Abstract
Purpose
To evaluate the performance of machine learning algorithms on organ-level classification of semistructured pathology reports, to incorporate surgical pathology monitoring into an automated imaging recommendation follow-up engine.
Materials and Methods
This retrospective study included 2013 pathology reports from patients who underwent abdominal imaging at a large tertiary care center between 2012 and 2018. The reports were labeled by two annotators as relevant to four abdominal organs: liver, kidneys, pancreas and/or adrenal glands, or none. Automated classification methods were compared: simple string matching, random forests, extreme gradient boosting, support vector machines, and two neural network architectures—convolutional neural networks and long short-term memory networks. Three methods from the literature were used to provide interpretability and qualitative validation of the learned network features.
Results
The neural networks performed well on the four-organ classification task (F1 score: 96.3% for convolutional neural network and 96.7% for long short-term memory vs 89.9% for support vector machines, 93.9% for extreme gradient boosting, 82.8% for random forests, and 75.2% for simple string matching). Multiple methods were used to visualize the decision-making process of the network, verifying that the networks used similar heuristics to a human annotator. The neural networks were able to classify, with a high degree of accuracy, pathology reports written in unseen formats, suggesting the networks had learned a generalizable encoding of the salient features.
Conclusion
Neural network–based approaches achieve high performance on organ-level pathology report classification, suggesting that it is feasible to use them within automated tracking systems.
© RSNA, 2019
Supplemental material is available for this article.
See also the commentary by Liu in this issue.
Summary
Neural network–based algorithms perform well in organ-level classification of multi-institution free-text pathology reports and learn features familiar to and understandable by humans.
Key Points
■ Neural network–based approaches achieve high performance on organ-level classification of free-text pathology reports.
■ The system qualitatively learned features similar to human annotators.
■ Similar approaches are likely feasible for use in a wide variety of clinical settings.
Introduction
Our institution currently uses an automated radiology recommendation tracking engine to increase the likelihood of follow-up completion after findings of possible cancer are detected in the abdomen or pelvis. The system extracts structured organ-level information from radiology reports, tracks follow-up within the health system, and automatically notifies the ordering physician of incomplete follow-up (1). To fully monitor recommended follow-up, it is also necessary to review surgical pathology reports, because some patients may proceed straight to biopsy rather than undergoing further imaging. An ideal automated system would identify when a pathology report is relevant to a previously discovered abnormal imaging finding (eg, a liver biopsy in a patient with a hepatic lesion suspicious for cancer previously depicted at CT), allowing clinicians to quickly hone in on relevant pathology reports and relevant sections within those reports. However, these reports often include large free-text segments and are structured differently due to interphysician and interinstitutional variability. Reports may often be relevant to multiple organs, or may describe multiple tissue samples, making it difficult to impose external structure from the top down. These limitations necessitate sophisticated approaches for classification and information extraction.
The field of natural language processing includes all algorithms designed to classify, cluster, or extract information from free text. The two general families of natural language processing algorithms are rule based and statistical (2). Many existing clinical systems rely on hand-engineered, rule-based approaches to process or preprocess text; under this paradigm, documents have to be split into sections, words have to be stemmed with specific algorithms, and external domain-specific lexicons are leveraged (2). However, statistical approaches, including machine learning algorithms, use properties of the data to learn how to perform end-to-end classification and information extraction from the raw text input, without requiring domain-specific rules or algorithms. Such approaches have become state of the art over the past few years in text classification and information extraction tasks (3,4).
In radiology, prior work has extracted specific clinical entities from radiology reports (5), and modeled latent topics in reports through unstructured clustering algorithms (6). Other approaches aim to extract follow-up recommendations from reports (7). Machine learning approaches, including support vector machines and deep neural network architectures, have also been used in other pathology report classification tasks (8,9).
We hypothesized that machine learning approaches, in particular neural networks, can outperform other approaches on organ-level pathology report classification with no domain-specific feature engineering. We present such a system, demonstrate its feasibility, and compare it to other approaches.
Materials and Methods
Data Collection
This was a retrospective study repurposing data collected for nonresearch purposes, approved by our institutional review board. A total of 2013 surgical pathology reports acquired between 2012 and 2018 were taken from our institution’s database. All of the patients in these reports had previously had an abnormal finding depicted at abdominal imaging that, in the reading radiologist’s opinion, required follow-up. The pathology had been obtained from adult patients of all sexes, came from multiple hospitals within a single health system, and were formatted with different structures, including entirely free-text reports. The reports included all major organ systems, including those not relevant to the abdomen (eg, brain biopsy, bone marrow biopsy, etc). The study was compliant with the Health Insurance Portability and Accountability Act.
Data Annotation
These reports were labeled by two annotators—one 4th-year medical student (J.M.S.) and one attending radiologist (T.S.C.)—as being relevant to any subset of the following abdominal organs: liver, pancreas, kidneys and/or adrenal glands, or none of the above. These organs were chosen because they represent the major categories of abnormal imaging findings followed by our tracking system. Reports were labeled as relevant to an organ if any tissue sample in the report contained tissue from that organ, or if the biopsy was performed to further work up a pathology of that organ (eg, a distant metastasis). Many of the reports had multiple tissue samples, so assigning each report a single label would not have been possible. For a separate experiment, reports were also labeled as being relevant to eight other organs—lungs, lymph nodes, peritoneum, ovaries, bladder, gallbladder, stomach, and small bowel—making 12 organs in total.
Model Evaluation
We treated the task as a multiclass and multilabel classification problem (ie, each report can be relevant to any subset of all labeled abdominal organs, including none). We evaluated the performance of different models on the four-organ classification task. Our simplest classifier used direct string matching; for example, a report was relevant to the liver if and only if it included some formulation of one of the strings “liver,” “hepato,” “hepatic,” et cetera (see Table 1 for details).
Table 1:
List of Search Terms Used for the String-Matching Algorithm with Each of the Four Major Organs

Note.—The space before “renal” and “liver” indicates any whitespace character.
We next tested various text classification algorithms based on n-gram term frequency–inverse document frequency features (10): support vector machines, extreme gradient boosting or XGBoost, and random forests. N-grams of size 1 to 6 were included as features based on preliminary testing. See Appendix E1 (supplement) for additional details about these algorithms and their implementation.
Last, we evaluated two neural network–based architectures—convolutional neural networks (CNNs) and long short-term memory (LSTM) networks (a common type of recurrent neural network used for processing sequential data, such as text or audio) (11)—both of which have shown promise in text classification (3,4). In both neural network models, documents were tokenized based on whitespace (we treated punctuation and other special characters such as parentheses as separate tokens, because they contain useful semantic content within reports), and tokens were embedded in a vector space by using pretrained Global Vectors for Word Representation, or GloVe (12). Parameters of the models, including network depth, units per layer, pretrained versus continually trained word vector embeddings, and convolutional filter size, were evaluated and compared to select the optimal models (13). Models were trained with the adaptive moment estimation (Adam) optimization algorithm (14) with binary cross-entropy as the loss function. Training both neural network models took approximately 15 minutes on a machine with one Nvidia GTX 1070 (Nvidia, Santa Clara, Calif) graphics processing unit, or GPU, and 2 hours on a machine with no GPUs. See Appendix E1 (supplement) for further details on model design decisions.
Optimal model parameters were selected by using 10-fold cross-validation on 1814 of the reports. The best models from each class were compared by using a test set of 199 unseen reports. Micro- and macro-averaged precision, recall, and F1 score (the harmonic mean of precision and recall; see Appendix E2 [supplement]) (14), as well as subset accuracy (the percentage of reports for which the exact set of relevant organs was correctly predicted) were calculated for each organ and averaged over all 10 runs. All software was written by using Python (version 3.6; https://www.python.org/). Publicly available machine learning packages (Keras, version 2.2.2; XGBoost, version 0.9; and scikit-learn, version 0.19.2) were used.
In a final experiment, we evaluated the performance of our best model on the more complex 12-organ classification task to demonstrate feasibility on a larger number of organ classes. Because there were many organs with few labeled examples, we felt that a full performance comparison of all models would be highly subject to the noise in this particular small dataset and thus be uninformative about the true performance of the models.
Interpretability
To confirm our system was learning to classify based on “true” generalizable features, and to ultimately improve user trust in the system, we conducted various experiments on interpretability. First, we identified the phrases in the report corpus that produced the maximal output from each of the convolutional filters of the CNN, which represent low-level features the system has learned are useful in classification (15). Second, we conducted gradient-weighted class activation mapping, or Grad-CAM (16), which uses networks gradients to identify input features that have the greatest effect on the classification outcome. In this way, we visualized words that influenced the network to make its decision. Third, for both networks, we performed word type–based occlusion sensitivity tests (15). We deleted all instances of various words from a given report and rechecked the prediction of the machine. Comparing occlusion of words with intuitively useful semantic content (eg, “liver,” “hepatic”) versus random word occlusion provides a coarse metric of word importance. Although it is difficult to provide quantitative measurements of interpretability, we do provide examples and qualitative interpretation of these methods on a subset of reports.
Results
Labeled Data
With regard to the four-organ classification task, the two human annotators agreed on the exact subset of organs on 1957 of 2013 pathology reports (97.2%). On the 12-organ classification task, the annotators agreed on 1815 of 2013 reports (90.2%). Conflicts were resolved through discussion between the two annotators to achieve consensus. In most cases, the conflicts were secondary to keystroke data entry errors rather than disagreement regarding the content of the pathology reports.
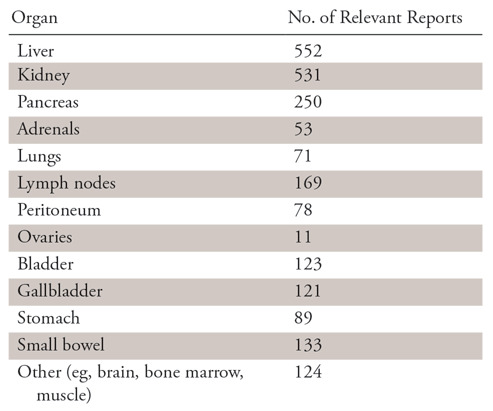
Of the 2013 pathology reports, 552 (27.4%) were labeled as being relevant to the liver, 531 (26.3%) to the kidneys, 250 (12.4%) to the pancreas, and 53 (2.6%) to the adrenal glands (Table 2). Most reports were relevant to exactly one (1290, 64.1%) or zero (676, 33.6%) of the four included abdominal organs. Some reports were relevant to two organs (45, 2.2%), whereas only two were relevant to three organs and none included all four. A more granular organ-level depiction of the pathology reports is shown in Table 2.
Table 2:
Number of Pathology Reports Labeled as Relevant to Each Organ
Neural Network Model Selection
Models were trained and validated on the four-organ classification task by using a grid search to identify optimal parameters of the architecture (see Appendix E1 [supplement]). The best-performing CNN used one convolutional layer, consisting of 200 filters of size 7, with rectified linear unit activation. This convolutional layer was followed by a global max-pooling layer, meant to identify the most salient location in the text for each learned feature. The output of this layer then projected through dense connections to a layer of size 4—one unit for each organ of interest. A sigmoid threshold function was applied to each of these four units, resulting in a probability between 0 and 1 for each organ. Deeper CNNs with two or more convolutional layers did not provide any benefit (see Appendix E1 [supplement]). The best LSTM used one bidirectional layer of 150 standard LSTM units in each direction. Use of more units resulted in overfitting to the training data (see Appendix E1 [supplement]). GloVe word embeddings pretrained on the Common Crawl dataset of web pages were used as input to both networks; no performance benefit was observed from continuing to train word embeddings during the experiments. A dropout rate of 0.5 was empirically found to perform well on both the CNN convolutional layer as well as the LSTM layer.
Performance Comparison
Table 3 displays the relative performance of the evaluated models on the four-organ classification task on the unseen test set of 199 reports, and Table 4 displays specific organ-level performance. The neural network approaches perform well on the task (F1 score: 96.3% [95% confidence interval {CI}: 93.0%, 99.6%] for CNN and 96.7% [95% CI: 93.6%, 99.8%] for LSTM, vs 89.9% [95% CI: 84.6%, 95.1%] for the support vector machine, 93.9% [95% CI: 89.7%, 98.0%] for XGBoost, 82.8% [95% CI: 76.2%, 89.4%] for random forests, and 75.2% [95% CI: 67.7%, 82.7%] for simple string matching). Subset accuracy scores (predicting the exact subset of relevant organs for a given pathology report) were 96.0% (95% CI: 93.3%, 98.7%) for LSTM and 96.0% (95% CI: 88.9%, 96.1%) for CNN versus 88.9% (95% CI: 84.6%, 93.2%) for the support vector machine, 92.5% (95% CI: 88.9%, 96.1%) for XGBoost, 83.4% (95% CI: 78.3%, 88.5%) for random forests, and 61.8% (95% CI: 55.1%, 68.4%) for simple string matching. Given the small volume of errors made by the best-performing models, we were unable to discern any qualitative or quantitative differences between the types of errors made by the LSTM and the CNN.
Table 3:
Relative Performance of the Final Evaluated Models on Test Data
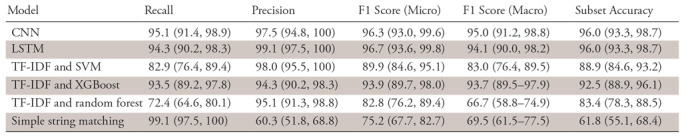
Note.—Data are percentages, with 95% confidence intervals in parentheses. CNN = convolutional neural network, LSTM = long short-term memory, SVM = support vector machine, TF-IDF = term frequency–inverse document frequency, XGBoost = extreme gradient boosting.
Table 4:
F1 Scores of the Different Models by Organ
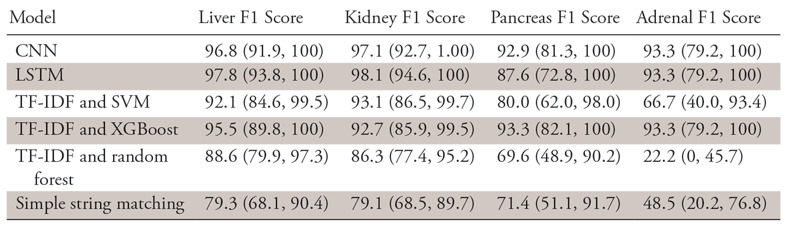
Note.—Data are percentages, with 95% confidence intervals in parentheses. CNN = convolutional neural network, LSTM = long short-term memory, SVM = support vector machine, TF-IDF = term frequency–inverse document frequency, XGBoost = extreme gradient boosting.
Last, we evaluated the performance of our best-performing model, the LSTM, on the more complex 12-organ classification task by changing the final dense layer from four to 12 output units, each representing a single organ, and retraining on the fully labeled data. The data for certain classes were far sparser (eg, only 11 reports were relevant to the ovaries). The LSTM achieves recall of 85.0% (95% CI: 80.2%, 89.8%), precision of 98.3% (95% CI: 96.6%, 100%), and F1 score of 91.1% (95% CI: 87.3%, 94.9%), with a subset accuracy of 85.6% (95% CI: 81.0%, 90.6%) on this task.
Interpretability
Neural network interpretability is an open and complex problem, but many methods have been developed to address the issue.
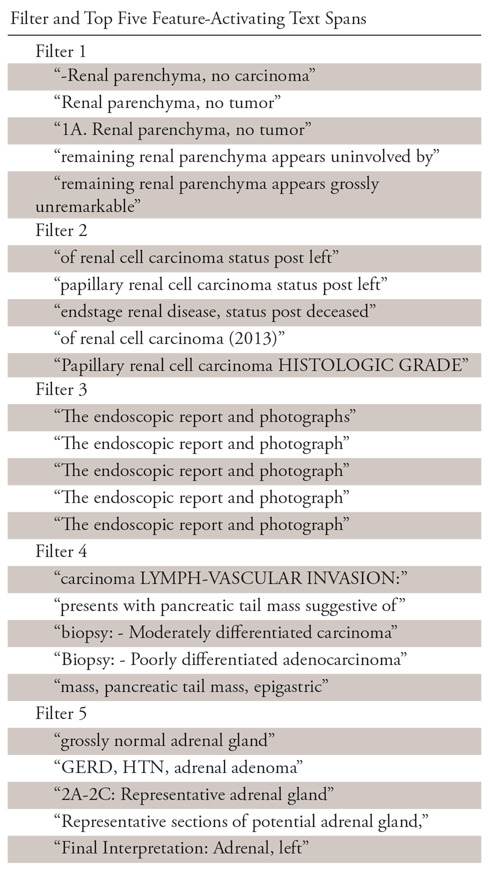
First, we examined the text spans in the documents that produced maximal output on each of the convolutional filters in the CNN. These loosely represent the learned features the network has found to be useful to solve the ultimate task. Table 5 shows the top five maximally activating phrases in the corpus for five randomly selected convolutional filters (note that whitespace and new line characters have been removed in this table for readability, so the spans may not be exactly seven “tokens” long). One can see that the features correspond to human-understandable clusters, which might reasonably be useful in classifying the documents. In a similar way, one can analyze the hidden states of the LSTM at each time step to interpret its learned features.
Table 5:
Maximally Activating Text Spans for Randomly Selected Features in the Convolutional Neural Network
Second, we used the Grad-CAM algorithm, which uses model gradients to calculate the impact of specific words and text subsections on the ultimate classification decision of the model. Figures 1 and 2 show randomly selected representative examples of this type of visualization. The colors, along a gradient from white to yellow, represent the impact each token in the document has on the ultimate decision of the network to assign a particular organ relevance to the document, normalized between zero and one over all tokens in entire pathology report. For ease of visualizing high-saliency tokens, normalized values less than 0.5 have been rounded to 0 in the figure. Although quantitative evaluation of interpretability is difficult, one can see qualitatively from these visualizations that the decision of the network is influenced by sensible parts of the document, and it is able to handle divergent document structures effectively.
Figure 1:

Image shows pathology report colored by normalized gradient-weighted class activation map for class Pancreas, representing salient text spans for classification of neural network.
Figure 2:

Image shows pathology report colored by normalized gradient-weighted class activation map for class Kidney, representing salient text spans for classification of neural network.
Third, results of occlusion experiments showed that removing all occurrences of the words used in the simple string-matching algorithm within a particular document often led to considerable changes in the prediction confidence of the models (eg, removing all occurrences of the words “liver,” “hepatic,” etc, led to frequent changes in model prediction). For instance, in one test run, 35 of the 53 instances classified as liver relevant would no longer be classified as such after the occlusion of liver-related words; similarly, the model changed its prediction on 18 of 21 pancreas reports, two of four adrenal reports, and two of 49 kidney reports after occlusion of relevant sets of words. We compared this with trials of occlusion of random words that occurred with similar frequency; the network did not change any of its predictions. This method, although coarse and based on imperfect occlusion, provides further evidence that the network is detecting human-understandable features and can be applied to either neural network–based model.
Discussion
We aimed to develop a system for organ-level pathology report classification, for use in a larger system of automated radiology recommendation follow-up tracking. We hypothesized that neural network–based algorithms would perform well on the task with minimal rule-based preprocessing of raw texts. We found that our hypothesis was correct, with both LSTM- and CNN-based neural network algorithms achieving approximately 95% accuracy and F1 scores on a highly varied corpus of multi-institution multiformat reports. Furthermore, we used methods for interpretability from the literature to evaluate the sensibility of our models’ learned parameters and found that the system qualitatively learned features similar to human annotators. Lastly, both neural network models were small enough to be trained and deployed within hours on machines without GPUs, making them feasible to use in a wide variety of clinical settings, even those without significant computational resources. These models are easily scalable to much larger corpora of labeled reports with no modifications.
We anticipate that this system will be useful within the context of the broader tracking engine, which aims to reduce missed follow-up of abnormal imaging findings. It is important to note that the use of the findings of this study within the broader tracking engine is designed to augment, rather than to replace, human monitors, by providing another independent “checker” to improve joint accuracy of the human-machine system and increase the overall efficiency of the human reviewer. Toward this end, one might incorporate the overall interpretability algorithms into the overall system by auto-populating the most salient word spans from new reports into the user interface along with the prediction, allowing readers to quickly judge whether the prediction of the machine was correct or incorrect. This would significantly decrease time spent opening and reading full pathology reports (the current procedure at our institution).
Large volumes of unstructured clinical text represent a huge store of information—not only for basic science usages, but also for clinical workflow optimization and quality improvement. Effective summarization and presentation of information contained within the chart holds great potential for improving care efficiency and quality. In general, end-to-end machine learning systems that take raw text as input often have significant advantages not only in performance, but also in algorithm deployment time over complex rule-based pipelines by minimizing time spent feature engineering or writing task-specific algorithms.
The “black box” nature of certain machine learning algorithms has become a popular topic of discussion as of late. In the clinical domain, where such algorithms will be used frequently by humans, interpretability is critical for system trust. Much study in the machine learning community has gone into improving the interpretability of these systems with methods such as Grad-CAM and occlusion sensitivity mapping. These systems, which have primarily been used in image-analysis systems, are equally applicable to natural language processing systems. In general, interpretability and so-called explainability are important concerns to keep in mind from the very beginning of the development process.
It is interesting to note that the string-matching algorithm performed relatively poorly, mostly due to false-positive classifications; it was not obvious to the authors that this would be the case a priori. From looking at misclassified examples, the most common errors resulted from reports in which patients’ clinical history was included in the note (eg, “patient with history of hepatocellular carcinoma” included as a summary statement in a bone marrow biopsy report). LSTM and CNN appear able to compensate for this effectively. The seven-word “receptive field” of the CNN likely allows it to detect surrounding context for words such as “liver,” which suggest whether the mention of “liver” is in the context of an unrelated clinical history or a specific tissue sample, whereas the LSTM can use its memory cells to perform the same task. Furthermore, CNNs and LSTMs may be more robust to individual heuristic failures than are rule-based approaches because they rely on the integration of information from hundreds of separate low-level features.
One limitation of our study was the small number of adrenal pathology reports in our sample. It is well known that datasets of a certain critical mass are necessary for machine learning algorithms to generalize effectively. Furthermore, any given 10-fold partition of our dataset may only include three or four adrenal reports, making it difficult to quantify and compare performance on this specific subtask. Another limitation was related to the use of noncontextual word embeddings. It has been shown that adding word vectors that incorporate character-level information or surrounding word context, such as Embeddings from Language Models (17), often improves performance on natural language processing tasks; this represents another avenue of future work. Lastly, all of these supervised learning algorithms require manually labeled training data, and thus each additional label categorization incurs a time and human labor cost to produce.
In summary, we provide evidence that end-to-end neural network architectures perform well on a clinical text-classification task with high levels of human interpretability. Such systems have the potential to improve information extraction and summarization in a wide variety of clinical contexts, toward the ultimate end of improving care quality and efficiency.
APPENDIX
Disclosures of Conflicts of Interest: J.M.S. disclosed no relevant relationships. C.M.C. disclosed no relevant relationships. D.L. disclosed no relevant relationships. H.M.Z. Activities related to the present article: institution received grant from Penn Center for Innovation. Activities not related to the present article: institution received grant from Eastern Cooperative Oncology Group and American College of Radiology Imaging Network (ECOG-ACRIN); author received payment for lectures including service on speakers bureaus from ACVR and Jefferson Health System. Other relationships: disclosed no relevant relationships. T.S.C. Activities related to the present article: disclosed no relevant relationships. Activities not related to the present article: is a board member of AUR, PRS, PRRS, and SIIM; has grants/grants pending with ACRIN, AUR, and Beryl Institute; received payment for lectures including service on speakers bureaus from AIRP VOICE program, Cornell University, Emory University, Thomas Jefferson University Hospital, and University of Wisconsin; received royalties from Osler Institute for lectures on cardiac imaging. Other relationships: disclosed no relevant relationships.
Abbreviations:
- CI
- confidence interval
- CNN
- convolutional neural network
- LSTM
- long short-term memory
References
- 1.Cook TS, Lalevic D, Sloan C, et al. Implementation of an automated radiology recommendation-tracking engine for abdominal imaging findings of possible cancer. J Am Coll Radiol 2017;14 (5):629–636. [DOI] [PubMed] [Google Scholar]
- 2.Pons E, Braun LMM, Hunink MGM, Kors JA. Natural language processing in radiology: a systematic review. Radiology 2016;279(2):329–343. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y. Convolutional neural networks for sentence classification. ArXiv [cs.CL] [preprint]. http://arxiv.org/abs/1408.5882. Posted 2014. Accessed October 4, 2018.
- 4.Nowak J, Taspinar A, Scherer R. LSTM Recurrent Neural Networks for Short Text and Sentiment Classification. In: International Conference on Artificial Intelligence and Soft Computing, 2017; 553–562. [Google Scholar]
- 5.Hassanpour S, Langlotz CP. Information extraction from multi-institutional radiology reports. Artif Intell Med 2016;66:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hassanpour S, Langlotz CP. Unsupervised topic modeling in a large free text radiology report repository. J Digit Imaging 2016;29(1):59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveira L, Tellis R, Qian Y, Trovato K, Mankovich G. Follow-up recommendation detection on radiology reports with incidental pulmonary nodules. Stud Health Technol Inform 2015;216:1028. [PubMed] [Google Scholar]
- 8.Gao S, Young MT, Qiu JX, et al. Hierarchical attention networks for information extraction from cancer pathology reports. J Am Med Inform Assoc 2017 Nov 16 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon H, Robinson S, Christian JB, Qiu JX, Tourassi GD. Filter pruning of convolutional neural networks for text classification: a case study of cancer pathology report comprehension. In: 2018 IEEE EMBS International Conference on Biomedical Health Informatics (BHI), 2018; 345–348. [Google Scholar]
- 10.Jones KS. A statistical interpretation of term specificity and its application in retrieval. J Doc 1972;28(1):11–21. [Google Scholar]
- 11.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput 1997;9(8):1735–1780. [DOI] [PubMed] [Google Scholar]
- 12.Pennington J. GloVe: Global Vectors for Word Representation. https://nlp.stanford.edu/projects/glove/. Accessed October 4, 2018.
- 13.Zhang Y, Wallace B. A sensitivity analysis of (and practitioners’ guide to) convolutional neural networks for sentence classification. ArXiv [cs.CL] [preprint]. http://arxiv.org/abs/1510.03820. Posted 2015. Accessed October 4, 2018.
- 14.Kingma DP, Ba J. Adam: a method for stochastic optimization. ArXiv [cs.LG] [preprint]. http://arxiv.org/abs/1412.6980. Posted 2014. Accessed October 4, 2018.
- 15.Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. ArXiv [cs.CV] [preprint]. http://arxiv.org/abs/1311.2901. Posted 2013. Accessed October 4, 2018.
- 16.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. ArXiv [cs.CV] [preprint]. http://arxiv.org/abs/1610.02391. Posted 2016. Accessed October 4, 2018.
- 17.Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. ArXiv [cs.CL] [preprint]. http://arxiv.org/abs/1802.05365. Posted 2018. Accessed October 4, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


