Abstract
Fatal familial insomnia (FFI), genetic Creutzfeldt–Jakob disease (gCJD), and Gerstmann–Sträussler–Scheinker (GSS) syndrome are neurodegenerative disorders linked to prion protein (PrP) mutations. The pathogenic mechanisms are not known, but increasing evidence points to mutant PrP misfolding and retention in the secretory pathway. We previously found that the D178N/M129 mutation associated with FFI accumulates in the Golgi of neuronal cells, impairing post-Golgi trafficking. In this study we further characterized the trafficking defect induced by the FFI mutation and tested the 178N/V129 variant linked to gCJD and a nine-octapeptide repeat insertion associated with GSS. We used transfected HeLa cells, embryonic fibroblasts and primary neurons from transgenic mice, and fibroblasts from carriers of the FFI mutation. In all these cell types, the mutant PrPs showed abnormal intracellular localizations, accumulating in the endoplasmic reticulum (ER) and Golgi. To test the efficiency of the membrane trafficking system, we monitored the intracellular transport of the temperature-sensitive vesicular stomatite virus glycoprotein (VSV-G), a well-established cargo reporter, and of endogenous procollagen I (PC-I). We observed marked alterations in secretory trafficking, with VSV-G accumulating mainly in the Golgi complex and PC-I in the ER and Golgi. A redacted version of mutant PrP with reduced propensity to misfold did not impair VSV-G trafficking, nor did artificial ER or Golgi retention of wild-type PrP; this indicates that both misfolding and intracellular retention were required to induce the transport defect. Pharmacological activation of Src family kinase (SFK) improved intracellular transport, suggesting that mutant PrP impairs secretory trafficking through corruption of SFK-mediated signaling.
Keywords: genetic prion disease, prion protein, protein misfolding, protein trafficking, Src family kinase
Abbreviations: AMPARs, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors; BSA, bovine serum albumin; CCD, charge-coupled device; CNS, central nervous system; DMEM, Dulbecco’s modified Eagle’s medium; EM, electron microscopy; ER, endoplasmic reticulum; FFI, fatal familial insomnia; gCJD, genetic Creutzfeldt–Jakob disease; GFP-GPI, GPI-anchored green fluorescent protein reporter; GPCR, G-protein-coupled receptors; GPI, glycosylphosphatidylinositol; GSS, Gerstmann–Sträussler–Scheinker; HFs, human fibroblasts; IF, immunofluorescence; KDEL-R, KDEL receptor; KO, knockout; LD, labeling density; mo, mouse; MEFs, mouse embryonic fibroblasts; MEM, minimum essential medium; moPrP, mouse PrP; PC-I, procollagen I; PDI, protein disulfide isomerase; PenStrep, penicillin/streptomycin; PG14, moPrP with a nine-octapeptide repeat insertion; PM, plasma membrane; PrP, prion protein; Rab GDP, dissociation inhibitor (GDI); RT, room temperature; SFK, Src family kinase; Tg, transgenic; VGCCs, voltage-gated calcium channels; VSV-G, vesicular stomatite virus glycoprotein; WT, wild-type; YAG, yttrium aluminum garnet
Genetic prion diseases are a rare group of fatal neurodegenerative disorders that include genetic Creutzfeldt–Jakob disease (gCJD), Gerstmann–Sträussler–Scheinker (GSS) syndrome, and fatal familial insomnia (FFI) (1). These disorders are inherited in an autosomal dominant fashion and are linked to mutations in the PRNP gene encoding the prion protein (PrP), on chromosome 20. More than 70 pathogenic PRNP variants have been reported, including protein-truncating variants, insertions or deletions of a repeated sequence encoding an octapeptide motif in the N-terminal region, and missense mutations (for an up-to-date list see http://www.cureffi.org/2015/01/13/list-of-reportedly-pathogenic-prnp-variants/).
PrP is a glycosylphosphatidylinositol (GPI) membrane-anchored glycoprotein of uncertain function, mainly expressed by neurons in the central nervous system (CNS) (2). It is synthesized in the endoplasmic reticulum (ER), where it undergoes several posttranslational modifications, including cleavage of the N-terminal signal peptide, facultative N-linked glycosylation, formation of a disulfide bond, and attachment of the GPI anchor after removal of a C-terminal hydrophobic peptide. During transit through the Golgi complex, PrP receives various additional modifications to its glycans and GPI anchor before being exported to the cell surface, where it resides in lipid rafts, membrane microdomains rich in cholesterol, and sphingolipids. From the cell surface PrP can be released into the extracellular space or internalized to an endosomal compartment from which it is either recycled to the plasma membrane (PM) or diverted to lysosomes for degradation.
Mouse (mo) PrP molecules carrying a nine-octapeptide repeat insertion associated with GSS (moPrP PG14) or the M/V129 polymorphic variants of the D178N mutations linked to FFI (moPrP D177N/M128) or gCJD (moPrP D177N/V128) misfold and aggregate soon after synthesis in the ER and are delayed in their biosynthetic maturation, resulting in their accumulation in transport organelles of the secretory pathway (3, 4, 5, 6, 7). We previously found that these mutants do not activate a maladaptive ER stress response (8, 9), but their intracellular retention impairs the trafficking and synaptic delivery of PrP-interacting proteins, including the α2δ-1 subunit of voltage-gated calcium channels (VGCCs) and the GluA2 subunit of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) (10, 11). This causes synaptic dysfunction, contributing to neuronal death in transgenic (Tg) mouse models.
Intracellular accumulation of mutant PrP may also have a more general effect on the neuronal trafficking machinery, as suggested by our previous study in which expression of the FFI mutant in neuroblastoma N2a cells was associated with reduced levels of functional Rab11 and impaired post-Golgi trafficking of a GPI-anchored green fluorescent protein reporter (GFP-GPI) (12). To further characterize the trafficking defect induced by the D177N/M128 mutation and test the effect of the PG14 and D177N/V128 mutants, we measured the efficiency of secretory transport of exogenous and endogenous cargos in different cell types, including transfected HeLa cells, embryonic fibroblasts and primary neurons from Tg mice, and human fibroblasts (HFs) from carriers of the FFI mutation. Secretory transport was impaired by each of the mutant PrPs, with the most marked alterations in cells expressing FFI PrP. Src family kinase (SFK) activation improved type I procollagen (PC-I) transport in FFI HFs, suggesting that mutant PrP may corrupt an SFK-mediated signaling circuit governing intracellular trafficking.
Results
Expression of mutant PrP impairs post-Golgi trafficking in HeLa cells
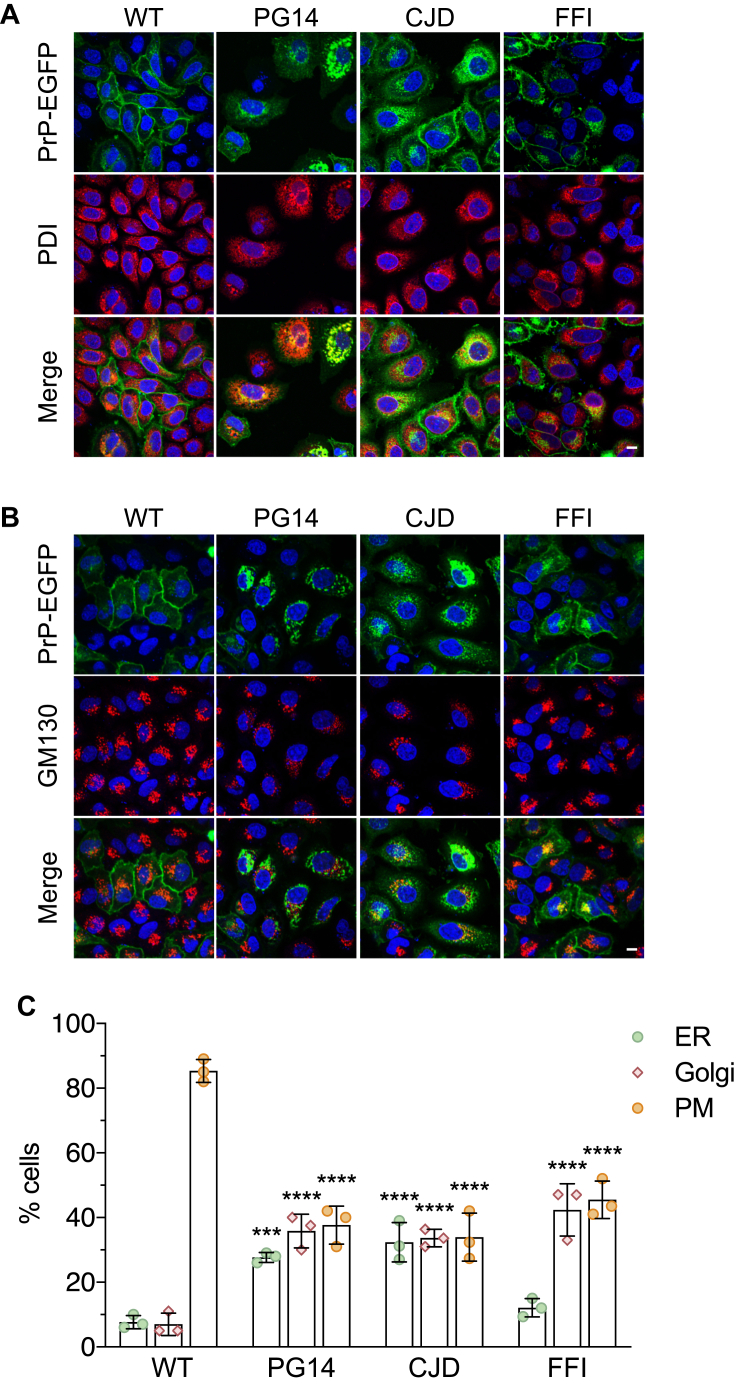
HeLa cells were transiently transfected with EGFP-tagged mouse PrP wild-type (WT), PG14, D177N/V128 (CJD), or D177N/M128 (FFI). After 48 h cells were fixed and immunostained with antibodies against protein disulfide isomerase (PDI) and GM130 (or giantin) to stain the ER and Golgi. Cells showing PrP-EGFP fluorescence on the PM with little or no colocalization with ER or Golgi markers, or intense intracellular fluorescence colocalizing with ER and Golgi markers and weak or no PM signal, were counted and expressed as percentages of the total. WT PrP was found at the PM in approximately 85% of cells; in contrast, 60 to 70% of the cells expressing each of the mutant PrPs had lower expression on the PM and greater intracellular staining pattern that largely colocalized with ER and Golgi makers (Fig. 1). Consistent with observations in neuronal cells (3, 4, 6, 10, 11, 12, 13), the mutants differed in their patterns of intracellular localization, with PG14 and CJD PrPs colocalizing with both ER and Golgi markers and the FFI PrP accumulating mostly in the Golgi (Fig. 1C). Results were similar when HeLa cells were transfected with nonfluorescent PrP constructs (see for example PG14 PrP in Fig. 3A and Fig. S1) (10, 11). This indicates that the abnormal intracellular localization was not a feature peculiar to the PrP-EGFP chimeras. However, to rule out any possible artifact due to the use of the fusion proteins, for the functional studies illustrated below we employed PrP molecules without the EGFP tag.
Figure 1.
Mutant PrPs localize abnormally in HeLa cells. HeLa cells were transfected with plasmids encoding WT, PG14, CJD, or FFI PrP-EGFP fusion protein. After 48 h cells were fixed, permeabilized, stained with anti-PDI (A) or anti-GM130 (B) polyclonal antibody followed by Alexa Fluor 647 (red)-conjugated anti-rabbit IgG secondary antibody, and reacted with Hoechst 33258 (blue) to stain the nuclei. Scale bar 10 μm. C, the number of cells showing PrP-EGFP on the plasma membrane (PM) with no or little colocalization with PDI or GM130/giantin, or intense colocalization with PDI (ER) or GM130/giantin (Golgi) was counted and expressed as a percentage of the total. Each bar indicates the mean ± SD of three independent experiments evaluating a total of 210 WT, 198 PG14, 191 CJD, and 204 FFI PrP-EGFP transfected cells. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 versus WT by two-way ANOVA, Tukey’s post-hoc test.
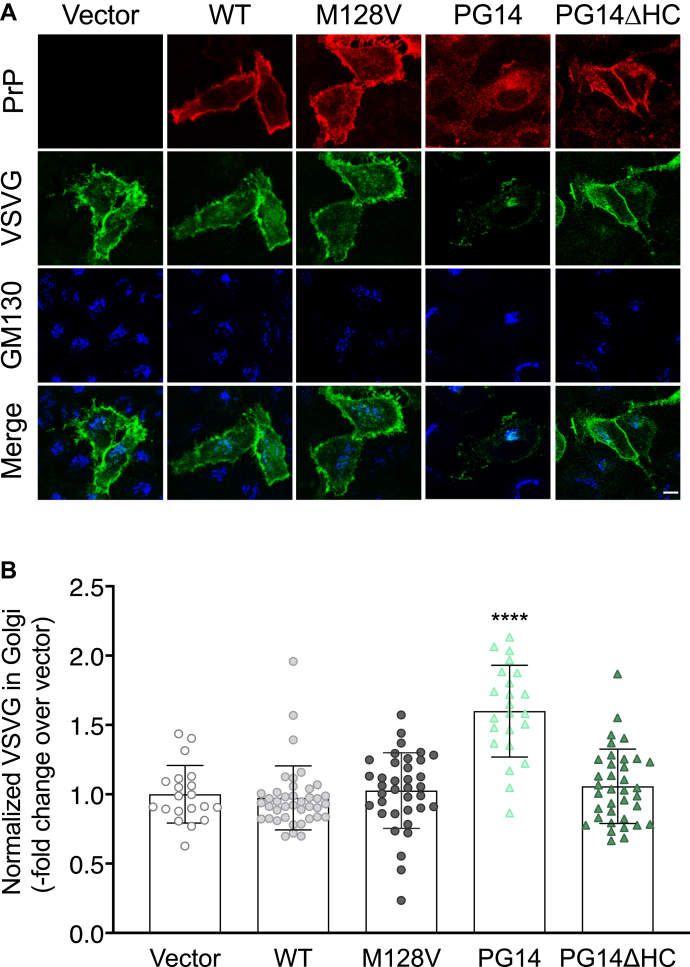
Figure 3.
Mutant PrP misfolding and aggregation are necessary to impair VSV-G transport.A, HeLa cells were transfected with plasmids encoding WT, M128V, PG14, or PG14ΔHC PrP. After 24 h at 35 °C cells were fixed, permeabilized, stained with polyclonal anti-GM130 and monoclonal anti-PrP 12B2 antibodies followed by Alexa Fluor-conjugated anti-IgG secondary antibodies. Cells were viewed with green excitation/emission settings to detect VSV-G and blue excitation/emission settings to detect GM130. Scale bar 10 μm. B, the normalized fluorescent density of VSV-G in the Golgi was measured and expressed as fold change over vector-transfected cells. Data are the mean ± SD of 20 to 40 cells. ∗∗∗∗p < 0.0001 versus vector by one-way ANOVA, Tukey’s post-hoc test.
To test the function of the secretory pathway, we monitored the intracellular transport of the temperature-sensitive vesicular stomatitis virus glycoprotein (VSV-G). VSV-G is a multisubunit transmembrane glycoprotein that is widely used to study the exocytic pathway. Secretory transport of VSV-G can be synchronized by sequential temperature shifts (14). At 40 °C VSV-G does not fold properly and is retained in the ER. After lowering the temperature to 32 °C, VSV-G folds and exits the ER, eventually reaching the PM.
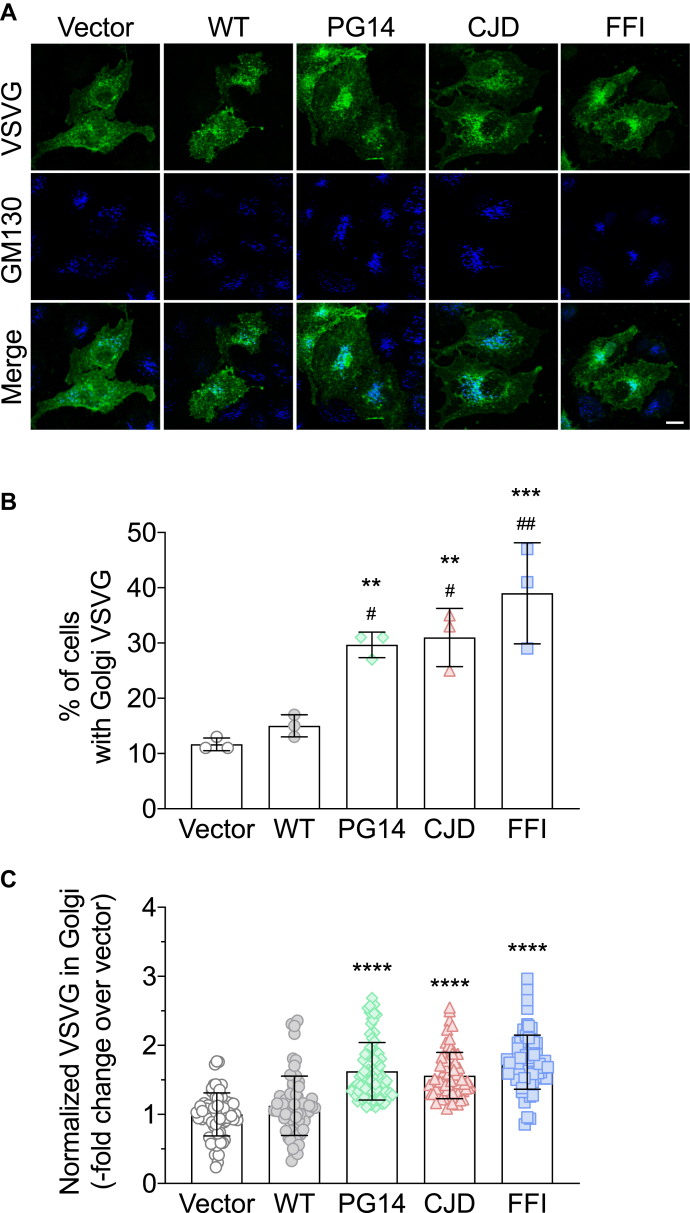
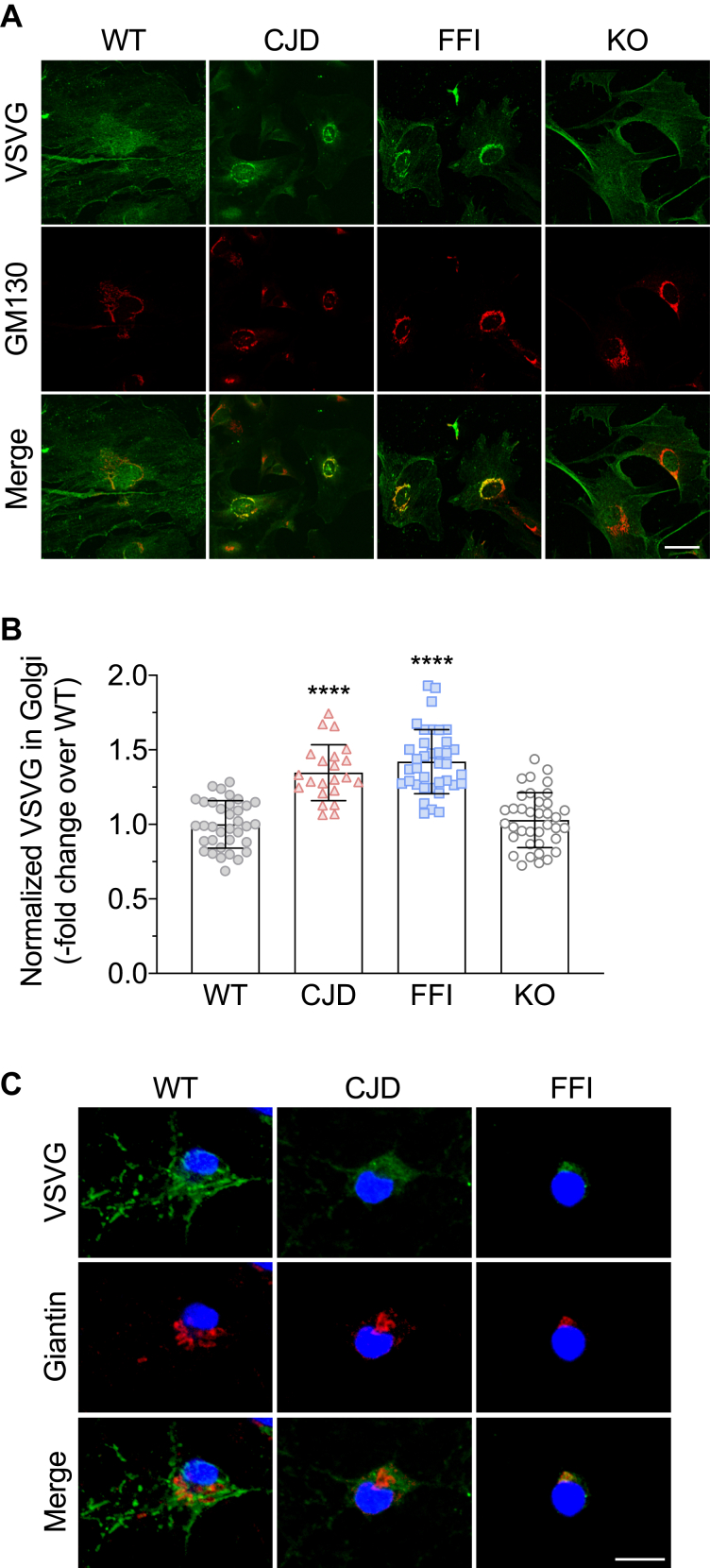
There is evidence that temperature can influence PrP folding in cultured cells (15), and our preliminary experiments showed that incubation at 40 °C caused WT PrP to accumulate intracellularly. Therefore we carried out the experiment at a constant temperature of 35 °C, which allowed efficient folding and cell surface expression of both VSV-G and WT PrP (see example in Fig. 3A). HeLa cells were transfected with the empty vector, or with plasmids encoding WT or each of the mutant PrP, and VSV-G-GFP. Cells were incubated at 35 °C for 24 h and analyzed by confocal microscopy after immunofluorescence (IF) staining with antibodies against PrP and the Golgi marker GM130 (Fig. 2A). The fluorescence of the VSV-G signal in the whole cell and in the GM130-positive area was measured in cells expressing PrP, identified by IF staining with anti-PrP antibody 12B2 (not shown). VSV-G was clearly accumulating in the Golgi of FFI cells in which most PrP localized in this organelle, but was also detectable in the Golgi of PG14 and CJD cells, where there was less PrP in the Golgi, and it colocalized mostly with PDI (see example in Fig. 3A, and Fig. S1). The percentage of cells accumulating VSV-G in the Golgi was significantly higher in mutant PrP than in vector- or WT PrP-transfected cells (Fig. 2B). In cells transfected with the mutant PrPs, the VSVG signal in the Golgi, normalized by the total VSV-G in the cell, was significantly more intense than in cells transfected with the empty vector or WT PrP (Fig. 2C).
Figure 2.
VSV-G accumulates in the Golgi of HeLa cells expressing mutant PrPs.A, HeLa cells were transfected with plasmids encoding WT, PG14, CJD or FFI PrP, and VSV-G-EGFP fusion protein. After 24 h at 35 °C cells were fixed, permeabilized, stained with polyclonal anti-GM130 and monoclonal anti-PrP 12B2 antibodies followed by Alexa Fluor-conjugated anti-IgG secondary antibodies. Cells were viewed with green excitation/emission settings to detect VSV-G and blue excitation/emission settings to detect GM130. Scale bar 10 μm. B, percentages of cells accumulating VSV-G in the Golgi. Data are the mean ± SD of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001 versus vector, and #p < 0.05, ##p < 0.01 versus WT by one-way ANOVA, Tukey’s post-hoc test. C, the normalized fluorescent density of VSV-G in the Golgi was measured in PrP-expressing cells and expressed as fold change over vector-transfected cells. Each bar indicates the mean ± SD of 65 to 80 cells from three independent experiments; ∗∗∗∗p < 0.0001 versus vector and WT by one-way ANOVA, Tukey’s post-hoc test.
This suggested that Golgi to PM trafficking was impaired in mutant PrP HeLa cells, in line with previous analysis in neuroblastoma N2a cells expressing FFI PrP (12). This was confirmed by the analysis of GFP-GPI and CD59-GFP, two GPI-anchored membrane proteins, which continuously cycle between the Golgi complex and the PM (16). PM expression of these proteins in HeLa cells transiently transfected with the mutant PrP was significantly reduced, particularly in cells expressing PG14 and FFI PrPs (Table 1).
Table 1.
Quantitative analysis of plasma membrane GFP-GPI and CD59-GFP in HeLa cells
| Construct | GFP-GPI (PM/total)a | p valueb | p valuec | CD59-GFP (PM/total)a | p valueb | p valuec |
|---|---|---|---|---|---|---|
| Empty vector | 1.47 ± 0.81 | - | - | 1.33 ± 1.12 | - | - |
| WT | 1.02 ± 0.66 | 0.0254 | - | 1.00 ± 0.65 | 0.1149 | - |
| PG14 | 0.55 ± 0.31 | 0.0001 | 0.0231 | 0.43 ± 0.15 | 0.0001 | 0.0074 |
| CJD | 0.93 ± 0.77 | 0.0077 | 0.9676 | 0.76 ± 0.43 | 0.0038 | 0.4597 |
| FFI | 0.50 ± 0.12 | 0.0001 | 0.0213 | 0.45 ± 0.18 | 0.0001 | 0.0028 |
Data are the mean ± SD of 20 to 32 cells.
Versus empty vector.
Versus WT by one-way ANOVA, Dunnett post-hoc test.
Intracellular PrP aggregation is necessary to impair VSV-G trafficking
To assess whether mutant PrP aggregation was required to impair secretory trafficking, we analyzed VSV-G transport in HeLa cells expressing a version of PG14 PrP with a deletion of amino acids 114 to 121 in the hydrophobic core (PG14/ΔHC). This molecule is less prone to misfolding and aggregation and is delivered to the cell surface more efficiently than its full-length counterpart (17). HeLa cells were cotransfected with VSV-G-GFP and PrP WT, PG14, or PG14ΔHC. We also tested moPrP with the M128V substitution homologous to the V129 polymorphic variant of human PrP, to model a natural, nonaggregating, and nonintracellularly retained variant (18). After 24 h incubation at 35 °C, cells were immunostained with antibodies against PrP and GM130, and the VSV-G-GFP signal intensity in the Golgi was analyzed in PrP-expressing cells (Fig. 3A). Compared with vector- and WT PrP-transfected cells, the Golgi VSV-G signal was significantly more intense in cells transfected with PG14 PrP, but not in those transfected with PG14/ΔHC or M128V PrP (Fig. 3B). Thus, aggregation and intracellular retention of mutant PrP were necessary to impair VSV-G transport.
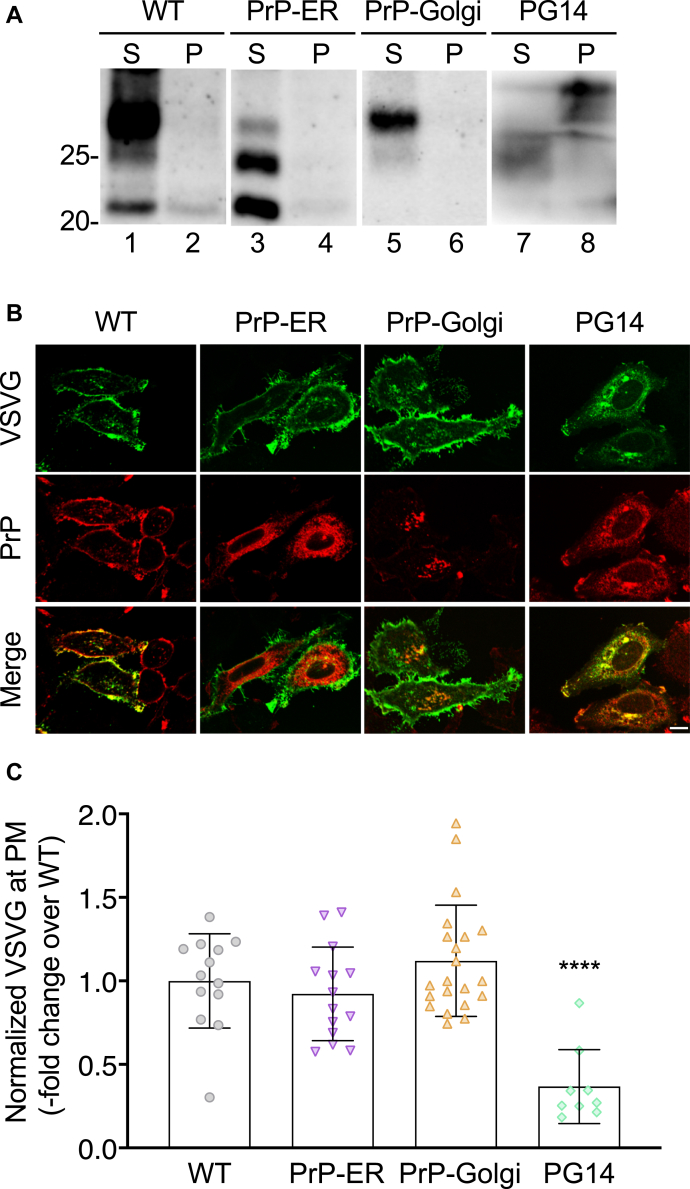
To assess whether PrP retention in transport organelles was sufficient to induce the trafficking defect, we analyzed VSV-G distribution in HeLa cells expressing WT PrP molecules artificially targeted to the ER or Golgi by replacing the GPI signal with the ER retention KDEL motif (PrP-ER) or with the transmembrane domain from the rubella virus envelop glycoprotein E2, which contains a Golgi-targeting signal (PrP-Golgi) (11). We had previously confirmed the predicted intracellular localizations of these PrP molecules by co-immunofluorescence staining with ER and Golgi markers (11) and demonstrate here that, unlike PG14 PrP, they do not form detergent-insoluble aggregates in HeLa cells (Fig. 4A). In cells expressing PrP-ER or PrP-Golgi, VSV-G was efficiently expressed on the PM, like in WT PrP-expressing cells (Fig. 4, B and C).
Figure 4.
Retention of nonaggregated PrP in the ER or Golgi does not impair VSV-G trafficking.A, HeLa cells were transfected with plasmids encoding WT, PrP-ER, PrP-Golgi, or PG14 PrP. After 24 h cells were lysed in nondenaturing detergents, and the cell lysates were ultracentrifuged at 186,000g for 45 min. PrP in the supernatant (S) and pellet (P) was analyzed by western blot. Molecular weight markers are in kDa. B, HeLa cells were transfected with plasmids encoding WT, PrP-ER, PrP-Golgi or PG14 PrP, and VSV-G-EGFP fusion protein. After 24 h at 35 °C cells were fixed, permeabilized, stained with monoclonal anti-PrP 12B2 antibody followed by Alexa Fluor-conjugated anti-IgG secondary antibody. Cells were viewed with green excitation/emission settings to detect VSV-G and redexcitation/emission settings to detect PrP. Scale bar 10 μm. C, the normalized fluorescent density of VSV-G at the plasma membrane (PM) was measured in PrP-expressing cells and expressed as the fold difference from WT PrP-transfected cells. Each bar indicates the mean ± SD of 9 to 21 cells; ∗∗∗∗p < 0.0001 versus WT by one-way ANOVA, Tukey’s post-hoc test.
These results indicate that PrP retention in the ER or Golgi is not sufficient to impair VSV-G trafficking and that mutation-induced misfolding and aggregation are required.
VSV-G transport is impaired in embryonic fibroblasts and primary hippocampal neurons from mutant PrP mice, but not in PrP knockout cells
To test whether the alterations in VSV-G transport documented in transiently transfected HeLa cells held true in other cell types, we analyzed VSV-G localization in mouse embryonic fibroblasts (MEFs) and primary hippocampal neurons from Tg mice expressing WT or mutant PrP under the control of the mouse PrP promoter (3, 4). We restricted this analysis to cells from Tg(CJD) and Tg(FFI) mice because they express similar levels of transgenic PrP—Tg(PG14) mice express less (19)—and are representative of PrP mutants accumulating preferentially in the ER (CJD) or Golgi (FFI) (3, 4). There was an increase in VSV-G signal density in the Golgi of the mutant MEFs compared with WT controls (Fig. 5, A and B). VSV-G distribution in MEFs from PrP knockout (KO) and WT mice was similar, indicating that loss of PrP function had no effect on the secretory transport of this cargo reporter (Fig. 5, A and B). VSV-G accumulated intracellularly also in mutant hippocampal neurons, with the majority colocalizing with the Golgi marker giantin, especially in FFI cells (Fig. 5C).
Figure 5.
VSV-G transport is impaired in primary cells from mutant PrP but not PrP knockout mice.A, MEFs from WT, CJD, FFI, and PrP knockout (KO) mice were infected with VSV at 32 °C for 1 h, washed, and cultured at 35 °C for 4 h. Cells were fixed and immunostained with monoclonal anti-VSV-G and polyclonal anti-GM130 antibodies, followed by Alexa Fluor-conjugated anti-IgG secondary antibodies. Cells were viewed with green excitation/emission settings to detect VSV-G and red excitation/emission settings to detect GM130. Scale bar 50 μm. B, the normalized fluorescent density of VSV-G in the Golgi was measured and expressed as fold change over WT. Each bar indicates the mean ± SD of 22 to 37 cells; ∗∗∗∗p < 0.0001 versus WT and KO by one-way ANOVA, Tukey’s post-hoc test. C, primary hippocampal neurons from WT, CJD, and FFI mice were infected with VSV at 32 °C for 45 min, washed and cultured at 35 °C for 2 h. Cells were fixed and immunostained with monoclonal anti-VSV-G and polyclonal anti-giantin antibodies, followed by Alexa Fluor-conjugated anti-IgG secondary antibodies. Cells were viewed with green excitation/emission settings to detect VSV-G and red excitation/emission settings to detect giantin. Scale bar 10 μm.
VSV-G and procollagen I (PC-I) accumulate in transport organelles of fibroblasts from carriers of the FFI mutation, and activation of Src family kinases reduces PC-I accumulation
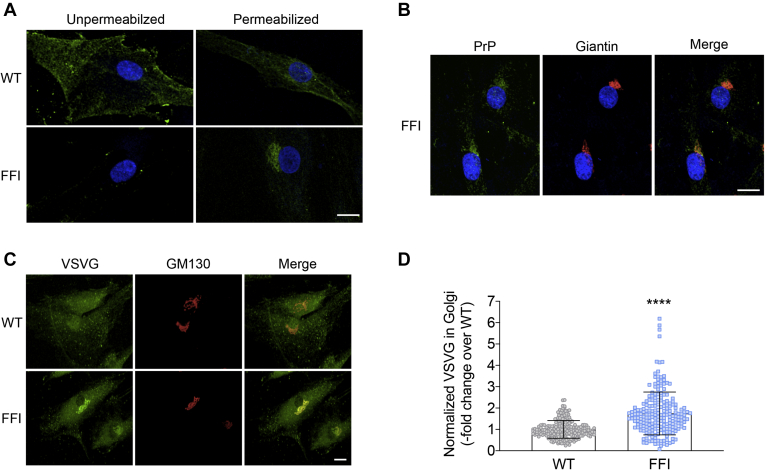
To test whether secretory trafficking was altered in a more pathophysiologically relevant cell model, we analyzed the steady-state distribution of VSV-G in human fibroblasts from carriers of the FFI mutation. First, we characterized PrP distribution by immunostaining the cells before or after permeabilization with Triton-X100. In HFs from noncarrier individuals (WT), PrP was mainly found on the PM where its distribution appeared patchy, consistent with the protein’s association with lipid rafts (Fig. 6A). In contrast, PrP was barely detectable on the PM of FFI HFs and was mainly found in intense perinuclear patches colocalizing with giantin (Fig. 6, A and B). Thus, PrP accumulated abnormally in the Golgi of fibroblasts from carriers of the FFI mutation, similar to observations in transgenic cells expressing the homologous mouse mutation (3, 6, 12).
Figure 6.
Abnormal intracellular PrP localization and impairment of VSV-G transport in human fibroblasts from carriers of the FFI mutation.A, to visualize PrP on the cell surface, human fibroblasts (HFs) from noncarriers (WT) and carriers of the PRNP D178N mutation (FFI) were immunostained with anti-PrP antibody 6H4 before fixation and application of Alexa Fluor 488 (green)-conjugated secondary antibody and staining with Hoechst 33258 (blue) to visualize the nuclei. To investigate the intracellular distribution of PrP, cells were fixed and permeabilized before immunostaining. B, FFI HFs were fixed, permeabilized, and immunostained with anti-PrP and anti-giantin antibodies, followed by Alexa Fluor-conjugated secondary antibodies. Cells were viewed with green excitation/emission settings to detect PrP, red excitation/emission settings to detect giantin, and UV excitation/emission settings to detect the nuclei. Scale bar 15 μm. C, HFs were infected with VSV at 32 °C for 1 h, washed, and cultured at 35 °C for 4 h. Cells were fixed and immunostained with monoclonal anti-VSV-G and polyclonal anti-GM130 antibodies, followed by Alexa Fluor-conjugated IgG secondary antibodies. Scale bar 10 μm. D, the normalized fluorescent density of VSV-G in the Golgi was measured and expressed as fold change over WT. Each bar indicates the mean ± SEM of 209 WT and 211 FFI cells (four independent lines each). ∗∗∗∗p < 0.0001 by unpaired t-test. Scale bar 10 μm.
To analyze the steady-state distribution of VSV-G, HFs were infected with VSV, cultured at 35 °C for 4 h, and immunostained with antibodies against VSV-G and GM130. VSV-G efficiently reached the PM of WT HFs, but accumulated in the Golgi of the mutant cells (Fig. 6, C and D); this is consistent with results in transiently transfected HeLa and MEFs and hippocampal neurons from Tg(FFI) mice.
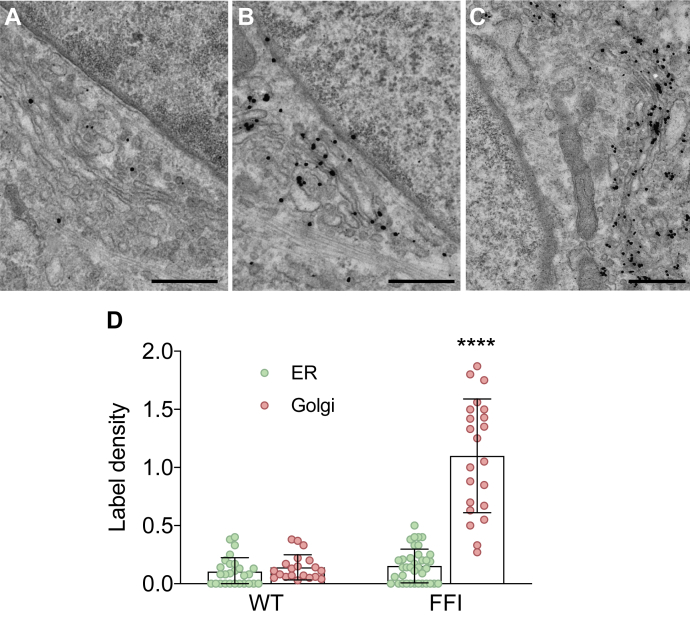
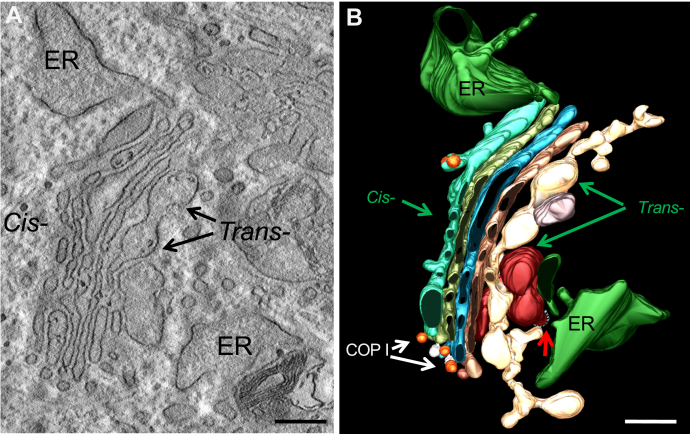
Immuno-electron microscopy (EM) confirmed the accumulation of VSV-G in the Golgi stacks of FFI but not WT HFs (Fig. 7). In FFI HFs we detected ultrastructural abnormalities of the ER and Golgi (Fig. 8A). Three-dimensional tomography showed ER swelling and continual distension of trans-Golgi cisternae (Fig. 8B). This suggested possible abnormalities in protein folding and export from the ER besides impaired exit from the Golgi.
Figure 7.
Electron microscopy localization of VSV-G in wild-type and FFI human fibroblasts. Cultures of fibroblasts from non-carriers (WT; A) and carriers of the PRNP D178N mutation (FFI; B and C) were infected with VSV at 32 °C for 1 h, washed, and cultured at 35 °C for 4 h. Cells were fixed and labeled with anti-VSV-G monoclonal antibody using the gold-enhance protocol. Scale bars 250 nm in A and B; 500 nm in C. D, quantification of gold particles in different cell compartments. Bars indicate the mean ± SD of 20 to 37 images. ∗∗∗∗p < 0.0001 versus WT by two-way ANOVA Bonferroni’s post-hoc test.
Figure 8.
Ultrastructural abnormalities of the ER and Golgi complex in a FFI fibroblast.A, electron micrograph of a FFI fibroblast. B, three-dimensional tomography reconstruction from virtual serial slices of the fibroblast shown in A. The dilated ER cisterna is colored green. The continuous distensions of trans-Golgi are in pale yellow/dark red, the cis-most cisterna of Golgi stack is light blue. COP I and clathrin-coated (red arrow) vesicles are indicated. Scale bars 200 nm.
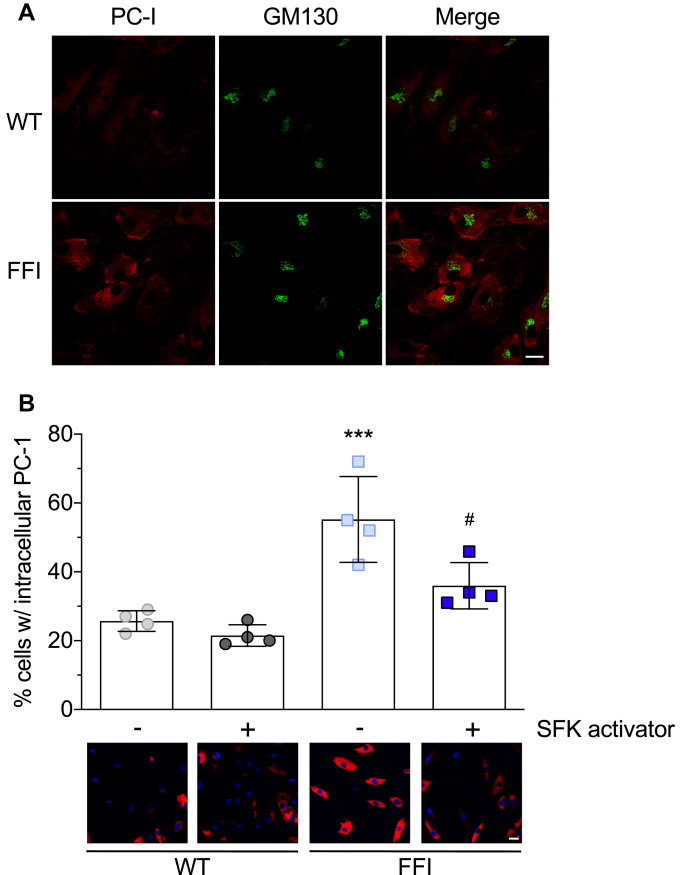
To assess this, we monitored the distribution of PC-I, a nondiffusible large cargo highly expressed by fibroblasts, whose biosynthesis and secretion are a complex process requiring a fully functional ER (20). Immunofluorescence showed a larger number of FFI HFs with intense intracellular PC-I staining than in WT cells and partial colocalization with the Golgi marker GM130 (Fig. 9A).
Figure 9.
Src activation partially rescues the trafficking impairment of procollagen-I in FFI fibroblasts.A, cultures of human fibroblasts from noncarriers (WT) and carriers of the PRNP D178N mutation (FFI) were fixed, permeabilized, and immunostained with anti-PC-I and anti-GM130 antibodies. After incubation with Alexa Fluor-conjugated IgG secondary antibodies, cells were viewed with red excitation/emission settings to detect PC-I and green excitation/emission settings to detect GM130. B, HFs were treated with 10 μM Src Family activator or vehicle. After 4 h cells were fixed and stained with anti-PC-I antibody (red) and reacted with Hoechst 33258 (blue) to stain the nuclei. The bar graph indicates the percentages of cells with intracellular accumulation of PC-I. Data are mean ± SD of ∼2000 cells from four separate experiments; ∗∗∗p < 0.001 versus WT vehicle; #p < 0.05 versus FFI vehicle by two-way ANOVA, Tukey’s post-hoc test. Scale bars 30 μm.
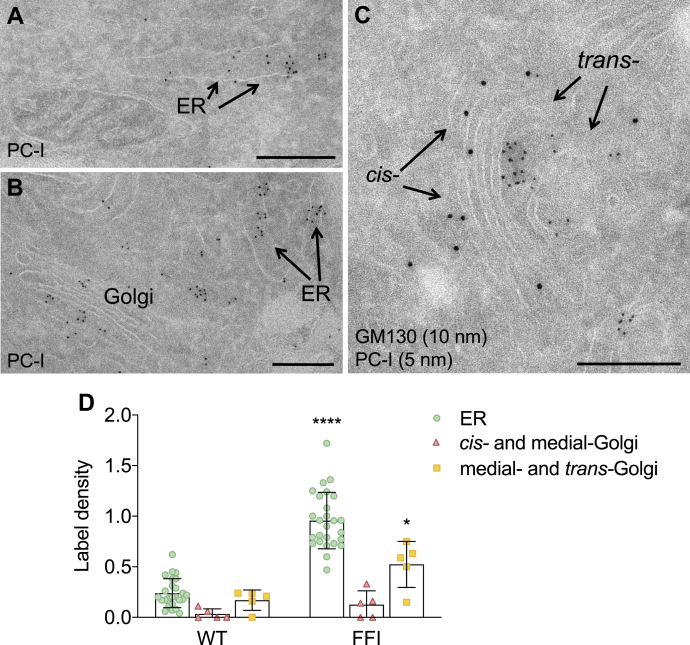
To characterize the localization of PC-I in the mutant cells better, we used immuno-gold labeling. Electron microscopy indicated that PC-I accumulated in the dilated ER cisternae (Fig. 10, A, B and D) and in the medial and trans Golgi stacks identified by lack of GM130 staining, which selectively marks the cis Golgi (21, 22) (Fig. 10, C and D). These data indicated impaired PC-I exit from both the ER and Golgi.
Figure 10.
Electron microscopy localization of procollagen-I in fibroblasts of carriers of the FFI mutation.A and B, immunogold staining on cryosections from FFI human fibroblasts using anti-PC-I, showing PC-I accumulation in dilated ER cisternae and in the Golgi. C, double immunogold staining using anti-PC-I (5 nm gold particles) and anti-GM130 antibodies (10 nm gold particles), showing accumulation of PC-I in the trans- but not in the cis- (GM130-positive) cisternae of the Golgi complex. Scale bars 500 nm. D, quantification of gold particles in different cell compartments. Bars indicate the mean ± SD of 5 to 26 images. ∗p < 0.05; ∗∗∗∗p < 0.0001 versus WT by two-way ANOVA Bonferroni’s post-hoc test.
Src family kinases mediate Golgi-based signaling governing secretory trafficking (23), and there is evidence that PrP participates in signaling involving the SFK Fyn (24). We reasoned that this signaling might be impaired in mutant PrP cells and tested whether pharmacological SFK activation rescued the PC-I trafficking defect. WT and FFI HFs were treated with 10 μM SFK activator for 4 h (25), then immunostained with anti-PC-I antibody. The treatment significantly reduced the number of mutant fibroblasts showing intracellular retention of PC-I, but had no effect on PC-I distribution in WT cells (Fig. 9B).
Discussion
We previously found that mutant PrP impairs the synaptic delivery of VGCC and AMPA receptor subunits through physical interaction and retention in the secretory pathway, leading to alterations in glutamatergic neurotransmission and increased sensitivity to excitotoxic cell death (10, 11). The present study indicates that besides impairing the PM delivery of proteins with which PrP interacts, expression of mutant PrP reduces the efficiency of secretory transport, as shown by the analysis of cargo reporters and endogenous PC-I in different cell types. It also indicates that mutant PrP misfolding is required to impair secretory trafficking and pharmacological activation of SFKs ameliorates the transport defect. These results support a pathogenic role of abnormal secretory trafficking in genetic prion diseases and suggest that boosting SFK signaling may be beneficial.
Subcellular localization of PG14, CJD, and FFI PrPs in HeLa cells confirmed previous observations in primary neurons and neuroblastoma N2a cells, indicating mutation-specific differences in intracellular PrP distribution (3, 4, 6, 10, 12). The PG14 and CJD mutants were mainly concentrated in the ER and Golgi, while FFI PrP was mostly in the Golgi, and this held true in FFI HFs, documenting for the first time abnormal intracellular PrP distribution in cells from carriers of a PRNP mutation.
VSV-G distribution at the steady state showed accumulation in the Golgi, pointing to inefficient transport out of this organelle. We previously documented alterations in post-Golgi trafficking in N2a cells expressing FFI PrP by the analysis of the trafficking dynamics of GFP-GPI, a model protein, which rapidly cycles between the Golgi and the PM (12). In HeLa cells expressing the FFI mutation, we found reduced PM expression of GFP-GPI and CD59-GFP, two GPI-anchored proteins used to monitor the efficiency of secretory transport. PM delivery of these reporters was also reduced in CJD and PG14 PrP-expressing HeLa, although less. Thus all three mutants impaired post-Golgi trafficking, but FFI PrP had the most marked effect, perhaps reflecting its preferential accumulation in the Golgi. We have in fact proposed that PrP accumulation in the Golgi activates a feedback loop involving the Rab GDP dissociation inhibitor (GDI)/Rab11 pathway that leads to further protein accumulation in this cell compartment (12). Alterations in post-Golgi vesicular transport were also documented in prion-infected cells (26) and attributed to PrPSc accumulation in recycling endosomes. Thus impaired delivery of membrane proteins to the cell surface may be a common pathogenic mechanism in genetic and acquired prion diseases.
EM analysis of FFI HFs showed swelling of the ER and continuous distensions of the trans-Golgi cisternae, indicative of protein accumulation in both compartments of the secretory pathway. This was confirmed by immuno-EM of PC-I, which showed accumulation in the dilated ER cisternae and in the medial and trans-Golgi stacks. Thus although in FFI neurons the ER appeared morphologically normal, with the Golgi volume selectively increased (3, 12), in professional secretory cells such as fibroblasts, which produce large amounts of extracellular matrix proteins such as collagen, the ER was enlarged too.
How does mutant PrP reduce secretory transport efficiency leading to protein accumulation in the ER and Golgi? VSV-G did not co-immunoprecipitate with PrP (data not shown), ruling out an interaction/retention mechanism like the one described for VGCCs and AMPARs. Analysis of VSV-G transport in PG14/ΔHC, PrP-ER, and PrP-Golgi expressing cells indicated that intracellular retention of nonaggregating PrP is not sufficient to induce the trafficking defect, pointing to a causal role of PrP misfolding and aggregation. Zavodszky and Hegde showed that moPrP C179A, an artificial constitutively misfolded mutant that cannot form a disulfide bond, interacts with ER-resident chaperones, including calnexin, BiP, Grp94, PDI, Erp57, and Erp72, and these interactions remain unaltered as PrP C179A slowly traffics to the Golgi and post-Golgi compartments (27). Although not as disruptive of PrP folding as the artificial C179A mutation, several disease-related PrP mutants, including D178N and octapeptide repeat insertions, boost the protein’s propensity to oligomerize in vitro (28, 29, 30, 31, 32) and misfold and aggregate in cells and transgenic mice (3, 4, 6, 13, 18, 33, 34, 35). Conceivably, like C179A, D178N and PG14 PrP too may sequester key ER components, reducing their availability for folding and transport of other cargos. In this regard, efficient folding and secretory transport of PC-I largely depend on some of the very same chaperones that are sequestered by PrP C179A such as BiP, Grp94, PDI, and calnexin (20).
PrP knockdown in zebrafish embryos causes loss of cell adhesion due to accumulation of E-cadherin in a post-Golgi compartment, and correct E-cadherin localization at the PM requires a PrP-mediated signal transduction mechanism involving activation of SFKs (36). This suggests that zebrafish PrP may serve a signaling function that regulates post-Golgi trafficking (37, 38). While we did not see any alteration in secretory trafficking in PrP KO MEFs—excluding a loss-of-function mechanism, our observation that pharmacological stimulation of SFK rescued PC-I trafficking in FFI HFs suggests that an SFK signaling governing secretory transport may be altered by mutant PrP.
An SFK-mediated signaling circuit has been described that coordinates Golgi to PM transport in mammalian cells (23). This is initiated by the KDEL receptor (KDEL-R), which, with its functional homology with G-protein-coupled receptors (GPCR), binds and activates the signaling heterotrimeric G-protein Gαq/11 (39). PrP is an agonistic ligand of the GPCR Adgrg6 (also known as Gpr126), and this accounts for its ability to promote myelin homeostasis in peripheral nerves (40). It is tempting to speculate that the homology between the KDEL-R and GPCRs implies that mutant PrP may engage in aberrant interactions with the KDEL-R, impairing the signaling mechanisms that regulate transport through the Golgi complex. This may lead to protein accumulation in the Golgi and, in turn, inhibit cargo exit from the ER (23).
In conclusion, our data indicate that mutant PrPs not only impair the trafficking and synaptic delivery of PrP-interacting cargoes, but also have a more general effect on protein transport, probably due to inappropriate activation of SFK-mediated signaling. Carriers of PRNP mutations do not manifest symptoms until adulthood (41), even though PrP is expressed from early embryogenesis and misfolded/aggregated mutant PrP starts accumulating early postnatally in the CNS, as inferred from studies in mice (3, 19, 42, 43, 44). This suggests that disease ensues when a critical toxic threshold is reached (45, 46, 47). Inhibition of secretory trafficking by mutant PrP may contribute to disease by exacerbating intracellular accumulation of PrP and its interacting partners, as well as other structurally challenged proteins with missense mutations, errors, and structural instability, whose cellular clearance may decline with age (48, 49).
Experimental procedures
Mice
The production of transgenic mice expressing wild-type, D177N/V128 and D177N/M128 mouse PrPs has already been reported (3, 4, 19). In this study we used transgenic mice of the Tg(WT-E1+/−), Tg(CJD-A66+/−) and Tg(FFI-26+/−) lines expressing PrP at ∼2X. These mice were originally generated on a C57BL/6J X CBA/J hybrid and were then bred with the Zurich I line of Prnp0/0 mice (50) with a pure C57BL/6J background (European Mouse Mutant Archive, Monterotondo, Rome, Italy; EM:01723). C57BL/6J mice were purchased from Charles River Laboratories.
Procedures involving animals and their care were conducted in conformity with the institutional guidelines at the Istituto di Ricerche Farmacologiche Mario Negri IRCCS in compliance with national (D.lgs 26/2014; Authorization no. 19/2008-A issued March 6, 2008 by Ministry of Health) and international laws and policies (EEC Council Directive 2010/63/UE; the NIH Guide for the Care and Use of Laboratory Animals, 2011 edition). They were reviewed and approved by the Mario Negri Institute Animal Care and Use Committee, which includes ad hoc members for ethical issues, and by the Italian Ministry of Health (Decreto no. 321/2015-PR and 212/2016-PR). Animal facilities meet international standards and are regularly checked by a certified veterinarian who is responsible for health monitoring, animal welfare supervision, experimental protocols, and review of procedures.
Antibodies and reagents
The following antibodies were used: mouse monoclonal anti-GM130 (Transduction Laboratories; 1:500); rabbit polyclonal anti-GM130 (23) (from A. De Matteis, Telethon Institute of Genetics and Medicine; 1:1000); rabbit polyclonal anti-giantin (Covance; 1:500) and rabbit polyclonal anti-PDI (Sigma Aldrich; 1:500); mouse monoclonal anti-VSV-G (clone P5D4, Sigma Aldrich; 1:100); rabbit polyclonal anti-VSV-G luminal domain (23) (from A. De Matteis; 1:100); mouse monoclonal anti-PC-I (Developmental Studies Hybridoma Bank, Iowa University; 1:100); mouse monoclonal anti-PrP antibodies 6H4 (Thermo Fisher Scientific; 1:500) and 12B2 (Central Veterinary Institute; 1:400 for immunofluorescence and 1:5000 for western blot). Secondary antibodies were conjugated with Alexa-488, Alexa-555, or Alexa-633 fluorophores (Invitrogen). The Src family activator (sc-3052) was from Santa Cruz Biotechnology.
Cells
Human HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with GlutaMAX (Invitrogen), 10% FBS, nonessential amino acids (Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/ml streptomycin (GIBCO) and maintained at 37 °C in 5% CO2/95% air.
Primary human fibroblasts from skin biopsies of noncarriers and carriers of the FFI mutation were cultured in Minimum Essential Medium (MEM) Eagle with 10% FBS, 2 mM glutamine, 100 U/ml penicillin/streptomycin (Gibco) and maintained at 37 °C in 5% CO2/95% air.
Primary mouse embryonic fibroblasts were obtained from 13.5- to 14-day-old embryos. Briefly, day E13.5 embryos from timed mating crosses were collected, and the head, heart, and liver were removed and discarded. Tissue was thoroughly minced and incubated with 0.05% trypsin in PBS for 10 min at 37 °C. Then 10 mg/ml DNAse I was added and incubated for other 10 min. Cells were dissociated by pipetting up and down 2 to 3 times and resuspended in DMEM supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin/streptomycin (Gibco). The cell suspension was passed through a 40-μm cell strainer and plated in a 100-mm tissue culture dish. Cells were incubated overnight; the medium was changed after 24 h and incubated for another 1 or 2 days until confluence.
Primary hippocampal neurons were obtained from 2- to 4-day-old pups, as described (51). Animals were euthanized and hippocampi were isolated under a surgical stereomicroscope. Tissues were digested for 30 min at 34 °C with papain (200 U/ml) in CNDM medium (5.8 mM MgCl2, 0.5 mM CaCl2, 3.2 mM HEPES, 0.2 mM NaOH, 30 mM K2SO4 and 90 mM Na2SO4. pH 7.4, 292 mOsm) supplemented with 0.4% glucose. Enzymatic activity was blocked with trypsin inhibitors (10 μg/ml, Sigma) in CNDM plus 0.4% glucose for 45 min at room temperature (RT). Mechanical dissociation was done in MEM (Invitrogen) supplemented with 10% FBS (HyperClone) and 0.4% glucose. 0.5 × 105 cells per cm2 were plated on polylysine-coated coverslips in the same medium with penicillin/streptomycin (PenStrep, Lonza, 100 U/ml). After attachment, culture medium was switched to Neurobasal-A (Invitrogen) supplemented with 2% B27 (Invitrogen), 200 mM glutamine, and 100 U/ml PenStrep. Medium was changed after 7 days in culture.
Transfections
pCDNA3.1(+) plasmids encoding WT (M128), M128V, D177N/M128, D177N/V128, PG14, and PG14/ΔHC PrPs containing the epitope for monoclonal antibody 3F4, and PrP-ER and PrP-Golgi have been described (6, 11, 17). The PrP-EGFP constructs were generated by inserting a monomerized version of EGFP containing a GS linker (GGGGS, repeated four times) at its 3’ end, after codon 34 of 3F4-tagged mouse PrP (12). The VSV-G-GFP, GFP-GPI, and CD59-GFP constructs have been described (12, 16, 23, 52). The cells were transfected with Fugene HD (Promega).
Detergent insolubility
To assay the detergent insolubility of PrP, cells cultured in 35-mm Petri dishes were lysed in 0.7 ml of 50 mM Tris pH 7.5, 150 mM NaCl, 0.5% sodium deoxycholate, and 0.5% Nonidet P-40 containing protease inhibitors (1X SigmaFast Protease Inhibitor Tablets) for 20 min at 4 °C, in agitation. They were then centrifuged at 16,000g for 2 min at 4 °C, and the supernatant was collected and ultracentrifuged at 186,000g for 40 min in a Beckman Optima Max-E ultracentrifuge at 4 °C. The pellet (P) was resuspended in Laemmli sample buffer; proteins in the supernatant (S) were precipitated by adding four volumes of ice-cold CH3OH. After overnight incubation at −20 °C, samples were centrifuged at 16,000g for 30 min at 4 °C. The supernatant was discarded and the precipitated proteins were resuspended in Laemmli sample buffer. PrP in the supernatant (S) and pellet (P) was analyzed by western blotting using antibody 12B2.
VSV-G transport
HeLa cells were cotransfected with plasmids encoding nonfluorescent PrP constructs and VSV-G-GFP (1:1 ratio) and incubated at 35 °C for 24 h before immunofluorescence analysis. MEFs, hippocampal neurons, and HFs were infected with ts-O45 VSV as previously described (53). Briefly, cells were incubated with VSV diluted in medium without serum for 1 h at 32 °C (the permissive temperature). Infection medium containing virus was removed and cells were incubated at 35 °C in complete medium for 4 h (MEF and HFs) or 2 h (hippocampal neurons).
Immunofluorescence
Cells were fixed in 4% paraformaldehyde (Sigma Aldrich) for 10 min at room temperature and incubated in blocking solution (0.05% saponin, 0.5% bovine serum albumin (BSA) and 50 mM NH4Cl in PBS) for 30 min at room temperature (RT). The cells were then incubated with the primary antibodies diluted in blocking solution, for 2 to 3 h at RT, or overnight at 4 °C, then washed three times in PBS, and incubated with a fluorescent conjugated anti-IgG secondary antibody for 1 h at RT.
For surface staining of PrP in HFs, live cells were incubated with anti-PrP primary antibody 6H4 (1:500) in Opti-MEM for 10 min at 37 °C. Cells were fixed in 4% paraformaldehyde and incubated in blocking solution (10% FBS, 5% BSA in PBS) for 1 h at RT. After three washes in PBS, cells were incubated with a biotinylated anti-IgG secondary antibody (1:200) for 1 h at RT, then reacted with Alexa Fluor 488-conjugated streptavidin (1:500) for 30 min at RT. For total PrP staining, HFs were fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS for 10 min, and blocked with 10% FBS, 5% BSA and 0.5% Triton X-100 in PBS for 2 h at RT, then incubated with anti-PrP antibody 6H4 overnight at 4 °C.
Quantitative fluorescence image analysis
Confocal images were acquired using a FV-500 Olympus laser confocal scanning system or a Zeiss LSM510 inverted confocal microscope system (Carl Zeiss). Fixed cells were analyzed using a 63X oil-immersion objective, maintaining the pinhole of the objective at 1 Airy unit. Images were acquired under nonsaturating conditions (pixel fluorescence below 255 arbitrary units) and analyzed with NIH ImageJ software. To determine the amount of VSV-G in the Golgi, the VSV-G fluorescence density was measured for the Golgi area of the cell identified by GM130 immunostaining and for the entire area of that cell excluding the Golgi, and the ratio was calculated (normalized VSV-G in the Golgi). To determine the amount of VSV-G at the PM, the VSV-G fluorescence density was measured on the cell edge and the rest of the cell, and the ratio was calculated (normalized VSV-G at PM). Data are expressed as the fold change relative to control.
Electron microscopy
Ultrastructural analysis
Cells were fixed with a 4% paraformaldehyde and 2.5% glutaraldehyde (EMS) mixture in 0.2 M sodium cacodylate pH 7.2, then incubated in a 1:1 mixture of 2% osmium tetraoxide and 3% potassium ferrocyanide for 1 h at RT, followed by six rinses in 0.2 M cacodylate buffer. Then the samples were sequentially treated with 0.3% thiocarbohydrazide in 0.2 M cacodylate buffer for 10 min and 1% OsO4 in 0.2 M cacodylate buffer (pH 6.9) for 30 min. Samples were rinsed with 0.1 M sodium cacodylate (pH 6.9) buffer to remove all traces of the yellow osmium fixative, washed in deionized water, treated with 1% uranyl acetate in water for 1 h, and washed in water again. The samples were subsequently dehydrated in ethanol then in acetone and embedded in epoxy resin at RT and polymerized for at least 72 h in a 60 °C oven. Embedded samples were then sectioned with a diamond knife (Diatome) using a Leica ultramicrotome. Sections were analyzed with a Tecnai 20 High Voltage EM (FEI, Thermo Fisher Scientific) operating at 200 kV.
Three-dimensional tomography
An ultramicrotome (Leica EM UC7; Leica Microsystems, Vienna) was used to cut 60-nm thick and 200-nm semithick serial sections. Sections were collected onto 1% Formvar films adhered to slot grids. Both sides of the grids were labeled with fiduciary 10 nm gold (PAG10, CMC). Tilt series were collected from the samples from ±65° with 1° increments at 200 kV in Tecnai 20 electron microscopes (FEI, Thermo Fisher Scientific). Tilt series were recorded at a magnification of 11,500×, using software supplied with the instrument. The nominal resolution in our tomograms was 4 nm, based on section thickness, the number of tilts, tilt increments, and tilt angle range. The IMOD 4.0.11 package was used to construct individual tomograms. Videos were made in IMOD and assembled in QuickTime Pro 7.5 (Apple).
Preembedding immuno-electron microscopy
Monolayers of human fibroblasts were cultured on glass-bottom microwell dishes (MatTek Corporation). After fixation with 0.05% glutaraldehyde and 4% paraformaldehyde in Hepes buffer (0.15 M, pH 7.4), the cells were washed three times with 4% paraformaldehyde and incubated in a wet chamber with a mouse primary antibody anti-VSV-G (Sigma Cat. No. V5507) (1:100), overnight at 4 °C. The cells were then washed with PBS and incubated with a nanogold-conjugated goat anti-mouse secondary antibody (Nanoprobes, Cat. No. 2002) (1:25) for 3 h at RT and the labeling was then intensified with a gold enhancement kit (Nanoprobes, Cat. No. 2113).
After postfixation with 1% glutaraldehyde in PBS for 10 min, the cells were incubated in a solution of 1% osmium tetroxide and 1.5% potassium ferrocyanide in cacodylate buffer 0.12 M, followed by 0.3% thiocarbohydrazide, and finally in 1% osmium tetroxide in cacodylate buffer 0.12 M.
After staining in uranyl acetate 1% overnight at 4 °C and dehydration in a graded series of ethanol solutions, the human fibroblasts were embedded in epoxy medium (Epoxy Embedding Medium kit; Sigma-Aldrich). Ultrathin (60 nm thick) sections of cell samples were then cut with a Leica EM UC6 ultramicrotome (Leica Microsystems) and observed with an energy filter transmission electron microscope (Libra120, Carl Zeiss NTS GmbH) equipped with an yttrium aluminum garnet (YAG) scintillator slow-scan charge-coupled device (CCD) camera (Sharp eye, TRS).
Immuno-labeling of cryosections
Cells were fixed, adding a mixture of 0.1 M PHEM buffer, 2% paraformaldehyde, 1% glutaraldehyde for 2 h, and kept in storage solution (0.1 M PHEM buffer and 0.5% paraformaldehyde in distilled water) overnight. After washing with 0.15 M glycine buffer in PBS, cells were embedded in 12% gelatin, cooled on ice and cut into 0.5-mm blocks in a cold room and infused with 2.3 M sucrose. Sections were then cut with an ultramicrotome (Leica EM UC7; Leica Microsystems, Vienna) and transferred onto formvar- and carbon-coated specimen grids. The grids were washed onto 100 μl droplets of PBS for 10 min and then washed again for 3 x 3 min with 0.02 M glycine in PBS, pH 7.4. The grids were incubated for 10 min in 0.5% acetylated BSA (BSA-c 10% in water, Aurion) in PBS to prevent unspecific labeling, then incubated for 2 h on 10 μl droplets of primary antibody (PC-I, from “DSHB” Developmental studies hybridoma bank) diluted 1:50. The grids were washed six times with 0.1% BSA-c in PBS. The primary antibody was monoclonal, and we used a rabbit anti-rat immunoglobulin antibody (bridge antibody, diluted 1:250, from “DAKO”) and then protein-A gold 10 nm (PAG10, CMC) diluted 1:50 in blocking solution for 20 min at RT. Next, sections were double-labeled with a rabbit polyclonal antibody against GM130 (23) diluted 1:100 for 2 h, then with protein-A gold 15 nm (PAG15, CMC) diluted 1:50 in blocking solution for 20 min at RT. The grids were rinsed six times with 0.1% BSA-c in PBS and postfixed with 1% glutaraldehyde in 0.15 M Hepes for 5 min. Finally, the grids were washed 5 x 1 min in distilled water and stained 10 min in 1.8% methyl cellulose plus 0.4% uranyl acetate, on ice. The grids were retrieved with a stainless steel loop onto a piece of Whatman 50 filter paper at an angle of 45°. After air-drying, the grids were examined under a Tecnai20 electron microscope (FEI, Thermo Fisher Scientific).
All morphometric analyses were performed using iTEM (TEM Imaging Platform, Olympus Soft Images Solution). Images were randomly selected and acquired at the same magnification. Then the text grid was placed over images and the number of gold particles (Ng) and test points (Np) over a structure was counted. The labeling density (LD or the number of gold/area; N/S) was estimated using the following formula: N/S = Ng/kNp, where the k coefficient depends on the test system (54).
Data availability
All relevant data are within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by grants from the Italian Ministry of Health (RF-2010-2314035), Telethon Italy (TCR08005; GGP12115; GGP10208), and “Fondazione Cariplo” (2012-0560). We thank Antonella De Matteis (Telethon Institute of Genetics and Medicine) for providing anti-VSV-G and anti-GM130 antibodies for immuno-EM and Roman Polishchuk (Telethon Institute of Genetics and Medicine) for the CD59-GFP plasmid. We are grateful to members of the Italian Fatal Familial Insomnia Family Association (AFIFF; https://www.afiff.net/) for skin biopsies.
Author contributions
E. R., V. C., M. P., D. O., E. Q., A. C., F. F., and G. V. B.: investigation; E. R., R. C.: writing—original draft; F. F., M. S. and R. C.: writing-review and editing; M. S. and R. C.: supervision; V. A., and I. R.: resources; M. S., and R. C.: conceptualization; M. S., I. R., and R. C.: funding acquisition.
Edited by Phyllis Hanson
Footnotes
Present address for Davide Ortolan: Unit on Ocular and Stem Cell Translational Research, National Eye Institute, NIH, Bethesda, Maryland, USA.
Supporting information
References
- 1.Kim M.-O., Takada L.T., Wong K., Forner S.A., Geschwind M.D. Genetic PrP prion diseases. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a033134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiesa R. The elusive role of the prion protein and the mechanism of toxicity in prion disease. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouybayoune I., Mantovani S., Del Gallo F., Bertani I., Restelli E., Comerio L., Tapella L., Baracchi F., Fernandez-Borges N., Mangieri M., Bisighini C., Beznoussenko G.V., Paladini A., Balducci C., Micotti E. Transgenic fatal familial insomnia mice indicate prion infectivity-independent mechanisms of pathogenesis and phenotypic expression of disease. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dossena S., Imeri L., Mangieri M., Garofoli A., Ferrari L., Senatore A., Restelli E., Balducci C., Fiordaliso F., Salio M., Bianchi S., Fioriti L., Morbin M., Pincherle A., Marcon G. Mutant prion protein expression causes motor and memory deficits and abnormal sleep patterns in a transgenic mouse model. Neuron. 2008;60:598–609. doi: 10.1016/j.neuron.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 5.Drisaldi B., Stewart R.S., Adles C., Stewart L.R., Quaglio E., Biasini E., Fioriti L., Chiesa R., Harris D.A. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J. Biol. Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- 6.Fioriti L., Dossena S., Stewart L.R., Stewart R.S., Harris D.A., Forloni G., Chiesa R. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J. Biol. Chem. 2005;280:11320–11328. doi: 10.1074/jbc.M412441200. [DOI] [PubMed] [Google Scholar]
- 7.Ivanova L., Barmada S., Kummer T., Harris D.A. Mutant prion proteins are partially retained in the endoplasmic reticulum. J. Biol. Chem. 2001;276:42409–42421. doi: 10.1074/jbc.M106928200. [DOI] [PubMed] [Google Scholar]
- 8.Quaglio E., Restelli E., Garofoli A., Dossena S., De Luigi A., Tagliavacca L., Imperiale D., Migheli A., Salmona M., Sitia R., Forloni G., Chiesa R. Expression of mutant or cytosolic PrP in transgenic mice and cells is not associated with endoplasmic reticulum stress or proteasome dysfunction. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Senatore A., Restelli E., Chiesa R. Synaptic dysfunction in prion diseases: A trafficking problem? Int. J. Cell Biol. 2013;2013:543803. doi: 10.1155/2013/543803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghirardini E., Restelli E., Morini R., Bertani I., Ortolan D., Perrucci F., Pozzi D., Matteoli M., Chiesa R. Mutant prion proteins increase calcium permeability of AMPA receptors, exacerbating excitotoxicity. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Senatore A., Colleoni S., Verderio C., Restelli E., Morini R., Condliffe S.B., Bertani I., Mantovani S., Canovi M., Micotti E., Forloni G., Dolphin A.C., Matteoli M., Gobbi M., Chiesa R. Mutant PrP suppresses glutamatergic neurotransmission in cerebellar granule neurons by impairing membrane delivery of VGCC alpha(2)delta-1 subunit. Neuron. 2012;74:300–313. doi: 10.1016/j.neuron.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massignan T., Biasini E., Lauranzano E., Veglianese P., Pignataro M., Fioriti L., Harris D.A., Salmona M., Chiesa R., Bonetto V. Mutant prion protein expression is associated with an alteration of the Rab GDP dissociation inhibitor alpha (GDI)/Rab11 pathway. Mol. Cell. Proteomics. 2010;9:611–622. doi: 10.1074/mcp.M900271-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashok A., Hegde R.S. Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dotti C.G., Simons K. Polarized sorting of viral glycoproteins to the axon and dendrites of hippocampal neurons in culture. Cell. 1990;62:63–72. doi: 10.1016/0092-8674(90)90240-f. [DOI] [PubMed] [Google Scholar]
- 15.Singh N., Zanusso G., Chen S.G., Fujioka H., Richardson S., Gambetti P., Petersen R.B. Prion protein aggregation reverted by low temperature in transfected cells carrying a prion protein gene mutation. J. Biol. Chem. 1997;272:28461–28470. doi: 10.1074/jbc.272.45.28461. [DOI] [PubMed] [Google Scholar]
- 16.Nichols B.J., Kenworthy A.K., Polishchuk R.S., Lodge R., Roberts T.H., Hirschberg K., Phair R.D., Lippincott-Schwartz J. Rapid cycling of lipid raft markers between the cell surface and Golgi complex. J. Cell Biol. 2001;153:529–541. doi: 10.1083/jcb.153.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biasini E., Tapella L., Restelli E., Pozzoli M., Massignan T., Chiesa R. The hydrophobic core region governs mutant prion protein aggregation and intracellular retention. Biochem. J. 2010;430:477–486. doi: 10.1042/BJ20100615. [DOI] [PubMed] [Google Scholar]
- 18.Lehmann S., Harris D.A. Mutant and infectious prion proteins display common biochemical properties in cultured cells. J. Biol. Chem. 1996;271:1633–1637. doi: 10.1074/jbc.271.3.1633. [DOI] [PubMed] [Google Scholar]
- 19.Chiesa R., Piccardo P., Ghetti B., Harris D.A. Neurological illness in transgenic mice expressing a prion protein with an insertional mutation. Neuron. 1998;21:1339–1351. doi: 10.1016/s0896-6273(00)80653-4. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa Y., Bächinger H.P. A molecular ensemble in the rER for procollagen maturation. Biochim. Biophys. Acta. 2013;1833:2479–2491. doi: 10.1016/j.bbamcr.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura N., Rabouille C., Watson R., Nilsson T., Hui N., Slusarewicz P., Kreis T.E., Warren G. Characterization of a cis-Golgi matrix protein, GM130. J. Cell Biol. 1995;131:1715–1726. doi: 10.1083/jcb.131.6.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Capone V., Clemente E., Restelli E., Di Clampi A., Sperduti S., Ornaghi F., Pietrangelo L., Protasi F., Chiesa R., Sallese M. PERK inhibition attenuates the abnormalities of the secretory pathway and the increased apoptotic rate induced by SIL1 knockdown in HeLa cells. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:3164–3180. doi: 10.1016/j.bbadis.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 23.Pulvirenti T., Giannotta M., Capestrano M., Capitani M., Pisanu A., Polishchuk R.S., San Pietro E., Beznoussenko G.V., Mironov A.A., Turacchio G., Hsu V.W., Sallese M., Luini A. A traffic-activated Golgi-based signalling circuit coordinates the secretory pathway. Nat. Cell Biol. 2008;10:912–922. doi: 10.1038/ncb1751. [DOI] [PubMed] [Google Scholar]
- 24.Mouillet-Richard S., Ermonval M., Chebassier C., Laplanche J.L., Lehmann S., Launay J.M., Kellermann O. Signal transduction through prion protein. Science. 2000;289:1925–1928. doi: 10.1126/science.289.5486.1925. [DOI] [PubMed] [Google Scholar]
- 25.Park S.-Y., Yang J.-S., Schmider A.B., Soberman R.J., Hsu V.W. Coordinated regulation of bidirectional COPI transport at the Golgi by CDC42. Nature. 2015;521:529–532. doi: 10.1038/nature14457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uchiyama K., Muramatsu N., Yano M., Usui T., Miyata H., Sakaguchi S. Prions disturb post-Golgi trafficking of membrane proteins. Nat. Commun. 2013;4:1846. doi: 10.1038/ncomms2873. [DOI] [PubMed] [Google Scholar]
- 27.Zavodszky E., Hegde R.S. Misfolded GPI-anchored proteins are escorted through the secretory pathway by ER-derived factors. Elife. 2019;8 doi: 10.7554/eLife.46740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu S., Yin S., Li C., Wong P., Chang B., Xiao F., Kang S.-C., Yan H., Xiao G., Tien P., Sy M.-S. Aggregation of prion protein with insertion mutations is proportional to the number of inserts. Biochem. J. 2007;403:343–351. doi: 10.1042/BJ20061592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Apetri A.C., Vanik D.L., Surewicz W.K. Polymorphism at residue 129 modulates the conformational conversion of the D178N variant of human prion protein 90-231. Biochemistry. 2005;44:15880–15888. doi: 10.1021/bi051455+. [DOI] [PubMed] [Google Scholar]
- 30.Moore R.A. Octapeptide repeat insertions increase the rate of protease-resistant prion protein formation. Protein Sci. 2006;15:609–619. doi: 10.1110/ps.051822606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S., Antony L., Hartmann R., Knaus K.J., Surewicz K., Surewicz W.K., Yee V.C. Conformational diversity in prion protein variants influences intermolecular beta-sheet formation. EMBO J. 2010;29:251–262. doi: 10.1038/emboj.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riek R., Wider G., Billeter M., Hornemann S., Glockshuber R., Wuthrich K. Prion protein NMR structure and familial human spongiform encephalopathies. Proc. Natl. Acad. Sci. U. S. A. 1998;95:11667–11672. doi: 10.1073/pnas.95.20.11667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tapella L., Stravalaci M., Bastone A., Biasini E., Gobbi M., Chiesa R. Epitope scanning indicates structural differences in brain-derived monomeric and aggregated mutant prion proteins related to genetic prion diseases. Biochem. J. 2013;454:417–425. doi: 10.1042/BJ20130563. [DOI] [PubMed] [Google Scholar]
- 34.Priola S.A., Chesebro B. Abnormal properties of prion protein with insertional mutations in different cell types. J. Biol. Chem. 1998;273:11980–11985. doi: 10.1074/jbc.273.19.11980. [DOI] [PubMed] [Google Scholar]
- 35.Daude N., Lehmann S., Harris D.A. Identification of intermediate steps in the conversion of a mutant prion protein to a scrapie-like form in cultured cells. J. Biol. Chem. 1997;272:11604–11612. doi: 10.1074/jbc.272.17.11604. [DOI] [PubMed] [Google Scholar]
- 36.Malaga-Trillo E., Solis G.P., Schrock Y., Geiss C., Luncz L., Thomanetz V., Stuermer C.A. Regulation of embryonic cell adhesion by the prion protein. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiesa R., Harris D.A. Fishing for prion protein function. PLoS Biol. 2009;7 doi: 10.1371/journal.pbio.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Málaga-Trillo E., Ochs K. Uncontrolled SFK-mediated protein trafficking in prion and Alzheimer’s disease. Prion. 2016;10:352–361. doi: 10.1080/19336896.2016.1221873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sallese M., Giannotta M., Luini A. Coordination of the secretory compartments via inter-organelle signalling. Semin. Cell Dev. Biol. 2009;20:801–809. doi: 10.1016/j.semcdb.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 40.Küffer A., Lakkaraju A.K.K., Mogha A., Petersen S.C., Airich K., Doucerain C., Marpakwar R., Bakirci P., Senatore A., Monnard A., Schiavi C., Nuvolone M., Grosshans B., Hornemann S., Bassilana F. The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature. 2016;536:464–468. doi: 10.1038/nature19312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minikel E.V., Vallabh S.M., Orseth M.C., Brandel J.-P., Haik S., Laplanche J.-L., Zerr I., Parchi P., Capellari S., Safar J., Kenny J., Fong J.C., Takada L.T., Ponto C., Hermann P. Age of onset in genetic prion disease and the design of preventive clinical trials. Neurology. 2018;93:e125–e134. doi: 10.1212/WNL.0000000000007745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiesa R., Drisaldi B., Quaglio E., Migheli A., Piccardo P., Ghetti B., Harris D.A. Accumulation of protease-resistant prion protein (PrP) and apoptosis of cerebellar granule cells in transgenic mice expressing a PrP insertional mutation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5574–5579. doi: 10.1073/pnas.97.10.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manson J., West J.D., Thomson V., McBride P., Kaufman M.H., Hope J. The prion protein gene: A role in mouse embryogenesis? Development. 1992;115:117–122. doi: 10.1242/dev.115.1.117. [DOI] [PubMed] [Google Scholar]
- 44.McKinley M.P., Hay B., Lingappa V.R., Lieberburg I., Prusiner S.B. Developmental expression of prion protein gene in brain. Dev. Biol. 1987;121:105–110. doi: 10.1016/0012-1606(87)90143-6. [DOI] [PubMed] [Google Scholar]
- 45.Chiesa R., Harris D.A. Prion diseases: What is the neurotoxic molecule? Neurobiol. Dis. 2001;8:743–763. doi: 10.1006/nbdi.2001.0433. [DOI] [PubMed] [Google Scholar]
- 46.Chiesa R., Piccardo P., Quaglio E., Drisaldi B., Si-Hoe S.L., Takao M., Ghetti B., Harris D.A. Molecular distinction between pathogenic and infectious properties of the prion protein. J. Virol. 2003;77:7611–7622. doi: 10.1128/JVI.77.13.7611-7622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biasini E., Medrano A.Z., Thellung S., Chiesa R., Harris D.A. Multiple biochemical similarities between infectious and non-infectious aggregates of a prion protein carrying an octapeptide insertion. J. Neurochem. 2008;104:1293–1308. doi: 10.1111/j.1471-4159.2007.05082.x. [DOI] [PubMed] [Google Scholar]
- 48.Hipp M.S., Kasturi P., Hartl F.U. The proteostasis network and its decline in ageing. Nat. Rev. Mol. Cell Biol. 2019;20:421–435. doi: 10.1038/s41580-019-0101-y. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Vicente M., Sovak G., Cuervo A.M. Protein degradation and aging. Exp. Gerontol. 2005;40:622–633. doi: 10.1016/j.exger.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 50.Bueler H., Fischer M., Lang Y., Bluethmann H., Lipp H.P., DeArmond S.J., Prusiner S.B., Aguet M., Weissmann C. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature. 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 51.Restelli E., Fioriti L., Mantovani S., Airaghi S., Forloni G., Chiesa R. Cell type-specific neuroprotective activity of untranslocated prion protein. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polishchuk R., Pentima A.D., Lippincott-Schwartz J. Delivery of raft-associated, GPI-anchored proteins to the apical surface of polarized MDCK cells by a transcytotic pathway. Nat. Cell Biol. 2004;6:297–307. doi: 10.1038/ncb1109. [DOI] [PubMed] [Google Scholar]
- 53.Mironov A.A., Beznoussenko G.V., Nicoziani P., Martella O., Trucco A., Kweon H.S., Di Giandomenico D., Polishchuk R.S., Fusella A., Lupetti P., Berger E.G., Geerts W.J., Koster A.J., Burger K.N., Luini A. Small cargo proteins and large aggregates can traverse the Golgi by a common mechanism without leaving the lumen of cisternae. J. Cell Biol. 2001;155:1225–1238. doi: 10.1083/jcb.200108073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lucocq J. Unbiased 3-D quantitation of ultrastructure in cell biology. Trends Cell Biol. 1993;3:354–358. doi: 10.1016/0962-8924(93)90106-b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the article.