Abstract
The surface modification of nanoparticles (NPs) using different ligands is a common strategy to increase NP–cell interactions. Here, dentin phosphophoryn-derived peptide (DSS) lignin nanoparticles (LNPs) are prepared and characterized, the cellular internalization of the DSS-functionalized LNPs (LNPs-DSS) into three different cancer cell lines is evaluated, and their efficacy with the widely used iRGD peptide is compared. It is shown that controlled extent of carboxylation of lignin improves the stability at physiological conditions of LNPs formed upon solvent exchange. Functionalization with DSS and iRGD peptides maintains the spherical morphology and moderate polydispersity of LNPs. The LNPs exhibit good cytocompatibility when cultured with PC3-MM2, MDA-MB-231, and A549 in the conventional 2D model and in the 3D cell spheroid morphology. Importantly, the 3D cell models reveal augmented internalization of peptide-functionalized LNPs and improve antiproliferative effects when the LNPs are loaded with a cytotoxic compound. Overall, LNPs-DSS show equal or even superior cellular internalization than the LNPs-iRGD, suggesting that DSS can also be used to enhance the cellular uptake of NPs into different types of cells, and release different cargos intracellularly.
Keywords: 3D cell culture, benzazulene, biofunctionalization, dentin phosphophoryn peptide, lignin nanoparticles
1. Introduction
Nanotechnology has been significantly applied for the development of nanomedicines for biomedical applications, such as cancer therapy.[1] Nanomedicines have the potential to overcome the limitations of the current chemotherapeutic drugs, such as poor water solubility, lack of specificity to reach the tumor site, and systemic side effects.[2,3] To achieve these benefits, several materials with different chemical compositions have been used to formulate nanomedicines, including inorganic nanoparticles (NPs),[4–9] liposomes,[10,11] polymeric NPs,[12–15] and hybrid nanocomposites.[16–18] However, polymers derived from natural and biorenewable sources have attracted increased interest for biomedical applications, because of their biodegradability and biocompatibility, as well as their availability in large scale and low cost.[19,20] Lignocellulosic materials are an example of biorenewable polymers derived from wood and plant sources, and are constituted by cellulose, hemicellulose, and lignin.[21,22] Unlike cellulose, lignin is still an underexploited natural source, despite of its availability, mainly due to the complex macromolecular structure, which is dependent on its source and extraction method. However, transforming raw lignin into NPs of uniform size and shape can overcome this problem. Lignin NPs (LNPs) with different sizes and shapes have been prepared using different approaches, such as antisolvent precipitation, interfacial crosslinking, polymerization, solvent exchange and sonication.[22] In addition to their application as reinforcing agents in nanocomposites, LNPs have been recently employed for drug delivery applications.[23–26] Moreover, LNPs present functional groups that can be chemically modified, and consequently, increase their application potential.[22,23]
In order to improve the accumulation of the nanomedicines at the tumor with increased therapeutic effects, the NPs can be modified to respond to certain stimuli (e.g., changes in pH, temperature, magnetic field, ultrasound intensity, and light)[27] or active-targeting ligands (e.g., antibodies, aptamers, and peptides).[28,29] Tumor homing or cell-penetrating peptides, such as RGD-containing peptides, are widely used as ligands to target NPs to different tumor tissues and cells.[30–32]
LNPs are considered to be biocompatible in the absence of active payloads.[24,25] Due to its environmentally friendly properties and its large availability in nature, we used lignin as a starting material for the development of functionalized LNPs for active targeting. In this study, we first characterized the lignin polymers recovered from the carboxylation reaction, and the optimal LNPs were chosen based on their stability at physiological conditions. Thereafter, the LNPs were functionalized with a Asp-Ser-Ser (DSS) polypeptide derived from dentin phosphophoryn (DPP), due to its ability to function as a cell-penetrating peptide.[33] For that, LNPs were conjugated to DSS, using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide (EDC/NHS) coupling chemistry, in order to obtain DSS-functionalized LNPs (LNPs-DSS). In addition, the cellular uptake of LNPs-DSS into cancer cells was compared to the uptake of iRGD-functionalized LNPs (LNPs-iRGD). After physicochemical characterization and cytocompatibility evaluation of the prepared LNPs, we compared the cellular uptake of peptide-functionalized LNPs, using 2D and 3D cell models made of three different cancer cell lines. Additionally, a poorly water-soluble cytotoxic compound, benzazulene (BZL, 1-methyl-8-(propan-2-yl)-4-(trifluoromethyl)-3H-benzo[cd]-azulen-3-one), was used as a model compound for testing the loading into LNPs, as well as the in vitro antiproliferation effect of the BZL-loaded LNPs, before and after peptide functionalization, against different cancer cell lines.
2. Results and Discussion
2.1. Characterization of the Carboxylated Lignin Polymers
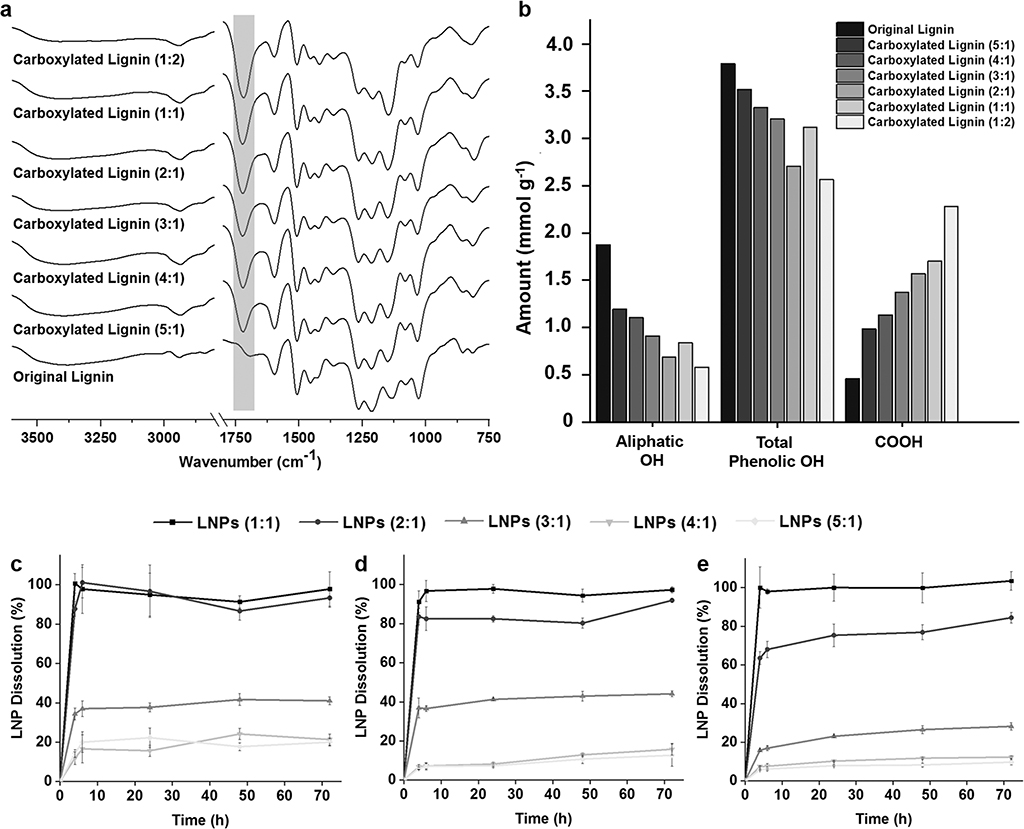
Carboxylated lignin was synthesized in order to increase the amount of free carboxyl groups for further conjugation reactions with the targeting ligands. For this, the hydroxyl groups on the original softwood kraft lignin structure were reacted with succinic anhydride, with 4-dimethylaminopyridine (DMAP) as a nucleophilic catalyst.[23] To find a balance between the lignin carboxylation degree and the long-term stability at physiological conditions, we tested different ratios of lignin:succinic anhydride for the reaction, as summarized in Table S1 (Supporting Information).
In order to confirm the success of the carboxylation reaction, Fourier transform infrared spectroscopy (FTIR) instrument with a horizontal attenuated total reflectance (ATR) accessory was used to analyze and compare the ATR–FTIR spectra of the prepared lignin polymers with the original lignin polymer (Figure 1a). All the IR absorption bands and respective type of vibration present on the lignin structure are detailed in Table S2 (Supporting Information). Apart from the typical bands present on both original and carboxylated lignin polymers, including the alcohol and phenol –OH (3500–3100 cm−1), the carbonyl groups (1600 cm−1) and the aromatic structure (1427–1512 cm−1), all the carboxylated lignin polymers exhibited a stronger adsorption band at ≈1720 cm−1 than the original lignin.[23,34] Stretching vibrations of C=O, which derive from the carbonyl situated at β-location that is also present in the original lignin polymer, can also contribute to this band. However, this band corresponds predominantly to the stretching vibrations of C=O on the free carboxyl groups on the carboxylated lignin structure. Moreover, it is possible to observe an increase on adsorption band at ≈1720 cm−1 according to the degree of carboxylation, suggesting that the reaction was successful. Additionally, the resulting carboxylated lignin polymers were also characterized using phosphorus-31 nuclear magnetic resonance (31P NMR) spectroscopy to quantitatively determine the carboxyl groups, as well as the aliphatic and phenolic –OH groups and hydrogen present on the lignin structures (Figure 1b). Before the carboxylation reaction, the amount of –COOH groups on the original lignin was 0.455 mmol g−1. However, the mass ratio of lignin:succinic anhydride and the amount of carboxyl groups after the reaction do not seem to be linearly correlated. Nevertheless, the amount of –COOH increased consistently from 0.985 mmol g−1 for the ratio 5:1 to 2.280 mmol g−1 for the ratio 1:2. This increase was generally accompanied by the decrease on the aliphatic –OH and total phenolic –OH groups, suggesting that the hydroxyl groups on the original lignin structure were effectively reacted with succinic anhydride.
Figure 1.
Characterization of the carboxylated lignin polymers by a) ATR−FTIR spectra and b) quantitative 31P NMR measurements. Evaluation of the long-term stability of LNPs prepared with the different carboxylated lignin by evaluating the dissolution rate of the LNPs after incubation of c) 50 μg mL−1, d) 200 μg mL−1, and e) 500 μg mL−1 in PBS buffer (pH 7.4) up to 72 h. Results are presented as mean ± s.d. (n = 3).
After the carboxylation reaction, LNPs were prepared with the carboxylated polymers, using the lignin:succinic anhydride in a ratio 1:2 [LNPs (1:2)], 1:1 [LNPs (1:1)], 2:1 [LNPs (2:1)], 3:1 [LNPs (3:1)], 4:1 [LNPs (4:1)], and 5:1 [LNPs (5:1)], by solvent exchange method described elsewhere.[23,24,35] The LNPs were formed during the dialysis process, where the carboxylated lignin molecules self-assembled into colloidal spheres as tetrahydrofuran (THF) was gradually replaced by deionized water. Afterward, 50 μg mL−1 (Figure 1c), 200 μg mL−1 (Figure 1d), and 500 μg mL−1 (Figure 1e) of LNPs were incubated in 1 × Dulbecco’s phosphate buffer saline (PBS; pH 7.4), mimicking the physiological pH, at 37 °C and up to 72 h. This dissolution experiment was done to evaluate the long-term stability of the LNPs at physiological conditions, and yield the ideal carboxylated lignin polymer to prepare the LNPs for further conjugation reactions and in vitro experiments. For that, the absorbance (λ = 380 nm) of the samples’ supernatant withdrawn at each time point was measured by UV–vis spectroscopy. The LNPs (1:2) were dissolved immediately after incubation (data not shown). For the three concentrations of LNPs tested, the LNPs (1:1) experienced a complete dissolution after 4 h. Both LNPs (2:1) and LNPs (3:1) showed a concentration-dependent dissolution overtime. After 48 h, the dissolution of the LNPs at 50, 200, and 500 μg mL−1 was ≈86.7 ± 4.7%, 80.3 ± 2.5%, and 76.9 ± 3.9% for the LNPs (2:1), and 41.7 ± 3.1%, 42.9 ± 2.5%, and 26.5 ± 2.1% for the LNPs (3:1), respectively. The same concentration-dependent trend for the dissolution of LNPs (4:1) and LNPs (5:1) was observed, but no statistically significant differences on the dissolution rate between LNPs (4:1) and LNPs (5:1) were detected, particularly for the two highest concentrations of LNPs. After 48 h, at 50, 200, and 500 μg mL−1, the dissolution of the LNPs (4:1) was ≈24.1 ± 2.8, 12.9 ± 0.8, 11.7 ± 0.4%, and 17.7 ± 2.0, 10.7 ± 2.3, and 8.2 ± 0.9% for the LNPs (5:1), respectively. Generally, the dissolution of the LNPs augmented according to the increase on the amount of carboxyl groups on the carboxylated lignin polymers. This dissolution behavior can be due to the conversion of carboxylic acids into carboxylates, because the sodium and potassium ions present in the PBS buffer in a basic environment react with the carboxylic acids. The carboxylic acids, with pKa values typically in the range of 3–5 in aqueous media at basic pH, will be ionized and converted into cation’s carboxylates, and therefore, enhance their water solubility at basic pH.[36] This effect is more pronounced when the amount of ions in the buffer is higher, i.e., when the concentration of LNPs is lower (50 μg mL−1), and therefore, the dissolution seems to be dependent on the LNP concentration in PBS buffer. Additionally, after a certain carboxylation degree, the dissolution rate appears to reach a constant value, as it can be observed for the LNPs (4:1) and LNP (5:1), suggesting that some degree of dissolution of LNPs will occur, regardless of the carboxylation degree after reaction. Based on these observations, we decided to select the LNPs (4:1) to carry out the in vitro studies, because the LNPs (4:1) present similar dissolution behavior to the LNPs (5:1) and lower dissolution than the other ratios (1:1, 2:1, and 3:1).
In summary, the ratio 4:1 of lignin:succinic anhydride was the best option, because it presents more than two times higher amount of –COOH (1.132 mmol g−1) than the original lignin, and exhibited a good stability at physiological conditions over 72 h.
2.2. Characterization of the Peptide-Functionalized LNPs
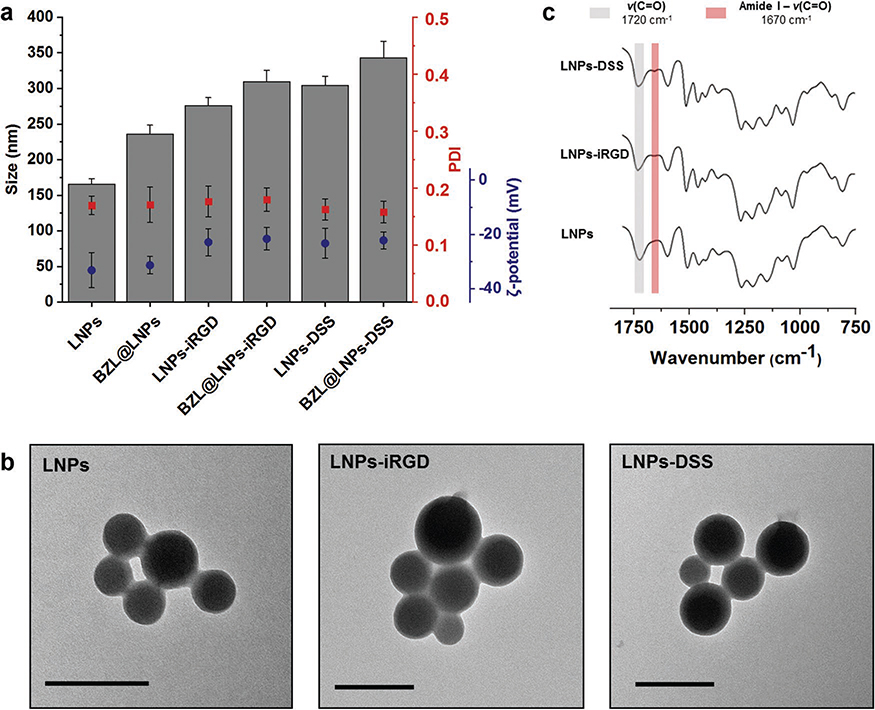
After synthesis and characterization of the carboxylated lignin, LNPs (4:1), henceforward simply referred as LNPs, were prepared using the same dialysis method previously used. Afterward, EDC/NHS coupling chemistry was carried out to conjugate the LNPs with equal molar amounts of both iRGD and DSS peptides, and consequently, the –COOH groups on the LNPs were covalently bonded to the –NH2 group on the peptides. Then, the LNPs were characterized for their average particle size (Z-average), polydispersity index (PDI), and average zeta (ζ)-potential by dynamic light scattering (DLS) and transmission electron microscopy (TEM), and ATR−FTIR (Figure 2a–c).
Figure 2.
Characterization of bare and BZL-loaded LNPs, before and after functionalization with iRGD and DSS by a) measuring average size, PDI, and ζ-potential of the LNPs, b) TEM images of the empty LNPs (scale bars are 200 nm), and c) ATR−FTIR spectra of the bare and functionalized LNPs. Error bars represent the mean ± s.d. (n ≥ 3).
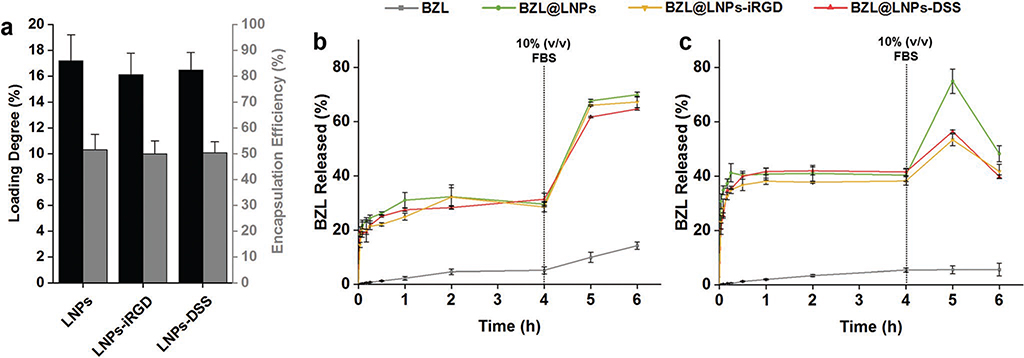
Additionally, the LNPs were loaded with a poorly water-soluble cytotoxic agent (BZL) as a model compound. BZL is an inhibitor of oncogenic Pim kinases that are often overexpressed in hematopoietic malignancies and in some solid tumors, including prostate and colon cancers, stimulating the cell survival and resistance against chemotherapy and radiation therapy.[37–39] Therefore, the Pim kinases family presents a target for pharmacological inhibition in cancer therapy, in which new small molecule inhibitors are being developed, such as the BZL.[38,40] Besides prostate cancer, BZL has also showed an in vitro inhibitory effect on other cancer cell lines, including breast cancer.[23,41] The BZL-loaded LNPs (BZL@ LNPs) were prepared using the same solvent exchange method, and subsequently functionalized with iRGD and DSS to yield BZL@LNPs-iRGD and BZL@LNPs-DSS, respectively. Then, the loading degree (LD) and encapsulation efficiency (EE) of BZL into LNPs was determined, and the release profile of BZL from LNPs was evaluated (Figure 3a–c).
Figure 3.
Evaluation of the a) loading degree (LD) and encapsulation efficiency (EE) of BZL into LNPs, LNPs-iRGD, and LNPs-DSS, and release profiles of pure BZL and BZL-loaded LNPs in b) HBSS–MES (pH 5.5) and c) HBSS–HEPES (pH 7.4), supplemented with 10% (v/v) FBS after 4 h, at 150 rpm and 37 °C for 6 h. Errors bars represent the mean ± s.d. (n ≥ 3).
Regarding the DLS characterization (Figure 2a), the bare LNPs presented an average size of 165 ± 7 nm, which increased after conjugation with iRGD and DSS to 275 ± 11 and 304 ± 13 nm, respectively. This difference in the size of the two peptide-functionalized LNPs can be ascribed to the length and molecular weight of the iRGD and DSS, in which the bigger and heavier peptide (DSS) gives a higher size for the LNPs than the smaller and lighter peptide (iRGD). After loading, the size of the BZL@LNPs slightly increased to 235 ± 12 nm. Moreover, the same trend on the average size was naturally observed after conjugation with peptides, in which the BZL@LNPs-iRGD showed 309 ± 7 nm and the BZL@LNPs-DSS exhibited 342 ± 7 nm. As for the size distribution (Figure 2a), the prepared LNPs and BZL@LNPs before and after functionalization exhibited PDI values lower than 0.20, suggesting a moderate dispersity of the LNPs. The size distribution and morphology of the LNPs was also verified by transmission electron microscopy (TEM, Figure 2b), where the particles showed an average size and moderate dispersity similar to the values found by DLS. Furthermore, the LNPs showed symmetric and spherical shape, due to the suitable interaction between the lignin and water during the process of solvent exchange. Moreover, the morphology of the LNPs did not change after conjugation with iRGD and DSS.
The stability of the bare and peptide-conjugated LNPs was also studied in cell culturing medium, supplemented with 10% (v/v) of fetal bovine serum (FBS) (Figure S1, Supporting Information). During the first minutes of incubation, the size of the bare LNPs increased around 100 nm and then was kept constant over time. However, the size of both peptide-functionalized LNPs increased ≈70 nm during the first 30 min of incubation, but decreased over time. This effect might be due to some protein adsorption that slowly detach from the LNP surface,[42] and consequently, their size decreased to the initial values, indicating a good stability of the peptide-decorated LNPs after interaction with the serum-containing media. Moreover, we previously prepared CPP-functionalized LNPs that were positively charged, and their size after incubation with cell culturing media containing 10% FBS increased 200 nm and then kept constant over time.[23] Furthermore, the negative surface charge of DSS or iRGD-functionalized LNPs might be responsible for less protein adsorption, and consequently, superior stability in serum-containing medium compared to the CPP-decorated LNPs.[43]
Regarding the surface charge (Figure 2a), the bare LNPs obtained a ζ-potential value of −33.2 ± 6.5 mV, similarly to the BZL@LNPs that showed −31.3 ± 3.2 mV. However, the ζ-potential of LNPs increased after conjugation with iRGD and DSS to −22.9 ± 5.0 and −23.4 ± 5.5 mV, respectively. Similarly, the ζ-potential of BZL@LNPs also increased, being −21.7 ± 4.2 mV for BZL@LNPs-iRGD and −22.2 ± 3.1 mV for BZL@LNPs-DSS. This suggested that the functionalization of the LNPs with both peptides occurred effectively, due to the decreased amount of –COOH groups on the LNPs’ surface and the overall charge of the side chains of the peptides. This was also confirmed by ATR−FTIR (Figure 2c), where the functional groups present on the surface of the particles were determined, and the spectra of the bare and peptide-functionalized LNPs was compared. Here, both LNPs-iRGD and LNPs-DSS showed a characteristic band near 1670 cm−1 that corresponds mostly to the C=O stretching vibrations of the amide bond (O=C–NH). Simultaneously, a slight decrease in the intensity of the band at 1720 cm−1 ascribed to the C=O stretching vibrations of the unconjugated –COOH groups was observed when compared to the bare LNPs. Overall, the changes in the obtained ζ-potential values for the bare and peptide-decorated LNPs along with the ATR−FTIR spectra indicated the success of the conjugation reactions.
The LD and EE of BZL into LNPs was determined by releasing the cargo after dissolving the LNPs in ethanol and analyzing the supernatant by high-performance liquid chromatography (HPLC, Figure 3a). The LD of BZL in the LNPs, LNPs-iRGD, and LNPs-DSS was 17.1 ± 2.0%, 16.1 ± 1.7%, and 16.5 ± 1.4%, respectively. Accordingly, the EE did not decrease significantly after the reactions, indicating that the cargo was not released during the conjugation reaction.
The in vitro release profiles of pure BZL, BZL@LNPs, BZL@ LNPs-iRGD, and BZL@LNPs-DSS, were conducted in two different buffers: Hank’s balanced salt solution (HBSS)–2-(N-morpholino)ethanesulfonic acid (MES) (HBSS–MES, pH 5.5) and HBSS–4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (HBSS–HEPES, pH 7.4), mimicking the tumor microenvironment and the physiological pH, respectively (Figure 3b,c). The release medium was supplemented with 10% FBS after 4 h to induce sink conditions during the release. The release of pure BZL at both pH was very low, being ≈5% after 4 h. However, the release profile of the BZL was greatly improved when loaded into the LNPs at both pH, compared to the pure BZL. After 4 h, the release of BZL from LNPs, LNPs-iRGD and LNPs-DSS reached plateaus around 30%, 28%, and 31% at pH 5.5, and 40%, 38%, and 41% at pH 7.4, respectively. The drug was released after 4 h, due to the dissolution of the LNPs in the release media containing sodium and potassium ions that increase the solubility of the LNPs. After 4 h, 10% FBS was added to increase the dissolution of the released drug, as the FBS is known to increase the solubility of some hydrophobic drugs, such as sorafenib.[44] Therefore, the release profiles of BZL from LNPs, LNPs-iRGD and LNPs-DSS at pH 5.5 were significantly improved after 6 h, being ≈70%, 67%, and 65%, respectively. However, at pH 7.4, the release of BZL increased after addition of 10% FBS, but decreased after 6 h, which can be due to the precipitation or degradation of BZL. The proposed nanosystems showed an enhancement of the dissolution rate of BZL that together with the higher stability of the BZL at acidic pH as in the tumor microenvironment make them suitable for cancer therapy applications.
2.3. Cytocompatibility of LNPs Using 2D and 3D Models
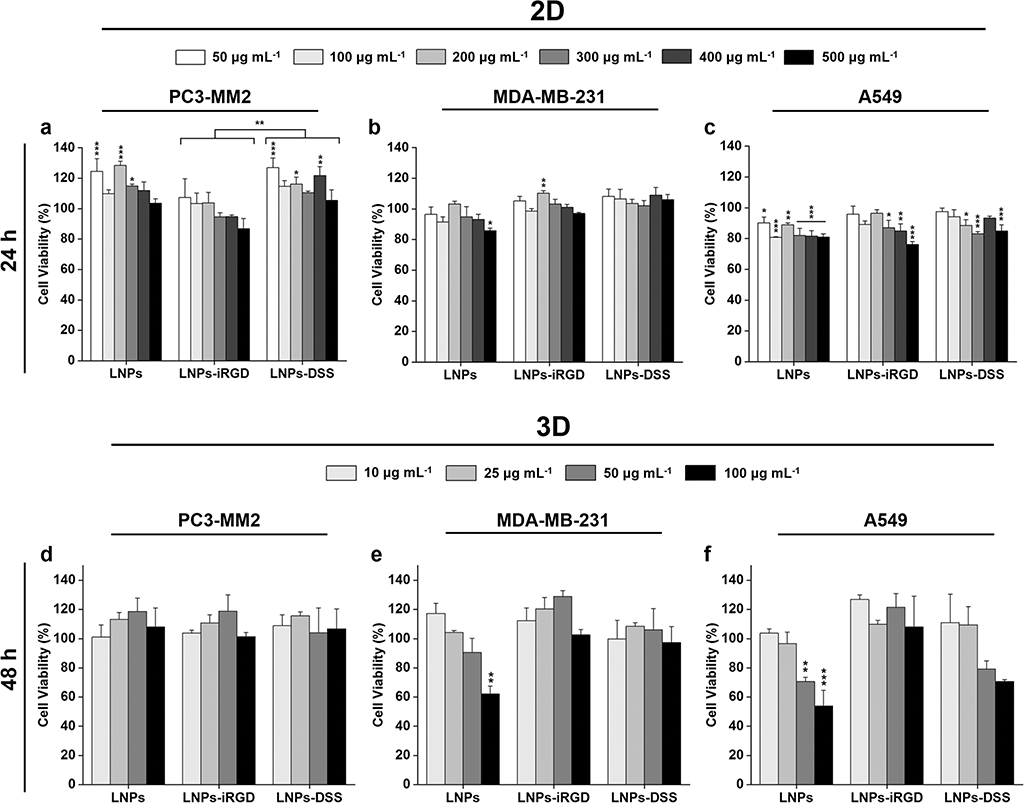
The cytotoxicity of the nanomedicines depends on the cell–NP interactions that relies on the different NPs’ properties, including size, shape, composition, and surface charge.[45,46] Therefore, we prepared both 2D cell model and 3D tumor cell spheroids to evaluate whether the modifications that were carried out on the LNPs affected their in vitro cytocompatibility. For that, different concentrations of bare and functionalized LNPs up to 500 μg mL−1 were incubated during 24 h using the 2D model with several human cancer cell lines: prostate cancer (PC3-MM2, Figure 4a), mammary carcinoma (MDA-MB-231, Figure 4b) and lung carcinoma (A549, Figure 4c), which were chosen to evaluate the potential cytotoxic effect of BZL in different types of cancer cells. Additionally, the 3D spheroid model was prepared using the bioprinting method, to study the cytocompatibility of the prepared LNPs up to 100 μg mL−1, during 48 h incubation (Figure 4d–f).
Figure 4.
Cell viability studies of LNPs using a 2D model of a) PC3-MM2, b) MDA-MB-231, and c) A549 cell lines after incubation with LNPs, LNPs-iRGD, and LNPs-DSS for 24 h at 37 °C, determined by the CellTiter-Glo luminescence assay, in which all data sets were compared to the positive control (cell medium). Cell viability studies of LNPs using 3D tumor spheroids of d) PC3-MM2, e) MDA-MB-231, and f) A549 cell lines after incubation with LNPs, LNPs-iRGD, and LNPs-DSS for 48 h at 37 °C, determined by the RealTime-Glo MT Cell Viability Assay in which all data sets were compared to the positive control (cell medium). The level of the significant differences was set at probabilities of *p < 0.05, **p < 0.01, and ***p < 0.001. Results are presented as mean ± s.d. (n ≥ 3).
Using the 2D cell model (Figure 4a–c), the LNPs before and after peptide functionalization showed a very good cytocompatibility after 24 h incubation with PC3-MM2 and MDA-MB-231 cells (Figure 4a,b), with more than 80% of cell viability. However, the LNPs presented in general higher toxicity toward A549 (Figure 4c) compared to the other two cell lines, particularly at the highest concentrations. This can be ascribed to the higher cellular uptake rate of LNPs into A549 compared to the other to cell lines (Figure 5), which can cause more toxicity. Additionally, the LNPs-DSS presented a general trend to be less cytotoxic than LNPs-iRGD for all the cell lines, being statistically significant for incubation with PC3-MM2.
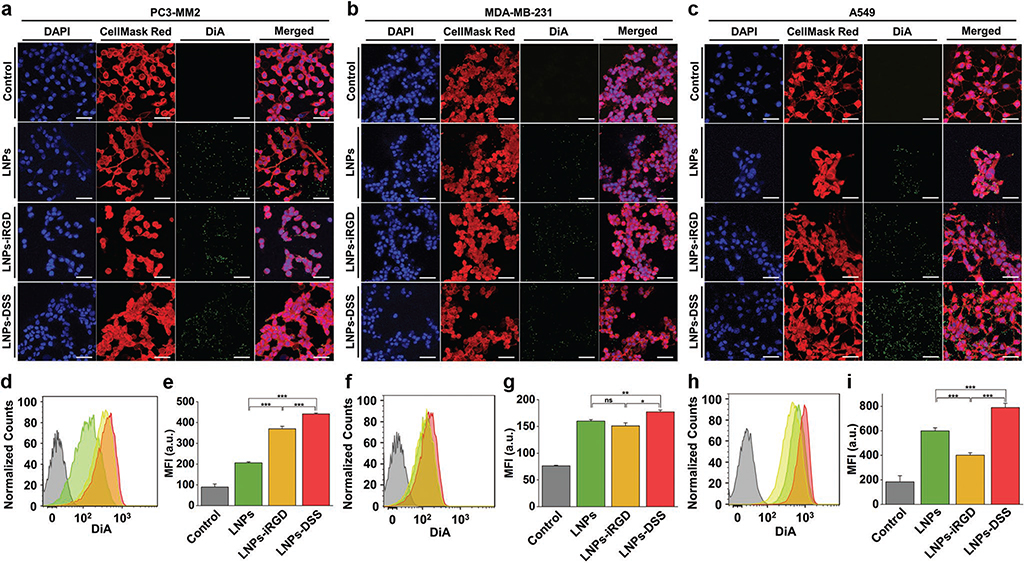
Figure 5.
In vitro cellular interaction studies using a 2D cell culture model: Confocal fluorescence microscopy images of a) PC3-MM2, b) MDA-MB-231, and c) A549 cell lines, after incubation with 100 μg mL−1 of bare and peptide-functionalized DiA@LNPs for 3 h at 37 °C. DAPI (blue), DiA (green), and CellMask Red (red) were used to stain the nucleus, the LNPs, and the cell membrane, respectively. The merged panels show the association of the LNPs with the cell membrane. Scale bars are 50 μm. Quantitative cellular uptake by flow cytometry, after incubation of 100 μg mL−1 of LNPs with d) PC3-MM2, f) MDA-MB-231, and h) A549, for 3 h at 37 °C in which the LNPs (green) LNPs-iRGD (yellow), and LNPs-DSS (red) were compared to the untreated control (gray). MFI quantification for e) PC3-MM2, g) MDA-MB-231, and i) A549 before and after treatment with the DiA@LNPs. At least 10 000 events were collected for each measurement. The error bars represent mean ± s.d. (n = 3). Statistical significance was set at probabilities of *p < 0.05, **p < 0.01, and ***p < 0.001, and ns is nonsignificant.
When a 3D tumor spheroid model was used (Figure 4d–f), the LNPs before and after peptide functionalization showed a very good cytocompatibility, even after a longer incubation time of 48 h, with cell viability rates higher than 80% for the concentrations tested. However, the bare LNPs showed significant toxicity at 100 μg mL−1 for MDA-MB-231 (Figure 4e), and 50 and 100 μg mL−1 for A549 (Figure 4f) cell lines. This can be due to a higher content on carboxyl groups of the bare LNPs compared to both peptide-functionalized LNPs that induces more toxicity to the cells, resulting in an impaired cell viability.[46,47] Therefore, the peptide-decorated LNPs were less toxic than the bare LNPs.
2.4. Cell−LNP Interaction Studies
In addition to the cytocompatibility, the physicochemical properties of the NPs can also affect the way that NPs interact with the cells. Different parameters, such as material composition, NP size and shape, hydrophobicity and hydrophilicity, and surface charge and functionalization can dictate their cellular uptake.[48,49] Additionally, the decoration of NPs with peptides can be carried out to promote an increased the cellular uptake of NPs into different cells and tissues.[30,31] Here, we intended to evaluate both qualitatively and quantitatively the cellular interaction/uptake of our LNPs with three different cell lines, by confocal microscopy and flow cytometry, respectively (Figures 5 and 6). For that, the LNPs were loaded with a highly stable fluorescent dye (DiA, 4-[4-(dihexadecylamino)-styryl]-N-methylpyridinium iodide), functionalized with iRGD and DSS, and incubated with PC3-MM2, MDA-MB-231, and A549 during 3 h.
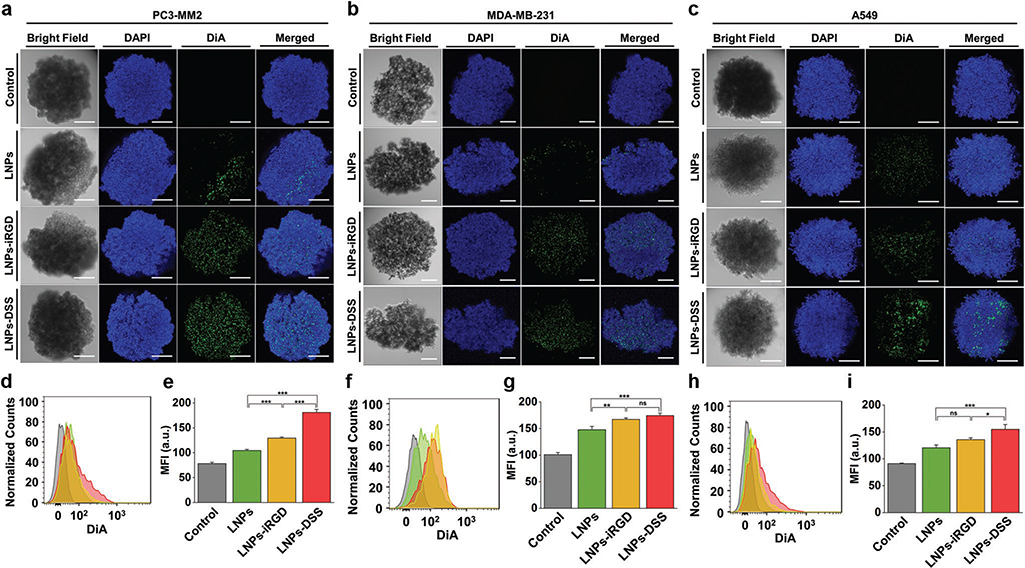
Figure 6.
In vitro cellular interaction studies using a 3D tumor spheroids: Confocal fluorescence microscopy images of a) PC3-MM2, b) MDA-MB-231, and c) A549 cell lines, after incubation with 100 μg mL−1 of bare and peptide-functionalized DiA@LNPs for 3 h at 37 °C. DAPI (blue) and DiA (green) were used to stain the nucleus and the LNPs, respectively. The merged panels show the association of the LNPs with the tumor spheroid. Scale bars are 200 μm. Quantitative cellular uptake by flow cytometry, after incubation of 100 μg mL−1 of LNPs with d) PC3-MM2, f) MDA-MB-231, and h) A549, for 3 h at 37 °C in which the LNPs (green) LNPs-iRGD (yellow), and LNPs-DSS (red) were compared to the untreated control (gray). MFI quantification for e) PC3-MM2, g) MDA-MB-231, and i) A549 before and after treatment with the DiA@LNPs. At least 5000 events were collected for each measurement. The error bars represent mean ± s.d. (n = 3). Statistical significance was set at probabilities of *p < 0.05, **p < 0.01, and ***p < 0.001, and ns is nonsignificant.
We started by studying the cellular uptake of the bare and peptide-functionalized LNPs using the conventional in vitro 2D cell culture model (Figure 5). The confocal microscopy images showed clearly an increased interaction of both LNPs-iRGD and LNPs-DSS with PC3-MM2 (Figure 5a), MDA-MB-231 (Figure 5b), and A549 (Figure 5c) cell lines. Then, the extracellular fluorescence was quenched with trypan blue, allowing the evaluation of the cellular uptake of the prepared LNPs using flow cytometry (Figure 5d–i). For the PC3-MM2, the LNPs-DSS showed an internalization rate slightly higher than LNPs-iRGD, and both peptide-functionalized LNPs showed a statistically significant higher cellular uptake than the bare LNPs (Figure 5d). These observations were also demonstrated and confirmed by the mean fluorescence intensity (MFI) values obtained during this analysis (Figure 5e). For MDA-MB-231 (Figure 5f,g) and A549 (Figure 5h,i), the LNPs-DSS showed more internalization than LNPs and LNPs-iRGD. However, the LNPs were more internalized than the LNPs-iRGD for A549. Some studies have reported that the high internalization of negatively charged NPs, such as –COOH modified NPs, can occur via nonspecific binding and clustering of NPs on cationic sites on the plasma membrane.[50] Furthermore, Guarnieri et al. showed that the cellular uptake by the cells of –COOH-modified silica NPs was higher than of the –NH2-modified silica NPs.[51] Therefore, the surface characteristics of the bare LNPs can lead to an increased cellular uptake by A549 compared to the LNPs-iRGD.
Conventionally, the in vitro cellular uptake of NPs is evaluated using a 2D model in a flat cell culturing surface, as discussed in section 2.3. However, this model does not represent the 3D organization and morphology of their native tissues, and the cell−cell interactions are not realistic, leading to a misinterpretation of the cellular uptake and a superior biological efficacy of the NPs comparing to the 3D culturing models.[52] Therefore, the cellular interactions/uptake of bare and peptide-functionalized LNPs with the different cell lines was also assessed using a 3D tumor spheroids of ≈600 μm of diameter (Figure 6), and the comparison with the conventional 2D model was evaluated. Observing the confocal microscopy images of the tumor spheroids, both peptide-decorated LNPs improved the interaction with the three cell lines, compared to the bare LNPs (Figure 6a–c). Moreover, the LNPs-DSS interact more than LNPs-iRGD with the PC3-MM2 and A549. In order to determine the cellular uptake, the tumor spheroids were disassembled after the incubation with the LNPs, and the extracellular fluorescence was quenched after incubating the cells with trypan blue before the flow cytometry analysis. Regarding the PC3-MM2, the cellular uptake of the LNPs-DSS was higher than the LNPs-iRGD and bare LNPs, which was statistically significant based on the MFI values obtained (Figure 6d,e). For the MDA-MB-231, both peptide-functionalized NPs showed an increased cellular internalization than the LNPs (Figure 6f,g). In the case of the A549, the same trend for the cellular internalization of LNPs was observed, where the LNPs-DSS showed an enhanced uptake compared to the LNPs-iRGD and LNPs (Figure 6h,i).
Comparing the two culturing models, the cellular internalization of the LNPs was expectably higher for the 2D cell model than for the 3D tumor spheroids, which was correlated with the MFI values obtained, i.e., higher MFI for the 2D-cultured cells. This can be due to the effect of sedimentation and diffusion of the NPs on the 2D cell culturing plate, as well as the stronger and more realistic cell–cell interactions in the 3D cell culturing model.[53] Additionally, the peptide-decorated LNPs were generally taken-up more efficiently by the three cell lines than the bare LNPs in both cell culturing models. Furthermore, in some cases, the LNPs-DSS showed a superior cellular internalization than the LNPs-iRGD. The DSS peptide derived from DPP is a repetitive DSS-rich domain that facilitates non-receptor-mediated endocytosis, acting as a cell penetrating peptide. DPP contains a conserved RGD domain in several species, but this DSS polypeptide was not mediated through integrin receptors.[33] Comparing to the iRGD that binds to the integrins overexpressed in the tumor endothelium and improves the tissue penetration in a tumor-specific and neuropilin-1-dependent manner,[54] the DSS peptide can improve the cellular uptake; however, neither clathrin nor the caveolae-mediated pathways was responsible for its internalization.[33] Moreover, the disruption of the intracellular transportation mechanisms affected the endocytic process, but the disruption of the microtubules and microfilaments involved in this process did not prevent the endocytosis of the DSS peptide.[33] Unlike iRGD peptide, the cellular internalization of the DSS peptide is not related with the amount of receptor/cellular target for its endocytosis. Therefore, the DSS peptide can also be used to enhance the cellular uptake of NPs into different types of cells via non-receptor-mediated endocytosis, releasing different cargos intracellularly for different therapeutic applications.
2.5. In Vitro Antiproliferative Studies
The dimensionality of the culturing method has been shown to affect the cytocompatibility and cellular uptake of the NPs, as well as the cellular response to the drug treatment.[55] Therefore, BZL-loaded LNPs and the free BZL were used to treat the cells and evaluate the growth inhibition effect of the BZL in 2D (Figure 7) and 3D (Figure 8) cell culturing models. Apart from its known role as an inhibitor of Pim kinases overexpressed in prostate cancer, BZL has been shown to be a potent cytotoxic compound against other cell lines.[23,41] Thus, pure BZL and BZL-loaded LNPs were incubated with PC3-MM2, MDA-MB-231 and A549, representing the prostate, breast and lung cancers, respectively.
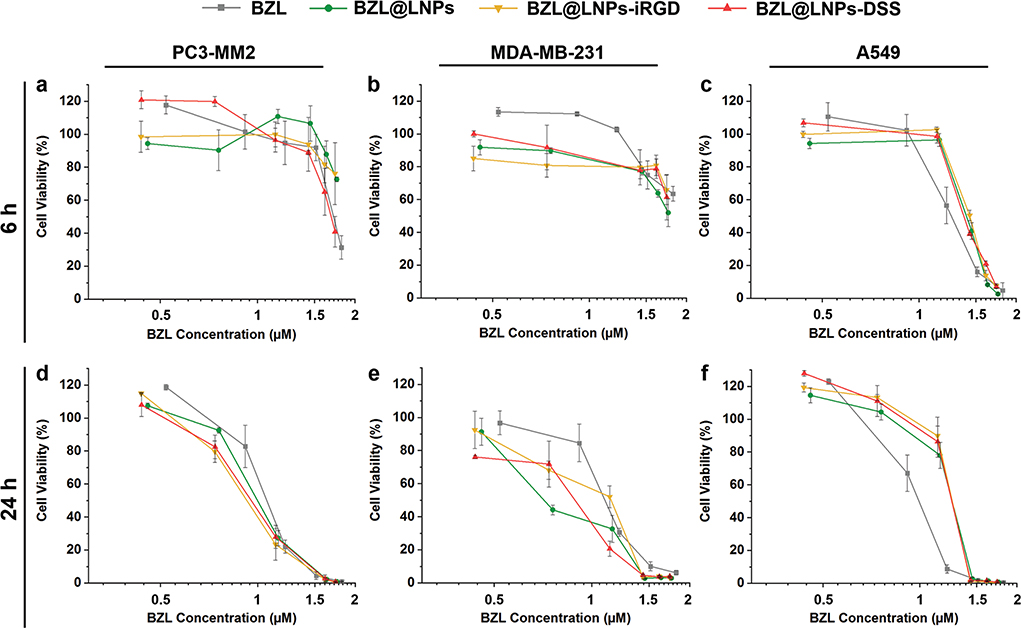
Figure 7.
Antiproliferative studies using a 2D cell culture model of a,d) PC3-MM2, b,e) MDA-MB-231, and c,f) A549 cell lines treated with pure BZL previously dissolved with 3% ethanol (v/v), and different concentrations of BZL-loaded LNPs, LNPs-iRGD, and LNPs-DSS in complete media, for a–c) 6 h and d–f) 24 h at 37 °C. Errors bars represent the mean ± s.d. (n ≥ 3).
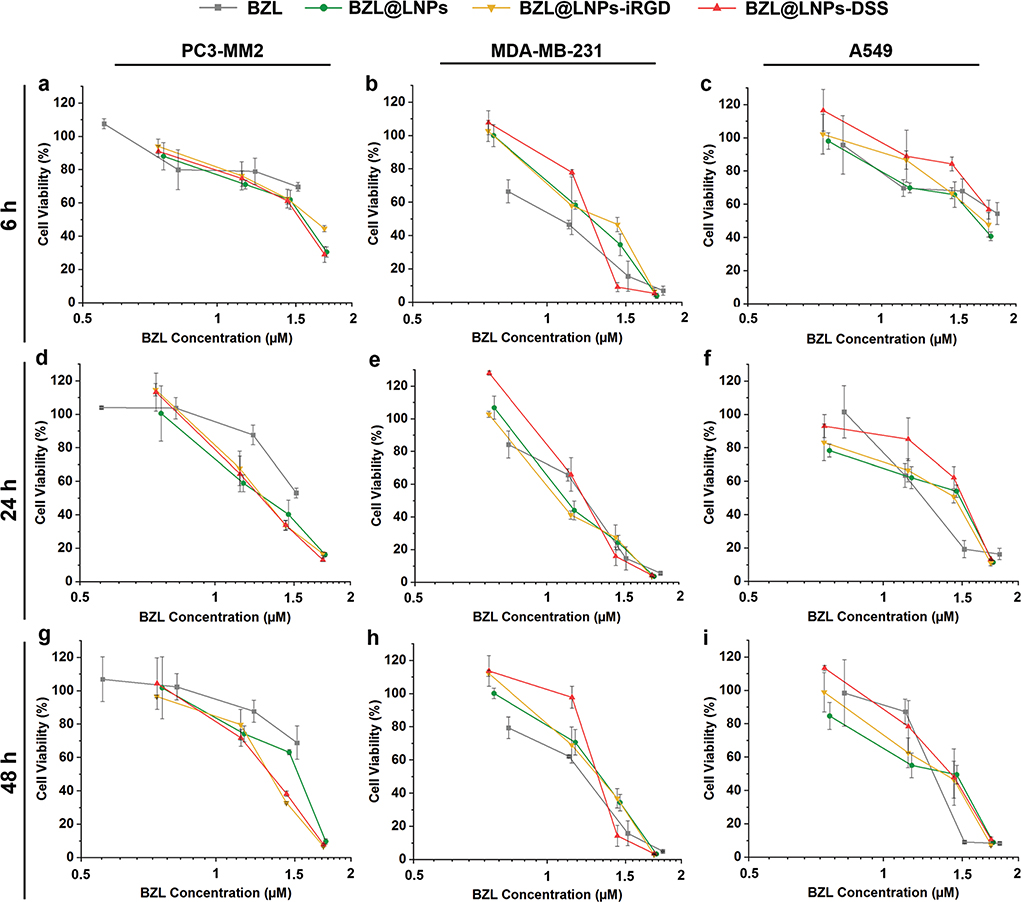
Figure 8.
Antiproliferative studies using 3D tumor spheroids of a,d,g) PC3-MM2, b,e,h) MDA-MB-231 and c,f,i) A549 cell lines treated with pure BZL previously dissolved with 3% ethanol (v/v), and different concentrations of BZL-loaded LNPs, LNPs-iRGD, and LNPs-DSS in complete media, for a–c) 6 h, d–f) 24 h, and g–i) 48 h at 37 °C. Errors bars represent the mean ± s.d. (n ≥ 3).
For the 2D model, LNP suspensions ranging from 5 to 100 μg mL−1 and similar concentrations of the free poorly water-soluble BZL previously dissolved with 3% (v/v) ethanol in cell media were incubated with the cells in a flat cell surface during 6 (Figure 7a–c) and 24 h (Figure 7d–f). Afterward, the inhibitory concentration (IC) values by 50% (IC50), 80% (IC80), and 90% (IC90) were determined after plotting a concentration response curve by nonlinear regression (Table S3, Supporting Information). For PC3-MM2, the BZL@LNPs-DSS reached the IC50 after 6 h of incubation, unlike the BZL@LNPs-iRGD and BZL@LNPs (Figure 7a). After 24 h, the inhibitory effect of all the BZL@LNPs presented similar trends toward PC3-MM2 (Figure 7d). Regarding MDA-MB-231, the incubation of the BZL@LNPs was not enough to observe 50% of growth inhibition after 6 h (Figure 7b), but after 24 h, the BZL@ LNPs, BZL@LNPs-iRGD, and BZL@LNPs-DSS reached IC90 values of 31.9 × 10−6, 35.5 × 10−6, and 31.4 × 10−6 m (Figure 7e). For A549, the same trend on the growth inhibition effect of pure BZL and BZL-loaded LNPs before and after peptide functionalization was observed after 6 and 24 h of incubation (Figure 7c,f).
Regarding the 3D tumor spheroids, the BZL@LNPs (concentrations ranging from 10 to 100 μg mL−1) and similar concentrations of free BZL, previously dissolved with 3% (v/v) ethanol, were incubated with spheroids during 6, 24, and 48 h (Figure 8), and the IC50, IC80, and IC90 were also determined after nonlinear regression and detailed in Table S4 (Supporting Information). Although the BZL@LNPs showed similar inhibition effect after incubation with PC3-MM2 (Figure 8a,d,g), the peptide-functionalized LNPs appeared to have an increased antiproliferative effect after 24 h of incubation. Here, the BZL@LNPs-DSS presented lower IC80 values (41.7 × 10−6 m) than BZL@LNPs-iRGD (44.0 × 10−6 m) and BZL@LNPs (49.3 × 10−6 m). The same trend in the inhibitory effect was observed for MDA-MB-231 (Figure 8b,e,h), where the BZL@LNPs-DSS presented inferior IC90 values (44.2 × 10−6 m) than BZL@LNPs-iRGD (47.5 × 10−6 m) and BZL@LNPs (51.5 × 10−6 m) after a long-term of exposure to the LNPs (48 h). Likewise, the IC90 values of both BZL@LNPs-DSS (60.1 × 10−6 m) and BZL@LNPs-iRGD (57.7 × 10−6 m) was lower than the bare BZL@LNPs (70.2 × 10−6 m) after 48 h of incubation with A459 (Figure 8i).
Unlike for 2D cell model, the peptide-functionalized BZL@ LNPs showed lower IC values than the bare BZL@LNPs when the 3D tumor spheroids model was used to test the antiproliferative effect of BZL. Moreover, the BZL@LNPs-DSS showed a superior efficacy than the BZL@LNPs-iRGD, as for MDA-MB-231 cell line. As expected, the antiproliferative effect of the BZL@LNPs was more pronounced toward the 2D cell model, with lower IC50 values for the 2D cell model compared to the IC values obtained for the 3D tumor spheroids. This can be due to the reduced proliferation rate of the cells in 3D culturing models compared to the 2D model, leading to a decreased antiproliferative effect of the cytotoxic compounds that act more potently on proliferating cells.[56]
In summary, all the BZL@LNPs showed an effective antiproliferative effect in all the cancer cell lines tested, without using any organic solvent (e.g., ethanol) to solubilize the BZL, because the solubility of BZL is improved when loaded into the LNPs. Moreover, the DSS-decorated BZL@LNPs exhibited a good and sustained antiproliferative effect toward the 3D tumor spheroids that are more biologically relevant.
3. Conclusion
In this study, all the LNPs showed low cytotoxicity at concentrations up to 500 μg mL−1, using the 2D model, and up to 100 μg mL−1 for peptide-functionalized LNPs or 50 μg mL−1 for bare LNPs, using the 3D tumor spheroids. Additionally, the peptide-functionalized LNPs showed higher cellular uptake than the bare LNPs, which was more pronounced when a 3D cell-culturing model was used. Moreover, the BZL@LNPs showed an improved antiproliferative effect after incubation with three different cancer cell lines cultured in 2D or 3D cell models. Overall, the functionalized LNPs showed interesting properties for drug delivery and biomedical applications, including biocompatibility and ability to load hydrophobic compounds. The LNPs-DSS also presented similar or increased cellular internalization and antiproliferative effect properties compared to LNPs-iRGD, suggesting that the DSS can also be conjugated to NPs as a targeting moiety to enhance the delivery of different cargos inside the cells.
4. Experimental Section
Materials and Cell Culturing:
BioPiva softwood kraft lignin was acquired from UPM Biochemicals (Finland). Benzazulene (BZL) was prepared in-house, as described elsewhere.[37] 6xDSS peptide was synthesized as described earlier at the University of Illinois at Chicago (USA).[33] The iRGD (CCRGDKGPDC) was purchased from GenicBio. 4-[4-(Dihexadecylamino)styryl]-N-methylpyridinium iodide (DiA) was acquired from Thermo Scientific, USA. Other materials and cell culturing procedures are detailed in Supporting Information.
Carboxylation of Lignin:
The original lignin material was carboxylated in order to increase the amount of carboxylic groups for further conjugation reactions, using the reaction chemistry described elsewhere.[23] Briefly, 100 mg of BioPiva softwood kraft lignin was reacted with succinic anhydride, in the presence of DMAP (20 mg) as catalyst, in a final volume of 40 mL of THF at room temperature (RT) for 48 h. Here, different mass ratios of lignin:succinic anhydride were reacted to synthesize different carboxylated lignin polymers, as summarized in Table S1 (Supporting Information).
After the reaction, the mixture was dialyzed against MilliQ-water using a dialysis bag (Spectra/Por 1 Standard RC Dry Dialysis Tubing, 12–14 kDa, Spectrum Labs, USA) during 48 h to remove the unreacted reagents, replacing the water periodically. Finally, the carboxylated lignin was freeze-dried and characterized with an FTIR instrument (Vertex 70, Bruker, USA) using a horizontal ATR accessory (MIRacle, PIKE Technologies, USA). The ATR-FTIR spectra were recorded at room temperature between 4000−650 cm−1 with a resolution of 4 cm−1 using OPUS 5.5 software. Additionally, aliphatic hydroxyl, phenolic hydroxyl, and carboxyl groups of original softwood kraft lignin and carboxylated lignins were analyzed by quantitative 31P NMR spectroscopy.[57] The sample preparation and analysis was carried out as described earlier, except that now the sample weight was 25 mg and 15 relaxation time was used in the inverse-gated pulse sequence.[58,59]
Preparation of Lignin Nanoparticles:
The LNPs were prepared by solvent exchange, as described elsewhere.[35] For this, the 1 mg of carboxylated lignin was dissolved with 1 mL THF and introduced into a dialysis bag (Spectra/Por 1 Standard RC Dry Dialysis Tubing, 12−14 kDa, Spectrum Labs, USA). The dialysis water was periodically replaced during 24 h and the dialysis was performed under slow stirring in a fume hood.
The BZL-loaded LNPs were prepared using the same method as described above. For that, 3 mg of carboxylated lignin were dissolved with 3 mL of THF containing 1 mg of BZL (mass ratio BZL:lignin of 1:3), and then introduced into a dialysis bag for the solvent exchange process, where the BZL-loaded LNPs were formed. The BZL loading into LNPs was determined by immersing the drug-loaded LNPs in ethanol for 30 min under vigorous stirring to degrade the LNPs. After centrifugation at 16 100 rcf for 5 min, the supernatant was collected to determine the BZL concentration by using HPLC (Agilent 1100 series, Agilent Technologies, USA). The experimental conditions used for quantification of the loaded BZL are described in the literature,[24] and detailed in Table S5 (Supporting Information).
Dissolution Experiments:
The dissolution experiments of the LNPs were done to evaluate their long-term stability at physiological conditions, after preparing LNPs from the different carboxylated lignin polymers. For that, 50, 200, and 500 μg mL−1 of LNPs were incubated in PBS pH 7.4, at 37 °C. Afterward, 100 μL of LNP suspension were withdrawn at determined time points (4, 6, 24, 48, and 72 h), and centrifuged at 16 100 rcf for 5 min. Finally, the absorbance of the supernatant at λ = 380 nm was measured by UV–vis spectroscopy using a Varioskan Flash plate reader (Thermo Fisher Scientific Inc., USA), and the dissolution of the LNPs was calculated using Equation (1)
| (1) |
To calculate the absorbance of the dissolved LNPs, the same concentration of LNPs was completely dissolved in ethanol, which corresponds to the maximum absorbance that the supernatant will have if dissolved at physiological conditions.
Characterization of LNPs:
LNPs were characterized for their average particle size (Z-average), PDI, and average ζ-potential by DLS using a Malvern Zetasizer Nano ZS instrument (Malvern Instruments Ltd, UK). For that, the samples were diluted in MilliQ-water (50 μg mL−1). To characterize their morphology and confirm their size distribution, the LNPs were visualized by TEM (Jeol JEM-1400, Jeol Ltd., Tokyo, Japan), using an acceleration voltage of 80 kV. For the sample preparation, a droplet of LNPs’ suspension was mounted on a carbon-coated copper grid, blotted using a filter paper, and then air-dried before analysis. Additionally, the LNPs were also characterized by ATR−FTIR.
The stability of LNPs was also evaluated in Dulbecco’s modified Eagle’s medium (DMEM), supplemented with 10% (v/v) FBS. For that, samples were withdrawn at 5, 15, 30, 60, 90, and 120 min after incubation of 300 μg mL−1 of LNPs with the cell culturing medium at 37 °C, and posteriorly diluted in water to evaluate changes on the size and ζ-potential of the LNPs over time. All the experiments were performed in triplicates.
Conjugation of the Peptides to LNPs:
The conjugation reaction between the –COOH groups of LNPs and –NH2 groups of DSS and iRGD peptides was performed in 10 × 10−3 m of MES buffer (pH 5.5), using EDC/NHS coupling chemistry. The molar amount of peptides used for the reaction was chosen taking in consideration the molecular weight of the peptides (Table S6, Supporting Information), and after optimization of the conjugation conditions. First, the carboxylic groups on the LNPs were activated by reacting 1 mg of LNPs in 400 μL of 100 × 10−3 m of MES buffer (pH 5.5), with 5 μL of EDC (40 × 10−3 m) and 4.6 mg of NHS (100 × 10−3 m), and the mixture was stirred for 1 h at room temperature in the dark. Then, the mixture was centrifuged to remove excess EDC/NHS, at 16 100 rcf for 5 min, and the LNPs were resuspended with 500 μL of PBS pH 7.4 containing 250 × 10−6 m of the DSS and iRGD, for 4 h, to prepare LNPs-DSS and LNPs-iRGD, respectively. Afterward, the peptide-functionalized LNPs were centrifuged and washed twice with MilliQ water. The same conditions were used for the reaction between the BZL@LNPs and the DSS and iRGD to prepare BZL@LNPs-DSS and BZL@LNPs-iRGD, respectively.
Drug Release Studies:
BZL was used to evaluate the in vitro release profiles of free BZL, BZL@LNPs, BZL@LNPs-iRGD, and BZL@LNPs-DSS, in two different buffers: HBSS–MES (pH 5.5) and HBSS–HEPES (pH 7.4), supplemented with 10% (v/v) of FBS after 4 h. For that, 30 μg of pure BZL and 100 μg of BZL-loaded LNPs were immersed in 10 mL of release media, and the samples were stirred at 150 rpm, at 37 °C. At scheduled time intervals (2, 5, 10, 15, 30, 60, 120, 240, 300, and 360 min), 200 μL of the release media were withdrawn and the same volume of fresh pre-heated release media was added, keeping the releasing volume constant. Afterward, the samples were centrifuged at 16 100 rcf for 5 min and the supernatant was collected and analyzed in HPLC. The amount of BZL released was determined by measuring the BZL concentration using the HPLC method detailed in Table S5 (Supporting Information). The average values were obtained from three replicates.
Preparation of the 3D Tumor Spheroids:
The 3D tumor spheroids were formed by the 3D bioprinting method. Initially, 1.5 mL of PC3-MM2, MDA-MB-231 and A549 cells were seeded in 6-well plates at a density of 400 000 cells per well and allowed to attach overnight. Afterward, the cells were treated with 50 μL of NanoShuttle-PL (Nano3D Biosciences Inc., Germany) and incubated for 8–10 h for the magnetization of the cells. Then, the cells were washed twice with PBS buffer (pH 7.4) and seeded in ultralow attachment 96-well plates at a density of 5000 (PC3-MM2) or 7000 (MDA-MB-231 and A549) cells per well, which was further placed atop the 96-well spheroid magnetic drive (Nano3D Biosciences Inc., Germany). The cells within the solution aggregated in the well plate, driven by the magnet, and the spheroids were cultured for 2 days.
Cytocompatibility Assessment:
For the 2D model, PC3-MM2, MDA-MB-231, and A549 cell lines were seeded in 96-well plates (PerkinElmer Inc., USA) at a density of 15 000 cells per well and allowed to attach overnight. Afterward, 100 μL of LNPs, LNPs-iRGD and LNPs-DSS suspensions in cell media at different concentrations (50, 100, 200, 300, 400, and 500 μg mL−1) were added to each well and the plates were incubated for 24 h at 37 °C. Incubations with cell media and 1% (v/v) Triton X-100 were used as a positive and negative controls, respectively. After that, the plates were equilibrated at room temperature for 30 min and the wells were washed once with 100 μL of HBSS–HEPES buffer. Then, 50 μL of CellTiter-Glo (Promega Corporation, USA) were added to 50 μL of HBSS–HEPES (pH 7.4) in each well. The plates were stirred for 2 min on an orbital shaker and then stabilized for 30 min at room temperature, protected from the light. Finally, the luminescence was measured using a Varioskan Flash plate reader. The number of viable cells in culture was quantified based on the amount of ATP produced by metabolically active cells.[12,24]
After preparation of the 3D tumor spheroids, using PC3-MM2, MDA-MB-231, and A549 cells, as described above, the spheroids were gently transferred to 96-well plates (PerkinElmer Inc., USA) to evaluate the cytocompatibility of LNPs. Then, 50 μL of LNPs, LNPs-iRGD and LNPs-DSS suspensions at different concentrations were mixed with 50 μL of NanoLuc luciferase and MT Cell Viability Substrate in complete cell culture medium, for 48 h at 37 °C. The final concentrations of NPs were 10, 25, 50, and 100 μg mL−1, and incubation with cell media was used as a positive control. The RealTime-Glo MT Cell Viability Assay is a nonlytic, homogeneous and bioluminescent method that allows to measure the cell viability in real time. The luminescence was measured using a Varioskan Flash plate reader, at the decided time point (48 h). The number of viable cells in culture was determined by measuring the reducing potential of cells, and thus metabolism. All the experiments were performed in triplicate.
Cell−LNP Interactions Using a 2D Cell Culture Model:
For these experiments, the LNPs were loaded with a hydrophobic dye, DiA, in a mass ratio 100:1 (LNPs:DiA), and the conjugation reactions with the peptides were done, as described for the empty LNPs. Afterward, the interactions between the PC3-MM2, MDA-MB-231 and A549 cell lines and DiA-loaded LNPs, LNPs-iRGD, and LNPs-DSS were qualitatively and quantitatively evaluated by confocal microscopy and flow cytometry, respectively.
For confocal microscopy, 200 μL of 50 000 cells per well were seeded in Lab-Tek 8-chamber slides (Thermo Fisher Scientific, USA) and allowed to attach overnight. After removing the cell culture medium, 200 μL of 100 μg mL−1 of LNP suspensions in PBS buffer (pH 7.4) were added to the cells and incubated for 3 h at 37 °C. Then, the cells were washed twice with PBS buffer, and the plasma membrane was stained by adding 200 μL of CellMask Red (5 μg mL−1; Invitrogen, USA) and incubated for 3 min at 37 °C. The excess of staining solution was washed once with fresh PBS buffer and the cells were fixed using 4% Paraformaldehyde (PFA) for 10 min at room temperature. Finally, the nuclei staining was done by adding 200 μL of DAPI-405 (2.8 μg mL−1; Thermo Scientific, USA) and incubated for 5 min at 37 °C. The localization of NPs was observed with a Leica SP5 inverted confocal microscope (Leica Microsystems, Germany), using a 63 × /1.2–0.6 oil immersion objective.
For flow cytometry analysis, 0.4 mL of cells were seeded in 24-well plates at a density of 100 000 cells per well and allowed to attach overnight. After removing the cell culture medium, the wells were washed once with PBS buffer (pH 7.4). Then, 0.3 mL of 100 μg mL−1 of LNP suspensions were incubated with the cells for 3 h at 37 °C. After removing the LNPs and washing the cells once with PBS buffer to remove the nonadherent LNPs, the cells were harvested with Gibco Versene Solution for 5 min (0.48 × 10−3 m; Thermo Scientific, USA). Subsequently, the cells were washed once with PBS buffer and suspended with PBS−ethylenediamine tetraacetic acid (EDTA) (pH 7.4) for flow cytometer analysis. Flow cytometry was performed with an LSR II flow cytometer (BD Biosciences, USA), using a laser excitation wavelength of 488 nm and an FACS Diva software. The data was analyzed using Flowjo VX software (Tree Star, Ashland, OR, USA) after collecting 20 000 events. All the experiments were performed in triplicate.
Cell−LNP Interactions Using a 3D Tumor Spheroids Model:
The interaction of the LNPs with 3D tumor spheroids of PC3-MM2, MDA-MB-231, and A549 cells was also qualitatively and quantitatively evaluated by confocal microscopy and flow cytometry, respectively. The 3D tumor spheroids were formed by the 3D bioprinting method, as described above. After removing the cell culture medium and washing the spheroids with PBS buffer, 100 μL of 100 μg mL−1 of LNPs’ suspension in PBS buffer (pH 7.4) were added to the cells and incubated at 37 °C for 3 h. Then, the cells were washed twice with PBS buffer to remove the nonadherent LNPs. For confocal microscopy, the spheroids were then fixed using 4% PFA for 24 h at 37 °C. Afterward, the spheroids were washed twice with PBS buffer, and the nuclei was posteriorly stained by adding 100 μL of DAPI-405 (2.8 μg mL−1) and incubated for 24 h at 37 °C. Finally, the cell spheroids were washed twice with PBS buffer and transferred to a Lab-Tek 8-chamber. The localization of NPs was observed with a Leica SP5 inverted confocal microscope (Leica Microsystems, Germany), equipped with a 20 × objective.
For flow cytometry, after incubation with the LNPs, the spheroids were harvested with trypsin-PBS-EDTA for 5 min, collected by centrifugation at 1600 rcf for 4 min, incubated with trypan blue (0.005% v/v) during 4 min, washed twice with PBS−EDTA (pH 7.4), and finally suspended with PBS−EDTA. Flow cytometry was performed with an LSR II flow cytometer (BD Biosciences, USA), using a laser excitation wavelength of 488 nm and an FACS Diva software. The data was analyzed by Flowjo VX software (Tree Star, Ashland, OR, USA) after collecting 5000 events. All the experiments were performed in triplicate.
In Vitro Anticancer Effect of the LNPs:
The cell growth inhibition of pure BZL and BZL@LNPs, BZL@LNPs-iRGD, BZL@LNPs-DSS after incubation with PC3-MM2, MDA-MB-231, and A549 cells was evaluated using 2D and 3D cell culture models.
For the 2D model, PC3-MM2, MDA-MB-231, and A549 cells were seeded in 96-well-plated at a density of 10 000 cells per well and incubated overnight at 37 °C to allow the cells to attach. After that, 100 μL of suspensions containing different concentrations of BZL-loaded LNPs (5, 10, 25, 50 75, and 100 μg mL−1) and similar concentrations of pure BZL previously dissolved with 3% (v/v) ethanol (3.3, 8.2, 16.4, 32.9, and 65.7 × 10−6 m) in cell media were added to each well, and incubated for 6 and 24 h. Incubations with cell media and 1% (v/v) Triton X-100 were used as a positive and negative controls, respectively. A specific control for pure BZL was also added with the same amount of ethanol used in the dilutions. After the incubation, the number of viable cells in culture was determined using the CellTiter-Glo luminescence cell viability assay kit (Promega Corporation, USA), and the luminescence was measured using a Varioskan Flash plate reader. All the experiments were performed at least in triplicate.
After transferring the 3D tumor spheroids (PC3-MM2, MDA-MB-231 and A549 cells) to 96-well plates, 50 μL of BZL-loaded LNPs suspensions at different concentrations and similar concentrations of pure BZL previously dissolved with 3% (v/v) ethanol were added to each well, and mixed with 50 μL of NanoLuc luciferase and MT Cell Viability Substrate in complete cell culture medium, up to 48 h at 37 °C. The final concentration of NPs was 10, 25, 50, and 100 μg mL−1 and the free BZL was 6.57, 16.4, 32.9, and 65.7 × 10−6 m, and incubation with cell media was used as a positive control. Finally, the luminescence was measured, using a Varioskan Flash plate reader, at certain time points (6, 24, and 48 h). The number of viable cells in culture was determined by measuring the reducing potential of cells, and thus, the metabolism activity. All the experiments were performed in triplicate.
Afterward, the IC50, IC80, and IC90 values after incubation of BZL and BZL-loaded LNPs, LNPs-iRGD, and LNPs-DSS with PC3-MM2, MDA-MB-231, and A549 cells were obtained by means of a concentration response curve by nonlinear regression using OriginPro 2018.
Statistical Analysis:
The measured values were expressed by mean ± standard deviation (s.d.). One-way analysis of variance (ANOVA) with Bonferroni posthoc test was used to evaluate the significant difference with probabilities set of *p < 0.05, **p < 0.01, and ***p < 0.001, using the GraphPad Prism (GraphPad software Inc., CA, USA).
Supplementary Material
Acknowledgements
P.F. acknowledges the Finnish Cultural Foundation for a research grant (decision no. 00170202). J.Y.-K. acknowledges Walter och Lisi Wahls Stiftelse för Naturvetenskaplig Forskning and the Academy of Finland (Grant No. 108376). A.G. acknowledges the grant from the National Institutes of Health (Grant No. DE028531). H.A.S. acknowledges financial support from the Sigrid Jusélius Foundation (decision no. 4704580), and the Helsinki Institute of Life Science Research Funds. The authors thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki, for providing laboratory facilities for TEM imaging.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Patrícia Figueiredo, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, FI-00014 Helsinki, Finland.
Mika H. Sipponen, School of Chemical Engineering, Department of Bioproducts and Biosystems, Aalto University, FI-00076 Aalto, Finland
Kalle Lintinen, Biohybrid Materials, Department of Bioproducts and Biosystems, Aalto University, FI-00076 Aalto, Finland.
Alexandra Correia, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, FI-00014 Helsinki, Finland.
Alexandros Kiriazis, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, FI-00014 Helsinki, Finland.
Jari Yli-Kauhaluoma, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, FI-00014 Helsinki, Finland.
Monika Österberg, School of Chemical Engineering, Department of Bioproducts and Biosystems, Aalto University, FI-00076 Aalto, Finland.
Anne George, Brodie Tooth Development Genetics and Regenerative Medicine, Research Laboratory Department of Oral Biology, University of Illinois, Chicago, IL 60612, USA.
Jouni Hirvonen, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, FI-00014 Helsinki, Finland.
Mauri A. Kostiainen, Biohybrid Materials, Department of Bioproducts and Biosystems, Aalto University, FI-00076 Aalto, Finland
Hélder A. Santos, Drug Research Program, Division of Pharmaceutical Chemistry and Technology, Faculty of Pharmacy, University of Helsinki, FI-00014 Helsinki, Finland Helsinki Institute of Life Science (HiLIFE), University of Helsinki, FI-00014 Helsinki, Finland.
References
- [1].Figueiredo P, Bauleth-Ramos T, Hirvonen J, Sarmento B, Santos HA, in Handbook of Nanomaterials for Cancer Theranostics (Ed: Conde J), Elsevier, Amsterdam, Netherlands: 2018, p. 1. [Google Scholar]
- [2].Wicki A, Witzigmann D, Balasubramanian V, Huwyler J, J. Controlled Release 2015, 200, 138. [DOI] [PubMed] [Google Scholar]
- [3].Mitra AK, Agrahari V, Mandal A, Cholkar K, Natarajan C, Shah S, Joseph M, Trinh HM, Vaishya R, Yang X, Hao Y, Khurana V, Pal D, J. Controlled Release 2015, 219, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rahikkala A, Pereira SAP, Figueiredo P, Passos MLC, Araújo ARTS, Saraiva MLMFS, Santos HA, Adv. Biosyst 2018, 2, 1800020. [Google Scholar]
- [5].Li W, Liu Z, Fontana F, Ding Y, Liu D, Hirvonen JT, Santos HA, Adv. Mater 2018, 30, 1703740. [DOI] [PubMed] [Google Scholar]
- [6].Janoniene A, Liu Z, Baranauskiene L, Mäkilä E, Ma M, Salonen J, Hirvonen J, Zhang H, Petrikaite V, Santos HA, ACS Appl. Mater. Interfaces 2017, 9, 13976. [DOI] [PubMed] [Google Scholar]
- [7].Zhang F, Correia A, Mäkilä E, Li W, Salonen J, Hirvonen JJ, Zhang H, Santos HA, ACS Appl. Mater. Interfaces 2017, 9, 10034. [DOI] [PubMed] [Google Scholar]
- [8].Liu Z, Balasubramanian V, Bhat C, Vahermo M, Mäkilä E, Kemell M, Fontana F, Janoniene A, Petrikaite V, Salonen J, Yli-Kauhaluoma J, Hirvonen J, Zhang H, Santos HA, Adv. Healthcare Mater 2017, 6, 1601009. [DOI] [PubMed] [Google Scholar]
- [9].Correia A, Shahbazi M-A, Mäkilä E, Almeida S, Salonen J, Hirvonen J, Santos HA, ACS Appl. Mater. Interfaces 2015, 7, 23197. [DOI] [PubMed] [Google Scholar]
- [10].Allen TM, Cullis PR, Adv. Drug Delivery Rev 2013, 65, 36. [DOI] [PubMed] [Google Scholar]
- [11].Sercombe L, Veerati T, Moheimani F, Wu SY, Sood AK, Hua S, Front. Pharmacol 2015, 6, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Figueiredo P, Balasubramanian V, Shahbazi M-A, Correia A, Wu D, Palivan CG, Hirvonen JT, Santos HA, Int. J. Pharm 2016, 511, 794. [DOI] [PubMed] [Google Scholar]
- [13].Herranz-Blanco B, Shahbazi M-A, Correia AR, Balasubramanian V, Kohout T, Hirvonen J, Santos HA, Adv. Healthcare Mater 2016, 5, 1904. [DOI] [PubMed] [Google Scholar]
- [14].Li W, Liu D, Zhang H, Correia A, Mäkilä E, Salonen J, Hirvonen J, Santos HA, Acta Biomater. 2017, 48, 238. [DOI] [PubMed] [Google Scholar]
- [15].Bauleth-Ramos T, Shahbazi M-A, Liu D, Fontana F, Correia A, Figueiredo P, Zhang H, Martins JP, Hirvonen JT, Granja P, Sarmento B, Santos HA, Adv. Funct. Mater 2017, 27, 1703303. [Google Scholar]
- [16].Zhang H, Liu D, Wang L, Liu Z, Wu R, Janoniene A, Ma M, Pan G, Baranauskiene L, Zhang L, Cui W, Petrikaite V, Matulis D, Zhao H, Pan J, Santos HA, Adv. Healthcare Mater 2017, 6, 1601406. [DOI] [PubMed] [Google Scholar]
- [17].Almeida PV, Shahbazi M-A, Correia A, Mäkilä E, Kemell M, Salonen J, Hirvonen J, Santos HA, Nanomedicine 2017, 12, 1401. [DOI] [PubMed] [Google Scholar]
- [18].Kong F, Zhang X, Zhang H, Qu X, Chen D, Servos M, Mäkilä E, Salonen J, Santos HA, Hai M, Weitz DA, Adv. Funct. Mater 2015, 25, 3330. [Google Scholar]
- [19].Wurm FR, Weiss CK, Front. Chem 2014, 2, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tschan MJL, Brulé E, Haquette P, Thomas CM, Polym. Chem 2012, 3, 836. [Google Scholar]
- [21].Upton BM, Kasko AM, Chem. Rev 2016, 116, 2275. [DOI] [PubMed] [Google Scholar]
- [22].Figueiredo P, Lintinen K, Hirvonen JT, Kostiainen MA, Santos HA, Prog. Mater. Sci 2018, 93, 233. [Google Scholar]
- [23].Figueiredo P, Ferro C, Kemell M, Liu Z, Kiriazis A, Lintinen K, Florindo HF, Yli-Kauhaluoma J, Hirvonen J, Kostiainen MA, Santos HA, Nanomedicine 2017, 12, 2581. [DOI] [PubMed] [Google Scholar]
- [24].Figueiredo P, Lintinen K, Kiriazis A, Hynninen V, Liu Z, Bauleth-Ramos T, Rahikkala A, Correia A, Kohout T, Sarmento B, Yli-Kauhaluoma J, Hirvonen J, Ikkala O, Kostiainen MA, Santos HA, Biomaterials 2017, 121, 97. [DOI] [PubMed] [Google Scholar]
- [25].Dai L, Liu R, Hu L-Q, Zou Z-F, Si C-L, ACS Sustainable Chem. Eng 2017, 5, 8241. [Google Scholar]
- [26].Sipponen MH, Lange H, Ago M, Crestini C, ACS Sustainable Chem. Eng 2018, 6, 9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mura S, Nicolas J, Couvreur P, Nat. Mater 2013, 12, 991. [DOI] [PubMed] [Google Scholar]
- [28].Adam DF, Sarah EC, Rihe L, Curr. Pharm. Des 2013, 19, 6315.23470005 [Google Scholar]
- [29].Bazak R, Houri M, El Achy S, Kamel S, Refaat T, J. Cancer Res. Clin. Oncol 2015, 141, 769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Teesalu T, Sugahara KN, Ruoslahti E, Front. Oncol 2013, 3, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Simon-Gracia L, Hunt H, Teesalu T, Molecules 2018, 23, 1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Silva S, Almeida AJ, Vale N, Biomolecules 2019, 9, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ravindran S, Snee PT, Ramachandran A, George A, J. Biol. Chem 2013, 288, 16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ahvazi B, Cloutier É, Wojciechowicz O, Ngo T-D, ACS Sustainable Chem. Eng 2016, 4, 5090. [Google Scholar]
- [35].Lievonen M, Valle-Delgado JJ, Mattinen M-L, Hult E-L, Lintinen K, Kostiainen MA, Paananen A, Szilvay GR, Setälä H, Österberg M, Green Chem. 2016, 18, 1416. [Google Scholar]
- [36].DeRuiter J, Princ. Drug Action 2005, 1, 1. [Google Scholar]
- [37].Aumüller IB, Yli-Kauhaluoma J, Org. Lett 2011, 13, 1670. [DOI] [PubMed] [Google Scholar]
- [38].Shah N, Pang B, Yeoh K-G, Thorn S, Chen CS, Lilly MB, Salto-Tellez M, Eur. J. Cancer 2008, 44, 2144. [DOI] [PubMed] [Google Scholar]
- [39].Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J, Haematologica 2010, 95, 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Swords R, Kelly K, Carew J, Nawrocki S, Mahalingam D, Sarantopoulos J, Bearss D, Giles F, Curr. Drug Targets 2011, 12, 2059. [DOI] [PubMed] [Google Scholar]
- [41].Kiriazis A, Vahakoski RL, Santio NM, Arnaudova R, Eerola SK, Rainio E-M, Aumüller IB, Yli-Kauhaluoma J, Koskinen PJ, PLoS One 2013, 8, e55409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Blundell E, Healey MJ, Holton E, Sivakumaran M, Manstana S, Platt M, Anal. Bioanal. Chem 2016, 408, 5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gessner A, Lieske A, Paulke BR, Müller RH, Eur. J. Pharm. Biopharm 2002, 54, 165. [DOI] [PubMed] [Google Scholar]
- [44].Wang C-F, Mäkilä EM, Kaasalainen MH, Liu D, Sarparanta MP, Airaksinen AJ, Salonen JJ, Hirvonen JT, Santos HA, Biomaterials 2014, 35, 1257. [DOI] [PubMed] [Google Scholar]
- [45].Gatoo MA, Naseem S, Arfat MY, Dar AM, Qasim K, Zubair S, BioMed Res. Int. 2014, 2014, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sukhanova A, Bozrova S, Sokolov P, Berestovoy M, Karaulov A, Nabiev I, Nanoscale Res. Lett. 2018, 13, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Thubagere A, Reinhard BM, ACS Nano 2010, 4, 3611. [DOI] [PubMed] [Google Scholar]
- [48].Zhu M, Nie G, Meng H, Xia T, Nel A, Zhao Y, Acc. Chem. Res 2013, 46, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Foroozandeh P, Aziz AA, Nanoscale Res. Lett 2018, 13, 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Verma A, Stellacci F, Small 2010, 6, 12. [DOI] [PubMed] [Google Scholar]
- [51].Guarnieri D, Malvindi MA, Belli V, Pompa PP, Netti P. J. J. o. N. R., J. Nanopart. Res 2014, 16, 2229. [Google Scholar]
- [52].Behzadi S, Vatan NM, Lema K, Nwaobasi D, Zenkov I, Abadi PPSS, Khan DA, Corbo C, Aghaverdi H, Farokhzad OC, Mahmoudi M, Adv. Biosyst 2018, 2, 1800046. [Google Scholar]
- [53].Cho EC, Zhang Q, Xia Y, Nat. Nanotechnol 2011, 6, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E, Science 2010, 328, 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Edmondson R, Broglie JJ, Adcock AF, Yang L, Assay Drug Dev. Technol 2014, 12, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fröhlich E, Artif. Cells, Nanomed., Biotechnol 2018, 46, 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Granata A, Argyropoulos DS, Agric J. Food Chem. 1995, 43, 1538. [Google Scholar]
- [58].Lintinen K, Latikka M, Sipponen MH, Ras RHA, Österberg M, Kostiainen MA, RSC Adv. 2016, 6, 31790. [Google Scholar]
- [59].Sipponen MH, Smyth M, Leskinen T, Johansson L-S, Österberg M, Green Chem. 2017, 19, 5831. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.