ABSTRACT
Tendons and ligaments are fibrous connective tissues vital to the transmission of force and stabilization of the musculoskeletal system. Arising in precise regions of the embryo, tendons and ligaments share many properties and little is known about the molecular differences that differentiate them. Recent studies have revealed heterogeneity and plasticity within tendon and ligament cells, raising questions regarding the developmental mechanisms regulating tendon and ligament identity. Here, we discuss recent findings that contribute to our understanding of the mechanisms that establish and maintain tendon progenitors and their differentiated progeny in the head, trunk and limb. We also review the extent to which these findings are specific to certain anatomical regions and model organisms, and indicate which findings similarly apply to ligaments. Finally, we address current research regarding the cellular lineages that contribute to tendon and ligament repair, and to what extent their regulation is conserved within tendon and ligament development.
KEY WORDS: Tendon, Ligament, Enthesis, Scx, Tenocyte, Fibrous connective tissue, Fgf
Summary: This Review summarizes the establishment and maintenance of tendon progenitors and their cellular lineages in different anatomical regions and organisms, and discusses the extent to which these processes are conserved between tendons and ligaments.
Introduction
Tendons and ligaments are closely related dense fibrous connective tissues that play vital roles in musculoskeletal mobility and stability. Tendons connect muscle to bone and facilitate body movement by transmitting tensile forces and storing elastic energy, whereas ligaments join bone to bone, stabilizing joints and guiding movement through the normal range of motion (Benjamin et al., 2008; Connizzo et al., 2013; Murchison et al., 2007). The functions of tendons and ligaments rely on their strong yet flexible structure of collagen fibrils, which are hierarchically organized and bundled by connective tissue sheaths. Though tendon and ligament are similarly composed of collagen 1 (Col1) fibrils, proteoglycans, elastin and glycoproteins, a few differences do exist. Col1 fibrils, which are aligned parallel to one another along the longitudinal axis in tendons, are multidirectional and less densely packed in ligaments to better support tensile load (Amiel et al., 1983; Amis, 2004; Jung et al., 2009; Tozer and Duprez, 2005) (Fig. 1).
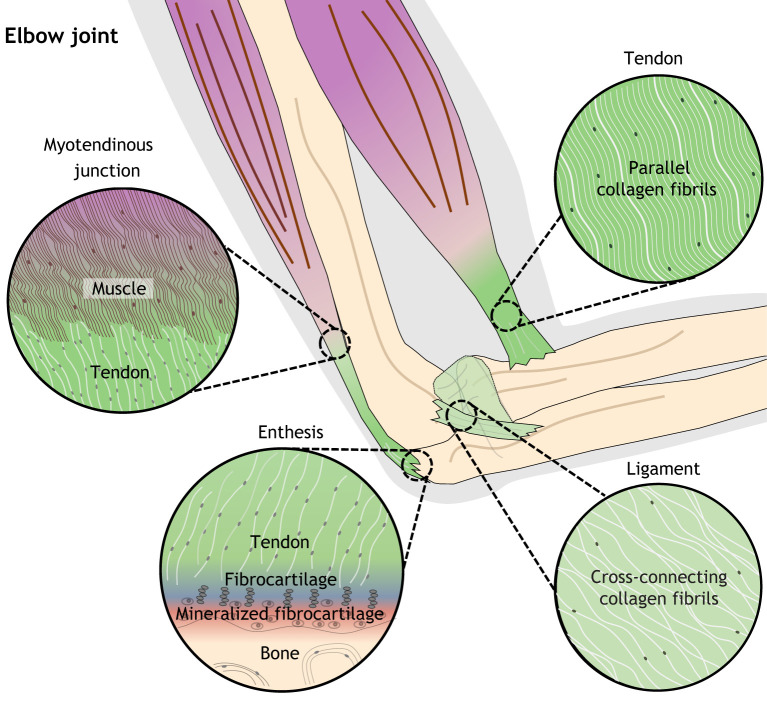
Fig. 1.
Tissue morphology of tendon, ligament, and musculoskeletal junctions of the human elbow. (Clockwise from top right) Tendons (which connect muscle to bone) and ligaments (which connect and stabilize bones) share similar properties but have slightly different orientation of collagen fibrils; whereas tendons have parallel arrangements of fibrils, ligament fibrils can overlap and cross. The tendon enthesis features graded, intermediate tissue types that facilitate the transmission of force from tendon to bone, whereas the myotendinous junction facilitates the joining of tendon and muscle through interactions of ECM proteins.
Individual tendons and ligaments have unique morphological properties adapted for specific roles. Remarkable variability exists in tendon and ligament shape, which can form cords, strap-like bands, flat ribbons, discs and fan-like structures to accommodate distinct mechanical and anatomical environments (Benjamin et al., 2008; Connizzo et al., 2013). Morphological diversity among tendons and ligaments is further enhanced through their variable association with synovial sheaths, bursae, fibrous retinacula and fat pads that function to lubricate, anchor, support and provide mechanosensory input (Benjamin et al., 2008). In addition, there is morphological variability along tendons, with fibrocartilage forming at bone insertions, sesamoid bones forming within regions that cross over joint surfaces, and myotendinous junctions where tendon unites with muscle (Fig. 1). Genome-wide expression analysis of porcine adult tendon and ligament tissue from distinct regions and subtypes suggests that morphological and functional differences largely correlate with variation in gene expression levels rather than activation of subtype-specific genes (Pearse et al., 2009). The extent to which embryonic factors are sufficient to establish location-specific morphology has not been directly investigated. Undoubtedly, morphological variation among tendon and ligament subtypes will be key to interpreting region-specific differences in development and repair.
Tendons and ligaments arise from distinct embryonic origins depending on their position along the rostro-caudal axis (Fig. 2). Tendons and ligaments in the head arise from cranial neural crest cells (CNCCs), whereas those in the neck and shoulder girdle are derived from lateral plate mesoderm, CNCCs and paraxial mesoderm (Durland et al., 2008; Evans and Noden, 2006; Köntges and Lumsden, 1996; Matsuoka et al., 2005; Nagashima et al., 2016; Valasek et al., 2010; Ziermann et al., 2018). Lineage tracing of CNCCs and mesoderm in the murine neck suggests that the embryonic origin of the tendon matches that of the bone to which it attaches (Heude et al., 2018). In the limb, tendons and ligaments originate from lateral plate mesoderm, whereas those in the trunk are derived from somites (Nassari et al., 2017). The extent to which the developmental origin of tendons and ligaments influences their mechanisms of formation and later properties is unclear.
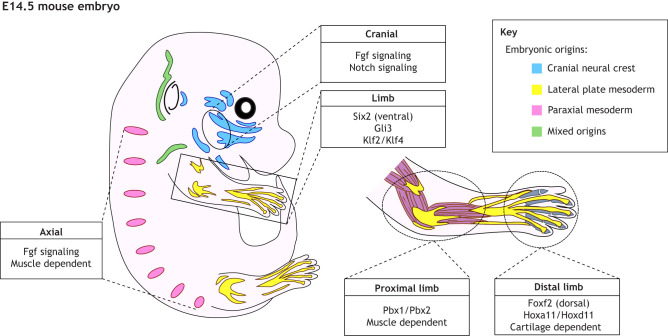
Fig. 2.
Embryonic origins and molecular regional regulators of tendon and ligament progenitors in the developing mouse. Tendons and ligaments are derived from mesenchyme of multiple embryonic origins with cranial tendons and ligaments originating from cranial neural crest cells (CNCCs; blue), limb tendons and ligaments from lateral plate mesoderm (yellow), axial tendons from paraxial mesoderm (pink), and neck and shoulder tendons and ligaments from a mixture of all three (green). In addition to their multiple origins, tendon and ligament progenitors require regional mechanisms for their establishment.
Fibroblastic-like cells, often called tenocytes and ligamentocytes, are the principal mature cell types within tendons and ligaments, respectively. The fundamental nature of tendon and ligament cell identity and their heterogeneity have yet to be determined. A major challenge in defining these cell types is in their seemingly strong potential to revert to a progenitor-like state or transdifferentiate into osteoblasts or chondrocytes (Howell et al., 2017; Oldfield and Evans, 2003; Tan et al., 2020; Wren et al., 2000). As both tendon and ligament pathologies are commonly associated with aberrant bone formation, there is a need to understand the factors that maintain tendon and ligament cell fate (Magne and Bougault, 2015).
In this Review, we focus primarily on the better-understood mechanism of tendon development, indicating where key findings are also applicable to ligaments. We discuss region-specific developmental mechanisms in the head, trunk and limb that induce and maintain tendon progenitors and regulate tendon cell differentiation and maturation. We also discuss current evidence in the field that points to heterogeneity and regional specificity within tendon cell populations. Finally, we compare tendon progenitors in development to adult stem and progenitor cells that contribute to tendon and tendon attachment repair.
Tendon and ligament development
The discovery of scleraxis (Scx), a basic helix-loop-helix transcription factor, as a marker of both progenitors and differentiated cells within tendon and ligament has led to the creation of powerful genetic tools for understanding their development (Cserjesi et al., 1995; Pryce et al., 2007; Schweitzer et al., 2001). The Scx-GFP reporter and Scx-Cre alleles in mice are routinely employed in mechanistic studies that address tendon progenitor patterning, differentiation and maintenance (Blitz et al., 2013; Pryce et al., 2007; Yoshimoto et al., 2017). Similarly, the Scx knockout mouse has underscored its necessity in tendon morphogenesis (Murchison et al., 2007; Roberts et al., 2019). In zebrafish, nonsense mutations in scxa and scxb lead to defects in cranial tendon and ligament morphogenesis, showing a conserved role for Scx in differentiation and maturation of tendons across vertebrates (Kague et al., 2019). Molecular studies indicate that Scx promotes tendon cell differentiation and maturation by transcriptionally activating a suite of tendon cell genes, including the major building blocks of the tendon extracellular matrix (ECM), such as Col1a1 and tenomodulin (Tnmd) (Leéjard et al., 2007; Shukunami et al., 2018).
Scx is expressed in all tendon and ligament progenitors, yet its requirement during development varies depending on the subtype and location of the tendons. Loss of Scx in mice preferentially disrupts formation of long-range, but not short-range, muscle-anchoring of tendons and ligaments in the limb and trunk (Murchison et al., 2007). This confounding aspect of the Scx knockout phenotype has been addressed in a recent study that more closely examined long-range tendon development. Development of both long-range and short-range tendons begins with formation of a tendon anlage that anchors to muscle and bone. In long-range tendons, this anlage subsequently undergoes rapid elongation that is fueled by recruitment of surrounding Scx+ progenitors into the tenogenic lineage. Scx, although not required for formation of the initial tendon anlagen, is necessary to recruit tendon progenitors for substantial extension during growth (Huang et al., 2019). Nevertheless, Scx remains the earliest known marker of tendon and ligament progenitors, it does not appear to be necessary for tendon cell fate determination.
Establishment of tendon progenitors
A central pathway key to establishment of tendon progenitors is transforming growth factor β (Tgfβ) signaling. Tgfβ2/3 ligands emanating from neighboring cartilage/muscle recruit undifferentiated mesenchymal cells to form tendon progenitors, and then maintain them by activating Scx expression in both mouse and chick (Havis et al., 2014; Pryce et al., 2009) (Fig. 3). Loss of Tgfb2/3 in mice leads to diminished Scx expression, considerable reduction in tendon progenitor numbers, and eventual widespread loss of cranial, limb and trunk tendons (Anthwal et al., 2008; Pryce et al., 2009). In addition, conditional loss of Tgfbr2 in mouse limb mesenchyme (via Prx1-Cre) results in depletion of limb tendons (Pryce et al., 2009). The dramatic reduction of tendon and ligament progenitors in mice with mutations in the Tgfβ signaling pathway is the result of lost cellular identity, either through de-differentiation or acquisition of different cell fates (Pryce et al., 2009).
Fig. 3.
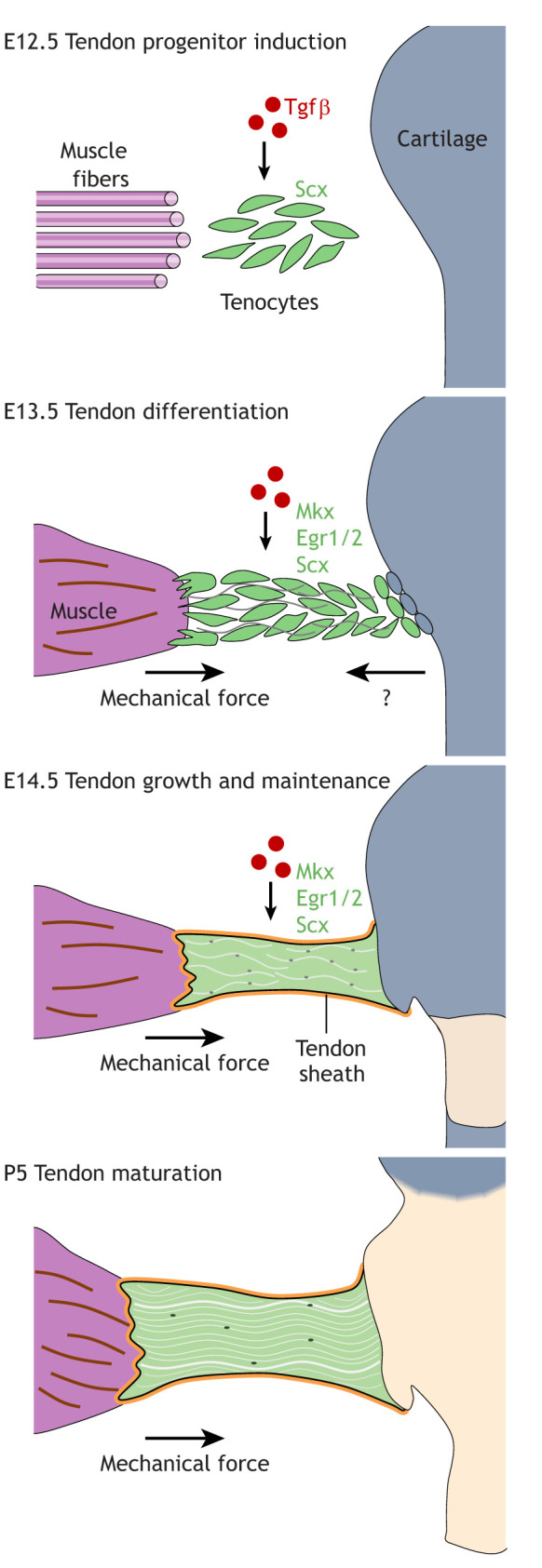
Tendon progenitor induction, differentiation, maintenance and maturation in the mouse limb. Induction of Scx in tendon progenitors relies on Tgfβ signaling during embryonic development. Later, tendon cells differentiate and multiply in the presence of mechanical stimulation from the muscle, as well as undetermined secreted factors from surrounding tissue, including cartilage. Various inputs are required for both the establishment and maintenance of the tendon cell fate. E, embryonic day; P, postnatal day.
Region-specific regulation of tendon progenitors
The distinct embryonic origins of cranial, limb and trunk tendons suggests the possibility that the establishment and differentiation of each population is uniquely regulated (Fig. 2). Although this has been difficult to address given the paucity of information on cranial tendon progenitors relative to those of the limb and trunk, growing evidence supports this idea. For example, fibroblast growth factor (Fgf) signaling from the adjacent myotome promotes the induction of axial tendon progenitors by driving Scx expression in mice and chicks (Brent et al., 2005, 2003; Brent and Tabin, 2004) (Fig. 2). Conversely, induction of cranial and distal limb (autopod) tendon progenitors is muscle independent, with muscle interaction only required later for maintenance of cellular identity (Bonnin et al., 2005; Chen and Galloway, 2014; Edom-Vovard and Duprez, 2004; Grenier et al., 2009; Grifone et al., 2008). In addition, interactions with cartilage are vital to the organization (and in some cases establishment) of tendon and ligament progenitors of the craniofacial and axial skeleton (Chen and Galloway, 2014).
Current research suggests that the relationship between Fgf signaling and Scx expression varies based on species and anatomical region. We have recently found that Fgfr2 promotes establishment of Scx+ tendon progenitor cells at tendon-bone insertion sites in the jaw (Roberts et al., 2019) (Fig. 2). Exogenous treatments that activate or block Fgf ligands, or its downstream effector Erk (Mapk1), in limb explants suggest that Fgf signaling promotes Scx expression in chick and inhibits Scx expression in mouse (Edom-Vovard et al., 2002; Havis et al., 2016, 2014). Conversely in zebrafish embryos, inhibition of Fgf signaling leads to loss of all Scx+ progenitors contributing to both tendon and ligament (Chen and Galloway, 2014). The reason for this species-specific difference is unclear, but the outcome of Fgf signaling on Scx expression may be related to the intensity and duration of the Fgf-Erk signal (Brent and Tabin, 2004) or, alternatively, differences in experimental approaches between species. It is also not yet clear what the specific role of Fgf signaling is in ligament, although the common embryonic origins and function of tendon and ligament would suggest that findings specific to tendons in various anatomical regions could be extrapolated to nearby ligaments. The limited available literature suggests that Fgf signaling enhances proliferation and differentiation in periodontal ligament cells in vitro by upregulating Scx and Tnmd while suppressing osteogenic differentiation through inhibition of Runx2 expression (An et al., 2015; Hyun et al., 2017). Defining a clearer role for Fgf signaling in tendon and ligament induction will require further analysis of Fgf-related mutant models.
Studies in the limb provide evidence that tendon progenitors of the same embryonic origin can have distinct regulatory programs depending on their location along the dorsal-ventral and proximal-distal axes. Whole-transcriptome expression profiling of limb-derived Scx-GFP+ cells during the early stages of development has identified Foxf2 and Six2 as tendon-enriched transcription factors with region-specific expression (Liu et al., 2015). Foxf2 expression is restricted to the dorsal tendons within the distal limb (autopod), whereas Six2 expression is enriched in ventral tendons of both the proximal (zeugopod) and distal (autopod) limb (Fig. 2). However, it is not clear whether this difference in expression is specific to Scx-expressing tendon and ligament populations (Liu et al., 2015). In the proximal limb (zeugopod), differentiation and growth of tendons is dependent on the muscle (Bonnin et al., 2005; Huang et al., 2015; Kardon, 1998; Schweitzer et al., 2001) (Fig. 2). In the autopod, however, the induction, differentiation and maintenance of tendons, progenitors of which are distinctly derived from a Six2 lineage, is instead dependent on the cartilage that prefigures the bone (Huang et al., 2015) (Fig. 2). The conclusion that development of autopod tendons is dependent on skeletal cues is underscored by the finding that autopod tendons are induced in supernumerary digits and fail to form in the absence of cartilage (Huang et al., 2015). Whether cartilage promotes tendon formation by secreting tenogenic factors or providing a source of progenitors with tenogenic potential is unknown; however, candidate signaling pathways for regulation of this process include Tgfβ, Fgf and Wnt (Pryce et al., 2009; Roberts et al., 2019; Yamamoto-Shiraishi and Kuroiwa, 2013).
Tendon cell maturation
Once tendon cells are specified, they function primarily in producing the tendon's ECM and establishing the patterning of collagen fibrils and their bundles (Birk et al., 1996) (Fig. 3). The most abundant structural protein in tendon is Col1, which comprises about 95% of the total collagen content. Expression of Col1a1 in tendon cells is regulated by multiple factors, including Scx, which directly binds to E-box sites within the Col1a1 promoter (Leéjard et al., 2007). Other transcription factors that are known to have functional roles in tendon development, such as mohawk (Mkx) and Egr1/2, are primarily required for tendon maturation, including matrix deposition and organization (Ito et al., 2010; Lejard et al., 2011; Liu et al., 2010) (Fig. 3). For example, loss of Mkx decreases Col1a1 expression and impairs collagen fibril formation, reducing tendon mass, but has no discernible effect on ligament development (Ito et al., 2010; Liu et al., 2010). Though expression of Mkx and Egr1/2 has been documented in ligament fibroblasts, it is not yet clear what their roles in ligament cell differentiation and maturation may be (Liu et al., 2010; Tamura et al., 2008).
As tendons mature, their respective fibroblasts arrange in longitudinal rows along the collagen fibrils, flatten and become less dense. Maturing tendon cells develop a complex network of cellular projections, including nanotubes that facilitate intercellular communications via gap junctions (Benjamin et al., 2008; Egerbacher et al., 2020; Kato et al., 2013). In zebrafish, the force of muscle contraction promotes release of Tgfβ, which is required for tendon progenitors to extend microtubule-rich projections (Subramanian et al., 2018). These projections, in turn, promote tendon cell maturation by regulating force-dependent expression of tendon- and ligament-specific ECM components, such as Thrombospondin 4b (Thbs4b) (Jelinsky et al., 2010; Subramanian et al., 2018; Subramanian and Schilling, 2014).
Tendon cell fate maintenance
In the mature tendon, tenocytes require continuous feedback to maintain their differentiated state and repress metaplasia into the osteochondral lineage. Feedback mechanisms that maintain tendon cell fate (and have been loosely implicated in ligament fibroblast maintenance) are discussed below, including growth factors, ECM components and mechanical force (Fig. 3). Tendon cell projections are also essential for cell fate maintenance, as their loss leads to the upregulation of scxa in zebrafish, suggesting a reversion to a de-differentiated state (Subramanian et al., 2018).
Growth factors
Tgfβ signaling is the primary growth factor pathway known to be involved in maintenance of tendon cell fate (Fig. 3). Targeted deletion of Tgfbr2 in tendon and ligament cells using Scx-Cre does not appear to affect cell fate during embryonic development. However, tendon and ligament cells in early postnatal conditional knockout mice lose expression of differentiation markers and reactivate genes associated with tendon progenitors at the earliest stages of embryonic tendon induction (Havis et al., 2014; Tan et al., 2020). The role of Tgfβr2 signaling is cell-autonomous in tendon cells, as targeted re-expression of Tgfbr2 in de-differentiated tendon cells is sufficient to reactivate differentiation. Together, this suggests that tendon cell fate is reversible and that tendon cell identity requires extrinsic growth factor signals for continuous maintenance.
ECM components
Proteoglycans within the tendon ECM control the bioavailability of growth factors, such as Tgfβ/Bmp, and play important roles in maintaining tendon cell fate. The metalloproteinases Adamts7 and Adamts12 are co-expressed in mouse hindlimb tendons starting in the late embryonic period and persist through adulthood to maintain tendon cell fate (Mead et al., 2018). Combined inactivation of Adamts7 and Adamts12 alters collagen fibrillogenesis and leads to progressive heterotopic ossification (HO) in juvenile mouse tendons (Mead et al., 2018). Prior to HO, tendon cells exhibit enhanced Bmp signaling and upregulate cartilage and bone markers. This suggests that Adamts7 and Adamts12 maintain the tendon cell fate by suppressing osteochondral lineage differentiation. Similarly, the small leucine-rich proteoglycans biglycan (Bgn) and fibromodulin (Fmod) promote tendon cell differentiation by repressing Bmp signaling, and their combined loss results in HO within the hindlimb tendons (Ameye et al., 2002; Bi et al., 2007). In a model for tendon HO caused by hyperactive Bmp signaling, ectopic bone is derived from Scx-lineage cells that aberrantly activate bone and cartilage markers (Agarwal et al., 2017). These studies show that tendon ECM preserves the tendon cell fate, in part by providing factors that inhibit bone formation, and strongly supports a role for ECM proteins in the maintenance of tendon cell fate.
Mechanical feedback
Mechanical signals are also necessary to maintain tendon cell fate during development and homeostasis (Arvind and Huang, 2017). Tendon cells respond to physical input in part through activation of the mechanoresponsive transcription factors Egr1/2 and Mkx. Increased load activates Egr1 and Mkx expression, and decreased load reduces their expression (Gaut et al., 2016; Kayama et al., 2016; Maeda et al., 2011). Downstream of mechanical signals, Egr1/2 and Mkx promote tendon cell fate through distinct mechanisms. Egr1 is sufficient to activate gene expression of Scx and Col1a1 during development, and to promote tendon cell fate during postnatal growth (Guerquin et al., 2013; Lejard et al., 2011). In postnatal tendons, Egr1 is enriched at the Tgfb2 promoter and the absence of Egr1 reduces Tgfb2 expression (Guerquin et al., 2013). This suggests that, in response to mechanical signals, Egr1 acts upstream of Tgfβ signaling to maintain tendon cell identity (Havis and Duprez, 2020). Although all of the aforementioned factors have also been found to be expressed in ligament, their roles in ligament cell maintenance remain unclear. However, a wealth of evidence supports the importance of mechanical signaling in controlling ligament cell fate, in part through regulation of Rho/Rock signaling in the ECM microenvironment (Meng et al., 2015; Yamamoto et al., 2018).
In Mkx knockout mice, mechanical stimuli fail to maintain tenogenic gene expression and hindlimb tendons exhibit HO (Kayama et al., 2016; Liu et al., 2019). These ossified sites are derived from aberrant differentiation of Scx-lineage cells into chondrocytes that then undergo endochondral-like bone formation similar to that observed in Adamts7, Adamts12, Bgn and Fmod mutant models. Selective inactivation of Mkx in postnatal Scx+ tendon cells recapitulates the HO phenotype (Liu et al., 2019). Although tendon HO is associated with increased Bmp signaling in the mouse models listed above, Hedgehog (Hh) signaling plays a crucial role in tendon ossification in Mkx mutant mice. Interestingly, many cells at the periphery of ossifying tendon nodules express both Scx-GFP and Sox9, similar to developing sesamoid bones and bone eminences (Blitz et al., 2013; Eyal et al., 2015, 2019) (Fig. 4). A chromatin immunoprecipitation assay for Mkx in rat tendon-derived cells identified enrichment of both tendon- and cartilage-related genes, suggesting a dual role for Mkx in transcriptional promotion of tendon cell differentiation, while preventing chondrogenic/osteogenic differentiation (Suzuki et al., 2016). Although these studies did not directly address ligaments, the mechanisms and signals are assumed to be recapitulated, as many human diseases featuring HO affect both types of tissue (Zhang et al., 2020).
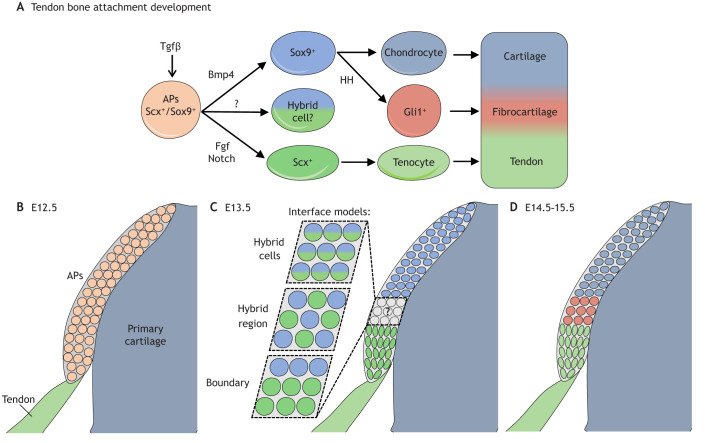
Fig. 4.
Regulation of cell fate during development of the tendon-bone attachment in the mouse limb. The enthesis is derived from attachment progenitors (APs), which have the potential to form endochondral bone, fibrocartilage and tendon. (A) Tgfβ is necessary to establish APs, which co-express Scx and Sox9. Once established, APs give rise to either Scx+ tenocytes, which make tendon, or Sox9+ chondrocytes, which make fibrocartilage and bone. Some APs may maintain co-expression of Scx and Sox9, and acquire a hybrid tenochondral phenotype. (B) The APs form as a secondary condensation atop the primary cartilage anlagen at the site of tendon insertion at E12.5. (C) By E13.5, the graded nature of the enthesis begins to emerge; however, it is not known if the intermediate tissue forming at the tendon-bone interface is made from hybrid tenochondral cell type (cells that co-express Scx and Sox9) and/or from discrete cell types (tenocytes and chondrocytes) that intermingle and/or are separated by a cellular boundary. (D) By E14.5-15.5, the graded enthesis has formed, connecting tendon to bone. E, embryonic day.
Tendon attachment
Tendon cell fate at the tendon-bone interface
Tendons and ligaments attach to bone through a specialized tissue called the enthesis, which functions to dissipate mechanical stress across the hard-soft tissue interface (Fig. 1). There are two major types of entheses: fibrous entheses, whereby tendons and ligaments attach or insert directly to the bone or periosteum, and fibrocartilaginous entheses, which are zonally organized into tendon or ligament, fibrocartilage, mineralized fibrocartilage and bone (Benjamin et al., 2006) (Fig. 4). In fibrocartilaginous entheses, this gradient in cell types, ECM and minerals is essential for the transmission of force during musculoskeletal movement (Lu and Thomopoulos, 2013). The intermediate properties of this tissue arise in development from attachment progenitors (APs), which are established at the tendon-bone interface through a little-understood mechanism requiring Tgfβ signaling (Blitz et al., 2013) (Fig. 4). Tgfbr2 is expressed in APs and its loss in the jaw (condyle and angular process) or limb (deltoid tuberosity) results in loss of these key tendon-bone attachment sites in mice (Anthwal et al., 2008; Blitz et al., 2013; Seo and Serra, 2007). Though the structure and function of tendon and ligament fibrocartilaginous entheses appear to be the same, it is unclear whether developmental mechanisms for enthesis establishment are conserved between these two types. However, it has been shown that Tgfβ signaling plays a crucial role in regulating bone resorption following traumatic injury to both tendon and ligament entheses, pointing to at least some conservation of enthesis development between the two tissue types (Doschak et al., 2005; Tang et al., 2009; Zhen et al., 2013).
Comparative transcriptomic analysis together with mouse genetics has identified that, in the appendicular skeleton, there are global and regional regulators that determine the correct location of APs along the bone's length. Spatial organization of APs throughout the limb requires Gli3, whereas proximal versus distal patterning of APs is regulated by Pbx1/Pbx2 and Hoxa11/Hoxd11, respectively (Eyal et al., 2019) (Fig. 2). Disruption in these global and regional regulators of AP patterning leads to mislocalization of the bone eminence attachment sites of both tendon and ligament. Global and regional regulators working together to fine-tune AP patterning demonstrates modularity in enthesis development.
APs co-express lineage commitment factors for tendon (Scx) and cartilage (Sox9) and have the potential to adopt either cell fate depending on their location along the tendon-bone axis (Fig. 4). During enthesis formation, APs near the tendon form fibroblasts of the tendon terminus, whereas those near the bone form chondrocytes, some of which then ossify into the bone eminence (Sugimoto et al., 2013) (Fig. 4). Loss of Scx or Sox9 disrupts tendon-bone attachment development throughout the skeleton (Blitz et al., 2013; Killian and Thomopoulos, 2016; Roberts et al., 2019; Sugimoto et al., 2013). Genome-wide transcriptomic and chromatin accessibility assays of the limb attachment cells early in their differentiation have shown that APs, as a population, are marked by a hybrid fibroblast-chondrocyte transcriptional profile with active enhancers shared between tenocytes or ligamentocytes and chondrocytes (Kult et al., 2021). This suggests that the graded structure of the enthesis is derived from attachment cells with a hybrid tendon-cartilage or ligament-cartilage cell fate (Fig. 4); however, transcriptional analysis at the single-cell level will be required to rule out the possibility that the graded structure results from intermingling of attachment cells of discrete types.
In recent years, several regulators of AP differentiation have been identified. The Krüppel-like transcription factors Klf2 and Klf4, which bind the proximal regulatory regions of genes expressed in attachment cells, such as Gli1 and Col5a1, are required for tendon AP differentiation in the limb (Kult et al., 2021). In periodontal ligaments, Klf5 has been implicated as a key regulator of progenitor cell proliferation and osteogenic differentiation via activation of both Fgf and Wnt signaling pathways (Gao et al., 2018). Whereas Klf family genes regulate the overall capacity of APs to differentiate, Bmp and Fgf signaling regulate the cell fate choice of APs (Fig. 4). In the limb, Bmp4 derived from the tendon tip induces APs to differentiate into chondrocytes, some of which then undergo endochondral-like ossification to form the bone eminence (Blitz et al., 2009). Conditional inactivation of Bmp4 using Scx-Cre blocks formation of the cartilage anlage prefiguring the bone eminence, despite normal tendon differentiation. Bmp2 and Bmp4 are also necessary for the endochondral ossification of APs into tendon-embedded sesamoid bones (Eyal et al., 2015). Although it is understood that Bmp promotes AP differentiation into chondrocytes, it is not clear what blocks Bmp-induced chondrogenesis in APs that differentiate into tendon cells.
During a similar time frame, APs also express the Bmp family member Gdf5. Gdf5 lineage tracing marks APs that contribute to linear growth of the enthesis and give rise to fibrocartilage (Dyment et al., 2015, 2014). The relationship between this Gdf5 lineage and APs that co-express Scx and Sox9 remains unresolved. However, in the developing joint Sox9+ cells give rise to Gdf5+ joint interzone progenitors that contribute to various joint tissues, including the menisci, cruciate ligaments, articular cartilage and intra-articular ligaments (Harada et al., 2007; Shwartz et al., 2016). It has been suggested that the onset and duration of Gdf5 signaling may instruct lineage divergence in the joint, and a similar mechanism may also occur in the enthesis. The one pathway currently known to promote tendon cell fate in APs is Fgf signaling (Fig. 4). We recently showed that perichondral Fgfr2 signaling, activated by Fgf2 from the tendon, promotes tendon cell differentiation in cranial APs by regulating Notch signaling (Roberts et al., 2019). Upon CNCC-specific deletion of Fgfr2, cranial APs lose Dll1-Notch2 signaling and undergo biased differentiation into chondrocytes over tenocytes. This indicates that Fgf signaling plays a key role in promoting the differentiation of tendon at the expense of cartilage in the face.
Hh signaling also plays an important role in the cellular complexity and maturation of tendon-bone and ligament-bone attachment cells. Sonic hedgehog (Shh) induces Gli1 expression in a subpopulation of newly differentiated attachment cells, with Gli1 expression maintained postnatally by Indian hedgehog (Ihh). These Gli1-expressing cells give rise to the mineralized fibrocartilage in the mature enthesis (Dyment et al., 2015; Felsenthal et al., 2018; Liu et al., 2013; Schwartz et al., 2015) (Fig. 4). Mouse genetics show that Hedgehog signaling is necessary for fibrocartilage mineralization: loss of the Hedgehog transmembrane effector smoothened using Scx-Cre greatly reduces the number of Gli1+ cells and inhibits their subsequent mineralization (Dyment et al., 2015; Liu et al., 2013; Schwartz et al., 2015). In addition, ablation of Gli1+ cells early in postnatal development results in loss of mineralized fibrocartilage (Schwartz et al., 2015). The lineage relationship between postnatal Gli1+ cells and embryonic APs that precede them differs depending on whether the enthesis remains stationary or migrates during bone growth. In stationary entheses, which are found at bone ends, Gli1+ cells are derived from APs; however, in migrating entheses found along the bone shaft, a separate population of Gli1+ cells replace APs (Felsenthal et al., 2018). The origin of Gli1+ cells in migrating entheses remains unclear, but the periosteum seems a likely source.
Tendon cell fate at the tendon-muscle interface
The structure and mechanical properties of the myotendinous junction (MTJ) rely on interaction between tendon and muscle via the ECM (Fig. 1). MTJ connections are unique to tendons because ligament, by definition, facilitates the joining of bones exclusively. During MTJ formation in Drosophila, myoblasts migrate to sites of integration and extend actin-based, filopodia-like structures to recognize tendon cells via thrombospondin-mediated cell adhesion (Liu and Geisbrecht, 2012). Interactions between Drosophila tendon and muscle are mediated by cell-cell contact through integrin heterodimers, as well as various ECM proteins that can be classified as force transmitting and structural collagens, scaffolding proteins or crosslinking proteins. Key ECM proteins that make up the MTJ include laminin, fibronectin, thrombospondins and collagens (Frolova et al., 2014; Subramanian and Schilling, 2015). Recent RNAi screens have expanded the predicted list of genes contributing to MTJ formation and highlight Tango1 and Fascin as key regulators of collagen deposition and actin bundling in filopodia, respectively (Camuglia et al., 2018; Tiwari et al., 2015).
It remains unclear to what extent insights made in Drosophila can be applied to vertebrate MTJ formation. In vertebrates, the migratory nature of cells contributing to the MTJ is known to differ based on both species and anatomical region. Avian hindlimb myoblasts elongate and adhere to tenocytes already present at attachment sites, yet in the avian cranium it is the tenocytes that must travel to meet the established muscle (Grenier et al., 2009; Kardon, 1998). In the zebrafish, cranial tenocytes appear to prefigure regions of future muscle attachment, with altered tenocyte migration in cyp26b1 mutants resulting in secondary mislocalization of muscle attachments (McGurk et al., 2017). It is unknown whether differences in MTJ patterning across regions and species are due to differing embryonic origins of tendon and muscle or other, potentially species-specific, factors.
During MTJ development in vertebrates, a careful balance between ECM deposition and remodeling is maintained in response to mechanical signals. This balance is achieved through reciprocal interactions between structural proteins, such as laminin and collagens, and matrix metalloproteinases (Brown, 2000; Matchett et al., 2019; Zhang et al., 2014). As previously discussed, both molecular and mechanical factors originating from the muscle play a vital role in inducing tendon progenitors in the axial skeleton and promoting tendon cell maturation in the limb and craniofacial complex. Mechanical signals also affect tendon cell morphogenesis at the MTJ through a Tgfβ-driven feedback loop that regulates the growth and branching of tenocyte projections (Subramanian et al., 2018). Studies in the zebrafish axial skeleton suggest that interactions of tendon and muscle are mediated by long, microtubule-rich cell projections originating from tenocytes that extend into the intersomitic space during early development of the MTJ (Ma et al., 2018; Subramanian et al., 2018). These projections likely increase the adhesion of tendon and muscle at the MTJ and provide support and structure to the muscle, a hypothesis supported by the phenotypes observed in dystroglycan 1 (dag1) mutants and col22a1 knockdown zebrafish, which model muscular dystrophy and feature increased muscle detachment (Charvet et al., 2013; Ma et al., 2018). Loss of function of transmembrane protein 2 (tmem2; also known as cemip2) in zebrafish also results in muscle fiber detachment, coupled with disorganization of key MTJ ECM proteins, such as laminin and fibronectin, indicating its important role in the regulation of muscle cell-ECM interactions that facilitate muscle-tendon attachment (Ryckebüsch et al., 2016). Transcription factors Tbx4 and Tbx5 originating in muscle connective tissue have also been implicated in the patterning of tendon and muscle in the mouse limb through modulation of N-cadherin (cadherin 2) and β-catenin expression (Hasson et al., 2010).
Tendon cell fate in repair
Tendons, ligaments and their entheses are commonly injured and, in adults, heal through the formation of a persistent fibrovascular scar that does not recapitulate the developmental process or mechanical properties of the native tissue (Galatz et al., 2015). Injured tendons and entheses heal through phases of inflammation, proliferation and remodeling (Thomopoulos et al., 2015). During inflammation, the release of growth factors and cytokines initiates vascularization and recruits immune cells and activates resident stem and progenitor cells. In the proliferative stage, resident stem and progenitor cells expand and synthesize a granulation tissue rich in fibroblasts, capillaries and an ECM network of Col3 (Juneja, 2013; Snedeker and Foolen, 2017). The granulation tissue then undergoes remodeling to form scar tissue that has reduced cellularity and vascularity as well as increased Col1 (Juneja, 2013; Snedeker and Foolen, 2017). As reparative strategies for tendon, ligament and enthesis, which are largely limited to surgical repair, fail to recreate the structural and functional characteristics of the native tissue that forms during development, there is a need to identify and harness the regenerative potential of endogenous stem and progenitor cells.
Tendon and ligament healing
As discussed, tendon and ligament development appears to share key morphological and functional characteristics. In fact, a frequently used surgical intervention to repair torn or injured ligaments is tendon auto- or allografting (Kan et al., 2016; Macaulay et al., 2012). For purposes of simplicity, we focus here on the larger body of current research pertaining to tendon healing with the hope that findings in this field can be extrapolated to improve understanding of ligament healing. Tendon stem and progenitor cells (TSPCs) were first identified through in vitro characterization of cells disassociated from the tendon proper (Bi et al., 2007). These cells were identified as expressing surface markers commonly associated with other stem cells, such as Sca1 (also known as Ly6a) and CD44, as well as tenogenic factors such as Scx. Tissue-isolated TSPCs also exhibit characteristic traits of stem cells, such as clonal serial passaging and multilineage potential in vitro, and give rise to tendon-like tissue upon engraftment. More recently, genetic lineage tracing after tendon injury has begun to characterize potential TSPCs in vivo. Emerging studies reveal that putative in vivo TSPCs that contribute to tendon repair following injury can arise from both the tendon proper and sheath, and that age is a major factor for whether healing is regenerative or fibrotic (Fig. 5).
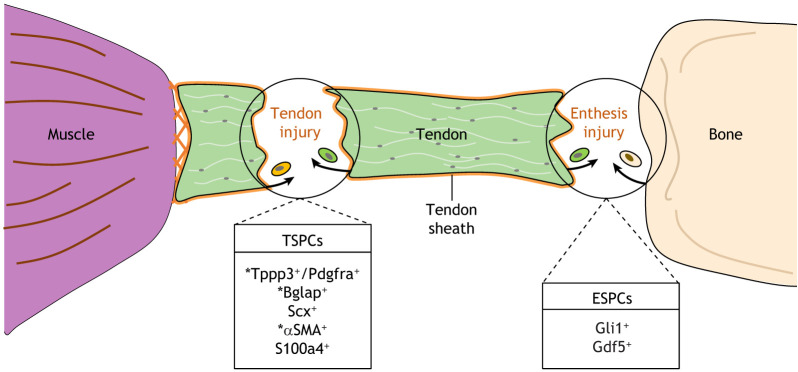
Fig. 5.
Cellular lineages contributing to tendon and enthesis repair in mice. Injuries to the tendon and enthesis heal through the formation of a persistent fibrovascular scar that does not recapitulate the native morphology or mechanical properties of the original tissue. Cells contributing to tendon repair originate from the tendon proper (green cell) and the tendon sheath (orange cell), whereas cells contributing to enthesis healing can be sourced from either tendon or bone (beige cell). A few cell-specific markers that characterize putative tendon stem/progenitor cells (TSPCs) or enthesis stem/progenitor cells (ESPCs) have been identified. Asterisks indicate markers found in tendon sheath-derived cells.
In neonatal mice, transection injury sustained to the Achilles tendon results in the recruitment of resident Scx-lineage tenocytes, which promote regenerative healing through new tendon formation (Howell et al., 2017). In adult mice, however, the same injury induces fibrotic healing whereby Scx-lineage cells remain quiescent and an extrinsic population of αSMA+ cells contribute to a permanent scar (Fig. 5). Similarly, following full thickness transection injury in the mouse patellar and supraspinatus tendons, αSMA-lineage cells are recruited, activate expression of Scx-GFP and become the main contributors to the healing response (Dyment et al., 2014; Yoshida et al., 2016). The αSMA+ cells that contribute to adult tendon repair are heterogeneous, arising from a subpopulation of resident tenocytes marked by S100a4, as well as from extrinsic progenitors originating from the tendon sheath (Ackerman et al., 2017, 2019; Best and Loiselle, 2019; Dyment et al., 2014, 2013). Since the αSMA-lineage contributes to multiple cell types within the tendon, only some of which turn on tenogenic genes during repair, αSMA is not a specific marker for TSPCs (Dyment et al., 2014; Harvey et al., 2019) (Fig. 5). Inducible Cre lines that more specifically trace lineages of the tendon sheath support the idea that the sheath may be a source of TSPCs during repair. An inducible Cre line driven by tubulin polymerization-promoting protein family member 3 (Tppp3), a developmental marker of tendon sheath cells, marks a population of Sca1+/Pdgfra+ cells that are recruited to the site of biopsy punch-injury of the patellar tendon where they activate Scx expression and give rise to tenocytes in a Pdgf-dependent manner (Harvey et al., 2019; Staverosky et al., 2009) (Fig. 5). Interestingly, Tppp3 expression overlaps with that of Bglap (osteocalcin), a marker typical of osteoblasts, in the sheath of some tendons. In these tendons, Bglap-lineage cells from the tendon sheath differentiate and contribute to the population of tenocytes activated during tendon repair (Wang et al., 2017). As Bglap and Tppp3 do not overlap in all tendons, there are likely regional differences in TSPC populations (Fig. 5). Alternatively, heterogeneity in TSPCs might also indicate different states of commitment along the differentiation pathway.
Evidence that TSPC populations differ in their capacity for repair comes from in vitro studies comparing the relative healing potential of putative TSPCs from the tendon proper versus the sheath. Tendon proper-derived TSPCs express higher levels of tenogenic markers and have greater potential to form tendon-like tissue, whereas sheath-derived TSPCs express pericyte-like markers, proliferate faster and have increased potential for myofibroblast differentiation (Cadby et al., 2014; Mienaltowski et al., 2013, 2014). Single-cell RNA-seq analysis is beginning to reveal the heterogeneity of cells within the tendon proper and tendon sheath; however, whether this is an output of diversity in TSPCs is unclear (De Micheli et al., 2020; Yin et al., 2016). Taken together, these studies suggest that TSPCs are heterogeneous depending on their anatomical location, tissue source (tendon proper versus sheath) and degree of lineage commitment.
Enthesis healing
Similar to tendons and ligaments, entheses heal via the contribution of multiple cell lineages. The putative enthesis stem and progenitor cells (ESPCs), primarily characterized by lineage tracing in repair models, appear to be distinct from TSPCs. For example, αSMA-lineage cells do not contribute to enthesis healing (Dyment et al., 2014; Moser et al., 2018) (Fig. 5). Evidence suggests that the reparative potential of intrinsic enthesis cells is age dependent. In adult mice, Scx-, Sox9- and Gli1-lineage cells, which mark tendon, ligament and fibrocartilage of the enthesis, show limited involvement during mature enthesis healing (Moser et al., 2018; Schwartz et al., 2017). In contrast, in juvenile mice following needle-punch injury, Gli1-lineage cells proliferate and contribute to immature enthesis healing through a regenerative process that more closely mimics normal development (Schwartz et al., 2017) (Fig. 5). This suggests that the adult enthesis healing process could potentially be enhanced by identifying methods to mimic the developmental process.
Although cells intrinsic to the adult enthesis have poor capacity for repair, recent studies in mice suggest that extrinsic cells from the adjacent bone and tendon are a source of ESPCs during enthesis healing. Following enthesis reconstruction surgery, in which tendon grafts are inserted into bone tunnels, Gdf5-lineage cells from the underlying bone proliferate, infiltrate the tendon graft, and give rise to a zonal tendon-bone attachment (Hagiwara et al., 2020) (Fig. 5). Following surgical repair of detached tendons, Axin2-lineage cells from the tendon proper also contribute to healing, although they promote formation of a fibrovascular scar rather than recapitulating the graded structure of the native attachment (Moser et al., 2018).
Conclusions and future directions
In this Review, we have discussed a number of mechanisms that regulate tendon and ligament cell fate in the context of both development and repair. Recent advances in our understanding of these mechanisms have opened up a number of interesting questions for future investigation. For example, mechanisms regulating tendon and ligament cell fate in development feature regional differences in the necessity of transcription factors and dependence on inductive signals from muscle versus cartilage. However, it is not yet known how regional and universal developmental programs are integrated to organize the musculoskeletal system, or the extent to which developmental insights learned in one anatomical region can be applied to others. Additionally, tendon cell identity shows evidence of plasticity and reversibility, but it is not yet fully understood how or what transcriptional factors can function to actively induce tendon or ligament cell fate while inhibiting osteoblast and chondrocyte cell fate. There is also a high likelihood that key members of these transcriptional networks have not yet been identified. Additional information is needed to determine the similarities and differences between tendon and ligament cells in a developmental, functional and reparative capacity. For example, can a close study of joint development reveal regulators that are unique to ligament development (Salva and Merrill, 2017; Smeeton et al., 2017)? To what extent are the factors regulating TSPCs conserved between anatomical regions and between organisms with differing regenerative potential? Can TSPCs harvested in one location be relied upon for tendon repair in another location? Answers to these questions will help determine whether repair and tissue engineering approaches need to be designed with regional specificity in mind.
Understanding tendon cell identity using single-cell sequencing technologies that allow for RNA expression profiling and chromatin accessibility will be a vital first step to begin to address many of the questions proposed here about tendon cell fate. For example, single-cell technologies will be able to determine whether the graded connective tissue within tendon-bone attachments arises from intermingling of canonical cell types (tenocyte, osteoblast, chondrocyte), hybrid cell types that feature mixed properties, or a combination of both. Single-cell technologies will allow for a better resolution of the similarities and differences between tendon and ligament cells, which will in turn affect our clinical approaches to injury and repair. A better understanding of the molecular relationship between these cell types would allow for in vivo studies that advance our knowledge of the mechanisms regulating development and repair of ligament and ligament-bone attachments.
Studies in model organisms such as mouse and zebrafish have significantly advanced our understanding of tendon cell differentiation and maintenance, yet we lack a complete picture of how tendon cell fate is regulated. By focusing research on human disorders that feature tendon and ligament defects, such as joint laxity, dislocations and contractures, it may be possible to identify key genes and mechanisms regulating the developmental process. Examples of such disorders include geleophysic dysplasia, which features joint contractures and shortened limbs due to tendon defects. Recent work has identified ADAMTSL2 as the likely affected gene, which had not traditionally been studied as a regulator of tendon and ligament cell identity (Hubmacher et al., 2019; Piccolo et al., 2019). Additionally, disorders such as progressive osseous heteroplasia (POH) (Kaplan et al., 1994) and McCune–Albright syndrome (Chapurlat and Orcel, 2008) feature metaplasia of bone and fibrous connective tissue caused by loss- and gain-of-function mutations in GNAS, respectively. The opposing phenotypes of these disorders (ossification of fibrous tissue in POH and fibrous metaplasia of bone in McCune–Albright syndrome) indicate a key role of GNAS in tendon cell identity and maintenance that is not yet fully understood (Shore et al., 2002; Weinstein et al., 1991). Mechanistic studies of congenital disorders with tendon and ligament defects such as these are likely to reveal new insights into tendon and ligament cell identity and development.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work was supported by the National Institutes of Health (R01DE025222 to A.E.M.; R35DE027550 to J.G.C.; R21DE029656 to J.G.C. and A.E.M., and T90DE021982 in support of L.B.). Deposited in PMC for release after 12 months.
References
- Ackerman, J. E., Best, K. T., O'Keefe, R. J. and Loiselle, A. E. (2017). Deletion of EP4 in S100a4-lineage cells reduces scar tissue formation during early but not later stages of tendon healing. Sci. Rep. 7, 8658. 10.1038/s41598-017-09407-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman, J. E., Nichols, A. E. C., Studentsova, V., Best, K. T., Knapp, E. and Loiselle, A. E. (2019). Cell non-autonomous functions of S100a4 drive fibrotic tendon healing. eLife 8, e45342. 10.7554/eLife.45342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal, S., Loder, S. J., Cholok, D., Peterson, J., Li, J., Breuler, C., Cameron Brownley, R., Hsin Sung, H., Chung, M. T., Kamiya, N.et al. (2017). Scleraxis-lineage cells contribute to ectopic bone formation in muscle and tendon. Stem Cells 35, 705-710. 10.1002/stem.2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameye, L., Aria, D., Jepsen, K., Oldberg, A., Xu, T. and Young, M. F. (2002). Abnormal collagen fibrils in tendons of biglycan/fibromodulin-deficient mice lead to gait impairment, ectopic ossification, and osteoarthritis. FASEB J. 16, 673-680. 10.1096/fj.01-0848com [DOI] [PubMed] [Google Scholar]
- Amiel, D., Frank, C., Harwood, F., Fronek, J. and Akeson, W. (1983). Tendons and ligaments: a morphological and biochemical comparison. J. Orthop. Res. 1, 257-265. 10.1002/jor.1100010305 [DOI] [PubMed] [Google Scholar]
- Amis, A. A. (2004). The Biomechanics of Ligaments. In Biomechanics and Biomaterials in Orthopedics (ed. Poitout D. G.), pp. 550-563. London: Springer London. [Google Scholar]
- An, S., Huang, X., Gao, Y., Ling, J., Huang, Y. and Xiao, Y. (2015). FGF-2 induces the proliferation of human periodontal ligament cells and modulates their osteoblastic phenotype by affecting Runx2 expression in the presence and absence of osteogenic inducers. Int. J. Mol. Med. 36, 705-711. 10.3892/ijmm.2015.2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthwal, N., Chai, Y. and Tucker, A. S. (2008). The role of transforming growth factor-β signalling in the patterning of the proximal processes of the murine dentary. Dev. Dyn. 237, 1604-1613. 10.1002/dvdy.21567 [DOI] [PubMed] [Google Scholar]
- Arvind, V. and Huang, A. H. (2017). Mechanobiology of limb musculoskeletal development. Ann. N. Y. Acad. Sci. 1409, 18-32. 10.1111/nyas.13427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin, M., Toumi, H., Ralphs, J. R., Bydder, G., Best, T. M. and Milz, S. (2006). Where tendons and ligaments meet bone: attachment sites (‘entheses’) in relation to exercise and/or mechanical load. J. Anat. 208, 471-490. 10.1111/j.1469-7580.2006.00540.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin, M., Kaiser, E. and Milz, S. (2008). Structure-function relationships in tendons: a review. J. Anat. 212, 211-228. 10.1111/j.1469-7580.2008.00864.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, K. T. and Loiselle, A. E. (2019). Scleraxis lineage cells contribute to organized bridging tissue during tendon healing and identify a subpopulation of resident tendon cells. FASEB J. 33, 8578-8587. 10.1096/fj.201900130RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, Y., Ehirchiou, D., Kilts, T. M., Inkson, C. A., Embree, M. C., Sonoyama, W., Li, L., Leet, A. I., Seo, B.-M., Zhang, L.et al. (2007). Identification of tendon stem/progenitor cells and the role of the extracellular matrix in their niche. Nat. Med. 13, 1219-1227. 10.1038/nm1630 [DOI] [PubMed] [Google Scholar]
- Birk, D. E., Hahn, R. A., Linsenmayer, C. Y. and Zycband, E. I. (1996). Characterization of collagen fibril segments from chicken embryo cornea, dermis and tendon. Matrix Biol. 15, 111-118. 10.1016/S0945-053X(96)90152-3 [DOI] [PubMed] [Google Scholar]
- Blitz, E., Viukov, S., Sharir, A., Shwartz, Y., Galloway, J. L., Pryce, B. A., Johnson, R. L., Tabin, C. J., Schweitzer, R. and Zelzer, E. (2009). Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev. Cell 17, 861-873. 10.1016/j.devcel.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz, E., Sharir, A., Akiyama, H. and Zelzer, E. (2013). Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development 140, 2680-2690. 10.1242/dev.093906 [DOI] [PubMed] [Google Scholar]
- Bonnin, M.-A., Laclef, C., Blaise, R., Eloy-Trinquet, S., Relaix, F., Maire, P. and Duprez, D. (2005). Six1 is not involved in limb tendon development, but is expressed in limb connective tissue under Shh regulation. Mech. Dev. 122, 573-585. 10.1016/j.mod.2004.11.005 [DOI] [PubMed] [Google Scholar]
- Brent, A. E. and Tabin, C. J. (2004). FGF acts directly on the somitic tendon progenitors through the Ets transcription factors Pea3 and Erm to regulate scleraxis expression. Development 131, 3885-3896. 10.1242/dev.01275 [DOI] [PubMed] [Google Scholar]
- Brent, A. E., Schweitzer, R. and Tabin, C. J. (2003). A somitic compartment of tendon progenitors. Cell 113, 235-248. 10.1016/S0092-8674(03)00268-X [DOI] [PubMed] [Google Scholar]
- Brent, A. E., Braun, T. and Tabin, C. J. (2005). Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development 132, 515-528. 10.1242/dev.01605 [DOI] [PubMed] [Google Scholar]
- Brown, N. H. (2000). Cell-cell adhesion via the ECM: integrin genetics in fly and worm. Matrix Biol. 19, 191-201. 10.1016/S0945-053X(00)00064-0 [DOI] [PubMed] [Google Scholar]
- Cadby, J. A., Buehler, E., Godbout, C., van Weeren, P. R. and Snedeker, J. G. (2014). Differences between the cell populations from the peritenon and the tendon core with regard to their potential implication in tendon repair. PLoS ONE 9, e92474-e92474. 10.1371/journal.pone.0092474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camuglia, J. M., Mandigo, T. R., Moschella, R., Mark, J., Hudson, C. H., Sheen, D. and Folker, E. S. (2018). An RNAi based screen in Drosophila larvae identifies fascin as a regulator of myoblast fusion and myotendinous junction structure. Skelet. Muscle 8, 12. 10.1186/s13395-018-0159-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapurlat, R. D. and Orcel, P. (2008). Fibrous dysplasia of bone and McCune-Albright syndrome. Best Practice Res. Clin. Rheumatol. 22, 55-69. 10.1016/j.berh.2007.11.004 [DOI] [PubMed] [Google Scholar]
- Charvet, B., Guiraud, A., Malbouyres, M., Zwolanek, D., Guillon, E., Bretaud, S., Monnot, C., Schulze, J., Bader, H. L., Allard, B.et al. (2013). Knockdown of col22a1 gene in zebrafish induces a muscular dystrophy by disruption of the myotendinous junction. Development 140, 4602-4613. 10.1242/dev.096024 [DOI] [PubMed] [Google Scholar]
- Chen, J. W. and Galloway, J. L. (2014). The development of zebrafish tendon and ligament progenitors. Development 141, 2035-2045. 10.1242/dev.104067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connizzo, B. K., Yannascoli, S. M. and Soslowsky, L. J. (2013). Structure-function relationships of postnatal tendon development: a parallel to healing. Matrix Biol. 32, 106-116. 10.1016/j.matbio.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserjesi, P., Brown, D., Ligon, K. L., Lyons, G. E., Copeland, N. G., Gilbert, D. J., Jenkins, N. A. and Olson, E. N. (1995). Scleraxis: a basic helix-loop-helix protein that prefigures skeletal formation during mouse embryogenesis. Development 121, 1099. [DOI] [PubMed] [Google Scholar]
- De Micheli, A. J., Swanson, J. B., Disser, N. P., Martinez, L. M., Walker, N. R., Oliver, D. J., Cosgrove, B. D. and Mendias, C. L. (2020). Single-cell transcriptomic analysis identifies extensive heterogeneity in the cellular composition of mouse Achilles tendons. Am. J. Physiol. Cell Physiol. 319, C885-C894. 10.1152/ajpcell.00372.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doschak, M. R., LaMothe, J. M., Cooper, D. M. L., Hallgrimsson, B., Hanley, D. A., Bray, R. C. and Zernicke, R. F. (2005). Bisphosphonates reduce bone mineral loss at ligament entheses after joint injury. Osteoarthrit. Cartilage 13, 790-797. 10.1016/j.joca.2005.04.015 [DOI] [PubMed] [Google Scholar]
- Durland, J. L., Sferlazzo, M., Logan, M. and Burke, A. C. (2008). Visualizing the lateral somitic frontier in the Prx1Cre transgenic mouse. J. Anat. 212, 590-602. 10.1111/j.1469-7580.2008.00879.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment, N. A., Liu, C.-F., Kazemi, N., Aschbacher-Smith, L. E., Kenter, K., Breidenbach, A. P., Shearn, J. T., Wylie, C., Rowe, D. W. and Butler, D. L. (2013). The paratenon contributes to scleraxis-expressing cells during patellar tendon healing. PLoS ONE 8, e59944-e59944. 10.1371/journal.pone.0059944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment, N. A., Hagiwara, Y., Matthews, B. G., Li, Y., Kalajzic, I. and Rowe, D. W. (2014). Lineage tracing of resident tendon progenitor cells during growth and natural healing. PLoS ONE 9, e96113. 10.1371/journal.pone.0096113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyment, N. A., Breidenbach, A. P., Schwartz, A. G., Russell, R. P., Aschbacher-Smith, L., Liu, H., Hagiwara, Y., Jiang, R., Thomopoulos, S., Butler, D. L.et al. (2015). Gdf5 progenitors give rise to fibrocartilage cells that mineralize via hedgehog signaling to form the zonal enthesis. Dev. Biol. 405, 96-107. 10.1016/j.ydbio.2015.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edom-Vovard, F. and Duprez, D. (2004). Signals regulating tendon formation during chick embryonic development. Dev. Dyn. 229, 449-457. 10.1002/dvdy.10481 [DOI] [PubMed] [Google Scholar]
- Edom-Vovard, F., Schuler, B., Bonnin, M.-A., Teillet, M.-A. and Duprez, D. (2002). Fgf4 positively regulates scleraxis and tenascin expression in chick limb tendons. Dev. Biol. 247, 351-366. 10.1006/dbio.2002.0707 [DOI] [PubMed] [Google Scholar]
- Egerbacher, M., Gabner, S., Battisti, S. and Handschuh, S. (2020). Tenocytes form a 3-D network and are connected via nanotubes. J. Anat. 236, 165-170. 10.1111/joa.13089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, D. J. R. and Noden, D. M. (2006). Spatial relations between avian craniofacial neural crest and paraxial mesoderm cells. Dev. Dyn. 235, 1310-1325. 10.1002/dvdy.20663 [DOI] [PubMed] [Google Scholar]
- Eyal, S., Blitz, E., Shwartz, Y., Akiyama, H., Schweitzer, R. and Zelzer, E. (2015). On the development of the patella. Development 142, 1831-1839. 10.1242/dev.121970 [DOI] [PubMed] [Google Scholar]
- Eyal, S., Kult, S., Rubin, S., Krief, S., Felsenthal, N., Pineault, K. M., Leshkowitz, D., Salame, T.-M., Addadi, Y., Wellik, D. M.et al. (2019). Bone morphology is regulated modularly by global and regional genetic programs. Development 146, dev167882. 10.1242/dev.167882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenthal, N., Rubin, S., Stern, T., Krief, S., Pal, D., Pryce, B. A., Schweitzer, R. and Zelzer, E. (2018). Development of migrating tendon-bone attachments involves replacement of progenitor populations. Development 145, dev165381. 10.1242/dev.165381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolova, E. G., Drazba, J., Krukovets, I., Kostenko, V., Blech, L., Harry, C., Vasanji, A., Drumm, C., Sul, P., Jenniskens, G. J.et al. (2014). Control of organization and function of muscle and tendon by thrombospondin-4. Matrix Biol. 37, 35-48. 10.1016/j.matbio.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatz, L. M., Gerstenfeld, L., Heber-Katz, E. and Rodeo, S. A. (2015). Tendon regeneration and scar formation: the concept of scarless healing. J. Orthop. Res. 33, 823-831. 10.1002/jor.22853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, J. Y., Yu, X. Q. and Wang, J. Q. (2018). [KLF5 modulates proliferation and osteogenic differentiation of human periodontal ligament cells subjected to cyclic tensile stress]. Shanghai kou qiang yi xue=Shanghai journal of stomatology 27, 28-33. [PubMed] [Google Scholar]
- Gaut, L., Robert, N., Delalande, A., Bonnin, M.-A., Pichon, C. and Duprez, D. (2016). EGR1 regulates transcription downstream of mechanical signals during tendon formation and healing. PLoS ONE 11, e0166237. 10.1371/journal.pone.0166237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier, J., Teillet, M.-A., Grifone, R., Kelly, R. G. and Duprez, D. (2009). Relationship between neural crest cells and cranial mesoderm during head muscle development. PLoS ONE 4, e4381-e4381. 10.1371/journal.pone.0004381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifone, R., Jarry, T., Dandonneau, M., Grenier, J., Duprez, D. and Kelly, R. G. (2008). Properties of branchiomeric and somite-derived muscle development in Tbx1 mutant embryos. Dev. Dyn. 237, 3071-3078. 10.1002/dvdy.21718 [DOI] [PubMed] [Google Scholar]
- Guerquin, M.-J., Charvet, B., Nourissat, G., Havis, E., Ronsin, O., Bonnin, M.-A., Ruggiu, M., Olivera-Martinez, I., Robert, N., Lu, Y.et al. (2013). Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. J. Clin. Invest. 123, 3564-3576. 10.1172/JCI67521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara, Y., Dyrna, F., Kuntz, A. F., Adams, D. J. and Dyment, N. A. (2020). Cells from a GDF5 origin produce zonal tendon-to-bone attachments following anterior cruciate ligament reconstruction. Ann. N. Y. Acad. Sci. 1460, 57-67. 10.1111/nyas.14250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada, M., Takahara, M., Zhe, P., Otsuji, M., Iuchi, Y., Takagi, M. and Ogino, T. (2007). Developmental failure of the intra-articular ligaments in mice with absence of growth differentiation factor 5. Osteoarthr. Cartilage 15, 468-474. 10.1016/j.joca.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Harvey, T., Flamenco, S. and Fan, C.-M. (2019). A Tppp3+Pdgfra+ tendon stem cell population contributes to regeneration and reveals a shared role for PDGF signalling in regeneration and fibrosis. Nat. Cell Biol. 21, 1490-1503. 10.1038/s41556-019-0417-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson, P., DeLaurier, A., Bennett, M., Grigorieva, E., Naiche, L. A., Papaioannou, V. E., Mohun, T. J. and Logan, M. P. O. (2010). Tbx4 and Tbx5 acting in connective tissue are required for limb muscle and tendon patterning. Dev. Cell 18, 148-156. 10.1016/j.devcel.2009.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havis, E. and Duprez, D. (2020). EGR1 transcription factor is a multifaceted regulator of matrix production in tendons and other connective tissues. Int. J. Mol. Sci. 21, 1664. 10.3390/ijms21051664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havis, E., Bonnin, M.-A., Olivera-Martinez, I., Nazaret, N., Ruggiu, M., Weibel, J., Durand, C., Guerquin, M.-J., Bonod-Bidaud, C., Ruggiero, F.et al. (2014). Transcriptomic analysis of mouse limb tendon cells during development. Development 141, 3683. 10.1242/dev.108654 [DOI] [PubMed] [Google Scholar]
- Havis, E., Bonnin, M.-A., Esteves de Lima, J., Charvet, B., Milet, C. and Duprez, D. (2016). TGFβ and FGF promote tendon progenitor fate and act downstream of muscle contraction to regulate tendon differentiation during chick limb development. Development 143, 3839-3851. 10.1242/dev.136242 [DOI] [PubMed] [Google Scholar]
- Heude, E., Tesarova, M., Sefton, E. M., Jullian, E., Adachi, N., Grimaldi, A., Zikmund, T., Kaiser, J., Kardon, G., Kelly, R. G.et al. (2018). Unique morphogenetic signatures define mammalian neck muscles and associated connective tissues. eLife 7, e40179. 10.7554/eLife.40179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, K., Chien, C., Bell, R., Laudier, D., Tufa, S. F., Keene, D. R., Andarawis-Puri, N. and Huang, A. H. (2017). Novel model of tendon regeneration reveals distinct cell mechanisms underlying regenerative and fibrotic tendon healing. Sci. Rep. 7, 45238. 10.1038/srep45238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. H., Riordan, T. J., Pryce, B., Weibel, J. L., Watson, S. S., Long, F., Lefebvre, V., Harfe, B. D., Stadler, H. S., Akiyama, H.et al. (2015). Musculoskeletal integration at the wrist underlies the modular development of limb tendons. Development 142, 2431-2441. 10.1242/dev.122374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A. H., Watson, S. S., Wang, L., Baker, B. M., Akiyama, H., Brigande, J. V. and Schweitzer, R. (2019). Requirement for scleraxis in the recruitment of mesenchymal progenitors during embryonic tendon elongation. Development 146, dev182782. 10.1242/dev.182782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubmacher, D., Taye, N., Balic, Z., Thacker, S., Adams, S. M., Birk, D. E., Schweitzer, R. and Apte, S. S. (2019). Limb- and tendon-specific Adamtsl2 deletion identifies a role for ADAMTSL2 in tendon growth in a mouse model for geleophysic dysplasia. Matrix Biol. 82, 38-53. 10.1016/j.matbio.2019.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun, S.-Y., Lee, J.-H., Kang, K.-J. and Jang, Y.-J. (2017). Effect of FGF-2, TGF-β-1, and BMPs on Teno/Ligamentogenesis and Osteo/Cementogenesis of human periodontal ligament stem cells. Mol. Cells 40, 550-557. 10.14348/molcells.2017.0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, Y., Toriuchi, N., Yoshitaka, T., Ueno-Kudoh, H., Sato, T., Yokoyama, S., Nishida, K., Akimoto, T., Takahashi, M., Miyaki, S.et al. (2010). The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. USA 107, 10538-10542. 10.1073/pnas.1000525107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky, S. A., Archambault, J., Li, L. and Seeherman, H. (2010). Tendon-selective genes identified from rat and human musculoskeletal tissues. J. Orthop. Res. 28, 289-297. 10.1002/jor.20999 [DOI] [PubMed] [Google Scholar]
- Juneja, S. C. (2013). Cellular distribution and gene expression profile during flexor tendon graft repair: a novel tissue engineering approach*. J. Tissue Eng. 4, 2041731413492741. 10.1177/2041731413492741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung, H.-J., Fisher, M. B. and Woo, S. L.-Y. (2009). Role of biomechanics in the understanding of normal, injured, and healing ligaments and tendons. Sports Med. Arthrosc. Rehabil. Ther. Technol. 1, 9. 10.1186/1758-2555-1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kague, E., Hughes, S. M., Lawrence, E. A., Cross, S., Martin-Silverstone, E., Hammond, C. L. and Hinits, Y. (2019). Scleraxis genes are required for normal musculoskeletal development and for rib growth and mineralization in zebrafish. FASEB J. 33, 9116-9130. 10.1096/fj.201802654RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, S.-L., Yuan, Z.-F., Ning, G.-Z., Yang, B., Li, H.-L., Sun, J.-C. and Feng, S.-Q. (2016). Autograft versus allograft in anterior cruciate ligament reconstruction: a meta-analysis with trial sequential analysis. Medicine (Baltim.) 95, e4936. 10.1097/MD.0000000000004936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, F. S., Craver, R., MacEwen, G. D., Gannon, F. H., Finkel, G., Hahn, G., Tabas, J., Gardner, R. J. and Zasloff, M. A. (1994). Progressive osseous heteroplasia: a distinct developmental disorder of. J. Bone Joint Surg. Am. Vol. 76, 425-436. 10.2106/00004623-199403000-00013 [DOI] [PubMed] [Google Scholar]
- Kardon, G. (1998). Muscle and tendon morphogenesis in the avian hind limb. Development 125, 4019-4032. [DOI] [PubMed] [Google Scholar]
- Kato, R., Ishihara, Y., Kawanabe, N., Sumiyoshi, K., Yoshikawa, Y., Nakamura, M., Imai, Y., Yanagita, T., Fukushima, H., Kamioka, H.et al. (2013). Gap-junction-mediated communication in human periodontal ligament cells. J. Dent. Res. 92, 635-640. 10.1177/0022034513489992 [DOI] [PubMed] [Google Scholar]
- Kayama, T., Mori, M., Ito, Y., Matsushima, T., Nakamichi, R., Suzuki, H., Ichinose, S., Saito, M., Marumo, K. and Asahara, H. (2016). Gtf2ird1-dependent mohawk expression regulates mechanosensing properties of the tendon. Mol. Cell. Biol. 36, 1297-1309. 10.1128/MCB.00950-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian, M. L. and Thomopoulos, S. (2016). Scleraxis is required for the development of a functional tendon enthesis. FASEB J. 30, 301-311. 10.1096/fj.14-258236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köntges, G. and Lumsden, A. (1996). Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development 122, 3229-3242. [DOI] [PubMed] [Google Scholar]
- Kult, S., Olender, T., Osterwalder, M., Markman, S., Leshkowitz, D., Krief, S., Blecher-Gonen, R., Ben-Moshe, S., Farack, L., Keren-Shaul, H.et al. (2021). Bi-fated tendon-to-bone attachment cells are regulated by shared enhancers and KLF transcription factors. eLife 10, e55361. 10.7554/eLife.55361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leéjard, V., Brideau, G., Blais, F., Salingcarnboriboon, R., Wagner, G., Roehrl, M. H. A., Noda, M., Duprez, D., Houillier, P. and Rossert, J. (2007). Scleraxis and NFATc regulate the expression of the pro-α1(I) collagen gene in tendon fibroblasts. J. Biol. Chem. 282, 17665-17675. 10.1074/jbc.M610113200 [DOI] [PubMed] [Google Scholar]
- Lejard, V., Blais, F., Guerquin, M.-J., Bonnet, A., Bonnin, M.-A., Havis, E., Malbouyres, M., Bidaud, C. B., Maro, G., Gilardi-Hebenstreit, P.et al. (2011). EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem. 286, 5855-5867. 10.1074/jbc.M110.153106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z. C. and Geisbrecht, E. R. (2012). “Importin” signaling roles for import proteins: the function of Drosophila importin-7 (DIM-7) in muscle-tendon signaling. Cell Adh. Migr. 6, 4-12. 10.4161/cam.19774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W., Watson, S. S., Lan, Y., Keene, D. R., Ovitt, C. E., Liu, H., Schweitzer, R. and Jiang, R. (2010). The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell. Biol. 30, 4797-4807. 10.1128/MCB.00207-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.-F., Breidenbach, A., Aschbacher-Smith, L., Butler, D. and Wylie, C. (2013). A role for Hedgehog signaling in the differentiation of the insertion site of the patellar tendon in the mouse. PLoS ONE 8, e65411. 10.1371/journal.pone.0065411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Xu, J., Liu, C.-F., Lan, Y., Wylie, C. and Jiang, R. (2015). Whole transcriptome expression profiling of mouse limb tendon development by using RNA-seq. J. Orthop. Res. 33, 840-848. 10.1002/jor.22886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Xu, J. and Jiang, R. (2019). Mkx-deficient mice exhibit Hedgehog signaling-dependent ectopic ossification in the Achilles tendons. J. Bone Miner. Res. 34, 557-569. 10.1002/jbmr.3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. H. and Thomopoulos, S. (2013). Functional attachment of soft tissues to bone: development, healing, and tissue engineering. Annu. Rev. Biomed. Eng. 15, 201-226. 10.1146/annurev-bioeng-071910-124656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, R. C., Jacobs, C. T., Sharma, P., Kocha, K. M. and Huang, P. (2018). Stereotypic generation of axial tenocytes from bipartite sclerotome domains in zebrafish. PLoS Genet. 14, e1007775. 10.1371/journal.pgen.1007775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay, A. A., Perfetti, D. C. and Levine, W. N. (2012). Anterior cruciate ligament graft choices. Sports Health 4, 63-68. 10.1177/1941738111409890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, T., Sakabe, T., Sunaga, A., Sakai, K., Rivera, A. L., Keene, D. R., Sasaki, T., Stavnezer, E., Iannotti, J., Schweitzer, R.et al. (2011). Conversion of mechanical force into TGF-β-mediated biochemical signals. Curr. Biol. 21, 933-941. 10.1016/j.cub.2011.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magne, D. and Bougault, C. (2015). What understanding tendon cell differentiation can teach us about pathological tendon ossification. Histol. Histopathol. 30, 901-910. [DOI] [PubMed] [Google Scholar]
- Matchett, E. F., Wang, S. and Crawford, B. D. (2019). Paralogues of Mmp11 and Timp4 interact during the development of the myotendinous junction in the zebrafish embryo. J. Dev. Biol. 7, 22. 10.3390/jdb7040022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, T., Ahlberg, P. E., Kessaris, N., Iannarelli, P., Dennehy, U., Richardson, W. D., McMahon, A. P. and Koentges, G. (2005). Neural crest origins of the neck and shoulder. Nature 436, 347. 10.1038/nature03837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk, P. D., Swartz, M. E., Chen, J. W., Galloway, J. L. and Eberhart, J. K. (2017). In vivo zebrafish morphogenesis shows Cyp26b1 promotes tendon condensation and musculoskeletal patterning in the embryonic jaw. PLoS Genet. 13, e1007112. 10.1371/journal.pgen.1007112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead, T. J., McCulloch, D. R., Ho, J. C., Du, Y., Adams, S. M., Birk, D. E. and Apte, S. S. (2018). The metalloproteinase-proteoglycans ADAMTS7 and ADAMTS12 provide an innate, tendon-specific protective mechanism against heterotopic ossification. JCI Insight 3, e92941. 10.1172/jci.insight.92941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, R., Song, M. and Pan, J. (2015). Rho is involved in periodontal tissue remodelling with experimental tooth movement in rats. Arch. Oral Biol. 60, 923-931. 10.1016/j.archoralbio.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Mienaltowski, M. J., Adams, S. M. and Birk, D. E. (2013). Regional differences in stem cell/progenitor cell populations from the mouse Achilles tendon. Tissue Eng. A 19, 199-210. 10.1089/ten.tea.2012.0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mienaltowski, M. J., Adams, S. M. and Birk, D. E. (2014). Tendon proper- and peritenon-derived progenitor cells have unique tenogenic properties. Stem Cell Res. Ther. 5, 86-86. 10.1186/scrt475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser, H. L., Doe, A. P., Meier, K., Garnier, S., Laudier, D., Akiyama, H., Zumstein, M. A., Galatz, L. M. and Huang, A. H. (2018). Genetic lineage tracing of targeted cell populations during enthesis healing. J. Orthop. Res. 36, 3275-3284. 10.1002/jor.24122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison, N. D., Price, B. A., Conner, D. A., Keene, D. R., Olson, E. N., Tabin, C. J. and Schweitzer, R. (2007). Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697-2708. 10.1242/dev.001933 [DOI] [PubMed] [Google Scholar]
- Nagashima, H., Sugahara, F., Watanabe, K., Shibata, M., Chiba, A. and Sato, N. (2016). Developmental origin of the clavicle, and its implications for the evolution of the neck and the paired appendages in vertebrates. J. Anat. 229, 536-548. 10.1111/joa.12502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassari, S., Duprez, D. and Fournier-Thibault, C. (2017). Non-myogenic contribution to muscle development and homeostasis: the role of connective tissues. Front. Cell Dev. Biol. 5, 22. 10.3389/fcell.2017.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield, S. F. and Evans, D. J. R. (2003). Tendon morphogenesis in the developing avian limb: plasticity of fetal tendon fibroblasts. J. Anat. 202, 153-164. 10.1046/j.1469-7580.2003.00145.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse, R. V., II, Esshaki, D., Tabin, C. J. and Murray, M. M. (2009). Genome-wide expression analysis of intra- and extraarticular connective tissue. J. Orthop. Res. 27, 427-434. 10.1002/jor.20774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo, P., Sabatino, V., Mithbaokar, P., Polishchuck, E., Law, S. K., Magraner-Pardo, L., Pons, T., Polishchuck, R. and Brunetti-Pierri, N. (2019). Geleophysic dysplasia: novel missense variants and insights into ADAMTSL2 intracellular trafficking. Mol. Genet. Metab. Rep. 21, 100504. 10.1016/j.ymgmr.2019.100504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce, B. A., Brent, A. E., Murchison, N. D., Tabin, C. J. and Schweitzer, R. (2007). Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Dev. Dyn. 236, 1677-1682. 10.1002/dvdy.21179 [DOI] [PubMed] [Google Scholar]
- Pryce, B. A., Watson, S. S., Murchison, N. D., Staverosky, J. A., Dünker, N. and Schweitzer, R. (2009). Recruitment and maintenance of tendon progenitors by TGFβ signaling are essential for tendon formation. Development 136, 1351-1361. 10.1242/dev.027342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, R. R., Bobzin, L., Teng, C. S., Pal, D., Tuzon, C. T., Schweitzer, R. and Merrill, A. E. (2019). FGF signaling patterns cell fate at the interface between tendon and bone. Development 146, dev170241. 10.1242/dev.170241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckebüsch, L., Hernandez, L., Wang, C., Phan, J. and Yelon, D. (2016). Tmem2 regulates cell-matrix interactions that are essential for muscle fiber attachment. Development 143, 2965-2972. 10.1242/dev.139485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salva, J. E. and Merrill, A. E. (2017). Signaling networks in joint development. Dev. Dyn. 246, 262-274. 10.1002/dvdy.24472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, A. G., Long, F. and Thomopoulos, S. (2015). Enthesis fibrocartilage cells originate from a population of Hedgehog-responsive cells modulated by the loading environment. Development 142, 196. 10.1242/dev.112714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, A. G., Galatz, L. M. and Thomopoulos, S. (2017). Enthesis regeneration: a role for Gli1+ progenitor cells. Development 144, 1159. 10.1242/dev.139303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer, R., Chyung, J. H., Murtaugh, L. C., Brent, A. E., Rosen, V., Olson, E. N., Lassar, A. and Tabin, C. J. (2001). Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 128, 3855-3866. [DOI] [PubMed] [Google Scholar]
- Seo, H.-S. and Serra, R. (2007). Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev. Biol. 310, 304-316. 10.1016/j.ydbio.2007.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore, E. M., Ahn, J., Jan de Beur, S., Li, M., Xu, M., Gardner, R. J. M. K., Zasloff, M. A., Whyte, M. P., Levine, M. A. and Kaplan, F. S. (2002). Paternally inherited inactivating mutations of the GNAS1 gene in progressive osseous heteroplasia. N Engl. J. Med. 346, 99-106. 10.1056/NEJMoa011262 [DOI] [PubMed] [Google Scholar]
- Shukunami, C., Takimoto, A., Nishizaki, Y., Yoshimoto, Y., Tanaka, S., Miura, S., Watanabe, H., Sakuma, T., Yamamoto, T., Kondoh, G.et al. (2018). Scleraxis is a transcriptional activator that regulates the expression of Tenomodulin, a marker of mature tenocytes and ligamentocytes. Sci. Rep. 8, 3155. 10.1038/s41598-018-21194-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwartz, Y., Viukov, S., Krief, S. and Zelzer, E. (2016). Joint development involves a continuous Influx of Gdf5-positive cells. Cell Rep. 15, 2577-2587. 10.1016/j.celrep.2016.05.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeeton, J., Askary, A. and Crump, J. G. (2017). Building and maintaining joints by exquisite local control of cell fate. Wiley Interdiscip. Rev. Dev. Biol. 6, wdev.245. 10.1002/wdev.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snedeker, J. G. and Foolen, J. (2017). Tendon injury and repair – a perspective on the basic mechanisms of tendon disease and future clinical therapy. Acta Biomater. 63, 18-36. 10.1016/j.actbio.2017.08.032 [DOI] [PubMed] [Google Scholar]
- Staverosky, J. A., Pryce, B. A., Watson, S. S. and Schweitzer, R. (2009). Tubulin polymerization-promoting protein family member 3, Tppp3, is a specific marker of the differentiating tendon sheath and synovial joints. Dev. Dyn. 238, 685-692. 10.1002/dvdy.21865 [DOI] [PubMed] [Google Scholar]
- Subramanian, A. and Schilling, T. F. (2014). Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. eLife 3, e02372. 10.7554/eLife.02372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A. and Schilling, T. F. (2015). Tendon development and musculoskeletal assembly: emerging roles for the extracellular matrix. Development 142, 4191-4204. 10.1242/dev.114777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, A., Kanzaki, L. F., Galloway, J. L. and Schilling, T. F. (2018). Mechanical force regulates tendon extracellular matrix organization and tenocyte morphogenesis through TGFbeta signaling. eLife 7, e38069. 10.7554/eLife.38069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto, Y., Takimoto, A., Akiyama, H., Kist, R., Scherer, G., Nakamura, T., Hiraki, Y. and Shukunami, C. (2013). Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 140, 2280. 10.1242/dev.096354 [DOI] [PubMed] [Google Scholar]
- Suzuki, H., Ito, Y., Shinohara, M., Yamashita, S., Ichinose, S., Kishida, A., Oyaizu, T., Kayama, T., Nakamichi, R., Koda, N.et al. (2016). Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc. Natl Acad. Sci. USA 113, 7840. 10.1073/pnas.1522054113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, I., Chaqour, B., Howard, P. S., Ikeo, T. and Macarak, E. J. (2008). Effect of fibroblast growth factor-1 on the expression of early growth response-1 in human periodontal ligament cells. J. Periodontal Res. 43, 305-310. 10.1111/j.1600-0765.2007.01030.x [DOI] [PubMed] [Google Scholar]
- Tan, G.-K., Pryce, B. A., Stabio, A., Brigande, J. V., Wang, C. J., Xia, Z., Tufa, S. F., Keene, D. R. and Schweitzer, R. (2020). Tgfβ signaling is critical for maintenance of the tendon cell fate. eLife 9, e52695. 10.7554/eLife.52695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, Y., Wu, X., Lei, W., Pang, L., Wan, C., Shi, Z., Zhao, L., Nagy, T. R., Peng, X., Hu, J.et al. (2009). TGF-beta1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 15, 757-765. 10.1038/nm.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomopoulos, S., Parks, W. C., Rifkin, D. B. and Derwin, K. A. (2015). Mechanisms of tendon injury and repair. J. Orthop. Res. 33, 832-839. 10.1002/jor.22806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari, P., Kumar, A., Das, R. N., Malhotra, V. and VijayRaghavan, K. (2015). A tendon cell specific RNAi screen reveals novel candidates essential for muscle tendon interaction. PLoS ONE 10, e0140976. 10.1371/journal.pone.0140976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozer, S. and Duprez, D. (2005). Tendon and ligament: development, repair and disease. Birth Defects Res. C Embryo Today Rev. 75, 226-236. 10.1002/bdrc.20049 [DOI] [PubMed] [Google Scholar]
- Valasek, P., Theis, S., Krejci, E., Grim, M., Maina, F., Shwartz, Y., Otto, A., Huang, R. and Patel, K. (2010). Somitic origin of the medial border of the mammalian scapula and its homology to the avian scapula blade. J. Anat. 216, 482-488. 10.1111/j.1469-7580.2009.01200.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y., Zhang, X., Huang, H., Xia, Y., Yao, Y., Mak, A. F.-T., Yung, P. S.-H., Chan, K.-M., Wang, L., Zhang, C.et al. (2017). Osteocalcin expressing cells from tendon sheaths in mice contribute to tendon repair by activating Hedgehog signaling. eLife 6, e30474. 10.7554/eLife.30474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, L. S., Shenker, A., Gejman, P. V., Merino, M. J., Friedman, E. and Spiegel, A. M. (1991). Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N Engl. J. Med. 325, 1688-1695. 10.1056/NEJM199112123252403 [DOI] [PubMed] [Google Scholar]
- Wren, T. A., Beaupré, G. S. and Carter, D. R. (2000). Tendon and ligament adaptation to exercise, immobilization, and remobilization. J. Rehabil. Res. Dev. 37, 217-224. [PubMed] [Google Scholar]
- Yamamoto-Shiraishi, Y.-I. and Kuroiwa, A. (2013). Wnt and BMP signaling cooperate with Hox in the control of Six2 expression in limb tendon precursor. Dev. Biol. 377, 363-374. 10.1016/j.ydbio.2013.02.023 [DOI] [PubMed] [Google Scholar]
- Yin, Z., Guo, J., Wu, T.-Y., Chen, X., Xu, L.-L., Lin, S.-E., Sun, Y.-X., Chan, K.-M., Ouyang, H. and Li, G. (2016). Stepwise differentiation of mesenchymal stem cells augments tendon-like tissue formation and defect repair in vivo. Stem Cells Transl. Med. 5, 1106-1116. 10.5966/sctm.2015-0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, R., Alaee, F., Dyrna, F., Kronenberg, M. S., Maye, P., Kalajzic, I., Rowe, D. W., Mazzocca, A. D. and Dyment, N. A. (2016). Murine supraspinatus tendon injury model to identify the cellular origins of rotator cuff healing. Connect. Tissue Res. 57, 507-515. 10.1080/03008207.2016.1189910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto, Y., Takimoto, A., Watanabe, H., Hiraki, Y., Kondoh, G. and Shukunami, C. (2017). Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci. Rep. 7, 45010. 10.1038/srep45010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Zhou, D., Wang, H. and Tan, J. (2020). Heterotopic ossification of tendon and ligament. J. Cell. Mol. Med. 24, 5428-5437. 10.1111/jcmm.15240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Lin, Z., Foolen, J., Schoen, I., Santoro, A., Zenobi-Wong, M. and Vogel, V. (2014). Disentangling the multifactorial contributions of fibronectin, collagen and cyclic strain on MMP expression and extracellular matrix remodeling by fibroblasts. Matrix Biol. 40, 62-72. 10.1016/j.matbio.2014.09.001 [DOI] [PubMed] [Google Scholar]
- Zhen, G., Wen, C., Jia, X., Li, Y., Crane, J. L., Mears, S. C., Askin, F. B., Frassica, F. J., Chang, W., Yao, J.et al. (2013). Inhibition of TGF-β signaling in mesenchymal stem cells of subchondral bone attenuates osteoarthritis. Nat. Med. 19, 704-712. 10.1038/nm.3143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziermann, J. M., Diogo, R. and Noden, D. M. (2018). Neural crest and the patterning of vertebrate craniofacial muscles. Genesis 56, e23097. 10.1002/dvg.23097 [DOI] [PubMed] [Google Scholar]