Abstract
Infectious diseases are a global health problem affecting billions of people. Developing rapid and sensitive diagnostic tools is key for successful patient management and curbing disease spread. Currently available diagnostics are very specific and sensitive but time-consuming and require expensive laboratory settings and well-trained personnel; thus, they are not available in resource-limited areas, for the purposes of large-scale screenings and in case of outbreaks and epidemics. Developing new, rapid, and affordable point-of-care diagnostic assays is urgently needed. This review focuses on CRISPR-based technologies and their perspectives to become platforms for point-of-care nucleic acid detection methods and as deployable diagnostic platforms that could help to identify and curb outbreaks and emerging epidemics. We describe the mechanisms and function of different classes and types of CRISPR-Cas systems, including pros and cons for developing molecular diagnostic tests and applications of each type to detect a wide range of infectious agents. Many Cas proteins (Cas3, Cas9, Cas12, Cas13, Cas14 etc.) have been leveraged to create highly accurate and sensitive diagnostic tools combined with technologies of signal amplification and fluorescent, potentiometric, colorimetric, lateral flow assay detection and other. In particular, the most advanced platforms -- SHERLOCK/v2, DETECTR, CARMEN or CRISPR-Chip -- enable detection of attomolar amounts of pathogenic nucleic acids with specificity comparable to that of PCR but with minimal technical settings. Further developing CRISPR-based diagnostic tools promises to dramatically transform molecular diagnostics, making them easily affordable and accessible virtually anywhere in the world. The burden of socially significant diseases, frequent outbreaks, recent epidemics (MERS, SARS and the ongoing COVID-19) and outbreaks of zoonotic viruses (African Swine Fever Virus etc.) urgently need the developing and distribution of express-diagnostic tools. Recently devised CRISPR-technologies represent the unprecedented opportunity to reshape epidemiological surveillance and molecular diagnostics.
Keywords: Molecular diagnostics, Molecular epidemiology, Viruses, Point-of-care (POC), One pot assays, Mobile phone microscopy, HIV, HPV, HBV, Tuberculosis, SARS-CoV-2, COVID-19
1. Introduction
Infections are one of the most daunting threats to humanity, responsible for an immense burden of disabilities and deaths [1]. Pandemics of influenza (Spanish flu, swine flu, bird flu), recent outbreaks of Ebola and Zika virus, deadly and wide-spread epidemics of MERS and SARS, as well as the ongoing pandemic of COVID-19, originated in China in 2019, sweep across continents and emerge as the most recent examples of widespread infections reported in this century [2], [3], [4], [5], [6].
Many infections can become chronic, often persisting through the infected person’s lifetime. Chronic infections, like chronic viral hepatitis (HBV, HCV), human immunodeficiency virus (HIV) infection, and tuberculosis, are widely distributed and admittedly are the most prolific infectious disease killers. These characteristics define the exceptional significance of chronic infections for the global health [7], [8].
Detecting the etiologic pathogens of infectious diseases is necessary for timely treatment, risk reduction for patients and caregivers, and prevention of further spread of the pathogen, especially for the emerging and re-emerging viral infections. One conventional method used for diagnosing infectious diseases is direct isolation of pathogen nucleic acids from biological samples and their detection by polymerase chain reaction (PCR) [9]. Using sequential doubling, PCR exponentially amplifies target templates, providing an opportunity to detect even single copies of pathogenic genomes. Due to its high accuracy and specificity, PCR is used in many biological and medical applications, including diagnosing virtually any human pathogen. However, PCR requires qualified personnel and expensive, highly sophisticated equipment, and often lacks standardized protocols, limiting its implementation in medical care settings. Importantly, PCR-diagnostics is time-consuming and cannot be used for rapid screening of large cohorts. The most recent COVID-19 viral infection was shown to be frequently asymptomatic or without any immediately discernible clinical symptoms, e.g. hyperthermia [10]. Thus, screening of people in crowded places (planes, hospitals etc.) and timely isolation of infected persons cannot be effectively performed with the use of infrared thermography.
On the other hand, mass population screening, rapidly controlling biological hazards, preventing infection spread, and diagnosing infectious diseases in remote areas are difficult to do with PCR and remain a big challenge for public health services worldwide.
According to WHO criteria (ASSURED CRITERIA), the ideal diagnostic assay for any pathogen should be cheap and accurate, provide rapid results, be applicable in point-of-care practice, and require little or no specialized equipment and technical assistance [11], [12]. To date, no assay fits all these requirements. Thus, inventing new, more effective methods of molecular diagnosis is urgently needed.
Discovery of the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated protein (Cas) (CRISPR-Cas) revolutionized biology and is already pushing health care systems to the era of precise molecular medicine. In 2020, Nobel prize in chemistry was awarded to Jennifer Doudna and Emmanuelle Charpentier for their breakthrough research on CRISPR that paved the way for gene editing and further emergence of the whole new field of CRISPR-diagnostics [13].
Using genetic engineering, CRISPR-Cas systems have been adapted for use in humans and are now being modified and enhanced at an extraordinary pace, enabling precise editing of virtually any DNA or RNA molecule in the body. CRISPR-Cas-based approaches are being tested to treat hereditary, infectious, and other diseases, as well as in methods of molecular visualization and other applications [14], [15], [16].
In 2016, CRISPR-Cas systems were first utilized to detect nucleic acids for molecular diagnostics [17]. Until then, a number of successful CRISPR-Cas-based approaches to detect and diagnose infectious and non-infectious diseases had been invented. The revolution of CRISPR-Cas editing may spread further into the area of molecular diagnostics and replace PCR in many applications. CRISPR-Cas diagnostic tools are characterized by sensitivity and specificity comparable to those of traditional PCR (or even outperforming it), but do not require sophisticated (and therefore expensive) equipment and have a very low estimated cost. Embedding CRISPR-Cas into molecular diagnostics may reshape the profile of global diagnostic and health care systems [18]. The existing gap between the need for rapid diagnostics and current technologies can well be bridged by CRISPR-diagnostics. COVID-19 pandemic has admittedly added the impetus for developing novel, accurate and sensitive express-tests.
In this paper, we review classes and types of CRISPR-Cas as the basis for the emerging CRISPR-diagnostic platforms, focusing on CRISPR-Cas systems already used for molecular diagnosis, and describe their properties, functions, and perspectives to become the ideal platforms for diagnosing infectious diseases and curbing disease outbreaks.
2. Brief nomenclature of CRISPR-Cas systems and their characteristics
CRISPR-Cas systems were first described 30 years ago in bacterial genomes [19]. Unique regions of DNA, later called spacers, were shown to be separated by short palindromic repeats in bacterial genomes. Small clusters of Cas genes, encoding proteins with nucleolytic activity, were frequently found located next to CRISPR repeat-spacer arrays [20]. Many years later, these palindromic repeats and Cas genes were shown to operate as a natural adaptive immune system providing defense against viral infections in bacteria and archaea. CRISPR-Cas function relies on effector Cas proteins and guiding CRISPR RNAs (crRNAs) [21]. CRISPR-Cas systems have been adapted to function in human and other mammalian cells. They can bind and cleave virtually any site of the target nucleic acid. CRISPR-Cas systems are partitioned into 2 distinct classes which are briefly described below (Table 1 ).
Table 1.
Brief summary of the key CRISPR-Cas systems used in gene editing.
| Class | Type | Effector protein | Target |
|---|---|---|---|
| Class 1 CRISPR-Cas systems | Type I | Multi-subunit complex (signature protein Cas3) | Single-stranded DNA[27], May exhibit collateral activity[50] |
| Type III | Multi-subunit complex (signature protein Cas10) | DNA/RNA[27] | |
| Type IV | Multi-subunit complex(signature protein Csf1) | Unknown | |
| Class 2 CRISPR-Cas systems | Type II | Cas9 | Double-stranded DNA [31] |
| Type V | Cas12 | Double-stranded DNA, Single-stranded DNAMay exhibit collateral activity [51] | |
| Cas14 | Single-stranded DNA, May exhibit collateral activity[46] | ||
| Type VI | Cas13 | Single-stranded RNA, May exhibit collateral activity [18] |
2.1. Class 1 systems
CRISPR-Cas class 1 systems comprise 3 types: I type, III type and IV type. Class I systems are characterized by multiple effector proteins.
CRISPR-Cas type I systems share the effector module Cascade, composed of Cas proteins in a complex with a crRNA molecule [22]. The Cascade complex recognizes protospacer adjacent motif (PAM) sequences and unwinds target DNA, thus enabling crRNA to interact with the complementary DNA strand. Recognition of the target site entails recruitment of Cas3 protein which cuts the DNA strand not bound by Cascade complex [23].
Functioning of CRISPR-Cas type III is based on a multi-subunit complex consisting of crRNA, Csm complexes in subtype III-A systems, and Cmr complexes in subtype III-B systems. These systems are characterized by the Cas10 protein. Cas10 has been recently shown to play a role in activating non-specific RNases Csm6 and Csx1. Target site recognition by CRISPR-Cas type III systems initiates polymerase activity of Cas10 protein followed by Cas10-mediated generation of cyclic oligo-(A)-nucleotides (cOA). Binding of cOA by Csm6 activates the RNase domain of Csm6, which destroys both target RNA (many CRISPR-Cas type III systems target RNA rather than DNA molecules) and other neighboring RNA molecules [24], [25].
CRISPR-Cas type IV systems are commonly found in plasmids but their function remain largely unknown [26].
2.2. Class 2 systems
Class 2 CRISPR-Cas systems include type II and the less common types V and VI, each possessing unique effector proteins [27], [28], [29]. Class 2 systems are characterized by less complex organization, as the effector module consists only of a large, multidomain, multifunctional protein.
Currently, class 2 CRISPR-Cas systems have been the ones most widely used in gene engineering due to their simplicity and highly effective gene editing. The CRISPR-Cas type II Cas9 protein targets the desired site of DNA by means of crRNA and trans-activating crRNA (tracrRNA) [30]. Jinek et al. created a chimeric RNA molecule (single-guide RNA or sgRNA) that combines crRNA and tracrRNA and simplifies CRISPR-Cas genome editing by reducing the 3-component system (Cas9 protein, crRNA, tracrRNA) to just 2 components (Cas9 protein and sgRNA). Cas9 protein is recruited to the target site by sgRNA and generates blunt-ended double-stranded breaks in the desired site of DNA [31]. Cas9 protein from Streptococcus pyogenes (SpCas9) is the most common choice in gene editing technology due to its highly effective on-target gene editing. However, frequent off-target activity of SpCas9, defined as cleavage of non-specific DNA genomic sequences similar to the target site, limits its use [14]. Non-specific targeting by Cas9 is based on its ability to tolerate mismatches between the sgRNA and DNA, allowing it to cut even very dissimilar sequences. More specific and clinically safe variants of CRISPR-Cas type II systems are genetically modified or evolved SpCas9 proteins (enhanced SpCas9 or eSpCas9 [32]; high-fidelity SpCas9-HF1 [33]; evoCas9 [34], HypaCas9 [35], sniper-Cas9 [36]) and certain orthologous Cas9 proteins from other species [37], [38], [39], [40]. The latter have more restrictive PAM sequences and thus exert fewer potential off-target sites and have intrinsically lower ability to unwind DNA mismatched with the sgRNA [38], [41], [42].
CRISPR-Cas type V systems include several subtypes (V-A, V-B, etc.); Cas12 proteins (Cas12a, Cas12b, etc.) are signature proteins of these systems [43]. Cas12a and Cas12b proteins cut the target sequences leaving sticky ends after DNA cleavage. Cas12a, also known as Cpf1, is often used in gene engineering. CRISPR-Cas type V systems require only the Cas protein and a crRNA to edit the target site. One of the advantages of using Cas12 instead of Cas9 for genetic engineering is its smaller size and lower tolerance of nucleotide mismatches between the target DNA and crRNA [44]. Sticky ends left after Cas12-mediated DNA cleavage are repaired via homologous recombination [45], a type of low-error repair, thus increasing accuracy of gene editing. More recently, Cas14 proteins, miniature Cas proteins (400–700 aa) of CRISPR-Cas type V systems, have been discovered [46]. Cas14 have been shown to destroy single-stranded DNA without requiring PAM. Some Cas12 and Cas14 proteins possess so-called collateral activity: after binding target DNA, the proteins start destroying any adjacent DNA in a non-specific manner [43].
CRISPR-Cas type VI systems encompass subtypes VI-A, VI-B, VI-C, and VI-D (also known as C2c2). The signature protein of CRISPR-Cas type VI is Cas13. The unique property of these systems is the ability to recognize single-stranded RNA molecules. Type VI Cas proteins bind target RNA using a guiding crRNA (no tracrRNA), introduce a blunt-ended break, and indiscriminately degrade any adjacent single-stranded RNA [47], [48].
Clearly, CRISPR-Cas systems are enormously diverse, varying in mechanisms of action, composition, and structure of their key elements [49]. In-depth analysis and characterization of new types and classes of CRISPR-Cas systems is on the forefront of world science, paving the way for creating new biological and diagnostic tools with the prospect to fundamentally transform the way we conceive health care systems. Nowadays, almost every type of CRISPR-Cas is being tested for the use of CRISPR-diagnostics and in different ways with the particular emphasis on type V and type VI CRISPR-Cas systems.
3. Detection of nucleic acids by CRISPR-Cas
A large set of different CRISPR-based methods used to detect nucleic acids has been recently described. Early technologies utilized the canonical Cas9 protein of type II CRISPR-Cas systems [52] or its modified nucleolytically null, or dead, Cas9 (dCas9) protein [53]. A huge leap toward developing CRISPR-based molecular diagnostics was the discovery of protein collateral activity of Cas12, Cas13, and Cas14, a property that can be harnessed to amplify the specific on-target signal [43]. Today, many modifications and improvements have been introduced into CRISPR-based molecular platforms relying on collateral activity of CRISPR-Cas type V and type VI proteins, but the general concept remains unaltered.
To date, CRISPR-Cas systems are routinely leveraged as tools for gene editing, epigenome remodeling, regulating gene transcription, and visualizing DNA/RNA sequences in living cells [53]. Another application of CRISPR-Cas as an instrument of molecular diagnostics proved possible only in recent years. Molecular methods for detecting nucleic acids based on CRISPR-Cas systems appear to be highly sensitive, specific, and capable of one-step detection of both RNA and DNA.
3.1. Detection of nucleic acids by CRISPR-Cas type I systems
Currently, there is a single study reporting the development of a CRISPR-diagnostic assay based on type I-E CRISPR systems. This assay was termed Cas3-Operated Nucleic Acid detection (CONAN) [50]. Yoshimi et al., demonstrated that Cas3 exhibits collateral activity with several types of PAM upon recognition of the target. Non-specific cleavage of single-stranded DNA probes upon activation of Cas3 provides a rapid and sensitive means for detecting nucleic acids. CONAN was developed in two formats, either with fluorescent detection by microplate reader or lateral flow assay.
3.2. Detection of nucleic acids by CRISPR-Cas type II systems
Zhang et al. (2017) created a diagnostic test based on two dCas9 proteins fused with split domains of luciferase enzyme [54]. Binding of two dCas9 proteins to the adjacent target DNA results in re-constitution of luciferase and emission of luminescent signal that can be readily detected by a luminometer. As a proof of concept, this technology was shown to detect Mycobacterium tuberculosis with high specificity and sensitivity [54].
In 2017, Wang et al. developed a multiplex method to detect human papillomavirus (HPV) by Cas9 protein targeting L1 and E6/E7 viral genes amplified using PCR (ctPCR) [55]. This technology employs a 3-step DNA detection protocol: (1) amplification of DNA using PCR; (2) nucleolytic cleavage of PCR amplicons by Cas9; and (3) amplification of cleaved fragments by PCR. The resulting PCR products are detected by gel electrophoresis or fluorescence. According to Wang and colleagues, this method increases sensitivity of the diagnostic test and helps to differentiate between HPV16 and HPV18 strains [55]. This test has been recently upgraded to provide quantitative detection of HPV16/18 using qPCR (ctPCR3.0) [56]. Another Cas9-based method of detection called Cas9/sgRNA-associated reverse PCR (CARP) [50] consists of three steps, namely (1) cleavage of target DNA at two sites by Cas9 endonuclease; (2) intermolecular and intramolecular ligation of the cleaved products by T4 DNA ligase and (3) PCR amplification of the ligated DNA. CARP successfully detected as little as 0.002 ng of HPV16 L1 gene and enabled specific discrimination of HPV16 and HPV18 subtypes.
Pardee and colleagues developed a technique called NASBA-CRISPR cleavage (NASBACC), which relies on the principle of toehold switch sensors and the ability of Cas9 protein to selectively cleave target DNA [17]. Toehold switch sensors represent programmed synthetic riboregulators able to control translation by binding trigger RNA. Riboregulators carry a hairpin structure that blocks translation in cis by sequestering the ribosome binding site and start codon. When a riboregulator is bound to complementary trigger RNA, the ribosome binding site and start codon are freed and translation is enabled. The RNA target is then amplified by NASBA so that the RNA of interest is reverse-transcribed and linked to the riboregulator. NASBA amplification system uses a battery of 3 enzymes: reverse transcriptase, RNase H, and T7 polymerase. In the NASBA method, target RNA is converted into a cDNA-RNA hybrid using specific primers; RNase H then destroys the RNA in this duplex. Next, a primer is added to the reaction mixture to produce synthetic templates recognized by T7 polymerase. The results of the reaction are detected by colorimetry [17]. This technology effectively detects Zika and dengue viruses with the sensitivity of 1–3 fM.
Another milestone in CRISPR diagnostics was the development of CRISPR-Chip [57]. CRISPR-Chip combines CRISPR principles with an electronic transistor made of graphene (a single layer of carbon atoms) [58]. CRISPR-Chip uses a dCas9 protein that binds, but does not cut, target DNA. dCas9 proteins are immobilized on graphene transistors. After adding DNA isolated from biological samples, dCas9 binds the target DNA, thereby altering the electric conductivity of graphene and the electric characteristics of the transistor. CRISPR-Chip is highly sensitive, able to detect as little as 1.7 fM of target DNA, and the procedure is extremely rapid, taking only 15 min. CRISPR-Chip has been used to detect genetic mutation in clinical samples from patients with Duchenne muscular dystrophy [57].
Methods of isothermal amplification are particularly important to performing molecular diagnosis in remote areas and in laboratories without specially trained personnel. CRISPR-Cas9-triggered nicking endonuclease-mediated strand displacement amplification (CRISDA) method uses a combination of CRISPR-Cas9 technology and isothermal amplification methods. It uses Cas9 nickases (Cas9 proteins with a mutation in one nucleolytic domain that makes the protein able to cut a single DNA strand), and single-strand displacement amplification (SDA) of target fragments followed by detection of the signal by fluorescent peptide nucleic acid invasion-mediated endpoint measurement. Intensity of the signal can be measured by a fluorimeter. CRISDA technology helps to detect target nucleic acids with specificity of 1 nucleotide [59].
Quan and colleagues described the finding low abundance sequences by hybridization (FLASH) method, developed to detect pathogens resistant to antimicrobial therapy [60]. FLASH uses a battery of sgRNAs together with Cas9 proteins that cut the gene of interest into small fragments suitable for further next-generation Illumina sequencing. DNA/cDNA ends are first blocked by phosphatase to prevent ligation with adapters utilized in the next step and then cut by Cas9. DNA of interest is nucleolytically cleaved resulting in fragments with non-blocked ends and thus can be linked with universal adapters, amplified by PCR, and sequenced. The authors confirmed the utility of this technology in models of pneumonia caused by gram-positive bacteria including S. aureus and of malaria-causing Plasmodium falciparum [60].
Similarly, the CRISPR-Cas9-triggered exponential amplification reaction (Cas-EXPAR) method combines the advantages of Cas9 to introduce site-specific cuts in target DNA with the isothermal EXPAR method with fluorescent signal detection [61]. Compared to other methods of isothermal amplification, such as NASBA, RCA, SDA, LAMP, or RPA, EXPAR is characterized by relatively high efficacy and speed of product amplification. Until now, EXPAR was not widely used in molecular diagnostics, and was applied mostly for detection of short microRNAs [62]. This limited utility was due to an inherent property of EXPAR that does not allow using extended DNA sequences as primers for amplification. However, CRISPR-Cas9 can be programmed to cut DNA molecules into fragments short enough to successfully implement EXPAR. Wild-type SpCas9 can target and cut only double-stranded DNA molecules, but using the so-called PAM-mers (templates of single-stranded DNA complementary to single-stranded templates with a PAM sequence) permits SpCas9 to recognize and cleave single-stranded RNA or DNA molecules. Reported sensitivity of Cas-EXPAR is ~ 1 amol–10 fmol, and the limit of detection fluctuates around 0.82 amol [61]. These values are comparable to those observed using PCR diagnostics. Due to the high specificity of Cas-EXPAR, this technology can be used to detect methylated DNA after bisulfite conversion, which changes all cytosines in DNA to uracil except for methylated cytosines (5-methylcytosine), which remain intact. Thus, a DNA site with cytosine converted to uracil during bisulfite conversion will not be recognized and cleaved by Cas9 protein when the sgRNA targets the original (non-converted) sequence. As a result, isothermal amplification of such templates by EXPAR proceeds with low efficacy. Conversely, 5-methylcytosine remains intact during bisulfite conversion; sites with 5-methylcytosine can thus be effectively recognized and cut by Cas9, effectively amplified by EXPAR, and detected using conventional fluorescence methods.
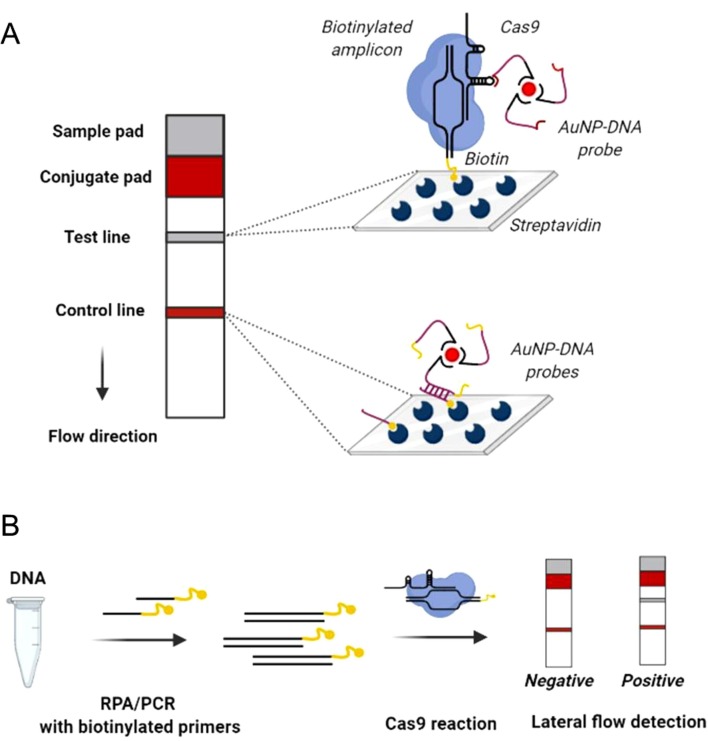
To avoid the use of many different nucleases, accessory proteins and expensive fluorescent probes, Wang with co-authors [63] combined lateral flow nucleic acid assay with a CRISPR-Cas9 tool. This technology, termed CRISPR-Cas9-mediated lateral flow nucleic acid assay (CASLFA), integrates the CRISPR-Cas9 system with a lateral flow device to detect target dsDNA (Fig. 1 ). The target is first amplified using PCR or isothermal amplification methods with biotinylated primers, incubated with CRISPR-Cas9 RNPs and then trickled onto the sample pad. At the surface of the conjugate pad, AuNP-DNA probes bind to a modified the sgRNA scaffold. In this technology, the stem-loop structure of sgRNA is extended by the insertion of additional nucleotides which are recognized by universal AuNPs-DNA probes. Next, biotinylated amplicons complexed with CRISPR-Cas9, flow to the test line and the whole mix gets captured by the streptavidin. This colored signal is visualized at the test line. At the control line, AuNP-DNA probes hybridize with the precoated streptavidin and generate a colored band. In summary, CASLFA is a simple method, consisting of PCR/RPA amplification step with biotinylated primers and a short (3–5 min) lateral flow assay that detects target DNA with high specificity and sensitivity (LOD is ≈ several gene copies). In conclusion, the authors envision several technical improvements in the CASLFA, such as multiplex detection of different targets by introducing primers with different labels and combination of CALSFA with microfluidic technology on an advanced diagnostic platform.
Fig. 1.
Schematics of CRISPR-Cas9-based CRISPR-diagnostic method CASLFA. (A) Structure of the lateral flow device. The lateral flow device consists of a sample pad where the isolate is applied, a conjugate pad with pre-assembled AuNP-DNA probes, a test line and a control line. At the test line, complexes of CRISPR-Cas with the target biotinylated DNA and AuNP-DNA probes, hybridized with the stem-loop region of sgRNA, interact with pre-coated streptavidin at the test pad to produce a visible signal. At the same time, AuNP-DNA probes move further and interact with streptavidin at the control line. AuNP-DNA probes contain three regions, namely (1) polyA-polyT (poly A used for labeling with Au and polyT as a linker); (2) purple area for hybridization with the embedded probe in the control line and (3) yellow area used for hybridization with the engineered stem-loop region in sgRNA. (B) Schematics of CASLFA procedure. Isolated DNA is amplified with biotinylated primers using RPA or PCR. Amplicons are mixed with CRISPR-Cas9 detection complex and DNA probes and, after short incubation, applied to the lateral flow device. The picture was created in BioRender. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Detection of nucleic acids by CRISPR-Cas type V and VI
In 2018, Doudna and colleagues presented the CRISPR-Cas diagnostic platform named DNA endonuclease-targeted CRISPR trans reporter (DETECTR) (Fig. 2 ) [51]. This method relies on collateral activity of Cas12a protein activated after recognition of target RNA by Cas12a. The authors demonstrated that Cas12a protein from Lachnospiraceae bacterium ND2006(LbCas12a) exhibits non-specific collateral activity and degrades all adjacent DNA molecules after recognizing target ssDNA or dsDNA. If the reaction with Cas12a protein and targeting crRNA is complemented by single-stranded DNA-reporters (probes) and then mixed with the biological sample, crRNA-dependent recognition of pathogenic nucleic acids by Cas12a turns on collateral activity that destroys DNA probes. DNA probes are designed similarly to conventional TaqMan probes, in which one end of the reporter is bound by a fluorophore and the opposite is linked to a quencher. Degradation of the DNA probes releases fluorophores and results in stable and strong fluorescent signal detected by a fluorimeter. Additionally, DETECTR has been combined with an isothermal pre-amplification step to enrich target sequences (RPA). RPA enhances analytical sensitivity of the diagnostic test and helps to avoid the need for sophisticated and expensive equipment.
Fig. 2.
Schematics of CRISPR-Cas DETECTR and OR-DETECTR diagnostic platform. (A) DETECTR pipeline. The DNA molecule is amplified using isothermal amplification RPA method followed by the addition of the Cas12a mix with sgRNA and fluorescent probes. Cas12a recognizes the target DNA and destroyes fluorescent probes by means of collateral activity to produce a fluorescent signal. (B) OR-DETECTR pipeline. CRISPR-Cas mix and RT-RPA mix are physically separated to avoid opening the tube and potential cross-contamination of the samples. The picture was created in BioRender.
Other orthologous proteins from different organisms - AsCas12a (Acidaminococcus sp.), FnCas12a (Franciella novicida), AaCas12b (Alycyclobacillus acidoterrestris) [43]– also possess collateral activity and can be used to create diagnostic platforms by the same principle as DETECTR.
DETECTR was provisionally used to detect HPV and differentiate between HPV16 and HPV18, the most pro-oncogenic types of HPV. In this setting, two crRNAs were designed to target the hypervariable V loop of the L1 gene, which differs by only 6 nucleotides betweenHPV16 and HPV18types. In cell culture, DETECTR effectively discriminated between these types of HPV. In crude DNA extracts, DETECTR identified HPV16 in 25 of 25 cases and HPV18 in 23 of 25 cases, provisionally determined by PCR. Notably, DETECTR analysis takes only 1 h to complete [51].
Also in 2018, Doudna et al. first characterized a highly diverse family of CRISPR-Cas systems similar to CRISPR-Cas type V [46]. In this system, the signature protein Cas14 PAM-independently cleaves single-stranded DNA molecules. Like Cas12 proteins, Cas14 exhibits collateral activity and cuts any adjacent DNA molecules, making Cas14 proteins useful in CRISPR-based diagnostics. Contrary to Cas12, Cas14 has lower tolerance to nucleotide mismatches between sgRNA and the target template; the internal seed-sequence is very sensitive to nucleotide mismatches, which greatly reduces on-target activity of Cas14. This property is instrumental for using Cas14 to detect single nucleotide polymorphisms (SNPs) in DNA. Using Cas14 in the DETECTR platform permitted identification of SNPs in human HERC2 gene, which is responsible for eye coloring [46].
More recently, type V CRISPR-Cas system was used to develop a high-throughput, all-solution phase detection system for detecting African Swine Fever Virus (ASFV). ASFV infects domestic pigs and wild boars, causing massive disease outbreaks with nearly 100% mortality rate. ASFV is another global threat affecting food industry, food supply and international trade. Cas12a proteins with crRNAs targeting ASFV DNA become activated and induced degradation of fluorescent ssDNA reporters by collateral activity. This test provided the means for very rapid (within 2 h) and accurate (detection limit of 1 pM) detection of ASFV, distinguishing closely related target DNA sequences [64]. Isothermal amplification increases sensitivity of the system enabling detection of femtomolar concentrations (detection limit of 100 fM) of DNA using a custom designed fluorometer. Coupling CRISPR-Cas12a assay with previously developed benchtop point-of-care system with a portable laser and mini spectrometer.
Overall, this technique is suitable for low resource settings and massive screening. According to the authors, the advanced AFSV system is inexpensive, automated, light weight and does not require skilled operators. Detecting ASFV using CRISPR-Cas12a is one of first examples that strengthen the unitality of CRISPR-Cas diagnostics for veterinary sciences.
Another group took advantage of Cas12a protein to develop the one-hour low-cost multipurpose highly efficient system (HOLMES) system [65]. Instead of isothermal amplification used by the DETECTR method for enriching target nucleic acids, HOLMES uses PCR. Using HOLMES, the authors detected DNA viruses like Japanese encephalitis virus and Aujeszky’s disease virus in 1–10 attomolaramounts [65]. However, the need for PCR amplification requires expensive equipment and adds an additional, time-consuming step.
The modified HOLMESv2 method uses isothermal amplification of nucleic acids by Loop-Mediated Isothermal Amplification (LAMP) and recognition of target double-stranded or single-stranded DNA templates by Cas12b protein followed by degradation of single-stranded DNA fluorescent probes by Cas12b collateral activity [66]. Peak collateral activity is reached within 10 min after recognition of target single-stranded DNA molecules by Cas12b, and shifts to 30 min when Cas12b recognizes double-stranded DNA. This property indicates the intricate differences between Cas12b and Cas12a, with the former protein being more active toward double-stranded DNA. Alicyclobacillus acidoterrestrisCas12b (AacCas12b) recognizes and cleaves target double-stranded DNA with adjacent PAM sequence 5ʹ-TTN-3ʹ and exhibits collateral activity [67]. Among target templates with different PAM regions, only double-stranded DNA with PAM 5ʹ-TTC-3ʹ and 5ʹ-TAC-3ʹsequences after cutting activates Cas12b collateral activity and the associated degradation of fluorescent DNA probes, indicating that not all double-stranded DNA sites recognized by Cas12b can effectively turn on collateral activity of the gene-editing protein. Upon binding to single-stranded DNA, collateral activity of Cas12b was shown to be PAM-independent, always activated after interaction with an appropriate target. The lowest concentration of the target single-stranded or double-stranded DNA detected by Cas12b was around 1 nM. Combining Cas12b and LAMP technologies provides specific detection of as little as 0,01 fM DNA, which is equivalent to the sensitivity of Cas12a. HOMLESv2 allows detection of trace amounts of both DNA and RNA as well, if the isolate is reverse transcribed during sample preparation. To reduce the time needed to perform RNA reverse transcription and amplification, the authors proposed using DNA polymerases with 5ʹ→3ʹ DNA polymerization and reverse transcriptase activity (e.g., DNA polymerase Bst) [68].
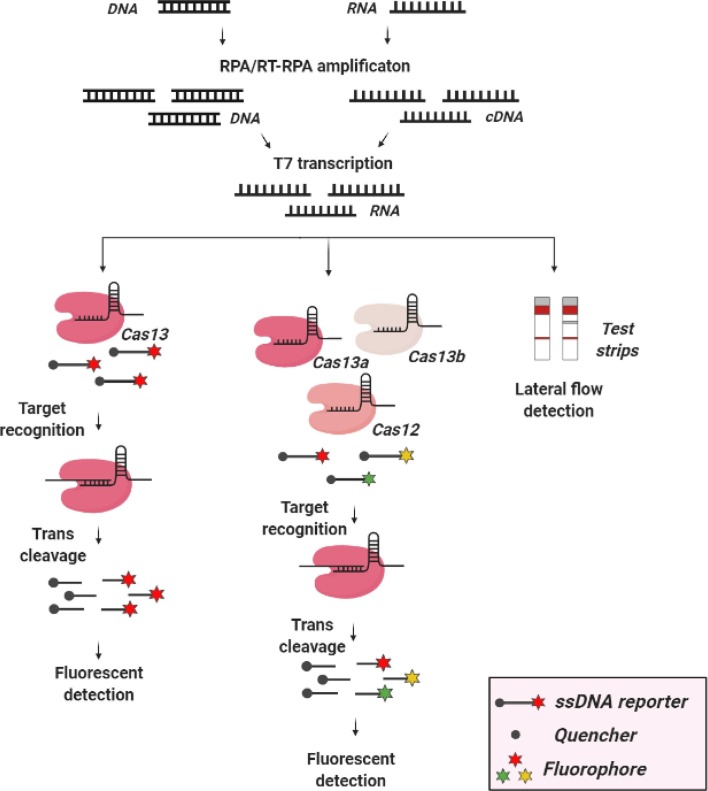
In 2017, Feng Zhang and colleagues presented SHERLOCK (Fig. 3 ), a diagnostic platform based on CRISPR-Cas type VI system [18], [69]. SHERLOCK is based on the same principles as DETECTR, but relies on activity of Cas13 nuclease from Leptotrichia wadei. Cas13 specifically recognizes and cleaves only RNA, rather than DNA like Cas12a, thus non-specifically degrading any neighboring RNA molecules. In vitro transcription of the isolate enables detection of DNA targets. Isothermal amplification RPA can be used to enrich target molecules and increase sensitivity. The amplified RNA fragments are mixed with Cas13 protein crRNA and fluorescent RNA probes. If the target molecules are present in the sample, Cas13 recognizes them via crRNA and indiscriminately cleaves (by collateral activity) fluorescent RNA probes, disrupting the interaction between the fluorophore and the quencher. The presence and intensity of the fluorescent signal thus indicate the amount of the target in the biological sample. The authors demonstrated that SHERLOCK detects Zika virus, dengue virus, various pathogenic bacteria, and SNPs in DNA with attomolar sensitivity. All components of the SHERLOCK reaction can be lyophilized and used after long storage periods without impacting the sensitivity and specificity of the test [18]. SHERLOCK had a major drawback: it was qualitative, not quantitative, but a year later, the authors presented the second version of the platform, SHERLOCKv2 (Fig. 3) [70]. SHERLOCKv2 can simultaneously detect targets in 4 different fluorescent channels, because Cas13 proteins from different organisms -- LwaCas13a (L. wadei), CcaCas13b (Apnocytophaga canimorsus Cc5), LbaCas13a (L. bacterium[strain NK4A179]), and PsmCas13b (Prevotella sp. MA2016)) --destroy adjacent RNA and DNA molecules preferentially at certain dinucleotide sites (AU, UC, AC, and GA, correspondingly). Generating probes enriched in different dinucleotides and linked to different fluorophores, the authors demonstrated the ability of SHERLOCKv2 to detect up to 4 targets. SHERLOCKv2 also qualitatively evaluates samples with the sensitivity of 2 attomols, optimized to result in strong correlation between signal intensity and concentration of the target. The sensitivity of the signal was dramatically increased over that of SHERLOCK by adding Csm6 protein to the reaction mix. Amplification of the signal becomes possible because collateral activity of LwaCas13a and PsmCas13b destroys RNA, generating hydroxylated 5ʹ-ends and linear homopolymers of adenine terminating with 2ʹ,3ʹ-cyclic phosphates. The latter activates Csm6, which destroys new probe molecules and amplifies the specific signal. By testing Csm6 proteins from many different species, the authors demonstrated that Csm6 from Enterococcus italicus and Lactobacillus salivarius were most efficiently activated by 2ʹ,3ʹ-cyclic phosphate ends. Finally, SHERLOCKv2 was engineered to produce a visual colorimetric readout on commercial lateral flow strips that do not require any special equipment. In this setting, the presence of the target is determined by visually inspecting the strips with different intensity of staining. The entire SHERLOCKv2 reaction is performed in a single step by directly applying the biological sample to the test strip without purifying and isolating nucleic acids. To conclude, SHERLOCKv2 is a highly sensitive quantitative diagnostic platform suitable for multiplex signal detection and visual/colorimetric detection on lateral flow strips [70].
Fig. 3.
Schematics of CRISPR-Cas diagnostic platforms SHERLOCK/v2. SHERLOCK/SHERLOCKv2 pipeline. DNA or RNA molecules are isothermally amplified using RPA or RT-RPA, correspondingly. DNA is transcribed into RNA using in vitro T7 transcription reaction. Cas13 recognizes target RNA molecules and cleaves fluorescent probes by means of collateral activity. Different nucleotide preferences of Cas13 proteins from different species can be used for preferential cleavage of fluorescent probes at specific dinucleotides. Thus, this method can be used for multiplex detection with designed probes. Alternatively, reaction results can be visualized on lateral flow strips using chromogenic reaction. The picture was created in BioRender.
The majority of CRISPR-diagnostic tools entail several pipetting steps, involving opening of reaction tubes with amplified product and high risk of sample contamination. This issue is elegantly addressed in the one-tube detection platform OR-DETECTR/OR-SHERLOCK technology (Fig. 2B) [71]. Here, the extracted sample is added to the RPA mix at the bottom of the centrifuge tube, while the DETECTR/SHERLOCK mix is physically separated on the inner side of the tube lid. Following RPA amplification, RPA components and CRISPR-Cas components are mixed by short centrifugation. The amplified target can then be recognized by CRISPR-Cas complexes. The signal can be visualized using lateral flow assay or fluorescence.
In an attempt to develop one pot assay, Arizti-Sanz with co-authors created a Streamlined Highlighting of Infections to Navigate Epidemics (SHINE) [72] procedure for sensitive and specific detection of nucleic acids from unextracted samples. They developed SHERLOCK procedure into a single-step reaction with optimal concentrations of monovalent salt, magnesium, pH and primer concentrations. Although combining of RT-RPA and SHERLOCK dramatically impairs sensitivity of the nucleic acid detection assay, addition of RNase H enhanced sensitivity of the test by increasing RT efficiency. Additionally, they developed a smartphone application that ensures unbiased detection and interpretation of the illuminated probes.
Similarly, Joung et al. [73] described a streamlined protocol for detection of SARS-CoV-2 in one pot that combines SHERLOCK testing with a simplified RNA extraction step (lysis and magnetic beads concentration of RNA without ethanol wash and elution steps) and LAMP isothermal amplification. One pot assay was possible using a thermostable Alicyclobacillus acidiphilus (AapCas12b) which can reportedly function at temperatures up to 65℃. However, AapCas12b did not perform well in one pot assays at 55 ℃ and higher. Joung with co-authors screened 94 additives and identified that addition of taurine improves reaction kinetics. This protocol is called SHERLOCK testing in one pot (STOP) and is optimized for SARS-CoV-2 detection (STOPCovid). The test can be performed at single temperature (60 ℃) within an hour. Sensitivity of STOPCovid outperforms CDC RT-qPCR test with the limit of detection (LOD) ≈33 copies of RNA per mL. Apparently, application of novel thermostable Cas proteins with improved performance and optimization of reaction mixtures may substantially improve sensitivity and speed of one pot CRISPR-diagnostics. In a different study, one pot assay called All-In-One Dual CRISPR-Cas12a (AIOD-CRISPR) [74] assay was developed for rapid and specific detection of nucleic acids with the minimal risk of amplicon cross-contamination. AIOD-CRISPR has one-pot reaction system consisting of RPA pre-amplification step and LbaCas12a detection mix. All reactions are carried out at 37℃, the process takes 40 min. AIOD-CRISPR LODs are 1.2 copies of DNA targets and 4.6 copies of RNA targets.
Myhrvold and colleagues paired the SHERLOCK protocol with a heating unextracted diagnostic samples to obliterate nuclease (HUDSON) method, aimed at detecting nucleic acids while avoiding the time-consuming step of nucleic acid isolation [75]. The HUDSON protocol was added to create field-deployable diagnostics, necessary in remote areas without developed infrastructure and laboratory diagnostics. Recent outbreaks of viral infections in Africa, South America and China highlight the urgent need for developing diagnostic tests that require minimal steps, reagents, and equipment. By heating, chemically inactivating ribonucleases, and lysing viral particles, HUDSON permits direct analysis of viral pathogens in whole blood, plasma, serum, urine, or saliva samples. HUDSON can be used for virtually any CRISPR-based diagnostic platform, permitting direct analysis of viral pathogens from bodily fluids.
Similarly, the HUDSON-SHERLOCK pipeline can detect several targets in one clinical sample [75]. Using pre-defined biological samples, the authors demonstrated that the test detected Dengue virus with high sensitivity (limit of detection was 45 copies of the viral genomes in 1 mL of whole blood and 1 copy of the viral genome in 1 mL of saliva) with the reported total turnaround time of less than 1 h [75]. Because viral diversity may affect sensitivity and specificity of the test, the authors then designed SHERLOCK crRNAs to discriminate between Zika virus and dengue virus. HUDSON-SHERLOCK successfully differentiated 4 serotypes of dengue and Zika viruses in biological samples from different regions of the world, mostly with 100% specificity, 100% sensitivity, and 100% concordance between the samples [75].
Dai and colleagues created E-CRISPR system which is equipped with electrochemical signal detection module utilizing Cas12a trans-cleavage activity and modified ssDNA reporter with methylene blue electrochemical tag attached to the sensor surface [80]. In the presence of target sequence, Cas12a proteins cleave ssDNA-MB reporter decreasing the level of electrochemical signal being transduced. The authors achieved picomolar sensitivity for detecting HPV16 and parvovirus B-19 nucleic acids. Moreover, authors devised an aptamer-based E-CRISPR cascade for protein detection. The platform utilizes aptamers for the protein of interest and Cas12a-crRNA designed to specifically target the aptamer. E-CRISPR is applied to evaluate the remaining concentration of aptamer in the sample. The authors confirmed the efficacy of the platform by TGF-β1 protein detection in clinical samples. Hence, E-CRISPR technology can potentially be used for wide variety of nucleic acid and proteins.
In addition to E-CRISPR, a number of new amplification-free testing strategies were designed. CRISPR-Cas12a-mediated interfacial cleaving of hairpin DNA reporter for electrochemical nucleic acid sensing [83] test uses immobilized interfacial hairpin DNA (hpDNA) electrochemical reporters which are cleaved by Cas12a upon recognition of the target DNA. HpDNA cleavage changes electrochemical properties of the biosensor resulting in a strong and specific signal. Compared to E-CRISPR, which uses a conventional linear ssDNA reporter as a sensing interface, hpDNA has an increased accessibility to Cas12a nucleases which dramatically improves electrochemical response change and analytical sensitivity of the method. As low as 30 pM of unamplified target DNA can be detected in ≈60 min.
The phenomenon of Au nanoparticle-assisted metal-enhanced fluorescence was utilized by Choi et al. [89] to develop CRISPR-Cas12a-based amplification-free test. In the developed AuNP nanosensor the amplification step was replaced with signal enhancement by metal-enhanced fluorescence of DNA-functionalized gold nanoparticles. The signal can be measured by fluorescence or colorimetry analysis with the LOD ranging from 1 fM to 100 pM.
A new CRISPR-Cas13-based assay that does not have a pre-amplification step has been recently reported by Fozouni with co-authors [92]. The authors managed to increase Cas13a activation and fluorescence intensity by combining multiple crRNAs for the target and by measuring fluorescence over time instead of the endpoint fluorescence. Combination of 2–3 crRNAs enables detection of as few as 30 copies/μL of target SARS-CoV-2 RNA. Applying several crRNAs also ensures specificity of the method and increases the chances to detect mutated RNA molecules. This assay uses a mobile phone camera with a low-cost laser illumination and collection of optics to detect the fluorescent signal and, most importantly, directly translates fluorescent signal into viral loads. Among the work required to introduce this test into the practice, it is necessary to introduce an extraction-free protocol, minimize the number of steps, improve sensitivity with optimized CRISPR-Cas components and reaction mixtures and, possibly, develop an embedded sensor that wirelessly sends the results to a mobile phone.
Another amplification-free strategy was implemented by Tian et al. [93] who developed an ultralocalized Cas13a assay, a isothermal one-step reaction (Fig. 4 ). In this assay, isolated sample together with Cas13a detection mix are trapped into cell-like-sized reactors. Confinement of the reaction mix enhances local concentrations of the target and CRISPR-Cas ribonucleoprotein complexes and provides > 10,000-fold increase in sensitivity of the test. The reaction accumulates a strong fluorescent signal in a pico-sized droplet from a single RNA molecule. Fluorescent microdroplets are detected and counted by fluorescent microscopy.
Fig. 4.
Pipeline and principle of ultralocalized Cas13a assay. (A) Schematics of ultralocalized Cas13a assay. Pico-sized droplets are mixed with target RNA and CRISPR-Cas13a detection reaction. Positive signal results in illumination of the droplets that can be counted by fluorescent microscope. (B) The principle of confinement effect on local concentration of target molecules. Upon decrease in the analytical volume, the local concentration of target molecules inversely increases. The picture was created in BioRender.
Finally, a recently developed CRISPR-diagnostic procedure CRISPR-ENHANCE [90] (Enhanced Analysis of Nucleic acids with CrRNA Extensions) employed genetically modified crRNAs with specific 7-mer 3′-extensions that achieve significantly higher sensitivity of CRISPR-Cas12a assays. Nguyen et al., demonstrated that CRISPR-ENHANCE can obviate the need for pre-amplification step by a simple modification of crRNAs. CRISPR-ENHANCE can be used in two different formats: (1) lateral flow assay and (2) fluorescent assay.
A plethora of new CRISPR-based diagnostic tools and approaches has been devised so far, but most of the early inventions relied on CRISPR-Cas9 type II systems, which themselves do not induce a strong, specific signal related to the presence of target nucleic acids in the sample. Instead, these technologies utilized pre-amplification of nucleic acids using PCR, an approach that compromises the vast perspectives of CRISPR diagnostics.
More complicated methods, such as CRISDA [59], work in completely isothermal conditions, but are expensive (requiring many enzymes) and time-consuming (total turnaround time for CRISDA is over 3–4 h). Although several methods, such as FLASH [60], can be applied in certain specific areas of molecular diagnostics, the majority of early technologies listed in Table 2 do not demonstrate any obvious advantages over PCR.
Table 2.
Types of CRISPR-Cas-based diagnostic tools, their applications and characteristics.
| Type of CRISPR system | Method | Protein | Target | Amplification | Detection | Model organism | Reported sensitivity |
|---|---|---|---|---|---|---|---|
| Type I-E | CONAN [50] | Cas3 | DNA | RT-LAMP | Lateral flow assay | SARS-CoV-2 | 1 copy |
| Type II | Chimeric dCas9-luciferase [54] | dCas | DNA | PCR | Luminescence | M. tuberculosis | ≈3 × 10−21 M |
| dCas9 (FISH) [76] | dCas9 | DNA | – | Fluorescence | S. aureus | 10 CFU/mL | |
| ctPCR [55] | Cas9 | DNA | PCR | Electrophoresis/qPCR | HPV 16/18 | ≈7 × 10−16 M | |
| CARP [77] | Cas9 | DNA | PCR | Electrophoresis/ qPCR | HPV 16/18 | 2 pg | |
| ctPCR3.0 [56] | Cas9 | DNA | qPCR | qPCR | HPV 16/18 | 2 ng | |
| NASBACC [17] | Cas9 | DNA | NASBA | Colorimetry | Zika virus, dengue virus | 1 × 10v15 M | |
| CRISPR-Chip [57] | Cas9 | DNA | – | Potentiometry | SNPs | 1.7 × 10−15 M | |
| CRISDA [59] | Cas9 nickases | DNA | SDA | Fluorescence | SNPs | ≈10−18-10−17 M | |
| CASLFA [63] | Cas9 | DNA | RPA or PCR | Lateral flow assay | African swine fever virus | 150 copies | |
| FLASH [60] | Cas9 | DNA | PCR | NGS | Antimicrobial resistance genes | ≈10−18-10−17 M | |
| CAS-EXPAR [61] | Cas9 | DNA, RNA | EXPAR | Fluorescence | L. monocytogenis | 0.82 × 10−18 M | |
| Type V | DETECTR [51] | Cas12a | DNA | RPA | Fluorescence | HPV 16/18 | ≈10−18-10−17 M |
| DETECTR [46], [78] | Cas14a | DNA | RPA | Fluorescence | – | 10−18 M | |
| OR-DETECTR [71] | Cas12a | RNA | RT-RPA | Fluorescence/Lateral flow assay | SARS-CoV-2, H1N1 | 1–2,5 copies/µL | |
| HOLMES [65] | Cas12a | DNA, RNA | PCR | Fluorescence | Japanese encephalitis virus, pseudorabies virus | ≈10−18-10−17 M | |
| HOLMESv2 [66] | Cas12b | DNA, RNA | LAMP | Fluorescence | Japanese encephalitis virus | 10−17 M | |
| CDetection [79] | AaCas12b | DNA | RPA | Fluorescence | HPV16/18 | 1 × 10−18 M | |
| E-CRISPR [80] | Cas12a | DNA, protein | – | Electrochemical | DNA: HPV16, parvovirus B19; Protein: TGFβ1 | 10−12 M | |
| CRISPR-Cas12a-NER [81] | LbCas12a | RNA | RT-RAA | Naked eye | SARS-CoV-2 | 10 copies | |
| CASdetec [82] | Cas12b | RNA | RT-RAA | Naked eye | SARS-CoV-2 | 1 × 104 copies/mL | |
| STOPCovid [73] | AapCas12b | RNA | RT–LAMP | Fluorescence/lateral flow assay | SARS-CoV-2 | 100 copies | |
| AIOD-CRISPR [74] | Lba Cas12a | RNA | RPA | Fluorescence, visual | SARS-CoV-2, HIV-1 | 11 copies- | |
| CRISPR-Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing [83] | Cas12a | DNA | – | Differential pulse voltammetry | HPV16/18 | 30 × 10−12 M | |
| CRISPR-FDS [84] | Cas12a | RNA | RT-RPA | Fluorescence | SARS-CoV-2 | 5 copies | |
| PGMs‐CRISPR [85] | Cas12a | RNA | RT-RAA | Glucose meter readout | SARS-CoV-2 | 1 copy | |
| opvCRISPR [86] | Cas12a | RNA | RT-LAMP | Fluorescent detection by naked eye | SARS-CoV-2 | 5 copies | |
| CODA [87] | Cas12a | RNA | RT-RPA | Fluorescent anisotropy | SARS-CoV-2 | 3 copy/μL | |
| CALIBURN [88] | Cas12a | RNA | RT-RPA | Fluorescence | SARS-CoV-2 | 5 viral copies per reaction | |
| CRISPR-Cas12a based nucleic acid biosensor [89] | Cas12a | DNA | – | Fluorescence | DNA | 0.34 × 10−15 M | |
| CRISPR-ENHANCE [90] | LbCas12a | RNA | Not required or coupled with LAMP/RT-LAMP | Fluorescence/Lateral flow assay | SARS-CoV-2 | 10−15 M | |
| CRISPR-MTB | Cas12a | DNA | RPA | Fluorescence | M. tuberculosis | 1 copy | |
| Type VI | SHERLOCK [18], [69] | Cas13a | DNA, RNA | RPA | Fluorescence | Viruses, bacteria, SNPs | 2 × 10−18 M |
| OR-SHERLOCK [71] | Cas13a | RNA | RPA | Fluorescence/Lateral flow assay | SARS-CoV-2, H1N1 | 1–2,5 copies/μL | |
| CREST | Cas13a | RNA | PCR | Fluorescence | SARS-CoV-2 | 200 copies/μL | |
| Microfluidic Ebola virus detection [91] | Cas13a | RNA | – | Microfluidic chip; Portable fluorimeter | Ebola virus | 20 pfu/mL | |
| SHINE [72] | Cas13a | RNA | RPA | Smartphone (in-tube fluorescence readout or lateral flow strip) | SARS-CoV-2 | 10 cp/μL | |
| CRISPR-Cas13 with mobile phone microscopy [92] | Cas13a | RNA | – | Fluorescence measurement by mobile phone camera with additional optics | SARS-CoV-2 | 30 copies/μL | |
| Ultralocalized Cas13a Assay [93] | LbuCas13a | RNA | – | Fluorescent microscopy (digital droplet readout) | SARS-CoV-2 | 6 copies/μL | |
| CARMEN-Cas13a | LwCas13a | DNA, RNA | PCR or RPA | Fluorescence | 169 viruses infecting humans | 10−18 M | |
| COMET chip CRISPR-Cas13a assay [94] | Cas13a | RNA | – | Electrochemical readout | RNA, miRNA | 50 × 10−18 M | |
| Type V + TypeVI + TypeIII | SHERLOCKv2 [70] | Cas13, Cas12a, Csm6 | DNA, RNA | RPA | Fluorescence/Lateral flow assay | Viruses, bacteria, SNPs | 8 × 10−21 M |
In contrast, DETECTR (Fig. 2), SHERLOCK/SHERLOCKv2 (Fig. 3), and CRISPR-Chip can be considered breakthroughs in molecular diagnostics, representing almost ideal diagnostic tests that can be used in completely isothermal conditions, with minimal equipment and hands-on training of the personnel. Workflow of DETECTR and SHERLOCK/v2 is described in Fig. 1. Especially important is that SHERLOCKv2 is already adopted for colorimetric and visual detection of pathogens on lateral flow strips, so that these tests can be utilized for mass screening and rapid diagnostics in virtually any geographic region [70]. High sensitivity and specificity of CRISPR-Cas systems extend their potential application beyond qualitative and quantitative detection of different pathogens to genotyping and detecting single SNPs in human genomes.
In 2020 Ackerman et al., developed the Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids (CARMEN) combined with Cas13-detection method (CARMEN-Cas13) (Fig. 5 ) [95]. CARMEN is based on a microfluidic technology that uses a collection of droplet emulsions with PCR-amplified or RPA-amplified inputs and CRISPR-Cas detection mixtures (Cas13 protein, specific crRNAs and reporters). Each experimental sample or detection mix is then combined with a solution-based fluorescent color code serving as an optical identifier.
Fig. 5.
Schematics of CARMEN-Cas13 detection method. (1) amplification of target nucleic acids and their emulsification with color codes. (2) Generation of emulsions with CRISPR-Cas detection systems. (3) Pooling of two mixes. (4) Loading of mixed emulsions into the chip. (5) Schematic representation of a droplet with target molecules (sample droplet) and a droplet with CRISPR/Cas detection mix (CRISPR detection droplet). After merging, CRISPR-Cas system interacts with the target molecules, producing a specific fluorescent signa. The picture was created in BioRender.
The color-coded solutions are then emulsified in fluorous oil, then pooled and sampled into microwell chips. By applying the electric field, droplets in the wells merge together to initiate detection reactions simultaneously in all wells. Moreover, the authors generated a set of 1,050 solution-based color codes and, subsequently, designed the massive-capacity chip that permits running over 4,500 tests per chip and substantially reduces both turnaround time and increases the throughput capacity. Sensitivity and specificity of CARMEN-Cas13 matched those of SHERLOCK. Reportedly, CARMEN-Cas13 allows simultaneous detection of 169 viruses infecting humans, thus representing a highly-multiplexed platform for rapid and sensitive detection of human pathogens. Ackerman et al., also demonstrated that CARMEN-Cas13 enabled identification of clinically relevant viral mutations in a multiplex format, providing comprehensive subtyping of human influenza A virus strains and detection of dozens of HIV drug-resistance mutations. CARMEN-Cas13 was also successful to detect Zika sequences.
Many CRISPR-tests with different readout were developed to provide rapid and accurate detection of pathogenic nucleic acids. Such technologies CRISPR-Cas-based system that allow reading with the naked eye (CRISPR-Cas12a-NER [81], opvCRISPR [86], CASdetec [82]), using electrochemical biosensors (E-CRISPR [80], COMET chip CRISPR-Cas13a assay [94], PGMs-CRISPR [85]), colorimetry (NASBACC [17], CRISPR-Cas coupled with DNA-functionalized nanoparticles [89]), potentiometry (CRISPR-Chip [57]), fluorescent microscopy (ultralocalized Cas13a assay [93]), optical detection of anisotropy (CODA [87]), fluorescent readers such as fluorimeters (CARMEN [95], CRISPR-ENHANCE [90], CRISPR-FDS [84], CALIBURN [88], STOPCovid [73], CDetection [79], CAS-EXPAR [61]), smartphones (mobile phone microscopy [92], SHINE [72]) or lateral flow assays.
In terms of the ongoing COVID-19 pandemic, first CRISPR-based diagnostic tools have been approved by FDA. SHERLOCKv2 developed by Zhang et al. [70], [96], became the first CRISPR-test admitted to the clinical use. It includes three steps, namely (1) 25-minute isothermal amplification of extracted nucleic acids by RPA, (2) 30-minutes detection by Cas13 and (3) 2-minute incubation required to visualize the results at paper dipsticks. Most recently, Broughton et al., reported DETECTR-based detection of SARS-CoV-2 RNA from swab extracts within 30–40 min with the results being visualized on lateral flow strips using FAM-biotin reporter molecules [97]. Before reaction, target RNA molecules are pre-amplified by isothermal RT-LAMP amplification. Comparison of CDC qRT-PCR assay and DETECTR lateral flow assay demonstrated that PCR-based method has an analytic performance of 1 copy per µl, whereas for DETECTR it values to 10 copies per µl. False-negative samples were possible in viral titers below the limit of detection for DETECTR. Concordance between DETECTR and PCR detection of SARS-CoV-2 was 95–100%. Similarly, Lucia et al., presented the results on rapid, ultrasensitive and portable detection of SARS-CoV-2 using LbCa12a-based method combined with RPA isothermal pre-amplification [98]. The parameters of the detection system reported by Lucia et al. [98], are comparable with DETECTR-based CRISPR method [97]. CARMEN-Cas13 was shown to be rapidly re-directed for clinically-relevant purposes [95]. As such, a new test to detect SARS-CoV-2 was rapidly incorporated into CARMEN-Cas13 platform, allowing analysis of > 400 samples per chip. Overall, CRISPR-Cas detection systems can benefit a lot when combined with highly-multiplexed and high-throughput CARMEN technology.
Although the described works provide exciting perspectives that may grow into a huge area of research and molecular diagnostics, many challenges stand in the way. Very encouraging results demonstrating specificity and sensitivity of assays similar to PCR should be tested in real-world situations. Many modifications to existing assays may be introduced to increase sensitivity of the tests and time required to perform diagnostic tests. Moreover, specificity of CRISPR diagnostics may also be a concern, as Cas proteins can tolerate nucleotide mismatches and recognize non-specific templates, compromising assay validity. It is also not quite clear how discrepant the results of such tests could be when handled by different personnel or with varying qualities of biological samples.
4. PCR v. CRISPR-Cas
Understanding the pros and cons of using CRISPR-Cas and PCR is important for defining the scope, advantages, and downsides of each method. Large-scale testing and comparative analysis of different PCR-based and CRISPR diagnostic systems in the real world is mandatory to move new methods into laboratory and field diagnostics. Introducing CRISPR diagnostic tools into practice may improve control over infectious diseases, prevent their rapid spread, introduce diagnostic testing to finally enter the era of precise and personalized medicine [99], expand the use of massive population screening, and better control outbreaks of infectious diseases, including outbreaks in remote geographic areas. Timely, inexpensive, high-quality diagnosis of infectious and non-infectious diseases by rapid and readily available tests will certainly have a pronounced positive impact on human health on the global level, reducing morbidity and mortality.
Amplification of nucleic acids by PCR, the central dogma of molecular diagnostics, has long been the only practical way to detect trace amounts of infectious pathogens in biological samples. PCR analysis requires expensive equipment and qualified personnel to run the tests and interpret the results. Many attempts have been undertaken in the past years to avoid PCR cycling and adopt amplification to isothermal conditions. However, all other methods (NASBA, LAMP, RPA, HAD) [100] suffered from serious flaws compromising their utility in molecular diagnostics; these flaws included low sensitivity, low specificity, complicated reactions, and high price. Embedding methods of isothermal amplification into CRISPR diagnostic platforms eliminates the shortcomings of both technologies, achieving rapid and highly specific results. Compared to PCR, SHERLOCK/SHERLOCKv2 and DETECTR add another level of specificity because these methods not only use specific primers during isothermal amplification, but also specifically recognize target templates via Cas-sg/crRNA complex.
Another important property to consider is the ability of CRISPR proteins to tolerate nucleotide mismatches between sg/crRNA and the target template [14]. PCR is very sensitive to mismatched nucleotides at the 3ʹ-end of primers, which dramatically reduces non-specific primer annealing and ensures accurate target amplification [101]. CRISPR proteins recognize and bind degenerate sequences with multiple mismatches, depending on the nature of the CRISPR protein. On the one hand, this property may compromise the validity of diagnostic tests, leading to false positive results. On the other, ignoring nucleotide mismatches allows CRISPR proteins to recognize naturally variable genetic sequences. For instance, this property may facilitate effective detection of genetically distinct mutant HIV genomes, hepatitis C genomes, etc. Notably, SHERLOCKv2 can avoid isothermal amplification [70]. The specific signal is amplified by a combination of Cas13a proteins and co-activating Csm6 proteins, which react to cyclic oligonucleotides generated after specific cleavage of the target.
Important properties of CRISPR diagnostics are simplicity, stability of components, no requirement for isolation of nucleic acids from biological samples (in the most advanced technologies), and isothermal conditions of each step, requiring minimal equipment.
Overall, CRISPR diagnostics will not completely replace PCR in daily laboratory diagnostics in the near future, but may fill gaps in the global health care system, providing an opportunity for mass population screening, better control of infectious outbreaks, wider distribution of diagnostic techniques, and field-deployable diagnostics. Still, rapid development of the CRISPR field and identification of novel CRISPR systems with new, exciting properties will dictate the development of CRISPR diagnostics -- and laboratory diagnostics in general -- in the coming years.
5. Potential applications of CRISPR diagnostics
As was briefly mentioned, the ideal diagnostic assay should provide accurate and sensitive identification of the pathogen while being affordable, portable, and able to distinguish different variants of the pathogen. Currently, no such test exists. Developing new tools and methods which meet (or attempt to meet) the requirements of the WHO ideal diagnostic test can completely reshape epidemiological surveillance and medical health care system for the majority of infectious and non-infectious diseases in the world [102].
Highly specific DNA-binding nucleases provide the promising platform to create new diagnostic tests for rapid and inexpensive detection of infectious agents or genetic mutations. A combination of CRISPR proteins and isothermal methods of amplification, such as NASBA or RPA, may enter laboratory and field diagnostics in the very near future. Novel CRISPR-diagnostic tools, their targets and applications to detect viral pathogens are summarized in Table 3 .
Table 3.
Infectious agents as diagnostic targets for CRISPR-Cas-diagnostics.
| Pathogen | Target | CRISPR-diagnostic |
|---|---|---|
| Socially significant infectious diseases | ||
| Tuberculosis | DNA, antibiotic resistance testing | Chimeric dCas9 luciferase (PC Reporter) [54], CRISPR-MTB [106] |
| HIV | RNA | AIOD-CRISPR [74], |
| HPV | DNA, virus typing | ctPCR [55], CARP [77], ctPCR3.0 [56], DETECTR, CDetection [79], CRISPR-Cas12a-Mediated Interfacial Cleaving of Hairpin DNA Reporter for Electrochemical Nucleic Acid Sensing [83] |
| HBV | DNA, genotyping | CRISPR-Cas13a [115] |
| EBV | DNA, genotyping | SHERLOCK [143] |
| HBV-HDV co-infection | RNA, genotyping | – |
| HCV | RNA, genotyping, drug resistance testing | – |
| Influenza virus | RNA | SHERLOCK [144], CRISPR-Cas13a [145] |
| Emerging and re-emerging infectious diseases | ||
| SARS-CoV | RNA | DETECTR, OR-DETECTR etc. |
| MERS-CoV | RNA | OR-DETECTR [71] |
| SARS-CoV-2 | RNA | SHERLOCKv2 (FDA approved) [96] DETECTR [71], OR-DETECTR, OR-SHERLOCK, AIOD-CRISPR, CRISPR-Cas12a-NER, SHINE, CRISPR-FDS, iSCAN, CONAN, CASdetec, VaNGuard, STOPCovid, CREST [146], PGMs‐CRISPR [85], CARMEN-Cas13[95], CRISPR-LbCas13 [98], opvCRISPR [86], CODA[87], CALIBURN [88] |
| Dengue virus | RNA | SHERLOCK, SHERLOCKv2, HUDSON + SHERLOCKv2 |
| Ebola virus | RNA | SHERLOCK EBOV assay [147], CRISPR-Cas13a [91] |
| Zika virus | RNA | SHERLOCK [18], SHERLOCKv2 [70], HUDSON + SHERLOCK [75], NASBACC [17] |
5.1. CRISPR-tools for socially significant infectious diseases
Below is an overview of some of the most socially significant infectious diseases whose treatment would be greatly improved if portable and affordable CRISPR-diagnostic platforms become available. In the world, morbidity of infectious diseases such as tuberculosis, HIV, and viral hepatitis, as well as the epidemiological situation associated with the burden of these diseases, can be greatly reduced by currently available therapeutics when CRISPR diagnostics are finally applied.
Tuberculosis is an airborne infectious disease caused by M. tuberculosis. According to the latest data, more than one quarter of the world’s population is infected. Causing over 1.3 million deaths per year, tuberculosis remains one of the leading infectious causes of death worldwide [103]. Real-time PCR analysis is a rapid and accurate method for diagnosing M. tuberculosis infection [104]. Moreover, PCR is widely used to detect drug resistance [105]. Zhang et al. created a new CRISPR-based approach for detecting M. tuberculosis DNA in clinical samples [54]. Highly sensitive and rapid Cas12a-based system (CRISPR-MTB) has also been recently adapted to identify M. tuberculosis [106] (Table 3).
HIV infection remains to be a major public health issue. >37,9 million people worldwide are infected [8], many in developing countries with limited access to highly active antiretroviral therapy (HAART) regimens [107]. Early diagnosis is key for successful treatment and prevention of the further spread of the infection, so a new, rapid assay detecting HIV RNA in patients’ blood samples is urgently needed.
Approximately 2.4 of 100,000 women die annually due to cervical cancer caused by HPV [108]. The gold standard for HPV diagnosis is viral DNA detection [109] and PAP testing [110]. There are many types of the virus, some of them characterized by their association with cervical cancer as highly oncogenic; HPV16 and HPV18 are responsible for most HPV-related cancers. Several CRISPR systems, including CARP [77] and DETECTR [51], were created for HPV diagnostics (Table 3). DETECTR enables sensitive, specific, and robust detection of the virus at attomolar concentrations and allows differentiation of viral subtypes.
Viral hepatitis B, D and C affect millions of people around the world [7], [111]. Hepatitis B virus (HBV), HBV + hepatitis D virus (HDV) co-infection and hepatitis C virus (HCV) infection is one of the principal causes of death among infectious viral diseases [112]; over 1.3 million people die annually due to consequences of HBV, HBV-HDV co-infection and HCV infection. Chronic viral hepatitis is usually asymptomatic until advanced liver injury develops, and>80–90% of patients are unaware that they are infected [113]. In low- and middle-income regions, most infected people are still not diagnosed, and many HBV, HBV-HDV co-infection and HCV carriers do not receive antiviral therapy [114]. No CRISPR-based diagnostic tests to detect HBV, HDV and HCV are yet available in clinic. However, CRISPR-Cas13a has been recently successfully employed to detect HBV DNA [115]. Rapid, sensitive, and specific point-of-care assays are urgently needed to detect viral DNA or RNA in blood samples, genotype the virus, and reveal drug resistance.
5.2. CRISPR-tools for emerging and re-emerging infectious diseases
Three million people are estimated to be at risk of dengue fever [116], caused by Dengue virus that is transmitted by mosquitos and widespread in Latin America and Asia [117]. Dengue fever symptoms resemble those of hemorrhagic fevers caused by Ebola virus and other viruses [118]. Thus, laboratory diagnosis of dengue fever is crucial for rapid management of patients with severe infection [119]. Dengue infection can be confirmed by isolating the virus and immunologically detecting viral protein Ns1 [120]. Real-time PCR analysis is widely used for Ebola virus diagnostics, but expensive equipment and well-trained personnel are required to obtain accurate results [121]. Most importantly, the situation in the areas of outbreaks requires urgent action to identify infected persons and curb disease spread. In this respect, PCR technologies are inferior to CRISPR-tools in terms of time required to get the results and that they cannot be readily deployable for field diagnostics, especially in remote areas. Developing new diagnostic tools will hopefully help to prevent further spread of infection and better inform patient treatment. The CRISPR based detection system NASBACC [17]can detect dengue virus, but SHERLOCKv2 [70] is the most sensitive system that differentiates the four viral serotypes with high efficacy.
Zika virus is transmitted by mosquitos and has been reported to lead to neurological defects in newborns whose mothers were infected during pregnancy. Accurate and rapid diagnosis is critical for timely treatment [122]. Several approaches were utilized for detecting the disease: NASBACC [17], SHERLOCKv2 [70] and HUDSON + SHERLOCK [75]. SHERLOCKv2 is more sensitive and robust, enabling femtomolar detection.
Coronaviral infections represent a daunting threat to the global health, appearing every 10–20 years with new outbreaks, diseasing thousands of people with relatively high death toll. The first case of severe acute respiratory syndrome (SARS) has been documented in November 2002 in Foshan, China [123]. During four months the disease spread into 27 countries, and a total of 8,096 cases were identified [124] . Since the outbreak, only the sporadic cases were reported. Bats are known to be a natural reservoir for a wide range of coronaviruses, but human-bat interactions are relatively rare, so an intermediate host is thought to be required to infect humans. However, it has been recently demonstrated that coronaviruses directly isolated from bats are able to infect human cells without prior adaptation [125], [126]. These data indicate that novel infections can emerge potentially leading to wide-spread outbreaks [5]. Middle East respiratory syndrome virus (MERS-CoV) was isolated from sputum of a man who died of acute pneumonia and renal failure in 2012 [127]. By April 2016, MERS has been detected in 27 countries, and 1,728 infection cases were confirmed [128] . MERS-CoV-like viruses are shown to inhabit dromedary camels [5], [129]. Thus, MERS-CoV zoonotic transmission is not rare, and MERS-CoV continues to infect humans due to the fact that dromedary camel are farmed and are in close proximity to humans [5].Both SARS and MERS viruses have high risk of nosocomial transmission [130], [131] and were spread from the initial outbreak location by infected individuals [132], [133], [134], [135]. PCR assays have been developed for SARS and MERS CoVs detection [136], [137]. New human Coronavirus infection emerged in December 2019 in Wuhan, China, and currently it is an ongoing outbreak that spread through the mainland of China and neighboring countries [6]. The first cases of novel coronavirus infection were reported in a fresh seafood market which distributed various mammals and exotic wildlife animals, including bats, snakes, rats, beavers etc. Such areas of crowded people, exotic wildlife and low-quality sanitary conditions represent high-risk places for emerging of novel zoonotic infections. The symptoms of the novel viral infection were identified as fever, dry cough, headache and pneumonia, altered sense of smell or taste (anosmia or hyposmia) [138], [139], [140], [141]. The infectious agent was identified as Coronavirus, designated SARS-CoV-2. At the current time, the pandemic of SARS-CoV-2 infection has already infected > 120 millions people worldwide [142].
Clinical features of pneumonia caused by SARS-CoV-2 were described shortly after the outbreak. Full-genome sequencing of SARS-CoV-2 revealed that it belongs to group 2B of the Betacoronaviridae family, possessing 70% similarity to SARS-CoV. The source of the virus still remains a matter of debate, although an initial report suggested snakes or pangolins as possible intermediate hosts [148], [149]. Nevertheless, another scientific group reported that virus potentially has a bat origin [150]. Due to the rapid spread of the virus, express diagnostics is essential for reducing the risk of further transmission. CRISPR based diagnostic tests would be helpful for effective identification, diagnosis and management of the infection. Since the outbreak and sequencing of the virus, a plethora of PCR-systems for detecting the pathogen were developed. However, PCR does not fit the relevant needs to control and curb the emerging outbreaks, as is noted throughout the review. The ideal tool would have been an express-method to unequivocally detect the pathogen and identified infected persons. Although the field of CRISPR-diagnostics is at the early stage of development, further advancement of such tests in the express-format holds a lot of promise both in the daily practice (point-of-care tests) and in epidemiological surveillance programs to prevent disease spread. While this manuscript was in preparation, several CRISPR-based methods for rapid and specific detection of SARS-CoV-2 were developed [97], [98]. The SHERLOCKv2 platform was for the first time approved for use by FDA [96]. The results of the first pilot study of SHERLOCK CRISPR SARS-CoV-2 test kit demonstrated accurate detection of the presense or absence of the SARS-CoV-2 in clinical samples. The results of SHERLOCK CRISPR SARS-CoV-2 test kit were 100% concordant with those of PCR test [151].
Field-deployable diagnostic tests that have low turnaround time and could be rapidly distributed for timely detection of viral pathogens can prevent global and regional spread given extremely rapid transportation systems and high contagiosity of emerging viral pathogens. Frequent (every 20–30 years, as reported by far) emergence of novel, highly contagious viral infections, proves the urgent need for developing field-deployable diagnostics which could have a profound effect on identification of new infectious cases and prevention of disease spread.
Rapid pathogen detection is a crucial part of molecular diagnostic. Recently described CRISPR-based methods for HPV, Zika virus, and dengue virus detection demonstrated high potency for using in diagnostic field. Although standard methods of nucleic acid amplification are highly effective, they require expensive instrumentation and well-trained lab personnel. Point-of-care assays based on CRISPR are a versatile detection platform for clinical diagnosis of infectious diseases, enabling rapid and robust detection of pathogens in limited-resource areas.
6. Conclusion
Although many milestones have been surpassed within several years, CRISPR diagnostics is rapidly advancing into the clinical use. In 2020, the first CRISPR diagnostic system (Sherlock™ CRISPR SARS-CoV-2 kit) received FDA emergency use authorization in the United States for SARS-CoV-2 diagnosis. Numerous obstacles may still hamper rapid translation of laboratory results into practice, including off-target recognition and potential false-positive results associated with the intricate property of CRISPR proteins to tolerate mismatched nucleotides, but the early results demonstrate that CRISPR-diagnostics are starting to conquer their place in modern health care system. Designing software-backed optimal sgRNAs with minimal potential off-target effects (e.g., CHOPCHOP [152]), and identifying or inventing novel CRISPR proteins with more practical properties or limited mismatch tolerance may solve this potential problem. Cas proteins can currently be adjusted by, for instance, direct evolution to change PAM sequence requirements (shorter/longer PAMs or PAMs with certain nucleotide preferences [38], [41]) or higher specificity (e.g., enSpCas9 [32]). Sg/crRNAs have also undergone many modifications, starting from shortened sg/crRNAs [153] to sg/crRNAs with engineered RNA hairpins [154]. Predicting and identifying novel CRISPR systems is currently the hottest topic in biology. One of the many examples of successful identification of CRISPR proteins with previously unknown properties is Cas13a (previously referred to as C2c2), which has transformed into a powerful molecular tool [18]. Application of CRISPR may go far beyond detection of nucleic acids or identification of clinically-relevant mutations, as several CRISPR technologies have been devised to identify proteins [80] and could be utilized for detecting small molecules [155] (e.g. toxic agents, opioids, drugs etc.). Practically, they may transform into new, more efficient and cheap approaches that could replace many conventional techniques, such as immunosorbent assays, chromatography etc.
Potential challenges mostly include the low quality of clinical sample, when specificity or sensitivity of the test could be compromised by the abundance of reaction inhibitors and interfering compounds. It is also unclear what types of the samples could be used for CRISPR-diagnostics, such as dry blood spot cards etc. Automation and high-throughput use of CRISPR-screening has just recently been addressed in a novel CARMEN-Cas13 technology, providing exciting results [95]. Optimization and simplification of the CRISPR-diagnostic procedure is also the key for their successful introduction into the practice. Eliminating an amplification step without substantially affecting sensitivity of the CRISPR-diagnostic system is important not only in terms of time, but also to reduce the risks of amplification bias and cross-contamination of the samples. In this respect, many technologies succeeded to minimize potential contamination issues by introducing one pot procedures (OR-DETECTR/OR-SHERLOCK, SHINE, AIOD) with thermostable Cas proteins or by using amplification-free strategies (E-CRISPR, ultralocalized assay, optimized assay with several crRNAs etc.). It is important that these simplified procedures also ensure minimal sample loss as they do not require several washing and pipetting steps. The majority of the currently developed CRISPR-diagnostic tests does not address the issue of multiplex detection (simultaneous detection of several targets), quantitative detection and unbiased interpretation of the results. Most importantly, CRISPR-diagnostics may provide the means for rapid, express-diagnostics, but the time currently required for performing the CRISPR-mediated identification of target nucleic acids is still far from being considered an «express method».
To conclude, using CRISPR-based methods to detect and qualitatively analyze infectious pathogens is a new reality in the field of molecular diagnostics. Developing new CRISPR tools and platforms for molecular diagnosis promises to reshape health care and improve epidemiological management on a global level.
Funding
The authors acknowledge the support of RFBR-DFG grant 20-515-12010 and GL 595/9-1; and RFBR grant 20-015-00442.
CRediT authorship contribution statement
Anastasiya Kostyusheva: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. Sergey Brezgin: Writing – review & editing. Yurii Babin: Resources. Irina Vasilyeva: Resources. Dieter Glebe: Writing – review & editing, Funding acquisition. Dmitry Kostyushev: Conceptualization, Visualization, Writing – original draft, Writing – review & editing, Funding acquisition. Vladimir Chulanov: Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Hwang H., Hwang B.-Y., Bueno J. Biomarkers in infectious diseases. Disease Markers. 2018;2018:8509127. doi: 10.1155/2018/8509127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kilbourne E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006;12:9. doi: 10.3201/eid1201.051254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moon S., et al. Will Ebola change the game? Ten essential reforms before the next pandemic. The report of the Harvard-LSHTM Independent Panel on the Global Response to Ebola. Lancet. 2015;386:2204–2221. doi: 10.1016/S0140-6736(15)00946-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gatherer D., Kohl A. Zika virus: a previously slow pandemic spreads rapidly through the Americas. J. Gen. Virol. 2016;97:269–273. doi: 10.1099/jgv.0.000381. [DOI] [PubMed] [Google Scholar]
- 5.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: Recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. Cohen, D. Normile. New SARS-like virus in China triggers alarm. (2020). [DOI] [PubMed]
- 7.World Health Organization Global hepatitis report, 2017 Who 2017 ISBN 978-92-4-156545-5.
- 8.UNAIDS. AIDS statistics—2018 fact sheet. UNAIDS website. unaids. org/en/resources/fact-sheet. Accessed May 31, (2019).
- 9.Watzinger F., Ebner K., Lion T. Detection and monitoring of virus infections by real-time PCR. Mol. Aspects Med. 2006;27:254–298. doi: 10.1016/j.mam.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shalhoub S., et al. IFN-α2a or IFN-β1a in combination with ribavirin to treat Middle East respiratory syndrome coronavirus pneumonia: a retrospective study. J. Antimicrob. Chemother. 2015;70:2129–2132. doi: 10.1093/jac/dkv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peeling R.W., Holmes K.K., Mabey D., Ronald A. Rapid tests for sexually transmitted infections (STIs): the way forward. Sex. Transm. Infect. 2006;82:v1–v6. doi: 10.1136/sti.2006.024265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.H. Kettler, K. White, S.J. Hawkes. Mapping the landscape of diagnostics for sexually transmitted infections: key findings and recommendations. (2004).
- 13.K. Davies. CRISPR Pioneers Doudna and Charpentier Win 2020 Nobel Prize for Chemistry. (2020).
- 14.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riordan S.M., Heruth D.P., Zhang L.Q., Ye S.Q. Application of CRISPR/Cas9 for biomedical discoveries. Cell Biosci. 2015;5:33. doi: 10.1186/s13578-015-0027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brezgin S., et al. Clearing of foreign episomal DNA from human cells by CRISPRa-mediated activation of cytidine deaminases. Int. J. Mol. Sci. 2020;21:6865. doi: 10.3390/ijms21186865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardee K., et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 18.Gootenberg J.S., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishino Y., Shinagawa H., Makino K., Amemura M., Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haft D.H., Selengut J., Mongodin E.F., Nelson K.E. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1 doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makarova K.S., Koonin E.V. Annotation and classification of CRISPR-Cas systems. Methods Mol. Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brouns S.J.J., et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayes R.P., et al. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016;530:499–503. doi: 10.1038/nature16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazlauskiene M., Kostiuk G., Venclovas C., Tamulaitis G., Siksnys V. A cyclic oligonucleotide signaling pathway in type III CRISPR-Cas systems. Science. 2017;357:605–609. doi: 10.1126/science.aao0100. [DOI] [PubMed] [Google Scholar]
- 25.Niewoehner O., et al. Type III CRISPR-Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017;548:543–548. doi: 10.1038/nature23467. [DOI] [PubMed] [Google Scholar]
- 26.Koonin E.V., Makarova K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2019;374:20180087. doi: 10.1098/rstb.2018.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova K.S., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shmakov S., et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017;15:169–182. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohanraju P., et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science (80-.) 2016;353:aad5147. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 30.Cong L., et al. Multiplex Genome Engineering Using CRISPR/VCas Systems. Science (80-.) 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jinek M., et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slaymaker I.M., et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleinstiver B.P., et al. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casini A., et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat. Biotechnol. 2018;36:265. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ikeda A., Fujii W., Sugiura K., Naito K. High-fidelity endonuclease variant HypaCas9 facilitates accurate allele-specific gene modification in mouse zygotes. Commun. Biol. 2019;2:1–7. doi: 10.1038/s42003-019-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J.K., et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018;9:3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esvelt K.M., et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1123. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C.M., Cradick T.J., Bao G. The Neisseria meningitidis CRISPR-Cas9 system enables specific genome editing in mammalian cells. Mol. Ther. 2016;24:645–654. doi: 10.1038/mt.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostyushev D., et al. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell. Mol. Life Sci. 2019;76:1779–1794. doi: 10.1007/s00018-019-03021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ran F.A., et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller M., et al. Streptococcus thermophilus CRISPR-Cas9 systems enable specific editing of the human genome. Mol. Ther. 2016;24:636–644. doi: 10.1038/mt.2015.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Amrani N., et al. NmeCas9 is an intrinsically high-fidelity genome-editing platform. Genome Biol. 2018;19:214. doi: 10.1186/s13059-018-1591-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makarova K.S., et al. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2019;1–17 doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D., et al. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat. Biotechnol. 2016;34:863–868. doi: 10.1038/nbt.3609. [DOI] [PubMed] [Google Scholar]
- 45.Bothmer A., et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 2017;8:13905. doi: 10.1038/ncomms13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrington L.B., et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abudayyeh O.O., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox D.B.T., et al. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makarova K.S., Wolf Y.I., Koonin E.V. Classification and nomenclature of CRISPR-Cas systems: where from here? Cris. J. 2018;1:325–336. doi: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.K. Yoshimi, et al. Rapid and accurate detection of novel coronavirus SARS-CoV-2 using CRISPR-Cas3. (2020).
- 51.Chen J.S., et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang H., La Russa M., Qi L.S. CRISPR/Cas9 in genome editing and beyond. Annu. Rev. Biochem. 2016;85:227–264. doi: 10.1146/annurev-biochem-060815-014607. [DOI] [PubMed] [Google Scholar]
- 53.Brezgin S., Kostyusheva A., Kostyushev D., Chulanov V. Dead cas systems: types, principles, and applications. Int. J. Mol. Sci. 2019;20:6041. doi: 10.3390/ijms20236041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y., et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth. Biol. 2017;6:211–216. doi: 10.1021/acssynbio.6b00215. [DOI] [PubMed] [Google Scholar]
- 55.Wang Q., Zhang B., Xu X., Long F., Wang J. CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method. Sci. Rep. 2018;8:14126. doi: 10.1038/s41598-018-32329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang B., Xia Q., Wang Q., Xia X., Wang J. Detecting and typing target DNA with a novel CRISPR-typing PCR (ctPCR) technique. Anal. Biochem. 2018;561:37–46. doi: 10.1016/j.ab.2018.09.012. [DOI] [PubMed] [Google Scholar]
- 57.Hajian R., et al. Detection of unamplified target genes via CRISPR–Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geim A.K., Novoselov K.S. World Scientific; 2010. The rise of graphene. in Nanoscience and technology: a collection of reviews from nature journals 11–19. [Google Scholar]
- 59.Zhou W., et al. A CRISPR–Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quan J., et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang M., Zhou X., Wang H., Xing D. Clustered regularly interspaced short palindromic Repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal. Chem. 2018;90:2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y., Zhang C. Sensitive detection of microRNA with isothermal amplification and a single-quantum-dot-based nanosensor. Anal. Chem. 2012;84:224–231. doi: 10.1021/ac202405q. [DOI] [PubMed] [Google Scholar]
- 63.Wang X., et al. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14:2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 64.He Q., et al. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens. Bioelectron. 2020;154 doi: 10.1016/j.bios.2020.112068. [DOI] [PubMed] [Google Scholar]
- 65.Li S.-Y., et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li L., et al. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 67.Yan W.X., et al. Functionally diverse type V CRISPR-Cas systems. Science (80-.) 2019;363:88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.L. Li, S. Li, J. Wang. CRISPR-Cas12b-assisted nucleic acid detection platform. bioRxiv 362889 (2018) doi:10.1101/362889.
- 69.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gootenberg J.S., et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Y., et al. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021;19:1–10. doi: 10.1186/s12967-021-02741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Arizti-Sanz J., et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020;11:1–9. doi: 10.1038/s41467-020-19097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Joung J., et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 2020;383:1492–1494. doi: 10.1056/NEJMc2026172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.X. Ding, K. Yin, Z. Li, C. Liu. All-in-One Dual CRISPR-Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. bioRxiv Prepr. Serv. Biol. 2020.03.19.998724 (2020) doi:10.1101/2020.03.19.998724.
- 75.Myhrvold C., et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guk K., et al. A facile, rapid and sensitive detection of MRSA using a CRISPR-mediated DNA FISH method, antibody-like dCas9/sgRNA complex. Biosens. Bioelectron. 2017;95:67–71. doi: 10.1016/j.bios.2017.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Zhang B., et al. Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique. Anal. Bioanal. Chem. 2018;410:2889–2900. doi: 10.1007/s00216-018-0873-5. [DOI] [PubMed] [Google Scholar]
- 78.Aquino-Jarquin G. CRISPR-Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomed. Nanotechnol. Biol. Med. 2019;18:428–431. doi: 10.1016/j.nano.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 79.Teng F., et al. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019;20:1–7. doi: 10.1186/s13059-019-1742-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dai Y., et al. Exploring the trans-cleavage activity of CRISPR-Cas12a (cpf1) for the development of a universal Electrochemical biosensor. Angew. Chemie Int. Ed. 2019;58:17399–17405. doi: 10.1002/anie.201910772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.X. Wang, et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. (2020). [DOI] [PMC free article] [PubMed]
- 82.Guo L., et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020;6:1–4. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang D., et al. CRISPR/Cas12a-Mediated interfacial cleaving of hairpin DNA reporter for electrochemical nucleic acid sensing. ACS Sens. 2020;5:557–562. doi: 10.1021/acssensors.9b02461. [DOI] [PubMed] [Google Scholar]
- 84.Huang Z., et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.D. Huang, et al. A CRISPR‐Cas12a‐Derived Biosensor Enabling Portable Personal Glucose Meter Readout for Quantitative Detection of SARS‐CoV‐2. Biotechnol. Bioeng. [DOI] [PubMed]
- 86.Wang R., et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021;172 doi: 10.1016/j.bios.2020.112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee C.Y., et al. Fluorescence polarization system for rapid COVID-19 diagnosis. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Y. Wang, et al. Development of a broadly applicable Cas12a-linked beam unlocking reaction for sensitive and specific detection of respiratory pathogens including SARS-CoV-2. ACS Chem. Biol. (2021). [DOI] [PubMed]
- 89.Choi J.-H., Lim J., Shin M., Paek S.-H., Choi J.-W. CRISPR-Cas12a-based nucleic acid amplification-free DNA biosensor via Au nanoparticle-assisted metal-enhanced fluorescence and colorimetric analysis. Nano Lett. 2020 doi: 10.1021/acs.nanolett.0c04303. [DOI] [PubMed] [Google Scholar]
- 90.Nguyen L.T., et al. CRISPR-ENHANCE: an enhanced nucleic acid detection platform using Cas12a. Methods. 2021 doi: 10.1016/j.ymeth.2021.02.001. [DOI] [PubMed] [Google Scholar]
- 91.Qin P., et al. Rapid and fully microfluidic Ebola virus detection with CRISPR-Cas13a. ACS Sens. 2019;4:1048–1054. doi: 10.1021/acssensors.9b00239. [DOI] [PubMed] [Google Scholar]
- 92.Fozouni P., et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184:323–333. doi: 10.1016/j.cell.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tian T., et al. An ultralocalized Cas13a assay enables universal and nucleic acid amplification-free single-molecule RNA diagnostics. ACS Nano. 2020 doi: 10.1021/acsnano.0c08165. [DOI] [PubMed] [Google Scholar]
- 94.Sheng Y., et al. A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens. Bioelectron. 2021;178 doi: 10.1016/j.bios.2021.113027. [DOI] [PubMed] [Google Scholar]
- 95.Ackerman C.M., et al. Massively multiplexed nucleic acid detection with Cas13. Nature. 2020;1–6 doi: 10.1038/s41586-020-2279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang F., Abudayyeh O.O., Gootenberg J.S. A protocol for detection of COVID-19 using CRISPR diagnostics. A Protoc. Detect. 2020 COVID-19 using Cris. diagnostics 8. [Google Scholar]
- 97.Broughton J.P., et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;1–5 doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.C. Lucia, P.-B. Federico, G.C. Alejandra. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv (2020).
- 99.Bodrova T.A., et al. Introduction into PPPM as a new paradigm of public health service: an integrative view. EPMA J. 2012;3:16. doi: 10.1186/1878-5085-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zanoli L.M., Spoto G. Isothermal amplification methods for the detection of nucleic acids in microfluidic devices. Biosensors. 2012;3:18–43. doi: 10.3390/bios3010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boyle B., Dallaire N., MacKay J. Evaluation of the impact of single nucleotide polymorphisms and primer mismatches on quantitative PCR. BMC Biotechnol. 2009;9:75. doi: 10.1186/1472-6750-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peeling R.W. Testing for sexually transmitted infections: a brave new world? Sexually Transmitted Infect. 2006;82:425–430. doi: 10.1136/sti.2005.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sulis G., Roggi A., Matteelli A., Raviglione M.C. Tuberculosis: epidemiology and control. Mediterr. J. Hematol. Infect. Dis. 2014;6 doi: 10.4084/MJHID.2014.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Salian N.V., Rish J.A., Eisenach K.D., Donald Cave M., Bates J.H. Polymerase chain reaction to detect Mycobacterium tuberculosis in histologic specimens. Am. J. Respir. Crit. Care Med. 1998;158:1150–1155. doi: 10.1164/ajrccm.158.4.9802034. [DOI] [PubMed] [Google Scholar]
- 105.Hristea A., et al. Detection of Mycobacterium tuberculosis resistance mutations to rifampin and isoniazid by real-time PCR. Indian J. Med. Microbiol. 2010;28:211. doi: 10.4103/0255-0857.66474. [DOI] [PubMed] [Google Scholar]
- 106.Ai J.-W., et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019;8:1361–1369. doi: 10.1080/22221751.2019.1664939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Moore R.D., Chaisson R.E. Natural history of HIV infection in the_era of combination antiretroviral therapy. Aids. 1999;13:1933–1942. doi: 10.1097/00002030-199910010-00017. [DOI] [PubMed] [Google Scholar]
- 108.Franco E.L., Duarte-Franco E., Ferenczy A. Cervical cancer: epidemiology, prevention and the role of human papillomavirus infection. CMAJ. 2001;164:1017–1025. [PMC free article] [PubMed] [Google Scholar]
- 109.Rozendaal L., et al. PCR-based high-risk HPV test in cervical cancer screening gives objective risk assessment of women with cytomorphologically normal cervical smears. Int. J. Cancer. 1996;68:766–769. doi: 10.1002/(SICI)1097-0215(19961211)68:6<766::AID-IJC13>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 110.Naucler P., et al. Human papillomavirus and Papanicolaou tests to screen for cervical cancer. N. Engl. J. Med. 2007;357:1589–1597. doi: 10.1056/NEJMoa073204. [DOI] [PubMed] [Google Scholar]
- 111.Chulanov V.P., et al. Hepatitis C can be cured: will hepatitis B become next? Ter. Arkh. 2017;89:4–13. doi: 10.17116/terarkh201789114-13. [DOI] [PubMed] [Google Scholar]
- 112.Papatheodoridis G., et al. Addressing barriers to the prevention, diagnosis and treatment of hepatitis B and C in the face of persisting fiscal constraints in Europe: report from a high level conference. J. Viral Hepat. 2016;23(Suppl 1):1–12. doi: 10.1111/jvh.12493. [DOI] [PubMed] [Google Scholar]
- 113.Razavi-Shearer D., et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol. Hepatol. 2018;3:383–403. doi: 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 114.Denniston M.M., Klevens R.M., McQuillan G.M., Jiles R.B. Awareness of infection, knowledge of hepatitis C, and medical follow-up among individuals testing positive for hepatitis C: National Health and Nutrition Examination Survey 2001–2008. Hepatology. 2012;55:1652–1661. doi: 10.1002/hep.25556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.S. Wang, et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin. Microbiol. Infect. (2020). [DOI] [PubMed]
- 116.Stanaway J.D., et al. The global burden of dengue: an analysis from the global burden of disease study 2013. Lancet Infect. Dis. 2016;16:712–723. doi: 10.1016/S1473-3099(16)00026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cafferata M.L., et al. Dengue epidemiology and burden of disease in Latin America and the Caribbean: a systematic review of the literature and meta-analysis. Value Heal. Reg. issues. 2013;2:347–356. doi: 10.1016/j.vhri.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 118.Benzine J.W., et al. Molecular diagnostic field test for point-of-care detection of ebola virus directly from blood. J. Infect. Dis. 2016;214:S234–S242. doi: 10.1093/infdis/jiw330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bhatt S., et al. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang S.M., Sekaran S.D. Early diagnosis of Dengue infection using a commercial Dengue Duo rapid test kit for the detection of NS1, IGM, and IGG. Am. J. Trop. Med. Hyg. 2010;83:690–695. doi: 10.4269/ajtmh.2010.10-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pang J., Chia P.Y., Lye D.C., Leo Y.S. Progress and challenges towards point-of-care diagnostic development for dengue. J. Clin. Microbiol. 2017;55:3339–3349. doi: 10.1128/JCM.00707-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Waggoner J.J., Pinsky B.A. Zika virus: diagnostics for an emerging pandemic threat. J. Clin. Microbiol. 2016;54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhong N.S., et al. Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003. Lancet. 2003;362:1353–1358. doi: 10.1016/S0140-6736(03)14630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.World Health Organization Epidemic and Pandemic Alert Response: Summary of probably SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Based Data as. 2003;31 [Google Scholar]
- 125.Ge X.Y., et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Menachery V.D., et al. SARS-like cluster of circulating bat coronavirus pose threat for human emergence HHS Public access. Nat. Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hijawi B., et al. Novel coronavirus infections in Jordan, April 2012: Epidemiological findings from a retrospective investigation. East. Mediterr. Heal. J. 2013;19:12–18. [PubMed] [Google Scholar]
- 128.Almaghrabi R.S., Omrani A.S. Middle East respiratory syndrome coronavirus (MERS-CoV) infection. Br. J. Hosp. Med. 2017;78:23–26. doi: 10.12968/hmed.2017.78.1.23. [DOI] [PubMed] [Google Scholar]
- 129.J.S.M. Sabir, et al. Co-circulation of three camel coronavirus We conducted surveillance for CoVs in dromedary camels in species and recombination of MERS-CoVs Saudi Arabia, the country most affected by MERS, from May 2014 to April 2015. Initially, in Saudi Arabia. Sci. Reports 1–6 (2015).
- 130.Chowell G., et al. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015;13:1–12. doi: 10.1186/s12916-015-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hunter J.C., et al. Transmission of middle east respiratory syndrome coronavirus infections in healthcare settings, abu dhabi. Emerg. Infect. Dis. 2016;22:647–656. doi: 10.3201/eid2204.151615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee N., Hul D., Wu A., Chan P., Atabay B. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;1 doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 133.Guan Y., et al. Molecular epidemiology of the novel CoV that causes SARS. Lancet. 2004;363:99–104. doi: 10.1016/S0140-6736(03)15259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wise J. Patient with new strain of coronavirus is treated in intensive care at London hospital. BMJ. 2012;345:6455. doi: 10.1136/bmj.e6455. [DOI] [PubMed] [Google Scholar]
- 135.Centers K. Middle east respiratory syndrome coronavirus outbreak in the Republic of Korea, 2015. Osong Public Heal. Res. Perspect. 2015;6:269–278. doi: 10.1016/j.phrp.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lu X., et al. Real-time reverse transcription-pcr assay panel for middle east respiratory syndrome coronavirus. J. Clin. Microbiol. 2014;52:67–75. doi: 10.1128/JCM.02533-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang P., et al. A rapid and specific assay for the detection of MERS-CoV. Front. Microbiol. 2018;9:1–9. doi: 10.3389/fmicb.2018.01101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wuhan Municipal Health Commision. Wuhan Municipal Health and Health Commission’s Briefing on the Current Pneumonia Epidemic Situation in Our City. http://wjw.wuhan.gov.cn/front/web/showDetail/2019123108989.
- 139.Begam N., Bashar M.A. Olfactory and taste disorders in patients with SARS-CoV-2 infection. Int. Arch. Otorhinolaryngol. 2020;24:e391–e392. doi: 10.1055/s-0040-1713142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Menni C., et al. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lechner M., et al. Anosmia and hyposmia in health-care workers with undiagnosed SARS-CoV-2 infection. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.World Health Organization. WHO Statement Regarding Cluster of Pneumonia Cases in Wuhan, ChinaAvailable online: https://www.who.int/china/news/detail/09-01-2020-who-statement-regarding-cluster-of-pneumonia-cases-in-wuhan-china.
- 143.Wu Y., Liu S.-X., Wang F., Zeng M.-S. Room temperature detection of plasma Epstein-Barr virus DNA with CRISPR–Cas13. Clin. Chem. 2019;65:591–592. doi: 10.1373/clinchem.2018.299347. [DOI] [PubMed] [Google Scholar]
- 144.Freije C.A., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu Y., et al. CRISPR-Cas13a nanomachine based simple technology for avian influenza A (H7N9) virus on-site detection. J. Biomed. Nanotechnol. 2019;15:790–798. doi: 10.1166/jbn.2019.2742. [DOI] [PubMed] [Google Scholar]
- 146.H. Rahimi, et al. CRISPR Systems for COVID-19 Diagnosis. ACS sensors acssensors.0c02312 (2021) doi:10.1021/acssensors.0c02312. [DOI] [PubMed]
- 147.Barnes K.G., et al. Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time. Nat. Commun. 2020;11:1–10. doi: 10.1038/s41467-020-17994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.W. Ji, W. Wang, X. Zhao, J. Zai, X. Li. Homologous recombination within the spike glycoprotein of the newly identified coronavirus 2019-nCoV may boost cross-species transmission from snake to human. (2019) doi:10.1002/jmv.25682.
- 149.T. Zhang, Q. Wu, Z. Zhang. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. (2020). [DOI] [PMC free article] [PubMed]
- 150.Peng Zhou, Xing-Lou Yang, Xian-Guang Wang, Ben Hu, Lei Zhang, Wei Zhang, Hao-Rui Si, Yan Zhu, Bei Li, Chao-Lin Huang, Hui-Dong Chen, Jing Chen, Yun Luo, Hua Guo, Ren-Di Jiang, Mei-Qin Liu, Ying Chen, X.-, Rui Shen, Xi Wang, Xiao-Shuang Zheng, Kai Zhao, Quan-Jiao Chen, F., Deng, Lin-Lin Liu, Bing Yan, Fa-Xian Zhan, Yan-Yi Wang, G.-F. X. & Shi, Z.-L. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in 2 humans and its potential bat origin. BioRxiv (2020).
- 151.Khan W.A., Barney R.E., Tsongalis G.J. CRISPR-cas13 enzymology rapidly detects SARS-CoV-2 fragments in a clinical setting. medRxiv. 2020 doi: 10.1016/j.jcv.2021.105019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Labun K., et al. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ren X., et al. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014;9:1151–1162. doi: 10.1016/j.celrep.2014.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kocak D.D., et al. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019;37:657. doi: 10.1038/s41587-019-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liang M., et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 2019;10:1–9. doi: 10.1038/s41467-019-11648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]