Abstract
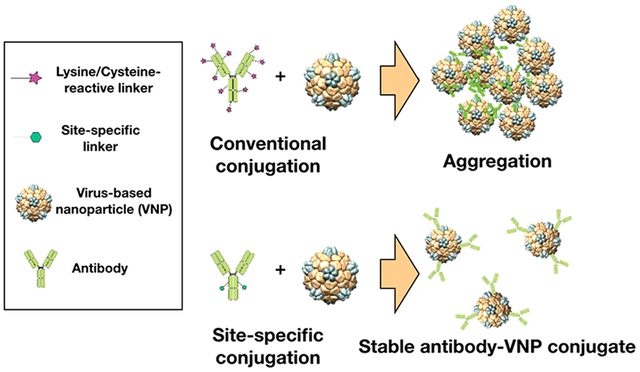
Amine/thiol-reactive chemistries are commonly used to conjugate antibodies to pharmaceuticals or nanoparticles. Yet, these conjugation strategies often result in unfavorable outcomes such as heterogeneous antibody display with hindered biological activity or aggregation due to multivalent interactions of the antibody and nanoparticles. Here, we report the application of a site-specific and enzymatically driven antibody conjugation strategy to functionalize virus-based nanoparticles (VNPs). Specifically, an azide-handle was introduced into the Fc region of a set of immunoglobulins using a two-step enzymatic reaction: (1) cleavage of N-linked glycan in the Fc region by a glycosidase and (2) conjugation of a chemically reactive linker (containing an azide functional handle) using a microbial transglutaminase. Conjugation of the azide-functional antibodies to several VNPs was achieved by making use of strain-promoted azide−alkyne cycloaddition. We report the conjugation of three immunoglobulin (IgG) isotypes (human IgG from sera, anti-CD47 Rat IgG2a, κ, and Trastuzumab recombinant humanized IgG1, κ) to the plant virus cowpea mosaic virus (CPMV) and the lysine mutant of tobacco mosaic virus (TMVlys) as well as bacteriophage Qβ. Site-specific conjugation resulted in stable and functional antibody-VNP conjugates. In stark contrast, the use of heterobifunctional linkers targeting thiols and amines on the antibodies and VNPs, respectively, led to aggregation due to nonspecific and multivalent coupling between the antibodies and VNPs. We demonstrate that antibody-VNP conjugates were functional, and Trastuzumab-displaying VNPs targeted HER2-positive SKOV-3 human ovarian cancer cells. This bioconjugation strategy adds to the portfolio of methods that can be used for designing functional antibody-VNP conjugates.
Graphical Abstract:
INTRODUCTION
Protein and antibody-nanoparticle conjugates have emerged in the pharmaceutical industry to develop safer and more effective drugs for a variety of applications including drug delivery, imaging, and immunotherapy.1 Small peptide and protein-based pharmaceuticals often suffer from fast renal clearance and protease degradation, which can be circumvented by conjugating them to nanoparticles to increase their bioavailability.2 Peptides, proteins, and antibodies have also been employed to provide nanoparticles with tissue specificity and promote the accumulation of their therapeutic payloads to diseased sites (e.g., cancer) while minimizing off-target cell toxicity.3,4
Both full length and fragment antibodies are able to bind to their target antigens with high specificity and affinity, making them ideal moieties to design targeted nanoparticles.5,6 Upon binding, antibodies may neutralize antigens or induce a pharmaceutically relevant biological response through the activation of their agonist/antagonist receptor. For example, Trastuzumab is a monoclonal antibody (mAb) approved by the FDA for the treatment of breast, gastric, and esophageal cancers by blocking human epidermal growth factor receptor (HER2) signaling and promoting apoptosis.7 The conjugation of Trastuzumab to a variety of nanoparticles, including iron oxide nanoparticles,8 gold nanoparticles,9 liposomes,10 poly- (lactic-co-glycolic) acid (PLGA) nanoparticles,11 mammalian adenovirus,12 and the plant virus potato virus X,13 has been reported.
Contemporary methods of antibody conjugation rely on the modification of side chains of amino acids such as lysine and cysteine.14 However, the relative abundance of these residues on the light and heavy chains of an antibody can result in its heterogeneous presentation and hindered biological activity. To compensate, high number of antibodies must be displayed on the surface of nanoparticles, which may lead to steric inhibition of antigen recognition as well as nanoparticle aggregation. To overcome these challenges, several alternative chemical conjugation techniques have been developed to target C-terminal or N-terminal groups, or to promote the S-alkylation of native disulfide bonds; these methods are reviewed elsewhere.15,16 Enzymatically driven conjugation techniques are another class of chemistries that can be used to promote site-directed conjugation yielding protein-drug conjugates.17–20 For example, a two-step enzymatic reaction has previously been developed to conjugate linkers at a privileged location (Gln295) on deglycosylated antibodies.20 Here, in a first reaction step, glycans on the heavy chains of antibodies are removed using peptide:N-glycosidase F (PNGase F); specifically, PNGase F cleaves the N-linked oligosaccharides between the innermost N-acetylglucosamine (GlcNAc) and asparagine (Asn297), rendering glutamine Gln295 enzymatically accessible. In a second step, microbial transglutaminase (MTGase) is used to catalyze the reaction between the deglycosylated γ-carboxyamide group of the antibody’s Gln295 residue and an amino group, for example, the side chain of lysines or an amine of a linker molecule. This two-step site-specific and enzymatically driven reaction has been used for conjugation of antibodies with fluorophores20 and antineoplastic drugs.18,19 Also, MTGase has been successfully applied to conjugate, for example, poly ethylene glycol (PEG) to various proteins with exposed Gln residues including apomyoglobin, α-lactalbumin, human growth hormone, and interleukin-2.17
Here we set out to apply this versatile bioconjugate chemistry to produce functional virus-based nanoparticles. Virus-based nanoparticles (VNPs) have been investigated for drug delivery, imaging, immunotherapy, biosensing, biocatalysis, among other applications targeting human health and agriculture as well as environmental applications.21,22 VNPs from plants and bacteriophages are noninfectious to humans and can be manufactured with high reproducibility and degree of monodispersity. Because we often observe aggregation of VNPs upon conjugating antibodies using contemporary strategies such as heterobifunctional linkers targeting amines/thiols, we set out to adapt the site-specific and enzymatically driven chemistry to yield functional and stable antibody-VNP conjugates. As a proof of concept, we used three VNPs as testbed: two plant viruses, namely the 30 nm-sized icosahedral cowpea mosaic virus (CPMV) and the 300 × 18 nm rod-shaped tobacco mosaic virus (TMVlys, a lysine added mutant23), and one bacteriophage, namely the virus-like particles (VLPs) from the 30 nm-sized icosahedral Qβ phage (VNPs are viral nanoparticles that contain their genome; VLPs are virus-like particles that lack their genome). We report successful conjugation of different antibody isotypes, including human IgG from sera (hIgG), anti-CD47 Rat IgG2a, κ (αCD47) investigated in macrophage-mediated oncolytic immunotherapy,24 and the FDA approved Trastuzumab recombinant humanized IgG1, κ.7
RESULTS AND DISCUSSION
Despite their popularity due to the ease of use in biomolecular conjugation, the application of amine/thiol reactive chemistries for antibody conjugation to VNPs can result in nanoparticle aggregation (Scheme 1A and Figure 1). Here hIgG isolated from sera was conjugated to TMVlys and Qβ by making use of two heterobifunctional linkers, maleimide-PEG8-NHS ester (SM(PEG)8) and PEGlayted N-succinimidyl S-acetylthioacetate (SAT(PEG)4) conjugated on VNPs and hIgG, respectively. In brief, the SAT(PEG)4 linker was first incubated with hIgG and the excess of unreacted linkers was removed by using desalting columns. The NHS ester group in SAT(PEG)4 linker reacts with primary amines on lysine residues exposed on the antibody and introduces a S-acetylthioacetate (SAT) group as shown in Scheme 1B. The SAT group prevents the sulfur atom from oxidation and was easily removed by treatment with hydroxylamine to expose sulfhydryl group (−SH) needed for conjugation. VNPs were modified with SM(PEG)8, resulting in the introduction of a maleimide functional group conjugated to solvent-exposed lysine side chains of the VNP’s surface. To conjugate the antibodies to the VNPs, thiol-reactive nanoparticles were incubated with the SAT-modified hIgG with deprotected and exposed −SH group. The reaction was allowed to proceed overnight. The conjugation resulted in precipitation of the nanoparticles and antibody-VNP conjugates (Figure 1A). The aggregates are visible as white pellets and aggregation was apparent within 30 min of the reaction. After overnight reaction, aggregates were briefly centrifuged at 6000 × g for 2 min, and the supernatant and pellet were analyzed by UV–vis. UV–vis measurement indicated that there were no VNPs in the supernatant; therefore, suspension of the pellet were made and analyzed by SDS-NuPAGE gel electrophoresis. SDS-NuPAGE gel electrophoresis revealed multiple high molecular weight bands (~150 kDa) in addition to the VNP coat proteins and free light and heavy chain of the antibodies in the hIgG-conjugated TMVlys samples (highlighted in Figure 1B). It should be noted that while these are denaturing gels, a reducing agent was not added which explains a mix of free light and heavy chain, but with the majority as full length antibody. Also, for the Qβ sample, dimers and multimers of the coat protein are apparent. The appearance of multiple high molecular weight bands may indicate that multiple coat proteins were conjugated to a single antibody chain or vice versa. In the case of conjugation of hIgG to Qβ, the cross-linking was severe, and the aggregates could not be resolved on the gel, that is, the protein aggregate remained in the gel pocket and showed no electrophoretic mobility (Figure 1B). Immunoglobulins such as IgG antibodies display multiple lysines and cysteines, all of which may react with the heterobifunctional linkers; such reaction may unselectively cross-link the antibodies and/or result in multiple linkers displayed per antibody. This initial aggregation and multivalent linker display is then likely to foster nanoparticle aggregation; we are targeting lysine side chains on TMVlys and Qβ; TMVlys and Qβ display 2130 and 720 lysine side chains on their surface available for conjugation, respectively.23,24
Scheme 1. Fabrication of Antibody-Conjugated VNP1.
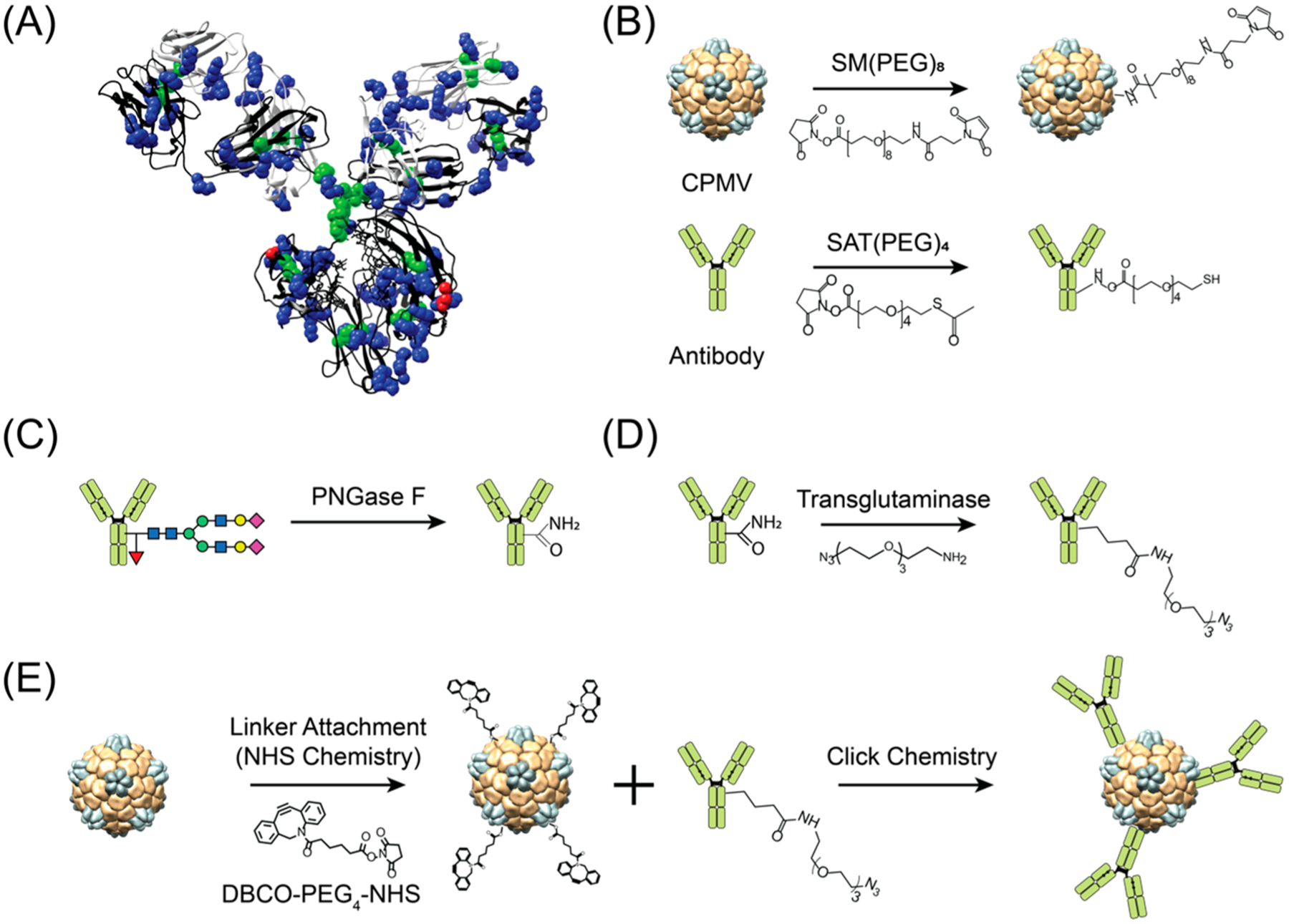
1(A) Structure of human IgG1 highlighting abundance of lysine (blue) and cysteine (green) and the location of Gln295 (red). (B) Lysine and cysteine targeted conjugation yields to heterogeneous antibody presentation. (C) Deglycosylation of Asn297 on heavy chains to make the adjacent Gln295 accessible, (D) site-specific attachment of a linker with azide (N3) group for conjugation via transglutaminase reaction, and (E) conjugation of azide-modified antibody to VNP/VLP. The structure of human IgG1 and cowpea mosaic virus (CPMV) were rendered using the UCSF Chimera software (PDB ID: 1IGY and 1NY7 for hIgG1 and CPMV, respectively).
Figure 1.
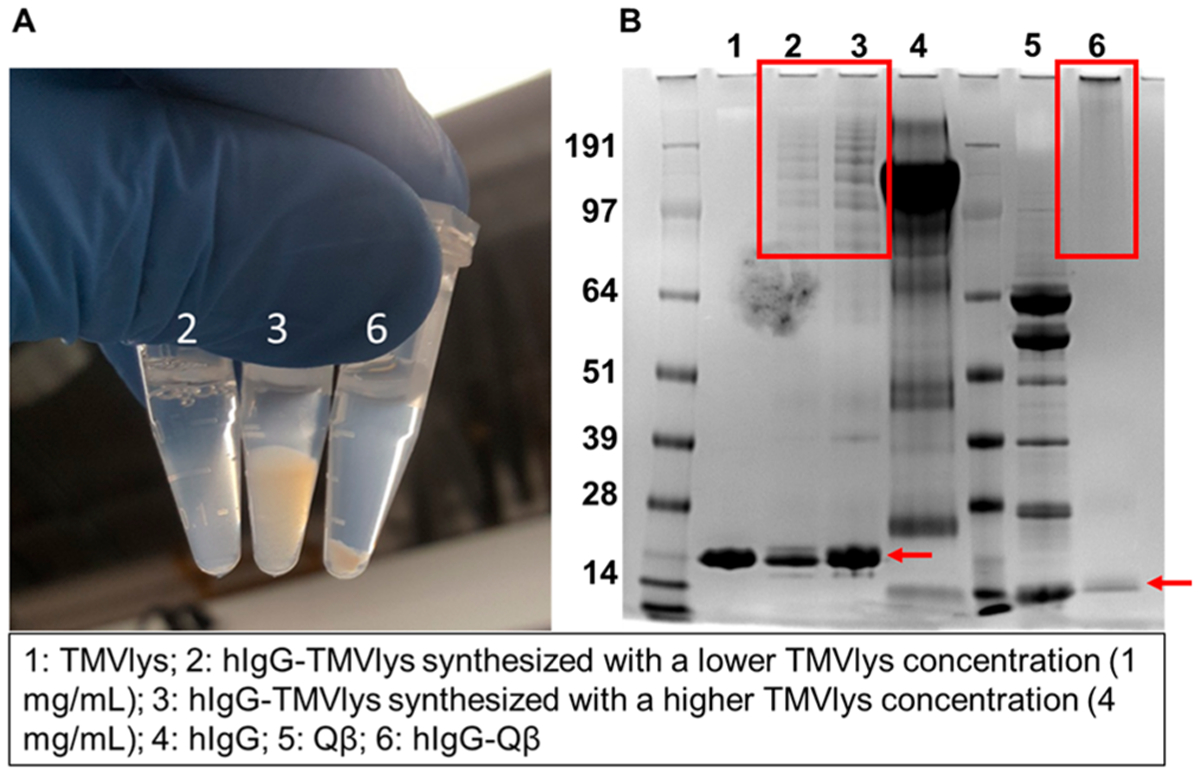
Chemical conjugation of antibodies to VNPs/VLPs using amine/thiol-reactive chemistries. (A) Photograph of antibodies conjugated to TMVlys and Qβ via amine and thiol-reactive chemistries. All particles are heavily agglomerated as evident by the formation of pellets. (B) Corresponding SDS-NuPAGE gel electrophoresis image. Red boxes highlight multiple high molecular weights bands for TMV-antibody conjugating indicating conjugation of multiple and/or cross-linking between antibodies and coat proteins. In the case of Qβ, the conjugated bands could not be resolved indicating high degree of cross-linking. Red arrows point to the coat proteins of TMVlys (~17 kDa) and Qβ (~14 kDa). It should be noted that a reducing agent was not added in the sample preparation; this explain the multiple banding for the Qβ samples showing coat proteins and dimers and multimers formed through disulfide bridges. Green arrows point to the light chain (~25 kDa) and heavy chain (~ 50 kDa) as well as the assembled IgG (again reducing agents were not added during sample preparation).
To overcome these challenges, we turned toward the aforementioned site-specific modification of antibodies followed by orthogonal coupling to VNPs. In brief, antibodies were deglycosylated using PNGase F followed by addition of an azide-functional handle using MTGase; we followed protocols previously reported in the literature;25 detailed procedures are outlined in Scheme 1B, and methods are described in the Experimental Procedures. Specifically, we used human IgG isolated from sera (hIgG), anti-CD47 Rat IgG2b, κ (α-CD47), and a recombinant humanized monoclonal IgG1 targeted against HER2 (Trastuzumab).
PNGase F cleaves glycans from Asn297 in the Fc region of the antibody; declycosylation exposes Gln295 and allows subsequent formation of isopeptide bonds between Gln295 and a substrate containing a primary amine group via transglutaminase catalysis.20 The deglycosylation of the antibodies following PNGase F treatment was monitored by SDS-NuPAGE gel electrophoresis (Figure 2A). The molecular weights of the native light and heavy chains of IgGs (150 kDa) are ~25 and ~50 kDa, respectively. While the electrophoretic mobility of the light chain of all three antibodies remained unchanged pre- and post- PNGase F treatment, PNGase F mediated deglycosylation resulted in a decrease in molecular weight of the heavy chains. This is as expected because the enzyme is targeting Asn297 on the heavy chain within the Fc region. The molecular weight of N-glycans varies depending on their structures and, in general, distributes between 1.8 to 3.0 kDa, which is large enough to differentiate electrophoretic mobility of successfully deglycosylated antibodies.26 As indicated by the lower molecular weight bands of the heavy chains of the IgGs in Figure 2A, glycans were effectively cleaved for each of the three antibody isotypes tested. It should be noted that deglycosylated hIgG in Figure 2A (far left image) exhibited multiple bands higher than the heavy chains. We attribute this phenomenon to the superposition of protein bands corresponding to the previously reported four different hIgG subclasses.27
Figure 2.
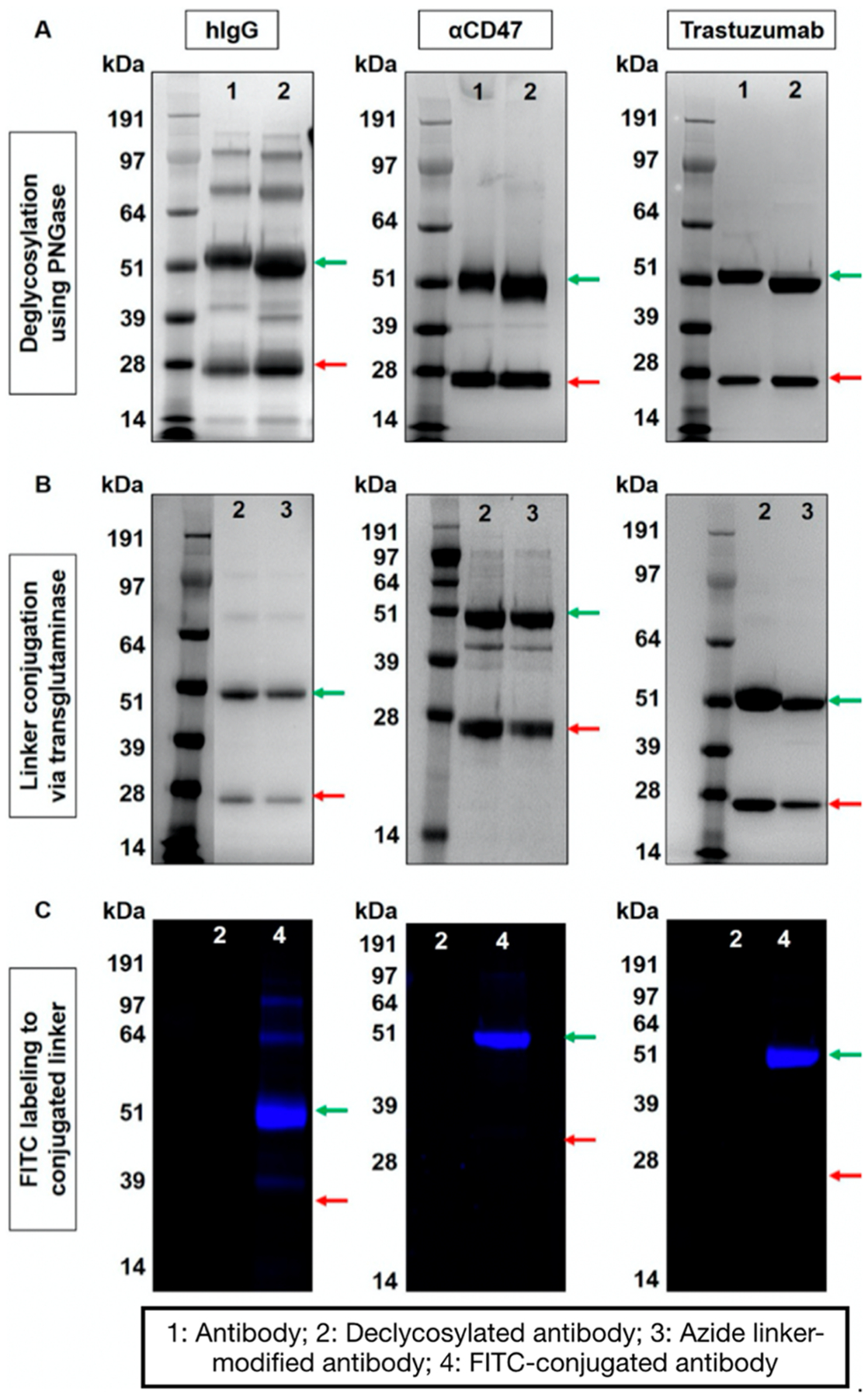
Characterization of site-specific antibody modification steps using gel electrophoresis. (A) IgGs pre- and postdeglycosylation using PNGase, (B) IgGs without and with azide-linker conjugated via transglutaminase, and (C) confirmation of the presence of the azide-linker via click reaction using FITC-PEG3-alkyne post transglutaminase reaction. Red and green arrows correspond to light and heavy chains of antibody (25 and 50 kDa), respectively.
Next the azido-PEG3-amine linker was introduced using MTGase; because the molecular weight of the linker is small (218 Da), the addition of the linker could not be verified using SDS-NuPAGE (Figure 2B). However, to confirm that the MTGase reaction was working and the linker was introduced, we chemically conjugated the azide-modified antibodies with an dibenzocyclooctyne-modified Fluorescein isothiocyanate (DBCO-PEG3-FITC) using copper-free, strain-promoted click chemistry. Fluorescent imaging of the antibody conjugated after electrophoretic separation using SDS-NuPAGE confirmed the successful and site-specific conjugation of the fluorophore to the heavy chain of all three tested antibodies (Figure 2C). The number of FITC molecules attached to each antibody was estimated from their UV-spectra (Supporting Information, Figure S1). We confirmed the successful conjugation of 1:1 FITC per hIgG and Trastuzumab, as compared to 0.7:1 FITC per α-CD47. This difference in linker conjugation efficiency may originate from isotype differences.
Next, azide-functional antibodies were conjugated to CPMV. CPMV possesses 300 surface exposed lysine side chains,28 which were reacted with a bifunctional DBCO-PEG4-NHS ester to enable the chemical conjugation of azide-modified antibodies via copper-free, strain-promoted click chemistry (Scheme 1B,C). Samples were denatured by SDS-NuPAGE to separate the large (L) and small (S) CPMV coat protein subunits, visualized as single bands at 42 kDa and 24 kDa, respectively (Figure 3A). Following the incubation of DBCO-modified CPMV with site-specific azide-modified antibodies, SDS-NuPAGE gels revealed the appearance of additional high molecular weight bands corresponding to the large and small coat proteins conjugated to the IgG (~174 and ~192 kDa). Note that samples for these gels were heated at 95 °C without a reducing agent such as 2-mercaptoethanol; therefore, the light and heavy chains of the IgG are not in their reduced state and appear as intact IgG on the gels. We further confirmed the conjugation of the IgG to CPMV using Western blot (Figure 3B) staining both for IgG and CPMV; the high molecular weight bands were positive for both, hIgG and CPMV, therefore confirming successful conjugation. In addition, we performed TEM imaging and immunogold staining using gold-labeled antihuman IgG antibodies. Imaging revealed intact CPMV particles maintaining their structural integrity. The colloidal gold (dark dots on images) bound exclusively to CPMV-hIgG, further confirming hIgG was indeed conjugated to CPMV (Figure 4A). It is also important to note that, unlike the nonspecific conjugation targeting amine/thiols, the site-specific conjugation reaction resulted in stable dispersion of the antibody-VNP conjugates; aggregation was not apparent. This was also consistent with size measurements using DLS (Supporting Information, Figure S2); DLS data show monodisperse particles with sizes of ~33 nm for CPMV-hIgG versus ~28 nm for CPMV; the increase in size is consistent with antibody display.
Figure 3.
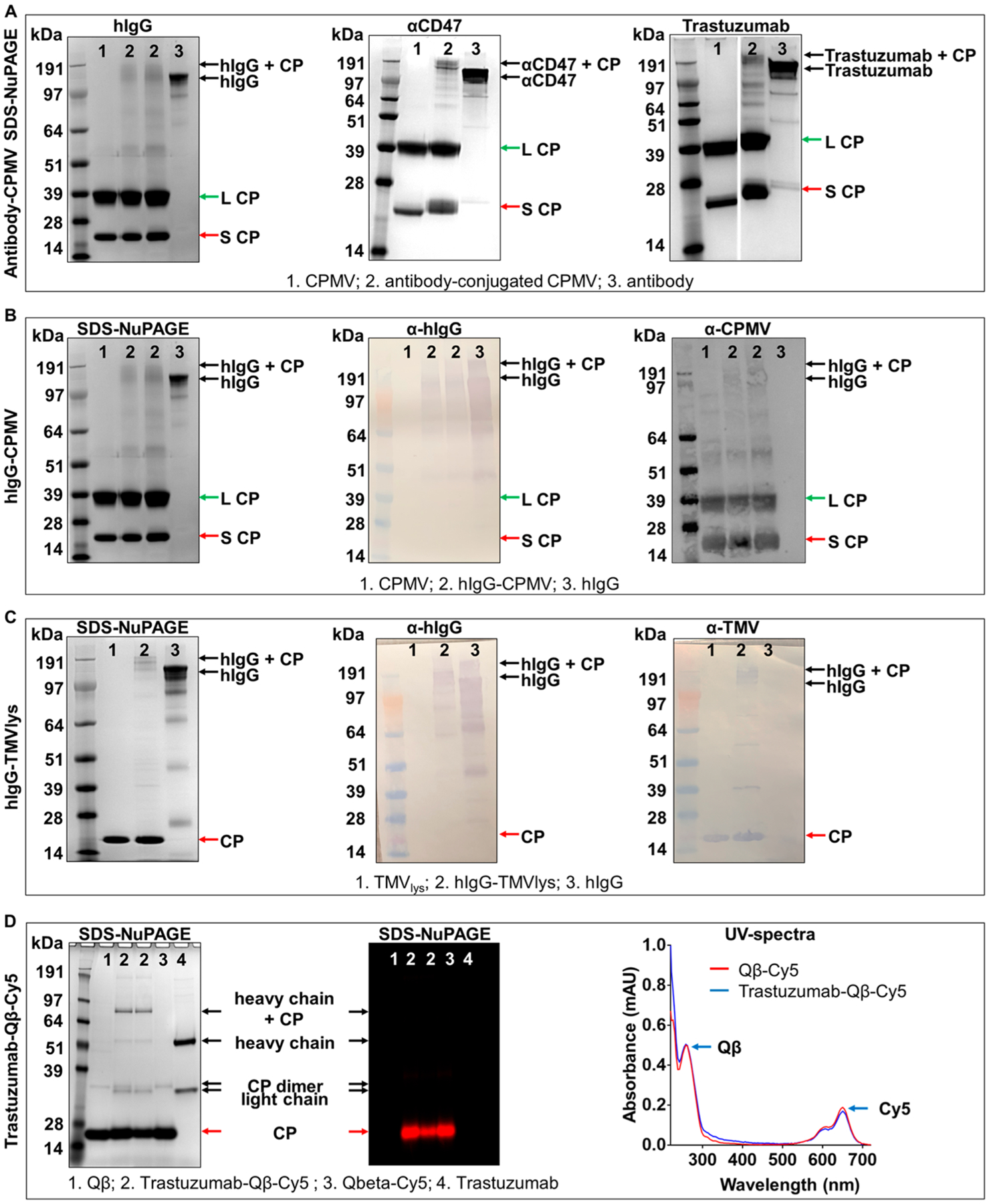
Conjugation of site-specifically modified antibodies to VNPs/VLPs via click chemistry. (A) SDS-NuPAGE gels of hIgG, αCD47, and Trastuzumab antibody conjugation to CPMV after Coomassie staining, (B) SDS-NuPAGE (left) of hIgG-CPMV and corresponding Western blot probed with anti-hIgG antibody (middle) and anti-CPMV antibody (right) (in Western blot additional high molecular weight bands appear in the CPMV sample; we attribute this to multimers of CPs; the Western blot is more sensitive than the SDS-NuPAGE and this is why these bands are not detected by SDS-NuPAGE). (C) SDS-NuPAGE (left) of hIgG-TMVlys and corresponding Western blot probed with anti-hIgG antibody (middle) and anti-TMVlys antibody (right). (D) SDS-NuPAGE of Cy5 labeled, Trastuzumab conjugated Qβ after Coomassie staining (left) and under red fluorescence (middle) to confirm Cy5 conjugation. Right panel corresponds to the UV-spectrum verification of Cy5 labeling onto Qβ.
Figure 4.
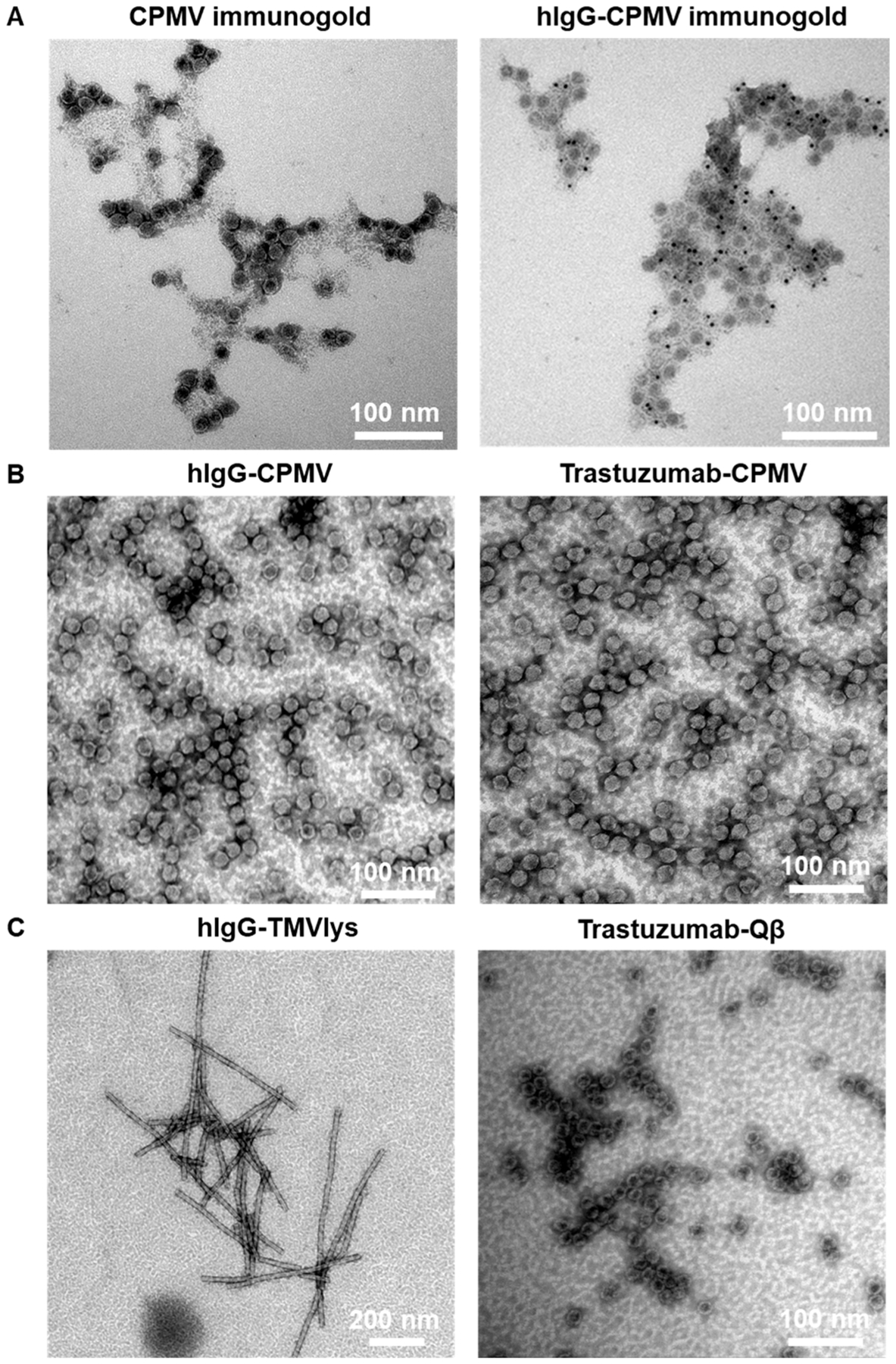
TEM images of antibody-conjugated VNPs. (A) Immunogold-staining of native CPMV and hIgG-CPMV hIgG. Six nm anti-hIgG labeled gold nanoparticles are visible as black dots only in the hIgG conjugated samples, (B) CPMV conjugated to hIgG (left) or Trastuzumab (right), (C) TMVlys conjugated to hIgG (left) and Qβ conjugated to Trastuzumab (right).
To demonstrate broad applicability, we next applied the conjugation scheme to TMV. As described above, successful conjugation of the hIgG (150 kDa) to TMVlys CP (17 kDa) was confirmed by SDS-NuPAGE electrophoresis and Western blot (Figure 3C). Lastly, we dually labeled the bacteriophage Qβ with Trastuzumab and Cy5 fluorophores (Figure 3D). Qβ nanoparticles were incubated with the equimolar of NHS-sulfoCy5 and NHS-PEG4-DBCO, followed by the conjugation with azide-modified Trastuzumab. As shown in Figure 3D, the presence of fluorescent Cy5 molecules on the surface of Qβ was confirmed by fluorescent imaging of SDS-NuPAGE gel and UV-spectroscopy.
In all cases studied, stable dispersion of antibody-VNP conjugates were formulated with no signs of aggregation. Therefore, we conclude that the site-specific, enzymatic functionalization of the antibodies followed by coupling to the VNPs is indeed an efficient method for the functionalization of VNPs. TEM imaging revealed intact particles (Figure 4). While it may appear that VNPs are clustered on the TEM grids, we attribute this to drying effects rather than aggregation.
Finally, to test whether the conjugated antibody maintain its functionality, we tested the interaction of Trastuzumab-functionalized Qβ with HER2+ cancer cells. Trastuzumab is a therapeutic antibody that targets human epidermal growth factor receptor-2 (HER2), which is expressed on aggressive cancer cells. Trastuzumab (sold under the brand name Herceptin) is a humanized recombinant monocolonal IgG1 that is FDA-approved for the treatment of HER2+ breast, gastric, and esophageal cancers. Furthermore, Trastuzumab therapy is in clinical development for treatment of women with ovarian cancer (clinicaltrials.gov: NCT00194714, NCT03342417). In our study, we chose the ovary adenocarcinoma SKOV-3 cell line, which is known to overexpress HER2.29 Here, we incubated SKOV-3 cells with 1:100 000 (cell: particles) using fluorescently tagged Qβ-Cy5 or Trastuzumab-Qβ-Cy5 particles for 1 h at 4 °C (Figure 5). Experiments were performed at 4 °C to investigate cell binding and minimize nonspecific cell uptake (which would occur at 37 °C). To minimize Fc receptor-derived nonspecific particle binding to the cells, we first blocked cells with an Fc receptor blocking solution for 10 min prior to the experiment. The incubation of SKOV-3 cells with Trastuzumab-Qβ-Cy5 resulted in ~50-fold increase in MFI compared to its counterpart. The display of Trastuzumab on Qβ conferred cell-targeting specificity; Trastuzumab-Qβ-Cy5 interacted efficiently with the cells; but Qβ-Cy5 was not. Overall, these results indicate that enzymatic modification of antibody followed by click chemistry conjugation to VNPs did not hinder the antibody functionalities yielding functional antibody-VNP conjugates.
Figure 5.
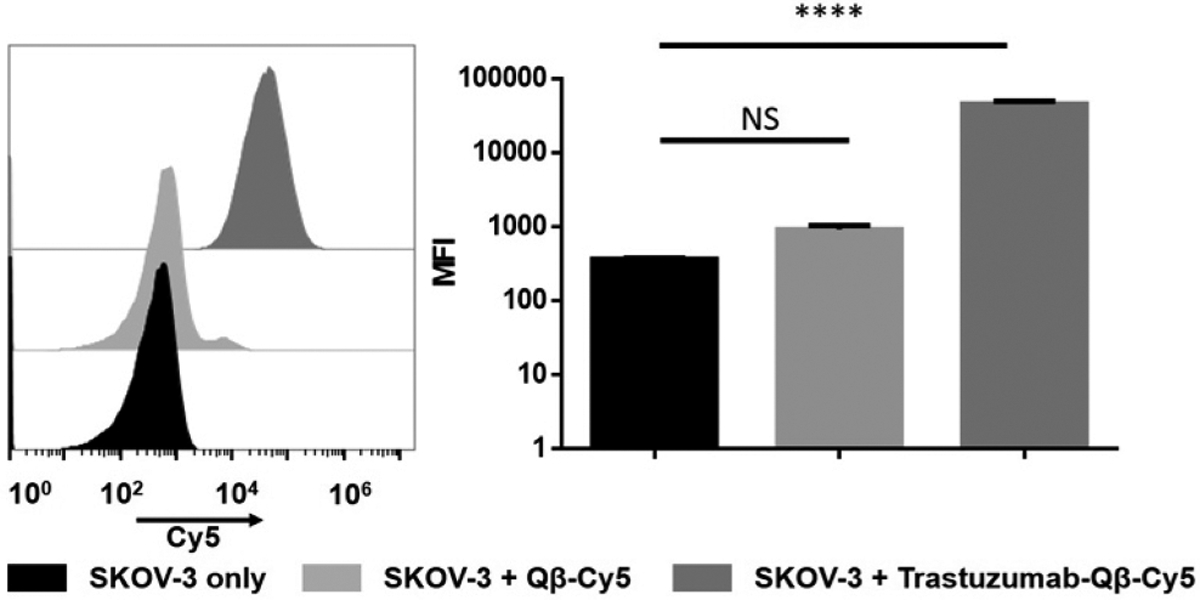
Flow cytometric binding assay of Trastuzumab-Qβ-Cy5 to SKOV-3 cells. Statistics were obtained using a one-way ANOVA test. **** corresponds to a p-value < 0.0001.
CONCLUSIONS
Antibody-VNP conjugates hold great promise in nanomedicine applications such as drug delivery, imaging, or immunotherapy. To date, most studies in which VNPs were utilized as targeted nanoparticle platforms made use of peptides to achieve target specificity, and there are only a few examples in which antibodies were conjugated.30 The conjugation of high molecular weight species, such as antibodies, can be challenging using nonspecific conjugation strategies; sample stability, in particular colloidal stability, must be maintained in particular for nanomedicine applications. While a large variety of antibody conjugation techniques are available, each has its strengths and limitations. Amine-reactive linking chemistries (e.g., NHS ester) are widely utilized due to the abundant presence of lysine residues in proteins including antibodies,31 which ensures fast and cost-effective conjugation. However, in some cases, generation of multiple conjugation sites in a protein for nanoparticle conjugation can lead to unwanted outcomes such as particle aggregation due to uncontrolled conjugation as shown in Figure 1.
Here we adapted site-specific transglutaminase-mediated antibody conjugation strategy; this chemistry has recently been reported to be effective when conjugating small molecule pharmaceutics;25,32 we extend its application to the conjugation of antibodies to VNPs. Importantly, we demonstrate that this method can be broadly applicable to different isotypes of antibodies and multiple VNP platforms. Site-specific modification of antibody enables researchers to generate one to two specific sites in Fc region, and make its conjugation to VNPs efficient even when conjugation has failed using other general conjugation methodologies. Furthermore, we show that clinically available drug, that is, Trastuzumab, can be efficiently modified by enzymatic reactions and trastuzumab-conjugated Qβ retained its targeting ability to HER2 receptor after conjugation. Site-specific antibody conjugation to VNPs is attractive because “off-the-shelf” antibodies can be utilized for the development of VNP-based therapeutics and contrast agents.
EXPERIMENTAL PROCEDURES
VNP/VLP Production.
CPMV and the well-established lysine-added TMV mutant were propagated in Burpee black-eyed pea (Vigna unguiculata)33 and tobacco (Nicotiana benthamiana)23 plants as previously described. Qβ virus-like nanoparticles (VLPs) were prepared in E. coli cells as previously described.24 USDA Permits (PPQ 526) were obtained for any work with plant viruses.
Antibody Deglycosylation Using Recombinant PNGase F: Removal of Glycans Attached to Heavy Chains of Antibody.
Glycans attached to the heavy chains of antibodies were removed using a recombinant peptide:N-glycosidase F (PNGase F) produced in Elizabethkingia miricola (Promega). Specifically, PNGase F cleaves N-linked oligosaccharides between the innermost N-acetylglucosamine (GlcNAc) and asparagines (Asn297). Ten milligrams of antibody was suspended in 1 mL of PBS and incubated with 5 μL of PNGase F (10 U μL−1) at 37 °C overnight (~16 h). PNGase F enzymes were removed using ultracentrifugal filter (Amicon, MWCO 100 kDa), and the concentrated deglycosylated antibodies were reconstituted in PBS at a final concentration of 20 mg mL−1.
Antibody-Linker Conjugation Mediated by MTGase: Linker Conjugation to Deglycosylated Antibody via Transglutaminase Reaction.
We conjugated a bifunctional linker, azido-PEG3-amine (BroadPharm, AZ101–100), to the glutamine 295 (Q295) on the heavy chains of antibodies making use of microbial transglutaminase (MTGase) (Ajinomoto, TI formula). Q295 was made accessible for MTGase reaction through glycans removal (see above). MTGase (100 mg mL−1) was dissolved in PBS prior to filtration through EMD Millipore Millex Sterile syringe filters with a pore size of 0.2 μm pores. Deglycosylated antibodies were incubated overnight with 10 mol equiv (eq) of azido-PEG3-amine (100 mg mL−1, dissolved in DMSO) and an equal volume of MTGase (100 mg mL−1) at 37 °C. MTGase was subsequently removed by either dialysis against PBS or by Amicon ultracentrifugal filtration. The transglutaminase mediated site-specific modification of antibodies was confirmed by the conjugation of an azide-reactive Fluorescein isothiocyanate (DBCO-PEG3-FITC, 1 mg mL−1 in DMSO) to antibodies (20 μL of 20 mg mL−1) modified with an azide-PEG4-NH2 linker in 20 mM potassium phosphate buffer (KP, pH 7.2). The click reaction was allowed to proceed for 24 h at room temperature prior to purification using Zeba spin filters (MWCO 40k). The conjugation of FITC to antibodies was observed by running the samples onto SDS NuPAGE gel electrophoresis as described below.
Antibody Conjugation to VNPs.
VNPs (1 mL at 2 mg mL−1) were suspended in their respective storage buffers (TMVlys, 10 mM potassium phosphate (KP); CPMV, 100 mM KP; Qβ, 1X PBS) and incubated for 2 h with 5 to 10 molar eq of DBCO-PEG4-NHS ester (BroadPharm, A134–10) per viral coat protein at room temperature. VNPs were pelleted down at 160 000 × g for 1 h over a 40% (w/v) sucrose cushion to remove excess linkers. The DBCO-modified VNPs were resuspended in 1 mL of their respective storage buffers and reacted overnight with 5 eq of azide-modified antibodies per particle at room temperature. CPMV and Qβ were purified by ultracentrifugation as described above. Antibody-conjugated TMVlys nanoparticles aggregate during the ultracentrifugation purification process; therefore, excess antibodies were removed using protein G or protein A HP spin traps (GE Healthcare).
Preparation of Trastuzumab-Conjugated Fluorescent Qβ.
The surface of native Qβ was incubated with equal molar ratio of DBCO-PEG4-NHS ester and sulfo-Cyanine5 NHS ester (Cy5, Lumiprobe) for 2 h at room temperature prior to centrifugation at 160 000 × g for 1 h over a 40% sucrose cushion to remove the excess dye and linker molecules. The modified Qβ nanoparticles were resuspended in 1 mL of PBS and mixed with azide-modified Trastuzumab antibody overnight at 4 °C overnight prior to purification by centrifugation as described above. Trastuzumab- and Cy5-conjugated Qβ was resuspended in 1 mL of PBS and kept at 4 °C until further analysis.
Immunogold Staining of Antibody-Conjugated VNPs and Transmission Electron Microscopy (TEM).
Native and human IgG conjugated CPMV (0.1 mg mL−1) were deposited onto Formvar copper grids coated with carbon film (Electron Microscopy Sciences) for 20 min and subsequently washed with 10 mM KP for 1 min. Grids were blocked in PBS supplemented with 1% (w/v) BSA and 0.1% (v/v) Tween-20 for 30 min, equilibrated in PBS supplemented with 0.1% (w/v) BSA and 0.1% (v/v) Tween-20 for 5 min, and incubated with goat antihuman IgG gold colloids (6 nm diameter, Jackson ImmunoResearch) for 2 h. The TEM grids were subsequently washed four times with 0.01% (v/v) Tween-20 PBS for 3 min, two times with 10 mM KP for 3 min, and three times with deionized water for 5 min. Grids were stained with 2% (w/v) uranyl acetate, and FEI Tecnai Spirit G2 BioTWIN transmission electron microscope was used to capture images of the samples at 80 kV.
Similarly, for negative stain TEM, Formvar copper grids coated with carbon film (Electron Microscopy Sciences) were glow discharged using the PELCO easiGlow operating system to render them more hydrophilic. Drops of CPMV, TMV, and Qβ (10 μL, 0.2 mg mL−1) were deposited onto the grids for 2 min at room temperature. The grids were then washed twice with deionized water for 30 s and subsequently stained twice with 2% (w/v) uranyl acetate for another 45 s. FEI Tecnai Spirit G2 BioTWIN transmission electron microscope was used to capture images of the samples at 80 kV.
UV–vis Spectroscopy.
The UV–vis spectra of VNPs and antibody were recorded using a NanoDrop spectrophotometer. CPMV and TMVlys were dispersed in 10 mM KP (pH 7.2) while Qβ and antibodies were dispersed in PBS (pH 7.2). The Beer–Lambert law (A = εcl, where A is absorbance, ε is the extinction coefficient, c is the protein concentration, and l is the path length) was used to calculate protein concentration and the number of dyes successfully conjugated. TMVlys: ε(260 nm) = 3.0 mL mg−1 cm−1, molecular weight of TMVlys = 39.4 × 106 g mol−1. CPMV: ε(260 nm) = 8.1 mL mg−1 cm−1, molecular weight of CPMV = 6 × 106 g mol−1. Antibodies: ε(280 nm) = 210 000 M−1 cm−1. Cy5: ε(647 nm) = 271 000 M−1 cm−1, molecular weight of Cy5 = 778 g mol−1. FITC ε(488 nm) = 73 000 M−1 cm−1. l = 0.1 cm.
BCA Protein Assay.
A Pierce BCA protein assay kit (Thermo Fisher) was used according to manufacturer’s recommendation to calculate the concentration of Qβ. To determine the concentration of the VNPs, UV–visible spectroscopy and the VNP-specific molecular extinction coefficient and Beer–Lambert law were used (see above).
Dynamic Light Scattering (DLS).
The hydrodynamic radii of native and conjugated VNPs (0.1 mg mL−1 in buffer 10 mM KP pH 7.4) were recorded using a Zetasizer Nano ZSP/Zen5600 instrument (Malvern Panalytical). The particle length was calculated as the weighted mean of the intensity distribution.
Denaturing Gel Electrophoresis SDS-NuPAGE.
VNPs or antibody samples were denatured 100 °C for 5 min in the presence of NuPAGE LDS sample buffer (1:4 dilution) (Thermo Fisher Scientific). Denatured particle samples were then separated in a 4–12% or 10% SDS-NuPAGE precast gels in 1× morpholinepropanesulfonic acid (MOPS) buffer (Thermo Fisher Scientific) at 200 V (120 mA) for 40 min. Gels were stained with Coomassie Blue and imaged with an FluorChem R (Protein simple) under white light and MultiFluor red light.
Cell Binding Assay Using Trastuzumab-Conjugated Fluorescent Qβ.
SKOV-3, a HER2+ cell line, was selected for binding experiment of Trastuzumab-conjugated Qβ. SKOV-3 cells were cultured in McCoy’s 5A medium supplemented with 10% FBS (v/v) and 1% (v/v) PenStrep until cells reached 70% confluency. A single cell suspension of SKOV-3 was prepared using enzyme-free, cell dissociation buffer (Gibco) and suspending at a stock concentration of 2 × 106 cells mL−1 in PBS supplemented with 1% (w/v) BSA and 2 mM EDTA. Cells were aliquoted in a tube containing 1 × 106 cells. To minimize antibody-derived nonspecific particle adsorption, cells were incubated with Fc receptor blocking solution (Human Trustain Fcx, BioLegend) for 10 min. Then Cy5-labeled as well as Cy5- or Trastuzumab-conjugated Qβ nanoparticles were incubated with SKOV-3 cells at the ratio of 1:100 000 (cell:VNP) for 1 h at 4 °C. Cells were subsequently rinsed twice with PBS and resuspended in 300 μL of PBS with 1% (w/v) BSA. Samples were analyzed using a BD Accuri C6 (BD Biosciences) and processed with the FlowJo software (https://www.flowjo.com/).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the following grants: CAREER DMR 1841848 from the National Science Foundation as well as R01 HL137674 and R01 CA202814 from the National Institutes of Health.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.bioconjchem.0c00118.
UV spectra of FITC-conjugated antibodies; hydrodynamic size change before and after antibody conjugation measured by DLS (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.bioconjchem.0c00118
The authors declare no competing financial interest.
Contributor Information
Jooneon Park, Department of NanoEngineering, University of California San Diego, La Jolla, California 92039, United States.
Paul L. Chariou, Department of Bioengineering, University of California San Diego, La Jolla, California 92039, United States
Nicole F. Steinmetz, Department of NanoEngineering, Department of Bioengineering, Department of Radiology, Moores Cancer Center, and Center for Nano- ImmunoEngineering, University of California San Diego, La Jolla, California 92039, United States.
REFERENCES
- (1).Spicer CD, Jumeaux C, Gupta B, and Stevens MM (2018) Peptide and protein nanoparticle conjugates: versatile platforms for biomedical applications. Chem. Soc. Rev 47, 3574–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Wu H, and Huang J (2018) Optimization of protein and peptide drugs based on the mechanisms of kidney clearance. Protein Pept. Lett 25, 514–521. [DOI] [PubMed] [Google Scholar]
- (3).Muhamad N, Plengsuriyakarn T, and Na-Bangchang K (2018) Application of active targeting nanoparticle delivery system for chemotherapeutic drugs and traditional/herbal medicines in cancer therapy: a systematic review. Int. J. Nanomed 13, 3921–3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bazak R, Houri M, Achy SE, Kamel S, and Refaat T (2015) Cancer active targeting by nanoparticles: a comprehensive review of literature. J. Cancer Res. Clin. Oncol 141, 769–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Birrer MJ, Moore KN, Betella I, and Bates RC (2019) Antibody-drug conjugate-based therapeutics: state of the science. J. Natl. Cancer Inst 111, 538–549. [DOI] [PubMed] [Google Scholar]
- (6).Carter T, Mulholland P, and Chester K (2016) Antibody-targeted nanoparticles for cancer treatment. Immunotherapy 8, 941–958. [DOI] [PubMed] [Google Scholar]
- (7).McKeage K, and Perry CM (2002) Trastuzumab. Drugs 62, 209–243. [DOI] [PubMed] [Google Scholar]
- (8).Truffi M, Colombo M, Sorrentino L, Pandolfi L, Mazzucchelli S, Pappalardo F, Pacini C, Allevi R, Bonizzi A, Corsi F, and Prosperi D (2018) Multivalent exposure of trastuzumab on iron oxide nanoparticles improves antitumor potential and reduces resistance in HER2-positive breast cancer cells. Sci. Rep 8, 6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Dziawer Ł, Majkowska-Pilip A, Gaweł D, Godlewska M, Pruszynski M, Jastrzebski J, Was B, and Bilewicz A (2019) Trastuzumab-modified gold nanoparticles labeled with 211At as a prospective tool for local treatment of HER2-positive breast cancer. Nanomaterials 9, 632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Park JW, Hong K, Kirpotin DB, Colbern G, Shalaby R, Baselga J, Shao Y, Nielsen UB, Marks JD, Moore D, Papahadjopoulos D, and Benz CC (2002) Anti-HER2 immunoliposomes: enhanced efficacy attributable to targeted delivery. Clin. Cancer Res 8, 1172–1181. [PubMed] [Google Scholar]
- (11).Colzani B, Pandolfi L, Hoti A, Iovene PA, Natalello A, Avvakumova S, Colombo M, and Prosperi D (2018) Investigation of antitumor activities of trastuzumab delivered by PLGA nanoparticles. Int. J. Nanomed 13, 957–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Kim P-H, Sohn J-H, Choi J-W, Jung Y, Kim SW, Haam S, and Yun C-O (2011) Active targeting and safety profile of PEG-modified adenovirus conjugated with herceptin. Biomaterials 32, 2314–2326. [DOI] [PubMed] [Google Scholar]
- (13).Esfandiari N, Arzanani MK, Soleimani M, Kohi-Habibi M, and Svendsen WE (2016) A new application of plant virus nanoparticles as drug delivery in breast cancer. Tumor Biol. 37, 1229–1236. [DOI] [PubMed] [Google Scholar]
- (14).Francis GE, Fisher D, Delgado C, Malik F, Gardiner A, and Neale D (1998) PEGylation of cytokines and other therapeutic proteins and peptides: the importance of biological optimization of coupling techniques. Int. J. Hematol 68, 1–18. [DOI] [PubMed] [Google Scholar]
- (15).Behrens CR, and Liu B (2014) Methods for site-specific drug conjugation to antibodies. mAbs 6, 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yamada K, and Ito Y (2019) Recent chemical approaches for site-specific conjugation of native antibodies: technologies toward next-generation antibody–drug conjugates. ChemBioChem 20, 2729–2737. [DOI] [PubMed] [Google Scholar]
- (17).Fontana A, Spolaore B, Mero A, and Veronese FM (2008) Site-specific modification and PEGylation of pharmaceutical proteins mediated by transglutaminase. Adv. Drug Delivery Rev 60, 13–28. [DOI] [PubMed] [Google Scholar]
- (18).Anami Y, Yamazaki CM, Xiong W, Gui X, Zhang N, An Z, and Tsuchikama K (2018) Glutamic acid–valine–citrulline linkers ensure stability and efficacy of antibody–drug conjugates in mice. Nat. Commun 9, 2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Anami Y, Xiong W, Gui X, Deng M, Zhang CC, Zhang N, An Z, and Tsuchikama K (2017) Enzymatic conjugation using branched linkers for constructing homogeneous antibody–drug conjugates with high potency. Org. Biomol. Chem 15, 5635–5642. [DOI] [PubMed] [Google Scholar]
- (20).Benjamin SR, Jackson CP, Fang S, Carlson DP, Guo Z, and Tumey LN (2019) Thiolation of Q295: site-specific conjugation of hydrophobic payloads without the need for genetic engineering. Mol. Pharmaceutics 16, 2795–2807. [DOI] [PubMed] [Google Scholar]
- (21).Wen AM, and Steinmetz NF (2016) Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem. Soc. Rev 45, 4074–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Eiben S, Koch C, Altintoprak K, Southan A, Tovar G, Laschat S, Weiss IM, and Wege C (2019) Plant virus-based materials for biomedical applications: Trends and prospects. Adv. Drug Delivery Rev 145, 96–118. [DOI] [PubMed] [Google Scholar]
- (23).Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, and Wege C (2013) TMV nanorods with programmed longitudinal domains of differently addressable coat proteins. Nanoscale 5, 3808. [DOI] [PubMed] [Google Scholar]
- (24).Prasuhn DE, Singh P, Strable E, Brown S, Manchester M, and Finn MG (2008) Plasma clearance of bacteriophage Qβ particles as a function of surface charge. J. Am. Chem. Soc 130, 1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Jeger S, Zimmermann K, Blanc A, Grünberg J, Honer M, Hunziker P, Struthers H, and Schibli R (2010) Site-specific and stoichiometric modification of antibodies by bacterial transglutaminase. Angew. Chem., Int. Ed 49, 9995–9997. [DOI] [PubMed] [Google Scholar]
- (26).Goldberg D, Bern M, North SJ, Haslam SM, and Dell A (2009) Glycan family analysis for deducing N-glycan topology from single MS. Bioinformatics 25, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Fasler S, Skvaril F, and Lutz HU (1988) Electrophoretic properties of human IgG and its subclasses on sodium dodecylsulfate-polyacrylamide gel electrophoresis and immunoblots. Anal. Biochem 174, 593–600. [DOI] [PubMed] [Google Scholar]
- (28).Chatterji A, Ochoa WF, Paine M, Ratna BR, Johnson JE, and Lin T (2004) New addresses on an addressable virus nanoblock: uniquely reactive Lys residues on cowpea mosaic virus. Chem. Biol 11, 855–863. [DOI] [PubMed] [Google Scholar]
- (29).Magnifico A, Albano L, Campaner S, Delia D, Castiglioni F, Gasparini P, Sozzi G, Fontanella E, Menard S, and Tagliabue E (2009) Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to Trastuzumab. Clin. Cancer Res 15, 2010. [DOI] [PubMed] [Google Scholar]
- (30).Aanei IL, Huynh T, Seo Y, and Francis MB (2018) Vascular cell adhesion molecule-targeted MS2 viral capsids for the detection of early-stage atherosclerotic plaques. Bioconjugate Chem. 29, 2526–2530. [DOI] [PubMed] [Google Scholar]
- (31).Rosen CB, and Francis MB (2017) Targeting the N terminus for site-selective protein modification. Nat. Chem. Biol 13, 697–705. [DOI] [PubMed] [Google Scholar]
- (32).Dickgiesser S, Deweid L, Kellner R, Kolmar H, and Rasche N (2019) Site-specific antibody–drug conjugation using microbial transglutaminase, in Enzyme-Mediated Ligation Methods (Nuijens T, and Schmidt M, Eds.) pp 135–149, Springer, New York. [DOI] [PubMed] [Google Scholar]
- (33).Wen AM, Shukla S, Saxena P, Aljabali AAA, Yildiz I, Dey S, Mealy JE, Yang AC, Evans DJ, Lomonossoff GP, and Steinmetz NF (2012) Interior engineering of a viral nanoparticle and its tumor homing properties. Biomacromolecules 13, 3990–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


