Abstract
The inflammasome initiates innate defense and inflammatory response by activating caspase-1 and pyroptotic cell death in myeloid cells1,2. It is comprised of an innate immune receptor/sensor, pro-caspase-1, and a common adaptor molecule, ASC (apoptotic speck-containing protein with a CARD). Consistent with their pro-inflammatory function, caspase-1, ASC and NLRP3 exacerbate autoimmunity during experimental autoimmune encephalomyelitis (EAE) by enhancing IL-1β and IL-18 secretion in myeloid cells3–6. Here we reveal an unexpected function of a DNA-binding inflammasome receptor, AIM2 (Absent in Melanoma 2)7–10, in T regulatory cells (Tregs) to restrain two models of autoimmunity (experimental autoimmune encephalomyelitis and T cell-mediated colitis) by studying whole-body and Treg-specific Aim2–/– mice. AIM2 is highly expressed by human and mouse Tregs, with its expression induced by TGF-β and its promoter occupied by transcription factors associated with Tregs, including Runx1, Ets1, Bcl11b and CREB. RNA-seq, biochemical and metabolic analyses revealed that AIM2 attenuates Akt-phosphorylation, mTOR, c-Myc and glycolysis, but promotes lipid oxidative phosphorylation in Tregs. Mechanistically, AIM2 interacts with the RACK1/PP2A-phosphatase complex to restrain Akt-phosphorylation. Lineage tracing demonstrates that AIM2 promotes the stability of Tregs during inflammation. While AIM2 is generally accepted as an inflammasome effector in myeloid cells, this report reveals a T cell-intrinsic role of AIM2 in restraining autoimmunity by diminishing Akt-mTOR signaling and altering immune-metabolism to enhance Treg stability.
Experimental autoimmune encephalomyelitis (EAE) was induced in wild type (WT), Aim2–/–, Asc–/–, and ICE–/– (caspase-1–/– and caspase-11–/–) mice after immunization with MOG35–55 peptide as described in Methods. Consistent with the literature, Asc–/– and ICE–/– mice had negligible EAE compared to WT controls4 (Fig. 1a). Unexpectedly, Aim2–/– mice developed more severe EAE with higher clinical score and disease incidence (Fig. 1a), increased pathology, demyelination and inflammatory immune cell infiltration in the spinal cord when compared to WT controls (Fig. 1b–d), while Asc–/– mice showed reduction in all of these parameters (Fig. 1b, c). These observations questioned the predicted, pro-inflammatory role of AIM2 through inflammasome activation in EAE. Indeed, IL-1β, IL-18, IL-6, and TNF cytokine levels were indistinguishable in the spinal cords of Aim2–/– and control mice (Fig. 1e).
Figure 1. Aim2–/– and Asc–/– mice have opposing responses to EAE.
a, EAE scoring of WT (n=46), Aim2–/– (n=45), Asc–/– (n=10) and ICE–/– (n=4) mice, three experiments. P value by two-way ANOVA and Holm-Sidak post-hoc test. b, Luxol fast blue and periodic acid-Schiff (LFB-PAS) staining of spinal cords. Scale bars indicated on the panels. WT (n=3), Aim2–/– (n=8) and Asc–/– (n=4) mice, day 22 of EAE. Representative of 3–8 mice/group, three experiments. c, Quantification of demyelination and inflammatory foci. WT (n=3), Aim2–/– (n=8) and Asc–/– (n=4) mice from (b). d. Infiltrating cells in spinal cords of WT and Aim2–/– mice, day 22 of EAE, n=6/group, two experiments. e, Spinal cord IL-1β, IL-18, IL-6, and TNFα analyzed by ELISA; WT (n=13) and Aim2–/– (n=16) for IL-1β (3 experiments), n=5 for other cytokines (2 experiments). f-g, Flow-cytometry of CD4+Foxp3+ Tregs (f), IFNγ+ or IL-17A+ CD4+ cells (g) in spinal cords of WT and Aim2–/– mice, days 14–15 during EAE, n=6/group, three experiments. h, RT-PCR of genes, n=3, 5 and 6 for WT, n=4, 6 and 8 for Aim2−/− samples, two experiments. i, IL-10 protein analyzed by ELISA; n=10 for WT and 11 for Aim2−/− samples, three experiments. j, Flow-cytometry of CD25+Foxp3+, Foxp3+IL-10+ and IL-17A+ CD4+ cells in WT and Aim2–/– spinal cords at days 22–23 of EAE, n=4 for WT, n=5 for Aim2−/− samples of CD25+Foxp3+ and IL-17A+ (2 experiments), n=13/group for Foxp3+IL-10+ (3 experiments). k, EAE scores of Rag1–/– mice that received WT (n=21) or Aim2–/– CD4+ T cells (n=19), three experiments. P value by two-way ANOVA and Holm -Sidak post-hoc test. l, Flow-cytometry of spinal cord CD4+Foxp3+ Tregs from (k), n=6/group, three experiments. Data are means ± SEM, P values: ns, not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, analyzed by two-sided t test unless specified.
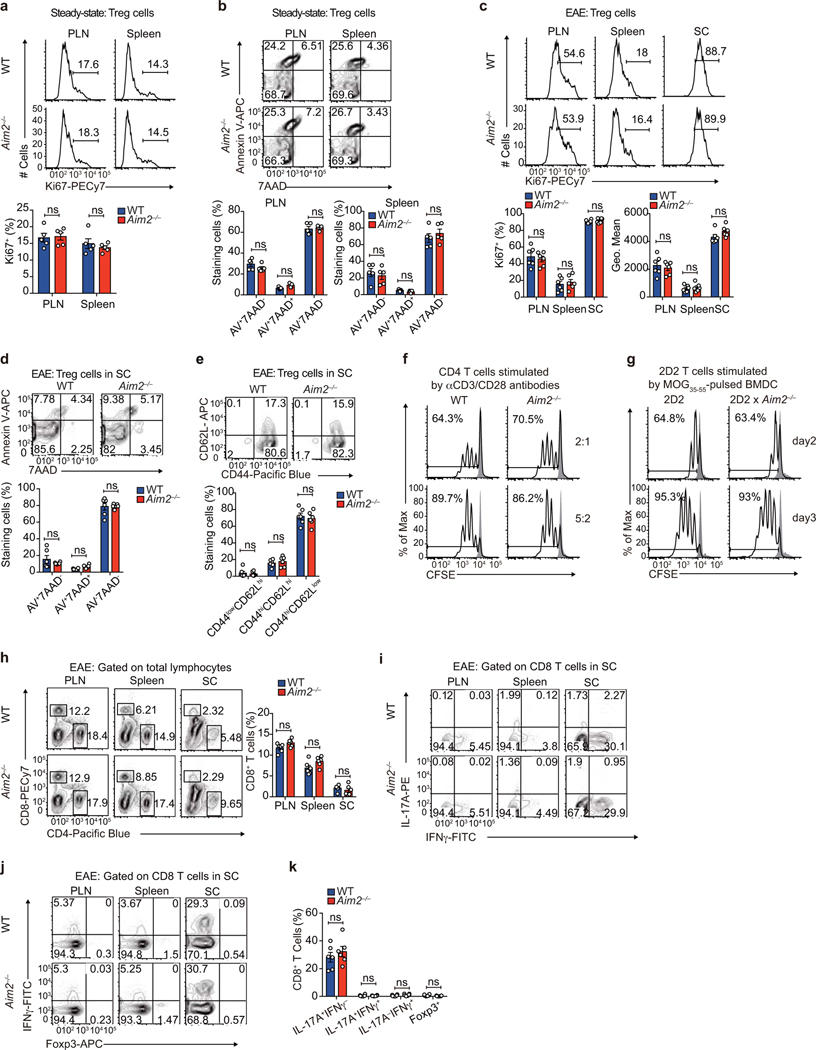
Th1 and Th17 cells promote whereas Tregs restrict EAE development11. Analysis of the CD4+ T cell population in spinal cords of diseased WT and Aim2–/– mice revealed that Aim2–/– mice had decreased Foxp3+ Tregs and increased IL-17A+ and IL-17A+IFNγ+ CD4 T cells during the early phase of EAE (Fig. 1f, g). Treg, Th1 and Th17 cells were not significantly different in the spleens and draining lymph nodes of WT and Aim2–/– mice (Extended Data Fig. 1a, b). Under steady state, Aim2–/– mice showed normal T cell homeostasis in thymus, spleen, and lymph nodes (Extended Data Fig. 1c–h). These findings indicate that AIM2-dependent changes in T cells occurred at sites of neuro-inflammation. Although Foxp3+ Tregs were reduced in spinal cords of Aim2–/– mice during EAE (Fig. 1f), WT and Aim2–/– Tregs proliferated, survived and expressed effector markers similarly, under steady state and during EAE (Extended Data Fig. 2a–e). During the late phase of EAE, Aim2 deletion led to reduced expression of Foxp3, Tgfβ1 and Il10 mRNA (Fig. 1h) and IL-10 protein (Fig. 1i) in spinal cords. Consistently, Foxp3+ Tregs were significantly decreased in spinal cords of Aim2–/– mice, while Th17 cells were increased (Fig. 1j). Aim2–/– CD4+ T cells proliferated normally in vitro (Extended Data Fig. 2f, g) and Aim2–/– CD8+ T cells were phenotypically normal during EAE (Extended Data Fig. 2h–k). We therefore posit that AIM2 likely controls CD4+ T cell function to mitigate EAE.
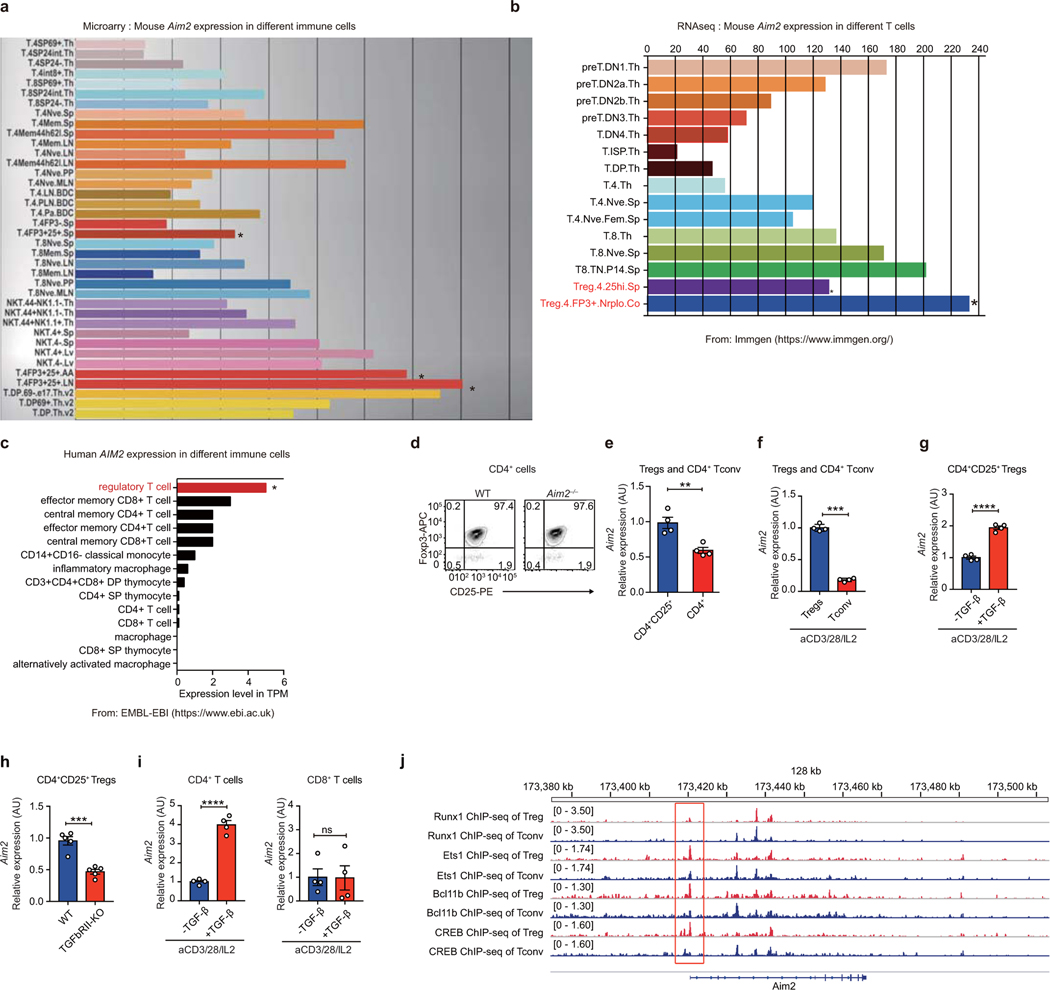
To assess CD4+ T cell intrinsic function of AIM2, we adoptively transferred CD4+ T cells from WT or Aim2–/– mice into Rag1–/– mice12 followed by EAE elicitation. Mice received Aim2–/– CD4+ T cells developed more severe EAE at an earlier time than those received WT CD4+ T cells (Fig. 1k), with concurrent reduced Tregs in the spinal cord (Fig. 1l). These findings suggest a previously unappreciated cell-intrinsic function of AIM2 in Tregs. High levels of AIM2 expression are detected in both mouse (https://www.immgen.org/)13 and human (https://www.ebi.ac.uk)14 Tregs (Extended Data Fig. 3a–c). We empirically confirmed that isolated Tregs expressed higher levels of Aim2 than CD4+ conventional T cells (Tconv) cells, with or without TCR activation (Extended Data Fig. 3d–f). Of interest, TGF-β, a factor that is vital for Treg cell generation and homeostasis15,, increased Aim2 expression in CD4+, but not in CD8+ T cells. Furthermore, genetic abrogation of TGF-β signaling in Tgfbr2 gene deletion mice reduced Aim2 expression in Tregs in vivo (Extended Data Fig. 3g–i). Analysis of ChIP-seq data sets (DRP003376)16 of Aim2 promoter in Tregs vs. Tconv cells showed that transcription factors Runx1, Est1, Bcl11b and CREB, that are known to bind the Foxp3 loci to regulate Treg stability16–18 also bound to the Aim2 promoter in Tregs more than Tconv cells (Extended Data Fig. 3j). These findings suggest that a Treg-specific molecular network favors Aim2 expression.
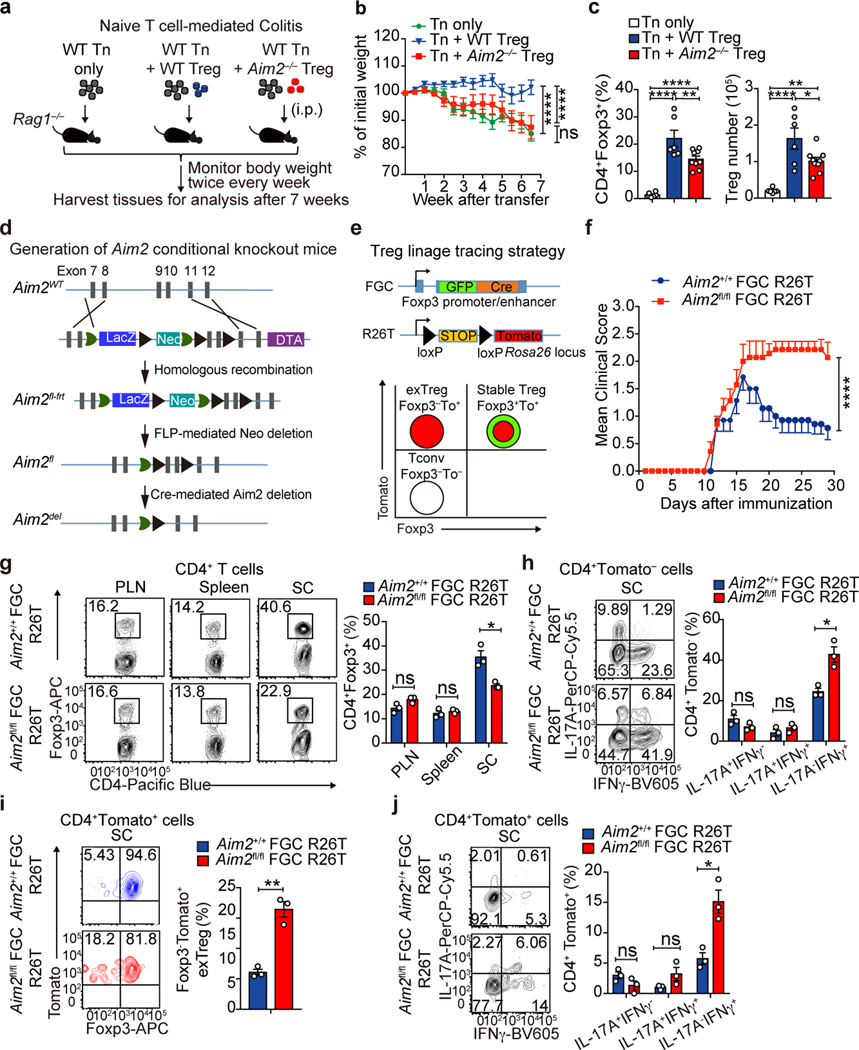
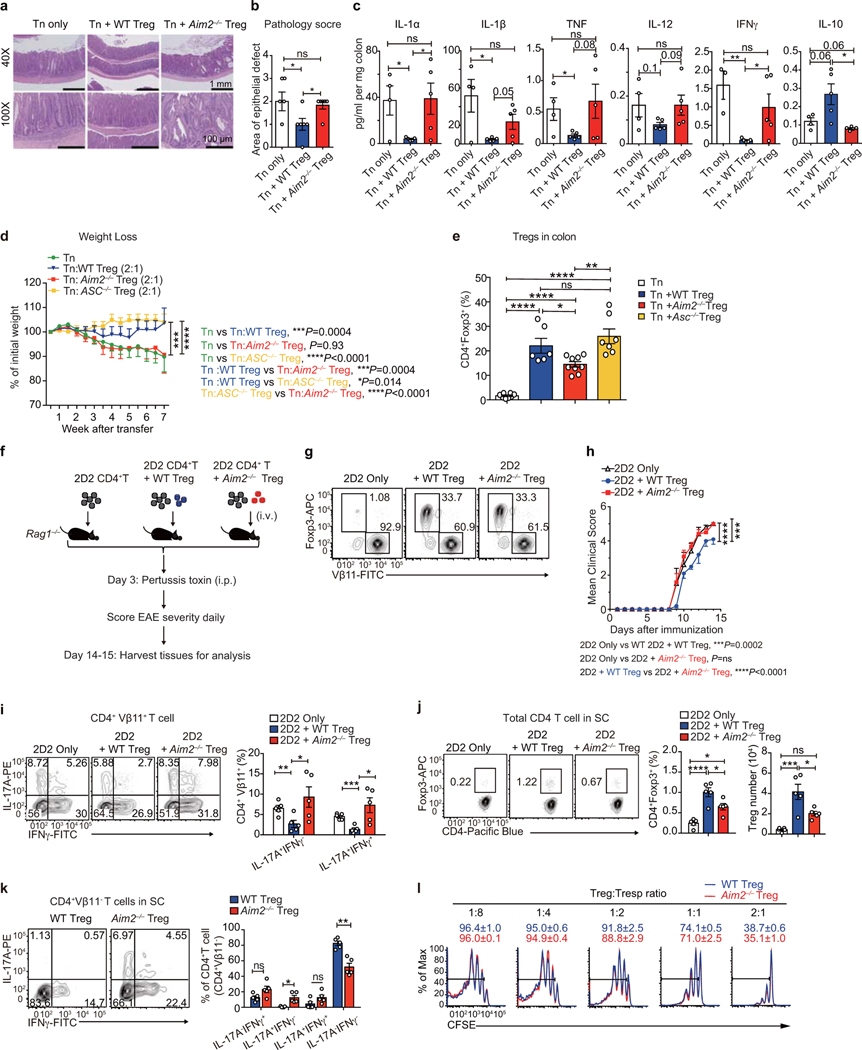
To assess the function of AIM2 in Tregs, we first studied a T cell-induced colitis model where transferred naïve T cells provoke colitis in Rag1–/– recipients, while the inclusion of Tregs offers protection19. CD4+CD45RBhi naïve T cells from WT mice were transferred alone or with sorted WT or Aim2–/– Tregs into Rag1–/– recipients (Fig. 2a). Transferred WT naïve T cells led to body weight decline of recipient mice (Fig. 2b) and intestinal pathology (Extended Data Fig. 4a, b). Transferred Aim2–/– Tregs in the colon were present at reduced frequency compared to WT Tregs (Fig. 2c). The inclusion of WT but not Aim2–/– Tregs mitigated weight loss (Fig. 2b), reduced pathology and suppressed the expression of pro-inflammatory cytokines IL-1α, IL-1β, TNF, IL-12 and IFNγ, but promoted anti-inflammatory IL-10 in colon explants (Extended Data Fig. 4a–c). To determine if the role of AIM2 in Tregs can be separated from its conventional role in inflammasome activation, we transferred Asc–/– Tregs in the colitis model. In contrast to Aim2–/– Tregs, Asc–/– Tregs had similar effects as WT Tregs (Extended Data Fig. 4d, e).
Figure 2. AIM2 stabilizes Tregs to restrain autoimmunity.
a, Schematic of WT CD4+CD45RBhi T cells (Tn)-induced colitis, with or without Tregs. b, Body weight of Rag1–/– mice received Tn (n=8), Tn+WT Treg (n=8), Tn+Aim2–/– Treg (n=9); two experiments, P value by two-way ANOVA and Holm -Sidak post-hoc test. c, Flow-cytometry of CD4+Foxp3+ Tregs in colons of Rag1–/– mice that received Tn (n=6), Tn+WT Treg (n=6), Tn+Aim2–/– Treg (n=9), 7 weeks after transfer, two experiments. P value by one-way ANOVA with Tukey’s multiple comparisons test. d, Schema for gene targeting to generate floxed Aim2 mice. e, Schema for generating Foxp3 Treg lineage tracing mice (upper). Tconv, stable Treg, and exTreg based on Foxp3 and Tomato expression (lower). f, EAE scores of mice of indicated genotypes (n=7/group). Two experiments, P value by two-way ANOVA and Holm-Sidak post-hoc test. g, Flow-cytometry of Foxp3 expression in CD4+ cells in PLN, spleen, and spinal cord (SC) of mice of indicated genotypes at day 28 of EAE, n=3 experiments. P value by multiple t test corrected by the Holm-Sadik method. h, Flow-cytometry of IFNγ and IL-17A production in CD4+Tomato− T cells in spinal cords of mice of indicated genotypes at day 28 of EAE, n=3 experiments. P value by Multiple t test corrected by Holm-Sadik method. i, Flow-cytometry of Foxp3 production in CD4+Tomato+ T cells in spinal cords of mice of indicated genotypes at day 28 of EAE, n=3 experiments. P value by unpaired t test. j, Flow-cytometry of IFNγ and IL-17A production in CD4+Tomato+ T cells in spinal cords of mice of indicated genotypes at day 28 of EAE, n=3 experiments. P value by Multiple t test corrected by Holm-Sadik method. Data are means ± SEM, P values: ns, not significant, *P<0.05, **P<0.01, ****P<0.0001.
We next examined the function of AIM2 in Tregs in the EAE model. Naïve CD4+ T cells from 2D2 mice that expressed a transgenic T cell receptor for the encephalitogenic autoantigen, MOG35–55 were transferred with sorted Tregs from either WT or Aim2–/– mice into Rag1–/– recipients. EAE was then elicited in these recipient mice (Extended Data Fig. 4f, g). Mice receiving 2D2 CD4+ T cells alone developed severe EAE (Extended Data Fig. 4h). Addition of WT Tregs modestly attenuated EAE development (Extended Data Fig. 4h) with a decrease of Th17 cells and an increase of Tregs in the spinal cord (Extended Data Fig. 4i, j). In contrast, Aim2–/– Tregs failed to reduce EAE (Extended Data Fig. 4g–j). Interestingly, transferred Aim2–/– Tregs gained expression of IL-17A but had reduced IL-17A–IFNγ– population (Extended Data Fig. 4k). These findings suggest a critical role for AIM2 in promoting Treg function in vivo. However, the in vitro suppressive function of Aim2–/– Tregs appeared normal (Extended Data Fig. 4l).
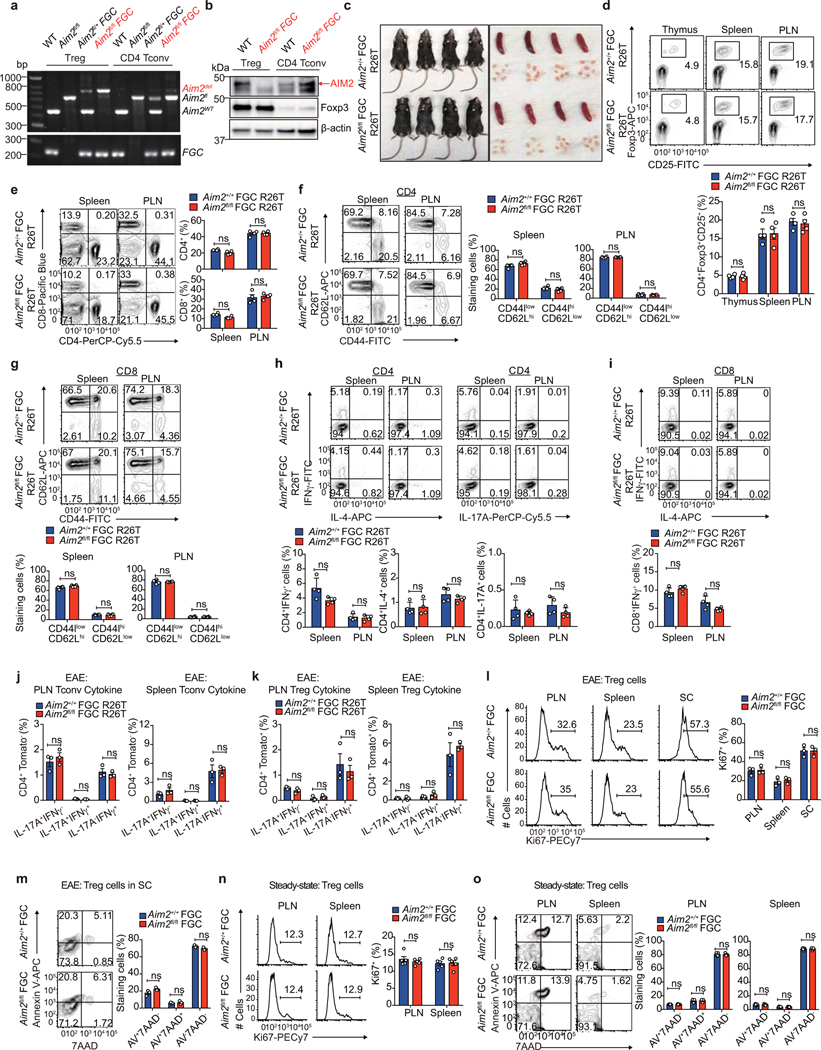
WT Tregs did not strongly suppress EAE in 2D2 mice likely due to robust T cell activation mediated by the transgenic TCR expressed in these mice. To better address the Treg-intrinsic role of AIM2, we generated mice with Treg cell-specific deletion by creating a mouse strain with a floxed Aim2 allele by gene-targeting (Fig. 2d). Mice with floxed Aim2 allele were bred with mice bearing Treg-specific Cre transgene Foxp3-GFP-Cre (FGC) and with mice bearing the Rosa26tdTomato (R26T) lineage tracing allele (Fig. 2e). Treg-specific deletion of Aim2 was confirmed by PCR and western blot (Extended Data Fig. 5a, b). Treg-specific Aim2–/– mice were normal without obvious defect under steady state (Extended Data Fig. 5c–i). EAE elicitation resulted in significantly higher clinical score in Treg-specific Aim2–/– mice compared to WT mice, accompanied by lower Foxp3+ Tregs and higher IFNγ+ CD4+ T cells in the spinal cord (Fig. 2g, h). Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice were used for lineage tracing based on the schematic depicted in Fig. 2e. Aim2 deletion led to elevated exTregs (Foxp3−Tomato+ cells that previously expressed Foxp3) and increased IFNγ expression by Tregs in the spinal cord (Fig. 2i, j), but not the spleen or draining lymph node during EAE (Extended Data Fig. 5j, k). Aim2-deficient Tregs proliferated and survived normally during EAE and under steady state (Extended Data Fig. 5l–o). These findings support a critical biologic role for AIM2 in promoting Treg stability.
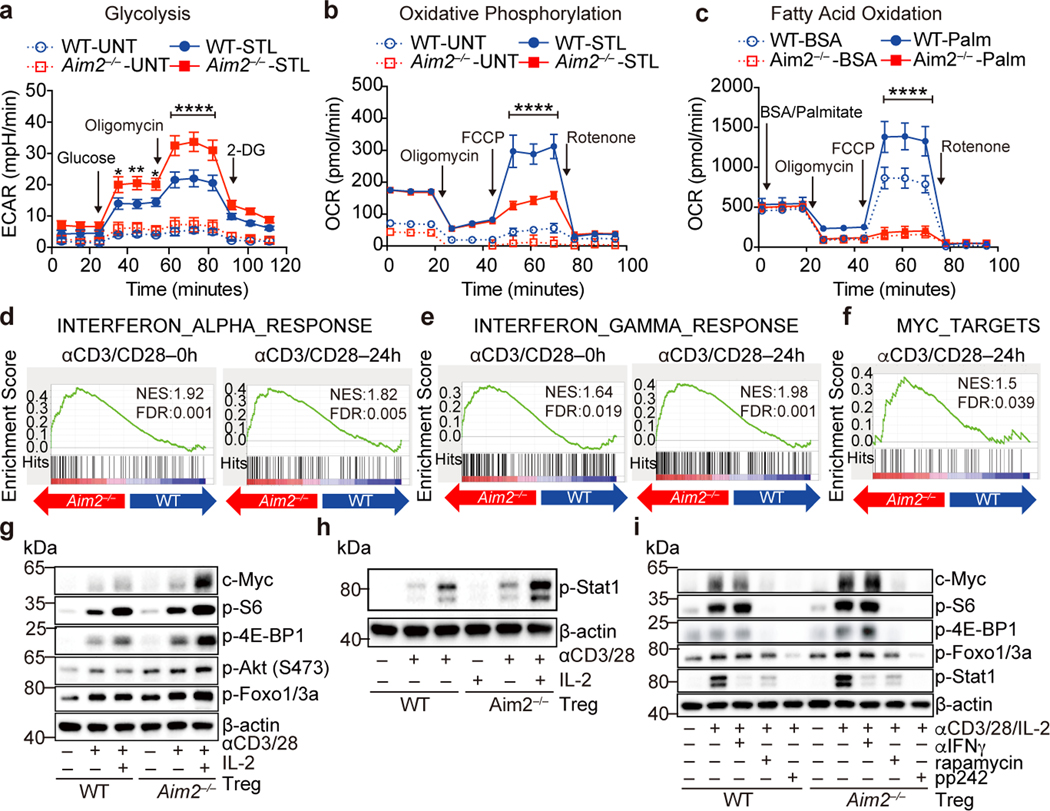
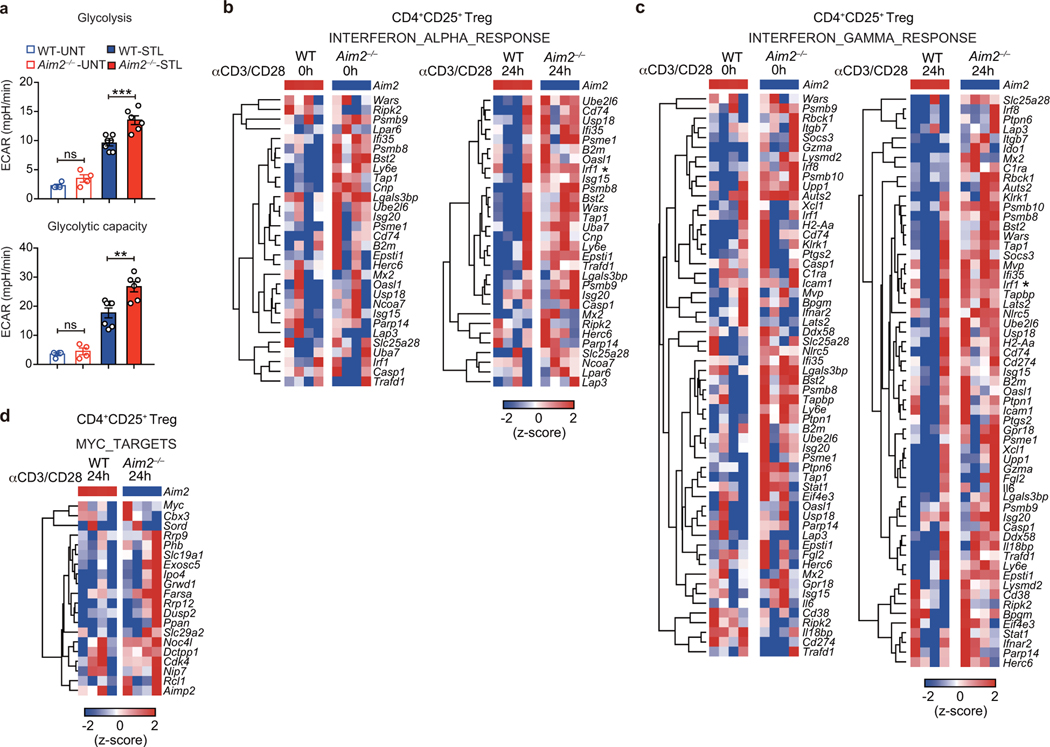
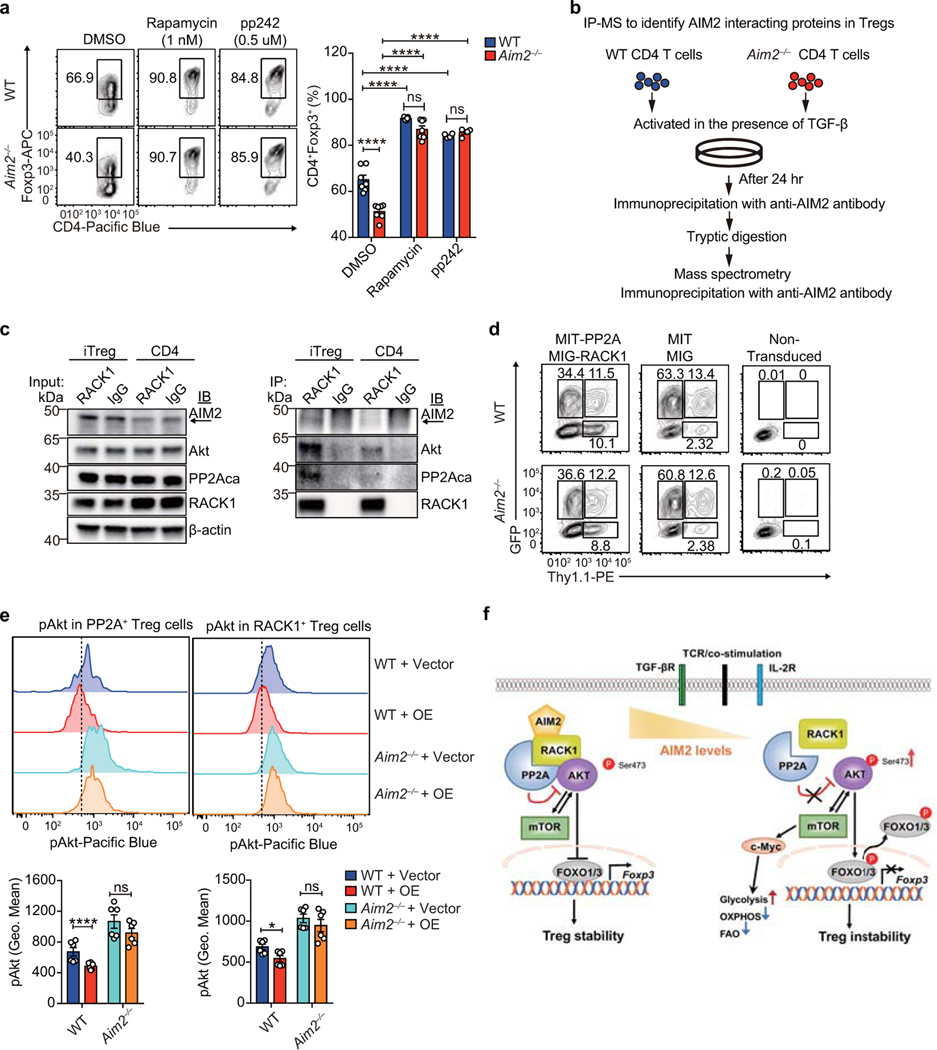
We next explored the mechanisms by which AIM2 regulates Tregs. Previous studies found that AIM2 could attenuate Akt activation in gastrointestinal epithelial cells20,21. Akt-mTOR signaling positively associates with glycolytic metabolism, which negatively impacts Treg cell function22. To investigate if Aim2 deletion altered Treg cell metabolism, WT or Aim2–/– Tregs were activated by TCR stimulation. Aim2–/– Tregs showed higher glycolytic activity assessed by extracellular acidification rate (ECAR) (Fig. 3a, quantified in Extended Data Fig. 6a) compared to WT Tregs. Aim2–/– Tregs also had reduced oxygen consumption rate (OCR) and fatty acid oxidation (FAO) (Fig. 3b, c), indicating impaired lipid oxidative phosphorylation and increased aerobic glycolysis23. Global transcriptional profiling by RNA-seq and gene-set enrichment analysis (GSEA) revealed that Aim2 deletion led to enhanced IFN-responsive signatures (Fig. 3d, e, Extended Data Fig. 6b, c), including IRF1 and IFNγ, both of which are known to attenuate Foxp3 expression and Treg function24,25, as well as enhanced Myc-dependent programming26 (Fig. 3f, Extended Data Fig. 6d). Consistently, Aim2–/– Tregs had increased expression of c-Myc protein26, hyper-phosphorylation of S6 and 4E-BP1 (an indicator of mTORC1 signaling), elevated levels of p-Akt and its downstream target p-Foxo1/3a (Fig. 3g), which can block Foxp3 expression27. Additionally, p-STAT1, a factor indicative of IFN signaling, was elevated in Aim2–/– Tregs (Fig. 3h). Enhanced mTOR, but not IFNγ, appeared to be central to, and causal for, the above-mentioned alterations in Aim2–/– T cells because treatment with pharmacological inhibitors of mTORC1 (rapamycin) or mTORC1/2 (pp242) blocked excessive c-Myc, p-S6, p-4E-BP1, p-Foxo1/3a and p-STAT1, while anti-IFNγ neutralization only blocked p-STAT1 (Fig. 3i).
Figure 3. AIM2 regulates immune metabolism, Akt-mTOR and IFN signaling in Tregs isolated in vivo.
a, Glycolytic activity of WT (n=7 experiments) and Aim2–/– (n=6 experiments) CD4+CD25+ Tregs with or without anti-CD3/CD28 plus IL-2 (500 U/ml), 24 hr. P value by two-way ANOVA. b, Oxidative consumption rate (OCR) of WT (n=3 experiments) and Aim2–/– (n=4 experiments) CD4+CD25+ Tregs with or without anti-CD3/CD28 plus IL-2 (500 U/ml) for 24 hr. P value by two-way ANOVA. c, Fatty acid oxidation (FAO) of WT (n=5 experiments) and Aim2–/– (n=6 experiments) CD4+CD25+ Tregs with or without anti-CD3/CD28 plus IL-2 (500 U/ml) for 24 hr. Cells were starved in substrate-limited medium and given only BSA or palmitate-BSA in FAO assay media and OCR was measured to indicate oxidation of fatty acids according to the Agilent FAO guideline. P value by two-way ANOVA. d-f, Enrichment of IFNα response signatures (d), IFNγ response signatures (e), and Myc-related targets (f) by Gene Set Enrichment Analysis (GSEA) of RNA-seq datasets from Aim2–/– and WT Tregs stimulated with anti-CD3/CD28 antibodies plus IL-2 (500 U/ml). g, Immunoblot analysis of indicated proteins in WT and Aim2–/– CD4+CD25+ Tregs with or without anti-CD3/CD28 plus IL-2 (500 U/ml) for 24 hr. Data are representative of three experiments. h, Immunoblot analysis of p-Stat1and β-actin in WT and Aim2–/– CD4+CD25+ Tregs treated as described in (g). Data are representative of three experiments. i, Immunoblot analysis of indicated proteins in WT and Aim2–/– CD4+CD25+ Tregs stimulated with anti-CD3/CD28 plus IL-2 (500 U/ml) in the presence of rapamycin (1 nM), pp242 (0.5 μM) or anti-IFNγ antibody (XMG1.2, 10 μg/ml) for 24 hr. Data are representative of three experiments. Data are means ± SEM, P values: *P<0.05, **P<0.01, ****P<0.0001.
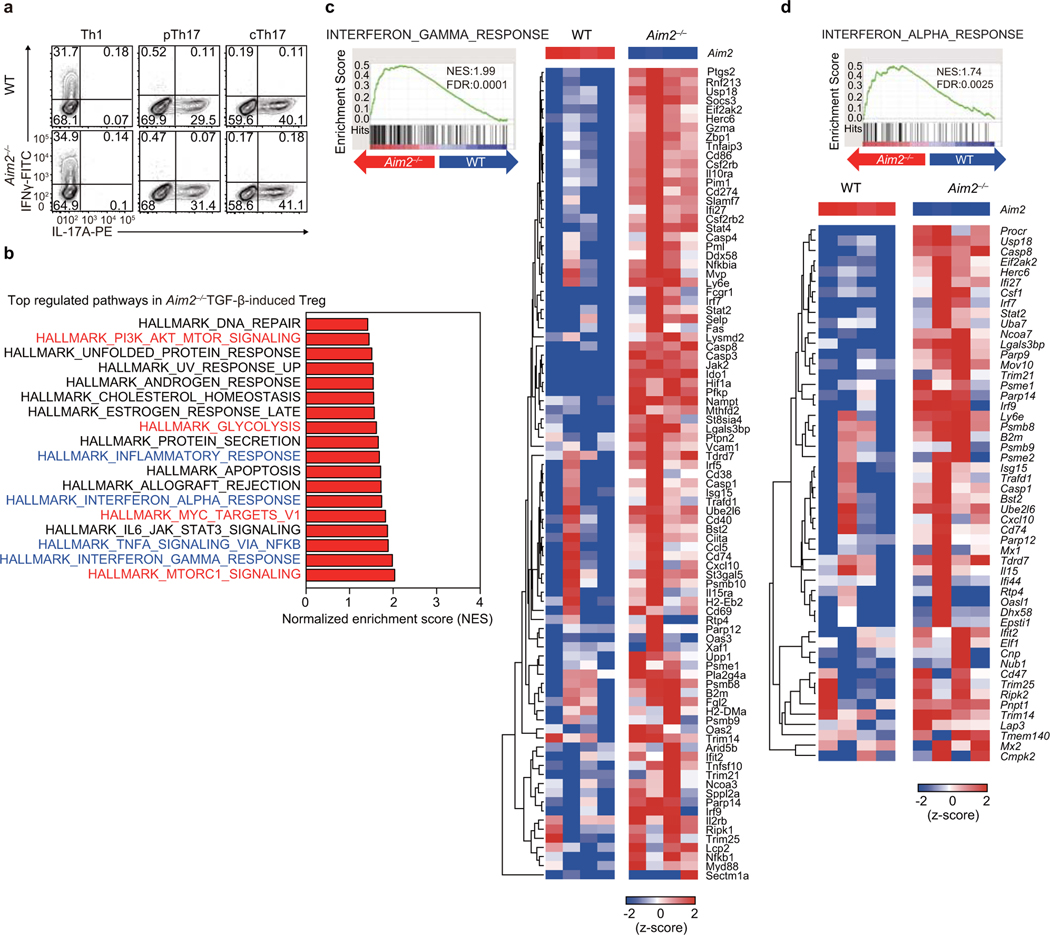
We then examined if AIM2 also controls de novo Treg generation by treating WT or Aim2–/– CD4+ T cells with TGF-β in vitro. Compared to WT cells, fewer Aim2–/– CD4+ T cells differentiated into Foxp3+ cells in the presence of TGF-β (Fig. 4a, b) while Th1 and Th17 cell differentiation were similar between WT and Aim2–/– cells (Fig. 4c, Extended Data Fig. 7a). To understand how AIM2 controls TGF-β-induced Treg cell generation, we performed RNA-seq analysis to compare gene expression profiles of WT and Aim2–/– CD4 T cells activated by TGF-β. The analysis revealed that pathways involving PI3K-Akt-mTOR, mTORC1, glycolysis, Myc and IFNs are among the top pathways preferentially up-regulated in TGF-β-induced Aim2–/– Tregs (Fig. 4d, Extended Data Fig. 7b–d, Extended Data Fig. 8a–d).
Figure 4. AIM2 promotes Treg in vitro and restrains Akt-mTOR via the RACK1/PP2A complex.
a, b, RT-PCR (a) and flow-cytometry (b) of Foxp3 in WT and Aim2–/– CD4+ T cells activated with the indicated amounts (a) or 2 ng/ml (b) of TGF-β for 4 days; n=5 experiments in a and n=4 experiments in b. c, Flow-cytometry of IFNγ+ or IL-17A+ CD4+ T cells of indicated genotypes, 4 days after differentiated under indicated polarizing conditions; n=4 experiments. d-g, WT and Aim2–/– CD4+ T cells were stimulated as in (b). Enrichment scores of indicated gene sets, based on RNA-seq datasets (d). ECAR (e) and OCR (f) levels during glycolysis, and OCR levels during FAO (g), by Seahorse analysis; n=10 experiments for e; n=5 experiments for f, and n=3 experiments for g; P values by two-way ANOVA (e left panel, f, g) or two-sided t-test (e right panel). h, immunoblotting of indicated proteins in WT and Aim2–/– CD4+ T cells stimulated as in (b) with indicated treatment for 24 hr. Representative of three experiments. i, Volcano plot of AIM2 interacting proteins by IP-MS analysis. Red indicates significantly enriched proteins (log2FC > 1; t-test adjusted P<0.05). j-k, The interactions of indicated proteins determined by immunoprecipitation using anti-AIM2 (j) or anti-Rack1 (k), in WT and Aim2–/– CD4+ T cells activated with TGF-β for 24 hr. Representative of three experiments. l, Flow-cytometry of p-Akt levels (Geo. Mean) in WT and Aim2–/– Tregs transduced with PP2A+RACK1 expressing vector (OE) or vector; n=6 experiments, P value by multiple unpaired t-test with Holm-Sidak method. m, Flow-cytometry of p-Akt in spinal cord Tregs from mice of indicated genotypes, 28 days after EAE induction, n=3 experiments. Data are means ± SEM, P values: ns, not significant, *P<0.05, **P<0.01, ***P<0.001 ****P<0.0001, analyzed by two-sided t test unless specified.
Since TGF-β-induced Treg cell differentiation is under metabolic control28, we investigated if AIM2 regulates metabolism during such a process. Indeed, TGF-β-induced Aim2–/– Tregs showed higher ECAR and glycolytic activity, but reduced OCR and FAO, when compared to WT cells (Fig. 4e–g), agreeing with what was observed in Tregs isolated from mice. In addition, Akt-mTOR signaling-related molecular markers, including c-Myc, p-S6, p-4E-BP1, p-Akt and p-Foxo1/3a, were upregulated in the absence of AIM2 (Fig. 4h). Biochemical analyses of CD4+ but not CD8+ T cells, revealed modestly enhanced Akt-mTOR signaling upon AIM2 deletion 24 hr after stimulation in vitro (Extended Data Fig. 9a, b). Additionally, Aim2 deletion consistently led to increased Akt/mTOR signaling in Tregs (Extended Data Figure 9c) but not Tconv cells nor CD8+ cells (Extended Data Figure 9d, e) at early time points, further highlighting a preferential role for AIM2 in controlling Treg function. The pharmacological mTOR inhibitors, rapamycin and pp242, neutralized these changes in CD4+ cells and reduced p-STAT1, while the addition of the anti-IFNγ antibody (XMG1.2) only decreased p-STAT1 (Fig. 4h). These results indicate that IFNγ lies downstream of mTOR. Importantly, rapamycin or pp242 treatment restored TGF-β-induced Treg cell differentiation to a normal level, suggesting that heightened mTOR activity due to Aim2 deletion accounts for defective TGF-β-induced Treg cell differentiation (Extended Data Fig. 10a). These findings suggest that AIM2 controls Treg cell generation and function via the unified mechanism of Akt-mTOR restriction.
To investigate the molecular mechanism by which AIM2 regulates Akt-mTOR, we performed an unbiased mass spectrometric analysis to identify AIM2 interacting proteins during Treg cell differentiation, using a mouse specific anti-AIM2 antibody for endogenous immunoprecipitation from WT and Aim2–/– lysates under TGF-β-induced Treg differentiation conditions (Extended Data Fig. 10b). One of the strongest AIM2-interacting proteins identified was RACK1 (Fig. 4i), which was validated by bi-directional endogenous co-immunoprecipitation (Fig. 4j, k). RACK1 promotes Akt de-phosphorylation by recruiting PP2A29, a phosphatase which dephosphorylates Akt and reduces mTOR signaling to promote Tregs30. Reduced RACK1-PP2A interaction is therefore expected to increase Akt phosphorylation. While the interaction of RACK1 with PP2A and Akt occurred normally in WT Tregs, this interaction was reduced in Aim2–/– Tregs (Fig. 4k). The specific interaction between AIM2 and RACK1/PP2A/Akt complex was observed in TGF-β-induced Treg condition, but not in conventional T cells (Extended Data Fig. 10c). To test the function of RACK1 and PP2A, we ectopically expressed both proteins in TGF-β-induced Treg cells and analyzed the phosphorylation of Akt (p-Akt). Ectopic expression of RACK1 and PP2A downregulated pAkt only in WT, but not Aim2–/– Tregs, indicating that the effects of RACK1/PP2A on pAkt is AIM2-dependent (Fig. 4l, Extended Data Fig. 10d–e). In agreement with the observed enhanced p-Akt in Aim2–/– Treg in vitro, p-Akt was elevated in Aim2–/– Tregs compared to WT controls in spinal cords during EAE (Fig. 4m). Therefore, hyperactivation of Akt in Aim2–/– Tregs in vitro and in vivo (Fig. 3g, Fig. 4h, m) can be attributed to an AIM2-dependent Akt de-phosphorylation by the RACK1/PP2A complex29.
In summary, AIM2 is an inflammasome effector in myeloid cells but its role in T cells has not been explored. This study discovers a previously unappreciated role of AIM2 in Tregs. It unveils a Treg cell-intrinsic, inflammasome-independent function of AIM2 that promotes Tregs to control autoimmunity, specifically in models of multiple sclerosis and inflammatory bowel disease. We demonstrated the mechanism of AIM2 function in Tregs at three levels. At the cellular level, lineage tracing indicates that AIM2 is needed for the stability of Tregs. At the molecular and metabolic levels, AIM2 attenuates the Akt-mTOR pathway to favor oxidative phosphorylation and fatty acid oxidation while mitigating glycolysis, thereby impacting immunometabolism profiles that favor Tregs. At the biochemical level AIM2 promotes the association of Akt with the RACK1/PP2A axis to restrain Akt activation, thus reprogramming immunometabolism to favor Treg cell function (model in Extended Data Fig. 10f).
Methods
Mice
All mice were housed and bred under specific pathogen–free conditions (temperature: 21–23 °C, humidity: 30–70%, 12-hour light/dark cycle) in the animal facility at the University of North Carolina at Chapel Hill. All sex- and age- matched (9–12 weeks) mouse experiments were approved by Institution Animal Care and Use Committee of the University of North Carolina. We complied with all relevant ethical regulations. WT (C57BL/6), Aim2–/–, Aim2+/+ FGC, Aim2fl/fl FGC, Aim2+/+ FGC R26T, Aim2fl/fl FGC R26T, Asc–/–, ICE–/–, Rag1–/–, 2D2 (MOG35–55-specific TCR transgenic), 2D2 × Aim2–/–, Tgfbr2fl/fl CD4Cre (TGFbRII-KO), and CD45.1 congenic wild type mice were generated on the C57BL/6 genetic background. Aim2fl/fl mice were generated using targeting vector (PRPGS00208_A_A10), which contained loxP sites flanking exon 7 and 12 of Aim2, purchased from Knock Out Mouse Project at Children’s Hospital Oakland Research Institute, the University of California at Davis (KOMP-CHORI) repository, for insertion into C57BL/6N embryonic stem cells by the UNC Animal Models Core. Chimeras were obtained from implanted C57BL/6-albino females and backcrossed first to Flpe+ strain to remove neomycin selection marker and then to C57BL/6J for 7 generations plus sibling crosses to produce homozygous flox/flox lines. Aim2fl/fl mice were then crossed with Foxp3-EGFP-Cre (FGC) transgenic mice, where a BAC transgenic construct encoding both EGFP and Cre under the control of Foxp3 promoter was inserted into mouse genome31, and Rosa26tdTomato knock-in mice32 (R26T; Jax Stock No: 007914) to generate Treg-specific deletion of Aim2 (Aim2fl/fl FGC or Aim2fl/fl FGC R26T) and control mice (Aim2+/+ FGC or Aim2+/+ FGC R26T). The sample sizes for all the mice experiments are determined based on the prevailing and widely accepted practice and similar designed experiments and results generated in the lab on the cellular immunology analyzes. No statistical method was used to pre-determine sample size.
Experimental autoimmune encephalomyelitis (EAE)
Sex- and age-matched (9 to 12 weeks old) WT and Aim2–/– mice were randomly allocated to be immunized subcutaneously (s.c.) with 200 μg of MOG35–55 peptide (MEVGWYRSPFSRVVHLYRNGK, GenemeSynthesis, Inc) emulsified in complete Freund’s adjuvant (CFA) (Sigma) containing heat-killed M. tuberculosis (Difco). In addition, the animals were administrated 200 ng of Pertussis Toxin (List Biological Laboratories) intra-peritoneally (i.p.) on days 0 and 2. The severity of EAE was monitored and graded in a blinded fashion on a clinical score of 0 to 5: 0 = No clinical signs; 1 = Limp tail; 2 = Para-paresis (weakness, incomplete paralysis of one or two hind limbs); 3 = Paraplegia (complete paralysis of two hind limbs); 4 = Paraplegia with forelimb weakness or paralysis; 5 = Moribund or death33.
Flow-cytometry and cell sorting
Lymphocytes were isolated from various lymphoid organs of age- and sex- matched mice of 8–12 weeks of age. Fluorescence-conjugated antibodies for CD4 (GK1.5), CD8 (53–6.7), CD45.1 (A20), CD45.2 (104), Vβ11 (RR3–15), CD25 (PC61.5), CD44 (IM7), CD62L (MEL-14), Ki67 (16A8), IFNγ (XMG1.2), IL-17A (TC11–18H10.1), IL-4 (11B11) and Annexin V were purchased from Biolegend. BV421 mouse anti-Akt (pS473) (M89–61) and 7AAD were purchased from BD Bioscience. The anti-Foxp3 antibody (FJK-16s) and Foxp3 staining kit (00–5523-00) were from eBioscience. For intracellular cytokine staining, lymphocytes were stimulated for 4 hr with 50 ng/mL of PMA (phorbol 12-myristate 13-acetate) and 1 mM ionomycin in the presence of brefeldin A. Stained cells were analyzed on LSRFortessa station or Canto (BD Biosciences) using FACSDiva software. A commercially available kit was used for intracellular cytokine staining in accordance with the manufacturer’s protocol (BD Biosciences). For cell sorting, CD4+ T cells or CD25+ Tregs were enriched by MACS and then stained with fluorescence-conjugated antibodies. Stained cells were either acquired on LSRII or LSR Fortessa (BD biosciences) or sorted on the Moflow cell sorter (Dako cytomation, Beckman Coulter) by the Flow Core Facility of University of North Carolina at Chapel Hill. FACS data were analyzed with FlowJo software (TreeStar). For the gating strategy for FACS analysis, see Supplementary Figure 2.
CD4+ T and Treg cell adoptive transfer in EAE
Total lymphocytes were isolated from spleens and peripheral lymph nodes of WT and Aim2–/–mice. Total CD4+ T cells or CD4+CD25+ Tregs were enriched by MACS and Moflow cell sorter. CD4+ T cells (5 × 106 per mouse) were introduced via retro-orbital injection into Rag1–/– female mice. One day later, the recipient mice were immunized with MOG35–55 to induce EAE as described above. To evaluate the function of Tregs in suppressing EAE, 2D2 CD4+ T cells alone (5 × 105 per mouse), 2D2 CD4+ T cells (5 × 105 per mouse) with WT or Aim2–/– Tregs (2 × 105 per mouse) were transferred into Rag1–/– mice via retro-orbital injection. One day later, the recipient mice were immunized with MOG35–55 to induce EAE as described above.
CD4+CD45RBhi T cell transfer colitis model
CD4+ T cells from WT mice were enriched by anti-CD4 (L3T4) magnetic beads (Miltenyi Biotec) and stained with anti-CD4 Pacific Blue, anti-CD25 PE and anti-CD45RB FITC reagents. Naïve CD4 (CD4+CD25−CD45RBhi) T cells were sorted by FACS. Tregs (CD4+CD25+) of WT, Aim2–/– and Asc–/– mice were sorted by FACS. 5 × 105 naïve T cells alone, or with 2 × 105 WT or Aim2–/– Tregs were transferred into Rag1−/− recipient mice by i.p. injection. The recipient mice were weighed twice every week to measure percentage of body weight change and major organs were harvested for analysis at the end of experiment.
In vitro T cell activation, differentiation, and proliferation
Lymphocytes were isolated from peripheral lymph nodes and spleens of age- and sex- matched mice and purified with CD4 microbeads (L3T4, Miltenyi Biotec). Purified CD4+ T cells were cultured in RPMI 1640 medium containing 10% FBS, 1% penicillin-streptomycin and 2.6 μl of β-mercaptoethanol and activated with plate-coated 2.5 μg/ml anti-CD3 (145–2c11, BioXCell) and 1 μg/ml anti-CD28 (37.51, BioXCell) antibodies. For Treg differentiation, designated doses of TGF-β (2 ng/ml) and IL-2 (40 U/ml) were added into the culture medium. Rapamycin and mTOR inhibitors (pp242) were used as indicated. For Th1 differentiation, 20 ng/mL IL-12 (Biolegend) and 20 μg/mL anti-IL-4 (11B11, BioXcell) were added to the culture. For pathogenic Th17 cell differentiation, 20 ng/mL IL-1β (Biolegend), 20 ng/mL IL-6 (Biolegend), 50 ng/mL IL-23 (Biolegend) and 10 μg/mL anti-IFNγ (XMG1.2, BioXcell) were added to the culture. For classical Th17 cell differentiation, 1 ng/mL TGF-β (Biolegend), 40 ng/mL IL-6 (Biolegend) and 10 μg/mL anti-IFNγ (XMG1.2, BioXcell) were added to the culture. For CFSE proliferation assay, a final concentration of 2 μM of carboxyfluorescein succinimidyl ester (CFSE) (C1157, Life Technologies) was used to label CD4+ T cells.
Ectopic expression of PP2A and RACK1 in Treg cells
To generate the retrovirus expressing PP2A and RACK1, we firstly cloned PP2A (OriGene Technologies, MR204384L4) and RACK1 (OriGene Technologies, MR204575L3) into retroviral vectors MSCV-IRES-Thy1.1 (MIT, Addgene #17442) and MSCV-IRES-GFP (MIG, Addgene #20672) respectively, and generated MIT-PP2A and MIG-RACK1 retrovirus in 293T cells by transient transfection. For retroviral transduction, isolated CD4+ T cells were stimulated with anti-CD3/CD28 in the presence of IL-2 (40 U/ml) and TGF-β (2 ng/ml) on day 0 and then transduced with indicated retroviruses containing 8 μg/ml polybrene (Sigma, H9268) by centrifuge at 1500 × g at 30 °C for 1.5 hr on day 1. Cells were harvested and analyzed by flow cytometry three days after retrovirus transduction.
In vitro Treg suppression
Tregs (suppressor) from CD45.1.1 WT mice and CD45.2.2 Aim2–/– mice and naive CD4 T cells (responder) from CD45.1.2 WT young mice were sorted by FACS. To assess the efficacy of Treg cell-mediated immune suppression in vitro, 1 × 105 sorted responder T cells were labeled with CFSE and mixed with varying amounts (as indicated) of Treg suppressor cells. Cell mixtures were stimulated with soluble CD3 antibody (0.125 μg/ml) in the presence of 4 × 105 irradiated (3000 cGy) T cell-depleted splenocytes as antigen-presenting cells. The proliferation of responder cells was assessed by CFSE dilution detected by flow-cytometric analysis 72 hr after stimulation.
Histology
For the assessment of tissue pathology, following an initial perfusion with PBS, mice were subsequently perfused trans-cardially with 4% paraformaldehyde and spinal cords were removed. Tissues were processed and blocked in paraffin wax. Transverse sections of the lumber spinal cord were stained with hematoxylin and eosin (H&E) or Luxol Fast Blue and periodic acid-Schiff (LFB-PAS). The number of inflammatory foci and total demyelination were measured using methods described previously34. Briefly, the numbers of inflammatory foci that contained at least 20 cells were counted within each H&E–stained section in a blinded fashion. Estimates were made of the number of foci, when foci coalesced. Areas of demyelination were assessed in LFB-PAS–stained sections. ImageJ software was used to manually trace the total cross-sectional area and the demyelinated area of each section. Total demyelination was expressed as a percentage of the total spinal cord area34. Colons were Swiss rolled, fixed in 10% neutral-buffered formalin and routinely paraffin embedded and processed. Five μm-thick colon sections were stained with H&E and evaluated by a board-certified veterinary pathologist (A.B.R.) in a blinded manner to perform semi-quantitatively scoring of histopathology. Histology scores represented the sum of each histological alterations as outlined here: inflammation, epithelial defects, area of inflammation, area of epithelial defect, crypt atrophy and dysplasia-neoplasia, by giving each parameter a separate score (0–4) for severity and extent as previously described35.
The enzyme-linked immunosorbent assay (ELISA)
Both spinal cord homogenates and colon-secreted cytokines were analyzed by Enzyme–Linked Immunosorbent Assay (ELISA, MCS00, R&D Systems, Minneapolis, MN) or by multiplex analyte assay using Luminex technology (EMD Millipore, Darmstadt, Germany) according to manufacturers’ protocols. IL-18 (Invitrogen BMS618–3), IL-6 (R&D DY406) and TNFα (Biolegend 430904) were measured using ELISA assays according to manufacturer’s instructions. For colon-secreted cytokines, excised colons were washed and flushed with PBS containing 2× penicillin/streptomycin. The distal-most 1 cm2 colon sections were cultured for 15 hr in RPMI media containing 2× penicillin/streptomycin at 37°C. Supernatants were collected, cleared of debris by centrifugation and assessed for cytokines by Luminex analyses.
Immuno-blotting, Immuno-precipitation (IP) and Mass Spectrometry (MS)
CD4+CD25+ Treg, TGF-β-induced Treg, CD4+ T and CD8+ T cells were lysed in RIPA buffer supplemented with complete proteinase inhibitor cocktail and PhoSTOP phosphatase inhibitors. Protein lysates were cleared of insoluble material through centrifugation and the resulting protein lysates were treated with sample buffer and subjected to SDS-PAGE. In brief, total proteins were wet transferred to 0.2 μm nitrocellulose membranes (BioRad Laboratories), which were blocked using 5% BSA in 1 × TBS-T buffer for 1 hr at room temperature. The membranes were incubated overnight using the following primary antibodies from Cell Signaling Technology (CST): anti-p-Akt (Ser473) (cat.no. 4060) (WB, 1:1000), anti-Akt (cat.no. 9272) (WB, 1:1000), anti-p-Foxo1/3a (cat.no. 9464) (WB, 1:1000), anti-p-4E-BP1 (cat.no. 2855) (WB, 1:1000), anti-p-S6 (Ser235/236) (cat.no. 4856) (WB, 1:1000), anti-c-Myc (cat.no. 5605) (WB, 1:1000), anti-p-Stat1 (Tyr 701) (cat.no. 9167), (WB, 1:1000) anti-AIM2 (cat.no. 63660) (WB, 1:1000) and anti-PP2A catalytic subunit (cat.no. 2259) (WB, 1:1000). The anti-RACK1 (sc-17754) (WB, 1:1000) and anti-actin HRP (sc-4778) (WB, 1:5000) reagents were from Santa Cruz Bioteconology. Membranes were washed in TBS-T and incubated with the following appropriate secondary antibodies from Jackson ImmunoResearch Laboratories: mouse anti-rabbit-HRP, light chain-specific (211–032-171) (WB, 1:10,000), and donkey anti-mouse HRP (715–035-151) (WB, 1:10,000). Protein bands were visualized following exposure of the membranes to ECL substrate solution (ThermoFisher) and quantified by densitometric analysis using Image Lab software. For gel source data, see Supplementary Figure 1.
For immuno-precipitation, WT and Aim2–/– CD4+ T cells differentiated under Treg polarizing and natural (IL-2 only) conditions for 24 hr were lysed with CHAPS lysis buffer (50 mM Tris HCl, pH 7.4, 120 mM NaCl, 0.3% CHAPS) and sonicated with Bioruptor PICO. Cell lysates were incubated with 50 μl magnetic protein A/G beads (Bio-Rad) conjugated with anti-AIM2 (CST, cat.no. 63660) (IP, 1:200) or anti-RACK1 (CST, cat.no. 5432s) (IP, 1:200) antibodies treated by dimethyl pimelimidate. After overnight incubation, beads were washed four times with lysis buffer. Associated protein was eluted by Laemmli sample buffer (Bio-Rad) and incubated at 95°C for 5 min. Eluted samples were separated by SDS-PAGE gel and analyzed by immuno-blotting.
For MS analysis, anti-AIM2 antibody immuno-precipitated proteins were eluted with buffer containing 8 M Urea, 50 mM Tris (pH 8.0), reduced with 5 mM DTT and alkylated with 15 mM iodoacetamide. Trypsin digestion was performed at room temperature overnight in 2 M urea buffer. The peptides were desalted on C18 stage-tips and dissolved in 0.1% formic acid. Peptides were loaded on an Acclain PepMap RSLC C18 Column (150 mm × 75 μm ID, C18, 2 μm, Thermo Fisher Scientific) and analyzed on a Q-Exactive HF-X coupled with Easy nanoLC 1200 (Thermo Fisher Scientific). Analytical separation of all tryptic peptides was achieved with a linear gradient of 5–30% buffer B over 29 min, 30–45% B over 6 min followed a ramp to 100% B in 1 min and 9 min wash periods with 100% buffer B, where buffer A was aqueous 0.1% formic acid and buffer B contained 80% acetonitrile and 0.1% formic acid. LC-MS experiments were performed in a data-dependent mode with full MS (externally calibrated to a mass accuracy of <5 ppm and a resolution of 60,000 at m/z 200) followed by high energy collision-activated dissociation-MS/MS of the top 15 most intense ions with a resolution of 15,000 at m/z 200. High energy collision-activated dissociation-MS/MS was used to dissociate peptides at a normalized collision energy (NCE) of 27 eV. Dynamic exclusion with 20.0 seconds was enabled. Then the mass spectra were processed and peptide identification was performed using the Andromeda search engine found in MaxQuant software version 1.6.0.16 (Max Planck Institute, Germany) against the UniProt mouse protein sequence database (UP000000589). Peptides were identified with a target-decoy approach using a combined database consisting of reverse protein sequences of the database. Up to two missed cleavages was allowed. Peptide identifications were reported by filtering reverse and contaminant entries and assigning to leading razor protein. Peptide inference and protein identification were filtered to maximum 1% and 1% false discovery rate, respectively. Data processing and statistical analysis were performed on Perseus (Version 1.6.0.7). A two sample t-test statistics was used with a P value < 0.05 to report statistically significant expression. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018638.
Glycolytic and mitochondrial respiration rate measurement
Extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR) were measured using the Seahorse Extracellular flux XF24e (Agilent) according to industry manuals. CD4+CD25+ Tregs were isolated from WT and Aim2–/– mice and then stimulated with anti-CD3/CD28 and IL-2 (500 U/ml) for 24 hr. For in vitro TGF-β-promoted Tregs, WT and Aim2–/– CD4+ T cells were stimulated with anti-CD3/CD28 in the presence of TGF-β (2 ng/ml) and IL-2 (40 U/ml) for 48 hr. Prior to metabolic flux analysis, Tregs were seeded at a density of 5 × 105 cells per well. ECAR profiles were assessed by real-time measurements at basal condition and after the addition of 10 mM glucose, 1 μM oligomycin and 20 mM 2-deoxyglucose (2-DG).
OCR profiles were assessed by real-time measurements at basal condition and after the addition of 1 μM oligomycin (Cat #75351), 4 μM FCCP (Cat #C2920), and 1 μM Rotenone (Cat#R8875), and all the reagents are from Sigma-Aldrich. Fatty acid oxidation (FAO) assay was performed where cells were starved in substrate-limited medium and given only BSA or palmitate-BSA (cat no. 102720–100) in FAO assay media. Then OCR was measured to indicate oxidation of fatty acids according to the Agilent FAO assay manual.
RNA preparation and real-time PCR
Total RNA was prepared from T cells using TRIzol reagent (Invitrogen) per manufacturer’s instructions and was reverse-transcribed into cDNA with iScript™ cDNA Synthesis Kit (Bio-Rad, cat.no. 1708891). The Taqman probes were purchased from Applied Biosystems and quantitative PCR was performed on the ABI9700 real-time PCR system with QuantStudio software (Applied Biosystems).
RNA sequencing (RNA-seq)
For RNA-seq analysis, total RNA was extracted from Tregs using the Direct-zol miniprep kit (Zymo Research, R2060). The RNA samples were first enriched by Oligo(dT) magnetic beads and used to construct BGISEQ-500 libraries. RNA-seq libraries were sequenced using the 50 bp single-end protocol (in vivo isolated Tregs) or 100 bp paired-end protocol (TGF-β-induced Tregs) via the BGISEQ-500 sequencer per manufacturer’s protocol. After filtering of adaptors and low quality reads, clean reads (>26 million reads per sample for in vivo isolated Tregs and >40 million reads per sample for TGF-β-induced Tregs) were mapped to the mouse reference genome using HISAT /Bowtie2 tool. Mapping results were stored in BAM files using SAMtools.
Total read counts at the gene level were summarized using featureCounts function in the Rsubread36 in R environment, with the R package biomaRt for gene and transcript mapping. The differential expression (DE) genes were analyzed by DESeq2 package37 with default settings using total read counts as input and the adjusted P value (padj) less than 0.05. Heatmaps of gene expression were generated based on zscore values of normalized expression matrix from DESeq2 analysis in Gene-E from Broad Institute (www.broadinstitute.org/GENE-E/).
Gene Set Enrichment Analysis (GSEA)
GSEA38 was performed using the Java application available from The Broad Institute (www.broadinstitute.org/gsea/). Gene set databases including Hallmarks (h.all.v6.1.symbols.gmt), KEGG (c2.cp.kegg.v6.1.symbols.gmt) and Reactome (c2.cp.reactome.v6.1.symbols.gmt) from the Molecular Signatures Database (MSigDB)39 were used in the analysis. One thousand gene set permutations were performed. An FDR cutoff of < 0.05 was used for enriched terms, as is recommended when performing permutations by gene set. R version 3.5.0 was used. The RNA-seq data are made available in the Gene Expression Omnibus repository at the National Center for Biotechnology Information under accession number GSE133019.
Statistical analysis
Data analysis were processed and represented by Prism (GraphPad, San Diego). Statistical significance was determined by two-sided Student’s t-test, two-way ANOVA followed by Tukey test or Holm–Sidak’s multiple-comparisons tests indicated in figures. A P value less than 0.05 (confidence interval of 95%) was considered significant. The asterisks in figures were used to indicate P values as follows: *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001. The exact P values are shown in the Source Data. The sample sizes n are stated in the figure legends to indicate biologically independent replicates used for statistical analyses.
Data availability
The RNA-seq data are made available in the Gene Expression Omnibus repository at the National Center for Biotechnology Information under accession number GSE133019. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018638. Source data are provided with this paper. All other data supporting the findings of this study are available from corresponding authors upon reasonable request.
Extended Data
Extended Data Figure 1. Aim2–/– mice show normal T cell homeostasis.
a-b, Flow-cytometry of CD4+Foxp3+ Tregs (a) and IFNγ-, IL-17A-producing CD4+ T cells (b) in the spleen and peripheral lymph node (PLN) of WT and Aim2–/– mice at day 14 of an EAE course. Representative results (left) and statistical analysis (right) of six experiments are shown. c, Flow-cytometry of CD4+Foxp3+ Tregs in the thymus, spleen, and PLN of WT and Aim2–/– mice. Representative results (left) and statistical analysis (right) of three experiments are shown. d, Total number of cells isolated from the thymus, spleen, and peripheral lymph node of WT and Aim2–/– mice. Experimental design and statistical analysis performed as described in (c). e-f, Flow-cytometry of naïve, effector/memory CD4+ (e) and CD8+ T cells (f) in the spleen and PLN of WT and Aim2–/– mice (n=3/ group) analyzed by CD44 and CD62L expression. Experimental design and statistical analysis performed as described in (c). g-h, Flow-cytometry of IFNγ-, IL-4- or IL-17A-producing CD4+ cells (g) and IFNγ-producing CD8+ T cells (h) in WT and Aim2–/– mice (n=3/ group). Experimental design and statistical analysis performed as described in (c). Representative (upper) and composite (lower) data are shown. Data are means ± SEM; P values: ns, not significant, per two-sided t-test.
Extended Data Figure 2. Aim2–/– mice show normal Treg proliferation, survival, and ratio of central/effector Tregs at steady state or during EAE in vivo, normal CD4 T cell proliferation in vitro, and normal CD8 T cells distribution and cytokine production during EAE.
a, Flow-cytometry of Ki67 to analyze proliferation of WT and Aim2–/– Tregs in the peripheral lymph node (PLN) and spleen at steady state. Representative results (upper) and statistical analysis (lower) of five experiments are shown. b, Apoptosis of WT and Aim2–/– Tregs in the PLN and spleen at steady state was analyzed by flow-cytometry using Annexin V and 7AAD staining. Representative results (upper) and statistical analysis (lower) of five experiments are shown. c, Flow-cytometry of Ki67 to analyze proliferation of WT and Aim2–/– Tregs in the PLN, spleen, and spinal cord (SC) during EAE. Representative results (upper) and statistical analysis (lower) of six experiments are shown. d, Apoptosis of WT and Aim2–/– Tregs in SC during EAE was analyzed by flow-cytometry using Annexin-V and 7AAD staining. Representative results (upper) and statistical analysis (lower) of six experiments are shown. e, Flow-cytometry of CD44 and CD62L in WT and Aim2–/– Tregs isolated from SC during EAE. Representative results (upper) and statistical analysis (lower) of six experiments are shown. f, Flow-cytometry of WT and Aim2–/– CD4+ T cell proliferation stimulated with different doses of anti-CD3/CD28, determined by CFSE dilution assay. Representative results of two independent experiments. g, Flow-cytometry of 2D2 and Aim2-deficient 2D2 (2D2 × Aim2–/–) CD4+ T cell proliferation stimulated with MOG35–55 peptide pulsed bone marrow-derived dendritic cells (BMDC) as determined by CFSE dilution assay. Representative of two independent experiments. h, Flow-cytometry of WT and Aim2–/– CD4+ or CD8+ T cells in the peripheral lymph node (PLN), spleen and spinal cord (SC) during EAE. Representative results (left) and statistical analysis (right) of six experiments are shown. i-k, Flow-cytometry of IFNγ-, IL-17A-producing (i) or Foxp3+, IFNγ-producing CD8+ T cells (j) in the PLN, spleen, and SC during EAE. Representative results (i, j) and statistical analysis (k) of six experiments are shown. Data are means ± SEM; P values: ns, not significant, by two-sided t-test.
Extended Data Figure 3. Aim2 is highly expressed in Tregs and its promoter is bound by Treg-related transcription factors.
a-b, Mouse Aim2 gene expression in different T cell subsets from publicly available gene microarray (a) and RNAseq (b) databases (https://www.immgen.org/). c, Human Aim2 gene expression in T cell and macrophage subsets from the Expression Atlas of EMBL-EBI (https://www.ebi.ac.uk/). d, Purity of isolated CD4+CD25+ Tregs from WT and Aim2–/– mice. Flow-cytometry of CD4+CD25+Foxp3+ Tregs shows that more than 97% of isolated Tregs are Foxp3+ cells. e, Aim2 expression was assessed from isolated CD4+CD25+ Tregs and CD4+ T cells. Cells were freshly isolated from pooled spleens and lymph nodes and purified by MACS beads. Aim2 mRNA expression was examined by real-time PCR; n=4 experiments. f, The mRNA expression of Aim2 in Tregs and CD4+ T cells stimulated with anti-CD3/CD28 plus IL-2 (500 U/ml) for 24 hr; n=4 experiments. g, The mRNA expression of Aim2 in Tregs stimulated with anti-CD3/CD28 plus IL-2 (500 U/ml) in the absence (−) or presence (+) of TGF-β (1 ng/ml) for 24 hr; n=4 experiments. h, Aim2 expression was assessed in freshly isolated CD4+CD25+ Tregs from WT and Tgfbr2fl/fl CD4Cre (TGFbRII-KO) mice. Aim2 mRNA expression was examined by real-time PCR; n=5 experiments. i, The mRNA expression of Aim2 in naïve CD4+ or CD8+ T cells stimulated with anti-CD3/CD28 plus IL-2 (40 U/ml) in the absence or presence of TGF-β (1 ng/ml) for 24 hr; n=4 experiments. j, ChIP-seq analysis of Runx1, Ets1, Bcl11b and CREB binding to the Aim2 promoter region in Tregs and CD4 Tconv cells (NCBI SRA database number: DRP003376). Data are means ± SEM; P values: ns, not significant, **P<0.01, ***P<0.001, ****P<0.0001.
Extended Data Figure 4. AIM2 is essential for Tregs to suppress T cell-mediated colitis and EAE.
a, H&E staining of colons from T cell-induced colitis mice transferred with WT CD4+CD45RBhi T cells (Tn) alone (n=5) or in combination with WT (n=6) or Aim2–/– (n=6) CD4+CD25+ Tregs, harvested 7 weeks after T cell transfer. Scale bars represent 1 mm for 40X and 100 μm for 100X. b, Statistical analysis of pathology score of colitis mice with biological replicates of each group is depicted in (a). Tn only: n=5; Tn+WT Treg: n=6; Tn+Aim2–/– Treg: n=6. c, Cytokine levels in the supernatants of colon tissue cultures from mice depicted in (a) measured by Millipore luminex assay, harvested 7 weeks after T cell transfer. Tn only: n=4; Tn+WT Treg: n=5; Tn+Aim2–/– Treg: n=5. d, Change of body weight of Rag1–/– recipients receiving WT naïve CD4+CD45RBhi T cells (Tn) alone or in combination with WT, Aim2–/– or Asc–/– CD4+CD25+ Tregs. Rag1–/– recipients of Tn (n=8), Tn+WT Treg (n=6), Tn+Aim2–/– Treg (n=9), Tn+Asc–/– Treg (n=7); composite of two independent experiments. P value by two-way ANOVA. e, Flow-cytometry of CD4+Foxp3+ Tregs in the colons of Rag1–/– recipients of Tn (n=8), Tn+WT Treg (n=6), Tn+Aim2–/– Treg (n=9), Tn+Asc–/– Treg (n=7), harvested 7 weeks after T cell transfer. P value by one-way ANOVA with Tukey’s multiple comparisons test. f, Schema of EAE induction in Rag1–/– mice transferred with either 2D2 CD4+ T cells alone or in combination with WT or Aim2–/– CD4+CD25+ Tregs. Lymphocytes and tissues were harvested 14–15 days after initial T cell transfer for further analysis. g, Flow-cytometry shows the distributions of 2D2 CD4+ T cells (Vβ11+) or Tregs (Foxp3+) before transfer to Rag1–/– recipient mice. h, Mean EAE clinical score of mice depicted in (f); n=5 mice per group. P value by two-way ANOVA and Holm -Sidak post-hoc test. Data are representative of three independent experiments. The difference between 2D2 alone and 2D2 with Aim2–/– Treg is not significant. i, Flow-cytometry of IFNγ+ or IL-17A+ CD4+Vβ11+ T cells in spinal cords (SC) from groups depicted in (f). Left, representative sample; right, composite data pooled of five mice per group from three independent experiments. j, Flow-cytometry of CD4+Foxp3+ Tregs from SC derived from mice depicted in (f).Left, representative sample; right, composite data pooled of five mice per group from three independent experiments. P value by one-way ANOVA with Tukey’s multiple comparisons test. k, Flow-cytometry of IFNγ+ or IL-17A+ CD4+Vβ11− T cells in SC from groups depicted in (f). Left, representative sample; right, composite data summarized from five biological replicates. l, CD25−CD44lowCD62Lhi naive CD4+ T cells (Tresp) were isolated from WT mice and labelled with CFSE. CD4+CD25+ Tregs were isolated from WT or Aim2–/– mice by FACS. Tresp and Tregs of different genotypes were mixed at indicated ratios and stimulated with anti-CD3 in the presence of irradiated antigen-presenting cells from mixed spleens and lymph nodes. The suppressive activity of Tregs was assessed by CFSE dilution of responder T cells (Tresp). Data are means ± SEM, P values: ns, not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, analyzed by two-sided t test unless specified.
Extended Data Figure 5. Aim2fl/fl FGC R26T mice show normal T cell homeostasis, normal Treg proliferation and survival at steady state and during EAE.
a, Genotyping of FACS-sorted Treg (CD4+CD25+GFP+) and CD4 Tconv (CD4+CD25−) cells showing efficient deletion of Aim2 genomic DNA in Tregs. The image is representative of three independent experiments. b, Immunoblot analysis of AIM2 protein in Tregs from Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice. The image is representative of three independent experiments. c, Images of Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice and corresponding lymphoid organs of spleen and lymph nodes. d, Flow-cytometry of CD4+Foxp3+CD25+Tregs in the thymus, spleen and peripheral lymph node (PLN) of Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice Representative results (upper) and statistical analysis (lower) of four experiments are shown. e, Flow-cytometry of CD4+ or CD8+ T cells in the spleen and PLN of Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice. Representative results (left) and statistical analysis (right) of four experiments are shown. f-g, Flow-cytometry of naïve, effector/memory CD4+ (f) and CD8+ T cells (g) in the spleen and PLN of Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice by assessing CD44 and CD62L expression. Representative results (left for f, upper for g) and statistical analysis (right for f, lower for g) of four experiments are shown. h-i, Flow-cytometry of IFNγ-, IL-4- or IL-17A-producing CD4+ cells (h) and IFNγ-producing CD8+ T cells (i) in Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice. Representative results (upper) and statistical analysis (lower) of four experiments are shown. j, Statistical summary of IFNγ-, IL-17A-producing CD4+Tomato− Tconv cells in the peripheral lymph node (PLN; left) and spleen (right) of Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice at day 28 of an EAE course. Composite data summarized from three biological replicates. k, Statistical summary of IFNγ-, IL-17A-producing CD4+Tomato+ Tregs in the PLN (left) and spleen (right) of Aim2+/+ FGC R26T and Aim2fl/fl FGC R26T mice at day 28 of EAE course. Composite data summarized from three biological replicates. l, Flow-cytometry of Ki67 to analyze proliferation of Aim2+/+ FGC and Aim2fl/fl FGC Tregs in the PLN, spleen, and spinal cord (SC) during EAE. Left, representative sample; right, composite data summarized from three biological replicates. m, Apoptosis of Aim2+/+ FGC and Aim2fl/fl FGC Tregs in SC during EAE was analyzed by flow-cytometry using Annexin-V and 7AAD staining. Left, representative sample; right, composite data summarized from three biological replicates. n, Flow-cytometry of Ki67 to analyze proliferation of Aim2+/+ FGC and Aim2fl/fl FGC Tregs in the PLN and spleen at steady state. Left, representative sample; right, composite data of five mice of two independent experiments. o, Apoptosis of Aim2+/+ FGC and Aim2fl/fl FGC Tregs in the PLN and spleen at steady state was analyzed by flow-cytometry using Annexin-V and 7AAD staining. Left, representative sample; right, composite data of five mice of two independent experiments. Data are means ± SEM, P values: ns, not significant, analyzed by two-sided t test unless specified.
Extended Data Figure 6. Enhanced glycolytic, interferon and Myc target signatures are found in Aim2–/–Tregs isolated in vivo.
a, Glycolytic activity of WT (n=7 biological replicates/group) and Aim2–/– (n=6 biological replicates/group) Tregs with or without stimulation by anti-CD3/CD28 plus IL-2 (500 U/ml) for 24 hr. Statistics of glycolysis (ECAR rate after glucose addition) and glycolytic capacity (maximal ECAR after subtracting the ECAR rate following 2-DG exposure) calculated from (Fig. 3a). Data are means ± SEM; P values: ns, not significant; **P<0.01, ***P<0.001, by two-sided t-test. b, Heatmap of IFNα response signature of RNA-seq data from Aim2–/– compared to WT Tregs stimulated with anti-CD3/CD28 plus IL-2 (500 U/ml) at indicated time points (0 or 24 hr). c, Heatmap of IFNγ response signature as described in (b). d, Heatmap of Myc target profiles as described in (b).
Extended Data Figure 7. Enhanced gene signature found in TGF-β-induced Aim2–/– Tregs using RNA-seq analysis.
a, Flow-cytometry of IFNγ+ or IL-17A+ CD4+ cells from WT and Aim2–/– mice after four days of differentiation under Th1, pathogenic Th17 (pTh17) and classic Th17 (cTh17) conditions respectively, as described in Fig. 4c. Data are representative of four independent experiments. b, Summary of top pathways positively enriched in anti-CD3/CD28 activated Aim2–/– CD4+ T cells in the presence of TGF-β (2 ng/ml) and IL-2 (40 U/ml) for 24 hr, by GSEA analysis of the RNA-seq dataset. c-d, Enrichment of IFNγ (c) and IFNα response pathways (d) by GSEA (left) and heatmap (right) of pathway related genes in Aim2–/– vs. WT CD4 T cells stimulated with anti-CD3/CD28 in the presence of TGF-β (2 ng/ml) and IL-2 (40 U/ml) for 24 hr.
Extended Data Figure 8. RNA-seq analysis reveals enhanced mTOR, Myc and glycolytic signatures in TGF-β-induced Aim2–/– Tregs.
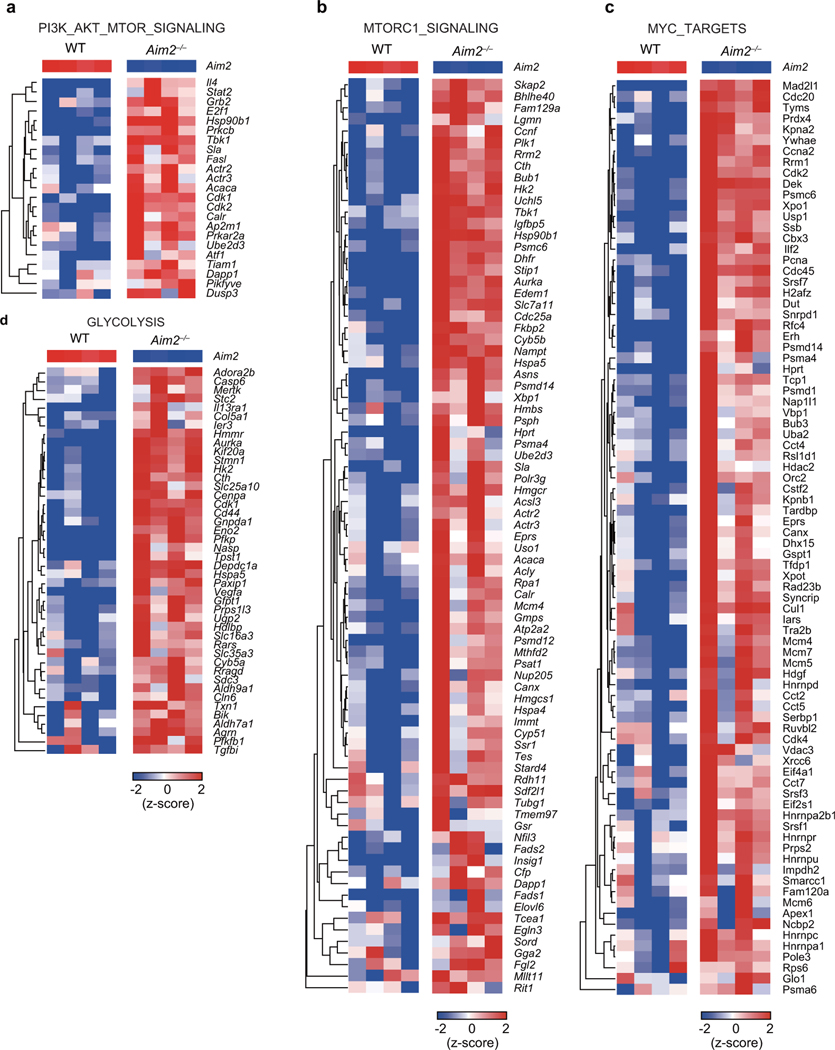
a, Heatmap of PI3K-Akt-mTORC related gene expression in WT and Aim2–/– CD4+ T cells stimulated with anti-CD3/CD28 in the presence of TGF-β (2 ng/ml) and IL-2 (40 U/ml) for 24 hr.
b, Heatmap of mTORC1 signaling related gene expression, with samples described in (a). c, Heatmap of Myc targets related gene expression, with samples described in (a). d, Heatmap of glycolysis related gene expression, with samples described in (a).
Extended Data Figure 9. Akt-mTOR signaling in WT and Aim2–/– Treg, CD4+ and CD8+ T cells.
a-b, Immunoblot analysis of p-Akt (S473), p-Foxo1/3a, p-S6, p-4E-BP1, c-Myc and β-actin in WT and Aim2–/– CD4+ T cells (a) or CD8+ T cells (b) stimulated with anti-CD3/CD28 plus IL-2 (40 U/ml) for 24 hr. c-e, Immunoblot analysis of p-Akt (S473), p-Foxo1/3a, p-S6, p-4E-BP1, c-Myc and β-actin in WT and Aim2–/– Tregs (c) stimulated with anti-CD3/CD28 plus IL-2 (500 U/ml), or CD4+ (d) and CD8+ (e) T cells stimulated with anti-CD3/CD28 plus IL-2 (40 U/ml) for indicated time points. Left: representative results; right: quantification for statistics by densitometric analysis using Image Lab software; n= 4 experiments (a-c); n=3 experiments (d,e); P values: ns, not significant, *P<0.05, **P<0.01, analyzed by two-sided paired t-test.
Extended Data Figure 10. AIM2 interacts with RACK1/PP2A/Akt complex and is critical to regulate Akt-mTOR signaling for Treg cell generation.
a, Flow-cytometry analysis of Foxp3 in WT and Aim2–/– CD4+ T cells stimulated by anti-CD3/CD28 plus IL-2 (40 U/ml) and TGF-β (2 ng/ml) and then cultured with DMSO (n=4 biological replicates/group), rapamycin (1 nM) (n=7 biological replicates/group), or pp242 (0.5 μM) (n=7 biological replicates/group) for 96 hr. Results are representative of three independent experiments. P value by one-way ANOVA with Tukey’s multiple comparisons test. b, Schema of immunoprecipitation–mass spectrometry (IP-MS) approach to identify AIM2 interacting proteins in TGF-β-induced Tregs. WT and Aim2–/– naïve CD4+ T cells were activated with anti-CD3/CD28 in the presence of TGF-β (2 ng/ml) and IL-2 (40 U/ml) for 24 hr and protein lysates from each group were collected for further IP-MS analysis. c, Interaction of AIM2 and RACK1 detected by immunoprecipitation using anti-RACK1 antibody or anti-IgG as control in TGF-β-induced Tregs and CD4+ T cells, and immunoblotted with different antibodies, including anti-PP2Aca, anti-Akt, anti-RACK1 and anti-AIM2. Arrow points to the AIM2 protein. Results are representative of three independent experiments. d, WT and Aim2–/– CD4 T cells were stimulated with anti-CD3 and CD28 plus IL-2 (40 U/ml) and TGF-β (2 ng/ml) for 24 hr and transduced either with MIT-PP2A and MIG-RACK1, or with MIT and MIG vector controls. The cells were harvested 3 days after virus transduction. The populations expressing PP2A (Thy1.1+), RACK1 (GFP+) and both (Thy1.1+GFP+) were identified by flow-cytometry. e, Flow-cytometry of p-Akt of WT and Aim2–/– Tregs that overexpressed PP2A (Thy1.1+) or RACK1 (GFP+) compared to corresponding vector controls. Representative FACS plots (upper) and statistical analysis (lower) of six experiments are shown. P value by multiple unpaired t-test with Holm-Sidak method. f, Model for AIM2 function shows AIM2 facilitates the interaction between RACK1 and PP2A-phosphatase causing de-phosphorylation of Akt to restrain the activity of mTOR pathway, therein promoting Foxp3 expression and Treg stability. Data are means ± SEM, P values: ns, not significant, *P<0.05, ****P<0.0001.
Supplementary Material
Acknowledgements:
The following funding supports are acknowledged: NIH (AI029564, CA156330, DK094779, AI097392, AI123193), the National Multiple Sclerosis Society (CA10068 to J.P.-Y.T and FG 1968-A-1 to W.C.). NIAID (AI067798 to J.P.-Y.T, H.G. and W.J.B.), the National Multiple Sclerosis Society (RG-1802-30483) and a Yang Family Biomedical Scholars Award to Y.Y.W. We received help from Gregory D. Sempowski (Duke University, Duke Human Vaccine Institute, Durham, NC, USA, Immune Monitoring Core for AI067798) for luminex assay and analysis. We thank Balfour Sartor (P01-DK094779) for advice regarding the colitis model, Nancy Fisher and Janet Dow (UNC Flow-cytometry facility support in part by P30 CA016086 Cancer Center Core Support Grant) for cell sorting, and Dale Cowley (UNC Animal Models Core for P30 CA016086) for generating Aim2f/f mice. This work benefitted from publicly available online database assembled by the ImmGen consortium and The European Bioinformatics Institute (EMBL-EBI) without prior permission. We thank Dr. Elizabeth Holley-Guthrie for genotyping mice.
Footnotes
Competing interests:
The authors declare no competing interests.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References:
- 1.Guo H, Callaway JB & Ting JP Inflammasomes: mechanism of action, role in disease, and therapeutics. Nature medicine 21, 677–687, doi: 10.1038/nm.3893 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broz P. & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol 16, 407–420, doi: 10.1038/nri.2016.58 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Gris D. et al. NLRP3 plays a critical role in the development of experimental autoimmune encephalomyelitis by mediating Th1 and Th17 responses. J Immunol 185, 974–981, doi: 10.4049/jimmunol.0904145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lalor SJ et al. Caspase-1-processed cytokines IL-1beta and IL-18 promote IL-17 production by gammadelta and CD4 T cells that mediate autoimmunity. J Immunol 186, 5738–5748, doi: 10.4049/jimmunol.1003597 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Shaw PJ et al. Cutting edge: critical role for PYCARD/ASC in the development of experimental autoimmune encephalomyelitis. J Immunol 184, 4610–4614, doi: 10.4049/jimmunol.1000217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin BN et al. T cell-intrinsic ASC critically promotes T(H)17-mediated experimental autoimmune encephalomyelitis. Nat Immunol 17, 583–592, doi: 10.1038/ni.3389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandes-Alnemri T, Yu JW, Datta P, Wu J. & Alnemri ES AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513, doi: 10.1038/nature07710 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandes-Alnemri T. et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat Immunol 11, 385–393, doi: 10.1038/ni.1859 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krieg AM AIMing 2 defend against intracellular pathogens. Nat Immunol 11, 367–369, doi: 10.1038/ni0510-367 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Rathinam VA et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat Immunol 11, 395–402, doi: 10.1038/ni.1864 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goverman J. Autoimmune T cell responses in the central nervous system. Nat Rev Immunol 9, 393–407, doi: 10.1038/nri2550 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reynolds JM, Martinez GJ, Chung Y. & Dong C. Toll-like receptor 4 signaling in T cells promotes autoimmune inflammation. Proc Natl Acad Sci U S A 109, 13064–13069, doi: 10.1073/pnas.1120585109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heng TS, Painter MW & Immunological Genome Project C. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094, doi: 10.1038/ni1008-1091 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Papatheodorou I. et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Res 46, D246–D251, doi: 10.1093/nar/gkx1158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Josefowicz SZ, Lu LF & Rudensky AY Regulatory T cells: mechanisms of differentiation and function. Annual review of immunology 30, 531–564, doi: 10.1146/annurev.immunol.25.022106.141623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitagawa Y. et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol 18, 173–183, doi: 10.1038/ni.3646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee W. & Lee GR Transcriptional regulation and development of regulatory T cells. Experimental & molecular medicine 50, e456, doi: 10.1038/emm.2017.313 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo CT & Li MO Transcriptional control of regulatory T cell development and function. Trends Immunol 34, 531–539, doi: 10.1016/j.it.2013.08.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mottet C, Uhlig HH & Powrie F. Cutting edge: cure of colitis by CD4+CD25+ regulatory T cells. J Immunol 170, 3939–3943 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Wilson JE et al. Inflammasome-independent role of AIM2 in suppressing colon tumorigenesis via DNA-PK and Akt. Nature medicine, doi: 10.1038/nm.3908 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Man SM et al. Critical Role for the DNA Sensor AIM2 in Stem Cell Proliferation and Cancer. Cell 162, 45–58, doi: 10.1016/j.cell.2015.06.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng H. & Chi H. mTOR signaling in the differentiation and function of regulatory and effector T cells. Curr Opin Immunol 46, 103–111, doi: 10.1016/j.coi.2017.04.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Procaccini C. et al. The Proteomic Landscape of Human Ex Vivo Regulatory and Conventional T Cells Reveals Specific Metabolic Requirements. Immunity 44, 406–421, doi: 10.1016/j.immuni.2016.01.028 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fragale A. et al. IFN regulatory factor-1 negatively regulates CD4+ CD25+ regulatory T cell differentiation by repressing Foxp3 expression. J Immunol 181, 1673–1682 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Overacre-Delgoffe AE et al. Interferon-gamma Drives Treg Fragility to Promote Anti-tumor Immunity. Cell 169, 1130–1141 e1111, doi: 10.1016/j.cell.2017.05.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R. et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882, doi: 10.1016/j.immuni.2011.09.021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merkenschlager M. & von Boehmer H. PI3 kinase signalling blocks Foxp3 expression by sequestering Foxo factors. J Exp Med 207, 1347–1350, doi: 10.1084/jem.20101156 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michalek RD et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol 186, 3299–3303, doi: 10.4049/jimmunol.1003613 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li G. et al. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nature communications 3, 667, doi: 10.1038/ncomms1675 (2012). [DOI] [PubMed] [Google Scholar]
- 30.Apostolidis SA et al. Phosphatase PP2A is requisite for the function of regulatory T cells. Nat Immunol 17, 556–564, doi: 10.1038/ni.3390 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Extended References:
- 31.Zhou X. et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med 205, 1983–1991, doi: 10.1084/jem.20080707 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Madisen L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience 13, 133–140, doi: 10.1038/nn.2467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller SD & Karpus WJ Experimental autoimmune encephalomyelitis in the mouse. Current protocols in immunology Chapter 15, Unit 15 11, doi: 10.1002/0471142735.im1501s77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsushita T, Yanaba K, Bouaziz JD, Fujimoto M. & Tedder TF Regulatory B cells inhibit EAE initiation in mice while other B cells promote disease progression. J Clin Invest 118, 3420–3430, doi: 10.1172/JCI36030 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erben U. et al. A guide to histomorphological evaluation of intestinal inflammation in mouse models. International journal of clinical and experimental pathology 7, 4557–4576 (2014). [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Y, Smyth GK & Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108, doi: 10.1093/nar/gkt214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liberzon A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740, doi: 10.1093/bioinformatics/btr260 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are made available in the Gene Expression Omnibus repository at the National Center for Biotechnology Information under accession number GSE133019. The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD018638. Source data are provided with this paper. All other data supporting the findings of this study are available from corresponding authors upon reasonable request.