Abstract
Spaceflight causes a decrease in skeletal muscle mass and strength. We set two murine experimental groups in orbit for 35 days aboard the International Space Station, under artificial earth-gravity (artificial 1 g; AG) and microgravity (μg; MG), to investigate whether artificial 1 g exposure prevents muscle atrophy at the molecular level. Our main findings indicated that AG onboard environment prevented changes under microgravity in soleus muscle not only in muscle mass and fiber type composition but also in the alteration of gene expression profiles. In particular, transcriptome analysis suggested that AG condition could prevent the alterations of some atrophy-related genes. We further screened novel candidate genes to reveal the muscle atrophy mechanism from these gene expression profiles. We suggest the potential role of Cacng1 in the atrophy of myotubes using in vitro and in vivo gene transductions. This critical project may accelerate the elucidation of muscle atrophy mechanisms.
Subject terms: Musculoskeletal system, Transcriptomics, Functional clustering, Gene ontology, Gene regulatory networks, Gene expression, RNA sequencing
Introduction
Gravity is the most constant factor affecting the entire process of evolution of organisms on Earth. As adapting to a changing environment is key for any organism’s survival, the constant mechanical stimulus of gravitational force has been shared by all organisms on Earth through evolution1. To elucidate how mammals respond to gravity, space experiments with mice are quintessential; however, they require specialized habitats applicable to the space environment.
Skeletal muscle is one of the most robustly plastic tissues that can adapt its structure and metabolism in response to various conditions2,3. Under disuse conditions such as prolonged time in bed, skeletal muscles undergo atrophy due to decreased protein synthesis and increased protein degradation4. Spaceflight is well-known to induce muscle atrophy and weakness in rats5,6 and mice7. In humans8–10, muscle atrophy during spaceflight can cause severe medical problems to astronauts upon return to Earth. Previous studies have shown that muscle atrophy and partial shift of muscle fibers from oxidative toward a more glycolytic phenotype were induced in space-flown rats and mice, as well as humans11. Therefore, there is a critical need for a deeper understanding of the molecular mechanisms responsible for muscle atrophy under microgravity and partial gravity (less than 1 g).
The effect of spaceflight on skeletal muscles has been examined in rats and mice, comparing a spaceflight group with a ground control group6,12,13. However, such studies did not consider various conditions, including space radiation, the microbial environment, lack of convection, and shock during the launch and return phases that differ between spaceflight and conventional control experiments. Comparing between the two control groups was needed to rule out the confounding factors originating from different housing conditions, such as the effect of short-term gravitational changes during launch and landing in spaceflight. Therefore, we developed the multiple artificial-gravity research system (MARS) and succeeded in housing mice under microgravity and an artificial 1 g onboard environment to study the effect of gravity on mammals14–16. In the first Mouse Habitat Unit-1 (MHU-1) mission, 12 mice were housed in the International Space Station (ISS) using MARS under either microgravity or artificial 1 g for 35 days in 2016. Each male mouse was reared in an individual cage to avoid unexpected fighting between mice, and all mice returned to the Earth alive. We set an artificial 1 g onboard environment induced by centrifuging cages as the novel control. This is because artificial 1 g onboard was a novel experimental group, we employed a comprehensive assessment against the effects of microgravity and artificial 1 g. Our findings from the MHU-1 mission were as follows; retinal apoptosis caused by spaceflight was prevented in artificial 1 g17; downregulation of transcription factors GATA1 and Tal1 (regulator of erythropoiesis) in the immune system was observed in microgravity18; and the physiological functions of the male reproductive organs, such as sperm function and offspring viability, were maintained in microgravity19.
In this study, we aimed to elucidate the effect of an artificial 1 g onboard environment on skeletal muscles at the molecular level. We investigated the histological characterization of muscle fibers and gene expression from the JAXA mission (MHU-1). We found that an artificial 1 g onboard environment prevented not only decreased muscle mass and change in fiber type composition but also the alteration of gene expression profiles in soleus muscle during spaceflight. Transcriptome analysis also suggested that AG condition could prevent the alterations of well-known atrophy-related genes. We further screened the candidate genes associated with muscle atrophy using these gene profiles. We identified Cacng1 that could play a functional role in myotubes atrophy by in vitro and in vivo gene transductions. Therefore, our project demonstrates the utility of spaceflight datasets using an artificial 1 g onboard environment for elucidating the basis of muscular adaptation during spaceflight.
Results and discussion
An artificial 1 g onboard environment prevents changes in muscle mass and the fiber type composition of the soleus muscle under microgravity
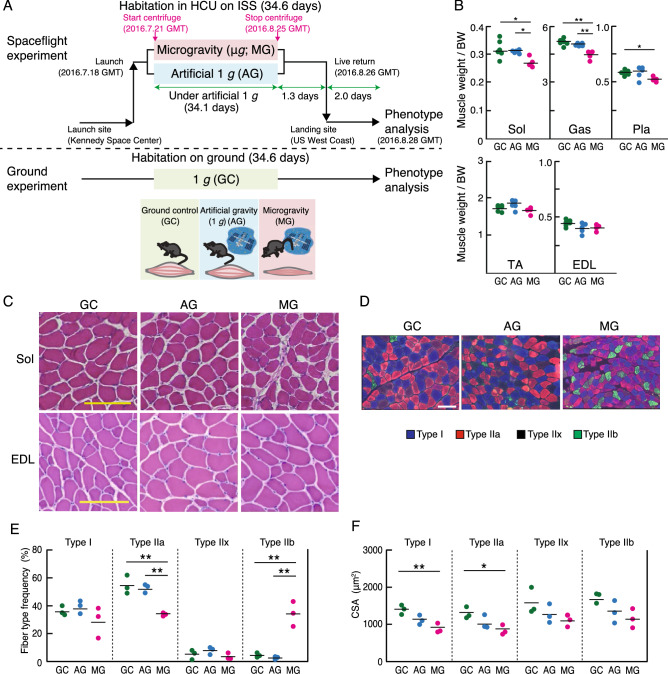
Twelve mice were housed onboard the ISS for 35 days. Half of them were housed under microgravity (MG), whereas the other 6 mice were housed under an artificial 1 g onboard environment (AG) induced by centrifuging (Fig. 1A). Since previous studies using ground control group did not distinguish the effect of microgravity from other various conditions such as space radiation and lack of convection, we examined whether artificial 1 g exposure prevents the reduction in muscle mass induced by spaceflight in mice. To compare the effects of microgravity and an artificial 1 g onboard environment on skeletal muscle, we measured the wet muscle mass of hindlimb skeletal muscles, namely soleus, gastrocnemius, plantaris, tibialis anterior (TA), and extensor digitorum longus (EDL) (Fig. 1B). The muscle weights were normalized using the body weight of each mouse. Previously, we reported that AG completely inhibited the effect of microgravity on soleus, which is the main postural muscle of hindlimb and gastrocnemius muscles, compared with MG and ground control (GC), which was housed on the ground for the same duration using similar cages and processed with the same method as spaceflight mice14. The weight of the plantaris muscle in MG was also significantly decreased compared with that of GC, although the weight of the plantaris muscle in MG tended to decrease compared with AG. The weight of the TA and EDL muscles showed no significant differences among the three groups. Hematoxylin–eosin (HE) staining of freshly-embedded soleus (predominantly slow-twitch fibers) and EDL (predominantly fast-twitch fibers) muscle sections, which are composed of different types of muscle fibers, showed the absence of major pathological anomalies, such as central nuclei, immune cell infiltration, and myofiber degeneration in all analyzed muscles from each experimental group: GC, AG, and MG (Fig. 1C). The previous studies have shown that the change in muscle fiber type composition in both type I and type II is observed in the soleus muscle after spaceflight in mice, rats, and humans11,20. Additionally, atrophy is reportedly more pronounced in type I than in type II fibers of the soleus muscle. Therefore, immunofluorescence staining of the different myosin heavy chain (MyHC) isoforms (i.e., types I, IIa, IIb) was performed for evaluating whether microgravity or artificial 1 g exposure in space affected muscle fiber type composition in the soleus muscle (Fig. 1D). The type IIx fibers were identified by the absence of immunoreaction during triple immunostaining. We quantified the fiber type frequency and cross-sectional area (CSA) of the myofibers by immunofluorescence staining of the soleus muscle (Fig. 1E, F). The ratio of type IIa fibers was reduced, while that of type IIb fibers increased in the soleus muscle under MG compared to that of under GC. The decrease of type IIa fibers ratio and the increase of type IIb fibers ratio were completely prevented in AG, implying that the change in fiber type composition induced by spaceflight was mainly caused by unloading due to microgravity. Additionally, the CSA significantly decreased in type I and type IIa fibers in MG compared with those in GC. These reductions in fibers were not significant when comparing GC and AG (Fig. 1F), suggesting that AG tends to prevent the reduction of CSA during spaceflight. These inflight 1 g control results suggested that an artificial 1 g onboard environment using centrifuge could prevent change in the fiber type composition and muscle atrophy during spaceflight in both the fast- and slow-twitch fibers of the soleus muscle. In contrast, no changes in the CSAs of EDL muscles were observed among the three groups (Supplementary Fig. 1). These results demonstrated that the postural muscles of lower limbs were highly responsive to microgravity. Our histological data supported previous findings of fiber composition change exclusively observed in the soleus muscle exposed to microgravity for 30 days using BION-M113. In addition, change in fiber type composition was observed in the soleus muscle, but not the EDL muscle, of mice housed onboard for as long as 91 days (MDS mission, 2009)12. Our findings showed that an artificial 1 g onboard environment is sufficient to prevent change in the fiber type composition, and has a potential to partially prevent muscle atrophy in the soleus muscle during spaceflight.
Figure 1.
Characterization of the hindlimb muscles among three experimental groups: GC, AG, and MG. (A) Overview of the MHU-1 study. Twelve male mice were launched by SpaceX-9 from the Kennedy Space Center in Florida on July 18, 2016. After arrival at the ISS, six of the mice were housed in HCU under microgravity (MG) for approximately 34.6 days, while the other six mice were housed in an artificial 1 g onboard environment (AG) induced by centrifugation for approximately 34.1 days during the approximate 37-day berthing period. After approximately 35 days of habitation on the ISS, all 12 mice were loaded into the Dragon capsule and kept under microgravity for approximately 1.3 days before undocking. The capsule subsequently splashed down in the Pacific Ocean near Southern California on August 26, 2016. The mice were then transported to a laboratory for behavioral tests and other analyses after approximately 2 days of undocking. Graphical representations of the study design using the multiple artificial-gravity research system (MARS). Three experimental groups, namely, ground control (GC), artificial 1 g onboard environment (AG), and microgravity (MG) in the ISS, are color-coded as green, blue, and red, respectively, in the following bar graphs. (B) Quantification of the change in individual skeletal muscle weights: soleus (Sol), gastrocnemius (Gas), plantaris (Pla), tibialis anterior (TA), and extensor digitorum longus (EDL), among GC (n = 6), AG (n = 5), and MG (n = 4) groups, normalized by body weight and graphed in the Tukey boxplot. P-value from Tukey’s test is indicated as follows: *P < 0.05 and **P < 0.01. (C) Hematoxylin–eosin staining of soleus and EDL sections. Scale bars indicate 100 μm. (D) Immunostaining of the myosin heavy chain using muscle fiber-specific antibodies (type I; blue, type IIa; red, type IIb; green) in the soleus muscle. The unstained fibers were predicted to be of type IIx fibers (black). Scale bars indicate 100 μm. (E, F) Frequency (E) and cross-sectional areas (CSA) (F) of each fiber type in the soleus muscles of the GC (n = 3), AG (n = 3), and MG (n = 3). P-value from Student’s t-test is indicated as follows: *P < 0.05 and **P < 0.01.
Artificial 1 g onboard environment prevents the alteration of gene expression under microgravity
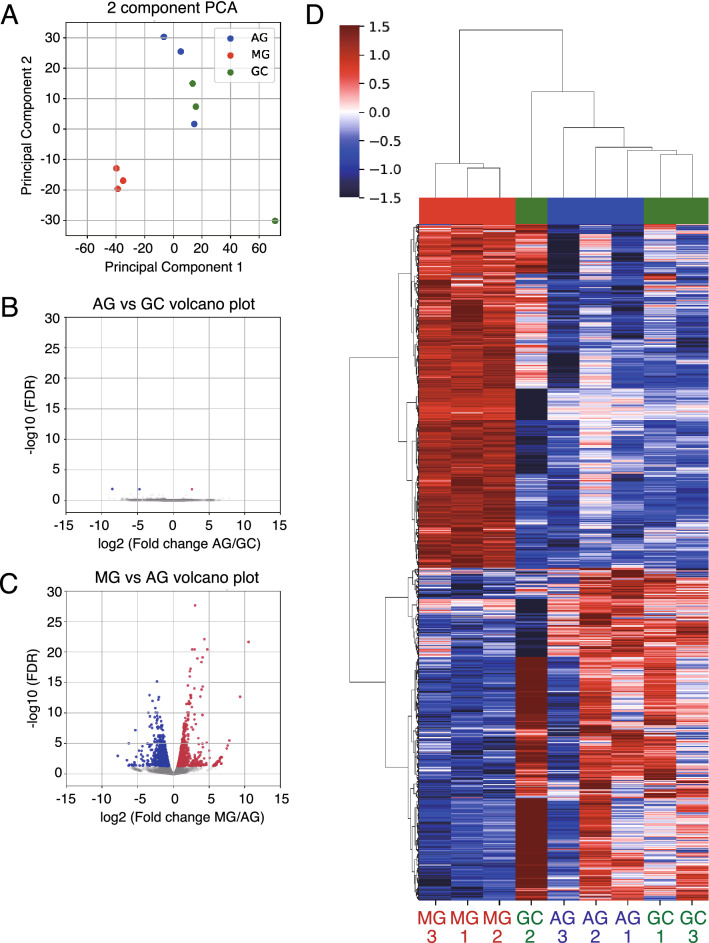
The previous studies have compared the global gene expression profile of the soleus muscle following microgravity exposure with that of the ground control group using microarray13 and RNA-seq21 techniques. However, as inflight 1 g exposed control groups have not been used in these studies, it remains to be elucidated whether the loading gravity is sufficient for maintaining the altered gene expression in the soleus muscle during spaceflight. A previous study demonstrated that the largest number of differentially expressed genes (DEGs) is found in the soleus muscle among nine different tissues in mice housed in the ISS for 37 days22. Therefore, in order to investigate skeletal muscle adaptation during spaceflight using the MARS platform, we focused on the soleus muscle, which is one of the representative antigravitational muscles. To investigate the comprehensive impact of gene expressions due to microgravity and artificial 1 g exposure in spaceflight, we performed transcriptome analysis of the soleus muscles from MG, AG, and GC mice (GC: n = 3; AG: n = 3; MG: n = 3). Principal component analysis (PCA) showed that MG mice’s gene expression profiles considerably differed from those of AG and GC mice (Fig. 2A). In addition, the gene expression profiles between AG and GC are relatively similar with the exception for one GC mouse (Fig. 2A). These results suggested that gravitational change could affect the muscle’s gene expression profile in mice under spaceflight for 35 days rather than other events, such as space radiation. Indeed, comparing the two control groups (i.e., GC and AG), only three genes, namely Nr4a3, Sfrp4, and Gm21541, were differentially regulated in the soleus muscle (Fig. 2B and Supplementary Table 1). The slight difference between these two control groups could be caused by different environmental factors, such as the effect of short-term gravitational changes during launch and landing in spaceflight. Comparing AG and MG, 1,720 genes were differentially regulated in the soleus muscle with 874 upregulated and 846 downregulated genes (Fig. 2C), although 1,992 genes were differentially regulated in the soleus muscle with 810 upregulated genes and 1,182 downregulated genes by comparing GC and MG (Supplementary Fig. 2). These results suggested microgravity may contribute to the change of comprehensive gene expressions. In other words, our results suggested that an artificial 1 g onboard environment ensured by centrifuging the cages prevented the alteration of gene expression under microgravity in the soleus muscle. We next focused on the subsequent analysis of genes differentially regulated in MG compared with AG. Hierarchical clustering was performed on DEGs of 1,720 genes (AG vs. MG) with all samples for GC, AG, and MG. The result could also confirm the similarity of gene expression profiles of the two control groups (GC and AG), compared with MG (Fig. 2D).
Figure 2.
Gene expression profiling in the soleus muscle of GC, AG, and MG. (A) PCA of DEGs in at least one comparison among the soleus muscle of GC (n = 3; green), AG (n = 3; blue), and MG (n = 3; red). (B, C) Volcano plots showing DEGs in the soleus muscle of AG and GC (B), and MG and AG (C). (D) Heat map and hierarchical clustering of the gene expression data in the soleus muscle of GC, AG, and MG showing 1,720 DEGs in AG and MG.
In summary, we demonstrated that an artificial 1 g onboard environment could prevent gene expression changes observed under microgravity, which suggested that gravitational change rather than other events such as space radiation largely affected the gene expression changes in the soleus muscle under spaceflight for 35 days. Besides, the GC experiment using the same cage unit as spaceflight conditions is acceptable for analyzing microgravity’s effect on mice’s muscles in spaceflight.
An artificial 1 g onboard environment could block alteration of atrogene expressions under microgravity
Muscle atrophy is mediated by transcription-dependent regulation of atrogenes (atrophy-related genes), whose expression is coordinately upregulated or downregulated in muscles during systemic wasting states, such as fasting, cancer cachexia, renal failure, and diabetes23. In particular, a previous study demonstrated that the E3 ubiquitin ligase MuRF1 (Trim63) null (KO) mice showed protection in ground-based models of muscle atrophy under multiple conditions, including denervation and immobilization24. However, the deletion of MuRF1 in mice did not prevent muscle atrophy during spaceflight, suggesting that muscle atrophy occurs through different mechanisms during spaceflight compared with those observed in ground-based models of atrophy21.
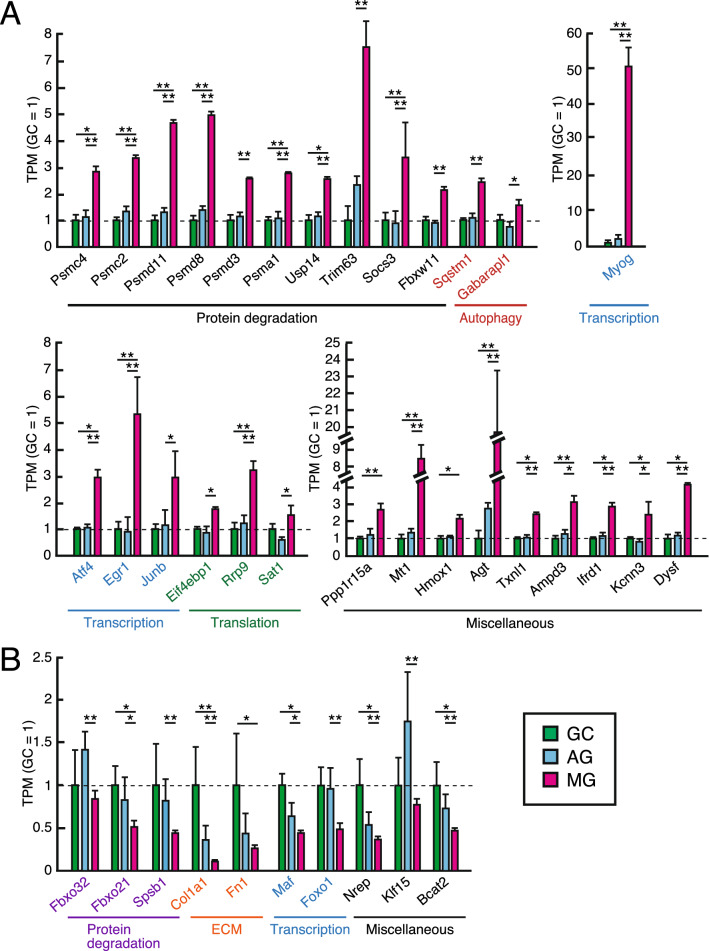
Hence, to elucidate the effects of microgravity and an artificial 1 g onboard environment on the gene expression profile of atrogenes, we identified genes under the regulation of microgravity by cross-referencing the list of atrogenes25–28. RNAseq analysis confirmed the differentially regulated atrogenes in MG compared with GC and/or AG (Fig. 3A, B). Consistent with muscle morphology, gene expressions involved in protein degradation mainly by the ubiquitin–proteasome system were significantly upregulated in MG (Fig. 3A). This finding includes the genes encoding for various subunits of the 26S proteasome, such as subunits of the 19S complex (Psmc4, Psmc2, Psmd11/Rpn6, Psmd8/Rpn12, and Psmd3), 20S core particle (Psma1, and Psmb3), de-ubiquitinating gene (Usp14), ubiquitin-fusion protein (Rps27a), and the ubiquitin E3 ligase complexes, Trim63 (MuRF1), Socs3, and F-box/WD repeat-containing protein 11 (Fbxw11). Although little is known about the substrates and functions of these E3 ligases in skeletal muscles, Fbxw11 (also known as β-TrCP2) plays a role in targeting IkBα for ubiquitin-dependent degradation29. In addition, autophagy-related target genes (P62/Sqstm1 and Gabarapl1), which are involved in protein degradation through the lysosomal/autophagy pathway, were upregulated in MG. In contrast, some of the atrogenes such as Atrogin-1/MAFbx (Fbxo32), Specific of Muscle Atrophy and Regulated by Transcription (SMART; Foxo21)28, splA/ryanodine receptor domain and SOCS box (Spsb1)26 were downregulated in MG compared with AG as shown in Fig. 3B.
Figure 3.
Differentially expressed genes of atrogenes in the soleus muscle of GC, AG, and MG. Bar diagrams showing the gene expression of atrogenes (atrophy-related genes) as significantly upregulated (A) and downregulated (B) in MG compared with GC and/or AG. FDR-corrected P-value from statistical test is indicated as follows: *FDR-corrected P < 0.05 and **FDR-corrected P < 0.01. GC (Green), AG (Blue), and MG (Red).
P311/Nrep, which is involved in smooth muscle differentiation30 and reported to be downregulated in skeletal muscle after denervation31, was downregulated in MG compared with AG and GC (Fig. 3B). Eif4ebp1, which inhibits translation initiation factors, was upregulated in MG compared with AG and could indicate a reduction in translation in muscle25. A decrease in mRNAs for ECM proteins, Col1a1, was also observed in MG compared with AG. The transcription factor Atf4, which promotes the expression of oxidative stress-responsive genes and is induced during muscle atrophy32, was induced in MG. Thioredoxin-like protein and Metallothionein-1 (Mt1), which is upregulated during rodent sarcopenia33, were significantly upregulated in MG compared with AG and GC.
Egr1 was significantly elevated in MG compared with GC, whose induction was significantly blocked in AG. However, the gene expression involved in ATP synthesis was not significantly changed among the three groups (Supplementary Table 2). Myogenin (Myog) is a muscle-specific transcriptional factor that is highly induced following muscle denervation22 and contributes to neurogenic muscle atrophy34. Myog was significantly increased in MG as compared with AG and GC. Foxo1 and Maf were significantly decreased in MG as compared with AG and GC. Although the Kruppel-like factor 15 (Klf15) mediates the inhibition of mTOR in glucocorticoid-induced muscle atrophy through the induction upregulation of branched-chain amino acid transaminase 2 (Bcat2)35, the expression of Klf15 and Bcat2 was downregulated in MG compared with AG. The increase of Klf15 was observed in AG compared with MG, despite no significant differences between GC and MG. This increased expression in AG compared with MG was also observed in Atrogin-1 (Fbxo 32), known as the target genes of Klf15.
In summary, the expressions of a set of atrogene were significantly changed in the soleus muscle under MG, and these altered expressions were blocked in that under AG. Atrogenes are known to be tightly and rapidly controlled, upregulated during atrophy, and returned to normal levels during recovery36. However, these results indicated that the expression of atrogenes was sustained in MG, although mice in MG were exposed to 1 g after returning to Earth. Conversely, some atrogenes such as Klf15 were significantly increased in AG compared with MG, although the expression in MG was comparable to that in GC. It is possible that short-term exposure of AG mice to microgravity during the return phase might affect the gene expression. In addition, the expression of autophagy-related gene Gabarapl1is known to increase transiently both during atrophy and recovery following disuse muscle atrophy37. Moreover, activation of protein degradation such as ubiquitin–proteasome system was also observed in soleus muscle during early reloading stages (18 h.) after disuse muscle atrophy in rats38. It is still possible that the genes upregulated in MG have comprised recovery-related genes. Taken together, these results indicated that a set of atrogene expressions previously reported by ground experiments was changed under microgravity for 35 days, and that an artificial 1 g onboard environment could prevent the alteration of most atrogene expressions during spaceflight.
Novel candidate genes associated with muscle atrophy were screened from gene expression profiles under spaceflight
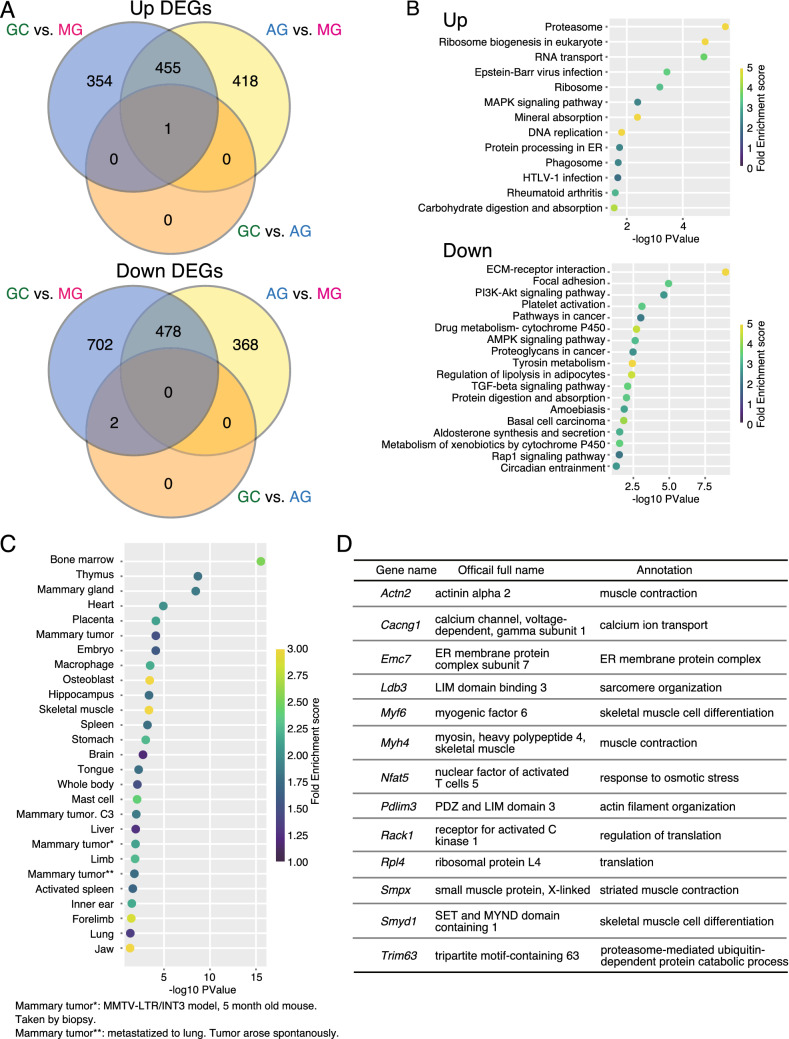
Novel candidate genes associated with muscle atrophy were detected through a combination of comparative and in silico approach. Among the 1,992 genes differentially regulated in GC and MG (810 upregulated and 1,182 downregulated) and the 1,720 genes differentially regulated in AG and MG (874 upregulated and 846 downregulated), 455 genes were upregulated and 478 genes were downregulated, as shown in the overlapping region between the two comparisons, GC vs. MG and AG vs. MG in the Venn diagrams (Fig. 4A). To identify the functional gene clusters, we performed enrichment analysis for 455 upregulated genes and 478 downregulated genes in MG compared with GC and AG in the overlapping region between these two comparisons. The upregulated genes in MG were enriched in the pathways of the proteasome, ribosome biogenesis, RNA transport, MAPK signaling, mineral absorption, DNA replication, and lysosome (Fig. 4B). In contrast, downregulated genes in MG were enriched in the pathways of ECM-receptor interaction, focal adhesion, PI3K-Akt signaling, AMPK signaling, and TGF-β signaling. Consistent with this, downregulation of TGF-β1 was reported in the soleus muscle of mice on the ISS39. Also, decreased PI3K-Akt signaling was observed in the short-term spaceflight response in the gastrocnemius muscle in a previous study40. The upregulation of the proteasome pathway and the downregulation of the PI3K-Akt signaling pathway, which are involved in protein degradation and protein synthesis, respectively, were observed during muscle atrophy in MG.
Figure 4.
Pathway analysis of changed genes between MG and GC, and MG and AG, and identifying novel candidate genes involved in muscle atrophy. (A) Venn diagram showing the number of upregulated (upper) and downregulated (lower) DEGs in each comparison between MG, GC, and AG. (B) Pathway analysis of upregulated (upper) and downregulated (lower) DEGs in the overlapping region of MG compared with GC and AG. (C) Tissue expression profiles of 418 upregulated DEGs in MG compared with AG. (D) The gene list showing skeletal muscle-related genes from Fig. 4C.
Since AG was the new research condition under an artificial 1 g onboard environment, we screened the potential candidate genes involved in muscle atrophy under microgravity by focusing on genes differentially regulated between AG and MG. A total of 418 upregulated genes were significantly changed only specifically in the comparison between AG and MG, but not in that between GC and MG. The previous studies investigating the research on skeletal muscles by comparing spaceflight groups with ground control groups6,12,13 could not validate the effect of various factors during spaceflight, such as space radiation. The comparison between spaceflight and inflight control groups, instead of conventional control experiments, may provide new insights into understanding the mechanism of muscle atrophy during spaceflight. As the regulatory genes that induce muscular atrophy were found in upregulated genes in the previous reports24, we focused on the genes that were upregulated for identifying the novel molecule(s) that are involved in muscular atrophy under microgravity. Upregulated genes were distributed into the tissue expression profiles provided by the enrichment analysis (Fig. 4C). Thirteen skeletal muscle-related genes listed in Fig. 4D were significantly regulated in MG compared to AG in the soleus muscle. The functions of eight of the thirteen genes identified herein have been reported in muscle as follows: muscle development (Actn2), fiber type (Myh4), regeneration (Nfat5), dystrophy (Ldb3), and muscle atrophy (Myf6, Trim63, Smyd1, and Rack1). Pdlim3 and Smpx were known not to affect the muscle when deficient (Pdlim3) or overexpressed in muscle (Smpx). Actn2, Ldb3 and Pdlim3 were known to be significantly upregulated in the cutaneous muscle of mice housed in the ISS for 91 days41, suggesting the potential role in diverse muscles during spaceflight. Myh4 is one of the fast specific genes coding for MyHC type IIb. Upregulation of Myh4 mRNA is consistent with the result that an increase in the ratio of type IIb fibers in MG was completely prevented in AG in the soleus muscle from MyHC staining, which is also reported in the soleus muscle of mice exposed to microgravity for 30 days on BION-M113. NFAT5 has a role in both myoblast migration and differentiation during skeletal muscle myogenesis42, suggesting muscle regeneration in response to recovery after a return. Although Myf6 is involved in differentiation as demonstrated by the temporal pattern of expression in embryos43,44, the role of Myf6 in adult skeletal muscle is still elusive. Myf6 was upregulated in both the soleus muscles of space-flown mice of the BION-M1 experiment13. Previous studies suggested that Myf6 is involved in the gene expression reprogramming in denervated and regenerating muscle45,46. Therefore, Myf6 could have a role in the response to muscle atrophy during spaceflight and/or recovery from microgravity. In addition, MuRF1 (Trim63) was increased and screened from this study. Although MuRF1 was upregulated in various ground-based models of muscle atrophy, muscle atrophy during spaceflight was not prevented in soleus muscle of MuRF1 KO mice in the previous study21.
On the other hand, the function of Cacng1 remains unclear in skeletal muscle. Cacng1 encodes γ1 subunit of the L-type Ca2+ channel, consisting of five subunits (α1S, α2, β, γ and δ) and is known as the dihydropyridine receptor (DHPR)47,48. DHPR is predominantly located in T-tables and interacts with the type 1 ryanodine sensitive Ca2+ release channel, RyR1, to release Ca2+ stored in the terminal cisternae of the sarcoplasmic reticulum for the initiation of contraction49. However, the biological significance of the DHPR is still unknown50. Therefore, we further validated whether Cacng1 implicates muscle atrophy in vitro.
Cacng1 induces a decrease in muscle fiber size in vitro and in vivo
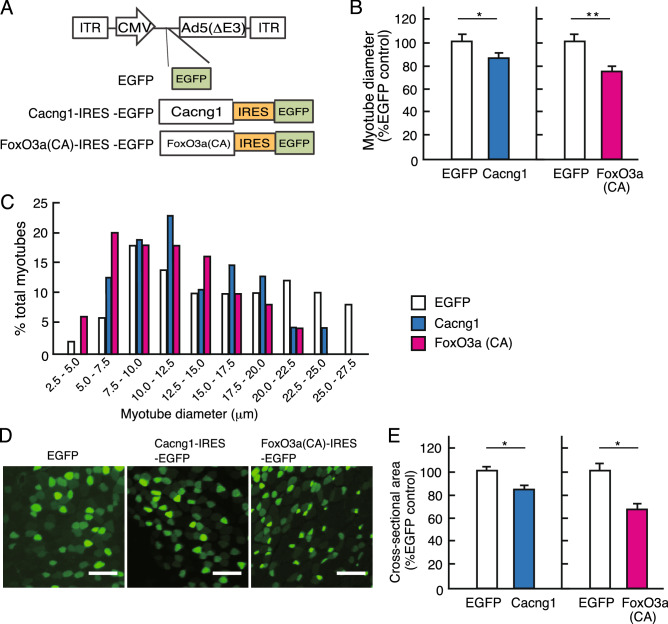
Based on in silico analyses, we identified Cacng1 as a novel candidate gene associated with muscle atrophy. Therefore, we further examined whether Cacng1 induces muscle atrophy in vitro. C2C12 myotubes were infected with adenoviral vectors expressing Cacng1 4 days after differentiation when the myotubes are of constant size, and myoblast fusion no longer occurs51 (Fig. 5A). Morphological examination of the cells expressing Cacng1 for 48 h. demonstrated a 15.2% reduction in mean myotube diameter and a leftward shift in myotube distribution compared with those expressing only EGFP control (Fig. 5B, C). Furthermore, when the myotubes were infected with a constitutively active mutant of FoxO3 (T32A, S252A, and S314A), FoxO3a (CA), a master regulator of muscle atrophy52, there was a 27.5% decrease in mean myotube diameter and a leftward shift in myotube distribution compared with those expressing only EGFP control (Fig. 5B, C). These findings demonstrate that the increased expression of Cacng1 has a key role in the atrophy of myotubes in vitro.
Figure 5.
Functional validation of candidate genes using culture cell and neonatal mice muscle. (A) Construction of adenovirus vectors, only EGFP, Cacng1-IRES-EGFP, and FoxO3a (constitutive active; CA)-IRES-EGFP. (B) Quantification of the myotube diameters of C2C12 myotubes expressing EGFP, Cacng1-IRES-EGFP, and FoxO3a(CA)-IRES-EGFP. P-value from Student’s t-test is indicated as follows: *P < 0.05 and **P < 0.01. (C) Frequency distribution of myotube diameters in C2C12 myotubes expressing EGFP, Cacng1-IRES-EGFP, or FoxO3a(CA)-IRES-EGFP. (D) Representative cross-sectional areas of TA muscles expressing EGFP (Green) after the transfection of EGFP, Cacng1-IRES-EGFP, or FoxO3a(CA)-IRES-EGFP. Scale bars indicate 50 μm. (E) Bar diagrams showing the cross-sectional area of TA muscles expressing EGFP. P-value from Student’s t-test is indicated as follows: * P < 0.05. Three independent infections were performed in three mice.
To determine whether the induction of Cacng1 expression influences fiber size in vivo, as it did in cultured myotubes, we conducted the gene transduction of Cacng1 in neonatal mice’s skeletal muscle. Fiber sizes were measured 5 days after the Cacng1-expressing adenovirus vector was injected into neonatal mice’s TA muscle. Muscle fibers expressing FoxO3a (CA) shown in Fig. 5D were much smaller than those expressing only EGFP control. Mean CSA was markedly reduced in three individual experiments. Cacng1 overexpression was reduced by 13.9% compared with the EGFP control, and FoxO3a overexpression was reduced by 32.2% compared with the EGFP control (Fig. 5D, E). These results suggested that Cacng1 increased the distribution of thinner myotubes in the muscles, probably independent of the fiber type, since the increased Cacng1 induces atrophy even in the fast-twitch TA muscle. Of note, the overexpression of Atrogin-1, a well-known atrogene involved in protein degradation, alone does not cause myotube or muscle atrophy52. Therefore, our findings suggest that Cacng1 induces a pathway such as protein degradation and other atrophy-related changes that lead to pathways necessary to account for such marked fiber shrinkage.
In our study, we observed that Cacng1 expression was significantly upregulated in mice exposed to microgravity compared with those exposed to an artificial 1 g onboard environment. We have shown that Cacng1 induced muscle atrophy in both C2C12 cells and mouse neonatal skeletal muscles. These experiments in this study are the first to implicate the role of the excessive expression of γ1 subunit of the L-type calcium channel in the control of muscle fiber size.
Prolonged muscle inactivity during bed rest, hindlimb unloading, immobilization, microgravity, or denervation result in significant muscle atrophy53–56. It is also known that direct electrical stimulation preserved or restored almost normal properties in denervated muscle fiber likely triggered by action potentials or prompted by the contracted function57,58. A previous report showed that the downregulation of α1S subunit of DHPR in adult skeletal muscle by RNAi underwent massive atrophy, and that decreasing α1S induced redistribution of the neuronal nitric oxide synthase (nNOS), activation of FoxO3A, increased expression of atrogenes, and initiated autophagosome formation50. Given that γ1 subunit of DHPR is supposed to exhibit an endogenous ligand similar to Ca2+ antagonists and minimize Ca2+ entry and Ca2+ release from the previous study49, increased expression of γ1 subunit could induce muscle atrophy through similar mechanisms to the downregulation of α1S subunit. Further studies to reveal the mechanisms of increased expression of Cacng1 to induce decreased fiber size are required to understand the contribution of Cacng1 in muscle atrophy during spaceflight and other muscle atrophy on the ground.
In summary, an important asset of our experimental design is the comprehensive comparison of mice housed under microgravity or an artificial 1 g onboard environment, as an ideal control group to minimize any housing conditions-related bias, other than gravitational change during spaceflight. In previous studies, the antigravitational soleus muscle has often been studied in ground-experimental models of unloading to investigate the signaling pathways involved in skeletal muscle atrophy59,60 and microgravity effects on skeletal muscle12,20 due to its known susceptibility to unloading and disuse. However, further studies were required to understand the functional role of these genes by the effect of actual microgravity. Our results demonstrated that an artificial 1 g onboard environment in the ISS could prevent morphological changes and the alteration of gene expression under microgravity in mammals for the first time. Although CSA tend to decrease in AG compared with GC, the gene expression profile and expression of atrogene indicates that the muscle property of AG is similar to that of GC, consistent with the maintained muscle mass. These results suggested that loading of 1 g could be effective for maintaining muscle during spaceflight for at least 35 days. In particular, to the best of our knowledge, this is the first study in which soleus muscles collected from space-flown mice under microgravity and an artificial 1 g onboard environment were analyzed using RNAseq. These gene profiles could provide a novel and comprehensive overview of the effect of gravity loss on the global gene expression in space-flown mice’s soleus muscles. Therefore, we emphasized that the use of an artificial 1 g onboard environment can serve as a novel approach for identifying gene clusters that could be regulated by microgravity or other various environmental factors during spaceflight, such as space radiation. Furthermore, this study demonstrated that Cacng1 is upregulated in atrophied soleus muscle during spaceflight, and the increased expression of Cacng1 decreased the diameter of the myotube in vitro and in vivo. These results suggested that Cacng1 might have a potential role in muscle atrophy during spaceflight.
However, our study has several limitations. First, it is necessary to consider the various possible conditions, such as the exposure against microgravity until splashdown and the post-landing effect of reloading due to the return of live animals. For example, given that AG mice were exposed to microgravity before splashdown for approximately 1.3 days because there was no centrifuge inside the rocket, it is possible that the short-term exposure of microgravity resulted in a significant increase in the gene expression of Klf15 and its target gene Atrogin-1 (Fbxo 32) in the soleus muscle of AG mice compared with that of MG mice (Fig. 3). Apart from the short-term microgravity exposure of the AG mice before splashdown, the post-landing effect of reloading on the MG mice, which lasted for approximately 2 days, should also be considered in explaining the differences in the gene expression profiles of the soleus muscle in AG and MG mice. Second, although we demonstrated that the gene expression profiles between AG and GC are relatively similar in the soleus muscle, this study only reflects that spaceflight for 35 days has a larger effect on mice under microgravity than other events such as space radiation. Future studies may need to study different time points, such as longer periods using the MARS platform to elucidate the space radiation effects in mammals. Third, our study has not determined whether disuse or microgravity contributed mainly to muscle atrophy under spaceflight.
Taken together, we could gain a foothold in accelerating the estimation of muscle atrophy mechanisms by these gene profiles. Although the current study was made possible by state-of-the-art devices that implement an artificial 1 g onboard environment in the ISS, future studies of mammals will validate the effect of long-term habitation under gravitational forces weaker than 1 g, which is meant to simulate the gravity of the Moon and Mars, known as partial gravity. As experiment methods for space biology continue to develop, future studies may more conclusively identify the underlying causes and offer strategies to prevent muscle atrophy.
Materials and methods
Mice
All experiments were approved by the Institutional Animal Care and Use Committee of the University of Tsukuba (No. 16-048), JAXA (Protocol Number: 016-014B), Explora Biolabs (Study Number: EB15-010A), and NASA (Protocol Number: NAS-15-004-Y1) and were conducted according to the guidelines and applicable laws in Japan and the United States of America and the ARRIVE guidelines. Mice were maintained under specific pathogen-free conditions throughout the space experiment. The JAXA mission, MHU-1, was described previously14. In brief, C57BL/6 J male mice (Stock #000,664) were purchased from Jackson Laboratories (USA) for the space experiment and Charles River Laboratories (Japan) for the ground experiment. The SpaceX Falcon 9 rocket (SpX9) was launched on June 18, 2016 (EDT) from the Kennedy Space Center (KSC). Inside the ISS, 12 mice (8 weeks old) were divided into two groups: artificial 1 g (AG) and microgravity (μg; MG) and housed for 34.6 days in Habitat Cage Units (HCUs) under μg conditions and for 34.1 days and under 1 g during the approximate 37-day berthing period. They were housed for 35 days in HCUs and then returned to Earth in specific pathogen-free conditions. All mice splashed down in the Pacific Ocean near the West Coast on August 26, 2016 and were then transported to the Explore Biolabs laboratory (San Diego) for behavioral observation and dissection 2 days later. The GC experiment that replicated the housing conditions of the flight experiment was conducted at the JAXA Tsukuba Space Center (TKSC) in Japan from August 27 to November 3, 2016. All mice were euthanized by inhalation of 2% isoflurane, and then dissected to collect tissue samples.
Histological staining of HE and immunostaining of the different muscle fiber types
The soleus muscles were immediately frozen in isopentane cooled with liquid nitrogen. Frozen tissues (8 μm in thickness) were mounted on glass slides and subjected to hematoxylin–eosin staining. Immunohistochemical analysis of the different muscle fiber types was performed using fiber type-specific antibodies (Type I; BA-D5, Type IIa; SC-71, Type IIb; BF-F8, the Developmental Studies Hybridoma Bank (DSHB)). The tissue sections were treated with a Mouse on Mouse (M.O.M.) fluorescein kit (Vector Laboratories) to block mouse IgG background. Immunodetection was performed using Alexa Fluor-conjugated secondary antibodies (Alexa Fluor 350 conjugated anti-mouse IgG2b, Alexa Fluor 488 conjugated anti-mouse IgG1, Alexa Fluor 555 conjugated anti-mouse IgM, Thermo Fisher). Images were captured and analyzed using a BIOREVO BZ-X800 microscope system (Keyence, Osaka, Japan). The number of immunoreactive muscle fibers and CSAs were determined using the hybrid cell count software by Keyence.
RNA sequencing
RNAseq was employed for transcriptome analysis. RNA was extracted from the soleus muscles of 3 mice in each group: AG, MG, and GC. Briefly, total RNA was isolated using the TRIZOL reagent (Thermo Fisher Scientific). RNA quality was controlled using an RNA 6000 Pico kit (Agilent, Santa Clara, CA, USA) from 100 frozen sections (8 μm in thickness) of the soleus muscle. A total amount of 50 ng total RNA was used for RNAseq library preparation using the NEBNext rRNA Depletion Kit and NEBNext Ultra Directional RNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA); 2 × 36 base paired-end sequencing was performed using NextSeq500 (Illumina, San Diego, CA) by Tsukuba i-Laboratory LLP (Tsukuba, Ibaraki, Japan). Sequence reads were mapped into the mouse genome (mm10) and quantified for annotated genes using the CLC Genomics Workbench (Version 11.0.2; Qiagen, Redwood City, CA, USA). The expression levels were calculated in transcripts per million (TPM)61. We excluded genes with less than 10 read counts in any sample. After filtering, the differential gene expression was calculated between conditions (GC vs. MG, AG vs. MG, GC vs. AG) with the FDR-corrected P < 0.05 and a 1.5-fold change cutoff using the empirical analysis of DGE tool (EdgeR) on the CLC Genomics Workbench software.
Enrichment analysis process
DAVID Bioinformatics Resources 6.862 was used for pathway analysis and for enrichment analysis of the tissue specificity database (UniProt; UP-Tissue) with P-value < 0.05.
Generation of the adenoviral vector
Three recombinant adenoviruses encoding the EGFP alone, mouse Cacng1 or FoxO3a (CA), a constitutively active (CA) form of mouse FoxO3a, fused with the IRES-EGFP cassette expressing the green fluorescent protein were generated by in vitro recombination techniques. Briefly, the IRES-EGFP cDNA was amplified from the pIRES2-EGFP vector (BD Biosciences Clontech) using the following primers: forward, 5′-CCCACTCGAGGCCCCTCTCCCTCC-3′; reverse, 5′-GGGTGATATCTTACTTGTACAGCTC-3′. Xho I and EcoRV restriction sites are underlined. The amplified fragment was inserted at the Xho I-EcoRV of the pENTR3C entry vector to generate the pENTR3C-IRES-EGFP entry vector. Cacng1 cDNA was cloned by RT–PCR from mouse soleus muscle using the following primers: forward, 5′-CCCAGGATCCGCCACCATGTCACAGACC-3′; reverse, 5′-GGGTCTCGAGCTAGTGCTCTGGCTC-3′. BamHI and Xho I restriction sites are underlined. The amplified fragment was inserted at the BamHI and Xho I sites of the pENTR3C-IRES-EGFP entry vector to generate the pENTR3C-Cacng1-IRES-EGFP vector. pcDNA3-Flag-mouse FoxO3a was a generous gift of Prof. A. Fukamizu (University of Tsukuba). The expression vectors for FoxO3a(CA) mutated at the three AKT phosphorylation sites (T32A, S252A, S314A) were constructed by introducing the mutations by PCR-based site-directed mutagenesis. Flag-FoxO3a-CA was cloned by RT–PCR from mouse soleus muscle using the following primers: forward, 5′-CCCCGAATCCGCCACCATGGATTATAAGGAC-3′; reverse, 5′-GAGGGGCAAACAACAGATGG-3′. EcoRI restriction sites are underlined. The amplified fragment was inserted at the EcoRI sites of the pENTR3C-IRES-EGFP entry vector to generate pENTR3C-Flag-FoxO3a(CA)-IRES-EGFP vector. Then, these inserts were transferred into the pAd/CMV/V5-DEST (Thermo Fisher Scientific) adenoviral expression vector using the ViraPower Adenoviral Gateway Expression Kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s instructions. All constructs were sequenced to confirm their identity. The recombinant plasmids obtained, pAd-EGFP, pAd-Cacng1-IRES-EGFP, and pAd-Flag-FoxO3a(CA)-IRES-EGFP, were linearized with Pac I (New England Biolabs Japan Inc., Tokyo, Japan) restriction enzyme, and ethanol-precipitated. They were then used for the transfection of 293A cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in Opti-MEM medium (Invitrogen) on collagen type I-coated dishes (Thermo Fisher Scientific) to produce the recombinant adenoviral vectors. 4–6 h. after incubation at 37 °C, the medium with the transfection mix was removed, and the growth medium was added. The transfected cells were monitored for EGFP expression and collected 7–14 days after transfection by scraping cells off the dishes and pelleting them along with any floating cells in the culture. After three cycles of freezing in liquid nitrogen and rapid thawing at 37 °C, the viral lysate was used to infect the new cells. One to two days later, the viruses were harvested as described above. This process was repeated 5 times; then, the viruses were purified by cesium chloride (CsCl) gradient ultracentrifugation followed by dialysis with three changes of buffer containing sodium chloride and magnesium chloride to remove CsCl. Viral titers were determined by a plaque-forming assay using the Adeno-X Rapid Titer Kit (Takara bio.) and 293 cells according to the manufacturer’s instructions.
Cell culture, adenoviral infection, and myotube analysis
The C2C12 myoblast cell line (ATCC; CRL-1772) was cultured in high-glucose Dulbecco’s modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum until the cells reached confluence. The medium was then replaced with DMEM 2% horse serum (HS; Thermo Fisher Scientific), the differentiation medium, and incubated for 4 days to induce myotube formation before proceeding with the experiments. For infection, the myotubes were incubated with adenovirus at a multiplicity of infection (MOI) of 250, 125, and 62.5 in the differentiation medium for 18 h., and then the medium was replaced with the differentiation medium without adenovirus. The infection efficiency was typically greater than 90%. Two days after infection, the cells were fixed in 4% paraformaldehyde/PBS for 10 min. and permeabilized in 0.1% Triton X-100/PBS for 10 min. The fixed cells were blocked for 1 h. in 2% BSA/PBS and incubated overnight at 4 °C with anti-myosin heavy chain (MHC) monoclonal antibody (1:100; MF20, DSHB). The antigens were visualized using appropriate secondary antibodies conjugated with Alexa Fluor 546 (1:200; Thermo Fisher Scientific), and the nuclei were labeled with Hoechst 33,342 (Molecular Probes). The cells were mounted with 20 mM Tris-HCl (pH 8.0), 90% glycerol. All images were captured by using a fluorescence microscope (BIOREVO BZ-9000, Keyence). The myotube diameter was quantified by measuring a total of > 100 tube diameters from ten random fields at 200 × magnification using ImageJ software (NIH, Frederick, MD, USA) as described51. All data are expressed as the mean ± SEM.
Adenoviral vector administration and fiber size measurements
Adenoviral vector administration and fiber size measurements were performed as previously described63. Briefly, 2- to 4-day-old ICR mice pups (Japan SLC Inc.) were anesthetized via hypothermia by placement on ice-cooled aluminum foil for a few minutes. Infectious particles (1–2 × 107) of Ad5 cytomegalovirus containing mouse Cacng1-IRES-EGFP, FoxO3a(CA)-IRES-EGFP, or EGFP cDNA diluted in a final volume of 10 μl in PBS were injected percutaneously into each skeletal muscle of the corresponding mice using an insulin syringe. The pups were reintroduced to the mother and kept in quarantine for 5 days. All pups survived after infection.
Statistics
All data are presented as the mean ± SEM. The statistical significance between GC, AG, and MG (Fig. 1B, E, F) were determined by Tukey’s test. Results with P < 0.05 were considered significant. The comparison between EGFP and Cacng1, and EGFP and FoxO3a(CA) were determined using Student’s t-test, and results with P < 0.05 were considered significant (Fig. 5B, E).
Supplementary Information
Acknowledgements
We would like to thank Ms. Miki Shimbo, Yuki Imamura, and Mr. Keigo Asano of the University of Tsukuba for the preparation of mouse samples, Ms. Akane Yumoto of JAXA and Ms. Hiromi Suzuki of the Japan Space Forum for coordinating the study, and Ms. Naoko Murakami and Ms. Rika Oshima of Advanced Engineering Services Co., Ltd. for their assistance in animal care. We also thank Prof. Shin’ichi Takeda of the National Institute of Neuroscience, National Center of Neurology and Psychiatry for supporting the design of space and skeletal muscle research. This work was supported by the Grant-in-Aid for the Japan Aerospace Exploration Agency (14YPTK-005512; S. Takahashi.), Grant-in-Aid for Scientific Research on Innovative Area from MEXT (18H04965; S. Takahashi.), Grand-in-Aid for Research Activity start-up from JSPS (17H06548; R.O.), and Grants-in-Aid for Scientific Research, JSPS KAKENHI (19H00966; S. Takahashi).
Author contributions
R.O., SI. F., T.K., M.M., and S.T. wrote the manuscript. R.O., R.S., T.H., H.T., C.K., S.S., M.K., S. F., Y.I., A.I. and A.T. conducted the experiments. SI. F., A.K., and M.M. performed RNAseq analysis. H.J., M.H., D.S., M.S., T.K., and S.T. prepared and analyzed the space mouse samples. J.T. and N.I. designed skeletal muscle research.
Data availability
All data that support the findings of this study are available from the corresponding author upon reasonable request. RNAseq data are deposited in the DDBJ database (The DNA Databank of Japan, https://www.ddbj.nig.ac.jp/) (accession number DRA010983).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Takashi Kudo, Email: t-kudo@md.tsukuba.ac.jp.
Satoru Takahashi, Email: satoruta@md.tsukuba.ac.jp.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-88392-4.
References
- 1.Volkmann D, Baluska F. Gravity: one of the driving forces for evolution. Protoplasma. 2006;229:143–148. doi: 10.1007/s00709-006-0200-4. [DOI] [PubMed] [Google Scholar]
- 2.Schiaffino S, Sandri M, Murgia M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 2007;22:269–278. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- 3.Frontera WR, Ochala J. Skeletal muscle: a brief review of structure and function. Calcif. Tissue Int. 2015;96:183–195. doi: 10.1007/s00223-014-9915-y. [DOI] [PubMed] [Google Scholar]
- 4.Baar K, Nader G, Bodine S. Resistance exercise, muscle loading/unloading and the control of muscle mass. Essays Biochem. 2006;42:61–74. doi: 10.1042/bse0420061. [DOI] [PubMed] [Google Scholar]
- 5.Desplanches D, Mayet MH, Ilyina-Kakueva EI, Sempore B, Flandrois R. Skeletal muscle adaptation in rats flown on Cosmos 1667. J. Appl. Physiol. 1990;1985(68):48–52. doi: 10.1152/jappl.1990.68.1.48. [DOI] [PubMed] [Google Scholar]
- 6.Ohira Y, et al. Rat soleus muscle fiber responses to 14 days of spaceflight and hindlimb suspension. J. Appl. Physiol. 1992;1985(73):51S–57S. doi: 10.1152/jappl.1992.73.2.S51. [DOI] [PubMed] [Google Scholar]
- 7.Harrison BC, et al. Skeletal muscle adaptations to microgravity exposure in the mouse. J. Appl. Physiol. 2003;1985(95):2462–2470. doi: 10.1152/japplphysiol.00603.2003. [DOI] [PubMed] [Google Scholar]
- 8.Edgerton VR, et al. Human fiber size and enzymatic properties after 5 and 11 days of spaceflight. J. Appl. Physiol. 1995;1985(78):1733–1739. doi: 10.1152/jappl.1995.78.5.1733. [DOI] [PubMed] [Google Scholar]
- 9.Ohira Y, et al. Myonuclear domain and myosin phenotype in human soleus after bed rest with or without loading. J. Appl. Physiol. 1999;1985(87):1776–1785. doi: 10.1152/jappl.1999.87.5.1776. [DOI] [PubMed] [Google Scholar]
- 10.Yamashita-Goto K, et al. Maximal and submaximal forces of slow fibers in human soleus after bed rest. J. Appl. Physiol. 2001;1985(91):417–424. doi: 10.1152/jappl.2001.91.1.417. [DOI] [PubMed] [Google Scholar]
- 11.Morey-Holton ER, Hill EL, Souza KA. Animals and spaceflight: from survival to understanding. J. Musculoskelet. Neuronal Interact. 2007;7:17–25. [PubMed] [Google Scholar]
- 12.Sandona D, et al. Adaptation of mouse skeletal muscle to long-term microgravity in the MDS mission. PLoS ONE. 2012;7:e33232. doi: 10.1371/journal.pone.0033232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gambara G, et al. Gene expression profiling in slow-type calf soleus muscle of 30 days space-flown mice. PLoS ONE. 2017;12:e0169314. doi: 10.1371/journal.pone.0169314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiba D, et al. Development of new experimental platform 'MARS'-multiple artificial-gravity research system-to elucidate the impacts of micro/partial gravity on mice. Sci. Rep. 2017;7:10837. doi: 10.1038/s41598-017-10998-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tominari T, et al. Hypergravity and microgravity exhibited reversal effects on the bone and muscle mass in mice. Sci. Rep. 2019;9:6614. doi: 10.1038/s41598-019-42829-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki T, et al. Nrf2 contributes to the weight gain of mice during space travel. Commun. Biol. 2020;3:496. doi: 10.1038/s42003-020-01227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao XW, et al. Impact of spaceflight and artificial gravity on the mouse retina: biochemical and proteomic analysis. Int. J. Mol. Sci. 2018 doi: 10.3390/ijms19092546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horie K, et al. Down-regulation of GATA1-dependent erythrocyte-related genes in the spleens of mice exposed to a space travel. Sci. Rep. 2019;9:7654. doi: 10.1038/s41598-019-44067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumura T, et al. Male mice, caged in the International Space Station for 35 days, sire healthy offspring. Sci. Rep. 2019;9:13733. doi: 10.1038/s41598-019-50128-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohira Y, et al. Gravitational unloading effects on muscle fiber size, phenotype and myonuclear number. Adv. Space Res. 2002;30:777–781. doi: 10.1016/s0273-1177(02)00395-2. [DOI] [PubMed] [Google Scholar]
- 21.Cadena SM, et al. Skeletal muscle in MuRF1 null mice is not spared in low-gravity conditions, indicating atrophy proceeds by unique mechanisms in space. Sci. Rep. 2019;9:9397. doi: 10.1038/s41598-019-45821-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da Silveira WA, et al. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact. Cell. 2020;183:1185–1201e1120. doi: 10.1016/j.cell.2020.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecker SH, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- 24.Bodine SC, et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 25.Sacheck JM, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–155. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- 26.Judge SM, et al. Genome-wide identification of FoxO-dependent gene networks in skeletal muscle during C26 cancer cachexia. BMC Cancer. 2014;14:997. doi: 10.1186/1471-2407-14-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayeuf-Louchart A, et al. Rev-erb-alpha regulates atrophy-related genes to control skeletal muscle mass. Sci. Rep. 2017;7:14383. doi: 10.1038/s41598-017-14596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milan G, et al. Regulation of autophagy and the ubiquitin-proteasome system by the FoxO transcriptional network during muscle atrophy. Nat. Commun. 2015;6:6670. doi: 10.1038/ncomms7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki H, et al. IkappaBalpha ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, betaTrCP1 and betaTrCP2. Biochem. Biophys. Res. Commun. 1999;256:127–132. doi: 10.1006/bbrc.1999.0289. [DOI] [PubMed] [Google Scholar]
- 30.Pan D, Zhe X, Jakkaraju S, Taylor GA, Schuger L. P311 induces a TGF-beta1-independent, nonfibrogenic myofibroblast phenotype. J. Clin. Invest. 2002;110:1349–1358. doi: 10.1172/JCI15614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magnusson C, Svensson A, Christerson U, Tagerud S. Denervation-induced alterations in gene expression in mouse skeletal muscle. Eur. J. Neurosci. 2005;21:577–580. doi: 10.1111/j.1460-9568.2005.03855.x. [DOI] [PubMed] [Google Scholar]
- 32.Harding HP, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 33.Summermatter S, et al. Blockade of metallothioneins 1 and 2 increases skeletal muscle mass and strength. Mol. Cell Biol. 2017 doi: 10.1128/MCB.00305-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moresi V, et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonaldo P, Sandri M. Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 2013;6:25–39. doi: 10.1242/dmm.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taillandier D, Polge C. Skeletal muscle atrogenes: from rodent models to human pathologies. Biochimie. 2019;166:251–269. doi: 10.1016/j.biochi.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 37.Kneppers A, et al. Coordinated regulation of skeletal muscle mass and metabolic plasticity during recovery from disuse. FASEB J. 2019;33:1288–1298. doi: 10.1096/fj.201701403RRR. [DOI] [PubMed] [Google Scholar]
- 38.Mirzoev TM. Skeletal muscle recovery from disuse atrophy: protein turnover signaling and strategies for accelerating muscle regrowth. Int. J. Mol. Sci. 2020 doi: 10.3390/ijms21217940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beheshti A, Ray S, Fogle H, Berrios D, Costes SV. A microRNA signature and TGF-beta1 response were identified as the key master regulators for spaceflight response. PLoS ONE. 2018;13:e0199621. doi: 10.1371/journal.pone.0199621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen DL, et al. Effects of spaceflight on murine skeletal muscle gene expression. J. Appl. Physiol. 2009;1985(106):582–595. doi: 10.1152/japplphysiol.90780.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neutelings T, et al. Skin physiology in microgravity: a 3-month stay aboard ISS induces dermal atrophy and affects cutaneous muscle and hair follicles cycling in mice. NPJ Microgravity. 2015;1:15002. doi: 10.1038/npjmgrav.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connor RS, Mills ST, Jones KA, Ho SN, Pavlath GK. A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 2007;120:149–159. doi: 10.1242/jcs.03307. [DOI] [PubMed] [Google Scholar]
- 43.Parker MH, Seale P, Rudnicki MA. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 2003;4:497–507. doi: 10.1038/nrg1109. [DOI] [PubMed] [Google Scholar]
- 44.Pownall ME, Gustafsson MK, Emerson CP., Jr Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2002;18:747–783. doi: 10.1146/annurev.cellbio.18.012502.105758. [DOI] [PubMed] [Google Scholar]
- 45.Weis J, Kaussen M, Calvo S, Buonanno A. Denervation induces a rapid nuclear accumulation of MRF4 in mature myofibers. Dev. Dyn. 2000;218:438–451. doi: 10.1002/1097-0177(200007)218:3<438::AID-DVDY1001>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Z, Bornemann A. MRF4 protein expression in regenerating rat muscle. J. Muscle Res. Cell Motil. 2001;22:311–316. doi: 10.1023/a:1013133616449. [DOI] [PubMed] [Google Scholar]
- 47.Arikkath J, Campbell KP. Auxiliary subunits: essential components of the voltage-gated calcium channel complex. Curr. Opin. Neurobiol. 2003;13:298–307. doi: 10.1016/s0959-4388(03)00066-7. [DOI] [PubMed] [Google Scholar]
- 48.Flucher BE, et al. The role of auxiliary dihydropyridine receptor subunits in muscle. J. Muscle Res. Cell Motil. 2005;26:1–6. doi: 10.1007/s10974-005-9000-2. [DOI] [PubMed] [Google Scholar]
- 49.Andronache Z, et al. The auxiliary subunit gamma 1 of the skeletal muscle L-type Ca2+ channel is an endogenous Ca2+ antagonist. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17885–17890. doi: 10.1073/pnas.0704340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietri-Rouxel F, et al. DHPR alpha1S subunit controls skeletal muscle mass and morphogenesis. EMBO J. 2010;29:643–654. doi: 10.1038/emboj.2009.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rommel C, et al. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 2001;3:1009–1013. doi: 10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 52.Sandri M, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams GR, Caiozzo VJ, Baldwin KM. Skeletal muscle unweighting: spaceflight and ground-based models. J. Appl. Physiol. 2003;1985(95):2185–2201. doi: 10.1152/japplphysiol.00346.2003. [DOI] [PubMed] [Google Scholar]
- 54.Borisov AB, Dedkov EI, Carlson BM. Interrelations of myogenic response, progressive atrophy of muscle fibers, and cell death in denervated skeletal muscle. Anat. Rec. 2001;264:203–218. doi: 10.1002/ar.1155. [DOI] [PubMed] [Google Scholar]
- 55.De-Doncker L, Kasri M, Picquet F, Falempin M. Physiologically adaptive changes of the L5 afferent neurogram and of the rat soleus EMG activity during 14 days of hindlimb unloading and recovery. J. Exp. Biol. 2005;208:4585–4592. doi: 10.1242/jeb.01931. [DOI] [PubMed] [Google Scholar]
- 56.Dedkov EI, Borisov AB, Carlson BM. Dynamics of postdenervation atrophy of young and old skeletal muscles: differential responses of fiber types and muscle types. J. Gerontol. A Biol. Sci. Med. Sci. 2003;58:984–991. doi: 10.1093/gerona/58.11.b984. [DOI] [PubMed] [Google Scholar]
- 57.Kern H, et al. Long-term denervation in humans causes degeneration of both contractile and excitation-contraction coupling apparatus, which is reversible by functional electrical stimulation (FES): a role for myofiber regeneration? J. Neuropathol. Exp. Neurol. 2004;63:919–931. doi: 10.1093/jnen/63.9.919. [DOI] [PubMed] [Google Scholar]
- 58.Squecco R, Kern H, Biral D, Rossini K, Francini F. Mechano-sensitivity of normal and long term denervated soleus muscle of the rat. Neurol. Res. 2008;30:155–159. doi: 10.1179/174313208X281028. [DOI] [PubMed] [Google Scholar]
- 59.Machida S, Booth FW. Changes in signalling molecule levels in 10-day hindlimb immobilized rat muscles. Acta Physiol. Scand. 2005;183:171–179. doi: 10.1111/j.1365-201X.2004.01395.x. [DOI] [PubMed] [Google Scholar]
- 60.Sugiura T, et al. Changes in PKB/Akt and calcineurin signaling during recovery in atrophied soleus muscle induced by unloading. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2005;288:R1273–1278. doi: 10.1152/ajpregu.00688.2004. [DOI] [PubMed] [Google Scholar]
- 61.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131:281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 62.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 63.Durbeej M, et al. Gene transfer establishes primacy of striated vs. smooth muscle sarcoglycan complex in limb-girdle muscular dystrophy. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8910–8915. doi: 10.1073/pnas.1537554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that support the findings of this study are available from the corresponding author upon reasonable request. RNAseq data are deposited in the DDBJ database (The DNA Databank of Japan, https://www.ddbj.nig.ac.jp/) (accession number DRA010983).