Abstract
Members of Dodonaea are broadly distributed across subtropical and tropical areas of southwest and southern China. This host provides multiple substrates that can be richly colonized by numerous undescribed fungal species. There is a severe lack of microfungal studies on Dodonaea in China, and consequently, the diversity, phylogeny and taxonomy of these microorganisms are all largely unknown. This paper presents two new genera and four new species in three orders of Dothideomycetes gathered from dead twigs of Dodonaea viscosa in Honghe, China. All new collections were made within a selected area in Honghe from a single Dodonaea sp. This suggests high fungal diversity in the region and the existence of numerous species awaiting discovery. Multiple gene sequences (non-translated loci and protein-coding regions) were analysed with maximum likelihood and Bayesian analyses. Results from the phylogenetic analyses supported placing Haniomyces dodonaeae gen. et sp. in the Teratosphaeriaceae family. Analysis of Rhytidhysteron sequences resulted in Rhytidhysteron hongheense sp. nov., while analysed Lophiostomataceae sequences revealed Lophiomurispora hongheensis gen. et sp. nov. Finally, phylogeny based on a combined dataset of pyrenochaeta-like sequences demonstrates strong statistical support for placing Quixadomyces hongheensis sp. nov. in Parapyrenochaetaceae. Morphological and updated phylogenetic circumscriptions of the new discoveries are also discussed.
Keywords: Ascomycota, Asexual morph, Capnodiales, Greater Mekong Subregion, Hysteriales, Pleosporales, Sexual morph, Yunnan
1. Introduction
Fungi are cosmopolitan, featuring a broad geographic distribution and high level of diversity compared to plants and other organisms [1]. 140,000 fungal species have been listed in Kirk [2], and one recent overview of global fungi and fungus-like taxa by Wijayawardene et al. [3] listed approximately 100,000 known taxa. However, both numbers represent less than 5% of global fungal estimates [4,5]. There is a need to bridge the gap between our understanding of these missing fungi and their diversity. Numerous diverse habitats and substrates remain unexplored. It has also been observed that several countries and regions are bountiful repositories of many missing fungi, such as northern Thailand [6]. Despite this, fungi in Asia are relatively understudied [5]. Even though the Greater Mekong Subregion (GMS) hosts a high level of biodiversity and forms an integral part of the Indo-Burma Biodiversity Hotspot, fungi from this region largely remain a mystery. Yunnan Province, China, as part of the GMS, is home to an extremely wide variety of ecosystems. Mycologists working in Yunnan have recently focused their attention on abundant “less-researched habitats” for fungal occurrences, including caves, forests, grasslands, lakes, karst landscapes and mountains; accordingly, there is a rich body of literature documenting novel discoveries across the region [7,8,9,10,11,12,13,14,15,16,17,18,19].
The Honghe Hani and Yi Autonomous Prefecture is in south-eastern Yunnan Province. The region features a mountainous topography, numerous limestone deposits and a south-eastward decreasing elevation gradient. Owing to its abundant precipitation and heat as well as its dramatic altitudinal range and varied flora, this region harbours a rich diversity of plant species [20,21]. Along the altitudinal gradient, vegetation from lower to higher elevations range from tropical and montane rain forests to monsoon evergreen, montane mossy evergreen and summit mossy evergreen broad-leaved forests [22]. This complex topography and climatic diversity are both significant contributors to local biodiversity richness [23]. Among publications documenting fungal encounters across Yunnan Province, ascomycetes are critically neglected when compared to the amount of research on basidiomycetes [24]. Regrettably, studies on microfungi in Honghe are virtually non-existent. Except Marasinghe et al. [25], we could not find a single detailed account of microfungi in Honghe based on both morphological and phylogenetic analyses.
Dodonaea viscosa is a perennial evergreen woody shrub belonging to the family Sapindaceae. It is drought- and pollution-resistant as well as capable of growing on poor soils and rocky sites. The plant can also easily inhabit open areas and secondary forests [26,27]. A fast-growing plant, it typically grows 1 to 3 m in height but on rare occasions can reach up to 8 m [28]. Dodonaea viscosa is believed to have originated from Australia [29], though it grows throughout tropical and subtropical countries, including the African, Asian, Northern American and Southern American continents [30,31,32]. Dodonaea viscosa is effective at performing sand dune fixation and controlling coastal erosion since its roots function as excellent soil binders [33]. It can also be used to reclaim marshes. It is also grown as an ornamental plant owing to its shiny foliage and pink–red winged fruit [33]. Moreover, it is a well-known topic in environmental impact studies to determine the growth and yield of crops based on the presence of D. viscosa [27,34] as well as study its capacity to increase resilience to pollution [35,36] and drought [37]. In traditional medicine systems, plant parts such as the stem, leaves, seeds, roots, bark and aerial parts are used for various treatments [38]. Hossain [39] reported that extract obtained from D. viscosa has shown significant antidiabetic, antimicrobial, insecticidal, antioxidant, cytotoxic, antifertility, anti-inflammatory, analgesic, anti-ulcer, antispasmodic, anti-diarrheal and detoxification properties [27].
This study is the second in a series comprising an exhaustive taxonomic effort to document the microfungi of Yunnan Province [24]. In this study, we collected fresh fungal specimens from dead woody twigs of Dodonaea species at the Centre for Mountain Futures (CMF), an applied research centre jointly managed by World Agroforestry (ICRAF) and the Kunming Institute of Botany, Chinese Academy of Sciences (CAS), in Honghe County of the Honghe Hani and Yi Autonomous Prefecture. Using morphology and multi-gene phylogenetic evidence retrieved from the gathered ascomycetes, we characterized two new genera and four new species in the orders Capnodiales, Hysteriales and Pleosporales from dead twigs of Dodonaea viscosa in Honghe.
2. Materials and Methods
2.1. Herbarium Material and Fungal Strains
Fresh fungal materials were gathered from dead twigs of Dodonaea viscosa at CMF in Honghe County (Yunnan Province, China UTM/WGS84: 48 Q 216849–217075 E, 2592645–2592856 N, 600–750 m above sea level) during the dry season (April 2020). The local environment is characterized by poor eroded soils, steep valleys and a subtropical monsoon climate. Specimens were transported to the laboratory in Ziploc bags. Single spore isolation was conducted in accordance with methods described in Wanasinghe et al. [40]. Germinated spores were individually placed on potato dextrose agar (PDA) plates and grown at 20 °C in daylight. Dry herbarium materials were stored in the herbarium of Cryptogams Kunming Institute of Botany, Academia Sinica (KUN-HKAS). Living cultures were deposited at the Kunming Institute of Botany Culture Collection (KUMCC), Kunming, China and duplicated at China General Microbiological Culture Collection Centre (CGMCC). MycoBank numbers were registered as outlined in MycoBank (http://www.MycoBank.org accessed on 11 November 2020).
2.2. Morphological Observations
The morphology of external and internal macro-/micro-structures were observed as described in Wanasinghe et al. [24]. Images were captured with a Canon EOS 600D digital camera fitted to a Nikon ECLIPSE Ni compound microscope. Measurements were made with the Tarosoft (R) Image Frame Work program, and images used for figures were processed with Adobe Photoshop CS5 Extended version 10.0 software (Adobe Systems, San José, CA, USA).
2.3. DNA Extraction, PCR Amplifications and Sequencing
The extraction of genomic DNA was performed in accordance with the methods of Wanasinghe et al. [24], using the Biospin Fungus Genomic DNA Extraction Kit-BSC14S1 (BioFlux, P.R. China) following the instructions of the manufacturer. The reference DNA for the polymerase chain reaction (PCR) was stored at 4 °C for regular use and duplicated at −20 °C for long-term storage. The primers and protocols used for the amplification are summarized in Table 1. The amplified PCR fragments were then sent to a private company for sequencing (BGI, Ltd. Shenzhen, P.R. China).
Table 1.
Genes/loci used in the study with PCR primers, references and protocols.
| Locus a | Primers b | PCR: Thermal Cycles: c (Annealing temp. in Bold) |
References |
|---|---|---|---|
| act | ACT-512F ACT2Rd |
(96 °C: 120 s, 52 °C: 60 s, 72 °C: 90 s) × 40 cycles | [41,42] |
| btub | TUB2Fw TUB4Rd |
(94 °C: 30 s, 56 °C: 45 s, 72 °C: 60 s) × 35 cycles | [43] |
| cal | CAL-235F CAL2Rd |
(96 °C: 120 s, 50 °C: 60 s, 72 °C: 90 s) × 40 cycles | [42,44] |
| ITS | ITS5 ITS4 |
(95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [45] |
| LSU | LR0R LR5 |
(95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [46,47] |
| rpb2 | fRPB2-5f fRPB2-7cR |
(94 °C: 60 s, 58 °C: 60 s, 72 °C: 90 s) × 40 cycles | [48] |
| fRPB2-414R | (96 °C: 120 s, 49 °C: 60 s, 72 °C: 90 s) × 40 cycles | [49] | |
| SSU | NS1 NS4 |
(95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [45] |
| tef1 | EF1-983F EF1-2218R |
(95 °C: 30 s, 55 °C:50 s, 72 °C: 90 s) × 35 cycles | [50,51] |
| EF1-728F EF-2 |
(96 °C: 120 s, 52 °C: 60 s, 72 °C: 90 s) × 40 cycles | [41,52] |
aact: actin; btub: β-tubulin; cal: calmodulin; ITS: part of rDNA 18S (3’ end), the first internal transcribed spacer (ITS1), the 5.8S rRNA gene, the second ITS region (ITS2), and part of the 28S rRNA (5’ end); LSU: large subunit (28S); rpb2: RNA polymerase II second largest subunit; SSU: small subunit rDNA (18S); tef1: translation elongation factor 1-alpha gene. b fRPB2-5f and fRPB2-414R were used only for Teratosphaeriaceae analysis. c All the PCR thermal cycles include initiation step of 95 °C: 5 min, and final elongation step of 72 °C: 10 min and final hold at 4 °C.
2.4. Molecular Phylogenetic Analyses
2.4.1. Sequence Alignment
Sequences featuring a high degree of similarity were determined from a BLAST search to identify the closest matches with taxa in Dothideomycetes and recently published data [49,53,54,55,56]. Initial alignments of the acquired sequence data were first completed using MAFFT v. 7 (http://mafft.cbrc.jp/alignment/server/index.html accessed on 18 January 2021) [57,58] and manually clarified in BioEdit v. 7.0.5.2 when indicated [59].
2.4.2. Phylogenetic Analyses
Single-locus data sets were scanned for topological incongruences between loci for members of the analyses. Conflict-free alignments were concatenated into a multi-locus alignment that underwent maximum-likelihood (ML) and Bayesian (BI) phylogenetic analyses. Evolutionary models for BI and ML were selected independently for every locus using MrModeltest v. 2.3 [60] under the Akaike Information Criterion (AIC) implemented in PAUP v. 4.0b10.
The CIPRES Science Gateway platform [61] was used to perform RAxML and Bayesian analyses. ML analyses were made with RAxML-HPC2 on XSEDE v. 8.2.10 [62] employing the GTR+GAMMA swap model with 1000 bootstrap repetitions.
MrBayes analyses were performed setting GTR+I+GAMMA for 2–5 million generations, sampling every 100 generations and ending the run automatically when standard deviation of split frequencies dropped below 0.01 with a burnin fraction of 0.25. ML bootstrap values equal or greater than 60% and Bayesian posterior probabilities (BYPPs) greater than 0.95 were placed above each node of every tree.
Phylograms were visualized with FigTree v1.4.0 program [63] and reassembled in Microsoft PowerPoint (2007) and Adobe Illustrator® CS5 (Version 15.0.0, Adobe®, San Jose, CA, USA). Finalized alignments and trees were deposited in TreeBASE, submission ID: S27699 (http://purl.org/phylo/treebase/phylows/study/TB2: S27699).
3. Results
3.1. Global Checklist of Fungi on Dodonaea Viscosa
Information for the global checklist (Table 2) was retrieved from the Agriculture Research Service Database generated by the United States Department of Agriculture (USDA) [64], related books and research papers. This checklist includes fungal species associated with Dodonaea viscosa and the countries from which they were recorded.
Table 2.
Checklist of fungi recorded from Dodonaea viscosa in worldwide.
| Phylum and Class | Order | Family | Species | Country | References |
|---|---|---|---|---|---|
| Ascomycota | |||||
| Dothideomycetes | Botryosphaeriales | Botryosphaeriaceae | Lasiodiplodia iraniensis | Australia | [65] |
| Macrophoma dodonaeae | India | [66] | |||
| Macrophomina phaseolina | Arizona | [67] | |||
| Capnodiales | Capnodiaceae | Antennariella californica | Fiji | [68] | |
| Mycosphaerellaceae | Cercospora dodonaeae | India | [69,70,71] | ||
| Cercospora sp. | Sierra Leone | [72] | |||
| Pseudocercospora dodonaeae | New Zealand | [73,74,75,76,77,78] | |||
| Pseudocercospora mitteriana | China | [79] | |||
| India | [69,71] | ||||
| Pakistan | [71,80] | ||||
| Teratosphaeriaceae | Haniomyces dodonaeae | China | This study | ||
| Hysteriales | Hysteriaceae | Rhytidhysteron hongheense | China | This study | |
| incertae sedis | Pseudoperisporiaceae | Episphaerella dodonaeae | Dominican Republic | [81] | |
| Ecuador | [82] | ||||
| Venezuela | [82] | ||||
| USA | [83] | ||||
| incertae sedis | Mycothyridium pakistanicum | Pakistan | [80] | ||
| Mycothyridium roosselianum | Pakistan | [80] | |||
| Patellariales | Patellariaceae | Tryblidaria pakistani | Pakistan | [80] | |
| Pleosporales | Coniothyriaceae | Coniothyrium sp. | Venezuela | [84] | |
| Corynesporascaceae | Corynespora cassiicola | India | [85] | ||
| Didymosphaeriaceae | Didymosphaeria oblitescens | Pakistan | [80] | ||
| Leptosphaeriaceae | Leptosphaeria dodonaeae | Eritrea | [86] | ||
| Lophiostomataceae | Lophiomurispora hongheensis | China | This study | ||
| Parapyrenochaetaceae | Quixadomyces hongheensis | China | This study | ||
| Pleosporaceae | Pleospora dodonaeae | Cyprus | [87] | ||
| Valsariales | Valsariaceae | Valsaria rubricosa | Pakistan | [80] | |
| Lecanoromycetes | Ostropales | Stictidaceae | Stictis marathwadensis | India | [88,89] |
| Leotiomycetes | Helotiales | Erysiphaceae | Oidium sp. | Iraq | [90] |
| Israel | [90] | ||||
| South Africa | [90] | ||||
| Zimbabwe | [91] | ||||
| Ovulariopsis erysiphoides | Ethiopia | [92] | |||
| Phyllactiniasp. | Ethiopia | [90] | |||
| Sawadaea bicornis | Germany | [93] | |||
| New Zealand | [74,90] | ||||
| South Africa | [90] | ||||
| Takamatsuella circinata | South Africa | [94] | |||
| Sordariomycetes | Diaporthales | Cytosporaceae | Cytospora sp. | USA | [95] |
| Glomerellales | Glomerellaceae | Colletotrichum gloeosporioides | India | [88] | |
| Meliolales | Meliolaceae | Meliola lyoni | Hawaii | [96,97,98,99] | |
| Hypocreales | Nectriaceae | Calonectria cylindrospora | USA | [100,101] | |
| Calonectria pauciramosa | Italy | [102] | |||
| Fusarium solani | Iran | [103] | |||
| Glomerellales | Plectosphaerellaceae | Verticillium dahliae | USA | [95] | |
| New Zealand | [74] | ||||
| Coronophorales | Scortechiniaceae | Tympanopsis lantanae | India | [104] | |
| Amphisphaeriales | Sporocadaceae | Monochaetia dodoneae | Ethiopia | [92] | |
| Pestalotia dodonaeae | Eritrea | [86] | |||
| Sarcostroma kennedyae | New Zealand | [74] | |||
| Seimatosporium kennedyae | New Zealand | [73] | |||
| Togniniales | Togniniaceae | Phaeoacremonium alvesii | Australia | [105,106,107,108] | |
| Phaeoacremonium italicum | Australia | [109] | |||
| Basidiomycota | |||||
| Agaricomycetes | Agaricales | Marasmiaceae | Campanella junghuhnii | Hawaii | [110] |
| Agaricales | incertae sedis | Dendrothele incrustans | New Zealand | [111] | |
| Bartheletiomycetes | Cantharellales | Ceratobasidiaceae | Rhizoctonia sp. | Italy | [112] |
| Hymenochaetales | Hymenochaetaceae | Arambarria cognata | Uruguay | [113] | |
| Fomitiporia australiensis | Australia | [114] | |||
| Phellinus melleoporus | Hawaii | [110] | |||
| Phellinus robustus | USA | [115] | |||
| Phellinus sonorae | USA | [116] | |||
| Schizoporaceae | Hyphodontia alutaria | Hawaii | [110] | ||
| Grandinia breviseta | Hawaii | [110] | |||
| Polyporales | Hyphodermataceae | Hyphoderma sphaeropedunculatum | Hawaii | [110] | |
| Pucciniomycetes | Pucciniales | incertae sedis | Uredo dodonaeae | Indonesia | [117] |
| Oomycota | |||||
| Peronosporomycetes | Peronosporales | Peronosporaceae | Phytophthora drechsleri | Australia | [118,119,120] |
| Phytophthora nicotianae | Italy | [121,122,123] | |||
| Phytophthora palmivora | Italy | [123] | |||
| Pythiaceae | Globisporangium debaryanum | New Zealand | [73,74] | ||
| Globisporangium irregulare | New Zealand | [74] | |||
| Globisporangium ultimum | New Zealand | [73] | |||
| Pythium inflatum | New Zealand | [73,74] | |||
| Pythium sp. | New Zealand | [73] | |||
| USA | [75] |
3.2. Phylogenetic Analyses
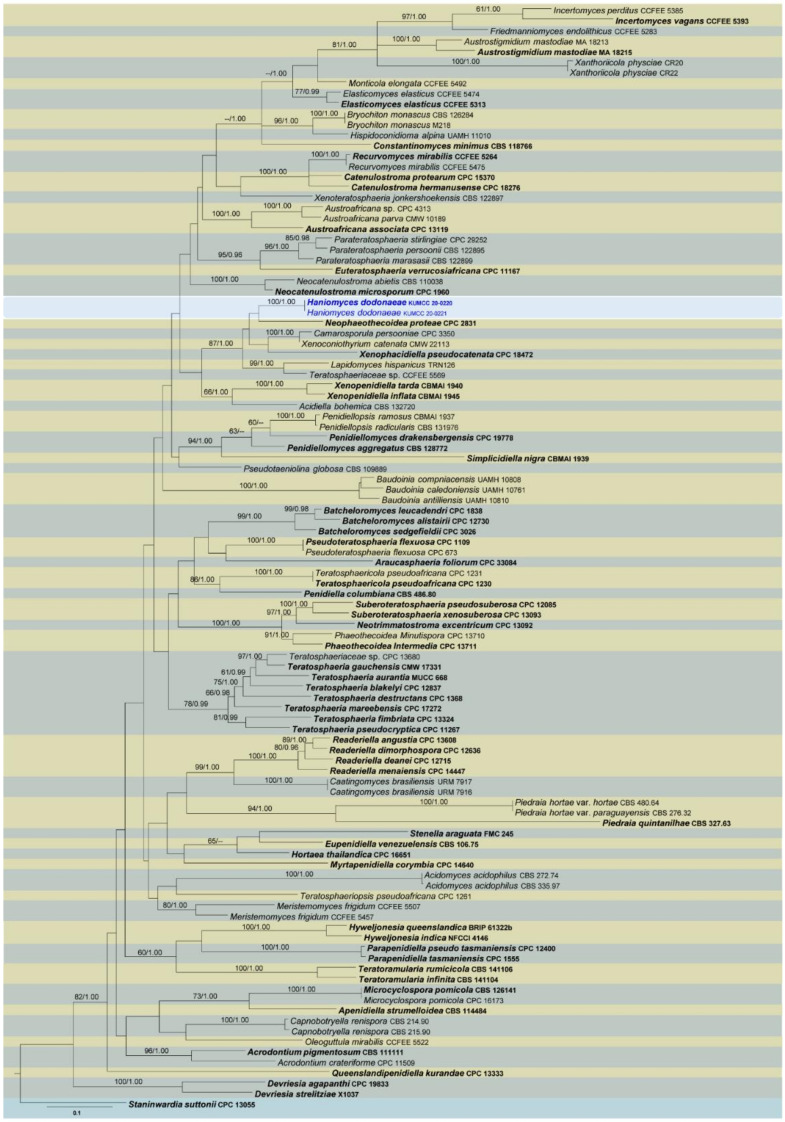
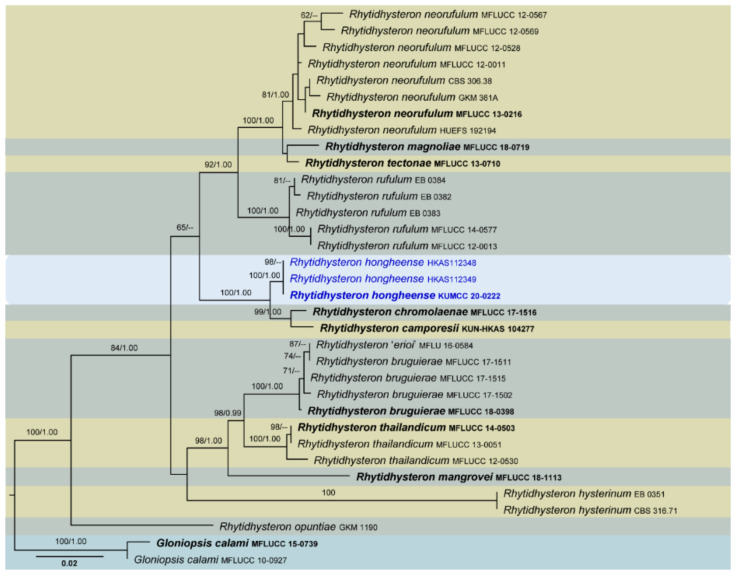
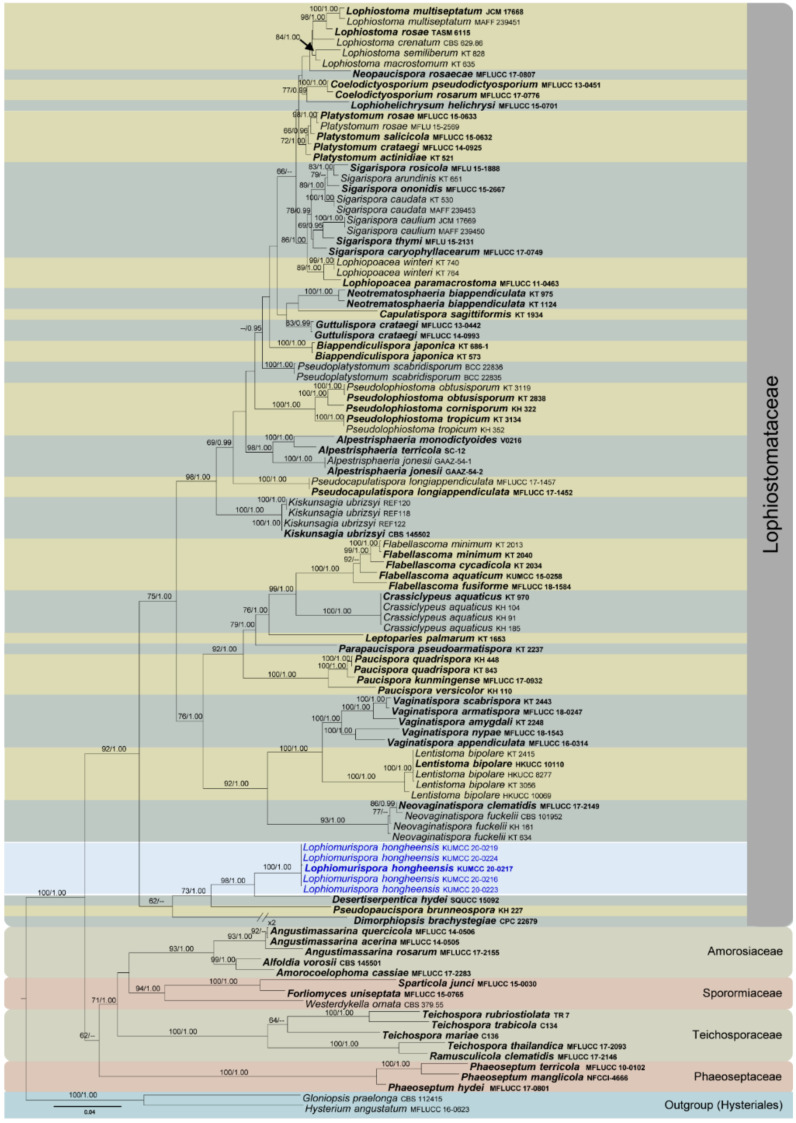
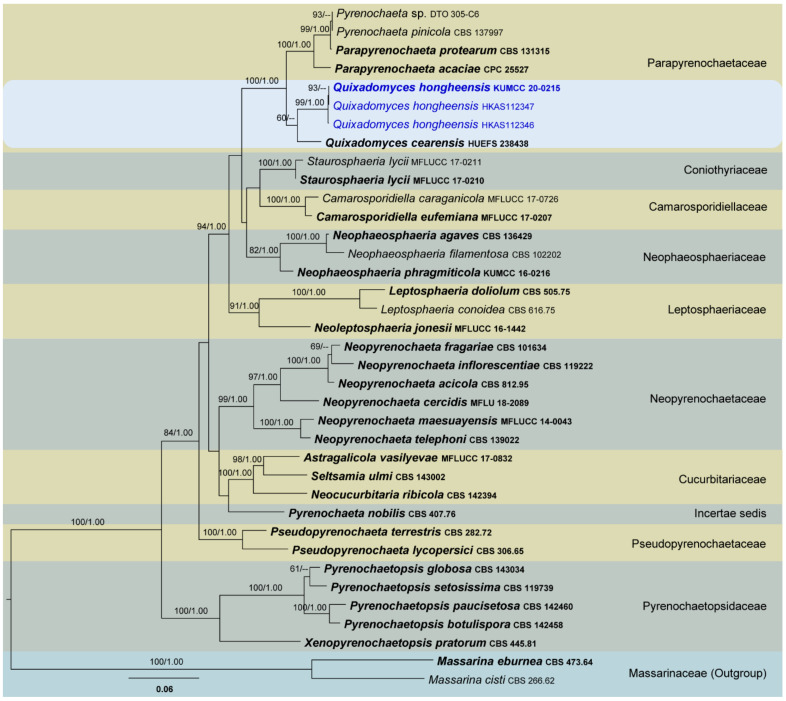
Four phylogenetic analyses were performed using the acquired sequences from GenBank (Table 3). The first is a phylogenetic overview of the genera treated in Teratosphaeriaceae (Figure 1), while the remaining three alignments represent the species in Rhytidhysteron (Figure 2), an overview of the phylogeny of the genera treated in Lophiostomataceae (Figure 3) and Parapyrenochaeta, and allied genera in Pleosporineae (Figure 4). Other details related to ML and BI analyses from different datasets are presented in Table 4. The acquired phylogenetic results are discussed where applicable in the notes below.
Table 3.
Taxa used in the phylogenetic analyses and their corresponding GenBank numbers.
| Species | Strain | GenBank Accession Numbers | Reference | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SSU | LSU | act | cal | ITS | rpb2 | tef1 | btub | |||
| Acidiella bohemica | CBS 132720 | - | KF901984 | - | - | - | KF902178 | - | - | [49] |
| Acidiella parva | CMW 10189 | - | KF901986 | KF903512 | KF902537 | KF901647 | KF902192 | KF903097 * | - | [49] |
| Acrodontium crateriforme | CPC 11509 | - | GU214682 | GU320413 | KX289011 | GU214682 | KX288404 | GU384425 * | - | [124,125] |
| Acrodontium pigmentosum | CBS 111111 | - | KX286963 | - | - | KX287275 | KX288412 | - | - | [125] |
| Alfoldia vorosii | CBS 145501 | MK589346 | MK589354 | - | - | JN859336 | - | MK599320 | - | [126] |
| Alpestrisphaeria jonesii | GZCC 16-0021 | KX687755 | KX687753 | - | - | KX687757 | - | KX687759 | - | [14] |
| Alpestrisphaeria jonesii | GZCC 16-0022 | KX687756 | KX687754 | - | - | KX687758 | - | KX687760 | - | [14] |
| Alpestrisphaeria monodictyoides | V0216 | MH160808 | - | - | MK503662 | - | - | [127] | ||
| Alpestrisphaeria terricola | SC-12H | JX985749 | JX985750 | - | - | JN662930 | - | - | [128] | |
| Amorocoelophoma cassiae | MFLUCC 17-2283 | NG_065775 | NG_066307 | - | - | NR_163330 | MK434894 | MK360041 | - | [127] |
| Angustimassarina acerina | MFLUCC 14-0505 | NG_063573 | KP888637 | - | - | NR_138406 | - | KR075168 | - | [129] |
| Angustimassarina quercicola | MFLUCC 14-0506 | NG_063574 | KP888638 | - | - | KP899133 | - | KR075169 | - | [129] |
| Angustimassarina rosarum | MFLUCC 17-2155 | MT226662 | MT214543 | - | - | MT310590 | MT394678 | MT394726 | - | [130] |
| Apenidiella strumelloidea | CBS 114484 | - | KF937229 | - | - | - | KF937266 | - | - | [49] |
| Araucasphaeria foliorum | CPC 33084 | - | MH327829 | - | - | MH327793 | - | - | - | [131] |
| Astragalicola vasilyevae | MFLUCC 17-0832 | MG829098 | MG828986 | - | - | NR_157504 | MG829248 | MG829193 | - | [130] |
| Austroafricana associata | CPC 13119 | - | KF901824 | KF903526 | KF902528 | KF901507 | KF902177 | KF903087 * | - | [49] |
| Austroafricana sp. | CPC 4313 | - | KF901813 | KF903460 | KF902527 | KF901498 | KF902186 | KF903086 * | - | [49] |
| Austrostigmidium mastodiae | MA 18215 | - | NG_057063 | - | - | - | - | - | - | [132] |
| Austrostigmidium mastodiae | MA 18213 | - | KP282862 | - | - | - | - | - | - | [132] |
| Batcheloromyces alistairii | CPC 12730 | - | KF937220 | - | - | - | KF937252 | - | - | [49] |
| Batcheloromyces leucadendri | CPC 1838 | - | KF937221 | - | - | - | KF937253 | - | - | [49] |
| Batcheloromyces sedgefieldii | CPC 3026 | - | KF937222 | - | - | - | KF937254 | - | - | [49] |
| Biappendiculispora japonica | KT 573 | AB618686 | AB619005 | - | - | LC001728 | - | LC001744 | - | [129,133] |
| Biappendiculispora japonica | KT 686-1 | AB618687 | AB619006 | - | - | LC001729 | - | LC001745 | - | [129,133] |
| Camarosporidiella caraganicola | MFLUCC 17-0726 | MF434300 | MF434212 | - | - | MF434125 | - | MF434388 | - | [134] |
| Camarosporidiella elongata | AFTOL-ID 1568 | DQ678009 | DQ678061 | - | - | - | DQ677957 | DQ677904 | - | [135] |
| Camarosporidiella eufemiana | MFLUCC 17-0207 | MF434321 | MF434233 | - | - | MF434145 | - | MF434408 | - | [134] |
| Camarosporula persooniae | CPC 3350 | - | JF770460 | - | - | - | KF937255 | - | - | [49,136] |
| Capulatispora sagittiformis | KT 1934 | AB618693 | AB369267 | - | - | AB369268 | - | LC001756 | - | [129,133] |
| Catenulostroma hermanusense | CPC 18276 | - | KF902089 | - | - | - | KF902197 | - | - | [49] |
| Catenulostroma protearum | CPC 15370 | - | KF902090 | - | - | - | KF902198 | - | - | [49] |
| Coelodictyosporium pseudodictyosporium | MFLUCC 13-0451 | - | KR025862 | - | - | KR025858 | - | - | - | [137] |
| Coelodictyosporium rosarum | MFLUCC 17-0776 | NG_063674 | NG_059056 | - | - | MG828875 | - | MG829195 | - | [130] |
| Coniothyrium palmarum | CBS 400.71 | EU754054 | JX681084 | - | - | MH860184 | KT389592 | - | KT389792 | [138] |
| Constantinomyces macerans | TRN 440 | - | KF310005 | - | - | NR_164011 | KF310081 | - | - | [139] |
| Constantinomyces minimus | CBS 118766 | - | KF310003 | - | - | NR_144957 | KF310077 | - | - | [139] |
| Crassiclypeus aquaticus | KH 91 | LC312469 | LC312527 | - | - | LC312498 | LC312585 | LC312556 | - | [140] |
| Crassiclypeus aquaticus | KH 104 | LC312470 | LC312528 | - | - | LC312499 | LC312586 | LC312557 | - | [140] |
| Crassiclypeus aquaticus | KH 185 | LC312471 | LC312529 | - | - | LC312500 | LC312587 | LC312558 | - | [140] |
| Crassiclypeus aquaticus | KT 970 | LC312472 | LC312530 | - | - | LC312501 | LC312588 | LC312559 | - | [140] |
| Desertiserpentica hydei | SQUCC 15092 | MW077163 | MW077156 | - | - | MW077147 | MW075773 | MW077163 | - | [54] |
| Devriesia agapanthi | CPC 19833 | - | JX069859 | - | - | - | KJ564346 | - | - | [49,141] |
| Devriesia strelitziae | X1037 | - | GU301810 | - | - | EU436763 | GU371738 | GU349049 * | - | [142] |
| Dimorphiopsis brachystegiae | CPC 22679 | - | KF777213 | - | - | KF777160 | - | - | - | [143] |
| Elasticomyces elasticus | CCFEE 5313 | - | KJ380894 | - | - | FJ415474 | - | - | - | [49,144] |
| Elasticomyces elasticus | CCFEE 5474 | - | KF309991 | - | - | - | KF310046 | - | - | [139] |
| Eupenidiella venezuelensis | CBS 106.75 | - | KF902163 | KF903393 | KF902540 | KF901802 | KF902202 | KF903100 * | - | [49] |
| Euteratosphaeria verrucosiafricana | CPC 11167 | - | - | - | - | DQ303056 | - | - | - | [139] |
| Flabellascoma aquaticum | KUMCC 15-0258 | MN304832 | NG_068307 | - | - | NR_166305 | MN328895 | MN328898 | - | [145] |
| Flabellascoma cycadicola | KT 2034 | LC312473 | LC312531 | - | - | LC312502 | LC312589 | LC312560 | - | [140] |
| Flabellascoma fusiforme | MFLUCC 18-1584 | - | NG_068308 | - | - | NR_166306 | - | MN328902 | - | [105] |
| Flabellascoma minimum | KT 2013 | LC312474 | LC312532 | - | - | LC312503 | LC312590 | LC312561 | - | [140] |
| Flabellascoma minimum | KT 2040 | LC312475 | LC312533 | - | - | LC312504 | LC312591 | LC312562 | - | [140] |
| Forliomyces uniseptata | MFLUCC 15-0765 | NG_061234 | NG_059659 | - | - | NR_154006 | - | KU727897 | - | [146] |
| Friedmanniomyces endolithicus | CCFEE 5199 | - | KF310007 | - | - | - | KF310093 | - | - | [139] |
| Friedmanniomyces endolithicus | CCFEE 5283 | - | KF310006 | - | - | - | KF310053 | - | - | [49] |
| Gloniopsis calami | MFLUCC 15-0739 | NG_063621 | NG_059715 | - | - | NR_164398 | - | KX671965 | - | [147] |
| Gloniopsis calami | MFLUCC 10-0927 | MN577426 | MN577415 | - | - | MN608546 | - | - | - | [148] |
| Gloniopsis praelonga | CBS 112415 | FJ161134 | FJ161173 | - | - | - | FJ161113 | FJ161090 | - | [149] |
| Guttulispora crataegi | MFLUCC 13-0442 | KP899125 | KP888639 | - | - | KP899134 | - | KR075161 | - | [129] |
| Guttulispora crataegi | MFLUCC 14-0993 | KP899126 | KP888640 | - | - | KP899135 | - | KR075162 | - | [129] |
| Haniomyces dodonaeae | KUMCC 20-0220 | MW264221 | MW264191 | MW256802 | MW256805 | MW264212 | MW269527 | MW256813 * | - | This study |
| Haniomyces dodonaeae | KUMCC 20-0221 | MW264222 | MW264192 | MW256803 | MW256806 | MW264213 | MW269528 | MW256814 * | - | This study |
| Hortaea thailandica | CPC 16651 | - | KF902125 | - | - | - | KF902206 | - | - | [49] |
| Hysterium angustatum | MFLUCC 16-0623 | MH535885 | MH535893 | - | - | - | MH535875 | FJ161096 | - | [149,150] |
| Hyweljonesia indica | NFCCI 4146 | - | NG_066398 | - | - | NR_164021 | - | - | - | [151] |
| Hyweljonesia queenslandica | BRIP 61322b | - | NG_059766 | - | - | NR_154095 | - | - | - | [152] |
| Incertomyces perditus | CCFEE 5385 | - | KF310008 | - | - | KF309977 | KF310083 | - | - | [139] |
| Incertomyces vagans | CCFEE 5393 | - | KF310009 | - | - | NR_154064 | KF310057 | - | - | [139] |
| Lapidomyces hispanicus | TRN126 | - | KF310016 | - | - | - | KF310076 | - | - | [139] |
| Lentistoma bipolare | HKUCC 10069 | LC312476 | LC312534 | - | - | LC312505 | LC312592 | LC312563 | - | [140] |
| Lentistoma bipolare | HKUCC 10110 | LC312477 | LC312535 | - | - | LC312506 | LC312593 | LC312564 | - | [140] |
| Lentistoma bipolare | HKUCC 8277 | LC312478 | LC312536 | - | - | LC312507 | LC312594 | LC312565 | - | [140] |
| Lentistoma bipolare | KT 2415 | LC312483 | LC312541 | - | - | LC312512 | LC312599 | LC312570 | - | [140] |
| Lentistoma bipolare | KT 3056 | LC312484 | LC312542 | - | - | LC312513 | LC312600 | LC312571 | - | [140] |
| Leptoparies palmarum | KT 1653 | LC312485 | LC312543 | - | - | LC312514 | LC312601 | LC312572 | - | [140] |
| Leptosphaeria conoidea | CBS 616.75 | JF740099 | JF740279 | - | - | JF740201 | KT389639 | - | KT389804 | [153] |
| Leptosphaeria doliolum | CBS 505.75 | NG_062778 | NG_068574 | - | - | NR_155309 | KY064035 | GU349069 | JF740144 | [154] |
| Lophiohelichrysum helichrysi | MFLUCC 15-0701 | KT333437 | KT333436 | - | - | KT333435 | - | KT427535 | - | [155] |
| Lophiopoacea paramacrostoma | MFLUCC 11-0463 | KP899122 | KP888636 | - | - | - | - | - | - | [129] |
| Lophiomurispora hongheensis | KUMCC 20-0217 | MW264225 | MW264195 | - | - | MW264216 | MW256808 | MW256817 | - | This study |
| Lophiomurispora hongheensis | KUMCC 20-0223 | MW264226 | MW264196 | - | - | MW264217 | MW256809 | MW256818 | - | This study |
| Lophiomurispora hongheensis | KUMCC 20-0216 | MW264227 | MW264197 | - | - | MW264218 | MW256810 | MW256819 | - | This study |
| Lophiomurispora hongheensis | KUMCC 20-0219 | MW264228 | MW264198 | - | - | MW264219 | MW256811 | MW256820 | - | This study |
| Lophiomurispora hongheensis | KUMCC 20-0224 | MW264229 | MW264199 | - | - | MW264220 | MW256812 | MW256821 | - | This study |
| Lophiopoacea winteri | KT 740 | AB618699 | AB619017 | - | - | JN942969 | JN993487 | LC001763 | - | [129,133,156] |
| Lophiopoacea winteri | KT 764 | AB618700 | AB619018 | - | - | JN942968 | JN993488 | LC001764 | - | [129,133,156] |
| Lophiostoma caulium | CBS 623.86 | GU296163 | GU301833 | - | - | - | GU371791 | - | - | [152] |
| Lophiostoma macrostomum | KT 635 | AB521731 | AB433273 | - | - | AB433275 | JN993484 | LC001752 | - | [129,133] |
| Lophiostoma multiseptatum | JCM 17668 | AB618684 | AB619003 | - | - | LC001726 | - | LC001742 | - | [129,133] |
| Lophiostoma multiseptatum | MAFF 239451 | AB618685 | AB619004 | - | - | LC001727 | - | LC001743 | - | [129,133] |
| Lophiostoma rosae | TASM 6115 | NG_065145 | NG_069558 | - | - | NR_158531 | - | MG829205 | - | [130] |
| Lophiostoma semiliberum | KT 828 | AB618696 | AB619014 | - | - | JN942970 | JN993489 | LC001759 | - | [129,133,156] |
| Massarina cisti | CBS 266.62 | AB797249 | AB807539 | - | - | LC014568 | FJ795464 | AB808514 | - | [157,158] |
| Massarina eburnea | CBS 473.64 | GU296170 | GU301840 | - | - | AF383959 | GU371732 | GU349040 | - | [143,159] |
| Meristemomyces frigidum | CCFEE 5457 | - | GU250389 | - | - | - | KF310066 | - | - | [49,144] |
| Meristemomyces frigidum | CCFEE 5507 | - | KF310013 | - | - | - | KF310067 | - | - | [139] |
| Monticola elongata | CCFEE 5492 | - | KF309994 | - | - | - | KF310065 | - | - | [139] |
| Myrtapenidiella corymbia | CPC 14640 | - | KF901838 | KF903558 | KF902558 | KF901517 | KF902227 | KF903119 * | - | [49] |
| Neocatenulostroma abietis | CBS 110038 | - | KF937226 | - | - | - | KF937263 | - | - | [49] |
| Neocatenulostroma microsporum | CPC 1960 | - | KF901814 | - | KF902561 | KF901499 | KF902232 | KF903122 * | - | [49] |
| Neocucurbitaria ribicola | CBS 142394 | MF795840 | MF795785 | - | - | MF795785 | MF795827 | MF795873 | MF795911 | [160] |
| Neoleptosphaeria jonesii | MFLUCC 16-1442 | NG_063625 | KY211870 | - | - | NR_152375 | - | KY211872 | - | [161] |
| Neopaucispora rosaecae | MFLUCC 17-0807 | NG_061293 | NG_059869 | - | - | MG828924 | - | MG829217 | - | [130] |
| Neophaeosphaeria agaves | CBS 136429 | - | KF777227 | - | - | NR_137833 | - | - | - | [143] |
| Neophaeosphaeria filamentosa | CBS 102202 | GQ387516 | GQ387577 | - | - | JF740259 | GU371773 | - | - | [162] |
| Neophaeosphaeria phragmiticola | KUMCC 16-0216 | MG837008 | MG837009 | - | - | - | - | MG838020 | - | [163] |
| Neophaeothecoidea proteae | CPC 2831 | - | KF937228 | - | - | - | KF937265 | - | - | [49] |
| Neopyrenochaeta acicola | CBS 812.95 | NG_065567 | GQ387602 | - | - | NR_160055 | LT623271 | - | LT623232 | [164] |
| Neopyrenochaeta cercidis | MFLU 18-2089 | NG_065769 | MK347932 | - | - | MK347718 | MK434908 | - | - | [127] |
| Neopyrenochaeta fragariae | CBS 101634 | GQ387542 | GQ387603 | - | - | LT623217 | LT623270 | - | LT623231 | [164] |
| Neopyrenochaeta inflorescentiae | CBS 119222 | - | EU552153 | - | - | EU552153 | LT623272 | - | LT623233 | [165] |
| Neopyrenochaeta maesuayensis | MFLUCC 14-0043 | - | MT183504 | - | - | NR_170043 | - | MT454042 | - | [166] |
| Neopyrenochaeta telephoni | CBS 139022 | - | NG_067485 | - | - | KM516291 | LT717685 | - | LT717678 | [154] |
| Neotrematosphaeria biappendiculata | KT 1124 | GU205256 | GU205227 | - | - | - | - | - | - | [129] |
| Neotrematosphaeria biappendiculata | KT 975 | GU205254 | GU205228 | - | - | - | - | - | - | [129] |
| Neotrimmatostroma excentricum | CPC 13092 | - | KF901840 | KF903534 | KF902562 | KF901518 | KF902236 | KF903123 * | - | [49] |
| Neovaginatispora clematidis | MFLUCC 17-2149 | MT226676 | MT214559 | - | - | MT310606 | - | MT394738 | - | [167] |
| Neovaginatispora fuckelii | CBS 101952 | FJ795496 | DQ399531 | - | - | - | FJ795472 | - | - | [158] |
| Neovaginatispora fuckelii | KH 161 | AB618689 | AB619008 | - | - | LC001731 | - | LC001749 | - | [129,133] |
| Neovaginatispora fuckelii | KT 634 | AB618690 | AB619009 | - | - | LC001732 | - | LC001750 | - | [129,133] |
| Oleoguttula mirabilis | CCFEE 5522 | - | KF310019 | - | - | - | KF310070 | - | - | [139] |
| Parapaucispora pseudoarmatispora | KT 2237 | LC100018 | LC100026 | - | - | LC100021 | - | LC100030 | - | [168] |
| Parapenidiella pseudo tasmaniensis | CPC 12400 | - | KF901844 | KF903562 | KF902589 | KF901522 | KF902265 | KF903152 * | - | [49] |
| Parapenidiella tasmaniensis | CPC 1555 | - | KF901843 | KF903451 | KF902587 | KF901521 | KF902263 | KF903150 * | - | [49] |
| Parapyrenochaeta acaciae | CPC 25527 | - | KX228316 | - | - | NR_155674 | LT717686 | - | LT717679 | [53] |
| Parapyrenochaeta protearum | CBS 131315 | - | JQ044453 | - | - | JQ044434 | LT717683 | - | LT717677 | [53] |
| Paucispora kunmingense | MFLUCC 17-0932 | MF173430 | NG_059829 | - | - | NR_156625 | MF173436 | MF173434 | - | [169] |
| Paucispora quadrispora | KH 448 | LC001720 | LC001722 | - | - | LC001733 | - | LC001754 | - | [129] |
| Paucispora quadrispora | KT 843 | AB618692 | AB619011 | - | - | LC001734 | - | LC001755 | - | [129,133] |
| Paucispora versicolor | KH 110 | LC001721 | AB918732 | - | - | AB918731 | - | LC001760 | - | [129,133] |
| Penidiella columbiana | CBS 486.80 | - | KF901965 | KF903587 | KF902594 | KF901630 | KF902272 | KF903158 * | - | [49] |
| Penidiellomyces aggregatus | CBS 128772 | - | NG_057905 | - | - | NR_137772 | - | - | - | [170] |
| Penidiellomyces drakensbergensis | CPC 19778 | - | NG_059482 | - | - | NR_111821 | - | - | - | [141] |
| Penidiellopsis radicularis | CBS 131976 | - | KU216314 | - | KU216292 | KT833148 | - | KU216339 * | - | [171] |
| Penidiellopsis ramosus | CBMAI 1937 | - | KU216317 | - | KU216295 | KT833151 | - | KU216342 * | - | [171] |
| Phaeoseptum carolshearerianum | NFCCI-4221 | MK307816 | MK307813 | - | - | MK307810 | MK309877 | MK309874 | - | [172] |
| Phaeoseptum hydei | MFLUCC 17-0801 | MT240624 | MT240623 | - | - | MT240622 | - | MT241506 | - | [40] |
| Phaeoseptum manglicola | NFCCI-4666 | MK307817 | MK307814 | - | - | MK307811 | MK309878 | MK309875 | - | [172] |
| Phaeoseptum terricola | MFLUCC 10-0102 | MH105780 | MH105779 | - | - | MH105778 | MH105782 | MH105781 | - | [163] |
| Phaeothecoidea Intermedia | CPC 13711 | - | KF902106 | KF903564 | KF902606 | KF901752 | KF902286 | KF903171 * | - | [49] |
| Phaeothecoidea Minutispora | CPC 13710 | - | KF902108 | KF903659 | KF902607 | KF901753 | KF902288 | KF903172 * | - | [49] |
| Piedraia hortae var. hortae | CBS 480.64 | - | KF901943 | - | - | - | KF902289 | - | - | [49] |
| Piedraia hortae var. paraguayensis | CBS 276.32 | - | KF901816 | - | - | - | - | - | - | [49] |
| Piedraia quintanilhae | CBS 327.63 | - | KF901957 | - | - | - | - | - | - | [49] |
| Platystomum actinidiae | KT 521 | JN941375 | JN941380 | - | - | JN942963 | JN993490 | LC001747 | - | [129,156] |
| Platystomum crataegi | MFLUCC 14-0925 | KT026113 | KT026109 | - | - | KT026117 | - | KT026121 | - | [129] |
| Platystomum rosae | MFLU 15-2569 | KY264750 | KY264746 | - | - | KY264742 | - | - | - | [173] |
| Platystomum rosae | MFLUCC 15-0633 | KT026115 | KT026111 | - | - | KT026119 | - | - | - | [129] |
| Platystomum salicicola | MFLUCC 15-0632 | KT026114 | KT026110 | - | - | KT026118 | - | - | - | [129] |
| Pseudolophiostoma cornisporum | KH 322 | LC312486 | LC312544 | - | - | LC312515 | LC312602 | LC312573 | - | [140] |
| Pseudolophiostoma obtusisporum | KT 2838 | LC312489 | LC312547 | - | - | LC312518 | LC312605 | LC312576 | - | [140] |
| Pseudolophiostoma obtusisporum | KT 3119 | LC312491 | LC312549 | - | - | LC312520 | LC312607 | LC312578 | - | [140] |
| Pseudolophiostoma tropicum | KH 352 | LC312492 | LC312550 | - | - | LC312521 | LC312608 | LC312579 | - | [140] |
| Pseudolophiostoma tropicum | KT 3134 | LC312493 | LC312551 | - | - | LC312522 | LC312609 | LC312580 | - | [140] |
| Pseudopaucispora brunneospora | KH 227 | LC312494 | LC312552 | - | - | LC312523 | LC312610 | LC312581 | - | [140] |
| Pseudoplatystomum scabridisporum | BCC 22835 | GQ925831 | GQ925844 | - | - | - | GU479830 | GU479857 | - | [174] |
| Pseudoplatystomum scabridisporum | BCC 22836 | GQ925832 | GQ925845 | - | - | - | GU479829 | GU479856 | - | [174] |
| Pseudopyrenochaeta lycopersici | CBS 306.65 | NG_062728 | MH870217 | - | - | NR_103581 | LT717680 | - | LT717674 | [154] |
| Pseudopyrenochaeta terrestris | CBS 282.72 | - | LT623216 | - | - | LT623228 | LT623287 | - | LT623246 | [53] |
| Pseudoteratosphaeria flexuosa | CPC 673 | - | KF902098 | KF903403 | KF902653 | KF901745 | KF902345 | KF903228 * | - | [49] |
| Pseudoteratosphaeria flexuosa | CPC 1109 | - | KF902110 | KF903421 | KF902654 | KF901755 | KF902346 | - | - | [49] |
| Pyrenochaeta nobilis | CBS 407.76 | DQ898287 | EU754206 | - | - | NR_103598 | DQ677991 | DQ677936 | MF795916 | [162] |
| Pyrenochaeta pinicola | CBS 137997 | - | KJ869209 | - | - | KJ869152 | LT717684 | - | KJ869249 | [175] |
| Pyrenochaeta sp. | DTO 305-C6 | - | KX171361 | - | - | KX147606 | - | - | - | [176] |
| Pyrenochaetopsis botulispora | CBS 142458 | - | LN907440 | - | - | LT592945 | LT593084 | - | LT593014 | [53] |
| Pyrenochaetopsis globosa | CBS 143034 | - | LN907418 | - | - | LT592934 | LT593072 | - | LT593003 | [53] |
| Pyrenochaetopsis paucisetosa | CBS 142460 | - | LN907336 | - | - | LT592897 | LT593035 | - | LT592966 | [53] |
| Pyrenochaetopsis setosissima | CBS 119739 | - | GQ387632 | - | - | LT623227 | LT623285 | - | LT623245 | [162] |
| Queenslandipenidiella kurandae | CPC 13333 | - | KF901860 | KF903538 | KF902663 | KF901538 | KF902356 | KF903238 * | - | [49] |
| Quixadomyces cearensis | HUEFS 238438 | - | NG_066409 | - | - | NR_160606 | - | - | - | [131] |
| Quixadomyces hongheensis | KUMCC 20-0215 | MW264223 | MW264193 | - | - | MW264214 | MW269529 | MW256815 | MW256804 | This study |
| Quixadomyces hongheensis | HKAS112346 | MW541833 | MW541822 | - | - | MW541826 | MW556136 | MW556134- | MW556137 | This study |
| Quixadomyces hongheensis | HKAS112347 | MW541834 | MW541823 | - | - | MW541827 | - | MW556135- | MW556138 | This study |
| Ramusculicola clematidis | MFLUCC 17-2146 | NG_070667 | MT214596 | - | - | MT310640 | MT394707 | MT394652 | - | [167] |
| Readeriella angustia | CPC 13608 | - | KF902114 | KF903566 | KF902669 | KF901759 | KF902364 | KF903246 * | - | [49] |
| Readeriella deanei | CPC 12715 | - | KF901864 | KF903583 | KF902673 | KF901542 | KF902368 | KF903250 * | - | [49] |
| Readeriella dimorphospora | CPC 12636 | - | KF901866 | KF903622 | KF902675 | KF901544 | KF902370 | KF903252 * | - | [49] |
| Readeriella menaiensis | CPC 14447 | - | KF901870 | KF903572 | KF902678 | KF901548 | KF902374 | KF903256 * | - | [49] |
| Recurvomyces mirabilis | CCFEE 5264 | - | GU250372 | - | - | - | KF310059 | - | - | [139,144] |
| Recurvomyces mirabilis | CCFEE 5475 | - | KC315876 | - | - | - | KF310060 | - | - | [139,144] |
| Rhytidhysteron bruguierae | MFLUCC 17-1502 | MN632464 | MN632453 | - | - | MN632458 | - | MN635662 | - | [55] |
| Rhytidhysteron bruguierae | MFLUCC 17-1515 | MN632463 | MN632452 | - | - | MN632457 | - | MN635661 | - | [55] |
| Rhytidhysteron bruguierae | MFLUCC 18-0398 | MN017901 | MN017833 | - | - | - | - | MN077056 | - | [172] |
| Rhytidhysteron bruguierae | MFLUCC 17-1511 | MN632465 | MN632454 | - | - | MN632459 | - | - | - | [55] |
| Rhytidhysteron camporesii | HKAS 104277 | MN429072 | - | - | MN429069 | - | MN442087 | - | [148] | |
| Rhytidhysteron chromolaenae | MFLUCC 17-1516 | NG_070139 | NG_068675 | - | - | MN632461 | - | MN635663 | - | [55] |
| Rhytidhysteron erioi | MFLU 16-0584 | - | MN429071 | - | - | MN429068 | - | MN442086 | - | [148] |
| Rhytidhysteron hongheense | KUMCC 20-0222 | MW264224 | MW264194 | - | - | MW264215 | MW256807 | MW256816 | - | This study |
| Rhytidhysteron hongheense | HKAS112348 | MW541831 | MW541820 | - | - | MW541824 | - | MW556132 | - | This study |
| Rhytidhysteron hongheense | HKAS112349 | MW541832 | MW541821 | - | - | MW541825 | - | MW556133 | - | This study |
| Rhytidhysteron hysterinum | EB 0351 | - | GU397350 | - | - | - | - | GU397340 | - | [149] |
| Rhytidhysteron hysterinum | CBS 316.71 | - | MH871912 | - | - | MH860141 | - | - | - | [154] |
| Rhytidhysteron magnoliae | MFLUCC 18-0719 | MN989382 | MN989384 | - | - | MN989383 | - | MN997309 | - | [177] |
| Rhytidhysteron mangrovei | MFLUCC 18-1113 | - | NG_067868 | - | - | NR_165548 | - | MK450030 | - | [178] |
| Rhytidhysteron neorufulum | MFLUCC 13-0216 | KU377571 | KU377566 | - | - | KU377561 | - | KU510400 | - | [177] |
| Rhytidhysteron neorufulum | GKM 361A | GU296192 | GQ221893 | - | - | - | - | - | - | [179] |
| Rhytidhysteron neorufulum | HUEFS 192194 | - | KF914915 | - | - | - | - | - | - | [180] |
| Rhytidhysteron neorufulum | MFLUCC 12-0528 | KJ418119 | KJ418117 | - | - | KJ418118 | - | - | - | [181] |
| Rhytidhysteron neorufulum | CBS 306.38 | AF164375 | FJ469672 | - | - | - | - | GU349031 | - | [142] |
| Rhytidhysteron neorufulum | MFLUCC 12-0011 | KJ418110 | KJ418109 | - | - | KJ206287 | - | - | - | [181] |
| Rhytidhysteron neorufulum | MFLUCC 12-0567 | KJ546129 | KJ526126 | - | - | KJ546124 | - | - | - | [181] |
| Rhytidhysteron neorufulum | MFLUCC 12-0569 | KJ546131 | KJ526128 | - | - | KJ546126 | - | - | - | [181] |
| Rhytidhysteron neorufulum | MFLUCC 14-0577 | KU377570 | KU377565 | - | - | KU377560 | - | KU510399 | - | [177] |
| Rhytidhysteron opuntiae | GKM 1190 | GQ221892 | - | - | - | - | GU397341 | - | [179] | |
| Rhytidhysteron rufulum | EB 0384 | GU397368 | GU397354 | - | - | - | - | - | - | [182] |
| Rhytidhysteron rufulum | EB 0382 | GU397367 | GU397352 | - | - | - | - | - | - | [182] |
| Rhytidhysteron rufulum | EB 0383 | GU397353 | - | - | - | - | - | - | [182] | |
| Rhytidhysteron rufulum | MFLUCC 12-0013 | KJ418113 | KJ418111 | - | - | KJ418112 | - | - | - | [181] |
| Rhytidhysteron tectonae | MFLUCC 13-0710 | KU712457 | KU764698 | - | - | KU144936 | - | KU872760 | - | [183] |
| Rhytidhysteron thailandicum | MFLUCC 13-0051 | MN509434 | - | - | MN509433 | - | MN509435 | - | [56] | |
| Rhytidhysteron thailandicum | MFLUCC 12-0530 | KJ546128 | KJ526125 | - | - | KJ546123 | - | - | - | [172] |
| Rhytidhysteron thailandicum | MFLUCC 14-0503 | KU377569 | KU377564 | - | - | KU377559 | - | KU497490 | - | [177] |
| Seltsamia ulmi | CBS 143002 | MF795794 | MF795794 | - | - | MF795794 | MF795836 | MF795882 | MF795918 | [160] |
| Sigarispora arundinis | KT 651 | AB618680 | AB618999 | - | - | JN942965 | JN993486 | LC001738 | - | [129,133] |
| Sigarispora caudata | MAFF 239453 | AB618681 | AB619000 | - | - | LC001723 | - | LC001739 | - | [129,133] |
| Sigarispora caulium | MAFF 239450 | AB618682 | AB619001 | - | - | LC001724 | - | LC001740 | - | [129,133] |
| Sigarispora caulium | JCM 17669 | AB618683 | AB619002 | - | - | LC001725 | - | LC001741 | - | [129,133] |
| Sigarispora ononidis | MFLUCC 15-2667 | KU243126 | KU243125 | - | - | KU243128 | - | KU243127 | - | [169] |
| Sigarispora rosicola | MFLU 15-1888 | NG_062116 | MG829080 | - | - | MG828968 | - | MG829240 | - | [130] |
| Simplicidiella nigra | CBMAI 1939 | - | KU216313 | - | KU216291 | KT833147 | - | KU216338 * | - | [171] |
| Sparticola junci | MFLUCC 15-0030 | NG_061235 | KU721765 | - | - | NR_154428 | KU727900 | KU727898 | - | [146] |
| Staninwardia suttonii | CPC 13055 | - | KF901874 | KF903517 | KF902693 | KF901552 | KF902392 | KF903270 * | - | [49] |
| Staurosphaeria lycii | MFLUCC 17-0210 | MF434372 | MF434284 | - | - | MF434196 | - | MF434458 | - | [134] |
| Staurosphaeria lycii | MFLUCC 17-0211 | MF434373 | MF434285 | - | - | MF434197 | - | MF434459 | - | [134] |
| Stenella araguata | FMC 245 | - | KF902168 | - | - | - | KF902393 | - | - | [49] |
| Suberoteratosphaeria pseudosuberosa | CPC 12085 | - | KF902144 | KF903508 | - | KF901786 | - | KF903275 * | - | [49] |
| Suberoteratosphaeria xenosuberosa | CPC 13093 | - | KF901879 | KF903584 | - | KF901557 | KF902402 | KF903280 * | - | [49] |
| Teichospora mariae | C136 | - | KU601581 | - | - | KU601581 | KU601595 | KU601611 | - | [184] |
| Teichospora rubriostiolata | TR 7 | - | KU601590 | - | - | KU601590 | KU601599 | KU601609 | - | [184] |
| Teichospora thailandica | MFLUCC 17-2093 | MT226708 | MT214597 | - | - | MT310641 | MT394708 | MT394653 | - | [167] |
| Teichospora trabicola | C 134 | - | KU601591 | - | - | KU601591 | KU601600 | KU601601 | - | [184] |
| Teratoramularia infinita | CBS 141104 | - | KX287249 | KX287828 | KX289125 | KX287545 | KX288710 | KX288107 * | - | [125] |
| Teratoramularia rumicicola | CBS 141106 | - | KX287255 | - | - | KX287550 | KX288716 | KX288113 * | - | [125] |
| Teratosphaeria aurantia | MUCC 668 | - | KF901884 | KF903578 | KF902700 | KF901561 | KF902409 | KF903284 * | - | [49] |
| Teratosphaeria blakelyi | CPC 12837 | - | KF901888 | KF903518 | KF902704 | KF901565 | KF902413 | KF903288 * | - | [49] |
| Teratosphaeria destructans | CPC 1368 | - | KF901898 | KF903447 | KF902716 | KF901574 | KF902427 | KF903301 * | - | [49] |
| Teratosphaeria fimbriata | CPC 13324 | - | KF901901 | KF903529 | KF902720 | KF901577 | KF902430 | KF903306 * | - | [49] |
| Teratosphaeria gauchensis | CMW 17331 | - | KF902148 | KF903521 | KF902729 | KF901790 | KF902439 | KF903315 * | - | [49] |
| Teratosphaeria mareebensis | CPC 17272 | - | KF901906 | KF903581 | KF902734 | KF901582 | KF902444 | KF903320 * | - | [49] |
| Teratosphaeria pseudocryptica | CPC 11267 | - | KF902032 | KF903598 | KF902760 | KF901687 | KF902472 | KF903348 * | - | [49] |
| Teratosphaeriaceae sp. | CPC 13680 | - | KF901921 | KF903657 | KF902765 | KF901597 | KF902477 | KF903353 * | - | [49] |
| Teratosphaeriaceae sp. | CCFEE 5569 | - | KF310015 | - | - | - | KF310071 | - | - | [139] |
| Teratosphaericola pseudoafricana | CPC 1231 | - | KF902045 | KF903435 | KF902782 | KF901699 | KF902499 | KF903370 * | - | [49] |
| Teratosphaericola pseudoafricana | CPC 1230 | - | KF902084 | KF903473 | KF902783 | KF901737 | KF902500 | KF903371 * | - | [49] |
| Teratosphaeriopsis pseudoafricana | CPC 1261 | - | KF902085 | KF903436 | KF902784 | KF901738 | KF902501 | KF903372 * | - | [49] |
| Vaginatispora amygdali | KT 2248 | LC312495 | LC312553 | - | - | LC312524 | LC312611 | LC312582 | - | [140] |
| Vaginatispora appendiculata | MFLUCC 16-0314 | KU743219 | KU743218 | - | - | KU743217 | - | KU743220 | - | [185] |
| Vaginatispora armatispora | MFLUCC 18-0247 | MK085058 | MK085060 | - | - | MK085056 | MK087669 | MK087658 | - | [146] |
| Vaginatispora nypae | MFLUCC 18-1543 | NG_065779 | NG_066313 | - | - | NR_163340 | MK434877 | MK360091 | - | [127] |
| Vaginatispora scabrispora | KT 2443 | LC312496 | LC312554 | - | - | LC312525 | LC312612 | LC312583 | - | [140] |
| Westerdykella ornata | CBS 379.55 | GU296208 | GU301880 | - | - | AY943045 | - | GU349021 | - | [142] |
| Xenopenidiella inflata | CBMAI 1945 | - | KU216337 | - | KU216312 | KT833171 | - | KU216359 * | - | [171] |
| Xenopenidiella tarda | CBMAI 1940 | - | KU216326 | - | KU216303 | KT833160 | - | KU216351 * | - | [171] |
| Xenophacidiella pseudocatenata | CPC 18472 | - | KF902092 | - | - | - | KF902508 | - | - | [49] |
| Xenopyrenochaetopsis pratorum | CBS 445.81 | NG_062792 | NG_057858 | - | - | NR_111623 | KT389671 | - | KT389846 | [186] |
GenBank accession numbers with * are resulting from EF1-728F and EF-2 primers and – means missing data or not used in the phylogenetic analyses. The newly generated sequences are indicated in bold.
Figure 1.
RAxML tree based on a combined dataset of partial LSU, ITS, rpb2, act, cal and tef1 DNA sequence analysis in Teratosphaeriaceae. The tree is rooted to Staninwardia suttonii (CPC 13055). Bootstrap support values for ML equal to or greater than 60%, Bayesian posterior probabilities (BYPP) equal to or greater than 0.95 are presented as ML/BI above nodes. Known genera are indicated with coloured blocks. Blue represents new isolates. The ex-type strains are indicated in bold. The scale bar presents the expected number of nucleotide substitutions per site.
Figure 2.
RAxML tree based on a combined dataset of partial SSU, LSU, ITS and tef1 DNA sequence analysis in Rhytidhysteron. The tree is rooted to Gloniopsis calami (MFLUCC 15-0739, MFLUCC 10-0927). Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are shown as ML/BI above the nodes. Known species are indicated with coloured blocks. Blue represents new isolates. The ex-type strains are indicated in bold. The scale bar represents the expected number of nucleotide substitutions per site.
Figure 3.
RAxML tree based on a combined dataset of partial SSU, LSU, ITS, tef1 and rpb2 DNA sequence analysis in Lophiostomataceae. The tree is rooted to Gloniopsis praelonga (CBS 112415) and Hysterium angustatum (MFLUCC 16-0623). Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are shown as ML/BI above the nodes. Known families and selected genera are indicated with coloured blocks. Blue represents new isolates. The ex-type strains are indicated in bold. The scale bar represents the expected number of nucleotide substitutions per site.
Figure 4.
RAxML tree based on a combined dataset of partial LSU, SSU, ITS, rpb2, tef1 and btub DNA sequence analysis in Pleosporineae. The tree is rooted to Massarina cisti (CBS 266.62) and M. eburnea (CBS 473.64). Bootstrap support values for ML equal to or greater than 60% and BYPP equal to or greater than 0.95 are shown as ML/BI above the nodes. Known families and the genus Quixadomyces are indicated with coloured blocks. Blue represents new isolates. The ex-type strains are indicated in bold. The scale bar represents the expected number of nucleotide substitutions per site.
Table 4.
Maximum-likelihood (ML) and Bayesian (BI) analyses results for each sequenced dataset.
| Analyses | Teratosphaeriaceae | Rhytidhysteron | Lophiostomataceae | Parapyrenochaeta | |
|---|---|---|---|---|---|
| Number of Taxa | 106 | 34 | 106 | 37 | |
| Gene regions | LSU, ITS, rpb2, act, cal and tef1 | SSU, LSU, ITS and tef1 | SSU, LSU, ITS, tef1 and rpb2 | LSU, SSU, ITS, rpb2, tef1 and btub | |
| Number of character positions (including gaps) | 3517 | 3667 | 4649 | 5510 | |
| ML optimization likelihood value | −50604.86449 | −10388.988691 | −42280.12689 | −27947.901235 | |
| Distinct alignment patterns in the matrix | 1973 | 739 | 2082 | 1710 | |
| Number of undetermined characters or gaps (%) | 48.76% | 30.69% | 27.07% | 38.18% | |
| Estimated base frequencies | A | 0.23693 | 0.241388 | 0.24893 | 0.245506 |
| C | 0.26813 | 0.244326 | 0.24732 | 0.244909 | |
| G | 0.283733 | 0.277859 | 0.267917 | 0.265204 | |
| T | 0.211207 | 0.236427 | 0.235833 | 0.244381 | |
| Substitution rates | AC | 1.498833 | 1.533268 | 1.549406 | 1.619926 |
| AG | 2.784366 | 2.507774 | 4.37387 | 4.391077 | |
| AT | 1.662835 | 1.340621 | 1.462392 | 1.995039 | |
| CG | 1.129905 | 1.029121 | 1.453674 | 1.225921 | |
| CT | 6.210175 | 6.529612 | 8.808274 | 8.980921 | |
| GT | 1.0 | 1.0 | 1.0 | 1.0 | |
| Proportion of invariable sites (I) | 0.416989 | 0.610823 | 0.453545 | 0.55191 | |
| Gamma distribution shape parameter (α) | 0.626612 | 0.475911 | 0.51454 | 0.443538 | |
| Number of generated trees in BI | 29861 | 3451 | 9001 | 951 | |
| Number of trees sampled in BI after 25% were discarded as burn-in | 22396 | 2589 | 6751 | 714 | |
| Final split frequency | 0.009999 | 0.009261 | 0.009977 | 0.007923 | |
| The total of unique site patterns | 1974 | 740 | 2084 | 1711 | |
3.3. Taxonomy of Fungi Colonising Dodonaea Viscosa Twigs
In the current study, two new genera and four novel species were found. These taxa are subsequently described below.
Class Dothideomycetes O.E. Erikss. and Winka, Myconet 1: 5 (1997)
Capnodiales Woron., Annales Mycologici 23: 177 (1925)
Teratosphaeriaceae Crous and U. Braun, Studies in Mycology 58: 8 (2007)
Haniomyces J.C. Xu gen. nov.
MycoBank: MB837991
Etymology: The generic epithet refers to the “Hani” ethnic group in Honghe County, Yunnan Province, China.
It is saprobic on dead twigs and branches in terrestrial habitats. Sexual morph: the ascomata is a scattered, immersed to semi-immersed, subglobose to conical or shaped irregularly, glabrous, brown to dark brown ostiolate. The ostiole is a short papillate, black, smooth periphysate. The peridium comprises cells of textura angularis. The hamathecium comprises numerous, filamentous, branched, septate, pseudoparaphyses. The asci are eight-spored, bitunicate, fissitunicate, clavate, with a pedicel, apically rounded with or without an ocular chamber. The ascospores overlap the biseriate, are ellipsoidal to sub-fusiform, hyaline, one-septate, with small to large guttules in each cell, with the ends remaining rounded, surrounded by a distinct mucilaginous sheath. Asexual morph: Coelomycetous. The conidiomata are sporodochial on PDA, globose, solitary or aggregated, semi-immersed, black, exuding yellow conidial masses. Conidiophores and conidiogenous cells were not observed in vitro. The conidia are solitary, aseptate, globose to ellipsoid, with the hyaline becoming medium to golden brown, and finely verruculose.
Type species: Haniomyces dodonaeae
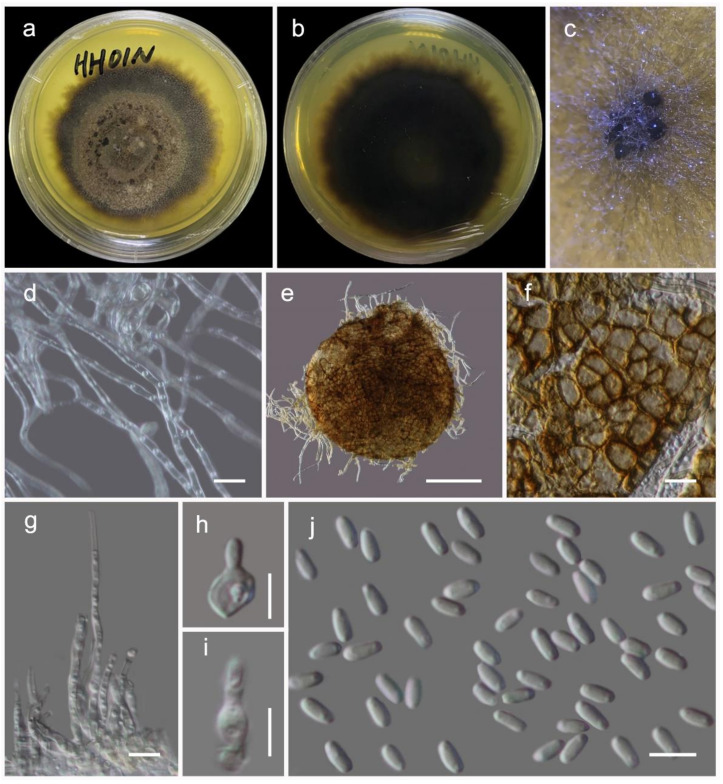
Haniomyces dodonaeae Wanas. and Mortimer sp. nov. (Figure 5)
Figure 5.
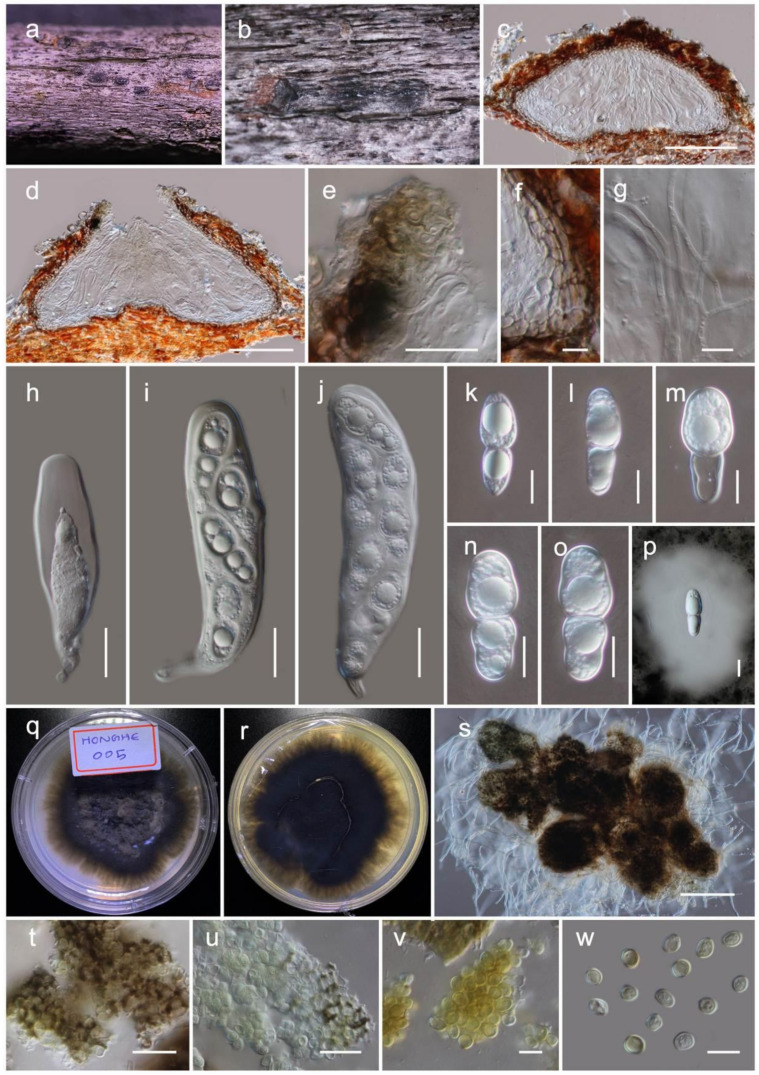
The sexual (HKAS110128, holotype) and asexual (KUMCC 20-0220, ex-type) morphs of Haniomyces dodonaeae. (a,b) ascomata on the dead woody twigs of Dodonaea viscosa; (c,d) vertical section of ascoma; (e) periphyses; (f) peridium; (g) pseudoparaphyses; (h–j) asci; (k–p) ascospores (p in Indian Ink); (q,r) colony on potato dextrose agar (PDA) (r from the bottom); (s) squashed pycnidia which were produced on PDA; (t) pycnidia wall; (u–w) conidia. Scale bars, (c,d) 100 µm; (e,h–j,t,u) 20 µm; (f,k–p,v,w) 10 µm; (s) 200 µm.
MycoBank: MB837997
Etymology: The specific epithet reflects the host genus Dodonaea.
Holotype: HKAS110128
It is saprobic on dead twigs of Dodonaea viscosa Jacq. (Sapindaceae). Sexual morph: the ascomata is a 150–200 μm high, 350–450 μm diam. (M = 165.4 × 390.3 µm, n = 5), scattered, semi-immersed to erumpent, subglobose to conical or shaped irregularly, flattened base, glabrous, brown to dark brown ostiolate, fused with host tissues. The ostiole is a short papillate, black and smooth, with hyaline periphyses (15–25 μm long, 1.5–2 μm wide). The peridium 5–10 µm wide at the base, 10–20 µm wide at sides, comprising 2–4 layers, outer layer pigmented, comprising reddish brown to dark brown, with thin-walled cells of textura angularis, and an inner layer composed of hyaline, loosen, cells of textura angularis. The hamathecium comprises numerous, 2–3 µm wide, filamentous, branched, septate, pseudoparaphyses. The asci are 110–130 × 25–35 µm (M = 118.5 × 31.2 µm, n = 20), eight-spored, bitunicate, fissitunicate, clavate, with a short pedicel (10–15 μm long), apically rounded with an ocular chamber. The ascospores 25–35 × 12–15 µm (M = 32.2 × 14.3 µm, n = 30), overlap the biseriate, are ellipsoidal to sub-fusiform, hyaline, one-septate, with the septum almost median, deeply constricted at the middle septum, with the upper cell wider than the lower cell, and are smooth-walled with small to large guttules in each cell, rounded at both ends and covered by a distinct mucilaginous sheath (30–50 µm, diam.). Asexual morph: Coelomycetous. The conidiomata are up to 250 μm diam., sporodochial on PDA, globose, solitary or aggregated, semi-immersed, black, exuding yellow conidial masses. Conidiophores and conidiogenous cells were not observed in vitro. The conidia are 5.5–7.5 × 4.5–5.5 µm (M = 6.4 × 5.4 µm, n = 30), solitary, aseptate, globose or ellipsoid, with the hyaline becoming medium to golden brown, and finely verruculose.
Culture characteristics: the colonies on PDA reached a 3 cm diameter after 2 weeks at 20 °C. They were circular has a serrate margin, whitish at the beginning, becoming brown at the centre and brownish green towards the margin after 4 weeks. They were slightly raised, and reverse blackish brown. The hyphae septate were branched, hyaline, thin, and smooth-walled.
Known distribution: Yunnan, China, on Dodonaea viscosa.
Material examined: China, Yunnan, Honghe Hani and Yi Autonomous Prefecture, Honghe County, 23.421068 N, 102.229128 E, 735 m, on dead twigs of Dodonaea viscosa, 22 April 2020, D.N. Wanasinghe, Honghe 005 (HKAS110128, holotype), ex-type living culture, KUMCC 20-0220, ibid. 23.419206 N, 102.231375 E, 618 m, Honghe 010 (HKAS110125, paratype), ex-paratype living culture, KUMCC 20-0221.
Hysteriales Lindau, Die Natürlichen Pflanzenfamilien nebst ihren Gattungen und wichtigeren Arten 1 (1): 265 (1897
Hysteriaceae Chevall., Flore Générale des Environs de Paris 1: 432 (1826)
Rhytidhysteron Speg., Anales de la Sociedad Científica Argentina 12 (4): 188 (1881)
Rhytidhysteron hongheenseWanas. sp. nov. (Figure 6)
Figure 6.
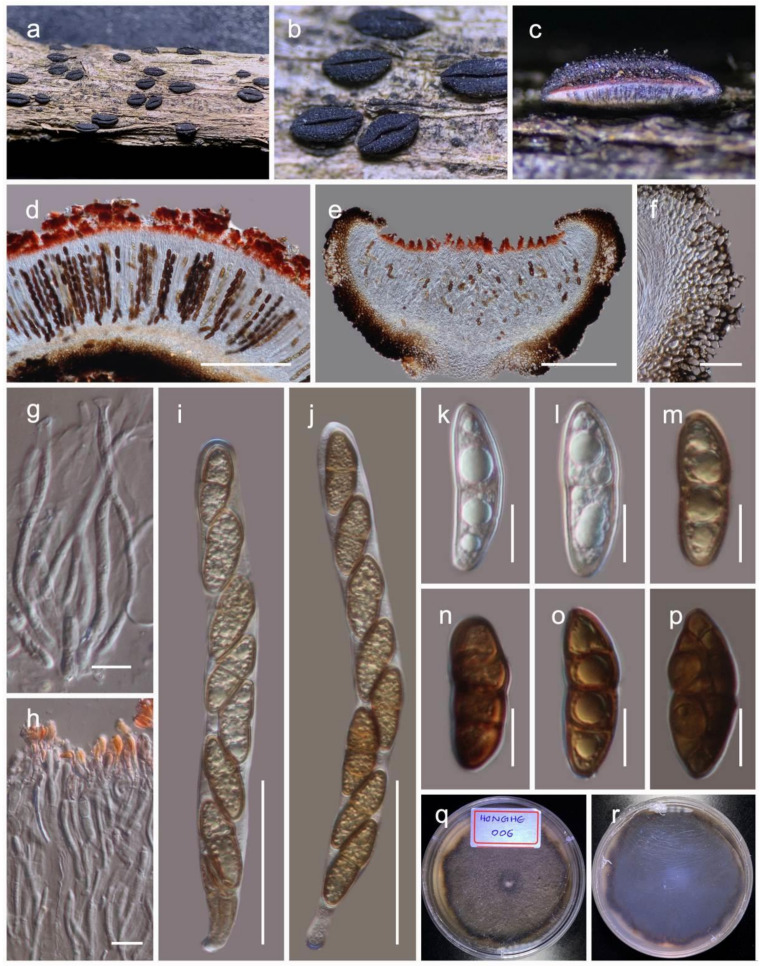
Rhytidhysteron hongheensis (HKAS110133, holotype). (a,b) Appearance of hysterothecia on the dead woody twigs of Dodonaea viscosa; (c,d) horizontal section of hysteriothecium; (e) vertical section of hysteriothecium; (f) cells of peridium; (g,h) pseudoparaphyses; (i,j) asci; (k–p) ascospores; (q,r) colony on PDA (r from the bottom). Scale bars, (d,e) 200 µm; (f,i,j) 50 µm; (g,h,k–p) 10 µm.
MycoBank: MB837992
Etymology: The specific epithet is derived from Honghe County, Yunnan Province, China.
Holotype: HKAS110133
It is aaprobic on dead twigs of Dodonaea Mill. (Sapindaceae). Sexual morph: The hystherothecia is 1200–2000 μm long × 350–500 high × 600–1000 µm diam. (M = 1590 × 410 × 840 µm, n = 10), arising singly or in small groups, sessile, and slightly erumpent from the substrate. The receptacle is cupulate, black, flat or slightly concave, with a slightly dentate margin. The excipulum are 70–100 µm wide, with the ectal excipulum narrow layered, deep, and thick-walled, with black cells of textura globulosa to textura angularis; the medullary excipulum is composed of narrow, long, thin-walled, hyaline to brown cells of textura angularis. The hamathecium are 2.5–4 µm wide, numerous, propoloid, pseudoparaphyses, exceeding asci in length, apically swollen, branched and reddish-orange pigmented. The branched apices form a layer on hymenium to develop pseudo-epithecium. The asci are 140–180 × 12–16 µm (M = 163.3 × 13.8 µm, n = 20), eight-spored, long cylindrical, short pedicellate, and is rounded at apex. The ascospores 20–33 × 9–13 µm (M = 28.2 × 11.2 µm, n = 30), overlap the uniseriate, are hyaline to light brown, one-septate, with wrinkled walls when young, becoming dark brown at maturity. They are ellipsoid with conical ends, regularly three-septate, and rarely muriform with one longitudinal septum, smooth walled, guttulate. Asexual morph: Undetermined.
Culture characteristics: Colonies on PDA reached a 4 cm diameter after 2 weeks at 20 °C. The colony was dense, circular, slightly raised, and the surface was smooth, with an undulated edge, with floccose which were greenish grey at the centre and brown towards margin from the top and reverse dark brown. The hyphae septate were branched, hyaline, thin, and smooth-walled.
Known distribution: Yunnan, China, on Dodonaea.
Material examined: China, Yunnan, Honghe Hani and Yi Autonomous Prefecture, Honghe County, 23.421068 N, 102.229128 E, 735 m, on dead twigs of Dodonaea, 22 April 2020, D.N. Wanasinghe, Honghe 006 (HKAS110133, holotype), ex-type culture, KUMCC 20-0222. ibid. on dead twigs of Dodonaea viscosa, 08 December 2020, DWH6-1 (HKAS112348). ibid. 07 December 2020, DWH7-2 (HKAS112349).
Pleosporales Luttr. ex M.E. Barr, Prodromus to class Loculoascomycetes: 67 (1987)
Lophiostomataceae Sacc., Sylloge Fungorum 2: 672 (1883)
Lophiomurispora Wanas. and Mortimer, gen. nov.
MycoBank: MB837993
Etymology: The generic epithet stems from the combined two words ‘‘lophio’’ and ‘‘murispora’’, referring to muriform ascospores in Lophiostomataceae.
It is saprobic on woody substrates in terrestrial habitats. Sexual morph: The ascomata is a solitary or gregarious, semi-immersed, erumpent through the host surface, coriaceous to carbonaceous, dark brown to black, globose to subglobose or conical ostiolate. The ostiole is a slit-like, central papillate, with or without a crest, opening by an apical, lysigenous pore or dehiscence, comprising hyaline periphyses or hyaline to lightly pigmented, pseudoparenchymatous cells. The peridium is broad at the apex and thinner at the base, comprising two strata with several layers of brown or lightly pigmented to hyaline cells of textura angularis to textura prismatica, fusing and indistinguishable from the host tissues. The hamathecium comprises many branched, septate, cellular pseudoparaphyses, located between and above the asci, embedded in a gelatinous matrix. The asci are eight-spored, bitunicate, fissitunicate, cylindric-clavate, pedicellate, and apically rounded, with an ocular chamber. The ascospores are uni- to bi-seriate, partially overlapping, and are hyaline when immature, becoming brown to dark brown when mature. They are ellipsoidal to fusiform, muriform, two-to-eight-transversely septate, with one-to-two-longitudinal septa, constricted at the central septum, with or without a mucilaginous sheath. Asexual morph: Coelomycetous. The conidiomata is pycnidial, phoma-like, solitary, gregarious, dark brown to black, immersed or slightly erumpent, coriaceous to carbonaceous, papillate or apapillate. The conidiomata wall is multi-layered, with three to four outer layers of brown-walled pseudoparenchymatous cells, with the inner most layer being thin and hyaline. The conidiophores are long, septate, and sparsely branched, which are formed from the inner most layer of the pycnidium wall. The conidiogenous cells are phialidic, cylindrical, hyaline, flexuous and smooth, with a short collarette. The conidia are hyaline, aseptate, straight to curved, ellipsoidal with rounded ends, thin-walled, smooth, and numerous.
Type species: Lophiomurispora hongheensis
Lophiomurispora hongheensis Wanas. sp. nov. (Figure 7 and Figure 8)
Figure 7.
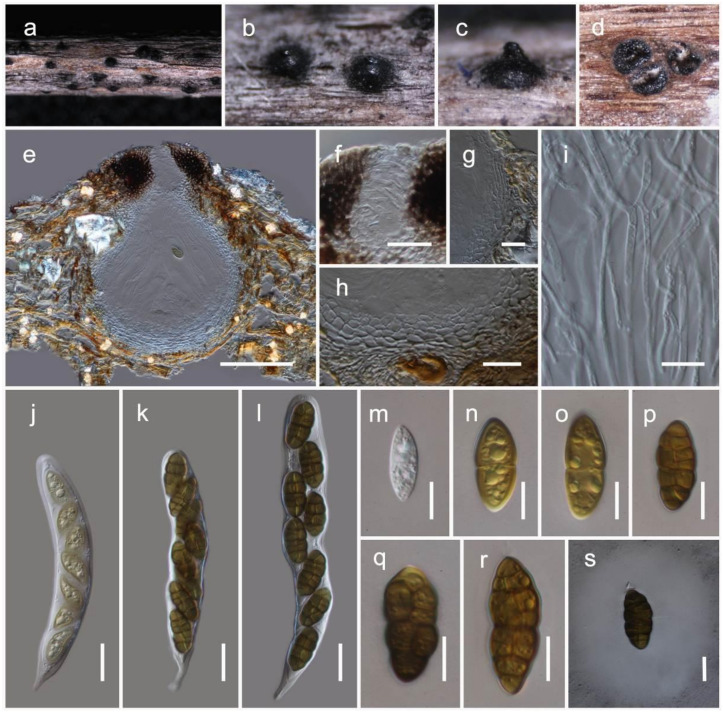
Sexual morph of Lophiomurispora hongheensis (HKAS110127, holotype). (a–c) Ascomata on the dead woody twigs of Dodonaea viscosa; (d) cross section of ascomata; (e) vertical section of ascoma; (f) closeup of ostiole; (g,h) peridium; (i) pseudoparaphyses; (j–l) asci; (m–s) ascospores (s in Indian Ink); Scale bars, (e) 100 µm; (f–h,j–l) 20 µm; (i,m–s) 10 µm.
Figure 8.
Asexual morph of Lophiomurispora hongheensis (KUMCC 20-0217, ex-type culture). (a,b) colony on PDA (b from the bottom); (c,d) immersed pycnidia in PDA (from the bottom); (e) pycnidia wall; (f–i) conidiophore; (j) conidia. Scale bars, (e–i) 10 µm; (j) 5 µm.
MycoBank: MB 837998
Etymology: The specific epithet is derived from Honghe County, the region of Yunnan Province in which this species was gathered.
Holotype: HKAS110127
It is saprobic on dead twigs of Dodonaea viscosa Jacq. (Sapindaceae) in terrestrial habitats. Sexual morph: The ascomata is a 280–360 μm high, 200–250 μm diam. (M = 318.6 × 232.7 µm, n = 5), scattered to gregarious, immersed, coriaceous, dark brown to black, globose to subglobose ostiolate. The ostiole is a 70–100 μm long, 40–80 μm diam. (M = 82.1 × 64.8 µm, n = 5), crest-like, central papillate, with a pore-like opening, comprising hyaline periphyses. The peridium is 20–30 μm wide at the base, 30–60 μm wide at the sides, broad at the apex, comprising two strata, with outer stratum composed of small, pale brown to brown, slightly flattened, thick-walled cells of textura angularis, fusing and indistinguishable from the host tissues. The inner stratum is composed of several layers with lightly pigmented to hyaline cells of textura angularis to textura prismatica. The hamathecium comprises 1–2 μm wide, branched, septate, cellular pseudoparaphyses, situated between and above the asci, embedded in a gelatinous matrix. The asci are 120–160 × 17–22 μm (M = 135.2 × 18.5 μm, n = 15), eight-spored, bitunicate, fissitunicate, cylindric-clavate, with a short pedicel, and is rounded at the apex, with an ocular chamber. The ascospores are 25–30 × 11–13 μm (M = 27.8 × 12 µm, n = 30), uni- to bi-seriate, overlapping, and are initially hyaline, turning brown at maturity. They are ellipsoidal to fusiform, muriform, four-to-eight-transversely septate, with one-to-two-longitudinal septa. They are slightly curved, deeply constricted at the central septum, slightly constricted at the remaining septa, conically rounded at the ends, and smooth-walled, with a distinct mucilaginous sheath. Asexual morph: Coelomycetous. The conidiomata is 1–1.5 mm diam. pycnidial, phoma-like, solitary, gregarious, dark brown to black, and immersed, with a sphaerical mass of slimy conidia oozing out at ostiolar apex. The conidiomata wall is multi-layered, with brown-walled pseudoparenchymatous cells, with a hyaline inner most layer. The conidiophores are 10–15 × 1.5–2.5 μm long (M = 12.4 × 2.1 µm, n = 15), septate and sparsely branched, which are formed from the inner most layer of the pycnidium wall. The conidiogenous cells are phialidic, cylindrical, hyaline, flexuous and smooth, with a short collarette. The conidia are 2.5–4 ×1.5–2 μm (M = 3 ×1.7 μm, n = 50), hyaline, aseptate, straight to curved, ellipsoidal with rounded ends, and are thin-walled, smooth-walled, and numerous.
Culture characteristics: the colonies on PDA reached a 4 cm diameter after 2 weeks at 20 °C. They were circular, had a serrate margin, and were whitish at the beginning, becoming greenish-brown 4 weeks later. They were slightly raised, and reverse dark brown. The hyphae septate were branched, hyaline, thin, and smooth-walled.
Known distribution: Yunnan, China, on Dodonaea viscosa.
Material examined: China, Yunnan, Honghe Hani and Yi Autonomous Prefecture, Honghe County, 23.421068 N, 102.229128 E, 735 m, on dead twigs of Dodonaea viscosa, 22 April 2020, D.N. Wanasinghe, Honghe 003 (HKAS110127, holotype), ex-type culture, KUMCC 20-0217, ibid. 23.419206 N, 102.231375 E, 618 m, Honghe 008 (HKAS110129, paratype), ex-paratype living culture, KUMCC 20-0223, ibid. 23 April 2020, ibid. DWHH07-1 (HKAS110130), living culture, KUMCC 20-0224, DWHH01 (HKAS110132), living culture, KUMCC 20-0216, ibid. DWHH04 3 (HKAS110131), living culture, KUMCC 20-0219.
Parapyrenochaetaceae Valenz-Lopez, Crous, Stchigel, Guarro and J.F. Cano, Studies in Mycology 90: 64 (2017)
Quixadomyces Cantillo and Gusmão, Persoonia 40: 317 (2018)
Quixadomyces hongheensis Wanas. sp. nov. (Figure 9)
Figure 9.
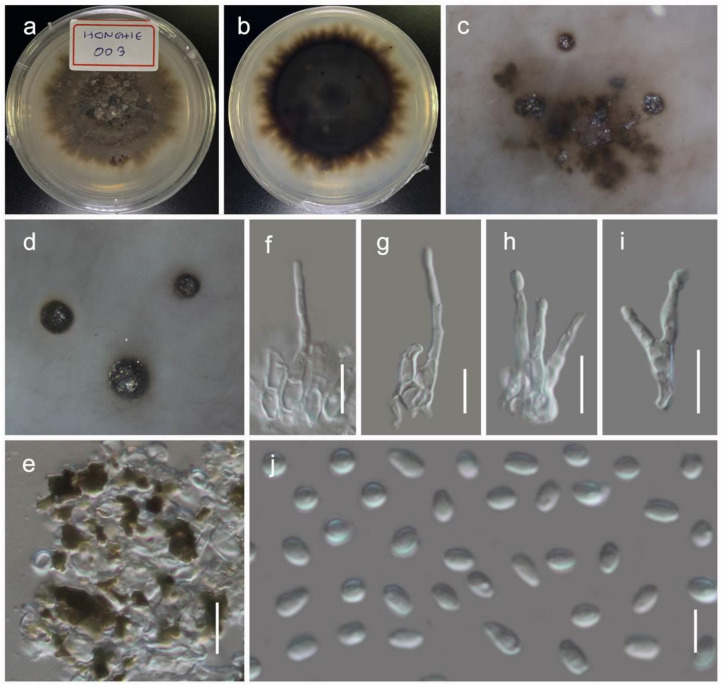
Quixadomyces hongheensis (KUMCC 20-0215, ex-type culture). (a,b) colony on PDA (b from the bottom); (c) pycnidia on PDA; (d) mycelia; (e) squashed pycnidia; (f) pycnidia wall; (g) paraphyses; (h,i) conidiophore; (j) conidia. Scale bars, (d,f,g) 10 µm; (e) 200 µm; (h–j) 5 µm.
MycoBank: MB837994
Etymology: The specific epithet is derived from Honghe County, Yunnan Province, China.
Holotype: HKAS110126
It is saprobic on dead twigs of Dodonaea viscosa Jacq. (Sapindaceae) in terrestrial habitats. Sexual morph: Undetermined. Asexual morph: Coelomycetous. The conidiomata is immersed to erumpent, solitary, globose, brown, from 200–300 μm diam, with a central ostiole, exuding a hyaline conidial mass. It has a wall of two to three layers of brown textura angularis. The paraphyses are 20–100 μm long, 2–3 μm wide, cylindrical, hyaline, septate, and smooth. The conidiophores are mostly reduced to conidiogenous cells. The conidiogenous cells are 5–8 × 3.5–5 μm (M = 6.4 × 3.1 µm, n = 15), lining the inner cavity, hyaline, smooth, are ampulliform to subcylindrical, and are phialidic with periclinal thickening. The conidia are 3–4.7 × 1.2–2 (M = 3.7 × 1.7 µm, n = 60) μm, solitary, hyaline, smooth, aseptate, and allantoid with obtuse ends.
Culture characteristics: The colonies on PDA reached a 4 cm diameter after 2 weeks at 20 °C. They were circular, had a serrate margin, and were greenish brown after 4 weeks. They were slightly raised, and reverse dark brown. The hyphae septate were branched, hyaline, thin, and smooth-walled.
Known distribution: Yunnan, China, on Dodonaea viscosa.
Material examined: China, Yunnan, Honghe Hani and Yi Autonomous Prefecture, Honghe County, 23.421068 N, 102.229128 E, 735 m, on dead twigs of Dodonaea viscosa, 22 April 2020, D.N. Wanasinghe, Honghe 01-N (HKAS110126, holotype), ex-type living culture, KUMCC 20-0215. 08 December 2020, HDW4-1 (HKAS112347). ibid. HDW4-3 (HKAS112346).
4. Discussion
Teratosphaeriaceae was introduced by Crous et al. [187]. Given that it is composed of 61 genera, it is regarded as one of the largest families in Dothideomycetes [188]. Members of this family are adapted to a broad range of life modes and can be saprobic, plant and human pathogenic, rock-inhabiting and endophytic; accordingly, they are widely distributed across varied terrain [49,136,139,188,189]. We have included representative sequence data of all available genera listed in Hongsanan et al. [188] for the phylogenetic analyses (except Davisoniella, Pachysacca and Placocrea, which lack DNA-based sequence data). Among them, Aulographina was grouped in Venturiales, and Leptomelanconium was related to Helotiales in the initial analysis. Therefore, they were excluded from the final analysis (Figure 1). In addition, representative taxa for Piedraia were included in the final dataset that were phylogenetically closely related to Teratosphaeriaceae. However, this genus is still considered a member in Piedraiaceae. The phylogeny generated herein (Figure 1) is congruent with those of other published studies to resolve intergeneric relationships in Teratosphaeriaceae [49,188]. In the combined LSU, ITS, rpb2, act, cal and tef1 data analysis, 58 clades are recognized from the ingroup taxa. Two strains from our new collections constitute a distinct monophyletic lineage (subclade 17, Figure 1) within the genera in Teratosphaeriaceae, which we introduce as a new genus.
The phylogeny (Figure 1) reveals a close relationship between two strains of the newly collected fungus (Haniomyces dodonaeae) to Camarosporula persooniae, Lapidomyces hispanicus, Neophaeothecoidea proteae, Teratosphaeriaceae sp. (CCFEE 5569), Xenoconiothyrium catenata and Xenophacidiella pseudocatenata, with 87% ML and 1.00 BYPP support values. Among them, only Camarosporula persooniae is reported from the sexual morph, and despite the high degree of phylogenetic similarity, these two species are morphologically dissimilar [136]. Neophaeothecoidea is more closely related to Haniomyces in the phylogenetic results, but this relationship lacks statistical support. In addition, Neophaeothecoidea is reported as a hyphomycete [188], whereas Haniomyces produces a coelomycetous asexual morph.
Out of the 61 genera listed in Teratosphaeriaceae, only 24 genera are described with sexual morphs. We suggest that the sexual morphs of these genera require further examination with increased collections to verify the accurate treatment of and relationships to remaining species. During asexual stages of Teratosphaeriaceae, most members are pathogenic, whereas they are non-pathogenic during sexual stages. This is an important distinction for identifying opportunistic pathogens, as members of this family can easily spread diseases between locations. The new taxon, Haniomyces dodonaeae, fits morphologically well into Teratosphaeriaceae by its periphysate ostiole and hyaline ascospores with a single septum in each. However, the dimensions of the asci and ascospores are significantly larger than the existing sexual reports of this family. The golden brown, ellipsoidal conidia of Haniomyces dodonaeae are similar to those in Neophaeothecoidea and Readeriella. Phylogenetically, Haniomyces dodonaeae has a close proximity to Neophaeothecoidea proteae. This relationship, however, is not strongly supported in the ML and BI analyses (Figure 1). Neophaeothecoidea proteae was originally isolated as a coelomycete (Phaeothecoidea proteae) based on its yeast-like growth in culture [190]; however, it is currently accounted for in a hyphomycetous genus. Comparison of the 805 base pairs (bp) across the LSU gene region of Haniomyces dodonaeae shows 17 bp (2.1%) differences exist in comparison with Neophaeothecoidea proteae. Similarly, comparison of the 356 bp of the rpb2 gene region shows 56 bp (15.73%) differences in comparison with Neophaeothecoidea proteae.
Rhytidhysteron was introduced by Spegazzini [191] to account for R. brasiliense and R. viride collected from southern Brazil in 1877 and 1880, respectively. Spegazzini [56] did not designate any type; therefore, Clements and Shear [192] designated R. brasiliense as the type species. Subsequently, few species were introduced to this genus based on morphological evidence [193,194,195,196]. In recent studies, more species have been introduced based on both morphology and DNA-based sequence data [55,56,172,177,178,183]. Presently, there are 23 species mentioned in Species Fungorum [197], including saprobic to weakly pathogenic taxa that grow on woody plants in terrestrial habitats [56,181]. Species of Rhytidhysteron are typically involved in wood degradation and occur primarily on the woody parts of a broad range of hosts [64,188].
We introduce a new species into Rhytidhysteron from a dead twig of Dodonaea sp. in Honghe, China, and its relationships with other species are presented based on multigene phylogenetic analyses (Figure 2). Our analysed molecular data generated phylogenies consistent with those of Mapook et al. [55] and Hyde et al. [56]. The novel species, Rhytidhysteron hongheense, is phylogenetically closely related to R. camporesii (KUN-HKAS 104277) and Rhytidhysteron chromolaenae (MFLUCC 17-1516), and these three constitute a strongly supported monophyletic clade. The ascospore and asci characteristics between the three species are similar, but the colour of hysterothecia in R. chromolaenae (green) is different from the other two (black). The pseudo-epithecium of R. camporesii is brown to purple, whereas it is reddish orange in R. hongheense. The significance of these morphological characteristics in species delineation should be further investigated in terms of phylogenetic signals. A pairwise comparison of 521 ITS (+5.8S) sequence data showed 31 (5.95%) bp differences between R. hongheense and R. camporesii as well 28 (5.37%) bp differences between R. hongheense and R. chromolaenae. Currently, there are only two Rhytidhysteron species, viz. Rhytidhysteron magnoliae and Rhytidhysteron thailandicum, reported from China [56,198], making this report the third of its kind from China and first from Honghe Prefecture.
Lophiostomataceae species are usually characterized by a slot-like ostiole on the top of the flattened neck, occurring mainly on twigs, stems or the bark of different woody and herbaceous plants in terrestrial, freshwater and marine environments as saprobes [129,140,188]. Thambugala et al. [129] undertook a comprehensive study of this family and accepted 16 genera. Subsequently, 12 new genera have been introduced by recent publications, and currently the family comprises 28 accepted genera [188]. The most recent multi-locus phylogenetic backbone tree to the family is presented in this study, including a novel genus (Lophiomurispora) found in Honghe County, Yunnan Province, China.
Lophiomurispora morphologically resembles Coelodictyosporium, Platystomum and Sigarispora with its crest-like ostiole and brown, multi-septate ascospores. However, these genera are revealed as phylogenetically distant in multi-gene phylogenetic analysis (Figure 3). Lophiomurispora has a close phylogenetic relationship to Desertiserpentica (Figure 3). However, Desertiserpentica is only known from its hyphomycetous asexual morph [54], whereas Lophiomurispora differs from Desertiserpentica by its coelomycetous asexual morph. Five strains of Lophiomurispora clustered in Lophiostomataceae as a strongly supported monophyletic clade (Figure 3) in both ML and BI of a concatenated SSU, LSU, ITS, tef1 and rpb2 dataset. All specimens were collected from dead twigs of Dodonaea viscosa at the Centre for Mountain Futures (CMF) in Honghe. There was no significant difference between morphological characteristics and DNA-based sequence comparisons between these collections. Therefore, we introduce them as different collections of Lophiomurispora hongheensis.
Parapyrenochaetaceae was proposed by Valenzuela-Lopez et al. [53] to accommodate three isolates which were previously recognized in Pyrenochaeta. They introduced the novel genus Parapyrenochaeta for P. acaciae (Pyrenochaeta acaciae), P. protearum (Pyrenochaeta protearum) and for the strain CBS 137997, formerly misidentified as Pyrenochaeta pinicola (re-identified as Parapyrenochaeta protearum). Later, Crous et al. [131] introduced Quixadomyces as another genus in Parapyrenochaetaceae to accommodate Quixadomyces cearensis. Therefore, there are currently two accepted genera in Parapyrenochaetaceae [3,188].
Crous et al. [131] introduced Quixadomyces for a fungus that was collected from Brazil on decaying bark. However, they did not observe the development of any internal structures. This fungus slightly resembles species in Pleosporales with its setose pycnidia [131,188]. In a multi-gene (concatenated LSU, SSU, ITS, rpb2, tef1 and btub) phylogenetic analysis, the ex-type strain of Quixadomyces cearensis (HUEFS 238438) clustered with two of our new strains as a monophyletic clade with poor bootstrap support (Figure 4). We introduce this isolate as a novel species belonging to this genus, Q. hongheensis. Based on the features of conidiogenous cells and conidia of Quixadomyces hongheensis, no substantial morphological differences exist to warrant two generic ranks. Therefore, this genus could potentially be reclassified as a synonym of Parapyrenochaeta in future studies. Because we did not perform extensive taxonomic reassessment using multiple fresh collections (especially sexual morphs of both genera), we will not attempt to synonymize any extant taxa.
Owing to lack of details on the internal structures of Quixadomyces cearensis, it is difficult to compare morphological characteristics such as conidiogenous cells and conidia between the new collection and this species. Lacking sufficient morphological evidence to perform accurate comparisons, we analysed nucleotide differences between these two strains. Comparing the 544 ITS (+5.8S) nucleotides of the two strains (HUEFS 238438 and KUMCC 20 0215) revealed 32 (5.88%) nucleotide differences. Therefore, it would seem prudent to treat our isolate as a new species in Quixadomyces as Q. hongheensis.
Nearly a century’s worth of taxonomic investigation into Dodonaea viscosa has yielded only 58 fungal records [Table 2]. These are mainly reported as saprobes or pathogens, but very few of these taxa are confirmed by both morphological and phylogenetic evidence. Many of these published records lack illustrations, descriptions or DNA sequence data, resulting in unclear taxonomic relationships. Even though Dodonaea viscosa is widely distributed across southwest and southern China, e.g., Fujian, Guangdong, Guangxi, Hainan, Sichuan and Yunnan [199], there is only one report for the fungus Pseudocercospora mitteriana on this host from China [124]. Previous taxonomic studies have suggested that increased collections might lead to the discovery of many new fungal species, and we, too, believe that Dodonaea is likely teeming with fungal diversity. More Dodonaea collections across different geographic regions are urgently needed, along with accompanying work in culture isolation, morphological description, DNA sequence analyses, phylogenetic relationship investigation, and accurate identification and classification. This study provides a case study for Dodonaea viscosa as a worthwhile host for the further study of microfungal associations and hints that it may potentially host numerous unknown fungal species.
Acknowledgments
Austin G. Smith at World Agroforestry (ICRAF), Kunming Institute of Botany, China, is thanked for English editing. Lu Wen Hua and Li Qin Xian are thanked for their invaluable assistance. We acknowledge Kunming Institute of Botany, Chinese Academy of Sciences for providing the laboratories and instruments for molecular work.
Author Contributions
Conceptualization, D.N.W.; resources, P.E.M. and J.X.; writing—original draft preparation, D.N.W.; writing—review and editing, P.E.M.; supervision, P.E.M. and J.X.; funding acquisition, P.E.M. and J.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Research Project, Agroforestry Systems for Restoration and Bio-industry Technology Development (Grant No. 2017YFC0505101), Ministry of Sciences and Technology of China (Grant No. 2017YFC0505100), CAS President’s International Fellowship Initiative (Grant No. 2019PC0008), the 64th batch of China Postdoctoral Science Foundation (Grant No. 2018M643549), Postdoctoral Fund from Human Resources and Social Security Bureau of Yunnan Province, NSFC project codes 41761144055 and 41771063.
Data Availability Statement
The datasets generated for this study can be found in the NCBI GenBank, MycoBank and TreeBASE.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Antonelli A., Fry C., Smith R.J., Simmonds M.S.J., Kersey P.J., Pritchard H.W., Abbo M.S., Acedo C., Adams J., Ainsworth A.M., et al. State of the World’s Plants and Fungi 2020. Royal Botanic Gardens, Kew; Richmond, UK: 2020. 100p. [DOI] [Google Scholar]
- 2.Kirk P.M. Catalogue of Life. [(accessed on 20 October 2020)]; Available online: http://www.catalogueoflife.org.
- 3.Wijayawardene N.N., Hyde K.D., Al-Ani L.K.T., Tedersoo L., Haelewaters D., Rajeshkumar K.C., Zhao R.L., Aptroot A., Leontyev D.V., Saxena R.K., et al. Outline of Fungi and fungus-like taxa. Mycosphere. 2020;11:1060–1456. doi: 10.5943/mycosphere/11/1/8. [DOI] [Google Scholar]
- 4.Hawksworth D.L., Lücking R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017;5 doi: 10.1128/microbiolspec.FUNK-0052-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyde K.D., Jeewon R., Chen Y.J., Bhunjun C.S., Calabon M.S., Jiang H.B., Lin C.G., Norphanphoun C., Sysouphanthong P., Pem D., et al. The numbers of fungi: Is the descriptive curve flattening? Fungal Divers. 2020;103:219–271. doi: 10.1007/s13225-020-00458-2. [DOI] [Google Scholar]
- 6.Hyde K., Norphanphoun C., Chen J., Dissanayake A., Doilom M., Hongsanan S., Jayawardena R., Jeewon R., Perera R.H., Thongbai B., et al. Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 2018;93:215–239. doi: 10.1007/s13225-018-0415-7. [DOI] [Google Scholar]
- 7.Wong M.K.M., Goh T.K., Hodgkiss I.J., Hyde K.D., Ranghoo V.M., Tsui C.K.M., Ho W.W.H., Wong W.S.W., Yuen T.K. Role of fungi in freshwater ecosystems. Biodivers. Conserv. 1998;7:1187–1206. doi: 10.1023/A:1008883716975. [DOI] [Google Scholar]
- 8.Fu-Qiang S., Xing-Jun T., Zhong-Qi L., Chang-Lin Y., Bin C., Jie-jie H., Jing Z. Diversity of filamentous fungi in organic layers of two forests in Zijin Mountain. J. For. Res. 2004;15:273–279. doi: 10.1007/BF02844951. [DOI] [Google Scholar]
- 9.Yuan J., Zheng X., Cheng F., Zhu X., Hou L., Li J., Zhang S. Fungal community structure of fallen pine and oak wood at different stages of decomposition in the Qinling Mountains, China. Sci. Rep. 2017;7:13866. doi: 10.1038/s41598-017-14425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao G.C., Zhao R.L. The higher microfungi from forests of Yunnan Province. Yunnan Science and Technology Press; Kunming, China: 2012. pp. 1–572. [Google Scholar]
- 11.Xing X.K., Chen J., Xu M.J., Lin W.H., Guo S.X. Fungal endophytes associated with Sonneratia (Sonneratiaceae) mangrove plants on the south coast of China. For. Pathol. 2011;41:334–340. doi: 10.1111/j.1439-0329.2010.00683.x. [DOI] [Google Scholar]
- 12.Ariyawansa H.A., Hyde K.D., Liu J.K., Wu S.P., Liu Z.Y. Additions to karst fungi 1: Botryosphaeria minutispermatia sp. nov., from Guizhou Province, China. Phytotaxa. 2016;275:35–44. doi: 10.11646/phytotaxa.275.1.4. [DOI] [Google Scholar]
- 13.Ariyawansa H.A., Hyde K.D., Tanaka K., Maharachchikumbura S.S.N., Al–Sadi A.M., Elgorban A.M., Liu Z.Y. Additions to karst fungi 3: Prosthemium sinense sp. nov., from Guizhou province, China. Phytotaxa. 2016;284:281–291. doi: 10.11646/phytotaxa.284.4.4. [DOI] [Google Scholar]
- 14.Ariyawansa H.A., Hyde K.D., Thambugala K.M., Maharachchikumbura S.S.N., Al–Sadi A.M., Liu Z.Y. Additions to karst fungi 2: Alpestrisphaeria jonesii from Guizhou Province, China. Phytotaxa. 2016;277:255–265. doi: 10.11646/phytotaxa.277.3.3. [DOI] [Google Scholar]
- 15.Hu D.M., Cai L., Hyde K.D. Three new ascomycetes from freshwater in China. Mycologia. 2012;104:1478–1489. doi: 10.3852/11-430. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z.F., Liu F., Zhou X., Liu X.Z., Liu S.J., Cai L. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia Mol. Phylogeny Evol. Fungi. 2017;39:1–31. doi: 10.3767/persoonia.2017.39.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De-Long Q., Liu L.L., Zhang X., Wen T.C., Kang J.C., Hyde K.D., Shen X.C., Li Q.R. Contributions to species of Xylariales in China-1 Durotheca species. Mycol. Prog. 2019;18:495–510. doi: 10.1007/s11557-018-1458-6. [DOI] [Google Scholar]
- 18.Bao D.F., McKenzie E.H.C., Bhat D.J., Hyde K.D., Luo Z.L., Shen H.W., Su H.Y. Acrogenospora (Acrogenosporaceae, Minutisphaerales) appears to be a very diverse genus. Front. Microbiol. 2020;11:1606. doi: 10.3389/fmicb.2020.01606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karunarathna S.C., Dong Y., Karasaki S., Tibpromma S., Hyde K.D., Lumyong S., Xu J.C., Sheng J., Mortimer P.E. Discovery of novel fungal species and pathogens on bat carcasses in a cave in Yunnan Province, China. Emerg. Microbes Infect. 2020;9:1554–1566. doi: 10.1080/22221751.2020.1785333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Myers N., Mittermeier R.A., Mittermeier C.G., da Fonseca G.A., Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 21.Hu W.Y., Shen Q. Evaluation on landscape stability of Yuanyang Hani terrace. Yunnan Geogr. Environ. Res. 2011;1:11–17. doi: 10.3969/j.issn.1001-7852.2011.01.003. [DOI] [Google Scholar]
- 22.Shui Y.M. Seed Plants of Honghe Region in SE Yunnan. Yunnan Science and Technology Press; Kunming, China: 2003. pp. 1–54. [Google Scholar]
- 23.Ju Y., Zhuo J.X., Liu B., Long C.L. Eating from the wild: Diversity of wild edible plants used by Tibetans in Shangri–la region, Yunnan, China. J. Ethnobiol. Ethnomed. 2013;1:28. doi: 10.1186/1746-4269-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wanasinghe D.N., Wijayawardene N.N., Xu J.C., Cheewangkoon R., Mortimer P.E. Taxonomic novelties in Magnolia-associated pleosporalean fungi in the Kunming Botanical Gardens (Yunnan, China) PLoS ONE. 2020;15:e0235855. doi: 10.1371/journal.pone.0235855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marasinghe D.S., Samarakoon M.C., Hongsanan S., Boonmee S., Mckenzie E.H.C. Iodosphaeria honghense sp. nov. (Iodosphaeriaceae, Xylariales) from Yunnan Province, China. Phytotaxa. 2019;420:273–282. doi: 10.11646/phytotaxa.420.4.3. [DOI] [Google Scholar]
- 26.Zeki H.F., Ajmi R.N., Ati E.M. Phytoremediation mechanisms of mercury (Hg) between some plants and soils in Baghdad City. Plant Arch. 2019;19:1395–1401. [Google Scholar]
- 27.Beshah F., Hunde Y., Getachew M., Bachheti R.K., Husen A., Bachheti A. Ethnopharmacological, phytochemistry and other potential applications of Dodonaea genus: A comprehensive review. Curr. Biotechnol. 2020;2:103–119. doi: 10.1016/j.crbiot.2020.09.002. [DOI] [Google Scholar]
- 28.Prakash N.K.U., Selvi C.R., Sasikala V., Dhanalakshmi S., Prakash S.B.U. Phytochemistry and bio-efficacy of a weed, Dodonaea viscosa. Int. J. Pharm. Pharm. Sci. 2012;4:509–512. [Google Scholar]
- 29.Al-Snafi P.A.E. A review on Dodonaea viscosa: A potential medicinal plant. IOSR J. Pharm. 2017;7:10–21. doi: 10.9790/3013-0702011021. [DOI] [Google Scholar]
- 30.Al-Aamri K.K., Hossain M.A. New prenylated flavonoids from the leaves of Dodonaea viscosa native to the Sultanate of Oman. Pacific Science Review A. Nat. Sci. Eng. 2016;18:53–61. doi: 10.1016/j.psra.2016.08.001. [DOI] [Google Scholar]
- 31.AL-Oraimi A.A., Hossain M.A. In vitro total flavonoids content and antimicrobial capacity of different organic crude extracts of D. viscosa. J. Biol. Active Prod. Nat. 2016;6:150–165. doi: 10.1080/22311866.2016.1188725. [DOI] [Google Scholar]
- 32.Christmas M.J., Biffin E., Lowe A.J. Measuring genome–wide genetic variation to reassess subspecies classifications in Dodonaea viscosa (Sapindaceae) Aust. J. Bot. 2018;66:287–297. doi: 10.1071/BT17046. [DOI] [Google Scholar]
- 33.Selvam V. Trees and Shrubs of the Maldives. FAO Regional Office for Asia and the Pacific, Thammada Press Co., Ltd.; Bangkok, Thailand: 2007. pp. 1–92. [Google Scholar]
- 34.Al-Jobori K.M.M., Ali S.A. Effect of Dodonaea viscosa Jacq. residues on growth and yield of mungbean (Vigna mungo L. Hepper) Afr. J. Biotechnol. 2014;13:2407–2413. doi: 10.5897/AJB2013.13569. [DOI] [Google Scholar]
- 35.Al-Jobori K.M.M., Ali S.A. Evaluation the effect of Dodonaea viscosa Jacq. residues on growth and yield of maize (Zea mays L.) Int. J. Adv. Res. 2014;2:514–521. [Google Scholar]
- 36.Al-Obaidy A.H.M.J., Jasim I.M., Al–Kubaisi A.R.A. Air pollution effects in some plant leaves morphological and anatomical characteristics within Baghdad City. Iraq. Eng. Technol. J. 2019;37:24–28. doi: 10.30684/etj.37.1C.13. [DOI] [Google Scholar]
- 37.Shtein I., Meir S., Riov J., Philosoph-hadas S. Interconnection of seasonal temperature, vascular traits, leaf anatomy and hydraulic performance in cut Dodonaea ‘Dana’ branches. Postharvest Biol. Technol. 2011;61:184–192. doi: 10.1016/j.postharvbio.2011.03.004. [DOI] [Google Scholar]
- 38.Rani M.S., Pippalla R.S., Mohan K. Dodonaea viscosa Linn-An overview. Asian J. Pharm. Res. Health Care. 2009;1:97–112. [Google Scholar]
- 39.Hossain M.A. Biological and phytochemicals review of Omani medicinal plant Dodonaea viscosa. J. King Saud Univ. Sci. 2019;31:1089–1094. doi: 10.1016/j.jksus.2018.09.012. [DOI] [Google Scholar]
- 40.Wanasinghe D.N., Mortimer P.E., Senwanna C., Cheewangkoon R. Saprobic Dothideomycetes in Thailand: Phaeoseptum hydei sp. nov., a new terrestrial ascomycete in Phaeoseptaceae. Phytotaxa. 2020;449:149–163. doi: 10.11646/phytotaxa.449.2.3. [DOI] [Google Scholar]
- 41.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 42.Groenewald J.Z., Nakashima C., Nishikawa J., Shin H.D., Park J.H., Jama A.N., Groenewald M., Braun U., Crous P.W. Species concepts in Cercospora: Spotting the weeds among the roses. Stud. Mycol. 2013;75:115–170. doi: 10.3114/sim0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woudenberg J.H., Aveskamp M.M., de Gruyter J., Spiers A.G., Crous P.W. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia. 2009;22:56–62. doi: 10.3767/003158509X427808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quaedvlieg W., Groenewald J.Z., de Jesús Yáñez-Morales M., Crous P.W. DNA barcoding of Mycosphaerella species of quarantine importance to Europe. Persoonia. 2012;29:101–115. doi: 10.3767/003158512X661282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White T.J., Bruns T., Lee S.J.W.T., Taylor J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols Appl. 1990;18:315–322. doi: 10.1016/b978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- 46.Rehner S.A., Samuels G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994;98:625–634. doi: 10.1016/S0953-7562(09)80409-7. [DOI] [Google Scholar]
- 47.Vilgalys R., Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung G.H., Sung J.M., Hywel-Jones N.L., Spatafora J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007;44:1204–1223. doi: 10.1016/j.ympev.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 49.Quaedvlieg W., Binder M., Groenewald J.Z., Summerell B.A., Carnegie A.J., Burgess T.I., Crous P.W. Introducing the Consolidated Species Concept to resolve species in the Teratosphaeriaceae. Persoonia. 2014;33:1–40. doi: 10.3767/003158514X681981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- 52.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valenzuela-Lopez N., Cano-Lira J.F., Guarro J., Sutton D.A., Wiederhold N., Crous P.W., Stchigel A.M. Coelomycetous Dothideomycetes with emphasis on the families Cucurbitariaceae and Didymellaceae. Stud. Mycol. 2018;90:1–69. doi: 10.1016/j.simyco.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maharachchikumbura S.S.N., Wanasinghe D.N., Cheewangkoon R., Al-Sadi A.M. Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Divers. 2021 doi: 10.1007/s13225-020-00467-1. in press. [DOI] [Google Scholar]
- 55.Mapook A., Hyde K.D., McKenzie E.H.C., Jones E.B.G., Bhat D.J., Jeewon R., Stadler M., Samarakoon M.C., Malaithong M., Tanunchai B., et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed) Fungal Divers. 2020;101:1–175. doi: 10.1007/s13225-020-00444-8. [DOI] [Google Scholar]
- 56.Hyde K.D., Dong Y., Phookamsak R., Jeewon R., Bhat D.J., Jones E., Liu N., Abeywickrama P.D., Mapook A., Wei D., et al. Fungal diversity notes 1151–1276: Taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 2020;100:5–277. doi: 10.1007/s13225-020-00439-5. [DOI] [Google Scholar]
- 57.Kuraku S., Zmasek C.M., Nishimura O., Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013;41:W22–W28. doi: 10.1093/nar/gkt389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh k., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 60.Nylander J.A.A., Wilgenbusch J.C., Warren D.L., Swofford D.L. AWTY (are we there yet?): A system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2008;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- 61.Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; Proceedings of the 2010 Gateway Computing Environments Workshop (GCE); New Orleans, LA, USA. 14 November 2010; pp. 1–8. [DOI] [Google Scholar]
- 62.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rambaut A. FigTree Version 1.4.0. [(accessed on 8 November 2020)]; Available online: http://tree.bio.ed.ac.uk/software/figtree/
- 64.Farr D.F., Rossman A.Y. Fungal Databases, U.S. National Fungus Collections, ARS, USDA. [(accessed on 22 February 2021)]; Available online: https://nt.ars-grin.gov/fungaldatabases/
- 65.Burgess T.I., Tan Y.P., Garnas J., Edwards J., Scarlett K.A., Shuttleworth L.A., Daniel R., Dann E.K., Parkinson L.E., Dinh Q., et al. Current status of the Botryosphaeriaceae in Australia. Australas. Plant Pathol. 2019;48:35–44. doi: 10.1007/s13313-018-0577-5. [DOI] [Google Scholar]
- 66.Mathur R.S. The Coelomycetes of India. Bishen Singh Mahendra Pal Singh; Delhi, India: 1979. 460p [Google Scholar]
- 67.Vandemark G.J., Martinez O., Pecina V., Alvarado M. Assessment of genetic relationships among isolates of Macrophomina phaseolina using a simplified AFLP technique and two different methods of analysis. Mycologia. 2000;92:656–664. doi: 10.1080/00275514.2000.12061206. [DOI] [Google Scholar]
- 68.Dingley J.M., Fullerton R.A., McKenzie E.H.C. Survey of Agricultural Pests and Diseases. FAO; Rome, Italy: 1981. 485p. p. 485. Technical Report Volume 2: Records of Fungi, Bacteria, Algae, and Angiosperms Pathogenic on Plants in Cook Islands, Fiji, Kiribati, Niue, Tonga, Tuvalu, and Western Samoa. [Google Scholar]
- 69. Kamal. Cercosporoid Fungi of INDIA. Bishen Singh Mahendra Pal Singh; Dehra Dun, India: 2010. 351p [Google Scholar]
- 70.Srivastava R.K., Srivastava N., Srivastava A.K. Additions to genus Cercospora from North-Eastern Uttar Pradesh. Proc. Natl. Acad. Sci. India. 1994;64:105–114. [Google Scholar]
- 71.Crous P.W., Braun U. Mycosphaerella and Its Anamorphs: 1. Names Published in Cercospora and Passalora. Centraalbureau voor Schimmelcultures; Utrecht, The Netherlands: 2003. 571p [Google Scholar]
- 72.Deighton F.C. Preliminary list of fungi and diseases of plants in Sierra Leone and list of fungi collected in Sierra Leone. Bull. Misc. Inf. 1936;1936:397–424. doi: 10.2307/4111838. [DOI] [Google Scholar]
- 73.Pennycook S.R. Plant Diseases Recorded in New Zealand. D.S.I.R.; Auckland, New Zealand: 1989. 958p 3 Vol. Pl. Dis. Div. [Google Scholar]
- 74.Gadgil P.D. Fungi on Trees and Shrubs in New Zealand. Fungal Diversity Press; Hong Kong: 2005. 437p Fungi of New Zealand (Volume 4) [Google Scholar]
- 75.Alfieri S.A., Jr., Langdon K.R., Wehlburg C., Kimbrough J.W. Index of Plant Diseases in Florida (Revised) Fla. Dept. Agric. Consum. Serv. Div. Plant Ind. Bull. 1984;11:1–389. [Google Scholar]
- 76.Boesewinkel H.J. Pseudocercospora dodonaeae sp. nov. and a note on powdery mildew of Dodonaea in New Zealand. Trans. Br. Mycol. Soc. 1981;77:453–455. [Google Scholar]
- 77.De Miranda B.E.C., Ferreira B.W., Alves J.L., de Macedo D.M., Barreto R.W. Pseudocercospora lonicerigena a leaf spot fungus on the invasive weed Lonicera japonica in Brazil. Australas. Plant Pathol. 2014;43:339–345. doi: 10.1007/s13313-014-0275-x. [DOI] [Google Scholar]
- 78.Osorio J.A., Wingfield M.J., De Beer Z.W., Roux J. Pseudocercospora mapelanensis sp. nov., associated with a fruit and leaf disease of Barringtonia racemosa in South Africa. Australas. Plant Pathol. 2015;44:349–359. doi: 10.1007/s13313-015-0357-4. [DOI] [Google Scholar]
- 79.Liu X.J., Guo Y.L., editors. Flora Fungorum Sinicorum. Science Press; Beijing, China: 1998. 474p Vol. 9. Pseudocercospora. [Google Scholar]
- 80.Ahmad S., Iqbal S.H., Khalid A.N. Fungi of Pakistan. Sultan Ahmad Mycological Society of Pakistan; Lahore, Pakistan: 1997. p. 248. [Google Scholar]
- 81.Ciferri R. Mycoflora Domingensis Integrata. Quaderno. 1961;19:1–539. [Google Scholar]
- 82.Dennis R.W.G. Fungus Flora of Venezuela and Adjacent Countries. Verlag von J. Cramer; Weinheim, Germany: 1970. p. 531. Kew Bulletin Additional Series III. [Google Scholar]
- 83.Farr M.L., Schoknecht J.D., Crane J.L. A Costa Rican black mildew found in Everglades National Park, Florida. Can. J. Bot. 2011;63:1983–1986. doi: 10.1139/b85-277. [DOI] [Google Scholar]
- 84.Urtiaga, R. Host index of plant diseases and disorders from Venezuela—Addendum. Unknown journal or publisher. 2004. 268p.
- 85.Singh K.P., Shukla R.S., Kumar S., Hussain A. A leaf-spot disease of Dodonaea viscosa caused by Corynespora cassiicola in India. Indian Phytopathol. 1982;35:325. [Google Scholar]
- 86.Castellani E., Ciferri R. Prodromus Mycoflorae Africae Orientalis Italicae. Istituto Agricolo Coloniale Italiano; Firenze, Italy: 1937. 167p [Google Scholar]
- 87.Georghiou G.P., Papadopoulos C. A Second List of Cyprus Fungi. Government of Cyprus, Department of Agriculture; Nicosia, Cyprus: 1957. 38p [Google Scholar]
- 88.Sarbhoy A.K., Lal G., Varshney J.L. Fungi of India (1967–71) Navyug Traders; New Delhi, India: 1971. 148p [Google Scholar]
- 89.Pande A. Ascomycetes of Peninsular India. Scientific Publishers (India); Jodhpur, India: 2008. 584p [Google Scholar]
- 90.Amano K. (Hirata) Host Range and Geographical Distribution of the Powdery Mildew Fungi. Japan Sci. Soc. Press; Tokyo, Japan: 1986. 741p [Google Scholar]
- 91.Whiteside J.O. A revised list of plant diseases in Rhodesia. Kirkia. 1966;5:87–196. [Google Scholar]
- 92.Castellani E., Ciferri R. Mycoflora Erythraea, Somala et Aethipica Suppl. 1. Atti Ist. Bot. Lab. Crittog. Univ.; Pavia, Italy: 1950. 52p [Google Scholar]
- 93.Boesewinkel H.J. Erysiphaceae of New Zealand. Sydowia. 1979;32:13–56. [Google Scholar]
- 94.Doidge E.M. The South African fungi and lichens to the end of 1945. Bothalia. 1950;5:1–1094. [Google Scholar]
- 95.French A.M. California Plant Disease Host Index. Calif. Dept. Food Agric.; Sacramento, CA, USA: 1989. 394p [Google Scholar]
- 96.Hansford C.G. The Meliolineae. A monograph. Beih. Sydowia. 1961;2:1–806. [Google Scholar]
- 97.Goos R.D., Anderson J.H. The Meliolaceae of Hawaii. Sydowia. 1972;26:73–80. [Google Scholar]
- 98.Raabe R.D., Conners I.L., Martinez A.P. Checklist of Plant Diseases in Hawaii. Hawaii Inst. Trop. Agric. Human Resources; Honolulu, HI, USA: 1981. 313p College of Tropical Agriculture and Human Resources, University of Hawaii. Information Text Series No. 22. [Google Scholar]
- 99.Stevens F.L. Hawaiian Fungi. Bernice P. Bishop Mus. Bull. 1925;19:1–189. [Google Scholar]
- 100.Crous P.W. Taxonomy and Pathology of Cylindrocladium (Calonectria) and Allied Genera. American Phytopathological Society; St. Paul, MN, USA: 2002. 278p [Google Scholar]
- 101.Schoch C.L., Crous P.W., Cronwright G., El-Gholl N.E., Wingfield B.D. Recombination in Calonectria morganii and phylogeny with other heterothallic small-spored Calonectria species. Mycologia. 2000;92:665–673. doi: 10.1080/00275514.2000.12061207. [DOI] [Google Scholar]
- 102.Polizzi G., Catara V. First report of leaf spot caused by Cylindrocladium pauciramosum on Acacia retinodes, Arbutus unedo, Feijoa sellowiana and Dodonaea viscosa in southern Italy. Plant Dis. 2001;85:803. doi: 10.1094/PDIS.2001.85.7.803C. [DOI] [PubMed] [Google Scholar]
- 103.Gerlach W., Ershad D. Beitrag zur Kenntnis der Fusarium—Und Cylindrocarpon-Arten in Iran. Nova Hedwigia. 1970;20:725–784. [Google Scholar]
- 104.Tilak S., Rao R. Contribution to our knowledge of ascomycetes of India. V. Mycopathol. Mycol. Appl. 1966;28:90–94. doi: 10.1007/BF02276033. [DOI] [PubMed] [Google Scholar]
- 105.Mostert L., Groenewald J.Z., Summerbell R.C., Robert V., Sutton D.A., Padhye A.A., Crous P.W. Species of Phaeoacremonium associated with infection in humans and environmental reservoirs in infected woody plants. J. Clin. Microbiol. 2005;43:1752–1767. doi: 10.1128/JCM.43.4.1752-1767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mostert L., Groenewald J.Z., Summerbell R.C., Gams W., Crous P.W. Taxonomy and pathology of Togninia (Diaporthales) and its Phaeoacremonium anamorphs. Stud. Mycol. 2006;54:1–115. doi: 10.3114/sim.54.1.1. [DOI] [Google Scholar]
- 107.Gramaje D., Mostert L., Groenewald J.Z., Crous P.W. Phaeoacremonium: From esca disease to phaeohyphomycosis. Fungal Biol. 2015;119:759–783. doi: 10.1016/j.funbio.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 108.Gramaje D., Leon M., Perez-Sierra A., Burgess T., Armengol J. New Phaeoacremonium species isolated from sandalwood trees in Western Australia. IMA Fungus. 2014;5:67–77. doi: 10.5598/imafungus.2014.05.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Spies C.F.J., Moyo P., Halleen F., Mostert L. Phaeoacremonium species diversity on woody hosts in the Western Cape Province of South Africa. Persoonia. 2018;40:26–62. doi: 10.3767/persoonia.2018.40.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gilbertson R.L., Bigelow D.M., Hemmes D.E., Desjardin D.E. Annotated check list of Wood-Rotting Basidiomycetes of Hawaii. Mycotaxon. 2002;82:215–239. [Google Scholar]
- 111.Nakasone K.K., Burdsall H.H., Jr. The genus Dendrothele (Agaricales, Basidiomycota) in New Zealand. N. Z. J. Bot. 2011;49:107–131. doi: 10.1080/0028825X.2010.512636. [DOI] [Google Scholar]
- 112.Aiello D., Guarnaccia V., Formica P.T., Hyakumachi M., Polizzi G. Occurence and characterisation of Rhizoctonia species causing diseases of ornamental plants in Italy. Eur. J. Plant Pathol. 2017;148:967–982. doi: 10.1007/s10658-017-1150-8. [DOI] [Google Scholar]
- 113.Pildain M.B., Perez G.A., Robledo G., Pappano D.B., Rajchenberg M. Arambarria the pathogen involved in canker rot of Eucalyptus, native trees wood rots and grapevine diseases in the southern hemisphere. For. Pathol. 2017;47:e12397. doi: 10.1111/efp.12397. [DOI] [Google Scholar]
- 114.Fischer M., Edwards J., Cunnington J.H., Pascoe I.G. Basidiomycetous pathogens on grapevine: A new species from Australia—Fomitiporia australiensis. Mycotaxon. 2005;92:85–96. [Google Scholar]
- 115.Gilbertson R.L., Martin K.J., Lindsey J.P. Annotated Check List and Host Index for Arizona Wood-Rotting Fungi. Volume 209. College of Agriculture, University of Arizona; Tucson, AZ, USA: 1974. pp. 1–48. [Google Scholar]
- 116.Gilbertson R.L. The genus Phellinus (Aphyllophorales: Hymenochaetaceae) in western North America. Mycotaxon. 1979;9:51–89. [Google Scholar]
- 117.Boedijn K.B. The Uredinales of Indonesia. Nova Hedwigia. 1959;1:463–496. [Google Scholar]
- 118.Hardy G.E., Sivasithamparam K. Phytophthora spp. associated with container- grown plants in nurseries in Western Australia. Plant Dis. 1988;72:435–437. [Google Scholar]
- 119.Shivas R.G. Fungal and bacterial diseases of plants in Western Australia. J. R. Soc. West. Aust. 1989;72:1–62. [Google Scholar]
- 120.Erwin D.C., Ribeiro O.K. Phytophthora Diseases Worldwide. APS Press; St. Paul, MN, USA: 1996. 562p [Google Scholar]
- 121.Mammella M.A., Cacciola S.O., Martin F., Schena L. Genetic characterization of Phytophthora nicotianae by the analysis of polymorphic regions of the mitochondrial DNA. Fungal Biol. 2011;115:432–442. doi: 10.1016/j.funbio.2011.02.018. [DOI] [PubMed] [Google Scholar]
- 122.Mammella M.A., Martin F.N., Cacciola S.O., Coffey M.D., Faedda R., Schena L. Analyses of the population structure in a global collection of Phytophthora nicotianae isolates inferred from mitochondrial and nuclear DNA sequences. Phytopathology. 2013;103:610–622. doi: 10.1094/PHYTO-10-12-0263-R. [DOI] [PubMed] [Google Scholar]
- 123.Jung T., Orlikowski L., Henricot B., Abad-Campos P., Aday A.G., Aguin Casal O., Bakonyi J., Cacciola S.O., Cech T., Chavarriaga D., et al. Widespread Phytophthora infestations in European nurseries put forest, semi-natural and horticultural ecosystems at high risk of Phytophthora diseases. For. Pathol. 2016;46:134–163. doi: 10.1111/efp.12239. [DOI] [Google Scholar]
- 124.Crous P.W., Schoch C.L., Hyde K.D., Wood A.R., Gueidan C., de Hoog G.S., Groenewald J.Z. Phylogenetic lineages in the Capnodiales. Stud. Mycol. 2009;64:17–4757. doi: 10.3114/sim.2009.64.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Videira S.I., Groenewald J.Z., Braun U., Shin H.D., Crous P.W. All that glitters is not Ramularia. Stud. Mycol. 2016;83:49–163. doi: 10.1016/j.simyco.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Crous P.W., Carnegie A.J., Wingfield M.J., Sharma R., Mughini G., Noordeloos M.E., Santini A., Shouche Y.S., Bezerra J.D.P., Dima B., et al. Fungal Planet description sheets: 868–950. Persoonia. 2019;42:291–473. doi: 10.3767/persoonia.2019.42.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jayasiri S.C., Hyde K.D., Jones E.B.G., McKenzie E.H.C., Jeewon R., Phillips A.J.L., Bhat D.J., Wanasinghe D.N., Liu J.K., Lu Y.Z., et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere. 2019;10:1–186. doi: 10.5943/mycosphere/10/1/1. [DOI] [Google Scholar]
- 128.Zhou Y., Gong G., Zhang S., Liu N., Wand L., Li P., Yu X. A new species of the genus Trematosphaeria from China. Mycol Prog. 2014;13:33–43. doi: 10.1007/s11557-013-0889-3. [DOI] [Google Scholar]
- 129.Thambugala K.M., Hyde K.D., Tanaka K., Tian Q., Wanasinghe D.N., Ariyawansa H.A., Jayasiri S.C., Boonmee S., Camporesi E., Hashimoto A., et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015;74:199–266. doi: 10.1007/s13225-015-0348-3. [DOI] [Google Scholar]
- 130.Wanasinghe D.N., Phukhamsakda C., Hyde K.D., Jeewon R., Lee H.B., Jones E.B.G., Tibpromma S., Tennakoon D.S., Dissanayake A.J., Jayasiri S.C., et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018;89:1–236. doi: 10.1007/s13225-018-0395-7. [DOI] [Google Scholar]
- 131.Crous P.W., Wingfield M.J., Burgess T.I., Hardy G., Gené J., Guarro J., Baseia I.G., García D., Gusmão L., Souza-Motta C.M. Fungal Planet description sheets: 716–784. Persoonia. 2018;40:239–392. doi: 10.3767/persoonia.2018.40.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Perez-Ortega S., Garrido-Benavent I., De Los Rios A. Austrostigmidium, a new austral genus of lichenicolous fungi close to rock-inhabiting meristematic fungi in Teratosphaeriaceae. Lichenologist. 2015;47:143–156. doi: 10.1017/S0024282915000031. [DOI] [Google Scholar]
- 133.Hirayama K., Tanaka K. Taxonomic revision of Lophiostoma and Lophiotrema based on reevaluation of morphological characters and molecular analyses. Mycoscience. 2011;52:401–412. doi: 10.1007/s10267-011-0126-3. [DOI] [Google Scholar]
- 134.Wanasinghe D.N., Hyde K.D., Jeewon R., Crous P.W., Wijayawardene N.N., Jones E.B.G., Bhat D.J., Phillips A.J.L., Groenewald J.Z., Dayarathne M.C., et al. Phylogenetic revision of Camarosporium (Pleosporineae, Dothideomycetes) and allied genera. Stud. Mycol. 2017;87:207–256. doi: 10.1016/j.simyco.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schoch C.L., Shoemaker R.A., Seifert K.A., Hambleton S., Spatafora J.W., Crous P.W. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia. 2006;98:1041–1052. doi: 10.3852/mycologia.98.6.1041. [DOI] [PubMed] [Google Scholar]
- 136.Crous P.W., Tanaka K., Summerell B.A., Groenewald J.Z. Additions to the Mycosphaerella complex. IMA Fungus. 2011;2:49–64. doi: 10.5598/imafungus.2011.02.01.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Liu J.K., Hyde K.D., Jones E.B.G., Ariyawansa H.A., Bhat D.J., Boonmee S., Maharachchikumbura S.S.N., Mckenzie E.H.C., Phookamsak R., Phukhamsakda C., et al. Fungal diversity notes 1–110: Taxonomic and phylogenetic contributions to fungal species. Fungal Divers. 2015;72:1–197. doi: 10.1007/s13225-015-0324-y. [DOI] [Google Scholar]
- 138.Verkley G.J., Dukik K., Renfurm R., Goker M., Stielow J.B. Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota) Persoonia. 2014;32:25–51. doi: 10.3767/003158514X679191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Egidi E., De Hoog G.S., Isola D., Onofri S., Quaedvlieg W., De Vries M., Verkley G.J.M., Stielow J.B., Zucconi L., Selbmann L. Phylogeny and taxonomy of meristematic rock–inhabiting black fungi in the Dothideomycetes based on multi–locus phylogenies. Fungal Divers. 2014;65:127–165. doi: 10.1007/s13225-013-0277-y. [DOI] [Google Scholar]
- 140.Hashimoto A., Hirayama K., Takahashi H., Matsumura M., Okada G., Chen C.Y., Huang J.W., Kakishima M., Ono T., Tanaka K. Resolving the Lophiostoma bipolare complex: Generic delimitations within Lophiostomataceae. Stud. Mycol. 2018;90:161–189. doi: 10.1016/j.simyco.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Crous P.W., Summerell B.A., Shivas R.G., Burgess T.I., Decock C.A., Dreyer L.L., Granke L.L., Guest D.I., Hardy G.E., Hausbeck M.K., et al. Fungal Planet description sheets: 107–127. Persoonia. 2012;28:138–182. doi: 10.3767/003158512X652633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Schoch C.L., Crous P.W., Groenewald J.Z., Boehm E.W., Burgess T.I., de Gruyter J., de Hoog G.S., Dixon L.J., Grube M., Gueidan C., et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009;64:1–15. doi: 10.3114/sim.2009.64.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Crous P.W., Wingfield M.J., Guarro J., Cheewangkoon R., van der Bank M., Swart W.J., Stchigel A.M., Cano-Lira J.F., Roux J., Madrid H., et al. Fungal Planet description sheets: 154–213. Persoonia. 2013;31:188–296. doi: 10.3767/003158513X675925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Selbmann L., de Hoog G.S., Zucconi L., Isola D., Ruisi S., van den Ende A.H., Ruibal C., De Leo F., Urzì C., Onofri S. Drought meets acid: Three new genera in a dothidealean clade of extremotolerant fungi. Stud Mycol. 2008;61:1–20. doi: 10.3114/sim.2008.61.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bao D.F., Su H.Y., Maharachchikumbura S.S.N., Liu J.K., Nalumpang S., Luo Z.L., Hyde K.D. Lignicolous freshwater fungi from China and Thailand: Multi-gene phylogeny reveals new species and new records in Lophiostomataceae. Mycosphere. 2019;10:1080–1099. doi: 10.5943/mycosphere/10/1/20. [DOI] [Google Scholar]
- 146.Phukhamsakda C., Ariyawansa H.A., Phillips A.L.J., Wanasinghe D.N., Bhat D.J., McKenzie E.H.C., Singtripop C., Camporesi E., Hyde K.D. Additions to Sporormiaceae: Introducing Two Novel Genera, Sparticola and Forliomyces, from Spartium. Cryptogam. Mycol. 2016;37:75–97. doi: 10.7872/crym/v37.iss1.2016.75. [DOI] [Google Scholar]
- 147.Hyde K., Hongsanan S., Jeewon R., Bhat D.J., Mckenzie E., Jones E., Phookamsak R., Ariyawansa H., Boonmee S., Zhao Q. Fungal diversity notes 367–490: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;80:1–270. doi: 10.1007/s13225-016-0373-x. [DOI] [Google Scholar]
- 148.Hyde K.D., de Silva N.I., Jeewon R., Bhat D.J., Phookamsak R., Doilom M., Boonmee S., Jayawardena R.S., Maharachchikumbura S.S.N., Senanayake I.C., et al. AJOM new records and collections of fungi: 1–100. AJOM. 2020;3:22–294. doi: 10.5943/ajom/3/1/3. [DOI] [Google Scholar]
- 149.Boehm E.W., Schoch C.L., Spatafora J.W. On the evolution of the Hysteriaceae and Mytilinidiaceae (Pleosporomycetidae, Dothideomycetes, Ascomycota) using four nuclear genes. Mycol. Res. 2009;113:461–479. doi: 10.1016/j.mycres.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 150.Jayasiri S.C., Hyde K.D., Jones E.B.G., Persoh D., Camporesi E., Kang J.C. Taxonomic novelties of hysteriform Dothideomycetes. Mycosphere. 2018;9:803–837. doi: 10.5943/mycosphere/9/4/8. [DOI] [Google Scholar]
- 151.Phookamsak R., Hyde K., Jeewon R., Bhat D.J., Jones E., Maharachchikumbura S., Raspé O., Karunarathna S., Wanasinghe D.N., Hongsanan S., et al. Fungal diversity notes 929–1035: Taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 2019;95:1–273. doi: 10.1007/s13225-019-00421-w. [DOI] [Google Scholar]
- 152.Crous P.W., Wingfield M.J., Burgess T.I., Hardy G.E., Crane C., Barrett S., Cano-Lira J.F., Le Roux J.J., Thangavel R., Guarro J., et al. Fungal Planet description sheets: 469–557. Persoonia. 2016;37:218–403. doi: 10.3767/003158516X694499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.De Gruyter J., Woudenberg J.H.C., Aveskamp M.M., Verkley G.J.M., Groenewald J.Z., Crous P.W. Redisposition of phoma–like anamorphs in Pleosporales. Stud. Mycol. 2013;75:1–36. doi: 10.3114/sim0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vu D., Groenewald M., de Vries M., Gehrmann T., Stielow B., Eberhardt U., Al-Hatmi A., Groenewald J.Z., Cardinali G., Houbraken J., et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ariyawansa H.A., Hyde K.D., Jayasiri S.C., Buyck B., Chethana K.W.T., Dai D.Q., Dai Y.C., Daranagama D.A., Jayawardena R.S., Lücking R., et al. Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015;75:27–274. doi: 10.1007/s13225-015-0346-5. [DOI] [Google Scholar]
- 156.Schoch C.L., Seifertb K.A., Huhndorfc S., Robertd V., Spougea J.L., Levesqueb C.A., Chenb W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tanaka K., Hirayama K., Yonezawa H., Sato G., Toriyabe A., Kudo H., Hashimoto A., Matsumura M., Harada Y., Kurihara Y., et al. Revision of the Massarineae (Pleosporales, Dothideomycetes) Stud. Mycol. 2015;82:75–136. doi: 10.1016/j.simyco.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Y., Wang H.K., Fournier J., Crous P.W., Jeewon R., Pointing S.B., Hyde K.D. Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers. 2009;38:225–251. [Google Scholar]
- 159.Liew E.C., Aptroot A., Hyde K.D. An evaluation of the monophyly of Massarina based on ribosomal DNA sequences. Mycologia. 2002;94:803–813. doi: 10.2307/3761695. [DOI] [PubMed] [Google Scholar]
- 160.Jaklitsch W.M., Checa J., Blanco M.N., Olariaga I., Tello S., Voglmayr H. A preliminary account of the Cucurbitariaceae. Stud. Mycol. 2018;90:71–118. doi: 10.1016/j.simyco.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wanasinghe D.N., Camporesi E., Hu D.M. Neoleptosphaeria jonesii sp. nov., a novel saprobic sexual species, in Leptosphaeriaceae. Mycosphere. 2016;7:1368–1377. doi: 10.5943/mycosphere/7/9/10. [DOI] [Google Scholar]
- 162.De Gruyter J., Aveskamp M.M., Woudenberg J.H., Verkley G.J., Groenewald J.Z., Crous P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009;113:508–519. doi: 10.1016/j.mycres.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 163.Hyde K.D., Chaiwan N., Norphanphoun C., Boonmee S., Camporesi E., Chethana K.W.T., Dayarathne M.C., de Silva N.I., Dissanayake A.J., Ekanayaka A.H., et al. Mycosphere notes 169–224. Mycosphere. 2018;9:271–430. doi: 10.5943/mycosphere/9/2/8. [DOI] [Google Scholar]
- 164.de Gruyter J., Woudenberg J.H., Aveskamp M.M., Verkley G.J., Groenewald J.Z., Crous P.W. Systematic reappraisal of species in Phoma section Paraphoma, Pyrenochaeta and Pleurophoma. Mycologia. 2010;102:1066–1081. doi: 10.3852/09-240. [DOI] [PubMed] [Google Scholar]
- 165.Marincowitz S., Crous P.W., Groenewald J.Z., Wingfield M.J. Microfungi Occurring on Proteaceae in the Fynbos. Evolutionary Phytopathology, CBS Fungal Biodiversity Centre; Utrecht, The Netherlands: 2008. pp. 1–166. CBS Biodiversity Series. [Google Scholar]
- 166.Li W.J., McKenzie E.H., Liu J.K.J., Bhat D.J., Dai D.Q., Camporesi E., Tian Q., Maharachchikumbura S.S.N., Luo Z.L., Shang Q.J., et al. Taxonomy and phylogeny of hyaline–spored coelomycetes. Fungal Divers. 2020;100:279–801. doi: 10.1007/s13225-020-00440-y. [DOI] [Google Scholar]
- 167.Phukhamsakda C., McKenzie E.H.C., Phillips A.J.L., Jones E.B.G., Bhat D.J., Stadler M., Bhunjun C.S., Wanasinghe D.N., Thongbai B., Camporesi E., et al. Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal Divers. 2020;102:1–203. doi: 10.1007/s13225-020-00448-4. [DOI] [Google Scholar]
- 168.Li G.J., Hyde K.D., Zhao R.L., Hongsanan S., Abdel–Aziz F.A., Abdel–Wahab M.A., Alvarado P., Alves–Silva G., Ammirati J.F., Ariyawansa H.A., et al. Fungal diversity notes 253–366: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237. doi: 10.1007/s13225-016-0366-9. [DOI] [Google Scholar]
- 169.Hyde K.D., Norphanphoun C., Abreu V.P., Bazzicalupo A., Chethana K.W.T., Clericuzio M., Dayarathne M.C., Dissanayake A.J., Ekanayaka A.H., He M., et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017;87:1–235. doi: 10.1007/s13225-017-0391-3. [DOI] [Google Scholar]
- 170.Crous P.W., Summerell B.A., Shivas R.G., Romberg M., Mel’nik V.A., Verkley G.J., Groenewald J.Z. Fungal Planet description sheets: 92–106. Persoonia. 2011;27:130–162. doi: 10.3767/003158511X617561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Duarte A.P.M., Attili–Angelis D., Baron N.C., Groenewald J.Z., Crous P.W., Pagnocca F.C. Riding with the ants. Persoonia. 2017;38:81–99. doi: 10.3767/003158517X693417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dayarathne M.C., Jones E.B.G., Maharachchikumbura S.S.N., Devadatha B., Sarma V.V., Khongphinitbunjong K., Chomnunti P., Hyde K.D. Morpho–molecular characterization of micro fungi associated with marine based habitats. Mycosphere. 2020;11:1–188. doi: 10.5943/mycosphere/11/1/1. [DOI] [Google Scholar]
- 173.Tibpromma S., Hyde K.D., Jeewon R., Maharachchikumbura S.S.N., Liu J.K., Bhat D.J., Jones E.B.G., McKenzie E.H.C., Camporesi E., Bulgakov T.S., et al. Fungal diversity notes 491–602: Taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2017;83:1–261. doi: 10.1007/s13225-017-0378-0. [DOI] [Google Scholar]
- 174.Suetrong S., Schoch C.L., Spatafora J.W., Kohlmeyer J., Volkmann-Kohlmeyer B., Sakayaroj J., Phongpaichit S., Tanaka K., Hirayama K., Jones E.B.G. Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 2009;64:155–173. doi: 10.3114/sim.2009.64.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Crous P.W., Shivas R.G., Quaedvlieg W., van der Bank M., Zhang Y., Summerell B.A., Guarro J., Wingfield M.J., Wood A.R., Alfenas A.C., et al. Fungal Planet description sheets: 214–280. Persoonia. 2014;32:184–306. doi: 10.3767/003158514X682395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Van Nieuwenhuijzen E.J., Houbraken J.A.M.P., Punt P.J., Roeselers G., Adan O.C.G., Samson R.A. The fungal composition of natural biofinishes on oil-treated wood. Fungal Biol Biotechnol. 2017;4:1–13. doi: 10.1186/s40694-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thambugala K.M., Hyde K.D., Eungwanichayapant P.D., Romero A.I., Liu Z.Y. Additions to the genus Rhytidhysteron in Hysteriaceae. Cryptogam Mycol. 2016;37:99–116. doi: 10.7872/crym/v37.iss1.2016.99. [DOI] [Google Scholar]
- 178.Kumar V., Cheewangkoon R., Thambugala K.M., Jones G.E.B., Brahmanage R.S., Doilom M., Jeewon R., Hyde K.D. Rhytidhysteron mangrovei (Hysteriaceae), a new species from mangroves in Phetchaburi Province, Thailand. Phytotaxa. 2019;401:166–178. doi: 10.11646/phytotaxa.401.3.2. [DOI] [Google Scholar]
- 179.Mugambi G.K., Huhndorf S.M. Parallel evolution of hysterothecial ascomata in ascolocularous fungi (Ascomycota, Fungi) Syst. Biodivers. 2009;7:453–464. doi: 10.1017/S147720000999020X. [DOI] [Google Scholar]
- 180.Almeida D.A.C., Gusmao L.F.P., Miller A.N. A new genus and three new species of hysteriaceous ascomycetes from the semiarid region of Brazil. Phytotaxa. 2014;176:298–308. doi: 10.11646/phytotaxa.176.1.28. [DOI] [Google Scholar]
- 181.Yacharoen S., Tian Q., Chomnunti P., Boonmee S., Chukeatirote E., Bhat J.D., Hyde K.D. Patellariaceae revisited. Mycosphere. 2015;6:290–326. doi: 10.5943/mycosphere/6/3/7. [DOI] [Google Scholar]
- 182.Boehm E.W., Mugambi G.K., Miller A.N., Huhndorf S.M., Marincowitz S., Spatafora J.W., Schoch C.L. A molecular phylogenetic reappraisal of the Hysteriaceae, Mytilinidiaceae and Gloniaceae (Pleosporomycetidae, Dothideomycetes) with keys to world species. Stud. Mycol. 2009;64:49–83. doi: 10.3114/sim.2009.64.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Doilom M., Dissanayake A.J., Wanasinghe D.N., Boonmee S., Liu J.K., Bhat D.J., Taylor J.E., Bahkali A.H., McKenzie E.H.C., Hyde K.D. Micro fungi on Tectona grandis (teak) in Northern Thailand. Fungal Divers. 2016;82:107–182. doi: 10.1007/s13225-016-0368-7. [DOI] [Google Scholar]
- 184.Jaklitsch W.M., Olariaga I., Voglmayr H. Teichospora and the Teichosporaceae. Mycol. Prog. 2016;15:31. doi: 10.1007/s11557-016-1171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wanasinghe D.N., Jones E.B.G., Dissanayake A.J., Hyde K.D. Saprobic Dothideomycetes in Thailand: Vaginatispora appendiculata sp. nov. (Lophiostomataceae) introduced based on morphological and molecular data. Stud. Fungi. 2016;1:56–68. doi: 10.5943/sif/1/1/5. [DOI] [Google Scholar]
- 186.Aveskamp M.M., de Gruyter J., Woudenberg J.H., Verkley G.J., Crous P.W. Highlights of the Didymellaceae: A polyphasic approach to characterize Phoma and related pleosporalean genera. Stud. Mycol. 2010;65:1–60. doi: 10.3114/sim.2010.65.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Crous P.W., Braun U., Groenewald J.Z. Mycosphaerella is polyphyletic. Stud. Mycol. 2007;58:1–32. doi: 10.3114/sim.2007.58.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hongsanan S., Hyde K.D., Phookamsak R., Wanasinghe D.N., McKenzie E.H.C., Sarma V.V., Boonmee S., Lücking R., Bhat D.J., Liu N.G. Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere. 2020;11:1553–2107. doi: 10.5943/mycosphere/11/1/13. [DOI] [Google Scholar]
- 189.Crous P.W. Taxonomy and phylogeny of the genus Mycosphaerella and its anamorphs. Fungal Divers. 2009;38:1–24. [Google Scholar]
- 190.Crous P.W., Summerell B.A., Mostert L., Groenewald J.Z. Host specificity and speciation of Mycosphaerella and Teratosphaeria species associated with leaf spots of Proteaceae. Persoonia. 2008;20:59–86. doi: 10.3767/003158508X323949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Spegazzini C. Fungi argentini additis nonnullis brasiliensibus montevideensibusque. Pugillus quartus (Continuacion). An. Soc. Cient. Argent. 1881;12:174–189. [Google Scholar]
- 192.Clements F.E., Shear C.L. The Genera of Fungi. Hafner Publishing Co.; New York, NY, USA: 1931. pp. 1–632. [Google Scholar]
- 193.Samuels G.J., Müller E. Life-history studies of Brazilian Ascomycetes. 7. Rhytidhysteron rufulum and the genus Eutryblidiella. Sydowia. 1979;32:277–292. [Google Scholar]
- 194.Sharma M.P., Rawla G.S. Ascomycetes new to India-III. Nova Hedwigia. 1985;42:81–90. [Google Scholar]
- 195.Barr M.E. Some dictyosporous genera and species of Pleosporales in North America. Mem. N. Y. Bot. Gard. 1990;62:1–92. [Google Scholar]
- 196.Magnes M. Weltmonographie der Triblidiaceae. Bibl. Mycol. 1997;165:1–177. [Google Scholar]
- 197.Species Fungorum. [(accessed on 25 February 2021)]; Available online: http://www.speciesfungorum.org/Names/Names.asp.
- 198.De Silva N.I., Tennakoon D.S., Thambugala K.M., Karunarathna S.C., Lumyong S., Hyde K.D. Morphology and multigene phylogeny reveal a new species and a new record of Rhytidhysteron (Dothideomycetes, Ascomycota) from China. AJOM. 2020;3:295–306. doi: 10.5943/ajom/3/1/4. [DOI] [Google Scholar]
- 199.Yu N.T., Xie H.M., Wang J.H., Liu S.B., Liu Z.X. First Report on the Molecular Identification of Phytoplasma (16SrI) Associated with Witches’ Broom on Dodonaea viscosa in China. Plant Dis. 2016;100:6:1232. doi: 10.1094/PDIS-10-15-1221-PDN. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the NCBI GenBank, MycoBank and TreeBASE.