Abstract
The PAX3-FOXO1 fusion protein is the key oncogenic driver in fusion positive rhabdomyosarcoma (FP-RMS), an aggressive soft tissue malignancy with a particularly poor prognosis. Identifying key downstream targets of PAX3-FOXO1 will provide new therapeutic opportunities for treatment of FP-RMS. Herein, we demonstrate that Forkhead Box F1 (FOXF1) transcription factor is uniquely expressed in FP-RMS and is required for FP-RMS tumorigenesis. The PAX3-FOXO1 directly binds to FOXF1 enhancers and induces FOXF1 gene expression. CRISPR/Cas9 mediated inactivation of either FOXF1 coding sequence or FOXF1 enhancers suppresses FP-RMS tumorigenesis even in the presence of PAX3-FOXO1 oncogene. Knockdown or genetic knockout of FOXF1 induces myogenic differentiation in PAX3-FOXO1-positive FP-RMS. Over-expression of FOXF1 decreases myogenic differentiation in primary human myoblasts. In FP-RMS tumor cells, FOXF1 protein binds chromatin near enhancers associated with FP-RMS gene signature. FOXF1 cooperates with PAX3-FOXO1 and E-box transcription factors MYOD1 and MYOG to regulate FP-RMS-specific gene expression. Altogether, FOXF1 functions downstream of PAX3-FOXO1 to promote FP-RMS tumorigenesis.
Introduction
Rhabdomyosarcoma (RMS) is a high-grade soft tissue neoplasm of mesenchymal origin and accounts for 3.5% of cancers among children aged 0-14 years and 2% of cancers among adolescents aged 15-19 years (1). Most RMS tumors can be classified into two subtypes: fusion-positive (FP-RMS) and fusion-negative (FN-RMS). FN-RMS is the most common subtype usually observed in younger children and is associated with good prognosis when localized. FN-RMS is characterized by the presence of a wide range of genetic aberrations, including mutations in TP53 (2), NRAS, KRAS, HRAS (3-5), PIK3CA, CTNNB1 and FGFR4 (6, 7). On the other hand, FP-RMS represent 20-30% of RMS, has a poor prognosis and steady incidence throughout childhood. The FP-RMS tumors are associated with translocation between FOXO1 gene on chromosome 13 and either PAX3 on chromosome 2 or PAX7 on chromosome 1, resulting in production of PAX3-FOXO1 or PAX7-FOXO1 fusion proteins (8). The presence or absence of fusion proteins is a prognostic indicator for the outcomes of RMS (9, 10). While PAX3-FOXO1 and PAX7-FOXO1 fusion proteins are the main drivers of FP-RMS carcinogenesis, key downstream regulators of these fusion proteins remain unclear.
The PAX3-FOXO1 functions predominantly as a transcriptional activator at gene enhancers (11, 12). Once bound to DNA, the PAX3-FOXO1 establishes super-enhancers by recruiting myogenic transcription factors MYOD1 and MYOG as well as MYCN, all of which are highly expressed in FP-RMS (12). Since MYOD1, MYOG and MYCN are also present in FN-RMS albeit at lower levels, it is unclear whether PAX3-FOXO1 requires transcriptional co-regulators that, like the PAX3-FOXO1 fusion itself, are unique for FP-RMS to regulate FP-RMS gene signature. Several signaling pathways are associated with FP-RMS tumorigenesis, including HGF/MET, IGF1R, ALK(13, 14). The receptors for HGF/MET, IGF1R and ALK signaling pathways have been identified as direct PAX3-FOXO1 transcriptional targets (11, 12). Unfortunately, clinical trials targeting these pathways in FP-RMS have demonstrated no improvement in patient survival (15-17) highlighting the need for new therapeutic approaches.
Gene expression profiling of RMS tumors has led to the discovery of a “gene signature” which clearly distinguishes fusion-positive from fusion-negative RMS (18-20). The FP-RMS gene signature is shaped by oncogenic PAX3-FOXO1 fusion protein, myogenic regulatory factors and additional transcriptional machinery which has yet to be characterized (7). We have identified the FOXF1, a member of the Forkhead box family of transcription factors, as one of the most highly and consistently upregulated genes in FP-RMS (18, 20-22). While it is known that FOXF1 is required for mesodermal differentiation toward endothelial and smooth muscle cell lineages (23-27), FOXF1 is not expressed in differentiated skeletal muscle cells, and its role in fusion-positive FP-RMS remains unknown.
In the present studies, we demonstrate that PAX3-FOXO1 directly activates FOXF1 via two distinct enhancers, causing aberrant FOXF1 expression in this skeletal muscle-like tumor cells. FOXF1 is essential for PAX3-FOXO1 oncogene to drive FP-RMS tumorigenesis and to inhibit myogenic differentiation. FOXF1 directly binds to FP-RMS-specific enhancers and cooperates with PAX3-FOXO1, MYOD1 and MYOG to regulate FP-RMS gene signature. Targeting of FOXF1 can expand therapeutic opportunities for patients with fusion-positive FP-RMS.
Materials and Methods
Cell lines and patient-derived RMS xenografts.
Cell lines RD, RH18 (fusion-positive) (28), and RH30 were a kind gift from Dr. Cripe (Nationwide Children’s, OH); RH4 and patient tumor RMS244 (29) were a kind gift from Dr. Khan (NCI, MD). Patient-derived RMS xenografts (PDX) were established by subcutaneous and/or intramuscular transplantation of freshly resected tumor tissue. 0.5x106 tumor cells were resuspended in 50% matrigel (Corning) and inoculated into quadriceps of 4-8-week-old NODscid IL2Rγnull mice. Tumor volumes were calculated as ½ (L x W x W) where L is the largest tumor diameter and W is tumor diameter perpendicular to L. Immunostaining of tissue sections was performed as described (30-32).
CRISPRi and CRISPR KO.
CRISPRi-mediated repression was performed by cloning gRNA into the lentiviral plasmid pLVhU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro (Addgene #71236). For CRISPR/Cas9-mediated knockout studies, gRNAs were cloned into the pSpCas9(BB)-2A-GFP (PX458) plasmid (Addgene #48138). RH18 WT cells were transiently transfected using Viafect Transfection Reagent (Promega) as described (33). GFP+ transfected cells were isolated by FACS. FACS analysis was performed as described (34-36). After clonal expansion, genomic DNA was isolated and a 339bp fragment encompassing the gRNA target site was PCR amplified and cloned into the pMINIT plasmid (NEB). A minimum of six colonies were picked and sequenced to identify the mutation at each allele.
RNAseq analysis.
RNAseq was performed using RH18 cells transduced with lentiviral plasmid pLVhU6-sgRNA hUbC-dCas9-KRAB-T2a-Puro containing empty or gRNA targeting exon 1 of FOXF1 as described (35). For analysis, reads were aligned to human assembly hg19 and differential gene expression performed using EBseq. qRT-PCR was performed as described (26, 31).
ChIP-Seq and HiChIP-Seq.
FOXF1 ChIP-Seq was performed as described (37) using human FP-RMS cell lines RH18 and RH4, and patient tumor RMS244 (29). Immunoprecipitation of FOXF1 was performed with a previously validated anti-FOXF1 antibody (R&D Systems) (37). FOXF1 ChIP-seq and previously published RMS ChIP-seq datasets (from GSE19063, GSE83726) were analyzed using BioWardrobe (SciDAP, https://scidap.com, Datirium, LLC) (38, 39). De novo motif discovery analysis on FOXF1 islands was performed using MEME-ChIP (40). Quantitative comparison between transcription factor datasets was performed using MAnorm (41). Identification of islands containing all TFs was performed using the HOMER merge Peaks function. To compare FOXF1 binding intensities between different clusters, we performed pairwise read density comparisons between each of the eight FOXF1 binding clusters ±500bp surrounding the center of the FOXF1 binding site using MAnorm. Differences in read densities are represented as p value using the Mann-Whitney-Wilcoxon (MWW) test. FOXF1 ChIP-seq read density profiles were generated using MAnorm and represent the average read density within cluster 1, 2, 6 or 8. H3K27ac ChIPseq data: GSE136799 (29), GSE116344 (42), GSE83728 (12), GSE84630 (43). H3K27ac and H3K27me3 PDX ChIPseq data (7). RH4 H3K27ac HiChIP data was obtained from GSE120770, and was processed as previously described (44). ChIPseq from St. Jude PDX models (7) was visualized using PeCanDataPortal https://pecan.stjude.cloud/. RH4 HiChIP data was visualized using Juicebox (45).
HSMM Growth and Differentiation Assays.
Human Skeletal Muscle Myoblasts (HSMM) were purchased from LONZA (Cat. #CC-2580). To establish stable FOXF1 overexpressing and control lines, HSMMs were transduced with pSD44-tagRFP or pSD44-FOXF1HA:tagRFP lentiviral constructs and maintained in vitro as described (46). All experiments were conducted on cells cultured less than five passages. Differentiation index and fusion index was performed by counting at least 10 random 20x images with a minimum of 100 nuclei counted.
Dual Luciferase Assays and Site Directed Mutagenesis.
ERRFI1 enhancer element was PCR amplified and cloned upstream of the minimal promoter of the luciferase reporter plasmid pGL4.23luc2/minP (Promega) as described (46, 47). Point mutations were introduced using the Q5 Site Directed Mutagenesis (NEB) according to the manufacturer’s instructions (48). The RD or RH4 tumor cells were transfected with a Renilla control plasmid and the respective pGL4.23 enhancer construct using Viafect transfection reagent (Promega). 48 hours after transfection, reporter activity was quantified using the Dual-Luciferase Reporter Assay System (Promega) as described (49). Enhancer activity is reported as Firefly luciferase activity normalized to Renilla luciferase.
Study Approval.
All animal studies were approved by Cincinnati Children’s Research Foundation Institutional Animal Care and Use Committee (protocol IACUC2016-0070). Utilization of human rhabdomyosarcoma samples was reviewed and approved by the Office for Research Compliance and Regulatory Affairs of the Cincinnati Children’s Hospital (2008-0021).
Results
FOXF1 is selectively expressed in fusion-positive FP-RMS.
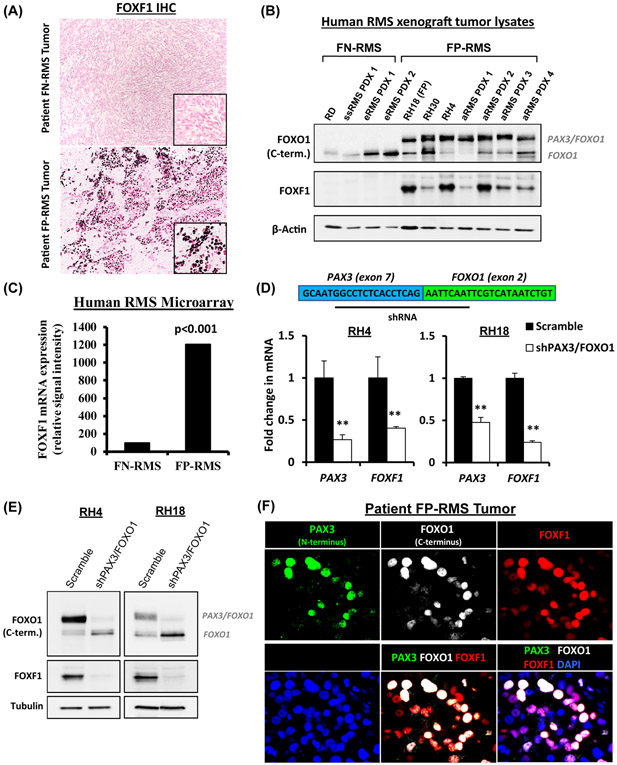
To examine FOXF1 protein expression in RMS, we performed immunohistochemistry and Western blot using surgically resected human RMS tumor specimens, patient-derived xenografts grown in immunocompromised mice and xenografts generated from human RMS cell lines RH18, RH30, RH4 and RD (Fig. 1A-B and Suppl. Fig. S1 and S2). FOXF1 was highly expressed in FP-RMS and correlated with the presence of the PAX3-FOXO1 fusion protein (Fig. 1A-B). In contrast, FOXF1 protein was not detected in FN-RMS (Fig. 1A-B and Suppl. Fig. S1 and S2). In addition, FOXF1 mRNA was increased only in tumors from patients with fusion-positive FP-RMS (Fig. 1C). To determine whether PAX3-FOXO1 fusion protein regulates FOXF1 expression, we used the shRNA spanning the PAX3 – FOXO1 exon junction to selectively knockdown PAX3-FOXO1 in fusion-positive FP-RMS cell lines, RH4 and RH18 (Fig. 1D). Efficient depletion of PAX3-FOXO1 was confirmed by qRT-PCR detecting N-terminal PAX3 mRNA (Fig. 1D) and by Western blot detecting PAX3-FOXO1 fusion protein (Fig. 1E). PAX3-FOXO1 knockdown decreased FOXF1 mRNA and protein in both RH4 and RH18 FP-RMS cells (Fig. 1D-E). In human FP-RMS tumors, FOXF1 staining co-localized with epitopes recognized by antibodies against N-terminal PAX3 and C-terminal FOXO1 (Fig. 1F). Altogether, FOXF1 is selectively expressed in fusion-positive FP-RMS and decreased after inactivation of the PAX3-FOXO1 fusion protein.
Figure 1. FOXF1 expressed in fusion-positive FP-RMS tumors.
(A) Immunostaining shows FOXF1 protein in tumors of patients with FP-RMS (n=13), but not with FN-RMS (n=11). (B) Western blots of tumor lysates show the presence of PAX3-FOXO1 and FOXF1 proteins in human RMS orthotopic xenografts. (C) Increased FOXF1 mRNA in fusion-positive FP-RMS is shown using gene expression microarrays of patient RMS tumors (ArrayExpress accession number E-MEXP-121). Groups are based on histology and fusion status. (D) Depletion of PAX3-FOXO1 decreases FOXF1 gene expression. Inhibition of PAX3-FOXO1 was performed using shRNA targeting the PAX3 and FOXO1 junction. qRT-PCR was performed, and values were normalized to β-actin (n=3). Data reported as mean ± SEM, ** p<0.01. (E) Depletion of PAX3-FOXO1 decreases FOXF1 protein levels. Western blots show efficient depletion of PAX3-FOXO1 fusion protein and FOXF1 protein. (F) FOXF1 co-localizes with PAX3-FOXO1 in FP-RMS tumors. Co-immunostaining of PAX3 (N-terminus), FOXO1 (C-terminus), and FOXF1 in a patient FP-RMS.
FOXF1 is a direct transcriptional target of PAX3-FOXO1.
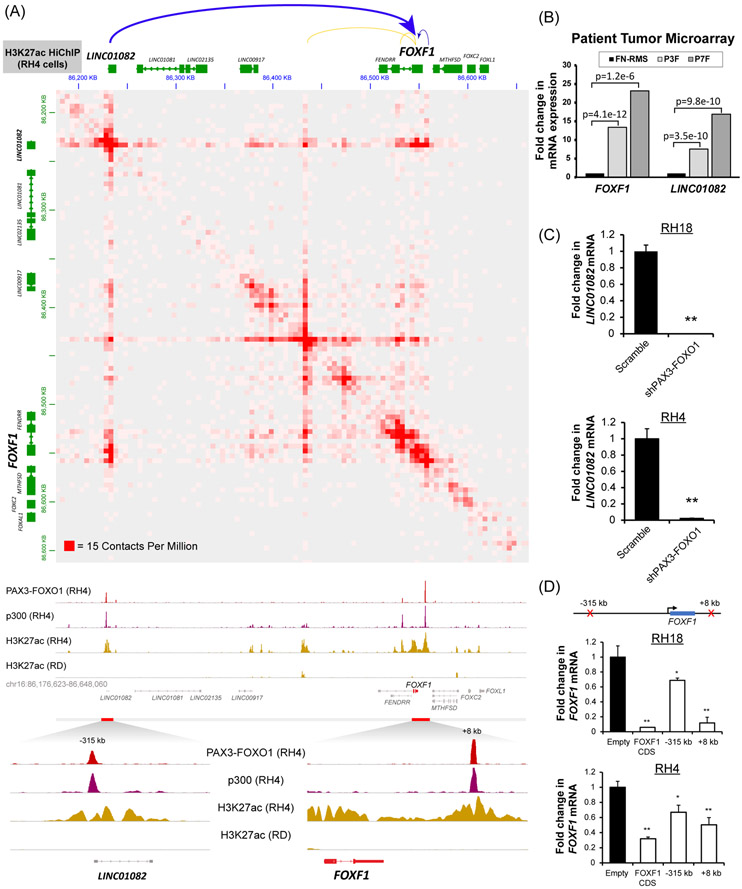
To map the active enhancers which loop to the FOXF1 gene, we analyzed H3K27ac HiChIP(50) and demonstrated the 3D contacts mediated by active chromatin in RH4 cells (Fig. 2A, upper panel). Previously published PAX3-FOXO1 ChIP-Seq data from RH4 cells (11) showed strong PAX3-FOXO1 binding peaks in the −315kb region upstream of the FOXF1 gene and in the +8kb region downstream from the FOXF1 3’UTR (Fig.2A, bottom panel). These binding sites showed features of active enhancers as supported by the presence of p300 and H3K27ac activating marks (Fig. 2A, bottom panel). HiChIP analysis showed direct interactions between the −315kb enhancer, the +8kb enhancer, and their target gene FOXF1 (Fig. 2A, upper panel). Of note, P300 and H3K27ac activating marks were present in FOXF1 enhancers in fusion-positive RH4 cells (FP-RMS), but not in fusion-negative RD cells (FN-RMS) (Fig. 2A). Furthermore, H3K27ac activating marks were found in FOXF1 enhancers in multiple other FP-RMS cell lines and patient tumors, but not in FN-RMS cell lines and patient tumors (Suppl. Fig. S3). Contrary to the activating marks, the polychrome repressive mark H3K27me3 was absent in the FOXF1 enhancers in FP-RMS cell lines and patient tumors (Suppl. Fig. S4), but were present in FOXF1 enhancers in normal skeletal muscles, FN-RMS cell lines and patient tumors (Suppl. Fig. S4). The −315kb PAX3-FOXO1 binding site was located near the transcriptional start site of non-coding RNA LINC01082, which activates the FOXF1 gene expression (51). LINC01082 mRNA was increased in fusion-positive FP-RMS compared to fusion-negative FP-RMS tumors (Fig. 2B). Expression of LINC01082 mirrored FOXF1 mRNA in primary FP-RMS tumors (Fig. 2B), patient-derived xenografts (Suppl. Fig. S5A), orthotopic xenograft tumors from FP-RMS cell lines (Suppl. Fig. S5A) and normal human tissues (Suppl. Fig. S5B). Knockdown of PAX3-FOXO1 resulted in a complete loss of LINC01082 (Fig. 2C), indicating that the PAX3-FOXO1 is essential for LINC01082 transcription. To test whether the −315kb or the +8kb PAX3-FOXO1-binding enhancers are required for FOXF1 gene expression, we used previously described CRISPR repression (CRISPRi) technique (52). This approach uses a nuclease-dead Cas9 (dCas9) fused to the potent Krüppel associated box (KRAB) repression domain which is directed to regulatory elements using a designed gRNA (52). Local repression of either the −315kb upstream or +8kb downstream enhancers significantly decreased FOXF1 mRNA in human FP-RMS cell lines (Fig. 2D). Thus, PAX3-FOXO1 activates the FOXF1 gene expression, at least in part, through the −315kb and the +8kb distal enhancer elements. Altogether, FOXF1 is a transcriptional target of PAX3-FOXO1 and is uniquely expressed in fusion-positive FP-RMS.
Figure 2. FOXF1 is a direct transcriptional target of the PAX3-FOXO1 protein.
(A) (top) HiChIP analysis shows long range chromatin interactions at the FOXF1 gene locus. Both the −315kb and the +8kb enhancers loop to the FOXF1 gene locus (top panel). H3K27ac HiChIP was performed using RH4 cells and is shown at 5kb resolution (GSE120770). ChIP-Seq shows binding of PAX3-FOXO1 fusion protein and p300 to DNA near the FOXF1 gene and LINC01082 in RH4 cells, but not in RD cells (bottom panel). Both enhancers are positive for H3K27ac. Data from GSE19063 and GSE83726. (B) Both the FOXF1 and the LINC01082 mRNA are increased in PAX3-FOXO1-positive (n=26) and PAX7-FOXO1-positive (n=7) RMS but not in fusion-negative FN-RMS (n=25). Data from microarray data GEO series accession number GSE66533 (80). (C) Depletion of PAX3-FOXO1 using shRNA decreased LINC01082 mRNA, shown by qRT-PCR using RH4 and RH18 cells. Values were normalized to β-actin (n=3). Data reported as mean ± SEM. (D) Repression of either −315kb upstream or +8kb downstream enhancers using CRISPRi decreased FOXF1 mRNA. RH4 and RH18 cells were transduced with a lentivirus containing a dCas9:KRAB and a gRNA targeting either −315kb upstream binding site or +8kb downstream binding site. The gRNA targeting the FOXF1 coding sequence (CDS) was used as a positive control. Values were normalized to β-actin (n=3). Data reported as mean ± SEM. * p<0.05; ** p<0.01.
FOXF1 is required for PAX3-FOXO1 to drive FP-RMS tumor growth.
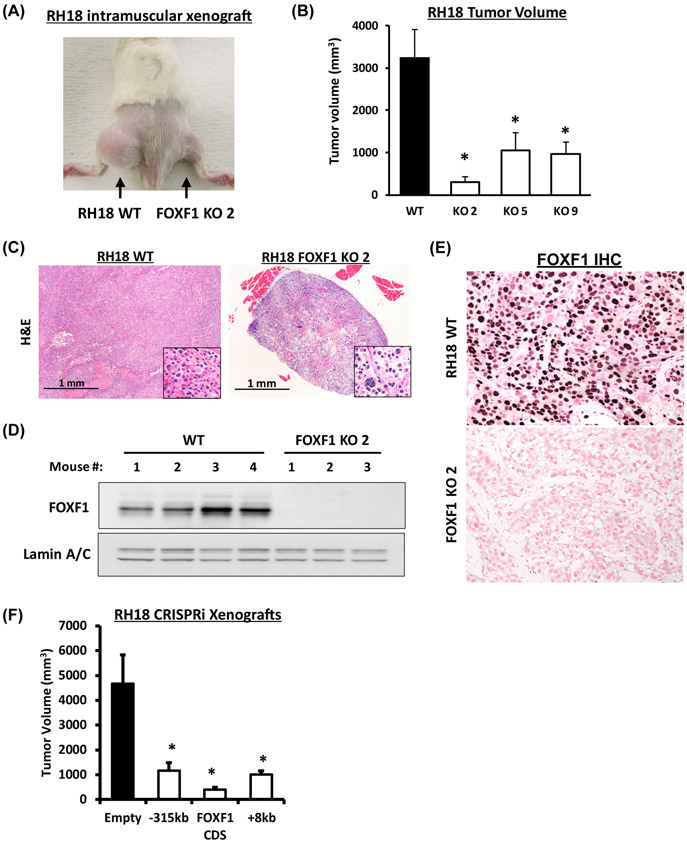
The importance of FOXF1 in PAX3-FOXO1-mediated tumorigenesis was tested using orthotopic FP-RMS mouse model. We generated stable FOXF1 knockout (FOXF1-KO) FP-RMS cell lines using CRISPR/Cas9-mediated gene editing in fusion-positive RH18 cells (Suppl. Fig. S6A). Four FOXF1-KO RH18 clones carried frameshift mutations on all FOXF1 alleles, leading to a complete absence of the FOXF1 protein (Suppl. Fig. S6B-C). The FOXF1-KO clones, number 2, 5 and 9, were used for the intramuscular injections into immunocompromised mice. Deletion of FOXF1 from fusion-positive RH18 tumor cells decreased FP-RMS tumor formation compared to parental RH18 cells shown for all three FOXF1-KO clones (Fig. 3A-B). Time-course studies demonstrated the time-dependent inhibition of tumor growth in FOXF1-knockout FP-RMS (Suppl. Fig. S7 A-B). FOXF1-KO tumors were hypo-cellular with increased stromal component (Fig. 3C). Western blot and immunostaining showed the total loss of FOXF1 protein in FOXF1-KO tumors (Fig. 3D-E). BrdU incorporation was decreased in FOXF1-KO tumors (Suppl. Fig. S7C), consistent with decreased cellular proliferation. Transient transfection studies of nocodazole-synchronized RH18 tumor cells with FOXF1-sensitive luciferase (LUC) reporter demonstrated that FOXF1 transcriptional activity was gradually increased from G1 to G2 phases of the cell cycle (Suppl. Fig. S8A-D). In addition, flow cytometry analyses showed that knockout of FOXF1 decreased the number of BrdU-positive and Ki67-positive RH18 tumor cells in vitro (Suppl. Fig. S9A-B). Thus, deletion of FOXF1 decreases cell proliferation and inhibits FP-RMS tumorigenesis driven by PAX3-FOXO1 oncogene.
Figure 3. FOXF1 is required for PAX3-FOXO1 to drive FP-RMS tumor growth.
(A) Xenograft tumors eight weeks after intramuscular inoculation of parental (RH18 WT) tumor cells into left flank or FOXF1-depleted RH18 (FOXF1 KO) tumor cells into right flank. (B) Tumor volumes were measured eight weeks after tumor cells inoculation. RH18 WT xenografts n=8; FOXF1KO xenografts using clones 2, 5, 9, n=4 per each cell line. Data reported as mean ± SEM. (C) H&E staining of FP-RMS tumors developed from parental RH18 WT and from FOXF1 KO tumor cells. (D) Western blot analysis of tumor lysates shows high levels of FOXF1 protein in WT RH18 tumors and complete loss of FOXF1 protein in FOXF1 KO tumors. Lamin A/C was used as a loading control. (E) Immunohistochemistry shows the expression of FOXF1 in parental RH18 WT tumors, but not in the FOXF1 KO tumors. (F) Repression of either −315kb upstream or +8kb downstream enhancers using CRISPRi inhibited FP-RMS tumor growth in orthotopic mouse model. RH18 cells were transduced with a lentivirus containing a dCas9:KRAB and a gRNA targeting either −315kb upstream or +8kb downstream enhancers. Repression of FOXF1 coding sequence (CDS) was used as a positive control. Generated stable cell lines were injected into the quadriceps of NSG mice and sacrificed after eight weeks (n=6 per group).
Since PAX3-FOXO1 increases FOXF1 gene expression via the −315kb and the +8kb enhancer elements, we tested whether inhibition of these enhancers would inhibit FP-RMS tumor growth in animal model. CRISPRi-mediated repression of either −315kb or +8kb enhancer elements in fusion-positive RH18 FP-RMS cells (Fig. 2D) significantly inhibited FP-RMS tumor growth (Fig. 3F). Altogether, the PAX3-FOXO1 fusion protein induces FP-RMS tumorigenesis by regulating FOXF1 through −315kb and +8kb enhancer elements.
Loss of FOXF1 induces spontaneous differentiation of FP-RMS tumor cells.
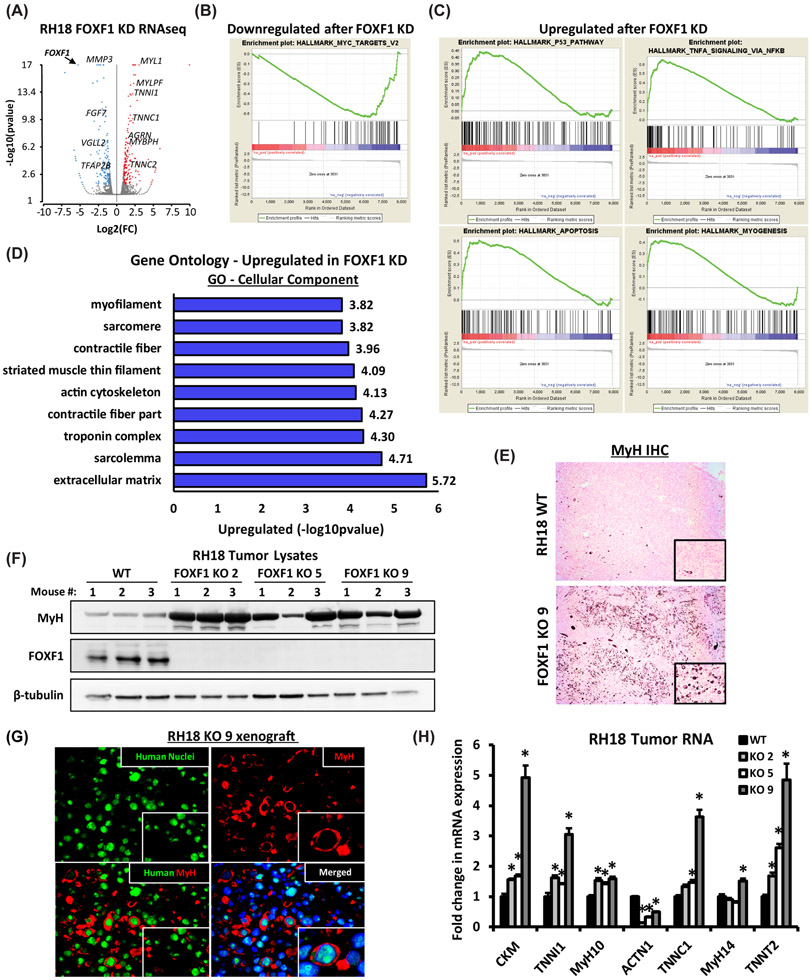
To determine molecular mechanisms through which FOXF1 regulates FP-RMS tumorigenesis, RNAseq was performed after FOXF1 knockdown in fusion-positive RH18 cells. The efficiency of FOXF1 knockdown was 90% (Suppl. Fig. S10A-B and Suppl. Table 1). A total of 150 genes were downregulated and 177 genes were upregulated compared to control RH18 tumor cells (Fig 4A and Suppl. Fig. S10C-D). Consistent with decreased tumorigenesis (Fig. 3A-B), knockdown of FOXF1 decreased expression of genes critical for tumor growth, invasion and metastasis including matrix metalloproteinase 3 (MMP3), fibroblast growth factor 7 (FGF7), transcription factor AP-2β (TFAP2β), vestigial-like family member 2 (VGLL2) (Fig. 4A, Suppl. Fig S10C), the latter of which is a known RMS oncogene (53-55). Expression of genes in the MYC network was downregulated (Fig. 4B), a finding consistent with decreased tumor cell proliferation in FOXF1-deficient FP-RMS. Furthermore, knockdown of FOXF1 was associated with gene signature of activated P53 pathway, apoptosis and myogenesis (Fig. 4C-D). The mRNAs of genes encoding mature skeletal muscle markers, including myosin light chain 1 (MYL1), troponin I1 (TNNI1), troponin C2 (TNNC2), and myosin binding protein H (MYBPH) were increased in FOXF1-deficient FP-RMS (Suppl. Fig. S10D). Thus, inhibition of FOXF1 increases expression of mature skeletal muscle genes in FP-RMS cells.
Figure 4. FOXF1 regulates expression of genes critical for FP-RMS tumor growth and metastasis.
(A) Volcano plot shows differentially expressed genes after FOXF1 knockdown in RH18 cells. (B) Gene Set Enrichment Analysis (GSEA) analysis shows downregulation of MYC gene network after FOXF1 knockdown. (C) GSEA analysis shows upregulation of P53 pathway, apoptosis and myogenesis after FOXF1 knockdown. (D) Gene ontology analysis of genes significantly upregulated after FOXF1 knockdown. (FC>1.5; p<0.05). (E) Loss of FOXF1 induced spontaneous differentiation of FP-RMS tumor cells in vivo. Deletion of FOXF1 (FOXF1 KO, clone 9) increased number of myosin heavy chain (MyH)-positive cells in orthotopic FP-RMS tumors compared to control RH18 WT cells. (F) Western blots show increased levels of MyH protein in micro-dissected FOXF1-KO tumors generated from three different clones (KO 2, KO 5 and K9) (n=3 mice per group). β-tubulin is used as a loading control. (G) MyH-positive cells are human in origin as shown by co-staining with a pan-human antibody using immunofluorescent staining of FP-RMS xenografts. (H) Deletion of FOXF1 increased mRNAs of mature skeletal muscle genes. qRT-PCR was performed using RNA isolated from micro-dissected FP-RMS tumors. Values normalized to β-actin. (n=4 mice per group). Data reported as mean ± SEM. * p<0.05.
Terminal myogenic differentiation of tumor cells has been reported in RMS patients following chemotherapy and radiation therapy (56, 57). To determine whether the loss of FOXF1 is sufficient to induce myogenic differentiation in tumor cells expressing PAX3-FOXO1 fusion protein, FP-RMS orthotopic xenograft tumors were used. CRISPR/Cas9-mediated deletion of FOXF1 dramatically increased the number of cells positive for myosin heavy chain (MyH) (Fig. 4E), a marker of differentiated muscle cells (58, 59). In protein lysates from FOXF1-defecient tumors, MyH was increased compared to tumors from parental cells (Fig. 4F). The MyH-positive cells in FOXF1-defecient FP-RMS xenografts were non-proliferative (Suppl. Fig. S11A) and were human in origin as shown by co-staining with a pan-human antibody (Fig. 4G). Increased expression of MyH in FOXF1-defecient xenograft tumors was associated with increased expression of mature skeletal muscle genes TNNI1, TNNC1, TNNT2, CKM, MyH10 and MyH14 as shown using qRT-PCR with human-specific primers (Fig. 4H). In contrast, ACTN1, a marker of undifferentiated myoblasts (60), was decreased in FOXF1-KO tumors (Fig. 4H). Finally, only single MyH-positive tumor cells were found in FP-RMS patient tumors and in FP-RMS patient-derived xenografts, and they were always negative for FOXF1 (Suppl. Fig. S11B-C). Altogether, these results demonstrate that inhibition of FOXF1 induces myogenic differentiation in PAX3-FOXO1-positive FP-RMS.
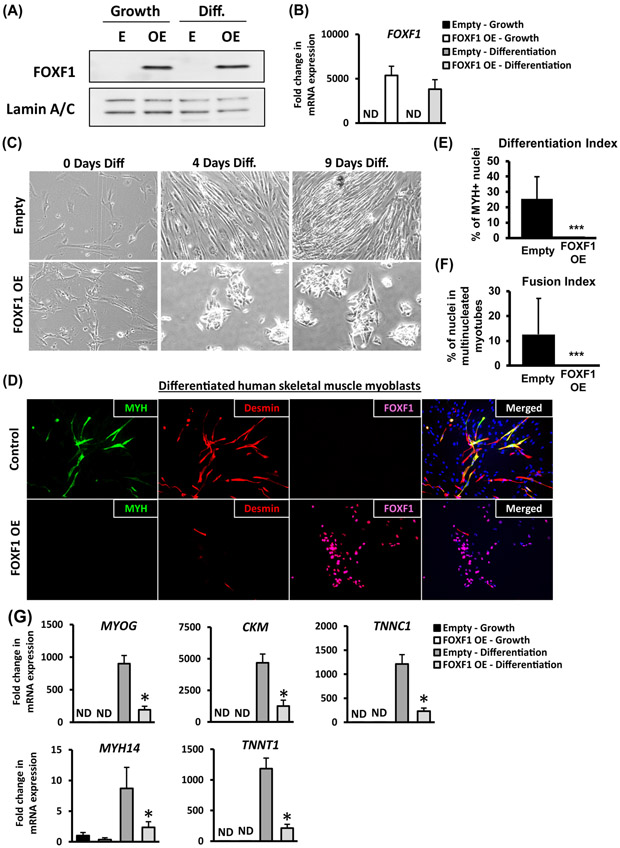
Ectopic expression of FOXF1 in human myoblasts inhibits myogenic differentiation.
To test whether FOXF1 is sufficient to inhibit myogenic differentiation in the absence of PAX3-FOXO1 fusion protein, we used an established human skeletal muscle myoblast (HSMM) primary culture system. FOXF1 is not expressed in normal human myoblasts, as shown by Western blot and qRT-PCR (Fig. 5A-B). FOXF1 was stably overexpressed using a lentivirus (FOXF1-OE) to maintain approximately equal FOXF1 levels during growth or differentiation of myoblasts in culture (Fig. 5A-B). Upon switching to differentiation media, control myoblasts formed elongated and fused myotubes, whereas FOXF1-OE myoblasts formed small clusters of cells with reduced cytoplasm (Fig. 5C). Immunostaining for MyH and Desmin showed a dramatic reduction in the number of differentiated FOXF1-OE myoblasts, with none of MyH-positive cells being found in cultures (Fig. 5D, bottom panels). Overexpression of FOXF1 in myoblasts completely abrogated their ability to differentiate in vitro as supported by reduction in differentiation index and fusion index (Fig. 5E-F). While control myoblasts increased expression of Troponin C1 (TNNC1), Myosin heavy chain 14 (MyH14), Troponin T1 (TNNT1), and Muscle Creatine Kinase (CKM) after differentiation, expression of these genes was decreased in FOXF1-OE myoblasts (Fig. 5G). Thus, FOXF1 inhibits differentiation of normal skeletal muscle myoblasts in vitro.
Figure 5. Ectopic expression of FOXF1 in primary human skeletal muscle myoblasts inhibits myogenic differentiation.
Primary human skeletal muscle myoblasts (HSMM) were transduced with empty (E) lentiviral plasmid or plasmid containing the human FOXF1 cDNA (OE). (A) Western blots show efficient over-expression of FOXF1 in HSMM (OE), but not in primary HSMM (E). Cell were cultured either in growth media (Growth) or in differentiation media (Diff.). Lamin A/C was used as a loading control. (B) FOXF1 mRNA in control HSMM (E) and in FOXF1 OE HSMM is shown by qRT-PCR. Values were normalized to β-actin. (C) Over-expression of FOXF1 in HSMM caused morphological changes during differentiation in vitro. (D) Immunostaining for MyH and Desmin showed a reduction in the number of MyH-positive and Desmin-positive FOXF1 OE myoblasts after 9 days in differentiation media. (E-F) Quantification of differentiation index and fusion index in control and FOXF1-expressing HSMM. A minimum of 100 nuclei were counted across three biological replicates and presented as mean ± SEM. ***p<0.001. (G) qRT-PCR shows the decrease in mRNAs of skeletal muscle differentiation markers using HSMM after 5 days in differentiation media. Values were normalized to β-actin (n=3). Data reported as mean ± SEM.
The proliferative capacity of FOXF1-OE myoblasts was assessed in growth media using a long-term outgrowth assay. While proliferation of control myoblasts began to slow down after several passages, FOXF1-OE cells maintained their proliferation at a rate similar to the first passage (Suppl. Fig. S12A). The number of proliferating cells was increased in FOXF1-OE myoblast cultures (Suppl. Fig. S12B, upper panels). When placed in differentiation media, both control and FOXF1-OE myoblasts had fully withdrawn from the cell cycle, as shown by the absence of the Ki-67-positive cells (Suppl. Fig. S12B, middle panels). After re-introduction of growth media, only FOXF1-OE myoblasts resumed the cell cycle, whereas control myoblasts remained differentiated (Suppl. Fig. S12B, bottom panels). Altogether, FOXF1 inhibits myogenic differentiation and increases proliferation capacity of primary skeletal muscle myoblasts.
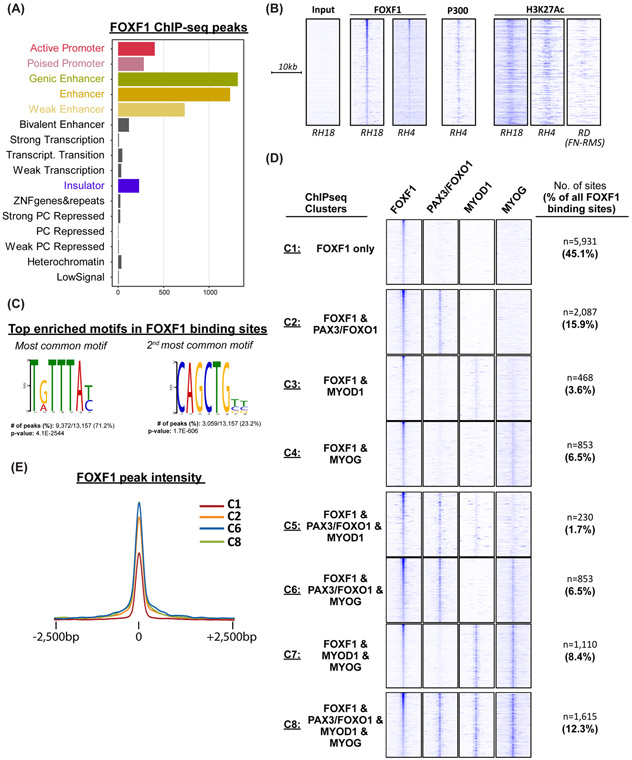
FOXF1 binds chromatin at active enhancer sites.
To identify molecular mechanisms whereby FOXF1 regulates FP-RMS tumorigenesis, we analyzed the genome-wide binding of FOXF1 protein to DNA by performing FOXF1 ChIP-seq in fusion-positive RH18 cells. The total of 85% of FOXF1-binding peaks were detected within gene introns (~44%) and in intergenic regions (~41%), the latter of which are defined as >20kb from the nearest gene (Suppl. Fig. S13A) To characterize the chromatin states within FOXF1-binding peaks, ChromHMM analysis was performed and demonstrated that FOXF1-binding sites were dominated by features of active enhancer elements with no enrichment for repressed chromatin (Fig. 6A and Suppl. Fig. S13B). The similar FOXF1-binding sites were also identified in a FOXF1 ChIP-seq using fusion-positive RH4 FP-RMS cells (Fig. 6B). In both fusion-positive RH18 and RH4 tumor cells, FOXF1 DNA-binding was associated with recruitment of P300 histone acetyl transferase and active enhancer mark H3K27ac (Fig. 6B). In contrast, in fusion-negative FN-RMS cell line RD, the H3K27ac marks were undetectable at the same DNA loci (Fig. 6B). Thus, FOXF1 binds to DNA enhancers that are active in fusion-positive FP-RMS but not in fusion-negative FN-RMS.
Figure 6. FOXF1 DNA-binding is associated with active enhancers and with binding of PAX3-FOXO1 and myogenic transcription factors.
(A) ChromHMM analysis of FOXF1 binding sites in fusion-positive RH18 cells. (B) FOXF1 binds to the enhancers that are active in FP-RMS but not in FN-RMS. Heatmaps of FOXF1, P300 and H3K27ac ChIPseq data in FP-RMS cell lines RH4, RH18 and FN-RMS cell line RD. (C) FOXF1 DNA-binding sites are enriched in Forkhead and E-box motifs. De novo motif analysis was performed using MEME-ChIP. (D) FOXF1 binding sites are shared with PAX3-FOXO1, MYOD1, and/or MYOG. The most common binding partner is PAX3-FOXO1. Out of all FOXF1 binding sites, 36% are also occupied by PAX3-FOXO1 (Cluster C2+C5+C6+C8). Each of the 13,157 FOXF1 binding sites were assigned to a single cluster based on the presence or absence of PAX3-FOXO1, MYOD1, and/or MYOG. Heatmaps are normalized for each transcription factor and the number of enhancers in each cluster are shown. (E) Tag density plot showing that FoxF1 signal is higher when co-bound with PAX3-FOXO1. Pairwise analysis of FOXF1 peak quality across clusters was performed. Enrichment of FOXF1 is highest in clusters C2, 6 and 8.
FOXF1 DNA-binding is associated with binding of PAX3-FOXO1 and myogenic transcription factors.
De novo motif discovery on FOXF1 DNA-binding peaks identified the canonical Forkhead TG/ATTTAT/C motif (37) as the most significantly enriched motif (Fig. 6C). The second most enriched motif in FOXF1 DNA-binding regions was the E-Box CAG/TCTGT/C motif (Fig. 6C), which is known to be enriched at PAX3-FOXO1, MYOD1 and MYOG DNA-binding sites in FP-RMS(11),(12). Using available ChIPseq datasets (11, 12), we determined whether any of these FP-RMS transcription factors were present at the FOXF1 binding sites. Out of all FOXF1 binding sites, 36% were also occupied by PAX3-FOXO1 (ChIPseq Clusters C2+C5+C6+C8; Fig. 6D and Suppl. Table 2), 26% were also occupied by MYOD1 (ChIPseq Clusters C3+C5+C7+C8 and Suppl. Table 3) and 34% were also occupied by MYOG (ChIPseq Clusters C4+C6+C7+C8) (Fig. 6D and Suppl. Table 4). Of these shared binding sites, 12.3% were occupied by all four transcription factors, FOXF1, PAX3-FOXO1, MYOD1, and MYOG (Fig. 6D, Suppl. Table 5). No significant changes in FOXF1-binding motif preferences (Suppl. Fig. S14A) or nearby gene expression levels (Suppl. Fig. S14B) were observed across these shared binding sites. Furthermore, chromatin states of FOXF1 only sites (Cluster C1) were similar to the rest of FOXF1 binding sites with no evidence of repressed chromatin (Suppl. Fig. S14C). To prioritize these clusters, we performed a pairwise analysis of FOXF1 peak quality across clusters. Interestingly, clusters 2, 6, and 8 had significantly higher enrichment at FOXF1 binding sites compared to other clusters (Fig. 6E). Since the common denominator between Clusters 2, 6, and 8 is the binding of PAX3-FOXO1, our data suggest that FOXF1 exhibits the strongest binding to FP-RMS enhancers in the presence of PAX3-FOXO1. Altogether, FOXF1 binds to FP-RMS enhancers in tandem with PAX3-FOXO1 fusion protein and E-box transcription factors. Interestingly, FOXF1 binding picks together with binding sites for MYOD1, MYOG, p300 and H3K27ac were present in the recently identified PAX3-FOXO1 enhancers (29) (Suppl. Fig. S15), implicating FOXF1 transcription factor in regulation of PAX3-FOXO1 expression.
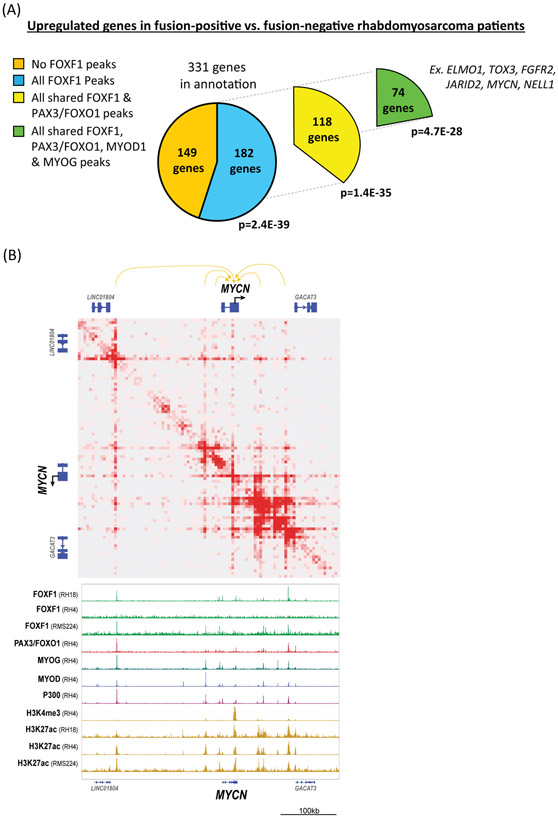
FOXF1 binds to the enhancers of genes associated with FP-RMS-specific gene signature.
To further determine the importance of FOXF1 DNA-binding for FP-RMS tumorigenesis, we performed analysis of genes proximal to FOXF1 binding sites. The FOXF1 ChIPseq peaks were annotated based on the nearest gene and were compared to fusion-positive FP-RMS gene signature identified previously (61). Out of 331 genes upregulated in FP-RMS (61), 182 genes (55%) had FOXF1 binding peaks (Fig. 7A and Suppl. Table 6). Out of these 182 genes, 118 genes (65%) had enhancers bound by both FOXF1 and PAX3-FOXO1. Interestingly, 74 genes from FP-RMS signature had enhancers bound by FOXF1, PAX3-PAXO1, MYOD1 and MYOG, including ELMO1, TOX3, FGFR2, MYCN, JARID2 (Fig. 7A). For example, MYCN gene, which is overexpressed in FP-RMS (8) (Fig. 4B) and associated with poor survival (62), had multiple enhancers looping to the MYCN gene locus as shown by HiChIP analysis for H3K27ac (Fig. 7B). MYCN enhancers with the strongest looping interactions were bound by FOXF1 together with PAX3-FOXO1, MYOD1 and MYOG, and were positive for p300 and H3K27ac activation marks (Fig. 7B). FGFR2 and ALK genes loci also contained enhancers simultaneously bound by FOXF1, PAX3-FOXO1, MYOD, MYOG, p300 and H3K27ac (Suppl. Fig. S16A-B). These enhancers were active in FP-RMS, but not in FN-RMS as supported by differential binding of p300 and H3K27ac to these enhancers (Suppl. Fig. S16A-B). Thus, FOXF1 binds with PAX3-FOXO1 and E-box myogenic transcription factors to the enhancers of genes associated with FP-RMS gene signature.
Figure 7. FOXF1 binds to enhancers of genes associated with FP-RMS gene signature.
(A) Annotated genes near FOXF1 binding sites overlap with genes signature specific to FP-RMS. Up-regulated FP-RMS gene set is from Davicioni et al. 2006 (18). Out of 331 genes upregulated in FP-RMS, 182 genes (55%) have FOXF1 binding peaks. Out of these 182 genes, 118 genes (65%) had enhancers bound by both FOXF1 and PAX3-FOXO1. 74 genes from FP-RMS signature had enhancers bound by FOXF1, PAX3-PAXO1, MYOD1 and MYOG. P-values are FDR B&F corrected. (B) HiChIP analysis identified long-distance interactions of enhancers with the MYCN gene locus shown at 5kb resolution. RH4 FP-RMS cells were used for HiChIP (top panel). ChIPseq shows binding of FOXF1, PAX3-FOXO1, MYOD1, MYOG, as well as p300 and H3K27ac at the MYCN enhancers (bottom panel). FOXF1 and H3K27ac ChIPseqs were performed using RH18, RH4 and RMS224 cells.
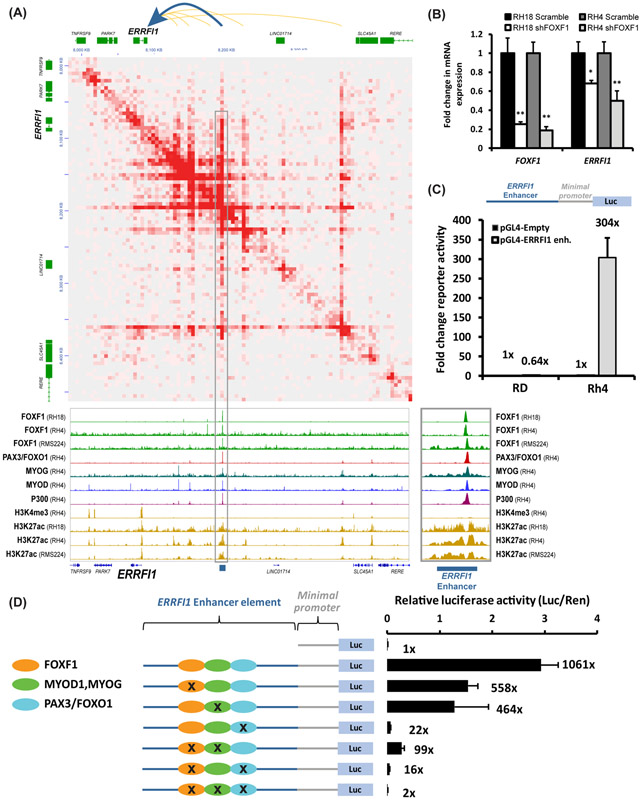
FOXF1 cooperates with PAX3-FOXO1 and E-box transcription factors to activate FP-RMS enhancers.
To determine the importance of the cooperative binding of these transcription factors on enhancer activity, we focused on the 3’ distal enhancer of ERRFI1, gene critical for tumor cell proliferation and chemoresistance (63). The 3’ ERRFI1 enhancer had overlapping binding sites for FOXF1, PAX3-FOXO1, MYOD1, and MYOG (Fig. 8A). This enhancer had active chromatin marks p300 and H3K27ac in fusion-positive FP-RMS (RH4) but not in fusion-negative FN-RMS (RD), and showed looping to the ERRFI1 gene locus (Fig. 8A). To determine whether FOXF1 increases ERRFI1 gene expression, shRNA was used to deplete FOXF1 in fusion-positive FP-RMS. Knockdown of FOXF1 decreased ERRFI1 mRNA in both RH18 and RH4 FP-RMS cell lines (Fig. 8B). Thus, FOXF1 induces ERRFI1 gene expression.
Figure 8. FOXF1 cooperates with PAX3-FOXO1 and E-box transcription factors to activate the ERRFI1 enhancer.
(A) HiChIP showed long-distance interactions of enhancers with the ERRFI1 gene in FP-RMS RH4 (top panel). ChIP-seq in RH18, RH4, and RMS224 fusion-positive RMS shows the binding of FOXF1, PAX3-FOXO1, MYOG and MYOD at the ERRFI1 enhancers (bottom panel). (B) Knockdown of FOXF1 decreased ERRFI1 mRNA in RH18 and RH4 cells as shown by qRT-PCR. mRNAs were normalized to β-actin mRNA (n=3). Data shown as mean ± SEM. (C) Cloning and validation of the −187kb ERRFI1 enhancer element. An 826bp fragment corresponding to the ERRFI1 enhancer element was cloned into pGL4.23 plasmid containing a minimal promoter driving the luciferase reporter. RD and RH4 cells were transfected with pGL4-ERRFI1, pGL4-Empty and renilla plasmids, and a dual luciferase assay was performed 48 hours. Normalized luciferase activity is shown as mean ± SEM (n=3). (D) Site-directed mutagenesis of Forkhead, E-Box, or PAX3 binding sites decreased activity of the −187kb ERRFI1 enhancer. Disruption of PAX3 binding site completely abolished the enhancer activity. Disruption of either FOXF1 or MYOD/MYOG binding sites decreased activity of the ERRFI1 enhancer, whereas disruption of both FOXF1 and MYOD/MYOG binding sites abolished ERRFI1 enhancer activity (n=3). Luciferase activity was normalized to renilla control.
Next, an 826bp DNA fragment encompassing the 3’ distal ERRFI1 enhancer element was cloned in front of a minimal promoter driving expression of the firefly luciferase (LUC) reporter. When transfected into fusion-positive RH4 cells, this enhancer increased LUC activity compared to reporter driven by the minimal promoter alone (Fig. 8C). The LUC reporter was inactive when transfected into the fusion-negative RD cells (Fig. 8C). Thus, the 3’ ERRFI1 enhancer construct retained FP-RMS-specific activity similar to the endogenous ERRFI1 gene locus. To dissect the contribution of each transcription factor binding site to the 3’ ERRFI1 enhancer activity, we generated a series of single and compound point mutants disrupting FOXF1, MYOD1/MYOG, and PAX3-FOXO1 binding sites (Fig. 8D). Point mutations in either FOXF1 or E-Box binding motifs decreased enhancer activity by 50% (Fig. 8D). Combined mutagenesis of the FOXF1 and the E-Box motifs reduced the enhancer activity by 97% compared to the WT enhancer (Fig. 8D), suggesting synergistic effects between FOXF1 and E-box transcription factors in activation of the ERRFI1 enhancer. Consistent with transcriptional synergy, FOXF1 protein physically bound to the MYOD protein as shown by immunoprecipitation (Suppl. Fig. S17). Although both FOXF1 and PAX3-FOXO1-binding peaks were present in the ERRFI1 enhancer (Fig. 8A), FOXF1 did not physically bind to the PAX3/FOXO1 fusion protein (Suppl. Fig. S17). Mutagenesis of the PAX3-FOXO1 consensus site alone or in combination with other motifs reduced enhancer activity (Fig. 8D). Altogether, FOXF1, PAX3-FOXO1 and E-box transcription factors cooperate via protein-DNA and protein-protein interactions to stimulate the ERRFI1 enhancer activity in FP-RMS tumors.
Discussion
In this study, we have identified the PAX3-FOXO1/ FOXF1 signaling cascade which is critical for FP-RMS tumorigenesis. Previous studies demonstrated that FOXF1 is capable to promote or inhibit cellular proliferation depending on tissue or cell specificity (33, 37, 64). FOXF1 induced cell cycle progression in endothelial cells during development of pulmonary vasculature (65, 66). FOXF1 promoted tumor growth and invasion in colorectal and breast tumors (67, 68) but inhibited cellular proliferation and tumor growth in colon and breast adenocarcinomas (69). Decreased cellular proliferation in FOXF1-defecient FP-RMS is consistent with previous studies demonstrating that FOXF1 stimulates RMS tumor growth through transcriptional repression of p21CIP1 (70). Interestingly, in the present studies we also found that in addition to its role in tumor cell proliferation, FOXF1 prevents myogenic differentiation in FP-RMS tumors. This effect is, most likely, independent of PAX3-FOXO1, since over-expression of FOXF1 decreased myogenic differentiation in primary skeletal muscle myoblasts.
Cell differentiation is regulated by lineage-specific transcription factors and disruption of these genes, by mutations or other mechanisms, can alter the differentiation process. While there is not a targeted differentiation therapy for rhabdomyosarcoma, it remains a promising therapeutic approach. Terminal differentiation of FP-RMS cells after chemotherapy has been reported in multiple studies (56, 57, 71) (72). These studies suggest the feasibility for directed FP-RMS differentiation similar to what has been achieved with retinoid-based differentiation therapy for acute promyelocytic leukemia (73), hepatocellular carcinoma (74), and squamous cell carcinoma (75). Our studies identified FOXF1 as a direct transcriptional target of PAX3-FOXO1 which can be explored pharmacologically or via gene therapy to achieve terminal differentiation of tumor cells in FP-RMS.
Rhabdomyosarcoma is characterized by the presence of markers of skeletal muscle differentiation and can be most reliably identified by transcription factors MYOD1 and MYOG (76). MYOD1 is expressed in normal skeletal muscle myoblasts. Upon committing to terminal differentiation, MYOD1 transcriptionally activates MYOG which induces cell cycle withdrawal, expression of skeletal muscle contractile machinery, and irreversible myogenic differentiation (77). Paradoxically, nearly every tumor cell in fusion-positive FP-RMS expresses MYOG (76, 78). This is in stark contrast to embryonal and spindle/sclerosing RMS which typically have scattered or focal positivity for MYOG (79). This feature of FP-RMS suggests that FP-RMS tumor cells require unique molecular mechanisms for antagonizing MYOG function and blocking differentiation. Since FOXF1 is co-expressed with MYOG in FP-RMS and often binds to the same DNA regulatory elements, it is possible that FOXF1 inhibits myogenic differentiation by interfering with MYOG function.
In summary, FOXF1 is a critical transcriptional target of the PAX3-FOXO1 oncogene in FP-RMS, which promotes tumor cell proliferation and protects tumor cells from terminal myogenic differentiation. These studies highlight a novel role for FOXF1 in the pathogenesis of FP-RMS and suggest that inhibiting FOXF1 can be promising approach for FP-RMS therapy.
Supplementary Material
Acknowledgements
We thank Tien Le for technical assistance. This work was supported by the NIH grants R01 CA142724 (TVK), R01 HL132849 (TVK), R01 HL141174 (VVK).
The work of TVK is supported by NIH (R01CA142724, R01HL132849), the work of VVK is supported by NIH (R01HL141174). AB is a co-founder of Datirium, LLC. Other authors declare no conflict of interest.
Footnotes
Competing interests
AB is a co-founder of Datirium, LLC, the developer of SciDAP data analysis platform. Other authors declare no conflict of interest.
References
- 1.Ognjanovic S, Linabery AM, Charbonneau B, Ross JA. Trends in childhood rhabdomyosarcoma incidence and survival in the United States, 1975-2005. Cancer. 2009;115(18):4218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor AC, Shu L, Danks MK, Poquette CA, Shetty S, Thayer MJ, et al. P53 mutation and MDM2 amplification frequency in pediatric rhabdomyosarcoma tumors and cell lines. Med Pediatr Oncol. 2000;35(2):96–103. [DOI] [PubMed] [Google Scholar]
- 3.Stratton MR, Fisher C, Gusterson BA, Cooper CS. Detection of point mutations in N-ras and K-ras genes of human embryonal rhabdomyosarcomas using oligonucleotide probes and the polymerase chain reaction. Cancer Res. 1989;49(22):6324–7. [PubMed] [Google Scholar]
- 4.Chen L, Shern JF, Wei JS, Yohe ME, Song YK, Hurd L, et al. Clonality and evolutionary history of rhabdomyosarcoma. PLoS Genet. 2015;11(3):e1005075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X, Stewart E, Shelat AA, Qu C, Bahrami A, Hatley M, et al. Targeting oxidative stress in embryonal rhabdomyosarcoma. Cancer Cell. 2013;24(6):710–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor JGt, Cheuk AT, Tsang PS, Chung JY, Song YK, Desai K, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119(11):3395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stewart E, McEvoy J, Wang H, Chen X, Honnell V, Ocarz M, et al. Identification of Therapeutic Targets in Rhabdomyosarcoma through Integrated Genomic, Epigenomic, and Proteomic Analyses. Cancer Cell. 2018;34(3):411–26 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4(2):216–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorensen PH, Lynch JC, Qualman SJ, Tirabosco R, Lim JF, Maurer HM, et al. PAX3-FKHR and PAX7-FKHR gene fusions are prognostic indicators in alveolar rhabdomyosarcoma: a report from the children's oncology group. J Clin Oncol. 2002;20(11):2672–9. [DOI] [PubMed] [Google Scholar]
- 10.Rudzinski ER, Anderson JR, Chi YY, Gastier-Foster JM, Astbury C, Barr FG, et al. Histology, fusion status, and outcome in metastatic rhabdomyosarcoma: A report from the Children's Oncology Group. Pediatr Blood Cancer. 2017;64(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao L, Yu Y, Bilke S, Walker RL, Mayeenuddin LH, Azorsa DO, et al. Genome-wide identification of PAX3-FKHR binding sites in rhabdomyosarcoma reveals candidate target genes important for development and cancer. Cancer Res. 2010;70(16):6497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gryder BE, Yohe ME, Chou HC, Zhang X, Marques J, Wachtel M, et al. PAX3-FOXO1 Establishes Myogenic Super Enhancers and Confers BET Bromodomain Vulnerability. Cancer Discov. 2017;7(8):884–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otabe O, Kikuchi K, Tsuchiya K, Katsumi Y, Yagyu S, Miyachi M, et al. MET/ERK2 pathway regulates the motility of human alveolar rhabdomyosarcoma cells. Oncol Rep. 2017;37(1):98–104. [DOI] [PubMed] [Google Scholar]
- 14.Taulli R, Scuoppo C, Bersani F, Accornero P, Forni PE, Miretti S, et al. Validation of met as a therapeutic target in alveolar and embryonal rhabdomyosarcoma. Cancer Res. 2006;66(9):4742–9. [DOI] [PubMed] [Google Scholar]
- 15.Schoffski P, Adkins D, Blay JY, Gil T, Elias AD, Rutkowski P, et al. An open-label, phase 2 study evaluating the efficacy and safety of the anti-IGF-1R antibody cixutumumab in patients with previously treated advanced or metastatic soft-tissue sarcoma or Ewing family of tumours. Eur J Cancer. 2013;49(15):3219–28. [DOI] [PubMed] [Google Scholar]
- 16.Pappo AS, Vassal G, Crowley JJ, Bolejack V, Hogendoorn PC, Chugh R, et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research Through Collaboration study. Cancer. 2014;120(16):2448–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schoffski P, Wozniak A, Leahy MG, Aamdal S, Rutkowski P, Bauer S, et al. The tyrosine kinase inhibitor crizotinib does not have clinically meaningful activity in heavily pre-treated patients with advanced alveolar rhabdomyosarcoma with FOXO rearrangement: European Organisation for Research and Treatment of Cancer phase 2 trial 90101 'CREATE'. Eur J Cancer. 2018;94:156–67. [DOI] [PubMed] [Google Scholar]
- 18.Davicioni E, Finckenstein FG, Shahbazian V, Buckley JD, Triche TJ, Anderson MJ. Identification of a PAX-FKHR gene expression signature that defines molecular classes and determines the prognosis of alveolar rhabdomyosarcomas. Cancer Res. 2006;66(14):6936–46. [DOI] [PubMed] [Google Scholar]
- 19.Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28(13):2151–8. [DOI] [PubMed] [Google Scholar]
- 20.Lae M, Ahn EH, Mercado GE, Chuai S, Edgar M, Pawel BR, et al. Global gene expression profiling of PAX-FKHR fusion-positive alveolar and PAX-FKHR fusion-negative embryonal rhabdomyosarcomas. J Pathol. 2007;212(2):143–51. [DOI] [PubMed] [Google Scholar]
- 21.Wachtel M, Dettling M, Koscielniak E, Stegmaier S, Treuner J, Simon-Klingenstein K, et al. Gene expression signatures identify rhabdomyosarcoma subtypes and detect a novel t(2;2)(q35;p23) translocation fusing PAX3 to NCOA1. Cancer Res. 2004;64(16):5539–45. [DOI] [PubMed] [Google Scholar]
- 22.Mercado GE, Xia SJ, Zhang C, Ahn EH, Gustafson DM, Lae M, et al. Identification of PAX3-FKHR-regulated genes differentially expressed between alveolar and embryonal rhabdomyosarcoma: focus on MYCN as a biologically relevant target. Genes Chromosomes Cancer. 2008;47(6):510–20. [DOI] [PubMed] [Google Scholar]
- 23.Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128(2):155–66. [DOI] [PubMed] [Google Scholar]
- 24.Kalinichenko VV, Zhou Y, Bhattacharyya D, Kim W, Shin B, Bambal K, et al. Haploinsufficiency of the Mouse Forkhead Box f1 Gene Causes Defects in Gall Bladder Development. J Biol Chem. 2002;277(14):12369–74. [DOI] [PubMed] [Google Scholar]
- 25.Ustiyan V, Bolte C, Zhang Y, Han L, Xu Y, Yutzey KE, et al. FOXF1 transcription factor promotes lung morphogenesis by inducing cellular proliferation in fetal lung mesenchyme. Dev Biol. 2018;443(1):50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren X, Ustiyan V, Guo M, Wang G, Bolte C, Zhang Y, et al. Postnatal Alveologenesis Depends on FOXF1 Signaling in c-KIT(+) Endothelial Progenitor Cells. Am J Respir Crit Care Med. 2019;200(9):1164–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, et al. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. J Biol Chem. 2013;288(40):28477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinson AR, Jones R, Crose LE, Belyea BC, Barr FG, Linardic CM. Human rhabdomyosarcoma cell lines for rhabdomyosarcoma research: utility and pitfalls. Front Oncol. 2013;3:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gryder BE, Wachtel M, Chang K, El Demerdash O, Aboreden NG, Mohammed W, et al. Miswired Enhancer Logic Drives a Cancer of the Muscle Lineage. iScience. 2020;23(5):101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren X, Zhang Y, Snyder J, Cross ER, Shah TA, Kalin TV, et al. Forkhead box M1 transcription factor is required for macrophage recruitment during liver repair. Mol Cell Biol. 2010;30(22):5381–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai Y, Balli D, Ustiyan V, Fulford L, Hiller A, Misetic V, et al. Foxm1 expression in prostate epithelial cells is essential for prostate carcinogenesis. J Biol Chem. 2013;288(31):22527–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ustiyan V, Wert SE, Ikegami M, Wang IC, Kalin TV, Whitsett JA, et al. Foxm1 transcription factor is critical for proliferation and differentiation of Clara cells during development of conducting airways. Dev Biol. 2012;370(2):198–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black M, Milewski D, Le T, Ren X, Xu Y, Kalinichenko VV, et al. FOXF1 Inhibits Pulmonary Fibrosis by Preventing CDH2-CDH11 Cadherin Switch in Myofibroblasts. Cell Rep. 2018;23(2):442–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Black M, Arumugam P, Shukla S, Pradhan A, Ustiyan V, Milewski D, et al. FOXM1 nuclear transcription factor translocates into mitochondria and inhibits oxidative phosphorylation. Mol Biol Cell. 2020;31(13):1411–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goda C, Balli D, Black M, Milewski D, Le T, Ustiyan V, et al. Loss of FOXM1 in macrophages promotes pulmonary fibrosis by activating p38 MAPK signaling pathway. PLoS Genet. 2020;16(4):e1008692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Ren X, Wang IC, Pradhan A, Zhang Y, Flood HM, et al. The FOXM1 inhibitor RCM-1 suppresses goblet cell metaplasia and prevents IL-13 and STAT6 signaling in allergen-exposed mice. Sci Signal. 2017;10(475). [DOI] [PubMed] [Google Scholar]
- 37.Bolte C, Flood HM, Ren X, Jagannathan S, Barski A, Kalin TV, et al. FOXF1 transcription factor promotes lung regeneration after partial pneumonectomy. Sci Rep. 2017;7(1):10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vallabh S, Kartashov AV, Barski A. Analysis of ChIP-Seq and RNA-Seq Data with BioWardrobe. Methods Mol Biol. 2018;1783:343–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kartashov AV, Barski A. BioWardrobe: an integrated platform for analysis of epigenomics and transcriptomics data. Genome Biol. 2015;16:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011;27(12):1696–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol. 2012;13(3):R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gryder BE, Wu L, Woldemichael GM, Pomella S, Quinn TR, Park PMC, et al. Chemical genomics reveals histone deacetylases are required for core regulatory transcription. Nat Commun. 2019;10(1):3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenente IM, Hayes MN, Ignatius MS, McCarthy K, Yohe M, Sindiri S, et al. Myogenic regulatory transcription factors regulate growth in rhabdomyosarcoma. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gryder BE, Khan J, Stanton BZ. Measurement of differential chromatin interactions with absolute quantification of architecture (AQuA-HiChIP). Nat Protoc. 2020;15(3):1209–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson JT, Turner D, Durand NC, Thorvaldsdottir H, Mesirov JP, Aiden EL. Juicebox.js Provides a Cloud-Based Visualization System for Hi-C Data. Cell Syst. 2018;6(2):256–8 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milewski D, Balli D, Ustiyan V, Le T, Dienemann H, Warth A, et al. FOXM1 activates AGR2 and causes progression of lung adenomas into invasive mucinous adenocarcinomas. PLoS Genet. 2017;13(12):e1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolte C, Ren X, Tomley T, Ustiyan V, Pradhan A, Hoggatt A, et al. Forkhead box F2 regulation of platelet-derived growth factor and myocardin/serum response factor signaling is essential for intestinal development. J Biol Chem. 2015;290(12):7563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng X-H, Black M, Ustiyan V, Le T, Fulford L, Sridharan A, et al. SPDEF Inhibits Prostate Carcinogenesis by Disrupting a Positive Feedback Loop in Regulation of the Foxm1 Oncogene. PLoS Genet. 2014;10(9):e1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kalin TV, Meliton L, Meliton AY, Zhu X, Whitsett JA, Kalinichenko VV. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am J Respir Cell Mol Biol. 2008;39(4):390–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gryder BE, Pomella S, Sayers C, Wu XS, Song Y, Chiarella AM, et al. Histone hyperacetylation disrupts core gene regulatory architecture in rhabdomyosarcoma. Nat Genet. 2019;51(12):1714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Szafranski P, Dharmadhikari AV, Wambach JA, Towe CT, White FV, Grady RM, et al. Two deletions overlapping a distant FOXF1 enhancer unravel the role of lncRNA LINC01081 in etiology of alveolar capillary dysplasia with misalignment of pulmonary veins. Am J Med Genet A. 2014;164A(8):2013–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, et al. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat Methods. 2015;12(12):1143–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alaggio R, Zhang L, Sung YS, Huang SC, Chen CL, Bisogno G, et al. A Molecular Study of Pediatric Spindle and Sclerosing Rhabdomyosarcoma: Identification of Novel and Recurrent VGLL2-related Fusions in Infantile Cases. Am J Surg Pathol. 2016;40(2):224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mehner C, Miller E, Khauv D, Nassar A, Oberg AL, Bamlet WR, et al. Tumor cell-derived MMP3 orchestrates Rac1b and tissue alterations that promote pancreatic adenocarcinoma. Mol Cancer Res. 2014;12(10):1430–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebauer M, Wachtel M, Niggli FK, Schafer BW. Comparative expression profiling identifies an in vivo target gene signature with TFAP2B as a mediator of the survival function of PAX3/FKHR. Oncogene. 2007;26(51):7267–81. [DOI] [PubMed] [Google Scholar]
- 56.Hakozaki M, Hojo H, Tajino T, Yamada H, Kikuchi S, Kikuta A, et al. Therapeutic cytodifferentiation in alveolar rhabdomyosarcoma without genetic change of the PAX3-FKHR chimeric fusion gene: a case study. Hum Cell. 2013;26(4):149–54. [DOI] [PubMed] [Google Scholar]
- 57.Smith LM, Anderson JR, Coffin CM. Cytodifferentiation and clinical outcome after chemotherapy and radiation therapy for rhabdomyosarcoma (RMS). Med Pediatr Oncol. 2002;38(6):398–404. [DOI] [PubMed] [Google Scholar]
- 58.Jothi M, Nishijo K, Keller C, Mal AK. AKT and PAX3-FKHR cooperation enforces myogenic differentiation blockade in alveolar rhabdomyosarcoma cell. Cell Cycle. 2012;11(5):895–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Skrzypek K, Kusienicka A, Trzyna E, Szewczyk B, Ulman A, Konieczny P, et al. SNAIL is a key regulator of alveolar rhabdomyosarcoma tumor growth and differentiation through repression of MYF5 and MYOD function. Cell Death Dis. 2018;9(6):643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feng Y, Ngu H, Alford SK, Ward M, Yin F, Longmore GD. alpha-actinin1 and 4 tyrosine phosphorylation is critical for stress fiber establishment, maintenance and focal adhesion maturation. Exp Cell Res. 2013;319(8):1124–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davicioni E, Anderson MJ, Finckenstein FG, Lynch JC, Qualman SJ, Shimada H, et al. Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group. Am J Pathol. 2009;174(2):550–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williamson D, Lu YJ, Gordon T, Sciot R, Kelsey A, Fisher C, et al. Relationship between MYCN copy number and expression in rhabdomyosarcomas and correlation with adverse prognosis in the alveolar subtype. J Clin Oncol. 2005;23(4):880–8. [DOI] [PubMed] [Google Scholar]
- 63.Cairns J, Fridley BL, Jenkins GD, Zhuang Y, Yu J, Wang L. Differential roles of ERRFI1 in EGFR and AKT pathway regulation affect cancer proliferation. EMBO Rep. 2018;19(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, et al. FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal. 2016;9(424):ra40. [DOI] [PubMed] [Google Scholar]
- 65.Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, et al. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circ Res. 2014;115(8):709–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pradhan A, Dunn A, Ustiyan V, Bolte C, Wang G, Whitsett JA, et al. The S52F FOXF1 Mutation Inhibits STAT3 Signaling and Causes Alveolar Capillary Dysplasia. Am J Respir Crit Care Med. 2019;200(8):1045–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang S, Yan S, Zhu S, Zhao Y, Yan J, Xiao Z, et al. FOXF1 Induces Epithelial-Mesenchymal Transition in Colorectal Cancer Metastasis by Transcriptionally Activating SNAI1. Neoplasia. 2018;20(10):996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nilsson G, Kannius-Janson M. Forkhead Box F1 promotes breast cancer cell migration by upregulating lysyl oxidase and suppressing Smad2/3 signaling. BMC Cancer. 2016;16:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lo PK, Lee JS, Liang X, Han L, Mori T, Fackler MJ, et al. Epigenetic inactivation of the potential tumor suppressor gene FOXF1 in breast cancer. Cancer Res. 2010;70(14):6047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milewski D, Pradhan A, Wang X, Cai Y, Le T, Turpin B, et al. FoxF1 and FoxF2 transcription factors synergistically promote rhabdomyosarcoma carcinogenesis by repressing transcription of p21(Cip1) CDK inhibitor. Oncogene. 2017;36(6):850–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.d'Amore ES, Tollot M, Stracca-Pansa V, Menegon A, Meli S, Carli M, et al. Therapy associated differentiation in rhabdomyosarcomas. Mod Pathol. 1994;7(1):69–75. [PubMed] [Google Scholar]
- 72.Kikuchi K, Tsuchiya K, Otabe O, Gotoh T, Tamura S, Katsumi Y, et al. Effects of PAX3-FKHR on malignant phenotypes in alveolar rhabdomyosarcoma. Biochem Biophys Res Commun. 2008;365(3):568–74. [DOI] [PubMed] [Google Scholar]
- 73.Iland HJ, Bradstock K, Supple SG, Catalano A, Collins M, Hertzberg M, et al. All-trans-retinoic acid, idarubicin, and IV arsenic trioxide as initial therapy in acute promyelocytic leukemia (APML4). Blood. 2012;120(8):1570–80; quiz 752. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Y, Guan DX, Shi J, Gao H, Li JJ, Zhao JS, et al. All-trans retinoic acid potentiates the chemotherapeutic effect of cisplatin by inducing differentiation of tumor initiating cells in liver cancer. J Hepatol. 2013;59(6):1255–63. [DOI] [PubMed] [Google Scholar]
- 75.Brewster AM, Lee JJ, Clayman GL, Clifford JL, Reyes MJ, Zhou X, et al. Randomized trial of adjuvant 13-cis-retinoic acid and interferon alfa for patients with aggressive skin squamous cell carcinoma. J Clin Oncol. 2007;25(15):1974–8. [DOI] [PubMed] [Google Scholar]
- 76.Rekhi B, Gupta C, Chinnaswamy G, Qureshi S, Vora T, Khanna N, et al. Clinicopathologic features of 300 rhabdomyosarcomas with emphasis upon differential expression of skeletal muscle specific markers in the various subtypes: A single institutional experience. Ann Diagn Pathol. 2018;36:50–60. [DOI] [PubMed] [Google Scholar]
- 77.Liu QC, Zha XH, Faralli H, Yin H, Louis-Jeune C, Perdiguero E, et al. Comparative expression profiling identifies differential roles for Myogenin and p38alpha MAPK signaling in myogenesis. J Mol Cell Biol. 2012;4(6):386–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dias P, Chen B, Dilday B, Palmer H, Hosoi H, Singh S, et al. Strong Immunostaining for Myogenin in Rhabdomyosarcoma Is Significantly Associated with Tumors of the Alveolar Subclass. The American Journal of Pathology. 2000;156(2):399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhao Z, Yin Y, Zhang J, Qi J, Zhang D, Ma Y, et al. Spindle cell/sclerosing rhabdomyosarcoma: case series from a single institution emphasizing morphology, immunohistochemistry and follow-up. Int J Clin Exp Pathol. 2015;8(11):13814–20. [PMC free article] [PubMed] [Google Scholar]
- 80.Sun W, Chatterjee B, Wang Y, Stevenson HS, Edelman DC, Meltzer PS, et al. Distinct methylation profiles characterize fusion-positive and fusion-negative rhabdomyosarcoma. Mod Pathol. 2015;28(9):1214–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.