Significance
In hematophagous female mosquitoes, each reproductive cycle is linked to a separate blood intake, serving as a foundation for the transmission of dangerous human diseases. During each reproductive cycle, female mosquitoes sequentially feed on carbohydrates and protein (blood). Metabolic flux is alternated to support the reproductive cyclicity. We have established that insulin-like peptides (ILPs), critical for regulating metabolism, are genetically controlled by juvenile hormone (JH) and 20-hydroxyecdysone (20E), the key hormones governing the reproduction of female mosquitoes. CRISPR gene-tagging experiments revealed that the JH and 20E pathways coordinate the production of ILPs. This study has uncovered the link between ILPs and JH and 20E pathways in controlling mosquito metabolism during reproduction of the Aedes aegypti mosquito.
Keywords: insulin, hormone, insect, metabolism, CRISPR-Cas9
Abstract
Female mosquitoes feed sequentially on carbohydrates (nectar) and proteins (blood) during each gonadotrophic cycle to become reproductively competent and effective disease vectors. Accordingly, metabolism is synchronized to support this reproductive cyclicity. However, regulatory pathways linking metabolism to reproductive cycles are not fully understood. Two key hormones, juvenile hormone (JH) and ecdysteroids (20-hydroxyecdysone, 20E, is the most active form) govern female mosquito reproduction. Aedes aegypti genome codes for eight insulin-like peptides (ILPs) that are critical for controlling metabolism. We examined the effects of the JH and 20E pathways on mosquito ILP expression to decipher regulation of metabolism in a reproducing female mosquito. Chromatin immunoprecipitation assays showed genomic interactions between ilp genes and the JH receptor, methoprene-tolerant, a transcription factor, Krüppel homolog 1 (Kr-h1), and two isoforms of the ecdysone response early gene, E74. The luciferase reporter assays showed that Kr-h1 activates ilps 2, 6, and 7, but represses ilps 4 and 5. The 20E pathway displayed the opposite effect in the regulation of ilps. E74B repressed ilps 2 and 6, while E74A activated ilps 4 and 5. Combining RNA interference, CRISPR gene tagging and enzyme-linked immunosorbent assay, we have shown that the JH and 20E regulate protein levels of all eight Ae. aegypti ILPs. Thus, we have established a regulatory axis between ILPs, JH, and 20E in coordination of metabolism during gonadotrophic cycles of Ae. aegypti.
Mosquito-borne diseases continue to emerge and reemerge globally (1). For some viral pathogens, the lack of vaccines places a greater emphasis on disease prevention through effective mosquito management and control (1, 2). The Aedes aegypti mosquito is the major vector involved in transmission of human arboviral diseases such as dengue, chikungunya, yellow fever, Zika virus, and others (3–6). The World Health Organization (WHO) and Centers for Disease Control and Prevention (CDC) state that mosquitoes, particularly those from the genera Aedes, Culex, and Anopheles, continue to be organisms of utmost concern in global public health.
Disease-vector female mosquitoes undergo a biphasic feeding pattern, shifting from carbohydrates (nectar) to proteins (vertebrate blood) for survivorship, reproduction, and pathogen transmission (7, 8). Interchanging titers of juvenile hormone (JH) and 20-hydroxyecdysone (20E) shape the mosquito reproductive cycle (8, 9). JH-III (JH) controls the posteclosion (PE) phase, during which a female mosquito undergoes preparatory development for subsequent blood feeding and egg maturation. 20E controls the postblood-meal (PBM) phase from blood feeding to oviposition. These two phases form the gonadotrophic cycle in anautogenous mosquitoes. Methoprene-tolerant (Met), a basic helix–loop–helix (bHLH)-Per-Arnt-Sim (PAS) transcription factor, has been identified as the JH receptor (10–12). Additionally, a steroid receptor coactivator (Taiman) forms a heterodimer with Met to transduce the JH signal (13, 14). In the nucleus, the JH receptor complex activates target genes directly by binding a 9-mer Met-motif (CACGC/TGA/GT/AG); it also represses target genes indirectly, which requires other downstream factors (15, 16). Krüppel-homolog 1 (Kr-h1) is a C2H2 zinc-finger transcription factor that mediates Met action, either activating or repressing JH target genes by direct interaction with their Kr-h1-binding motifs (17, 18). The 9-mer sequence variations have been identified in Ae. aegypti as the Kr-h1-binding motif, which are similar to those first reported in Bombyx mori (16, 19).
A heterodimer of the ecdysone receptor (EcR) and ultraspiracle (USP) mediates 20E signaling to trigger a vast transcriptional program (20). The 20E receptor complex regulates target genes by means of either pseudopalindromic response elements resembling inverted repeats of AGGTCA separated by a single nucleotide spacer or downstream responsive genes (21). The ecdysone-inducible early gene E74 (Eip74EF) encodes an ETS (erythroblast transformation specific) transcription factor that mediates the 20E response in gene regulation. In mosquitoes, there are two isoforms, E74A and E74B, that act by differentially activating or repressing 20E-responsive genes (22, 23). They share a common C-terminal ETS DNA-binding domain and bind to a consensus core motif C/AGGAA, but have unique N-terminal sequences (23, 24). Moreover, E74 isoforms are differentially expressed in the Ae. aegypti mosquito (23).
Insulin-like peptides (ILPs) are pleiotropic peptide hormones encoded by multigene families that regulate metabolism, growth, reproduction, and longevity (25). The insulin signaling pathway is evolutionarily conserved (26). As many as 1 to 38 ILPs have been identified in insects (26). Even different mosquito species have varying numbers of ILPs (25). The model insect Drosophila melanogaster has eight ILPs (DILPs) (26). The production and release of DILPs are controlled by several different mechanisms, including nutrient sensing, neuropeptides, neurotransmitters or factors originating from the intestine, and adipocytes (26). After release, the action of DILPs can be modulated by hemolymph proteins that act either as protective carriers or competitive inhibitors (26). However, a species-specific and stage-dependent outcome with the control of the whole set of ILPs remains to be investigated.
Like D. melanogaster, the Ae. aegypti genome carries eight ilps that are expressed in the brain and other tissues (27). CRISPR-Cas9 analyses have shown that ILPs in this mosquito play different roles in body size determination and lipid metabolism (28, 29). Previous studies have revealed the involvement of JH and 20E pathways in the coordination of lipid and carbohydrate metabolism during the mosquito gonadotrophic cycle (30, 31). Here, we identified the functional link of JH and 20E pathways with eight ILPs in the control of mosquito metabolism.
Results
Tissue Distribution and Expression Patterns of ILPs in the Female Ae. aegypti Mosquito.
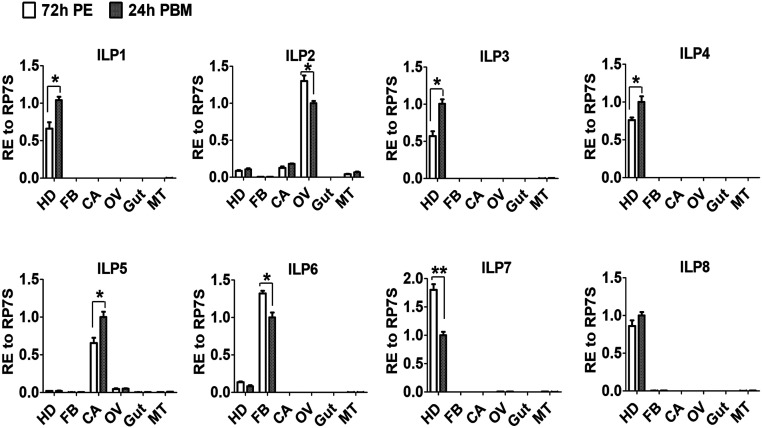
We first determined the tissue localization of all eight Aedes ILP transcripts by means of quantitative real-time PCR (qPCR) analysis. This revealed differential distribution of ILP transcripts in female mosquito tissues. ILPs 1, 3, 4, 7, and 8 were found to be brain specific. However, three other ILPs showed distribution in other tissues: ILP2 transcripts were enriched in the ovary, ILP5 in the carcass void of the fat body, and ILP6 in the isolated fat body (Fig. 1). Next, we collected female mosquito tissues at five time points during the gonadotrophic cycle: 12 h and 48 h PE and at 12 h, 24 h, and 48 h PBM. Transcript levels of ILPs 4 and 7 were high at 12 h PE and lower thereafter; the ILP6 transcript had its peak at 48 h PE. Transcript levels of five other ILPs were higher during the PBM phase (SI Appendix, Fig. S1). The ILP2 transcript exhibited a peak at 12 h PBM, and ILPs 1, 3, 5, and 8 at 24 h PBM (SI Appendix, Fig. S1). These expression patterns of the ilp genes suggest a possible link to the hormonal regulatory axis governed by JH and 20E.
Fig. 1.
Tissue distribution of ILP transcription in female Ae. aegypti mosquitoes. Relative expression levels of ilp genes in the head (HD), isolated fat body (FB), carcass (CA, abdominal wall without the fat body), ovary (OV), gut, and Malpighian tube (MT) at 72 h PE and 24 h PBM. Relative expression levels in abundant group at 24 h PBM are represented as 1, with corresponding adjustments in other values. Data represent three biological replicates with 30 individuals in each and are shown as mean ± SEM *P < 0.05, **P < 0.01.
JH and 20E Pathways Are Involved in the Regulation of ilp Gene Expression.
Lipids and glycogen are the major energy reserves stored in the mosquito fat body, an insect analog of the vertebrate liver and adipose tissue (32, 33). JH and 20E have been implicated in the expression control of expression of genes coding for carbohydrate and lipid metabolic enzymes in the female Ae. aegypti (30, 31). To investigate the possibility of a direct effect of JH and 20E on the expression of metabolic enzyme genes, we conducted chromatin immunoprecipitation (ChIP) analysis. This revealed no genomic interaction between the JH (Met and Kr-h1) or the 20E (EcR and E74) pathway and selected metabolic genes (SI Appendix, Figs. S2–S4). Therefore, we hypothesized that JH and 20E might be involved in the cross-talk with other regulators of metabolism.
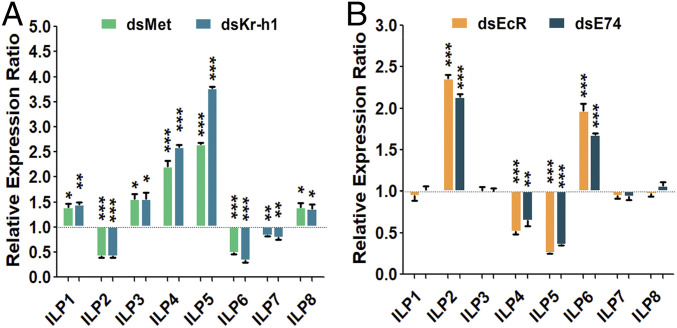
The role of insulin and ILPs in regulating metabolism is well established (25, 26). To determine a possible interaction of ILPs with the JH pathway in female mosquitoes, we first performed topical JH treatment. JH III (a 0.3-μL aliquot of 1 μg/mL) or acetone (control) was applied onto the abdomens of female mosquitoes at 6 h PE, when the level of this hormone is very low, and then the transcript levels of each of the eight ILPs was measured using RT-qPCR at 24 h post-JH application. The transcripts for ILP2, ILP6, and ILP7 were significantly higher, while those for ILP1, ILP3, ILP4, ILP5, and ILP8 were lower in JH III-treated mosquitoes than in controls (SI Appendix, Fig. S5A). The effect of Met RNAi was opposite: transcripts of ILP2, ILP6, and ILP7 were down-regulated and those of ILP1, ILP3, ILP4, ILP5, and ILP8 were up-regulated (Fig. 2A). Studies have shown that Kr-h1 is the key factor that mediates Met action in both activation and repression of JH-regulated genes (16, 18). After Kr-h1 RNAi, transcripts of ILP2, ILP6, and ILP7 were down-regulated, while ILP1, ILP3, ILP4, ILP5, and ILP8 were up-regulated in a manner similar to that of Met RNAi (Fig. 2A).
Fig. 2.
Comparative analysis of ILP transcript abundance in dsMet, dsKr-h1, dsEcR, and dsE74 RNAi female mosquitoes. (A) RNAi knockdowns of Met and Kr-h1 repressed ILP 2, 6, and 7 transcripts but activated the other five ILPs. (B) Knockdowns of EcR and E74 elevated ILP 2 and 6 transcripts, down-regulated ILP 4 and 5, while displaying no significant effect on the other ILPs. dsLuc (RNAi-luciferase) was used as control. Data represent three biological replicates with 30 individuals in each and are shown as mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001.
To investigate whether ilps were transcriptionally responsive to the 20E pathway, we performed RNAi of EcR and E74 and measured ILP transcript levels using RT-qPCR. Levels of ILP2 and ILP6 transcripts increased while those of ILP4 and ILP5 reduced in response to the EcR knockdown (Fig. 2B). Similar results were observed for RNAi of E74, demonstrating the indispensable role of E74 in conveying the 20E-EcR/USP signaling in regulating the ilp genes (Fig. 2B). However, except for ILP4, the brain-specific ILPs 1, 3, 7, and 8 were unaffected, suggesting that 20E signaling modulates mainly those expressed in peripheral tissue ILPs (Fig. 2B). It is also possible that the brain-specific ILPs lack responsiveness due to a poor penetrance of dsRNAs into the mosquito brain.
Injection of 20E (0.3-μL aliquot of 500 ng/mL) induced a decline of the ILP2 transcript levels and an elevation of transcripts of ILP4 and ILP5 (SI Appendix, Fig. S5B). No response to 20E was detected for transcripts of ILPs 1, 3, 7, and 8. The effect of exogenous 20E on ILP6 expression was not as clear cut as that elicited by EcR or E74 knockdown (SI Appendix, Fig. S5B). Notably, the other factors of the insulin pathway displayed no changes in transcription after either of the above-mentioned RNAi treatments (SI Appendix, Fig. S6). Taken together, these data suggest that the JH and 20E influence ilp gene expression in the reproducing female mosquitoes.
Regulatory Interactions of ilp Gene Promoters with Met and Kr-h1.
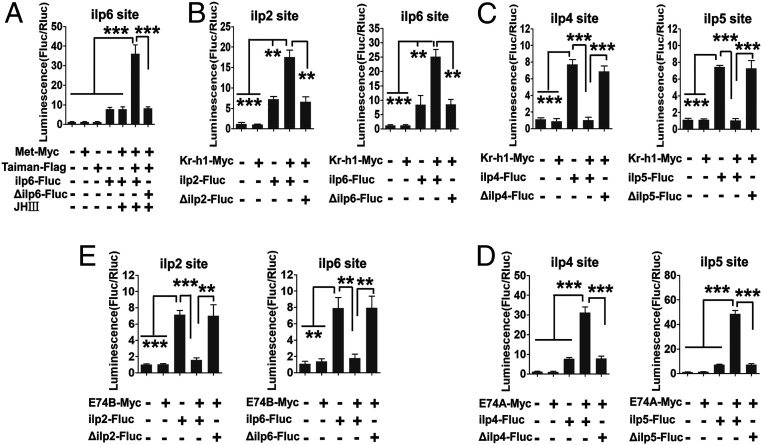
A single Met binding site was found only in the ilp6 gene promoter (SI Appendix, Fig. S7). However, Kr-h1 binding motifs were identified in the 5′ upstream regulatory regions of all eight ilp genes (SI Appendix, Fig. S7). To determine whether Met and/or Kr-h1 confer their binding on ilps, we performed ChIP analysis followed by qPCR to specifically amplify Met- and Kr-h1-bound regions. The Met binding was detected only at the ilp6 gene promoter, and Met RNAi depletion diminished this binding enrichment, confirming the authenticity of the Met binding (SI Appendix, Fig. S8). Kr-h1 binding was enriched not only at JH-activated genes—ilps 2, 6, and 7—but also at JH-inhibited genes—ilps 4, 5, and ilp8-ilp1-ilp3. Kr-h1 RNAi abolished these binding enrichments (SI Appendix, Fig. S9). To gain further confirmation of Met and Kr-h1 interaction with ilp genes, the dual-luciferase assay was performed in D. melanogaster S2 cells. The 1-kb 5′ upstream regulatory regions of the ilp genes harboring the Met and/or Kr-h1 binding motif sites were subcloned into the firefly luciferase reporter vector pGL3basic (ilp-Fluc). When the ilp6-Fluc plasmid was cotransfected with the pAc-Met-Myc and pAc-Taiman-Flag expression plasmids into S2 cells, the luciferase activity in the presence of JH III was more than 4.5-fold higher than in the control group transfected with only ilp6-Fluc (Fig. 3A). Next, we cotransfected reporter plasmids for each of eight ilps together with the pAc-Kr-h1-Myc expression vector. An elevation of luciferase activity was observed after cotransfection of ilp2-Fluc, ilp6-Fluc, and ilp7-Fluc with the pAc-Kr-h1-Myc expression vector; in contrast, there was a reduction after cotransfection of ilp4-Fluc, ilp5-Fluc, or ilp8-1-3-Fluc with pAc-Kr-h1-Myc (Fig. 3 B and C and SI Appendix, Fig. S10A). These responses are consistent with results from the JH treatment assay (SI Appendix, Fig. S5A). Furthermore, these changes in the luciferase activity were abolished after mutation of their binding motifs in ilp promoters (Fig. 3 A–C and SI Appendix, Fig. S10A).
Fig. 3.
Met, Kr-h1, E74A, and E74B regulate the transcription of ilp genes. (A) Luciferase reporter assay after cotransfection of expression vectors pAc-Met-Myc and pAc-Taiman-Flag, along with the reporter construct and JH III, indicates that Met is an activator of ilp6 gene transcription. (B) Luciferase reporter assay after cotransfection of the expression vector pAc-Kr-h1-Myc and reporter constructs points out the activation role of Kr-h1 in the transcription of ilps 2 and 6. (C) Luciferase reporter assay shows that Kr-h1 is a repressor of the transcription of ilps 4 and 5. (D) Luciferase reporter assay after cotransfection of expression vectors pAc-E74A-Myc and reporter constructs indicates that E74A is a transcription activator of ilp4 and ilp5 genes. (E) Luciferase reporter assay after cotransfection of expression vectors pAc-E74B-Myc and reporter constructs indicates E74B as a repressor of ilps 2 and 6 transcription. Treatments with the empty expression vector and no input DNA and motif mutation served as controls. Data represent six replicates and are shown as mean ± SEM **P < 0.01, ***P < 0.001.
The Regulatory Relationships of the ilps Genes with EcR and E74 Transcription Factors.
The ChIP assays were conducted to elucidate whether there is any interaction of EcR with ilp gene promoters. This yielded no binding reaction between EcR and any ilp gene throughout 3 kb of their 5′ upstream regulatory regions, indicating that the regulation of all eight ilp genes by EcR is indirect (SI Appendix, Fig. S11). Unlike the switch in two isoforms E74, the other ecdysone-inducible early gene, broad-complex (BR-C) and Eip75 (E75), have multiple protein isoforms and complex regulatory actions (34, 35). Thus, we used the ChIP assay to examine a possible interaction of E74A and E74B to ilp gene promoters. The E74A genomic binding was identified at the promoters of ilp4 and ilp5 genes, but not detectable at ilp8-ilp1-ilp3, ilp2, ilp6, and ilp7 gene promoters (SI Appendix, Fig. S12). We then tested the E74B isoform for binding to promoters of ilp genes. Such binding was enriched at the promoters of ilp2 and ilp6 genes but was not observed at any other ilp genes (SI Appendix, Fig. S13). Silencing of either E74A or E74B by their respective RNAi abolished binding enrichments of the respective ilp gene promoters (SI Appendix, Figs. S12 and S13).
To further confirm these results, a dual-luciferase assay was performed. When the pAc-E74A-Myc expression plasmid was cotransfected along with either ilp4-Fluc or ilp5-Fluc reporter plasmids, the luciferase activity was significantly higher than the control groups and ilp6-Fluc (Fig. 3D and SI Appendix, Fig. S10B). In contrast when pAc-E74B-Myc was cotransfected with either ilp2-Fluc or ilp6-Fluc, the luciferase activity was significantly lower than the empty vector and ilp5-Fluc controls (Fig. 3E and SI Appendix, Fig. S10B). Mutations of E74 sequence motifs in tested ilp gene promoters abolished the luciferase activity changes (Fig. 3 D and E and SI Appendix, Fig. S10B). Thus, these analyses indicate that the 20E/EcR pathway stimulates ilp4 and ilp5 genes through E74A, while suppressing ilp2 and ilp6 through E74B.
Regulation of ILP Protein Dynamics by the JH and 20E Pathways.
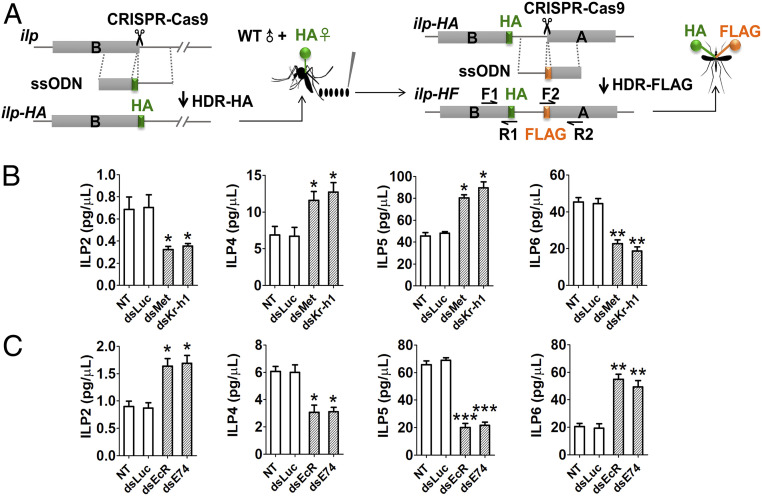
In the experiments described above, we established that both JH and 20E pathways are involved in regulation of ilp gene expression. Effects of these pathways on ILP protein dynamics were further characterized. Insect hemolymph is a dynamic tissue and functions as an extension of intracellular environments transporting hormones (36). For total ILP levels or circulating ILP levels in hemolymph, CRISPR-Cas9-mediated homology-directed repair (HDR) and single-stranded oligodeoxynucleotides (ssODNs) were used to fuse hemagglutinin (HA) and FLAG tags into exons encoding the B- and A- chains in each of eight ilp genes (Fig. 4A). A mixture of guide RNA, Cas9 protein and ssODNs for HA tagging was delivered into wild-type (WT) eggs. The positive females with HA tag were crossed with WT males, and their offspring eggs were used for FLAG tagging. The insertion rate was recorded and verified by means of genomic PCR and Sanger DNA sequencing (SI Appendix, Figs. S14 and S15). Enzyme-linked immunosorbent assay (ELISA) was then performed for epitope-tagged ILP quantification using commercially available HA and FLAG monoclonal antibodies (SI Appendix, Supplemental Materials and Methods). The total content of ILPs ranged from 3 to 80 pg/mg, and their concentration in hemolymph varied from 0.2 to 90 pg/μL (Fig. 4 B and C and SI Appendix, Figs. S16 and S17). These analyses demonstrate that the JH and 20E pathways modulate ILP production and secretion, restricting them to appropriate amounts required during PE and PBM phases of the mosquito reproductive cycle.
Fig. 4.
CRISPR-Cas9-mediated gene tagging for the determination of hemolymph ILP levels. (A) Diagram indicates ILP HDR of ssODN donor bearing HA tag inserted into sequences encoding the last codon of B chain amino terminuses of each ilp gene to generate HA-tagged mosquito lines (ilp-HA). Then the eggs from HA-tagged females were used for second tagging. ssODN donor bearing FLAG tag was inserted into sequences encoding the first codon of A chain amino terminuses of each ilp gene to generate HA and FLAG double-tagged mosquito lines (ilp-HF). (B) Hemolymph levels of ILPs in response to JH signaling. Hemolymph ILP-HA/FLAG content (pg/μL) was determined using ELISA in tagged females after RNAi Met or RNAi Kr-h1 treatments and respective controls. (C) Hemolymph levels of ILPs in response to 20E signaling. Hemolymph ILP-HA/FLAG content (pg/μL) was determined using ELISA in tagged females after RNAi EcR or RNAi E74 treatments and respective controls. Data represent three biological replicates with five individuals in each and are shown as mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001.
The JH and 20E Pathways Antagonistically Affect FoxO Localization in the Fat Body.
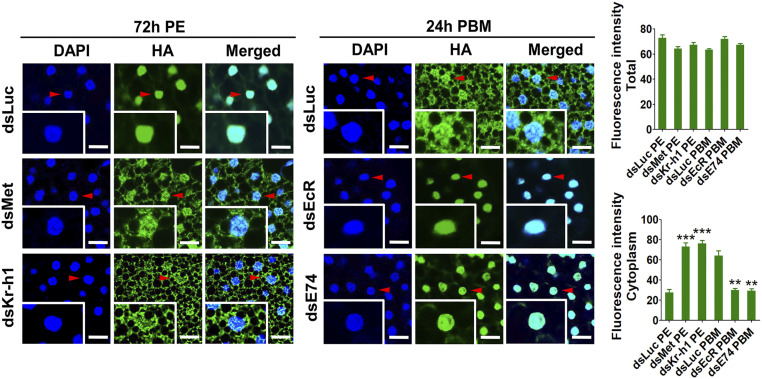
The insect fat body serves as a central organ for metabolism and nutrient reserve storage (37). Forkhead box O (FoxO) is one of the final transcription factors in the insulin pathway. ILPs bind to the insulin receptor (InR) and activate a signal transduction pathway and affect FoxO phosphorylation and nuclear localization (38). CRISPR-Cas9 was used to insert HA tag into the carboxy terminus of the endogenous FoxO, as previously described (28). To examine whether JH and 20E transduce their ILP effects on FoxO, we observed HA-tagged FoxO localization in fat body cells of female mosquitoes after RNAi knockdown of factors mediated by JH and 20E actions (Fig. 5 and SI Appendix, Fig. S18A). In controls, HA-tagged FoxO demonstrated nuclear localization at 72 h PE, although it was predominantly cytoplasmic at 24 h PBM, indicating the shift in FoxO activity during the vitellogenic cycle (Fig. 5). The loss of either Met or Kr-h1 by RNAi in female mosquitoes with HA-tagged FoxO at 72 h PE resulted in its cytoplasmic retention (Fig. 5). In contrast, RNAi knockdown of EcR and E74 at 24 h PBM caused HA-tagged FoxO nuclear translocation (Fig. 5).
Fig. 5.
FoxO subcellular localization in fat body cells. Confocal microscopic (Leica SP5) images of the fat body cells showing the FoxO-HA (green) and the DAPI signal (blue) (Scale bar: 25 μm.) Insets represent images of single cells. Arrowheads indicate nuclei. The mean fluorescence intensity of green signals indicates total and cytoplasmic FoxO-HA levels. Data represent three independent biological replicates with five images in each replicate and are shown as mean ± SEM **P < 0.01, ***P < 0.001.
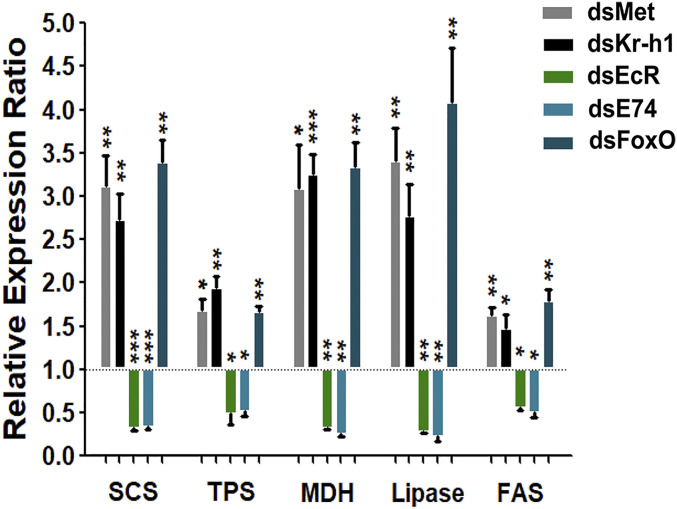
To study FoxO genomic interaction with metabolic genes, we examined promoter regions of several such genes for the presence of FoxO binding sites: succinyl-coA synthetase (SCS), trehalose-6-phosphate synthase (TPS), malate dehydrogenase (MDH), lipase, and fatty acid synthases (FAS) (SI Appendix, Fig. S19). After revealing the presence of such genomic binding sites, the ChIP assay was performed to confirm the interactions of FoxO with these metabolic enzyme genes. Indeed, the FoxO-binding enrichment was observed at the promoters of these metabolic enzyme genes, which was abolished by FoxO RNAi silencing (SI Appendix, Figs. S19 and S18B). Moreover, FoxO RNAi elevated the transcripts of these metabolic enzyme genes as well as Met- or Kr-h1-RNAi, while opposite to the effects of EcR- or E74-RNAi (Fig. 6).
Fig. 6.
Comparative analysis of transcript abundance of metabolic enzyme genes in dsMet, dsKr-h1, dsEcR, dsE74, and dsFoxO RNAi female mosquitoes. RNAi knockdowns of Met, Kr-h1, and FoxO elevated transcripts of the metabolic enzyme genes: succinyl-coA synthetase (SCS), trehalose-6-phosphate synthase (TPS), malate dehydrogenase (MDH), lipase, and fatty acid synthases (FAS). Knockdowns of EcR and E74 repressed them. dsLuc (RNAi-luciferase) was used as control. Data represent three biological replicates and are shown as mean ± SEM *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The insulin signaling system is inherently complex (26). Peptide hormones and other factors affect the activity of ILPs in the regulation of multiple outcomes, such as regulation of metabolism, body size determination, development, and reproduction (25, 26). Mechanistic understanding of the transcriptional regulation of individual ilps is critical for revealing their actions in mosquito gonadotrophic cycles. Our present study identified that JH and 20E, the two principal hormones controlling mosquito gonadotrophic cycles, regulate ILPs, coordinating metabolism with reproduction in the disease vector mosquito Ae. aegypti. Moreover, our study has revealed that the JH and 20E pathways act differentially in determining the expression of the ilp genes. This is achieved through a direct physical interaction of JH and 20E pathway factors with promoters of ilp genes. Our previous CRISPR-Cas9 studies showed that ilp2, ilp6, and ilp8 mutants are characterized by diminished fat reserves (28, 29). Depletion of either ilp4, ilp5, or ilp7 by CRISPR-Cas9 leads to mosquito reproductive defects related to lipid homeostasis, promoting lipid storage (28, 29). Here, we have demonstrated that ilp2, ilp6, and ilp7 are positively regulated by the JH pathway, and Kr-h1 activates the expression of these genes by directly interacting with their promoters. In contrast, the 20E pathway factor E74B inhibits the expression of ilp2 and ilp6 genes by directly interacting with their promoters. The situation is reversed in the regulation of ilp4 and ilp5 gene expression. These ilps are down-regulated by the JH pathway factor Kr-h1 and up-regulated by E74A through a direct interaction of these factors with promoters of these genes. Thus, both the JH and 20E pathways are involved in the expression control of the ilp genes, achieving coordination of their differential actions during the gonadotrophic mosquito cycle. Furthermore, we found that Met only provokes the fat body ilp6 expression by direct binding to this ilp gene promoter. Thus, some regulatory proteins are specific to one ilp gene, some proteins act more widely, and ilp genes can be regulated by several factors simultaneously or at different times.
There are examples showing that expression of ilp genesis regulated by JH and 20E in several insect species (39–43). Juvenile hormone regulates vitellogenin (Vg) gene expression through the ILP signaling pathway in the red flour beetle, Tribolium castaneum (42). Ecdysone-inducible insulin growth factor (IGF)-like peptides in B. mori (BIGFLP) and D. melanogaster (DILP6) are predominantly expressed in the fat body (44, 45).
Insect ILPs differ in their secretion patterns (26, 45). We generated CRISPR-Cas9 epitope-tagged ILPs in Ae. aegypti to determine each ILP protein abundance. Our experimental design included tagging each of two ILP chains with either HA or FLAG. This allowed very accurate measurements of ILP protein levels using ELISA. The whole-body levels and the hemolymph circulating levels are different among ILPs. Although ILPs displayed different abnormalities in their response to RNAi of Met, Kr-h1, EcR, or E74, the set of eight ILPs was mobilized to tightly control the insulin signaling. Thus, we revealed the elements that make up the mechanisms for insulin expression, secretion, and signaling to mediate specific physiological traits in female reproducing mosquitoes.
Insect fat body is the central organ for energy metabolism, accumulating and releasing lipids and carbohydrates to meet the needs of the developing or reproducing organism. These actions are controlled by several hormones, including ILPs (33). The FoxO transcription factor is a major target of insulin action (46, 47).
In D. melanogaster, 20E prompts nuclear translocation of the fat body FoxO inhibiting larval growth (48). Reduced 20E signaling by RNAi-EcR in the larval fat body leads to increased triacyl glyceride levels (49). During pupal development, however, 20E signaling promotes adiposity (50). In female adult mosquitoes, we found the differential FoxO localization in fat body cells during two phases of the gonadotrophic cycle; FoxO was located in nuclei at 72 h PE, when the JH level is high, and in the cytoplasm at 24 h PBM, when the 20E titer is high. Knockdowns of Met and Kr-h1 caused FoxO retardation in the cytoplasm, while those of EcR and E74 resulted in nuclear localization. Thus, the JH and 20E pathways act in opposite ways affecting the FoxO cellular localization and, therefore, its activity. FoxO binds and activates the Drosophila acid lipase 4 (dLip4) and regulates gluconeogenesis through the activation of phosphoenolpyruvate carboxykinase (Pepck) expression (51, 52). We demonstrated FoxO binding to promoters of several carbohydrate and lipid metabolic enzyme genes in the Ae. aegypti.
In conclusion, we have identified a regulatory axis between ILPs and the JH and 20E pathways coordinating metabolism during gonadotrophic cycles of the disease vector, Ae. aegypti.
Materials and Methods
A detailed description of the materials and methods used in this paper is provided in SI Appendix, Supplemental Materials and Methods). Ae. aegypti mosquitoes and their genetics were used. RNAi, CRISPR-Cas9 knockin, gene-tagging, hormonal treatment (JH/20E), ChIP, cell luciferase, ELISA, and immunofluorescence were performed. dsRNA injection, mosquito embryo microinjection, and cell culture were previously described.
CRISPR-Cas9-Mediated Epitope Tagging.
CRISPR-Cas9-mediated HDR was used to generate gene tagging. ssODN donors (199 bases) were designed (SI Appendix, Table S1) and synthesized as Ultramer DNA oligos (Integrated DNA Technologies) containing the 27-base HA-tag sequence or 24-base FLAG-tag sequence, flanked by homologous arms of about 86 bases. sgRNAs were designed as N21GG rule (SI Appendix, Table S1), synthesized using the MEGAscript T7 Transcription Kit (Ambion), and purified using the MEGAclear Transcription Clean-Up Kit (Ambion) following the manufacturer’s protocol. Cas9 protein with nuclear localization signal was purchased (PNA Bio) and stored as 1-mg/mL reconstitutions. Preblastoderm-stage embryos were lined on filter paper, desiccated slightly, transferred onto glass slides, and microinjected with a mixture of ssODNs (125 ng/μL), Cas9 protein (333 ng/μL), and sgRNAs (40 ng/μL). Embryos were injected into the posterior pole at an angle of 10 to 25°. Injected embryos were incubated at 27 °C and 80% humidity to recover for 5 d. They were later hatched and raised to adults. Then females were crossed with wild-type males for oviposition. The females were used for PCR verification. Offspring eggs from positive females were injected for next tagging as mentioned above. The hatchlings were raised to adults. These adults (female) were subjected to RNAi (render ovary abnormalities), direct PCR (dPCR), and ELISA analysis. See SI Appendix, Table S1 for a list of sgRNAs and ssODN donors sequences.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01AI036959 (to A.S.R.).
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2023470118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Ramírez A. L., van den Hurk A. F., Meyer D. B., Ritchie S. A., Searching for the proverbial needle in a haystack: Advances in mosquito-borne arbovirus surveillance. Parasit. Vectors 11, 320 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olson M. F., et al., Sugar feeding patterns for Aedes aegypti and Culex quinquefasciatus (Diptera: Culicidae) mosquitoes in South Texas. J. Med. Entomol. 57, 1111–1119 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett A. D. T., Higgs S., Yellow fever: A disease that has yet to be conquered. Annu. Rev. Entomol. 52, 209–229 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Bhatt S., et al., The global distribution and burden of dengue. Nature 496, 504–507 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsetsarkin K. A., Chen R., Weaver S. C., Interspecies transmission and chikungunya virus emergence. Curr. Opin. Virol. 16, 143–150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNeil C. J., Shetty A. K., Zika virus: A serious global health threat. J. Trop. Pediatr. 63, 242–248 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Foster W. A., Mosquito sugar feeding and reproductive energetics. Annu. Rev. Entomol. 40, 443–474 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Attardo G. M., Hansen I. A., Raikhel A. S., Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem. Mol. Biol. 35, 661–675 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Roy S., et al., Regulation of gene expression patterns in mosquito reproduction. PLoS Genet. 11, e1005450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashok M., Turner C., Wilson T. G., Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc. Natl. Acad. Sci. U.S.A. 95, 2761–2766 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zou Z., et al., Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, E2173–E2181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles J. P., et al., Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc. Natl. Acad. Sci. U.S.A. 108, 21128–21133 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M., Mead E. A., Zhu J., Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc. Natl. Acad. Sci. U.S.A. 108, 638–643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z., Xu J., Sheng Z., Sui Y., Palli S. R., Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J. Biol. Chem. 286, 8437–8447 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha T. T., et al., Hairy and Groucho mediate the action of juvenile hormone receptor Methoprene-tolerant in gene repression. Proc. Natl. Acad. Sci. U.S.A. 113, E735–E743 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saha T. T., et al., Synergistic action of the transcription factors Krüppel homolog 1 and Hairy in juvenile hormone/Methoprene-tolerant-mediated gene-repression in the mosquito Aedes aegypti. PLoS Genet. 15, e1008443 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pecasse F., Beck Y., Ruiz C., Richards G., Krüppel-homolog, a stage-specific modulator of the prepupal ecdysone response, is essential for Drosophila metamorphosis. Dev. Biol. 221, 53–67 (2000). [DOI] [PubMed] [Google Scholar]
- 18.Ojani R., Fu X., Ahmed T., Liu P., Zhu J., Krüppel homologue 1 acts as a repressor and an activator in the transcriptional response to juvenile hormone in adult mosquitoes. Insect Mol. Biol. 27, 268–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayukawa T., et al., Krüppel Homolog 1 inhibits insect metamorphosis via direct transcriptional repression of Broad-Complex, a pupal specifier gene. J. Biol. Chem. 291, 1751–1762 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King-Jones K., Thummel C. S., Nuclear receptors–a perspective from Drosophila. Nat. Rev. Genet. 6, 311–323 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Devarakonda S., Harp J. M., Kim Y., Ozyhar A., Rastinejad F., Structure of the heterodimeric ecdysone receptor DNA-binding complex. EMBO J. 22, 5827–5840 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher J. C., D’Avino P. P., Thummel C. S., A steroid-triggered switch in E74 transcription factor isoforms regulates the timing of secondary-response gene expression. Proc. Natl. Acad. Sci. U.S.A. 94, 4582–4586 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun G., Zhu J., Li C., Tu Z., Raikhel A. S., Two isoforms of the early E74 gene, an Ets transcription factor homologue, are implicated in the ecdysteroid hierarchy governing vitellogenesis of the mosquito, Aedes aegypti. Mol. Cell. Endocrinol. 190, 147–157 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Burtis K. C., Thummel C. S., Jones C. W., Karim F. D., Hogness D. S., The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell 61, 85–99 (1990). [DOI] [PubMed] [Google Scholar]
- 25.Sharma A., Nuss A. B., Gulia-Nuss M., Insulin-like peptide signaling in mosquitoes: The road behind and the road ahead. Front. Endocrinol. (Lausanne) 10, 166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nässel D. R., Vanden Broeck J., Insulin/IGF signaling in Drosophila and other insects: Factors that regulate production, release and post-release action of the insulin-like peptides. Cell. Mol. Life Sci. 73, 271–290 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riehle M. A., Fan Y., Cao C., Brown M. R., Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: Expression, cellular localization, and phylogeny. Peptides 27, 2547–2560 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Ling L., Kokoza V. A., Zhang C., Aksoy E., Raikhel A. S., MicroRNA-277 targets insulin-like peptides 7 and 8 to control lipid metabolism and reproduction in Aedes aegypti mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 114, E8017–E8024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ling L., Raikhel A. S., Serotonin signaling regulates insulin-like peptides for growth, reproduction, and metabolism in the disease vector Aedes aegypti. Proc. Natl. Acad. Sci. U.S.A. 115, E9822–E9831 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou Y., et al., Temporal coordination of carbohydrate metabolism during mosquito reproduction. PLoS Genet. 11, e1005309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X., et al., Hormone and receptor interplay in the regulation of mosquito lipid metabolism. Proc. Natl. Acad. Sci. U.S.A. 114, E2709–E2718 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanhandel E., Metabolism of nutrients in the adult mosquito. Mosq. News 44, 573–579 (1984). [Google Scholar]
- 33.Arrese E. L., Soulages J. L., Insect fat body: Energy, metabolism, and regulation. Annu. Rev. Entomol. 55, 207–225 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiBello P. R., Withers D. A., Bayer C. A., Fristrom J. W., Guild G. M., The Drosophila Broad-Complex encodes a family of related proteins containing zinc fingers. Genetics 129, 385–397 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz J., Mane-Padros D., Zou Z., Raikhel A. S., Distinct roles of isoforms of the heme-liganded nuclear receptor E75, an insect ortholog of the vertebrate Rev-erb, in mosquito reproduction. Mol. Cell. Endocrinol. 349, 262–271 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hillyer J. F., Pass G., The insect circulatory system: Structure, function, and evolution. Annu. Rev. Entomol. 65, 121–143 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Raikhel A. S., “Vitellogenesis of disease vectors, from Physiology to genes” in Biology of Disease Vectors, Marquardt W. C., Ed. (Elsevier Academic Press, 2005), pp. 329–345. [Google Scholar]
- 38.Accili D., Arden K. C., FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 117, 421–426 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Okamoto N., et al., An ecdysteroid-inducible insulin-like growth factor-like peptide regulates adult development of the silkmoth Bombyx mori. FEBS J. 276, 1221–1232 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Okamoto N., et al., A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17, 885–891 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slaidina M., Delanoue R., Gronke S., Partridge L., Léopold P., A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev. Cell 17, 874–884 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheng Z., Xu J., Bai H., Zhu F., Palli S. R., Juvenile hormone regulates vitellogenin gene expression through insulin-like peptide signaling pathway in the red flour beetle, Tribolium castaneum. J. Biol. Chem. 286, 41924–41936 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto R., Bai H., Dolezal A. G., Amdam G., Tatar M., Juvenile hormone regulation of Drosophila aging. BMC Biol. 11, 85 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizoguchi A., Okamoto N., Insulin-like and IGF-like peptides in the silkmoth Bombyx mori: Discovery, structure, secretion, and function. Front. Physiol. 4, 217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okamoto N., Yamanaka N., Nutrition-dependent control of insect development by insulin-like peptides. Curr. Opin. Insect Sci. 11, 21–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barthel A., Schmoll D., Unterman T. G., FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 16, 183–189 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Lee S., Dong H. H., FoxO integration of insulin signaling with glucose and lipid metabolism. J. Endocrinol. 233, R67–R79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colombani J., et al., Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310, 667–670 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Kamoshida Y., et al., Ecdysone receptor (EcR) suppresses lipid accumulation in the Drosophila fat body via transcription control. Biochem. Biophys. Res. Commun. 421, 203–207 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Francis V. A., Zorzano A., Teleman A. A., dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr. Biol. 20, 1799–1808 (2010). [DOI] [PubMed] [Google Scholar]
- 51.Mattila J., Hietakangas V., Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 207, 1231–1253 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vihervaara T., Puig O., dFOXO regulates transcription of a Drosophila acid lipase. J. Mol. Biol. 376, 1215–1223 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.