Abstract
The journey of cancer cells from a primary tumor to distant sites is a multi-step process that involves cellular reprogramming, the breaking or breaching of physical barriers and the preparation of a pre-metastatic niche for colonization. The loss of adhesion between cells, cytoskeletal remodeling, the reduction in size and change in cell shape, the destruction of the extracellular matrix, and the modification of the tumor microenvironment facilitate migration and invasion into surrounding tissues. The promotion of vascular leakiness enables intra- and extravasation, while angiogenesis and immune suppression help metastasizing cells become established in the new site. Tumor-derived exosomes have long been known to harbor microRNAs (miRNAs or miRs) that help prepare secondary sites for metastasis; however, their roles in the early and intermediate steps of the metastatic cascade are only beginning to be characterized. The present review article presents a summary and discussion of the miRNAs that form part of colorectal cancer (CRC)-derived exosomal cargoes and which play distinct roles in epithelial to mesenchymal plasticity and metastatic organotropism. First, an overview of epithelial-to-mesenchymal transition (EMT), metastatic organotropism, as well as exosome biogenesis, cargo sorting and uptake by recipient cells is presented. Lastly, the potential of these exosomal miRNAs as prognostic biomarkers for metastatic CRC, and the blocking of these as a possible therapeutic intervention is discussed.
Keywords: exosomes, microRNA, exosomal miRNA, exomiRs, exosome cargo, colorectal cancer, metastasis, metastatic cascade
1. Introduction
Colorectal cancer (CRC) remains the third most commonly diagnosed type of cancer globally, only following breast and lung cancer. In addition, it is now the second most common cause of cancer-related mortality following lung cancer (1). Metastatic spread, notably to the liver, accounts for ~70% of deaths, and ~25% of cases are metastatic at the time of diagnosis (2,3). While the majority of primary tumors can be removed, only a small fraction of those diagnosed with metastatic CRC are eligible for upfront surgery. The five-year survival rate for patients with metastatic CRC is dismal at only 12%, compared to 64% for CRC in general (4). Furthermore, a considerable proportion of those with localized disease may also have recurrence as distant metastases (5), due to residual cancer cells that migrate out of the localized tumor towards distant sites. Arguably, metastasis is the major cause of mortality among patients with CRC, and understanding this multi-step process may provide insight on the steps where therapeutic intervention could be possible.
The journey of metastatic cells from the primary tumor to distant sites of metastases is an arduous one and requires the breaking and breaching of barriers before a new site is colonized. Up until the last decade, a number of the mechanisms that contribute to metastatic dissemination remained obscure. The epithelial-to-mesenchymal (EMT) program is now considered key to understanding invasion and metastasis and explains the physical and ultrastructural changes cells have to undergo to break away from the primary tumor and migrate towards a new metastatic niche. More importantly, it is now recognized that a tumor has to modify its microenvironment for it to thrive and colonize distant sites (6).
The discovery of exosomes as purveyors of metastatic spread helped explain the incompletely understood concept of metastatic organotropism and the preparation of a pre-metastatic niche (7). Guided by connexins and integrins (7,8) and through their cargo of functional biomolecules, they can instigate an entire program of angiogenesis and immune suppression to recipient cells in the new site via horizontal transfer (9), thus ensuring survival of metastasizing cells. In recent years, it has become apparent that the role of exosomes is not limited to organotropism of primary tumor cells. They affect both local and distant environments and are involved in multiple steps of the metastatic cascade. Among their multiple cargoes, microRNAs (miRNAs or miRs) play critical roles in metastatic success. Exosomal miRNAs (exomiRs), released by both tumor and stromal cells, have been shown to affect multiple hallmarks of cancer.
In the present review, key processes contributing to metastatic spread, including EMT, organotropism and the preparation of the pre-metastatic niche are discussed. Furthermore, a discussion of exosome biogenesis and cargo sorting is included to link the initial steps of the metastatic cascade with those of colonization of distant sites. Lastly, the role of exosomal miRNAs identified from CRC tumor and stromal cells in the metastatic continuum is discussed along with insight on the mechanisms through which they ensure the survival of metastasizing tumors.
2. Epithelial-to-mesenchymal transition
EMT is a biological process wherein adherent epithelial cells convert to a migratory mesenchymal phenotype. Epithelial cells are characterized by cell-cell and cell-matrix connections that allow them to function as barriers by forming organized interconnected sheets of cells. These intercellular adhesions prevent movement and establish apical, lateral and basal membrane domains, with cellular components displaying apico-basal polarity. On the other hand, mesenchymal cells lack intercellular junctions and therefore have no clear apical and lateral membranes, no apico-basal polarized distribution of cell components, and are essentially loose motile cells that are capable of invasion (10).
While EMT is required for key developmental processes, such as embryogenesis and organ formation (type 1 EMT), as well as wound healing and tissue regeneration (type 2 EMT), it is also known to play a role in tumor dissemination and metastasis in epithelium-derived carcinomas (type 3 EMT) (11). EMT provides tumor cells with the ability to dissociate from the primary tumor to ultimately metastasize to distant organs. In fact, EMT, as evidenced by significant changes in the levels of EMT-related markers, has been shown to be associated with a poor prognosis in several types of cancer, including lung (12), esophageal (13), pancreatic (14), breast (15) and CRC (16,17).
The term 'transition' in EMT refers to its transient and reversible nature, with the converse process termed mesenchymal-to-epithelial transition (MET). The plasticity of epithelial cells allows them to transition into mesenchymal cells then back to an epithelial phenotype, either fully at either end of the spectrum or only partially, wherein they express both mesenchymal and epithelial characteristics (18). Core phenotypic changes in EMT include the loss of intercellular junctions and apical-basal polarity, cytoskeletal remodeling, the acquisition of cell protrusions and extracellular matrix degradation. Gene expression and signaling pathways are also reprogrammed, further contributing to the EMT process.
Loss of adhesion and apical-basal polarity
Epithelial cells are initially attached to their neighboring cells through cell-cell adhesion complexes mediated by E-cadherin (19,20). EMT begins with the dissolution of epithelial cell-cell contacts essential for basement membrane integrity, such as tight junctions (at the apical surface of cells), adherens junctions (basal to the apical tight junctions), desmosomes (anchored to the intermediate filaments in the cytoskeleton) and gap junctions (channels between cells for passage of ions and small molecules), resulting in the downregulation of their associated proteins, claudins and occludins, cadherins, desmoplakin and plakophilin and connexins, respectively (21,22). The loss of E-cadherin leads to the translocation of β-catenin to the nucleus, where it can contribute to EMT through wingless-type MMTV integration site family (Wnt) signaling (23,24).
The disintegration of cell-cell junctions is also accompanied by the disruption of cell polarity complexes, consequently resulting in epithelial cells losing their apical-basal polarity and acquiring a front-rear polarity (25). In mammals, three protein complexes establish apical-basal cell polarity and membrane identity through their mutually antagonistic interactions (26). Partitioning-defective (PAR) complex [PAR6, PAR3 and atypical protein kinase C (aPKC)] (27) and Crumbs (CRB) complex [CRB, protein associated with Lin-7 1 (Pals 1) and Pals 1-associated tight-junction protein (PATJ)] (28) localize at the apical domain. Here, the PAR complex, which is required for tight junction formation (29), is stabilized by the CRB complex. The Scribble (Scrib) complex [Scrib, Disc large (dlg) and lethal giant larvae (lgl)] localize at the basolateral domain, opposite the PAR complex (30). During EMT, inducers such as Zinc finger E-box-binding homeobox (ZEB) and Zinc finger protein SNAI1 (commonly known as Snail) transcription factors disrupt the assembly and suppress the expression of these polarity complexes (26). This is enhanced by the downregulation of E-cadherin, which prevents the recruitment of Scrib complexes to the basolateral membrane (31). The overall loss of contact with the basement membrane reduces epithelial cell adhesion to eventually facilitate movement.
Cytoskeletal remodeling and extracellular matrix degradation
Cells reorganize their cytoskeletal architecture to enable migration. Peripheral F-actin fibers are replaced by stress fibers, and extracellular matrix (ECM) adhesion molecules such as integrins, paxillin, and focal adhesion kinase localize at the tips (32-34). Intermediate filaments, such as cytokeratins are replaced by vimentin, changing the cellular morphology from cuboidal or columnar to fibroblastic or spindle-shaped (35,36). The cell membrane projections [lamellipodia (thin sheet-like region at the leading edge of migrating cells) and filopodia (spike-like extensions at the edge of lamellipodia)] permit directional movement of the cell across its surroundings. Invadopodia, together with the increased expression of matrix metalloproteinases (MMP-1, -2, -3, -7, -9, -13 and -14), degrade the ECM, allowing the cells to detach from each other, penetrate the basement membrane, and invade the stroma and surrounding tissues (25,32,37).
Reversal of EMT
Once mesenchymal cells reach the metastatic site, they may undergo EMT reversal or MET. Cells revert to an epithelial state, resulting in secondary tumors that resemble the histopathological phenotype of the primary tumor (11). MET may occur in the local environment of the distant organ, due to the absence of heterotypic signals that induced EMT at the primary tumor site (11). Additionally, tumor cells would need to induce signaling pathways to resume proliferation during colonization of the foreign environment (38,39).
Transcriptional reprogramming during EMT
EMT is driven by a marked change in transcriptional programming. The modulation of gene expression during EMT involves several signal transduction pathways and transcription factors that reprogram the cell towards a mesenchymal phenotype. The changes in gene expression are responsible for phenotypic manifestations, such as the loss of adhesion and polarity, and an increase in migration and invasiveness.
Several transcription factors that control EMT are dysregulated or aberrantly expressed in CRC. These include SNAI1/SNAI2 (Snail and Slug), ZEB1/ZEB2 (40,41) in the ZEB family, twist family basic helix-loop-helix transcription factors (TWIST) Twist1/Twist2 (42,43) and the forkhead box (FOX) family (44-46) of transcription factors. SNAIL and SLUG are responsible for the repression of E-cadherin (47,48). Slug is also required for Twist1-mediated EMT (48). Twist1 and Zeb1 are also known to drive the loss of E-cadherin and a pro-metastatic cellular phenotype of enhanced invasion (49). These transcription factors have been shown to be associated with cell migration, tumor progression and metastatic potential in CRC (25,50,51).
Multiple signal transduction pathways have been implicated as modulators of the EMT transcriptional program. Transforming growth factor β (TGF-β) is one of the key inducers of EMT in CRC, as well as other types of cancer. Mothers against decapentaplegic homolog (SMAD) family member 4 (SMAD4) is a pro-epithelial factor whose loss leads to heightened EMT through aberrant signal transducer and activator of transcription 3 (STAT3) activation and increased signaling via the TGF-β/bone morphogenetic protein (BMP) pathways (52). Apart from the SMAD pathways, TGF-β also signals via SMAD-independent pathways, such as the Ras/mitogen-activated protein kinase (MAPK) and phosphoinositide 3-kinase (PI3K)/Akt pathways (53,54).
The Wnt/β-catenin pathway is another crucial driver of EMT in CRC. The canonical Wnt pathway leads to SNAIL upregulation and SLUG stabilization. Consequently, Wnt3a expression is associated with mesenchymal markers in clinical samples, and Wnt inhibition can partially reverse EMT expression. In addition, the non-canonical Wnt pathway has also been implicated in metastatic CRC (55).
Transcriptional reprogramming during EMT drives the characteristic switch towards mesenchymal marker expression. The common markers of EMT are typically proteins involved in the architecture of cell junctions and the cytoskeleton, controlling cell morphology and interactions with neighboring cells, as well as the extracellular environment, key physical features that are altered in EMT. Thus, the EMT transcriptional program capacitates the cancer cell for metastatic spread by altering the expression of key structural proteins to facilitate pro-metastatic characteristics such as loss of adhesion, cytoskeletal remodeling, migration and invasion.
One hallmark of EMT is the cadherin switch from E-cadherin to N-cadherin. The epithelial marker, E-cadherin, is a cell-surface protein that mediates cell-cell adhesion, and also binds β-catenin at its cytoplasmic domain. The downregulation of E-cadherin results in the loss of adhesion, an important phenotype for cells progressing towards metastasis, and also frees β-catenin for activation of its signaling cascade. On the other hand, N-cadherin, which is upregulated in mesenchymal cells, facilitates dynamic adhesion contacts crucial for migration, and confers an affinity for other N-cadherin-expressing mesenchymal cells. Thus, the E-to-N-cadherin switch results in the loss of adhesion and increased migration (25).
Claudins and occludins are crucial cell-cell adhesion proteins found in tight junctions, and are subsequently downregulated in EMT to dissolve epithelial apical-basal polarity (56). Conversely, the N-cadherin ligand, neural cell adhesion molecule (NCAM), is upregulated in EMT, modulating the activity of N-cadherin-associated receptor tyrosine kinases, such as Fyn to facilitate focal adhesions (57).
As regards cytoskeletal proteins, EMT is marked by the downregulation of cytokeratin and the upregulation of vimentin, which are both components of intermediate filaments (IFs). The altered composition of IFs facilitates differential trafficking of organelles and proteins, as well as enhanced cellular motility (56). Finally, cell-matrix interactions are also modulated via the altered regulation of integrins, upregulation of fibronectin, and increased expression and secretion of matrix metalloproteinases MMP2 and MMP9 (25).
Cancer stem cells (CSCs) and EMT
Similar to normal stem cells, CSCs also have the capacity for self-renewal and differentiation, and can initiate new tumors, such as in seeding metastatic lesions, or in cancer recurrence following therapy. CSCs are distinguishable by specific expression marker profiles, and can apparently arise from the acquisition of oncogenic alterations in normal stem cells, or from the dedifferentiation of differentiated cancer cells (58). In fact, pro-stemness factors are often proto-oncogenic molecules that are dysregulated in cancer. Thus, CSCs are often viewed as an important component of emerging therapeutic resistance and subsequent progression and metastasis, as CSCs that escape treatment are able to expand and initiate new lesions at a later stage.
EMT has also been implicated in the emergence and maintenance of CSCs. Cancer cells that have undergone EMT often exhibit a CSC phenotype, and conversely, CSCs often express EMT markers, signifying a mesenchymal shift. The close association between EMT and CSCs underscores the important role of both in the overall process of metastatic spread. TGF-β/Smad signaling is a key link, as it modulates both EMT and stemness phenotypes (59).
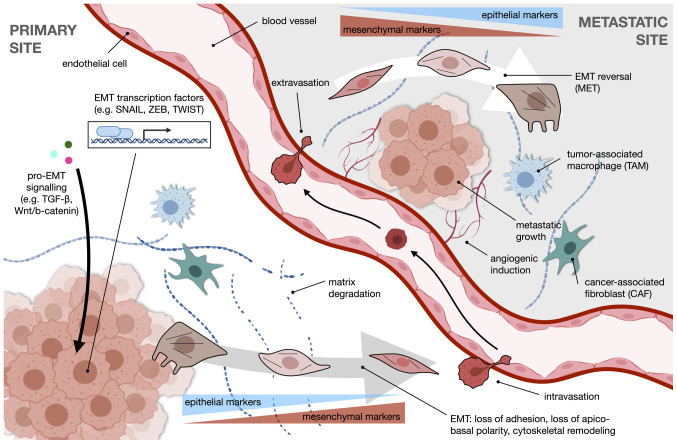
A summary of the overall process of EMT in the context of metastasis is illustrated in Fig. 1. Pro-EMT signaling, such as through the TGF-β or Wnt/β-catenin pathways, drives transcriptional reprogramming facilitated by EMT transcription factors which include SNAIL, ZEB and TWIST. The cancer cell undergoes changes in morphology towards a more motile mesenchymal phenotype, accompanied by a switch in expression from epithelial to mesenchymal markers. With enhanced ECM degradation, the cell is free to intravasate into the circulation and extravasate at a distant metastatic site. Once there, the cell reverts to an epithelial-like phenotype. Angiogenic induction and the action of stromal cells, such as tumor-associated macrophages (TAMs) or cancer-associated fibroblasts (CAFs) further modify the niche to favor metastatic growth.
Figure 1.
An overview of EMT and the metastatic process. Transcriptional reprogramming during EMT, a core component of the metastatic cascade, is driven by pro-EMT signaling pathways, such as TGF-β and Wnt/β-catenin, and EMT transcription factors of the SNAIL, ZEB, and TWIST families, among others. The resulting physical changes help cells assume a motile phenotype. This shift is accompanied by the downregulation of epithelial markers and the upregulation of mesenchymal markers, as well as an enhanced capability for degradation of the extracellular matrix. The primed metastatic cell intravasates and travels through the circulation to the distant metastatic site, where it extravasates and undergoes EMT reversal, or MET. Angiogenic induction is crucial for establishing metastasis. Stromal cells in the TME, such as TAMs and CAFs, modulate EMT and help sculpt the metastatic niche. EMT, epithelial-mesenchymal transition; TGF-β, transforming growth factor β; ZEB, Zinc finger E-box-binding homeobox; MET, mesenchymal-epithelial transition; TAMs, tumor-associated macrophages; CAFs, cancer-associated fibroblasts.
Organotropism, EMT and the pre-metastatic niche
Tumors are known to have a predisposition to metastasize to specific organs. This phenomenon, however, remained poorly understood for almost a century. The intrinsic properties of cancer cells, including genes and pathways implicated in the colonization of new metastatic niches, were invoked and constituted the predominant explanation for the phenomenon known as metastatic organotropism (60-62). Indeed, EMT itself has been shown to mediate metastatic colonization and metastatic organotropism both directly and indirectly. In pancreatic ductal adenocarcinoma (PDAC) models, the loss of p120ctn, binding partner and stabilizer of E-cadherin at adherens junctions, has been shown to cause a shift in the metastatic load from the liver to the lung. Furthermore, this could be reversed by the transfection of PDAC cells with p120ctn isoform 1A (63). EMT has also been shown to affect organ-specific metastasis by influencing metabolic reprogramming (64,65) and by regulating the tumor immune microenvironment (66). Whether these can be extrapolated to other epithelial tumors, such as CRC warrants further investigation.
While events that render the pre-metastatic niche favorable for dissemination and the growth of tumor cells upon arrival were being characterized, among these being angiogenesis and immunosuppression (67-69), the question of what dictates metastasis to specific organs remained unanswered. The discovery that exosomal integrins in tumor-released exosomes directed organ-specific colonization helped explain the induction of the pre-metastatic niche (7). Together with integrins, connexins, proteins that function as an integral component of channels at gap junctions, were found to be embedded in exosomal membranes (70). Furthermore, resident stromal cells in metastatic target organs have been shown to internalize these exosomes and their cargo, further influencing expression of genes implicated in the preparation of the pre-metastatic niche (7).
3. Exosomes
Exosomes are spherical membrane-enclosed nanovesicles secreted by all types of living cells into the extracellular milieu, notably in abundance by tumor cells. They have been detected in a range of biological fluids, including blood, urine, saliva, breast milk and pleural effusions (71-73), with a substantial amount of up to 1012 exosomes/ml of body fluid (~0.1% volume) in physiological conditions (74). Exosomes are important mediators of intercellular communications, capable of transferring functional genetic cargo to modify neighboring and distant recipient cells (75-78).
Exosome biogenesis
Exosomes can be differentiated from other extracellular vesicles based on their mode of biogenesis: microvesicles (100 to 1,000 nm) originate from the cell surface and are formed through the direct outward budding of the plasma membrane (79), while apoptotic bodies (800 to 5,000 nm) are generated from the outward blebbing and fragmentation of the cell membrane as cells undergo apoptosis (80). Exosomes (40-120 nm in diameter) are initially formed as intraluminal vesicles (ILVs) within the endosomal system. In this pathway, the invagination of early endosomes during maturation sequesters cytoplasmic genetic material, such as RNA, DNA, lipids and proteins into ILVs. The late endosomes, now referred to as multivesicular bodies (MVBs), are either targeted to the lysosomes for degradation, serve as temporary storage compartments, or fuse with the plasma membrane, releasing ILVs into the extracellular space as exosomes (81-83).
The formation of intraluminal vesicles occurs via multiple mechanisms. The most well-described pathway is dependent on the endosomal sorting complex required for transport (ESCRT) budding machinery, where ESCRT-0 recognizes and forms domains of ubiquitylated proteins, which are later confined when the endosomal membrane is deformed by the ESCRT-I and ESCRT-II complexes. ESCRT-III then cleaves the bud neck to form intraluminal vesicles (84). On the other hand, the syndecan-syntenin-ALG-2-interacting protein X (ALIX) pathway recognizes any material bound to the heparan sulfate of syndecan. Syndecans are transmembrane proteins whose cytosolic domain can bind to and recruit syntenin, a soluble cytoplasmic protein, to the plasma membrane. Syntenin also binds to ALIX, an accessory protein to the ESCRT-III, in turn linking the syndecans to the ESCRT complexes for ILV formation (85,86). Alternatively, membrane budding can occur through an ESCRT-independent pathway involving the sphingolipid ceramide. Ceramide induces the coalescence of lipid microdomains found in the endosomal membrane, and its cone-shaped structure causes spontaneous negative curvature between the membrane leaflets, resulting in the inward budding of endosomes to produce ILVs (87). A recent study also reported another ESCRT-independent pathway using Rab31, which prevents the fusion of the MVB to the lysosome to instead promote ILV secretion (88). In these pathways, Rab guanosine triphosphatases (Rab GTPases) mediate the fusion of MVBs with the plasma membrane. Rab27a and Rab27b influence the localization (89), whereas Rab35 regulates the docking and tethering (90) of the MVBs at the plasma membrane for subsequent secretion of the intraluminal vesicles into the extracellular fluid as exosomes.
Exosome cargo and cargo sorting
The major cargo residing within exosomes are proteins, lipids, RNA and a minimal amount of DNA. The contents of exosomes are highly heterogeneous, depending on the tissue, cell type and physiological or pathological context. Notably, exosome profiles do not fully reflect that of parent cells, signifying a selective mechanism for sorting cellular genetic material into exosomes (9,91,92). There is also evidence to indicate that some RNA transcripts can be found exclusively in exosomes and not in the cells of origin (9,92) which suggests that certain genes are destined for export and communication with other cells (93).
Deep sequencing has revealed a variety of exosomal RNA cargo, including mRNAs, miRNAs, long non-coding RNAs (lncRNAs), ribosomal RNAs, transfer RNAs, piwi-interacting RNAs, small nuclear RNAs, small nucleolar RNAs and even circular RNAs, each present at varying levels of abundance (74,91,94-96). Among these, miRNAs are generally the most predominant RNAs encapsulated in exosomes (91,96). Several mechanisms regulating the selective export of miRNAs into exosomes have been proposed, from intrinsic sequences present in the 3′ end of miRNAs to cellular lipids and proteins that guide their incorporation into exosomes. First, heterogeneous nuclear ribonucleoproteins (hnRNPs) directly bind to 4-bp sequence motifs in the 3′ end of miRNA sequences to package them into exosomes. Sumoylated hnRNP A2/B1 can recognize the RNA motif GGAG in miR-198 and miR-601 (97), while hnRNP-Q can recognize the GGCU motif in miR-3470a, miR-194-2-3p, miR-6981-5p, miR-690 and miR-365-2-5p (98). Other members of the hnRNP family, such as hnRNPA1 and hnRNPC (97) and miRNAs enriched with the GUUG motif (99) were also sorted into exosomes, although their cognate sequence motifs and RNA binding proteins, respectively, have not yet been identified. Second, 3′ end post-transcriptional modifications can also influence cargo loading. The RNA sequencing of B cells and urine samples has revealed that adenylated miRNAs are retained in the cells, while those that are uridylated are preferentially sorted into exosomes (100). Third, membrane lipids involved in EV biogenesis also control the loading of miRNAs into exosomes, indicative of the ceramide-dependent secretory pathway independent of the ESCRT machinery (101). The overexpression of neural sphingomyelinase 2 (nSMase2), which regulates ceramide biosynthesis, has been shown to increase the quantity of miRNA exported into exosomes, whereas nSMase2 inhibition reduces the quantity of exosomal miRNAs (102). Similarly, the inhibition of sphingomyelin phosphodiesterase 3 (SMPD3), which catalyzes the hydrolysis of sphingomyelin to ceramide, has been shown to result in a concentration-dependent increase in cellular miR-638 levels and a decrease in exosomal miR-638 levels, using the SW480 human colorectal and HuH-7 human hepatocellular cancer cell line (103). In patients with CRC, the expression levels of miR-638 have also been found to be downregulated in serum exosomes and to be associated with a poor overall and disease-free survival (104). Fourth, Argonaute 2 (Ago2), a key component of the RNA-induced silencing complex (RISC) appears to play a role in exosomal miRNA secretion. The knockdown of Ago2 has been shown to decrease the expression of miRNAs (miR-451, miR-150 and miR-142-3p) that are most commonly exported from different cell types (105). In this regard, KRAS mutations have also been implicated in miRNA export, negatively regulating the secretion of miRNAs into exosomes, since downstream MEK/ERK signaling inhibits the association of Ago2 with multivesicular bodies (106). Notably, all essential RISC components, such as Dicer, transactivation response RNA-binding protein (TRBP) and Ago2 are present in cancer-derived exosomes and this enables processing of pre-miRNAs into mature miRNAs within the nanovesicles, unlike exosomes derived from normal cells (107).
Exosome release and uptake in cancer cells
Apart from cell state, exosome secretion is influenced by several factors, such as cellular stress, heat, ischemia, pH, the loss of cellular attachment and the accumulation of intracellular calcium (108). Of note, low pH levels and hypoxia are common features of solid tumors. The low pH of the tumor microenvironment results in increased exosome release and subsequent uptake by recipient cells, which explains the abundance and effects of exosomes in cancer (109). Moreover, hypoxia increases the release of exosomes, which can potentially facilitate angiogenesis and metastasis, further sustaining the growth and survival of cancer cells (110).
Exosome internalization by recipient cells highlights the critical role of exosomes in cell-to-cell crosstalk, with the unique advantage of targeting specific locations compared to cytokines and hormones in the systemic circulation. Exosomes can transfer genetic information to neighboring or distant cells through three principal mechanisms: i) Direct fusion of the exosomal lipid membrane with the cellular membrane of recipient cells, releasing the exosome cargo into the cytosol (111); ii) proteins on the exosomal membrane serve as ligands for receptors on the surface of recipient cells; and iii) endocytosis either mediated by the clathrin protein, which is associated with engulfment of partner receptors, or micropinocytosis, which is associated with membrane ruffles that are induced by receptor tyrosine kinases (112). The specificity of these receptor-ligand interactions suggests that exosomes are targeted to particular cells (7).
Tumor-derived exosomes and cancer progression
Exosomes were once considered to serve only as a means of cellular waste disposal when they were first described for removal of plasma membrane proteins during reticulocyte maturation (81,113). Later on, Raposo et al (114) isolated exosomes from B lymphocytes and demonstrated their involvement in antigen presentation, capable of inducing an immune response. Exosomal messenger RNAs were then found to be internalized and translated into functional proteins (9,92), while exosomal miRNAs and lncRNAs can regulate the translation of target mRNAs (75,107,115,116) in recipient cells, concretely establishing a role in intercellular communication. This links exosomes to several biological processes as well as disease pathogenesis.
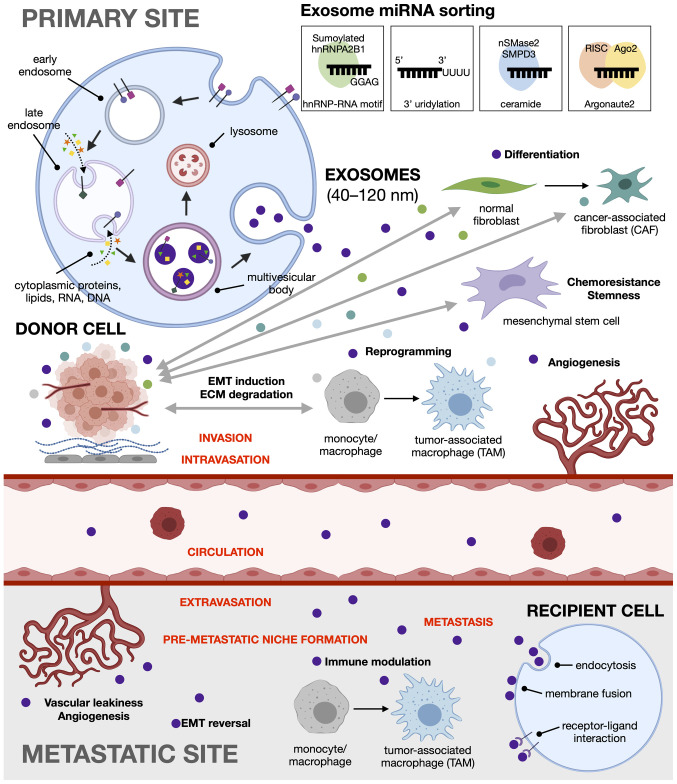
In the context of cancer, cumulative evidence suggests that exosomes can promote tumorigenesis through the horizontal transfer of oncogenic material to recipient cells. Likewise, cancer cells can utilize exosomes to discard tumor-suppressive genetic material not beneficial for tumor growth so as to increase their own oncogenicity (117). For instance, in CRC, miR-100 is a tumor suppressor that inhibits cellular migration and invasion by targeting Leucine-rich repeat-containing G protein-coupled receptor 5 (Lgr5) (118). Mutant KRAS CRC cells have been reported to secrete exosomes enriched in miR-100 as a strategy to sustain low intracellular levels (117). Tumor-derived exosomes have been shown to induce (119) or suppress (120) the immune response, promote the formation of a pre-metastatic tumor niche (7,121), regulate angiogenesis (122), enhance migration (76) and cell proliferation (77), and induce epithelial to mesenchymal transition (123), among others (Fig. 2).
Figure 2.
Tumor-and stroma-derived exosomes as major drivers of EMT and pre-metastatic niche formation. Exosomes are formed via the endocytic pathway. In the primary tumor site, both cancer cells and stromal cells in the tumor microenvironment release exosomal miRNA to promote cancer metastasis. Tumor-derived exosomes can reprogram fibroblasts, macrophages, mesenchymal stem cells, and endothelial cells, as well as induce epithelial to mesenchymal transition to enable cell migration and invasion. At the metastatic site, the exosomes participate in pre-metastatic niche formation through immune modulation and angiogenesis, and induce EMT reversal or mesenchymal to epithelial transition to facilitate colonization of the foreign environment. EMT, epithelial-mesenchymal transition; miRNA, microRNA.
Exosomes can also promote tumor resistance by encapsulating drugs and their metabolites into exosomes for export, as a drug efflux mechanism (124). Similarly, drug-resistant cancer cells can induce chemoresistance in other cancer cells through exosome-mediated transfer of efflux transporters (78).
4. CRC exosomal miRNAs implicated in EMT and organotropism
Various miRNAs can inhibit EMT progression by directly targeting components of the EMT regulatory pathways. CRC exosomes may be enriched with oncogenic miRNAs that downregulate EMT inhibitors. Alternatively, tumor suppressive miRNAs that downregulate inducers of EMT may themselves be downregulated or disposed of in CRC exosomes. Exosomal cargo can both originate from and be delivered to either tumor cells or cells in the tumor microenvironment, enhancing the capacity for metastasis by both driving EMT in tumor cells and influencing the properties of the microenvironment. Moreover, miRNAs carried by serum exosomes can be delivered to sites distant from the originating tumor, further extending metastatic potential. The promotion of EMT by altered regulation of exosomal miRNAs results in expression of characteristic mesenchymal markers and enhanced phenotypic features of pro-metastatic cells.
Reciprocal transfer of exomiRs between CRC cells and CAFs modulate the metastatic phenotype
CAFs are essential components of the TME, with roles in matrix deposition and remodeling (125). Up to 80% of stromal fibroblasts in a tumor are believed to acquire an activated phenotype and become CAFs (126). These cells can modulate tumor progression by releasing cytokines such as TGF-β, tumor necrosis factor α (TNF-α), and interleukin (IL)-6 and IL-8 (127). Tumor cells and CAFs in the TME maintain extensive reciprocal signaling interactions (125). Recently, primary CRC cells, as well as HCT116 cells were shown to release exosomes containing miR-10b which are taken up by surrounding fibroblasts. miR-10b was found to increase TGF-β and α-smooth muscle actin (α-SMA) through the inhibition of PI3K activity, which is indicative of fibroblast activation into pro-tumorigenic CAFs (127). In a similar manner, CAFs have been shown to deliver exosomal miRNA to tumor cells. In patients with CRC, CAF-derived exosomes have been found to be enriched with miR-92a, which is delivered to CRC cells (128). The uptake of exosomal miR-92a enhances EMT and metastatic capability by downregulating F-Box and WD repeat domain containing 7 (FBXW7) and modulator of apoptosis 1 (MOAP1), and by activating the Wnt/β-catenin pathway (128). Of note, cellular miR-92a has also been characterized as an oncogenic miRNA promoting migration, F-actin remodeling and EMT marker expression in CRC cells via the downregulation of neurofibromin (NF2)/Merlin, a tumor suppressor gene that controls contact-dependent inhibition, adhesion and migration (129). Other miR-92a targets include Dickkopf-3 (Dkk-3) and claudin-11, resulting in enhanced angiogenesis and disruption of tight junctions, respectively (130). In patients with CRC, the loss of claudin-11 has been shown to be associated with increased metastasis and a poor prognosis (131). In the CRC microenvironment, CAFs were previously shown to deliver miR-21 to cancer cells in exosomes. miR-21 was identified as the most abundant and most enriched miRNA in an exosomal cancer-associated fibroblast signature that also includes miRNAs 329, 181a, 199b, 382 and 215. Orthotopic xenografts derived from fibroblasts overexpressing miR-21 exhibited an increased liver metastases relative to the controls (132).
The exosomal cargo of CAFs in CRC may also include the lncRNA H19. H19 can attenuate the inhibitory effects of miR-141 and can then activate the Wnt/β-catenin pathway to promote stemness (133). Thus, exosomal delivery of miRNAs and/or lncRNAs from CAFs may drive EMT through redundant mechanisms. This is in addition to other forms of stromal-tumor crosstalk that may exist in parallel, notably the secretion of CAF-derived soluble factors such as TGF-β and juxtacrine signaling (134,135). Increasing evidence suggests that CAFs are heterogenous and may assume different functional roles (136,137). It is highly likely that different groups of CAFs are involved in exosomal miRNA exchange with tumor cells.
CRC-derived exosomal miRNA in cytoskeletal remodeling, loss of adhesion and ECM degradation
In patients with metastatic CRC, serum exosomal miR-106b-3p has been found to be upregulated and correlated with metastatic progression (138). Cellular and exosomal miR-106b-3p has been found to enhance migration and invasion, and drive pro-EMT expression, as well as promote lung metastasis in a mouse xenograft model, by targeting the tumor suppressor, deleted in liver cancer 1 (DLC-1) (138). DLC-1 is a GTPase activating protein (GAP), thus a negative regulator, for Rho GTPases, which regulate actin remodeling and thus cellular motility (139).
Exosomal miR-106b-3p from highly metastatic CRC cells also has profound effects on cell adhesion. They are released to less metastatic CRC cells and are then able to upregulate N-cadherin and downregulate E-cadherin protein expression. This is also achieved via the downregulation of DLC-1, effectively promoting metastasis in situ via coordinated effects on both cell adhesion and cytoskeletal rearrangement (138). Exosomes harboring miR-210 are also secreted by adherent HCT-8 colon cancer cells and cause them to detach and grow in suspension, indicative of metastatic capacity and anoikis resistance. Furthermore, these detached cells have been shown to be E-cadherin-negative and vimentin-positive, in contrast to adherent colonies that are E-cadherin-positive and vimentin-negative (123).
Exosomal miR-1246 in CRC, on the other hand, has been linked to the degradation of the ECM. Gain-of-function mutations in the p53 gene found in CRC cells have been shown to increase miR-1246 levels in exosomes, which are in turn able to reprogram macrophages into TAMs, the major component of tumor-infiltrating immune cells (140-142). The TAM phenotype is a classic phenotype of solid tumors with poor prognosis, and is characterized by heightened stimulation of ECM degradation, as well as enhanced migratory and invasive capacities (142).
Exosomal miRNAs and EMT reversal/EMT inhibition in CRC
Tumor-derived exosomal miRNAs may promote EMT reversal or MET by suppressing effectors, inducers, transcription factors, and other players involved in EMT. An increased expression of miR-200c and miR-141 in exosomes is indicative of MET in CRC cells. In a previous study, upon treatment with the drug decitabine, SW480 (primary CRC) cells did not exhibit any significant differences, while the metastatic cell lines SW620 (derived from lymph node metastasis) and SW620/OxR (derived from lymph node metastasis with acquired resistance to oxaliplatin) exhibited decreased migration and invasion properties. This was accompanied by the upregulation of E-cadherin and exosomal miR-200c and miR-141, which together suggest the acquisition of epithelial characteristics through the reversal of EMT (143).
CRC-derived exosomal miR-200c, miR-141 and miR-429 can also inhibit EMT by directly targeting the ZEB family in endothelial cells. In a previous study, in 3D spheroid models, co-culture with naïve CRC cells did not demonstrate disruption of lymphatic (exomiR-200c) (144) and blood (exomiR-200c, -141 and -429) (145) endothelial cell layers. By contrast, co-culture with 5-fluorouracil-resistant CRC cells which release exosomes without these miRNAs, resulted in enhanced disruption of endothelial cell layers. This suggests that exosomal miR-200c, -141 and -429 contribute to maintaining epithelial barrier integrity and prevents EMT, which explains the increased metastasis in chemoresistant CRC (144,145).
Another tumor suppressor exosomal miRNA is miR-1255b-5p, which has been found to be an EMT inhibitory miRNA downregulated in serum exosomes of CRC patients (146). CRC-derived exosomal miR-1255b-5p, which downregulates human telomerase reverse transcriptase (hTERT) and inhibits Wnt/β-catenin signaling, has been found to be downregulated under hypoxic conditions, providing a link between hypoxic regulation, telomere maintenance and EMT (146).
CRC exosomal miRNAs, EMT and CSCs
CSC and EMT plasticity can also be modulated by miRNA action, involving the deregulation of key tumor suppressor miRNAs, such as miR-200c, miR-203 and miR-183 repressed by TGF-β/Zeb1 (147); and the let-7 downregulation of high mobility group A2 (HMGA2), implicated in TGF-β/Smad/SNAIL, SLUG transcriptional activation (148).
As exosomes are considered to play a role in CSC homeostasis (149), exosomal miRNA cargoes are also implicated in modulating stemness. Bone marrow-derived mesenchymal stem cells have been found to produce exosomes enriched with miR-142-3p, which enlarges the colorectal CSC population by targeting Numb, an inhibitor of Notch signaling (150). Certain CSC-associated miRNAs have been found to be possible CRC tissue and serum biomarkers, particularly miR-18a as a metastatic serum marker (151). Exosomal miR-92a-3p secreted from CAFs has also been found to promote both stemness and EMT in CRC by downregulating FBXW7 and MOAP1 (128). Exosomal miR-128-3p is a tumor suppressor that targets B lymphoma Mo-MLV insertion region 1 homolog (Bmi1), upregulating E-cadherin and inhibiting EMT, as well as the multidrug resistance-associated protein 5 (MRP5) drug transporter, re-sensitizing resistant cells to oxaliplatin (152). Drug resistance is another phenotype closely associated with both EMT and CSCs, particularly given the important role of CSCs in mediating tumor recurrence and metastasis. CAF-derived exosomes have similarly been implicated in promoting chemoresistance through the priming of CSCs (153).
Downregulation or disposal of tumor suppressor exomiRs enable invasion and metastasis in CRC
The downregulation of tumor suppressive exosomal miRNAs, as well as disposal via cargo sorting are also resorted to by CRC cells to promote invasion and metastasis. The tumor suppressive miR-149 and miR-96-5p are both downregulated in exosomes of CRC cells, while expression levels of glypican-1 (GPC1), its direct in vitro and in vivo target, and which induces EMT and promotes invasion, is increased (131).
The tumor suppressive miR-486-5p is downregulated in CRC tissues, in part because of promoter hypermethylation. miR-486-5p is a negative regulator of pleiomorphic adenoma gene-like 2 (PLAGL2), a transcription factor for β-catenin and insulin-like growth factor 2 (IGF2) with roles in promoting proliferation, cell survival and metastasis, as well as decreasing E-cadherin and increasing N-cadherin expression. Interestingly, miR-486-5p was found to be particularly enriched in plasma exosomes (154), suggesting that preferential exomiR cargo loading may be at play to evade its tumor suppressive effects.
Similar observations have been reported for the tumor suppressive miR-8073 and miR-193a. While miR-8073 has invariant expression intracellularly and in exosomes of normal colorectal cells, it is preferentially sorted into exosomes at up to 60 times the concentration found inside CRC cells. Thus, its oncogenic mRNA targets are effectively de-repressed. These include, among others, FOXM1, which is involved in cancer growth and metastasis, as well as Methyl-CpG-binding domain protein 3 (MBD3) which is known to induce pluripotent stem cells (155). In metastatic colon cancer in the liver, tumor cells selectively sort miR-193a out of cells via exosomes. Thus, its direct target inside the cell, Caprin1, is de-repressed and leads to G1 cell cycle arrest, and consequently, cell proliferation. Likewise, miR-193a has been found to be enriched in the exosomal fraction of serum from patients with CRC, particularly in advanced stages of the disease with higher risks of metastasis (156).
Exosomal miRNAs in the preparation of the pre-metastatic niche
The journey of metastatic cells continues way after they have detached from the primary tumor, remodeled their cytoskeletal architecture, and breached tissue boundaries. The remaining steps of the metastatic cascade are fraught with further hurdles that include intravasation into the circulation, extravasation into the secondary site, and preparation of the pre-metastatic niche prior to colonization. The latter entails forming new blood vessels as well as immune-proofing of the new microenvironment.
Vascular leakiness and angiogenesis
Tumor growth and metastasis depend on blood vessels for the supply of oxygen and nutrients, for the removal of waste products, and as routes for cancer cells to be able to migrate to a different site (157). However, to stimulate new vessel growth, there must be a balance between activators and inhibitors of angiogenesis (158). Tumor-derived exosomes can shuttle such cargo, including miRNAs which can target anti-angiogenic and pro-angiogenic genes, between cancer cells and endothelial cells (159).
Among the pro-angiogenic exosomal miRNAs in CRC are miR-25-3p, miR-92a, miR-1229, miR-183-5p and miR-1246. CRC cells are able to transfer the metastasis-promoting miR-25-3p to endothelial cells via exosome transfer. By targeting Kruppel-like factor (KLF)2, vascular endothelial growth factor (VEGF) receptor 2 (VEGFR2) expression is upregulated, promoting angiogenesis (160). Targeting KLF4, on the other hand, regulates endothelial integrity through the activation of tight junction proteins, such as claudin, occludin and zonula occludens-1 (ZO-1) (161). CRC exosome-mediated transfer of miR-25-3p is thus able to induce vascular leakiness and can promote pre-metastatic niche formation in secondary sites, such as the liver and lungs. miR-25-3p has also been shown to be enriched in the circulating exosomes of CRC patients with metastasis when compared to those from patients with non-metastatic CRC (162).
Exosomal miR-92a-3p facilitates tumor angiogenesis by inducing partial EMT in endothelial cells (130). Exosomes derived from colon cancer cells and from plasma derived from murine xenograft models which were enriched with miR-92a-3p have been found to stimulate tube formation in human umbilical vein endothelial cells (HUVECs) upon transfer. miR-92a-3p promotes angiogenesis through the downregulation of Dkk-3 and claudin-11 (130,163).
CRC-derived exosomal miR-1229 promotes tube formation in HUVECs through the inhibition of homeodomain-interacting protein kinase 2 (HIPK2) and the subsequent activation of the VEGF pathway. In patients with CRC, serum exosomes harbor increased levels of miR-1229 which correlate with tumor size, lymphatic metastasis, 'tumor, nodes, metastases' (TNM) stage and poor survival (164). The upregulation of miR-183-5p in CRC cell-derived exosomes has been found to enhance angiogenesis by the repression of FOXO1 (165). A pro-angiogenic role has also been demonstrated for miR-1246, which has been found to be contained in microvesicles secreted by CRC cells, and activates Smad1/5/8 signaling via the direct targeting of promyelocytic leukemia (PML) mRNA (166).
Anti-angiogenic exosomal miRNAs in CRC include miR-126, miR-125a-3p and miR-125a-5p. miR-126 has been reported to target VEGF, an activator of angiogenesis (167) as well as its negative regulators, such as Sprouty-related, EVH1 domain-containing protein 1 (SPRED1) (168,169). However, in vitro studies on CRC have yielded contradicting results, wherein both the overexpression (170) and silencing (171) of miR-126 have been shown to lead to neo-vessel formation. Nonetheless, patients with metastatic CRC exhibit low miR-126 expression levels which are associated with poor survival, confirming the tumor suppressive role of miR-126 in CRC (167). Increased levels of extracellular miR-126 in the plasma of patients with metastatic CRC could also be a predictive biomarker for resistance to anti-angiogenic treatment using bevacizumab, a monoclonal anti-VEGF antibody (172).
miR-125a-3p targets fucosyltransferase (FUT)5 and FUT6, which regulate the PI3K/Akt signaling pathway. The overexpression of miR-125a-3p has been shown to result in the downregulation of FUT5 and FUT6, subsequently inhibiting the proliferation, migration, invasion and angiogenesis of CRC cells (173). The significant upregulation of miR-125a-3p levels in plasma exosomes is being considered as a useful biomarker for the detection of early-stage colon cancer (174).
miR-125a-5p is a known tumor suppressor in CRC, since it directly targets: i) VEGFA, resulting in reduced tube formation in HUVECs and a suppressed cell proliferation, migration and invasion in CRC (175); ii) Tafazzin (TAZ), a key transducer of the Hippo tumor-suppressor pathway, resulting in inhibited migration, invasion and EMT (176); and iii) B-cell lymphoma 2 (Bcl-2), Bcl-2-like protein 12 (BCL2L12) and myeloid cell leukemia 1 (Mcl-1), resulting in the inhibition of cell proliferation and the promotion of apoptosis (177) in colon cancer cells. However, as a biomarker, miR-125a-5p is barely detectable in plasma exosomes due to its low expression in CRC tissues (174).
Exosomal miRNAs and immunosuppression in the pre-metastatic niche
Tumor-derived exosomes act as intercellular messengers between cancer cells and immune cells to either activate or inhibit immune response and/or escape recognition by the immune system. In tumors, an immunosuppressive microenvironment is mainly induced by inflammation (178). CRC-derived exosomes harboring miR-203 have been shown to differentiate monocytes into TAMs following internalization (121). It has been hypothesized that tumor-derived exosomal miR-203 targets suppressor of cytokine signaling 3 (SOCS3) (179), which is crucial for the activation of M1 macrophages characterized by a pro-inflammatory phenotype. Consequently, the loss of SOCS3 expression in the macrophages results in their anti-inflammatory M2 TAM characteristics (180). In line with this finding, circulating miR-203 has been found to promote liver metastasis in murine xenograft models, suggesting their role in hepatic pre-metastatic niche formation. In patients with CRC, a high miR-203 expression in serum exosomes and a low miR-203 expression in tumor tissues has been shown to be associated with metastasis and a poor prognosis (121).
CRC cell-derived exosomal miR-934 has been found to induce M2 polarization in macrophages, enabling the induced TAMs to promote liver metastasis via secretion of the chemokine C-X-C motif chemokine ligand 13 (CXCL13) to remodel the premetastatic niche (181). Exosomal miR-25, miR-130b and miR-425 have been similarly implicated in TAM polarization and C-X-C motif chemokine 12 (CXCL12)/C-X-C motif chemokine receptor 4 (CXCR4)-dependent liver metastasis (182). On the other hand, CRC cell-derived exosomes have been found to diminish the migration of THP-1 monocytes in vitro via the delivery of let-7d, which can downregulate the chemokine C-C motif chemokine ligand 7 (CCL7) (183), suggesting that exosomal miRNAs can aid immune evasion by interfering with immune cell chemotaxis. A list of exosomal miRNAs that have been implicated in different steps of the metastatic cascade is presented in Table I.
Table I.
Summary of exosomal miRNAs and their affected metastatic processes in CRC.
| Exosomal miRNA (target, if identified) | Process affected | Author, year of publication | (Refs.) |
|---|---|---|---|
| Reciprocal transfer between CRC cells and CAFs
| |||
| miR-10b (PIK3CA) | CAF activation via inhibition of PI3K | Dai et al, 2018 | (127) |
| miR-92a (FBXW7, MOAP1) | Wnt/β-catenin activation | Hu et al, 2019 | (128) |
| miR-21 | Enhanced liver metastasis | Bhome et al, 2017 | (132) |
|
| |||
| Cytoskeletal remodeling, loss of adhesion and ECM degradation
| |||
| miR-106b-3p (DLC-1) | Cytoskeletal rearrangement, adhesion through DLC-1 RhoGAP; cadherin switch | Liu et al, 2020 | (138) |
| miR-210 | Induced EMT and anoikis resistance | Bigagli et al, 2016 | (123) |
| miR-1246 | Macrophage reprogramming into TAM | Cooks et al, 2018 | (142) |
|
| |||
| EMT reversal (MET) or EMT inhibition
| |||
| miR-200c, miR-141 | Upregulated in decitabine-treated CRC cells, resulting in pro-epithelial phenotype in metastatic CRC cells | Tanaka et al, 2015 | (143) |
| miR-200c, miR-141, miR-429 | Downregulated in exosomes from 5FU-resistant | Holzner et al, 2016 | (145) |
| (ZEB transcription factors) | CRC cells, resulting in enhanced endothelial disruption | Senfter et al, 2015 | (144) |
| miR-1255b-5p (hTERT) | Downregulated in hypoxia; inhibits Wnt signaling | Zhang et al, 2020 | (146) |
|
| |||
| Maintenance of CSCs
| |||
| miR-142-3p (Numb) | Relieves inhibition of Notch signaling, enlarging the CSC population | Li and Li, 2018 | (150) |
| miR-92a-3p (FBXW7, MOAP1) | Promotes stemness and EMT in CRC cells | Hu et al, 2019 | (128) |
| miR-128-3p (Bmi1) (MRP5) | Upregulation of E-cadherin, EMT inhibition Sensitization to oxaliplatin | Liu et al, 2019 | (138) |
|
| |||
| Downregulation or disposal of tumor suppressor exomiRs
| |||
| miR-149, miR-96-5p (GPC1) | Downregulated in CRC exosomes, increasing pro-EMT GPC-1 expression | Li et al, 2017 | (131) |
| miR-486-5p (PLAGL2) | Disposed in CRC exosomes, promoting β-catenin and IGF2 pathways | Liu et al, 2018 | (154) |
| miR-8073 (FOXM1, MBD3, CCND1, KLK10, CASP2) | Disposed in exosomes, derepressing pro-oncogenic target genes | Mizoguchi et al, 2018 | (155) |
| miR-193a (Caprin1) | Disposed in exosomes, promoting cell cycle progression and proliferation | Teng et al, 2017 | (156) |
|
| |||
| Vascular leakiness and angiogenesis
| |||
| miR-25-3p (KLF2, KLF4) | Promotes vascular permeability and angiogenesis in endothelial cells | Zeng et al, 2018 | (162) |
| miR-92a-3p (Dkk-3) | Wnt/β-catenin activation | Yamada et al, 2013 | (163) |
| (claudin-11) | Tight junction disruption | Yamada et al, 2019 | (130) |
| miR-1229 (HIPK2) | VEGF pathway activation | Hu et al, 2019 | (164) |
| miR-183-5p (FOXO1) | Enhances angiogenesis | Shang et al, 2020 | (165) |
| miR-1246 (PML) | Activation of Smad1/5/8 signaling | Yamada et al, 2014 | (166) |
| miR-126 | Anti-angiogenic (VEGF inhibition) | Ebrahimi et al, 2015 | (167) |
| Pro-angiogenic signaling | Hansen et al, 2011 | (170) | |
| miR-125a-3p (FUT5, FUT6) | Upregulated in plasma exosomes; PI3K/Akt regulation; tumor suppressive miRNA | Wang et al, 2017; | (174) |
| Liang et al, 2017 | (173) | ||
| miR-125a-5p (VEGFA) | Downregulated in plasma exosomes; anti-angiogenic and tumor suppressive | Wang et al, 2017; | (174) |
| Yang et al, 2018 | (175) | ||
|
| |||
| Modulation or suppression of the immune system
| |||
| miR-203 (SOCS3) | Pro-M2/TAM monocyte differentiation | Takano et al, 2017 | (121) |
| miR-934 (PTEN) | Pro-M2/TAM polarization, CXCL13 secretion directing liver metastasis | Zhao et al, 2020 | (181) |
| miR-25, miR-130b, miR-425 (PTEN) | Pro-M2/TAM polarization in CXCL12/CXCR4-dependent liver metastasis | Wang et al, 2020 | (182) |
| let-7d (CCL7) | Inhibition of monocyte migration (immune evasion by interfering with chemotaxis) | Noh et al, 2020 | (183) |
5. Conclusion and perspectives
Much has been achieved in terms of identifying the morphological and structural changes that accompany EMT, the signaling pathways involved, and the transcriptional reprogramming that has to take place to bring about these changes. Investigations into the contributions of the tumor microenvironment to cancer progression has provided further insight on the mechanisms through which tumor cells can modify stromal cells, and vice versa, to break free from the primary site en route to its metastatic destination. More importantly, the discovery of exosomes and their cargo was pivotal in elucidating the mechanisms of tumor-stroma crosstalk, regulation of genes involved in metastatic dissemination, metastatic organotropism, and preparation of the pre-metastatic niche. The functional elucidation of individual exomiRs, in particular, clarified the mechanisms through which invading and metastasizing cells are able to breach barriers and overcome sheer stress in the circulation to enable metastasis to distant sites.
Exosomes are an ideal source of disease biomarkers since they contain genetic material representative of the parental tumor (92,140) and the lipid bilayer membrane protects exosome cargo from nuclease degradation and unfavorable storage conditions (184,185). Cancer cells also secrete significantly more exosomes than normal cells (186), resulting in the enrichment of cancer-derived exosomes in all types of biological fluids, making them easy to obtain in a non-invasive or minimally invasive manner. For instance, serum exosome levels of seven miRNAs (let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223 and miR-23a) have been found to be increased in patients with CRC compared with the healthy controls, and the expression levels significantly decreased following the surgical resection of tumors (140). Exosomal miR-19a has also been identified as a serum biomarker for recurrence of CRC. Compared with healthy individuals, circulating exosomal miR-19a was more abundant in patients with CRC, regardless of stage of the disease, and was associated with a poorer patient prognosis (187). Additionally, tumor-derived exosomes may be useful biomarkers to predict future sites of organ metastasis. Integrins on the exosome surface have been reported to bind to specific cell types to prepare them as pre-metastatic niches (7).
Given their role in disease pathogenesis, exosomes can serve as therapeutic targets, either by inhibiting exosome formation, release, and uptake or by targeting bioactive cargo that can contribute to tumor metastasis. Exosomes can also serve as therapeutic agents, as unlike common drug delivery vehicles, such as liposomes and polymer nanoparticles, exosomes have minimal immunogenicity and toxicity and can be modified with synthetic peptides to carry small molecule drugs for targeting specific cells and tissues. Furthermore, unlike monoclonal antibodies (mAbs), which are used as targeted drug delivery vehicles, smaller iterations of therapeutic mAbs, such as fragment antibodies, domain antibodies and nanobodies can themselves be part of the exosomal cargo that can be internalized by recipient cells. Lastly, the exomiRs implicated in metastasis can themselves be potential targets for antagomiRs loaded onto exosomes.
Acknowledgments
The figures presented herein were created with BioRender. com on a student plan premium license.
Funding Statement
The present study was funded by grants from the Philippine Council for Health Research and Development (grant no. FP150025), the University of the Philippines System (OVPAA-EIDR code 06-008), the University of the Philippines Diliman Office of the Vice Chancellor for Research and Development (grant project no. 181809 PNSE), and the National Institute of Molecular Biology and Biotechnology, University of the Philippines Diliman (in-house grant).
Availability of data and materials
Not applicable.
Authors' contributions
All three authors (JMCD, AGGU and RLG) contributed equally in organizing and writing the present review article. JMCD, AGGU and RLG confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021 Feb 4; doi: 10.3322/caac.21660. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Augestad KM, Bakaki PM, Rose J, Crawshaw BP, Lindsetmo RO, Dørum LM, Koroukian SM, Delaney CP. Metastatic spread pattern after curative colorectal cancer surgery. A retrospective, longitudinal analysis. Cancer Epidemiol. 2015;39:734–744. doi: 10.1016/j.canep.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Van Cutsem E, Cervantes A, Nordlinger B. Metastatic colorectal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii1–iii9. doi: 10.1093/annonc/mdu260. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 5.Nozawa H, Kawai K, Hata K, Tanaka T, Nishikawa T, Otani K, Sasaki K, Kaneko M, Emoto S, Murono K. High-risk stage II colorectal cancers carry an equivalent risk of peritoneal recurrence to stage III. In Vivo. 2018;32:1235–1240. doi: 10.21873/invivo.11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hutchinson L. Understanding metastasis. Nat Rev Clin Oncol. 2015;12:247. doi: 10.1038/nrclinonc.2015.71. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soares AR, Martins-Marques T, Ribeiro-Rodrigues T, Ferreira JV, Catarino S, Pinho MJ, Zuzarte M, Isabel Anjo S, Manadas BPG, Sluijter J, et al. Gap junctional protein Cx43 is involved in the communication between extracellular vesicles and mammalian cells. Sci Rep. 2015;5:13243. doi: 10.1038/srep13243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 10.Fristrom D. The cellular basis of epithelial morphogenesis. A review Tissue Cell. 1988;20:645–690. doi: 10.1016/0040-8166(88)90015-8. [DOI] [PubMed] [Google Scholar]
- 11.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soltermann A, Tischler V, Arbogast S, Braun J, Probst-Hensch N, Weder W, Moch H, Kristiansen G. Prognostic significance of epithelial-mesenchymal and mesenchymal-epithelial transition protein expression in non-small cell lung cancer. Clin Cancer Res. 2008;14:7430–7437. doi: 10.1158/1078-0432.CCR-08-0935. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Chen L, Deng H, Xu B, Li M, Zheng X, Wu C, Jiang J. Epithelial-to-mesenchymal transition in human esophageal cancer associates with tumor progression and patient's survival. Int J Clin Exp Pathol. 2014;7:6943–6949. [PMC free article] [PubMed] [Google Scholar]
- 14.Handra-Luca A, Hong SM, Walter K, Wolfgang C, Hruban R, Goggins M. Tumour epithelial vimentin expression and outcome of pancreatic ductal adenocarcinomas. Br J Cancer. 2011;104:1296–1302. doi: 10.1038/bjc.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DiMeo TA, Anderson K, Phadke P, Feng C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell self-renewal and epithelial-mesenchymal transition in basal-like breast cancer. Cancer Res. 2009;69:5364–5373. doi: 10.1158/0008-5472.CAN-08-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shioiri M, Shida T, Koda K, Oda K, Seike K, Nishimura M, Takano S, Miyazaki M. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br J Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spaderna S, Schmalhofer O, Hlubek F, Berx G, Eger A, Merkel S, Jung A, Kirchner T, Brabletz T. A transient, EMT-Linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131:830–840. doi: 10.1053/j.gastro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida-Noro C, Suzuki N, Takeichi M. Molecular nature of the calcium-dependent cell-cell adhesion system in mouse teratocarcinoma and embryonic cells studied with a monoclonal antibody. Dev Biol. 1984;101:19–27. doi: 10.1016/0012-1606(84)90112-X. [DOI] [PubMed] [Google Scholar]
- 20.Frixen UH, Behrens J, Sachs M, Eberle G, Voss B, Warda A, Löchner D, Birchmeier W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J Cell Biol. 1991;113:173–185. doi: 10.1083/jcb.113.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: Direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116(Pt 10):1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 22.Vandewalle C, Comijn J, De Craene B, Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F, Berx G. SIP1/ZEB2 induces EMT by repressing genes of different epithelial cell-cell junctions. Nucleic Acids Res. 2005;33:6566–6578. doi: 10.1093/nar/gki965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maretzky T, Reiss K, Ludwig A, Buchholz J, Scholz F, Proksch E, de Strooper B, Hartmann D, Saftig P. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and -catenin translocation. Proc Natl Acad Sci USA. 2005;102:9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb Perspect Biol. 2010;2:a002915. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno-Bueno G, Portillo F, Cano A. Transcriptional regulation of cell polarity in EMT and cancer. Oncogene. 2008;27:6958–6969. doi: 10.1038/onc.2008.346. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki A, Yamanaka T, Hirose T, Manabe N, Mizuno K, Shimizu M, Akimoto K, Izumi Y, Ohnishi T, Ohno S. Atypical protein kinase C is involved in the evolutionarily conserved par protein complex and plays a critical role in establishing epithelia-specific junctional structures. J Cell Biol. 2001;152:1183–1196. doi: 10.1083/jcb.152.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B. The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol. 2002;157:161–172. doi: 10.1083/jcb.200109010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 30.Bilder D, Li M, Perrimon N. Cooperative regulation of cell polarity and growth by drosophila tumor suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 31.Navarro C, Nola S, Audebert S, Santoni MJ, Arsanto JP, Ginestier C, Marchetto S, Jacquemier J, Isnardon D, Le Bivic A, et al. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene. 2005;24:4330–4339. doi: 10.1038/sj.onc.1208632. [DOI] [PubMed] [Google Scholar]
- 32.Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED, Thompson EW. Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J Cell Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 33.Haynes J, Srivastava J, Madson N, Wittmann T, Barber DL. Dynamic actin remodeling during epithelial-mesenchymal transition depends on increased moesin expression. Mol Biol Cell. 2011;22:4750–4764. doi: 10.1091/mbc.e11-02-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 35.Micalizzi DS, Farabaugh SM, Ford HL. Epithelial-mesenchymal transition in cancer: Parallels between normal development and tumor progression. J Mammary Gland Biol Neoplasia. 2010;15:117–134. doi: 10.1007/s10911-010-9178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagan R, Martín I, Alonso A, Llobera M, Vilaró S. Vimentin filaments follow the preexisting cytokeratin network during epithelial-mesenchymal transition of cultured neonatal rat hepatocytes. Exp Cell Res. 1996;222:333–344. doi: 10.1006/excr.1996.0043. [DOI] [PubMed] [Google Scholar]
- 37.Chen WT. Proteolytic activity of specialized surface protrusions formed at rosette contact sites of transformed cells. J Exp Zool. 1989;251:167–185. doi: 10.1002/jez.1402510206. [DOI] [PubMed] [Google Scholar]
- 38.Tsai JH, Yang J. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 2013;27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang GJ, Zhou T, Tian HP, Liu ZL, Xia SS. High expression of ZEB1 correlates with liver metastasis and poor prognosis in colorectal cancer. Oncol Lett. 2013;5:564–568. doi: 10.3892/ol.2012.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kahlert C, Lahes S, Radhakrishnan P, Dutta S, Mogler C, Herpel E, Brand K, Steinert G, Schneider M, Mollenhauer M, et al. Overexpression of ZEB2 at the invasion front of colorectal cancer is an independent prognostic marker and regulates tumor invasion in vitro. Clin Cancer Res. 2011;17:7654–7663. doi: 10.1158/1078-0432.CCR-10-2816. [DOI] [PubMed] [Google Scholar]
- 42.Gomez I, Peña C, Herrera M, Muñoz C, Larriba MJ, Garcia V, Dominguez G, Silva J, Rodriguez R, Garcia de Herreros A, et al. TWIST1 is expressed in colorectal carcinomas and predicts patient survival. PLoS One. 2011;6:e18023. doi: 10.1371/journal.pone.0018023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, Jin GZ, Liu K, Dong H, Yu H, Duan JC, Li Z, Dong W, Cong WM, Yang JH. Twist2 is a valuable prognostic biomarker for colorectal cancer. World J Gastroenterol. 2013;19:2404–2411. doi: 10.3748/wjg.v19.i15.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Wu J, Wei P, Xu Y, Zhuo C, Wang Y, Li D, Cai S. Overexpression of forkhead Box C2 promotes tumor metastasis and indicates poor prognosis in colon cancer via regulating epithelial-mesenchymal transition. Am J Cancer Res. 2015;5:2022–2034. [PMC free article] [PubMed] [Google Scholar]
- 45.Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 are promising prognostic biomarkers and novel targets of tumor-suppressive miR-342 in human colorectal cancer. Clin Cancer Res. 2016;22:4947–4957. doi: 10.1158/1078-0432.CCR-16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li D, Li Q, Zhuo C, Xu Y, Cai S. Contribution of FOXC1 to the progression and metastasis and prognosis of human colon cancer. J Clin Oncol. 2015;33(Suppl 3):S636. doi: 10.1200/jco.2015.33.3_suppl.636. [DOI] [Google Scholar]
- 47.Peinado H, Ballestar E, Esteller M, Cano A. Snail Mediates E-cadherin repression by the recruitment of the Sin3A/Histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol Cell Biol. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bolos V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: A comparison with Snail and E47 repressors. J Cell Sci. 2003;116(Pt 3):499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 49.Caramel J, Papadogeorgakis E, Hill L, Browne GJ, Richard G, Wierinckx A, Saldanha G, Osborne J, Hutchinson P, Tse G, et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell. 2013;24:466–480. doi: 10.1016/j.ccr.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 50.Vu T, Datta P. Regulation of EMT in colorectal cancer: A culprit in metastasis. Cancers (Basel) 2017;9:171. doi: 10.3390/cancers9120171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat Cell Biol. 2019;21:102–112. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- 52.Zhao S, Venkatasubbarao K, Lazor JW, Sperry J, Jin C, Cao L, Freeman JW. Inhibition of STAT3 Tyr705 Phosphorylation by Smad4 suppresses transforming growth factor beta-mediated invasion and metastasis in pancreatic cancer cells. Cancer Res. 2008;68:4221–4228. doi: 10.1158/0008-5472.CAN-07-5123. [DOI] [PubMed] [Google Scholar]
- 53.Zavadil J, Bitzer M, Liang D, Yang YC, Massimi A, Kneitz S, Piek E, Bottinger EP. Genetic programs of epithelial cell plasticity directed by transforming growth factor-beta. Proc Natl Acad Sci USA. 2001;98:6686–6691. doi: 10.1073/pnas.111614398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkes MC, Mitchell H, Penheiter SG, Doré JJ, Suzuki K, Edens M, Sharma DK, Pagano RE, Leof EB. Transforming growth factor-beta activation of phosphatidylinositol 3-Kinase is independent of Smad2 and Smad3 and regulates fibroblast responses via p21-Activated Kinase-2. Cancer Res. 2005;65:10431–10440. doi: 10.1158/0008-5472.CAN-05-1522. [DOI] [PubMed] [Google Scholar]
- 55.Gujral TS, Chan M, Peshkin L, Sorger PK, Kirschner MW, MacBeath G. A Noncanonical Frizzled2 pathway regulates epithelial-mesenchymal transition and metastasis. Cell. 2014;159:844–856. doi: 10.1016/j.cell.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang RY, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci. 2012;125(Pt 19):4417–4422. doi: 10.1242/jcs.099697. [DOI] [PubMed] [Google Scholar]
- 57.Lehembre F, Yilmaz M, Wicki A, Schomber T, Strittmatter K, Ziegler D, Kren A, Went P, Derksen PW, Berns A, et al. NCAM-induced focal adhesion assembly: A functional switch upon loss of E-cadherin. EMBO J. 2008;27:2603–2615. doi: 10.1038/emboj.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 60.Müller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 61.Chang Q, Bournazou E, Sansone P, Berishaj M, Gao SP, Daly L, Wels J, Theilen T, Granitto S, Zhang X, et al. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia. 2013;15:848–862. doi: 10.1593/neo.13706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cox TR, Rumney RMH, Schoof EM, Perryman L, Høye AM, Agrawal A, Bird D, Latif NA, Forrest H, Evans HR, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Reichert M, Bakir B, Moreira L, Pitarresi JR, Feldmann K, Simon L, Suzuki K, Maddipati R, Rhim AD, Schlitter AM, et al. Regulation of epithelial plasticity determines metastatic organotropism in pancreatic cancer. Dev Cell. 2018;45:696–711.e8. doi: 10.1016/j.devcel.2018.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shaul YD, Freinkman E, Comb WC, Cantor JR, Tam WL, Thiru P, Kim D, Kanarek N, Pacold ME, Chen WW, et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim NH, Cha YH, Lee J, Lee SH, Yang JH, Yun JS, Cho ES, Zhang X, Nam M, Kim N, et al. Snail reprograms glucose metabolism by repressing phosphofructokinase PFKP allowing cancer cell survival under metabolic stress. Nat Commun. 2017;8:14374. doi: 10.1038/ncomms14374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-Induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 67.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hiratsuka S, Nakamura K, Iwai S, Murakami M, Itoh T, Kijima H, Shipley JM, Senior RM, Shibuya M. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell. 2002;2:289–300. doi: 10.1016/S1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 69.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimaoka M, Kawamoto E, Gaowa A, Okamoto T, Park E. Connexins and integrins in exosomes. Cancers. 2019;11:106. doi: 10.3390/cancers11010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu T, Zhang Q, Zhang J, Li C, Miao YR, Lei Q, Li Q, Guo AY. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D89–D93. doi: 10.1093/nar/gky985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simpson RJ, Jensen SS, Lim JWE. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 74.Li M, Zeringer E, Barta T, Schageman J, Cheng A, Vlassov AV. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130502. doi: 10.1098/rstb.2013.0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baroni S, Romero-Cordoba S, Plantamura I, Dugo M, D'Ippolito E, Cataldo A, Cosentino G, Angeloni V, Rossini A, Daidone MG, Iorio MV. Exosome-mediated delivery of miR-9 induces cancer-associated fibroblast-like properties in human breast fibroblasts. Cell Death Dis. 2016;7:e2312. doi: 10.1038/cddis.2016.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Demory Beckler M, Higginbotham JN, Franklin JL, Ham AJ, Halvey PJ, Imasuen IE, Whitwell C, Li M, Liebler DC, Coffey RJ. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol Cell Proteomics. 2013;12:343–355. doi: 10.1074/mcp.M112.022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lucchetti D, Calapà F, Palmieri V, Fanali C, Carbone F, Papa A, De Maria R, De Spirito M, Sgambato A. Differentiation affects the release of exosomes from colon cancer cells and their ability to modulate the behavior of recipient cells. Am J Pathol. 2017;187:1633–1647. doi: 10.1016/j.ajpath.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 78.Lv MM, Zhu XY, Chen WX, Zhong SL, Hu Q, Ma TF, Zhang J, Chen L, Tang JH, Zhao JH. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumor Biol. 2014;35:10773–10779. doi: 10.1007/s13277-014-2377-z. [DOI] [PubMed] [Google Scholar]
- 79.Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. doi: 10.1182/blood.V94.11.3791. [DOI] [PubMed] [Google Scholar]
- 80.Crescitelli R, Lässer C, Szabó TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles. 2013;2:20677. doi: 10.3402/jev.v2i0.20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pan BT, Teng K, Wu C, Adam M, Johnstone RM. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J Cell Biol. 1985;101:942–948. doi: 10.1083/jcb.101.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 83.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. The biogenesis and functions of exosomes. Traffic. 2002;3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 84.Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464:864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 86.Roucourt B, Meeussen S, Bao J, Zimmermann P, David G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015;25:412–428. doi: 10.1038/cr.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 88.Wei D, Zhan W, Gao Y, Huang L, Gong R, Wang W, Zhang R, Wu Y, Gao S, Kang T. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 2021;31:157–177. doi: 10.1038/s41422-020-00409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(19-30):1–13. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 90.Hsu C, Morohashi Y, Yoshimura S, Manrique-Hoyos N, Jung S, Lauterbach MA, Bakhti M, Grønborg M, Möbius W, Rhee J, et al. Regulation of exosome secretion by Rab35 and its GTPase-activating proteins TBC1D10A-C. J Cell Biol. 2010;189:223–232. doi: 10.1083/jcb.200911018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pérez-Boza J, Lion M, Struman I. Exploring the RNA landscape of endothelial exosomes. RNA. 2018;24:423–435. doi: 10.1261/rna.064352.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amorim MG, Valieris R, Drummond RD, Pizzi MP, Freitas VM, Sinigaglia-Coimbra R, Calin GA, Pasqualini R, Arap W, Silva IT, et al. A total transcriptome profiling method for plasma-derived extracellular vesicles: Applications for liquid biopsies. Sci Rep. 2017;7:14395. doi: 10.1038/s41598-017-14264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jenjaroenpun P, Kremenska Y, Nair VM, Kremenskoy M, Joseph B, Kurochkin IV. Characterization of RNA in exosomes secreted by human breast cancer cell lines using next-generation sequencing. PeerJ. 2013;1:e201. doi: 10.7717/peerj.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yuan T, Huang X, Woodcock M, Du M, Dittmar R, Wang Y, Tsai S, Kohli M, Boardman L, Patel T, Wang L. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. doi: 10.1038/srep19413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M. The RNA-Binding Protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling MicroRNA sorting. Cell Rep. 2016;17:799–808. doi: 10.1016/j.celrep.2016.09.031. [DOI] [PubMed] [Google Scholar]
- 99.Gao T, Shu J, Cui J. A systematic approach to RNA-associated motif discovery. BMC Genomics. 2018;19:146. doi: 10.1186/s12864-018-4528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koppers-Lalic D, Hackenberg M, Bijnsdor p IV, van Eijndhoven MAJ, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, et al. Nontemplated Nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 101.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of MicroRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T. Neutral Sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic MicroRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kubota S, Chiba M, Watanabe M, Sakamoto M, Watanabe N. Secretion of small/microRNAs including miR-638 into extracellular spaces by sphingomyelin phosphodiesterase 3. Oncol Rep. 2015;33:67–73. doi: 10.3892/or.2014.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yan S, Dang G, Zhang X, Jin C, Qin L, Wang Y, Shi M, Huang H, Duan Q. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget. 2017;8:72220–72226. doi: 10.18632/oncotarget.19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Guduric-Fuchs J, O'Connor A, Camp B, O'Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McKenzie AJ, Hoshino D, Hong NH, Cha DJ, Franklin JL, Coffey RJ, Patton JG, Weaver AM. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 2016;15:978–987. doi: 10.1016/j.celrep.2016.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Melo SA, Sugimoto H, O'Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. Cancer exosomes perform cell-independent MicroRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–20090. doi: 10.1074/jbc.M301642200. [DOI] [PubMed] [Google Scholar]
- 109.Parolini I, Federici C, Raggi C, Lugini L, Palleschi S, De Milito A, Coscia C, Iessi E, Logozzi M, Molinari A, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Park JE, Tan HS, Datta A, Lai RC, Zhang H, Meng W, Lim SK, Sze SK. Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol Cell Proteomics. 2010;9:1085–1099. doi: 10.1074/mcp.M900381-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci. 2011;124(Pt 3):447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 112.Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J Biol Chem. 2014;289:22258–22267. doi: 10.1074/jbc.M114.588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Harding C, Heuser J, Stahl P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol. 1983;97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183:1161–1172. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle-mediated transfer of long non-coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio. 2014;4:458–467. doi: 10.1016/j.fob.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cha DJ, Franklin JL, Dou Y, Liu Q, Higginbotham JN, Demory Beckler M, Weaver AM, Vickers K, Prasad N, Levy S, et al. KRAS-dependent sorting of miRNA to exosomes. Elife. 2015;4:e07197. doi: 10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou MK, Liu XJ, Zhao ZG, Cheng YM. MicroRNA-100 functions as a tumor suppressor by inhibiting Lgr5 expression in colon cancer cells. Mol Med Rep. 2015;11:2947–2952. doi: 10.3892/mmr.2014.3052. [DOI] [PubMed] [Google Scholar]
- 119.Gastpar R, Gehrmann M, Bausero MA, Asea A, Gross C, Schroeder JA, Multhoff G. Heat shock protein 70 surface-positive tumor exosomes stimulate migratory and cytolytic activity of natural killer cells. Cancer Res. 2005;65:5238–5247. doi: 10.1158/0008-5472.CAN-04-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huber V, Fais S, Iero M, Lugini L, Canese P, Squarcina P, Zaccheddu A, Colone M, Arancia G, Gentile M, et al. Human colorectal cancer cells induce T-Cell death through release of proapoptotic microvesicles: Role in immune escape. Gastroenterology. 2005;128:1796–1804. doi: 10.1053/j.gastro.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 121.Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, et al. Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget. 2017;8:78598–78613. doi: 10.18632/oncotarget.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang Z, Feng Y. Exosomes derived from hypoxic colorectal cancer cells promote angiogenesis through Wnt4-Induced β-catenin signaling in endothelial cells. Oncol Res. 2017;25:651–661. doi: 10.3727/096504016X14752792816791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bigagli E, Luceri C, Guasti D, Cinci L. Exosomes secreted from human colon cancer cells influence the adhesion of neighboring metastatic cells: Role of microRNA-210. Cancer Biol Ther. 2016;17:1062–1069. doi: 10.1080/15384047.2016.1219815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shedden K, Xie XT, Chandaroy P, Chang YT, Rosania GR. Expulsion of small molecules in vesicles shed by cancer cells: Association with gene expression and chemosensitivity profiles. Cancer Res. 2003;63:4331–4337. [PubMed] [Google Scholar]
- 125.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR, Hunter T, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020;20:174–186. doi: 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sappino AP, Skalli O, Jackson B, Schürch W, Gabbiani G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer. 1988;41:707–712. doi: 10.1002/ijc.2910410512. [DOI] [PubMed] [Google Scholar]
- 127.Dai G, Yao X, Zhang Y, Gu J, Geng Y, Xue F, Zhang J. Colorectal cancer cell-derived exosomes containing miR-10b regulate fibroblast cells via the PI3K/Akt pathway. Bull Cancer. 2018;105:336–349. doi: 10.1016/j.bulcan.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 128.Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR, Song FY, Wang FF, Zhu XH, Liao WJ, et al. CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer. 2019;18:91. doi: 10.1186/s12943-019-1019-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alcantara KMM, Garcia RL. MicroRNA-92a promotes cell proliferation, migration and survival by directly targeting the tumor suppressor gene NF2 in colorectal and lung cancer cells. Oncol Rep. 2019;41:2103–2116. doi: 10.3892/or.2019.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yamada NO, Heishima K, Akao Y, Senda T. Extracellular vesicles containing MicroRNA-92a-3p facilitate partial endothelial-mesenchymal transition and angiogenesis in endothelial cells. Int J Mol Sci. 2019;20:4406. doi: 10.3390/ijms20184406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li J, Zhou C, Ni S, Wang S, Ni C, Yang P, Ye M. Methylated claudin-11 associated with metastasis and poor survival of colorectal cancer. Oncotarget. 2017;8:96249–96262. doi: 10.18632/oncotarget.21997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bhome R, Goh RW, Bullock MD, Pillar N, Thirdborough SM, Mellone M, Mirnezami R, Galea D, Veselkov K, Gu Q, et al. Exosomal microRNAs derived from colorectal cancer-associated fibroblasts: Role in driving cancer progression. Aging (Albany NY) 2017;9:2666–2694. doi: 10.18632/aging.101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 135.Kojima Y, Acar A, Eaton EN, Mellody KT, Scheel C, Ben-Porath I, Onder TT, Wang ZC, Richardson AL, Weinberg RA, Orimo A. Autocrine TGF- and stromal cell-derived factor-1 (SDF-1) signaling drives the evolution of tumor-promoting mammary stromal myofibroblasts. Proc Natl Acad Sci USA. 2010;107:20009–20014. doi: 10.1073/pnas.1013805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Philippeos C, Telerman SB, Oulès B, Pisco AO, Shaw TJ, Elgueta R, Lombardi G, Driskell RR, Soldin M, Lynch MD, Watt FM. Spatial and single-cell transcriptional profiling identifies functionally distinct human dermal fibroblast subpopulations. J Invest Dermatol. 2018;138:811–825. doi: 10.1016/j.jid.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Croft AP, Campos J, Jansen K, Turner JD, Marshall J, Attar M, Savary L, Wehmeyer C, Naylor AJ, Kemble S, et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570:246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu H, Liu Y, Sun P, Leng K, Xu Y, Mei L, Han P, Zhang B, Yao K, Li C, et al. Colorectal cancer-derived exosomal miR-106b-3p promotes metastasis by down-regulating DLC-1 expression. Clin Sci (Lond) 2020;134:419–434. doi: 10.1042/CS20191087. [DOI] [PubMed] [Google Scholar]
- 139.Kim TY, Vigil D, Der CJ, Juliano RL. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 2009;28:77–83. doi: 10.1007/s10555-008-9167-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, Gunji T, Ohta H, Okamoto H, Sonoda H, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wei C, Li Y, Huang K, Li G, He M. Exosomal miR-1246 in body fluids is a potential biomarker for gastrointestinal cancer. Biomark Med. 2018;12:1185–1196. doi: 10.2217/bmm-2017-0440. [DOI] [PubMed] [Google Scholar]
- 142.Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, Forshew T, Appella E, Gorgoulis VG, Harris CC. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. 2018;9:771. doi: 10.1038/s41467-018-03224-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tanaka S, Hosokawa M, Ueda K, Iwakawa S. Effects of decitabine on invasion and exosomal expression of miR-200c and miR-141 in oxaliplatin-resistant colorectal cancer cells. Biol Pharm Bull. 2015;38:1272–1279. doi: 10.1248/bpb.b15-00129. [DOI] [PubMed] [Google Scholar]
- 144.Senfter D, Holzner S, Kalipciyan M, Staribacher A, Walzl A, Huttary N, Krieger S, Brenner S, Jäger W, Krupitza G, et al. Loss of miR-200 family in 5-fluorouracil resistant colon cancer drives lymphendothelial invasiveness in vitro. Hum Mol Genet. 2015;24:3689–3698. doi: 10.1093/hmg/ddv113. [DOI] [PubMed] [Google Scholar]
- 145.Holzner S, Senfter D, Stadler S, Staribacher A, Nguyen CH, Gaggl A, Geleff S, Huttary N, Krieger S, Jäger W, et al. Colorectal cancer cell-derived microRNA200 modulates the resistance of adjacent blood endothelial barriers in vitro. Oncol Rep. 2016;36:3065–3071. doi: 10.3892/or.2016.5114. [DOI] [PubMed] [Google Scholar]
- 146.Zhang X, Bai J, Yin H, Long L, Zheng Z, Wang Q, Chen F, Yu X, Zhou Y. Exosomal miR-1255b-5p targets human telomerase reverse transcriptase in colorectal cancer cells to suppress epithelial-to-mesenchymal transition. Mol Oncol. 2020;14:2589–2608. doi: 10.1002/1878-0261.12765. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147.Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- 148.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 149.Xu J, Liao K, Zhou W. Exosomes regulate the transformation of cancer cells in cancer stem cell homeostasis. Stem Cells Int. 2018;2018:4837370. doi: 10.1155/2018/4837370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Li H, Li F. Exosomes from BM-MSCs increase the population of CSCs via transfer of miR-142-3p. Br J Cancer. 2018;119:744–755. doi: 10.1038/s41416-018-0254-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Farace C, Pisano A, Griñan-Lison C, Solinas G, Jiménez G, Serra M, Carrillo E, Scognamillo F, Attene F, Montella A, et al. Deregulation of cancer-stem-cell-associated miRNAs in tissues and sera of colorectal cancer patients. Oncotarget. 2020;11:116–130. doi: 10.18632/oncotarget.27411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, Wang L, Li P, Zhao Y, Duan W, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer. 2019;18:43. doi: 10.1186/s12943-019-0981-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 153.Hu Y, Yan C, Mu L, Huang K, Li X, Tao D, Wu Y, Qin J. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One. 2015;10:e0125625. doi: 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu X, Chen X, Zeng K, Xu M, He B, Pan Y, Sun H, Pan B, Xu X, Xu T, et al. DNA-methylation-mediated silencing of miR-486-5p promotes colorectal cancer proliferation and migration through activation of PLAGL2/IGF2/β-catenin signal pathways. Cell Death Dis. 2018;9:1037. doi: 10.1038/s41419-018-1105-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mizoguchi A, Takayama A, Arai T, Kawauchi J, Sudo H. MicroRNA-8073: Tumor suppressor and potential therapeutic treatment. PLoS One. 2018;13:e0209750. doi: 10.1371/journal.pone.0209750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, Deng Z, Kumar A, Zhang L, Merchant ML, et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun. 2017;8:14448. doi: 10.1038/ncomms14448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Folkman J. Tumor angiogenesis: Therapeutic implications. N Engl J Med. 1971;285:1182–1186. doi: 10.1056/NEJM197111182852108. [DOI] [PubMed] [Google Scholar]
- 158.Dameron KM, Volpert OV, Tainsky MA, Bouck N. Control of angiogenesis in fibroblasts by p53 regulation of thrombospondin-1. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 159.Soheilifar MH, Grusch M, Neghab HK, Amini R, Maadi H, Saidijam M, Wang Z. Angioregulatory microRNAs in colorectal cancer. Cancers (Basel) 2019;12:71. doi: 10.3390/cancers12010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bhattacharya R, SenBanerjee S, Lin Z, Mir S, Hamik A, Wang P, Mukherjee P, Mukhopadhyay D, Jain MK. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–28851. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- 161.Ma J, Wang P, Liu Y, Zhao L, Li Z, Xue Y. Krüppel-Like Factor 4 regulates blood-tumor barrier permeability via ZO-1, Occludin and Claudin-5. J Cell Physiol. 2014;229:916–926. doi: 10.1002/jcp.24523. [DOI] [PubMed] [Google Scholar]
- 162.Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yamada N, Nakagawa Y, Tsujimura N, Kumazaki M, Noguchi S, Mori T, Hirata I, Maruo K, Akao Y. Role of intracellular and extracellular MicroRNA-92a in colorectal cancer. Transl Oncol. 2013;6:482–492. doi: 10.1593/tlo.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, Chen Q. Exosomal miR-1229 derived from colorectal cancer cells promotes angiogenesis by targeting HIPK2. Int J Biol Macromol. 2019;132:470–477. doi: 10.1016/j.ijbiomac.2019.03.221. [DOI] [PubMed] [Google Scholar]
- 165.Shang A, Wang X, Gu C, Liu W, Sun J, Zeng B, Chen C, Ji P, Wu J, Quan W, et al. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging (Albany NY) 2020;12:8352–8371. doi: 10.18632/aging.103145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yamada N, Tsujimura N, Kumazaki M, Shinohara H, Taniguchi K, Nakagawa Y, Naoe T, Akao Y. Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochim Biophys Acta. 2014;1839:1256–1272. doi: 10.1016/j.bbagrm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 167.Ebrahimi F, Gopalan V, Wahab R, Lu CT, Smith RA, Lam AK. Deregulation of miR-126 expression in colorectal cancer pathogenesis and its clinical significance. Exp Cell Res. 2015;339:333–341. doi: 10.1016/j.yexcr.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 168.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. MiR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 2010;464:1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hansen TF, Andersen CL, Nielsen BS, Spindler KL, Sørensen FB, Lindebjerg J, Brandslund I, Jakobsen A. Elevated microRNA-126 is associated with high vascular endothelial growth factor receptor 2 expression levels and high microvessel density in colorectal cancer. Oncol Lett. 2011;2:1101–1106. doi: 10.3892/ol.2011.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhang Y, Wang X, Xu B, Wang B, Wang Z, Liang Y, Zhou J, Hu J, Jiang B. Epigenetic silencing of miR-126 contributes to tumor invasion and angiogenesis in colorectal cancer. Oncol Rep. 2013;30:1976–1984. doi: 10.3892/or.2013.2633. [DOI] [PubMed] [Google Scholar]
- 172.Hansen TF, Carlsen AL, Heegaard NH, Sørensen FB, Jakobsen A. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer. 2015;112:624–629. doi: 10.1038/bjc.2014.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Liang L, Gao C, Li Y, Sun M, Xu J, Li H, Jia L, Zhao Y. MiR-125a-3p/FUT5-FUT6 axis mediates colorectal cancer cell proliferation, migration, invasion and pathological angiogenesis via PI3K-Akt pathway. Cell Death Dis. 2017;8:e2968. doi: 10.1038/cddis.2017.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang J, Yan F, Zhao Q, Zhan F, Wang R, Wang L, Zhang Y, Huang X. Circulating exosomal miR-125a-3p as a novel biomarker for early-stage colon cancer. Sci Rep. 2017;7:4150. doi: 10.1038/s41598-017-04386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yang X, Qiu J, Kang H, Wang Y, Qian J. MiR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag Res. 2018;10:5839–5853. doi: 10.2147/CMAR.S161990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Tang L, Zhou L, Wu S, Shi X, Jiang G, Niu S, Ding D. MiR-125a-5p inhibits colorectal cancer cell epithelial-mesenchymal transition, invasion and migration by targeting TAZ. Onco Targets Ther. 2019;12:3481–3489. doi: 10.2147/OTT.S191247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Tong Z, Liu N, Lin L, Guo X, Yang D, Zhang Q. MiR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed Pharmacother. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 178.Katoh H, Wang D, Daikoku T, Sun H, Dey SK, DuBois RN. CXCR2-expressing myeloid-derived suppressor cells are essential to promote colitis-associated tumorigenesis. Cancer Cell. 2013;24:631–644. doi: 10.1016/j.ccr.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ru P, Steele R, Hsueh EC, Ray RB. Anti-miR-203 Upregulates SOCS3 expression in breast cancer cells and enhances cisplatin chemosensitivity. Genes Cancer. 2011;2:720–727. doi: 10.1177/1947601911425832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Liu Y, Stewart KN, Bishop E, Marek CJ, Kluth DC, Rees AJ, Wilson HM. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J Immunol. 2008;180:6270–6278. doi: 10.4049/jimmunol.180.9.6270. [DOI] [PubMed] [Google Scholar]
- 181.Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13:156. doi: 10.1186/s13045-020-00991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020;474:36–52. doi: 10.1016/j.canlet.2020.01.005. [DOI] [PubMed] [Google Scholar]
- 183.Noh GT, Kwon J, Kim J, Park M, Choi DW, Cho KA, Woo SY, Oh BY, Lee KY, Lee RA. Verification of the role of exosomal microRNA in colorectal tumorigenesis using human colorectal cancer cell lines. PLoS One. 2020;15:e0242057. doi: 10.1371/journal.pone.0242057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. 2014;3:23743. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. MiRNA in plasma exosome is stable under different storage conditions. Molecules. 2014;19:1568–1575. doi: 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal MicroRNA: A diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 187.Matsumura T, Sugimachi K, Iinuma H, Takahashi Y, Kurashige J, Sawada G, Ueda M, Uchi R, Ueo H, Takano Y, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer. 2015;113:275–281. doi: 10.1038/bjc.2015.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.