Abstract
Persistent activation of the latent transcription factor STAT3 is observed in gastric tumor epithelial and immune cells and is associated with a poor patient prognosis. Although targeting STAT3-activating upstream kinases offers therapeutically viable targets with limited specificity, direct inhibition of STAT3 remains challenging. Here we provide functional evidence that myeloid-specific hematopoietic cell kinase (HCK) activity can drive STAT3-dependent epithelial tumor growth in mice and is associated with alternative macrophage activation alongside matrix remodeling and tumor cell invasion. Accordingly, genetic reduction of HCK expression in bone marrow–derived cells or systemic pharmacologic inhibition of HCK activity suppresses alternative macrophage polarization and epithelial STAT3 activation, and impairs tumor growth. These data validate HCK as a molecular target for the treatment of human solid tumors harboring excessive STAT3 activity.
Introduction
The latent transcription factor STAT3 is activated in a majority of gastric cancers, and positively correlates with disease progression, metastasis, and reduced overall patient survival (1). Molecular reconciliation of these observations is afforded by excessive STAT3 activity in tumor cells promoting transcription of genes that mediate cancer hallmark characteristics, including cell survival, proliferation, angiogenesis, invasion, and metastasis (2). STAT3 is also excessively activated in tumor-associated myeloid cells and inhibits antitumor immunity through immunosuppressive IL10, which further sustains STAT3 activation in a feed-forward loop (2).
Hematopoietic cell kinase (HCK) is one of nine SRC-family tyrosine kinases and is primarily expressed in myeloid cells (3). HCK gene amplifications and copy number gains occur in >50% of human gastric cancers and are associated with poor prognosis (4, 5), but the functional significance of this observation remains unknown. In colon cancer, we have defined a tumor cell–extrinsic role for HCK-dependent signaling in macrophages that promotes their polarization toward a wound-healing/immunosuppressive alternatively activated endotype with alternative activation and enhances expression of the STAT3-activating cytokines IL6 and IL11 in tumor cells and tumor-associated macrophages (TAM). HCK-dependent signaling coincided with increased STAT3 activity in the tumor epithelium and the formation of larger tumors (6). Conversely, genetic ablation or pharmacologic inhibition of HCK activity suppressed colon tumor growth (6).
Here, we genetically delineate a causal relationship between HCK signaling in TAMs and STAT3-dependent gastric tumor growth in mice. Combining genetic and pharmacologic approaches, we establish the therapeutic benefit of targeting HCK activity in myeloid cells as an effective means of interfering with STAT3-dependent tumor formation and progression.
Materials and Methods
Mice and treatments
Age- and sex-matched wild-type (WT), homozygous HckCA/CA (7), Rag1KO (8), Gp130F/F (9), HckKO (10), and heterozygous Gp130F/+ mice were maintained in specific pathogen–free facilities at the Ludwig Institute for Cancer Research, the Walter and Eliza Hall Institute of Medical Research, La Trobe University, and the Austin Hospital, Australia.
The HCK inhibitor RK20449 (11) was dissolved in 12% Captisol and administered twice daily (i.p., 30 mg/kg) for 3 weeks (6). For adoptive transfer experiments, 8-week-old lethally irradiated recipient mice received 5 × 106 donor bone marrow cells via tail-vein injection (6). Tumor burden on whole-mounted excised stomachs was assessed as described previously (12). Tumors and the adjacent “unaffected” normal antrum were processed separately for molecular and histologic analysis.
Immune cell isolation and flow cytometry
Tumors were cut into 1-mm pieces and digested in Collagenase/Dispase (Roche) and DNase I (Roche) in Ca2+- and Mg2+-free Hank medium plus 10% FCS for 30 minutes at 37°C under continuous rotation. Subsequently, samples were vortexed for 30 seconds to dissociate immune cells, and the resulting cell suspension was filtered and washed in PBS plus 10% FCS. Following incubation with Fc block (Thermo Fisher Scientific) on ice for 10 minutes, cells were incubated with fluorophore-conjugated primary antibodies (Supplementary Table S1) for 20 minutes on ice in the dark, washed twice, and resuspended in PBS supplemented with 10% FCS. Purification and quantification of TAMs, identified as F4/80+ subpopulation of CD45+CD11b+Ly6G−Ly6C−SiglecF− cells, was performed and analyzed on the BD Fortessax20 and Aria cell sorter as described previously (6). Dead cells were identified by propidium iodide staining and excluded from analysis. Analysis of all experiments was performed using compensated data with FlowJo software (Version 10).
RNA extraction and qRT-PCR analysis
RNA extraction from whole tissues and FACS-purified macrophages was performed using RNeasy Mini/Micro kits (Qiagen) according to the manufacturer's instructions. RNA (2 μg) was used to generate cDNA with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's instructions. Quantitative RT-PCR analysis was performed on duplicate samples with TaqMan Real-Time PCR Master mix (Life Technologies) using the Viia7 Real-Time PCR System (Life Technologies) for 40 cycles (95°C for 15 seconds, 60°C/1 minute) and following an initial holding stage (50°C/2 minutes, 95°C/10 minutes). 18S or Gapdh were used as housekeepers and fold changes in gene expression were obtained using the 2−ΔΔCt method (13).
TaqMan probes used were mouse 18s (Mm04277571_s1), Gapdh (Mm99999915_g1), Il6 (Mm00446190_m1), Il11 (Mm00434162_m1), Socs3 (Mm00545913_s1), Mmp2 (Mm00439498_m1), Mmp9 (Mm00442991_m1), Il10 (Mm01288386_m1), Arg1 (Mm00475988_m1), and Vegfα (Mm00437306_m1) from Thermo Fisher Scientific.
Western blotting
Protein lysates were prepared and resolved on 10% SDS-polyacrylamide gels (6). Following dry transfer, polyvinylidene difluoride membranes were blocked for 1 hour at room temperature in Odyssey Blocking Buffer (LI-COR Biosciences) and incubated overnight at 4°C with antibodies against pY-STAT3/STAT3 or actin (Supplementary Table S1). Blots were then incubated with fluorescent-conjugated secondary antibodies for 1 hour at room temperature and visualized using the Odyssey Infrared Imaging System (LI-COR Biosciences).
IHC and Masson Trichrome quantification
Paraffin-embedded sections were boiled in citrate buffer (pH 6 for 15 minutes) and incubated with 3% H2O2 for 20 minutes to block endogenous peroxidases. Sections were blocked in 10% normal goat serum for 1 hour at room temperature before incubation in primary antibody against F4/80 at 4°C in a humidified chamber overnight (Supplementary Table S1). Biotinylated secondary antibody from the Avidin Biotin Complex ABC Kit (Vector Laboratories) was used according to the manufacturer's instructions. Antigen visualization was achieved using 3,3-Diaminobenzine (DAB, DAKO). Images were collected and analyzed with Aperio ImageScope v11.2.0.780 software. Quantification of Masson Trichrome staining at the tumor stroma boundary (% blue staining) was performed using the color threshold function in ImageJ.
Matrigel Boyden chamber assay
Bone marrow–derived macrophages (BMDM) were seeded at 1 × 105 cells/insert in a Matrigel invasion chamber (BD Biosciences) supplemented with a CSF-1 gradient for 16 hours before fixation in 4% paraformaldehyde. Some chambers were supplemented with 30 nmol/L RK20449. Invasion of BMDM was determined by enumerating cells at the distal side of membranes in 10 representative fields at 20× magnification, and these numbers were normalized to the total number of cells inoculated in the control wells. Assays were performed in triplicate in at least three independent experiments.
Gelatin degradation assay
BMDMs (2 × 105) were seeded in 35-mm glass-bottom dishes coated with Cy3-labeled gelatin (QCM Gelatin Invadopodia Assay; Millipore) in medium supplemented with 120 ng/mL CSF1 (kind gift from Richard Stanley, Albert Einstein College of Medicine, Bronx, NY) and 20 ng/mL IL4 (Miltenyi Biotec) and, where indicated, treated with either DMSO, 2 μmol/L SU6656 (Sigma), 30 nmol/L RK20449 (11), or 5 μmol/L GM6001 (Sigma). Cells were incubated in 37°C, 5% CO2 for 24 hours, then fixed, stained for F-actin with Alexa 488-phalloidin, and mounted in Prolong Gold (Thermo Fisher Scientific) with DAPI. Samples were imaged with five representative fields at 10× magnification and degraded area (corrected for cell area), as indicated by the absence of Cy3 signal, was quantified relative to total area by thresholding using ImageJ.
3D coculture invasion assay
The BMDM coculture invasion assay was conducted with tumor cell spheroids generated from the Mycoplasma-free Py8119 cell line (ATCC CRL-3278; received in 2015 and authenticated by the ATCC) as described previously (14). For each experiment, freshly thawed cells were maintained in culture for approximately 20 days. Individual spheroids were cocultured with 1 × 105 BMDMs for 3 days before embedding into Matrigel, and phase-contrast imaging was used to determine the starting area of the spheroids (day 0). After 7 days, the degree of invasion was measured in ImageJ by subtracting the area of the tumor spheroid at day 0 from the area at day 7 (ΔD7–D0), defined by the extent of the “halo” of invading cells that had dispersed away from the original spheroid into the matrix. At least 3 tumor spheroids for each condition were assessed in three independent experiments.
Study approval
All animal studies were approved and conducted in accordance with the Animal Ethics Committees of the Ludwig Institute for Cancer Research, the Walter and Eliza Hall Institute of Medical Research, and Austin Life Science.
Statistical analysis
All experiments were performed at least twice with a minimum of 3 age- and sex-matched mice per group. The specific number of animals used per cohort is indicated in the respective figure legends.
Unless otherwise stated, comparisons between mean values were performed with a two-tailed Student t test using Prism 7 software (GraphPad). A P value of <0.05 was considered statistically significant.
Results
Constitutive HCK activity in BMDMs promotes gastric cancer through STAT3 signaling
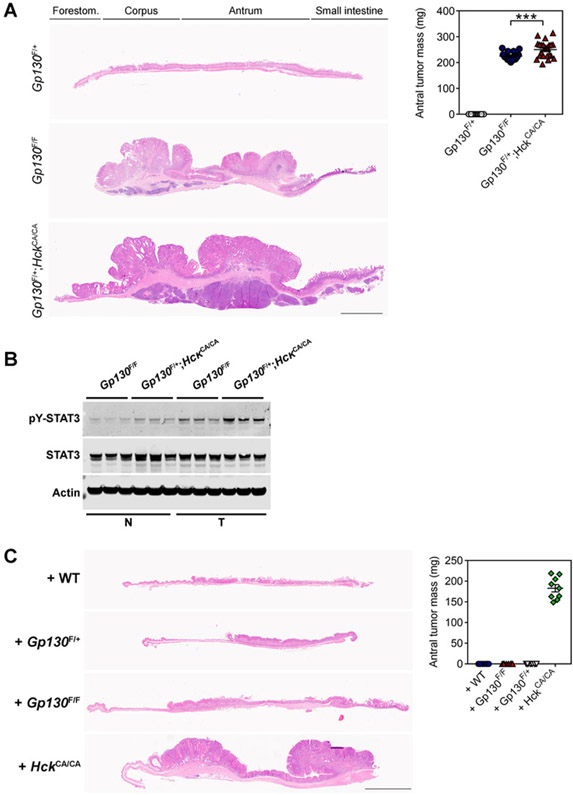
To investigate a functional link between HCK and STAT3 signaling in gastric cancer, we utilized the Gp130F/F mouse model, where disruption of the Socs3-dependent negative feedback loop results in excessive STAT3 signaling upon engagement of the shared Gp130 receptor by IL6 family cytokines (15). All homozygous Gp130F/F mice spontaneously develop intestinal-type gastric adenomatous tumors that are dependent on continuous excessive STAT3 activity in the gastric epithelium (9, 16, 17) but not in immune cells, as Gp130F/F mice reconstituted with WT or lymphocyte-deficient Rag1KO bone marrow still develop tumors (Supplementary Fig. S1A). In contrast, Gp130F/+ mice remain tumor-free as a result of insufficient STAT3 activity (ref. 9; Fig. 1A). All Gp130F/+;HckCA/CA mice harboring a homozygous mutation encoding a constitutively active isoform of HCK (ref. 6; HckCA/CA) developed gastric tumors. Tumors in 1-year-old Gp130F/+; HckCA/CA mice were larger than in age-matched Gp130F/F littermates, and coincided with increased abundance of phosphorylated and transcriptionally activated pY-STAT3 (Fig. 1A and B). Compared with tumors from Gp130F/F littermates, we also observed increased expression of genes associated with STAT3 activation (i.e., the cytokines Il6, Il11) and STAT3 activity (i.e., the target gene Socs3) in tumors and in FACS-purified TAMs of Gp130F/+;HckCA/CA mice (Supplementary Fig. S1B and S1C).
Figure 1.
Constitutive HCK activity in bone marrow–derived cells enhances STAT3-dependent tumor growth. A, Representative hematoxylin and eosin (H&E)-stained stomach cross-sections collected from 1-year-old Gp130F/+, Gp130F/F, and Gp130F/+;HckCA/CA mice. Total antral tumor mass is shown, and each symbol represents an individual mouse. Forestom., forestomach. Scale bar, 3 mm. B, Western blot analysis for phosphorylated and total STAT3 protein isoforms on normal stomach (N) and tumor cell (T) lysates of 1-year-old Gp130F/F and Gp130F/+;HckCA/CA mice. Actin was used as a loading control. Each lane represents an individual mouse. C, Representative H&E-stained stomach cross-sections collected from 1-year-old Gp730F/+ recipient mice that were reconstituted at 8 weeks of age with bone marrow of the indicated genotypes. Tumor burden was quantified as in A in individual mice. Scale bar, 3 mm. Data represent mean ± SEM, P values from unpaired Student t test (***, P < 0.001). All experiments were performed at least twice. Please also refer to Supplementary Fig. S1.
To confirm that tumor development in Gp130F/+;HckCA/CA mice was a consequence of excessive tumor-extrinsic (i.e., hematopoietic cell–derived) HCK activity, we adoptively transferred bone marrow of HckCA/CA mice into tumor-free Gp130F/+ animals. At 1 year of age, these recipients had developed gastric tumors, whereas Gp130F/+ recipients reconstituted with either WT, Gp130F/+, or Gp130F/F bone marrow remained tumor-free (Fig. 1C). Tumors of Gp13F/+ recipients reconstituted with HckCA/CA bone marrow showed elevated pY-STAT3 (Supplementary Fig. S1D), which correlated with upregulation of Il6, Il11, and Socs3 gene expression when compared with the unaffected antrum of Gp130F/+ recipients reconstituted with WT bone marrow (Supplementary Fig. S1E). These results suggest that increased HCK activity in hematopoietic cells drives tumor initiation and development, most likely by increasing STAT3 activity in the gastric epithelium and emerging tumors.
Constitutive HCK activation in myeloid cells promotes gastric tumor invasion
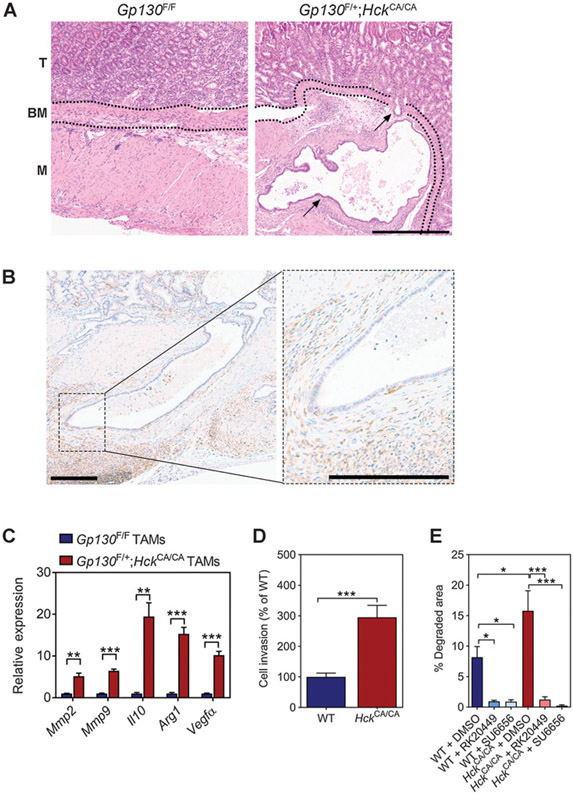
Given that tumors in Gp130F/+ mice with excessive HCK activity were larger than those in their Gp130F/F littermates, we hypothesized that these tumors had acquired other characteristics of neoplastic transformation. Indeed, all tumors of 22 Gp130F/+;HckCA/CA mice showed epithelial cell invasion through the basement membrane, which was not observed in tumors of the 15 age-matched Gp130F/F littermates (Fig. 2A). Tumors of Gp130F/+;HckCA/CA mice also showed increased collagen deposition at the tumor/stroma boundary compared with Gp130F/F animals (Supplementary Fig. S2A). Epithelial cells at the invasive front of Gp130F/+;HckCA/CA tumors were surrounded by F4/80+ TAMs (Fig. 2B). Although flow cytometry analysis revealed a comparable abundance of TAMs in tumors of Gp130F/F and Gp130F/+;HckCA/CA mice, we observed a higher percentage of CD206+ alternatively activated macrophages in Gp130F/+;HckCA/CA tumors (Supplementary Fig. S2B). TAMs of Gp130F/+;HckCA/CA mice also displayed increased expression of STAT3-responsive genes associated with matrix degradation (Mmp2, Mmp9) and alternative macrophage polarization (Il10, Arg1, Vegfα; Fig. 2C).
Figure 2.
Excessive HCK activation promotes tumor cell invasion in Gp130F/+;HckCA/CA-mutant mice. A, Representative hematoxylin and eosin (H&E)–stained stomach cross-sections collected from 1-year-old Gp130F/F and Gp130F/+;HckCA/CA mice. BM, basement membrane; M, muscularis; T, tumor. Arrows indicate tumor cells that have invaded through the basement membrane into the muscularis. Scale bar, 300 μm. B, Representative immunostaining for F4/80+ macrophages surrounding the invasive front of a Gp130F/+;HckCA/CA tumor. Scale bar, 200 μm. C, qRT-PCR analysis on TAMs isolated from Gp130F/F and Gp130F/+;HckCA/CA mice for genes associated with matrix remodeling and alternative macrophage polarization. n = 4 mice per cohort, P values from unpaired Student t test (**, P < 0.01; ***, P < 0.001). D, Invasion of WT and HckCA/CA BMDMs through a modified Matrigel Boyden chamber assay and cultured in the presence of CSF1. Data were normalized to WT, and P values were calculated from one-way ANOVA with Bonferroni post hoc analysis (***, P < 0.001). E, Invasion of WT and HckCA/CA BMDMs through a Cy3-conjugated gelatin matrix. Where indicated, cells were treated with 2 μmol/L DMSO, 30 nmol/L RK20449, or 2 μmol/L SU6656. P values from one-way ANOVA with Bonferroni post hoc analysis (*, P < 0.05; ***, P < 0.001). All data represent mean ± SEM. All experiments were performed at least twice. Please also refer to Supplementary Fig. S2.
To show that excessive HCK activity enhances the capacity of macrophages to invade and degrade extracellular matrix, we assessed the activity of WT and HckCA/CA BMDMs in a Matrigel-coated Boyden chamber assay and found enhanced invasive capacity of HckCA/CA macrophages compared with their WT counterparts (Fig. 2D). Treatment with the HCK-specific small-molecule inhibitor RK20449 was as effective as treatment with the pan-SFK inhibitor SU6656 at reducing the capacity of macrophages of either genotype to degrade matrix (Fig. 2E). To assess the contribution of MMPs in matrix degradation, we treated WT and HckCA/CA BMDMs with the pan-MMP inhibitor GM6001. We observed that matrix degradation by HckCA/CA BMDMs, but not by WT BMDMs, still occurred in the presence of GM6001 (Supplementary Fig. S2C).
We then used a three-dimensional spheroid coculture system to assess whether HckCA/CA macrophages had an increased ability to support tumor cell invasion into the surrounding matrix. As shown previously, tumor cells comprising the spheroid fail to invade matrix in the absence of BMDMs (14). However, tumor cell invasion readily occurred when spheroids contained WT BMDMs and further increased in cocultures with HckCA/CA BMDMs (Supplementary Fig. S2D). Thus, excessive HCK activation in macrophages enhances their ability to remodel and degrade matrix and to support tumor cell invasion.
Inhibition of HCK attenuates STAT3 signaling and gastric tumor growth
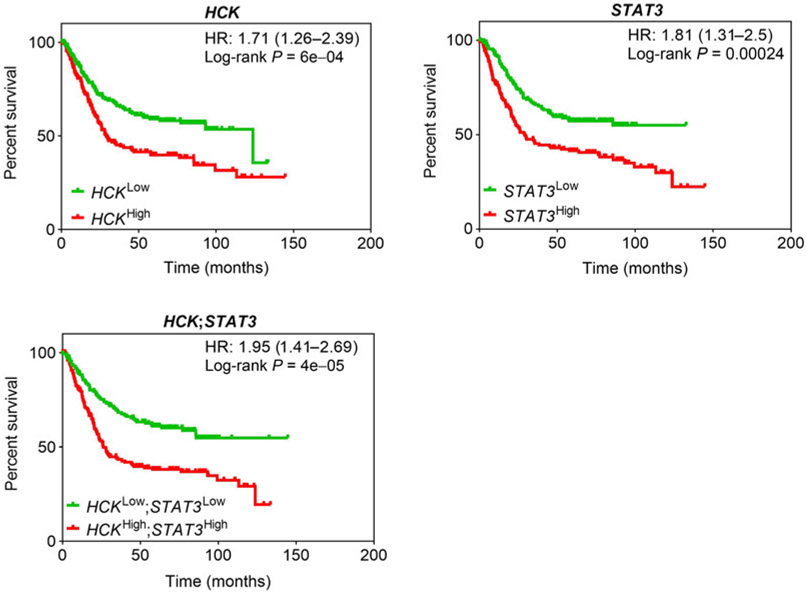
To correlate our preclinical observations with consequences of aberrant HCK and STAT3 activation in gastric cancer in humans, we assessed the survival probability of patients with intestinal-type gastric cancer whose tumors highly expressed HCK and STAT3, either independently of each other or simultaneously. We exploited the Kaplan–Meier plot database (18) and separated 320 patients according to median gene expression. We observed a poorer outcome for patients with either HCKHigh- or STAT3High-expressing tumors compared with individuals with HCKLow- or STAT3Low-expressing tumors (Fig. 3). Consistent with our observation that HCK lies functionally upstream of the IL6/11-Gp130-STAT3 signaling cascade in mice (19, 20), we detected only a minimal increase in hazard ratio (HR) when the low versus high expression criteria for HCK and STAT3 were combined.
Figure 3.
Increased HCK expression confers a poorer overall survival in patients with human intestinal-type gastric cancer. Overall survival analysis of HCK, STAT3, and HCK; STAT3 high- versus low-expressing intestinal-type gastric cancer patients (n = 320) stratified at the median level of gene expression from the manually curated Kaplan–Meier plot database (18). Please also refer to Supplementary Fig. S3.
We also compared the survival probability of patients according to their HCK expression with that for macrophage markers associated with alternative polarization (e.g., IL10, IL13, CD163) and abundance (CD68). We observed a reduced overall survival probability for patients with IL10High, IL13High, CD163High, or CD68High-expressing tumors compared with their low-expressing counterparts (Supplementary Fig. S3). However, combining expression of HCK with that of these markers individually did not significantly affect the HR. This correlative observation is consistent with our findings in mice that excessive HCK activation promotes alternative macrophage polarization and associated tumor progression.
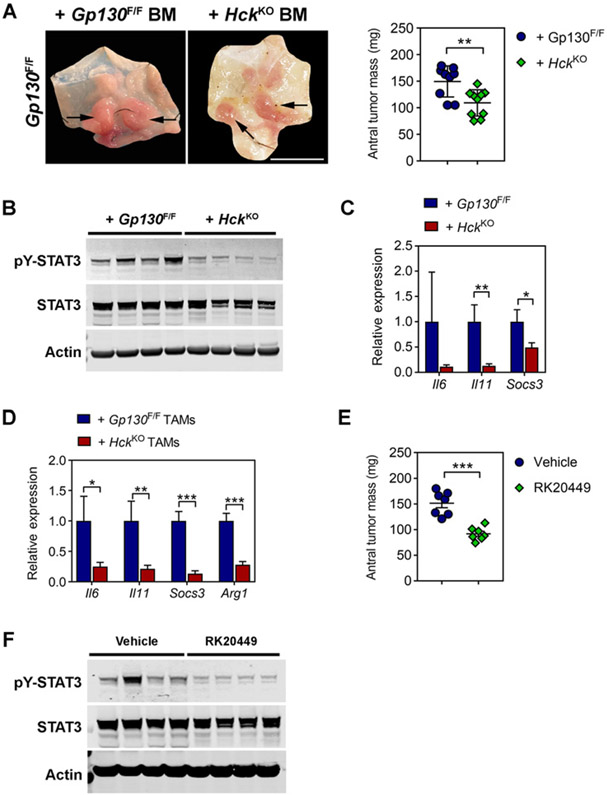
We next assessed the therapeutic benefit of deleting the Hck gene as an indirect way to impair STAT3 signaling. We adoptively transferred bone marrow from homozygous Hck knockout (HckKO) mice into tumor-bearing 8-week-old Gp130F/F mice. Twelve weeks later, we observed smaller tumors in mice reconstituted with HckKO bone marrow compared with recipient mice of Gp130F/F bone marrow (Fig. 4A). This observation coincided with reduced pY-STAT3 and reduced expression of Il6, Il11, and Socs3 in tumors of mice reconstituted with HckKO bone marrow (Fig. 4B and C). Similarly, these genes, alongside Arg1 as a prototypical gene of the alternatively activated macrophage endotype, were also reduced in TAMs of HckKO bone marrow recipients, suggesting impaired alternative macrophage activation (Fig. 4D). We validated these observations in a therapeutic setting, where we treated tumor-bearing 16-week-old Gp130F/F mice with the small-molecule HCK kinase inhibitor RK20449 (6) for 3 weeks and observed a significant reduction in total tumor burden, which coincided with decreased pY-STAT3 in these tumors (Fig. 4E and F).
Figure 4.
Genetic ablation or pharmacologic inhibition of HCK signaling impairs STAT3-dependent gastric tumor growth. A, Representative whole-mount images of stomachs opened along the outer curvature and collected from Gp130F/F bone marrow chimeras 12 weeks after their reconstitution with either Gp130F/F or HckKO bone marrow (BM) at 8 weeks of age. Scale bar, 2 cm. Arrows indicate tumors. Total antral tumor mass for each individual mouse is also shown. B, Western blot analysis for phosphorylated and total STAT3 protein isoforms on tumor lysates prepared from Gp130F/F bone marrow recipients analyzed in A. Actin was used as a loading control. Each lane represents an individual mouse. C, qRT-PCR analysis for Il6, Il11, and Socs3 expression in whole tumors of Gp130F/F bone marrow chimeras analyzed in A. n = 8 mice per cohort. D, qRT-PCR gene expression analysis of TAMs isolated from Gp130F/F bone marrow chimeras analyzed in A. n ≥ 6 mice per cohort. E, Total antral tumor mass of Gp130F/F mice following systemic administration of RK20449 (30 mg/kg) or vehicle (12% Captisol) at 16 weeks of age for 3 weeks. F, Western blot analysis for phosphorylated and total STAT3 protein isoforms on tumor lysates prepared from Gp130F/F mice treated for 3 consecutive weeks with RK20449 (30 mg/kg) or vehicle (12% Captisol) at 16 weeks of age. Actin was used as a loading control. Each lane represents an individual mouse. Data represent mean ± SEM, P values from unpaired Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). All experiments were performed at least twice.
Collectively, our results demonstrate that inhibition of HCK signaling in bone marrow–derived cells provides a therapeutic opportunity to reduce protumorigenic STAT3 activity in the tumor epithelium and to suppress tumor progression in a preclinical model of gastric cancer.
Discussion
STAT3 is overexpressed or excessively activated in most solid malignancies and represents a therapeutic target for cancer treatment (21). We have identified a mechanism whereby myeloid cell–specific HCK signaling confers excessive STAT3 activity in the gastric epithelium and promotes tumor development and invasion. Accordingly, either genetic reduction of Hck expression or pharmacologic inhibition of its activity impairs the growth of STAT3-dependent tumors by interrupting crosstalk between myeloid and epithelial cancer cells. Given the association between poor patient prognosis and persistent STAT3 activation, as well as its frequent occurrence in colon (22), stomach (23), and other solid malignancies, our findings raise potential therapeutic opportunities for patients with excessive tumor-associated HCK activity or elevated HCK gene expression (4, 6).
The findings here align with our previous suggestion that excessive STAT3 activity in carcinogen-induced colon tumors in HckCA/CA mice promotes their growth (6). Both in the colon and stomach, these observations were underpinned by increased Il6, Il11, and Socs3 gene expression in tumors and TAMs of mice reconstituted with bone marrow harboring the HckCA/CA allele. Accordingly, tumor burden in either organ was reduced following genetic ablation or pharmacologic inhibition of HCK activity and correlated with reduced Il6, Il11, and Socs3 expression. In TAMs with excessive HCK activity, we also observed increased expression of the STAT3-activating ligands Vegfα and Il10, suggesting that increased HCK signaling may instigate and sustain a feed-forward loop between innate immune and tumor epithelial cells to retain excessively activated STAT3 in the latter cell type. Although our analysis does not distinguish between the relative contribution of the STAT3-activating cytokines IL6, IL10, and IL11 to tumor growth, we have previously shown that genetic ablation of the Il11rα receptor subunit, but not the Il6 ligand, completely prevents tumor development in Gp130F/F mice (24). In addition to its indirect effect described here, HCK is also likely to modulate Gp130-dependent STAT3 signaling in a cell-autonomous manner, because HCK binds to the acidic domain of and coimmunoprecipitates with Gp130 (19, 20). Meanwhile, the Gp130 activating cytokines IL6 and leukemia-inhibitory factor stimulate HCK activity in multiple myeloma and embryonic stem cells (19, 20). Thus, excessive HCK activity in tumor-associated myeloid cells, alongside the associated induction and maintenance of their alternatively activated endotype, may also be maintained through an additional autocrine feed-forward mechanism based on excessive IL6/IL11 production by TAMs (Supplementary Fig. S4). Our observation of comparable association between patient survival and simultaneously elevated HCK and STAT3 expression indicates that HCK and STAT3 lie in the same functional, albeit not necessarily biochemical, pathway during gastric cancer progression. Consistent with our observations in mice, we also find similar HRs between HCK expression in patients with gastric cancer and expression of the alternative macrophage activation markers IL10, IL13, and CD163.
Here we reveal in vivo evidence that aberrant HCK activity in macrophages promotes tumor invasion in advanced stage cancer. Our evidence is supported by observations whereby HCK facilitates podosome formation and extracellular matrix degradation by macrophages (25, 26) to enable formation of tunnels in the stroma that facilitate escape of tumor cells independent of their own matrix-degrading ability (27). Likewise, macrophages are obligate partners for cancer cell migration and metastasis (28) and can stimulate the invasion of gastric and colorectal cancer cells in vitro (29). Indeed, HckKO macrophages fail to migrate through Matrigel and to promote tumor cell invasion (30). Conversely, we observed invading tumor cells alongside macrophages in the gastric submucosa of Gp130F/+;HckCA/CA mice but not in their Gp130F/F littermates, consistent with the alternatively activated endotype of Gp130F/+;HckCA/CA TAMs.
Despite the persistence of STAT3 activation in a majority of human cancers, direct inhibition of its activity has proven challenging due to dose-limiting toxicity. Global inhibition of STAT3 impacts normal cellular function and health (31, 32) and genetic ablation of Stat3 in hematopoietic cells predisposes mice to leukocytosis and chronic enterocolitis (33). For this reason, targeting upstream activating kinases such as HCK offers an alternative approach for indirect suppression of STAT3-dependent tumor growth, without incurring detrimental phenotypes (6). Indeed, our results demonstrate that inhibition of HCK in innate immune cells is sufficient to interfere with autocrine/paracrine signaling mechanisms that retain constitutive STAT3 activation within tumors, resulting in impaired growth. Future studies are required to determine whether inhibition of HCK also mediates STAT3 independent antitumor activities and facilitates a more effective antitumor immune response.
Supplementary Material
Acknowledgments
This work was made possible through the Victorian State Government Operational Infrastructure Support; the National Health and Medical Research Council (NHMRC) of Australia project grants 1025239, 1079257, 1081373, and 1092788; and the RFA-UD from La Trobe University. M. Ernst also received funding from Ludwig Cancer Research. A.R. Poh received funding from an Australian Post-Graduate Award PhD Scholarship and the Cancer Therapeutics CTx PhD Top-Up Scholarship. Research supported by the 2018 Debbie's Dream Foundation-Stupid Strong-AACR Gastric Cancer Research Fellowship, in memory of Candace Netzer, Grant Number 18-40-41-POH. A.R. Dwyer received funding from an Australian Post-Graduate Award PhD Scholarship and University of Western Australia PhD Top-Up Scholarship. R.J.J. O'Donoghue received funding from a Victoria Cancer Agency Early Career Seed Grant ESG13041. T.L. Putoczki is a Victorian Cancer Agency Fellow. M. Ernst and A.R. Poh are Research Fellows of the NHMRC.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Reprints and Subscriptions To order reprints of this article or to subscribe to the journal, contact the AACR Publications Department at pubs@aacr.org.
References
- 1.Deng JY, Sun D, Liu XY, Pan Y, Liang H. STAT-3 correlates with lymph node metastasis and cell survival in gastric cancer. World J Gastroenterol 2010;16:5380–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol 2007;7: 41–51. [DOI] [PubMed] [Google Scholar]
- 3.Ziegler SF, Marth JD, Lewis DB, Perlmutter RM. Novel protein-tyrosine kinase gene (hck) preferentially expressed in cells of hematopoietic origin. Mol Cell Biol 1987;7:2276–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubo T, Kuroda Y, Shimizu H, Kokubu A, Okada N, Hosoda F, et al. Resequencing and copy number analysis of the human tyrosine kinase gene family in poorly differentiated gastric cancer. Carcinogenesis 2009;30:1857–64. [DOI] [PubMed] [Google Scholar]
- 5.Kiyose S, Nagura K, Tao H, Igarashi H, Yamada H, Goto M, et al. Detection of kinase amplifications in gastric cancer archives using fluorescence in situ hybridization. Pathol Int 2012;62:477–84. [DOI] [PubMed] [Google Scholar]
- 6.Poh AR, Love CG, Masson F, Preaudet A, Tsui C, Whitehead L,et al. Inhibition of hematopoietic cell kinase activity suppresses myeloid cell-mediated colon cancer progression. Cancer Cell 2017;31:563–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst M, Inglese M, Scholz GM, Harder KW, Clay FJ, Bozinovski S, et al. Constitutive activation of the Src family kinase Hck results in spontaneous pulmonary inflammatory and an enhanced innate immune response. J Exp Med 2002;196:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell 1992; 68:869–77. [DOI] [PubMed] [Google Scholar]
- 9.Jenkins BJ, Grail D, Nheu T, Najdovska M, Wang B, Waring P, et al. Hyper-activation of Stat3 in gp130 mutant mice promotes gastric hyperproliferation and desensitizes TGF-beta signaling. Nat Med 2005;11:845–52. [DOI] [PubMed] [Google Scholar]
- 10.Lowell CA, Soriano P, Varmus HE. Functional overlap in the src gene family: inactivation of hck and fgr impairs natural immunity. Genes Dev 1994;8:387–98. [DOI] [PubMed] [Google Scholar]
- 11.Saito Y, Yuki H, Kuratani M, Hashizume Y, Takagi S, Honma T, et al. A pyrrolo-pyrimidine derivative targets human primary AML stem cells in vivo. Sci Transl Med 2013;5:181ra52. [DOI] [PubMed] [Google Scholar]
- 12.Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest 2008;118: 1727–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25: 402–8. [DOI] [PubMed] [Google Scholar]
- 14.Dwyer AR, Ellies LG, Holme AL, Pixley FJ. A three-dimensional co-culture system to investigate macrophage-dependent tumor cell invasion. J Biol Methods 2016;3:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poh AR, O'Donoghue RJ, Ernst M, Putoczki TL. Mouse models for gastric cancer: matching models to biological questions. J Gastroenterol Hepatol 2016;31:1257–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alorro MG, Pierce TP, Eissmann MF, Dijkstra C, Dickins RA, Ernst M, et al. Generation of an inducible mouse model to reversibly silence Stat3. Genesis 2017;55:e23023. [DOI] [PubMed] [Google Scholar]
- 17.Stuart E, Buchert M, Putoczki T, Thiem S, Farid R, Elzer J, et al. Therapeutic inhibition of Jak activity inhibits progression of gastrointestinal tumors in mice. Mol Cancer Ther 2014;13:468. [DOI] [PubMed] [Google Scholar]
- 18.Szasz AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 2016;7:49322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallek M, Neumann C, Schäffer M, Danhauser-Riedl S, von Bubnoff N, de Vos G, et al. Signal transduction of interleukin-6 involves tyrosine phosphorylation of multiple cytosolic proteins and activation of Src-family kinases Fyn, Hck and Lyn in multiple myeloma cells. Exp Hematol 1997;25:1367–77. [PubMed] [Google Scholar]
- 20.Schaeffer M, Schneiderbauer M, Weidler S, Tavares R, Warmuth M, de Vos G, et al. Signaling through a novel domain of gp130 mediates cell proliferation and activation of Hck and Erk kinases. Mol Cell Biol 2001;21:8068–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh J, Chand A, Gough D, Ernst M. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat Rev Cancer 2019;19:82–96. [DOI] [PubMed] [Google Scholar]
- 22.Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia 2005;7:545–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Judd LM, Menheniott TR, Ling H, Jackson CB, Howlett M, Kalantzis A, et al. Inhibition of the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and in vivo. PLoS One 2014;9:e95993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Putoczki T, Thiem S, Loving A, Busuttil RA, Wilson NJ, Ziegler PK, et al. Interleukin-11 is the dominant IL-6 family cytokine during gastrointestinal tumourigenesis and can be targeted therapeutically. Cancer Cell 2013;24:257–71. [DOI] [PubMed] [Google Scholar]
- 25.Park H, Dovas A, Hanna S, Lastrucci C, Cougoule C, Guiet R, et al. Tyrosine phosphorylation of Wiskott-Aldrich syndrome protein (WASP) by Hck regulates macrophage function. J Biol Chem 2014;289:7897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cougoule C, Le Cabec V, Poincloux R, Al Saati T, Mège JL, Tabouret G, et al. Three-dimensional migration of macrophages requires Hck for podosome organization and extracellular matrix-proteolysis. Blood 2010;115:1444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poh AR, O'Donoghue RJJ, Ernst M. Hematopoietic cell kinase (HCK) as a therapeutic target in immune and cancer cells. Oncotarget 2015;6:15752–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leek R, Lewis CE, Whitehouse R, Greenall M, Clarke J, Harris AL. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res 1996;56:4625–9. [PubMed] [Google Scholar]
- 29.Cardoso AP, Pinto ML, Pinto AT, Oliveira MI, Pinto MT, Gonçalves R, et al. Macrophages stimulate gastric and colorectal cancer invasion through EGFR Y (1086), c-Src, Erk1/2 and Akt phosphorylation and smallGTPase activity. Oncogene 2014;33:2123–33. [DOI] [PubMed] [Google Scholar]
- 30.Guiet R, Van Goethem E, Cougoule C, Balor S, Valette A, Al Saati T, et al. The process of macrophage migration promotes matrix metalloproteinase-independent invasion by tumour cells. J Immunol 2011;187:3806–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim CP, Fu XY. Multiple roles of STAT3 in cardiovascular inflammatory responses. Prog Mol Biol Transl Sci 2012;106:63–73. [DOI] [PubMed] [Google Scholar]
- 32.Wong AL, Soo RA, Tan DS, Lee SC, Lim JS, Marban PC, et al. Phase I and biomarker study of OPB-51602, a novel signal transducer and activator of transcription (STAT) 3 inhibitor, in patients with refractory solid malignancies. Ann Oncol 2015;26:998–1005. [DOI] [PubMed] [Google Scholar]
- 33.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med 2005;11:1314–21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.