Abstract
Meiosis generates genetic variation through homologous recombination (HR) that is harnessed during breeding. HR occurs in the context of meiotic chromosome axes and the synaptonemal complex. To study the role of axis remodelling in crossover (CO) formation in a crop species, we characterized mutants of the axis-associated protein ASY1 and the axis-remodelling protein PCH2 in Brassica rapa. asy1 plants form meiotic chromosome axes that fail to synapse. CO formation is almost abolished, and residual chiasmata are proportionally enriched in terminal chromosome regions, particularly in the nucleolar organizing region (NOR)-carrying chromosome arm. pch2 plants show impaired ASY1 loading and remodelling, consequently achieving only partial synapsis, which leads to reduced CO formation and loss of the obligatory CO. PCH2-independent chiasmata are proportionally enriched towards distal chromosome regions. Similarly, in Arabidopsis pch2, COs are increased towards telomeric regions at the expense of (peri-) centromeric COs compared with the wild type. Taken together, in B. rapa, axis formation and remodelling are critical for meiotic fidelity including synapsis and CO formation, and in asy1 and pch2 CO distributions are altered. While asy1 plants are sterile, pch2 plants are semi-sterile and thus PCH2 could be an interesting target for breeding programmes.
Keywords: ASY1, Brassica rapa, crossover, meiosis, meiotic chromosome axis remodelling, meiotic recombination, PCH2, synaptonemal complex
ASY1 and PCH2 are critical for meiotic axis remodelling as well as synaptonemal complex and crossover formation in Brassica rapa. Residual crossovers in asy1 and pch2 plants show altered patterning.
Introduction
Meiosis is a specialized cell division in sexually reproducing organisms shuffling maternal and paternal genomes through homologous recombination (HR) and independent assortment of homologous chromosomes.
HR is initiated by programmed formation of SPO11-catalysed DNA double-strand breaks (DSBs) (Keeney and Kleckner, 1995; Keeney et al., 1997; Neale et al., 2005; Pan et al., 2011). DSBs undergo a series of transitions, being repaired differently (Osman et al., 2011; Mercier et al., 2015; Wang and Copenhaver, 2018) into either a crossover (CO; reciprocal genetic exchange between homologous chromosomes) or a non-crossover (NCO), using as repair template either the sister chromatid or the homologous chromosome exchanging only short stretches of DNA. Erroneous meiotic DSB formation or repair may lead to univalent formation or chromosome breakage and thus genome instability, reducing fertility. In plants <10% of DSBs are repaired as COs, and DSBs/COs occur heterogeneously along chromosomes (Muyt et al., 2009; Mercier et al., 2015) limiting genetic variation generated during each meiotic cycle. In many crop species, COs are restricted to chromosome ends, inhibiting access to traits residing in ‘cold’ regions or creating ‘linkage drag’; therefore, modulating CO number and distribution is of interest for plant breeding (Phillips et al., 2013; Choulet et al., 2014; Li et al., 2015; Demirci et al., 2017; Lambing and Heckmann, 2018; Dreissig et al., 2019).
CO formation is tightly controlled in the following ways: (i) interhomologue bias, where recombination is favoured between homologous chromosomes; (ii) CO assurance, where at least one ‘obligate’ CO forms per chromosome essential for faithful chromosome segregation; and (iii) CO interference, where formation of one CO reduces the probability of another close by, thereby hampering CO clustering and limiting CO number per chromosome (Zickler and Kleckner, 2015). In Arabidopsis, at least two CO classes exist: ~85% of COs are catalysed by the ZMM proteins (MER3, HEI10, ZIP4, SHOC1, PTD, MSH4, and MSH5) and MLH1–MLH3 (Higgins et al., 2004, 2008b; Chen et al., 2005; Mercier et al., 2005; Wijeratne et al., 2006; Chelysheva et al., 2007, 2012; Macaisne et al., 2008), generating interference-sensitive class I COs; and part of the remaining ~15% are MUS81- or FANCD2-dependent interference-insensitive class II COs (Berchowitz et al., 2007; Higgins et al., 2008a; Kurzbauer et al., 2018).
Concurrently with HR, the meiotic nucleus undergoes extensive reorganization of the chromatin. Following S phase, sister chromatids are linked by cohesins and, during leptotene, are organized in chromatin loops tethered to a linear proteinaceous structure called the meiotic chromosome axis (Kleckner, 2006). In Arabidopsis, this structure comprises, for example, ASY1, ASY3, ASY4, SMC3, and REC8 (Caryl et al., 2000; Cai et al., 2003; Chelysheva et al., 2005; Lam et al., 2005; Ferdous et al., 2012; Chambon et al., 2018). DSBs form at preferred sites (hotspots) in the chromatin loops which are then tethered to the axes where they are repaired as COs or NCOs (Panizza et al., 2011). Following DSB formation, a proteinaceous structure, called the synaptonemal complex (SC), starts to form during zygotene by progressively polymerizing between homologous chromosome axes physically connecting them and promoting CO formation (Page and Hawley, 2004).
The Arabidopsis meiotic axis-associated protein ASY1 (Caryl et al., 2000; Armstrong et al., 2002; Sanchez-Moran et al., 2007) is a HORMA domain-containing protein (yeast HOP1, mouse HORMAD1/2, or rice PAIR2; Hollingsworth et al., 1990; Caryl et al., 2000; Nonomura et al., 2004; Fukuda et al., 2010) required for synapsis and HR. In different organisms it is involved in the interhomologue bias (Martinez-Perez and Villeneuve, 2005; Niu et al., 2005; Sanchez-Moran et al., 2007; Carballo et al., 2008; Kim et al., 2010). In plants, asy1 or pair2 mutants display asynapsis and univalents due to reduced chiasma formation (Ross et al., 1997; Caryl et al., 2000; Nonomura et al., 2004). In addition to axis and SC formation per se, dynamic regulation of these structures is critical for CO formation. Concomitant with ZYP1 loading, ASY1 becomes depleted from synapsed regions in a PCH2-dependent manner (Wojtasz et al., 2009; Chen et al., 2014; Lambing et al., 2015). PCH2 is a conserved AAA-ATPase with diverse functions in different organisms. Initially reported as a checkpoint protein in yeast (San-Segundo and Roeder, 1999) and other species (Bhalla and Dernburg, 2005; Joyce and McKim, 2009), it has also been implicated in numerous meiotic processes, including DSB/CO formation, interhomologue bias, interference, axis morphogenesis, synapsis, and inhibition of DSB formation at rDNA borders (Börner et al., 2008; Joshi et al., 2009., 2015; Wojtasz et al., 2009; Zanders and Alani, 2009; Vader et al., 2011; Zanders et al., 2011; Farmer et al., 2012; Miao et al., 2013; Lambing et al., 2015; Subramanian et al., 2016). In plants, Arabidopsis PCH2 is critical for axis remodelling as well as SC and CO formation (Lambing et al., 2015; Yang et al., 2020a, b) while rice PCH2 (CRC) is an integral component of the SC essential for DSB and CO formation as well as ASY1 assembly (Miao et al., 2013). Interestingly, in Arabidopsis pch2, CO rates are altered in some chromosome regions (Lambing et al., 2015).
To study the role of axis remodelling in a crop species, we characterized asy1 and pch2 mutants in Brassica rapa (2n=20). asy1 plants form axes but SC formation is defective, leading to a reduction in CO frequency. Interestingly, residual chiasmata are proportionally increased towards chromosome ends and are enriched in the major 45S rDNA-carrying chromosome; in particular, chiasmata form within or close to the 45S rDNA locus while this is not the case in the wild type (WT) or in pch2. pch2 plants form a partial SC while axes are not remodelled; that is, ASY1 loading during leptotene is reduced and ASY1 is not depleted from synapsed regions, leading to a reduction in CO frequency. PCH2-independent chiasmata are more skewed towards terminal chromosome regions. To corroborate this cytological observation, in Arabidopsis CO rates were established in different genetic intervals, revealing increased CO rates in telomeric chromosome regions at the expense of decreased (peri-)centromeric CO rates in pch2 compared with the WT. We conclude that ASY1 and PCH2 are critical for SC and CO formation during meiosis in B. rapa and that asy1 and pch2 show altered CO patterning. Due to pch2 showing only semi-sterility, it could potentially be an interesting target in breeding programmes to redistribute COs.
Materials and methods
Plant material
Brassica rapa cultivar R-o-18 and Arabidopsis ecotype Columbia (Col-0) were used as the WTs. The following B. rapa mutant lines were received from RevGenUK (https://www.jic.ac.uk/research-impact/technology-research-platforms/reverse-genetics/): asy1-13 (JI32391-B), asy1-14 (JI31044-B), pch2-9 (JI32373-B), and pch2-12 (JI32174-B). The following Arabidopsis T-DNA insertion lines (Alonso et al., 2003) in the Col-0 background were obtained from the T-DNA mutant collection at the Salk Institute Genomics Analysis Laboratory (SIGnAL, http://signal.salk.edu/cgi-bin/tdnaexpress) via the NASC (http://arabidopsis.info/): asy1-4 (SALK_046272) (Lambing et al., 2020), shoc1-1 (SALK_057589) (Macaisne et al., 2008), mus81-2 (SALK_107515) (Higgins et al., 2008a), and pch2-1 (Sail_1187_C06) (Lambing et al., 2015). Plants were grown in greenhouses under 16 h day/8 h night, at 16 °C/14 °C (B. rapa) and 21 °C/18 °C (Arabidopsis) day/night temperatures.
Genotyping
In B. rapa, PCR-based genotyping was performed using derived cleaved amplified polymorphic sequences (dCAPS) (Neff et al., 1998) with primers selected using the dCAPS Finder 2.0 tool (http://helix.wustl.edu/dcaps/) (Neff et al., 2002). A list of primers and restriction enzymes is shown in Supplementary Table S1. After PCR amplification, the resulting amplicons were digested with corresponding restriction enzymes at 37 °C overnight and resolved on 2.5% agarose gels. Primers used to genotype Arabidopsis T-DNA mutants are indicated in Supplementary Table S1.
RNA extraction and reverse transcription–PCR
Total RNA of sample tissues was extracted with an RNeasy Plant Mini Kit (Qiagen) performing DNase digestion with the RNase-Free DNase Set (Qiagen). Reverse transcription was performed in a 20 µl reaction employing 750 ng of total RNA with a Tetro cDNA synthesis kit (Bioline) using oligo(dT) primers. Expression of Actin was evaluated as technical control for the integrity of the RNA/cDNA. A 5 µl aliquot of undiluted cDNA was used as template for 36 (PCH2-12) and 28 (Actin) PCR cycles. Primers used are listed in Supplementary Table S1.
Cytology and microscopy
Cytological procedures were carried out as described (Armstrong et al., 2009) with minor modifications. 5S (pCT4.2; Campell et al., 1992) and 45S (pTa71; Gerlach and Bedbrook, 1979) rDNA fluorescence in situ hybridization (FISH) probes were labelled by nick translation with Texas Red and Atto488 (NT labelling kits, Jena Biosciences). Chromosome spreading for immunostaining in B. rapa was done using five anthers per slide; digestion was done during 8 min in a moist chamber at 37 °C, disrupting the material with a brass rod after the first 4 min; and spreading was done with 1.5% lipsol. Immunostaining in Arabidopsis was performed on acid chromosome spreads from male meiocytes as described (Lambing et al., 2020). The following antibodies and dilutions were used: anti-ASY1 (rabbit/rat, Sanchez-Moran et al., 2007, 1:500), anti-ZYP1 (rat/guinea pig, Higgins et al., 2005, 1:500), anti-SMC3 (rat, Ferdous et al., 2012, 1:300), anti-ASY3 (rabbit, Ferdous et al., 2012, 1:200), anti-MLH1 (for B. rapa rabbit, Jackson et al., 2006; for Arabidopsis rabbit, Chelysheva et al., 2010, 1:200), and anti-HEI10 (rat, Lambing et al., 2015, 1:200). ASY1 intensity in B. rapa asynapsed versus synapsed regions was measured as described (Lambing et al., 2015). Quantification of ASY1 signal intensity in Arabidopsis leptotene nuclei was performed according to Ziolkowski et al. (2017). In brief, Arabidopsis images were acquired as z-stacks of 10 images with an optical section of 0.2 µm each. The maximum fluorescence intensity projection for each cell was generated, and total signal intensity was quantified using ImageJ. A region adjacent to the chromatin was used to quantify the background level that was deducted from the total signal intensity. Each signal intensity was normalized by the mean WT signal intensity for comparison. Quantification of ASY1 signal intensity in B. rapa leptotene nuclei was performed in a similar way, but images were acquired as a single plane, and measurements were done with the corresponding microscope software. Images were acquired with a Nikon Eclipse 90i fluorescence microscope equipped with a Nikon DS-Qi1Mc digital camera and NIS-Elements-F software, or a Nikon Eclipse Ni-E equipped with a Nikon DS-Qi2 camera and NIS-Elements-AR version 4.60 software (Nikon, Tokyo, Japan). Images were processed with GIMP 2.10 (www.gimp.org).
Recombination measurement at genetic intervals
Recombination measurements for the intervals I5b, I2f, I2g, I5a, I5d, and I5c using fluorescent pollen (Berchowitz and Copenhaver, 2008) are from the study of Lambing et al. (2015); 420 and 5.11 are seed-based fluorescent assays (Melamed-Bessudo et al., 2005; Wu et al., 2015). Fluorescent seeds and recombination measurements were calculated using Cell Profiler as previously described (Ziolkowski et al., 2015). CEN3 is a pollen-based fluorescent assay (Berchowitz and Copenhaver, 2008; Yelina et al., 2012). Fluorescent pollen was manually recorded under an epifluorescence microscope. and recombination measurements were calculated as previously reported (Lambing et al., 2015).
Results
Isolation of Brassica rapa asy1 and pch2
The B. rapa genome encodes ASY1 (Bra004222) and PCH2 (Bra013827) on chromosomes A07 and A01, respectively. One region spanning ~1 kb within ASY1 and within PCH2 were screened in a B. rapa R-o-18 TILLING (targeting induced local lesions in genomes) population (Stephenson et al., 2010) by the RevGenUK TILLING service (https://www.jic.ac.uk/research-impact/technology-research-platforms/reverse-genetics/) to isolate putative asy1 and pch2 loss-of-function alleles.
For ASY1, two lines with predicted premature stop codons were selected (Fig. 1A): asy1-13 (1255C>T, Q143>Stop) and asy1-14 (819G>A, W72>Stop) predicted to encode 24% and 12% of the native protein length, respectively. Sequencing analysis in each line confirmed the presence of a single nucleotide polymorphism (SNP).
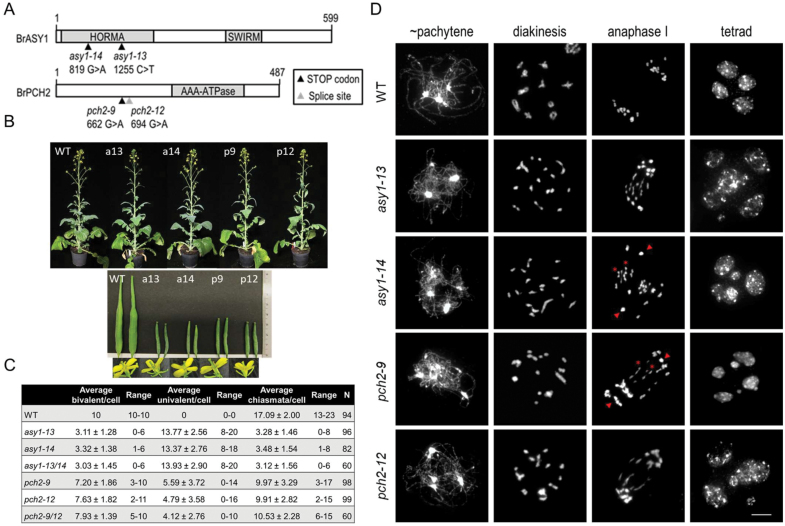
Fig. 1.
asy1 and pch2 plants display reduced fertility due to defects during meiosis. (A) Schematic representation of Brassica rapa ASY1 and PCH2 including mutations analysed (arrowheads). (B) B. rapa plant phenotypes (upper panel) including siliques and flowers (lower panel): WT, asy1-13 (a13), asy1-14 (a14), pch2-9 (p9), and pch2-12 (p12). (C) Average, SD, and range of bivalents, univalents, and chiasmata per cell in analysed lines. n=number of cells; a minimum of two independent plants per genotype. (D) B. rapa WT, asy1, and pch2 male meiotic chromosome spreads. In both mutants, synapsis and chiasma formation are impaired, leading to the occurrence of univalents at diakinesis that results in unequal chromosome segregation and unbalanced tetrads. Note, univalents in the mutants could migrate complete to a pole (arrowheads) or separate chromatids (asterisks) during anaphase I. DNA is counterstained with DAPI and shown in grey. Scale bar=10µm.
For PCH2, one line with a predicted premature stop codon and a second line with an SNP in an intron–exon boundary were selected (Fig. 1A): pch2-9 (662G>A, W143>Stop), predicted to encode 29% of the native protein length, and pch2-12 (694G>A, intron4–exon4 boundary). SNPs were confirmed by sequencing. Reverse transcription–PCR revealed the presence of alternative splice variants in pch2-12 (Supplementary Fig. S1). Although the presence of the PCH2 WT transcript cannot be excluded, pch2-12 probably results in a null mutation as pch2-12 is allelic to pch2-9 in terms of meiotic fidelity (see below). PCH2 expression is found in flower buds (containing cells undergoing meiosis) and also in leaves, similar to the expression pattern found for other meiotic genes (Caryl et al., 2000; Grelon et al., 2001).
For all lines, three backcrosses to the WT (R-o-18) were performed in order to decrease the secondary mutation load. Data presented correspond to plants from the third backcross generation.
asy1 and pch2 plants have reduced fertility due to defects in meiosis
All plants exhibited normal vegetative growth and development (Fig. 1B). However, plants homozygous for any of the SNPs showed sterile anthers, short siliques, and reduced seed set, while corresponding heterozygous or WT TILLING plants were similar to the WT (Fig. 1B; Supplementary Table S2). To explore whether reduced fertility was due to meiotic defects, we performed male meiotic chromosome spread analysis. During WT meiosis (Fig. 1D; Supplementary Fig. S2a), unpaired chromosomes appear as thin threads during leptotene. During zygotene, chromosomes start to align and synapse, reaching full synapsis visible as thick threads at pachytene. At diakinesis, homologous chromosomes are visible as bivalents physically connected by chiasmata (cytological manifestation of COs). At metaphase I, 10 bivalents align at the equatorial plate and 10 homologous chromosomes migrate to opposite poles during anaphase I. During the second meiotic division, chromosomes align at metaphase II and 10 chromatids separate to opposite poles during anaphase II, producing four haploid products in a balanced tetrad.
In asy1, thick chromatin threads indicative of synapsis were not observed (Fig. 1D). During diakinesis and metaphase I, most chromosomes appeared as univalents, indicating a failure to form COs. During anaphase I, univalents either segregated to one pole or showed precocious separation of sister chromatids (Fig. 1D), leading to unbalanced tetrads and micronuclei. The WT showed invariably 10 bivalents with a mean of 17.1±2 chiasmata per cell, while asy1 showed univalents in all cells. Reduced chiasma values for both asy1 lines (3.28±1.46 and 3.48±1.54 chiasmata per cell for asy1-13 and asy1-14, respectively) were not significantly different (Student’s t-test, P=0.39, n=96 asy1-13 and n=82 asy1-14) (Fig. 1C). Typically, bivalents formed rods (one chiasma). Only 12.4% of cells were found with one ring bivalent (at least one chiasma on each chromosome arm) and just 2.2% of cells had two ring bivalents (22/178 and 4/178 cells, respectively). No difference was found in asy1-13/14 compared with each single mutant, confirming that the mutations are allelic (Fig. 1C; Supplementary Fig. S2c; Supplementary Table S2). Moreover, heterozygous ASY1 plants pollinated with WT pollen produced long siliques with WT seed levels, while WT-pollinated homozygous asy1 plants produced short siliques with zero seeds, suggesting that female meiosis is also defective. TILLING plants, WT or heterozygous for the mutations, showed WT-like meiosis (Supplementary Fig. S2b). Together, reduced fertility in asy1 is based on defective synapsis and CO formation, and unbalanced chromosome segregation.
In pch2 plants, cells with complete synapsis were never observed. At diakinesis/metaphase I, 89% of cells showed a mixture of bivalents and univalents (0–16 univalents per cell) that led to unbalanced tetrads and micronuclei (Fig. 1D). Univalents during anaphase I either separated chromatids or migrated to one pole (Fig. 1D). WT or heterozygous TILLING plants for PCH2 showed WT-like meiosis (Supplementary Fig. S2b). The mean bivalent number was 7.20±1.86 (range 3–10) with an average of 9.97±3.29 (range 3–17) chiasmata per cell in pch2-9, and 7.63±1.82 (range 2–11) bivalents with an average of 9.91±2.82 (range 2–15) chiasmata per cell in pch2-12 (Fig. 1C). Chiasma values for both lines were not significantly different (Student’s t-test, P=0.89, n=98 pch2-9 and n=99 pch2-12). Similar to asy1, ring bivalents co-existed with univalents. Notably, 46% of all cells showed ≥10 chiasmata per cell (a number that would be sufficient for all chromosome pairs to acquire at least one obligate CO), together with univalents. An allelism test crossing revealed no difference between pch2-9/12 and pch2-9 or pch2-12 in terms of chiasma formation (Fig. 1C; Supplementary Fig. S2c), supporting that both mutations are allelic. However, in pch2-12, despite three backcrosses, a slight reduction in fertility was found among segregating families independent of PCH2 (Supplementary Table S2). Thus, we focused our analysis on pch2-9.
Increased chiasmata in the major 45S rDNA locus-carrying chromosome in asy1
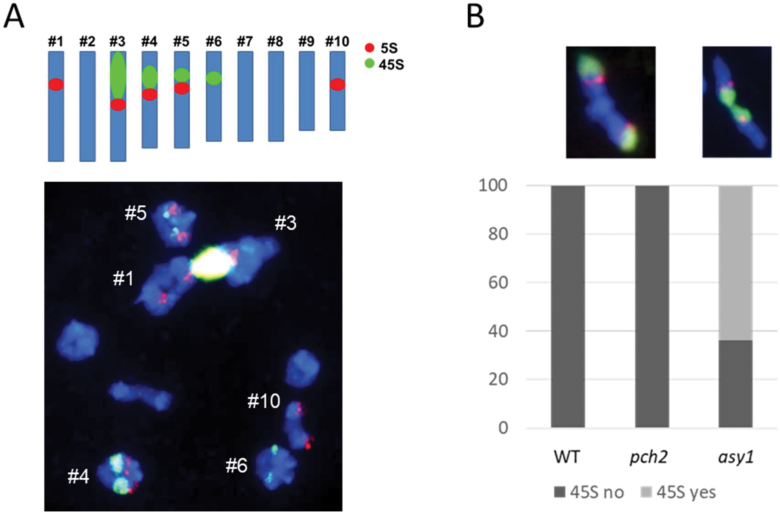
To identify whether in asy1 bivalents formed preferentially between any particular chromosome pair(s), we performed FISH with 5S and 45S rDNA probes (Fig. 2). The number of 5S and 45S signals in B. rapa differs among varieties (Fukui et al., 1998; Snowdon et al., 2002; Koo et al., 2004; Lim et al., 2005; Mun et al., 2010; Xiong and Pires, 2011; Perumal et al., 2017) and has already been discussed (e.g. Hasterok et al., 2006; Boutte et al., 2020). In B. rapa R-o-18, we found four 45S and five 5S FISH signals, and assigned them to the chromosomes organized from the largest to the smallest, thereby distinguishing chromosome pairs #1 (largest chromosome, carrying 5S signals), #3 [large block of 45S, the nucleolar organizing region (NOR), and small 5S, probably corresponding to chromosome A03], #4 and #5 (both containing 5S and 45S, with a larger 45S locus in #4), #6 (45S only), and #10 (smallest chromosome, 5S only) (Fig. 2A).
Fig. 2.
The major 45S rDNA locus-carrying chromosome arm shows increased chiasmata in asy1 but not in pch2 or the WT. (A) Top: B. rapa chromosomes organized by size with 5S and 45S rDNA clusters detected by FISH. Bottom: a representative WT cell after FISH. (B) Percentage of #3 rod bivalents with a chiasma in the 45S-carrying arm (‘45S yes’, example FISH image right) or in the opposite arm (‘45S no’, example FISH image left). Total number of rod bivalents scored: WT 41, pch2 16, asy1 36. FISH images: 5S (red), 45S (green); DNA is counterstained with DAPI and shown in blue.
In asy1, except for #3, all discernible chromosomes including the largest and the smallest appeared as univalents at similar frequencies (79–86%) (Supplementary Table S3). However, #3 which possesses the NOR with the largest 45S block, appeared as univalent in only 37% of cells. From the 63% #3 bivalents, 84% were rods and 16% rings; and from the #3 rod bivalents, in 64% of asy1 cells the chiasma was cytologically associated with the 45S rDNA; that is, the chiasma was found either inside the NOR or distal to it (Fig. 2B). In contrast to asy1, in WT and pch2 #3 rod bivalents, the single chiasma was never associated with the NOR but instead was invariably formed in the opposite arm (Fig. 2B).
Chromosome axes form in asy1 but fail to assemble a synaptonemal complex
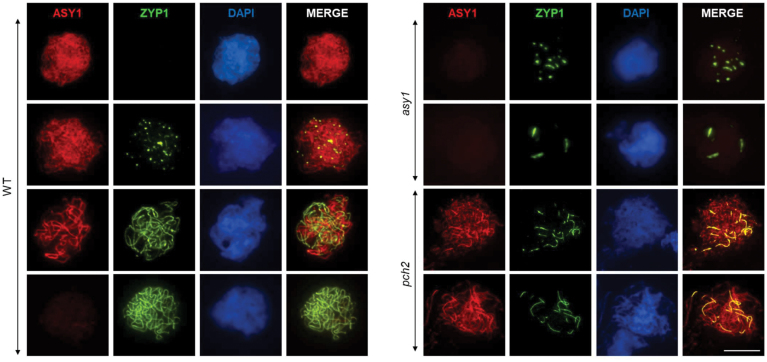
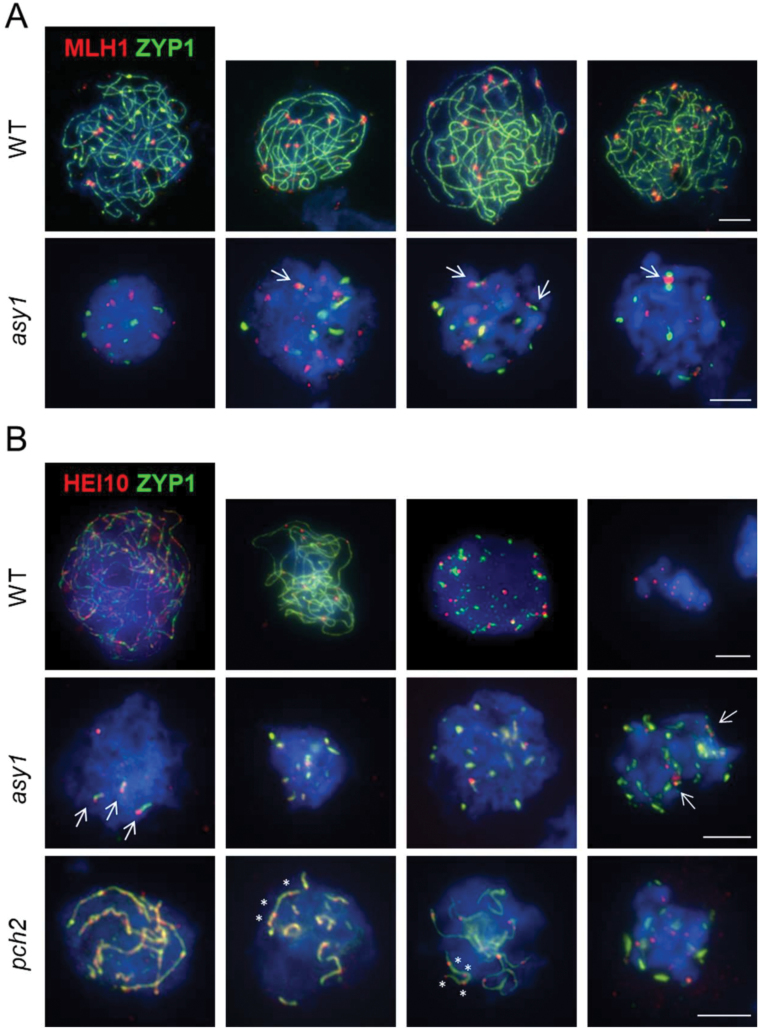
The asy1 mutants encode <25% of the native protein length, thus having a truncated or deleted HORMA domain and closure motif, critical for ASY1 localization (West et al., 2019; Yang et al., 2020b). To determine if the truncated ASY1 protein localizes to the axes and an SC forms in the asy1 cells, we performed ASY1 and ZYP1 (transverse filament protein of the SC; Higgins et al., 2005) immunolocalization. In the WT (Fig. 3), ASY1 localized to chromosome axes during leptotene. Once ASY1 was fully polymerized, ZYP1 initially formed foci and then short stretches which progressively elongated until all homologous chromosomes were fully synapsed. In asy1 (Fig. 3), ASY1 could not be detected. ZYP1 predominantly formed foci (from <5 to >20, possibly in a stage-dependent manner) and rarely elongated beyond forming short stretches, indicating that SC formation is largely impaired. In late pachytene-/early diplotene-like cells, ZYP1 formed aggregates probably due to its inability to assemble the SC (Zickler and Kleckner, 1999).
Fig. 3.
Synapsis is defective in asy1 and pch2. Immunolocalization of ASY1 (red) and ZYP1 (green) in the WT, asy1, and pch2. In asy1, ASY1 is not detected and ZYP1 forms foci in zygotene–pachytene cells and aggregates in late pachytene–diplotene cells. In pch2, different from the WT, ASY1 does not get depleted from the axes and appears highly abundant in limited synapsed regions co-localizing with ZYP1. DNA is counterstained with DAPI and shown in blue. Scale bar=10µm.
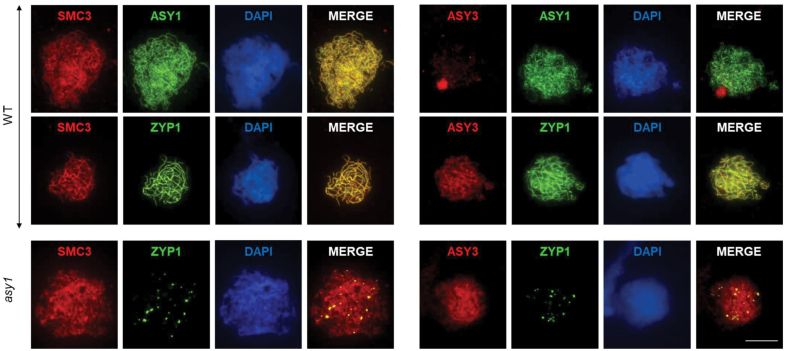
To analyse axis morphogenesis in asy1, we performed immunolocalization of the cohesin subunit SMC3 (Lam et al., 2005) and the coiled-coil protein ASY3 (Ferdous et al., 2012). In the WT (Fig. 4), SMC3 was present from leptotene to pachytene, co-localizing initially with ASY1 and later on with ZYP1. Upon synapsis, ASY1 signals became faint while SMC3 persisted, gradually thickening as synapsis proceeded. ASY3 initially localized to the nucleolus during leptotene similar to Arabidopsis (Ferdous et al., 2012) before it co-localized with ASY1 and then with ZYP1; however, ASY3 signals were patchier and less linear than those of ZYP1. In asy1 (Fig. 4), both SMC3 and ASY3 localized to the axis following WT dynamics. Thus, ASY1-independent axes form but are insufficient to support SC formation, resulting in reduced CO formation.
Fig. 4.
ASY3 and SMC3 localize to chromosome axes in asy1. Immunolocalization of SMC3 (red, left) and ASY3 (red, right) together with ASY1 (green, WT top) and ZYP1 (green, WT bottom and asy1). ASY3 and SMC3 display similar dynamics in WT and asy1 cells. DNA is counterstained with DAPI and shown in blue. Scale bar=10µm.
Class I and class II crossovers are reduced in asy1
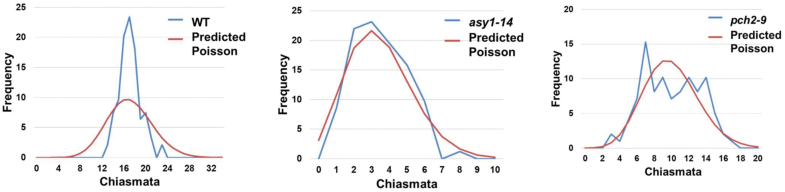
CO formation was reduced but not abolished in asy1. From 178 asy1 cells analysed, only one cell showed 20 univalents, whereas in all others between one and eight chiasmata were observed. To evaluate the nature of the remaining COs in asy1, we analysed the chiasma frequency distribution per cell. In the WT, the majority of COs are sensitive to interference, which leads to a non-random numerical distribution between cells. As a result, the mean chiasma frequency significantly deviates from a Poisson distribution [χ 2(R-o-18)=46.32, P<0.0001, n=94] (Fig. 5). However, in asy1, the chiasma frequency per cell was not significantly different from a Poisson distribution [χ 2(asy1-14)=7.79, P=0.25, n=82] (Fig. 5), suggesting that residual ASY1-independent COs are randomly distributed between cells.
Fig. 5.
Chiasma frequency distribution follows a Poisson distribution in asy1 and pch2. Chiasma frequency distribution (blue) and predicted Poisson distribution (red) for the WT, asy1-14, and pch2-9. In the mutants, chiasma frequency distribution does not significantly deviate from a Poisson-predicted distribution.
To better understand CO formation in asy1, we performed immunolocalization of the class I CO marker MLH1 (Jackson et al., 2006) together with ZYP1. We scored on average 9.44±0.72 (n=37) and 8.7±4.79 (n=10) MLH1 foci per cell in the WT and asy1, respectively (Fig. 6A). The MLH1 foci number per cell was highly variable in asy1 (1–15) but not in the WT (8–11). While in the WT MLH1 foci typically overlapped with ZYP1, in asy1 only an average of 3.9 MLH1 foci co-localized with residual ZYP1 foci (range 0–9). This manifested in different ways: ZYP1 stretches appeared to originate from MLH1 foci, two MLH1 foci were ‘bridging’ a ZYP1 stretch, or an MLH1 focus formed between two ZYP1 foci (Fig. 6A). Assuming that only ZYP1-associated MLH1 foci or a subset of these are CO competent, at least some of the residual chiasmata in asy1 could be ZMM-dependent class I COs.
Fig. 6.
Class I CO formation in asy1 and pch2. Immunolocalization of MLH1 (A, red) and HEI10 (B, red) together with ZYP1 (green) in the WT, asy1, and pch2. In asy1, a subset of MLH1/HEI10 foci co-localize with ZYP1 foci/short stretches (arrows). In pch2, several HEI10 signals appear close to each other in a single ZYP1 stretch (asterisks). Note, to depict HEI10 dynamics in the WT, cells are shown ranging from zygotene (left) to diakinesis (right), whereas in all other cases representative examples of cells used for quantification are shown. DNA is counterstained with DAPI and shown in blue. Scale bars=5µm.
We also immunolocalized HEI10 (Chelysheva et al., 2012; Wang et al., 2012) together with ZYP1. In the WT, numerous small HEI10 foci found during early pachytene progressively fade, leaving on average 11.31±1.71 bright HEI10 foci per cell (n=27, range 8–15) during diplotene–diakinesis (Fig. 6B). In asy1, this number was significantly lower and more variable (9±3.3, n=19, range 4–14). This variability could be attributed to the difficulty in cytologically defining meiotic prophase stages in asy1 and it is possible that some of the observed HEI10 foci will not mature in CO sites. However, as in the WT, in asy1, HEI10 foci typically co-localized with ZYP1. Similar to MLH1, short ZYP1 stretches originated from HEI10 foci or two HEI10 foci bridged a ZYP1 stretch. Altogether, our data suggest that interference-sensitive COs are designated in asy1, but the synaptic defect compromises CO maturation. As a result, an obligate CO fails to form between all chromosome pairs, and those COs that do mature exhibit a random distribution.
Next, we asked whether class II COs also form in the absence of ASY1 by taking advantage of available Arabidopsis resources. We crossed Arabidopsis asy1 either with shoc1 (involved in class I CO formation; Macaisne et al., 2008, 2011) or with mus81 (involved in class II CO formation; Berchowitz et al., 2007; Higgins et al., 2008a), and compared the chiasma frequency in these lines. Under our growth conditions, Arabidopsis asy1 showed 2.3±0.88 bivalents and 2.77±1.22 chiasmata per cell (Table 1), shoc1 showed 2.38±1.04 bivalents and 2.79±1.41 chiasmata per cell, and mus81 invariably showed five bivalents. In asy1/shoc1 and in asy1/mus81, fewer chiasmata formed than in asy1; in asy1/shoc1, we found a reduction of ~72% and in asy1/mus81 a reduction of ~21% compared with asy1. This suggests that residual COs in asy1 are both class I and II.
Table 1.
ASY1-independent CO formation in Arabidopsis thaliana
| Average bivalent/cell | Range | Average chiasmata/cell | Range | Average ring/cell | Range | n | |
|---|---|---|---|---|---|---|---|
| asy1 | 2.30±0.88 | 1–4 | 2.77±1.22 | 1–5 | 0.47±0.63 | 0–2 | 30 |
| shoc1 | 2.38±1.04 | 0–5 | 2.79±1.41 | 0–7 | 0.41±0.61 | 0–2 | 34 |
| mus81 | 5±0 | 5–5 | 30 | ||||
| asy1/shoc1 | 0.69±0.69 | 0–2 | 0.78±0.91 | 0–3 | 0.09±0.30 | 0–1 | 32 |
| asy1/mus81 | 2.14±1.01 | 1–4 | 2.18±1.09 | 1–5 | 0.04±0.19 | 0–1 | 28 |
Average, SD, and range of bivalents, chiasmata, and ring bivalents per cell in Arabidopsis asy1, shoc1, mus81 single mutants and asy1/shoc1 and asy1/mus81 double mutants. Note: the bivalent frequency between asy1 and asy1/mus81 was not significantly different, while the number of ring bivalents in asy1/mus81 was reduced, causing the observed reduction in chiasma frequency. n=number of cells.
Axis remodelling is defective in pch2
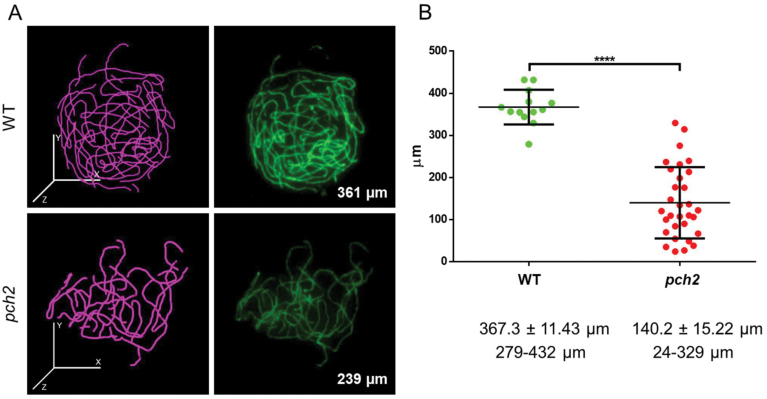
In pch2, ASY1 loaded onto chromosomes and, once ASY1 was fully polymerized, ZYP1 appeared initially as foci and later on as stretches. However, full ZYP1 polymerization was never observed (Fig. 3). Indeed, measuring ZYP1 extension in WT and pch2-9 nuclei revealed a large variation in the extension of the SC across pch2 cells and, on average, a reduction of 62% in SC length (Mann–Whitney–Wilcoxon test, P<0.0001, n=13 WT and n=31 pch2) (Fig. 7).
Fig. 7.
Compromised SC formation in pch2. (A) 3D reconstruction of the SC with simple neurite tracer (ImageJ) (pink, left) of representative pachytene cells immunolabelled with ZYP1 (green, right) in the WT and pch2. SC length is indicated in the bottom right corner. (B) SC measurements in individual WT and pch2 cells. Average, SD, and range are indicated.
Furthermore, in contrast to the WT, in pch2 ASY1 remained brightly stained following ZYP1 installation (Fig. 3), suggesting that ASY1 is not depleted from chromosome axes at synapsed regions. In various organisms including Arabidopsis, PCH2 is needed for the programmed removal of ASY1 during zygotene from chromosome axes concomitant with SC installation (Börner et al., 2008; Martinez-Perez et al., 2008; Wojtasz et al., 2009; Lambing et al., 2015). To validate this observation, we measured ASY1 intensity (Supplementary Table S4) and found that in the WT there was a 55% reduction in ASY1 intensity in synapsed versus asynapsed regions, whereas in pch2 there was a 21% increase. This increase in pch2 is probably based on the juxtaposition of the two homologous chromosome axes, leading to an increase in signal intensity per pixel.
Interestingly, in asynapsed regions, ASY1 intensity was consistently lower in pch2 than in the WT. In Arabidopsis, PCH2 is involved not only in ASY1 removal, but also in its loading (Yang et al., 2020b). To check whether ASY1 loading is also defective in pch2, we measured ASY1 intensity in leptotene whole nuclei. Despite substantial variation in ASY1 intensity values among slides/experiments, ASY1 intensity was consistently lower in pch2 than in the WT (range 14–76%). To corroborate this cytological observation, we also immunostained ASY1 in Arabidopsis WT and pch2, and found that ASY1 signal intensity was significantly reduced at leptotene in Arabidopsis pch2 (Mann–Whitney–Wilcoxon test, P=4.01e-03, n=24 Col-0 and n=24 pch2) (Supplementary Fig. S3). In summary, our data indicate that PCH2 in B. rapa is important for initial ASY1 loading during leptotene and programmed removal of ASY1 from chromosome axes concomitant with SC installation during zygotene.
Crossover patterning in pch2: distalized crossover at the expense of interstitial crossover
In pch2, chiasma frequency was highly variable and in ~50% of cells the obligatory CO was lost. To better understand how CO control is altered, we analysed CO frequency distribution. We found that chiasma frequency in pch2 did not significantly deviate from a Poisson-predicted distribution [χ 2(pch2-9)=14.70, P=0.26, n=98)] (Fig. 5), revealing a random distribution of chiasmata between cells. To identify whether interference-sensitive class I COs form in pch2, we performed immunolocalization of HEI10 together with ZYP1 (Fig. 6B). In pch2-9, we found an average of 10.5±3.15 (range 4–15) HEI10 foci during diplotene–diakinesis. These values are comparable with the average chiasma number in pch2-9 (9.97±3.29, range 3–17). Interestingly, we found nuclei where several HEI10 foci localized in close proximity onto one ZYP1 stretch (Fig. 6B, asterisks) together with ZYP1 stretches devoid of HEI10 foci. Thus, a majority of COs in pch2 showing a random distribution according a Poisson-predicted distribution probably belong to class I COs (marked by HEI10 foci).
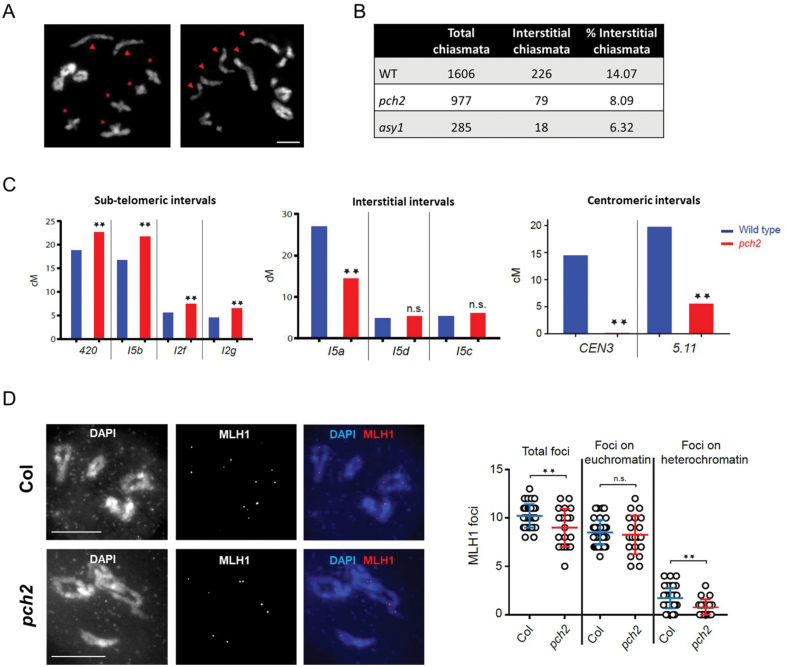
We next asked whether residual chiasmata in pch2 are positioned differently compared with the WT. We scored the number of terminal versus interstitial chiasmata and found that the percentage of interstitial chiasmata among all chiasmata was 8% in pch2 and 14% in the WT (Fig. 8A, B). Similarly, in asy1, among residual chiasmata, only 6% were interstitial while the majority were found towards chromosome ends (Fig. 8B). Thus, cytologically residual chiasmata in pch2 and asy1 are proportionally more frequently located in terminal chromosome regions compared with the WT.
Fig. 8.
Distalization of chiasmata in pch2 and in asy1. (A) Representative example of B. rapa diakinesis with a majority of chiasmata being interstitial (asterisks) in the WT (left) and distal (arrowheads) in pch2 (right). DNA is counterstained with DAPI and shown in grey. Scale bar=5 µm. (B) Quantification of interstitial versus total chiasmata in B. rapa WT, pch2, and asy1 reveals reduced interstitial CO frequency in the mutants. (C) Recombination frequency in Arabidopsis at genetic intervals defined with genes coding for fluorescent proteins. Genetic intervals are classed based on their locations on the chromosomes. Asterisks represent statistically significant differences (generalized linear model) and ‘n.s.’ means non-significant. (D) Left: staining of MLH1 (red) and DAPI (blue) at diakinesis in Arabidopsis WT and pch2. Scale bars=10 µM. Right: plots showing total MLH1 foci count, MLH1 foci count on euchromatin, and foci count on heterochromatin per cell. Error bars represent the SD. Asterisks represent statistically significant differences (Mann–Whitney–Wilcoxon test) and ‘n.s.’ means non-significant.
To corroborate our cytological finding on altered chiasma patterning in pch2 with proportionally increased terminal and decreased interstitial chiasmata, we asked whether in Arabidopsis pch2 a similar situation occurs. We re-analysed published recombination data from six genetic intervals (Lambing et al., 2015) and extended this study by analysing recombination frequency in three more genetic intervals. We classed the genetic intervals based on their chromosomal locations as subtelomeric, interstitial, and centromeric. We found that subtelomeric intervals show increased recombination frequency, while interstitial intervals show few changes, and centromeric intervals show a drastic reduction in recombination frequency in pch2 (Fig. 8C). These data suggest that COs are repressed in the heterochromatic centromeric regions and that they are distalized in the absence of PCH2. Immunostaining of MLH1 revealed that total MLH1 foci are reduced in pch2 (10.4 versus 9.0, Mann–Whitney–Wilcoxon test, P<0.01, n=41 WT and n=20 pch2) and that the reduction is mostly seen on the DAPI-dense regions that are representative of the heterochromatic (peri-)centromeric regions (1.7 versus 0.8, Mann–Whitney–Wilcoxon test, P<0.001, n=41 WT and n=20 pch2) (Fig. 8D). Together, these data reveal that PCH2 is required for CO patterning control.
Discussion
In this work, we have characterized asy1 and pch2 in B. rapa. Both mutants display reduced chiasma formation and univalents at metaphase I that lead to unbalanced gametes and reduced fertility, highlighting the importance of the axis and SC morphogenesis for CO formation. Depletion of ASY1 or PCH2 strongly impacts SC formation, resulting in differential patterning of residual chiasmata being proportionally enriched towards chromosome ends.
ASY1-independent meiotic chromosome axes are insufficient for synaptonemal complex and crossover formation
Our analyses indicate that asy1-13 and asy1-14 are allelic and probably represent null mutations. Similar to asy1 in other plant species (Armstrong et al., 2002; Nonomura et al., 2004), B. rapa asy1 is largely, if not completely, asynaptic (Figs 1D, 3). ZYP1 forms foci and occasionally short stretches which are commonly distorted in appearance (Fig. 3), probably representing ZYP1 aggregates (Zickler and Kleckner, 1999) or polycomplexes forming at sites where ZYP1 polymerization/extension along lateral elements is impaired due to a defect in ASY1-dependent interhomologue bias and recombination progression (Higgins et al., 2005).
Despite the absence of an SC, residual chiasmata form in B. rapa asy1 (Fig. 1C, D). This is reminiscent of Arabidopsis and the corresponding hop1 mutant in budding yeast (Hollingsworth and Johnson, 1993; Ross et al., 1997) but contrasts with rice pair2 (Nonomura et al., 2004). Axis mutants asy3 or asy4 in Arabidopsis also show reduced COs (but to a lesser extent than asy1), and asy1 is epistatic to them in terms of chiasma frequency (Ferdous et al., 2012; Chambon et al., 2018), suggesting that ASY1 is needed upstream from ASY3/ASY4 to regulate DMC1 dynamics and thus interhomologue recombination (Sanchez-Moran et al., 2007). Moreover, ASY4 is required for axial organization of ASY1 and ASY3 in Arabidopsis (Chambon et al., 2018), and ASY3 for correct ASY1 localization in Arabidopsis, maize, and rice (Wang et al., 2011; Ferdous et al., 2012; Lee et al., 2015), while cytologically ASY3 and the cohesin SMC3 localize correctly in asy1 (Sanchez-Moran et al., 2007; Ferdous et al., 2012). Similarly, in B. rapa, SMC3 and ASY3 display WT-like axes localization in asy1 (Fig. 4), suggesting WT-like dynamics of the core axis-associated proteins despite subsequent synaptic and CO defects.
In Arabidopsis axis mutants, the majority of the remaining COs are dependent on the class I CO (ZMM) pathway similar to the WT; for example, in asy1, MLH1 foci form on bivalents (Lambing et al., 2020), in asy3/msh4 double mutants chiasma formation is abolished (Ferdous et al., 2012), or in asy4/zip4 and asy4/msh5 double mutants bivalent formation is reduced by ~95% compared with asy4 (Chambon et al., 2018). In B. rapa asy1, both HEI10 and MLH1 foci numbers scored around pachytene-/diplotene-like stages were highly variable compared with the WT, exceeding the number of cytologically scored chiasmata. This observation could be attributed to different causes. First, HEI10 foci numbers vary significantly through prophase I and therefore variation could stem from the difficulty in selecting late prophase cells due to complete absence of synapsis. Second, interestingly, in Arabidopsis asy1, dmc1 or haploid meiosis MLH1 foci were reported on univalent chromosomes, possibly representing sites of intersister repair (Cifuentes et al., 2013; Lambing et al., 2020). Similarly, HEI10 has been indicated to mark intersister events (Voelkel-Meiman et al., 2012) and it follows initial WT dynamics in mutants with severely impaired CO formation such as the ZMM mutant mer3 (Wang et al., 2012). Third, non-co-localizing HEI10 and MLH1 foci were found in B. napus allohaploids (Grandont et al. 2014). Finally, MLH1 in Sordaria is important for interlock resolution (Storlazzi et al., 2010), and although the presence of MLH1 at interlocks has not been shown, this could arise in asy1 due to defective SC polymerization. Assuming that only ZYP1-associated ZMM foci or a subset of these are CO competent (Ferdous et al., 2012), then probably only a proportion of observed foci will mature into class I CO sites.
In B. rapa asy1, chiasmata were strongly reduced but not abolished, and cytologically foci corresponding to class I CO markers HEI10 and MLH1 formed, suggesting that residual chiasmata may arise via the class I CO pathway. In support of this, by using available Arabidopsis resources, we showed that a majority of ASY1-independent chiasmata are dependent on the ZMM pathway, while a subset is also MUS81 dependent, indicative of class II CO formation (Table 1). Interestingly, as in the WT, roughly similar proportions of class I and class II COs arise in the Arabidopsis asy1 mutant, suggesting that both CO pathways are similarly dependent on ASY1. In future, to determine if the same applies in B. rapa, it will be interesting to evaluate CO formation in asy1/msh4 (class I CO-defective mutant with ~4 residual chiasmata; Blary et al., 2018).
Residual chiasmata in asy1 are proportionally enriched in terminal chromosome regions and on the major 45S rDNA-carrying chromosome
Residual chiasmata in B. rapa asy1 are proportionally enriched towards chromosome ends and, except for the major 45S rDNA-carrying chromosome, all discernible chromosomes appear as univalents with similar frequencies, suggesting no positive correlation between chromosome size and chiasma formation.
Terminal dominance of residual chiasmata was also found in Arabidopsis asy1 and further axis-associated mutants such as asy3 and asy4 (Sanchez-Moran et al., 2007; Ferdous et al., 2012; Chambon et al., 2018). It has been proposed that this could be a consequence of recombination initiating by telomeric regions (Sanchez-Moran et al., 2007) and that ASY1 could be antagonizing this telomere-led recombination (Lambing et al., 2020). This highlights the key role of axis components in regulating CO frequency and distribution.
Notably, residual chiasmata showed a strong bias towards the 45S rDNA-carrying chromosome and, in particular, towards the 45S rDNA chromosome end (Fig. 2). Due to limited cytological resolution, we cannot distinguish whether chiasmata involved ribosomal repeats or rather formed in non-ribosomal repeat-free regions distal to the 45S rDNA or intermingled with it. We prefer that CO enrichment is found in non-ribosomal repeat-free chromosome ends, as bivalents were typically linked via what seems to be cytologically a very terminal chiasma, and also because we could not see any signs of repair/chromosome segregation problems which would be likely to arise if recombination would occur within highly repetitive regions due to the possibility of non-allelic exchanges (Sasaki et al., 2010). This would agree with recent data from Arabidopsis indicating that exclusion of ASY1 from the NOR in chromosomes 2 and 4 early during prophase is implicated in restricting DSB formation and thus HR in the NOR (Sims et al., 2019). Nonetheless, chiasma enrichment towards the 45S rDNA-carrying chromosome (arm) has also been observed in Arabidopsis asy1 (Sanchez Moran et al., 2001; Lambing et al., 2020). Thus, in asy1, the rDNA probably promotes CO formation in the 45S rDNA-carrying chromosome (arm) (Sanchez Moran et al., 2001; Lambing et al., 2020).
pch2 plants fail to remodel ASY1 and only achieve partial synapsis leading to reduced chiasma formation
Both pch2-9 and pch2-12 appear allelic in terms of meiotic behaviour and probably represent null mutants. However, despite three backcrosses, compared with the WT, a slight reduction in fertility was found in pch2-12 segregating families independent of PCH2, suggesting secondary mutation load impacting overall plant fertility. Thus, data were acquired on pch2-9.
In B. rapa pch2, full synapsis was never observed (Figs 1D, 3) despite substantial variation in SC extension (based on ZYP1 immunolocalization) and, on average, SC length was ~62% reduced compared with the WT (Fig. 7). This reduction in SC length is similar to pch2 in Arabidopsis (~68%; Lambing et al., 2015) but differs from rice pch2/crc1, where SC formation is completely abolished (Miao et al., 2013). Notably, in rice pch2, no chiasmata form, while in B. rapa pch2 chiasmata are reduced by ~40% and in Arabidopsis pch2 by ~30%. Moreover, observed chiasma numbers are variable across cells, and often univalents occurred in cells with at least 10 chiasmata, suggesting a defect in CO assurance. Overall, despite species-specific differences, PCH2 in plants is critical for WT levels of synapsis and CO formation.
Brassica rapa pch2 plants form a partial SC while axes are not remodelled; that is, ASY1 loading during leptotene is reduced and ASY1 is not depleted from synapsed regions. This dual function of PCH2 in B. rapa is consistent with data from Arabidopsis and budding yeast (Börner et al., 2008; Lambing et al., 2015; Yang et al., 2020a, b), showing that PCH2 is critical for chromosomal localization of ASY1 as well as for depletion of ASY1 from synapsed regions. In Arabidopsis, in addition to PCH2, the axis proteins ASY3 and ASY4 are also critical for WT dynamics and correct localization of ASY1; for example, in asy4, ASY1 is not depleted from synapsed regions and ASY3 is involved in correct ASY1 recruitment to the chromosome axes (Ferdous et al., 2012; Chambon et al., 2018). Thus, a complex interplay between axis components and associated proteins exists, and not only axis formation but also axis morphogenesis is critical for HR, including DSB repair, template choice, SC formation and, ultimately, CO formation.
Redistribution of chiasmata in pch2
Residual COs in B. rapa pch2, contrary to Arabidopsis (Lambing et al., 2015), do not significantly deviate from a Poisson-predicted distribution which could be interpreted as suggesting that they are interference insensitive, typical for class II COs. However, the fact that the chiasma frequency distribution does not differ from a Poisson distribution only means that chiasma formation in the mutant has a high random component. In fact, HEI10 foci marking putative class I CO sites appear in pch2 with similar numbers as cytologically scored chiasmata (minimum CO number), with some nuclei having HEI10 foci closely spaced along ZYP1 stretches and at the same time ZYP1 stretches devoid of HEI10 foci (Fig. 6). Possibly maturation of designated COs might be compromised in B. rapa pch2 due to the defect in chromosome axis remodelling, resulting in a CO deficit as in Arabidopsis pch2 (Lambing et al., 2015). Notably, the average reduction in SC length (62%) is larger than the average reduction in chiasmata (~40%), suggesting that similar to Arabidopsis (Lambing et al., 2015), the mean reduction in CO frequency was not coordinated with that in SC length. A similar phenotype has been observed in Arabidopsis mutants for the kinesin PSS1 and the E1 enzyme of the neddylation complex AXR1 (Duroc et al., 2014; Jahns et al., 2014), both being strongly defective for synapsis, showing univalents at metaphase I, and with chiasma frequencies similar to the WT but COs being redistributed with closely spaced HEI10 foci along residual synapsed regions.
Furthermore, PCH2-independent chiasmata are cytologically skewed towards terminal chromosome regions in B. rapa. To support this cytological observation, in Arabidopsis pch2, CO rates were established in different genetic intervals, revealing increased CO rates in telomeric chromosome regions at the expense of decreased (peri-)centromeric CO rates compared with the WT (Fig. 8). Thus, considering terminal initiation of SC formation (Hurel et al., 2018), recombination events on limited extended ZYP1 stretches in pch2 might be proportionally enriched in distal regions, explaining the skewed chiasma distribution.
Altogether, in B. rapa, a majority of residual PCH2-independent COs are likely to be ZMM-dependent class I COs that are redistributed, being proportionally enriched towards chromosome ends.
Conclusions
Altering CO frequency and distribution is of special interest for plant breeding, since in many crop species CO numbers are limited and skewed towards chromosome ends, limiting recombination and access to naturally available genetic variation and creating linkage drag.
We have analysed mutants of axis-associated ASY1 and axis-remodelling PCH2. Both mutants show reduced CO formation due to defects in meiosis, but interestingly COs are redistributed and at least pch2 plants produce some viable seeds. Considering this altered patterning of chiasmata that arise via the class I and class II CO pathways in asy1 and pch2, it would be interesting to determine whether in combination with HEI10 overexpression (increasing class I CO) or hyperrec mutants (increasing class II COs) (Lambing et al., 2017). ASY1-/PCH2-independent chiasmata could be increased, thus potentially increasing bivalent formation and seed setting while maintaining an altered recombination pattern. In any case, considering their general influence on DSB and CO formation including DNA repair template choice, axis-associated or remodelling proteins such as ASY1 or PCH2 are interesting targets to modify meiotic recombination landscapes in the context of plant breeding.
Supplementary data
The following supplementary data are available at JXB online.
Table S1. Primers used in this study.
Table S2. Plant fertility in the B. rapa WT, and asy1 and pch2 mutants.
Table S3. Relative occurrence (%) of 5S/45S-labelled chromosomes as either univalents or bivalents.
Table S4. ASY1 immunofluorescence relative signal intensity in WT and pch2 cells.
Fig. S1. Alternative splicing of PCH2 in pch2-12.
Fig. S2. Male meiotic chromosome behaviour in B. rapa.
Fig. S3. Localization defect of ASY1 in Arabidopsis pch2.
Acknowledgements
We would like to acknowledge Jana Lorenz, Franziska Hartmann, and Steve Price for excellent technical assistance, Karen Staples at UoB and the gardeners at IPK for horticultural support, and all lab members for fruitful discussions. CL and IRH would like to thank Mathilde Grelon for sharing the MLH1 antibody. We would also like to thank the Bundesministerium für Bildung und Forschung (grant FKZ-031B0188), the IPK Gatersleben, the Biotechnology and Biological Sciences Research Council UK (grants BB/M004902/1, BB/J007633/1, and BB/M014908/), the European Union’s Seventh Framework Programme FP7/2007-2013 (grant KBBE-2009-222883), and the European Union’s Marie Sklodowska-Curie Innovative Training Network MEICOM (grant 765212-H2020-MSCA-ITN-2017) for financial support.
Glossary
Abbreviations
- CO
crossover
- DSB
DNA double-strand break
- FISH
fluorescence in situ hybridization
- HR
homologous recombination
- NCO
non-crossover
- SC
synaptonemal complex
- WT
wild type
Author contributions
MC, CL, and MP-P performed the experiments; MC, CL, MP-P, and KO analysed the data; SJA, IRH, ES-M, FCHF, and SH acquired funding; MC and SH drafted the manuscript; all authors edited and approved the final text.
Data availability
All data supporting the findings of this study are available within the paper and within its supplementary data published online.
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Caryl AP, Jones GH, Franklin FC. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. Journal of Cell Science 115, 3645–3655. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Sanchez-Moran E, Franklin FCH. 2009. Cytological analysis of arabidopsis thaliana meiotic chromosomes. In: Keeney S, ed. Meiosis: volume 2, cytological methods. Totowa, NJ: Humana Press, 131–145. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. 2008. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nature Protocols 3, 41–50. [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. 2007. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genetics 3, e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF. 2005. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science 310, 1683–1686. [DOI] [PubMed] [Google Scholar]
- Blary A, Gonzalo A, Eber F, et al. 2018. FANCM limits meiotic crossovers in Brassica crops. Frontiers in Plant Science 9, 368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Barot A, Kleckner N. 2008. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proceedings of the National Academy of Sciences, USA 105, 3327–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutte J, Maillet L, Chaussepied T, et al. 2020. Genome size variation and comparative genomics reveal intraspecific diversity in Brassica rapa. Frontiers in Plant Science 11, 577536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Dong F, Edelmann RE, Makaroff CA. 2003. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. Journal of Cell Science 116, 2999–3007. [DOI] [PubMed] [Google Scholar]
- Campell BR, Song Y, Posch TE, Cullis CA, Town CD. 1992. Sequence and organization of 5S ribosomal RNA-encoding genes of Arabidopsis thaliana. Gene 112, 225–228. [DOI] [PubMed] [Google Scholar]
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS. 2008. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 132, 758–770. [DOI] [PubMed] [Google Scholar]
- Caryl AP, Armstrong SJ, Jones GH, Franklin FC. 2000. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71. [DOI] [PubMed] [Google Scholar]
- Chambon A, West A, Vezon D, et al. 2018. Identification of ASYNAPTIC4, a component of the meiotic chromosome axis. Plant Physiology 178, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Diallo S, Vezon D, et al. 2005. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. Journal of Cell Science 118, 4621–4632. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M. 2007. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genetics 3, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Grandont L, Vrielynck N, le Guin S, Mercier R, Grelon M. 2010. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenetic and Genome Research 129, 143–153. [DOI] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, Lemhemdi A, Vrielynck N, Le Guin S, Novatchkova M, Grelon M. 2012. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genetics 8, e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Jomaa A, Ortega J, Alani EE. 2014. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proceedings of the National Academy of Sciences, USA 111, E44–E53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Zhang W, Timofejeva L, Gerardin Y, Ma H. 2005. The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. The Plant Journal 43, 321–334. [DOI] [PubMed] [Google Scholar]
- Choulet F, Alberti A, Theil S, et al. 2014. Structural and functional partitioning of bread wheat chromosome 3B. Science 345, 1249721. [DOI] [PubMed] [Google Scholar]
- Cifuentes M, Rivard M, Pereira L, Chelysheva L, Mercier R. 2013. Haploid meiosis in Arabidopsis: double-strand breaks are formed and repaired but without synapsis and crossovers. PLoS One 8, e72431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci S, van Dijk AD, Sanchez Perez G, Aflitos SA, de Ridder D, Peters SA. 2017. Distribution, position and genomic characteristics of crossovers in tomato recombinant inbred lines derived from an interspecific cross between Solanum lycopersicum and Solanum pimpinellifolium. The Plant Journal 89, 554–564. [DOI] [PubMed] [Google Scholar]
- Dreissig S, Mascher M, Heckmann S. 2019. Variation in recombination rate is shaped by domestication and environmental conditions in barley. Molecular Biology and Evolution 36, 2029–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Lemhemdi A, Larchevêque C, Hurel A, Cuacos M, Cromer L, Horlow C, Armstrong SJ, Chelysheva L, Mercier R. 2014. The kinesin AtPSS1 promotes synapsis and is required for proper crossover distribution in meiosis. PLoS Genetics 10, e1004674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer S, Hong EJ, Leung WK, Argunhan B, Terentyev Y, Humphryes N, Toyoizumi H, Tsubouchi H. 2012. Budding yeast Pch2, a widely conserved meiotic protein, is involved in the initiation of meiotic recombination. PLoS One 7, e39724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdous M, Higgins JD, Osman K, et al. 2012. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genetics 8, e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Daniel K, Wojtasz L, Toth A, Höög C. 2010. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Experimental Cell Research 316, 158–171. [DOI] [PubMed] [Google Scholar]
- Fukui K, Nakayama S, Ohmido N, Yoshiaki H, Yamabe M. 1998. Quantitative karyotyping of three diploid Brassica species by imaging methods and localization of 45s rDNA loci on the identified chromosomes. Theoretical and Applied Genetics 96, 325–330. [DOI] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. 1979. Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Research 7, 1869–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandont L, Cuñado N, Coriton O, Huteau V, Eber F, Chèvre AM, Grelon M, Chelysheva L, Jenczewski E. 2014. Homoeologous chromosome sorting and progression of meiotic recombination in Brassica napus: ploidy does matter! The Plant Cell 26, 1448–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G. 2001. AtSPO11-1 is necessary for efficient meiotic recombination in plants. The EMBO Journal 20, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen WK, Maluszynska J. 2006. Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Annals of Botany 97, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Armstrong SJ, Franklin FC, Jones GH. 2004. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes & Development 18, 2557–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FC, Jones GH. 2008a. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. The Plant Journal 54, 152–162. [DOI] [PubMed] [Google Scholar]
- Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FC. 2005. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes & Development 19, 2488–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Vignard J, Mercier R, Pugh AG, Franklin FC, Jones GH. 2008b. AtMSH5 partners AtMSH4 in the class I meiotic crossover pathway in Arabidopsis thaliana, but is not required for synapsis. The Plant Journal 55, 28–39. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, Byers B. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61, 73–84. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Johnson AD. 1993. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. Genetics 133, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurel A, Phillips D, Vrielynck N, Mézard C, Grelon M, Christophorou N. 2018. A cytological approach to studying meiotic recombination and chromosome dynamics in Arabidopsis thaliana male meiocytes in three dimensions. The Plant Journal 95, 385–396. [DOI] [PubMed] [Google Scholar]
- Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FC. 2006. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. The EMBO Journal 25, 1315–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahns MT, Vezon D, Chambon A, Pereira L, Falque M, Martin OC, Chelysheva L, Grelon M. 2014. Crossover localisation is regulated by the neddylation posttranslational regulatory pathway. PLoS Biology 12, e1001930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Barot A, Jamison C, Börner GV. 2009. Pch2 links chromosome axis remodeling at future crossover sites and crossover distribution during yeast meiosis. PLoS Genetics 5, e1000557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi N, Brown MS, Bishop DK, Börner GV. 2015. Gradual implementation of the meiotic recombination program via checkpoint pathways controlled by global DSB levels. Molecular Cell 57, 797–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce EF, McKim KS. 2009. Drosophila PCH2 is required for a pachytene checkpoint that monitors double-strand-break-independent events leading to meiotic crossover formation. Genetics 181, 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384. [DOI] [PubMed] [Google Scholar]
- Keeney S, Kleckner N. 1995. Covalent protein–DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proceedings of the National Academy of Sciences, USA 92, 11274–11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. 2010. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 143, 924–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. 2006. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194. [DOI] [PubMed] [Google Scholar]
- Koo DH, Plaha P, Lim YP, Hur Y, Bang JW. 2004. A high-resolution karyotype of Brassica rapa ssp. pekinensis revealed by pachytene analysis and multicolor fluorescence in situ hybridization. Theoretical and Applied Genetics 109, 1346–1352. [DOI] [PubMed] [Google Scholar]
- Kurzbauer MT, Pradillo M, Kerzendorfer C, et al. 2018. Arabidopsis thaliana FANCD2 promotes meiotic crossover formation. The Plant Cell 30, 415–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam WS, Yang X, Makaroff CA. 2005. Characterization of Arabidopsis thaliana SMC1 and SMC3: evidence that AtSMC3 may function beyond chromosome cohesion. Journal of Cell Science 118, 3037–3048. [DOI] [PubMed] [Google Scholar]
- Lambing C, Franklin FC, Wang CR. 2017. Understanding and manipulating meiotic recombination in plants. Plant Physiology 173, 1530–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C, Heckmann S. 2018. Tackling plant meiosis: from model research to crop improvement. Frontiers in Plant Science 9, 829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C, Kuo PC, Tock AJ, Topp SD, Henderson IR. 2020. ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proceedings of the National Academy of Sciences, USA 117, 13647–13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambing C, Osman K, Nuntasoontorn K, et al. 2015. Arabidopsis PCH2 mediates meiotic chromosome remodeling and maturation of crossovers. PLoS Genetics 11, e1005372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Kao YH, Ku JC, Lin CY, Meeley R, Jan YS, Wang CJ. 2015. The axial element protein DESYNAPTIC2 mediates meiotic double-strand break formation and synaptonemal complex assembly in maize. The Plant Cell 27, 2516–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li L, Yan J. 2015. Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nature Communications 6, 6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KB, de Jong H, Yang TJ, et al. 2005. Characterization of rDNAs and tandem repeats in the heterochromatin of Brassica rapa. Molecules and Cells 19, 436–444. [PubMed] [Google Scholar]
- Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R. 2008. SHOC1, an XPF endonuclease-related protein, is essential for the formation of class I meiotic crossovers. Current Biology 18, 1432–1437. [DOI] [PubMed] [Google Scholar]
- Macaisne N, Vignard J, Mercier R. 2011. SHOC1 and PTD form an XPF–ERCC1-like complex that is required for formation of class I crossovers. Journal of Cell Science 124, 2687–2691. [DOI] [PubMed] [Google Scholar]
- Martinez-Perez E, Schvarzstein M, Barroso C, Lightfoot J, Dernburg AF, Villeneuve AM. 2008. Crossovers trigger a remodeling of meiotic chromosome axis composition that is linked to two-step loss of sister chromatid cohesion. Genes & Development 22, 2886–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E, Villeneuve AM. 2005. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes & Development 19, 2727–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamed-Bessudo C, Yehuda E, Stuitje AR, Levy AA. 2005. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. The Plant Journal 43, 458–466. [DOI] [PubMed] [Google Scholar]
- Mercier R, Jolivet S, Vezon D, et al. 2005. Two meiotic crossover classes cohabit in arabidopsis: one is dependent on MER3, whereas the other one is not. Current Biology 15, 692–701. [DOI] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. 2015. The molecular biology of meiosis in plants. Annual Review of Plant Biology 66, 297–327. [DOI] [PubMed] [Google Scholar]
- Miao C, Tang D, Zhang H, Wang M, Li Y, Tang S, Yu H, Gu M, Cheng Z. 2013. Central region component1, a novel synaptonemal complex component, is essential for meiotic recombination initiation in rice. The Plant Cell 25, 2998–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Seol YJ, et al. 2010. Sequence and structure of Brassica rapa chromosome A3. Genome Biology 11, R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyt A, Mercier R, Mézard C, Grelon M. 2009. Meiotic recombination and crossovers in plants. Genome Dynamics 5, 14–25. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. 2005. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. The Plant Journal 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Neff MM, Turk E, Kalishman M. 2002. Web-based primer design for single nucleotide polymorphism analysis. Trends in Genetics 18, 613–615. [DOI] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. 2005. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Molecular Biology of the Cell 16, 5804–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura KI, Nakano M, Murata K, Miyoshi K, Eiguchi M, Miyao A, Hirochika H, Kurata N. 2004. An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Molecular Genetics and Genomics 271, 121–129. [DOI] [PubMed] [Google Scholar]
- Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC. 2011. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytologist 190, 523–544. [DOI] [PubMed] [Google Scholar]
- Page SL, Hawley RS. 2004. The genetics and molecular biology of the synaptonemal complex. Annual Review of Cell and Developmental Biology 20, 525–558. [DOI] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, et al. 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383. [DOI] [PubMed] [Google Scholar]
- Perumal S, Waminal NE, Lee J, Lee J, Choi BS, Kim HH, Grandbastien MA, Yang TJ. 2017. Elucidating the major hidden genomic components of the A, C, and AC genomes and their influence on Brassica evolution. Scientific Reports 7, 17986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips D, Wnetrzak J, Nibau C, Barakate A, Ramsay L, Wright F, Higgins JD, Perry RM, Jenkins G. 2013. Quantitative high resolution mapping of HvMLH3 foci in barley pachytene nuclei reveals a strong distal bias and weak interference. Journal of Experimental Botany 64, 2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FC, Jones GH. 1997. Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Research 5, 551–559. [DOI] [PubMed] [Google Scholar]
- Sanchez Moran E, Armstrong SJ, Santos JL, Franklin FC, Jones GH. 2001. Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Research 9, 121–128. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran E, Santos JL, Jones GH, Franklin FC. 2007. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & Development 21, 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo PA, Roeder GS. 1999. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97, 313–324. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. 2010. Genome destabilization by homologous recombination in the germ line. Nature Reviews. Molecular Cell Biology 11, 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J, Copenhaver GP, Schlögelhofer P. 2019. Meiotic DNA repair in the nucleolus employs a nonhomologous end-joining mechanism. The Plant Cell 31, 2259–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowdon RJ, Friedrich T, Friedt W, Köhler W. 2002. Identifying the chromosomes of the A- and C-genome diploid Brassica species B. rapa (syn. campestris) and B. oleracea in their amphidiploid B. napus. Theoretical and Applied Genetics 104, 533–538. [DOI] [PubMed] [Google Scholar]
- Stephenson P, Baker D, Girin T, Perez A, Amoah S, King GJ, Østergaard L. 2010. A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biology 10, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Gargano S, Ruprich-Robert G, Falque M, David M, Kleckner N, Zickler D. 2010. Recombination proteins mediate meiotic spatial chromosome organization and pairing. Cell 141, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian VV, MacQueen AJ, Vader G, Shinohara M, Sanchez A, Borde V, Shinohara A, Hochwagen A. 2016. Chromosome synapsis alleviates mek1-dependent suppression of meiotic DNA repair. PLoS Biology 14, e1002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Blitzblau HG, Tame MA, Falk JE, Curtin L, Hochwagen A. 2011. Protection of repetitive DNA borders from self-induced meiotic instability. Nature 477, 115–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Moustafa SS, Lefrançois P, Villeneuve AM, MacQueen AJ. 2012. Full-length synaptonemal complex grows continuously during meiotic prophase in budding yeast. PLoS Genetics 8, e1002993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Miao C, Hu Q, Lu T, Cheng Z. 2012. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genetics 8, e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Wang M, Tang D, Shen Y, Qin B, Li M, Cheng Z. 2011. PAIR3, an axis-associated protein, is essential for the recruitment of recombination elements onto meiotic chromosomes in rice. Molecular Biology of the Cell 22, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Copenhaver GP. 2018. Meiotic recombination: mixing it up in plants. Annual Review of Plant Biology 69, 577–609. [DOI] [PubMed] [Google Scholar]
- West AMV, Rosenberg SC, Ur SN, et al. 2019. A conserved filamentous assembly underlies the structure of the meiotic chromosome axis. eLife 8, e40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijeratne AJ, Chen C, Zhang W, Timofejeva L, Ma H. 2006. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Molecular Biology of the Cell 17, 1331–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasz L, Daniel K, Roig I, et al. 2009. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLoS Genetics 5, e1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Rossidivito G, Hu T, Berlyand Y, Poethig RS. 2015. Traffic lines: new tools for genetic analysis in Arabidopsis thaliana. Genetics 200, 35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Pires JC. 2011. Karyotype and identification of all homoeologous chromosomes of allopolyploid Brassica napus and its diploid progenitors. Genetics 187, 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hu B, Portheine SM, Chuenban P, Schnittger A. 2020a. State changes of the HORMA protein ASY1 are mediated by an interplay between its closure motif and PCH2. Nucleic Acids Research 48, 11521–11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Sofroni K, Wijnker E, et al. 2020. b. The Arabidopsis Cdk1/Cdk2 homolog CDKA;1 controls chromosome axis assembly during plant meiosis. The EMBO Journal 39, e101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelina NE, Choi K, Chelysheva L, et al. 2012. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genetics 8, e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, Alani E. 2009. The pch2Delta mutation in baker’s yeast alters meiotic crossover levels and confers a defect in crossover interference. PLoS Genetics 5, e1000571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanders S, Sonntag Brown M, Chen C, Alani E. 2011. Pch2 modulates chromatid partner choice during meiotic double-strand break repair in Saccharomyces cerevisiae. Genetics 188, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1999. Meiotic chromosomes: integrating structure and function. Annual Review of Genetics 33, 603–754. [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harbor Perspectives in Biology 7, a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Berchowitz LE, Lambing C, et al. 2015. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. eLife 4, e03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziolkowski PA, Underwood CJ, Lambing C, et al. 2017. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes & Development 31, 306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the paper and within its supplementary data published online.