predicted protein-protein, transcription factor-gene and miRNA-mRNA interactions related to autophagy in human [2506]. ARN contains data from 26 resources, including an in-house extensive manual curation, the dataset of a ChIP-MS study [658], ADB and ELM. As of June 2020, a total of more than 15,000 proteins and 800 miRNAs and lncRNAs are included in ARN, including 38 core autophagy proteins with more than 500,000 transcriptional, post-transcriptional and post-translational interactions. Importantly, all autophagy-related proteins are linked to major signaling pathways. A flexible—in terms of both content and format—download functionality enables users to locally use the ARN data under the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The autophagy regulatory network resource is publicly available online at the URL http://autophagyregulation.org.
e. Prediction of Atg8-family interacting proteins
Being central components of the autophagic core machinery, Atg8-family members (e.g., LC3 and GABARAP subfamilies in mammals) and their interactome have attracted substantial interest [658, 2507, 2508]. During the last decade, a number of proteins have been shown to interact with Atg8 homologs via a short linear peptide; depending on context, different research groups have described this peptide as the LIR [423], the LC3 recognition sequence (LRS) [1020], or the AIM [2509]. Two independent efforts resulted in the first online available tools for identification of these motifs (LIR-motifs for brevity) in combination with other sequence features, which may signify interesting targets for further validation (see below).
f. The iLIR server
The iLIR server is a specialized web server that scans an input sequence for the presence of a degenerate version of LIR, the extended LIR-motif (xLIR) [2510]. Currently, the server also reports additional matches to the “canonical” LIR motif (WxxL), described by the simple regular expression x(2)-[WFY]-x(2)-[LIV]. A position-specific scoring matrix (PSSM) based on validated instances of the LIR motif has also been compiled, demonstrating that many of the false positive hits (i.e., spurious matches to the xLIR motif) are eliminated when a PSSM score >15 is sought. In addition, iLIR also overlays the aforementioned results to segments that reside in or are adjacent to disordered regions and are likely to form stabilizing interactions upon binding to another globular protein as predicted by the ANCHOR package [2511]. A combination of an xLIR match with a high PSSM score (>13) and/or an overlap with an ANCHOR segment gives reliable predictions [2510]. It is worth mentioning that, intentionally, iLIR does not provide explicit predictions of functional LIR motifs but rather displays all the above information accompanied by a graphical depiction of query matches to known protein domains and motifs; it is up to the user to interpret the iLIR output. As mentioned in the original iLIR publication, a limitation of this tool is that it does not handle any noncanonical LIR motifs at present. The iLIR server was jointly developed by the University of Warwick and University of Cyprus and is freely available online at the URL http://repeat.biol.ucy.ac.cy/iLIR. A similar web-based AIM prediction tool termed high-fidelity AIM (hfAIM) was also developed by scientists at the Weizmann Institute and Ghent University [2512], and is freely available online at the URL http://bioinformatics.psb.ugent.be/hfAIM/.
iLIR database: Using the iLIR server, a database of putative LIR-containing proteins (LIRCPs) has been created. The iLIR database (https://ilir.warwick.ac.uk) lists all the putative canonical LIRCPs identified in silico in the proteomes of eight model organisms combined with a Gene Ontology/GO term analysis. Additionally, a curated text-mining analysis of the literature suggests novel putative LIRCPs in mammals that have not previously been associated with autophagy [2513].
iLIR@viral: The iLIR@viral database (http://ilir.uk/virus/) lists all the putative canonical LIR motifs identified in viral proteins, using the iLIR server. Curated text-mining analysis of the literature suggests the presence of novel putative LIRCPs in viruses [2514].
g. The Eukaryotic Linear Motif resource (ELM)
The Eukaryotic Linear Motif resource [2515] is a generic resource for examining functional sites in proteins in the form of short linear motifs, which have been manually curated from the literature. Sophisticated filters based on known (or predicted) query features (such as taxonomy, subcellular localization, structural context) are used to narrow down the results lists, which can be very long lists of potential matches due to the short lengths of ELMs. This resource has incorporated four entries related to the LIR-motif (since May 2014; http://elm.eu.org/infos/news.html), while another three are being evaluated as candidate ELM additions (Table 3). Again, the ELM resource displays matches to any motifs and users are left with the decision as to which of them are worth studying further. ELM is developed/maintained by a consortium of European groups coordinated by the European Molecular Biology Laboratory and is freely available online at the URL http://elm.eu.org.
h. Molecular modeling of interactions between Atg8-family proteins and LIR-containing proteins
The availability of several sets of experimental data on LIR-containing proteins, the 3-dimensional structure of their complexes, and sequence-based predictors such as iLIR [2513], has been providing the foundations to apply molecular modeling and simulations to the study of the complexes between members of the Atg8-family proteins and LIR-containing proteins. This class of methods can help autophagy research at different levels: i) to provide information on the role of the residues N- and C-terminal from the core LIR motifs for which coordinates are often missing in the available experimental structures; ii) as a guide for experiments to suggest the residue to mutate to validate structure-based hypotheses; iii) to provide a structure-based rationale of available experimental data and shed light on determinants of specificity towards different members of the Atg8-family proteins; and iv) to help in the identification of the best LIR-containing candidates for experimental validation in case of multi-domain proteins with several predicted LIRs [1258, 2516, 2517]. In approaching modeling and simulations studies of the Atg8-family protein-LIR complexes, it is important to have a careful design of the modeling and simulation protocol, selection of the physical model (i.e., force field) to employ to describe the complex structure and dynamics, and use, where possible, multiple models with different conformations of the LIR-containing region in the Atg8-family protein binding pockets to avoid limitation due to the sampling of the conformational space accessible to classical molecular dynamics simulations.
i. The ncRNA-associated cell death database (ncRDeathDB)
The noncoding RNA (ncRNA)-associated cell death database (ncRDeathDB) [2518], most recently developed at the Harbin Medical University (Harbin, China) and Shantou University Medical College (Shantou, China), documents a total of more than 4,600 ncRNA-mediated programmed cell death entries. Compared to previous versions of the miRDeathDB [2519–2521], the ncRDeathDB further collected a large amount of published data describing the roles of diverse ncRNAs (including microRNA, long noncoding RNA/lncRNA and small nucleolar RNA/snoRNA) in programmed cell death for the purpose of archiving comprehensive ncRNA-associated cell death interactions. The current version of ncRDeathDB provides an all-inclusive bioinformatics resource on information detailing the ncRNA-mediated cell death system and documents 4,615 ncRNA-mediated programmed cell death entries (including 1,817 predicted entries) involving 12 species, as well as 2,403 apoptosis-associated entries, 2,205 autophagy-associated entries and 7 necrosis-associated entries. The ncRDeathDB also integrates a variety of useful tools for analyzing RNA-RNA and RNA-protein binding sites and for network visualization. This resource will help researchers to visualize and navigate current knowledge of the noncoding RNA component of cell death and autophagy, to uncover the generic organizing principles of ncRNA-associated cell death systems, and to generate valuable biological hypotheses. The ncRNA-associated cell death interactions resource is publicly available online at the URL http://www.rna-society.org/ncrdeathdb.
j. Predicting impact for autophagy-related gene copy number alterations in cancer
Autophagy is tumor suppressive, yet can also exert pro-survival effects once tumors have been established. The HAPTRIG R tool developed at UCSD uses a curated data set of autophagy genes to predict the functional impact of increases and decreases of genes in the autophagy pathway in cancer [2522]. This tool is useful for determining deficiencies in autophagy among tumor types, as well as for individual tumors within a tumor type. The tool also can prioritize which genes most influence autophagy within a dataset based on protein-protein interactions and haploinsufficiency data. These prioritized genes can then be the subject of further experimentation. The Shiny application of this tool is available at the URL https://delaney.shinyapps.io/haptrig_single_pathway_networks.
k. KFERQ finder
There is a growing interest in studying CMA due to its fundamental regulatory role in the physiopathology of diverse cellular processes [2523]. Substrate selectivity is one of the main features of CMA, which relies on the recognition by HSPA8 of KFERQ-like motifs in the sequence of the proteins to be degraded [2524]. Therefore, a reliable, quick and high-throughput method has been developed to find these motifs. This tool (KFERQ finder) allows the identification of KFERQ-like motifs in any given protein of the human, mouse and rat proteomes using their Uniprot ID. Furthermore, multiple proteins can be analyzed uploading the Uniprot IDs in a .csv file, and, finally, the search can be also performed in protein sequences [1809]. The KFERQ finder is available at the URL http://tinyurl.com/kferq.
l. Autophagy to Disease (ATD)
Autophagy to Disease (ATD) is a comprehensive bioinformatics resource for deciphering the association of autophagy and diseases. The Liao group developed ATD (http://auto2disease.nwsuaflmz.com) to archive autophagy-associated diseases. This resource provides a bioinformatics annotation system about genes, chemicals, autophagy and human diseases by extracting results from previous studies with text mining technology. Based on ATD, some classes of disease tend to be related with autophagy, including respiratory diseases, cancer, urogenital diseases and digestive system diseases. In addition, some classes of autophagy-related diseases have a strong association among each other and constitute modules. Furthermore, by extracting autophagy-disease-related genes from ATD, a novel algorithm was generated, Optimized Random Forest with Label model, to predict potential autophagy-disease-related genes. This bioinformatics annotation system about autophagy and human diseases may provide a basic resource for the further detection of the molecular mechanisms of autophagy as they relate to disease.
m. LysoQuant
A seven-layer convolutional network with U-Net architecture was trained by Molinari’s lab to perform segmentation and classification of individual lysosomes from confocal images with human-level accuracy. This approach, termed LysoQuant, offers quantitative analyses of lysosome number, size, shape, position and occupancy with cargo (i.e., proteins or organelles to be cleared from cells). These parameters eventually inform on activity of lysosome-driven pathways including autophagy at the molecular level and on consequences of genetic or environmental modifications [2525]. LysoQuant is freely available at http://www.imaging.irb.usi.ch/lysoquant.
3. Mathematical models of autophagy dynamics
The idea of using mathematical modeling to characterize the population dynamics of autophagosomes and other vesicles involved in autophagy (e.g., autolysosomes) was discussed as early as 1975 [1529, 1530]. However, realization of this idea occurred only much later, after methods became available to precisely monitor changing autophagic vesicle populations in individual cells [2526, 2527]. Present and increasing opportunities to generate quantitative data make further modeling work timely, as do compelling needs to better understand the spatiotemporal dynamics of the subcellular structures affected by and mediating autophagy as well as the system-level behaviors of the molecular networks that regulate autophagy, which contain numerous potential drug targets relevant for diverse diseases [2528]. Because even simple mathematical models have proven to be powerful aids for reasoning about biological systems [2529], we strongly encourage greater use of mathematical modeling in studies of autophagy.
In recent years, several autophagy-relevant mathematical models have been developed and analyzed to study a range of subjects, including the cell fate decision between autophagy and apoptosis [2530–2532], the role of feedback loops in cellular regulatory networks and the possibility of bifurcations in qualitative system-level behavior [2533–2535], autophagy-related gene expression dynamics [2536], mitophagy [2537, 2538], pexophagy [2539], and the design of drug interventions for manipulating autophagy [2540].
Mathematical modeling can be, and is, pursued through a rich variety of techniques [2541], and new methods, together with enabling software tools [2542], continue to emerge regularly. The method that one selects for a particular study should be well-matched to the question(s) being asked; the appropriate level of abstraction is invariably context-dependent. Methods specialized for modeling dynamic compartments [2543] and biomolecular site dynamics [2544] may be of special interest in autophagy studies.
Although these modeling processes carry limitations in terms of complexity and portraiture of the realistic biological phenomenon, they can simultaneously be used to study a biological system where the goal is to unveil the underlying principles that are veiled at different levels of description. Various types of mathematical models can be used to study the autophagy process that includes ordinary, partial and stochastic differential equations. Ordinary differential equations/ODE are the simplest form to model a biological system where the focus is to study autophagy dynamics with respect to change in the protein/metabolite concentration [2530, 2532, 2540, 2545–2548]. Partial differential equations/PDE can be an important approach to model autophagy-dependent processes such as autophagy-dependent motility, an area yet to be explored in autophagy. To study the randomness imposed by the generation and variability of different stresses and continuous fluctuations in cellular energy levels, stochastic modeling techniques can be applied [2527, 2549]. Autophagy dynamics can also be studied in a discrete-based approach using agent-based modeling [2526, 2537]. Another useful modeling tool is petri net (place/transition) [2550], which is capable of modeling both discrete and continuous types of autophagy in cellular biochemical reactions [2551].
In brief, some of the most important, general guidelines for good modeling practice are as follows: Whenever possible, model development and analysis should be tightly integrated with experimental efforts [2552], and model analysis should be directed at generating non-obvious insights and testable predictions, not simply at reproducing phenomenology. Of course, models have purposes beyond prediction, for example, in capturing knowledge and providing explanations, in exposing knowledge gaps, and in determining the logical consequences of assumptions [2553–2555]. Models should be made shareable and reusable—for this purpose, standardized model-definition formats [2556, 2557], means for encoding simulation protocols [2558, 2559], and online databases [2560] have been developed. The problem of estimating the values of model parameters is an incessant concern of modelers. Some have recommended that this task is best accomplished through curve fitting versus direct measurement [2561, 2562]. In any case, uncertainties of parameter estimates and model predictions should be quantified, which is, arguably, best accomplished via Bayesian methods [2563, 2564]. These methods are not always practical because of their computational expense; however, alternative, less computationally expensive approaches are available [2565]. Reproducibility of modeling, a growing concern [2566, 2567], is enhanced when general-purpose software compatible with established standards is used for simulations, curve fitting, uncertainty quantification, etc.
For the beginner, excellent, fairly comprehensive introductions to systems biology modeling are available [2568, 2569], and short courses are also available [2570].
As one specific example, mathematical models minimizing the membrane bending energy show that phagophore expansion, which elongates the length of the energetically expensive phagophore edge, is sufficient to drive remodeling of the initially flat phagophore into a curved shape [2571]. Furthermore, geometric considerations indicate that several hundred or thousands of vesicles are required to form a single autophagosome [2572]. The absence of comparable vesicle numbers implies that vesicles provide a minor autophagosomal membrane source.
3. Conclusions and Future Perspectives
There is no question that research on the topic of autophagy has expanded dramatically since the publication of the first set of guidelines [3]. To help keep track of the field we have published a glossary of autophagy-related molecules and processes [2573, 2574], and now include the glossary as part of these guidelines.
With this continued influx of new researchers, we think it is critical to try to define standards for the field. Accordingly, we have highlighted the uses and caveats of an expanding set of recommended methods for monitoring autophagy in a wide range of systems (Table 4). Importantly, investigators need to determine whether they are evaluating levels of early or late autophagic compartments, or autophagic flux. If the question being asked is whether a particular condition changes autophagic flux (i.e., the rate of delivery of autophagy substrates to lysosomes or the vacuole, followed by degradation and efflux), then assessment of steady state levels of autophagosomes (e.g., by counting GFP-LC3 puncta, monitoring the amount of LC3-II without examining turnover, or by single time point electron micrographs) is not sufficient as an isolated approach. In this case it is also necessary to directly measure the flux of autophagosomes and/or autophagy cargo (e.g., in wild-type cells compared to autophagy-deficient cells, the latter generated by treatment with an autophagy inhibitor or resulting from ATG gene knockdowns or knockouts). Collectively, we strongly recommend the use of multiple assays whenever possible, rather than relying on the results from a single method.
As a final reminder, we stated at the beginning of this article that this set of guidelines is not meant to be a formulaic compilation of rules, because the appropriate assays depend in part on the question being asked and the system being used. Rather, these guidelines are presented primarily to emphasize key issues that need to be addressed such as the difference between measuring autophagy components, and flux or substrate clearance; they are not meant to constrain imaginative approaches to monitoring autophagy. Indeed, it is hoped that new methods for monitoring autophagy will continue to be developed, and new findings may alter our view of the current assays. This is a dynamic field, much like the process of autophagy, and we need to remain flexible in the standards we apply.
For those on the move, a Quick Guide to autophagy is provided below.
Glossary
2-D08: An inhibitor of protein SUMOylation that induces autophagy-mediated cancer cell death [2673].
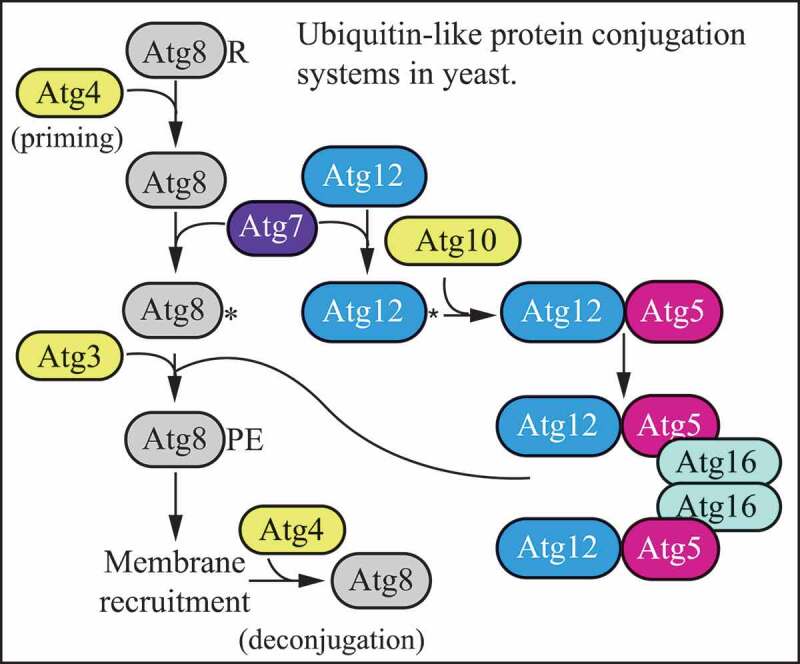
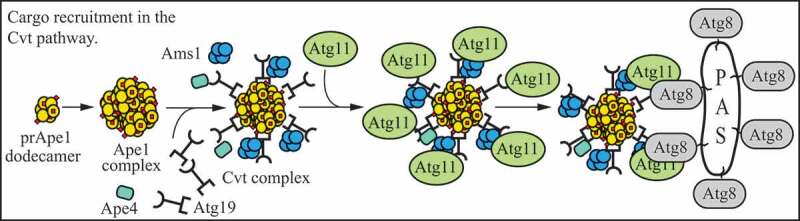
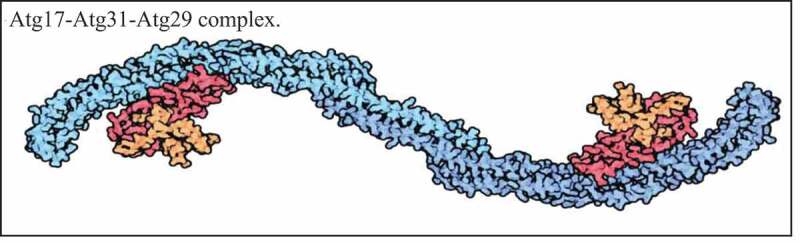
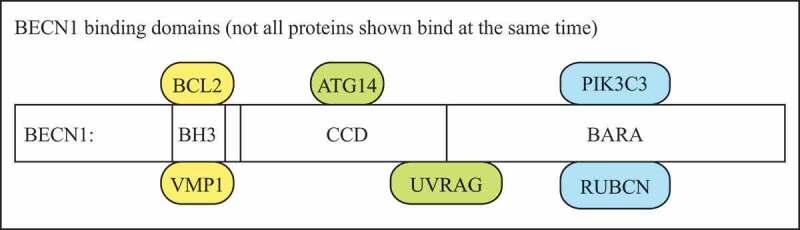
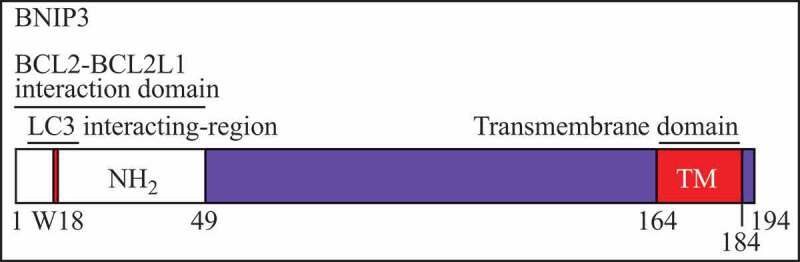
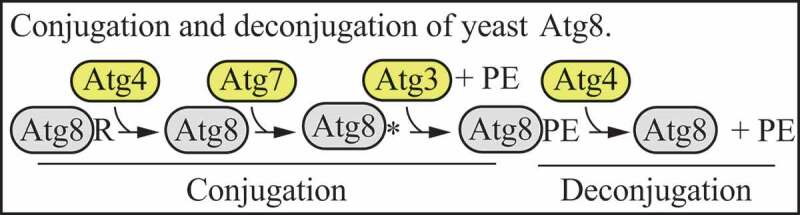
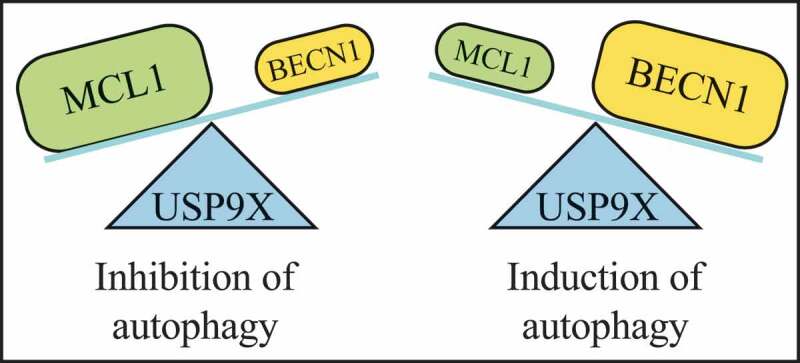
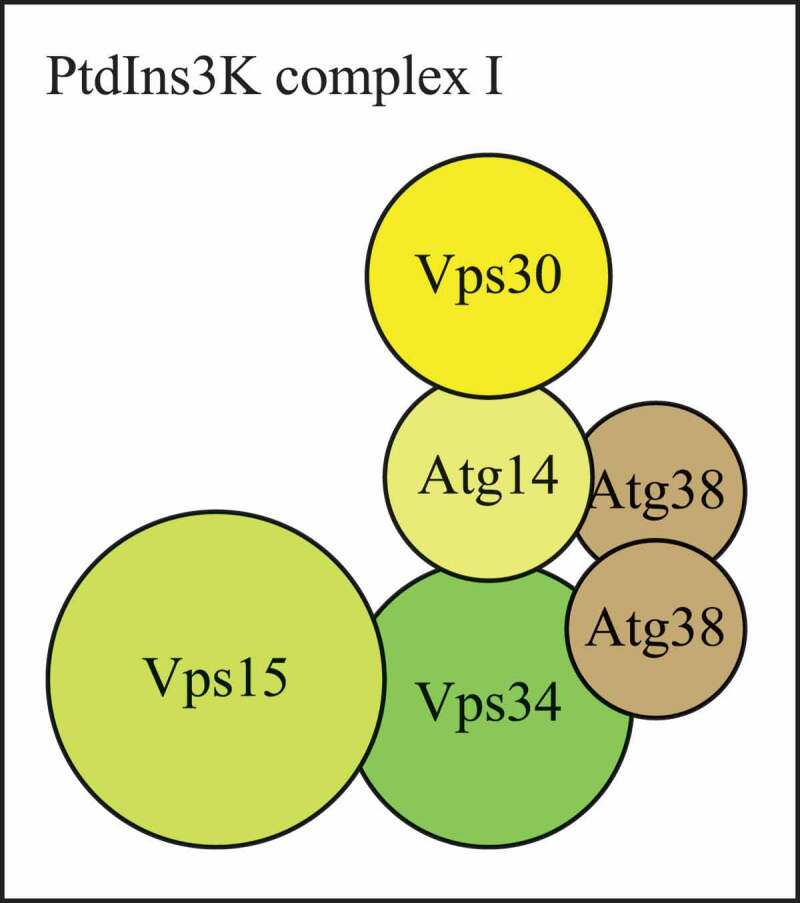
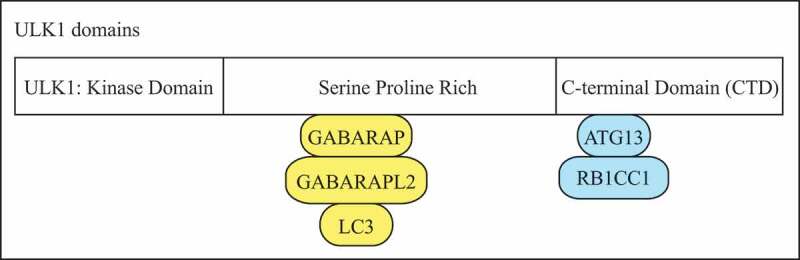
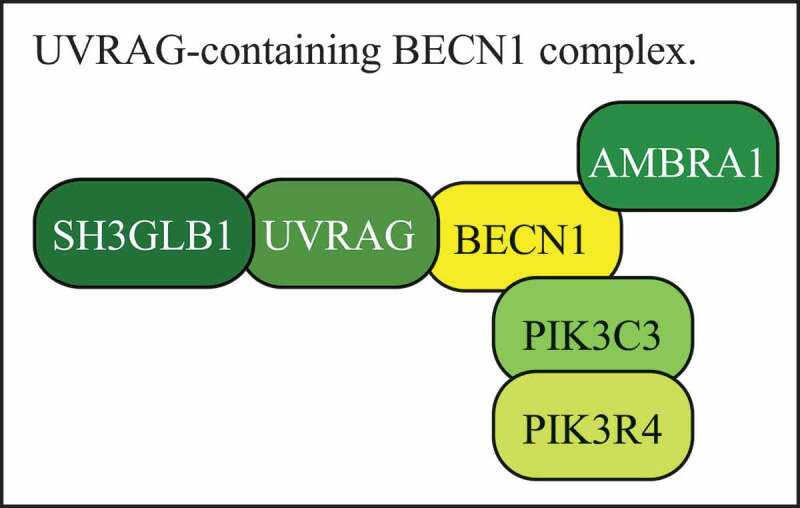
2-Methoxyestradiol (2-ME): 2-ME is a natural metabolite of estrogen that prevents angiogenesis and tumor progression. 2-ME regulates autophagy through mechanisms that involve both ROS production [2674] and MAPK/JNK-DRAM pathway activation [2675].
3-MA: See 3-methyladenine.
3-Methyladenine (3-MA): An inhibitor of class I PI3K and class III PtdIns3K, which results in autophagy inhibition due to suppression of class III PtdIns3K [435], but may under some conditions show the opposite effect [436, 437, 1749]. At concentrations >10 mM 3-MA inhibits other kinases such as AKT (Ser473), MAPK/p38 (Thr180/Tyr182) and MAPK/JNK (Thr183/Tyr185) [2676].
3BDO (3-benzyl-5-[[2-nitrophenoxy} methyl]–dihydrofuran-2[3h]-one): A novel MTOR activator that occupies the rapamycin-binding site and blocks the interaction between rapamycin and FKBP1A, and then activates the MTOR signaling pathway to inhibit autophagy initiation [2677].
11ʹ-deoxyverticillin A (C42): An epipolythiodioxopiperazine fungal secondary metabolite that is used as an anticancer drug; it triggers apoptotic and necrotic cell death, and enhances autophagy through the action of PARP1 and RIPK1 [2678].
12-ylation: The modification of substrates by covalent conjugation to ATG12, first used to describe the autocatalytic conjugation of ATG12 to ATG3 [2679]. See also LC3/GABARAP-ylation.
14-3-3ç: See YWHAZ.
AAK1 (AP2 associated kinase 1): A cellular serine-threonine protein kinase that functions as a key regulator of clathrin-mediated endocytosis and interacts with LC3B [2680].
ABT737: A BH3 mimetic that competitively disrupts the interaction between BECN1 and BCL2 or BCL2L1, thus inducing autophagy [2681]. It should be noted, however, that by its inhibitory action on the anti-apoptotic BCL2 family members, ABT737 also leads to apoptosis [2682].
ABTL0812 (2-hydroxylinoleic acid): ABTL0812 is in clinical development for the treatment of endometrial and lung cancer. ABTL0812 shows anticancer activity in several animal models of cancer by stimulating autophagy-mediated cancer cell death. This effect occurs via activating PPARA (peroxisome proliferator activated receptor alpha) and PPARG, as well as via triggering the ER stress-related response. Both effects synergize to inhibit the AKT-MTORC1 axis [2683, 2684].
ACBD5 (acyl-CoA binding domain containing 5): ACBD5 is the human ortholog of fungal KpAtg37. ACBD5 localizes on peroxisomes and, via its interaction with the VAPA/B ER proteins, mediates physical contact between peroxisomes and the ER in humans [2685]; ACBD5 is required for pexophagy [458, 2686]. See also Atg37.
ACBP (acyl-CoA-binding type 2 protein): ACBP transports acyl-CoA esters, and is involved in the elongation of fatty acid. In the filamentous fungus Aspergillus oryzae, ACBP moves in the cytoplasm in a microtubule-dependent manner, and is transported to vacuoles via the autophagy machinery [2687].
Acetyl-coenzyme A: A central energy metabolite that represses autophagy if present in the cytosol [2688, 2689].
Acinus: A protein that in Drosophila regulates both endocytosis and autophagy; the acn mutant is defective in autophagosome maturation, whereas stabilization of endogenous Acn by mutation of its caspase cleavage site [2690] or by CDK5-mediated phosphorylation [2691], or overexpression of Acn leads to excessive autophagy [2692]. Note that Acn can also induce DNA condensation or fragmentation after its activation by CASP3 in apoptotic cells.
ACSL1 (acyl-CoA synthetase long chain family member 1): An isozyme of the long-chain fatty-acid-coenzyme A ligase family that converts fatty acids to acyl-CoAs. It is the major long-chain mammalian ACSL isoform in the heart. The knockdown of the ACSL1 gene indirectly impairs cardiac autophagy through MTORC1 activation [2693].
ActA: A L. monocytogenes protein that recruits the Arp2/3 complex and other actin-associated components to the cell surface to evade recognition by xenophagy; this effect is independent of bacterial motility [2694].
ACY-1215/Ricolinostat: ACY-1215 is an orally bio-available, selective HDAC6 inhibitor (with an IC50 in the nanomolar range in vitro), which abrogates the autophagic clearance of aggresomes. Consequently, ACY-1215 treatment results in accumulation of misfolded protein aggregates, as reported in multiple myeloma and mantle cell lymphoma [2577, 2695].
Adaptophagy: Selective degradation of signaling adaptors downstream of TLRs or similar types of receptor families [2696].
ADH5/GSNOR (alcohol dehydrogenase 5 [class III], chi polypeptide): A denitrosylase that catalyzes the reduction of the S-nitrosylated form of glutathione formed by the exchange of NO with S-nitrosylated proteins. ADH5/GSNOR expression declines during age in mammals and affects the efficiency of selective mitophagy of damaged mitochondria, being, in this manner, implicated in cell senescence and aging [1011, 1012].
ADIPOQ (adiponectin, C1Q and collagen domain containing): An adipocytokine that induces cytotoxic autophagy by activating the STK11 tumor suppressor [2697].
ADNP (activity-dependent neuroprotective homeobox): A protein that interacts with LC3B and shows an increased expression in lymphocytes from schizophrenia patients [1662]. When mutated de novo, ADNP causes an autism spectrum disorder, the ADNP syndrome, exhibiting intellectual disabilities (see also https://www.orpha.net/consor/cgi-bin/OC_Exp.php?lng=EN&Expert=404448).
Aduk (Another Drosophila Unc-51-like kinase): A ULK3 homolog that is required for normal lifespan and autophagy induction in response to chemical stress but not nutrient or developmental cues [2698].
AEG-1: See MTDH.
AEN/ISG20L1 (apoptosis enhancing nuclease): A protein that localizes to nucleolar and perinucleolar regions of the nucleus, which regulates autophagy associated with genotoxic stress; transcription of AEN is regulated by TP53-family members [2699].
AFIM (ATG5 [five] interacting motif): A peptide motif forming an α-helical structure that interacts with ATG5, which has a conserved sequence, W-x(3)-I-x(3)-L-x(2)-R-x(2)-[QE] [882].
AGER/RAGE (advanced glycosylation end-product specific receptor): A member of the immunoglobulin gene superfamily that binds the HMGB1 (high mobility group box 1) chromatin binding protein [2700], AGER overexpression enhances autophagy and reduces apoptosis. This can occur in response to ROS, resulting in the upregulation of autophagy and the concomitant downregulation of apoptosis, favoring tumor cell survival in response to anticancer treatments that increase ROS production. In addition, autophagy is involved in the removal of advanced glycation end products, further linking this process to age-related diseases [2701]. See also HMGB1.
Aggrephagy: The selective removal of aggregates by a autophagy-like process [1092].
AGS3: See GPSM1.
Aggresome: An aggregation of misfolded proteins formed by a highly regulated process mediated by HDAC6 or BAG3 [1838, 2702]. This process requires protein transport by a dynein motor and microtubule integrity. Aggresomes form at the microtubule-organizing center and are surrounded by a cage of the intermediate filament protein VIM (vimentin). Note that not all proteins that aggregate and form filaments like HTT or MAPT form aggresomes. HDAC6 recognizes ubiquitin chains through a UBD domain. HDAC6 interacts with both Lys48 and Lys63 linkages, with preference for Lys63. Proteostat is a molecular probe that emits a strong fluorescent signal upon binding to β-sheet structures in misfolded protein aggregates [2703]. Proteostat staining colocalizes with LC3B puncta and can therefore be used to stain intracellular protein aggregates as well as aggresomes [2704].
AHA (L-azidohomoalanine): An amino acid analog used for labeling newly synthesized protein and monitoring autophagic protein degradation [1018].
AICAR (aminoimidazole-4-carboxamide riboside): Cell permeable nucleotide analog that is an activator of AMPK; inhibits autophagy [666] through mechanisms that are not related to its effect on AMPK [679, 2705].
AICD (APP intracellular domain): The C-terminal fragment of APP (amyloid beta precursor protein) derived from gamma-secretase cleavage. This fragment has been described as a transcription factor [2706]. AICD controls PINK1-mediated mitophagy by upregulating its transcription in a FOXO3-dependent manner [2707].
AIM (Atg8-family interacting motif): A short peptide motif that allows interaction with Atg8 by binding at the LIR/AIM docking site [2509]. See also WXXL motif, LIR/LRS and LDS.
AKT/PKB (AKT serine/threonine kinase): A serine/threonine kinase that negatively regulates autophagy in some cellular systems.
Alfy: See WDFY3.
ALIS (aggresome-like induced structures): These structures may function as protein storage compartments and are cleared by autophagy [418]. SQSTM1 may regulate their formation and autophagic degradation [419]. See also DALIS.
Allophagy: The selective degradation of sperm components by autophagy; this process occurs in C. elegans [1109–1112]. See also post-fertilization sperm mitophagy.
ALLO-1 (ALLOphagy defective): ALLO-1 is a C. elegans protein acting as an autophagy receptor required for LGG-1 recruitment and degradation of sperm-inherited organelles [1116].
ALOX5 (arachidonate 5-lipoxygenase): See lipoxygenases.
ALOX15 (arachidonate 15-lipoxygenase): See lipoxygenases.
ALR: See autophagic lysosome reformation.
ALS2/alsin (alsin Rho guanine nucleotide exchange factor ALS2): A guanine nucleotide exchange factor for the small GTPase RAB5 that regulates endosome and autophagosome fusion and trafficking; loss of ALS2 accounts for juvenile recessive ALS, juvenile primary lateral sclerosis, and infantile-onset ascending hereditary spastic paralysis [2708, 2709].
ALS-FTD (amyotrophic lateral sclerosis-frontotemporal dementia): See C9orf72.
AMBRA1 (autophagy and beclin 1 regulator 1): A positive regulator of autophagy and a mitophagy receptor. AMBRA1 interacts with both BECN1 and ULK1, modulating their activity [703, 741, 1956]. Also, a role in both PRKN-dependent and -independent mitophagy has been described for AMBRA1 [1174], and its mitophagy function is controlled by the E3 ubiquitin ligase HUWE1 and the CHUK/IKKα kinase [2516]. AMBRA1 activity is regulated by dynamic interactions with DDB1 and ELOB/TCEB2 (elongin B), the adaptor proteins of the E3 ubiquitin ligase complexes containing CUL4 (cullin 4) and CUL5, respectively [2710]. Finally, AMBRA1 is the autophagy adaptor linking this process to cell proliferation, by negatively regulating the oncogene MYC through the latter’s phosphorylation status [2711].
AMFR/gp78 (autocrine motility factor receptor): An ER-associated E3 ubiquitin ligase that degrades the MFN (mitofusin) mitochondrial fusion proteins and induces mitophagy [2712].
Amino acid analogs: Mimics of natural amino acids that induce the formation of protein aggregates and activate the autophagic flux in an NFKB-dependent manner [1107].
Amiodarone: An FDA-approved antiarrhythmic drug that induces autophagic flux via AMPK- and AKT-mediated MTOR inhibition [2713]. Amiodarone exerts severe side effects that include lung and liver toxicity where amiodarone-induced autophagy plays context- and organ-specific roles [2713–2715].
Amphisome (AM): Intermediate compartment formed by the fusion of an autophagosome with an endosome (this compartment can be considered a type of autophagic vacuole and may be equivalent to a late autophagosome, and as such has a single limiting membrane); the amphisome has not yet fused with a lysosome [2716]. Amphisomes can also fuse with the plasma membrane to release the autophagic cargo (exosomal pathway). See also exophagy.
AMPK (AMP-activated protein kinase): A sensor of energy level that is activated by an increase in the AMP:ATP ratio via the STK11/LKB1 kinase. Phosphorylates the MTORC1 subunit RPTOR to cause induction of autophagy. AMPK also activates the TSC1/2 complex (thus inhibiting RHEB), and binds and directly phosphorylates (and activates) ULK1 as part of the ULK1 kinase complex, which includes ATG13, ATG101 and RB1CC1 [672, 673]. The yeast homolog of AMPK is Snf1 [666, 2717]. Conversely, ULK1 can phosphorylate AMPK through a negative feedback loop [710]. AMPK is a heterotrimeric enzyme composed of the PRKAA1/AMPKα1 or PRKAA2/AMPKα2 subunit, the PRKAB1/AMPKβ1 or PRKAB2/AMPKβ2 subunit and the PRKAG1/AMPKγ1, PRKAG2/AMPKγ2 or PRKAG3/AMPKγ subunits.
Ams1/α-mannosidase: A resident vacuolar hydrolase that is a cargo of the Cvt pathway; Ams1 forms an oligomer in the cytosol similar to prApe1 [2718]. See also Atg11 and Atg19.
AMSH1/3: Two Arabidopsis deubiquitinating enzymes that have been linked to plant autophagy [2719, 2720].
AN4171/BapH: An effector of Aspergillus nidulans RAB11 that connects exocytic membranes with basal autophagy. AN4171/BapH contains BAR and PH domains; the PH domain is essential for membrane targeting [2200].
Antimycin A (AMA): A mitophagy inducer; antimycin A inhibits mitochondrial electron transport chain complex III, leading to increased ROS generation and a relatively limited dissipation of mitochondrial transmembrane potential; therefore, it is often used in combination with oligomycin [1224]. See also oligomycin.
Antiviral innate immune response: A process of establishing an antiviral state through DDX58/retinoic acid-inducible gene I-like receptor-mediated production of IFNs (interferons) in virus-infected cells. The IFNs ultimately trigger the expression of IFN-stimulated genes that suppress viral replication [2721]. Autophagy suppresses the antiviral innate immune response by targeting the adaptor proteins of DDX58-IFN signaling [2722, 2723].
ANXA5/annexin V (annexin A5): ANXA5 is one member of the family of calcium and phospholipid-binding proteins. Early ellipsometry studies that focused on PS demonstrate that adsorption of proteins is calcium-dependent and is completely reversible upon calcium depletion. Treatments that alter membrane phospholipids and/or their charge will alter ANXA5 binding. Cells undergoing apoptosis lose plasma membrane asymmetry, such that high levels of PS become exposed on the cell surface. These cells can be recognized by staining with ANXA5, which binds to PS with high affinity. Increased ANXA5 binding to the outer mitochondrial membrane has been related to the induction of mitophagy. The molecules responsible for ANXA5 binding to mitochondrial membranes are not extensively studied. Diminished mitochondrial OGDH (oxoglutarate dehydrogenase) complex/alpha-ketoglutarate dehydrogenase complex may increase the levels of various metabolites prior to the OGDH complex. For example, loss of FH (fumarate hydratase) activity causes accumulation of intracellular fumarate, which can directly modify cysteine residues to form 2-succinocysteine through succination, and this could alter the surface charge on the mitochondria. The OGDH complex can also modify lysine groups by succinylation, which changes the charge and shape of molecules [1198].
AP2 (adaptor related protein complex 2): AP2 is a heterotetrameric clathrin adaptor complex comprised of four subunits (AP2A/α, AP2B1/β2, AP2M1/μ2, AP2S1/σ2). Apart from its canonical function in endocytosis, AP2 facilitates retrograde transport of signaling autophagosomes containing active NTRK2/neurotrophin receptor TrkB in developing neurons. This mechanism involves the direct binding of a LIR motif within the AP2A subunit of AP2 to LC3 and complex formation of AP2 with DCTN1/p150Glued [2724].
AP4 (adaptor related protein complex 4): AP4 is a clathrin-independent heterotetrameric adaptor complex comprised of four subunits (AP4E1/ε, AP4B1/β4, AP4M1/µ4, AP4S1/σ4). Mutations in all four subunits can lead to complicated forms of hereditary spastic paraparesis [2725–2727]. AP4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation [2728], and AP4 loss leads to depletion of ATG9A and disrupted autophagosome biogenesis in the distal axon [2729].
AP5 (adaptor related protein complex 5): AP5 is a clathrin independent heterotetrameric adaptor complex consisting of 4 subunits (AP5Z1/ζ, AP5B1/β5, AP5M1/μ5, and AP5S1/σ5). Mutation in the AP5Z1/ζ subunit results in hereditary spastic paraparesis [2730]. AP5 is involved in Golgi retrieval and autophagic degradation [2731, 2732].
APC (activated protein C): APC (PROC that has been activated by thrombin) modulates cardiac metabolism and augments autophagy in the ischemic heart by inducing the activation of AMPK in a mouse model of ischemia/reperfusion injury [2733].
Ape1 (aminopeptidase I): A resident vacuolar hydrolase that can be delivered in its precursor form (prApe1) to the vacuole through either the cytoplasm-to-vacuole targeting (Cvt) pathway or autophagy, in vegetative or starvation conditions, respectively [177]. The propeptide of prApe1 is removed upon vacuolar delivery, providing a convenient way to monitor localization of the protein and the functioning of these pathways, although it must be noted that delivery involves a receptor and scaffold so that its transit involves a type of selective autophagy even in starvation conditions. See also Atg11, Atg19 and cytoplasm-to-vacuole targeting pathway.
Ape1 complex/prApe1 complex: A large protein complex comprised of multiple prApe1 dodecamers localized in the cytosol [180].
Ape4: A vacuolar aspartyl aminopeptidase that binds the Atg19 receptor and is transported to the vacuole through the Cvt pathway [2734].
Apicoplast: Multimembrane nonphotosynthetic plastid present in several apicomplexan parasites. ATG8 is recruited to its outermost membrane using the same conjugation machinery as for its recruitment to the phagophore membrane, and there it plays an essential role for maintaining the homeostasis of this organelle, which is clearly distinct from the canonical degradative autophagy pathway [2735].
APMA (autophagic macrophage activation): A collection of autophagy-related processes in cells of the reticulo-endothelial system. APMA includes (1) convergence of phagocytosis and the autophagic machinery, (2) enhanced microbicidal properties of autolysosomes in comparison to standard phagolysosomes, (3) autophagic modulation of pathogen recognition receptor signaling, (4) cooperation between immunity-related GTPases and ATG proteins in attacking parasitophorus vacuoles, and (5) enhanced antigen presentation. APMA is thus recognized as a complex outcome of autophagy stimulation in macrophages, representing a unique composite process that brings about a heightened state of immunological activation [2736].
APOL9 (apolipoprotein L 9): A phosphatidylethanolamine-binding mouse protein, which interacts preferentially with the lipidated form of LC3/GABARAP proteins, associates with microtubules and localizes to mitochondria and lysosomes in autophagy-arrested cells [2737].
Apoptosis: Apoptosis is a programmed cell death process that controls autophagy. Conversely, autophagy can also regulate apoptosis [2738]. See also caspases.
Appressorium: A specialized infection structure produced by pathogenic fungi to rupture the outer layer of their host and gain entry to host cells. In plant pathogenic fungi, such as the rice blast fungus M. oryzae, formation of appressoria follows autophagy in conidia and recycling of the spore contents to the developing infection cell [373, 2197].
AR-12 (OSU-03012): A broad-specificity anti-viral celecoxib-derivative that stimulates autophagosome formation and viral protein degradation [2580].
ARD1: See NAA10.
Are1: See Ayr1.
Are2: See Ayr1.
Arl1: A small GTP-binding protein involved in formation of the autophagosome in yeast [2739].
ARRB1/β-arrestin-1 (arrestin beta 1): Members of the arrestin/beta-arrestin protein family are thought to participate in agonist-mediated desensitization of G-protein-coupled receptors and cause specific dampening of cellular responses to stimuli such as hormones, neurotransmitters, or sensory signals. ARRB1 is a cytosolic protein and acts as a cofactor in the ADRBK/BARK (adrenergic, beta, receptor kinase)-mediated desensitization of beta-adrenergic receptors. Besides the central nervous system, it is expressed at high levels in peripheral blood leukocytes, and thus the ADRBK/beta-arrestin system is thought to play a major role in regulating receptor-mediated immune functions. This protein plays a neuroprotective role in the context of cerebral ischemia through regulating BECN1-dependent autophagosome formation [2740].
ARHI: See DIRAS3.
ARIH1/HHARI/UBCH7BP (ariadne RBR E3 ubiquitin protein ligase 1): An E3 ligase that belongs to the RBR family (including PRKN) that is involved in the regulation of mitophagy in a PINK1-dependent manner. ARIH1-dependent mitophagy plays a key role in the response of cancer cells to chemotherapy [2741].
Arl8: A small GTPase binding partner of the homotypic fusion and vacuole protein sorting (HOPS) tethering complex that positively regulates lysosomal fusion events including autophagosome-lysosome fusions [2742].
ARN5187: Lysosomotropic compound with dual inhibitory activity against the circadian regulator NR1D2/REV-ERBβ and autophagy. Although ARN5187 and CQ have similar lysosomotropic potency and are equivalent with regard to autophagy inhibition, ARN5187 has a significantly improved in vitro anticancer activity [2582].
Arsenic trioxide: A compound used currently to treat leukemia, which triggers cell death and autophagy [2743].
ASB10 (ankyrin repeat and SOCS box containing 10): The ASB family of proteins mediate ubiquitination of protein substrates via their SOCS box and as such have been implicated as negative regulators of cell signaling. ASB10 colocalizes with aggresome biomarkers and pre-autophagic structures and may form ALIS [2744].
AT-101: (-)-Gossypol, a polyphenolic compound from cotton seed that induces mitophagy via the mitochondrial permeability transition pore in human glioblastoma cells and in the filamentous fungus Podospora anserina [2745, 2746].
AtEH/Pan1: The Arabidopsis EH proteins (AtEH1/Pan1 and AtEH2/Pan1) are components of the endocytic TPLATE complex, which is essential for endocytosis. Both proteins are homologs of the yeast Arp2/3 complex activator, Pan1. The depletion of AtEH1/Pan1 impairs autophagosome formation and pollen development [2747].
ATF4 (activating transcription factor 4): A transcription factor that is induced by hypoxia, amino acid starvation and ER stress during the integrated stress response. The induction of ATF4 depends on translational control mediated by the EIF2S1/eIF2α pathway and involves a mechanism involving upstream open reading frames that promote translation of ATF4 in the face of global reduction of translation [2748]. ATF4 is involved in the unfolded protein response, playing a critical role in stress adaptation and progression of neuro- and retinal degeneration [2749, 2750]; ATF4 binds to a cAMP response element binding site in the LC3B promoter, resulting in upregulation of LC3B [2751], and also directs an autophagy gene transcriptional program in response to amino acid depletion and ER stress [473, 570].
ATF5 (activating transcription factor 5): A transcription factor that is upregulated by the BCR-ABL protein tyrosine kinase, an autophagy repressor, through the PI3K-AKT pathway that inhibits FOXO4, a repressor of ATF5 transcription; one of the targets of ATF5 is MTOR [2752].
ATF6: ATF6 is a 90-kDa type II transmembrane protein of the ER, that is released from HSPA5/Grp78 binding upon ER stress and translocates to the Golgi apparatus where it is cleaved by site 1 and site 2 proteases. Subsequently, the activated ATF6 is translocated to the nucleus where it functions in transactivating the DDIT3 gene and thus contributes to induction of those autophagy genes whose expression depends on DDIT3.
Atg (autophagy-related): Abbreviation used for most of the components of the protein machinery that are involved in selective and nonselective macroautophagy and in selective microautophagy [2753].
Atg1: A serine/threonine protein kinase that functions in recruitment and release of other Atg proteins from the PAS [2754]. The functional homologs in more complex eukaryotes are ULK1 and ULK2, and in C. elegans is UNC-51.
Atg2: A protein that interacts with Atg18 via an N-terminal domain [2755] and binds PtdIns3P via an APT1 domain in a calcium-dependent manner [2756]. Atg2 also interacts with Atg9, and in atg2∆ mutant cells Atg9 accumulates primarily at the PAS [2757]. Atg2 tethers the ER to the phagophore along with Atg18 [2758]; Atg2 is able to transfer lipids from one membrane to the other via a hydrophobic channel similar to Vps13, which may contribute to expansion of the phagophore [2759–2762].
Atg3: A ubiquitin-conjugating enzyme (E2) analog that conjugates Atg8-family proteins to phosphatidylethanolamine (PE) after activation of the C-terminal residue by Atg7 [2763, 2764]. ATG3 can also be conjugated to ATG12 in more complex eukaryotes [2679]. See also 12-ylation.
Atg4: A cysteine protease that processes Atg8-family proteins by removing the amino acid residue(s) that are located on the C-terminal side of what will become the ultimate glycine. Atg4 also removes PE from Atg8-family proteins in a step referred to as “deconjugation” [288]. Mammals have four ATG4 proteins (ATG4A to ATG4D) [2765], but ATG4B appears to be the most relevant for autophagy and has the broadest range of activity for all of the Atg8 homologs [237, 2766]. C. elegans has 2 proteins, atg-4.1 and atg-4.2, which perform partially redundant, but distinct functions in conjugation and deconjugation, with different phenotypes concerning the biogenesis and accumulation of autophagosomes [471]. See also deconjugation.
Atg5: A protein containing ubiquitin folds that is part of the Atg12–Atg5-Atg16 complex, which acts in part as an E3 ligase for Atg8-family protein conjugation to PE [2767].
Atg6: See Vps30.
Atg7: A ubiquitin activating (E1) enzyme homolog that activates both Atg8-family proteins and Atg12 in an ATP-dependent process, leading to the lipidation of Atg8-family proteins [2768, 2769]. In addition to a full-length ATG7, several human tissues also express an isoform encoding an ATG7 protein that lacks the binding site with LC3 and is unable to facilitate its lipidation [2770].
Atg8: A ubiquitin-like protein that is conjugated to PE; involved in cargo recruitment into phagophores, and biogenesis of autophagosomes. Autophagosomal size is regulated by the amount of Atg8 [149], possibly by controlling the membrane asymmetry of the phagophore [2771]. Because Atg8 is selectively enclosed into autophagosomes, its breakdown allows measurement of the rate of autophagy. Mammals have several Atg8 homologs that are members of the LC3 and GABARAP subfamilies, which are also involved in autophagosome formation and/or maturation [32, 199, 204, 914]. The C. elegans homologs are LGG-1 and LGG-2.
Atg9: A transmembrane protein that may act as a lipid carrier for expansion of the phagophore. In mammalian cells, ATG9A localizes to the trans-Golgi network and endosomes, whereas in fungi this protein localizes in part to peripheral sites (termed Atg9 reservoirs or tubulovesicular clusters) that are localized near the mitochondria, and to the PAS [807, 2772]. Mouse ATG9A has functions independent of autophagy, such as those in innate immune response and cell death [2773, 2774].Whereas mammalian ATG9A is ubiquitously expressed, ATG9B is almost exclusively expressed in the placenta and pituitary gland [2775]. See also Atg9 peripheral sites/structures.
Atg9 peripheral sites/structures: In yeast, these are peri-mitochondrial sites where Atg9 localizes, which are distinct from the phagophore assembly site [807, 808]. The Atg9 peripheral sites may be the precursors of the phagophore.
Atg10: A ubiquitin conjugating (E2) enzyme analog that conjugates Atg12 to Atg5 [2776].
ATG10S (short ATG10): A short protein isoform of ATG10 with 184 amino acids, in which 36 amino acids (encoded in exon 4 of the ATG10 transcript variant 3 sequence in human chromosome 5) are lost via alternative splicing of ATG10 mRNA. ATG10S functions in autolysosome formation by promoting autophagosome fusion to lysosomes, and inactivation of type III interferon transcription by binding to the promoter of IFNL2/type III interferon 2 [2437, 2777, 2778] (M.-q. Zhang, Q. Zhao, J.-p. Zhang, unpublished data).
Atg11: A scaffold protein that acts in selective types of autophagy including the Cvt pathway, mitophagy and pexophagy. Atg11 binds Atg19, K. phaffii/P. pastoris Atg30 (PpAtg30) and Atg32 as part of its role in specific cargo recognition. Atg11 also binds Atg9 and is needed for its movement to the PAS [2779]. Atg11 in conjunction with receptor-bound targets may activate Atg1 kinase activity during selective autophagy [2780]. Homologs of Atg11 include RB1CC1 in mammals (Atg11 has significant homology to the C terminus of RB1CC1, although the latter does not appear to function as an Atg11 ortholog) [2781, 2782], EPG-7 in C. elegans [2783], and ATG11 in Arabidopsis [2781], as well as some sequence and functional homology to HTT [2784].
Atg12: A ubiquitin-like protein that modifies an internal lysine of Atg5 by covalently binding via its C-terminal glycine [2767]. In mouse and human cells, ATG12 also forms a covalent bond with ATG3, and this conjugation event plays a role in mitochondrial homeostasis [2679]. The C. elegans homolog is LGG-3.
Atg13: A component of the Atg1 complex that is needed for Atg1 kinase activity. Atg13 is highly phosphorylated in a PKA- and TOR-dependent manner in nutrient-rich conditions. During starvation-induced autophagy in yeast, Atg13 is partially dephosphorylated. In mammalian cells, at least MTOR and ULK1 phosphorylate ATG13. The decreased phosphorylation of Atg13/ATG13 that results from TOR/MTOR inhibition is partly offset in terms of the change in molecular mass by the ULK1-dependent phosphorylation that occurs upon ULK1 activation [745, 2785]. The C. elegans ortholog is EPG-1.
Atg14: A component of the class III PtdIns3K complex that is necessary for the complex to function in autophagy [2786]. ATG14 also tethers to the ER-mitochondria contact site and facilitates lipophagy-mediated ER stress regulation [2787]. Also known as ATG14/ATG14L/BARKOR in mammals [799], or EPG-8 in C. elegans [2788].
Atg15: A yeast vacuolar protein that contains a lipase/esterase active site motif and is needed for the breakdown of autophagic and Cvt bodies within the vacuole lumen (as well as MVB-derived and other subvacuolar vesicles) and the turnover of lipid droplets [2789–2791].
Atg16: A component of the Atg12–Atg5-Atg16 complex. Atg16 dimerizes to form a large complex [2792]. There are two mammalian homologs, ATG16L1 and ATG16L2; mutations in either of the corresponding genes correspond to risk alleles associated with Crohn disease [2793, 2794].
Atg17: A yeast protein that is part of the Atg1 kinase complex. Atg17 is not essential for autophagy, but modulates the magnitude of the response; smaller autophagosomes are formed in the absence of Atg17 [148, 743]. In yeast, Atg17 exists as part of a stable ternary complex that includes Atg31 and Atg29; this complex functions as a dimer [2795–2797]. The functional counterpart of this complex in mammalian cells may be RB1CC1.
Atg18: A yeast protein that binds to PtdIns3P (and PtdIns[3,5]P2) via its WD40 β-propeller domain. Atg18 interacts with Atg2, and in atg18∆ cells Atg9 accumulates primarily at the PAS. Atg18 has additional nonautophagic functions, such as in retrograde transport from the vacuole to the Golgi complex, and in the regulation of PtdIns(3,5)P2 synthesis; the latter function affects the vacuole’s role in osmoregulation [831]. See also WIPI.
Atg19: A receptor for the Cvt pathway that binds Atg11, Atg8 and the propeptide of precursor aminopeptidase I. Atg19 is also a receptor for Ams1/α-mannosidase, another Cvt pathway cargo [2798, 2799]. See also Ams1.
Atg20/Snx42: A yeast PtdIns3P-binding sorting nexin that is part of the Atg1 kinase complex and associates with Snx4/Atg24 [2800]. Atg20 is a hybrid protein mixing IDPRs with the PX-BAR domain-containing architecture [2801]. Atg20 is involved in the Cvt pathway and pexophagy, and facilitates nonselective autophagy. M. oryzae Snx41 (MoSnx41) is homologous to both yeast Atg20 and Snx41, and carries out functions in both pexophagy and nonautophagy vesicular trafficking [2203].
Atg21: A yeast PtdIns3P binding protein that is a homolog of, and partially redundant with, Atg18 [443]. See also WIPI.
Atg22: A yeast vacuolar amino acid permease that is required for efflux after autophagic breakdown of proteins [2802, 2803].
Atg23: A yeast peripheral membrane protein that associates and transits with Atg9 [809, 2804, 2805]. Atg23 (together with Atg27) contributes to the efficient formation of the Atg9 peripheral sites/structures, and thus affects Atg9 trafficking to the PAS [2806, 2807]. See also Atg9, Atg27.
Atg24: See Snx4.
Atg25: A coiled-coil protein required for macropexophagy in H. polymorpha [2808].
Atg26: A sterol glucosyltransferase that is required for micro- and macropexophagy in K. phaffii/P. pastoris, but not in S. cerevisiae [2809, 2810].
Atg27: A yeast integral membrane protein that is required for the movement of Atg9 to the PAS; together with Atg23 it contributes to the efficient formation of the Atg9 peripheral sites/structures [2806, 2807]. The absence of Atg27 results in a reduced number of autophagosomes under autophagy-inducing conditions [2811]. See also Atg9, Atg23.
Atg28: A coiled-coil protein involved in micro- and macropexophagy in K. phaffii/P. pastoris [2812].
Atg29: A yeast protein required for efficient nonselective autophagy in fungi. Part of the yeast Atg17-Atg31-Atg29 complex that functions at the PAS for protein recruitment [2795–2797, 2813]. See also Atg17, Atg31.
Atg30: A pexophagy receptor required for the recognition of peroxisomes during micro- and macropexophagy. KpAtg30 localizes to the membrane of peroxisomes in K. phaffii/P. pastoris, and binds the peroxisomal membrane proteins KpPex3 and PpPex14 [1068]. KpAtg30 also interacts with KpAtg37, the peroxisomal acyl-CoA-binding protein, which regulates the phosphorylation status of KpAtg30, as well as to KpAtg17, KpAtg8 and the scaffold protein KpAtg11, that link KpAtg30 to the core autophagy machinery [458, 2814, 2815]. See also Atg37.
Atg31: A yeast protein required for nonselective autophagy in fungi. Part of the yeast Atg17-Atg31-Atg29 complex that functions at the PAS for protein recruitment and initiation of phagophore formation [2795–2797, 2816]. See also Atg17, Atg29.
Atg32: A mitochondrial outer membrane protein that is required for mitophagy in yeast. Atg32 binds Atg8 and Atg11 preferentially during mitophagy-inducing conditions [1049, 1050]. See also BCL2L13. See also casein kinase 2.
Atg33: A mitochondrial outer membrane protein that is required for mitophagy in yeast [1048].
Atg34: A protein that functions as a receptor for import of Ams1/α-mannosidase during autophagy (i.e., under starvation conditions) in yeast [2817]. This protein was initially referred to as Atg19-B based on predictions from in silico studies [2818].
Atg35: The Atg35 protein relocates to the peri-nuclear structure and specifically regulates MIPA formation during micropexophagy; the atg35∆ mutant is able to form pexophagosomes during macropexophagy [2819].
Atg36: Atg36 is a pexophagy receptor, which localizes to the membrane of peroxisomes in S. cerevisiae. Atg36 binds Atg8 and the scaffold protein Atg11 that links receptors for selective types of autophagy to the core autophagy machinery [2820].
Atg37: KpAtg37 is a conserved acyl-CoA-binding protein that is required specifically for pexophagy in K. phaffii/P. pastoris at the stage of phagophore formation [458]. KpAtg37 regulates the interaction between the pexophagy receptor, KpAtg30, and the KpHrr25 kinase required for the recruitment of the selective autophagy scaffold protein, KpAtg11, as well as KpAtg8 and consequently the core autophagy machinery, to KpAtg30 [2814]. KpAtg37 interacts also with the peroxin KpPex3 [458]. See also ACBD5.
Atg38: Atg38 physically interacts with Atg14 and Vps34 via its N terminus. Atg38 is required for autophagy as an integral component of the PtdIns3K complex I in yeast, and Atg38 functions as a linker connecting the Vps15-Vps34 and Vps30/Atg6-Atg14 subcomplexes to facilitate complex I formation [2821].
Atg39: A receptor for selective autophagic degradation of nuclear membrane in yeast [1334].
Atg40: A receptor that functions in yeast reticulophagy [1334]. See also RETREG1.
Atg41/Icy2: A small protein that interacts with the transmembrane protein Atg9, and plays a role in autophagosome formation. Atg41 expression increases substantially under autophagy-inducing conditions; its upregulation is required for efficient autophagy, and it is regulated by the transcription factor Gcn4 [2822]. See also Atg9.
Atg42/Ybr139w: A resident soluble vacuolar glycoprotein acting as a serine carboxypeptidase, induced in nitrogen-poor conditions or following rapamycin treatment; partially redundant with Prc1/carboxypeptidase Y. Atg42 plays a role in the breakdown of autophagic bodies in the vacuole lumen and in the maintenance of cytosolic amino acid pools under conditions of nitrogen starvation [2823].
Atg43: A mitochondrial outer membrane protein in S. pombe that functions as a mitophagy receptor.
Atg44: A mitochondrial fission factor that functions in mitophagy.
ATG101: An ATG13-binding protein conserved in various eukaryotes but not in S. cerevisiae. Forms a stable complex with ULK1/2-ATG13-RB1CC1 (i.e., not nutrient-dependent) required for autophagy and localizes to the phagophore [2824, 2825]. Note that the official name for this protein in rodents is 9430023L20Rik, and in C. elegans it is EPG-9.
Atglistatin (ASTAT): N’-[4ʹ-(dimethylamino)[1,1ʹ-biphenyl]-3-yl]-N,N-dimethyl-urea is a potent, selective, and competitive inhibitor of PNPLA2/ATGL. This peptide is highly selective for PNPLA2 and does not inhibit other lipases, thus serving as a novel therapeutic tool to modulate lipolysis [2826]. See also PNPLA2.
ATI1/2 (ATG8-interacting protein 1/2): Two closely related ATG8-binding transmembrane proteins in Arabidopsis, which are unique to plants and define a stress-induced and ER-associated compartment that may function in a direct, Golgi-independent, ER-to-vacuole trafficking pathway [2827]. AT2G45980/ATI1 and AT4G00355/ATI2 interact with ER-associated AT1G48410/AGO1/ARGONAUTE 1, and are involved in its degradation in response to the viral suppressor of RNA-silencing protein P0 [2828]. ATI1 is also found in plastids following abiotic stress where it interacts with both ATG8 and plastid-localized proteins to act in their delivery to the central vacuole in an ATG5-dependent manner [1245, 2829].
ATL3 (atlastin GTPase 3): An ER-resident LC3-binding protein that functions in mammalian reticulophagy [1372].
ATM/ataxia telangiectasia mutated (ATM serine/threonine kinase): A protein kinase that is activated in the cytosol and activates TSC2 via the STK11/LKB1-AMPK cascade in response to elevated ROS, resulting in inhibition of MTOR and activation of autophagy [1208, 1209]. In addition, ATM is known as a DNA damage checkpoint kinase responsible for maintenance of genome integrity [2830]. ATM also regulates autophagy by sustaining the levels and activity of ATG4C in cancer stem cells [2831]. The interaction between ATM and autophagy is required to promote senescence in response to the 20A G-quadruplex ligand [2832]. See also senescence.
ATP2A2/SERCA: A sarco/endoplasmic reticulum type 2 calcium ATPase, a member of a family of calcium pumps important in intracellular calcium signaling. ATP2A/SERCA activity is regulated by the ER-localized autophagy protein EPG-3/VMP1 to modulate ER contacts with other organelles [2833].
ATP6V0A1 (ATPase H+ transporting V0 subunit a1): A component of the vacuolar proton ATPase that mediates acidification of intracellular organelles, including the lysosomal lumen, and regulates autophagosome-to-lysosome fusion [2834]. Genomic variability at the ATP6V0A1 locus has been associated with an increased risk of PD [2835].
ATP13A2 (ATPase type 13A2): A transmembrane lysosomal type 5 P-type ATPase that is mutated in recessive familial atypical parkinsonism, with effects on lysosomal function [2836]. Loss of ATP13A2 function inhibits the clearance of dysfunctional mitochondria [2837].
Ats-1 (Anaplasma translocated substrate-1): A type IV secretion effector of the obligatory intracellular bacterium Anaplasma phagocytophilum that binds BECN1 and induces autophagosome formation; the autophagosomes traffic to, and fuse with, A. phagocytophilum-containing vacuoles, delivering autophagic cargoes into the vacuole, which can serve as nutrients for bacterial growth [2838, 2839].
ATRA (all-trans retinoic acid): A signaling molecule derived from vitamin A that actives autophagy and cell differentiation as demonstrated in leukemia cells [577, 2743, 2840, 2841].
ATTEC (autophagosome-tethering compound): A small molecule that binds a disease-causing protein and LC3, linking the former to a phagophore [2842]
AtTSPO (Arabidopsis thaliana TSPO-related): An ER- and Golgi-localized polytopic membrane protein transiently induced by abiotic stresses. AtTSPO binds ATG8 and heme in vivo and may be involved in scavenging of cytosolic porphyrins through selective autophagy [2843].
ATXN3 (ataxin 3): A deubiquitinating enzyme that positively regulates autophagy [2844, 2845]). ATXN3 contains a polyglutamine repeat that, when expanded, causes the neurodegenerative disorder Machado-Joseph disease also known as spinocerebellar ataxia type-3 (SCA-3). Autophagy is dysregulated in this disease [2846].
AUTEN-67 (autophagy enhancer-67): An inhibitor of MTMR14, which enhances autophagy [2847].
Autogramins: Small-molecule autophagy inhibitors, which selectively target the cholesterol transport protein GRAMD1A (GRAM domain containing 1A), an essential protein for biogenesis of autophagosomes, via competing with cholesterol to limit the binding to the StAR-related lipid transfer (StART) domain, thereby inhibiting the cholesterol transfer activity of GRAMD1A [2848, 2849].
Autolysosome (AL): A degradative compartment formed by the fusion of an autophagosome (or initial autophagic vacuole/AVi) or amphisome with a lysosome (also called degradative autophagic vacuole/AVd). Upon completion of degradation the autolysosome can become a residual body [2716, 2850], or the autolysosomal membrane can be recycled to generate mature lysosomes during autophagic flux. This regenerative process, referred to as autophagic lysosome reformation (ALR), relies on the scission of extruded autolysosomal membrane tubules by the mechanoenzyme DNM2 (dynamin 2) [781, 2851].
Autophagic body (AB): The single-membrane vesicle present within the vacuole lumen that results from the fusion of an autophagosome with a vacuole limiting membrane. In S. cerevisiae, autophagic bodies can be stabilized by the addition of the proteinase B inhibitor PMSF to the medium or by the deletion of the PEP4 or ATG15 genes. Visualization of the accumulating autophagic bodies by differential interference contrast using light microscopy is a convenient, but not easily quantified, method to follow autophagy [135], whereas visualization by TEM allows the measurement of their size and number.
Autophagic lysosome reformation (ALR): A self-regulating tubulation process in which the autophagic generation of nutrients reactivates MTOR, suppresses autophagy and allows for the regeneration of lysosomes that were consumed as autolysosomes [781]. See also autolysosome.
Autophagic cell death: A historically ambiguous term describing cell death with morphological features of increased autophagic vacuoles. This term is best reserved for cell death contexts in which specific molecular methods, rather than only pharmacological or correlative methods, are used to demonstrate increased cell survival following inhibition of autophagy [1757].
Autophagic stress: A pathological situation in which induction of autophagy exceeds the cellular capacity to complete lysosomal degradation and recycling of constituents; may involve a combination of bioenergetics, acidification and microtubule-dependent trafficking deficits, to which neurons may be particularly vulnerable [16].
Autophagic vacuole: A term typically used for mammalian cells that collectively refers to autophagic structures at all stages of maturation. We recommend using this term when the specific identity of autophagosomes, amphisomes and autolysosomes are not distinguished.
AutophagamiR: A term to describe miRNAs that function in the regulation of autophagy [2852].
Autophagolysosome (APL): A degradative compartment formed by the fusion of an LC3-containing phagosome (see LAP) or an autophagosome that has sequestered a partial or complete phagosome with a lysosome. In contrast to a phagolysosome, formation of the autophagolysosome involves components of the autophagic machinery. Note that this term is not interchangeable with “autophagosome” or “autolysosome” [1480]. See also autolysosome and phagosome.
Autophagoproteasome: A cytosolic membrane-bound compartment denoted by a limiting single, double or multiple membrane, which contains both LC3 and UPS antigens. The autophagoproteasome may be derived from the inclusion of ubiquitin-proteasome structures within either early or late autophagosomes containing cytoplasmic material at various stages of degradation [100]. Thus, the concept of the autophagoproteasome refers to an ultrastructural organelle. From a functional point of view the autophagoproteasome may feature empowered substrate clearance through the concomitant activity of both autophagy and the proteasome. In fact, the autophagoproteasome contains both LC3 and UPS antigens, which are detected by TEM and show co-immunoprecipitation, as well as proteasome enzymatic activity [2853]. In other cases the proteasome component itself may be destroyed under the process later defined as proteaphagy.
Autophagosome (AP): A cytosolic membrane-bound compartment denoted by a limiting double membrane (also referred to as initial autophagic vacuole, AVi, or early autophagosome). The early autophagosome contains cytoplasmic inclusions and organelles that are morphologically unchanged because the compartment has not fused with a lysosome and lacks proteolytic enzymes. Notably, the double-membrane structure may not be apparent with certain types of fixatives. Although in most cases the term autophagosome refers to a double-membrane compartment, the late autophagosome or late autophagic vacuole (AVl) may also appear to have a single membrane (also referred to as an intermediate or intermediate/degradative autophagic vacuole, AVi/d) [2716, 2850].
Autophagy: This term summarizes all processes in which intracellular material is degraded within the lysosome/vacuole and where the macromolecular constituents are recycled.
Autophagy: A journal devoted to research in the field of autophagy (http://www.tandfonline.com/toc/kaup20/current#.VdzKoHjN5xu).
Autophagy adaptor: An Atg8-family-interacting protein that is not itself a cargo for autophagy.
Autophagy-dependent cell death: A type of RCD that depends on the autophagic machinery [1266].
Autophagy India Network: The autophagy and lysosome researchers of India meet annually to promote collaboration and share research ideas (https://autophagyindianetwork.weebly.com/).
Autophagy receptor: An Atg8-family-interacting protein that targets specific cargo for degradation and is itself degraded by autophagy (e.g., SQSTM1, NBR1, OPTN, Atg19) [2854].
Autophagy receptor: An Atg8-family-interacting protein that targets specific cargo for degradation and is itself degraded by autophagy (e.g., SQSTM1, NBR1, OPTN, Atg19) [2854].
Autophagy-like vesicles (ALVs): Double-membraned vesicles (70–400 nm) that accumulate in cells infected by a number of different viruses. These vesicles also have been referred to as compound-membrane vesicles (CMVs) or as double-membraned vesicles (DMVs).
Autosis: A form of autophagy-dependent cell death that requires Na+,K+-ATPase activity (in addition to the autophagy machinery) [2638]. Morphologically, autosis has increased numbers of autophagosomes and autolysosomes, and nuclear convolution during its early stages, followed by focal swelling of the perinuclear space. Autosis occurs in response to various types of stress including starvation and hypoxia-ischemia.
Axonal autophagy pathway: Neurons are polarized cells consisting of a highly extended long axon. One widely-accepted model is that mature lysosomes are mainly distributed in the soma, so that newly formed autophagosomes in the distal axon need to be transported in a retrograde manner into the soma for degradation [880, 881]. However, studies suggest a second pathway through which degradative lysosomes are delivered into axons. This pathway has been characterized by applying a set of fluorescent probes that selectively label active forms of lysosomal hydrolases in developing and mature neurons in microfluidic devices. Soma-derived degradative lysosomes rapidly influx into distal axons and target to autophagosomes for local degradation [2855, 2856]. Disrupting axon-targeted delivery of degradative lysosomes induces axonal autophagic stress. Thus, the axon is also an active compartment for local degradation of autophagosomes.
Ayr1: A triacylglycerol lipase involved in autophagy in yeast [2857]. Enzymes that participate in the metabolism of lipid droplets including Dga1 and Lro1 (acyltransferases involved in triacylglycerol synthesis) and Are1/2 (Acyl-CoA:sterol acyltransferases) that generate the major components of lipid droplets, triacylglycerols and steryl esters, are required for efficient autophagy. Deletion of the genes encoding Yeh1 (a steryl ester hydrolase), Ayr1 or Ldh1 (an enzyme with esterase and triacylglycerol lipase activities) also partially blocks autophagy. Finally, Ice2 and Ldb16, integral membrane proteins that participate in formation of ER-lipid droplet contact sites that may be involved in lipid transfer between these sites are also needed for efficient autophagy.
AZD8055: A novel ATP-competitive inhibitor of MTOR kinase activity. AZD8055 shows excellent selectivity against all class I PI3K isoforms and other members of the PI3K-like kinase family. Treatment with AZD8055 inhibits MTORC1 and MTORC2 and prevents feedback to AKT [1945].
Azithromycin: A well-known antibiotic that blocks autophagy through inhibition of V-ATPase activity and subsequent blocking of autophagosome-lysosome fusion [2858].
β-oxidation: A multi-step process by which fatty acids are broken down to produce energy (ATP) in mitochondria. Deficiency in selective autophagy is associated with suppression of lipid oxidation. Positive-stranded RNA viruses such as DENV utilize the autophagy-dependent lipid metabolism for their efficient replication [2859, 2860].
Bafilomycin A1 (BAFA1/BAF): An inhibitor of the V-type ATPase as well as certain P-type ATPases that prevents acidification and alters the membrane potential of certain compartments; treatment with bafilomycin A1 ultimately results in a block in fusion of autophagosomes with lysosomes, thus preventing the maturation of autophagosomes into autolysosomes [218, 219, 303]. Bafilomycin A1 can also inhibit some forms of non-canonical autophagy [760, 2861]. Note that the abbreviation for bafilomycin A1 is not “BFA,” as the latter is the standard abbreviation for brefeldin A; nor should BAF be confused with the abbreviation for the caspase inhibitor boc-asp(o-methyl)fluoremethylketone. See also concanamycin A.
BAG3 (BCL2-associated athanogene 3): A stress-induced co-chaperone that utilizes the specificity of HSP70 molecular chaperones toward non-native proteins as the basis for targeted, ubiquitin-independent autophagic degradation in mammalian cells (“BAG3-mediated selective macroautophagy”); BAG3 is induced by stress and during cell aging, and interacts with HSP70 and dynein to target misfolded protein substrates to aggresomes, leading to their selective degradation [1838, 1839]. BAG3 also interacts with HSPB6 and HSPB8 to target substrates for chaperone-assisted selective autophagy via a ubiquitin-dependent mechanism [1828].
BAG6/BAT3 (BCL2-associated athanogene 6): BAG6 tightly controls autophagy by modulating EP300 intracellular localization, affecting the accessibility of EP300 to its substrates, TP53 and ATG7. In the absence of BAG6 or when this protein is located exclusively in the cytosol, autophagy is abrogated, ATG7 is hyperacetylated, TP53 acetylation is abolished, and EP300 accumulates in the cytosol, indicating that of BAG6 regulates the nuclear localization of EP300 [2862].
BapH: See AN4171.
BAPTA-AM (1,2-bis[2-aminophenoxy]ethane-N,N,N′,N′-tetraacetic acid tetrakis[acetoxymethyl ester]): BAPTA-AM is a potent cell permeant calcium-chelating agent that is an analog of BAPTA with fast, high-affinity calcium-buffering properties, typically applied in the 10-20 µM range. BAPTA-AM suppresses starvation-, rapamycin- and resveratrol-induced autophagic flux [1983, 1985, 1991]). BAPTA-AM is useful for manipulation of intracellular free calcium levels [2863–2865]; however, once inside cells, BAPTA-AM can also exert calcium-independent effects such as inhibition of the Na+/K+ ATPase [2002, 2003].
BARA (β-α repeated, autophagy-specific): A domain at the C terminus of Vps30/Atg6 that is required for targeting PtdIns3K complex I to the PAS [2866]. The BARA domain is also found at the C terminus of BECN1.
Barkor: See ATG14.
Basal autophagy: Constitutive autophagic degradation that proceeds in the absence of any overt stress or stimulus. Basal autophagy is important for the clearance of damaged proteins and organelles in normal cells (especially fully differentiated, nondividing cells).
BATS (Barkor/Atg14[L] autophagosome targeting sequence) domain: A protein domain within ATG14 that is required for the recruitment of the class III PtdIns3K to LC3-containing puncta during autophagy induction; the predicted structure of the BATS domain suggests that it senses membrane curvature [824].
Bck1: A MAPKKK downstream of Pkc1 and upstream of Mkk1/2 and Slt2 that controls cell integrity in response to cell wall stress; Bck1 is required for pexophagy [1045] and mitophagy [747]. See also Slt2 and Hog1.
BCL2-family proteins: There are 3 general classes of BCL2 proteins: anti-apoptotic proteins include BCL2, BCL2L1/Bcl-XL, BCL2L2/BCL-W and MCL1 that inhibit autophagy; the pro-apoptotic BH3-only proteins include BAD, BIK, PMAIP1/NOXA, BBC3/PUMA and BCL2L11/Bim/BimEL that induce autophagy; and the pro-apoptotic effector proteins BAX and BAK1. Interaction of BCL2 with BECN1 prevents the association of the latter with the class III PtdIns3K; however, anti-apoptotic BCL2 proteins require BAX and BAK1 to modulate autophagy [2867].
BCL2L13/BCL-RAMBO (BCL2 like 13): BCL2L13 is a mammalian homolog of Atg32, which is located in the mitochondrial outer membrane and has an LC3-interacting region. BCL2L13 induces mitochondrial fission and mitophagy [2868]. See also Atg32.
BCL10 (BCL10 immune signaling adaptor): The adaptor protein BCL10 is a critically important mediator of T cell receptor (TCR)-to-NFKB signaling. After association with the receptor SQSTM1, BCL10 is degraded upon TCR engagement. Selective autophagy of BCL10 is a pathway-intrinsic homeostatic mechanism that modulates TCR signaling to NFKB in effector T cells [2869].
BDNF (brain derived neurotrophic factor): BDNF is a member of the nerve growth factor family of proteins that plays a key role in neuronal survival [2870–2872]. BDNF can promote neuronal survival via modulation of autophagy through the MTORC1 pathway [2873]. Under conditions where apoptosis is still functional, BDNF can promote neuronal survival by promoting autophagy, and activating the MTORC2-PI3K-AKT pathway in a neurotoxic cell model of mitochondrial complex II inhibition (R. Sathyanarayanan and M.M. Srinivas Bharath, unpublished data).
BEC-1: The C. elegans ortholog of BECN1.
Beclin 1: See BECN1.
BECN1/Beclin 1 (beclin 1): A mammalian homolog of yeast Vps30/Atg6 that forms part of the class III PtdIns3K complex involved in activating autophagy [2874]. BECN1 interacts with many proteins including BCL2, VMP1, ATG14, UVRAG, PIK3C3 and RUBCN through its BH3, flexible helical, coiled-coil, and BARA domains; the latter two domains are also referred to as the evolutionarily conserved domain (ECD) based on high sequence conservation of this region [2875]. The C. elegans ortholog is BEC-1.
BECN1s (BECN1 short isoform): A splice variant of BECN1 that lacks the sequence corresponding to exons 10 and 11; BECN1s associates with the mitochondrial outer membrane and is required for mitophagy [2876] BECN1s can bind ATG14 and activate PIK3C3/VPS34, but does not bind UVRAG.
BECN2/Beclin 2 (beclin 2): A mammalian-specific homolog of yeast Vps30/Atg6 that forms part of the class III PtdIns3K complex involved in activating autophagy and that also functions in the endolysosomal degradation of several cellular G protein-coupled receptors (GPCRs; independently of the class III PtdIns3K complex) [2877] and a virally encoded GPCR [2878].
Betulinic acid: Betulinic acid and its derivatives activate autophagy as a rescue mechanism to deal with damaged mitochondria [314, 1901, 1902, 2879]; however, betulinic acid impairs lysosomal integrity and converts autophagy into a detrimental process, leading to accumulation of nonfunctional autolysosomes that can be detected over a long time frame [314].
BH domain: BCL2 homology domain [2880]. There are 4 domains of homology, consisting of BH1, BH2, BH3 and BH4.
BH3 domain: A BCL2 homology (BH) domain that is found in all BCL2 family proteins, whether they are pro-apoptotic or anti-apoptotic. A BH3 domain is also present in BECN1 and binds to a hydrophobic surface groove on anti-apoptotic BCL2 proteins such as human BCL2, BCL2L1/bcl-xL, BCL2L2/BCL-W and MCL1, as well as γ-herpesvirus 68 M11 [2881].
BH3-only proteins: A series of proteins that contain a BH3 domain (but not any other BCL2 homology domains). Several BH3-only proteins (BAD, BIK, PMAIP1/NOXA, BBC3/PUMA and BCL2L11/Bim/BimEL) can competitively disrupt the inhibitory interaction between BCL2 and BECN1, which is also a BH3-only protein [2881], to allow the latter to act as an allosteric activator of PtdIns3K and to activate autophagy.
Bif-1: See SH3GLB1.
BIPASS (BAG-instructed proteasomal-to-autophagosomal switch and sorting): Upon proteasomal impairment, cells switch to autophagy to ensure proper clearance of substrates (the proteasome-to-autophagy switch). Following this proteasome impairment, increasing the BAG3:BAG1 ratio ensures the initiation of BIPASS [2882].
BIRC2/cIAP1/hiap-2 (baculoviral IAP repeat containing 2): Canonically regarded as an anti-apoptotic protein, BIRC2 is also involved in regulating mitophagy during serum starvation by association with ubiquitin [2883].
BIRC5/Survivin (baculoviral IAP repeat containing 5): As a member of the inhibitor-of-apoptosis proteins (IAPs) family, BIRC5 interacts with ATG7 and the ATG12–ATG5 conjugate to interrupt the formation of the ATG12–ATG5-ATG16L1 protein complex, thus limiting autophagy. This BIRC5-directed autophagy inhibition maintains DNA integrity in human cancer and mouse embryonic fibroblast cells [821]. BIRC5 can bind to LC3 via a canonical LIR, but its mechanism of autophagy inhibition does not utilize this interaction [2884].
BIRC6/BRUCE (baculoviral IAP repeat containing 6): An anti-apoptotic protein and an E2 ubiquitin enzyme that directly ubiquitinates substrates without E3 ubiquitin ligases. BIRC6 is required to complete fusion of autophagosomes with lysosomes; BIRC6 forms a complex with STX17 and SNAP29 [2885].
BIX01294: An inhibitor of EHMT2/G9a histone methyltransferase (that introduces H3K9me2 and H3K27me3 repressive marks) that triggers autophagy or autophagy-associated cell death in several tumor cells, including human glioma cells as well as glioma stem-like cells (GSCs) isolated from an established cell line and patient-derived cultures [2886–2889]. EHMT2 binds to the promoters of autophagy- (LC3B, WIPI1) and differentiation-related (GFAP, TUBB3) genes in GSCs; BIX01294 treatment upregulates the expression of these genes in GSCs by counteracting EHMT2 activity [2889].
BLOC1S1/GCN5L1 (biogenesis of lysosomal organelles complex 1 subunit 1): A component of the mitochondrial acetyltransferase activity that modulates mitophagy and mitochondrial biogenesis [2890].
BNIP3 (BCL2 interacting protein 3): Identified in a yeast two-hybrid screen as interacting through its N-terminal 40 amino acids with BCL2 and adenovirus E1B [2891]. Previously classified as a pro-apoptotic protein, BNIP3 promotes mitophagy through direct interaction with LC3B-II mediated by a conserved LIR motif that overlaps with its BCL2-interacting region [2892, 2893]. BNIP3 also modulates mitochondrial fusion through inhibitory interactions with OPA1 via its carboxy terminal 10 amino acids [2894]. BNIP3 is transcriptionally regulated by HIF1A [2895], E2Fs [2896], FOXO3 [662], TP53 [2897] and NFKB [2898] and is most highly expressed in adult heart and liver [2899, 2900].
BNIP3L/NIX (BCL2 interacting protein 3 like): Identified as a BNIP3 homolog, BNIP3L is required for mitophagy in red blood cells [2170, 2171], three factors-induced reprogramming [2901], and neuronal differentiation in the mouse retina [1151]. As with BNIP3, BNIP3L is hypoxia-inducible and also interacts with LC3B-II and GABARAP through a conserved LIR motif in its amino terminus [284]. BNIP3L also interacts with RHEB at the mitochondria and the LC3-BNIP3L-RHEB complex promotes mitochondrial turnover and efficient mitochondrial function [2902]. In CCCP-induced mitophagy, BNIP3L is required for the activation of autophagy by promoting CCCP-induced mitochondrial depolarization and ROS generation, which inhibits MTORC1 [2903].
Bre5: A cofactor for the deubiquitinating enzyme Ubp3. See also Ubp3.
C/EBPβ: See CEBPB.
C9orf72: C9orf72 plays an important role in the regulation of endosomal trafficking, and interacts with RAB proteins involved in autophagy and endocytic transport. C9orf72 contains a DENN (differentially expressed in normal and neoplasia)-like domain, suggesting that it may function as a GDP-GTP exchange factor for a RAB GTPase, similar to other DENN proteins. The normal function of C9orf72 remains unknown, but it is highly conserved and expressed in many tissues, including the cerebellum and cortex. Hexanucleotide (GGGGCC) repeat expansions in a noncoding region of the C9orf72 gene are the major cause of familial ALS and frontotemporal dementia [2904]. Recent studies suggest that C9orf72 is an important regulator of autophagic functions [715, 1300, 2905–2908] including lipophagy via TFEB and its cofactor CARM1 [1299]. See also CARM1 and TFEB.
C12orf5: See TIGAR.
C12orf44: See ATG101
Ca-P60A/dSERCA: The Drosophila ER calcium-translocating ATPase. Inhibition of Ca-P60A with bafilomycin A1 blocks autophagosome-lysosome fusion [303].
CA7-4: A lncRNA that is upregulated by a high concentration of glucose. CA7-4 facilitates endothelial autophagy and apoptosis as a competing endogenous RNA (ceRNA) by decoying MIR877-3P and MIR5680, promoting the expression of CTNNBIP1 and DPP4, decreasing the level of CTNNB1 and increasing AMPK phosphorylation [2909].
Cad96Ca/Stit/Stitcher (Cadherin 96Ca): A Drosophila receptor tyrosine kinase that is orthologous to the human proto-oncogene RET. Cad96Ca suppresses autophagy in epithelial tissues through Akt1-TORC1 signaling in parallel to InR (Insulin-like receptor). This endows epithelial tissues with starvation resistance and anabolic development during nutritional stress [2910].
Caf4: A component of the mitochondrial fission complex that is recruited to degrading mitochondria to facilitate mitophagy-specific fission [2911].
CAL-101: A small molecule inhibitor of the PIK3CD/p110δ subunit of class 1A phosphoinositide 3-kinase; treatment of multiple myeloma cells results in autophagy induction [2912].
Calcineurin: See PPP3.
CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2): An autophagy receptor that binds to the bacterial ubiquitin coat and Atg8-family proteins to target invasive bacteria, including S. typhimurium and Streptococcus pyogenes for autophagosomal sequestration [1470]. CALCOCO2 also regulates autophagosome maturation through its interaction with TOM1-bound MYO6 (myosin VI) on endosomes, and LC3A/B or GABARAPL2 on autophagosomes [2913, 2914].
CALM (calmodulin): A calcium-receptor protein in all eukaryotic cells containing four EF-hand calcium-binding sites. CALM transduces signals mediated by transient changes in the concentration of free calcium within the cytosol and the nucleus, upon binding and regulating a large number of enzymes, channels, signaling, adaptor and structural proteins. CALM also regulates a minor group of target proteins in its calcium-free form. Autophagy is among the multiple cellular functions regulated by CALM through modulating key proteins involved in this process [2915].
Calpain-dependent autophagy: A form of caspase-independent programmed cell death that occurs in neurons during normal development and in certain neurological diseases [2916].
Calpains: A class of calcium-dependent, non-lysosomal cysteine proteases that cleaves and inactivates ATG5 and the ATG12–ATG5 conjugate, hence establishing a link between reduced calcium concentrations and induction of autophagy [2917].
Calpeptin: A potent calpain inhibitor compound that induces autophagy, including increased production of autophagosomes [2918], in an MTOR-independent manner [1951].
CALR (calreticulin): A chaperone that is mainly associated with the ER lumen, where it performs important functions such as calcium buffering, and participates in protein folding and maturation of, as well as antigen loading on, MHC molecules [2919]. An extracellular role for CALR has emerged where it acts as an “eat me” signal on the surface of cancer cells [2920]. Importantly, in the context of Hyp-PDT, autophagy suppresses CALR surface exposure by reducing ER-associated proteotoxicity [1724, 1729, 2921]. Disruption of LAMP2A also affects CALR surface exposure [1729].
CaMKKβ: See CAMKK2.
CAMKK2 (calcium/calmodulin-dependent protein kinase kinase 2, beta): Activates OGT and AMPK in response to an increase in the cytosolic calcium concentration [2922], resulting in the induction of autophagy [1007, 1984].
Cannabidiol (CBD): A major nonpsychoactive constituent of cannabis, recently considered an antineoplastic compound on the basis of its in vitro and in vivo activity against tumor cells of different histogenesis. CBD is able to induce autophagy. In particular, CBD induces programmed cell death in breast cancer cells by coordinating the cross-talk between apoptosis and autophagy [2923]. In leukemic cells CBD was revealed to induce autophagy and cell death by directly targeting mitochondria [2924].
Canonical autophagy: Engagement of the hierarchically ordered activity of ATG proteins at the phagopore for the formation of double-membraned autophagosomes. The non-canonical use, in contrast, does not require the organized intervention of all ATG proteins, does not necessarily initiate from a single source, and a set of ATG proteins can be recruited to a pre-existing membrane that is different from the phagophore [2925].
CAPNS1 (calpain, small subunit 1): The regulatory subunit of micro- and millicalpain; CAPNS1-deficient cells are autophagy defective and display a substantial increase in apoptotic cell death [2926].
CAPZA1 (capping actin protein of muscle Z-line alpha subunit 1): The α subunit of F-actin capping protein that regulates actin polymerization and cell motility via binding to the barbed ends of actin filaments; CAPZA1 inhibits LAMP1 expression via binding to the LRP1 intracellular domain (LRP1-ICD) in the nuclei. The binding of CAPZA1 to the LRP1-ICD prevents LRP1-ICD binding to the LAMP1 proximal promoter. Thus, in CAPZA1-overexpressing gastric epithelial cells infected with H. pylori, autolysosome formation is inhibited [2927]. See also LRP1.
CAR3 (carbonic anhydrase 3): An isoform of carbonic anhydrase that interacts with BAG3 and suppresses CASA in skeletal muscle cells [2928].
Cardiolipin: A phospholipid found in bacteria and mitochondria that regulates transmembrane protein complexes including the mitochondrial respiratory chain. Cardiolipin is translocated to the outer mitochondrial membrane during mitochondrial injury and binds Atg8-family proteins to trigger mitophagy [201, 1190].
CARM1 (coactivator associated arginine methyltransferase 1): CARM1, a histone-arginine methyltransferase, is a coactivator of TFEB. The AMPK-SKP2-CARM1 signaling axis is involved in the transciptional regulation of autophagy [980]. The C9orf72-CARM1 axis regulates lipophagy induction after nutrient starvation [1299].
CASA (chaperone-assisted selective autophagy): A selective macroautophagy pathway that utilizes the co-chaperone BAG3 or its Drosophila homolog stv/starvin to direct the ubiquitination and lysosomal degradation of substrates through the action of HSPA8, HSPB8, STUB1/CHIP (or other ubiquitin ligases), SYNPO2/myopodin, and SQSTM1 [1828, 1837]. CASA routes substrates to the lysosome via an autophagosome. The process is thought to take place at the microtubule organizing center to which the CASA complex is transported in a retrograde manner by dynein associated directly with a PxxP motif of BAG3 [1840, 2929]. If HSPB8-BAG3 are displaced by the co-chaperone BAG1, the cargoes are routed by HSPA8-STUB1 to the proteasome. CASA should not be confused with CMA, which also uses chaperones for lysosome-dependent degradation.
Casein kinase 2 (CK2): CK2 is a serine/threonine protein kinase complex that is involved in many cellular activities. CK2 phosphorylates Atg32 and induces mitophagy in yeast [2930]. See also Atg32.
Caspases (cysteine-dependent aspartate-directed proteases): A class of cysteine proteases that play essential roles in apoptosis (formerly called programmed cell death type I) and inflammation. Several pro-apoptotic caspases directly interact with and cleave essential ATG proteins, resulting in the inhibition of autophagy [611, 2931]. For example, CASP3 and CASP8 cleave BECN1 and inhibit autophagy [2932, 2933]. The active form of CASP8 is essential for upregulation of apoptosis in some types of autophagy-deficient cells under conditions of nutrient limitation [2934].
CASR (calcium sensing receptor): A class C G-protein coupled receptor that senses extracellular levels of calcium ion, with a primary function in calcium homeostasis regulation. CASR stimulation is associated with increased autophagy [1563, 2594, 2595, 2935], whereas its inhibition is linked to decreased autophagy [2936–2938].
CCCP (carbonyl cyanide m-chlorophenyl hydrazone): Protonophore and uncoupler of oxidative phosphorylation in mitochondria; stimulates general autophagy [2593] and mitochondrial degradation inducing mitophagic activity [339].
CCDC88A/GIV (coiled-coil domain containing 88A): A guanine nucleotide exchange factor for GNAI3 that acts to downregulate autophagy [2939]. CCDC88A disrupts the GPSM1-GNAI3 complex in response to growth factors, releasing the G protein from the phagophore or autophagosome membrane; GNAI3-GTP also activates the class I PI3K, thus inhibiting autophagy. See also GNAI3.
CCI-779 (temsirolimus): A water-soluble rapamycin ester that induces autophagy.
CCND1 (cyclin D1): CCND1 functions as a regulator of cyclin-dependent kinases, and is a protein required for progression through the G1 phase of the cell cycle. CCND1 is overexpressed in various cancers, including hepatocellular carcinoma, and is selectively recruited and degraded in a process mediated by SQSTM1 after ubiquitination [2940, 2941].
CCNY (cyclin Y): A substrate of AMPK that interacts with and activates CDK16 to promote autophagy [762]. See also CDK16.
CCPG1 (cell cycle progression 1): An ER-resident LC3- and RB1CC1-binding protein that functions in ER stress-induced mammalian reticulophagy [1376].
CD5L (CD5 molecule like): A 40-kDa soluble protein, expressed and secreted mainly by tissue macrophages, that controls macrophage inflammatory responses and polarization through the activation of autophagy. CD5L induces macrophage autophagy through the CD36 receptor [2942–2944].
CD38 (CD38 molecule): CD38 is a powerful disease marker for human leukemias and myelomas and is involved in the development of many disease such as chronic lymphocytic leukemia, type II diabetes mellitus, atherosclerosis. As a multifunctional enzyme, CD38 can cleave NAD+ and NADP+ to generate cyclic ADP-ribose/cADPR and NAADP, which regulate autophagy by lysosomal calcium release events that promote fusion of the lysosome with autophagosomes [2945, 2946].
Cdc48: Yeast homolog of VCP that is a type II AAA+-ATPase that extracts ubiquitinated proteins from the membrane as part of the ER-associated protein degradation pathway and during ER homeotypic fusion [2947], but is also required for nonselective autophagy [2948]. Disease-associated mutant forms of Cdc48 are also targeted for autophagic degradation in plants and yeast [1359]. See also Shp1 and VCP.
CD46: A cell-surface glycoprotein that interacts with the scaffold protein GOPC to mediate an immune response to invasive pathogens including Neisseria and Group A Streptococcus. Interaction of pathogens via the Cyt1 cytosolic tail induces autophagy, which involves GOPC binding to BECN1. CD46 is also used as a cellular receptor by several pathogens [2949].
CDC42BPA/MRCKA/MRCKα (CDC42 binding protein kinase alpha): An actin-myosin regulatory kinase required for TP53-dependent autophagy induced by MDM2/HDM2 inhibition [2950].
Cdk5 (cyclin dependent kinase 5): An unusual member of the cyclin-dependent kinase family, having no known function in the cell cycle, that is primarily active in postmitotic neurons. CDK5-mediated downregulation of autophagy is associated with neurodegeneration primarily through overactivation of the innate immune response [2951], and CDK5-mediated autophagy has been linked with PD [2952, 2953]. This protein regulates autophagy by phosphorylation of different substrates such as PIK3C3/VPS34 [411], SH3GLB1/endophilin B1 [2940, 2952] and ACIN1/acinus [2691].
CDK16 (cyclin dependent kinase 16): CDK16, a member of the PCTAIRE kinase family, associates with CCNY (cyclin Y), which is a substrate of AMPK. Phosphorylation of CCNY at S326 stimulates interaction with CDK16 and activates the kinase, which promotes autophagy. CDK16 seems not to have a major impact on the cell cycle; rather, it is associated with cellular differentiation and with intracellular transport processes [762].
CDKN1A/p21/Cip1 (cyclin dependent kinase inhibitor 1A): A cyclin-dependent kinase inhibitor that is associated with the induction of autophagy in melanoma cells upon exposure to a telomeric G-quadruplex stabilizing agent [2954].
CDKN1B/p27/Kip1 (cyclin dependent kinase inhibitor 1B): A cyclin-dependent kinase inhibitor that is phosphorylated and stabilized by an AMPK-dependent process and stimulates autophagy [2955].
CDKN2A (cyclin dependent kinase inhibitor 2A): The CDKN2A locus encodes two overlapping tumor suppressors that do not share reading frame: p16INK4a and p14ARF. The p14ARF tumor suppressor protein (p19ARF in mouse) can localize to mitochondria and induce autophagy. Tumor-derived mutant forms of p14ARF that do not affect the p16INK4a coding region are impaired for autophagy induction, thus implicating this activity in tumor suppression by this commonly mutated locus [2956]. This gene also encodes a lower molecular weight variant called smARF. See also smARF.
CEBPB/C/EBPβ (CCAAT enhancer binding protein beta): A transcription factor that regulates several autophagy genes; CEBPB is induced in response to starvation, and the protein levels display a diurnal rhythm [989].
Cell differentiation: This is a process through which a cell commits to becoming a more specialized cell type having a distinct form and a specific function(s). Autophagy is activated during the differentiation of various normal and cancerous cells, as revealed, for example, in adipocytes, erythrocytes, lymphocytes and leukemia cells [636, 2957].
CEP-1 (C. elegans P-53-like protein): See TP53.
Ceramide: Ceramide is a bioactive sphingolipid, which plays a mitochondrial receptor role to recruit LC3-II-associated phagophores to mitochondria for degradation in response to ceramide stress and DNM1L-mediated mitochondrial fission; the direct binding between ceramide and LC3-II involves F52 and I35 residues of LC3B [896, 901]. Ceramide is also a key synthetic building block that occupies the central metabolic branchpoint for anabolic production of sphingolipids such as sphingomyelin, ceramide-1-phosphate, and glucosylceramide, which are needed for complex glycosphingolipid synthesis (including gangliosides) [885, 887].
Ceramide-1-phosphate (C1P): C1P is a bioactive sphingolipid that is synthesized by CERK (ceramide kinase) in the trans-Golgi and promotes some of the same anti-apoptotic responses as sphingosine-1-phosphate. Exogenous C1P treatment of cells or elevation of C1P at the Golgi apparatus by downregulation of CPTP (ceramide-1-phosphate transfer protein) induces eicosanoid production initiated by PLA2G4/cytoplasmic phospholipase A2 as well as autophagy in various cells, and inflammasome assembly in surveillance cells that drives pro-inflammatory interleukin release [2958–2960]. It is noteworthy that UVRAG contains a putative C1P binding site predicted to enhance membrane interaction [2958, 2961]. See also lipid transfer proteins.
CFATG (Club Francophone de l’AuTophaGie): A club created in 2011 under the impetus of Patrice Codogno to bring together the French scientific community around the theme of autophagy and to favor synergy between scientists coming from different backgrounds. A dedicated website (www.cfatg.org) has been created and a national meeting is organized every year since 2011.
CFLAR/c-FLIP: An inactive CASP8 homolog that inhibits death receptor-mediated apoptosis and necroptosis. In addition, CFLAR and its viral homologs bind to ATG3 and inhibit autophagy [1965]. CFLAR-mediated autophagy modulation is also observed in T cells [2962]. Moreover, v-FLIP of the Kaposi sarcoma-associated herpesvirus (KSHV) inhibits autophagy during viral latency [2963].
CGAS/MB21D1 (cyclic GMP-AMP synthase): A cytosolic sensor that produces cGMP to initiate IFN production via STING1/TMEM173 upon binding microbial DNA [2964]. MB21D1 also binds to BECN1, releasing RUBCN, resulting in the induction of autophagy to eliminate cytosolic pathogens and cytosolic DNA; the latter serves to downregulate the immune response to prevent overactivation.
CH5132799 (PA-799): An inhibitor of class I PI3Ks and particularly PIK3CA/PI3Kα. The role of the PI3K-AKT-MTOR signaling pathway in autophagy inhibition is a well established [2965].
Chaperone-mediated autophagy (CMA): An autophagic process in most mammalian cells, and some birds and reptile species by which proteins containing a particular pentapeptide motif related to KFERQ are transported across the lysosomal membrane and degraded [1809, 2966, 2967]. The translocation process requires the action of the integral membrane protein LAMP2A and both cytosolic and lumenal HSPA8 [1816, 2968].
CHEK2 (checkpoint kinase 2): The serine/threonine kinase CHEK2 is a key component of the DNA damage response. Autophagy is involved in proper CHEK2 activation, whereas its inhibition results in CHEK2 inactivation through a proteasome-dependent mechanism [2832, 2969].
CHKB (choline kinase beta): A kinase involved in phosphatidylcholine synthesis; mutations in CHKB cause mitochondrial dysfunction leading to mitophagy and megaconial congenital muscular dystrophy [2970].
Chloroquine (CQ): CQ and its derivatives (such as 3-hydroxychloroquine) raise the lysosomal pH and ultimately inhibit the fusion between autophagosomes and lysosomes [302], thus preventing the maturation of autophagosomes into autolysosomes, and blocking a late step of autophagy [302, 2971].
CHMP1A (charged multivesicular body protein 1A): CHMP1A is a member of the CHMP family of proteins that are involved in multivesicular body sorting of proteins to the interiors of lysosomes. CHMP1A regulates the autophagic turnover of plastid constituents in A. thaliana [1246].
Chlorophagy: A selective autophagy pathway to degrade chloroplasts in plant cells. In leaves of A. thaliana, starvation or aging induces autophagic degradation of chloroplast stromal protein-containing vesicles termed Rubisco-containing bodies (RCBs) [1241]. Damaged chloroplasts are eliminated in their entirety via microautophagic incorporation into the vacuole [1243, 1244].
Cholesterol: A sterol synthesized by all animal cells, which is an essential structural component of cell membranes and a precursor for the biosynthesis of steroid hormones, bile acid and vitamin D. Increased cholesterol levels enhance autophagosome formation by promoting the oxidative inactivation of ATG4, but impair lysosomal fusion ability by altering RAB7A and SNAREs content and distribution in lysosome membranes [2972, 2973].
CHOP: See DDIT3.
Chromatophagy: A form of autophagy that involves nuclear chromatin/DNA leakage captured by autophagosomes or autolysosomes [1248].
Chromosomal instability: Abnormal chromosomal segregation during cell division leads to chromosomal instability, one of the hallmarks of cancer. Inhibition of autophagy and lysosome acidification capacity in mitotic cells induces chromosomal instability, which can be followed by the formation of toroidal nuclei in daughter cells [2974].
Ciliophagy: Degradation by autophagy of cilia components and proteins involved in the process of ciliogenesis (formation of primary cilia) [2975–2977]. Ciliophagy can modulate ciliogenesis positively or negatively depending on whether the subset of proteins degraded via autophagosomes are activators or inhibitors of the formation of primary cilia [2978]. See also primary cilium.
CIP2A/KIAA1524 (cellular inhibitor of PP2A): KIAA1524/CIP2A suppresses MTORC1-associated PPP2/PP2A activity in an allosteric manner thereby stabilizing the phosphorylation of MTORC1 substrates and inhibiting autophagy. CIP2A can be degraded by autophagy in an SQSTM1-dependent manner [2979].
circ-DNMT1 (hsa_circRNA_102439): A circRNA that stimulates autophagy and increases breast cancer cell proliferation [2980].
circEIF6 (hsa_circ_0060060): A circRNA that promotes cisplatin-induced autophagy in papillary thyroid carcinoma and anaplastic thyroid carcinoma cells [2981].
circ-GATAD2A (circular RNA GATA zinc finger domain containing 2A): A circRNA that inhibits autophagy and promotes the replication of H1N1 [2982].
circHECTD1: A circRNA that inhibits astrocyte activation via autophagy [2983].
circHECW2: A circRNA that contributes to the nonautophagic role of ATG5 in endothelial-mesenchymal transition [2984].
circHIPK2: A circRNA that regulates astrocyte activation via the interplay between autophagy and ER stress [2985].
circHIPK3 (circular homeodomain interacting protein kinase 3): A circRNA prognostic factor that regulates autophagy in lung cancer [2986].
circRNA.2837: A circRNA that regulates neuronal autophagy and acts as a competing endogenous RNA/ceRNA [2987].
circRNA ACR (autophagy-related circular RNA): A circRNA that represses myocardial infarction by inhibiting autophagy [2988].
CircRNAs (circular RNAs): CircRNAs as novel endogenous noncoding RNAs exhibit cell-type-specific and tissue-specific patterns. These covalently closed RNAs are implicated in diseases and play crucial roles in autophagy regulation [2989–2991].
ciRS‐7: A circRNA that inhibits autophagy in esophageal squamous cell carcinoma [2992].
CISD2/NAF-1 (CDGSH iron sulfur domain 2): An integral membrane component that associates with the ITPR complex; CISD2 binds BCL2 at the ER, and is required for BCL2 to bind BECN1, resulting in the inhibition of autophagy [2993]. CISD2 was reported to be associated with the ER, but the majority of the protein is localized at mitochondria, and mutations in CISD2 are associated with Wolfram syndrome 2; accelerated autophagy in cisd2−/− mice may cause mitochondrial degradation, leading to neuron and muscle degeneration [2994].
Cka (Connector of kinase to AP-1): A scaffold protein that is a core component of the striatin-interacting phosphatase and kinase (STRIPAK) complex in Drosophila, that is essential for autophagosome transport in neurons. Cka mediates attachment of autophagosomes to the dynein-dynactin transport machinery through direct binding to Atg8a [2147].
CLEAR (coordinated lysosomal expression and regulation) gene network: A regulatory pathway involving TFEB, which regulates the biogenesis and function of the lysosome and associated pathways including autophagy [969]. See also PPP3 and TFEB.
CLEC16A (C-type lectin domain family 16, member A): See Ema.
Clg1: A yeast cyclin-like protein that interacts with Pho85 to induce autophagy by inhibiting Sic1 [2995].
CLN3 (CLN3 lysosomal/endosomal transmembrane protein, battenin): An endosomal/lysosomal protein whose deficiency causes inefficient autolysosome clearance and accumulation of autofluorescent lysosomal storage material and ATP5G/subunit c (ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C) [2996, 2997]. In human, recessive CLN3 mutations cause juvenile neuronal ceroid lipofuscinosis (JNCL; Batten disease). Recessive CLN3 mutations have also been reported in cases of autophagic vacuolar myopathy and non-syndromic retinal disease [2998, 2999].
CLN7: See MFSD8.
Clockophagy: A type of selective autophagy that degradates the key circadian clock protein ARNTL/BMAL1 (aryl hydrocarbon receptor nuclear translocator-like) during RSL3-induced ferroptosis [1249].
CMA: See chaperone-mediated autophagy.
COG (conserved oligomeric Golgi) complex: A cytosolic tethering complex that functions in the fusion of vesicles within the Golgi complex, but also participates in autophagy and facilitates the delivery of Atg8 and Atg9 to the PAS [3000].
Complete autophagy: Also termed productive autophagy, this refers to the entire process of autophagy, from formation of a phagophore, maturation of an autophagosome encompassing the cargo, delivery of the cargo to lysosomes/vacuole and subsequent degradation and efflux [3001, 3002]. See also incomplete autophagy.
Concanamycin A: Vacuolar-type H+-translocating ATPase inhibitor that raises vacuolar pH and impedes hydrolase activity in this cellular compartment [3003]. See also bafilomycin A1.
Connexins: See gap junction proteins/connexins.
Cordycepin (3ʹ-deoxyadenosine): A bioactive compound that activates autophagy, independently of the MTOR pathway [3004].
CORM (CO-releasing molecule): Carbon monoxide, partly through activation of autophagy, exerts cardioprotective effects in a mouse model of metabolic syndrome-induced myocardial dysfunction [3005].
Cornification: A form of programmed cell death during which many intracellular proteins are degraded by autophagy while others are covalently cross-linked [1043]. Cornified cells remain connected by intercellular junctions and function as mechanically resistant components of the epidermis, hair and nails.
Corynoxine/Cory: An oxindole alkaloid isolated from Uncaria rhynchophylla (Miq.) Jacks (Gouteng in Chinese) that is a Chinese herb, which acts as an MTOR-dependent autophagy inducer [3006].
Corynoxine B/Cory B: An isomer of corynoxine, also isolated from the Chinese herb Uncaria rhynchophylla (Miq.) Jacks that acts as a BECN1-dependent autophagy inducer [3007].
COST Action TRANSAUTOPHAGY: The European Cooperation in Science and Technology site for autophagy research (http://cost-transautophagy.eu) [3008].
COST1 (CONSTITUTIVELY STRESSED 1): A plant-specific DUF641/COST-family protein that negatively regulates autophagy by direct interaction with the key autophagy adaptor ATG8E [3009]. The cost1 mutant has strong drought tolerance with constitutive induction of autophagy and broad expression of normally stress-responsive genes, as well as great retardation of plant growth and development, thus controlling the tradeoff between plant growth and stress tolerance.
CpATG8: A Cryphonectria parasitica homolog of Atg8 [3010].
CpdA: A nonsteroidal selective NR3C1/glucocorticoid receptor modulator with anti-inflammatory actions that can work independently of NR3C1 in macrophages. Herein, the autophagy receptor SQSTM1 but not NR3C1 mediates the anti-inflammatory action of CpdA. SQSTM1 target gene upregulation by CpdA involves a mechanism whereby the NFE2L2 transcription factor is recruited to its promoter. CpdA can be useful to include as a positive control of an autophagy-promoting agent in assays, alongside an enhancement of gene and protein levels of SQSTM1 [3011].
Crinophagy: Selective degradation of secretory granules by fusion with the lysosome, independent of autophagy [120, 1251, 1255, 1256]. See also SINGD and zymophagy.
CRM1: See XPO1.
CRYAB/HSPB5 (crystallin alpha B): Member of the small heat shock protein family abundantly expressed in lens, cardiac and skeletal muscle and involved in the maintaining of cytoskeletal integrity by chaperoning DES (desmin) and VIM (vimentin). Its R120G missense mutation causes cataract, cardiomyopathy and myofibrillar myopathy. At the cellular level CRYABR120G leads to protein aggregation and activates an NFKB-dependent autophagy [1107].
Cryptides: Peptides with a cryptic biological function that are released from cytoplasmic proteins by partial degradation or processing through autophagy (e.g., neoantimocrobial peptide released from ribosomal protein FAU/RPS30) [2058].
CSNK2 (casein kinase 2): A serine/threonine protein kinase that disrupts the BECN1-BCL2 complex to induce autophagy [3012]. CSNK2 also phosphorylates ATG16L1, in particular on Ser139, to positively regulate autophagy. See also PPP1.
Csp37: A mitochondrial outer membrane protein used in C. albicans as an indicator of mitophagy [3013].
Ctl1: A multi-transmembrane protein in the fission yeast Schizosaccharomyces pombe that binds to Atg9 and is required for autophagosome formation [3013].
CTLH complex: See Gid complex.
CTNS (cystinosin, lysosomal cystine transporter): CTNS is a proton-driven lysosomal cotransporter that actively exports cystine out of the lysosomes. The functional loss of CTNS, as encountered in the lysosomal disease cystinosis due to recessive mutations in the CTNS gene, leads to lysosomal dysfunction. This impairs the cellular clearance of autophagosomes containing damaged mitochondria, causing oxidative stress, disruption of tight junction integrity and abnormal signaling events leading to epithelial dysfunction in kidney tubules [3014, 3015].
Ctr9: A component of the yeast Paf1 complex (Paf1C) that downregulates the expression of ATG11 and ATG32 and represses mitophagy in growing conditions [3016].
CTR9 (CTR9 homolog, Paf1/RNA polymerase II complex component): A component of the mammalian Paf1-RNA polymerase II complex that regulates mitophagy through a PINK1- and PRKN-dependent pathway [3016]. See also Ctr9.
CTSD (cathepsin D): CTSD is an endolysosomal enzyme that exhibits pepsin-like activity and plays a role in protein turnover and in the proteolytic activation of hormones and growth factors [3017].
Cue5: A yeast receptor similar to mammalian SQSTM1 that binds ubiquitin through its CUE domain, and Atg8 via its C-terminal AIM [627]. Some Cue5-dependent substrates are ubiquitinated by Rsp5. See also CUET.
CUET (Cue5/TOLLIP): A family of autophagy receptor proteins containing a CUE domain that are involved in autophagic clearance of protein aggregates. See also Cue5 [627].
CUP-5 (coelomocyte uptake defective mutant-5): The ortholog of human MCOLN1, in C. elegans CUP-5 localizes to lysosomes, and is required for endo-lysosomal transport, lysosomal degradation [3018–3020], and proteolytic degradation in autolysosomes [3021].
CUPS (compartment for unconventional protein secretion): A compartment located near ER exit sites that is involved in the secretion of Acb1; Grh1 is localized to the CUPS membrane, and Atg8 and Atg9 are subsequently recruited under starvation conditions [3022]. Atg8 and Atg9 function in Acb1 secretion, but rapamycin-induced autophagy does not result in CUPS formation.
Curcumin: Major bioactive constituent of turmeric and popular dietary supplement [3023]. Curcumin is a natural polyphenol from turmeric herbs. Depending on the context, curcumin can induce or inhibit autophagy in a variety of in vitro and in vivo models [3024, 3025].
Cvt body: The single-membrane vesicle present inside the vacuole lumen that results from the fusion of a Cvt vesicle with the vacuole [180].
Cvt complex: A cytosolic protein complex consisting primarily of prApe1 dodecamers in the form of an Ape1 complex that are bound to the Atg19 receptor. This complex may also contain Ams1 and Ape4, but prApe1 is the predominant component [180].
Cvt pathway: See cytoplasm-to-vacuole targeting (Cvt) pathway.
Cvt vesicle: The double-membrane sequestering vesicle of the Cvt pathway [180].
CXCR4 (C-X-C chemokine receptor 4): CXCR4 belongs to the GPCR superfamily of proteins, the largest class of integral membrane proteins. It is involved in hematopoietic stem cell migration and in T-cell entry for HIV-1 infection [3026].
CYBB/NOX2 (cytochrome b-245 beta chain): The activity of this enzyme is essential for LAP [801].
CYP46A1)/cholesterol 24-hydroxylase (cytochrome P450 family 46 subfamily A member 1): A key enzyme of brain cholesterol metabolism; its overexpression in the brain activates autophagy in the context of neurodegenerative diseases [3027].
Cysmethynil: A small-molecule inhibitor of ICMT (isoprenylcysteine carboxyl methyltransferase); treatment of PC3 cells causes an increase in LC3-II and cell death with autophagic features [3028].
Cystatin C (CysC): A cysteine protease inhibitor [3029]. Injections of CysC to the substantia nigra of SNCAA53T transgenic mice results in a significant increase in VEGF, NR4A2/NURR1 and LC3B, and a decrease in SNCA and cleaved CASP3 in different brain regions. CysC-induced VEGF attenuates 6-OHDA-lesioned PC12 cell degeneration by regulating p-PRKCA-p-MAPK/ERK-NR4A2 signaling and inducing enhanced autophagy. In addition, VEGF-mediated angiogenesis is markedly enhanced in the conditioned media of 6-OHDA-lesioned PC12 cells with CysC-overexpression, whereas blockage of autophagy downregulates VEGF expression and associated angiogenesis. Therefore, CysC exerts a neuroprotective effect by regulating VEGF-mediated autophagy.
Cytoplasm-to-vacuole targeting (Cvt) pathway: A constitutive, biosynthetic pathway in yeast that transports resident hydrolases to the vacuole through a selective autophagy-like process [3030]. See also Ams1, Ape1, Ape4 and Atg19.
DAF-2 (abnormal dauer formation): Encodes the C. elegans INSR (insulin receptor)/IGF1R (insulin like growth factor 1 receptor) homolog that acts through a conserved PI3K pathway to negatively regulate the activity of DAF-16/FOXO and limit life span. DAF-2 inhibits autophagy by a mechanism that remains to be elucidated [367, 3031, 3032]. Reduced INS-DAF-2 signaling in daf-2 mutant C. elegans protects against SNCA/α-synuclein-mediated rupture of endomembranes [3033].
DAF-16: A C. elegans FOXO transcription factor ortholog.
DALIS (dendritic cell aggresome-like induced structures): Large polyubiquitinated protein aggregates formed in dendritic cells. These are similar to aggresomes, but they do not localize to the microtubule-organizing center. DALIS are transient in nature and small DALIS have the ability to move and form larger aggregates; they require proteasome activity to clear them [421]. See also ALIS.
DAMP (danger/damage-associated molecular pattern): DAMPs are endogenous molecules released by dead or damaged cells that bind to pattern recognition receptors (DDX58/RIG-I-like receptors [RLRs] Toll-like receptors [TLRs], NOD-like receptors [NLRs] and C-type lectin receptors [CLRs]) of the innate surveillance response system to activate the inflammatory response and autophagy [3034]. “Non-self” or exogenous molecules such as viral RNA that activate the same receptors are termed pathogen-associated molecular patterns (PAMPs) or microorganism-associated molecular patterns (MAMPs), the latter of which are derived from any microbe, not necessarily pathogenic [3035].
DAP (death associated protein): A conserved phosphoprotein that is a substrate of MTOR and inhibits autophagy; inhibition of MTOR results in dephosphorylation of DAP and inhibition of autophagy, thus limiting the magnitude of the autophagic response [3036].
DAPK1 (death associated protein kinase 1): A kinase that phosphorylates Thr119 of BECN1 to activate it by causing dissociation from BCL2L1/Bcl-xL and BCL2, thus activating autophagy [3037].
DAPK3 (death associated protein kinase 3): See Sqa.
DAXX/DAP6 (death domain associated protein): DAXX interacts with SQSTM1 to promote SQSTM1 phase condensation and puncta formation [3038].
DBeQ: A reversible ATP-competitive inhibitor of VCP/p97. DBeQ inhibits the degradation of ubiquitinated proteins, the ERAD pathway, and the maturation of autophagosomes [3039].
DBI/ACBP (diazepam binding inhibitor, acyl-CoA binding protein): A phylogenetically conserved protein that is released from cells during autophagy. DBI affects intracellular lipid mechanisms but also acts as a paracrine and endocrine regulator of autophagy and general metabolism at the whole-body level, notably as a stimulator of appetite [3040].
DCN (decorin): An archetypical member of the small leucine rich proteoglycans that functions as a soluble pro-autophagic and pro-mitophagic signal. DCN acts as a partial agonist for KDR/VEGFR2 and MET for endothelial cell autophagy and tumor cell mitophagy, respectively. DCN elicits these processes in a PEG3-dependent manner to induce endothelial cell autophagy, and in a TCHP/mitostatin-dependent manner for tumor cell mitophagy. It is postulated that induction of these fundamental cellular programs underlies the oncostatic and angiostatic properties of DCN [3041]. DCN itself is responsive to autophagic stimuli, as starvation significantly increases cardiac Dcn mRNA and protein. Moreover, DCN induction is required for proper autophagy, as measured by LC3-II formation and puncta formation, in the murine heart [3042].
Dcp-1 (death caspase-1): A Drosophila caspase that localizes to mitochondria and positively regulates autophagic flux [3043].
Dcp2/DCP2 (decapping mRNA 2): A decapping enzyme involved in the downregulation of ATG transcripts [3044]. See also Dhh1.
DCT-1: The C. elegans homolog of BNIP3 and BNIP3L, which functions downstream of PINK-1 and PDR-1 to regulate mitophagy under conditions of oxidative stress [2122].
DDIT3/CHOP (DNA damage inducible transcript 3): DDIT3 is a member of the CEBP family of transcription factors that appears to have a dual role in both inducing apoptosis and limiting autophagy by the promotion of the transcription of autophagy genes through cooperation with ATF4 and/or CEBPB during ER stress. DDIT3 is involved in the formation, elongation and function of the phagophore.
DDIT4/DIG2/RTP801/REDD1 (DNA damage inducible transcript 4): The DDIT4 protein is notably synthesized in response to stress hormones, glucocorticoids and adrenaline or hypoxia, and inhibits MTOR, resulting in the induction of autophagy and enhanced cell survival [1691, 3045]. Additionally, induction of the DDIT4-MTOR-autophagy axis is correlated with NETosis-driven IL1B autoinflammation [1691, 1693]. See also NETosis.
Deconjugation: The Atg4/ATG4-dependent cleavage of lipidated Atg8-family proteins (e.g., Atg8–PE, LC3-II) that releases the protein from PE (illustrated for the nascent yeast protein that contains a C-terminal arginine). The liberated Atg8-family proteins can subsequently go through another round of conjugation. Mammalian ATG4 also deconjugates LC3 from proteins. See also 12-ylation. Atg8*, activated Atg8.
Decorin: See DCN.
Decoupled signaling: When limited for an auxotrophic requirement, yeast cells fail to induce the expression of autophagy genes even when growing slowly, which contributes to decreased cell viability [3046].
DEDD (death effector domain containing): DEDD acts as an endogenous suppressor of tumor growth and metastasis through the epithelial-mesenchymal transition process [3047, 3048]. DEDD binds directly to the class III PtdIns3K core complex to stabilize PIK3C3 and promote the interaction of this enzyme with BECN1 to induce autophagy activation as well as subsequent degradation of SNAI and TWIST, two master modulators of the epithelial-mesenchymal transition.
Deferiprone (DFP): Deferiprone is an iron chelator that has strongly induces mitophagy in a PINK1- and PRKN-independent manner [460].
Deforolimus (AP39573): This chemical promotes autophagy through inhibition of MTORC1 [3049].
DEGS1 (delta 4-desaturase, sphingolipid 1): The enzyme that catalyzes the last step of ceramide biosynthesis converting dihydroceramides into ceramides. DEGS1 inhibition enhances dihydroceramide levels and induces autophagy, as well as autophagy-mediated cancer cell death. See also dihydroceramide.
Desat1: A Drosophila lipid desaturase that localizes to autophagosomes under starvation conditions; the Desat mutant is defective in autophagy induction [3050].
DFCP1: See ZFYVE1.
DFP: See deferiprone.
Dga1: See Ayr1.
Dhh1: An RCK member of the RNA-binding DExD/H-box proteins involved in mRNA decapping and translational regulation; Dhh1 in S. cerevisiae and Vad1 in Cryptococcus neoformans bind certain ATG transcripts, leading to the recruitment of the Dcp2 decapping enzyme and mRNA degradation to suppress autophagy in nutrient-rich conditions [3044]. Conversely, Dhh1 switches its role to become a positive regulator, coordinating with an EIF4E-binding protein, Eap1, and is required for efficient translation of Atg1 and Atg13 induced by nitrogen starvation [3051]. See also Dcp2 and Eap1.
Diacylglycerol: A lipid second messenger that contributes to autophagic targeting of Salmonella-containing vacuoles [3052].
DIG2: See DDIT4.
Dihydroceramide: A metabolite that constitutes the penultimate step of ceramide biosynthesis. Dihydroceramide has been implicated in the regulation of autophagy [905] and specifically in autophagy-mediated cell death [906]. See also DEGS1.
DIRAS3 (DIRAS family GTPase 3): A protein that interacts with BECN1, displacing BCL2 and blocking BECN1 dimer formation, thus promoting the interaction of BECN1 with PIK3C3 and ATG14, resulting in autophagy induction [3053]. DIRAS3 is a weight loss target gene in adipose stem/progenitor cells, inducing autophagy [3054] and protecting against premature senescence [3055].
DMVs (double-membraned vesicles): See autophagy-like vesicles.
DNautophagy: An autophagic process by which DNA is transported across the lysosomal membrane and degraded [1395]. The translocation process is mediated by lysosomal integral membrane proteins LAMP2C and SIDT2 [1395]. See also LAMP2C, RNautophagy and SIDT2.
Dnm1: A dynamin-related GTPase that is required for both mitochondrial and peroxisomal fission. Dnm1 is recruited to degrading mitochondria by Atg11, or to degrading peroxisomes by both Atg11 and Atg36 (or PpAtg30), to mediate mitophagy- or pexophagy-specific fission [2911, 3056]. See also DNM1L.
DNM1L/Drp1 (dynamin 1 like): The mammalian homolog of yeast Dnm1. PRKA-mediated phosphorylation of rat DNM1L on Ser656 (Ser637 in humans) prevents both mitochondrial fission and some forms of mitophagy in neurons [3057]. See also Dnm1.
DNM2 (dynamin 2): DNM2 is recruited to extruded autolysosomal membranes during the process of autophagic lysosome reformation and catalyzes their scission, promoting the regeneration of nascent protolysosomes during autophagic flux [2851]. See also autophagic lysosome reformation.
dom (domino): A Drosophila SWI2/SNF2 chromatin remodeling protein. A loss-of-function mutation at the dom locus synergizes with genotypes depressed in autophagy pathway activity [3058].
Dopamine: A neurotransmitter whose accumulation outside vesicles induces autophagy and cell degeneration [3059].
Dopamine receptor: The function of dopamine is mediated by dopamine receptors (DRs). There are five different DR subtypes, named DRD1 to DRD5. Activation of DRD5 induces autophagic cell death in human cancer cells, and inhibition of DRD4 impedes autophagic flux in glioblastoma stem cells [3060, 3061]. Activation of DRD3, but not DRD2, induces autophagy, while maintaining protein synthesis [3062].
DOR: See TP53INP2.
Doryphagy: The selective degradation of centriolar satellites (CS) by autophagy. Selectivity to this process is provided by the LIR-mediated interaction between the CS component PCM1 and GABARAP/GABARAPL2 [1258]. This process regulates centrosomal composition and stability, thereby maintaining functional centrosomes required for proper cell division.
DQ-BSA (dequenched bovine serum albumin): A derivative of BSA that is heavily labeled by fluorescent dyes, leading to self-quenching. After being endocytosed by cells, it is trafficked by the endosomal system to the lysosome for degradation, which releases smaller fragments of the fluorophore as cleavage products. This degradation produces a fluorescent signal that is proportional to the proteolytic activity of the cellular lysosomes and thus can serve as a readout for functional endocytosis and lysosomal hydrolase activity [2388].
DRAM1 (DNA damage regulated autophagy modulator 1): DRAM1 is a small hydrophobic protein with six transmembrane domains colocalizing with GOLGB1/giantin and GOLGA2/GM130, and also in early and late endosomes and lysosomes, colocalizing with EEA1 and LAMP2 [3063], and interacting with BAX [3064]. DRAM1 gene expression is induced by TP53 in response to DNA damage that results in cell death by a mechanism that involves autophagy, which is induced in a DRAM1-dependent manner [2675, 3065]. DRAM1 gene expression is induced by NFKB in response to mycobacterial infection, and is required for host defense by selective autophagy [2435].
DRAM2/TMEM77 (DNA damage regulated autophagy modulator 2): DRAM2 is closely related to DRAM1 and contributes to induction of autophagy [3066]. Overexpression of DRAM2 induces cytosolic GPF-LC3 puncta and increases the level of endogenous LC3-II, whereas its silencing interferes with starvation-induced autophagy [3067]. DRAM2 interacts with BECN1, UVRAG, LAMP1 and LAMP2, and enhances phagosome maturation and antimicrobial activity during mycobacterial infection [3068]. Defects in the DRAM2 gene cause a type of retinal dystrophy. DRAM2 has been linked to HOTAIRM1-mediated regulation of autophagy. See also HOTAIRM1.
DRAM3: See TMEM150B.
drpr/draper: A Drosophila homolog of the C. elegans engulfment receptor CED-1 that is required for autophagy associated with cell death during salivary gland degradation, but not for starvation-induced autophagy in the fat body [3069].
Drs: See SRPX.
Dsk2: A nuclear-enriched ubiquitin-like polyubiquitin-binding protein in budding yeast, which transports ubiquitinated proteins to the proteasome for degradation. In budding yeast, Dsk2 is dispensable for proteasome-dependent degradation of mutated HTT with polyQ expansion (HTT103Q), but it is required for efficient clearance of HTT103Q through autophagy [1099]. In A. thaliana, DSK2 acts as a ubiquitin receptor for autophagy-mediated degradation of AT1G19350/BES1, a transcription factor mediating plant steroid hormone brassinosteroid regulation of plant growth and stress response [3070].
DsRed tetramer mouse: Tetrameric DsRed, a mammalian derivative of red fluorescent protein of the coral Discosoma was generated originally for imaging applications. DsRed tetramers form aggregates, and the transgenic mouse expressing DsRed tetramers was generated as a spontaneous in vivo cardiac fibrosis and heart failure model caused by chronic autophagy and proteasome degradation insufficiency. Skeletal muscles of this mouse tolerate the toxicity of DsRed aggregation, suggesting that additional factors such as oxidative stress and renewal capability determine tissue vulnerability to proteopathy [3071].
DUSP4/MKP2 (dual specificity phosphatase 4): A member of the dual specificity protein phosphatase subfamily, which is induced in a MAPK1/3-dependent manner and establishes feedback inhibition. Inhibition of histone methyltransferase EHMT2/G9a induces autophagic cell death in head and neck squamous cell carcinoma/HNSCC via a DUSP4-dependent MAPK1/3 inactivation mechanism [2888]. Overexpression of DUSP4 induced by MAPK1/3 positively regulates AKT-MTOR signaling resulting in impaired autophagy in hearts of a mouse model of LMNA (lamin A/C) cardiomyopathy [3072]. See also MAPK1.
E2F1: A mammalian transcription factor that upregulates the expression of BNIP3, LC3, ULK1 and DRAM1 directly, and ATG5 indirectly [938]. E2F1 plays a role during DNA damage- and hypoxia-induced autophagy.
E64d/Pepstatin: An inhibitor of aspartic and cysteine proteases that can block late steps of autophagy through the reduction of lysosomal degradation [2352].
Eap1: A eukaryotic translation initiation factor 4 E (EIF4E) binding protein that interacts with Dhh1 to promote ATG1 and ATG13 mRNA translation under nitrogen-starvation conditions [3051]. See also Dhh1.
EAT (early autophagy targeting/tethering) domain: The C-terminal domain of Atg1, which is able to tether vesicles [3073]. This part of the protein also contains the binding site for Atg13.
EAT-2 (EATing: abnormal pharyngeal pumping): A ligand-gated ion channel subunit closely related to the non-alpha subunit of nicotinic acetylcholine receptors, which functions to regulate the rate of pharyngeal pumping in C. elegans. eat-2 loss-of-function mutants are dietary restricted and require autophagy for the extension of life span [3031, 3074, 3075].
EDTP: See MTMR14.
EEA1 (early endosome antigen 1): A RAB5 effector used as a common marker for early endosome vesicles.
EEF1A1/EF1A/eF1α (eukaryotic translation elongation factor 1 alpha 1): Multifunctional member of the family of G-proteins with different cellular variants. The lysosomal variant of this protein acts coordinately with GFAP at the lysosomal membrane to modulate the stability of the CMA translocation complex. Release of membrane bound EEF1A1 in a GTP-dependent manner promotes disassembly of the translocation complex and consequently reduces CMA activity [3076].
eF1α: See EEF1A1.
EGFR (epidermal growth factor receptor): A tyrosine kinase receptor that negatively regulates autophagy through PI3K, AKT, and MTOR modulation [775].
EGO (exit from rapamycin-induced growth arrest) complex: The Meh1/Ego1, Ego2, Slm4/Ego3, Gtr1 and Gtr2 proteins form a pentameric complex that mediates amino acid signals to control TORC1, and positively regulates microautophagy in yeast [3077, 3078].
eIF2α kinase: See EIF2S1 kinase.
EIF2AK2/PKR (eukaryotic translation initiation factor 2 alpha kinase 2): A mammalian EIF2S1/EIF2 alpha kinase that induces autophagy in response to viral infection [836].
EIF2AK3/PERK (eukaryotic translation initiation factor 2 alpha kinase 3): A mammalian EIF2S1/EIF2 alpha kinase that induces autophagy in response to ER stress [473, 918] and mycolactone exposure [2021].
EIF2AK4/GCN2 (eukaryotic translation initiation factor 2 alpha kinase 4): A mammalian EIF2S1/EIF2 alpha kinase that induces autophagy in response to mycolactone exposure [2021].
EIF2S1 (eukaryotic translation initiation factor 2 subunit alpha): An initiation factor that is involved in stress-induced translational regulation of autophagy.
EIF2S1/eIF2α kinase: There are four mammalian EIF2S1/EIF2 alpha kinases that respond to different types of stress, inducing a so-called integrated stress response. EIF2AK2 and EIF2AK3 induce autophagy in response to virus infection and ER stress, respectively [918, 3079], and/or mycolactone exposure [2021]. See also Gcn2, EIF2AK2 and EIF2AK3.
EIF5A (eukaryotic translation initiation factor 5A): A translation factor that is hypusinated, and promotes translation elongation, and regulates the synthesis of TFEB [958]. See also spermidine.
ELA11: An 11-residue FURIN-cleaved fragment of mature APELA/ELABELA (apelin receptor early endogenous ligand), a hormone that mediates endoderm development and heart morphogenesis [3080, 3081]. This peptide inhibits autophagy in murine and cell models of renal ischemia/reperfusion injury [3082].
Elaiophylin: A natural compound late-stage autophagy inhibitor that results in lysosomal membrane permeabilization and decreased cell viability [3083]. See also LMP.
ELAVL1/HuR (ELAVL like RNA binding protein 1): This RNA-binding protein regulates SQSTM1 expression at the post-transcriptional level by binding to SQSTM1 mRNA and favoring its translation when autophagy is triggered [359, 586].
ema (endosomal maturation defective): ema is required for phagophore expansion and for efficient mitophagy in Drosophila fat body cells. It is a transmembrane protein that relocalizes from the Golgi to phagophores following starvation [3084]. The vertebrate ortholog CLEC16A regulates mitophagy and is a susceptibility locus for many autoimmune disorders [3085, 3086].
Embryoid bodies/EBs: Three-dimensional aggregates of pluripotent stem cells including embryonic stem cells and induced pluripotent stem cells.
EMC6/TMEM93 (ER membrane protein complex subunit 6): A novel ER-localized transmembrane protein, which interacts with both RAB5A and BECN1 and colocalizes with the omegasome marker ZFYVE1/DFCP1 [3087]. EMC6 enhances autophagosome formation when overexpressed.
EndoA/SH3GL2/Endophilin A1 (Endophilin A): Phosphorylation of the serine residue 75 in Drosophila EndoA (an ortholog of human SH3GL2) promotes autophagy by creating highly curved membranes that attract Atg3 onto the phagophore [3088]. In mammals, SH3GL2 interacts with FBXO32 (F-box protein 32), and both proteins colocalize transiently with phagophores and are necessary for autophagosome formation [3089].
Endorepellin: The anti-angiogenic C-terminal cleavage product of HSPG2/perlecan. Endorepellin engages KDR/VEGFR2 and ITGA2/α2β1 integrin in a novel mechanism termed dual receptor antagonism for achieving endothelial cell specificity and function. Endorepellin evokes endothelial cell autophagy downstream of KDR and in a PEG3-dependent manner [3090], and protracted endothelial cell mitochondria depolarization downstream of KDR with a concurrent induction and colocalization of TCHP/mitostatin and PRKN [3091].
Endosomal microautophagy (e-MI): A form of autophagy in which cytosolic proteins are sequestered into late endosomes/MVBs through a microautophagy-like process. Sequestration can be nonselective or can occur in a selective manner mediated by HSPA8. This process differs from chaperone-mediated autophagy as it does not require substrate unfolding, and it is independent of the CMA receptor LAMP2A [1823]. This process occurs during MVB formation and requires the ESCRT-I and ESCRT-III protein machinery; accordingly, this can be referred to as ESCRT-mediated sorting at the endosome. See also endosome and multivesicular body.
Endosome: The endosomal compartments receive molecules engulfed from the extracellular space and are also in communication with the Golgi apparatus. The endosomal system can be viewed as a series of compartments starting with the early endosome. From there, cargos can be recycled back to the plasma membrane; however, more typically, internalized cargo is transported to the late endosome/MVB. These latter compartments can fuse with lysosomes. Endosomal maturation from early endosomes is a dynamic process that involves a progressive reduction in lumenal pH. In mammalian cells, early and/or multivesicular endosomes fuse with autophagosomes to generate amphisomes.
EP300/p300 (E1A binding protein p300): An acetyltransferase that inhibits autophagy by acetylating ATG5, ATG7, ATG12 and/or LC3 [1005]. EP300 is also involved in the GLI3-dependent transcriptional activation of VMP1 in cancer cells [967]. See also GLI3.
EPAC1: See RAPGEF3.
EPAC2: See RAPGEF4.
EPAS1/HIF2A/Hif-2α (endothelial PAS domain protein 1): Part of a dimeric transcription factor in which the α subunit is regulated by oxygen; the hydroxylated protein is degraded by the proteasome. EPAS1 activation in mouse liver augments peroxisome turnover by pexophagy, and the ensuing deficiency in peroxisomal function encompasses major changes in the lipid profile that are reminiscent of peroxisomal disorders [1207].
Ependymoma: Ependymoma is a prodigious pediatric brain tumor. Children with ependymoma have high mortality rates because ependymoma challenges all forms of conventional therapy. An oncogenic role of nuclear pore protein/nucleoporin TPR (translocated promoter region, nuclear basket protein) in regulating HSF1 (heat shock transcription factor 1) mRNA trafficking, maintaining MTORC1 activity to phosphorylate ULK1 and preventing autophagy induction in ependymoma has been described [3092].
epg (ectopic PGL granules) mutants: C. elegans mutants that are defective in the autophagic degradation of PGL-1, SEPA-1 and/or SQST-1 [965]. The EPG-3, EPG-7, EPG-8 and EPG-9 proteins are homologs of VMP1, Atg11/RB1CC1, ATG14 and ATG101, respectively, whereas EPG-1 may be a homolog of ATG13 [1734].
EPG-1: The highly divergent homolog of Atg13 in C. elegans. EPG-1 directly interacts with the C. elegans Atg1 homolog UNC-51 [3093]. See also Atg13.
EPG-2: A nematode-specific coiled-coil protein that functions as a scaffold protein mediating the autophagic degradation of PGL granule in C. elegans. EPG-2 directly interacts with SEPA-1 and LGG-1. EPG-2 itself is also degraded by autophagy [965].
EPG-3: A metazoan-specific autophagy protein that is the homolog of human VMP1. EPG-3/VMP1 are involved in an early step of autophagosome formation [965].
EPG-4: An ER-localized transmembrane protein that is the homolog of human EI24/PIG8. EPG-4 is conserved in multicellular organisms, but not in yeast. EPG-4 functions in the progression of omegasomes to autophagosomes [965]. EI24 also mediates the crosstalk between the UPS and autophagy by mediating the autophagic degradation of RING domain E3 ligases in human cells [3094, 3095].
EPG-5: A novel autophagy protein that is conserved in multicellular organisms. EPG-5 regulates lysosome degradative capacity and thus could be involved in other pathways that terminate at this organelle [965]. Mutations in the human EPG5 gene lead to Vici syndrome [3096].
EPG-6: A WD40 repeat PtdIns3P-binding protein that directly interacts with ATG-2 [845]. EPG-6 is the C. elegans functional homolog of yeast Atg18 and probably of mammalian WDR45/WIPI4. EPG-6 is required for the progression of omegasomes to autophagosomes. See also Atg18.
EPG-7: A scaffold protein mediating the autophagic degradation of the C. elegans SQSTM1 homolog SQST-1 [2783]. EPG-7 interacts with SQST-1 and also with multiple ATG proteins. EPG-7 itself is degraded by autophagy.
EPG-8: An essential autophagy protein that functions as the homolog of yeast Atg14 in C. elegans [2788]. EPG-8 is a coiled-coil protein and directly interacts with the C. elegans BECN1 homolog BEC-1. See also Atg14.
EPG-9: A protein with significant homology to mammalian ATG101 in C. elegans [3097]. EPG-9 directly interacts with EPG-1/Atg13. See also ATG101.
EPG-11: An arginine methyltransferase in C. elegans that is the homolog of PRMT1 [3098]. EPG-11 regulates the association of PGL granules with EPG-2 and LGG-1 puncta. EPG-11 directly methylates arginine residues in the RGG domain of PGL-1 and PGL-3.
EPM2A/laforin (EPM2A glucan phosphatase, laforin): A member of the dual specificity protein phosphatase family that acts as a positive regulator of autophagy probably by inhibiting MTOR, as EPM2A deficiency causes increased MTOR activity [3099]. Mutations in the genes encoding EPM2A or the putative E3-ubiquitin ligase NHLRC1/malin, which form a complex, are associated with the majority of defects causing Lafora disease, a type of progressive neurodegeneration. EPM2A is well conserved among mammals [3100], and may also act as a regulator of autophagy, considering that an impairment of EPM2A-mediated autophagy may lead to cell death [3099, 3101]. See also NHLRC1.
ER-mitochondria contact sites: The contact sites that the ER forms with mitochondria in mammalian cells. Also known as mitochondria-associated membranes (MAMs) when biochemically isolated by fractionation. ER-mitochondria contact sites regulate exchange of lipids and calcium between organelles and are implicated in autophagosome formation. See also mitochondria-associated ER membranes.
ER-phagy: See reticulophagy.
ERAD (endoplasmic reticulum-associated degradation): A process that can operate in parallel to ERLAD (including reticulophagy, ER-exit site microautophagy and vesicular transport) to degrade misfolded proteins from the ER. In ERAD, misfolded proteins are translocated across the ER membrane, polyubiquitinated, and degraded by the 26S proteasome [1378, 3102].
ERAS (ES cell-expressed Ras): A small GTPase of the RAS family involved both in PRKN-dependent and -independent mitophagy. Unlike the situation in humans, ERAS is constitutively expressed in domestic animals [3103].
ERBB2/HER2/EGFR2 (erb-b2 receptor tyrosine kinase 2): A transmembrane receptor tyrosine kinase that is amplified in approximately 20% of breast cancers. ERBB2 binds to BECN1 and inhibits autophagy [2659, 2660].
ERES (ER exit sites): The COPII-coated subcompartments that specialize in loading and exporting cargo bound for the Golgi apparatus from the ER [3104–3106].
ERGIC (ER-Golgi intermediate compartment): A donor compartment that produces small vesicles that are active for LC3 lipidation [1540].
ERK1: See MAPK3.
ERK2: See MAPK1.
ERLAD (ER-to-lysosome-associated degradation): Processes activated to deliver ERAD-resistant misfolded proteins to endolysosomes for clearance. In ERLAD, misfolded proteins are segregated in ER subdomains or in ER exit sites and are delivered to lysosomal compartments via reticulophagy [1380] and vesicular transport [1379], where an involvement of the ER-resident LC3-binding protein RETREG1 has been shown, or by mechanistically ill-defined ER-exit site microautophagy [1388]. See also RETREG1.
ERMES (ER-mitochondria encounter structure): A complex connecting the endoplasmic reticulum and the mitochondrial outer membrane in yeast. The core components of ERMES are the mitochondrial outer membrane proteins Mdm10 and Mdm34, the ER membrane protein Mmm1, and the peripheral membrane protein Mdm12. ERMES plays an important role in yeast mitophagy presumably by supporting the membrane lipid supply for the growing phagophore membrane [3107]. ERMES involvement in mitophagy requires the ubiquitination of Mdm34 by the E3 ligase Rsp5 [3108]. See also Rsp5 and Mdm34.
ERN1/IRE1α (endoplasmic reticulum to nucleus signaling 1): A component of the unfolded protein response (ER stress response). Deletion of Ern1 in podocytes leads to reduction of autophagy and induction of injury in these cells [3109].
Etf-1 (Ehrlichia translocated factor-1): A type IV secretion effector of the obligatory intracellular bacterium Ehrlichia chaffeensis that binds RAB5-GTP and the autophagy-initiating class III PtdIns3K complex containing PIK3C3/VPS34, and BECN1, and induces RAB5-regulated autophagosome formation; the immature autophagosomes traffic to, and fuse with, E. chaffeensis-containing vacuoles, delivering autophagic cargoes into the vacuole, which can serve as nutrients for bacterial growth [3110–3112].
Everolimus/RAD001 (40-O-[2-hydroxyethyl]): An orally administered MTOR inhibitor that is a derivative of rapamycin that induces autophagy.
ESC8: A autophagy inducer that bears a cationic estradiol moiety and causes downregulation of p-MTOR and its downstream effectors including p-RPS6KB [3113].
ESCRT (endosomal sorting complex required for transport): ESCRT is composed of multiple subcomplexes. The ESCRT complexes are required for the formation of MVBs, membrane damage repair and phagophore closure [360, 3114]. Components of the ESCRT machinery are also involved in endosomal microautophagy (e-MI, type 3) [1806, 1851]. See also phagophore closure.
EVA1A/FAM176A/TMEM166 (eva-1 homolog A, regulator of programmed cell death): An integral membrane protein that induces autophagy and cell death when overexpressed [3115, 3116]. EVA1A interacts with the WD repeats of ATG16L1 through its C terminus and promotes ATG12–ATG5-ATG16L1 complex recruitment to the phagophore membrane, and enhances the formation of the autophagosome [3115]. See also TMEM166.
EX-527: SIRT1 inhibitor.
EXOC2/SEC5L1 (exocyst complex component 2): A component of the exocyst complex; EXOC2 binds RALB, BECN1, MTORC1, ULK1 and PIK3C3 under nutrient-rich conditions and prevents these components from interacting with EXOC8/EXO84, thus inhibiting autophagy [3117]. See also RALB and EXOC8.
EXOC8/EXO84 (exocyst complex component 8): A component of the exocyst complex, and an effector of RALB that is involved in nucleation and/or expansion of the phagophore; EXOC8 binds RALB under nutrient-poor conditions, and stimulates the formation of a complex that includes ULK1 and the class III PtdIns3K [3117]. See also RALB and EXOC2.
Exocyst: An octameric complex that helps in tethering secretory vesicles to the plasma membrane. The exocyst also plays a role during autophagosome biogenesis by regulating Atg9 trafficking [3118].
Exophagy: A process in yeast and mammalian cells that is used for protein secretion that is independent of the secretory pathway (i.e., unconventional secretion), and dependent on Atg proteins and the Golgi protein Grh1; Acb1 (acyl-coenzyme A-binding protein) uses this route for delivery to the cell surface [2476, 2477, 3119]. See also secretory autophagy.
EZR (Ezrin): As a member of the EZR (ezrin)-RDX (radixin)-MSN (moesin) (ERM) family, EZR is a membrane-bound cytoskeleton linker protein, which has been associated with poor outcome in several cancer types [3120]. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR pathway [3121].
FABP1 (fatty acid binding protein 1): FABP1 constitutes 5% of cytosolic proteins in the liver that localize to lysosomes, and negatively regulates autophagic flux and lipid metabolism in hepatic steatosis [3122].
FADD (Fas associated via death domain): An adaptor protein transmitting apoptotic signals from death receptors. FADD also suppressess autophagy by downregulating expression of the small GTP binding protein RHEB and subsequent MTOR activation in human breast adenocarcinoma cells [3123]. FADD also interacts with the ATG12–ATG5 complex to limit excessive autophagy during T cell proliferation [3124]. See also RHEB.
FAM48A: See SUPT20H.
FAM134B: See RETREG1.
FAM134B2: An N-terminal truncated isoform of RETREG1/FAM134B. FAM134B2 is a predominant isoform in the liver and is induced during starvation through the induction of CEBPB [3125].
FAM176A: See EVA1A.
Fasudil: A ROCK (Rho-associated coiled-coil containing protein kinase) inhibitor that enhances autophagy [3126].
Far11: A MAP kinase target that is involved in the dephosphorylation of Atg13 and the induction of autophagy [3127]. Far11 interacts with Pph21, Pph22 and Pph3 and may coordinate different cellular stress responses by regulating phosphatase activity.
Far complex: The Far complex consists of Far3, Far7, Far8, Vps64/Far9, Far10, and Far11, and is essential for Ppg1 to prevent phosphorylation of Atg32 [3128].
Fascaplysin: A marine sponge (Fascaplysinopsis sp.)-derived product with anti-cancer activities. Fascaplysin induces NUPR1/p8-dependent but MTOR signaling-independent autophagy in vascular endothelial cells [3129].
FBXW7/Sel10/hCDC4/hAgo (F-box and WD repeat domain containing 7): FBXW7 is a member of the F-box protein family, which functions as the substrate recognition component of the SCF E3 ubiquitin ligase and is a positive regulator of autophagy through inhibition of MTOR; knockdown of FBXW7 inhibits autophagic flux in SMB-S15 cells [3130]. In addition, GSK3B-mediated phosphorylation of MCL1 induces axonal autophagy, which promotes axonal degeneration. Phosphorylated MCL1 is ubiquitinated by FBXW7 and degraded by the proteasome, which is necessary for the induction of axonal autophagy. GSK3B-MCL1 signaling to regulate autophagy might be important for the successful completion of axonal degeneration [3131]. See also GSK3B and MCL1.
FBXW11/β-TrCP2 (F-box and WD repeat domain containing 11): BTRC/β-TrCP1 (beta-transducin repeat containing E3 ubiquitin protein ligase) and FBXW11/β-TrCP2 are paralogs that regulate several cellular physiological processes. FBXW11 activates MTORC1 by preferentially degrading the MTORC1 inhibitors DEPTOR and DDIT4/REDD1 which leads to the inhibition of autophagy and cell growth. FBXW11 also ubiquitinates and degrades the AMPK kinase-phosphorylated BTRC under glucose-deprived conditions.
Ferritinophagy: The selective degradation of ferritin through an autophagy-like process [1264] that involves a specificity receptor, NCOA4.
FEZ1/zygin I (fasciculation and elongation protein zeta 1): FEZ1 interacts with ULK1 or UVRAG, and forms a trimeric complex with either component by also binding SCOC [3132]. FEZ1 appears to be a negative regulator of autophagy when it is bound only to ULK1, and this inhibition is relieved upon formation of the trimeric complex containing SCOC. Similarly, the SCOC-FEZ1-UVRAG complex is inhibitory; dissociation of UVRAG under starvation conditions allows the activation of the class III PtdIns3K complex. See also SCOC.
FGFR1 (fibroblast growth factor receptor 1): FGFR1 is a member of the FGFR family, which comprises four receptor tyrosine kinases that cooperate with extracellular FGFs (fibroblast growth factors) in the transduction of signals influencing organogenesis, angiogenesis, metabolism and tissue repair. FGFR1 abnormalities recur in multiple developmental and acquired diseases including Hartsfield syndrome, Kallmann syndrome, nonsyndromic hypogonadotropic hypogonadism, Pfeiffer syndrome, and osteoglophonic dysplasia, and appear to induce autophagy [3133].
FIP200: See RB1CC1.
FIG4 (FIG4 phosphoinositide 5-phosphatase): A phospholipid phosphatase that controls the levels of the PtdIns(3,5)P2 phosphoinositide. Loss of FIG4 causes a decrease of PtdIns(3,5)P2 levels, enlargement of late endosomes and lysosomes, and cytosolic vacuolization [3134]. Recessive FIG4 mutations in humans are responsible for Yunis-Varón syndrome, familial epilepsy with polymicrogyria, and Charcot-Marie-Tooth type 4J neuropathy. In addition, FIG4 haploinsufficiency may be a risk factor for ALS.
FIP-gts: A fungal immunomodulatory protein from Ganoderma tsugae, FIP-gts induces autophagy, which leads to caspase-independent cell death [3135].
Fis1: A component of the mitochondrial fission complex. Fis1 also plays a role in peroxisomal fission by recruiting Dnm1 to peroxisomes; it interacts with Atg11 to facilitate mitophagy- and pexophagy-specific fission [2911, 3056]. See also Dnm1.
FKBP1A (FKBP prolyl isomerase 1A): An immunophilin that forms a complex with rapamycin and inhibits MTOR.
FKBP4/FKBP52 (FKBP prolyl isomerase 4): An immunophilin that forms a complex with FK506 and rapamycin; FKBP4 physiologically localizes with the endolysosomal system in human and rodent brain neurons. This immunophilin frequently colocalizes with pathological MAPT/tau in the autophagy-endolysosomal system in Alzheimer brain neurons [3136].
FKBP5/FKBP51 (FKBP prolyl isomerase 5): An immunophilin that forms a complex with FK506 and rapamycin; FKBP5 promotes autophagy in irradiated melanoma cells, thus enhancing resistance to radiation therapy [3137]. FKBP5 also associates with BECN1 and shows synergistic effects with antidepressants on autophagy in cells, mice and humans, possibly explaining its requirement in antidepressant action [3138].
FKBP8/FKBP38 (FKBP prolyl isomerase 8): An immunophilin that contains a single transmembrane domain at the C terminus and localizes predominantly to the mitochondrial outer membrane. FKBP8 is a noncanonical FKBP and peptidyl-prolyl cis-trans isomerase because its enzymatic activity is triggered only when it is bound to calcium-bound CALM (calmodulin). FKBP8 is an inhibitor of MTOR that is antagonized by RHEB in response to growth factors or nutrients [3139]. Additionally, FKBP8 acts as a mitophagy receptor in PRKN-independent mitophagy [196]. FKBP8 contains a LIR motif in its N terminus, which mediates strong binding to LC3A, and thus recruits LC3A to damaged mitochondria. Notably, FKBP8 escapes from degradation when mitophagy is activated.
FKBP12: See FKBP1A.
FKBP38: See FKBP8.
FKBP51: See FKBP5.
FKBP52: See FKBP4.
FLCN (folliculin): A tumor suppressor mutated in Birt-Hogg-Dubé syndrome [3140]. FLCN interacts with GABARAP, and this association is modulated by the presence of either FNIP1 (folliculin interacting protein 1) or FNIP2. ULK1 can induce FLCN phosphorylation, which modulates the FLCN-FNIP-GABARAP interaction [712]. FLCN is also linked to MTOR modulation through its interaction with the RRAG GTPases on lysosomes [3141, 3142].
Fluspirilene: A diphenylbutylpiperidine typical antipsychotic drug, used for the treatment of schizophrenia. Fluspirilene reduces intracellular calcium-dependent CAPN1 (calpain 1) activity and leads to autophagy induction by preventing CAPN1-mediated cleavage of ATG5 [2917].
FM 4-64: A lipophilic dye that primarily stains endocytic compartments and the yeast vacuole limiting membrane.
FNBP1L (formin binding protein 1 like): An F-BAR-containing protein that interacts with ATG3 and is required for the autophagy-dependent clearance of S. typhimurium, but not other types of autophagy [3143].
FNIP1 (folliculin interacting protein 1): An interactor with the tumor suppressor FLCN. FNIP1 [658] and its homolog FNIP2 [712] can also interact with GABARAP.
FOXO1 (forkhead box O1): A mammalian transcription factor that regulates autophagy independent of transcriptional control; the cytosolic form of FOXO1 is acetylated after dissociation from SIRT2, and binds ATG7 to allow induction of autophagy in response to oxidative stress or starvation [3144]. FOXO1 can also be deacetylated by SIRT1, which leads to upregulation of RAB7 and increased autophagic flux [3145]. The C. elegans ortholog is DAF-16. See also SIRT1.
FOXO3 (forkhead box O3): A transcription factor that stimulates autophagy through transcriptional control of autophagy-related genes [984, 3146]. The C. elegans ortholog is DAF-16.
Frataxin: See FXN.
Fsc1: A type I transmembrane protein localizing to the vacuole membrane in the fission yeast S. pombe; required for the fusion of autophagosomes with vacuoles [3147].
Fullerene C60 nanocrystals: A type of water-suspended carbon nanomaterial that activates autophagy as a chemosensitization mechanism [3148]. Fullerene C60 nanocrystals induce cytoprotective autonomous CAMK2A/CaMKIIα activity. Inhibition of CAMK2A activity enhances blocking of autophagic degradation through lysosomal dysfunction induction, leading to an increase in fullerene C60 nanocrystals-elicited cytotoxicity [3149].
FUNDC1 (FUN14 domain containing 1): A mitochondrial outer membrane protein that functions as a receptor for hypoxia-induced mitophagy [1169]. FUNDC1 contains a LIR and binds LC3, and plays a role in disposal of paternal mitochondria in the post-fertilization embryo [3150].
FUS (FUS RNA binding protein): A DNA/RNA binding protein involved in DNA repair, gene transcription, and RNA splicing. FUS has also been implicated in tumorigenesis and RNA metabolism, and multiple missense and nonsense mutations in FUS are associated with ALS. Autophagy reduces FUS-positive stress granules [1281].
FXN (frataxin): A nuclear-encoded protein involved in iron-sulfur cluster protein biogenesis. Reduced expression of the C. elegans homolog, FRH-1, activates autophagy in an evolutionarily conserved manner [2121].
FYCO1 (FYVE and coiled-coil domain autophagy adaptor 1): A protein that interacts with LC3, PtdIns3P and RAB7 to move autophagosomes toward the lysosome through microtubule plus end-directed transport [3151].
Gαi3: See GNAI3.
GABA (γ‐aminobutyric acid): GABA inhibits the selective autophagy pathways mitophagy and pexophagy through Sch9, leading to oxidative stress, which can be mitigated by the Tor1 inhibitor rapamycin [3152].
GABARAP (GABA type A receptor-associated protein): A homolog of Atg8 and LC3 [786, 3153]. The GABARAP family includes GABARAP, GABARAPL1/Atg8L/GEC1, and GABARAPL2/GATE-16/GEF2. The GABARAP proteins are involved in autophagosome formation and cargo recruitment [32, 199].
GADD34: See PPP1R15A.
GAIP: See RGS19.
Gap junction proteins/connexins: Multispan membrane proteins that directly connect the cytoplasm of adjacent cells through the formation of gap junction channels composed by the docking of two hemi-channels or gap junctions at the plasma membrane. In invertebrates, these channels are constituted by innexins, whereas in vertebrates they are formed by GJ (gap junction protein)/connexin proteins [3154]. GJs act as endogenous inhibitors of autophagosome formation by directly interacting and sequestering at the plasma membrane essential ATG proteins required for autophagosome biogenesis [3155, 3156]. At the same time, gap junction proteins/connexins themselves are degraded through autophagy, contributing to their rapid turnover [3157].
GAS5 (growth arrest specific 5): A lncRNA that is a positive regulator of autophagy through microRNA sponging [3158]. Depletion of GAS5 decreases autophagy in non-small-cell lung carcinoma cells [3159], and knockdown of GAS5 in epithelial cells suppresses the expression of LC3-II, ATG3, and ATG12–ATG5 complex formation, whereas the levels of SQSTM1 are promoted [3160].
GATA1: A hematopoietic GATA transcription factor, expressed in erythroid precursors, megakaryocytes, eosinophils, and mast cells, that provides the differentiating cells with the requisite autophagy machinery and lysosomal components to ensure high-fidelity generation of erythrocytes [981]. See also ZFPM1/FOG1.
GATE-16: See GABARAP.
Gaucher disease (GD): Caused by mutations in the gene encoding GBA (glucosylceramidase beta), Gaucher disease is the most common of the lysosomal storage disorders, and GBA mutations in the heterozygous state can increase susceptibility to PD and other neurodegenerative synucleinopathies [3161–3163]. See also GBA.
GBA/GBA1/GCase/glucocerebrosidase (glucosylceramidase beta): A lysosomal enzyme that breaks down glucosylceramide and glucosylsphingosine to glucose and ceramide, or glucose and sphingosine, respectively. Mutations cause Gaucher disease and are associated with increased risk of PD. Loss of GBA is also associated with impaired autophagy and failure to clear dysfunctional mitochondria, which accumulate in the cell [3164]. See also Gaucher disease.
GBM Studiengruppe Autophagie (German Autophagy Association): See the organization website (http://autophagie-gbm.de/)
GCA (grancalcin): GCA activates TRAF6 ubiquitin ligase activity to induce Lys63 ubiquitination of ULK1, a crucial regulator of autophagy, resulting in its stabilization and activation [649].
Gcn2: A mammalian and yeast EIF2S1/eIF2α serine/threonine kinase that causes the activation of the integrated stress response, including induction of Gcn4 in response to amino acid depletion, thus positively regulating autophagy [3079]. Gcn2 is homologous to EIF2AK4 in humans. See also Gcn4.
Gcn4: A yeast transcriptional activator that controls the synthesis of amino acid biosynthetic genes and positively regulates autophagy in response to amino acid depletion [3079]. Gcn4 directly targets ATG1 and ATG41 to promote transcription during nitrogen starvation [910, 2822]. See also Gcn2.
GCN5L1: See BLOC1S1.
GEEC (GPI-enriched endocytic compartments) pathway: A form of clathrin-independent endocytosis that contributes membrane for phagophore expansion [3165].
Genotoxin-induced targeted autophagy (GTA): A selective autophagy pathway that is induced specifically in response to genotoxic stress in S. cerevisiae. GTA is dependent on the core autophagy machinery in yeast, as well as Atg11 and the DNA damage response kinases Mec1/ATR and Tel1/ATM [1093].
GFAP (glial fibrillary acid protein): Intermediate filament protein ubiquitously distributed in all cell types that bears functions beyond filament formation. Monomeric and dimeric forms of this protein associate with the cytosolic side of the lysosomal membrane and contribute to modulating the stability of the CMA translocation complex in a GTP-dependent manner coordinated with EEF1A/eF1α also at the lysosomal membrane [3076].
GFER/ERV1 (growth factor, augmenter of liver regeneration): A flavin adenine dinucleotide-dependent sulfhydryl oxidase that is part of a disulfide redox system in the mitochondrial intermembrane space, and is also present in the cytosol and nucleus. Downregulation of GFER results in elevated levels of the mitochondrial fission GTPase DNM1L, and decreased mitophagy [3166].
Ghrelin: An endogenous small peptide produced from the GHRL gene that activates autophagy and mediates caloric restriction-induced autophagy in rat cortical neurons [3167]. Ghrelin also modulates autophagy in metabolic, cardiac and neurodegenerative disorders associated with impaired autophagy [3168]. See also NPY.
GID (glucose induced degradation deficient) complex: A highly evolutionary conserved ubiquitin ligase that targets key enzymes of gluconeogenesis (Fbp1, Mdh2, Pck1) for polyubiquitination and subsequent proteasomal degradation in S. cerevisiae [3169–3172]. Individual subunits of the yeast Gid complex are conserved throughout the eukaryotic kingdom and Vid30/Gid1, Rmd5/Gid2, Vid24/Gid4, Vid28/Gid5, Gid7, Gid8 and Fyv10/Gid9 [3173] have their closest human orthologs in RANBP9 (RAN binding protein 9)-RANBP10, RMND5A (required for meiotic nuclear division 5 homolog A)-RMND5B, GID4 (GID complex subunit 4 homolog), ARMC8 (armadillo repeat containing 8), MKLN1 (muskelin 1) or WDR26 (WD repeat domain 26), GID8 (GID complex subunit 8 homolog) and MAEA (macrophage erythroblast attacher), respectively. These subunits are also part of the human equivalent to the yeast GID complex (CTLH complex) [3174–3176]. The vertebrate GID-complex regulates AMPK activity through ubiquitination [3177].
GILT: See IFI30.
Ginkgolic acids: Ginkgolic acids are a group of alkyl phenols found in crude extracts of Ginkgo biloba leaves, an ancient gymnosperm species. The incubation of tumoral cells with ginkgolic acids blocks the sumo pathway and, as a consequence, induces autophagy-mediated cancer cell death [2673].
GIV/Girdin: See CCDC88A.
GLI3 (GLI family zinc finger 3): A C2H2 type of zinc finger transcription factor that plays a role in the transcriptional activation of VMP1 during the induction of autophagy by the oncogene KRAS [2673]. See also EP300.
Glutamoptosis: Cell death induced during nutritional imbalance due to the inhibition of autophagy mediated by glutaminolysis [3178, 3179].
Glycophagy (glycogen autophagy): The selective sequestration of glycogen and subsequent vacuolar hydrolysis of glycogen to produce glucose; this can occur by a micro- or macroautophagic process and has been reported in mammalian newborns and adult cardiac tissues as well as filamentous fungi [58, 2184, 2185, 3180–3182].
GMI: A fungal immunomodulatory protein from Ganoderma microsporum, GMI induces autophagic cell death via calcium-TP53 and AKT-MTOR signaling pathways [3183].
GNAI3 (G protein subunit alpha i3): A heterotrimeric G protein that activates autophagy in the GDP-bound (inactive) form, and inhibits it when bound to GTP (active state) [3184, 3185]. See also GPSM1, RGS19, MAPK1/3 and CCDC88A.
Golgi membrane-associated degradation (GOMED): A non-canonical Golgi membrane-mediated process that is activated by the disruption of PtdIns4P-dependent anterograde trafficking, which occurs in both yeast and mammals [3186].
GOPC/PIST/FIG/CAL (golgi associated PDZ and coiled-coil motif containing): GOPC interacts with BECN1, and the SNARE protein STX6 (syntaxin 6). GOPC can induce autophagy via a CD46-Cyt-1 domain-dependent pathway following pathogen invasion [2949].
GORASP2/GRASP55 (golgi reassembly stacking protein 2): A Golgi apparatus peripheral membrane protein originally identified as a Golgi apparatus stacking protein in the medial-trans Golgi apparatus. Under growth condition, GORASP2 is modified by a reversible, cytosolic glycosylation called O-GlcNAcylation, and plays a critical role in Golgi apparatus stack and ribbon formation by forming trans oligomers [3187]. Upon energy or nutrient deprivation, GORASP2 is de-O-GlcNAcylated and partially targeted to the autophagosome-lysosome interface by interacting with LC3 and LAMP2. GORASP2 facilitates autophagosome-lysosome fusion by physically linking the two membrane organelles and by recruiting the UVRAG-containing PtdIns3K complex to autophagosomes [3188, 3189]. See also secretory autophagy.
Gossypol: See AT-101.
Gp78: See AMFR.
GPSM1/AGS3 (G protein signaling modulator 1): A guanine nucleotide dissociation inhibitor for GNAI3 that promotes autophagy by keeping GNAI3 in an inactive state [2939]. GPSM1 directly binds LC3 and recruits GNAI3 to phagophores or autophagosomes under starvation conditions to promote autophagosome biogenesis and/or maturation. See also GNAI3.
GRAMD1A (GRAM domain containing protein 1A): Upon starvation, the cholesterol transfer protein GRAMD1A accumulates during autophagosome initiation, which has been found to affect cholesterol distribution, a process required for autophagosome biogenesis [2848].
Granulophagy: The process of bulk autophagic degradation of mRNP granules, particularly stress granules, although autophagic degradation of P-granules and P-bodies in C. elegans and human cells has also been described [1275, 1276]. Granulophagy of stress granules has been characterized in S. cerevisiae and mammalian cells and is dependent on Cdc48/VCP/p97 in addition to the core autophagic machinery. Mutations in VCP associated with various neurodegenerative diseases also impair granulophagy [1274]. SQSTM1 facilitates granulophagy of stress granules in human cells [1275, 1277, 3190]. See also granulostasis and MSP.
Granulostasis: The process of chaperone-mediated quality control of RNP granules. This process involves the HSPB8/BAG3/HSP70 chaperone complex [1278, 1279] and autophagy factors such as Cdc48/VCP/p97 and SQSTM1, which mediate granulophagy of aberrant RNP granules. Impairment of granulostasis through mutations or chemical inhibition of granulostasis factors leads to the formation of aberrant RNP granules that may cause disease [3191, 3192]. See also granulophagy.
GRB2 (growth factor receptor bound protein 2): GRB2 binds EGFR and contains one SH2 domain and two SH3 domains, working as a signal adaptor protein. The two SH3 domains bind to proline-rich regions of other proteins, and its SH2 domain binds phosphorylated-tyrosine sequences. GRB2 promotes degradation of mutant HTT by augmenting autophagy in a Huntington disease cell model [3193, 3194].
GRN/PGRN (granulin precursor): GRN is an 88-kDa secreted multi-functional glycoprotein. Human GRN is composed of 7.5 repeats of a highly conserved twelve-cysteine granulin motif. GRN loss-of-function mutations cause frontotemporal dementia and neuronal ceroid lipfuscinosis, type 11 (CLN11). GRN is trafficked to the lysosome amd processed into stable, bio-active 6-kDa granulin proteins [3195]. Deficiency of GRN and granulins impairs autophagy and lysosome function [3196, 3197].
GSK3/GSK-3 (glycogen synthase kinase 3): A regulator of autophagy. GSK3A and GSK3B are highly similar isoforms. GSK3 may act positively by inhibiting MTORC1 through the activation of TSC1/2 and by activating ULK1 through KAT5 [3198]. GSK3 modulates protein aggregation through the phosphorylation of the autophagy receptor NBR1 [2664]. GSK3, however, is also reported to be a negative regulator of autophagy. GSK3-mediated phosphorylation of MCL1 induces axonal autophagy, which promotes axonal degeneration. Phosphorylated MCL1 is ubiquitinated by the FBXW7 ubiquitin ligase and degraded by the proteasome [3131]. GSK3 also inhibits autophagy independently of MTORC1 through phosphorylation and cytosolic retention of the transcription factor TFEB [3199, 3200]. See also FBXW7, KAT5, MCL1 and TFEB.
Guanine quadruplex (G4) ligands: A range of compounds that recognize unusual nucleic acid structures formed by guanine-rich sequences; some of them activate autophagy [2832, 2954].
H1-2/HIST1H1C (H1.2 linker histone, cluster member): A variant of linker histone H1, which regulates autophagic gene expression in the retina of diabetic rodents and high-glucose-cultured cells [3201].
H2AX/H2AFX (H2A.X variant histone): Histone H2AX phosphorylation on a serine located four residues from the C terminus (producing γH2AX) is a sensitive marker for DNA double-strand breaks [3202].
HA15: A new thiazol benzenesulfonamide compound that directly targets HSPA5/GRP78 to induce a strong ER stress and cancer cell death by concomitant autophagy and apoptosis mechanisms [3203, 3204].
HDAC4 (histone deacetylase 4): A deacetylase distributed in both the cytoplasm and nuclei that interacts with MAP1S and the aggregation-prone mutant HTT protein (mHTT) that causes Huntington disease; this interaction regulates the MAP1S-mediated autophagic turnover of mHTT. Spermidine reduces the distribution of HDAC4 in the cytosol and enhances MAP1S-mediated autophagy to prolong lifespan and prevent liver fibrosis and hepatocellular carcinomas [956, 3205].
HDAC6 (histone deacetylase 6): A microtubule-associated deacetylase that interacts with ubiquitinated proteins. HDAC6 stimulates autophagosome-lysosome fusion by promoting the remodeling of F actin, and the quality control function of autophagy, and also acts as a mediator between autophagy and the UPS [1024, 1025, 3206, 3207]. HDAC6 is also a biomarker of aggresomes [1781].
HDAC10 (histone deacetylase 10): A lysine deacetylase that promotes lysosomal exocytosis supporting drug resistance in aggressive tumor cells [3208, 3209].
HDX-MS (hydrogen deuterium exchange mass spectrometry): A mass-spectrometry-based method, which can be used to examine protein-protein, protein-lipid, protein-nucleic acid, and protein-chemical interactions, as well as to estimate regions of intrinsic disorder. HDX-MS measures rate of solvent exchange at the amide positions of the protein backbone through the isotopic exchange of hydrogen with deuterium. When using this procedure, it is highly recommended to follow the guidelines established by the HDX-MS community [3210].
HER2: See ERBB2.
Hfl1: A vacuole membrane protein that interacts with Atg8 and mediates the lipidation-independent vacuolar functions of Atg8 in both fission yeast and budding yeast; Hfl1 uses a non-canonical type of Atg8-interacting motif, termed a helical AIM, to interact with Atg8 [3211].
HIF1A/HIF-1α (hypoxia inducible factor 1 subunit alpha): A dimeric transcription factor in which the α subunit is regulated by oxygen and other stimuli, including inflammation [3212] and increases in tonicity [3213]; the hydroxylated protein is degraded by the proteasome. HIF1A-mediated expression of BNIP3 results in the disruption of the BCL2-BECN1 interaction, thus inducing autophagy [3214, 3215]. HIF1A also regulates xenophagic degradation of intracellular E. coli [3216].
Histone H3K56 acetylation: H3K56ac is one of the histone modifications governing gene expression and regulation. H3K56ac plays a primary role in maintaining genomic stability and the DNA damage response [3217]. The role of this mark in regulating the autophagy epigenome has also been demonstrated by POLR2/RNA polymerase 2 occupancy in H3K56ac regions [986]. H3K56ac could influence the expression of mitophagy-related genes including PINK1, PRKN and AKT. In the neurotoxic model of mitochondrial complex II inhibition, elevated H3K56ac is associated with autophagy/mitophagy genes (R. Sathyanarayanan and M.M. Srinivas Bharath, unpublished data).
HK2 (hexokinase 2): The enzyme responsible for phosphorylation of glucose at the beginning of glycolysis; during glucose starvation, HK2 switches from a glycolytic role and directly binds to and inhibits MTORC1 to induce autophagy [3218].
HLH-30: C. elegans ortholog of the helix-loop-helix transcription factor TFEB.
HMGA1 (high mobility group AT-hook 1): An architectural chromatin protein, whose overexpression is a common feature of several malignant neoplasias, and that has a causal role in cancer initiation and progression. In skin cancer and HeLa cells, HMGA1 knockdown increases autophagosome formation by constraining the activity of the MTOR pathway, and transcriptionally upregulating ULK1, without inducing a proportionate increase in autophagosome maturation. The autophagosome accumulation induced by HMGA1 depletion is associated with a decrease in cell proliferation and viability [922]. HMGA1 is able to activate the PI3K-AKT signaling pathway, and thus MTORC1, also in pancreatic adenocarcinoma cells, ultimately inducing anoikis resistance [3219].
HMGB1 (high mobility group box 1): A chromatin-associated nuclear protein that translocates out of the nucleus in response to stress such as ROS; HMGB1 binds to BECN1, displacing BCL2, thus promoting autophagy and inhibiting apoptosis [398]. In addition, autophagy promotes the release of HMGB1 from the nucleus and the cell, and extracellular HMGB1 can further induce autophagy through binding AGER [3220, 3221]. In a different context, autophagy-deficiency in hepatocytes can also trigger the active secretion of HMGB1 via an NFE2L2-CASP1 signaling pathway. Released HMGB1 promotes ductular reaction, a repair/regeneration response to liver injury, and tumor progression, in the absence of autophagy function [3222]. See also AGER.
Hog1: A yeast MAPK involved in hyperosmotic stress, which is a homolog of mammalian MAPK/p38; Hog1 is required for mitophagy, but not other types of selective autophagy or nonselective autophagy [747]. See also MAPK, Pbs2 and Slt2.
HMOX1 (heme oxygenase 1): An inducible enzyme involved in heme metabolism and cell protection from oxidative damage. HMOX1 is required for activation of nonselective autophagy and mitophagy in response to oxidative stress [3223].
HOPS (homotypic fusion and vacuole protein sorting): The HOPS complex is a six-subunit tethering complex that is required for the fusion of autophagosomes, late endosomes and biosynthetic vesicles with lysosomes from yeast to mammals [3224–3226].
HOTAIRM1 (HOXA transcript antisense RNA, myeloid-specific 1): A lncRNA that is downregulated in acute promyelocytic leukemia/APL patients. HOTAIRM1 regulates autophagy by acting as a microRNA sponge of MIR20A, MIR106B, MIR125B and their targets ULK1, E2F1 and DRAM2. Knocking down HOTAIRM1 can inhibit ATRA-triggered autophagosome formation [3227]. See also DRAM2.
hrm (hermes): A Drosophila proton-coupled pyruvate transporter that is required for autophagy associated with cell death during salivary gland degradation [3228].
Hrr25: A protein kinase (homologous to CSNK1D/E) regulating diverse cellular processes such as DNA repair and vesicular trafficking. Hrr25 phosphorylates the C terminus of Atg19, which is essential for Atg19 binding to Atg11 and subsequent Cvt vesicle formation [3229]. Hrr25 also phosphorylates Atg36, and this phosphorylation is required for the interaction of Atg36 with Atg11 (and subsequent pexophagy) [3230], and with Atg34 [3231]. The KpHrr25 ortholog in K. phaffii/P. pastoris phosphorylates KpAtg30 [2814], for subsequent KpAtg11 recruitment and selective autophagy initiation. See also Atg11.
HS1BP3 (HCLS1 binding protein 3): A negative regulator of autophagy. HS1BP3 regulates autophagy by modulating the phosphatidic acid content of the ATG16L1-positive autophagosome precursor membranes through PLD1 (phospholipase D1) activity and localization [3232].
HSC70: See HSPA8.
Hsp83 (Heat shock protein 83): The Drosophila ortholog of human HSP90AA1/HSP90. Loss of Hsp83 results in reduced proteasomal activity and enhanced autophagic flux that is dependent on the Drosophila effector caspase Dcp-1 [3233].
HSPA1A: The major cytosolic stress-inducible version of the HSP70 family. This protein localizes to the lysosomal lumen in cancer cells, and pharmacological inhibition leads to lysosome dysfunction and inhibition of autophagy [3234].
HSPA5/GRP78/BiP (heat shock protein family A (Hsp70) member 5): A master regulator of the UPR. This chaperone, maintaining ER structure and homeostasis, can also facilitate autophagy [3235]. Cellular stresses such as cytosolic DNA induce N-terminal arginylation of HSPA5/BiP (arginylated HSPA5/R-Bip), which is selectively recognized by the ZZ domain of SQSTM1 and targeted to phagophores together with SQSTM1 and associated cargoes [463, 3236, 3237]. See also SQSTM1.
HSPA8/HSC70 (heat shock protein family A (Hsp70) member 8): This multifunctional cytosolic chaperone is the constitutive member of the HSP70 family of chaperones and participates in targeting of cytosolic proteins to lysosomes for their degradation via chaperone-mediated autophagy [3238]. The cytosolic form of the protein also regulates the dynamics of the CMA receptor, whereas the lumenal form (lys-HSPA8) is required for substrate translocation across the membrane [3239]. This chaperone plays a role in the targeting of aggregated proteins (in a KFERQ-independent manner) for degradation through chaperone-assisted selective autophagy [1828], and in KFERQ-dependent targeting of cytosolic proteins to late endosomes for microautophagy [1823]. See also chaperone-assisted selective autophagy, chaperone-mediated autophagy, and endosomal microautophagy.
HSP70 (heat shock protein 70): The major cytosolic heat shock-inducible member of the HSP70 family. This form accumulates in the lysosomal lumen in cancer cells. HSP70 is also a biomarker of aggresomes [3240]. See also HSPA1A.
HSP90: See HSP90AA1.
HSP90-ROF1 complex: AT4G24690/NBR1 targets HSP90 and AT3G25230/ROF1, a member of the FKBP family, and mediates their degradation by autophagy, which represses the expression of heat shock proteins regulated by the AT2G26150/HSFA2 transcription factor and the response to heat stress in Arabidopsis [3241] (V.P. Thirumalaikumar, M. Gorka, M. Schulz, C. Masclaux-Daubresse, A. Sampathkumar, A. Skirycz, R.D. Vierstra and S. Balazadeh, unpublished data). See also NBR1.
HSP90AA1/HSP90/HSPC1 (heat shock protein 90 alpha family class A member 1): A cytosolic chaperone that is also located in the lysosome lumen. The cytosolic form helps to stabilize BECN1, and promotes autophagy [3242]. The lysosomal form of HSP90AA1 contributes to the stabilization of LAMP2A during its lateral mobility in the lysosomal membrane [3243].
HSPB1/HSP27 (heat shock protein family B (small) member 1): A stress-responsive molecular chaperone that interacts with the oligomerization domain of SQSTM1. Neuropathy-causing mutations in HSPB1 lead to autophagy impairment by disrupting the interaction with SQSTM1 [3244].
HSPB8/HSP22 (heat shock protein family B (small) member 8): A stress-induced chaperone that associates with BAG3 forming the CASA complex [1828, 1841, 3245, 3246] with HSPA8-STUB1, DNAJB6 and SQSTM1. Once formed and cargo is recognized, STUB1 (an E3-ubiquitin ligase) ubiquitinates the substrate allowing its SQSTM1-mediated insertion into the phagophore. At variance with CMA, which routes cargoes directly to lysosomes, CASA routes substrates to the lysosome via an autophagosome. The process is thought to take place at the microtubule organizing center to which the CASA complex is transported in a retrograde manner by dynein associated directly with a PxxP motif of BAG3 [1840, 2929]. If HSPB8-BAG3 are displaced by the co-chaperone BAG1, the cargoes are routed by HSPA8-STUB1 to the proteasome.
HSPC1: See HSP90AA1.
HTRA2/Omi (HtrA serine peptidase 2): A nuclear-encoded mitochondrial serine protease that was reported to degrade HAX1, a BCL2 family-related protein, to allow autophagy induction [3247]. In this study, knockdown of HTRA2, or the presence of a protease-defective mutant form, results in decreased basal autophagy that may lead to neurodegeneration. Separate studies, however, indicate that mitochondrial HTRA2 plays a role in mitochondrial quality control; in this case loss of the protein leads to increased autophagy and in particular mitophagy [3248–3250].
HTT (huntingtin): Expanded polyglutamine repeats in mutant HTT (repeat number beyond 36) lead to Huntington disease, a neurodegenerative disorder involving loss of striatal medium spiny neurons [1649, 3251, 3252]. The mutant HTT protein fragment can be degraded by autophagy via adaptor proteins such as WDFY3/Alfy and upregulated by HTT acetylation at K444 and K9, and phosphorylation at S13 by IKK [1024, 3251, 3253–3257]. In addition, targeting the autophagic pathway is beneficial in multiple models of HD [1951, 2036, 3251, 3258, 3259]. HTT can also directly regulate autophagy either by modulating autophagosome axonal transport in neurons via its interaction with DYNC1I (dynein cytoplasmic 1 intermediate chain) [3260, 3261] or as a scaffold for selective autophagy [2784, 3262].
Hypersensitive response: A rapid and locally restricted form of programmed cell death as part of the plant immune response to pathogen attack. The hypersensitive response is activated by different immune receptors upon recognition of pathogen-derived effector proteins, and can be positively regulated by autophagy [1760, 1764, 3263].
HYPK (huntingtin interacting protein K): HYPK was identified as an HTT-interacting intrinsically disordered protein with chaperone-like activity, which activates autophagy in a HD cell model [3264].
IAPP (islet amyloid polypeptide): A 37 amino acid polypeptide derived from processing of an 89 amino acid precursor, which is coexpressed with INS (insulin) by pancreatic β-cells. IAPP aggregation is implicated in the pathogenesis of type 2 diabetes. Autophagy regulates IAPP levels through SQSTM1-dependent lysosomal degradation [3265–3267].
iC-MA (immune cell-mediated autophagy): IL2-activated natural killer cell- and T cell-induced autophagy [3268].
Ice2: See Ayr1.
ICP0 (infected cell protein 0): An immediate-early gene product of herpes simplex virus 1 that functions as a promiscuous transactivator of genes introduced into the cells by infection or transfection. This protein carries a zinc-binding RING finger domain and functions as an E3 ubiquitin ligase targeting for degradation proteins that are hostile to the cell. ICP0 localizes in the nucleus immediately after its production and translocates to the cytoplasm, after accomplishing its nuclear functions. The autophagy receptors SQSTM1 and OPTN are downregulated by cytoplasmic ICP0, in a proteasome-dependent manner [1422].
ICP34.5: A neurovirulence gene product encoded by the herpes simplex virus type 1 that blocks EIF2S1-EIF2AK2 induction of autophagy [3079]. ICP34.5-dependent inhibition of autophagy depends upon its ability to bind to BECN1 [1418].
IcsB: A T3SS effector found in Shigella spp. that acylates the protein CHMP5 and enables escape of secreting S. flexneri cells from LC3-positive vacuoles [3269, 3270]. IcsB is also proposed to prevent recognition of VirG by ATG5. See also VirG.
IDP (Intrinsically disordered protein): A protein that does not possess unique structure and exists as a highly dynamic ensemble of interconverting conformations [3271–3273]. IDPs are very common in nature [3274] and have numerous biological functions that complement the functional repertoire of ordered proteins [3275–3278]. Many proteins involved in autophagy are IDPs [3279, 3280].
IDPR (intrinsically disordered protein region): A protein region without unique structure that may be biologically important. IDPRs are considered as a source of functional novelty [3281], and they are common sites of protein-protein interactions [3282] and posttranslational modifications [3283].
IFI30/GILT (IFI30 lysosomal thiol reductase): A thiol reductase that controls ROS levels; in the absence of IFI30 there is an increase in oxidative stress that results in the upregulation of autophagy [3284].
IFT (intraflagellar transport): Bidirectional transport of multimolecular complexes, called IFT particles, that shuttle ciliary cargo along axonemal microtubules through the association with molecular motors (i.e., kinesins and dyneins). The IFT system plays an essential role in the assembly and function of motile cilia and flagella as well as in the modulation of cilium-dependent signaling pathways at the primary cilium. Moreover, some IFT proteins have been involved in the regulation of vesicular trafficking with relevant implications in different vesicle-based processes, including autophagy. In ciliated cells, IFT20 (intraflagellar transport 20) and IFT88 contribute to autophagosome formation by promoting ATG16L1 recruitment to and into the primary cilium under starvation conditions [3285]. In non-ciliated lymphocytes, IFT20 controls lysosome biogenesis and function, and therefore lysosomal degradation of autophagic cargo, by modulating the TFEB-driven transcriptional program and regulating IGF2R/cation-independent mannose-6-phosphate receptor recycling [3286]; this lysosome-related function of IFT20 is conserved in ciliated cells. See also ciliophagy and primary cilium.
IKBKE (Inhibitor of nuclear factor kappa B kinase subunit epsilon): IKBKE is frequently amplified or overexpressed in a number of human cancers, in particular of the breast, and is implicated in their tumorigenic process. It induces the autophagic process, when overexpressed, and oncogenic pathways typically involved in breast cancer, namely ERBB2 and PI3K-AKT-MTOR, also rely on its activity to control autophagy and, in turn, cancer cell proliferation [2680].
IKK (IκB kinase): An activator of the classical NFKB pathway composed of three subunits (CHUK/IKKα/IKK1, IKBKB/IKKβ/IKK2, IKBKG/IKKγ/NEMO) that are required for optimal induction of autophagy in human and mouse cells [3287].
IKKβ/ird5 (I-kappaB kinase β): Drosophila homolog of IKBKB/IKKβ that is selectively degraded by autophagy through its interacting partner key/kenny [2162]. See also key.
iLIR: A web resource for prediction of Atg8-family-interacting proteins (http://repeat.biol.ucy.ac.cy/iLIR) [2510].
iLIR database: A database listing all the putative canonical LIR-motif-containing proteins identified in silico in the proteomes of eight model organisms combined with a Gene Ontology (GO) term analysis (https://ilir.warwick.ac.uk) [2513].
iLIR@viral: A database listing all the putative canonical LIR motifs identified in viral proteins (http://ilir.uk/virus/) [2513].
ILK (integrin linked kinase): ILK is involved with integrin-mediated signal transduction. This enzyme is activated by autophagy during TGFB-induced epithelial-mesenchymal transition [3288].
Iml1 complex: A protein complex containing Iml1, Npr2 and Npr3 that regulates non-nitrogen-starvation-induced autophagosome formation; the complex partially localizes to the PAS [3289]. See also non-nitrogen-starvation (NNS)-induced autophagy.
Immunoamphisomes: An organelle derived from the fusion of endosomes/phagosomes with autophagosomes that regulate dendritic cell-mediated innate and adaptive immune responses [3290].
Immunophagy: A sum of diverse immunological functions of autophagy [3291].
IMPA (inositol monophosphatase): An enzyme that regulates the inositol 1,4,5-triphosphate (IP3) levels. Inhibition of IMPA stimulates autophagy independent of MTOR [1976].
Incomplete autophagy: A process involving activation of autophagy but without increasing autophagic degradation. Some RNA viruses and bacteria utilize this mechanism to facilitate their replication [3001].
Inflammasome: The inflammasome is an intracellular inflammatory machinery that can switch on the inflammatory response of tissues to a variety of stimuli. Autophagy and several types of the inflammasome such as AIM2 and NLRP3 inflammasomes are functionally interconnected. This crosstalk between autophagy and the inflammasome machineries plays important roles in the control of cell homeostatic processes including cell metabolism, organelle maintenance, clearance of pathogens, and inflammatory response [3292–3295].
Inflammation: A stereotyped and protective immune, vascular and tissue response to exogenous or endogenous harmful stimuli that is spontaneously extinguished and resolved after elimination or termination of the damage by means of the bioaction of several proteins and lipid mediators [3296–3298]. The cellular degradative pathway of autophagy has a fundamental role in controlling the elimination of inflammatory insults and simultaneously in contributing to immune cell development and in limiting inflammatory pathologies [3299].
InlK: An internalin family protein on the surface of L. monocytogenes that recruits vault ribonucleoprotein particles to escape xenophagy [3300].
Innate immune surveillance: Recognition and response system for the sensing of DAMPs, including pathogens and products of somatically mutated genes. Innate surveillance responses include activation of autophagy to degrade DAMPs [3034].
INSR (insulin receptor): Upregulation of INSR induces protective autophagy and assists in mitigating poly(Q)-mediated neurodegeneration in Drosophila [3301].
Integrated stress response: A highly conserved adaptation to stress centered upon phosphorylation of the alpha subunit of EIF2S1/eIF2α [3301]. Four EIF2S1 kinases sense distinct stress conditions: EIF2AK1/HRI, EIF2AK2/PKR, EIF2AK3/PERK and EIF2AK4/GCN2. Phosphorylation of EIF2S1 inhibits global translation, conserving cellular resources and facilitating reprogramming of gene expression. Simultaneously, p-EIF2S1 directs preferential translation of a subset of “stress response” mRNAs, including ATF4, via a delayed translation reinitiation that allows ribosome scanning through inhibitory upstream open reading frames (uORFs) [2748].
Ionomycin: A calcium ionophore that releases calcium from intracellular stores, thus increasing the intracytosolic level of this ion [3302].
IP3K2 (inositol 1,4,5-trisphosphate kinase 2): A target of mir-14 that regulates calcium signaling and autophagy in Drosophila salivary glands [3303]. See also mir-14.
IP3R: See ITPR.
IRGM (immunity related GTPase M): Involved in the auto-phagic control of intracellular pathogens [3304]. IRGM interacts with core autophagy proteins ULK1, BECN1 and ATG16L1 to govern antimicrobial autophagy [3305], and limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective degradation by autophagy [3306]. IRGM is a common target of RNA viruses to subvert autophagy [3307]. In mouse, there are three IRGM homologs named IRGM1, IRGM2, and IRGM3. Mouse IRGM1 has the most extensive shared functions with human IRGM.
IRGM1 (immunity-related GTPase family M member 1): See IRGM.
Irs4: Irs4 and Tax4 localize to the PAS under autophagy-inducing conditions in yeast and play a role in the recruitment of Atg17 [3308]. These proteins have partially overlapping functions and are required for efficient nonselective autophagy and pexophagy.
Isolation membrane: See phagophore.
ITM2A (integral membrane protein 2A): A target of PRKA/PKA-CREB that interacts with the V-ATPase and interferes with autophagic flux [3309].
ITPR1/2/3 (inositol 1,4,5-trisphosphate receptor type 1/2/3): A large tetrameric intracellular calcium-release channel present in the ER and Golgi apparatus membranes that is responsible for the initiation/propagation of intracellular calcium signals augmenting free calcium concentrations in the cytosol and/or organelles such as mitochondria [2001]. The ITPR is activated by inositol 1,4,5-trisphosphate/IP3 produced in response to extracellular agonists. Many proteins regulate the properties of ITPR including anti-apoptotic BCL2-family proteins and BECN1 [3310, 3311]. The ITPR can inhibit autophagy by scaffolding BECN1 as well as by driving ATP production through transfer of calcium from the ER to mitochondria, and consequent stimulation of mitochondrial metabolism [668, 1976, 2063, 3312], whereas BECN1-dependent sensitization of ITPR-mediated calcium release (e.g., in response to starvation) can promote autophagic flux [1983]. The latter process is a direct effect of BECN1 on ITPRs and appears not to be mediated via ITPR-associated BCL2 recruiting BECN1 [1983]. Under the conditions of physiological starvation, ITPR-mediated calcium release activates the CAMK2-OGT-ULK cascade that initiates autophagy [1007]. ITPRs and calcium signaling not only contribute to driving autophagic flux upon starvation but also in response to chemical MTOR inhibitors such as rapamycin [1985], to resveratrol [1991] and to chemical inducers of ER stress such as the proline analog L-azetidine-2-carboxylic acid [1992]. An integrated discussion on the complex role of intracellular calcium signaling in autophagy has been provided elsewhere [1994].
JNK1: See MAPK8.
Jumpy: See MTMR14.
JUN/c-Jun/JunB (Jun proto-oncogene, AP-1 transcription factor subunit): A mammalian transcription factor that inhibits starvation-induced autophagy [3313].
Karyoptosis: A form of CASP3-independent cell death proposed to be directly associated with the nucleophagy process, leading to cell atrophy, nuclear degeneration and DNA damage [1750].
KAT5/TIP60 (lysine acetyltransferase 5): In response to growth factor deprivation, KAT5 is phosphorylated and activated by GSK3 and then acetylates and activates ULK1 [3198].
Kcs1: A yeast inositol hexakisphosphate/heptakisphosphate kinase; the kcs1∆ strain has a decrease in autophagy that may be associated with an incorrect localization of the PAS [3314].
KDM1A/LSD1 (lysine demethylase 1A): A mammalian demethylase that modulates autophagy via multiple mechanisms including modulation of MTORC1 activity and SQSTM1 stability [3315–3317].
KDM2B (lysine demethylase 2B): A histone lysine demethylase that promotes H3K4 demethylation. The downregulation of KDM2B in gastric cancer cells induces autophagy and inhibits cell proliferation via PI3K-AKT-MTOR inhibition and MAPK1/3 activation [3318].
KDM4A (lysine demethylase 4A): A mammalian demethylase that regulates the expression of a subset of ATG genes [912, 913]. See also Rph1.
KEAP1 (kelch like ECH associated protein 1): An E3 ubiquitin ligase responsible for the degradation of transcription factor NFE2L2/NRF2 and the NFKB activator IKBKB/IKKβ. KEAP1 is a substrate for SQSTM1-dependent sequestration. SQSTM1 influences oxidative stress-related gene transcription and regulates the NFKB pathway via its interaction with KEAP1 [596, 3319, 3320]. See also NFE2L2.
key/kenny: Drosophila homolog of IKBKG/IKKγ that interacts with Drosophila Atg8a via a LIR motif and is selectively degraded by autophagy. key is the selective autophagy receptor for the degradation of the IKK complex (key and IKKβ/ird5) [2162]. See also IKKβ.
KFSDA pentapeptide (“KFERQ” mutant): A mutant form of the KFERQ-like recognition motif as a negative reference substrate peptide in CMA activity assays. Mutagenesis of the CMA recognition motif is based on altering the physical properties of amino acids in the targeting motif [3321]. The resultant changes in individual amino acid are: “KFERQ” to “KFSDA” (“KFERQ” mutant): First and second amino acids: unchanged; third amino acid: E (acidic) to S (polar); fourth amino acid: R (basic) to D (acidic); fifth amino acid: Q (polar) to A (hydrophobic). An immunoprecipitation study shows that a CMA substrate carrying a KFSDA mutant peptide does not bind HSPA8, a key step prior to CMA degradation [1814].
KIAA0226: See RUBCN.
KIAA1524: see CIP2A.
KIFC3 (kinesin family member C3): A minus-end directed motor protein, member of the C-kinesin family, highly enriched in lysosomes and required for the motility of the lysosomal compartment [3322].
KillerRed: A red fluorescent protein that produces a high amount of superoxide upon excitation. The construct with a mitochondria targeting sequence (mitoKillerRed) can be used to induce mitochondria damage and subsequent mitophagy [462, 1173].
KL (klotho): A protein involved in the crosstalk between autophagy and apoptosis. A reduction of KL intensifies oxidative stress and inflammatory response, resulting in genomic instability, p-EIF2A/eIF2α-mediated ER stress, retardation of autophagy, and induction of apoptosis [3323, 3324].
KLHL20 (kelch like family member 20): A substrate adaptor of CUL3 (cullin 3) ubiquitin ligase that targets autophosphorylated ULK1 and phagophore-localized PIK3C3/VPS34 and BECN1 for degradation to contribute to autophagy termination [648].
Klp98A (Kinesin-like protein at 98A): Drosophila homolog of KIF16B, which promotes microbutule plus end-directed transport of autophagic vesicles toward the cell periphery as well as autophagosome-lysosome fusion, through interactions with Atg8a and Rab14 [3325].
Knockdown: An experimental technique to reduce protein level without altering the endogenous gene encoding that protein, through the means of short DNA or RNA oligonucleotides (miRNA, RNAi, shRNA, siRNA) that are complementary to the corresponding mRNA transcript.
Knockout: Targeted inactivation of an endogenous genetic locus (or multiple loci) via homologous recombination or gene targeting technology.
KPT-185: A small molecule selective inhibitor of nuclear export, that blocks XPO1-mediated nuclear export. KPT-185 is able to induce apoptosis in mantle cell lymphoma, and autophagy declines post-treatment [3326].
KRAS: KRAS is the critical genetic driver of pancreatic ductal adenocarcinoma (PDAC) initiation and progression, and is essential for maintenance of PDAC tumorigenic growth. Suppression of KRAS induces autophagic flux through activating phosphorylation of AMPK and upregulation of total BECN1 [3327].
Ku-0063794: A catalytic MTOR inhibitor that increases autophagic flux to a greater level than allosteric inhibitors such as rapamycin; short-term treatment with Ku-0063794 can inhibit both MTORC1 and MTORC2, but the effects on flux are due to the former [453]. See also WYE-354.
KU55933: An inhibitor of the class III PtdIns3K, which inhibits autophagosome formation at concentrations not affecting the class I PI3K [331]. Also inhibits ATM.
LACRT (lacritin): A prosecretory mitogen primarily in tears and saliva that transiently accelerates autophagic flux in stressed cells [3328]. Lacritin targets heparanase-deglycanated SDC1 (syndecan 1) on the cell surface [3329], and accelerates flux by stimulating the acetylation of FOXO3 as a novel ligand for ATG101 and by promoting the coupling of stress acetylated FOXO1 with ATG7 [3330].
Laforin: See EPM2A.
Lalistat (LSTAT): 4-(Piperidin-1-yl)-1,2,5-thiadiazol-3-yl morpholine-4-carboxylate is a potent and selective LIP (lipase, lysosomal acid type) inhibitor. Lalistat exhibits no discernible activity against LPL (lipoprotein lipase) and PNLIP (pancreatic lipase) [3331].
LAMP1 (lysosomal associated membrane protein 1): LAMP1 shares common functions with LAMP2 [1829], is largely used as marker of late autophagic and endocytic organelles [3332], and is a target of TFEB. Activation of the TFEB-mediated autophagy-lysosome pathway greatly increases LAMP1 levels [3333].
LAMP2 (lysosomal associated membrane protein 2): A widely expressed and abundant single-span lysosomal membrane protein. Three spliced variants of the LAMP2 gene have been described. Knockout of the entire gene results in altered intracellular vesicular trafficking, defective lysosomal biogenesis, inefficient autophagosome clearance and alterations in intracellular cholesterol metabolism [1829, 1830, 3334]. In human, deficiency of LAMP2 causes a cardioskeletal autophagic vacuolar myopathy, called Danon disease [3335].
LAMP2A (lysosomal associated membrane protein 2A): One of the spliced variants of the LAMP2 gene that functions as a lysosomal membrane receptor for chaperone-mediated autophagy [1816]. LAMP2A forms multimeric complexes that allow translocation of substrates across the lysosome membrane [3243]. Regulation of LAMP2A is partly achieved by dynamic movement into and out of lipid microdomains in the lysosomal membrane [3239].
LAMP2C (lysosomal associated membrane protein 2C): One of the spliced variants of the LAMP2 gene that functions as a lysosomal membrane receptor for RNautophagy and DNautophagy [1395–1398]. LAMP2C also perturbs CMA and autophagy in mammalian cells [1832, 1833].
LAMP5 (lysosomal associated membrane protein family member 5): A LAMP family member that is specifically and highly expressed in patients with MLL leukemia. LAMP5 is a novel autophagic suppressor that interacts with ATG5. Knockdown of LAMP5 expression dramatically enhances the protein level of ATG5 but not the mRNA level [3336].
LANDO: See LC3-associated endocytosis.
LAP: See LC3-associated phagocytosis.
LAP-like non-canonical autophagy (LNCA): Non-canonical autophagy with properties similar to LAP, including single-membrane LC3 conjugation. Whereas LAP is specific to phagosomes [247], LNCA describes a similar phenomenon involving other types of organelles (e.g., macropinosomes [244]), entotic bodies [244] and endocytic vacuoles in exocrine secretory cells [760]. See also LC3-associated phagocytosis.
LAPosome (LAP-engaged phagosome or LC3-associated phagosome): A structure used for conjugation of LC3 to single-membrane phagosomes. LC3 on LAPosomes promotes phagosome maturation and processing of the engulfed cargo [801, 2446]. In mammalian cells and C. elegans, LAPosomes are observed during midbody remnant degradation and cell corpse clearance [244, 758, 2450, 2451].
Late nucleophagy: A process in which bulk nucleoplasm is delivered to the vacuole after prolonged periods of nitrogen starvation and subsequently degraded within the vacuole lumen [1081].
LC3: See MAP1LC3.
LC3/GABARAP-ylation: The modification of protein substrates by covalent conjugation to LC3/GABARAP, first used to describe the autocatalytic conjugation of LC3/GABARAP to ATG3 [262]. This process is counteracted by ATG4 proteases. See also deconjugation and 12-ylation.
LC3-associated endocytosis (LANDO): An LC3-dependent process that requires ATG5 and RUBCN, but is independent of RB1CC1 [2458].
LC3-associated phagocytosis (LAP): Phagocytosis that involves the conjugation of LC3 to single-membrane phagosomes, a process that promotes phagosome maturation [247, 801, 2446]. Signaling by pattern recognition receptors such as TLRs [247, 3337] CLEC7A/Dectin-1 [3338–3340], CLEC6A/Dectin-2 [3341] and ITGAM/Mac-1/CR3/integrin αmβ2 [904], IgG receptors such as FCGR/FcγR [3342] and receptors recognizing dead cells such as TIMD4/Tim4 [245] is required for LAP and leads to the recruitment of the BECN1 complex to phagosomes. See also NADPH oxidase.
LCN2/NGAL (lipocalin 2): First isolated from the supernatant of human neutrophils, LCN2 has a major role in the innate immune response, and in human cancer. In oral cancer, LCN2 expression is downregulated and the silencing of LCN2 suppresses autophagy via the MTOR pathway [3343].
Ldb16: See Ayr1.
Ldh1: See Ayr1.
LDS (LIR/AIM-docking site): Two conserved hydrophobic pockets on the surface of Atg8-family proteins into which bind the bulky hydrophobic residues found in WXXL-type AIM/LIR sequences [658, 1359]. See also AIM, LIR, and WXXL motif.
LGALS3 (galectin 3): LGALS3 is a galactose-specific lectin that recognizes and relocates to damaged endo-lysosomal vesicles. Therefore, LGALS3 is used as a reporter for endosomal membrane rupture in a so called “galectin puncta assay” [3344]. Together with TRIM16, LGALS3 recruits the core autophagy initiating factors ATG16L1 and ULK1 to clear damaged endosomes [3345].
LGALS8 (galectin 8): A carbohydrate binding protein that recognizes damaged membranes resulting from Salmonella infection and subsequently recruits the cargo receptor CALCOCO2 [1468]. LGALS8 consists of 2 separate carbohydrate-recognition domains (CRDs; N-CRD and C-CRD); the N-CRD binds to the galactose exposed from the damaged membrane, whereas the C-CRD binds specifically to the CALCOCO2 autophagy receptor [3346].
LGG-1: A C. elegans homolog of Atg8.
LGG-2: A C. elegans homolog of Atg8.
LGG-3: A C. elegans homolog of Atg12.
Licochalcone: Licochalcone A (LicA) is present in the root of Glycyrrhiza inflata Batalin (licorice). LicA-induced early autophagy is reliant on the induction of autophagosomes, the conversion of LC3-I to LC3-II, the induction of ATG5, ATG7, ATG12 and BECN1, and the inhibition of BCL2 [3347].
Lipid peroxidation: Lipid peroxidation is one of the major consequences of oxidative stress, characterized by raised levels of its subproducts, such as thiobarbituric acid-reactive substances (TBARS) [3348]. 4-hydroxynonenal (4-HNE), is a TBARS that induces autophagy [3349].
Lipid rafts: Supramolecular structures of the cell membranes that contain combinations of glycosphingolipids, sphingomyelin, cholesterol and proteins organized in glycolipoprotein microdomains. Lipid rafts are involved in the regulation of signal transduction and in autophagosome formation [908] and, through activation of the NADPH oxidase CYBB/Nox2, also LAP [904, 3350].
Lipid transfer proteins: Formation and maturation of autophagosomes requires mobilization of various lipids to produce and remodel these double-membrane organelles. Atg2/ATG2 is a lipid transfer protein [2759–2761, 3351] (for a more detailed description, see Atg2). Other lipid transfer proteins such as CERT1 (ceramide transporter 1) [3352], PITP (phosphatidyl inositol transfer protein) [3353], and CPTP (ceramide-1-phosphate transfer protein) [2958, 2959] have also been implicated as regulators of autophagy. See also Atg2.
Liponecrosis: A lipotoxicity-related mode of regulated cell death in which different autophagy-related processes within budding yeast have opposing roles; the non-selective autophagy pathway for massive degradation of various cellular organelles and macromolecules executes liponecrosis [1755], whereas the cargo-specific mitophagy pathway for elimination of damaged and dysfunctional mitochondria protects yeast cells from liponecrotic death [1755, 1756]. Note that liponecrosis is an aging-associated cell death mode whose age-related onset in yeast can be postponed by certain aging-decelerating dietary and pharmacological interventions [3354, 3355].
Lipophagy: Selective degradation of lipid droplets by lysosomes contributing to lipolysis (breakdown of triglycerides into free fatty acids). In mammals, this selective degradation has been described to occur via autophagy (macrolipophagy) [1288], whereas in yeast, microlipophagy of cellular lipid stores has also been described. This process is distinct from the PNPLA5-dependent mobilization of lipid droplets as contributors of lipid precursors to phagophore membranes. An association of SQSTM1 with lipid turnover reveals a novel pathway for the breakdown of lipid droplets. Accordingly, lipophagy promotes the clearance of lipids from myocytes and switches to an alternative, SQSTM1-mediated, lysosomal-independent pathway in the context of chronic lipid overload [3356].
Liposomes: These are artificial vesicles that are categorized by their size into three groups: small unilamellar vesicles (SUVs, 20-100 nm), large unilamellar vesicles (LUVs, 100-1000 nm), and giant unilamellar vesicles (GUVs, 1-200 μm).
Lipoxygenases: A family of iron-containing enzymes that catalyze the dioxygenation of polyunsaturated fatty acids in different carbon atoms. Two specific lipoxygenases, ALOX5 and ALOX15, inhibit IFNG-induced autophagy in macrophages [783].
LIR/LRS (LC3-interacting region): This term refers to the WXXL-like sequences (consensus sequence [W/F/Y]-X-X-[I/L/V]) found in proteins that bind to the Atg8/LC3/GABARAP-family proteins (see also AIM and WXXL-motif) [496]. The core LIR residues interact with two hydrophobic pockets of the ubiquitin-like domain of the Atg8 homologs, which are known collectively as the LIR/AIM docking site (LDS). The functional LIR has been classified as an IDPR-based short linear motif [3357] because it is always located in an intrinsically disordered region of an autophagy-related protein. See also LDS.
LITAF (lipopolysaccharide induced TNF factor): An activator of inflammatory cytokine secretion in monocytes that has other functions in different cell types; LITAF is a positive regulator of autophagy in B cells [3358]. LITAF associates with autophagosomes, and controls the expression of LC3B.
Lithium: This drug is primarily used as a psychiatric medication. Lithium inhibits IMPA and promotes autophagy by increasing the levels of BECN1-containing PIK3C3/VPS34 complexes [1976].
LKB1: See STK11.
LMNB1 (lamin B1): A protein of the nuclear lamina that directly binds LC3 molecules and directs the process of nucleophagy in mammalian cells [1335].
LMP (lysosome membrane permeabilization): The process by which lysosomal membranes become disrupted through the action of lysosomotropic agents, detergents or toxins [3359]. LMP blocks lysosomal activity and thus autophagy, and induces the release of lysosomal content to the cytoplasm including cathepsins that can induce cell death [1611, 3360].
LNCA: See LAP-like non-canonical autophagy.
LON2 (LON protease 2): A protease localized to the peroxisome matrix that impedes pexophagy in Arabidopsis [2343].
Long-lived protein degradation (LLPD): Autophagy is a primary mechanism used by cells to degrade long-lived proteins, and a corresponding assay can be used to monitor autophagic flux [4, 25]; a useful abbreviation is LLPD [688].
LONP1 (lon peptidase 1, mitochondrial): Mitochondrial matrix protease that reduces PINK1 accumulation. A reduction of LONP1 induces mitophagy by increasing PINK1 accumulation. LONP1 protein is reduced in fibroblasts from PD patients (with or without LRRK2 mutations), inducing mitophagy [1006].
LPS (lipopolysaccharide): An endotoxin derived from the outer membrane of Gram-negative bacteria [3361]. LPS treatment leads to a dramatic increase in autophagosome formation and autophagic flux in different cell types [3362, 3363].
LpSpl: Legionella pneumophila sphingosine-1-phosphate lyase, has SGPL1 activity and prevents an increase in sphingosine levels in infected host cells, thereby inhibiting autophagy during macrophage infection.
Lro1: See Ayr1.
Lucanthone: An anti-schistosome compound that inhibits a late stage of autophagy; treatment results in deacidification of lysosomes and the accumulation of autophagosomes [3364].
LRBA (LPS responsive beige-like anchor protein): A human protein that facilitates fusion between autophagosomes and the late endosomes. LRBA-deficient B cells have a significantly reduced ability to induce autophagy in response to starvation [3365].
LRP1/TM219/IGFBP3R1 (LDL receptor related protein 1): A member of the low-density lipoprotein receptor family of proteins and the receptor for VacA exotoxin produced by H. pylori to induce autophagy; after binding of VacA to LRP1, the LRP1-ICD is translocated to the nucleus. Nuclear translocated LRP1-ICD enhances LAMP1 expression by binding to the proximal LAMP1 promoter region, leading to autolysosome formation [2927]. LRP1 contains a di-arginine motif and GFBP3 signaling module that activates processing of proCASP8, inducing CASP8-dependent autophagy; blocking LRP1 signaling leads to inhibition of autophagy [3366]. See also CAPZA1.
LRPPRC (leucine rich pentatricopeptide repeat containing): A mitochondrion-associated protein that binds BCL2 and PRKN to control the initiation of general autophagy and mitophagy [3367, 3368].
LRRK2 (leucine rich repeat kinase 2): A large multidomain, membrane-associated kinase and GTPase whose PD-associated mutations affect the regulation of autophagy and mitophagy, potentially through derepression of ULK1 or mitochondrial calcium dysregulation [264, 3369, 3370]. LRRK2 is also recruited to stressed lysosomes (e.g., after CQ treatment), where it then recruits and phosphorylates its substrate, small RAB GTPases [3371], to maintain lysosomal homeostasis [3372].
LRS (LC3 recognition sequence): See LIR/LRS.
LRSAM1 (leucine rich repeat and sterile alpha motif containing 1): A human leucine-rich repeat protein that potentially interacts with GABARAPL2; knockdown of LRSAM1 results in a defect in anti-Salmonella autophagy [3373].
LS2 (Life Sciences Switzerland) Section Autophagy: See https://www.ls2.ch for more information.
Lst1: A component of the COPII-cargo adaptor complex Lst1-Sec23 that interacts with the reticulophagy receptor Atg40 to allow specific domains of the ER to be sequestered within phagophores [1382].
Ltn1: See Rkr1.
LY294002: An inhibitor of phosphoinositide 3-kinases and PtdIns3K that inhibits autophagy [3374].
LYNUS (lysosomal nutrient sensing): A complex including MTORC1 and the V-ATPase located on the lysosomal surface that senses nutrient conditions [1296]. The LYNUS complex regulates TFEB activity.
Lys05: A dimeric CQ derivative that accumulates in the lysosome and inhibits autophagy [3375, 3376].
Lysophagy: The autophagic removal of damaged lysosomes [1321, 3313, 3314].
Lysosome: A degradative organelle in more complex eukaryotes that compartmentalizes a range of hydrolytic enzymes and maintains a highly acidic pH. A primary lysosome is a relatively small compartment that has not yet participated in a degradation process, whereas secondary lysosomes are sites of present or past digestive activity. The secondary lysosomes include autolysosomes and telolysosomes. Autolysosomes/early secondary lysosomes are larger compartments actively engaged in digestion, whereas telolysosomes/late secondary lysosomes do not have significant digestive activity and contain residues of previous digestions. Both may contain material of either autophagic or heterophagic origin.
Lysosomotropism: The property of some compounds (such as weak bases) to diffuse across the lysosomal membrane and to accumulate in the acidic interior of lysosomes. Once inside the lysosome, these compounds become protonated, and, due to the charge, incapable of diffusing back into the cytosol. Such compounds are referred to as lysosomotropic [3377].
Macroautophagy: The forms of autophagy that involve phagophores and autophagosomes, distinguishing it from other major forms of autophagy that do not involve these compartments, such as microautophagy and CMA. Autophagy can be selective and nonselective. For selective autophagy, the autophagosomes can be termed according to their content (for instance “mitophagosome” for autophagosomes that selectively sequester mitochondria).
MAGEA3 (MAGE family member A3): MAGEA3 and MAGEA6 form a complex with the E3 ligase TRIM28, resulting in the degradation of AMPK and the subsequent increase in MTOR activity, which in turn causes a downregulation of autophagy [3378]. See also TRIM28.
MAP1L C3/LC3 (microtubule associated protein 1 light chain 3): A homolog of yeast Atg8, which is frequently used as a phagophore or autophagosome marker. Cytosolic LC3-I is conjugated to phosphatidylethanolamine to become phagophore-, autophagosome- or LAPosome-associated LC3-II [365]. The LC3 family includes LC3A, LC3B, LC3B2 and LC3C. These proteins are involved in the biogenesis of auto-phagosomes, and in cargo recruitment [199]. Vertebrate LC3 is regulated by phosphorylation of the N-terminal helical region by PRKA/PKA [293]. The LC3A gene has six exons, which encode two variants: LC3A-V1 formed by the inclusion of exons 3 to 6, which translate into 121 amino acids (NM_032514.4); LC3A-V2 is formed by the inclusion of exons 1, 2, and 4 to 6, which translates into 125 amino acids. LC3A-V1 is frequently inactivated in cancer by promoter methylation [3379, 3380].
MAP1S (microtubule associated protein 1S): A ubiquitously distributed homolog of the neuron-specific MAP1A and MAP1B with which LC3 was originally copurified. It is required for autophagosome trafficking along microtubular tracks [3381, 3382].
MAP3K7/MEKK7/TAK1 (mitogen-activated protein kinase kinase kinase 7): A MAP3K activated by a variety of extracellular stimuli such as LPS, IL1, TNF, TGFB, and TNFSF10/TRAIL. MAP3K7 plays a central role in TLR-activation of NFKB and AP-1. MAP3K7 is required for TNFSF10-induced activation of AMPK and also activates MAPK/JNK, both of which are required for microbial pathogen-induced autophagy [3383–3385]. MAP3K7 is required for optimal autophagy induction by multiple stimuli [3386], and it phosphorylates SQSTM1 at multiple sites [3387, 3388]. See also RPS6KB1.
MAP4K3/GLK3 (mitogen-activated protein kinase kinase kinase kinase 3): An amino-acid dependent activator of MTORC1. MAP4K3 physically interacts with TFEB, and, under conditions of amino acid satiety, MAP4K3 phosphorylates TFEB at serine 3 to suppress autophagy; MAP4K3 inhibition of TFEB is upstream of MTORC1 inhibition, as MAP4K3 phosphorylation of TFEB at serine 3 is required for MTORC1’s inhibitory phosphorylation of TFEB at serine 211 [3389]. Hence, MAP4K3 inhibition is sufficient to induce productive autophagy.
MAPK1 (mitogen-activated protein kinase 1): A kinase that along with MAPK3 phosphorylates and stimulates RGS19/Gα-interacting protein/GAIP, which is a GTPase activating protein (GAP) for the trimeric GNAI3 protein that activates autophagy [3390], and which may be involved in BECN1-independent autophagy [118]. Constitutively active MAPK1/3 also traffics to mitochondria to activate mitophagy [1138].
MAPK3: See MAPK1.
MAPK8/JNK1: A stress-activated kinase that phosphorylates BCL2 at Thr69, Ser70 and Ser87, causing its dissociation from BECN1, thus inducing autophagy [853].
MAPK8IP1/JIP1 (mitogen-activated protein kinase 8 interacting protein 1): A LIR-containing LC3-binding protein that mediates the retrograde movement of RAB7-positive autophagosomes in axons [3391]. Movement toward the proximal axon involves activation of dynein, whereas binding of LC3 to MAPK8IP1 prevents activation of kinesin. The DUSP1/MKP1 phosphatase may dephosphorylate Ser421, promoting binding to dynein.
MAPK9/JNK2: A stress-activated kinase that prevents the accumulation of acidic compartments in cells undergoing autophagic flux, thus keeping stressed cells alive [3392].
MAPK14 (mitogen-activated protein kinase 14): A signaling component that negatively regulates the interaction of ATG9 and SUPT20H/FAM48A, and thus inhibits autophagy. In addition, MAPK14-mediated phosphorylation of ATG5 at T75 negatively regulates autophagosome formation [3393]. The widely used pyridinyl imidazole class inhibitors of MAPK14 including SB202190 interfere with autophagy in a MAPK/p38-independent manner and should not be used to monitor the role of this signaling pathway in autophagy [3394, 3395]. The yeast homolog is Hog1. See also Hog1.
MAPK15/ERK7/ERK8 (mitogen-activated protein kinase 15): MAPK15 is a LIR-containing protein that interacts with LC3B, GABARAP and GABARAPL1 [272]. This kinase is localized in the cytoplasm and can be recruited to autophagic membranes through its binding to Atg8-family proteins. MAPK15 responds to starvation stimuli by self-activating through phosphorylation on its T-E-Y motif, and its activation contributes to the regulation of autophagy. MAPK15 also interacts with components of the ULK complex and controls ULK1/2 activity to regulate early phases of the autophagic process [3396].
MAPKAPK2 (MAPK activated protein kinase 2): MAPKAPK2 is a Ser/Thr protein kinase downstream of MAPK/p38. Its activation contributes to starvation-induced autophagy by phosphorylating BECN1/Beclin 1 [2658]. See also BECN1.
MAPKAPK3 (MAPK activated protein kinase 3): MAPKAPK3 shares a similar function with MAPKAPK2 in autophagy [2658]. See also MAPKAPK2 and BECN1.
Matrine: A natural compound extract from traditional Chinese medicine that inhibits autophagy by elevating lysosomal pH and interfering with the maturation of lysosomal proteases [3397].
MB21D1: See CGAS.
MCL1 (MCL1 apoptosis regulator, BCL2 family member): A member of the BCL2 family of proteins, which reciprocally controls BECN1 proteasomal degradation and thus protein levels through competitive binding to the USP9X deubiquitinating enzyme [3398, 3399]. GSK3B-mediated phosphorylation of MCL1 induces axonal autophagy [3131]. See also FBXW7, GSK3B and USP9X.
MCOLN1/TRPML1 (mucolipin 1): A lysosomal channel belonging to the transient receptor potential gene family that is permeable to several cations including calcium [3400, 3401]. MCOLN1-mediated calcium release from lysosomes can modulate autophagy in a multistep manner. Acute MCOLN1 activation induces autophagosome biogenesis by increasing the generation of PtdIns3P through the activation of PIK3C3/VPS34, and the recruitment of essential PtdIns3P-binding proteins to the nascent phagophore in a TFEB-independent manner, whereas prolonged channel activation induces TFEB nuclear translocation, triggered by its PPP3/calcineurin-dependent dephosphorylation [2000, 3402]. In addition, MCOLN1 mediates calcium-CALM (calmodulin)-dependent MTORC1 reactivation in conditions of prolonged starvation [1999, 3403]. MCOLN1 also localizes to late endosomes [3404].
MDC (monodansylcadaverine): A lysosomotropic autofluorescent compound that accumulates in acidic compartments of live cells such as autolysosomes, and also labels (but is not specific for) autophagosomes [2, 1866]. MDC also accumulates in hydrophobic environments, which stimulates fluorescent light emission (solvent polarity probe) and labels late autophagosomes in formaldehyde-fixed cells [1603].
MDH (monodansylpentane): A structural homolog of MDC that has lost its lysosomotropic properties but accumulates in hydrophobic environments and labels late autophagosomes in live and formaldehyde-fixed cells [1603].
MDK-ALK axis: MDK (midkine) is a growth factor for which increased levels are associated with a poor prognosis in malignant tumors. MDK promotes resistance to cannabinoid-evoked autophagy-mediated cell death via stimulation of ALK (ALK receptor tyrosine kinase). Targeting of the MDK-ALK axis could help to improve the efficacy of antitumoral therapies based on the stimulation of autophagy-mediated cancer cell death [3405, 3406].
Mdm10: A component of the ERMES complex in yeast that is required for mitophagy. See also ERMES [3107].
Mdm12: A component of the ERMES complex in yeast. Mdm12 colocalizes with Atg32-Atg11 and is required for mitophagy. See also Atg11, Atg32, and ERMES [2911, 3107].
Mdm34: A component of the ERMES complex in yeast. Mdm34 colocalizes with Atg32-Atg11 and is required for mitophagy. The ubiquitination of Mdm34 by the E3 ligase Rsp5 is also required for efficient mitophagy. See also Atg11, Atg32, Rsp5 and ERMES [2911, 3107, 3108].
Mdv1: A component of the mitochondrial fission complex that plays a role in mediating mitophagy-specific fission [2911]. See also Dnm1.
MEFV/TRIM20/pyrin/Mediterannean fever (MEFV innate immunity regulator, pyrin): The gene encoding MEFV is a site of polymorphisms associated with familial Mediterranean fever; MEFV/TRIM20 acts as a receptor for selective autophagy of several inflammasome components [3407].
Mega-autophagy: The final lytic process during developmental programmed cell death in plants that involves tonoplast permeabilization and rupture, resulting in the release of hydrolases from the vacuole, followed by rapid disintegration of the protoplast at the time of cell death [2349, 3408, 3409]. This term has also been used to refer to the rupture of the yeast vacuole during sporulation, which results in the destruction of cellular material, including nuclei that are not used to form spores [3410].
Megaphagosomes: Very large (5-10 μm) double-membraned, autophagy-related vesicles that accumulate in cells infected by coxsackievirus and, possibly, influenza virus [260].
Melatonin (N-acetyl-5-methoxy tryptamine): A sleep-wake cycle regulating and antioxidant hormone that inhibits autophagy in animal models of fibrosis [2612], cancer [2613] and acute organ failure [2614].
Membrane curvature: Membrane curvature is the geometrical measure or characterization of the physical bending of membranes to accommodate various cell morphology changes as well as the formation of membrane-bound transport intermediates such as spherical vesicles or tubules. An increase in membrane curvature facilitates insertion of ATG3 into membranes [2106]. Bilayer curvature modulates Atg8-family protein-mediated model phagophore elongation [552].
Membrane fission/scission and membrane fusion: Topological transformations of the membrane, which involve nonbilayer states of the membrane molecules [9, 10]. Membrane scission (also called membrane fission) refers to the process where one membrane separates into two distinct membranes and occurs during closure of the phagophore. Membrane fusion refers to the reverse change where two separate bilayers merge and form a single continuous bilayer, as seen for example with the fusion of the autophagosome outer membrane with a vacuole or lysosome.
Mcr (Merlin): A Drosophila complement-related protein that is required for autophagy in neighboring cells during salivary gland degradation and wound healing [3411].
Metformin: An anti-diabetic drug that induces autophagy through AMPK pathway activation [3412].
MFN2 (mitofusin 2): An outer mitochondrial membrane GTPase with dual, mutually exclusive roles in mitochondrial fusion and mitophagy; upon phosphorylation by PINK1 kinase in damaged mitochondria, MFN2 is functionally transformed from being fusogenic to a mitochondrial binding protein for PRKN, thus promoting sequestering and targeting the damaged organelle for degradation [3413, 3414]. MFN2 is also localized at the ER membrane and modulates ER-mitochondria tethering; the absence of MFN2 leads to impaired autophagy [3415, 3416].
MFSD8/CLN7 (major facilitator superfamily domain containing 8): A polytopic endosomal/lysosomal membrane glycoprotein of unknown function. Mutations in the MFSD8/CLN7 gene lead to the CLN7 disease (MIM 610951) with late infantile phenotype, which belongs to the neuronal ceroid lipofuscinoses (NCLs) [3417]. In an mfsd8/cln7 knockout mouse model, accumulation of autofluorescent lipopigments, lysosomal dysfunction and impaired autophagy are observed [3418].
MG132: Reversible inhibitor of the chymotrypsin- and caspase-like activities of the proteasome that induces the formation of protein aggregates and activates the autophagic flux in an NFKB-dependent manner [1107].
MGEA5: See OGA.
Microautophagy: An autophagic process involving direct uptake of cytosol, inclusions (e.g., glycogen) and organelles (e.g., ribosomes, peroxisomes) at the lysosome/vacuole by protrusion, invagination or septation of the sequestering organelle membrane.
MIPA (micropexophagic apparatus): A curved double-membrane structure formed by the PAS that may serve as a scaffold for completion of the sequestration of peroxisomes during micropexophagy; fusion with the vacuolar sequestering membranes encloses the organelles within an intralumenal vesicle [3419]. See also vacuolar sequestering membranes.
mir-14 (mir-14 stem loop): A microRNA that is necessary and sufficient for autophagy in Drosophila salivary glands [3303].
MIR9 (microRNA 9): A human miRNA that influences autophagy by inhibiting SIRT1 expression in chondrocytes [3420].
MIR29 (microRNA 29): A miRNA that knocks down LAMTOR1/p18 and leads to limited MTORC1 recruitment to lysosomes when overexpressed in retinal pigment epithelial cells. Consequently, MIR29 enhances autophagy, which aids in removal of protein aggregates in the RPE cells [3421].
MIR21 (microRNA 21): A miRNA that is overexpressed in almost all types of solid tumors and is involved in cancer chemoresistance. MIR21 modulates autophagy and the sensitivity of tumor cells towards drugs that induce autophagy [3422].
Mir31 (microRNA 31): A mouse miRNA that targets PPP2/PP2A to inhibit IFNG-induced autophagy in macrophages during mycobacterial infection [783]. See also Mir155.
MIR95: A human miRNA that inhibits autophagy and blocks lysosome function via repression of SUMF1 [332].
MIR101: A human miRNA that inhibits autophagy and the expression of STMN1, RAB5A and ATG4D [334].
Mir155: A mouse miRNA that targets PPP2/PP2A to inhibit IFNG-induced autophagy in macrophages during mycobacterial infection [783]. Human MIR155 counteracts the autophagic flux in chondrocytes by downregulating various autophagy factors (ULK1, FOXO3, ATG14, ATG5, ATG3, GABARAPL1, and LC3) [3423]. See also Mir31.
MIR205: A microRNA precursor that impairs the autophagic flux in castration-resistant prostate cancer cells by downregulating the lysosome-associated proteins RAB27A and LAMP3 [3424].
Mir224 (microRNA 224): Mir224 is an oncogenic microRNA in hepatocellular cellular carcinoma (HCC) through targeting Smad4. Mir224 is selectively recruited and degraded by the autophagy degradative machinery; however, the underlying mechanism remains unclear [3425, 3426].
MIR4465: A microRNA that, in HEK293, HeLa, and SH-SY5Y cells, inhibits the expression of PTEN, upregulates phosphorylated AKT and inhibits autophagy by activating MTOR [3427].
MIRLET7 (microRNA let-7): A microRNA capable of modulating autophagy [3428–3432]. Mirlet7 microRNA activates autophagy in primary neurons in culture, in adult-born olfactory bulb neurons and mammalian brain and peripheral tissue by coordinately downregulating the amino acid sensing pathway to prevent MTORC1 activation [3428, 3429]. However, in vascular smooth muscle cells pretreated with the HsMIRLET7G isoform, an inhibition of autophagy has been reported [3432]. Mirlet7/let-7 is also able to modulate autophagy in neurodegenerative disorders associated with an autophagic impairment [3430, 3431]. In Alzheimer adrenal pheochromocytoma PC12 and human neuroblastoma SK-N-SH cell models, MIRLET7A isoform overexpression further upregulates autophagy induced by treatment with β-amyloid protein 40 [3430]. Moreover, it has been reported that the Mirlet7f isoform is also able to activate autophagy in the brain of an in vivo mouse model of spinocerebellar ataxia-type 3/Machado-Joseph disease, whereas an anti-Mirlet7 decreases autophagy in a Huntington disease mouse model [3428]. Nevertheless, in a C. elegans model of Parkinson disease expressing human SNCA, it has been reported that a loss of the let-7 miRNA leads to an increase in autophagy [3431].
MITF (melanocyte inducing transcription factor): A transcription factor belonging to the microphthalmia/transcription factor E (MiT/TFE) family, along with TFEB and TFE3; MITF binds to symmetrical DNA sequences (E-boxes; 5-CACGTG-3), and regulates lysosomal biogenesis and autophagy (including the genes BCL2, UVRAG, ATG16L1, ATG9B, GABARAPL1, and WIPI1). MITF shares a common mechanism of regulation with TFEB and TFE3; MITF can partially compensate when TFEB is lost upon specific stimuli or in specific cell types [977, 3433]. See also TFEB.
mito-QC: A mitophagy reporter construct expressed in mouse and Drosophila models that constitutively and ubiquitously expresses a mitochondrial outer membrane-localized mCherry-GFP tag. This model allows visualization of mitochondrial quality control (QC) by mitophagy, in addition to mitochondrial network architecture in vivo [38, 466].
Mitochondria-associated ER membranes (MAMs): Functional domains that reversibly connect the ER to mitochondria with a significant role in the maintenance of calcium homeostasis and mitochondria biogenesis; these membranes also participate in autophagosome formation [908]. See also ER-mitochondria sites.
Mitochondrial spheroid: A mitochondrial structure formed in PRKN-deficient cells treated with a mitochondrial uncoupler (such as CCCP) [1135, 1136]. Under this condition, mitophagy fails to occur and a damaged mitochondrion can transform into a spheroid containing cytosolic components in the newly formed lumen. PRKN and MFNs can reciprocally regulate mitophagy and mitochondrial spheroid formation.
Mitolysosome: This term refers to the product of a mitophagosome fused with a lysosome [3434, 3435].
Mitophagic body: The single-membrane vesicle present inside the vacuole lumen following the fusion of a mitophagosome with a vacuole.
Mitophagosome: An autophagosome containing mitochondria and no more than a small amount of other cytoplasmic components, as observed during selective macromitophagy [55, 1113].
Mitophagy: The selective autophagic sequestration and degradation of mitochondria; can occur by a micro- or autophagic process [3436].
Mitostatin: See TCHP.
MitoTimer: A mitochondria-targeted time-sensitive fluorescent protein for which emission shifts from green to red. Constructs that encode an inducible form of the protein can be used to study mitochondrial turnover [1177, 1178, 1180], and constructs that encode a constitutively active form of the protein can be used to study mitochondrial structure, oxidative stress and mitophagy [1179, 1181–1187].
MitoTracker® Dyes: Commercially available fluorescent probes that can stain mitochondria in both fixed or living samples [3437]; these can be dependent or independent of the mitochondrial membrane potential.
Mkk1/2: A MAPKK downstream of Bck1 that is required for mitophagy and pexophagy in yeast [747]. See also Bck1 and Slt2.
MLN4924: An inhibitor of NAE1 (NEDD8 activating enzyme E1 subunit 1) that is required for CUL (cullin)-RING E3 ligase activation; treatment with MLN4924 induces autophagy through the accumulation of the MTOR inhibitory protein DEPTOR [2620].
Mmm1: A component of the ERMES complex in yeast that is required for mitophagy. See also ERMES [3107].
MoAtg24: MoAtg24 is directly involved in mitophagy in a way similar to the function of yeast Atg32 [1069]. During the pathogenesis process, carbon starvation induces the breakdown of the mitochondrial network and leads to more punctate mitochondria in M. oryzae. This nutrient-based regulation of organellar dynamics precedes MoAtg24-mediated mitophagy, which is essential for proper biotrophic development and invasive growth of M. oryzae in planta [3438].
MoMkk1/2 (Magnaporthe oryzae Mkk1/2): Homologs of S. cerevisiae Mkk1/2 (MAP kinase kinases) that are phosphorylated by MoAtg1 to activate the cell wall integrity pathway under conditions of ER stress, which is essential for virulence during infection [2195]. MoMkk1 Ser115 is an identified phosphorylation site.
Mon1-Ccz1: A dimeric guanine-nucleotide exchange factor for the GTPase Ypt7 in yeast [3439] and RAB7 in metazoan cells [3440, 3441]; the complex has a third subunit in human cells [35]. Mon1-Ccz1 is recruited by Atg8 and PtdIns3P to the autophagosomal surface, and activates Ypt7/RAB7 for fusion with the vacuole/lysosome [3442].
Monensin: A polyether antibiotic, which interferes with autophagosome-lysosome fusion and consequently inhibits autophagy [3443].
MoRF (molecular recognition feature): A short (10-70 residues) IDPR in proteins, including Atg20, that undergo a disorder-to-order transition upon binding to its partners [2801, 3281, 3444–3447].
MORN2 (MORN repeat containing 2): MORN2 is a membrane occupation and recognition nexus (MORN)-motif protein that was identified in mouse testis. The gene localizes on chromosome 17E3, spanning approximately 7 kb; Morn2 contains 669 nucleotides of open reading frame, and encodes 79 amino acids [3448]. MORN domains have the sequence GKYQGQWQ. MORN2 promotes the recruitment of LC3 in LAP, and MORN2 co-immunoprecipitates with LC3 [2452].
MRAP (MCU regulating acidic patch): An electronegatively charged surface region on the matrix-residing amino terminal domain of MCU (mitochondrial calcium uniporter) [3449]. Binding of divalent cations such as Mg2+ and Ca2+ to MRAP disrupts MCU assembly and function [3449]. Inhibition of MCU perturbs cellular bioenergetics and promotes autophagy [3450].
MREG (melanoregulin): A cargo sorting protein that associates with MAP1LC3 in LC3- associated phagocytosis [2383, 3451].
MRT68921: A small molecule inhibitor of ULK1 and ULK2 kinases that blocks autophagy in cells [3452].
MSP: Multiple system proteinopathy (an inherited pleiotropic degenerative disorder of skeletal muscle, bone, and the central nervous system) that is thought to be due to abnormal granulophagy (autophagic degradation of RNP granules). Five different MSP subtypes are known thus far, three due to mutations in RNA-binding proteins and two (MSP1 and MSP4) due to mutations in autophagic proteins (VCP/p97 and SQSTM1, respectively) [3190]. See also granulophagy.
mt-Keima: A mitochondrially-targeted version of the coral Keima protein that has a pH-sensitive fluorescence spectrum, useful for monitoring mitophagy events [1036].
MTDH/AEG-1 (metadherin): An oncogenic protein that induces noncanonical (BECN1- and class III PtdIns3K-independent) autophagy as a cytoprotective mechanism [3453].
MTM-3: A C. elegans myotubularin lipid phosphatase that is an ortholog of human MTMR3 and MTMR4; MTM-3 acts upstream of EPG-5 to catalyze the turnover of PtdIns3P and promote autophagosome maturation [3454].
MTM1 (myotubularin 1): A PtdIns3P and PtdIns(3,5)P2 3-phosphatase [3455]. Mutations affecting MTM1 lead to myotubular myopathy and alteration of autophagy.
MTMR3 (myotubularin related protein 3): This protein localizes to the phagophore and negatively regulates autophagy. See also MTMR14 [3456].
MTMR6 (myotubularin related protein 6): A PtdIns 3-phosphatase; knockdown of MTMR6 increases the level of LC3-II [3457].
MTMR7 (myotubularin related protein 7): A PtdIns 3-phosphatase; knockdown of MTMR7 increases the level of LC3-II [3457].
MTMR8 (myotubularin related protein 8): A phosphoinositide phosphatase with activity toward PtdIns3P and PtdIns(3,5)P2; MTMR8 in a complex with MTMR9 inhibits autophagy based on the formation of WIPI1 puncta [3458].
MTMR9 (myotubularin related protein 9): A catalytically inactive myotubularin that increases the activity of other members of the MTMR family and controls their substrate specificity; MTMR8-MTMR9 preferentially dephosphorylates PtdIns3P and thus inhibits autophagy [3458].
MTMR13: See SBF2.
MTMR14/Jumpy (myotubularin related protein 14): A member of the myotubularin family that is a PtdIns 3-phosphatase; knockdown increases autophagic activity [3457, 3459]. MTMR14 regulates the interaction of WIPI1 with the phagophore. The Drosophila homolog is EDTP.
MTOR (mechanistic target of rapamycin kinase): The mammalian ortholog of TOR. Together with its binding partners it forms either MTOR complex 1 (MTORC1) or MTOR complex 2 (MTORC2). See also TORC1 and TORC2.
MTORC1/2 (MTOR complex 1/2): See TORC1 and TORC2.
MUL1 (mitochondrial E3 ubiquitin protein ligase 1): A mitochondria-targeted E3 ligase that mediates the ubiquitination and the subsequent degradation of MFN2, thus acting as a negative regulator of PRKN-mediated neuronal mitophagy [3460, 3461]. Chronic mitochondrial stress is associated with major neurodegenerative diseases; thus, the recovery of those mitochondria constitutes a critical step of energy maintenance in early stages of neurodegeneration. The MUL1-MFN2 pathway acts as an early checkpoint to maintain mitochondrial integrity by regulating mitochondrial morphology and interplay with the endoplasmic reticulum (ER). This mechanism ensures that degradation of stressed mitochondria through mitophagy is restrained in neurons under early stress conditions. Failure of the MUL1-MFN2 pathway activates mitophagy to eliminate damaged mitochondria.
Multivesicular body (MVB)/multivesicular endosome: An endosome containing multiple 50- to 80-nm vesicles that are derived from invagination of the limiting membrane. Under some conditions the MVB contains hydrolytic enzymes in which case it may be considered to be a lysosome or autolysosome with ongoing microautophagy.
Multivesicular body sorting pathway: A process in which proteins are sequestered into vesicles within the endosome through the invagination of the limiting membrane. This process is usually, but not always, dependent upon ubiquitin tags on the cargo and serves as one means of delivering integral membrane proteins destined for degradation into the vacuole/lysosome lumen. ESCRT complexes are required for the formation of MVBs, for autophagosome maturation [3462], and they cooperate with autophagy to repair membrane damage to their compartment inflicted by pathogenic mycobacteria [1322].
Mycolactone: The exotoxin virulence factor of Mycobacterium ulcerans [3463] that acts as an inhibitor of SEC61-dependent translocation of polypeptides into the endoplasmic reticulum [2022]. Mycolactone induces autophagy by a mechanism that involves the integrated stress response and is dependent on its ability to block protein translocation [2020, 2021].
MYO1C (myosin IC): A class I myosin that functions as an actin motor protein essential for the trafficking of cholesterol-rich lipid rafts from intracellular storage compartments to the plasma membrane; MYO1C is important for efficient autophagosome-lysosome fusion [3464].
MYO6 (myosin VI): A unique, minus-end directed actin motor protein required for autophagosome maturation and fusion with a lysosome via delivery of early endosomes to autophagosomes; mediated by the interaction of MYO6 with the alternative ESCRT-0 protein TOM1 [1471, 3465].
N1,N3-bis(2-[2,4,6-triaminopyrimidin-5-yl]ethyl)isophthalimidamide (AQAMAN): A bisamidine-based small molecule that can prevent expanded polyglutamine (polyQ) protein aggregation and dissociate preformed polyQ aggregates. The cytoprotective effect of AQAMAN on polyQ toxicity depends on autophagy activation [3466].
NAA10/ARD1 (N-alpha-acetyltransferase 10, NatA catalytic subunit): A protein that interacts with and stabilizes TSC2 by acetylation, resulting in repression of MTOR and induction of autophagy [3467].
NACC1/NAC1 (nucleus accumbens associated 1): A transcription factor that increases the expression and cytosolic levels of HMGB1 in response to stress, thereby increasing autophagy activity [3468].
NAD+: A small natural molecule necessary for cellular energy homeostasis that is involved in broad cellular pathways, including aging and neurodegeneration. NAD+ regulates both nonselective autophagy and specific autophagy, such as mitophagy, through the NAD+-dependent sirtuins, SARM1, CD38 and other proteins [3469, 3470].
NADPH oxidases: These enzymes contribute to autophagic targeting of Salmonella in leukocytes and epithelial cells through the generation of reactive oxygen species [3342]. The CYBB/NOX2 NADPH oxidase in macrophages is required for LC3-associated phagocytosis.
NAF-1: See CISD2.
NAMPT/visfatin (nicotinamide phosphoribosyltransferase): NAMPT is a protein that catalyzes the condensation of nicotinamide with 5-phosphoribosyl-1-pyrophosphate to yield nicotinamide mononucleotide, one step in the biosynthesis of nicotinamide adenine dinucleotide. The protein belongs to the nicotinic acid phosphoribosyltransferase (NAPRTase) family and is thought to be involved in many important biological processes, including metabolism, stress response and aging. NAMPT promotes neuronal survival through inducing autophagy via regulating the TSC2-MTOR-RPS6KB1 signaling pathway in a SIRT1-dependent manner during cerebral ischemia [3471].
Nanoparticles (NPs): Small particles in the size range of 1–100 nm that can modulate autophagy [1919, 3472, 3473] and have been used for autophagy monitoring [3474, 3475]. Furthermore, nanoparticles are able to efficiently deliver autophagy inducers/inhibitors into cells [3476, 3477].
NAPA/αSNAP (NSF attachment protein alpha): A key regulator of SNARE-mediated vesicle fusion. Loss of NAPA promotes noncanonical autophagy in human epithelial cell by interrupting ER-Golgi vesicle trafficking and triggering Golgi fragmentation [3478].
NBR1 (NBR1 autophagy cargo receptor): A selective substrate of autophagy with structural similarity to SQSTM1. NRB1 functions as a receptor that binds ubiquitinated proteins and LC3 to allow the degradation of the former by an autophagy-like process [496]. NBR1 shows specificity for substrates including peroxisomes [459] and ubiquitinated aggregates [496]. Phosphorylation of NBR1 by GSK3A/B prevents the aggregation of ubiquitinated proteins [2664]. Arabidopsis AT4G24690/NBR1 is involved in regulating abiotic and biotic stresses [1354, 3479].
NCKAP1/NAP1 (NCK associated protein 1): NCKAP1 is a component of the SCAR/WAVE complex, which activates the ARP2/3 complex and actin branching. In plants, AT2G35110/NAP1 regulates mechanical pressure-induced autophagosome formation. The T-DNA knockout mutant of NAP1 is more susceptible to low nutrient and high salt stresses [3480, 3481].
NCOA4 (nuclear receptor coactivator 4): A selective cargo receptor that is involved in iron homeostasis through the recycling of ferritin by autophagy [1264]. See also ferritinophagy.
NDP52: See CALCOCO2.
Necroptosis: A form of regulated necrotic cell death [3482]; induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance [3483]. Suppression of autophagic flux contributes to RIPK1-RIPK3 interaction and necroptosis of cardiomyocytes [3484].
NEDD4 (NEDD4 E3 ubiquitin protein ligase): An E3 ubiquitin protein ligase that interacts with Atg8-family proteins and forms a complex with ULK1 and BECN1. NEDD4 increases the stability of BECN1 and promotes autophagy as well as various infections-induced xenophagy by mediating K6- and K27-linkage ubiquitination of BECN1 [3485].
NETosis: NETosis is a specialized form of neutrophil cell death that is characterized by the release of decondensed nuclear chromatin and neutrophil granule contents to the extracellular space; autophagy regulates NET formation. See also DDIT4.
NFAT5/TonEBP (nuclear factor of activated T cells 5): The transcription factor NFAT5 can be activated by hypertonic stress [3486] in addition to other stimuli [3487] such as inflammation [3488]. NFAT5 does not directly promote the expression of genes related to autophagy [3489], but facilitates autolysosomal formation and targeting of cargo to autolysosomes [3213].
NFATC2/NFAT1 (nuclear factor of activated T cells 2): The transcription factor NFATC2 can be activated by ROS and binds directly to the Lamp2a proximal promoter region, leading to an increase of LAMP2A expression and CMA activation, which is required for a proper T cell activation through degradation of negative regulators, ITCH and RCAN [3490].
NFE2L2/NRF2 (nuclear factor, erythroid 2 like 2): A stress responsive transcription factor that regulates a complex program of cytoprotective responses, including the expression of the autophagy-related genes Sqstm1, Calcoco2, Ulk1, Atg2b, Atg4d, Atg5, Atg7 and Gabarapl1 and the CMA-related gene Lamp2a [140, 419, 596, 597, 987, 988, 3320, 3491]. NFE2L2 is inactivated by KEAP1. See also KEAP1.
NFKB/NF-κB (nuclear factor kappa B): NFKB activates MTOR to inhibit autophagy upon TNF stimulation [3492]. In contrast, the RELA subunit of NFKB induces BECN1 gene expression in T cells [3493], and NFKB enhances BAG3 and HSPB8 expression to induce aggrephagy [1107, 3494].
NH4Cl (ammonium chloride): A weak base that is protonated in acidic compartments and neutralizes them; inhibits the clearance of autophagosomes and amphisomes.
NHLRC1/EPM2B/malin (NHL repeat containing E3 ubiquitin protein ligase 1): A putative E3-ubiquitin ligase, which forms a complex with EPM2A/laforin. Recessive mutations in the genes EPM2A, or NHLRC1/EMP2B are found in the majority of cases of Lafora disease, a very rare type of progressive neurodegeneration associated with impaired autophagy [3495].
NIBAN1/FAM129/C1orf24 (niban apoptosis regulator 1): NIBAN1 is highly expressed in a number of human cancers, in particular of the thyroid, and it may play a dual role in the regulation of autophagy in thyroid cells. While NIBAN1 can increase baseline autophagy in response to nutrient and growth factor depletion, it can inhibit autophagy in thyroid carcinoma cells in the presence of oncogenes, such as RET/PTC1 fusion. Therefore, NIBAN1 regulates autophagy in thyroid cells in an oncogene-dependent manner, possibly through the AKT-MTOR axis [3496].
NID-1 (novel inducer of cell death 1): A small molecule that induces activation of an ATG5- and CTSL-dependent cell death process reminiscent of autophagy [2453].
NIPSNAP1/2 (nipsnap homolog 1/2): Two small mitochondrial matrix proteins, NIPSNAP1 and NIPSNAP2, that act as “eat-me” signals for damaged mitochondria. NIPSNAP1 and NIPSNAP2 accumulate on the mitochondrial outer membrane following mitochondrial depolarization, recruiting autophagy receptors and adaptors, as well as human Atg8-family proteins to facilitate mitophagy [3497].
Nitric oxide: A gas and a messenger that has complex regulatory roles in autophagy, depending on its concentration and the cell type [455, 3498–3500].
NIX: See BNIP3L.
NLRX1 (NLR family member X1): The only mitochondrial member of the NLR family that is characterized by structural homology in a central nucleotide-binding oligomerization domain, a C-terminal leucine-rich repeat domain, and an N-terminal effector domain. NLRX1 promotes autophagy and mitophagy by serving as a scaffold protein to recruit ATG12–ATG5 and ATG16L1. NLRX1 also contains a LIR and directly associates with LC3 [3501–3504]. See also TUFM.
NME4/NDPKD/NM23-H4 (NME/NM23 nucleoside diphosphate kinase 4): NME4 is the group I member of the NDPK/NME family. It forms large homohexameric complexes localized in mitochondria. NME4 has two different topologies [3505, 3506]. In healthy mitochondria, NME4 is in the topology with nucleoside diphosphate kinase activity and is mainly bound to the mitochondrial inner membrane. When the mitochondria membrane potential collapses and mitochondria depolarize, NME4 becomes nucleoside diphosphate kinase-inactive but gains intermembrane phospholipid transfer activity. It is then localized in the mitochondrial intermembrane space where it binds simultaneously to both the mitochondrial inner and outer membrane. In this topology, NME4 mediates the redistribution of cardiolipin to the mitochondrial outer membrane, which functions as a signal for the elimination of damaged mitochondria via mitophagy [1189].
NOD (nucleotide-binding oligomerization domain): An intracellular peptidoglycan (or pattern recognition) receptor that senses bacteria and induces autophagy, involving ATG16L1 recruitment to the plasma membrane during bacterial cell invasion [1487]. NOD1 causes direct inflammasome NLRP3 scaffold protein interaction with LC3, inducing autophagy flux [1489].
Non-nitrogen-starvation (NNS)-induced autophagy: A type of autophagy that is induced when yeast cells are shifted from rich to minimal medium in the presence of a respiratory carbon source such as lactate; this process is controlled in part by the Iml1 complex/SEACIT (SEh1-Associated subComplex Inhibiting TORC1), which consists of Iml1, Npr2 and Npr3, and which functions as a GTPase activator complex for the Rag GTPase Gtr1 [3289, 3507, 3508].
Noncanonical autophagy: A functional autophagy pathway that only uses a subset of the characterized ATG proteins to generate an autophagosome. BECN1-independent [118, 2486], and ATG5-ATG7-independent [31] forms of autophagy have been reported.
Nordic Autophagy Society (NAS): The NAS (https://nordicautophagy.org/) acts to promote autophagy-related research in its nine membership countries (Norway, Sweden, Denmark, Finland, Iceland, Estonia, Latvia, Lithuania, The Netherlands). The society aims to increase the interaction of Nordic autophagy researchers with researchers from other countries, by, among others, providing lab exchange and conference grants, and by being open for all nationalities to become members (also from outside the nine membership countries). Moreover, Nordic Autophagy Conferences are held yearly (since 2012), and have featured prominent invited speakers from Europe, USA, and Japan. The NAS developed from the Nordic Autophagy Network.
NPC1 (NPC intracellular cholesterol transporter 1): Defects in autophagy in the lysosomal storage disease Niemann-Pick type C1 disease due to pathogenic NPC1 gene variants cause cholesterol accumulation in hepatocytes and neural cells and defective xenophagy in macrophages [3509–3511].
NPY (neuropeptide Y): An endogenous neuropeptide produced mainly by the hypothalamus that mediates caloric restriction-induced autophagy [3512] and autophagy stimulation induced by ghrelin in rat cortical neurons [3167]. Along these lines, different neuropeptides (such as SST [somatostatin], ADCYAP1 [adenylate cyclase activating polypeptide 1], TAAC1/substance P [tachykinin precursor 1] and others) have been described to control autophagy, likely being involved in the outcome of many pathological conditions, including neurodegeneration, metabolic disorders, and cancer [3513, 3514]. See also ghrelin.
NR1D1/Rev-erba (nuclear receptor subfamily 1 group D member 1): A nuclear receptor that modulates autophagy and lysosomal biogenesis in a tissue-specific manner.
NR1H4/FXR (nuclear receptor subfamily 1 group H member 4): A ligand-activated transcription factor that plays a role in lipophagy [921]. NR1H4 is a bile acid nuclear receptor and is necessary and sufficient to suppress lipophagy by the mechanisms of either a genomic competition with PPARA/PPARα/NR1C1 or inhibition of the CREB-CRTC2 complex, which leads to the altered expression of many autophagy-related genes [936].
NRBF2 (nuclear receptor binding factor 2): NRBF2 is the mammalian homolog of yeast Atg38, and is a binding partner of the BECN1-PIK3C3 complex; NRBF2 is required for the assembly of the ATG14-BECN1-PIK3C3/VPS34-PIK3R4/VPS15 complex and regulates autophagy [3515, 3516]. nrbf2 knockout mice display impaired ATG14-linked PIK3C3 lipid kinase activity and impaired autophagy.
NSP2: A nonstructural protein of chikungunya virus that interacts with human CALCOCO2 (but not the mouse ortholog) to promote viral replication. In contrast, binding of SQSTM1 to ubiquitinated capsid leads to viral degradation through autophagy [3517].
NT219: A highly efficient IGF1 signaling inhibitor that protects nematodes from toxic protein aggregation [3518]. NT219 reduces autophagy in cultured cells as detected by the accumulation of SQSTM1 foci in treated cells [3519].
Nt-R/Nt-Arg/arginylated substrates: Proteins bearing the amino-terminal arginine residue that are degraded by autophagy. Binding of Nt-R substrates to the ZZ domain of SQSTM1 stimulates SQSTM1 aggregation and autophagy [463]. Known Nt-R substrates (including R-HSPA5/R-Bip, R-CALR/CRT, R-PDI, and R-CDC6) are generated through N-terminal arginylation [3236, 3520].
NTRK1/TrkA (neurotrophic receptor tyrosine kinase): A receptor tyrosine kinase for NGF (nerve growth factor). Genetic mutations in NTRK1 cause congenital insensitivity to pain with anhidrosis (CIPA). Some CIPA mutations cause mental retardation because they induce protein aggregation and inhibit autophagic flux in neurons [3521].
Nucleolar stress: Perturbation of the nucleolar structure and function (e.g., by chemotherapeutic drugs, genetic approaches, RNAi and others) triggers this cellular stress response, which is characterized by impaired ribosome biogenesis. Nucleolar stress can be propagated in a TP53-dependent and -independent manner. Nucleolar stress emerges as a stress signal for autophagy [2027].
Nucleolus: Subnuclear, membrane free compartment and site for ribosome biogenesis. Also, a hub in the cellular stress response.
Nuclear pore complex (NPC): A nano-pore located on the nuclear membrane that consists of a conserved set of ~30 different proteins, termed nucleoporins, and serves as a gatekeeper for the exchange of materials between the cytoplasm and nucleus [3522]. Studies have revealed transcriptional factors passing through the NPC entering the nucleus that control autophagy. The identification of closely controlled transcription factors (such as TFEB and ZKSCAN3), microRNAs and histone marks (especially acetylated Lys16 of histone 4 [H4K16ac] and dimethylated H3K9 [H3K9me2]) associated with the autophagic process suggest an attractive conceptual framework to understand the short-term transcriptional response and potential long-term responses to autophagy [3523].
Nucleophagy: The selective autophagic degradation of the nucleus or parts of the nucleus. Depletion of the nuclear pore protein TPR induces LC3-II nuclear translocation. TPR, LC3-II, LMNB1, and chromatin nuclear budding complexes are formed, which induce nucleophagy, in both TPR-depleted or rapamycin-treated brain tumor cells [3092].
Nucleus-vacuole junction (NVJ): A junction formed by the interaction between Nvj1, a membrane protein of the outer nuclear membrane, and Vac8 of the vacuole membrane, that are necessary for PMN/micronucleophagy [1079]. See also piecemeal microautophagy of the nucleus.
NUFIP1 (nuclear FMR1 interacting protein 1): An autophagy receptor for ribosomes during starvation-induced ribophagy [3524].
Nup159: A component of the NPC in yeast that acts as a receptor for selective degradation of NPCs [3525].
NUPR1/p8 (nuclear protein 1, transcriptional regulator): A transcriptional regulator that controls autophagy by repressing the transcriptional activity of FOXO3 [3526].
Nvj1: An outer nuclear envelope protein that interacts with Vac8 to form nucleus-vacuole junctions during the process of PMN/micronucleophagy [1079].
NVP-BGT226 (8-[6-methoxy-pyridin-3-yl]-3-methyl-1-[4-piperazin-1-yl-3-trifluoromethyl-phenyl]-1,3-dihydroimidazo[4,5-c]quinolin-2-one maleate): A class I PI3K and MTOR dual inhibitor that induces autophagy [3527].
NVT (Nbr1-mediated vacuolar targeting): A pathway used for the delivery of cytosolic hydrolases (Lap2 and Ape2) into the vacuole in S. pombe that involves interaction with Nbr1 and relies on the ESCRT machinery [1851].
OATL1: See TBC1D25.
OCRL (OCRL inositol polyphosphate-5-phosphatase): A PtdIns(4,5)P2 5-phospatase. OCRL knowdown reduces autophagy flux by impairing the activity of MCOLN1/TRPML1 [3528].
OCT3: See POU5F1.
OCT4: See POU5F1.
OGA/MGEA5/NCOAT/oga-1 (O-GlcNAcase): OGA removes the O-GlcNAc modification and regulates the autophagy machinery by countering the action of OGT [3529].
OGT/ogt-1 (O-linked N-acetylglucosamine [GlcNAc] transferase): OGT is a nutrient-dependent signaling transferase that regulates the autophagy machinery by adding the O-GlcNAc modification. Similar to phosphorylation, this modification is involved in signaling [3529, 3530]. OGT is required for glucagon-stimulated liver autophagy and metabolic adaptation to starvation. Upon glucagon-induced calcium signaling, CAMK2 phosphorylates OGT, which in turn promotes O-GlcNAc modification and activation of ULK proteins by potentiating AMPK-dependent phosphorylation [1007]. Furthermore, O-GlcNAc modification of SNAP29 regulates autophagosome maturation [3531].
Oligomycin: A mitophagy inducer used in combination with AMA to promote a potent mitochondrial depolarization; oligomycin inhibits F1Fo-ATP synthase, and its reverse hydrolysis activity that is activated as a compensatory mechanism when AMA is used alone [1224].
Omegasome: ZFYVE1-containing structures located at the ER that are involved in autophagosome formation during amino acid starvation [884].
Omi: See HTRA2.
Oncophagy: A general term describing cancer-related autophagy [3532].
OPTN (optineurin): An autophagy receptor that functions in the elimination of Salmonella, bulk protein aggregates and mitochondria; OPTN has an N-terminal TBK1-binding motf, a LIR and a ubiquitin-binding UBAN domain, allowing it to link tagged bacteria and ubiquitinated mitochondria and protein aggregates to the autophagy machinery [1140, 1472, 3533–3537]. Phosphorylation of OPTN by TBK1 not only increases its affinity for LC3 but also promotes its binding ability to ubiquitinated proteins [1472, 3535, 3538]. OPTN may function together with CALCOCO2 and TAX1BP1/CALCOCO3. OPTN can also regulate autophagosome maturation [2913]. OPTN mutations are associated with ALS [3539]. See also CALCOCO2, TAX1BP1 and TBK1.
Organellophagy: General terminology for autophagic processes selective for organelles such as the peroxisome, mitochondrion, nucleus, and portions of the ER [1067, 3540].
Oxiapoptophagy: A type of cell death induced by oxysterols that involves OXIdation + APOPTOsis + autophagy [1344, 1345]. See also oxysterols.
Oxidized phospholipids: Oxidized phospholipids induce autophagy, and in ATG7-deficient keratinocytes and melanocytes the levels of phospholipid oxidation are elevated [3541, 3542].
Oxysterols: Oxysterols are oxide derivatives of cholesterol. They are formed by auto-oxidation, enzymatic oxidation or by both processes (http://www.lipidhome.co.uk/lipids/simple/chol-der/index.htm). Some oxysterols (including 7-ketocholesterol, 7β-hydroxycholesterol and 24[S]-hydroxycholesterol) can induce a complex type of cell death named oxiapoptophagy [1343–1345]. See also oxiapoptophagy.
P0: A plant virus-encoded F-box protein that targets AGO1/ARGONAUTE1 to autophagy in order to suppress RNA silencing [1391].
p8: See NUPR1.
p14ARF: See CDKN2A.
p27/p27Kip1: See CDKN1B.
p38α: See MAPK14.
p38IP: See SUPT20H.
p53: See TP53.
p62: see SQSTM1.
p66SHC: See SHC1.
p97: See VCP.
Pacer: See RUBCNL.
Paclitaxel (PTX): A chemotherapeutic drug used to treat a number of different cancer types. Paclitaxel act as a microtubule stabilizer that inhibits autophagy through induction of inhibitory phosphorylation of PIK3C3/VPS34 at T159 and blocking the fusion of autophagosomes with lysosomes [3543].
Paf1: A component of the yeast Paf1 complex (Paf1C) that downregulates the expression of ATG11 and ATG32 and represses mitophagy in growing conditions [3016].
PAF1 (PAF1 homolog, Paf1/RNA polymerase II complex component): A component of the mammalian Paf1/RNA polymerase II complex that regulates mitophagy through a PINK1-PRKN-dependent pathway [3016]. See also Paf1.
Paf1C: The yeast polymerase-associated factor 1 complex that is composed of multiple subunits and suppresses glucose starvation-induced autophagy. Deletion of the genes encoding two components of Paf1C, PAF1 and CTR9, increase ATG32 and ATG11 expression and facilitate mitophagy activity [3016].
PARK2/parkin: See PRKN.
PARK7/DJ-1 (Parkinsonism associated deglycase): An oncogene product whose loss of function is associated with PD; overexpression suppresses autophagy through the MAPK8/JNK pathway [3544].
Parkin: See PRKN.
PARL (presenilin associated rhomboid like): The mammalian ortholog of Drosophila rho-7 (rhomboid-7), a mitochondrial intramembrane protease; regulates the stability and localization of PINK1 [3545–3547]. A missense mutation in the N terminus has been identified in some patients with PD [3548]. See also PINK1.
Paraptosis: A form of nonapoptotic programmed cell death characterized by cytoplasmic vacuolation and resistance to apoptosis inhibitors [1783].
PARP1 (poly[ADP-ribose] polymerase 1): A nuclear enzyme involved in DNA damage repair; doxorubicin-induced DNA damage elicits an autophagic response that is dependent on PARP1 [3549]. In conditions of oxidative stress, PARP1 promotes autophagy through the STK11/LKB1-AMPK-MTOR pathway [3550].
PAS: See phagophore assembly site.
PAWR/par-4 (pro-apoptotic WT1 regulator): A cancer selective apoptosis-inducing tumor suppressor protein that functions as a positive regulator of autophagy when overexpressed [3551, 3552].
PBPE: A selective and high affinity ligand of the microsomal antiestrogen-binding site (AEBS). PBPE induces protective autophagy in cancer cells through an AEBS-mediated accumulation of zymostenol (5α-cholest-8-en-3β-ol) [2031, 3553].
Pbs2: A yeast MAPKK upstream of Hog1 that is required for mitophagy [747].
Pcl1: A yeast cyclin that activates Pho85 to stimulate autophagy by inhibiting Sic1 [2995].
Pcl5: A yeast cyclin that activates Pho85 to inhibit autophagy through degradation of Gcn4 [2995].
PCYT1A (phosphate cytidylyltransferase 1, choline, alpha): A rate-limiting enzyme in the Kennedy pathway for the synthesis of phosphatidylcholine. Activation of PCYT1A and increased de novo choline phospholipid production are found in cancer cells undergoing drug-induced autophagy. The loss of PCYT1A activity results in the inability of cells to maintain autophagosome biogenesis [3554].
PDCD6/ALG-2 (programmed cell death 6): An EF-hand calcium-binding protein that is a lysosomal calcium sensor, which enables the calcium-dependent movement of lysosomes to the perinuclear region where autophagosomes accumulate following induction of autophagy. This retrograde lysosomal transport involves the signaling through the lysosomal-localized phosphoinositide PtdIns(3,5)P2, the lysosomal calcium channel MCOLN1/TRPML1 and dynein; PDCD6 is a direct binding partner to both MCOLN1 and dynein [3555].
PDCD6IP (programmed cell death 6 interacting protein): PDCD6IP is an ESCRT-associated protein that interacts with the ATG12–ATG3 conjugate to promote basal autophagy [3556]. See also 12-ylation.
PDPK1/PDK1 (3-phosphoinositide dependent protein kinase 1): An activator of AKT. Recruited to the plasma membrane and activated by PtdIns(3,4,5)P3 which is generated by the class I PI3K.
PEA15/PED (proliferation and apoptosis adaptor protein 15): A death effector domain-containing protein that modulates MAPK8 in glioma cells to promote autophagy [3557].
PEBP1/RKIP (phosphatidylethanolamine binding protein 1): PEBP1 inhibits autophagy through the modulation of LC3 lipidation and MTORC1 signaling [3558]. This protein also shows a functional linkage with some autophagy gene products such as WDR45, PIK3CB or PIK3C3 during the development of prostate cancer [3559]
PEG3 (paternally expressed 3): A DCN (decorin)- and endorepellin-induced, genomically imprinted tumor suppressor gene that is required for autophagy in endothelial cells [3041]. PEG3 colocalizes with and physically binds to canonical autophagic markers such as BECN1 and LC3. Moreover, loss of PEG3 ablates the DCN- or endorepellin-mediated induction of BECN1 or MAP1LC3A; basal expression of BECN1 mRNA and BECN1 protein requires PEG3 as well as autophagic flux (measured by LC3-II formation and concurrent THBS1 (thrombospondin 1) expression [3560]. Mechanistically, PEG3 is upstream of TFEB and is required for proper TFEB expression and nuclear localization in endothelial cells [3561]. See also DCN and endorepellin.
PER1 (period circadian clock 1): Per1 is a gene in the clock gene family that includes members of the basic helix-loop-helix-PAS (PER-ARNT-SIM) transcription factor family. The PER1 protein has a central role in driving circadian rhythm over a period of approximately 24 h. During periods of injury in the brain such as stroke, autophagy in the hippocampus can be depressed during the absence of PER1 that may increase injury in the brain, suggesting a protective pathway with PER1 and autophagy [3562].
Peripheral structures: See Atg9 peripheral structures.
PERK: See EIF2AK3.
PES/pifithrin-µ (2-phenylethynesulfonamide): A small molecule inhibitor of HSPA1A/HSP70-1/HSP72; PES interferes with lysosomal function, causing a defect in autophagy and chaperone-mediated autophagy [3234].
peup (peroxisome unusual positioning): Mutants isolated in A. thaliana that accumulate aggregated peroxisomes [2344]. The peup1, peup2 and peup4 mutants correspond to mutations in ATG3, ATG18A and ATG7.
PEX2 (peroxisomal biogenesis factor 2: A peroxisomal ring finger E3 ubiquitin ligase that mediates pexophagy [1204].
PEX13: An integral membrane protein on the peroxisome that regulates peroxisomal matrix protein import during peroxisome biogenesis and plays a role in both virophagy and mitophagy [3563].
Pexophagic body: The single-membrane vesicle present inside the vacuole lumen following the fusion of a pexophagosome with a vacuole.
Pexophagosome: An autophagosome containing peroxisomes, but largely excluding other cytoplasmic components; a pexophagosome forms during macropexophagy [3564].
Pexophagy: A selective type of autophagy involving the sequestration and degradation of peroxisomes; it can occur by a micro- or macroautophagy-like process (micro- or macropexophagy) [179].
PexRD54: Effector of Phytophthora infestans that specifically binds host autophagy protein ATG8CL of the Atg8-family of proteins to stimulate autophagosome formation [2350].
PFKFB3 (6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3): PFKFB3 controls the synthesis and degradation of fructose-2,6-bisphosphate, a key regulator of glycolysis in eukaryotes. The inhibition of PFKFB3 decreases glucose uptake and promotes autophagy in cancer cells [3565].
PGAM5 (PGAM family member 5, mitochondrial serine/threonine protein phosphatase): PGAM5 is a protein phosphatase responsible for the activation of DNM1L in fed cells [878] and for the activation of the mitophagy receptor FUNDC1 in mitophagy [1196]. See also STX17.
PGRP (peptidoglycan recognition protein): A cytosolic Drosophila protein that induces autophagy in response to invasive L. monocytogenes [3566].
Phagolysosome: The product of a single-membrane phagosome fusing directly with a lysosome in a process that does not involve macroautophagy (we include this definition here simply for clarification relative to autolysosome, autophagosome and autophagolysosome) [1480].
Phagophore (PG): Membrane cisterna that has been implicated in an initial event during formation of the autophagosome. Thus, the phagophore may be the initial sequestering compartment of macroautophagy [3567]. The phagophore has previously been referred to as the “isolation membrane” [7].
Phagophore assembly site (PAS): A perivacuolar compartment or location that is involved in the formation of Cvt vesicles, autophagosomes and other sequestering compartments used in macroautophagy and related processes in fungi. The PAS may supply membranes during the formation of the sequestering vesicles or may be an organizing center where most of the autophagic machinery resides, at least transiently. The PAS or its equivalent is yet to be defined in mammalian cells [242, 3568].
Phagophore closure: Sealing of the phagophore to generate an autophagosome. Vps21/RAB5 in yeast was first reported to be required for phagophore closure [3569]. The ESCRT complex in both yeast and mammalian cells is also required for phagophore closure [360, 3114]. Furthermore, Vps21/RAB5 controls the interaction between Atg17 and the ESCRT subunit Snf7 to recruit the ESCRT complex to the phagophore for closure [3114]. Also see RAB5 and ESCRT.
Phagosome: A single-membrane vesicle formed in the cytoplasm of a cell, containing a phagocytosed particle enclosed within a part of the cell membrane. A phagosome can fuse directly with a lysosome, independently from the autophagic machinery, to form a phagolysosome (PL). During LC3-associated phagocytosis, an LC3-decorated phagosome can fuse with a lysosome, and in this case the fusion product is called an autophagolysosome (APL). In a separate scenario, a phagosome may be engulfed within an autophagosome; when such a structure fuses with a lysosome, the fusion product is again called an autophagolysosome [245, 1480, 2446, 3570]. Note that when an autophagosome that does not contain a phagosome fuses with a lysosome, the product is an autolysosome (AL). See also autolysosome and autophagolysosome.
PHB2 (prohibitin 2): PHB2 is a highly conserved inner mitochondrial membrane scaffold protein that acts as mitophagy receptor required for PRKN-mediated mitophagy. PHB2 facilitates PINK1 (PTEN induced kinase 1) stabilization on, and PRKN/Parkin recruitment to, mitochondria, and interacts with LC3 after rupture of the outer mitochondrial membrane [1176, 3571].
Phenothiazine: An FDA-approved antipsychotic phenothiazine derivative that induces autophagy through AKT-mediated MTOR inhibition [1979, 1980], and also by modulating the WNT-CTNNB1/β-catenin signaling pathway [1981].
Pho8: A yeast vacuolar phosphatase that acts upon 3ʹ nucleotides generated by Rny1 to generate nucleosides [3572]. A modified form of Pho8, Pho8∆60, is used in an enzymatic assay for monitoring autophagy in yeast. See also Rny1. See also Pho8∆60 assay.
Pho23: A component of the yeast Rpd3L histone deacetylase complex that negatively regulates the expression of ATG9 and other ATG genes [917].
Pho80: A yeast cyclin that activates Pho85 to inhibit autophagy in response to high phosphate levels [2995].
Pho8∆60 assay: An enzymatic assay used to monitor autophagy in yeast. Deletion of the N-terminal cytosolic tail and transmembrane domain of Pho8 prevents the protein from entering the secretory pathway; the cytosolic mutant form is delivered to the vacuole via autophagy, where proteolytic removal of the C-terminal propeptide by Prb1 generates the active enzyme [355, 356, 1038].
Pho85: A multifunctional cyclin-dependent kinase that interacts with at least ten different cyclins or cyclin-like proteins to regulate the cell cycle and responses to nutrient levels. Pho85 acts to negatively and positively regulate autophagy, depending on its binding to specific cyclins [2995]. See also Clg1, Pcl1, Pcl5, Pho80 and Sic1.
Phosphatidylinositol 3-kinase (PtdIns3K): A family of enzymes that add a phosphate group to the 3ʹ hydroxyl on the inositol ring of phosphatidylinositol. The 3ʹ phosphorylating lipid kinase isoforms are subdivided into three classes (I-III) and the class I enzymes are further subdivided into class IA and IB. The class III phosphatidylinositol 3-kinases (see PIK3C3 and Vps34) are stimulatory for autophagy, whereas class I enzymes (referred to as phosphoinositide 3-kinases, PI3Ks) are inhibitory [3573]. The class II PtdIns3K substantially contributes to PtdIns3P generation and autophagy in Pik3c3 knockout MEFs, also functioning as a positive factor for autophagy induction [3574]. In yeast, Vps34 is the catalytic subunit of the PtdIns3K complex. There are two yeast PtdIns3K complexes, both of which contain Vps34, Vps15 (a regulatory kinase), and Vps30/Atg6. Complex I includes Atg14 and Atg38 and is involved in autophagy, whereas complex II contains Vps38 and is involved in the vacuolar protein sorting (Vps) pathway. The X-ray crystal structure of yeast complex II has a Y-shape, where one arm of the Y is the regulatory/adaptor arm and the other arm is the catalytic arm [800]. The kinase domains of Vps15 and Vps34 engage each other at the tip of the catalytic arm. The regulatory/adaptor arm has elements from Vps30, Vps38 and the C-terminal region of Vps15, and the Vps30 BARA domain in this arm is critical for activity of the kinase on membranes. Electron microscopy suggests that mammalian complex II has a similar organization, except that the catalytic arm shows extensive conformational diversity [3575–3577]. See also phosphoinositide 3-kinase.
Phosphatidylinositol-3-phosphate (PtdIns3P): The product of the PtdIns3K and the target of myotubularin phosphatases. PtdIns3P is present at the PAS, and is involved in the recruitment of components of the autophagic machinery. It is important to note that PtdIns3P is also generated at the endosome (e.g., by the yeast PtdIns3K complex II). Additionally, FYVE-domain probes block PtdIns3P-dependent signaling, presumably by sequestering the molecule away from interactions with downstream effectors or preventing its interconversion by additional kinases [3578]. Thus, general PtdIns3P probes such as GFP-tagged FYVE and PX domains are generally not good markers for the autophagy-specific pool of this phosphoinositide.
Phosphatidylinositol-3,5-bisphosphate (PtdIns[3,5]P2): This molecule is generated by PIKFYVE (phosphoinositide kinase, FYVE finger containing) and targeted by several myotubularin phosphatases, and is abundant at the membrane of the late endosome. Its function is relevant for the replication of intracellular pathogens such as the bacteria Salmonella [3579], and ASFV [3580]. PtdIns(3,5)P2 also plays a role in regulating autophagy [3581]. In addition, PtdIns(3,5)P2 plays a crucial role in lysosome homeostasis where it is required for lysosome fission, autophagosome-lysosome fusion, and trafficking of molecules into lysosomes, events that are essential for autophagy [3582].
Phosphatidylinositol-4,5-bisphosphate (PtdIns[4,5]P2): Phosphoinositide lipid generated by both the type I canonical phosphinositide-4-phosphate 5-kinases (PI4P5Ks) and the type II non-canonical PI4P5Ks. PtdIns(4,5)P2 function has been implicated in many stages of the autophagic process [3583, 3584].
Phosphatidylserine (PS): This glycerophospholipid contains two fatty acyl hydrocarbon chains attached via ester linkages to the first and second carbons of glycerol, along with serine attached via a phosphodiester linkage to the third carbon of the glycerol. In healthy cells, PS is actively restricted to the cytosolic (inner) side of the cell plasma membrane by ATP-requiring flippases. This localization for PS becomes disrupted when cells undergo autophagy or apoptosis due to scramblases catalyzing rapid PS transbilayer exchange between the two sides of the membrane [3585]. The exposure of PS in the extracellular (outer) surface of the cell acts as a signal for macrophages to engulf the cells, and autophagy mediates PS exposure and phagosome degradation during apoptosis [3586]. PS exposure is also involved in axonal autophagy. GSK3B-mediated phosphorylation of MCL1 regulates autophagy to promote axonal degeneration. The GSK3B-MCL1 pathway affects ATP production locally in degenerating axons, and the exposure of PS as an “eat-me” signal for phagocytes that degrade these axons; defects in this process result in the failed engulfment of axonal debris in vivo [3585]. See also FBXS7, GSK3B and MCL1.
Phosphoinositide 3-kinase (PI3K): The class I family of enzymes that add a phosphate group to the 3ʹ hydroxyl on the inositol ring of phosphoinositides. PI3K activity results in the activation of MTOR and the inhibition of autophagy.
Phosphoinositide-4-phosphate 5-kinases (PI4P5Ks): The type II non-canonical family of lipid kinases that phosphorylate the 4-position of the minor lipid phosphatidylinositol-5-phosphate (PtdIns5P) to generate a small yet significant pool of PtdIns(4,5)P2 at intracellular locations. The PI5P4Ks are required for autophagosome-lysosome fusion [3587].
Phosphoinositides (PI) or inositol phosphates: These are membrane phospholipids that control vesicular traffic and physiology. There are several different PIs generated by quick interconversions by phosphorylation/dephosphorylation at different positions of their inositol ring by a number of kinases and phosphatases. The presence of a particular PI participates in conferring membrane identity to an organelle.
Phosphorylated ubiquitin/p-S65-Ub: Phosphorylated ubiquitin is essential for PINK1-PRKN-mediated mitophagy and plays a dual role in the initial activation and recruitment of PRKN to damaged mitochondria (reviewed in [1194]). Specific antibodies can be used to faithfully detect PINK1-PRKN-dependent mitophagy at early steps [1191, 1193]; however, the exact functions of p-S65-Ub during the different phases of mitophagy remain unclear.
Phycocyanin: Phycocyanin promotes autophagy-mediated cell death by inhibiting PI3K-AKT-MTOR signaling pathways in pancreatic cancer cells [3588].
Piecemeal microautophagy of the nucleus (PMN)/micronucleophagy: A process in which portions of the yeast nuclear membrane and nucleoplasm are invaginated into the vacuole, scissioned off from the remaining nuclear envelope and degraded within the vacuole lumen [157, 1077]. This process is dependent on Vac8 and Nvj1. See also nucleus-vacuole junction.
PI3Kγ: See PIK3CG.
PI4K2A/PI4KIIα (phosphatidylinositol 4-kinase type 2 alpha): A lipid kinase that generates PtdIns4P, which plays a role in autophagosome-lysosome fusion [3589]. PI4K2A is recruited to autophagosomes through an interaction with GABARAP or GABARAPL2 (but the protein does not bind LC3).
PIK3C2B/PI3KC2β (phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta): A ubiquitously expressed class II phosphatidylinositol 3-kinase and phosphoinositide 3-kinase isoform that is recruited to late endosomes and lysosomes in serum-starved cells to locally produce phosphatidylinositol-3,4-bisphosphate (PtdIns[3,4]P2), resulting in repression of nutrient signaling via MTORC1. PIK3C2B-depleted cells in addition to elevated MTORC1 activity accumulate LC3-positive autophagosomes and undegraded SQSTM1, suggesting that protein turnover via the autophagy-lysosome pathway requires PIK3C2B activity [3590].
PIK3C3 (phosphatidylinositol 3-kinase catalytic subunit type 3): The mammalian homolog of yeast Vps34, a class III PtdIns3K that generates PtdIns3P, which is required for autophagy [3573]. In mammalian cells there are at least three PtdIns3K complexes that include PIK3C3/VPS34, PIK3R4/VPS15 and BECN1, and combinations of ATG14, UVRAG, AMBRA1, SH3GLB1 and/or RUBCN. See also phosphatidylinositol 3-kinase.
PIK3CB/p110β (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta): A catalytic subunit of the class IA phosphoinositide 3-kinase; this subunit plays a positive role in autophagy induction that is independent of MTOR or AKT, and instead acts through the generation of PtdIns3P, possibly by acting as a scaffold for the recruitment of phosphatases that act on PtdIns(3,4,5)P3 or by recruiting and activating PIK3C3 [3591].
PIK3CG/PI3Kγ/p110γ (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma): Catalytic subunit of the only class IB PI3K member; this subunit is enriched in specific cell subtypes, such as leukocytes and cardiomyocytes. In the heart, PIK3CG is activated under stress conditions and inhibits autophagy by triggering the AKT-MTOR-ULK1 signaling pathway. In hearts from mice treated with doxorubicin, PIK3CG is activated downstream of TLR9 by the mitochondrial DNA contained within autolysosomes, and drives a feedback inhibition of autophagy, ultimately contributing to doxorubicin-induced cardiomyopathy [2583]. See also TLR9.
PIK3R4/p150/VPS15 (phosphoinositide-3-kinase regulatory subunit 4): The mammalian homolog of yeast Vps15, PIK3R4 is a core component of all complexes containing PIK3C3 and is required for autophagy [3592]. PIK3R4 interacts with the kinase domain of PIK3C3, to regulate its activity and also functions as a scaffold for binding to NRBF2 and ATG14 [3515, 3516]. While PIK3R4 is classified as a protein serine/threonine kinase, it possesses an atypical catalytic domain and lacks catalytic activity, at least in vitro (J. Murray, personal communication). PIK3R4 also interacts with RAB GTPases, including RAB5 [3593] that may be responsible for recruitment of PIK3C3-PIK3R4-complexes to sites of autophagosome formation.
PINK1/PARK6 (PTEN induced kinase 1): A mitochondrial protein kinase (mutated in autosomal recessive forms of PD) that is normally processed in a membrane potential-dependent manner to maintain mitochondrial structure and function [82, 3546], suppressing the need for mitophagy [1137]. Upon mitochondrial depolarization, mitochondrial import blockade, accumulation of unfolded proteins in the mitochondrial matrix or ablation of the inner membrane protease PARL, PINK1 is stabilized and activated, phosphorylating ubiquitin and PRKN for full activation and recruitment of PRKN (reviewed in [1194]) to facilitate mitophagy [1155, 3545, 3594–3597]. Processed PINK1 can also interact with VCP/p97 to regulate PRKA activation [1742], with indirect effects on autophagy. See also PARL and phosphorylated ubiquitin/p-S65-Ub.
PKA (protein kinase A): A serine/threonine kinase that negatively regulates autophagy in yeast [3598]; composed of the Tpk1/2/3 catalytic and Bcy1 regulatory (inhibitory) subunits. The mammalian PKA homolog, PRKA, directly phosphorylates LC3 to negatively regulate phagophore recruitment [293]. Bacterial toxins that activate mammalian PRKA can also inhibit autophagy [3599]. In addition, cAMP inducers, such as β2-adrenergic agonists (D.A.P. Gonçalves, personal communication), CALC/calcitonin gene-related peptide (J. Machado, personal communication) and forskolin plus isobutilmethylxantine (W.A. Silveira, personal communication), block the conversion of LC3-I to LC3-II in C2C12 myotubes and adult skeletal muscles. Phosphorylation of the fission modulator DNM1L by mitochondrially-localized PRKA blocks mitochondrial fragmentation and autophagy induced by loss of endogenous PINK1 or by exposure to a neurotoxin in neuronal cell cultures [3057]. Conversely, forskolin treatment in glucose-starved cancer cells, specifically NIH3T3 KRAS transformed mouse fibroblasts, MDA-MB-231 (breast cancer cells) and Mia Paca2 (pancreatic cancer cells), is able to sustain cell survival by increasing mitochondrial activity, intracellular ATP levels, mitochondrial interconnection and glutamine metabolism as well as autophagy [3600, 3601]. Indeed, as measured by MDC staining and LC3 localization and cleavage, forskolin induces a PRKA-dependent appearance of autophagosomes. Such appearance has been confirmed by combined treatment with forskolin and CQ, with the PRKA/PKA inhibitor H89 as well as upon the knockdown of the catalytic PRKACA/PKAα subunit, all causing an inhibition of the positive effect of forskolin on autophagy. Importantly such a positive effect of PRKA on autophagy is also observed in starved and detached cancer cells. Indeed, in this harsh conditions, endogenous PRKA activation favors cancer cell survival by activating mitochondrial function, glutamine metabolism and autophagy [3601]. See also DNM1L.
PKB: See AKT.
Pkc1: A yeast serine/threonine kinase involved in the cell wall integrity pathway upstream of Bck1; required for pexophagy and mitophagy [747]. See also Bck1 and Slt2.
PKCδ: See PRKCD.
PKR: See EIF2AK2.
PLA2G4A/cPLA2 (phospholipase A2 group IVA): A cytoplasmic phospholipase that can translocate to lysosomes upon activation, where it can mediate cleavage of lysosomal membrane phospholipids to generate lysosphospholipids. Accumulation of lysophospholipids affects membrane structure and properties, leading to LMP [1606] See also lysosomal membrane permeabilization.
Plasmodium-associated autophagy-related (PAAR) response: An intracellular host cell response towards an infection with Plasmodium sporozoites, clearly differing from canonical autophagy, typical xenophagy and LAP [3602, 3603].
Plastolysome: A plant plastid that transforms into a lytic compartment, with acid phosphatase activity, engulfing and digesting cytoplasmic regions in particular cell types and under particular developmental processes [1282–1285].
PLD1 (phospholipase D1): PLD1 is a membrane-associated enzyme that enhances autophagic flux by promoting vesicular fusion through phosphatidic acid generation and recruitment of PIP5K1/PtdIns4P 5-kinase to mediate fusion and inner membrane curvature [3604]. Blockade of PLD1 blocks autophagic flux and increases the aggregation and retention of proteins such as MAPT/tau and polyglutamines [3605]. PLD1 also has putative roles as a downstream regulator of PIK3C3/VPS34 and, paradoxically, RHEB-mediated activation of MTOR [3605, 3606].
PLEKHG5 (pleckstrin homology and RhoGEF domain containing G5): A guanine nucleotide exchange factor activating RAB26 for the removal of synaptic vesicles by autophagy. Disruption of this pathway in mice results in swollen axon terminals in motoneurons leading to a motoneuron disease with late onset. Mutations in the human PLEKHG5 gene have been linked to several forms of motoneuron disease [3607]. See also RAB26.
PLEKHM1: An autophagic adaptor protein that contains a LIR motif, which directs binding to all of the LC3/GABARAP proteins. PLEKHM1 also interacts with GTP-bound RAB7 and the HOPS complex. PLEKHM1 is present on the cytosolic face of late endosomes, autophagosomes, amphisomes and lysosomes, and serves to coordinate endocytic and autophagic pathway convergence at, and fusion with, the lysosome [3608].
PLGA-aNPs: Poly(DL-lactide-co-glycolide)/PLGA acidic nanoparticles/aNPs are approved by the US Food and Drug Administration and have been reported to (1) traffic to lysosomes and (2) act on the lysosomal pH.
PLK1 (polo like kinase 1): A serine/threonine kinase that inhibits MTORC1’s lysosomal association and enhances autophagy under conditions of nutrient starvation and sufficiency in mammalian cells and C. elegans. Whereas PLK1 is widely recognized as a controller of mitosis, it controls autophagy during interphase [3609].
PMT7: A phloroglucinol derivative used as a chemotherapeutic drug to target glycolytic cancer cells [3610].
PND (programmed nuclear destruction): A yeast cell death-related process that occurs during gametogenesis involving a noncanonical type of vacuole-dependent degradation [3410].
PNPLA1 (patatin like phospholipase domain containing 1): An enzyme that plays a key role in epidermal omega-O-acylceramide synthesis and localizes on the surface of lipid droplets. Mutations in the PNPLA1 gene are associated with autosomal recessive congenital ichthyosis (ARCI). Mutant or knocked down PNPLA1 leads to decreased autophagic flux and impaired lipophagy [1310] in fibroblasts.
PNPLA2/ATGL (patatin like phospholipase domain containing 2): A lipase that catalyzes the catabolism of triglycerides in lipid droplets. PNPLA2 acts to hydrolyze large lipid droplets [1312] and acts upstream of SIRT1 to promote lipophagy [1311]. PNPLA2 also interacts with LC3, which facilitates PNPLA2 targeting to lipid droplets [3611].
PNPLA3 (patatin like phospholipase domain containing 3): A triglyceride lipase/acyl transferase enzyme mainly expressed in the liver. The I148M variant of the protein represents a genetic risk factor for developing nonalcoholic fatty liver disease (NAFLD) and leads to decreased autophagic flux and reduced lipophagy in the human hepatocyte cell line HepG2 [1313].
PNPLA5 (patatin like phospholipase domain containing 5): A lipase that mobilizes neutral lipid stores (e.g., triglycerides in lipid droplets) to enhance autophagic capacity of the cell by contributing lipid precursors for membrane biogenesis (thus enhancing autophagic capacity) and signaling [3612]. This process should not be confused with the process of lipophagy, which is the uptake of lipid droplets for triglyceride degradation in autolysosomes.
PNPLA8/calcium-independent phospholipase A2γ/iPLA2γ (patatin like phospholipase domain containing 8): Global deletion of Pnpla8 leads to mitochondrial damage and enhanced autophagy in podocytes [3613].
PNS (peri-nuclear structure): A punctate structure in K. phaffii/P. pastoris marked by Atg35, which requires Atg17 for recruitment and is involved in micropexophagy; the PNS may be identical to the PAS [2819].
Podocytes: Podocytes are post-mitotic highly differentiated epithelial cells with limited capacity for self renewal. Podocytes are located in the glomerulus, a network of capillaries, which constitutes the filtering unit of the kidney. The blood is filtered across the glomerular capillary walls including the podocytes. Podocytes wrap around capillaries; thus, they are extracapillary pericytes, essential to the survival of the underlying glomerular endothelium. The healthy glomerulus filters water-soluble, non-protein-bound metabolic byproducts into the urine but prevents the passage of ALB (albumin) and other larger essential molecules. Podocytes contribute to such permeability-selectivity (or permselectivity) of the glomerulus. Dysfunction or loss of podocytes leads to urinary ALB leakage and eventually to loss of glomeruli and subsequent kidney failure. Podocytes exhibit a remarkably high autophagy flux at baseline that protects them in a number of pathological conditions. Variations of podocyte autophagy are found in diseases, the mechanisms of which remain to be determined.
Polyphenol: A class of plant phytochemicals that have been described as autophagy regulators in different disease models, such as neurodegenerative disease (reviewed in ref. [3614]) including PD [3615] and cancer (reviewed in ref. [3616]).
PolyQ: Polyglutamine or glutamine-rich repeats whose presence leads to the formation of cytoplasmic aggregates [627,715]. Expansions of polyQ repeats occur in HD and have been found in certain viruses [1427], although their potential pathogenic role in the latter is not clear. See also HTT, Dsk2, and AQAMAN [3466]).
Post-fertilization sperm mitophagy: Ubiquitin- and autophagy (SQSTM1)-dependent mechanism for selective degradation of paternal, sperm-borne mitochondria in the mammalian zygote, promoting clonal, maternal inheritance of mitochondrial DNA [3617]. See also allophagy.
POU5F1/Oct3/Oct4 (POU class 5 homeobox 1): A transcription factor in embryonic stem cells associated with their pluripotency, proliferative potential and self-renewal capacity [3618]; basal autophagy is needed to maintain cancer stem cell pluripotency, and knockdown of POU5F1 inhibits autophagy [3619].
PP242: A pharmacological catalytic kinase inhibitor of TOR; inhibits TORC1 and TORC2.
PPAN (peter pan homolog): A ribosome biogenesis factor regulating the maturation of the 60S ribosomal precursor. Loss of PPAN triggers TP53-independent nucleolar stress, abrogates mitochondrial homeostasis and enhances autophagic flux [2028, 3620].
PPARs (peroxisome proliferator activated receptors): Ligand-activated transcription factors, members of the nuclear receptor superfamily, consisting of 3 isotypes: PPARA/PPARα/NR1C1 (peroxisome proliferator activated receptor alpha), PPARD/PPARδ/NR1C2, and PPARG/PPARγ/NR1C3 [1202]. PPAR-mediated signaling pathways regulate, or are regulated by, molecules involved in autophagy [3621, 3622].
Ppg1: Ppg1 is a PP2A-like protein phosphatase that counteracts CK2-mediated phosphorylation of the mitophagy receptor Atg32 to inhibit mitophagy in yeast. See also casein kinase 2 and Far complex [3128].
PPI (protein-protein interaction): Proper biological activity of many proteins depends on physical interactions with other proteins. Specific PPI has a functional objective. Therefore, complete understanding of protein function requires consideration of proteins in the context of their binding partners [3623, 3624]. Often, interactions between proteins and protein complexes are presented in a form of large densely connected networks (PPI networks). Such network-based representation of PPIs provide the means for a more complete understanding of physiological and pathogenic mechanisms [3625].
PPM1D/Wip1 (protein phosphatase, Mg2+/Mn2+ dependent 1D): A protein phosphatase that negatively regulates ATM and autophagy [3626].
PPP1 (protein phosphatase 1): A serine/threonine protein phosphatase that regulates ATG16L1 by dephosphorylation of CSNK2-modified Ser139 to inhibit autophagy. See also CSNK2 [3012].
PPP1R13L/iASPP (protein phosphatase 1 regulatory subunit 13 like): PPP1R13L is a TP53 inhibitor that is usually upregulated in human cancers. Known as an important regulator of epidermal homeostasis, PPP1R13L also inhibits autophagy in keratinocytes. TP53BP2/ASPP-depletion in these cells results in elevated levels of LC3B indicating the activation of autophagy. PPP1R13L levels can serve as an indicator of the level of autophagy in keratinocytes [3627].
PPP1R15A/GADD34 (protein phosphatase 1 regulatory subunit 15A): A protein that is upregulated by growth arrest and DNA damage; PPP1R15A binds to and dephosphorylates TSC2, leading to MTOR suppression and autophagy induction [3628].
PPP2 (protein phosphatase 2): A serine/threonine protein phosphatase that negatively regulates autophagy via dephosphorylation of BECN1 at Ser90 [3629].
PPP2R5A (protein phosphatase 2, regulatory subunit B’ alpha): B56 subunit of PPP2/PP2A, a phosphatase that binds to and dephosphorylates GSK3B at Ser9 to make it active and thus activate autophagy [783].
PPP3/calcineurin (protein phosphatase 3): A calcium-dependent phosphatase. In response to a calcium pulse via the lysosomal calcium channel MCOLN1, PPP3 dephosphorylates Ser142 and Ser211 of TFEB, leading to nuclear localization and upregulation of the CLEAR network [2000]. See also CLEAR and TFEB.
prApe1 (precursor Ape1): See Ape1.
Pre-autophagosomal structure (PAS): See phagophore assembly site.
Primary cilium: A single, non-motile appendage that emerges, with few exceptions (e.g., in hematopoietic cells), from the surface of most vertebrate cell types. Autophagy-related proteins localize at cilia or periciliary regions: Both mature autophagosome markers and autophagy proteins acting on the initial steps of autophagosome formation show discrete puncta at basal bodies and cilia axonemes [3285]. The primary cilium functions as a specialized signaling device involved in the transduction of multiple signaling pathways (e.g., Hedgehog, WNT, PDGFR, NOTCH, TGFB and other GPCR-associated pathways) [3630]. Defects in the assembly and function of primary cilia are associated with genetic diseases, collectively known as ciliopathies [3631]. Ciliopathy proteins are involved in the regulation of autophagy: 1) INPP5E (inositol polyphosphate-5-phosphatase E), which localizes both at primary cilia and at lysosomes, is required for autophagosome-lysosome fusion [3632]; 2) FLCN (folliculin), which is localized both at primary cilia and lysosomes, physically interacts with autophagic proteins (e.g., GABARAP and ULK1), playing a positive role in autophagy [712]; 3) RPGRIP1L (RPGRIP1 like) positively regulates proteasomal activity at the ciliary base and autophagy, independently of each other [2978, 3633]. A reciprocal interaction between primary cilia and autophagy has emerged, and common players have only started to be identified. The primary cilium is a site of autophagosome formation, where components of the autophagic machinery are recruited and initiate autophagy in response to chemical and mechanical stimuli, or during neuroectodermal differentiation [3285, 3634, 3635]. In turn, autophagy controls ciliogenesis and cilia length, even though the outcome of the autophagic degradation on primary cilia is cell specific and context dependent [2975, 3633, 3636–3638]. See also ciliophagy and intraflagellar transport.
PRKA (protein kinase cAMP-dependent): The mammalian homolog of yeast PKA. See also PKA.
PRKCD/PKCδ (protein kinase C delta): PRKCD regulates MAPK8 activation. PRKCD also activates NADPH oxidases, which are required for antibacterial autophagy [3052].
PRKD1 (protein kinase D1): A serine/threonine kinase that activates PIK3C3/VPS34 by phosphorylation; recruited to phagophore membranes [3639].
PRKDC (protein kinase, DNA-activated, catalytic subunit): A positive regulator of autophagy via phosphorylation of PRKAG1/AMPKγ1 [652].
PRKN (parkin RBR E3 ubiquitin protein ligase): An E3 ubiquitin ligase (mutated in autosomal recessive forms of PD) that is recruited from the cytosol to mitochondria following mitochondrial depolarization, mitochondrial import blockade or accumulation of unfolded proteins in the mitochondrial matrix or ablation of the rhomboid protease PARL, to promote their clearance by mitophagy [339, 3545, 3594–3596]. PINK1-dependent phosphorylation of Ser65 in the ubiquitin-like domain of PRKN and in ubiquitin itself (see phosphorylated ubiquitin/p-S65-Ub) promotes activation and recruitment of PRKN to mitochondria (reviewed in ref. [1194]) [1155], and USP8 deubiquitination of K6-linked ubiquitin on PRKN to promote its efficient recruitment [3640].
Programmed cell death (PCD): Regulated self-destruction of a cell. Type I is associated with apoptosis and is marked by cytoskeletal breakdown and condensation of cytoplasm and chromatin followed by fragmentation. Type II is associated with autophagy and is characterized by the presence of autophagic vacuoles (autophagosomes) that sequester organelles. Type III is marked by the absence of nuclear condensation, and the presence of a necrotic morphology with swelling of cytoplasmic organelles (oncosis). These categories of cell death are based on morphological criteria, and the Nomenclature Committee on Cell Death now recommends the use of terms that are more precise and refer to different types of RCD [1759].
PROM1/CD133 (prominin 1): PROM1 is a transmembrane glycoprotein, residing cytosolically in retinal pigment epithelium that gets upregulated along with an increase in autophagy induced by stress signals. Knockout of Prom1 results in SQSTM1 accumulation and decreased autophagosome trafficking [2403].
PROPPINs (β-propellers that bind phosphoinositides): A WD40-protein family conserved from yeast to human [3641]. These proteins fold as seven-bladed β-propellers, and each blade contains four antiparallel β-strands. With two lipid binding sites at the circumference of their propeller they bind PtdIns3P and PtdIns(3,5)P2 [3642–3644]. The S. cerevisiae PROPPINs are Atg18, Atg21 and Hsv2, and the mammalian counterparts are termed WIPIs.
Proteaphagy: The autophagic degradation of the 26S proteasomes, which is mediated by two routes; bulk, nonselective degradation under starvation conditions, and selective degradation of inactive proteasomes [1352–1355, 1357]. Extensive polyubiquitination of the proteasome accompanies proteaphagy in yeast, plants, and mammals.
Proteasome: A large, multi-subunit proteolytic complex responsible for degrading individual proteins tagged with polyubiquitin chains; consists of a barrel-shaped core protease containing six proteolytic active sites, capped with a regulatory particle that contains activities for substrate recognition, deubiquitination, unfolding and delivery into the core particle chamber [3645].
Proteasome storage granules (PSGs): Membraneless cytoplasmic condensates containing proteasome core proteases and regulatory particles that form in yeast, mammals and plants in response to carbon starvation; sequestration into these structures protects proteasomes from autophagic degradation [1358].
Protein translation: Nutrient deprivation and some other stress stimuli that promote autophagy inhibit MTOR and limit protein synthesis. Reduced translation of essential autophagy proteins and their degradation under such stress conditions contributes to the limitation of autophagy amplification [3646].
Proteostasis: Proteostasis is the maintenance of proteome homeostasis, thereby regulating protein translation, folding, trafficking, subcellular localization, and degradation. Proteostasis interacts with essential components of the autophagy-lysosomal pathways, thus regulating autophagy and mitophagy, and also participates in tumorigenesis or cancer progression [3647].
Proto-lysosomes: Vesicles derived from autolysosomes that mature into lysosomes during autophagic lysosome reformation [781]. See also autophagic lysosome reformation.
Protophagy: Autophagy-like processes in microbial populations. The term summarizes all self-destructing patterns in prokaryotic colonies including bacterial cannibalism, autolysis, programmed cell death, and other processes, in which a part of the colony is lysed and consumed by neighboring prokaryotic cells to recycle matter and energy [3648].
PSEN1/2 (presenilin 1/2): Catalytic components of the γ-secretase complex. Both PSEN1 and PSEN2 are necessary for correct autophagy in a γ-secretase-independent manner [3649]. Moreover, mutations in PSEN1 and PSEN2 (linked to familial forms of AD) result in the accumulation of autophagosomes. In the presence of PSEN1 mutations, one of the V-ATPase subunits does not target properly to the lysosome, leading to defective lysosomal acidification and calcium deregulation [80, 81, 3649]. PSEN2 mutations instead cause a reduced cytosolic calcium signal that induces a defective recruitment of RAB7 to autophagosomes, blocking fusion of the latter with lysosomes [2589, 3416]. Recent studies in a tractable model system, D. discoideum, suggest that both the human PSEN1 protein, and its D. discoideum ortholog, function to regulate phagosomal proteolysis, as well as autophagosomal acidification and autophagic flux through a non-catalytic mechanism [1768, 1769].
Psp2: An RGG motif protein that positively regulates autophagy through promoting the translation of ATG1 and ATG13. During nitrogen starvation conditions, unmethylated Psp2 interacts with the 5′ UTR of ATG1 and ATG13 mRNA in an RGG motif-dependent manner in conjunction with translation initiation factors. The switch of this regulation is arginine methylation controlled by TOR signaling through Hmt1 [3650].
PT21: A potential PIK3C3/VPS34 kinase inhibitor. Autophagy initiation is regulated by the activity of the class III PtdIns3K complex, which can be inhibited by PT21, which promotes the degradation of these complexes [3651].
Ptc2/3: Redundant PP2C phosphatases that interact with and are involved in the dephosphorylation of the Atg1 kinase complex in S. cerevisiae, to promote autophagy as well as the Cvt pathway [3652].
PTEN (phosphatase and tensin homolog): A 3ʹ phosphoinositide phosphatase that dephosphorylates PtdIns(3,4,5)P3, thereby inhibiting PDPK1/PDK1 and AKT activity.
PTEN-L: A long isoform of PTEN with the addition of 173 amino acids at the N terminus, which functions as a negative regulator of mitophagy by dephosphorylating p-Ser65-Ub and p-Ser65-PRKN via its protein phosphatase activity [1192].
PTK2B/PYK2 (protein tyrosine kinase 2 beta): A non-receptor kinase of the PTK2/FAK family, involved in several cellular processes, including oxidative stress, calcium homeostasis, proliferation, cytoskeleton organization, and cell motility. PTK2B has tumor suppressive functions in prostate cancer cells, and the expression of a PTK2B kinase-dead mutant in the prostate epithelial EPN cell line sensitizes the cells to the pro-autophagic effects of the polyphenol resveratrol, and induces an increase in the number and size of autophagosomes as well as an enlargement of the lysosomal compartment [3653]. A deeper connection between PTK2B and autophagy regulation is suggested by the observation that RB1CC1, a known inhibitor of PTK2B, is a regulatory partner of the ULK–ATG13–RB1CC1 complex [3654].
PTM (posttranslational modification): After biosynthesis, many proteins undergo covalent modifications that are often catalyzed by special enzymes that recognize specific target sequences in particular proteins. PTMs provide dramatic extension of the structures, properties, and physico-chemical diversity of amino acids, thereby diversifying structures and functions of proteins [3655]. There are more than 300 physiological PTMs [3656]. Some PTMs (e.g., phosphorylation, acetylation, glycosylation, etc.) are reversible by the action of specific deconjugating enzymes. The interplay between modifying and demodifying enzymes allows for rapid and economical control of protein function [3655]. PTMs clearly play a role in regulating the autophagy machinery [998, 3657].
PTP4A3 (protein tyrosine phosphatase 4A3): A plasma membrane- and endosome-localized prenylated protein phosphatase that stimulates autophagy; PTP4A3 is also an autophagic substrate [3658].
PTPRS/PTPσ (protein tyrosine phosphatase receptor type S): A dual domain protein tyrosine phosphatase that antagonizes the action of the class III PtdIns3K; loss of PTPRS results in hyperactivation of basal and induced autophagy [3659].
PULKA (p-ULK1 assay): This acronym describes the analysis of Ser317 phosphorylated (activated) ULK1 puncta by fluorescence microscopy [3660].
PUX7/8/9/13 (Plant UBX domain-containing protein 7/8/9/13): A family of plant ubiquitin regulatory X (UBX) domain-containing proteins that act as autophagy receptors for inhibited or mutant forms of CDC48 in Arabidopsis; they bind the UDS of ATG8 via a UIM-like sequence [1359]. See also Cdc48, UDS and UIM.
PYK2: See PTK2B.
Quercetin (3,5,7,3′,4′-pentahydroxyflavone): A broadly studied phenolic compound, belonging to the class of flavonoids, sub-class flavonols. Quercetin is the most abundant flavonol in food and it is largely used as a dietary supplement [3661]. The multifaceted outcomes of quercetin in cancer cell lines indicate its capacity to induce protective and non-protective forms of autophagy [3662]. In addition, quercetin, in association with dasatinib, has been recognized as a senolytic compound [3663], providing a tool for studying how basal, selective autophagy can act as an antisenescence mechanism [3664].
RAB1: See Ypt1.
RAB2: A small GTPase that is required for ER-to-Golgi transport. RAB2 is also required for the fusion between autophagosomes and lysosomes through interaction with the HOPS complex [3665, 3666] and for the formation of autophagosomes and lysosomes [3667]. Knockout of RAB2 in the U2OS cell line results in a significant reduction in LC3B levels both under autophagy-stimulated and -unstimulated conditions. RAB2 modulates ULK1 activity to generate signals for autophagy initiation [3667].
RAB4A: This small GTPase was previously called HRES-1/Rab4, as it is encoded by the antisense strand of the HRES-1 human endogenous retroviral locus in region q42 of human chromosome 1 [3668]. It has been recently designated as RAB4A to distinguish it from RAB4B on human chromosome 19. RAB4A regulates the endocytic recycling of surface proteins, such as CD4, CD247/CD3ζ, and CD2AP, and TFRC/CD71, which control signal transduction through the immunological synapse in human T lymphocytes [3668, 3669]. Among these proteins, CD4 and CD247 are targeted by RAB4A for lysosomal degradation via autophagy [3668–3670]. Beyond T lymphocytes, RAB4A generally promotes the formation of LC3+ autophagosomes and the accumulation of mitochondria during autophagy [3671]. During accelerated autophagy, RAB4A also promotes the lysosomal degradation of intracellular proteins, such as DNM1L that initiates the fission and turnover of mitochondria [1573, 3672]. Thus, RAB4A-mediated depletion of DNM1L selectively inhibits mitophagy and causes the accumulation of mitochondria in patients and mice with lupus [3670]. The formation of interconnected mitochondrial tubular networks is enhanced by constitutively active RAB4AQ72L upon starvation, which may contribute to the retention of mitochondria during autophagy [3671].
RAB5: A small GTPase that is required for early endosome biogenesis. RAB5 is also involved in ESCRT-mediated sequestration of mitochondria [3673] and closure of the phagophore [3114]. See also phagophore closure.
RAB6: Small RAS family GTPase required for delivery of lysosomal hydrolases and normal localization of InR/insulin receptor in Drosophila [3674].
RAB7: A small GTPase of the RAS oncogene family functioning in transport from early to late endosomes and from late endosomes to lysosomes [3675]. RAB7 is also needed for the clearance of autophagic compartments, most likely for the fusion of amphisomes with lysosomes [1510, 1511]. RAB7 recruitment to autophagosomes, needed for their fusion with lysosomes, is calcium regulated [3416]. In RAB7 knockout mammalian cells, autolysosomes, rather than autophagosomes, may accumulate [3676]. Organelles positive for lysosomal markers may also label for RAB7 [3677], indicating hybrid vesicles [3678] as well as the dynamic and interconnected nature of trafficking organelles. Mutations in the RAB7A gene cause Charcot-Marie-Tooth type 2B (CMT2B) disease, a dominant axonal peripheral neuropathy. In neurons, RAB7A also controls neuronal-specific processes such as NTF (neurotrophin) trafficking and signaling, neurite outgrowth and neuronal migration. All CMT2B-causing RAB7A mutants cause a reduced autophagic flux and inhibit basal and starvation-induced autophagy, suggesting that alteration of the autophagic flux could be responsible for neurodegeneration [271]. The yeast homolog is Ypt7. See also PSEN2.
RAB8: A small GTPase of the RAS oncogene family. RAB8A functions in secretory autophagy [1700], whereas RAB8B plays a role in degradative autophagy [3679].
RAB9: A member of the Ras superfamily of small RAB GTPases involved in the transport between late endosomes and the trans Golgi network. A growing body of evidence indicates that mitochondria can be eliminated through a RAB9-dependent, but LC3-independent, noncanonical autophagy pathway that utilizes autophagosomes derived from the trans-Golgi and late endosomes. This pathway is essential during erythrocyte maturation or in stress conditions such as ischemia/reperfusion injury in the heart [3680, 3681]. RAB9 may also play a role in the formation of group A Streptococcus (GAS)-containing autophagosome-like vacuoles [3682].
RAB10: A small GTPase that binds to the autophagy receptor OPTN, promotes OPTN accumulation on depolarized mitochondria and facilitates mitophagy. RAB10 is a substrate of the PD-linked kinase LRRK2 [3371]. Phosphorylation of RAB10 by LRRK2 interferes with RAB10 binding to OPTN and mitophagy [3683].
RAB11: A small GTPase that localizes on multivesicular bodies (MVBs), recycling endosomes and late endosomes [3684]. RAB11 plays an important role both at the early and late stages of autophagy [3685, 3686]. RAB11 is required for autophagosome formation; ULK1 and ATG9 localize in part to RAB11-positive recycling endosomes [3685]. Upon autophagy induction, RAB11 promotes the fusion between endosomes and autophagosomes removing HOOK from mature late endosomes, which is a negative regulator of endosome maturation, and translocates to autophagosomes [3687]. See also TBC1D9B and TBC1D14.
RAB12: A small GTPase that controls degradation of the amino acid transporter SLC36A4 (solute carrier family 36 member 4)/PAT4 and indirectly regulates MTORC1 activity and autophagy [3688].
RAB13: A small GTPase that is required for autophagosome formation. The active form of RAB13 promotes its interaction with GRB2 (growth factor receptor bound protein 2), which in turn activates AMPK, leading to MTOR inhibition and the induction of autophagy [3689].
RAB21: A small GTPase that is required for autophagosome-lysosome fusion. Starvation induces RAB21 activity that promotes VAMP8 trafficking to the lysosome, where VAMP8 is needed to mediate fusion. See also SBF2 [3690].
RAB24: A small GTPase with unusual characteristics that associates with autophagic vacuoles and is needed for the clearance of autolysosomes under basal conditions [3691, 3692].
RAB26: A small GTPase enriched on synaptic vesicles that interacts with ATG16L1 in its GTP-bound form and directs synaptic vesicles to autophagosomes [3693]. RAB26 is activated by PLEKHG5, a GEF, which has been linked to motoneuron disease [3607]. See also PLEKHG5.
RAB32: A small GTPase that localizes to the ER, and enhances autophagosome formation under basal conditions [3694].
RAB33B: A small GTPase of the medial Golgi complex that binds ATG16L1 and plays a role in autophagosome maturation by regulating fusion with lysosomes [3695]. RAB33B is a target of TBC1D25/OATL1, which functions as a GAP [3696].
RAB37: A small GTPase that functions as a key organizer of autophagosomal membrane biogenesis. RAB37 interacts with ATG5 and promotes autophagosome formation by modulating ATG12–ATG5-ATG16L1 complex assembly [3697, 3698].
RABG3B: A RAB GTPase that functions in the differentiation of tracheary elements of the Arabidopsis xylem through its role in autophagy; this protein is a homolog of RAB7/Ypt7 [1762].
RACK1 (receptor for activated C kinase 1): During autophagy ATG5 is bound by RACK1 to initiate the formation of autophagosomes. Knockdown of RACK1 or inhibition of the RACK1-ATG5 interaction inhibits autophagy. Therefore, RACK1, as an interacting partner of ATG5, could be one a reliable marker to study autophagy through colocalization [3699].
RAD001: See everolimus.
RAG: See RRAG.
RAGE: See AGER.
RAL: An RRAS-like subfamily in the RAS family, RAL small GTPases typically function downstream of the RRAS effector RALGDS/RalGEF and are inhibited by RALGAP, a heterodimeric GAP structurally analogous to TSC1/2 that functions as a GAP for RHEB [3700, 3701]. The RAL subfamily includes mammalian RALA and RALB, Drosophila Rala, and C. elegans RAL-1. Mammalian RALB regulates exocytosis, the immune response and an anabolic/catabolic switch. In nutrient-rich conditions RALB-GTP binds EXOC2/Sec5 and EXOC8/Exo84, and through the latter associates with MTORC1 to promote anabolic metabolism [3702]. Under starvation conditions RALB-GTP nucleates phagophore formation through assembly of a ULK1-BECN1-PIK3C3 complex, also via interaction with the EXOC8/Exo84 protein [3117]. Although RALB direct activation and indirect inactivation (through MTORC1) of autophagy appears contradictory, RALB may function as a critical anabolic/catabolic switch in response to global and local nutrient contexts. RALB may be an analog of yeast Sec4 [3703]. See also EXOC2, Sec4/RAB40B and EXOC8.
RALGAP: A heterodimeric complex consisting of catalytic alpha and regulatory beta subunits, RALGAP inactivates RAL small GTPases. RALGAP is structurally analogous to the TSC1/2 GAP, and like TSC1/2 is phosphorylated and inhibited by AKT [3700, 3704]. An additional partner of the RALGAP complex, NKIRAS1/kappaB-Ras, also inhibits RAL function [3705].
RANS (required for autophagy induced under non-nitrogen-starvation conditions) domain: Also referred to as domain of unknown function 3608 (DUF3608; PFAM: PF12257, http://pfam.xfam.org/family/PF12257), this sequence in Iml1 is required for non-nitrogen starvation-induced autophagy [3289]. This domain is spread throughout the eukaryotes (see for example, http://pfam.xfam.org/family/PF1tabview=tab72257#tabview=tab7) and frequently reported in combination with a DEP (Dishevelled, Egl-10, and Pleckstrin) domain (PFAM: PF00610), which is also the case with Iml1 [3289]. See also non-nitrogen starvation (NNS)-induced autophagy.
Rapamycin/sirolimus: Allosteric TOR (in particular, TOR complex 1) inhibitor, which induces autophagy. TOR complex 2 is much less sensitive to inhibition by rapamycin.
RAPGEF3/EPAC1 (Rap guanine nucleotide exchange factor 3): RAPGEF3/EPAC1 and its downstream effector RAP2B are recruited to the Staphylococcus aureus-containing phagosome and involved in regulation of pore-forming toxin α-hemolysin (Hla)-induced autophagy through CAPN (calpain) activation [3706, 3707]. In cardiomyocytes, RAPGEF3, acting through the CAMK2B/CaMKKβ-AMPK pathway, promotes autophagy. Deletion of Rapgef3 in mice protects against ADRB/β-adrenergic receptor-induced cardiac remodeling and autophagy [3708].
RAPGEF4/EPAC2: Oleate stimulates autophagy in pancreatic β-cells by reducing intracellular cAMP. The effect of oleate is mediated by RAPGEF4/EPAC2, as oleate-mediated autophagy can be potentiated or suppressed by RAPGEF4 knockdown or activation, respectively [3709].
RAPTOR: See RPTOR.
Ras: See RRAS.
RASSF1/RASSF1A (Ras association domain family member 1): RASSF1 is a tumor suppressor and frequently inactivated by promoter hypermethylation in multiple types of cancers. RASSF1 enhances autophagy initiation by suppressing PI3K-AKT-MTOR through the Hippo pathway-regulatory component MST1 and promotes autophagy maturation by recruiting autophagosomes on RASSF1-stabilized acetylated microtubules through MAP1S [3710].
RavZ: A Legionella pathogen effector protein that is part of a survival strategy employed by intracellular pathogens to evade clearance through autophagic recognition [3711]. RavZ consists of a catalytic domain, a membrane-targeting domain, and flexible N- and C-terminal regions [3712]; the latter allows recognition of Atg8-family proteins via three LIR motifs (two at the N terminus and one at the C terminus) [1968]. RavZ specifically localizes on phagophores and irreversibly deconjugates Atg8-family proteins from phosphatidylethanolamine; RavZ functions as a cysteine protease that competes with the host protease ATG4B and cleaves the amide bond before the C-terminal glycine [1967]. RavZ-cleaved Atg8-family proteins cannot be relipidated nor utilized in the autophagy pathway [1968].
RB1-E2F1 (RB transcriptional corepressor 1-E2 transcription factor 1): RB1 is a tumor suppressor that promotes growth arrest, and protects against apoptosis. E2F1 regulates the transition from the G1 to the S phase in the cell cycle, and is a pro-apoptotic member of the E2F transcription family. In addition to controlling the cell cycle and apoptosis, the interaction between RB1 and E2F1 regulates autophagy; RB1 and E2F1 downregulate and upregulate BCL2, respectively, resulting in the induction of autophagy or apoptosis [937].
RB1CC1/FIP200 (RB1 inducible coiled-coil 1): A putative mammalian functional counterpart of yeast Atg17 and Atg11 [1376, 2782, 3713, 3714]. RB1CC1 is a component of the ULK1 complex [699]. In addition, RB1CC1 interacts with other proteins in several signaling pathways, suggesting the possibility of autophagy-independent functions, and a potential role in linking other cellular functions and signaling pathways to autophagy.
RCB (Rubisco-containing body): Generated by the budding of chloroplast, stromal protein-containing material into distinct autophagic vesicles [1241].
RCD: See regulated cell death.
Reactive oxygen species (ROS): Reactive molecules that contain oxygen, including hydrogen peroxide, the hydroxyl radical OH·, and the superoxide radical O2·−. Hydrogen peroxide transiently inhibits delipidation of LC3 by ATG4, which is permissive for starvation-induced autophagy [767]. Superoxide is essential for triggering injury-induced mitochondrial fission and mitophagy [1137]. ROS are also essential for induction of LAP [2446].
ref(2)P: The Drosophila homolog of SQSTM1.
Regulated cell death (RCD): A form of cell death that results from the activation of one or more signal transduction pathways. RCD insures organismal homeostasis in both physiological and pathological conditions, being involved in two opposed scenarios. First, RCD can occur in the absence of any exogenous perturbation in physiological conditions. This form is related to physiological programs during development or tissue turnover and hence is referred to as programmed cell death (PCD).The second scenario is caused by perturbations in intracellular or extracellular microenvironments, and is refered to as stress-driven RCD [1266].
Residual body: A lysosome that contains indigestible material such as lipofuscin [3715].
ReSiN (redox-responsive silica nanoprobe): A biocompatible, plant cell wall-penetrable, biothiol-responsive silica nanoprobe for the fluorescence imaging of starvation-induced vesicle trafficking [3716].
Resveratrol: An allosteric activator of SIRT1 and inhibitor of several other cellular proteins [2627] that induces autophagy [3717].
Reticulophagy: The selective degradation of ER by an autophagy-like process [3718]. Autophagy counterbalances ER expansion during the unfolded protein response. Activation of the UPR in yeast induces reticulophagy.
RETREG1/FAM134B (reticulophagy regulator 1): An ER-resident receptor that functions in reticulophagy through interaction with LC3 and GABARAP [74]. RETREG1 also participates in ER-to-lysosome-associated degradation (ERLAD) of proteasome-resistant misfolded proteins from the ER [1379, 1380]. See also ERLAD.
RGS19/GAIP (regulator of G protein signaling 19): A GTPase activating protein that inactivates GNAI3 (converting it to the GDP-bound form) and stimulates autophagy [3719]. See also GNAI3.
RHEB (Ras homolog, mTORC1 binding): A small GTP-binding protein that activates MTOR when it is in the GTP-bound form [377].
RHOT1/Miro1 (ras homolog family member T1): Member of the mitochondrial Rho family of proteins implicated in the regulation of mitochondrial trafficking [3720, 3721]. RHOT1 is an early target of PRKN ubiquitination and degradation after mitochondrial damage [3722, 3723], which is proposed to be a mechanism for uncoupling mitochondria from the transport machinery. In addition, RHOT1 is implicated in the regulation of mitophagy [3724].
RHOT2/Miro2 (ras homolog family member T2): Member of the mitochondrial Rho family of proteins which has been implicated in the regulation of mitophagy by recruiting and serving as a docking site for PRKN on damaged mitochondria [3724].
Ribophagy: The selective sequestration and degradation of ribosomes by a autophagy-like process [1389].
Ribosomopathy: A diverse class of diseases connected to impaired ribosome biogenesis. Patients carry haploinsufficiency mutations in ribosomal proteins or ribosome biogenesis factors. Common phenotypes include anemia, craniofacial cartilage defects and an elevated cancer risk. Classically, TP53 activation is observed as a consequence of nucleolar stress induction. Recent studies report an implication of autophagy in ribosomopathy models [2027].
RILP (Rab interacting lysosomal protein): A cytoplasmic dynein adaptor protein that controls autophagy progression and transport in more complex eukaryotic neurons as well as nonneuronal cells. RILP interacts sequentially through distinct sites with ATG5 on the phagophore, LC3 on the autophagosome, and RAB7 on the late endosome/amphisome. Dynein displaces ATG5 from autophagosomes post-closure, to ensure transport of only fully formed structures. MTOR inhibition upregulates RILP expression, dramatically stimulating its recruitment to autophagosomes and subsequent autophagic clearance [3725].
Rim15: A yeast kinase that regulates transcription factors in response to nutrients. Rim15 positively regulates autophagy and is negatively regulated by several upstream kinases including TOR, PKA, Sch9 and Pho85 [2995, 3726].
RIPK1 (receptor interacting serine/threonine kinase 1): RIPK1 inhibits basal autophagy independent of its kinase function, through activation of MAPK1/3 and inhibition of TFEB [3727].
Rkr1: A yeast ubiquitin ligase that antagonizes ribophagy [1390].
RNautophagy: An autophagic process by which RNA is transported across the lysosomal membrane and degraded [1397]. The translocation process is mediated by lysosomal integral membrane proteins LAMP2C and SIDT2 [1397, 1400]. See also DNautophagy, LAMP2C and SIDT2.
RNASET2/RNS2 (ribonuclease T2): A conserved class II RNase of the T2 family that localizes to the lumen of the ER (or an ER-related structure) and vacuole in Arabidopsis, and to lysosomes in zebrafish and nematodes; RNASET2 is involved in rRNA turnover, and rns2 mutants display constitutive autophagy, likely due to a defect in nucleoside homeostasis [3728–3731].
RNF216 (ring finger protein 216): An E3 ubiquitin ligase that mediates the ubiquitination and the subsequent degradation of BECN1, thus acting as a negative regulator of autophagy [3732].
Rny1: A yeast vacuolar RNase that hydrolyzes RNA that has been delivered to the vacuole via autophagy into 3ʹ nucleotides [3572]. See also Pho8.
ROCK (Rho associated coiled-coil containing protein kinase): ROCK is a serine/threonine kinase and a major downstream effector of the small GTPase RHOA. The ROCK family consists of two isoforms, ROCK1 and ROCK2. ROCK1 activates BECN1-mediated autophagy initiation through phosphorylating BECN1 in the starvation-stimulated autophagic response [3733, 3734] or doxorubicin-induced autophagic response in mouse hearts [3735]. In addition, due to the impact of ROCKs on actin cytoskeleton dynamics, ROCK activity can activate or inhibit autophagy through multiple mechanisms depending on the cellular context [3734].
Rpd3: A yeast histone deacetylase that negatively regulates the expression of ATG8 [2011]. See also Sin3/SIN3 and Ume6.
Rph1: A histone demethylase that negatively regulates the expression of ATG7; demethylase activity is not required for transcriptional repression [912, 913].
RPN10: A component of the 26S proteasome lid. In its free form, RPN10 acts as a receptor that links ubiquitinated (inactive) 26S proteasomes to ATG8 during proteaphagy in Arabidopsis [1352]. Binding to ATG8 is mediated by a UIM-like sequence [1359]. See also proteaphagy, UIM, and UDS.
RPS6KB1/p70S6 kinase/S6K1 (ribosomal protein S6 kinase B1): A substrate of MTORC1, in mammalian cells RPS6KB1/2 inhibits INSR (insulin receptor), which in turn causes a reduction in the activity of the class I PI3K and subsequently MTORC1; this may represent a feedback loop to help maintain basal levels of autophagy [1876, 1974]. Conversely, under conditions of long-term starvation RPS6KB1/2 levels may fall sufficiently to allow reactivation of MTORC1 to prevent excessive autophagy. In Drosophila, the RPS6KB1/2 ortholog S6k may act in a more direct manner to positively regulate autophagy [377]. In human cells, RPS6KB1 is indispensable for autophagosome maturation during stress conditions [3736]. RPS6KB1 interacts with MAP3K7 and suppresses its activity; RPS6KB1 suppression activates MAP3K7 resulting in subsequent AMPK activation, leading to autophagy induction [3737–3740]. See also MAP3K7.
RPS6KB2: See RPS6KB1.
RPTOR/raptor (regulatory associated protein of MTOR complex 1): A component of MTORC1. RPTOR interacts with ULK1, allowing MTORC1 to phosphorylate both ULK1 and ATG13, and thus repress ULK1 kinase activity and autophagy [698, 700, 701]. This interaction also permits a negative feedback loop to operate, whereby ULK1 phosphorylates RPTOR to inhibit MTORC1 activity [711, 718].
RRAG (Ras related GTP binding): A GTPase that activates MTORC1 in response to amino acids [3741]. There are RRAGA, B, C and D isoforms.
RRAS/RAS (RAS related): The small GTPase RRAS is an oncogene involved in the regulation of several cellular signaling pathways. RRAS can upregulate or downregulate autophagy through distinct signaling pathways that depend on the cellular contexts [3742].
Rsp5: A yeast E3 ubiquitin ligase that is responsible for the autophagic clearance of certain cytosolic proteins via Cue5 [627]. Rsp5 also triggers mitophagy through the ubiquitination of the ERMES component Mdm34 [3108]. See also Cue5, ERMES and Mdm34.
RTN3 (reticulon 3): A receptor that functions in starvation-induced mammalian reticulophagy [1371].
RUBCN/Rubicon/KIAA0226 (rubicon autophagy regulator): RUBCN is part of a PtdIns3K complex (RUBCN-UVRAG-BECN1-PIK3C3-PIK3R4) that localizes to the late endosome/lysosome, inhibits autophagy, and promotes LC3-associated phagocytosis [798, 801].
RUBCNL/C13orf18/Pacer (rubicon like autophagy enhancer): A component of the PtdIns3K and UVRAG-HOPS complexes that targets them to the autophagosome to positively regulate autophagosome maturation; RUBCNL binds to STX17 on the autophagosome [2667]. RUBCNL acts in an opposite manner to that of RUBCN. MTORC1 phosphorylates RUBCNL at S157 to negatively regulate its activity in autophagosome maturation [3743]. RUBCNL appears to be an autophagy protein involved in ALS pathogenesis [3744].
RUNX2 (RUNX family transcription factor 2): A transcriptional regulator essential for osteoblastic cell differentiation, RUNX2 promotes autophagy and autophagosome trafficking in metastatic cancer breast cells by increasing acetylation of the α-tubulin subunits of microtubules [3745].
RYR1/2/3 (ryanodine receptor 1/2/3): RYRs are the second major group of large intracellular calcium-release channels besides ITPRs that are located at the ER (or sarcoplasmic reticulum in muscle cells). Spontaneous RYR-mediated calcium-release events suppress basal autophagic flux at the level of the lysosomes in a variety of cell systems [3746].
SAHA/vorinostat (suberoylanilide hydroxamic acid): An HDAC inhibitor that induces autophagy [3747]; however, SAHA/vorinostat treatment has also been reported to suppress autophagy (e.g., see ref. [3748]), suggesting context dependency. For example, the presence of the TP53 protein is essential for the suppression of SAHA-induced autophagy in several tumor cell lines [3749]. See also TP53/p53.
Saikosaponin d: An ATP2A/SERCA inhibitor that induces autophagy and autophagy-dependent cell death in apoptosis-defective cells [1909].
Sanguinarine (SNG): A benzophenanthridine alkaloid isolated from Sanguinaria canadensis, that exhibits anticancer activity against a wide variety of cancer cells, both in vitro and in vivo [3750]. SNG is highly effective in suppressing the growth of human malignant glioma cells via inducing autophagic cell death, which is associated with ROS-dependent upregulation of MAPK1/3 activity [3751].
SASP (senescence-associated secretory phenotype): The secretome of senescent cells. In the case of autophagy-deficient prematurely senescent murine melanocytes, the SASP includes oxidized phospholipid signaling mediators [3752].
SBF2/MTMR13 (SET binding factor 2): A catalytically inactive myotubularin that is also a RAB21 guanine nucleotide exchange factor (GEF) required with RAB21 for autophagosome-lysosome fusion. Starvation induces SBF2 RAB21 GEF activity that promotes VAMP8 trafficking to the lysosome, where VAMP8 is needed to mediate fusion. See also RAB21 [3690]. The Drosophila homolog is Sbf.
Sch9: A yeast kinase that functions in parallel with PKA to negatively regulate autophagy. Sch9 appears to function in parallel with TOR, but is also downstream of the TOR kinase [3726].
SCOC (short coiled-coil protein): A protein in the Golgi that interacts with FEZ1 in a complex with either ULK1 or UVRAG; the ternary complex with ULK1 promotes autophagy, whereas the complex with UVRAG has a negative effect by sequestering the latter from the BECN1-containing PtdIns3K complex [3132]. See also FEZ1.
SEA (Seh1-associated) protein complex: A complex found in yeast that includes the Seh1 nucleoporin and the COPII component Sec13 (also a nucleoporin), in addition to Npr2 and Npr3, and four other relatively uncharacterized proteins; the SEA complex associates with the vacuole, potentially acting as a membrane coat and is involved in protein trafficking, amino acid biogenesis, and the starvation response including autophagy [3753].
Seasonality-dependent autophagy: The autophagy pathway is influenced by seasonality in reproduction. The highest intensity of gene and protein expression is observed in the period after reproductive activity, which reflects cell protection and control of the normal spermatogenesis process [3754].
Sec1: Functions with the plasma membrane SNAREs Sso1, Sso2 and Sec9 to form the site for vesicle-mediated exocytosis; as with Sso1/Sso2 and Sec9, temperature sensitive sec1 mutations also abrogate autophagic delivery of GFP-Atg8 [3755]. See also Sso1/Sso2.
Sec2: A guanine nucleotide exchange factor for Sec4 that normally functions in exocytosis. Upon the induction of autophagy, Sec2 function is diverted to promote membrane delivery to the PAS [3703].
Sec4: A Rab family GTPase that normally functions in exocytosis; under autophagy-inducing conditions yeast Sec4 is needed for the anterograde movement of Atg9 to the PAS [3703]. The mammalian homolog is RAB40B.
SEC5L1: See EXOC2.
Sec9: Plasma membrane SNARE light chain that forms a complex with Sso1/Sso2 to generate the target complex of vesicle exocytosis; as with Sso1/Sso2, loss of Sec9 function blocks autophagy at an early stage by disrupting targeting of Atg9 to the Atg9 peripheral sites and PAS [3755]. See also Sso1/Sso2 and Atg9 peripheral sites/structures.
Sec18: Homolog of mammalian NSF, an ATPase globally responsible for SNARE disassembly. Loss of function inhibits SNARE-dependent early and late events of autophagy (i.e., vesicular delivery of Atg9 to the Atg9 peripheral sites and PAS [3755] and fusion of autophagosomes with the vacuole [3756]). See also Atg9 peripheral sites/structures.
Sec22: A vesicle SNARE involved in ER and Golgi transport; mutations in Sec22 also block Atg9 trafficking to the Atg9 peripheral sites and PAS. Crosslinking experiments suggest Sec22 may be the v-SNARE responsible for the autophagy functions of the ordinarily plasma membrane Sso1/Sso2-Sec9 t-SNARE complex [3755]. See also Sso1/Sso2 and Atg9 peripheral sites/structures.
SEC24C (SEC24 homolog C, COPII coat complex component): A mammalian paralog of yeast Lst1, which forms a COPII cargo adaptor complex with Sec23, which is essential for reticulophagy [1382]. COPII plays a role in providing membrane for autophagosome biogenesis.
SEC62: An ER-resident LC3-binding protein that functions in mammalian reticulophagy during recovery from ER stress [1375, 3757].
Secretory autophagy: A biosynthetic mode of autophagy that occurs in mammalian cells [1700, 3758]. Secretory autophagy depends on the ATG proteins, RAB8A and the Golgi protein GORASP2/GRASP55, and is used for the extracellular delivery (via unconventional secretion) of proteins such as the cytokines IL1B and IL18, and HMGB1. ATG4B has also been described to be required for secretory autophagy in two different cell types: epithelial cells of the inner ear during its development, and Paneth cells [3759–3761]. In the intestine, Paneth cells use secretory autophagy to secrete lysozyme in order to maintain host defense during bacterial invasion. This process is activated in Paneth cells by the ER stress response pathway and signals from the innate immune system; secretory autophagy is defective in mice carrying a Crohn disease-associated variant in Atg16l1 [3762, 3763]. See also exophagy, GORASP2.
SEFAGIA (Sociedad Española de Autofagia): Started as an intiative of Jose Luis Crespo and Patricia Boya, the Spanish Autophagy Society has held annual meetings since 2013. See https://autofagia.org.
Selenite: A naturally occurring sulfate mineral that induces mitophagy through a mechanism that involves the E3 ubiquitin-protein ligase MUL1 (mitochondrial E3 ubiquitin protein ligase 1) [3764].
Senescence: Cellular senescence is a terminal arrest of cell proliferation. Depending on the context, autophagy can act as an activator or a repressor of senescence [3664, 3765]. For example, autophagy contributes to the senescence onset in cancer cells exposed to the 20A G-quadruplex ligand [2832].
SENP3 (SUMO specific peptidase 3): A SUMO2- and SUMO3-specific deSUMOylation enzyme that regulates both mitophagy and autophagy [3766].
SEPA-1 (suppressor of ectopic P granule in autophagy mutants-1): A C. elegans protein that is involved in the selective degradation of P granules through an autophagy-like process [1276]. SEPA-1 self-oligomerizes and functions as the receptor for the accumulation of PGL-1 and PGL-3 aggregates. SEPA-1 directly binds PGL-3 and LGG-1.
Septins: Septins are GTP-binding proteins that assemble into nonpolar filaments (characterized as unconventional cytoskeleton), often acting as scaffolds for the recruitment of other proteins. Septin cages form in response to infection by Shigella, and serve to recruit autophagy components such as SQSTM1 and LC3 [3767]. In yeast, septins form ring-like structures around Atg8 and are involved in autophagosome biogenesis [810]. Under autophagy conditions, septins form multiple puncta that colocalize with several organelles such as mitochondria, the Golgi apparatus, plasma membrane, and endosomes [3768].
Sequestration membrane: See Phagophore.
SERPINA1/A1AT (serpin family A member 1): SERPINA1 is the most abundant circulating protease inhibitor and is synthesized in the liver. A point mutation in the SERPINA1 gene alters protein folding of the gene product, making it aggregation prone; the proteasomal and autophagic pathways mediate degradation of mutant SERPINA1 [3769].
sesB (stress-sensitive B): A Drosophila mitochondrial adenine nucleotide translocase that negatively regulates autophagic flux, possibly by increasing cytosolic ATP levels [3043]. See also Dcp-1.
SESN2 (sestrin 2): A stress-inducible protein that reduces oxidative stress, inhibits MTORC1 and induces autophagy, also acting as an AMPK activator [3770, 3771]. SESN2 physically associates with ULK1 and SQSTM1, promotes ULK1-dependent phosphorylation of SQSTM1, and facilitates autophagic degradation of SQSTM1 targets such as KEAP1 [2668, 3491]. SESN2-mediated ULK1 activation also upregulates mitophagy through BECN1 phosphorylation and mitochondrial translocation of PRKN [3772]. SESN2-bound SQSTM1 promotes its association with ubiquitinated mitochondria, thereby activating mitophagy [3773]. SESN2 suppresses MTORC1 in response to diverse stresses including DNA damage [3774], ER stress [3775], nutritional stress [1293, 3776] or energetic stress [3777].
SET/IPP2A2 (SET nuclear proto-oncogene): SET is a regulator of intracellular redox state by controlling APEX1/APE1 (apurinic/apyrimidinic endodeoxyribonuclease 1) localization with an impact on autophagy and cell survival [3778].
SEX chlorophagy (starch excess-associated chloroplast autophagy): A vacuolar degradation process for clearance of dysfunctional chloroplasts in plants caused by excessive leaf starch accumulation, which is independent of several classical ATG genes, such as ATG5, ATG6 and ATG7 [3779].
SH3BP4 (SH3 domain binding protein 4): SH3BP4 is a negative regulator of amino acid-RRAG GTPase-MTORC1 signaling. SH3BP4 binds to the inactive RRAG GTPase complex through its Src homology 3 (SH3) domain under conditions of amino acid starvation and inhibits the formation of an active RRAG GTPase complex [3780]. SH3BP4 is also a WNT signaling target gene acting as a negative feedback regulator of WNT signaling through modulating CTNNB1/β-catenin’s subcellular localization through its ZU5 domain [3781].
SH3GL2/Endophilin A1 (SH3 domain containing GRB2 like 2, endophilin A1): See EndoA.
SH3GLB1/Bif-1 (SH3 domain containing GRB2 like, endophilin B1): A protein that interacts with BECN1 via UVRAG and is required for autophagy. SH3GLB1 has a BAR domain that may be involved in deforming the membrane as part of autophagosome biogenesis [3782]. SH3GLB1 activity is regulated by phosphorylation at residue T145, which in starved neurons occurs via CDK5 [2952]. SH3GLB1 regulates autophagic degradation of EGFR [3783], NTRK1 [2952], and CHRNA1 [2940, 3784]. Turnover of CHRNA1 is coregulated by TRIM63 and CDK5 [2940, 3784].
SH3P2/AT4G34660 (SH3 Domain-Containing Protein 2): A. thaliana SH3P2 contains a BAR domain and functions in mediating autophagosome formation. Particularly, SH3P2 forms a complex with the PtdIns3K components and interacts with ATG8 [2586]. Upon autophagic induction, SH3P2 is translocated to the phagophore membrane, whereas knockdown of SH3P2 may disrupt the delivery of autophagosomes into the vacuole in plant cells.
SH3PX1 (SH3 and PX domain containing 1): The ortholog of human SNX9, SNX18, and SNX33. In Drosophila adult midgut, SH3PX1 restrains intestinal stem cell division through an endocytosis-autophagy network that includes shi/dynamin, Rab5, Rab7, Atg1, Atg5, Atg6, Atg7, Atg8a, Atg9, Atg12, Atg16 and Syx17. Upon SH3PX1-dependent autophagy loss of function, the ligand-bound Egfr will be recycled to the cell surface via Rab11-endosomes rather than degraded via autophagosomes. This hyperactivates rl/MAPK/ERK and autonomously stimulates intestinal stem cell proliferation [2144, 3785].
SHC1/p66SHC (SHC adaptor protein 1): The 66-kDa isoform of the Src homology 2 domain containing (SHC) family of protein adaptors. Mitochondria-associated SHC1 primes B lymphocytes for autophagy by inducing an ROS-dependent dissipation of the mitochondrial transmembrane potential, and ensures degradation of depolarized mitochondria through its ability to interact with LC3-II and active AMPK [3786].
Shear stress: Shear stress is induced by the blood flow in the circulatory system, and by the urinary flow in the kidney tubules. Shear stress-dependent autophagy is atheroprotective in endothelial cells [3787] and regulates epithelial cell size and ATP production in kidney proximal tubules [3634].
SHH (sonic hedgehog signaling molecule): A ligand of the sonic hedgehog pathway. Activation of this pathway suppresses IFNG-induced autophagy in macrophages during mycobacterial infection [783].
Shp1/Ubx1: A yeast Ubx (ubiquitin regulatory x)-domain protein that is needed for the formation of autophagosomes during nonselective autophagy; Shp1 binds Cdc48 and Atg8–PE, and may be involved in extracting the latter during phagophore expansion [2948].
Sic1: A yeast cyclin-dependent kinase inhibitor that blocks the activity of Cdc28-Clb kinase complexes to control entry into the S phase of the cell cycle. Sic1 is a negative regulator of autophagy that inhibits Rim15 [2995].
SIDT2 (SID1 transmembrane family member 2): The lysosomal vertebrate ortholog of the C. elegans RNA channel SID-1. SIDT2 is a transmembrane protein that mainly localizes to the lysosomal membrane and mediates the transport of nucleic acids into lysosomes in the processes of RNautophagy and DNautophagy [1399–1401]. See also LAMP2C.
Signalphagy: A type of autophagy that degrades active signaling proteins [3788].
Sin3/SIN3: Part of the Rpd3L regulatory complex including Rpd3 and Ume6 in yeast, which downregulates transcription of ATG8 in growing conditions [2011]. In mammalian cells knockdown of both SIN3A (SIN3 transcription regulator family member A) and SIN3B is needed to allow increased expression of LC3. See also Rpd3 and Ume6.
Sirolimus: An immunosuppressant also referred to as rapamycin. See also rapamycin.
SIRT1 (sirtuin 1): A NAD+-dependent protein deacetylase that is activated by caloric restriction or glucose deprivation; SIRT1 can induce autophagy through the deacetylation of autophagy-related proteins and/or FOXO transcription factors [3789]. Deacetylation of K49 and K51 of nuclear LC3 leads to localization in the cytosol and association with phagophores [1004]. See also SIRT2.
SIRT2 (sirtuin 2): A NAD+-dependent protein deacetylase sharing homology with SIRT1 that is involved in neurodegeneration and might play a role in autophagy activation through regulation of the acetylation state of FOXO1 [3144]. Under prolonged stress, the SIRT2-dependent regulation of FOXO1 acetylation is impaired, and acetylated FOXO1 can bind to ATG7 in the cytoplasm and directly affect autophagy.
SIRT3 (sirtuin 3): A mitochondrial NAD+-dependent protein deacetylase sharing homology with SIRT1, which is responsible for deacetylation of mitochondrial proteins and modulation of mitophagy [3790, 3791].
SIRT5: A mitochondrial SIRT1 homolog with NAD+-dependent protein desuccinylase/demalonylase activity; SIRT5 modulates ammonia-induced autophagy [3792].
SIRT6: A member of the sirtuin family with nuclear localization, that is associated with chromatin and promotes the repair of DNA. The involvement of SIRT6 in senescence has been proposed, possibly by the modulation of IGF-AKT signaling; a role for SIRT6 in autophagy linked to senescence has been determined [3793].
SIRT7: A member of the sirtuin family that is highly expressed in the nucleus/nucleolus where it interacts with POLR1 (RNA polymerase I) as well as with histones. Many lines of evidence point to a role for SIRT7 in oncogenic transformation and tumor growth. The involvement of SIRT7 in autophagy was recently suggested in a model of acute cardiovascular injury, where loss of SIRT7 activates autophagy in cardiac fibroblasts [3793].
SLAPs (spacious Listeria-containing phagosomes): SLAPs can be formed by L. monocytogenes during infection of macrophages or fibroblasts if bacteria are not able to escape into the cytosol [3794]. SLAPs are thought to be immature autophagosomes in that they bear LC3 but are not acidic and do not contain lysosomal degradative enzymes. The pore-forming toxin listeriolysin O is essential for SLAPs formation and is thought to create small pores in the SLAP membrane that prevent acidification by the V-ATPase. SLAP-like structures have been observed in a model of chronic L. monocytogenes infection in immunocompromised mice [3795], suggesting that autophagy may contribute to the establishment/maintenance of chronic infection.
SLC1A5 (solute carrier family 1 member 5): A high affinity, Na+-dependent transporter for L-glutamine; a block of transport activity leads to inhibition of MTORC1 signaling and the subsequent activation of autophagy [452]. See also SLC7A5.
SLC3A2/CD98hc/4F2hc (solute carrier family 3 member 2): A transmembrane protein that acts as a heterodimerization partner and a chaperone for a group of L-type amino acid transporters from the SLC7 family by mediating their recruitment to the plasma membrane. These heterodimeric amino acid transporters have broad substrate specificity and regulate MTOR and autophagy [3796]. See also SLC7A5.
SLC7A5 (solute carrier family 7 member 5): A bidirectional transporter that allows the simultaneous efflux of L-glutamine and influx of L-leucine; this transporter works in conjunction with SLC1A5 to regulate MTORC1 [452].
SLC9A3R1 (SLC9A3 regulator 1): A scaffold protein that competes with BCL2 for binding to BECN1, thus promoting autophagy [3797].
SLC25A1 (solute carrier family 25 member 1): This protein maintains mitochondrial activity and promotes the movement of citrate from the mitochondria to the cytoplasm, providing cytosolic acetyl-coenzyme A. Inhibition of SLC25A1 results in the activation of autophagy and mitophagy [3798].
SLC38A9 (solute carrier family 38 member 9): A multi-spanning membrane protein that localizes to the lysosome as part of the RRAG-Ragulator complex. SLC38A9 functions as a transceptor (transporter-receptor) to link amino acid status with MTORC1 activity [3798–3800].
Slg1/Wsc1: A yeast cell surface sensor in the Slt2 MAPK pathway that is required for mitophagy [3800]. See also Slt2.
SLR (sequestosome 1/p62-like receptor): Proteins that act as autophagy receptors, and in proinflammatory or other types of signaling [3801].
Slt2: A yeast MAPK that is required for pexophagy and mitophagy [747]. See also Pkc1, Bck1 and Mkk1/2.
smARF (short mitochondrial ARF): A small isoform of CDKN2A/p19ARF that results from the use of an alternate translation initiation site, which localizes to mitochondria and disrupts the membrane potential, leading to a massive increase in autophagy and cell death [3802].
SNAP29 (synaptosome associated protein 29): A SNARE protein required for fusion of the completed autophagosome with a lysosome in metazoans [870, 871, 874]. SNAP29 is cleaved upon enterovirus infection, leading to impaired autophagosome-lysosome fusion [3803, 3804].
SNAPIN (SNAP associated protein): An adaptor protein involved in dynein-mediated late endocytic transport; SNAPIN is needed for the delivery of endosomes from distal processes to lysosomes in the neuronal soma, allowing maturation of autolysosomes [206].
SNCA/α-synuclein (synuclein alpha): A presynaptic protein relevant for PD pathogenesis due to the presence of several mutations and its deposition into Lewy bodies (intracellular protein and lipid aggregates), characteristic of this disease. SNCA degradation in neuronal cells involves the autophagy-lysosomal pathway via autophagy and chaperone-mediated autophagy [3805]. Conversely, SNCA accumulation over time might impair autophagy function, and an inhibitory interaction of SNCA with HMGB1 has been reported [3806]. This interaction can be reversed by the natural autophagy inducer corynoxine B. Similarly, in human T lymphocytes the aggregated form of SNCA, once generated, can be degraded by autophagy, whereas interfering with this pathway can result in the abnormal accumulation of SNCA. Hence, SNCA can be considered as an autophagy-related marker of peripheral blood lymphocytes [2226]. SNCA also impairs ferritinophagy in retinal pigment epithelial cells by inhibiting lysosomal function [3807]. Lysosomal dysfunction might be a result of SNCA-mediated rupture of lysosomes [3808, 3809], which can be suppressed by reduced INS-DAF-2 signaling in daf-2 mutant C. elegans [3033].
Snx4/Atg24: A yeast PtdIns3P-binding sorting nexin studied in H. sapiens, Trypanosoma brucei, yeast, and other fungi. It is involved in membrane trafficking, protein sorting and recycling and is important for mitophagy, pexophagy and the Cvt pathway [380, 1069, 2800, 3564, 3810–3812]. Through its interaction with Atg20 it is part of the Atg1 kinase complex. Snx4/Atg24 is also involved in recycling from early endosomes. In the filamentous fungus M. oryzae, Atg24 is required for mitophagy [1069].
SNX18 (sorting nexin 18): A PX-BAR domain-containing protein involved in phagophore expansion [3813].
SOD1/Cu,Zn-superoxide dismutase (superoxide dismutase 1): An enzyme responsible for conversion of the ROS superoxide into hydrogen peroxide. Numerous point mutations in SOD1 are associated with 20% of familial ALS. Evidence suggests that autophagy can degrade mutated SOD1 in vitro. Activated autophagy is observed in the spinal cord of SOD1G93A mice, and the G85R and G93A mutated forms lead to protein aggregation and activate autophagic flux in an NFKB-dependent manner [1107], indicating a possible role of mutated SOD1-induced autophagy in the pathogenesis of ALS.
SopF: A Salmonella T3SS effector that blocks the V-ATPase-ATG16L1 axis that critically mediates autophagic recognition of intracellular pathogens [1476]. SopF is an ADP-ribosyltransferase that modifies Q124 of ATP6V0C to prevent its interaction with, and recruitment of, ATG16L1 to bacteria-containing vacuoles.
Sorafenib: An antitumoral inhibitor of tyrosine kinase receptors whose sustained administration induces a shift from early induction of survival autophagic processes to apoptosis [2634].
SORT1 (sortilin 1): A related member of the VPS10 sorting receptor family that transports proteins across the plasma membrane or subcellular compartments. SORT1 transports soluble proteases into lysosomes. [3814].
SOX2 (SRY-box transcription factor 2): A key stem cell maintanence factor that promotes the development of squamous cell carcinomas. SOX2 promotes the degradation of an innate immune sensing adaptor, STING1, to encourage tumor evasion from immunosurveillance [3815].
SpeB: A cysteine protease secreted by Streptococcus pyogenes that degrades autophagy components at the bacterial surface, leading to autophagy escape [3816]. The lack of SpeB allows capture and killing of cytoplasmic S. pyogenes by the autophagy system [175, 3816].
Spautin-1 (specific and potent autophagy inhibitor-1): An inhibitor of USP10 and USP13, identified in a screen for inhibitors of autophagy, which promotes the degradation of the PIK3C3/VSP34-BECN1 complex [2635].
Spermidine: A natural polyamine that induces autophagy through the inhibition of histone acetylases such as EP300 [955, 1635], the inhibition of HDAC4 [956], and hypusination of the translation initiation factor EIF5A, which in turn controls the translation of TFEB [958].
SPG11 (SPG11 vesicle trafficking associated, spatacsin): Mutations in SPG11 are associated with a complicated form of hereditary spastic paraparesis (SPG11). SPG11 interacts with AP5 and ZFYVE26/spastizin to form a complex [3817]. SPG11 has a role in ALR, and protein loss results in the accumulation of autophagosomes and enlarged lysosomes in SPG11-mutated cells [3818]. See also autophagic lysosome reformation and ZFYVE26.
Sphingolipids: Sphingolipids are a major class of lipids characterized by the presence of the long-chain aliphatic aminoalcohol sphingosine (2-amino-4-trans-octadecene-1,3-diol) or sphinganine (2-amino-octadecane-1,3-diol). Sphingomyelin, the most abundant cellular sphingolipid, inhibits autophagosome formation [812], whereas its metabolites including ceramide and sphingosine-1-phosphate are positive regulators of autophagy [886, 3819–3822].
Sphingosine-1-phosphate (S1P): S1P is a signaling sphingolipid synthesized by SPHK (sphingosine kinase), an enzyme ubiquitously found in the cytosol [3823], endoplasmic reticulum or nuclear membranes [3824] of various types of cells. S1P is a key component of the “sphingolipid rheostat” that regulates cell fate by inducing autophagy to counter the death-promoting effects of ceramide [891, 3820, 3825], as well as important autophagy-dependent cell processes [888, 3826, 3827]. See also SPHK2.
SPHK2 (sphingosine kinase 2): SPHK2 phosphorylates sphingosine to S1P to maintain balance in sphingolipid metabolites. In mammals, there are two major isoforms of SPHK, SPHK1 and SPHK2. SPHK2 in neurons could activate autophagy and elicit neuroprotection against ischemic injury. Notably, SPHK2-mediated autophagy is independent of S1P. SPHK2 interacts with BCL2 via its BH3 domain to dissociate the BECN1-BCL2 complex, thereby promoting the release of BECN1 to activate autophagy [3828–3830].
SPM (specialized pro-resolving mediators): Super family of endogenous bioactive lipids formed in cells by the metabolism of omega-6 and omega-3 polyunsaturated fatty acids that actively promote the resolution of inflammation and restoration of tissue homeostasis [3831, 3832]. SPM induce autophagy by promoting LC3-I to LC3-II processing and the degradation of SQSTM1 as well as the formation of LC3-positive autophagosomes and autophagic vesicles [3833].
SPNS/spinster: A putative lysosomal efflux carbohydrate transporter required for autophagic lysosome reformation [3834].
SPP1/OPN/osteopontin (secreted phosphoprotein 1): SPP1 is an autophagy-enhancing glycoprotein that is widely expressed by bone, immune cells, smooth muscle, epithelial and endothelial cells, neurons, adipocytes and Kupffer cells. Nasal administration of recombinant SPP1 increases the expression of BECN1 and LC3 in neurons after subarachnoid hemorrhage, and attenuates early brain injury [3835].
SPRED2 (sprouty-related EVH1 domain containing 2): SPRED2 was initially described as an inhibitor of the receptor tyrosine kinase-RAS-RAF-MAPK pathway and more recently as an interaction partner of NBR1, SQSTM1, and LC3 both in transfected cells as well as in mouse heart lysates [563].
Spt4-Spt5: A transcription factor complex in S. cerevisiae that downregulates the expression of ATG8 and ATG41 during active growth; upon starvation, this inhibition is removed via Spt5 phosphorylation by the Sgv1/Bur1-Bur2 kinase [3836]. The mammalian homologs are SUPT4H1 and SUPT5H, and the kinase homolog is P-TEFb.
Sqa (spaghetti-squash activator): A myosin light chain kinase-like protein that is a substrate of Atg1 in Drosophila, and is required for starvation-induced autophagosome formation; the mammalian homolog DAPK3 is also involved in ATG9 trafficking [716].
SQST-1: The C. elegans homolog of SQSTM1.
SQSTM1/p62 (sequestosome 1): An autophagy receptor that links ubiquitinated proteins to LC3. SQSTM1 accumulates in cells when autophagy is inhibited. SQSTM1 interaction with LC3 requires a WXXL or a LIR motif analogous to the interaction of Atg8 with Atg19 [119]. SQSTM1 also interacts with HDAC6 to regulate microtubule acetylation and autophagosome turnover [3837]. The ZZ domain of SQSTM1 recognizes arginylated (Nt-R) substrates and mediates autophagic degradation of misfolded proteins [463, 3236, 3237, 3838]. The oligomerization of SQSTM1 via the N-terminal PB1 domain is a critical process for its function as an autophagy receptor [3237, 3839–3841]. In addition, SQSTM1 forms condensates with ubiquitinated proteins via the C-terminal UBA domain [3842, 3843]. In AML, SQSTM1 is essential for cell growth and survival, presumably due to autophagic degradation of mitochondria and ubiquinated proteins [569, 3844]. Finally, SQSTM1 is targeted for degradation during enterovirus infection [3804, 3845] to generate a fragment that is dominant-negative against the function of the native protein [3846]. See also HDAC6, LIR/LRS and Nt-R.
SRC (SRC proto-oncogene, non-receptor tyrosine kinase): A member of the family of non-receptor tyrosine kinases. Autophagy promotes the activation of SRC, and SRC phosphorylation of CTNNB1/β-catenin at Y654 in TGFB-induced epithelial-mesenchymal transition [3288].
SRPX/Drs (sushi repeat containing protein X-linked): An apoptosis-inducing tumor suppressor that is involved in the maturation of autophagosomes [3847].
SseL: A Salmonella deubiquitinating enzyme secreted by a type III secretion system; deubiquitination of aggregates and ALIS decreases host macrophage autophagic flux and results in an environment more favorable to bacterial replication [3848].
SSi6 (C23H30N4O7): A semisynthetic compound produced from structural modification made to the natural product [6]-gingerol (C17H26O4, CAS Number: 23513-14-6), by the nucleophilic addition of a 2,4-dinitrophenylhydrazine reagent. This compound can induce ROS in vitro, which leads to the activation of autophagy in the first hour of treatment followed by caspase-dependent apoptosis in longer treatment times in the triple-negative cell line MDA-MB-231 [3849].
Ssk1: A yeast component of the Hog1 signaling cascade that is required for mitophagy [747]. See also Hog1.
Sso1/Sso2: Highly homologous plasma membrane syntaxins (SNAREs) of S. cerevisiae involved in exocytosis; the Sso1/Sso2 proteins also control the movement of Atg9 to the Atg9 peripheral sites and PAS during autophagy and the Cvt pathway [3755].
STAT3 (signal transducer and activator of transcription 3): A transcription factor that also functions in the cytosol as a suppressor of autophagy [3850]. STAT3 binds EIF2AK2/PKR and inhibits the phosphorylation of EIF2S1.
Stationary phase lipophagy: A type of lipophagy that occurs in yeast cells entering quiescence [1856, 3851].
STED (stimulated emission depletion) nanoscopy: A powerful super-resolution instrument for capturing abundant nanoscopic spatial details. Super-resolution microscopy techniques offer subdiffraction limited resolution that is ten-fold better-quality compared to conventional confocal microscopy, and STED nanoscopy has contributed to new findings in autophagy [3092].
STIM1/2 (stromal interaction molecule 1/2): An ER calcium sensor. STIM1/2 is degraded as a result of proteasome inhibition-induced autophagy, whereas STIM2 is also degraded through ER stress-induced autophagy. Thus, proteasome inhibition or reduction of ER stress may modify the dendrite arbor, implicating dysfunction of the store-operated calcium channel at the early stage of neurodegeneration [3852, 3853].
STING1/STING/TMEM173: A highly conserved intracellular pattern recognition receptor that detects cytosolic nucleic acids and in turn activates type I interferon responses. STING1 activates anti-microbial autophagy downstream of bacterial and viral infections in mammals and Drosophila [1384, 3854, 3855]. STING1 is hypersensitive in autophagy-deficient cells and is linked to increased cell death in atg16l1-deleted intestinal epithelial cells [3856, 3857]. STING1 itself can also activate autophagic processes from the ERGIC in an IRF3-independent manner [1384, 3858]. STING1 possesses LIRs and interacts with LC3 to activate autophagy, resulting in self-degradation. STING1 also induces ATG5-dependent non-canonical autophagy. Poly (dA:dT) and cGAMP are activators of the STING1 pathway, and they increase LC3-II conversion. See also SOX2.
STK3/MST2 (serine/threonine kinase 3): The mammalian homolog of the Ste20/hpo (hippo) kinase, which can phosphorylate LC3B on Thr50; this modification is needed for the fusion of autophagosomes with lysosomes [3859].
STK4/MST1 (serine/threonine kinase 4): As with STK3, STK4 can phosphorylate LC3B, and is needed for the fusion of autophagosomes with lysosomes [3859]. STK4 also phosphorylates Thr108 of BECN1, promoting the interaction of BECN1 with BCL2 or BCL2L1, inhibiting autophagy [3860].
STK11/LKB1 (serine/threonine kinase 11): A kinase that is upstream of, and activates, AMPK [2955]. STK11/LKB1 regulates starvation-induced autophagy at the organismal level [2417].
STK26/MST4 (serine/threonine kinase 26): STK26 activates MAPK/ERK to induce cell growth and transformation. STK26 also phosphorylates ATG4B at serine residue 383, which stimulates ATG4B activity and increases autophagic flux [3861].
STK38/NDR1/trc (serine/threonine kinase 38): A kinase that promotes autophagy initiation in mammals and Drosophila by associating with BECN1/Atg6 [3862].
STO-609: STO-609 (7-oxo-7H-benzo[de]benzo[4,5]imidazo[2,1-a]isoquinoline-3-carboxylic acid) inhibits both the CAMKK1/CaMKKα and the CAMKK2/CaMKKβ isoforms, but CAMKK2 is more sensitive to this inhibitor. CAMKKs phosphorylate and activate CAMK1/CaMKI and CAMK4/CaMKIV, which directly activate transcription factors [3863–3865].
Stress-induced nascent granule degradation (SINGD): An autophagy-independent crinophagic pathway that counters autophagy through localized activation of MTORC1 in INS (insulin)-producing β cells. The SINGD pathway leads to the depletion of secretory granules together with the inhibition of autophagy, thus protecting against unwanted INS release during fasting [120]. Erroneous activation of the SINGD pathway contributes to β-cell failure in type 2 diabetes [1256]. See also crinophagy.
STX3 (syntaxin 3): A Qa SNARE plasma membrane syntaxin involved in the intracellular trafficking pathway for secretory autophagy [3866].
STX5 (syntaxin 5): A Golgi-localized SNARE protein involved in vesicular transport of lysosomal hydrolases, a process that is critical for lysosome biogenesis; STX5 is thus needed for the later stages of autophagy [2858].
STX12/STX13/STX14 (syntaxin 12): A genetic modifier of mutant CHMP2B in frontotemporal dementia that is required for autophagosome maturation; STX12 interacts with VTI1A [3867].
STX17 (syntaxin 17): An autophagosomal SNARE protein required for fusion of the completed autophagosome with an endosome or lysosome in metazoans [870, 871]. STX17 also recruits PtdIns3K complex to the ER-mitochondria contact sites through interaction with the complex component ATG14 [872, 873], but the interaction between STX17 and ATG14 is also important for autophagosome-lysosome fusion [3868]. In fed cells, the interaction of STX17 with ATG14 is prevented by MAP1B/LC1 with phosphorylation at Thr217, and starvation causes the dephosphorylation of MAP1B, allowing STX17 to dissociate from MAP1B and associate with ATG14 [3869]. In autophagosome formation, STX17 interacts not only with the PtdIns3K complex but also with several ULK1 complex components [877, 3870]. During autophagosome maturation, the PtdIns3K complex remains associated with autophagosomes via STX17 with the help of RUBCNL/C13orf18/Pacer [3743]. For autophagosome fusion with lysosomes, STX17 recruits the HOPS complex [3225, 3226] perhaps through interaction with the SM protein VPS33 [3871]. STX17’s cognate was originally reported to be VAMP8, but other studies suggest that its cognate is probably VAMP7 [3871, 3872]. STX17 function is intricately regulated by interactions with many binding proteins such as ULK1 [3870], IRGM [3873], ANXA2 [3874] and the IAP family member BIRC6/BRUCE [2885]. STX17 also participates in PRKN-dependent and -independent mitophagy [878, 879], fusion of mitochondria-derived vesicles with lysosomes [3875], mitochondrial fission by interacting with DNM1L and PGAM5 [873, 878], and lipid droplet formation [3876]. After autophagosome-lysosome fusion, STX17 is retrieved from autolysosomal membranes in a TLR9-induced clathrin-dependent vesicle budding process [3528]. See also PGAM5.
STYK1 (serine/threonine/tyrosine kinase 1): A member of the receptor tyrosine kinase (RTK) family that acts as a pro-autophagic molecule by promoting the assembly of the class I PI3K complex and its kinase activity; STYK1 also phosphorylates BECN1 as a substrate [3877].
Sui2: The yeast homolog of EIF2S1/eIF2α.
Sulforaphane: A bioactive isothiocyanate from Brassicaceae plants that induces autophagy in a variety of in vitro and in vivo models [1905, 2636, 2637, 3878–3880].
Sulforaphene: Sulforaphene (LFS-01) is obtained from Raphanus sativus, a medicinal plant, and it concomitantly induces both mitophagy and apoptosis in lymphoma cells through selective upregulation of SQSTM1 [3881].
SUPT20H/FAM48A (SPT20 homolog, SAGA complex component): A protein that interacts with the C-terminal domain of ATG9; this interaction is negatively regulated by MAPK14 [3882].
Sunitinib: An autofluorescent multitarget tyrosine kinase inhibitor with lysosomotropic properties; sunitinib interferes with autophagic flux by blocking trafficking to lysosomes [3883]. In contrast to lower (tolerable) doses that are associated with impeded autophagy, cytotoxic doses of sunitinib trigger autophagy [3884, 3885].
Symbiophagy: A process in which invertebrates such as the coralline demosponge Astrosclera willeyana degrade part of their symbiotic bacterial community, as part of a biomineralization pathway that generates the sponge skeleton [2293].
Synaptic autophagy: The selective autophagic degradation of synaptic components (including synaptic proteins, synaptic vesicles, postsynaptic receptors and organelles such as mitochondria) at the synapse to safeguard synaptic and neuronal homeostasis [3886–3893].
Synj (Synaptojanin): Synj, the Drosophila homolog of human SYNJ1 is required for autophagy. A mutation in the SAC1 phosphatase domain of Synj leads to the accumulation of Atg18a/WIPI2 on nascent synaptic autophagosomes, blocking autophagosome maturation at Drosophila synapses and in neurites of human patient induced pluripotent stem cell-derived neurons [3894].
Syx13 (Syntaxin 13): The Drosophila homolog of human STX12 that is required for autophagosome maturation [3867].
TAB2 (TGF-beta activated kinase 1 [MAP3K7] binding protein 2): MAP3K7-binding protein that, in the MAP3K7-free state, binds to BECN1 and inhibits autophagy [3895, 3896]. Upon autophagy induction, TAB2 dissociates from BECN1 and binds MAP3K7. The TAB2-containing MAP3K7 complex also promotes autophagy through multiple signaling cascades.
TAB3 (TGF-beta activated kinase 1 [MAP3K7] binding protein 3): TAB2 homolog, with cellular functions similar to TAB2. See also TAB2.
TAG (Targeting by AutophaGy proteins): targeting of IFN-induced effectors onto pathogen-containing vacuoles by the autophagic ubiquitin-like conjugation system, including ATG7, ATG3, and the ATG12–ATG5-ATG16L1 complex [2468, 3897].
TAK1: See MAP3K7.
TAKA (transport of Atg9 after knocking out ATG1) assay: An epistasis analysis that examines the localization of Atg9-GFP in a double mutant, where one of the mutations is a deletion of ATG1 [106]. In atg1∆ mutants, Atg9-GFP is restricted primarily to the PAS; if the second mutation results in a multiple puncta phenotype, the corresponding protein is presumably required for anterograde transport of Atg9 from peripheral sites to the PAS [1088]. This analysis can be combined with localization of RFP-Ape1 to determine if any of the Atg9-GFP puncta reach the PAS, in which case that punctum would colocalize with the RFP-Ape1 PAS marker.
Tamoxifen (TAM): A triphenylethylenic compound widely used for the management of estrogen receptor-positive breast cancers. This drug is a dual modulator of ESR (estrogen receptor) and a high affinity ligand of the microsomal antiestrogen binding site (AEBS). TAM induces protective autophagy in cancer cells through an AEBS-mediated accumulation of zymostenol (5α-cholest-8-en-3β-ol) [2031, 3553, 3898]. TAM is considered as an agonist of membrane G protein-coupled estrogen receptors (GPER). TAM was reported to induce autophagy in leukemic T cells (Jurkat), negative for both ESR1/ERα and ESR2/ERβ nuclear estrogen receptors, in a GPER-dependent manner [3899].
TARDBP/TDP-43 (TAR DNA binding protein): A DNA/RNA binding protein that stabilizes Atg7 mRNA [3900]. Mutations in TARDBP are found in ALS, and deposition of this protein is a characteristic neuropathology in ALS-FTD.
TASCC (TOR-autophagy spatial coupling compartment): A compartment located at the trans-Golgi where autolysosomes and MTOR accumulate during RRAS-induced senescence to provide spatial coupling of protein secretion (anabolism) with degradation (catabolism); for example, amino acids generated from autophagy would quickly reactivate MTOR, whereas autophagy would be rapidly induced via MTOR inhibition when nutrients are again depleted [782].
TAX1BP1/CALCOCO3 (Tax1 binding protein 1): An autophagy receptor that contains a SKIP carboxyl homology (SKICH) domain, a LIR motif and a C-terminal double zinc-finger ubiquitin binding domain [3901]. TAX1BP1 interacts with ubiquitinated substrates, such as S. typhimurium, and recruits LC3-positive autophagosomal membranes [1471, 3465, 3902]. TAX1BP1 also regulates autophagosome maturation [3903].
Tax4: See Irs4 [3308].
TBC1D7 (TBC1 domain family member 7): This protein is the third functional subunit of the TSC1-TSC2 complex upstream of MTORC1. Loss of function of TBC1D7 results in an increase of MTORC1 signaling, delayed induction of autophagy and enhancement of cell growth under poor growth conditions [3904]. Mutations in TBC1D7 have been associated with intellectual disability, macrocrania, and delayed autophagy [3905, 3906].
TBC1D9B (TBC1 domain family member 9B): This protein was found to interact with several members of the mammalian Atg8 homologs, including LC3 in mammalian cells [3907]. The Atg8-family protein interaction domain in TBC1D9B is different from the LIR previously detected in other LC3-interacting molecules. TBC1D9B can be found in colocalization with LC3 in autophagosomes. When the expression of TBC1D9B is inhibited, cellular autophagy activity is reduced, suggesting that this molecule may positively regulate autophagic flux. TBC1D9B can interact with RAB11A, RAB11B and RAB4A, but has a GAP activity only for RAB11A in the presence of Mg2+ [3908]. See also RAB11.
TBC1D14 (TBC1 domain family member 14): TBC1D14 colocalizes and interacts with ULK1, and upon overexpression causes tubulation of ULK1-positive endosomes, inhibiting autophagosome formation [3685]. TBC1D14 binds activated RAB11, but does not function as a GAP. TBC1D14 localizes to the Golgi complex during amino acid starvation. See also RAB11.
TBC1D25/OATL1 (TBC1 domain family member 25): A Tre2-Bub2-Cdc16 (TBC) domain-containing GAP for RAB33B; TBC1D25 is recruited to the outer membrane of phagophores and autophagosomes [3909] via direct interaction with the Atg8 family proteins (via a LIR/LRS-like sequence), and it regulates the interaction of autophagosomes with lysosomes by inactivating RAB33B [3695]. Overexpression of TBC1D25 inhibits autophagosome maturation at a step prior to fusion, suggesting that it might interfere with a tethering/docking function of RAB33B. See also RAB33B and LIR/LRS.
TBK1 (TANK binding kinase 1): A serine/threonine protein kinase that is similar to IKK involved in the activation of NFKB [3910]. TBK1 binds and directly phosphorylates OPTN at Ser177 (in humans) within the LIR, increasing the affinity of the latter for LC3 [1472].
TCHP/mitostatin (trichoplein keratin filament binding): A DCN (decorin)-inducible tumor suppressor gene that functions in, and is required for, tumor cell mitophagy. TCHP/mitostatin responds to DCN as well as canonical cues (e.g., nutrient deprivation and rapamycin) for mitophagic induction. DCN regulates mitostatin in a PPARGC1A/PGC-1α-dependent manner. Moreover, DCN-induced mitophagy is entirely dependent on TCHP for angiogenic inhibition [3911].
TDP-43: See TARDBP.
TECPR1 (tectonin beta-propeller repeat containing 1): A protein that interacts with ATG5 and WIPI2, and localizes to the phagophore (localization is dependent on WIPI2); TECPR1 is needed for phagophore formation during autophagic elimination of Shigella, but not for starvation-induced autophagy [3912]. TECPR1 also localizes to autophagosomes that target other pathogenic microbes such as group A Streptococcus, to depolarized mitochondria and to protein aggregates, suggesting a general role in selective autophagy. TECPR1 also plays a role in fusion of the autophagosome with the lysosome by competing with ATG16L1 to bind ATG5 and PtdIns3P, recruiting ATG5 to the lysosome membrane [3913].
TECPR2: A WD repeat- and TECPR domain-containing protein that plays a role in autophagy; mutation of TECPR2 results in a form of monogenic hereditary spastic paraparesis [3914, 3915].
TEX264 (testis expressed gene 264): Colocalization of TEX264, an ER-resident protein, with LC3B, WIPI2, RB1CC1 and LAMP1 confirm its role in autophagy. In addition, TEX264-depleted HeLa cells show impaired reticulophagy, validating TEX264 as a reticulophagy receptor [464, 1373, 3916]. See also reticulophagy.
TFE3 (transcription factor binding to IGHM enhancer 3): A transcription factor belonging to the microphthalmia/transcription factor E (MiT/TFE) family, along with TFEB and MITF [977, 3433]. See also TFEB and MITF.
TFEB (transcription factor EB): A transcription factor that positively regulates the expression of genes involved in lysosomal biogenesis (those in the CLEAR network [969]), and also several of those involved in autophagy (including UVRAG, WIPI, MAP1LC3B and ATG9B); the use of a common transcription factor allows the coordinated expression of genes whose products are involved in the turnover of cytoplasm [949]. TFEB and MTORC1 can form a regulatory loop in which TFEB can promote, but itself can be suppressed by, MTORC1 activation, which may result in oscillating autophagy functions and pathological consequences [3913]. See also CLEAR and PPP3.
TGA9/AT1G08320 (TGACG [TGA] motif-binding protein 9): A transcription factor that positively regulates the expression of genes involved in autophagy in A. thaliana [3917].
TGFB1/TGF-β (transforming growth factor beta 1): A cytokine that activates autophagy through the SMAD and MAPK8 pathways. TGFB1 induces the expression of several ATG genes including BECN1.
TGFB2-OT1: A newly discovered long noncoding RNA (lncRNA) derived from the 3ʹ UTR of TGFB2. The level of TGFB2-OT1 is markedly increased by LPS and oxidized low-density lipoprotein, two vascular endothelial cell (VEC) inflammation triggers. TGFB2-OT1 can regulate autophagy in VECs by sequestering MIR3960, MIR4488, and MIR4459, and increasing the levels of downstream target genes, including CERS1, NAT8L, ATG13 and LARP1 [3918].
TGM2/TG2/TGase 2 (transglutaminase 2): An enzyme that catalyzes the formation of an isopeptide bond between a free amine group (e.g., protein- or peptide-bound lysine) and the acyl group at the end of the side chain of protein- or peptide-bound glutamine (protein crosslinking); TGM2 interacts with SQSTM1 and is involved in the autophagic clearance of ubiquitinated proteins [1212, 3919].
Thapsigargin: A cell-permeable inhibitor of ATP2A/SERCA. Thapsigargin inhibits autophagy as a non-competitive inhibitor of ATP2A, which inhibits the fusion of autophagosomes with lysosomes and leads to the accumulation of autophagosomes in the cytosol [2640].
THC (∆9-tetrahydrocannabinol): The main psychoactive component of the hemp plant Cannabis sativa. The anticancer activity of THC in several animal models of cancer relies on its ability to stimulate autophagy-mediated cancer cell death. This effect occurs via THC binding to cannabinoid receptors, and the subsequent triggering of an ER stress-related response, which leads in turn to the inhibition of the AKT-MTORC1 axis [3920–3922].
Thiamet G (2-ethylimino-5-[hydroxymethyl]-1,3a,5,6,7,7a-hexahydropyrano[3,2-d][1,3]thiazole-6,7-diol): A potent (Ki = 2 nM) small molecule inhibitor that selectively inhibits OGA (O-GlcNAcase), the sole enzyme catalyzing the removal of O-GlcNAc from nuclear and cytoplasmic proteins. Treatment of cells and tissues with Thiamet G results in increased levels of global cellular O-GlcNAc [3923]. This orally available compound stimulates autophagy in neuroblastoma N2a cells, primary rat neurons, and mouse brain through an MTOR-independent pathway without obvious toxicity [3924].
TIGAR/C12orf5 (TP53 induced glycolysis regulatory phosphatase): A protein that modulates glycolysis, causing an increase in NADPH, which results in a lower ROS level; this reduces the sensitivity to oxidative stress and apoptosis, but also has the effect of lowering the level of autophagy [3925].
Timosaponin A-III: A medicinal saponin that induces a type of autophagy with some features that are distinct from rapamycin-induced autophagy [3926].
Tlg2: A yeast endocytic SNARE light chain involved in early stages of the Cvt pathway [1089] and in autophagosome membrane formation [3755]. Deletion of TLG2 results in a modest impairment in Atg9 delivery to the PAS.
TLR (toll like receptor): A family of receptors that induces autophagy following binding to a variety of pathogen- and damage-associated molecules as ligands. TLR2 mediates SQSTM1-dependent activation of autophagy, which plays a protective role in DEN-induced tumorigenesis through clearing of intracellularly accumulated ROS and SQSTM1 aggregates [3927, 3928]. DAMPs release from injured liver cells activates TLR4 signaling, which initiates or supports senescence and autophagy to retard genotoxic carcinogen-induced hepatocellular carcinoma initiation and progression via removing accumulated ROS, repairing DNA damage and decreasing SQSTM1 aggregates [3929, 3930]. Moreover, TLR2 activity is critically involved in the pathogenesis of cardiac dysfunction and fibrosis via mediating extracellular HMGB1 (functions as a DAMP molecule)-dependent autophagy suppression in failing hearts [3931].
TLR9 (toll like receptor 9): This isoform of the TLR family translocates from the endoplasmic reticulum to the lysosomal membrane upon induction of autophagy. It is activated by unmethylated CpG DNA sequences, including not only bacterial genomes and viral DNA, but also mitochondrial DNA (mtDNA). Mitochondrial DNA that escapes from autophagy activates TLR9 resulting in cardiac inflammation and heart failure [3932]. Furthermore, mtDNA-activated TLR9 triggers PIK3CG and the downstream AKT-MTOR-ULK1 signaling pathway, eventually resulting in feedback inhibition of autophagy and cardiomyopathy [2583].
TM9SF1 (transmembrane 9 superfamily member 1): A protein with 9 transmembrane domains that induces autophagy when overexpressed [3933].
TM9SF4 (transmembrane 9 superfamily member 4): TM9SF4 facilitates inactivation of MTOR under starvation conditions to promote autophagy induction [3934].
TMED2 (transmembrane p24 trafficking protein 2): TMED2 is thought to be a cargo receptor involved in vesicular protein trafficking; it mainly functions in the early secretory pathway but also in post-Golgi membranes. Diseases associated with TMED2 include Charcot-Marie-Tooth disease, axonal, type 2L, and Crohn disease. An important paralog of this gene is TMED7. Overexpression of TMED2 triggers autophagy and is likely to be involved in autophagy-dependent secretion [3935].
TMEM41B (transmembrane protein 41B): A protein that is required for the formation of autophagosomes [3936–3938]. TMEM41B is structurally related to VMP1 and physically and functionally interacts with VMP1. See also VMP1.
TMEM59 (transmembrane protein 59): A type-I transmembrane protein able to induce an unconventional autophagic process involving LC3 labeling of single-membrane endosomes through direct interaction with the C-terminal WD40 domain of ATG16L1 [3939]. Binding between TMEM59 and ATG16L1 is impaired by the T300A mutation encoded by a single-nucleotide polymorphism (rs2241880) that increases the risk of Crohn disease [3940].
TMEM74 (transmembrane protein 74): An integral membrane protein that induces autophagy when overexpressed [3115, 3116]. TMEM74 promotes autophagy via interactions with ATG16L1 and ATG9A [3941].
TMEM150B/DRAM3 (transmembrane protein 150B): TMEM150B/DRAM3 is a transmembrane protein related to DRAM1 and DRAM2, which enhances autophagic flux and cell survival under glucose-starvation conditions [3942, 3943].
TMEM166: See EVA1A.
TMEM173: See STING1.
TNFAIP3/A20 (TNF alpha induced protein 3): An E3 ubiquitin ligase that also functions as a deubiquitinating enzyme that removes K63-linked ubiquitin from BECN1, thus limiting autophagy induction in response to TLR signaling [3944]. In contrast, TNFAIP3 restricts MTOR signaling, acting as a positive factor to promote autophagy in CD4 T cells [3945]. TNFAIP3/A20 interacts with the WD40 domain of ATG16L1 to control intestinal homeostasis [3946].
TNFSF10/TRAIL (TNF superfamily member 10): Induces autophagy by activating AMPK, thus inhibiting MTORC1 during lumen formation.
TOLLIP (toll interacting protein): A vertebrate ubiquitin-binding receptor protein similar to yeast Cue5 that contains a CUE domain and plays a role in the autophagic removal of protein aggregates [627]. TOLLIP regulates innate immune responses to intracellular pathogens in the lung [3947, 3948]. See also Cue5, CUET and TOM1.
TOM1 (target of myb1 membrane trafficking protein): A MYO6 (myosin VI) cargo adaptor protein that mediates the delivery of endosomes to lysosomes. TOM1 functions in autophagosome maturation and fusion with lysosomes, and binds TOLLIP [3465]. See also TOLLIP.
TOR (target of rapamycin): A serine/threonine protein kinase that negatively regulates yeast autophagy. Present in two complexes, TORC1 and TORC2. TORC1 is particularly sensitive to inhibition by rapamycin. TORC1 regulates autophagy in part through Tap42-protein phosphatase 2A, and also by phosphorylating Atg13 and Atg1.
TORC1 (TOR complex I): A rapamycin-sensitive protein complex of TOR that includes at least Tor1 or Tor2 (MTOR), Kog1 (RPTOR), Lst8 (MLST8), and Tco89 [3949]. MTORC1 also includes DEPTOR and AKT1S1/PRAS40 [3950]. In mammalian cells, sensitivity to rapamycin is conferred by RPTOR. TORC1 directly regulates autophagy, and, at the lysosomes, MTORC1 targets TPCN2 inhibiting its channel activity. Inhibition of MTORC1 promotes autophagy and protects cardiomyocytes from necroptosis by a TFEB-dependent mechanism [3951].
TORC2 (TOR complex II): A relatively rapamycin-insensitive protein complex of TOR that includes at least Tor2 (MTOR), Avo1 (MAPKAP1/SIN1), Avo2, Avo3 (RICTOR), Bit61, Lst8 (MLST8) and Tsc11; MTORC2 also includes FKBP8/FKBP38, and PRR5/Protor-1 [3949, 3950, 3952]. A critical difference in terms of components relative to TORC1 is the replacement of RPTOR by RICTOR. TORC2 is primarily involved with regulation of the cytoskeleton, but this complex functions to positively regulate autophagy during amino acid starvation [3953]. Finally, studies also support the idea that TORC2 activity is required to sustain autophagosome biogenesis [3954], whereas it exerts an inhibitory effect on CMA [3955], suggesting that a switch in TORC2 substrates may contribute to coordinating the activity of these two types of autophagy.
Torin1: A selective catalytic ATP-competitive MTOR inhibitor that directly inhibits both TORC1 and TORC2 [802].
TP53/p53 (tumor protein 53): A tumor suppressor. Nuclear TP53 activates autophagy, at least in part, by stimulating AMPK and DRAM1, whereas cytoplasmic TP53 inhibits autophagy [2120]. The presence of TP53 protein was found to be essential for the suppression of SAHA-induced autophagy in several tumor cell lines [3749]. TP53 may also mediate autophagy induction upon proteasome inhibition [3956]. Conversely, the TP53-dependent increase of AMPK activity is the major factor responsible for autophagy disruption in melanoma cells (likely inducing excessive accumulation of autophagosomes that are unable to be degraded), although the MTOR pathway unrelated to the TP53-AMPK axis also plays a role [3957]. Note that the official name for this protein in rodents is TRP53. The TP53 C. elegans ortholog, cep-1, also regulates autophagy [2119, 2121]. See also SAHA/vorinostat.
TP53INP1 (tumor protein p53 inducible nuclear protein 1): A stress-response protein that promotes TP53 transcriptional activity; cells lacking TP53INP1 display reduced basal and stress-induced autophagy, whereas its overexpression enhances autophagic flux [3958]. TP53INP1 interacts directly with LC3 via a functional LIR, and stimulates autophagosome formation [3958]. Cells lacking TP53INP1 display reduced mitophagy; TP53INP1 interacts with PRKN and PINK1, and thus could be a recognition molecule involved in mitophagy [3959].
TP53INP2/DOR (tumor protein p53 inducible nuclear protein 2): A mammalian and Drosophila regulatory protein that shuttles from the nucleus, passing through the nucleolus, to reach the cytosol [3960]; the nuclear protein interacts with deacetylated LC3 [1004] and GABARAPL2, and stimulates autophagosome formation [3961]. TP53INP2 also interacts with GABARAP and VMP1, and is needed for the recruitment of BECN1 and LC3 to phagophores. TP53INP2 translocates from the nucleus to phagophores during autophagy induction and binds VMP1 and LC3 directly [3962]. In addition, TP53INP2 modulates muscle mass in mice through the regulation of autophagy [3963], and regulates adiposity by activating CTNNB1/β-catenin in preadipocytes through autophagy-dependent sequestration of GSK3B [3964].
TPCN/two-pore channel (two pore segment channel): TPCNs are endolysosomal cation channels that maintain the proton gradient and membrane potential of endosomal and lysosomal membranes. TPCN2 mediates lysosomal Ca2+ release and physically interacts with MTOR, providing bidirectional control between the two. On the one hand, MTORC1 inhibits TPCN2, whereby channel opening can be provoked by nutrient starvation [1997] and rapamycin, which provokes lysosomal TPCN2-mediated Ca2+ release [1998]. On the other hand, TPCN2 regulates MTOR activity and reactivation, and autophagic flux in skeletal muscle [1997, 3965].
TPR (translocated promoter region, nuclear basket protein): TPR is a component of the nuclear pore complex that presumably localizes at intranuclear filaments or nuclear baskets. Nuclear pore complex components, including TPR, are jointly referred to as nucleoporins. TPR was originally identified as the oncogenic activator of the MET and NTRK1/trk proto-oncogenes. Knockdown of TPR facilitates autophagy. TPR depletion is not only responsible for TP53 nuclear accumulation, which activates the TP53-induced autophagy modulator DRAM, but also contributes to HSF1 and HSP70 mRNA trafficking, and transcriptional regulation of ATG7 and ATG12 [3522, 3966]. See also ependymoma.
TRAF2 (TNF receptor associated factor 2): An E3 ubiquitin ligase that plays an essential role in mitophagy in unstressed cardiac myocytes, as well as those treated with TNF or CCCP [1226].
TRAF6 (TNF receptor associated factor 6): An E3 ubiquitin ligase that ubiquitinates BECN1 to induce TLR4-triggered autophagy in macrophages [3944].
TRAIL: See TNFSF10.
Transgenic: Harboring genetic material of another species/organism or extra copies of an endogenous gene, usually gained through transfer by genetic engineering.
Transmitophagy/transcellular mitophagy: A process in which axonal mitochondria are degraded in a cell-nonautonomous mechanism within neighboring cells [1237].
TRAPPII (transport protein particle II): A guanine nucleotide exchange factor for Ypt1 and perhaps Ypt31/32 that functions in autophagy in yeast [3967]. TRAPPII is composed of Bet3, Bet5, Trs20, Trs23, Trs31, Trs33 and the unique subunits Trs65, Trs120 and Trs130.
TRAPPIII (transport protein particle III): A guanine nucleotide exchange factor for Ypt1 that functions in autophagy in yeast [2202]. TRAPPIII is composed of Bet3, Bet5, Trs20, Trs23, Trs31, Trs33 and a unique subunit, Trs85.
Trehalose: A small disaccharide, which induces autophagy and autophagy flux in cells in an MTOR-independent manner [2036]. In hepatic cells, trehalose induces autophagy through a pseudo-starvation response by inhibiting SLC2A [3968]. In macrophages, trehalose activates the PIKFYVE-MCOLN1/TRPML1-TFEB pathway to induce autophagy, autophagy flux and also xenophagy flux against Mycobacterium tuberculosis. Additionally, in primary macrophages, the synthetic analog trehalose-6,6´-dibehenate promotes lysosome biogenesis and autophagy, through MYD88-mediated activation of the PtdIns3K pathway, which contributes to its antimicrobial properties against M. tuberculosis [3969].
TRIB3 (tribbles pseudokinase 3): A pseudokinase that plays a crucial role in the mechanism by which various anticancer agents (and specifically cannabinoids, the active components of marijuana and their derived products) activate autophagy in cancer cells. Cannabinoids elicit an ER stress-related response that leads to the upregulation of TRIB3 whose interaction with AKT impedes the activation of this kinase, thus leading to a decreased phosphorylation of TSC2 and AKT1S1/PRAS40. These events trigger the inhibition of MTORC1 and the induction of autophagy [3921]. Conversely, TRIB3 binding to SQSTM1 via its UBA and LIR motifs interferes with autophagic flux, in particular of ubiquitinated proteins, and also reduces the efficiency of the UPS, resulting in tumor and fibrosis progression due to the accumulation of tumor- and fibrosis-promoting factors [3920, 3970–3972].
Trichostatin A: An inhibitor of class I and class II HDACs that induces autophagy [3973].
Triclosan: Induces autophagy via the AMPK-ULK1 and MAPK/JNK-MAPK/ERK-MAPK/p38 pathways independent of MTOR in a dose-dependent manner in HeLa cells and in macrophages, and may play a role in the clearance of MAPT/tau oligomers in HEK293 cells [3974, 3975].
TRIM5/TRIM5α (tripartite motif containing 5): A proposed selective autophagy receptor for cell context-specific xenophagy of incoming retroviruses [3660, 3976, 3977]. TRIM5 binds retroviral capsids [3660, 3978] and separately interacts with multiple autophagy factors including SQSTM1, and with Atg8-family proteins via a helical LIR-motif located in the central coiled-coil region of the tripartite-motif [3979]. This helical-LIR has the consensus [DEST]-X3-[WFY]-X6-[LIVQ].
TRIM16 (tripartite motif containing 16): TRIM16 interacts with LGALS3 and recognizes endomembrane damage with mobilization of the core autophagy regulators ATG16L1, ULK1, and BECN1 to protect cells from lysosomal damage and Mycobacterium tuberculosis invasion [3345]. TRIM16 controls assembly and degradation of protein aggregates by modulating the SQSTM1-NFE2L2 axis and autophagy [3980].
TRIM17 (tripartite motif containing 17): TRIM17 recruits MCL1 to BECN1 and negatively regulates autophagy [3660, 3981].
TRIM20: See MEFV.
TRIM21: An antigen in autoimmune diseases such as systemic lupus erythematosus, and Sjögren syndrome, TRIM21 is a receptor for selective autophagy of IRF3 dimers, a key transcriptional activator of type I interferon responses [3407].
TRIM28 (tripartite motif containing 28): TRIM28 is an E3 ligase that is part of a ubiquitin ligase complex that targets PRKAA1, leading to ubiquitination and proteasomal degradation in part through the upregulation of MTOR activity [3378]. See also MAGEA3.
TRIM32 (tripartite motif containing 32): TRIM32 is an E3 ubiquitin ligase that interacts with AMBRA1 and ULK1 to stimulate ULK1 activity in muscle cells upon atrophy induction via unanchored K63-linked polyubiquitin chains [73]. TRIM32 mutations lead to muscular dystrophy.
TRIM37 (tripartite motif containing 37): TRIM37 gene mutations cause mulibrey (muscle-liver-brain-eye) nanism, a severe growth disorder, and predispose to tumor development. Loss of TRIM37 induces autophagy in an MTORC1-dependent manner [3982].
TRIM50 (tripartite motif containing 50): TRIM50 is a cytoplasmic E3 ubiquitin ligase [3983], which interacts and colocalizes with SQSTM1 and promotes the formation and clearance of aggresome-associated polyubiquitinated proteins through HDAC6-mediated interaction and acetylation [3984, 3985].
TRIM63/MURF-1 (tripartite motif containing 63): Muscle-specific atrophy-related E3 ubiquitin ligase [3986, 3987] that cooperates with SH3GLB1 to regulate autophagic degradation of CHRNA1 in skeletal muscle, particularly upon muscle-atrophy induction [3784].
TRPC4 (transient receptor potential cation channel subfamily C member 4): A cation channel in human umbilical vascular endothelial cells; upregulation of TRPC4 increases the intracellular Ca2+ concentration resulting in activation of CAMKK2, which leads to MTOR inhibition and the induction of autophagy [2641].
TRPML1: See MCOLN1.
Trs85: A component of the TRAPPIII complex that is required specifically for autophagy [1062].
Trs130: A component of the TRAPPII complex that is required for the transport of Atg8 and Atg9 to the PAS [3967].
TSC1/2 (tuberous sclerosis 1/2): A stable heterodimer (composed of TSC1/hamartin and TSC2/tuberin) inhibited by AKT and MAPK1/3 (phosphorylation causes dissociation of the dimer), and activated by AMPK. TSC1/2 acts as a GAP for RHEB, thus inhibiting MTORC1.
TSPO (translocator protein): TSPO is a mitochondrial protein that interacts with VDAC1 to modulate the efficiency of mitophagy [3988].
Tubastatin A: An HDAC inhibitor that interferes with lysosomes, thereby also affecting autophagy [3208, 3209].
TUBGCP4 (tubulin gamma complex associated protein 4): TUBGCP4 can inhibit autophagy by competing with ATG3 to interact with ATG7, thus interfering with lipidation of LC3B in the retina [2396].
Tubulovesicular autophagosome (TVA): Cationic lipoplex and polyplex carriers used for nonviral gene delivery enter mammalian cells by endocytosis and fuse with autophagosomes, generating large tubulovesicular structures (tubulovesicular autophagosomes) that immunostain for LC3; these structures do not fuse efficiently with lysosomes and interfere with gene expression [2109].
Tubulovesicular cluster (TVC): A structure identified morphologically in yeast that corresponds to the Atg9 peripheral sites [808]. See also Atg9 peripheral sites/structures.
TUFM (Tu translation elongation factor, mitochondrial): An autophagy-promoting protein that resides in mitochondria. TUFM associates with NLRX1 to establish a protein scaffold recruiting ATG12–ATG5, ATG16L1, and LC3 [3501, 3504, 3989]. See also NLRX1.
TXA1: A thioxanthone that decreases the viability of human melanoma cells by modulation of autophagy and which may serve as a lead compound for the development of autophagy modulators with antitumor activity [3990]. Importantly, TXA1 was previously shown to be an inhibitor of the drug-efflux pump ABCB1/P-glycoprotein [3991].
U0126: U0126 is a selective inhibitor of MAP2K/MEK, which specifically inhibits the ability of MAP2K to phosphorylate MAPK/ERK [3992].
UBE2N (ubiquitin conjugating enzyme E2 N): A ubiquitin-conjugating enzyme involved in PRKN-mediated mitophagy [3993, 3994]. UBE2N activity may be only partly redundant with that of UBE2L3, UBE2D2 and UBE2D3, as it is also involved during later steps of mitophagy.
Ubiquitin: A 76-amino acid protein that is conjugated to lysine residues. Ubiquitin is traditionally considered part of the ubiquitin-proteasome system, and polyubiquitin chains tag proteins for degradation; however, ubiquitin is also linked to various types of autophagy including aggrephagy (see SQSTM1 and NBR1). Lysine linkage-specific monoclonal antibodies, which are commercially available, can be used to investigate the degradation pathway usage [3995]. Proteins covalently tagged with polyubiquitin chains via K48 are mostly destined for proteasomal degradation, whereas proteins tagged with K63-linked ubiquitin are predominantly degraded via the autophagy pathway. In addition, phosphorylated forms of ubiquitin have been identified including p-S65-Ub, which is specifically generated during PINK1-PRKN-mediated mitophagy. Potentially, several PTMs of the modifier ubiquitin may turn out to be highly relevant and specific for distinct forms of selective autophagy (reviewed in [1194]).
Ubp3: A yeast deubiquitinating enzyme that forms a complex with Bre5 and is required for proteaphagy [1358] and ribophagy [1389]. Conversely, the Ubp3-Bre5 complex inhibits mitophagy [3996].
UBQLN (ubiquilin): Ubiquilins are receptor proteins that deliver ubiquitinated substrates to the proteasome. Ubiquilins may aid in the incorporation of protein aggregates into autophagosomes, and also promote the maturation of autophagosomes at the stage of fusion with lysosomes [3997, 3998].
Ubx5: A ubiquitin regulatory X (UBX) domain-containing protein from yeast that acts as an autophagy receptor for mutant forms of Cdc48; it binds the UDS of Atg8 via a UIM-like sequence [1359]. See also Cdc48, UDS, UIM and VCP.
UDS (UIM-docking site): A binding domain on Arabidopsis ATG8 that interacts with UIM-like sequences from the proteaphagy receptor RPN10, and multiple other proteins [1359]. This site is also present in yeast and human Atg8-family proteins. See also RPN10.
UIM (ubiquitin-interacting motif)-like sequences: UIM-like sequences participate in high-affinity binding to an alternative (i.e., non-LDS) Atg8-family protein interaction site [1359]. See also UDS.
ULK family (unc-51 like autophagy activating kinase): The ULK proteins are homologs of yeast Atg1. In mammalian cells the family consists of five members, ULK1, ULK2, ULK3, ULK4 and STK36/ULK5. ULK1 and ULK2 are required for autophagy, and ULK3 for oncogene-induced senescence [805, 3999, 4000]. See also Atg1. Figure modified from Fig. 2 of ref. [697].
ULK1.DN: A dominant-negative form of ULK1 expressing only the C-terminal domain of the kinase, that inhibits autophagy via competitive inhibition of endogenous ULK1 in vitro and in vivo [645, 646].
ULK-101: A potent and selective small molecule inhibitor of ULK1/ULK2 that reduces basal and induced autophagy in cancer cells and sensitizes to nutrient stress [4001].
Ume6: A component of the Rpd3L complex that binds to the URS1 sequence in the ATG8 promoter and downregulates transcription in growing conditions [2011]. See also Rpd3 and Sin3/SIN3.
UNC-51: The C. elegans Atg1/ULK1/ULK2 homolog. UNC-51 is an autophagy-associated serine/threonine kinase that functions to regulate axon outgrowth and elongation [4002]. RNAi knockdown of unc-51 is associated with accumulation of a human SNCA-GFP fusion in a C. elegans PD model [4003]. See also Atg1.
UNC13D: A regulator of endocytic maturation and endosomal function that binds to the SNARE proteins STX7 and VAMP8 [4003] and whose downregulation induces TFEB-mediated upregulation of autophagic genes and autophagy [4004].
UPR (unfolded protein response): A coordinated process to adapt to ER stress, providing a mechanism to buffer fluctuations in the unfolded protein load. The activation of this pathway is often related with autophagy. UPR-mediated autophagy positively regulates replication of viruses such as hepatitis C virus and foot-and-mouth disease virus by inhibitory action on innate immunity [2722].
UPRmt (mitochondrial unfolded protein response): An inducible stress response activated to maintain mitochondrial protein homeostasis upon folding stress that is also connected to induction of mitophagy [3596, 4005].
USP8 (ubiquitin specific peptidase 8): A deubiquitinating enzyme that removes K6-linked ubiquitin chains from PRKN to promote its recruitment to depolarized mitochondria and mitophagy [3640]. Inhibition of USP8 ameliorates PD phenotypes caused by Pink1 deficiency in Drosophila [4006].
USP9X (ubiquitin specific peptidase 9 X-linked): A deubiquitinating enzyme that regulates BECN1 proteasomal degradation. Competitive binding between BECN1 and MCL1 to USP9X mediates the reciprocal inverse regulation of the two proteins [3398, 3399]. See also MCL1.
USP14 (ubiquitin specific peptidase 14): A proteasome-associated deubiquitinase. USP14 is a substrate of autophagy, and interacts with LC3 and SQSTM1 [606]. In autophagy-deficient cells, USP14 regulates DNA repair, an effect that can be blocked by inhibition of USP14 or AKT; AKT phosphorylates USP14 on Ser432, and USP14 is constitutively active in PTEN-deficient cells [2670].
USP15 (ubiquitin specific peptidase 15): A deubiquitinating enzyme that antagonizes PRKN-mediated mitophagy [4007]. See also USP30.
USP24 (ubiquitin specific peptidase 24): A deubiquitinating enzyme that negatively regulates autophagy by affecting ULK1 ubiquitination and protein stability. The USP24 gene is located in the PARK10 locus suggesting potential involvement in PD [4008].
USP30: A deubiquitinating enzyme that antagonizes PRKN-mediated mitophagy [4009, 4010] and PEX2-mediated pexophagy [1205, 1206]. USP30 is also a substrate of PRKN and is subject to proteasome-mediated degradation. See also USP15.
USP35: A deubiquitinating enzyme that antagonizes PRKN-mediated mitophagy [4011].
USP36: A deubiquitinating enzyme that negatively regulates selective autophagy in Drosophila and human cells [4012].
UVRAG (UV radiation resistance associated): A Vps38 homolog that can be part of the class III PtdIns3K complex. UVRAG functions in several ways to regulate autophagy: 1) It disrupts BECN1 dimer formation and forms a heterodimer that activates autophagy. 2) It binds to SH3GLB1 to allow activation of class III PtdIns3K to stimulate autophagy. 3) It interacts with the class C Vps/HOPS proteins involved in fusion of autophagosomes or amphisomes with the lysosome. 4) It competes with ATG14 for binding to BECN1, thus directing the class III PtdIns3K to function in the maturation step of autophagy [797]. MTORC1 phosphorylates UVRAG to inhibit autophagy [2671]. In contrast, MTORC1 can also phosphorylate UVRAG to stimulate PIK3C3 activity and autophagic lysosome reformation [2672]. UVRAG also has an autophagy-independent function, interacting with membrane fusion machinery to facilitate the cellular entry of enveloped viruses [4013].
Vac8: A yeast peripheral vacuolar membrane protein, implicated in vacuolar inheritance, the Cvt pathway, pexophagy and the formation of NVJs [157, 745, 4014, 4015]. Vac8 binding to the nuclear membrane protein Nvj1 promotes the formation of NVJs and PMN. Vac8 binding to Atg13 is required for the Cvt pathway and efficient autophagy. In addition, Vac8 binding to Atg13 localizes the Atg1 initiation complex to the vacuolar periphery for PAS formation. See also nucleus-vacuole junction, Nvj1 and PMN.
Vacuolar cell death: One of the two major types of cell death in plants (another type is necrosis), wherein the content of the dying cell is gradually engulfed by growing lytic vacuoles without loss of protoplast turgor, and culminates in vacuolar collapse [1761]. Vacuolar cell death is commonly observed during plant development, for example in the embryo-suspensor and xylem elements, and critically depends on autophagy [1763]. Atg1-dependent but Atg7-independent autophagy is also required for vacuolar cell death in Dictyostelium development [4016, 4017].
Vacuolar compartment (VAC): The T. gondii equivalent of the lysosome [2366, 2367].
Vacuolar H+-translocating ATPase (V-ATPase): A ubiquitously expressed proton pump that is responsible for acidifying lysosomes and the yeast or plant vacuole, and therefore is important for the normal progression of autophagy. Inhibitors of the V-ATPase (e.g., bafilomycin A1) are efficient autophagy inhibitors [218, 219]. Upon bacteria-induced vacuolar damage, the V-ATPase recruits ATG16L1 to the bacteria-residing vacuole to initiate LC3 lipidation [1476].
Vacuolar sequestering membranes (VSM): Extensions/protrusions of the vacuole limiting membrane along the surface of peroxisomes that occur during macropexophagy [4014].
Vacuole: The fungal and plant equivalent of the lysosome; this organelle also carries out storage and osmoregulatory functions [4018]. The bona fide plant equivalent of the lysosome is the lytic vacuole.
Vacuole import and degradation (Vid): A degradative pathway in yeast in which a specific protein(s) is sequestered into small (30- to 50-nm) single-membrane cytosolic vesicles that fuse with the vacuole allowing the contents to be degraded in the lumen [1407, 1411]. This process has been characterized for the catabolite-induced degradation of the gluconeogenic enzyme Fbp1 (fructose-1,6-bisphosphatase) in the presence of glucose, and sequestration is thought to involve translocation into the completed vesicle. An alternate pathway for degradation of Fbp1 by the ubiquitin-proteasome system has also been described [4019].
Vacuolin-1: A small chemical that potently and reversibly inhibits the fusion between autophagosomes or endosomes with lysosomes by activating RAB5A [2647].
Valinomycin: A K+ ionophore that destroys the electrochemical gradient across the mitochondrial membrane and is widely used as a stimulator of mitophagy, similar to CCCP [4020].
Vam3: A yeast syntaxin homolog needed for the fusion of autophagosomes with the vacuole [4021].
VAMP3 (vesicle associated membrane protein 3): A SNARE protein that facilitates the fusion of MVBs with autophagosomes to generate amphisomes [4022].
VAMP7 (vesicle associated membrane protein 7): VAMP7 is a SNARE protein that colocalizes with ATG16L1-positive vesicles and phagophores, and is required, along with STX7 (syntaxin 7), STX8 (syntaxin 8) and VTI1B, for autophagosome formation [4023]. VAMP7 is also involved in the maturation of autophagosomes by facilitating fusion with a lysosome [4022].
VAMP8 (vesicle associated membrane protein 8): A SNARE protein that, in conjunction with VTI1B, is needed for the fusion of autophagosomes with lysosomes [4024].
VAP27 (Vesicle-Associated Protein): Plant homologs of VAPA. The Arabidopsis VAP27 family contains 10 members (VAP27-1 to VAP27-10). VAP27-1 interact with AtEH/Pan1 and regulates autophagosome biogenesis. Plants with reduced expression of VAP27-1 and VAP27-3 are more susceptible to low nutrient stress [2747]. See also AtEH/Pan1.
VAPA (VAMP associated protein A): VAPA and VAPB, are generally involved in forming ER contacts with other membranes by directly interacting with the FFAT motif. VAPA/B directly interact with RB1CC1 and ULK1 at the phagophore initiation stage and stabilize the ULK1-RB1CC1 complex on the ER. VAPA/B also interact with WIPI2 to tether the ER-phagophore for phagophore expansion. Depletion of VAPA/B impairs autophagosome formation [4025].
VAPB (VAMP associated protein B and C): See VAPA.
VCP/p97 (valosin containing protein): A type II AAA+-ATPase that is a protein segregase required for autophagosome maturation under basal conditions or when the proteasomal system is impaired; mutations of VCP result in the accumulation of immature, acidified autophagic vacuoles that contain ubiquitinated substrates [4026–4028]. VCP is also required for granulophagy, lysophagy and mitophagy [1153, 1274, 4029]. See also Cdc48.
VDAC2 (voltage dependent anion channel 2): VDAC2 can inhibit autophagy through stabilizing the interaction between BECN1 and BCL2L1 [4030].
Verteporfin: An FDA-approved drug that is used in photodynamic therapy; however, it inhibits the formation of autophagosomes in vivo without light activation [2648].
Vesicophagy: Sequestration and degradation of hormone-containing secretory vesicles by phagophores in endocrine cells. Different from crinophagy or zymophagy, the process is dependent on autophagy proteins ATG5, ATG7, and BECN1, but not VMP1 [4031].
VHL (von Hippel-Lindau tumor suppressor): VHL serves as the substrate recognition subunit of a ubiquitin ligase that targets the α subunit of the heterodimeric transcription factor HIF1 for degradation. This interaction requires the hydroxylation of HIF1A on one or both of two conserved prolyl residues by members of the EGLN family of prolyl hydroxylases [4032].
VirA: A T3SS effector found in Shigella spp. whose Rab GTPase activating protein function enables escape of secreting S. flexneri cells from LC3-positive vacuoles [3269, 4033]. See also VirG and IcsB.
VirG/IcsA: A Shigella protein that is required for intracellular actin-based motility. VirG binds ATG5, which induces xenophagy; IcsB, a protein secreted by the type III secretion system, competitively blocks this interaction [4034]. See also IcsB and VirA.
Virophagy: The selective autophagic clearance of viruses [1432]. Virophagy is a form of xenophagy in which virions and viral components are captured and directed to phagophores for lysosomal degradation [1458].
VMP1 (vacuole membrane protein 1): A multispanning membrane protein that is required for autophagy [964, 4035]. VMP1 regulates the levels of PtdIns3P [4036], binding of the ATG12–ATG5-ATG16L1 complex, lipidation of LC3 [4037], and membrane contact sites between the ER and other organelles including phagophores [966]. VMP1 also interacts with the structurally related ER protein TMEM41B. See also TMEM41B.
Vps1: A dynamin-like GTPase required for peroxisomal fission. Vps1 interacts with Atg11 and Atg36 on peroxisomes that are being targeted for degradation by pexophagy [3056]. See also Dnm1.
Vps11: A member of the core subunit of the HOPS and class C core vacuole/endosome tethering (CORVET) complexes, originally found in yeast but also conserved in more complex eukaryotes [4038]. These complexes are important for correct endolysosomal trafficking, as well as the trafficking of black pigment cell organelles, melanosomes; zebrafish Vps11 is involved in maintaining melanosome integrity, possibly through an autophagy-dependent mechanism [4039].
VPS13A: Mutations in this protein leads to chorea-acanthocytosis, a rare neurodegenerative disease. This protein is a lipid transfer protein that regulates the interface between mitochondria-endosomes and mitochondria-ER and affects the degradative capacity of lysosomes [4040, 4041]. Disease mutations that limit binding of the yeast Vps13 APT1 domain to PtdIns3P result in loss of function [4042]. This mutation also reduces the binding of the APT1 domain of VPS13A to PtdIns3P and PtdIns5P [4043].
VPS13D: A protein that is required for autophagy and that regulates mitochondrial size [4044].
VPS15: See PIK3R4.
Vps30/Atg6: A component of the class III PtdIns3K complex. Vps30/Atg6 forms part of two distinct yeast complexes (I and II) that are required for the Atg and Vps pathways, respectively. See also BECN1 and phosphatidylinositol 3-kinase [2786].
Vps34: The yeast phosphatidylinositol 3-kinase; the lipid kinase catalytic component of the PtdIns3K complex I and II [3573]. See also phosphatidylinositol 3-kinase and PIK3C3.
Vps38: A yeast component of the class III PtdIns3K complex II, which directs it to function in the vacuolar protein sorting pathway.
Vps41: A major component of the HOPS complex responsible for the tethering of late endosomes and autophagosomes to lysosomes [3226, 3608, 4045]. This protein also directly interacts with Rab7 and Arl8 to coordinate the interaction of motor proteins for vesicular transport [4046, 4047]. Human VPS41 exhibits a protective role against neurodegeneration in both transgenic C. elegans and in SH-SY5Y neuroblastoma cell models of PD [4048, 4049].
VTC (vacuolar transporter chaperone): A complex composed of Vtc1, Vtc2, Vtc3 and Vtc4 that is required for microautophagy in yeast [4050].
Vti1: A yeast soluble SNARE that, together with Sec18/NSF, is needed for the fusion of autophagosomes with the vacuole [3756]. In mammalian cells, the SNARE proteins VAMP8 and VTI1B mediate the fusion of antimicrobial and canonical autophagosomes with lysosomes [4024].
VTRNA1-1 (vault RNA 1-1): A small non-coding RNA that negatively regulates selective autophagy by binding SQSTM1 and interfering with its oligomerization [4051].
WAC (WW domain containing adaptor with coiled-coil): A positive regulator of autophagy that interacts with BECN1, WAC also negatively regulates the UPS [3132].
WAPL (WAPL cohesion release factor): Accessory subunit of the cohesin complex involved in the resolution of sister chromatids during mitotic entry, which is necessary for correct chromosome partition during mitotic exit. WAPL binds to SQSTM1 and is enriched in mitotic fractions after inhibition of lysosome function during cell division [2974].
WDFY3/ALFY (WD repeat and FYVE domain containing 3): A scaffold protein that targets cytosolic protein aggregates and damaged mitochondria for selective autophagic degradation [4052, 4053]. WDFY3 interacts directly with ATG5 [3255], GABARAP proteins [202] and SQSTM1 [4054].
WDR45/WIPI4 (WD repeat domain 45): See WIPI.
WHAMM (WASP homolog associated with actin, golgi membranes and microtubules): A nucleation-promoting factor that directs the activity of the actin related protein 2/3 complex subunits (ARPC) to function in autophagosome formation [4055]. WHAMM colocalizes with LC3, ZFYVE1 and SQSTM1 and acts in autophagosome biogenesis through a mechanism dependent on actin comet tail formation. WHAMM also instigates autolysosome tubulation by promoting actin polymerization during autophagic lysosome reformation [4056]. See also autophagic lysosome reformation.
WIPI (WD repeat domain, phosphoinositide interacting): The WIPI proteins are putative mammalian homologs of yeast Atg18 and Atg21. There are four WIPI proteins in mammalian cells. WIPI1/WIPI49 and WIPI2 localize with LC3 and bind PtdIns3P. WIPI2 is required for starvation-induced autophagy [837]. WDR45/WIPI4 is also involved in autophagy. In humans, WDR45 is localized on the X-chromosome and so far only de novo loss-of-function mutations are described. Heterozygous and somatic mutations cause neurodegeneration with brain iron accumulation [4057], whereas hemizygous mutations result in early-onset epileptic encephalopathy [4058]. Impaired autophagy has been shown in lymphoblastoid cell lines derived from affected patients, showing abnormal colocalization of LC3-II and ATG9A. Furthermore, lymphoblastoid cell lines from affected subjects, show increased levels of LC3-II, even under normal conditions [4059]. Surprisingly, complete wdr45 knockout mice develop normally, but show neurodegeneration, as of nine months of age, thereby indicating overlapping activity of the four WIPI proteins in mammals [4060]. WDR45/WIPI4 appears to be the member of the mammalian WIPI protein family that binds ATG2 [658, 845].
WNK1 (WNK lysine deficient protein kinase 1): The protein kinase WNK1 is an inhibitor of autophagy, as well as the upstream class III PtdIns3K complex. WNK1 physically interacts with the PtdIns3K component UVRAG [4061].
WNT family: Cysteine-rich glycosylated secreted proteins that determine multiple cellular functions such as neuronal development, angiogenesis, tumor growth, and stem cell proliferation. Signaling pathways of WNT such as those that involve CTNNB1/beta-catenin can suppress autophagy [4062, 4063].
WNT5A: A ligand of the WNT signaling pathway. Activation of the WNT5A-CTNNB1 pathway suppresses IFNG-induced autophagy in macrophages during mycobacterial infection [783].
Wortmannin (WM): An inhibitor of PI3K and PtdIns3K; wortmannin inhibits autophagy due to the downstream effect on PtdIns3K [3374].
WXXL motif: An amino acid sequence present in proteins that allows an interaction with Atg8/LC3/GABARAP proteins; the consensus is [W/F/Y]-X-X-[I/L/V]. Also see AIM and LIR/LRS [2509].
WYE-354: A catalytic MTOR inhibitor that increases autophagic flux to a greater level than allosteric inhibitors such as rapamycin (and may be used to induce autophagy in cell lines that are resistant to rapamycin and its derivatives); short-term treatment with WYE-354 can inhibit both MTORC1 and MTORC2, but the effects on flux are due to the former [453]. See also Ku-0063794.
Xanthohumol: Prenylated chalconoid that blocks VCP/p97 and inhibits the maturation of autophagosomes. Xanthohumol impairs autophagosome maturation and leads to accumulation of LC3-II [4064].
XBP1 (X-box binding protein 1): A component of the ER stress response that may activate autophagy. The XBP1 yeast ortholog is Hac1 [4065].
Xenophagy: Cell-autonomous innate immunity defense, whereby cells eliminate intracellular microbes (e.g., bacteria, fungi, parasites and/or viruses) by sequestration into phagophores with subsequent delivery to the lysosome [4066].
Xestospongin B: An antagonist of the ITPR that dissociates the inhibitory interaction between ITPR and BECN1 and induces autophagy [4067]. Xestopongin B additionally works as a competitive inhibitor of ITPR, blocking calcium release from the ER and inducing MTORC1-independent and AMPK-dependent autophagy [668].
XIAP (X-linked inhibitor of apoptosis): XIAP is a ubiquitin ligase downstream of NOD2. It is involved in the antimicrobial process of LC3-associated phagocytosis. XIAP deficiency is associated with defective antimicrobial activity in macrophages and intestinal inflammation [3511]. XIAP also regulates autophagy through TP53 and SQSTM1 [4068, 4069].
XPO1/CRM1 (exportin 1): XPO1 is phosphorylated by STK38 (serine/threonine kinase 38) and regulates the nuclear exit of BECN1 and YAP1 (Yes1 associated transcriptional regulator); XPO1 is actively involved in efflux of more than 200 proteins. Inhibition of the XPO1 nuclear export pathway by leptomycin B (a compound produced by Streptomyces) blocks the nuclear export of BECN1 leading to interference in nutrient-deprivation induced autophagy and the suppression of mammary cell tumorigenesis [4070, 4071].
Xrn1: An exoribonuclease that hydrolyzes RNA in the 5ʹ to 3ʹ direction. Xrn1 functions as a post-transcriptional negative regulator of autophagy in yeast [4072]. Some viruses target XRN1 for downregulation to facilitate autophagy-dependent replication.
YBR139W: See Atg42.
Yeh1: See Ayr1.
Ykt6: A prenylated vesicle SNARE involved in Golgi transport and fusion with the vacuole (including Cvt vesicle delivery to the vacuole [4073]); in yeast and metazoan cells Ykt6/YKT6 is required on autophagosomes as the R-SNARE for fusion with lysosomes and vacuoles [507, 4073]. In Drosophila, Ykt6 may act as a cofactor with Syx17 [4074, 4075]. One temperature sensitive ykt6 mutation also prevents closure of the yeast phagophore [3755], whereas another only blocks autophagosome-vacuole fusion in yeast [507].
Ymr1: A yeast PtdIns3P-specific phosphatase involved in autophagosome maturation [4076, 4077]. Ymr1 is an ortholog of myotubularins. See also MTM-3, MTM1, MTMR3, phosphatidylinositol-3-phosphate, phosphatidylinositol 3,5-bisphosphate, and SBF2.
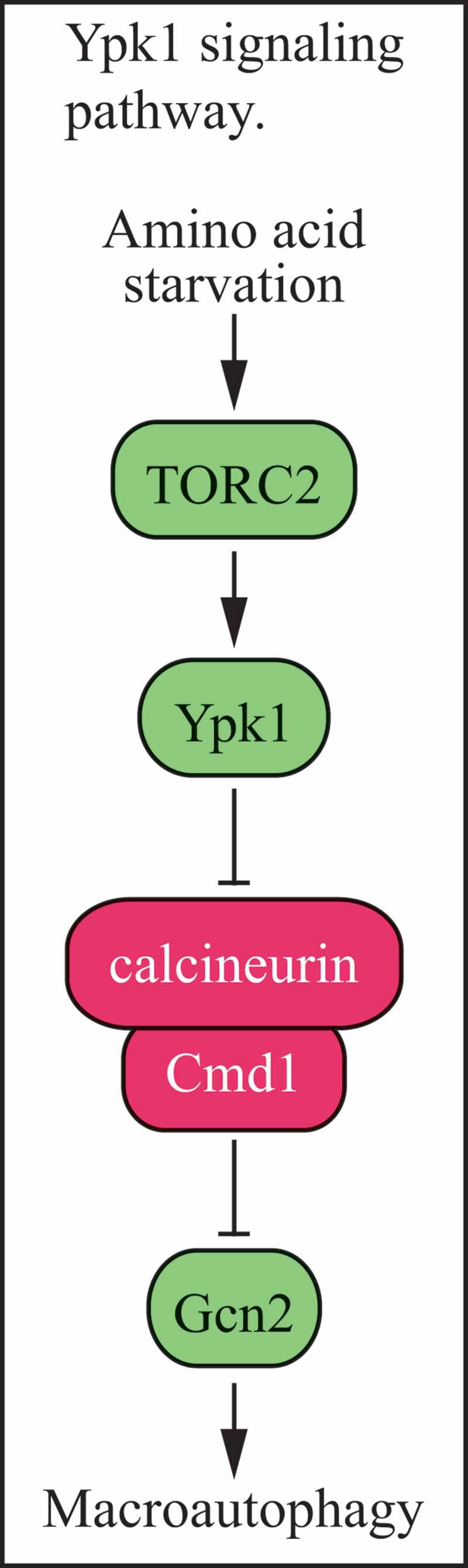
Ypk1: A downstream effector of TORC2 that stimulates autophagy under conditions of amino acid depletion [3953]. TORC2 activation of Ypk1 results in inhibition of the PPP3/calcineurin-Cmd1/calmodulin phosphatase, which otherwise dephosphorylates and inhibits Gcn2, a positive regulator of autophagy. See also Gcn2.
Ypt1: A yeast GTPase that functions in several forms of autophagy [2202]. Ypt1 is needed for correct localization of Atg8 to the PAS. The mammalian homolog, RAB1, is required for autophagosome formation and for autophagic targeting of Salmonella [4078, 4079]. See also TRAPPIII.
Ypt6: A small GTP-binding protein involved in formation of the autophagosome in yeast [2739].
Ypt7: A yeast homolog of mammalian RAB7, needed for the fusion of autophagosomes with the vacuole.
YWHAZ/14-3-3ζ (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta): A member of the 14-3-3 family of proteins that inhibits autophagy; direct interaction with PIK3C3 negatively regulates kinase activity, and this interaction is disrupted by starvation or C2-ceramide [2739].
ZFPM1/FOG1 (zinc finger protein, FOG family member 1): A cofactor of GATA1, a positive regulator of autophagy gene transcription [981]. See also GATA1.
ZFYVE1/DFCP1 (zinc finger FYVE-type containing 1): A PtdIns3P-binding protein that localizes to the omegasome [884]. Knockdown of ZFYVE1 does not result in an autophagy-defective phenotype.
ZFYVE26/spastizin/SPG15 (zinc finger FYVE-type containing 26): Mutations in ZFYVE26 result in a complicated form of hereditary spastic paraparesis (SPG15); this protein interacts with the autophagy complex BECN1-UVRAG-RUBCN and is required for autophagosome maturation [4080]. ZFYVE26 interacts also with the AP5 complex and is involved in autophagic lysosome reformation [3818]. The loss of ZFYVE26 function results in the accumulation of autophagosomes and enlarged lysosomes. See also AP5, autophagic lysosome reformation and SPG11.
ZIPK: See Sqa.
ZKSCAN3/ZNF306 (zinc finger with KRAB and SCAN domains 3): A zinc finger family transcription factor harboring Kruppel-associated box and SCAN domains that functions as a master transcriptional repressor of autophagy and lysosome biogenesis. ZKSCAN3 represses the transcription of more than 60 genes integral to, or regulatory for, autophagy and lysosome biogenesis and/or function and a subset of these genes, including MAP1LC3B and WIPI2, are its direct targets. Starvation and torin1 treatment induce translocation of ZKSCAN3 from the nucleus to the cytoplasm [985].
Zoledronic acid: A bisphosphonate that induces autophagy and may result in autophagic cell death in prostate and breast cancer cells [985, 4081]. Zoledronic acid-induced autophagy is inhibited by anti-oxidants; hence autophagy can also be modulated by oxidative stress [4081].
Zymophagy: The selective degradation of activated zymogen granules by an autophagy-like process that is dependent on VMP1, SQSTM1 and the ubiquitin protease USP9X [1497]. See also crinophagy.
Quick guide
Whenever possible, use more than one assay to monitor autophagy.
Whenever possible, include flux measurements for autophagy (e.g., using timed inhibitor studies to estimate cargo degradation rates or using tandem fluorochrome assays for maturation such as RFP-EGFP-LC3 or, preferably, cargo-specific variations thereof).
Whenever possible, use genetic inhibition of autophagy to complement studies with nonspecific pharmacological inhibitors such as 3-MA.
For analysis of genetic inhibition, a minimum of two ATG genes (including for example BECN1, ATG7, LC3/GABARAP or ULK1) should be targeted to help ensure the phenotype is due to inhibition of autophagy.
When monitoring GFP-LC3 puncta formation, provide quantification, ideally in the form of number of puncta per cell. In cells that are difficult to transfect, endogenous LC3 puncta can be monitored by immunofluorescence.
Whenever possible, use antibodies that detect specific isoforms of LC3 and GABARAP proteins.
For the interpretation of decreased SQSTM1 levels, measure mRNA levels and use strong lysis reagents (such as SDS), proteasome inhibitors and a pan-caspase inhibitor to ensure that the reduced SQSTM1 amount is not due to reduced biosynthesis, accumulation of detergent-resistant aggregates that are not solubilized in Triton X-100, proteasomal degradation or a caspase-induced cleavage of the protein.
Whenever possible, monitor autophagic responses using both short-term and long-term assays.
Figure 5.
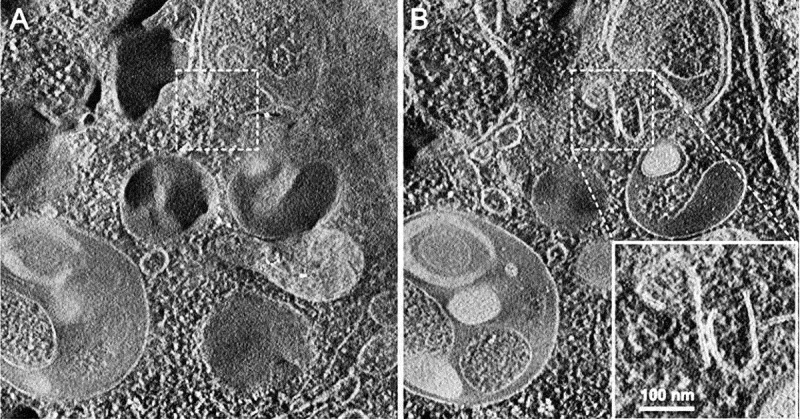
Cryoelectron microscopy can be used as a three-dimensional approach to monitor the autophagic process. Computed sections of an electron tomogram of the autophagic vacuole-rich cytoplasm in a hemophagocyte of a semi-thin section after high-pressure freezing preparation. The dashed area is membrane-free (A) but tomography reveals newly formed or degrading membranes with a parallel stretch (B). Image published previously [4082] and provided by M. Schneider and P. Walter
Acknowledgments
In a rapidly expanding and highly dynamic field such as autophagy, it is possible that some authors who should have been included on this manuscript have been missed. D.J.K. extends his apologies to researchers in the field of autophagy who, due to oversight or any other reason, could not be included on this manuscript.
Funding Statement
This work was supported by the National Institute of General Medical Sciences [GM131919]. Due to space and other limitations, it is not possible to include all other sources of financial support.
Footnotes
This table is not meant to be complete, as there are many compounds and genetic methods that regulate autophagy, and new ones are routinely being discovered. See also [2575] for a partial listing of chemical and phytochemical modulators.
Contact the T. Bozkurt lab at Imperial College London.
Obtained from http://elm.eu.org
This table is not meant to provide a comprehensive list.
Disclaimer
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the U.S. Food and Drug Administration and the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure statement
No potential conflict of interest was reported by the corresponding author.
References
- 1.Klionsky DJ, Abdalla FC, Abeliovich H, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012. Apr;8(4):445–544. PubMed PMID: 22966490; PubMed Central PMCID: PMC3404883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 2016;12(1):1–222. PubMed PMID: 26799652; PubMed Central PMCID: PMCPMC4835977. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klionsky DJ, Abeliovich H, Agostinis P, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008. Feb;4(2):151–75. PubMed PMID: 18188003; PubMed Central PMCID: PMC2654259. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klionsky DJ, Cuervo AM, Seglen PO.. Methods for monitoring autophagy from yeast to human. Autophagy. 2007. May-Jun;3(3):181–206. PubMed PMID: 17224625; eng. [DOI] [PubMed] [Google Scholar]
- 5.Xia HG, Najafov A, Geng J, et al. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol. 2015. Aug 31;210(5):705–16. PubMed PMID: 26323688; PubMed Central PMCID: PMC4555813. doi: 10.1083/jcb.201503044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eskelinen E-L, Reggiori F, Baba M, et al. Seeing is believing: the impact of electron microscopy on autophagy research. Autophagy. 2011. Sep 1;7(9):935–56. PubMed PMID: 21566462; Eng. [DOI] [PubMed] [Google Scholar]
- 7.Klionsky DJ. The autophagosome is overrated! [Editorial]. Autophagy. 2011. Apr;7(4):353–4. PubMed PMID: 21258205; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seglen PO. Regulation of autophagic protein degradation in isolated liver cells. In: Glaumann H, Ballard FJ, editors. Lysosomes: their role in protein breakdown. London: Academic Press; 1987. p. 369–414. [Google Scholar]
- 9.Knorr RL, Lipowsky R, Dimova R.. Autophagosome closure requires membrane scission. Autophagy. 2015. Nov 2;11(11):2134–2137. PubMed PMID: 26466816; PubMed Central PMCID: PMCPMC4824592. doi: 10.1080/15548627.2015.1091552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knorr RL, Mizushima N, Dimova R.. Fusion and scission of membranes: Ubiquitous topological transformations in cells. Traffic. 2017. Nov;18(11):758–761. PubMed PMID: 28799689. doi: 10.1111/tra.12509. [DOI] [PubMed] [Google Scholar]
- 11.Gordon PB, Seglen PO.. Prelysosomal convergence of autophagic and endocytic pathways. Biochem Biophys Res Commun. 1988. Feb 29;151(1):40–7. PubMed PMID: 3126737; eng. [DOI] [PubMed] [Google Scholar]
- 12.Lucocq JM, Hacker C.. Cutting a fine figure: on the use of thin sections in electron microscopy to quantify autophagy. Autophagy. 2013. Sep;9(9):1443–8. PubMed PMID: 23881027. doi: 10.4161/auto.25570. [DOI] [PubMed] [Google Scholar]
- 13.Kovács J, Fellinger E, Karpati AP, et al. Morphometric evaluation of the turnover of autophagic vacuoles after treatment with Triton X-100 and vinblastine in murine pancreatic acinar and seminal vesicle epithelial cells. Virchows Arch B Cell Pathol Incl Mol Pathol. 1987;53(3):183–90. PubMed PMID: 2888237; eng. [DOI] [PubMed] [Google Scholar]
- 14.Kovács J, Fellinger E, Karpati PA, et al. The turnover of autophagic vacuoles: evaluation by quantitative electron microscopy. Biomed Biochim Acta. 1986;45(11–12):1543–7. PubMed PMID: 3579875; eng. [PubMed] [Google Scholar]
- 15.Kovács J, Laszlo L, Kovács AL.. Regression of autophagic vacuoles in pancreatic acinar, seminal vesicle epithelial, and liver parenchymal cells: a comparative morphometric study of the effect of vinblastine and leupeptin followed by cycloheximide treatment. Exp Cell Res. 1988. Jan;174(1):244–51. PubMed PMID: 3335225; eng. [DOI] [PubMed] [Google Scholar]
- 16.Chu CT. Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol. 2006. May;65(5):423–32. PubMed PMID: 16772866; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fass E, Shvets E, Degani I, et al. Microtubules support production of starvation-induced autophagosomes but not their targeting and fusion with lysosomes. J Biol Chem. 2006. Nov 24;281(47):36303–16. PubMed PMID: 16963441; eng. [DOI] [PubMed] [Google Scholar]
- 18.Kovács AL, Reith A, Seglen PO.. Accumulation of autophagosomes after inhibition of hepatocytic protein degradation by vinblastine, leupeptin or a lysosomotropic amine. Exp Cell Res. 1982. Jan;137(1):191–201. PubMed PMID: 7056284; eng. [DOI] [PubMed] [Google Scholar]
- 19.Bestebroer J, V’Kovski P, Mauthe M, et al. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic. 2013. Oct;14(10):1029–41. PubMed PMID: 23837619. doi: 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo SM, Ge ZJ, Wang ZW, et al. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci U S A. 2013. Aug 6;110(32):13038–43. PubMed PMID: 23878233; PubMed Central PMCID: PMC3740871. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Politi Y, Gal L, Kalifa Y, et al. Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev Cell. 2014. May 12;29(3):305–20. PubMed PMID: 24823375. doi: 10.1016/j.devcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Toth S, Nagy K, Palfia Z, et al. Cellular autophagic capacity changes during azaserine-induced tumour progression in the rat pancreas. Up-regulation in all premalignant stages and down-regulation with loss of cycloheximide sensitivity of segregation along with malignant transformation. Cell Tissue Res. 2002. Sep;309(3):409–16. PubMed PMID: 12195297; eng. doi: 10.1007/s00441-001-0506-7. [DOI] [PubMed] [Google Scholar]
- 23.Loos B, Engelbrecht AM.. Cell death: a dynamic response concept. Autophagy. 2009. Jul;5(5):590–603. PubMed PMID: 19363298; eng. [DOI] [PubMed] [Google Scholar]
- 24.Seglen PO, Gordon PB, Grinde B, et al. Inhibitors and pathways of hepatocytic protein degradation. Acta Biol Med Ger. 1981;40(10–11):1587–98. PubMed PMID: 7342604. [PubMed] [Google Scholar]
- 25.Luhr M, Saetre F, Engedal N.. The long-lived protein degradation assay: an efficient method for quantitative determination of the autophagic flux of endogenous proteins in adherent cell lines. bio-protocol. 2018;8(9). doi: 10.21769/BioProtoc.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ktistakis NT, Andrews S, Long J.. What is the advantage of a transient precursor in autophagosome biogenesis?. Autophagy. 2011. Jan;7(1):118–22. PubMed PMID: 20935487; eng. doi: 10.1083/jcb.200803137. [DOI] [PubMed] [Google Scholar]
- 27.Beugnet A, Tee AR, Taylor PM, et al. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem J. 2003. Jun1;372(Pt 2):555–66. PubMed PMID: 12611592; PubMed Central PMCID: PMC1223408. eng. doi: 10.1042/BJ20021266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kovács AL, Réz G, Pálfia Z, et al. Autophagy in the epithelial cells of murine seminal vesicle in vitro. Formation of large sheets of nascent isolation membranes, sequestration of the nucleus and inhibition by wortmannin and 3-methyladenine. Cell Tissue Res. 2000. Nov;302(2):253–61. PubMed PMID: 11131136. [DOI] [PubMed] [Google Scholar]
- 29.Stavoe AK, Hill SE, Hall DH, et al. KIF1A/UNC-104 Transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev Cell. 2016. Jul 25;38(2):171–85. PubMed PMID: 27396362; PubMed Central PMCID: PMCPMC4961624. doi: 10.1016/j.devcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mizushima N, Yoshimori T.. How to interpret LC3 immunoblotting. Autophagy. 2007. Nov-Dec;3(6):542–5. PubMed PMID: 17611390; eng. [DOI] [PubMed] [Google Scholar]
- 31.Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009. Oct 1;461(7264):654–8. doi: 10.1038/nature08455. PubMed PMID: 19794493; eng. [DOI] [PubMed] [Google Scholar]
- 32.Szalai P, Hagen LK, Saetre F, et al. Autophagic bulk sequestration of cytosolic cargo is independent of LC3, but requires GABARAPs. Exp Cell Res. 2015. Apr 10;333(1):21–38. PubMed PMID: 25684710. doi: 10.1016/j.yexcr.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Engedal N, Seglen PO.. Autophagy of cytoplasmic bulk cargo does not require LC3. Autophagy. 2016;12(2):439–41. PubMed PMID: 26237084; PubMed Central PMCID: PMCPMC4836025. doi: 10.1080/15548627.2015.1076606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen TN, Padman BS, Usher J, et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome-lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J Cell Biol. 2016. Dec 19;215(6):857–874. PubMed PMID: 27864321; PubMed Central PMCID: PMCPMC5166504. doi: 10.1083/jcb.201607039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaites LP, Paulo JA, Huttlin EL, et al. Systematic analysis of human cells lacking ATG8 proteins uncovers roles for GABARAPs and the CCZ1/MON1 regulator C18orf8/RMC1 in macroautophagic and selective autophagic flux. Mol Cell Biol. 2018. Jan 1;38(1). PubMed PMID: 29038162; PubMed Central PMCID: PMCPMC5730722. doi: 10.1128/MCB.00392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galluzzi L, Pietrocola F, Levine B, et al. Metabolic control of autophagy. Cell. 2014. Dec 4;159(6):1263–1276. PubMed PMID: 25480292. doi: 10.1016/j.cell.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loos B, Engelbrecht AM, Lockshin RA, et al. The variability of autophagy and cell death susceptibility: Unanswered questions. Autophagy. 2013. Sep;9(9):1270–85. PubMed PMID: 23846383; PubMed Central PMCID: PMC4026026. doi: 10.4161/auto.25560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McWilliams TG, Prescott AR, Allen GF, et al. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016. Aug 1;214(3):333–45. PubMed PMID: 27458135; PubMed Central PMCID: PMCPMC4970326. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hombrebueno JR, Cairns L, Dutton LR, et al. Uncoupled turnover disrupts mitochondrial quality control in diabetic retinopathy. JCI Insight. 2019. Dec 5;4(23). PubMed PMID: 31661466. doi: 10.1172/jci.insight.129760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calvo-Garrido J, Carilla-Latorre S, Mesquita A, et al. A proteolytic cleavage assay to monitor autophagy in Dictyostelium discoideum. Autophagy. 2011. Sep 1;7(9):1063–8. PubMed PMID: 21876387; eng. doi: 10.4161/auto.7.9.16629. [DOI] [PubMed] [Google Scholar]
- 41.Chu CT, Plowey ED, Dagda RK, et al. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 2009;453:217–49. PubMed PMID: 19216909; PubMed Central PMCID: PMC2669321. eng. doi: 10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geng J, Klionsky DJ.. Determining Atg protein stoichiometry at the phagophore assembly site by fluorescence microscopy. Autophagy. 2010. Jan;6(1):144–7. PubMed PMID: 20131413; PubMed Central PMCID: PMC2841983. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grander D, Kharaziha P, Laane E, et al. Autophagy as the main means of cytotoxicity by glucocorticoids in hematological malignancies. Autophagy. 2009. Nov;5(8):1198–200. PubMed PMID: 19855186; eng. [DOI] [PubMed] [Google Scholar]
- 44.He C, Klionsky DJ.. Analyzing autophagy in zebrafish. Autophagy. 2010. Jul 19;6(5). PubMed PMID: 20495344; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanki T, Kang D, Klionsky DJ.. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy. 2009. Nov;5(8):1186–9. PubMed PMID: 19806021; PubMed Central PMCID: PMC2850110. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klionsky DJ. Autophagy: lower eukaryotes and non-mammalian systems, part A. In: Klionsky DJ, editor. Methods enzymol. Vol. 451. Amsterdam: Academic Press/Elsevier; 2008. 2009 2 3 eng. [Google Scholar]
- 47.Klionsky DJ. Autophagy in disease and clinical applications, Part C. In: Klionsky DJ, editor. Methods enzymol Vol. 453. Amsterdam: Academic Press/Elsevier; 2008. [Google Scholar]
- 48.Klionsky DJ. Autophagy in mammalian systems, part B. In: Klionsky DJ, editor. Methods enzymol. Vol. 452. Amsterdam: Academic Press/Elsevier; 2008. [Google Scholar]
- 49.Raju D, Jones NL.. Methods to monitor autophagy in H. pylori vacuolating cytotoxin A (VacA)-treated cells [Review]. Autophagy. 2010. Jan;6(1):138–43. PubMed PMID: 19875940; eng. [DOI] [PubMed] [Google Scholar]
- 50.Seglen PO, Brinchmann MF.. Purification of autophagosomes from rat hepatocytes. Autophagy. 2010. May 21;6(4):542–7. PubMed PMID: 20505360; Eng. [DOI] [PubMed] [Google Scholar]
- 51.Swanlund JM, Kregel KC, Oberley TD.. Investigating autophagy: quantitative morphometric analysis using electron microscopy. Autophagy. 2010. Feb;6(2):270–7. PubMed PMID: 19923921; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welter E, Thumm M, Krick R.. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy. 2010. Aug;6(6):794–7. PubMed PMID: 20523132; eng. [DOI] [PubMed] [Google Scholar]
- 53.Xu F, Liu XH, Zhuang FL, et al. Analyzing autophagy in Magnaporthe oryzae. Autophagy. 2011. May;7(5):525–30. PubMed PMID: 21317549; eng. [DOI] [PubMed] [Google Scholar]
- 54.Zhang J, Ney PA.. Reticulocyte mitophagy: monitoring mitochondrial clearance in a mammalian model. Autophagy. 2010. Apr;6(3):405–8. PubMed PMID: 20200480; eng. [DOI] [PubMed] [Google Scholar]
- 55.Zhu J, Dagda RK, Chu CT.. Monitoring mitophagy in neuronal cell cultures. Methods Mol Biol. 2011;793:325–39. doi: 10.1007/978-1-61779-328-8_21. PubMed PMID: 21913110; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luhr M, Szalai P, Engedal N.. The Lactate dehydrogenase sequestration assay - a simple and reliable method to determine bulk autophagic sequestration activity in mammalian cells. J vis exp. 2018. Jul;27(137). PubMed PMID: 30102280; PubMed Central PMCID: PMCPMC6126555. doi: 10.3791/57971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klionsky DJ, Bruford EA, Cherry JM, et al. In the beginning there was babble. Autophagy. 2012. Aug;8(8):1165–7. PubMed PMID: 22836666; PubMed Central PMCID: PMC3625114. doi: 10.4161/auto.20665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotoulas OB, Kalamidas SA, Kondomerkos DJ.. Glycogen autophagy. Microsc Res Tech. 2004. May 1;64(1):10–20. PubMed PMID: 15287014. doi: 10.1002/jemt.20046. [DOI] [PubMed] [Google Scholar]
- 59.Kotoulas OB, Kalamidas SA, Kondomerkos DJ.. Glycogen autophagy in glucose homeostasis. Pathol Res Pract. 2006;202(9):631–8. PubMed PMID: 16781826. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Singh PK, Singh S.. Changing shapes of glycogen-autophagy nexus in neurons: perspective from a rare epilepsy. Front Neurol. 2015;6:14. PubMed PMID: 25699013; PubMed Central PMCID: PMC4316721. doi: 10.3389/fneur.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lincoln C, Long J, Yamaguchi J, et al. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994. Dec;6(12):1859–76. PubMed PMID: 7866029; PubMed Central PMCID: PMCPMC160567. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cui Y, Cao W, He Y, et al. A whole-cell electron tomography model of vacuole biogenesis in arabidopsis root cells. Nat Plants. 2019. Jan;5(1):95–105. PubMed PMID: 30559414. doi: 10.1038/s41477-018-0328-1. [DOI] [PubMed] [Google Scholar]
- 63.Yla-Anttila P, Vihinen H, Jokitalo E, et al. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009;452:143–64. PubMed PMID: 19200881; eng. doi: 10.1016/S0076-6879(08)03610-0. [DOI] [PubMed] [Google Scholar]
- 64.Cardenal-Munoz E, Arafah S, Lopez-Jimenez AT, et al. Mycobacterium marinum antagonistically induces an autophagic response while repressing the autophagic flux in a TORC1- and ESX-1-dependent manner. PLoS Pathog. 2017. Apr;13(4):e1006344. PubMed PMID: 28414774; PubMed Central PMCID: PMCPMC5407849. doi: 10.1371/journal.ppat.1006344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eskelinen E-L. Maturation of autophagic vacuoles in mammalian cells. Autophagy. 2005. Apr;1(1):1–10. PubMed PMID: 16874026; eng. [DOI] [PubMed] [Google Scholar]
- 66.Eskelinen E-L. To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells [Evaluation Studies Research Support, Non-U.S. Gov’t]. Autophagy. 2008. Feb;4(2):257–60. PubMed PMID: 17986849; eng. [DOI] [PubMed] [Google Scholar]
- 67.Eskelinen E-L, Kovacs AL.. Double membranes vs. lipid bilayers, and their significance for correct identification of macroautophagic structures. Autophagy. 2011. Sep 1;7(9):931–2. PubMed PMID: 21642767; Eng. [DOI] [PubMed] [Google Scholar]
- 68.Biazik J, Yla-Anttila P, Vihinen H, et al. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy. 2015;11(3):439–51. PubMed PMID: 25714487; PubMed Central PMCID: PMC4502653. doi: 10.1080/15548627.2015.1017178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Berg TO, Fengsrud M, Stromhaug PE, et al. Isolation and characterization of rat liver amphisomes. Evidence for fusion of autophagosomes with both early and late endosomes. J Biol Chem. 1998. Aug 21;273(34):21883–92. PubMed PMID: 9705327; eng. [DOI] [PubMed] [Google Scholar]
- 70.Eskelinen E-L. Macroautophagy in mammalian cells. In: Saftig P, editor. Lysosomes. Georgetown, TX: LandesBioscience/Eurekah.com; 2005. [Google Scholar]
- 71.Turturici G, Tinnirello R, Sconzo G, et al. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Physiol. 2014. Apr 1;306(7):C621–33. PubMed PMID: 24452373. doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 72.Eskelinen E-L. Fine structure of the autophagosome. In: Deretic V, editor. Autophagosome and phagosome. Methods in molecular biology. Vol. 445. Totowa, NJ: Humana Press; 2008. p. 11–28. [DOI] [PubMed] [Google Scholar]
- 73.Di Rienzo M, Antonioli M, Fusco C, et al. Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci Adv. 2019. May;5(5):eaau8857. PubMed PMID: 31123703; PubMed Central PMCID: PMCPMC6527439. doi: 10.1126/sciadv.aau8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015. Jun 18;522(7556):354–8. PubMed PMID: 26040720. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 75.Zielke S, Meyer N, Mari M, et al. Loperamide, pimozide, and STF-62247 trigger autophagy-dependent cell death in glioblastoma cells. Cell Death Dis. 2018. Sep 24;9(10):994. PubMed PMID: 30250198; PubMed Central PMCID: PMCPMC6155211. doi: 10.1038/s41419-018-1003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang DS, Lee JH, Nixon RA.. Monitoring autophagy in Alzheimer’s disease and related neurodegenerative diseases. Methods Enzymol. 2009;453:111–44. PubMed PMID: 19216904. doi: 10.1016/S0076-6879(08)04006-8. [DOI] [PubMed] [Google Scholar]
- 77.Yokota S, Himeno M, Kato K.. Immunocytochemical localization of acid phosphatase in rat liver. Cell Struct Funct. 1989. Apr;14(2):163–71. PubMed PMID: 2743419. [DOI] [PubMed] [Google Scholar]
- 78.Boland B, Kumar A, Lee S, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008. Jul 2;28(27):6926–37. PubMed PMID: 18596167; PubMed Central PMCID: PMC2676733. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nixon RA, Wegiel J, Kumar A, et al. Extensive involvement of autophagy in Alzheimer disease:an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005. Feb;64(2):113–22. PubMed PMID: 15751225; eng. [DOI] [PubMed] [Google Scholar]
- 80.Lee JH, McBrayer MK, Wolfe DM, et al. Presenilin 1 maintains lysosomal Ca homeostasis via TRPML1 by regulating vATpase-mediated lysosome acidification. Cell Rep. 2015. Aug 19. PubMed PMID: 26299959. doi: 10.1016/j.celrep.2015.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee JH, Yu WH, Kumar A, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010. Jun 25;141(7):1146–58. PubMed PMID: 20541250; eng. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chu CT. A pivotal role for PINK1 and autophagy in mitochondrial quality control: implications for Parkinson disease. Hum Mol Genet. 2010. Apr 15;19(R1):R28–37. PubMed PMID: 20385539; PubMed Central PMCID: PMC2875056. eng. doi: 10.1093/hmg/ddq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee S, Sato Y, Nixon RA.. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J Neurosci. 2011. May 25;31(21):7817–30. PubMed PMID: 21613495; PubMed Central PMCID: PMC3351137. doi: 10.1523/JNEUROSCI.6412-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maday S, Wallace KE, Holzbaur EL.. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J Cell Biol. 2012. Feb 20;196(4):407–17. PubMed PMID: 22331844; PubMed Central PMCID: PMCPMC3283992. doi: 10.1083/jcb.201106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Largeau C, Legouis R.. Correlative Light and Electron Microscopy to Analyze LC3 Proteins in Caenorhabditis elegans Embryo. Methods Mol Biol. 2019;1880:281–293. PubMed PMID: 30610704. doi: 10.1007/978-1-4939-8873-0_18. [DOI] [PubMed] [Google Scholar]
- 86.Zhan L, Chen S, Li K, et al. Autophagosome maturation mediated by Rab7 contributes to neuroprotection of hypoxic preconditioning against global cerebral ischemia in rats. Cell Death Dis. 2017. Jul 20;8(7):e2949. PubMed PMID: 28726776; PubMed Central PMCID: PMCPMC5550874. doi: 10.1038/cddis.2017.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rabouille C, Strous GJ, Crapo JD, et al. The differential degradation of two cytosolic proteins as a tool to monitor autophagy in hepatocytes by immunocytochemistry. J Cell Biol. 1993. Feb;120(4):897–908. PubMed PMID: 8432730; PubMed Central PMCID: PMC2200086. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kovács AL, Pálfia Z, Réz G, et al. Sequestration revisited: integrating traditional electron microscopy, de novo assembly and new results. Autophagy. 2007;3:655–662. [DOI] [PubMed] [Google Scholar]
- 89.Gao W, Kang JH, Liao Y, et al. Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment. J Biol Chem. 2010. Jan 8;285(2):1371–83. PubMed PMID: 19910472; PubMed Central PMCID: PMC2801263. eng. doi: 10.1074/jbc.M109.054197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lajoie P, Guay G, Dennis JW, et al. The lipid composition of autophagic vacuoles regulates expression of multilamellar bodies. J Cell Sci. 2005. May 1;118(Pt 9):1991–2003. PubMed PMID: 15840653. doi: 10.1242/jcs.02324. [DOI] [PubMed] [Google Scholar]
- 91.Pentchev PG, Comly ME, Kruth HS, et al. Group C Niemann-Pick disease: faulty regulation of low-density lipoprotein uptake and cholesterol storage in cultured fibroblasts. FASEB J. 1987. Jul;1(1):40–5. PubMed PMID: 3609608. doi: 10.1096/fasebj.1.1.3609608. [DOI] [PubMed] [Google Scholar]
- 92.Garcia-Sanz P, Orgaz L, Bueno-Gil G, et al. N370S-GBA1 mutation causes lysosomal cholesterol accumulation in Parkinson’s disease. Mov Disord. 2017. Oct;32(10):1409–1422. PubMed PMID:28779532. doi: 10.1002/mds.27119. [DOI] [PubMed] [Google Scholar]
- 93.Garcia-Sanz P, Orgaz L, Fuentes JM, et al. Cholesterol and multilamellar bodies: lysosomal dysfunction in GBA-Parkinson disease. Autophagy. 2018;14(4):717–718. PubMed PMID: 29368986; PubMed Central PMCID: PMCPMC5959320. doi: 10.1080/15548627.2018.1427396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shahmoradian SH, Lewis AJ, Genoud C, et al. Lewy pathology in Parkinson’s disease consists of crowded organelles and lipid membranes. Nat Neurosci. 2019. Jul;22(7):1099–1109. PubMed PMID: 31235907. doi: 10.1038/s41593-019-0423-2. [DOI] [PubMed] [Google Scholar]
- 95.Cenedella RJ. Cholesterol synthesis inhibitor U18666A and the role of sterol metabolism and trafficking in numerous pathophysiological processes. Lipids. 2009. Jun;44(6):477–87. PubMed PMID: 19440746. doi: 10.1007/s11745-009-3305-7. [DOI] [PubMed] [Google Scholar]
- 96.Elgner F, Ren H, Medvedev R, et al. The intracellular cholesterol transport inhibitor U18666A inhibits the exosome-dependent release of mature hepatitis C virus. J Virol. 2016. Dec 15;90(24):11181–11196. PubMed PMID: 27707921; PubMed Central PMCID: PMCPMC5126375. doi: 10.1128/JVI.01053-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.King MA, Ganley IG, Flemington V.. Inhibition of cholesterol metabolism underlies synergy between mTOR pathway inhibition and chloroquine in bladder cancer cells. Oncogene. 2016. Aug 25;35(34):4518–28. PubMed PMID: 26853465; PubMed Central PMCID: PMCPMC5000518 interest. doi: 10.1038/onc.2015.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mayhew TM. Quantitative immunoelectron microscopy: alternative ways of assessing subcellular patterns of gold labeling. Methods Mol Biol. 2007;369:309–29. PubMed PMID: 17656757; eng. [DOI] [PubMed] [Google Scholar]
- 99.Mayhew TM, Lucocq JM, Griffiths G.. Relative labelling index: a novel stereological approach to test for non-random immunogold labelling of organelles and membranes on transmission electron microscopy thin sections. J Microsc. 2002. Feb;205(Pt 2):153–64. PubMed PMID: 11879430; eng. [DOI] [PubMed] [Google Scholar]
- 100.Isidoro C, Biagioni F, Giorgi FS, et al. The role of autophagy on the survival of dopamine neurons. Curr Top Med Chem. 2009;9(10):869–79. PubMed PMID: 19754403. [PubMed] [Google Scholar]
- 101.Avin-Wittenberg T, Baluska F, Bozhkov PV, et al. Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot. 2018. Mar 14;69(6):1335–1353. PubMed PMID: 29474677. doi: 10.1093/jxb/ery069. [DOI] [PubMed] [Google Scholar]
- 102.Kuzuoglu-Ozturk D, Cebeci Yalcinkaya O, Akpinar BA, et al. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta. 2012. Oct;236(4):1081–92. PubMed PMID: 22569921. doi: 10.1007/s00425-012-1657-3. [DOI] [PubMed] [Google Scholar]
- 103.Schmid D, Pypaert M, Münz C.. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007. Jan;26(1):79–92. PubMed PMID: 17182262; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Runwal G, Stamatakou E, Siddiqi FH, et al. LC3-positive structures are prominent in autophagy-deficient cells. Sci Rep. 2019. Jul 12;9(1):10147. PubMed PMID: 31300716; PubMed Central PMCID: PMCPMC6625982. doi: 10.1038/s41598-019-46657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ponpuak M, Mandell MA, Kimura T, et al. Secretory autophagy. Curr Opin Cell Biol. 2015. Aug;35:106–16. PubMed PMID: 25988755; PubMed Central PMCID: PMC4529791. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Subramani S, Malhotra V.. Non-autophagic roles of autophagy-related proteins. EMBO Rep. 2013. Feb;14(2):143–51. PubMed PMID: 23337627; PubMed Central PMCID: PMC3566844. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Addis R, Campesi I, Fois M, et al. Human umbilical endothelial cells (HUVECs) have a sex: characterisation of the phenotype of male and female cells. Biol Sex Differ. 2014;5(1):18. PubMed PMID: 25535548; PubMed Central PMCID: PMCPMC4273493. doi: 10.1186/s13293-014-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Campesi I, Occhioni S, Capobianco G, et al. Sex-specific pharmacological modulation of autophagic process in human umbilical artery smooth muscle cells. Pharmacol Res. 2016. Nov;113(Pt A):166–174. PubMed PMID: 27521838. doi: 10.1016/j.phrs.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 109.Campesi I, Straface E, Occhioni S, et al. Protein oxidation seems to be linked to constitutive autophagy: a sex study. Life Sci. 2013. Aug 6;93(4):145–52. doi: 10.1016/j.lfs.2013.06.001. PubMed PMID: 23770210. [DOI] [PubMed] [Google Scholar]
- 110.Cosper PF, Leinwand LA.. Cancer causes cardiac atrophy and autophagy in a sexually dimorphic manner. Cancer Res. 2011. Mar 1;71(5):1710–20. doi: 10.1158/0008-5472.CAN-10-3145. PubMed PMID: 21163868; PubMed Central PMCID: PMCPMC3049989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Du L, Hickey RW, Bayir H, et al. Starving neurons show sex difference in autophagy. J Biol Chem. 2009. Jan 23;284(4):2383–96. doi: 10.1074/jbc.M804396200. PubMed PMID: 19036730; PubMed Central PMCID: PMCPMC2629091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saito T, Asai K, Sato S, et al. Proof of myocardial autophagy by combining antigen retrieval and the avidin-biotin peroxidase complex method. Int J Cardiol. 2013. Oct 12;168(5):4843–4. doi: 10.1016/j.ijcard.2013.07.032. PubMed PMID: 23871334. [DOI] [PubMed] [Google Scholar]
- 113.Kovács J. Regression of autophagic vacuoles in seminal vesicle cells following cycloheximide treatment. Exp Cell Res. 1983. Mar;144(1):231–4. PubMed PMID: 6840208; eng. [DOI] [PubMed] [Google Scholar]
- 114.Réz G, Csak J, Fellinger E, et al. Time course of vinblastine-induced autophagocytosis and changes in the endoplasmic reticulum in murine pancreatic acinar cells: a morphometric and biochemical study. Eur J Cell Biol. 1996. Dec;71(4):341–50. PubMed PMID: 8980904; eng. [PubMed] [Google Scholar]
- 115.Kovács AL, Grinde B, Seglen PO.. Inhibition of autophagic vacuole formation and protein degradation by amino acids in isolated hepatocytes. Exp Cell Res. 1981. Jun;133(2):431–6. PubMed PMID: 7238609; eng. [DOI] [PubMed] [Google Scholar]
- 116.Mortimore GE, Hutson NJ, Surmacz CA.. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci U S A. 1983. Apr;80(8):2179–83. PubMed PMID: 6340116; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mortimore GE, Lardeux BR, Adams CE.. Regulation of microautophagy and basal protein turnover in rat liver. Effects of short-term starvation. J Biol Chem. 1988. Feb 15;263(5):2506–12. PubMed PMID: 3257493; eng. [PubMed] [Google Scholar]
- 118.Zhu JH, Horbinski C, Guo F, et al. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007. Jan;170(1):75–86. PubMed PMID: 17200184; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bjørkøy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005. Nov 21;171(4):603–14. PubMed PMID: 16286508; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goginashvili A, Zhang Z, Erbs E, et al. Insulin granules. Insulin secretory granules control autophagy in pancreatic beta cells. Science. 2015. Feb 20;347(6224):878–82. doi: 10.1126/science.aaa2628. PubMed PMID: 25700520. [DOI] [PubMed] [Google Scholar]
- 121.Orvedahl A, Sumpter R, Jr., Xiao G, et al. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011. Dec 1;480(7375):113–7. doi: 10.1038/nature10546. PubMed PMID: 22020285; PubMed Central PMCID: PMC3229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Razi M, Tooze SA.. Correlative light and electron microscopy. Methods Enzymol. 2009;452:261–75. doi: 10.1016/S0076-6879(08)03617-3. PubMed PMID: 19200888; eng. [DOI] [PubMed] [Google Scholar]
- 123.Shu X, Lev-Ram V, Deerinck TJ, et al. A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol. 2011. Apr;9(4):e1001041. doi: 10.1371/journal.pbio.1001041. PubMed PMID: 21483721; PubMed Central PMCID: PMC3071375. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Giulivi C, Sarcansky M, Rosenfeld E, et al. The photodynamic effect of rose bengal on proteins of the mitochondrial inner membrane. Photochem Photobiol. 1990. Oct;52(4):745–51. doi: 10.1111/j.1751-1097.1990.tb08676.x. PubMed PMID: 2089421. [DOI] [PubMed] [Google Scholar]
- 125.Castillo K, Rojas-Rivera D, Lisbona F, et al. BAX inhibitor-1 regulates autophagy by controlling the IRE1α/JNK branch of the unfolded protein response. EMBO J. 2011;30:4465–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hayashi-Nishino M, Fujita N, Noda T, et al. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2010. Feb;6(2):301–3. PubMed PMID: 20104025; eng. [DOI] [PubMed] [Google Scholar]
- 127.Yla-Anttila P, Vihinen H, Jokitalo E, et al. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009. Nov;5(8):1180–5. PubMed PMID: 19855179; eng. [DOI] [PubMed] [Google Scholar]
- 128.Zhuang X, Chung KP, Cui Y, et al. ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc Natl Acad Sci U S A. 2017. Jan 17;114(3):E426–E435. doi: 10.1073/pnas.1616299114. PubMed PMID: 28053229; PubMed Central PMCID: PMCPMC5255614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lai LTF, Yu C, Wong JSK, et al. Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy. 2019. Jul 16:in press. doi: 10.1080/15548627.2019.1639300. PubMed PMID: 31276439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duke EM, Razi M, Weston A, et al. Imaging endosomes and autophagosomes in whole mammalian cells using correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM). Ultramicroscopy. 2014. Aug;143:77–87. doi: 10.1016/j.ultramic.2013.10.006. PubMed PMID: 24238600; PubMed Central PMCID: PMC4045213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ciuffa R, Lamark T, Tarafder AK, et al. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 2015. May 5;11(5):748–58. doi: 10.1016/j.celrep.2015.03.062. PubMed PMID: 25921531. [DOI] [PubMed] [Google Scholar]
- 132.Hurley JH, Nogales E.. Next-generation electron microscopy in autophagy research. Curr Opin Struct Biol. 2016. Dec;41:211–216. doi: 10.1016/j.sbi.2016.08.006. PubMed PMID: 27614295; PubMed Central PMCID: PMCPMC5154772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Massey AC, Kaushik S, Sovak G, et al. Consequences of the selective blockage of chaperone-mediated autophagy. Proc Natl Acad Sci U S A. 2006. Apr 11;103(15):5805–10. PubMed PMID: 16585521; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Felszeghy S, Viiri J, Paterno JJ, et al. Loss of NRF-2 and PGC-1alpha genes leads to retinal pigment epithelium damage resembling dry age-related macular degeneration. Redox Biol. 2019. Jan;20:1–12. PubMed PMID: 30253279; PubMed Central PMCID: PMCPMC6156745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baba M, Osumi M, Ohsumi Y.. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995. Dec;20(6):465–71. PubMed PMID: 8825067; eng. [DOI] [PubMed] [Google Scholar]
- 136.Rez G, Meldolesi J.. Freeze-fracture of drug-induced autophagocytosis in the mouse exocrine pancreas. Lab Invest. 1980. Sep;43(3):269–77. PubMed PMID: 7401637; eng. [PubMed] [Google Scholar]
- 137.Fengsrud M, Erichsen ES, Berg TO, et al. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol. 2000. Dec;79(12):871–82. PubMed PMID: 11152279; eng. [DOI] [PubMed] [Google Scholar]
- 138.Punnonen E-L, Pihakaski K, Mattila K, et al. Intramembrane particles and filipin labelling on the membranes of autophagic vacuoles and lysosomes in mouse liver. Cell Tissue Res. 1989. Nov;258(2):269–76. PubMed PMID: 2582478; eng. [DOI] [PubMed] [Google Scholar]
- 139.Dickey JS, Gonzalez Y, Aryal B, et al. Mito-tempol and dexrazoxane exhibit cardioprotective and chemotherapeutic effects through specific protein oxidation and autophagy in a syngeneic breast tumor preclinical model. PLoS One. 2013;8(8):e70575. doi: 10.1371/journal.pone.0070575. PubMed PMID: 23940596; PubMed Central PMCID: PMC3734284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rao VA, Klein SR, Bonar SJ, et al. The antioxidant transcription factor Nrf2 negatively regulates autophagy and growth arrest induced by the anticancer redox agent mitoquinone. J Biol Chem. 2010. Nov 5;285(45):34447–59. doi: 10.1074/jbc.M110.133579. PubMed PMID: 20805228; PubMed Central PMCID: PMC2966059. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. [DOI] [PubMed] [Google Scholar]
- 142.Krick R, Muhe Y, Prick T, et al. Piecemeal microautophagy of the nucleus: genetic and morphological traits. Autophagy. 2009. Feb;5(2):270–2. PubMed PMID: 19182523; eng. [DOI] [PubMed] [Google Scholar]
- 143.Meschini S, Condello M, Calcabrini A, et al. The plant alkaloid voacamine induces apoptosis-independent autophagic cell death on both sensitive and multidrug resistant human osteosarcoma cells. Autophagy. 2008. Nov;4(8):1020–33. PubMed PMID: 18838862; eng. [DOI] [PubMed] [Google Scholar]
- 144.Proikas-Cezanne T, Robenek H.. Freeze-fracture replica immunolabelling reveals human WIPI-1 and WIPI-2 as membrane proteins of autophagosomes. J Cell Mol Med. 2011. Sep;15(9):2007–10. doi: 10.1111/j.1582-4934.2011.01339.x. PubMed PMID: 21564513; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hirsimaki Y, Hirsimaki P, Lounatmaa K.. Vinblastine-induced autophagic vacuoles in mouse liver and Ehrlich ascites tumor cells as assessed by freeze-fracture electron microscopy. Eur J Cell Biol. 1982. Jun;27(2):298–301. PubMed PMID: 7117273; eng. [PubMed] [Google Scholar]
- 146.Kovacs J, Rez G, Kovacs AL, et al. Autophagocytosis: freeze-fracture morphology, effects of vinblastine and influence of transcriptional and translational inhibitors. Acta Biol Med Ger. 1982;41(1):131–5. PubMed PMID: 7113544; eng. [PubMed] [Google Scholar]
- 147.Backues SK, Chen D, Ruan J, et al. Estimating the size and number of autophagic bodies by electron microscopy. Autophagy. 2014. Jan;10(1):155–64. doi: 10.4161/auto.26856. PubMed PMID: 24270884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cheong H, Yorimitsu T, Reggiori F, et al. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005. Jul;16(7):3438–53. doi: 10.1091/mbc.E04-10-0894. PubMed PMID: 15901835; PubMed Central PMCID: PMC1165424. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Xie Z, Nair U, Klionsky DJ.. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008. Aug;19(8):3290–8. doi: 10.1091/mbc.E07-12-1292. PubMed PMID: 18508918; PubMed Central PMCID: PMC2488302. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Colasuonno F, Borghi R, Niceforo A, et al. Senescence-associated ultrastructural features of long-term cultures of induced pluripotent stem cells (iPSCs). Aging (Albany NY). 2017. Oct 23;9(10):2209–2222. doi: 10.18632/aging.101309. PubMed PMID: 29064821; PubMed Central PMCID: PMCPMC5680563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McDonald KL, Webb RI.. Freeze substitution in 3 hours or less. J Microsc. 2011. Sep;243(3):227–33. doi: 10.1111/j.1365-2818.2011.03526.x. PubMed PMID: 21827481. [DOI] [PubMed] [Google Scholar]
- 152.Reipert S, Goldammer H, Richardson C, et al. Agitation Modules: Flexible Means to Accelerate Automated Freeze Substitution. J Histochem Cytochem. 2018. Dec;66(12):903–921. doi: 10.1369/0022155418786698. PubMed PMID: 29969056; PubMed Central PMCID: PMCPMC6262506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kovács AL, Vellai T, Müller F.. Autophagy in Caenorhabditis elegans. In: Klionsky DJ, editor. Autophagy. Georgetown, Texas: Landes Bioscience; 2004. p. 217–23. [Google Scholar]
- 154.Sigmond T, Feher J, Baksa A, et al. Qualitative and quantitative characterization of autophagy in Caenorhabditis elegans by electron microscopy. Methods Enzymol. 2008;451:467–91. doi: 10.1016/S0076-6879(08)03228-X. PubMed PMID: 19185736; eng. [DOI] [PubMed] [Google Scholar]
- 155.Electron microscopy of model systems. In: Müller-Reichert T, editor. Methods cell biol. Vol. 96. 2010. 2010 09 28. [Google Scholar]
- 156.Nowikovsky K, Reipert S, Devenish RJ, et al. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007. Sep;14(9):1647–56. doi: 10.1038/sj.cdd.4402167. PubMed PMID: 17541427. [DOI] [PubMed] [Google Scholar]
- 157.Roberts P, Moshitch-Moshkovitz S, Kvam E, et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003. Jan;14(1):129–41. PubMed PMID: 12529432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Howard V, Reed MG.. Unbiased stereology; three dimensional measurement in microscopy. U Bios Scientific Publishers; 1998. [Google Scholar]
- 159.Weibel ER. Practical Methods for Biological Morphometry. In: Stereological Methods. Vol. 1. Academic Press, New York; 1979. [Google Scholar]
- 160.Williams MA. Quantitative methods in biology: Practical methods in electron microscopy. Vol. 6. Amsterdam, New York, Oxford: North-Holland Publishing Company; 1977. [Google Scholar]
- 161.Eskelinen EL. Fine structure of the autophagosome. Methods Mol Biol. 2008;445:11–28. doi: 10.1007/978-1-59745-157-4_2. PubMed PMID: 18425441. [DOI] [PubMed] [Google Scholar]
- 162.Kovacs AL. A simple method to estimate the number of autophagic elements by electron microscopic morphometry in real cellular dimensions. Biomed Res Int. 2014;2014:578698. doi: 10.1155/2014/578698. PubMed PMID: 25105130; PubMed Central PMCID: PMC4106081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Xie Z, Nair U, Geng J, et al. Indirect estimation of the area density of Atg8 on the phagophore. Autophagy. 2009. Feb;5(2):217–20. PubMed PMID: 19088501; PubMed Central PMCID: PMC2941343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kovacs AL, Laszlo L, Fellinger E, et al. Combined effects of fasting and vinblastine treatment on serum insulin level, the size of autophagic-lysosomal compartment, protein content and lysosomal enzyme activities of liver and exocrine pancreatic cells of the mouse. Comp biochem physiol B Comp biochem. 1989;94(3):505–10. PubMed PMID: 2695284; eng. [DOI] [PubMed] [Google Scholar]
- 165.Punnonen EL, Reunanen H.. Effects of vinblastine, leucine, and histidine, and 3-methyladenine on autophagy in Ehrlich ascites cells. Exp Mol Pathol. 1990. Feb;52(1):87–97. PubMed PMID: 2307216; eng. [DOI] [PubMed] [Google Scholar]
- 166.Griffiths G. Fine structure immunocytochemistry Heidelberg, Germany: Springer-Verlag; 1993. [Google Scholar]
- 167.Reyes FC, Chung T, Holding D, et al. Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell. 2011. Feb;23(2):769–84. doi: 10.1105/tpc.110.082156. PubMed PMID: 21343414; PubMed Central PMCID: PMC3077793. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dunn WA, Jr., Cregg JM, Kiel JAKW, et al. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005. Jul;1(2):75–83. PubMed PMID: 16874024. [DOI] [PubMed] [Google Scholar]
- 169.Wang K, Klionsky DJ.. Mitochondria removal by autophagy. Autophagy. 2011. Mar;7(3):297–300. PubMed PMID: 21252623; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Belanger M, Rodrigues PH, Dunn WA, Jr., et al. Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy. 2006. Jul-Sep;2(3):165–70. PubMed PMID: 16874051; eng. [DOI] [PubMed] [Google Scholar]
- 171.Birmingham CL, Brumell JH.. Autophagy recognizes intracellular Salmonella enterica serovar Typhimurium in damaged vacuoles. Autophagy. 2006. Jul-Sep;2(3):156–8. PubMed PMID: 16874057; eng. [DOI] [PubMed] [Google Scholar]
- 172.Colombo MI, Gutierrez MG, Romano PS.. The two faces of autophagy: Coxiella and Mycobacterium. Autophagy. 2006. Jul-Sep;2(3):162–4. PubMed PMID: 16874070; eng. [DOI] [PubMed] [Google Scholar]
- 173.Ogawa M, Sasakawa C.. Shigella and autophagy. Autophagy. 2006. Jul-Sep;2(3):171–4. PubMed PMID: 16874102; eng. [DOI] [PubMed] [Google Scholar]
- 174.Vergne I, Singh S, Roberts E, et al. Autophagy in immune defense against Mycobacterium tuberculosis. Autophagy. 2006. Jul-Sep;2(3):175–8. PubMed PMID: 16874111; eng. [DOI] [PubMed] [Google Scholar]
- 175.Yoshimori T. Autophagy vs. Group A Streptococcus. Autophagy. 2006. Jul-Sep;2(3):154–5. PubMed PMID: 16874113; eng. [DOI] [PubMed] [Google Scholar]
- 176.Gorbunov NV, McDaniel DP, Zhai M, et al. Autophagy and mitochondrial remodelling in mouse mesenchymal stromal cells challenged with Staphylococcus epidermidis. J Cell Mol Med. 2015. May;19(5):1133–50. doi: 10.1111/jcmm.12518. PubMed PMID: 25721260; PubMed Central PMCID: PMC4420615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lynch-Day MA, Klionsky DJ.. The Cvt pathway as a model for selective autophagy [Review]. FEBS Lett. 2010. Apr 2;584(7):1359–66. doi: 10.1016/j.febslet.2010.02.013. PubMed PMID: 20146925; PubMed Central PMCID: PMC2843786. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Birmingham CL, Canadien V, Gouin E, et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007. Sep-Oct;3(5):442–51. PubMed PMID: 17568179; eng. [DOI] [PubMed] [Google Scholar]
- 179.Klionsky DJ. Protein transport from the cytoplasm into the vacuole. J Membr Biol. 1997. May 15;157(2):105–15. PubMed PMID: 9151652. [DOI] [PubMed] [Google Scholar]
- 180.Baba M, Osumi M, Scott SV, et al. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol. 1997. Dec 29;139(7):1687–95. PubMed PMID: 9412464; PubMed Central PMCID: PMC2132654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Dini L, Pagliara P, Carla EC.. Phagocytosis of apoptotic cells by liver: a morphological study [Review]. Microsc Res Tech. 2002. Jun 15;57(6):530–40. doi: 10.1002/jemt.10107. PubMed PMID: 12112436; eng. [DOI] [PubMed] [Google Scholar]
- 182.Kroemer G, El-Deiry WS, Golstein P, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death [Guideline]. Cell Death Differ. 2005. Nov;12:1463–7. doi: 10.1038/sj.cdd.4401724. PubMed PMID: 16247491; eng. [DOI] [PubMed] [Google Scholar]
- 183.Nagy P, Varga A, Kovács AL, et al. How and why to study autophagy in Drosophila: It’s more than just a garbage chute. Methods. 2015;75:151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rez G, Palfia Z, Fellinger E.. Occurrence and inhibition by cycloheximide of apoptosis in vinblastine-treated murine pancreas. A role for autophagy? Acta Biol Hung. 1991; 42 (1–3): 133–40. PubMed PMID: 1844306; eng. [PubMed] [Google Scholar]
- 185.Giammarioli AM, Gambardella L, Barbati C, et al. Differential effects of the glycolysis inhibitor 2-deoxy-D-glucose on the activity of pro-apoptotic agents in metastatic melanoma cells, and induction of a cytoprotective autophagic response. int J cancer J Inter du cancer. 2012. Sep 12;131:E337–47. doi: 10.1002/ijc.26420. PubMed PMID: 21913183; Eng. [DOI] [PubMed] [Google Scholar]
- 186.Cheng XT, Xie YX, Zhou B, et al. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J Cell Biol. 2018. Sep 3;217(9):3127–3139. doi: 10.1083/jcb.201711083. PubMed PMID: 29695488; PubMed Central PMCID: PMCPMC6123004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cheng XT, Xie YX, Zhou B, et al. Revisiting LAMP1 as a marker for degradative autophagy-lysosomal organelles in the nervous system. Autophagy. 2018;14(8):1472–1474. doi: 10.1080/15548627.2018.1482147. PubMed PMID: 29940787; PubMed Central PMCID: PMCPMC6103665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Xie Y, Zhou B, Lin MY, et al. Progressive endolysosomal deficits impair autophagic clearance beginning at early asymptomatic stages in fALS mice. Autophagy. 2015;11(10):1934–6. doi: 10.1080/15548627.2015.1084460. PubMed PMID: 26290961; PubMed Central PMCID: PMCPMC4824580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xie Y, Zhou B, Lin MY, et al. Endolysosomal Deficits Augment Mitochondria Pathology in Spinal Motor Neurons of Asymptomatic fALS Mice. Neuron. 2015. Jul 15;87(2):355–70. doi: 10.1016/j.neuron.2015.06.026. PubMed PMID: 26182418; PubMed Central PMCID: PMCPMC4511489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Karanasios E. Correlative Live-Cell Imaging and Super-Resolution Microscopy of Autophagy. Methods Mol Biol. 2019;1880:231–242. doi: 10.1007/978-1-4939-8873-0_15. PubMed PMID: 30610701. [DOI] [PubMed] [Google Scholar]
- 191.Kenny SJ, Chen X, Ge L, et al. Super-resolution microscopy unveils FIP200-scaffolded, cup-shaped organization of mammalian autophagic initiation machinery. bioRxiv. 2019. 10.1101/712828. [DOI] [Google Scholar]
- 192.Sou YS, Tanida I, Komatsu M, et al. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J Biol Chem. 2006. Feb 10;281(6):3017–24. doi: 10.1074/jbc.M505888200. PubMed PMID: 16303767; eng. [DOI] [PubMed] [Google Scholar]
- 193.Le Grand JN, Chakrama FZ, Seguin-Py S, et al. GABARAPL1 (GEC1): Original or copycat? Autophagy. 2011. Oct 1;7(10):1098–107. PubMed PMID: 21597319; Eng. [DOI] [PubMed] [Google Scholar]
- 194.Hemelaar J, Lelyveld VS, Kessler BM, et al. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem. 2003. Dec 19;278(51):51841–50. doi: 10.1074/jbc.M308762200. PubMed PMID: 14530254. [DOI] [PubMed] [Google Scholar]
- 195.Tanida I, Sou YS, Ezaki J, et al. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J Biol Chem. 2004. Aug 27;279(35):36268–76. doi: 10.1074/jbc.M401461200. PubMed PMID: 15187094. [DOI] [PubMed] [Google Scholar]
- 196.Bhujabal Z, Birgisdottir AB, Sjottem E, et al. FKBP8 recruits LC3A to mediate Parkin-independent mitophagy. EMBO Rep. 2017. Jun;18(6):947–961. doi: 10.15252/embr.201643147. PubMed PMID: 28381481; PubMed Central PMCID: PMCPMC5452039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Koukourakis MI, Kalamida D, Giatromanolaki A, et al. Autophagosome proteins LC3A, LC3B and LC3C have distinct subcellular distribution kinetics and expression in cancer cell lines. PLoS One. 2015;10(9):e0137675. doi: 10.1371/journal.pone.0137675. PubMed PMID: 26378792; PubMed Central PMCID: PMCPMC4574774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kabeya Y, Mizushima N, Yamamoto A, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004. Jun 1;117(Pt 13):2805–12. doi: 10.1242/jcs.01131. PubMed PMID: 15169837; eng. [DOI] [PubMed] [Google Scholar]
- 199.Weidberg H, Shvets E, Shpilka T, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010. Jun 2;29(11):1792–802. doi: 10.1038/emboj.2010.74. PubMed PMID: 20418806; PubMed Central PMCID: PMC2885923. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sugawara K, Suzuki NN, Fujioka Y, et al. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells. 2004. Jul;9(7):611–8. doi: 10.1111/j.1356-9597.2004.00750.x. PubMed PMID: 15265004. [DOI] [PubMed] [Google Scholar]
- 201.Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013. Oct;15(10):1197–205. doi: 10.1038/ncb2837. PubMed PMID: 24036476; PubMed Central PMCID: PMC3806088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Lystad AH, Ichimura Y, Takagi K, et al. Structural determinants in GABARAP required for the selective binding and recruitment of ALFY to LC3B-positive structures. EMBO Rep. 2014. Mar 25;15:557–565. doi: 10.1002/embr.201338003. PubMed PMID: 24668264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.von Muhlinen N, Akutsu M, Ravenhill BJ, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol Cell. 2012. Nov 9;48(3):329–42. doi: 10.1016/j.molcel.2012.08.024. PubMed PMID: 23022382; PubMed Central PMCID: PMC3510444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Huang W-P, Scott SV, Kim J, et al. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem. 2000. Feb 25;275(8):5845–51. PubMed PMID: 10681575; eng. [DOI] [PubMed] [Google Scholar]
- 205.Nash Y, Schmukler E, Trudler D, et al. DJ-1 deficiency impairs autophagy and reduces alpha-synuclein phagocytosis by microglia. J Neurochem. 2017. Dec;143(5):584–594. doi: 10.1111/jnc.14222. PubMed PMID: 28921554. [DOI] [PubMed] [Google Scholar]
- 206.Cai Q, Lu L, Tian J-H, et al. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010. Oct 6;68(1):73–86. doi: 10.1016/j.neuron.2010.09.022. PubMed PMID: 20920792; PubMed Central PMCID: PMC2953270. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Castino R, Fiorentino I, Cagnin M, et al. Chelation of lysosomal iron protects dopaminergic SH-SY5Y neuroblastoma cells from hydrogen peroxide toxicity by precluding autophagy and Akt dephosphorylation. Toxicol Sci. 2011. Jul 8:523–41. doi: 10.1093/toxsci/kfr179. PubMed PMID: 21742779; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Michiorri S, Gelmetti V, Giarda E, et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010. Jun;17(6):962–74. doi: 10.1038/cdd.2009.200. PubMed PMID: 20057503; eng. [DOI] [PubMed] [Google Scholar]
- 209.Abdul Rahim SA, Dirkse A, Oudin A, et al. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br J Cancer. 2017. Sep 5;117(6):813–825. doi: 10.1038/bjc.2017.263. PubMed PMID: 28797031; PubMed Central PMCID: PMCPMC5590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Altmann C, Hardt S, Fischer C, et al. Progranulin overexpression in sensory neurons attenuates neuropathic pain in mice: Role of autophagy. Neurobiol Dis. 2016. Dec;96:294–311. doi: 10.1016/j.nbd.2016.09.010. PubMed PMID: 27629805. [DOI] [PubMed] [Google Scholar]
- 211.Rehorova M, Vargova I, Forostyak S, et al. A Combination of Intrathecal and Intramuscular Application of Human Mesenchymal Stem Cells Partly Reduces the Activation of Necroptosis in the Spinal Cord of SOD1(G93A) Rats. Stem Cells Transl Med. 2019. Jun;8(6):535–547. doi: 10.1002/sctm.18-0223. PubMed PMID: 30802001; PubMed Central PMCID: PMCPMC6525562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Yang DS, Stavrides P, Mohan PS, et al. Reversal of autophagy dysfunction in the TgCRND8 mouse model of Alzheimer’s disease ameliorates amyloid pathologies and memory deficits. Brain. 2011. Jan;134(Pt 1):258–77. doi: 10.1093/brain/awq341. PubMed PMID: 21186265; PubMed Central PMCID: PMC3009842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Liu S, Li Y, Choi HMC, et al. Lysosomal damage after spinal cord injury causes accumulation of RIPK1 and RIPK3 proteins and potentiation of necroptosis. Cell Death Dis. 2018. May 1;9(5):476. doi: 10.1038/s41419-018-0469-1. PubMed PMID: 29686269; PubMed Central PMCID: PMCPMC5913300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sarkar C, Zhao Z, Aungst S, et al. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy. 2014. Nov 11;10:2208–22. doi: 10.4161/15548627.2014.981787. PubMed PMID: 25484084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Padman BS, Bach M, Lucarelli G, et al. The protonophore CCCP interferes with lysosomal degradation of autophagic cargo in yeast and mammalian cells. Autophagy. 2013. Nov 1;9(11):1862–75. doi: 10.4161/auto.26557. PubMed PMID: 24150213. [DOI] [PubMed] [Google Scholar]
- 216.Engedal N, Torgersen ML, Guldvik IJ, et al. Modulation of intracellular calcium homeostasis blocks autophagosome formation. Autophagy. 2013. Oct;9(10):1475–90. doi: 10.4161/auto.25900. PubMed PMID: 23970164. [DOI] [PubMed] [Google Scholar]
- 217.Jahreiss L, Menzies FM, Rubinsztein DC.. The itinerary of auto-phagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic. 2008. Apr;9(4):574–87. doi: 10.1111/j.1600-0854.2008.00701.x. PubMed PMID: 18182013; PubMed Central PMCID: PMC2329914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Klionsky DJ, Elazar Z, Seglen PO, et al. Does bafilomycin A1 block the fusion of auto-phagosomes with lysosomes? [Editorial]. Autophagy. 2008. Oct;4(7):849–950. PubMed PMID: 18758232; eng. [DOI] [PubMed] [Google Scholar]
- 219.Yamamoto A, Tagawa Y, Yoshimori T, et al. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between auto-phagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998. Feb;23(1):33–42. PubMed PMID: 9639028; eng. [DOI] [PubMed] [Google Scholar]
- 220.Ahlberg J, Berkenstam A, Henell F, et al. Degradation of short and long lived proteins in isolated rat liver lysosomes. Effects of pH, temperature, and proteolytic inhibitors. J Biol Chem. 1985. May 10;260(9):5847–54. PubMed PMID: 3988775; eng. [PubMed] [Google Scholar]
- 221.Yoon YH, Cho KS, Hwang JJ, et al. Induction of lysosomal dilatation, arrested autophagy, and cell death by chloroquine in cultured ARPE-19 cells. Invest Ophthalmol Vis Sci. 2010. Nov;51(11):6030–7. doi: 10.1167/iovs.10-5278. PubMed PMID: 20574031. [DOI] [PubMed] [Google Scholar]
- 222.Juhasz G. Interpretation of bafilomycin, pH neutralizing or protease inhibitor treatments in autophagic flux experiments: novel considerations. Autophagy. 2012. Dec;8(12):1875–6. PubMed PMID: 22874642; PubMed Central PMCID: PMC3541311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Li M, Khambu B, Zhang H, et al. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem. 2013. Dec 13;288(50):35769–80. doi: 10.1074/jbc.M113.511212. PubMed PMID: 24174532; PubMed Central PMCID: PMC3861628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Thomas G, Hall MN.. TOR signalling and control of cell growth. Curr Opin Cell Biol. 1997. Dec;9(6):782–7. PubMed PMID: 9425342. [DOI] [PubMed] [Google Scholar]
- 225.Seglen PO, Grinde B, Solheim AE.. Inhibition of the lysosomal pathway of protein degradation in isolated rat hepatocytes by ammonia, methylamine, chloroquine and leupeptin. Eur J Biochem/FEBS. 1979. Apr 2;95(2):215–25. PubMed PMID: 456353. [DOI] [PubMed] [Google Scholar]
- 226.Yoshimori T, Yamamoto A, Moriyama Y, et al. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991. Sep 15;266(26):17707–12. PubMed PMID: 1832676. [PubMed] [Google Scholar]
- 227.Artero-Castro A, Perez-Alea M, Feliciano A, et al. Disruption of the ribosomal P complex leads to stress-induced autophagy. Autophagy. 2015;11(9):1499–519. doi: 10.1080/15548627.2015.1063764. PubMed PMID: 26176264; PubMed Central PMCID: PMCPMC4590587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Bahr BA, Wisniewski ML, Butler D.. Positive lysosomal modulation as a unique strategy to treat age-related protein accumulation diseases. Rejuvenation Res. 2012. Apr;15(2):189–97. doi: 10.1089/rej.2011.1282. PubMed PMID: 22533430; PubMed Central PMCID: PMCPMC3332372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.McLeland CB, Rodriguez J, Stern ST.. Autophagy monitoring assay: qualitative analysis of MAP LC3-I to II conversion by immunoblot. Methods Mol Biol. 2011;697:199–206. doi: 10.1007/978-1-60327-198-1_21. PubMed PMID: 21116969; eng. [DOI] [PubMed] [Google Scholar]
- 230.Chakrama FZ, Seguin-Py S, Le Grand JN, et al. GABARAPL1 (GEC1) associates with autophagic vesicles. Autophagy. 2010. May 22;6(4):495–505. PubMed PMID: 20404487; Eng. [DOI] [PubMed] [Google Scholar]
- 231.Maynard S, Ghosh R, Wu Y, et al. GABARAP is a determinant of apoptosis in growth-arrested chicken embryo fibroblasts. J Cell Physiol. 2015. Jul;230(7):1475–88. doi: 10.1002/jcp.24889. PubMed PMID: 25514832. [DOI] [PubMed] [Google Scholar]
- 232.Kim J, Huang W-P, Klionsky DJ.. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001. Jan 8;152(1):51–64. PubMed PMID: 11149920; PubMed Central PMCID: PMC2193654. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lopez-Otin C, Marino G.. Tagged ATG8-Coding Constructs for the In Vitro and In Vivo Assessment of ATG4 Activity. Methods Enzymol. 2017;587:189–205. doi: 10.1016/bs.mie.2016.11.001. PubMed PMID: 28253955. [DOI] [PubMed] [Google Scholar]
- 234.Shu CW, Drag M, Bekes M, et al. Synthetic substrates for measuring activity of autophagy proteases: autophagins (Atg4). Autophagy. 2010. Oct;6(7):936–47. doi: 10.4161/auto.6.7.13075. PubMed PMID: 20818167; PubMed Central PMCID: PMC3039740. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Li M, Chen X, Ye Q-Z, et al. A High-throughput FRET-based Assay for Determination of Atg4 Activity. Autophagy. 2012;8:401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ketteler R, Seed B.. Quantitation of autophagy by luciferase release assay [Evaluation Studies Research Support, Non-U.S. Gov’t]. Autophagy. 2008. Aug;4(6):801–6. PubMed PMID: 18641457; PubMed Central PMCID: PMC2910585. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Li M, Hou Y, Wang J, et al. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem. 2011. Mar 4;286(9):7327–38. doi: 10.1074/jbc.M110.199059. PubMed PMID: 21177865; PubMed Central PMCID: PMC3044989. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Klionsky DJ. For the last time, it is GFP-Atg8, not Atg8-GFP (and the same goes for LC3). Autophagy. 2011;7:1093–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mizushima N, Yamamoto A, Matsui M, et al. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004. Mar;15(3):1101–11. PubMed PMID: 14699058; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Tanida I, Minematsu-Ikeguchi N, Ueno T, et al. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 2005. Jul;1(2):84–91. PubMed PMID: 16874052; eng. [DOI] [PubMed] [Google Scholar]
- 241.Matsui Y, Takagi H, Qu X, et al. Distinct roles of autophagy in the heart during ischemia and reperfusion: roles of AMP-activated protein kinase and Beclin 1 in mediating autophagy. Circ Res. 2007. Mar 30;100(6):914–22. PubMed PMID: 17332429; eng. [DOI] [PubMed] [Google Scholar]
- 242.Suzuki K, Kirisako T, Kamada Y, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001. Nov 1;20(21):5971–81. doi: 10.1093/emboj/20.21.5971. PubMed PMID: 11689437; PubMed Central PMCID: PMC125692. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hanson HH, Kang S, Fernandez-Monreal M, et al. LC3-dependent intracellular membrane tubules induced by gamma-protocadherins A3 and B2: a role for intraluminal interactions. J Biol Chem. 2010. Jul 2;285(27):20982–92. doi: 10.1074/jbc.M109.092031. PubMed PMID: 20439459; PubMed Central PMCID: PMC2898317. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Florey O, Kim SE, Sandoval CP, et al. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol. 2011;13(11):1335–43. doi: 10.1038/ncb2363. PubMed PMID: 22002674; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Martinez J, Almendinger J, Oberst A, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc Natl Acad Sci U S A. 2011. Oct 18;108(42):17396–401. doi: 10.1073/pnas.1113421108. PubMed PMID: 21969579; PubMed Central PMCID: PMCPMC3198353. eng. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 246.Choi J, Park S, Biering SB, et al. The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity. 2014. Jun 19;40(6):924–35. doi: 10.1016/j.immuni.2014.05.006. PubMed PMID: 24931121; PubMed Central PMCID: PMC4107903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007. Dec 20;450(7173):1253–7. doi: 10.1038/nature06421. PubMed PMID: 18097414; eng. [DOI] [PubMed] [Google Scholar]
- 248.Sanjuan MA, Milasta S, Green DR.. Toll-like receptor signaling in the lysosomal pathways [Review]. Immunol Rev. 2009. Jan;227(1):203–20. doi: 10.1111/j.1600-065X.2008.00732.x. PubMed PMID: 19120486; eng. [DOI] [PubMed] [Google Scholar]
- 249.Ushio H, Ueno T, Kojima Y, et al. Crucial role for autophagy in degranulation of mast cells. J Allergy Clin Immunol. 2011. May;127(5):1267–76 e6. doi: 10.1016/j.jaci.2010.12.1078. PubMed PMID: 21333342. [DOI] [PubMed] [Google Scholar]
- 250.Ishibashi K, Uemura T, Waguri S, et al. Atg16L1, an essential factor for canonical autophagy, participates in hormone secretion from PC12 cells independently of autophagic activity. Mol Biol Cell. 2012. Aug;23(16):3193–202. doi: 10.1091/mbc.E12-01-0010. PubMed PMID: 22740627; PubMed Central PMCID: PMC3418313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.DeSelm CJ, Miller BC, Zou W, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev Cell. 2011. Nov 15;21(5):966–74. doi: 10.1016/j.devcel.2011.08.016. PubMed PMID: 22055344; PubMed Central PMCID: PMC3244473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Patel KK, Miyoshi H, Beatty WL, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013. Dec 11;32(24):3130–44. doi: 10.1038/emboj.2013.233. PubMed PMID: 24185898; PubMed Central PMCID: PMC3981139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Dupont N, Lacas-Gervais S, Bertout J, et al. Shigella phagocytic vacuolar membrane remnants participate in the cellular response to pathogen invasion and are regulated by autophagy. Cell Host Microbe. 2009. Aug 20;6(2):137–49. doi: 10.1016/j.chom.2009.07.005. PubMed PMID: 19683680; eng. [DOI] [PubMed] [Google Scholar]
- 254.Cottam EM, Maier HJ, Manifava M, et al. Coronavirus nsp6 proteins generate auto-phagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011. Nov;7(11):1335–47. doi: 10.4161/auto.7.11.16642. PubMed PMID: 21799305; PubMed Central PMCID: PMC3242798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Reggiori F, Monastyrska I, Verheije MH, et al. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010. Jun 25;7(6):500–8. doi: 10.1016/j.chom.2010.05.013. PubMed PMID: 20542253; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Sharma M, Bhattacharyya S, Nain M, et al. Japanese encephalitis virus replication is negatively regulated by autophagy and occurs on LC3-I- and EDEM1-containing membranes. Autophagy. 2014. Sep;10(9):1637–51. doi: 10.4161/auto.29455. PubMed PMID: 25046112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.English L, Chemali M, Duron J, et al. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009. May;10(5):480–7. doi: 10.1038/ni.1720. PubMed PMID: 19305394; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Beale R, Wise H, Stuart A, et al. A LC3-interacting motif in the influenza A virus M2 protein is required to subvert autophagy and maintain virion stability. Cell Host Microbe. 2014. Feb 12;15(2):239–47. doi: 10.1016/j.chom.2014.01.006. PubMed PMID: 24528869; PubMed Central PMCID: PMC3991421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Bello-Morales R, Lopez-Guerrero JA.. Extracellular Vesicles in Herpes Viral Spread and Immune Evasion. Front Microbiol. 2018;9:2572. doi: 10.3389/fmicb.2018.02572. PubMed PMID: 30410480; PubMed Central PMCID: PMCPMC6209645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Kemball CC, Alirezaei M, Flynn CT, et al. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol. 2010. Dec;84(23):12110–24. doi: 10.1128/JVI.01417-10. PubMed PMID: 20861268; PubMed Central PMCID: PMC2976412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Alirezaei M, Flynn CT, Wood MR, et al. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe. 2012. Mar 15;11(3):298–305. doi: 10.1016/j.chom.2012.01.014. PubMed PMID: 22423969; PubMed Central PMCID: PMC3308121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Agrotis A, von Chamier L, Oliver H, et al. Human ATG4 autophagy proteases counteract attachment of ubiquitin-like LC3/GABARAP proteins to other cellular proteins. J Biol Chem. 2019. Aug 23;294(34):12610–12621. doi: 10.1074/jbc.AC119.009977. PubMed PMID: 31315929; PubMed Central PMCID: PMCPMC6709618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ichimura Y, Imamura Y, Emoto K, et al. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J Biol Chem. 2004. Sep 24;279(39):40584–92. doi: 10.1074/jbc.M405860200. PubMed PMID: 15277523. [DOI] [PubMed] [Google Scholar]
- 264.Plowey ED, Cherra SJ, 3rd, Liu YJ, et al. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008. May;105(3):1048–56. doi: 10.1111/j.1471-4159.2008.05217.x. PubMed PMID: 18182054; PubMed Central PMCID: PMC2361385. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Nicotra G, Mercalli F, Peracchio C, et al. Autophagy-active beclin-1 correlates with favourable clinical outcome in non-Hodgkin lymphomas. Mod Pathol. 2010. Jul;23(7):937–50. doi: 10.1038/modpathol.2010.80. PubMed PMID: 20473282; eng. [DOI] [PubMed] [Google Scholar]
- 266.Tanida I, Ueno T, Kominami E.. LC3 and autophagy. Methods Mol Biol. 2008;445:77–88. [DOI] [PubMed] [Google Scholar]
- 267.Gros F, Arnold J, Page N, et al. Macroautophagy is deregulated in murine and human lupus T lymphocytes. Autophagy. 2012. Jul 1;8(7):1113–23. doi: 10.4161/auto.20275. PubMed PMID: 22522825; PubMed Central PMCID: PMC3429547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Welinder C, Ekblad L.. Coomassie staining as loading control in Western blot analysis. J Proteome Res. 2011. Mar 4;10(3):1416–9. doi: 10.1021/pr1011476. PubMed PMID: 21186791. [DOI] [PubMed] [Google Scholar]
- 269.Rocha-Martins M, Njaine B, Silveira MS.. Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and Western blot throughout rat retinal development. PLoS One. 2012;7(8):e43028. doi: 10.1371/journal.pone.0043028. PubMed PMID: 22916200; PubMed Central PMCID: PMCPMC3423434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Colecchia D, Rossi M, Sasdelli F, et al. MAPK15 mediates BCR-ABL1-induced autophagy and regulates oncogene-dependent cell proliferation and tumor formation. Autophagy 2015;11(10):1790–802. doi: 10.1080/15548627.2015.1084454. PubMed PMID: 26291129; PubMed Central PMCID: PMCPMC4824572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Colecchia D, Stasi M, Leonardi M, et al. Alterations of autophagy in the peripheral neuropathy Charcot-Marie-Tooth type 2B. Autophagy. 2018;14(6):930–941. doi: 10.1080/15548627.2017.1388475. PubMed PMID: 29130394; PubMed Central PMCID: PMCPMC6103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Colecchia D, Strambi A, Sanzone S, et al. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy. 2012. Dec;8(12):1724–40. doi: 10.4161/auto.21857. PubMed PMID: 22948227; PubMed Central PMCID: PMC3541284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Colella AD, Chegenii N, Tea MN, et al. Comparison of Stain-Free gels with traditional immunoblot loading control methodology. Anal Biochem. 2012. Nov 15;430(2):108–10. doi: 10.1016/j.ab.2012.08.015. PubMed PMID: 22929699. [DOI] [PubMed] [Google Scholar]
- 274.Ghosh R, Gilda JE, Gomes AV.. The necessity of and strategies for improving confidence in the accuracy of western blots. Expert Rev Proteomics. 2014. Oct;11(5):549–60. doi: 10.1586/14789450.2014.939635. PubMed PMID: 25059473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Yan L, Vatner DE, Kim SJ, et al. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci U S A. 2005. Sep 27;102(39):13807–12. doi: 10.1073/pnas.0506843102. PubMed PMID: 16174725; PubMed Central PMCID: PMC1224362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Russ DW, Boyd IM, McCoy KM, et al. Muscle-specificity of age-related changes in markers of autophagy and sphingolipid metabolism. Biogerontology. 2015. Aug 22. doi: 10.1007/s10522-015-9598-4. PubMed PMID: 26296420. [DOI] [PubMed] [Google Scholar]
- 277.Russ DW, Krause J, Wills A, et al. “SR stress” in mixed hindlimb muscles of aging male rats. Biogerontology. 2012. Oct;13(5):547–55. doi: 10.1007/s10522-012-9399-y. PubMed PMID: 22955580. [DOI] [PubMed] [Google Scholar]
- 278.Simonovitch S, Schmukler E, Bespalko A, et al. Impaired Autophagy in APOE4 Astrocytes. J Alzheimers Dis. 2016;51(3):915–27. doi: 10.3233/JAD-151101. PubMed PMID: 26923027. [DOI] [PubMed] [Google Scholar]
- 279.He H, Dang Y, Dai F, et al. Post-translational modifications of three members of the human MAP1LC3 family and detection of a novel type of modification for MAP1LC3B. J Biol Chem. 2003. Aug 1;278(31):29278–87. PubMed PMID: 12740394; eng. [DOI] [PubMed] [Google Scholar]
- 280.Shpilka T, Weidberg H, Pietrokovski S, et al. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011. Jul 27;12(7):226. doi: 10.1186/gb-2011-12-7-226. PubMed PMID: 21867568; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Zois CE, Koukourakis MI.. Radiation-induced autophagy in normal and cancer cells: towards novel cytoprotection and radio-sensitization policies?. Autophagy. 2009. May;5(4):442–50. PubMed PMID: 19164950; eng. [DOI] [PubMed] [Google Scholar]
- 282.Klionsky DJ. Location, location, location? No. Catalog number. Autophagy. 2009;5:441. [Google Scholar]
- 283.Xin Y, Yu L, Chen Z, et al. Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics. 2001. Jun 15;74(3):408–13. doi: 10.1006/geno.2001.6555. PubMed PMID: 11414770; eng. [DOI] [PubMed] [Google Scholar]
- 284.Novak I, Kirkin V, McEwan DG, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010. Jan;11(1):45–51. doi: 10.1038/embor.2009.256. PubMed PMID: 20010802; PubMed Central PMCID: PMC2816619. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Schwarten M, Mohrluder J, Ma P, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009. Jul;5(5):690–8. PubMed PMID: 19363302. [DOI] [PubMed] [Google Scholar]
- 286.Gassmann M, Grenacher B, Rohde B, et al. Quantifying Western blots: pitfalls of densitometry. Electrophoresis. 2009. Jun;30(11):1845–55. doi: 10.1002/elps.200800720. PubMed PMID: 19517440; eng. [DOI] [PubMed] [Google Scholar]
- 287.Agrotis A, Pengo N, Burden JJ, et al. Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy. 2019. Jun;15(6):976–997. doi: 10.1080/15548627.2019.1569925. PubMed PMID: 30661429; PubMed Central PMCID: PMCPMC6526816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Kirisako T, Ichimura Y, Okada H, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol. 2000. Oct 16;151(2):263–76. PubMed PMID: 11038174; PubMed Central PMCID: PMC2192639. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Chung T, Phillips AR, Vierstra RD.. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A AND ATG12B loci. Plant J. 2010. May;62(3):483–93. doi: 10.1111/j.1365-313X.2010.04166.x. PubMed PMID: 20136727; eng. [DOI] [PubMed] [Google Scholar]
- 290.Chung T, Suttangkakul A, Vierstra RD.. The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 2009. Jan;149(1):220–34. doi: 10.1104/pp.108.126714. PubMed PMID: 18790996; PubMed Central PMCID: PMC2613746. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wang W, Chen Z, Billiar TR, et al. The carboxyl-terminal amino acids render pro-human LC3B migration similar to lipidated LC3B in SDS-PAGE. PLoS One. 2013;8(9):e74222. doi: 10.1371/journal.pone.0074222. PubMed PMID: 24040206; PubMed Central PMCID: PMCPMC3769297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Kovsan J, Bluher M, Tarnovscki T, et al. Altered autophagy in human adipose tissues in obesity. J Clin Endocrinol Metab. 2011. Feb;96(2):E268–77. doi: 10.1210/jc.2010-1681. PubMed PMID: 21047928; eng. [DOI] [PubMed] [Google Scholar]
- 293.Cherra SJ, III, Kulich SM, Uechi G, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010. Aug 23;190(4):533–9. doi: 10.1083/jcb.201002108. PubMed PMID: 20713600; PubMed Central PMCID: PMC2928022. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Gao Z, Gammoh N, Wong PM, et al. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy. 2010. Jan;6(1):126–37. PubMed PMID: 20061800. [DOI] [PubMed] [Google Scholar]
- 295.Harris VM. Protein detection by Simple Western analysis. Methods Mol Biol. 2015;1312:465–8. doi: 10.1007/978-1-4939-2694-7_47. PubMed PMID: 26044028. [DOI] [PubMed] [Google Scholar]
- 296.Voeten RLC, Ventouri IK, Haselberg R, et al. Capillary Electrophoresis: Trends and Recent Advances. Anal Chem. 2018. Feb 6;90(3):1464–1481. doi: 10.1021/acs.analchem.8b00015. PubMed PMID: 29298038; PubMed Central PMCID: PMCPMC5994730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Beekman C, Janson AA, Baghat A, et al. Use of capillary Western immunoassay (Wes) for quantification of dystrophin levels in skeletal muscle of healthy controls and individuals with Becker and Duchenne muscular dystrophy. PLoS One. 2018;13(4):e0195850. doi: 10.1371/journal.pone.0195850. PubMed PMID: 29641567; PubMed Central PMCID: PMCPMC5895072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Pierzynowska K, Gaffke L, Cyske Z, et al. Genistein induces degradation of mutant huntingtin in fibroblasts from Huntington’s disease patients. Metab Brain Dis. 2019. Jun;34(3):715–720. doi: 10.1007/s11011-019-00405-4. PubMed PMID: 30850940; PubMed Central PMCID: PMCPMC6520327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Saetre F, Hagen LK, Engedal N, et al. Novel steps in the autophagic-lysosomal pathway. FEBS J. 2015. Jun;282(11):2202–14. doi: 10.1111/febs.13268. PubMed PMID: 25779646. [DOI] [PubMed] [Google Scholar]
- 300.Degtyarev M, De Maziere A, Orr C, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol. 2008. Oct 6;183(1):101–16. doi: 10.1083/jcb.200801099. PubMed PMID: 18838554; PubMed Central PMCID: PMC2557046. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Ju JS, Varadhachary AS, Miller SE, et al. Quantitation of “autophagic flux” in mature skeletal muscle. Autophagy. 2010. Oct;6(7):929–35. doi: 10.4161/auto.6.7.12785. PubMed PMID: 20657169; PubMed Central PMCID: PMC3039739. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Mauthe M, Orhon I, Rocchi C, et al. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14(8):1435–1455. doi: 10.1080/15548627.2018.1474314. PubMed PMID: 29940786; PubMed Central PMCID: PMCPMC6103682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Mauvezin C, Nagy P, Juhasz G, et al. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. PubMed PMID: 25959678; PubMed Central PMCID: PMC4428688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Xie R, Nguyen S, McKeehan WL, et al. Acetylated microtubules are required for fusion of auto-phagosomes with lysosomes. BMC Cell Biol. 2010;11:89. doi: 10.1186/1471-2121-11-89. PubMed PMID: 21092184; PubMed PMID: 21092184; PubMed Central PMCID: PMC2995476. eng [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Morell C, Bort A, Vara-Ciruelos D, et al. Up-Regulated Expression of LAMP2 and Autophagy Activity during Neuroendocrine Differentiation of Prostate Cancer LNCaP Cells. PLoS One. 2016;11(9):e0162977. doi: 10.1371/journal.pone.0162977. PubMed PMID: 27627761; PubMed Central PMCID: PMCPMC5023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Gonzalez-Polo RA, Boya P, Pauleau AL, et al. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci. 2005. Jul 15;118:3091–102. PubMed PMID: 15985464; eng. [DOI] [PubMed] [Google Scholar]
- 307.Cuervo AM, Stefanis L, Fredenburg R, et al. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science. 2004. Aug 27;305(5688):1292–5. doi: 10.1126/science.1101738. PubMed PMID: 15333840; eng. [DOI] [PubMed] [Google Scholar]
- 308.Trincheri NF, Follo C, Nicotra G, et al. Resveratrol-induced apoptosis depends on the lipid kinase activity of Vps34 and on the formation of autophagolysosomes. Carcinogenesis. 2008. Feb;29(2):381–9. doi: 10.1093/carcin/bgm271. PubMed PMID: 18048384; eng. [DOI] [PubMed] [Google Scholar]
- 309.Rubinsztein DC, Cuervo AM, Ravikumar B, et al. In search of an “autophagomometer” [Editorial]. Autophagy. 2009. Jul;5(5):585–9. PubMed PMID: 19411822; eng. [DOI] [PubMed] [Google Scholar]
- 310.Sarkar S, Ravikumar B, Rubinsztein DC.. Autophagic clearance of aggregate-prone proteins associated with neurodegeneration. Methods Enzymol. 2009;453:83–110. doi: 10.1016/S0076-6879(08)04005-6. PubMed PMID: 19216903; eng. [DOI] [PubMed] [Google Scholar]
- 311.Sarkar S, Korolchuk V, Renna M, et al. Methodological considerations for assessing autophagy modulators: a study with calcium phosphate precipitates. Autophagy. 2009. Apr;5(3):307–13. PubMed PMID: 19182529; eng. [DOI] [PubMed] [Google Scholar]
- 312.du Toit A, Hofmeyr JS, Gniadek TJ, et al. Measuring autophagosome flux. Autophagy. 2018;14(6):1060–1071. doi: 10.1080/15548627.2018.1469590. PubMed PMID: 29909716; PubMed Central PMCID: PMCPMC6103398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Martins WK, Severino D, Souza C, et al. Rapid screening of potential autophagic inductor agents using mammalian cell lines. Biotechnol J. 2013. Jun;8(6):730–7. doi: 10.1002/biot.201200306. PubMed PMID: 23420785. [DOI] [PubMed] [Google Scholar]
- 314.Martins WK, Costa ET, Cruz MC, et al. Parallel damage in mitochondrial and lysosomal compartments promotes efficient cell death with autophagy: The case of the pentacyclic triterpenoids. Sci Rep. 2015;5:12425. doi: 10.1038/srep12425. PubMed PMID: 26213355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Martins WK, Gomide AB, Costa ET, et al. Membrane damage by betulinic acid provides insights into cellular aging. Biochim Biophys Acta Gen Subj. 2017. Jan;1861(1 Pt A):3129–3143. doi: 10.1016/j.bbagen.2016.10.018. PubMed PMID: 27773704. [DOI] [PubMed] [Google Scholar]
- 316.Martins WK, Santos NF, Rocha CS, et al. Parallel damage in mitochondria and lysosomes is an efficient way to photoinduce cell death. Autophagy. 2019. Feb;15(2):259–279. doi: 10.1080/15548627.2018.1515609. PubMed PMID: 30176156; PubMed Central PMCID: PMCPMC6333451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Tsubone TM, Martins WK, Pavani C, et al. Enhanced efficiency of cell death by lysosome-specific photodamage. Sci Rep. 2017. Jul 27;7(1):6734. doi: 10.1038/s41598-017-06788-7. PubMed PMID: 28751688; PubMed Central PMCID: PMCPMC5532215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Hakonarson H, Grant SF, Bradfield JP, et al. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007. Aug 2;448(7153):591–4. doi: 10.1038/nature06010. PubMed PMID: 17632545. [DOI] [PubMed] [Google Scholar]
- 319.Pandey R, Bakay M, Hain HS, et al. CLEC16A regulates splenocyte and NK cell function in part through MEK signaling. PLoS One. 2018;13(9):e0203952. doi: 10.1371/journal.pone.0203952. PubMed PMID: 30226884; PubMed Central PMCID: PMCPMC6143231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Pandey R, Bakay M, Hain HS, et al. The autoimmune disorder susceptibility gene CLEC16A restrains NK Cell function in YTS NK cell line and Clec16a knockout mice. Front Immunol. 2019;10:68. doi: 10.3389/fimmu.2019.00068. PubMed PMID: 30774629; PubMed Central PMCID: PMCPMC6367972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Martin-Maestro P, Gargini R, AS A, et al. Mitophagy failure in fibroblasts and iPSC-derived neurons of Alzheimer’s disease-associated presenilin 1 mutation. Front Mol Neurosci. 2017;10:291. doi: 10.3389/fnmol.2017.00291. PubMed PMID: 28959184; PubMed Central PMCID: PMCPMC5603661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Martin-Maestro P, Gargini R, Perry G, et al. PARK2 enhancement is able to compensate mitophagy alterations found in sporadic Alzheimer’s disease. Hum Mol Genet. 2016. Feb 15;25(4):792–806. doi: 10.1093/hmg/ddv616. PubMed PMID: 26721933; PubMed Central PMCID: PMCPMC4743695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Schmukler E, Wolfson E, Elazar Z, et al. Continuous treatment with FTS confers resistance to apoptosis and affects autophagy. PLoS One. 2017;12(2):e0171351. doi: 10.1371/journal.pone.0171351. PubMed PMID: 28151959; PubMed Central PMCID: PMCPMC5289601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Shintani T, Klionsky DJ.. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004. Jul 16;279(29):29889–94. PubMed PMID: 15138258; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Barmada SJ, Serio A, Arjun A, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014. Aug;10(8):677–85. doi: 10.1038/nchembio.1563. PubMed PMID: 24974230; PubMed Central PMCID: PMCPMC4106236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Tsvetkov AS, Arrasate M, Barmada S, et al. Proteostasis of polyglutamine varies among neurons and predicts neurodegeneration. Nat Chem Biol. 2013. Sep;9(9):586–92. doi: 10.1038/nchembio.1308. PubMed PMID: 23873212; PubMed Central PMCID: PMC3900497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Loos B, du Toit A, Hofmeyr JH. Defining and measuring autophagosome flux-concept and reality. Autophagy. 2014. Oct 30;10:2087–96. doi: 10.4161/15548627.2014.973338. PubMed PMID: 25484088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Khuansuwan S, Barnhill LM, Cheng S, et al. A novel transgenic zebrafish line allows for in vivo quantification of autophagic activity in neurons. Autophagy. 2019. Aug;15(8):1322–1332. doi: 10.1080/15548627.2019.1580511. PubMed PMID: 30755067; PubMed Central PMCID: PMCPMC6613892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.du Toit A, De Wet S, Hofmeyr JS, et al. The precision control of autophagic flux and vesicle dynamics-a micropattern approach. Cells. 2018. Aug 3;7(8). doi: 10.3390/cells7080094. PubMed PMID: 30081508; PubMed Central PMCID: PMCPMC6116198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Farkas T, Hoyer-Hansen M, Jaattela M.. Identification of novel autophagy regulators by a luciferase-based assay for the kinetics of autophagic flux. Autophagy. 2009. Oct;5(7):1018–25. PubMed PMID: 19652534; eng. [DOI] [PubMed] [Google Scholar]
- 331.Farkas T, Daugaard M, Jaattela M.. Identification of small molecule inhibitors of phosphatidylinositol 3-kinase and autophagy. J Biol Chem. 2011. Sep 19;286:38904–12. doi: 10.1074/jbc.M111.269134. PubMed PMID: 21930714; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Frankel LB, Di Malta C, Wen J, et al. A non-conserved miRNA regulates lysosomal function and impacts on a human lysosomal storage disorder. Nat Commun. 2014;5:5840. doi: 10.1038/ncomms6840. PubMed PMID: 25524633. [DOI] [PubMed] [Google Scholar]
- 333.Frankel LB, Lund AH.. MicroRNA regulation of autophagy. Carcinogenesis. 2012. Nov;33(11):2018–25. doi: 10.1093/carcin/bgs266. PubMed PMID: 22902544. [DOI] [PubMed] [Google Scholar]
- 334.Frankel LB, Wen J, Lees M, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011. Sep 13:4628–41. doi: 10.1038/emboj.2011.331. PubMed PMID: 21915098; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Nguyen HT, Dalmasso G, Muller S, et al. Crohn’s disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014. Feb;146(2):508–19. doi: 10.1053/j.gastro.2013.10.021. PubMed PMID: 24148619. [DOI] [PubMed] [Google Scholar]
- 336.Szyniarowski P, Corcelle-Termeau E, Farkas T, et al. A comprehensive siRNA screen for kinases that suppress macroautophagy in optimal growth conditions. Autophagy. 2011. Aug 1;7(8):892–903. PubMed PMID: 21508686; eng. [DOI] [PubMed] [Google Scholar]
- 337.Yeganeh B, Ghavami S, Kroeker AL, et al. Suppression of influenza A virus replication in human lung epithelial cells by noncytotoxic concentrations bafilomycin A1. Am J Physiol Lung Cell Mol Physiol. 2015. Feb 1;308(3):L270–86. doi: 10.1152/ajplung.00011.2014. PubMed PMID: 25361566; PubMed Central PMCID: PMCPMC4338931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Iwata J, Ezaki J, Komatsu M, et al. Excess peroxisomes are degraded by autophagic machinery in mammals. J Biol Chem. 2006. Feb17;281(7):4035–41. PubMed PMID: 16332691. [DOI] [PubMed] [Google Scholar]
- 339.Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008. Dec 1;183(5):795–803. doi: 10.1083/jcb.200809125. PubMed PMID: 19029340; PubMed Central PMCID: PMC2592826. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Nogalska A, Terracciano C, D’Agostino C, et al. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009. Sep;118(3):407–13. doi: 10.1007/s00401-009-0564-6. PubMed PMID: 19557423; eng. [DOI] [PubMed] [Google Scholar]
- 341.Ivanova S, Repnik U, Bojic L, et al. Lysosomes in apoptosis. Methods Enzymol. 2008;442:183–99. doi: 10.1016/S0076-6879(08)01409-2. PubMed PMID: 18662570. [DOI] [PubMed] [Google Scholar]
- 342.Chahory S, Keller N, Martin E, et al. Light induced retinal degeneration activates a caspase-independent pathway involving cathepsin D. Neurochem Int. 2010. Oct;57(3):278–87. doi: 10.1016/j.neuint.2010.06.006. PubMed PMID: 20558223. [DOI] [PubMed] [Google Scholar]
- 343.Padron-Barthe L, Courta J, Lepretre C, et al. Leukocyte Elastase Inhibitor, the precursor of L-DNase II, inhibits apoptosis by interfering with caspase-8 activation. Biochim Biophys Acta. 2008. Oct;1783(10):1755–66. doi: 10.1016/j.bbamcr.2008.06.018. PubMed PMID: 18674571. [DOI] [PubMed] [Google Scholar]
- 344.Kimura S, Noda T, Yoshimori T.. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007. Sep 21;3(5):452–460. PubMed PMID: 17534139; Eng. [DOI] [PubMed] [Google Scholar]
- 345.Gutierrez MG, Saka HA, Chinen I, et al. Protective role of autophagy against Vibrio cholerae cytolysin, a pore-forming toxin from V. cholerae. Proc Natl Acad Sci USA 2007. Feb 6;104(6):1829–34. PubMed PMID: 17267617; eng [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Hosokawa N, Hara Y, Mizushima N.. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006. May 15;580(11):2623–9. PubMed PMID: 16647067; eng. [DOI] [PubMed] [Google Scholar]
- 347.Guedes A, Ludovico P, Sampaio-Marques B.. Caloric restriction alleviates alpha-synuclein toxicity in aged yeast cells by controlling the opposite roles of Tor1 and Sir2 on autophagy. Mech Ageing Dev. 2017. Jan;161(Pt B):270–276. doi: 10.1016/j.mad.2016.04.006. PubMed PMID: 27109470. [DOI] [PubMed] [Google Scholar]
- 348.Pinar M, Pantazopoulou A, Peñalva MA.. Live-cell imaging of Aspergillus nidulans autophagy: RAB1 dependence, Golgi independence and ER involvement. Autophagy. 2013. Jul;9(7):1024–43. doi: 10.4161/auto.24483. PubMed PMID: 23722157; PubMed Central PMCID: PMC3722313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Sampaio-Marques B, Guedes A, Vasilevskiy I, et al. alpha-Synuclein toxicity in yeast and human cells is caused by cell cycle re-entry and autophagy degradation of ribonucleotide reductase 1. Aging cell. 2019. Aug;18(4):e12922. doi: 10.1111/acel.12922. PubMed PMID: 30977294; PubMed Central PMCID: PMCPMC6612645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Suttangkakul A, Li F, Chung T, et al. The ATG1/13 protein kinase complex is both a regulator and a substrate of autophagic recycling in Arabidopsis. Plant Cell. 2011;23:3761–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Ni HM, Bockus A, Wozniak AL, et al. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011. Feb;7(2):188–204. PubMed PMID: 21107021; PubMed Central PMCID: PMC3039769. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Balgi AD, Fonseca BD, Donohue E, et al. Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One. 2009;4(9):e7124. doi: 10.1371/journal.pone.0007124. PubMed PMID: 19771169; PubMed Central PMCID: PMC2742736. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Patterson GH, Lippincott-Schwartz J.. Selective photolabeling of proteins using photoactivatable GFP. Methods. 2004. Apr;32(4):445–50. PubMed PMID: 15003607; eng. [DOI] [PubMed] [Google Scholar]
- 354.Hamacher-Brady A, Brady NR, Gottlieb RA.. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006. Oct 6;281(40):29776–87. PubMed PMID: 16882669; eng. [DOI] [PubMed] [Google Scholar]
- 355.Klionsky DJ. Monitoring autophagy in yeast: the Pho8∆60 assay. Methods Mol Biol. 2007;390:363–71. PubMed PMID: 17951700; eng. [DOI] [PubMed] [Google Scholar]
- 356.Noda T, Klionsky DJ.. The quantitative Pho8∆60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. PubMed PMID: 19185711; eng. [DOI] [PubMed] [Google Scholar]
- 357.Dauphinee AN, Cardoso C, Dalman K, et al. Chemical screening pipeline for identification of specific plant autophagy modulators. Plant Physiol. 2019. Nov;181(3):855–866. doi: 10.1104/pp.19.00647. PubMed PMID: 31488572; PubMed Central PMCID: PMCPMC6836817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Shvets E, Elazar Z.. Autophagy-independent incorporation of GFP-LC3 into protein aggregates is dependent on its interaction with p62/SQSTM1. Autophagy. 2008. Nov;4(8):1054–6. doi: 10.4161/auto.6823. PubMed PMID: 18776740. [DOI] [PubMed] [Google Scholar]
- 359.Viiri J, Amadio M, Marchesi N, et al. Autophagy activation clears ELAVL1/HuR-mediated accumulation of SQSTM1/p62 during proteasomal inhibition in human retinal pigment epithelial cells. PLoS One. 2013;8(7):e69563. doi: 10.1371/journal.pone.0069563. PubMed PMID: 23922739; PubMed Central PMCID: PMCPMC3726683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Takahashi Y, He H, Tang Z, et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat Commun. 2018. Jul 20;9(1):2855. doi: 10.1038/s41467-018-05254-w. PubMed PMID: 30030437; PubMed Central PMCID: PMCPMC6054611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Los GV, Encell LP, McDougall MG, et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem Biol. 2008. Jun 20;3(6):373–82. doi: 10.1021/cb800025k. PubMed PMID: 18533659. [DOI] [PubMed] [Google Scholar]
- 362.Takahashi Y, Liang X, Hattori T, et al. VPS37A directs ESCRT recruitment for phagophore closure. J Cell Biol. 2019. Oct 7;218(10):3336–3354. doi: 10.1083/jcb.201902170. PubMed PMID: 31519728; PubMed Central PMCID: PMCPMC6781443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Divakaruni AS, Wiley SE, Rogers GW, et al. Thiazolidinediones are acute, specific inhibitors of the mitochondrial pyruvate carrier. Proc Natl Acad Sci U S A. 2013. Apr 2;110(14):5422–7. doi: 10.1073/pnas.1303360110. PubMed PMID: 23513224; PubMed Central PMCID: PMCPMC3619368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Badr CE, Wurdinger T, Nilsson J, et al. Lanatoside C sensitizes glioblastoma cells to tumor necrosis factor-related apoptosis-inducing ligand and induces an alternative cell death pathway. Neuro Oncol. 2011. Nov;13(11):1213–24. doi: 10.1093/neuonc/nor067. PubMed PMID: 21757445; PubMed Central PMCID: PMC3199161. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000. Nov 1;19(21):5720–8. PubMed PMID: 11060023; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Sampaio-Marques B, Felgueiras C, Silva A, et al. SNCA (alpha-synuclein)-induced toxicity in yeast cells is dependent on sirtuin 2 (Sir2)-mediated mitophagy. Autophagy. 2012. Oct;8(10):1494–509. doi: 10.4161/auto.21275. PubMed PMID: 22914317. [DOI] [PubMed] [Google Scholar]
- 367.Meléndez A, Tallóczy Z, Seaman M, et al. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003. Sep 5;301(5638):1387–91. PubMed PMID: 12958363; eng. [DOI] [PubMed] [Google Scholar]
- 368.Otto GP, Wu MY, Kazgan N, et al. Macroautophagy is required for multicellular development of the social amoeba Dictyostelium discoideum. J Biol Chem 2003. May 16;278(20):17636–45. PubMed PMID: 12626495; eng. [DOI] [PubMed] [Google Scholar]
- 369.Kikuma T, Ohneda M, Arioka M, et al. Functional analysis of the ATG8 homologue Aoatg8 and role of autophagy in differentiation and germination in Aspergillus oryzae. Eukaryot Cell. 2006. Aug;5(8):1328–36. doi: 10.1128/EC.00024-06. PubMed PMID: 16896216; PubMed Central PMCID: PMC1539149. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Liu XH, Liu TB, Lin FC.. Monitoring autophagy in Magnaporthe oryzae. Methods Enzymol. 2008;451:271–94. doi: 10.1016/S0076-6879(08)03219-9. PubMed PMID: 19185727; eng. [DOI] [PubMed] [Google Scholar]
- 371.Nolting N, Bernhards Y, Poggeler S.. SmATG7 is required for viability in the homothallic ascomycete Sordaria macrospora. Fungal Genet biol FG & B. 2009. Aug;46(8):531–42. doi: 10.1016/j.fgb.2009.03.008. PubMed PMID: 19351563; eng. [DOI] [PubMed] [Google Scholar]
- 372.Pinan-Lucarre B, Paoletti M, Dementhon K, et al. Autophagy is induced during cell death by incompatibility and is essential for differentiation in the filamentous fungus Podospora anserina. Mol Microbiol. 2003. Jan;47(2):321–33. PubMed PMID: 12519185; eng. [DOI] [PubMed] [Google Scholar]
- 373.Veneault-Fourrey C, Barooah M, Egan M, et al. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006. Apr 28;312(5773):580–3. doi: 10.1126/science.1124550. PubMed PMID: 16645096; eng. [DOI] [PubMed] [Google Scholar]
- 374.Baghdiguian S, Martinand-Mari C, Mangeat P.. Using Ciona to study developmental programmed cell death. Semin Cancer Biol. 2007. Apr;17(2):147–53. doi: 10.1016/j.semcancer.2006.11.005. PubMed PMID: 17197195; eng. [DOI] [PubMed] [Google Scholar]
- 375.Denton D, Shravage B, Simin R, et al. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009. Nov 3;19(20):1741–6. doi: 10.1016/j.cub.2009.08.042. PubMed PMID: 19818615; PubMed Central PMCID: PMC2783269. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Rusten TE, Lindmo K, Juhasz G, et al. Programmed autophagy in the Drosophila fat body is induced by ecdysone through regulation of the PI3K pathway. Dev Cell. 2004. Aug;7(2):179–92. PubMed PMID: 15296715; eng. [DOI] [PubMed] [Google Scholar]
- 377.Scott RC, Schuldiner O, Neufeld TP.. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004. Aug;7(2):167–78. PubMed PMID: 15296714; eng. [DOI] [PubMed] [Google Scholar]
- 378.Yoshimoto K, Hanaoka H, Sato S, et al. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell. 2004. Nov;16(11):2967–83. PubMed PMID: 15494556; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Li F, Chung T, Pennington JG, et al. Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell. 2015. May;27(5):1389–408. doi: 10.1105/tpc.15.00158. PubMed PMID: 25944100; PubMed Central PMCID: PMC4456646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Brennand A, Rico E, Rigden DJ, et al. ATG24 represses autophagy and differentiation and is essential for homeostasy of the flagellar pocket in trypanosoma brucei. PLoS One. 2015;10(6):e0130365. doi: 10.1371/journal.pone.0130365. PubMed PMID: 26090847; PubMed Central PMCID: PMC4474607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Li FJ, Shen Q, Wang C, et al. A role of autophagy in Trypanosoma brucei cell death. Cell Microbiol. 2012. Aug;14(8):1242–56. doi: 10.1111/j.1462-5822.2012.01795.x. PubMed PMID: 22463696. [DOI] [PubMed] [Google Scholar]
- 382.Schmidt RS, Butikofer P.. Autophagy in Trypanosoma brucei: amino acid requirement and regulation during different growth phases. PLoS One. 2014;9(4):e93875. doi: 10.1371/journal.pone.0093875. PubMed PMID: 24699810; PubMed Central PMCID: PMC3974859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Besteiro S, Williams RA, Morrison LS, et al. Endosome sorting and autophagy are essential for differentiation and virulence of Leishmania major. J Biol Chem. 2006. Apr 21;281(16):11384–96. doi: 10.1074/jbc.M512307200. PubMed PMID: 16497676; eng. [DOI] [PubMed] [Google Scholar]
- 384.Williams RA, Tetley L, Mottram JC, et al. Cysteine peptidases CPA and CPB are vital for autophagy and differentiation in Leishmania mexicana. Mol Microbiol. 2006. Aug;61(3):655–74. doi: 10.1111/j.1365-2958.2006.05274.x. PubMed PMID: 16803590; eng. [DOI] [PubMed] [Google Scholar]
- 385.Williams RA, Woods KL, Juliano L, et al. Characterization of unusual families of ATG8-like proteins and ATG12 in the protozoan parasite Leishmania major. Autophagy. 2009. Feb;5(2):159–72. PubMed PMID: 19066473; PubMed Central PMCID: PMC2642932. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Alvarez VE, Kosec G, Sant’Anna C, et al. Autophagy is involved in nutritional stress response and differentiation in Trypanosoma cruzi. J Biol Chem. 2008. Feb 8;283(6):3454–64. doi: 10.1074/jbc.M708474200. PubMed PMID: 18039653; eng. [DOI] [PubMed] [Google Scholar]
- 387.Schoijet AC, Sternlieb T, Alonso GD.. The Phosphatidylinositol 3-kinase Class III Complex Containing TcVps15 and TcVps34 Participates in Autophagy in Trypanosoma cruzi. J Eukaryot Microbiol. 2017. May;64(3):308–321. doi: 10.1111/jeu.12367. PubMed PMID: 27603757. [DOI] [PubMed] [Google Scholar]
- 388.He C, Bartholomew CR, Zhou W, et al. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy. 2009. May;5(4):520–6. PubMed PMID: 19221467; PubMed Central PMCID: PMC2754832. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Banerjee M, Huang Y, Ouseph MM, et al. Autophagy in platelets. Methods Mol Biol. 2019;1880:511–528. doi: 10.1007/978-1-4939-8873-0_32. PubMed PMID: 30610718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Ouseph MM, Huang Y, Banerjee M, et al. Autophagy is induced upon platelet activation and is essential for hemostasis and thrombosis. Blood. 2015. Sep 3;126(10):1224–33. doi: 10.1182/blood-2014-09-598722. PubMed PMID: 26209658; PubMed Central PMCID: PMCPMC4559933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Elsasser A, Vogt AM, Nef H, et al. Human hibernating myocardium is jeopardized by apoptotic and autophagic cell death. J Am Coll Cardiol. 2004. Jun 16;43(12):2191–9. PubMed PMID: 15193679; eng. [DOI] [PubMed] [Google Scholar]
- 392.Knaapen MW, Davies MJ, De Bie M, et al. Apoptotic versus autophagic cell death in heart failure. Cardiovasc Res. 2001. Aug 1;51(2):304–12. PubMed PMID: 11470470; eng. [DOI] [PubMed] [Google Scholar]
- 393.Koike M, Shibata M, Waguri S, et al. Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol. 2005. Dec;167(6):1713–28. PubMed PMID: 16314482; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Kostin S, Pool L, Elsasser A, et al. Myocytes die by multiple mechanisms in failing human hearts. Circ Res. 2003. Apr 18;92(7):715–24. PubMed PMID: 12649263; eng. [DOI] [PubMed] [Google Scholar]
- 395.Motori E, Puyal J, Toni N, et al. Inflammation-induced alteration of astrocyte mitochondrial dynamics requires autophagy for mitochondrial network maintenance. Cell Metab. 2013. Dec 3;18(6):844–59. doi: 10.1016/j.cmet.2013.11.005. PubMed PMID: 24315370. [DOI] [PubMed] [Google Scholar]
- 396.Ost A, Svensson K, Ruishalme I, et al. Attenuated mTOR signaling and enhanced autophagy in adipocytes from obese patients with type 2 diabetes. Mol Med. 2010. Jul-Aug;16(7–8):235–46. doi: 10.2119/molmed.2010.00023. PubMed PMID: 20386866; PubMed Central PMCID: PMC2896460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Perez-Perez ME, Florencio FJ, Crespo JL.. Inhibition of target of rapamycin signaling and stress activate autophagy in Chlamydomonas reinhardtii. Plant Physiol. 2010. Apr;152(4):1874–88. doi: 10.1104/pp.109.152520. PubMed PMID: 20107021; PubMed Central PMCID: PMC2850011. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Tang D, Kang R, Livesey KM, et al. Endogenous HMGB1 regulates autophagy. J Cell Biol. 2010. Sep 6;190(5):881–92. doi: 10.1083/jcb.200911078. PubMed PMID: 20819940; PubMed Central PMCID: PMC2935581. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Decuypere J-P, Welkenhuyzen K, Luyten Y, et al.IP3 receptor-mediated Ca2+ signaling and autophagy induction are interrelated. Autophagy. 2011;7:1472–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Gniadek TJ, Warren G.. WatershedCounting3D: a new method for segmenting and counting punctate structures from confocal image data. Traffic. 2007. Apr;8(4):339–46. doi: 10.1111/j.1600-0854.2007.00538.x. PubMed PMID: 17319897; eng. [DOI] [PubMed] [Google Scholar]
- 401.Anwar T, Liu X, Suntio T, et al. ER-targeted beclin 1 supports autophagosome biogenesis in the absence of ULK1 and ULK2 Kinases. Cells. 2019. May 17;8(5). doi: 10.3390/cells8050475. PubMed PMID: 31108943; PubMed Central PMCID: PMCPMC6562811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Jackson W, Yamada M, Moninger T, et al. Visualization and quantitation of abundant macroautophagy in virus-infected cells by confocal three-dimensional fluorescence imaging. J Virol Methods. 2013. Oct;193(1):244–50. doi: 10.1016/j.jviromet.2013.06.018. PubMed PMID: 23792686; PubMed Central PMCID: PMCPMC3735810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Lamb CA, Joachim J, Tooze SA.. Quantifying autophagic structures in mammalian cells using confocal microscopy. Methods Enzymol. 2017;587:21–42. doi: 10.1016/bs.mie.2016.09.051. PubMed PMID: 28253957. [DOI] [PubMed] [Google Scholar]
- 404.Xu Y, Yuan J, Lipinski MM.. Live imaging and single-cell analysis reveal differential dynamics of autophagy and apoptosis. Autophagy. 2013. Sep;9(9):1418–30. doi: 10.4161/auto.25080. PubMed PMID: 23748697; PubMed Central PMCID: PMC4026027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Wu Z, Zhao J, Qiu M, et al. CRISPR/Cas9 mediated GFP knock-in at the MAP1LC3B locus in 293FT cells is better for bona fide monitoring cellular autophagy. Biotechnol J. 2018. Nov;13(11):e1700674. doi: 10.1002/biot.201700674. PubMed PMID: 29673078. [DOI] [PubMed] [Google Scholar]
- 406.Kamentsky L, Jones TR, Fraser A, et al. Improved structure, function and compatibility for cellprofiler: modular high-throughput image analysis software. Bioinformatics. 2011. Apr 15;27(8):1179–80. doi: 10.1093/bioinformatics/btr095. PubMed PMID: 21349861; PubMed Central PMCID: PMC3072555. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Wu JQ, Pollard TD.. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005. Oct 14;310(5746):310–4. PubMed PMID: 16224022; eng. [DOI] [PubMed] [Google Scholar]
- 408.Geng J, Baba M, Nair U, et al. Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol. 2008. Jul 14;182(1):129–40. doi: 10.1083/jcb.200711112. PubMed PMID: 18625846; PubMed Central PMCID: PMC2447896. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Brady NR, Hamacher-Brady A, Yuan H, et al. The autophagic response to nutrient deprivation in the HL-1 cardiac myocyte is modulated by Bcl-2 and sarco/endoplasmic reticulum calcium stores. FEBS J. 2007. Jun;274(12):3184–97. PubMed PMID: 17540004; eng. [DOI] [PubMed] [Google Scholar]
- 410.Qadir MA, Kwok B, Dragowska WH, et al. Macroautophagy inhibition sensitizes tamoxifen-resistant breast cancer cells and enhances mitochondrial depolarization. Breast Cancer Res Treat. 2008. Dec;112(3):389–403. doi: 10.1007/s10549-007-9873-4. PubMed PMID: 18172760; eng. [DOI] [PubMed] [Google Scholar]
- 411.Furuya T, Kim M, Lipinski M, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010. May 28;38(4):500–11. doi: 10.1016/j.molcel.2010.05.009. PubMed PMID: 20513426; PubMed Central PMCID: PMC2888511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Dolloff NG, Ma X, Dicker DT, et al. Spectral imaging-based methods for quantifying autophagy and apoptosis. Cancer Biol Ther. 2011. Aug 15;12(4):349–56. PubMed PMID: 21757995; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Mastorci K, Montico B, Fae DA, et al. Phospholipid scramblase 1 as a critical node at the crossroad between autophagy and apoptosis in mantle cell lymphoma. Oncotarget. 2016. Jul 5;7(27):41913–41928. doi: 10.18632/oncotarget.9630. PubMed PMID: 27248824; PubMed Central PMCID: PMCPMC5173105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Lee HK, Lund JM, Ramanathan B, et al. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007. Mar 9;315(5817):1398–401. PubMed PMID: 17272685; eng. [DOI] [PubMed] [Google Scholar]
- 415.Phadwal K, Alegre-Abarrategui J, Watson AS, et al. A novel method for autophagy detection in primary cells: Impaired levels of macroautophagy in immunosenescent T cells. Autophagy. 2012;8:677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Davey HM, Hexley P.. Red but not dead? Membranes of stressed Saccharomyces cerevisiae are permeable to propidium iodide. Environ Microbiol. 2011. Jan;13(1):163–71. doi: 10.1111/j.1462-2920.2010.02317.x. PubMed PMID: 21199254. [DOI] [PubMed] [Google Scholar]
- 417.Kuma A, Matsui M, Mizushima N.. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy. 2007. Jul;3(4):323–328. PubMed PMID: 17387262; Eng. [DOI] [PubMed] [Google Scholar]
- 418.Szeto J, Kaniuk NA, Canadien V, et al. ALIS are stress-induced protein storage compartments for substrates of the proteasome and autophagy. Autophagy. 2006. Jul-Sep;2(3):189–99. PubMed PMID: 16874109; eng. [DOI] [PubMed] [Google Scholar]
- 419.Fujita K, Maeda D, Xiao Q, et al. Nrf2-mediated induction of p62 controls Toll-like receptor-4-driven aggresome-like induced structure formation and autophagic degradation. Proc Natl Acad Sci U S A. 2011. Jan 25;108(4):1427–32. doi: 10.1073/pnas.1014156108. PubMed PMID: 21220332; PubMed Central PMCID: PMC3029726. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Kaniuk NA, Kiraly M, Bates H, et al. Ubiquitinated-protein aggregates form in pancreatic [beta]-cells during diabetes-induced oxidative stress and are regulated by autophagy. Diabetes. 2007. Apr;56(4):930–9. PubMed PMID: 17395740; eng. [DOI] [PubMed] [Google Scholar]
- 421.Pierre P. Dendritic cells, DRiPs, and DALIS in the control of antigen processing. Immunol Rev. 2005. Oct;207:184–90. PubMed PMID: 16181336; eng. [DOI] [PubMed] [Google Scholar]
- 422.Calvo-Garrido J, Escalante R.. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy. 2010. Jan;6(1):100–9. PubMed PMID: 20009561; eng. [DOI] [PubMed] [Google Scholar]
- 423.Pankiv S, Høyvarde Clausen T, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007. Jun 19;282:24131–24145. PubMed PMID: 17580304; Eng. [DOI] [PubMed] [Google Scholar]
- 424.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006. Jun15;441(7095):885–9. PubMed PMID: 16625204. [DOI] [PubMed] [Google Scholar]
- 425.Komatsu M, Waguri S, Chiba T, et al.Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006. Jun15;441(7095):880–4. PubMed PMID: 16625205. [DOI] [PubMed] [Google Scholar]
- 426.Bjorkoy G, Lamark T, Johansen T.. p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy. 2006. Apr-Jun;2(2):138–9. PubMed PMID: 16874037; eng. [DOI] [PubMed] [Google Scholar]
- 427.Matsumoto G, Wada K, Okuno M, et al. Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins. Mol Cell. 2011. Oct 21;44(2):279–89. doi: 10.1016/j.molcel.2011.07.039. PubMed PMID: 22017874; eng. [DOI] [PubMed] [Google Scholar]
- 428.Lerner C, Bitto A, Pulliam D, et al. Reduced mammalian target of rapamycin activity facilitates mitochondrial retrograde signaling and increases life span in normal human fibroblasts. Aging cell. 2013. Dec;12(6):966–77. doi: 10.1111/acel.12122. PubMed PMID: 23795962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Napoli E, Liu S, Marsilio I, et al. Lipid-based DNA/siRNA transfection agents disrupt neuronal bioenergetics and mitophagy. Biochem J. 2017. Nov 10;474(23):3887–3902. doi: 10.1042/BCJ20170632. PubMed PMID: 29025974. [DOI] [PubMed] [Google Scholar]
- 430.Zahedi-Amiri A, Sequiera GL, Dhingra S, et al. Influenza a virus-triggered autophagy decreases the pluripotency of human-induced pluripotent stem cells. Cell Death Dis. 2019. Apr 18;10(5):337. doi: 10.1038/s41419-019-1567-4. PubMed PMID: 31000695; PubMed Central PMCID: PMCPMC6472374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Sher AA, Glover KKM, Coombs KM.. Zika Virus Infection Disrupts Astrocytic Proteins Involved in Synapse Control and Axon Guidance. Front Microbiol. 2019;10:596. doi: 10.3389/fmicb.2019.00596. PubMed PMID: 30984137; PubMed Central PMCID: PMCPMC6448030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Köchl R, Hu XW, Chan EYW, et al. Microtubules facilitate autophagosome formation and fusion of auto-phagosomes with endosomes. Traffic. 2006. Feb;7(2):129–45. PubMed PMID: 16420522; eng. [DOI] [PubMed] [Google Scholar]
- 433.Eng KE, Panas MD, Karlsson Hedestam GB, et al. A novel quantitative flow cytometry-based assay for autophagy. Autophagy. 2010. Jul 19;6(5):634–41. PubMed PMID: 20458170; Eng. [DOI] [PubMed] [Google Scholar]
- 434.Ciechomska IA, Tolkovsky AM.. Non-autophagic GFP-LC3 puncta induced by saponin and other detergents. Autophagy. 2007;3:586–590. [DOI] [PubMed] [Google Scholar]
- 435.Seglen PO, Gordon PB.. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982. Mar;79(6):1889–92. PubMed PMID: 6952238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Dembitz V, Lalic H, Visnjic D.. 5-Aminoimidazole-4-carboxamide ribonucleoside-induced autophagy flux during differentiation of monocytic leukemia cells. Cell Death Discov. 2017;3:17066. doi: 10.1038/cddiscovery.2017.66. PubMed PMID: 28975042; PubMed Central PMCID:PMCPMC5624282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Wu YT, Tan HL, Shui G, et al. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem. 2010. Apr 2;285(14):10850–61. doi: 10.1074/jbc.M109.080796. PubMed PMID: 20123989; PubMed Central PMCID: PMC2856291. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Guimar[a]es CA, Benchimol M, Amarante-Mendes GP, et al. Alternative programs of cell death in developing retinal tissue. J Biol Chem. 2003. Oct 24;278(43):41938–46. doi: 10.1074/jbc.M306547200. PubMed PMID: 12917395; eng. [DOI] [PubMed] [Google Scholar]
- 439.Bampton ET, Goemans CG, Niranjan D, et al. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy. 2005. Apr;1(1):23–36. PubMed PMID: 16874023; eng. [DOI] [PubMed] [Google Scholar]
- 440.Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010. Feb 26;32(2):227–39. doi: 10.1016/j.immuni.2009.12.006. PubMed PMID: 20171125; PubMed Central PMCID: PMC2996467. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Tormo D, Checinska A, Alonso-Curbelo D, et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer cell. 2009. Aug 4;16(2):103–14. doi: 10.1016/j.ccr.2009.07.004. PubMed PMID: 19647221; PubMed Central PMCID: PMC2851205. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Tamura N, Oku M, Sakai Y.. Atg8 regulates vacuolar membrane dynamics in a lipidation-independent manner in Pichia pastoris. J Cell Sci. 2010. Dec 1;123(Pt 23):4107–16. doi: 10.1242/jcs.070045. PubMed PMID: 21045113; eng. [DOI] [PubMed] [Google Scholar]
- 443.Stromhaug PE, Reggiori F, Guan J, et al. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol Biol Cell. 2004. Aug;15(8):3553–66. PubMed PMID: 15155809; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Zhang J, Nadtochiy SM, Urciuoli WR, et al. The cardioprotective compound cloxyquin uncouples mitochondria and induces autophagy. Am J Physiol Heart Circ Physiol. 2016. Jan 1;310(1):H29–38. doi: 10.1152/ajpheart.00926.2014. PubMed PMID: 26519034; PubMed Central PMCID: PMCPMC4796459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Mareninova OA, Jia W, Gretler SR, et al. Transgenic expression of GFP-LC3 perturbs autophagy in exocrine pancreas and acute pancreatitis responses in mice. Autophagy. 2020. Jan 16:1–14. doi: 10.1080/15548627.2020.1715047. PubMed PMID: 31942816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Baens M, Noels H, Broeckx V, et al. The dark side of EGFP: defective polyubiquitination. PLoS One. 2006;1:e54. doi: 10.1371/journal.pone.0000054. PubMed PMID: 17183684; PubMed Central PMCID: PMC1762387. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Al-Younes HM, Al-Zeer MA, Khalil H, et al. Autophagy-independent function of MAP-LC3 during intracellular propagation of Chlamydia trachomatis. Autophagy. 2011. Aug 1;7(8):814–28. PubMed PMID: 21464618; eng. [DOI] [PubMed] [Google Scholar]
- 448.Cali T, Galli C, Olivari S, et al. Segregation and rapid turnover of EDEM1 by an autophagy-like mechanism modulates standard ERAD and folding activities. Biochem Biophys Res Commun. 2008. Jul 4;371(3):405–10. doi: 10.1016/j.bbrc.2008.04.098. PubMed PMID: 18452703; eng. [DOI] [PubMed] [Google Scholar]
- 449.Shroff H, Galbraith CG, Galbraith JA, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci U S A. 2007. Dec 18;104(51):20308–13. doi: 10.1073/pnas.0710517105. PubMed PMID: 18077327; PubMed Central PMCID: PMC2154427. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Tanida I, Ueno T, Uchiyama Y.. A super-ecliptic, pHluorin-mKate2, tandem fluorescent protein-tagged human LC3 for the monitoring of mammalian autophagy. PLoS One. 2014;9(10):e110600. doi: doi:s 10.1371/journal.pone.0110600. PubMed PMID: 25340751; PubMed Central PMCID: PMCPMC4207750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Lee JH, Rao MV, Yang DS, et al. Transgenic expression of a ratiometric autophagy probe specifically in neurons enables the interrogation of brain autophagy in vivo. Autophagy. 2019. Mar;15(3):543–557. doi: 10.1080/15548627.2018.1528812. PubMed PMID: 30269645; PubMed Central PMCID: PMCPMC6351128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Nicklin P, Bergman P, Zhang B, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009. Feb 6;136(3):521–34. doi: 10.1016/j.cell.2008.11.044. PubMed PMID: 19203585; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Nyfeler B, Bergman P, Triantafellow E, et al. Relieving autophagy and 4EBP1 from rapamycin resistance. Mol Cell Biol. 2011. Jul;31(14):2867–76. doi: 10.1128/MCB.05430-11. PubMed PMID: 21576371; PubMed Central PMCID: PMC3133392. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Singh K, Sharma A, Mir MC, et al. Autophagic flux determines cell death and survival in response to Apo2L/ TRAIL (dulanermin). Mol Cancer. 2014;13:70. doi: 10.1186/1476-4598-13-70. PubMed PMID: 24655592; PubMed Central PMCID: PMC3998041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Sarkar S, Korolchuk VI, Renna M, et al. Complex inhibitory effects of nitric oxide on autophagy. Mol Cell. 2011. Jul 8;43(1):19–32. doi: 10.1016/j.molcel.2011.04.029. PubMed PMID: 21726807; PubMed Central PMCID: PMC3149661. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Khmelinskii A, Keller PJ, Bartosik A, et al. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol. 2012. Jun 24;30(7):708–14. doi: 10.1038/nbt.2281. PubMed PMID: 22729030. [DOI] [PubMed] [Google Scholar]
- 457.Wen RH, Stanar P, Tam B, et al. Autophagy in Xenopus laevis rod photoreceptors is independently regulated by phototransduction and misfolded RHO(P23H). Autophagy. 2019. Nov;15(11):1970–1989. doi: 10.1080/15548627.2019.1596487. PubMed PMID: 30975014; PubMed Central PMCID: PMCPMC6844500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Nazarko TY, Ozeki K, Till A, et al. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J Cell Biol. 2014. Feb 17;204(4):541–57. doi: 10.1083/jcb.201307050. PubMed PMID: 24535825; PubMed Central PMCID: PMC3926955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Deosaran E, Larsen KB, Hua R, et al. NBR1 acts as an autophagy receptor for peroxisomes. J Cell Sci. 2013. Feb 15;126(Pt 4):939–52. doi: 10.1242/jcs.114819. PubMed PMID: 23239026. [DOI] [PubMed] [Google Scholar]
- 460.Allen GF, Toth R, James J, et al. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013. Dec;14(12):1127–35. doi: 10.1038/embor.2013.168. PubMed PMID: 24176932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Kim SJ, Syed GH, Khan M, et al. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A. 2014. Apr 29;111(17):6413–8. doi: 10.1073/pnas.1321114111. PubMed PMID: 24733894; PubMed Central PMCID: PMC4035934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Wang Y, Nartiss Y, Steipe B, et al. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy. 2012. Oct;8(10):1462–76. doi: 10.4161/auto.21211. PubMed PMID: 22889933. [DOI] [PubMed] [Google Scholar]
- 463.Zhang Y, Mun SR, Linares JF, et al. ZZ-dependent regulation of p62/SQSTM1 in autophagy. Nat Commun. 2018. Oct 22;9(1):4373. doi: 10.1038/s41467-018-06878-8. PubMed PMID: 30349045; PubMed Central PMCID: PMCPMC6197226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Chino H, Hatta T, Natsume T, et al. Intrinsically Disordered Protein TEX264 Mediates ER-phagy. Mol Cell. 2019. Jun 6;74(5):909–921 e6. doi: 10.1016/j.molcel.2019.03.033. PubMed PMID: 31006538. [DOI] [PubMed] [Google Scholar]
- 465.Liang JR, Lingeman E, Ahmed S, et al. Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol. 2018. Oct 1;217(10):3354–3367. doi: 10.1083/jcb.201804185. PubMed PMID: 30143524; PubMed Central PMCID: PMCPMC6168278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Lee JJ, Sanchez-Martinez A, Zarate AM, et al. Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. J Cell Biol. 2018. May 7;217(5):1613–1622. doi: 10.1083/jcb.201801044. PubMed PMID: 29500189; PubMed Central PMCID: PMCPMC5940313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Mijaljica D, Rosado CJ, Devenish RJ, et al. Biosensors for monitoring autophagy. In: Serra PA, editor. Biosensors-emerging materials and applications croatia: intech; 2011. p. 383–400. [Google Scholar]
- 468.Rosado CJ, Mijaljica D, Hatzinisiriou I, et al. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast [Evaluation Studies Research Support, Non-U.S. Gov’t]. Autophagy. 2008. Feb;4(2):205–13. PubMed PMID: 18094608; eng. [DOI] [PubMed] [Google Scholar]
- 469.Mukherjee R, Chakrabarti O.. Ubiquitin-mediated regulation of the E3 ligase GP78 by MGRN1 in trans affects mitochondrial homeostasis. J Cell Sci. 2016. Feb 15;129(4):757–73. doi: 10.1242/jcs.176537. PubMed PMID: 26743086. [DOI] [PubMed] [Google Scholar]
- 470.Kobayashi S, Patel J, Zhao F, et al. Novel Dual-Fluorescent Mitophagy Reporter Reveals a Reduced Mitophagy Flux in Type 1 Diabetic Mouse Heart. J Am Osteopath Assoc. 2020. Jul 1;120(7):446–455. doi: 10.7556/jaoa.2020.072. PubMed PMID: 32598458. [DOI] [PubMed] [Google Scholar]
- 471.Hill SE, Kauffman KJ, Krout M, et al. Maturation and clearance of auto-phagosomes in neurons depends on a specific cysteine protease isoform, ATG-4.2. Dev Cell. 2019. Apr 22;49(2):251–266 e8. doi: 10.1016/j.devcel.2019.02.013. PubMed PMID: 30880001; PubMed Central PMCID: PMCPMC6482087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Chudakov DM, Matz MV, Lukyanov S, et al. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010. Jul;90(3):1103–63. doi: 10.1152/physrev.00038.2009. PubMed PMID: 20664080; eng. [DOI] [PubMed] [Google Scholar]
- 473.Luhr M, Torgersen ML, Szalai P, et al. The kinase PERK and the transcription factor ATF4 play distinct and essential roles in autophagy resulting from tunicamycin-induced ER stress. J Biol Chem. 2019. May 17;294(20):8197–8217. doi: 10.1074/jbc.RA118.002829. PubMed PMID: 30926605; PubMed Central PMCID: PMCPMC6527152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Zhou C, Zhong W, Zhou J, et al. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy. 2012. Aug;8(8):1215–26. doi: 10.4161/auto.20284. PubMed PMID: 22647982. [DOI] [PubMed] [Google Scholar]
- 475.Tanida I, Ueno T, Uchiyama Y.. Use of pHlurorin-mKate2-human LC3 to Monitor Autophagic Responses. Methods Enzymol. 2017;587:87–96. doi: 10.1016/bs.mie.2016.09.054. PubMed PMID: 28253978. [DOI] [PubMed] [Google Scholar]
- 476.Lie PPY, Nixon RA.. Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol Dis. 2019. Feb;122:94–105. doi: 10.1016/j.nbd.2018.05.015. PubMed PMID: 29859318; PubMed Central PMCID: PMCPMC6381838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Cossarizza A, Chang HD, Radbruch A, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur J Immunol. 2017. Oct;47(10):1584–1797. doi: 10.1002/eji.201646632. PubMed PMID: 29023707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Patterson GH, Knobel SM, Sharif WD, et al. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997. Nov;73(5):2782–90. PubMed PMID: 9370472; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Oliva Trejo JA, Tanida I, Suzuki C, et al. Characterization of starvation-induced autophagy in cerebellar Purkinje cells of pHluorin-mKate2-human LC3B transgenic mice. Sci Rep. 2020. Jun 15;10(1):9643. doi: 10.1038/s41598-020-66370-6. PubMed PMID: 32541814; PubMed Central PMCID: PMCPMC7295967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Yazawa R, Nishida Y, Aoyama S, et al. Establishment of a system for screening autophagic flux regulators using a modified fluorescent reporter and CRISPR/Cas9. Biochem Biophys Res Commun. 2019. Aug 27;516(3):686–692. doi: 10.1016/j.bbrc.2019.06.129. PubMed PMID: 31253397. [DOI] [PubMed] [Google Scholar]
- 481.Magidson V, Khodjakov A.. Circumventing photodamage in live-cell microscopy. Methods Cell Biol. 2013;114:545–60. doi: 10.1016/B978-0-12-407761-4.00023-3. PubMed PMID: 23931522; PubMed Central PMCID: PMCPMC3843244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Bogdanov AM, Kudryavtseva EI, Lukyanov KA.. Anti-fading media for live cell GFP imaging. PLoS One. 2012;7(12):e53004. doi: 10.1371/journal.pone.0053004. PubMed PMID: 23285248; PubMed Central PMCID: PMCPMC3528736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Coffey EE, Beckel JM, Laties AM, et al. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 2014. Mar 28;263:111–24. doi: 10.1016/j.neuroscience.2014.01.001. PubMed PMID: 24418614; PubMed Central PMCID: PMCPMC4028113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Guha S, Coffey EE, Lu W, et al. Approaches for detecting lysosomal alkalinization and impaired degradation in fresh and cultured RPE cells: evidence for a role in retinal degenerations. Exp Eye Res. 2014. Sep;126:68–76. doi: 10.1016/j.exer.2014.05.013. PubMed PMID: 25152362; PubMed Central PMCID: PMCPMC4143779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Ramachandra Rao S, Pfeffer BA, Mas Gomez N, et al. Compromised phagosome maturation underlies RPE pathology in cell culture and whole animal models of Smith-Lemli-Opitz Syndrome. Autophagy. 2018;14(10):1796–1817. doi: 10.1080/15548627.2018.1490851. PubMed PMID: 29979914; PubMed Central PMCID: PMCPMC6135634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Zhou J, Lin J, Zhou C, et al. Cytotoxicity of red fluorescent protein DsRed is associated with the suppression of Bcl-xL translation. FEBS Lett. 2011. Mar 9;585(5):821–7. doi: 10.1016/j.febslet.2011.02.013. PubMed PMID: 21320495; eng. [DOI] [PubMed] [Google Scholar]
- 487.Wen Y, Zand B, Ozpolat B, et al. Antagonism of tumoral prolactin receptor promotes autophagy-related cell death. Cell Rep. 2014. Apr 24;7(2):488–500. doi: 10.1016/j.celrep.2014.03.009. PubMed PMID: 24703838; PubMed Central PMCID: PMC4038960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Loos B, Genade S, Ellis B, et al. At the core of survival: autophagy delays the onset of both apoptotic and necrotic cell death in a model of ischemic cell injury. Exp Cell Res. 2011. Jun 10;317(10):1437–53. doi: 10.1016/j.yexcr.2011.03.011. PubMed PMID: 21420401; eng. [DOI] [PubMed] [Google Scholar]
- 489.de la Calle C, Joubert PE, Law HK, et al. Simultaneous assessment of autophagy and apoptosis using multispectral imaging cytometry. Autophagy. 2011. Sep 1;7(9):1045–51. PubMed PMID: 21606680; eng. [DOI] [PubMed] [Google Scholar]
- 490.Degtyarev M, Reichelt M, Lin K.. Novel quantitative autophagy analysis by organelle flow cytometry after cell sonication. PLoS One. 2014;9(1):e87707. doi: 10.1371/journal.pone.0087707. PubMed PMID: 24489953; PubMed Central PMCID: PMC3906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Hundeshagen P, Hamacher-Brady A, Eils R, et al. Concurrent detection of autolysosome formation and lysosomal degradation by flow cytometry in a high-content screen for inducers of autophagy. BMC Biol. 2011;9:38. doi: 10.1186/1741-7007-9-38. PubMed PMID: 21635740; PubMed Central PMCID: PMC3121655. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Shvets E, Fass E, Elazar Z.. Utilizing flow cytometry to monitor autophagy in living mammalian cells. Autophagy. 2008. Jul;4(5):621–8. PubMed PMID: 18376137; eng. [DOI] [PubMed] [Google Scholar]
- 493.Gannage M, Dormann D, Albrecht R, et al. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009. Oct 22;6(4):367–80. doi: 10.1016/j.chom.2009.09.005. PubMed PMID: 19837376; PubMed Central PMCID: PMC2774833. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Kaizuka T, Morishita H, Hama Y, et al. An Autophagic Flux Probe that Releases an Internal Control. Mol Cell. 2016. Nov 17;64(4):835–849. doi: 10.1016/j.molcel.2016.09.037. PubMed PMID: 27818143. [DOI] [PubMed] [Google Scholar]
- 495.Kaminskyy V, Abdi A, Zhivotovsky B.. A quantitative assay for the monitoring of autophagosome accumulation in different phases of the cell cycle. Autophagy. 2011. Jan;7(1):83–90. PubMed PMID: 20980814; eng. [DOI] [PubMed] [Google Scholar]
- 496.Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009. Feb 27;33(4):505–16. doi: 10.1016/j.molcel.2009.01.020. PubMed PMID: 19250911; eng. [DOI] [PubMed] [Google Scholar]
- 497.Larsen KB, Lamark T, Overvatn A, et al. A reporter cell system to monitor autophagy based on p62/SQSTM1. Autophagy. 2010. Aug;6(6):784–93. PubMed PMID: 20574168; eng. [DOI] [PubMed] [Google Scholar]
- 498.Wilson RB, Murphy RF.. Flow-cytometric analysis of endocytic compartments. Methods Cell Biol. 1989;31:293–317. doi: 10.1016/s0091-679x(08)61616-7. PubMed PMID: 2674626. [DOI] [PubMed] [Google Scholar]
- 499.Cossarizza A, Ceccarelli D, Masini A.. Functional heterogeneity of an isolated mitochondrial population revealed by cytofluorometric analysis at the single organelle level. Exp Cell Res. 1996. Jan 10;222(1):84–94. doi: 10.1006/excr.1996.0011. PubMed PMID: 8549677. [DOI] [PubMed] [Google Scholar]
- 500.Dhandayuthapani S, Via LE, Thomas CA, et al. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol Microbiol. 1995. Sep;17(5):901–12. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. PubMed PMID: 8596439. [DOI] [PubMed] [Google Scholar]
- 501.Koga H, Kaushik S, Cuervo AM.. Altered lipid content inhibits autophagic vesicular fusion. FASEB J. 2010. Aug;24(8):3052–65. doi: 10.1096/fj.09-144519. PubMed PMID: 20375270; PubMed Central PMCID: PMCPMC2909278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Picot S, Morga B, Faury N, et al. A study of autophagy in hemocytes of the Pacific oyster, Crassostrea gigas. Autophagy. 2019. Oct;15(10):1801–1809. doi: 10.1080/15548627.2019.1596490. PubMed PMID: 30939979; PubMed Central PMCID: PMCPMC6735588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 503.Rong Y, Liu W, Wang J, et al. Neural stem cell-derived small extracellular vesicles attenuate apoptosis and neuroinflammation after traumatic spinal cord injury by activating autophagy. Cell Death Dis. 2019. Apr 18;10(5):340. doi: 10.1038/s41419-019-1571-8. PubMed PMID: 31000697; PubMed Central PMCID: PMCPMC6472377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Guo S, Liang Y, Murphy SF, et al. A rapid and high content assay that measures cyto-ID-stained autophagic compartments and estimates autophagy flux with potential clinical applications. Autophagy. 2015;11(3):560–72. doi: 10.1080/15548627.2015.1017181. PubMed PMID: 25714620; PubMed Central PMCID: PMCPMC4502761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Skah S, Richartz N, Duthil E, et al. cAMP-mediated autophagy inhibits DNA damage-induced death of leukemia cells independent of p53. Oncotarget. 2018. Jul 13;9(54):30434–30449. doi: 10.18632/oncotarget.25758. PubMed PMID: 30100998; PubMed Central PMCID: PMCPMC6084393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Bas L, Papinski D, Licheva M, et al. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol. 2018. Oct 1;217(10):3656–3669. doi: 10.1083/jcb.201804028. PubMed PMID: 30097514; PubMed Central PMCID: PMCPMC6168255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Gao J, Reggiori F, Ungermann C.. A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J Cell Biol. 2018. Oct 1;217(10):3670–3682. doi: 10.1083/jcb.201804039. PubMed PMID: 30097515; PubMed Central PMCID: PMCPMC6168247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Matsui T, Jiang P, Nakano S, et al. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol. 2018. Aug 6;217(8):2633–2645. doi: 10.1083/jcb.201712058. PubMed PMID: 29789439; PubMed Central PMCID: PMCPMC6080929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Huang JJ, Li HR, Huang Y, et al. Beclin 1 expression: a predictor of prognosis in patients with extranodal natural killer T-cell lymphoma, nasal type. Autophagy. 2010. Aug;6(6):777–83. PubMed PMID: 20639699; eng. [DOI] [PubMed] [Google Scholar]
- 510.Lefort S, Joffre C, Kieffer Y, et al. Inhibition of autophagy as a new means of improving chemotherapy efficiency in high-LC3B triple-negative breast cancers. Autophagy. 2014;10(12):2122–42. doi: 10.4161/15548627.2014.981788. PubMed PMID: 25427136; PubMed Central PMCID: PMCPMC4502743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Sivridis E, Giatromanolaki A, Zois C, et al. The “stone-like” pattern of autophagy in human epithelial tumors and tumor-like lesions: an approach to the clinical outcome. Autophagy. 2010. Aug;6(6):830–3. PubMed PMID: 20622525; eng. [DOI] [PubMed] [Google Scholar]
- 512.Giatromanolaki A, Koukourakis MI, Koutsopoulos A, et al. High Beclin 1 expression defines a poor prognosis in endometrial adenocarcinomas. Gynecol Oncol. 2011. Oct;123(1):147–51. doi: 10.1016/j.ygyno.2011.06.023. PubMed PMID: 21741077; eng. [DOI] [PubMed] [Google Scholar]
- 513.Sivridis E, Giatromanolaki A, Liberis V, et al. Autophagy in endometrial carcinomas and prognostic relevance of ‘stone-like’ structures (SLS): what is destined for the atypical endometrial hyperplasia? Autophagy. 2011. Jan;7(1):74–82. PubMed PMID: 21099253; eng. [DOI] [PubMed] [Google Scholar]
- 514.Chen Y, Lu Y, Lu C, et al. Beclin-1 expression is a predictor of clinical outcome in patients with esophageal squamous cell carcinoma and correlated to hypoxia-inducible factor (HIF)-1alpha expression. Pathol Oncol Res. 2009. Sep;15(3):487–93. doi: 10.1007/s12253-008-9143-8. PubMed PMID: 19130303; PubMed Central PMCID: PMC2791489. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Sakakura K, Takahashi H, Kaira K, et al. Immunological significance of the accumulation of autophagy components in oral squamous cell carcinoma. Cancer Sci. 2015. Jan;106(1):1–8. doi: 10.1111/cas.12559. PubMed PMID: 25338734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Wan XB, Fan XJ, Chen MY, et al. Elevated Beclin 1 expression is correlated with HIF-1[a] in predicting poor prognosis of nasopharyngeal carcinoma. Autophagy. 2010. Apr;6(3):395–404. PubMed PMID: 20150769; eng. [DOI] [PubMed] [Google Scholar]
- 517.Ding ZB, Shi YH, Zhou J, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res. 2008. Nov 15;68(22):9167–75. doi: 10.1158/0008-5472.CAN-08-1573. PubMed PMID: 19010888; eng. [DOI] [PubMed] [Google Scholar]
- 518.Shi YH, Ding ZB, Zhou J, et al. Prognostic significance of Beclin 1-dependent apoptotic activity in hepatocellular carcinoma. Autophagy. 2009. Apr;5(3):380–2. PubMed PMID: 19145109; eng. [DOI] [PubMed] [Google Scholar]
- 519.Pirtoli L, Cevenini G, Tini P, et al. The prognostic role of Beclin 1 protein expression in high-grade gliomas. Autophagy. 2009. Oct;5(7):930–6. PubMed PMID: 19556884; eng. [DOI] [PubMed] [Google Scholar]
- 520.Karpathiou G, Sivridis E, Koukourakis MI, et al. Light-chain 3A autophagic activity and prognostic significance in non-small cell lung carcinomas. Chest. 2011. Jul;140(1):127–34. doi: 10.1378/chest.10-1831. PubMed PMID: 21148243; eng. [DOI] [PubMed] [Google Scholar]
- 521.Fujii S, Mitsunaga S, Yamazaki M, et al. Autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome. Cancer Sci. 2008. Sep;99(9):1813–9. doi: 10.1111/j.1349-7006.2008.00893.x. PubMed PMID: 18616529; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Giatromanolaki A, Koukourakis MI, Harris AL, et al. Prognostic relevance of light chain 3 (LC3A) autophagy patterns in colorectal adenocarcinomas. J Clin Pathol. 2010. Oct;63(10):867–72. doi: 10.1136/jcp.2010.079525. PubMed PMID: 20876316; eng. [DOI] [PubMed] [Google Scholar]
- 523.Koukourakis MI, Giatromanolaki A, Sivridis E, et al. Beclin 1 over- and underexpression in colorectal cancer: distinct patterns relate to prognosis and tumour hypoxia. Br J Cancer. 2010. Oct 12;103(8):1209–14. doi: 10.1038/sj.bjc.6605904. PubMed PMID: 20842118; PubMed Central PMCID: PMC2967071. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Li BX, Li CY, Peng RQ, et al. The expression of beclin 1 is associated with favorable prognosis in stage IIIB colon cancers. Autophagy. 2009. Apr;5(3):303–6. PubMed PMID: 19066461; eng. [DOI] [PubMed] [Google Scholar]
- 525.Giatromanolaki AN, St Charitoudis G, Bechrakis NE, et al. Autophagy patterns and prognosis in uveal melanomas. Mod Pathol. 2011. Aug;24(8):1036–45. doi: 10.1038/modpathol.2011.63. PubMed PMID: 21499230; eng. [DOI] [PubMed] [Google Scholar]
- 526.Sivridis E, Koukourakis MI, Mendrinos SE, et al. Beclin-1 and LC3A expression in cutaneous malignant melanomas: a biphasic survival pattern for beclin-1. Melanoma Res. 2011. Jun;21(3):188–95. doi: 10.1097/CMR.0b013e328346612c. PubMed PMID: 21537144; eng. [DOI] [PubMed] [Google Scholar]
- 527.Long M, McWilliams TG.. Monitoring autophagy in cancer: from bench to bedside. Semin Cancer Biol. 2019. Jul 15. doi: 10.1016/j.semcancer.2019.05.016. PubMed PMID: 31319163. [DOI] [PubMed] [Google Scholar]
- 528.Langer R, Neppl C, Keller MD, et al. Expression analysis of autophagy related markers LC3B, p62 and HMGB1 indicate an autophagy-independent negative prognostic impact of high p62 expression in pulmonary squamous cell carcinomas. Cancers (Basel). 2018. Aug 21;10(9). doi: 10.3390/cancers10090281. PubMed PMID: 30134604; PubMed Central PMCID: PMCPMC6162479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Schlafli AM, Adams O, Galvan JA, et al. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget. 2016. Jun 28;7(26):39544–39555. doi: 10.18632/oncotarget.9647. PubMed PMID: 27250032; PubMed Central PMCID: PMCPMC5129952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.McShane LM, Altman DG, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst. 2005. Aug 17;97(16):1180–4. doi: 10.1093/jnci/dji237. PubMed PMID: 16106022; eng. [DOI] [PubMed] [Google Scholar]
- 531.Hou YJ, Dong LW, Tan YX, et al. Inhibition of active autophagy induces apoptosis and increases chemosensitivity in cholangiocarcinoma. Lab Invest. 2011. Aug;91(8):1146–57. doi: 10.1038/labinvest.2011.97. PubMed PMID: 21647092; eng. [DOI] [PubMed] [Google Scholar]
- 532.Kuwahara Y, Oikawa T, Ochiai Y, et al. Enhancement of autophagy is a potential modality for tumors refractory to radiotherapy. Cell Death Dis. 2011;2:e177. doi: 10.1038/cddis.2011.56. PubMed PMID: 21716292; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.O’Donovan TR, O’Sullivan GC, McKenna SL.. Induction of autophagy by drug-resistant esophageal cancer cells promotes their survival and recovery following treatment with chemotherapeutics. Autophagy. 2011. May;7(5):509–24. PubMed PMID: 21325880; PubMed Central PMCID: PMC3127212. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Ginet V, Pittet MP, Rummel C, et al. Dying neurons in thalamus of asphyxiated term newborns and rats are autophagic. Ann Neurol. 2014. Nov;76(5):695–711. doi: 10.1002/ana.24257. PubMed PMID: 25146903. [DOI] [PubMed] [Google Scholar]
- 535.Xie C, Ginet V, Sun Y, et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy. 2016;12(2):410–23. doi: 10.1080/15548627.2015.1132134. PubMed PMID: 26727396; PubMed Central PMCID: PMCPMC4835980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Cui J, Bai XY, Shi S, et al. Age-related changes in the function of autophagy in rat kidneys. Age (Omaha). 2011. Apr 1;10.1007/s11357-011-9237-1. doi: 10.1007/s11357-011-9237-1. PubMed PMID: 21455601; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Tamura H, Shibata M, Koike M, et al. Atg9A protein, an autophagy-related membrane protein, is localized in the neurons of mouse brains. J Histochem Cytochem. 2010. May;58(5):443–53. doi: 10.1369/jhc.2010.955690. PubMed PMID: 20124090; PubMed Central PMCID: PMC2857816. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Yoshimura K, Shibata M, Koike M, et al. Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy. 2006. Jul-Sep;2(3):200–8. PubMed PMID: 16874114; eng. [DOI] [PubMed] [Google Scholar]
- 539.Zhan L, Liu L, Li K, et al. Neuroprotection of hypoxic postconditioning against global cerebral ischemia through influencing posttranslational regulations of heat shock protein 27 in adult rats. Brain Pathol. 2017. Nov;27(6):822–838. doi: 10.1111/bpa.12472. PubMed PMID: 27936516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Marinelli S, Nazio F, Tinari A, et al. Schwann cell autophagy counteracts the onset and chronification of neuropathic pain. Pain. 2014. Jan;155(1):93–107. doi: 10.1016/j.pain.2013.09.013. PubMed PMID: 24041962. [DOI] [PubMed] [Google Scholar]
- 541.Adolph TE, Tomczak MF, Niederreiter L, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013. Nov 14;503(7475):272–6. doi: 10.1038/nature12599. PubMed PMID: 24089213; PubMed Central PMCID: PMC3862182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Gorbunov NV, Kiang JG.. Autophagy-Mediated Innate Defense Mechanism in Crypt Paneth Cells Responding to Impairment of Small Intestine Barrier after Total-Body Gamma-Photon Irradiation. In: Gorbunov NV, editor. Autophagy: Principles, Regulation and Roles in Disease. Hauppauge, NY: NOVA SCIENCE PUBLISHERS, INC; 2011. p. 61–84. [Google Scholar]
- 543.Thachil E, Hugot JP, Arbeille B, et al. Abnormal activation of autophagy-induced crinophagy in Paneth cells from patients with Crohn’s disease. Gastroenterology. 2012. May;142(5):1097–1099 e4. doi: 10.1053/j.gastro.2012.01.031. PubMed PMID: 22285936. [DOI] [PubMed] [Google Scholar]
- 544.Sepe S, Nardacci R, Fanelli F, et al. Expression of Ambra1 in mouse brain during physiological and Alzheimer type aging. Neurobiol Aging. 2014. Jan;35(1):96–108. doi: 10.1016/j.neurobiolaging.2013.07.001. PubMed PMID: 23910655. [DOI] [PubMed] [Google Scholar]
- 545.Rosso P, Moreno S, Fracassi A, et al. Nerve growth factor and autophagy: effect of nasal anti-NGF-antibodies administration on Ambra1 and Beclin-1 expression in rat brain. Growth Factors. 2015;33(5–6):401–9. doi: 10.3109/08977194.2015.1122002. PubMed PMID: 26728403. [DOI] [PubMed] [Google Scholar]
- 546.Morais RD, Thome RG, Lemos FS, et al. Autophagy and apoptosis interplay during follicular atresia in fish ovary: a morphological and immunocytochemical study. Cell Tissue Res. 2012. Feb;347(2):467–78. doi: 10.1007/s00441-012-1327-6. PubMed PMID: 22314847. [DOI] [PubMed] [Google Scholar]
- 547.Shibata M, Yoshimura K, Furuya N, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009. May 1;382(2):419–23. doi: 10.1016/j.bbrc.2009.03.039. PubMed PMID: 19285958; eng. [DOI] [PubMed] [Google Scholar]
- 548.Komatsu M, Waguri S, Koike M, et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–63. [DOI] [PubMed] [Google Scholar]
- 549.Lazzeri G, Biagioni F, Fulceri F, et al. mTOR modulates methamphetamine-induced toxicity through cell clearing systems. Oxid Med Cell Longev. 2018;2018:6124745. doi: 10.1155/2018/6124745. PubMed PMID: 30647813; PubMed Central PMCID: PMCPMC6311854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 550.Saito T, Asai K, Sato S, et al. Autophagic vacuoles in cardiomyocytes of dilated cardiomyopathy with initially decompensated heart failure predict improved prognosis. Autophagy. 2016;12(3):579–87. doi: 10.1080/15548627.2016.1145326. PubMed PMID: 26890610; PubMed Central PMCID: PMCPMC4836017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Vega-Rubin-de-Celis S, Wiemann S.. Autophagy LC3 HiBiT reporter assay system demonstrates mTORC1 regulation of autophagic flux. Promega Rep. 2018. [Google Scholar]
- 552.Landajuela A, Hervas JH, Anton Z, et al. Lipid Geometry and Bilayer Curvature Modulate LC3/GABARAP-Mediated Model Autophagosomal Elongation. Biophys J. 2016. Jan 19;110(2):411–422. doi: 10.1016/j.bpj.2015.11.3524. PubMed PMID: 26789764; PubMed Central PMCID: PMCPMC4724631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Germain M, Nguyen AP, Le Grand JN, et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011. Jan 19;30(2):395–407. doi: 10.1038/emboj.2010.327. PubMed PMID: 21139567; PubMed Central PMCID: PMC3025469. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Ladoire S, Chaba K, Martins I, et al. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy. 2012. Aug;8(8):1175–84. doi: 10.4161/auto.20353. PubMed PMID: 22647537; PubMed Central PMCID: PMCPMC3973657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Rosenfeldt MT, Nixon C, Liu E, et al. Analysis of macroautophagy by immunohistochemistry. Autophagy. 2012. Jun;8(6):963–9. doi: 10.4161/auto.20186. PubMed PMID: 22562096; PubMed Central PMCID: PMCPMC3427261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Zhu L, Zhu Y, Han S, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019. May 16;10(6):383. doi: 10.1038/s41419-019-1585-2. PubMed PMID: 31097692; PubMed Central PMCID: PMCPMC6522595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Bartlett BJ, Isakson P, Lewerenz J, et al. p62, Ref(2)P and ubiquitinated proteins are conserved markers of neuronal aging, aggregate formation and progressive autophagic defects. Autophagy. 2011. Jun;7(6):572–83. PubMed PMID: 21325881; PubMed Central PMCID: PMC3127048. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Komatsu M, Wang QJ, Holstein GR, et al. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A. 2007. Sep 4;104(36):14489–94. PubMed PMID: 17726112; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Masiero E, Agatea L, Mammucari C, et al. Autophagy is required to maintain muscle mass [Research Support, Non-U.S. Gov’t]. Cell Metab. 2009. Dec;10(6):507–15. doi: 10.1016/j.cmet.2009.10.008. PubMed PMID: 19945408; eng. [DOI] [PubMed] [Google Scholar]
- 560.Nezis IP, Simonsen A, Sagona AP, et al. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008. Mar 24;180(6):1065–71. doi: 10.1083/jcb.200711108. PubMed PMID: 18347073; PubMed Central PMCID: PMC2290837. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Wang QJ, Ding Y, Kohtz DS, et al. Induction of autophagy in axonal dystrophy and degeneration. J Neurosci. 2006. Aug 2;26(31):8057–68. PubMed PMID: 16885219; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Zhang H, Chang JT, Guo B, et al. Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy. 2015;11(1):9–27. doi: 10.1080/15548627.2014.1003478. PubMed PMID: 25569839; PubMed Central PMCID: PMC4502811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Ullrich M, Assmus B, Augustin AM, et al. SPRED2 deficiency elicits cardiac arrhythmias and premature death via impaired autophagy. J Mol Cell Cardiol. 2019. Apr;129:13–26. doi: 10.1016/j.yjmcc.2019.01.023. PubMed PMID: 30771306. [DOI] [PubMed] [Google Scholar]
- 564.Navarro-Yepes J, Anandhan A, Bradley E, et al. Inhibition of protein ubiquitination by paraquat and 1-methyl-4-phenylpyridinium impairs ubiquitin-dependent protein degradation pathways. Mol Neurobiol. 2016. Oct;53(8):5229–51. doi: 10.1007/s12035-015-9414-9. PubMed PMID: 26409479; PubMed Central PMCID: PMCPMC4842169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.El-Khoury V, Pierson S, Szwarcbart E, et al. Disruption of autophagy by the histone deacetylase inhibitor MGCD0103 and its therapeutic implication in B-cell chronic lymphocytic leukemia. Leukemia. 2014. Aug;28(8):1636–46. doi: 10.1038/leu.2014.19. PubMed PMID: 24418989; PubMed Central PMCID: PMC4131250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Guo B, Huang X, Zhang P, et al. Genome-wide screen identifies signaling pathways that regulate autophagy during Caenorhabditis elegans development. EMBO Rep. 2014. Jun;15(6):705–13. doi: 10.1002/embr.201338310. PubMed PMID: 24764321; PubMed Central PMCID: PMCPMC4197881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Aqbi HF, Tyutyunyk-Massey L, Keim RC, et al. Autophagy-deficient breast cancer shows early tumor recurrence and escape from dormancy. Oncotarget. 2018. Apr 24;9(31):22113–22122. doi: 10.18632/oncotarget.25197. PubMed PMID: 29774126; PubMed Central PMCID: PMCPMC5955162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Nakaso K, Yoshimoto Y, Nakano T, et al. Transcriptional activation of p62/A170/ZIP during the formation of the aggregates: possible mechanisms and the role in Lewy body formation in Parkinson’s disease. Brain Res. 2004. Jun 25;1012(1–2):42–51. PubMed PMID: 15158159; eng. [DOI] [PubMed] [Google Scholar]
- 569.Trocoli A, Bensadoun P, Richard E, et al. p62/SQSTM1 upregulation constitutes a survival mechanism that occurs during granulocytic differentiation of acute myeloid leukemia cells. Cell Death Differ. 2014. Jul 18. doi: 10.1038/cdd.2014.102. PubMed PMID: 25034783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.B’Chir W, Maurin AC, Carraro V, et al. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013. Sep;41(16):7683–99. doi: 10.1093/nar/gkt563. PubMed PMID: 23804767; PubMed Central PMCID: PMC3763548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Cnop M, Abdulkarim B, Bottu G, et al. RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes. 2014. Jun;63(6):1978–93. doi: 10.2337/db13-1383. PubMed PMID: 24379348. [DOI] [PubMed] [Google Scholar]
- 572.Kumsta C, Chang JT, Schmalz J, et al. Hormetic heat stress and HSF-1 induce autophagy to improve survival and proteostasis in C. elegans. Nat Commun. 2017. Feb 15;8:14337. doi: 10.1038/ncomms14337. PubMed PMID: 28198373; PubMed Central PMCID: PMCPMC5316864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Lapierre LR, De Magalhaes Filho CD, McQuary PR, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. doi: 10.1038/ncomms3267. PubMed PMID: 23925298; PubMed Central PMCID: PMC3866206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Colosetti P, Puissant A, Robert G, et al. Autophagy is an important event for megakaryocytic differentiation of the chronic myelogenous leukemia K562 cell line. Autophagy. 2009. Nov;5(8):1092–8. PubMed PMID: 19786835; eng. [DOI] [PubMed] [Google Scholar]
- 575.Toepfer N, Childress C, Parikh A, et al. Atorvastatin induces autophagy in prostate cancer PC3 cells through activation of LC3 transcription. Cancer Biol Ther. 2011. Oct 15;12(8):691–9. doi: 10.4161/cbt.12.8.15978. PubMed PMID: 21768780; eng. [DOI] [PubMed] [Google Scholar]
- 576.Zheng Q, Su H, Ranek MJ, et al. Autophagy and p62 in cardiac proteinopathy. Circ Res. 2011. Jul 22;109(3):296–308. doi: 10.1161/CIRCRESAHA.111.244707. PubMed PMID: 21659648; PubMed Central PMCID: PMC3142307. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Trocoli A, Mathieu J, Priault M, et al. ATRA-induced upregulation of Beclin 1 prolongs the life span of differentiated acute promyelocytic leukemia cells. Autophagy. 2011. Oct;7(10):1108–14. doi: 10.4161/auto.7.10.16623. PubMed PMID: 21691148; PubMed Central PMCID: PMC3242613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Kato M, Ospelt C, Gay RE, et al. Dual role of autophagy in stress-induced cell death in rheumatoid arthritis synovial fibroblasts. Arthritis Rheumatol. 2014. Jan;66(1):40–8. doi: 10.1002/art.38190. PubMed PMID: 24449574. [DOI] [PubMed] [Google Scholar]
- 579.Kim JH, Hong SK, Wu PK, et al. Raf/MEK/ERK can regulate cellular levels of LC3B and SQSTM1/p62 at expression levels. Exp Cell Res. 2014. Oct 1;327(2):340–52. doi: 10.1016/j.yexcr.2014.08.001. PubMed PMID: 25128814; PubMed Central PMCID: PMC4164593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.B’Chir W, Chaveroux C, Carraro V, et al. Dual role for CHOP in the crosstalk between autophagy and apoptosis to determine cell fate in response to amino acid deprivation. Cell Signal. 2014. Jul;26(7):1385–91. doi: 10.1016/j.cellsig.2014.03.009. PubMed PMID: 24657471. [DOI] [PubMed] [Google Scholar]
- 581.Sahani MH, Itakura E, Mizushima N.. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014. Mar 1;10(3):431–41. doi: 10.4161/auto.27344. PubMed PMID: 24394643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Jamart C, Naslain D, Gilson H, et al. Higher activation of autophagy in skeletal muscle of mice during endurance exercise in the fasted state. Am J Physiol Endocrinol Metab. 2013. Oct 15;305(8):E964–74. doi: 10.1152/ajpendo.00270.2013. PubMed PMID: 23964069. [DOI] [PubMed] [Google Scholar]
- 583.Sanchez AM, Bernardi H, Py G, et al. Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol. 2014. Oct 15;307(8):R956–69. doi: 10.1152/ajpregu.00187.2014. PubMed PMID: 25121614. [DOI] [PubMed] [Google Scholar]
- 584.Stingele S, Stoehr G, Peplowska K, et al. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. PubMed PMID: 22968442; PubMed Central PMCID: PMC3472693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Tang YC, Williams BR, Siegel JJ, et al. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011. Feb 18;144(4):499–512. doi: 10.1016/j.cell.2011.01.017. PubMed PMID: 21315436; PubMed Central PMCID: PMC3532042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Marchesi N, Thongon N, Pascale A, et al. Autophagy Stimulus Promotes Early HuR Protein Activation and p62/SQSTM1 Protein Synthesis in ARPE-19 Cells by Triggering Erk1/2, p38(MAPK), and JNK Kinase Pathways. Oxid Med Cell Longev. 2018;2018:4956080. doi: 10.1155/2018/4956080. PubMed PMID: 29576851; PubMed Central PMCID: PMCPMC5822911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Penna F, Costamagna D, Pin F, et al. Autophagic degradation contributes to muscle wasting in cancer cachexia. Am J Pathol. 2013. Apr;182(4):1367–78. doi: 10.1016/j.ajpath.2012.12.023. PubMed PMID: 23395093. [DOI] [PubMed] [Google Scholar]
- 588.BenYounes A, Tajeddine N, Tailler M, et al. A fluorescence-microscopic and cytofluorometric system for monitoring the turnover of the autophagic substrate p62/SQSTM1 [Research Support, Non-U.S. Gov’t]. Autophagy. 2011. Aug 1;7(8):883–91. PubMed PMID: 21460612; eng. [DOI] [PubMed] [Google Scholar]
- 589.Chang Y-Y, Neufeld TP.. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009. Apr;20(7):2004–14. doi: 10.1091/mbc.E08-12-1250. PubMed PMID: 19225150; PubMed Central PMCID: PMC2663935. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Jiang Y, Zhu J, Wu L, et al. Tetracycline inhibits local inflammation induced by cerebral ischemia via modulating autophagy. PLoS One. 2012;7(11):e48672. doi: 10.1371/journal.pone.0048672. PubMed PMID: 23144925; PubMed Central PMCID: PMC3492486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Moullan N, Mouchiroud L, Wang X, et al. Tetracyclines Disturb Mitochondrial Function across Eukaryotic Models: A Call for Caution in Biomedical Research. Cell Rep. 2015. Mar 17;10(10):1681–1691. doi: 10.1016/j.celrep.2015.02.034. PubMed PMID: 25772356; PubMed Central PMCID: PMCPMC4565776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Bjorkoy G, Lamark T, Pankiv S, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–97. doi: 10.1016/S0076-6879(08)03612-4. PubMed PMID: 19200883; eng. [DOI] [PubMed] [Google Scholar]
- 593.Moscat J, Diaz-Meco MT.. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009. Jun 12;137(6):1001–4. doi: 10.1016/j.cell.2009.05.023. PubMed PMID: 19524504; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Duran A, Amanchy R, Linares JF, et al. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol Cell. 2011. Oct 7;44(1):134–46. doi: 10.1016/j.molcel.2011.06.038. PubMed PMID: 21981924; PubMed Central PMCID: PMC3190169. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Gonzalez Y, Aryal B, Chehab L, et al. Atg7- and Keap1-dependent autophagy protects breast cancer cell lines against mitoquinone-induced oxidative stress. Oncotarget. 2014. Mar 30;5(6):1526–37. PubMed PMID: 24681637; PubMed Central PMCID: PMC4039229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Komatsu M, Kurokawa H, Waguri S, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat Cell Biol. 2010. Mar;12(3):213–23. doi: 10.1038/ncb2021. PubMed PMID: 20173742; eng. [DOI] [PubMed] [Google Scholar]
- 597.Jain A, Lamark T, Sjottem E, et al. p62/SQSTM1 is a target gene for transcription factor NRF2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J Biol Chem. 2010. Jul 16;285(29):22576–91. doi: 10.1074/jbc.M110.118976. PubMed PMID: 20452972; PubMed Central PMCID: PMC2903417. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Korolchuk VI, Menzies FM, Rubinsztein DC.. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010. Apr 2;584(7):1393–8. doi: 10.1016/j.febslet.2009.12.047. PubMed PMID: 20040365. [DOI] [PubMed] [Google Scholar]
- 599.Cecarini V, Bonfili L, Cuccioloni M, et al. The fine-tuning of proteolytic pathways in Alzheimer’s disease. Cell Mol Life Sci.: CMLS. 2016. Sep;73(18):3433–51. doi: 10.1007/s00018-016-2238-6. PubMed PMID: 27120560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 600.Bardag-Gorce F, Francis T, Nan L, et al. Modifications in p62 occur due to proteasome inhibition in alcoholic liver disease. Life Sci 2005. Sep 30;77(20):2594–602. PubMed PMID: 15964033; eng. [DOI] [PubMed] [Google Scholar]
- 601.Myeku N, Figueiredo-Pereira ME.. Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. J Biol Chem. 2011. Jun 24;286(25):22426–40. doi: 10.1074/jbc.M110.149252. PubMed PMID: 21536669; PubMed Central PMCID: PMC3121389. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Korolchuk VI, Mansilla A, Menzies FM, et al. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol Cell. 2009. Feb 27;33(4):517–27. doi: 10.1016/j.molcel.2009.01.021. PubMed PMID: 19250912; PubMed Central PMCID: PMC2669153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Yoshiyama Y, Higuchi M, Zhang B, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007. Feb 1;53(3):337–51. doi: 10.1016/j.neuron.2007.01.010. PubMed PMID: 17270732. [DOI] [PubMed] [Google Scholar]
- 604.Akwa Y, Gondard E, Mann A, et al. Synaptic activity protects against AD and FTD-like pathology via autophagic-lysosomal degradation. Mol Psychiatry. 2018. Jun;23(6):1530–1540. doi: 10.1038/mp.2017.142. PubMed PMID: 28696431; PubMed Central PMCID: PMCPMC5641448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Sharma A, Alswillah T, Kapoor I, et al. USP14 is a deubiquitinase for Ku70 and critical determinant of non-homologous end joining repair in autophagy and PTEN-deficient cells. Nucleic Acids Res. 2020. Jan 24;48(2):736–747. doi: 10.1093/nar/gkz1103. PubMed PMID: 31740976; PubMed Central PMCID: PMC7145659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Sharma A, Alswillah T, Singh K, et al. USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy. 2018;14(11):1976–1990. doi: 10.1080/15548627.2018.1496877. PubMed PMID: 29995557; PubMed Central PMCID: PMCPMC6152509; PubMed Central PMCID: PMC6152509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Dantuma NP, Lindsten K, Glas R, et al. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000. May;18(5):538–43. doi: 10.1038/75406. PubMed PMID: 10802622. [DOI] [PubMed] [Google Scholar]
- 608.Monick MM, Powers LS, Walters K, et al. Identification of an autophagy defect in smokers’ alveolar macrophages. J Iimmunol. 2010. Nov 1;185(9):5425–35. doi: 10.4049/jimmunol.1001603. PubMed PMID: 20921532; PubMed Central PMCID: PMC3057181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Vallelian F, Deuel JW, Opitz L, et al. Proteasome inhibition and oxidative reactions disrupt cellular homeostasis during heme stress. Cell Death Differ. 2015. Apr;22(4):597–611. doi: 10.1038/cdd.2014.154. PubMed PMID: 25301065; PubMed Central PMCID: PMC4356336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Long J, Garner TP, Pandya MJ, et al. Dimerisation of the UBA domain of p62 inhibits ubiquitin binding and regulates NF-kappaB signalling. J Mol Biol. 2010. Feb 12;396(1):178–94. doi: 10.1016/j.jmb.2009.11.032. PubMed PMID: 19931284. [DOI] [PubMed] [Google Scholar]
- 611.Norman JM, Cohen GM, Bampton ET.. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy. 2010. Nov;6(8):1042–56. PubMed PMID: 21121091; eng. [DOI] [PubMed] [Google Scholar]
- 612.Lelouard H, Schmidt EK, Camosseto V, et al. Regulation of translation is required for dendritic cell function and survival during activation. J Cell Biol. 2007. Dec 31;179(7):1427–39. doi: 10.1083/jcb.200707166. PubMed PMID: 18166652; PubMed Central PMCID: PMC2373495. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Cecarini V, Bonfili L, Cuccioloni M, et al. Wild type and mutant amyloid precursor proteins influence downstream effects of proteasome and autophagy inhibition. Biochim Biophys Acta. 2014. Feb;1842(2):127–34. doi: 10.1016/j.bbadis.2013.11.002. PubMed PMID: 24215712. [DOI] [PubMed] [Google Scholar]
- 614.Schmidt EK, Clavarino G, Ceppi M, et al. SUnSET, a nonradioactive method to monitor protein synthesis [Research Support, Non-U.S. Gov’t]. Nat Methods. 2009. Apr;6(4):275–7. doi: 10.1038/nmeth.1314. PubMed PMID: 19305406; eng. [DOI] [PubMed] [Google Scholar]
- 615.Lim J, Kim HW, Youdim MB, et al. Binding preference of p62 towards LC3-ll during dopaminergic neurotoxin-induced impairment of autophagic flux. Autophagy. 2011. Jan;7(1):51–60. PubMed PMID: 21045561; eng. [DOI] [PubMed] [Google Scholar]
- 616.Fouillet A, Levet C, Virgone A, et al. ER stress inhibits neuronal death by promoting autophagy. Autophagy. 2012. Jun;8(6):915–26. doi: 10.4161/auto.19716. PubMed PMID: 22660271; PubMed Central PMCID: PMC3427257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Waguri S, Komatsu M.. Biochemical and morphological detection of inclusion bodies in autophagy-deficient mice. Methods Enzymol. 2009;453:181–96. doi: 10.1016/S0076-6879(08)04009-3. PubMed PMID: 19216907; eng. [DOI] [PubMed] [Google Scholar]
- 618.Hocking LJ, Lucas GJ, Daroszewska A, et al. Domain-specific mutations in sequestosome 1 (SQSTM1) cause familial and sporadic Paget’s disease. Hum Mol Genet. 2002. Oct 15;11(22):2735–9. PubMed PMID: 12374763; eng. [DOI] [PubMed] [Google Scholar]
- 619.Kara NZ, Toker L, Agam G, et al. Trehalose induced antidepressant-like effects and autophagy enhancement in mice. Psychopharmacology (Berl). 2013. Sep;229(2):367–75. doi: 10.1007/s00213-013-3119-4. PubMed PMID: 23644913. [DOI] [PubMed] [Google Scholar]
- 620.Nimmerjahn F, Milosevic S, Behrends U, et al. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003. May;33(5):1250–9. PubMed PMID: 12731050; eng. [DOI] [PubMed] [Google Scholar]
- 621.Guha P, Tyagi R, Chowdhury S, et al. IPMK Mediates Activation of ULK Signaling and Transcriptional Regulation of Autophagy Linked to Liver Inflammation and Regeneration. Cell Rep. 2019. Mar 5;26(10):2692–2703 e7. doi: 10.1016/j.celrep.2019.02.013. PubMed PMID: 30840891; PubMed Central PMCID: PMCPMC6494083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 622.Beasley CL, Pennington K, Behan A, et al. Proteomic analysis of the anterior cingulate cortex in the major psychiatric disorders: Evidence for disease-associated changes. Proteomics. 2006. Jun;6(11):3414–25. doi: 10.1002/pmic.200500069. PubMed PMID: 16637010. [DOI] [PubMed] [Google Scholar]
- 623.Behan AT, Byrne C, Dunn MJ, et al. Proteomic analysis of membrane microdomain-associated proteins in the dorsolateral prefrontal cortex in schizophrenia and bipolar disorder reveals alterations in LAMP, STXBP1 and BASP1 protein expression. Mol Psychiatry. 2009. Jun;14(6):601–13. doi: 10.1038/mp.2008.7, PubMed PMID: 18268500. [DOI] [PubMed] [Google Scholar]
- 624.Chetcuti A, Adams LJ, Mitchell PB, et al. Microarray gene expression profiling of mouse brain mRNA in a model of lithium treatment. Psychiatr Genet. 2008. Apr;18(2):64–72. doi: 10.1097/YPG.0b013e3282fb0051. PubMed PMID: 18349697. [DOI] [PubMed] [Google Scholar]
- 625.Focking M, Dicker P, English JA, et al. Common proteomic changes in the hippocampus in schizophrenia and bipolar disorder and particular evidence for involvement of cornu ammonis regions 2 and 3. Arch gen psychiatry. 2011. May;68(5):477–88. doi: 10.1001/archgenpsychiatry.2011.43. PubMed PMID: 21536977. [DOI] [PubMed] [Google Scholar]
- 626.Nielsen J, Hoffert JD, Knepper MA, et al. Proteomic analysis of lithium-induced nephrogenic diabetes insipidus: mechanisms for aquaporin 2 down-regulation and cellular proliferation. Proc Natl Acad Sci U S A. 2008. Mar 4;105(9):3634–9. doi: 10.1073/pnas.0800001105. PubMed PMID: 18296634; PubMed Central PMCID: PMC2265122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Lu K, Psakhye I, Jentsch S.. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell. 2014. Jul 31;158(3):549–63. doi: 10.1016/j.cell.2014.05.048. PubMed PMID: 25042851. [DOI] [PubMed] [Google Scholar]
- 628.Mesquita A, Cardenal-Munoz E, Dominguez E, et al. Autophagy in Dictyostelium: Mechanisms, regulation and disease in a simple biomedical model. Autophagy. 2017. Jan 2;13(1):24–40. doi: 10.1080/15548627.2016.1226737. PubMed PMID: 27715405; PubMed Central PMCID: PMCPMC5240833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Sampaio-Marques B, Pereira H, Santos AR, et al. Caloric restriction rescues yeast cells from alpha-synuclein toxicity through autophagic control of proteostasis. Aging (Albany NY). 2018. Dec 7;10(12):3821–3833. doi: 10.18632/aging.101675. PubMed PMID: 30530923; PubMed Central PMCID: PMCPMC6326672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Baker B, Geng S, Chen K, et al. Alteration of Lysosome Fusion and Low-grade Inflammation Mediated by Super-low-dose Endotoxin. J Biol Chem. 2015. Mar 6;290(10):6670–8. doi: 10.1074/jbc.M114.611442. PubMed PMID: 25586187; PubMed Central PMCID: PMC4358298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 631.Li T, Hu J, Li L.. Characterization of Tollip protein upon Lipopolysaccharide challenge. Mol Immunol. 2004. May;41(1):85–92. doi: 10.1016/j.molimm.2004.03.009. PubMed PMID: 15140579. [DOI] [PubMed] [Google Scholar]
- 632.Mitra S, Traughber CA, Brannon MK, et al. Ubiquitin interacts with the Tollip C2 and CUE domains and inhibits binding of Tollip to phosphoinositides. J Biol Chem. 2013. Sep 6;288(36):25780–91. doi: 10.1074/jbc.M113.484170. PubMed PMID: 23880770; PubMed Central PMCID: PMCPMC3764785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Chen K, Yuan R, Zhang Y, et al. Tollip Deficiency Alters Atherosclerosis and Steatosis by Disrupting Lipophagy. J Am Heart Assoc. 2017. Apr 10;6(4). doi: 10.1161/JAHA.116.004078. PubMed PMID: 28396568; PubMed Central PMCID: PMCPMC5532987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 634.Chen K, Yuan R, Geng S, et al. Toll-interacting protein deficiency promotes neurodegeneration via impeding autophagy completion in high-fat diet-fed ApoE(-/-) mouse model. Brain Behav Immun. 2017. Jan;59:200–210. doi: 10.1016/j.bbi.2016.10.002. PubMed PMID: 27720815; PubMed Central PMCID: PMCPMC5154796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 635.Tian Z, Wang C, Hu C, et al. Autophagic-lysosomal inhibition compromises ubiquitin-proteasome system performance in a p62 dependent manner in cardiomyocytes. PLoS One. 2014;9(6):e100715. doi: 10.1371/journal.pone.0100715. PubMed PMID: 24959866; PubMed Central PMCID: PMCPMC4069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Mizushima N, Levine B.. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010. Sep;12(9):823–30. doi: 10.1038/ncb0910-823. PubMed PMID: 20811354; PubMed Central PMCID: PMC3127249. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Maloverjan A, Piirsoo M, Michelson P, et al. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway [Research Support, Non-U.S. Gov’t]. Exp Cell Res. 2010. Feb 15;316(4):627–37. doi: 10.1016/j.yexcr.2009.10.018. PubMed PMID: 19878745; eng. [DOI] [PubMed] [Google Scholar]
- 638.Young ARJ, Narita M, Ferreira M, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009. Apr 1;23(7):798–803. doi: 10.1101/gad.519709. PubMed PMID: 19279323; PubMed Central PMCID: PMC2666340. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Chan EY, Tooze SA.. Evolution of Atg1 function and regulation [Review]. Autophagy. 2009. Aug;5(6):758–65. PubMed PMID: 19411825; eng. [DOI] [PubMed] [Google Scholar]
- 640.Chan EYW, Kir S, Tooze SA.. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007. Aug 31;282(35):25464–74. doi:M703663200[pii] doi: 10.1074/jbc.M703663200. PubMed PMID: 17595159; eng. [DOI] [PubMed] [Google Scholar]
- 641.Fuqua JD, Mere CP, Kronemberger A, et al. ULK2 is essential for degradation of ubiquitinated protein aggregates and homeostasis in skeletal muscle. FASEB J. 2019. Nov;33(11):11735–11745. doi: 10.1096/fj.201900766R. PubMed PMID: 31361156; PubMed Central PMCID: PMCPMC6902739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Lee EJ, Tournier C.. The requirement of uncoordinated 51-like kinase 1 (ULK1) and ULK2 in the regulation of autophagy. Autophagy. 2011. Jul;7(7):689–95. doi: 10.4161/auto.7.7.15450. PubMed PMID: 21460635; PubMed Central PMCID: PMCPMC3149696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Ro SH, Jung CH, Hahn WS, et al. Distinct functions of Ulk1 and Ulk2 in the regulation of lipid metabolism in adipocytes. Autophagy. 2013. Dec;9(12):2103–14. doi: 10.4161/auto.26563. PubMed PMID: 24135897; PubMed Central PMCID: PMCPMC4028344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Shukla S, Patric IR, Patil V, et al. Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem. 2014. Aug 8;289(32):22306–18. doi: 10.1074/jbc.M114.567032. PubMed PMID: 24923441; PubMed Central PMCID: PMCPMC4139240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 645.Balke D, Tatenhorst L, Dambeck V, et al. AAV-Mediated Expression of Dominant-Negative ULK1 Increases Neuronal Survival and Enhances Motor Performance in the MPTP Mouse Model of Parkinson’s Disease. Mol Neurobiol. 2019. Aug 24. doi: 10.1007/s12035-019-01744-0. PubMed PMID: 31446549. [DOI] [PubMed] [Google Scholar]
- 646.Chan EYW, Longatti A, McKnight NC, et al. Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism [Research Support, Non-U.S. Gov’t]. Mol Cell Biol. 2009. Jan;29(1):157–71. doi: 10.1128/MCB.01082-08. PubMed PMID: 18936157; PubMed Central PMCID: PMC2612494. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Joo JH, Dorsey FC, Joshi A, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Mol Cell. 2011. Aug 19;43(4):572–85. doi: 10.1016/j.molcel.2011.06.018. PubMed PMID: 21855797; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Liu CC, Lin YC, Chen YH, et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol Cell. 2016. Jan 7;61(1):84–97. doi: 10.1016/j.molcel.2015.11.001. PubMed PMID: 26687681. [DOI] [PubMed] [Google Scholar]
- 649.Han SH, Korm S, Han YG, et al. GCA links TRAF6-ULK1-dependent autophagy activation in resistant chronic myeloid leukemia. Autophagy. 2019. Dec;15(12):2076–2090. doi: 10.1080/15548627.2019.1596492. PubMed PMID: 30929559; PubMed Central PMCID: PMCPMC6844495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Samari HR, Moller MT, Holden L, et al. Stimulation of hepatocytic AMP-activated protein kinase by okadaic acid and other autophagy-suppressive toxins. Biochem J. 2005. Mar 1;386(Pt 2):237–44. doi: 10.1042/BJ20040609. PubMed PMID: 15461583; PubMed Central PMCID: PMC1134787. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Carling D, Mayer FV, Sanders MJ, et al. AMP-activated protein kinase: nature’s energy sensor. Nat Chem Biol. 2011. Aug;7(8):512–8. doi: 10.1038/nchembio.610. PubMed PMID: 21769098; eng. [DOI] [PubMed] [Google Scholar]
- 652.Puustinen P, Keldsbo A, Corcelle-Termeau E, et al. DNA-dependent protein kinase regulates lysosomal AMP-dependent protein kinase activation and autophagy. Autophagy. 2020. Jan 26:1–18. doi: 10.1080/15548627.2019.1710430. PubMed PMID: 31983282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Dando I, Donadelli M, Costanzo C, et al. Cannabinoids inhibit energetic metabolism and induce AMPK-dependent autophagy in pancreatic cancer cells. Cell Death Dis. 2013;4:e664. doi: 10.1038/cddis.2013.151. PubMed PMID: 23764845; PubMed Central PMCID: PMC3698539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Hawley SA, Ross FA, Chevtzoff C, et al. Use of cells expressing gamma subunit variants to identify diverse mechanisms of AMPK activation. Cell Metab. 2010. Jun 9;11(6):554–65. doi: 10.1016/j.cmet.2010.04.001. PubMed PMID: 20519126; PubMed Central PMCID: PMC2935965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 655.Gross AS, Zimmermann A, Pendl T, et al. Acetyl-CoA carboxylase 1-dependent lipogenesis promotes autophagy downstream of AMPK. J Biol Chem. 2019. Aug 9;294(32):12020–12039. doi: 10.1074/jbc.RA118.007020. PubMed PMID: 31209110; PubMed Central PMCID: PMCPMC6690696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Nwadike C, Williamson LE, Gallagher LE, et al. AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms. Mol Cell Biol. 2018. May 15;38(10). doi: 10.1128/MCB.00023-18. PubMed PMID: 29507183; PubMed Central PMCID: PMCPMC5954193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Ha S, Jeong SH, Yi K, et al. Phosphorylation of p62 by AMP-activated protein kinase mediates autophagic cell death in adult hippocampal neural stem cells. J Biol Chem. 2017. Aug 18;292(33):13795–13808. doi: 10.1074/jbc.M117.780874. PubMed PMID: 28655770; PubMed Central PMCID: PMCPMC5566532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Behrends C, Sowa ME, Gygi SP, et al. Network organization of the human autophagy system. Nature. 2010. Jul 1;466(7302):68–76. doi: 10.1038/nature09204. PubMed PMID: 20562859; PubMed Central PMCID: PMC2901998. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Chiacchiera F, Matrone A, Ferrari E, et al. p38alpha blockade inhibits colorectal cancer growth in vivo by inducing a switch from HIF1alpha- to FoxO-dependent transcription. Cell Death Differ. 2009. Sep;16(9):1203–14. doi: 10.1038/cdd.2009.36. PubMed PMID: 19343039; eng. [DOI] [PubMed] [Google Scholar]
- 660.Kovács AL, Seglen PO.. Inhibition of hepatocytic protein degradation by methylaminopurines and inhibitors of protein synthesis. Biochim Biophys Acta. 1981. Aug 17;676(2):213–20. PubMed PMID: 7260116; eng. [DOI] [PubMed] [Google Scholar]
- 661.Liu HY, Han J, Cao SY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009. Nov 6;284(45):31484–92. doi: 10.1074/jbc.M109.033936. PubMed PMID: 19758991; PubMed Central PMCID: PMC2781544. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.Mammucari C, Milan G, Romanello V, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007. Dec;6(6):458–71. doi: 10.1016/j.cmet.2007.11.001. PubMed PMID: 18054315; eng. [DOI] [PubMed] [Google Scholar]
- 663.Mihaylova MM, Vasquez DS, Ravnskjaer K, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011. May 13;145(4):607–21. doi: 10.1016/j.cell.2011.03.043. PubMed PMID: 21565617; PubMed Central PMCID: PMC3117637. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Pfisterer SG, Mauthe M, Codogno P, et al. Ca2+/calmodulin-dependent kinase (CaMK) signaling via CaMKI and AMP-activated protein kinase contributes to the regulation of WIPI-1 at the onset of autophagy. Mol Pharmacol. 2011. Sep 6;80:1066–75. doi: 10.1124/mol.111.071761. PubMed PMID: 21896713; Eng. [DOI] [PubMed] [Google Scholar]
- 665.Rodgers JT, Lerin C, Gerhart-Hines Z, et al. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008. Jan 9;582(1):46–53. doi: 10.1016/j.febslet.2007.11.034. PubMed PMID: 18036349; PubMed Central PMCID: PMC2275806. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 666.Samari HR, Seglen PO.. Inhibition of hepatocytic autophagy by adenosine, aminoimidazole-4-carboxamide riboside, and N6-mercaptopurine riboside. Evidence for involvement of amp-activated protein kinase. J Biol Chem. 1998. Sep 11;273(37):23758–63. PubMed PMID: 9726984; eng. [DOI] [PubMed] [Google Scholar]
- 667.Sanchez AM, Csibi A, Raibon A, et al. AMPK promotes skeletal muscle autophagy through activation of Forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2011. Oct 17. doi: 10.1002/jcb.23399. PubMed PMID: 22006269; Eng. [DOI] [PubMed] [Google Scholar]
- 668.Cardenas C, Miller RA, Smith I, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Cell. 2010. Jul 23;142(2):270–83. doi: 10.1016/j.cell.2010.06.007. PubMed PMID: 20655468; PubMed Central PMCID: PMC2911450. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 669.Dalle Pezze P, Ruf S, Sonntag AG, et al. A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun. 2016. Nov 21;7:13254. doi: 10.1038/ncomms13254. PubMed PMID: 27869123; PubMed Central PMCID: PMCPMC5121333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Inoki K, Zhu T, Guan KL.. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003. Nov 26;115(5):577–90. PubMed PMID: 14651849; eng. [DOI] [PubMed] [Google Scholar]
- 671.Egan D, Kim J, Shaw RJ, et al. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011. Jun;7(6):643–4. PubMed PMID: 21460621; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 672.Egan DF, Shackelford DB, Mihaylova MM, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Science. 2011. Jan 28;331(6016):456–61. doi: 10.1126/science.1196371. PubMed PMID: 21205641; PubMed Central PMCID: PMC3030664. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 673.Kim J, Kundu M, Viollet B, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1 [Research Support, N.I.H., Extramural]. Nat Cell Biol. 2011. Feb;13(2):132–41. doi: 10.1038/ncb2152. PubMed PMID: 21258367; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Sharma A, Singh K, Mazumder S, et al. BECN1 and BIM interactions with MCL-1 determine fludarabine resistance in leukemic B cells. Cell Death Dis. 2013;4:e628. doi: 10.1038/cddis.2013.155. PubMed PMID: 23681223; PubMed Central PMCID: PMC3674362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.Zhou G, Myers R, Li Y, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001. Oct;108(8):1167–74. doi: 10.1172/JCI13505. PubMed PMID: 11602624; PubMed Central PMCID: PMC209533. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 676.Emerling BM, Viollet B, Tormos KV, et al. Compound C inhibits hypoxic activation of HIF-1 independent of AMPK. FEBS Lett. 2007. Dec 11;581(29):5727–31. doi: 10.1016/j.febslet.2007.11.038. PubMed PMID: 18036344; PubMed Central PMCID: PMC2169511. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Vucicevic L, Misirkic M, Janjetovic K, et al. Compound C induces protective autophagy in cancer cells through AMPK inhibition-independent blockade of Akt/mTOR pathway. Autophagy. 2011. Jan;7(1):40–50. PubMed PMID: 20980833; eng. [DOI] [PubMed] [Google Scholar]
- 678.Ezquerro S, Mocha F, Fruhbeck G, et al. Ghrelin Reduces TNF-alpha-Induced Human Hepatocyte Apoptosis, Autophagy, and Pyroptosis: Role in Obesity-Associated NAFLD. J Clin Endocrinol Metab. 2019. Jan 1;104(1):21–37. doi: 10.1210/jc.2018-01171. PubMed PMID: 30137403. [DOI] [PubMed] [Google Scholar]
- 679.Meley D, Bauvy C, Houben-Weerts JH, et al. AMP-activated protein kinase and the regulation of autophagic proteolysis. J Biol Chem. 2006. Nov 17;281(46):34870–9. doi: 10.1074/jbc.M605488200. PubMed PMID: 16990266; eng. [DOI] [PubMed] [Google Scholar]
- 680.Grotemeier A, Alers S, Pfisterer SG, et al. AMPK-independent induction of autophagy by cytosolic Ca2+ increase. Cell Signal. 2010. Jun;22(6):914–25. doi: 10.1016/j.cellsig.2010.01.015. PubMed PMID: 20114074; eng. [DOI] [PubMed] [Google Scholar]
- 681.Williams T, Forsberg LJ, Viollet B, et al. Basal autophagy induction without AMP-activated protein kinase under low glucose conditions. Autophagy. 2009. Nov;5(8):1155–65. PubMed PMID: 19844161; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Beck-Wodl S, Harzer K, Sturm M, et al. Homozygous TBC1 domain-containing kinase (TBCK) mutation causes a novel lysosomal storage disease - a new type of neuronal ceroid lipofuscinosis (CLN15)? Acta Neuropathol Commun. 2018. Dec 27;6(1):145. doi: 10.1186/s40478-018-0646-6. PubMed PMID: 30591081; PubMed Central PMCID: PMCPMC6307319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 683.Liu Y, Yan X, Zhou T.. TBCK influences cell proliferation, cell size and mTOR signaling pathway. PLoS One. 2013;8(8):e71349. doi: 10.1371/journal.pone.0071349. PubMed PMID: 23977024; PubMed Central PMCID: PMCPMC3747267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Li W, Han M, Guan KL.. The leucine-rich repeat protein SUR-8 enhances MAP kinase activation and forms a complex with Ras and Raf. Genes Dev. 2000. Apr 15;14(8):895–900. PubMed PMID: 10783161; PubMed Central PMCID: PMCPMC316541. [PMC free article] [PubMed] [Google Scholar]
- 685.Sieburth DS, Sun Q, Han M.. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell. 1998. Jul 10;94(1):119–30. PubMed PMID: 9674433. [DOI] [PubMed] [Google Scholar]
- 686.Xie CM, The Sun Y.. MTORC1-mediated autophagy is regulated by the FBXW7-SHOC2-RPTOR axis. Autophagy. 2019. Aug;15(8):1470–1472. doi: 10.1080/15548627.2019.1609864. PubMed PMID: 31010381; PubMed Central PMCID: PMCPMC6613887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Xie CM, Tan M, Lin XT, et al. The FBXW7-SHOC2-Raptor Axis Controls the Cross-Talks between the RAS-ERK and mTORC1 Signaling Pathways. Cell Rep. 2019. Mar 12;26(11):3037–3050 e4. doi: 10.1016/j.celrep.2019.02.052. PubMed PMID: 30865892; PubMed Central PMCID: PMCPMC6503676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 688.Shang L, Chen S, Du F, et al. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A. 2011. Mar 22;108(12):4788–93. doi: 10.1073/pnas.1100844108. PubMed PMID: 21383122; PubMed Central PMCID: PMC3064373. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Gwinn DM, Shackelford DB, Egan DF, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Mol Cell. 2008. Apr 25;30(2):214–26. doi: 10.1016/j.molcel.2008.03.003. PubMed PMID: 18439900; PubMed Central PMCID: PMC2674027. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 690.Guha P, Snyder SH.. Noncatalytic functions of IPMK are essential for activation of autophagy and liver regeneration. Autophagy. 2019. Aug;15(8):1473–1474. doi: 10.1080/15548627.2019.1615305. PubMed PMID: 31066329; PubMed Central PMCID: PMCPMC6613895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Li Z, Tian X, Ji X, et al. ULK1-ATG13 and their mitotic phospho-regulation by CDK1 connect autophagy to cell cycle. PLoS Biol. 2020. Jun;18(6):e3000288. doi: 10.1371/journal.pbio.3000288. PubMed PMID: 32516310; PubMed Central PMCID: PMCPMC7282624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Liu Z, Sin KWT, Ding H, et al. p38beta MAPK mediates ULK1-dependent induction of autophagy in skeletal muscle of tumor-bearing mice. Cell Stress. 2018. Oct 10;2(11):311–324. doi: 10.15698/cst2018.11.163. PubMed PMID: 31225455; PubMed Central PMCID: PMCPMC6551802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Alers S, Löffler AS, Paasch F, et al. Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy. 2011. Dec 1;7(12):1424–1433. PubMed PMID: 22024743; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Bach M, Larance M, James DE, et al. The serine/threonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J. 2011. Dec 1;440(2):283–91. doi: 10.1042/BJ20101894. PubMed PMID: 21819378. [DOI] [PubMed] [Google Scholar]
- 695.Egan DF, Chun MG, Vamos M, et al. Small Molecule Inhibition of the Autophagy Kinase ULK1 and Identification of ULK1 Substrates. Mol Cell. 2015. Jul 16;59(2):285–97. doi: 10.1016/j.molcel.2015.05.031. PubMed PMID: 26118643; PubMed Central PMCID: PMCPMC4530630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 696.Lazarus MB, Novotny CJ, Shokat KM.. Structure of the human autophagy initiating kinase ULK1 in complex with potent inhibitors. ACS Chem Biol. 2015. Jan 16;10(1):257–61. doi: 10.1021/cb500835z. PubMed PMID: 25551253; PubMed Central PMCID: PMCPMC4301081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Dorsey FC, Rose KL, Coenen S, et al. Mapping the phosphorylation sites of Ulk1. J Proteome Res. 2009. Nov;8(11):5253–63. doi: 10.1021/pr900583m. PubMed PMID: 19807128. [DOI] [PubMed] [Google Scholar]
- 698.Ganley IG, Lam du H, Wang J, et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009. May 1;284(18):12297–305. doi: 10.1074/jbc.M900573200. PubMed PMID: 19258318; PubMed Central PMCID: PMC2673298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 699.Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008. May 5;181(3):497–510. doi:jcb.200712064 [pii] doi: 10.1083/jcb.200712064. PubMed PMID: 18443221; PubMed Central PMCID: PMC2364687. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 700.Hosokawa N, Hara T, Kaizuka T, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009. Apr;20(7):1981–91. doi: 10.1091/mbc.E08-12-1248. PubMed PMID: 19211835; PubMed Central PMCID: PMC2663915. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 701.Jung CH, Jun CB, Ro S-H, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009. Apr;20(7):1992–2003. doi: 10.1091/mbc.E08-12-1249. PubMed PMID: 19225151; PubMed Central PMCID: PMC2663920. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Park JM, Jung CH, Seo M, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12(3):547–64. doi: 10.1080/15548627.2016.1140293. PubMed PMID: 27046250; PubMed Central PMCID: PMCPMC4835982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Di Bartolomeo S, Corazzari M, Nazio F, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010. Oct 4;191(1):155–68. doi: 10.1083/jcb.201002100. PubMed PMID: 20921139; PubMed Central PMCID: PMC2953445. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Ma X, Zhang S, He L, et al. MTORC1-mediated NRBF2 phosphorylation functions as a switch for the class III PtdIns3K and autophagy. Autophagy. 2017. Mar 4;13(3):592–607. doi: 10.1080/15548627.2016.1269988. PubMed PMID: 28059666; PubMed Central PMCID: PMCPMC5361594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Russell RC, Tian Y, Yuan H, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013. Jul;15(7):741–50. doi: 10.1038/ncb2757. PubMed PMID: 23685627; PubMed Central PMCID: PMC3885611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 706.Park JM, Seo M, Jung CH, et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy. 2018;14(4):584–597. doi: 10.1080/15548627.2017.1422851. PubMed PMID: 29313410; PubMed Central PMCID: PMCPMC5959323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Zhou C, Ma K, Gao R, et al. Regulation of mATG9 trafficking by Src- and ULK1-mediated phosphorylation in basal and starvation-induced autophagy. Cell Res. 2017. Feb;27(2):184–201. doi: 10.1038/cr.2016.146. PubMed PMID: 27934868; PubMed Central PMCID: PMCPMC5339848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Pengo N, Agrotis A, Prak K, et al. A reversible phospho-switch mediated by ULK1 regulates the activity of autophagy protease ATG4B. Nat Commun. 2017. Aug 18;8(1):294. doi: 10.1038/s41467-017-00303-2. PubMed PMID: 28821708; PubMed Central PMCID: PMCPMC5562857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.Alsaadi RM, Losier TT, Tian W, et al. ULK1-mediated phosphorylation of ATG16L1 promotes xenophagy, but destabilizes the ATG16L1 Crohn’s mutant. EMBO Rep. 2019. Jul;20(7):e46885. doi: 10.15252/embr.201846885. PubMed PMID: 31267703; PubMed Central PMCID: PMCPMC6607016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 710.Löffler AS, Alers S, Dieterle AM, et al. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy. 2011. Jul;7(7):696–706. PubMed PMID: 21460634; eng. [DOI] [PubMed] [Google Scholar]
- 711.Dunlop EA, Hunt DK, Acosta-Jaquez HA, et al. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy. 2011. Jul;7(7):737–47. PubMed PMID: 21460630; PubMed Central PMCID: PMC3149699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Dunlop EA, Seifan S, Claessens T, et al. FLCN, a novel autophagy component, interacts with GABARAP and is regulated by ULK1 phosphorylation. Autophagy. 2014. Oct 1;10(10):1749–60. doi: 10.4161/auto.29640. PubMed PMID: 25126726; PubMed Central PMCID: PMC4198360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 713.Li TY, Sun Y, Liang Y, et al. ULK1/2 Constitute a Bifurcate Node Controlling Glucose Metabolic Fluxes in Addition to Autophagy. Mol Cell. 2016. May 5;62(3):359–370. doi: 10.1016/j.molcel.2016.04.009. PubMed PMID: 27153534. [DOI] [PubMed] [Google Scholar]
- 714.Lim J, Lachenmayer ML, Wu S, et al. Proteotoxic stress induces phosphorylation of p62/SQSTM1 by ULK1 to regulate selective autophagic clearance of protein aggregates. PLoS Genet. 2015;11(2):e1004987. doi: 10.1371/journal.pgen.1004987. PubMed PMID: 25723488; PubMed Central PMCID: PMCPMC4344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.Sellier C, Campanari ML, Julie Corbier C, et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016. Jun 15;35(12):1276–97. doi: 10.15252/embj.201593350. PubMed PMID: 27103069; PubMed Central PMCID: PMCPMC4910533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Tang HW, Wang YB, Wang SL, et al. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J. 2011. Feb 16;30(4):636–51. doi: 10.1038/emboj.2010.338. PubMed PMID: 21169990; PubMed Central PMCID: PMC3041946. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Xu J, Fotouhi M, McPherson PS.. Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep. 2015. Jun;16(6):709–18. doi: 10.15252/embr.201440006. PubMed PMID: 25925668; PubMed Central PMCID: PMCPMC4467855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Jung CH, Seo M, Otto NM, et al. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy. 2011. Oct 1;7(10):1212–21. PubMed PMID: 21795849; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Gan W, Zhang C, Siu KY, et al. ULK1 phosphorylates Sec23A and mediates autophagy-induced inhibition of ER-to-Golgi traffic. BMC Cell Biol. 2017. May 10;18(1):22. doi: 10.1186/s12860-017-0138-8. PubMed PMID: 28486929; PubMed Central PMCID: PMCPMC5424413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 720.Joo JH, Wang B, Frankel E, et al. The noncanonical role of ULK/ATG1 in ER-to-Golgi trafficking is essential for cellular homeostasis. Mol Cell. 2016. May 19;62(4):491–506. doi: 10.1016/j.molcel.2016.04.020. PubMed PMID: 27203176; PubMed Central PMCID: PMCPMC4993601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 721.Konno H, Konno K, Barber GN.. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013. Oct 24;155(3):688–98. doi: 10.1016/j.cell.2013.09.049. PubMed PMID: 24119841; PubMed Central PMCID: PMCPMC3881181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Hwang SH, Bang S, Kang KS, et al. ULK1 negatively regulates Wnt signaling by phosphorylating Dishevelled. Biochem Biophys Res Commun. 2019. Jan 1;508(1):308–313. doi: 10.1016/j.bbrc.2018.11.139. PubMed PMID: 30497781. [DOI] [PubMed] [Google Scholar]
- 723.Jeong YT, Simoneschi D, Keegan S, et al. The ULK1-FBXW5-SEC23B nexus controls autophagy. eLife. 2018. Dec 31;7. doi: 10.7554/eLife.42253. PubMed PMID: 30596474; PubMed Central PMCID: PMCPMC6351106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 724.Li R, Yuan F, Fu W, et al. Serine/threonine kinase Unc-51-like kinase-1 (Ulk1) phosphorylates the co-chaperone cell division cycle protein 37 (Cdc37) and thereby disrupts the stability of Cdc37 client proteins. J Biol Chem. 2017. Feb 17;292(7):2830–2841. doi: 10.1074/jbc.M116.762443. PubMed PMID: 28073914; PubMed Central PMCID: PMCPMC5314178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Rajesh S, Bago R, Odintsova E, et al. Binding to syntenin-1 protein defines a new mode of ubiquitin-based interactions regulated by phosphorylation. J Biol Chem. 2011. Nov 11;286(45):39606–14. doi: 10.1074/jbc.M111.262402. PubMed PMID: 21949238; PubMed Central PMCID: PMCPMC3234783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 726.Kim K, Park SG, Park BC, et al. Serine 389 phosphorylation of 3-phosphoinositide-dependent kinase 1 by UNC-51-like kinase 1 affects its ability to regulate Akt and p70 S6kinase. BMB Rep. 2020. Apr 22. PubMed PMID: 32317083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 727.Saleiro D, Mehrotra S, Kroczynska B, et al. Central role of ULK1 in type I interferon signaling. Cell Rep. 2015. Apr 28;11(4):605–17. doi: 10.1016/j.celrep.2015.03.056. PubMed PMID: 25892232; PubMed Central PMCID: PMCPMC4477687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Shibata S, Ishizawa K, Wang Q, et al. ULK1 Phosphorylates and Regulates Mineralocorticoid Receptor. Cell Rep. 2018. Jul 17;24(3):569–576. doi: 10.1016/j.celrep.2018.06.072. PubMed PMID: 30021155. [DOI] [PubMed] [Google Scholar]
- 729.Wang B, Maxwell BA, Joo JH, et al. ULK1 and ULK2 regulate stress granule disassembly through phosphorylation and activation of VCP/p97. Mol Cell. 2019. May 16;74(4):742–757 e8. doi: 10.1016/j.molcel.2019.03.027. PubMed PMID: 30979586; PubMed Central PMCID: PMCPMC6859904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Yuan F, Jin X, Li D, et al. ULK1 phosphorylates Mad1 to regulate spindle assembly checkpoint. Nucleic Acids Res. 2019. Sep 5;47(15):8096–8110. doi: 10.1093/nar/gkz602. PubMed PMID: 31291454; PubMed Central PMCID: PMCPMC6736072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 731.Zhao P, Wong KI, Sun X, et al. TBK1 at the Crossroads of Inflammation and Energy Homeostasis in Adipose Tissue. Cell. 2018. Feb 8;172(4):731–743 e12. doi: 10.1016/j.cell.2018.01.007. PubMed PMID: 29425491; PubMed Central PMCID: PMCPMC5808582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Tyra LK, Nandi N, Tracy C, et al. Yorkie growth-promoting activity is limited by Atg1-mediated phosphorylation. Dev Cell. 2020. Mar 9;52(5):605–616 e7. doi: 10.1016/j.devcel.2020.01.011. PubMed PMID: 32032548; PubMed Central PMCID: PMCPMC7105283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Mao L, Zhan YY, Wu B, et al. ULK1 phosphorylates Exo70 to suppress breast cancer metastasis. Nat Commun. 2020. Jan 8;11(1):117. doi: 10.1038/s41467-019-13923-7. PubMed PMID: 31913283; PubMed Central PMCID: PMCPMC6949295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 734.Wu W, Wang X, Berleth N, et al. The autophagy-initiating kinase ULK1 controls RIPK1-mediated cell death. Cell Rep. 2020. Apr 21;31(3):107547. doi: 10.1016/j.celrep.2020.107547. PubMed PMID: 32320653. [DOI] [PubMed] [Google Scholar]
- 735.Wu W, Tian W, Hu Z, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014. May;15(5):566–75. doi: 10.1002/embr.201438501. PubMed PMID: 24671035; PubMed Central PMCID: PMCPMC4210082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Brunn GJ, Hudson CC, Sekulic A, et al. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997. Jul 4;277(5322):99–101. PubMed PMID: 9204908; eng. [DOI] [PubMed] [Google Scholar]
- 737.Erlich S, Alexandrovich A, Shohami E, et al. Rapamycin is a neuroprotective treatment for traumatic brain injury. Neurobiol Dis. 2007. Apr;26(1):86–93. PubMed PMID: 17270455; eng. [DOI] [PubMed] [Google Scholar]
- 738.Lavieu G, Scarlatti F, Sala G, et al. Regulation of autophagy by sphingosine kinase 1 and its role in cell survival during nutrient starvation. J Biol Chem. 2006. Mar 31;281(13):8518–27. PubMed PMID: 16415355; eng. [DOI] [PubMed] [Google Scholar]
- 739.Yip CK, Murata K, Walz T, et al. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol Cell. 2010. Jun 11;38(5):768–74. doi: 10.1016/j.molcel.2010.05.017. PubMed PMID: 20542007; PubMed Central PMCID: PMC2887672. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 740.Roux PP, Shahbazian D, Vu H, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007. May 11;282(19):14056–64. doi: 10.1074/jbc.M700906200. PubMed PMID: 17360704; PubMed Central PMCID: PMCPMC3618456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Nazio F, Strappazzon F, Antonioli M, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013. Apr;15(4):406–16. doi: 10.1038/ncb2708. PubMed PMID: 23524951. [DOI] [PubMed] [Google Scholar]
- 742.Cheong H, Nair U, Geng J, et al. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008. Feb;19(2):668–81. doi: 10.1091/mbc.E07-08-0826. PubMed PMID: 18077553; PubMed Central PMCID: PMC2230592. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 743.Kabeya Y, Kamada Y, Baba M, et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005. May;16(5):2544–53. doi: 10.1091/mbc.E04-08-0669. PubMed PMID: 15743910; PubMed Central PMCID: PMC1087256. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 744.Kamada Y, Funakoshi T, Shintani T, et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000. Sep 18;150(6):1507–13. PubMed PMID: 10995454; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 745.Scott SV, Nice DC, III, Nau JJ, et al. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem. 2000. Aug 18;275(33):25840–9. doi: 10.1074/jbc.M002813200. PubMed PMID: 10837477; eng. [DOI] [PubMed] [Google Scholar]
- 746.Miller-Fleming L, Cheong H, Antas P, et al. Detection of Saccharomyces cerevisiae Atg13 by western blot. Autophagy. 2014. Mar;10(3):514–7. doi: 10.4161/auto.27707. PubMed PMID: 24430166; PubMed Central PMCID: PMC4077888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 747.Mao K, Wang K, Zhao M, et al. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol. 2011. May 16;193(4):755–67. doi: 10.1083/jcb.201102092. PubMed PMID: 21576396; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 748.Yeh YY, Wrasman K, Herman PK.. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics. 2010. Jul;185(3):871–82. doi: 10.1534/genetics.110.116566. PubMed PMID: 20439775; PubMed Central PMCID: PMC2907206. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Kim M, Park HL, Park HW, et al. Drosophila Fip200 is an essential regulator of autophagy that attenuates both growth and aging. Autophagy. 2013. Aug;9(8):1201–13. doi: 10.4161/auto.24811. PubMed PMID: 23819996; PubMed Central PMCID: PMC3748192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Nagy P, Karpati M, Varga A, et al. Atg17/FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in Drosophila. Autophagy. 2014. Mar;10(3):453–67. doi: 10.4161/auto.27442. PubMed PMID: 24419107; PubMed Central PMCID: PMC4077884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Shang L, AMP Wang X.. K and mTOR coordinate the regulation of Ulk1 and mammalian autophagy initiation. Autophagy. 2011. Aug;7(8):924–6. PubMed PMID: 21521945. [DOI] [PubMed] [Google Scholar]
- 752.Singh K, Matsuyama S, Drazba JA, et al. Autophagy-dependent senescence in response to DNA damage and chronic apoptotic stress. Autophagy. 2012;8(2):236-51. doi: 10.4161/auto.8.2.18600. PMID: 22240589; PMCID: PMC3336077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 753.Karanasios E, Stapleton E, Manifava M, et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci. 2013. Nov 15;126(Pt 22):5224–38. doi: 10.1242/jcs.132415. PubMed PMID: 24013547. [DOI] [PubMed] [Google Scholar]
- 754.Karanasios E, Walker SA, Okkenhaug H, et al. Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles. Nat Commun. 2016. Aug 11;7:12420. doi: 10.1038/ncomms12420. PubMed PMID: 27510922; PubMed Central PMCID: PMCPMC4987534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Follo C, Barbone D, Richards WG, et al. Autophagy initiation correlates with the autophagic flux in 3D models of mesothelioma and with patient outcome. Autophagy. 2016. Jul 2;12(7):1180–94. . PubMed PMID: 27097020; PubMed Central PMCID: PMCPMC4990992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 756.Follo C, Cheng Y, Richards WG, et al. Inhibition of autophagy initiation potentiates chemosensitivity in mesothelioma. Mol Carcinog. 2018. Mar;57(3):319–332. doi: 10.1002/mc.22757. PubMed PMID: 29073722. [DOI] [PubMed] [Google Scholar]
- 757.Li W, Zou W, Yang Y, et al. Autophagy genes function sequentially to promote apoptotic cell corpse degradation in the engulfing cell. J Cell Biol. 2012. Apr 2;197(1):27–35. doi: 10.1083/jcb.201111053. PubMed PMID: 22451698; PubMed Central PMCID: PMC3317810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Fazeli G, Trinkwalder M, Irmisch L, et al. C. elegans midbodies are released, phagocytosed and undergo LC3-dependent degradation independent of macroautophagy. J Cell Sci. 2016. Oct 15;129(20):3721–3731. doi: 10.1242/jcs.190223. PubMed PMID: 27562069; PubMed Central PMCID: PMCPMC5087666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 759.Ruck A, Attonito J, Garces KT, et al. The Atg6/Vps30/Beclin 1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy. 2011. Apr;7(4):386–400. PubMed PMID: 21183797; PubMed Central PMCID: PMC3108013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 760.De Faveri F, Chvanov M, Voronina S, et al. LAP-like non-canonical autophagy and evolution of endocytic vacuoles in pancreatic acinar cells. Autophagy. 2020. Jul;16(7):1314–1331. doi: 10.1080/15548627.2019.1679514. PubMed PMID: 31651224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 761.Cunha LD, Yang M, Carter R, et al. LC3-Associated Phagocytosis in Myeloid Cells Promotes Tumor Immune Tolerance. Cell. 2018. Oct 4;175(2):429–441 e16. doi: 10.1016/j.cell.2018.08.061. PubMed PMID: 30245008; PubMed Central PMCID: PMCPMC6201245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 762.Dohmen M, Krieg S, Agalaridis G, et al. AMPK-dependent activation of the Cyclin Y/CDK16 complex controls autophagy. Nat Commun. 2020. Feb 25;11(1):1032. doi: 10.1038/s41467-020-14812-0. PubMed PMID: 32098961; PubMed Central PMCID: PMCPMC7042329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 763.Cap M, Stepanek L, Harant K, et al. Cell differentiation within a yeast colony: metabolic and regulatory parallels with a tumor-affected organism. Mol Cell. 2012. May 25;46(4):436–48. doi: 10.1016/j.molcel.2012.04.001. PubMed PMID: 22560924. [DOI] [PubMed] [Google Scholar]
- 764.Djavaheri-Mergny M, Amelotti M, Mathieu J, et al. NF-[kappa]B activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem. 2006. Oct 13;281(41):30373–82. PubMed PMID: 16857678; eng. [DOI] [PubMed] [Google Scholar]
- 765.Liu Z, Lenardo MJ.. Reactive oxygen species regulate autophagy through redox-sensitive proteases. Dev Cell. 2007. Apr;12(4):484–5. PubMed PMID: 17419989; eng. [DOI] [PubMed] [Google Scholar]
- 766.Scarlatti F, Bauvy C, Ventruti A, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004. Apr 30;279(18):18384–91. PubMed PMID: 14970205; eng. [DOI] [PubMed] [Google Scholar]
- 767.Scherz-Shouval R, Shvets E, Fass E, et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007. Apr 4;26(7):1749–60. PubMed PMID: 17347651; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 768.Pereira O, Teixeira A, Sampaio-Marques B, et al. Signalling mechanisms that regulate metabolic profile and autophagy of acute myeloid leukaemia cells. J Cell Mol Med. 2018. Oct;22(10):4807–4817. doi: 10.1111/jcmm.13737. PubMed PMID: 30117681; PubMed Central PMCID: PMCPMC6156238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Sujobert P, Poulain L, Paubelle E, et al. Co-activation of AMPK and mTORC1 induces cytotoxicity in acute myeloid leukemia. Cell Rep. 2015. Jun 9;11(9):1446–57. doi: 10.1016/j.celrep.2015.04.063. PubMed PMID: 26004183. [DOI] [PubMed] [Google Scholar]
- 770.Datan E, Shirazian A, Benjamin S, et al. mTOR/p70S6K signaling distinguishes routine, maintenance-level autophagy from autophagic cell death during influenza A infection. Virology. 2014. Mar;452-453:175–190. doi: 10.1016/j.virol.2014.01.008. PubMed PMID: 24606695; PubMed Central PMCID: PMCPMC4005847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Zeng X, Kinsella TJ.. Mammalian target of rapamycin and S6 kinase 1 positively regulate 6-thioguanine-induced autophagy. Cancer Res. 2008. Apr 1;68(7):2384–90. doi: 10.1158/0008-5472.CAN-07-6163. PubMed PMID: 18381446; eng. [DOI] [PubMed] [Google Scholar]
- 772.Lee JW, Nam H, Kim LE, et al. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy. 2019. May;15(5):753–770. doi: 10.1080/15548627.2018.1556946. PubMed PMID: 30523761; PubMed Central PMCID: PMCPMC6526818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Losier TT, Russell RC.. Bacterial outer membrane vesicles trigger pre-activation of a xenophagic response via AMPK. Autophagy. 2019. Aug;15(8):1489–1491. doi: 10.1080/15548627.2019.1618640. PubMed PMID: 31107135; PubMed Central PMCID: PMCPMC6613880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 774.Wang RC, Wei Y, An Z, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012. Nov 16;338(6109):956–9. doi: 10.1126/science.1225967. PubMed PMID: 23112296; PubMed Central PMCID: PMC3507442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 775.Wei Y, Zou Z, Becker N, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell. 2013. Sep 12;154(6):1269–84. doi: 10.1016/j.cell.2013.08.015. PubMed PMID: 24034250; PubMed Central PMCID: PMC3917713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Yasugi M, Takigawa N, Ochi N, et al. Everolimus prolonged survival in transgenic mice with EGFR-driven lung tumors. Exp Cell Res. 2014. Aug 15;326(2):201–9. doi: 10.1016/j.yexcr.2014.04.012. PubMed PMID: 24768699. [DOI] [PubMed] [Google Scholar]
- 777.Castets P, Lin S, Rion N, et al. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013. May 7;17(5):731–44. doi: 10.1016/j.cmet.2013.03.015. PubMed PMID: 23602450. [DOI] [PubMed] [Google Scholar]
- 778.Castets P, Ruegg MA.. MTORC1 determines autophagy through ULK1 regulation in skeletal muscle. Autophagy. 2013. Sep;9(9):1435–7. doi: 10.4161/auto.25722. PubMed PMID: 23896646. [DOI] [PubMed] [Google Scholar]
- 779.Wang Z, Zhou L, Zheng X, et al. Autophagy protects against PI3K/Akt/mTOR-mediated apoptosis of spinal cord neurons after mechanical injury. Neurosci Lett. 2017. Aug 24;656:158–164. doi: 10.1016/j.neulet.2017.07.036. PubMed PMID: 28739349. [DOI] [PubMed] [Google Scholar]
- 780.Bernard M, Dieude M, Yang B, et al. Autophagy fosters myofibroblast differentiation through MTORC2 activation and downstream upregulation of CTGF. Autophagy. 2014. Dec 12:0. doi: 10.4161/15548627.2014.981786. PubMed PMID: 25495560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.Yu L, McPhee CK, Zheng L, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010. Jun 17;465(7300):942–6. doi: 10.1038/nature09076. PubMed PMID: 20526321; PubMed Central PMCID: PMC2920749. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Narita M, Young AR, Arakawa S, et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science. 2011. May 20;332(6032):966–70. doi: 10.1126/science.1205407. PubMed PMID: 21512002; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 783.Holla S, Kurowska-Stolarska M, Bayry J, et al. Selective inhibition of IFNG-induced autophagy by Mir155- and Mir31-responsive WNT5A and SHH signaling. Autophagy. 2014. Feb;10(2):311–30. doi: 10.4161/auto.27225. PubMed PMID: 24343269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Joshi A, Iyengar R, Joo JH, et al. Nuclear ULK1 promotes cell death in response to oxidative stress through PARP1. Cell Death Differ. 2016. Feb;23(2):216–30. doi: 10.1038/cdd.2015.88. PubMed PMID: 26138443; PubMed Central PMCID: PMCPMC4716304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 785.Hamamichi S, Rivas RN, Knight AL, et al. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson’s disease model. Proc Natl Acad Sci U S A. 2008. Jan 15;105(2):728–33. doi: 10.1073/pnas.0711018105. PMCID:PubMed PMIDPMCPMC2206604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 786.Okazaki N, Yan J, Yuasa S, et al. Interaction of the Unc-51-like kinase and microtubule-associated protein light chain 3 related proteins in the brain: possible role of vesicular transport in axonal elongation. Brain Res Mol Brain Res. 2000. Dec 28;85(1–2):1–12. PubMed PMID: 11146101; eng. [DOI] [PubMed] [Google Scholar]
- 787.Tomoda T, Kim JH, Zhan C, et al. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 2004. Mar 1;18(5):541–58. doi: 10.1101/gad.1151204:15014045; PubMed Central PMCID: PMC374236. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 788.Loh SH, Francescut L, Lingor P, et al. Identification of new kinase clusters required for neurite outgrowth and retraction by a loss-of-function RNA interference screen. Cell Death Differ. 2008. Feb;15(2):283–98. doi: 10.1038/sj.cdd.4402258. PubMed PMID: 18007665; eng. [DOI] [PubMed] [Google Scholar]
- 789.Mochizuki H, Toda H, Ando M, et al. Unc-51/ATG1 controls axonal and dendritic development via kinesin-mediated vesicle transport in the Drosophila brain. PLoS One. 2011;6(5):e19632. doi: 10.1371/journal.pone.0019632. PubMed PMID: 21589871; PubMed Central PMCID: PMC3093397. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Wairkar YP, Toda H, Mochizuki H, et al. Unc-51 controls active zone density and protein composition by downregulating ERK signaling. J Neurosci. 2009. Jan 14;29(2):517–28. doi: 10.1523/JNEUROSCI.3848-08.2009. PubMed PMID: 19144852; PubMed Central PMCID: PMC2741695. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 791.Zhou X, Babu JR, da Silva S, et al. Unc-51-like kinase 1/2-mediated endocytic processes regulate filopodia extension and branching of sensory axons. Proc Natl Acad Sci U S A. 2007. Apr 3;104(14):5842–7. doi: 10.1073/pnas.0701402104. PubMed PMID: 17389358; PubMed Central PMCID: PMC1851579. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 792.Ribas VT, Schnepf B, Challagundla M, et al. Early and sustained activation of autophagy in degenerating axons after spinal cord injury. Brain Pathol. 2015. Mar;25(2):157–70. doi: 10.1111/bpa.12170. PubMed PMID: 25040536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.Schu PV, Takegawa K, Fry MJ, et al. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993. Apr 2;260(5104):88–91. doi: 10.1126/science.8385367. PubMed PMID: 8385367. [DOI] [PubMed] [Google Scholar]
- 794.Volinia S, Dhand R, Vanhaesebroeck B, et al. A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 1995. Jul 17;14(14):3339–48. PubMed PMID: 7628435; PubMed Central PMCID: PMCPMC394401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 795.Itakura E, Kishi C, Inoue K, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008. Dec;19(12):5360–72. doi: 10.1091/mbc.E08-01-0080. PubMed PMID: 18843052; PubMed Central PMCID: PMC2592660. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 796.Kihara A, Noda T, Ishihara N, et al. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001. Feb 5;152(3):519–30. PubMed PMID: 11157979; PubMed Central PMCID: PMC2196002. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Liang C, Feng P, Ku B, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006. Jul;8(7):688–99. doi: ncb1426 [pii] doi: 10.1038/ncb1426. PubMed PMID: 16799551; eng. [DOI] [PubMed] [Google Scholar]
- 798.Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009. Apr;11(4):385–96. doi: 10.1038/ncb1846. PubMed PMID: 19270696; eng. [DOI] [PubMed] [Google Scholar]
- 799.Sun Q, Fan W, Chen K, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase [Research Support, Non-U.S. Gov’t]. Proc Natl Acad Sci U S A. 2008. Dec 9;105(49):19211–6. doi: 10.1073/pnas.0810452105. PubMed PMID: 19050071; PubMed Central PMCID: PMC2592986. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Rostislavleva K, Soler N, Ohashi Y, et al. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science. 2015. Oct 9;350(6257):aac7365. doi: 10.1126/science.aac7365. PubMed PMID: 26450213; PubMed Central PMCID: PMCPMC4601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 801.Martinez J, Malireddi RK, Lu Q, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat Cell Biol. 2015. Jul;17(7):893–906. doi: 10.1038/ncb3192. PubMed PMID: 26098576; PubMed Central PMCID: PMCPMC4612372. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 802.Thoreen CC, Kang SA, Chang JW, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009. Mar 20;284(12):8023–32. doi: 10.1074/jbc.M900301200. PubMed PMID: 19150980; PubMed Central PMCID: PMC2658096. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 803.Yan Y, Flinn RJ, Wu H, et al. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J. 2009. Feb 1;417(3):747–55. doi: 10.1042/BJ20081865. PubMed PMID: 18957027; PubMed Central PMCID: PMC2652830. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 804.Ohashi Y, Tremel S, Williams RL.. VPS34 complexes from a structural perspective. J Lipid Res. 2019. Feb;60(2):229–241. doi: 10.1194/jlr.R089490. PubMed PMID: 30397185; PubMed Central PMCID: PMCPMC6358306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 805.Young ARJ, Chan EYW, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006. Sep 15;119(Pt 18):3888–900. PubMed PMID: 16940348; eng. [DOI] [PubMed] [Google Scholar]
- 806.Yamaguchi J, Suzuki C, Nanao T, et al. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy. 2018;14(5):764–777. doi: 10.1080/15548627.2017.1314897. PubMed PMID: 28513333; PubMed Central PMCID: PMCPMC6070006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 807.Reggiori F, Shintani T, Nair U, et al. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005. Jul;1(2):101–9. PubMed PMID: 16874040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Mari M, Griffith J, Rieter E, et al. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010. Sep 20;190(6):1005–22. doi: 10.1083/jcb.200912089. PubMed PMID: 20855505; PubMed Central PMCID: PMC3101592. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 809.Reggiori F, Tucker KA, Stromhaug PE, et al. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev cell 2004. Jan;6(1):79–90. PubMed PMID: 14723849. [DOI] [PubMed] [Google Scholar]
- 810.Barve G, Sridhar S, Aher A, et al. Septins are involved at the early stages of macroautophagy in S. cerevisiae. J Cell Sci. 2018. Feb 22;131(4). doi: 10.1242/jcs.209098. PubMed PMID: 29361537; PubMed Central PMCID: PMCPMC5868950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.Imai K, Hao F, Fujita N, et al. Atg9A trafficking through the recycling endosomes is required for autophagosome formation. J Cell Sci. 2016. Oct 15;129(20):3781–3791. doi: 10.1242/jcs.196196. PubMed PMID: 27587839. [DOI] [PubMed] [Google Scholar]
- 812.Corcelle-Termeau E, Vindelov SD, Hamalisto S, et al. Excess sphingomyelin disturbs ATG9A trafficking and autophagosome closure. Autophagy. 2016. May 3;12(5):833–49. doi: 10.1080/15548627.2016.1159378. PubMed PMID: 27070082; PubMed Central PMCID: PMCPMC4854555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Mizushima N, Kuma A, Kobayashi Y, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003. May 1;116(Pt 9):1679–88. PubMed PMID: 12665549; eng. [DOI] [PubMed] [Google Scholar]
- 814.Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001. Feb 19;152(4):657–68. PubMed PMID: 11266458; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.Mikhaylova O, Stratton Y, Hall D, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer cell. 2012. Apr 17;21(4):532–46. doi: 10.1016/j.ccr.2012.02.019. PubMed PMID: 22516261; PubMed Central PMCID: PMC3331999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 816.Thompson AR, Doelling JH, Suttangkakul A, et al. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005. Aug;138(4):2097–110. doi: 10.1104/pp.105.060673. PubMed PMID: 16040659; PubMed Central PMCID: PMC1183398. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Yousefi S, Perozzo R, Schmid I, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006. Oct;8(10):1124–32. PubMed PMID: 16998475; eng. [DOI] [PubMed] [Google Scholar]
- 818.Maskey D, Yousefi S, Schmid I, et al. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. PubMed PMID: 23945651; PubMed Central PMCID: PMC3753548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 819.Haller M, Hock AK, Giampazolias E, et al. Ubiquitination and proteasomal degradation of ATG12 regulates its proapoptotic activity. Autophagy. 2014;10(12):2269–78. doi: 10.4161/15548627.2014.981914. PubMed PMID: 25629932; PubMed Central PMCID: PMCPMC4502749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 820.Li SP, He JD, Wang Z, et al. miR-30b inhibits autophagy to alleviate hepatic ischemia-reperfusion injury via decreasing the Atg12-Atg5 conjugate. World J Gastroenterol. 2016. May 14;22(18):4501–14. doi: 10.3748/wjg.v22.i18.4501. PubMed PMID: 27182160; PubMed Central PMCID: PMCPMC4858632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Lin TY, Chan HH, Chen SH, et al. BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy. 2019. Oct 15:1–18. doi: 10.1080/15548627.2019.1671643. PubMed PMID: 31612776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 822.Bec N, Bonhoure A, Henry L, et al. Proteasome 19S RP and translation preinitiation complexes are secreted within exosomes upon serum starvation. Traffic. 2019. Jul;20(7):516–536. doi: 10.1111/tra.12653 PubMed PMID: 31042005. [DOI] [PubMed] [Google Scholar]
- 823.Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009. Apr;11(4):468–76. doi: 10.1038/ncb1854. PubMed PMID: 19270693; PubMed Central PMCID: PMC2664389. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 824.Fan W, Nassiri A, Zhong Q.. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci U S A. 2011. May 10;108(19):7769–74. doi: 10.1073/pnas.1016472108. PubMed PMID: 21518905; PubMed Central PMCID: PMC3093500. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010. Aug 23;190(4):511–21. doi: 10.1083/jcb.200911141. PubMed PMID: 20713597; PubMed Central PMCID: PMC2928018. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Ravikumar B, Moreau K, Jahreiss L, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010. Aug;12(8):747–57. doi: 10.1038/ncb2078. PubMed PMID: 20639872; PubMed Central PMCID: PMC2923063. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 827.Tian W, Alsaadi R, Guo Z, et al. An antibody for analysis of autophagy induction. Nat Methods. 2020. Feb;17(2):232–239. doi: 10.1038/s41592-019-0661-y. PubMed PMID: 31768061. [DOI] [PubMed] [Google Scholar]
- 828.Scrivo A, Codogno P, Bomont P.. Gigaxonin E3 ligase governs ATG16L1 turnover to control autophagosome production. Nat Commun. 2019. Feb 15;10(1):780. doi: 10.1038/s41467-019-08331-w. PubMed PMID: 30770803; PubMed Central PMCID: PMCPMC6377711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 829.Lassen KG, Kuballa P, Conway KL, et al. Atg16L1 T300A variant decreases selective autophagy resulting in altered cytokine signaling and decreased antibacterial defense. Proc Natl Acad Sci U S A. 2014. May 27;111(21):7741–6. doi: 10.1073/pnas.1407001111. PubMed PMID: 24821797; PubMed Central PMCID: PMCPMC4040621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Murthy A, Li Y, Peng I, et al. A Crohn’s disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014. Feb 27;506(7489):456–62. doi: 10.1038/nature13044. PubMed PMID: 24553140. [DOI] [PubMed] [Google Scholar]
- 831.Guan J, Stromhaug PE, George MD, et al. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell. 2001. Dec;12(12):3821–38. PubMed PMID: 11739783; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832.Barth H, Meiling-Wesse K, Epple UD, et al. Autophagy and the cytoplasm to vacuole targeting pathway both require Aut10p. FEBS Lett. 2001. Nov 9;508(1):23–8. PubMed PMID: 11707261; eng. [DOI] [PubMed] [Google Scholar]
- 833.Proikas-Cezanne T, Waddell S, Gaugel A, et al. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004. Dec 16;23(58):9314–25. doi: 10.1038/sj.onc.1208331. PubMed PMID: 15602573; eng. [DOI] [PubMed] [Google Scholar]
- 834.Monastyrska I, Klionsky DJ.. Autophagy in organelle homeostasis: peroxisome turnover. Mol Aspects Med. 2006. Oct-Dec;27(5–6):483–94. PubMed PMID: 16973210; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 835.Nair U, Klionsky DJ.. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005. Dec 23;280(51):41785–8. PubMed PMID: 16230342; eng. [DOI] [PubMed] [Google Scholar]
- 836.Tallóczy Z, Virgin HW, IV, Levine B.. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy. 2006. Jan-Mar;2(1):24–9. PubMed PMID: 16874088. [DOI] [PubMed] [Google Scholar]
- 837.Polson HE, de Lartigue J, Rigden DJ, et al. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy. 2010. May 16;6(4):506–522. PubMed PMID: 20505359; Eng. [DOI] [PubMed] [Google Scholar]
- 838.Proikas-Cezanne T, Ruckerbauer S, Stierhof YD, et al. Human WIPI-1 puncta-formation: A novel assay to assess mammalian autophagy. FEBS Lett. 2007. Jun 27;581:3396–404. PubMed PMID: 17618624; Eng. [DOI] [PubMed] [Google Scholar]
- 839.Scacioc A, Schmidt C, Hofmann T, et al. Structure based biophysical characterization of the PROPPIN Atg18 shows Atg18 oligomerization upon membrane binding. Sci Rep. 2017. Oct 25;7(1):14008. doi: 10.1038/s41598-017-14337-5. PubMed PMID: 29070817; PubMed Central PMCID: PMCPMC5656675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Gopaldass N, Fauvet B, Lashuel H, et al. Membrane scission driven by the PROPPIN Atg18. EMBO J. 2017. Nov 15;36(22):3274–3291. doi: 10.15252/embj.201796859. PubMed PMID: 29030482; PubMed Central PMCID: PMCPMC5686546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 841.Itakura E, Mizushima N.. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010. Aug;6(6):764–76. PubMed PMID: 20639694; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.Mauthe M, Jacob A, Freiberger S, et al. Resveratrol-mediated autophagy requires WIPI-1 regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy. 2011. Dec 1;7(12):1448–1461. PubMed PMID: 22082875; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 843.Bakula D, Mueller AJ, Proikas-Cezanne T.. WIPI beta-propellers function as scaffolds for STK11/LKB1-AMPK and AMPK-related kinase signaling in autophagy. Autophagy. 2018;14(6):1082–1083. doi: 10.1080/15548627.2017.1382784. PubMed PMID: 28976799; PubMed Central PMCID: PMCPMC6103416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Bakula D, Muller AJ, Zuleger T, et al. WIPI3 and WIPI4 beta-propellers are scaffolds for LKB1-AMPK-TSC signalling circuits in the control of autophagy. Nat Commun. 2017. May 31;8:15637. doi: 10.1038/ncomms15637. PubMed PMID: 28561066; PubMed Central PMCID: PMCPMC5460038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845.Lu Q, Yang P, Huang X, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to auto-phagosomes. Dev Cell. 2011. Aug 16;21(2):343–57. doi: 10.1016/j.devcel.2011.06.024. PubMed PMID: 21802374; eng. [DOI] [PubMed] [Google Scholar]
- 846.Cao Y, Klionsky DJ.. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007. Oct;17(10):839–49. doi: 10.1038/cr.2007.78. PubMed PMID: 17893711; eng. [DOI] [PubMed] [Google Scholar]
- 847.Yang Z, Klionsky DJ.. Mammalian autophagy: core molecular machinery and signaling regulation [Review]. Curr Opin Cell Biol. 2010. Apr;22(2):124–31. doi: 10.1016/j.ceb.2009.11.014. PubMed PMID: 20034776; PubMed Central PMCID: PMC2854249. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 848.Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005. Sep 23;122(6):927–39. PubMed PMID: 16179260; eng. [DOI] [PubMed] [Google Scholar]
- 849.Erlich S, Mizrachy L, Segev O, et al. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy. 2007. Nov-Dec;3(6):561–8. doi: 10.4161/auto.4713. PubMed PMID: 17643073. [DOI] [PubMed] [Google Scholar]
- 850.Oberstein A, Jeffrey PD, Shi Y.. Crystal structure of the Bcl-XL-Beclin 1 peptide complex: Beclin 1 is a novel BH3-only protein. J Biol Chem. 2007. Apr 27;282(17):13123–32. doi: 10.1074/jbc.M700492200. PubMed PMID: 17337444. [DOI] [PubMed] [Google Scholar]
- 851.Shiloh R, Gilad Y, Ber Y, et al. Non-canonical activation of DAPK2 by AMPK constitutes a new pathway linking metabolic stress to autophagy. Nat Commun. 2018. May 1;9(1):1759. doi: 10.1038/s41467-018-03907-4. PubMed PMID: 29717115; PubMed Central PMCID: PMCPMC5931534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 852.Zalckvar E, Berissi H, Mizrachy L, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009. Mar;10(3):285–92. doi: 10.1038/embor.2008.246. PubMed PMID: 19180116; PubMed Central PMCID: PMC2658558. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.Wei Y, Pattingre S, Sinha S, et al. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008. Jun 20;30(6):678–88. doi: 10.1016/j.molcel.2008.06.001. PubMed PMID: 18570871; PubMed Central PMCID: PMC2478643. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 854.Wei Y, Sinha S, Levine B.. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. 2008. Oct;4(7):949–51. PubMed PMID: 18769111; PubMed Central PMCID: PMC2677707. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 855.Fernandez AF, Sebti S, Wei Y, et al. Disruption of the beclin 1-BCL2 autophagy regulatory complex promotes longevity in mice. Nature. 2018. Jun;558(7708):136–140. doi: 10.1038/s41586-018-0162-7. PubMed PMID: 29849149; PubMed Central PMCID: PMCPMC5992097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 856.Lossi L, Gambino G, Ferrini F, et al. Posttranslational regulation of BCL2 levels in cerebellar granule cells: A mechanism of neuronal survival. Dev Neurobiol. 2009. Nov;69(13):855–70. doi: 10.1002/dneu.20744. PubMed PMID: 19672954; eng. [DOI] [PubMed] [Google Scholar]
- 857.Lossi L, Gambino G, Salio C, et al. Autophagy regulates the post-translational cleavage of BCL-2 and promotes neuronal survival. ScientificWorldJournal. 2010;10:924–9. doi: 10.1100/tsw.2010.82. PubMed PMID: 20495771; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 858.Scarlatti F, Maffei R, Beau I, et al. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008. Aug;15(8):1318–29. doi: 10.1038/cdd.2008.51. PubMed PMID: 18421301; eng. [DOI] [PubMed] [Google Scholar]
- 859.Sok SP, Arshad NM, Azmi MN, et al. The apoptotic effect of 1’S-1ʹ-Acetoxychavicol Acetate (ACA) enhanced by inhibition of non-canonical autophagy in human non-small cell lung cancer cells. PLoS One. 2017;12(2):e0171329. doi: 10.1371/journal.pone.0171329. PubMed PMID: 28158287; PubMed Central PMCID: PMCPMC5291426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860.Kang R, Zeh HJ, Lotze MT, et al. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011. Apr;18(4):571–80. doi: 10.1038/cdd.2010.191. PubMed PMID: 21311563; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 861.Kihara A, Kabeya Y, Ohsumi Y, et al. Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2001. Apr;2(4):330–5. doi: 10.1093/embo-reports/kve061. PubMed PMID: 11306555; PubMed Central PMCID: PMC1083858. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Amritraj A, Peake K, Kodam A, et al. Increased activity and altered subcellular distribution of lysosomal enzymes determine neuronal vulnerability in Niemann-Pick type C1-deficient mice. Am J Pathol. 2009. Dec;175(6):2540–56. doi: 10.2353/ajpath.2009.081096. PubMed PMID: 19893049; PubMed Central PMCID: PMC2789601. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 863.McKnight NC, Zhenyu Y.. Beclin 1, an essential component and master regulator of PI3K-III in health and disease. Curr Pathobiol Rep. 2013. Dec 1;1(4):231–238. doi: 10.1007/s40139-013-0028-5. PubMed PMID: 24729948; PubMed Central PMCID: PMCPMC3979578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 864.Castino R, Bellio N, Follo C, et al. Inhibition of PI3k class III-dependent autophagy prevents apoptosis and necrosis by oxidative stress in dopaminergic neuroblastoma cells. Toxicol Sci. 2010. Sep;117(1):152–62. doi: 10.1093/toxsci/kfq170. PubMed PMID: 20525898; eng. [DOI] [PubMed] [Google Scholar]
- 865.Yue Z, Horton A, Bravin M, et al. A novel protein complex linking the delta 2 glutamate receptor and autophagy: implications for neurodegeneration in lurcher mice. Neuron. 2002. Aug 29;35(5):921–33. PubMed PMID: 12372286; eng. [DOI] [PubMed] [Google Scholar]
- 866.Luo S, Rubinsztein DC.. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ. 2010. Feb;17(2):268–77. doi: 10.1038/cdd.2009.121. PubMed PMID: 19713971; PubMed Central PMCID: PMC2894406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867.Furuya N, Yu J, Byfield M, et al. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy. 2005. Apr;1(1):46–52. PubMed PMID: 16874027. [DOI] [PubMed] [Google Scholar]
- 868.Kim J, Kim YC, Fang C, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013. Jan 17;152(1–2):290–303. doi: 10.1016/j.cell.2012.12.016. PubMed PMID: 23332761; PubMed Central PMCID: PMC3587159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Nemazanyy I, Montagnac G, Russell RC, et al. Class III PI3K regulates organismal glucose homeostasis by providing negative feedback on hepatic insulin signalling. Nat Commun. 2015. Sep 21;6:8283. doi: 10.1038/ncomms9283. PubMed PMID: 26387534; PubMed Central PMCID: PMCPMC4579570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Takats S, Nagy P, Varga A, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013. May 13;201(4):531–9. doi: 10.1083/jcb.201211160. PubMed PMID: 23671310; PubMed Central PMCID: PMC3653357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Itakura E, Kishi-Itakura C, Mizushima N.. The hairpin-type tail-anchored SNARE syntaxin 17 targets to auto-phagosomes for fusion with endosomes/lysosomes. Cell. 2012. Dec 7;151(6):1256–69. doi: 10.1016/j.cell.2012.11.001. PubMed PMID: 23217709. [DOI] [PubMed] [Google Scholar]
- 872.Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013. Mar 21;495(7441):389–93. doi: 10.1038/nature11910. PubMed PMID: 23455425. [DOI] [PubMed] [Google Scholar]
- 873.Arasaki K, Shimizu H, Mogari H, et al. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev Cell. 2015. Feb 9;32(3):304–17. doi: 10.1016/j.devcel.2014.12.011. PubMed PMID: 25619926. [DOI] [PubMed] [Google Scholar]
- 874.Morelli E, Ginefra P, Mastrodonato V, et al. Multiple functions of the SNARE protein Snap29 in autophagy, endocytic, and exocytic trafficking during epithelial formation in Drosophila. Autophagy. 2014;10(12):2251–68. doi: 10.4161/15548627.2014.981913. PubMed PMID: 25551675; PubMed Central PMCID: PMC4502674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 875.Tsuboyama K, Koyama-Honda I, Sakamaki Y, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016. Nov 25;354(6315):1036–1041. doi: 10.1126/science.aaf6136. PubMed PMID: 27885029. [DOI] [PubMed] [Google Scholar]
- 876.Arasaki K, Mikami Y, Shames SR, et al. Legionella effector Lpg1137 shuts down ER-mitochondria communication through cleavage of syntaxin 17. Nat Commun. 2017. May 15;8:15406. doi: 10.1038/ncomms15406. PubMed PMID: 28504273; PubMed Central PMCID: PMCPMC5440676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 877.Kumar S, Gu Y, Abudu YP, et al. Phosphorylation of syntaxin 17 by TBK1 controls autophagy initiation. Dev Cell. 2019. Apr 8;49(1):130–144 e6. doi: 10.1016/j.devcel.2019.01.027. PubMed PMID: 30827897; PubMed Central PMCID: PMCPMC6907693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 878.Sugo M, Kimura H, Arasaki K, et al. Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO J. 2018. Nov 2;37(21). doi: 10.15252/embj.201798899. PubMed PMID: 30237312; PubMed Central PMCID: PMCPMC6213275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 879.Xian H, Yang Q, Xiao L, et al. STX17 dynamically regulated by Fis1 induces mitophagy via hierarchical macroautophagic mechanism. Nat Commun. 2019. May 3;10(1):2059. doi: 10.1038/s41467-019-10096-1. PubMed PMID: 31053718; PubMed Central PMCID: PMCPMC6499814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 880.Cheng XT, Zhou B, Lin MY, et al. Axonal auto-phagosomes recruit dynein for retrograde transport through fusion with late endosomes. J Cell Biol. 2015. May 11;209(3):377–86. doi: 10.1083/jcb.201412046. PubMed PMID: 25940348; PubMed Central PMCID: PMCPMC4427784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 881.Cheng XT, Zhou B, Lin MY, et al. Axonal auto-phagosomes use the ride-on service for retrograde transport toward the soma. Autophagy. 2015;11(8):1434–6. doi: 10.1080/15548627.2015.1062203. PubMed PMID: 26102591; PubMed Central PMCID: PMCPMC4590659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 882.Kim JH, Hong SB, Lee JK, et al. Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. Autophagy. 2015;11(1):75–87. doi: 10.4161/15548627.2014.984276. PubMed PMID: 25484072; PubMed Central PMCID: PMCPMC4502675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 883.Chen D, Zhong Q.. A tethering coherent protein in autophagosome maturation. Autophagy. 2012. Jun;8(6):985–6. doi: 10.4161/auto.20255. PubMed PMID: 22617511; PubMed Central PMCID: PMC3427267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 884.Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008. Aug 25;182(4):685–701. doi: 10.1083/jcb.200803137. PubMed PMID: 18725538; PubMed Central PMCID: PMC2518708. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 885.Karunakaran I, van Echten-Deckert G.. Sphingosine 1-phosphate - A double edged sword in the brain. Biochim Biophys Acta Biomembr. 2017. Sep;1859(9 Pt B):1573–1582. doi: 10.1016/j.bbamem.2017.03.008. PubMed PMID: 28315304. [DOI] [PubMed] [Google Scholar]
- 886.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018. Jan;18(1):33–50. doi: 10.1038/nrc.2017.96. PubMed PMID: 29147025; PubMed Central PMCID: PMCPMC5818153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 887.van Echten-Deckert G, Alam S.. Sphingolipid metabolism - an ambiguous regulator of autophagy in the brain. Biol Chem. 2018. Jul 26;399(8):837–850. doi: 10.1515/hsz-2018-0237. PubMed PMID: 29908127. [DOI] [PubMed] [Google Scholar]
- 888.Mitroi DN, Karunakaran I, Graler M, et al. SGPL1 (sphingosine phosphate lyase 1) modulates neuronal autophagy via phosphatidylethanolamine production. Autophagy. 2017. May 4;13(5):885–899. doi: 10.1080/15548627.2017.1291471. PubMed PMID: 28521611; PubMed Central PMCID: PMCPMC5446076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 889.Moruno Manchon JF, Uzor NE, Dabaghian Y, et al. Cytoplasmic sphingosine-1-phosphate pathway modulates neuronal autophagy. Sci Rep. 2015. Oct 19;5:15213. doi: 10.1038/srep15213. PubMed PMID: 26477494; PubMed Central PMCID: PMCPMC4609990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 890.Bamborschke D, Pergande M, Becker K, et al. A novel mutation in sphingosine-1-phosphate lyase causing congenital brain malformation. Brain Dev. 2018. Jun;40(6):480–483. doi: 10.1016/j.braindev.2018.02.008. PubMed PMID: 29501407. [DOI] [PubMed] [Google Scholar]
- 891.Taniguchi M, Kitatani K, Kondo T, et al. Regulation of autophagy and its associated cell death by “sphingolipid rheostat”: reciprocal role of ceramide and sphingosine 1-phosphate in the mammalian target of rapamycin pathway. J Biol Chem. 2012. Nov 16;287(47):39898–910. doi: 10.1074/jbc.M112.416552. PubMed PMID: 23035115; PubMed Central PMCID: PMC3501064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 892.Justice MJ, Petrusca DN, Rogozea AL, et al. Effects of lipid interactions on model vesicle engulfment by alveolar macrophages. Biophys J. 2014. Feb 4;106(3):598–609. doi: 10.1016/j.bpj.2013.12.036. PubMed PMID: 24507600; PubMed Central PMCID: PMC3944992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 893.Tagaya M, Arasaki K.. Regulation of mitochondrial dynamics and autophagy by the mitochondria-associated membrane. Adv Exp Med Biol. 2017;997:33–47. doi: 10.1007/978-981-10-4567-7_3. PubMed PMID: 28815520. [DOI] [PubMed] [Google Scholar]
- 894.Guenther GG, Peralta ER, Rosales KR, et al. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci U S A. 2008. Nov 11;105(45):17402–7. doi: 10.1073/pnas.0802781105. PubMed PMID: 18981422; PubMed Central PMCID: PMC2582319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 895.Pattingre S, Bauvy C, Levade T, et al. Ceramide-induced autophagy: to junk or to protect cells? Autophagy. 2009. May;5(4):558–60. PubMed PMID: 19337026; PubMed Central PMCID: PMC3501009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 896.Sentelle RD, Senkal CE, Jiang W, et al. Ceramide targets auto-phagosomes to mitochondria and induces lethal mitophagy. Nat Chem Biol. 2012. Oct;8(10):831–8. doi: 10.1038/nchembio.1059. PubMed PMID: 22922758; PubMed Central PMCID: PMC3689583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 897.Dany M, Gencer S, Nganga R, et al. Targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML. Blood. 2016. Oct 13;128(15):1944–1958. doi: 10.1182/blood-2016-04-708750. PubMed PMID: 27540013; PubMed Central PMCID: PMCPMC5064718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 898.Jiang W, Ogretmen B.. Ceramide stress in survival versus lethal autophagy paradox: ceramide targets auto-phagosomes to mitochondria and induces lethal mitophagy. Autophagy. 2013. Feb 1;9(2):258–9. doi: 10.4161/auto.22739. PubMed PMID: 23182807; PubMed Central PMCID: PMC3552895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 899.Jiang W, Ogretmen B.. Autophagy paradox and ceramide. Biochim Biophys Acta. 2014. May;1841(5):783–92. doi: 10.1016/j.bbalip.2013.09.005. PubMed PMID: 24055889; PubMed Central PMCID: PMC3960371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 900.Thomas RJ, Oleinik N, Panneer Selvam S, et al. HPV/E7 induces chemotherapy-mediated tumor suppression by ceramide-dependent mitophagy. EMBO Mol Med. 2017. Aug;9(8):1030–1051. doi: 10.15252/emmm.201607088. PubMed PMID: 28606997; PubMed Central PMCID: PMCPMC5538428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 901.Oleinik N, Kim J, Roth BM, et al. Mitochondrial protein import is regulated by p17/PERMIT to mediate lipid metabolism and cellular stress. Sci Adv. 2019. Sep;5(9):eaax1978. doi: 10.1126/sciadv.aax1978. PubMed PMID: 31535025; PubMed Central PMCID: PMCPMC6739097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 902.Lepine S, Allegood JC, Park M, et al. Sphingosine-1-phosphate phosphohydrolase-1 regulates ER stress-induced autophagy. Cell Death Differ. 2011. Feb;18(2):350–61. doi: 10.1038/cdd.2010.104. PubMed PMID: 20798685; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 903.Cervia D, Assi E, De Palma C, et al. Essential role for acid sphingomyelinase-inhibited autophagy in melanoma response to cisplatin. Oncotarget. 2016. May 3;7(18):24995–5009. doi: 10.18632/oncotarget.8735. PubMed PMID: 27107419; PubMed Central PMCID: PMCPMC5041885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 904.Gluschko A, Herb M, Wiegmann K, et al. The beta2 Integrin Mac-1 Induces Protective LC3-Associated Phagocytosis of Listeria monocytogenes. Cell Host Microbe. 2018. Mar 14;23(3):324–337 e5. doi: 10.1016/j.chom.2018.01.018. PubMed PMID: 29544096. [DOI] [PubMed] [Google Scholar]
- 905.Signorelli P, Munoz-Olaya JM, Gagliostro V, et al. Dihydroceramide intracellular increase in response to resveratrol treatment mediates autophagy in gastric cancer cells. Cancer Lett. 2009. Sep 18;282(2):238–43. doi: 10.1016/j.canlet.2009.03.020 PubMed PMID: 19394759. [DOI] [PubMed] [Google Scholar]
- 906.Hernandez-Tiedra S, Fabrias G, Davila D, et al. Dihydroceramide accumulation mediates cytotoxic autophagy of cancer cells via autolysosome destabilization. Autophagy. 2016. Nov;12(11):2213–2229. doi: 10.1080/15548627.2016.1213927. PubMed PMID: 27635674; PubMed Central PMCID: PMCPMC5103338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 907.Matarrese P, Garofalo T, Manganelli V, et al. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy. 2014. May 1;10(5):750–65. doi: 10.4161/auto.27959. PubMed PMID: 24589479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 908.Garofalo T, Matarrese P, Manganelli V, et al. Evidence for the involvement of lipid rafts localized at the ER-mitochondria associated membranes in autophagosome formation. Autophagy. 2016. Jun 2;12(6):917–35. doi: 10.1080/15548627.2016.1160971. PubMed PMID: 27123544; PubMed Central PMCID: PMCPMC4922444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 909.Russ DW, Wills AM, Boyd IM, et al. Weakness, SR function and stress in gastrocnemius muscles of aged male rats. Exp Gerontol. 2014. Feb;50:40–4. doi: 10.1016/j.exger.2013.11.018. PubMed PMID: 24316040. [DOI] [PubMed] [Google Scholar]
- 910.Bernard A, Jin M, Xu Z, et al. A large-scale analysis of autophagy-related gene expression identifies new regulators of autophagy.Autophagy. 2015. Nov 2;11(11):2114–2122. doi: 10.1080/15548627.2015.1099796. PubMed PMID: 26649943; PubMed Central PMCID: PMCPMC4824583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 911.Barany I, Berenguer E, Solis MT, et al. Autophagy is activated and involved in cell death with participation of cathepsins during stress-induced microspore embryogenesis in barley. J Exp Bot. 2018. Mar 14;69(6):1387–1402. doi: 10.1093/jxb/erx455. PubMed PMID: 29309624; PubMed Central PMCID: PMCPMC6019037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 912.Bernard A, Jin M, Gonzalez-Rodriguez P, et al. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol. 2015. Mar 2;25(5):546–55. doi: 10.1016/j.cub.2014.12.049. PubMed PMID: 25660547; PubMed Central PMCID: PMC4348152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 913.Bernard A, Klionsky DJ.. Rph1 mediates the nutrient-limitation signaling pathway leading to transcriptional activation of autophagy. Autophagy. 2015. Apr 3;11(4):718–9. doi: 10.1080/15548627.2015.1018503. PubMed PMID: 25751780; PubMed Central PMCID: PMC4502745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 914.Kirisako T, Baba M, Ishihara N, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol. 1999. Oct 18;147(2):435–46. PubMed PMID: 10525546; PubMed Central PMCID: PMC2174223. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 915.Nara A, Mizushima N, Yamamoto A, et al. SKD1 AAA ATPase-dependent endosomal transport is involved in autolysosome formation. Cell Struct Funct. 2002. Feb;27(1):29–37. PubMed PMID: 11937716; eng. [DOI] [PubMed] [Google Scholar]
- 916.Tsuyuki S, Takabayashi M, Kawazu M, et al. Detection of WIPI1 mRNA as an indicator of autophagosome formation. Autophagy. 2014. Mar;10(3):497–513. doi: 10.4161/auto.27419. PubMed PMID: 24384561; PubMed Central PMCID: PMCPMC4077887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 917.Jin M, He D, Backues SK, et al. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014. Jun 16;24(12):1314–22. doi: 10.1016/j.cub.2014.04.048. PubMed PMID: 24881874; PubMed Central PMCID: PMC4169046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 918.Kouroku Y, Fujita E, Tanida I, et al. ER stress (PERK/eIF2[alpha] phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007. Feb;14(2):230–9. PubMed PMID: 16794605; eng. [DOI] [PubMed] [Google Scholar]
- 919.Xiong X, Tao R, DePinho RA, et al. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012. Nov 9;287(46):39107–14. doi: 10.1074/jbc.M112.412569. PubMed PMID: 22992773; PubMed Central PMCID: PMC3493951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 920.Moussay E, Kaoma T, Baginska J, et al. The acquisition of resistance to TNFalpha in breast cancer cells is associated with constitutive activation of autophagy as revealed by a transcriptome analysis using a custom microarray. Autophagy. 2011. Jul;7(7):760–70. PubMed PMID: 21490427; eng. [DOI] [PubMed] [Google Scholar]
- 921.Lee JM, Wagner M, Xiao R, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014. Dec 4;516(7529):112–5. doi: 10.1038/nature13961. PubMed PMID: 25383539; PubMed Central PMCID: PMC4267857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 922.Conte A, Paladino S, Bianco G, et al. High mobility group A1 protein modulates autophagy in cancer cells. Cell Death Differ. 2017. Nov;24(11):1948–1962. doi: 10.1038/cdd.2017.117. PubMed PMID: 28777374; PubMed Central PMCID: PMCPMC5635219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 923.Mitroulis I, Kourtzelis I, Kambas K, et al. Regulation of the autophagic machinery in human neutrophils [Research Support, Non-U.S. Gov’t]. Eur J Immunol. 2010. May;40(5):1461–72. doi: 10.1002/eji.200940025. PubMed PMID: 20162553; eng. [DOI] [PubMed] [Google Scholar]
- 924.Rodriguez-Muela N, Germain F, Marino G, et al. Autophagy promotes survival of retinal ganglion cells after optic nerve axotomy in mice. Cell Death Differ. 2012. Jan;19(1):162–9. doi: 10.1038/cdd.2011.88. PubMed PMID: 21701497; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 925.Vázquez P, Arroba AI, Cecconi F, et al. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. 2012;8:187–99. [DOI] [PubMed] [Google Scholar]
- 926.Rouschop KM, van den Beucken T, Dubois L, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010. Jan 4;120(1):127–41. doi: 10.1172/JCI40027. PubMed PMID: 20038797; PubMed Central PMCID: PMC2798689. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 927.Haim Y, Blüher M, Slutsky N, et al. Elevated autophagy gene expression in adipose tissue of obese humans: A potential noncell- cycle-dependent function of E2F1. Autophagy. 2015;11:2074–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 928.Zhang J, Wang J, Lee YM, et al. Proteomic profiling of de novo protein synthesis in starvation-induced autophagy using Bioorthogonal noncanonical amino acid tagging. Methods Enzymol. 2017;588:41–59. doi: 10.1016/bs.mie.2016.09.075. PubMed PMID: 28237112. [DOI] [PubMed] [Google Scholar]
- 929.Pourpirali S, Valacca C, Merlo P, et al. Prolonged pseudohypoxia targets Ambra1 mRNA to P-bodies for translational repression. PLoS One. 2015;10(6):e0129750. doi: 10.1371/journal.pone.0129750. PubMed PMID: 26086269; PubMed Central PMCID: PMCPMC4473010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 930.Las G, Serada SB, Wikstrom JD, et al. Fatty acids suppress autophagic turnover in beta-cells. J Biol Chem. 2011. Dec 9;286(49):42534–44. doi: 10.1074/jbc.M111.242412. PubMed PMID: 21859708; PubMed Central PMCID: PMC3234912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 931.Woldt E, Sebti Y, Solt LA, et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013. Aug;19(8):1039–46. doi: 10.1038/nm.3213. PubMed PMID: 23852339; PubMed Central PMCID: PMC3737409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 932.Huang G, Zhang F, Ye Q, et al. The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Rev-erbalpha and indirectly via Cebpb/(C/ebpbeta) in zebrafish. Autophagy. 2016. Aug 2;12(8):1292–309. doi: 10.1080/15548627.2016.1183843. PubMed PMID: 27171500; PubMed Central PMCID: PMCPMC4968235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 933.Ferder IC, Fung L, Ohguchi Y, et al. Meiotic gatekeeper STRA8 suppresses autophagy by repressing Nr1d1 expression during spermatogenesis in mice. PLoS Genet. 2019. May;15(5):e1008084. doi: 10.1371/journal.pgen.1008084. PubMed PMID: 31059511; PubMed Central PMCID: PMCPMC6502318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 934.Chandra V, Bhagyaraj E, Nanduri R, et al. NR1D1 ameliorates Mycobacterium tuberculosis clearance through regulation of autophagy. Autophagy. 2015. Nov 2;11(11):1987–1997. doi: 10.1080/15548627.2015.1091140. PubMed PMID: 26390081; PubMed Central PMCID: PMCPMC4824569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 935.Luo R, Su LY, Li G, et al. Activation of PPARA-mediated autophagy reduces Alzheimer disease-like pathology and cognitive decline in a murine model. Autophagy. 2020. Jan;16(1):52–69. doi: 10.1080/15548627.2019.1596488, PubMed PMID: 30898012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 936.Seok S, Fu T, Choi SE, et al. Transcriptional regulation of autophagy by an FXR-CREB axis. Nature. 2014. Dec 4;516(7529):108–11. doi: 10.1038/nature13949. PubMed PMID: 25383523; PubMed Central PMCID: PMC4257899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 937.Jiang H, Martin V, Gomez-Manzano C, et al. The RB-E2F1 pathway regulates autophagy. Cancer Res. 2010. Oct 15;70(20):7882–93. doi: 0008-5472.CAN-10-1604 [pii] doi: 10.1158/0008-5472.CAN-10-1604. PubMed PMID: 20807803; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 938.Polager S, Ofir M, Ginsberg D.. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008. Aug 14;27(35):4860–4. doi: onc2008117 [pii] doi: 10.1038/onc.2008.117. PubMed PMID: 18408756; eng. [DOI] [PubMed] [Google Scholar]
- 939.Gorski SM, Chittaranjan S, Pleasance ED, et al. A SAGE approach to discovery of genes involved in autophagic cell death. Curr Biol CB. 2003. Feb 18;13(4):358–63. PubMed PMID: 12593804; eng. [DOI] [PubMed] [Google Scholar]
- 940.Lee C-Y, Clough EA, Yellon P, et al. Genome-wide analyses of steroid- and radiation-triggered programmed cell death in Drosophila. Curr Biol. 2003. Feb 18;13(4):350–7. PubMed PMID: 12593803; eng. [DOI] [PubMed] [Google Scholar]
- 941.Denton D, Shravage B, Simin R, et al. Larval midgut destruction in Drosophila: not dependent on caspases but suppressed by the loss of autophagy. Autophagy. 2010. Jan;6(1):163–5. PubMed PMID: 20009534; PubMed Central PMCID: PMC2819273. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 942.Franzetti E, Huang ZJ, Shi YX, et al. Autophagy precedes apoptosis during the remodeling of silkworm larval midgut. Apoptosis. 2012;17:305–24. [DOI] [PubMed] [Google Scholar]
- 943.Tian L, Ma L, Guo E, et al. 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy. 2013. Aug;9(8):1172–87. doi: 10.4161/auto.24731. PubMed PMID: 23674061; PubMed Central PMCID: PMC3748190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 944.Montali A, Romanelli D, Cappellozza S, et al. Timing of autophagy and apoptosis during posterior silk gland degeneration in Bombyx mori. Arthropod Struct Dev. 2017. Jul;46(4):518–528. doi: 10.1016/j.asd.2017.05.003. PubMed PMID: 28549564. [DOI] [PubMed] [Google Scholar]
- 945.Juhasz G, Puskas LG, Komonyi O, et al. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007. Jun;14(6):1181–90. PubMed PMID: 17363962; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 946.Erdi B, Nagy P, Zvara A, et al. Loss of the starvation-induced gene Rack1 leads to glycogen deficiency and impaired autophagic responses in Drosophila. Autophagy. 2012. Jul 1;8(7):1124–35. doi: 10.4161/auto.20069. PubMed PMID: 22562043; PubMed Central PMCID: PMC3429548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 947.Barth JM, Szabad J, Hafen E, et al. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ. 2011. Jun;18(6):915–24. doi: 10.1038/cdd.2010.157. PubMed PMID: 21151027; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 948.O’Rourke EJ, Ruvkun G.. MXL-3 and HLH-30 transcriptionally link lipolysis and autophagy to nutrient availability. Nat Cell Biol. 2013. Jun;15(6):668–76. doi: 10.1038/ncb2741. PubMed PMID: 23604316; PubMed Central PMCID: PMC3723461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 949.Settembre C, Di Malta C, Polito VA, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011. Jun 17;332(6036):1429–33. doi: 10.1126/science.1204592. PubMed PMID: 21617040; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 950.Lecker SH, Jagoe RT, Gilbert A, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004. Jan;18(1):39–51. doi: 10.1096/fj.03-0610com. PubMed PMID: 14718385; eng. [DOI] [PubMed] [Google Scholar]
- 951.Phillips AR, Suttangkakul A, Vierstra RD.. The ATG12-conjugating enzyme ATG10 Is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics. 2008. Mar;178(3):1339–53. doi: 10.1534/genetics.107.086199. PubMed PMID: 18245858; PubMed Central PMCID: PMC2278079. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 952.Seiliez I, Gutierrez J, Salmeron C, et al. An in vivo and in vitro assessment of autophagy-related gene expression in muscle of rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol Part B Biochem Mol Bio. 2010. Nov;157(3):258–66. doi: 10.1016/j.cbpb.2010.06.011. PubMed PMID: 20601058; eng. [DOI] [PubMed] [Google Scholar]
- 953.Alshudukhi AA, Zhu J, Huang D, et al. Lipin-1 regulates Bnip3-mediated mitophagy in glycolytic muscle. FASEB J. 2018. Dec;32(12):6796–6807. doi: 10.1096/fj.201800374. PubMed PMID: 29939786; PubMed Central PMCID: PMCPMC6219840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 954.Sandri M. Autophagy in health and disease. 3. Involvement of autophagy in muscle atrophy. Am J Physiol Cell Physiol. 2010. Jun;298(6):C1291–7. doi: 10.1152/ajpcell.00531.2009. PubMed PMID: 20089936; eng. [DOI] [PubMed] [Google Scholar]
- 955.Eisenberg T, Knauer H, Schauer A, et al. Induction of autophagy by spermidine promotes longevity [Research Support, Non-U.S. Gov’t]. Nat Cell Biol. 2009. Nov;11(11):1305–14. doi: 10.1038/ncb1975. PubMed PMID: 19801973; eng. [DOI] [PubMed] [Google Scholar]
- 956.Yue F, Li W, Zou J, et al. Spermidine Prolongs Lifespan and Prevents Liver Fibrosis and Hepatocellular Carcinoma by Activating MAP1S-Mediated Autophagy. Cancer Res. 2017. Jun 1;77(11):2938–2951. . PubMed PMID: 28386016; PubMed Central PMCID: PMCPMC5489339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 957.Allaire M, Rautou P-E, Codogno P, et al. Autophagy in liver diseases: Time for translation?. J Hepatol. 2019. May;70(5):985–998. doi: 10.1016/j.jhep.2019.01.026. PubMed PMID: 30711404.. [DOI] [PubMed] [Google Scholar]
- 958.Zhang H, Alsaleh G, Feltham J, et al. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol Cell. 2019. Oct 3;76(1):110–125 e9. doi: 10.1016/j.molcel.2019.08.005. PubMed PMID: 31474573; PubMed Central PMCID: PMCPMC6863385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 959.Eisenberg T, Abdellatif M, Schroeder S, et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat Med. 2016. Dec;22(12):1428–1438. doi: 10.1038/nm.4222. PubMed PMID: 27841876; PubMed Central PMCID: PMCPMC5806691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 960.Xu W, Cai SY, Zhang Y, et al. Melatonin enhances thermotolerance by promoting cellular protein protection in tomato plants. J Pineal Res. 2016. Nov;61(4):457–469. doi: 10.1111/jpi.12359. PubMed PMID: 27484733. [DOI] [PubMed] [Google Scholar]
- 961.Cai SY, Zhang Y, Xu YP, et al. HsfA1a upregulates melatonin biosynthesis to confer cadmium tolerance in tomato plants. J Pineal Res. 2017. Mar;62(2):e12387. doi: 10.1111/jpi.12387. PubMed PMID: 28095626. [DOI] [PubMed] [Google Scholar]
- 962.Wang Y, Cai S, Yin L, et al. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy. 2015. Nov 2;11(11):2033–2047. doi: 10.1080/15548627.2015.1098798. PubMed PMID: 26649940; PubMed Central PMCID: PMCPMC4824577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 963.Wang Y, Cao JJ, Wang KX, et al. BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 2019. Feb;179(2):671–685. doi: 10.1104/pp.18.01028. PubMed PMID: 30482787; PubMed Central PMCID: PMCPMC6426427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 964.Ropolo A, Grasso D, Pardo R, et al. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem. 2007. Dec 21;282(51):37124–33. doi: 10.1074/jbc.M706956200. PubMed PMID: 17940279; eng. [DOI] [PubMed] [Google Scholar]
- 965.Tian Y, Li Z, Hu W, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell. 2010. Jun 11;141(6):1042–55. doi: 10.1016/j.cell.2010.04.034. PubMed PMID: 20550938; eng. [DOI] [PubMed] [Google Scholar]
- 966.Tabara LC, Escalante R.. VMP1 Establishes ER-Microdomains that Regulate Membrane Contact Sites and Autophagy. PLoS One. 2016;11(11):e0166499. doi: 10.1371/journal.pone.0166499. PubMed PMID: 27861594; PubMed Central PMCID: PMCPMC5115753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 967.Re AE Lo, Fernandez-Barrena MG, Almada LL, et al. Novel AKT1-GLI3-VMP1 pathway mediates KRAS oncogene-induced autophagy in cancer cells. J Biol Chem. 2012. Jul 20;287(30):25325–34. doi: 10.1074/jbc.M112.370809. PubMed PMID: 22535956; PubMed Central PMCID: PMC3408195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 968.Sardiello M, Palmieri M, di Ronza A, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009. Jul 24;325(5939):473–7. doi: 10.1126/science.1174447. PubMed PMID: 19556463; eng. [DOI] [PubMed] [Google Scholar]
- 969.Palmieri M, Impey S, Kang H, et al. Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum Mol Genet. 2011. Oct 1;20(19):3852–66. doi: 10.1093/hmg/ddr306. PubMed PMID: 21752829; eng. [DOI] [PubMed] [Google Scholar]
- 970.Martina JA, Chen Y, Gucek M, et al. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012. Jun;8(6):903–14. doi: 10.4161/auto.19653. PubMed PMID: 22576015; PubMed Central PMCID: PMC3427256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 971.Settembre C, Zoncu R, Medina DL, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012. Mar 7;31(5):1095–108. doi: 10.1038/emboj.2012.32. PubMed PMID: 22343943; PubMed Central PMCID: PMC3298007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 972.Vega-Rubin-de-Celis S, Pena-Llopis S, Konda M, et al. Multistep regulation of TFEB by MTORC1. Autophagy. 2017. Mar 4;13(3):464–472. doi: 10.1080/15548627.2016.1271514. PubMed PMID: 28055300; PubMed Central PMCID: PMCPMC5361595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 973.Palmieri M, Pal R, Nelvagal HR, et al. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017. Feb 6;8:14338. doi: 10.1038/ncomms14338. PubMed PMID: 28165011; PubMed Central PMCID: PMCPMC5303831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 974.Napolitano G, Esposito A, Choi H, et al. mTOR-dependent phosphorylation controls TFEB nuclear export. Nat Commun. 2018. Aug 17;9(1):3312. doi: 10.1038/s41467-018-05862-6. PubMed PMID: 30120233; PubMed Central PMCID: PMCPMC6098152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 975.Silvestrini MJ, Johnson JR, Kumar AV, et al. Nuclear export inhibition enhances HLH-30/TFEB activity, autophagy, and lifespan. Cell Rep. 2018. May 15;23(7):1915–1921. doi: 10.1016/j.celrep.2018.04.063. PubMed PMID: 29768192; PubMed Central PMCID: PMCPMC5991088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 976.Li L, Friedrichsen HJ, Andrews S, et al. A TFEB nuclear export signal integrates amino acid supply and glucose availability. Nat Commun. 2018. Jul 11;9(1):2685. doi: 10.1038/s41467-018-04849-7. PubMed PMID: 29992949; PubMed Central PMCID: PMCPMC6041281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 977.Nezich CL, Wang C, Fogel AI, et al. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J Cell Biol. 2015. Aug 3;210(3):435–50. doi: 10.1083/jcb.201501002. PubMed PMID: 26240184; PubMed Central PMCID: PMC4523611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 978.Perera RM, Stoykova S, Nicolay BN, et al. Transcriptional control of autophagy-lysosome function drives pancreatic cancer metabolism. Nature. 2015. Aug 20;524(7565):361–5. doi: 10.1038/nature14587. PubMed PMID: 26168401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 979.Zhang J, Wang J, Zhou Z, et al. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy. 2018;14(6):1043–1059. doi: 10.1080/15548627.2018.1447290. PubMed PMID: 30059277; PubMed Central PMCID: PMCPMC6103407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 980.Shin HJ, Kim H, Oh S, et al. AMPK-SKP2-CARM1 signalling cascade in transcriptional regulation of autophagy. Nature. 2016. Jun 23;534(7608):553–7. doi: 10.1038/nature18014. PubMed PMID: 27309807; PubMed Central PMCID: PMCPMC5568428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 981.Kang YA, Sanalkumar R, O’Geen H, et al. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012. Jan;32(1):226–39. doi: 10.1128/MCB.06166-11. PubMed PMID: 22025678; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 982.Brigger D, Proikas-Cezanne T, Tschan MP.. WIPI-dependent autophagy during neutrophil differentiation of NB4 acute promyelocytic leukemia cells. Cell Death Dis. 2014. Jul 3;5:e1315. doi: 10.1038/cddis.2014.261. PubMed PMID: 24991767; PubMed Central PMCID: PMCPMC4123064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 983.Jin J, Britschgi A, Schlafli AM, et al. Low autophagy (ATG) gene expression is associated with an immature AML blast cell phenotype and can be restored during AML differentiation therapy. Oxid Med Cell Longev. 2018;2018:1482795. doi: 10.1155/2018/1482795. PubMed PMID: 29743969; PubMed Central PMCID: PMCPMC5878891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 984.Zhao J, Brault JJ, Schild A, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007. Dec;6(6):472–83. doi:S1550-4131(07)00339-7 [pii] doi: 10.1016/j.cmet.2007.11.004. PubMed PMID: 18054316; eng. [DOI] [PubMed] [Google Scholar]
- 985.Chauhan S, Goodwin JG, Chauhan S, et al. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell. 2013. Apr 11;50(1):16–28. doi: 10.1016/j.molcel.2013.01.024. PubMed PMID: 23434374; PubMed Central PMCID: PMC3628091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 986.Peeters JGC, Picavet LW, Coenen S, et al. Transcriptional and epigenetic profiling of nutrient-deprived cells to identify novel regulators of autophagy. Autophagy. 2019. Jan;15(1):98–112. doi: 10.1080/15548627.2018.1509608. PubMed PMID: 30153076; PubMed Central PMCID: PMCPMC6287694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 987.Pajares M, Jimenez-Moreno N, Garcia-Yague AJ, et al. Transcription factor NFE2L2/NRF2 is a regulator of macroautophagy genes. Autophagy. 2016. Oct 2;12(10):1902–1916. doi: 10.1080/15548627.2016.1208889. PubMed PMID: 27427974; PubMed Central PMCID: PMCPMC5079676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 988.Pajares M, Rojo AI, Arias E, et al. Transcription factor NFE2L2/NRF2 modulates chaperone-mediated autophagy through the regulation of LAMP2A. Autophagy. 2018;14(8):1310–1322. doi: 10.1080/15548627.2018.1474992. PubMed PMID: 29950142; PubMed Central PMCID: PMCPMC6103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 989.Ma D, Panda S, Lin JD.. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 2011. Nov 16;30(22):4642–51. doi: 10.1038/emboj.2011.322. PubMed PMID: 21897364; PubMed Central PMCID: PMC3243590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 990.Brest P, Lapaquette P, Souidi M, et al. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn’s disease [Research Support, Non-U.S. Gov’t]. Nat Genet. 2011. Mar;43(3):242–5. doi: 10.1038/ng.762. PubMed PMID: 21278745; eng. [DOI] [PubMed] [Google Scholar]
- 991.Meenhuis A, van Veelen PA, de Looper H, et al. MiR-17/20/93/106 promote hematopoietic cell expansion by targeting sequestosome 1-regulated pathways in mice. Blood. 2011. Jul 28;118(4):916–25. doi: 10.1182/blood-2011-02-336487. PubMed PMID: 21628417; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 992.Roccaro AM, Sacco A, Jia X, et al. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010. Sep 2;116(9):1506–14. doi: 10.1182/blood-2010-01-265686. PubMed PMID: 20519629; PubMed Central PMCID: PMC2938840. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 993.Engedal N, Zerovnik E, Rudov A, et al. From Oxidative Stress Damage to Pathways, Networks, and Autophagy via MicroRNAs. Oxid Med Cell Longev. 2018;2018:4968321. doi: 10.1155/2018/4968321. PubMed PMID: 29849898; PubMed Central PMCID: PMCPMC5932428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 994.Colangelo T, Polcaro G, Ziccardi P, et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis. 2016. Feb 25;7:e2108. doi: 10.1038/cddis.2016.29. PubMed PMID: 26913599; PubMed Central PMCID: PMCPMC4849155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 995.Martinet W, De Meyer GR, Andries L, et al. In situ detection of starvation-induced autophagy. J Histochem Cytochem. 2006. Jan;54(1):85–96. PubMed PMID: 16148314; eng. [DOI] [PubMed] [Google Scholar]
- 996.Banreti A, Sass M, Graba Y.. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013. Jun 1;9(6):819–29. doi: 10.4161/auto.23908. PubMed PMID: 23466676; PubMed Central PMCID: PMC3672293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 997.Jin M, Klionsky DJ.. Regulation of autophagy: Modulation of the size and number of auto-phagosomes. FEBS Lett. 2014. Aug 1;588(15):2457–2463. doi: 10.1016/j.febslet.2014.06.015. PubMed PMID: 24928445; PubMed Central PMCID: PMC4118767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 998.Feng Y, Yao Z, Klionsky DJ.. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015. Jun;25(6):354–363. doi: 10.1016/j.tcb.2015.02.002. PubMed PMID: 25759175; PubMed Central PMCID: PMC4441840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 999.Xie Y, Kang R, Sun X, et al. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy. 2015;11(1):28–45. doi: 10.4161/15548627.2014.984267. PubMed PMID: 25484070; PubMed Central PMCID: PMC4502723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000.Jin S, Zhang X, Miao Y, et al. m(6)A RNA modification controls autophagy through upregulating ULK1 protein abundance. Cell Res. 2018. Sep;28(9):955–957. doi: 10.1038/s41422-018-0069-8. PubMed PMID: 30046135; PubMed Central PMCID: PMCPMC6123428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001.Pietrocola F, Marino G, Lissa D, et al. Pro-autophagic polyphenols reduce the acetylation of cytoplasmic proteins. Cell cycle. 2012. Oct 15;11(20):3851–60. doi: 10.4161/cc.22027. PubMed PMID: 23070521; PubMed Central PMCID: PMC3495827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1002.Marino G, Pietrocola F, Madeo F, et al. Caloric restriction mimetics: natural/physiological pharmacological autophagy inducers. Autophagy. 2014. Nov 2;10(11):1879–82. doi: 10.4161/auto.36413. PubMed PMID: 25484097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1003.Madeo F, Pietrocola F, Eisenberg T, et al. Caloric restriction mimetics: towards a molecular definition. Nat Rev Drug Discov. 2014. Oct;13(10):727–40. doi: 10.1038/nrd4391. PubMed PMID: 25212602. [DOI] [PubMed] [Google Scholar]
- 1004.Huang R, Xu Y, Wan W, et al. Deacetylation of Nuclear LC3 Drives Autophagy Initiation under Starvation. Mol Cell. 2015. Jan 13. doi: 10.1016/j.molcel.2014.12.013. PubMed PMID: 25601754. [DOI] [PubMed] [Google Scholar]
- 1005.Lee IH, Finkel T.. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009. Mar 6;284(10):6322–8. doi: 10.1074/jbc.M807135200. PubMed PMID: 19124466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1006.Yakhine-Diop SMS, Niso-Santano M, Rodriguez-Arribas M, et al. Impaired mitophagy and protein acetylation levels in fibroblasts from parkinson’s disease patients. Mol Neurobiol. 2019. Apr;56(4):2466–2481. doi: 10.1007/s12035-018-1206-6. PubMed PMID: 30032424. [DOI] [PubMed] [Google Scholar]
- 1007.Ruan HB, Ma Y, Torres S, et al. Calcium-dependent O-GlcNAc signaling drives liver autophagy in adaptation to starvation. Genes Dev. 2017. Aug 15;31(16):1655–1665. doi: 10.1101/gad.305441.117. PubMed PMID: 28903979; PubMed Central PMCID: PMCPMC5647936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1008.Pyo KE, Kim CR, Lee M, et al. ULK1 O-GlcNAcylation is crucial for activating VPS34 via ATG14L during autophagy initiation. Cell Rep. 2018. Dec 4;25(10):2878–2890 e4. doi: 10.1016/j.celrep.2018.11.042. PubMed PMID: 30517873. [DOI] [PubMed] [Google Scholar]
- 1009.Montagna C, Rizza S, Maiani E, et al. To eat, or NOt to eat: S-nitrosylation signaling in autophagy. FEBS J. 2016. Nov;283(21):3857–3869. doi: 10.1111/febs.13736. PubMed PMID: 27083138. [DOI] [PubMed] [Google Scholar]
- 1010.Sadhu A, Moriyasu Y, Acharya K, et al. Nitric oxide and ROS mediate autophagy and regulate Alternaria alternata toxin-induced cell death in tobacco BY-2 cells. Sci Rep. 2019. Jun 20;9(1):8973. doi: 10.1038/s41598-019-45470-y. PubMed PMID: 31222105; PubMed Central PMCID: PMCPMC6586778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1011.Rizza S, Cardaci S, Montagna C, et al. S-nitrosylation drives cell senescence and aging in mammals by controlling mitochondrial dynamics and mitophagy. Proc Natl Acad Sci U S A. 2018. Apr 10;115(15):E3388–E3397. doi: 10.1073/pnas.1722452115. PubMed PMID: 29581312; PubMed Central PMCID: PMCPMC5899480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1012.Rizza S, Filomeni G.. Denitrosylate and live longer: how ADH5/GSNOR links mitophagy to aging. Autophagy. 2018;14(7):1285–1287. doi: 10.1080/15548627.2018.1475818. PubMed PMID: 30029585; PubMed Central PMCID: PMCPMC6103690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1013.Vandiver MS, Paul BD, Xu R, et al. Sulfhydration mediates neuroprotective actions of parkin. Nat Commun. 2013;4:1626. doi: 10.1038/ncomms2623. PubMed PMID: 23535647; PubMed Central PMCID: PMCPMC3622945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1014.Sun Y, Lu F, Yu X, et al. Exogenous H2S promoted USP8 sulfhydration to regulate mitophagy in the hearts of db/db mice. Aging Dis. 2020;11:in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1015.Aroca A, Benito JM, Gotor C, et al. Persulfidation proteome reveals the regulation of protein function by hydrogen sulfide in diverse biological processes in Arabidopsis. J Exp Bot. 2017. Oct 13;68(17):4915–4927. doi: 10.1093/jxb/erx294. PubMed PMID: 28992305; PubMed Central PMCID: PMCPMC5853657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1016.Wang X, Wu R, Liu Y, et al. m(6)A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy. 2019. Aug 26:1–15. doi: 10.1080/15548627.2019.1659617. PubMed PMID: 31451060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1017.Pattingre S, Petiot A, Codogno P.. Analyses of Gα-interacting protein and activator of G-protein-signaling-3 functions in macroautophagy. Methods Enzymol. 2004;390:17–31. PubMed PMID: 15488168; eng. [DOI] [PubMed] [Google Scholar]
- 1018.Zhang J, Wang J, Ng S, et al. Development of a novel method for quantification of autophagic protein degradation by AHA labeling. Autophagy. 2014. May 1;10(5):901–12. doi: 10.4161/auto.28267. PubMed PMID: 24675368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1019.Wang J, Zhang J, Lee YM, et al. Nonradioactive quantification of autophagic protein degradation with L-azidohomoalanine labeling. Nat Protoc. 2017. Dec;12(2):279–288. doi: 10.1038/nprot.2016.160. PubMed PMID: 28079880. [DOI] [PubMed] [Google Scholar]
- 1020.Ichimura Y, Kumanomidou T, Sou YS, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J Biol Chem. 2008. Aug 15;283(33):22847–57. doi: 10.1074/jbc.M802182200. PubMed PMID: 18524774; eng. [DOI] [PubMed] [Google Scholar]
- 1021.Kabuta T, Furuta A, Aoki S, et al. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008. Aug 29;283(35):23731–8. doi: 10.1074/jbc.M801918200. PubMed PMID: 18550537; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1022.Saitoh Y, Fujikake N, Okamoto Y, et al. p62 plays a protective role in the autophagic degradation of polyglutamine protein oligomers in polyglutamine disease model flies. J Biol Chem. 2015. Jan 16;290(3):1442–53. doi: 10.1074/jbc.M114.590281. PubMed PMID: 25480790; PubMed Central PMCID: PMC4340391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1023.Ding WX, Ni HM, Gao W, et al. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol. 2007. Aug;171(2):513–24. PubMed PMID: 17620365; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1024.Iwata A, Riley BE, Johnston JA, et al. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005. Dec 2;280(48):40282–92. PubMed PMID: 16192271; eng. [DOI] [PubMed] [Google Scholar]
- 1025.Pandey UB, Nie Z, Batlevi Y, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007. Jun 14;447(7146):859–63. PubMed PMID: 17568747; eng. [DOI] [PubMed] [Google Scholar]
- 1026.Tomek K, Wagner R, Varga F, et al. Blockade of fatty acid synthase induces ubiquitination and degradation of phosphoinositide-3-kinase signaling proteins in ovarian cancer. Mol Cancer Res. 2011. Nov 28:1767–79. doi: 10.1158/1541-7786.MCR-10-0467. PubMed PMID: 21970855; Eng. [DOI] [PubMed] [Google Scholar]
- 1027.Fuertes G, Martin De Llano JJ, Villarroya A, et al. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem J. 2003. Oct 1;375(Pt 1):75–86. doi: 10.1042/BJ20030282. PubMed PMID: 12841850; PubMed Central PMCID: PMCPMC1223664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1028.Zimmermann AC, Zarei M, Eiselein S, et al. Quantitative proteomics for the analysis of spatio-temporal protein dynamics during autophagy. Autophagy. 2010. Nov;6(8):1009–16. PubMed PMID: 20603599; eng. [DOI] [PubMed] [Google Scholar]
- 1029.Kristensen AR, Schandorff S, Hoyer-Hansen M, et al. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008. Dec;7(12):2419–28. doi: 10.1074/mcp.M800184-MCP200. PubMed PMID: 18687634; eng. [DOI] [PubMed] [Google Scholar]
- 1030.Furuya N, Kanazawa T, Fujimura S, et al. Leupeptin-induced appearance of partial fragment of betaine homocysteine methyltransferase during autophagic maturation in rat hepatocytes. J Biochem (Tokyo). 2001. Feb;129(2):313–20. PubMed PMID: 11173534; eng. [DOI] [PubMed] [Google Scholar]
- 1031.Ueno T, Ishidoh K, Mineki R, et al. Autolysosomal membrane-associated betaine homocysteine methyltransferase. Limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem. 1999. May 21;274(21):15222–9. PubMed PMID: 10329731; eng. [DOI] [PubMed] [Google Scholar]
- 1032.Overbye A, Saetre F, Hagen LK, et al. Autophagic activity measured in whole rat hepatocytes as the accumulation of a novel BHMT fragment (p10), generated in amphisomes by the asparaginyl proteinase, legumain. Autophagy. 2011. Sep;7(9):1011–27. PubMed PMID: 21610319; PubMed Central PMCID: PMC3210315. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1033.Seglen PO, Overbye A, Saetre F.. Sequestration assays for mammalian autophagy. Methods Enzymol. 2009;452:63–83. doi: 10.1016/S0076-6879(08)03605-7. PubMed PMID: 19200876; eng. [DOI] [PubMed] [Google Scholar]
- 1034.Mercer CA, Kaliappan A, Dennis PB.. Macroautophagy-dependent, intralysosomal cleavage of a betaine homocysteine methyltransferase fusion protein requires stable multimerization. Autophagy. 2008. Feb;4(2):185–94. PubMed PMID: 18059170; eng. [DOI] [PubMed] [Google Scholar]
- 1035.Taylor GS, Long HM, Haigh TA, et al. A role for intercellular antigen transfer in the recognition of EBV-transformed B cell lines by EBV nuclear antigen-specific CD4+ T cells. J Iimmunol. 2006. Sep 15;177(6):3746–56. PubMed PMID: 16951335; eng. [DOI] [PubMed] [Google Scholar]
- 1036.Katayama H, Kogure T, Mizushima N, et al. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem biol. 2011. Aug 26;18(8):1042–52. doi: 10.1016/j.chembiol.2011.05.013. PubMed PMID: 21867919; eng. [DOI] [PubMed] [Google Scholar]
- 1037.An H, Harper JW.. Systematic analysis of ribophagy in human cells reveals bystander flux during selective autophagy. Nat Cell Biol. 2018. Feb;20(2):135–143. doi: 10.1038/s41556-017-0007-x. PubMed PMID: 29230017; PubMed Central PMCID: PMCPMC5786475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1038.Klionsky DJ, Emr SD.. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 1989. Aug;8(8):2241–50. PubMed PMID: 2676517; PubMed Central PMCID: PMC401154. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1039.Venerando R, Miotto G, Kadowaki M, et al. Multiphasic control of proteolysis by leucine and alanine in the isolated rat hepatocyte. Am J Physiol. 1994. Feb;266(2):C455–61. PubMed PMID: 8141260; eng. [DOI] [PubMed] [Google Scholar]
- 1040.Häussinger D, Hallbrucker C, vom Dahl S, et al. Cell swelling inhibits proteolysis in perfused rat liver. Biochem J. 1990. Nov 15;272(1):239–42. PubMed PMID: 2264828; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1041.vom Dahl S, Häussinger D.. Cell hydration and proteolysis control in liver. Biochem J. 1995. Dec 15;312:988–9. PubMed PMID: 8554549; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1042.Vincow ES, Merrihew G, Thomas RE, et al. The PINK1-Parkin pathway promotes both mitophagy and selective respiratory chain turnover in vivo. Proc Natl Acad Sci U S A. 2013. Apr 16;110(16):6400–5. doi: 10.1073/pnas.1221132110. PubMed PMID: 23509287; PubMed Central PMCID: PMC3631677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1043.Jaeger K, Sukseree S, Zhong S, et al. Cornification of nail keratinocytes requires autophagy for bulk degradation of intracellular proteins while sparing components of the cytoskeleton. Apoptosis. 2019. Feb;24(1–2):62–73. doi: 10.1007/s10495-018-1505-4. PubMed PMID: 30552537; PubMed Central PMCID: PMCPMC6373260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1044.Reggiori F, Monastyrska I, Shintani T, et al. The actin cytoskeleton is required for selective types of autophagy, but not nonspecific autophagy, in the yeast Saccharomyces cerevisiae [Research Support, N.I.H., Extramural]. Mol Biol Cell. 2005. Dec;16(12):5843–56. doi: 10.1091/mbc.E05-07-0629, PubMed PMID: 16221887; PubMed Central PMCID: PMC1289426. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1045.Manjithaya R, Jain S, Farre JC, et al. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol. 2010. Apr 19;189(2):303–10. doi: 10.1083/jcb.200909154. PubMed PMID: 20385774; PubMed Central PMCID: PMC2856896. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1046.Journo D, Mor A, Abeliovich H.. Aup1-mediated regulation of Rtg3 during mitophagy. J Biol Chem. 2009. Dec 18;284(51):35885–95. doi: 10.1074/jbc.M109.048140. PubMed PMID: 19840933; PubMed Central PMCID: PMC2791017. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1047.Kanki T, Klionsky DJ.. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008. Nov 21;283(47):32386–93. doi: 10.1074/jbc.M802403200. PubMed PMID: 18818209; PubMed Central PMCID: PMC2583303. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1048.Kanki T, Wang K, Baba M, et al. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol Biol Cell. 2009. Nov;20(22):4730–8. doi: 10.1091/mbc.E09-03-0225. PubMed PMID: 19793921; PubMed Central PMCID: PMC2777103. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1049.Kanki T, Wang K, Cao Y, et al. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009. Jul;17(1):98–109. doi: 10.1016/j.devcel.2009.06.014. PubMed PMID: 19619495; PubMed Central PMCID: PMC2746076. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1050.Okamoto K, Kondo-Okamoto N, Ohsumi Y.. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev Cell. 2009. Jul;17(1):87–97. doi: 10.1016/j.devcel.2009.06.013. PubMed PMID: 19619494; eng. [DOI] [PubMed] [Google Scholar]
- 1051.Kolitsida P, Abeliovich H.. Methods for studying mitophagy in yeast. Methods Mol Biol. 2019;1880:669–678. doi: 10.1007/978-1-4939-8873-0_44. PubMed PMID: 30610730. [DOI] [PubMed] [Google Scholar]
- 1052.Werner A, Herzog B, Voigt O, et al. NBR1 is involved in selective pexophagy in filamentous ascomycetes and can be functionally replaced by a tagged version of its human homolog. Autophagy. 2019. Jan;15(1):78–97. doi: 10.1080/15548627.2018.1507440. PubMed PMID: 30081713; PubMed Central PMCID: PMCPMC6287692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1053.Sakai Y, Koller A, Rangell LK, et al. Peroxisome degradation by microautophagy in Pichia pastoris: identification of specific steps and morphological intermediates. J Cell Biol. 1998. May 4;141(3):625–36. PubMed PMID: 9566964; PubMed Central PMCID: PMC2132739. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1054.Nazarko TY, Nicaud JM, Sibirny AA.. Observation of the Yarrowia lipolytica peroxisome-vacuole dynamics by fluorescence microscopy with a single filter set. Cell Biol Int. 2005;29:65–70. [DOI] [PubMed] [Google Scholar]
- 1055.Roetzer A, Gratz N, Kovarik P, et al. Autophagy supports Candida glabrata survival during phagocytosis. Cell Microbiol. 2010. Feb;12(2):199–216. doi: 10.1111/j.1462-5822.2009.01391.x. PubMed PMID: 19811500; PubMed Central PMCID: PMC2816358. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1056.Bormann C, Sahm H.. Degradation of microbodies in relation to activities of alcohol oxidase and catalase in Candida boidinii. Arch Microbiol. 1978. Apr 27;117(1):67–72. PubMed PMID: 678013; eng. [DOI] [PubMed] [Google Scholar]
- 1057.Clare DA, Duong MN, Darr D, et al. Effects of molecular oxygen on detection of superoxide radical with nitroblue tetrazolium and on activity stains for catalase. Anal Biochem 1984. Aug1;140(2):532–7. PubMed PMID: 6091498. [DOI] [PubMed] [Google Scholar]
- 1058.Vachova L, Kucerova H, Devaux F, et al. Metabolic diversification of cells during the development of yeast colonies. Environ Microbiol. 2009. Feb;11(2):494–504. doi: 10.1111/j.1462-2920.2008.01789.x. PubMed PMID: 19196279. [DOI] [PubMed] [Google Scholar]
- 1059.Hutchins MU, Veenhuis M, Klionsky DJ.. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999. Nov;112:4079–87. PubMed PMID: 10547367. [DOI] [PubMed] [Google Scholar]
- 1060.Mukaiyama H, Oku M, Baba M, et al. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002. Jan;7(1):75–90. PubMed PMID: 11856375; eng. [DOI] [PubMed] [Google Scholar]
- 1061.Tuttle DL, Dunn WA, Jr.. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995. Jan;108(Pt 1):25–35. PubMed PMID: 7738102; eng. [DOI] [PubMed] [Google Scholar]
- 1062.Nazarko TY, Huang J, Nicaud JM, et al. Trs85 is required for macroautophagy, pexophagy and cytoplasm to vacuole targeting in Yarrowia lipolytica and Saccharomyces cerevisiae. Autophagy. 2005. Apr;1(1):37–45. PubMed PMID: 16874038; PubMed Central PMCID: PMC1828867. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1063.Veenhuis M, Douma A, Harder W, et al. Degradation and turnover of peroxisomes in the yeast Hansenula polymorpha induced by selective inactivation of peroxisomal enzymes. Arch Microbiol. 1983. Jun;134(3):193–203. PubMed PMID: 6351780; eng. [DOI] [PubMed] [Google Scholar]
- 1064.Monosov EZ, Wenzel TJ, Luers GH, et al. Labeling of peroxisomes with green fluorescent protein in living P. pastoris cells. J Histochem Cytochem. 1996. Jun;44(6):581–9. PubMed PMID: 8666743; eng. [DOI] [PubMed] [Google Scholar]
- 1065.Wiemer EA, Wenzel T, Deerinck TJ, et al. Visualization of the peroxisomal compartment in living mammalian cells: dynamic behavior and association with microtubules. J Cell Biol 1997. Jan 13;136(1):71–80. PubMed PMID: 9008704; PubMed Central PMCID: PMC2132450. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1066.Monastyrska I, van der Heide M, Krikken AM, et al. Atg8 is essential for macropexophagy in Hansenula polymorpha. Traffic. 2005. Jan;6(1):66–74. doi: 10.1111/j.1600-0854.2004.00252.x. PubMed PMID: 15569246; eng. [DOI] [PubMed] [Google Scholar]
- 1067.Devenish RJ, Prescott M, Turcic K, et al. Monitoring organelle turnover in yeast using fluorescent protein tags. Methods Enzymol. 2008;451:109–31. doi: 10.1016/S0076-6879(08)03209-6. PubMed PMID: 19185717; eng. [DOI] [PubMed] [Google Scholar]
- 1068.Farre JC, Manjithaya R, Mathewson RD, et al. PpAtg30 tags peroxisomes for turnover by selective autophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Dev Cell. 2008. Mar;14(3):365–76. doi: 10.1016/j.devcel.2007.12.011. PubMed PMID: 18331717; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1069.He Y, Deng YZ, Naqvi NI.. Atg24-assisted mitophagy in the foot cells is necessary for proper asexual differentiation in Magnaporthe oryzae. Autophagy. 2013. Nov 1;9(11):1818–27. doi: 10.4161/auto.26057. PubMed PMID: 23958498. [DOI] [PubMed] [Google Scholar]
- 1070.Kiššová I, Deffieu M, Manon S, et al. Uth1p is involved in the autophagic degradation of mitochondria. J Biol Chem. 2004. Sep 10;279(37):39068–74. PubMed PMID: 15247238. [DOI] [PubMed] [Google Scholar]
- 1071.Kiššová I, Salin B, Schaeffer J, et al. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. [DOI] [PubMed] [Google Scholar]
- 1072.Kanki T, Klionsky DJ.. The molecular mechanism of mitochondria autophagy in yeast. Mol Microbiol. 2010. Feb;75(4):795–800. doi: 10.1111/j.1365-2958.2009.07035.x. PubMed PMID: 20487284; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1073.Tal R, Winter G, Ecker N, et al. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J Biol Chem. 2007. Feb 23;282(8):5617–24. doi: 10.1074/jbc.M605940200. PubMed PMID: 17166847; eng. [DOI] [PubMed] [Google Scholar]
- 1074.Abeliovich H. Stationary-phase mitophagy in respiring Saccharomyces cerevisiae. Antioxid Redox Signal. 2011. May 15;14(10):2003–11. doi: 10.1089/ars.2010.3807. PubMed PMID: 21194383; eng. [DOI] [PubMed] [Google Scholar]
- 1075.Eiyama A, Kondo-Okamoto N, Okamoto K.. Mitochondrial degradation during starvation is selective and temporally distinct from bulk autophagy in yeast. FEBS Lett. 2013. Jun 19;587(12):1787–92. doi: 10.1016/j.febslet.2013.04.030. PubMed PMID: 23660403. [DOI] [PubMed] [Google Scholar]
- 1076.Aksam EB, Koek A, Kiel JAKW, et al. A peroxisomal lon protease and peroxisome degradation by autophagy play key roles in vitality of Hansenula polymorpha cells. Autophagy. 2007. Mar-Apr;3(2):96–105. PubMed PMID: 17172804; eng. [DOI] [PubMed] [Google Scholar]
- 1077.Krick R, Muehe Y, Prick T, et al. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell. 2008. Oct;19(10):4492–505. doi: 10.1091/mbc.E08-04-0363. PubMed PMID: 18701704; PubMed Central PMCID: PMC2555948. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1078.Farre JC, Krick R, Subramani S, et al. Turnover of organelles by autophagy in yeast. Curr Opin Cell Biol. 2009. Aug;21(4):522–30. doi: 10.1016/j.ceb.2009.04.015. PubMed PMID: 19515549; PubMed Central PMCID: PMC2725217. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1079.Kvam E, Goldfarb DS.. Structure and function of nucleus-vacuole junctions: outer-nuclear-membrane targeting of Nvj1p and a role in tryptophan uptake. J Cell Sci. 2006. Sep 1;119(Pt 17):3622–33. doi: 10.1242/jcs.03093. PubMed PMID: 16912077; eng. [DOI] [PubMed] [Google Scholar]
- 1080.Millen JI, Krick R, Prick T, et al. Measuring piecemeal microautophagy of the nucleus in Saccharomyces cerevisiae [Research Support, U.S. Gov’t, Non-P.H.S.]. Autophagy. 2009. Jan;5(1):75–81. PubMed PMID: 18989095; eng. [DOI] [PubMed] [Google Scholar]
- 1081.Mijaljica D, Prescott M, Devenish RJ.. A late form of nucleophagy in Saccharomyces cerevisiae. PLoS One. 2012;7(6):e40013. doi: 10.1371/journal.pone.0040013. PubMed PMID: 22768199; PubMed Central PMCID: PMC3386919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1082.Shoji JY, Kikuma T, Arioka M, et al. Macroautophagy-mediated degradation of whole nuclei in the filamentous fungus Aspergillus oryzae. PLoS One. 2010;5(12):e15650. doi: 10.1371/journal.pone.0015650. PubMed PMID: 21187926; PubMed Central PMCID: PMC3004950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1083.He M, Kershaw MJ, Soanes DM, et al. Infection-associated nuclear degeneration in the rice blast fungus Magnaporthe oryzae requires non-selective macro-autophagy. PLoS One. 2012;7(3):e33270. doi: 10.1371/journal.pone.0033270. PubMed PMID: 22448240; PubMed Central PMCID: PMC3308974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1084.Maheshwari R. Nuclear behavior in fungal hyphae. FEMS Microbiol Lett. 2005. Aug 1;249(1):7–14. doi: 10.1016/j.femsle.2005.06.031. PubMed PMID: 16002240. [DOI] [PubMed] [Google Scholar]
- 1085.Shoji J-y, Craven KD.. Autophagy in basal hyphal compartments: A green strategy of great recyclers. Fungal Biol Rev. 2011;25:79–83. [Google Scholar]
- 1086.Voigt O, Poggeler S.. Autophagy genes Smatg8 and Smatg4 are required for fruiting-body development, vegetative growth and ascospore germination in the filamentous ascomycete Sordaria macrospora. Autophagy. 2013. Jan;9(1):33–49. doi: 10.4161/auto.22398. PubMed PMID: 23064313; PubMed Central PMCID: PMC3542216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1087.Yorimitsu T, Klionsky DJ.. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005. Apr;16(4):1593–605. doi: 10.1091/mbc.E04-11-1035. PubMed PMID: 15659643; PubMed Central PMCID: PMC1073644. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1088.Shintani T, Huang W-P, Stromhaug PE, et al. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002. Dec;3(6):825–37. PubMed PMID: 12479808; PubMed Central PMCID: PMC2737732. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1089.Abeliovich H, Darsow T, Emr SD.. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999. Nov 1;18(21):6005–16. doi: 10.1093/emboj/18.21.6005. PubMed PMID: 10545112; PubMed Central PMCID: PMC1171666. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1090.Muller M, Schmidt O, Angelova M, et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. eLife. 2015. Apr 22;4:e07736. doi: 10.7554/eLife.07736. PubMed PMID: 25902403; PubMed Central PMCID: PMCPMC4424281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1091.Abeliovich H, Zarei M, Rigbolt KT, et al. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat Commun. 2013;4:2789. doi: 10.1038/ncomms3789. PubMed PMID: 24240771; PubMed Central PMCID: PMC3909740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1092.Overbye A, Fengsrud M, Seglen PO.. Proteomic analysis of membrane-associated proteins from rat liver auto-phagosomes. Autophagy. 2007;3:300–322. [DOI] [PubMed] [Google Scholar]
- 1093.Eapen VV, Waterman DP, Bernard A, et al. A pathway of targeted autophagy is induced by DNA damage in budding yeast. Proc Natl Acad Sci U S A. 2017. Feb 14;114(7):E1158–E1167. doi: 10.1073/pnas.1614364114. PubMed PMID: 28154131; PubMed Central PMCID: PMCPMC5320992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1094.Petroi D, Popova B, Taheri-Talesh N, et al. Aggregate clearance of alpha-synuclein in Saccharomyces cerevisiae depends more on autophagosome and vacuole function than on the proteasome. J Biol Chem. 2012. Aug 10;287(33):27567–79. doi: 10.1074/jbc.M112.361865. PubMed PMID: 22722939; PubMed Central PMCID: PMC3431624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1095.Shahpasandzadeh H, Popova B, Kleinknecht A, et al. Interplay between sumoylation and phosphorylation for protection against alpha-synuclein inclusions. J Biol Chem. 2014. Nov 7;289(45):31224–40. doi: 10.1074/jbc.M114.559237. PubMed PMID: 25231978; PubMed Central PMCID: PMC4223324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1096.Kleinknecht A, Popova B, Lazaro DF, et al. C-terminal tyrosine residue modifications modulate the protective phosphorylation of serine 129 of alpha-synuclein in a yeast model of Parkinson’s disease. PLoS Genet. 2016. Jun;12(6):e1006098. doi: 10.1371/journal.pgen.1006098. PubMed PMID: 27341336; PubMed Central PMCID: PMCPMC4920419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1097.Wafa K, MacLean J, Zhang F, et al. Characterization of growth suppressive functions of a splice variant of cyclin D2. PLoS One. 2013;8(1):e53503. doi: 10.1371/journal.pone.0053503. PubMed PMID: 23326442; PubMed Central PMCID: PMC3542336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1098.Ju JS, Miller SE, Jackson E, et al. Quantitation of selective autophagic protein aggregate degradation in vitro and in vivo using luciferase reporters. Autophagy. 2009. May;5(4):511–9. PubMed PMID: 19305149; PubMed Central PMCID: PMC2992796. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1099.Chuang KH, Liang F, Higgins R, et al. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol Biol Cell. 2016. Jul 1;27(13):2025–36. doi: 10.1091/mbc.E16-01-0026. PubMed PMID: 27170182; PubMed Central PMCID: PMCPMC4927277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1100.Higgins R, Kabbaj MH, Hatcher A, et al. The absence of specific yeast heat-shock proteins leads to abnormal aggregation and compromised autophagic clearance of mutant Huntingtin proteins. PLoS One. 2018;13(1):e0191490. doi: 10.1371/journal.pone.0191490. PubMed PMID: 29346421; PubMed Central PMCID: PMCPMC5773196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1101.Hohn A, Sittig A, Jung T, et al. Lipofuscin is formed independently of macroautophagy and lysosomal activity in stress-induced prematurely senescent human fibroblasts. Free Radic Biol Med. 2012. Nov 1;53(9):1760–9. doi: 10.1016/j.freeradbiomed.2012.08.591. PubMed PMID: 22982048. [DOI] [PubMed] [Google Scholar]
- 1102.Jung T, Hohn A, Catalgol B, et al. Age-related differences in oxidative protein-damage in young and senescent fibroblasts. Arch Biochem Biophys. 2009. Mar 1;483(1):127–35. doi: 10.1016/j.abb.2008.12.007. PubMed PMID: 19135972. [DOI] [PubMed] [Google Scholar]
- 1103.Li L, Wang ZV, Hill JA, et al. New autophagy reporter mice reveal dynamics of proximal tubular autophagy. J Am Soc Nephrol. 2014. Feb;25(2):305–15. doi: 10.1681/ASN.2013040374. PubMed PMID: 24179166; PubMed Central PMCID: PMCPMC3904563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1104.Arotcarena ML, Bourdenx M, Dutheil N, et al. Transcription factor EB overexpression prevents neurodegeneration in experimental synucleinopathies. JCI Insight. 2019. Aug 22;4(16). doi: 10.1172/jci.insight.129719. PubMed PMID: 31434803; PubMed Central PMCID: PMCPMC6777809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1105.Decressac M, Mattsson B, Weikop P, et al. TFEB-mediated autophagy rescues midbrain dopamine neurons from alpha-synuclein toxicity. Proc Natl Acad Sci U S A. 2013. May 7;110(19):E1817–26. doi: 10.1073/pnas.1305623110. PubMed PMID: 23610405; PubMed Central PMCID: PMCPMC3651458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1106.Torra A, Parent A, Cuadros T, et al. Overexpression of TFEB drives a pleiotropic neurotrophic effect and prevents Parkinson’s disease-related neurodegeneration. Mol Ther. 2018. Jun 6;26(6):1552–1567. doi: 10.1016/j.ymthe.2018.02.022. PubMed PMID: 29628303; PubMed Central PMCID: PMCPMC5986717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1107.Nivon M, Fort L, Muller P, et al. NFkappaB is a central regulator of protein quality control in response to protein aggregation stresses via autophagy modulation. Mol Biol Cell. 2016. Jun 1;27(11):1712–27. doi: 10.1091/mbc.E15-12-0835. PubMed PMID: 27075172; PubMed Central PMCID: PMCPMC4884063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1108.Fuentealba RA, Marasa J, Diamond MI, et al. An aggregation sensing reporter identifies leflunomide and teriflunomide as polyglutamine aggregate inhibitors. Hum Mol Genet 2012;21:664–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1109.Al Rawi S, Louvet-Vallee S, Djeddi A, et al. Allophagy: a macroautophagic process degrading spermatozoid-inherited organelles. Autophagy. 2012. Mar;8(3):421–3. doi: 10.4161/auto.19242. PubMed PMID: 22361582; PubMed Central PMCID: PMCPMC3337843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1110.Sato M, Sato K.. Maternal inheritance of mitochondrial DNA: degradation of paternal mitochondria by allogeneic organelle autophagy, allophagy. Autophagy. 2012. Mar;8(3):424–5. doi: 10.4161/auto.19243. PubMed PMID: 22302002. [DOI] [PubMed] [Google Scholar]
- 1111.Al Rawi S, Louvet-Vallee S, Djeddi A, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011. Oct 27;334:1144–7. doi: 10.1126/science.1211878. PubMed PMID: 22033522; Eng. [DOI] [PubMed] [Google Scholar]
- 1112.Sato M, Sato K.. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011. Nov 25;334(6059):1141–4. doi: 10.1126/science.1210333. PubMed PMID: 21998252; eng. [DOI] [PubMed] [Google Scholar]
- 1113.Kim I, Rodriguez-Enriquez S, Lemasters JJ.. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007. Jun 15;462(2):245–53. doi: 10.1016/j.abb.2007.03.034. PubMed PMID: 17475204; PubMed Central PMCID: PMC2756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1114.Djeddi A, Al Rawi S, Deuve JL, et al. Sperm-inherited organelle clearance in C. elegans relies on LC3-dependent autophagosome targeting to the pericentrosomal area. Development. 2015. May 1;142(9):1705–16. doi: 10.1242/dev.117879. PubMed PMID: 25922527. [DOI] [PubMed] [Google Scholar]
- 1115.Manil-Segalen M, Lefebvre C, Jenzer C, et al. The C. elegans LC3 acts downstream of GABARAP to degrade auto-phagosomes by interacting with the HOPS subunit VPS39. Dev Cell. 2014. Jan 13;28(1):43–55. doi: 10.1016/j.devcel.2013.11.022. PubMed PMID: 24374177. [DOI] [PubMed] [Google Scholar]
- 1116.Sato M, Sato K, Tomura K, et al. The autophagy receptor ALLO-1 and the IKKE-1 kinase control clearance of paternal mitochondria in Caenorhabditis elegans. Nat Cell Biol. 2018. Jan;20(1):81–91. doi: 10.1038/s41556-017-0008-9. PubMed PMID: 29255173. [DOI] [PubMed] [Google Scholar]
- 1117.Molina P, Lim Y, Boyd L.. Ubiquitination is required for the initial removal of paternal organelles in C. elegans. Dev Biol. 2019. Sep 15;453(2):168–179. doi: 10.1016/j.ydbio.2019.05.015. PubMed PMID: 31153831; PubMed Central PMCID: PMCPMC6685074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1118.Zhou Q, Li H, Li H, et al. Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science. 2016. Jul 22;353(6297):394–9. doi: 10.1126/science.aaf4777. PubMed PMID: 27338704; PubMed Central PMCID: PMCPMC5469823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1119.Wang Y, Zhang Y, Chen L, et al. Kinetics and specificity of paternal mitochondrial elimination in Caenorhabditis elegans. Nat Commun. 2016. Sep 1;7:12569. doi: 10.1038/ncomms12569. PubMed PMID: 27581092; PubMed Central PMCID: PMCPMC5025750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1120.Ding WX, Yin XM.. Mitophagy: mechanisms, pathophysiological roles, and analysis. Biol Chem. 2012. Jul;393(7):547–64. doi: 10.1515/hsz-2012-0119. PubMed PMID: 22944659; PubMed Central PMCID: PMC3630798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1121.Ding WX, Li M, Chen X, et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010. Nov;139(5):1740–52. doi: 10.1053/j.gastro.2010.07.041. PubMed PMID: 20659474; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1122.Dong H, Cheung SH, Liang Y, et al. “Stainomics”: identification of mitotracker labeled proteins in mammalian cells. Electrophoresis. 2013. Jul;34(13):1957–64. doi: 10.1002/elps.201200557. PubMed PMID: 23595693. [DOI] [PubMed] [Google Scholar]
- 1123.Mauro-Lizcano M, Esteban-Martinez L, Seco E, et al. New method to assess mitophagy flux by flow cytometry. Autophagy. 2015;11(5):833–43. doi: 10.1080/15548627.2015.1034403. PubMed PMID: 25945953; PubMed Central PMCID: PMC4509449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1124.Presley AD, Fuller KM, Arriaga EA.. MitoTracker Green labeling of mitochondrial proteins and their subsequent analysis by capillary electrophoresis with laser-induced fluorescence detection. J chromatograp B Anal Technol Biomed Life Sci 2003. Aug5;793(1):141–50. PubMed PMID: 12880861. [DOI] [PubMed] [Google Scholar]
- 1125.Keij JF, Bell-Prince C, Steinkamp JA.. Staining of mitochondrial membranes with 10-nonyl acridine orange, MitoFluor Green, and MitoTracker Green is affected by mitochondrial membrane potential altering drugs. Cytometry. 2000. Mar1;39(3):203–10. PubMed PMID: 10685077. [DOI] [PubMed] [Google Scholar]
- 1126.Poot M, Zhang YZ, Kramer JA, et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J Histochem Cytochem Off J Histochem Soc 1996. Dec;44(12):1363–72. PubMed PMID: 8985128; [DOI] [PubMed] [Google Scholar]
- 1127.Canonico B, Cesarini E, Salucci S, et al. Defective autophagy, mitochondrial clearance and lipophagy in niemann-pick type B lymphocytes. PLoS One. 2016;11(10):e0165780. doi: 10.1371/journal.pone.0165780. PubMed PMID: 27798705; PubMed Central PMCID: PMCPMC5087958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1128.Geisler S, Holmstrom KM, Skujat D, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010. Feb;12(2):119–31. doi: 10.1038/ncb2012. PubMed PMID: 20098416; eng. [DOI] [PubMed] [Google Scholar]
- 1129.Geisler S, Holmstrom KM, Treis A, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010. Oct;6(7):871–8. PubMed PMID: 20798600; eng. [DOI] [PubMed] [Google Scholar]
- 1130.Zhou Y, Long Q, Wu H, et al. Topology-dependent, bifurcated mitochondrial quality control under starvation. Autophagy. 2019. Jul 4:1–13. doi: 10.1080/15548627.2019.1634944. PubMed PMID: 31234709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1131.Demers-Lamarche J, Guillebaud G, Tlili M, et al. Loss of Mitochondrial Function Impairs Lysosomes. J Biol Chem. 2016. May 6;291(19):10263–76. doi: 10.1074/jbc.M115.695825. PubMed PMID: 26987902; PubMed Central PMCID: PMCPMC4858975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1132.Ouellet M, Guillebaud G, Gervais V, et al. A novel algorithm identifies stress-induced alterations in mitochondrial connectivity and inner membrane structure from confocal images. PLoS Comput Biol. 2017. Jun;13(6):e1005612. doi: 10.1371/journal.pcbi.1005612. PubMed PMID: 28640814; PubMed Central PMCID: PMCPMC5501662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1133.Zhu JH, Guo F, Shelburne J, et al. Localization of phosphorylated ERK/MAP kinases to mitochondria and auto-phagosomes in Lewy body diseases. Brain Pathol. 2003. Oct;13(4):473–81. doi: 10.1111/j.1750-3639.2003.tb00478.x. PubMed PMID: 14655753; PubMed Central PMCID: PMCPMC1911206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1134.Diot A, Hinks-Roberts A, Lodge T, et al. A novel quantitative assay of mitophagy: Combining high content fluorescence microscopy and mitochondrial DNA load to quantify mitophagy and identify novel pharmacological tools against pathogenic heteroplasmic mtDNA. Pharmacol Res. 2015. Jul 18;100:24–35. doi: 10.1016/j.phrs.2015.07.014. PubMed PMID: 26196248. [DOI] [PubMed] [Google Scholar]
- 1135.Ding WX, Guo F, Ni HM, et al. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem. 2012. Dec 7;287(50):42379–88. doi: 10.1074/jbc.M112.413682. PubMed PMID: 23095748; PubMed Central PMCID: PMC3516781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1136.Ding WX, Li M, Biazik JM, et al. Electron microscopic analysis of a spherical mitochondrial structure. J Biol Chem. 2012. Dec 7;287(50):42373–8. doi: 10.1074/jbc.M112.413674. PubMed PMID: 23093403; PubMed Central PMCID: PMC3516780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1137.Dagda RK, Cherra SJ, III, Kulich SM, et al. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009. May 15;284(20):13843–55. doi:M808515200[pii] doi: 10.1074/jbc.M808515200. PubMed PMID: 19279012; PubMed Central PMCID: PMC2679485. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1138.Dagda RK, Zhu J, Kulich SM, et al. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson’s disease. Autophagy. 2008. Aug;4(6):770–82. PubMed PMID: 18594198; PubMed Central PMCID: PMC2574804. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1139.Boya P, Gonzalez-Polo RA, Casares N, et al. Inhibition of macroautophagy triggers apoptosis. Mol Cell Biol 2005. Feb;25(3):1025–40. PubMed PMID: 15657430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1140.Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015. Aug 20;524(7565):309–314. doi: 10.1038/nature14893. PubMed PMID: 26266977; PubMed Central PMCID: PMCPMC5018156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1141.Um JH, Kim YY, Finkel T, et al. Sensitive measurement of mitophagy by flow cytometry using the pH-dependent fluorescent reporter mt-keima. J vis exp. 2018. Aug 12(138). doi: 10.3791/58099. PubMed PMID: 30148491; PubMed Central PMCID: PMCPMC6126785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1142.Wong YC, Ysselstein D, Krainc D.. Mitochondria-lysosome contacts regulate mitochondrial fission via RAB7 GTP hydrolysis. Nature. 2018. Feb 15;554(7692):382–386. doi: 10.1038/nature25486. PubMed PMID: 29364868; PubMed Central PMCID: PMCPMC6209448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1143.Wong YC, Kim S, Peng W, et al. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 2019. Jun;29(6):500–513. doi: 10.1016/j.tcb.2019.02.004. PubMed PMID: 30898429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1144.Wong YC, Peng W, Krainc D.. Lysosomal regulation of inter-mitochondrial contact fate and motility in Charcot-Marie-Tooth type 2. Dev Cell. 2019. Aug 5;50(3):339–354 e4. doi: 10.1016/j.devcel.2019.05.033. PubMed PMID: 31231042; PubMed Central PMCID: PMCPMC6726396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1145.Sun N, Yun J, Liu J, et al. Measuring in vivo mitophagy. Mol Cell. 2015. Nov 19;60(4):685–96. doi: 10.1016/j.molcel.2015.10.009. PubMed PMID: 26549682; PubMed Central PMCID: PMCPMC4656081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1146.McWilliams TG, Prescott AR, Villarejo-Zori B, et al. A comparative map of macroautophagy and mitophagy in the vertebrate eye. Autophagy. 2019. Jul;15(7):1296–1308. doi: 10.1080/15548627.2019.1580509. PubMed PMID: 30786807; PubMed Central PMCID: PMCPMC6613837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1147.Cornelissen T, Vilain S, Vints K, et al. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. eLife. 2018. May 29;7. doi: 10.7554/eLife.35878. PubMed PMID: 29809156; PubMed Central PMCID: PMCPMC6008047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1148.Kim YY, Um JH, Yoon JH, et al. Assessment of mitophagy in mt-Keima Drosophila revealed an essential role of the PINK1-Parkin pathway in mitophagy induction in vivo. FASEB J. 2019. Sep;33(9):9742–9751. doi: 10.1096/fj.201900073R. PubMed PMID: 31120803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1149.Reipert S, Berry J, Hughes MF, et al. Changes of mitochondrial mass in the hemopoietic stem cell line FDCP-mix after treatment with etoposide: a correlative study by multiparameter flow cytometry and confocal and electron microscopy. Exp Cell Res. 1995. Dec;221(2):281–8. doi: 10.1006/excr.1995.1376. PubMed PMID: 7493625. [DOI] [PubMed] [Google Scholar]
- 1150.Wilfinger N, Austin S, Scheiber-Mojdehkar B, et al. Novel p53-dependent anticancer strategy by targeting iron signaling and BNIP3L-induced mitophagy. Oncotarget. 2016. Jan 12;7(2):1242–61. doi: 10.18632/oncotarget.6233. PubMed PMID: 26517689; PubMed Central PMCID: PMCPMC4811457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1151.Esteban-Martinez L, Sierra-Filardi E, McGreal RS, et al. Programmed mitophagy is essential for the glycolytic switch during cell differentiation. EMBO J. 2017. Jun 14;36(12):1688–1706. doi: 10.15252/embj.201695916. PubMed PMID: 28465321; PubMed Central PMCID: PMCPMC5470043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1152.Valente AJ, Maddalena LA, Robb EL, et al. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017. Apr;119(3):315–326. doi: 10.1016/j.acthis.2017.03.001. PubMed PMID: 28314612. [DOI] [PubMed] [Google Scholar]
- 1153.Tanaka A, Cleland MM, Xu S, et al. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J Cell Biol. 2010. Dec 27;191(7):1367–80. doi: 10.1083/jcb.201007013. PubMed PMID: 21173115; PubMed Central PMCID: PMC3010068. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1154.Yoshii SR, Kishi C, Ishihara N, et al. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011. Jun 3;286(22):19630–40. doi: 10.1074/jbc.M110.209338. PubMed PMID: 21454557; PubMed Central PMCID: PMC3103342. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1155.Shiba-Fukushima K, Imai Y, Yoshida S, et al. PINK1-mediated phosphorylation of the Parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Sci Rep. 2012;2:1002. doi: 10.1038/srep01002. PubMed PMID: 23256036; PubMed Central PMCID: PMC3525937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1156.Giulivi C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA. 2010. Dec 1;304(21):2389–96. doi: 10.1001/jama.2010.1706. PubMed PMID: 21119085; PubMed Central PMCID: PMCPMC3915058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1157.Amadoro G, Corsetti V, Florenzano F, et al. AD-linked, toxic NH2 human tau affects the quality control of mitochondria in neurons. Neurobiol Dis. 2014. Feb;62:489–507. doi: 10.1016/j.nbd.2013.10.018. PubMed PMID: 24411077. [DOI] [PubMed] [Google Scholar]
- 1158.Napoli E, Song G, Wong S, et al. Altered bioenergetics in primary dermal fibroblasts from adult carriers of the FMR1 premutation before the onset of the neurodegenerative disease fragile X-associated tremor/ataxia syndrome. Cerebellum. 2016. Oct;15(5):552–64. doi: 10.1007/s12311-016-0779-8. PubMed PMID: 27089882; PubMed Central PMCID: PMCPMC5014718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1159.Fujisawa Y, Napoli E, Wong S, et al. Impact of a novel homozygous mutation in nicotinamide nucleotide transhydrogenase on mitochondrial DNA integrity in a case of familial glucocorticoid deficiency. BBA Clin. 2015. Jun 1;3:70–78. doi: 10.1016/j.bbacli.2014.12.003. PubMed PMID: 26309815; PubMed Central PMCID: PMCPMC4545511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1160.Park YS, Choi SE, Koh HC.. PGAM5 regulates PINK1/Parkin-mediated mitophagy via DRP1 in CCCP-induced mitochondrial dysfunction. Toxicol Lett. 2018. Mar 1;284:120–128. doi: 10.1016/j.toxlet.2017.12.004. PubMed PMID: 29241732. [DOI] [PubMed] [Google Scholar]
- 1161.Lu X, Altshuler-Keylin S, Wang Q, et al. Mitophagy controls beige adipocyte maintenance through a Parkin-dependent and UCP1-independent mechanism. Sci Signal. 2018. Apr 24;11(527). doi: 10.1126/scisignal.aap8526. PubMed PMID: 29692364; PubMed Central PMCID: PMCPMC6410368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1162.Chang TK, Shravage BV, Hayes SD, et al. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013. Sep;15(9):1067–78. doi: 10.1038/ncb2804. PubMed PMID: 23873149; PubMed Central PMCID: PMC3762904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1163.Hsieh CH, Shaltouki A, Gonzalez AE, et al. Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic parkinson’s disease. Cell Stem Cell. 2016. Dec 1;19(6):709–724. doi: 10.1016/j.stem.2016.08.002. PubMed PMID: 27618216; PubMed Central PMCID: PMCPMC5135570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1164.Monzio Compagnoni G, Kleiner G, Bordoni A, et al. Mitochondrial dysfunction in fibroblasts of Multiple System Atrophy. Biochim Biophys Acta Mol Basis Dis. 2018. Dec;1864(12):3588–3597. doi: 10.1016/j.bbadis.2018.09.018. PubMed PMID: 30254015. [DOI] [PubMed] [Google Scholar]
- 1165.Pickrell AM, Youle RJ.. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015. Jan 21;85(2):257–73. doi: 10.1016/j.neuron.2014.12.007. PubMed PMID: 25611507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1166.East DA, Fagiani F, Crosby J, et al. PMI: a DeltaPsim independent pharmacological regulator of mitophagy. Chem Biol. 2014. Nov 20;21(11):1585–96. doi: 10.1016/j.chembiol.2014.09.019. PubMed PMID: 25455860; PubMed Central PMCID: PMC4245710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1167.Vannini N, Campos V, Girotra M, et al. The NAD-booster nicotinamide riboside potently stimulates hematopoiesis through increased mitochondrial clearance. Cell Stem Cell. 2019. Mar 7;24(3):405–418 e7. doi: 10.1016/j.stem.2019.02.012. PubMed PMID: 30849366. [DOI] [PubMed] [Google Scholar]
- 1168.Fang EF, Kassahun H, Croteau DL, et al. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016. Oct 11;24(4):566–581. doi: 10.1016/j.cmet.2016.09.004. PubMed PMID: 27732836; PubMed Central PMCID: PMCPMC5777858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1169.Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012. Feb;14(2):177–85. doi: 10.1038/ncb2422. PubMed PMID: 22267086. [DOI] [PubMed] [Google Scholar]
- 1170.Bellot G, Garcia-Medina R, Gounon P, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009. May;29(10):2570–81. doi: 10.1128/MCB.00166-09. PubMed PMID: 19273585; PubMed Central PMCID: PMC2682037. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1171.Zhang J, Ney PA.. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009. Jul;16(7):939–46. doi: 10.1038/cdd.2009.16. PubMed PMID: 19229244; PubMed Central PMCID: PMCPMC2768230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1172.Schiavi A, Maglioni S, Palikaras K, et al. Iron-starvation-induced mitophagy mediates lifespan extension upon mitochondrial stress in C. elegans. Curr Biol. 2015. Jul 20;25(14):1810–22. doi: 10.1016/j.cub.2015.05.059. PubMed PMID: 26144971. [DOI] [PubMed] [Google Scholar]
- 1173.Yang JY, Yang WY.. Spatiotemporally controlled initiation of Parkin-mediated mitophagy within single cells. Autophagy. 2011. Oct;7(10):1230–8. doi: 10.4161/auto.7.10.16626. PubMed PMID: 22011618; eng. [DOI] [PubMed] [Google Scholar]
- 1174.Strappazzon F, Nazio F, Corrado M, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2014. Sep 12;22:419–32. doi: 10.1038/cdd.2014.139. PubMed PMID: 25215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1175.Chakraborty J, von Stockum S, Marchesan E, et al. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol Med. 2018. Nov;10(11). doi: 10.15252/emmm.201809014. PubMed PMID: 30249595; PubMed Central PMCID: PMCPMC6220287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1176.Wei Y, Chiang WC, Sumpter R, Jr., et al. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell. 2017. Jan 12;168(1–2):224–238 e10. doi: 10.1016/j.cell.2016.11.042. PubMed PMID: 28017329; PubMed Central PMCID: PMCPMC5235968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1177.Ferree AW, Trudeau K, Zik E, et al. MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy. 2013. Nov 1;9(11):1887–96. doi: 10.4161/auto.26503. PubMed PMID: 24149000; PubMed Central PMCID: PMCPMC4028338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1178.Hernandez G, Thornton C, Stotland A, et al. MitoTimer: a novel tool for monitoring mitochondrial turnover. Autophagy. 2013. Nov 1;9(11):1852–61. doi: 10.4161/auto.26501. PubMed PMID: 24128932; PubMed Central PMCID: PMCPMC4028337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1179.Laker RC, Xu P, Ryall KA, et al. A novel MitoTimer reporter gene for mitochondrial content, structure, stress, and damage in vivo. J Biol Chem. 2014. Apr 25;289(17):12005–15. doi: 10.1074/jbc.M113.530527. PubMed PMID: 24644293; PubMed Central PMCID: PMCPMC4002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1180.Martin-Maestro P, Gargini R, Garcia E, et al. Slower dynamics and aged mitochondria in sporadic Alzheimer’s disease. Oxid Med Cell Longev. 2017;2017:9302761. doi: 10.1155/2017/9302761. PubMed PMID: 29201274; PubMed Central PMCID: PMCPMC5672147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1181.Laker RC, Drake JC, Wilson RJ, et al. Ampk phosphorylation of Ulk1 is required for targeting of mitochondria to lysosomes in exercise-induced mitophagy. Nat Commun. 2017. Sep 15;8(1):548. doi: 10.1038/s41467-017-00520-9. PubMed PMID: 28916822; PubMed Central PMCID: PMCPMC5601463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1182.Call JA, Wilson RJ, Laker RC, et al. Ulk1-mediated autophagy plays an essential role in mitochondrial remodeling and functional regeneration of skeletal muscle. Am J Physiol Cell Physiol. 2017. Jun 1;312(6):C724–C732. doi: 10.1152/ajpcell.00348.2016. PubMed PMID: 28356270; PubMed Central PMCID: PMCPMC5494591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1183.Perry HM, Huang L, Wilson RJ, et al. Dynamin-related protein 1 deficiency promotes recovery from AKI. J Am Soc Nephrol. 2018. Jan;29(1):194–206. doi: 10.1681/ASN.2017060659. PubMed PMID: 29084809; PubMed Central PMCID: PMCPMC5748924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1184.Wilson RJ, Drake JC, Cui D, et al. Mitochondrial protein S-nitrosation protects against ischemia reperfusion-induced denervation at neuromuscular junction in skeletal muscle. Free Radic Biol Med. 2018. Mar;117:180–190. doi: 10.1016/j.freeradbiomed.2018.02.006. PubMed PMID: 29432799; PubMed Central PMCID: PMCPMC5896769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1185.Wilson RJ, Drake JC, Cui D, et al. Voluntary running protects against neuromuscular dysfunction following hindlimb ischemia-reperfusion in mice. J Appl Physiol (1985). 2019 Jan 1;126(1):193–201. doi: 10.1152/japplphysiol.00358.2018. PubMed PMID: 30433863; PubMed Central PMCID: PMCPMC6383643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1186.Wilson RJ, Drake JC, Cui D, et al. Conditional MitoTimer reporter mice for assessment of mitochondrial structure, oxidative stress, and mitophagy. Mitochondrion. 2019. Jan;44:20–26. doi: 10.1016/j.mito.2017.12.008. PubMed PMID: 29274400; PubMed Central PMCID: PMCPMC6387589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1187.Xu P, Damschroder D, Zhang M, et al. Atg2, Atg9 and Atg18 in mitochondrial integrity, cardiac function and healthspan in Drosophila. J Mol Cell Cardiol. 2019. Feb;127:116–124. doi: 10.1016/j.yjmcc.2018.12.006. PubMed PMID: 30571977; PubMed Central PMCID: PMCPMC6533900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1188.Chu CT. Mechanisms of selective autophagy and mitophagy: Implications for neurodegenerative diseases. Neurobiol Dis. 2019. Feb;122:23–34. doi: 10.1016/j.nbd.2018.07.015. PubMed PMID: 30030024; PubMed Central PMCID: PMCPMC6396690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1189.Kagan VE, Jiang J, Huang Z, et al. NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death Differ. 2016. Jul;23(7):1140–51. doi: 10.1038/cdd.2015.160. PubMed PMID: 26742431; PubMed Central PMCID: PMCPMC4946882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1190.Anton Z, Landajuela A, Hervas JH, et al. Human Atg8-cardiolipin interactions in mitophagy: Specific properties of LC3B, GABARAPL2 and GABARAP. Autophagy. 2016. Dec;12(12):2386–2403. doi: 10.1080/15548627.2016.1240856. PubMed PMID: 27764541; PubMed Central PMCID: PMCPMC5172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1191.Fiesel FC, Ando M, Hudec R, et al. (Patho-)physiological relevance of PINK1-dependent ubiquitin phosphorylation. EMBO Rep. 2015. Jul 10;16:1114–30. doi: 10.15252/embr.201540514. PubMed PMID: 26162776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1192.Wang L, Cho YL, Tang Y, et al. PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy. Cell Res. 2018. Aug;28(8):787–802. doi: 10.1038/s41422-018-0056-0. PubMed PMID: 29934616; PubMed Central PMCID: PMCPMC6082900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1193.Shiba-Fukushima K, Ishikawa KI, Inoshita T, et al. Evidence that phosphorylated ubiquitin signaling is involved in the etiology of Parkinson’s disease. Hum Mol Genet. 2017. Aug 15;26(16):3172–3185. doi: 10.1093/hmg/ddx201. PubMed PMID: 28541509. [DOI] [PubMed] [Google Scholar]
- 1194.Herhaus L, Dikic I.. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015. Aug 12. doi: 10.15252/embr.201540891. PubMed PMID: 26268526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1195.Koyano F, Okatsu K, Kosako H, et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 2014. Jun 5;510(7503):162–6. doi: 10.1038/nature13392. PubMed PMID: 24784582. [DOI] [PubMed] [Google Scholar]
- 1196.Chen G, Han Z, Feng D, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol Cell. 2014. May 8;54(3):362–77. doi: 10.1016/j.molcel.2014.02.034. PubMed PMID: 24746696. [DOI] [PubMed] [Google Scholar]
- 1197.Wu H, Xue D, Chen G, et al. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014. Oct 1;10(10):1712–25. doi: 10.4161/auto.29568. PubMed PMID: 25126723; PubMed Central PMCID: PMCPMC4198357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1198.Banerjee K, Munshi S, Xu H, et al. Mild mitochondrial metabolic deficits by alpha-ketoglutarate dehydrogenase inhibition cause prominent changes in intracellular autophagic signaling: Potential role in the pathobiology of Alzheimer’s disease. Neurochem Int. 2016. Jun;96:32–45. . PubMed PMID: 26923918; PubMed Central PMCID: PMCPMC4860123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1199.Lemasters JJ. Variants of mitochondrial autophagy: Types 1 and 2 mitophagy and micromitophagy (Type 3). Redox Biol. 2014;2:749–54. doi: 10.1016/j.redox.2014.06.004. PubMed PMID: 25009776; PubMed Central PMCID: PMC4085350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1200.Manjithaya R, Nazarko TY, Farre JC, et al. Molecular mechanism and physiological role of pexophagy. FEBS Lett. 2010. Apr 2;584(7):1367–73. doi: 10.1016/j.febslet.2010.01.019. PubMed PMID: 20083110; PubMed Central PMCID: PMC2843806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1201.Till A, Lakhani R, Burnett SF, et al. Pexophagy: the selective degradation of peroxisomes. Int J Cell Biol. 2012;2012:512721. doi: 10.1155/2012/512721. PubMed PMID: 22536249; PubMed Central PMCID: PMC3320016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1202.Michalik L, Auwerx J, Berger JP, et al. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev. 2006. Dec;58(4):726–41. doi: 10.1124/pr.58.4.5. PubMed PMID: 17132851. [DOI] [PubMed] [Google Scholar]
- 1203.D’Orio B, Fracassi A, Ceru MP, et al. Targeting PPARalpha in Alzheimer’s Disease. Curr Alzheimer Res. 2018. Feb 22;15(4):345–354. doi: 10.2174/1567205014666170505094549. PubMed PMID: 28474570. [DOI] [PubMed] [Google Scholar]
- 1204.Sargent G, van Zutphen T, Shatseva T, et al. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J Cell Biol. 2016. Sep 12;214(6):677–90. doi: 10.1083/jcb.201511034. PubMed PMID: 27597759; PubMed Central PMCID: PMCPMC5021090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1205.Riccio V, Demers N, Hua R, et al. Deubiquitinating enzyme USP30 maintains basal peroxisome abundance by regulating pexophagy. J Cell Biol. 2019. Mar 4;218(3):798–807. doi: 10.1083/jcb.201804172. PubMed PMID: 30700497; PubMed Central PMCID: PMCPMC6400567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1206.Marcassa E, Kallinos A, Jardine J, et al. Dual role of USP30 in controlling basal pexophagy and mitophagy. EMBO Rep. 2018. Jul;19(7). doi: 10.15252/embr.201745595. PubMed PMID: 29895712; PubMed Central PMCID: PMCPMC6030704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1207.Walter KM, Schonenberger MJ, Trotzmuller M, et al. Hif-2alpha promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab. 2014. Nov 4;20(5):882–97. doi: 10.1016/j.cmet.2014.09.017. PubMed PMID: 25440060. [DOI] [PubMed] [Google Scholar]
- 1208.Alexander A, Cai SL, Kim J, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci USA 2010. Feb 16;107:4153–8. doi: 0913860107 [pii] doi: 10.1073/pnas.0913860107. PubMed PMID: 20160076; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1209.Tripathi DN, Chowdhury R, Trudel LJ, et al. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proc Natl Acad Sci U S A. 2013. Aug 6;110(32):E2950–7. doi: 10.1073/pnas.1307736110. PubMed PMID: 23878245; PubMed Central PMCID: PMC3740898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1210.Zhang J, Kim J, Alexander A, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat Cell Biol. 2013. Oct;15(10):1186–96. doi: 10.1038/ncb2822. PubMed PMID: 23955302; PubMed Central PMCID: PMC3789865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1211.Zhang J, Tripathi DN, Jing J, et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015. Sep 7;17:1259–69. doi: 10.1038/ncb3230. PubMed PMID: 26344566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1212.D’Eletto M, Farrace MG, Rossin F, et al. Type 2 transglutaminase is involved in the autophagy-dependent clearance of ubiquitinated proteins. Cell Death Differ. 2012. Jul;19(7):1228–38. doi: 10.1038/cdd.2012.2. PubMed PMID: 22322858; PubMed Central PMCID: PMC3374086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1213.Luiken JJ, van den Berg M, Heikoop JC, et al. Autophagic degradation of peroxisomes in isolated rat hepatocytes. FEBS Lett. 1992. Jun8;304(1):93–7. PubMed PMID: 1618306. [DOI] [PubMed] [Google Scholar]
- 1214.Nardacci R, Sartori C, Stefanini S.. Selective autophagy of clofibrate-induced rat liver peroxisomes. Cytochemistry and immunocytochemistry on tissue specimens and on fractions obtained by Nycodenz density gradient centrifugation. Cell Mol Biol. 2000. Nov;46(7):1277–90. PubMed PMID: 11075957. [PubMed] [Google Scholar]
- 1215.Yokota S. Formation of auto-phagosomes during degradation of excess peroxisomes induced by administration of dioctyl phthalate [Research Support, Non-U.S. Gov’t]. Eur J Cell Biol. 1993. Jun;61(1):67–80. PubMed PMID: 8223709; eng. [PubMed] [Google Scholar]
- 1216.Huybrechts SJ, Van Veldhoven PP, Brees C, et al. Peroxisome dynamics in cultured mammalian cells [Research Support, Non-U.S. Gov’t]. Traffic. 2009. Nov;10(11):1722–33. doi: 10.1111/j.1600-0854.2009.00970.x. PubMed PMID: 19719477; eng. [DOI] [PubMed] [Google Scholar]
- 1217.Jiang L, Hara-Kuge S, Yamashita S, et al. Peroxin Pex14p is the key component for coordinated autophagic degradation of mammalian peroxisomes by direct binding to LC3-II. Genes Cells. 2015. Jan;20(1):36–49. doi: 10.1111/gtc.12198. PubMed PMID: 25358256. [DOI] [PubMed] [Google Scholar]
- 1218.Fujiki Y, Okumoto K, Mukai S, et al. Peroxisome biogenesis in mammalian cells. Front Physiol. 2014;5:307. doi: 10.3389/fphys.2014.00307. PubMed PMID: 25177298; PubMed Central PMCID: PMCPMC4133648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1219.Hara-Kuge S, Fujiki Y.. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes [Research Support, Non-U.S. Gov’t]. Exp Cell Res. 2008. Nov 15;314(19):3531–41. doi: 10.1016/j.yexcr.2008.09.015. PubMed PMID: 18848543; eng. [DOI] [PubMed] [Google Scholar]
- 1220.Yamashita S, Abe K, Tatemichi Y, et al. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy. 2014. Sep;10(9):1549–64. doi: 10.4161/auto.29329. PubMed PMID: 25007327; PubMed Central PMCID: PMCPMC4206534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1221.Yamashita S, Fujiki Y.. Assessing Pexophagy in Mammalian Cells. Methods Mol Biol. 2017;1595:243–248. doi: 10.1007/978-1-4939-6937-1_23. PubMed PMID: 28409468. [DOI] [PubMed] [Google Scholar]
- 1222.Lee JY, Nagano Y, Taylor JP, et al. Disease-causing mutations in parkin impair mitochondrial ubiquitination, aggregation, and HDAC6-dependent mitophagy [Research Support, N.I.H., Extramural]. J Cell Biol. 2010. May 17;189(4):671–9. doi: 10.1083/jcb.201001039. PubMed PMID: 20457763; PubMed Central PMCID: PMC2872903. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1223.Zhou Y, Long Q, Wu H, et al. Topology-dependent, bifurcated mitochondrial quality control under starvation. Autophagy. 2020. Mar;16(3):562–574. doi: 10.1080/15548627.2019.1634944. PubMed PMID: 31234709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1224.Georgakopoulos ND, Wells G, Campanella M.. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017. Jan 19;13(2):136–146. doi: 10.1038/nchembio.2287. PubMed PMID: 28103219. [DOI] [PubMed] [Google Scholar]
- 1225.Kondapalli C, Kazlauskaite A, Zhang N, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012. May;2(5):120080. doi: 10.1098/rsob.120080. PubMed PMID: 22724072; PubMed Central PMCID: PMC3376738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1226.Yang KC, Ma X, Liu H, et al. Tumor necrosis factor receptor-associated factor 2 mediates mitochondrial autophagy. Circ Heart Fail. 2014. Oct 22;8:175–87. doi: 10.1161/CIRCHEARTFAILURE.114.001635. PubMed PMID: 25339503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1227.Whitworth AJ, Pallanck LJ.. PINK1/Parkin mitophagy and neurodegeneration-what do we really know in vivo? Curr Opin Genet Dev. 2017. Jun;44:47–53. doi: 10.1016/j.gde.2017.01.016. PubMed PMID: 28213158. [DOI] [PubMed] [Google Scholar]
- 1228.Chan NC, Salazar AM, Pham AH, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011. May 1;20(9):1726–37. doi: 10.1093/hmg/ddr048. PubMed PMID: 21296869; PubMed Central PMCID: PMC3071670. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1229.Martinez A, Lectez B, Ramirez J, et al. Quantitative proteomic analysis of Parkin substrates in Drosophila neurons. Mol Neurodegener. 2017. Apr 11;12(1):29. doi: 10.1186/s13024-017-0170-3. PubMed PMID: 28399880; PubMed Central PMCID: PMCPMC5387213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1230.Okatsu K, Saisho K, Shimanuki M, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010. Aug;15(8):887–900. doi: 10.1111/j.1365-2443.2010.01426.x. PubMed PMID: 20604804; PubMed Central PMCID: PMC2970908. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1231.McWilliams TG, Prescott AR, Montava-Garriga L, et al. Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metab. 2018. Feb 6;27(2):439–449 e5. doi: 10.1016/j.cmet.2017.12.008. PubMed PMID: 29337137; PubMed Central PMCID: PMCPMC5807059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1232.Ivatt RM, Sanchez-Martinez A, Godena VK, et al. Genome-wide RNAi screen identifies the Parkinson disease GWAS risk locus SREBF1 as a regulator of mitophagy. Proc Natl Acad Sci U S A. 2014. Jun 10;111(23):8494–9. doi: 10.1073/pnas.1321207111. PubMed PMID: 24912190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1233.Kim KY, Stevens MV, Akter MH, et al. Parkin is a lipid-responsive regulator of fat uptake in mice and mutant human cells. J Clin Invest. 2011. Sep;121(9):3701–12. doi: 10.1172/JCI44736. PubMed PMID: 21865652; PubMed Central PMCID: PMC3171101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1234.Klinkenberg M, Gispert S, Dominguez-Bautista JA, et al. Restriction of trophic factors and nutrients induces PARKIN expression. Neurogenetics. 2012. Feb;13(1):9–21. doi: 10.1007/s10048-011-0303-8. PubMed PMID: 22028146; PubMed Central PMCID: PMC3274670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1235.Parganlija D, Klinkenberg M, Dominguez-Bautista J, et al. Loss of PINK1 impairs stress-induced autophagy and cell survival. PLoS One. 2014;9(4):e95288. doi: 10.1371/journal.pone.0095288. PubMed PMID: 24751806; PubMed Central PMCID: PMC3994056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1236.Lyamzaev KG, Nepryakhina OK, Saprunova VB, et al. Novel mechanism of elimination of malfunctioning mitochondria (mitoptosis): formation of mitoptotic bodies and extrusion of mitochondrial material from the cell. Biochim Biophys Acta. 2008. Jul-Aug;1777(7–8):817–25. doi: 10.1016/j.bbabio.2008.03.027. PubMed PMID: 18433711; eng. [DOI] [PubMed] [Google Scholar]
- 1237.Davis CH, Kim KY, Bushong EA, et al. Transcellular degradation of axonal mitochondria. Proc Natl Acad Sci U S A. 2014. Jul 1;111(26):9633–8. doi: 10.1073/pnas.1404651111. PubMed PMID: 24979790; PubMed Central PMCID: PMC4084443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1238.Zhu JH, Gusdon AM, Cimen H, et al. Impaired mitochondrial biogenesis contributes to depletion of functional mitochondria in chronic MPP+ toxicity: dual roles for ERK1/2. Cell Death Dis. 2012. May 24;3:e312. doi: 10.1038/cddis.2012.46. PubMed PMID: 22622131; PubMed Central PMCID: PMCPMC3366080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1239.Wang KZ, Zhu J, Dagda RK, et al. ERK-mediated phosphorylation of TFAM downregulates mitochondrial transcription: implications for Parkinson’s disease. Mitochondrion. 2014. Jul;17:132–40. doi: 10.1016/j.mito.2014.04.008. PubMed PMID: 24768991; PubMed Central PMCID: PMCPMC4134365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1240.Ezaki J, Kominami E, Ueno T.. Peroxisome degradation in mammals. IUBMB life. 2011. Nov;63(11):1001–8. doi: 10.1002/iub.537. PubMed PMID: 21990012; eng. [DOI] [PubMed] [Google Scholar]
- 1241.Ishida H, Yoshimoto K, Izumi M, et al. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008. Sep;148(1):142–55. doi: 10.1104/pp.108.122770. PubMed PMID: 18614709; PubMed Central PMCID: PMC2528122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1242.Wada S, Ishida H, Izumi M, et al. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009. Feb;149(2):885–93. doi: 10.1104/pp.108.130013. PubMed PMID: 19074627; PubMed Central PMCID: PMC2633819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1243.Izumi M, Ishida H, Nakamura S, et al. Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell. 2017. Feb;29(2):377–394. doi: 10.1105/tpc.16.00637. PubMed PMID: 28123106; PubMed Central PMCID: PMCPMC5354188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1244.Nakamura S, Hidema J, Sakamoto W, et al. Selective elimination of membrane-damaged chloroplasts via microautophagy. Plant Physiol. 2018. Jul;177(3):1007–1026. doi: 10.1104/pp.18.00444. PubMed PMID: 29748433; PubMed Central PMCID: PMCPMC6052986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1245.Michaeli S, Honig A, Levanony H, et al. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell. 2014. Oct;26(10):4084–101. doi: 10.1105/tpc.114.129999. PubMed PMID: 25281689; PubMed Central PMCID: PMC4247578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1246.Spitzer C, Li F, Buono R, et al. The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell. 2015. Feb 3;27:391–402. doi: 10.1105/tpc.114.135939. PubMed PMID: 25649438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1247.Azzopardi M, Farrugia G, Balzan R.. Cell-cycle involvement in autophagy and apoptosis in yeast. Mech Ageing Dev. 2017. Jan;161(Pt B):211–224. doi: 10.1016/j.mad.2016.07.006. PubMed PMID: 27450768. [DOI] [PubMed] [Google Scholar]
- 1248.Changou CA, Chen YR, Xing L, et al. Arginine starvation-associated atypical cellular death involves mitochondrial dysfunction, nuclear DNA leakage, and chromatin autophagy. Proc Natl Acad Sci U S A. 2014. Sep 30;111(39):14147–52. doi: 10.1073/pnas.1404171111. PubMed PMID: 25122679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1249.Yang M, Chen P, Liu J, et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv. 2019. Jul;5(7):eaaw2238. doi: 10.1126/sciadv.aaw2238. PubMed PMID: 31355331; PubMed Central PMCID: PMCPMC6656546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1250.Scotton C, Bovolenta M, Schwartz E, et al. Deep RNA profiling identified CLOCK and molecular clock genes as pathophysiological signatures in collagen VI myopathy. J Cell Sci. 2016. Apr 15;129(8):1671–84. doi: 10.1242/jcs.175927. PubMed PMID: 26945058; PubMed Central PMCID: PMCPMC4852766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1251.Smith RE, Farquhar MG.. Lysosome function in the regulation of the secretory process in cells of the anterior pituitary gland. J Cell Biol. 1966. Nov 1;31(2):319–47. PubMed PMID: 19866704; PubMed Central PMCID: PMC2107048. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1252.Orci L, Junod A, Pictet R, et al. Granulolysis in a cells of endocrine pancreas in spontaneous and experimental diabetes in animals. J Cell Biol. 1968. Aug;38(2):462–6. doi: 10.1083/jcb.38.2.462. PubMed PMID: 5664218; PubMed Central PMCID: PMCPMC2107480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1253.Orci L, Ravazzola M, Amherdt M, et al. Insulin, not C-peptide (proinsulin), is present in crinophagic bodies of the pancreatic B-cell. J Cell Biol. 1984. Jan;98(1):222–8. doi: 10.1083/jcb.98.1.222. PubMed PMID: 6368567; PubMed Central PMCID: PMCPMC2112993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1254.Halban PA, Wollheim CB.. Intracellular degradation of insulin stores by rat pancreatic islets in vitro. An alternative pathway for homeostasis of pancreatic insulin content. J Biol Chem. 1980. Jul 10;255(13):6003–6. PubMed PMID: 6993463. [PubMed] [Google Scholar]
- 1255.Csizmadia T, Lorincz P, Hegedus K, et al. Molecular mechanisms of developmentally programmed crinophagy in Drosophila. J Cell Biol. 2018. Jan 2;217(1):361–374. doi: 10.1083/jcb.201702145. PubMed PMID: 29066608; PubMed Central PMCID: PMCPMC5748974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1256.Pasquier A, Vivot K, Erbs E, et al. Lysosomal degradation of newly formed insulin granules contributes to beta cell failure in diabetes. Nat Commun. 2019. Jul 25;10(1):3312. doi: 10.1038/s41467-019-11170-4. PubMed PMID: 31346174; PubMed Central PMCID: PMCPMC6658524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1257.Tollenaere MA, Mailand N, Bekker-Jensen S.. Centriolar satellites: key mediators of centrosome functions. Cellular Mol Life Sci.: CMLS. 2015. Jan;72(1):11–23. doi: 10.1007/s00018-014-1711-3. PubMed PMID: 25173771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1258.Holdgaard SG, Cianfanelli V, Pupo E, et al. Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites. Nat Commun. 2019. Sep 13;10(1):4176. doi: 10.1038/s41467-019-12094-9. PubMed PMID: 31519908; PubMed Central PMCID: PMCPMC6744468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1259.Ott C, Konig J, Hohn A, et al. Reduced autophagy leads to an impaired ferritin turnover in senescent fibroblasts. Free Radic Biol Med. 2016. Dec;101:325–333. doi: 10.1016/j.freeradbiomed.2016.10.492. PubMed PMID: 27789294. [DOI] [PubMed] [Google Scholar]
- 1260.Ott C, Konig J, Hohn A, et al. Macroautophagy is impaired in old murine brain tissue as well as in senescent human fibroblasts. Redox Biol. 2016. Dec;10:266–273. doi: 10.1016/j.redox.2016.10.015. PubMed PMID: 27825071; PubMed Central PMCID: PMCPMC5099282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1261.Asano T, Komatsu M, Yamaguchi-Iwai Y, et al. Distinct mechanisms of ferritin delivery to lysosomes in iron-depleted and iron-replete cells. Mol Cell Biol. 2011. May;31(10):2040–52. doi: 10.1128/MCB.01437-10. PubMed PMID: 21444722; PubMed Central PMCID: PMC3133360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1262.Bauckman KA, Haller E, Flores I, et al. Iron modulates cell survival in a Ras- and MAPK-dependent manner in ovarian cells. Cell Death Dis. 2013;4:e592. doi: 10.1038/cddis.2013.87. PubMed PMID: 23598404; PubMed Central PMCID: PMC3668627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1263.Sturm B, Goldenberg H, Scheiber-Mojdehkar B.. Transient increase of the labile iron pool in HepG2 cells by intravenous iron preparations. Eur J Biochem FEBS 2003. Sep;270(18):3731–8. PubMed PMID: 12950256. [DOI] [PubMed] [Google Scholar]
- 1264.Mancias JD, Wang X, Gygi SP, et al. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature. 2014. Mar 30;509:105–9. doi: 10.1038/nature13148. PubMed PMID: 24695223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1265.Xie Y, Hou W, Song X, et al. Ferroptosis: process and function. Cell Death Differ. 2016. Mar;23(3):369–79. doi: 10.1038/cdd.2015.158. PubMed PMID: 26794443; PubMed Central PMCID: PMCPMC5072448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1266.Galluzzi L, Vitale I, Aaronson SA, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018. Mar;25(3):486–541. . PubMed PMID: 29362479; PubMed Central PMCID: PMCPMC5864239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1267.Hou W, Xie Y, Song X, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy. 2016. Aug 2;12(8):1425–8. doi: 10.1080/15548627.2016.1187366. PubMed PMID: 27245739; PubMed Central PMCID: PMCPMC4968231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1268.Bauckman KA, Mysorekar IU.. Ferritinophagy drives uropathogenic Escherichia coli persistence in bladder epithelial cells. Autophagy. 2016. Mar 22:0. doi: 10.1080/15548627.2016.1160176. PubMed PMID: 27002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1269.Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012. May 25;149(5):1060–72. doi: 10.1016/j.cell.2012.03.042. PubMed PMID: 22632970; PubMed Central PMCID: PMCPMC3367386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1270.Dixon SJ, Patel DN, Welsch M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. eLife. 2014. May 20;3:e02523. doi: 10.7554/eLife.02523. PubMed PMID: 24844246; PubMed Central PMCID: PMCPMC4054777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1271.Wu Z, Geng Y, Lu X, et al. Chaperone-mediated autophagy is involved in the execution of ferroptosis. Proc Natl Acad Sci U S A. 2019. Feb 19;116(8):2996–3005. doi: 10.1073/pnas.1819728116. PubMed PMID: 30718432; PubMed Central PMCID: PMCPMC6386716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1272.Zhang Z, Yao Z, Wang L, et al. Activation of ferritinophagy is required for the RNA-binding protein ELAVL1/HuR to regulate ferroptosis in hepatic stellate cells. Autophagy. 2018;14(12):2083–2103. doi: 10.1080/15548627.2018.1503146. PubMed PMID: 30081711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1273.Zhang Z, Guo M, Li Y, et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy. 2019. Nov 11:1–24. doi: 10.1080/15548627.2019.1687985. PubMed PMID: 31679460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1274.Buchan JR, Kolaitis RM, Taylor JP, et al. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013. Jun 20;153(7):1461–74. doi: 10.1016/j.cell.2013.05.037. PubMed PMID: 23791177; PubMed Central PMCID: PMC3760148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1275.Guo H, Chitiprolu M, Gagnon D, et al. Autophagy supports genomic stability by degrading retrotransposon RNA. Nat Commun. 2014. Nov 4;5:5276. doi: 10.1038/ncomms6276. PubMed PMID: 25366815. [DOI] [PubMed] [Google Scholar]
- 1276.Zhang Y, Yan L, Zhou Z, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009. Jan 23;136(2):308–21. doi: 10.1016/j.cell.2008.12.022. PubMed PMID: 19167332; eng. [DOI] [PubMed] [Google Scholar]
- 1277.Chitiprolu M, Jagow C, Tremblay V, et al. A complex of C9ORF72 and p62 uses arginine methylation to eliminate stress granules by autophagy. Nat Commun. 2018. Jul 18;9(1):2794. doi: 10.1038/s41467-018-05273-7. PubMed PMID: 30022074; PubMed Central PMCID: PMCPMC6052026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1278.Ganassi M, Mateju D, Bigi I, et al. A surveillance function of the HSPB8-BAG3-HSP70 chaperone complex ensures stress granule integrity and dynamism. Mol Cell. 2016. Sep 1;63(5):796–810. doi: 10.1016/j.molcel.2016.07.021. PubMed PMID: 27570075. [DOI] [PubMed] [Google Scholar]
- 1279.Mateju D, Franzmann TM, Patel A, et al. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017. Jun 14;36(12):1669–1687. doi: 10.15252/embj.201695957. PubMed PMID: 28377462; PubMed Central PMCID: PMCPMC5470046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1280.Li YR, King OD, Shorter J, et al. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013. Apr 29;201(3):361–72. doi: 10.1083/jcb.201302044. PubMed PMID: 23629963; PubMed Central PMCID: PMCPMC3639398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1281.Ryu HH, Jun MH, Min KJ, et al. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging. 2014. Dec;35(12):2822–31. doi: 10.1016/j.neurobiolaging.2014.07.026. PubMed PMID: 25216585. [DOI] [PubMed] [Google Scholar]
- 1282.Nagl W. ‘‘Plastolysomes’ - Plastids involved in the autolysis of the embryo-suspensor in Phaseolus. Zeitschrift fur Pflanzenphysiologie 1977;85:45–51. [Google Scholar]
- 1283.Gartner PJ, Nagl W.. Acid phosphatase activity in plastids (plastolysomes) of senescing embryo-suspensor cells. Planta. 1980. Jan;149(4):341–9. doi: 10.1007/BF00571168. PubMed PMID: 24306370. [DOI] [PubMed] [Google Scholar]
- 1284.van Doorn WG, Kirasak K, Sonong A, et al. Do plastids in Dendrobium cv. Lucky Duan petals function similar to auto-phagosomes and autolysosomes? Autophagy. 2011. Jun7(6):584–97. PubMed PMID: 21460624. [DOI] [PubMed] [Google Scholar]
- 1285.Parra-Vega V, Corral-Martinez P, Rivas-Sendra A, et al. Formation and excretion of autophagic plastids (plastolysomes) in Brassica napus embryogenic microspores. Front Plant Sci. 2015;6:94. doi: 10.3389/fpls.2015.00094. PubMed PMID: 25745429; PubMed Central PMCID: PMC4333807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1286.Gonzalez-Melendi P, Uyttewaal M, Morcillo CN, et al. A light and electron microscopy analysis of the events leading to male sterility in Ogu-INRA CMS of rapeseed (Brassica napus). J Exp Bot. 2008;59(4):827–38. doi: 10.1093/jxb/erm365. PubMed PMID: 18349052. [DOI] [PubMed] [Google Scholar]
- 1287.Newcomb EH. Fine structure of protein-storing plastids in bean root tips. J Cell Biol. 1967. Apr;33(1):143–63. PubMed PMID: 6033932; PubMed Central PMCID: PMC2107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1288.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009. Apr 30;458(7242):1131–5. doi: 10.1038/nature07976. PubMed PMID: 19339967; PubMed Central PMCID: PMC2676208. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1289.Koenig U, Fobker M, Lengauer B, et al. Autophagy facilitates secretion and protects against degeneration of the Harderian gland. Autophagy. 2015;11(2):298–313. doi: 10.4161/15548627.2014.978221. PubMed PMID: 25484081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1290.Shi Y, Han JJ, Tennakoon JB, et al. Androgens promote prostate cancer cell growth through induction of autophagy. Mol Endocrinol. 2013. Feb;27(2):280–95. doi: 10.1210/me.2012-1260. PubMed PMID: 23250485; PubMed Central PMCID: PMC3683804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1291.O’Rourke EJ, Soukas AA, Carr CE, et al. C. elegans major fats are stored in vesicles distinct from lysosome-related organelles. Cell Metab. 2009. Nov;10(5):430–5. doi: 10.1016/j.cmet.2009.10.002. PubMed PMID: 19883620; PubMed Central PMCID: PMC2921818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1292.Inokuchi-Shimizu S, Park EJ, Roh YS, et al. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J Clin Invest. 2014. Aug 1;124(8):3566–78. doi: 10.1172/JCI74068. PubMed PMID: 24983318; PubMed Central PMCID: PMC4109552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1293.Lee JH, Budanov AV, Talukdar S, et al. Maintenance of metabolic homeostasis by Sestrin2 and Sestrin3. Cell Metab. 2012. Sep 5;16(3):311–21. doi: 10.1016/j.cmet.2012.08.004. PubMed PMID: 22958918; PubMed Central PMCID: PMC3687365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1294.Settembre C, De Cegli R, Mansueto G, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013. Jun;15(6):647–58. doi: 10.1038/ncb2718. PubMed PMID: 23604321; PubMed Central PMCID: PMC3699877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1295.Cuervo AM. Preventing lysosomal fat indigestion. Nat Cell Biol. 2013. Jun;15(6):565–7. doi: 10.1038/ncb2778. PubMed PMID: 23728462. [DOI] [PubMed] [Google Scholar]
- 1296.Settembre C, Fraldi A, Medina DL, et al. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013. May;14(5):283–96. doi: 10.1038/nrm3565. PubMed PMID: 23609508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1297.Chao X, Wang S, Zhao K, et al. Impaired TFEB-mediated lysosome biogenesis and autophagy promote chronic ethanol-induced liver injury and steatosis in mice. Gastroenterology. 2018. Sep;155(3):865–879 e12. doi: 10.1053/j.gastro.2018.05.027. PubMed PMID: 29782848; PubMed Central PMCID: PMCPMC6120772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1298.Yadav N, Cheng D, Richard S, et al. CARM1 promotes adipocyte differentiation by coactivating PPARgamma. EMBO Rep. 2008. Feb;9(2):193–8. doi: 10.1038/sj.embor.7401151. PubMed PMID: 18188184; PubMed Central PMCID: PMCPMC2246418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1299.Liu Y, Wang T, Ji YJ, et al. A C9orf72-CARM1 axis regulates lipid metabolism under glucose starvation-induced nutrient stress. Genes Dev. 2018. Nov 1;32(21–22):1380–1397. doi: 10.1101/gad.315564.118. PubMed PMID: 30366907; PubMed Central PMCID: PMCPMC6217731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1300.Ugolino J, Ji YJ, Conchina K, et al. Loss of C9orf72 enhances autophagic activity via deregulated mTOR and TFEB signaling. PLoS Genet. 2016. Nov;12(11):e1006443. doi: 10.1371/journal.pgen.1006443. PubMed PMID: 27875531; PubMed Central PMCID: PMCPMC5119725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1301.Mizielinska S, Isaacs AM.. C9orf72 amyotrophic lateral sclerosis and frontotemporal dementia: gain or loss of function? Curr Opin Neurol. 2014. Oct;27(5):515–23. doi: 10.1097/WCO.0000000000000130. PubMed PMID: 25188012; PubMed Central PMCID: PMCPMC4165481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1302.Jiang J, Cleveland DW.. Bidirectional transcriptional inhibition as therapy for ALS/FTD caused by repeat expansion in C9orf72. Neuron. 2016. Dec 21;92(6):1160–1163. doi: 10.1016/j.neuron.2016.12.008. PubMed PMID: 28009271. [DOI] [PubMed] [Google Scholar]
- 1303.Fernandez AF, Barcena C, Martinez-Garcia GG, et al. Autophagy couteracts weight gain, lipotoxicity and pancreatic beta-cell death upon hypercaloric pro-diabetic regimens. Cell Death Dis. 2017. Aug 3;8(8):e2970. doi: 10.1038/cddis.2017.373. PubMed PMID: 28771229; PubMed Central PMCID: PMCPMC5596561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1304.O’Rourke EJ, Kuballa P, Xavier R, et al. omega-6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013. Feb 15;27(4):429–40. doi: 10.1101/gad.205294.112. PubMed PMID: 23392608; PubMed Central PMCID: PMCPMC3589559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1305.Visvikis O, Ihuegbu N, Labed SA, et al. Innate host defense requires TFEB-mediated transcription of cytoprotective and antimicrobial genes. Immunity. 2014. Jun 19;40(6):896–909. doi: 10.1016/j.immuni.2014.05.002. PubMed PMID: 24882217; PubMed Central PMCID: PMCPMC4104614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1306.Jeong SJ, Kim S, Park JG, et al. Prdx1 (peroxiredoxin 1) deficiency reduces cholesterol efflux via impaired macrophage lipophagic flux. Autophagy. 2018;14(1):120–133. doi: 10.1080/15548627.2017.1327942. PubMed PMID: 28605287; PubMed Central PMCID: PMCPMC5846566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1307.Chiang PM, Ling J, Jeong YH, et al. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci U S A. 2010. Sep 14;107(37):16320–4. doi: 10.1073/pnas.1002176107. PubMed PMID: 20660762; PubMed Central PMCID: PMC2941284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1308.Heck MV, Azizov M, Stehning T, et al. Dysregulated expression of lipid storage and membrane dynamics factors in Tia1 knockout mouse nervous tissue. Neurogenetics. 2014. May;15(2):135–44. doi: 10.1007/s10048-014-0397-x. PubMed PMID: 24659297; PubMed Central PMCID: PMC3994287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1309.Popovic D, Akutsu M, Novak I, et al. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol Cell Biol. 2012. May;32(9):1733–44. doi: 10.1128/MCB.06717-11. PubMed PMID: 22354992; PubMed Central PMCID: PMC3347240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1310.Onal G, Kutlu O, Ozer E, et al. Impairment of lipophagy by PNPLA1 mutations causes lipid droplet accumulation in primary fibroblasts of Autosomal Recessive Congenital Ichthyosis patients. J Dermatol Sci. 2019. Jan;93(1):50–57. doi: 10.1016/j.jdermsci.2018.11.013. PubMed PMID: 30655104. [DOI] [PubMed] [Google Scholar]
- 1311.Sathyanarayan A, Mashek MT, Mashek DG.. ATGL promotes autophagy/lipophagy via SIRT1 to control hepatic lipid droplet catabolism. Cell Rep. 2017. Apr 4;19(1):1–9. doi: 10.1016/j.celrep.2017.03.026. PubMed PMID: 28380348; PubMed Central PMCID: PMCPMC5396179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1312.Schott MB, Weller SG, Schulze RJ, et al. Lipid droplet size directs lipolysis and lipophagy catabolism in hepatocytes. J Cell Biol. 2019. Oct 7;218(10):3320–3335. doi: 10.1083/jcb.201803153. PubMed PMID: 31391210; PubMed Central PMCID: PMCPMC6781454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1313.Negoita F, Blomdahl J, Wasserstrom S, et al. PNPLA3 variant M148 causes resistance to starvation-mediated lipid droplet autophagy in human hepatocytes. J Cell Biochem. 2019. Jan;120(1):343–356. doi: 10.1002/jcb.27378. PubMed PMID: 30171718. [DOI] [PubMed] [Google Scholar]
- 1314.Tatsumi T, Takayama K, Ishii S, et al. Forced lipophagy reveals that lipid droplets are required for early embryonic development in mouse. Development. 2018. Feb 23;145(4). doi: 10.1242/dev.161893. PubMed PMID: 29475974. [DOI] [PubMed] [Google Scholar]
- 1315.Kaushik S, Cuervo AM.. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat Cell Biol. 2015. Jun;17(6):759–70. doi: 10.1038/ncb3166. PubMed PMID: 25961502; PubMed Central PMCID: PMCPMC4449813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1316.Ma D, Molusky MM, Song J, et al. Autophagy deficiency by hepatic FIP200 deletion uncouples steatosis from liver injury in NAFLD. Mol Endocrinol. 2013. Oct;27(10):1643–54. doi: 10.1210/me.2013-1153. PubMed PMID: 23960084; PubMed Central PMCID: PMCPMC4061382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1317.Kim KH, Jeong YT, Oh H, et al. Autophagy deficiency leads to protection from obesity and insulin resistance by inducing Fgf21 as a mitokine. Nat Med. 2013. Jan;19(1):83–92. doi: 10.1038/nm.3014. PubMed PMID: 23202295. [DOI] [PubMed] [Google Scholar]
- 1318.Li Y, Chao X, Wang S, et al. Role of mechanistic target of rapamycin and autophagy in alcohol-induced adipose atrophy and liver injury. Am J Pathol. 2020. Jan;190(1):158–175. doi: 10.1016/j.ajpath.2019.09.023. PubMed PMID: 31733185; PubMed Central PMCID: PMCPMC6940593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1319.Hung YH, Chen LM, Yang JY, et al. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun. 2013;4:2111. doi: 10.1038/ncomms3111. PubMed PMID: 23817530. [DOI] [PubMed] [Google Scholar]
- 1320.Maejima I, Takahashi A, Omori H, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013. Aug 28;32(17):2336–47. doi: 10.1038/emboj.2013.171. PubMed PMID: 23921551; PubMed Central PMCID: PMC3770333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1321.Raben N, Takikita S, Pittis MG, et al. Deconstructing Pompe disease by analyzing single muscle fibers: to see a world in a grain of sand. Autophagy. 2007. Nov-Dec;3(6):546–52. doi: 10.4161/auto.4591. PubMed PMID: 17592248. [DOI] [PubMed] [Google Scholar]
- 1322.Lopez-Jimenez AT, Cardenal-Munoz E, Leuba F, et al. The ESCRT and autophagy machineries cooperate to repair ESX-1-dependent damage at the Mycobacterium-containing vacuole but have opposite impact on containing the infection. PLoS Pathog. 2018. Dec;14(12):e1007501. doi: 10.1371/journal.ppat.1007501. PubMed PMID: 30596802; PubMed Central PMCID: PMCPMC6329560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1323.Thumm M, Simons M.. Myelinophagy: Schwann cells dine in. J Cell Biol. 2015. Jul 6;210(1):9–10. doi: 10.1083/jcb.201506039. PubMed PMID: 26150387; PubMed Central PMCID: PMCPMC4494007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1324.Jessen KR, Mirsky R.. The repair Schwann cell and its function in regenerating nerves. J Physiol. 2016. Jul 1;594(13):3521–31. doi: 10.1113/JP270874. PubMed PMID: 26864683; PubMed Central PMCID: PMCPMC4929314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1325.Jang SY, Shin YK, Park SY, et al. Autophagic myelin destruction by Schwann cells during Wallerian degeneration and segmental demyelination. Glia. 2016. May;64(5):730–42. doi: 10.1002/glia.22957. PubMed PMID: 26712109. [DOI] [PubMed] [Google Scholar]
- 1326.Gomez-Sanchez JA, Carty L, Iruarrizaga-Lejarreta M, et al. Schwann cell autophagy, myelinophagy, initiates myelin clearance from injured nerves. J Cell Biol. 2015. Jul 6;210(1):153–68. doi: 10.1083/jcb.201503019. PubMed PMID: 26150392; PubMed Central PMCID: PMCPMC4494002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1327.Coccurello R, Nazio F, Rossi C, et al. Effects of caloric restriction on neuropathic pain, peripheral nerve degeneration and inflammation in normometabolic and autophagy defective prediabetic Ambra1 mice. PLoS One. 2018;13(12):e0208596. doi: 10.1371/journal.pone.0208596. PubMed PMID: 30532260; PubMed Central PMCID: PMCPMC6287902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1328.Haidar M, Timmerman V.. Autophagy as an emerging common pathomechanism in inherited peripheral neuropathies. Front Mol Neurosci. 2017;10:143. doi: 10.3389/fnmol.2017.00143. PubMed PMID: 28553203; PubMed Central PMCID: PMCPMC5425483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1329.Park YE, Hayashi YK, Bonne G, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009. Aug;5(6):795–804. doi: 10.4161/auto.8901. PubMed PMID: 19550147. [DOI] [PubMed] [Google Scholar]
- 1330.Liu ML, Yao MC.. Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryot Cell. 2012. Apr;11(4):494–506. doi: 10.1128/EC.05296-11. PubMed PMID: 22366125; PubMed Central PMCID: PMC3318292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1331.Akinduro O, Sully K, Patel A, et al. Constitutive Autophagy and Nucleophagy during Epidermal Differentiation. J Invest Dermatol. 2016. Jul;136(7):1460–1470. doi: 10.1016/j.jid.2016.03.016. PubMed PMID: 27021405. [DOI] [PubMed] [Google Scholar]
- 1332.Kvam E, Goldfarb DS.. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007. Mar-Apr;3(2):85–92. doi: 10.4161/auto.3586. PubMed PMID: 17204844. [DOI] [PubMed] [Google Scholar]
- 1333.Rello-Varona S, Lissa D, Shen S, et al. Autophagic removal of micronuclei. Cell cycle. 2012. Jan 1;11(1):170–6. doi: 10.4161/cc.11.1.18564. PubMed PMID: 22185757. [DOI] [PubMed] [Google Scholar]
- 1334.Mochida K, Oikawa Y, Kimura Y, et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015. Jun 18;522(7556):359–62. doi: 10.1038/nature14506. PubMed PMID: 26040717. [DOI] [PubMed] [Google Scholar]
- 1335.Dou Z, Xu C, Donahue G, et al. Autophagy mediates degradation of nuclear lamina. Nature. 2015. Nov 5;527(7576):105–9. doi: 10.1038/nature15548. PubMed PMID: 26524528; PubMed Central PMCID: PMCPMC4824414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1336.De Meyer GR, Grootaert MO, Michiels CF, et al. Autophagy in vascular disease. Circ Res. 2015. Jan 30;116(3):468–79. doi: 10.1161/CIRCRESAHA.116.303804. PubMed PMID: 25634970. [DOI] [PubMed] [Google Scholar]
- 1337.Bar-Yosef T, Damri O, Agam G.. Dual role of autophagy in diseases of the central nervous system. Front Cell Neurosci. 2019;13:196. doi: 10.3389/fncel.2019.00196. PubMed PMID: 31191249; PubMed Central PMCID: PMCPMC6548059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1338.Brown AJ, Jessup W.. Oxysterols and atherosclerosis. Atherosclerosis. 1999. Jan;142(1):1–28. PubMed PMID: 9920502. [DOI] [PubMed] [Google Scholar]
- 1339.Zarrouk A, Vejux A, Mackrill J, et al. Involvement of oxysterols in age-related diseases and ageing processes. Ageing Res Rev. 2014. Nov;18:148–62. doi: 10.1016/j.arr.2014.09.006. PubMed PMID: 25305550. [DOI] [PubMed] [Google Scholar]
- 1340.He C, Zhu H, Zhang W, et al. 7-Ketocholesterol induces autophagy in vascular smooth muscle cells through Nox4 and Atg4B. Am J Pathol. 2013. Aug;183(2):626–37. doi: 10.1016/j.ajpath.2013.04.028. PubMed PMID: 23770348; PubMed Central PMCID: PMC3730774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1341.Martinet W, Schrijvers DM, Timmermans JP, et al. Interactions between cell death induced by statins and 7-ketocholesterol in rabbit aorta smooth muscle cells. Br J Pharmacol. 2008. Jul;154(6):1236–46. doi: 10.1038/bjp.2008.181. PubMed PMID: 18469840; PubMed Central PMCID: PMC2483392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1342.Nury T, Zarrouk A, Ragot K, et al. 7-Ketocholesterol is increased in the plasma of X-ALD patients and induces peroxisomal modifications in microglial cells: Potential roles of 7-ketocholesterol in the pathophysiology of X-ALD. J Steroid Biochem Mol Biol. 2017. May;169:123–136. doi: 10.1016/j.jsbmb.2016.03.037 PubMed PMID: 27041118. [DOI] [PubMed] [Google Scholar]
- 1343.Monier S, Samadi M, Prunet C, et al.Impairment of the cytotoxic and oxidative activities of 7 beta-hydroxycholesterol and 7-ketocholesterol by esterification with oleate. Biochem Biophys Res Commun. 2003. Apr11;303(3):814–24. PubMed PMID: 12670484. [DOI] [PubMed] [Google Scholar]
- 1344.Nury T, Zarrouk A, Mackrill JJ, et al. Induction of oxiapoptophagy on 158N murine oligodendrocytes treated by 7-ketocholesterol-, 7beta-hydroxycholesterol-, or 24(S)-hydroxycholesterol: Protective effects of alpha-tocopherol and docosahexaenoic acid (DHA; C22:6 n-3). Steroids. 2015. Jul;99(Pt B):194–203. doi: 10.1016/j.steroids.2015.02.003. PubMed PMID: 25683890. [DOI] [PubMed] [Google Scholar]
- 1345.Nury T, Zarrouk A, Vejux A, et al. Induction of oxiapoptophagy, a mixed mode of cell death associated with oxidative stress, apoptosis and autophagy, on 7-ketocholesterol-treated 158N murine oligodendrocytes: impairment by alpha-tocopherol. Biochem Biophys Res Commun. 2014. Apr 11;446(3):714–9. doi: 10.1016/j.bbrc.2013.11.081. PubMed PMID: 24299956. [DOI] [PubMed] [Google Scholar]
- 1346.Sghaier R, Zarrouk A, Nury T, et al. Biotin attenuation of oxidative stress, mitochondrial dysfunction, lipid metabolism alteration and 7beta-hydroxycholesterol-induced cell death in 158N murine oligodendrocytes. Free Radic Res. 2019. May;53(5):535–561. doi: 10.1080/10715762.2019.1612891. PubMed PMID: 31039616. [DOI] [PubMed] [Google Scholar]
- 1347.Vejux A, Abed-Vieillard D, Hajji K, et al. 7-Ketocholesterol and 7beta-hydroxycholesterol: In vitro and animal models used to characterize their activities and to identify molecules preventing their toxicity.Biochem Pharmacol. 2019. Oct 3:113648. doi: 10.1016/j.bcp.2019.113648. PubMed PMID: 31586589. [DOI] [PubMed] [Google Scholar]
- 1348.Zheng K, Li Y, Wang S, et al. Inhibition of autophagosome-lysosome fusion by ginsenoside Ro via the ESR2-NCF1-ROS pathway sensitizes esophageal cancer cells to 5-fluorouracil-induced cell death via the CHEK1-mediated DNA damage checkpoint. Autophagy. 2016. Sep;12(9):1593–613. doi: 10.1080/15548627.2016.1192751. PubMed PMID: 27310928; PubMed Central PMCID: PMCPMC5082787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1349.Redmann M, Benavides GA, Berryhill TF, et al. Inhibition of autophagy with bafilomycin and chloroquine decreases mitochondrial quality and bioenergetic function in primary neurons. Redox Biol. 2017. Apr;11:73–81. doi: 10.1016/j.redox.2016.11.004. PubMed PMID: 27889640; PubMed Central PMCID: PMCPMC5124357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1350.Hsu SPC, Kuo JS, Chiang HC, et al. Temozolomide, sirolimus and chloroquine is a new therapeutic combination that synergizes to disrupt lysosomal function and cholesterol homeostasis in GBM cells. Oncotarget. 2018. Jan 23;9(6):6883–6896. doi: 10.18632/oncotarget.23855. PubMed PMID: 29467937; PubMed Central PMCID: PMCPMC5805523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1351.Bello-Perez M, Pereiro P, Coll J, et al. Zebrafish C-reactive protein isoforms inhibit SVCV replication by blocking autophagy through interactions with cell membrane cholesterol. Sci Rep. 2020. Jan 17;10(1):566. doi: 10.1038/s41598-020-57501-0. PubMed PMID: 31953490; PubMed Central PMCID: PMCPMC6969114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1352.Marshall RS, Li F, Gemperline DC, et al. Autophagic degradation of the 26s proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in arabidopsis. Mol Cell. 2015. Jun 18;58(6):1053–66. doi: 10.1016/j.molcel.2015.04.023. PubMed PMID: 26004230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1353.Marshall RS, McLoughlin F, Vierstra RD.. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 2016. Aug 9;16(6):1717–1732. doi: 10.1016/j.celrep.2016.07.015. PubMed PMID: 27477278. [DOI] [PubMed] [Google Scholar]
- 1354.Ustun S, Hafren A, Liu Q, et al. Bacteria exploit autophagy for proteasome degradation and enhanced virulence in plants. Plant Cell. 2018. Mar;30(3):668–685. doi: 10.1105/tpc.17.00815. PubMed PMID: 29500318; PubMed Central PMCID: PMCPMC5894834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1355.Waite KA, De-La Mota-Peynado A, Vontz G, et al. Starvation induces proteasome autophagy with different pathways for core and regulatory particles. J Biol Chem. 2016. Feb 12;291(7):3239–53. doi: 10.1074/jbc.M115.699124. PubMed PMID: 26670610; PubMed Central PMCID: PMCPMC4751371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1356.Nemec AA, Howell LA, Peterson AK, et al. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J Biol Chem. 2017. Dec 29;292(52):21466–21480. doi: 10.1074/jbc.M117.817999. PubMed PMID: 29109144; PubMed Central PMCID: PMCPMC5766950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1357.Cohen-Kaplan V, Livneh I, Avni N, et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc Natl Acad Sci U S A. 2016. Nov 22;113(47):E7490–E7499. doi: 10.1073/pnas.1615455113. PubMed PMID: 27791183; PubMed Central PMCID: PMCPMC5127335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1358.Marshall RS, Vierstra RD.. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. eLife. 2018. Apr 6;7. doi: 10.7554/eLife.34532. PubMed PMID: 29624167; PubMed Central PMCID: PMCPMC5947986. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 1359.Marshall RS, Hua Z, Mali S, et al. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019. Apr 18;177(3):766–781 e24. doi: 10.1016/j.cell.2019.02.009. PubMed PMID: 30955882; PubMed Central PMCID: PMCPMC6810650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1360.Marshall RS, Vierstra RD.. Dynamic regulation of the 26s proteasome: from synthesis to degradation. Front Mol Biosci. 2019;6:40. doi: 10.3389/fmolb.2019.00040. PubMed PMID: 31231659; PubMed Central PMCID: PMCPMC6568242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1361.Li J, Breker M, Graham M, et al. AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLoS Genet. 2019. Nov;15(11):e1008387. doi: 10.1371/journal.pgen.1008387. PubMed PMID: 31738769; PubMed Central PMCID: PMCPMC6886873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1362.Hamasaki M, Noda T, Baba M, et al. Starvation triggers the delivery of the endoplasmic reticulum to the vacuole via autophagy in yeast. Traffic. 2005. Jan;6(1):56–65. doi: 10.1111/j.1600-0854.2004.00245.x. PubMed PMID: 15569245. [DOI] [PubMed] [Google Scholar]
- 1363.Yorimitsu T, Nair U, Yang Z, et al. Endoplasmic reticulum stress triggers autophagy. J Biol Chem. 2006. Oct 6;281(40):30299–304. doi: 10.1074/jbc.M607007200. PubMed PMID: 16901900; PubMed Central PMCID: PMC1828866. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1364.Schuck S, Gallagher CM, Walter P.. ER-phagy mediates selective degradation of endoplasmic reticulum independently of the core autophagy machinery. J Cell Sci. 2014. Sep 15;127(Pt 18):4078–88. doi: 10.1242/jcs.154716. PubMed PMID: 25052096; PubMed Central PMCID: PMC4163648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1365.Bernales S, Schuck S, Walter P.. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007. May-Jun;3(3):285–7. PubMed PMID: 17351330. [DOI] [PubMed] [Google Scholar]
- 1366.Bolender RP, Weibel ER.. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of threatment. J Cell Biol. 1973. Mar;56(3):746–61. PubMed PMID: 4569312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1367.Grumati P, Dikic I, Stolz A.. ER-phagy at a glance. J Cell Sci. 2018. Sep 3;131(17). doi: 10.1242/jcs.217364. PubMed PMID: 30177506. [DOI] [PubMed] [Google Scholar]
- 1368.Wilkinson S. ER-phagy: shaping up and destressing the endoplasmic reticulum. FEBS J. 2019. Jul;286(14):2645–2663. doi: 10.1111/febs.14932. PubMed PMID: 31116513; PubMed Central PMCID: PMCPMC6772018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1369.Loi M, Fregno I, Guerra C, et al. Eat it right: ER-phagy and recovER-phagy. Biochem Soc Trans. 2018. Jun 19;46(3):699–706. doi: 10.1042/BST20170354. PubMed PMID: 29802216; PubMed Central PMCID: PMCPMC6008593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1370.Wilkinson S. Emerging Principles of Selective ER Autophagy. J Mol Biol. 2020. Jan 3;432(1):185–205. doi: 10.1016/j.jmb.2019.05.012. PubMed PMID: 31100386; PubMed Central PMCID: PMCPMC6971691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1371.Grumati P, Morozzi G, Holper S, et al. Full length RTN3 regulates turnover of tubular endoplasmic reticulum via selective autophagy. eLife. 2017. Jun 15;6. doi: 10.7554/eLife.25555. PubMed PMID: 28617241; PubMed Central PMCID: PMCPMC5517149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1372.Chen Q, Xiao Y, Chai P, et al. ATL3 Is a tubular ER-phagy receptor for GABARAP-mediated selective autophagy. Curr Biol. 2019. Mar 4;29(5):846–855 e6. doi: 10.1016/j.cub.2019.01.041. PubMed PMID: 30773365. [DOI] [PubMed] [Google Scholar]
- 1373.An H, Ordureau A, Paulo JA, et al. TEX264 is an endoplasmic reticulum-resident ATG8-interacting protein critical for ER remodeling during nutrient stress. Mol Cell. 2019. Jun 6;74(5):891–908 e10. doi: 10.1016/j.molcel.2019.03.034. PubMed PMID: 31006537; PubMed Central PMCID: PMCPMC6747008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1374.Fumagalli F, Noack J, Bergmann TJ, et al. Corrigendum: Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016. Dec 23;19(1):76. doi: 10.1038/ncb3451. PubMed PMID: 28008182. [DOI] [PubMed] [Google Scholar]
- 1375.Fumagalli F, Noack J, Bergmann TJ, et al. Translocon component Sec62 acts in endoplasmic reticulum turnover during stress recovery. Nat Cell Biol. 2016. Nov;18(11):1173–1184. doi: 10.1038/ncb3423. PubMed PMID: 27749824. [DOI] [PubMed] [Google Scholar]
- 1376.Smith MD, Harley ME, Kemp AJ, et al. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell. 2018. Jan 22;44(2):217–232 e11. doi: 10.1016/j.devcel.2017.11.024. PubMed PMID: 29290589; PubMed Central PMCID: PMCPMC5791736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1377.Lipatova Z, Segev N. A. role for macro-ER-phagy in ER quality control. PLoS Genet. 2015. Jul;11(7):e1005390. doi: 10.1371/journal.pgen.1005390. PubMed PMID: 26181331; PubMed Central PMCID: PMC4504476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1378.Fregno I, Molinari M.. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit Rev Biochem Mol Biol. 2019. Apr;54(2):153–163. doi: 10.1080/10409238.2019.1610351. PubMed PMID: 31084437. [DOI] [PubMed] [Google Scholar]
- 1379.Fregno I, Fasana E, Bergmann TJ, et al. ER-to-lysosome-associated degradation of proteasome-resistant ATZ polymers occurs via receptor-mediated vesicular transport. EMBO J. 2018. Sep 3;37(17). doi: 10.15252/embj.201899259. PubMed PMID: 30076131; PubMed Central PMCID: PMCPMC6120659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1380.Forrester A, De Leonibus C, Grumati P, et al. A selective ER-phagy exerts procollagen quality control via a Calnexin-FAM134B complex. EMBO J. 2019. Jan 15;38(2). doi: 10.15252/embj.201899847. PubMed PMID: 30559329; PubMed Central PMCID: PMCPMC6331724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1381.Cunningham CN, Williams JM, Knupp J, et al. Cells deploy a two-pronged strategy to rectify misfolded proinsulin aggregates. Mol Cell. 2019. Aug 8;75(3):442–456 e4. doi: 10.1016/j.molcel.2019.05.011. PubMed PMID: 31176671; PubMed Central PMCID: PMCPMC6688957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1382.Cui Y, Parashar S, Zahoor M, et al. A COPII subunit acts with an autophagy receptor to target endoplasmic reticulum for degradation. Science. 2019. Jul 5;365(6448):53–60. doi: 10.1126/science.aau9263. PubMed PMID: 31273116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1383.Chiramel AI, Best SM.. Role of autophagy in Zika virus infection and pathogenesis. Virus Res. 2018. Aug 2;254:34–40. doi: 10.1016/j.virusres.2017.09.006. PubMed PMID: 28899653; PubMed Central PMCID: PMCPMC5844781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1384.Moretti J, Roy S, Bozec D, et al. STING senses microbial viability to orchestrate stress-mediated autophagy of the endoplasmic reticulum. Cell. 2017. Nov 2;171(4):809–823 e13. doi: 10.1016/j.cell.2017.09.034. PubMed PMID: 29056340; PubMed Central PMCID: PMCPMC5811766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1385.Peng Y, Shapiro SL, Banduseela VC, et al. Increased transport of acetyl-CoA into the endoplasmic reticulum causes a progeria-like phenotype. Aging cell. 2018. Oct;17(5):e12820. doi: 10.1111/acel.12820. PubMed PMID: 30051577; PubMed Central PMCID: PMCPMC6156544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1386.Farrugia MA, Puglielli L.. Nepsilon-lysine acetylation in the endoplasmic reticulum - a novel cellular mechanism that regulates proteostasis and autophagy. J Cell Sci. 2018. Nov 16;131(22). doi: 10.1242/jcs.221747. PubMed PMID: 30446507; PubMed Central PMCID: PMCPMC6262770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1387.Peng Y, Kim MJ, Hullinger R, et al. Improved proteostasis in the secretory pathway rescues Alzheimer’s disease in the mouse. Brain. 2016. Mar;139(Pt 3):937–52. doi: 10.1093/brain/awv385. PubMed PMID: 26787453; PubMed Central PMCID: PMCPMC4805081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1388.Omari S, Makareeva E, Roberts-Pilgrim A, et al. Noncanonical autophagy at ER exit sites regulates procollagen turnover. Proc Natl Acad Sci U S A. 2018. Oct 23;115(43):E10099–E10108. doi: 10.1073/pnas.1814552115. PubMed PMID: 30287488; PubMed Central PMCID: PMCPMC6205486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1389.Kraft C, Deplazes A, Sohrmann M, et al. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008. May;10(5):602–10. doi: 10.1038/ncb1723. PubMed PMID: 18391941; eng. [DOI] [PubMed] [Google Scholar]
- 1390.Ossareh-Nazari B, Nino CA, Bengtson MH, et al. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J Cell Biol. 2014. Mar 17;204(6):909–17. doi: 10.1083/jcb.201308139. PubMed PMID: 24616224; PubMed Central PMCID: PMC3998797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1391.Derrien B, Baumberger N, Schepetilnikov M, et al. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc Natl Acad Sci U S A. 2012. Sep 25;109(39):15942–6. doi: 10.1073/pnas.1209487109. PubMed PMID: 23019378; PubMed Central PMCID: PMC3465452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1392.Kobayashi H, Shoji K, Kiyokawa K, et al. VCP machinery mediates autophagic degradation of empty Argonaute. Cell Rep. 2019. Jul 30;28(5):1144–1153 e4. doi: 10.1016/j.celrep.2019.07.003. PubMed PMID: 31365860. [DOI] [PubMed] [Google Scholar]
- 1393.Gibbings D, Mostowy S, Jay F, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol. 2012. Dec;14(12):1314–21. doi: 10.1038/ncb2611. PubMed PMID: 23143396; PubMed Central PMCID: PMC3771578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1394.Zhang P, Zhang H.. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO Rep. 2013. Jun;14(6):568–76. doi: 10.1038/embor.2013.53. PubMed PMID: 23619095; PubMed Central PMCID: PMC3674441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1395.Fujiwara Y, Kikuchi H, Aizawa S, et al. Direct uptake and degradation of DNA by lysosomes. Autophagy. 2013. Aug;9(8):1167–71. doi: 10.4161/auto.24880. PubMed PMID: 23839276; PubMed Central PMCID: PMC3748189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1396.Fujiwara Y, Hase K, Wada K, et al. An RNautophagy/DNautophagy receptor, LAMP2C, possesses an arginine-rich motif that mediates RNA/DNA-binding. Biochem Biophys Res Commun. 2015. May 1;460(2):281–6. doi: 10.1016/j.bbrc.2015.03.025. PubMed PMID: 25772617. [DOI] [PubMed] [Google Scholar]
- 1397.Fujiwara Y, Furuta A, Kikuchi H, et al. Discovery of a novel type of autophagy targeting RNA. Autophagy. 2013. Mar;9(3):403–9. doi: 10.4161/auto.23002. PubMed PMID: 23291500; PubMed Central PMCID: PMC3590259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1398.Hase K, Fujiwara Y, Kikuchi H, et al. RNautophagy/DNautophagy possesses selectivity for RNA/DNA substrates. Nucleic Acids Res. 2015. Jul 27;43(13):6439–49. doi: 10.1093/nar/gkv579. PubMed PMID: 26038313; PubMed Central PMCID: PMC4513860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1399.Aizawa S, Contu VR, Fujiwara Y, et al. Lysosomal membrane protein SIDT2 mediates the direct uptake of DNA by lysosomes. Autophagy. 2017. Jan 2;13(1):218–222. doi: 10.1080/15548627.2016.1248019. PubMed PMID: 27846365; PubMed Central PMCID: PMCPMC5245770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1400.Aizawa S, Fujiwara Y, Contu VR, et al. Lysosomal putative RNA transporter SIDT2 mediates direct uptake of RNA by lysosomes. Autophagy. 2016;12(3):565–78. . PubMed PMID: 27046251; PubMed Central PMCID: PMCPMC4836006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1401.Contu VR, Hase K, Kozuka-Hata H, et al. Lysosomal targeting of SIDT2 via multiple YxxPhi motifs is required for SIDT2 function in the process of RNautophagy. J Cell Sci. 2017. Sep 1;130(17):2843–2853. doi: 10.1242/jcs.202481. PubMed PMID: 28724756. [DOI] [PubMed] [Google Scholar]
- 1402.Brown CR, Chiang H-L.. A selective autophagy pathway that degrades gluconeogenic enzymes during catabolite inactivation. Commun Integr Biol. 2009;2(2):177–83. PubMed PMID: 19513275; PubMed Central PMCID: PMC2686377. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1403.Regelmann J, Schule T, Josupeit FS, et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways [Research Support, Non-U.S. Gov’t]. Mol Biol Cell. 2003. Apr;14(4):1652–63. doi: 10.1091/mbc.E02-08-0456. PubMed PMID: 12686616; PubMed Central PMCID: PMC153129. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1404.Schork SM, Thumm M, Wolf DH.. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995. Nov 3;270(44):26446–50. PubMed PMID: 7592860; eng. [DOI] [PubMed] [Google Scholar]
- 1405.Schule T, Rose M, Entian KD, et al. Ubc8p functions in catabolite degradation of fructose-1, 6-bisphosphatase in yeast. EMBO J. 2000. May 15;19(10):2161–7. doi: 10.1093/emboj/19.10.2161. PubMed PMID: 10811607; PubMed Central PMCID: PMC384366. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1406.Hung GC, Brown CR, Wolfe AB, et al. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004. Nov19;279(47):49138–50. PubMed PMID: 15358789. [DOI] [PubMed] [Google Scholar]
- 1407.Chiang H-L, Schekman R, Hamamoto S.. Selective uptake of cytosolic, peroxisomal, and plasma membrane proteins into the yeast lysosome for degradation. J Biol Chem. 1996. Apr 26;271(17):9934–41. PubMed PMID: 8626630; eng. [DOI] [PubMed] [Google Scholar]
- 1408.Huang PH, Chiang H-L.. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997. Feb 24;136(4):803–10. PubMed PMID: 9049246; PubMed Central PMCID: PMC2132494. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1409.Alibhoy AA, Giardina BJ, Dunton DD, et al. Vid30 is required for the association of Vid vesicles and actin patches in the vacuole import and degradation pathway. Autophagy. 2012;8:29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1410.Brown CR, Wolfe AB, Cui D, et al. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem. 2008. Sep 19;283(38):26116–27. doi: 10.1074/jbc.M709922200. PubMed PMID: 18660504; PubMed Central PMCID: PMC2533773. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1411.Chiang MC, Chiang H-L.. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol. 1998. Mar 23;140(6):1347–56. PubMed PMID: 9508768; PubMed Central PMCID: PMC2132677. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1412.Vida TA, Emr SD.. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995. Mar;128(5):779–92. PubMed PMID: 7533169; PubMed Central PMCID: PMC2120394. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1413.Brown CR, Hung GC, Dunton D, et al. The TOR complex 1 is distributed in endosomes and in retrograde vesicles that form from the vacuole membrane and plays an important role in the vacuole import and degradation pathway. J Biol Chem. 2010. Jul 23;285(30):23359–70. doi: 10.1074/jbc.M109.075143. PubMed PMID: 20457600; PubMed Central PMCID: PMC2906328. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1414.Brown CR, Dunton D, Chiang H-L.. The vacuole import and degradation pathway utilizes early steps of endocytosis and actin polymerization to deliver cargo proteins to the vacuole for degradation. J Biol Chem. 2010. Jan 8;285(2):1516–28. doi: 10.1074/jbc.M109.028241. PubMed PMID: 19892709; PubMed Central PMCID: PMC2801277. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1415.McLean JE, Wudzinska A, Datan E, et al. Flavivirus NS4A-induced autophagy protects cells against death and enhances virus replication. J Biol Chem. 2011. Jun 24;286(25):22147–59. doi: 10.1074/jbc.M110.192500. PubMed PMID: 21511946; PubMed Central PMCID: PMC3121359. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1416.Lee YR, Lei HY, Liu MT, et al. Autophagic machinery activated by dengue virus enhances virus replication. Virology. 2008. May 10;374(2):240–8. doi: 10.1016/j.virol.2008.02.016. PubMed PMID: 18353420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1417.Mao Y, Da L, Tang H, et al. Hepatitis B virus X protein reduces starvation-induced cell death through activation of autophagy and inhibition of mitochondrial apoptotic pathway. Biochem Biophys Res Commun. 2011. Nov 11;415(1):68–74. doi: 10.1016/j.bbrc.2011.10.013. PubMed PMID: 22020078; eng. [DOI] [PubMed] [Google Scholar]
- 1418.Orvedahl A, Alexander D, Talloczy Z, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Cell Host Microbe. 2007. Mar 15;1(1):23–35. doi:10.1016/j.chom.2006.12.001. PubMed PMID: 18005679; eng. [DOI] [PubMed] [Google Scholar]
- 1419.Alexander DE, Ward SL, Mizushima N, et al. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007. Nov;81(22):12128–34. doi: 10.1128/JVI.01356-07. PubMed PMID: 17855538; PubMed Central PMCID: PMC2169004. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1420.Leib DA, Alexander DE, Cox D, et al. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009. Dec;83(23):12164–71. doi:10.1128/JVI.01676-09. PubMed PMID: 19759141; PubMed Central PMCID: PMC2786728. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1421.Yordy B, Iijima N, Huttner A, et al. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012. Sep 13;12(3):334–45. doi: 10.1016/j.chom.2012.07.013. PubMed PMID: 22980330; PubMed Central PMCID: PMC3454454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1422.Waisner H, Kalamvoki M.. The ICP0 protein of herpes simplex virus 1 (HSV-1) downregulates major autophagy adaptor proteins Sequestosome 1 and Optineurin during the early stages of HSV-1 infection. J Virol. 2019. Nov 1;93(21). doi: 10.1128/JVI.01258-19. PubMed PMID: 31375597; PubMed Central PMCID: PMCPMC6803258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1423.Liang C, E X, Jung JU. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy. 2008. Apr;4(3):268–72. PubMed PMID: 17993780. [DOI] [PubMed] [Google Scholar]
- 1424.Hernaez B, Cabezas M, Munoz-Moreno R, et al. A179L, a new viral Bcl2 homolog targeting Beclin 1 autophagy related protein. Curr Mol Med. 2013. Feb;13(2):305–16. PubMed PMID: 23228131. [PubMed] [Google Scholar]
- 1425.Alonso C, Galindo I, Cuesta-Geijo MA, et al. African swine fever virus-cell interactions: from virus entry to cell survival. Virus Res. 2013. Apr;173(1):42–57. doi: 10.1016/j.virusres.2012.12.006. PubMed PMID: 23262167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1426.Galindo I, Hernaez B, Diaz-Gil G, et al. A179L, a viral Bcl-2 homologue, targets the core Bcl-2 apoptotic machinery and its upstream BH3 activators with selective binding restrictions for Bid and Noxa. Virology. 2008. Jun 5;375(2):561–72. doi: 10.1016/j.virol.2008.01.050. PubMed PMID: 18329683; PubMed Central PMCID: PMC2572728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1427.Schein CH. Polyglutamine Repeats in Viruses. Mol Neurobiol. 2019. May;56(5):3664–3675. doi: 10.1007/s12035-018-1269-4. PubMed PMID: 30182336; PubMed Central PMCID: PMCPMC6399083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1428.Nardacci R, Ciccosanti F, Marsella C, et al. Role of autophagy in HIV infection and pathogenesis. J Intern Med. 2017. May;281(5):422–432. . PubMed PMID: 28139864. [DOI] [PubMed] [Google Scholar]
- 1429.Leymarie O, Lepont L, Berlioz-Torrent C.. Canonical and Non-Canonical Autophagy in HIV-1 Replication Cycle. Viruses. 2017. Sep 23;9(10). doi: 10.3390/v9100270. PubMed PMID: 28946621; PubMed Central PMCID: PMCPMC5691622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1430.Nardacci R, Amendola A, Ciccosanti F, et al. Autophagy plays an important role in the containment of HIV-1 in nonprogressor-infected patients. Autophagy. 2014. Jul;10(7):1167–78. doi: 10.4161/auto.28678. PubMed PMID: 24813622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1431.Moy RH, Gold B, Molleston JM, et al. Antiviral autophagy restrictsRift Valley fever virus infection and is conserved from flies to mammals. Immunity. 2014. Jan 16;40(1):51–65. doi: 10.1016/j.immuni.2013.10.020. PubMed PMID: 24374193; PubMed Central PMCID: PMCPMC3951734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1432.Sumpter R, Jr., Sirasanagandla S, Fernandez AF, et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell. 2016. May 5;165(4):867–81. doi: 10.1016/j.cell.2016.04.006. PubMed PMID: 27133164; PubMed Central PMCID: PMCPMC4881391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1433.Paludan C, Schmid D, Landthaler M, et al. Endogenous MHC class II processing of a viral nuclear antigen after autophagy [Research Support, Non-U.S. Gov’t]. Science. 2005. Jan 28;307(5709):593–6. doi: 10.1126/science.1104904. PubMed PMID: 15591165; eng. [DOI] [PubMed] [Google Scholar]
- 1434.Bhattacharjee S, Bose P, Patel K, et al. Transcriptional and epigenetic modulation of autophagy promotes EBV oncoprotein EBNA3C induced B-cell survival. Cell Death Dis. 2018. May 22;9(6):605. doi: 10.1038/s41419-018-0668-9. PubMed PMID: 29789559; PubMed Central PMCID: PMCPMC5964191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1435.Lee DY, Sugden B.. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. 2008. May 1;27(20):2833–42. doi: 10.1038/sj.onc.1210946. PubMed PMID: 18037963. [DOI] [PubMed] [Google Scholar]
- 1436.Pujals A, Favre L, Pioche-Durieu C, et al. Constitutive autophagy contributes to resistance to TP53-mediated apoptosis in Epstein-Barr virus-positive latency III B-cell lymphoproliferations. Autophagy. 2015;11(12):2275–87. doi: 10.1080/15548627.2015.1115939. PubMed PMID: 26565591; PubMed Central PMCID: PMCPMC4835200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1437.Fotheringham JA, Raab-Traub N.. Epstein-Barr virus latent membrane protein 2 induces autophagy to promote abnormal acinus formation. J Virol. 2015. Jul;89(13):6940–4. doi: 10.1128/JVI.03371-14. PubMed PMID: 25878108; PubMed Central PMCID: PMCPMC4468476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1438.Hung CH, Chen LW, Wang WH, et al. Regulation of autophagic activation by Rta of Epstein-Barr virus via the extracellular signal-regulated kinase pathway. J Virol. 2014. Oct;88(20):12133–45. doi: 10.1128/JVI.02033-14. PubMed PMID: 25122800; PubMed Central PMCID: PMCPMC4178756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1439.McFadden K, Hafez AY, Kishton R, et al. Metabolic stress is a barrier to Epstein-Barr virus-mediated B-cell immortalization. Proc Natl Acad Sci U S A. 2016. Feb 9;113(6):E782–90. doi: 10.1073/pnas.1517141113. PubMed PMID: 26802124; PubMed Central PMCID: PMCPMC4760815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1440.Montespan C, Marvin SA, Austin S, et al. Multi-layered control of Galectin-8 mediated autophagy during adenovirus cell entry through a conserved PPxY motif in the viral capsid. PLoS Pathog. 2017. Feb;13(2):e1006217. doi: 10.1371/journal.ppat.1006217. PubMed PMID: 28192531; PubMed Central PMCID: PMCPMC5325606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1441.Hurwitz SN, Cheerathodi MR, Nkosi D, et al. Tetraspanin CD63 bridges autophagic and endosomal processes to regulate exosomal secretion and intracellular signaling of Epstein-Barr Virus LMP1. J Virol. 2018. Mar 1;92(5). doi: 10.1128/JVI.01969-17. PubMed PMID: 29212935; PubMed Central PMCID: PMCPMC5809724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1442.Panyasrivanit M, Khakpoor A, Wikan N, et al. Co-localization of constituents of the dengue virus translation and replication machinery with amphisomes. J Gen Virol. 2009. Feb;90(Pt 2):448–56. doi: 10.1099/vir.0.005355-0. PubMed PMID: 19141455. [DOI] [PubMed] [Google Scholar]
- 1443.Khakpoor A, Panyasrivanit M, Wikan N, et al. A role for autophagolysosomes in dengue virus 3 production in HepG2 cells. J Gen Virol. 2009. May;90(Pt 5):1093–103. doi: 10.1099/vir.0.007914-0. PubMed PMID: 19264601. [DOI] [PubMed] [Google Scholar]
- 1444.Zhang H, Monken CE, Zhang Y, et al. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006. Apr-Jun;2(2):91–5. PubMed PMID: 16874104; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1445.Heaton NS, Randall G.. Dengue virus and autophagy. Viruses. 2011. Aug;3(8):1332–41. doi: 10.3390/v3081332. PubMed PMID: 21994782; PubMed Central PMCID: PMC3185800. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1446.Lee YR, Kuo SH, Lin CY, et al. Dengue virus-induced ER stress is required for autophagy activation, viral replication, and pathogenesis both in vitro and in vivo. Sci Rep. 2018. Jan 11;8(1):489. doi: 10.1038/s41598-017-18909-3. PubMed PMID: 29323257; PubMed Central PMCID: PMCPMC5765116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1447.Dreux M, Gastaminza P, Wieland SF, et al. The autophagy machinery is required to initiate hepatitis C virus replication [Research Support, N.I.H., Extramural]. Proc Natl Acad Sci U S A. 2009. Aug 18;106(33):14046–51. doi: 10.1073/pnas.0907344106. PubMed PMID: 19666601; PubMed Central PMCID: PMC2729017. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1448.Cao B, Parnell LA, Diamond MS, et al. Inhibition of autophagy limits vertical transmission of Zika virus in pregnant mice. J Exp Med. 2017. Aug 7;214(8):2303–2313. doi: 10.1084/jem.20170957. PubMed PMID: 28694387; PubMed Central PMCID: PMCPMC5551583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1449.Abernathy E, Mateo R, Majzoub K, et al. Differential and convergent utilization of autophagy components by positive-strand RNA viruses. PLoS Biol. 2019. Jan;17(1):e2006926. doi: 10.1371/journal.pbio.2006926. PubMed PMID: 30608919; PubMed Central PMCID: PMCPMC6334974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1450.Liang Q, Luo Z, Zeng J, et al. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell. 2016. Nov 3;19(5):663–671. doi: 10.1016/j.stem.2016.07.019. PubMed PMID: 27524440; PubMed Central PMCID: PMCPMC5144538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1451.Nombela I, Requena-Platek R, Morales-Lange B, et al. Rainbow trout red blood cells exposed to viral hemorrhagic septicemia virus up-regulate antigen-processing mechanisms and MHC I&II, CD86, and CD83 antigen-presenting cell markers. Cells. 2019. Apr 27;8(5):386. doi: 10.3390/cells8050386. PubMed PMID: 31035565; PubMed Central PMCID: PMCPMC6562805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1452.Webster P. Cytoplasmic bacteria and the autophagic pathway [Review]. Autophagy. 2006. Jul-Sep;2(3):159–61. PubMed PMID: 16874112; eng. [DOI] [PubMed] [Google Scholar]
- 1453.Dubuisson JF, Swanson MS.. Mouse infection by Legionella, a model to analyze autophagy [Research Support, N.I.H., Extramural Review]. Autophagy. 2006. Jul-Sep;2(3):179–82. PubMed PMID: 16874080; PubMed Central PMCID: PMC1774947. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1454.Jordan TX, Randall G.. Manipulation or capitulation: virus interactions with autophagy. Microbes Infect. 2011. Oct 24;in press. doi: 10.1016/j.micinf.2011.09.007. PubMed PMID: 22051604; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1455.Knodler LA, Celli J.. Eating the strangers within: host control of intracellular bacteria via xenophagy [Research Support, N.I.H., Intramural]. Cell Microbiol. 2011. Sep;13(9):1319–27. doi: 10.1111/j.1462-5822.2011.01632.x. PubMed PMID: 21740500; PubMed Central PMCID: PMC3158265. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1456.Levine B, Mizushima N, Virgin HW.. Autophagy in immunity and inflammation. Nature. 2011. Jan 20;469(7330):323–35. doi: 10.1038/nature09782. PubMed PMID: 21248839; PubMed Central PMCID: PMC3131688. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1457.Deretic V. Autophagy in immunity and cell-autonomous defense against intracellular microbes. Immunol Rev. 2011. Mar;240(1):92–104. doi: 10.1111/j.1600-065X.2010.00995.x. PubMed PMID: 21349088; PubMed Central PMCID: PMC3057454. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1458.Dong X, Levine B.. Autophagy and viruses: adversaries or allies? J Innate Immun. 2013;5(5):480–93. doi: 10.1159/000346388. PubMed PMID: 23391695; PubMed Central PMCID: PMC3790331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1459.Cardenal-Munoz E, Barisch C, Lefrancois LH, et al. When dicty met myco, a (Not So) romantic story about one amoeba and its intracellular pathogen. Front Cell Infect Microbiol. 2017;7:529. doi: 10.3389/fcimb.2017.00529. PubMed PMID: 29376033; PubMed Central PMCID: PMCPMC5767268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1460.Dunn JD, Bosmani C, Barisch C, et al. Eat prey, live: dictyostelium discoideum as a model for cell-autonomous defenses. Front Immunol. 2017;8:1906. doi: 10.3389/fimmu.2017.01906. PubMed PMID: 29354124; PubMed Central PMCID: PMCPMC5758549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1461.Leary AY, Savage Z, Tumtas Y, et al. Contrasting and emerging roles of autophagy in plant immunity. Curr Opin Plant Biol. 2019. Dec;52:46–53. doi: 10.1016/j.pbi.2019.07.002. PubMed PMID: 31442734. [DOI] [PubMed] [Google Scholar]
- 1462.Wang C, Symington JW, Mysorekar IU.. ATG16L1 and pathogenesis of urinary tract infections. Autophagy. 2012. Nov;8(11):1693–4. doi: 10.4161/auto.21600. PubMed PMID: 22874553; PubMed Central PMCID: PMC3494604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1463.Choy A, Roy CR.. Autophagy and bacterial infection: an evolving arms race. Trends Microbiol. 2013. Sep;21(9):451–6. doi: 10.1016/j.tim.2013.06.009. PubMed PMID: 23880062; PubMed Central PMCID: PMC3839292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1464.Mostowy S, Cossart P.. Bacterial autophagy: restriction or promotion of bacterial replication? Trends Cell Biol. 2012. Jun;22(6):283–91. doi: 10.1016/j.tcb.2012.03.006. PubMed PMID: 22555009. [DOI] [PubMed] [Google Scholar]
- 1465.Andersson AM, Andersson B, Lorell C, et al. Autophagy induction targeting mTORC1 enhances Mycobacterium tuberculosis replication in HIV co-infected human macrophages. Sci Rep. 2016. Jun 15;6:28171. doi: 10.1038/srep28171. PubMed PMID: 27302320; PubMed Central PMCID: PMCPMC4908603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1466.Dinic M, Lukic J, Djokic J, et al. Lactobacillus fermentum Postbiotic-induced Autophagy as Potential Approach for Treatment of Acetaminophen Hepatotoxicity. Front Microbiol. 2017;8:594. doi: 10.3389/fmicb.2017.00594. PubMed PMID: 28428777; PubMed Central PMCID: PMCPMC5382196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1467.Kageyama S, Omori H, Saitoh T, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella [Research Support, Non-U.S. Gov’t]. Mol Biol Cell. 2011. Jul 1;22(13):2290–300. doi: 10.1091/mbc.E10-11-0893. PubMed PMID: 21525242; PubMed Central PMCID: PMC3128531. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1468.Thurston TL, Wandel MP, von Muhlinen N, et al. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012. Feb 16;482(7385):414–8. doi: 10.1038/nature10744. PubMed PMID: 22246324; PubMed Central PMCID: PMC3343631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1469.Zheng YT, Shahnazari S, Brech A, et al. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J Iimmunol. 2009. Nov 1;183(9):5909–16. doi: 10.4049/jimmunol.0900441. PubMed PMID: 19812211; eng. [DOI] [PubMed] [Google Scholar]
- 1470.Thurston TL, Ryzhakov G, Bloor S, et al. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol. 2009. Nov;10(11):1215–21. doi: 10.1038/ni.1800. PubMed PMID: 19820708; eng. [DOI] [PubMed] [Google Scholar]
- 1471.Tumbarello DA, Manna PT, Allen M, et al. The autophagyrReceptor TAX1BP1 and the molecular motor myosin VI are required for clearance of Salmonella Typhimurium by autophagy. PLoS Pathog. 2015;11:e1005174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1472.Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011. Jul 8;333(6039):228–33. doi: 10.1126/science.1205405. PubMed PMID: 21617041; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1473.Lopez-Montero N, Ramos-Marques E, Risco C, et al. Intracellular Salmonella induces aggrephagy of host endomembranes in persistent infections. Autophagy. 2016. Oct 2;12(10):1886–1901. doi: 10.1080/15548627.2016.1208888 PubMed PMID: 27485662; PubMed Central PMCID: PMCPMC5079681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1474.Benjamin JL, Sumpter R, Jr., Levine B, et al. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013. Jun 12;13(6):723–34. doi: 10.1016/j.chom.2013.05.004. PubMed PMID: 23768496; PubMed Central PMCID: PMCPMC3755484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1475.Ammanathan V, Mishra P, Chavalmane AK, et al. Restriction of intracellular Salmonella replication by restoring TFEB-mediated xenophagy. Autophagy. 2019. Nov 19:1–14. doi: 10.1080/15548627.2019.1689770. PubMed PMID: 31744366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1476.Xu Y, Zhou P, Cheng S, et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cell. 2019. Jul 25;178(3):552–566 e20. doi: 10.1016/j.cell.2019.06.007. PubMed PMID: 31327526. [DOI] [PubMed] [Google Scholar]
- 1477.Zhang H, Wu F, Wang X, et al. The two C. elegans ATG-16 homologs have partially redundant functions in the basal autophagy pathway. Autophagy. 2013. Dec;9(12):1965–74. PubMed PMID: 24185444; PubMed Central PMCID: PMC4028341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1478.Fletcher K, Ulferts R, Jacquin E, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018. Feb 15;37(4). doi: 10.15252/embj.201797840. PubMed PMID: 29317426; PubMed Central PMCID: PMCPMC5813257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1479.Shahnazari S, Brumell JH.. Mechanisms and consequences of bacterial targeting by the autophagy pathway. Curr Opin Microbiol. 2011. Feb;14(1):68–75. doi: 10.1016/j.mib.2010.11.001. PubMed PMID: 21112809; eng. [DOI] [PubMed] [Google Scholar]
- 1480.Klionsky DJ, Eskelinen EL, Deretic V.. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes … wait, I’m confused. Autophagy. 2014. Apr;10(4):549–51. doi: 10.4161/auto.28448. PubMed PMID: 24657946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1481.Li X, He S, Zhou X, et al. Lyn delivers bacteria to lysosomes for eradication through TLR2-initiated autophagy related phagocytosis. PLoS Pathog. 2016. Jan;12(1):e1005363. doi: 10.1371/journal.ppat.1005363. PubMed PMID: 26735693; PubMed Central PMCID: PMCPMC4703367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1482.Li X, Ye Y, Zhou X, et al. Atg7 enhances host defense against infection via downregulation of superoxide but upregulation of nitric oxide. J Iimmunol. 2015. Feb 1;194(3):1112–21. doi: 10.4049/jimmunol.1401958. PubMed PMID: 25535282; PubMed Central PMCID: PMC4409144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1483.Ye Y, Tan S, Zhou X, et al. Inhibition of p-IkappaBalpha ubiquitylation by autophagy-related gene 7 to regulate inflammatory responses to bacterial infection. J Infect Dis. 2015. May 28;212:1816–26. doi: 10.1093/infdis/jiv301. PubMed PMID: 26022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1484.Yuan K, Huang C, Fox J, et al. Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci. 2012. Jan 15;125(Pt 2):507–15. doi: 10.1242/jcs.094573. PubMed PMID: 22302984; PubMed Central PMCID: PMC3283879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1485.Irving AT, Mimuro H, Kufer TA, et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. 2014. May 14;15(5):623–35. doi: 10.1016/j.chom.2014.04.001. PubMed PMID: 24746552. [DOI] [PubMed] [Google Scholar]
- 1486.Kaparakis-Liaskos M, Ferrero RL.. Immune modulation by bacterial outer membrane vesicles. Nat Rev Immunol. 2015. Jun;15(6):375–87. doi: 10.1038/nri3837. PubMed PMID: 25976515. [DOI] [PubMed] [Google Scholar]
- 1487.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010. Jan;11(1):55–62. doi: 10.1038/ni.1823. PubMed PMID: 19898471; eng. [DOI] [PubMed] [Google Scholar]
- 1488.Chaudhary A, Kamischke C, Leite M, et al. beta-Barrel outer membrane proteins suppress mTORC2 activation and induce autophagic responses. Sci Signal. 2018. Nov 27;11(558). doi: 10.1126/scisignal.aat7493. PubMed PMID: 30482849. [DOI] [PubMed] [Google Scholar]
- 1489.Hayrabedyan S, Todorova K, Jabeen A, et al. Sertoli cells have a functional NALP3 inflammasome that can modulate autophagy and cytokine production. Sci Rep. 2016. Jan 8;6:18896. doi: 10.1038/srep18896. PubMed PMID: 26744177; PubMed Central PMCID: PMCPMC4705529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1490.Keestra-Gounder AM, Tsolis RM.. NOD1 and NOD2: Beyond Peptidoglycan Sensing. Trends Immunol. 2017. Oct;38(10):758–767. doi: 10.1016/j.it.2017.07.004. PubMed PMID: 28823510; PubMed Central PMCID: PMCPMC5624830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1491.Nabatov AA, Hatzis P, Rouschop KM, et al. Hypoxia inducible NOD2 interacts with 3-O-sulfogalactoceramide and regulates vesicular homeostasis. Biochim Biophys Acta. 2013. Nov;1830(11):5277–86. doi: 10.1016/j.bbagen.2013.07.017. PubMed PMID: 23880069. [DOI] [PubMed] [Google Scholar]
- 1492.Casassa AF, Vanrell MC, Colombo MI, et al. Autophagy plays a protective role against Trypanosoma cruzi infection in mice. Virulence. 2019. Dec;10(1):151–165. doi: 10.1080/21505594.2019.1584027. PubMed PMID: 30829115; PubMed Central PMCID: PMCPMC6550547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1493.Collins CA, De Maziere A, van Dijk S, et al. Atg5-independent sequestration of ubiquitinated mycobacteria [Research Support, Non-U.S. Gov’t]. PLoS Pathog. 2009. May;5(5):e1000430. doi: 10.1371/journal.ppat.1000430. PubMed PMID: 19436699; PubMed Central PMCID: PMC2673685. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1494.Moreau K, Lacas-Gervais S, Fujita N, et al. Autophagosomes can support Yersinia pseudotuberculosis replication in macrophages. Cell Microbiol. 2010. Aug;12(8):1108–23. doi: 10.1111/j.1462-5822.2010.01456.x. PubMed PMID: 20180800; eng. [DOI] [PubMed] [Google Scholar]
- 1495.Chandra P, Ghanwat S, Matta SK, et al. Mycobacterium tuberculosis inhibits RAB7 recruitment to selectively modulate autophagy flux in macrophages. Sci Rep. 2015. Nov 6;5:16320. doi: 10.1038/srep16320. PubMed PMID: 26541268; PubMed Central PMCID: PMCPMC4635374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1496.Chandra P, Kumar D.. Selective autophagy gets more selective: Uncoupling of autophagy flux and xenophagy flux in Mycobacterium tuberculosis-infected macrophages. Autophagy. 2016;12(3):608–9. doi: 10.1080/15548627.2016.1139263. PubMed PMID: 27046255; PubMed Central PMCID: PMCPMC4836011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1497.Grasso D, Ropolo A, Re A Lo, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011. Mar 11;286(10):8308–24. doi: 10.1074/jbc.M110.197301. PubMed PMID: 21173155; PubMed Central PMCID: PMC3048716. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1498.Wang S, Ni HM, Chao X, et al. Critical role of TFEB-mediated lysosomal biogenesis in alcohol-induced pancreatitis in mice and humans. Cell Mol Gastroenterol Hepatol. 2020;10(1):59–81. doi: 10.1016/j.jcmgh.2020.01.008. PubMed PMID: 31987928; PubMed Central PMCID: PMCPMC7210479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1499.Mareninova OA, Hermann K, French SW, et al. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J Clin Invest. 2009. Nov;119(11):3340–55. doi: 10.1172/JCI38674. PubMed PMID: 19805911; PubMed Central PMCID: PMCPMC2769194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1500.Wang S, Ni HM, Chao X, et al. Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis. Autophagy. 2019. Nov;15(11):1954–1969. doi: 10.1080/15548627.2019.1596486. PubMed PMID: 30894069; PubMed Central PMCID: PMCPMC6844531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1501.Seglen PO, Gordon PB, Tolleshaug H, et al. Use of [3H]raffinose as a specific probe of autophagic sequestration. Exp Cell Res. 1986. Jan;162(1):273–7. PubMed PMID: 3940229; eng. [DOI] [PubMed] [Google Scholar]
- 1502.Kopitz J, Kisen GO, Gordon PB, et al. Nonselective autophagy of cytosolic enzymes by isolated rat hepatocytes. J Cell Biol. 1990. Sep;111(3):941–53. PubMed PMID: 2391370; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1503.Seglen PO, Luhr M, Mills IG, et al. Macroautophagic cargo sequestration assays. Methods. 2015. Mar;75:25–36. doi: 10.1016/j.ymeth.2014.12.021. PubMed PMID: 25576638. [DOI] [PubMed] [Google Scholar]
- 1504.Gordon PB, Seglen PO.. Autophagic sequestration of [14C]sucrose, introduced into rat hepatocytes by reversible electro-permeabilization. Exp Cell Res. 1982. Nov142(1):1–14. PubMed PMID: 7140848. [DOI] [PubMed] [Google Scholar]
- 1505.Kjos I, Borg Distefano M, Saetre F, et al. Rab7b modulates autophagic flux by interacting with Atg4B. EMBO Rep. 2017. Oct;18(10):1727–1739. doi: 10.15252/embr.201744069. PubMed PMID: 28835545; PubMed Central PMCID: PMCPMC5623852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1506.Luhr M, Szalai P, Saetre F, et al. A simple cargo sequestration assay for quantitative measurement of nonselective autophagy in cultured cells. Methods Enzymol. 2017;587:351–364. doi: 10.1016/bs.mie.2016.09.064. PubMed PMID: 28253965. [DOI] [PubMed] [Google Scholar]
- 1507.Boland B, Smith DA, Mooney D, et al. Macroautophagy is not directly involved in the metabolism of amyloid precursor protein. J Biol Chem. 2010. Nov 26;285(48):37415–26. doi: 10.1074/jbc.M110.186411. PubMed PMID: 20864542; PubMed Central PMCID: PMC2988347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1508.Nair U, Thumm M, Klionsky DJ, et al. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy. 2011. Dec 1;7(12):1546–1550. PubMed PMID: 22108003; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1509.Plomp PJ, Gordon PB, Meijer AJ, et al. Energy dependence of different steps in the autophagic-lysosomal pathway. J Biol Chem. 1989. Apr 25;264(12):6699–704. PubMed PMID: 2708336; eng. [PubMed] [Google Scholar]
- 1510.Gutierrez MG, Munafo DB, Beron W, et al. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004. Jun1;117(Pt 13):2687–97. PubMed PMID: 15138286; eng. [DOI] [PubMed] [Google Scholar]
- 1511.Jager S, Bucci C, Tanida I, et al. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004. Sep15;117(Pt 20):4837–48. doi: 10.1242/jcs.01370 jcs.01370 [pii]. PubMed PMID: 15340014; eng. [DOI] [PubMed] [Google Scholar]
- 1512.Rodriguez-Enriquez S, Kim I, Currin RT, et al. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006. Jan-Mar;2(1):39–46. PubMed PMID: 16874071; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1513.Lorenz H, Hailey DW, Lippincott-Schwartz J.. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat Methods. 2006. Mar;3(3):205–10. PubMed PMID: 16489338; eng. [DOI] [PubMed] [Google Scholar]
- 1514.Korsnes MS, Korsnes R.. Single-cell tracking of A549 lung cancer cells exposed to a marine toxin reveals correlations in pedigree tree profiles. Front Oncol. 2018;8:260. doi: 10.3389/fonc.2018.00260. PubMed PMID: 30023341; PubMed Central PMCID: PMCPMC6039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1515.Korsnes MS, Kolstad H, Kleiveland CR, et al. Autophagic activity in BC3H1 cells exposed to yessotoxin. Toxicol In Vitro. 2016. Apr;32:166–80. doi: 10.1016/j.tiv.2015.12.010. PubMed PMID: 26743762. [DOI] [PubMed] [Google Scholar]
- 1516.Takats S, Toth S, Szenci G, et al. Investigating Non-selective autophagy in Drosophila. Methods Mol Biol. 2019;1880:589–600. doi: 10.1007/978-1-4939-8873-0_38. PubMed PMID: 30610724. [DOI] [PubMed] [Google Scholar]
- 1517.McNeil PL, Murphy RF, Lanni F, et al. A method for incorporating macromolecules into adherent cells. J Cell Biol. 1984. Apr;98(4):1556–64. PubMed PMID: 6201494; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1518.Kim J, Huang WP, Stromhaug PE, et al. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002. Jan 4;277(1):763–73. doi: 10.1074/jbc.M109134200. PubMed PMID: 11675395; PubMed Central PMCID: PMC2754695. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1519.Velikkakath AK, Nishimura T, Oita E, et al. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell. 2012. Mar;23(5):896–909. doi: 10.1091/mbc.E11-09-0785. PubMed PMID: 22219374; PubMed Central PMCID: PMC3290647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1520.Seglen PO, Gordon PB.. Amino acid control of autophagic sequestration and protein degradation in isolated rat hepatocytes. J Cell Biol. 1984. Aug;99(2):435–44. doi: 10.1083/jcb.99.2.435. PubMed PMID: 6746735; PubMed Central PMCID: PMCPMC2113269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1521.Kovács AL, Laszlo L, Kovács J.. Effect of amino acids and cycloheximide on changes caused by vinblastine, leupeptin and methylamine in the autophagic/lysosomal system of mouse hepatocytes in vivo. Exp Cell Res. 1985. Mar;157(1):83–94. PubMed PMID: 3972014; eng. [DOI] [PubMed] [Google Scholar]
- 1522.Swanson MS, Byrne BG, Dubuisson JF.. Kinetic analysis of autophagosome formation and turnover in primary mouse macrophages. Methods Enzymol. 2009;452:383–402. doi: 10.1016/S0076-6879(08)03623-9. PubMed PMID: 19200894; eng. [DOI] [PubMed] [Google Scholar]
- 1523.Urban J, Soulard A, Huber A, et al. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol Cell. 2007. Jun 8;26(5):663–74. doi: 10.1016/j.molcel.2007.04.020. PubMed PMID: 17560372; eng. [DOI] [PubMed] [Google Scholar]
- 1524.Jomain-Baum M, Garber AJ, Farber E, et al. The effect of cycloheximide on the interaction between mitochondrial respiration and gluconeogenesis in guinea pig and rat liver. J Biol Chem. 1973. Mar 10;248(5):1536–43. PubMed PMID: 4348543; eng. [PubMed] [Google Scholar]
- 1525.Garber AJ, Jomain-Baum M, Salganicoff L, et al. The effects of cycloheximide on energy transfer in rat and guinea pig liver mitochondria. J Biol Chem. 1973. Mar 10;248(5):1530–5. PubMed PMID: 4144389; eng. [PubMed] [Google Scholar]
- 1526.Mora R, Dokic I, Kees T, et al. Sphingolipid rheostat alterations related to transformation can be exploited for specific induction of lysosomal cell death in murine and human glioma. Glia. 2010. Aug 15;58(11):1364–83. doi: 10.1002/glia.21013. PubMed PMID: 20607862; eng. [DOI] [PubMed] [Google Scholar]
- 1527.Bright NA, Lindsay MR, Stewart A, et al. The relationship between lumenal and limiting membranes in swollen late endocytic compartments formed after wortmannin treatment or sucrose accumulation. Traffic. 2001. Sep;2(9):631–42. PubMed PMID: 11555417; eng. [DOI] [PubMed] [Google Scholar]
- 1528.Deter RL. Quantitative characterization of dense body, autophagic vacuole, and acid phosphatase-bearing particle populations during the early phases of glucagon-induced autophagy in rat liver. J Cell Biol. 1971. Mar;48(3):473–89. PubMed PMID: 5553081; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1529.Deter RL. Analog modeling of glucagon-induced autophagy in rat liver. I. Conceptual and mathematical model of telolysosome-autophagosome-autolysosome interaction. Exp Cell Res. 1975. Aug;94(1):122–6. PubMed PMID: 1193121; eng. [DOI] [PubMed] [Google Scholar]
- 1530.Deter RL. Analog modeling of glucagon-induced autophagy in rat liver. II. Evaluation of iron labeling as a means for identifying telolysosome, autophagosome and autolysosome populations.Exp Cell Res. 1975. Aug;94(1):127–39. PubMed PMID: 172336; eng. [DOI] [PubMed] [Google Scholar]
- 1531.Deter RL, Baudhuin P, de Duve C.. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967. Nov;35(2):C11–6. PubMed PMID: 6055998; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1532.Deter RL, de Duve C.. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J Cell Biol. 1967. May;33(2):437–49. PubMed PMID: 4292315; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1533.Stromhaug PE, Berg TO, Fengsrud M, et al. Purification and characterization of auto-phagosomes from rat hepatocytes. Biochem J. 1998. Oct;15;335(Pt 2):217–24. PubMed PMID: 9761717; PubMed Central PMCID: PMC1219772. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1534.Deter RL. Electron microscopic evaluation of subcellular fractions obtained by ultracentrifugation. In: Hayat MA, editor. Principles and Techniques of Electron Microscopy. Vol. 3. New York: Van Nostrand Reinhold Co; 1973. p. 199–235. [Google Scholar]
- 1535.Marzella L, Ahlberg J, Glaumann H.. Isolation of autophagic vacuoles from rat liver: morphological and biochemical characterization. J Cell Biol. 1982. Apr;93(1):144–54. PubMed PMID: 7068752; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1536.Wattiaux R, Wattiaux-De Coninck S, Ronveaux-Dupal M-F, et al. Isolation of rat liver lysosomes by isopycnic centrifugation in a metrizamide gradient. J Cell Biol. 1978. Aug;78(2):349–68. PubMed PMID: 211139; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1537.Rodriguez-Navarro JA, Rodriguez L, Casarejos MJ, et al. Trehalose ameliorates dopaminergic and tau pathology in parkin deleted/tau overexpressing mice through autophagy activation. Neurobiol Dis. 2010. Sep;39(3):423–38. doi: 10.1016/j.nbd.2010.05.014. PubMed PMID: 20546895; eng. [DOI] [PubMed] [Google Scholar]
- 1538.Ge L, Melville D, Zhang M, et al. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife. 2013. Aug 6;2:e00947. doi: 10.7554/eLife.00947. PubMed PMID: 23930225; PubMed Central PMCID: PMCPMC3736544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1539.Ge L, Zhang M, Schekman R.. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. eLife. 2014. Nov 28;3:e04135. doi: 10.7554/eLife.04135. PubMed PMID: 25432021; PubMed Central PMCID: PMCPMC4270069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1540.Ge L, Wilz L, Schekman R.. Biogenesis of autophagosomal precursors for LC3 lipidation from the ER-Golgi intermediate compartment. Autophagy. 2015;11(12):2372–4. doi: 10.1080/15548627.2015.1105422. PubMed PMID: 26565421; PubMed Central PMCID: PMCPMC4835199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1541.Shima T, Kirisako H, Nakatogawa H.. COPII vesicles contribute to autophagosomal membranes. J Cell Biol. 2019. May 6;218(5):1503–1510. doi: 10.1083/jcb.201809032. PubMed PMID: 30787039; PubMed Central PMCID: PMCPMC6504894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1542.Ge L, Zhang M, Kenny SJ, et al. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017. Sep;18(9):1586–1603. doi: 10.15252/embr.201744559. PubMed PMID: 28754694; PubMed Central PMCID: PMCPMC5579361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1543.He C, Sumpter R, Jr., Levine B.. Exercise induces autophagy in peripheral tissues and in the brain. Autophagy. 2012. Oct;8(10):1548–51. doi: 10.4161/auto.21327. PubMed PMID: 22892563; PubMed Central PMCID: PMC3463459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1544.Munoz-Galdeano T, Reigada D, Del Aguila A, et al. Cell specific changes of autophagy in a mouse model of contusive spinal cord injury. Front Cell Neurosci. 2018;12:164. doi: 10.3389/fncel.2018.00164. PubMed PMID: 29946241; PubMed Central PMCID: PMCPMC6005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1545.Mizushima N, Yoshimori T, Levine B.. Methods in mammalian autophagy research. Cell. 2010. Feb 5;140(3):313–26. doi: 10.1016/j.cell.2010.01.028. PubMed PMID: 20144757; PubMed Central PMCID: PMC2852113. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1546.Iwai-Kanai E, Yuan H, Huang C, et al. A method to measure cardiac autophagic flux in vivo [Research Support, N.I.H., Extramural]. Autophagy. 2008. Apr;4(3):322–9. PubMed PMID: 18216495; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1547.Zhu H, Tannous P, Johnstone JL, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007. Jul;117(7):1782–93. doi: 10.1172/JCI27523. PubMed PMID: 17607355; PubMed Central PMCID: PMC1890995. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1548.Castillo K, Valenzuela V, Matus S, et al. Measurement of autophagy flux in the nervous system in vivo. Cell Death Dis. 2013;4:e917. doi: 10.1038/cddis.2013.421. PubMed PMID: 24232093; PubMed Central PMCID: PMC3847309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1549.Matus S, Valenzuela V, Hetz C.. A new method to measure autophagy flux in the nervous system. Autophagy. 2014. Apr;10(4):710–4. doi: 10.4161/auto.28434. PubMed PMID: 24717689; PubMed Central PMCID: PMC4091163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1550.Leiva-Rodriguez T, Romeo-Guitart D, Marmolejo-Martinez-Artesero S, et al. ATG5 overexpression is neuroprotective and attenuates cytoskeletal and vesicle-trafficking alterations in axotomized motoneurons. Cell Death Dis. 2018. May 24;9(6):626. doi: 10.1038/s41419-018-0682-y. PubMed PMID: 29799519; PubMed Central PMCID: PMCPMC5967323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1551.Castillo K, Nassif M, Valenzuela V, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy. 2013. Sep;9(9):1308–20. doi: 10.4161/auto.25188. PubMed PMID: 23851366. [DOI] [PubMed] [Google Scholar]
- 1552.Chiarelli R, Agnello M, Roccheri MC.. Sea urchin embryos as a model system for studying autophagy induced by cadmium stress. Autophagy. 2011. Sep 1;7(9):1028–34. PubMed PMID: 21628995; eng. [DOI] [PubMed] [Google Scholar]
- 1553.Morici G, Agnello M, Spagnolo F, et al. Confocal microscopy study of the distribution, content and activity of mitochondria during Paracentrotus lividus development. J Microsc. 2007. Nov;228(Pt 2):165–73. doi: 10.1111/j.1365-2818.2007.01860.x. PubMed PMID: 17970916; eng. [DOI] [PubMed] [Google Scholar]
- 1554.McWilliams TG, Ganley IG.. Investigating mitophagy and mitochondrial morphology in vivo using mito-QC: A comprehensive guide. Methods Mol Biol. 2019;1880:621–642. doi: 10.1007/978-1-4939-8873-0_41. PubMed PMID: 30610727. [DOI] [PubMed] [Google Scholar]
- 1555.Martinet W, De Meyer GR, Andries L, et al. Detection of autophagy in tissue by standard immunohistochemistry: possibilities and limitations. Autophagy. 2006. Jan-Mar;2(1):55–7. PubMed PMID: 16874065; eng. [DOI] [PubMed] [Google Scholar]
- 1556.Holt SV, Wyspianska B, Randall KJ, et al. The development of an immunohistochemical method to detect the autophagy-associated protein LC3-II in human tumor xenografts. Toxicol Pathol. 2011. Apr;39(3):516–23. doi: 10.1177/0192623310396903. PubMed PMID: 21441228; eng. [DOI] [PubMed] [Google Scholar]
- 1557.Kimura S, Fujita N, Noda T, et al. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. PubMed PMID: 19200872; eng. [DOI] [PubMed] [Google Scholar]
- 1558.Dehay B, Bove J, Rodriguez-Muela N, et al. Pathogenic lysosomal depletion in Parkinson’s disease. J Neurosci. 2010. Sep 15;30(37):12535–44. doi: 10.1523/JNEUROSCI.1920-10.2010. PubMed PMID: 20844148; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1559.Daniels BH, McComb RD, Mobley BC, et al. LC3 and p62 as diagnostic markers of drug-induced autophagic vacuolar cardiomyopathy: a study of 3 cases. Am J Surg Pathol. 2013. Jul;37(7):1014–21. doi: 10.1097/PAS.0b013e3182863fa8. PubMed PMID: 23681079. [DOI] [PubMed] [Google Scholar]
- 1560.Hiniker A, Daniels BH, Lee HS, et al. Comparative utility of LC3, p62 and TDP-43 immunohistochemistry in differentiation of inclusion body myositis from polymyositis and related inflammatory myopathies. Acta Neuropathol Commun. 2013;1(1):29. doi: 10.1186/2051-5960-1-29. PubMed PMID: 24252466; PubMed Central PMCID: PMC3893502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1561.Lee HS, Daniels BH, Salas E, et al. Clinical utility of LC3 and p62 immunohistochemistry in diagnosis of drug-induced autophagic vacuolar myopathies: a case-control study. PLoS One. 2012;7(4):e36221. doi: 10.1371/journal.pone.0036221. PubMed PMID: 22558391; PubMed Central PMCID: PMC3338695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1562.Lastres-Becker I, Garcia-Yague AJ, Scannevin RH, et al. Repurposing the NRF2 activator dimethyl fumarate as therapy against synucleinopathy in Parkinson’s disease. Antioxid Redox Signal. 2016. Jul 10;25(2):61–77. doi: 10.1089/ars.2015.6549. PubMed PMID: 27009601; PubMed Central PMCID: PMCPMC4943471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1563.Chung YC, Lim JH, Oh HM, et al. Calcimimetic restores diabetic peripheral neuropathy by ameliorating apoptosis and improving autophagy. Cell Death Dis. 2018. Nov 26;9(12):1163. doi: 10.1038/s41419-018-1192-7. PubMed PMID: 30478254; PubMed Central PMCID: PMCPMC6255917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1564.Huang HC, Chen L, Zhang HX, et al. Autophagy promotes peripheral nerve regeneration and motor recovery following sciatic nerve crush injury in rats. J Mol Neurosci. 2016. Apr;58(4):416–23. doi: 10.1007/s12031-015-0672-9. PubMed PMID: 26738732; PubMed Central PMCID: PMCPMC4829621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1565.Hamada K, Terauchi A, Nakamura K, et al. Aberrant calcium signaling by transglutaminase-mediated posttranslational modification of inositol 1,4,5-trisphosphate receptors. Proc Natl Acad Sci U S A. 2014. Sep 23;111(38):E3966–75. doi: 10.1073/pnas.1409730111. PubMed PMID: 25201980; PubMed Central PMCID: PMC4183345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1566.Rodriguez-Muela N, Koga H, Garcia-Ledo L, et al. Balance between autophagic pathways preserves retinal homeostasis. Aging cell. 2013. Jun;12(3):478–88. doi: 10.1111/acel.12072. PubMed PMID: 23521856; PubMed Central PMCID: PMC3655122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1567.Esteban-Martinez L, Boya P.. Autophagic flux determination in vivo and ex vivo. Methods. 2015;75:79–86. [DOI] [PubMed] [Google Scholar]
- 1568.Gomez-Sintes R, Villarejo-Zori B, Serrano-Puebla A, et al. Standard assays for the study of autophagy in the ex vivo retina. Cells. 2017. Oct 22;6(4). doi: 10.3390/cells6040037. PubMed PMID: 29065501; PubMed Central PMCID: PMCPMC5755496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1569.McMahon J, Huang X, Yang J, et al. Impaired autophagy in neurons after disinhibition of mammalian target of rapamycin and its contribution to epileptogenesis. J Neurosci. 2012. Nov 7;32(45):15704–14. doi: 10.1523/JNEUROSCI.2392-12.2012. PubMed PMID: 23136410; PubMed Central PMCID: PMC3501684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1570.Messing L, Decker JM, Joseph M, et al. Cascade of tau toxicity in inducible hippocampal brain slices and prevention by aggregation inhibitors. Neurobiol Aging. 2013. May;34(5):1343–1354. doi: 10.1016/j.neurobiolaging.2012.10.024. PubMed PMID: 23158765; PubMed Central PMCID: PMCPMC4984976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1571.Herrando-Grabulosa M, Casas C, Aguilera J.. The C-terminal domain of tetanus toxin protects motoneurons against acute excitotoxic damage on spinal cord organotypic cultures. J Neurochem. 2013. Jan;124(1):36–44. doi: 10.1111/jnc.12062. PubMed PMID: 23106494. [DOI] [PubMed] [Google Scholar]
- 1572.Follo C, Cheng Y, Richards WG, et al. Autophagy facilitates the release of immunogenic signals following chemotherapy in 3D models of mesothelioma. Mol Carcinog. 2019. Oct;58(10):1754–1769. doi: 10.1002/mc.23050. PubMed PMID: 31215708. [DOI] [PubMed] [Google Scholar]
- 1573.Gomes LC, Di Benedetto G, Scorrano L.. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011. May;13(5):589–98. doi: 10.1038/ncb2220. PubMed PMID: 21478857; PubMed Central PMCID: PMC3088644. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1574.Grumati P, Coletto L, Sabatelli P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration.Nat Med. 2010. Nov;16(11):1313–20. doi: 10.1038/nm.2247. PubMed PMID: 21037586; eng. [DOI] [PubMed] [Google Scholar]
- 1575.Zecchini S, Giovarelli M, Perrotta C, et al. Autophagy controls neonatal myogenesis by regulating the GH-IGF1 system through a NFE2L2- and DDIT3-mediated mechanism. Autophagy. 2019. Jan;15(1):58–77. doi: 10.1080/15548627.2018.1507439. PubMed PMID: 30081710; PubMed Central PMCID: PMCPMC6287695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1576.Bloemberg D, McDonald E, Dulay D, et al. Autophagy is altered in skeletal and cardiac muscle of spontaneously hypertensive rats. Acta Physiol (Oxf). 2014. Feb;210(2):381–91. doi: 10.1111/apha.12178. PubMed PMID: 24119246. [DOI] [PubMed] [Google Scholar]
- 1577.Ogata T, Oishi Y, Higuchi M, et al. Fasting-related autophagic response in slow- and fast-twitch skeletal muscle. Biochem Biophys Res Commun. 2010. Mar 26;394(1):136–40. doi: 10.1016/j.bbrc.2010.02.130. PubMed PMID: 20184860. [DOI] [PubMed] [Google Scholar]
- 1578.Yamada E, Bastie CC, Koga H, et al. Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep. 2012. May 31;1(5):557–69. doi: 10.1016/j.celrep.2012.03.014. PubMed PMID: 22745922; PubMed Central PMCID: PMC3383827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1579.Pare MF, Baechler BL, Fajardo VA, et al. Effect of acute and chronic autophagy deficiency on skeletal muscle apoptotic signaling, morphology, and function. Biochim Biophys Acta Mol Cell Res. 2017. Apr;1864(4):708–718. doi: 10.1016/j.bbamcr.2016.12.015. PubMed PMID: 27993671. [DOI] [PubMed] [Google Scholar]
- 1580.He C, Bassik MC, Moresi V, et al. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature. 2012. Jan 26;481(7382):511–5. doi: 10.1038/nature10758. PubMed PMID: 22258505; PubMed Central PMCID: PMC3518436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1581.Haspel J, Shaik RS, Ifedigbo E, et al. Characterization of macroautophagic flux in vivo using a leupeptin-based assay [Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Autophagy. 2011. Jun;7(6):629–42. PubMed PMID: 21460622; PubMed Central PMCID: PMC3127049. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1582.Kominami E, Hashida S, Khairallah EA, et al. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J Biol Chem. 1983. May 25;258(10):6093–100. PubMed PMID: 6133857; eng. [PubMed] [Google Scholar]
- 1583.Bell RM, Mocanu MM, Yellon DM.. Retrograde heart perfusion: the Langendorff technique of isolated heart perfusion. J Mol Cell Cardiol. 2011. Jun50(6):940–50. doi: 10.1016/j.yjmcc.2011.02.018. PubMed PMID: 21385587. [DOI] [PubMed] [Google Scholar]
- 1584.Huang C, Andres AM, Ratliff EP, et al. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011;6(6):e20975. doi: 10.1371/journal.pone.0020975. PubMed PMID: 21687634; PubMed Central PMCID: PMC3110820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1585.Gottlieb RA, Finley KD, Mentzer RM, Jr.. Cardioprotection requires taking out the trash. Basic Res Cardiol. 2009. Mar;104(2):169–80. doi: 10.1007/s00395-009-0011-9. PubMed PMID: 19242643; PubMed Central PMCID: PMC3661679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1586.Kaludercic N, Maiuri MC, Kaushik S, et al. Comprehensive autophagy evaluation in cardiac diseases models. Cardiovasc Res. 2019. Aug 27. doi: 10.1093/cvr/cvz233. PubMed PMID: 31504266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1587.Nakashima A, Tsuda S, Kusabiraki T, et al. Current understanding of autophagy in pregnancy. Int J Mol Sci. 2019. May 11;20(9). doi: 10.3390/ijms20092342. PubMed PMID: 31083536; PubMed Central PMCID: PMCPMC6539256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1588.Oh SY, Roh CR.. Autophagy in the placenta. Obstet Gynecol Sci. 2017. May; 60(3):241–259. doi: 10.5468/ogs.2017.60.3.241. PubMed PMID: 28534010; PubMed Central PMCID: PMCPMC5439273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1589.Avagliano L, Virgili E, Garo C, et al. Autophagy and human parturition: evaluation of LC3 expression in placenta from spontaneous or medically induced onset of labor. Biomed Res Int. 2013;2013:689768. doi: 10.1155/2013/689768. PubMed PMID: 23956998; PubMed Central PMCID: PMC3730383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1590.Hung TH, Hsieh TT, Chen SF, et al. Autophagy in the human placenta throughout gestation. PLoS One. 2013;8(12):e83475. doi: 10.1371/journal.pone.0083475. PubMed PMID: 24349516; PubMed Central PMCID: PMC3862763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1591.Signorelli P, Avagliano L, Virgili E, et al. Autophagy in term normal human placentas. Placenta. 2011. Jun;32(6):482–5. doi: 10.1016/j.placenta.2011.03.005. PubMed PMID: 21459442. [DOI] [PubMed] [Google Scholar]
- 1592.Hung TH, Chen SF, Lo LM, et al. Increased autophagy in placentas of intrauterine growth-restricted pregnancies. PLoS One. 2012;7(7):e40957. doi: 10.1371/journal.pone.0040957. PubMed PMID: 22815878; PubMed Central PMCID: PMC3397998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1593.Chang YL, Wang TH, Chang SD, et al. Increased autophagy in the placental territory of selective intrauterine growth-restricted monochorionic twins. Prenat Diagn. 2013. Feb;33(2):187–90. doi: 10.1002/pd.4040. PubMed PMID: 23288835. [DOI] [PubMed] [Google Scholar]
- 1594.Oh SY, Choi SJ, Kim KH, et al. Autophagy-related proteins, LC3 and Beclin-1, in placentas from pregnancies complicated by preeclampsia. Reprod Sci. 2008. Nov;15(9):912–20. doi: 10.1177/1933719108319159. PubMed PMID: 19050324. [DOI] [PubMed] [Google Scholar]
- 1595.Avagliano L, Danti L, Doi P, et al. Autophagy in placentas from acidotic newborns: an immunohistochemical study of LC3 expression. Placenta. 2013. Nov;34(11):1091–4. doi: 10.1016/j.placenta.2013.09.004. PubMed PMID: 24070620. [DOI] [PubMed] [Google Scholar]
- 1596.Mellen MA, de la Rosa EJ, Boya P.. Autophagy is not universally required for phosphatidyl-serine exposure and apoptotic cell engulfment during neural development. Autophagy. 2009. Oct;5(7):964–72. PubMed PMID: 19587526; eng. [DOI] [PubMed] [Google Scholar]
- 1597.Amato R, Catalani E, Dal Monte M, et al. Autophagy-mediated neuroprotection induced by octreotide in an ex vivo model of early diabetic retinopathy. Pharmacol Res. 2018. Feb;128:167–178. doi: 10.1016/j.phrs.2017.09.022. PubMed PMID: 28970178. [DOI] [PubMed] [Google Scholar]
- 1598.Cammalleri M, Locri F, Catalani E, et al. The beta adrenergic receptor blocker propranolol counteracts retinal dysfunction in a mouse model of oxygen induced retinopathy: restoring the balance between apoptosis and autophagy. Front Cell Neurosci. 2017;11:395. doi: 10.3389/fncel.2017.00395. PubMed PMID: 29375312; PubMed Central PMCID: PMCPMC5770647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1599.Perry CN, Kyoi S, Hariharan N, et al. Novel methods for measuring cardiac autophagy in vivo. Methods Enzymol. 2009;453:325–42. doi: 10.1016/S0076-6879(08)04016-0. PubMed PMID: 19216914; PubMed Central PMCID: PMC3658837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1600.Munafo DB, Colombo MI.. A novel assay to study autophagy: regulation of autophagosome vacuole size by amino acid deprivation. J Cell Sci. 2001. Oct;114(Pt 20):3619–29. PubMed PMID: 11707514; eng. [DOI] [PubMed] [Google Scholar]
- 1601.Carloni S, Buonocore G, Balduini W.. Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol Dis. 2008. Dec;32(3):329–39. doi: 10.1016/j.nbd.2008.07.022. PubMed PMID: 18760364. [DOI] [PubMed] [Google Scholar]
- 1602.Carloni S, Girelli S, Scopa C, et al. Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy. 2010. Apr;6(3):366–77. PubMed PMID: 20168088. [DOI] [PubMed] [Google Scholar]
- 1603.Niemann A, Baltes J, Elsasser HP.. Fluorescence properties and staining behavior of monodansylpentane, a structural homologue of the lysosomotropic agent monodansylcadaverine. J Histochem Cytochem. 2001. Feb;49(2):177–85. PubMed PMID: 11156686; eng. [DOI] [PubMed] [Google Scholar]
- 1604.Jung H, Leal-Ekman JS, Lu Q, et al. Atg14 protects the intestinal epithelium from TNF-triggered villus atrophy. Autophagy. 2019. Nov;15(11):1990–2001. doi: 10.1080/15548627.2019.1596495. PubMed PMID: 30894050; PubMed Central PMCID: PMCPMC6844524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1605.Subirada PV, Paz MC, Ridano ME, et al. Effect of Autophagy Modulators on Vascular, Glial, and Neuronal Alterations in the Oxygen-Induced Retinopathy Mouse Model. Front Cell Neurosci. 2019;13:279. doi: 10.3389/fncel.2019.00279. PubMed PMID: 31297049; PubMed Central PMCID: PMCPMC6608561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1606.Sarkar C, Jones JW, Hegdekar N, et al. PLA2G4A/cPLA2-mediated lysosomal membrane damage leads to inhibition of autophagy and neurodegeneration after brain trauma. Autophagy. 2020. Mar;16(3):466–485. doi: 10.1080/15548627.2019.1628538. PubMed PMID: 31238788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1607.Li Y, Jones JW, MCC H, et al. cPLA2 activation contributes to lysosomal defects leading to impairment of autophagy after spinal cord injury. Cell Death Dis. 2019. Jul 11;10(7):531. doi: 10.1038/s41419-019-1764-1. PubMed PMID: 31296844; PubMed Central PMCID: PMCPMC6624263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1608.Ginet V, Puyal J, Clarke PG, et al. Enhancement of autophagic flux after neonatal cerebral hypoxia-ischemia and its region-specific relationship to apoptotic mechanisms. Am J Pathol. 2009. Nov;175(5):1962–74. doi: 10.2353/ajpath.2009.090463. PubMed PMID: 19815706; PubMed Central PMCID: PMC2774060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1609.Penas C, Font-Nieves M, Fores J, et al. Autophagy, and BiP level decrease are early key events in retrograde degeneration of motoneurons. Cell Death Differ. 2011. Oct;18(10):1617–27. doi: 10.1038/cdd.2011.24. PubMed PMID: 21436843; PubMed Central PMCID: PMC3172115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1610.Colacurcio DJ, Nixon RA.. Disorders of lysosomal acidification-The emerging role of v-ATPase in aging and neurodegenerative disease. Ageing Res Rev. 2016. Dec;32:75–88. doi: 10.1016/j.arr.2016.05.004. PubMed PMID: 27197071; PubMed Central PMCID: PMCPMC5112157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1611.Rodriguez-Muela N, Hernandez-Pinto AM, Serrano-Puebla A, et al. Lysosomal membrane permeabilization and autophagy blockade contribute to photoreceptor cell death in a mouse model of retinitis pigmentosa. Cell Death Differ. 2014. Dec 12. doi: 10.1038/cdd.2014.203. PubMed PMID: 25501597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1612.Uchiyama Y. Autophagic cell death and its execution by lysosomal cathepsins. Arch Histol Cytol. 2001. Aug;64(3):233–46. PubMed PMID: 11575420; eng. [DOI] [PubMed] [Google Scholar]
- 1613.Marino ML, Fais S, Djavaheri-Mergny M, et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 2010. Oct 21;1:e87. doi: 10.1038/cddis.2010.67. PubMed PMID: 21368860; PubMed Central PMCID: PMCPMC3035900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1614.Udelnow A, Kreyes A, Ellinger S, et al. Omeprazole inhibits proliferation and modulates autophagy in pancreatic cancer cells. PLoS One. 2011;6(5):e20143. doi: 10.1371/journal.pone.0020143. PubMed PMID: 21629657; PubMed Central PMCID: PMC3101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1615.McWilliams TG, Barini E, Pohjolan-Pirhonen R, et al. Phosphorylation of Parkin at serine 65 is essential for its activation in vivo. Open Biol. 2018. Nov 7;8(11). doi: 10.1098/rsob.180108. PubMed PMID: 30404819; PubMed Central PMCID: PMCPMC6282074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1616.Shang D, Wang L, Klionsky DJ, et al. Sex differences in autophagy-mediated diseases: toward precision medicine. Autophagy. 2020. Apr 17:in press. doi: 10.1080/15548627.2020.1752511. PubMed PMID: 32264724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1617.Weber SM, Levitz SM.. Chloroquine interferes with lipopolysaccharide-induced TNF-alpha gene expression by a nonlysosomotropic mechanism [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.]. J Iimmunol. 2000. Aug 1;165(3):1534–40. PubMed PMID: 10903761; eng. [DOI] [PubMed] [Google Scholar]
- 1618.Ryzhikov M, Ehlers A, Steinberg D, et al. Diurnal Rhythms Spatially and Temporally Organize Autophagy. Cell Rep. 2019. Feb 12;26(7):1880–1892 e6. doi: 10.1016/j.celrep.2019.01.072. PubMed PMID: 30759397; PubMed Central PMCID: PMCPMC6442472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1619.Akagi Y, Isaka Y, Akagi A, et al. Transcriptional activation of a hybrid promoter composed of cytomegalovirus enhancer and beta-actin/beta-globin gene in glomerular epithelial cells in vivo. Kidney Int. 1997. Apr;51(4):1265–9. PubMed PMID: 9083295; eng. [DOI] [PubMed] [Google Scholar]
- 1620.Kimura T, Takabatake Y, Takahashi A, et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J Am Soc Nephrol. 2011. May;22(5):902–13. doi: 10.1681/ASN.2010070705. PubMed PMID: 21493778; PubMed Central PMCID: PMC3083312. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1621.Hartleben B, Godel M, Meyer-Schwesinger C, et al. Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest. 2010. Apr 1;120(4):1084–96. doi: 10.1172/JCI39492. PubMed PMID: 20200449; PubMed Central PMCID: PMC2846040. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1622.Cudjoe EK, Jr., Saleh T, Hawkridge AM, et al. Proteomics insights into autophagy. Proteomics. 2017. Oct;17(20). doi: 10.1002/pmic.201700022. PubMed PMID: 28902446. [DOI] [PubMed] [Google Scholar]
- 1623.Wong YK, Zhang J, Hua ZC, et al. Recent advances in quantitative and chemical proteomics for autophagy studies. Autophagy. 2017. Sep 2;13(9):1472–1486. doi: 10.1080/15548627.2017.1313944. PubMed PMID: 28820289; PubMed Central PMCID: PMC5612287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1624.McLoughlin F, Augustine RC, Marshall RS, et al. Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants. 2018. Dec;4(12):1056–1070. doi: 10.1038/s41477-018-0299-2. PubMed PMID: 30478358. [DOI] [PubMed] [Google Scholar]
- 1625.Schneider JL, Suh Y, Cuervo AM.. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014. Sep 2;20(3):417–32. doi: 10.1016/j.cmet.2014.06.009. PubMed PMID: 25043815; PubMed Central PMCID: PMCPMC4156578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1626.Le Guerroue F, Eck F, Jung J, et al. Autophagosomal content profiling reveals an LC3C-dependent piecemeal mitophagy pathway. Mol Cell. 2017. Nov 16;68(4):786–796 e6. doi: 10.1016/j.molcel.2017.10.029. PubMed PMID: 29149599. [DOI] [PubMed] [Google Scholar]
- 1627.Zhang T, Shen S, Qu J, et al. Global analysis of cellular protein flux quantifies the selectivity of basal autophagy. Cell Rep. 2016. Mar 15;14(10):2426–39. doi: 10.1016/j.celrep.2016.02.040. PubMed PMID: 26947064; PubMed Central PMCID: PMCPMC5470642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1628.Mellacheruvu D, Wright Z, Couzens AL, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013. Aug;10(8):730–6. doi: 10.1038/nmeth.2557. PubMed PMID: 23921808; PubMed Central PMCID: PMCPMC3773500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1629.Stryeck S, Birner-Gruenberger R, Madl T.. Integrative metabolomics as emerging tool to study autophagy regulation. Microb Cell. 2017. Jul 13;4(8):240–258. doi: 10.15698/mic2017.08.584. PubMed PMID: 28845422; PubMed Central PMCID: PMCPMC5568430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1630.Galdieri L, Zhang T, Rogerson D, et al. Protein acetylation and acetyl coenzyme a metabolism in budding yeast. Eukaryot Cell. 2014. Dec;13(12):1472–83. doi: 10.1128/EC.00189-14. PubMed PMID: 25326522; PubMed Central PMCID: PMCPMC4248685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1631.Hartl FU, Bracher A, Hayer-Hartl M.. Molecular chaperones in protein folding and proteostasis. Nature. 2011. Jul 20;475(7356):324–32. doi: 10.1038/nature10317. PubMed PMID: 21776078. [DOI] [PubMed] [Google Scholar]
- 1632.Bartlett AI, Radford SE.. An expanding arsenal of experimental methods yields an explosion of insights into protein folding mechanisms. Nat Struct Mol Biol. 2009. Jun;16(6):582–8. doi: 10.1038/nsmb.1592. PubMed PMID: 19491935. [DOI] [PubMed] [Google Scholar]
- 1633.Madeo F, Eisenberg T, Buttner S, et al. Spermidine: a novel autophagy inducer and longevity elixir. Autophagy. 2010. Jan;6(1):160–2. doi: 10.4161/auto.6.1.10600. PubMed PMID: 20110777. [DOI] [PubMed] [Google Scholar]
- 1634.Morselli E, Marino G, Bennetzen MV, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011. Feb 21;192(4):615–29. doi: 10.1083/jcb.201008167. PubMed PMID: 21339330; PubMed Central PMCID: PMCPMC3044119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1635.Pietrocola F, Lachkar S, Enot DP, et al. Spermidine induces autophagy by inhibiting the acetyltransferase EP300. Cell Death Differ. 2015. Dec 19;22:509–16. doi: 10.1038/cdd.2014.215. PubMed PMID: 25526088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1636.Saiki S, Sasazawa Y, Fujimaki M, et al. A metabolic profile of polyamines in parkinson disease: A promising biomarker. Ann Neurol. 2019. Aug;86(2):251–263. doi: 10.1002/ana.25516. PubMed PMID: 31155745; PubMed Central PMCID: PMCPMC6772170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1637.Gu H, Du J, Carnevale Neto F, et al. Metabolomics method to comprehensively analyze amino acids in different domains. Analyst. 2015. Apr 21;140(8):2726–34. doi 10.1039/c4an02386b. PubMed PMID: 25699545; PubMed Central PMCID: PMCPMC4380628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1638.Meijer AJ, Lorin S, Blommaart EF, et al. Regulation of autophagy by amino acids and MTOR-dependent signal transduction. Amino Acids. 2015. Oct;47(10):2037–63. doi: 10.1007/s00726-014-1765-4. PubMed PMID: 24880909; PubMed Central PMCID: PMCPMC4580722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1639.Kofeler HC, Fauland A, Rechberger GN, et al. Mass spectrometry based lipidomics: an overview of technological platforms. Metabolites. 2012. Jan 5;2(1):19–38. doi: 10.3390/metabo2010019. PubMed PMID: 24957366; PubMed Central PMCID: PMCPMC3901195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1640.Cao Y, Wen J, Li Y, et al. Uric acid and sphingomyelin enhance autophagy in iPS cell-originated cardiomyocytes through lncRNA MEG3/miR-7-5p/EGFR axis. Artif Cells Nanomed Biotechnol. 2019. Dec;47(1):3774–3785. doi: 10.1080/21691401.2019.1667817. PubMed PMID: 31559872. [DOI] [PubMed] [Google Scholar]
- 1641.Li X, Qi J, Zhu Q, et al. The role of androgen in autophagy of granulosa cells from PCOS. Gynecol Endocrinol. 2019. Aug;35(8):669–672. doi: 10.1080/09513590.2018.1540567. PubMed PMID: 31056990. [DOI] [PubMed] [Google Scholar]
- 1642.Qian X, Li X, Cai Q, et al. Phosphoglycerate Kinase 1 Phosphorylates Beclin1 to Induce Autophagy. Mol Cell. 2017. Mar 2;65(5):917–931 e6. doi: 10.1016/j.molcel.2017.01.027. PubMed PMID: 28238651; PubMed Central PMCID: PMCPMC5389741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1643.Vandrovcova J, Anaya F, Kay V, et al. Disentangling the role of the tau gene locus in sporadic tauopathies. Curr Alzheimer Res. 2010. Dec;7(8):726–34. PubMed PMID: 20704554. [DOI] [PubMed] [Google Scholar]
- 1644.Chen YS, Chen SD, Wu CL, et al. Induction of sestrin2 as an endogenous protective mechanism against amyloid beta-peptide neurotoxicity in primary cortical culture. Exp Neurol. 2014. Mar;253:63–71. doi: 10.1016/j.expneurol.2013.12.009. PubMed PMID: 24368194. [DOI] [PubMed] [Google Scholar]
- 1645.Ulamek-Koziol M, Kocki J, Bogucka-Kocka A, et al. Autophagy, mitophagy and apoptotic gene changes in the hippocampal CA1 area in a rat ischemic model of Alzheimer’s disease. Pharmacol Rep. 2017. Dec;69(6):1289–1294. doi: 10.1016/j.pharep.2017.07.015. PubMed PMID: 29128811. [DOI] [PubMed] [Google Scholar]
- 1646.Ulamek-Koziol M, Kocki J, Bogucka-Kocka A, et al. Dysregulation of autophagy, mitophagy, and apoptotic genes in the medial temporal lobe cortex in an ischemic model of Alzheimer’s disease. J Alzheimers Dis. 2016. Jul 27;54(1):113–21. doi: 10.3233/JAD-160387. PubMed PMID: 27472881; PubMed Central PMCID: PMCPMC5008226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1647.Tofaris GK, Spillantini MG.. Physiological and pathological properties of alpha-synuclein. Cell Mol Life Sci. 2007. Sep;64(17):2194–201. doi: 10.1007/s00018-007-7217-5. PubMed PMID: 17605001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1648.Wanker EE. Protein aggregation and pathogenesis of Huntington’s disease: mechanisms and correlations. Biol Chem. 2000. Sep-Oct;381(9–10):937–42. doi: 10.1515/BC.2000.114. PubMed PMID: 11076024. [DOI] [PubMed] [Google Scholar]
- 1649.Croce KR, Yamamoto A.. A role for autophagy in Huntington’s disease. Neurobiol Dis. 2019. Feb;122:16–22. doi: 10.1016/j.nbd.2018.08.010. PubMed PMID: 30149183; PubMed Central PMCID: PMCPMC6364695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1650.Sandri M, Coletto L, Grumati P, et al. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J Cell Sci. 2013. Dec1;126\(Pt 23):5325–33. doi: 10.1242/jcs.114041. PubMed PMID: 24293330. [DOI] [PubMed] [Google Scholar]
- 1651.Margeta M. Autophagy Defects in Skeletal Myopathies. Annu Rev Pathol. 2020. Jan 24;15:261–285. doi: 10.1146/annurev-pathmechdis-012419-032618. PubMed PMID: 31594457. [DOI] [PubMed] [Google Scholar]
- 1652.Bentmann E, Haass C, Dormann D.. Stress granules in neurodegeneration–lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma. FEBS J. 2013. Sep;280(18):4348–70. doi: 10.1111/febs.12287. PubMed PMID: 23587065. [DOI] [PubMed] [Google Scholar]
- 1653.Simonovitch S, Schmukler E, Masliah E, et al. The effects of APOE4 on mitochondrial dynamics and proteins in vivo. J Alzheimers Dis. 2019;70(3):861–875. doi: 10.3233/JAD-190074. PubMed PMID: 31306119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1654.Puyal J, Ginet V, Clarke PG.. Multiple interacting cell death mechanisms in the mediation of excitotoxicity and ischemic brain damage: a challenge for neuroprotection. Prog Neurobiol. 2013. Jun;105:24–48. doi: 10.1016/j.pneurobio.2013.03.002. PubMed PMID: 23567504. [DOI] [PubMed] [Google Scholar]
- 1655.Grishchuk Y, Ginet V, Truttmann AC, et al. Beclin 1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy. 2011. Oct;7(10):1115–31. doi: 10.4161/auto.7.10.16608. PubMed PMID: 21646862. [DOI] [PubMed] [Google Scholar]
- 1656.Menzies FM, Fleming A, Caricasole A, et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron. 2017. Mar 8;93(5):1015–1034. doi: 10.1016/j.neuron.2017.01.022. PubMed PMID: 28279350. [DOI] [PubMed] [Google Scholar]
- 1657.Comerota MM, Tumurbaatar B, Krishnan B, et al. Near infrared light treatment reduces synaptic levels of toxic tau oligomers in two transgenic mouse models of human tauopathies. Mol Neurobiol. 2019. May;56(5):3341–3355. doi: 10.1007/s12035-018-1248-9. PubMed PMID: 30120733; PubMed Central PMCID: PMCPMC6476871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1658.Scarffe LA, Stevens DA, Dawson VL, et al. Parkin and PINK1: much more than mitophagy. Trends in neurosciences. 2014. Jun;37(6):315–324. doi: 10.1016/j.tins.2014.03.004. PubMed PMID: 24735649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1659.Salminen A, Kaarniranta K, Haapasalo A, et al. Emerging role of p62/sequestosome-1 in the pathogenesis of Alzheimer’s disease. Prog Neurobiol. 2012. Jan;96(1):87–95. doi: 10.1016/j.pneurobio.2011.11.005. PubMed PMID: 22138392. [DOI] [PubMed] [Google Scholar]
- 1660.Seidel K, Brunt ER, de Vos RA, et al. The p62 antibody reveals various cytoplasmic protein aggregates in spinocerebellar ataxia type 6. Clin Neuropathol. 2009. Sep-Oct;28(5):344–9. PubMed PMID: 19788049. [DOI] [PubMed] [Google Scholar]
- 1661.Harada H, Warabi E, Matsuki T, et al. Deficiency of p62/Sequestosome 1 causes hyperphagia due to leptin resistance in the brain. J Neurosci. 2013. Sep 11;33(37):14767–77. doi: 10.1523/JNEUROSCI.2954-12.2013. PubMed PMID: 24027277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1662.Merenlender-Wagner A, Malishkevich A, Shemer Z, et al. Autophagy has a key role in the pathophysiology of schizophrenia. Mol Psychiatry. 2015. Dec 24;20:126–32. doi: 10.1038/mp.2013.174. PubMed PMID: 24365867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1663.Dresner E, Agam G, Gozes I.. Activity-dependent neuroprotective protein (ADNP) expression level is correlated with the expression of the sister protein ADNP2: deregulation in schizophrenia. Eur Neuropsychopharmacol. 2011. May;21(5):355–61. doi: 10.1016/j.euroneuro.2010.06.004. PubMed PMID: 20598862. [DOI] [PubMed] [Google Scholar]
- 1664.Lotze MT, Maranchie J, Appleman L.. Inhibiting autophagy: a novel approach for the treatment of renal cell carcinoma. Cancer J. 2013. Jul-Aug;19(4):341–7. doi: 10.1097/PPO.0b013e31829da0d6. PubMed PMID: 23867516. [DOI] [PubMed] [Google Scholar]
- 1665.Chude CI, Amaravadi RK.. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int J Mol Sci. 2017. Jun 16;18(6). doi: 10.3390/ijms18061279. PubMed PMID: 28621712; PubMed Central PMCID: PMCPMC5486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1666.Maes H, Kuchnio A, Peric A, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer cell. 2014. Aug 11;26(2):190–206. doi: 10.1016/j.ccr.2014.06.025. PubMed PMID: 25117709. [DOI] [PubMed] [Google Scholar]
- 1667.Maycotte P, Aryal S, Cummings CT, et al. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012. Feb 1;8(2):200–12. doi: 10.4161/auto.8.2.18554. PubMed PMID: 22252008; PubMed Central PMCID: PMCPMC3336076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1668.Ebrahimi-Fakhari D, Wahlster L, Hoffmann GF, et al. Emerging role of autophagy in pediatric neurodegenerative and neurometabolic diseases. Pediatr Res. 2014. Jan;75(1–2):217–26. doi: 10.1038/pr.2013.185. PubMed PMID: 24165736. [DOI] [PubMed] [Google Scholar]
- 1669.Lee KM, Hwang SK, Lee JA.. Neuronal autophagy and neurodevelopmental disorders. Exp Neurobiol. 2013. Sep;22(3):133–42. doi: 10.5607/en.2013.22.3.133. PubMed PMID: 24167408; PubMed Central PMCID: PMC3807000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1670.Yasin SA, Ali AM, Tata M, et al. mTOR-dependent abnormalities in autophagy characterize human malformations of cortical development: evidence from focal cortical dysplasia and tuberous sclerosis. Acta Neuropathol. 2013. Aug;126(2):207–18. doi: 10.1007/s00401-013-1135-4. PubMed PMID: 23728790. [DOI] [PubMed] [Google Scholar]
- 1671.Nishino I. Autophagic vacuolar myopathy. Semin Pediatr Neurol. 2006. Jun;13(2):90–5. doi: 10.1016/j.spen.2006.06.004. PubMed PMID: 17027858. [DOI] [PubMed] [Google Scholar]
- 1672.Girolamo F, Lia A, Amati A, et al. Overexpression of autophagic proteins in the skeletal muscle of sporadic inclusion body myositis. Neuropathol Appl Neurobiol. 2013. Dec;39(7):736–49. doi: 10.1111/nan.12040. PubMed PMID: 23452291. [DOI] [PubMed] [Google Scholar]
- 1673.Temiz P, Weihl CC, Pestronk A.. Inflammatory myopathies with mitochondrial pathology and protein aggregates. J Neurol Sci. 2009. Mar 15;278(1–2):25–9. doi: 10.1016/j.jns.2008.11.010. PubMed PMID: 19101700. [DOI] [PubMed] [Google Scholar]
- 1674.Maugeri N, Campana L, Gavina M, et al. Activated platelets present high mobility group box 1 to neutrophils, inducing autophagy and promoting the extrusion of neutrophil extracellular traps. J Thromb Haemost. 2014. Dec;12(12):2074–88. doi: 10.1111/jth.12710. PubMed PMID: 25163512. [DOI] [PubMed] [Google Scholar]
- 1675.Lunemann JD, Schmidt J, Schmid D, et al. Beta-amyloid is a substrate of autophagy in sporadic inclusion body myositis. Ann Neurol. 2007. May;61(5):476–83. doi: 10.1002/ana.21115. PubMed PMID: 17469125. [DOI] [PubMed] [Google Scholar]
- 1676.Screen M, Raheem O, Holmlund-Hampf J, et al. Gene expression profiling in tibial muscular dystrophy reveals unfolded protein response and altered autophagy. PLoS One. 2014;9(3):e90819. doi: 10.1371/journal.pone.0090819. PubMed PMID: 24618559; PubMed Central PMCID: PMC3949689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1677.Brady S, Squier W, Sewry C, et al. A retrospective cohort study identifying the principal pathological features useful in the diagnosis of inclusion body myositis. BMJ open. 2014;4(4):e004552. doi: 10.1136/bmjopen-2013-004552. PubMed PMID: 24776709; PubMed Central PMCID: PMC4010816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1678.Yu Z, Ma J, Li X, et al. Autophagy defects and related genetic variations in renal cell carcinoma with eosinophilic cytoplasmic inclusions. Sci Rep. 2018. Jul 2;8(1):9972. doi: 10.1038/s41598-018-28369-y. PubMed PMID: 29967346; PubMed Central PMCID: PMCPMC6028630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1679.Pan Q, Gao C, Chen Y, et al. Update on the role of autophagy in systemic lupus erythematosus: A novel therapeutic target. Biomed Pharmacother. 2015. Apr;71:190–3. doi: 10.1016/j.biopha.2015.02.017. PubMed PMID: 25960235. [DOI] [PubMed] [Google Scholar]
- 1680.An N, Chen Y, Wang C, et al. Chloroquine autophagic inhibition rebalances Th17/Treg-mediated immunity and ameliorates systemic lupus erythematosus. Cell Physiol Biochem. 2017;44(1):412–422. doi: 10.1159/000484955. PubMed PMID: 29141242. [DOI] [PubMed] [Google Scholar]
- 1681.Alessandri C, Barbati C, Vacirca D, et al. T lymphocytes from patients with systemic lupus erythematosus are resistant to induction of autophagy. FASEB J. 2012. Nov;26(11):4722–32. doi: 10.1096/fj.12-206060. PubMed PMID: 22835828; PubMed Central PMCID: PMCPMC3475261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1682.Tchetina EV, Poole AR, Zaitseva EM, et al. Differences in Mammalian target of rapamycin gene expression in the peripheral blood and articular cartilages of osteoarthritic patients and disease activity. Arthritis. 2013;2013:461486. doi: 10.1155/2013/461486. PubMed PMID: 23864948; PubMed Central PMCID: PMC3707211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1683.Mitroulis I, Kourtzelis I, Kambas K, et al. Evidence for the involvement of mTOR inhibition and basal autophagy in familial Mediterranean fever phenotype. Hum Immunol. 2011. Feb;72(2):135–8. doi: 10.1016/j.humimm.2010.11.006. PubMed PMID: 21081147. [DOI] [PubMed] [Google Scholar]
- 1684.Bachetti T, Chiesa S, Castagnola P, et al. Autophagy contributes to inflammation in patients with TNFR-associated periodic syndrome (TRAPS). Ann Rheum Dis. 2013. Jun;72(6):1044–52. doi: 10.1136/annrheumdis-2012-201952. PubMed PMID: 23117241. [DOI] [PubMed] [Google Scholar]
- 1685.Mi S, Li Z, Yang HZ, et al. Blocking IL-17A promotes the resolution of pulmonary inflammation and fibrosis via TGF-beta1-dependent and -independent mechanisms. J Iimmunol. 2011. Sep 15;187(6):3003–14. doi: 10.4049/jimmunol.1004081. PubMed PMID: 21841134. [DOI] [PubMed] [Google Scholar]
- 1686.Liu H, Mi S, Li Z, et al. Interleukin 17A inhibits autophagy through activation of PIK3CA to interrupt the GSK3B-mediated degradation of BCL2 in lung epithelial cells. Autophagy. 2013. May;9(5):730–42. doi: 10.4161/auto.24039. PubMed PMID: 23514933; PubMed Central PMCID: PMCPMC3669182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1687.Wang K, Zhang T, Lei Y, et al. Identification of ANXA2 (annexin A2) as a specific bleomycin target to induce pulmonary fibrosis by impeding TFEB-mediated autophagic flux. Autophagy. 2018;14(2):269–282. doi: 10.1080/15548627.2017.1409405. PubMed PMID: 29172997; PubMed Central PMCID: PMCPMC5902212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1688.Remijsen Q, Vanden Berghe T, Wirawan E, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011. Feb;21(2):290–304. doi: 10.1038/cr.2010.150. PubMed PMID: 21060338; PubMed Central PMCID: PMC3193439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1689.Mitroulis I, Kambas K, Chrysanthopoulou A, et al. Neutrophil extracellular trap formation is associated with IL-1beta and autophagy-related signaling in gout. PLoS One. 2011;6(12):e29318. doi: 10.1371/journal.pone.0029318. PubMed PMID: 22195044; PubMed Central PMCID: PMC3241704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1690.Apostolidou E, Skendros P, Kambas K, et al. Neutrophil extracellular traps regulate IL-1beta-mediated inflammation in familial Mediterranean fever. Ann Rheum Dis. 2016. Jan;75(1):269–77. doi: 10.1136/annrheumdis-2014-205958. PubMed PMID: 25261578. [DOI] [PubMed] [Google Scholar]
- 1691.Skendros P, Chrysanthopoulou A, Rousset F, et al. Regulated in development and DNA damage responses 1 (REDD1) links stress with IL-1beta-mediated familial Mediterranean fever attack through autophagy-driven neutrophil extracellular traps. J Allergy Clin Immunol. 2017. Nov;140(5):1378–1387 e13. doi: 10.1016/j.jaci.2017.02.021. PubMed PMID: 28342915. [DOI] [PubMed] [Google Scholar]
- 1692.Papagoras C, Chrysanthopoulou A, Mitsios A, et al. Autophagy inhibition in adult-onset Still’s disease: still more space for hydroxychloroquine? Clin Exp Rheumatol. 2017. Nov-Dec;35Suppl 108(6):133–134. PubMed PMID: 29148405. [PubMed] [Google Scholar]
- 1693.Angelidou I, Chrysanthopoulou A, Mitsios A, et al. REDD1/autophagy pathway is associated with neutrophil-driven IL-1beta inflammatory response in active ulcerative colitis. J Iimmunol. 2018. Jun 15;200(12):3950–3961. doi: 10.4049/jimmunol.1701643. PubMed PMID: 29712770. [DOI] [PubMed] [Google Scholar]
- 1694.Kambas K, Mitroulis I, Apostolidou E, et al. Autophagy mediates the delivery of thrombogenic tissue factor to neutrophil extracellular traps in human sepsis. PLoS One. 2012;7(9):e45427. doi: 10.1371/journal.pone.0045427. PubMed PMID: 23029002; PubMed Central PMCID: PMC3446899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1695.Stakos DA, Kambas K, Konstantinidis T, et al. Expression of functional tissue factor by neutrophil extracellular traps in culprit artery of acute myocardial infarction. Eur Heart J. 2015. Jun 7;36(22):1405–14. doi: 10.1093/eurheartj/ehv007. PubMed PMID: 25660055; PubMed Central PMCID: PMCPMC4458286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1696.Chrysanthopoulou A, Kambas K, Stakos D, et al. Interferon lambda1/IL-29 and inorganic polyphosphate are novel regulators of neutrophil-driven thromboinflammation. J Pathol. 2017. Sep;243(1):111–122. doi: 10.1002/path.4935. PubMed PMID: 28678391. [DOI] [PubMed] [Google Scholar]
- 1697.Chrysanthopoulou A, Mitroulis I, Apostolidou E, et al. Neutrophil extracellular traps promote differentiation and function of fibroblasts. J Pathol. 2014. Jul;233(3):294–307. doi: 10.1002/path.4359. PubMed PMID: 24740698. [DOI] [PubMed] [Google Scholar]
- 1698.Maugeri N, Capobianco A, Rovere-Querini P, et al. Platelet microparticles sustain autophagy-associated activation of neutrophils in systemic sclerosis. Sci Transl Med. 2018. Jul 25;10(451). doi: 10.1126/scitranslmed.aao3089. PubMed PMID: 30045975. [DOI] [PubMed] [Google Scholar]
- 1699.Manfredi AA, Rovere-Querini P, D’Angelo A, et al. Low molecular weight heparins prevent the induction of autophagy of activated neutrophils and the formation of neutrophil extracellular traps. Pharmacol Res. 2017. Sep;123:146–156. doi: 10.1016/j.phrs.2016.08.008. PubMed PMID: 28161237. [DOI] [PubMed] [Google Scholar]
- 1700.Dupont N, Jiang S, Pilli M, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011. Nov30;30(23):4701–11. doi: 10.1038/emboj.2011.398. PubMed PMID: 22068051; PubMed Central PMCID: PMC3243609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1701.Kambas K, Chrysanthopoulou A, Vassilopoulos D, et al. Tissue factor expression in neutrophil extracellular traps and neutrophil derived microparticles in antineutrophil cytoplasmic antibody associated vasculitis may promote thromboinflammation and the thrombophilic state associated with the disease. Ann Rheum Dis. 2014. Jul 19;73:1854–63. doi: 10.1136/annrheumdis-2013-203430. PubMed PMID: 23873874. [DOI] [PubMed] [Google Scholar]
- 1702.Berton G. Editorial: Gigantism: a new way to prolong neutrophil life. J Leukoc Biol. 2014. Oct;96(4):505–6. doi: 10.1189/jlb.3CE0214-107R. PubMed PMID: 25271292. [DOI] [PubMed] [Google Scholar]
- 1703.Dyugovskaya L, Berger S, Polyakov A, et al. The development of giant phagocytes in long-term neutrophil cultures. J Leukoc Biol. 2014. Oct;96(4):511–21. doi 10.1189/jlb.0813437. PubMed PMID: 24577569. [DOI] [PubMed] [Google Scholar]
- 1704.Masini M, Bugliani M, Lupi R, et al. Autophagy in human type 2 diabetes pancreatic beta cells. Diabetologia. 2009. Jun;52(6):1083–6. doi: 10.1007/s00125-009-1347-2. PubMed PMID: 19367387. [DOI] [PubMed] [Google Scholar]
- 1705.Mizukami H, Takahashi K, Inaba W, et al. Involvement of oxidative stress-induced DNA damage, endoplasmic reticulum stress, and autophagy deficits in the decline of beta-cell mass in Japanese type 2 diabetic patients. Diabetes care. 2014. Jul;37(7):1966–1974. doi: 10.2337/dc13-2018. PubMed PMID: 24705612. [DOI] [PubMed] [Google Scholar]
- 1706.Kosacka J, Kern M, Kloting N, et al. Autophagy in adipose tissue of patients with obesity and type 2 diabetes. Mol Cell Endocrinol. 2015. Jul 5;409:21–32. doi: 10.1016/j.mce.2015.03.015. PubMed PMID: 25818883. [DOI] [PubMed] [Google Scholar]
- 1707.Stienstra R, Haim Y, Riahi Y, et al. Autophagy in adipose tissue and the beta cell:implications for obesity and diabetes. Diabetologia. 2014. Aug;57(8):1505–16. doi: 10.1007/s00125-014-3255-3. PubMed PMID: 24795087. [DOI] [PubMed] [Google Scholar]
- 1708.Tatsuno K, Yamazaki T, Hanlon D, et al. Extracorporeal photochemotherapy induces bona fide immunogenic cell death. Cell Death Dis. 2019. Aug 2;10(8):578. doi: 10.1038/s41419-019-1819-3. PubMed PMID: 31371700; PubMed Central PMCID: PMCPMC6675789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1709.Garg AD, Krysko DV, Verfaillie T, et al. A novel pathway combining calreticulin exposure and ATP secretion in immunogenic cancer cell death. EMBO J. 2012. Mar 7;31(5):1062–79. doi: 10.1038/emboj.2011.497. PubMed PMID: 22252128; PubMed Central PMCID: PMC3298003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1710.Bacellar IO, Tsubone TM, Pavani C, et al. Photodynamic Efficiency: From Molecular Photochemistry to Cell Death. Int J Mol Sci. 2015. Aug 31;16(9):20523–59. doi: 10.3390/ijms160920523. PubMed PMID: 26334268; PubMed Central PMCID: PMCPMC4613217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1711.Dewaele M, Martinet W, Rubio N, et al. Autophagy pathways activated in response to PDT contribute to cell resistance against ROS damage. J Cell Mol Med. 2011. Jun;15(6):1402–14. doi: 10.1111/j.1582-4934.2010.01118.x. PubMed PMID: 20626525; PubMed Central PMCID: PMCPMC4373339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1712.Du L, Jiang N, Wang G, et al. Autophagy inhibition sensitizes bladder cancer cells to the photodynamic effects of the novel photosensitizer chlorophyllin e4. J Photochem Photobiol B. 2014. Apr 5;133:1–10. doi: 10.1016/j.jphotobiol.2014.02.010. PubMed PMID: 24650577. [DOI] [PubMed] [Google Scholar]
- 1713.Kessel D, Reiners JJ, Jr.. Effects of combined lysosomal and mitochondrial photodamage in a non-small-cell lung cancer cell line: the role of paraptosis. Photochem Photobiol. 2017. Nov;93(6):1502–1508. doi: 10.1111/php.12805. PubMed PMID: 28696570; PubMed Central PMCID: PMCPMC5693656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1714.Kessel DH, Price M, Reiners JJ, Jr.. ATG7 deficiency suppresses apoptosis and cell death induced by lysosomal photodamage. Autophagy. 2012. Sep;8(9):1333–41. doi: 10.4161/auto.20792. PubMed PMID: 22889762; PubMed Central PMCID: PMCPMC3442880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1715.Lihuan D, Jingcun Z, Ning J, et al. Photodynamic therapy with the novel photosensitizer chlorophyllin f induces apoptosis and autophagy in human bladder cancer cells. Lasers Surg Med. 2014. Apr;46(4):319–34. doi: 10.1002/lsm.22225. PubMed PMID: 24464873. [DOI] [PubMed] [Google Scholar]
- 1716.Galluzzi L, Kepp O, Kroemer G.. Enlightening the impact of immunogenic cell death in photodynamic cancer therapy. EMBO J. 2012. Mar 7;31(5):1055–7. doi: 10.1038/emboj.2012.2. PubMed PMID: 22252132; PubMed Central PMCID: PMC3298006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1717.Panzarini E, Inguscio V, Fimia GM, et al. Rose Bengal Acetate PhotoDynamic Therapy (RBAc-PDT) induces exposure and release of damage-associated molecular patterns (DAMPs) in human HeLa cells. PLoS One 2014;9:e105778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1718.Santin G, Bottone MG, Malatesta M, et al. Regulated forms of cell death are induced by the photodynamic action of the fluorogenic substrate, Hypocrellin B-acetate. J Photochem Photobiol B. 2013. Aug 5;125:90–7. doi: 10.1016/j.jphotobiol.2013.05.006. PubMed PMID: 23770337. [DOI] [PubMed] [Google Scholar]
- 1719.Maes H, Rubio N, Garg AD, et al. Autophagy: shaping the tumor microenvironment and therapeutic response. Trends Mol Med. 2013. Jul;19(7):428–46. doi: 10.1016/j.molmed.2013.04.005. PubMed PMID: 23714574. [DOI] [PubMed] [Google Scholar]
- 1720.Garg AD, Krysko DV, Vandenabeele P, et al. The emergence of phox-ER stress induced immunogenic apoptosis. Oncoimmunology. 2012. Aug 1;1(5):786–788. doi: 10.4161/onci.19750. PubMed PMID: 22934283; PubMed Central PMCID: PMC3429595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1721.Garg AD, Martin S, Golab J, et al. Danger signalling during cancer cell death: origins, plasticity and regulation. Cell Death Differ. 2014. Jan;21(1):26–38. doi: 10.1038/cdd.2013.48. PubMed PMID: 23686135; PubMed Central PMCID: PMC3858605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1722.Kroemer G, Galluzzi L, Kepp O, et al. Immunogenic cell death in cancer therapy. Annu Rev Immunol. 2013;31:51–72. doi: 10.1146/annurev-immunol-032712-100008. PubMed PMID: 23157435. [DOI] [PubMed] [Google Scholar]
- 1723.Dudek AM, Garg AD, Krysko DV, et al. Inducers of immunogenic cancer cell death. Cytokine Growth Factor Rev. 2013. Aug;24(4):319–33. doi: 10.1016/j.cytogfr.2013.01.005. PubMed PMID: 23391812. [DOI] [PubMed] [Google Scholar]
- 1724.Garg AD, Dudek AM, Ferreira GB, et al. ROS-induced autophagy in cancer cells assists in evasion from determinants of immunogenic cell death. Autophagy. 2013. Sep;9(9):1292–307. doi: 10.4161/auto.25399. PubMed PMID: 23800749. [DOI] [PubMed] [Google Scholar]
- 1725.Yamazaki T, Kirchmair A, Sato A, et al. Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat Immunol. 2020. Oct;21(10):1160–1171. doi: 10.1038/s41590-020-0751-0. PubMed PMID: 32747819. [DOI] [PubMed] [Google Scholar]
- 1726.Michaud M, Martins I, Sukkurwala AQ, et al. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science. 2011. Dec 16;334(6062):1573–7. doi: 10.1126/science.1208347. PubMed PMID: 22174255. [DOI] [PubMed] [Google Scholar]
- 1727.Bian S, Sun X, Bai A, et al. P2X7 integrates PI3K/AKT and AMPK-PRAS40-mTOR signaling pathways to mediate tumor cell death. PLoS One. 2013;8(4):e60184. doi: 10.1371/journal.pone.0060184. PubMed PMID: 23565201; PubMed Central PMCID: PMC3615040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1728.Di Virgilio F. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol Sci. 2007. Sep;28(9):465–72. doi: 10.1016/j.tips.2007.07.002. PubMed PMID: 17692395. [DOI] [PubMed] [Google Scholar]
- 1729.Garg AD, Dudek AM, Agostinis P.. Calreticulin surface exposure is abrogated in cells lacking, chaperone-mediated autophagy-essential gene, LAMP2A. Cell Death Dis. 2013;4:e826. doi: 10.1038/cddis.2013.372. PubMed PMID: 24091669; PubMed Central PMCID: PMC3824681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1730.Poon AH, Chouiali F, Tse SM, et al. Genetic and histologic evidence for autophagy in asthma pathogenesis. J Allergy Clin Immunol. 2012. Feb;129(2):569–71. doi: 10.1016/j.jaci.2011.09.035. PubMed PMID: 22040902; PubMed Central PMCID: PMCPMC3268897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1731.Martin LJ, Gupta J, Jyothula SS, et al. Functional variant in the autophagy-related 5 gene promotor is associated with childhood asthma. PLoS One. 2012;7(4):e33454. doi: 10.1371/journal.pone.0033454. PubMed PMID: 22536318; PubMed Central PMCID: PMCPMC3335039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1732.McAlinden KD, Deshpande DA, Ghavami S, et al. Autophagy activation in asthma airways remodeling. Am J Respir Cell Mol Biol. 2019. May;60(5):541–553. doi: 10.1165/rcmb.2018-0169OC. PubMed PMID: 30383396; PubMed Central PMCID: PMCPMC6503620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1733.Orenstein SJ, Kuo SH, Tasset I, et al. Interplay of LRRK2 with chaperone-mediated autophagy. Nat Neurosci. 2013. Apr;16(4):394–406. doi: 10.1038/nn.3350. PubMed PMID: 23455607; PubMed Central PMCID: PMC3609872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1734.Napolitano G, Johnson JL, He J, et al. Impairment of chaperone-mediated autophagy leads to selective lysosomal degradation defects in the lysosomal storage disease cystinosis. EMBO Mol Med. 2015. Feb;7(2):158–74. doi: 10.15252/emmm.201404223. PubMed PMID: 25586965; PubMed Central PMCID: PMC4328646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1735.Venugopal B, Mesires NT, Kennedy JC, et al. Chaperone-mediated autophagy is defective in mucolipidosis type IV. J Cell Physiol. 2009. May;219(2):344–53. doi: 10.1002/jcp.21676. PubMed PMID: 19117012. [DOI] [PubMed] [Google Scholar]
- 1736.Franch HA. Pathways of proteolysis affecting renal cell growth. Curr Opin Nephrol Hypertens. 2002. Jul;11(4):445–50. PubMed PMID: 12105396. [DOI] [PubMed] [Google Scholar]
- 1737.Sooparb S, Price SR, Shaoguang J, et al. Suppression of chaperone-mediated autophagy in the renal cortex during acute diabetes mellitus. Kidney Int. 2004. Jun;65(6):2135–44. doi: 10.1111/j.1523-1755.2004.00639.x, PubMed PMID: 15149326. [DOI] [PubMed] [Google Scholar]
- 1738.Chen ZH, Kim HP, Sciurba FC, et al. Egr-1 regulates autophagy in cigarette smoke-induced chronic obstructive pulmonary disease. PLoS One. 2008;3(10):e3316. doi: 10.1371/journal.pone.0003316. PubMed PMID: 18830406; PubMed Central PMCID: PMC2552992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1739.Qaisiya M, Mardesic P, Pastore B, et al. The activation of autophagy protects neurons and astrocytes against bilirubin-induced cytotoxicity. Neurosci Lett. 2017. Nov 20;661:96–103. doi: 10.1016/j.neulet.2017.09.056. PubMed PMID: 28965934. [DOI] [PubMed] [Google Scholar]
- 1740.Wasko MC, Hubert HB, Lingala VB, et al. Hydroxychloroquine and risk of diabetes in patients with rheumatoid arthritis. JAMA. 2007. Jul 11;298(2):187–93. doi: 10.1001/jama.298.2.187. PubMed PMID: 17622600. [DOI] [PubMed] [Google Scholar]
- 1741.Merlini L, Nishino I, Consortium for Autophagy in Muscular D. 201st ENMC International Workshop: Autophagy in muscular dystrophies–translational approach, 1–3 November 2013, Bussum, The Netherlands. Neuromuscular disorders: NMD. 2014 Jun;24(6):546–61. doi: 10.1016/j.nmd.2014.03.009. PubMed PMID: 24746377. [DOI] [PubMed] [Google Scholar]
- 1742.Wang KZQ, Steer E, Otero PA, et al. PINK1 Interacts with VCP/p97 and Activates PKA to Promote NSFL1C/p47 Phosphorylation and Dendritic Arborization in Neurons. eNeuro. 2018. Nov-Dec;5(6). doi: 10.1523/ENEURO.0466-18.2018. PubMed PMID: 30783609; PubMed Central PMCID: PMCPMC6377406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1743.Denton D, Kumar S.. Autophagy-dependent cell death. Cell Death Differ. 2019. Mar;26(4):605–616. doi: 10.1038/s41418-018-0252-y. PubMed PMID: 30568239; PubMed Central PMCID: PMCPMC6460387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1744.Doherty J, Baehrecke EH.. Life, death and autophagy. Nat Cell Biol. 2018. Oct;20(10):1110–1117. doi: 10.1038/s41556-018-0201-5. PubMed PMID: 30224761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1745.Ramos-Torres A, Bort A, Morell C, et al. The pepper’s natural ingredient capsaicin induces autophagy blockage in prostate cancer cells. Oncotarget. 2016. Jan 12;7(2):1569–83. doi: 10.18632/oncotarget.6415. PubMed PMID: 26625315; PubMed Central PMCID: PMCPMC4811481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1746.Beaulaton J, Lockshin RA.. Ultrastructural study of the normal degeneration of the intersegmental muscles of Anthereae polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J Morphol. 1977. Oct;154(1):39–57. doi: 10.1002/jmor.1051540104. PubMed PMID: 915948; eng. [DOI] [PubMed] [Google Scholar]
- 1747.Clarke PG. Developmental cell death: morphological diversity and multiple mechanisms. Anatomy and embryology. 1990;181(3):195–213. PubMed PMID: 2186664; eng. [DOI] [PubMed] [Google Scholar]
- 1748.Assuncao Guimaraes C, Linden R.. Programmed cell deaths. Apoptosis and alternative deathstyles. Eur J Biochem/FEBS. 2004. May;271(9):1638–50. doi: 10.1111/j.1432-1033.2004.04084.x. PubMed PMID: 15096203. [DOI] [PubMed] [Google Scholar]
- 1749.Guimaraes CA, Benchimol M, Amarante-Mendes GP, et al. Alternative programs of cell death in developing retinal tissue. J Biol Chem. 2003. Oct 24;278(43):41938–46. doi: 10.1074/jbc.M306547200. PubMed PMID: 12917395. [DOI] [PubMed] [Google Scholar]
- 1750.Napoletano F, Baron O, Vandenabeele P, et al. Intersections between Regulated Cell Death and Autophagy. Trends Cell Biol. 2019. Apr;29(4):323–338. doi: 10.1016/j.tcb.2018.12.007. PubMed PMID: 30665736. [DOI] [PubMed] [Google Scholar]
- 1751.Baron O, Fanto M.. Karyoptosis: A novel type of cell death caused by chronic autophagy inhibition. Autophagy. 2018;14(4):722–723. doi: 10.1080/15548627.2018.1434372. PubMed PMID: 29388501; PubMed Central PMCID: PMCPMC5959325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1752.Barthet VJA, Ryan KM.. Autophagy in neurodegeneration: can’t digest it, spit it out! Trends Cell Biol. 2018. Mar;28(3):171–173. doi: 10.1016/j.tcb.2018.01.001. PubMed PMID: 29395716. [DOI] [PubMed] [Google Scholar]
- 1753.Yu H, Yin S, Zhou S, et al. Magnolin promotes autophagy and cell cycle arrest via blocking LIF/Stat3/Mcl-1 axis in human colorectal cancers. Cell Death Dis. 2018. Jun 13;9(6):702. doi: 10.1038/s41419-018-0660-4. PubMed PMID: 29899555; PubMed Central PMCID: PMCPMC5999973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1754.Yu H, Qiu Y, Pang X, et al. Lycorine promotes autophagy and apoptosis via TCRP1/Akt/mTOR axis inactivation in human hepatocellular carcinoma. Mol Cancer Ther. 2017. Dec;16(12):2711–2723. doi: 10.1158/1535-7163.MCT-17-0498. PubMed PMID: 28974556. [DOI] [PubMed] [Google Scholar]
- 1755.Richard VR, Beach A, Piano A, et al. Mechanism of liponecrosis, a distinct mode of programmed cell death. Cell cycle. 2014;13(23):3707–26. doi: 10.4161/15384101.2014.965003. PubMed PMID: 25483081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1756.Sheibani S, Richard VR, Beach A, et al. Macromitophagy, neutral lipids synthesis, and peroxisomal fatty acid oxidation protect yeast from “liponecrosis”, a previously unknown form of programmed cell death. Cell cycle. 2014;13(1):138–47. PubMed PMID: 24196447; PubMed Central PMCID: PMC3925724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1757.Tang D, Kang R, Berghe TV, et al. The molecular machinery of regulated cell death. Cell Res. 2019. May;29(5):347–364. doi: 10.1038/s41422-019-0164-5. PubMed PMID: 30948788; PubMed Central PMCID: PMCPMC6796845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1758.Galluzzi L, Aaronson SA, Abrams J, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes [Research Support, N.I.H., Intramural Research Support, Non-U.S. Gov’t Review]. Cell Death Differ. 2009. Aug;16(8):1093–107. doi: 10.1038/cdd.2009.44. PubMed PMID: 19373242; PubMed Central PMCID: PMC2757140. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1759.Galluzzi L, Bravo-San Pedro JM, Vitale I, et al. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 2015. Jan;22(1):58–73. doi: 10.1038/cdd.2014.137. PubMed PMID: 25236395; PubMed Central PMCID: PMC4262782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1760.Minina EA, Bozhkov PV, Hofius D.. Autophagy as initiator or executioner of cell death. Trends Plant Sci. 2014. Aug 22. doi: 10.1016/j.tplants.2014.07.007. PubMed PMID: 25156061. [DOI] [PubMed] [Google Scholar]
- 1761.van Doorn WG, Beers EP, Dangl JL, et al. Morphological classification of plant cell deaths. Cell Death Differ. 2011. Aug;18(8):1241–6. doi: 10.1038/cdd.2011.36. PubMed PMID: 21494263; PubMed Central PMCID: PMC3172093. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1762.Kwon SI, Cho HJ, Jung JH, et al. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J Cell Mol Biol 2010. Jul 23;64:151–64. . PubMed PMID: 20659276; Eng. [DOI] [PubMed] [Google Scholar]
- 1763.Minina EA, Filonova LH, Fukada K, et al. Autophagy and metacaspase determine the mode of cell death in plants. J Cell Biol. 2013. Dec 23;203(6):917–27. doi: 10.1083/jcb.201307082. PubMed PMID: 24344187; PubMed Central PMCID: PMC3871426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1764.Hofius D, Schultz-Larsen T, Joensen J, et al. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009. May 15;137(4):773–83. doi: 10.1016/j.cell.2009.02.036. PubMed PMID: 19450522. [DOI] [PubMed] [Google Scholar]
- 1765.Giusti C, Tresse E, Luciani MF, et al. Autophagic cell death: analysis in Dictyostelium [Research Support, Non-U.S. Gov’t Review]. Biochim Biophys Acta. 2009. Sep;1793(9):1422–31. doi: 10.1016/j.bbamcr.2008.12.005. PubMed PMID: 19133302; eng. [DOI] [PubMed] [Google Scholar]
- 1766.Luciani MF, Giusti C, Harms B, et al. Atg1 allows second-signaled autophagic cell death in Dictyostelium. Autophagy. 2011. May;7(5):501–8. PubMed PMID: 21301205; eng. [DOI] [PubMed] [Google Scholar]
- 1767.Uchikawa T, Yamamoto A, Inouye K.. Origin and function of the stalk-cell vacuole in Dictyostelium. Dev Biol. 2011. Apr 1;352(1):48–57. doi: 10.1016/j.ydbio.2011.01.014. PubMed PMID: 21256841; eng. [DOI] [PubMed] [Google Scholar]
- 1768.Sharma D, Otto G, Warren EC, et al. Gamma secretase orthologs are required for lysosomal activity and autophagic degradation in Dictyostelium discoideum, independent of PSEN (presenilin) proteolytic function. Autophagy. 2019. Aug;15(8):1407–1418. doi: 10.1080/15548627.2019.1586245. PubMed PMID: 30806144; PubMed Central PMCID: PMCPMC6613883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1769.Ludtmann MH, Otto GP, Schilde C, et al. An ancestral non-proteolytic role for presenilin proteins in multicellular development of the social amoeba Dictyostelium discoideum. J Cell Sci. 2014. Apr1;127(Pt 7):1576–84. doi: 10.1242/jcs.140939. PubMed PMID: 24463814; PubMed Central PMCID: PMCPMC3970561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1770.Denton D, Xu T, Dayan S, et al. Dpp regulates autophagy-dependent midgut removal and signals to block ecdysone production. Cell Death Differ. 2019. Mar;26(4):763–778. doi: 10.1038/s41418-018-0154-z. PubMed PMID: 29959404; PubMed Central PMCID: PMCPMC6460390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1771.Xu T, Nicolson S, Denton D, et al. Distinct requirements of Autophagy-related genes in programmed cell death. Cell Death Differ. 2015. Nov;22(11):1792–802. . PubMed PMID: 25882046; PubMed Central PMCID: PMCPMC4648326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1772.Berry DL, Baehrecke EH.. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007. Dec 14;131(6):1137–48. doi: 10.1016/j.cell.2007.10.048. PubMed PMID: 18083103; PubMed Central PMCID: PMC2180345. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1773.Daish TJ, Mills K, Kumar S.. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell. 2004. Dec;7(6):909–15. doi: 10.1016/j.devcel.2004.09.018. PubMed PMID: 15572132. [DOI] [PubMed] [Google Scholar]
- 1774.Mills K, Daish T, Harvey KF, et al. The Drosophila melanogaster Apaf-1 homologue ARK is required for most, but not all, programmed cell death. J Cell Biol. 2006. Mar 13;172(6):809–15. doi: 10.1083/jcb.200512126. PubMed PMID: 16533943; PubMed Central PMCID: PMCPMC2063725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1775.Denton D, Aung-Htut MT, Lorensuhewa N, et al. UTX coordinates steroid hormone-mediated autophagy and cell death.Nat Commun. 2013;4:2916. doi: 10.1038/ncomms3916. PubMed PMID: 24336022; PubMed Central PMCID: PMCPMC3973156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1776.Koenig U, Robenek H, Barresi C, et al. Cell death induced autophagy contributes to terminal differentiation of skin and skin appendages. Autophagy. 2019. Aug 4:1–14. doi: 10.1080/15548627.2019.1646552. PubMed PMID: 31379249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1777.Jung S, Choe S, Woo H, et al. Autophagic death of neural stem cells mediates chronic stress-induced decline of adult hippocampal neurogenesis and cognitive deficits. Autophagy. 2020. Mar;16(3):512–530. doi: 10.1080/15548627.2019.1630222. PubMed PMID: 31234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1778.Zhou B, Liu J, Kang R, et al. Ferroptosis is a type of autophagy-dependent cell death. Semin Cancer Biol. 2019. Mar 14. doi: 10.1016/j.semcancer.2019.03.002. PubMed PMID: 30880243. [DOI] [PubMed] [Google Scholar]
- 1779.Bai Y, Meng L, Han L, et al. Lipid storage and lipophagy regulates ferroptosis. Biochem Biophys Res Commun. 2019. Jan 22;508(4):997–1003. doi: 10.1016/j.bbrc.2018.12.039.PubMed PMID: 30545638. [DOI] [PubMed] [Google Scholar]
- 1780.Song X, Zhu S, Chen P, et al. AMPK-Mediated BECN1 Phosphorylation Promotes Ferroptosis by Directly Blocking System Xc(-) Activity. Curr Biol. 2018. Aug 6;28(15):2388–2399 e5. doi: 10.1016/j.cub.2018.05.094. PubMed PMID: 30057310; PubMed Central PMCID: PMCPMC6081251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1781.Kang R, Zhu S, Zeh HJ, et al. BECN1 is a new driver of ferroptosis. Autophagy. 2018;14(12):2173–2175. doi: 10.1080/15548627.2018.1513758. PubMed PMID: 30145930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1782.Gao H, Bai Y, Jia Y, et al. Ferroptosis is a lysosomal cell death process. Biochem Biophys Res Commun. 2018. Sep 10;503(3):1550–1556. doi: 10.1016/j.bbrc.2018.07.078. PubMed PMID: 30031610. [DOI] [PubMed] [Google Scholar]
- 1783.Sperandio S, de Belle I, Bredesen DE.. An alternative, nonapoptotic form of programmed cell death.Proceedings of the National Academy of Sciences of the United States of America. 2000. Dec 19;97(26):14376–81. doi: 10.1073/pnas.97.26.14376. PubMed PMID: 11121041; PubMed Central PMCID: PMCPMC18926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1784.Kar R, Singha PK, Venkatachalam MA, et al. A novel role for MAP1 LC3 in nonautophagic cytoplasmic vacuolation death of cancer cells. Oncogene. 2009. Jul 16;28(28):2556–68. doi: 10.1038/onc.2009.118. PubMed PMID: 19448671; PubMed Central PMCID: PMCPMC2717022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1785.Wang WB, Feng LX, Yue QX, et al. Paraptosis accompanied by autophagy and apoptosis was induced by celastrol, a natural compound with influence on proteasome, ER stress and Hsp90. J Cell Physiol. 2012. May;227(5):2196–206. doi: 10.1002/jcp.22956. PubMed PMID: 21866552. [DOI] [PubMed] [Google Scholar]
- 1786.Singha PK, Pandeswara S, Venkatachalam MA, et al. Manumycin A inhibits triple-negative breast cancer growth through LC3-mediated cytoplasmic vacuolation death. Cell Death Dis. 2013. Jan 17;4:e457. doi: 10.1038/cddis.2012.192. PubMed PMID: 23328664; PubMed Central PMCID: PMCPMC3563980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1787.Ram BM, Ramakrishna G.. Endoplasmic reticulum vacuolation and unfolded protein response leading to paraptosis like cell death in cyclosporine A treated cancer cervix cells is mediated by cyclophilin B inhibition. Biochim Biophys Acta. 2014. Nov;1843(11):2497–512. doi: 10.1016/j.bbamcr.2014.06.020. PubMed PMID: 25003316. [DOI] [PubMed] [Google Scholar]
- 1788.Zheng H, Dong Y, Li L, et al. Novel Benzo[a]quinolizidine Analogs Induce Cancer Cell Death through Paraptosis and Apoptosis. J Med Chem. 2016. May 26;59(10):5063–76. doi: 10.1021/acs.jmedchem.6b00484. PubMed PMID: 27077446. [DOI] [PubMed] [Google Scholar]
- 1789.Mi X, Wang C, Sun C, et al. Xanthohumol induces paraptosis of leukemia cells through p38 mitogen activated protein kinase signaling pathway. Oncotarget. 2017. May 9;8(19):31297–31304. doi: 10.18632/oncotarget.16185. PubMed PMID: 28415750; PubMed Central PMCID: PMCPMC5458208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1790.Nedungadi D, Binoy A, Pandurangan N, et al. 6-Shogaol induces caspase-independent paraptosis in cancer cells via proteasomal inhibition. Exp Cell Res. 2018. Mar 15;364(2):243–251. doi: 10.1016/j.yexcr.2018.02.018. PubMed PMID: 29462602. [DOI] [PubMed] [Google Scholar]
- 1791.Binoy A, Nedungadi D, Katiyar N, et al. Plumbagin induces paraptosis in cancer cells by disrupting the sulfhydryl homeostasis and proteasomal function. Chem Biol Interact. 2019. Sep 1;310:108733. doi: 10.1016/j.cbi.2019.108733. PubMed PMID: 31276663. [DOI] [PubMed] [Google Scholar]
- 1792.Wang L, Gundelach JH, Bram RJ.. Cycloheximide promotes paraptosis induced by inhibition of cyclophilins in glioblastoma multiforme. Cell Death Dis. 2017. May 18;8(5):e2807. doi: 10.1038/cddis.2017.217. PubMed PMID: 28518150; PubMed Central PMCID: PMCPMC5520731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1793.Tsai TL, Wang HC, Hung CH, et al. Wheat germ agglutinin-induced paraptosis-like cell death and protective autophagy is mediated by autophagy-linked FYVE inhibition. Oncotarget. 2017. Oct 31;8(53):91209–91222. doi: 10.18632/oncotarget.20436. PubMed PMID: 29207637; PubMed Central PMCID: PMCPMC5710917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1794.Zhang C, Jiang Y, Zhang J, et al. 8-p-Hdroxybenzoyl Tovarol Induces Paraptosis Like Cell Death and Protective Autophagy in Human Cervical Cancer HeLa Cells. Int J Mol Sci. 2015. Jul 2;16(7):14979–96. doi: 10.3390/ijms160714979. PubMed PMID: 26147427; PubMed Central PMCID: PMCPMC4519883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1795.Han H, Chou CC, Li R, et al. Chalcomoracin is a potent anticancer agent acting through triggering Oxidative stress via a mitophagy- and paraptosis-dependent mechanism. Sci Rep. 2018. Jun 22;8(1):9566. doi: 10.1038/s41598-018-27724-3. PubMed PMID: 29934599; PubMed Central PMCID: PMCPMC6014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1796.Cadwell K, Debnath J.. Beyond self-eating: The control of nonautophagic functions and signaling pathways by autophagy-related proteins. J Cell Biol. 2018. Mar 5;217(3):813–822. doi: 10.1083/jcb.201706157. PubMed PMID: 29237720; PubMed Central PMCID: PMCPMC5839790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1797.Galluzzi L, Green DR.. Autophagy-Independent Functions of the Autophagy Machinery. Cell. 2019. Jun 13;177(7):1682–1699. doi: 10.1016/j.cell.2019.05.026. PubMed PMID: 31199916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1798.Galluzzi L, Vitale I, Abrams JM, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2011. Jul 15. doi: 10.1038/cdd.2011.96. PubMed PMID: 21760595; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1799.Kaushik S, Bandyopadhyay U, Sridhar S, et al.Chaperone-mediated autophagy at a glance. J Cell Sci. 2011. Feb 15;124(Pt 4):495–9. doi:124/4/495[pii] doi: 10.1242/jcs.073874. PubMed PMID: 21282471; PubMed Central PMCID: PMC3031365. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1800.Arias E, Cuervo AM.. Chaperone-mediated autophagy in protein quality control. Curr Opinion Cell Biol 2010. Nov 18;23:184–9. doi:S0955-0674(10)00180-8 [pii] doi: 10.1016/j.ceb.2010.10.009. PubMed PMID: 21094035; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1801.Li W, Dou J, Yang J, et al. Targeting Chaperone-Mediated Autophagy for Disease Therapy. Current Pharmacol Rep. 2018;4:261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1802.Li W, Yang Q, Mao Z.. Chaperone-mediated autophagy: machinery, regulation and biological consequences. Cell Mol Life Sci. 2011. Mar;68(5):749–63. doi: 10.1007/s00018-010-0565-6. PubMed PMID: 20976518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1803.Patel B, Cuervo AM.. Methods to study chaperone-mediated autophagy. Methods. 2015. Mar;75:133–40. doi: 10.1016/j.ymeth.2015.01.003. PubMed PMID: 25595300; PubMed Central PMCID: PMCPMC4355229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1804.Yabu T, Imamura S, Mohammed MS, et al. Differential gene expression of HSC70/HSP70 in yellowtail cells in response to chaperone-mediated autophagy. FEBS J. 2011. Feb;278(4):673–85. doi: 10.1111/j.1742-4658.2010.07989.x. PubMed PMID: 21205201. [DOI] [PubMed] [Google Scholar]
- 1805.Lescat L, Herpin A, Mourot B, et al. CMA restricted to mammals and birds: myth or reality? Autophagy. 2018;14(7):1267–1270. doi: 10.1080/15548627.2018.1460021. PubMed PMID: 29929419; PubMed Central PMCID: PMCPMC6103740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1806.Mukherjee A, Patel B, Koga H, et al. Selective endosomal microautophagy is starvation-inducible in Drosophila. Autophagy. 2016. Nov;12(11):1984–1999. doi: 10.1080/15548627.2016.1208887. PubMed PMID: 27487474; PubMed Central PMCID: PMCPMC5103356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1807.Eisermann DJ, Wenzel U, Fitzenberger E.. Inhibition of chaperone-mediated autophagy prevents glucotoxicity in the Caenorhabditis elegans mev-1 mutant by activation of the proteasome. Biochem Biophys Res Commun. 2017. Feb 26;484(1):171–175. doi: 10.1016/j.bbrc.2017.01.043. PubMed PMID: 28089866. [DOI] [PubMed] [Google Scholar]
- 1808.Lescat L, Veron V, Mourot B, et al. Chaperone-Mediated Autophagy in the light of evolution: insight from fish. Mol Biol Evol. 2020. May 21. doi: 10.1093/molbev/msaa127. PubMed PMID: 32437540. [DOI] [PubMed] [Google Scholar]
- 1809.Kirchner P, Bourdenx M, Madrigal-Matute J, et al. Proteome-wide analysis of chaperone-mediated autophagy targeting motifs. PLoS Biol. 2019. May;17(5):e3000301. doi: 10.1371/journal.pbio.3000301. PubMed PMID: 31150375; PubMed Central PMCID: PMCPMC6561683 of the submitted manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1810.Gough NR, Hatem CL, Fambrough DM.. The family of LAMP-2 proteins arises by alternative splicing from a single gene: characterization of the avian LAMP-2 gene and identification of mammalian homologs of LAMP-2b and LAMP-2c. DNA Cell Biol. 1995. Oct;14(10):863–7. doi: 10.1089/dna.1995.14.863. PubMed PMID: 7546292. [DOI] [PubMed] [Google Scholar]
- 1811.Kaushik S, Cuervo AM.. Methods to monitor chaperone-mediated autophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Methods Enzymol. 2009;452:297–324. doi: 10.1016/S0076-6879(08)03619-7. PubMed PMID: 19200890; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1812.Dice JF. Peptide sequences that target cytosolic proteins for lysosomal proteolysis [Research Support, U.S. Gov’t, P.H.S Review]. Trends Biochem Sci. 1990. Aug;15(8):305–9. PubMed PMID: 2204156; eng. [DOI] [PubMed] [Google Scholar]
- 1813.Bonhoure A, Vallentin A, Martin M, et al. Acetylation of translationally controlled tumor protein promotes its degradation through chaperone-mediated autophagy. Eur J Cell Biol. 2017. Mar;96(2):83–98. doi: 10.1016/j.ejcb.2016.12.002. PubMed PMID: 28110910. [DOI] [PubMed] [Google Scholar]
- 1814.Ho PW, Leung CT, Liu H, et al. Age-dependent accumulation of oligomeric SNCA/alpha-synuclein from impaired degradation in mutant LRRK2 knockin mouse model of Parkinson disease: role for therapeutic activation of chaperone-mediated autophagy (CMA). Autophagy. 2020. Feb;16(2):347–370. doi: 10.1080/15548627.2019.1603545. PubMed PMID: 30983487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1815.Cuervo AM, Dice JF, Knecht E.. A population of rat liver lysosomes responsible for the selective uptake and degradation of cytosolic proteins. J Biol Chem. 1997. Feb 28;272(9):5606–15. PubMed PMID: 9038169; eng. [DOI] [PubMed] [Google Scholar]
- 1816.Cuervo AM, Dice JF.. A receptor for the selective uptake and degradation of proteins by lysosomes [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.]. Science. 1996. Jul 26;273(5274):501–3. PubMed PMID: 8662539; eng. [DOI] [PubMed] [Google Scholar]
- 1817.Cuervo AM, Dice JF.. Unique properties of lamp2a compared to other lamp2 isoforms [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.]. J Cell Sci. 2000. Dec;113:4441–50. PubMed PMID: 11082038; eng. [DOI] [PubMed] [Google Scholar]
- 1818.Finn PF, Mesires NT, Vine M, et al. Effects of small molecules on chaperone-mediated autophagy. Autophagy. 2005. Oct-Dec;1(3):141–5. PubMed PMID: 16874031; eng. [DOI] [PubMed] [Google Scholar]
- 1819.Bandyopadhyay U, Kaushik S, Varticovski L, et al. The chaperone-mediated autophagy receptor organizes in dynamic protein complexes at the lysosomal membrane. Mol Cell Biol. 2008. Sep;28(18):5747–63. doi:MCB.02070-07 [pii] doi: 10.1128/MCB.02070-07. PubMed PMID: 18644871; PubMed Central PMCID: PMC2546938. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1820.Aniento F, Emans N, Griffiths G, et al. Cytoplasmic dynein-dependent vesicular transport from early to late endosomes [In Vitro Research Support, Non-U.S. Gov’t]. J Cell Biol. 1993. Dec;123(6 Pt 1):1373–87. PubMed PMID: 8253838; PubMed Central PMCID: PMC2290907. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1821.Salvador N, Aguado C, Horst M, et al. Import of a cytosolic protein into lysosomes by chaperone-mediated autophagy depends on its folding state [Research Support, Non-U.S. Gov’t]. J Biol Chem. 2000. Sep 1;275(35):27447–56. doi: 10.1074/jbc.M001394200. PubMed PMID: 10862611; eng. [DOI] [PubMed] [Google Scholar]
- 1822.Koga H, Martinez-Vicente M, Macian F, et al. A photoconvertible fluorescent reporter to track chaperone-mediated autophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Nat Commun. 2011;2:386. doi: 10.1038/ncomms1393. PubMed PMID: 21750540; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1823.Sahu R, Kaushik S, Clement CC, et al. Microautophagy of cytosolic proteins by late endosomes [Research Support, N.I.H., Extramural]. Develop cell. 2011. Jan 18; 20(1):131–9. doi: 10.1016/j.devcel.2010.12.003. PubMed PMID: 21238931; PubMed Central PMCID: PMC3025279. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1824.Wang Y, Martinez-Vicente M, Kruger U, et al. Tau fragmentation, aggregation and clearance: the dual role of lysosomal processing. Hum Mol Genet. 2009. Nov 1;18(21):4153–70. doi: 10.1093/hmg/ddp367. PubMed PMID: 19654187; PubMed Central PMCID: PMCPMC2758146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1825.Caballero B, Wang Y, Diaz A, et al. Interplay of pathogenic forms of human tau with different autophagic pathways. Aging cell. 2018. Feb;17(1). doi: 10.1111/acel.12692. PubMed PMID: 29024336; PubMed Central PMCID: PMCPMC5770880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1826.Morozova K, Clement CC, Kaushik S, et al. Structural and Biological Interaction of hsc-70 Protein with Phosphatidylserine in Endosomal Microautophagy. J Biol Chem. 2016. Aug 26;291(35):18096–106. doi: 10.1074/jbc.M116.736744. PubMed PMID: 27405763; PubMed Central PMCID: PMCPMC5000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1827.Uytterhoeven V, Lauwers E, Maes I, et al. Hsc70-4 Deforms Membranes to Promote Synaptic Protein Turnover by Endosomal Microautophagy. Neuron. 2015. Nov 18;88(4):735–48. doi: 10.1016/j.neuron.2015.10.012. PubMed PMID: 26590345. [DOI] [PubMed] [Google Scholar]
- 1828.Arndt V, Dick N, Tawo R, et al. Chaperone-assisted selective autophagy is essential for muscle maintenance [Research Support, Non-U.S. Gov’t]. Curr Biol. 2010. Jan 26;20(2):143–8. doi: 10.1016/j.cub.2009.11.022. PubMed PMID: 20060297; eng. [DOI] [PubMed] [Google Scholar]
- 1829.Eskelinen EL, Schmidt CK, Neu S, et al. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004. Jul;15(7):3132–45. doi: 10.1091/mbc.E04-02-0103. PubMed PMID: 15121881; PubMed Central PMCID: PMC452571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1830.Eskelinen EL, Illert AL, Tanaka Y, et al. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002. Sep;13(9):3355–68. doi: 10.1091/mbc.E02-02-0114. PubMed PMID: 12221139; PubMed Central PMCID: PMC124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1831.Huynh KK, Eskelinen EL, Scott CC, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 2007. Jan 24;26(2):313–24. doi: 10.1038/sj.emboj.7601511. PubMed PMID: 17245426; PubMed Central PMCID: PMC1783450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1832.Perez L, McLetchie S, Gardiner GJ, et al. LAMP-2C Inhibits MHC Class II Presentation of Cytoplasmic Antigens by Disrupting Chaperone-Mediated Autophagy. J Iimmunol. 2016. Mar 15; 196(6):2457–65. doi: 10.4049/jimmunol.1501476. PubMed PMID: 26856698; PubMed Central PMCID: PMCPMC4779666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1833.Perez L, Sinn AL, Sandusky GE, et al. Melanoma LAMP-2C Modulates Tumor Growth and Autophagy. Front Cell Dev Biol. 2018;6:101. doi: 10.3389/fcell.2018.00101. PubMed PMID: 30211163; PubMed Central PMCID: PMCPMC6123356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1834.Furuta A, Kikuchi H, Fujita H, et al. Property of lysosomal storage disease associated with midbrain pathology in the central nervous system of lamp-2-deficient mice. Am J Pathol. 2015. Jun;185(6):1713–23. doi: 10.1016/j.ajpath.2015.02.015. PubMed PMID: 25998250. [DOI] [PubMed] [Google Scholar]
- 1835.Rothaug M, Stroobants S, Schweizer M, et al. LAMP-2 deficiency leads to hippocampal dysfunction but normal clearance of neuronal substrates of chaperone-mediated autophagy in a mouse model for Danon disease. Acta Neuropathol Commun. 2015;3:6. doi: 10.1186/s40478-014-0182-y. PubMed PMID: 25637286; PubMed Central PMCID: PMC4359523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1836.Sarparanta J, Jonson PH, Golzio C, et al. Mutations affecting the cytoplasmic functions of the co-chaperone DNAJB6 cause limb-girdle muscular dystrophy. Nat Genet. 2012. Feb 26;44(4):450–5, S1-2. doi: 10.1038/ng.1103. PubMed PMID: 22366786; PubMed Central PMCID: PMCPMC3315599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1837.Ulbricht A, Eppler FJ, Tapia VE, et al. Cellular mechanotransduction relies on tension-induced and chaperone-assisted autophagy. Curr Biol. 2013. Mar 4;23(5):430–5. doi 10.1016/j.cub.2013.01.064. PubMed PMID: 23434281. [DOI] [PubMed] [Google Scholar]
- 1838.Gamerdinger M, Kaya AM, Wolfrum U, et al. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011. Feb;12(2):149–56. doi: 10.1038/embor.2010.203. PubMed PMID: 21252941; PubMed Central PMCID: PMCPMC3049430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1839.Gamerdinger M, Hajieva P, Kaya AM, et al. Protein quality control during aging involves recruitment of the macroautophagy pathway by BAG3. EMBO J. 2009. Apr 8;28(7):889–901. doi: 10.1038/emboj.2009.29. PubMed PMID: 19229298; PubMed Central PMCID: PMCPMC2647772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1840.Sturner E, Behl C.. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front Mol Neurosci. 2017;10:177. doi: 10.3389/fnmol.2017.00177. PubMed PMID: 28680391; PubMed Central PMCID: PMCPMC5478690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1841.Cristofani R, Crippa V, Rusmini P, et al. Inhibition of retrograde transport modulates misfolded protein accumulation and clearance in motoneuron diseases. Autophagy. 2017. Aug 3;13(8):1280–1303. doi: 10.1080/15548627.2017.1308985. PubMed PMID: 28402699; PubMed Central PMCID: PMCPMC5584856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1842.Adriaenssens E, Tedesco B, Mediani L, et al. BAG3 Pro209 mutants associated with myopathy and neuropathy relocate chaperones of the CASA-complex to aggresomes. Sci Rep. 2020. May 29;10(1):8755. doi: 10.1038/s41598-020-65664-z. PubMed PMID: 32472079; PubMed Central PMCID: PMCPMC7260189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1843.Evangelinos M, Martzoukou O, Chorozian K, et al. BsdA(Bsd2) -dependent vacuolar turnover of a misfolded version of the UapA transporter along the secretory pathway: prominent role of selective autophagy. Mol Microbiol. 2016. Jun;100(5):893–911. doi: 10.1111/mmi.13358. PubMed PMID: 26917498. [DOI] [PubMed] [Google Scholar]
- 1844.Oku M, Sakai Y.. Three Distinct Types of Microautophagy Based on Membrane Dynamics and Molecular Machineries. Bioessays. 2018. Jun;40(6):e1800008. doi: 10.1002/bies.201800008. PubMed PMID: 29708272. [DOI] [PubMed] [Google Scholar]
- 1845.Xie Z, Klionsky DJ.. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007. Oct;9(10):1102–9. doi: 10.1038/ncb1007-1102. PubMed PMID: 17909521. [DOI] [PubMed] [Google Scholar]
- 1846.Mukaiyama H, Baba M, Osumi M, et al. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. ?Mol Biol Cell 2004. Jan;15(1):58–70. PubMed PMID: 13679515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1847.Salum TT, Tsil’mer K, Vikhalemm TE, et al. [Features of temperature dependence of the Na+,K+-ATPase reaction in normal and tumorous brain tissue]. Ukr Biokhim Zh. 1989. Jul-Aug;61(4):65–9. PubMed PMID: 2555948. [PubMed] [Google Scholar]
- 1848.Vevea JD, Garcia EJ, Chan RB, et al. Role for Lipid Droplet Biogenesis and Microlipophagy in Adaptation to Lipid Imbalance in Yeast. Dev Cell. 2015. Dec 7;35(5):584–599. doi: 10.1016/j.devcel.2015.11.010. PubMed PMID: 26651293; PubMed Central PMCID: PMCPMC4679156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1849.Oku M, Maeda Y, Kagohashi Y, et al. Evidence for ESCRT- and clathrin-dependent microautophagy. J Cell Biol. 2017. Oct 2;216(10):3263–3274. doi: 10.1083/jcb.201611029. PubMed PMID: 28838958; PubMed Central PMCID: PMCPMC5626533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1850.Chanoca A, Kovinich N, Burkel B, et al. Anthocyanin Vacuolar Inclusions Form by a Microautophagy Mechanism. Plant Cell. 2015. Sep;27(9):2545–59. doi: 10.1105/tpc.15.00589. PubMed PMID: 26342015; PubMed Central PMCID: PMCPMC4815043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1851.Liu XM, Sun LL, Hu W, et al. ESCRTs Cooperate with a Selective Autophagy Receptor to Mediate Vacuolar Targeting of Soluble Cargos. Mol Cell. 2015. Sep 17;59(6):1035–42. doi: 10.1016/j.molcel.2015.07.034. PubMed PMID: 26365378. [DOI] [PubMed] [Google Scholar]
- 1852.Rahman MA, Terasawa M, Mostofa MG, et al. The TORC1-Nem1/Spo7-Pah1/lipin axis regulates microautophagy induction in budding yeast. Biochem Biophys Res Commun. 2018. Oct 2;504(2):505–512. doi: 10.1016/j.bbrc.2018.09.011. PubMed PMID: 30201264. [DOI] [PubMed] [Google Scholar]
- 1853.Hatakeyama R, De Virgilio C.. TORC1 specifically inhibits microautophagy through ESCRT-0. Curr Genet. 2019. Oct;65(5):1243–1249. doi: 10.1007/s00294-019-00982-y. PubMed PMID: 31041524; PubMed Central PMCID: PMCPMC6744375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1854.Seo AY, Lau PW, Feliciano D, et al. AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. eLife. 2017. Apr 10;6. doi: 10.7554/eLife.21690. PubMed PMID: 28394250; PubMed Central PMCID: PMCPMC5407857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1855.Tsuji T, Fujimoto M, Tatematsu T, et al. Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. eLife. 2017. Jun 7;6. doi: 10.7554/eLife.25960. PubMed PMID: 28590904; PubMed Central PMCID: PMCPMC5462540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1856.Wang CW, Miao YH, Chang YS.. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol. 2014. Aug 4;206(3):357–66. doi: 10.1083/jcb.201404115. PubMed PMID: 25070953; PubMed Central PMCID: PMC4121974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1857.Sattler T, Mayer A.. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation [Research Support, Non-U.S. Gov’t]. J Cell Biol. 2000. Oct 30;151(3):529–38. PubMed PMID: 11062255; PubMed Central PMCID: PMC2185593. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1858.Thery C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750. doi: 10.1080/20013078.2018.1535750. PubMed PMID: 30637094; PubMed Central PMCID: PMCPMC6322352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1859.Exosomes Vidal M.: Revisiting their role as “garbage bags”. Traffic. 2019. Nov;20(11):815–828. doi: 10.1111/tra.12687. PubMed PMID: 31418976. [DOI] [PubMed] [Google Scholar]
- 1860.Paglin S, Hollister T, Delohery T, et al. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001. Jan 15;61(2):439–44. PubMed PMID: 11212227; eng. [PubMed] [Google Scholar]
- 1861.Florez-McClure ML, Linseman DA, Chu CT, et al. The p75 neurotrophin receptor can induce autophagy and death of cerebellar Purkinje neurons. J Neurosci. 2004. May 12;24(19):4498–509. PubMed PMID: 15140920; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1862.Moriyasu Y, Ohsumi Y.. Autophagy in tobacco suspension-cultured cells in response to sucrose starvation. Plant Physiol. 1996. Aug;111(4):1233–1241. PubMed PMID: 12226358; PubMed Central PMCID: PMC161001. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1863.Wolfe DM, Lee JH, Kumar A, et al. Autophagy failure in Alzheimer’s disease and the role of defective lysosomal acidification. Eur J Neurosci. 2013. Jun;37(12):1949–1961. doi: 10.1111/ejn.12169. PubMed PMID: 23773064; PubMed Central PMCID: PMC3694736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1864.Thome MP, Filippi-Chiela EC, Villodre ES, et al. Ratiometric analysis of Acridine Orange staining in the study of acidic organelles and autophagy. J Cell Sci. 2016. Dec 15;129(24):4622–4632. doi: 10.1242/jcs.195057. PubMed PMID: 27875278. [DOI] [PubMed] [Google Scholar]
- 1865.Bashmail HA, Alamoudi AA, Noorwali A, et al. Thymoquinone synergizes gemcitabine anti-breast cancer activity via modulating its apoptotic and autophagic activities. Sci Rep. 2018. Aug 3;8(1):11674. doi: 10.1038/s41598-018-30046-z. PubMed PMID: 30076320; PubMed Central PMCID: PMCPMC6076303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1866.Biederbick A, Kern HF, Elsasser HP.. Monodansylcadaverine (MDC) is a specific in vivo marker for autophagic vacuoles. Eur J Cell Biol. 1995. Jan;66(1):3–14. PubMed PMID: 7750517; eng. [PubMed] [Google Scholar]
- 1867.Hoyer-Hansen M, Bastholm L, Mathiasen IS, et al. Vitamin D analog EB1089 triggers dramatic lysosomal changes and Beclin 1-mediated autophagic cell death. Cell Death Differ. 2005. Oct;12(10):1297–309. PubMed PMID: 15905882; eng. [DOI] [PubMed] [Google Scholar]
- 1868.Fogel JL, Thein TZ, Mariani FV.. Use of LysoTracker to detect programmed cell death in embryos and differentiating embryonic stem cells. J vis exp. 2012. (68). doi: 10.3791/4254. PubMed PMID: 23092960; PubMed Central PMCID: PMC3490301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1869.Freundt EC, Czapiga M, Lenardo MJ.. Photoconversion of Lysotracker Red to a green fluorescent molecule [Letter Research Support, N.I.H., Intramural]. Cell Res. 2007. Nov;17(11):956–8. doi: 10.1038/cr.2007.80. PubMed PMID: 17893709; eng. [DOI] [PubMed] [Google Scholar]
- 1870.Oeste CL, Seco E, Patton WF, et al. Interactions between autophagic and endo-lysosomal markers in endothelial cells. Histochem Cell Biol. 2013. May;139(5):659–70. doi: 10.1007/s00418-012-1057-6. PubMed PMID: 23203316. [DOI] [PubMed] [Google Scholar]
- 1871.Rubinsztein DC, Gestwicki JE, Murphy LO, et al. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007. Apr;6(4):304–12. PubMed PMID: 17396135; eng. [DOI] [PubMed] [Google Scholar]
- 1872.Funderburk SF, Wang QJ, Yue Z.. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review]. Trends Cell Biol. 2010. Jun;20(6):355–62. doi: 10.1016/j.tcb.2010.03.002. PubMed PMID: 20356743; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1873.Levine B, Sinha S, Kroemer G.. Bcl-2 family members: dual regulators of apoptosis and autophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review]. Autophagy. 2008. Jul;4(5):600–6. PubMed PMID: 18497563; PubMed Central PMCID: PMC2749577. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1874.Simonsen A, Tooze SA.. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes [Review]. J Cell Biol. 2009. Sep 21;186(6):773–82. doi: 10.1083/jcb.200907014. PubMed PMID: 19797076; PubMed Central PMCID: PMC2753151. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1875.Pyo JO, Jang MH, Kwon YK, et al. Essential roles of Atg5 and FADD in autophagic cell death: dissection of autophagic cell death into vacuole formation and cell death [Research Support, Non-U.S. Gov’t]. J Biol Chem. 2005. May 27;280(21):20722–9. doi: 10.1074/jbc.M413934200. PubMed PMID: 15778222; eng. [DOI] [PubMed] [Google Scholar]
- 1876.Petiot A, Ogier-Denis E, Blommaart EF, et al. Distinct classes of phosphatidylinositol 3ʹ-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem. 2000. Jan 14;275(2):992–8. PubMed PMID: 10625637; eng. [DOI] [PubMed] [Google Scholar]
- 1877.Harris J, Hartman M, Roche C, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. J Biol Chem. 2011. Mar 18;286(11):9587–97. doi: 10.1074/jbc.M110.202911. PubMed PMID: 21228274; PubMed Central PMCID: PMC3058966. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1878.Crisan TO, Plantinga TS, van de Veerdonk FL, et al. Inflammasome-independent modulation of cytokine response by autophagy in human cells [Research Support, Non-U.S. Gov’t]. PLoS One. 2011;6(4):e18666. doi: 10.1371/journal.pone.0018666. PubMed PMID: 21490934; PubMed Central PMCID: PMC3072416. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1879.Kleinnijenhuis J, Oosting M, Plantinga TS, et al. Autophagy modulates the Mycobacterium tuberculosis-induced cytokine response [Research Support, Non-U.S. Gov’t]. Immunology. 2011. Nov;134(3):341–8. doi: 10.1111/j.1365-2567.2011.03494.x. PubMed PMID: 21978003; PubMed Central PMCID: PMC3209573. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1880.Peral de Castro C, Jones SA, Ni Cheallaigh C, et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Iimmunol. 2012. Oct 15;189(8):4144–53. doi: 10.4049/jimmunol.1201946. PubMed PMID: 22972933. [DOI] [PubMed] [Google Scholar]
- 1881.Herbst S, Schaible UE, Schneider BE.. Interferon gamma activated macrophages kill mycobacteria by nitric oxide induced apoptosis. PLoS One. 2011. May 2;6(5):e19105. doi: 10.1371/journal.pone.0019105. PubMed PMID: 21559306; PubMed Central PMCID: PMCPMC3085516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1882.Dowdle WE, Nyfeler B, Nagel J, et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol. 2014. Nov;16(11):1069–79. doi: 10.1038/ncb3053. PubMed PMID: 25327288. [DOI] [PubMed] [Google Scholar]
- 1883.Ronan B, Flamand O, Vescovi L, et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol. 2014. Dec;10(12):1013–9. doi: 10.1038/nchembio.1681. PubMed PMID: 25326666. [DOI] [PubMed] [Google Scholar]
- 1884.Isosaki M. Inhibition of wortmannin activities by amino compounds. Biochem Biophys Res Commun. 2004. Nov 26;324(4):1406–12. doi: 10.1016/j.bbrc.2004.09.200. PubMed PMID: 15504370. [DOI] [PubMed] [Google Scholar]
- 1885.Yuan H, Barnes KR, Weissleder R, et al. Covalent reactions of wortmannin under physiological conditions. Chem Biol. 2007. Mar;14(3):321–8. doi: 10.1016/j.chembiol.2007.02.007. PubMed PMID: 17379147. [DOI] [PubMed] [Google Scholar]
- 1886.Chen J, Chen MX, Fogo AB, et al. mVps34 deletion in podocytes causes glomerulosclerosis by disrupting intracellular vesicle trafficking. J Am Soc Nephrol. 2013. Feb;24(2):198–207. doi: 10.1681/ASN.2012010101. PubMed PMID: 23291473; PubMed Central PMCID: PMC3559479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1887.Kovács J. Morphometric study of the effect of leupeptin, vinblastine, estron acetate and cycloheximide on the autophagic vacuole-lysosomal compartments in mouse seminal vesicle cells. Virchows Arch B Cell Pathol Incl Mol Pathol. 1983;42(1):83–93. PubMed PMID: 6132491; eng. [DOI] [PubMed] [Google Scholar]
- 1888.Papadopoulos T, Pfeifer U.. Regression of rat liver autophagic vacuoles by locally applied cycloheximide. Lab Invest. 1986. Jan;54(1):100–7. PubMed PMID: 3941538; eng. [PubMed] [Google Scholar]
- 1889.Rumpelt HJ, Albring M, Thoenes W.. Prevention of D-galactosamine-induced hepatocellular autophagocytosis by cycloheximide. Virchows Arch B Cell Pathol. 1974;16(2):195–203. PubMed PMID: 4216140; eng. [DOI] [PubMed] [Google Scholar]
- 1890.Kovács AL, Kovács J.. Autophagocytosis in mouse seminal vesicle cells in vitro. Temperature dependence and effects of vinblastine and inhibitors of protein synthesis. Virchows Arch B Cell Pathol Incl Mol Pathol. 1980;32(2):97–104. PubMed PMID: 6102826; eng. [DOI] [PubMed] [Google Scholar]
- 1891.Rodemann HP, Dittmann K, Toulany M.. Radiation-induced EGFR-signaling and control of DNA-damage repair [Research Support, Non-U.S. Gov’t Review]. Int J Radiat Biol.2007. Nov-Dec;83(11–12):781–91. doi: 10.1080/09553000701769970. PubMed PMID: 18058366; eng. [DOI] [PubMed] [Google Scholar]
- 1892.Chaachouay H, Ohneseit P, Toulany M, et al. Autophagy contributes to resistance of tumor cells to ionizing radiation [Research Support, Non-U.S. Gov’t]. Radiother Oncol. 2011. Jun;99(3):287–92. doi: 10.1016/j.radonc.2011.06.002. PubMed PMID: 21722986; eng. [DOI] [PubMed] [Google Scholar]
- 1893.Apel A, Herr I, Schwarz H, et al. Blocked autophagy sensitizes resistant carcinoma cells to radiation therapy [Research Support, Non-U.S. Gov’t]. Cancer Res. 2008. Mar 1;68(5):1485–94. doi: 10.1158/0008-5472.CAN-07-0562. PubMed PMID: 18316613; eng. [DOI] [PubMed] [Google Scholar]
- 1894.Hounjet J, Habets R, Schaaf MB, et al. The anti-malarial drug chloroquine sensitizes oncogenic NOTCH1 driven human T-ALL to gamma-secretase inhibition. Oncogene. 2019. Jul;38(27):5457–5468. doi: 10.1038/s41388-019-0802-x. PubMed PMID: 30967635. [DOI] [PubMed] [Google Scholar]
- 1895.Eng CH, Yu K, Lucas J, et al. Ammonia derived from glutaminolysis is a diffusible regulator of autophagy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Sci Signal. 2010;3(119):ra31. doi: 10.1126/scisignal.2000911. PubMed PMID: 20424262; eng. [DOI] [PubMed] [Google Scholar]
- 1896.Seglen PO, Gordon PB.. Effects of lysosomotropic monoamines, diamines, amino alcohols, and other amino compounds on protein degradation and protein synthesis in isolated rat hepatocytes [In Vitro Research Support, Non-U.S. Gov’t]. Mol Pharmacol. 1980. Nov;18(3):468–75. PubMed PMID: 7464813; eng. [PubMed] [Google Scholar]
- 1897.Cheong H, Lindsten T, Wu J, et al. Ammonia-induced autophagy is independent of ULK1/ULK2 kinases [Research Support, N.I.H., Extramural]. Proc Natl Acad Sci U S A. 2011. Jul 5;108(27):11121–6. doi: 10.1073/pnas.1107969108. PubMed PMID: 21690395; PubMed Central PMCID: PMC3131371. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1898.Li Z, Ji X, Wang W, et al. Ammonia Induces Autophagy through Dopamine Receptor D3 and MTOR. PLoS One. 2016;11(4):e0153526. doi: 10.1371/journal.pone.0153526. PubMed PMID: 27077655; PubMed Central PMCID: PMCPMC4831814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1899.Pellegrini P, Strambi A, Zipoli C, et al. Acidic extracellular pH neutralizes the autophagy-inhibiting activity of chloroquine: implications for cancer therapies. Autophagy. 2014. Apr;10(4):562–71. doi: 10.4161/auto.27901. PubMed PMID: 24492472; PubMed Central PMCID: PMC3984580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1900.Fischer S, Ronellenfitsch MW, Thiepold AL, et al. Hypoxia enhances the antiglioma cytotoxicity of B10, a glycosylated derivative of betulinic acid. PLoS One. 2014;9(4):e94921. doi: 10.1371/journal.pone.0094921. PubMed PMID: 24743710; PubMed Central PMCID: PMC3990545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1901.Gonzalez P, Mader I, Tchoghandjian A, et al. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ. 2012. Aug;19(8):1337–46. doi: 10.1038/cdd.2012.10. PubMed PMID: 22343715; PubMed Central PMCID: PMC3392623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1902.Potze L, Mullauer FB, Colak S, et al. Betulinic acid-induced mitochondria-dependent cell death is counterbalanced by an autophagic salvage response. Cell Death Dis. 2014;5:e1169. doi: 10.1038/cddis.2014.139. PubMed PMID: 24722294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1903.Chen Y, Sun R, Wang B.. Monolayer behavior of binary systems of betulinic acid and cardiolipin: thermodynamic analyses of Langmuir monolayers and AFM study of Langmuir-Blodgett monolayers. J Colloid Interface Sci. 2011. Jan 1;353(1):294–300. doi: 10.1016/j.jcis.2010.09.019. PubMed PMID: 20888569. [DOI] [PubMed] [Google Scholar]
- 1904.Gao M, Lau PM, Kong SK.. Mitochondrial toxin betulinic acid induces in vitro eryptosis in human red blood cells through membrane permeabilization. Arch Toxicol. 2014Mar;88(3):755–68. doi: 10.1007/s00204-013-1162-x. PubMed PMID: 24241250. [DOI] [PubMed] [Google Scholar]
- 1905.Milczarek M, Wiktorska K, Mielczarek L, et al. Autophagic cell death and premature senescence: New mechanism of 5-fluorouracil and sulforaphane synergistic anticancer effect in MDA-MB-231 triple negative breast cancer cell line. Food Chem Toxicol. 2018. Jan;111:1–8. doi: 10.1016/j.fct.2017.10.056. PubMed PMID: 29104175. [DOI] [PubMed] [Google Scholar]
- 1906.Hsieh MJ, Chien SY, Lin JT, et al. Polyphyllin G induces apoptosis and autophagy cell death in human oral cancer cells. Phytomedicine. 2016. Dec 1;23(13):1545–1554. doi: 10.1016/j.phymed.2016.09.004. PubMed PMID: 27823618. [DOI] [PubMed] [Google Scholar]
- 1907.Hsieh MJ, Lin CW, Chiou HL, et al. Dehydroandrographolide, an iNOS inhibitor, extracted from Andrographis paniculata (Burm.f.) Nees, induces autophagy in human oral cancer cells. Oncotarget. 2015. Oct 13;6(31):30831–49. doi: 10.18632/oncotarget.5036. PubMed PMID: 26356821; PubMed Central PMCID: PMCPMC4741571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1908.Law BYK, Mok SWF, Chen J, et al. N-Desmethyldauricine Induces Autophagic Cell Death in Apoptosis-Defective Cells via Ca(2+) Mobilization. Front Pharmacol. 2017;8:388. doi: 10.3389/fphar.2017.00388. PubMed PMID: 28670281; PubMed Central PMCID: PMCPMC5472688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1909.Wong VK, Li T, Law BY, et al. Saikosaponin-d, a novel SERCA inhibitor, induces autophagic cell death in apoptosis-defective cells. Cell Death Dis. 2013;4:e720. doi: 10.1038/cddis.2013.217. PubMed PMID: 23846222; PubMed Central PMCID: PMC3730398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1910.Xu SW, Law BY, Mok SW, et al. Autophagic degradation of epidermal growth factor receptor in gefitinib-resistant lung cancer by celastrol. Int J Oncol. 2016. Oct;49(4):1576–88. doi: 10.3892/ijo.2016.3644. PubMed PMID: 27498688. [DOI] [PubMed] [Google Scholar]
- 1911.Tsuyoshi H, Wong VKW, Han Y, et al. Saikosaponin-d, a calcium mobilizing agent, sensitizes chemoresistant ovarian cancer cells to cisplatin-induced apoptosis by facilitating mitochondrial fission and G2/M arrest. Oncotarget. 2017. Nov 21;8(59):99825–99840. doi: 10.18632/oncotarget.21076. PubMed PMID: 29245943; PubMed Central PMCID: PMCPMC5725134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1912.Law BY, Chan WK, Xu SW, et al. Natural small-molecule enhancers of autophagy induce autophagic cell death in apoptosis-defective cells. Sci Rep. 2014. Jul 1;4:5510. doi: 10.1038/srep05510. PubMed PMID: 24981420; PubMed Central PMCID: PMCPMC4076737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1913.Law BY, Mok SW, Chan WK, et al. Hernandezine, a novel AMPK activator induces autophagic cell death in drug-resistant cancers. Oncotarget. 2016. Feb 16;7(7):8090–104. doi: 10.18632/oncotarget.6980. PubMed PMID: 26811496; PubMed Central PMCID: PMCPMC4884978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1914.Law BYK, Gordillo-Martinez F, Qu YQ, et al. Thalidezine, a novel AMPK activator, eliminates apoptosis-resistant cancer cells through energy-mediated autophagic cell death. Oncotarget. 2017. May 2;8(18):30077–30091. doi: 10.18632/oncotarget.15616. PubMed PMID: 28404910; PubMed Central PMCID: PMCPMC5444727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1915.Zhou X, Qu YQ, Zheng Z, et al. Novel dauricine derivatives suppress cancer via autophagy-dependent cell death. Bioorg Chem. 2019. Mar;83:450–460. doi: 10.1016/j.bioorg.2018.10.074. PubMed PMID: 30448723. [DOI] [PubMed] [Google Scholar]
- 1916.Wong VKW, Zeng W, Chen J, et al. Tetrandrine, an Activator of Autophagy, Induces Autophagic Cell Death via PKC-alpha Inhibition and mTOR-Dependent Mechanisms. Front Pharmacol. 2017;8:351. doi: 10.3389/fphar.2017.00351. PubMed PMID: 28642707; PubMed Central PMCID: PMCPMC5462963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1917.Teng JF, Qin DL, Mei QB, et al. Polyphyllin VI, a saponin from Trillium tschonoskii Maxim. induces apoptotic and autophagic cell death via the ROS triggered mTOR signaling pathway in non-small cell lung cancer. Pharmacol Res. 2019. Sep;147:104396. doi: 10.1016/j.phrs.2019.104396. PubMed PMID: 31404628. [DOI] [PubMed] [Google Scholar]
- 1918.Wei P, Zhang L, Lu Y, et al. C60(Nd) nanoparticles enhance chemotherapeutic susceptibility of cancer cells by modulation of autophagy [Research Support, Non-U.S. Gov’t]. Nanotechnology. 2010. Dec 10;21(49):495101. doi: 10.1088/0957-4484/21/49/495101. PubMed PMID: 21071824; eng. [DOI] [PubMed] [Google Scholar]
- 1919.Mohammadinejad R, Moosavi MA, Tavakol S, et al. Necrotic, apoptotic and autophagic cell fates triggered by nanoparticles. Autophagy. 2019. Jan;15(1):4–33. doi: 10.1080/15548627.2018.1509171. PubMed PMID: 30160607; PubMed Central PMCID: PMCPMC6287681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1920.Lee DH, Goldberg AL.. Proteasome inhibitors: valuable new tools for cell biologists [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. Review]. Trends Cell Biol. 1998. Oct;8(10):397–403. PubMed PMID: 9789328; eng. [DOI] [PubMed] [Google Scholar]
- 1921.Mehdi S. Cell-penetrating inhibitors of calpain [Review]. Trends Biochem Sci. 1991. Apr;16(4):150–3. PubMed PMID: 1877091; eng. [DOI] [PubMed] [Google Scholar]
- 1922.Holen I, Gordon PB, Seglen PO.. Inhibition of hepatocytic autophagy by okadaic acid and other protein phosphatase inhibitors [Research Support, Non-U.S. Gov’t]. Eur J Biochem/FEBS. 1993. Jul 1;215(1):113–22. PubMed PMID: 8393787; eng. [DOI] [PubMed] [Google Scholar]
- 1923.Sasaki K, Murata M, Yasumoto T, et al. Affinity of okadaic acid to type-1 and type-2A protein phosphatases is markedly reduced by oxidation of its 27-hydroxyl group. Biochem J. 1994. Mar 1;298:259–62. PubMed PMID: 8135728; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1924.Robinson DG, Albrecht S, Moriyasu Y.. The V-ATPase inhibitors concanamycin A and bafilomycin A lead to Golgi swelling in tobacco BY-2 cells [Research Support, Non-U.S. Gov’t]. Protoplasma. 2004. Dec;224(3–4):255–60. doi: 10.1007/s00709-004-0070-6. PubMed PMID: 15614486; eng. [DOI] [PubMed] [Google Scholar]
- 1925.Zoncu R, Bar-Peled L, Efeyan A, et al. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H-ATPase [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.]. Science. 2011. Nov 4;334(6056):678–83. doi: 10.1126/science.1207056. PubMed PMID: 22053050; PubMed Central PMCID: PMC3211112. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1926.Zhang CS, Jiang B, Li M, et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014. Sep 2;20(3):526–40. doi: 10.1016/j.cmet.2014.06.014. PubMed PMID: 25002183. [DOI] [PubMed] [Google Scholar]
- 1927.Wu YC, Wu WK, Li Y, et al. Inhibition of macroautophagy by bafilomycin A1 lowers proliferation and induces apoptosis in colon cancer cells. Biochem Biophys Res Commun. 2009. May 1;382(2):451–6. doi: 10.1016/j.bbrc.2009.03.051. PubMed PMID: 19289106; eng. [DOI] [PubMed] [Google Scholar]
- 1928.Vats S, Manjithaya R.. A reversible autophagy inhibitor blocks autophagosome-lysosome fusion by preventing Stx17 loading onto auto-phagosomes. Mol Biol Cell. 2019. Aug 1;30(17):2283–2295. doi: 10.1091/mbc.E18-08-0482. PubMed PMID: 31188703; PubMed Central PMCID: PMCPMC6743457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1929.Ostenfeld MS, Hoyer-Hansen M, Bastholm L, et al. Anti-cancer agent siramesine is a lysosomotropic detergent that induces cytoprotective autophagosome accumulation [Research Support, Non-U.S. Gov’t]. Autophagy. 2008. May;4(4):487–99. PubMed PMID: 18305408; eng. [DOI] [PubMed] [Google Scholar]
- 1930.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007. Jan 18;117:326–36. PubMed PMID: 17235397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1931.Garcia-Garcia A, Anandhan A, Burns M, et al. Impairment of Atg5-dependent autophagic flux promotes paraquat- and MPP(+)-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol Sci. 2013. Nov;136(1):166–82. doi: 10.1093/toxsci/kft188. PubMed PMID: 23997112; PubMed Central PMCID: PMC3829573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1932.Maclean KH, Dorsey FC, Cleveland JL, et al. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008. Jan;118(1):79–88. doi: 10.1172/JCI33700. PubMed PMID: 18097482; PubMed Central PMCID: PMC2148253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1933.Poole B, Ohkuma S.. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol. 1981. Sep;90(3):665–9. PubMed PMID: 6169733; PubMed Central PMCID: PMC2111912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1934.Matsuoka K, Higuchi T, Maeshima M, et al. A vacuolar-type H+-ATPase in a nonvacuolar organelle is required for the sorting of soluble vacuolar protein precursors in tobacco cells. Plant Cell. 1997. Apr;9(4):533–546. doi: 10.1105/tpc.9.4.533. PubMed PMID: 12237363; PubMed Central PMCID: PMC156937. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1935.Yano K, Yanagisawa T, Mukae K, et al. Dissection of autophagy in tobacco BY-2 cells under sucrose starvation conditions using the vacuolar H(+)-ATPase inhibitor concanamycin A and the autophagy-related protein Atg8. Plant Signal Behav. 2015;10(11):e1082699. doi: 10.1080/15592324.2015.1082699. PubMed PMID: 26368310; PubMed Central PMCID: PMCPMC4883836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1936.Arstila AU, Nuuja IJ, Trump BF.. Studies on cellular autophagocytosis. Vinblastine-induced autophagy in the rat liver. Exp Cell Res. 1974. Aug;87(2):249–52. PubMed PMID: 4415937; eng. [DOI] [PubMed] [Google Scholar]
- 1937.Hirsimaki Y, Arstila AU, Trump BF.. Autophagocytosis: in vitro induction by microtuble poisons. Exp Cell Res. 1975. Apr;92(1):11–4. PubMed PMID: 1169154; eng. [DOI] [PubMed] [Google Scholar]
- 1938.Réz G, Fellinger E, Reti M, et al. Time course of quantitative morphological changes of the autophagic-lysosomal compartment of murine seminal vesicle epithelial cells under the influence of vinblastine. J Submicrosc Cytol Pathol. 1990. Oct;22(4):529–34. PubMed PMID: 2282639; eng. [PubMed] [Google Scholar]
- 1939.Oliva O, Réz G, Pálfia Z, et al. Dynamics of vinblastine-induced autophagocytosis in murine pancreatic acinar cells: influence of cycloheximide post-treatments. Exp Mol Pathol. 1992. Feb;56(1):76–86. PubMed PMID: 1547871; eng. [DOI] [PubMed] [Google Scholar]
- 1940.Dominguez-Martin E, Cardenal-Munoz E, King JS, et al. Methods to Monitor and Quantify Autophagy in the Social Amoeba Dictyostelium discoideum. Cells. 2017. Jul 3;6(3). doi: 10.3390/cells6030018. PubMed PMID: 28671610; PubMed Central PMCID: PMCPMC5617964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1941.Mejlvang J, Olsvik H, Svenning S, et al. Starvation induces rapid degradation of selective autophagy receptors by endosomal microautophagy. J Cell Biol. 2018. Oct 1;217(10):3640–3655. doi: 10.1083/jcb.201711002. PubMed PMID: 30018090; PubMed Central PMCID: PMCPMC6168274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1942.Feldman ME, Apsel B, Uotila A, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2 [Research Support, Non-U.S. Gov’t]. PLoS Biol. 2009. Feb 10;7(2):e38. doi: 10.1371/journal.pbio.1000038. PubMed PMID: 19209957; PubMed Central PMCID: PMC2637922. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1943.Fleming A, Noda T, Yoshimori T, et al. Chemical modulators of autophagy as biological probes and potential therapeutics [Research Support, Non-U.S. Gov’t Review]. Nat Chem Biol. 2011. Jan;7(1):9–17. doi: 10.1038/nchembio.500. PubMed PMID: 21164513; eng. [DOI] [PubMed] [Google Scholar]
- 1944.Yu K, Toral-Barza L, Shi C, et al. Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 2009. Aug 1;69(15):6232–40. doi: 10.1158/0008-5472.CAN-09-0299. PubMed PMID: 19584280; eng. [DOI] [PubMed] [Google Scholar]
- 1945.Chresta CM, Davies BR, Hickson I, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010. Jan 1;70(1):288–98. doi: 10.1158/0008-5472.CAN-09-1751. PubMed PMID: 20028854; eng. [DOI] [PubMed] [Google Scholar]
- 1946.Roscic A, Baldo B, Crochemore C, et al. Induction of autophagy with catalytic mTOR inhibitors reduces huntingtin aggregates in a neuronal cell model [Research Support, Non-U.S. Gov’t]. J Neurochem. 2011. Oct;119(2):398–407. doi: 10.1111/j.1471-4159.2011.07435.x. PubMed PMID: 21854390; eng. [DOI] [PubMed] [Google Scholar]
- 1947.Fan QW, Cheng C, Hackett C, et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Sci Signal. 2010;3(147):ra81. doi: 10.1126/scisignal.2001017. PubMed PMID: 21062993; PubMed Central PMCID: PMC3001107. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1948.Yang L, Li P, Fu S, et al. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Cell Metab. 2010. Jun 9;11(6):467–78. doi: 10.1016/j.cmet.2010.04.005. PubMed PMID: 20519119; PubMed Central PMCID: PMC2881480. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1949.Yamamoto A, Yue Z.. Autophagy and its normal and pathogenic States in the brain. Annu Rev Neurosci. 2014. Jul 8;37:55–78. doi: 10.1146/annurev-neuro-071013-014149. PubMed PMID: 24821313. [DOI] [PubMed] [Google Scholar]
- 1950.Tsvetkov AS, Miller J, Arrasate M, et al. A small-molecule scaffold induces autophagy in primary neurons and protects against toxicity in a Huntington disease model [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Proc Natl Acad Sci U S A. 2010. Sep 28;107(39):16982–7. doi: 10.1073/pnas.1004498107. PubMed PMID: 20833817; PubMed Central PMCID: PMC2947884. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1951.Williams A, Sarkar S, Cuddon P, et al. Novel targets for Huntington’s disease in an mTOR-independent autophagy pathway [Research Support, Non-U.S. Gov’t]. Nat Chem Biol. 2008. May;4(5):295–305. doi: 10.1038/nchembio.79. PubMed PMID: 18391949; PubMed Central PMCID: PMC2635566. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1952.Palomo GM, Cerrato T, Gargini R, et al. Silencing of frataxin gene expression triggers p53-dependent apoptosis in human neuron-like cells [Research Support, Non-U.S. Gov’t]. Hum Mol Genet. 2011. Jul 15;20(14):2807–22. doi: 10.1093/hmg/ddr187. PubMed PMID: 21531789; eng. [DOI] [PubMed] [Google Scholar]
- 1953.Bolinches-Amoros A, Molla B, Pla-Martin D, et al. Mitochondrial dysfunction induced by frataxin deficiency is associated with cellular senescence and abnormal calcium metabolism. Front Cell Neurosci. 2014;8:124. doi: 10.3389/fncel.2014.00124. PubMed PMID: 24860428; PubMed Central PMCID: PMC4026758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1954.Sakagami H, Kawase M, Wakabayashi H, et al. Factors that affect the type of cell death induced by chemicals. Autophagy. 2007. Sep-Oct;3(5):493–5. PubMed PMID: 17611389; eng. [DOI] [PubMed] [Google Scholar]
- 1955.Doelling JH, Walker JM, Friedman EM, et al. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S. Research Support, U.S. Gov’t, P.H.S.]. J Biol Chem. 2002. Sep 6;277(36):33105–14. doi: 10.1074/jbc.M204630200. PubMed PMID: 12070171; eng. [DOI] [PubMed] [Google Scholar]
- 1956.Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007. Jun 28;447(7148):1121–5. PubMed PMID: 17589504; eng. [DOI] [PubMed] [Google Scholar]
- 1957.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004. Dec 23;432(7020):1032–6. PubMed PMID: 15525940; eng. [DOI] [PubMed] [Google Scholar]
- 1958.Hwang S, Maloney NS, Bruinsma MW, et al. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012. Apr 19;11(4):397–409. doi: 10.1016/j.chom.2012.03.002. PubMed PMID: 22520467; PubMed Central PMCID: PMC3348177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1959.Zhu H, Wu H, Liu X, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells [Research Support, N.I.H., Extramural Research Support, U.S. Gov’t, Non-P.H.S.]. Autophagy. 2009. Aug;5(6):816–23. PubMed PMID: 19535919; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1960.Hamacher-Brady A, Brady NR, Logue SE, et al. Response to myocardial ischemia/reperfusion injury involves Bnip3 and autophagy. Cell Death Differ. 2007. Jan;14(1):146–57. PubMed PMID: 16645637; eng. [DOI] [PubMed] [Google Scholar]
- 1961.Poeck H, Besch R, Maihoefer C, et al. 5ʹ-Triphosphate-siRNA: turning gene silencing and Rig-I activation against melanoma [Research Support, Non-U.S. Gov’t]. Nat Med. 2008. Nov;14(11):1256–63. doi: 10.1038/nm.1887. PubMed PMID: 18978796; eng. [DOI] [PubMed] [Google Scholar]
- 1962.Delgado MA, Elmaoued RA, Davis AS, et al. Toll-like receptors control autophagy [Research Support, N.I.H., Extramural]. EMBO J. 2008. Apr 9;27(7):1110–21. doi: 10.1038/emboj.2008.31. PubMed PMID: 18337753; PubMed Central PMCID: PMC2323261. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1963.Pua HH, Dzhagalov I, Chuck M, et al. A critical role for the autophagy gene Atg5 in T cell survival and proliferation [Research Support, N.I.H., Extramural]. J Exp Med. 2007. Jan 22;204(1):25–31. doi: 10.1084/jem.20061303. PubMed PMID: 17190837; PubMed Central PMCID: PMC2118420. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1964.Miller BC, Zhao Z, Stephenson LM, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Autophagy. 2008. Apr;4(3):309–14. PubMed PMID: 18188005; eng. [DOI] [PubMed] [Google Scholar]
- 1965.Lee JS, Li Q, Lee JY, et al. FLIP-mediated autophagy regulation in cell death control [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Nat Cell Biol. 2009. Nov;11(11):1355–62. doi: 10.1038/ncb1980. PubMed PMID: 19838173; PubMed Central PMCID: PMC2802862. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1966.Kyei GB, Dinkins C, Davis AS, et al. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. J Cell Biol. 2009. Jul 27;186(2):255–68. doi: 10.1083/jcb.200903070. PubMed PMID: 19635843; PubMed Central PMCID: PMC2717652. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1967.Choy A, Dancourt J, Mugo B, et al. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science. 2012. Nov 23;338(6110):1072–6. doi: 10.1126/science.1227026. PubMed PMID: 23112293; PubMed Central PMCID: PMCPMC3682818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1968.Kwon DH, Kim S, Jung YO, et al. The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane. Autophagy. 2017. Jan 2;13(1):70–81. doi: 10.1080/15548627.2016.1243199. PubMed PMID: 27791457; PubMed Central PMCID: PMCPMC5240826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1969.David S, Rusniok C, Mentasti M, et al. Multiple major disease-associated clones of Legionella pneumophila have emerged recently and independently. Genome Res. 2016. Nov;26(11):1555–1564. doi: 10.1101/gr.209536.116. PubMed PMID: 27662900; PubMed Central PMCID: PMCPMC5088597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1970.Gomez-Valero L, Buchrieser C.. Intracellular parasitism, the driving force of evolution of Legionella pneumophila and the genus Legionella. Genes Immun. 2019. May;20(5):394–402. doi: 10.1038/s41435-019-0074-z. PubMed PMID: 31053752. [DOI] [PubMed] [Google Scholar]
- 1971.Rolando M, Escoll P, Nora T, et al. Legionella pneumophila S1P-lyase targets host sphingolipid metabolism and restrains autophagy. Proc Natl Acad Sci U S A. 2016. Feb 16;113(7):1901–1906. doi: 10.1073/pnas.1522067113. PubMed PMID: 26831115; PubMed Central PMCID: PMCPMC4763766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1972.Kimball SR, Siegfried BA, Jefferson LS.. Glucagon represses signaling through the mammalian target of rapamycin in rat liver by activating AMP-activated protein kinase. J Biol Chem. 2004. Dec 24;279(52):54103–9. PubMed PMID: 15494402; eng. [DOI] [PubMed] [Google Scholar]
- 1973.Blommaart EF, Luiken JJ, Blommaart PJ, et al. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J Biol Chem 1995. Feb 3;270(5):2320–6. PubMed PMID: 7836465. [DOI] [PubMed] [Google Scholar]
- 1974.Klionsky DJ, Meijer AJ, Codogno P, et al. Autophagy and p70S6 kinase. Autophagy. 2005. Apr;1(1):59–61. PubMed PMID: 16874035. [DOI] [PubMed] [Google Scholar]
- 1975.Noda T, Ohsumi Y.. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998. Feb 13;273(7):3963–6. PubMed PMID: 9461583. [DOI] [PubMed] [Google Scholar]
- 1976.Sarkar S, Floto RA, Berger Z, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol 2005. Sep 26;170(7):1101–11. PubMed PMID: 16186256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1977.Renna M, Jimenez-Sanchez M, Sarkar S, et al. Chemical inducers of autophagy that enhance the clearance of mutant proteins in neurodegenerative diseases. J Biol Chem. 2010. Apr 9;285(15):11061–7. doi: 10.1074/jbc.R109.072181. PubMed PMID: 20147746; PubMed Central PMCID: PMC2856980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1978.Zhang L, Yu J, Pan H, et al. Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc Natl Acad Sci USA. 2007;104:19023–19028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1979.Shin SY, Lee KS, Choi YK, et al. The antipsychotic agent chlorpromazine induces autophagic cell death by inhibiting the Akt/mTOR pathway in human U-87MG glioma cells. Carcinogenesis. 2013. Sep;34(9):2080–9. doi: 10.1093/carcin/bgt169. PubMed PMID: 23689352. [DOI] [PubMed] [Google Scholar]
- 1980.Wu CH, Bai LY, Tsai MH, et al. Pharmacological exploitation of the phenothiazine antipsychotics to develop novel antitumor agents-A drug repurposing strategy. Sci Rep. 2016. Jun 9;6:27540. doi: 10.1038/srep27540. PubMed PMID: 27277973; PubMed Central PMCID: PMCPMC4899727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1981.Chu CW, Ko HJ, Chou CH, et al. Thioridazine Enhances P62-Mediated Autophagy and Apoptosis Through Wnt/beta-Catenin Signaling Pathway in Glioma Cells. Int J Mol Sci. 2019. Jan 22;20(3). doi: 10.3390/ijms20030473. PubMed PMID: 30678307; PubMed Central PMCID: PMCPMC6386927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1982.Medeiros HCD, Colturato-Kido C, Ferraz LS, et al. AMPK activation induced by promethazine increases NOXA expression and Beclin-1 phosphorylation and drives autophagy-associated apoptosis in chronic myeloid leukemia. Chem Biol Interact. 2020. Jan 5;315:108888. doi: 10.1016/j.cbi.2019.108888. PubMed PMID: 31682805. [DOI] [PubMed] [Google Scholar]
- 1983.Decuypere JP, Welkenhuyzen K, Luyten T, et al. Ins(1,4,5)P3 receptor-mediated Ca2+ signaling and autophagy induction are interrelated. Autophagy. 2011. Dec;7(12):1472–89. doi: 10.4161/auto.7.12.17909. PubMed PMID: 22082873; PubMed Central PMCID: PMCPMC3327615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1984.Hoyer-Hansen M, Bastholm L, Szyniarowski P, et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-[b], and Bcl-2 [Research Support, Non-U.S. Gov’t]. Mol Cell. 2007. Jan 26;25(2):193–205. doi: 10.1016/j.molcel.2006.12.009. PubMed PMID: 17244528; eng. [DOI] [PubMed] [Google Scholar]
- 1985.Decuypere JP, Kindt D, Luyten T, et al. mTOR-Controlled Autophagy Requires Intracellular Ca(2+) Signaling. PLoS One. 2013;8(4):e61020. doi: 10.1371/journal.pone.0061020. PubMed PMID: 23565295; PubMed Central PMCID: PMC3614970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1986.Kary C. Liver autophagy’s sweet side. Nat Cell Biol. 2018. Mar;20(3):224. doi: 10.1038/s41556-018-0059-6. PubMed PMID: 29476155. [DOI] [PubMed] [Google Scholar]
- 1987.Szalai P, Parys JB, Bultynck G, et al. Nonlinear relationship between ER Ca(2+) depletion versus induction of the unfolded protein response, autophagy inhibition, and cell death. Cell Calcium. 2018. Dec;76:48–61. doi: 10.1016/j.ceca.2018.09.005. PubMed PMID: 30261424. [DOI] [PubMed] [Google Scholar]
- 1988.Wong VKW, Qiu C, Xu SW, et al. Ca(2+) signalling plays a role in celastrol-mediated suppression of synovial fibroblasts of rheumatoid arthritis patients and experimental arthritis in rats. Br J Pharmacol. 2019. Aug;176(16):2922–2944. doi: 10.1111/bph.14718. PubMed PMID: 31124139; PubMed Central PMCID: PMCPMC6637043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1989.Qu YQ, Gordillo-Martinez F, Law BYK, et al. 2-Aminoethoxydiphenylborane sensitizes anti-tumor effect of bortezomib via suppression of calcium-mediated autophagy. Cell Death Dis. 2018. Mar 2;9(3):361. doi: 10.1038/s41419-018-0397-0. PubMed PMID: 29500417; PubMed Central PMCID: PMCPMC5834458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1990.Decuypere JP, Bultynck G, Parys JB.. A dual role for Ca(2+) in autophagy regulation. Cell Calcium 2011;50:242–50. [DOI] [PubMed] [Google Scholar]
- 1991.Luyten T, Welkenhuyzen K, Roest G, et al. Resveratrol-induced autophagy is dependent on IP3Rs and on cytosolic Ca(2). Biochim Biophys Acta Mol Cell Res. 2017. Jun;1864(6):947–956. doi: 10.1016/j.bbamcr.2017.02.013. PubMed PMID: 28254579. [DOI] [PubMed] [Google Scholar]
- 1992.Roest G, Hesemans E, Welkenhuyzen K, et al. The ER Stress Inducer l-Azetidine-2-Carboxylic Acid Elevates the Levels of Phospho-eIF2alpha and of LC3-II in a Ca(2+)-Dependent Manner. Cells. 2018. Nov 30;7(12). doi: 10.3390/cells7120239. PubMed PMID: 30513588; PubMed Central PMCID: PMCPMC6316609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1993.Tuncer S, Sade-Memisoglu A, Keskus AG, et al. Enhanced expression of HNF4alpha during intestinal epithelial differentiation is involved in the activation of ER stress. FEBS J. 2020. Jun;287(12):2504–2523. doi: 10.1111/febs.15152. PubMed PMID: 31762160. [DOI] [PubMed] [Google Scholar]
- 1994.Bootman MD, Chehab T, Bultynck G, et al. The regulation of autophagy by calcium signals: Do we have a consensus? Cell Calcium. 2018. Mar;70:32–46. doi: 10.1016/j.ceca.2017.08.005. PubMed PMID: 28847414. [DOI] [PubMed] [Google Scholar]
- 1995.Pereira GJ, Antonioli M, Hirata H, et al. Glutamate induces autophagy via the two-pore channels in neural cells. Oncotarget. 2017. Feb 21;8(8):12730–12740. doi: 10.18632/oncotarget.14404. PubMed PMID: 28055974; PubMed Central PMCID: PMCPMC5355049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1996.Pereira GJ, Hirata H, Fimia GM, et al. Nicotinic acid adenine dinucleotide phosphate (NAADP) regulates autophagy in cultured astrocytes [Research Support, Non-U.S. Gov’t]. J Biol Chem. 2011. Aug 12;286(32):27875–81. doi: 10.1074/jbc.C110.216580. PubMed PMID: 21610076; PubMed Central PMCID: PMC3151033. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1997.Cang C, Zhou Y, Navarro B, et al. mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state. Cell. 2013. Feb 14;152(4):778–90. doi: 10.1016/j.cell.2013.01.023. PubMed PMID: 23394946; PubMed Central PMCID: PMC3908667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1998.Ogunbayo OA, Duan J, Xiong J, et al. mTORC1 controls lysosomal Ca(2+) release through the two-pore channel TPC2. Sci Signal. 2018. Apr 10;11(525). doi: 10.1126/scisignal.aao5775. PubMed PMID: 29636391; PubMed Central PMCID: PMCPMC6055479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1999.Sun X, Yang Y, Zhong XZ, et al. A negative feedback regulation of MTORC1 activity by the lysosomal Ca(2+) channel MCOLN1 (mucolipin 1) using a CALM (calmodulin)-dependent mechanism. Autophagy. 2018;14(1):38–52. doi: 10.1080/15548627.2017.1389822. PubMed PMID: 29460684; PubMed Central PMCID: PMCPMC5846559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2000.Medina DL, Di Paola S, Peluso I, et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015. Mar;17(3):288–99. doi: 10.1038/ncb3114. PubMed PMID: 25720963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2001.Bootman MD, Bultynck G.. Fundamentals of cellular calcium signaling: a primer. Cold Spring Harb Perspect Biol. 2020. Jan 2;12(1). doi: 10.1101/cshperspect.a038802. PubMed PMID: 31427372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2002.Smith NA, Kress BT, Lu Y, et al. Fluorescent Ca(2+) indicators directly inhibit the Na,K-ATPase and disrupt cellular functions. Sci Signal. 2018. Jan 30;11(515). doi: 10.1126/scisignal.aal2039. PubMed PMID: 29382785; PubMed Central PMCID: PMCPMC6190706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2003.Bootman MD, Allman S, Rietdorf K, et al. Deleterious effects of calcium indicators within cells; an inconvenient truth. Cell Calcium. 2018. Jul;73:82–87. doi: 10.1016/j.ceca.2018.04.005. PubMed PMID: 29689523. [DOI] [PubMed] [Google Scholar]
- 2004.Shoji-Kawata S, Sumpter R, Leveno M, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013. Feb 14;494(7436):201–6. doi: 10.1038/nature11866. PubMed PMID: 23364696; PubMed Central PMCID: PMC3788641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2005.Su M, Mei Y, Sanishvili R, et al. Targeting gamma-herpesvirus 68 Bcl-2-mediated down-regulation of autophagy. J Biol Chem. 2014. Mar 21;289(12):8029–40. doi: 10.1074/jbc.M113.515361. PubMed PMID: 24443581; PubMed Central PMCID: PMC3961636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2006.Winter G, Hazan R, Bakalinsky AT, et al. Caffeine induces macroautophagy and confers a cytocidal effect on food spoilage yeast in combination with benzoic acid [Research Support, Non-U.S. Gov’t]. Autophagy. 2008. Jan;4(1):28–36. PubMed PMID: 17952024; eng. [DOI] [PubMed] [Google Scholar]
- 2007.Saiki S, Sasazawa Y, Imamichi Y, et al. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy. 2011. Feb;7(2):176–87. PubMed PMID: 21081844; PubMed Central PMCID: PMC3039768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2008.Tsabar M, Eapen VV, Mason JM, et al. Caffeine impairs resection during DNA break repair by reducing the levels of nucleases Sae2 and Dna2. Nucleic Acids Res. 2015. Aug 18;43(14):6889–901. doi: 10.1093/nar/gkv520. PubMed PMID: 26019182; PubMed Central PMCID: PMC4538808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2009.Fu J, Shao CJ, Chen FR, et al. Autophagy induced by valproic acid is associated with oxidative stress in glioma cell lines. Neuro Oncol. 2010. Apr;12(4):328–40. doi: 10.1093/neuonc/nop005. PubMed PMID: 20308311; PubMed Central PMCID: PMC2940599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2010.Robert T, Vanoli F, Chiolo I, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011. Mar 3;471(7336):74–9. doi: 10.1038/nature09803. PubMed PMID: 21368826; PubMed Central PMCID: PMC3935290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2011.Bartholomew CR, Suzuki T, Du Z, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A. 2012. Jul 10;109(28):11206–10. doi: 10.1073/pnas.1200313109. PubMed PMID: 22733735; PubMed Central PMCID: PMC3396506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2012.Yi C, Ma M, Ran L, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012. Apr 27;336(6080):474–7. doi: 10.1126/science.1216990. PubMed PMID: 22539722. [DOI] [PubMed] [Google Scholar]
- 2013.Koukourakis MI, Giatromanolaki A, Fylaktakidou K, et al. SMER28 is a mTOR-independent small molecule enhancer of autophagy that protects mouse bone marrow and liver against radiotherapy. Invest New Drugs. 2018. Oct;36(5):773–781. doi: 10.1007/s10637-018-0566-0. PubMed PMID: 29387992. [DOI] [PubMed] [Google Scholar]
- 2014.Xie N, Zhong L, Liu L, et al. Autophagy contributes to dasatinib-induced myeloid differentiation of human acute myeloid leukemia cells. Biochem Pharmacol. 2014. May 1;89(1):74–85. doi: 10.1016/j.bcp.2014.02.019. PubMed PMID: 24607273. [DOI] [PubMed] [Google Scholar]
- 2015.Romeo-Guitart D, Fores J, Herrando-Grabulosa M, et al. Neuroprotective Drug for Nerve Trauma Revealed Using Artificial Intelligence. Sci Rep. 2018. Jan 30;8(1):1879. doi: 10.1038/s41598-018-19767-3. PubMed PMID: 29382857; PubMed Central PMCID: PMCPMC5790005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2016.Romeo-Guitart D, Leiva-Rodriguez T, Fores J, et al. Improved Motor Nerve Regeneration by SIRT1/Hif1a-Mediated Autophagy. Cells. 2019. Oct 30;8(11). doi: 10.3390/cells8111354. PubMed PMID: 31671642; PubMed Central PMCID: PMCPMC6912449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2017.Romeo-Guitart D, Fores J, Navarro X, et al. Boosted regeneration and reduced denervated muscle atrophy by neuroheal in a pre-clinical model of lumbar root avulsion with delayed reimplantation. Sci Rep. 2017. Sep 20;7(1):12028. doi: 10.1038/s41598-017-11086-3. PubMed PMID: 28931824; PubMed Central PMCID: PMCPMC5607317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2018.Romeo-Guitart D, Leiva-Rodriguez T, Espinosa-Alcantud M, et al. SIRT1 activation with neuroheal is neuroprotective but SIRT2 inhibition with AK7 is detrimental for disconnected motoneurons. Cell Death Dis. 2018. May 1;9(5):531. doi: 10.1038/s41419-018-0553-6. PubMed PMID: 29748539; PubMed Central PMCID: PMCPMC5945655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2019.Mishra P, Rai S, Manjithaya R.. A novel dual luciferase based high throughput assay to monitor autophagy in real time in yeast S. cerevisiae. Biochem Biophys Rep. 2017. Sep;11:138–146. doi: 10.1016/j.bbrep.2017.07.008. PubMed PMID: 28955778; PubMed Central PMCID: PMCPMC5614714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2020.Gama JB, Ohlmeier S, Martins TG, et al. Proteomic analysis of the action of the Mycobacterium ulcerans toxin mycolactone: targeting host cells cytoskeleton and collagen. PLoS Negl Trop Dis. 2014. Aug;8(8):e3066. doi: 10.1371/journal.pntd.0003066. PubMed PMID: 25101965; PubMed Central PMCID: PMCPMC4125307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2021.Ogbechi J, Hall BS, Sbarrato T, et al. Inhibition of Sec61-dependent translocation by mycolactone uncouples the integrated stress response from ER stress, driving cytotoxicity via translational activation of ATF4. Cell Death Dis. 2018. Mar 14;9(3):397. doi: 10.1038/s41419-018-0427-y. PubMed PMID: 29540678; PubMed Central PMCID: PMCPMC5852046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2022.Hall BS, Hill K, McKenna M, et al. The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog. 2014. Apr;10(4):e1004061. doi: 10.1371/journal.ppat.1004061. PubMed PMID: 24699819; PubMed Central PMCID: PMCPMC3974873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2023.Capela C, Dossou AD, Silva-Gomes R, et al. Genetic variation in autophagy-related genes influences the risk and phenotype of buruli ulcer. PLoS Negl Trop Dis. 2016. Apr;10(4):e0004671. doi: 10.1371/journal.pntd.0004671. PubMed PMID: 27128681; PubMed Central PMCID: PMCPMC4851401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2024.Qi X, Man SM, Malireddi RK, et al. Cathepsin B modulates lysosomal biogenesis and host defense against Francisella novicida infection. J Exp Med. 2016. Sep 19;213(10):2081–97. doi: 10.1084/jem.20151938. PubMed PMID: 27551156; PubMed Central PMCID: PMCPMC5030800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2025.Katagiri N, Kuroda T, Kishimoto H, et al. The nucleolar protein nucleophosmin is essential for autophagy induced by inhibiting Pol I transcription. Sci Rep. 2015;5:8903. doi: 10.1038/srep08903. PubMed PMID: 25754892; PubMed Central PMCID: PMC4354046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2026.Kreiner G, Bierhoff H, Armentano M, et al. A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell Death Differ. 2013. Nov;20(11):1455–64. doi: 10.1038/cdd.2013.66. PubMed PMID: 23764776; PubMed Central PMCID: PMC3792439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2027.Pfister AS. Emerging role of the nucleolar stress response in autophagy. Front Cell Neurosci. 2019;13:156. doi: 10.3389/fncel.2019.00156. PubMed PMID: 31114481; PubMed Central PMCID: PMCPMC6503120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2028.Dannheisig DP, Beck E, Calzia E, et al. Loss of Peter Pan (PPAN) Affects Mitochondrial Homeostasis and Autophagic Flux. Cells. 2019. Aug 14;8(8). doi: 10.3390/cells8080894. PubMed PMID: 31416196; PubMed Central PMCID: PMCPMC6721654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2029.Furuya N, Liang XH, Levine B.. Autophagy and cancer. In: Klionsky DJ, editor. Autophagy. Georgetown, TX: Landes Bioscience; 2004. p. 241–255. [Google Scholar]
- 2030.de Medina P, Paillasse MR, Segala G, et al. Importance of cholesterol and oxysterols metabolism in the pharmacology of tamoxifen and other AEBS ligands. Chem Phys Lipids. 2011;164(6):432–7. doi: 10.1016/j.chemphyslip.2011.05.005. PubMed PMID: 21641337. [DOI] [PubMed] [Google Scholar]
- 2031.de Medina P, Payre B, Boubekeur N, et al. Ligands of the antiestrogen-binding site induce active cell death and autophagy in human breast cancer cells through the modulation of cholesterol metabolism [Research Support, Non-U.S. Gov’t]. Cell Death Differ. 2009. Oct;16(10):1372–84. doi: 10.1038/cdd.2009.62. PubMed PMID: 19521424; eng. [DOI] [PubMed] [Google Scholar]
- 2032.Sarkar S, Perlstein EO, Imarisio S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat Chem Biol. 2007. Jun;3(6):331–8. PubMed PMID: 17486044; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2033.Savolainen MH, Richie CT, Harvey BK, et al. The beneficial effect of a prolyl oligopeptidase inhibitor, KYP-2047, on alpha-synuclein clearance and autophagy in A30P transgenic mouse. Neurobiol Dis. 2014. Aug;68:1–15. doi: 10.1016/j.nbd.2014.04.003. PubMed PMID: 24746855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2034.Svarcbahs R, Julku U, Kilpelainen T, et al. New tricks of prolyl oligopeptidase inhibitors - A common drug therapy for several neurodegenerative diseases. Biochem Pharmacol. 2019. Mar;161:113–120. doi: 10.1016/j.bcp.2019.01.013. PubMed PMID: 30660495. [DOI] [PubMed] [Google Scholar]
- 2035.Svarcbahs R, Julku UH, Norrbacka S, et al. Removal of prolyl oligopeptidase reduces alpha-synuclein toxicity in cells and in vivo. Sci Rep. 2018. Jan 24;8(1):1552. doi: 10.1038/s41598-018-19823-y. PubMed PMID: 29367610; PubMed Central PMCID: PMCPMC5784134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2036.Sarkar S, Davies JE, Huang Z, et al. Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and [a]-synuclein [Research Support, Non-U.S. Gov’t]. J Biol Chem. 2007. Feb 23;282(8):5641–52. doi: 10.1074/jbc.M609532200. PubMed PMID: 17182613; eng. [DOI] [PubMed] [Google Scholar]
- 2037.Kruger U, Wang Y, Kumar S, et al. Autophagic degradation of tau in primary neurons and its enhancement by trehalose. Neurobiol Aging. 2012. Dec 12;33:2291–305. doi: 10.1016/j.neurobiolaging.2011.11.009. PubMed PMID: 22169203; Eng. [DOI] [PubMed] [Google Scholar]
- 2038.Rusmini P, Cortese K, Crippa V, et al. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy. 2019. Apr;15(4):631–651. doi: 10.1080/15548627.2018.1535292. PubMed PMID: 30335591; PubMed Central PMCID: PMCPMC6526812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2039.Tien NT, Karaca I, Tamboli IY, et al. Trehalose Alters Subcellular Trafficking and the Metabolism of the Alzheimer-associated Amyloid Precursor Protein. J Biol Chem. 2016. May 13;291(20):10528–40. doi: 10.1074/jbc.M116.719286. PubMed PMID: 26957541; PubMed Central PMCID: PMCPMC4865903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2040.Yoon YS, Cho ED, Jung Ahn W, et al. Is trehalose an autophagic inducer? Unraveling the roles of non-reducing disaccharides on autophagic flux and alpha-synuclein aggregation. Cell Death Dis. 2017. Oct 5;8(10):e3091. doi: 10.1038/cddis.2017.501. PubMed PMID: 28981090; PubMed Central PMCID: PMCPMC5682667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2041.Kruse KB, Brodsky JL, McCracken AA.. Characterization of an ERAD gene as VPS30/ATG6 reveals two alternative and functionally distinct protein quality control pathways: one for soluble Z variant of human alpha-1 proteinase inhibitor (A1PiZ) and another for aggregates of A1PiZ. Mol Biol Cell. 2006. Jan;17(1):203–12. doi: 10.1091/mbc.e04-09-0779. PubMed PMID: 16267277; PubMed Central PMCID: PMCPMC1345659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2042.Gossner G, Choi M, Tan L, et al. Genistein-induced apoptosis and autophagocytosis in ovarian cancer cells. Gynecol Oncol. 2007. Apr;105(1):23–30. doi: 10.1016/j.ygyno.2006.11.009. PubMed PMID: 17234261. [DOI] [PubMed] [Google Scholar]
- 2043.Nakamura Y, Yogosawa S, Izutani Y, et al. A combination of indol-3-carbinol and genistein synergistically induces apoptosis in human colon cancer HT-29 cells by inhibiting Akt phosphorylation and progression of autophagy. Mol Cancer. 2009. Nov 12;8:100. doi: 10.1186/1476-4598-8-100. PubMed PMID: 19909554; PubMed Central PMCID: PMCPMC2784428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2044.Prietsch RF, Monte LG, da Silva FA, et al. Genistein induces apoptosis and autophagy in human breast MCF-7 cells by modulating the expression of proapoptotic factors and oxidative stress enzymes. Mol Cell Biochem. 2014. May;390(1–2):235–42. doi: 10.1007/s11010-014-1974-x. PubMed PMID: 24573886. [DOI] [PubMed] [Google Scholar]
- 2045.Suzuki R, Kang Y, Li X, et al. Genistein potentiates the antitumor effect of 5-Fluorouracil by inducing apoptosis and autophagy in human pancreatic cancer cells. Anticancer Res. 2014. Sep;34(9):4685–92. PubMed PMID: 25202045; PubMed Central PMCID: PMCPMC4240628. [PMC free article] [PubMed] [Google Scholar]
- 2046.Castro L, Gao X, Moore AB, et al. A High Concentration of Genistein Induces Cell Death in Human Uterine Leiomyoma Cells by Autophagy. Expert Opin Environ Biol. 2016;5(Suppl 1). doi: 10.4172/2325-9655.S1-003. PubMed PMID: 27512718; PubMed Central PMCID: PMCPMC4976942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2047.Pierzynowska K, Gaffke L, Hac A, et al. Correction of huntington’s disease phenotype by genistein-induced autophagy in the cellular model. Neuromolecular Med. 2018. Mar;20(1):112–123. doi: 10.1007/s12017-018-8482-1. PubMed PMID: 29435951; PubMed Central PMCID: PMCPMC5834590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2048.Pierzynowska K, Podlacha M, Gaffke L, et al. Autophagy-dependent mechanism of genistein-mediated elimination of behavioral and biochemical defects in the rat model of sporadic Alzheimer’s disease. Neuropharmacology. 2019. Apr;148:332–346. doi: 10.1016/j.neuropharm.2019.01.030. PubMed PMID: 30710571. [DOI] [PubMed] [Google Scholar]
- 2049.Pierzynowska K, Gaffke L, Cyske Z, et al. Autophagy stimulation as a promising approach in treatment of neurodegenerative diseases. Metab Brain Dis. 2018. Aug;33(4):989–1008. doi: 10.1007/s11011-018-0214-6. PubMed PMID: 29542037; PubMed Central PMCID: PMCPMC6060747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2050.Pierzynowska K, Gaffke L, Podlacha M, et al. Mucopolysaccharidosis and autophagy: controversies on the contribution of the process to the pathogenesis and possible therapeutic applications. Neuromolecular Med. 2019. Aug 1. doi: 10.1007/s12017-019-08559-1. PubMed PMID: 31372809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2051.Moskot M, Montefusco S, Jakobkiewicz-Banecka J, et al. The phytoestrogen genistein modulates lysosomal metabolism and transcription factor EB (TFEB) activation. J Biol Chem. 2014. Jun 13;289(24):17054–69. doi: 10.1074/jbc.M114.555300. PubMed PMID: 24770416; PubMed Central PMCID: PMCPMC4059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2052.Lee KY, Kim JR, Choi HC.. Genistein-induced LKB1-AMPK activation inhibits senescence of VSMC through autophagy induction. Vascul Pharmacol. 2016. Jun;81:75–82. doi: 10.1016/j.vph.2016.02.007. PubMed PMID: 26924458. [DOI] [PubMed] [Google Scholar]
- 2053.Zhang H, Yang X, Pang X, et al. Genistein protects against ox-LDL-induced senescence through enhancing SIRT1/LKB1/AMPK-mediated autophagy flux in HUVECs. Mol Cell Biochem. 2019. May;455(1–2): 127–134. doi: 10.1007/s11010-018-3476-8. PubMed PMID: 30443855. [DOI] [PubMed] [Google Scholar]
- 2054.Bai X, Bai A, Honda JR, et al. Alpha-1-Antitrypsin Enhances Primary Human Macrophage Immunity Against Non-tuberculous Mycobacteria. Front Immunol. 2019;10:1417. doi: 10.3389/fimmu.2019.01417. PubMed PMID: 31293581; PubMed Central PMCID: PMCPMC6606736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2055.Bai X, Feldman NE, Chmura K, et al. Inhibition of nuclear factor-kappa B activation decreases survival of Mycobacterium tuberculosis in human macrophages. PLoS One. 2013;8(4):e61925. doi: 10.1371/journal.pone.0061925. PubMed PMID: 23634218; PubMed Central PMCID: PMCPMC3636238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2056.Hilchie AL, Wuerth K, Hancock RE.. Immune modulation by multifaceted cationic host defense (antimicrobial) peptides. Nat Chem Biol. 2013. Dec;9(12):761–8. doi: 10.1038/nchembio.1393. PubMed PMID: 24231617. [DOI] [PubMed] [Google Scholar]
- 2057.Alonso S, Pethe K, Russell DG, et al. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc Natl Acad Sci U S A. 2007. Apr 3;104(14):6031–6. doi: 10.1073/pnas.0700036104. PubMed PMID: 17389386; PubMed Central PMCID: PMCPMC1851611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2058.Ponpuak M, Davis AS, Roberts EA, et al. Delivery of cytosolic components by autophagic adaptor protein p62 endows auto-phagosomes with unique antimicrobial properties [Research Support, N.I.H., Extramural]. Immunity. 2010. Mar 26;32(3):329–41. doi: 10.1016/j.immuni.2010.02.009. PubMed PMID: 20206555; PubMed Central PMCID: PMC2846977. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2059.Bera A, Singh S, Nagaraj R, et al. Induction of autophagic cell death in Leishmania donovani by antimicrobial peptides. Mol Biochem Parasitol. 2003. Mar;127(1):23–35. doi: 10.1016/s0166-6851(02)00300-6. PubMed PMID: 12615333. [DOI] [PubMed] [Google Scholar]
- 2060.Rekha RS, Rao Muvva SS, Wan M, et al. Phenylbutyrate induces LL-37-dependent autophagy and intracellular killing of Mycobacterium tuberculosis in human macrophages. Autophagy. 2015;11(9):1688–99. doi: 10.1080/15548627.2015.1075110. PubMed PMID: 26218841; PubMed Central PMCID: PMCPMC4590658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2061.Koshkina NV, Briggs K, Palalon F, et al. Autophagy and enhanced chemosensitivity in experimental pancreatic cancers induced by noninvasive radiofrequency field treatment. Cancer. 2014. Feb 15;120(4):480–91. doi: 10.1002/cncr.28453. PubMed PMID: 24496866; PubMed Central PMCID: PMC3916783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2062.Suresh SN, Chavalmane AK, Pillai M, et al. Modulation of Autophagy by a Small Molecule Inverse Agonist of ERRalpha Is Neuroprotective. Front Mol Neurosci. 2018;11:109. doi: 10.3389/fnmol.2018.00109. PubMed PMID: 29686608; PubMed Central PMCID: PMCPMC5900053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2063.Vicencio JM, Ortiz C, Criollo A, et al. The inositol 1,4,5-trisphosphate receptor regulates autophagy through its interaction with Beclin 1 [Research Support, Non-U.S. Gov’t]. Cell Death Differ. 2009. Jul;16(7):1006–17. doi: 10.1038/cdd.2009.34. PubMed PMID: 19325567; eng. [DOI] [PubMed] [Google Scholar]
- 2064.Dayan F, Bilton RL, Laferriere J, et al. Activation of HIF-1alpha in exponentially growing cells via hypoxic stimulation is independent of the Akt/mTOR pathway [Research Support, Non-U.S. Gov’t]. J Cell Physiol. 2009. Jan;218(1):167–74. doi: 10.1002/jcp.21584. PubMed PMID: 18781596; eng. [DOI] [PubMed] [Google Scholar]
- 2065.Yamashita S, Yurimoto H, Murakami D, et al. Lag-phase autophagy in the methylotrophic yeast Pichia pastoris [Research Support, Non-U.S. Gov’t]. Genes Cells Devot Mol Cell Mech 2009. Jul;14(7):861–70. doi: 10.1111/j.1365-2443.2009.01316.x. PubMed PMID: 19549169; eng. [DOI] [PubMed] [Google Scholar]
- 2066.van Zutphen T, Baerends RJ, Susanna KA, et al. Adaptation of Hansenula polymorpha to methanol: a transcriptome analysis [Research Support, Non-U.S. Gov’t]. BMC genomics. 2010;11:1. doi: 10.1186/1471-2164-11-1. PubMed PMID: 20044946; PubMed Central PMCID: PMC2827406. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2067.Moriyasu Y, Hattori M, Jauh G-Y, et al. Alpha tonoplast intrinsic protein is specifically associated with vacuole membrane involved in an autophagic process [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.]. Plant Cell Physiol. 2003. Aug;44(8):795–802. PubMed PMID: 12941871; eng. [DOI] [PubMed] [Google Scholar]
- 2068.Inoue Y, Suzuki T, Hattori M, et al. AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells [Research Support, Non-U.S. Gov’t]. Plant Cell Physiol. 2006. Dec;47(12):1641–52. doi: 10.1093/pcp/pcl031. PubMed PMID: 17085765; eng. [DOI] [PubMed] [Google Scholar]
- 2069.Yano K, Suzuki T, Moriyasu Y.. Constitutive autophagy in plant root cells [Comment]. Autophagy. 2007. Jul-Aug;3(4):360–2. PubMed PMID: 17426438; eng. [DOI] [PubMed] [Google Scholar]
- 2070.Gordon PB, Kisen GO, Kovacs AL, et al. Experimental characterization of the autophagic-lysosomal pathway in isolated rat hepatocytes. Biochem Soc Symp. 1989;55:129–43. PubMed PMID: 2619764; eng. [PubMed] [Google Scholar]
- 2071.Poli A, Gordon PB, Schwarze PE, et al. Effects of insulin and anchorage on hepatocytic protein metabolism and amino acid transport. J Cell Sci. 1981. Apr;48:1–18. PubMed PMID: 7024288; eng. [DOI] [PubMed] [Google Scholar]
- 2072.Schliess F, Reissmann R, Reinehr R, et al. Involvement of integrins and Src in insulin signaling toward autophagic proteolysis in rat liver. J Biol Chem. 2004. May 14;279(20):21294–301. PubMed PMID: 14985360; eng. [DOI] [PubMed] [Google Scholar]
- 2073.vom Dahl S, Stoll B, Gerok W, et al. Inhibition of proteolysis by cell swelling in the liver requires intact microtubular structures. Biochem J. 1995. Jun 1;308(Pt 2):529–36. PubMed PMID: 7772037; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2074.vom Dahl S, Dombrowski F, Schmitt M, et al. Cell hydration controls autophagosome formation in rat liver in a microtubule-dependent way downstream from p38MAPK activation. Biochem J. 2001. Feb 15;354(Pt 1):31–6. PubMed PMID: 11171076; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2075.Shyu YJ, Liu H, Deng X, et al. Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques. 2006. Jan;40(1):61–6. PubMed PMID: 16454041. [DOI] [PubMed] [Google Scholar]
- 2076.Colson YL, Grinstaff MW.. Biologically responsive polymeric nanoparticles for drug delivery. Adv Mater. 2012. Jul 24;24(28):3878–86. doi: 10.1002/adma.201200420. PubMed PMID: 22988558. [DOI] [PubMed] [Google Scholar]
- 2077.Stern ST, Adiseshaiah PP, Crist RM.. Autophagy and lysosomal dysfunction as emerging mechanisms of nanomaterial toxicity. Part Fibre Toxicol. 2012. Jun 14;9:20. doi: 10.1186/1743-8977-9-20. PubMed PMID: 22697169; PubMed Central PMCID: PMCPMC3441384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2078.Anozie UC, Dalhaimer P.. Molecular links among non-biodegradable nanoparticles, reactive oxygen species, and autophagy. Adv Drug Deliv Rev. 2017. Dec 1;122:65–73. doi: 10.1016/j.addr.2017.01.001. PubMed PMID: 28065863. [DOI] [PubMed] [Google Scholar]
- 2079.Gallud A, Kloditz K, Ytterberg J, et al. Cationic gold nanoparticles elicit mitochondrial dysfunction: a multi-omics study. Sci Rep. 2019. Mar 13;9(1):4366. doi: 10.1038/s41598-019-40579-6. PubMed PMID: 30867451; PubMed Central PMCID: PMCPMC6416392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2080.Hulea L, Markovic Z, Topisirovic I, et al. Biomedical Potential of mTOR Modulation by Nanoparticles. Trends Biotechnol. 2016. May;34(5):349–353. doi: 10.1016/j.tibtech.2016.01.005. PubMed PMID: 26900005. [DOI] [PubMed] [Google Scholar]
- 2081.Trudeau KM, Colby AH, Zeng J, et al. Lysosome acidification by photoactivated nanoparticles restores autophagy under lipotoxicity. J Cell Biol. 2016. Jul 4;214(1):25–34. doi: 10.1083/jcb.201511042. PubMed PMID: 27377248; PubMed Central PMCID: PMCPMC4932370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2082.Zeng J, Shirihai OS, Grinstaff MW.. Degradable Nanoparticles Restore Lysosomal pH and Autophagic Flux in Lipotoxic Pancreatic Beta Cells. Adv Healthc Mater. 2019. Jun;8(12):e1801511. doi: 10.1002/adhm.201801511. PubMed PMID: 30698920. [DOI] [PubMed] [Google Scholar]
- 2083.Zeng J, Martin A, Han X, et al. Biodegradable PLGA nanoparticles restore lysosomal acidity and protect neural PC-12 cells against mitochondrial toxicity. Ind Eng Chem Res. 2019;58:13910–13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2084.Bourdenx M, Daniel J, Genin E, et al. Nanoparticles restore lysosomal acidification defects: Implications for Parkinson and other lysosomal-related diseases. Autophagy. 2016;12(3):472–83. doi: 10.1080/15548627.2015.1136769. PubMed PMID: 26761717; PubMed Central PMCID: PMCPMC4835967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2085.Sadhu A, Ghosh I, Moriyasu Y, et al. Role of cerium oxide nanoparticle-induced autophagy as a safeguard to exogenous H2O2-mediated DNA damage in tobacco BY-2 cells. Mutagenesis. 2018. Apr 13;33(2):161–177. doi: 10.1093/mutage/gey004. PubMed PMID: 29506140. [DOI] [PubMed] [Google Scholar]
- 2086.Liu Y, Yu H, Zhang X, et al. The protective role of autophagy in nephrotoxicity induced by bismuth nanoparticles through AMPK/mTOR pathway. Nanotoxicology. 2018. Aug;12(6):586–601. doi: 10.1080/17435390.2018.1466932. PubMed PMID: 29732938. [DOI] [PubMed] [Google Scholar]
- 2087.Tomic S, Janjetovic K, Mihajlovic D, et al. Graphene quantum dots suppress proinflammatory T cell responses via autophagy-dependent induction of tolerogenic dendritic cells. Biomaterials. 2017. Nov;146:13–28. doi: 10.1016/j.biomaterials.2017.08.040. PubMed PMID: 28892752. [DOI] [PubMed] [Google Scholar]
- 2088.Markovic ZM, Ristic BZ, Arsikin KM, et al. Graphene quantum dots as autophagy-inducing photodynamic agents. Biomaterials. 2012. Oct;33(29):7084–92. doi: 10.1016/j.biomaterials.2012.06.060. PubMed PMID: 22795854. [DOI] [PubMed] [Google Scholar]
- 2089.Moosavi MA, Sharifi M, Ghafary SM, et al. Photodynamic N-TiO2 nanoparticle treatment induces controlled ROS-mediated autophagy and terminal differentiation of leukemia cells. Sci Rep. 2016. Oct 4;6:34413. doi: 10.1038/srep34413. PubMed PMID: 27698385; PubMed Central PMCID: PMCPMC5048164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2090.Liu R, Colby AH, Gilmore D, et al. Nanoparticle tumor localization, disruption of autophagosomal trafficking, and prolonged drug delivery improve survival in peritoneal mesothelioma. Biomaterials. 2016. Sep;102:175–86. doi: 10.1016/j.biomaterials.2016.06.031. PubMed PMID: 27343465; PubMed Central PMCID: PMCPMC4948582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2091.Wu L, Zhang Y, Zhang C, et al. Tuning cell autophagy by diversifying carbon nanotube surface chemistry. ACS Nano. 2014. Mar 25;8(3):2087–99. doi: 10.1021/nn500376w. PubMed PMID: 24552177; PubMed Central PMCID: PMCPMC5586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2092.Zhang Y, Zheng F, Yang T, et al. Tuning the autophagy-inducing activity of lanthanide-based nanocrystals through specific surface-coating peptides. Nat Mater. 2012. Sep;11(9):817–26. doi: 10.1038/nmat3363. PubMed PMID: 22797828. [DOI] [PubMed] [Google Scholar]
- 2093.Seleverstov O, Zabirnyk O, Zscharnack M, et al. Quantum dots for human mesenchymal stem cells labeling. A size-dependent autophagy activation. Nano Lett. 2006. Dec;6(12):2826–32. PubMed PMID: 17163713; eng. [DOI] [PubMed] [Google Scholar]
- 2094.Zabirnyk O, Yezhelyev M, Seleverstov O.. Nanoparticles as a novel class of autophagy activators. Autophagy. 2007. May-Jun;3(3):278–81. doi: 10.4161/auto.3916. PubMed PMID: 17351332. [DOI] [PubMed] [Google Scholar]
- 2095.Cohignac V, Landry MJ, Ridoux A, et al. Carbon nanotubes, but not spherical nanoparticles, block autophagy by a shape-related targeting of lysosomes in murine macrophages. Autophagy. 2018;14(8):1323–1334. doi: 10.1080/15548627.2018.1474993. PubMed PMID: 29938576; PubMed Central PMCID: PMCPMC6103705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2096.Ma X, Wu Y, Jin S, et al. Gold nanoparticles induce autophagosome accumulation through size-dependent nanoparticle uptake and lysosome impairment. ACS Nano 2011. Nov 22;5(11):8629–39. doi: 10.1021/nn202155y. PubMed PMID: 21974862. [DOI] [PubMed] [Google Scholar]
- 2097.Zhang JQ, Zhou W, Zhu SS, et al. Persistency of enlarged autolysosomes underscores nanoparticle-induced autophagy in hepatocytes. Small. 2017. Feb;13(7). doi: 10.1002/smll.201602876. PubMed PMID: 27925395. [DOI] [PubMed] [Google Scholar]
- 2098.Wang J, Li Y, Duan J, et al. Silica nanoparticles induce autophagosome accumulation via activation of the EIF2AK3 and ATF6 UPR pathways in hepatocytes. Autophagy. 2018;14(7):1185–1200. doi: 10.1080/15548627.2018.1458174. PubMed PMID: 29940794; PubMed Central PMCID: PMCPMC6103719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2099.Liu HL, Zhang YL, Yang N, et al. A functionalized single-walled carbon nanotube-induced autophagic cell death in human lung cells through Akt-TSC2-mTOR signaling. Cell Death Dis. 2011. May 19;2:e159. doi: 10.1038/cddis.2011.27. PubMed PMID: 21593791; PubMed Central PMCID: PMCPMC3122114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2100.Chen GY, Meng CL, Lin KC, et al. Graphene oxide as a chemosensitizer: diverted autophagic flux, enhanced nuclear import, elevated necrosis and improved antitumor effects. Biomaterials. 2015. Feb;40:12–22. doi: 10.1016/j.biomaterials.2014.11.034. PubMed PMID: 25498801. [DOI] [PubMed] [Google Scholar]
- 2101.Ha SW, Weitzmann MN, Beck GR, Jr.. Bioactive silica nanoparticles promote osteoblast differentiation through stimulation of autophagy and direct association with LC3 and p62. ACS Nano. 2014. Jun 24;8(6):5898–910. doi: 10.1021/nn5009879. PubMed PMID: 24806912; PubMed Central PMCID: PMCPMC4076025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2102.Lin YX, Qiao SL, Wang Y, et al. An in situ intracellular self-assembly strategy for quantitatively and temporally monitoring autophagy. ACS Nano. 2017. Feb 28;11(2):1826–1839. doi: 10.1021/acsnano.6b07843. PubMed PMID: 28112893. [DOI] [PubMed] [Google Scholar]
- 2103.Lin YX, Wang Y, Qiao SL, et al.“In vivo self-assembled” nanoprobes for optimizing autophagy-mediated chemotherapy. Biomaterials. 2017. Oct;141:199–209. doi: 10.1016/j.biomaterials.2017.06.042. PubMed PMID: 28689116. [DOI] [PubMed] [Google Scholar]
- 2104.Choi KM, Nam HY, Na JH, et al. A monitoring method for Atg4 activation in living cells using peptide-conjugated polymeric nanoparticles. Autophagy. 2011. Sep;7(9):1052–62. doi: 10.4161/auto.7.9.16451. PubMed PMID: 21610316. [DOI] [PubMed] [Google Scholar]
- 2105.Neumann S, Pucadyil TJ, Schmid SL.. Analyzing membrane remodeling and fission using supported bilayers with excess membrane reservoir. Nat Protoc. 2013. Jan;8(1):213–22. doi: 10.1038/nprot.2012.152. PubMed PMID: 23288321; PubMed Central PMCID: PMCPMC4753780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2106.Hervas JH, Landajuela A, Anton Z, et al. Human ATG3 binding to lipid bilayers: role of lipid geometry, and electric charge. Sci Rep. 2017. Nov 15;7(1):15614. doi: 10.1038/s41598-017-15057-6. PubMed PMID: 29142222; PubMed Central PMCID: PMCPMC5688168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2107.King JS, Veltman DM, Insall RH.. The induction of autophagy by mechanical stress. Autophagy. 2011. Dec 1;7(12):1490–9. PubMed PMID: 22024750; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2108.Grodzki AC, Giulivi C, Lein PJ.. Oxygen tension modulates differentiation and primary macrophage functions in the human monocytic THP-1 cell line. PLoS One. 2013;8(1):e54926. doi: 10.1371/journal.pone.0054926. PubMed PMID: 23355903; PubMed Central PMCID: PMCPMC3552948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2109.Roberts R, Al-Jamal WT, Whelband M, et al. Autophagy and formation of tubulovesicular auto-phagosomes provide a barrier against nonviral gene delivery. Autophagy. 2013. May;9(5):667–82. doi: 10.4161/auto.23877. PubMed PMID: 23422759; PubMed Central PMCID: PMC3669178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2110.Kovacs AL, Zhang H.. Role of autophagy in Caenorhabditis elegans. FEBS Lett. 2010. Apr 2;584(7):1335–41. doi: 10.1016/j.febslet.2010.02.002. PubMed PMID: 20138173; eng. [DOI] [PubMed] [Google Scholar]
- 2111.Wu F, Li Y, Wang F, et al. Differential function of the two Atg4 homologues in the aggrephagy pathway in Caenorhabditis elegans. J Biol Chem. 2012. Aug 24;287(35):29457–67. doi: 10.1074/jbc.M112.365676. PubMed PMID: 22767594; PubMed Central PMCID: PMC3436130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2112.Morselli E, Maiuri MC, Markaki M, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. PubMed PMID: 21364612; PubMed Central PMCID: PMC3032517. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2113.Samara C, Syntichaki P, Tavernarakis N.. Autophagy is required for necrotic cell death in Caenorhabditis elegans. Cell Death Differ. 2008. Jan;15(1):105–12. doi: 10.1038/sj.cdd.4402231. PubMed PMID: 17901876; eng. [DOI] [PubMed] [Google Scholar]
- 2114.Alberti A, Michelet X, Djeddi A, et al. The autophagosomal protein LGG-2 acts synergistically with LGG-1 in dauer formation and longevity in C. elegans. Autophagy. 2010. Jul 5;6(5):622–633. PubMed PMID: 20523114; Eng. [DOI] [PubMed] [Google Scholar]
- 2115.Kang C, You YJ, Avery L.. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007. Sep 1;21(17):2161–71. PubMed PMID: 17785524; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2116.Evans TC. Transformation and microinjection. Pasadena, CA: WormBook; 2006. [Google Scholar]
- 2117.Chapin HC, Okada M, Merz AJ, et al. Tissue-specific autophagy responses to aging and stress in C. elegans. Aging (Albany NY). 2015. Jun;7(6):419–34. doi: 10.18632/aging.100765. PubMed PMID: 26142908; PubMed Central PMCID: PMCPMC4505168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2118.Springhorn A, Hoppe T.. Western blot analysis of the autophagosomal membrane protein LGG-1/LC3 in Caenorhabditis elegans. Methods Enzymol 2019;619:319–336. doi: 10.1016/bs.mie.2018.12.034. PubMed PMID: 30910027. [DOI] [PubMed] [Google Scholar]
- 2119.Tasdemir E, Maiuri MC, Galluzzi L, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008. Jun;10(6):676–87. doi: 10.1038/ncb1730. PubMed PMID: 18454141; PubMed Central PMCID: PMC2676564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2120.Tavernarakis N, Pasparaki A, Tasdemir E, et al. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy. 2008. Oct;4(7):870–3. PubMed PMID: 18728385; eng. [DOI] [PubMed] [Google Scholar]
- 2121.Schiavi A, Torgovnick A, Kell A, et al. Autophagy induction extends lifespan and reduces lipid content in response to frataxin silencing in C. elegans. Exp Gerontol. 2013. Feb;48(2):191–201. doi: 10.1016/j.exger.2012.12.002. PubMed PMID: 23247094; PubMed Central PMCID: PMC3572394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2122.Palikaras K, Lionaki E, Tavernarakis N.. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015. May 28;521(7553):525–8. doi: 10.1038/nature14300. PubMed PMID: 25896323. [DOI] [PubMed] [Google Scholar]
- 2123.Fostel JL, Benner Coste L, Jacobson LA.. Degradation of transgene-coded and endogenous proteins in the muscles of Caenorhabditis elegans. Biochem Biophys Res Commun. 2003. Dec 5;312(1):173–7. doi: 10.1016/j.bbrc.2003.09.248. PubMed PMID: 14630038. [DOI] [PubMed] [Google Scholar]
- 2124.Lehmann S, Shephard F, Jacobson LA, et al. Integrated control of protein degradation in C. elegans muscle. Worm. 2012. Jul 1;1(3):141–50. doi: 10.4161/worm.20465. PubMed PMID: 23457662; PubMed Central PMCID: PMCPMC3583358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2125.Etheridge T, Oczypok EA, Lehmann S, et al. Calpains mediate integrin attachment complex maintenance of adult muscle in Caenorhabditis elegans. PLoS Genet. 2012. Jan;8(1):e1002471. doi: 10.1371/journal.pgen.1002471. PubMed PMID: 22253611; PubMed Central PMCID: PMCPMC3257289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2126.Szewczyk NJ, Hartman JJ, Barmada SJ, et al. Genetic defects in acetylcholine signalling promote protein degradation in muscle cells of Caenorhabditis elegans. J Cell Sci. 2000. Jun;113 (Pt 11):2003–10. PubMed PMID: 10806111. [DOI] [PubMed] [Google Scholar]
- 2127.Gaffney CJ, Shephard F, Chu J, et al. Degenerin channel activation causes caspase-mediated protein degradation and mitochondrial dysfunction in adult C. elegans muscle. J Cachexia Sarcopenia Muscle. 2016. May;7(2):181–92. doi: 10.1002/jcsm.12040. PubMe;d PMID: 27493871; PubMed Central PMCID: PMCPMC4864282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2128.Lehmann S, Bass JJ, Szewczyk NJ.. Knockdown of the C. elegans kinome identifies kinases required for normal protein homeostasis, mitochondrial network structure, and sarcomere structure in muscle. Cell Commun Signal. 2013. Sep 23;11:71. doi: 10.1186/1478-811X-11-71. PubMed PMID: 24060339; PubMed Central PMCID: PMCPMC3849176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2129.Lehmann S, Bass JJ, Barratt TF, et al. Functional phosphatome requirement for protein homeostasis, networked mitochondria, and sarcomere structure in C. elegans muscle. J Cachexia Sarcopenia Muscle. 2017. Aug;8(4):660–672. doi: 10.1002/jcsm.12196. PubMed PMID: 28508547; PubMed Central PMCID: PMCPMC5566650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2130.Chang JT, Kumsta C, Hellman AB, et al. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. eLife. 2017. Jul 4;6. doi: 10.7554/eLife.18459. PubMed PMID: 28675140; PubMed Central PMCID: PMCPMC5496740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2131.Brown WR, Hubbard SJ, Tickle C, et al. The chicken as a model for large-scale analysis of vertebrate gene function [Research Support, Non-U.S. Gov’t Review]. Nat Rev Genet. 2003. Feb;4(2):87–98. doi: 10.1038/nrg998. PubMed PMID: 12560806; eng. [DOI] [PubMed] [Google Scholar]
- 2132.Mellen MA, de la Rosa EJ, Boya P.. The autophagic machinery is necessary for removal of cell corpses from the developing retinal neuroepithelium. Cell Death Differ. 2008. Aug;15(8):1279–90. doi: 10.1038/cdd.2008.40. PubMed PMID: 18369370; eng. [DOI] [PubMed] [Google Scholar]
- 2133.Aburto MR, Sanchez-Calderon H, Hurle JM, et al. Early otic development depends on autophagy for apoptotic cell clearance and neural differentiation. Cell Death Dis. 2012;3:e394. doi: 10.1038/cddis.2012.132. PubMed PMID: 23034329; PubMed Central PMCID: PMC3481121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2134.Wang L, Rodrigues NA, Wu Y, et al. Pleiotropic action of AP-1 in v-Src-transformed cells. J Virol. 2011. Jul;85(13):6725–35. doi: 10.1128/JVI.01013-10. PubMed PMID: 21507983; PubMed Central PMCID: PMC3126506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2135.Baba TW, Giroir BP, Humphries EH.. Cell lines derived from avian lymphomas exhibit two distinct phenotypes. Virology. 1985. Jul 15;144(1):139–51. PubMed PMID: 2998040; eng. [DOI] [PubMed] [Google Scholar]
- 2136.Perez-Martin M, Perez-Perez ME, Lemaire SD, et al. Oxidative stress contributes to autophagy induction in response to endoplasmic reticulum stress in chlamydomonas reinhardtii. Plant Physiol. 2014. Oct;166(2):997–1008. doi: 10.1104/pp.114.243659. PubMed PMID: 25143584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2137.Perez-Perez ME, Couso I, Crespo JL.. Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy. 2012. Mar;8(3):376–88. doi: 10.4161/auto.18864. PubMed PMID: 22302003. [DOI] [PubMed] [Google Scholar]
- 2138.Tran QG, Yoon HR, Cho K, et al. Dynamic interactions between auto-phagosomes and lipid droplets in chlamydomonas reinhardtii. Cells. 2019. Aug 28;8(9). doi: 10.3390/cells8090992. PubMed PMID: 31466295; PubMed Central PMCID: PMCPMC6769876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2139.Couso I, Perez-Perez ME, Martinez-Force E, et al. Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J Exp Bot. 2018. Mar 14;69(6):1355–1367. doi: 10.1093/jxb/erx372. PubMed PMID: 29053817; PubMed Central PMCID: PMCPMC6018900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2140.Heredia-Martinez LG, Andres-Garrido A, Martinez-Force E, et al. Chloroplast Damage Induced by the Inhibition of Fatty Acid Synthesis Triggers Autophagy in Chlamydomonas. Plant Physiol. 2018. Nov;178(3):1112–1129. doi: 10.1104/pp.18.00630. PubMed PMID: 30181343; PubMed Central PMCID: PMCPMC6236622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2141.Xu T, Kumar S, Denton D.. Characterization of autophagic responses in drosophila melanogaster. Methods Enzymol. 2017;588:445–465. doi: 10.1016/bs.mie.2016.09.089. PubMed PMID: 28237115. [DOI] [PubMed] [Google Scholar]
- 2142.Mulakkal NC, Nagy P, Takats S, et al. Autophagy in Drosophila: from historical studies to current knowledge. Biomed Res Int. 2014;2014:273473. doi: 10.1155/2014/273473. PubMed PMID: 24949430; PubMed Central PMCID: PMCPMC4052151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2143.Lorincz P, Mauvezin C, Juhasz G.. Exploring Autophagy in Drosophila. Cells. 2017. Jul 12;6(3). doi: 10.3390/cells6030022. PubMed PMID: 28704946; PubMed Central PMCID: PMCPMC5617968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2144.Zhang P, Holowatyj AN, Roy T, et al. An SH3PX1-dependent endocytosis-autophagy network restrains intestinal stem cell proliferation by counteracting EGFR-ERK signaling. Dev Cell. 2019. May 20;49(4):574–589 e5. doi: 10.1016/j.devcel.2019.03.029. PubMed PMID: 31006650; PubMed Central PMCID: PMCPMC6542281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2145.Nelson C, Baehrecke EH.. Autophagy and cell death in the fly. Methods Enzymol. 2014;545:181–99. doi: 10.1016/B978-0-12-801430-1.00008-1. PubMed PMID: 25065891. [DOI] [PubMed] [Google Scholar]
- 2146.Mauvezin C, Ayala C, Braden CR, et al. Assays to monitor autophagy in Drosophila. Methods. 2014. Jun 15;68(1):134–9. doi: 10.1016/j.ymeth.2014.03.014. PubMed PMID: 24667416; PubMed Central PMCID: PMC4048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2147.Neisch AL, Neufeld TP, Hays TS.. A STRIPAK complex mediates axonal transport of auto-phagosomes and dense core vesicles through PP2A regulation. J Cell Biol. 2017. Feb;216(2):441–461. doi: 10.1083/jcb.201606082. PubMed PMID: 28100687; PubMed Central PMCID: PMCPMC5294782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2148.Denton D, Chang TK, Nicolson S, et al. Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ. 2012. Aug;19(8):1299–307. doi: 10.1038/cdd.2012.43. PubMed PMID: 22555456; PubMed Central PMCID: PMCPMC3392632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2149.Shelly S, Lukinova N, Bambina S, et al. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus [Research Support, N.I.H., Extramural]. Immunity. 2009. Apr 17;30(4):588–98. doi: 10.1016/j.immuni.2009.02.009. PubMed PMID: 19362021; PubMed Central PMCID: PMC2754303. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2150.Juhasz G, Hill JH, Yan Y, et al. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008. May 19;181(4):655–66. doi: 10.1083/jcb.200712051. PubMed PMID: 18474623; PubMed Central PMCID: PMC2386105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2151.Nagy P, Hegedus K, Pircs K, et al. Different effects of Atg2 and Atg18 mutations on Atg8a and Atg9 trafficking during starvation in Drosophila. FEBS Lett. 2014. Jan 31;588(3):408–13. doi: 10.1016/j.febslet.2013.12.012. PubMed PMID: 24374083; PubMed Central PMCID: PMCPMC3928829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2152.Shravage BV, Hill JH, Powers CM, et al. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013. Mar;140(6):1321–9. doi: 10.1242/dev.089490. PubMed PMID: 23406899; PubMed Central PMCID: PMC3585664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2153.Melani M, Valko A, Romero NM, et al. Zonda is a novel early component of the autophagy pathway in Drosophila. Mol Biol Cell. 2017. Nov 1;28(22):3070–3081. doi: 10.1091/mbc.E16-11-0767. PubMed PMID: 28904211; PubMed Central PMCID: PMCPMC5662263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2154.Nezis IP, Shravage BV, Sagona AP, et al. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. J Cell Biol. 2010. Aug 23;190(4):523–31. doi: 10.1083/jcb.201002035. PubMed PMID: 20713604; PubMed Central PMCID: PMC2928014. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2155.Robin M, Issa AR, Santos CC, et al. Drosophila p53 integrates the antagonism between autophagy and apoptosis in response to stress. Autophagy. 2019. May;15(5):771–784. doi: 10.1080/15548627.2018.1558001. PubMed PMID: 30563404; PubMed Central PMCID: PMCPMC6526837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2156.Kim M, Semple I, Kim B, et al. Drosophila Gyf/GRB10 interacting GYF protein is an autophagy regulator that controls neuron and muscle homeostasis. Autophagy. 2015. Aug 3;11(8):1358–72. doi: 10.1080/15548627.2015.1063766. PubMed PMID: 26086452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2157.Pircs K, Nagy P, Varga A, et al. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One. 2012;7(8):e44214. doi: 10.1371/journal.pone.0044214. PubMed PMID: 22952930; PubMed Central PMCID: PMC3432079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2158.Wyers F, Dru P, Simonet B, et al. Immunological cross-reactions and interactions between the Drosophila melanogaster ref(2)P protein and sigma rhabdovirus proteins. J Virol. 1993. Jun; 67(6):3208–16. PubMed PMID: 7684462; PubMed Central PMCID: PMCPMC237660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2159.Hindle S, Afsari F, Stark M, et al. Dopaminergic expression of the Parkinsonian gene LRRK2-G2019S leads to non-autonomous visual neurodegeneration, accelerated by increased neural demands for energy. Hum Mol Genet. 2013. Jun 1; 22(11):2129–40. doi: 10.1093/hmg/ddt061. PubMed PMID: 23396536; PubMed Central PMCID: PMC3652415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2160.Anding AL, Baehrecke EH.. Vps15 is required for stress induced and developmentally triggered autophagy and salivary gland protein secretion in Drosophila. Cell Death Differ. 2014. Oct 24. doi: 10.1038/cdd.2014.174. PubMed PMID: 25342466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2161.Hou YC, Chittaranjan S, Barbosa SG, et al. Effector caspase Dcp-1 and IAP protein Bruce regulate starvation-induced autophagy during Drosophila melanogaster oogenesis [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.]. J Cell Biol. 2008. Sep 22;182(6):1127–39. doi: 10.1083/jcb.200712091. PubMed PMID: 18794330; PubMed Central PMCID: PMC2542474. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2162.Tusco R, Jacomin AC, Jain A, et al. Kenny mediates selective autophagic degradation of the IKK complex to control innate immune responses. Nat Commun. 2017. Nov 2;8(1):1264. doi: 10.1038/s41467-017-01287-9. PubMed PMID: 29097655; PubMed Central PMCID: PMCPMC5668318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2163.Marinkovic D, Zhang X, Yalcin S, et al. Foxo3 is required for the regulation of oxidative stress in erythropoiesis. J Clin Invest. 2007. Aug;117(8):2133–44. doi: 10.1172/JCI31807. PubMed PMID: 17671650; PubMed Central PMCID: PMC1934587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2164.McIver SC, Kang YA, DeVilbiss AW, et al. The exosome complex establishes a barricade to erythroid maturation. Blood. 2014. Oct 2;124(14):2285–97. doi: 10.1182/blood-2014-04-571083. PubMed PMID: 25115889; PubMed Central PMCID: PMC4183988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2165.Fujiwara T, O’Geen H, Keles S, et al. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009. Nov 25;36(4):667–81. doi: 10.1016/j.molcel.2009.11.001. PubMed PMID: 19941826; PubMed Central PMCID: PMC2784893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2166.Welch JJ, Watts JA, Vakoc CR, et al. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 2004. Nov 15;104(10):3136–47. doi: 10.1182/blood-2004-04-1603. PubMed PMID: 15297311. [DOI] [PubMed] [Google Scholar]
- 2167.Yu M, Riva L, Xie H, et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009. Nov 25;36(4):682–95. doi: 10.1016/j.molcel.2009.11.002. PubMed PMID: 19941827; PubMed Central PMCID: PMC2800995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2168.Kundu M, Lindsten T, Yang CY, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008. Aug 15; 112(4):1493–502. doi: 10.1182/blood-2008-02-137398. PubMed PMID: 18539900; PubMed Central PMCID: PMC2515143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2169.Mortensen M, Ferguson DJ, Edelmann M, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci U S A. 2010. Jan 12; 107(2):832–7. doi: 10.1073/pnas.0913170107. PubMed PMID: 20080761; PubMed Central PMCID: PMC2818953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2170.Sandoval H, Thiagarajan P, Dasgupta SK, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008. Jul 10;454(7201):232–5. doi: 10.1038/nature07006. PubMed PMID: 18454133; PubMed Central PMCID: PMC2570948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2171.Schweers RL, Zhang J, Randall MS, et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci U S A. 2007. Dec 4;104(49):19500–5. doi: 0708818104 [pii] doi: 10.1073/pnas.0708818104. PubMed PMID: 18048346; PubMed Central PMCID: PMC2148318. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2172.Zhang X, Camprecios G, Rimmele P, et al. FOXO3-mTOR metabolic cooperation in the regulation of erythroid cell maturation and homeostasis. Am J Hematol. 2014. Oct;89(10):954–63. doi: 10.1002/ajh.23786. PubMed PMID: 24966026; PubMed Central PMCID: PMCPMC4201594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2173.Lithanatudom P, Wannatung T, Leecharoenkiat A, et al. Enhanced activation of autophagy in beta-thalassemia/Hb E erythroblasts during erythropoiesis. Ann Hematol. 2011. Jul;90(7):747–58. doi: 10.1007/s00277-010-1152-5. PubMed PMID: 21221583. [DOI] [PubMed] [Google Scholar]
- 2174.Lupo F, Tibaldi E, Matte A, et al. A new molecular link between defective autophagy and erythroid abnormalities in chorea-acanthocytosis. Blood. 2016. Dec 22;128(25):2976–2987. doi: 10.1182/blood-2016-07-727321. PubMed PMID: 27742708; PubMed Central PMCID: PMCPMC5179337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2175.Beneduce E, Matte A, De Falco L, et al. Fyn kinase is a novel modulator of erythropoietin signaling and stress erythropoiesis. Am J Hematol. 2019. Jan;94(1):10–20. doi: 10.1002/ajh.25295. PubMed PMID: 30252956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2176.Josefsen L, Droce A, Sondergaard TE, et al. Autophagy provides nutrients for nonassimilating fungal structures and is necessary for plant colonization but not for infection in the necrotrophic plant pathogen Fusarium gaminearum. Autophagy. 2012;8:326–37. [DOI] [PubMed] [Google Scholar]
- 2177.Nadal M, Gold SE.. The autophagy genes ATG8 and ATG1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol Plant Pathol. 2010. Jul;11(4):463–78. doi: 10.1111/j.1364-3703.2010.00620.x. PubMed PMID: 20618705; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2178.Pollack JK, Harris SD, Marten MR.. Autophagy in filamentous fungi [Research Support, U.S. Gov’t, Non-P.H.S Review]. Fungal Genet biol FG & B. 2009. Jan;46(1):1–8. doi: 10.1016/j.fgb.2008.10.010. PubMed PMID: 19010432; eng. [DOI] [PubMed] [Google Scholar]
- 2179.Richie DL, Fuller KK, Fortwendel J, et al. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus [Research Support, N.I.H., Extramural]. Eukaryot Cell. 2007. Dec;6(12):2437–47. doi: 10.1128/EC.00224-07. PubMed PMID: 17921348; PubMed Central PMCID: PMC2168250. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2180.Voigt O, Poggeler S.. Self-eating to grow and kill: autophagy in filamentous ascomycetes. Appl Microbiol Biotechnol. 2013. Nov;97(21):9277–90. doi: 10.1007/s00253-013-5221-2. PubMed PMID: 24077722. [DOI] [PubMed] [Google Scholar]
- 2181.Kim Y, Islam N, Moss BJ, et al. Autophagy induced by rapamycin and carbon-starvation have distinct proteome profiles in Aspergillus nidulans. Biotechnol Bioeng. 2011. Nov;108(11):2705–15. doi: 10.1002/bit.23223. PubMed PMID: 21618477. [DOI] [PubMed] [Google Scholar]
- 2182.Ren W, Zhang Z, Shao W, et al. The autophagy-related gene BcATG1 is involved in fungal development and pathogenesis in Botrytis cinerea. Mol Plant Pathol. 2017. Feb;18(2):238–248. doi: 10.1111/mpp.12396. PubMed PMID: 26972592; PubMed Central PMCID: PMCPMC6638273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2183.Pinan-Lucarre B, Balguerie A, Clave C.. Accelerated cell death in Podospora autophagy mutants [Research Support, Non-U.S. Gov’t]. Eukaryot Cell. 2005. Nov;4(11):1765–74. doi: 10.1128/EC.4.11.1765-1774.2005. PubMed PMID: 16278443; PubMed Central PMCID: PMC1287858. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2184.Deng YZ, Naqvi NI.. A vacuolar glucoamylase, Sga1, participates in glycogen autophagy for proper asexual differentiation in Magnaporthe oryzae. Autophagy. 2010. May;6(4):455–61. doi: 10.4161/auto.6.4.11736. PubMed PMID: 20383057. [DOI] [PubMed] [Google Scholar]
- 2185.Deng YZ, Ramos-Pamplona M, Naqvi NI.. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy. 2009. Jan;5(1):33–43. PubMed PMID: 19115483. [DOI] [PubMed] [Google Scholar]
- 2186.Ren W, Liu N, Sang C, et al. The Autophagy Gene BcATG8 Regulates the Vegetative Differentiation and Pathogenicity of Botrytis cinerea. Appl Environ Microbiol. 2018. Jun 1;84(11). doi: 10.1128/AEM.02455-17. PubMed PMID: 29572212; PubMed Central PMCID: PMCPMC5960959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2187.Knuppertz L, Hamann A, Pampaloni F, et al. Identification of autophagy as a longevity-assurance mechanism in the aging model Podospora anserina. Autophagy. 2014. May;10(5):822–34. doi: 10.4161/auto.28148. PubMed PMID: 24584154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2188.Asakura M, Ninomiya S, Sugimoto M, et al. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare [Research Support, Non-U.S. Gov’t]. Plant Cell. 2009. Apr;21(4):1291–304. doi: 10.1105/tpc.108.060996. PubMed PMID: 19363139; PubMed Central PMCID: PMC2685618. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2189.Liu XH, Lu JP, Zhang L, et al. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis [Research Support, Non-U.S. Gov’t]. Eukaryot Cell. 2007. Jun;6(6):997–1005. doi: 10.1128/EC.00011-07. PubMed PMID: 17416896; PubMed Central PMCID: PMC1951528. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2190.Nguyen LN, Bormann J, Le GT, et al. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection [Research Support, Non-U.S. Gov’t]. Fungal Genet biol FG & B. 2011. Mar;48(3):217–24. doi: 10.1016/j.fgb.2010.11.004. PubMed PMID: 21094265; eng. [DOI] [PubMed] [Google Scholar]
- 2191.Duan Z, Chen Y, Huang W, et al. Linkage of autophagy to fungal development, lipid storage and virulence in Metarhizium robertsii. Autophagy. 2013. Apr;9(4):538–49. doi: 10.4161/auto.23575. PubMed PMID: 23380892; PubMed Central PMCID: PMC3627669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2192.Sun G, Qi X, Wilson RA.. A Feed-Forward Subnetwork Emerging from Integrated TOR- and cAMP/PKA-Signaling Architecture Reinforces Magnaporthe oryzae Appressorium Morphogenesis. Mol Plant Microbe Interact. 2019. May;32(5):593–607. doi: 10.1094/MPMI-10-18-0287-R. PubMed PMID: 30431400. [DOI] [PubMed] [Google Scholar]
- 2193.Sun G, Elowsky C, Li G, et al. TOR-autophagy branch signaling via Imp1 dictates plant-microbe biotrophic interface longevity. PLoS Genet. 2018. Nov;14(11):e1007814. doi: 10.1371/journal.pgen.1007814. PubMed PMID: 30462633; PubMed Central PMCID: PMCPMC6281275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2194.Yin Z, Chen C, Yang J, et al. Histone acetyltransferase MoHat1 acetylates autophagy-related proteins MoAtg3 and MoAtg9 to orchestrate functional appressorium formation and pathogenicity in Magnaporthe oryzae. Autophagy. 2019. Jul;15(7):1234–1257. doi: 10.1080/15548627.2019.1580104. PubMed PMID: 30776962; PubMed Central PMCID: PMCPMC6613890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2195.Yin Z, Feng W, Chen C, et al. Shedding light on autophagy coordinating with cell wall integrity signaling to govern pathogenicity of Magnaporthe oryzae. Autophagy. 2019. Jul 24:1–17. doi: 10.1080/15548627.2019.1644075. PubMed PMID: 31313634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2196.Deng YZ, Ramos-Pamplona M, Naqvi NI.. Methods for functional analysis of macroautophagy in filamentous fungi [Research Support, Non-U.S. Gov’t]. Methods Enzymol. 2008;451:295–310. doi: 10.1016/S0076-6879(08)03220-5. PubMed PMID: 19185728; eng. [DOI] [PubMed] [Google Scholar]
- 2197.Kershaw MJ, Talbot NJ.. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease.Proc Natl Acad Sci U S A. 2009. Sep 15;106(37):15967–72. doi: 10.1073/pnas.0901477106. PubMed PMID: 19717456; PubMed Central PMCID: PMC2747227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2198.Liu TB, Liu XH, Lu JP, et al. The cysteine protease MoAtg4 interacts with MoAtg8 and is required for differentiation and pathogenesis in Magnaporthe oryzae. Autophagy. 2010. Jan;6(1):74–85. PubMed PMID: 19923912. [DOI] [PubMed] [Google Scholar]
- 2199.Peñalva MA, Galindo A, Abenza JF, et al. Searching for gold beyond mitosis: Mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist. 2012. Jan 1;2(1):2–14. doi: 10.4161/cl.19304. PubMed PMID: 22645705; PubMed Central PMCID: PMC3355971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2200.Pinar M, Peñalva MA.. Aspergillus nidulans BapH is a RAB11 effector that connects membranes in the Spitzenkorper with basal autophagy. Mol Microbiol. 2017. Nov;106(3):452–468. doi: 10.1111/mmi.13777. PubMed PMID: 28857357. [DOI] [PubMed] [Google Scholar]
- 2201.Lipatova Z, Belogortseva N, Zhang XQ, et al. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A. 2012. May 1;109(18):6981–6. doi: 10.1073/pnas.1121299109. PubMed PMID: 22509044; PubMed Central PMCID: PMC3344974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2202.Lynch-Day MA, Bhandari D, Menon S, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010. Apr 27;107(17):7811–6. doi: 1000063107 [pii] doi: 10.1073/pnas.1000063107. PubMed PMID: 20375281; PubMed Central PMCID: PMC2867920. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2203.Deng Y, Qu Z, Naqvi NI.. The role of snx41-based pexophagy in magnaporthe development. PLoS One. 2013;8(11):e79128. doi: 10.1371/journal.pone.0079128. PubMed PMID: 24302988; PubMed Central PMCID: PMC3841179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2204.Zhong K, Li X, Le X, et al. MoDnm1 dynamin mediating peroxisomal and mitochondrial fission in complex with MoFis1 and MoMdv1 is important for development of functional appressorium in Magnaporthe oryzae. PLoS Pathog. 2016. Aug;12(8):e1005823. doi: 10.1371/journal.ppat.1005823. PubMed PMID: 27556292; PubMed Central PMCID: PMCPMC4996533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2205.Zhu XM, Liang S, Shi HB, et al. VPS9 domain-containing proteins are essential for autophagy and endocytosis in Pyricularia oryzae. Environ Microbiol. 2018. Apr;20(4):1516–1530. doi: 10.1111/1462-2920.14076. PubMed PMID: 29468804. [DOI] [PubMed] [Google Scholar]
- 2206.Li X, Gao C, Li L, et al. MoEnd3 regulates appressorium formation and virulence through mediating endocytosis in rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2017. Jun;13(6):e1006449. doi: 10.1371/journal.ppat.1006449. PubMed PMID: 28628655; PubMed Central PMCID: PMCPMC5491321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2207.Piggott N, Cook MA, Tyers M, et al. Genome-wide fitness profiles reveal a requirement for autophagy during yeast fermentation. Genes Genomes Genetics. 2011;1:353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2208.Cebollero E, Gonzalez R.. Induction of autophagy by second-fermentation yeasts during elaboration of sparkling wines. Appl Environ Microbiol. 2006. Jun;72(6):4121–7. doi: 10.1128/AEM.02920-05. PubMed PMID: 16751523; PubMed Central PMCID: PMC1489611. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2209.Marks VD, Ho Sui SJ, Erasmus D, et al. Dynamics of the yeast transcriptome during wine fermentation reveals a novel fermentation stress response [Research Support, Non-U.S. Gov’t]. FEMS Yeast Res. 2008. Feb;8(1):35–52. doi: 10.1111/j.1567-1364.2007.00338.x. PubMed PMID: 18215224; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2210.Mendes-Ferreira A, Sampaio-Marques B, Barbosa C, et al. Accumulation of non-superoxide anion reactive oxygen species mediates nitrogen-limited alcoholic fermentation by Saccharomyces cerevisiae. Appl Environ Microbiol. 2010. Dec;76(24):7918–24. doi: 10.1128/AEM.01535-10. PubMed PMID: 20952643; PubMed Central PMCID: PMC3008223. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2211.Rossignol T, Dulau L, Julien A, et al. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast. 2003. Dec;20(16):1369–85. doi: 10.1002/yea.1046. PubMed PMID: 14663829; eng. [DOI] [PubMed] [Google Scholar]
- 2212.Teixeira MC, Raposo LR, Mira NP, et al. Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol [Research Support, Non-U.S. Gov’t]. Appl Environ Microbiol. 2009. Sep;75(18):5761–72. doi: 10.1128/AEM.00845-09. PubMed PMID: 19633105; PubMed Central PMCID: PMC2747848. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2213.Yoshikawa K, Tanaka T, Furusawa C, et al. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 2009. Feb;9(1):32–44. doi: 10.1111/j.1567-1364.2008.00456.x. PubMed PMID: 19054128; eng. [DOI] [PubMed] [Google Scholar]
- 2214.Hazan R, Levine A, Abeliovich H.. Benzoic acid, a weak organic acid food preservative, exerts specific effects on intracellular membrane trafficking pathways in Saccharomyces cerevisiae. Appl Environ Microbiol. 2004. Aug;70(8):4449–57. doi: 10.1128/AEM.70.8.4449-4457.2004. PubMed PMID: 15294772; PubMed Central PMCID: PMC492424. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2215.Singletary K, Milner J.. Diet, autophagy, and cancer: a review [Review]. Cancer Epidemiol Biomarkers Prev. 2008. Jul;17(7):1596–610. doi: 10.1158/1055-9965.EPI-07-2917. PubMed PMID: 18628411; eng. [DOI] [PubMed] [Google Scholar]
- 2216.Su CL, Chen FN, Won SJ.. Involvement of apoptosis and autophagy in reducing mouse hepatoma ML-1 cell growth in inbred BALB/c mice by bacterial fermented soybean products. Food Chem Toxicol. 2011. Jan;49(1):17–24. doi: 10.1016/j.fct.2010.08.017. PubMed PMID: 20732379; eng. [DOI] [PubMed] [Google Scholar]
- 2217.Abeliovich H, Gonzalez R.. Autophagy in food biotechnology. Autophagy. 2009. Oct;5(7):925–9. PubMed PMID: 19556866; eng. [DOI] [PubMed] [Google Scholar]
- 2218.Berger B, Abdalla FC, Cruz-Landim C.. Effect of narcosis with CO2 on the ovarian development in queens of Apis mellifera (Hymenoptera, Apini). Sociobiology. 2005;45:261–70. [Google Scholar]
- 2219.ECM Silva-Zacarin, Tomaino GA, Brocheto-Braga MR, et al. Programmed cell death in the larval salivary glands of Apis mellifera (Hymenoptera, Apidae). J Biosci. 2007. Mar;32(2):309–28. PubMed PMID: 17435323; eng. [DOI] [PubMed] [Google Scholar]
- 2220.Gregorc A, Bowen ID.. Programmed cell death in the honey-bee (Apis mellifera L.) larvae midgut. Cell Biol Int. 1997. Mar;21(3):151–8. doi: 10.1006/cbir.1997.0127. PubMed PMID: 9151991; eng. [DOI] [PubMed] [Google Scholar]
- 2221.Navajas M, Migeon A, Alaux C, et al. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC genomics. 2008;9:301. doi: 10.1186/1471-2164-9-301. PubMed PMID: 18578863; PubMed Central PMCID: PMC2447852. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2222.Kimura T, Takabatake Y, Takahashi A, et al. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res. 2013. Jan 1;73(1):3–7. doi: 10.1158/0008-5472.CAN-12-2464. PubMed PMID: 23288916. [DOI] [PubMed] [Google Scholar]
- 2223.Takahashi A, Kimura T, Takabatake Y, et al. Autophagy guards against cisplatin-induced acute kidney injury. Am J Pathol. 2012. Feb;180(2):517–25. doi: 10.1016/j.ajpath.2011.11.001. PubMed PMID: 22265049. [DOI] [PubMed] [Google Scholar]
- 2224.Rodriguez A, Gomez-Ambrosi J, Catalan V, et al. The ghrelin O-acyltransferase-ghrelin system reduces TNF-alpha-induced apoptosis and autophagy in human visceral adipocytes. Diabetologia. 2012. Nov;55(11):3038–50. doi: 10.1007/s00125-012-2671-5. PubMed PMID: 22869322. [DOI] [PubMed] [Google Scholar]
- 2225.Ramachandran N, Munteanu I, Wang P, et al. VMA21 deficiency prevents vacuolar ATPase assembly and causes autophagic vacuolar myopathy. Acta Neuropathol. 2013. Mar;125(3):439–57. doi: 10.1007/s00401-012-1073-6. PubMed PMID: 23315026. [DOI] [PubMed] [Google Scholar]
- 2226.Colasanti T, Vomero M, Alessandri C, et al. Role of alpha-synuclein in autophagy modulation of primary human T lymphocytes. Cell Death Dis. 2014;5:e1265. doi: 10.1038/cddis.2014.211. PubMed PMID: 24874737; PubMed Central PMCID: PMC4047919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2227.Spruessel A, Steimann G, Jung M, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques. 2004. Jun;36(6):1030–7. PubMed PMID: 15211754; eng. [DOI] [PubMed] [Google Scholar]
- 2228.Espina V, Edmiston KH, Heiby M, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics. 2008. Oct;7(10):1998–2018. doi: 10.1074/mcp.M700596-MCP200. PubMed PMID: 18667411; PubMed Central PMCID: PMC2559936. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2229.Barth S, Glick D, Macleod KF.. Autophagy: assays and artifacts. J Pathol. 2010. Jun;221(2):117–24. doi: 10.1002/path.2694. PubMed PMID: 20225337; PubMed Central PMCID: PMC2989884. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2230.Domart MC, Esposti DD, Sebagh M, et al. Concurrent induction of necrosis, apoptosis, and autophagy in ischemic preconditioned human livers formerly treated by chemotherapy. J Hepatol. 2009. Nov;51(5):881–9. doi: 10.1016/j.jhep.2009.06.028. PubMed PMID: 19765849. [DOI] [PubMed] [Google Scholar]
- 2231.Vanhorebeek I, Gunst J, Derde S, et al. Insufficient activation of autophagy allows cellular damage to accumulate in critically ill patients. J Clin Endocrinol Metab. 2011. Apr;96(4):E633–45. doi: 10.1210/jc.2010-2563. PubMed PMID: 21270330; eng. [DOI] [PubMed] [Google Scholar]
- 2232.Jahania SM, Sengstock D, Vaitkevicius P, et al. Activation of the homeostatic intracellular repair response during cardiac surgery. J Am Coll Surg. 2013. Apr;216(4):719–26; discussion 726–9. doi: 10.1016/j.jamcollsurg.2012.12.034. PubMed PMID: 23415552; PubMed Central PMCID: PMC3724756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2233.Singh KK, Yanagawa B, Quan A, et al. Autophagy gene fingerprint in human ischemia and reperfusion. J Thorac Cardiovasc Surg. 2014. Mar;147(3):1065–1072 e1. doi: 10.1016/j.jtcvs.2013.04.042. PubMed PMID: 23778083. [DOI] [PubMed] [Google Scholar]
- 2234.Gao F, Li G, Liu C, et al. Autophagy regulates testosterone synthesis by facilitating cholesterol uptake in Leydig cells. J Cell Biol. 2018. Jun 4;217(6):2103–2119. doi: 10.1083/jcb.201710078. PubMed PMID: 29618492; PubMed Central PMCID: PMCPMC5987723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2235.Nyman E, Brannmark C, Palmer R, et al. A hierarchical whole-body modeling approach elucidates the link between in vitro insulin signaling and in vivo glucose homeostasis. J Biol Chem. 2011. Jul 22;286(29):26028–41. doi: 10.1074/jbc.M110.188987. PubMed PMID: 21572040; PubMed Central PMCID: PMC3138269. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2236.Faraj J, Bodas M, Pehote G, et al. Novel cystamine-core dendrimer-formulation rescues DeltaF508-CFTR and inhibits Pseudomonas aeruginosa infection by augmenting autophagy. Expert Opin Drug Deliv. 2019. Feb;16(2):177–186. doi: 10.1080/17425247.2019.1575807. PubMed PMID: 30732491. [DOI] [PubMed] [Google Scholar]
- 2237.Shrestha CL, Assani KD, Rinehardt H, et al. Cysteamine-mediated clearance of antibiotic-resistant pathogens in human cystic fibrosis macrophages. PLoS One. 2017;12(10):e0186169. doi: 10.1371/journal.pone.0186169. PubMed PMID: 28982193; PubMed Central PMCID: PMCPMC5642023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2238.Brockman SM, Bodas M, Silverberg D, et al. Dendrimer-based selective autophagy-induction rescues DeltaF508-CFTR and inhibits Pseudomonas aeruginosa infection in cystic fibrosis. PLoS One. 2017;12(9):e0184793. doi: 10.1371/journal.pone.0184793. PubMed PMID: 28902888; PubMed Central PMCID: PMCPMC5597233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2239.Krause K, Kopp BT, Tazi MF, et al. The expression of Mirc1/Mir17-92 cluster in sputum samples correlates with pulmonary exacerbations in cystic fibrosis patients. J Cyst Fibros. 2018. Jul;17(4):454–461. doi: 10.1016/j.jcf.2017.11.005. PubMed PMID: 29241629; PubMed Central PMCID: PMCPMC5995663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2240.Assani K, Shrestha CL, Rinehardt H, et al. AR-13 reduces antibiotic-resistant bacterial burden in cystic fibrosis phagocytes and improves cystic fibrosis transmembrane conductance regulator function. J Cyst Fibros. 2019. Sep;18(5):622–629. doi: 10.1016/j.jcf.2018.10.010. PubMed PMID: 30366849. [DOI] [PubMed] [Google Scholar]
- 2241.Buzgariu W, Chera S, Galliot B.. Methods to investigate autophagy during starvation and regeneration in hydra. Methods Enzymol. 2008;451:409–37. doi: 10.1016/S0076-6879(08)03226-6. PubMed PMID: 19185734; eng. [DOI] [PubMed] [Google Scholar]
- 2242.Chera S, de Rosa R, Miljkovic-Licina M, et al. Silencing of the hydra serine protease inhibitor Kazal1 gene mimics the human SPINK1 pancreatic phenotype. J Cell Sci. 2006. Mar 1;119(Pt 5):846–57. doi: 10.1242/jcs.02807. PubMed PMID: 16478786; eng. [DOI] [PubMed] [Google Scholar]
- 2243.Galliot B. Autophagy and self-preservation: a step ahead from cell plasticity?. Autophagy. 2006. Jul-Sep;2(3):231–3. PubMed PMID: 16874084; eng. [DOI] [PubMed] [Google Scholar]
- 2244.Chera S, Buzgariu W, Ghila L, et al. Autophagy in Hydra: a response to starvation and stress in early animal evolution. Biochim Biophys Acta. 2009. Sep;1793(9):1432–43. doi: 10.1016/j.bbamcr.2009.03.010. PubMed PMID: 19362111; eng. [DOI] [PubMed] [Google Scholar]
- 2245.Dixit NS, Shravage BV, Ghaskadbi S.. Identification and characterization of the autophagy-related genes Atg12 and Atg5 in hydra. . 2017;61(6–7):389–395. doi: 10.1387/ijdb.160461sg. PubMed PMID: 28695958. [DOI] [PubMed] [Google Scholar]
- 2246.Tiscornia G, Vivas EL, Izpisua Belmonte JC.. Diseases in a dish: modeling human genetic disorders using induced pluripotent cells. Nat Med. 2011. Dec;17(12):1570–6. doi: 10.1038/nm.2504. PubMed PMID: 22146428. [DOI] [PubMed] [Google Scholar]
- 2247.Karagiannis P, Takahashi K, Saito M, et al. Induced pluripotent stem cells and their use in human models of disease and development. Physiol Rev. 2019. Jan 1;99(1):79–114. doi: 10.1152/physrev.00039.2017. PubMed PMID: 30328784. [DOI] [PubMed] [Google Scholar]
- 2248.Schondorf DC, Aureli M, McAllister FE, et al. iPSC-derived neurons from GBA1-associated Parkinson’s disease patients show autophagic defects and impaired calcium homeostasis. Nat Commun. 2014. Jun 6;5:4028. doi: 10.1038/ncomms5028. PubMed PMID: 24905578. [DOI] [PubMed] [Google Scholar]
- 2249.di Domenico A, Carola G, Calatayud C, et al. Patient-specific iPSC-derived astrocytes contribute to non-cell-autonomous neurodegeneration in Parkinson’s disease. Stem Cell Reports. 2019. Feb 12;12(2):213–229. doi: 10.1016/j.stemcr.2018.12.011. PubMed PMID: 30639209; PubMed Central PMCID: PMCPMC6372974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2250.Marrone L, Drexler HCA, Wang J, et al. FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 2019. Jul;138(1):67–84. doi: 10.1007/s00401-019-01998-x. PubMed PMID: 30937520; PubMed Central PMCID: PMCPMC6570784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2251.Seibler P, Burbulla LF, Dulovic M, et al. Iron overload is accompanied by mitochondrial and lysosomal dysfunction in WDR45 mutant cells. Brain. 2018. Oct 1;141(10):3052–3064. doi: 10.1093/brain/awy230. PubMed PMID: 30169597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2252.Lin DS, Huang YW, Ho CS, et al. Oxidative insults and mitochondrial DNA mutation promote enhanced autophagy and mitophagy compromising cell viability in pluripotent cell model of mitochondrial disease. Cells. 2019. Jan 17;8(1). doi: 10.3390/cells8010065. PubMed PMID: 30658448; PubMed Central PMCID: PMCPMC6356288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2253.Gilkerson RW, De Vries RL, Lebot P, et al. Mitochondrial autophagy in cells with mtDNA mutations results from synergistic loss of transmembrane potential and mTORC1 inhibition. Hum Mol Genet. 2012. Mar 1;21(5):978–90. doi: 10.1093/hmg/ddr529. PubMed PMID: 22080835; PubMed Central PMCID: PMCPMC3277306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2254.Kwon Y, Kim JW, Jeoung JA, et al. Autophagy is pro-senescence when seen in close-up, but anti-senescence in long-shot. Mol Cells. 2017. Sep 30;40(9):607–612. doi: 10.14348/molcells.2017.0151. PubMed PMID: 28927262; PubMed Central PMCID: PMCPMC5638768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2255.Sala-Mercado JA, Wider J, Undyala VV, et al. Profound cardioprotection with chloramphenicol succinate in the swine model of myocardial ischemia-reperfusion injury. Circulation. 2010. Sep 14;122(11 Suppl):S179–84. doi: 10.1161/CIRCULATIONAHA.109.928242. PubMed PMID: 20837911; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2256.Botting KJ, McMillen IC, Forbes H, et al. Chronic hypoxemia in late gestation decreases cardiomyocyte number but does not change expression of hypoxia-responsive genes. J Am Heart Assoc. 2014. Aug;3(4). doi: 10.1161/JAHA.113.000531. PubMed PMID: 25085511; PubMed Central PMCID: PMC4310356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2257.Wang KC, Brooks DA, Summers-Pearce B, et al. Low birth weight activates the renin-angiotensin system, but limits cardiac angiogenesis in early postnatal life. Physiol Rep. 2015. Feb 1;3(2). doi: 10.14814/phy2.12270. PubMed PMID: 25649246; PubMed Central PMCID: PMC4393187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2258.Zhang S, Regnault TR, Barker PL, et al. Placental adaptations in growth restriction. Nutrients. 2015. Jan;7(1):360–89. doi: 10.3390/nu7010360. PubMed PMID: 25580812; PubMed Central PMCID: PMC4303845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2259.Derde S, Vanhorebeek I, Guiza F, et al. Early parenteral nutrition evokes a phenotype of autophagy deficiency in liver and skeletal muscle of critically ill rabbits. Endocrinology. 2012. May;153(5):2267–76. doi: 10.1210/en.2011-2068. PubMed PMID: 22396453. [DOI] [PubMed] [Google Scholar]
- 2260.Gunst J, Derese I, Aertgeerts A, et al. Insufficient autophagy contributes to mitochondrial dysfunction, organ failure, and adverse outcome in an animal model of critical illness. Crit Care Med. 2013. Jan;41(1):182–94. doi: 10.1097/CCM.0b013e3182676657. PubMed PMID: 23222264. [DOI] [PubMed] [Google Scholar]
- 2261.Hermans G, Casaer MP, Clerckx B, et al. Effect of tolerating macronutrient deficit on the development of intensive-care unit acquired weakness: a subanalysis of the EPaNIC trial. Lancet Respir Med. 2013. Oct;1(8):621–9. doi: 10.1016/S2213-2600(13)70183-8. PubMed PMID: 24461665. [DOI] [PubMed] [Google Scholar]
- 2262.Lopez-Alonso I, Aguirre A, Gonzalez-Lopez A, et al. Impairment of autophagy decreases ventilator-induced lung injury by blockade of the NF-kappaB pathway. Am J Physiol Lung Cell Mol Physiol. 2013. Jun 15;304(12):L844–52. doi: 10.1152/ajplung.00422.2012. PubMed PMID: 23585228. [DOI] [PubMed] [Google Scholar]
- 2263.Sun Y, Li C, Shu Y, et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal. 2012. Feb 21;5(212):ra16. doi: 10.1126/scisignal.2001931. PubMed PMID: 22355189. [DOI] [PubMed] [Google Scholar]
- 2264.Liu Y, Xue X, Zhang H, et al. Neuronal-targeted TFEB rescues dysfunction of the autophagy-lysosomal pathway and alleviates ischemic injury in permanent cerebral ischemia. Autophagy. 2019. Mar;15(3):493–509. doi: 10.1080/15548627.2018.1531196. PubMed PMID: 30304977; PubMed Central PMCID: PMCPMC6351122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2265.Zhang X, Yan H, Yuan Y, et al. Cerebral ischemia-reperfusion-induced autophagy protects against neuronal injury by mitochondrial clearance. Autophagy. 2013. Sep;9(9):1321–33. doi: 10.4161/auto.25132. PubMed PMID: 23800795. [DOI] [PubMed] [Google Scholar]
- 2266.Wen YD, Sheng R, Zhang LS, et al. Neuronal injury in rat model of permanent focal cerebral ischemia is associated with activation of autophagic and lysosomal pathways. Autophagy. 2008. Aug;4(6):762–9. doi: 10.4161/auto.6412. PubMed PMID: 18567942. [DOI] [PubMed] [Google Scholar]
- 2267.Ginet V, Spiehlmann A, Rummel C, et al. Involvement of autophagy in hypoxic-excitotoxic neuronal death. Autophagy. 2014. May;10(5):846–60. doi: 10.4161/auto.28264. PubMed PMID: 24674959; PubMed Central PMCID: PMCPMC5119065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2268.Belibi F, Zafar I, Ravichandran K, et al. Hypoxia-inducible factor-1alpha (HIF-1alpha) and autophagy in polycystic kidney disease (PKD). Am J Physiol Renal Physiol. 2011. May;300(5):F1235–43. doi: 10.1152/ajprenal.00348.2010. PubMed PMID: 21270095; PubMed Central PMCID: PMC3094047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2269.Gukovskaya AS, Gorelick FS, Groblewski GE, et al. Recent insights into the pathogenic mechanism of pancreatitis: role of acinar cell organelle disorders. Pancreas. 2019. Apr;48(4):459–470. doi: 10.1097/MPA.0000000000001298. PubMed PMID: 30973461; PubMed Central PMCID: PMCPMC6461375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2270.Sobolewska A, Motyl T, Gajewska M.. Role and regulation of autophagy in the development of acinar structures formed by bovine BME-UV1 mammary epithelial cells. Eur J Cell Biol. 2011. Oct;90(10):854–64. doi: 10.1016/j.ejcb.2011.06.007. PubMed PMID: 21868124; eng. [DOI] [PubMed] [Google Scholar]
- 2271.Motyl T, Gajewska M, Zarzynska J, et al. Regulation of autophagy in bovine mammary epithelial cells. Autophagy. 2007. Sep-Oct;3(5):484–6. PubMed PMID: 17592247; eng. [DOI] [PubMed] [Google Scholar]
- 2272.Sobolewska A, Gajewska M, Zarzynska J, et al. IGF-I, EGF, and sex steroids regulate autophagy in bovine mammary epithelial cells via the mTOR pathway. Eur J Cell Biol. 2009. Feb;88(2):117–30. doi: 10.1016/j.ejcb.2008.09.004. PubMed PMID: 19013662; eng. [DOI] [PubMed] [Google Scholar]
- 2273.Tettamanti G, Casartelli M.. Cell death during complete metamorphosis. Philos Trans R Soc London, Ser B. 2019. Oct 14;374(1783):20190065. doi: 10.1098/rstb.2019.0065. PubMed PMID: 31438818; PubMed Central PMCID: PMCPMC6711292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2274.Facey CO, Lockshin RA.. The execution phase of autophagy associated PCD during insect metamorphosis. Apoptosis. 2010. Jun;15(6):639–52. doi: 10.1007/s10495-010-0499-3. PubMed PMID: 20405221; eng. [DOI] [PubMed] [Google Scholar]
- 2275.Malagoli D, Abdalla FC, Cao Y, et al. Autophagy and its physiological relevance in arthropods: Current knowledge and perspectives. Autophagy. 2010. Jul 1;6(5):575–88. PubMed PMID: 20458176; Eng. [DOI] [PubMed] [Google Scholar]
- 2276.Mpakou VE, Nezis IP, Stravopodis DJ, et al. Programmed cell death of the ovarian nurse cells during oogenesis of the silkmoth Bombyx mori [Research Support, Non-U.S. Gov’t]. Dev Growth Differ. 2006. Sep;48(7):419–28. doi: 10.1111/j.1440-169X.2006.00878.x. PubMed PMID: 16961589; eng. [DOI] [PubMed] [Google Scholar]
- 2277.Mpakou VE, Nezis IP, Stravopodis DJ, et al. Different modes of programmed cell death during oogenesis of the silkmoth Bombyx mori [Research Support, Non-U.S. Gov’t]. Autophagy. 2008. Jan;4(1):97–100. PubMed PMID: 17986869; eng. [DOI] [PubMed] [Google Scholar]
- 2278.Sumithra P, Britto CP, Krishnan M.. Modes of cell death in the pupal perivisceral fat body tissue of the silkworm Bombyx mori L. Cell Tissue Res. 2010. Feb;339(2):349–58. doi: 10.1007/s00441-009-0898-3. PubMed PMID: 19949813; eng. [DOI] [PubMed] [Google Scholar]
- 2279.Tettamanti G, Grimaldi A, Casartelli M, et al. Programmed cell death and stem cell differentiation are responsible for midgut replacement in Heliothis virescens during prepupal instar. Cell Tissue Res. 2007. Nov;330(2):345–59. doi: 10.1007/s00441-007-0449-8. PubMed PMID: 17661086; eng. [DOI] [PubMed] [Google Scholar]
- 2280.Romanelli D, Casartelli M, Cappellozza S, et al. Roles and regulation of autophagy and apoptosis in the remodelling of the lepidopteran midgut epithelium during metamorphosis. Sci Rep. 2016. Sep 9;6:32939. doi: 10.1038/srep32939. PubMed PMID: 27609527; PubMed Central PMCID: PMCPMC5016986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2281.Khoa DB, Takeda M.. Expression of autophagy 8 (Atg8) and its role in the midgut and other organs of the greater wax moth, Galleria mellonella, during metamorphic remodelling and under starvation. Insect Mol Biol. 2012. Oct;21(5):473–87. doi: 10.1111/j.1365-2583.2012.01152.x. PubMed PMID: 22830988. [DOI] [PubMed] [Google Scholar]
- 2282.Gai Z, Zhang X, Islam M, et al. Characterization of Atg8 in lepidopteran insect cells. Arch Insect Biochem Physiol 2013;84:57–77. [DOI] [PubMed] [Google Scholar]
- 2283.Dai Y, Li K, Wu W, et al. Steroid hormone 20-hydroxyecdysone induces the transcription and complex assembly of V-ATPases to facilitate autophagy in Bombyx mori. Insect Biochem Mol Biol. 2020. Jan;116:103255. doi: 10.1016/j.ibmb.2019.103255. PubMed PMID: 31654713. [DOI] [PubMed] [Google Scholar]
- 2284.Wu W, Luo M, Li K, et al. Cholesterol derivatives induce dephosphorylation of the histone deacetylases Rpd3/HDAC1 to upregulate autophagy. Autophagy. 2020. Feb 12:1–17. doi: 10.1080/15548627.2020.1725376. PubMed PMID: 32013726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2285.Goncu E, Parlak O.. Some autophagic and apoptotic features of programmed cell death in the anterior silk glands of the silkworm, Bombyx mori. Autophagy. 2008. Nov;4(8):1069–72. PubMed PMID: 18838861; eng. [DOI] [PubMed] [Google Scholar]
- 2286.Zhou S, Zhou Q, Liu Y, et al. Two Tor genes in the silkworm Bombyx mori. Insect Mol Biol. 2010. Dec;19(6):727–35. doi: 10.1111/j.1365-2583.2010.01026.x. PubMed PMID: 20609020; eng. [DOI] [PubMed] [Google Scholar]
- 2287.Zhang X, Hu ZY, Li WF, et al. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol Biol. 2009;10:50. doi: 10.1186/1471-2199-10-50. ]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2288.Romanelli D, Casati B, Franzetti E, et al. A molecular view of autophagy in Lepidoptera. Biomed Res Int. 2014;2014:902315. doi: 10.1155/2014/902315. PubMed PMID: 25143951; PubMed Central PMCID: PMC4124216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2289.Li Q, Deng X, Huang Z, et al. Expression of autophagy-related genes in the anterior silk gland of the silkworm (Bombyx mori) during metamorphosis. Can J Zool 2011;89:1019–26. [Google Scholar]
- 2290.Casati B, Terova G, Cattaneo AG, et al. Molecular cloning, characterization and expression analysis of ATG1 in the silkworm, Bombyx mori. Gene. 2012. Dec 15;511(2):326–37. doi: 10.1016/j.gene.2012.09.086. PubMed PMID: 23041082. [DOI] [PubMed] [Google Scholar]
- 2291.Godefroy N, Hoa C, Tsokanos F, et al. Identification of autophagy genes in Ciona intestinalis: a new experimental model to study autophagy mechanism. Autophagy. 2009. Aug;5(6):805–15. PubMed PMID: 19502774. [DOI] [PubMed] [Google Scholar]
- 2292.Martinand-Mari C, Vacelet J, Nickel M, et al. Cell death and renewal during prey capture and digestion in the carnivorous sponge Asbestopluma hypogea (Porifera: Poecilosclerida). J Exp Biol. 2012. Nov 15;215\(Pt 22):3937–43. doi: 10.1242/jeb.072371. PubMed PMID: 22899530. [DOI] [PubMed] [Google Scholar]
- 2293.Jackson DJ, Worheide G.. Symbiophagy and biomineralization in the “living fossil” Astrosclera willeyana. Autophagy 2014. Mar;10(3):408–15. doi: 10.4161/auto.27319. PubMed PMID: 24343243; PubMed Central PMCID: PMC4077880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2294.Galasso C, D’Aniello S, Sansone C, et al. Identification of cell death genes in sea urchin paracentrotus lividus and their expression patterns during embryonic development. Genome Biol Evol. 2019. Feb 1;11(2):586–596. doi: 10.1093/gbe/evz020. PubMed PMID: 30698765; PubMed Central PMCID: PMCPMC6394757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2295.Moore MN, Allen JI, McVeigh A, et al. Lysosomal and autophagic reactions as predictive indicators of environmental impact in aquatic animals. Autophagy. 2006. Jul-Sep;2(3):217–20. doi: 10.4161/auto.2663. PubMed PMID: 16874099. [DOI] [PubMed] [Google Scholar]
- 2296.Sforzini S, Moore MN, Oliveri C, et al. Role of mTOR in autophagic and lysosomal reactions to environmental stressors in molluscs. Aquat Toxicol. 2018. Feb;195:114–128. doi: 10.1016/j.aquatox.2017.12.014. PubMed PMID: 29306034. [DOI] [PubMed] [Google Scholar]
- 2297.Moreau P, Moreau K, Segarra A, et al. Autophagy plays an important role in protecting Pacific oysters from OsHV-1 and Vibrio aestuarianus infections. Autophagy. 2015;11(3):516–26. doi: 10.1080/15548627.2015.1017188. PubMed PMID: 25714877; PubMed Central PMCID: PMCPMC4502751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2298.Balbi T, Cortese K, Ciacci C, et al. Autophagic processes in Mytilus galloprovincialis hemocytes: Effects of Vibrio tapetis. Fish Shellfish Immunol. 2018. Feb;73:66–74. doi: 10.1016/j.fsi.2017.12.003. PubMed PMID: 29208501. [DOI] [PubMed] [Google Scholar]
- 2299.Tanguy M, Gauthier-Clerc S, Pellerin J, et al. The immune response of Mytilus edulis hemocytes exposed to Vibrio splendidus LGP32 strain: A transcriptomic attempt at identifying molecular actors. Fish Shellfish Immunol. 2018. Mar;74:268–280. doi: 10.1016/j.fsi.2017.12.038. PubMed PMID: 29305989. [DOI] [PubMed] [Google Scholar]
- 2300.Wang L, Song X, Song L.. The oyster immunity. Dev Comp Immunol. 2018. Mar;80:99–118. doi: 10.1016/j.dci.2017.05.025. PubMed PMID: 28587860. [DOI] [PubMed] [Google Scholar]
- 2301.Fuess LE, Pinzon CJ, Weil E, et al. Life or death: disease-tolerant coral species activate autophagy following immune challenge. Proc Biol Sci. 2017. Jun 14;284(1856). doi: 10.1098/rspb.2017.0771. PubMed PMID: 28592676; PubMed Central PMCID: PMCPMC5474081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2302.Zhao MR, Meng C, Xie XL, et al. Characterization of microRNAs by deep sequencing in red claw crayfish Cherax quadricarinatus haematopoietic tissue cells after white spot syndrome virus infection. Fish Shellfish Immunol. 2016. Dec;59:469–483. doi: 10.1016/j.fsi.2016.11.012. PubMed PMID: 27825947. [DOI] [PubMed] [Google Scholar]
- 2303.He Y, Sun Y, Zhang X.. Noncoding miRNAs bridge virus infection and host autophagy in shrimp in vivo. FASEB J. 2017. Jul;31(7):2854–2868. doi: 10.1096/fj.201601141RR. PubMed PMID: 28330853. [DOI] [PubMed] [Google Scholar]
- 2304.Yang W, Liu C, Xu Q, et al. Beclin-1 is involved in the regulation of antimicrobial peptides expression in Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol. 2019. Jun;89:207–216. doi: 10.1016/j.fsi.2019.03.065. PubMed PMID: 30936045. [DOI] [PubMed] [Google Scholar]
- 2305.Wang M, Lee JS, Li Y.. Global proteome profiling of a marine copepod and the mitigating effect of ocean acidification on mercury toxicity after multigenerational exposure. Environ Sci Technol. 2017. May 16;51(10):5820–5831. doi: 10.1021/acs.est.7b01832. PubMed PMID: 28414453. [DOI] [PubMed] [Google Scholar]
- 2306.Sforzini S, Oliveri C, Barranger A, et al. Effects of fullerene C60 in blue mussels: Role of mTOR in autophagy related cellular/tissue alterations. Chemosphere. 2019. Dec 23;246:125707. doi: 10.1016/j.chemosphere.2019.125707. PubMed PMID: 31891845. [DOI] [PubMed] [Google Scholar]
- 2307.Thome RG, Santos HB, Arantes FP, et al. Dual roles for autophagy during follicular atresia in fish ovary. Autophagy. 2009. Jan; 5(1):117–9. PubMed PMID: 19011378; eng. [DOI] [PubMed] [Google Scholar]
- 2308.Santos HB, Thome RG, Arantes FP, et al. Ovarian follicular atresia is mediated by heterophagy, autophagy, and apoptosis in Prochilodus argenteus and Leporinus taeniatus. Theriogenology 2008. Dec;70(9):1449–60. doi: 10.1016/j.theriogenology.2008.06.091. PubMed PMID: 18701155; eng. [DOI] [PubMed] [Google Scholar]
- 2309.Santos HB, Sato Y, Moro L, et al. Relationship among follicular apoptosis, integrin beta1 and collagen type IV during early ovarian regression in the teleost Prochilodus argenteus after induced spawning. Cell Tissue Res. 2008. Apr;332(1):159–70. doi: 10.1007/s00441-007-0540-1. PubMed PMID: 18193286; eng. [DOI] [PubMed] [Google Scholar]
- 2310.Santos HB, Rizzo E, Bazzoli N, et al. Ovarian regression and apoptosis in the South American teleost Leporinus taeniatus Lutken (Characiformes, Anostomidae) from the São Francisco Basin. 2005;67:1446–1459. [Google Scholar]
- 2311.Couve E, Schmachtenberg O.. Autophagic activity and aging in human odontoblasts. J Dent Res. 2011. Apr;90(4):523–8. doi: 10.1177/0022034510393347. PubMed PMID: 21212314; eng. [DOI] [PubMed] [Google Scholar]
- 2312.Zhuang H, Hu D, Singer D, et al. Local anesthetics induce autophagy in young permanent tooth pulp cells. Cell Death Discov. 2015;1:15024. doi: 10.1038/cddiscovery.2015.24. PubMed PMID: 27551457; PubMed Central PMCID: PMCPMC4979463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2313.An Y, Liu W, Xue P, et al. Increased autophagy is required to protect periodontal ligament stem cells from apoptosis in inflammatory microenvironment. J Clin Periodontol. 2016. Jul;43(7):618–25. doi: 10.1111/jcpe.12549. PubMed PMID: 26990245. [DOI] [PubMed] [Google Scholar]
- 2314.Vescarelli E, Pilloni A, Dominici F, et al. Autophagy activation is required for myofibroblast differentiation during healing of oral mucosa. J Clin Periodontol. 2017. Oct;44(10):1039–1050. doi: 10.1111/jcpe.12767. PubMed PMID: 28646601. [DOI] [PubMed] [Google Scholar]
- 2315.Olson PD, Tkach VV.. Advances and trends in the molecular systematics of the parasitic Platyhelminthes. Adv Parasitol. 2005;60:165–243. doi: 10.1016/S0065-308X(05)60003-6. PubMed PMID: 16230104. [DOI] [PubMed] [Google Scholar]
- 2316.Collins JJ, 3rd. Platyhelminthes. Curr Biol. 2017. Apr 3;27(7):R252–R256. doi: 10.1016/j.cub.2017.02.016. PubMed PMID: 28376328. [DOI] [PubMed] [Google Scholar]
- 2317.Threadgold LT, Arme C.. Electron microscope studies of Fasciola hepatica. XI. Autophagy and parenchymal cell function. Exp Parasitol. 1974. Jun;35(3):389–405. doi: 10.1016/0014-4894(74)90045-9. PubMed PMID: 4363771. [DOI] [PubMed] [Google Scholar]
- 2318.Bogitsh BJ. Cytochemistry of gastrodermal autophagy following starvation in Schistosoma mansoni. J Parasitol. 1975. Apr;61(2):237–48. PubMed PMID: 1127552. [PubMed] [Google Scholar]
- 2319.Clarkson J, Erasmus DA.. Schistosoma mansoni: an in vivo study of drug-induced autophagy in the gastrodermis. J Helminthol. 1984. Mar;58(1):59–68. doi: 10.1017/s0022149x00028066. PubMed PMID: 6325532. [DOI] [PubMed] [Google Scholar]
- 2320.Richards KS, Arme C, Bridges JF.. Echinococcus granulosus equinus: variation in the germinal layer of murine hydatids and evidence of autophagy. Parasitology. 1984. Aug;89(Pt 1):35–47. doi: 10.1017/s0031182000001116. PubMed PMID: 6472884. [DOI] [PubMed] [Google Scholar]
- 2321.McCullough JS, Fairweather I.. The structure, composition, formation and possible functions of calcareous corpuscles in Trilocularia acanthiaevulgaris Olsson 1867 (Cestoda, Tetraphyllidea). Parasitol Res. 1987;74(2):175–82. doi: 10.1007/bf00536030. PubMed PMID: 3438298. [DOI] [PubMed] [Google Scholar]
- 2322.Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006. Dec;26(24):9220–31. doi: 10.1128/MCB.01453-06. PubMed PMID: 17030611; PubMed Central PMCID: PMCPMC1698520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2323.Ahn CS, Kim JG, Bae YA, et al. Fasciclin-calcareous corpuscle binary complex mediated protein-protein interactions in Taenia solium metacestode. Parasit Vectors. 2017. Sep 20;10(1):438. doi: 10.1186/s13071-017-2359-2. PubMed PMID: 28931431; PubMed Central PMCID: PMCPMC5606126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2324.Loos JA, Caparros PA, Nicolao MC, et al. Identification and pharmacological induction of autophagy in the larval stages of Echinococcus granulosus: an active catabolic process in calcareous corpuscles. Int J Parasitol. 2014. Jun;44(7):415–27. doi: 10.1016/j.ijpara.2014.02.007. PubMed PMID: 24703869. [DOI] [PubMed] [Google Scholar]
- 2325.Berriman M, Haas BJ, LoVerde PT, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009. Jul 16;460(7253):352–8. doi: 10.1038/nature08160. PubMed PMID: 19606141; PubMed Central PMCID: PMCPMC2756445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2326.Tsai IJ, Zarowiecki M, Holroyd N, et al. The genomes of four tapeworm species reveal adaptations to parasitism. Nature. 2013. Apr 4;496(7443):57–63. doi 10.1038/nature12031. PubMed PMID: 23485966; PubMed Central PMCID: PMCPMC3964345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2327.Zheng H, Zhang W, Zhang L, et al. The genome of the hydatid tapeworm Echinococcus granulosus. Nat Genet. 2013. Oct;45(10):1168–75. doi: 10.1038/ng.2757. PubMed PMID: 24013640. [DOI] [PubMed] [Google Scholar]
- 2328.International Helminth Genomes C. Comparative genomics of the major parasitic worms. Nat Genet. 2019. Jan;51(1):163–174. doi: 10.1038/s41588-018-0262-1. PubMed PMID: 30397333; PubMed Central PMCID: PMCPMC6349046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2329.Cumino AC, Lamenza P, Denegri GM.. Identification of functional FKB protein in Echinococcus granulosus: its involvement in the protoscolicidal action of rapamycin derivates and in calcium homeostasis. Int J Parasitol. 2010. May;40(6):651–61. doi: 10.1016/j.ijpara.2009.11.011. PubMed PMID: 20005877. [DOI] [PubMed] [Google Scholar]
- 2330.Loos JA, Nicolao MC, Cumino AC.. Metformin promotes autophagy in Echinococcus granulosus larval stage. Mol Biochem Parasitol. 2018. Sep;224:61–70. doi: 10.1016/j.molbiopara.2018.07.003. PubMed PMID: 30017657. [DOI] [PubMed] [Google Scholar]
- 2331.Nicolao MC, Loos JA, Rodriguez Rodrigues C, et al. Bortezomib initiates endoplasmic reticulum stress, elicits autophagy and death in Echinococcus granulosus larval stage. PLoS One. 2017;12(8):e0181528. doi: 10.1371/journal.pone.0181528. PubMed PMID: 28817601; PubMed Central PMCID: PMCPMC5560652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2332.Gonzalez-Estevez C. Autophagy in freshwater planarians. Methods Enzymol. 2008;451:439–65. doi: 10.1016/S0076-6879(08)03227-8. PubMed PMID: 19185735; eng. [DOI] [PubMed] [Google Scholar]
- 2333.Gonzalez-Estevez C, Felix DA, Aboobaker AA, et al. Gtdap-1 promotes autophagy and is required for planarian remodeling during regeneration and starvation. Proc Natl Acad Sci U S A. 2007. Aug 14;104(33):13373–8. doi: 10.1073/pnas.0703588104. PubMed PMID: 17686979; PubMed Central PMCID: PMC1948951. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2334.Toyooka K, Moriyasu Y, Goto Y, et al. Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy. 2006. Apr-Jun;2(2):96–106. PubMed PMID: 16874101; eng. [DOI] [PubMed] [Google Scholar]
- 2335.Corral-Martinez P, Parra-Vega V, Segui-Simarro JM.. Novel features of Brassica napus embryogenic microspores revealed by high pressure freezing and freeze substitution: evidence for massive autophagy and excretion-based cytoplasmic cleaning. J Exp Bot. 2013. Jul;64(10):3061–75. doi: 10.1093/jxb/ert151. PubMed PMID: 23761486. [DOI] [PubMed] [Google Scholar]
- 2336.Le Bars R, Marion J, Le Borgne R, et al. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun. 2014;5:4121. doi: 10.1038/ncomms5121. PubMed PMID: 24947672. [DOI] [PubMed] [Google Scholar]
- 2337.Shin KD, Lee HN, Chung T.. A revised assay for monitoring autophagic flux in Arabidopsis thaliana reveals involvement of AUTOPHAGY-RELATED9 in autophagy. Mol Cells. 2014. May;37(5):399–405. doi: 10.14348/molcells.2014.0042. PubMed PMID: 24805779; PubMed Central PMCID: PMC4044311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2338.Wojciechowska N, Smugarzewska I, Marzec-Schmidt K, et al. Occurrence of autophagy during pioneer root and stem development in Populus trichocarpa. Planta. 2019. Dec;250(6):1789–1801. doi: 10.1007/s00425-019-03265-5. PubMed PMID: 31451904. [DOI] [PubMed] [Google Scholar]
- 2339.Wojciechowska N, Marzec-Schmidt K, Kalemba EM, et al. Autophagy counteracts instantaneous cell death during seasonal senescence of the fine roots and leaves in Populus trichocarpa. BMC Plant Biol. 2018. Oct 29;18(1):260. doi: 10.1186/s12870-018-1439-6. PubMed PMID: 30373512; PubMed Central PMCID: PMCPMC6206944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2340.Svenning S, Lamark T, Krause K, et al. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy. 2011. Sep 1;7(9):993–1010. PubMed PMID: 21606687; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2341.Zientara-Rytter K, Lukomska J, Moniuszko G, et al. Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy. 2011. Oct 1;7(10):1145–58. PubMed PMID: 21670587; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2342.Minina EA, Sanchez-Vera V, Moschou PN, et al. Autophagy mediates caloric restriction-induced lifespan extension in Arabidopsis. Aging cell. 2013. Apr;12(2):327–9. doi: 10.1111/acel.12048. PubMed PMID: 23331488. [DOI] [PubMed] [Google Scholar]
- 2343.Farmer LM, Rinaldi MA, Young PG, et al. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. Plant Cell. 2013. Oct;25(10):4085–100. doi: 10.1105/tpc.113.113407. PubMed PMID: 24179123; PubMed Central PMCID: PMC3877801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2344.Shibata M, Oikawa K, Yoshimoto K, et al. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013. Dec;25(12):4967–83. doi: 10.1105/tpc.113.116947. PubMed PMID: 24368788; PubMed Central PMCID: PMCPMC3903999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2345.Fahy D, Sanad M, Duscha K, et al. Impact of salt stress, cell death, and autophagy on peroxisomes: quantitative and morphological analyses using small fluorescent probe N-BODIPY. Sci Rep. 2017. Feb 1;7:39069. doi: 10.1038/srep39069. PubMed PMID: 28145408; PubMed Central PMCID: PMCPMC5286434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2346.Landrum M, Smertenko A, Edwards R, et al. BODIPY probes to study peroxisome dynamics in vivo. Plant J. 2010. May;62(3):529–38. doi: 10.1111/j.1365-313X.2010.04153.x. PubMed PMID: 20113442. [DOI] [PubMed] [Google Scholar]
- 2347.Eapen D, Barroso ML, Campos ME, et al. A no hydrotropic response root mutant that responds positively to gravitropism in Arabidopsis. Plant Physiol. 2003. Feb;131(2):536–46. doi: 10.1104/pp.011841. PubMed PMID: 12586878; PubMed Central PMCID: PMCPMC166830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2348.Jimenez-Nopala G, Salgado-Escobar AE, Cevallos-Porta D, et al. Autophagy mediates hydrotropic response in Arabidopsis thaliana roots. Plant Sci. 2018. Jul;272:1–13. doi: 10.1016/j.plantsci.2018.03.026. PubMed PMID: 29807580. [DOI] [PubMed] [Google Scholar]
- 2349.van Doorn WG, Papini A.. Ultrastructure of autophagy in plant cells: a review. Autophagy. 2013. Dec;9(12):1922–36. doi: 10.4161/auto.26275. PubMed PMID: 24145319. [DOI] [PubMed] [Google Scholar]
- 2350.Dagdas YF, Belhaj K, Maqbool A, et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife. 2016. Jan 14;5. doi: 10.7554/eLife.10856. PubMed PMID: 26765567; PubMed Central PMCID: PMCPMC4775223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2351.Dagdas YF, Pandey P, Tumtas Y, et al. Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. eLife. 2018. Jun 22;7. doi: 10.7554/eLife.37476. PubMed PMID: 29932422; PubMed Central PMCID: PMCPMC6029844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2352.Moriyasu Y, Inoue Y.. Use of protease inhibitors for detecting autophagy in plants. Methods Enzymol. 2008;451:557–80. doi: 10.1016/S0076-6879(08)03232-1. PubMed PMID: 19185740. [DOI] [PubMed] [Google Scholar]
- 2353.Takatsuka C, Inoue Y, Matsuoka K, et al. 3-methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol 2004. Mar;45(3):265–74. PubMed PMID: 15047874. [DOI] [PubMed] [Google Scholar]
- 2354.Besteiro S, Brooks CF, Striepen B, et al. Autophagy protein Atg3 is essential for maintaining mitochondrial integrity and for normal intracellular development of Toxoplasma gondii tachyzoites. PLoS Pathog. 2011;7(12):e1002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2355.Calvo-Garrido J, Carilla-Latorre S, Kubohara Y, et al. Autophagy in Dictyostelium: genes and pathways, cell death and infection. Autophagy. 2010. Aug;6(6):686–701. PubMed PMID: 20603609; eng. [DOI] [PubMed] [Google Scholar]
- 2356.Tung SM, Unal C, Ley A, et al. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell Microbiol. 2010. Jun;12(6):765–80. doi: 10.1111/j.1462-5822.2010.01432.x. PubMed PMID: 20070309; eng. [DOI] [PubMed] [Google Scholar]
- 2357.0Bozzaro S, Eichinger L.. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets. 2011. Jun;12(7):942–54. PubMed PMID: 21366522; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2358.Schlegel M, Hülsmann N.. Protists – A textbook example for a paraphyletic taxon. Org Divers Evol 2007;7:166–172. [Google Scholar]
- 2359.Kitamura K, Kishi-Itakura C, Tsuboi T, et al. Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite Plasmodium falciparum. PLoS One. 2012;7(8):e42977. doi: 10.1371/journal.pone.0042977. PubMed PMID: 22900071; PubMed Central PMCID: PMC3416769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2360.Barquilla A, Crespo JL, Navarro M.. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci U S A. 2008. Sep 23;105(38):14579–84. doi: 10.1073/pnas.0802668105. PubMed PMID: 18796613; PubMed Central PMCID: PMC2567229. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2361.Vanrell MC, Losinno AD, Cueto JA, et al. The regulation of autophagy differentially affects Trypanosoma cruzi metacyclogenesis. PLoS Negl Trop Dis. 2017. Nov;11(11):e0006049. doi: 10.1371/journal.pntd.0006049. PubMed PMID: 29091711; PubMed Central PMCID: PMCPMC5683653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2362.Li FJ, He CY.. Acidocalcisome is required for autophagy in Trypanosoma brucei. Autophagy. 2014;10(11):1978–88. doi: 10.4161/auto.36183. PubMed PMID: 25484093; PubMed Central PMCID: PMCPMC4502762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2363.Hain AU, Bartee D, Sanders NG, et al. Identification of an Atg8-Atg3 protein-protein interaction inhibitor from the medicines for Malaria Venture Malaria Box active in blood and liver stage Plasmodium falciparum parasites. J Med Chem. 2014. Jun 12;57(11):4521–31. doi: 10.1021/jm401675a. PubMed PMID: 24786226; PubMed Central PMCID: PMC4059259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2364.Hain AU, Weltzer RR, Hammond H, et al. Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J Struct Biol. 2012. Dec;180(3):551–62. doi: 10.1016/j.jsb.2012.09.001. PubMed PMID: 22982544; PubMed Central PMCID: PMC3496014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2365.Navale R, Atul, Allanki AD, et al. Characterization of the autophagy marker protein Atg8 reveals atypical features of autophagy in Plasmodium falciparum. PLoS One. 2014;9(11):e113220. doi: 10.1371/journal.pone.0113220. PubMed PMID: 25426852; PubMed Central PMCID: PMC4245143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2366.Parussini F, Coppens I, Shah PP, et al. Cathepsin L occupies a vacuolar compartment and is a protein maturase within the endo/exocytic system of Toxoplasma gondii. Mol Microbiol. 2010. Jun;76(6):1340–57. doi: 10.1111/j.1365-2958.2010.07181.x. PubMed PMID: 20444089; PubMed Central PMCID: PMCPMC2909120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2367.Miranda K, Pace DA, Cintron R, et al. Characterization of a novel organelle in Toxoplasma gondii with similar composition and function to the plant vacuole. Mol Microbiol. 2010. Jun;76(6):1358–75. doi: 10.1111/j.1365-2958.2010.07165.x. PubMed PMID: 20398214; PubMed Central PMCID: PMCPMC2907454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2368.Nguyen HM, El Hajj H, El Hajj R, et al. Toxoplasma gondii autophagy-related protein ATG9 is crucial for the survival of parasites in their host. Cell Microbiol. 2017. Jun;19(6). doi: 10.1111/cmi.12712. PubMed PMID: 27992947. [DOI] [PubMed] [Google Scholar]
- 2369.Di Cristina M, Dou Z, Lunghi M, et al. Toxoplasma depends on lysosomal consumption of auto-phagosomes for persistent infection. Nat Microbiol. 2017. Jun 19;2:17096. doi: 10.1038/nmicrobiol.2017.96. PubMed PMID: 28628099; PubMed Central PMCID: PMCPMC5527684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2370.Kannan G, Di Cristina M, Schultz AJ, et al. Role of toxoplasma gondii chloroquine resistance transporter in bradyzoite viability and digestive vacuole maintenance. mBio. 2019. Aug 6;10(4). doi: 10.1128/mBio.01324-19. PubMed PMID: 31387907; PubMed Central PMCID: PMCPMC6686041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2371.Morais P, Lamas J, Sanmartin ML, et al. Resveratrol induces mitochondrial alterations, autophagy and a cryptobiosis-like state in scuticociliates. Protist. 2009. Nov;160(4):552–64. doi: 10.1016/j.protis.2009.04.004. PubMed PMID: 19640787; eng. [DOI] [PubMed] [Google Scholar]
- 2372.Yakisich JS, Kapler GM.. The effect of phosphoinositide 3-kinase inhibitors on programmed nuclear degradation in Tetrahymena and fate of surviving nuclei. Cell Death Differ. 2004. Oct;11(10):1146–9. doi: 10.1038/sj.cdd.4401473. PubMed PMID: 15257301; eng. [DOI] [PubMed] [Google Scholar]
- 2373.Akematsu T, Pearlman RE, Endoh H.. Gigantic macroautophagy in programmed nuclear death of Tetrahymena thermophila [Research Support, Non-U.S. Gov’t]. Autophagy. 2010. Oct;6(7):901–11. doi: 10.4161/auto.6.7.13287. PubMed PMID: 20798592; PubMed Central PMCID: PMC3039737. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2374.Schoijet AC, Miranda K, Girard-Dias W, et al. A Trypanosoma cruzi phosphatidylinositol 3-kinase (TcVps34) is involved in osmoregulation and receptor-mediated endocytosis. J Biol Chem. 2008. Nov 14;283(46):31541–50. doi: 10.1074/jbc.M801367200. PubMed PMID: 18801733; PubMed Central PMCID: PMCPMC2581564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2375.Thorgaard GH, Bailey GS, Williams D, et al. Status and opportunities for genomics research with rainbow trout. Comp Biochem Physiol Part B Biochem Mol Bio. 2002. Dec;133(4):609–46. PubMed PMID: 12470823; eng. [DOI] [PubMed] [Google Scholar]
- 2376.Govoroun M, Le Gac F, Guiguen Y.. Generation of a large scale repertoire of Expressed Sequence Tags (ESTs) from normalised rainbow trout cDNA libraries. BMC genomics. 2006;7:196. doi: 10.1186/1471-2164-7-196. PubMed PMID: 16887034; PubMed Central PMCID: PMC1564016. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2377.Rexroad CE, III, Lee Y, Keele JW, et al. Sequence analysis of a rainbow trout cDNA library and creation of a gene index. Cytogenet Genome Res. 2003;102(1–4): 347–54. doi: 10.1159/000075773. PubMed PMID: 14970727; eng. [DOI] [PubMed] [Google Scholar]
- 2378.Rise ML, von Schalburg KR, Brown GD, et al. Development and application of a salmonid EST database and cDNA microarray: data mining and interspecific hybridization characteristics. Genome Res. 2004. Mar;14(3):478–90. doi: 10.1101/gr.1687304. PubMed PMID: 14962987; PubMed Central PMCID: PMC353236. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2379.Salem M, Rexroad CE, III, Wang J, et al. Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC genomics. 2010;11:564. doi: 10.1186/1471-2164-11-564. PubMed PMID: 20942956; PubMed Central PMCID: PMC3091713. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2380.Seiliez I, Gabillard J-C, Riflade M, et al. Amino acids downregulate the expression of several autophagy-related genes in rainbow trout myoblasts. Autophagy. 2012;8:364–75. [DOI] [PubMed] [Google Scholar]
- 2381.Seiliez I, Gabillard JC, Skiba-Cassy S, et al. An in vivo and in vitro assessment of TOR signaling cascade in rainbow trout (Oncorhynchus mykiss). Am J Physiol Regul Integr Comp Physiol. 2008. Jul;295(1):R329–35. doi: 10.1152/ajpregu.00146.2008. PubMed PMID: 18434442; eng. [DOI] [PubMed] [Google Scholar]
- 2382.Polakof S, Panserat S, Craig PM, et al. The metabolic consequences of hepatic AMP-kinase phosphorylation in rainbow trout. PLoS One. 2011;6(5):e20228. doi: 10.1371/journal.pone.0020228. PubMed PMID: 21625448; PubMed Central PMCID: PMC3098864. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2383.Frost LS, Mitchell CH, Boesze-Battaglia K.. Autophagy in the eye: implications for ocular cell health. Exp Eye Res. 2014. Jul;124:56–66. doi: 10.1016/j.exer.2014.04.010. PubMed PMID: 24810222; PubMed Central PMCID: PMCPMC4156154. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2384.Muniz-Feliciano L, Doggett TA, Zhou Z, et al. RUBCN/rubicon and EGFR regulate lysosomal degradative processes in the retinal pigment epithelium (RPE) of the eye. Autophagy. 2017;13(12):2072–2085. doi: 10.1080/15548627.2017.1380124. PubMed PMID: 28933590; PubMed Central PMCID: PMCPMC5788552. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2385.Zhang Y, Cross SD, Stanton JB, et al. Early AMD-like defects in the RPE and retinal degeneration in aged mice with RPE-specific deletion of Atg5 or Atg7. Mol Vis. 2017;23:228–241. PubMed PMID: 28465655; PubMed Central PMCID: PMCPMC5398883. eng. [PMC free article] [PubMed] [Google Scholar]
- 2386.Zhao C, Yasumura D, Li X, et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011. Jan;121(1):369–83. doi: 10.1172/jci44303. PubMed PMID: 21135502; PubMed Central PMCID: PMCPMC3007156. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2387.Huang J, Gu S, Chen M, et al. Abnormal mTORC1 signaling leads to retinal pigment epithelium degeneration. Theranostics. 2019;9(4):1170–1180. doi: 10.7150/thno.26281. PubMed PMID: 30867823; PubMed Central PMCID: PMCPMC6401408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2388.Frost LS, Dhingra A, Reyes-Reveles J, et al. The Use of DQ-BSA to Monitor the Turnover of Autophagy-Associated Cargo. Methods Enzymol. 2017;587:43–54. doi: 10.1016/bs.mie.2016.09.052. PubMed PMID: 28253971; PubMed Central PMCID: PMCPMC5338641. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2389.Frost LS, Lopes VS, Bragin A, et al. The contribution of melanoregulin to microtubule-associated protein 1 light chain 3 (LC3) associated phagocytosis in retinal pigment epithelium. Mol Neurobiol. 2015. Dec;52(3):1135–1151. doi: 10.1007/s12035-014-8920-5. PubMed PMID: 25301234; PubMed Central PMCID: PMCPMC5531606. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2390.Dhingra A, Bell BA, Peachey NS, et al. Microtubule-associated protein 1 light chain 3B, (LC3B) is necessary to maintain lipid-mediated homeostasis in the retinal pigment epithelium. Front Cell Neurosci. 2018;12:351. doi: 10.3389/fncel.2018.00351. PubMed PMID: 30349463; PubMed Central PMCID: PMCPMC6186781. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2391.Baltazar GC, Guha S, Lu W, et al. Acidic nanoparticles are trafficked to lysosomes and restore an acidic lysosomal pH and degradative function to compromised ARPE-19 cells. PLoS One. 2012;7(12):e49635. doi: 10.1371/journal.pone.0049635. PubMed PMID: 23272048; PubMed Central PMCID: PMCPMC3525582. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2393.Guha S, Liu J, Baltazar G, et al. Rescue of compromised lysosomes enhances degradation of photoreceptor outer segments and reduces lipofuscin-like autofluorescence in retinal pigmented epithelial cells. Adv Exp Med Biol. 2014;801:105–11. doi: 10.1007/978-1-4614-3209-8_14.PubMed PMID: 24664687; PubMed Central PMCID: PMCPMC4163923. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2394.Lu W, Campagno KE, Tso HY, et al. Oral delivery of the P2Y12 receptor antagonist ticagrelor prevents loss of photoreceptors in an ABCA4-/- mouse model of retinal degeneration. Invest Ophthalmol Vis Sci. 2019. Jul 1;60(8):3046–3053. doi: 10.1167/iovs.19-27241. PubMed PMID: 31319418; PubMed Central PMCID: PMCPMC6640265. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2395.Lu W, Gomez NM, Lim JC, et al. The P2Y12 receptor antagonist ticagrelor reduces lysosomal pH and autofluorescence in retinal pigmented epithelial cells from the ABCA4(-/-) mouse model of retinal degeneration. Front Pharmacol. 2018;9:242. doi: 10.3389/fphar.2018.00242. PubMed PMID: 29725296; PubMed Central PMCID: PMCPMC5917064. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2396.Li Z, Li H, Xu X, et al. Haploinsufficiency of GCP4 induces autophagy and leads to photoreceptor degeneration due to defective spindle assembly in retina. Cell Death Differ. 2020. Feb;27(2):556–572. doi: 10.1038/s41418-019-0371-0. PubMed PMID: 31209365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2397.Xu X, Shang D, Cheng H, et al. Gene essentiality of Tubgcp4: dosage effect and autophagy regulation in retinal photoreceptors. Autophagy. 2019. Oct;15(10):1834–1837. doi: 10.1080/15548627.2019.1647023. PubMed PMID: 31345090; PubMed Central PMCID: PMCPMC6735468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2398.Ruggiero L, Connor MP, Chen J, et al. Diurnal, localized exposure of phosphatidylserine by rod outer segment tips in wild-type but not Itgb5-/- or Mfge8-/- mouse retina. Proc Natl Acad Sci U S A. 2012. May 22;109(21):8145–8. doi: 10.1073/pnas.1121101109. PubMed PMID: 22566632; PubMed Central PMCID: PMCPMC3361434. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2399.Ferguson TA, Green DR.. Autophagy and phagocytosis converge for better vision. Autophagy. 2014. Jan;10(1):165–7. doi: 10.4161/auto.26735. PubMed PMID: 24220227; PubMed Central PMCID: PMC4028322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2400.Kim JY, Zhao H, Martinez J, et al. Noncanonical autophagy promotes the visual cycle. Cell. 2013. Jul 18;154(2):365–76. doi: 10.1016/j.cell.2013.06.012. PubMed PMID: 23870125; PubMed Central PMCID: PMCPMC3744125. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2401.Dhingra A, Alexander D, Reyes-Reveles J, et al. Microtubule-Associated Protein 1 Light Chain 3 (LC3) Isoforms in RPE and Retina. Adv Exp Med Biol. 2018;1074:609–616. doi: 10.1007/978-3-319-75402-4_74. PubMed PMID: 29721994; eng. [DOI] [PubMed] [Google Scholar]
- 2402.Yu B, Egbejimi A, Dharmat R, et al. Phagocytosed photoreceptor outer segments activate mTORC1 in the retinal pigment epithelium. Sci Signal. 2018. May 29;11(532). doi: 10.1126/scisignal.aag3315. PubMed PMID: 29844054; PubMed Central PMCID: PMCPMC6198651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2403.Bhattacharya S, Yin J, Winborn CS, et al. Prominin-1 is a novel regulator of autophagy in the human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2017. Apr 1;58(4):2366–2387. doi: 10.1167/iovs.16-21162. PubMed PMID: 28437526; PubMed Central PMCID: PMCPMC5403116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2404.Chiarelli R, Agnello M, Bosco L, et al. Sea urchin embryos exposed to cadmium as an experimental model for studying the relationship between autophagy and apoptosis. Mar Environ Res. 2014. Feb;93:47–55. doi: 10.1016/j.marenvres.2013.06.001. PubMed PMID: 23838188. [DOI] [PubMed] [Google Scholar]
- 2405.Umemiya R, Matsuo T, Hatta T, et al. Cloning and characterization of an autophagy-related gene, ATG12, from the three-host tick Haemaphysalis longicornis. Insect Biochem Mol Biol. 2007. Sep;37(9):975–84. doi: 10.1016/j.ibmb.2007.05.006. PubMed PMID: 17681237; eng. [DOI] [PubMed] [Google Scholar]
- 2406.Kawano S, Umemiya-Shirafuji R, Boldbaatar D, et al. Cloning and characterization of the autophagy-related gene 6 from the hard tick, Haemaphysalis longicornis. Parasitol Res. 2011;109(5):1341–9. doi: 10.1007/s00436-011-2429-x. [DOI] [PubMed] [Google Scholar]
- 2407.Umemiya-Shirafuji R, Galay RL, Maeda H, et al. Expression analysis of autophagy-related genes in the hard tick Haemaphysalis longicornis. Vet Parasitol. 2014. Mar 17;201(1–2): 169–75. doi: 10.1016/j.vetpar.2014.01.024. PubMed PMID: 24556037. [DOI] [PubMed] [Google Scholar]
- 2408.Umemiya-Shirafuji R, Matsuo T, Liao M, et al. Increased expression of ATG genes during nonfeeding periods in the tick Haemaphysalis longicornis. Autophagy. 2010. May 16;6(4):473–81. PubMed PMID: 20404490; Eng. [DOI] [PubMed] [Google Scholar]
- 2409.de la Fuente J, Kocan KM, Almazan C, et al. RNA interference for the study and genetic manipulation of ticks. Trends Parasitol. 2007. Sep;23(9):427–33. doi: 10.1016/j.pt.2007.07.002. PubMed PMID: 17656154. [DOI] [PubMed] [Google Scholar]
- 2410.Ayllón N, Villar V, Galindo RC, et al. Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet. 2015;11(3):e1005120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2411.Genomic Resources Development C, Contreras M, de la Fuente J, et al. Genomic resources notes accepted 1 April 2014–31 May 2014. Mol Ecol Resour. 2014. Sep;14(5):1095. doi: 10.1111/1755-0998.12298. PubMed PMID: 24976445. [DOI] [PubMed] [Google Scholar]
- 2412.Lee E, Koo Y, Ng A, et al. Autophagy is essential for cardiac morphogenesis during vertebrate development. Autophagy. 2014. Apr;10(4):572–87. doi: 10.4161/auto.27649. PubMed PMID: 24441423; PubMed Central PMCID: PMC4091146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2413.Sasaki T, Lian S, Qi J, et al. Aberrant autolysosomal regulation is linked to the induction of embryonic senescence: differential roles of Beclin 1 and p53 in vertebrate Spns1 deficiency. PLoS Genet. 2014. Jun;10(6):e1004409. doi: 10.1371/journal.pgen.1004409. PubMed PMID: 24967584; PubMed Central PMCID: PMC4072523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2414.Varga M, Fodor E, Vellai T.. Autophagy in zebrafish. Methods. 2015. Mar;75:172–80. doi: 10.1016/j.ymeth.2014.12.004. PubMed PMID: 25498006. [DOI] [PubMed] [Google Scholar]
- 2415.Komoike Y, Shimojima K, Liang JS, et al. A functional analysis of GABARAP on 17p13.1 by knockdown zebrafish. J Hum Genet. 2010. Mar;55(3):155–62. doi: 10.1038/jhg.2010.1. PubMed PMID: 20111057; eng. [DOI] [PubMed] [Google Scholar]
- 2416.Hu Z, Zhang J, Zhang Q.. Expression pattern and functions of autophagy-related gene atg5 in zebrafish organogenesis. Autophagy. 2011. Dec;7(12):1514–27. doi: 10.4161/auto.7.12.18040. PubMed PMID: 22082871; PubMed Central PMCID: PMCPMC3288024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2417.Mans LA, Querol Cano L, van Pelt J, et al. The tumor suppressor LKB1 regulates starvation-induced autophagy under systemic metabolic stress. Sci Rep. 2017. Aug 4;7(1):7327. doi: 10.1038/s41598-017-07116-9. PubMed PMID: 28779098; PubMed Central PMCID: PMCPMC5544676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2418.Masud S, Prajsnar TK, Torraca V, et al. Macrophages target Salmonella by Lc3-associated phagocytosis in a systemic infection model. Autophagy. 2019. May;15(5):796–812. doi: 10.1080/15548627.2019.1569297. PubMed PMID: 30676840; PubMed Central PMCID: PMCPMC6526873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2419.Dowling JJ, Low SE, Busta AS, et al. Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum Mol Genet. 2010. Jul 1;19(13):2668–81. doi: 10.1093/hmg/ddq153. PubMed PMID: 20400459; PubMed Central PMCID: PMC2883342. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2420.Makky K, Tekiela J, Mayer AN.. Target of rapamycin (TOR) signaling controls epithelial morphogenesis in the vertebrate intestine. Dev Biol. 2007. Mar 15;303(2):501–13. doi: 10.1016/j.ydbio.2006.11.030. PubMed PMID: 17222402; PubMed Central PMCID: PMC2715143. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2421.Schulte-Merker S, Stainier DY.. Out with the old, in with the new: reassessing morpholino knockdowns in light of genome editing technology. Development. 2014. Aug;141(16):3103–4. doi: 10.1242/dev.112003. PubMed PMID: 25100652. [DOI] [PubMed] [Google Scholar]
- 2422.Stainier DYR, Raz E, Lawson ND, et al. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017. Oct;13(10):e1007000. doi: 10.1371/journal.pgen.1007000. PubMed PMID: 29049395; PubMed Central PMCID: PMCPMC5648102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2423.Lai JKH, Gagalova KK, Kuenne C, et al. Induction of interferon-stimulated genes and cellular stress pathways by morpholinos in zebrafish. Dev Biol. 2019. Oct 1;454(1):21–28. doi: 10.1016/j.ydbio.2019.06.008. PubMed PMID: 31201802; PubMed Central PMCID: PMCPMC6717701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2424.Mastrodonato V, Beznoussenko G, Mironov A, et al. A genetic model of CEDNIK syndrome in zebrafish highlights the role of the SNARE protein Snap29 in neuromotor and epidermal development. Sci Rep. 2019. Feb 4;9(1):1211. doi: 10.1038/s41598-018-37780-4. PubMed PMID: 30718891; PubMed Central PMCID: PMCPMC6361908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2425.Zhang R, Varela M, Vallentgoed W, et al. The selective autophagy receptors Optineurin and p62 are both required for zebrafish host resistance to mycobacterial infection. PLoS Pathog. 2019. Feb;15(2):e1007329. doi: 10.1371/journal.ppat.1007329. PubMed PMID: 30818338; PubMed Central PMCID: PMCPMC6413957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2426.Meneghetti G, Skobo T, Chrisam M, et al. The epg5 knockout zebrafish line: a model to study Vici syndrome. Autophagy. 2019. Aug;15(8):1438–1454. doi: 10.1080/15548627.2019.1586247. PubMed PMID: 30806141; PubMed Central PMCID: PMCPMC6613882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2427.Liu K, Petree C, Requena T, et al. Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front Cell Dev Biol. 2019;7:13. doi: 10.3389/fcell.2019.00013. PubMed PMID: 30886848; PubMed Central PMCID: PMCPMC6409501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2428.Hernandez-Moreno D, Blazquez M, Andreu-Sanchez O, et al. Acute hazard of biocides for the aquatic environmental compartment from a life-cycle perspective. Sci Total Environ. 2019. Mar 25;658:416–423. doi: 10.1016/j.scitotenv.2018.12.186. PubMed PMID: 30579199. [DOI] [PubMed] [Google Scholar]
- 2429.Moreau K, Fleming A, Imarisio S, et al. PICALM modulates autophagy activity and tau accumulation. Nat Commun. 2014;5:4998. doi: 10.1038/ncomms5998. PubMed PMID: 25241929; PubMed Central PMCID: PMC4199285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2430.Zhang Y, Nguyen DT, Olzomer EM, et al. Rescue of Pink1 Deficiency by Stress-Dependent Activation of Autophagy. Cell Chem Biol. 2017. Apr 20;24(4):471–480 e4. doi: 10.1016/j.chembiol.2017.03.005. PubMed PMID: 28366621. [DOI] [PubMed] [Google Scholar]
- 2431.Ruparelia AA, McKaige EA, Williams C, et al. Metformin rescues muscle function in BAG3 myofibrillar myopathy models, 2021 Autophagy, in press. doi: 10.1080/15548627.2020.1833500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2432.Ruparelia AA, Oorschot V, Ramm G, et al. FLNC myofibrillar myopathy results from impaired autophagy and protein insufficiency. Hum Mol Genet. 2016. Jun 1;25(11):2131–2142. doi: 10.1093/hmg/ddw080. PubMed PMID: 26969713. [DOI] [PubMed] [Google Scholar]
- 2433.Ruparelia AA, Oorschot V, Vaz R, et al. Zebrafish models of BAG3 myofibrillar myopathy suggest a toxic gain of function leading to BAG3 insufficiency. Acta Neuropathol. 2014. Dec;128(6):821–33. doi: 10.1007/s00401-014-1344-5. PubMed PMID: 25273835. [DOI] [PubMed] [Google Scholar]
- 2434.Mostowy S, Boucontet L, Mazon Moya MJ, et al. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS Pathog. 2013;9(9):e1003588. doi: 10.1371/journal.ppat.1003588. PubMed PMID: 24039575; PubMed Central PMCID: PMC3764221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2435.van der Vaart M, Korbee CJ, Lamers GE, et al. The DNA damage-regulated autophagy modulator DRAM1 links mycobacterial recognition via TLP-MYD88 to authophagic defense. Cell Host Microbe. 2014. Jun 11;15(6):753–67. doi: 10.1016/j.chom.2014.05.005. PubMed PMID: 24922577. [DOI] [PubMed] [Google Scholar]
- 2436.Espin-Palazon R, Martinez-Lopez A, Roca FJ, et al. TNFalpha impairs rhabdoviral clearance by inhibiting the host autophagic antiviral response. PLoS Pathog. 2016. Jun;12(6):e1005699. doi: 10.1371/journal.ppat.1005699. PubMed PMID: 27351838; PubMed Central PMCID: PMCPMC4924823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2437.Li YC, Zhang MQ, Zhang JP.. Opposite effects of two human ATG10 isoforms on replication of a HCV sub-genomic replicon are mediated via regulating autophagy flux in zebrafish. Front Cell Infect Microbiol. 2018;8:109. doi: 10.3389/fcimb.2018.00109. PubMed PMID: 29670865; PubMed Central PMCID: PMCPMC5893791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2438.Garcia-Valtanen P, Ortega-Villaizan Mdel M, Martinez-Lopez A, et al. Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy. 2014. Sep;10(9):1666–80. doi: 10.4161/auto.29557. PubMed PMID: 25046110; PubMed Central PMCID: PMCPMC4206542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2439.Varga M, Sass M, Papp D, et al. Autophagy is required for zebrafish caudal fin regeneration. Cell Death Differ. 2014. Apr;21(4):547–56. doi: 10.1038/cdd.2013.175. PubMed PMID: 24317199; PubMed Central PMCID: PMC3950318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2440.Benato F, Skobo T, Gioacchini G, et al. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy. 2013. Apr;9(4):476–95. doi: 10.4161/auto.23278. PubMed PMID: 23348054; PubMed Central PMCID: PMC3627665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2441.Skobo T, Benato F, Grumati P, et al. Zebrafish ambra1a and ambra1b knockdown impairs skeletal muscle development. PLoS One. 2014;9(6):e99210. doi: 10.1371/journal.pone.0099210. PubMed PMID: 24922546; PubMed Central PMCID: PMC4055674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2442.Driever W, Rangini Z.. Characterization of a cell line derived from zebrafish (Brachydanio rerio) embryos. In Vitro Cell Dev Biol Anim. 1993. Sep;29A(9):749–54. doi: 10.1007/bf02631432. PubMed PMID: 8407719. [DOI] [PubMed] [Google Scholar]
- 2443.Liu L, Zhu B, Wu S, et al. Spring viraemia of carp virus induces autophagy for necessary viral replication. Cell Microbiol. 2015. Apr;17(4):595–605. doi: 10.1111/cmi.12387. PubMed PMID: 25376386. [DOI] [PubMed] [Google Scholar]
- 2444.Bello-Perez M, Falco A, Novoa B, et al. Hydroxycholesterol binds and enhances the anti-viral activities of zebrafish monomeric c-reactive protein isoforms. PLoS One. 2019;14(1):e0201509. doi: 10.1371/journal.pone.0201509. PubMed PMID: 30653529; PubMed Central PMCID: PMCPMC6336239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2445.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. PubMed PMID: 19200873. [DOI] [PubMed] [Google Scholar]
- 2446.Herb M, Gluschko A, Schramm M.. LC3-associated phagocytosis - The highway to hell for phagocytosed microbes. Semin Cell Dev Biol. 2019. Apr 29. doi: 10.1016/j.semcdb.2019.04.016. PubMed PMID: 31029766. [DOI] [PubMed] [Google Scholar]
- 2447.Sil P, Muse G, Martinez J.. A ravenous defense: canonical and non-canonical autophagy in immunity. Curr Opin Immunol. 2018. Feb;50:21–31. doi: 10.1016/j.coi.2017.10.004. PubMed PMID: 29125936; PubMed Central PMCID: PMCPMC5857463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2448.Henault J, Martinez J, Riggs JM, et al. Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes. Immunity. 2012. Dec 14;37(6):986–97. doi: 10.1016/j.immuni.2012.09.014. PubMed PMID: 23219390; PubMed Central PMCID: PMC3786711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2449.Santarino IB, Viegas MS, Domingues NS, et al. Involvement of the p62/NRF2 signal transduction pathway on erythrophagocytosis. Sci Rep. 2017. Jul 19;7(1):5812. doi: 10.1038/s41598-017-05687-1. PubMed PMID: 28724916; PubMed Central PMCID: PMCPMC5517431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2450.Fazeli G, Wehman AM.. Safely removing cell debris with LC3-associated phagocytosis. Biol Cell. 2017. Oct;109(10):355–363. doi: 10.1111/boc.201700028. PubMed PMID: 28755428. [DOI] [PubMed] [Google Scholar]
- 2451.Fazeli G, Stetter M, Lisack JN, et al. C. elegans blastomeres clear the corpse of the second polar body by LC3-associated phagocytosis. Cell Rep. 2018. May 15;23(7):2070–2082. doi: 10.1016/j.celrep.2018.04.043. PubMed PMID: 29768205. [DOI] [PubMed] [Google Scholar]
- 2452.Abnave P, Mottola G, Gimenez G, et al. Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe. 2014. Sep 10;16(3):338–50. doi: 10.1016/j.chom.2014.08.002. PubMed PMID: 25211076. [DOI] [PubMed] [Google Scholar]
- 2453.Varma H, Gangadhar NM, Letso RR, et al. Identification of a small molecule that induces ATG5-and-cathepsin-l-dependent cell death and modulates polyglutamine toxicity. Exp Cell Res. 2013. Jul 15;319(12):1759–73. doi: 10.1016/j.yexcr.2013.03.019. PubMed PMID: 23588206; PubMed Central PMCID: PMC3700633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2454.Jacquin E, Leclerc-Mercier S, Judon C, et al. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy. 2017. May 4;13(5):854–867. doi: 10.1080/15548627.2017.1287653. PubMed PMID: 28296541; PubMed Central PMCID: PMCPMC5446083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2455.Jiang P, Mizushima N.. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015. Mar;75:13–8. doi: 10.1016/j.ymeth.2014.11.021. PubMed PMID: 25484342. [DOI] [PubMed] [Google Scholar]
- 2456.Yang CS, Lee JS, Rodgers M, et al. Autophagy protein Rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012. Mar 15;11(3):264–76. doi: 10.1016/j.chom.2012.01.018. PubMed PMID: 22423966; PubMed Central PMCID: PMCPMC3616771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2457.Martinez J, Cunha LD, Park S, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016. May 5;533(7601):115–9. doi: 10.1038/nature17950. PubMed PMID: 27096368; PubMed Central PMCID: PMCPMC4860026. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2458.Heckmann BL, Teubner BJW, Tummers B, et al. LC3-associated endocytosis facilitates beta-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell. 2019. Jul 25;178(3):536–551 e14. doi: 10.1016/j.cell.2019.05.056. PubMed PMID: 31257024; PubMed Central PMCID: PMCPMC6689199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2459.Rai S, Arasteh M, Jefferson M, et al. The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy. 2019. Apr;15(4):599–612. doi: 10.1080/15548627.2018.1534507. PubMed PMID: 30403914; PubMed Central PMCID: PMCPMC6526875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2460.Kong-Hap MA, Mouammine A, Daher W, et al. Regulation of ATG8 membrane association by ATG4 in the parasitic protist Toxoplasma gondii. Autophagy. 2013. Sep;9(9):1334–48. doi: 10.4161/auto.25189. PubMed PMID: 23748741. [DOI] [PubMed] [Google Scholar]
- 2461.Jayabalasingham B, Voss C, Ehrenman K, et al. Characterization of the ATG8-conjugation system in 2 Plasmodium species with special focus on the liver stage: possible linkage between the apicoplastic and autophagic systems? Autophagy. 2014. Feb;10(2):269–84. doi: 10.4161/auto.27166. PubMed PMID: 24342964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2462.Tomlins AM, Ben-Rached F, Williams RA, et al. Plasmodium falciparum ATG8 implicated in both autophagy and apicoplast formation. Autophagy. 2013. Oct;9(10):1540–52. doi: 10.4161/auto.25832. PubMed PMID: 24025672. [DOI] [PubMed] [Google Scholar]
- 2463.Mizushima N, Sahani MH.. ATG8 localization in apicomplexan parasites: apicoplast and more? Autophagy. 2014. Sep;10(9):1487–94. doi: 10.4161/auto.32183. PubMed PMID: 25102412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2464.Voss C, Ehrenman K, Mlambo G, et al. Overexpression of Plasmodium berghei ATG8 by Liver Forms Leads to Cumulative Defects in Organelle Dynamics and to Generation of Noninfectious Merozoites. mBio. 2016. Jun 28;7(3). doi: 10.1128/mBio.00682-16. PubMed PMID: 27353755; PubMed Central PMCID: PMCPMC4937212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2465.Leveque MF, Berry L, Cipriano MJ, et al. autophagy-related protein ATG8 has a noncanonical function for apicoplast inheritance in Toxoplasma gondii. mBio. 2015. Oct 27;6(6):e01446–15. doi: 10.1128/mBio.01446-15. PubMed PMID: 26507233; PubMed Central PMCID: PMCPMC4626856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2466.Nguyen HM, Liu S, Daher W, et al. Characterisation of two Toxoplasma PROPPINs homologous to Atg18/WIPI suggests they have evolved distinct specialised functions. PLoS One. 2018;13(4):e0195921. doi: 10.1371/journal.pone.0195921. PubMed PMID: 29659619; PubMed Central PMCID: PMCPMC5901921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2467.Bansal P, Tripathi A, Thakur V, et al. Autophagy-related protein ATG18 regulates apicoplast biogenesis in apicomplexan parasites. mBio. 2017. Oct 31;8(5). doi: 10.1128/mBio.01468-17. PubMed PMID: 29089429; PubMed Central PMCID: PMCPMC5666157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2468.Park S, Choi J, Biering SB, et al. Targeting by AutophaGy proteins (TAG): Targeting of IFNG-inducible GTPases to membranes by the LC3 conjugation system of autophagy. Autophagy. 2016. Jul 2;12(7):1153–67. doi: 10.1080/15548627.2016.1178447. PubMed PMID: 27172324; PubMed Central PMCID: PMCPMC4990996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2469.Sasai M, Sakaguchi N, Ma JS, et al. Essential role for GABARAP autophagy proteins in interferon-inducible GTPase-mediated host defense. Nat Immunol. 2017. Aug;18(8):899–910. doi: 10.1038/ni.3767. PubMed PMID: 28604719. [DOI] [PubMed] [Google Scholar]
- 2470.Haldar AK, Piro AS, Pilla DM, et al. The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to Chlamydia- and Toxoplasma-containing vacuoles and host resistance. PLoS One. 2014;9(1):e86684. doi: 10.1371/journal.pone.0086684. PubMed PMID: 24466199; PubMed Central PMCID: PMC3895038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2471.Ohshima J, Lee Y, Sasai M, et al. Role of mouse and human autophagy proteins in IFN-gamma-induced cell-autonomous responses against Toxoplasma gondii. J Iimmunol. 2014. Apr 1;192(7):3328–35. doi: 10.4049/jimmunol.1302822. PubMed PMID: 24563254. [DOI] [PubMed] [Google Scholar]
- 2472.Zhao YO, Khaminets A, Hunn JP, et al. Disruption of the Toxoplasma gondii parasitophorous vacuole by IFNgamma-inducible immunity-related GTPases (IRG proteins) triggers necrotic cell death. PLoS Pathog. 2009. Feb;5(2):e1000288. doi: 10.1371/journal.ppat.1000288. PubMed PMID: 19197351; PubMed Central PMCID: PMC2629126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2473.Meunier E, Dick MS, Dreier RF, et al. Caspase-11 activation requires lysis of pathogen-containing vacuoles by IFN-induced GTPases. Nature. 2014. May 15;509(7500):366–70. doi: 10.1038/nature13157. PubMed PMID: 24739961. [DOI] [PubMed] [Google Scholar]
- 2474.Taguchi Y, Imaoka K, Kataoka M, et al. Yip1A, a novel host factor for the activation of the IRE1 pathway of the unfolded protein response during Brucella infection. PLoS Pathog. 2015. Mar;11(3):e1004747. doi: 10.1371/journal.ppat.1004747. PubMed PMID: 25742138; PubMed Central PMCID: PMC4350842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2475.Starr T, Child R, Wehrly TD, et al. Selective subversion of autophagy complexes facilitates completion of the Brucella intracellular cycle. Cell Host Microbe. 2012. Jan 19;11(1):33–45. doi: 10.1016/j.chom.2011.12.002. PubMed PMID: 22264511; PubMed Central PMCID: PMC3266535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2476.Manjithaya R, Anjard C, Loomis WF, et al. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J Cell Biol. 2010. Feb 22;188(4):537–46. doi: 10.1083/jcb.200911149. PubMed PMID: 20156962; PubMed Central PMCID: PMCPMC2828923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2477.Duran JM, Anjard C, Stefan C, et al. Unconventional secretion of Acb1 is mediated by auto-phagosomes. J Cell Biol. 2010. Feb 22;188(4):527–36. doi:jcb.200911154 [pii] doi:10.1083/jcb. 200911154. PubMed PMID: 20156967; PubMed Central PMCID: PMC2828925. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2478.Zhang M, Kenny SJ, Ge L, et al. Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. eLife. 2015. Nov 2;4. doi: 10.7554/eLife.11205. PubMed PMID: 26523392; PubMed Central PMCID: PMCPMC4728131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2479.Gee HY, Noh SH, Tang BL, et al. Rescue of DeltaF508-CFTR trafficking via a GRASP-dependent unconventional secretion pathway. Cell. 2011. Sep 2;146(5):746–60. doi: 10.1016/j.cell.2011.07.021. PubMed PMID: 21884936. [DOI] [PubMed] [Google Scholar]
- 2480.Curwin AJ, Brouwers N, Alonso YAM, et al. ESCRT-III drives the final stages of CUPS maturation for unconventional protein secretion. eLife. 2016. Apr 26;5. doi: 10.7554/eLife.16299. PubMed PMID: 27115345; PubMed Central PMCID: PMCPMC4868542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2481.Noh SH, Gee HY, Kim Y, et al. Specific autophagy and ESCRT components participate in the unconventional secretion of CFTR. Autophagy. 2018;14(10):1761–1778. doi: 10.1080/15548627.2018.1489479. PubMed PMID: 29969945; PubMed Central PMCID: PMCPMC6135621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2482.Robinson SM, Tsueng G, Sin J, et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014. Apr;10(4):e1004045. doi: 10.1371/journal.ppat.1004045. PubMed PMID: 24722773; PubMed Central PMCID: PMCPMC3983045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2483.Nowag H, Guhl B, Thriene K, et al. Macroautophagy proteins assist epstein barr virus production and get incorporated into the virus particles. EBioMedicine. 2014. Dec;1(2–3):116–25. doi: 10.1016/j.ebiom.2014.11.007. PubMed PMID: 26137519; PubMed Central PMCID: PMCPMC4457436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2484.Chen YH, Du W, Hagemeijer MC, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015. Feb 12;160(4):619–630. doi: 10.1016/j.cell.2015.01.032. PubMed PMID: 25679758; PubMed Central PMCID: PMCPMC6704014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2485.Mehta P, Henault J, Kolbeck R, et al. Noncanonical autophagy: one small step for LC3, one giant leap for immunity. Curr Opin Immunol. 2014. Feb;26:69–75. doi: 10.1016/j.coi.2013.10.012. PubMed PMID: 24556403. [DOI] [PubMed] [Google Scholar]
- 2486.Scarlatti F, Maffei R, Beau I, et al. Non-canonical autophagy: an exception or an underestimated form of autophagy? Autophagy. 2008. Nov 16;4(8):1083–5. doi:7068 [pii]. PubMed PMID: 18849663; eng. [DOI] [PubMed] [Google Scholar]
- 2487.Takeshita F, Kobiyama K, Miyawaki A, et al. The non-canonical role of Atg family members as suppressors of innate antiviral immune signaling. Autophagy. 2008. Jan;4(1):67–9. PubMed PMID: 17921696. [DOI] [PubMed] [Google Scholar]
- 2488.Deretic V, Jiang S, Dupont N.. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 2012. Aug;22(8):397–406. doi: 10.1016/j.tcb.2012.04.008. PubMed PMID: 22677446; PubMed Central PMCID: PMC3408825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2489.Cleyrat C, Darehshouri A, Steinkamp MP, et al. Mpl traffics to the cell surface through conventional and unconventional routes. Traffic. 2014. Sep;15(9):961–82. doi: 10.1111/tra.12185. PubMed PMID: 24931576; PubMed Central PMCID: PMC4141020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2490.Hughes T, Rusten TE.. Origin and evolution of self-consumption: autophagy. Adv Exp Med Biol. 2007;607:111–8. doi: 10.1007/978-0-387-74021-8_9. PubMed PMID: 17977463. [DOI] [PubMed] [Google Scholar]
- 2491.Kiel JA. Autophagy in unicellular eukaryotes. Philos Trans R Soc London, Ser B. 2010. Mar 12;365(1541):819–30. doi: 10.1098/rstb.2009.0237. PubMed PMID: 20124347; PubMed Central PMCID: PMC2817228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2492.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997. Sep 1;25(17):3389–402. PubMed PMID: 9254694; PubMed Central PMCID: PMC146917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2493.Pertsemlidis A, Fondon JW, III.. Having a BLAST with bioinformatics (and avoiding BLASTphemy). Genome Biol. 2001;2(10):REVIEWS2002. PubMed PMID: 11597340; PubMed Central PMCID: PMC138974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2494.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999. Feb;12(2):85–94. PubMed PMID: 10195279. [DOI] [PubMed] [Google Scholar]
- 2495.Duszenko M, Ginger ML, Brennand A, et al. Autophagy in protists. Autophagy. 2011. Feb;7(2):127–58. PubMed PMID: 20962583; PubMed Central PMCID: PMC3039767. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2496.Rigden DJ, Michels PA, Ginger ML.. Autophagy in protists: Examples of secondary loss, lineage-specific innovations, and the conundrum of remodeling a single mitochondrion. Autophagy. 2009. Aug;5(6):784–94. PubMed PMID: 19483474. [DOI] [PubMed] [Google Scholar]
- 2497.Katsani KR, Irimia M, Karapiperis C, et al. Functional genomics evidence unearths new moonlighting roles of outer ring coat nucleoporins. Sci Rep. 2014;4:4655. doi: 10.1038/srep04655. PubMed PMID: 24722254; PubMed Central PMCID: PMC3983603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2498.Mei Y, Su M, Soni G, et al. Intrinsically disordered regions in autophagy proteins. Proteins. 2014. Apr;82(4):565–78. doi: 10.1002/prot.24424. PubMed PMID: 24115198; PubMed Central PMCID: PMC3949125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2499.Promponas VJ, Ouzounis CA, Iliopoulos I.. Experimental evidence validating the computational inference of functional associations from gene fusion events: a critical survey. Brief Bioinform. 2014. May;15(3):443–54. doi: 10.1093/bib/bbs072. PubMed PMID: 23220349; PubMed Central PMCID: PMC4017328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2500.Promponas VJ, Iliopoulos I, Ouzounis CA.. Annotation inconsistencies beyond sequence similarity-based function prediction - phylogeny and genome structure. Stand Genomic Sci. 2015;10:108. doi: 10.1186/s40793-015-0101-2. PubMed PMID: 26594309; PubMed Central PMCID: PMCPMC4653902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2501.Deng W, Ma L, Zhang Y, et al. THANATOS: an integrative data resource of proteins and post-translational modifications in the regulation of autophagy. Autophagy. 2018;14(2):296–310. doi: 10.1080/15548627.2017.1402990. PubMed PMID: 29157087; PubMed Central PMCID: PMCPMC5902229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2502.Chen LL, Wang YB, Song JX, et al. Phosphoproteome-based kinase activity profiling reveals the critical role of MAP2K2 and PLK1 in neuronal autophagy. Autophagy. 2017;13(11):1969–1980. doi: 10.1080/15548627.2017.1371393. PubMed PMID: 28933595; PubMed Central PMCID: PMCPMC5788482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2503.Zhang Y, Xie Y, Liu W, et al. DeepPhagy: a deep learning framework for quantitatively measuring autophagy activity in Saccharomyces cerevisiae. Autophagy. 2019. Jun 20:1–15. doi: 10.1080/15548627.2019.1632622. PubMed PMID: 31204567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2504.Li S, Shui K, Zhang Y, et al. CGDB: a database of circadian genes in eukaryotes. Nucleic Acids Res. 2017. Jan 4;45(D1):D397–D403. doi: 10.1093/nar/gkw1028. PubMed PMID: 27789706; PubMed Central PMCID: PMCPMC5210527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2505.Homma K, Suzuki K, Sugawara H.. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 2011. Jan;39(Database issue):D986–90. doi: 10.1093/nar/gkq995. PubMed PMID: 20972215; PubMed Central PMCID: PMC3013813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2506.Turei D, Foldvari-Nagy L, Fazekas D, et al. Autophagy Regulatory Network - a systems-level bioinformatics resource for studying the mechanism and regulation of autophagy. Autophagy. 2015;11(1):155–65. doi: 10.4161/15548627.2014.994346. PubMed PMID: 25635527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2507.Birgisdottir AB, Lamark T, Johansen T.. The LIR motif - crucial for selective autophagy. J Cell Sci. 2013. Aug1;126(Pt 15):3237–47. doi: 10.1242/jcs.126128. PubMed PMID: 23908376. [DOI] [PubMed] [Google Scholar]
- 2508.Wild P, McEwan DG, Dikic I.. The LC3 interactome at a glance. J Cell Sci. 2014. Jan1;127(Pt 1):3–9. doi: 10.1242/jcs.140426. PubMed PMID: 24345374. [DOI] [PubMed] [Google Scholar]
- 2509.Noda NN, Ohsumi Y, Inagaki F.. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010. Jan 17;584:1379–85. doi:S0014-5793(10)00037-2 [pii] doi: 10.1016/j.febslet.2010.01.018. PubMed PMID: 20083108; Eng. [DOI] [PubMed] [Google Scholar]
- 2510.Kalvari I, Tsompanis S, Mulakkal NC, et al. iLIR: A web resource for prediction of Atg8-family interacting proteins. Autophagy. 2014. May;10(5):913–25. doi: 10.4161/auto.28260. PubMed PMID: 24589857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2511.Dosztanyi Z, Meszaros B, Simon I.. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009. Oct 15;25(20):2745–6. doi: 10.1093/bioinformatics/btp518. PubMed PMID: 19717576; PubMed Central PMCID: PMC2759549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2512.Xie Q, Tzfadia O, Levy M, et al. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy. 2016. May 3;12(5):876–87. doi: 10.1080/15548627.2016.1147668. PubMed PMID: 27071037; PubMed Central PMCID: PMCPMC4854547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2513.Jacomin AC, Samavedam S, Promponas V, et al. iLIR database: A web resource for LIR motif-containing proteins in eukaryotes. Autophagy. 2016. Oct 2;12(10):1945–1953. doi: 10.1080/15548627.2016.1207016. PubMed PMID: 27484196; PubMed Central PMCID: PMCPMC5079668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2514.Jacomin AC, Samavedam S, Charles H, et al. iLIR@viral: A web resource for LIR motif-containing proteins in viruses. Autophagy. 2017. Oct 3;13(10):1782–1789. doi: 10.1080/15548627.2017.1356978. PubMed PMID: 28806134; PubMed Central PMCID: PMCPMC5640201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2515.Dinkel H, Van Roey K, Michael S, et al. The eukaryotic linear motif resource ELM: 10 years and counting. Nucleic Acids Res. 2014. Jan;42(Database issue):D259–66. doi: 10.1093/nar/gkt1047. PubMed PMID: 24214962; PubMed Central PMCID: PMC3964949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2516.Di Rita A, Peschiaroli A, P DA, et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKalpha. Nat Commun. 2018. Sep 14;9(1):3755. doi: 10.1038/s41467-018-05722-3. PubMed PMID: 30217973; PubMed Central PMCID: PMCPMC6138665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2517.Jatana N, Ascher DB, Pires DEV, et al. Human LC3 and GABARAP subfamily members achieve functional specificity via specific structural modulations. Autophagy. 2020. Feb;16(2):239–255. doi: 10.1080/15548627.2019.1606636. PubMed PMID: 30982432; PubMed Central PMCID: PMCPMC6984608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2518.Wu D, Huang Y, Kang JJ, et al. ncRDeathDB: a comprehensive bioinformatics resource for deciphering network organization of the ncRNA-mediated cell death system. Autophagy. 2015;11:1917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2519.Li Y, Zhuang L, Wang Y, et al. Connect the dots: a systems level approach for analyzing the miRNA-mediated cell death network. Autophagy. 2013. Mar;9(3):436–9. doi: 10.4161/auto.23096. PubMed PMID: 23322033; PubMed Central PMCID: PMC3590271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2520.Xu J, Li YH.. miRDeathDB: a database bridging microRNAs and the programmed cell death. Cell Death Differ. 2012. Sep;19(9):1571. doi: 10.1038/cdd.2012.87. PubMed PMID: 22743998; PubMed Central PMCID: PMC3422482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2521.Xu J, Wang Y, Tan X, et al. MicroRNAs in autophagy and their emerging roles in crosstalk with apoptosis. Autophagy. 2012. Jun;8(6):873–82. doi: 10.4161/auto.19629. PubMed PMID: 22441107; PubMed Central PMCID: PMC3427253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2522.Delaney JR, Patel CB, Willis KM, et al. Haploinsufficiency networks identify targetable patterns of allelic deficiency in low mutation ovarian cancer. Nat Commun. 2017. Feb 15;8:14423. doi: 10.1038/ncomms14423. PubMed PMID: 28198375; PubMed Central PMCID: PMCPMC5316854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2523.Kaushik S, Cuervo AM.. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018. Jun;19(6):365–381. doi: 10.1038/s41580-018-0001-6. PubMed PMID: 29626215; PubMed Central PMCID: PMCPMC6399518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2524.Dice JF, Chiang HL, Spencer EP, et al. Regulation of catabolism of microinjected ribonuclease A. Identification of residues 7–11 as the essential pentapeptide. J Biol Chem. 1986. May 25;261(15):6853–9. PubMed PMID: 3700419. [PubMed] [Google Scholar]
- 2525.Morone D, Marazza A, Bergmann TJ, et al. Deep learning approach for quantification of organelles and misfolded polypeptide delivery within degradative compartments. Mol Biol Cell. 2020. Jul 1;31(14):1512–1524. doi: 10.1091/mbc.E20-04-0269. PubMed PMID: 32401604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2526.Börlin CS, Lang V, Hamacher-Brady A, et al. Agent-based modeling of autophagy reveals emergent regulatory behavior of spatio-temporal autophagy dynamics. Cell Commun Signal. 2014. Sep 10;12:56. doi: 10.1186/s12964-014-0056-8. PubMed PMID: 25214434; PubMed Central PMCID: PMCPMC4172826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2527.Martin KR, Barua D, Kauffman AL, et al. Computational model for autophagic vesicle dynamics in single cells. Autophagy. 2013. Jan;9(1):74–92. doi: 10.4161/auto.22532. PubMed PMID: 23196898; PubMed Central PMCID: PMC3542220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2528.Rubinsztein DC, Codogno P, Levine B.. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012. Sep;11(9):709–30. doi: 10.1038/nrd3802. PubMed PMID: 22935804; PubMed Central PMCID: PMCPMC3518431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2529.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002. Jan;2(1):28–36. doi: 10.1038/nri700. PubMed PMID: 11905835. [DOI] [PubMed] [Google Scholar]
- 2530.Kapuy O, Vinod PK, Mandl J, et al. A cellular stress-directed bistable switch controls the crosstalk between autophagy and apoptosis. Mol BioSyst. 2013. Feb 2;9(2):296–306. doi: 10.1039/c2mb25261a. PubMed PMID: 23223525. [DOI] [PubMed] [Google Scholar]
- 2531.Liu B, Oltvai ZN, Bayir H, et al. Quantitative assessment of cell fate decision between autophagy and apoptosis. Sci Rep. 2017. Dec 14;7(1):17605. doi: 10.1038/s41598-017-18001-w. PubMed PMID: 29242632; PubMed Central PMCID: PMCPMC5730598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2532.Tavassoly I, Parmar J, Shajahan-Haq AN, et al. Dynamic modeling of the interaction between autophagy and apoptosis in mammalian cells. CPT Pharmacometrics Syst Pharmacol. 2015. Apr;4(4):263–72. doi: 10.1002/psp4.29. PubMed PMID: 26225250; PubMed Central PMCID: PMC4429580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2533.Han K, Kim J, Choi M.. Autophagy mediates phase transitions from cell death to life. Heliyon. 2015. Sep;1(1):e00027. doi:ARTN e00027 doi: 10.1016/j.heliyon.2015.e00027. PubMed PMID: WOS:000432001200001; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2534.Jin H, Lei J.. A hybrid model of molecular regulation and population dynamics for yeast autophagy. J Theor Biol. 2016. Aug 7;402:45–53. doi: 10.1016/j.jtbi.2016.04.019. PubMed PMID: 27103581. [DOI] [PubMed] [Google Scholar]
- 2535.Szymańska P, Martin KR, MacKeigan JP, et al. Computational analysis of an autophagy/translation switch based on mutual inhibition of MTORC1 and ULK1. PLoS One. 2015;10(3):e0116550. doi: 10.1371/journal.pone.0116550. PubMed PMID: 25761126; PubMed Central PMCID: PMCPMC4356596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2536.Bolliet V, Labonne J, Olazcuaga L, et al. Modeling of autophagy-related gene expression dynamics during long term fasting in European eel (Anguilla anguilla). Sci Rep. 2017. Dec 20;7(1):17896. doi: 10.1038/s41598-017-18164-6. PubMed PMID: 29263413; PubMed Central PMCID: PMCPMC5738402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2537.Dalmasso G, Marin Zapata PA, Brady NR, et al. Agent-based modeling of mitochondria links sub-cellular dynamics to cellular homeostasis and heterogeneity. PLoS One. 2017;12(1):e0168198. doi: 10.1371/journal.pone.0168198. PubMed PMID: 28060865; PubMed Central PMCID: PMCPMC5217980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2538.Hoffman TE, Barnett KJ, Wallis L, et al. A multimethod computational simulation approach for investigating mitochondrial dynamics and dysfunction in degenerative aging. Aging cell. 2017. Dec;16(6):1244–1255. doi: 10.1111/acel.12644. PubMed PMID: WOS:000418387600004; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2539.Brown AI, Rutenberg AD.. A model of autophagy size selectivity by receptor clustering on peroxisomes. 2. 2017. May 19;5:14. doi: ARTN 14 doi: 10.3389/fphy.2017.00014. PubMed PMID: WOS:000403720300001; English. [DOI] [Google Scholar]
- 2540.Shirin A, Klickstein IS, Feng S, et al. Prediction of optimal drug schedules for controlling autophagy. Sci Rep. 2019. Feb 5;9(1):1428. doi: 10.1038/s41598-019-38763-9. PubMed PMID: 30723233; PubMed Central PMCID: PMCPMC6363771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2541.Munsky B, Hlavacek WS, Tsimring LS, editors. Quantitative Biology: Theory, Computational Methods, and Models. Cambridge, MA: The MIT Press; 2018. [Google Scholar]
- 2542.Bergmann FT, Hoops S, Klahn B, et al. COPASI and its applications in biotechnology. J Biotechnol. 2017. Nov 10;261:215–220. doi: 10.1016/j.jbiotec.2017.06.1200. PubMed PMID: 28655634; PubMed Central PMCID: PMCPMC5623632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2543.Binder B, Goede A, Berndt N, et al. A conceptual mathematical model of the dynamic self-organisation of distinct cellular organelles. PLoS One. 2009. Dec 30;4(12):e8295. doi: 10.1371/journal.pone.0008295. PubMed PMID: 20041124; PubMed Central PMCID: PMCPMC2795802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2544.Chylek LA, Harris LA, Tung C-S, et al. Rule-based modeling: a computational approach for studying biomolecular site dynamics in cell signaling systems. Wiley Interdiscip Rev Syst Biol Med. 2014. Jan-Feb;6(1):13–36. doi: 10.1002/wsbm.1245. PubMed PMID: 24123887; PubMed Central PMCID: PMCPMC3947470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2545.Kapuy O, Papp D, Vellai T, et al. Systems-Level Feedbacks of NRF2 Controlling Autophagy upon Oxidative Stress Response. Antioxidants (Basel). 2018. Mar 5;7(3). doi: 10.3390/antiox7030039. PubMed PMID: 29510589; PubMed Central PMCID: PMCPMC5874525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2546.Kapuy O, Vinod PK, Banhegyi G.. mTOR inhibition increases cell viability via autophagy induction during endoplasmic reticulum stress - An experimental and modeling study. FEBS Open Bio. 2014;4:704–13. doi: 10.1016/j.fob.2014.07.006. PubMed PMID: 25161878; PubMed Central PMCID: PMCPMC4141208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2547.Schwartz-Roberts JL, Cook KL, Chen C, et al. Interferon regulatory factor-1 signaling regulates the switch between autophagy and apoptosis to determine breast cancer cell fate. Cancer Res. 2015. Mar 15;75(6):1046–55. doi: 10.1158/0008-5472.CAN-14-1851. PubMed PMID: 25576084; PubMed Central PMCID: PMCPMC4359953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2548.Ouzounoglou E, Kalamatianos D, Emmanouilidou E, et al. In silico modeling of the effects of alpha-synuclein oligomerization on dopaminergic neuronal homeostasis. BMC Syst Biol. 2014. May 13;8:54. doi: 10.1186/1752-0509-8-54. PubMed PMID: 24885905; PubMed Central PMCID: PMCPMC4062111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2549.Holczer M, Marton M, Kurucz A, et al. A Comprehensive Systems Biological Study of Autophagy-Apoptosis Crosstalk during Endoplasmic Reticulum Stress. Biomed Res Int. 2015;2015:319589. doi: 10.1155/2015/319589. PubMed PMID: 25984530; PubMed Central PMCID: PMCPMC4423012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2550.Petri C. Kommunikation mit Automaten. Technische Universitat Darmstadt, Germany: 1962. [Google Scholar]
- 2551.Scheidel J, Amstein L, Ackermann J, et al. In Silico Knockout Studies of Xenophagic Capturing of Salmonella. PLoS Comput Biol. 2016. Dec;12(12):e1005200. doi: 10.1371/journal.pcbi.1005200. PubMed PMID: 27906974; PubMed Central PMCID: PMCPMC5131900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2552.Janes KA, Chandran PL, Ford RM, et al. An engineering design approach to systems biology. Integr Biol. 2017. Jul 17;9(7):574–583. doi: 10.1039/c7ib00014f. PubMed PMID: 28590470; PubMed Central PMCID: PMCPMC6534349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2553.Gunawardena J. Models in biology: ‘accurate descriptions of our pathetic thinking’. BMC Biol. 2014. Apr 30;12:29. doi: 10.1186/1741-7007-12-29. PubMed PMID: 24886484; PubMed Central PMCID: PMCPMC4005397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2554.Lander AD. The edges of understanding. BMC Biol. 2010. Apr 12;8:40. doi: 10.1186/1741-7007-8-40. PubMed PMID: 20385033; PubMed Central PMCID: PMCPMC2864098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2555.Tyson JJ, Novák B.. Models in biology: lessons from modeling regulation of the eukaryotic cell cycle. BMC Biol. 2015. Jul 1;13:46. doi: 10.1186/s12915-015-0158-9. PubMed PMID: 26129844; PubMed Central PMCID: PMCPMC4486427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2556.Hucka M, Bergmann FT, Hoops S, et al. The Systems Biology Markup Language (SBML): language specification for Level 3 Version 1 Core. J Integr Bioinform. 2015. Sep 4;12(2):266. doi: 10.2390/biecoll-jib-2015-266. PubMed PMID: 26528564; PubMed Central PMCID: PMCPMC5451324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2557.Zhang F, Meier-Schellersheim M.. SBML Level 3 package: Multistate, Multicomponent and Multicompartment Species, Version 1, Release 1. J Integr Bioinform. 2018. Apr 20;15(1):20170077. doi: 10.1515/jib-2017-0077. PubMed PMID: 29676994; PubMed Central PMCID: PMCPMC6167033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2558.Ewald R, Uhrmacher AM.. SESSL: a domain-specific language for simulation experiments. Acm T Model Comput S. 2014. Feb;24(2):11. doi: Artn 11 doi: 10.1145/2567895. PubMed PMID: WOS:000334526100005; English. [DOI] [Google Scholar]
- 2559.Waltemath D, Adams R, Bergmann FT, et al. Reproducible computational biology experiments with SED-ML—the Simulation Experiment Description Markup Language. BMC Syst Biol. 2011. Dec 15;5:198. doi: 10.1186/1752-0509-5-198. PubMed PMID: 22172142; PubMed Central PMCID: PMCPMC3292844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2560.Chelliah V, Juty N, Ajmera I, et al. BioModels: ten-year anniversary. Nucleic Acids Res. 2015. Jan;43(Database issue):D542–8. doi: 10.1093/nar/gku1181. PubMed PMID: 25414348; PubMed Central PMCID: PMCPMC4383975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2561.Azeloglu EU, Iyengar R.. Good practices for building dynamical models in systems biology. Sci Signal. 2015. Apr 7;8(371):fs8. doi: 10.1126/scisignal.aab0880. PubMed PMID: 25852187. [DOI] [PubMed] [Google Scholar]
- 2562.Gutenkunst RN, Waterfall JJ, Casey FP, et al. Universally sloppy parameter sensitivities in systems biology models. PLOS Comput Biol. 2007. Oct;3(10):1871–78. doi: 10.1371/journal.pcbi.0030189. PubMed PMID: 17922568; PubMed Central PMCID: PMCPMC2000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2563.Kirk PDW, Babtie AC, Stumpf MPH.. Systems biology (un)certainties. Science. 2015. Oct 23;350(6259):386–8. doi: 10.1126/science.aac9505. PubMed PMID: 26494748. [DOI] [PubMed] [Google Scholar]
- 2564.Wilkinson DJ. Bayesian methods in bioinformatics and computational systems biology. Brief Bioinform. 2007. Mar;8(2):109–16. doi: 10.1093/bib/bbm007. PubMed PMID: 17430978. [DOI] [PubMed] [Google Scholar]
- 2565.Kreutz C, Raue A, Kaschek D, et al. Profile likelihood in systems biology. FEBS J. 2013. Jun;280(11):2564–71. doi: 10.1111/febs.12276. PubMed PMID: 23581573. [DOI] [PubMed] [Google Scholar]
- 2566.Hellerstein JL, Gu S, Choi K, et al. Recent advances in biomedical simulations: a manifesto for model engineering. F1000Res. 2019;8:261. doi: 10.12688/f1000research.15997.1. PubMed PMID: 30881691; PubMed Central PMCID: PMCPMC6406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2567.Medley JK, Goldberg AP, Karr JR.. Guidelines for reproducibly building and simulating systems biology models. IEEE Trans Biomed Eng. 2016. Oct;63(10):2015–20. doi: 10.1109/TBME.2016.2591960. PubMed PMID: 27429432; PubMed Central PMCID: PMCPMC5131863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2568.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits, Second Edition. 2nd Edition ed. Boca Raton, FL: Chapman and Hall/CRC; 2019. [Google Scholar]
- 2569.Voit EO. A First Course in Systems Biology. 2nd Edition ed. New York: Garland Science; 2017. [Google Scholar]
- 2570.Resnekov O, Munsky B, Hlavacek WS.. Perspective on the q-bio Summer School and Conference: 2007–2014 and beyond. Quant Biol. 2014. Mar 1;2(1):54–58. doi: 10.1007/s40484-014-0029-3. PubMed PMID: 27595041; PubMed Central PMCID: PMCPMC5008235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2571.Knorr RL, Dimova R, Lipowsky R.. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One. 2012;7(3):e32753. doi: 10.1371/journal.pone.0032753. PubMed PMID: 22427874; PubMed Central PMCID: PMCPMC3299685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2572.Agudo-Canalejo J, Knorr RL.. Formation of auto-phagosomes coincides with relaxation of membrane curvature. Methods Mol Biol. 2019;1880:173–188. doi: 10.1007/978-1-4939-8873-0_10. PubMed PMID: 30610696. [DOI] [PubMed] [Google Scholar]
- 2573.Klionsky DJ, Baehrecke EH, Brumell JH, et al. A comprehensive glossary of autophagy-related molecules and processes (2nd) edition). Autophagy. 2011. Nov 1;7(11):1273–94. PubMed PMID: 21997368; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2574.Klionsky DJ, Codogno P, Cuervo AM, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010. May 16;6(4):438–448. PubMed PMID: 20484971; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2575.Moosavi MA, Haghi A, Rahmati M, et al. Phytochemicals as potent modulators of autophagy for cancer therapy. Cancer Lett. 2018. Jun 28;424:46–69. doi: 10.1016/j.canlet.2018.02.030. PubMed PMID: 29474859. [DOI] [PubMed] [Google Scholar]
- 2576.Quintana M, Bilbao A, Comas-Barcelo J, et al. Identification of benzo[cd]indol-2(1H)-ones as novel Atg4B inhibitors via a structure-based virtual screening and a novel AlphaScreen assay. Eur J Med Chem. 2019. Sep 15;178:648–666. doi: 10.1016/j.ejmech.2019.05.086. PubMed PMID: 31226656. [DOI] [PubMed] [Google Scholar]
- 2577.Santo L, Hideshima T, Kung AL, et al. Preclinical activity, pharmacodynamic, and pharmacokinetic properties of a selective HDAC6 inhibitor, ACY-1215, in combination with bortezomib in multiple myeloma. Blood. 2012. Mar 15;119(11):2579–89. doi: 10.1182/blood-2011-10-387365. PubMed PMID: 22262760; PubMed Central PMCID: PMCPMC3337713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2578.Garcia-Echeverria C. Allosteric and ATP-competitive kinase inhibitors of mTOR for cancer treatment. Bioorg Med Chem Lett. 2010. Aug 1;20(15):4308–12. doi: 10.1016/j.bmcl.2010.05.099. PubMed PMID: 20561789. [DOI] [PubMed] [Google Scholar]
- 2579.Rosich L, Xargay-Torrent S, Lopez-Guerra M, et al. Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. clin cancer res off j am assoc cancer res. 2012. Oct 1;18(19):5278–89. doi: 10.1158/1078-0432.CCR-12-0351. PubMed PMID: 22879389. [DOI] [PubMed] [Google Scholar]
- 2580.Booth L, Roberts JL, Ecroyd H, et al. AR-12 inhibits multiple chaperones concomitant with stimulating autophagosome formation collectively preventing virus replication. J Cell Physiol. 2016. Oct;231(10):2286–302. doi: 10.1002/jcp.25431. PubMed PMID: 27187154; PubMed Central PMCID: PMCPMC6327852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2581.Anguiano J, Garner TP, Mahalingam M, et al. Chemical modulation of chaperone-mediated autophagy by retinoic acid derivatives. Nat Chem Biol. 2013. Jun;9(6):374–82. doi: 10.1038/nchembio.1230. PubMed PMID: 23584676; PubMed Central PMCID: PMC3661710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2582.De Mei C, Ercolani L, Parodi C, et al.Dual inhibition of REV-ERBbeta and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene. 2015. May 14;34(20):2597–608. doi: 10.1038/onc.2014.203. PubMed PMID: 25023698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2583.Li M, Sala V, De Santis MC, et al. Phosphoinositide 3-kinase gamma inhibition protects from anthracycline cardiotoxicity and reduces tumor growth. Circulation. 2018. Aug 14;138(7):696–711. doi: 10.1161/CIRCULATIONAHA.117.030352. PubMed PMID: 29348263. [DOI] [PubMed] [Google Scholar]
- 2584.Fujita N, Hayashi-Nishino M, Fukumoto H, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008. Nov;19(11):4651–9. doi: 10.1091/mbc.E08-03-0312. PubMed PMID: 18768752; PubMed Central PMCID: PMC2575160. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2585.Robke L, Laraia L, Carnero Corrales MA, et al. Phenotypic identification of a novel autophagy inhibitor chemotype targeting lipid kinase VPS34. Angew Chem Int Ed Engl. 2017. Jul 3;56(28):8153–8157. doi: 10.1002/anie.201703738. PubMed PMID: 28544137. [DOI] [PubMed] [Google Scholar]
- 2586.Zhuang X, Wang H, Lam SK, et al. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell. 2013. Nov;25(11):4596–615. doi: 10.1105/tpc.113.118307. PubMed PMID: 24249832; PubMed Central PMCID: PMCPMC3875738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2587.Ansari MY, Ahmad N, Haqqi TM.. Butein activates autophagy through AMPK/TSC2/ULK1/mTOR pathway to inhibit IL-6 expression in IL-1beta stimulated human chondrocytes. Cell Physiol Biochem. 2018;49(3):932–946. doi: 10.1159/000493225. PubMed PMID: 30184535. [DOI] [PubMed] [Google Scholar]
- 2588.Lyamzaev KG, Tokarchuk AV, Panteleeva AA, et al. Induction of autophagy by depolarization of mitochondria. Autophagy. 2018;14(5):921–924. doi: 10.1080/15548627.2018.1436937. PubMed PMID: 29458285; PubMed Central PMCID: PMCPMC6070013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2589.Filadi R, Pizzo P.. Defective autophagy and Alzheimer’s disease: is calcium the key? Neural Regen Res. 2019. Dec;14(12):2081–2082. doi: 10.4103/1673-5374.262584. PubMed PMID: 31397341; PubMed Central PMCID: PMCPMC6788238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2590.Williams RS, Cheng L, Mudge AW, et al. A common mechanism of action for three mood-stabilizing drugs. Nature. 2002. May 16;417(6886):292–5. doi: 10.1038/417292a. PubMed PMID: 12015604. [DOI] [PubMed] [Google Scholar]
- 2591.Parzych K, Saavedra-Garcia P, Valbuena GN, et al. The coordinated action of VCP/p97 and GCN2 regulates cancer cell metabolism and proteostasis during nutrient limitation. Oncogene. 2019. Apr;38(17):3216–3231. doi: 10.1038/s41388-018-0651-z. PubMed PMID: 30626938; PubMed Central PMCID: PMCPMC6756015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2592.Anderson DJ, Le Moigne R, Djakovic S, et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer cell. 2015. Nov 9;28(5):653–665. doi: 10.1016/j.ccell.2015.10.002. PubMed PMID: 26555175; PubMed Central PMCID: PMCPMC4941640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2593.Kane MS, Paris A, Codron P, et al. Current mechanistic insights into the CCCP-induced cell survival response. Biochem Pharmacol. 2018. Feb;148:100–110. doi: 10.1016/j.bcp.2017.12.018. PubMed PMID: 29277693. [DOI] [PubMed] [Google Scholar]
- 2594.Mattar P, Bravo-Sagua R, Tobar N, et al. Autophagy mediates calcium-sensing receptor-induced TNFalpha production in human preadipocytes. Biochim Biophys Acta Mol Basis Dis. 2018. Nov;1864(11):3585–3594. doi: 10.1016/j.bbadis.2018.08.020. PubMed PMID: 30251678. [DOI] [PubMed] [Google Scholar]
- 2595.Lim JH, Kim HW, Kim MY, et al. Cinacalcet-mediated activation of the CaMKKbeta-LKB1-AMPK pathway attenuates diabetic nephropathy in db/db mice by modulation of apoptosis and autophagy. Cell Death Dis. 2018. Feb 15;9(3):270. doi: 10.1038/s41419-018-0324-4. PubMed PMID: 29449563; PubMed Central PMCID: PMCPMC5833853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2596.Vanrell MC, Cueto JA, Barclay JJ, et al. Polyamine depletion inhibits the autophagic response modulating Trypanosoma cruzi infectivity. Autophagy. 2013. Jul;9(7):1080–93. doi: 10.4161/auto.24709. PubMed PMID: 23697944; PubMed Central PMCID: PMC3722317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2597.D’Eliseo D, Di Renzo L, Santoni A, et al. Docosahexaenoic acid (DHA) promotes immunogenic apoptosis in human multiple myeloma cells, induces autophagy and inhibits STAT3 in both tumor and dendritic cells. Genes Cancer. 2017. Jan;8(1–2): 426–437. doi: 10.18632/genesandcancer.131. PubMed PMID: 28435516; PubMed Central PMCID: PMCPMC5396621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2598.Jing K, Song KS, Shin S, et al. Docosahexaenoic acid induces autophagy through p53/AMPK/mTOR signaling and promotes apoptosis in human cancer cells harboring wild-type p53. Autophagy. 2011. Nov;7(11):1348–58. doi: 10.4161/auto.7.11.16658. PubMed PMID: 21811093; PubMed Central PMCID: PMCPMC3242799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2599.Mildenberger J, Johansson I, Sergin I, et al. N-3 PUFAs induce inflammatory tolerance by formation of KEAP1-containing SQSTM1/p62-bodies and activation of NFE2L2. Autophagy. 2017. Oct 3;13(10):1664–1678. doi: 10.1080/15548627.2017.1345411. PubMed PMID: 28820283; PubMed Central PMCID: PMCPMC5640206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2600.Zhou X, Yue GG, Chan AM, et al. Eriocalyxin B, a novel autophagy inducer, exerts anti-tumor activity through the suppression of Akt/mTOR/p70S6K signaling pathway in breast cancer. Biochem Pharmacol. 2017. Oct 15;142:58–70. doi: 10.1016/j.bcp.2017.06.133. PubMed PMID: 28669564. [DOI] [PubMed] [Google Scholar]
- 2601.Yamamura T, Ohsaki Y, Suzuki M, et al. Inhibition of Niemann-Pick-type C1-like1 by ezetimibe activates autophagy in human hepatocytes and reduces mutant alpha1-antitrypsin Z deposition. Hepatology. 2014. Apr;59(4):1591–9. doi: 10.1002/hep.26930. PubMed PMID: 24214142. [DOI] [PubMed] [Google Scholar]
- 2602.Kim SH, Kim G, Han DH, et al. Ezetimibe ameliorates steatohepatitis via AMP activated protein kinase-TFEB-mediated activation of autophagy and NLRP3 inflammasome inhibition. Autophagy. 2017Oct 3;13(10):1767–1781. doi: 10.1080/15548627.2017.1356977. PubMed PMID: 28933629; PubMed Central PMCID: PMCPMC5640190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2603.Liu FT, Yang YJ, Wu JJ, et al. Fasudil, a Rho kinase inhibitor, promotes the autophagic degradation of A53T alpha-synuclein by activating the JNK 1/Bcl-2/beclin 1 pathway. Brain Res. 2016. Feb 1;1632:9–18. doi: 10.1016/j.brainres.2015.12.002. PubMed PMID: 26683082. [DOI] [PubMed] [Google Scholar]
- 2604.Gao H, Hou F, Dong R, et al. Rho-Kinase inhibitor fasudil suppresses high glucose-induced H9c2 cell apoptosis through activation of autophagy. Cardiovasc Ther. 2016. Oct;34(5):352–9. doi: 10.1111/1755-5922.12206. PubMed PMID: 27333569. [DOI] [PubMed] [Google Scholar]
- 2605.Koch JC, Tonges L, Barski E, et al. ROCK2 is a major regulator of axonal degeneration, neuronal death and axonal regeneration in the CNS. Cell Death Dis. 2014. May 15;5:e1225. doi: 10.1038/cddis.2014.191. PubMed PMID: 24832597; PubMed Central PMCID: PMCPMC4047920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2606.Prieto-Dominguez N, Garcia-Mediavilla MV, Sanchez-Campos S, et al. Autophagy as a molecular target of flavonoids underlying their protective effects in human disease. Curr Med Chem. 2018;25(7):814–838. doi: 10.2174/0929867324666170918125155. PubMed PMID: 28925866. [DOI] [PubMed] [Google Scholar]
- 2607.Zheng Q, Li Z, Zhou S, et al. Heparin-binding hemagglutinin of mycobacterium tuberculosis is an inhibitor of autophagy. Front Cell Infect Microbiol. 2017;7:33. doi: 10.3389/fcimb.2017.00033. PubMed PMID: 28224118; PubMed Central PMCID: PMCPMC5293787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2608.Song W, Zukor H, Liberman A, et al. Astroglial heme oxygenase-1 and the origin of corpora amylacea in aging and degenerating neural tissues. Exp Neurol. 2014. Apr;254:78–89. doi: 10.1016/j.expneurol.2014.01.006. PubMed PMID: 24440642; PubMed Central PMCID: PMC4020524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2609.Song W, Zukor H, Lin SH, et al. Unregulated brain iron deposition in transgenic mice over-expressing HMOX1 in the astrocytic compartment. J Neurochem. 2012. Oct;123(2):325–36. doi: 10.1111/j.1471-4159.2012.07914.x. PubMed PMID: 22881289. [DOI] [PubMed] [Google Scholar]
- 2610.Garcia-Martinez JM, Moran J, Clarke RG, et al. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem J. 2009. Jul 1;421(1):29–42. doi: 10.1042/BJ20090489. PubMed PMID: 19402821; PubMed Central PMCID: PMC2708931. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2611.Bosc D, Vezenkov L, Bortnik S, et al. A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B. Sci Rep. 2018. Aug 3;8(1):11653. doi: 10.1038/s41598-018-29900-x. PubMed PMID: 30076329; PubMed Central PMCID: PMCPMC6076261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2612.San-Miguel B, Crespo I, Sanchez DI, et al. Melatonin inhibits autophagy and endoplasmic reticulum stress in mice with carbon tetrachloride-induced fibrosis. J Pineal Res. 2015. Sep;59(2):151–62. doi: 10.1111/jpi.12247. PubMed PMID: 25958928. [DOI] [PubMed] [Google Scholar]
- 2613.Ordoñez R, Fernandez A, Prieto-Dominguez N, et al. Ceramide metabolism regulates autophagy and apoptotic cell death induced by melatonin in liver cancer cells. J Pineal Res. 2015. Sep;59(2):178–89. doi: 10.1111/jpi.12249. PubMed PMID: 25975536; PubMed Central PMCID: PMCPMC4523438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2614.San-Miguel B, Crespo I, Vallejo D, et al. Melatonin modulates the autophagic response in acute liver failure induced by the rabbit hemorrhagic disease virus. J Pineal Res. 2014. Apr;56(3):313–21. doi: 10.1111/jpi.12124. PubMed PMID: 24499270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2615.Kanamori H, Naruse G, Yoshida A, et al. Metformin enhances autophagy and provides cardioprotection in delta-sarcoglycan deficiency-induced dilated cardiomyopathy. Circ Heart Fail. 2019. Apr;12(4):e005418. doi: 10.1161/CIRCHEARTFAILURE.118.005418. PubMed PMID: 30922066. [DOI] [PubMed] [Google Scholar]
- 2616.Wang Y, Yang Z, Zheng G, et al. Metformin promotes autophagy in ischemia/reperfusion myocardium via cytoplasmic AMPKalpha1 and nuclear AMPKalpha2 pathways. Life Sci. 2019. May 15;225:64–71. doi: 10.1016/j.lfs.2019.04.002. PubMed PMID: 30953640. [DOI] [PubMed] [Google Scholar]
- 2617.Zilinyi R, Czompa A, Czegledi A, et al. The cardioprotective effect of metformin in doxorubicin-induced cardiotoxicity: the role of autophagy. Molecules. 2018. May 15;23(5). doi: 10.3390/molecules23051184. PubMed PMID: 29762537; PubMed Central PMCID: PMCPMC6100061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2618.Pal R, Xiong Y, Sardiello M.. Abnormal glycogen storage in tuberous sclerosis complex caused by impairment of mTORC1-dependent and -independent signaling pathways. Proc Natl Acad Sci U S A. 2019. Feb 19;116(8):2977–2986 . doi: 10.1073/pnas.1812943116. PubMed PMID: 30728291; PubMed Central PMCID: PMCPMC6386676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2619.Soucy TA, Smith PG, Milhollen MA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009. Apr 9;458(7239):732–6. doi: 10.1038/nature07884. PubMed PMID: 19360080. [DOI] [PubMed] [Google Scholar]
- 2620.Luo Z, Yu G, Lee HW, et al. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012. Jul 1;72(13):3360–71. doi: 10.1158/0008-5472.CAN-12-0388. PubMed PMID: 22562464. [DOI] [PubMed] [Google Scholar]
- 2621.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008. Oct 1;68(19):8022–30. doi: 10.1158/0008-5472.CAN-08-1385. PubMed PMID: 18829560; eng. [DOI] [PubMed] [Google Scholar]
- 2622.Liu TJ, Koul D, LaFortune T, et al. NVP-BEZ235, a novel dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor, elicits multifaceted antitumor activities in human gliomas. Mol Cancer Ther. 2009. Aug;8(8):2204–10. doi:10.1158/1535-7163. MCT-09-0160. PubMed PMID: 19671762; PubMed Central PMCID: PMC2752877. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2623.Wang F, Bonam SR, Schall N, et al. Blocking nuclear export of HSPA8 after heat shock stress severely alters cell survival. Sci Rep. 2018. Nov 14;8(1):16820. doi: 10.1038/s41598-018-34887-6. PubMed PMID: 30429537; PubMed Central PMCID: PMCPMC6235846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2624.Page N, Schall N, Strub JM, et al. The spliceosomal phosphopeptide P140 controls the lupus disease by interacting with the HSC70 protein and via a mechanism mediated by gammadelta T cells. PLoS One. 2009;4(4):e5273. doi: 10.1371/journal.pone.0005273. PubMed PMID: 19390596; PubMed Central PMCID: PMCPMC2669294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2625.Macri C, Wang F, Tasset I, et al. Modulation of deregulated chaperone-mediated autophagy by a phosphopeptide. Autophagy. 2015;11(3):472–86. doi: 10.1080/15548627.2015.1017179. PubMed PMID: 25719862; PubMed Central PMCID: PMCPMC4502742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2626.Jabir MS, Sulaiman GM, Taqi ZJ, et al. Iraqi propolis increases degradation of IL-1beta and NLRC4 by autophagy following Pseudomonas aeruginosa infection. Microbes Infect. 2018. Feb;20(2):89–100. doi: 10.1016/j.micinf.2017.10.007. PubMed PMID: 29104144. [DOI] [PubMed] [Google Scholar]
- 2627.Pirola L, Frojdo S.. Resveratrol: one molecule, many targets. IUBMB life. 2008. May;60(5):323–32. doi: 10.1002/iub.47. PubMed PMID: 18421779. [DOI] [PubMed] [Google Scholar]
- 2628.Vingtdeux V, Giliberto L, Zhao H, et al. AMP-activated protein kinase signaling activation by resveratrol modulates amyloid-beta peptide metabolism. J Biol Chem. 2010. Mar 19;285(12):9100–13. doi: 10.1074/jbc.M109.060061. PubMed PMID: 20080969; PubMed Central PMCID: PMC2838330. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2629.Puissant A, Auberger P.. AMPK- and p62/SQSTM1-dependent autophagy mediate Resveratrol-induced cell death in chronic myelogenous leukemia. Autophagy. 2010. Jul 1;6(5):655–7. PubMed PMID: 20458181; Eng. [DOI] [PubMed] [Google Scholar]
- 2630.Vingtdeux V, Chandakkar P, Zhao H, et al. Novel synthetic small-molecule activators of AMPK as enhancers of autophagy and amyloid-[b] peptide degradation. FASEB J. 2011. Jan;25(1):219–31. doi: 10.1096/fj.10-167361. PubMed PMID: 20852062; PubMed Central PMCID: PMC3005419. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2631.Dyczynski M, Yu Y, Otrocka M, et al. Targeting autophagy by small molecule inhibitors of vacuolar protein sorting 34 (Vps34) improves the sensitivity of breast cancer cells to Sunitinib. Cancer Lett. 2018. Oct 28;435:32–43. doi: 10.1016/j.canlet.2018.07.028. PubMed PMID: 30055290. [DOI] [PubMed] [Google Scholar]
- 2632.Tang F, Hu P, Yang Z, et al. SBI0206965, a novel inhibitor of Ulk1, suppresses non-small cell lung cancer cell growth by modulating both autophagy and apoptosis pathways. Oncol Rep. 2017. Jun;37(6):3449–3458. doi: 10.3892/or.2017.5635. PubMed PMID: 28498429. [DOI] [PubMed] [Google Scholar]
- 2633.Dite TA, Langendorf CG, Hoque A, et al. AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965. J Biol Chem. 2018. Jun 8;293(23):8874–8885. doi: 10.1074/jbc.RA118.003547. PubMed PMID: 29695504; PubMed Central PMCID: PMCPMC5995511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2634.Rodriguez-Hernandez MA, Gonzalez R, de la Rosa AJ, et al. Molecular characterization of autophagic and apoptotic signaling induced by sorafenib in liver cancer cells. J Cell Physiol. 2018. Jan;234(1):692–708. doi: 10.1002/jcp.26855. PubMed PMID: 30132846. [DOI] [PubMed] [Google Scholar]
- 2635.Liu J, Xia H, Kim M, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011. Sep 30;147(1):223–34. doi: 10.1016/j.cell.2011.08.037. PubMed PMID: 21962518; PubMed Central PMCID: PMC3441147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2636.Milczarek M, Wiktorska K, Mielczarek L, et al. Corrigendum to ‘Autophagic cell death and premature senescence: New mechanism of 5-fluorouracil and sulforaphane synergistic anticancer effect in MDA-MB-231 triple negative breast cancer cell line’ Food Chem Toxicol. Food Chem Toxicol. 111 (2018) 1–8. 2018. Aug;118:972. doi: 10.1016/j.fct.2018.04.037. PubMed PMID: 29731172. [DOI] [PubMed] [Google Scholar]
- 2637.Yang F, Wang F, Liu Y, et al. Sulforaphane induces autophagy by inhibition of HDAC6-mediated PTEN activation in triple negative breast cancer cells. Life Sci. 2018. Nov 15;213:149–157. doi: 10.1016/j.lfs.2018.10.034. PubMed PMID: 30352240. [DOI] [PubMed] [Google Scholar]
- 2638.Liu Y, Shoji-Kawata S, Sumpter RM, Jr., et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013. Dec 17;110(51):20364–71. doi: 10.1073/pnas.1319661110. PubMed PMID: 24277826; PubMed Central PMCID: PMC3870705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2639.Gordon PB, Holen I, Fosse M, et al. Dependence of hepatocytic autophagy on intracellularly sequestered calcium [In Vitro Research Support, Non-U.S. Gov’t]. J Biol Chem. 1993. Dec 15;268(35):26107–12. PubMed PMID: 8253727; eng. [PubMed] [Google Scholar]
- 2640.Ganley IG, Wong PM, Gammoh N, et al. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell. 2011. Jun 24;42(6):731–43. doi: 10.1016/j.molcel.2011.04.024. PubMed PMID: 21700220; PubMed Central PMCID: PMC3124681. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2641.Zhang L, Dai F, Cui L, et al. Novel role for TRPC4 in regulation of macroautophagy by a small molecule in vascular endothelial cells. Biochim Biophys Acta. 2015. Feb;1853(2):377–87. doi: 10.1016/j.bbamcr.2014.10.030. PubMed PMID: 25476892. [DOI] [PubMed] [Google Scholar]
- 2642.Rodrigues D, Viotto AC, Checchia R, et al. Mechanism of Aloe Vera extract protection against UVA: shelter of lysosomal membrane avoids photodamage. Photochem Photobiol Sci Off J Eur Photochem Assoc Eur Soc Photobiol. 2016. Mar;15(3):334–50. doi: 10.1039/c5pp00409h. PubMed PMID: 26815913. [DOI] [PubMed] [Google Scholar]
- 2643.Casarejos MJ, Solano RM, Gomez A, et al. The accumulation of neurotoxic proteins, induced by proteasome inhibition, is reverted by trehalose, an enhancer of autophagy, in human neuroblastoma cells. Neurochem Int. 2011. Mar;58(4):512–20. doi: 10.1016/j.neuint.2011.01.008. PubMed PMID: 21232572; eng. [DOI] [PubMed] [Google Scholar]
- 2644.Fernandez-Estevez MA, Casarejos MJ, Lopez Sendon J, et al. Trehalose reverses cell malfunction in fibroblasts from normal and Huntington’s disease patients caused by proteosome inhibition. PLoS One. 2014;9(2):e90202. doi: 10.1371/journal.pone.0090202. PubMed PMID: 24587280; PubMed Central PMCID: PMCPMC3934989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2645.Lotfi P, Tse DY, Di Ronza A, et al. Trehalose reduces retinal degeneration, neuroinflammation and storage burden caused by a lysosomal hydrolase deficiency. Autophagy. 2018;14(8):1419–1434. doi: 10.1080/15548627.2018.1474313. PubMed PMID: 29916295; PubMed Central PMCID: PMCPMC6103706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2646.Carpenter JE, Jackson W, Benetti L, et al. Autophagosome formation during varicella-zoster virus Infection following endoplasmic reticulum stress and the unfolded protein response. J Virol. 2011. Sep;85(18):9414–24. doi: 10.1128/JVI.00281-11. PubMed PMID: 21752906; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2647.Lu Y, Dong S, Hao B, et al. Vacuolin-1 potently and reversibly inhibits autophagosome-lysosome fusion by activating RAB5A. Autophagy. 2014;10(11):1895–905. doi: 10.4161/auto.32200. PubMed PMID: 25483964; PubMed Central PMCID: PMC4502727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2648.Donohue E, Tovey A, Vogl AW, et al. Inhibition of autophagosome formation by the benzoporphyrin derivative verteporfin. J Biol Chem 2011;286:7290–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2649.de Munck E, Palomo V, Munoz-Saez E, et al. Small GSK-3 Inhibitor Shows Efficacy in a Motor Neuron Disease Murine Model Modulating Autophagy. PLoS One. 2016;11(9):e0162723. doi: 10.1371/journal.pone.0162723. PubMed PMID: 27631495; PubMed Central PMCID: PMCPMC5025054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2650.Bago R, Malik N, Munson MJ, et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J. 2014. Nov 1;463(3):413–27. doi: 10.1042/BJ20140889. PubMed PMID: 25177796; PubMed Central PMCID: PMCPMC4209782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2651.Korsnes MS. Yessotoxin as a tool to study induction of multiple cell death pathways. Toxins (Basel). 2012. Jul;4(7):568–79. doi: 10.3390/toxins4070568. PubMed PMID: 22852069; PubMed Central PMCID: PMCPMC3407893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2652.Kijanska M, Dohnal I, Reiter W, et al. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy. 2010. Nov;6(8):1168–78. PubMed PMID: 20953146. [DOI] [PubMed] [Google Scholar]
- 2653.Papinski D, Schuschnig M, Reiter W, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014. Feb 6;53(3):471–83. doi: 10.1016/j.molcel.2013.12.011. PubMed PMID: 24440502; PubMed Central PMCID: PMC3978657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2654.Weerasekara VK, Panek DJ, Broadbent DG, et al. Metabolic-stress-induced rearrangement of the 14-3-3zeta interactome promotes autophagy via a ULK1- and AMPK-regulated 14-3-3zeta interaction with phosphorylated Atg9. Mol Cell Biol. 2014. Dec;34(24):4379–88. doi: 10.1128/MCB.00740-14. PubMed PMID: 25266655; PubMed Central PMCID: PMCPMC4248729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2655.Kamada Y, Yoshino K, Kondo C, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010. Feb;30(4):1049–58. doi: 10.1128/MCB.01344-09. PubMed PMID: 19995911; PubMed Central PMCID: PMC2815578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2656.Stephan JS, Yeh YY, Ramachandran V, et al. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009. Oct 6;106(40):17049–54. doi: 10.1073/pnas.0903316106. PubMed PMID: 19805182; PubMed Central PMCID: PMC2761351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2657.Wold MS, Lim J, Lachance V, et al. ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington’s disease models. Mol Neurodegener. 2016. Dec 9;11(1):76. doi: 10.1186/s13024-016-0141-0. PubMed PMID: 27938392; PubMed Central PMCID: PMCPMC5148922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2658.Wei Y, An Z, Zou Z, et al. The stress-responsive kinases MAPKAPK2/MAPKAPK3 activate starvation-induced autophagy through Beclin 1 phosphorylation. eLife. 2015;4. doi: 10.7554/eLife.05289. PubMed PMID: 25693418; PubMed Central PMCID: PMC4337728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2659.Vega-Rubin-de-Celis S, Zou Z, Fernandez AF, et al. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc Natl Acad Sci U S A. 2018. Apr 17;115(16):4176–4181. doi: 10.1073/pnas.1717800115. PubMed PMID: 29610308; PubMed Central PMCID: PMCPMC5910832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2660.Wang BJ, Her GM, Hu MK, et al. ErbB2 regulates autophagic flux to modulate the proteostasis of APP-CTFs in Alzheimer’s disease. Proc Natl Acad Sci U S A. 2017. Apr 11;114(15):E3129–E3138. doi: 10.1073/pnas.1618804114. PubMed PMID: 28351972; PubMed Central PMCID: PMCPMC5393216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2661.Kratter IH, Zahed H, Lau A, et al. Serine 421 regulates mutant huntingtin toxicity and clearance in mice. J Clin Invest. 2016. Sep 1;126(9):3585–97. doi: 10.1172/JCI80339. PubMed PMID: 27525439; PubMed Central PMCID: PMCPMC5004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2662.Nave BT, Ouwens M, Withers DJ, et al. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J. 1999. Dec1;344Pt 2:427–31. PubMed PMID: 10567225; PubMed Central PMCID: PMC1220660. [PMC free article] [PubMed] [Google Scholar]
- 2663.Peterson RT, Beal PA, Comb MJ, et al. FKBP12-rapamycin-associated protein (FRAP) autophosphorylates at serine 2481 under translationally repressive conditions. J Biol Chem 2000. Mar10;275(10):7416–23. PubMed PMID: 10702316. [DOI] [PubMed] [Google Scholar]
- 2664.Nicot AS, Lo Verso F, Ratti F, et al. Phosphorylation of NBR1 by GSK3 modulates protein aggregation. Autophagy. 2014. Jun;10(6):1036–53. doi: 10.4161/auto.28479. PubMed PMID: 24879152; PubMed Central PMCID: PMC4091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2665.Rosner M, Fuchs C, Siegel N, et al. Functional interaction of mammalian target of rapamycin complexes in regulating mammalian cell size and cell cycle. Hum Mol Genet. 2009. Sep 1;18(17):3298–310. doi: 10.1093/hmg/ddp271. PubMed PMID: 19505958; PubMed Central PMCID: PMC2722991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2666.Shin S, Wolgamott L, Yu Y, et al. Glycogen synthase kinase (GSK)-3 promotes p70 ribosomal protein S6 kinase (p70S6K) activity and cell proliferation. Proc Natl Acad Sci U S A. 2011. Nov 22;108(47):E1204–13. doi: 10.1073/pnas.1110195108. PubMed PMID: 22065737; PubMed Central PMCID: PMC3223461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2667.Cheng X, Ma X, Ding X, et al. Pacer mediates the function of class III PI3K and HOPS complexes in autophagosome maturation by engaging Stx17. Mol Cell. 2017. Mar 16;65(6):1029–1043 e5. doi: 10.1016/j.molcel.2017.02.010. PubMed PMID: 28306502. [DOI] [PubMed] [Google Scholar]
- 2668.Ro SH, Semple IA, Park H, et al. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J. 2014. Jul 4. doi: 10.1111/febs.12905. PubMed PMID: 25040165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2669.Roczniak-Ferguson A, Petit CS, Froehlich F, et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012. Jun 12;5(228):ra42. doi: 10.1126/scisignal.2002790. PubMed PMID: 22692423; PubMed Central PMCID: PMCPMC3437338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2670.Wang Z, Gong Y, Peng B, et al. MRE11 UFMylation promotes ATM activation. Nucleic Acids Res. 2019. May 7;47(8):4124–4135. doi: 10.1093/nar/gkz110.PubMed PMID: 30783677; PubMed Central PMCID: PMCPMC6486557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2671.Kim YM, Jung CH, Seo M, et al. mTORC1 phosphorylates UVRAG to negatively regulate autophagosome and endosome maturation. Mol Cell. 2015. Jan 22;57(2):207–18. doi: 10.1016/j.molcel.2014.11.013. PubMed PMID: 25533187; PubMed Central PMCID: PMC4304967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2672.Munson MJ, Allen GF, Toth R, et al. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J. 2015. Jul 2. doi: 10.15252/embj.201590992. PubMed PMID: 26139536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2673.Lorente M, Garcia-Casas A, Salvador N, et al. Inhibiting SUMO1-mediated SUMOylation induces autophagy-mediated cancer cell death and reduces tumour cell invasion via RAC1. J Cell Sci. 2019. Oct 22;132(20). doi: 10.1242/jcs.234120. PubMed PMID: 31578236; PubMed Central PMCID: PMCPMC6826015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2674.Chen Y, Azad MB, Gibson SB.. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009. Jul;16(7):1040–52. doi: 10.1038/cdd.2009.49. PubMed PMID: 19407826. [DOI] [PubMed] [Google Scholar]
- 2675.Lorin S, Borges A, Ribeiro Dos Santos L, et al. c-Jun NH2-terminal kinase activation is essential for DRAM-dependent induction of autophagy and apoptosis in 2-methoxyestradiol-treated Ewing sarcoma cells. Cancer Res. 2009. Sep 1;69(17):6924–31. doi: 10.1158/0008-5472.CAN-09-1270. PubMed PMID: 19706754. [DOI] [PubMed] [Google Scholar]
- 2676.Xue L, Fletcher GC, Tolkovsky AM.. Autophagy is activated by apoptotic signalling in sympathetic neurons: an alternative mechanism of death execution. Mol Cell Neurosci. 1999. Sep;14(3):180–98. PubMed PMID: 10576889; eng. [DOI] [PubMed] [Google Scholar]
- 2677.Ge D, Han L, Huang S, et al. Identification of a novel MTOR activator and discovery of a competing endogenous RNA regulating autophagy in vascular endothelial cells. Autophagy. 2014. Jun;10(6):957–71. doi: 10.4161/auto.28363. PubMed PMID: 24879147; PubMed Central PMCID: PMCPMC4091179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2678.Zhang N, Chen Y, Jiang R, et al.PARP and RIP 1 are required for autophagy induced by 11ʹ-deoxyverticillin A, which precedes caspase-dependent apoptosis. Autophagy. 2011. Jun;7(6):598–612. PubMed PMID: 21460625. [DOI] [PubMed] [Google Scholar]
- 2679.Radoshevich L, Murrow L, Chen N, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010. Aug 20;142(4):590–600. doi: 10.1016/j.cell.2010.07.018. PubMed PMID: 20723759; PubMed Central PMCID: PMC2925044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2680.Leonardi M, Perna E, Tronnolone S, et al. Activated kinase screening identifies the IKBKE oncogene as a positive regulator of autophagy. Autophagy. 2019. Feb;15(2):312–326. doi: 10.1080/15548627.2018.1517855. PubMed PMID: 30289335; PubMed Central PMCID: PMCPMC6333447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2681.Maiuri MC, Criollo A, Tasdemir E, et al. BH3-only proteins and BH3 mimetics induce autophagy by competitively disrupting the interaction between Beclin 1 and Bcl-2/Bcl-X(L). Autophagy. 2007. Jul-Aug;3(4):374–6. PubMed PMID: 17438366; eng. [DOI] [PubMed] [Google Scholar]
- 2682.Oltersdorf T, Elmore SW, Shoemaker AR, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005. Jun 2;435(7042):677–81. doi: 10.1038/nature03579. PubMed PMID: 15902208. [DOI] [PubMed] [Google Scholar]
- 2683.Erazo T, Lorente M, Lopez-Plana A, et al. The New Antitumor Drug ABTL0812 Inhibits the Akt/mTORC1 Axis by Upregulating Tribbles-3 Pseudokinase. clin cancer res off j am assoc cancer res. 2016. May 15;22(10):2508–19. doi: 10.1158/1078-0432.CCR-15-1808. PubMed PMID: 26671995. [DOI] [PubMed] [Google Scholar]
- 2684.Felip I, Moiola CP, Megino-Luque C, et al. Therapeutic potential of the new TRIB3-mediated cell autophagy anticancer drug ABTL0812 in endometrial cancer. Gynecol Oncol. 2019. May;153(2):425–435. doi: 10.1016/j.ygyno.2019.03.002. PubMed PMID: 30853360. [DOI] [PubMed] [Google Scholar]
- 2685.Costello JL, Castro IG, Hacker C, et al. ACBD5 and VAPB mediate membrane associations between peroxisomes and the ER. J Cell Biol. 2017. Feb;216(2):331–342. doi: 10.1083/jcb.201607055. PubMed PMID: 28108524; PubMed Central PMCID: PMCPMC5294785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2686.Nazarko TY. Atg37 regulates the assembly of the pexophagic receptor protein complex. Autophagy. 2014. Jul;10(7):1348–9. doi: 10.4161/auto.29073. PubMed PMID: 24905344; PubMed Central PMCID: PMC4203562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2687.Kawaguchi K, Kikuma T, Higuchi Y, et al. Subcellular localization of acyl-CoA binding protein in Aspergillus oryzae is regulated by autophagy machinery. Biochem Biophys Res Commun. 2016. Nov 4;480(1):8–12. doi: 10.1016/j.bbrc.2016.10.018. PubMed PMID: 27725156. [DOI] [PubMed] [Google Scholar]
- 2688.Eisenberg T, Schroeder S, Andryushkova A, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014. Mar 4;19(3):431–44. doi: 10.1016/j.cmet.2014.02.010. PubMed PMID: 24606900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2689.Marino G, Pietrocola F, Eisenberg T, et al. Regulation of autophagy by cytosolic acetyl-coenzyme a. Mol Cell. 2014. Mar 6;53(5):710–25. doi: 10.1016/j.molcel.2014.01.016. PubMed PMID: 24560926. [DOI] [PubMed] [Google Scholar]
- 2690.Nandi N, Tyra LK, Stenesen D, et al. Acinus integrates AKT1 and subapoptotic caspase activities to regulate basal autophagy. J Cell Biol. 2014. Oct 27;207(2):253–68. doi: 10.1083/jcb.201404028. PubMed PMID: 25332163; PubMed Central PMCID: PMC4210446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2691.Nandi N, Tyra LK, Stenesen D, et al.Stress-induced Cdk5 activity enhances cytoprotective basal autophagy in Drosophila melanogaster by phosphorylating acinus at serine(437). eLife. 2017. Dec 11;6. doi: 10.7554/eLife.30760. PubMed PMID: 29227247; PubMed Central PMCID: PMCPMC5760206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2692.Haberman AS, Akbar MA, Ray S, et al. Drosophila acinus encodes a novel regulator of endocytic and autophagic trafficking. Development. 2010. Jul;137(13):2157–66. doi: 10.1242/dev.044230. PubMed PMID: 20504956; PubMed Central PMCID: PMC2882135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2693.Grevengoed TJ, Cooper DE, Young PA, et al. Loss of long-chain acyl-CoA synthetase isoform 1 impairs cardiac autophagy and mitochondrial structure through mechanistic target of rapamycin complex 1 activation. FASEB J. 2015. Nov;29(11):4641–53. doi: 10.1096/fj.15-272732. PubMed PMID: 26220174; PubMed Central PMCID: PMCPMC4608904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2694.Yoshikawa Y, Ogawa M, Hain T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009. Oct;11(10):1233–40. doi: 10.1038/ncb1967. PubMed PMID: 19749745. [DOI] [PubMed] [Google Scholar]
- 2695.Vekaria PH, Kumar A, Subramaniam D, et al. Functional cooperativity of p97 and histone deacetylase 6 in mediating DNA repair in mantle cell lymphoma cells. Leukemia. 2019. Jul;33(7):1675–1686. doi: 10.1038/s41375-018-0355-y. PubMed PMID: 30664664; PubMed Central PMCID: PMCPMC6730676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2696.Till A, Lipinski S, Ellinghaus D, et al. Autophagy receptor CALCOCO2/NDP52 takes center stage in Crohn disease. Autophagy. 2013. Aug;9(8):1256–7. doi: 10.4161/auto.25483. PubMed PMID: 23820297; PubMed Central PMCID: PMC3748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2697.Chung SJ, Nagaraju GP, Nagalingam A, et al. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy. 2017. Aug 3;13(8):1386–1403. doi: 10.1080/15548627.2017.1332565. PubMed PMID: 28696138; PubMed Central PMCID: PMCPMC5584870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2698.Braden CR, Neufeld TP.. Atg1-independent induction of autophagy by the Drosophila Ulk3 homolog, ADUK. FEBS J. 2016. Nov;283(21):3889–3897. doi: 10.1111/febs.13906. PubMed PMID: 27717182; PubMed Central PMCID: PMCPMC5123689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2699.Eby KG, Rosenbluth JM, Mays DJ, et al. ISG20L1 is a p53 family target gene that modulates genotoxic stress-induced autophagy. Mol cancer 2010;9:95. doi: 1476-4598-9-95 [pii] doi: 10.1186/1476-4598-9-95. PubMed PMID: 20429933; PubMed Central PMCID: PMC2873442. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2700.Kang R, Tang D, Livesey KM, et al. The Receptor for Advanced Glycation End-products (RAGE) protects pancreatic tumor cells against oxidative injury. Antioxid Redox Signal. 2011. Oct 15;15(8):2175–84. doi: 10.1089/ars.2010.3378. PubMed PMID: 21126167; PubMed Central PMCID: PMC3166176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2701.Uchiki T, Weikel KA, Jiao W, et al. Glycation-altered proteolysis as a pathobiologic mechanism that links dietary glycemic index, aging, and age-related disease (in nondiabetics). Aging cell. 2012. Feb;11(1):1–13. doi: 10.1111/j.1474-9726.2011.00752.x. PubMed PMID: 21967227; PubMed Central PMCID: PMCPMC3257376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2702.Johnston JA, Ward CL, Kopito RR.. Aggresomes: a cellular response to misfolded proteins. J Cell Biol. 1998. Dec 28;143(7):1883–98. PubMed PMID: 9864362; PubMed Central PMCID: PMC2175217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2703.Kwon YT, Ciechanover A.. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. 2017. Nov;42(11):873–886. doi: 10.1016/j.tibs.2017.09.002. PubMed PMID: 28947091. [DOI] [PubMed] [Google Scholar]
- 2704.Shen D, Coleman J, Chan E, et al. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem Biophys. 2011. Jul;60(3):173–85. doi: 10.1007/s12013-010-9138-4. PubMed PMID: 21132543; PubMed Central PMCID: PMCPMC3112480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2705.Viana R, Aguado C, Esteban I, et al. Role of AMP-activated protein kinase in autophagy and proteasome function. Biochem Biophys Res Commun. 2008. May 9;369(3):964–8. 10.1016/j.bbrc.2008.02.126. PubMed PMID: 18328803; eng. [DOI] [PubMed] [Google Scholar]
- 2706.Pardossi-Piquard R, Petit A, Kawarai T, et al. Presenilin-dependent transcriptional control of the Abeta-degrading enzyme neprilysin by intracellular domains of betaAPP and APLP. Neuron. 2005. May 19;46(4):541–54. doi: 10.1016/j.neuron.2005.04.008. PubMed PMID: 15944124. [DOI] [PubMed] [Google Scholar]
- 2707.Goiran T, Duplan E, Chami M, et al. beta-amyloid precursor protein intracellular domain controls mitochondrial function by modulating phosphatase and tensin homolog-induced kinase 1 transcription in cells and in Alzheimer mice models. Biol Psychiatry. 2018. Mar 1;83(5):416–427. doi: 10.1016/j.biopsych.2017.04.011. PubMed PMID: 28587718. [DOI] [PubMed] [Google Scholar]
- 2708.Hadano S, Otomo A, Kunita R, et al. Loss of ALS2/Alsin exacerbates motor dysfunction in a SOD1-expressing mouse ALS model by disturbing endolysosomal trafficking. PLoS One. 2010;5(3):e9805. doi: 10.1371/journal.pone.0009805. PubMed PMID: 20339559; PubMed Central PMCID: PMC2842444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2709.Otomo A, Kunita R, Suzuki-Utsunomiya K, et al. Defective relocalization of ALS2/alsin missense mutants to Rac1-induced macropinosomes accounts for loss of their cellular function and leads to disturbed amphisome formation. FEBS Lett. 2011. Mar 9;585(5):730–6. doi: 10.1016/j.febslet.2011.01.045. PubMed PMID: 21300063. [DOI] [PubMed] [Google Scholar]
- 2710.Antonioli M, Albiero F, Nazio F, et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. 2014. Dec 22;31(6):734–46. doi: 10.1016/j.devcel.2014.11.013. PubMed PMID: 25499913. [DOI] [PubMed] [Google Scholar]
- 2711.Cianfanelli V, Fuoco C, Lorente M, et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol. 2015. Jan;17(1):20–30. doi: 10.1038/ncb3072. PubMed PMID: 25438055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2712.Fu M, St-Pierre P, Shankar J, et al. Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Mol Biol Cell. 2013. Apr;24(8):1153–62. doi: 10.1091/mbc.E12-08-0607. PubMed PMID: 23427266; PubMed Central PMCID: PMC3623636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2713.Lee KY, Oh S, Choi YJ, et al. Activation of autophagy rescues amiodarone-induced apoptosis of lung epithelial cells and pulmonary toxicity in rats. Toxicol Sci. 2013. Nov;136(1):193–204. doi: 10.1093/toxsci/kft168. PubMed PMID: 23912912. [DOI] [PubMed] [Google Scholar]
- 2714.Mahavadi P, Knudsen L, Venkatesan S, et al. Regulation of macroautophagy in amiodarone-induced pulmonary fibrosis. J Pathol Clin Res. 2015. Oct;1(4):252–63. doi: 10.1002/cjp2.20. PubMed PMID: 27499909; PubMed Central PMCID: PMCPMC4939895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2715.Lin CW, Chen YS, Lin CC, et al. Amiodarone as an autophagy promoter reduces liver injury and enhances liver regeneration and survival in mice after partial hepatectomy. Sci Rep. 2015. Oct 30;5:15807. doi: 10.1038/srep15807. PubMed PMID: 26515640; PubMed Central PMCID: PMCPMC4626804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2716.Seglen PO, Berg TO, Blankson H, et al. Structural aspects of autophagy. Adv Exp Med Biol. 1996;389:103–11. PubMed PMID: 8860999; eng. [DOI] [PubMed] [Google Scholar]
- 2717.Meijer AJ, Codogno P.. AMP-activated protein kinase and autophagy. Autophagy. 2007. May-Jun;3(3):238–40. doi: 3710 [pii]. PubMed PMID: 17224623; eng. [DOI] [PubMed] [Google Scholar]
- 2718.Hutchins MU, Klionsky DJ.. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem. 2001. Jun 8;276(23):20491–8. doi: 10.1074/jbc.M101150200. PubMed PMID: 11264288; PubMed Central PMCID: PMCPMC2754691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2719.Katsiarimpa A, Anzenberger F, Schlager N, et al. The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell. 2011. Aug;23(8):3026–40. doi: 10.1105/tpc.111.087254. PubMed PMID: 21810997; PubMed Central PMCID: PMC3180808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2720.Katsiarimpa A, Kalinowska K, Anzenberger F, et al. The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell. 2013. Jun;25(6):2236–52. doi: 10.1105/tpc.113.113399. PubMed PMID: 23800962; PubMed Central PMCID: PMC3723623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2721.Freed EO, Gale M, Jr.. Antiviral innate immunity: editorial overview. J Mol Biol. 2014. Mar 20;426(6):1129–32. doi: 10.1016/j.jmb.2014.01.005. PubMed PMID: 24462565; PubMed Central PMCID: PMCPMC6943832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2722.Ke PY, Chen SS.. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011. Jan 4;121(1):37–56. doi: 10.1172/JCI41474. PubMed PMID: 21135505; PubMed Central PMCID: PMC3007134. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2723.Shrivastava S, Raychoudhuri A, Steele R, et al. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011. Feb;53(2):406–14. doi: 10.1002/hep.24073. PubMed PMID: 21274862; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2724.Kononenko NL, Classen GA, Kuijpers M, et al. Retrograde transport of TrkB-containing auto-phagosomes via the adaptor AP-2 mediates neuronal complexity and prevents neurodegeneration. Nat Commun. 2017. Apr 7;8:14819. doi: 10.1038/ncomms14819. PubMed PMID: 28387218; PubMed Central PMCID: PMCPMC5385568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2725.Verkerk AJ, Schot R, Dumee B, et al. Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am J Hum Genet. 2009. Jul;85(1):40–52. doi: 10.1016/j.ajhg.2009.06.004. PubMed PMID: 19559397; PubMed Central PMCID: PMCPMC2706965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2726.Moreno-De-Luca A, Helmers SL, Mao H, et al. Adaptor protein complex-4 (AP-4) deficiency causes a novel autosomal recessive cerebral palsy syndrome with microcephaly and intellectual disability. J Med Genet. 2011. Feb;48(2):141–4. doi: 10.1136/jmg.2010.082263. PubMed PMID: 20972249; PubMed Central PMCID: PMCPMC3150730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2727.Abou Jamra R, Philippe O, Raas-Rothschild A, et al. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet. 2011. Jun 10;88(6):788–795. doi: 10.1016/j.ajhg.2011.04.019. PubMed PMID: 21620353; PubMed Central PMCID: PMCPMC3113253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2728.Mattera R, Park SY, De Pace R, et al. AP-4 mediates export of ATG9A from the trans-Golgi network to promote autophagosome formation. Proc Natl Acad Sci U S A. 2017. Dec 12;114(50):E10697–E10706. doi: 10.1073/pnas.1717327114. PubMed PMID: 29180427; PubMed Central PMCID: PMCPMC5740629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2729.Ivankovic D, Drew J, Lesept F, et al. Axonal autophagosome maturation defect through failure of ATG9A sorting underpins pathology in AP-4 deficiency syndrome. Autophagy. 2020. Mar;16(3):391–407. doi: 10.1080/15548627.2019.1615302. PubMed PMID: 31142229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2730.Slabicki M, Theis M, Krastev DB, et al. A genome-scale DNA repair RNAi screen identifies SPG48 as a novel gene associated with hereditary spastic paraplegia. PLoS Biol. 2010. Jun 29;8(6):e1000408. doi: 10.1371/journal.pbio.1000408. PubMed PMID: 20613862; PubMed Central PMCID: PMCPMC2893954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2731.Hirst J, Itzhak DN, Antrobus R, et al. Role of the AP-5 adaptor protein complex in late endosome-to-Golgi retrieval. PLoS Biol. 2018. Jan;16(1):e2004411. doi: 10.1371/journal.pbio.2004411. PubMed PMID: 29381698; PubMed Central PMCID: PMCPMC5806898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2732.Khundadze M, Ribaudo F, Hussain A, et al. A mouse model for SPG48 reveals a block of autophagic flux upon disruption of adaptor protein complex five. Neurobiol Dis. 2019. Jul;127:419–431. doi: 10.1016/j.nbd.2019.03.026. PubMed PMID: 30930081. [DOI] [PubMed] [Google Scholar]
- 2733.Costa R, Morrison A, Wang J, et al. Activated protein C modulates cardiac metabolism and augments autophagy in the ischemic heart. J Thromb Haemost. 2012. Sep;10(9):1736–44. doi: 10.1111/j.1538-7836.2012.04833.x. PubMed PMID: 22738025; PubMed Central PMCID: PMC3433592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2734.Yuga M, Gomi K, Klionsky DJ, et al. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011. Apr 15;286(15):13704–13. doi: 10.1074/jbc.M110.173906. PubMed PMID: 21343297; PubMed Central PMCID: PMC3075714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2735.Besteiro S. Autophagy in apicomplexan parasites. Curr Opin Microbiol. 2017. Dec;40:14–20. doi: 10.1016/j.mib.2017.10.008. PubMed PMID: 29096193. [DOI] [PubMed] [Google Scholar]
- 2736.Deretic V, Levine B.. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009. Jun 18;5(6):527–49. doi: 10.1016/j.chom.2009.05.016. PubMed PMID: 19527881; PubMed Central PMCID: PMCPMC2720763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2737.Thekkinghat AA, Yadav KK, Rangarajan PN.. Apolipoprotein L9 interacts with LC3/GABARAP and is a microtubule-associated protein with a widespread subcellular distribution. Biol Open. 2019;8(9):bio045930. 10.1101/671065. PubMed PMID: 31515254; PubMed Central PMCID: PMCPMC6777357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2738.Rubinstein AD, Kimchi A.. Life in the balance - a mechanistic view of the crosstalk between autophagy and apoptosis. J Cell Sci. 2012. Nov 15;125(Pt 22):5259–68. doi: 10.1242/jcs.115865. PubMed PMID: 23377657. [DOI] [PubMed] [Google Scholar]
- 2739.Yang S, Rosenwald AG.. Autophagy in Saccharomyces cerevisiae requires the monomeric GTP-binding proteins, Arl1 and Ypt6. Autophagy. 2016. Oct 2;12(10):1721–1737. doi: 10.1080/15548627.2016.1196316. PubMed PMID: 27462928; PubMed Central PMCID: PMCPMC5079543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2740.Wang P, Xu TY, Wei K, et al.ARRB1/beta-arrestin-1 mediates neuroprotection through coordination of BECN1-dependent autophagy in cerebral ischemia. Autophagy. 2014. Jun25;10(9):1535–48. PubMed PMID: 24988431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2741.Villa E, Proics E, Rubio-Patino C, et al. Parkin-Independent Mitophagy Controls Chemotherapeutic Response in Cancer Cells. Cell Rep. 2017. Sep 19;20(12):2846–2859. doi: 10.1016/j.celrep.2017.08.087. PubMed PMID: 28930681. [DOI] [PubMed] [Google Scholar]
- 2742.Boda A, Lorincz P, Takats S, et al. Drosophila Arl8 is a general positive regulator of lysosomal fusion events. Biochim Biophys Acta Mol Cell Res. 2019. Apr;1866(4):533–544. doi: 10.1016/j.bbamcr.2018.12.011. PubMed PMID: 30590083. [DOI] [PubMed] [Google Scholar]
- 2743.Moosavi MA, Djavaheri-Mergny M.. Autophagy: New Insights into Mechanisms of Action and Resistance of Treatment in Acute Promyelocytic leukemia. Int J Mol Sci. 2019. Jul 20;20(14). doi: 10.3390/ijms20143559. PubMed PMID: 31330838; PubMed Central PMCID: PMCPMC6678259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2744.Keller KE, Yang YF, Sun YY, et al. Ankyrin repeat and suppressor of cytokine signaling box containing protein-10 is associated with ubiquitin-mediated degradation pathways in trabecular meshwork cells. Mol Vis. 2013;19:1639–55. PubMed PMID: 23901248; PubMed Central PMCID: PMC3724959. [PMC free article] [PubMed] [Google Scholar]
- 2745.Meyer N, Zielke S, Michaelis JB, et al. AT 101 induces early mitochondrial dysfunction and HMOX1 (heme oxygenase 1) to trigger mitophagic cell death in glioma cells. Autophagy. 2018;14(10):1693–1709. doi: 10.1080/15548627.2018.1476812. PubMed PMID: 29938581; PubMed Central PMCID: PMCPMC6135628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2746.Warnsmann V, Meyer N, Hamann A, et al. A novel role of the mitochondrial permeability transition pore in (-)-gossypol-induced mitochondrial dysfunction. Mech Ageing Dev. 2018. Mar;170:45–58. doi: 10.1016/j.mad.2017.06.004. PubMed PMID: 28684269. [DOI] [PubMed] [Google Scholar]
- 2747.Wang P, Pleskot R, Zang J, et al. Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun. 2019. Nov 13;10(1):5132. doi: 10.1038/s41467-019-12782-6. PubMed PMID: 31723129; PubMed Central PMCID: PMCPMC6853982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2748.Harding HP, Novoa I, Zhang Y, et al. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000. Nov;6(5):1099–108. doi: 10.1016/s1097-2765(00)00108-8. PubMed PMID: 11106749. [DOI] [PubMed] [Google Scholar]
- 2749.Gully JC, Sergeyev VG, Bhootada Y, et al. Up-regulation of activating transcription factor 4 induces severe loss of dopamine nigral neurons in a rat model of Parkinson’s disease. Neurosci Lett. 2016. Aug 3;627:36–41. doi: 10.1016/j.neulet.2016.05.039. PubMed PMID: 27233218; PubMed Central PMCID: PMCPMC6052763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2750.Bhootada Y, Kotla P, Zolotukhin S, et al. Limited ATF4 expression in degenerating retinas with ongoing ER stress promotes photoreceptor survival in a mouse model of autosomal dominant retinitis pigmentosa. PLoS One. 2016;11(5):e0154779. doi: 10.1371/journal.pone.0154779. PubMed PMID: 27144303; PubMed Central PMCID: PMCPMC4856272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2751.Rzymski T, Milani M, Pike L, et al. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010. Aug 5;29(31):4424–35. doi: 10.1038/onc.2010.191. PubMed PMID: 20514020. [DOI] [PubMed] [Google Scholar]
- 2752.Sheng Z, Ma L, Sun JE, et al. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011. Sep 8;118(10):2840–8. doi: 10.1182/blood-2010-12-322537. PubMed PMID: 21715304; PubMed Central PMCID: PMC3172800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2753.Klionsky DJ, Cregg JM, Dunn WA, Jr., et al. A unified nomenclature for yeast autophagy-related genes. Dev cell 2003. Oct;5(4):539–45. PubMed PMID: 14536056. [DOI] [PubMed] [Google Scholar]
- 2754.Matsuura A, Tsukada M, Wada Y, et al. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997. Jun 19;192(2):245–50. doi:S0378-1119(97)00084-X [pii]. PubMed PMID: 9224897; eng. [DOI] [PubMed] [Google Scholar]
- 2755.Romanyuk D, Polak A, Maleszewska A, et al. Human hAtg2A protein expressed in yeast is recruited to preautophagosomal structure but does not complement autophagy defects of atg2Delta strain. Acta Biochim Pol. 2011;58(3):365–74. PubMed PMID: 21887408. [PubMed] [Google Scholar]
- 2756.Kaminska J, Rzepnikowska W, Polak A, et al. Phosphatidylinositol-3-phosphate regulates response of cells to proteotoxic stress. Int J Biochem Cell Biol. 2016. Oct;79:494–504. doi: 10.1016/j.biocel.2016.08.007. PubMed PMID: 27498190. [DOI] [PubMed] [Google Scholar]
- 2757.Shintani T, Suzuki K, Kamada Y, et al.Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem. 2001. Aug 10;276(32):30452–60. doi: 10.1074/jbc.M102346200M102346200 [pii].PubMed PMID: 11382761; eng. [DOI] [PubMed] [Google Scholar]
- 2758.Gomez-Sanchez R, Rose J, Guimaraes R, et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol. 2018. Aug 6;217(8):2743–2763. doi: 10.1083/jcb.201710116. PubMed PMID: 29848619; PubMed Central PMCID: PMCPMC6080931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2759.Maeda S, Otomo C, Otomo T.. The autophagic membrane tether ATG2A transfers lipids between membranes. eLife. 2019. Jul 4;8. doi: 10.7554/eLife.45777. PubMed PMID: 31271352; PubMed Central PMCID: PMCPMC6625793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2760.Osawa T, Kotani T, Kawaoka T, et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol. 2019. Apr;26(4):281–288. doi: 10.1038/s41594-019-0203-4. PubMed PMID: 30911189. [DOI] [PubMed] [Google Scholar]
- 2761.Valverde DP, Yu S, Boggavarapu V, et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol. 2019. Jun 3;218(6):1787–1798. doi: 10.1083/jcb.201811139. PubMed PMID: 30952800; PubMed Central PMCID: PMCPMC6548141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2762.Kotani T, Kirisako H, Koizumi M, et al. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci U S A. 2018. Oct 9;115(41):10363–10368. doi: 10.1073/pnas.1806727115. PubMed PMID: 30254161; PubMed Central PMCID: PMCPMC6187169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2763.Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000. Nov 23;408(6811):488–92. doi: 10.1038/35044114. PubMed PMID: 11100732; eng. [DOI] [PubMed] [Google Scholar]
- 2764.Schlumpberger M, Schaeffeler E, Straub M, et al. AUT1, a gene essential for autophagocytosis in the yeast Saccharomyces cerevisiae. J Bacteriol. 1997. Feb;179(4):1068–76. PubMed PMID: 9023185; PubMed Central PMCID: PMC178799. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2765.Fernandez AF, Lopez-Otin C.. The functional and pathologic relevance of autophagy proteases. J Clin Invest. 2015. Jan;125(1):33–41. doi: 10.1172/JCI73940. PubMed PMID: 25654548; PubMed Central PMCID: PMCPMC4382236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2766.Tanida I, Sou YS, Minematsu-Ikeguchi N, et al. Atg8L/Apg8L is the fourth mammalian modifier of mammalian Atg8 conjugation mediated by human Atg4B, Atg7 and Atg3. FEBS J. 2006. Jun;273(11):2553–62. doi: 10.1111/j.1742-4658.2006.05260.x. PubMed PMID: 16704426; eng. [DOI] [PubMed] [Google Scholar]
- 2767.Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature. 1998. Sep 24;395(6700):395–8. doi: 10.1038/26506. PubMed PMID: 9759731; eng. [DOI] [PubMed] [Google Scholar]
- 2768.Kim J, Dalton VM, Eggerton KP, et al. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999. May;10(5):1337–51. PubMed PMID: 10233148; PubMed Central PMCID: PMC25275. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2769.Tanida I, Mizushima N, Kiyooka M, et al. Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999. May;10(5):1367–79. PubMed PMID: 10233150; PubMed Central PMCID: PMC25280. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2770.Ogmundsdottir MH, Fock V, Sooman L, et al. A short isoform of ATG7 fails to lipidate LC3/GABARAP. Sci Rep. 2018. Sep 26;8(1):14391. doi: 10.1038/s41598-018-32694-7. PubMed PMID: 30258106; PubMed Central PMCID: PMCPMC6158294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2771.Knorr RL, Nakatogawa H, Ohsumi Y, et al. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids. PLoS One. 2014;9(12):e115357. doi: 10.1371/journal.pone.0115357. PubMed PMID: 25522362; PubMed Central PMCID: PMCPMC4270758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2772.Noda T, Kim J, Huang W-P, et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000. Feb 7;148(3):465–80. PubMed PMID: 10662773; PubMed Central PMCID: PMC2174799. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2773.Saitoh T, Fujita N, Hayashi T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009. Dec 8;106(49):20842–6. doi: 10.1073/pnas.0911267106. PubMed PMID: 19926846; PubMed Central PMCID: PMCPMC2791563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2774.Imagawa Y, Saitoh T, Tsujimoto Y.. Vital staining for cell death identifies Atg9a-dependent necrosis in developmental bone formation in mouse. Nat Commun. 2016. Nov 4;7:13391. doi: 10.1038/ncomms13391. PubMed PMID: 27811852; PubMed Central PMCID: PMCPMC5097171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2775.Yamada T, Carson AR, Caniggia I, et al. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005. May 6;280(18):18283–90. doi: 10.1074/jbc.M413957200. PubMed PMID: 15755735; eng. [DOI] [PubMed] [Google Scholar]
- 2776.Shintani T, Mizushima N, Ogawa Y, et al. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J. 1999. Oct 1;18(19):5234–41. doi: 10.1093/emboj/18.19.5234. PubMed PMID: 10508157; PubMed Central PMCID: PMC1171594. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2777.Zhao Q, Hu ZY, Zhang JP, et al. Dual Roles of Two Isoforms of Autophagy-related Gene ATG10 in HCV-Subgenomic replicon Mediated Autophagy Flux and Innate Immunity. Sci Rep. 2017. Sep 12;7(1):11250. doi: 10.1038/s41598-017-11105-3. PubMed PMID: 28900156; PubMed Central PMCID: PMCPMC5595887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2778.Zhang MQ, Li JR, Peng ZG, et al. Differential effects of autophagy-related 10 protein on hcv replication and autophagy flux are mediated by its cysteine(44) and cysteine(135). Front Immunol. 2018;9:2176. doi: 10.3389/fimmu.2018.02176. PubMed PMID: 30319633; PubMed Central PMCID: PMCPMC6165859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2779.Kim J, Kamada Y, Stromhaug PE, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001. Apr16;153(2):381–96. PubMed PMID: 11309418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2780.Kamber RA, Shoemaker CJ, Denic V.. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell. 2015. Aug 6;59(3):372–81. doi: 10.1016/j.molcel.2015.06.009. PubMed PMID: 26166702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2781.Li F, Chung T, Vierstra RD.. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014. Feb;26(2):788–807. doi: 10.1105/tpc.113.120014. PubMed PMID: 24563201; PubMed Central PMCID: PMCPMC3967041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2782.Turco E, Witt M, Abert C, et al. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol Cell. 2019. Apr 18;74(2):330–346 e11. doi: 10.1016/j.molcel.2019.01.035. PubMed PMID: 30853400; PubMed Central PMCID: PMCPMC6477179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2783.Lin L, Yang P, Huang X, et al. The scaffold protein EPG-7 links cargo-receptor complexes with the autophagic assembly machinery. J Cell Biol. 2013. Apr 1;201(1):113–29. doi: 10.1083/jcb.201209098. PubMed PMID: 23530068; PubMed Central PMCID: PMC3613692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2784.Ochaba J, Lukacsovich T, Csikos G, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A. 2014. Nov 25;111(47):16889–94. doi: 10.1073/pnas.1420103111. PubMed PMID: 25385587; PubMed Central PMCID: PMCPMC4250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2785.Funakoshi T, Matsuura A, Noda T, et al. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997. Jun 19;192(2):207–13. doi: 10.1016/s0378-1119(97)00031-0 [pii]. PubMed PMID: 9224892; eng. [DOI] [PubMed] [Google Scholar]
- 2786.Kametaka S, Okano T, Ohsumi M, et al. Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem 1998. Aug 28;273(35):22284–91. PubMed PMID: 9712845; eng. [DOI] [PubMed] [Google Scholar]
- 2787.Mukhopadhyay S, Schlaepfer IR, Bergman BC, et al. ATG14 facilitated lipophagy in cancer cells induce ER stress mediated mitoptosis through a ROS dependent pathway. Free Radical biol Med. 2017. Mar;104:199–213. doi: 10.1016/j.freeradbiomed.2017.01.007. PubMed PMID: 28069524. [DOI] [PubMed] [Google Scholar]
- 2788.Yang P, Zhang H.. The coiled-coil domain protein EPG-8 plays an essential role in the autophagy pathway in C. elegans. Autophagy. 2011. Feb;7(2):159–65. PubMed PMID: 21116129. [DOI] [PubMed] [Google Scholar]
- 2789.Epple UD, Suriapranata I, Eskelinen E-L, et al. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J Bacteriol. 2001. Oct;183(20):5942–55. doi: 10.1128/JB.183.20.5942-5955.2001. PubMed PMID: 11566994; PubMed Central PMCID: PMC99673. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2790.Teter SA, Eggerton KP, Scott SV, et al. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J Biol Chem. 2001. Jan 19;276(3):2083–7. doi: 10.1074/jbc.C000739200C000739200 [pii]. PubMed PMID: 11085977; PubMed Central PMCID: PMC2749705. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2791.van Zutphen T, Todde V, de Boer R, et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014. Jan;25(2):290–301. doi: 10.1091/mbc.E13-08-0448. PubMed PMID: 24258026; PubMed Central PMCID: PMC3890349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2792.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999. Jul 15;18(14):3888–96. doi: 10.1093/emboj/18.14.3888. PubMed PMID: 10406794; PubMed Central PMCID: PMC1171465. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2793.Massey DC, Parkes M.. Genome-wide association scanning highlights two autophagy genes, ATG16L1 and IRGM, as being significantly associated with Crohn’s disease. Autophagy. 2007. Nov-Dec;3(6):649–51. doi: 5075 [pii]. PubMed PMID: 17921695; eng. [DOI] [PubMed] [Google Scholar]
- 2794.Yang SK, Hong M, Zhao W, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014. Jan;63(1):80–7. doi: 10.1136/gutjnl-2013-305193. PubMed PMID: 23850713. [DOI] [PubMed] [Google Scholar]
- 2795.Chew LH, Setiaputra D, Klionsky DJ, et al. Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy. 2013. Oct;9(10):1467–74. doi: 10.4161/auto.25687. PubMed PMID: 23939028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2796.Mao K, Chew LH, Inoue-Aono Y, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A. 2013. Jul 30;110(31):E2875–84. doi: 10.1073/pnas.1300064110. PubMed PMID: 23858448; PubMed Central PMCID: PMC3732952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2797.Mao K, Chew LH, Yip CK, et al. The role of Atg29 phosphorylation in PAS assembly. Autophagy. 2013. Dec;9(12):2178–9. doi: 10.4161/auto.26740. PubMed PMID: 24141181; PubMed Central PMCID: PMC4028347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2798.Leber R, Silles E, Sandoval IV, et al.Yol082p, a novel CVT protein involved in the selective targeting of aminopeptidase I to the yeast vacuole. J Biol Chem. 2001. Aug 3;276(31):29210–7. doi: 10.1074/jbc.M101438200 M101438200 [pii]. PubMed PMID: 11382752; eng. [DOI] [PubMed] [Google Scholar]
- 2799.Scott SV, Guan J, Hutchins MU, et al. Cvt19 is a receptor for the cytoplasm-to-vacuole targeting pathway. Mol Cell. 2001. Jun;7(6):1131–41. doi: S1097-2765(01)00263-5 [pii]. PubMed PMID: 11430817; PubMed Central PMCID: PMC2767243. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2800.Nice DC, Sato TK, Stromhaug PE, et al. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J Biol Chem. 2002. Aug 16;277(33):30198–207. doi: 10.1074/jbc.M204736200 M204736200 [pii]. PubMed PMID: 12048214; PubMed Central PMCID: PMC2754692. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2801.Popelka H, Damasio A, Hinshaw JE, et al. Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc Natl Acad Sci U S A. 2017. Nov 21;114(47):E10112–E10121. doi: 10.1073/pnas.1708367114. PubMed PMID: 29114050; PubMed Central PMCID: PMCPMC5703286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2802.Suriapranata I, Epple UD, Bernreuther D, et al. The breakdown of autophagic vesicles inside the vacuole depends on Aut4p. J Cell Sci. 2000. Nov;113:4025–33. PubMed PMID: 11058089; eng. [DOI] [PubMed] [Google Scholar]
- 2803.Yang Z, Huang J, Geng J, et al. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell. 2006. Dec;17(12):5094–104. doi:E06-06-0479 [pii] doi: 10.1091/mbc.E06-06-0479. PubMed PMID: 17021250; PubMed Central PMCID: PMC1679675. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2804.Legakis JE, Yen W-L, Klionsky DJ.. A cycling protein complex required for selective autophagy. Autophagy. 2007. Sep-Oct;3(5):422–32. doi: 4129 [pii]. PubMed PMID: 17426440; eng. [DOI] [PubMed] [Google Scholar]
- 2805.Tucker KA, Reggiori F, Dunn WA, Jr., et al. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J Biol Chem. 2003. Nov 28;278(48):48445–52. doi: 10.1074/jbc.M309238200 M309238200 [pii]. PubMed PMID: 14504273; PubMed Central PMCID: PMC1705954. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2806.Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012. Jul 23;198(2):219–33. doi: 10.1083/jcb.201202061. PubMed PMID: 22826123; PubMed Central PMCID: PMCPMC3410421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2807.Backues SK, Orban DP, Bernard A, et al. Atg23 and Atg27 act at the early stages of Atg9 trafficking in S. cerevisiae. Traffic. 2015. Feb;16(2):172–90. doi: 10.1111/tra.12240. PubMed PMID: 25385507; PubMed Central PMCID: PMCPMC4305007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2808.Monastyrska I, Kiel JAKW, Krikken AM, et al. The Hansenula polymorpha ATG25 gene encodes a novel coiled-coil protein that is required for macropexophagy. Autophagy. 2005. Jul;1(2):92–100. PubMed PMID: 16874036. [DOI] [PubMed] [Google Scholar]
- 2809.Cao Y, Klionsky DJ.. Atg26 is not involved in autophagy-related pathways in Saccharomyces cerevisiae. Autophagy. 2007. Jan 7;3(1):17–20. PubMed PMID: 17012830. [DOI] [PubMed] [Google Scholar]
- 2810.Yamashita S, Oku M, Wasada Y, et al.PI4P-signaling pathway for the synthesis of a nascent membrane structure in selective autophagy. J Cell Biol 2006. Jun5;173(5):709–17. PubMed PMID: 16754956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2811.Yen W-L, Legakis JE, Nair U, et al. Atg27 is required for autophagy-dependent cycling of Atg9. Mol Biol Cell. 2007. Feb;18(2):581–93. doi:E06-07-0612[pii] 10.1091/mbc.E06-07-0612. PubMed PMID: 17135291; PubMed Central PMCID: PMC1783788. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2812.Stasyk OV, Stasyk OG, Mathewson RD, et al. Atg28, a novel coiled-coil protein involved in autophagic degradation of peroxisomes in the methylotrophic yeast Pichia pastoris. Autophagy. 2006. Jan-Mar;2(1):30–8. PubMed PMID: 16874081. [DOI] [PubMed] [Google Scholar]
- 2813.Kawamata T, Kamada Y, Suzuki K, et al. Characterization of a novel autophagy-specific gene, ATG29. Biochem Biophys Res Commun. 2005. Dec 30;338(4):1884–9. doi:S0006-291X(05)02441-1 [pii] doi: 10.1016/j.bbrc.2005.10.163. PubMed PMID: 16289106; eng. [DOI] [PubMed] [Google Scholar]
- 2814.Zientara-Rytter K, Ozeki K, Nazarko TY, et al. Pex3 and Atg37 compete to regulate the interaction between the pexophagy receptor, Atg30, and the Hrr25 kinase. Autophagy. 2018;14(3):368–384. doi: 10.1080/15548627.2017.1413521. PubMed PMID: 29260977; PubMed Central PMCID: PMCPMC5915033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2815.Farre JC, Burkenroad A, Burnett SF, et al. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013. May;14(5):441–9. doi: 10.1038/embor.2013.40. PubMed PMID: 23559066; PubMed Central PMCID: PMCPMC3642380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2816.Kabeya Y, Kawamata T, Suzuki K, et al. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2007. May 4;356(2):405–10. doi: S0006-291X(07)00437-8 [pii] doi: 10.1016/j.bbrc.2007.02.150. PubMed PMID: 17362880; eng. [DOI] [PubMed] [Google Scholar]
- 2817.Watanabe Y, Noda NN, Kumeta H, et al. Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J Biol Chem. 2010. Sep 24;285(39):30026–33. doi: 10.1074/jbc.M110.143545. PubMed PMID: 20659891; PubMed Central PMCID: PMC2943322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2818.Meijer WH, van der Klei IJ, Veenhuis M, et al. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–16. [DOI] [PubMed] [Google Scholar]
- 2819.Nazarko VY, Nazarko TY, Farre JC, et al. Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy. 2011. Apr;7(4):375–85. doi: 10.4161/auto.7.4.14369. PubMed PMID: 21169734; PubMed Central PMCID: PMCPMC3127218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2820.Motley AM, Nuttall JM, Hettema EH.. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012. Jun 29;31(13):2852–68. doi: 10.1038/emboj.2012.151. PubMed PMID: 22643220; PubMed Central PMCID: PMC3395097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2821.Araki Y, Ku WC, Akioka M, et al. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol. 2013. Oct 28;203(2):299–313. doi: 10.1083/jcb.201304123. PubMed PMID: 24165940; PubMed Central PMCID: PMC3812978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2822.Yao Z, Delorme-Axford E, Backues SK, et al. Atg41/Icy2 regulates autophagosome formation. Autophagy. 2015;11(12):2288–99. doi: 10.1080/15548627.2015.1107692. PubMed PMID: 26565778; PubMed Central PMCID: PMCPMC4835205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2823.Parzych KR, Ariosa A, Mari M, et al. A newly characterized vacuolar serine carboxypeptidase, Atg42/Ybr139w, is required for normal vacuole function and the terminal steps of autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2018. May 1;29(9):1089–1099. doi: 10.1091/mbc.E17-08-0516. PubMed PMID: 29514932; PubMed Central PMCID: PMCPMC5921575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2824.Hosokawa N, Sasaki T, Iemura S, et al. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy. 2009. Oct;5(7):973–9. doi: 9296 [pii]. PubMed PMID: 19597335; eng. [DOI] [PubMed] [Google Scholar]
- 2825.Mercer CA, Kaliappan A, Dennis PB.. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy. 2009. Jul;5(5):649–62. doi: 8249 [pii]. PubMed PMID: 19287211; eng. [DOI] [PubMed] [Google Scholar]
- 2826.Cerk IK, Salzburger B, Boeszoermenyi A, et al. A peptide derived from G0/G1 switch gene 2 acts as noncompetitive inhibitor of adipose triglyceride lipase. J Biol Chem. 2014. Nov 21;289(47):32559–70. doi: 10.1074/jbc.M114.602599. PubMed PMID: 25258314; PubMed Central PMCID: PMCPMC4239610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2827.Honig A, Avin-Wittenberg T, Ufaz S, et al. A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell. 2012. Jan;24(1):288–303. doi: 10.1105/tpc.111.093112. PubMed PMID: 22253227; PubMed Central PMCID: PMC3289568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2828.Michaeli S, Clavel M, Lechner E, et al. The viral F-box protein P0 induces an ER-derived autophagy degradation pathway for the clearance of membrane-bound AGO1. Proc Natl Acad Sci U S A. 2019. Nov 5;116(45):22872–22883. doi: 10.1073/pnas.1912222116. PubMed PMID: 31628252; PubMed Central PMCID: PMCPMC6842623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2829.Sjogaard IMZ, Bressendorff S, Prestel A, et al. The transmembrane autophagy cargo receptors ATI1 and ATI2 interact with ATG8 through intrinsically disordered regions with distinct biophysical properties. Biochem J. 2019. Feb 5;476(3):449–465. doi: 10.1042/BCJ20180748. PubMed PMID: 30642888. [DOI] [PubMed] [Google Scholar]
- 2830.Kastan MB, Bartek J.. Cell-cycle checkpoints and cancer. Nature. 2004. Nov 18;432(7015):316–23. doi: 10.1038/nature03097. PubMed PMID: 15549093. [DOI] [PubMed] [Google Scholar]
- 2831.Antonelli M, Strappazzon F, Arisi I, et al. ATM kinase sustains breast cancer stem-like cells by promoting ATG4C expression and autophagy. Oncotarget. 2017. Mar 28;8(13):21692–21709. doi: 10.18632/oncotarget.15537. PubMed PMID: 28423511; PubMed Central PMCID: PMCPMC5400616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2832.Beauvarlet J, Bensadoun P, Darbo E, et al. Modulation of the ATM/autophagy pathway by a G-quadruplex ligand tips the balance between senescence and apoptosis in cancer cells. Nucleic Acids Res. 2019. Apr 8;47(6):2739–2756. doi: 10.1093/nar/gkz095. PubMed PMID: 30759257; PubMed Central PMCID: PMCPMC6451122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2833.Zhao YG, Zhang H.. The ER-localized autophagy protein EPG-3/VMP1 regulates ER contacts with other organelles by modulating ATP2A/SERCA activity. Autophagy. 2018;14(2):362–363. doi: 10.1080/15548627.2017.1415591. PubMed PMID: 29494262; PubMed Central PMCID: PMCPMC5902242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2834.Hsin IL, Sheu GT, Jan MS, et al. Inhibition of lysosome degradation on autophagosome formation and responses to GMI, an immunomodulatory protein from Ganoderma microsporum. Br J Pharmacol. 2012. Nov;167(6):1287–300. doi: 10.1111/j.1476-5381.2012.02073.x. PubMed PMID: 22708544; PubMed Central PMCID: PMCPMC3504994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2835.Chang D, Nalls MA, Hallgrimsdottir IB, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017. Oct;49(10):1511–1516. doi: 10.1038/ng.3955. PubMed PMID: 28892059; PubMed Central PMCID: PMCPMC5812477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2836.Dehay B, Ramirez A, Martinez-Vicente M, et al. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc Natl Acad Sci U S A. 2012. Jun 12;109(24):9611–6. doi: 10.1073/pnas.1112368109. PubMed PMID: 22647602; PubMed Central PMCID: PMC3386132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2837.Gusdon AM, Zhu J, Van Houten B, et al. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol Dis. 2012. Mar;45(3):962–72. doi: 10.1016/j.nbd.2011.12.015. PubMed PMID: 22198378; PubMed Central PMCID: PMC3291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2838.Niu H, Rikihisa Y.. Ats-1: a novel bacterial molecule that links autophagy to bacterial nutrition. Autophagy. 2013. May;9(5):787–8. doi: 10.4161/auto.23693. PubMed PMID: 23388398; PubMed Central PMCID: PMC3669189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2839.Niu H, Xiong Q, Yamamoto A, et al. Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc Natl Acad Sci U S A. 2012. Dec 18;109(51):20800–7. doi: 10.1073/pnas.1218674109. PubMed PMID: 23197835; PubMed Central PMCID: PMC3529060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2840.Isakson P, Bjoras M, Boe SO, et al. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010. Sep 30;116(13):2324–31. doi: 10.1182/blood-2010-01-261040. PubMed PMID: 20574048. [DOI] [PubMed] [Google Scholar]
- 2841.Orfali N, McKenna SL, Cahill MR, et al. Retinoid receptor signaling and autophagy in acute promyelocytic leukemia. Exp Cell Res. 2014. May 15;324(1):1–12. doi: 10.1016/j.yexcr.2014.03.018. PubMed PMID: 24694321; PubMed Central PMCID: PMC4047711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2842.Li Z, Wang C, Wang Z, et al. Allele-selective lowering of mutant HTT protein by HTT-LC3 linker compounds. Nature. 2019. Nov;575(7781):203–209. doi: 10.1038/s41586-019-1722-1. PubMed PMID: 31666698. [DOI] [PubMed] [Google Scholar]
- 2843.Vanhee C, Zapotoczny G, Masquelier D, et al. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell. 2011. Feb;23(2):785–805. doi: 10.1105/tpc.110.081570. PubMed PMID: 21317376; PubMed Central PMCID: PMC3077796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2844.Herzog LK, Kevei E, Marchante R, et al. The Machado-Joseph disease deubiquitylase ataxin-3 interacts with LC3C/GABARAP and promotes autophagy. Aging cell. 2020. Jan;19(1):e13051. doi: 10.1111/acel.13051. PubMed PMID: 31625269; PubMed Central PMCID: PMCPMC6974715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2845.Ashkenazi A, Bento CF, Ricketts T, et al. Polyglutamine tracts regulate autophagy. Autophagy. 2017. Sep 2;13(9):1613–1614. doi: 10.1080/15548627.2017.1336278. PubMed PMID: 28722507; PubMed Central PMCID: PMCPMC5612341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2846.Sittler A, Muriel MP, Marinello M, et al. Deregulation of autophagy in postmortem brains of Machado-Joseph disease patients. Neuropathology. 2018. Apr;38(2):113–124. doi: 10.1111/neup.12433. PubMed PMID: 29218765. [DOI] [PubMed] [Google Scholar]
- 2847.Papp D, Kovacs T, Billes V, et al. AUTEN-67, an autophagy-enhancing drug candidate with potent antiaging and neuroprotective effects. Autophagy. 2016;12(2):273–86. doi: 10.1080/15548627.2015.1082023. PubMed PMID: 26312549; PubMed Central PMCID: PMCPMC4835959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2848.Laraia L, Friese A, Corkery DP, et al. The cholesterol transfer protein GRAMD1A regulates autophagosome biogenesis. Nat Chem Biol. 2019. Jul;15(7):710–720. doi: 10.1038/s41589-019-0307-5. PubMed PMID: 31222192. [DOI] [PubMed] [Google Scholar]
- 2849.Wu YW, Waldmann H.. Toward the role of cholesterol and cholesterol transfer protein in autophagosome biogenesis. Autophagy. 2019. Dec;15(12):2167–2168. doi: 10.1080/15548627.2019.1666595. PubMed PMID: 31512558; PubMed Central PMCID: PMCPMC6844521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2850.Dunn WA, Jr. Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990. Jun;110(6):1923–33. PubMed PMID: 2351689; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2851.Schulze RJ, Weller SG, Schroeder B, et al. Lipid droplet breakdown requires dynamin 2 for vesiculation of autolysosomal tubules in hepatocytes. J Cell Biol. 2013. Oct 28;203(2):315–26. doi: 10.1083/jcb.201306140. PubMed PMID: 24145164; PubMed Central PMCID: PMC3812963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2852.Gundara JS, Robinson BG, Sidhu SB.. Evolution of the “autophagamiR”. Autophagy. 2011. Dec;7(12):1553–4. PubMed PMID: 22024754; PubMed Central PMCID: PMC3288028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2853.Lenzi P, Lazzeri G, Biagioni F, et al. The autophagoproteasome a novel cell clearing organelle in baseline and stimulated conditions. Front Neuroanat. 2016;10:78. doi: 10.3389/fnana.2016.00078. PubMed PMID: 27493626; PubMed Central PMCID: PMCPMC4955296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2854.Mijaljica D, Nazarko TY, Brumell JH, et al. Receptor protein complexes are in control of autophagy. Autophagy. 2012. Nov;8(11):1701–5. doi: 10.4161/auto.21332. PubMed PMID: 22874568; PubMed Central PMCID: PMC3494607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2855.Farfel-Becker T, Roney JC, Cheng XT, et al. Neuronal soma-derived degradative lysosomes are continuously delivered to distal axons to maintain local degradation capacity. Cell Rep. 2019. Jul 2;28(1):51–64 e4. doi: 10.1016/j.celrep.2019.06.013. PubMed PMID: 31269450; PubMed Central PMCID: PMCPMC6696943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2856.Farfel-Becker T, Roney JC, Cheng XT, et al. The secret life of degradative lysosomes in axons: delivery from the soma, enzymatic activity, and local autophagic clearance. Autophagy. 2020. Jan;16(1):167–168. doi: 10.1080/15548627.2019.1669869. PubMed PMID: 31533518; PubMed Central PMCID: PMCPMC6984450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2857.Shpilka T, Welter E, Borovsky N, et al. Lipid droplets and their component triglycerides and steryl esters regulate autophagosome biogenesis. EMBO J. 2015. Aug 13;34(16):2117–31. doi: 10.15252/embj.201490315. PubMed PMID: 26162625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2858.Renna M, Schaffner C, Brown K, et al. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J Clin Invest. 2011. Sep;121(9):3554–63. doi: 10.1172/JCI46095. PubMed PMID: 21804191; PubMed Central PMCID: PMCPMC3163956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2859.Saito T, Kuma A, Sugiura Y, et al. Autophagy regulates lipid metabolism through selective turnover of NCoR1. Nat Commun. 2019. Apr 5;10(1):1567. doi: 10.1038/s41467-019-08829-3. PubMed PMID: 30952864; PubMed Central PMCID: PMCPMC6450892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2860.Heaton NS, Randall G.. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010. Nov 18;8(5):422–32. doi: 10.1016/j.chom.2010.10.006. PubMed PMID: 21075353; PubMed Central PMCID: PMC3026642. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2861.Florey O, Gammoh N, Kim SE, et al. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2015;11(1):88–99. doi: 10.4161/15548627.2014.984277. PubMed PMID: 25484071; PubMed Central PMCID: PMCPMC4502810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2862.Sebti S, Prebois C, Perez-Gracia E, et al. BAT3 modulates p300-dependent acetylation of p53 and autophagy-related protein 7 (ATG7) during autophagy. Proc Natl Acad Sci U S A. 2014. Mar 18;111(11):4115–20. doi: 10.1073/pnas.1313618111. PubMed PMID: 24591579; PubMed Central PMCID: PMC3964035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2863.Kowaltowski AJ, Smaili SS, Russell JT, et al. Elevation of resting mitochondrial membrane potential of neural cells by cyclosporin A, BAPTA-AM, and bcl-2. Am J Physiol Cell Physiol. 2000. Sep;279(3):C852–9. doi: 10.1152/ajpcell.2000.279.3.C852. PubMed PMID: 10942734. [DOI] [PubMed] [Google Scholar]
- 2864.Tang Q, Jin MW, Xiang JZ, et al. The membrane permeable calcium chelator BAPTA-AM directly blocks human ether a-go-go-related gene potassium channels stably expressed in HEK 293 cells. Biochem Pharmacol. 2007. Dec 3;74(11):1596–607. doi: 10.1016/j.bcp.2007.07.042. PubMed PMID: 17826747. [DOI] [PubMed] [Google Scholar]
- 2865.Chi Y, Li K, Yan Q, et al. Nonsteroidal anti-inflammatory drug flufenamic acid is a potent activator of AMP-activated protein kinase. J Pharmacol Exp Ther. 2011. Oct;339(1):257–66. doi: 10.1124/jpet.111.183020. PubMed PMID: 21765041. [DOI] [PubMed] [Google Scholar]
- 2866.Noda NN, Kobayashi T, Adachi W, et al. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J Biol Chem. 2012. May 11;287(20):16256–66. doi: 10.1074/jbc.M112.348250. PubMed PMID: 22437838; PubMed Central PMCID: PMC3351336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2867.Lindqvist LM, Heinlein M, Huang DC, et al. Prosurvival Bcl-2 family members affect autophagy only indirectly, by inhibiting Bax and Bak. Proc Natl Acad Sci U S A. 2014. Jun 10;111(23):8512–7. doi: 10.1073/pnas.1406425111. PubMed PMID: 24912196; PubMed Central PMCID: PMC4060681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2868.Murakawa T, Yamaguchi O, Hashimoto A, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. doi: 10.1038/ncomms8527. PubMed PMID: 26146385; PubMed Central PMCID: PMC4501433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2869.Paul S, Kashyap AK, Jia W, et al. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity. 2012. Jun 29;36(6):947–58. doi: 10.1016/j.immuni.2012.04.008. PubMed PMID: 22658522; PubMed Central PMCID: PMC3389288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2870.Baydyuk M, Xie Y, Tessarollo L, et al. Midbrain-derived neurotrophins support survival of immature striatal projection neurons. J Neurosci. 2013. Feb 20;33(8):3363–9. doi: 10.1523/JNEUROSCI.3687-12.2013. PubMed PMID: 23426664; PubMed Central PMCID: PMCPMC3711532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2871.Ortega F, Perez-Sen R, Morente V, et al. P2X7, NMDA and BDNF receptors converge on GSK3 phosphorylation and cooperate to promote survival in cerebellar granule neurons. Cell Mol Life Sci. 2010. May;67(10):1723–33. doi: 10.1007/s00018-010-0278-x. PubMed PMID: 20146080; PubMed Central PMCID: PMCPMC2858808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2872.Brito V, Puigdellivol M, Giralt A, et al. Imbalance of p75(NTR)/TrkB protein expression in Huntington’s disease: implication for neuroprotective therapies. Cell Death Dis. 2013. Apr 18;4:e595. doi: 10.1038/cddis.2013.116. PubMed PMID: 23598407; PubMed Central PMCID: PMCPMC3641339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2873.Smith ED, Prieto GA, Tong L, et al. Rapamycin and interleukin-1beta impair brain-derived neurotrophic factor-dependent neuron survival by modulating autophagy. J Biol Chem. 2014. Jul 25;289(30):20615–29. doi: 10.1074/jbc.M114.568659. PubMed PMID: 24917666; PubMed Central PMCID: PMCPMC4110274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2874.Liang X, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B.. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. [DOI] [PubMed] [Google Scholar]
- 2875.Mei Y, Glover K, Su M, et al. Conformational flexibility of BECN1: Essential to its key role in autophagy and beyond. Protein Sci. 2016. Oct;25(10):1767–85. doi: 10.1002/pro.2984. PubMed PMID: 27414988; PubMed Central PMCID: PMCPMC5029530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2876.Cheng B, Xu A, Qiao M, et al. BECN1s, a short splice variant of BECN1, functions in mitophagy. Autophagy. 2015;11:2048–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2877.He C, Wei Y, Sun K, et al. Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell. 2013. Aug 29;154(5):1085–99. doi: 10.1016/j.cell.2013.07.035. PubMed PMID: 23954414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2878.Dong X, Cheng A, Zou Z, et al. Endolysosomal trafficking of viral G protein-coupled receptor functions in innate immunity and control of viral oncogenesis. Proc Natl Acad Sci U S A. 2016. Mar 15;113(11):2994–9. doi: 10.1073/pnas.1601860113. PubMed PMID: 26929373; PubMed Central PMCID: PMCPMC4801257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2879.Yang LJ, Chen Y, He J, et al. Betulinic acid inhibits autophagic flux and induces apoptosis in human multiple myeloma cells in vitro. Acta Pharmacol Sin. 2012. Dec;33(12):1542–8. doi: 10.1038/aps.2012.102. PubMed PMID: 23064721; PubMed Central PMCID: PMC4001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2880.Yin XM, Oltvai ZN, Korsmeyer SJ.. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature. 1994. May 26;369(6478):321–3. doi: 10.1038/369321a0. PubMed PMID: 8183370. [DOI] [PubMed] [Google Scholar]
- 2881.Sinha S, Levine B.. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene. 2008. Dec;27Suppl 1:S137–48. doi: 10.1038/onc.2009.51. PubMed PMID: 19641499; PubMed Central PMCID: PMCPMC2731580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2882.Minoia M, Boncoraglio A, Vinet J, et al. BAG3 induces the sequestration of proteasomal clients into cytoplasmic puncta: Implications for a proteasome-to-autophagy switch. Autophagy. 2014. Jul 10;10(9):1603–21. PubMed PMID: 25046115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2883.Mukhopadhyay S, Naik PP, Panda PK, et al. Serum starvation induces anti-apoptotic cIAP1 to promote mitophagy through ubiquitination. Biochem Biophys Res Commun. 2016. Oct 28;479(4):940–946. doi: 10.1016/j.bbrc.2016.09.143. PubMed PMID: 27693792. [DOI] [PubMed] [Google Scholar]
- 2884.Humphry NJ, Wheatley SP.. Survivin inhibits excessive autophagy in cancer cells but does so independently of its interaction with LC3. Biol Open. 2018. Oct 22;7(10). doi: 10.1242/bio.037374. PubMed PMID: 30348810; PubMed Central PMCID: PMCPMC6215416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2885.Ebner P, Poetsch I, Deszcz L, et al. The IAP family member BRUCE regulates autophagosome-lysosome fusion. Nat Commun. 2018. Feb 9;9(1):599. doi: 10.1038/s41467-018-02823-x. PubMed PMID: 29426817; PubMed Central PMCID: PMCPMC5807552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2886.Kim Y, Kim YS, Kim DE, et al. BIX-01294 induces autophagy-associated cell death via EHMT2/G9a dysfunction and intracellular reactive oxygen species production. Autophagy. 2013. Dec;9(12):2126–39. doi: 10.4161/auto.26308. PubMed PMID: 24322755. [DOI] [PubMed] [Google Scholar]
- 2887.Artal-Martinez de Narvajas A, Gomez TS, Zhang JS, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013. Oct;33(20):3983–93. doi: 10.1128/MCB.00813-13. PubMed PMID: 23918802; PubMed Central PMCID: PMCPMC3811684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2888.Li KC, Hua KT, Lin YS, et al. Inhibition of G9a induces DUSP4-dependent autophagic cell death in head and neck squamous cell carcinoma. Mol Cancer. 2014. Jul 15;13:172. doi: 10.1186/1476-4598-13-172. PubMed PMID: 25027955; PubMed Central PMCID: PMCPMC4107555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2889.Ciechomska IA, Przanowski P, Jackl J, et al. BIX01294, an inhibitor of histone methyltransferase, induces autophagy-dependent differentiation of glioma stem-like cells. Sci Rep. 2016. Dec 9;6:38723. doi: 10.1038/srep38723. PubMed PMID: 27934912; PubMed Central PMCID: PMCPMC5146656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2890.Webster BR, Scott I, Han K, et al. Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J Cell Sci. 2013. Nov 1;126(Pt 21):4843–9. doi: 10.1242/jcs.131300. PubMed PMID: 24006259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2891.Boyd JM, Malstrom S, Subramanian T, et al. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell. 1994. Oct21;79(2):341–51. PubMed PMID: 7954800. [DOI] [PubMed] [Google Scholar]
- 2892.Hanna RA, Quinsay MN, Orogo AM, et al. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012. Jun 1;287(23):19094–104. doi: 10.1074/jbc.M111.322933. PubMed PMID: 22505714; PubMed Central PMCID: PMC3365942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2893.Chourasia AH, Boland ML, Macleod KF.. Mitophagy and cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. PubMed PMID: 25810907; PubMed Central PMCID: PMC4373087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2894.Landes T, Emorine LJ, Courilleau D, et al. The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Rep. 2010. Jun;11(6):459–65. doi: 10.1038/embor.2010.50. PubMed PMID: 20436456; PubMed Central PMCID: PMC2892319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2895.Kasper LH, Boussouar F, Boyd K, et al. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 2005. Nov 16;24(22):3846–58. doi: 10.1038/sj.emboj.7600846. PubMed PMID: 16237459; PubMed Central PMCID: PMC1283945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2896.Tracy K, Dibling BC, Spike BT, et al. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007. Sep;27(17):6229–42. doi: 10.1128/MCB.02246-06. PubMed PMID: 17576813; PubMed Central PMCID: PMC1952167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2897.Feng X, Liu X, Zhang W, et al. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011. Aug 17;30(16):3397–415. doi: 10.1038/emboj.2011.248. PubMed PMID: 21792176; PubMed Central PMCID: PMC3160666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2898.Shaw J, Yurkova N, Zhang T, et al. Antagonism of E2F-1 regulated Bnip3 transcription by NF-kappaB is essential for basal cell survival. Proc Natl Acad Sci U S A. 2008. Dec 30;105(52):20734–9. doi: 10.1073/pnas.0807735105. PubMed PMID: 19088195; PubMed Central PMCID: PMC2603431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2899.Diwan A, Krenz M, Syed FM, et al. Inhibition of ischemic cardiomyocyte apoptosis through targeted ablation of Bnip3 restrains postinfarction remodeling in mice. J Clin Invest. 2007. Oct;117(10):2825–33. doi: 10.1172/JCI32490. PubMed PMID: 17909626; PubMed Central PMCID: PMC1994631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2900.Glick D, Zhang W, Beaton M, et al. BNip3 regulates mitochondrial function and lipid metabolism in the liver. Mol Cell Biol. 2012. Jul;32(13):2570–84. doi: 10.1128/MCB.00167-12. PubMed PMID: 22547685; PubMed Central PMCID: PMC3434502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2901.Xiang G, Yang L, Long Q, et al. BNIP3L-dependent mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy. 2017. Sep 2;13(9):1543–1555. doi: 10.1080/15548627.2017.1338545. PubMed PMID: 28722510; PubMed Central PMCID: PMCPMC5612220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2902.Melser S, Chatelain EH, Lavie J, et al. Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metab. 2013. May 7;17(5):719–30. doi: 10.1016/j.cmet.2013.03.014. PubMed PMID: 23602449. [DOI] [PubMed] [Google Scholar]
- 2903.Ding WX, Ni HM, Li M, et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J Biol Chem. 2010. Sep 3;285(36):27879–90. doi: 10.1074/jbc.M110.119537. PubMed PMID: 20573959; PubMed Central PMCID: PMCPMC2934655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2904.Farg MA, Sundaramoorthy V, Sultana JM, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014. Jul 1;23(13):3579–95. doi: 10.1093/hmg/ddu068. PubMed PMID: 24549040; PubMed Central PMCID: PMC4049310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2905.Sullivan PM, Zhou X, Robins AM, et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016. May 18;4(1):51. doi: 10.1186/s40478-016-0324-5. PubMed PMID: 27193190; PubMed Central PMCID: PMCPMC4870812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2906.Webster CP, Smith EF, Bauer CS, et al. The C9orf72 protein interacts with Rab1a and the ULK1 complex to regulate initiation of autophagy. EMBO J. 2016. Aug 1;35(15):1656–76. doi: 10.15252/embj.201694401. PubMed PMID: 27334615; PubMed Central PMCID: PMCPMC4969571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2907.Yang M, Liang C, Swaminathan K, et al. A C9ORF72/SMCR8-containing complex regulates ULK1 and plays a dual role in autophagy. Sci Adv. 2016. Sep;2(9):e1601167. doi: 10.1126/sciadv.1601167. PubMed PMID: 27617292; PubMed Central PMCID: PMCPMC5010369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2908.Cali CP, Patino M, Tai YK, et al. C9orf72 intermediate repeats are associated with corticobasal degeneration, increased C9orf72 expression and disruption of autophagy. Acta Neuropathol. 2019. Nov;138(5):795–811. doi: 10.1007/s00401-019-02045-5. PubMed PMID: 31327044; PubMed Central PMCID: PMCPMC6802287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2909.Zhao X, Su L, He X, et al. Long noncoding RNA CA7-4 promotes autophagy and apoptosis via sponging MIR877-3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy. 2020. Jan;16(1):70–85. doi: 10.1080/15548627.2019.1598750. PubMed PMID: 30957640; PubMed Central PMCID: PMCPMC6984615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2910.O’Farrell F, Wang S, Katheder N, et al. Two-tiered control of epithelial growth and autophagy by the insulin receptor and the ret-like receptor, stitcher. PLoS Biol. 2013. Jul;11(7):e1001612. doi: 10.1371/journal.pbio.1001612. PubMed PMID: 23935447; PubMed Central PMCID: PMC3720245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2911.Mao K, Wang K, Liu X, et al. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013. Jul 15;26(1):9–18. doi: 10.1016/j.devcel.2013.05.024. PubMed PMID: 23810512; PubMed Central PMCID: PMC3720741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2912.Ikeda H, Hideshima T, Fulciniti M, et al. PI3K/p110delta is a novel therapeutic target in multiple myeloma. Blood. 2010. May 26;116:1460–8. doi: blood-2009-06-222943 [pii] doi: 10.1182/blood-2009-06-222943. PubMed PMID: 20505158; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2913.Verlhac P, Gregoire IP, Azocar O, et al. Autophagy receptor NDP52 regulates pathogen-containing autophagosome maturation. Cell Host Microbe. 2015. Apr 8;17(4):515–25. doi: 10.1016/j.chom.2015.02.008. PubMed PMID: 25771791. [DOI] [PubMed] [Google Scholar]
- 2914.Hu S, Guo Y, Wang Y, et al. Structure of Myosin VI/Tom1 complex reveals a cargo recognition mode of Myosin VI for tethering. Nat Commun. 2019. Aug 1;10(1):3459. doi: 10.1038/s41467-019-11481-6. PubMed PMID: 31371777; PubMed Central PMCID: PMCPMC6673701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2915.Berchtold MW, Villalobo A.. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta. 2014. Feb;1843(2):398–435. doi: 10.1016/j.bbamcr.2013.10.021. PubMed PMID: 24188867. [DOI] [PubMed] [Google Scholar]
- 2916.Yagami T, Yamamoto Y, Koma H.. Pathophysiological roles of intracellular proteases in neuronal development and neurological diseases. Mol Neurobiol. 2019. May;56(5):3090–3112. doi: 10.1007/s12035-018-1277-4. PubMed PMID: 30097848. [DOI] [PubMed] [Google Scholar]
- 2917.Xia HG, Zhang L, Chen G, et al. Control of basal autophagy by calpain1 mediated cleavage of ATG5. Autophagy. 2010. Jan;6(1):61–6. doi: 10.4161/auto.6.1.10326 [pii]. PubMed PMID: 19901552; PubMed Central PMCID: PMC2883879. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2918.Watchon M, Yuan KC, Mackovski N, et al. Calpain inhibition is protective in Machado-Joseph disease zebrafish due to induction of autophagy. J Neurosci. 2017. Aug 9;37(32):7782–7794. doi: 10.1523/JNEUROSCI.1142-17.2017. PubMed PMID: 28687604; PubMed Central PMCID: PMCPMC6596655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2919.Zitvogel L, Kepp O, Senovilla L, et al. Immunogenic tumor cell death for optimal anticancer therapy: the calreticulin exposure pathway. Clin Cancer Res. 2010. Jun 15;16(12):3100–4. doi: 10.1158/1078-0432.CCR-09-2891. PubMed PMID: 20421432. [DOI] [PubMed] [Google Scholar]
- 2920.Obeid M, Tesniere A, Ghiringhelli F, et al. Calreticulin exposure dictates the immunogenicity of cancer cell death. Nat Med. 2007. Jan;13(1):54–61. doi: 10.1038/nm1523. PubMed PMID: 17187072. [DOI] [PubMed] [Google Scholar]
- 2921.Garg AD, Agostinis P.. ER stress, autophagy and immunogenic cell death in photodynamic therapy-induced anti-cancer immune responses. Photochem Photobiol Sci. 2014. Mar;13(3):474–87. doi: 10.1039/c3pp50333j. PubMed PMID: 24493131. [DOI] [PubMed] [Google Scholar]
- 2922.Hurley RL, Anderson KA, Franzone JM, et al. The Ca2+/calmodulin-dependent protein kinase kinases are AMP-activated protein kinase kinases. J Biol Chem. 2005. Aug 12;280(32):29060–6. doi: 10.1074/jbc.M503824200. PubMed PMID: 15980064; eng. [DOI] [PubMed] [Google Scholar]
- 2923.Shrivastava A, Kuzontkoski PM, Groopman JE, et al. Cannabidiol induces programmed cell death in breast cancer cells by coordinating the cross-talk between apoptosis and autophagy. Mol Cancer Ther. 2011. Jul;10(7):1161–72. doi: 10.1158/1535-7163.MCT-10-1100. PubMed PMID: 21566064. [DOI] [PubMed] [Google Scholar]
- 2924.Olivas-Aguirre M, Torres-Lopez L, Valle-Reyes JS, et al. Cannabidiol directly targets mitochondria and disturbs calcium homeostasis in acute lymphoblastic leukemia. Cell Death Dis. 2019. Oct 14;10(10):779. doi: 10.1038/s41419-019-2024-0. PubMed PMID: 31611561; PubMed Central PMCID: PMCPMC6791884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2925.Codogno P, Mehrpour M, Proikas-Cezanne T.. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011. Dec 14;13(1):7–12. doi: 10.1038/nrm3249. PubMed PMID: 22166994. [DOI] [PubMed] [Google Scholar]
- 2926.Demarchi F, Bertoli C, Copetti T, et al. Calpain is required for macroautophagy in mammalian cells. J Cell Biol. 2006. Nov 20;175(4):595–605. doi:jcb.200601024[pii] doi: 10.1083/jcb.200601024. PubMed PMID: 17101693; PubMed Central PMCID: PMC2064596. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2927.Tsugawa H, Mori H, Matsuzaki J, et al. CAPZA1 determines the risk of gastric carcinogenesis by inhibiting Helicobacter pylori CagA-degraded autophagy. Autophagy. 2019. Feb;15(2):242–258. doi: 10.1080/15548627.2018.1515530. PubMed PMID: 30176157; PubMed Central PMCID: PMCPMC6333452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2928.Du A, Huang S, Zhao X, et al. Suppression of CHRN endocytosis by carbonic anhydrase CAR3 in the pathogenesis of myasthenia gravis. Autophagy. 2017;13(11):1981–1994. doi: 10.1080/15548627.2017.1375633. PubMed PMID: 28933591; PubMed Central PMCID: PMCPMC5788490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2929.Gamerdinger M, Carra S, Behl C.. Emerging roles of molecular chaperones and co-chaperones in selective autophagy: focus on BAG proteins. J Mol Med (Berl). 2011. Dec;89(12):1175–82. doi: 10.1007/s00109-011-0795-6. PubMed PMID: 21818581. [DOI] [PubMed] [Google Scholar]
- 2930.Kanki T, Kurihara Y, Jin X, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013. Sep;14(9):788–94. doi: 10.1038/embor.2013.114. PubMed PMID: 23897086; PubMed Central PMCID: PMCPMC3790056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2931.Tsapras P, Nezis IP.. Caspase involvement in autophagy. Cell Death Differ. 2017. Aug;24(8):1369–1379. doi: 10.1038/cdd.2017.43. PubMed PMID: 28574508; PubMed Central PMCID: PMCPMC5520455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2932.Zhu Y, Zhao L, Liu L, et al. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell. 2010. May;1(5):468–77. doi: 10.1007/s13238-010-0048-4. PubMed PMID: 21203962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2933.Li H, Wang P, Sun Q, et al. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res. 2011. May 15;71(10):3625–34. doi: 10.1158/0008-5472.CAN-10-4475. PubMed PMID: 21444671; PubMed Central PMCID: PMC3096685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2934.Allavena G, Cuomo F, Baumgartner G, et al. Suppressed translation as a mechanism of initiation of CASP8 (caspase 8)-dependent apoptosis in autophagy-deficient NSCLC cells under nutrient limitation. Autophagy. 2018;14(2):252–268. doi: 10.1080/15548627.2017.1405192. PubMed PMID: 29165042; PubMed Central PMCID: PMCPMC5902222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2935.Chi J, Wang L, Zhang X, et al. Activation of calcium-sensing receptor-mediated autophagy in angiotensinII-induced cardiac fibrosis in vitro. Biochem Biophys Res Commun. 2018. Mar 4;497(2):571–576. doi: 10.1016/j.bbrc.2018.02.098. PubMed PMID: 29452090. [DOI] [PubMed] [Google Scholar]
- 2936.Peng X, Wei C, Li HZ, et al. NPS2390, a selective calcium-sensing receptor antagonist controls the phenotypic modulation of hypoxic human pulmonary arterial smooth muscle cells by regulating autophagy. J Transl Int Med. 2019. Jun;7(2):59–68. doi: 10.2478/jtim-2019-0013. PubMed PMID: 31380238; PubMed Central PMCID: PMCPMC6661874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2937.Liu L, Wang C, Lin Y, et al. Suppression of calcium sensing receptor ameliorates cardiac hypertrophy through inhibition of autophagy. Mol Med Rep. 2016. Jul;14(1):111–20. doi: 10.3892/mmr.2016.5279. PubMed PMID: 27176663; PubMed Central PMCID: PMCPMC4918534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2938.Liu L, Wang C, Sun D, et al. Calhex(2)(3)(1) Ameliorates cardiac hypertrophy by inhibiting cellular autophagy in vivo and in vitro. Cell Physiol Biochem. 2015;36(4):1597–612. doi: 10.1159/000430322. PubMed PMID: 26159880. [DOI] [PubMed] [Google Scholar]
- 2939.Garcia-Marcos M, Ear J, Farquhar MG, et al. A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals. Mol Biol Cell. 2011. Mar;22(5):673–86. doi: 10.1091/mbc.E10-08-0738. PubMed PMID: 21209316; PubMed Central PMCID: PMC3046063. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2940.Wild LWild F, Khan MM, Straka T, et al. Progress of endocytic CHRN to autophagic degradation is regulated by RAB5-GTPase and T145 phosphorylation of SH3GLB1 at mouse neuromuscular junctions in vivo. Autophagy. 2016. Dec;12(12):2300–2310. doi: 10.1080/15548627.2016.1234564. PubMed PMID: 27715385; PubMed Central PMCID: PMCPMC5173261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2941.Wu SY, Lan SH, Liu HS.. Degradative autophagy selectively regulates CCND1 (cyclin D1) and MIR224, two oncogenic factors involved in hepatocellular carcinoma tumorigenesis. Autophagy. 2019. Apr;15(4):729–730. doi: 10.1080/15548627.2019.1569918. PubMed PMID: 30646811; PubMed Central PMCID: PMCPMC6526824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2942.Sanjurjo L, Amezaga N, Aran G, et al. The human CD5L/AIM-CD36 axis: A novel autophagy inducer in macrophages that modulates inflammatory responses. Autophagy. 2015;11(3):487–502. doi: 10.1080/15548627.2015.1017183. PubMed PMID: 25713983; PubMed Central PMCID: PMCPMC4502645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2943.Sanjurjo L, Aran G, Roher N, et al. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J Leukoc Biol. 2015. Aug;98(2):173–84. doi: 10.1189/jlb.3RU0215-074R. PubMed PMID: 26048980. [DOI] [PubMed] [Google Scholar]
- 2944.Sanjurjo L, Aran G, Tellez E, et al. CD5L Promotes M2 Macrophage Polarization through Autophagy-Mediated Upregulation of ID3. Front Immunol. 2018;9:480. doi: 10.3389/fimmu.2018.00480. PubMed PMID: 29593730; PubMed Central PMCID: PMCPMC5858086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2945.Zhang Y, Xu M, Xia M, et al. Defective autophagosome trafficking contributes to impaired autophagic flux in coronary arterial myocytes lacking CD38 gene. Cardiovasc Res. 2014. Apr 1;102(1):68–78. doi: 10.1093/cvr/cvu011. PubMed PMID: 24445604; PubMed Central PMCID: PMCPMC3958620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2946.Xiong J, Xia M, Xu M, et al. Autophagy maturation associated with CD38-mediated regulation of lysosome function in mouse glomerular podocytes. J Cell Mol Med. 2013. Dec;17(12):1598–607. doi: 10.1111/jcmm.12173. PubMed PMID: 24238063; PubMed Central PMCID: PMCPMC3914646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2947.Latterich M, Frohlich KU, Schekman R.. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995. Sep 22;82(6):885–93. PubMed PMID: 7553849. [DOI] [PubMed] [Google Scholar]
- 2948.Krick R, Bremer S, Welter E, et al. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol. 2010. Sep 20;190(6):965–73. doi:jcb.201002075[pii] doi: 10.1083/jcb.201002075. PubMed PMID: 20855502; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2949.Joubert PE, Meiffren G, Gregoire IP, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009. Oct 22;6(4):354–66. doi: S1931-3128(09)00315-1 [pii] doi: 10.1016/j.chom.2009.09.006. PubMed PMID: 19837375; eng. [DOI] [PubMed] [Google Scholar]
- 2950.Celano SL, Yco LP, Kortus MG, et al. Identification of Kinases Responsible for p53-Dependent Autophagy. iScience. 2019. May 31;15:109–118. doi: 10.1016/j.isci.2019.04.023. PubMed PMID: 31048145; PubMed Central PMCID: PMCPMC6495467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2951.Shukla AK, Spurrier J, Kuzina I, et al. Hyperactive Innate Immunity Causes Degeneration of Dopamine Neurons upon Altering Activity of Cdk5. Cell Rep. 2019. Jan 2;26(1):131–144 e4. doi: 10.1016/j.celrep.2018.12.025. PubMed PMID: 30605670; PubMed Central PMCID: PMCPMC6442473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2952.Wong AS, Lee RH, Cheung AY, et al. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson’s disease. Nat Cell Biol. 2011. May;13(5):568–79. doi: 10.1038/ncb2217. PubMed PMID: 21499257. [DOI] [PubMed] [Google Scholar]
- 2953.Su LY, Li H, Lv L, et al. Melatonin attenuates MPTP-induced neurotoxicity via preventing CDK5-mediated autophagy and SNCA/alpha-synuclein aggregation. Autophagy. 2015;11(10):1745–59. doi: 10.1080/15548627.2015.1082020. PubMed PMID: 26292069; PubMed Central PMCID: PMCPMC4824603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2954.Orlotti NI, Cimino-Reale G, Borghini E, et al. Autophagy acts as a safeguard mechanism against G-quadruplex ligand-mediated DNA damage. Autophagy. 2012. Aug;8(8):1185–96. doi: 10.4161/auto.20519. PubMed PMID: 22627293. [DOI] [PubMed] [Google Scholar]
- 2955.Liang J, Shao SH, Xu ZX, et al. The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat Cell Biol. 2007. Feb;9(2):218–24. doi: ncb1537 [pii] doi: 10.1038/ncb1537. PubMed PMID: 17237771; eng. [DOI] [PubMed] [Google Scholar]
- 2956.Budina-Kolomets A, Hontz RD, Pimkina J, et al. A conserved domain in exon 2 coding for the human and murine ARF tumor suppressor protein is required for autophagy induction. Autophagy. 2013. Oct;9(10):1553–65. doi: 10.4161/auto.25831. PubMed PMID: 23939042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2957.Baechler BL, Bloemberg D, Quadrilatero J.. Mitophagy regulates mitochondrial network signaling, oxidative stress, and apoptosis during myoblast differentiation. Autophagy. 2019. Sep;15(9):1606–1619. doi: 10.1080/15548627.2019.1591672. PubMed PMID: 30859901; PubMed Central PMCID: PMCPMC6693454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2958.Mishra SK, Gao YG, Deng Y, et al. CPTP: A sphingolipid transfer protein that regulates autophagy and inflammasome activation. Autophagy. 2018;14(5):862–879. doi: 10.1080/15548627.2017.1393129. PubMed PMID: 29164996; PubMed Central PMCID: PMCPMC6070007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2959.Simanshu DK, Kamlekar RK, Wijesinghe DS, et al. Non-vesicular trafficking by a ceramide-1-phosphate transfer protein regulates eicosanoids. Nature. 2013. Aug 22;500(7463):463–7. doi: 10.1038/nature12332. PubMed PMID: 23863933; PubMed Central PMCID: PMCPMC3951269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2960.Qi HY, Daniels MP, Liu Y, et al. A cytosolic phospholipase A2-initiated lipid mediator pathway induces autophagy in macrophages. J Iimmunol. 2011. Nov 15;187(10):5286–92. doi: 10.4049/jimmunol.1004004. PubMed PMID: 22003202; PubMed Central PMCID: PMCPMC3208068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2961.Ward KE, Bhardwaj N, Vora M, et al. The molecular basis of ceramide-1-phosphate recognition by C2 domains. J Lipid Res. 2013. Mar;54(3):636–48. doi: 10.1194/jlr.M031088. PubMed PMID: 23277511; PubMed Central PMCID: PMCPMC3617939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2962.He MX, He YW.. A role for c-FLIP(L) in the regulation of apoptosis, autophagy, and necroptosis in T lymphocytes. Cell Death Differ. 2013. Feb;20(2):188–97. doi: 10.1038/cdd.2012.148. PubMed PMID: 23175183; PubMed Central PMCID: PMCPMC3554340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2963.Leidal AM, Cyr DP, Hill RJ, et al. Subversion of autophagy by Kaposi’s sarcoma-associated herpesvirus impairs oncogene-induced senescence. Cell Host Microbe. 2012. Feb 16;11(2):167–80. doi: 10.1016/j.chom.2012.01.005. PubMed PMID: 22341465. [DOI] [PubMed] [Google Scholar]
- 2964.Liang Q, Seo GJ, Choi YJ, et al. Crosstalk between the cGAS DNA sensor and beclin-1 autophagy protein shapes innate antimicrobial immune responses. Cell Host Microbe. 2014. Feb 12;15(2):228–38. doi: 10.1016/j.chom.2014.01.009. PubMed PMID: 24528868; PubMed Central PMCID: PMC3950946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2965.Ohwada J, Ebiike H, Kawada H, et al. Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799. Bioorg Med Chem Lett. 2011. Mar 15;21(6):1767–72. doi: 10.1016/j.bmcl.2011.01.065. PubMed PMID: 21316229. [DOI] [PubMed] [Google Scholar]
- 2966.Cuervo AM. Chaperone-mediated autophagy: selectivity pays off. Trends Endocrinol Metab 2010. Oct 24;21:142–50. doi: S1043-2760(09)00162-3 [pii] doi: 10.1016/j.tem.2009.10.003. PubMed PMID: 19857975; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2967.Dice J. Chaperone-mediated autophagy. Autophagy. 2007;3:295–9. [DOI] [PubMed] [Google Scholar]
- 2968.Agarraberes F, Terlecky S, Dice J.. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2969.Liu EY, Xu N, O’Prey J, et al. Loss of autophagy causes a synthetic lethal deficiency in DNA repair. Proc Natl Acad Sci U S A. 2015. Jan 20;112(3):773–8. doi: 10.1073/pnas.1409563112. PubMed PMID: 25568088; PubMed Central PMCID: PMCPMC4311830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2970.Mitsuhashi S, Hatakeyama H, Karahashi M, et al. Muscle choline kinase beta defect causes mitochondrial dysfunction and increased mitophagy. Hum Mol Genet. 2011. Oct 1;20(19):3841–51. doi: 10.1093/hmg/ddr305. PubMed PMID: 21750112; PubMed Central PMCID: PMC3168292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2971.Fedorko M. Effect of chloroquine on morphology of cytoplasmic granules in maturing human leukocytes--an ultrastructural study. J Clin Invest. 1967. Dec;46(12):1932–42. doi: 10.1172/JCI105683. PubMed PMID: 6073998; PubMed Central PMCID: PMC292946. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2972.Barbero-Camps E, Roca-Agujetas V, Bartolessis I, et al. Cholesterol impairs autophagy-mediated clearance of amyloid beta while promoting its secretion. Autophagy. 2018;14(7):1129–1154. doi: 10.1080/15548627.2018.1438807. PubMed PMID: 29862881; PubMed Central PMCID: PMCPMC6103708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2973.Fraldi A, Annunziata F, Lombardi A, et al. Lysosomal fusion and SNARE function are impaired by cholesterol accumulation in lysosomal storage disorders. EMBO J. 2010. Nov 3;29(21):3607–20. doi: 10.1038/emboj.2010.237. PubMed PMID: 20871593; PubMed Central PMCID: PMCPMC2982760. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2974.Almacellas E, Pelletier J, Day C, et al. Lysosomal degradation ensures accurate chromosomal segregation to prevent chromosomal instability. Autophagy. 2020. Jun 23:1–18. doi: 10.1080/15548627.2020.1764727. PubMed PMID: 32573315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2975.Lam HC, Cloonan SM, Bhashyam AR, et al. Histone deacetylase 6-mediated selective autophagy regulates COPD-associated cilia dysfunction. J Clin Invest. 2013. Dec;123(12):5212–30. doi: 10.1172/JCI69636. PubMed PMID: 24200693; PubMed Central PMCID: PMCPMC3859407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2976.Lee J, Yi S, Kang YE, et al. Defective ciliogenesis in thyroid hurthle cell tumors is associated with increased autophagy. Oncotarget. 2016. Nov 29;7(48):79117–79130. doi: 10.18632/oncotarget.12997. PubMed PMID: 27816963; PubMed Central PMCID: PMCPMC5346702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2977.Cloonan SM, Lam HC, Ryter SW, et al. “Ciliophagy”: The consumption of cilia components by autophagy. Autophagy. 2014. Mar;10(3):532–4. doi: 10.4161/auto.27641. PubMed PMID: 24401596; PubMed Central PMCID: PMCPMC4077895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2978.Morleo M, Franco B.. The Autophagy-Cilia Axis: An Intricate Relationship. Cells. 2019. Aug 15;8(8). doi: 10.3390/cells8080905. PubMed PMID: 31443299; PubMed Central PMCID: PMCPMC6721705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2979.Puustinen P, Rytter A, Mortensen M, et al. CIP2A oncoprotein controls cell growth and autophagy through mTORC1 activation. J Cell Biol. 2014. Mar 3;204(5):713–27. doi: 10.1083/jcb.201304012. PubMed PMID: 24590173; PubMed Central PMCID: PMC3941044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2980.Du WW, Yang W, Li X, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018. Nov;37(44):5829–5842. doi: 10.1038/s41388-018-0369-y. PubMed PMID: 29973691. [DOI] [PubMed] [Google Scholar]
- 2981.Liu F, Zhang J, Qin L, et al. Circular RNA EIF6 (Hsa_circ_0060060) sponges miR-144-3p to promote the cisplatin-resistance of human thyroid carcinoma cells by autophagy regulation. Aging (Albany NY). 2018. Dec 12;10(12):3806–3820. doi: 10.18632/aging.101674. PubMed PMID: 30540564; PubMed Central PMCID: PMCPMC6326687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2982.Yu T, Ding Y, Zhang Y, et al. Circular RNA GATAD2A promotes H1N1 replication through inhibiting autophagy. Vet Microbiol. 2019. Apr;231:238–245. doi: 10.1016/j.vetmic.2019.03.012. PubMed PMID: 30955816. [DOI] [PubMed] [Google Scholar]
- 2983.Han B, Zhang Y, Zhang Y, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. 2018;14(7):1164–1184. doi: 10.1080/15548627.2018.1458173. PubMed PMID: 29938598; PubMed Central PMCID: PMCPMC6103660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2984.Yang L, Han B, Zhang Y, et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. 2018;14(3):404–418. doi: 10.1080/15548627.2017.1414755. PubMed PMID: 29260931; PubMed Central PMCID: PMCPMC5915020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2985.Huang R, Zhang Y, Han B, et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017. Oct 3;13(10):1722–1741. doi: 10.1080/15548627.2017.1356975. PubMed PMID: 28786753; PubMed Central PMCID: PMCPMC5640207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2986.Chen X, Mao R, Su W, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKalpha signaling in STK11 mutant lung cancer. Autophagy. 2019. Jun 28:1–13. doi: 10.1080/15548627.2019.1634945. PubMed PMID: 31232177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2987.Zhou ZB, Niu YL, Huang GX, et al. Silencing of circRNA.2837 plays a protective role in sciatic nerve injury by sponging the miR-34 family via regulating neuronal autophagy. Mol Ther Nucleic Acids. 2018. Sep 7;12:718–729. doi: 10.1016/j.omtn.2018.07.011. PubMed PMID: 30098504; PubMed Central PMCID: PMCPMC6088565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2988.Zhou LY, Zhai M, Huang Y, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/ FAM65B pathway. Cell Death Differ. 2019. Jul;26(7):1299–1315. doi: 10.1038/s41418-018-0206-4. PubMed PMID: 30349076; PubMed Central PMCID: PMCPMC6748144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2989.Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019. Nov;20(11):675–691. doi: 10.1038/s41576-019-0158-7. PubMed PMID: 31395983. [DOI] [PubMed] [Google Scholar]
- 2990.Li X, Yang L, Chen LL.. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018. Aug 2;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. PubMed PMID: 30057200. [DOI] [PubMed] [Google Scholar]
- 2991.Zhang J, Wang P, Wan L, et al. The emergence of noncoding RNAs as Heracles in autophagy. Autophagy. 2017. Jun 3;13(6):1004–1024. doi: 10.1080/15548627.2017.1312041. PubMed PMID: 28441084; PubMed Central PMCID: PMCPMC5486373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2992.Meng L, Liu S, Ding P, et al. Circular RNA ciRS-7 inhibits autophagy of ESCC cells by functioning as miR-1299 sponge to target EGFR signaling. J Cell Biochem. 2020. Feb;121(2):1039–1049. doi: 10.1002/jcb.29339. PubMed PMID: 31490018. [DOI] [PubMed] [Google Scholar]
- 2993.Chang NC, Nguyen M, Germain M, et al. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010. Feb 3;29(3):606–18. doi: 10.1038/emboj.2009.369. PubMed PMID: 20010695; PubMed Central PMCID: PMC2830692. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2994.Chen YF, Kao CH, Chen YT, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009. May 15;23(10):1183–94. doi: 10.1101/gad.1779509. PubMed PMID: 19451219; PubMed Central PMCID: PMC2685531. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2995.Yang Z, Geng J, Yen W-L, et al. Positive or negative regulatory roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae Mol Cell. 2010;38:250–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2996.Cao Y, Espinola JA, Fossale E, et al. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem. 2006. Jul 21;281(29):20483–93. doi: 10.1074/jbc.M602180200. PubMed PMID: 16714284. [DOI] [PubMed] [Google Scholar]
- 2997.Chandrachud U, Walker MW, Simas AM, et al. Unbiased cell-based screening in a neuronal cell model of batten disease highlights an interaction between Ca2+ homeostasis, autophagy, and CLN3 protein function. J Biol Chem. 2015. Jun 5;290(23):14361–80. doi: 10.1074/jbc.M114.621706. PubMed PMID: 25878248; PubMed Central PMCID: PMC4505505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2998.Cortese A, Tucci A, Piccolo G, et al. Novel CLN3 mutation causing autophagic vacuolar myopathy. Neurology. 2014. Jun 10;82(23):2072–6. doi: 10.1212/WNL.0000000000000490. PubMed PMID: 24827497; PubMed Central PMCID: PMC4118497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2999.Wang F, Wang H, Tuan HF, et al. Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014. Mar;133(3):331–45. doi: 10.1007/s00439-013-1381-5. PubMed PMID: 24154662; PubMed Central PMCID: PMC3945441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3000.Yen W-L, Shintani T, Nair U, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010. Jan 11;188(1):101–14. doi: jcb.200904075 [pii] doi: 10.1083/jcb.200904075. PubMed PMID: 20065092; PubMed Central PMCID: PMC2812853. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3001.Sir D, Chen WL, Choi J, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008. Oct;48(4):1054–61. doi: 10.1002/hep.22464. PubMed PMID: 18688877; PubMed Central PMCID: PMCPMC2562598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3002.Shoji-Kawata S, Levine B.. Autophagy, antiviral immunity, and viral countermeasures. Biochim Biophys Acta. 2009. Sep;1793(9):1478–84. doi: 10.1016/j.bbamcr.2009.02.008. PubMed PMID: 19264100; PubMed Central PMCID: PMCPMC2739265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3003.Drose S, Bindseil KU, Bowman EJ, et al. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 1993. Apr 20;32(15):3902–6. doi: 10.1021/bi00066a008. PubMed PMID: 8385991. [DOI] [PubMed] [Google Scholar]
- 3004.Marcelo A, Brito F, Carmo-Silva S, et al. Cordycepin activates autophagy through AMPK phosphorylation to reduce abnormalities in Machado-Joseph disease models. Hum Mol Genet. 2019. Jan 1;28(1):51–63. doi: 10.1093/hmg/ddy328. PubMed PMID: 30219871. [DOI] [PubMed] [Google Scholar]
- 3005.Lancel S, Montaigne D, Marechal X, et al. Carbon monoxide improves cardiac function and mitochondrial population quality in a mouse model of metabolic syndrome. PLoS One. 2012;7(8):e41836. doi: 10.1371/journal.pone.0041836. PubMed PMID: 22870253; PubMed Central PMCID: PMC3411569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3006.Chen LL, Song JX, Lu JH, et al. Corynoxine, a natural autophagy enhancer, promotes the clearance of alpha-synuclein via Akt/mTOR pathway. J Neuroimmune Pharmacol. 2014. Feb 13:380–7. doi: 10.1007/s11481-014-9528-2. PubMed PMID: 24522518. [DOI] [PubMed] [Google Scholar]
- 3007.Lu JH, Tan JQ, Durairajan SS, et al. Isorhynchophylline, a natural alkaloid, promotes the degradation of alpha-synuclein in neuronal cells via inducing autophagy. Autophagy. 2012. Jan;8(1):98–108 (see also the erratum in Autophagy 2012;8:864-6). doi: 10.4161/auto.8.1.18313. PubMed PMID: 22113202. [DOI] [PubMed] [Google Scholar]
- 3008.Casas C, Codogno P, Pinti M, et al. TRANSAUTOPHAGY: European network for multidisciplinary research and translation of autophagy knowledge. Autophagy. 2016;12(3):614–7. doi: 10.1080/15548627.2016.1140294. PubMed PMID: 27046256; PubMed Central PMCID: PMCPMC4836001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3009.Bao Y, Song WM, Wang P, et al. COST1 regulates autophagy to control plant drought tolerance. Proc Natl Acad Sci U S A. 2020. Mar 31;117(13):7482–7493. doi: 10.1073/pnas.1918539117. PubMed PMID: 32170020; PubMed Central PMCID: PMCPMC7132278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3010.Shi L, Wang J, Quan R, et al. CpATG8, a homolog of yeast autophagy protein ATG8, is required for pathogenesis and hypovirus accumulation in the chest blight fungus. Front Cell Infect Microbiol. 2019;9:222. doi: 10.3389/fcimb.2019.00222. PubMed PMID: 31355148; PubMed Central PMCID: PMCPMC6635641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3011.Mylka V, Deckers J, Ratman D, et al. The autophagy receptor SQSTM1/p62 mediates anti-inflammatory actions of the selective NR3C1/glucocorticoid receptor modulator compound A (CpdA) in macrophages. Autophagy. 2018;14(12):2049–2064. doi: 10.1080/15548627.2018.1495681. PubMed PMID: 30215534; PubMed Central PMCID: PMCPMC6984772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3012.Song H, Pu J, Wang L, et al. ATG16L1 phosphorylation is oppositely regulated by CSNK2/casein kinase 2 and PPP1/protein phosphatase 1 which determines the fate of cardiomyocytes during hypoxia/reoxygenation. Autophagy. 2015. Jun 17;11:1308–25. doi: 10.1080/15548627.2015.1060386. PubMed PMID: 26083323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3013.Cui L, Zhao H, Yin Y, et al. Function of Atg11 in non-selective autophagy and selective autophagy of Candida albicans. Biochem Biophys Res Commun. 2019. Sep 3;516(4):1152–1158. doi: 10.1016/j.bbrc.2019.06.148. PubMed PMID: 31284951. [DOI] [PubMed] [Google Scholar]
- 3014.Festa BP, Chen Z, Berquez M, et al. Impaired autophagy bridges lysosomal storage disease and epithelial dysfunction in the kidney. Nat Commun. 2018. Jan 11;9(1):161. doi: 10.1038/s41467-017-02536-7. PubMed PMID: 29323117; PubMed Central PMCID: PMCPMC5765140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3015.Luciani A, Festa BP, Chen Z, et al. Defective autophagy degradation and abnormal tight junction-associated signaling drive epithelial dysfunction in cystinosis. Autophagy. 2018;14(7):1157–1159. doi: 10.1080/15548627.2018.1446625. PubMed PMID: 29806776; PubMed Central PMCID: PMCPMC6103718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3016.Zheng L, Shu WJ, Li YM, et al. The Paf1 complex transcriptionally regulates the mitochondrial-anchored protein Atg32 leading to activation of mitophagy. Autophagy. 2019. Sep 19:in press. doi: 10.1080/15548627.2019.1668228. PubMed PMID: 31525119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3017.Stoka V, Turk V, Turk B.. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res Rev. 2016. Dec;32:22–37. doi: 10.1016/j.arr.2016.04.010. PubMed PMID: 27125852. [DOI] [PubMed] [Google Scholar]
- 3018.Campbell EM, Fares H.. Roles of CUP-5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis. BMC Cell Biol. 2010;11:40. doi: 10.1186/1471-2121-11-40. PubMed PMID: 20540742; PubMed Central PMCID: PMC2891664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3019.Fares H, Greenwald I.. Regulation of endocytosis by CUP-5, the Caenorhabditis elegans mucolipin-1 homolog. Nat Genet. 2001. May;28(1):64–8. doi: 10.1038/88281. PubMed PMID: 11326278. [DOI] [PubMed] [Google Scholar]
- 3020.Hersh BM, Hartwieg E, Horvitz HR.. The Caenorhabditis elegans mucolipin-like gene cup-5 is essential for viability and regulates lysosomes in multiple cell types. Proc Natl Acad Sci U S A. 2002. Apr 2;99(7):4355–60. doi: 10.1073/pnas.062065399. PubMed PMID: 11904372; PubMed Central PMCID: PMC123652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3021.Sun T, Wang X, Lu Q, et al. CUP-5, the C. elegans ortholog of the mammalian lysosomal channel protein MLN1/TRPML1, is required for proteolytic degradation in autolysosomes. Autophagy. 2011. Nov;7(11):1308–15. doi: 10.4161/auto.7.11.17759. PubMed PMID: 21997367. [DOI] [PubMed] [Google Scholar]
- 3022.Bruns C, McCaffery JM, Curwin AJ, et al. Biogenesis of a novel compartment for autophagosome-mediated unconventional protein secretion. J Cell Biol. 2011. Dec 12;195(6):979–92. doi: 10.1083/jcb.201106098. PubMed PMID: 22144692; PubMed Central PMCID: PMC3241719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3023.Yeung AWK, Horbanczuk M, Tzvetkov NT, et al. Curcumin: total-scale analysis of the scientific literature. Molecules. 2019. Apr 9;24(7). doi: 10.3390/molecules24071393. PubMed PMID: 30970601; PubMed Central PMCID: PMCPMC6480685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3024.Shakeri A, Cicero AFG, Panahi Y, et al. Curcumin: A naturally occurring autophagy modulator. J Cell Physiol. 2019. May;234(5):5643–5654. doi: 10.1002/jcp.27404. PubMed PMID: 30239005. [DOI] [PubMed] [Google Scholar]
- 3025.Maiti P, Scott J, Sengupta D, et al. Curcumin and solid lipid curcumin particles induce autophagy, but inhibit mitophagy and the PI3K-Akt/mTOR Pathway in Cultured Glioblastoma Cells. Int J Mol Sci. 2019. Jan 18;20(2). doi: 10.3390/ijms20020399. PubMed PMID: 30669284; PubMed Central PMCID: PMCPMC6359162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3026.Wescott MP, Kufareva I, Paes C, et al. Signal transmission through the CXC chemokine receptor 4 (CXCR4) transmembrane helices. Proc Natl Acad Sci U S A. 2016. Aug 30;113(35):9928–33. doi: 10.1073/pnas.1601278113. PubMed PMID: 27543332; PubMed Central PMCID: PMCPMC5024644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3027.Nobrega C, Mendonca L, Marcelo A, et al. Restoring brain cholesterol turnover improves autophagy and has therapeutic potential in mouse models of spinocerebellar ataxia. Acta Neuropathol. 2019. Nov;138(5):837–858. doi: 10.1007/s00401-019-02019-7. PubMed PMID: 31197505. [DOI] [PubMed] [Google Scholar]
- 3028.Wang M, Tan W, Zhou J, et al. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008. Jul 4;283(27):18678–84. doi: M801855200 [pii] doi: 10.1074/jbc.M801855200. PubMed PMID: 18434300; eng. [DOI] [PubMed] [Google Scholar]
- 3029.Zou J, Chen Z, Wei X, et al. Cystatin C as a potential therapeutic mediator against Parkinson’s disease via VEGF-induced angiogenesis and enhanced neuronal autophagy in neurovascular units. Cell Death Dis. 2017. Jun 1;8(6):e2854. doi: 10.1038/cddis.2017.240. PubMed PMID: 28569795; PubMed Central PMCID: PMCPMC5520899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3030.Harding TM, Morano KA, Scott SV, et al. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1995. Nov;131(3):591–602. PubMed PMID: 7593182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3031.Hansen M, Chandra A, Mitic LL, et al. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet. 2008. Feb;4(2):e24. doi: 10.1371/journal.pgen.0040024. PubMed PMID: 18282106; PubMed Central PMCID: PMC2242811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3032.Lapierre LR, Gelino S, Melendez A, et al. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr Biol. 2011. Sep 27;21(18):1507–14. doi: 10.1016/j.cub.2011.07.042. PubMed PMID: 21906946; PubMed Central PMCID: PMC3191188. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3033.Sandhof CA, Hoppe SO, Druffel-Augustin S, et al. Reducing INS-IGF1 signaling protects against non-cell autonomous vesicle rupture caused by SNCA spreading. Autophagy. 2019. Jul 29:1–22. doi: 10.1080/15548627.2019.1643657. PubMed PMID: 31354022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3034.Netea-Maier RT, Plantinga TS, Van De Veerdonk FL, et al. Modulation of inflammation by autophagy: consequences for human disease. Autophagy. 2016. Jul 29;12:245–60. doi: 10.1080/15548627.2015.1071759. PubMed PMID: 26222012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3035.Mills KH. TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol. 2011. Nov 18;11(12):807–22. doi: 10.1038/nri3095. PubMed PMID: 22094985. [DOI] [PubMed] [Google Scholar]
- 3036.Koren I, Reem E, Kimchi A.. DAP1, a novel substrate of mTOR, negatively regulates autophagy. Curr Biol. 2010. May 26;20:1093–8. doi: S0960-9822(10)00520-8 [pii] doi: 10.1016/j.cub.2010.04.041. PubMed PMID: 20537536; Eng. [DOI] [PubMed] [Google Scholar]
- 3037.Inbal B, Bialik S, Sabanay I, et al. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002. Apr 29;157(3):455–68. doi: 10.1083/jcb.200109094 jcb.200109094 [pii]. PubMed PMID: 11980920; PubMed Central PMCID: PMC2173279. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3038.Yang Y, Willis TL, Button RW, et al. Cytoplasmic DAXX drives SQSTM1/p62 phase condensation to activate Nrf2-mediated stress response. Nat Commun. 2019. Aug 21;10(1):3759. doi: 10.1038/s41467-019-11671-2. PubMed PMID: 31434890; PubMed Central PMCID: PMCPMC6704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3039.Chou TF, Brown SJ, Minond D, et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A. 2011. Mar 22;108(12):4834–9. doi: 10.1073/pnas.1015312108. PubMed PMID: 21383145; PubMed Central PMCID: PMCPMC3064330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3040.Bravo-San Pedro JM, Sica V, Martins I, et al. Acyl-CoA-binding protein is a lipogenic factor that triggers food intake and obesity. Cell Metab. 2019. Oct 1;30(4):754–767 e9. doi: 10.1016/j.cmet.2019.07.010. PubMed PMID: 31422903. [DOI] [PubMed] [Google Scholar]
- 3041.Buraschi S, Neill T, Goyal A, et al. Decorin causes autophagy in endothelial cells via Peg3. Proc Natl Acad Sci U S A. 2013. Jul 9;110(28):E2582–91. doi: 10.1073/pnas.1305732110. PubMed PMID: 23798385; PubMed Central PMCID: PMC3710796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3042.Gubbiotti MA, Neill T, Frey H, et al. Decorin is an autophagy-inducible proteoglycan and is required for proper in vivo autophagy. Matrix Biol. 2015. Oct;48:14–25. doi: 10.1016/j.matbio.2015.09.001. PubMed PMID: 26344480; PubMed Central PMCID: PMCPMC4661125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3043.DeVorkin L, Go NE, Hou Y-CC, et al. The Drosophila effector caspase Dcp-1 regulates mitochondrial dynamics and autophagic flux via SesB. J Cell Biol. 2014. May 26;205(4):477–492. doi: 10.1083/jcb.201303144. PubMed PMID: 24862573; PubMed Central PMCID: PMC4033768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3044.Hu G, McQuiston T, Bernard A, et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nat Cell Biol. 2015. Jul;17(7):930–42. doi: 10.1038/ncb3189. PubMed PMID: 26098573; PubMed Central PMCID: PMC4528364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3045.Molitoris JK, McColl KS, Swerdlow S, et al. Glucocorticoid elevation of dexamethasone-induced gene 2 (Dig2/RTP801/REDD1) protein mediates autophagy in lymphocytes. J Biol Chem. 2011. Aug 26;286(34):30181–9. doi: 10.1074/jbc.M111.245423. PubMed PMID: 21733849; PubMed Central PMCID: PMC3191057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3046.Slavov N, Botstein D.. Decoupling nutrient signaling from growth rate causes aerobic glycolysis and deregulation of cell size and gene expression. Mol Biol Cell. 2013. Jan;24(2):157–68. doi: 10.1091/mbc.E12-09-0670. PubMed PMID: 23135997; PubMed Central PMCID: PMC3541962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3047.Lv Q, Hua F, Hu ZW.. DEDD, a novel tumor repressor, reverses epithelial-mesenchymal transition by activating selective autophagy. Autophagy. 2012. Nov;8(11):1675–6. doi: 10.4161/auto.21438. PubMed PMID: 22874565; PubMed Central PMCID: PMCPMC3494596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3048.Lv Q, Wang W, Xue J, et al. DEDD interacts with PI3KC3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012. Jul 1;72(13):3238–50. doi: 10.1158/0008-5472.CAN-11-3832. PubMed PMID: 22719072. [DOI] [PubMed] [Google Scholar]
- 3049.Mita M, Sankhala K, Abdel-Karim I, et al. Deforolimus (AP23573) a novel mTOR inhibitor in clinical development. Expert Opin Investig Drugs. 2008. Dec;17(12):1947–54. doi: 10.1517/13543780802556485. PubMed PMID: 19012509. [DOI] [PubMed] [Google Scholar]
- 3050.Kohler K, Brunner E, Guan XL, et al. A combined proteomic and genetic analysis identifies a role for the lipid desaturase Desat1 in starvation-induced autophagy in Drosophila. Autophagy. 2009. Oct;5(7):980–90. PubMed PMID: 19587536. [DOI] [PubMed] [Google Scholar]
- 3051.Liu X, Yao Z, Jin M, et al. Dhh1 promotes autophagy-related protein translation during nitrogen starvation. PLoS Biol. 2019. Apr;17(4):e3000219. doi: 10.1371/journal.pbio.3000219. PubMed PMID: 30973873; PubMed Central PMCID: PMCPMC6459490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3052.Shahnazari S, Yen W-L, Birmingham CL, et al. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010. Aug 19;8(2):137–46. doi: S1931-3128(10)00219-2 [pii] doi: 10.1016/j.chom.2010.07.002. PubMed PMID: 20674539; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3053.Lu Z, Baquero MT, Yang H, et al. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy. 2014. Jun;10(6):1071–92. doi: 10.4161/auto.28577. PubMed PMID: 24879154; PubMed Central PMCID: PMC4091169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3054.Ejaz A, Mitterberger MC, Lu Z, et al. Weight loss upregulates the small GTPase DIRAS3 in human white adipose progenitor cells, which negatively regulates adipogenesis and activates autophagy via Akt-mTOR inhibition. EBioMedicine. 2016. Apr;6:149–161. doi: 10.1016/j.ebiom.2016.03.030. PubMed PMID: 27211557; PubMed Central PMCID: PMCPMC4856797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3055.Ejaz A, Mattesich M, Zwerschke W.. Silencing of the small GTPase DIRAS3 induces cellular senescence in human white adipose stromal/progenitor cells. Aging (Albany NY). 2017. Mar 17;9(3):860–879. doi: 10.18632/aging.101197. PubMed PMID: 28316325; PubMed Central PMCID: PMCPMC5391236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3056.Mao K, Liu X, Feng Y, et al. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy. 2014. Apr;10(4):652–61. doi: 10.4161/auto.27852. PubMed PMID: 24451165; PubMed Central PMCID: PMC4091152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3057.Dagda RK, Gusdon AM, Pien I, et al. Mitochondrially localized PKA reverses mitochondrial pathology and dysfunction in a cellular model of Parkinson’s disease. Cell Death Differ. 2011. Dec;18(12):1914–23. doi: 10.1038/cdd.2011.74. PubMed PMID: 21637291; PubMed Central PMCID: PMC3177020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3058.Kwon MH, Callaway H, Zhong J, et al. A targeted genetic modifier screen links the SWI2/SNF2 protein domino to growth and autophagy genes in Drosophila melanogaster. G3 (Bethesda). 2013. May;3(5):815–25. doi: 10.1534/g3.112.005496. PubMed PMID: 23550128; PubMed Central PMCID: PMC3656729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3059.Gomez-Santos C, Ferrer I, Santidrian AF, et al. Dopamine induces autophagic cell death and alpha-synuclein increase in human neuroblastoma SH-SY5Y cells. J Neurosci Res. 2003. Aug 1;73(3):341–50. doi: 10.1002/jnr.10663. PubMed PMID: 12868068. [DOI] [PubMed] [Google Scholar]
- 3060.Leng ZG, Lin SJ, Wu ZR, et al. Activation of DRD5 (dopamine receptor D5) inhibits tumor growth by autophagic cell death. Autophagy. 2017. Aug 3;13(8):1404–1419. doi: 10.1080/15548627.2017.1328347. PubMed PMID: 28613975; PubMed Central PMCID: PMCPMC5584849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3061.Dolma S, Selvadurai HJ, Lan X, et al. Inhibition of Dopamine Receptor D4 Impedes Autophagic Flux, Proliferation, and Survival of Glioblastoma Stem Cells. Cancer cell. 2016. Jun 13;29(6):859–873. doi: 10.1016/j.ccell.2016.05.002. PubMed PMID: 27300435; PubMed Central PMCID: PMCPMC5968455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3062.Barroso-Chinea P, Luis-Ravelo D, Fumagallo-Reading F, et al. DRD3 (dopamine receptor D3) but not DRD2 activates autophagy through MTORC1 inhibition preserving protein synthesis. Autophagy. 2019. Oct 2:1–17. doi: 10.1080/15548627.2019.1668606. PubMed PMID: 31538542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3063.Valbuena A, Castro-Obregon S, Lazo PA.. Downregulation of VRK1 by p53 in response to DNA damage is mediated by the autophagic pathway. PLoS One. 2011;6(2):e17320. doi: 10.1371/journal.pone.0017320. PubMed PMID: 21386980; PubMed Central PMCID: PMC3046209. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3064.Guan JJ, Zhang XD, Sun W, et al. DRAM1 regulates apoptosis through increasing protein levels and lysosomal localization of BAX. Cell Death Dis. 2015. Jan 29;6:e1624. doi: 10.1038/cddis.2014.546. PubMed PMID: 25633293; PubMed Central PMCID: PMCPMC4669745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3065.Crighton D, Wilkinson S, O’Prey J, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006. Jul 14;126(1):121–34. doi: 10.1016/j.cell.2006.05.034. PubMed PMID: 16839881; eng. [DOI] [PubMed] [Google Scholar]
- 3066.O’Prey J, Skommer J, Wilkinson S, et al. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell cycle. 2009. Jul 15;8(14):2260–5. doi: 10.4161/cc.8.14.9050. PubMed PMID: 19556885. [DOI] [PubMed] [Google Scholar]
- 3067.Yoon JH, Her S, Kim M, et al. The expression of damage-regulated autophagy modulator 2 (DRAM2) contributes to autophagy induction. Mol Biol Rep. 2012. Feb;39(2):1087–93. doi: 10.1007/s11033-011-0835-x. PubMed PMID: 21584698. [DOI] [PubMed] [Google Scholar]
- 3068.Kim JK, Lee HM, Park KS, et al. MIR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy. 2017. Feb;13(2):423–441. doi: 10.1080/15548627.2016.1241922. PubMed PMID: 27764573; PubMed Central PMCID: PMCPMC5324854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3069.McPhee CK, Logan MA, Freeman MR, et al. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010. Jun 24;465(7301):1093–U159. doi: Doi 10.1038/Nature09127. PubMed PMID: ISI:000279056900056; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3070.Nolan TM, Brennan B, Yang M, et al. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell. 2017. Apr 10;41(1):33–46 e7. doi: 10.1016/j.devcel.2017.03.013. PubMed PMID: 28399398; PubMed Central PMCID: PMCPMC5720862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3071.Chen TH, Chen MR, Chen TY, et al. Cardiac fibrosis in mouse expressing DsRed tetramers involves chronic autophagy and proteasome degradation insufficiency. Oncotarget. 2016. Aug 23;7(34):54274–54289. doi: 10.18632/oncotarget.11026. PubMed PMID: 27494843; PubMed Central PMCID: PMCPMC5342341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3072.Choi JC, Wu W, Muchir A, et al. Dual specificity phosphatase 4 mediates cardiomyopathy caused by lamin A/C (LMNA) gene mutation. J Biol Chem. 2012. Nov 23;287(48):40513–24. doi: 10.1074/jbc.M112.404541. PubMed PMID: 23048029; PubMed Central PMCID: PMCPMC3504766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3073.Ragusa MJ, Stanley RE, Hurley JH.. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012. Dec 21;151(7):1501–12. doi: 10.1016/j.cell.2012.11.028. PubMed PMID: 23219485; PubMed Central PMCID: PMC3806636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3074.Jia K, Levine B.. Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy. 2007. Nov-Dec;3(6):597–9. PubMed PMID: 17912023. [DOI] [PubMed] [Google Scholar]
- 3075.Toth ML, Sigmond T, Borsos E, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008. Apr;4(3):330–8. PubMed PMID: 18219227. [DOI] [PubMed] [Google Scholar]
- 3076.Bandyopadhyay U, Sridhar S, Kaushik S, et al. Identification of regulators of chaperone-mediated autophagy. Mol Cell. 2010. Aug 27;39(4):535–47. doi: 10.1016/j.molcel.2010.08.004. PubMed PMID: 20797626; PubMed Central PMCID: PMC2945256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3077.Nicastro R, Sardu A, Panchaud N, et al. The Architecture of the Rag GTPase Signaling Network. Biomolecules. 2017. Jun 30;7(3). doi: 10.3390/biom7030048. PubMed PMID: 28788436; PubMed Central PMCID: PMCPMC5618229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3078.Zhang T, Peli-Gulli MP, Zhang Z, et al. Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases. Sci Adv. 2019. Sep;5(9):eaax8164. doi: 10.1126/sciadv.aax8164. PubMed PMID: 31579828; PubMed Central PMCID: PMCPMC6760929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3079.Talloczy Z, Jiang W, HWt Virgin, et al. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci U S A. 2002. Jan 8;99(1):190–5. doi: 10.1073/pnas.012485299. PubMed PMID: 11756670; PubMed Central PMCID: PMC117537. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3080.Chng SC, Ho L, Tian J, et al. ELABELA: a hormone essential for heart development signals via the apelin receptor. Dev Cell. 2013. Dec 23;27(6):672–80. doi: 10.1016/j.devcel.2013.11.002. PubMed PMID: 24316148. [DOI] [PubMed] [Google Scholar]
- 3081.Pauli A, Norris ML, Valen E, et al. Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science. 2014. Feb 14;343(6172):1248636. doi: 10.1126/science.1248636. PubMed PMID: 24407481; PubMed Central PMCID: PMCPMC4107353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3082.Chen H, Wang L, Wang W, et al. ELABELA and an ELABELA Fragment Protect against AKI. J Am Soc Nephrol. 2017. Sep;28(9):2694–2707. doi: 10.1681/ASN.2016111210. PubMed PMID: 28583915; PubMed Central PMCID: PMCPMC5576937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3083.Zhao X, Fang Y, Yang Y, et al. Elaiophylin, a novel autophagy inhibitor, exerts antitumor activity as a single agent in ovarian cancer cells. Autophagy. 2015. Apr 20;11:1849–63. doi: 10.1080/15548627.2015.1017185. PubMed PMID: 25893854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3084.Kim S, Naylor SA, DiAntonio A.. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc Natl Acad Sci U S A. 2012. May 1;109(18):E1072–81. doi: 10.1073/pnas.1120320109. PubMed PMID: 22493244; PubMed Central PMCID: PMC3344964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3085.Berge T, Leikfoss IS, Harbo HF.. From identification to characterization of the multiple sclerosis susceptibility gene CLEC16A. Int J Mol Sci. 2013;14(3):4476–97. doi: 10.3390/ijms14034476. PubMed PMID: 23439554; PubMed Central PMCID: PMC3634488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3086.Soleimanpour SA, Gupta A, Bakay M, et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell. 2014. Jun 19;157(7):1577–90. doi: 10.1016/j.cell.2014.05.016. PubMed PMID: 24949970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3087.Li Y, Zhao Y, Hu J, et al. A novel ER-localized transmembrane protein, EMC6, interacts with RAB5A and regulates cell autophagy. Autophagy. 2013. Feb 1;9(2):150–63. doi: 10.4161/auto.22742. PubMed PMID: 23182941; PubMed Central PMCID: PMC3552880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3088.Soukup SF, Kuenen S, Vanhauwaert R, et al. A LRRK2-Dependent EndophilinA Phosphoswitch Is Critical for Macroautophagy at Presynaptic Terminals. Neuron. 2016. Nov 23;92(4):829–844. doi: 10.1016/j.neuron.2016.09.037. PubMed PMID: 27720484. [DOI] [PubMed] [Google Scholar]
- 3089.Murdoch JD, Rostosky CM, Gowrisankaran S, et al. Endophilin-A Deficiency Induces the Foxo3a-Fbxo32 Network in the Brain and Causes Dysregulation of Autophagy and the Ubiquitin-Proteasome System. Cell Rep. 2016. Oct 18;17(4):1071–1086. doi: 10.1016/j.celrep.2016.09.058. PubMed PMID: 27720640; PubMed Central PMCID: PMCPMC5080600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3090.Poluzzi C, Casulli J, Goyal A, et al. Endorepellin evokes autophagy in endothelial cells. J Biol Chem. 2014. Jun 6;289(23):16114–28. doi: 10.1074/jbc.M114.556530. PubMed PMID: 24737315; PubMed Central PMCID: PMC4047384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3091.Neill T, Andreuzzi E, Wang ZX, et al. Endorepellin remodels the endothelial transcriptome toward a pro-autophagic and pro-mitophagic gene signature. J Biol Chem. 2018. Aug 3;293(31):12137–12148. doi: 10.1074/jbc.RA118.002934. PubMed PMID: 29921586; PubMed Central PMCID: PMCPMC6078466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3092.Dewi FRP, Jiapaer S, Kobayashi A, et al. Nucleoporin TPR (translocated promoter region, nuclear basket protein) upregulation alters MTOR-HSF1 trails and suppresses autophagy induction in ependymoma. Autophagy. 2020. Mar 24:in press. doi: 10.1080/15548627.2020.1741318. PubMed PMID: 32207633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3093.Tian E, Wang F, Han J, et al. epg-1 functions in autophagy-regulated processes and may encode a highly divergent Atg13 homolog in C. elegans. Autophagy. 2009. Jul;5(5):608–15. doi: 8624 [pii]. PubMed PMID: 19377305; eng. [DOI] [PubMed] [Google Scholar]
- 3094.Devkota S, Jeong H, Kim Y, et al. Functional characterization of EI24-induced autophagy in the degradation of RING-domain E3 ligases. Autophagy. 2016. Nov;12(11):2038–2053. doi: 10.1080/15548627.2016.1217371. PubMed PMID: 27541728; PubMed Central PMCID: PMCPMC5103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3095.Nam T, Han JH, Devkota S, et al. Emerging paradigm of crosstalk between autophagy and the ubiquitin-proteasome system. Mol Cells. 2017. Dec 31;40(12):897–905. doi: 10.14348/molcells.2017.0226. PubMed PMID: 29237114; PubMed Central PMCID: PMCPMC5750708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3096.Cullup T, Kho AL, Dionisi-Vici C, et al. Recessive mutations in EPG5 cause Vici syndrome, a multisystem disorder with defective autophagy. Nat Genet. 2013. Jan;45(1):83–7. doi: 10.1038/ng.2497. PubMed PMID: 23222957; PubMed Central PMCID: PMC4012842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3097.Liang Q, Yang P, Tian E, et al. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy. 2012. Oct;8(10):1426–33. doi: 10.4161/auto.21163. PubMed PMID: 22885670. [DOI] [PubMed] [Google Scholar]
- 3098.Li S, Yang P, Tian E, et al. Arginine methylation modulates autophagic degradation of PGL granules in C. elegans. Mol Cell. 2013. Nov 7;52(3):421–33. doi: 10.1016/j.molcel.2013.09.014. PubMed PMID: 24140420. [DOI] [PubMed] [Google Scholar]
- 3099.Aguado C, Sarkar S, Korolchuk VI, et al. Laforin, the most common protein mutated in Lafora disease, regulates autophagy. Hum Mol Genet. 2010. Jul 15;19(14):2867–76. doi: ddq190 [pii] doi: 10.1093/hmg/ddq190. PubMed PMID: 20453062; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3100.Ganesh S, Agarwala KL, Amano K, et al. Regional and developmental expression of Epm2a gene and its evolutionary conservation. Biochem Biophys Res Commun. 2001. May 25;283(5):1046–53. doi: 10.1006/bbrc.2001.4914. PubMed PMID: 11355878. [DOI] [PubMed] [Google Scholar]
- 3101.Jain N, Mishra R, Ganesh S.. FoxO3a-mediated autophagy is down-regulated in the laforin deficient mice, an animal model for Lafora progressive myoclonus epilepsy. Biochem Biophys Res Commun. 2016. May 27;474(2):321–327. doi: 10.1016/j.bbrc.2016.04.094. PubMed PMID: 27107699. [DOI] [PubMed] [Google Scholar]
- 3102.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012. Dec 7;151(6):1163–7. doi: 10.1016/j.cell.2012.11.012. PubMed PMID: 23217703; PubMed Central PMCID: PMCPMC3521611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3103.Roperto S, Russo V, De Falco F, et al. FUNDC1-mediated mitophagy in bovine papillomavirus-infected urothelial cells. Vet Microbiol. 2019. Jul;234:51–60. doi: 10.1016/j.vetmic.2019.05.017. PubMed PMID: 31213272. [DOI] [PubMed] [Google Scholar]
- 3104.Budnik A, Stephens DJ.. ER exit sites--localization and control of COPII vesicle formation. FEBS Lett. 2009. Dec 3;583(23):3796–803. doi: 10.1016/j.febslet.2009.10.038. PubMed PMID: 19850039. [DOI] [PubMed] [Google Scholar]
- 3105.Raote I, Malhotra V.. Protein transport by vesicles and tunnels. J Cell Biol. 2019. Mar 4;218(3):737–739. doi: 10.1083/jcb.201811073. PubMed PMID: 30718263; PubMed Central PMCID: PMCPMC6400553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3106.Kurokawa K, Nakano A.. The ER exit sites are specialized ER zones for the transport of cargo proteins from the ER to the Golgi apparatus. J Biochem. 2019. Feb 1;165(2):109–114. doi: 10.1093/jb/mvy080. PubMed PMID: 30304445. [DOI] [PubMed] [Google Scholar]
- 3107.Bockler S, Westermann B.. Mitochondrial ER contacts are crucial for mitophagy in yeast. Dev Cell. 2014. Feb 24;28(4):450–8. doi: 10.1016/j.devcel.2014.01.012. PubMed PMID: 24530295. [DOI] [PubMed] [Google Scholar]
- 3108.Belgareh-Touze N, Cavellini L, Cohen MM.. Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy. Autophagy. 2017. Jan 2;13(1):114–132. doi: 10.1080/15548627.2016.1252889. PubMed PMID: 27846375; PubMed Central PMCID: PMCPMC5240830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3109.Kaufman DR, Papillon J, Larose L, et al. Deletion of inositol-requiring enzyme-1alpha in podocytes disrupts glomerular capillary integrity and autophagy. Mol Biol Cell. 2017. Jun 15;28(12):1636–1651. doi: 10.1091/mbc.E16-12-0828. PubMed PMID: 28428258; PubMed Central PMCID: PMCPMC5469607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3110.Lin M, Liu H, Xiong Q, et al. Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy. 2016. Nov;12(11):2145–2166. doi: 10.1080/15548627.2016.1217369. PubMed PMID: 27541856; PubMed Central PMCID: PMCPMC5103349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3111.Rikihisa Y. Role and function of the type IV secretion system in Anaplasma and Ehrlichia species. Curr Top Microbiol Immunol. 2017;413:297–321. doi: 10.1007/978-3-319-75241-9_12. PubMed PMID: 29536364. [DOI] [PubMed] [Google Scholar]
- 3112.Rikihisa Y. Subversion of RAB5-regulated autophagy by the intracellular pathogen Ehrlichia chaffeensis. Small GTPases. 2019. Sep;10(5):343–349. doi: 10.1080/21541248.2017.1332506. PubMed PMID: 28650718; PubMed Central PMCID: PMCPMC6748376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3113.Sinha S, Roy S, Reddy BS, et al. A lipid-modified estrogen derivative that treats breast cancer independent of estrogen receptor expression through simultaneous induction of autophagy and apoptosis. Mol Cancer Res. 2011. Mar;9(3):364–74. doi: 10.1158/1541-7786.MCR-10-0526. PubMed PMID: 21289296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3114.Zhou F, Wu Z, Zhao M, et al. Rab5-dependent autophagosome closure by ESCRT. J Cell Biol. 2019. Jun 3;218(6):1908–1927. doi: 10.1083/jcb.201811173. PubMed PMID: 31010855; PubMed Central PMCID: PMCPMC6548130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3115.Wang L, Yu C, Lu Y, et al. TMEM166, a novel transmembrane protein, regulates cell autophagy and apoptosis. Apoptosis. 2007. Aug;12(8):1489–502. doi: 10.1007/s10495-007-0073-9. PubMed PMID: 17492404; eng. [DOI] [PubMed] [Google Scholar]
- 3116.Yu C, Wang L, Lv B, et al. TMEM74, a lysosome and autophagosome protein, regulates autophagy. Biochem Biophys Res Commun. 2008. May 2;369(2):622–9. doi: S0006-291X(08)00324-0 [pii] doi: 10.1016/j.bbrc.2008.02.055. PubMed PMID: 18294959; eng. [DOI] [PubMed] [Google Scholar]
- 3117.Bodemann BO, Orvedahl A, Cheng T, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011. Jan 21;144(2):253–67. doi: S0092-8674(10)01436-4 [pii] doi: 10.1016/j.cell.2010.12.018. PubMed PMID: 21241894; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3118.Singh S, Kumari R, Chinchwadkar S, et al. Exocyst Subcomplex Functions in Autophagosome Biogenesis by Regulating Atg9 Trafficking. J Mol Biol. 2019. Jul 12;431(15):2821–2834. doi: 10.1016/j.jmb.2019.04.048. PubMed PMID: 31103773; PubMed Central PMCID: PMCPMC6698439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3119.Abrahamsen H, Stenmark H.. Protein secretion: unconventional exit by exophagy. Curr Biol. 2010. May 11;20(9):R415–8. doi: S0960-9822(10)00292-7 [pii] doi: 10.1016/j.cub.2010.03.011. PubMed PMID: 20462486; eng. [DOI] [PubMed] [Google Scholar]
- 3120.Clucas J, Valderrama F.. ERM proteins in cancer progression. J Cell Sci. 2015. Mar 15;128(6):1253. doi: 10.1242/jcs.170027. PubMed PMID: 25774052. [DOI] [PubMed] [Google Scholar]
- 3121.Qureshi-Baig K, Kuhn D, Viry E, et al. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy. 2019. Nov 27:1–17. doi: 10.1080/15548627.2019.1687213. PubMed PMID: 31775562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3122.Pi H, Liu M, Xi Y, et al. Long-term exercise prevents hepatic steatosis: a novel role of FABP1 in regulation of autophagy-lysosomal machinery. FASEB J. 2019. Nov;33(11):11870–11883. doi: 10.1096/fj.201900812R. PubMed PMID: 31366243; PubMed Central PMCID: PMCPMC6902714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3123.He L, Ren Y, Zheng Q, et al. Fas-associated protein with death domain (FADD) regulates autophagy through promoting the expression of Ras homolog enriched in brain (Rheb) in human breast adenocarcinoma cells. Oncotarget. 2016. Apr 26;7(17):24572–84. doi: 10.18632/oncotarget.8249. PubMed PMID: 27013580; PubMed Central PMCID: PMCPMC5029724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3124.Bell BD, Leverrier S, Weist BM, et al. FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc Natl Acad Sci U S A. 2008. Oct 28;105(43):16677–82. doi: 10.1073/pnas.0808597105. PubMed PMID: 18946037; PubMed Central PMCID: PMCPMC2575479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3125.Kohno S, Shiozaki Y, Keenan AL, et al. An N-terminal-truncated isoform of FAM134B (FAM134B-2) regulates starvation-induced hepatic selective ER-phagy. Life Sci Alliance. 2019. Jun;2(3). doi: 10.26508/lsa.201900340. PubMed PMID: 31101736; PubMed Central PMCID: PMCPMC6526285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3126.Iorio F, Bosotti R, Scacheri E, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc Natl Acad Sci U S A. 2010. Aug 17;107(33):14621–6. doi: 1000138107 [pii] doi: 10.1073/pnas.1000138107. PubMed PMID: 20679242; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3127.Lisa-Santamaria P, Jimenez A, Revuelta JL.. The protein factor-arrest 11 (Far11) is essential for the toxicity of human caspase-10 in yeast and participates in the regulation of autophagy and the DNA damage signaling. J Biol Chem. 2012. Aug 24;287(35):29636–47. doi: 10.1074/jbc.M112.344192. PubMed PMID: 22782902; PubMed Central PMCID: PMC3436162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3128.Furukawa K, Fukuda T, Yamashita SI, et al. The PP2A-like protein phosphatase Ppg1 and the far complex cooperatively counteract CK2-mediated phosphorylation of Atg32 to inhibit mitophagy. Cell Rep. 2018. Jun 19;23(12):3579–3590. doi: 10.1016/j.celrep.2018.05.064. PubMed PMID: 29925000. [DOI] [PubMed] [Google Scholar]
- 3129.Meng N, Mu X, Lv X, et al. Autophagy represses fascaplysin-induced apoptosis and angiogenesis inhibition via ROS and p8 in vascular endothelia cells. Biomed Pharmacother. 2019. Jun;114:108866. doi: 10.1016/j.biopha.2019.108866. PubMed PMID: 30999113. [DOI] [PubMed] [Google Scholar]
- 3130.Xu Y, Tian C, Sun J, et al. FBXW7-induced MTOR degradation forces autophagy to counteract persistent prion infection. Mol Neurobiol. 2016. Jan;53(1):706–719. doi: 10.1007/s12035-014-9028-7. PubMed PMID: 25579381. [DOI] [PubMed] [Google Scholar]
- 3131.Wakatsuki S, Tokunaga S, Shibata M, et al. GSK3B-mediated phosphorylation of MCL1 regulates axonal autophagy to promote Wallerian degeneration. J Cell Biol. 2017. Feb;216(2):477–493. doi: 10.1083/jcb.201606020. PubMed PMID: 28053206; PubMed Central PMCID: PMCPMC5294778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3132.McKnight NC, Jefferies HB, Alemu EA, et al. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 2012. Apr 18;31(8):1931–46. doi: 10.1038/emboj.2012.36. PubMed PMID: 22354037; PubMed Central PMCID: PMC3343327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3133.Palumbo P, Petracca A, Maggi R, et al. A novel dominant-negative FGFR1 variant causes Hartsfield syndrome by deregulating RAS/ERK1/2 pathway. Eur J Hum Genet. 2019. Jul;27(7):1113–1120. doi: 10.1038/s41431-019-0350-4. PubMed PMID: 30787447; PubMed Central PMCID: PMCPMC6777633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3134.Vaccari I, Carbone A, Previtali SC, et al. Loss of Fig4 in both Schwann cells and motor neurons contributes to CMT4J neuropathy. Hum Mol Genet. 2015. Jan 15;24(2):383–96. doi: 10.1093/hmg/ddu451. PubMed PMID: 25187576; PubMed Central PMCID: PMC4275070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3135.Jinn TR, Wu CM, Tu WC, et al. Functional expression of FIP-gts, a fungal immunomodulatory protein from Ganoderma tsugae in Sf21 insect cells. Biosci Biotechnol Biochem. 2006. Nov;70(11):2627–34. doi: 10.1271/bbb.60232. PubMed PMID: 17090952. [DOI] [PubMed] [Google Scholar]
- 3136.Meduri G, Guillemeau K, Dounane O, et al. Caspase-cleaved Tau-D(421) is colocalized with the immunophilin FKBP52 in the autophagy-endolysosomal system of Alzheimer’s disease neurons. Neurobiol Aging. 2016. Oct;46:124–37. doi: 10.1016/j.neurobiolaging.2016.06.017. PubMed PMID: 27479154. [DOI] [PubMed] [Google Scholar]
- 3137.Romano S, D’Angelillo A, Pacelli R, et al. Role of FK506-binding protein 51 in the control of apoptosis of irradiated melanoma cells. Cell Death Differ. 2010. Jan;17(1):145–57. doi: cdd2009115 [pii] doi: 10.1038/cdd.2009.115. PubMed PMID: 19696786; eng. [DOI] [PubMed] [Google Scholar]
- 3138.Gassen NC, Hartmann J, Zschocke J, et al. Association of FKBP51 with priming of autophagy pathways and mediation of antidepressant treatment response: evidence in cells, mice, and humans. PLoS Med. 2014. Nov;11(11):e1001755. doi: 10.1371/journal.pmed.1001755. PubMed PMID: 25386878; PubMed Central PMCID: PMC4227651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3139.Bai X, Ma D, Liu A, et al. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science. 2007. Nov 9;318(5852):977–80. doi: 10.1126/science.1147379. PubMed PMID: 17991864. [DOI] [PubMed] [Google Scholar]
- 3140.Nickerson ML, Warren MB, Toro JR, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dube syndrome. Cancer cell. 2002. Aug;2(2):157–64. PubMed PMID: 12204536. [DOI] [PubMed] [Google Scholar]
- 3141.Petit CS, Roczniak-Ferguson A, Ferguson SM.. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013. Sep 30;202(7):1107–22. doi: 10.1083/jcb.201307084. PubMed PMID: 24081491; PubMed Central PMCID: PMC3787382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3142.Tsun ZY, Bar-Peled L, Chantranupong L, et al. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013. Nov 21;52(4):495–505. doi: 10.1016/j.molcel.2013.09.016. PubMed PMID: 24095279; PubMed Central PMCID: PMC3867817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3143.Huett A, Ng A, Cao Z, et al. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J Iimmunol. 2009. Apr 15;182(8):4917–30. doi: 182/8/4917 [pii] doi: 10.4049/jimmunol.0803050. PubMed PMID: 19342671; PubMed Central PMCID: PMC2752416. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3144.Zhao Y, Yang J, Liao W, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010. Jul;12(7):665–75. doi: ncb2069 [pii] doi: 10.1038/ncb2069. PubMed PMID: 20543840; eng. [DOI] [PubMed] [Google Scholar]
- 3145.Hariharan N, Maejima Y, Nakae J, et al. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010. Dec 10;107(12):1470–82. doi: 10.1161/CIRCRESAHA.110.227371. PubMed PMID: 20947830; PubMed Central PMCID: PMC3011986. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3146.Attaix D, Bechet D.. FoxO3 controls dangerous proteolytic liaisons. Cell Metab. 2007. Dec;6(6):425–7. doi: S1550-4131(07)00340-3 [pii] [DOI] [PubMed] [Google Scholar]
- 3147.Sun LL, Li M, Suo F, et al. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 2013;9(8):e1003715. doi: 10.1371/journal.pgen.1003715. PubMed PMID: 23950735; PubMed Central PMCID: PMC3738441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3148.Zhang Q, Yang W, Man N, et al. Autophagy-mediated chemosensitization in cancer cells by fullerene C60 nanocrystal. Autophagy. 2009. Nov;5(8):1107–17. doi: 10.4161/auto.5.8.9842. PubMed PMID: 19786831. [DOI] [PubMed] [Google Scholar]
- 3149.Xu J, Wang H, Hu Y, et al. Inhibition of CaMKIIalpha Activity Enhances Antitumor Effect of Fullerene C60 Nanocrystals by Suppression of Autophagic Degradation. Adv Sci (Weinh). 2019. Apr 17;6(8):1801233. doi: 10.1002/advs.201801233. PubMed PMID: 31016106; PubMed Central PMCID: PMCPMC6468974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3150.Lim Y, Rubio-Pena K, Sobraske PJ, et al. Fndc-1 contributes to paternal mitochondria elimination in C. elegans. Dev Biol. 2019. Oct 1;454(1):15–20. doi: 10.1016/j.ydbio.2019.06.016. PubMed PMID: 31233739; PubMed Central PMCID: PMCPMC6717525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3151.Pankiv S, Alemu EA, Brech A, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport [Research Support, Non-U.S. Gov’t]. J Cell Biol. 2010. Jan 25;188(2):253–69. doi: 10.1083/jcb.200907015. PubMed PMID: 20100911; PubMed Central PMCID: PMC2812517. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3152.Lakhani R, Vogel KR, Till A, et al. Defects in GABA metabolism affect selective autophagy pathways and are alleviated by mTOR inhibition. EMBO Mol Med. 2014. Apr;6(4):551–66. doi: 10.1002/emmm.201303356. PubMed PMID: 24578415; PubMed Central PMCID: PMC3992080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3153.Tanida I, Tanida-Miyake E, Ueno T, et al. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001. Jan 19;276(3):1701–6. doi: 10.1074/jbc.C000752200 C000752200 [pii]. PubMed PMID: 11096062; eng. [DOI] [PubMed] [Google Scholar]
- 3154.Saez JC, Berthoud VM, Branes MC, et al. Plasma membrane channels formed by connexins: their regulation and functions. Physiol Rev. 2003. Oct;83(4):1359–400. doi: 10.1152/physrev.00007.2003. PubMed PMID: 14506308. [DOI] [PubMed] [Google Scholar]
- 3155.Bejarano E, Yuste A, Patel B, et al. Connexins modulate autophagosome biogenesis. Nat Cell Biol. 2014. May;16(5):401–14. doi: 10.1038/ncb2934. PubMed PMID: 24705551; PubMed Central PMCID: PMCPMC4008708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3156.Iyyathurai J, Decuypere JP, Leybaert L, et al. Connexins: substrates and regulators of autophagy. BMC Cell Biol. 2016. May 24;17Suppl 1:20. doi: 10.1186/s12860-016-0093-9. PubMed PMID: 27229147; PubMed Central PMCID: PMCPMC4896244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3157.Lichtenstein A, Minogue PJ, Beyer EC, et al. Autophagy: a pathway that contributes to connexin degradation. J Cell Sci. 2011. Mar 15;124(Pt 6):910–20. doi: 10.1242/jcs.073072. PubMed PMID: 21378309; PubMed Central PMCID: PMCPMC3048889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3158.Gu J, Wang Y, Wang X, et al. Effect of the LncRNA GAS5-MiR-23a-ATG3 axis in regulating autophagy in patients with breast cancer. Cell Physiol Biochem. 2018;48(1):194–207. doi: 10.1159/000491718. PubMed PMID: 30007957. [DOI] [PubMed] [Google Scholar]
- 3159.Zhang N, Yang GQ, Shao XM, et al. GAS5 modulated autophagy is a mechanism modulating cisplatin sensitivity in NSCLC cells. Eur Rev Med Pharmacol Sci. 2016. Jun;20(11):2271–7. PubMed PMID: 27338051. [PubMed] [Google Scholar]
- 3160.Li L, Huang C, He Y, et al. Knockdown of Long Non-Coding RNA GAS5 Increases miR-23a by Targeting ATG3 Involved in Autophagy and Cell Viability. Cell Physiol Biochem. 2018;48(4):1723–1734. doi: 10.1159/000492300. PubMed PMID: 30078013. [DOI] [PubMed] [Google Scholar]
- 3161.Mata IF, Samii A, Schneer SH, et al. Glucocerebrosidase gene mutations: a risk factor for Lewy body disorders. Arch Neurol. 2008. Mar;65(3):379–82. doi: 10.1001/archneurol.2007.68. PubMed PMID: 18332251; PubMed Central PMCID: PMC2826203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3162.Mitsui J, Mizuta I, Toyoda A, et al. Mutations for Gaucher disease confer high susceptibility to Parkinson disease. Arch Neurol. 2009. May;66(5):571–6. doi: 10.1001/archneurol.2009.72. PubMed PMID: 19433656. [DOI] [PubMed] [Google Scholar]
- 3163.Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009. Oct 22;361(17):1651–61. doi: 10.1056/NEJMoa0901281. PubMed PMID: 19846850; PubMed Central PMCID: PMC2856322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3164.Osellame LD, Rahim AA, Hargreaves IP, et al. Mitochondria and quality control defects in a mouse model of Gaucher disease--links to Parkinson’s disease. Cell Metab. 2013. Jun 4;17(6):941–53. doi: 10.1016/j.cmet.2013.04.014. PubMed PMID: 23707074; PubMed Central PMCID: PMC3678026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3165.Moreau K, Rubinsztein DC.. The plasma membrane as a control center for autophagy. Autophagy. 2012. May 1;8(5):861–3. doi: 10.4161/auto.20060. PubMed PMID: 22617437; PubMed Central PMCID: PMC3378426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3166.Todd LR, Damin MN, Gomathinayagam R, et al. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Mol Biol Cell. 2010. Apr 1;21(7):1225–36. doi: 10.1091/mbc.E09-11-0937. PubMed PMID: 20147447; PubMed Central PMCID: PMC2847526. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3167.Ferreira-Marques M, Aveleira CA, Carmo-Silva S, et al. Caloric restriction stimulates autophagy in rat cortical neurons through neuropeptide Y and ghrelin receptors activation. Aging (Albany NY). 2016. Jul;8(7):1470–84. doi: 10.18632/aging.100996. PubMed PMID: 27441412; PubMed Central PMCID: PMCPMC4993343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3168.Cecarini V, Bonfili L, Cuccioloni M, et al. Effects of Ghrelin on the Proteolytic Pathways of Alzheimer’s Disease Neuronal Cells. Mol Neurobiol. 2016. Jul;53(5):3168–3178. doi: 10.1007/s12035-015-9227-x. PubMed PMID: 26033219. [DOI] [PubMed] [Google Scholar]
- 3169.Santt O, Pfirrmann T, Braun B, et al. The yeast GID complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism [Research Support, Non-U.S. Gov’t]. Mol Biol Cell. 2008. Aug;19(8):3323–33. doi: 10.1091/mbc.E08-03-0328. PubMed PMID: 18508925; PubMed Central PMCID: PMC2488282. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3170.Liu H, Pfirrmann T.. The Gid-complex: an emerging player in the ubiquitin ligase league. Biol Chem. 2019. Oct 25;400(11):1429–1441. doi: 10.1515/hsz-2019-0139. PubMed PMID: 30893051. [DOI] [PubMed] [Google Scholar]
- 3171.Braun B, Pfirrmann T, Menssen R, et al. Gid9, a second RING finger protein contributes to the ubiquitin ligase activity of the Gid complex required for catabolite degradation. FEBS Lett. 2011. Dec 15;585(24):3856–61. doi: 10.1016/j.febslet.2011.10.038. PubMed PMID: 22044534. [DOI] [PubMed] [Google Scholar]
- 3172.Chen SJ, Wu X, Wadas B, et al. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science. 2017. Jan 27;355(6323). doi: 10.1126/science.aal3655. PubMed PMID: 28126757; PubMed Central PMCID: PMCPMC5457285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3173.Menssen R, Schweiggert J, Schreiner J, et al. Exploring the topology of the Gid complex, the E3 ubiquitin ligase involved in catabolite-induced degradation of gluconeogenic enzymes. J Biol Chem. 2012. Jul 20;287(30):25602–14. doi: 10.1074/jbc.M112.363762. PubMed PMID: 22645139; PubMed Central PMCID: PMCPMC3408164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3174.Texier Y, Toedt G, Gorza M, et al. Elution profile analysis of SDS-induced subcomplexes by quantitative mass spectrometry. Mol Cell Proteomics. 2014. May;13(5):1382–91. doi: 10.1074/mcp.O113.033233. PubMed PMID: 24563533; PubMed Central PMCID: PMCPMC4014293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3175.Pfirrmann T, Villavicencio-Lorini P, Subudhi AK, et al. RMND5 from Xenopus laevis is an E3 ubiquitin-ligase and functions in early embryonic forebrain development. PLoS One. 2015;10(3):e0120342. doi: 10.1371/journal.pone.0120342. PubMed PMID: 25793641; PubMed Central PMCID: PMCPMC4368662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3176.Lampert F, Stafa D, Goga A, et al. The multi-subunit GID/CTLH E3 ubiquitin ligase promotes cell proliferation and targets the transcription factor Hbp1 for degradation. eLife. 2018. Jun 18;7. doi: 10.7554/eLife.35528. PubMed PMID: 29911972; PubMed Central PMCID: PMCPMC6037477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3177.Liu H, Ding J, Kohnlein K, et al. The GID ubiquitin ligase complex is a regulator of AMPK activity and organismal lifespan. Autophagy. 2019. Dec 3:1–17. doi: 10.1080/15548627.2019.1695399. PubMed PMID: 31795790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3178.Villar VH, Duran RV.. Glutamoptosis: A new cell death mechanism inhibited by autophagy during nutritional imbalance. Autophagy. 2017. Jun 3;13(6):1078–1079. doi: 10.1080/15548627.2017.1299315. PubMed PMID: 28296535; PubMed Central PMCID: PMCPMC5486366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3179.Villar VH, Nguyen TL, Delcroix V, et al. mTORC1 inhibition in cancer cells protects from glutaminolysis-mediated apoptosis during nutrient limitation. Nat Commun. 2017. Jan 23;8:14124. doi: 10.1038/ncomms14124. PubMed PMID: 28112156; PubMed Central PMCID: PMCPMC5264013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3180.Kalamidas SA, Kotoulas OB.. Glycogen autophagy in newborn rat hepatocytes. Histol Histopathol. 2000. Oct;15(4):1011–8. PubMed PMID: 11005224. [DOI] [PubMed] [Google Scholar]
- 3181.Mellor KM, Varma U, Stapleton DI, et al. Cardiomyocyte glycophagy is regulated by insulin and exposure to high extracellular glucose. Am J Physiol Heart Circ Physiol. 2014. Apr 15;306(8):H1240–5. doi: 10.1152/ajpheart.00059.2014. PubMed PMID: 24561860. [DOI] [PubMed] [Google Scholar]
- 3182.Delbridge LMD, Mellor KM, Taylor DJ, et al. Myocardial stress and autophagy: mechanisms and potential therapies. Nat Rev Cardiol. 2017. Jul;14(7):412–425. doi: 10.1038/nrcardio.2017.35. PubMed PMID: 28361977; PubMed Central PMCID: PMCPMC6245608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3183.Li CH, Ko JL, Ou CC, et al. The protective role of GMI, an immunomodulatory protein from ganoderma microsporum, on 5-fluorouracil-induced oral and intestinal mucositis. Integr Cancer Ther. 2019. Jan-Dec;18:1534735419833795. doi: 10.1177/1534735419833795. PubMed PMID: 30879354; PubMed Central PMCID: PMCPMC6423674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3184.Ogier-Denis E, Couvineau A, Maoret JJ, et al. A heterotrimeric Gi3-protein controls autophagic sequestration in the human colon cancer cell line HT-29. J Biol Chem. 1995. Jan 6;270(1):13–6. PubMed PMID: 7814364; eng. [DOI] [PubMed] [Google Scholar]
- 3185.Ogier-Denis E, Houri JJ, Bauvy C, et al. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J Biol Chem. 1996. Nov 8;271(45):28593–600. PubMed PMID: 8910489; eng. [DOI] [PubMed] [Google Scholar]
- 3186.Yamaguchi H, Arakawa S, Kanaseki T, et al. Golgi membrane-associated degradation pathway in yeast and mammals. EMBO J. 2016. Sep 15;35(18):1991–2007. doi: 10.15252/embj.201593191. PubMed PMID: 27511903; PubMed Central PMCID: PMCPMC5282831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3187.Xiang Y, Wang Y.. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010. Jan 25;188(2):237–51. doi: 10.1083/jcb.200907132. PubMed PMID: 20083603; PubMed Central PMCID: PMCPMC2812519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3188.Zhang X, Wang L, Ireland SC, et al. GORASP2/GRASP55 collaborates with the PtdIns3K UVRAG complex to facilitate autophagosome-lysosome fusion. Autophagy. 2019. Oct;15(10):1787–1800. doi: 10.1080/15548627.2019.1596480. PubMed PMID: 30894053; PubMed Central PMCID: PMCPMC6735621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3189.Zhang X, Wang L, Lak B, et al. GRASP55 senses glucose deprivation through O-GlcNAcylation to promote autophagosome-lysosome fusion. Dev Cell. 2018. Apr 23;45(2):245–261 e6. doi: 10.1016/j.devcel.2018.03.023. PubMed PMID: 29689198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3190.Taylor JP. Multisystem proteinopathy: intersecting genetics in muscle, bone, and brain degeneration. Neurology. 2015. Aug 25;85(8):658–60. doi: 10.1212/WNL.0000000000001862. PubMed PMID: 26208960. [DOI] [PubMed] [Google Scholar]
- 3191.Alberti S, Carra S.. Quality Control of Membraneless Organelles. J Mol Biol. 2018. Nov 2;430(23):4711–4729. doi: 10.1016/j.jmb.2018.05.013. PubMed PMID: 29758260. [DOI] [PubMed] [Google Scholar]
- 3192.Alberti S, Mateju D, Mediani L, et al. Granulostasis: protein quality control of RNP granules. Front Mol Neurosci. 2017;10:84. doi: 10.3389/fnmol.2017.00084. PubMed PMID: 28396624; PubMed Central PMCID: PMCPMC5367262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3193.Baksi S, Bagh S, Sarkar S, et al. Systemic study of a natural feedback loop in Huntington’s disease at the onset of neurodegeneration. Biosystems. 2016. Dec;150:46–51. doi: 10.1016/j.biosystems.2016.08.012. PubMed PMID: 27587340. [DOI] [PubMed] [Google Scholar]
- 3194.Baksi S, Jana NR, Bhattacharyya NP, et al. Grb2 is regulated by foxd3 and has roles in preventing accumulation and aggregation of mutant huntingtin. PLoS One. 2013;8(10):e76792. doi: 10.1371/journal.pone.0076792. PubMed PMID: 24116161; PubMed Central PMCID: PMCPMC3792889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3195.Holler CJ, Taylor G, Deng Q, et al. Intracellular proteolysis of progranulin generates stable, lysosomal granulins that are haploinsufficient in patients with frontotemporal dementia caused by GRN mutations. eNeuro. 2017. Jul-Aug;4(4). doi: 10.1523/ENEURO.0100-17.2017. PubMed PMID: 28828399; PubMed Central PMCID: PMCPMC5562298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3196.Chang MC, Srinivasan K, Friedman BA, et al. Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med. 2017. Sep 4;214(9):2611–2628. doi: 10.1084/jem.20160999. PubMed PMID: 28778989; PubMed Central PMCID: PMCPMC5584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3197.Ward ME, Chen R, Huang HY, et al. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med. 2017. Apr 12;9(385). doi: 10.1126/scitranslmed.aah5642. PubMed PMID: 28404863; PubMed Central PMCID: PMCPMC5526610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3198.Lin SY, Li TY, Liu Q, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012. Apr 27;336(6080):477–81. doi: 10.1126/science.1217032. PubMed PMID: 22539723. [DOI] [PubMed] [Google Scholar]
- 3199.Parr C, Carzaniga R, Gentleman SM, et al. Glycogen synthase kinase 3 inhibition promotes lysosomal biogenesis and autophagic degradation of the amyloid-beta precursor protein. Mol Cell Biol. 2012. Nov;32(21):4410–8. doi: 10.1128/MCB.00930-12. PubMed PMID: 22927642; PubMed Central PMCID: PMCPMC3486153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3200.Marchand B, Arsenault D, Raymond-Fleury A, et al. Glycogen synthase kinase-3 (GSK3) inhibition induces prosurvival autophagic signals in human pancreatic cancer cells. J Biol Chem. 2015. Feb 27;290(9):5592–605. doi: 10.1074/jbc.M114.616714. PubMed PMID: 25561726; PubMed Central PMCID: PMCPMC4342473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3201.Wang W, Wang Q, Wan D, et al. Histone HIST1H1C/H1.2 regulates autophagy in the development of diabetic retinopathy. Autophagy. 2017. May 4;13(5):941–954. doi: 10.1080/15548627.2017.1293768. PubMed PMID: 28409999; PubMed Central PMCID: PMCPMC5446066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3202.Bonner WM, Redon CE, Dickey JS, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008. Dec;8(12):957–67. doi: 10.1038/nrc2523. PubMed PMID: 19005492; PubMed Central PMCID: PMCPMC3094856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3203.Cerezo M, Lehraiki A, Millet A, et al. Compounds Triggering ER Stress Exert Anti-Melanoma Effects and Overcome BRAF Inhibitor Resistance. Cancer cell. 2016. Jun 13;29(6):805–819. doi: 10.1016/j.ccell.2016.04.013. PubMed PMID: 27238082. [DOI] [PubMed] [Google Scholar]
- 3204.Cerezo M, Rocchi S.. New anti-cancer molecules targeting HSPA5/BIP to induce endoplasmic reticulum stress, autophagy and apoptosis. Autophagy. 2017. Jan 2;13(1):216–217. doi: 10.1080/15548627.2016.1246107. PubMed PMID: 27791469; PubMed Central PMCID: PMCPMC5240825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3205.Yue F, Li W, Zou J, et al. Blocking the association of HDAC4 with MAP1S accelerates autophagy clearance of mutant Huntingtin. Aging (Albany NY). 2015. Oct;7(10):839–53. doi: 10.18632/aging.100818. PubMed PMID: 26540094; PubMed Central PMCID: PMCPMC4637209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3206.Lee JY, Koga H, Kawaguchi Y, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010. Mar 3;29(5):969–80. doi: 10.1038/emboj.2009.405. PubMed PMID: 20075865; PubMed Central PMCID: PMC2837169. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3207.Cecarini V, Bonfili L, Cuccioloni M, et al. Crosstalk between the ubiquitin-proteasome system and autophagy in a human cellular model of Alzheimer’s disease. Biochim Biophys Acta. 2012. Nov;1822(11):1741–51. doi: 10.1016/j.bbadis.2012.07.015. PubMed PMID: 22867901. [DOI] [PubMed] [Google Scholar]
- 3208.Ridinger J, Koeneke E, Kolbinger FR, et al. Dual role of HDAC10 in lysosomal exocytosis and DNA repair promotes neuroblastoma chemoresistance. Sci Rep. 2018. Jul 3;8(1):10039. doi: 10.1038/s41598-018-28265-5. PubMed PMID: 29968769; PubMed Central PMCID: PMCPMC6030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3209.Oehme I, Linke JP, Bock BC, et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci U S A. 2013. Jul 9;110(28):E2592–601. doi: 10.1073/pnas.1300113110. PubMed PMID: 23801752; PubMed Central PMCID: PMCPMC3710791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3210.Masson GR, Burke JE, Ahn NG, et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat Methods. 2019. Jul;16(7):595–602. doi: 10.1038/s41592-019-0459-y. PubMed PMID: 31249422; PubMed Central PMCID: PMCPMC6614034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3211.Liu XM, Yamasaki A, Du XM, et al. Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein. eLife. 2018. Nov 19;7. doi: 10.7554/eLife.41237. PubMed PMID: 30451685; PubMed Central PMCID: PMCPMC6279349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3212.Dehne N, Brune B.. HIF-1 in the inflammatory microenvironment. Exp Cell Res. 2009. Jul 1;315(11):1791–7. doi: 10.1016/j.yexcr.2009.03.019. PubMed PMID: 19332053. [DOI] [PubMed] [Google Scholar]
- 3213.Neubert P, Weichselbaum A, Reitinger C, et al. HIF1A and NFAT5 coordinate Na(+)-boosted antibacterial defense via enhanced autophagy and autolysosomal targeting. Autophagy. 2019. Nov;15(11):1899–1916. doi: 10.1080/15548627.2019.1596483. PubMed PMID: 30982460; PubMed Central PMCID: PMCPMC6844503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3214.Bohensky J, Shapiro IM, Leshinsky S, et al. HIF-1 regulation of chondrocyte apoptosis: induction of the autophagic pathway. Autophagy. 2007. May-Jun;3(3):207–14. doi: 3708 [pii]. PubMed PMID: 17224629; eng. [DOI] [PubMed] [Google Scholar]
- 3215.Mellor HR, Harris AL.. The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev. 2007. Dec; 26 (3–4): 553–66. doi: 10.1007/s10555-007-9080-0. PubMed PMID: 17805942; eng. [DOI] [PubMed] [Google Scholar]
- 3216.Mimouna S, Bazin M, Mograbi B, et al. HIF1A regulates xenophagic degradation of adherent and invasive Escherichia coli (AIEC). Autophagy. 2014;10(12):2333–45. doi: 10.4161/15548627.2014.984275. PubMed PMID: 25484075; PubMed Central PMCID: PMC4502747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3217.Yuan J, Pu M, Zhang Z, et al. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell cycle. 2009. Jun 1;8(11):1747–53. doi: 10.4161/cc.8.11.8620. PubMed PMID: 19411844; PubMed Central PMCID: PMCPMC2776713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3218.Roberts DJ, Miyamoto S.. Hexokinase II integrates energy metabolism and cellular protection: Akting on mitochondria and TORCing to autophagy. Cell Death Differ. 2015. Feb;22(2):248–57. doi: 10.1038/cdd.2014.173. PubMed PMID: 25323588; PubMed Central PMCID: PMC4291497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3219.Liau SS, Jazag A, Ito K, et al. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer. 2007. Mar 26;96(6):993–1000. doi: 10.1038/sj.bjc.6603654. PubMed PMID: 17342093; PubMed Central PMCID: PMCPMC2360112. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3220.Tang D, Kang R, Cheh CW, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010. Sep 23;29(38):5299–310. doi: 10.1038/onc.2010.261. PubMed PMID: 20622903; PubMed Central PMCID: PMC2945431. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3221.Thorburn J, Horita H, Redzic J, et al. Autophagy regulates selective HMGB1 release in tumor cells that are destined to die [Research Support, N.I.H., Extramural]. Cell Death Differ. 2009. Jan;16(1):175–83. doi: 10.1038/cdd.2008.143. PubMed PMID: 18846108; PubMed Central PMCID: PMC2605182. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3222.Khambu B, Huda N, Chen X, et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J Clin Invest. 2018. Jun 1;128(6):2419–2435. doi: 10.1172/JCI91814. PubMed PMID: 29558368; PubMed Central PMCID: PMCPMC5983330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3223.Suliman HB, Keenan JE, Piantadosi CA.. Mitochondrial quality-control dysregulation in conditional HO-1(-/-) mice. JCI Insight. 2017. Feb 9;2(3):e89676. doi: 10.1172/jci.insight.89676. PubMed PMID: 28194437; PubMed Central PMCID: PMCPMC5291731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3224.Wang CW, Stromhaug PE, Kauffman EJ, et al. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003. Dec 8;163(5):973–85. doi: 10.1083/jcb.200308071. PubMed PMID: 14662743; PubMed Central PMCID: PMCPMC1705953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3225.Takats S, Pircs K, Nagy P, et al. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014. Apr;25(8):1338–54. doi: 10.1091/mbc.E13-08-0449. PubMed PMID: 24554766; PubMed Central PMCID: PMCPMC3982998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3226.Jiang P, Nishimura T, Sakamaki Y, et al. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014. Apr;25(8):1327–37. doi: 10.1091/mbc.E13-08-0447. PubMed PMID: 24554770; PubMed Central PMCID: PMCPMC3982997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3227.Chen ZH, Wang WT, Huang W, et al. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017. Feb;24(2):212–224. doi: 10.1038/cdd.2016.111. PubMed PMID: 27740626; PubMed Central PMCID: PMCPMC5299705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3228.Velentzas PD, Zhang L, Das G, et al. The proton-coupled monocarboxylate transporter hermes is necessary for autophagy during cell death. Dev Cell. 2018. Nov 5;47(3):281–293 e4. doi: 10.1016/j.devcel.2018.09.015. PubMed PMID: 30318245; PubMed Central PMCID: PMCPMC6219939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3229.Pfaffenwimmer T, Reiter W, Brach T, et al. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 2014. Aug;15(8):862–70. doi: 10.15252/embr.201438932. PubMed PMID: 24968893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3230.Tanaka C, Tan LJ, Mochida K, et al. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J Cell Biol. 2014. Oct 13;207(1):91–105. doi: 10.1083/jcb.201402128. PubMed PMID: 25287303; PubMed Central PMCID: PMC4195827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3231.Mochida K, Ohsumi Y, Nakatogawa H.. Hrr25 phosphorylates the autophagic receptor Atg34 to promote vacuolar transport of alpha-mannosidase under nitrogen starvation conditions. FEBS Lett. 2014. Nov 3;588(21):3862–9. doi: 10.1016/j.febslet.2014.09.032. PubMed PMID: 25281559. [DOI] [PubMed] [Google Scholar]
- 3232.Holland P, Knaevelsrud H, Soreng K, et al. HS1BP3 negatively regulates autophagy by modulation of phosphatidic acid levels. Nat Commun. 2016. Dec 22;7:13889. doi: 10.1038/ncomms13889. PubMed PMID: 28004827; PubMed Central PMCID: PMCPMC5412012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3233.Choutka C, DeVorkin L, Go NE, et al. Hsp83 loss suppresses proteasomal activity resulting in an upregulation of caspase-dependent compensatory autophagy.. Autophagy. 2017. Sep 2;13(9):1573–1589. doi: 10.1080/15548627.2017.1339004. PubMed PMID: 28806103; PubMed Central PMCID: PMCPMC5612217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3234.Leu JI, Pimkina J, Frank A, et al. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009. Oct 9;36(1):15–27. doi: 10.1016/j.molcel.2009.09.023. PubMed PMID: 19818706; PubMed Central PMCID: PMC2771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3235.Li J, Ni M, Lee B, et al. The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ. 2008. Sep;15(9):1460–71. doi: 10.1038/cdd.2008.81. PubMed PMID: 18551133; PubMed Central PMCID: PMC2758056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3236.Cha-Molstad H, Sung KS, Hwang J, et al. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nat Cell Biol. 2015. Jul;17(7):917–29. doi: 10.1038/ncb3177. PubMed PMID: 26075355; PubMed Central PMCID: PMCPMC4490096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3237.Kwon DH, Park OH, Kim L, et al. Insights into degradation mechanism of N-end rule substrates by p62/SQSTM1 autophagy adapter. Nat Commun. 2018. Aug 17;9(1):3291. doi: 10.1038/s41467-018-05825-x. PubMed PMID: 30120248; PubMed Central PMCID: PMCPMC6098011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3238.Chiang HL, Terlecky SR, Plant CP, et al. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins [Research Support, U.S. Gov’t, P.H.S.]. Science. 1989. Oct 20;246(4928):382–5. PubMed PMID: 2799391; eng. [DOI] [PubMed] [Google Scholar]
- 3239.Kaushik S, Massey AC, Cuervo AM.. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 2006. Aug 17;25:3921–3933. PubMed PMID: 16917501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3240.Garcia-Mata R, Gao YS, Sztul E.. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002. Jun;3(6):388–96. PubMed PMID: 12010457. [DOI] [PubMed] [Google Scholar]
- 3241.Thirumalaikumar VP, Wagner M, Balazadeh S, et al. Autophagy is responsible for the accumulation of proteogenic dipeptides in response to heat stress in Arabidopsis thaliana. FEBS J. 2020. Apr 17. doi: 10.1111/febs.15336. PubMed PMID: 32301545. [DOI] [PubMed] [Google Scholar]
- 3242.Xu C, Liu J, Hsu LC, et al. Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J. 2011. Aug;25(8):2700–10. doi: 10.1096/fj.10-167676. PubMed PMID: 21543763; PubMed Central PMCID: PMC3136344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3243.Bandhyopadhyay U, Kaushik S, Vartikovsky L, et al. Dynamic organization of the receptor for chaperone-mediated autophagy at the lysosomal membrane. Mol Cell Biol. 2008;28:5747–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3244.Haidar M, Asselbergh B, Adriaenssens E, et al. Neuropathy-causing mutations in HSPB1 impair autophagy by disturbing the formation of SQSTM1/p62 bodies.. Autophagy. 2019. Jun;15(6):1051–1068. doi: 10.1080/15548627.2019.1569930. PubMed PMID: 30669930; PubMed Central PMCID: PMCPMC6526868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3245.Cicardi ME, Cristofani R, Rusmini P, et al. Tdp-25 routing to autophagy and proteasome ameliorates its aggregation in amyotrophic lateral sclerosis target cells. Sci Rep. 2018. Aug 17;8(1):12390. doi: 10.1038/s41598-018-29658-2. PubMed PMID: 30120266; PubMed Central PMCID: PMCPMC6098007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3246.Crippa V, Sau D, Rusmini P, et al. The small heat shock protein B8 (HspB8) promotes autophagic removal of misfolded proteins involved in amyotrophic lateral sclerosis (ALS). Hum Mol Genet. 2010. Sep 1;19(17):3440–56. doi: 10.1093/hmg/ddq257. PubMed PMID: 20570967. [DOI] [PubMed] [Google Scholar]
- 3247.Li B, Hu Q, Wang H, et al. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ. 2010. May 14;17:1773–84. doi: cdd201055 [pii] doi 10.1038/cdd.2010.55. PubMed PMID: 20467442; Eng. [DOI] [PubMed] [Google Scholar]
- 3248.Cilenti L, Ambivero CT, Ward N, et al. Inactivation of Omi/HtrA2 protease leads to the deregulation of mitochondrial Mulan E3 ubiquitin ligase and increased mitophagy. Biochim Biophys Acta. 2014. Jul;1843(7):1295–307. doi: 10.1016/j.bbamcr.2014.03.027. PubMed PMID: 24709290. [DOI] [PubMed] [Google Scholar]
- 3249.Kang S, Fernandes-Alnemri T, Alnemri ES.. A novel role for the mitochondrial HTRA2/OMI protease in aging. Autophagy. 2013. Mar;9(3):420–1. doi: 10.4161/auto.22920. PubMed PMID: 23242108; PubMed Central PMCID: PMC3590264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3250.Kang S, Louboutin JP, Datta P, et al. Loss of HtrA2/Omi activity in non-neuronal tissues of adult mice causes premature aging. Cell Death Differ. 2013. Feb;20(2):259–69. doi: 10.1038/cdd.2012.117. PubMed PMID: 22976834; PubMed Central PMCID: PMC3554338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3251.Ravikumar B, Duden R, Rubinsztein DC.. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002. May 1;11(9):1107–17. doi: 10.1093/hmg/11.9.1107. PubMed PMID: 11978769. [DOI] [PubMed] [Google Scholar]
- 3252.Franich NR, Basso M, Andre EA, et al. Striatal mutant huntingtin protein levels decline with age in homozygous huntington’s disease knock-in mouse models. J Huntingtons Dis. 2018;7(2):137–150. doi: 10.3233/JHD-170274. PubMed PMID: 29843246; PubMed Central PMCID: PMCPMC6002862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3253.Yamamoto A, Cremona ML, Rothman JE.. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol. 2006. Feb 27;172(5):719–31. doi: 10.1083/jcb.200510065. PubMed PMID: 16505167; PubMed Central PMCID: PMCPMC2063704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3254.Jeong H, Then F, Melia TJ, Jr., et al. Acetylation targets mutant huntingtin to auto-phagosomes for degradation. Cell. 2009. Apr 3;137(1):60–72. doi: 10.1016/j.cell.2009.03.018. PubMed PMID: 19345187; PubMed Central PMCID: PMCPMC2940108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3255.Filimonenko M, Isakson P, Finley KD, et al. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. Mol Cell. 2010. Apr 23;38(2):265–79. doi: 10.1016/j.molcel.2010.04.007. PubMed PMID: 20417604; PubMed Central PMCID: PMC2867245. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3256.Thompson LM, Aiken CT, Kaltenbach LS, et al. IKK phosphorylates Huntingtin and targets it for degradation by the proteasome and lysosome. J Cell Biol. 2009. Dec 28;187(7):1083–99. doi: 10.1083/jcb.200909067. PubMed PMID: 20026656; PubMed Central PMCID: PMCPMC2806289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3257.Ochaba J, Fote G, Kachemov M, et al. IKKbeta slows Huntington’s disease progression in R6/1 mice. Proc Natl Acad Sci U S A. 2019. May 28;116(22):10952–10961. doi: 10.1073/pnas.1814246116. PubMed PMID: 31088970; PubMed Central PMCID: PMCPMC6561205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3258.Al-Ramahi I, Giridharan SSP, Chen YC, et al. Inhibition of PIP4Kgamma ameliorates the pathological effects of mutant huntingtin protein. eLife. 2017. Dec 26;6. doi: 10.7554/eLife.29123. PubMed PMID: 29256861; PubMed Central PMCID: PMCPMC5743427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3259.Tsunemi T, Ashe TD, Morrison BE, et al. PGC-1alpha rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med. 2012. Jul 11;4(142):142ra97. doi: 10.1126/scitranslmed.3003799. PubMed PMID: 22786682; PubMed Central PMCID: PMCPMC4096245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3260.Wong YC, Holzbaur EL.. The regulation of autophagosome dynamics by huntingtin and HAP1 is disrupted by expression of mutant huntingtin, leading to defective cargo degradation. J Neurosci. 2014. Jan 22;34(4):1293–305. doi: 10.1523/JNEUROSCI.1870-13.2014. PubMed PMID: 24453320; PubMed Central PMCID: PMCPMC3898289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3261.Caviston JP, Ross JL, Antony SM, et al. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc Natl Acad Sci U S A. 2007. Jun 12;104(24):10045–50. doi: 10.1073/pnas.0610628104. PubMed PMID: 17548833; PubMed Central PMCID: PMCPMC1891230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3262.Rui YN, Xu Z, Patel B, et al. Huntingtin functions as a scaffold for selective macroautophagy. Nat Cell Biol. 2015. Mar;17(3):262–75. doi: 10.1038/ncb3101. PubMed PMID: 25686248; PubMed Central PMCID: PMCPMC4344873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3263.Coll NS, Smidler A, Puigvert M, et al. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 2014. Sep;21(9):1399–408. doi: 10.1038/cdd.2014.50. PubMed PMID: 24786830; PubMed Central PMCID: PMC4131171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3264.Choudhury KR, Bucha S, Baksi S, et al. Chaperone-like protein HYPK and its interacting partners augment autophagy. Eur J Cell Biol. 2016. Jun-Jul; 95 (6–7): 182–94. doi: 10.1016/j.ejcb.2016.03.003. PubMed PMID: 27067261. [DOI] [PubMed] [Google Scholar]
- 3265.Kim J, Cheon H, Jeong YT, et al. Amyloidogenic peptide oligomer accumulation in autophagy-deficient beta cells induces diabetes. J Clin Invest. 2014. Aug;124(8):3311–24. doi: 10.1172/JCI69625. PubMed PMID: 25036705; PubMed Central PMCID: PMC4109549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3266.Rivera JF, Costes S, Gurlo T, et al. Autophagy defends pancreatic beta cells from human islet amyloid polypeptide-induced toxicity. J Clin Invest. 2014. Aug;124(8):3489–500. doi: 10.1172/JCI71981. PubMed PMID: 25036708; PubMed Central PMCID: PMC4109537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3267.Shigihara N, Fukunaka A, Hara A, et al. Human IAPP-induced pancreatic beta cell toxicity and its regulation by autophagy. J Clin Invest. 2014. Aug;124(8):3634–44. doi: 10.1172/JCI69866. PubMed PMID: 25036706; PubMed Central PMCID: PMC4109539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3268.Lotze MT, Buchser WJ, Liang X.. Blocking the interleukin 2 (IL2)-induced systemic autophagic syndrome promotes profound antitumor effects and limits toxicity. Autophagy. 2012. Aug;8(8):1264–6. doi: 10.4161/auto.20752. PubMed PMID: 22660171. [DOI] [PubMed] [Google Scholar]
- 3269.Campbell-Valois FX, Sachse M, Sansonetti PJ, et al. Escape of actively secreting shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. mBio. 2015. May 26;6(3):e02567–14. doi: 10.1128/mBio.02567-14. PubMed PMID: 26015503; PubMed Central PMCID: PMCPMC4447254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3270.Liu W, Zhou Y, Peng T, et al. N(epsilon)-fatty acylation of multiple membrane-associated proteins by Shigella IcsB effector to modulate host function. Nat Microbiol. 2018. Sep;3(9):996–1009. doi: 10.1038/s41564-018-0215-6. PubMed PMID: 30061757; PubMed Central PMCID: PMCPMC6466622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3271.Dunker AK, Lawson JD, Brown CJ, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. PubMed PMID: 11381529. [DOI] [PubMed] [Google Scholar]
- 3272.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002. Oct;27(10):527–33. PubMed PMID: 12368089. [DOI] [PubMed] [Google Scholar]
- 3273.Uversky VN, Gillespie JR, Fink AL.. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000. Nov 15;41(3):415–27. PubMed PMID: 11025552. [DOI] [PubMed] [Google Scholar]
- 3274.Peng Z, Yan J, Fan X, et al. Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci. 2015. Jan;72(1):137–51. doi: 10.1007/s00018-014-1661-9. PubMed PMID: 24939692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3275.De Guzman RN, Wojciak JM, Martinez-Yamout MA, et al. CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding. Biochemistry. 2005. Jan 18;44(2):490–7. doi: 10.1021/bi048161t. PubMed PMID: 15641773. [DOI] [PubMed] [Google Scholar]
- 3276.Dunker AK, Brown CJ, Lawson JD, et al. Intrinsic disorder and protein function. Biochemistry. 2002. May 28;41(21):6573–82. PubMed PMID: 12022860. [DOI] [PubMed] [Google Scholar]
- 3277.Dunker AK, Silman I, Uversky VN, et al. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008. Dec;18(6):756–64. doi: 10.1016/j.sbi.2008.10.002. PubMed PMID: 18952168. [DOI] [PubMed] [Google Scholar]
- 3278.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005. Jun 13;579(15):3346–54. doi: 10.1016/j.febslet.2005.03.072. PubMed PMID: 15943980. [DOI] [PubMed] [Google Scholar]
- 3279.Peng Z, Xue B, Kurgan L, et al. Resilience of death: intrinsic disorder in proteins involved in the programmed cell death. Cell Death Differ. 2013. Sep;20(9):1257–67. doi: 10.1038/cdd.2013.65. PubMed PMID: 23764774; PubMed Central PMCID: PMC3741502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3280.Popelka H, Uversky VN, Klionsky DJ.. Identification of Atg3 as an intrinsically disordered polypeptide yields insights into the molecular dynamics of autophagy-related proteins in yeast.. Autophagy. 2014. Jun;10(6):1093–104. doi: 10.4161/auto.28616. PubMed PMID: 24879155; PubMed Central PMCID: PMC4091170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3281.van der Lee R, Buljan M, Lang B, et al. Classification of intrinsically disordered regions and proteins. Chem Rev. 2014. Jul 9;114(13):6589–631. doi: 10.1021/cr400525m. PubMed PMID: 24773235; PubMed Central PMCID: PMC4095912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3282.Uversky VN. Intrinsic disorder-based protein interactions and their modulators. Curr Pharm Des. 2013;19(23):4191–213. PubMed PMID: 23170892. [DOI] [PubMed] [Google Scholar]
- 3283.Pejaver V, Hsu WL, Xin F, et al. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014. Aug;23(8):1077–93. doi: 10.1002/pro.2494. PubMed PMID: 24888500; PubMed Central PMCID: PMC4116656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3284.Chiang HS, Maric M.. Lysosomal thiol reductase negatively regulates autophagy by altering glutathione synthesis and oxidation. Free Radic Biol Med. 2011. Aug 1;51(3):688–99. doi: 10.1016/j.freeradbiomed.2011.05.015. PubMed PMID: 21640818. [DOI] [PubMed] [Google Scholar]
- 3285.Pampliega O, Orhon I, Patel B, et al. Functional interaction between autophagy and ciliogenesis. Nature. 2013. Oct 10;502(7470):194–200. doi: 10.1038/nature12639. PubMed PMID: 24089209; PubMed Central PMCID: PMCPMC3896125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3286.Finetti F, Cassioli C, Cianfanelli V, et al. The intraflagellar transport protein IFT20 controls lysosome biogenesis by regulating the post-Golgi transport of acid hydrolases. Cell Death Differ. 2020. Jan;27(1):310–328. doi: 10.1038/s41418-019-0357-y. PubMed PMID: 31142807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3287.Criollo A, Senovilla L, Authier H, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010. Feb 3;29(3):619–31. doi: emboj2009364 [pii] doi: 10.1038/emboj.2009.364. PubMed PMID: 19959994; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3288.Pang M, Wang H, Rao P, et al. Autophagy links beta-catenin and Smad signaling to promote epithelial-mesenchymal transition via upregulation of integrin linked kinase. Int J Biochem Cell Biol. 2016. Jul;76:123–34. doi: 10.1016/j.biocel.2016.05.010. PubMed PMID: 27177845. [DOI] [PubMed] [Google Scholar]
- 3289.Wu X, Tu BP.. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol Biol Cell. 2011. Nov;22(21):4124–33. doi: 10.1091/mbc.E11-06-0525. PubMed PMID: 21900499; PubMed Central PMCID: PMC3204073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3290.Blanchet FP, Moris A, Nikolic DS, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010. May 28;32(5):654–69. doi: S1074-7613(10)00160-3 [pii] doi: 10.1016/j.immuni.2010.04.011. PubMed PMID: 20451412; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3291.Deretic V. Autophagy in innate and adaptive immunity. Trends Immunol. 2005. Oct;26(10):523–8. doi: S1471-4906(05)00206-1 [pii] doi: 10.1016/j.it.2005.08.003. PubMed PMID: 16099218; eng. [DOI] [PubMed] [Google Scholar]
- 3292.Piippo N, Korhonen E, Hytti M, et al. Hsp90 inhibition as a means to inhibit activation of the NLRP3 inflammasome. Sci Rep. 2018. Apr 30;8(1):6720. doi: 10.1038/s41598-018-25123-2. PubMed PMID: 29712950; PubMed Central PMCID: PMCPMC5928092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3293.Piippo N, Korkmaz A, Hytti M, et al. Decline in cellular clearance systems induces inflammasome signaling in human ARPE-19 cells. Biochim Biophys Acta. 2014. Dec;1843(12):3038–46. doi: 10.1016/j.bbamcr.2014.09.015. PubMed PMID: 25268952. [DOI] [PubMed] [Google Scholar]
- 3294.Yuan X, Bhat OM, Meng N, et al. Protective role of autophagy in Nlrp3 inflammasome activation and medial thickening of mouse coronary arteries. Am J Pathol. 2018. Dec;188(12):2948–2959. doi: 10.1016/j.ajpath.2018.08.014. PubMed PMID: 30273598; PubMed Central PMCID: PMCPMC6334256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3295.Seveau S, Turner J, Gavrilin MA, et al. Checks and balances between autophagy and inflammasomes during infection. J Mol Biol. 2018. Jan 19;430(2):174–192. doi: 10.1016/j.jmb.2017.11.006. PubMed PMID: 29162504; PubMed Central PMCID: PMCPMC5766433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3296.Dennis EA, Norris PC.. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015. Aug;15(8):511–23. doi: 10.1038/nri3859. PubMed PMID: 26139350; PubMed Central PMCID: PMCPMC4606863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3297.Nathan C, Ding A.. Nonresolving inflammation. Cell. 2010. Mar 19;140(6):871–82. doi: 10.1016/j.cell.2010.02.029. PubMed PMID: 20303877. [DOI] [PubMed] [Google Scholar]
- 3298.Chiurchiu V, Leuti A, Maccarrone M.. Bioactive lipids and chronic inflammation: managing the fire within. Front Immunol. 2018;9:38. doi: 10.3389/fimmu.2018.00038. PubMed PMID: 29434586; PubMed Central PMCID: PMCPMC5797284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3299.Matsuzawa-Ishimoto Y, Hwang S, Cadwell K.. Autophagy and Inflammation. Annu Rev Immunol. 2018. Apr 26;36:73–101. doi: 10.1146/annurev-immunol-042617-053253. PubMed PMID: 29144836. [DOI] [PubMed] [Google Scholar]
- 3300.Dortet L, Mostowy S, Samba-Louaka A, et al. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011. Aug;7(8):e1002168. doi: 10.1371/journal.ppat.1002168. PubMed PMID: 21829365; PubMed Central PMCID: PMC3150275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3301.Raj K, Tissue-specific Sarkar S.. upregulation of Drosophila insulin receptor (InR) mitigates poly(q)-mediated neurotoxicity by restoration of cellular transcription machinery. Mol Neurobiol. 2019. Feb;56(2):1310–1329. doi: 10.1007/s12035-018-1160-3. PubMed PMID: 29881950. [DOI] [PubMed] [Google Scholar]
- 3302.Ravikumar B, Sarkar S, Davies JE, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev. 2010;90(4):1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 3303.Nelson C, Ambros V, Baehrecke EH.. miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Mol Cell. 2014. Nov 6;56(3):376–88. doi: 10.1016/j.molcel.2014.09.011. PubMed PMID: 25306920; PubMed Central PMCID: PMCPMC4252298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3304.Singh SB, Davis AS, Taylor GA, et al. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006. Sep 8;313(5792):1438–41. PubMed PMID: 16888103; eng [DOI] [PubMed] [Google Scholar]
- 3305.Chauhan S, Mandell MA, Deretic V.. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015. May 7;58(3):507–21. doi: 10.1016/j.molcel.2015.03.020. PubMed PMID: 25891078; PubMed Central PMCID: PMCPMC4427528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3306.Mehto S, Jena KK, Nath P, et al. The Crohn’s disease risk factor IRGM limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective autophagy. Mol Cell. 2019. Feb 7;73(3):429–445 e7. doi: 10.1016/j.molcel.2018.11.018. PubMed PMID: 30612879; PubMed Central PMCID: PMCPMC6372082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3307.Gregoire IP, Richetta C, Meyniel-Schicklin L, et al. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011. Dec;7(12):e1002422. doi: 10.1371/journal.ppat.1002422. PubMed PMID: 22174682; PubMed Central PMCID: PMCPMC3234227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3308.Bugnicourt A, Mari M, Reggiori F, et al. Irs4p and Tax4p: two redundant EH domain proteins involved in autophagy. Traffic. 2008. May;9(5):755–69. doi: 10.1111/j.1600-0854.2008.00715.x. PubMed PMID: 18298591. [DOI] [PubMed] [Google Scholar]
- 3309.Namkoong S, Lee KI, Lee JI, et al. The integral membrane protein ITM2A, a transcriptional target of PKA-CREB, regulates autophagic flux via interaction with the vacuolar ATPase. Autophagy. 2015;11(5):756–68. doi: 10.1080/15548627.2015.1034412. PubMed PMID: 25951193; PubMed Central PMCID: PMC4502675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3310.Ivanova H, Vervliet T, Monaco G, et al. Bcl-2-protein family as modulators of IP3 receptors and other organellar Ca(2+) channels. Cold Spring Harb Perspect Biol. 2020; 12(4):a035089. doi: 10.1101/cshperspect.a035089. PubMed PMID: 31501195; PubMed Central PMCID: PMC7111250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3311.Bittremieux M, Parys JB, Pinton P, et al. ER functions of oncogenes and tumor suppressors: Modulators of intracellular Ca(2+) signaling. Biochim Biophys Acta. 2016. Jun;1863(6 Pt B):1364–78. doi: 10.1016/j.bbamcr.2016.01.002. PubMed PMID: 26772784. [DOI] [PubMed] [Google Scholar]
- 3312.Filadi R, Leal NS, Schreiner B, et al. TOM70 sustains cell bioenergetics by promoting IP3R3-mediated ER to mitochondria Ca(2+) transfer. Curr Biol. 2018. Feb 5;28(3):369–382 e6. doi: 10.1016/j.cub.2017.12.047. PubMed PMID: 29395920. [DOI] [PubMed] [Google Scholar]
- 3313.Yogev O, Goldberg R, Anzi S, et al. Jun proteins are starvation-regulated inhibitors of autophagy. Cancer Res. 2010. Mar 15;70(6):2318–27. doi: 10.1158/0008-5472.CAN-09-3408. PubMed PMID: 20197466. [DOI] [PubMed] [Google Scholar]
- 3314.Taylor R, Jr., Chen PH, Chou CC, et al. KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy. 2012. Sep;8(9):1300–11. doi: 10.4161/auto.20681. PubMed PMID: 22889849; PubMed Central PMCID: PMC3442877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3315.Ambrosio S, Sacca CD, Amente S, et al. Lysine-specific demethylase LSD1 regulates autophagy in neuroblastoma through SESN2-dependent pathway. Oncogene. 2017. Nov 30;36(48):6701–6711. doi: 10.1038/onc.2017.267. PubMed PMID: 28783174; PubMed Central PMCID: PMCPMC5717079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3316.Abdel-Aziz AK, Pallavicini I, Ceccacci E, et al. Tuning mTORC1 activity dictates the response of acute myeloid leukemia to LSD1 inhibition. Haematologica. 2019. Sep 19. doi: 10.3324/haematol.2019.224501. PubMed PMID: 31537694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3317.Chao A, Lin CY, Chao AN, et al. Lysine-specific demethylase 1 (LSD1) destabilizes p62 and inhibits autophagy in gynecologic malignancies. Oncotarget. 2017. Sep 26;8(43):74434–74450. doi: 10.18632/oncotarget.20158. PubMed PMID: 29088798; PubMed Central PMCID: PMCPMC5650353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3318.Zhao E, Tang C, Jiang X, et al. Inhibition of cell proliferation and induction of autophagy by KDM2B/FBXL10 knockdown in gastric cancer cells. Cell Signal. 2017. Aug;36:222–229. doi: 10.1016/j.cellsig.2017.05.011. PubMed PMID: 28506929. [DOI] [PubMed] [Google Scholar]
- 3319.Lee DF, Kuo HP, Liu M, et al. KEAP1 E3 ligase-mediated downregulation of NF-kappaB signaling by targeting IKKbeta. Mol Cell. 2009. Oct 9;36(1):131–40. doi: 10.1016/j.molcel.2009.07.025. PubMed PMID: 19818716; PubMed Central PMCID: PMC2770835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3320.Stepkowski TM, Kruszewski MK.. Molecular cross-talk between the NRF2/KEAP1 signaling pathway, autophagy, and apoptosis. Free Radic Biol Med. 2011. May 1;50(9):1186–95. doi: 10.1016/j.freeradbiomed.2011.01.033. PubMed PMID: 21295136. [DOI] [PubMed] [Google Scholar]
- 3321.Orenstein SJ, Cuervo AM.. Chaperone-mediated autophagy: molecular mechanisms and physiological relevance. Semin Cell Dev Biol. 2010. Sep;21(7):719–26. doi: 10.1016/j.semcdb.2010.02.005. PubMed PMID: 20176123; PubMed Central PMCID: PMCPMC2914824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3322.Bejarano E, Murray JW, Wang X, et al. Defective recruitment of motor proteins to autophagic compartments contributes to autophagic failure in aging. Aging cell. 2018. Aug;17(4):e12777. doi: 10.1111/acel.12777. PubMed PMID: 29845728; PubMed Central PMCID: PMCPMC6052466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3323.Mytych J, Solek P, Tabecka-Lonczynska A, et al. Klotho-mediated changes in shelterin complex promote cytotoxic autophagy and apoptosis in amitriptyline-treated hippocampal neuronal cells. Mol Neurobiol. 2019. Oct;56(10):6952–6963. doi: 10.1007/s12035-019-1575-5. PubMed PMID: 30945158. [DOI] [PubMed] [Google Scholar]
- 3324.Mytych J, Solek P, Koziorowski M.. Klotho modulates ER-mediated signaling crosstalk between prosurvival autophagy and apoptotic cell death during LPS challenge. Apoptosis. 2019. Feb;24(1–2):95–107. doi: 10.1007/s10495-018-1496-1. PubMed PMID: 30357572. [DOI] [PubMed] [Google Scholar]
- 3325.Mauvezin C, Neisch AL, Ayala CI, et al. Coordination of autophagosome-lysosome fusion and transport by a Klp98A-Rab14 complex in Drosophila. J Cell Sci. 2016. Mar 1;129(5):971–82. doi: 10.1242/jcs.175224. PubMed PMID: 26763909; PubMed Central PMCID: PMCPMC4813314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3326.Zhang K, Wang M, Tamayo AT, et al. Novel selective inhibitors of nuclear export CRM1 antagonists for therapy in mantle cell lymphoma. Exp Hematol. 2013. Jan;41(1):67–78 e4. doi: 10.1016/j.exphem.2012.09.002. PubMed PMID: 22986101. [DOI] [PubMed] [Google Scholar]
- 3327.Bryant KL, Stalnecker CA, Zeitouni D, et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat Med. 2019. Apr;25(4):628–640. doi: 10.1038/s41591-019-0368-8. PubMed PMID: 30833752; PubMed Central PMCID: PMCPMC6484853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3328.Feng MM, Baryla J, Liu H, et al. Cytoprotective effect of lacritin on human corneal epithelial cells exposed to benzalkonium chloride in vitro. Curr Eye Res. 2014. Jun;39(6):604–10. doi: 10.3109/02713683.2013.859275. PubMed PMID: 24401093; PubMed Central PMCID: PMC4371594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3329.Ma P, Beck SL, Raab RW, et al. Heparanase deglycanation of syndecan-1 is required for binding of the epithelial-restricted prosecretory mitogen lacritin. J Cell Biol. 2006. Sep 25;174(7):1097–106. doi: 10.1083/jcb.200511134. PubMed PMID: 16982797; PubMed Central PMCID: PMC1666580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3330.Wang N, Zimmerman K, Raab RW, et al. Lacritin rescues stressed epithelia via rapid forkhead box O3 (FOXO3)-associated autophagy that restores metabolism. J Biol Chem. 2013. Jun 21;288(25):18146–61. doi: 10.1074/jbc.M112.436584. PubMed PMID: 23640897; PubMed Central PMCID: PMC3689958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3331.Tuohetahuntila M, Molenaar MR, Spee B, et al. Lysosome-mediated degradation of a distinct pool of lipid droplets during hepatic stellate cell activation. J Biol Chem. 2017. Jul 28;292(30):12436–12448. doi: 10.1074/jbc.M117.778472. PubMed PMID: 28615446; PubMed Central PMCID: PMCPMC5535019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3332.Wiersma VI, van Ziel AM, Vazquez-Sanchez S, et al. Granulovacuolar degeneration bodies are neuron-selective lysosomal structures induced by intracellular tau pathology. Acta Neuropathol. 2019. Dec;138(6):943–970. doi: 10.1007/s00401-019-02046-4. PubMed PMID: 31456031; PubMed Central PMCID: PMCPMC6851499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3333.Li C, Wang X, Li X, et al. Proteasome inhibition activates autophagy-lysosome pathway associated with TFEB dephosphorylation and nuclear translocation. Front Cell Dev Biol 2019;7:170. doi: 10.3389/fcell.2019.00170. PubMed PMID: 31508418; PubMed Central PMCID: PMCPMC6713995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3334.Tanaka Y, Guhde G, Suter A, et al. Accumulation of autophagic vacuoles and cardiomyopathy in Lamp-2-deficient mice. Nature. 2000;406:902–906. [DOI] [PubMed] [Google Scholar]
- 3335.Nishino I, Fu J, Tanji K, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000. Aug 24;406(6798):906–10. doi: 10.1038/35022604. PubMed PMID: 10972294. [DOI] [PubMed] [Google Scholar]
- 3336.Wang WT, Han C, Sun YM, et al. Activation of the lysosome-associated membrane protein LAMP5 by DOT1L serves as a bodyguard for MLL fusion oncoproteins to evade degradation in leukemia. clin cancer res off j am assoc cancer res. 2019. May 1;25(9):2795–2808. doi: 10.1158/1078-0432.CCR-18-1474. PubMed PMID: 30651276. [DOI] [PubMed] [Google Scholar]
- 3337.Hayashi K, Taura M, Iwasaki A.. The interaction between IKKalpha and LC3 promotes type I interferon production through the TLR9-containing LAPosome. Sci Signal. 2018. May 1;11(528). doi: 10.1126/scisignal.aan4144. PubMed PMID: 29717061; PubMed Central PMCID: PMCPMC6462218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3338.Ma J, Becker C, Lowell CA, et al. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem. 2012. Oct 5;287(41):34149–56. doi: 10.1074/jbc.M112.382812. PubMed PMID: 22902620; PubMed Central PMCID: PMCPMC3464523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3339.Ma J, Becker C, Reyes C, et al. Cutting edge: FYCO1 recruitment to dectin-1 phagosomes is accelerated by light chain 3 protein and regulates phagosome maturation and reactive oxygen production. J Iimmunol. 2014. Feb 15;192(4):1356–60. doi: 10.4049/jimmunol.1302835. PubMed PMID: 24442442; PubMed Central PMCID: PMCPMC3966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3340.Tam JM, Mansour MK, Khan NS, et al. Dectin-1-dependent LC3 recruitment to phagosomes enhances fungicidal activity in macrophages. J Infect Dis. 2014. Dec 1;210(11):1844–54. doi: 10.1093/infdis/jiu290. PubMed PMID: 24842831; PubMed Central PMCID: PMCPMC4271056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3341.Lamprinaki D, Beasy G, Zhekova A, et al. LC3-associated phagocytosis is required for dendritic cell inflammatory cytokine response to gut commensal yeast Saccharomyces cerevisiae. Front Immunol. 2017;8:1397. doi: 10.3389/fimmu.2017.01397. PubMed PMID: 29118762; PubMed Central PMCID: PMCPMC5661120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3342.Huang J, Canadien V, Lam GY, et al. Activation of antibacterial autophagy by NADPH oxidases [Research Support, Non-U.S. Gov’t]. Proc Natl Acad Sci U S A. 2009. Apr 14;106(15):6226–31. doi: 10.1073/pnas.0811045106. PubMed PMID: 19339495; PubMed Central PMCID: PMC2664152. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3343.Monisha J, Roy NK, Padmavathi G, et al. NGAL is downregulated in oral squamous cell carcinoma and leads to increased survival, proliferation, migration and chemoresistance. Cancers (Basel). 2018. Jul 10;10(7). doi: 10.3390/cancers10070228. PubMed PMID: 29996471; PubMed Central PMCID: PMCPMC6071146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3344.Aits S, Kricker J, Liu B, et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11(8):1408–24. doi: 10.1080/15548627.2015.1063871. PubMed PMID: 26114578; PubMed Central PMCID: PMCPMC4590643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3345.Chauhan S, Kumar S, Jain A, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell. 2016. Oct 10;39(1):13–27. doi: 10.1016/j.devcel.2016.08.003. PubMed PMID: 27693506; PubMed Central PMCID: PMCPMC5104201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3346.Kim BW, Hong SB, Kim JH, et al. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat Commun. 2013;4:1613. doi: 10.1038/ncomms2606. PubMed PMID: 23511477. [DOI] [PubMed] [Google Scholar]
- 3347.Tsai JP, Lee CH, Ying TH, et al. Licochalcone A induces autophagy through PI3K/Akt/mTOR inactivation and autophagy suppression enhances Licochalcone A-induced apoptosis of human cervical cancer cells. Oncotarget. 2015. Oct 6;6(30):28851–66. doi: 10.18632/oncotarget.4767. PubMed PMID: 26311737; PubMed Central PMCID: PMCPMC4745696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3348.Esterbauer H. Estimation of peroxidative damage. A critical review. Pathol Biol. 1996. Jan;44(1):25–8. PubMed PMID: 8734296. [PubMed] [Google Scholar]
- 3349.Dodson M, Wani WY, Redmann M, et al. Regulation of autophagy, mitochondrial dynamics, and cellular bioenergetics by 4-hydroxynonenal in primary neurons. Autophagy. 2017;13(11):1828–1840. doi: 10.1080/15548627.2017.1356948. PubMed PMID: 28837411; PubMed Central PMCID: PMCPMC5788494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3350.Zhang Y, Li X, Carpinteiro A, et al. Acid sphingomyelinase amplifies redox signaling in Pseudomonas aeruginosa-induced macrophage apoptosis. J Iimmunol. 2008. Sep 15;181(6):4247–54. doi: 10.4049/jimmunol.181.6.4247. PubMed PMID: 18768882. [DOI] [PubMed] [Google Scholar]
- 3351.Osawa T, Noda NN.. Atg2: A novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci. 2019. Jun;28(6):1005–1012. doi: 10.1002/pro.3623. PubMed PMID: 30993752; PubMed Central PMCID: PMCPMC6511744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3352.Lee AJ, Roylance R, Sander J, et al. CERT depletion predicts chemotherapy benefit and mediates cytotoxic and polyploid-specific cancer cell death through autophagy induction. J Pathol. 2012. Feb;226(3):482–94. doi: 10.1002/path.2998. PubMed PMID: 21953249. [DOI] [PubMed] [Google Scholar]
- 3353.Mao D, Lin G, Tepe B, et al. VAMP associated proteins are required for autophagic and lysosomal degradation by promoting a PtdIns4P-mediated endosomal pathway. Autophagy. 2019. Jul;15(7):1214–1233. doi: 10.1080/15548627.2019.1580103. PubMed PMID: 30741620; PubMed Central PMCID: PMCPMC6613884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3354.Arlia-Ciommo A, Leonov A, Beach A, et al. Caloric restriction delays yeast chronological aging by remodeling carbohydrate and lipid metabolism, altering peroxisomal and mitochondrial functionalities, and postponing the onsets of apoptotic and liponecrotic modes of regulated cell death. Oncotarget. 2018. Mar 23;9(22):16163–16184. doi: 10.18632/oncotarget.24604. PubMed PMID: 29662634; PubMed Central PMCID: PMCPMC5882325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3355.Arlia-Ciommo A, Leonov A, Mohammad K, et al. Mechanisms through which lithocholic acid delays yeast chronological aging under caloric restriction conditions. Oncotarget. 2018. Oct 9;9(79):34945–34971. doi: 10.18632/oncotarget.26188. PubMed PMID: 30405886; PubMed Central PMCID: PMCPMC6201858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3356.Lam T, Harmancey R, Vasquez H, et al. Reversal of intramyocellular lipid accumulation by lipophagy and a p62-mediated pathway. Cell Death Discov 2016;2:16061. doi: 10.1038/cddiscovery.2016.61. PubMed PMID: 27625792; PubMed Central PMCID: PMCPMC4993124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3357.Popelka H, Klionsky DJ.. Analysis of the native conformation of the LIR/AIM motif in the Atg8/LC3/GABARAP-binding proteins. Autophagy. 2015;11(12):2153–9. doi: 10.1080/15548627.2015.1111503. PubMed PMID: 26565669; PubMed Central PMCID: PMCPMC4835208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3358.Bertolo C, Roa S, Sagardoy A, et al. LITAF, a BCL6 target gene, regulates autophagy in mature B-cell lymphomas. Br J Haematol. 2013. Sep;162(5):621–30. doi: 10.1111/bjh.12440. PubMed PMID: 23795761; PubMed Central PMCID: PMC4111142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3359.Boya P. Lysosomal function and dysfunction: mechanism and disease. Antioxid Redox Signal. 2012. Sep 1;17(5):766–74. doi: 10.1089/ars.2011.4405. PubMed PMID: 22098160. [DOI] [PubMed] [Google Scholar]
- 3360.Gabande-Rodriguez E, Boya P, Labrador V, et al. High sphingomyelin levels induce lysosomal damage and autophagy dysfunction in Niemann Pick disease type A. Cell Death Differ. 2014. Jun;21(6):864–75. doi: 10.1038/cdd.2014.4. PubMed PMID: 24488099; PubMed Central PMCID: PMC4013520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3361.Kagan JC. Lipopolysaccharide Detection across the Kingdoms of Life. Trends Immunol. 2017. Oct;38(10):696–704. doi: 10.1016/j.it.2017.05.001. PubMed PMID: 28551077; PubMed Central PMCID: PMCPMC5624813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3362.Chen M, Liu J, Yang W, et al. Lipopolysaccharide mediates hepatic stellate cell activation by regulating autophagy and retinoic acid signaling. Autophagy. 2017;13(11):1813–1827. doi: 10.1080/15548627.2017.1356550. PubMed PMID: 29160747; PubMed Central PMCID: PMCPMC5788469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3363.Doyle A, Zhang G, Abdel Fattah EA, et al. Toll-like receptor 4 mediates lipopolysaccharide-induced muscle catabolism via coordinate activation of ubiquitin-proteasome and autophagy-lysosome pathways. FASEB J. 2011. Jan;25(1):99–110. doi: 10.1096/fj.10-164152. PubMed PMID: 20826541; PubMed Central PMCID: PMCPMC3005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3364.Carew JS, Espitia CM, Esquivel JA, II, et al. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis [Research Support, Non-U.S. Gov’t]. J Biol Chem. 2011. Feb 25;286(8):6602–13. doi: 10.1074/jbc.M110.151324. PubMed PMID: 21148553; PubMed Central PMCID: PMC3057822. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3365.Martinez Jaramillo C, Trujillo-Vargas CM.. LRBA in the endomembrane system. Colomb Med (Cali). 2018. Sep 30;49(3):236–243. doi: 10.25100/cm.v49i2.3802. PubMed PMID: 30410199; PubMed Central PMCID: PMCPMC6220489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3366.Joyce S, Nour AM.. Blocking transmembrane219 protein signaling inhibits autophagy and restores normal cell death. PLoS One 2019;14(6):e0218091. doi: 10.1371/journal.pone.0218091. PubMed PMID: 31220095; PubMed Central PMCID: PMCPMC6586287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3367.Zou J, Yue F, Jiang X, et al. Mitochondrion-associated protein LRPPRC suppresses the initiation of basal levels of autophagy via enhancing Bcl-2 stability. Biochem J. 2013. Sep 15;454(3):447–57. doi: 10.1042/BJ20130306. PubMed PMID: 23822101; PubMed Central PMCID: PMC3778712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3368.Zou J, Yue F, Li W, et al. Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PLoS One. 2014;9(4):e94903. doi: 10.1371/journal.pone.0094903. PubMed PMID: 24722279; PubMed Central PMCID: PMC3983268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3369.Alegre-Abarrategui J, Christian H, Lufino MM, et al. LRRK2 regulates autophagic activity and localizes to specific membrane microdomains in a novel human genomic reporter cellular model. Hum Mol Genet. 2009. Nov 1;18(21):4022–34. doi: 10.1093/hmg/ddp346. PubMed PMID: 19640926; PubMed Central PMCID: PMC2758136. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3370.Verma M, Callio J, Otero PA, et al. Mitochondrial Calcium Dysregulation Contributes to Dendrite Degeneration Mediated by PD/LBD-Associated LRRK2 Mutants. J Neurosci. 2017. Nov 15;37(46):11151–11165. doi: 10.1523/JNEUROSCI.3791-16.2017. PubMed PMID: 29038245; PubMed Central PMCID: PMCPMC5688524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3371.Steger M, Tonelli F, Ito G, et al. Phosphoproteomics reveals that Parkinson’s disease kinase LRRK2 regulates a subset of Rab GTPases. eLife. 2016. Jan 29;5. doi: 10.7554/eLife.12813. PubMed PMID: 26824392; PubMed Central PMCID: PMCPMC4769169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3372.Eguchi T, Kuwahara T, Sakurai M, et al. LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proc Natl Acad Sci U S A. 2018. Sep 25;115(39):E9115–E9124. doi: 10.1073/pnas.1812196115. PubMed PMID: 30209220; PubMed Central PMCID: PMCPMC6166828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3373.Ng ACY, Eisenberg JM, Heath RJW, et al. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci USA 2011. Jun 29;108:4631–4638. PubMed PMID: 20616063; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3374.Blommaart EF, Krause U, Schellens JP, et al. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem FEBS 1997. Jan 15;243(1–2):240–6. PubMed PMID: 9030745. [DOI] [PubMed] [Google Scholar]
- 3375.McAfee Q, Zhang Z, Samanta A, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012. May 22;109(21):8253–8. doi: 10.1073/pnas.1118193109. PubMed PMID: 22566612; PubMed Central PMCID: PMC3361415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3376.Amaravadi RK, Winkler JD.. Lys05: a new lysosomal autophagy inhibitor. Autophagy. 2012. Sep;8(9):1383–4. doi: 10.4161/auto.20958. PubMed PMID: 22878685; PubMed Central PMCID: PMC3442884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3377.Villamil Giraldo AM, Appelqvist H, Ederth T, et al. Lysosomotropic agents: impact on lysosomal membrane permeabilization and cell death. Biochem Soc Trans. 2014. Oct;42(5):1460–4. doi: 10.1042/BST20140145. PubMed PMID: 25233432. [DOI] [PubMed] [Google Scholar]
- 3378.Pineda CT, Ramanathan S, Fon Tacer K, et al. Degradation of AMPK by a Cancer-Specific Ubiquitin Ligase. Cell. 2015. Feb 12;160(4):715–28. doi: 10.1016/j.cell.2015.01.034. PubMed PMID: 25679763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3379.Bai H, Inoue J, Kawano T, et al. A transcriptional variant of the LC3A gene is involved in autophagy and frequently inactivated in human cancers. Oncogene. 2012. Oct 4;31(40):4397–408. doi: 10.1038/onc.2011.613. PubMed PMID: 22249245. [DOI] [PubMed] [Google Scholar]
- 3380.Nassar M, Samaha H, Ghabriel M, et al. LC3A silencing hinders aggresome vimentin cage clearance in primary choroid plexus carcinoma. Sci Rep. 2017. Aug 14;7(1):8022. doi: 10.1038/s41598-017-07403-5. PubMed PMID: 28808307; PubMed Central PMCID: PMCPMC5556083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3381.Mann SS, Hammarback JA.. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J Biol Chem. 1994. Apr 15;269(15):11492–7. PubMed PMID: 7908909. [PubMed] [Google Scholar]
- 3382.Xie R, Nguyen S, McKeehan K, et al. Microtubule-associated protein 1S (MAP1S) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. J Biol Chem. 2011. Mar 25;286(12):10367–77. doi: 10.1074/jbc.M110.206532. PubMed PMID: 21262964; PubMed Central PMCID: PMC3060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3383.Liu W, Jiang Y, Sun J, et al. Activation of TGF-beta-activated kinase 1 (TAK1) restricts Salmonella Typhimurium growth by inducing AMPK activation and autophagy. Cell Death Dis. 2018. May 1;9(5):570. doi: 10.1038/s41419-018-0612-z. PubMed PMID: 29752434; PubMed Central PMCID: PMCPMC5948208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3384.Sheng T, Sun Y, Sun J, et al. Role of TGF-beta-activated kinase 1 (TAK1) activation in H5N1 influenza A virus-induced c-Jun terminal kinase activation and virus replication. Virology. 2019. Nov;537:263–271. doi: 10.1016/j.virol.2019.09.004. PubMed PMID: 31539775. [DOI] [PubMed] [Google Scholar]
- 3385.Liu W, Zhuang J, Jiang Y, et al. Toll-like receptor signalling cross-activates the autophagic pathway to restrict Salmonella Typhimurium growth in macrophages. Cell Microbiol. 2019. Dec;21(12):e13095. doi: 10.1111/cmi.13095. PubMed PMID: 31392811. [DOI] [PubMed] [Google Scholar]
- 3386.Herrero-Martin G, Hoyer-Hansen M, Garcia-Garcia C, et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009. Mar 18;28(6):677–85. doi: 10.1038/emboj.2009.8. PubMed PMID: 19197243; PubMed Central PMCID: PMC2666037. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3387.Hashimoto K, Simmons AN, Kajino-Sakamoto R, et al. TAK1 regulates the Nrf2 antioxidant system through modulating p62/SQSTM1. Antioxid Redox Signal. 2016. Dec 10;25(17):953–964. doi: 10.1089/ars.2016.6663. PubMed PMID: 27245349; PubMed Central PMCID: PMCPMC5144887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3388.Kehl SR, Soos BA, Saha B, et al. TAK1 converts Sequestosome 1/p62 from an autophagy receptor to a signaling platform. EMBO Rep. 2019. Sep;20(9):e46238. doi: 10.15252/embr.201846238. PubMed PMID: 31347268; PubMed Central PMCID: PMCPMC6726904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3389.Hsu CL, Lee EX, Gordon KL, et al. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun. 2018. Mar 5;9(1):942. doi: 10.1038/s41467-018-03340-7. PubMed PMID: 29507340; PubMed Central PMCID: PMCPMC5838220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3390.Ogier-Denis E, Pattingre S, El Benna J, et al. Erk1/2-dependent phosphorylation of Galpha-interacting protein stimulates its GTPase accelerating activity and autophagy in human colon cancer cells. J Biol Chem. 2000. Dec 15;275(50):39090–5. doi: 10.1074/jbc.M006198200M006198200[pii]. PubMed PMID: 10993892; eng. [DOI] [PubMed] [Google Scholar]
- 3391.Fu MM, Nirschl JJ, Holzbaur EL.. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of auto-phagosomes. Dev Cell. 2014. Jun 9;29(5):577–90. doi: 10.1016/j.devcel.2014.04.015. PubMed PMID: 24914561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3392.Raciti M, Lotti LV, Valia S, et al. JNK2 is activated during ER stress and promotes cell survival. Cell Death Dis. 2012;3:e429. doi: 10.1038/cddis.2012.167. PubMed PMID: 23171849; PubMed Central PMCID: PMC3542603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3393.Keil E, Hocker R, Schuster M, et al. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ. 2013. Feb;20(2):321–32. doi: 10.1038/cdd.2012.129. PubMed PMID: 23059785; PubMed Central PMCID: PMC3554344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3394.Menon MB, Dhamija S, Kotlyarov A, et al. The problem of pyridinyl imidazole class inhibitors of MAPK14/p38alpha and MAPK11/p38beta in autophagy research. Autophagy. 2015. Aug 3;11(8):1425–7. doi: 10.1080/15548627.2015.1059562. PubMed PMID: 26061537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3395.Menon MB, Kotlyarov A, Gaestel M.. SB202190-induced cell type-specific vacuole formation and defective autophagy do not depend on p38 MAP kinase inhibition. PLoS One. 2011;6(8):e23054. doi: 10.1371/journal.pone.0023054. PubMed PMID: 21853067; PubMed Central PMCID: PMC3154272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3396.Colecchia D, Dapporto F, Tronnolone S, et al. MAPK15 is part of the ULK complex and controls its activity to regulate early phases of the autophagic process. J Biol Chem. 2018. Oct 12;293(41):15962–15976. doi: 10.1074/jbc.RA118.002527. PubMed PMID: 30131341; PubMed Central PMCID: PMCPMC6187625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3397.Wang Z, Zhang J, Wang Y, et al. Matrine, a novel autophagy inhibitor, blocks trafficking and the proteolytic activation of lysosomal proteases. Carcinogenesis. 2013. Jan;34(1):128–38. doi: 10.1093/carcin/bgs295. PubMed PMID: 23002236. [DOI] [PubMed] [Google Scholar]
- 3398.Elgendy M, Ciro M, Abdel-Aziz AK, et al. Beclin 1 restrains tumorigenesis through Mcl-1 destabilization in an autophagy-independent reciprocal manner. Nat Commun. 2014. Dec 4;5:5637. doi: 10.1038/ncomms6637. PubMed PMID: 25472497. [DOI] [PubMed] [Google Scholar]
- 3399.Elgendy M, Minucci S.. A novel autophagy-independent, oncosuppressive function of BECN1: Degradation of MCL1. Autophagy. 2015;11(3):581–2. doi: 10.1080/15548627.2015.1029836. PubMed PMID: 25837021; PubMed Central PMCID: PMCPMC4502650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3400.Fliniaux I, Germain E, Farfariello V, et al. TRPs and Ca(2+) in cell death and survival. Cell Calcium. 2018. Jan;69:4–18. doi: 10.1016/j.ceca.2017.07.002. PubMed PMID: 28760561. [DOI] [PubMed] [Google Scholar]
- 3401.Di Paola S, Scotto-Rosato A, Medina DL.. TRPML1: The Ca((2+))retaker of the lysosome. Cell Calcium. 2018. Jan;69:112–121. doi: 10.1016/j.ceca.2017.06.006. PubMed PMID: 28689729. [DOI] [PubMed] [Google Scholar]
- 3402.Scotto Rosato A, Montefusco S, Soldati C, et al. TRPML1 links lysosomal calcium to autophagosome biogenesis through the activation of the CaMKKbeta/VPS34 pathway. Nat Commun. 2019. Dec 10;10(1):5630. doi: 10.1038/s41467-019-13572-w. PubMed PMID: 31822666; PubMed Central PMCID: PMCPMC6904751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3403.Li RJ, Xu J, Fu C, et al. Regulation of mTORC1 by lysosomal calcium and calmodulin. eLife. 2016. Oct 27;5. 10.7554/eLife.19360 PubMed PMID: 27787197; PubMed Central PMCID: PMCPMC5106211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3404.Venkatachalam K, Wong CO, Zhu MX.. The role of TRPMLs in endolysosomal trafficking and function. Cell Calcium. 2015. Jul;58(1):48–56. doi: 10.1016/j.ceca.2014.10.008. PubMed PMID: 25465891; PubMed Central PMCID: PMCPMC4412768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3405.Lorente M, Torres S, Salazar M, et al. Stimulation of ALK by the growth factor midkine renders glioma cells resistant to autophagy-mediated cell death. Autophagy. 2011. Sep;7(9):1071–3. PubMed PMID: 21593591. [DOI] [PubMed] [Google Scholar]
- 3406.Lorente M, Torres S, Salazar M, et al. Stimulation of the midkine/ALK axis renders glioma cells resistant to cannabinoid antitumoral action. Cell Death Differ. 2011. Jun;18(6):959–73. doi: 10.1038/cdd.2010.170. PubMed PMID: 21233844; PubMed Central PMCID: PMC3131933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3407.Kimura T, Jain A, Choi SW, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015. Sep 14;210(6):973–89. doi: 10.1083/jcb.201503023. PubMed PMID: 26347139; PubMed Central PMCID: PMCPMC4576868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3408.van Doorn WG, Woltering EJ.. Many ways to exit? Cell death categories in plants. Trends Plant Sci. 2005. Mar;10(3):117–22. doi: 10.1016/j.tplants.2005.01.006. PubMed PMID: 15749469. [DOI] [PubMed] [Google Scholar]
- 3409.Bagniewska-Zadworna A, Byczyk J, Eissenstat DM, et al. Avoiding transport bottlenecks in an expanding root system: xylem vessel development in fibrous and pioneer roots under field conditions. Am J Bot. 2012. Sep;99(9):1417–26. doi: 10.3732/ajb.1100552. PubMed PMID: 22917946. [DOI] [PubMed] [Google Scholar]
- 3410.Eastwood MD, Cheung SW, Lee KY, et al. Developmentally programmed nuclear destruction during yeast gametogenesis. Dev Cell. 2012. Jul 17;23(1):35–44. doi: 10.1016/j.devcel.2012.05.005. PubMed PMID: 22727375. [DOI] [PubMed] [Google Scholar]
- 3411.Lin L, Rodrigues F, Kary C, et al. Complement-Related Regulates Autophagy in Neighboring Cells. Cell. 2017. Jun 29;170(1):158–171e8. 10.1016/j.cell.2017.06.018. PubMed PMID: 28666117; PubMed Central PMCID: PMCPMC5533186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3412.Kourelis TV, Siegel RD.. Metformin and cancer: new applications for an old drug. Med Oncol. 2012. Jun;29(2):1314–27. doi: 10.1007/s12032-011-9846-7. PubMed PMID: 21301998. [DOI] [PubMed] [Google Scholar]
- 3413.Chen Y, Dorn GW, PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013. Apr 26;340(6131):471–5. doi: 10.1126/science.1231031. PubMed PMID: 23620051; PubMed Central PMCID: PMCPMC3774525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3414.Gong G, Song M, Csordas G, et al. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015. Dec 4;350(6265):aad2459. doi: 10.1126/science.aad2459. PubMed PMID: 26785495; PubMed Central PMCID: PMCPMC4747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3415.Zhao T, Huang X, Han L, et al. Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem. 2012. Jul 6;287(28):23615–25. doi: 10.1074/jbc.M112.379164. PubMed PMID: 22619176; PubMed Central PMCID: PMCPMC3390636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3416.Fedeli C, Filadi R, Rossi A, et al. PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca(2+) homeostasis. Autophagy. 2019. Dec;15(12):2044–2062. doi: 10.1080/15548627.2019.1596489. PubMed PMID: 30892128; PubMed Central PMCID: PMCPMC6844518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3417.Kousi M, Siintola E, Dvorakova L, et al. Mutations in CLN7/MFSD8 are a common cause of variant late-infantile neuronal ceroid lipofuscinosis. Brain. 2009. Mar;132\(Pt 3):810–9. doi: 10.1093/brain/awn366. PubMed PMID: 19201763. [DOI] [PubMed] [Google Scholar]
- 3418.Brandenstein L, Schweizer M, Sedlacik J, et al. Lysosomal dysfunction and impaired autophagy in a novel mouse model deficient for the lysosomal membrane protein Cln7. Hum Mol Genet. 2016. Feb 15;25(4):777–91. doi: 10.1093/hmg/ddv615. PubMed PMID: 26681805. [DOI] [PubMed] [Google Scholar]
- 3419.Oku M, Warnecke D, Noda T, et al. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J 2003. Jul 1;22(13):3231–41. PubMed PMID: 12839986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3420.Gu R, Liu N, Luo S, et al. MicroRNA-9 regulates the development of knee osteoarthritis through the NF-kappaB1 pathway in chondrocytes. Medicine (Baltimore). 2016. Sep;95(36):e4315. doi: 10.1097/MD.0000000000004315. PubMed PMID: 27603333; PubMed Central PMCID: PMCPMC5023855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3421.Cai J, Zhang H, Zhang YF, et al. MicroRNA-29 enhances autophagy and cleanses exogenous mutant alphaB-crystallin in retinal pigment epithelial cells. Exp Cell Res. 2019. Jan 1;374(1):231–248. doi: 10.1016/j.yexcr.2018.11.028. PubMed PMID: 30513336. [DOI] [PubMed] [Google Scholar]
- 3422.Seca H, Lima RT, Lopes-Rodrigues V, et al. Targeting miR-21 induces autophagy and chemosensitivity of leukemia cells. Curr Drug Targets 2013. Sep;14(10):1135–43. PubMed PMID: 23834154. [DOI] [PubMed] [Google Scholar]
- 3423.D’Adamo S, Alvarez-Garcia O, Muramatsu Y, et al. MicroRNA-155 suppresses autophagy in chondrocytes by modulating expression of autophagy proteins. Osteoarthritis Cartilage. 2016. Jun;24(6):1082–91. doi: 10.1016/j.joca.2016.01.005. PubMed PMID: 26805019; PubMed Central PMCID: PMCPMC4875787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3424.Pennati M, Lopergolo A, Profumo V, et al. miR-205 impairs the autophagic flux and enhances cisplatin cytotoxicity in castration-resistant prostate cancer cells. Biochem Pharmacol. 2014. Feb 15;87(4):579–97. doi: 10.1016/j.bcp.2013.12.009. PubMed PMID: 24370341. [DOI] [PubMed] [Google Scholar]
- 3425.Lan SH, Wu SY, Zuchini R, et al. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014. Feb;59(2):505–17. doi: 10.1002/hep.26659. PubMed PMID: 23913306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3426.Lan SH, Wu SY, Zuchini R, et al. Autophagy-preferential degradation of MIR224 participates in hepatocellular carcinoma tumorigenesis. Autophagy. 2014. Sep;10(9):1687–9. doi: 10.4161/auto.29959. PubMed PMID: 25068270; PubMed Central PMCID: PMCPMC4206546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3427.Tao Z, Feng C, Mao C, et al. MiR-4465 directly targets PTEN to inhibit AKT/mTOR pathway-mediated autophagy. Cell Stress Chaperones. 2019. Jan;24(1):105–113. doi: 10.1007/s12192-018-0946-6. PubMed PMID: 30421325; PubMed Central PMCID: PMCPMC6363616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3428.Dubinsky AN, Dastidar SG, Hsu CL, et al. Let-7 coordinately suppresses components of the amino acid sensing pathway to repress mTORC1 and induce autophagy. Cell Metab. 2014. Oct 7;20(4):626–38. 10.1016/j.cmet.2014.09.001 PubMed PMID: 25295787; PubMed Central PMCID: PMCPMC4245205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3429.Petri R, Pircs K, Jonsson ME, et al. let-7 regulates radial migration of new-born neurons through positive regulation of autophagy. EMBO J. 2017. May 15;36(10):1379–1391. doi: 10.15252/embj.201695235. PubMed PMID: 28336683; PubMed Central PMCID: PMCPMC5430214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3430.Gu H, Li L, Cui C, et al. Overexpression of let-7a increases neurotoxicity in a PC12 cell model of Alzheimer’s disease via regulating autophagy. Exp Ther Med. 2017. Oct;14(4):3688–3698. doi: 10.3892/etm.2017.4977. PubMed PMID: 29042965; PubMed Central PMCID: PMCPMC5639351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3431.Shamsuzzama, Kumar L, Nazir A.. Modulation of Alpha-synuclein Expression and Associated Effects by MicroRNA Let-7 in Transgenic C. elegans. Front Mol Neurosci. 2017;10:328. doi: 10.3389/fnmol.2017.00328. PubMed PMID: 29081733; PubMed Central PMCID: PMCPMC5645510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3432.Ding Z, Wang X, Schnackenberg L, et al. Regulation of autophagy and apoptosis in response to ox-LDL in vascular smooth muscle cells, and the modulatory effects of the microRNA hsa-let-7 g. Int J Cardiol. 2013. Sep 30;168(2):1378–85. doi: 10.1016/j.ijcard.2012.12.045. PubMed PMID: 23305858. [DOI] [PubMed] [Google Scholar]
- 3433.Martina JA, Diab HI, Lishu L, et al. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal. 2014. Jan 21;7(309):ra9. doi: 10.1126/scisignal.2004754. PubMed PMID: 24448649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3434.Eid N, Ito Y, Otsuki Y.. Triggering of Parkin Mitochondrial Translocation in Mitophagy: Implications for Liver Diseases. Front Pharmacol. 2016;7:100. doi: 10.3389/fphar.2016.00100. PubMed PMID: 27199746; PubMed Central PMCID: PMCPMC4850158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3435.Eid N, Ito Y, Horibe A, et al.Ethanol-Induced Mitochondrial Damage in Sertoli Cells is Associated with Parkin Overexpression and Activation of Mitophagy. Cells. 2019. Mar 25;8(3). doi: 10.3390/cells8030283. PubMed PMID: 30934625; PubMed Central PMCID: PMCPMC6468925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3436.Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005. Spring;8(1):3–5. PubMed PMID: 15798367. [DOI] [PubMed] [Google Scholar]
- 3437.Chazotte B. Labeling mitochondria with MitoTracker dyes. Cold Spring Harb Protoc. 2011. Aug 1;2011(8):990–2. doi: 10.1101/pdb.prot5648. PubMed PMID: 21807856. [DOI] [PubMed] [Google Scholar]
- 3438.Kou Y, He Y, Qiu J, et al. Mitochondrial dynamics and mitophagy are necessary for proper invasive growth in rice blast. Mol Plant Pathol 2019. Aug;20(8):1147–1162. doi: 10.1111/mpp.12822. PubMed PMID: 31218796; PubMed Central PMCID: PMCPMC6640187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3439.Nordmann M, Cabrera M, Perz A, et al. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010. Sep 28;20(18):1654–9. doi: 10.1016/j.cub.2010.08.002. PubMed PMID: 20797862. [DOI] [PubMed] [Google Scholar]
- 3440.Hegedus K, Takats S, Boda A, et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell. 2016. Oct 15;27(20):3132–3142. 10.1091/mbc.E16-03-0205. PubMed PMID: 27559127; PubMed Central PMCID: PMCPMC5063620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3441.Gerondopoulos A, Langemeyer L, Liang JR, et al. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012. Nov 20;22(22):2135–9. 10.1016/j.cub.2012.09.020. PubMed PMID: 23084991; PubMed Central PMCID: PMCPMC3502862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3442.Gao J, Langemeyer L, Kummel D, et al. Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. eLife. 2018. Feb 15;7. 10.7554/eLife.31145. PubMed PMID: 29446751; PubMed Central PMCID: PMCPMC5841931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3443.Choi HS, Jeong EH, Lee TG, et al. Autophagy inhibition with monensin enhances cell cycle arrest and apoptosis induced by mTOR or epidermal growth factor receptor inhibitors in lung cancer cells. Tuberc Respir Dis (Seoul) 2013. Jul;75(1):9–17. doi: 10.4046/trd.2013.75.1.9. PubMed PMID: 23946753; PubMed Central PMCID: PMCPMC3741474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3444.Oldfield CJ, Cheng Y, Cortese MS, et al. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005. Sep 20;44(37):12454–70. doi: 10.1021/bi050736e. PubMed PMID: 16156658. [DOI] [PubMed] [Google Scholar]
- 3445.Mohan A, Oldfield CJ, Radivojac P, et al. Analysis of molecular recognition features (MoRFs). J Mol Biol. 2006. Oct 6;362(5):1043–59. doi: 10.1016/j.jmb.2006.07.087. PubMed PMID: 16935303. [DOI] [PubMed] [Google Scholar]
- 3446.Cheng Y, Oldfield CJ, Meng J, et al. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007. Nov 27;46(47):13468–77. 10.1021/bi7012273 PubMed PMID: 17973494; PubMed Central PMCID: PMCPMC2570644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3447.Yan J, Dunker AK, Uversky VN, et al. Molecular recognition features (MoRFs) in three domains of life. Mol Biosyst. 2016. Mar;12(3):697–710. 10.1039/c5mb00640f. PubMed PMID: 26651072. [DOI] [PubMed] [Google Scholar]
- 3448.Choi YJ, Hwang KC, Park JY, et al. Identification and characterization of a novel mouse and human MOPT gene containing MORN-motif protein in testis. Theriogenology. 2010. Feb;73(3):273–81. doi: 10.1016/j.theriogenology.2009.09.010. PubMed PMID: 19913896. [DOI] [PubMed] [Google Scholar]
- 3449.Lee SK, Shanmughapriya S, Mok MCY, et al. Structural insights into mitochondrial calcium uniporter regulation by divalent cations. Cell Chem Biol. 2016. Sep 22;23(9):1157–1169. doi: 10.1016/j.chembiol.2016.07.012 PubMed PMID: 27569754; PubMed Central PMCID: PMCPMC5035232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3450.Tomar D, Dong Z, Shanmughapriya S, et al. MCUR1 is a scaffold factor for the MCU complex function and promotes mitochondrial bioenergetics. Cell Rep. 2016. May 24;15(8):1673–85. doi: 10.1016/j.celrep.2016.04.050. PubMed PMID: 27184846; PubMed Central PMCID: PMCPMC4880542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3451.Frost LS, Lopes VS, Bragin A, et al. The contribution of melanoregulin to microtubule-associated protein 1 light chain 3 (LC3) associated phagocytosis in retinal pigment epithelium. Mol Neurobiol. 2015. Oct 10;52:1135–1151. 10.1007/s12035-014-8920-5. PubMed PMID: 25301234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3452.Petherick KJ, Conway OJ, Mpamhanga C, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J Biol Chem. 2015. May 1;290(18):11376–83. doi: 10.1074/jbc.C114.627778. PubMed PMID: 25833948; PubMed Central PMCID: PMC4416842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3453.Bhutia SK, Kegelman TP, Das SK, et al. Astrocyte elevated gene-1 induces protective autophagy. Proc Natl Acad Sci USA 2010. Dec 2;107:22243–8. doi: 10.1073/pnas.1009479107. PubMed PMID: 21127263; Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3454.Wu Y, Cheng S, Zhao H, et al. PI3P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 2014. Sep;15(9):973–81. 10.15252/embr.201438618. PubMed PMID: 25124690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3455.Al-Qusairi L, Prokic I, Amoasii L, et al. Lack of myotubularin (MTM1) leads to muscle hypotrophy through unbalanced regulation of the autophagy and ubiquitin-proteasome pathways. FASEB J. 2013. Aug;27(8):3384–94. 10.1096/fj.12-220947. PubMed PMID: 23695157. [DOI] [PubMed] [Google Scholar]
- 3456.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, et al. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic. 2010. Jan 6;11:468–78. doi: TRA1034 [pii] doi: 10.1111/j.1600-0854.2010.01034.x. PubMed PMID: 20059746; Eng. [DOI] [PubMed] [Google Scholar]
- 3457.Vergne I, Roberts E, Elmaoued RA, et al. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J. 2009. Aug 5;28(15):2244–58. emboj2009159 [pii] doi: 10.1038/emboj.2009.159. PubMed PMID: 19590496; PubMed Central PMCID: PMC2726690. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3458.Zou J, Zhang C, Marjanovic J, et al. Myotubularin-related protein (MTMR) 9 determines the enzymatic activity, substrate specificity, and role in autophagy of MTMR8. Proc Natl Acad Sci U S A. 2012. Jun 12;109(24):9539–44. 10.1073/pnas.1207021109. PubMed PMID: 22647598; PubMed Central PMCID: PMC3386095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3459.Hnia K, Kretz C, Amoasii L, et al. Primary T-tubule and autophagy defects in the phosphoinositide phosphatase Jumpy/MTMR14 knockout mice muscle. Adv Biol Regul. 2012. Jan;52(1):98–107. doi: 10.1016/j.advenzreg.2011.09.007. PubMed PMID: 21930146. [DOI] [PubMed] [Google Scholar]
- 3460.Yun J, Puri R, Yang H, et al. MUL1 acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. eLife. 2014. Jun 4;3:e01958. doi: 10.7554/eLife.01958. PubMed PMID: 24898855; PubMed Central PMCID: PMCPMC4044952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3461.Puri R, Cheng XT, Lin MY, et al. Mul1 restrains Parkin-mediated mitophagy in mature neurons by maintaining ER-mitochondrial contacts. Nat Commun. 2019. Aug 13;10(1):3645. doi: 10.1038/s41467-019-11636-5. PubMed PMID: 31409786; PubMed Central PMCID: PMCPMC6692330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3462.Rusten TE, Vaccari T, Lindmo K, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007. Oct 23;17(20):1817–25. doi: 10.1016/j.cub.2007.09.032. PubMed PMID: 17935992. [DOI] [PubMed] [Google Scholar]
- 3463.Yotsu RR, Suzuki K, Simmonds RE, et al. Buruli Ulcer: a Review of the Current Knowledge. Curr Trop Med Rep. 2018;5(4):247–256. doi: 10.1007/s40475-018-0166-2. PubMed PMID: 30460172; PubMed Central PMCID: PMCPMC6223704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3464.Brandstaetter H, Kishi-Itakura C, Tumbarello DA, et al. Loss of functional MYO1C/myosin 1c, a motor protein involved in lipid raft trafficking, disrupts autophagosome-lysosome fusion. Autophagy. 2014;10(12):2310–23. doi: 10.4161/15548627.2014.984272. PubMed PMID: 25551774; PubMed Central PMCID: PMC4502697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3465.Tumbarello DA, Waxse BJ, Arden SD, et al. Autophagy receptors link myosin VI to auto-phagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nat Cell Biol. 2012. Oct;14(10):1024–35. doi: 10.1038/ncb2589. PubMed PMID: 23023224; PubMed Central PMCID: PMC3472162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3466.Hong H, Koon AC, Chen ZS, et al. AQAMAN, a bisamidine-based inhibitor of toxic protein inclusions in neurons, ameliorates cytotoxicity in polyglutamine disease models. J Biol Chem. 2019. Feb 22;294(8):2757–2770. 10.1074/jbc.RA118.006307. PubMed PMID: 30593503; PubMed Central PMCID: PMCPMC6393596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3467.Kuo HP, Lee DF, Chen CT, et al. ARD1 stabilization of TSC2 suppresses tumorigenesis through the mTOR signaling pathway. Sci Signaling 2010;3(108):ra9. doi: 3/108/ra9 [pii] doi: 10.1126/scisignal.2000590. PubMed PMID: 20145209; PubMed Central PMCID: PMC2874891. eng.10.1126/scisignal. 2000590.PubMed PMID: 20145209; PubMed Central PMCID: PMC2874891. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3468.Zhang Y, Cheng Y, Ren X, et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene. 2012. Feb 23;31(8):1055–64. 10.1038/onc.2011.290. PubMed PMID: 21743489; PubMed Central PMCID: PMC3275651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3469.Fang EF, Scheibye-Knudsen M, Brace LE, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014. May 8;157(4):882–896. 10.1016/j.cell.2014.03.026. PubMed PMID: 24813611; PubMed Central PMCID: PMCPMC4625837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3470.Fang EF. Mitophagy and NAD(+) inhibit Alzheimer disease. Autophagy. 2019. Jun;15(6):1112–1114. doi: 10.1080/15548627.2019.1596497. PubMed PMID: 30922179; PubMed Central PMCID: PMCPMC6526831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3471.Wang P, Guan YF, Du H, et al. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. 2012. Jan;8(1):77–87. doi: 10.4161/auto.8.1.18274. PubMed PMID: 22113203. [DOI] [PubMed] [Google Scholar]
- 3472.Jin R, Liu L, Zhu W, et al. Iron oxide nanoparticles promote macrophage autophagy and inflammatory response through activation of toll-like Receptor-4 signaling. Biomaterials. 2019. May;203:23–30. 10.1016/j.biomaterials.2019.02.026 PubMed PMID: 30851490. [DOI] [PubMed] [Google Scholar]
- 3473.Wan HY, Chen JL, Zhu X, et al. Titania-coated gold nano-bipyramids for blocking autophagy flux and sensitizing cancer cells to proteasome inhibitor-induced death. Adv Sci (Weinh). 2018. Mar;5(3):1700585. 10.1002/advs.201700585 PubMed PMID: 29593960; PubMed Central PMCID: PMCPMC5867123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3474.Lin YX, Wang Y, Wang H.. Recent advances in nanotechnology for autophagy detection. Small. 2017. Sep;13(33). 10.1002/smll.201700996 PubMed PMID: 28677891. [DOI] [PubMed] [Google Scholar]
- 3475.Zhang C, Ren J, He J, et al. Long-term monitoring of tumor-related autophagy in vivo by Fe3O4NO. nanoparticles Biomaterials. 2018. Oct;179:186–198. doi: 10.1016/j.biomaterials.2018.07.004. PubMed PMID: 30037455. [DOI] [PubMed] [Google Scholar]
- 3476.Shi Z, Chen X, Zhang L, et al. FA-PEG decorated MOF nanoparticles as a targeted drug delivery system for controlled release of an autophagy inhibitor. Biomater Sci. 2018. Sep 25;6(10):2582–2590. 10.1039/c8bm00625c. PubMed PMID: 30151542. [DOI] [PubMed] [Google Scholar]
- 3477.Gong C, Hu C, Gu F, et al. Co-delivery of autophagy inhibitor ATG7 siRNA and docetaxel for breast cancer treatment. J Control Release. 2017. Nov 28;266:272–286. doi: 10.1016/j.jconrel.2017.09.042. PubMed PMID: 28987884. [DOI] [PubMed] [Google Scholar]
- 3478.Naydenov NG, Harris G, Morales V, et al. Loss of a membrane trafficking protein alphaSNAP induces non-canonical autophagy in human epithelia. Cell cycle. 2012. Dec 15;11(24):4613–25. 10.4161/cc.22885. PubMed PMID: 23187805; PubMed Central PMCID: PMC3562306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3479.Hafren A, Macia JL, Love AJ, et al. Selective autophagy limits cauliflower mosaic virus infection by NBR1-mediated targeting of viral capsid protein and particles. Proc Natl Acad Sci U S A. 2017. Mar 7;114(10):E2026–E2035. doi: 10.1073/pnas.1610687114 PubMed PMID: 28223514; PubMed Central PMCID: PMCPMC5347569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3480.Wang P, Richardson C, Hawes C, et al. Arabidopsis NAP1 regulates the formation of auto-phagosomes. Curr Biol. 2016. Aug 8;26(15):2060–2069. 10.1016/j.cub.2016.06.008 PubMed PMID: 27451899. [DOI] [PubMed] [Google Scholar]
- 3481.Hohfeld J. Autophagy: press and push for destruction. Curr Biol. 2016. Aug 8;26(15):R703–R705. 10.1016/j.cub.2016.06.017. PubMed PMID: 27505239. [DOI] [PubMed] [Google Scholar]
- 3482.Hitomi J, Christofferson DE, Ng A, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008. Dec 26;135(7):1311–23. doi: 10.1016/j.cell.2008.10.044. PubMed PMID: 19109899; PubMed Central PMCID: PMC2621059. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3483.Bonapace L, Bornhauser BC, Schmitz M, et al. Induction of autophagy-dependent necroptosis is required for childhood acute lymphoblastic leukemia cells to overcome glucocorticoid resistance [Research Support, Non-U.S. Gov’t]. J Clin Invest. 2010. Apr 1;120(4):1310–23. doi: 10.1172/JCI39987. PubMed PMID: 20200450; PubMed Central PMCID: PMC2846044. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3484.Ogasawara M, Yano T, Tanno M, et al. Suppression of autophagic flux contributes to cardiomyocyte death by activation of necroptotic pathways. J Mol Cell Cardiol. 2017. Jul;108:203–213. doi: 10.1016/j.yjmcc.2017.06.008. PubMed PMID: 28647341. [DOI] [PubMed] [Google Scholar]
- 3485.Pei G, Buijze H, Liu H, et al. The E3 ubiquitin ligase NEDD4 enhances killing of membrane-perturbing intracellular bacteria by promoting autophagy. Autophagy. 2017;13(12):2041–2055. doi: 10.1080/15548627.2017.1376160. PubMed PMID: 29251248; PubMed Central PMCID: PMCPMC5788543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3486.Aramburu J, Drews-Elger K, Estrada-Gelonch A, et al. Regulation of the hypertonic stress response and other cellular functions by the Rel-like transcription factor NFAT5 Biochem Pharm. 2006. Nov 30;72(11):1597–604. doi: 10.1016/j.bcp.2006.07.002. PubMed PMID: 16904650. [DOI] [PubMed] [Google Scholar]
- 3487.Halterman JA, Kwon HM, Wamhoff BR.. Tonicity-independent regulation of the osmosensitive transcription factor TonEBP (NFAT5). Am J Physiol Cell Physiol. 2012. Jan 1;302(1):C1–8. 10.1152/ajpcell.00327.2011 PubMed PMID: 21998140; PubMed Central PMCID: PMCPMC3328893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3488.Aramburu J, Lopez-Rodriguez C.. Regulation of Inflammatory Functions of Macrophages and T Lymphocytes by NFAT5. Front Immunol. 2019;10:535. 10.3389/fimmu.2019.00535 PubMed PMID: 30949179; PubMed Central PMCID: PMCPMC6435587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3489.Liu C, Choi H, Johnson ZI, et al. Lack of evidence for involvement of TonEBP and hyperosmotic stimulus in induction of autophagy in the nucleus pulposus. Sci Rep. 2017. Jul 3;7(1):4543. doi: 10.1038/s41598-017-04876-2. PubMed PMID: 28674405; PubMed Central PMCID: PMCPMC5495809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3490.Valdor R, Mocholi E, Botbol Y, et al. Chaperone-mediated autophagy regulates T cell responses through targeted degradation of negative regulators of T cell activation. Nat Immunol. 2014. Nov;15(11):1046–54. doi: 10.1038/ni.3003.. PubMed PMID: 25263126; PubMed Central PMCID: PMCPMC4208273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3491.Bae SH, Sung SH, Oh SY, et al. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013. Jan 8;17(1):73–84. doi: 10.1016/j.cmet.2012.12.002. PubMed PMID: 23274085. [DOI] [PubMed] [Google Scholar]
- 3492.Djavaheri-Mergny M, Amelotti M, Mathieu J, et al. Regulation of autophagy by NFkappaB transcription factor and reactives oxygen species. Autophagy. 2007. Jul-Aug;3(4):390–2. PubMed PMID: 17471012; eng. [DOI] [PubMed] [Google Scholar]
- 3493.Copetti T, Demarchi F, Schneider C.. p65/RelA binds and activates the beclin 1 promoter. Autophagy. 2009. Aug;5(6):858–9. 10.4161/auto.8822 PubMed PMID: 19458474. [DOI] [PubMed] [Google Scholar]
- 3494.Wattin M, Gaweda L, Muller P, et al.Modulation of protein quality control and proteasome to autophagy switch in immortalized myoblasts from duchenne muscular dystrophy patients. Int J Mol Sci. 2018. Jan 7;19(1). 10.3390/ijms19010178 PubMed PMID: 29316663; PubMed Central PMCID: PMCPMC5796127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3495.Criado O, Aguado C, Gayarre J, et al. Lafora bodies and neurological defects in malin-deficient mice correlate with impaired autophagy. Hum Mol Genet. 2012. Apr 1;21(7):1521–33. 10.1093/hmg/ddr590 PubMed PMID: 22186026. [DOI] [PubMed] [Google Scholar]
- 3496.Nozima BH, Mendes TB, Pereira G, et al. FAM129A regulates autophagy in thyroid carcinomas in an oncogene-dependent manner. Endocr Relat Cancer. 2019. Jan 1;26(1):227–238. 10.1530/ERC-17-0530 PubMed PMID: 30400008. [DOI] [PubMed] [Google Scholar]
- 3497.Princely Abudu Y, Pankiv S, Mathai BJ, et al. NIPSNAP1 and NIPSNAP2 Act as “Eat Me” Signals for Mitophagy. Dev Cell. 2019. May 20;49(4):509–525e12. 10.1016/j.devcel.2019.03.013 PubMed PMID: 30982665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3498.Cervia D, Perrotta C, Moscheni C, et al. Nitric oxide and sphingolipids control apoptosis and autophagy with a significant impact on Alzheimer’s disease. J Biol Regulator Hom Agents 2013. Apr-Jun;27(2 Suppl):11–22. PubMed PMID: 24813312. [PubMed] [Google Scholar]
- 3499.Rabkin SW. Nitric oxide-induced cell death in the heart: the role of autophagy. Autophagy. 2007. Jul-Aug;3(4):347–9. PubMed PMID: 17438363. [DOI] [PubMed] [Google Scholar]
- 3500.Zang L, He H, Ye Y, et al. Nitric oxide augments oridonin-induced efferocytosis by human histocytic lymphoma U937 cells via autophagy and the NF-kappaB-COX-2-IL-1beta pathway. Free Radic Res. 2012. Oct;46(10):1207–19. doi: 10.3109/10715762.2012.700515. PubMed PMID: 22670565. [DOI] [PubMed] [Google Scholar]
- 3501.Lei Y, Wen H, Yu Y, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012. Jun 29;36(6):933–46. doi: 10.1016/j.immuni.2012.03.025. PubMed PMID: 22749352; PubMed Central PMCID: PMCPMC3397828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3502.Lei Y, Kansy BA, Li J, et al. EGFR-targeted mAb therapy modulates autophagy in head and neck squamous cell carcinoma through NLRX1-TUFM protein complex. Oncogene. 2016. Sep 8;35(36):4698–707. doi: 10.1038/onc.2016.11. PubMed PMID: 26876213; PubMed Central PMCID: PMCPMC5257174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3503.Zhang Y, Yao Y, Qiu X, et al. Listeria hijacks host mitophagy through a novel mitophagy receptor to evade killing. Nat Immunol. 2019. Apr;20(4):433–446. doi: 10.1038/s41590-019-0324-2. PubMed PMID: 30804553. [DOI] [PubMed] [Google Scholar]
- 3504.Wang K, Ma H, Liu H, et al. The Glycoprotein and Nucleocapsid Protein of Hantaviruses Manipulate Autophagy Flux to Restrain Host Innate Immune Responses. Cell Rep. 2019. May 14;27(7):2075–2091 e5. doi: 10.1016/j.celrep.2019.04.061. PubMed PMID: 31091447. [DOI] [PubMed] [Google Scholar]
- 3505.Schlattner U, Tokarska-Schlattner M, Epand RM, et al. Mitochondrial NM23-H4/NDPK-D: a bifunctional nanoswitch for bioenergetics and lipid signaling. Naunyn Schmiedebergs Arch Pharmacol. 2015. Feb;388(2):271–8. doi: 10.1007/s00210-014-1047-4. PubMed PMID: 25231795. [DOI] [PubMed] [Google Scholar]
- 3506.Schlattner U, Tokarska-Schlattner M, Epand RM, et al. NME4/nucleoside diphosphate kinase D in cardiolipin signaling and mitophagy. Lab Invest. 2018. Feb;98(2):228–232. doi: 10.1038/labinvest.2017.113. PubMed PMID: 29035377. [DOI] [PubMed] [Google Scholar]
- 3507.Panchaud N, Peli-Gulli MP, De Virgilio C.. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci Signal. 2013. May 28;6(277):ra42. doi: 10.1126/scisignal.2004112. PubMed PMID: 23716719. [DOI] [PubMed] [Google Scholar]
- 3508.Panchaud N, Peli-Gulli MP, De Virgilio C.. SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell cycle. 2013. Sep 15;12(18):2948–52. doi: 10.4161/cc.26000. PubMed PMID: 23974112; PubMed Central PMCID: PMCPMC3875668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3509.Sarkar S, Carroll B, Buganim Y, et al. Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Rep. 2013. Dec 12;5(5):1302–15. doi: 10.1016/j.celrep.2013.10.042. PubMed PMID: 24290752; PubMed Central PMCID: PMCPMC3957429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3510.Maetzel D, Sarkar S, Wang H, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports. 2014. Jun 3;2(6):866–80. doi: 10.1016/j.stemcr.2014.03.014. PubMed PMID: 24936472; PubMed Central PMCID: PMCPMC4050353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3511.Schwerd T, Pandey S, Yang HT, et al. Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn’s disease. Gut. 2017. Jun;66(6):1060–1073. doi: 10.1136/gutjnl-2015-310382. PubMed PMID: 26953272; PubMed Central PMCID: PMCPMC5532464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3512.Aveleira CA, Botelho M, Carmo-Silva S, et al. Neuropeptide Y stimulates autophagy in hypothalamic neurons. Proc Natl Acad Sci U S A. 2015. Mar 31;112(13):E1642–51. doi: 10.1073/pnas.1416609112. PubMed PMID: 25775546; PubMed Central PMCID: PMC4386327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3513.Catalani E, De Palma C, Perrotta C, et al.Current evidence for a role of neuropeptides in the regulation of autophagy. Biomed Res Int. 2017;2017:5856071. doi: 10.1155/2017/5856071. PubMed PMID: 28593174; PubMed Central PMCID: PMCPMC5448050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3514.Cervia D, Catalani E, Casini G.. Neuroprotective peptides in retinal disease. J Clin Med. 2019. Aug 1;8(8). doi: 10.3390/jcm8081146. PubMed PMID: 31374938; PubMed Central PMCID: PMCPMC6722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3515.Cao Y, Wang Y, Abi Saab WF, et al. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J. 2014. Jul 15;461(2):315–22. doi: 10.1042/BJ20140515. PubMed PMID: 24785657; PubMed Central PMCID: PMC4180102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3516.Lu J, He L, Behrends C, et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5:3920. doi: 10.1038/ncomms4920. PubMed PMID: 24849286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3517.Judith D, Mostowy S, Bourai M, et al. Species-specific impact of the autophagy machinery on Chikungunya virus infection. EMBO Rep. 2013. Jun;14(6):534–44. doi: 10.1038/embor.2013.51. PubMed PMID: 23619093; PubMed Central PMCID: PMC3674439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3518.El-Ami T, Moll L, Carvalhal Marques F, et al. A novel inhibitor of the insulin/IGF signaling pathway protects from age-onset, neurodegeneration-linked proteotoxicity. Aging cell. 2014. Feb;13(1):165–74. doi: 10.1111/acel.12171. PubMed PMID: 24261972; PubMed Central PMCID: PMCPMC4326862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3519.Moll L, Ben-Gedalya T, Reuveni H, et al. The inhibition of IGF-1 signaling promotes proteostasis by enhancing protein aggregation and deposition. FASEB J. 2016. Apr;30(4):1656–69. doi: 10.1096/fj.15-281675. PubMed PMID: 26722006. [DOI] [PubMed] [Google Scholar]
- 3520.Ji CH, Kim HY, Heo AJ, et al. The N-degron pathway mediates ER-phagy. Mol Cell. 2019. Sep 5;75(5):1058–1072 e9. doi: 10.1016/j.molcel.2019.06.028. PubMed PMID: 31375263. [DOI] [PubMed] [Google Scholar]
- 3521.Franco ML, Melero C, Sarasola E, et al. Mutations in TrkA causing congenital insensitivity to pain with anhidrosis (CIPA) induce misfolding, aggregation, and mutation-dependent neurodegeneration by dysfunction of the autophagic flux. J Biol Chem. 2016. Oct 7;291(41):21363–21374. doi: 10.1074/jbc.M116.722587. PubMed PMID: 27551041; PubMed Central PMCID: PMCPMC5076807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3522.Funasaka T, Tsuka E, Wong RW.. Regulation of autophagy by nucleoporin Tpr. Sci Rep. 2012;2:878. doi: 10.1038/srep00878. PubMed PMID: 23170199; PubMed Central PMCID: PMC3501823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3523.Fullgrabe J, Klionsky DJ, Joseph B.. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014. Jan;15(1):65–74. doi: 10.1038/nrm3716. PubMed PMID: 24326622. [DOI] [PubMed] [Google Scholar]
- 3524.Wyant GA, Abu-Remaileh M, Frenkel EM, et al. NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science. 2018. May 18;360(6390):751–758. doi: 10.1126/science.aar2663. PubMed PMID: 29700228; PubMed Central PMCID: PMCPMC6020066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3525.Lee CW, Wilfling F, Ronchi P, et al. Selective autophagy degrades nuclear pore complexes. Nat Cell Biol. 2020. Feb22(2):159–166. doi: 10.1038/s41556-019-0459-2. PubMed PMID: 32029894. [DOI] [PubMed] [Google Scholar]
- 3526.Kong DK, Georgescu SP, Cano C, et al. Deficiency of the transcriptional regulator p8 results in increased autophagy and apoptosis, and causes impaired heart function. Mol Biol Cell. 2010. Apr;21(8):1335–49. doi: 10.1091/mbc.E09-09-0818. PubMed PMID: 20181828; PubMed Central PMCID: PMC2854092. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3527.Chang KY, Tsai SY, Wu CM, et al. Novel phosphoinositide 3-kinase/mTOR dual inhibitor, NVP-BGT226, displays potent growth-inhibitory activity against human head and neck cancer cells in vitro and in vivo. clin cancer res off j am assoc cancer res. 2011. Nov 15;17(22):7116–26. doi: 10.1158/1078-0432.CCR-11-0796. PubMed PMID: 21976531. [DOI] [PubMed] [Google Scholar]
- 3528.De Leo MG, Staiano L, Vicinanza M, et al. Autophagosome-lysosome fusion triggers a lysosomal response mediated by TLR9 and controlled by OCRL. Nat Cell Biol. 2016. Aug;18(8):839–850. doi: 10.1038/ncb3386. PubMed PMID: 27398910; PubMed Central PMCID: PMCPMC5040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3529.Wang P, Lazarus BD, Forsythe ME, et al. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc Natl Acad Sci U S A. 2012. Oct 23;109(43):17669–74. doi: 10.1073/pnas.1205748109. PubMed PMID: 22988095; PubMed Central PMCID: PMC3491483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3530.Yang X, Qian K.. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017. Jul;18(7):452–465. 10.1038/nrm.2017.22. PubMed PMID: 28488703; PubMed Central PMCID: PMCPMC5667541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3531.Guo B, Liang Q, Li L, et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol. 2014. Dec;16(12):1215–26. 10.1038/ncb3066. PubMed PMID: 25419848. [DOI] [PubMed] [Google Scholar]
- 3532.Gundara JS, Zhao J, Robinson BG, et al. Oncophagy: harnessing regulation of autophagy in cancer therapy. Endocr Relat Cancer. 2012. Dec;19(6):R281–95. doi: 10.1530/ERC-12-0325. PubMed PMID: 23082009. [DOI] [PubMed] [Google Scholar]
- 3533.Wong YC, Holzbaur EL.. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014. Oct 21;111(42):E4439–48. doi: 10.1073/pnas.1405752111. PubMed PMID: 25294927; PubMed Central PMCID: PMCPMC4210283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3534.Wong YC, Holzbaur EL.. Temporal dynamics of PARK2/parkin and OPTN/optineurin recruitment during the mitophagy of damaged mitochondria. Autophagy. 2015;11(2):422–4. doi: 10.1080/15548627.2015.1009792. PubMed PMID: 25801386; PubMed Central PMCID: PMCPMC4502688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3535.Richter B, Sliter DA, Herhaus L, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A. 2016. Apr 12;113(15):4039–44. doi: 10.1073/pnas.1523926113. PubMed PMID: 27035970; PubMed Central PMCID: PMCPMC4839414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3536.Korac J, Schaeffer V, Kovacevic I, et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J Cell Sci. 2013. Jan15;126\(Pt 2):580–92. 10.1242/jcs.114926. PubMed PMID: 23178947; PubMed Central PMCID: PMCPMC3654196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3537.Li F, Xie X, Wang Y, et al. Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat Commun. 2016. Sep 13;7:12708. 10.1038/ncomms12708. PubMed PMID: 27620379; PubMed Central PMCID: PMCPMC5027247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3538.Li F, Xu D, Wang Y, et al. Structural insights into the ubiquitin recognition by OPTN (optineurin) and its regulation by TBK1-mediated phosphorylation. Autophagy. 2018;14(1):66–79. 10.1080/15548627.2017.1391970. PubMed PMID: 29394115; PubMed Central PMCID: PMCPMC5846504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3539.Maruyama H, Morino H, Ito H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010. May 13;465(7295):223–6. 10.1038/nature08971. PubMed PMID: 20428114. [DOI] [PubMed] [Google Scholar]
- 3540.Mijaljica D. Autophagy in 2020 and beyond: eating our way into a healthy future. Autophagy. 2010. Jan6(1):194–6. PubMed PMID: 20110773. [DOI] [PubMed] [Google Scholar]
- 3541.Zhang CF, Gruber F, Ni C, et al. Suppression of autophagy dysregulates the antioxidant response and causes premature senescence of melanocytes. J Invest Dermatol. 2015. May;135(5):1348–57. 10.1038/jid.2014.439. PubMed PMID: 25290687. [DOI] [PubMed] [Google Scholar]
- 3542.Zhao Y, Zhang CF, Rossiter H, et al. Autophagy is induced by UVA and promotes removal of oxidized phospholipids and protein aggregates in epidermal keratinocytes. J Invest Dermatol. 2013. Jun;133(6):1629–37. 10.1038/jid.2013.26. PubMed PMID: 23340736. [DOI] [PubMed] [Google Scholar]
- 3543.Veldhoen RA, Banman SL, Hemmerling DR, et al. The chemotherapeutic agent paclitaxel inhibits autophagy through two distinct mechanisms that regulate apoptosis. Oncogene. 2013. Feb 7;32(6):736–46. 10.1038/onc.2012.92. PubMed PMID: 22430212. [DOI] [PubMed] [Google Scholar]
- 3544.Ren H, Fu K, Mu C, et al. DJ-1, a cancer and Parkinson’s disease associated protein, regulates autophagy through JNK pathway in cancer cells. Cancer Lett 2010. May 24;297:101–8. doi: 10.1016/j.canlet.2010.05.001. PubMed PMID: 20510502; Eng. [DOI] [PubMed] [Google Scholar]
- 3545.Meissner C, Lorenz H, Hehn B, et al. Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy. Autophagy. 2015. Jun 23;11:1484–98. 10.1080/15548627.2015.1063763. PubMed PMID: 26101826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3546.Jin SM, Lazarou M, Wang C, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010. Nov 29;191(5):933–42. 10.1083/jcb.201008084. PubMed PMID: 21115803; PubMed Central PMCID: PMC2995166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3547.Meissner C, Lorenz H, Weihofen A, et al. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J Neurochem. 2011. Jun;117(5):856–67. doi: 10.1111/j.1471-4159.2011.07253.x. PubMed PMID: 21426348. [DOI] [PubMed] [Google Scholar]
- 3548.Shi G, Lee JR, Grimes DA, et al. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum Mol Genet. 2011. May 15;20(10):1966–74. 10.1093/hmg/ddr077. PubMed PMID: 21355049. [DOI] [PubMed] [Google Scholar]
- 3549.Munoz-Gamez JA, Rodriguez-Vargas JM, Quiles-Perez R, et al. PARP-1 is involved in autophagy induced by DNA damage. Autophagy. 2009. Jan 1;5(1):61–74. PubMed PMID: 19001878; eng. [DOI] [PubMed] [Google Scholar]
- 3550.Huang Q, Shen HM.. To die or to live: the dual role of poly(ADP-ribose) polymerase-1 in autophagy and necrosis under oxidative stress and DNA damage. Autophagy. 2009. Feb;5(2):273–6. PubMed PMID: 19139632. [DOI] [PubMed] [Google Scholar]
- 3551.Wang LJ, Chen PR, Hsu LP, et al. Concomitant induction of apoptosis and autophagy by prostate apoptosis response-4 in hypopharyngeal carcinoma cells. Am J Pathol. 2014. Feb;184(2):418–30. doi: 10.1016/j.ajpath.2013.10.012. PubMed PMID: 24418097. [DOI] [PubMed] [Google Scholar]
- 3552.Thayyullathil F, Rahman A, Pallichankandy S, et al. ROS-dependent prostate apoptosis response-4 (Par-4) up-regulation and ceramide generation are the prime signaling events associated with curcumin-induced autophagic cell death in human malignant glioma. FEBS Open Bio 2014;4:763–76. 10.1016/j.fob.2014.08.005. PubMed PMID: 25349781; PubMed Central PMCID: PMC4208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3553.Silvente-Poirot S, Poirot M.. Cholesterol metabolism and cancer: the good, the bad and the ugly. Curr Opin Pharmacol. 2012. Dec;12(6):673–6. 10.1016/j.coph.2012.10.004. PubMed PMID: 23103112. [DOI] [PubMed] [Google Scholar]
- 3554.Andrejeva G, Gowan S, Lin G, et al. De novo phosphatidylcholine synthesis is required for autophagosome membrane formation and maintenance during autophagy. Autophagy. 2019. Sep 13:1–17. 10.1080/15548627.2019.1659608. PubMed PMID: 31517566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3555.Li X, Rydzewski N, Hider A, et al. A molecular mechanism to regulate lysosome motility for lysosome positioning and tubulation. Nat Cell Biol. 2016. Apr;18(4):404–17. 10.1038/ncb3324. PubMed PMID: 26950892; PubMed Central PMCID: PMCPMC4871318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3556.Murrow L, Malhotra R, Debnath J.. ATG12-ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat Cell Biol. 2015. Mar;17(3):300–10. 10.1038/ncb3112. PubMed PMID: 25686249; PubMed Central PMCID: PMC4344874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3557.Bock BC, Tagscherer KE, Fassl A, et al. The PEA-15 protein regulates autophagy via activation of JNK. J Biol Chem. 2010. Jul 9;285(28):21644–54. 10.1074/jbc.M109.096628. PubMed PMID: 20452983; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3558.Noh HS, Hah YS, Zada S, et al. PEBP1, a RAF kinase inhibitory protein, negatively regulates starvation-induced autophagy by direct interaction with LC3. Autophagy. 2016. Nov;12(11):2183–2196. doi: 10.1080/15548627.2016.1219013. PubMed PMID: 27540684; PubMed Central PMCID: PMCPMC5103343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3559.Ahmed M, Lai TH, Zada S, et al. Functional linkage of RKIP to the epithelial to mesenchymal transition and autophagy during the development of prostate cancer. Cancers (Basel). 2018. Aug 16;10(8). doi: 10.3390/cancers10080273. PubMed PMID: 30115852; PubMed Central PMCID: PMCPMC6115972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3560.Torres A, Gubbiotti MA, Iozzo RV.. Decorin-inducible Peg3 Evokes Beclin 1-mediated autophagy and thrombospondin 1-mediated angiostasis. J Biol Chem. 2017. Mar 24;292(12):5055–5069. 10.1074/jbc.M116.753632. PubMed PMID: 28174297; PubMed Central PMCID: PMCPMC5377817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3561.Neill T, Sharpe C, Owens RT, et al. Decorin-evoked paternally expressed gene 3 (PEG3) is an upstream regulator of the transcription factor EB (TFEB) in endothelial cell autophagy. J Biol Chem. 2017. Sep 29;292(39):16211–16220. doi: 10.1074/jbc.M116.769950. PubMed PMID: 28798237; PubMed Central PMCID: PMCPMC5625051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3562.Rami A, Fekadu J, Rawashdeh O.. The Hippocampal Autophagic Machinery is Depressed in the Absence of the Circadian Clock Protein PER1 that may Lead to Vulnerability During Cerebral Ischemia. Curr Neurovasc Res. 2017;14(3):207–214. doi: 10.2174/1567202614666170619083239. PubMed PMID: 28625127. [DOI] [PubMed] [Google Scholar]
- 3563.Lee MY, Sumpter R, Jr., Zou Z, et al. Peroxisomal protein PEX13 functions in selective autophagy. EMBO Rep. 2017. Jan;18(1):48–60. doi: 10.15252/embr.201642443. PubMed PMID: 27827795; PubMed Central PMCID: PMCPMC5210156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3564.Ano Y, Hattori T, Oku M, et al. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. ?Mol Biol Cell 2005. Feb;16(2):446–57. PubMed PMID: 15563611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3565.Klarer AC, O’Neal J, Imbert-Fernandez Y, et al. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) induces autophagy as a survival mechanism. Cancer Metab. 2014. Jan 23;2(1):2. doi: 10.1186/2049-3002-2-2. PubMed PMID: 24451478; PubMed Central PMCID: PMCPMC3913946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3566.Yano T, Mita S, Ohmori H, et al. Autophagic control of listeria through intracellular innate immune recognition in drosophila. Nat Immunol. 2008. Aug;9(8):908–16. doi: 10.1038/ni.1634. PubMed PMID: 18604211; PubMed Central PMCID: PMC2562576. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3567.Seglen PO, Gordon PB, Holen I.. Non-selective autophagy. Semin Cell Biol. 1990. Dec;1(6):441–8. PubMed PMID: 2103895; eng. [PubMed] [Google Scholar]
- 3568.He C, Klionsky DJ.. Atg9 trafficking in autophagy-related pathways. Autophagy. 2007. May-Jun;3(3):271–4. PubMed PMID: 17329962; eng. [DOI] [PubMed] [Google Scholar]
- 3569.Zhou F, Zou S, Chen Y, et al. A Rab5 GTPase module is important for autophagosome closure. PLoS Genet. 2017. Sep;13(9):e1007020. doi: 10.1371/journal.pgen.1007020. PubMed PMID: 28934205; PubMed Central PMCID: PMCPMC5626503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3570.Amer AO, Swanson MS.. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell Microbiol 2005. Jun;7(6):765–78. PubMed PMID: 15888080; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3571.Yan C, Gong L, Chen L, et al. PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy. 2020. Mar;16(3):419–434. 10.1080/15548627.2019.1628520. PubMed PMID: 31177901; PubMed Central PMCID: PMCPMC6999623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3572.Huang H, Kawamata T, Horie T, et al. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO J. 2015. Jan 14;34(2):154–68. doi: 10.15252/embj.201489083. PubMed PMID: 25468960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3573.Meijer AJ, Klionsky DJ.. Vps34 is a phosphatidylinositol 3-kinase, not a phosphoinositide 3-kinase. Autophagy. 2011. Jun;7(6):563–4. PubMed PMID: 21278489; PubMed Central PMCID: PMC3625115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3574.Devereaux K, Dall’Armi C, Alcazar-Roman A, et al. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS One. 2013;8(10):e76405. doi: 10.1371/journal.pone.0076405. PubMed PMID: 24098492; PubMed Central PMCID: PMC3789715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3575.Stjepanovic G, Baskaran S, Lin MG, et al.Vps34 kinase domain dynamics regulate the autophagic PI 3-kinase complex. Mol Cell. 2017. Aug 3;67(3):528–534 e3. doi: 10.1016/j.molcel.2017.07.003 PubMed PMID: 28757208; PubMed Central PMCID: PMCPMC5573195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3576.Ma M, Liu JJ, Li Y, et al.Cryo-EM structure and biochemical analysis reveal the basis of the functional difference between human PI3KC3-C1 and -C2. Cell Res. 2017. Aug27(8):989–1001. doi: 10.1038/cr.2017.94 PubMed PMID: 28731030; PubMed Central PMCID: PMCPMC5539356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3577.Baskaran S, Carlson LA, Stjepanovic G, et al.Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. eLife. 2014. Dec 9;3. doi: 10.7554/eLife.05115 PubMed PMID: 25490155; PubMed Central PMCID: PMCPMC4281882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3578.Byfield MP, Murray JT, Backer JM.. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem. 2005. Sep 23;280(38):33076–82. doi: 10.1074/jbc.M507201200. PubMed PMID: 16049009. [DOI] [PubMed] [Google Scholar]
- 3579.Roppenser B, Grinstein S, Brumell JH.. Modulation of host phosphoinositide metabolism during Salmonella invasion by the type III secreted effector SopB. Methods Cell Biol. 2012;108:173–86. doi: 10.1016/B978-0-12-386487-1.00009-2. PubMed PMID: 22325603. [DOI] [PubMed] [Google Scholar]
- 3580.Cuesta-Geijo MA, Galindo I, Hernaez B, et al. Endosomal maturation, Rab7 GTPase and phosphoinositides in African swine fever virus entry. PLoS One. 2012;7(11):e48853. doi: 10.1371/journal.pone.0048853. PubMed PMID: 23133661; PubMed Central PMCID: PMC3486801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3581.Jin N, Mao K, Jin Y, et al. Roles for PI(3,5)P2 in nutrient sensing through TORC1. Mol Biol Cell. 2014. Jan 29;25:1171–85. doi: 10.1091/mbc.E14-01-0021. PubMed PMID: 24478451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3582.Sharma G, Guardia CM, Roy A, et al. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy. 2019. Oct;15(10):1694–1718. doi: 10.1080/15548627.2019.1586257. PubMed PMID: 30806145; PubMed Central PMCID: PMCPMC6735543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3583.Tan X, Thapa N, Choi S, et al. Emerging roles of PtdIns(4,5)P2–beyond the plasma membrane. J Cell Sci. 2015. Nov 15;128(22):4047–56. doi: 10.1242/jcs.175208. PubMed PMID: 26574506; PubMed Central PMCID: PMCPMC4712784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3584.Tan X, Thapa N, Liao Y, et al. PtdIns(4,5)P2 signaling regulates ATG14 and autophagy. Proc Natl Acad Sci U S A. 2016. Sep 27;113(39):10896–901. doi: 10.1073/pnas.1523145113. PubMed PMID: 27621469; PubMed Central PMCID: PMCPMC5047215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3585.Wakatsuki S, Araki T.. Specific phospholipid scramblases are involved in exposure of phosphatidylserine, an “eat-me” signal for phagocytes, on degenerating axons. Commun Integr Biol. 2017;10(2):e1296615. doi: 10.1080/19420889.2017.1296615. PubMed PMID: 28451058; PubMed Central PMCID: PMCPMC5398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3586.Jenzer C, Simionato E, Largeau C, et al. Autophagy mediates phosphatidylserine exposure and phagosome degradation during apoptosis through specific functions of GABARAP/LGG-1 and LC3/LGG-2. Autophagy. 2019. Feb;15(2):228–241. doi: 10.1080/15548627.2018.1512452. PubMed PMID: 30160610; PubMed Central PMCID: PMCPMC6333449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3587.Lundquist MR, Goncalves MD, Loughran RM, et al. Phosphatidylinositol-5-phosphate 4-kinases regulate cellular lipid metabolism by facilitating autophagy. Mol Cells. 2018. May 3;70(3):531–544 e9. doi: 10.1016/j.molcel.2018.03.037. PubMed PMID: 29727621; PubMed Central PMCID: PMCPMC5991623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3588.Liao G, Gao B, Gao Y, et al. Phycocyanin inhibits tumorigenic potential of pancreatic cancer cells: role of apoptosis and autophagy. Sci Rep. 2016. Oct 3;6:34564. doi: 10.1038/srep34564. PubMed PMID: 27694919; PubMed Central PMCID: PMCPMC5046139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3589.Wang H, Sun HQ, Zhu X, et al. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc Natl Acad Sci U S A. 2015. Jun 2;112(22):7015–20. doi: 10.1073/pnas.1507263112. PubMed PMID: 26038556; PubMed Central PMCID: PMC4460452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3590.Marat AL, Wallroth A, Lo WT, et al. mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate. Science. 2017. Jun 2;356(6341):968–972. doi: 10.1126/science.aaf8310. PubMed PMID: 28572395. [DOI] [PubMed] [Google Scholar]
- 3591.Dou Z, Chattopadhyay M, Pan JA, et al. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol. 2010. Nov 15;191(4):827–43. doi: 10.1083/jcb.201006056. PubMed PMID: 21059846; PubMed Central PMCID: PMC2983054. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3592.Lindmo K, Brech A, Finley KD, et al. The PI 3-kinase regulator Vps15 is required for autophagic clearance of protein aggregates. Autophagy. 2008. May;4(4):500–6. PubMed PMID: 18326940. [DOI] [PubMed] [Google Scholar]
- 3593.Murray JT, Panaretou C, Stenmark H, et al. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic. 2002. Jun;3(6):416–27. PubMed PMID: 12010460. [DOI] [PubMed] [Google Scholar]
- 3594.Bertolin G, Ferrando-Miguel R, Jacoupy M, et al. The TOMM machinery is a molecular switch in PINK1 and PARK2/PARKIN-dependent mitochondrial clearance. Autophagy. 2013. Nov 1;9(11):1801–17. doi: 10.4161/auto.25884. PubMed PMID: 24149440. [DOI] [PubMed] [Google Scholar]
- 3595.Greene AW, Grenier K, Aguileta MA, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012. Apr;13(4):378–85. doi: 10.1038/embor.2012.14. PubMed PMID: 22354088; PubMed Central PMCID: PMC3321149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3596.Jin SM, Youle RJ.. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013. Nov 1;9(11):1750–7. doi: 10.4161/auto.26122. PubMed PMID: 24149988; PubMed Central PMCID: PMC4028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3597.Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010. Jan 5;107(1):378–83. doi: 10.1073/pnas.0911187107. PubMed PMID: 19966284; PubMed Central PMCID: PMC2806779. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3598.Budovskaya YV, Stephan JS, Reggiori F, et al. The Ras/cAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem. 2004. May 14;279(20):20663–71. doi: 10.1074/jbc.M400272200 PubMed PMID: 15016820; PubMed Central PMCID: PMC1705971. eng [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3599.Shahab S, Namolovan A, Mogridge J, et al. Bacterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy. 2011;7:957–65. [DOI] [PubMed] [Google Scholar]
- 3600.Palorini R, De Rasmo D, Gaviraghi M, et al. Oncogenic K-ras expression is associated with derangement of the cAMP/PKA pathway and forskolin-reversible alterations of mitochondrial dynamics and respiration. Oncogene. 2013. Jan 17;32(3):352–62. doi: 10.1038/onc.2012.50. PubMed PMID: 22410778. [DOI] [PubMed] [Google Scholar]
- 3601.Palorini R, Votta G, Pirola Y, et al. Protein kinase A activation promotes cancer cell resistance to glucose starvation and anoikis. PLoS Genet. 2016. Mar;12(3):e1005931. doi: 10.1371/journal.pgen.1005931. PubMed PMID: 26978032; PubMed Central PMCID: PMCPMC4792400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3602.Agop-Nersesian C, Niklaus L, Wacker R, et al. Host cell cytosolic immune response during Plasmodium liver stage development. FEMS Microbiol Rev 2018. May 1;42(3):324–334. doi: 10.1093/femsre/fuy007. PubMed PMID: 29529207; PubMed Central PMCID: PMCPMC5995216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3603.Niklaus L, Agop-Nersesian C, Schmuckli-Maurer J, et al. Deciphering host lysosome-mediated elimination of Plasmodium berghei liver stage parasites. Sci Rep. 2019. May 28;9(1):7967. doi: 10.1038/s41598-019-44449-z. PubMed PMID: 31138850; PubMed Central PMCID: PMCPMC6538699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3604.Jenkins GM, Frohman MA.. Phospholipase D: a lipid centric review. Cell Mol Life Sci. 2005. Oct;62(19–20):2305–16. doi: 10.1007/s00018-005-5195-z. PubMed PMID: 16143829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3605.Dall’Armi C, Hurtado-Lorenzo A, Tian H, et al. The phospholipase D1 pathway modulates macroautophagy. Nat Commun. 2010;1:142. doi: 10.1038/ncomms1144. PubMed PMID: 21266992; PubMed Central PMCID: PMCPMC3328354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3606.Yoon MS. Vps34 and PLD1 take center stage in nutrient signaling: their dual roles in regulating autophagy. Cell Commun Signal. 2015. Nov 21;13:44. doi: 10.1186/s12964-015-0122-x. PubMed PMID: 26589724; PubMed Central PMCID: PMCPMC4654845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3607.Luningschror P, Binotti B, Dombert B, et al. Plekhg5-regulated autophagy of synaptic vesicles reveals a pathogenic mechanism in motoneuron disease. Nat Commun. 2017. Oct 30;8(1):678. doi: 10.1038/s41467-017-00689-z. PubMed PMID: 29084947; PubMed Central PMCID: PMCPMC5662736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3608.McEwan DG, Popovic D, Gubas A, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015. Jan 8;57(1):39–54. doi: 10.1016/j.molcel.2014.11.006. PubMed PMID: 25498145. [DOI] [PubMed] [Google Scholar]
- 3609.Ruf S, Heberle AM, Langelaar-Makkinje M, et al. PLK1 (polo like kinase 1) inhibits MTOR complex 1 and promotes autophagy. Autophagy. 2017. Mar 4;13(3):486–505. doi: 10.1080/15548627.2016.1263781. PubMed PMID: 28102733; PubMed Central PMCID: PMCPMC5361591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3610.Broadley K, Larsen L, Herst PM, et al. The novel phloroglucinol PMT7 kills glycolytic cancer cells by blocking autophagy and sensitizing to nutrient stress [Research Support, Non-U.S. Gov’t]. J Cell Biochem. 2011. Jul;112(7):1869–79. doi: 10.1002/jcb.23107. PubMed PMID: 21433059; eng. [DOI] [PubMed] [Google Scholar]
- 3611.Martinez-Lopez N, Garcia-Macia M, Sahu S, et al. Autophagy in the CNS and periphery coordinate lipophagy and lipolysis in the brown adipose tissue and liver. Cell Metab. 2016. Jan 12;23(1):113–27. doi: 10.1016/j.cmet.2015.10.008. PubMed PMID: 26698918; PubMed Central PMCID: PMCPMC4715637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3612.Dupont N, Chauhan S, Arko-Mensah J, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014. Mar 17;24(6):609–20. doi: 10.1016/j.cub.2014.02.008. PubMed PMID: 24613307; PubMed Central PMCID: PMC4016984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3613.Elimam H, Papillon J, Kaufman DR, et al. Genetic ablation of calcium-independent phospholipase A2gamma induces glomerular injury in mice. J Biol Chem. 2016. Jul 8;291(28):14468–82. doi: 10.1074/jbc.M115.696781. PubMed PMID: 27226532; PubMed Central PMCID: PMCPMC4938171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3614.Bhullar KS, Rupasinghe HP.. Polyphenols: multipotent therapeutic agents in neurodegenerative diseases. Oxid Med Cell Longev. 2013;2013:891748. doi: 10.1155/2013/891748. PubMed PMID: 23840922; PubMed Central PMCID: PMC3690243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3615.Macedo D, Tavares L, McDougall GJ, et al. (Poly)phenols protect from alpha-synuclein toxicity by reducing oxidative stress and promoting autophagy. Hum Mol Genet. 2015. Mar 15;24(6):1717–32. doi: 10.1093/hmg/ddu585. PubMed PMID: 25432533. [DOI] [PubMed] [Google Scholar]
- 3616.Hasima N, Ozpolat B.. Regulation of autophagy by polyphenolic compounds as a potential therapeutic strategy for cancer. Cell Death Dis. 2014;5:e1509. doi: 10.1038/cddis.2014.467. PubMed PMID: 25375374; PubMed Central PMCID: PMC4260725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3617.Song WH, Yi YJ, Sutovsky M, et al. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc Natl Acad Sci U S A. 2016. Sep 6;113(36):E5261–70. doi: 10.1073/pnas.1605844113. PubMed PMID: 27551072; PubMed Central PMCID: PMCPMC5018771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3618.Cai S, Geng S, Jin F, et al. POU5F1/Oct-4 expression in breast cancer tissue is significantly associated with non-sentinel lymph node metastasis. BMC Cancer. 2016. Mar 1;16:175. doi: 10.1186/s12885-015-1966-6. PubMed PMID: 26931354; PubMed Central PMCID: PMCPMC4774000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3619.Sharif T, Martell E, Dai C, et al. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy. 2017. Feb;13(2):264–284. doi: 10.1080/15548627.2016.1260808. PubMed PMID: 27929731; PubMed Central PMCID: PMCPMC5324853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3620.Pfister AS, Keil M, Kuhl M.. The Wnt target protein Peter Pan defines a novel p53-independent nucleolar stress-response pathway. J Biol Chem. 2015. Apr 24;290(17):10905–18. doi: 10.1074/jbc.M114.634246. PubMed PMID: 25759387; PubMed Central PMCID: PMCPMC4409253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3621.Laplante M, Sabatini DM.. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013. Apr 15;126(Pt 8):1713–9. doi: 10.1242/jcs.125773. PubMed PMID: 23641065; PubMed Central PMCID: PMC3678406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3622.Palomer X, Capdevila-Busquets E, Botteri G, et al. PPARbeta/delta attenuates palmitate-induced endoplasmic reticulum stress and induces autophagic markers in human cardiac cells. Int J Cardiol. 2014. Jun 1;174(1):110–8. doi: 10.1016/j.ijcard.2014.03.176. PubMed PMID: 24767130. [DOI] [PubMed] [Google Scholar]
- 3623.Pawson T, Nash P.. Protein-protein interactions define specificity in signal transduction. Genes dev 2000. May 1;14(9):1027–47. PubMed PMID: 10809663. [PubMed] [Google Scholar]
- 3624.Phizicky EM, Fields S.. Protein-protein interactions: methods for detection and analysis. Microbiol Rev 1995. Mar;59(1):94–123. PubMed PMID: 7708014; PubMed Central PMCID: PMC239356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3625.Safari-Alighiarloo N, Taghizadeh M, Rezaei-Tavirani M, et al. Protein-protein interaction networks (PPI) and complex diseases. Gastroenterol Hepatol Bed Bench. 2014. Mar;7(1):17–31. PubMed PMID: 25436094; PubMed Central PMCID: PMC4017556. [PMC free article] [PubMed] [Google Scholar]
- 3626.Le Guezennec X, Brichkina A, Huang YF, et al. Wip1-dependent regulation of autophagy, obesity, and atherosclerosis. Cell Metab. 2012. Jul 3;16(1):68–80. doi: 10.1016/j.cmet.2012.06.003. PubMed PMID: 22768840. [DOI] [PubMed] [Google Scholar]
- 3627.Chikh A, Sanza P, Raimondi C, et al. iASPP is a novel autophagy inhibitor in keratinocytes. J Cell Sci. 2014. Jul 15;127\(Pt 14):3079–93. doi: 10.1242/jcs.144816. PubMed PMID: 24777476. [DOI] [PubMed] [Google Scholar]
- 3628.Uddin MN, Ito S, Nishio N, et al. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem Biophys Res Commun. 2011. Mar 22;407:692–8. doi: 10.1016/j.bbrc.2011.03.077. PubMed PMID: 21439266; Eng. [DOI] [PubMed] [Google Scholar]
- 3629.Fujiwara N, Usui T, Ohama T, et al. Regulation of beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3). J Biol Chem. 2016. May 13;291(20):10858–66. doi: 10.1074/jbc.M115.704908. PubMed PMID: 26994142; PubMed Central PMCID: PMCPMC4865930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3630.Pala R, Alomari N, Nauli SM.. Primary cilium-dependent signaling mechanisms. Int J Mol Sci. 2017. Oct 28;18(11). doi: 10.3390/ijms18112272. PubMed PMID: 29143784; PubMed Central PMCID: PMCPMC5713242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3631.Reiter JF, Leroux MR.. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017. Sep;18(9):533–547. doi: 10.1038/nrm.2017.60. PubMed PMID: 28698599; PubMed Central PMCID: PMCPMC5851292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3632.Hasegawa J, Iwamoto R, Otomo T, et al. Autophagosome-lysosome fusion in neurons requires INPP5E, a protein associated with Joubert syndrome. EMBO J. 2016. Sep 1;35(17):1853–67. doi: 10.15252/embj.201593148. PubMed PMID: 27340123; PubMed Central PMCID: PMCPMC5007553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3633.Struchtrup A, Wiegering A, Stork B, et al. The ciliary protein RPGRIP1L governs autophagy independently of its proteasome-regulating function at the ciliary base in mouse embryonic fibroblasts. Autophagy. 2018;14(4):567–583. doi: 10.1080/15548627.2018.1429874. PubMed PMID: 29372668; PubMed Central PMCID: PMCPMC5959336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3634.Orhon I, Dupont N, Zaidan M, et al. Primary-cilium-dependent autophagy controls epithelial cell volume in response to fluid flow. Nat Cell Biol. 2016. Jun;18(6):657–67. doi: 10.1038/ncb3360. PubMed PMID: 27214279. [DOI] [PubMed] [Google Scholar]
- 3635.Jang J, Wang Y, Lalli MA, et al. Primary cilium-autophagy-Nrf2 (PAN) axis activation commits human embryonic stem cells to a neuroectoderm fate. Cell. 2016. Apr 7;165(2):410–20. doi: 10.1016/j.cell.2016.02.014. PubMed PMID: 27020754. [DOI] [PubMed] [Google Scholar]
- 3636.Tang Z, Lin MG, Stowe TR, et al. Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature. 2013. Oct 10;502(7470):254–7. doi: 10.1038/nature12606. PubMed PMID: 24089205; PubMed Central PMCID: PMCPMC4075283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3637.Wang S, Livingston MJ, Su Y, et al. Reciprocal regulation of cilia and autophagy via the MTOR and proteasome pathways. Autophagy. 2015. Apr 3;11(4):607–16. doi: 10.1080/15548627.2015.1023983. PubMed PMID: 25906314; PubMed Central PMCID: PMCPMC4502771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3638.Liu ZQ, Lee JN, Son M, et al. Ciliogenesis is reciprocally regulated by PPARA and NR1H4/FXR through controlling autophagy in vitro and in vivo. Autophagy. 2018;14(6):1011–1027. doi: 10.1080/15548627.2018.1448326. PubMed PMID: 29771182; PubMed Central PMCID: PMCPMC6103415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3639.Eisenberg-Lerner A, Kimchi A.. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ. 2012. May;19(5):788–97. doi: 10.1038/cdd.2011.149. PubMed PMID: 22095288; PubMed Central PMCID: PMC3321617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3640.Durcan TM, Tang MY, Perusse JR, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO J. 2014. Sep 12;ww:2473–91. doi: 10.15252/embj.201489729. PubMed PMID: 25216678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3641.Moravcevic K, Oxley CL, Lemmon MA.. Conditional peripheral membrane proteins: facing up to limited specificity. Structure. 2012. Jan 11;20(1):15–27. doi: 10.1016/j.str.2011.11.012. PubMed PMID: 22193136; PubMed Central PMCID: PMC3265387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3642.Baskaran S, Ragusa MJ, Boura E, et al. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012. Aug 10;47(3):339–48. doi: 10.1016/j.molcel.2012.05.027. PubMed PMID: 22704557; PubMed Central PMCID: PMC3595537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3643.Krick R, Busse RA, Scacioc A, et al. Structural and functional characterization of the two phosphoinositide binding sites of PROPPINs, a beta-propeller protein family. Proc Natl Acad Sci U S A. 2012. Jul 24;109(30):E2042–9. doi: 10.1073/pnas.1205128109. PubMed PMID: 22753491; PubMed Central PMCID: PMC3409749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3644.Watanabe Y, Kobayashi T, Yamamoto H, et al. Structure-based analyses reveal distinct binding sites for Atg2 and phosphoinositides in Atg18. J Biol Chem. 2012. Sep 14;287(38):31681–90. doi: 10.1074/jbc.M112.397570. PubMed PMID: 22851171; PubMed Central PMCID: PMC3442503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3645.Finley D, Prado MA.. The Proteasome and Its Network: Engineering for Adaptability. Cold Spring Harb Perspect Biol. 2020. Jan 2;12(1). doi: 10.1101/cshperspect.a033985. PubMed PMID: 30833452; PubMed Central PMCID: PMCPMC6829053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3646.Allavena G, Boyd C, Oo KS, et al. Suppressed translation and ULK1 degradation as potential mechanisms of autophagy limitation under prolonged starvation. Autophagy. 2016. Nov;12(11):2085–2097. doi: 10.1080/15548627.2016.1226733. PubMed PMID: 27629431; PubMed Central PMCID: PMCPMC5103336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3647.Dong Z, Cui H.. The autophagy-lysosomal pathways and their emerging roles in modulating proteostasis in tumors. Cells. 2018. Dec 20;8(1). doi: 10.3390/cells8010004. PubMed PMID: 30577555; PubMed Central PMCID: PMCPMC6356230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3648.Starokadomskyy P, Dmytruk KV.. A bird’s-eye view of autophagy. Autophagy. 2013. Jul;9(7):1121–6. doi: 10.4161/auto.24544. PubMed PMID: 23615436; PubMed Central PMCID: PMC3722328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3649.Neely KM, Green KN, Laferla FM.. Presenilin is necessary for efficient proteolysis through the autophagy-lysosome system in a gamma-secretase-independent manner. J Neurosci. 2011. Feb 23;31(8):2781–91. doi: 10.1523/JNEUROSCI.5156-10.2010. PubMed PMID: 21414900; PubMed Central PMCID: PMC3064964. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3650.Yin Z, Liu X, Ariosa A, et al. Psp2, a novel regulator of autophagy that promotes autophagy-related protein translation. Cell Res. 2019. Dec;29(12):994–1008. doi: 10.1038/s41422-019-0246-4. PubMed PMID: 31666677; PubMed Central PMCID: PMCPMC6951345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3651.Miller S, Tavshanjian B, Oleksy A, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010. Mar 26;327(5973):1638–42. doi: 10.1126/science.1184429. PubMed PMID: 20339072; PubMed Central PMCID: PMCPMC2860105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3652.Memisoglu G, Eapen VV, Yang Y, et al. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proc Natl Acad Sci U S A. 2019. Jan 29;116(5):1613–1620. doi: 10.1073/pnas.1817078116. PubMed PMID: 30655342; PubMed Central PMCID: PMCPMC6358665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3653.Conte A, Kisslinger A, Procaccini C, et al. Convergent Effects of Resveratrol and PYK2 on Prostate Cells. Int J Mol Sci. 2016. Sep 13;17(9). doi: 10.3390/ijms17091542. PubMed PMID: 27649143; PubMed Central PMCID: PMCPMC5037816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3654.Ueda H, Abbi S, Zheng C, et al. Suppression of Pyk2 kinase and cellular activities by FIP200. J Cell Biol. 2000. Apr 17;149(2):423–30. doi: 10.1083/jcb.149.2.423. PubMed PMID: 10769033; PubMed Central PMCID: PMCPMC2175150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3655.Walsh CT, Garneau-Tsodikova S, Gatto GJ, Jr.. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 2005. Dec 1;44(45):7342–72. doi: 10.1002/anie.200501023. PubMed PMID: 16267872. [DOI] [PubMed] [Google Scholar]
- 3656.Witze ES, Old WM, Resing KA, et al. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007. Oct;4(10):798–806. doi: 10.1038/nmeth1100. PubMed PMID: 17901869. [DOI] [PubMed] [Google Scholar]
- 3657.Popelka H, Klionsky DJ.. Posttranslationally-modified structures in the autophagy machinery: an integrative perspective. FEBS J. 2015. Jun 23;282:3474–88. doi: 10.1111/febs.13356. PubMed PMID: 26108642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3658.Huang YH, Al-Aidaroos AQ, Yuen HF, et al. A role of autophagy in PTP4A3-driven cancer progression. Autophagy. 2014. Oct 1;10(10):1787–800. doi: 10.4161/auto.29989. PubMed PMID: 25136802; PubMed Central PMCID: PMC4198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3659.Martin KR, Xu Y, Looyenga BD, et al. Identification of PTPsigma as an autophagic phosphatase. J Cell Sci. 2011. Mar 1;124(Pt 5):812–9. doi: 10.1242/jcs.080341. PubMed PMID: 21303930; PubMed Central PMCID: PMC3039021. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3660.Mandell MA, Jain A, Arko-Mensah J, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014. Aug 25;30(4):394–409. doi: 10.1016/j.devcel.2014.06.013. PubMed PMID: 25127057; PubMed Central PMCID: PMC4146662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3661.Andres S, Pevny S, Ziegenhagen R, et al. Safety aspects of the use of quercetin as a dietary supplement. Mol Nutr Food Res 2018. Jan;62(1). doi: 10.1002/mnfr.201700447. PubMed PMID: 29127724. [DOI] [PubMed] [Google Scholar]
- 3662.Russo M, Russo GL.. Autophagy inducers in cancer. Biochem Pharmacol. 2018. Jul;153:51–61. doi: 10.1016/j.bcp.2018.02.007. PubMed PMID: 29438677. [DOI] [PubMed] [Google Scholar]
- 3663.Zhu Y, Tchkonia T, Pirtskhalava T, et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging cell. 2015. Aug;14(4):644–58. doi: 10.1111/acel.12344. PubMed PMID: 25754370; PubMed Central PMCID: PMCPMC4531078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3664.Kang C, Elledge SJ.. How autophagy both activates and inhibits cellular senescence. Autophagy. 2016. May 3;12(5):898–9. doi: 10.1080/15548627.2015.1121361. PubMed PMID: 27129029; PubMed Central PMCID: PMCPMC4854549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3665.Fujita N, Huang W, Lin TH, et al. Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. eLife. 2017. Jan 7;6. doi: 10.7554/eLife.23367. PubMed PMID: 28063257; PubMed Central PMCID: PMCPMC5249261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3666.Lorincz P, Toth S, Benko P, et al. Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol. 2017. Jul 3;216(7):1937–1947. doi: 10.1083/jcb.201611027. PubMed PMID: 28483915; PubMed Central PMCID: PMCPMC5496615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3667.Ding X, Jiang X, Tian R, et al. RAB2 regulates the formation of autophagosome and autolysosome in mammalian cells. Autophagy. 2019. Oct;15(10):1774–1786. doi: 10.1080/15548627.2019.1596478. PubMed PMID: 30957628; PubMed Central PMCID: PMCPMC6735470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3668.Nagy G, Ward J, Mosser DD, et al. Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem. 2006. Nov 10;281(45):34574–91. doi: 10.1074/jbc.M606301200. PubMed PMID: 16935861. [DOI] [PubMed] [Google Scholar]
- 3669.Fernandez DR, Telarico T, Bonilla E, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Iimmunol. 2009. Feb 15;182(4):2063–73. doi: 10.4049/jimmunol.0803600. PubMed PMID: 19201859; PubMed Central PMCID: PMC2676112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3670.Caza TN, Fernandez DR, Talaber G, et al. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann Rheum Dis. 2014. Oct;73(10):1888–97. doi: 10.1136/annrheumdis-2013-203794. PubMed PMID: 23897774; PubMed Central PMCID: PMC4047212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3671.Talaber G, Miklossy G, Oaks Z, et al. HRES-1/Rab4 promotes the formation of LC3(+) auto-phagosomes and the accumulation of mitochondria during autophagy. PLoS One. 2014;9(1):e84392. doi: 10.1371/journal.pone.0084392. PubMed PMID: 24404161; PubMed Central PMCID: PMC3880286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3672.Weidberg H, Shvets E, Elazar Z.. Biogenesis and cargo selectivity of auto-phagosomes. Annual Rev Biochem. 2011;80:125–156. PubMed PMID: 21548784. [DOI] [PubMed] [Google Scholar]
- 3673.Hammerling BC, Najor RH, Cortez MQ, et al. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat Commun. 2017. Jan 30;8:14050. doi: 10.1038/ncomms14050. PubMed PMID: 28134239; PubMed Central PMCID: PMCPMC5290275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3674.Ayala CI, Kim J, Neufeld TP.. Rab6 promotes insulin receptor and cathepsin trafficking to regulate autophagy induction and activity in Drosophila. J Cell Sci. 2018. Sep 7;131(17). doi: 10.1242/jcs.216127. PubMed PMID: 30111579; PubMed Central PMCID: PMCPMC6140324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3675.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009. Aug;10(8):513–525. PubMed PMID: 19603039; eng. [DOI] [PubMed] [Google Scholar]
- 3676.Kuchitsu Y, Homma Y, Fujita N, et al. Rab7 knockout unveils regulated autolysosome maturation induced by glutamine starvation. J Cell Sci. 2018. Apr 6;131(7). doi: 10.1242/jcs.215442. PubMed PMID: 29514857. [DOI] [PubMed] [Google Scholar]
- 3677.Keeling E, DS Chatelet, DA Johnston, et al. Oxidative stress and dysfunctional intracellular traffic linked to an unhealthy diet results in impaired cargo transport in the retinal pigment epithelium (RPE). Mol Nutr Food Res. 2019. Aug;63(15):e1800951. doi: 10.1002/mnfr.201800951. PubMed PMID: 30835933. [DOI] [PubMed] [Google Scholar]
- 3678.Rink J, Ghigo E, Kalaidzidis Y, et al. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005. Sep 9;122(5):735–749. doi: 10.1016/j.cell.2005.06.043. PubMed PMID: 16143105; eng. [DOI] [PubMed] [Google Scholar]
- 3679.Pilli M, Arko-Mensah J, Ponpuak M, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012. Aug 24;37(2):223–234. PubMed PMID: 22921120; PubMed Central PMCID: PMC3428731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3680.Honda S, Arakawa S, Nishida Y, et al. Ulk1-mediated Atg5-independent macroautophagy mediates elimination of mitochondria from embryonic reticulocytes. Nat Commun. 2014. Jun 4;5:4004. PubMed PMID: 24895007. [DOI] [PubMed] [Google Scholar]
- 3681.Saito T, Nah J, Oka SI, et al. An alternative mitophagy pathway mediated by Rab9 protects the heart against ischemia. J Clin Invest. 2019. Feb 1;129(2):802–819. doi: 10.1172/JCI122035. PubMed PMID: 30511961; PubMed Central PMCID: PMCPMC6355232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3682.Nozawa T, Aikawa C, Goda A, et al. The small GTPases Rab9A and Rab23 function at distinct steps in autophagy during Group A Streptococcus infection. Cell Microbiol. 2012. Aug;14(8):1149–1165. doi: 10.1111/j.1462-5822.2012.01792.x. PubMed PMID: 22452336. [DOI] [PubMed] [Google Scholar]
- 3683.Wauters F, Cornelissen T, Imberechts D, et al. LRRK2 mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy. 2020. Feb;16(2):203–222. doi: 10.1080/15548627.2019.1603548. PubMed PMID: 30945962; PubMed Central PMCID: PMCPMC6984591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3684.Kelly EE, Horgan CP, McCaffrey MW.. Rab11 proteins in health and disease. Biochem Soc Trans. 2012. Dec 1;40(6):1360–1367. PubMed PMID: 23176481. [DOI] [PubMed] [Google Scholar]
- 3685.Longatti A, Lamb CA, Razi M, et al. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol. 2012. May 28;197(5):659–675. doi: 10.1083/jcb.201111079. PubMed PMID: 22613832; PubMed Central PMCID: PMC3365497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3686.Fader CM, Sanchez D, Furlan M, et al. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008. Feb;9(2):230–250. doi: 10.1111/j.1600-0854.2007.00677.x. PubMed PMID: 17999726; eng. [DOI] [PubMed] [Google Scholar]
- 3687.Szatmari Z, Kis V, Lippai M, et al. Rab11 facilitates cross-talk between autophagy and endosomal pathway through regulation of Hook localization. Molecular Biol Cell. 2014. Feb;25(4):522–531. doi: 10.1091/mbc.E13-10-0574. PubMed PMID: 24356450; PubMed Central PMCID: PMCPMC3923643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3688.Matsui T, Fukuda M. Rab12 regulates mTORC1 activity and autophagy through controlling the degradation of amino-acid transporter PAT4. EMBO Rep. 2013. May;14(5):450–457. PubMed PMID: 23478338; PubMed Central PMCID: PMC3642374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3689.Zhang L, Dai F, Cui L, et al. Up-regulation of the active form of small GTPase Rab13 promotes macroautophagy in vascular endothelial cells. Biochim Biophys Acta Mol Cell Res. 2017. Apr;1864(4):613–624. doi: 10.1016/j.bbamcr.2017.01.003. PubMed PMID: 28087344. [DOI] [PubMed] [Google Scholar]
- 3690.Jean S, Cox S, Nassari S, et al. Starvation-induced MTMR13 and RAB21 activity regulates VAMP8 to promote autophagosome-lysosome fusion. EMBO Rep. 2015. Mar;16(3):297–311. PubMed PMID: 25648148; PubMed Central PMCID: PMC4364869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3691.Munafo DB, Colombo MI.. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic. 2002. Jul;3(7):472–482. PubMed PMID: 12047555; eng. [DOI] [PubMed] [Google Scholar]
- 3692.Yla-Anttila P, Mikkonen E, Otteby KE, et al. RAB24 facilitates clearance of autophagic compartments during basal conditions. Autophagy. 2015. Sep 1;11:1833–1848. doi: 10.1080/15548627.2015.1086522. PubMed PMID: 26325487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3693.Binotti B, Pavlos NJ, Riedel D, et al. The GTPase Rab26 links synaptic vesicles to the autophagy pathway. eLife. 2015. Feb 2;4:e05597. doi: 10.7554/eLife.05597. PubMed PMID: 25643395; PubMed Central PMCID: PMCPMC4337689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3694.Hirota Y, Tanaka Y.. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions [Research Support, Non-U.S. Gov’t]. Cell Mol Life Sci. 2009. Sep;66(17):2913–2932. PubMed PMID: 19593531; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3695.Itoh T, Fujita N, Kanno E, et al. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008. Jul;19(7):2916–2925. PubMed PMID: 18448665; PubMed Central PMCID: PMC2441679. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3696.Itoh T, Kanno E, Uemura T, et al. OATL1, a novel autophagosome-resident Rab33B-GAP, regulates autophagosomal maturation [Research Support, Non-U.S. Gov’t]. J Cell Biol. 2011. Mar 7;192(5):839–853. doi: 10.1083/jcb.201008107. PubMed PMID: 21383079; PubMed Central PMCID: PMC3051816. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3697.Song Y, Shang D, Cheng H, et al. The small GTPase RAB37 functions as an organizer for autophagosome biogenesis. Autophagy. 2018;14(4):727–729. doi: 10.1080/15548627.2018.1434374. PubMed PMID: 29388490; PubMed Central PMCID: PMCPMC5959331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3698.Sheng Y, Song Y, Li Z, et al. RAB37 interacts directly with ATG5 and promotes autophagosome formation via regulating ATG5-12-16 complex assembly. Cell Death Differ. 2018. May;25(5):918–934. doi: 10.1038/s41418-017-0023-1. PubMed PMID: 29229996; PubMed Central PMCID: PMCPMC5943352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3699.Ye X, Zhou XJ, Zhang H.. Exploring the role of autophagy-related gene 5 (ATG5) yields important insights into autophagy in autoimmune/autoinflammatory diseases. Front Immunol. 2018;9:2334. PubMed PMID: 30386331; PubMed Central PMCID: PMCPMC6199349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3700.Chen XW, Leto D, Xiong T, et al. A Ral GAP complex links PI 3-kinase/Akt signaling to RalA activation in insulin action. Mol Biol Cell. 2011. Jan 1;22(1):141–152. PubMed PMID: 21148297; PubMed Central PMCID: PMC3016972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3701.Gentry LR, Martin TD, Reiner DJ, et al. Ral small GTPase signaling and oncogenesis: more than just 15minutes of fame. Biochim Biophys Acta. 2014. Dec;1843(12):2976–2988. PubMed PMID: 25219551; PubMed Central PMCID: PMC4201770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3702.Martin TD, Chen XW, Kaplan RE, et al. Ral and Rheb GTPase activating proteins integrate mTOR and GTPase signaling in aging, autophagy, and tumor cell invasion. Mol Cell. 2014. Jan 23;53(2):209–220. PubMed PMID: 24389102; PubMed Central PMCID: PMC3955741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3703.Geng J, Nair U, Yasumura-Yorimitsu K, et al. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2010. Jul 1;21(13):2257–2269. doi: 10.1091/mbc.E09-11-0969. PubMed PMID: 20444978; PubMed Central PMCID: PMC2893989. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3704.Shirakawa R, Fukai S, Kawato M, et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J Biol Chem. 2009. Aug 7;284(32):21580–21588. doi: 10.1074/jbc.M109.012112. PubMed PMID: 19520869; PubMed Central PMCID: PMC2755882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3705.Oeckinghaus A, Postler TS, Rao P, et al. kappaB-Ras proteins regulate both NF-kappaB-dependent inflammation and Ral-dependent proliferation. Cell Rep. 2014. Sep 25;8(6):1793–1807. PubMed PMID: 25220458; PubMed Central PMCID: PMC4177457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3706.Mestre MB, Colombo MI.. cAMP and EPAC are key players in the regulation of the signal transduction pathway involved in the alpha-hemolysin autophagic response. PLoS Pathog. 2012;8(5):e1002664. PubMed PMID: 22654658; PubMed Central PMCID: PMCPMC3359991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3707.Mestre MB, Colombo MI.. Staphylococcus aureus promotes autophagy by decreasing intracellular cAMP levels. Autophagy. 2012. Dec;8(12):1865–1867. doi: 10.4161/auto.22161. PubMed PMID: 23047465; PubMed Central PMCID: PMCPMC3541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3708.Laurent AC, Bisserier M, Lucas A, et al. Exchange protein directly activated by cAMP 1 promotes autophagy during cardiomyocyte hypertrophy. Cardiovasc Res. 2015. Jan 1;105(1):55–64. doi: 10.1093/cvr/cvu242. PubMed PMID: 25411381. [DOI] [PubMed] [Google Scholar]
- 3709.Chu KY, O’Reilly L, Mellet N, et al. Oleate disrupts cAMP signaling, contributing to potent stimulation of pancreatic beta-cell autophagy. J Biol Chem. 2019. Jan 25;294(4):1218–1229. doi: 10.1074/jbc.RA118.004833. PubMed PMID: 30518550; PubMed Central PMCID: PMCPMC6349129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3710.Li W, Yue F, Dai Y, et al. Suppressor of hepatocellular carcinoma RASSF1A activates autophagy initiation and maturation. Cell Death Differ. 2019. Aug;26(8):1379–1395. doi: 10.1038/s41418-018-0211-7. PubMed PMID: 30315205; PubMed Central PMCID: PMCPMC6748129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3711.Thakur A, Mikkelsen H, Jungersen G.. Intracellular Pathogens: host Immunity and Microbial Persistence Strategies. J Immunol Res. 2019;2019:1356540. doi: 10.1155/2019/1356540. PubMed PMID: 31111075; PubMed Central PMCID: PMCPMC6487120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3712.Horenkamp FA, Kauffman KJ, Kohler LJ, et al. The legionella anti-autophagy effector RavZ targets the autophagosome via PI3P- and curvature-sensing motifs. Dev Cell. 2015. Sep 14;34(5):569–576. doi: 10.1016/j.devcel.2015.08.010. PubMed PMID: 26343456; PubMed Central PMCID: PMCPMC4594837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3713.Vargas JNS, Wang C, Bunker E, et al. Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell. 2019. Apr 18;74(2):347–362e6. doi: 10.1016/j.molcel.2019.02.010. PubMed PMID: 30853401; PubMed Central PMCID: PMCPMC6642318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3714.Ravenhill BJ, Boyle KB, von Muhlinen N, et al. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol Cell. 2019. Apr 18;74(2):320–329e6. doi: 10.1016/j.molcel.2019.01.041. PubMed PMID: 30853402; PubMed Central PMCID: PMCPMC6477152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3715.Punnonen EL, Reunanen H, Hirsimaki P, et al. Filipin labelling and intramembrane particles on the membranes of early and later autophagic vacuoles in Ehrlich ascites cells. Virchows Archiv B Cell Pathol Mol Pathol. 1988;54(5):317–326. PubMed PMID: 2451345; eng. [DOI] [PubMed] [Google Scholar]
- 3716.Huang XC, Inoue-Aono Y, Moriyasu Y, et al. Plant cell wall-penetrable, redox-responsive silica nanoprobe for the imaging of starvation-induced vesicle trafficking. Anal Chem. 2016. Oct 18;88(20):10231–10236. doi: 10.1021/acs.analchem.6b02920. PubMed PMID: 27673337. [DOI] [PubMed] [Google Scholar]
- 3717.Opipari AJ, Tan L, Boitano AE, Sorenson DR, Aurora A, Liu JR.. Resveratrol-induced autophagocytosis in ovarian cancer cells. Cancer Res. 2004;15:696–703. [DOI] [PubMed] [Google Scholar]
- 3718.Klionsky DJ, Cuervo AM, Dunn WA, Jr., et al. How shall I eat thee?. Autophagy. 2007. Sep-Oct;3(5):413–416. PubMed PMID: 17568180; eng. [DOI] [PubMed] [Google Scholar]
- 3719.Ogier-Denis E, Petiot A, Bauvy C, et al. Control of the expression and activity of the Galpha-interacting protein (GAIP) in human intestinal cells. J Biol Chem. 1997. Sep 26;272(39):24599–24603. PubMed PMID: 9305927; eng. [DOI] [PubMed] [Google Scholar]
- 3720.Fransson S, Ruusala A, Aspenstrom P.. The atypical Rho GTPases Miro-1 and Miro-2 have essential roles in mitochondrial trafficking. Biochem Biophys Res Commun. 2006. Jun 2;344(2):500–510. doi: 10.1016/j.bbrc.2006.03.163. PubMed PMID: 16630562. [DOI] [PubMed] [Google Scholar]
- 3721.Lopez-Domenech G, Covill-Cooke C, Ivankovic D, et al. Miro proteins coordinate microtubule- and actin-dependent mitochondrial transport and distribution. Embo J. 2018. Feb 1;37(3):321–336. PubMed PMID: 29311115; PubMed Central PMCID: PMCPMC5793800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3722.Wang X, Winter D, Ashrafi G, et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell. 2011. Nov 11;147(4):893–906. doi: 10.1016/j.cell.2011.10.018. PubMed PMID: 22078885; PubMed Central PMCID: PMCPMC3261796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3723.Birsa N, Norkett R, Wauer T, et al. Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014. May 23;289(21):14569–14582. doi: 10.1074/jbc.M114.563031. PubMed PMID: 24671417; PubMed Central PMCID: PMCPMC4031514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3724.Safiulina D, Kuum M, Choubey V, et al. Miro proteins prime mitochondria for Parkin translocation and mitophagy. Embo J. 2019. Jan 15;38(2). doi: 10.15252/embj.201899384. PubMed PMID: 30504269; PubMed Central PMCID: PMCPMC6331716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3725.Khobrekar NV, Quintremil S, Dantas TJ, et al. The dynein adaptor RILP controls neuronal autophagosome biogenesis, transport, and clearance. Dev Cell. 2020. Apr 20;53(2):141–153e4. doi: 10.1016/j.devcel.2020.03.011. PubMed PMID: 32275887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3726.Yorimitsu T, Zaman S, Broach JR, et al. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007. Oct;18(10):4180–4189. Doi: 10.1091/mbc.E07-05-0485. PubMed PMID: 17699586; PubMed Central PMCID: PMC1995722. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3727.Yonekawa T, Gamez G, Kim J, et al. RIP1 negatively regulates basal autophagic flux through TFEB to control sensitivity to apoptosis. EMBO Rep. 2015. Jun;16(6):700–708. PubMed PMID: 25908842; PubMed Central PMCID: PMC4467854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3728.Hillwig MS, Contento AL, Meyer A, et al. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci U S A. 2011. Jan 18;108(3):1093–1098. PubMed PMID: 21199950; PubMed Central PMCID: PMC3024651. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3729.Haud N, Kara F, Diekmann S, et al. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc Natl Acad Sci U S A. 2011. Jan 18;108(3):1099–1103. doi: 10.1073/pnas.1009811107. PubMed PMID: 21199949; PubMed Central PMCID: PMC3024650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3730.Liu Y, Zou W, Yang P, et al. Autophagy-dependent ribosomal RNA degradation is essential for maintaining nucleotide homeostasis during C. elegans development. eLife. 2018. Aug 13;7. doi: 10.7554/eLife.36588. PubMed PMID: 30102152; PubMed Central PMCID: PMCPMC6101943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3731.Floyd BE, Mugume Y, Morriss SC, et al. Localization of RNS2 ribonuclease to the vacuole is required for its role in cellular homeostasis. Planta. 2017. Apr;245(4):779–792. doi: 10.1007/s00425-016-2644-x. PubMed PMID: 28025674. [DOI] [PubMed] [Google Scholar]
- 3732.Xu C, Feng K, Zhao X, et al. Regulation of autophagy by E3 ubiquitin ligase RNF216 through BECN1 ubiquitination. Autophagy. 2014. Nov 11;10:2239–2250. PubMed PMID: 25484083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3733.Gurkar AU, Chu K, Raj L, et al. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun. 2013;4:2189. doi: 10.1038/ncomms3189. PubMed PMID: 23877263; PubMed Central PMCID: PMCPMC3740589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3734.Shi J, Surma M, Yang Y, et al. Disruption of both ROCK1 and ROCK2 genes in cardiomyocytes promotes autophagy and reduces cardiac fibrosis during aging. Faseb J. 2019. Jun;33(6):7348–7362. PubMed PMID: 30848941; PubMed Central PMCID: PMCPMC6529334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3735.Shi J, Surma M, Wei L.. Disruption of ROCK1 gene restores autophagic flux and mitigates doxorubicin-induced cardiotoxicity. Oncotarget. 2018. Feb 27;9(16):12995–13008. PubMed PMID: 29560126; PubMed Central PMCID: PMCPMC5849190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3736.Hac A, Domachowska A, Narajczyk M, et al. S6K1 controls autophagosome maturation in autophagy induced by sulforaphane or serum deprivation. Eur J Cell Biol. 2015. Oct;94(10):470–481. doi: 10.1016/j.ejcb.2015.05.001. PubMed PMID: 26054233. [DOI] [PubMed] [Google Scholar]
- 3737.Xu X, Sun J, Song R, et al. Inhibition of p70 S6 kinase (S6K1) activity by A77 1726, the active metabolite of leflunomide, induces autophagy through TAK1-mediated AMPK and JNK activation. Oncotarget. 2017. May 2;8(18):30438–30454. doi: 10.18632/oncotarget.16737. PubMed PMID: 28389629; PubMed Central PMCID: PMCPMC5444754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3738.Sun J, Mu Y, Jiang Y, et al. Inhibition of p70 S6 kinase activity by A77 1726 induces autophagy and enhances the degradation of superoxide dismutase 1 (SOD1) protein aggregates. Cell Death Dis. 2018. Mar 14;9(3):407. PubMed PMID: 29540819; PubMed Central PMCID: PMCPMC5851998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3739.Selman C, Tullet JM, Wieser D, et al. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science. 2009. Oct 2;326(5949):140–144. doi: 10.1126/science.1177221. PubMed PMID: 19797661; PubMed Central PMCID: PMCPMC4954603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3740.Aguilar V, Alliouachene S, Sotiropoulos A, et al. S6 kinase deletion suppresses muscle growth adaptations to nutrient availability by activating AMP kinase. Cell Metab. 2007. Jun;5(6):476–487. doi: 10.1016/j.cmet.2007.05.006. PubMed PMID: 17550782. [DOI] [PubMed] [Google Scholar]
- 3741.Kim E, Goraksha-Hicks P, Li L, et al. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008. Aug;10(8):935–945. PubMed PMID: 18604198; PubMed Central PMCID: PMC2711503. Eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3742.White E. Exploiting the bad eating habits of Ras-driven cancers. Genes Dev. 2013. Oct 1;27(19):2065–2071. doi: 10.1101/gad.228122.113. PubMed PMID: 24115766; PubMed Central PMCID: PMC3850091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3743.Cheng X, Ma X, Zhu Q, et al. Pacer is a mediator of mTORC1 and GSK3-TIP60 signaling in regulation of autophagosome maturation and lipid metabolism. Mol Cell. 2019. Feb 21;73(4):788–802e7. PubMed PMID: 30704899. [DOI] [PubMed] [Google Scholar]
- 3744.Beltran S, Nassif M, Vicencio E, et al. Network approach identifies Pacer as an autophagy protein involved in ALS pathogenesis. Mol Neurodegener. 2019. Mar 27;14(1):14. doi: 10.1186/s13024-019-0313-9. PubMed PMID: 30917850; PubMed Central PMCID: PMCPMC6437924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3745.Tandon M, Othman AH, Ashok V, et al. The role of Runx2 in facilitating autophagy in metastatic breast cancer cells. J Cell Physiol. 2018. Jan;233(1):559–571. PubMed PMID: 28345763. [DOI] [PubMed] [Google Scholar]
- 3746.Vervliet T, Pintelon I, Welkenhuyzen K, et al. Basal ryanodine receptor activity suppresses autophagic flux. Biochem Pharmacol. 2017. May 15;132:133–142. doi: 10.1016/j.bcp.2017.03.011. PubMed PMID: 28322744. [DOI] [PubMed] [Google Scholar]
- 3747.Shao Y, Gao Z, Marks PA, et al. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2004. Dec 28;101(52):18030–18035. PubMed PMID: 15596714; PubMed Central PMCID: PMC539807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3748.Stankov MV, El Khatib M, Kumar Thakur B, et al. Histone deacetylase inhibitors induce apoptosis in myeloid leukemia by suppressing autophagy. Leukemia. 2014. Mar;28(3):577–588. PubMed PMID: 24080946; PubMed Central PMCID: PMC3947652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3749.Frohlich LF, Mrakovcic M, Smole C, et al. Molecular mechanism leading to SAHA-induced autophagy in tumor cells: evidence for a p53-dependent pathway. Cancer Cell Int. 2016;16(1):68. doi: 10.1186/s12935-016-0343-0. PubMed PMID: 27601937; PubMed Central PMCID: PMCPMC5011867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3750.Galadari S, Rahman A, Pallichankandy S, et al. Molecular targets and anticancer potential of sanguinarine-a benzophenanthridine alkaloid. Phytomedicine. 2017. Oct 15;34:143–153. PubMed PMID: 28899497. [DOI] [PubMed] [Google Scholar]
- 3751.Pallichankandy S, Rahman A, Thayyullathil F, et al. ROS-dependent activation of autophagy is a critical mechanism for the induction of anti-glioma effect of sanguinarine. Free Radic Biol Med. 2015. Dec;89:708–720. PubMed PMID: 26472194. [DOI] [PubMed] [Google Scholar]
- 3752.Ni C, Narzt MS, Nagelreiter IM, et al. Autophagy deficient melanocytes display a senescence associated secretory phenotype that includes oxidized lipid mediators. Int J Biochem Cell Biol. 2016. Dec;81(Pt B):375–382. PubMed PMID: 27732890. [DOI] [PubMed] [Google Scholar]
- 3753.Dokudovskaya S, Waharte F, Schlessinger A, et al. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol Cell Proteomics. 2011. Jun;10(6):M110006478. doi: 10.1074/mcp.M110.006478. PubMed PMID: 21454883; PubMed Central PMCID: PMC3108837. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3754.Tabecka-Lonczynska A, Mytych J, Solek P, et al. Autophagy as a consequence of seasonal functions of testis and epididymis in adult male European bison (Bison bonasus, Linnaeus 1758). Cell Tissue Res. 2020. Mar;379(3):613–624. doi: 10.1007/s00441-019-03111-w. PubMed PMID: 31705214. [DOI] [PubMed] [Google Scholar]
- 3755.Nair U, Jotwani A, Geng J, et al. SNARE proteins are required for macroautophagy. Cell. 2011. Jul 22;146(2):290–302. doi: 10.1016/j.cell.2011.06.022. PubMed PMID: 21784249; PubMed Central PMCID: PMC3143362. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3756.Ishihara N, Hamasaki M, Yokota S, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001. Nov;12(11):3690–3702. PubMed PMID: 11694599; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3757.Loi M, Raimondi A, Morone D, et al. ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat Commun. 2019. Nov 7;10(1):5058. doi: 10.1038/s41467-019-12991-z. PubMed PMID: 31699981; PubMed Central PMCID: PMCPMC6838186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3758.Jiang S, Dupont N, Castillo EF, et al. Secretory versus degradative autophagy: unconventional secretion of inflammatory mediators. J Innate Immun. 2013;5(5):471–479. PubMed PMID: 23445716; PubMed Central PMCID: PMC3723810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3759.Marino G, Fernandez AF, Cabrera S, et al. Autophagy is essential for mouse sense of balance [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t]. J Clin Invest. 2010. Jul 1;120(7):2331–2344. PubMed PMID: 20577052; PubMed Central PMCID: PMC2898610. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3760.Cabrera S, Fernandez AF, Marino G, et al. ATG4B/autophagin-1 regulates intestinal homeostasis and protects mice from experimental colitis. Autophagy. 2013. Aug;9(8):1188–1200. doi: 10.4161/auto.24797. PubMed PMID: 23782979; PubMed Central PMCID: PMCPMC3748191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3761.Cabrera S, Marino G, Fernandez AF, et al. Autophagy, proteases and the sense of balance. Autophagy. 2010. Oct;6(7):961–963. doi: 10.4161/auto.6.7.13065. PubMed PMID: 20724821; eng. [DOI] [PubMed] [Google Scholar]
- 3762.Bel S, Hooper LV.. Secretory autophagy of lysozyme in Paneth cells. Autophagy. 2018;14(4):719–721. doi: 10.1080/15548627.2018.1430462. PubMed PMID: 29388875; PubMed Central PMCID: PMCPMC5959324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3763.Bel S, Pendse M, Wang Y, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017. Sep 8;357(6355):1047–1052. doi: 10.1126/science.aal4677. PubMed PMID: 28751470; PubMed Central PMCID: PMCPMC5702267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3764.Li J, Qi W, Chen G, et al. Mitochondrial outer-membrane E3 ligase MUL1 ubiquitinates ULK1 and regulates selenite-induced mitophagy. Autophagy. 2015;11(8):1216–1229. doi: 10.1080/15548627.2015.1017180. PubMed PMID: 26018823; PubMed Central PMCID: PMCPMC4590677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3765.Gewirtz DA. Autophagy, senescence and tumor dormancy in cancer therapy. Autophagy. 2009. Nov;5(8):1232–1234. doi: 10.4161/auto.5.8.9896. PubMed PMID: 19770583. [DOI] [PubMed] [Google Scholar]
- 3766.Liu K, Guo C, Lao Y, et al. A fine-tuning mechanism underlying self-control for autophagy: deSUMOylation of BECN1 by SENP3. Autophagy. 2019. Aug 2:1–16. doi: 10.1080/15548627.2019.1647944. PubMed PMID: 31373534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3767.Mostowy S, Bonazzi M, Hamon MA, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010. Nov 18;8(5):433–444. PubMed PMID: 21075354; eng. [DOI] [PubMed] [Google Scholar]
- 3768.Barve G, Sanyal P, Manjithaya R.. Septin localization and function during autophagy. Curr Genet. 2018. Oct;64(5):1037–1041. doi: 10.1007/s00294-018-0834-8. PubMed PMID: 29651536. [DOI] [PubMed] [Google Scholar]
- 3769.Hidvegi T, Ewing M, Hale P, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010. Jul 9;329(5988):229–232. PubMed PMID: 20522742. [DOI] [PubMed] [Google Scholar]
- 3770.Lee JH, Budanov AV, Karin M.. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013. Dec 3;18(6):792–801. doi: 10.1016/j.cmet.2013.08.018. PubMed PMID: 24055102; PubMed Central PMCID: PMC3858445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3771.Ho A, Cho CS, Namkoong S, et al. Biochemical basis of sestrin physiological activities. Trends Biochem Sci. 2016. Jul;41(7):621–632. doi: 10.1016/j.tibs.2016.04.005. PubMed PMID: 27174209; PubMed Central PMCID: PMCPMC4930368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3772.Kumar A, Shaha C.. SESN2 facilitates mitophagy by helping Parkin translocation through ULK1 mediated Beclin1 phosphorylation. Sci Rep. 2018. Jan 12;8(1):615. doi: 10.1038/s41598-017-19102-2. PubMed PMID: 29330382; PubMed Central PMCID: PMCPMC5766514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3773.Kim MJ, Bae SH, Ryu JC, et al. SESN2/sestrin2 suppresses sepsis by inducing mitophagy and inhibiting NLRP3 activation in macrophages. Autophagy. 2016. Aug 2;12(8):1272–91. doi: 10.1080/15548627.2016.1183081. PubMed PMID: 27337507; PubMed Central PMCID: PMCPMC4968237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3774.Budanov AV, Karin M.. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008. Aug 8;134(3):451–60. doi: 10.1016/j.cell.2008.06.028. PubMed PMID: 18692468; PubMed Central PMCID: PMC2758522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3775.Park HW, Park H, Ro SH, et al. Hepatoprotective role of Sestrin2 against chronic ER stress. Nat Commun. 2014;5:4233. doi: 10.1038/ncomms5233. PubMed PMID: 24947615; PubMed Central PMCID: PMC4074707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3776.Wolfson RL, Chantranupong L, Saxton RA, et al. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016. Jan 1;351(6268):43–8. doi: 10.1126/science.aab2674. PubMed PMID: 26449471; PubMed Central PMCID: PMCPMC4698017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3777.Ben-Sahra I, Dirat B, Laurent K, et al. Sestrin2 integrates Akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ. 2013. Apr;20(4):611–9. doi: 10.1038/cdd.2012.157. PubMed PMID: 23238567; PubMed Central PMCID: PMC3595485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3778.Ouchida AT, Uyemura VT, Queiroz AL, et al. SET protein accumulation prevents cell death in head and neck squamous cell carcinoma through regulation of redox state and autophagy. Biochim Biophys Acta Mol Cell Res. 2019. Apr;1866(4):623–637. doi: 10.1016/j.bbamcr.2019.01.005. PubMed PMID: 30658075. [DOI] [PubMed] [Google Scholar]
- 3779.Wang Y, Zheng X, Yu B, et al. Disruption of microtubules in plants suppresses macroautophagy and triggers starch excess-associated chloroplast autophagy. Autophagy. 2015;11(12):2259–74. doi: 10.1080/15548627.2015.1113365. PubMed PMID: 26566764; PubMed Central PMCID: PMCPMC4835195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3780.Kim YM, Stone M, Hwang TH, et al. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol Cell. 2012. Jun 29;46(6):833–46. doi: 10.1016/j.molcel.2012.04.007. PubMed PMID: 22575674; PubMed Central PMCID: PMCPMC3389276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3781.Antas P, Novellasdemunt L, Kucharska A, et al. SH3BP4 regulates intestinal stem cells and tumorigenesis by modulating beta-catenin nuclear localization. Cell Rep. 2019. Feb 26;26(9):2266–2273 e4. doi: 10.1016/j.celrep.2019.01.110. PubMed PMID: 30811977; PubMed Central PMCID: PMCPMC6391711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3782.Takahashi Y, Coppola D, Matsushita N, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007. Oct;9(10):1142–51. doi: 10.1038/ncb1634. PubMed PMID: 17891140; PubMed Central PMCID: PMC2254521. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3783.Zhang C, Li A, Zhang X, et al. A novel TIP30 protein complex regulates EGF receptor signaling and endocytic degradation. J Biol Chem. 2011. Mar 18;286(11):9373–81. doi: 10.1074/jbc.M110.207720. PubMed PMID: 21252234; PubMed Central PMCID: PMC3058969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3784.Khan MM, Strack S, Wild F, et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 2014. Jan;10(1):123–36. doi: 10.4161/auto.26841. PubMed PMID: 24220501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3785.Zhang P, Holowatyj AN, Ulrich CM, et al. Tumor suppressive autophagy in intestinal stem cells controls gut homeostasis. Autophagy. 2019. Sep;15(9):1668–1670. doi: 10.1080/15548627.2019.1633863. PubMed PMID: 31213134; PubMed Central PMCID: PMCPMC6693466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3786.Onnis A, Cianfanelli V, Cassioli C, et al. The pro-oxidant adaptor p66SHC promotes B cell mitophagy by disrupting mitochondrial integrity and recruiting LC3-II. Autophagy. 2018;14(12):2117–2138. doi: 10.1080/15548627.2018.1505153. PubMed PMID: 30109811; PubMed Central PMCID: PMCPMC6984773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3787.Vion AC, Kheloufi M, Hammoutene A, et al. Autophagy is required for endothelial cell alignment and atheroprotection under physiological blood flow. Proc Natl Acad Sci U S A. 2017. Oct 10;114(41):E8675–E8684. doi: 10.1073/pnas.1702223114. PubMed PMID: 28973855; PubMed Central PMCID: PMCPMC5642679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3788.Belaid A, Ndiaye PD, Klionsky DJ, et al. Signalphagy: Scheduled signal termination by macroautophagy. Autophagy. 2013. Aug 13;9(10):1629–30. PubMed PMID: 24004837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3789.Lee IH, Cao L, Mostoslavsky R, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008. Mar 4;105(9):3374–9. doi: 10.1073/pnas.0712145105. PubMed PMID: 18296641; PubMed Central PMCID: PMC2265142. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3790.Webster BR, Scott I, Traba J, et al. Regulation of autophagy and mitophagy by nutrient availability and acetylation. Biochim Biophys Acta. 2014. Apr 4;1841(4):525–34. doi: 10.1016/j.bbalip.2014.02.001. PubMed PMID: 24525425; PubMed Central PMCID: PMC3969632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3791.Pi H, Xu S, Reiter RJ, et al. SIRT3-SOD2-mROS-dependent autophagy in cadmium-induced hepatotoxicity and salvage by melatonin. Autophagy. 2015. Jul 3;11(7):1037–51. doi: 10.1080/15548627.2015.1052208. PubMed PMID: 26120888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3792.Polletta L, Vernucci E, Carnevale I, et al. SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy. 2015;11(2):253–70. doi: 10.1080/15548627.2015.1009778. PubMed PMID: 25700560; PubMed Central PMCID: PMC4502726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3793.Takasaka N, Araya J, Hara H, et al. Autophagy induction by SIRT6 through attenuation of insulin-like growth factor signaling is involved in the regulation of human bronchial epithelial cell senescence. J Iimmunol. 2014. Feb 1;192(3):958–68. doi: 10.4049/jimmunol.1302341. PubMed PMID: 24367027. [DOI] [PubMed] [Google Scholar]
- 3794.Birmingham CL, Canadien V, Kaniuk NA, et al. Listeriolysin O allows Listeria monocytogenes replication in macrophage vacuoles [Research Support, Non-U.S. Gov’t]. Nature. 2008. Jan 17;451(7176):350–4. doi: 10.1038/nature06479. PubMed PMID: 18202661; eng. [DOI] [PubMed] [Google Scholar]
- 3795.Bhardwaj V, Kanagawa O, Swanson PE, et al. Chronic Listeria infection in SCID mice: requirements for the carrier state and the dual role of T cells in transferring protection or suppression [Research Support, U.S. Gov’t, P.H.S.]. J Iimmunol. 1998. Jan 1;160(1):376–84. PubMed PMID: 9551994; eng. [PubMed] [Google Scholar]
- 3796.Digomann D, Kurth I, Tyutyunnykova A, et al. The CD98 heavy chain is a marker and regulator of head and neck squamous cell carcinoma radiosensitivity. clin cancer res off j am assoc cancer res. 2019. May 15;25(10):3152–3163. doi: 10.1158/1078-0432.CCR-18-2951. PubMed PMID: 30670494. [DOI] [PubMed] [Google Scholar]
- 3797.Liu H, Ma Y, He HW, et al. SLC9A3R1 stimulates autophagy via BECN1 stabilization in breast cancer cells. Autophagy. 2015. Jul 28;11:2323–34. doi: 10.1080/15548627.2015.1074372. PubMed PMID: 26218645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3798.Catalina-Rodriguez O, Kolukula VK, Tomita Y, et al. The mitochondrial citrate transporter, CIC, is essential for mitochondrial homeostasis. Oncotarget. 2012. Oct;3(10):1220–35. PubMed PMID: 23100451; PubMed Central PMCID: PMC3717962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3799.Rebsamen M, Pochini L, Stasyk T, et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature. 2015. Mar 26;519(7544):477–81. doi: 10.1038/nature14107. PubMed PMID: 25561175; PubMed Central PMCID: PMC4376665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3801.Deretic V, Saitoh T, Akira S.. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013. Oct;13(10):722–37. doi: 10.1038/nri3532. PubMed PMID: 24064518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3802.Reef S, Zalckvar E, Shifman O, et al. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006. May 19;22(4):463–75. doi: 10.1016/j.molcel.2006.04.014. PubMed PMID: 16713577; eng. [DOI] [PubMed] [Google Scholar]
- 3803.Mohamud Y, Shi J, Qu J, et al. Enteroviral infection inhibits autophagic flux via disruption of the SNARE complex to enhance viral replication. Cell Rep. 2018. Mar 20;22(12):3292–3303. doi: 10.1016/j.celrep.2018.02.090. PubMed PMID: 29562184. [DOI] [PubMed] [Google Scholar]
- 3804.Corona AK, Saulsbery HM, Corona Velazquez AF, et al. Enteroviruses remodel autophagic trafficking through regulation of host SNARE proteins to promote virus replication and cell exit. Cell Rep. 2018. Mar 20;22(12):3304–3314. doi: 10.1016/j.celrep.2018.03.003. PubMed PMID: 29562185; PubMed Central PMCID: PMCPMC5894509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3805.Batelli S, Peverelli E, Rodilossi S, et al. Macroautophagy and the proteasome are differently involved in the degradation of alpha-synuclein wild type and mutated A30P in an in vitro inducible model (PC12/TetOn). Neuroscience. 2011. Nov 10;195:128–37. doi: 10.1016/j.neuroscience.2011.08.030. PubMed PMID: 21906659; PubMed Central PMCID: PMC3188703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3806.Song JX, Lu JH, Liu LF, et al. HMGB1 is involved in autophagy inhibition caused by SNCA/alpha-synuclein overexpression: a process modulated by the natural autophagy inducer corynoxine B. Autophagy. 2014. Jan;10(1):144–54. doi: 10.4161/auto.26751. PubMed PMID: 24178442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3807.Baksi S, Singh N.. alpha-Synuclein impairs ferritinophagy in the retinal pigment epithelium: Implications for retinal iron dyshomeostasis in Parkinson’s disease. Sci Rep. 2017. Oct 9;7(1):12843. doi: 10.1038/s41598-017-12862-x. PubMed PMID: 28993630; PubMed Central PMCID: PMCPMC5634503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3808.Freeman D, Cedillos R, Choyke S, et al. Alpha-synuclein induces lysosomal rupture and cathepsin dependent reactive oxygen species following endocytosis. PLoS One. 2013;8(4):e62143. doi: 10.1371/journal.pone.0062143. PubMed PMID: 23634225; PubMed Central PMCID: PMCPMC3636263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3809.Flavin WP, Bousset L, Green ZC, et al. Endocytic vesicle rupture is a conserved mechanism of cellular invasion by amyloid proteins. Acta Neuropathol. 2017. Oct;134(4):629–653. doi: 10.1007/s00401-017-1722-x. PubMed PMID: 28527044. [DOI] [PubMed] [Google Scholar]
- 3810.Haft CR, de la Luz Sierra M, Barr VA, et al. Identification of a family of sorting nexin molecules and characterization of their association with receptors. Mol Cell Biol. 1998. Dec;18(12):7278–87. doi: 10.1128/mcb.18.12.7278. PubMed PMID: 9819414; PubMed Central PMCID: PMCPMC109309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3811.Traer CJ, Rutherford AC, Palmer KJ, et al. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol. 2007. Dec;9(12):1370–80. doi: 10.1038/ncb1656. PubMed PMID: 17994011. [DOI] [PubMed] [Google Scholar]
- 3812.Ma M, Burd CG, Chi RJ.. Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic. 2017. Feb;18(2):134–144. doi: 10.1111/tra.12462. PubMed PMID: 28026081; PubMed Central PMCID: PMCPMC5262529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3813.Knaevelsrud H, Soreng K, Raiborg C, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol. 2013. Jul 22;202(2):331–49. doi: 10.1083/jcb.201205129. PubMed PMID: 23878278; PubMed Central PMCID: PMC3718966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3814.Canuel M, Korkidakis A, Konnyu K, et al. Sortilin mediates the lysosomal targeting of cathepsins D and H. Biochem Biophys Res Commun. 2008. Aug 22;373(2):292–7. doi: 10.1016/j.bbrc.2008.06.021. PubMed PMID: 18559255. [DOI] [PubMed] [Google Scholar]
- 3815.Tan YS, Sansanaphongpricha K, Xie Y, et al. Mitigating SOX2-potentiated immune escape of head and neck squamous cell carcinoma with a STING-inducing nanosatellite vaccine. clin cancer res off j am assoc cancer res. 2018. Sep 1;24(17):4242–4255. doi: 10.1158/1078-0432.CCR-17-2807. PubMed PMID: 29769207; PubMed Central PMCID: PMCPMC6125216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3816.Barnett TC, Liebl D, Seymour LM, et al. The globally disseminated M1T1 clone of group A Streptococcus evades autophagy for intracellular replication. Cell Host Microbe. 2013. Dec 11;14(6):675–82. doi: 10.1016/j.chom.2013.11.003. PubMed PMID: 24331465; PubMed Central PMCID: PMC3918495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3817.Hirst J, Borner GH, Edgar J, et al. Interaction between AP-5 and the hereditary spastic paraplegia proteins SPG11 and SPG15. Mol Biol Cell. 2013. Aug;24(16):2558–69. doi: 10.1091/mbc.E13-03-0170. PubMed PMID: 23825025; PubMed Central PMCID: PMCPMC3744948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3818.Chang J, Lee S, Blackstone C.. Spastic paraplegia proteins spastizin and spatacsin mediate autophagic lysosome reformation. J Clin Invest. 2014. Dec;124(12):5249–62. doi: 10.1172/JCI77598. PubMed PMID: 25365221; PubMed Central PMCID: PMCPMC4348974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3819.Ghidoni R, Houri JJ, Giuliani A, et al. The metabolism of sphingo(glyco)lipids is correlated with the differentiation-dependent autophagic pathway in HT-29 cells. Eur J Biochem/FEBS. 1996. Apr 15;237(2):454–9. PubMed PMID: 8647085; eng. [DOI] [PubMed] [Google Scholar]
- 3820.Lavieu G, Scarlatti F, Sala G, et al. Is autophagy the key mechanism by which the sphingolipid rheostat controls the cell fate decision? Autophagy. 2007. Jan-Feb;3(1):45–47. PubMed PMID: 17035732; eng. [DOI] [PubMed] [Google Scholar]
- 3821.Young MM, Wang HG.. Sphingolipids as Regulators of Autophagy and Endocytic Trafficking. Adv Cancer Res. 2018;140:27–60. doi: 10.1016/bs.acr.2018.04.008. PubMed PMID: 30060813. [DOI] [PubMed] [Google Scholar]
- 3822.Tommasino C, Marconi M, Ciarlo L, et al. Autophagic flux and autophagosome morphogenesis require the participation of sphingolipids. Apoptosis. 2015. May;20(5):645–57. doi: 10.1007/s10495-015-1102-8. PubMed PMID: 25697338. [DOI] [PubMed] [Google Scholar]
- 3823.Chakraborty P, Vaena SG, Thyagarajan K, et al. Pro-survival lipid sphingosine-1-phosphate metabolically programs T cells to limit anti-tumor activity. Cell Rep. 2019. Aug 13;28(7):1879–1893 e7. doi: 10.1016/j.celrep.2019.07.044. PubMed PMID: 31412253; PubMed Central PMCID: PMCPMC6889821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3824.Panneer Selvam S, De Palma RM, Oaks JJ, et al. Binding of the sphingolipid S1P to hTERT stabilizes telomerase at the nuclear periphery by allosterically mimicking protein phosphorylation. Sci Signal. 2015. Jun 16;8(381):ra58. doi: 10.1126/scisignal.aaa4998. PubMed PMID: 26082434; PubMed Central PMCID: PMCPMC4492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3825.Chang CL, Ho MC, Lee PH, et al. S1P(5) is required for sphingosine 1-phosphate-induced autophagy in human prostate cancer PC-3 cells. Am J Physiol Cell Physiol. 2009. Aug;297(2):C451–8. doi: 10.1152/ajpcell.00586.2008. PubMed PMID: 19474291. [DOI] [PubMed] [Google Scholar]
- 3826.Dai L, Bai A, Smith CD, et al. ABC294640, a novel sphingosine kinase 2 inhibitor, induces oncogenic virus-infected cell autophagic death and represses tumor growth. Mol Cancer Ther. 2017. Dec;16(12):2724–2734. doi: 10.1158/1535-7163.MCT-17-0485. PubMed PMID: 28939554; PubMed Central PMCID: PMCPMC5716930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3827.Orsini M, Chateauvieux S, Rhim J, et al. Sphingolipid-mediated inflammatory signaling leading to autophagy inhibition converts erythropoiesis to myelopoiesis in human hematopoietic stem/progenitor cells. Cell Death Differ. 2019. Sep;26(9):1796–1812. doi: 10.1038/s41418-018-0245-x. PubMed PMID: 30546074; PubMed Central PMCID: PMCPMC6748125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3828.Sheng R, Zhang TT, Felice VD, et al. Preconditioning stimuli induce autophagy via sphingosine kinase 2 in mouse cortical neurons. J Biol Chem. 2014. Jul 25;289(30):20845–57. doi: 10.1074/jbc.M114.578120. PubMed PMID: 24928515; PubMed Central PMCID: PMCPMC4110292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3829.Song DD, Zhang TT, Chen JL, et al. Sphingosine kinase 2 activates autophagy and protects neurons against ischemic injury through interaction with Bcl-2 via its putative BH3 domain. Cell Death Dis. 2017. Jul 6;8(7):e2912. doi: 10.1038/cddis.2017.289. PubMed PMID: 28682313; PubMed Central PMCID: PMCPMC5550846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3830.Song DD, Zhou JH, Sheng R.. Regulation and function of sphingosine kinase 2 in diseases. Histol Histopathol. 2018. May;33(5):433–445. doi: 10.14670/HH-11-939. PubMed PMID: 29057430. [DOI] [PubMed] [Google Scholar]
- 3831.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014. Jun 5;510(7503):92–101. doi: 10.1038/nature13479. PubMed PMID: 24899309; PubMed Central PMCID: PMCPMC4263681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3832.Leuti A, Maccarrone M, Chiurchiu V.. Proresolving lipid mediators: endogenous modulators of oxidative stress. Oxid Med Cell Longev. 2019;2019:8107265. doi: 10.1155/2019/8107265. PubMed PMID: 31316721; PubMed Central PMCID: PMCPMC6604337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3833.Prieto P, Rosales-Mendoza CE, Terron V, et al. Activation of autophagy in macrophages by pro-resolving lipid mediators. Autophagy. 2015;11(10):1729–44. doi: 10.1080/15548627.2015.1078958. PubMed PMID: 26506892; PubMed Central PMCID: PMCPMC4824594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3834.Rong Y, McPhee C, Deng S, et al. Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation. Proc Natl Acad Sci U S A. 2011. May 10;108(19):7826–31. doi: 10.1073/pnas.1013800108. PubMed PMID: 21518918; PubMed Central PMCID: PMC3093520. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3835.Sun C, Enkhjargal B, Reis C, et al. Osteopontin-enhanced autophagy attenuates early brain injury via FAK-ERK pathway and improves long-term outcome after subarachnoid hemorrhage in rats. Cells 2019. Aug 27;8(9). doi: 10.3390/cells8090980. PubMed PMID: 31461955; PubMed Central PMCID: PMCPMC6769958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3836.Wen X, Gatica D, Yin Z, et al. The transcription factor Spt4-Spt5 complex regulates the expression of ATG8 and ATG41. Autophagy. 2020. Sep 8:in press. doi: 10.1080/15548627.2019.1659573. PubMed PMID: 31462158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3837.Chen Q, Yue F, Li W, et al. Potassium bisperoxo (1,10-phenanthroline) oxovanadate (bpV(phen)) induces apoptosis and pyroptosis and disrupts the P62-HDAC6 interaction to suppress the acetylated microtubule-dependent degradation of auto-phagosomes. J Biol Chem. 2015. Sep 11;290:26051–8. doi: 10.1074/jbc.M115.653568. PubMed PMID: 26363065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3838.Zhang Y, Mun SR, Linares JF, et al. Mechanistic insight into the regulation of SQSTM1/p62. Autophagy. 2019. Apr;15(4):735–737. doi: 10.1080/15548627.2019.1569935. PubMed PMID: 30653391; PubMed Central PMCID: PMCPMC6526835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3839.Itakura E, Mizushima N.. p62 Targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J Cell Biol. 2011. Jan 10;192(1):17–27. doi: 10.1083/jcb.201009067. PubMed PMID: 21220506; PubMed Central PMCID: PMCPMC3019556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3840.Wurzer B, Zaffagnini G, Fracchiolla D, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife. 2015. Sep 28;4:e08941. doi: 10.7554/eLife.08941. PubMed PMID: 26413874; PubMed Central PMCID: PMCPMC4684078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3841.Kwon DH, Kim L, Song HK.. pH-dependent regulation of SQSTM1/p62 during autophagy. Autophagy. 2019. Jan;15(1):180–181. doi: 10.1080/15548627.2018.1532264. PubMed PMID: 30290711; PubMed Central PMCID: PMCPMC6287675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3842.Sun D, Wu R, Zheng J, et al. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018. Apr;28(4):405–415. doi: 10.1038/s41422-018-0017-7. PubMed PMID: 29507397; PubMed Central PMCID: PMCPMC5939046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3843.Zaffagnini G, Savova A, Danieli A, et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018. Mar 1;37(5). doi: 10.15252/embj.201798308. PubMed PMID: 29343546; PubMed Central PMCID: PMCPMC5830917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3844.Nguyen TD, Shaid S, Vakhrusheva O, et al. Loss of the selective autophagy receptor p62 impairs murine myeloid leukemia progression and mitophagy. Blood. 2019. Jan 10;133(2):168–179. doi: 10.1182/blood-2018-02-833475. PubMed PMID: 30498063. [DOI] [PubMed] [Google Scholar]
- 3845.Shi J, Wong J, Piesik P, et al. Cleavage of sequestosome 1/p62 by an enteroviral protease results in disrupted selective autophagy and impaired NFKB signaling. Autophagy. 2013. Oct;9(10):1591–603. doi: 10.4161/auto.26059. PubMed PMID: 23989536. [DOI] [PubMed] [Google Scholar]
- 3846.Shi J, Fung G, Piesik P, et al. Dominant-negative function of the C-terminal fragments of NBR1 and SQSTM1 generated during enteroviral infection. Cell Death Differ. 2014. Sep;21(9):1432–41. doi: 10.1038/cdd.2014.58. PubMed PMID: 24769734; PubMed Central PMCID: PMCPMC4131175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3847.Tambe Y, Yamamoto A, Isono T, et al. The drs tumor suppressor is involved in the maturation process of autophagy induced by low serum. Cancer Lett. 2009. Sep 28;283(1):74–83. doi: 10.1016/j.canlet.2009.03.028. PubMed PMID: 19368996. [DOI] [PubMed] [Google Scholar]
- 3848.Mesquita FS, Thomas M, Sachse M, et al. The Salmonella deubiquitinase SseL inhibits selective autophagy of cytosolic aggregates. PLoS Pathog. 2012;8(6):e1002743. doi: 10.1371/journal.ppat.1002743. PubMed PMID: 22719249; PubMed Central PMCID: PMC3375275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3849.Luna-Dulcey L, Tomasin R, Naves MA, et al. Autophagy-dependent apoptosis is triggered by a semi-synthetic [6]-gingerol analogue in triple negative breast cancer cells. Oncotarget. 2018. Jul 20;9(56):30787–30804. doi: 10.18632/oncotarget.25704. PubMed PMID: 30112107; PubMed Central PMCID: PMCPMC6089392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3850.Shen S, Niso-Santano M, Adjemian S, et al. Cytoplasmic STAT3 represses autophagy by inhibiting PKR activity. Mol Cell. 2012. Dec 14;48(5):667–80. doi: 10.1016/j.molcel.2012.09.013. PubMed PMID: 23084476. [DOI] [PubMed] [Google Scholar]
- 3851.Wang CW. Stationary phase lipophagy as a cellular mechanism to recycle sterols during quiescence. Autophagy. 2014;10(11):2075–6. doi: 10.4161/auto.36137. PubMed PMID: 25484090; PubMed Central PMCID: PMC4502705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3852.Kuang XL, Liu Y, Chang Y, et al. Inhibition of store-operated calcium entry by sub-lethal levels of proteasome inhibition is associated with STIM1/STIM2 degradation. Cell Calcium. 2016. Apr;59(4):172–80. doi: 10.1016/j.ceca.2016.01.007. PubMed PMID: 26960935. [DOI] [PubMed] [Google Scholar]
- 3853.Zhou J, Song J, Wu S.. Autophagic degradation of stromal interaction molecule 2 mediates disruption of neuronal dendrites by endoplasmic reticulum stress. J Neurochem. 2019. Nov;151(3):351–369. doi: 10.1111/jnc.14712. PubMed PMID: 31038732. [DOI] [PubMed] [Google Scholar]
- 3854.Liu Y, Gordesky-Gold B, Leney-Greene M, et al. Inflammation-induced, STING-dependent autophagy restricts zika virus infection in the drosophila brain. Cell Host Microbe. 2018. Jul 11;24(1):57–68 e3. doi: 10.1016/j.chom.2018.05.022. PubMed PMID: 29934091; PubMed Central PMCID: PMCPMC6173519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3855.Watson RO, Bell SL, MacDuff DA, et al. The cytosolic sensor cGAS detects mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe. 2015. Jun 10;17(6):811–819. doi: 10.1016/j.chom.2015.05.004. PubMed PMID: 26048136; PubMed Central PMCID: PMCPMC4466081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3856.Aden K, Tran F, Ito G, et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J Exp Med. 2018. Nov 5;215(11):2868–2886. doi: 10.1084/jem.20171029. PubMed PMID: 30254094; PubMed Central PMCID: PMCPMC6219748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3857.Prabakaran T, Bodda C, Krapp C, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018. Apr 13;37(8). doi: 10.15252/embj.201797858. PubMed PMID: 29496741; PubMed Central PMCID: PMCPMC5897779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3858.Gui X, Yang H, Li T, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019. Mar;567(7747):262–266. doi: 10.1038/s41586-019-1006-9. PubMed PMID: 30842662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3859.Wilkinson DS, Jariwala JS, Anderson E, et al. Phosphorylation of LC3 by the Hippo Kinases STK3/STK4 Is Essential for Autophagy. Mol Cell. 2015. Jan 8;57(1):55–68. doi: 10.1016/j.molcel.2014.11.019. PubMed PMID: 25544559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3860.Maejima Y, Kyoi S, Zhai P, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013. Nov;19(11):1478–88. doi: 10.1038/nm.3322. PubMed PMID: 24141421; PubMed Central PMCID: PMC3823824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3861.Huang T, Kim CK, Alvarez AA, et al. MST4 phosphorylation of ATG4B regulates autophagic activity, tumorigenicity, and radioresistance in glioblastoma. Cancer cell. 2017. Dec 11;32(6):840–855 e8. doi: 10.1016/j.ccell.2017.11.005. PubMed PMID: 29232556; PubMed Central PMCID: PMCPMC5734934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3862.Joffre C, Dupont N, Hoa L, et al. The pro-apoptotic STK38 kinase is a new beclin1 partner positively regulating autophagy. Curr Biol. 2015. Oct 5;25(19):2479–92. doi: 10.1016/j.cub.2015.08.031. PubMed PMID: 26387716; PubMed Central PMCID: PMCPMC4598746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3863.Tokumitsu H, Inuzuka H, Ishikawa Y, et al. STO-609, a specific inhibitor of the Ca(2+)/calmodulin-dependent protein kinase kinase. J Biol Chem. 2002. May 3;277(18):15813–8. doi: 10.1074/jbc.M201075200. PubMed PMID: 11867640. [DOI] [PubMed] [Google Scholar]
- 3864.Anderson KA, Ribar TJ, Lin F, et al. Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab. 2008. May;7(5):377–88. doi: 10.1016/j.cmet.2008.02.011. PubMed PMID: 18460329. [DOI] [PubMed] [Google Scholar]
- 3865.Monteiro P, Gilot D, Langouet S, et al. Activation of the aryl hydrocarbon receptor by the calcium/calmodulin-dependent protein kinase kinase inhibitor 7-oxo-7H-benzimidazo[2,1-a]benz[de]isoquinoline-3-carboxylic acid (STO-609). Drug Metab Dispos. 2008. Dec;36(12):2556–63. doi: 10.1124/dmd.108.023333. PubMed PMID: 18755850. [DOI] [PubMed] [Google Scholar]
- 3866.Kimura T, Jia J, Claude-Taupin A, et al. Cellular and molecular mechanism for secretory autophagy. Autophagy. 2017. Jun 3;13(6):1084–1085. doi: 10.1080/15548627.2017.1307486. PubMed PMID: 28368721; PubMed Central PMCID: PMCPMC5486376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3867.Lu Y, Zhang Z, Sun D, et al. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol Cell. 2013. Oct 24;52(2):264–71. doi: 10.1016/j.molcel.2013.08.041. PubMed PMID: 24095276; PubMed Central PMCID: PMC3825790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3868.Diao J, Liu R, Rong Y, et al. ATG14 promotes membrane tethering and fusion of auto-phagosomes to endolysosomes. Nature. 2015. Apr 23;520(7548):563–6. doi: 10.1038/nature14147. PubMed PMID: 25686604; PubMed Central PMCID: PMCPMC4442024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3869.Arasaki K, Nagashima H, Kurosawa Y, et al. MAP1B-LC1 prevents autophagosome formation by linking syntaxin 17 to microtubules. EMBO Rep. 2018. Aug;19(8). doi: 10.15252/embr.201745584. PubMed PMID: 29925525; PubMed Central PMCID: PMCPMC6073212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3870.Wang C, Wang H, Zhang D, et al. Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy. Nat Commun. 2018. Aug 28;9(1):3492. doi: 10.1038/s41467-018-05449-1. PubMed PMID: 30154410; PubMed Central PMCID: PMCPMC6113293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3871.Saleeb RS, Kavanagh DM, Dun AR, et al. A VPS33A-binding motif on syntaxin 17 controls autophagy completion in mammalian cells. J Biol Chem. 2019. Mar 15;294(11):4188–4201. doi: 10.1074/jbc.RA118.005947. PubMed PMID: 30655294; PubMed Central PMCID: PMCPMC6422071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3872.Tian X, Zheng P, Zhou C, et al. DIPK2A promotes STX17- and VAMP7-mediated autophagosome-lysosome fusion by binding to VAMP7B. Autophagy. 2019. Jul 4:1–14. doi: 10.1080/15548627.2019.1637199. PubMed PMID: 31251111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3873.Kumar S, Jain A, Farzam F, et al. Mechanism of Stx17 recruitment to auto-phagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol. 2018. Mar 5;217(3):997–1013. doi: 10.1083/jcb.201708039. PubMed PMID: 29420192; PubMed Central PMCID: PMCPMC5839791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3874.Bustos V, Pulina MV, Bispo A, et al. Phosphorylated Presenilin 1 decreases beta-amyloid by facilitating autophagosome-lysosome fusion. Proc Natl Acad Sci U S A. 2017. Jul 3;114(27):7148–7153. doi: 10.1073/pnas.1705240114. PubMed PMID: 28533369; PubMed Central PMCID: PMCPMC5502640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3875.McLelland GL, Lee SA, McBride HM, et al. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J Cell Biol. 2016. Aug 1;214(3):275–91. doi: 10.1083/jcb.201603105. PubMed PMID: 27458136; PubMed Central PMCID: PMCPMC4970327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3876.Kimura H, Arasaki K, Ohsaki Y, et al. Syntaxin 17 promotes lipid droplet formation by regulating the distribution of acyl-CoA synthetase 3. J Lipid Res. 2018. May;59(5):805–819. doi: 10.1194/jlr.M081679. PubMed PMID: 29549094; PubMed Central PMCID: PMCPMC5928434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3877.Zhou C, Qian X, Hu M, et al. STYK1 promotes autophagy through enhancing the assembly of autophagy-specific class III phosphatidylinositol 3-kinase complex I. Autophagy. 2019. Nov 7:1–21. doi: 10.1080/15548627.2019.1687212. PubMed PMID: 31696776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3878.Herman-Antosiewicz A, Johnson DE, Singh SV.. Sulforaphane causes autophagy to inhibit release of cytochrome C and apoptosis in human prostate cancer cells. Cancer Res. 2006. Jun 1;66(11):5828–35. doi: 10.1158/0008-5472.CAN-06-0139. PubMed PMID: 16740722. [DOI] [PubMed] [Google Scholar]
- 3879.Vyas AR, Hahm ER, Arlotti JA, et al. Chemoprevention of prostate cancer by d,l-sulforaphane is augmented by pharmacological inhibition of autophagy. Cancer Res. 2013. Oct 1;73(19):5985–95. doi: 10.1158/0008-5472.CAN-13-0755. PubMed PMID: 23921360; PubMed Central PMCID: PMCPMC3790864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3880.Lee JH, Jeong JK, Park SY.. Sulforaphane-induced autophagy flux prevents prion protein-mediated neurotoxicity through AMPK pathway. Neuroscience. 2014. Oct 10;278:31–9. doi: 10.1016/j.neuroscience.2014.07.072. PubMed PMID: 25130556. [DOI] [PubMed] [Google Scholar]
- 3881.Wang H, Wang F, Wu S, et al. Traditional herbal medicine-derived sulforaphene promotes mitophagic cell death in lymphoma cells through CRM1-mediated p62/SQSTM1 accumulation and AMPK activation. Chem Biol Interact. 2018. Feb 1;281:11–23. doi: 10.1016/j.cbi.2017.12.017. PubMed PMID: 29247643. [DOI] [PubMed] [Google Scholar]
- 3882.Webber JL, Tooze SA.. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010. Jan 6;29(1):27–40. doi: 10.1038/emboj.2009.321. PubMed PMID: 19893488; PubMed Central PMCID: PMC2808369. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3883.Lopergolo A, Nicolini V, Favini E, et al. Synergistic cooperation between sunitinib and cisplatin promotes apoptotic cell death in human medullary thyroid cancer. J Clin Endocrinol Metab. 2014. Feb;99(2):498–509. doi: 10.1210/jc.2013-2574. PubMed PMID: 24276455. [DOI] [PubMed] [Google Scholar]
- 3884.Abdel-Aziz AK, Abdel-Naim AB, Shouman S, et al. From resistance to sensitivity: insights and implications of biphasic modulation of autophagy by sunitinib. Front Pharmacol. 2017;8:718. doi: 10.3389/fphar.2017.00718. PubMed PMID: 29066973; PubMed Central PMCID: PMCPMC5641351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3885.Elgendy M, Abdel-Aziz AK, Renne SL, et al. Dual modulation of MCL-1 and mTOR determines the response to sunitinib. J Clin Invest. 2017. Jan 3;127(1):153–168. doi: 10.1172/JCI84386. PubMed PMID: 27893461; PubMed Central PMCID: PMCPMC5199697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3886.Nikoletopoulou V, Sidiropoulou K, Kallergi E, et al. Modulation of autophagy by BDNF underlies synaptic plasticity. Cell Metab. 2017. Jul 5;26(1):230–242 e5. doi: 10.1016/j.cmet.2017.06.005. PubMed PMID: 28683289. [DOI] [PubMed] [Google Scholar]
- 3887.Okerlund ND, Schneider K, Leal-Ortiz S, et al. Bassoon controls presynaptic autophagy through Atg5. Neuron. 2017. Feb 22;93(4):897–913 e7. doi: 10.1016/j.neuron.2017.01.026. PubMed PMID: 28231469. [DOI] [PubMed] [Google Scholar]
- 3888.Liang Y, Sigrist S.. Autophagy and proteostasis in the control of synapse aging and disease. Curr Opin Neurobiol. 2018. Feb;48:113–121. doi: 10.1016/j.conb.2017.12.006. PubMed PMID: 29274917. [DOI] [PubMed] [Google Scholar]
- 3889.Liang Y. Emerging concepts and functions of autophagy as a regulator of synaptic components and plasticity. Cells. 2019. Jan 9;8(1). doi: 10.3390/cells8010034. PubMed PMID: 30634508; PubMed Central PMCID: PMCPMC6357011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3890.Rowland AM, Richmond JE, Olsen JG, et al. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006. Feb 8;26(6):1711–20. doi: 10.1523/JNEUROSCI.2279-05.2006. PubMed PMID: 16467519; PubMed Central PMCID: PMCPMC6793639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3891.Shehata M, Matsumura H, Okubo-Suzuki R, et al. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J Neurosci. 2012. Jul 25;32(30):10413–22. doi: 10.1523/JNEUROSCI.4533-11.2012. PubMed PMID: 22836274; PubMed Central PMCID: PMCPMC6703735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3892.Ashrafi G, Schlehe JS, LaVoie MJ, et al. Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. J Cell Biol. 2014. Sep 1;206(5):655–70. doi: 10.1083/jcb.201401070. PubMed PMID: 25154397; PubMed Central PMCID: PMCPMC4151150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3893.Hoffmann S, Orlando M, Andrzejak E, et al. Light-activated ROS production induces synaptic autophagy. J Neurosci. 2019. Mar 20;39(12):2163–2183. doi: 10.1523/JNEUROSCI.1317-18.2019. PubMed PMID: 30655355; PubMed Central PMCID: PMCPMC6433757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3894.Vanhauwaert R, Kuenen S, Masius R, et al. The SAC1 domain in synaptojanin is required for autophagosome maturation at presynaptic terminals. EMBO J. 2017. May 15;36(10):1392–1411. doi: 10.15252/embj.201695773. PubMed PMID: 28331029; PubMed Central PMCID: PMCPMC5430236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3895.Criollo A, Niso-Santano M, Malik SA, et al. Inhibition of autophagy by TAB2 and TAB3. EMBO J. 2011. Dec 14;30(24):4908–20. doi: 10.1038/emboj.2011.413. PubMed PMID: 22081109; PubMed Central PMCID: PMC3243630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3896.Takaesu G, Kobayashi T, Yoshimura A.. TGFbeta-activated kinase 1 (TAK1)-binding proteins (TAB) 2 and 3 negatively regulate autophagy. J Biochem. 2012. Feb;151(2):157–66. doi: 10.1093/jb/mvr123. PubMed PMID: 21976705. [DOI] [PubMed] [Google Scholar]
- 3897.Biering SB, Choi J, Halstrom RA, et al. Viral Replication Complexes Are Targeted by LC3-Guided Interferon-Inducible GTPases. Cell Host Microbe. 2017. Jul 12;22(1):74–85 e7. doi: 10.1016/j.chom.2017.06.005. PubMed PMID: 28669671; PubMed Central PMCID: PMCPMC5591033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3898.Nagahara Y, Takeyoshi M, Sakemoto S, et al. Novel tamoxifen derivative Ridaifen-B induces Bcl-2 independent autophagy without estrogen receptor involvement. Biochem Biophys Res Commun. 2013. Jun 14;435(4):657–63. doi: 10.1016/j.bbrc.2013.05.040. PubMed PMID: 23688426. [DOI] [PubMed] [Google Scholar]
- 3899.Torres-Lopez L, Maycotte P, Linan-Rico A, et al. Tamoxifen induces toxicity, causes autophagy, and partially reverses dexamethasone resistance in Jurkat T cells. J Leukoc Biol. 2019. May;105(5):983–998. doi: 10.1002/JLB.2VMA0818-328R. PubMed PMID: 30645008. [DOI] [PubMed] [Google Scholar]
- 3900.Bose JK, Huang CC, Shen CK.. Regulation of autophagy by neuropathological protein TDP-43. J Biol Chem. 2011. Dec 30;286(52):44441–8. doi: 10.1074/jbc.M111.237115. PubMed PMID: 22052911; PubMed Central PMCID: PMC3247982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3901.Fu T, Liu J, Wang Y, et al. Mechanistic insights into the interactions of NAP1 with the SKICH domains of NDP52 and TAX1BP1. Proc Natl Acad Sci U S A. 2018. Dec 11;115(50):E11651–E11660. doi: 10.1073/pnas.1811421115. PubMed PMID: 30459273; PubMed Central PMCID: PMCPMC6294882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3902.Newman AC, Scholefield CL, Kemp AJ, et al. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-kappaB signalling. PLoS One. 2012;7(11):e50672. doi: 10.1371/journal.pone.0050672. PubMed PMID: 23209807; PubMed Central PMCID: PMC3510188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3903.Petkova DS, Verlhac P, Rozieres A, et al. Distinct contributions of autophagy receptors in measles virus replication. Viruses. 2017. May 22;9(5). doi: 10.3390/v9050123. PubMed PMID: 28531150; PubMed Central PMCID: PMCPMC5454435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3904.Dibble CC, Elis W, Menon S, et al. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012. Aug 24;47(4):535–46. doi: 10.1016/j.molcel.2012.06.009. PubMed PMID: 22795129; PubMed Central PMCID: PMC3693578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3905.Alfaiz AA, Micale L, Mandriani B, et al. TBC1D7 mutations are associated with intellectual disability, macrocrania, patellar dislocation, and celiac disease. Hum Mutat. 2014. Apr;35(4):447–51. doi: 10.1002/humu.22529. PubMed PMID: 24515783. [DOI] [PubMed] [Google Scholar]
- 3906.Capo-Chichi JM, Tcherkezian J, Hamdan FF, et al. Disruption of TBC1D7, a subunit of the TSC1-TSC2 protein complex, in intellectual disability and megalencephaly. J Med Genet. 2013. Nov;50(11):740–4. doi: 10.1136/jmedgenet-2013-101680. PubMed PMID: 23687350. [DOI] [PubMed] [Google Scholar]
- 3907.Liao Y, Li M, Chen X, et al. Interaction of TBC1D9B with mammalian ATG8 homologues regulates autophagic flux. Sci Rep. 2018. Sep 10;8(1):13496. doi: 10.1038/s41598-018-32003-2. PubMed PMID: 30202024; PubMed Central PMCID: PMCPMC6131546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3908.Gallo LI, Liao Y, Ruiz WG, et al. TBC1D9B functions as a GTPase-activating protein for Rab11a in polarized MDCK cells. Mol Biol Cell. 2014. Nov 15;25(23):3779–97. doi: 10.1091/mbc.E13-10-0604. PubMed PMID: 25232007; PubMed Central PMCID: PMCPMC4230784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3909.Hirano S, Uemura T, Annoh H, et al. Differing susceptibility to autophagic degradation of two LC3-binding proteins: SQSTM1/p62 and TBC1D25/OATL1. Autophagy. 2016;12(2):312–26. doi: 10.1080/15548627.2015.1124223. PubMed PMID: 26902585; PubMed Central PMCID: PMCPMC4836008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3910.Pomerantz JL, Baltimore D.. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J. 1999. Dec 1;18(23):6694–704. doi: 10.1093/emboj/18.23.6694. PubMed PMID: 10581243; PubMed Central PMCID: PMC1171732. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3911.Neill T, Torres A, Buraschi S, et al. Decorin induces mitophagy in breast carcinoma cells via peroxisome proliferator-activated receptor gamma coactivator-1alpha (PGC-1alpha) and mitostatin. J Biol Chem. 2014. Feb 21;289(8):4952–68. doi: 10.1074/jbc.M113.512566. PubMed PMID: 24403067; PubMed Central PMCID: PMC3931056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3912.Ogawa M, Yoshikawa Y, Kobayashi T, et al. A tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011. May 19;9(5):376–89. doi: 10.1016/j.chom.2011.04.010. PubMed PMID: 21575909; eng. [DOI] [PubMed] [Google Scholar]
- 3913.Zhang H, Yan S, Khambu B, et al. Dynamic MTORC1-TFEB feedback signaling regulates hepatic autophagy, steatosis and liver injury in long-term nutrient oversupply. Autophagy. 2018;14(10):1779–1795. doi: 10.1080/15548627.2018.1490850. PubMed PMID: 30044707; PubMed Central PMCID: PMCPMC6135624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3914.Oz-Levi D, Ben-Zeev B, Ruzzo EK, et al. Mutation in TECPR2 reveals a role for autophagy in hereditary spastic paraparesis. Am J Hum Genet. 2012. Dec 7;91(6):1065–72. doi: 10.1016/j.ajhg.2012.09.015. PubMed PMID: 23176824; PubMed Central PMCID: PMC3516605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3915.Oz-Levi D, Gelman A, Elazar Z, et al. TECPR2: a new autophagy link for neurodegeneration. Autophagy. 2013. May;9(5):801–2. doi: 10.4161/auto.23961. PubMed PMID: 23439247; PubMed Central PMCID: PMC3669195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3916.Delorme-Axford E, Popelka H, Klionsky DJ.. TEX264 is a major receptor for mammalian reticulophagy. Autophagy. 2019. Oct;15(10):1677–1681. doi: 10.1080/15548627.2019.1646540. PubMed PMID: 31362563; PubMed Central PMCID: PMCPMC6735500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3917.Wang P, Nolan TM, Yin Y, et al. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy. 2020. Jan;16(1):123–139. doi: 10.1080/15548627.2019.1598753. PubMed PMID: 30909785; PubMed Central PMCID: PMCPMC6984607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3918.Huang S, Lu W, Ge D, et al. A new microRNA signal pathway regulated by long noncoding RNA TGFB2-OT1 in autophagy and inflammation of vascular endothelial cells. Autophagy. 2015;11(12):2172–83. doi: 10.1080/15548627.2015.1106663. PubMed PMID: 26565952; PubMed Central PMCID: PMCPMC4835209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3919.D’Eletto M, Farrace MG, Falasca L, et al. Transglutaminase 2 is involved in autophagosome maturation. Autophagy. 2009. Nov;5(8):1145–54. PubMed PMID: 19955852. [DOI] [PubMed] [Google Scholar]
- 3920.Salazar M, Carracedo A, Salanueva IJ, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009. May;119(5):1359–72. PubMed PMID: 19425170; PubMed Central PMCID: PMC2673842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3921.Salazar M, Lorente M, Garcia-Taboada E, et al. The pseudokinase tribbles homologue-3 plays a crucial role in cannabinoid anticancer action. Biochim Biophys Acta. 2013. Oct;1831(10):1573–8. doi: 10.1016/j.bbalip.2013.03.014. PubMed PMID: 23567453. [DOI] [PubMed] [Google Scholar]
- 3922.Velasco G, Sanchez C, Guzman M.. Towards the use of cannabinoids as antitumour agents. Nat Rev Cancer. 2012. Jun;12(6):436–44. doi: 10.1038/nrc3247. PubMed PMID: 22555283. [DOI] [PubMed] [Google Scholar]
- 3923.Yuzwa SA, Macauley MS, Heinonen JE, et al. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 2008. Aug;4(8):483–90. doi: 10.1038/nchembio.96. PubMed PMID: 18587388. [DOI] [PubMed] [Google Scholar]
- 3924.Zhu Y, Shan X, Safarpour F, et al. Pharmacological Inhibition of O-GlcNAcase Enhances Autophagy in Brain through an mTOR-Independent Pathway. ACS Chem Neurosci. 2018. Jun 20;9(6):1366–1379. doi: 10.1021/acschemneuro.8b00015. PubMed PMID: 29460617. [DOI] [PubMed] [Google Scholar]
- 3925.Bensaad K, Cheung EC, Vousden KH.. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009. Oct 7;28(19):3015–26. doi: 10.1038/emboj.2009.242. PubMed PMID: 19713938; PubMed Central PMCID: PMC2736014. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3926.Lok CN, Sy LK, Liu F, et al. Activation of autophagy of aggregation-prone ubiquitinated proteins by timosaponin A-III. J Biol Chem. 2011. Sep 9;286(36):31684–96. doi: 10.1074/jbc.M110.202531. PubMed PMID: 21757721; PubMed Central PMCID: PMC3173142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3927.Lin H, Yan J, Wang Z, et al. Loss of immunity-supported senescence enhances susceptibility to hepatocellular carcinogenesis and progression in Toll-like receptor 2-deficient mice. Hepatology. 2013. Jan;57(1):171–82. doi: 10.1002/hep.25991. PubMed PMID: 22859216. [DOI] [PubMed] [Google Scholar]
- 3928.Lin H, Hua F, Hu ZW.. Autophagic flux, supported by toll-like receptor 2 activity, defends against the carcinogenesis of hepatocellular carcinoma. Autophagy. 2012. Dec, 8(12):1859–61. doi: 10.4161/auto.22094. PubMed PMID: 22996042; PubMed Central PMCID: PMCPMC3541305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3929.Wang Z, Yan J, Lin H, et al. Toll-like receptor 4 activity protects against hepatocellular tumorigenesis and progression by regulating expression of DNA repair protein Ku70 in mice. Hepatology 2013. May;57(5):1869–81. doi: 10.1002/hep.26234. PubMed PMID: 23299825. [DOI] [PubMed] [Google Scholar]
- 3930.Wang Z, Lin H, Hua F, et al. Repairing DNA damage by XRCC6/KU70 reverses TLR4-deficiency-worsened HCC development via restoring senescence and autophagic flux. Autophagy. 2013. Jun 1;9(6):925–7. doi: 10.4161/auto.24229. PubMed PMID: 23518600; PubMed Central PMCID: PMCPMC3672303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3931.Wu RN, Yu TY, Zhou JC, et al. Targeting HMGB1 ameliorates cardiac fibrosis through restoring TLR2-mediated autophagy suppression in myocardial fibroblasts. Int J Cardiol. 2018. Sep 15;267:156–162. doi: 10.1016/j.ijcard.2018.04.103. PubMed PMID: 29957254. [DOI] [PubMed] [Google Scholar]
- 3932.Oka T, Hikoso S, Yamaguchi O, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012. May 10;485(7397):251–5. doi: 10.1038/nature10992. PubMed PMID: 22535248; PubMed Central PMCID: PMCPMC3378041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3933.He P, Peng Z, Luo Y, et al. High-throughput functional screening for autophagy-related genes and identification of TM9SF1 as an autophagosome-inducing gene. Autophagy. 2009. Jan 1;5(1):52–60. PubMed PMID: 19029833; eng. [DOI] [PubMed] [Google Scholar]
- 3934.Sun L, Meng Z, Zhu Y, et al. TM9SF4 is a novel factor promoting autophagic flux under amino acid starvation. Cell Death Differ. 2018. Feb;25(2):368–379. doi: 10.1038/cdd.2017.166. PubMed PMID: 29125601; PubMed Central PMCID: PMCPMC5762850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3935.Maurel M, Obacz J, Avril T, et al. Control of anterior GRadient 2 (AGR2) dimerization links endoplasmic reticulum proteostasis to inflammation. EMBO Mol Med. 2019. Jun;11(6). doi: 10.15252/emmm.201810120. PubMed PMID: 31040128; PubMed Central PMCID: PMCPMC6554669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3936.Morita K, Hama Y, Izume T, et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol. 2018. Nov 5;217(11):3817–3828. doi: 10.1083/jcb.201804132. PubMed PMID: 30093494; PubMed Central PMCID: PMCPMC6219718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3937.Shoemaker CJ, Huang TQ, Weir NR, et al. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol. 2019. Apr;17(4):e2007044. doi: 10.1371/journal.pbio.2007044. PubMed PMID: 30933966; PubMed Central PMCID: PMCPMC6459555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3938.Moretti F, Bergman P, Dodgson S, et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 2018. Sep;19(9). doi: 10.15252/embr.201845889. PubMed PMID: 30126924; PubMed Central PMCID: PMCPMC6123663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3939.Boada-Romero E, Letek M, Fleischer A, et al. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 2013. Feb 20;32(4):566–82. doi: 10.1038/emboj.2013.8. PubMed PMID: 23376921; PubMed Central PMCID: PMC3579146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3940.Boada-Romero E, Serramito-Gomez I, Sacristan MP, et al. The T300A Crohn’s disease risk polymorphism impairs function of the WD40 domain of ATG16L1. Nat Commun. 2016. Jun 8;7:11821. doi: 10.1038/ncomms11821. PubMed PMID: 27273576; PubMed Central PMCID: PMCPMC4899871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3941.Sun Y, Chen Y, Zhang J, et al. TMEM74 promotes tumor cell survival by inducing autophagy via interactions with ATG16L1 and ATG9A. Cell Death Dis. 2017. Aug 31;8(8):e3031. doi: 10.1038/cddis.2017.370. PubMed PMID: 29048433; PubMed Central PMCID: PMCPMC5596558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3942.Mrschtik M, O’Prey J, Lao LY, et al. DRAM-3 modulates autophagy and promotes cell survival in the absence of glucose. Cell Death Differ. 2015. Oct;22(10):1714–26. doi: 10.1038/cdd.2015.26. PubMed PMID: 25929859; PubMed Central PMCID: PMCPMC4563785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3943.Mrschtik M, O’Prey J, Lao LY, et al. DRAM-3 modulates autophagy and promotes cell survival in the absence of glucose. Cell Death Differ. 2017. Aug;24(8):1470. doi: 10.1038/cdd.2017.57. PubMed PMID: 28665403; PubMed Central PMCID: PMCPMC5520458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3944.Shi CS, Kehrl JH.. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1 [Research Support, N.I.H., Intramural]. Autophagy. 2010. Oct;6(7):986–7. doi: 10.4161/auto.6.7.13288. PubMed PMID: 20798608; PubMed Central PMCID: PMC3039745. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3945.Matsuzawa Y, Oshima S, Takahara M, et al. TNFAIP3 promotes survival of CD4 T cells by restricting MTOR and promoting autophagy. Autophagy. 2015. Jul 3;11(7):1052–62. doi: 10.1080/15548627.2015.1055439. PubMed PMID: 26043155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3946.Slowicka K, Serramito-Gomez I, Boada-Romero E, et al. Physical and functional interaction between A20 and ATG16L1-WD40 domain in the control of intestinal homeostasis. Nat Commun. 2019. Apr 23;10(1):1834. doi: 10.1038/s41467-019-09667-z. PubMed PMID: 31015422; PubMed Central PMCID: PMCPMC6478926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3947.Shah JA, Emery R, Lee B, et al. TOLLIP deficiency is associated with increased resistance to Legionella pneumophila pneumonia. Mucosal Immunol. 2019. Nov;12(6):1382–1390. doi: 10.1038/s41385-019-0196-7. PubMed PMID: 31462698; PubMed Central PMCID: PMCPMC6824992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3948.Shah JA, Vary JC, Chau TT, et al. Human TOLLIP regulates TLR2 and TLR4 signaling and its polymorphisms are associated with susceptibility to tuberculosis. J Iimmunol. 2012. Aug 15;189(4):1737–46. doi: 10.4049/jimmunol.1103541. PubMed PMID: 22778396; PubMed Central PMCID: PMCPMC3428135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3949.Jacinto E. What controls TOR? IUBMB life. 2008. Aug;60(8):483–96. doi: 10.1002/iub.56. PubMed PMID: 18493947; eng. [DOI] [PubMed] [Google Scholar]
- 3950.Peterson TR, Laplante M, Thoreen CC, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009. May 29;137(5):873–86. doi: 10.1016/j.cell.2009.03.046. PubMed PMID: 19446321; PubMed Central PMCID: PMC2758791. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3951.Abe K, Yano T, Tanno M, et al. mTORC1 inhibition attenuates necroptosis through RIP1 inhibition-mediated TFEB activation. Biochim Biophys Acta Mol Basis Dis. 2019. Dec 1;1865(12):165552. doi: 10.1016/j.bbadis.2019.165552. PubMed PMID: 31499159. [DOI] [PubMed] [Google Scholar]
- 3952.Pearce LR, Huang X, Boudeau J, et al. Identification of Protor as a novel Rictor-binding component of mTOR complex-2. Biochem J. 2007. Aug 1;405(3):513–22. doi: BJ20070540 [pii] doi: 10.1042/BJ20070540. PubMed PMID: 17461779; PubMed Central PMCID: PMC2267312. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3953.Vlahakis A, Graef M, Nunnari J, et al. TOR complex 2-Ypk1 signaling is an essential positive regulator of the general amino acid control response and autophagy. Proc Natl Acad Sci U S A. 2014. Jul 22;111(29):10586–91. doi: 10.1073/pnas.1406305111. PubMed PMID: 25002487; PubMed Central PMCID: PMC4115538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3954.Renna M, Bento CF, Fleming A, et al. IGF-1 receptor antagonism inhibits autophagy. Hum Mol Genet. 2013. Nov 15;22(22):4528–44. doi: 10.1093/hmg/ddt300. PubMed PMID: 23804751; PubMed Central PMCID: PMC3889807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3955.Arias E, Koga H, Diaz A, et al. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol Cell. 2015. Jul 16;59(2):270–84. doi: 10.1016/j.molcel.2015.05.030. PubMed PMID: 26118642; PubMed Central PMCID: PMC4506737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3956.Bonfili L, Cuccioloni M, Cecarini V, et al. Ghrelin induces apoptosis in colon adenocarcinoma cells via proteasome inhibition and autophagy induction. Apoptosis. 2013. Oct;18(10):1188–200. doi: 10.1007/s10495-013-0856-0. PubMed PMID: 23632965. [DOI] [PubMed] [Google Scholar]
- 3957.Zecchini S, Proietti Serafini F, Catalani E, et al. Dysfunctional autophagy induced by the pro-apoptotic natural compound climacostol in tumour cells. Cell Death Dis. 2018. Dec 19;10(1):10. doi: 10.1038/s41419-018-1254-x. PubMed PMID: 30584259; PubMed Central PMCID: PMCPMC6315039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3958.Sancho A, Duran J, Garcia-Espana A, et al. DOR/Tp53inp2 and Tp53inp1 constitute a metazoan gene family encoding dual regulators of autophagy and transcription. PLoS One. 2012;7(3):e34034. doi: 10.1371/journal.pone.0034034. PubMed PMID: 22470510; PubMed Central PMCID: PMC3314686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3959.Seillier M, Peuget S, Gayet O, et al. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ. 2012. Sep;19(9):1525–35. doi: 10.1038/cdd.2012.30. PubMed PMID: 22421968; PubMed Central PMCID: PMC3422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3960.Mauvezin C, Sancho A, Ivanova S, et al. DOR undergoes nucleo-cytoplasmic shuttling, which involves passage through the nucleolus. FEBS Lett. 2012. Sep 21;586(19):3179–86. doi: 10.1016/j.febslet.2012.06.032. PubMed PMID: 22750142. [DOI] [PubMed] [Google Scholar]
- 3961.Mauvezin C, Orpinell M, Francis VA, et al. The nuclear cofactor DOR regulates autophagy in mammalian and Drosophila cells. EMBO Rep. 2010. Jan;11(1):37–44. doi: 10.1038/embor.2009.242. PubMed PMID: 20010805; PubMed Central PMCID: PMCPMC2816618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3962.Nowak J, Archange C, Tardivel-Lacombe J, et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol Biol Cell. 2009. Feb;20(3):870–81. doi: 10.1091/mbc.E08-07-067; PubMed PMID: 19056683. PubMed Central PMCID: PMC2633384. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3963.Sala D, Ivanova S, Plana N, et al. Autophagy-regulating TP53INP2 mediates muscle wasting and is repressed in diabetes. J Clin Invest. 2014. May 1;124(5):1914–27. doi: 10.1172/JCI72327. PubMed PMID: 24713655; PubMed Central PMCID: PMC4001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3964.Romero M, Sabate-Perez A, Francis VA, et al. TP53INP2 regulates adiposity by activating beta-catenin through autophagy-dependent sequestration of GSK3beta. Nat Cell Biol. 2018. Apr;20(4):443–454. doi: 10.1038/s41556-018-0072-9. PubMed PMID: 29593329. [DOI] [PubMed] [Google Scholar]
- 3965.Lin PH, Duann P, Komazaki S, et al. Lysosomal two-pore channel subtype 2 (TPC2) regulates skeletal muscle autophagic signaling. J Biol Chem. 2015. Feb 6;290(6):3377–89. doi: 10.1074/jbc.M114.608471. PubMed PMID: 25480788; PubMed Central PMCID: PMC4319008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3966.Kobayashi A, Hashizume C, Dowaki T, et al. Therapeutic potential of mitotic interaction between the nucleoporin Tpr and aurora kinase A. Cell cycle. 2015;14(9):1447–58. doi: 10.1080/15384101.2015.1021518. PubMed PMID: 25789545; PubMed Central PMCID: PMCPMC4614903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3967.Zou S, Chen Y, Liu Y, et al. Trs130 participates in autophagy through GTPases Ypt31/32 in Saccharomyces cerevisiae. Traffic. 2013. Feb;14(2):233–46. doi: 10.1111/tra.12024. PubMed PMID: 23078654; PubMed Central PMCID: PMC3538905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3968.DeBosch BJ, Heitmeier MR, Mayer AL, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal. 2016. Feb 23;9(416):ra21. doi: 10.1126/scisignal.aac5472. PubMed PMID: 26905426; PubMed Central PMCID: PMCPMC4816640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3969.Pahari S, Negi S, Aqdas M, et al. Induction of autophagy through CLEC4E in combination with TLR4: an innovative strategy to restrict the survival of Mycobacterium tuberculosis. Autophagy. 2020. Jun;16(6):1021–1043. doi: 10.1080/15548627.2019.1658436. PubMed PMID: 31462144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3970.Hua F, Li K, Yu JJ, et al. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;6:7951. doi: 10.1038/ncomms8951. PubMed PMID: 26268733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3971.Salazar M, Carracedo A, Salanueva IJ, et al. TRB3 links ER stress to autophagy in cannabinoid anti-tumoral action. Autophagy. 2009. Oct, 5(7):1048–9. PubMed PMID: 19652543. [DOI] [PubMed] [Google Scholar]
- 3972.Zhang XW, Zhou JC, Peng D, et al. Disrupting the TRIB3-SQSTM1 interaction reduces liver fibrosis by restoring autophagy and suppressing exosome-mediated HSC activation. Autophagy. 2019. Jul 9:1–15. doi: 10.1080/15548627.2019.1635383. PubMed PMID: 31286822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3973.Francisco R, Perez-Perarnau A, Cortes C, et al. Histone deacetylase inhibition induces apoptosis and autophagy in human neuroblastoma cells. Cancer Lett. 2012. May 1;318(1):42–52. doi: 10.1016/j.canlet.2011.11.036. PubMed PMID: 22186300. [DOI] [PubMed] [Google Scholar]
- 3974.Wang C, Yu Z, Shi X, et al. Triclosan enhances the clearing of pathogenic intracellular salmonella or candida albicans but disturbs the intestinal microbiota through mTOR-independent autophagy. Front Cell Infect Microbiol. 2018;8:49. doi: 10.3389/fcimb.2018.00049. PubMed PMID: 29515975; PubMed Central PMCID: PMCPMC5826388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3975.Lo CH, Lim CK, Ding Z, et al. Targeting the ensemble of heterogeneous tau oligomers in cells: A novel small molecule screening platform for tauopathies. Alzheimers Dement. 2019. Nov;15(11):1489–1502. doi: 10.1016/j.jalz.2019.06.4954. PubMed PMID: 31653529; PubMed Central PMCID: PMCPMC7038631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3976.Ribeiro CM, Sarrami-Forooshani R, Setiawan LC, et al. Receptor usage dictates HIV-1 restriction by human TRIM5alpha in dendritic cell subsets. Nature. 2016. Dec 15;540(7633):448–452. doi: 10.1038/nature20567. PubMed PMID: 27919079. [DOI] [PubMed] [Google Scholar]
- 3977.Imam S, Talley S, Nelson RS, et al. TRIM5alpha degradation via autophagy is not required for retroviral restriction. J Virol. 2016. Jan 13;90(7):3400–10. doi: 10.1128/JVI.03033-15. PubMed PMID: 26764007; PubMed Central PMCID: PMCPMC4794682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3978.Sebastian S, Luban J.. TRIM5alpha selectively binds a restriction-sensitive retroviral capsid. Retrovirology. 2005. Jun 20;2:40. doi: 10.1186/1742-4690-2-40. PubMed PMID: 15967037; PubMed Central PMCID: PMCPMC1166576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3979.Keown JR, Black MM, Ferron A, et al. A helical LC3-interacting region mediates the interaction between the retroviral restriction factor Trim5alpha and mammalian autophagy-related ATG8 proteins. J Biol Chem. 2018. Nov 23;293(47):18378–18386. doi: 10.1074/jbc.RA118.004202. PubMed PMID: 30282803; PubMed Central PMCID: PMCPMC6254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3980.Jena KK, Kolapalli SP, Mehto S, et al. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018. Sep 14;37(18). doi: 10.15252/embj.201798358. PubMed PMID: 30143514; PubMed Central PMCID: PMCPMC6138442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3981.Mandell MA, Jain A, Kumar S, et al. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J Cell Sci. 2016. Oct 1;129(19):3562–3573. doi: 10.1242/jcs.190017. PubMed PMID: 27562068; PubMed Central PMCID: PMCPMC5087653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3982.Wang W, Xia Z, Farre JC, et al. TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis. Autophagy. 2018;14(9):1574–1585. doi: 10.1080/15548627.2018.1463120. PubMed PMID: 29940807; PubMed Central PMCID: PMCPMC6135569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3983.Micale L, Fusco C, Augello B, et al. Williams-Beuren syndrome TRIM50 encodes an E3 ubiquitin ligase. Eur J Human Genet. 2008. Sep;16(9):1038–49. doi: 10.1038/ejhg.2008.68. PubMed PMID: 18398435; PubMed Central PMCID: PMC2680067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3984.Fusco C, Micale L, Augello B, et al. HDAC6 mediates the acetylation of TRIM50. Cell Signal. 2014. Feb;26(2):363–9. doi: 10.1016/j.cellsig.2013.11.036. PubMed PMID: 24308962. [DOI] [PubMed] [Google Scholar]
- 3985.Fusco C, Micale L, Egorov M, et al. The E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and promotes the sequestration and clearance of ubiquitinated proteins into the aggresome. PLoS One. 2012;7(7):e40440. doi: 10.1371/journal.pone.0040440. PubMed PMID: 22792322; PubMed Central PMCID: PMC3392214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3986.Bodine SC, Latres E, Baumhueter S, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001. Nov 23;294(5547):1704–8. doi: 10.1126/science.1065874. PubMed PMID: 11679633. [DOI] [PubMed] [Google Scholar]
- 3987.Centner T, Yano J, Kimura E, et al. Identification of muscle specific ring finger proteins as potential regulators of the titin kinase domain. J Mol Biol. 2001. Mar 2;306(4):717–26. doi: 10.1006/jmbi.2001.4448. PubMed PMID: 11243782. [DOI] [PubMed] [Google Scholar]
- 3988.Gatliff J, East D, Crosby J, et al. TSPO interacts with VDAC1 and triggers a ROS-mediated inhibition of mitochondrial quality control. Autophagy. 2014;10(12):2279–96. doi: 10.4161/15548627.2014.991665. PubMed PMID: 25470454; PubMed Central PMCID: PMC4502750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3989.Huang JH, Liu CY, Wu SY, et al. NLRX1 facilitates histoplasma capsulatum-Induced LC3-associated phagocytosis for cytokine production in macrophages. Front Immunol. 2018;9:2761. doi: 10.3389/fimmu.2018.02761. PubMed PMID: 30559741; PubMed Central PMCID: PMCPMC6286976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3990.Lima RT, Sousa D, Paiva AM, et al. Modulation of autophagy by a thioxanthone decreases the viability of melanoma cells. Molecules. 2016. Oct 10;21(10). doi: 10.3390/molecules21101343. PubMed PMID: 27735867; PubMed Central PMCID: PMCPMC6274546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3991.Palmeira A, Vasconcelos MH, Paiva A, et al. Dual inhibitors of P-glycoprotein and tumor cell growth: (re)discovering thioxanthones. Biochem Pharmacol. 2012. Jan 1;83(1):57–68. doi: 10.1016/j.bcp.2011.10.004. PubMed PMID: 22044878. [DOI] [PubMed] [Google Scholar]
- 3992.DeSilva DR, Jones EA, Favata MF, et al. Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. J Immunol 1998. May 1;160(9):4175–81. PubMed PMID: 9574517. [PubMed] [Google Scholar]
- 3993.Geisler S, Vollmer S, Golombek S, et al. UBE2N, UBE2L3 and UBE2D2/3 ubiquitin-conjugating enzymes are essential for parkin-dependent mitophagy. J Cell Sci. 2014. Jun 6;127:3280–3293. doi: 10.1242/jcs.146035. PubMed PMID: 24906799. [DOI] [PubMed] [Google Scholar]
- 3994.Fiesel FC, Moussaud-Lamodiere EL, Ando M, et al. A specific subset of E2 ubiquitin-conjugating enzymes regulate Parkin activation and mitophagy differently. J Cell Sci. 2014. Aug 15;127\(Pt 16):3488–504. doi: 10.1242/jcs.147520. PubMed PMID: 24928900; PubMed Central PMCID: PMC4132391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3995.Newton K, Matsumoto ML, Wertz IE, et al. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008. Aug 22;134(4):668–78. doi: 10.1016/j.cell.2008.07.039. PubMed PMID: 18724939. [DOI] [PubMed] [Google Scholar]
- 3996.Muller M, Kotter P, Behrendt C, et al. Synthetic quantitative array technology identifies the Ubp3-Bre5 deubiquitinase complex as a negative regulator of mitophagy. Cell Rep. 2015. Feb 24;10(7):1215–25. doi: 10.1016/j.celrep.2015.01.044. PubMed PMID: 25704822. [DOI] [PubMed] [Google Scholar]
- 3997.Rothenberg C, Srinivasan D, Mah L, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010. Aug 15;19(16):3219–32. doi: 10.1093/hmg/ddq231. PubMed PMID: 20529957; PubMed Central PMCID: PMCPMC2908472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3998.Wu JJ, Cai A, Greenslade JE, et al. ALS/FTD mutations in UBQLN2 impede autophagy by reducing autophagosome acidification through loss of function. Proc Natl Acad Sci U S A. 2020. Jun 30;117(26):15230–15241. doi: 10.1073/pnas.1917371117. PubMed PMID: 32513711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3999.Chan EY, Kir S, Tooze SA.. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem. 2007. Aug 31;282(35):25464–74. doi: 10.1074/jbc.M703663200. PubMed PMID: 17595159; eng. [DOI] [PubMed] [Google Scholar]
- 4000.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opinion Cell Biol 2010. Jan 5;22:132–139. doi: 10.1016/j.ceb.2009.12.004. PubMed PMID: 20056399; Eng. [DOI] [PubMed] [Google Scholar]
- 4001.Martin KR, Celano SL, Solitro AR, et al. A potent and selective ULK1 inhibitor suppresses autophagy and sensitizes cancer cells to nutrient stress. iScience. 2018. Oct 26;8:74–84. doi: 10.1016/j.isci.2018.09.012. PubMed PMID: 30292171; PubMed Central PMCID: PMCPMC6172447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4002.Ogura K, Wicky C, Magnenat L, et al. Caenorhabditis elegans unc-51 gene required for axonal elongation encodes a novel serine/threonine kinase. Genes Dev. 1994. Oct 15;8(20):2389–400. doi: 10.1101/gad.8.20.2389. PubMed PMID: 7958904. [DOI] [PubMed] [Google Scholar]
- 4003.He J, Johnson JL, Monfregola J, et al. Munc13-4 interacts with syntaxin 7 and regulates late endosomal maturation, endosomal signaling, and TLR9-initiated cellular responses. Mol Biol Cell. 2016. Feb 1;27(3):572–87. doi: 10.1091/mbc.E15-05-0283. PubMed PMID: 26680738; PubMed Central PMCID: PMCPMC4751605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4004.Zhang J, He J, Johnson JL, et al. Cross-regulation of defective endolysosome trafficking and enhanced autophagy through TFEB in UNC13D deficiency. Autophagy. 2019. Oct;15(10):1738–1756. doi: 10.1080/15548627.2019.1596475. PubMed PMID: 30892133; PubMed Central PMCID: PMCPMC6735675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4005.Munch C. The different axes of the mammalian mitochondrial unfolded protein response. BMC Biol. 2018. Jul 26;16(1):81. doi: 10.1186/s12915-018-0548-x. PubMed PMID: 30049264; PubMed Central PMCID: PMCPMC6060479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4006.von Stockum S, Sanchez-Martinez A, Corra S, et al. Inhibition of the deubiquitinase USP8 corrects a Drosophila PINK1 model of mitochondria dysfunction. Life Sci Alliance. 2019. Apr;2(2). doi: 10.26508/lsa.201900392. PubMed PMID: 30988163; PubMed Central PMCID: PMCPMC6467245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4007.Cornelissen T, Haddad D, Wauters F, et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014. May 22. doi: 10.1093/hmg/ddu244. PubMed PMID: 24852371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4008.Thayer JA, Awad O, Hegdekar N, et al. The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability. Autophagy. 2020. Jan;16(1):140–153. doi: 10.1080/15548627.2019.1598754. PubMed PMID: 30957634; PubMed Central PMCID: PMCPMC6984603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4009.Bingol B, Tea JS, Phu L, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014. Jun 19;510(7505):370–5. doi: 10.1038/nature13418. PubMed PMID: 24896179. [DOI] [PubMed] [Google Scholar]
- 4010.Liang JR, Martinez A, Lane JD, et al. USP30 deubiquitylates mitochondrial Parkin substrates and restricts apoptotic cell death. EMBO Rep. 2015. May;16(5):618–27. doi: 10.15252/embr.201439820. PubMed PMID: 25739811; PubMed Central PMCID: PMCPMC4428036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4011.Wang Y, Serricchio M, Jauregui M, et al. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015. Apr 3;11(4):595–606. doi: 10.1080/15548627.2015.1034408. PubMed PMID: 25915564; PubMed Central PMCID: PMCPMC4502823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4012.Taillebourg E, Gregoire I, Viargues P, et al. The deubiquitinating enzyme USP36 controls selective autophagy activation by ubiquitinated proteins. Autophagy. 2012. May 1;8(5):767–79. doi: 10.4161/auto.19381. PubMed PMID: 22622177. [DOI] [PubMed] [Google Scholar]
- 4013.Pirooz SD, He S, Zhang T, et al. UVRAG is required for virus entry through combinatorial interaction with the class C-Vps complex and SNAREs. Proc Natl Acad Sci U S A. 2014. Feb 18;111(7):2716–21. doi: 10.1073/pnas.1320629111. PubMed PMID: 24550300; PubMed Central PMCID: PMC3932887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4014.Oku M, Nishimura T, Hattori T, et al. Role of Vac8 in formation of the vacuolar sequestering membrane during micropexophagy. Autophagy. 2006. Oct-Dec;2(4):272–9. PubMed PMID: 16874085. [DOI] [PubMed] [Google Scholar]
- 4015.Gatica D, Damasio A, Pascual C, et al. The carboxy terminus of yeast Atg13 binds phospholipid membrane via motifs that overlap with the Vac8-interacting domain. Autophagy. 2020. Jun;16(6):1007–1020. doi: 10.1080/15548627.2019.1648117. PubMed PMID: 31352862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4016.Kosta A, Roisin-Bouffay C, Luciani MF, et al. Autophagy gene disruption reveals a non-vacuolar cell death pathway in Dictyostelium. J Biol Chem. 2004. Nov 12;279(46):48404–9. doi: 10.1074/jbc.M408924200. PubMed PMID: 15358773. [DOI] [PubMed] [Google Scholar]
- 4017.Yamada Y, Cyclic Schaap P.. AMP induction of Dictyostelium prespore gene expression requires autophagy. Dev Biol. 2019. Aug 15;452(2):114–126. doi: 10.1016/j.ydbio.2019.04.017. PubMed PMID: 31051160; PubMed Central PMCID: PMCPMC6598861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4018.Klionsky DJ, Herman PK, Emr SD.. The fungal vacuole: composition, function, and biogenesis. Microbiol Rev 1990. Sep;54(3):266–92. PubMed PMID: 2215422; PubMed Central PMCID: PMC372777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4019.Hoffman M, Chiang H-L.. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996. Aug;143(4):1555–66. PubMed PMID: 8844145; PubMed Central PMCID: PMC1207420. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4020.Zhang C, Lee S, Peng Y, et al. PINK1 triggers autocatalytic activation of Parkin to specify cell fate decisions. Curr Biol. 2014. Aug 18;24(16):1854–65. doi: 10.1016/j.cub.2014.07.014. PubMed PMID: 25088558; PubMed Central PMCID: PMC4143385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4021.Darsow T, Rieder SE, Emr SD.. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J Cell Biol. 1997. Aug 11;138(3):517–29. PubMed PMID: 9245783; PubMed Central PMCID: PMC2141632. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4022.Fader CM, Sanchez DG, Mestre MB, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways [Research Support, Non-U.S. Gov’t]. Biochim Biophys Acta. 2009. Dec;1793(12):1901–16. doi: 10.1016/j.bbamcr.2009.09.011. PubMed PMID: 19781582; eng. [DOI] [PubMed] [Google Scholar]
- 4023.Moreau K, Ravikumar B, Renna M, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011. Jul 22;146(2):303–17. doi: 10.1016/j.cell.2011.06.023. PubMed PMID: 21784250; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4024.Furuta N, Fujita N, Noda T, et al. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical auto-phagosomes with lysosomes. Mol Biol Cell. 2010. Mar;21(6):1001–10. doi: 10.1091/mbc.E09-08-0693. PubMed PMID: 20089838; PubMed Central PMCID: PMC2836953. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4025.Zhao YG, Liu N, Miao G, et al. The ER contact proteins VAPA/B interact with multiple autophagy proteins to modulate autophagosome biogenesis. Curr Biol. 2018. Apr 23;28(8):1234–1245 e4. doi: 10.1016/j.cub.2018.03.002. PubMed PMID: 29628370. [DOI] [PubMed] [Google Scholar]
- 4026.Ju JS, Fuentealba RA, Miller SE, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol. 2009. Dec 14;187(6):875–88. doi: 10.1083/jcb.200908115. PubMed PMID: 20008565; PubMed Central PMCID: PMC2806317. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4027.Tresse E, Salomons FA, Vesa J, et al. VCP/p97 is essential for maturation of ubiquitin-containing auto-phagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010. Feb;6(2):217–27. PubMed PMID: 20104022; PubMed Central PMCID: PMC2929010. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4028.Arhzaouy K, Strucksberg KH, Tung SM, et al. Heteromeric p97/p97R155C complexes induce dominant negative changes in wild-type and autophagy 9-deficient Dictyostelium strains. PLoS One. 2012;7(10):e46879. doi: 10.1371/journal.pone.0046879. PubMed PMID: 23056506; PubMed Central PMCID: PMCPMC3463532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4029.Papadopoulos C, Kirchner P, Bug M, et al. VCP/p97 cooperates with YOD1, UBXD1 and PLAA to drive clearance of ruptured lysosomes by autophagy. EMBO J. 2017. Jan 17;36(2):135–150. doi: 10.15252/embj.201695148. PubMed PMID: 27753622; PubMed Central PMCID: PMCPMC5242375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4030.Yuan J, Zhang Y, Sheng Y, et al. MYBL2 guides autophagy suppressor VDAC2 in the developing ovary to inhibit autophagy through a complex of VDAC2-BECN1-BCL2L1 in mammals. Autophagy. 2015;11(7):1081–98. doi: 10.1080/15548627.2015.1040970. PubMed PMID: 26060891; PubMed Central PMCID: PMCPMC4590641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4031.Yamamoto S, Kuramoto K, Wang N, et al. Autophagy differentially regulates insulin production and insulin sensitivity. Cell Rep. 2018. Jun 12;23(11):3286–3299. doi: 10.1016/j.celrep.2018.05.032. PubMed PMID: 29898399; PubMed Central PMCID: PMCPMC6054876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4032.Kaelin WG, Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer. 2008. Nov;8(11):865–73. doi: 10.1038/nrc2502. PubMed PMID: 18923434. [DOI] [PubMed] [Google Scholar]
- 4033.Dong N, Zhu Y, Lu Q, et al. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012. Aug 31;150(5):1029–41. doi: 10.1016/j.cell.2012.06.050. PubMed PMID: 22939626. [DOI] [PubMed] [Google Scholar]
- 4034.Ogawa M, Yoshimori T, Suzuki T, et al. Escape of intracellular Shigella from autophagy. Science. 2005. Feb 4;307(5710):727–31. doi: 10.1126/science.1106036. PubMed PMID: 15576571. [DOI] [PubMed] [Google Scholar]
- 4035.Vaccaro MI, Ropolo A, Grasso D, et al. A novel mammalian trans-membrane protein reveals an alternative initiation pathway for autophagy. Autophagy. 2008. Apr 1;4(3):388–90. [pii]. PubMed PMID: 18253086; eng. [DOI] [PubMed] [Google Scholar]
- 4036.Calvo-Garrido J, King JS, Munoz-Braceras S, et al. Vmp1 regulates PtdIns3P signaling during autophagosome formation in Dictyostelium discoideum. Traffic. 2014. Nov;15(11):1235–46. doi: 10.1111/tra.12210. PubMed PMID: 25131297. [DOI] [PubMed] [Google Scholar]
- 4037.Molejon MI, Ropolo A, Re AL, et al. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci Rep. 2013;3:1055. doi: 10.1038/srep01055. PubMed PMID: 23316280; PubMed Central PMCID: PMC3542764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4038.Nickerson DP, Brett CL, Merz AJ.. Vps-C complexes: gatekeepers of endolysosomal traffic. Current opinion in cell biology. 2009. Aug;21(4):543–51. doi: 10.1016/j.ceb.2009.05.007. PubMed PMID: 19577915; PubMed Central PMCID: PMC2807627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4039.Clancey LF, Beirl AJ, Linbo TH, et al. Maintenance of melanophore morphology and survival is cathepsin and vps11 dependent in zebrafish. PLoS One. 2013;8(5):e65096. doi: 10.1371/journal.pone.0065096. PubMed PMID: 23724125; PubMed Central PMCID: PMC3664566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4040.Munoz-Braceras S, Tornero-Ecija AR, Vincent O, et al. VPS13A is closely associated with mitochondria and is required for efficient lysosomal degradation. Dis Model Mech. 2019. Feb 22;12(2). doi: 10.1242/dmm.036681. PubMed PMID: 30709847; PubMed Central PMCID: PMCPMC6398486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4041.Kumar N, Leonzino M, Hancock-Cerutti W, et al. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol. 2018. Oct 1;217(10):3625–3639. doi: 10.1083/jcb.201807019. PubMed PMID: 30093493; PubMed Central PMCID: PMCPMC6168267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4042.Rzepnikowska W, Flis K, Kaminska J, et al. Amino acid substitution equivalent to human chorea-acanthocytosis I2771R in yeast Vps13 protein affects its binding to phosphatidylinositol 3-phosphate. Hum Mol Genet. 2017. Apr 15;26(8):1497–1510. doi: 10.1093/hmg/ddx054. PubMed PMID: 28334785; PubMed Central PMCID: PMCPMC5393151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4043.Kolakowski D, Kaminska J, Zoladek T.. The binding of the APT1 domains to phosphoinositides is regulated by metal ions in vitro. Biochim Biophys Acta Biomembr. 2020. Sep 1;1862(9):183349. doi: 10.1016/j.bbamem.2020.183349. PubMed PMID: 32407779. [DOI] [PubMed] [Google Scholar]
- 4044.Anding AL, Wang C, Chang TK, et al. Vps13D Encodes a Ubiquitin-Binding Protein that Is Required for the Regulation of Mitochondrial Size and Clearance. Curr Biol CB 2018. Jan 22;28(2):287–295 e6. doi: 10.1016/j.cub.2017.11.064. PubMed PMID: 29307555; PubMed Central PMCID: PMCPMC5787036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4045.Huotari J, Helenius A.. Endosome maturation. EMBO J. 2011. Aug 31;30(17):3481–500. doi: 10.1038/emboj.2011.286. PubMed PMID: 21878991; PubMed Central PMCID: PMCPMC3181477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4046.Khatter D, Raina VB, Dwivedi D, et al. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex on lysosomes. J Cell Sci. 2015. May 1;128(9):1746–61. doi: 10.1242/jcs.162651. PubMed PMID: 25908847; PubMed Central PMCID: PMCPMC4432227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4047.Pu J, Schindler C, Jia R, et al. BORC, a multisubunit complex that regulates lysosome positioning. Dev Cell. 2015. Apr 20;33(2):176–88. doi: 10.1016/j.devcel.2015.02.011. PubMed PMID: 25898167; PubMed Central PMCID: PMCPMC4788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4048.Ruan Q, Harrington AJ, Caldwell KA, et al. VPS41, a protein involved in lysosomal trafficking, is protective in Caenorhabditis elegans and mammalian cellular models of Parkinson’s disease. Neurobiol Dis. 2010. Feb;37(2):330–8. doi: 10.1016/j.nbd.2009.10.011. PubMed PMID: 19850127; PubMed Central PMCID: PMCPMC2818321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4049.Harrington AJ, Yacoubian TA, Slone SR, et al. Functional analysis of VPS41-mediated neuroprotection in Caenorhabditis elegans and mammalian models of Parkinson’s disease. J Neurosci Off J Soc Neurosci 2012. Feb 8;32(6):2142–53. doi: 10.1523/JNEUROSCI.2606-11.2012. PubMed PMID: 22323726; PubMed Central PMCID: PMCPMC6621695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4050.Uttenweiler A, Schwarz H, Neumann H, et al. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol Biol Cell. 2007. Jan;18(1):166–75. doi: 10.1091/mbc.E06-08-0664. PubMed PMID: 17079729; PubMed Central PMCID: PMC1751332. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4051.Horos R, Buscher M, Kleinendorst R, et al. The Small Non-coding Vault RNA1-1 Acts as a Riboregulator of Autophagy. Cell. 2019. Feb 21;176(5):1054–1067 e12. doi: 10.1016/j.cell.2019.01.030. PubMed PMID: 30773316. [DOI] [PubMed] [Google Scholar]
- 4052.Simonsen A, Birkeland HC, Gillooly DJ, et al. Alfy, a novel FYVE-domain-containing protein associated with protein granules and autophagic membranes. J Cell Sci. 2004. Aug 15;117\(Pt 18):4239–51. doi: 10.1242/jcs.01287jcs.01287 [pii]. PubMed PMID: 15292400; eng. [DOI] [PubMed] [Google Scholar]
- 4053.Napoli E, Song G, Panoutsopoulos A, et al. Beyond autophagy: a novel role for autism-linked Wdfy3 in brain mitophagy. Sci Rep. 2018. Jul 27;8(1):11348. doi: 10.1038/s41598-018-29421-7. PubMed PMID: 30054502; PubMed Central PMCID: PMCPMC6063930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4054.Clausen TH, Lamark T, Isakson P, et al. p62/SQSTM1 and ALFY interact to facilitate the formation of p62 bodies/ALIS and their degradation by autophagy. Autophagy. 2010. Apr;6(3):330–44. PubMed PMID: 20168092; eng. [DOI] [PubMed] [Google Scholar]
- 4055.Kast DJ, Zajac AL, Holzbaur EL, et al. WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism. Curr Biol. 2015. Jun 29;25(13):1791–7. doi: 10.1016/j.cub.2015.05.042. PubMed PMID: 26096974; PubMed Central PMCID: PMC4489997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4056.Dai A, Yu L, Wang HW.. WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat Commun. 2019. Aug 16;10(1):3699. doi: 10.1038/s41467-019-11694-9. PubMed PMID: 31420534; PubMed Central PMCID: PMCPMC6697732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4057.Haack TB, Hogarth P, Kruer MC, et al. Exome sequencing reveals de novo WDR45 mutations causing a phenotypically distinct, X-linked dominant form of NBIA. Am J Hum Genet. 2012. Dec 7;91(6):1144–9. doi: 10.1016/j.ajhg.2012.10.019. PubMed PMID: 23176820; PubMed Central PMCID: PMC3516593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4058.Abidi A, Mignon-Ravix C, Cacciagli P, et al. Early-onset epileptic encephalopathy as the initial clinical presentation of WDR45 deletion in a male patient. Eur J Human Genet. 2015. Jul 15. doi: 10.1038/ejhg.2015.159. PubMed PMID: 26173968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4059.Saitsu H, Nishimura T, Muramatsu K, et al. De novo mutations in the autophagy gene WDR45 cause static encephalopathy of childhood with neurodegeneration in adulthood. Nat Genet. 2013. Apr;45(4):445–9, 449e1. doi: 10.1038/ng.2562. PubMed PMID: 23435086. [DOI] [PubMed] [Google Scholar]
- 4060.Biagosch CA, Hensler S, Kühn R, et al. ALEN-mediated mutagenesis as a tool to generate disease models for diseases caused by dominant de novo mutations. Eur J Human Gen EJHG 2014;22:153. [Google Scholar]
- 4061.Gallolu Kankanamalage S, Lee AY, Wichaidit C, et al. Multistep regulation of autophagy by WNK1. Proc Natl Acad Sci U S A. 2016. Dec 13;113(50):14342–14347. doi: 10.1073/pnas.1617649113. PubMed PMID: 27911840; PubMed Central PMCID: PMCPMC5167150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4062.Maiese K, Chong ZZ, Shang YC, et al. Targeting disease through novel pathways of apoptosis and autophagy. Expert Opin Ther Targets. 2012. Dec;16(12):1203–14. doi: 10.1517/14728222.2012.719499. PubMed PMID: 22924465; PubMed Central PMCID: PMC3500415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4063.Petherick KJ, Williams AC, Lane JD, et al. Autolysosomal beta-catenin degradation regulates Wnt-autophagy-p62 crosstalk. EMBO J. 2013. Jul 3;32(13):1903–16. doi: 10.1038/emboj.2013.123. PubMed PMID: 23736261; PubMed Central PMCID: PMC3981178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4064.Sasazawa Y, Kanagaki S, Tashiro E, et al. Xanthohumol impairs autophagosome maturation through direct inhibition of valosin-containing protein. ACS Chem Biol. 2012. May 18;7(5):892–900. doi: 10.1021/cb200492h. PubMed PMID: 22360440. [DOI] [PubMed] [Google Scholar]
- 4065.Kaser A, Blumberg RS.. Endoplasmic reticulum stress in the intestinal epithelium and inflammatory bowel disease. Semin Immunol. 2009. Jun;21(3):156–63. doi: 10.1016/j.smim.2009.01.001. PubMed PMID: 19237300; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4066.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005. Jan 28;120(2):159–62. PubMed PMID: 15680321. [DOI] [PubMed] [Google Scholar]
- 4067.Criollo A, Maiuri MC, Tasdemir E, et al. Regulation of autophagy by the inositol trisphosphate receptor [Research Support, Non-U.S. Gov’t]. Cell Death Differ. 2007. May;14(5):1029–39. doi: 10.1038/sj.cdd.4402099. PubMed PMID: 17256008; eng. [DOI] [PubMed] [Google Scholar]
- 4068.Huang X, Wu Z, Mei Y, et al. XIAP inhibits autophagy via XIAP-Mdm2-p53 signalling. EMBO J. 2013. Aug 14;32(16):2204–16. doi: 10.1038/emboj.2013.133. PubMed PMID: 23749209; PubMed Central PMCID: PMCPMC3746193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4069.Huang X, Wang XN, Yuan XD, et al. XIAP facilitates breast and colon carcinoma growth via promotion of p62 depletion through ubiquitination-dependent proteasomal degradation. Oncogene. 2019. Feb;38(9):1448–1460. doi: 10.1038/s41388-018-0513-8. PubMed PMID: 30275562. [DOI] [PubMed] [Google Scholar]
- 4070.Martin AP, Jacquemyn M, Lipecka J, et al. STK38 kinase acts as XPO1 gatekeeper regulating the nuclear export of autophagy proteins and other cargoes. EMBO Rep. 2019. Nov 5;20(11):e48150. doi: 10.15252/embr.201948150. PubMed PMID: 31544310; PubMed Central PMCID: PMCPMC6832005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4071.Liang XH, Yu J, Brown K, et al. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer res 2001. Apr 15;61(8):3443–9. PubMed PMID: 11309306. [PubMed] [Google Scholar]
- 4072.Delorme-Axford E, Abernathy E, Lennemann NJ, et al. The exoribonuclease Xrn1 is a post-transcriptional negative regulator of autophagy. Autophagy. 2018;14(5):898–912. doi: 10.1080/15548627.2018.1441648. PubMed PMID: 29465287; PubMed Central PMCID: PMCPMC6070002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4073.Kweon Y, Rothe A, Conibear E, et al. Ykt6p is a multifunctional yeast R-SNARE that is required for multiple membrane transport pathways to the vacuole, Mol Biol Cell. 2003 May;14(5):1868–81. doi: 10.1091/mbc.E02-10-0687. PubMed PMID: 12802061; PubMed Central PMCID: PMC165083. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4074.Takats S, Glatz G, Szenci G, et al. Non-canonical role of the SNARE protein Ykt6 in autophagosome-lysosome fusion. PLoS Genet. 2018. Apr;14(4):e1007359. doi: 10.1371/journal.pgen.1007359. PubMed PMID: 29694367; PubMed Central PMCID: PMCPMC5937789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4075.Kriegenburg F, Bas L, Gao J, et al. The multi-functional SNARE protein Ykt6 in autophagosomal fusion processes. Cell cycle. 2019. Mar - Apr;18(6–7):639–651. doi: 10.1080/15384101.2019.1580488. PubMed PMID: 30836834; PubMed Central PMCID: PMCPMC6464585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4076.Cebollero E, van der Vaart A, Zhao M, et al. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012. Sep 11;22(17):1545–53. doi: 10.1016/j.cub.2012.06.029. PubMed PMID: 22771041; PubMed Central PMCID: PMC3615650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4077.Cheng J, Fujita A, Yamamoto H, et al. Yeast and mammalian auto-phagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun. 2014;5:3207. doi: 10.1038/ncomms4207. PubMed PMID: 24492518. [DOI] [PubMed] [Google Scholar]
- 4078.Huang J, Birmingham CL, Shahnazari S, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy. 2011. Jan;7(1):17–26. PubMed PMID: 20980813; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4079.Zoppino FC, Militello RD, Slavin I, et al. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites [Research Support, Non-U.S. Gov’t]. Traffic. 2010. Sep;11(9):1246–61. doi: 10.1111/j.1600-0854.2010.01086.x. PubMed PMID: 20545908; eng. [DOI] [PubMed] [Google Scholar]
- 4080.Vantaggiato C, Crimella C, Airoldi G, et al. Defective autophagy in spastizin mutated patients with hereditary spastic paraparesis type 15. Brain. 2013. Oct;136(Pt 10):3119–39. doi: 10.1093/brain/awt227. PubMed PMID: 24030950; PubMed Central PMCID: PMC3784282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4081.Khandelwal VK, Mitrofan LM, Hyttinen JM, et al. Oxidative stress plays an important role in zoledronic acid-induced autophagy. Physiol Res 2014;63 Suppl 4:S601–12. PubMed PMID: 25669691. [DOI] [PubMed] [Google Scholar]
- 4082.Schneider EM, Lorezn M, Walther P.. Autophagy as a hallmark of hemophagocytic diseases In: Gorbunov N, editor. Autophagy: Principles, Regulation and Roles in Disease: Nova Science Publishers; 2012. [Google Scholar]
- 4083.Ryhanen T, Hyttinen JM, Kopitz J, et al. Crosstalk between Hsp70 molecular chaperone, lysosomes and proteasomes in autophagy-mediated proteolysis in human retinal pigment epithelial cells. J Cell Mol Med. 2009. Sep;13(9B):3616–31. doi: 10.1111/j.1582-4934.2008.00577.x. PubMed PMID: 19017362; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4084.Cheong JK, Zhang F, Chua PJ, et al. Casein kinase 1alpha-dependent feedback loop controls autophagy in RAS-driven cancers. J Clin Invest. 2015. Apr;125(4):1401–18. doi: 10.1172/JCI78018. PubMed PMID: 25798617; PubMed Central PMCID: PMCPMC4396475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4085.Amadoro G, Corsetti V, Florenzano F, et al. Morphological and bioenergetic demands underlying the mitophagy in post-mitotic neurons: the pink-parkin pathway. Front Aging Neurosci. 2014;6:18. doi: 10.3389/fnagi.2014.00018. PubMed PMID: 24600391; PubMed Central PMCID: PMC3927396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4086.Kang C, Avery L.. Systemic regulation of starvation response in Caenorhabditis elegans. Genes Dev. 2009. Jan 1;23(1):12–7. doi: 10.1101/gad.1723409. PubMed PMID: 19136622; PubMed Central PMCID: PMC2632168. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]


