Abstract
Historically, technology has been central to new discoveries in biology and progress in medicine. Among various technologies, microtechnologies, in particular, have had a prominent role in the revolution experienced by the life sciences in the last few decades, which will surely continue in the years to come. In this Perspective, we illustrate how microtechnologies, with a focus on microfluidics, have evolved in trends/waves to tackle the boundary of knowledge in the life sciences. We provide illustrative examples of technology-enabled biological breakthroughs and their current and future use in clinics. Finally, we take a closer look at the translational process to understand why the incorporation of new micro-scale technologies in medicine has been comparatively slow so far.
I. INTRODUCTION
The important role that technology plays in advancing biology and medicine may not be obvious on the surface. However, we believe that almost every major milestone in biology and medicine has happened on account of or was triggered by new technology, transforming the way one “sees” or “detects” the biological world.
Historically, technological development has been a key enabler of new discoveries and a driver of rapid growth in the life sciences. Antonie van Leeuwenhoek is commonly known as “the father of microbiology,” as his initial hobby of grinding lenses to observe tiny objects led him to document bacteria, red blood cells, and blood capillaries for the first time. Linus Pauling changed biology by interpreting sickle cell anemia in terms of its molecular basis as an evolutionary adaptation, but it was the electrophoresis instrument developed by physical chemist Arne Tiselius that enabled his discoveries. Similarly, recombinant DNA and cloning are central to modern biology, but they were not possible without the now ubiquitous polymerase chain reactions (PCRs), invented by Kary Mullis. The laser, the mass spectrometer, and magnetic resonance imaging all opened doors to modern science.
The progress in biology and medicine enabled by the contribution of new technologies has significantly accelerated in the last few decades and has centered around two main areas of development: novel bioassays and new instrumentation. Well-known examples of the former are CRISPR gene editing and RNA interference, while the latter ranges from pipettors to flow cytometers. Both assays and instruments are essential and often intrinsically linked but here we choose to concentrate mainly on the latter, and particularly on microfluidic and similar micro-scale technologies, which have been a major driver of progress in the last few decades. The microfluidics community celebrates 30 years of developments since the first proposal of lab-on-a-chip devices. At the same time, we see how the recent COVID-19 pandemic has leveraged so many of these past developments and drives further technological concepts while urgently demanding clinical maturation. Therefore, we think the time is pertinent now to reflect on the technological transformations for the life sciences that resulted in the world we live in today.
In 1990, only three decades ago, biological discovery could leverage far fewer tools of considerably lower performance. Essentially, nothing about the human genome was known at the time and the human genome project had just been launched. For example, to find the BRCA1 gene associated with breast cancer, Mary-Claire King and colleagues used Southern blotting to genotype 173 different markers within 23 extended families to identify a region on chromosome 17q21.1 Without the possibility of sequencing, chromosomal location was one of the few ways available to start the painstaking work of locating the gene through further linkage analysis, chromosome walking and cDNA libraries.1 Today, genome-wide association studies (GWAS) leverage DNA microarrays and increasingly also next-generation sequencing (NGS) technologies to test hundreds of thousands to millions of genetic variations of many individuals to pinpoint genotype–phenotype associations, which have so far identified over 55 000 unique loci for nearly 5000 diseases and traits.2 As another example, Michael Houghton and colleagues required almost a decade of work to identify the hepatitis C virus, finally achieved in 1989 through a blind cDNA immunoscreening approach. At the time, reverse transcription was just arising, and RNA analysis was mostly done by Northern blotting, which was of low sensitivity, quantitatively inaccurate, and time consuming. Sequencing a new virus within weeks of its emergence, as was the case recently with SARS-CoV-2,3 was unthinkable at the time. A further case in point is protein analysis, which in 1989 mostly relied on protein sequencing through automated Edman degradation, which then required 1 h per amino acid residue.4 Additionally, previous sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) separation and identification of non-target proteins was a laborious process that required significant amounts of time and funds. Today, matrix-assisted laser desorption/ionization (MALDI) can process >100 000 samples per day for protein identification, and new technologies are rapidly emerging for protein sequencing.
Here, by discussing such examples of technological development and their impact, we argue that technology was, is, and will continue to be inexorable in fundamental biological research and clinical progress. Additionally, we hope to bring attention to the essential role of the technologist in medicine and biology, a role that we believe is, unfortunately, not yet fully defined. One approach is to bring principles from engineering and physics to the direct discovery of biological processes—with the goal of transforming biology into a basic, quantitative science. We would like to emphasize an approach that we think is at least as important: applying engineering to the creation of new and unique tools, which will accelerate new discoveries and enable rapid growth in biological research and applications. Furthermore, we would like to bring a fresh perspective to a new generation of students and researchers, in the hope that medical and life science students will truly appreciate the role of technology as well as benefit from an increased technological training in their curriculum. At the same time, students in the physical sciences will hopefully realize that they can make an impact in medicine and biology and be more exposed to both during their training. To this aim, in Sec. II of this Perspective, we offer a non-exhaustive overview of many of the developments that microtechnologies and microfluidics have seen in the last three decades, focusing on the technical aspects and on the process of replacing older technologies by new ones. In Sec. III, we provide examples of how these technological developments have translated into key discoveries in fundamental biological research and new clinical diagnostic solutions. Finally, in Sec. IV, we give an overview of the translational process up to clinical use and illustrate why and how the process lets through so few of the promising technologies that we see in academic publications.
II. A BRIEF OVERVIEW OF THE HISTORY, CHANGES, AND NEEDS OF MICROTECHNOLOGIES FOR THE LIFE SCIENCES
The first half of the 20th century can be considered a golden age of physics. Since the middle of the century, the development of new technologies and methods to address biological problems increasingly opened the doors for equally revolutionary transformations in the life sciences. Advances in x-ray diffraction methods, notably led by Franklin and Wilkins, were fundamental for Watson and Crick to propose the DNA double helix in the early 1950s. The first immunoassays also date from that decade, initially in the form of radioactive isotopes and later using fluorogenic reporters or enzyme signal amplification. The 1960s saw the development of gel electrophoresis to separate biomolecules based on physical/chemical differences, which led to the development of Southern blotting to separate DNA strands, Northern blotting for RNA, and Western blotting for proteins. Gel electrophoresis was also essential for Frederick Sanger and colleagues to develop the Sanger method or “dideoxy” chain-termination method to accurately sequence long stretches of DNA in 1977. In 1986, Applied Biosystems commercialized the first sequencing instrument based on the Sanger method (model ABI 370). A few years earlier, in 1983, the polymerase chain reaction for DNA amplification was invented, which was followed by real-time or quantitative PCR (qPCR) a decade later, using intercalating fluorescent dyes or specific labeled probes.5 The late 1980s also saw the rise of capillary electrophoresis for DNA separation, owing to faster times and a higher resolution, as the high surface to volume ratio enables heat to dissipate more efficiently, allowing higher voltages.6 Similar trends in the development of chromatography and flow injection analysis, to improve performance and reduce time through smaller scales, led to the concept of a total analysis system proposed by Manz to integrate and automate all analysis steps from sample preparation to detection.7 This soon led to an ever increasing interest in micro-scale devices and the physical properties and manipulation of fluids at this scale.
Here, we provide a glimpse of the historical developments of this era of micro-scale technological developments, since approximately 1990. We do this by overviewing key technological breakthroughs, in Fig. 1, and their respective evolution over time in a landscape of alternative, overlapping, or even synergistic technologies, in Fig. 2. We divide technologies into three main and loosely defined application areas: technologies for oligonucleotide (DNA/RNA) analysis, technologies for protein analysis, and technologies for single-cell analysis and microstructured cell culture, such as organs-on-a-chip. We see technological progress as occurring in waves of emergence, maturation, consolidation, and eventually replacement by new emergent technologies. When appropriate, we try to emphasize these transitions, additionally illustrated in Fig. 2. As the developments described here are nothing short of frenetic and vastly divergent, we recognize that we will inevitably overlook relevant topics, technologies, and historical facts. Where considered appropriate, we have tried to focus on those technologies that have been most successful at reaching commercial use and/or clinical impact.
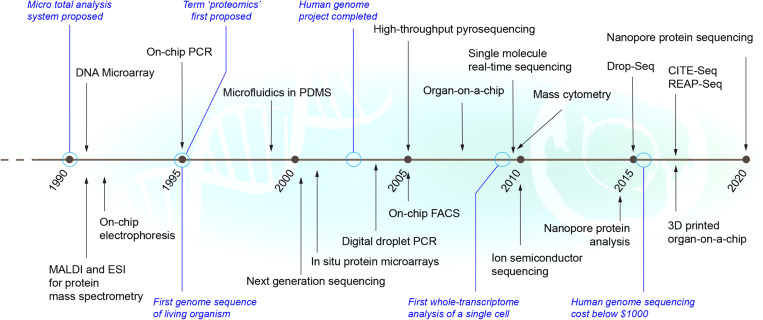
FIG. 1.
Timeline of key life sciences and medical micro-scale technologies and associated milestones. Black: new technologies with arrows pointing to approximate year of first publication and/or release. Blue: important associated events or milestones.
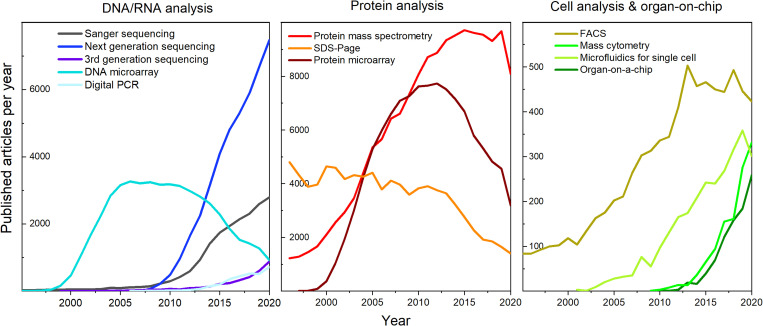
FIG. 2.
Trends in technological development: evolution of publications per year on selected technologies in three application areas: DNA/RNA analysis, protein analysis, and cell analysis comprising organ-on-a-chip devices.
A. Nucleic acid analysis: From microarrays to third-generation sequencing
In the early 1990s, advances in microfabrication techniques made it possible to integrate known macroscopic functionalities into microdevices, initially fabricated out of glass and silicon. Well-known examples were on-chip electrophoresis8 and on-chip PCR, soon followed by isothermal DNA amplification methods.9 The integration of the multiple manual steps of the standard methods (e.g., extraction, purification, PCR, and detection) in one device enabled significant gains in speed and reliability and reduced the amount of reagent used.10,11 Initial systems for PCR mostly relied on flow-through chips with serpentine channels that pass through three constant temperature zones or heaters necessary for thermal cycling.12 However, it soon became clear that diluting and partitioning the initial sample to isolate single template molecules provided many advantages [Fig. 3(a)]. The binary signal measurements improved quantitative accuracy, avoided artifacts due to impaired amplification, and improved the sensitivity of rare species, no longer competing for amplification reagents.13 Thus, digital PCR was born, initially employing microwells or microfluidic valves. This limited the number of individual partitions per sample, which also negatively affected the limit of detection, and the fabricated platform was costly to run compared to traditional qPCR. It was only in the late 2000s that developments in droplet microfluidics, including T-junctions, flow-focusing, or step emulsifications, made droplet digital PCR a simpler and more practical method,14 with each generated picoliter-sized microdroplet thermocycled to perform single-molecule digital PCR. Since then, the technology has seen an exponential increase in interest for technological and biological applications, reflected in the number of publications over the last ten years (Fig. 2). In Sec. IV, we further expand on how this technology has transitioned to commercial products.
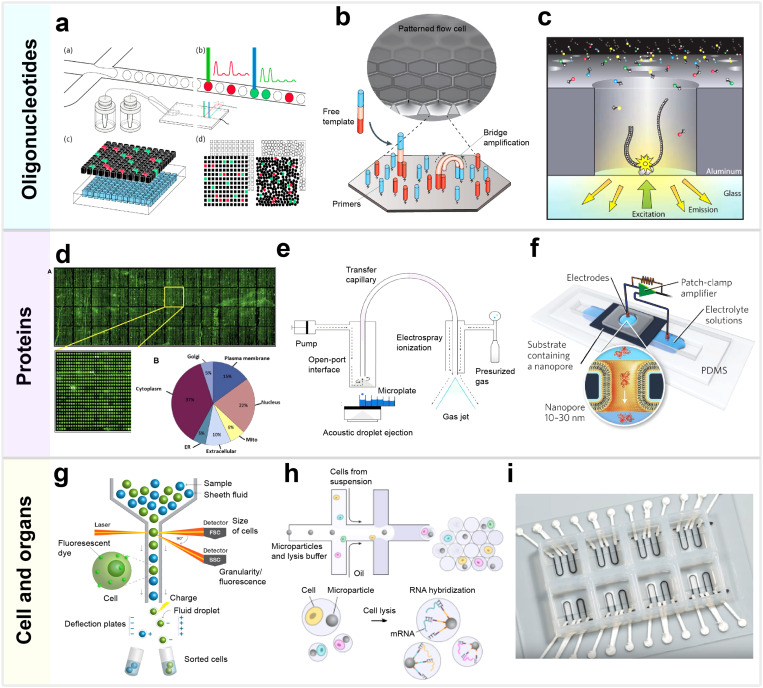
FIG. 3.
Selected technologies for the analysis of oligonucleotides, proteins, and cells: (a) Digital PCR, including droplet digital PCR, micro-well partitioning. and planar imaging of PCR products. Reproduced with permission from Liao and Huang, Micromachines 8(8), 1–7 (2017). Copyright 2017 MDPI. (b) Patterned flow cell molecular principle of next-generation sequencing with Illumina's dye sequencing method,. Adapted with permission from Goodwin et al., Nat. Rev. Genet. 17(6), 333–351 (2016). Copyright 2016 Springer Nature. (c) The zero-mode waveguide nanostructure of single-molecule real-time sequencing with nucleotides being incorporated into the DNA by the bottom-linked polymerase. Reproduced with permission from Eid et al., Science 323(5910), 133–138 (2009). Copyright 2009 The American Association for the Advancement of Science. (d) Protein microarray with 16 368 recombinant proteins and their cellular distribution. Reproduced with permission from Uzoma and Zhu, Genomics Proteomics Bioinforma. 11(1), 18–28 (2013). Copyright 2013 Elsevier. (e) Mass spectrometer with acoustic droplet ejection and electrospray ionization. Adapted with permission from Dirico et al., Med. Chem. Lett. 11, 1101–1110 (2020). Copyright 2020 American Chemical Society. (f) Device with a nanopore to measure resistive pulses from the translocation of individual proteins. Reproduced with permission from Yusko et al., Nat. Nanotechnol. 12(4), 360–367 (2017). Copyright 2017 Springer Nature. (g) The principle of fluorescence-activated cell sorting. (h) Initial steps of single-cell RNA-seq workflow with Drop-seq. Adapted with permission from Macosko et al., Cell 161(5), 1202–1214 (2015). Copyright 2015 Elsevier. (i) 3D-printed and multimaterial cardiac organ-on-a-chip device. Reproduced with permission from Lind et al., Nat. Mater. 16(3), 303–308 (2017). Copyright 2017 Springer Nature.
The 1990s also saw the development of DNA microarrays, consisting of spotted or in situ synthesized single stranded oligonucleotides on flat surfaces, allowing to hybridize and detect specific DNA sequences. This resulted from merging photolithography techniques and biochemistry, allowing to manipulate molecules on solid surfaces with light-directed spatially addressable chemical synthesis. The first high-density microarray was developed by Affymetrix in 1994. Progress in photolithography and surface chemistry led to an increasing density of synthesis sites, lower production costs, and an open source mindset for fabrication, resulting in a wave of constructed in-house devices by most major university and research laboratory in the early 2000s.15 Amplifying the DNA or RNA with PCR prior to microarray enabled measuring thousands of genes simultaneously. In 2005, Roche's CYP450 AmpliChip was the first microarray-based test approved by the FDA, soon followed by Agendia's Mammaprint.16 The enthusiasm in this decade can be seen in the sharp increase in microarray-related publications, as microarrays promised to revolutionize functional genomics and disease molecular diagnostics (Fig. 2).17 However, microarrays faced the competition of a parallel development in sequencing technologies. Eventually, some of the DNA microarrays’ inherent limitations, such as suboptimal quantification for low/highly expressed genes, a need for a priori knowledge of the sequence of interest, and cross-hybridization artifacts, made sequencing more attractive for many applications. Transcriptomics mostly transitioned to sequence-based methods (RNA-seq) in the last decade, resulting in a significant drop in DNA microarray use (Fig. 2). However, new and exciting applications also employ forms of microarrays, one example being spatially resolved transcriptomics, which can analyze the spatial and heterogeneous expression of RNA in tumor microenvironments by capturing RNA locally before sequencing.18
In contrast to microarray methods, sequence-based approaches directly determine the nucleic acid sequence of analyzed DNA (or cDNA) molecules. The first generation of sequencing methods is based on the “chain-termination” technique. It uses modified fluorescent nucleotides (ddNTPs) to stop the replication process, and the lengths of the accumulated sequences can be used to determine the position of each nucleotide through electrophoresis. Sanger sequencers allowed the simultaneous sequencing of hundreds of samples. By 1995, Craig Venter and colleagues had completed the first genome of a free-living organism, the bacterium Haemophilus influenzae, using Sanger sequencing. The same method drove the Human Genome Project in the 1990s and early 2000s, which in turn resulted in considerable progress in sequencing technologies. Switching from gel to capillary-based electrophoresis, increased throughput with parallel sampling, and 1 kb base pair generation per reaction have made Sanger sequencing considerably cheaper than it used to be. As a result, Sanger sequencing is still considered the gold standard for low–medium throughput sequencing and its overall use has continued increasing, although it has been replaced by next-generation sequencing for high-throughput applications (Fig. 2).
In the late 1990s, the groundwork for a new generation of high-throughput sequencing technologies was established with the invention of pyrosequencing19 and DNA colony sequencing.20 In the year 2000, Lynx Therapeutics launched Massive Parallel Signature Sequencing, and in 2004 the first commercial pyrosequencer was launched by 454 Life Sciences, which offered at a six times lower price than Sanger sequencing. These new instruments were classified as second-generation or next-generation sequencing (NGS). In pyrosequencing, nucleotide addition is detected by the emission of photons following a chain reaction initiated by pyrophosphate release. The sequencing instrument became possible by combining microfluidics, micro-well array technologies, and DNA-coated beads.21 Ion semiconductor sequencing is based on the same concept as pyrosequencing, but measures pH changes caused by hydrogen release, avoiding using light for measurements. Ion semiconductor sequencing was developed by Ion Torrent Systems Inc., and the first benchtop instrument was released in 2010. Another prominent technology is Illumina's dye sequencing, in which complementary oligonucleotides are bound to the bottom of nanowells in a microfluidic flow cell [Fig. 3(b)]. DNA fragments are first amplified by hybridizing to two oligonucleotides forming a bridge that is then PCR amplified. By repeating the process, a cluster of bound copies of the original DNA is formed locally. Next, a primer attaches to the generated forward strands and a polymerase adds fluorescently tagged dNTP in sequential rounds and reads. In 2012, Illumina launched the HiSeq 2500 instrument, able to sequence a whole human genome within 24 h.20 NGS offers massive throughput of up to billions of reads per run and has revolutionized genomics, transcriptomics,22 and epigenomics in the last decade, resulting in an exponential increase of NGS-based publications (Fig. 2), and is increasingly accelerating diagnosis, prognosis, and biomarker discovery, as discussed in Sec. III. Technological development also led to genome sequencing costs reducing from more than $10 million in 2005 to close to the long anticipated $1000 cost per genome in 201623 (by mid-2020, the estimated cost was $689). Importantly, the first step of any NGS protocol before actual sequencing consists of a library preparation (DNA fragments of defined length and NGS-compatible oligomer sequences on both ends), which is often still done manually. This has been identified as a possible bottleneck24 as NGS transitions from research purposes to clinical diagnostics, driving the development of new microfluidic solutions to tackle this problem [e.g., Fluidigm, Juno (2019) and Oxford Nanopore, VolTRAX (2019)].
Although NGS has revolutionized genomics and algorithm processing has significantly improved, genome assembly is still challenging as it is based on short reads (a few hundred bps). For example, NGS genome coverage is uneven and sometimes partially non-covered in guanine-cytosine poor or rich sequences (termed GC bias), and the mapping of repetitive elements is difficult. Thus, a third generation of sequencing technologies emerged, primarily to enable reading long single molecules. This has been driven by two main technologies: single-molecule real-time sequencing (SMRT) and nanopore sequences. SMRT uses millions of “tiny” wells, acting as zero-mode waveguides, in order to detect light emitted by a released fluorophore when a nucleotide incorporates into a single DNA molecule with the aid of a polymerase attached to the well bottom [Fig. 3(c)].25 This technology was first commercialized by Pacific Biosciences in 2011. In nanopore sequencing, a nanopore is inserted into electrical resisting membranes of synthetic polymer and an electric potential is applied across the membrane, creating a current through the nanopore. When a DNA molecule passes through the nanopore, the electric current is disrupted, with each nucleotide providing a characteristic electric signature. An enzyme unzips the double strand at the entrance of the nanopore and the measurement occurs in real-time and is suitable for entire strands of 100s of kbs. Nanopores can be obtained with the self-assembling of biological molecules, such as bacterial transmembrane channels inserted in lipid bilayers (e.g., mutated α-hemolysin and mycobacterial porin), or through lithography-based processes. The latter includes membranes of solid-state materials such as SiO2, SiN, HfO2, graphene, and MoS2, released from glass or Si substrates by etching and with pores opened with ion bombardment, focused electrode beam drilling, or electrochemical reactions.26 The concept of nanopore sequencing was first described in a publication in 1996,27 but only in 2005 did a nanopore achieve single-nucleobase discrimination in a DNA strand.28 Oxford Nanopore released its first benchtop instrument MinION to early users in 2014.29 Development is still ongoing as error rates are higher than for NGS, but this third generation is already favored for addressing important questions concerning large genomic alterations, such as structural variants, resulting in a very fast increase in its use in research (Fig. 2).
B. Protein analysis: Microarrays and mass spectroscopy
Although DNA microarrays and developments in sequencing promised to fully open the doors to understanding gene expression, the abundance of mRNA in a cell was soon discovered to poorly correlate with the actual abundance of the translated protein.30,31 Additionally, post-translational modifications (PTMs) and post-translational structural processing are revealed only by direct protein-level analysis.32 New technologies were thus necessary to analyze the hundreds of thousands of protein variants in a human cell.33 This task was hardly addressable with low-throughput and labor-intensive 2D gel electrophoresis (particularly SDS-PAGE), chromatography, and the slow Edman degradation technique.4
By directly adapting DNA microarray manufacturing and know-how, protein microarrays were developed, initially consisting of inkjet-printed antibody spots,34 complementary to a panel of target proteins to be detected simultaneously. This mimics the concept of Enzyme-linked Immunosorbent Assay (ELISA) immunoassays with detection through labeled reporters (e.g., secondary antibodies) in a highly parallelized way at the micro-scale [Fig. 3(d)]. Unfortunately, while nucleic acid arrays are extremely stable, proteins have varying stabilities and microarrays are thus very vulnerable to individual and uncontrolled protein degradation. Additionally, the large amounts of antibodies require specific cell culture, protein purification steps, and transfer to surface chemistries, with varying yields and protein integrity assurance.31 These limitations drove the development of alternative technologies, in particular, to obtain protein microarrays directly from the translation of DNA on the solid surface, followed by protein immobilized in situ:35 Because they are DNA-based until translated, the protein in situ array (PISA) first reported in 2001 by He and Taussig36 and the nucleic acid programmable protein array (NAPPA) from 200435 can be stored long term and made into protein microarrays on demand. The obtained proteins do not reach the complexity of capture antibodies but can be used to study protein–protein interactions or antibody biomarkers. An additional disadvantage of microarrays, cross-contamination issues due to the diffusion of species, also led to the development of microarrays with tens of thousands of nanowells.37 Despite these developments, the use of protein microarrays has steadily decreased in the last ten years to the detriment of mass spectroscopy (Fig. 2). This is due to limitations still inherent to protein microarrays: limited availability of antibodies for protein detection, the need for antibody pairs to bind to different epitopes, and challenges in obtaining uniform spots from inkjet printing, among others.34 Nonetheless, applications such as detecting low abundance proteins remain advantageous, as capture is not significantly affected by non-target proteins in this case.
Protein analysis through mass spectrometry is another successful technique in which proteins in ionized form and in gas phase are injected and accelerated in an electric or magnetic field in a time-of-flight unit [Fig. 3(e)]. The results of the mass-to-charge ratio measured are provided in the form of mass spectra or fingerprints that can be compared with databases. Although mass spectrometers were available decades earlier, producing protein ions was a challenge before 1991, when two methods compatible with the amount of protein extracted from cell lysis and gel electrophoresis were first commercialized:4 electrospray ionization (ESI)38 for high mass proteins and matrix-assisted laser desorption/ionization (MALDI),39 which are faster and less affected by contaminants and additives. These developments enabled a throughput in the analysis of proteins unforeseeable until then and gave rise to the concept of proteomics, the analysis of all proteins produced by a genome, in 1995.4 Large improvements in sample preparation techniques and the performance of mass spectrometers resulted in a dramatic increase in the acceptance of protein identification in the 2000s (Fig. 2). Today, protein mass spectrometry is the gold standard for protein sequencing, with a limit of detection of 0.1 fmol and a dynamic range of 104–105, and is complemented with a strong peptide database and dedicated software for identification and quantitation.40 Despite this, some applications are still challenging or out of the scope of mass spectrometry. Ion suppression can mask the presence of low abundance proteins and mass spectrometry does not offer single-molecule resolution. In fact, the limit of detection is not compatible with low abundance protein identification in single-cell analysis, as millions of cells are still required.40 Even more ambitious tasks, such as single-molecule resolution, seem to be out of the scope of mass spectrometry.
In the last years, new technological concepts have emerged for protein sequencing, analogous to DNA sequencing. One of them, tunnel current analysis, measures tunneling currents in angstrom–nanometer gaps between electrodes. First measurements of short peptides were demonstrated in 2014, with two electrodes coated with a layer of recognition molecules.41 Other technologies leverage nanopores analogous to those used for DNA sequencing to deduce the shape and the fingerprint of single proteins through modulations of the blockade current as the protein translocates through the pore42 [Fig. 3(f)]. A true sequencing technology using ionic current detection is challenging, as it requires the discernment of subtle molecular differences among all 20 amino acids, something that Ouldali et al. first demonstrated to be feasible in 2020.43 Finally, electrical charge measurements of single proteins (and DNA strands) at the sub-elementary level were achieved by Ruggeri et al.44 This is accomplished by transiently confining the molecules in electrostatic fluidic traps of sub-μm scale etched in silicon oxide. Such a system may be complementary to sequencing methods, providing information on protein folded and disordered states and non-uniform charge distribution, potentially allowing to probe 3D structures and monitor structural changes in real time. Overall, these recent developments show a trend of increasingly promising technologies that will very likely revolutionize proteomics in the next few decades.
C. Cell analysis: Flow cytometry, single cells, and organs-on-a-chip
Technological progress, particularly in microtechnologies, has made it possible to develop structures and fluidic functions at the scale of single or few cells, helping to ask novel, interesting, and pertinent biological questions. The analysis of single cells provides important information that conventional bulk methods miss with population-averaged results.51 Examples are cell–cell interaction dynamics, cell migration studies, single-cell genomics, and cell signaling.
Flow cytometry was one of the first technologies able to perform single-cell measurements,52 initially commercialized in the early 1970s. A true precursor of microfluidic methods, flow cytometry focuses cells in a liquid using laminar sheath flows that surround the cell-containing stream (hydrodynamic focusing), resulting in a stream of one cell at a time that is read by a laser beam [Fig. 3(g)]. The forward and side scatter of the laser provide relevant information about size and structural properties. With the development of monoclonal antibodies and fluorescent dyes, it became possible to target surface proteins, and by 1990, flow cytometers could measure seven fluorescence signals simultaneously. Today, some cytometers allow up to 32 parameters for cell counting, sorting, and biomarker detection. The fluorescence-activated cell sorter (FACS) has been particularly successful. It is a type of flow cytometer that breaks the stream of cells into droplets after the fluorescence measurement. These droplets contain one or no cell and become charged according to their prior fluorescence measurement, allowing downstream separation through electrostatic deflection. FACS allows studies of cell phenotype, proliferation, cycle, viability, signaling, and intracellular cytokine secretion, among others. Conventional FACS is not suitable for small samples with less than a few hundred thousand cells. However, the concept was miniaturized first for particles and bacteria by Fu et al. in 199953 and for the more fragile mammalian cells by Genoptix, Inc., in 2005.54
An important drawback of FACS is that it is limited in the number of measurable parameters due to the spectral overlap of fluorophore signals. To tackle this issue, mass cytometry was conceived, combining flow cytometry with mass spectrometry, and first commercialized in 2009 (CyTOF, DVS Sciences). It is a destructive method in which antibody–isotope conjugates bind to the cells, which in turn are vaporized and sent to the time-of-flight unit. This produces large amounts of data with 40+ multiple parameters per individual cell, although throughput is still lower than for FACS (∼1000 cells/s) and cells cannot be retrieved for downstream analysis. Applications include phenotype characterization, intracellular cytokine determination, and intracellular signaling states, among many others.55
FACS point measurements per cell are less well suited for studies involving single-cell interactions with their surrounding environment, such as secretion dynamics, cell–cell interactions, affinity studies, or cell mechanics. The last two decades have seen a formidable rate of innovation on microfluidic devices for cell biology, including the automated isolation of cells for individual growth studies, forced cell deformation through microchannels, and micropillar arrays to study cell–surface interactions.56 Microfluidics provides precise control of geometries, surface chemistry, and flows, creating on demand spatiotemporal microenvironments for single-cell studies. One of the first such commercial platforms was Fluidigm C1 launched in 2012. It was able to isolate single cells in individual reaction chambers for staining, studying viability, surface markers, and reporter genes before lysing for sequencing. Additionally, the development of droplet microfluidics combined with high-throughput sequencing has resulted in systems such as Drop-Seq, inDrop, and Chromium 10× for single-cell transcriptome analysis.57 Here, cells are compartmentalized in nanoliter-sized droplets together with beads containing barcoded primers able to capture mRNA and identify the cell after lysis and pooling [Fig. 3(h)]. Reverse transcription PCR is followed by sequencing to obtain a gene expression map of single cells and understand tissues at the level of individual cells. These techniques are able to capture >10 000 transcripts from 1000 to 4000 genes.57 More recently, a simultaneous analysis of both mRNA and protein expression has become possible with methods such as CITE-Seq58 and REAP-Seq,59 first proposed in 2017. These leverage droplet-based RNA-Seq but make additional use of DNA barcode labeled antibodies specific for surface proteins of the cell. As compared to the first single-cell transcriptome in 2009 using NGS,22 today these methods allow massive parallel throughput of up to 80 000 cells per run in less than 20 min, although the whole process from library preparation to bioinformatic analysis can still take a couple of days.
Beyond single-cell analysis, microfluidic platforms for cell manipulation and culture have seen an exponential development, particularly since the introduction of porous polydimethylsiloxane (PDMS) devices.60 The development of on-chip cell-cultures led to a transition from 2D to 3D microenvironments to better mimic physiological conditions, resulting in a converge of microfabrication and tissue engineering know-how in the form of organs-on-a-chip. Such devices recreate tissue–tissue responses, organ-level functions, and biological tissue barriers, compatible with human cell lines, thus potentially becoming an alternative to animal models. Since the late 2000s, models for multiple organs haven been shown, including the lung, heart, kidney, arteries, brain, and bone, among others. Nonetheless, a true and relevant simulation or organ function needs to mimic a very large number of physiological conditions, including adequate geometrical structuring, shear forces, molecular gradients, and a sufficiently complex arrangement of cellular patterns.61 A step toward systemic whole body physiological process has been the combination of several organ functions in the form of multi-organ-on-a-chip, first proposed in 2010 through combining liver and intestine slices.62 Finally, the rapid development of biological 3D printing in the last few years has also made it possible to fully print an organ-on-a-chip, a cardiac microphysiological device [Fig. 3(i)],50 thus completely avoiding multi-step lithographic processes. Organs-on-a-chip is still a nascent technology, and their true impact on physiological modeling, drug discovery, and toxicity research will likely be seen in the years to come.
III. MICROTECHNOLOGY-ENABLED BREAKTHROUGHS IN FUNDAMENTAL AND CLINICAL RESEARCH
The technological breakthroughs reviewed in Sec. II set the groundwork for a revolution in our fundamental understanding of the life sciences from a molecular, cellular, and physiological perspective. Additionally, the resulting amassed knowledge has already impacted the clinical practice, although the true impact is likely only starting now as new technologies and methods prove their validity and utility for medical care. The enormous transformations in fundamental research and the clinic resulting from microtechnologies for the life sciences cannot be thoroughly reviewed in any single article and much less in a short perspective such as this one. In the following, we briefly illustrate part of the impact that four selected technologies have had in enabling discovery and applications.
Next-generation sequencing has redefined the resolution of molecular analysis, enabling previously unimaginable breakthroughs in cancer genomics, rare inherited disorders, prenatal testing, metagenomics, and microbiome analysis, to name a few. Through genome-wide association studies (GWAS), markers in the genomes of many people are scanned to find genetic variations associated with disease, from cancer to osteoporosis;63 and human traits, from personality64 to male-pattern baldness.65 The number of newly identified disease-associated genes has grown exponentially since the emergence of NGS,66 including rare inherited disorders, which previously represented a diagnostic odyssey for affected families. Additionally, NGS has uncovered the relevance of mosaic and de novo mutations and the wide phenotypic spectrum of most genes. This is laying the groundwork for an incoming era of personalized medicine and customized treatment strategies. Furthermore, NGS has shed light on cancer genome heterogeneity,67 as tumors are composed of cell subpopulations with genetic alterations such as single-nucleotide variants, insertions/deletions, or copy number alterations. NGS is helping to underpin the biological mechanisms of cancer clonal evolution, allowing evolutionary frameworks to better understand cancer progression and failure of therapy.68 Thus, NGS is increasingly used to guide the selection of targeted therapies, taking into account personal tumoral genomes, particularly types of advanced cancer, paving the way for precision oncology. The number of alterations with a corresponding targeted therapy has been growing steadily, and recent clinical trials to guide cancer therapy for advanced malignancies are showing that NGS testing can have an impact on the response rate and the survival of patients.69 As clinical evaluation is still underway, routine upfront NGS testing is still uncommon in clinical practice. However, a clear case of direct clinical impact has occurred in the area of non-invasive prenatal testing for Trisomy 21, 18, or 13 from simple blood samples. By sequencing cell-free DNA in the maternal plasma, it is now possible to determine the number of chromosomes and thus detect aneuploidies, and even inherited diseases. This has, in many cases, replaced invasive diagnostic techniques such as amniocentesis that incur a risk (albeit small) of miscarriage.
Flow cytometry has also been key to better understanding cancer, enabling the study of circulating tumor cells in blood, cell isolation to generate tumor models, and the study of tumor microenvironments. The isolation and identification of cancer stem-like cells from surgically removed tumors, likely responsible for developed drug resistance, is and will continue to be important in identifying targets for treatment.70 Similarly, flow cytometry is used to analyze 3D tumor cultures (tumor organoids/spheroids), showing, for example, how allogeneic T and NK cells infiltrate spheroids and kill tumor cells through immune-mediated cell apoptosis.71 Additionally, flow cytometry has been essential to evaluate new cancer immunotherapies, such as chimeric antigen receptor (CAR) T-cell therapies. CAR T-cells, genetically engineered to recognize cancer cells, are transforming the way cancer tumors are treated, the outcome being influenced by the proliferation and persistence of these cells in blood, measured with flow cytometry.72 Besides cancer, flow cytometry has largely impacted areas such as immunology and the development of vaccines. Indeed, understanding immunogenicity and vaccine efficacy requires monitoring the frequency of immune cell populations and their differentiation and activation status upon exposure to specific antigens.
Microfluidics-based single-cell technologies have made it possible to investigate immune function, therapeutic resistance, or neurological disease mechanisms, to name a few applications. Wide ranged examples include studies of time-dependent cytokine secretion and transcription factor activity from single macrophage cells,73 cell lineage tracking with transcriptional signature measurements for the study of multigenerational development at the single-cell level,74 or multidrug resistance studies in adult acute myeloid leukemia, allowing assays to distinguish multidrug resistance variability among individual leukemic blasts.75 The long sought after objective of understanding the extremely complex human nervous system has started to be tackled. For instance, single-cell RNA-seq has revealed that oligodendrocytes could be classified into classes based on their molecular signatures and microglia can be distinguished from very similar perivascular macrophages.76
Finally, organs-on-a-chip are emerging as a crucial tool in areas such as the discovery and validation of biomarkers, the study of pharmacokinetic (PK) analysis, drug toxicity screening, and tumor modeling. Cell lines and animal models are inadequate to predict pathophysiology of human disease and patient-specific sensitivities, as fundamental interspecies differences affect the estimated predictions for safety margins, toxicity, and efficacy. Qualitative predictions of drug toxicity responses are being generated with fluid transfer between organ-on-a-chip devices. Recently, it has been shown that such systems predict PK parameters for orally administered nicotine and intravenously injected cisplatin by using multi-organ chips such as gut, liver, and kidney, in some cases matching reported patient data.77
Additionally, organ-on-a-chip technologies are being used to study cardiac and liver toxicity causes of post-approval drug withdrawal and drug failure due to lack of efficiency or poorly understood action mechanisms.78 Other applications can be as surprising as an in vitro model for smoke-induced injury, consisting of a microfluidic lung model that inhales/exhales smoke from cigarettes, showing similar physiological and inflammatory reactions to clinical studies and enabling the discovery of several associated biomarkers.79 Finally, brain-on-a-chip platforms are becoming essential in helping to tackle complex disease mechanisms. Modeling Alzheimer's disease has shown that amyloid-b accumulations in cortical neurons resulted in anomalies in neurotransmitter signaling pathways,80 while an in vitro model of epilepsy could reproduce aspects of spontaneous firing activity depending on the expression of specific receptors.81 Organ-on-a-chip technologies are in the midst of enormous developments that will likely lead to vast research and clinical impact. Current challenges include mimicking complex 3D microenvironments and multicellular tissue constructs and leveraging stem cell technologies.82
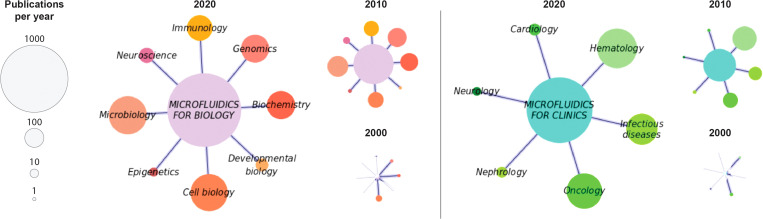
Figure 4 illustrates the application areas of microfluidic technologies in 2020, 2010, and 2000 in the form of networks of research applications in biology and in clinical studies based on PubMed queries (summing relevant terms associated with each application). The size of the nodes represents the number of publications that year. As seen, application publications involving microfluidics have grown extraordinarily in the last 20 years. Strong areas of fundamental research are genomics, biochemistry, cell biology, microbiology, and immunology. Current clinical studies of microfluidics are dominated by hematology, infectious diseases, and oncology.
FIG. 4.
Focus on microfluidic applications: prominent application areas addressed by microfluidics in the life sciences (left) and in clinical research (right). Data are presented for 2020, 2010, and 2000. Node size indicates number of publications that year according to a PubMed search.
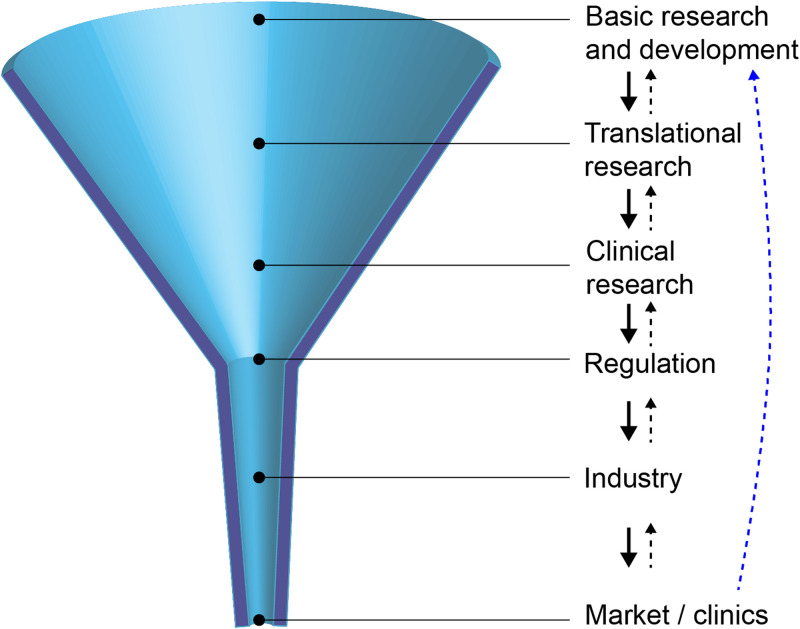
IV. MICROTECHNOLOGIES REACHING CLINICAL DIAGNOSTICS: THE FUNNEL GETS NARROWER
As seen so far, microtechnologies easily intersect with the life sciences to develop applications, which sometimes make their way to clinical diagnostics. However, the path to reach clinical use is a multi-step and long process that most technologies fail to complete successfully. The general objective of the translational process is to transform a method or a tool into a routine diagnostic platform that is safe and commercially available. In Fig. 5, we illustrate this process, in a simplified manner, as a funnel with some basic and idealized steps from basic research to market/clinics. The narrowing shape of the funnel illustrates both the progressive reduction in technologies successfully completing the steps on the way toward market and the gradual shift from the many possibilities offered by initial research concepts to mature well-defined products addressing specific applications. We try to highlight here how each step can represent a hurdle in the path toward the clinics, all of them entailing multiple challenges and obstacles. This makes the translation process a research, entrepreneurial, clinical, legal, and marketing quest.
FIG. 5.
The translational bottleneck: laboratory-developed technologies must go through technical, regulatory, and commercial transitions before reaching the clinics. A thorough understanding and feedback from the following steps (black dashed line) and the end clinical application (blue dashed line) is critical for success.
Initial basic research, often performed within an academic setting, generates novel methods on a conceptual level, typically providing a sufficiently high degree of performance to confirm that the principle is scientifically solid. The “end product” of such a process is generally a scientific publication. Although preliminary clinical trials sometimes happen, most proposed new technologies end at this stage. In fact, research laboratories are often intrinsically biased toward producing new ideas, concepts, and technologies, even if the end-user or clinical need are not precisely defined yet.83 In many cases, the new technologies solve issues at the laboratory/researcher level, and this is particularly the case in fundamental biological research, but more often than not the original method is not directly scalable in the industry or efficient in clinical practice. Well-known examples of barriers to scalability in microfluidics are the use of polydimethylsiloxane (PMDS), convenient for low-scale laboratory prototyping but unsuitable for automated mass production84 and liquid degassing or clogging,85 often deemed acceptable for research devices but constituting a hurdle for further translation.
Translational research bridges the gap between basic science and an early-stage prototype aimed either at further clinical research or directly to market for non-medical settings. We focus here on the former case, with an example of the latter being a research tool with no clinical use. Even when basic research shows strong potential, the maturity phase still constitutes a major bottleneck. Some of the reasons are as follows: (1) A deep understanding of the clinical environment is needed at the early stage of technological development, as besides providing a clear performance improvement, new technology needs to be user-friendly, compatible with clinical workflows, of reasonable cost, and apt for automation.83 Early-stage technologies that promise superior performance commonly fail to result in products with equal or superior operation efficiency compared to established technologies. (2) Adequate management of the technological maturity, including regular evaluation of the technology status, risk assessment, and decisions on funding. Funding agencies increasingly nudge researchers to estimate and consider technological maturity at an early stage. As an example, the European Commission included technology readiness levels in its Horizon 2020 program. (3) An increasingly saturated patent landscape.83 For example, thousands of patents now include the word microfluidic, many of them being very generic, making it difficult, for example, for start-ups, to operate in associated domains without potentially incurring patent infringements.83 (4) Successful post-invention funding through angel investors, bootstrapping, corporations, venture capital firms, and/or subsidies. (5) The absence of industry standards84 that help improve communication between customers and suppliers, streamline manufacturing processes, and fulfill regulatory requirements. (6) An entrepreneurial action with suitable collaborators, e.g., dedicated to the diagnostics industry and able to manage reimbursement, government compliance, regulations, or medical affairs. (7) High upfront costs for the mass production of disposable devices through replication processes.
With the resulting prototype, clinical research studies are undertaken to assess performance evaluation and support the intended clinical application. Performance evaluation reports must demonstrate scientific validity and both analytical and clinical performance. Our focus here are in vitro diagnostics (IVD), loosely defined as tests performed outside the human body and thus covering most of the technologies and applications seen so far. For IVD, clinical trials are generally much simpler to conduct than for drug or medical devices, as clinical performance studies are generally observational, and the results obtained are not used for patient management or treatment decisions. Nonetheless, appropriate study design must avoid selection and verification biases and claims must be context-based, e.g., to diagnose a target condition, to screen a certain population, to obtain a risk assessment, to perform prognosis, to monitor changes in a patient's condition, or to serve as companion diagnostics. The clinical study must also be aligned with regulatory requirements. For example, the FDA requires clinical trials to be performed at three or more clinical sites.
The regulatory control phase aims to ensure the safety and effectiveness of the medical devices used for patient diagnosis. While commercial technologies for fundamental research face less restrictions and supervision, the clinical regulatory process can be highly time consuming and costly, including submission fees and the funding of preclinical and clinical trials. This can put small laboratories and companies at a disadvantage compared to large corporations with more resources, thereby being detrimental to clinical innovation. In the United States, the regulation of IVD falls under two categories: (1) FDA-cleared or approved devices, which can undergo a 510(k) submission to obtain clearance, typically the case for IVD, or a more stringent and rigorous approval process following the submission of a premarket approval. The FDA categorizes diagnostic tests based on the level of complexity: waived complexity (e.g., pregnancy test), moderate complexity (e.g., when use of a microscope is necessary), and high complexity (e.g., NGS). (2) Laboratory-developed tests (LDTs), offered for clinical use by the single laboratory in which they were developed, manufactured, and validated. LDTs are regulated by the Clinical Laboratory Improvement Amendment program of the Centers for Medicare & Medicaid Services (CMS). LDTs are widely used by molecular diagnostics laboratories, as they often prefer to develop the tests themselves rather than pay high prices to large companies with FDA-approved tests. For example, most NGS-based genetic tests are still performed as LDTs. Nonetheless, the FDA has expressed concerns about the overall quality of LDTs, while LDT regulations are becoming costly and insurance companies in some cases favor FDA-approved tests. In the European Union, the CE mark for in vitro diagnostic devices is managed by the In Vitro Diagnostic Regulation (IVDR) regulation since 2017.
A new technology can perform well, have a strong clinical validation, and receive regulatory approval, and yet be unsuccessful in the market. Particularly in clinical settings, the new technology must present a strong advantage in overall performance and/or cost compared to existing well-established technologies. Incremental advances are generally insufficient, and the introduction of disruptive technologies often only succeeds when they find a market that does not currently exist.86 Projects often fail or result in no market-fit product due to not integrating perceptions/insights of end-users early enough.87 If the IVD is expensive, it may find a higher rate of success if associated with clinical decisions of high cost (e.g., administering second-line therapy) or a health risk (e.g., surgery). Additionally, new guidelines and agreed practices need to be developed for any new technology, but particularly for complex tools, such as NGS. In this case, the creation of network infrastructures can help link initiatives, develop collaborative projects, and contribute to nation-level databases, taking into consideration privacy, data protection, and associated legal and ethical aspects. Closely related to this is the adequate training of clinicians, laboratory, and information technology staff.
As an illustration of this multi-step process from concept to clinical use, we consider the example of droplet digital PCR (ddPCR), described in Sec. II, diving into some of the turns of events that led to its current commercialization. Even though associated developments had been presented before, it was in 1999 that Vogelstein and Kinzler coined the term digital PCR, demonstrating selective rare mutation detection in colorectal cancer.13 However, this technology was still expensive, limited by the number of partitions, and faced the strong competition of real-time PCR.14 It was only a few years later that its association with droplet microfluidics was able to address most of these limitations. New start-ups were formed, notably RainDance Technologies in 2004 and QuantaLife in 2008. Originally relying on a single U.S. patent, Quantalife aimed to rapidly increase its intellectual property (IP) portfolio and launched the first commercial ddPCR instrument in 2010, the Droplet Digital system, soon rebranded as QX100 as the company was acquired by BioRad the following year. RainDance launched its commercial platform, the RainDrop, in 2012 but was eventually also acquired by BioRad in 2017. As a result of these company acquisitions and the resulting securing of much of the IP portfolio regarding ddPCR and associated technologies, BioRad established itself as the dominant player for ddPCR in the market. An example of local pre-FDA clinical use of this technology was the development of a test for EGFR and KRAS genes by the Dana-Farber/Brigham and Women's Cancer Center in Boston, followed by performing a clinical trial involving 180 patients of non-small cell lung cancer.88 The gains in turnaround time compared with tissue biopsies and the observed high predictive value led to both institutions starting to offer this test to eligible patients in routine clinics. BioRad's second-generation instrument, the QX200, received the CE IVD mark in 2016. In 2019, their QXDx AutoDG system was the first ddPCR system to be FDA-cleared, as a test for monitoring chronic myeloid leukemia treatment response. This was almost two decades after the technological concept was first presented. In 2020, with the onset of the COVID-19 pandemic, both instruments received emergency use authorization by the FDA for SARS-CoV-2 PCR testing, a rapid authorization procedure that allows unapproved products to be used in cases of public health emergencies.
As seen, the path from basic research to the clinics is often non-linear and winding, involving successful scientific demonstration, clinical validation, right timing, associated technological landscape, business initiative, IP strength, market competition, company acquisitions, and regulatory restrictions. As a result, the process typically lasts years or even decades. Therefore, although some technologies have already percolated to the clinics, we are hopeful that the true impact to the medical field promised by the technological developments illustrated here is yet and soon to come.
V. CONCLUDING REMARKS AND THE CASE OF COVID-19
Three decades of micro-scale technological developments for the life sciences had passed as the COVID-19 pandemic hit most of the world in early 2020. And yet, at a first glance, it might seem that the response to the pandemic was essentially led by long-established technologies: real-time PCR and antibody tests, such as ELISA and lateral flow immunoassays, all necessary to establish current and past infection. A more attentive look, however, reveals that the new virus would have been confronted very differently 30 years ago. NGS was essential to rapidly sequence and identify the virus causing the pandemic within a few weeks of the first cases being reported.3 It also played a crucial role in closely monitoring viral spread, determineing sources of infection and routes of transmission, detect the emergence of new mutant variants (e.g., B.1.1.7 and 501.V2), screening targets for new therapies, and helping in vaccine development. The FDA also granted Emergency Use Authorization for a NGS-based diagnostic, the first such authorization for NGS. Cartridge-based PCR benchtop instruments also became essential for rapid diagnosis, taking minutes instead of hours or days, examples being Abbot's ID Now or Cepheid's GeneXpert. Flow cytometry has been used to identify and characterize T-cells involved in the immune system response to the virus and to better understand their targets,89 while mass cytometry has revealed how the virus leads to immunosuppression and dysfunction of immune mechanisms.90 Even lung-on-a-chip models are being used to better understand immune cell response, alveolar barrier injury, and inflammation.91 Additionally, the field responded to the pandemic with multiple new proposed microtechnologies to increase testing capacity and performance, from innovative lateral flow sensors, through viral electrochemical sensors to magnetic bead capture in labs-on-a-chip.92 Furthermore, initiatives such as the XPRIZE Pandemic Response Challenge have been launched to incentivize collaboration and competition to accelerate measures to fight the pandemic.
Despite this, the COVID-19 pandemic shows how routine molecular diagnostics still relies largely on conventional immunoassays93 and PCR. Section IV, illustrating the long path to clinical use, is an attempt to explain why the many developments of the last few decades have not fully catalyzed into the promised revolution. However, a decrease in sequencing costs and large population studies will likely have important implications for precision medicine in the future. Some countries have already started ambitious implementation programs for NGS. These include an external quality assessment of somatic variants and allele frequencies, baseline performance by characterizing tumor cell lines, an extension to pre-analytical conditions and clinical interpretation, and the creation of a national intention to establish network data programs to monitor the quality of testing in clinical practice.94
Additionally, point-of-care diagnostics is increasingly reaching non-laboratory and remote resource-limited settings, reducing the need for instruments and operators of current ELISA and PCR-based diagnosis.95 Often, these are diagnostic platforms integrated with smartphones, leveraging widely available high-resolution cameras, processing power, and internet networks.
In these and other aspects, microtechnologies will continue to make their way into fundamental research and particularly clinical diagnostics. The dynamic nature of the medical market forces medical device producers to constantly invest in research and development. However, as microtechnologies often need to adapt to specific needs in fundamental research, non-commercial solutions or creative custom adaptations will continue being sought after.
We thus hope that this Perspective reflects the essential nature of technological progress to enable progress in biological research, and the key role played by technologists in the life sciences, from basic research all the way to industrial translation, a role that we believe is not yet being given enough attention.
ACKNOWLEDGMENTS
We acknowledge funding by European Research Council-PoC CellProbe Grant (No. 842790) and the Swiss National Science Foundation—Spark Program (No. CRSK-2_190877). We thank Dr. A. Kashyap, Dr. A. Fomitcheva Khartchenko, Dr. M. Serra (Harvard Medical School), and L. Rudin for comments on the manuscript. We would also like to thank Dr. E. Delamarche and Dr. H. Riel for continuous support.
DATA AVAILABILITY
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1.Hurst J. H., “Pioneering geneticist Mary-Claire King receives the 2014 Lasker–Koshland special achievement award in medical science,” J. Clin. Invest. 124(10), 4148–4151 (2014). 10.1172/JCI78507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loos R. J. F., “15 years of genome-wide association studies and no signs of slowing down,” Nat. Commun. 11(1), 10–12 (2020). 10.1038/s41467-020-19653-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G. F., and Tan W., “A novel coronavirus from patients with pneumonia in China, 2019,” N. Engl. J. Med. 382(8), 727–733 (2020). 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henzel W. J., Watanabe C., and Stults J. T., “Protein identification: The origins of peptide mass fingerprinting,” J. Am. Soc. Mass Spectrom. 14(9), 931–942 (2003). 10.1016/S1044-0305(03)00214-9 [DOI] [PubMed] [Google Scholar]
- 5.Heid C. A., Stevens J., Livak K. J., and Williams P. M., “Real time quantitative PCR,” Exp. Mol. Med. 6, 986–994 (1996). 10.1016/b978-012372185-3/50024-9 [DOI] [PubMed] [Google Scholar]
- 6.Durney B. C., Crihfield C. L., and Holland L. A., “Capillary electrophoresis applied to DNA: Determining and harnessing sequence and structure to advance bioanalyses (2009-2014),” Anal. Bioanal. Chem. 407(23), 6923–6938 (2017). 10.1007/s00216-015-8703-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manz A., Graber N., and Widmer H. M., “Miniaturized total chemical analysis systems: A novel concept for chemical sensing,” Sens. Actuators B Chem. B1, 244–248 (1990). 10.1016/0925-4005(90)80209-I [DOI] [Google Scholar]
- 8.Harrison D. J., Manz A., Lüdi H., Widmer H. M., and Fan Z., “Capillary electrophoresis and sample injection systems integrated on a planar glass chip,” Anal. Chem. 64(17), 1926–1932 (1992). 10.1021/ac00041a030 [DOI] [Google Scholar]
- 9.Ahmad F. and Hashsham S. A., “Miniaturized nucleic acid amplification systems for rapid and point-of-care diagnostics: A review,” Anal. Chim. Acta 733, 1–15 (2012). 10.1016/j.aca.2012.04.031 [DOI] [PubMed] [Google Scholar]
- 10.Woolley A. T., Hadley D., Landre P., deMello A. J., Mathies R. A., and Northrup M. A., “Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device,” Anal. Chem. 68(23), 4081–4086 (1996). 10.1021/ac960718q [DOI] [PubMed] [Google Scholar]
- 11.Giordano B. C., Ferrance J., Swedberg S., Hühmer A. F. R., and Landers J. P., “Polymerase chain reaction in polymeric microchips: DNA amplification in less than 240 s,” Anal. Biochem. 291(1), 124–132 (2001). 10.1006/abio.2000.4974 [DOI] [PubMed] [Google Scholar]
- 12.Kopp M. U., Mello A. J., and Manz A., “Chemical amplification: Continuous-flow PCR on a chip,” Science 280(5366), 1046–1048 (1998). 10.1126/science.280.5366.1046 [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein B. and Kinzler K. W., “Digital PCR,” Proc. Natl. Acad. Sci. U.S.A. 96(16), 9236–9241 (1999). 10.1073/pnas.96.16.9236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morley A. A., “Digital PCR: A brief history,” Biomol. Detect. Quantif. 1(1), 1–2 (2014). 10.1016/j.bdq.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lenoir T. and Giannella E., “The emergence and diffusion of DNA microarray technology,” J. Biomed. Discov. Collab. 1(1), 1–39 (2006). 10.1186/1747-5333-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X., Quigg R., Zhou J., Gu W., Rao P., and Reed E., “Clinical utility of microarrays: Current status, existing challenges and future outlook,” Curr. Genomics 9(7), 466–474 (2008). 10.2174/138920208786241199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiltgen M. and Tilz G. P., “DNA microarray analysis: Principles and clinical impact,” Hematology 12(4), 271–287 (2007). 10.1080/10245330701283967 [DOI] [PubMed] [Google Scholar]
- 18.Ståhl P. L., Salmén F., Vickovic S., Lundmark A., Navarro J. F., Magnusson J., Giacomello S., Asp M., Westholm J. O., Huss M., Mollbrink A., Linnarsson S., Codeluppi S., Borg Å., Pontén F., Costea P. I., Sahlén P., Mulder J., Bergmann O., Lundeberg J., and Frisén J., “Visualization and analysis of gene expression in tissue sections by spatial transcriptomics,” Science 353(6294), 78–82 (2016). 10.1126/science.aaf2403 [DOI] [PubMed] [Google Scholar]
- 19.Ronaghi M., Karamohamed S., Pettersson B., Uhlén M., and Nyrén P., “Real-time DNA sequencing using detection of pyrophosphate release,” Anal. Biochem. 242(1), 84–89 (1996). 10.1006/abio.1996.0432 [DOI] [PubMed] [Google Scholar]
- 20.Barba M., Czosnek H., and Hadidi A., “Historical perspective, development and applications of next-generation sequencing in plant virology,” Viruses 6(1), 106–136 (2013). 10.3390/v6010106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margulies M., Egholm M., Altman W. E., Attiya S., Bader J. S., Bemben L. A., Berka J., Braverman M. S., Chen Y. J., Chen Z., Dewell S. B., Du L., Fierro J. M., Gomes X. V., Godwin B. C., He W., Helgesen S., Ho C. H., Irzyk G. P., Jando S. C., Alenquer M. L. I., Jarvie T. P., Jirage K. B., Kim J. B., Knight J. R., Lanza J. R., Leamon J. H., Lefkowitz S. M., Lei M., Li J., Lohman K. L., Lu H., Makhijani V. B., McDade K. E., McKenna M. P., Myers E. W., Nickerson E., Nobile J. R., Plant R., Puc B. P., Ronan M. T., Roth G. T., Sarkis G. J., Simons J. F., Simpson J. W., Srinivasan M., Tartaro K. R., Tomasz A., Vogt K. A., Volkmer G. A., Wang S. H., Wang Y., Weiner M. P., Yu P., Begley R. F., and Rothberg J. M., “Genome sequencing in microfabricated high-density picolitre reactors,” Nature 437(7057), 376–380 (2005). 10.1038/nature03959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B. B., Siddiqui A., Lao K., and Surani M. A., “MRNA-Seq whole-transcriptome analysis of a single cell,” Nat. Methods 6(5), 377–382 (2009). 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- 23.Wetterstrand K., see www.genome.gov/sequencingcostsdata for “DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP).”
- 24.Hess J. F., Kohl T. A., Kotrová M., Rönsch K., Paprotka T., Mohr V., Hutzenlaub T., Brüggemann M., Zengerle R., Niemann S., and Paust N., “Library preparation for next generation sequencing: A review of automation strategies,” Biotechnol. Adv. 41, 107537 (2020). 10.1016/j.biotechadv.2020.107537 [DOI] [PubMed] [Google Scholar]
- 25.Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., Bibillo A., Bjornson K., Chaudhuri B., Christians F., Cicero R., Clark S., Dalal R., DeWinter A., Dixon J., Foquet M., Gaertner A., Hardenbol P., Heiner C., Hester K., Holden D., Kearns G., Kong X., Kuse R., Lacroix Y., Lin S., Lundquist P., Ma C., Marks P., Maxham M., Murphy D., Park I., Pham T., Phillips M., Roy J., Sebra R., Shen G., Sorenson J., Tomaney A., Travers K., Trulson M., Vieceli J., Wegener J., Wu D., Yang A., Zaccarin D., Zhao P., Zhong F., Korlach J., and Turner S., “Real-time DNA sequencing from single polymerase molecules,” Science 323(5910), 133–138 (2009). 10.1126/science.1162986 [DOI] [PubMed] [Google Scholar]
- 26.Xue L., Yamazaki H., Ren R., Wanunu M., Ivanov A. P., and Edel J. B., “Solid-state nanopore sensors,” Nat. Rev. Mater. 5(12), 931–951 (2020). 10.1038/s41578-020-0229-6 [DOI] [Google Scholar]
- 27.Kasianowicz J. J., Brandin E., Branton D., and Deamer D. W., “Characterization of individual polynucleotide molecules using a membrane channel,” Proc. Natl. Acad. Sci. U.S.A. 93(24), 13770–13773 (1996). 10.1073/pnas.93.24.13770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashkenasy N., Sánchez-Quesada J., Bayley H., and Ghadiri M. R., “Recognizing a single base in an individual DNA strand: A step toward DNA sequencing in nanopores,” Angew. Chem. Int. Ed. 44(9), 1401–1404 (2005). 10.1002/anie.200462114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jain M., Olsen H. E., Paten B., and Akeson M., “The Oxford nanopore MinION: Delivery of nanopore sequencing to the genomics community,” Genome Biol. 17(1), 239 (2016). 10.1186/s13059-016-1103-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gygi S. P., Rochon Y., Franza B. R., and Aebersold R., “Correlation between protein and MRNA abundance in yeast,” Mol. Cell. Biol. 19(3), 1720–1730 (1999). 10.1128/MCB.19.3.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell P., “A perspective on protein microarrays,” Nat. Biotechnol. 20, 225–229 (2002). 10.1038/nbt0302-225 [DOI] [PubMed] [Google Scholar]
- 32.Timp W. and Timp G., “Beyond mass spectrometry, the next step in proteomics,” Sci. Adv. 6(2), 1–17 (2020). 10.1126/sciadv.aax8978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aebersold R., Agar J. N., Amster I. J., Baker M. S., Bertozzi C. R., Boja E. S., Costello C. E., Cravatt B. F., Fenselau C., Garcia B. A., Ge Y., Gunawardena J., Hendrickson R. C., Hergenrother P. J., Huber C. G., Ivanov A. R., Jensen O. N., Jewett M. C., Kelleher N. L., Kiessling L. L., Krogan N. J., Larsen M. R., Loo J. A., Ogorzalek Loo R. R., Lundberg E., MacCoss M. J., Mallick P., Mootha V. K., Mrksich M., Muir T. W., Patrie S. M., Pesavento J. J., Pitteri S. J., Rodriguez H., Saghatelian A., Sandoval W., Schlüter H., Sechi S., Slavoff S. A., Smith L. M., Snyder M. P., Thomas P. M., Uhlén M., Van Eyk J. E., Vidal M., Walt D. R., White F. M., Williams E. R., Wohlschlager T., Wysocki V. H., Yates N. A., Young N. L., and Zhang B., “How many human proteoforms are there?” Nat. Chem. Biol. 14(3), 206–214 (2018). 10.1038/nchembio.2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereiro I., Cors J. F., Pané S., Nelson B. J., and Kaigala G. V., “Underpinning transport phenomena for the patterning of biomolecules,” Chem. Soc. Rev. 48(5), 1236–1254 (2019). 10.1039/C8CS00852C [DOI] [PubMed] [Google Scholar]
- 35.Ramachandran N., Hainsworth E., Bhullar B., Eisenstein S., Rosen B., Lau A. Y., Walter J. C., and LaBaer J., “Self-assembling protein microarrays,” Science 305(5680), 86–90 (2004). 10.1126/science.1097639 [DOI] [PubMed] [Google Scholar]
- 36.He M. and Taussig M. J., “Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method),” Nucleic Acids Res. 29(15), e73–e73 (2001). 10.1093/nar/29.15.e73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takulapalli B. R., Qiu J., Magee D. M., Kahn P., Brunner A., Barker K., Means S., Miersch S., Bian X., Mendoza A., Festa F., Syal K., Park J. G., Labaer J., and Wiktor P., “High density diffusion-free nanowell arrays,” J. Proteome Res. 11(8), 4382–4391 (2012). 10.1021/pr300467q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenn J. B., Mann M., Meng C. K., Wong S. F., and Whitehouse C. M., “Electrospray ionization for mass spectrometry of large biomolecules,” Science 246(4926), 64–71 (1989). 10.1126/science.2675315 [DOI] [PubMed] [Google Scholar]
- 39.Karas M. and Hillenkamp F., “Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons,” Anal. Chem. 60(20), 2299–2301 (1988). 10.1021/ac00171a028 [DOI] [PubMed] [Google Scholar]
- 40.Restrepo-pérez L., Joo C., and Dekker C., “Paving the way to single-molecule protein sequencing,” Nat. Nanotechnol. 13, 786–796 (2018). 10.1038/s41565-018-0236-6 [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y., Ashcroft B., Zhang P., Liu H., Sen S., Song W., Im J., Gyarfas B., Manna S., Biswas S., Borges C., and Lindsay S., “Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling,” Nat. Nanotechnol. 9, 466–473 (2014). 10.1038/nnano.2014.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yusko E. C., Bruhn B. R., Eggenberger O. M., Houghtaling J., Rollings R. C., Walsh N. C., Nandivada S., Pindrus M., Hall A. R., Sept D., Li J., Kalonia D. S., and Mayer M., “Real-time shape approximation and fingerprinting of single proteins using a nanopore,” Nat. Nanotechnol. 12(4), 360–367 (2017). 10.1038/nnano.2016.267 [DOI] [PubMed] [Google Scholar]
- 43.Ouldali H., Sarthak K., Ensslen T., Piguet F., Manivet P., Pelta J., Behrends J. C., Aksimentiev A., and Oukhaled A., “Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore,” Nat. Biotechnol. 38, 176–181 (2020). 10.1038/s41587-019-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruggeri F., Zosel F., Mutter N., Rozycka M., Wojtas M., Ozyhar A., Schuler B., and Krishnan M., “Single-molecule electrometry,” Nat. Nanotechnol. 12(5), 488–495 (2017). 10.1038/nnano.2017.26 [DOI] [PubMed] [Google Scholar]
- 45.Liao P. and Huang Y., “Digital PCR: Endless frontier of ‘divide and conquer,’” Micromachines 8(8), 1–7 (2017). 10.3390/mi8080231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goodwin S., McPherson J. D., and McCombie W. R., “Coming of age: Ten years of next-generation sequencing technologies,” Nat. Rev. Genet. 17(6), 333–351 (2016). 10.1038/nrg.2016.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzoma I. and Zhu H., “Interactome mapping: Using protein microarray technology to reconstruct diverse protein networks,” Genomics Proteomics Bioinf. 11(1), 18–28 (2013). 10.1016/j.gpb.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dirico K. J., Hua W., Liu C., Tucker J. W., Ratnayake A. S., Flanagan M. E., Troutman M. D., Noe M. C., and Zhang H., “Ultra-high-throughput acoustic droplet ejection-open port interface-mass spectrometry for parallel medicinal chemistry,” Med. Chem. Lett. 11, 1101–1110 (2020). 10.1021/acsmedchemlett.0c00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A. R., Kamitaki N., Martersteck E. M., Trombetta J. J., Weitz D. A., Sanes J. R., Shalek A. K., Regev A., and McCarroll S. A., “Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets,” Cell 161(5), 1202–1214 (2015). 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lind J. U., Busbee T. A., Valentine A. D., Pasqualini F. S., Yuan H., Yadid M., Park S. J., Kotikian A., Nesmith A. P., Campbell P. H., Vlassak J. J., Lewis J. A., and Parker K. K., “Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing,” Nat. Mater. 16(3), 303–308 (2017). 10.1038/nmat4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altschuler S. J. and Wu L. F., “Cellular heterogeneity: Do differences make a difference?” Cell 141(4), 559–563 (2010). 10.1016/j.cell.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonner W. A., Hulett H. R., Sweet R. G., and Herzenberg L. A., “Fluorescence activated cell sorting,” Rev. Sci. Instrum. 43(3), 404–409 (1972). 10.1063/1.1685647 [DOI] [PubMed] [Google Scholar]
- 53.Fu A. Y., Spence C., Scherer A., Arnold F. H., and Quake S. R., “A microfabricated fluorescence-activated cell sorter,” Nat. Biotechnol. 17(11), 1109–1111 (1999). 10.1038/15095 [DOI] [PubMed] [Google Scholar]
- 54.Wang M. M., Tu E., Raymond D. E., Yang J. M., Zhang H., Hagen N., Dees B., Mercer E. M., Forster A. H., Kariv I., Marchand P. J., and Butler W. F., “Microfluidic sorting of mammalian cells by optical force switching,” Nat. Biotechnol. 23, 83–87 (2005). 10.1038/nbt1050 [DOI] [PubMed] [Google Scholar]
- 55.Zhang T., Warden A. R., Li Y., and Ding X., “Progress and applications of mass cytometry in sketching immune landscapes,” Clin. Transl. Med. 10(6), 1–25 (2020). 10.1002/ctm2.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dahl J. B., Lin J.-M. G., Muller S. J., and Kumar S., “Microfluidic strategies for understanding the mechanics of cells and cell-mimetic systems,” Annu. Rev. Chem. Biomol. Eng. 6, 293–317 (2015). 10.1146/annurev-chembioeng-061114-123407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X., Li T., Liu F., Chen Y., Yao J., Li Z., Huang Y., and Wang J., “Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-Seq systems,” Mol. Cell 73(1), 130–142 (2019). 10.1016/j.molcel.2018.10.020 [DOI] [PubMed] [Google Scholar]
- 58.Stoeckius M., Hafemeister C., Stephenson W., Houck-loomis B., Chattopadhyay P. K., Swerdlow H., Satija R., and Smibert P., “Simultaneous epitope and transcriptome measurement in single cells,” Nat. Methods 14(9), 865–868 (2017). 10.1038/nmeth.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peterson V. M., Zhang K. X., Kumar N., Wong J., Li L., Wilson C., Moore R., Mcclanahan T. K., Sadekova S., and Klappenbach J. A., “Multiplexed quantification of proteins and transcripts in single cells,” Nat. Biotechnol. 35(10), 936–939 (2017). 10.1038/nbt.3973 [DOI] [PubMed] [Google Scholar]
- 60.Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., “Rapid prototyping of microfluidic systems in poly(dimethylsiloxane),” Anal. Chem. 70(23), 4974–4984 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- 61.Wu Q., Liu J., Wang X., Feng L., Wu J., Zhu X., and Wen W., “Organ-on-a-chip : Recent breakthroughs and future prospects,” Biomed. Eng. Online 19, 1–19 (2020). 10.1186/s12938-020-0752-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Midwoud P. M., Merema M. T., Verpoorte E., and Groothuis G. M. M., “A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices,” Lab Chip 10(20), 2278–2286 (2010). 10.1039/c0lc00043d [DOI] [PubMed] [Google Scholar]
- 63.Al-Barghouthi B. M. and Farber C. R., “Dissecting the genetics of osteoporosis using systems approaches,” Trends Genet. 35(1), 55–67 (2019). 10.1016/j.tig.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo M.-T., Hinds D. A., Tung J. Y., Franz C., Fan C.-C., Wang Y., Smeland O. B., Schork A., Holland D., Kauppi K., Sanyal N., Escott-Price V., Smith D. J., O’Donovan M., Stefansson H., Bjornsdottir G., Thorgeirsson T. E., Stefansson K., McEvoy L. K., Dale A. M., Andreassen O. A., and Chen C.-H., “Genome-wide analyses for personality traits identify six genomic loci and show correlations with psychiatric disorders,” Nat. Genet. 49(1), 152–156 (2017). 10.1038/ng.3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pirastu N., Joshi P. K., De Vries P. S., Cornelis M. C., McKeigue P. M., Keum N., Franceschini N., Colombo M., Giovannucci E. L., Spiliopoulou A., Franke L., North K. E., Kraft P., Morrison A. C., Esko T., and Wilson J. F., “GWAS for male-pattern baldness identifies 71 susceptibility loci explaining 38% of the risk,” Nat. Commun. 8(1), 1–9 (2017). 10.1038/s41467-017-01490-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandez-Marmiesse A., Gouveia S., and Couce M. L., “NGS technologies as a turning point in rare disease research, diagnosis and treatment,” Curr. Med. Chem. 25, 404–432 (2018). 10.2174/0929867324666170718101946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang J., Späth S. S., Marjani S. L., Zhang W., and Pan X., “Characterization of cancer genomic heterogeneity by next-generation sequencing advances precision medicine in cancer treatment,” Precis. Clin. Med. 1(1), 29–48 (2018). 10.1093/pcmedi/pby007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Turajilic S., Sottoriva A., Graham T., and Swanton C., “Resolving genetic heterogeneity in cancer,” Nat. Rev. Genet. 20, 404–416 (2019). 10.1038/s41576-019-0114-6 [DOI] [PubMed] [Google Scholar]
- 69.Colomer R., Mondejar R., Romero-Laorden N., Alfranca A., Sanchez-Madrid F., and Quintela-Fandino M., “When should we order a next generation sequencing test in a patient with cancer?” EClinicalMedicine 25, 1–9 (2020). 10.1016/j.eclinm.2020.100487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Masciale V., Grisendi G., Banchelli F., D’Amico R., Maiorana A., Sighinolfi P., Stefani A., Morandi U., Dominici M., and Aramini B., “Isolation and identification of cancer stem-like cells in adenocarcinoma and squamous cell carcinoma of the lung: A pilot study,” Front. Oncol. 9, 1–12 (2019). 10.3389/fonc.2019.01394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Courau T., Bonnereau J., Chicoteau J., Bottois H., Remark R., Miranda L. A., Toubert A., Blery M., Aparicio T., Allez M., and Bourhis L. L., “Cocultures of human colorectal tumor spheroids with immune cells reveal the therapeutic potential of MICA/B and NKG2A targeting for cancer treatment,” J. Immunother. Cancer 9, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Demaret J., Varlet P., Trauet J., Beauvais D., Grossemy A., Yakoub-agha F. H. I., and Labalette M., “Monitoring CAR T-cells using flow cytometry,” Cytom. Part B Clin. Cytom. 100, 1–7 (2020). 10.1002/cyto.b.21941 [DOI] [PubMed] [Google Scholar]
- 73.Junkin M., Kaestli A. J., Cheng Z., Jordi C., Albayrak C., Hoffmann A., and Tay S., “High-content quantification of single-cell immune dynamics,” Cell Rep. 15(2), 411–422 (2016). 10.1016/j.celrep.2016.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kimmerling R. J., Lee Szeto G., Li J. W., Genshaft A. S., Kazer S. W., Payer K. R., De Riba Borrajo J., Blainey P. C., Irvine D. J., Shalek A. K., and Manalis S. R., “A microfluidic platform enabling single-cell RNA-Seq of multigenerational lineages,” Nat. Commun. 7, 1–7 (2016). 10.1038/ncomms10220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Khamenehfar A., Gandhi M. K., Chen Y., Hogge D. E., and Li P. C. H., “Dielectrophoretic microfluidic chip enables single-cell measurements for multidrug resistance in heterogeneous acute myeloid leukemia patient samples,” Anal. Chem. 88(11), 5680–5688 (2016). 10.1021/acs.analchem.5b04446 [DOI] [PubMed] [Google Scholar]
- 76.Zeisel A., Muñoz-Manchado A. B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., Rolny C., Castelo-Branco G., Hjerling-Leffler J., and Linnarsson S., “Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-Seq,” Science 347(6226), 1138–1142 (2015). 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- 77.Herland A., Maoz B. M., Das D., Somayaji M. R., Prantil-Baun R., Novak R., Cronce M., Huffstater T., Jeanty S. S. F., Ingram M., Chalkiadaki A., Benson Chou D., Marquez S., Delahanty A., Jalili-Firoozinezhad S., Milton Y., Sontheimer-Phelps A., Swenor B., Levy O., Parker K. K., Przekwas A., and Ingber D. E., “Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips,” Nat. Biomed. Eng. 4(4), 421–436 (2020). 10.1038/s41551-019-0498-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang B., Korolj A., Fook B., Lai L., and Radisic M., “Advances in organ-on-a-chip engineering,” Nat. Rev. Mater. 3, 257–278 (2018). 10.1038/s41578-018-0034-7 [DOI] [Google Scholar]
- 79.Benam K. H., Novak R., Nawroth J., Hirano-Kobayashi M., Ferrante T. C., Choe Y., Prantil-Baun R., Weaver J. C., Bahinski A., Parker K. K., and Ingber D. E., “Matched-comparative modeling of normal and diseased human airway responses using a microengineered breathing lung chip,” Cell Syst. 3(5), 456–466 (2016). 10.1016/j.cels.2016.10.003 [DOI] [PubMed] [Google Scholar]
- 80.Deleglise B., Magnifico S., Duplus E., Vaur P., Soubeyre V., Belle M., Vignes M., Viovy J. L., Jacotot E., Peyrin J. M., and Brugg B., “Β-Amyloid induces a dying-back process and remote trans-synaptic alterations in a microfluidic-based reconstructed neuronal network,” Acta Neuropathol. Commun. 2(1), 1–9 (2014). 10.1186/s40478-014-0145-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gullo F., Manfredi I., Lecchi M., Casari G., Wanke E., and Becchetti A., “Multi-electrode array study of neuronal cultures expressing nicotinic (Β2-V287L subunits, linked to autosomal dominant nocturnal frontal lobe epilepsy. An in vitro model of spontaneous epilepsy,” Front. Neural Circuits 8, 1–12 (2014). 10.3389/fncir.2014.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frimat J.-P. and Luttge R., “The need for physiological micro-nanofluidic systems of the brain,” Front. Bioeng. Biotechnol. 7, 1–11 (2019). 10.3389/fbioe.2019.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shields C. W., Ohiri K. A., Szott L. M., and López G. P., “Translating microfluidics: Cell separation technologies and their barriers to commercialization,” Cytom. Part B Clin. Cytom. 92(2), 115–125 (2017). 10.1002/cyto.b.21388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reyes D. R., van Heeren H., Guha S., Herbertson L., Tzannis A. P., Ducrée J., Bissig H., and Becker H., “Accelerating innovation and commercialization through standardization of microfluidic-based medical devices,” Lab Chip 21(1), 9–21 (2021). 10.1039/D0LC00963F [DOI] [PubMed] [Google Scholar]
- 85.Pereiro I., Fomitcheva Khartchenko A., Petrini L., and Kaigala G. V., “Nip the bubble in the bud: A guide to avoid gas nucleation in microfluidics,” Lab Chip 19(14), 2296–2314 (2019). 10.1039/C9LC00211A [DOI] [PubMed] [Google Scholar]
- 86.Christensen C. M., The Innovator’s Dilemma When New Technologies Cause Great Firms to Fail (Harvard Business School Press, Boston, MA, 1997). [Google Scholar]
- 87.Béjean M. and Siqueira E., “Organizing Medtech innovation with concept maturity levels,” in Proceedings of the 12th International Joint Conference on Biomedical Engineering Systems and Technologies (SCITEPRESS, 2019), pp. 621–626. 10.5220/0007697006210626 [DOI] [Google Scholar]
- 88.Sacher A. G., Paweletz C., Dahlberg S. E., Alden R. S., O’Connell A., Feeney N., Mach S. L., Jänne P. A., and Oxnard G. R., “Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer,” JAMA Oncol. 2(8), 1014–1022 (2016). 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., and Sette A., “Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals,” Cell 181(7), 1489–1501 (2020). 10.1016/j.cell.2020.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang W., Su B., Pang L., Qiao L., Feng Y., Ouyang Y., Guo X., Shi H., Wei F., Su X., Yin J., Jin R., and Chen D., “High-dimensional immune profiling by mass cytometry revealed immunosuppression and dysfunction of immunity in COVID-19 patients,” Cell. Mol. Immunol. 17(6), 650–652 (2020). 10.1038/s41423-020-0447-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang M., Wang P., Luo R., Wang Y., Li Z., Guo Y., Yao Y., Li M., Tao T., Chen W., Han J., Liu H., Cui K., Zhang X., Zheng Y., and Qin J., “Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system,” Adv. Sci. 8(3), 2002928 (2020). 10.1002/advs.202002928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tymm C., Zhou J., Tadimety A., Burklund A., and Zhang J. X. J., “Scalable COVID-19 detection enabled by lab-on-chip biosensors,” Cell. Mol. Bioeng. 13(4), 313–329 (2020). 10.1007/s12195-020-00642-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pereiro I., Fomitcheva-Khartchenko A., and Kaigala G. V., “Shake it or shrink it: Mass transport and kinetics in surface bioassays using agitation and microfluidics,” Anal. Chem. 92(15), 10187–10195 (2020). 10.1021/acs.analchem.0c01625 [DOI] [PubMed] [Google Scholar]
- 94.Valckenborgh E. V., Hébrant A., Antoniou A., Hoof W. V., Bussel J. V., Pauwels P., Salgado R., Doren W. V., Waeytens A., and Bulcke M. V. D., “Roadbook for the implementation of next-generation sequencing in clinical practice in oncology and hemato-oncology in Belgium,” Arch. Public Heal. 76(49), 1–7 (2018). 10.1186/s13690-018-0295-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shrivastava S., Trung T. Q., and Lee N. E., “Recent progress, challenges, and prospects of fully integrated mobile and wearable point-of-care testing systems for self-testing,” Chem. Soc. Rev. 49, 1812–1866 (2020). 10.1039/C9CS00319C [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.