Abstract
Epigenetic regulation is critical for proper bone development. Evidence from a large body of published literature informs us that microRNAs (miRNAs) are important epigenetic factors that control many aspects of bone development, homeostasis, and repair processes. These small non-coding RNAs function at the post-transcriptional level to suppress expression of specific target genes. Many target genes may be affected by one miRNA resulting in alteration in cellular pathways and networks. Therefore, changes in levels or activity of a specific miRNA (e.g. via genetic mutations, disease scenarios, or by over-expression or inhibition strategies in vitro or in vivo) can lead to substantial changes in cell processes including proliferation, metabolism, apoptosis and differentiation. In this review, Section 1 briefly covers general background information on processes that control bone development as well as the biogenesis and function of miRNAs. In Section 2, we discuss the importance of miRNAs in skeletal development based on findings from in vivo mouse models and human clinical reports. Section 3 focuses on describing more recent data from the last three years related to miRNA regulation of osteoblast differentiation in vitro. Some of these studies also involve utilization of an in vivo rodent model to study the effects of miRNA modulation in scenarios of osteoporosis, bone repair or ectopic bone formation. In Section 4, we provide some recent information from studies analyzing the potential of miRNA-mediated crosstalk in bone and how exosomes containing miRNAs from one bone cell may affect the differentiation or function of another bone cell type. We then conclude by summarizing where the field currently stands with respect to miRNA-mediated regulation of osteogenesis and how information gained from developmental processes can be instructive in identifying potential therapeutic miRNA targets for the treatment of certain bone conditions.
Keywords: microRNA, miRNA, Osteoblast, Osteoblast differentiation, Osteogenesis, Bone development, Skeletal development, Epigenetics
1. Introduction: bone development and microRNAs
Bone development begins by the establishment of mesenchymal stem cell condensations that prefigure the shape, size and location of mature bone elements [1,2]. In addition to these skeletal patterning events, mesenchymal cells within these condensations receive signals to differentiate toward either cartilage-forming chondrocytes or bone-forming osteoblasts. The end result of a complex, tightly-controlled progenitor cell differentiation program is the generation of bone tissue consisting of a unique mineralized extracellular matrix (ECM).
Bone formation that transitions via a cartilaginous template is referred to as endochondral ossification and occurs during limb development [3-5]. Cells within this cartilage template terminally differentiate to form large hypertrophic chondrocytes that regulate mineralization of the surrounding ECM and induce vessel invasion. These events lead to the formation of a primary ossification center, and eventually cancellous bone following replacement of cartilage tissue by bone ECM components. Until recently, it was generally accepted that endochondral bone-forming osteoblasts are derived from progenitor cells lining or within blood vessels as well as from progenitor cells of the adjacent perichondrium [6]. However, it has now been established in recent years that hypertrophic chondrocytes, or a subset of progenitor cells within hypertrophic cartilage, are a significant source of osteoblasts during endochondral ossification [7-10]. Coupling of chondrogenesis and osteogenesis is also apparent in the formation of cortical bone of the limbs. Mature cortical bone is derived from the bone collar region in the perichondrium of developing limbs. While this process generally involves differentiation of progenitor cells directly to osteoblasts, hypertrophic chondrocytes have been proposed to play an important role in regulating “perichondrial osteogenesis” [3,4,6,11]. In addition to endochondral bone formation, other bones in the body are generated without the requirement of a cartilage template and involve cells within mesenchymal condensations differentiating directly to osteoblasts. This process, called intramembranous ossification, occurs in some bones of the cranium, and parts of the mandible and clavicle [12].
Toward the end phases of endochondral or intramembranous bone development, some osteoblasts will give rise to osteocytes that are found embedded deep within the mineralized bone ECM. Osteocytes can communicate with adjacent cells (including surrounding osteocytes, osteoblasts, osteoclasts, endothelial cells) via cytoplasmic extensions that occupy tiny canals called canaliculi [13]. These cells play critical mechano-sensing roles to control bone formation and homeostasis. Specifically, there is evidence that osteocytes can regulate the differentiation and function of bone-forming osteoblasts as well as the bone-resorbing osteoclasts [14]. In addition, crosstalk between osteoblasts and osteoclasts also occurs thereby increasing the complexity controlling bone development and turnover [15].
With respect to molecular regulation of osteoblast differentiation during endochondral or intramembranous bone formation, many important players have been identified. These include key transcription factors that are critical for chondrocyte or osteoblast formation (i.e. SOX9, RUNX2, respectively), homeodomain proteins that control various stages of osteoblast differentiation [16-19], growth factors (including FGFs, IGFs, VEGF, BMPs and other TGF-β superfamily members) as well as other signaling pathways (Wnt/β-catenin, Hedgehog, PTHrP, etc) [3-6]. It is also apparent that bone formation is regulated by epigenetic factors that can function at the level of transcription or translation to alter gene or protein expression. Examples of epigenetic regulators include histone modifying enzymes (HDACs, HACs), enzymes that control DNA methylation (DNMTs, TETs), long non-coding RNAs and microRNAs (miRNAs) [20-22].
Since the first discovery of a miRNA in Caenorhabditis elegans over 25 years ago [23], many more have now been identified in cells of humans, rodents, flies, viruses, plants and other species. In the current miRBase website (http://www.mirbase.org), 1917 mature miRNAs have been identified in humans and 1234 in mice. Mature non-coding miRNAs are commonly 19–24 nucleotides (nt) in length and are derived from larger precursor RNAs. Genes encoding miRNAs (predominantly located in intergenic regions or within introns of protein-coding genes) are first transcribed as large primary precursors (pri-miRNAs). In some cases, miRNA-encoding genes may be clustered (i.e. adjacently located within 10 Kb of each other as per miRBase definition) and transcribed in a polycistronic manner. Primary miRNA transcripts are processed in the nucleus by a Drosha-containing complex and the resulting precursor miRNAs (pre-miRNA) are transported to the cytoplasm and processed further by a Dicer-containing complex to form a short, mature miRNA duplex containing a 5p and 3p strand [24]. Commonly, one of these strands is functional whereby it will bind via its seed sequence (positions 2–8 of the mature miRNA strand) to a complementary region within the 3′UTR of a target mRNA. This interaction occurs within the RNA-induced silencing complex (RISC), the end result being either degradation of the target mRNA or inhibition of mRNA translation [25,26]. Fig. 1 depicts the stages of miRNA transcription, processing, and interaction with a target mRNA. Compared to short interfering RNAs (siR-NAs) that are generated exogenously, the level of miRNA-induced target suppression is quite modest. This is because the entire sequence of siR-NAs bind with high specificity (100% complementarity) to one mRNA target resulting in robust suppression via mRNA cleavage, while miRNAs interact with target mRNAs via imperfect pairing, with the exception of the seed sequence interaction [27]. However, unlike siRNAs, miRNAs have the ability to target tens to even hundreds of mRNAs within a given cell type [28], thereby resulting in modulation of many cellular pathways and networks. Complexity is enhanced by the fact that multiple miRNAs may compete to bind to a specific target mRNA.
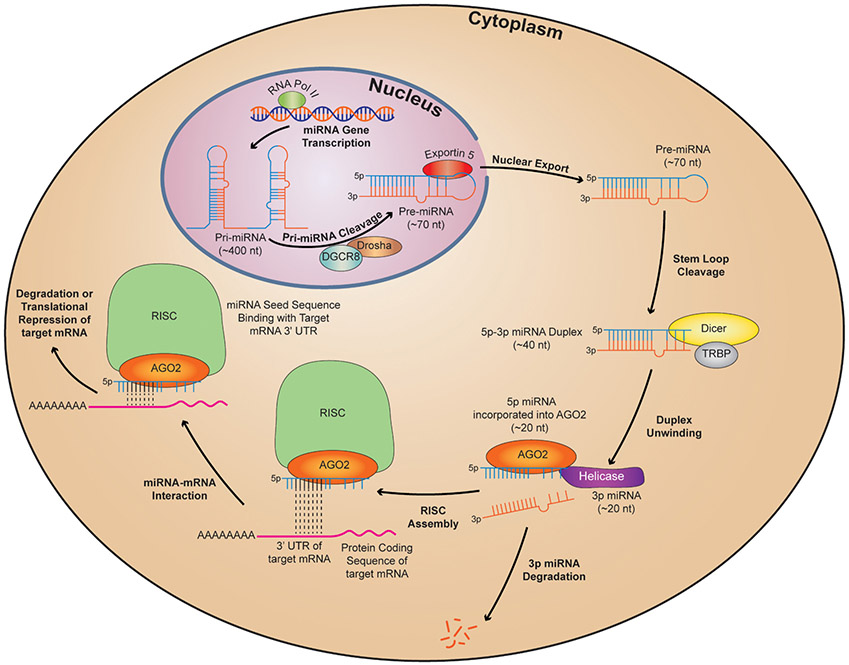
Fig. 1.
Biogenesis, processing and function of microRNAs in the cell. In the nucleus, RNA polymerase II (RNA pol II)-mediated transcription results in generation of primary miRNA transcripts which are processed by a complex containing Drosha and DiGeorge Critical Region 8 (DGCR8) to form stem-loop precursor miRNAs (pre-miRNA). Pre-miRNAs are exported out of the nucleus by Exportin 5 and, in the cytoplasm, are processed further by a complex containing Dicer and TAR RNA Binding Protein (TRBP). Following unwinding of mature miRNA duplexes, one functional strand (here shown as the 5p strand) enters the RNA-induced Silencing Complex (RISC) where it binds to a specific region of the 3′UTR of its target mRNA. Complementary binding via the miRNA seed sequence is shown with black dotted lines. The outcome of miRNA-mediated binding is either mRNA degradation or suppression of mRNA translation.
Although miRNAs account for only 1–5% of the human genome [29], up to 60% of protein-coding genes may be modulated by miRNAs [30] thereby rendering these non-coding RNAs as major epigenetic regulators in the cell. While the majority of miRNAs carry out their function in the cytoplasm, many miRNAs have also been localized to other organelles including the nucleoli, processing bodies and mitochondria [31]. It has also been demonstrated that miRNAs can exist inside extracellular vesicles such as exosomes and that cells can communicate with each other via exosomal delivery of miRNAs [32,33]. Overall, a massive body of published research indicates that miRNAs play important roles in development, homeostasis, turnover, disease and repair of many different tissue types. Specifically, miRNAs have been shown to regulate a wide range of cellular processes including proliferation, cytoskeleton formation, apoptosis, growth factor signaling, metabolism, cell differentiation, and many others.
In this review, we will discuss some in vivo findings that highlight the importance of miRNAs in regulating skeletal development with an emphasis on osteogenesis. In Section 2, some information describing the importance of miRNAs in skeletal development via in vivo mouse models and human clinical reports is similar to that included in our previously published review on miRNAs in orthopaedic research [34]. We believe that such details are also important in the context of this review as well. However, new information is described in Section 2 that was not included in our previous review including details on Prx1-Cre and CD11b-Cre deletion of Dicer in mice, the effects of deleting miR-181 and miR-182 in vivo, a recent report describing a new human neomorphic mutation in miR-140 [35], and a newly published manuscript describing the smallest human deletion mutation in 1q24 containing a microRNA cluster that is associated with skeletal phenotypes [36]. Given some recently published reviews on miRNAs regulating bone formation [34,37-39] we have focused Section 3 of this review on findings published predominantly within the last three years with respect to miRNA regulation of osteoblast differentiation. Preference was given to discussing studies where a miRNA target was validated and/or a specific pathway or cellular process was identified to be regulated by the miRNA of interest. While bone-resorbing osteoclasts are also important in the regulation of bone development, information on how miRNAs affect these cells will be covered by another review in this Special Edition. However, this review will include details of some studies showing that miRNAs in exosomes derived from either osteoclasts or osteocytes can regulate osteoblast differentiation/function, thus highlighting the complex miRNA-mediated crosstalk that likely occurs between different cell types in bone.
2. The importance of miRNAs in skeletal development: lessons from in vivo findings
a). Modulation of miRNA processing via Cre-driver lines in skeletal cells
Conditional transgenic mice devoid of proteins involved in miRNA processing have been generated. While the function of these proteins is not completely restricted to regulating miRNAs, findings from these mice (which contain substantially lower levels of functional miRNAs in specific cell types) suggest an important role for these non-coding RNAs in controlling cellular processes involved in proper skeletal development.
Deletion of the pre-miRNA processing enzyme, Dicer, in osteochon-droprogenitor cells via Prx-1-Cre mice resulted in formation of smaller limbs due, in part, to increased cell death during early limb bud development. Interestingly, there did not appear to be defects in basic patterning or in overall cartilage and bone differentiation within these smaller limbs [40]. However, when Dicer was eliminated specifically in chondrocytes by crossing Dicer floxed mice with Col2a1-Cre driver mice, growth plate analysis revealed modest defects in chondrocyte differentiation which subsequently resulted in reduced length and width of the developing long bones [41]. Given the cross-talk between hypertrophic cartilage and bone collar/cortical bone formation, it would have been interesting to determine if there were any defects in matrix mineralization and bone production in the limbs of these mice. Thorough analysis of post-natal bone formation was not possible given that these mice normally die around the time of weaning. Following Dicer deletion in osteoprogenitor cells via Col1a1-Cre mice, skeletal examination of E14.5 embryos revealed compromised ECM mineralization in cartilage and osseous tissues as well as an overall significant reduction in bone tissue [42]. Whether this phenotype represents delayed rather than inhibition of ossification was not concluded from these studies given that embryo survival was compromised after E14.5, which may have been partly due to the lack of marrow cavity formation which support hematopoiesis. This study also showed that when Dicer was deleted specifically in mature osteocalcin-producing osteoblasts (Ocn-Cre), a post-natal increase in long bone and vertebral bone mass (but not cranial bone mass) was found [42]. These findings suggest an overall positive role for Dicer-generated miRNAs in regulating osteoblast differentiation during embryonic bone development, particularly bones that are generated by endochondral ossification. On the other hand, the Ocn-Cre findings suggest that miRNAs may generally function to suppress bone formation during post-natal bone development and turnover. However, conditional knock-out of Dicer via Runx2-Cre revealed growth retardation, low bone mass and impaired bone formation rate in post-natal mice [43]. A recent study also revealed decreased cortical bone mass in young and adult mice following inducible post-natal ablation of Dicer in osterix (Sp7)-producing cells (Sp7-CreERT2; Dicerflox/flox) [44]. It was also shown that Dgcr8 (DiGeorge syndrome critical region 8) deletion in osteopro-genitors (Col1a1-Cre) resulted in enhanced bone formation due, in part, to decreased miR-22 and enhanced osteocalcin transcripts [45]. Dgcr8 is a critical component of the pri-miRNA processing complex via its interaction with Drosha. Taken together, it is apparent that the timing and the choice of Cre-driver model to induce deletion of miRNA processing proteins are critical factors in determining the outcome of miRNA deficiency on bone formation.
Dicer has also been deleted predominantly (but not specifically) in murine osteoclasts via CD11b-Cre [46] or Cathepsin K-Cre [47] approaches. In both cases, increased post-natal bone mass was found due to impaired osteoclastogenesis. Whether bone mass or mineralization was affected at the embryonic level was not investigated in these studies. The latter study by Mizoguchi et al [47] also reported no changes in post-natal bone mass when Dicer was removed by Col1a1-Cre mediated deletion. This result is intriguing given that Gaur et al reported embryonic lethality when this enzyme was knocked-out using the same 2.3 kb collagen type I promoter Cre-mice [42]. Similar defects in osteoclast differentiation and function were noted when Dgcr8 was deleted in osteoclasts using Cathepsin-Cre mice [48]. These mice also displayed growth retardation as well as increased bone mass.
b). miRNAs regulating human skeletal development
Data from clinical reports have demonstrated the importance of miRNAs in regulating human skeletal development. One study found that a mutation within the 3′UTR of HDAC6 disrupts the miR-433 binding site resulting in a dominant X-linked chondrodysplasia [49]. Deletion or duplication of the miR-17~92 cluster was reported to cause short stature, digit abnormalities, microcephaly and abnormal facial features [50,51]. Additional findings show that deletion of miR-17~92 specifically in murine Col1α2-expressing cells resulted in smaller bones as well as a reduction in periosteal bone formation following mechanical loading [52].
A recent report identified a mutation within the gene encoding miR-140, one of the most studied miRNAs in cartilage. The resulting production of neomorphic mutant miR-140-5p caused a number of skeletal defects in family members including short stature, brachydactyly, premature degeneration of intervertebral discs and delayed epiphyseal ossification of the hip and knee [35]. Previous findings from miR-140 knock-out mice [53,54] revealed shortened limbs as a result of defects in endochondral ossification due to accelerated hypertrophic chondrocyte differentiation.
Deletions within specific regions of chromosome 1q24 appear to cause a range of skeletal phenotypes and cognitive disabilities [55-58]. Skeletal issues include short stature, microcephaly, brachydactyly and, in some cases, a marked delay in bone age. Interestingly, the various deletions reported contain the clustered miRNAs, miR-199a and miR-214, which are located within a long non-coding RNA (lncRNA) transcript called DNM3OS. Further evidence that heterozygous deletion of these non-coding RNAs may be responsible for the skeletal phenotype comes from a newly published study reporting the smallest 1q24 microdeletion to date (94 Kb) in the genome of two patients [36]. In addition, transgenic mice devoid of Dnm3os, and hence the miR-199a~214 cluster, presented with similar skeletal phenotypes to that reported in human patients [59]. While one cannot rule out the possibility that Dnm3os may function as a lncRNA independent of its role in serving as a miRNA precursor, it is likely that the miRNAs themselves play a role in regulating the skeletal phenotype. In fact, a number of published studies have reported a functional role for miR-199a or miR-214 in regulating osteogenesis in vitro or in vivo.
c). miRNAs with functional roles in regulating bone formation in vivo
Altered expression of other miRNAs in mice has revealed functional roles in regulating bone formation in vivo. For example, expression of members of the miR-34 family (miRs-34a, b, c) were found to increase during osteoblast differentiation of murine calvarial cells [60]. This study also showed that when miR-34b and miR-34c were deleted in Col1a1-producing cells in mice, increased bone mass was observed during embryonic development and increased bone mass accrual was observed post-natally. The opposite was found when miR-34c was over-expressed in murine Col1a1-producing cells. One of the mechanisms proposed for the negative effects of miR-34 on bone development was via targeting and suppression of SATB2 (special AT-rich sequence-binding protein 2). In agreement with this work, miR-34a was found to suppress osteoblast differentiation of human MSCs in vitro (in part via targeting Jagged 1) and reduce formation of bone following subcutaneous transfer of hMSC-loaded ceramic beads in SCID mice [61]. Interestingly, when miR-34a was over-expressed predominantly in osteoclasts, osteoclastogenesis and bone resorption was suppressed and provided some protection against ovariectomy (OVX)-induced bone loss [62]. These findings demonstrate the functional divergence between miR-34 family members but also highlight the fact that modulation of miRNAs in different cell types within the same tissue can result in different biological outcomes.
Expression of miR-206, regarded as more of a muscle-specific miRNA, was found to be one of a number of miRNAs downregulated during BMP-2-induced osteogenic differentiation of C2C12 cells [63]. This study also showed that conditional over-expression of miR-206 in Col1a1-positive cells resulted in low bone mass in mice due, in part, to targeting and suppression of connexin 43 (Cx43). It has also been reported that when members of the miR-181 family were globally deleted in mice, those that survived were smaller in size [64,65]. This suggests a potential role for these miRNA paralogs in controlling growth plate and bone development. As will be discussed more in this review, miR-181a/b has been shown to enhance osteogenesis in vitro.
A recent report showed that miR-182 inhibits osteoclastogenesis in vitro and that myeloid-specific (LysM-Cre) deletion of miR-182 in mice results in increased post-natal trabecular bone mass [66]. These mice were apparently protected from bone loss associated with OVX and inflammatory arthritis. Mechanistically, miR-182 was found to target protein kinase double stranded RNA dependent (PKR) and regulate IFN-β signaling. As expected, this study also showed that in vivo over-expression of miR-182 in osteoclasts resulted in low trabecular bone mass. Cortical bone mass was not affected by miR-182 modulation and it is not clear if trabecular bone mass was affected at earlier embryonic stages of bone development. In agreement with these negative effects on bone mass, previous studies have shown that miR-182 (in part via targeting FoxO1) suppresses osteoblast differentiation in vitro and impairs bone formation in vivo in zebrafish [67]. Another study showed that suppression of miR-182 enhanced osteoblast differentiation in vitro and apparently induced the effects of alendronate in combating osteoporosis in rats via regulating Rap1/MAPK signaling [68].
In vitro studies found that miR-21 appeared to enhance both osteogenesis and osteoclastogenesis [69,70]. Interestingly, global knock-out of miR-21 in mice [71] did not appear to affect bone development, which may be due to functional redundancy by other miRNAs expressed during development. However, miR-21 deficiency promoted trabecular bone mass with age and also prevented OVX-induced bone loss during aging due, in part, to suppressed osteoclast function. In this case, as has been reported for many transgenic mice, a post-natal phenotype exists following aging or a disease/injury challenge, even though a developmental/baseline phenotype is absent.
3. Recent research on miRNAs regulating osteoblast differentiation
Given the large body of published studies and some recent reviews on osteogenic regulation by miRNAs, this section will cover research findings reported within the last three years (2017 to present). PubMed search keywords included “miRNA or microRNA and osteoblastogenesis or osteogenesis”. Preference was given to studies that determined a miRNA target gene and/or cellular pathway modulated by the miRNA. Fig. 2 lists the osteo-enhancing and osteo-suppressing miRNAs from this PubMed search as well some functional miRNAs discussed in Section 2 of this review. Tables 1 and 2 provide more information on the findings from our literature search covering the last three years. Table 1 lists miRNAs reported to enhance osteoblast differentiation in vitro when over-expressed while Table 2 lists miRNAs that suppress in vitro osteogenesis. Additional information provided in the Tables include the cell types used to study osteogenesis in vitro and, where applicable, the endogenous expression patterns of the miRNA of interest during in vitro osteogenesis, as well as the in vivo animal model utilized. The majority of these studies involved altering miRNA activity using either mimics or antagomirs whereas those that utilized lentiviral approaches are highlighted. We believe it is important to emphasize the mode of miRNA modulation given a report suggesting that transient transfection of miRNA mimics often results in supraphysiological levels of over-expression that may lead to non-specific changes in gene expression [72].
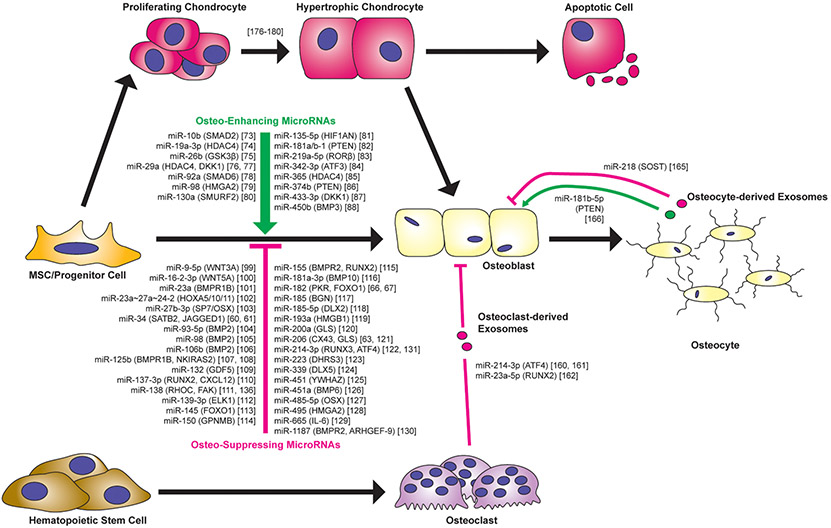
Fig. 2.
MicroRNAs regulating osteoblast differentiation and function. Listed are the osteo-enhancing and osteo-suppressing miRNAs included in Section 3 in addition to other miRNAs discussed in Sections 2 and 4 of this review. Confirmed miRNA targets are shown in parenthesis. Note that while long bone formation involves endochondral ossification, via formation of hypertrophic chondrocytes, all studies identifying functional miRNAs utilized in vitro osteogenesis assays that mimic intramembranous ossification whereby stem/progenitor cells differentiate directly to osteoblasts. Also shown are miRNAs present in exosomes of osteoclasts or osteocytes that may function in regulating osteoblast differentiation or function. See Tables 1 and 2 for a list of the various stem/progenitor cell types used to determine the function of the majority of miRNAs listed in this figure. Note: not included in this figure is the reference to a report suggesting that let-7a-5p, present in osteoclast-derived exosomes, may enhance hypertrophic chondrocyte differentiation [164]. Note: while osteoblasts certainly secrete exosomes, we did not identify published studies reporting exosome-derived miRNAs from osteoblasts directly regulating other specific bone-related cell types. Therefore, we did not include osteoblast-derived exosomes in this figure given that we cannot support this depiction with a specific published study.
Table 1.
MicroRNAs that enhance osteoblast differentiation in vitro.
| Osteo-enhancing miRNA |
Endogenous expression during OG |
miRNA target |
Progenitor cell type | Animal model | Reference |
|---|---|---|---|---|---|
| miR-10b |  |
SMAD2 | Human ASC | Heterotopic ossification | Li et al. [73] |
| miR-19a-3p |  |
HDAC4 | Human BMSC | NA | Chen et al. [74] |
| miR-26b | 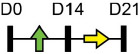 |
GSK3β | Rat BMSC | NA | Hu et al. [75]* |
| miR-29a | 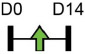 |
HDAC4 | Human BMSC | NA | Tan et al. [76] |
| miR-29a | NA | DKK1 | Mouse primary Cal Pre-OB | Ovariectomy-induced osteoporosis | Lee et al. [77] |
| miR-92a | NA | SMAD6 | Mouse BMSC | NA | Yan et al. [78] |
| miR-98 | 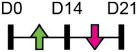 |
HMGA2 | Human BMSC | NA | Gao et al. [79] |
| miR-130a | 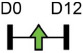 |
SMURF2 | Mouse BMSC | Age-related osteoporosis | Lin et al. [80] |
| miR-135-5p | 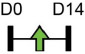 |
HIF1AN | MC3T3-E1 | NA | Yin et al. [81] |
| miR-181a/b-1 | 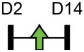 |
PTEN | Human DDC | NA | Zheng et al. [82]* |
| miR-219a-5p | 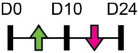 |
RORβ | Mouse Cal OB | NA | Aquino-Martinez et al. [83] |
| miR-342-3p | 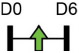 |
ATF3 | Human BMSC, MC3T3-E1 | NA | Han et al. [84] |
| miR-365 | NA | HDAC4 | MC3T3-E1 | NA | Xu et al. [85] |
| miR-374b | NA | PTEN | Mouse BMSC | Tibial plateau fracture | Ge et al. [86] |
| miR-433-3p | NA | DKK1 | Human FOB 1.19, Rat ROS17/2.8 | Ovariectomy-induced osteoporosis | Tang et al. [87]* |
| miR-450b | 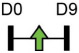 |
BMP3 | Human ASC | Heterotopic ossification | Fan et al. [88]* |
Legend: this list was generated from a PubMed literature search (2017–2020) to determine miRNAs that play a functional role in regulating osteoblast differentiation. This list contains miRNAs reported to enhance osteogenesis following over-expression in vitro. The second column shows endogenous expression patterns of the miRNA during in vitro osteogenesis assays: pink arrows represent reduced expression, green arrows denote increased expression and yellow arrows translate to unchanged expression at specific time points of osteogenic differentiation. MiRNA target genes and progenitor cell types used to study in vitro osteogenesis are also shown. Wherever applicable, the in vivo rodent model used to further test the effects of miRNA modulation on bone formation is also shown. Those studies marked with an asterisk (*) used lentiviral technology to modulate miRNA expression. All other studies utilized miRNA mimics or antagomirs. Abbreviations: ASC = adipose-derived mesenchymal stem/stromal cells; BMSC = bone marrow-derived mesenchymal stem/stromal cells; Cal OB = calvarial-derived osteoblasts; Cal pre-OB = calvarial-derived pre-osteoblasts; DDC = dedifferentiated chondrocytes (or cartilage progenitor cells); FOB = fetal osteoblasts; NA = not analyzed; OG = osteogenesis; ROS = rat osteosarcoma cells.
Table 2.
MicroRNAs that suppress osteoblast differentiation in vitro.
| Osteo-suppressing miRNA |
Endogenous expression during OG | miRNA target | Progenitor cell type | Animal model | Reference |
|---|---|---|---|---|---|
| miR-9-5p | 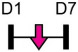 |
WNT3A | Rat BMSC | NA | Zhang et al. [99] |
| miR-16-2-3p | 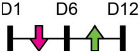 |
WNT5A | Human BMSC | NA | Duan et al. [100] |
| miR-23a | NA | BMPR1B | Human PDLSC | NA | Zhang et al. [101]* |
| miR-23a–27a–24-2 | 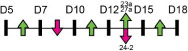 |
HOXA5/10/11 | MC3T3-E1 | Age-related osteoporosis | Godfrey et al. [102]* |
| miR-27b-3p | 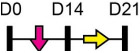 |
SP7/OSX | Human MSMSC | Heterotopic ossification | Peng et al. [103] |
| miR-93-5p | 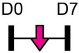 |
BMP2 | Human BMSC | NA | Zhang et al. [104] |
| miR-98 | NA | BMP2 | Human BMSC | NA | Zhang et al. [105] |
| miR-106b | 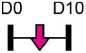 |
BMP2 | Human PMSC | Glucocorticoid-induced osteoporosis | Liu et al. [106] |
| miR-125b | 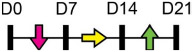 |
BMPR1B | Human BMSC | Bone defect | Wang et al. [107]* |
| miR-125b | 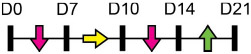 |
NKIRAS2 | Human PDLSC | NA | Xue et al. [108]* |
| miR-132 | 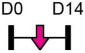 |
GDF5 | Human PDLSC | NA | Xu et al. [109] |
| miR-137-3p | NA | RUNX2, CXCL12 | Rat BMSC | Steroid-induced osteonecrosis of the femoral head | Kong et al. [110]* |
| miR-138 | 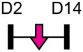 |
RHOC | Human DDC | NA | Zheng et al. [111]* |
| miR-139-3p | NA | ELK1 | MC3T3-E1 | NA | Wang et al. [112] |
| miR-145 | 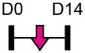 |
FOXO1 | Human ASC | NA | Hao et al. [113] |
| miR-150 | 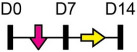 |
GPNMB | Mouse BMSC | Age-related osteoporosis | Moussa et al. [114] |
| miR-155 | 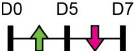 |
BMPR2, RUNX2 | C2C12, MEF | Heterotopic ossification | Liu et al. [115] |
| miR-181a-3p | 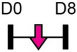 |
BMP10 | Human BMSC | NA | Tao et al. [116] |
| miR-185 | NA | BGN | MC3T3-E1, mouse BMSC | Ovariectomy-induced osteoporosis | Cui et al. [117] |
| miR-185-5p | 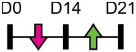 |
DLX2 | MC3T3-E1 | NA | Chang et al. [118] |
| miR-193a | 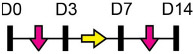 |
HMGB1 | Human BMSC | NA | Wang et al. [119] |
| miR-200a |  |
GLS | Human BMSC | NA | Lv et al. [120] |
| miR-206 |  |
GLS | Human BMSC | NA | Chen et al. [121] |
| miR-214-3p | NA | RUNX3 | Human MSMSC | Heterotopic ossification | Peng et al. [122] |
| miR-223 |  |
DHRS3 | Human BMSC | NA | Zhang et al. [123] |
| miR-339 | 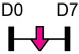 |
DLX5 | Human BMSC | NA | Zhou et al. [124] |
| miR-451 | 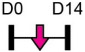 |
YWHAZ | Cal Pre-OB | Ovariectomy-induced osteoporosis | Pan et al. [125] |
| miR-451a | NA | BMP6 | Mouse BMSC, MC3T3-E1 | Ovariectomy-induced osteoporosis | Lu et al. [126] |
| miR-485-5p | 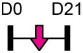 |
SP7/OSX | Rat BMSC | NA | Zhang et al. [127] |
| miR-495 | NA | HMGA2 | Mouse Cal OB | Bone defect | Tian et al. [128] |
| miR-665 | 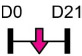 |
IL-6 | Human ASC | NA | Wu et al. [129] |
| miR-1187 |  |
BMPR2, ARHGEF-9 | Mouse Cal OB | Ovariectomy-induced osteoporosis | John et al. [130] |
Legend: this list was generated from a PubMed literature search (2017–2020) to determine miRNAs that play a functional role in regulating osteoblast differentiation. This list contains miRNAs reported to suppress osteogenesis following over-expression in vitro. The second column shows endogenous expression patterns of the miRNA during in vitro osteogenesis assays: pink arrows represent reduced expression, green arrows denote increased expression and yellow arrows translate to no significant changes in expression at specific time points of osteogenic differentiation. MiRNA target genes and progenitor cell types used to study in vitro osteogenesis are also shown. Wherever applicable, the in vivo rodent model used to further test the effects of miRNA modulation on bone formation is also shown. Those studies marked with an asterisk (*) used lentiviral technology to modulate miRNA expression. All other studies utilized miRNA mimics or antagomirs. Abbreviations: ASC = adipose-derived mesenchymal stem/stromal cells; BMSC = bone marrow-derived mesenchymal stem/stromal cells; Cal OB = calvarial-derived osteoblasts; Cal Pre-OB = calvarial-derived pre-osteoblasts; DDC = dedifferentiated chondrocytes (or cartilage progenitor cells); MEF = mouse embryonic fibroblasts; MSMSC = maxillary sinus membrane stem cells; NA = not analyzed; OG = osteogenesis; PDLSC = periodontal mesenchymal stem cells; PMSC = placenta-derived mesenchymal stem cells.
Table 1 shows that, where applicable, the endogenous expression of miRNAs reported to have enhancing activity appears to increase during in vitro osteogenesis, particularly at the early phases of differentiation induction. Additional over-expression of each miRNA listed in Table 1 [73-88] in a range of rodent or human progenitor cell lines enhanced the osteogenic program as shown by increased mineralized matrix formation (commonly detected by Alizarin red staining) and increased expression of osteoblast-related genes when compared to control cultures. A number of these osteo-enhancing miRNAs were found to target and suppress other epigenetic regulators including HDAC4 or HMGA2 [74,76,79,85]. Another common mechanism to enhance osteoblast differentiation is via miRNA-mediated suppression of Wnt inhibitors (i.e. DKK1 or GSK3β) [75,77,87] or via targeting negative regulators of RUNX2 (i.e. SMAD6 or SMURF2) [78,80]. Less commonly reported miRNA-mediated mechanisms to enhance bone formation include regulation of retinoic acid receptor-related orphan receptor beta (Rorβ). This transcription factor, which is a negative regulator of bone, was reported to be a direct target of miR-219-5p, thereby partly explaining the mechanism by which this miRNA enhances osteogenesis [83]. Two reports in Table 1 [82,86] show that miRNA-mediated targeting of PTEN (phosphatase and tensin homolog) results in enhanced osteogenesis. PTEN functions as the primary negative regulator of PI3K/AKT signaling in the cell [89] thereby affecting a number of cellular processes. Also, it was previously reported that mice lacking PTEN have higher bone mass and improved fracture healing [90,91], providing further evidence that mechanisms to suppress PTEN may have positive effects on bone formation. In addition to confirming PTEN protein suppression by over-expression of the miR-181a/b-1 cluster, research from our laboratory also showed that PI3K/AKT signaling was indeed increased during early phases of osteogenic differentiation following miR-181a/b-1 over-expression [82]. In this study, we also carried out RNA-Seq following over-expression of miR-181a/b-1 or a non-silencing control RNA during osteogenesis and found, via pathway analyses, that a number of cell processes related to mitochondrial metabolism were increased. We subsequently showed by Seahorse technology that mitochondrial respiration was elevated during osteogenesis by miR-181a/b-1 and that enhancing PI3K/AKT signaling may be partly responsible for these metabolic changes. Preliminary unpublished data from our laboratory has also shown enhanced endochondral ulnar fracture healing in mice following lentiviral delivery of miR-181a/b-1 to the fracture site. While it is known that glycolysis is a major metabolic process during osteoblast differentiation, a number of other studies clearly show that mitochondrial respiration is also critical for proper bone formation [92-97]. Research in our lab is currently focused on determining other miR-181a/b-1 target genes or pathways regulated by this miRNA cluster that may be responsible for regulating mitochondrial respiration. Interestingly, another in vitro study showed that miR-181a over-expression enhanced osteogenesis via repression of TGF-β signaling [98]. Undoubtedly, miR-181a/b paralogs will likely target a number of genes in the cell that will subsequently modulate multiple cellular pathways to regulate osteoblast differentiation.
Table 2 shows that endogenous expression of all miRNAs reported to have negative effects on osteoblast differentiation decreased during in vitro osteogenesis assays, particularly during the earlier phases of differentiation. The suppressive effect of these miRNAs on osteoblast differentiation was demonstrated following their over-expression by mimics or virus-mediated transduction [99-130]. Common target genes of a number of these miRNAs were found to be known activators of osteoblast differentiation including BMP2 [104-106], other BMPs [116,126], BMP receptors [101,107,115,130], specific WNTs [99,100] or transcription factors RUNX2 [110,115] and Osterix/Sp7 [103,127]. In agreement with the osteo-inhibitory function of miR-214-3p via targeting ATF4 [131], Table 2 lists a more recent study demonstrating that this miRNA can also target and suppress RUNX3 [122]. With respect to modulating cellular metabolism, two independent studies reported miRNA-mediated targeting and suppression of glutaminase (which regulates glutamine metabolism) was partly responsible for decreased osteogenesis [120,121]. This work is in agreement with a recent report showing a positive role for glutaminase in regulating osteoblast differentiation [132].
Research from our laboratory showed that miR-138 inhibited osteogenesis, in part, via targeting and suppression of RHOC [111]. This was the first report implicating a role for this small GTPase in regulating osteoblast differentiation. We also found that a major effect of miR-138 over-expression and RHOC suppression was inhibition of actin polymerization. Another miRNA listed in Table 2, miR-1187, was also shown to suppress osteogenesis by inhibiting actin cytoskeletal rearrangement [130]. Together, these findings agree with previous studies showing that suppression of actin cytoskeleton formation has negative effects on osteoblast differentiation [133-135]. The inhibitory effect of miR-138 on osteogenesis was also shown previously and this study identified focal adhesion kinase (FAK) as a target of miR-138 [136]. Interestingly, previous studies have shown that cells defective in either RHOC or FAK are less invasive/metastatic [137,138]. A recent in vivo study showed that delivery of miR-138 antagomirs could enhance bone formation in a murine model of multiple myeloma [139]. While these findings are encouraging and support the in vitro data on miR-138 in regulating osteogenesis, long-term effects of inhibiting a miRNA with reported tumor suppressive activity should be considered. On the other hand, miR-138 over-expression may be a useful strategy to attempt to inhibit pathological bone formation. Indeed, preliminary, unpublished data from our laboratory showed a reduction in trauma-induced heterotopic bone formation in mice following lentiviral delivery of miR-138. Current studies are focused on investigating these findings further.
Determining the function and mechanism of a specific miRNA in regulating in vitro osteogenesis can be a useful first-step approach to justify pursuing in vivo studies in rodents. Such pre-clinical studies are strengthened by the fact that many miRNAs are conserved between rodent and human. Tables 1 and 2 contain information on studies that utilized models of bone loss, bone repair or ectopic bone formation to determine how increasing or inhibiting miRNA activity can affect bone formation in these scenarios. In addition, two of these studies also reported age-related changes in miRNAs whereby miR-130a levels were lower in BMSCs from older mice [80] while miR-219a-5p expression was decreased in bone samples from old mice or from aged humans when compared to young controls [83]. A number of other recent studies have also reported age-related changes in miRNAs in bone or bone marrow cells as a consequence of age [140-145]. Therefore, identifying miRNAs that are not only functional in regulating osteogenesis, but also appear to be regulated with age, or even in bone disease scenarios, would further improve the discovery of effective miRNA targets to treat low bone mass or enhance bone repair.
4. miRNA-mediated crosstalk in bone
It is now well-established that intercellular communication between different cell types via exosomes, microvesicles or matrix vesicles is important for proper regulation of bone development, turnover and repair [146-153]. Exosomes are extracellular vesicles (EVs) with an average diameter of ~100 nm and contain protein, DNA, RNA and other components depending on the cell type from which they are derived. They originate from the endosomal pathway via the formation of multivesicular bodies (MVBs). Exosome-containing MVBs can fuse with the plasma membrane resulting in exosome release from the cell [154]. The discovery that exosomes containing miRNAs are present in circulation has led to many studies aimed at identifying miRNA biomarkers associated with various bone diseases [155-157]. In addition, there are new research endeavors exploring the possibility of exosome-derived miRNAs in mediating intercellular signaling between bone cells (osteoblasts, osteoclasts, osteocytes) and other cell types involved in bone formation (e.g. BMSCs, periosteal progenitors, hypertrophic chondrocytes).
A recent study reported that expression of miR-31a-5p was higher in BMSC-derived exosomes from aged rats compared to young rats [140]. This miRNA negatively affects osteoblast differentiation but promotes osteoclast differentiation and bone resorption, in part, via targeting SATB2 and RhoA, respectively. Inhibition of miR-31a-5p apparently prevented bone loss and decreased osteoclast activity in vivo. Overall, their findings suggest that miR-31a-5p is a modulator of the bone marrow microenvironment to influence both osteoblast and osteoclast differentiation during aging and that BMSC-derived exosomes may be a significant source of this miRNA. Another study found that bone marrow-derived EVs from aged mice had higher levels of the miR-183 cluster (miR-96~183) compared to EVs isolated from young animals [142]. The negative effects of miR-183 on BMSC osteogenesis was also demonstrated in this work. These findings suggest that enriched miRNAs within BMSC-derived EVs may interact with other BMSCs in the bone marrow microenvironment to reduce their osteogenic potential. In general, it also very likely that the contents of BMSC-derived EVs may also influence the function of surrounding osteoblasts and osteoclasts to affect bone metabolism [158].
Emerging evidence also suggests that contents of exosomes derived from differentiated bone cells can regulate other cells types involved in bone formation/turnover. One study, showed that exosomes derived from a mineralizing osteoblast cell line (MC3T3) can promote osteoblast differentiation of the ST2 cell line [159]. While the transfer of miRNA cargo from exosomes was not examined in this work, exosome treatment was found to significantly alter miRNA profiles in the recipient cells. Other in-depth studies have demonstrated that exosomal miR-214 derived from osteoclasts could be transferred to osteoblasts to inhibit osteoblast activity and bone formation [160,161]. Recent work has also shown that miR-23a-5p from osteoclast-derived exosomes can suppress osteoblast differentiation, in part, by targeting Runx2 [162].
Further evidence of miRNA-mediated osteoblast-osteoclast communication comes from research showing accumulation of miR-125b within osteoblast-derived matrix vesicles in bone [163]. The authors of this work suggested that miR-125b could be released into the bone marrow microenvironment to suppress osteoclastogenesis and bone resorption. A recent report suggests that let-7a-5p from osteoclast-derived exosomes can enhance the expression of hypertrophic genes in the chondrocyte ATDC5 cell line [164]. These findings imply that miRNA-mediated intercellular communication between osteoclasts and chondrocytes may influence terminal hypertrophic chondrocyte differentiation. Another study demonstrated that exosomes derived from a common osteocyte cell line (Ocy454) could be taken up by osteoblastic MC3T3 cells resulting in a marked decrease in osteogenic potential of these cells [165]. This study also showed that myostatin treatment altered miRNA profiles in these osteocyte-derived exosomes and that, in particular, reduced expression of miR-218 was partly responsible for the negative effects on osteogenesis. A novel mechanism controlling muscle-bone communication was postulated from these findings. Further indication of possible miRNA-mediated crosstalk between osteocytes and osteoblasts was described in a recent study showing that miR-181b-5p in osteocyte-derived exosomes enhances osteogenesis of human periodontal ligament stem cells [166]. Interestingly, similar to our research, this miR-181 family member was also shown to target PTEN and enhance PI3K/AKT signaling [82].
However, it should be noted that the majority of these studies demonstrating exosomal miRNA-mediated cell-cell communication have been performed exclusively in vitro using cell lines rather than primary cells. While the concept of miRNA-mediated intercellular communication via EVs is intriguing and certainly possible given the proximity of the different cell types involved in regulating bone development and homeostasis, more research is needed to elucidate the mechanism of exosome transfer and miRNA uptake and to conclusively demonstrate this in vivo. One study suggests that an interaction between ephrinA2 and EphA2 facilitates the recognition of osteoclast exosomes by osteoblasts [161]. Most likely, other processes will be necessary to permit miRNA transfer via exosomes within the bone microenvironment and basic science research is currently ongoing to better understand this phenomenon in other systems [167,168]. In vitro studies using fluorescently-labeled exosomes or miRNAs has indeed shown that transfer to osteoblasts is possible [160,161,169]. Also, it was demonstrated that fluorescently-labeled prostate cancer cell-derived exosomes enriched in miR-141-3p could home to bone in mice to induce osteoblastic bone metastasis due to the pro-osteogenic function of miR-141a-3p [169]. While these studies are certainly rigorous and informative, more sophisticated imaging technologies [170] would be required to fully characterize exosome uptake and transfer of miRNAs to cells in the bone microenvironment in vivo.
5. Summary and perspectives
The importance of miRNA-mediated epigenetic regulation in controlling skeletal development and homeostasis is clearly evident. There is now a vast array of published reports demonstrating how osteoblast differentiation can be modulated in vitro by either over-expressing or inhibiting a specific miRNA. In many of these studies, the functional effect of targeting the miRNA of interest has been further confirmed in vivo utilizing rodent models of osteoporosis, bone fracture repair or heterotopic ossification, for example. It is therefore apparent that understanding how miRNAs regulate bone developmental processes will aid in the design of new therapeutic strategies to treat bone conditions.
These small non-coding RNAs are recognized as attractive therapeutic targets due to their size, known sequence and the fact that they can target multiple genes to subsequently alter cellular pathways and networks. This function is particularly relevant in the context of more complex diseases via the ability to target “interactomes”. In fact, a number of Phase I/II human clinical trials are underway toward testing the effects of miRNA mimics or antagomirs in vivo to treat specific diseases including cancers. While new research endeavors to improve the stability and cellular uptake of mimics/antagomirs are underway, additional efforts are needed to target cells specifically to the bone microenvironment. Indeed, a few studies have reported some success in the design of peptides/nanoparticles to target osteoclasts or osteoblasts in vivo [171-174]. Perhaps also attempts to better understand the biology behind why exosomes derived from certain cancer cells appear to home to bone [169] would also be advantageous toward designing strategies to predominantly target bone cells.
Many miRNAs known to regulate osteoblast differentiation have been discovered via expression profiling during in vitro osteogenesis using microarray or bulk RNA-Seq approaches. Other candidates have been identified from analysis of RNA extracted directly from bone or bone marrow. While useful information has been gained by such approaches, the ability to now determine expression of miRNAs at the single cell level in vitro or in vivo will aid in determining either new miRNA candidates to pursue and/or confirm those already shown to regulate bone cell function to then justify further studies. The introduction of miRNA-mRNA co-sequencing at the single cell level [175] will also aid in the development of regulatory networks that exist in different bone cells. These technologies may be useful in determining how miRNAs and regulatory networks change with age in specific bone cells, for example. Findings from such analyses could aid in determining miRNAs or miRNA-regulated pathways that could be targeted to combat age-related bone loss or to enhance bone repair that is often deficient as a consequence of aging.
However, what should be done with this ever-increasing list of miRNAs that have been reported to have similar functions in either enhancing or suppressing osteogenesis? In this review, we have covered only a fraction of the published literature on miRNAs regulating osteoblast differentiation. Also, additional miRNA candidates that have been shown to regulate terminal hypertrophic chondrocyte differentiation [176-180] may also turn out to be potential targets to enhance bone formation and repair given the requirement of hypertrophic cartilage in these endochondral processes. It may be that a large-scale project would be required to systematically test and directly compare the function of many of these miRNAs in vitro and in vivo to generate a consensus toward determining the most promising candidates to pursue with respect to modulating osteogenesis. From such studies, we may also learn that targeting multiple miRNAs may be better than modulating one in certain scenarios. Also, given that therapeutic strategies will likely result in miRNA delivery to multiple cell types within the bone microenvironment, the function of specific miRNAs regulating osteoblasts should also be determined in osteoclasts and other bone-associated cell types as well. This is particularly important given that miRNA-mediated cross-talk between bone cells may also represent a significant mode of regulation to control bone formation, turnover and repair.
Acknowledgements
NIH grant funding, R01AR075730 and R21AR077203 (to AM).
References
- [1].Karsenty G, The complexities of skeletal biology, Nature 423 (2003) 316–318. [DOI] [PubMed] [Google Scholar]
- [2].Mariani FV, Martin GR, Deciphering skeletal patterning: clues from the limb, Nature 423 (2003) 319–325. [DOI] [PubMed] [Google Scholar]
- [3].Kronenberg HM, Developmental regulation of the growth plate, Nature 423 (2003) 332–336. [DOI] [PubMed] [Google Scholar]
- [4].Langhans MT, Alexander PG, Tuan RS, Skeletal development (Chapter 28), in: Moody SA (Ed.), Principles of Developmental Genetics, Second ed., Academic Press, 2015, pp. 505–530. [Google Scholar]
- [5].Karsenty G, Wagner EF, Reaching a genetic and molecular understanding of skeletal development, Dev. Cell 2 (2002) 389–406. [DOI] [PubMed] [Google Scholar]
- [6].Karaplis AC (2008) Embryonic development of bone and regulation of intramembranous and endochondral bone formation (chapter 3). in Principles of Bone Biology (Bilezikian John P., L. G. R., Martin T. John ed.), Third ed., Academic Press, pp. 53–84. [Google Scholar]
- [7].Aghajanian P, Mohan S, The art of building bone: emerging role of chondrocyte-to-osteoblast transdifferentiation in endochondral ossification, Bone Res 6 (2018) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhou X, von der Mark K, Henry S, Norton W, Adams H, de Crombrugghe B, Chondrocytes transdifferentiate into osteoblasts in endochondral bone during development, postnatal growth and fracture healing in mice, PLoS Genet. 10 (2014), e1004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yang L, Tsang KY, Tang HC, Chan D, Cheah KS, Hypertrophic chondrocytes can become osteoblasts and osteocytes in endochondral bone formation, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 12097–12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Park J, Gebhardt M, Golovchenko S, Perez-Branguli F, Hattori T, Hartmann C, Zhou X, deCrombrugghe B, Stock M, Schneider H, von der Mark K, Dual pathways to endochondral osteoblasts: a novel chondrocyte-derived osteoprogenitor cell identified in hypertrophic cartilage, Biol Open 4 (2015) 608–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].St-Jacques B, Hammerschmidt M, McMahon AP, Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation, Genes Dev. 13 (1999) 2072–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hall B, The embryonic development of bone, Am. Sci 76 (1988) 174–181. [Google Scholar]
- [13].Bonewald LF, The amazing osteocyte, J. Bone Miner. Res 26 (2011) 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dallas SL, Prideaux M, Bonewald LF, The osteocyte: an endocrine cell … and more, Endocr. Rev 34 (2013) 658–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sims NA, Vrahnas C, Regulation of cortical and trabecular bone mass by communication between osteoblasts, osteocytes and osteoclasts, Arch. Biochem. Biophys 561 (2014) 22–28. [DOI] [PubMed] [Google Scholar]
- [16].Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB, Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene, Mol. Cell. Biol 24 (2004) 9248–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Isaac J, Erthal J, Gordon J, Duverger O, Sun HW, Lichtler AC, Stein GS, Lian JB, Morasso MI, DLX3 regulates bone mass by targeting genes supporting osteoblast differentiation and mineral homeostasis in vivo, Cell Death Differ. 21 (2014) 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gordon JA, Hassan MQ, Koss M, Montecino M, Selleri L, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex, Cells Tissues Organs 194 (2011) 146–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rux DR, Wellik DM, Hox genes in the adult skeleton: novel functions beyond embryonic development, Dev. Dyn 246 (2017) 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Park-Min KH, Epigenetic regulation of bone cells, Connect. Tissue Res 58 (2017) 76–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huynh NP, Anderson BA, Guilak F, McAlinden A, Emerging roles for long noncoding RNAs in skeletal biology and disease, Connect. Tissue Res 58 (2017) 116–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bartel DP, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell 116 (2004) 281–297. [DOI] [PubMed] [Google Scholar]
- [23].Lee RC, Feinbaum RL, Ambros V, The C elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14, Cell 75 (1993) 843–854. [DOI] [PubMed] [Google Scholar]
- [24].Ha M, Kim VN, Regulation of microRNA biogenesis, Nat. Rev. Mol. Cell Biol 15 (2014) 509–524. [DOI] [PubMed] [Google Scholar]
- [25].Eulalio A, Huntzinger E, Izaurralde E, Getting to the root of miRNA-mediated gene silencing, Cell 132 (2008) 9–14. [DOI] [PubMed] [Google Scholar]
- [26].Djuranovic S, Nahvi A, Green R, miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay, Science 336 (2012) 237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lam JK, Chow MY, Zhang Y, and Leung SW (2015) siRNA versus miRNA as therapeutics for gene silencing. Mol Ther Nucleic Acids 4, e252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bartel DP, MicroRNAs: target recognition and regulatory functions, Cell 136 (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E, Phylogenetic shadowing and computational identification of human microRNA genes, Cell 120 (2005) 21–24. [DOI] [PubMed] [Google Scholar]
- [30].Friedman RC, Farh KK, Burge CB, Bartel DP, Most mammalian mRNAs are conserved targets of microRNAs, Genome Res. 19 (2009) 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Leung AK, The whereabouts of microRNA actions: cytoplasm and beyond, Trends Cell Biol. 25 (2015) 601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Desrochers LM, Antonyak MA, Cerione RA, Extracellular vesicles: satellites of information transfer in cancer and stem cell biology, Dev. Cell 37 (2016) 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schwarzenbach H, Gahan PB, MicroRNA shuttle from cell-to-cell by exosomes and its impact in cancer, Noncoding RNA 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].McAlinden A, Im GI, MicroRNAs in orthopaedic research: disease associations, potential therapeutic applications, and perspectives, J. Orthop. Res 36 (2018) 33–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Grigelioniene G, Suzuki HI, Taylan F, Mirzamohammadi F, Borochowitz ZU, Ayturk UM, Tzur S, Horemuzova E, Lindstrand A, Weis MA, Grigelionis G, Hammarsjö A, Marsk E, Nordgren A, Nordenskjöld M, Eyre DR, Warman ML, Nishimura G, Sharp PA, Kobayashi T, Gain-of-function mutation of microRNA-140 in human skeletal dysplasia, Nat. Med 25 (2019) 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shepherdson JL, Zheng H, Amarillo IE, McAlinden A, and Shinawi M (2020) Delineation of the 1q24.3 microdeletion syndrome provides further evidence for the potential role of non-coding RNAs in regulating the skeletal phenotype. Bone 142, 115705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Liu J, Dang L, Wu X, Li D, Ren Q, Lu A, and Zhang G (2019) microRNA-mediated regulation of bone remodeling: a brief review. JBMR Plus 3, e10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Moghaddam T, Neshati Z, Role of microRNAs in osteogenesis of stem cells, J. Cell. Biochem 120 (2019) 14136–14155. [DOI] [PubMed] [Google Scholar]
- [39].Huang C, Geng J, Jiang S, MicroRNAs in regulation of osteogenic differentiation of mesenchymal stem cells, Cell Tissue Res. 368 (2017) 229–238. [DOI] [PubMed] [Google Scholar]
- [40].Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ, The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb, Proc. Natl. Acad. Sci. U. S. A 102 (2005) 10898–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kobayashi T, Lu J, Cobb BS, Rodda SJ, McMahon AP, Schipani E, Merkenschlager M, Kronenberg HM, Dicer-dependent pathways regulate chondrocyte proliferation and differentiation, Proc. Natl. Acad. Sci. U. S. A 105 (2008) 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gaur T, Hussain S, Mudhasani R, Parulkar I, Colby JL, Frederick D, Kream BE, van Wijnen AJ, Stein JL, Stein GS, Jones SN, Lian JB, Dicer inactivation in osteoprogenitor cells compromises fetal survival and bone formation, while excision in differentiated osteoblasts increases bone mass in the adult mouse, Dev. Biol 340 (2010) 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu P, Baumgart M, Groth M, Wittmann J, Jack HM, Platzer M, Tuckermann JP, Baschant U, Dicer ablation in osteoblasts by Runx2 driven cre-loxP recombination affects bone integrity, but not glucocorticoid-induced suppression of bone formation, Sci. Rep 6 (2016) 32112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bendre A, Moritz N, Vaananen V, Maatta JA, Dicer1 ablation in osterix positive bone forming cells affects cortical bone homeostasis, Bone 106 (2018) 139–147. [DOI] [PubMed] [Google Scholar]
- [45].Choi YJ, Jeong S, Yoon KA, Sung HJ, Cho HS, Kim DW, Cho JY, Deficiency of DGCR8 increases bone formation through downregulation of miR-22 expression, Bone 103 (2017) 287–294. [DOI] [PubMed] [Google Scholar]
- [46].Sugatani T, Hruska KA, Impaired micro-RNA pathways diminish osteoclast differentiation and function, J. Biol. Chem 284 (2009) 4667–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y, Noda M, Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption, J. Cell. Biochem 109 (2010) 866–875. [DOI] [PubMed] [Google Scholar]
- [48].Sugatani T, Hildreth III BE, Toribio RE, Malluche HH, Hruska KA, Expression of DGCR8-dependent microRNAs is indispensable for osteoclastic development and bone-resorbing activity, J. Cell. Biochem 115 (2014) 1043–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Simon D, Laloo B, Barillot M, Barnetche T, Blanchard C, Rooryck C, Marche M, Burgelin I, Coupry I, Chassaing N, Gilbert-Dussardier B, Lacombe D, Grosset C, Arveiler B, A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia, Hum. Mol. Genet 19 (2010) 2015–2027. [DOI] [PubMed] [Google Scholar]
- [50].de Pontual L, Yao E, Callier P, Faivre L, Drouin V, Cariou S, Van Haeringen A, Genevieve D, Goldenberg A, Oufadem M, Manouvrier S, Munnich A, Vidigal JA, Vekemans M, Lyonnet S, Henrion-Caude A, Ventura A, Amiel J, Germline deletion of the miR-17 approximately 92 cluster causes skeletal and growth defects in humans, Nat. Genet 43 (2011) 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hemmat M, Rumple MJ, Mahon LW, Strom CM, Anguiano A, Talai M, Nguyen A, Boyar FZ, Short stature, digit anomalies and dysmorphic facial features are associated with the duplication of miR-17–92 cluster, Mol. Cytogenet 7 (2014) 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mohan S, Wergedal JE, Das S, Kesavan C, Conditional disruption of miR17-92 cluster in collagen type I-producing osteoblasts results in reduced periosteal bone formation and bone anabolic response to exercise, Physiol. Genomics 47 (2015) 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H, MicroRNA-140 plays dual roles in both cartilage development and homeostasis, Genes Dev. 24 (2010) 1173–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nakamura Y, Inloes JB, Katagiri T, Kobayashi T, Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling, Mol. Cell. Biol 31 (2011) 3019–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Burkardt DD, Rosenfeld JA, Helgeson ML, Angle B, Banks V, Smith WE, Gripp KW, Moline J, Moran RT, Niyazov DM, Stevens CA, Zackai E, Lebel RR, Ashley DG, Kramer N, Lachman RS, Graham JM Jr., Distinctive phenotype in 9 patients with deletion of chromosome 1q24-q25, Am. J. Med. Genet. A 155A (2011) 1336–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chatron N, Haddad V, Andrieux J, Désir J, Boute O, Dieux A, Baumann B, Drunat S, Gérard M, Bonnet C, Leheup B, Till M, Rossi M, Flori E, Alembik Y, Stewart H, McParland J, Bernardini L, Castelluccio P, Roos L, Tümer Z, Fagan K, Hackett A, Bain N, van Haeringen A, Ruivenkamp C, Benzacken B, Sanlaville D, Edery P, Aboura A, and Schluth-Bolard C (2015) Refinement of genotype-phenotype correlation in 18 patients carrying a 1q24q25 deletion. Am J Med Genet A 167a, 1008–1017. [DOI] [PubMed] [Google Scholar]
- [57].Ashraf T, Collinson MN, Fairhurst J, Wang R, Wilson LC, Foulds N, Two further patients with the 1q24 deletion syndrome expand the phenotype: A possible role for the miR199-214 cluster in the skeletal features of the condition, Am. J. Med. Genet. A 167A (2015) 3153–3160. [DOI] [PubMed] [Google Scholar]
- [58].Lefroy H, Fox O, Javaid MK, Makaya T, Shears DJ, 1q24 deletion syndrome. Two cases and new insights into genotype-phenotype correlations, Am. J. Med. Genet. A 176 (2018) 2004–2008. [DOI] [PubMed] [Google Scholar]
- [59].Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T, Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice, Dev. Dyn 237 (2008) 3738–3748. [DOI] [PubMed] [Google Scholar]
- [60].Wei J, Shi Y, Zheng L, Zhou B, Inose H, Wang J, Guo XE, Grosschedl R, Karsenty G, miR-34s inhibit osteoblast proliferation and differentiation in the mouse by targeting SATB2, J. Cell Biol 197 (2012) 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen L, Holmstrøm K, Qiu W, Ditzel N, Shi K, Hokland L, Kassem M, MicroRNA-34a inhibits osteoblast differentiation and in vivo bone formation of human stromal stem cells, Stem Cells 32 (2014) 902–912. [DOI] [PubMed] [Google Scholar]
- [62].Krzeszinski JY, Wei W, Huynh H, Jin Z, Wang X, Chang TC, Xie XJ, He L, Mangala LS, Lopez-Berestein G, Sood AK, Mendell JT, Wan Y, miR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2, Nature 512 (2014) 431–435. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [63].Inose H, Ochi H, Kimura A, Fujita K, Xu R, Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, Saito K, Nakamura T, Siomi H, Ito H, Arai Y, Shinomiya K, Takeda S, A microRNA regulatory mechanism of osteoblast differentiation, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 20794–20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Williams A, Henao-Mejia J, Harman CC, Flavell RA, miR-181 and metabolic regulation in the immune system, Cold Spring Harb. Symp. Quant. Biol 78 (2013) 223–230. [DOI] [PubMed] [Google Scholar]
- [65].Henao-Mejia J, Williams A, Goff LA, Staron M, Licona-Limon P, Kaech SM, Nakayama M, Rinn JL, Flavell RA, The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis, Immunity 38 (2013) 984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Inoue K, Deng Z, Chen Y, Giannopoulou E, Xu R, Gong S, Greenblatt MB, Mangala LS, Lopez-Berestein G, Kirsch DG, Sood AK, Zhao L, Zhao B, Bone protection by inhibition of microRNA-182, Nat. Commun 9 (2018) 4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Kim KM, Park SJ, Jung SH, Kim EJ, Jogeswar G, Ajita J, Rhee Y, Kim CH, Lim SK, miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1, J. Bone Miner. Res 27 (2012) 1669–1679. [DOI] [PubMed] [Google Scholar]
- [68].Pan BL, Tong ZW, Li SD, Wu L, Liao JL, Yang YX, Li HH, Dai YJ, Li JE, and Pan L (2018) Decreased microRNA-182-5p helps alendronate promote osteoblast proliferation and differentiation in osteoporosis via the Rap1/MAPK pathway. Biosci Rep 38. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [69].Yang N, Wang G, Hu C, Shi Y, Liao L, Shi S, Cai Y, Cheng S, Wang X, Liu Y, Tang L, Ding Y, Jin Y, Tumor necrosis factor alpha suppresses the mesenchymal stem cell osteogenesis promoter miR-21 in estrogen deficiency-induced osteoporosis, J. Bone Miner. Res 28 (2013) 559–573. [DOI] [PubMed] [Google Scholar]
- [70].Sugatani T, Vacher J, Hruska KA, A microRNA expression signature of osteodastogenesis, Blood 117 (2011) 3648–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hu CH, Sui BD, Du FY, Shuai Y, Zheng CX, Zhao P, Yu XR, and Jin Y (2017) miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci Rep 7, 43191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jin HY, Gonzalez-Martin A, Miletic AV, Lai M, Knight S, Sabouri-Ghomi M, Head SR, Maeauley MS, Rickert RC, Xiao C, Transfection of microRNA mimics should be used with caution, Front. Genet 6 (2015) 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Li H, Fan J, Fan L, Li T, Yang Y, Xu H, Deng L, Li J, Li T, Weng X, Wang S, Chunhua Zhao R, MiRNA-10b reciprocally stimulates osteogenesis and inhibits adipogenesis partly through the TGF-β/SMAD2 signaling pathway, Aging Dis. 9 (2018) 1058–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chen R, Q. H, Tong Y, Liao F, Hu X, Qiu Y, Liao Y. (2019) MiRNA-19a-3p alleviates the progression of osteoporosis by targeting HDAC4 to promote the osteogenic differentiation of hMSCs. Biochem. Biophys. Res. Commun 516, 666–672. [DOI] [PubMed] [Google Scholar]
- [75].Hu H, Z. C, Zhang P, Liu Y, Jiang Y, Wu E, Xue H, Liu C, Li Z. (2019) miR-26b modulates OA induced BMSC osteogenesis through regulating GSK3β/β-catenin pathway. Exp. Mol. Pathol 107, 158–164. [DOI] [PubMed] [Google Scholar]
- [76].Tan K, P. Y, Guo P. (2018) MiR-29a promotes osteogenic differentiation of mesenchymal stem cells via targeting HDAC4. Eur. Rev. Med. Pharmacol. Sci 22, 3318–3326. [DOI] [PubMed] [Google Scholar]
- [77].Lee EJ, Kim SM, Choi B, Kim EY, Chung YH, Lee EJ, Yoo B, Lee CK, Hong S, Kim BJ, Koh JM, Kim SH, Kim YG, Chang EJ, Interleukin-32 gamma stimulates bone formation by increasing miR-29a in osteoblastic cells and prevents the development of osteoporosis, Sci. Rep 7 (2017) 40240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yan X, W. H, Li Y, Jiang Y, Shao Q, Xu W. (2018) MicroRNA-92a overexpression promotes the osteogenic differentiation of bone mesenchymal stem cells by impeding Smad6-mediated runt-related transcription factor 2 degradation. Mol. Med. Rep 17, 7821–7826. [DOI] [PubMed] [Google Scholar]
- [79].Gao XL, C. M, Ai GG, Hu YB. (2018) Mir-98 reduces the expression of HMGA2 and promotes osteogenic differentiation of mesenchymal stem cells. Eur. Rev. Med. Pharmacol. Sci 22, 3311–3317. [DOI] [PubMed] [Google Scholar]
- [80].Lin Z, H. H, Wang M, Liang J. (2019) MicroRNA-130a controls bone marrow mesenchymal stem cell differentiation towards the osteoblastic and adipogenic fate. Cell Prolif. 52, e12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yin N, Z. L, Ding L, Yuan J, Du L, Pan M, Xue F, Xiao H. (2019) MiR-135-5p promotes osteoblast differentiation by targeting HIF1AN in MC3T3-E1 cells. Cell Mol Biol Lett 24, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zheng H, Liu J, Tycksen E, Nunley R, McAlinden A, MicroRNA-181a/b-1 over-expression enhances osteogenesis by modulating PTEN/PI3K/AKT signaling and mitochondrial metabolism, Bone 123 (2019) 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Aquino-Martinez R, F. J, Weivoda MM, Negley BA, Onken JL, Thicke BS, Fulcer MM, Fraser DG, van Wijnen AJ, Khosla S, Monroe DG. (2019) miR-219a-5p regulates Rorβ during osteoblast differentiation and in age-related bone loss. J. Bone Miner. Res 34, 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Han Y, Z. K, Hong Y, Wang J, Liu Q, Zhang Z, Xia H, Tang Y, Li T, Li L, Xue Y, Hong W. (2018) miR-342-3p promotes osteogenic differentiation via targeting ATF3. FEBS Lett. 592, 4051–4065. [DOI] [PubMed] [Google Scholar]
- [85].Xu D, G. Y, Hu N, Wu L, Chen Q. (2017) miR-365 Ameliorates Dexamethasone-Induced Suppression of Osteogenesis in MC3T3-E1 Cells by Targeting HDAC4. Int J Mol Sci 18, 977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ge JB, L. J, Hong HY, Sun YJ, Li Y, Zhang CM. (2018) MiR-374b promotes osteogenic differentiation of MSCs by degrading PTEN and promoting fracture healing. Eur. Rev. Med. Pharmacol. Sci 22, 3303–3310. [DOI] [PubMed] [Google Scholar]
- [87].Tang X, Lin J, Wang G, Lu J, MicroRNA-433-3p promotes osteoblast differentiation through targeting DKK1 expression, PLoS One 12 (2017), e0179860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Fan L, F. J, Liu Y, Li T, Xu H, Yang Y, Deng L, Li H, Zhao RC. (2018) miR-450b promotes osteogenic differentiation in vitro and enhances bone formation in vivo by targeting BMP3. Stem Cells Dev. 27, 600–611. [DOI] [PubMed] [Google Scholar]
- [89].Chen CY, Chen J, He L, Stiles BL, PTEN: tumor suppressor and metabolic regulator, Front Endocrinol (Lausanne) 9 (2018) 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Liu X, Bruxvoort KJ, Zylstra CR, Liu J, Cichowski R, Faugere MC, Bouxsein ML, Wan C, Williams BO, Clemens TL, Lifelong accumulation of bone in mice lacking Pten in osteoblasts, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Burgers TA, Hoffmann MF, Collins CJ, Zahatnansky J, Alvarado MA, Morris MR, Sietsema DL, Mason JJ, Jones CB, Ploeg HL, Williams BO, Mice lacking pten in osteoblasts have improved intramembranous and late endochondral fracture healing, PLoS One 8 (2013), e63857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Shum LC, White NS, Mills BN, Bentley KL, Eliseev RA, Energy metabolism in mesenchymal stem cells during osteogenic differentiation, Stem Cells Dev. 25 (2016) 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Dobson PF, Dennis EP, Hipps D, Reeve A, Laude A, Bradshaw C, Stamp C, Smith A, Deehan DJ, Turnbull DM, Greaves LC, Mitochondrial dysfunction impairs osteogenesis, increases osteoclast activity, and accelerates age related bone loss, Sci. Rep 10 (2020) 11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Li Q, Gao Z, Chen Y, Guan MX, The role of mitochondria in osteogenic, adipogenic and chondrogenic differentiation of mesenchymal stem cells, Protein Cell 8 (2017) 439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Heinz S, Freyberger A, Lawrenz B, Schladt L, Schmuck G, Ellinger-Ziegelbauer H, Mechanistic investigations of the mitochondrial complex I inhibitor rotenone in the context of pharmacological and safety evaluation, Sci. Rep 7 (2017) 45465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Shares BH, Busch M, White N, Shum L, Eliseev RA, Active mitochondria support osteogenic differentiation by stimulating beta-catenin acetylation, J. Biol. Chem 293 (2018) 16019–16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Pietila M, Lehtonen S, Narhi M, Hassinen IE, Leskela HV, Aranko K, Nordstrom K, Vepsalainen A, Lehenkari P, Mitochondrial function determines the viability and osteogenic potency of human mesenchymal stem cells, Tissue Eng Part C Methods 16 (2010) 435–445. [DOI] [PubMed] [Google Scholar]
- [98].Bhushan R, Grunhagen J, Becker J, Robinson PN, Ott CE, Knaus P, miR-181a promotes osteoblastic differentiation through repression of TGF-beta signaling molecules, Int. J. Biochem. Cell Biol 45 (2013) 696–705. [DOI] [PubMed] [Google Scholar]
- [99].Zhang HG, W. X, Zhao H, Zhou CN. (2019) MicroRNA-9-5p promotes osteoporosis development through inhibiting osteogenesis and promoting adipogenesis via targeting Wnt3a. Eur. Rev. Med. Pharmacol. Sci 23, 456–463. [DOI] [PubMed] [Google Scholar]
- [100].Duan L, Z. H, Xiong Y, Tang X, Yang Y, Hu Z, Li C, Chen S, Yu X. (2018) miR-16-2* interferes with WNT5A to regulate osteogenesis of mesenchymal stem cells. Cell. Physiol. Biochem 51, 1087–1102. [DOI] [PubMed] [Google Scholar]
- [101].Zhang Y, L. S, Yuan S, Zhang H, Liu J. (2019) MicroRNA-23a inhibits osteogenesis of periodontal mesenchymal stem cells by targeting bone morphogenetic protein signaling. Arch. Oral Biol 102. [DOI] [PubMed] [Google Scholar]
- [102].Godfrey TC, W. B, Beloti MM, Kemper AG, Ferraz EP, Roy B, Rehan M, Afreen LH, Km E, Lengner CJ, Hassan Q. (2018) The microRNA-23a cluster regulates the developmental HoxA cluster function during osteoblast differentiation. J. Biol. Chem 293, 17646–17660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Peng W, Z. S, Li X, Weng J, Chen S. (2017) miR-27b-3p suppressed osteogenic differentiation of maxillary sinus membrane stem cells by targeting Sp7. Implant. Dent 26, 492–499. [DOI] [PubMed] [Google Scholar]
- [104].Zhang Y, W. Q, Ding WB, Zhang LL, Wang HC, Zhu YJ, He W, Chai YN, Liu YW. (2017) Increased microRNA-93-5p inhibits osteogenic differentiation by targeting bone morphogenetic protein-2. PLoS One 12, e0182678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhang GP, Z. J, Zhu CH, Lin L, Wang J, Zhang HJ, Li J, Yu XG, Zhao ZS, Dong W, Liu GB. (2017) MicroRNA-98 regulates osteogenic differentiation of human bone mesenchymal stromal cells by targeting BMP2. J. Cell. Mol. Med 21, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Liu K, J. Y, Zhang W, Fu X, Zhao H, Zhou X, Tao Y, Yang H, Zhang Y, Zen K, Zhang C, Li D, Shi Q. (2017) Silencing miR-106b accelerates osteogenesis of mesenchymal stem cells and rescues against glucocorticoid-induced osteoporosis by targeting BMP2. Bone 97, 130–138. [DOI] [PubMed] [Google Scholar]
- [107].Wang H, X. Z, Hou T, Li Z, Huang K, Gong J, Zhou W, Tang K, Xu J, Dong S. (2017) MiR-125b regulates the osteogenic differentiation of human mesenchymal stem cells by targeting BMPR1b. Cell. Physiol. Biochem 41, 530–542. [DOI] [PubMed] [Google Scholar]
- [108].Xue N, Q. L, Zhang G, Zhang Y. (2018) miRNA-125b regulates osteogenic differentiation of periodontal ligament cells through NKIRAS2/NF-κB pathway. Cell. Physiol. Biochem 48, 1771–1781. [DOI] [PubMed] [Google Scholar]
- [109].Xu Y, R. C, Zhao X, Wang W, Zhang N. (2019) microRNA-132 inhibits osteogenic differentiation of periodontal ligament stem cells via GDF5 and the NF-κB signaling pathway. Pathol Res Pract 215, 152722. [DOI] [PubMed] [Google Scholar]
- [110].Kong L, Z. R, Wang M, Wang W, Xu J, Chai Y, Guan J, Kang Q. (2020) Silencing microRNA-137-3p, which targets RUNX2 and CXCL12 prevents steroid-induced osteonecrosis of the femoral head by facilitating osteogenesis and angiogenesis. Int. J. Biol. Sci 16, 655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Zheng H, R. D, Anderson BA, Tycksen E, Nunley R, McAlinden A. (2018) MicroRNA-138 inhibits osteogenic differentiation and mineralization of human dedifferentiated chondrocytes by regulating RhoC and the actin cytoskeleton. JBMR Plus 3, e10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Wang Y, W. K, Hu Z, Zhou H, Zhang L, Wang H, Li G, Zhang S, Cao X, Shi F. (2018) MicroRNA-139-3p regulates osteoblast differentiation and apoptosis by targeting ELK1 and interacting with long noncoding RNA ODSM. Cell Death Dis. 9, 1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Hao W, L. H, Zhou L, Sun Y, Su H, Ni J, He T, Shi P, Wang X. (2018) MiR-145 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells through targeting FoxO1. Exp Biol Med (Maywood) 243, 386–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Moussa FM, C. B, Sondag GR, DeSanto M, Obri MS, McDermott SE, Safadi FF. (2020) The role of miR-150 regulates bone cell differentiation and function. Bone. [DOI] [PubMed] [Google Scholar]
- [115].Liu H, Z. L, Yuan T, Chen S, Zhou Y, An L, Guo Y, Fan M, Li Y, Sun Y, Li W, Shi Q, Weng Y. (2018) MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int. J. Mol. Med 41, 3379–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Tao G, M. P, Guan H, Jiang M, Chu T, Zhong C, Liu J. (2019) Effect of miR-181a-3p on osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting BMP10. Artif Cells Nanomed Biotechnol 47, 4159–4164. [DOI] [PubMed] [Google Scholar]
- [117].Cui Q, X. J, Yu M, Wang Y, Xu J, Gu Y, Nan X, Ma W, Liu H, Zhao H. (2019) Mmu-miR-185 depletion promotes osteogenic differentiation and suppresses bone loss in osteoporosis through the Bgn-mediated BMP/Smad pathway. Cell Death Dis. 10, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Chang H, W. Y, Liu H, Nan X, Wong S, Peng S, Gu Y, Zhao H, Feng H. (2017) Mutant Runx2 regulates amelogenesis and osteogenesis through a miR-185-5p-Dlx2 axis. Cell Death Dis. 8, 3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Wang SN, Z. X, Yu B, Wang BW. (2018) miR-193a inhibits osteogenic differentiation of bone marrow-derived stroma cell via targeting HMGB1. Biochem. Biophys. Res. Cornmun 503, 536–543. [DOI] [PubMed] [Google Scholar]
- [120].Lv R, P. X, Song L, Sun Q, Guo C, Zou S, Zhou Q. (2019) MicroRNA-200a-3p accelerates the progression of osteoporosis by targeting glutaminase to inhibit osteogenic differentiation of bone marrow mesenchymal stem cells. Biomed. Pharmacother 116, 108960. [DOI] [PubMed] [Google Scholar]
- [121].Chen Y, Y. Y, Fan XL, Lin P, Yang H, Chen XZ, Xu XD. (2019) miR-206 inhibits osteogenic differentiation of bone marrow mesenchymal stem cells by targetting glutaminase. Biosci Rep 39, BSR20181108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Peng W, Z. S, Chen J, Wang J, Rong Q, Chen S. (2019) Hsa_circRNA_33287 promotes the osteogenic differentiation of maxillary sinus membrane stem cells via miR-214-3p/Runx3. Biomed. Pharmacother 109, 1709–1717. [DOI] [PubMed] [Google Scholar]
- [123].Zhang S, L. Y, Zheng Z, Zeng X, Liu D, Wang C, Ting K. (2018) MicroRNA-223 suppresses osteoblast differentiation by inhibiting DHRS3. Cell. Physiol. Biochem 47, 667–679. [DOI] [PubMed] [Google Scholar]
- [124].Zhou J, N. H, Liu P, Wang Z, Yao B, Yang L. (2019) Down-regulation of miR-339 promotes differentiation of BMSCs and alleviates osteoporosis by targeting DLX5. Eur. Rev. Med. Pharmacol. Sci 23, 29–36. [DOI] [PubMed] [Google Scholar]
- [125].Pan J, H. C, Chen G, Cai Z, Zhang Z. (2018) MicroRNA-451 blockade promotes osteoblastic differentiation and skeletal anabolic effects by promoting YWHAZ-mediated RUNX2 protein stabilization. Medchemcomm 9, 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [126].Lu XD, H. W, Liu YX. (2019) Suppression of miR-451a accelerates osteogenic differentiation and inhibits bone loss via Bmp6 signaling during osteoporosis. Biomed. Pharmacother 120, 109378. [DOI] [PubMed] [Google Scholar]
- [127].Zhang SY, G. F, Peng CG, Zheng CJ, Wu MF. (2018) miR-485-5p promotes osteoporosis via targeting Osterix. Eur. Rev. Med. Pharmacol. Sci 22, 4792–4799. [DOI] [PubMed] [Google Scholar]
- [128].Tian Z, Z. H, Xu Y, Bai J. (2017) MicroRNA-495 inhibits new bone regeneration via targeting high mobility group AT-hook 2 (HMGA2). Med. Sci. Monit 23, 4689–4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wu R, R. J, Sun Y, Liu M, Sha Z, Fan C, Wu Q. (2018) Long non-coding RNA HIF1A-AS2 facilitates adipose-derived stem cells (ASCs) osteogenic differentiation through miR-665/IL6 axis via PI3K/Akt signaling pathway. Stem Cell Res Ther 9, 348. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [130].John AA, P. R, Kureel J, Singh D. (2018) Identification of novel microRNA inhibiting actin cytoskeletal rearrangement thereby suppressing osteoblast differentiation. J Mol Med (Berl) 96, 427–444. [DOI] [PubMed] [Google Scholar]
- [131].Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, Cao H, Wu H, Song J, Pan X, Sun Q, Ling S, Li Y, Zhu M, Zhang P, Peng S, Xie X, Tang T, Hong A, Bian Z, Bai Y, Lu A, Li Y, He F, Zhang G, Li Y, miR-214 targets ATF4 to inhibit bone formation, Nat. Med 19 (2013) 93–100. [DOI] [PubMed] [Google Scholar]
- [132].Yu Y, Newman H, Shen L, Sharma D, Hu G, Mirando AJ, Zhang H, Knudsen E, Zhang GF, Hilton MJ, and Karner CM (2019) Glutamine metabolism regulates proliferation and lineage allocation in skeletal stem cells. Cell Metab 29, 966–978.e964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS, Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment, Dev. Cell 6 (2004) 483–495. [DOI] [PubMed] [Google Scholar]
- [134].Sonowal H, Kumar A, Bhattacharyya J, Gogoi PK, Jaganathan BG, Inhibition of actin polymerization decreases osteogeneic differentiation of mesenchymal stem cells through p38 MAPK pathway, J. Biomed. Sci 20 (2013) 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Mathieu PS, Loboa EG, Cytoskeletal and focal adhesion influences on mesenchymal stem cell shape, mechanical properties, and differentiation down osteogenic, adipogenic, and chondrogenic pathways, Tissue Eng Part B Rev 18 (2012) 436–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Eskildsen T, Taipaleenmaki H, Stenvang J, Abdallah BM, Ditzel N, Nossent AY, Bak M, Kauppinen S, Kassem M, MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo, Proc. Natl. Acad. Sci. U. S. A 108 (2011) 6139–6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Chan KT, Cortesio CL, Huttenlocher A, FAK alters invadopodia and focal adhesion composition and dynamics to regulate breast cancer invasion, J. Cell Biol 185 (2009) 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Hakem A, Sanchez-Sweatman O, You-Ten A, Duncan G, Wakeham A, Khokha R, Mak TW, RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis, Genes Dev. 19 (2005) 1974–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Tsukamoto S, Lovendorf MB, Park J, Salem KZ, Reagan MR, Manier S, Zavidij O, Rahmat M, Huynh D, Takagi S, Kawano Y, Kokubun K, Thrue CA, Nagano K, Petri A, Roccaro AM, Capelletti M, Baron R, Kauppinen S, Ghobrial IM, Inhibition of microRNA-138 enhances bone formation in multiple myeloma bone marrow niche, Leukemia 32 (2018) 1739–1750. [DOI] [PubMed] [Google Scholar]
- [140].Xu R, Shen X, Si Y, Fu Y, Zhu W, Xiao T, Fu Z, Zhang P, Cheng J, Jiang H, MicroRNA-31a-5p from aging BMSCs links bone formation and resorption in the aged bone marrow microenvironment, Aging Cell 17 (2018), e12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, Liao EY, Luo XH, MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation, J. Clin. Invest 125 (2015) 1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Davis C, Dukes A, Drewry M, Helwa I, Johnson MH, Isales CM, Hill WD, Liu Y, Shi X, Fulzele S, Hamrick MW, MicroRNA-183-5p increases with age in bone-derived extracellular vesicles, suppresses bone marrow stromal (stem) cell proliferation, and induces stem cell senescence, Tissue Eng Part A 23 (2017) 1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Liu H, Liu Q, Wu XP, He HB, Fu L, MiR-96 regulates bone metabolism by targeting osterix, Clin. Exp. Pharmacol. Physiol 45 (2018) 602–613. [DOI] [PubMed] [Google Scholar]
- [144].Periyasamy-Thandavan S, Burke J, Mendhe B, Kondrikova G, Kolhe R, Hunter M, Isales CM, Hamrick MW, Hill WD, Fulzele S, MicroRNA-141-3p negatively modulates SDF-1 expression in age-dependent pathophysiology of human and murine bone marrow stromal cells, J. Gerontol. A Biol. Sci. Med. Sci 74 (2019) 1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Kranjc T, Ostanek B, Marc J, Bone microRNAs and ageing, Curr. Pharm. Biotechnol 18 (2017) 210–220. [DOI] [PubMed] [Google Scholar]
- [146].Qin Y, Wang L, Gao Z, Chen G, Zhang C, Bone marrow stromal/stem cell-derived extracellular vesicles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo, Sci. Rep 6 (2016) 21961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Liu J, Li D, Wu X, Dang L, Lu A, Zhang G, Bone-derived exosomes, Curr. Opin. Pharmacol 34 (2017) 64–69. [DOI] [PubMed] [Google Scholar]
- [148].Xie Y, Chen Y, Zhang L, Ge W, Tang P, The roles of bone-derived exosomes and exosomal microRNAs in regulating bone remodelling, J. Cell. Mol. Med 21 (2017) 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Yuan FL, Wu QY, Miao ZN, Xu MH, Xu RS, Jiang DL, Ye JX, Chen FH, Zhao MD, Wang HJ, Li X, Osteoclast-derived extracellular vesicles: novel regulators of osteoclastogenesis and osteoclast-osteoblasts communication in bone remodeling, Front. Physiol 9 (2018) 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Gao M, Gao W, Papadimitriou JM, Zhang C, Gao J, Zheng M, Exosomes-the enigmatic regulators of bone homeostasis, Bone Res 6 (2018) 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Lu J, Wang QY, Sheng JG, Exosomes in the repair of bone defects: next-generation therapeutic tools for the treatment of nonunion, Biomed. Res. Int 2019 (2019) 1983131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Masaoutis C, Theocharis S, The role of exosomes in bone remodeling: implications for bone physiology and disease, Dis. Markers 2019 (2019) 9417914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Liu M, Sun Y, Zhang Q, Emerging role of extracellular vesicles in bone remodeling, J. Dent. Res 97 (2018) 859–868. [DOI] [PubMed] [Google Scholar]
- [154].Kalluri R, LeBleu VS, The biology, function, and biomedical applications of exosomes, Science 367 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Wood SL, Brown JE, Personal medicine and bone metastases: biomarkers, micro-RNAs and bone metastases, Cancers (Basel) 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Donati S, Ciuffi S, Palmini G, Brandi ML, Circulating miRNAs: a new opportunity in bone fragility, Biomolecules 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bottani M, Banfi G, Lombardi G, The clinical potential of circulating miRNAs as biomarkers: present and future applications for diagnosis and prognosis of age-associated bone diseases, Biomolecules (2020) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Lyu H, Xiao Y, Guo Q, Huang Y, Luo X, The role of bone-derived exosomes in regulating skeletal metabolism and extraosseous diseases, Front Cell Dev Biol 8 (2020) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Cui Y, Luan J, Li H, Zhou X, Han J, Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression, FEBS Lett. 590 (2016) 185–192. [DOI] [PubMed] [Google Scholar]
- [160].Li D, Liu J, Guo B, Liang C, Dang L, Lu C, He X, Cheung HY, Xu L, Lu C, He B, Liu B, Shaikh AB, Li F, Wang L, Yang Z, Au DW, Peng S, Zhang Z, Zhang BT, Pan X, Qian A, Shang P, Xiao L, Jiang B, Wong CK, Xu J, Bian Z, Liang Z, Guo DA, Zhu H, Tan W, Lu A, Zhang G, Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation, Nat. Cornmun 7 (2016) 10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Sun W, Zhao C, Li Y, Wang L, Nie G, Peng J, Wang A, Zhang P, Tian W, Li Q, Song J, Wang C, Xu X, Tian Y, Zhao D, Xu Z, Zhong G, Han B, Ling S, Chang YZ, Li Y, Osteoclast-derived microRNA-containing exosomes selectively inhibit osteoblast activity, Cell Discov 2 (2016) 16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Yang JX, X. P, Li YS, Wen T, Yang XC. (2020) Osteoclast-derived miR-23a-5p-containing exosomes inhibit osteogenic differentiation by regulating Runx2. Cell. Signal 70, 109504. [DOI] [PubMed] [Google Scholar]
- [163].Minamizaki T, Nakao Y, Irie Y, Ahmed F, Itoh S, Sarmin N, Yoshioka H, Nobukiyo A, Fujimoto C, Niida S, Sotomaru Y, Tanimoto K, Kozai K, Sugiyama T, Bonnelye E, Takei Y, Yoshiko Y, The matrix vesicle cargo miR-125b accumulates in the bone matrix, inhibiting bone resorption in mice, Cornmun Biol 3 (2020) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Dai J, D. R, Han X, Li J, Gong X, Bai Y, Kang F, Liang M, Zeng F, Hou Z, Dong S. (2020) Osteoclast-derived exosomal let-7a-5p targets Smad2 to promote the hypertrophic differentiation of chondrocytes. Am J Physiol Cell Physiol. [DOI] [PubMed] [Google Scholar]
- [165].Qin Y, P. Y, Zhao W, Pan J, Ksiezak-Reding H, Cardozo C, Wu Y, Divieti Pajevic P, Bonewald LF, Bauman WA, Qin W. (2017) Myostatin inhibits osteoblastic differentiation by suppressing osteocyte-derived exosomal microRNA-218: a novel mechanism in muscle-bone communication. J. Biol. Chem 292, 11021–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Lv PY, Gao PF, Tian GJ, Yang YY, Mo FF, Wang ZH, Sun L, Kuang MJ, Wang Y, Osteocyte-derived exosomes induced by mechanical strain promote human periodontal ligament stem cell proliferation and osteogenic differentiation via the miR-181b-5p/PTEN/AKT signaling pathway, Stem Cell Res Ther 11 (2020) 295. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [167].Joshi BS, de Beer MA, Giepmans BNG, Zuhorn IS, Endocytosis of extracellular vesicles and release of their cargo from endosomes, ACS Nano 14 (2020) 4444–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Zhang Y, Liu Y, Liu H, Tang WH, Exosomes: biogenesis, biologic function and clinical potential, Cell Biosci 9 (2019) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Ye Y, Li SL, Ma YY, Diao YJ, Yang L, Su MQ, Li Z, Ji Y, Wang J, Lei L, Fan WX, Li LX, Xu Y, Hao XK, Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer, Oncotarget 8 (2017) 94834–94849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Chuo ST, Chien JC, Lai CP, Imaging extracellular vesicles: current and emerging methods, J. Biomed. Sci 25 (2018) 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Liu J, Dang L, Li D, Liang C, He X, Wu H, Qian A, Yang Z, Au DW, Chiang MW, Zhang BT, Han Q, Yue KK, Zhang H, Lv C, Pan X, Xu J, Bian Z, Shang P, Tan W, Liang Z, Guo B, Lu A, Zhang G, A delivery system specifically approaching bone resorption surfaces to facilitate therapeutic modulation of microRNAs in osteoclasts, Biomaterials 52 (2015) 148–160. [DOI] [PubMed] [Google Scholar]
- [172].Cai M, Yang L, Zhang S, Liu J, Sun Y, Wang X, A bone-resorption surface-targeting nanoparticle to deliver anti-miR214 for osteoporosis therapy, Int. J. Nanomedicine 12 (2017) 7469–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Dang L, Liu J, Li F, Wang L, Li D, Guo B, He X, Jiang F, Liang C, Liu B, Badshah SA, He B, Lu J, Lu C, Lu A, Zhang G, Targeted delivery systems for molecular therapy in skeletal disorders, Int. J. Mol. Sci 17 (2016) 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Sun Y, Ye X, Cai M, Liu X, Xiao J, Zhang C, Wang Y, Yang L, Liu J, Li S, Kang C, Zhang B, Zhang Q, Wang Z, Hong A, Wang X, Osteoblast-targeting-peptide modified nanoparticle for siRNA/microRNA delivery, ACS Nano 10 (2016) 5759–5768. [DOI] [PubMed] [Google Scholar]
- [175].Wang N, Zheng J, Chen Z, Liu Y, Dura B, Kwak M, Xavier-Ferrucio J, Lu YC, Zhang M, Roden C, Cheng J, Krause DS, Ding Y, Fan R, Lu J, Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation, Nat. Commun 10 (2019) 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Zhao C, Miao Y, Cao Z, Shi J, Li J, Kang F, Dou C, Xie Z, Xiang Q, Dong S, MicroRNA-29b regulates hypertrophy of murine mesenchymal stem cells induced toward chondrogenesis, J. Cell. Biochem (2019), 10.1002/jcb.28161. [DOI] [PubMed] [Google Scholar]
- [177].Li P, Wei X, Guan Y, Chen Q, Zhao T, Sun C, Wei L, MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development, FASEB J. 28 (2014) 3930–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Vonk LA, Kragten AH, Dhert WJ, Saris DB, Creemers LB, Overexpression of hsa-miR-148a promotes cartilage production and inhibits cartilage degradation by osteoarthritic chondrocytes, Osteoarthr. Cartil 22 (2014) 145–153. [DOI] [PubMed] [Google Scholar]
- [179].Chen W, Sheng P, Huang Z, Meng F, Kang Y, Huang G, Zhang Z, Liao W, Zhang Z, MicroRNA-381 regulates chondrocyte hypertrophy by inhibiting histone deacetylase 4 expression, Int. J. Mol. Sci 17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Lee JM, Ko JY, Kim HY, Park JW, Guilak F, Im GI, miR-892b inhibits hypertrophy by targeting KLF10 in the chondrogenesis of mesenchymal stem cells, Mol Ther Nucleic Acids 17 (2019) 310–322. [DOI] [PMC free article] [PubMed] [Google Scholar]