Here, Viktorovskaya et al. sought to understand how the functions of three essential histone chaperones are coordinated during gene transcription. Using genetic and biochemical approaches, this work sheds light on previously unknown connections between FACT activity and the functions of Spt6 and Spn1.
Keywords: Spt6, Spn1, FACT, histone chaperones, transcription, chromatin
Abstract
Histone chaperones are critical for controlling chromatin integrity during transcription, DNA replication, and DNA repair. Three conserved and essential chaperones, Spt6, Spn1/Iws1, and FACT, associate with elongating RNA polymerase II and interact with each other physically and/or functionally; however, there is little understanding of their individual functions or their relationships with each other. In this study, we selected for suppressors of a temperature-sensitive spt6 mutation that disrupts the Spt6-Spn1 physical interaction and that also causes both transcription and chromatin defects. This selection identified novel mutations in FACT. Surprisingly, suppression by FACT did not restore the Spt6-Spn1 interaction, based on coimmunoprecipitation, ChIP, and mass spectrometry experiments. Furthermore, suppression by FACT bypassed the complete loss of Spn1. Interestingly, the FACT suppressor mutations cluster along the FACT-nucleosome interface, suggesting that they alter FACT-nucleosome interactions. In agreement with this observation, we showed that the spt6 mutation that disrupts the Spt6-Spn1 interaction caused an elevated level of FACT association with chromatin, while the FACT suppressors reduced the level of FACT-chromatin association, thereby restoring a normal Spt6-FACT balance on chromatin. Taken together, these studies reveal previously unknown regulation between histone chaperones that is critical for their essential in vivo functions.
An enduring quest in the field of gene expression is to understand the function and coordination of the multitude of proteins that dynamically associate with RNAPII during transcription initiation, elongation, and termination (Schier and Taatjes 2020). Many of these proteins are required for transcription to overcome the repressive effects of nucleosomes, by helping to maintain normal chromatin structure after the passage of RNAPII or by the modification of histone proteins.
Histone chaperones comprise one class of factor essential for transcription by directly modulating histone-DNA interactions in an ATP-independent fashion. While the mechanisms and roles of some histone chaperones are well understood (for example, see English et al. 2006), the functions of most chaperones are not well defined (Hammond et al. 2017; Warren and Shechter 2017). One mystery is why so many histone chaperones are required during transcription elongation, when at least eight histone chaperones associate with elongating RNAPII. These include three that are conserved and essential for viability and that are the focus of our studies: Spt6, Spn1/Iws1, and FACT.
Spt6 and Spn1 directly interact with each other, and this interaction is important for their function in both yeast and mammalian cells (Yoh et al. 2008; Diebold et al. 2010; McDonald et al. 2010). However, the reasons for this interaction are not well understood, and there are several distinctions between Spt6 and Spn1. For example, in yeast, depletion of Spt6 results in massive changes in the specificity of transcription initiation (Doris et al. 2018), while depletion of Spn1 has little known effect on initiation specificity (Reim et al. 2020). In addition, Spt6 is required for the levels of certain histone modifications, including H3K36me2 and H3K36me3 (Carrozza et al. 2005; Chu et al. 2006; Youdell et al. 2008), while Spn1 is not needed for the level of these modifications but rather is required for their normal distribution on chromatin (Reim et al. 2020). Finally, while both Spt6 and Spn1 interact directly with histones (Bortvin and Winston 1996; McCullough et al. 2015; Li et al. 2018), only Spt6 has been shown to interact directly with RNAPII (Sdano et al. 2017; Vos et al. 2018). Thus, both proteins play vital roles, yet the nature of these roles and how they connect to each other remains to be determined.
Spt6 also shares many functional similarities with FACT, another conserved and essential histone chaperone (Duina 2011). Both Spt6 and FACT are believed to facilitate transcription of RNAPII through nucleosomes and/or to ensure nucleosome reassembly after the passage of RNAPII. In addition, both regulate core histone levels (Jeronimo et al. 2019) as well as the deposition of histone H2A.Z (Jeronimo et al. 2015). Furthermore, both are required for the specificity of transcription initiation (Cheung et al. 2008). Despite their related functions, Spt6 and FACT clearly have independent, nonredundant roles, since both are essential for viability in many types of cells, as well as differing with respect to interactions with nucleosomes and patterns of association with chromatin (Mayer et al. 2010; Duina 2011; McCullough et al. 2015; Pathak et al. 2018). While one of the main mechanisms for recruitment of Spt6 to chromatin is by its interaction with RNAPII (Mayer et al. 2010; Sdano et al. 2017; Dronamraju et al. 2018), the recruitment mechanism for FACT is unknown and likely occurs by multiple means, including interactions with histones (Hodges et al. 2017; Cucinotta et al. 2019), histone H2B ubiquitylation (Fleming et al. 2008; Murawska et al. 2020), and by the recognition of an altered nucleosome structure (Martin et al. 2018).
Thus, studies of Spt6, Spn1, and FACT have revealed intriguing relationships between the three, raising the question of how they functionally interact during transcription. In this study, we have addressed this issue in Saccharomyces cerevisiae, starting with spt6-YW, an spt6 mutation that was designed to disrupt the Spt6-Spn1 interaction (Diebold et al. 2010). We first show that spt6-YW does severely impair the Spt6-Spn1 interaction. Then, to understand the requirement for this interaction, we isolate extragenic suppressors of spt6-YW and identify mutations in genes encoding several known transcription elongation and chromatin factors. Notably, the suppressors include novel changes in FACT, as well as in Spt5, a conserved and essential elongation factor. By genetic and biochemical approaches, we show that the mechanisms by which alterations of FACT and Spt5 suppress spt6-YW are distinct, as altered FACT bypasses the requirement for Spn1 while suppression by altered Spt5 is Spn1-dependent. Nevertheless, both classes suppress the chromatin defects caused by spt6-YW. Finally, we show that the FACT suppressors cluster on its nucleosome binding surface, weaken FACT-histone interactions, and restore a balance between the levels of Spt6 and FACT associated with chromatin. Taken together, our studies reveal a network of interactions between essential histone chaperones and other conserved factors.
Results
Suppressors of spt6-YW identify important transcription elongation factors
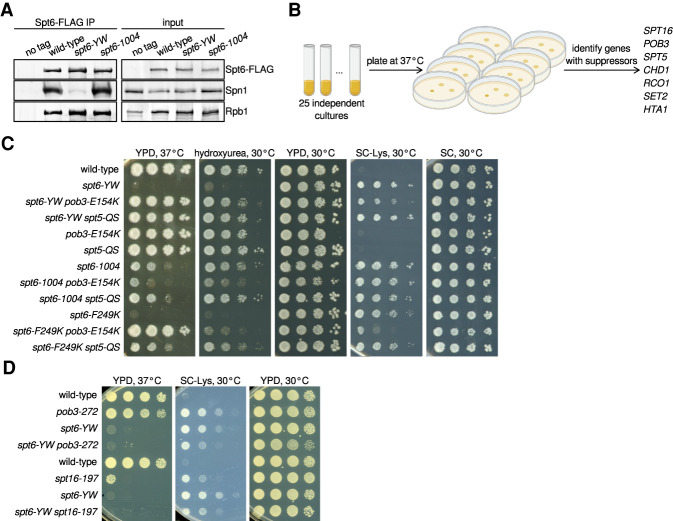
To investigate interactions between the histone chaperones Spt6 and Spn1, we used spt6-YW, a mutation predicted to impair the Spt6-Spn1 interaction due to changes of two conserved Spt6 residues (Y255A, W257A) on the interface of Spt6 with Spn1 (Diebold et al. 2010; McDonald et al. 2010). The spt6-YW mutation causes strong mutant phenotypes, including temperature-sensitive growth at 37°C and sensitivity to DNA-damaging agents (Diebold et al. 2010). By coimmunoprecipitation (co-IP), we observed a dramatic decrease in Spt6-Spn1 interaction in an spt6-YW mutant, while the interaction of Spt6 with Rpb1 was largely unaffected (Fig. 1A). This Spt6-Spn1 defect was specific for spt6-YW, as it was unaffected in an spt6-1004 mutant, which contains a mutation outside of the Spn1 binding site (Kaplan et al. 2003). Thus, spt6-YW disrupts the conserved Spt6-Spn1 interaction, providing the opportunity to study its requirement in vivo.
Figure 1.
Mutations in SPT16/POB3 and SPT5 suppress spt6-YW in an allele-specific fashion. (A) Western blots showing the levels of Spn1, Rpb1, and Spt6-FLAG in Spt6-FLAG immunoprecipitation (IP) samples and the corresponding inputs from untagged control (FY87), wild-type (FY3276), spt6-YW (FY3277), and spt6-1004 (FY3283) strains. Spn1, Rpb1, and Spt6 were detected using anti-Spn1, 8WG16 anti-Rpb1, and anti-FLAG antibodies, respectively (Supplemental Table S5). (B) A schematic showing the isolation of spt6-YW suppressors, with genes identified as extragenic suppressors listed at the right. (C,D) Analysis of genetic interactions between spt6, spt5, pob3, and spt16 mutations. Strains were grown to saturation in YPD, serially diluted 10-fold, spotted on the indicated media, and grown at the indicated temperature.
As a genetic approach to understand the roles for Spt6-Spn1 interaction, we selected for suppressors of the spt6-YW temperature-sensitive phenotype (Fig. 1B). We isolated 28 independent revertants that allowed growth at 37°C and identified the causative mutations for each by a combination of whole-genome sequencing and genetic tests (Materials and Methods). These analyses identified extragenic suppressors in seven genes (17/28 mutants) (Fig. 1B), as well as intragenic suppressors and strains disomic for chromosome 16, the location of Spn1 (Supplemental Table S1).
All seven genes identified by the extragenic suppressors encode factors that regulate transcription elongation and chromatin (Fig. 1B; Supplemental Table S1). Among these suppressors, the most surprising results were the identification of mutations in three essential genes: SPT16, POB3, and SPT5. Spt16 and Pob3 are the two subunits of the histone chaperone FACT (Gurova et al. 2018), and Spt5 is a conserved elongation factor that dimerizes with Spt4, directly controls RNAPII processivity, and has recently been implicated in controlling chromatin structure (Hartzog and Fu 2013; Crickard et al. 2017; Ehara et al. 2019). The identification of spt6-YW suppressors in SPT16, POB3, and SPT5 was unprecedented, as previously isolated mutations in these genes conferred similar, rather than opposite, phenotypes compared with spt6 mutations (for example, Swanson and Winston 1992; Kaplan et al. 2003; Cheung et al. 2008; Jeronimo et al. 2015; McCullough et al. 2015; Pathak et al. 2018).
Although the analysis of the FACT and Spt5 suppressors is the focus of our studies, we first briefly summarize our findings for the other suppressors identified. The remaining extragenic suppressors—SET2, RCO1, CHD1, and HTA1—were previously identified as suppressors of either spt6 mutations or mutations that impair related elongation factors (Simic et al. 2003; Keogh et al. 2005; Chu et al. 2006; Biswas et al. 2007; Quan and Hartzog 2010; McCullough et al. 2015; Lee et al. 2018). For SET2, RCO1, and CHD1, which are not essential for viability, we found that complete deletions also suppressed spt6-YW, showing that suppression is conferred by loss of function (Supplemental Fig. S1A; Supplemental Table S2). The fourth gene, HTA1, is one of two genes encoding histone H2A, and its isolation fits with previous studies that identified H2A and H2B mutants as suppressors of a similar spt6 mutation, spt6-F249K (McCullough et al. 2015). The intragenic class of spt6-YW suppressors contained three independent isolates that each contained a P231L amino acid change in Spt6 in addition to the original Y255A and W257A substitutions. P231 is located just outside of the region of Spt6 previously cocrystallized with Spn1 (Diebold et al. 2010; McDonald et al. 2010); the proximity of P231 to the Spt6-Spn1 interface suggests that it suppresses spt6-YW by strengthening Spt6-Spn1 interactions. Finally, the eight disomic suppressors each contained an extra copy of chromosome 16, where SPN1 is located. To test whether suppression was caused by the second copy of SPN1, we supplied spt6-YW mutants with SPN1 on a centromeric plasmid and found suppression of spt6-YW temperature sensitivity (Supplemental Fig. S1B). This result supports the idea that the temperature-sensitive defect in the spt6-YW mutant is caused by the impaired Spt6-Spn1 interaction.
Mutational changes in FACT and Spt5 suppress spt6-YW in an allele-specific fashion
Given the novel genetic suppression of spt6-YW by pob3, spt16, and spt5 mutations, we asked whether the genetic interactions were allele-specific, which would suggest that suppression occurs by the alteration of specific molecular properties of these factors rather than by a general reduction of function. To test for allele-specific suppression, we first combined either pob3-E154K, the strongest FACT suppressor, or our one spt5 suppressor, spt5-QS, with two other spt6 mutations, spt6-F249K and spt6-1004 (Supplemental Tables S1, S2). The spt6-F249K mutation, like spt6-YW, impairs the Spt6-Spn1 interaction (McDonald et al. 2010), and we found that the temperature sensitivity of spt6-F249K was suppressed by both pob3-E154K and spt5-QS (Fig. 1C). In contrast, the spt6-1004 mutation does not impair the Spt6-Spn1 interaction (Fig. 1A); its temperature-sensitive phenotype became more severe with pob3-E154K and it was not suppressed by spt5-QS. We also combined spt6-YW with previously isolated pob3, spt16, spt4, or spt5 alleles and we did not observe suppression of any spt6-YW phenotypes (Fig. 1D; Supplemental Table S2). In fact, some spt4 and spt5 alleles caused double-mutant lethality when combined with spt6-YW. Together, these results demonstrate a high degree of allele-specificity, indicating a distinctive functional relationship between mutations that impair the Spt6-Spn1 interaction and their suppressors.
Spt6, Spn1, FACT, and Spt5 functionally interact to modulate nucleosome organization in vivo
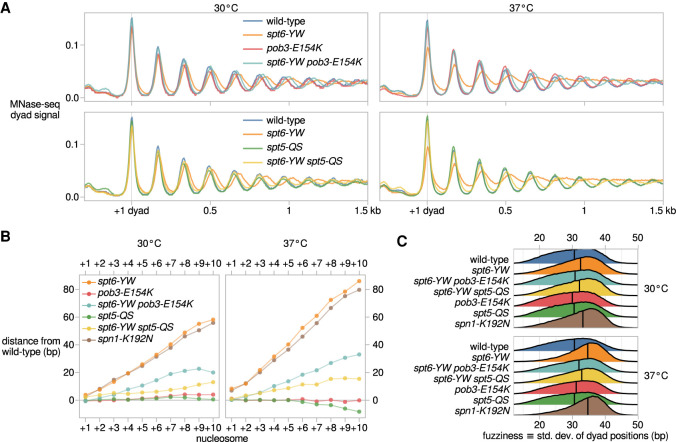
Spt6 and FACT are required for normal nucleosome positioning and occupancy (Ivanovska et al. 2011; Perales et al. 2013; van Bakel et al. 2013; Doris et al. 2018; Jeronimo et al. 2019). Spt5 has also been implicated in contact with nucleosomes during transcription, and our spt5-QS suppressor mutation maps proximal to the nucleosome interacting region of Spt5 (Ehara et al. 2019). Therefore, we investigated the effect of spt6-YW and its suppressors on nucleosome organization. To do this, we performed micrococcal nuclease sequencing (MNase-seq) on a set of wild-type and mutant strains grown both at 30°C and after a shift to nonpermissive temperature, 37°C. The wild-type strain showed the expected nucleosome pattern over genes, with a nucleosome-depleted region upstream of transcription start sites (TSSs) and a regularly phased nucleosome array downstream (Fig. 2A). In spt6-YW, the median distance between adjacent nucleosome dyads increased from the wild-type value of 165 bp to 169 bp at 30°C and to 171 bp at 37°C, manifesting as a progressive 3′ shift of nucleosomes over genes (Fig. 2A,B; Supplemental Fig. S2A). The spt6-YW mutation also caused increases in “nucleosome fuzziness”; i.e., the variability of nucleosome positions within the population, which we quantified as the standard deviation of dyad positions within each region occupied by a nucleosome. Median nucleosome fuzziness increased from 30.6 bp in wild type to 32.5 bp in spt6-YW at 30°C, and to 34.6 bp at 37°C (Fig. 2C). From these results, we conclude that the Spt6-Spn1 interaction controls internucleosome distance and variability in nucleosome positioning genome-wide.
Figure 2.
Spt6, FACT, Spt5, and Spn1 functionally interact to modulate nucleosome organization in vivo. (A) Average MNase-seq dyad signal over 3086 nonoverlapping verified coding genes aligned by the 30°C wild-type +1 nucleosome dyad, for wild-type (FY87), spt6-YW (FY3223), pob3-E154K (FY3206), spt6-YW pob3-E154K (FY3205), spt5-QS (FY3273), and spt6-YW spt5-QS (FY3274) strains grown at 30°C or with a shift to 37°C. Values are the mean of the mean library-size normalized coverage over the genes considered, over at least two replicates. (B) Mean differences in nucleosome position between mutant and wild type, quantified from MNase-seq data. Nucleosomes are grouped based on their position in the nucleosome array of a gene, with the +1 nucleosome defined as the nucleosome region with midpoint position immediately 3′ of the TSS. (C) Distributions of nucleosome fuzziness, defined as the standard deviation of MNase-seq dyad positions within a nucleosome region. Vertical lines indicate median values of each distribution.
Strikingly, both the pob3-E154K and spt5-QS mutations suppressed the spt6-YW defects in internucleosome distance and nucleosome fuzziness (Fig. 2A–C; Supplemental Fig. S2A). The spt5-QS suppressor rescued spt6-YW internucleosome distances to a greater degree than pob3-E154K (Fig. 2B), while pob3-E154K rescued spt6-YW nucleosome fuzziness to a greater degree than spt5-QS (Fig. 2C). As the nucleosome organization defects of spt6-YW are likely caused by the loss of Spt6-Spn1 interaction, we investigated this further by analyzing the temperature-sensitive spn1-K192N mutant, which has reduced interactions of Spn1 with Spt6 and RNAPII (Zhang et al. 2008). MNase-seq of spn1-K192N revealed nucleosome positioning and fuzziness defects similar to those of spt6-YW (Fig. 2B,C; Supplemental Fig. S2B). Additionally, the temperature sensitivity phenotype of spn1-K192N was suppressed by both pob3-E154K and spt5-QS (Supplemental Table S2). These results, then, suggest that Spt6 and Spn1 are together required for normal chromatin structure by a mechanism that can be modulated by either FACT or Spt5.
The spt6-YW mutation alters transcription genome-wide
Previous studies showed that Spt6 is required for transcriptional integrity, as when it is depleted there are widespread changes in initiation specificity and RNA levels (Cheung et al. 2008; Uwimana et al. 2017; Doris et al. 2018). To study the contribution of the Spt6-Spn1 interaction in transcription, we performed transcription start site-sequencing (TSS-seq) (Arribere and Gilbert 2013; Malabat et al. 2015; Doris et al. 2018) to quantitatively identify the 5′ ends of capped and polyadenylated transcripts. This analysis was carried out for wild-type and spt6-YW strains grown at 30°C and after a shift to 37°C. The previously studied spt6-1004 mutant was included for comparison as a condition when Spt6 protein is depleted (Doris et al. 2018). In contrast to spt6-1004, in an spt6-YW mutant, Spt6 protein was stable, both at 30°C and after a shift to 37°C (Supplemental Fig. S3A). Also in contrast to spt6-1004, spt6-YW did not alter the level of H3K36me2/me3, a histone modification known to regulate transcription (Carrozza et al. 2005; Chu et al. 2006; Li et al. 2007; Youdell et al. 2008; Gopalakrishnan et al. 2019). Thus, any transcriptional changes observed in spt6-YW would be independent of changes in the level of either Spt6 protein or H3K36 methylation.
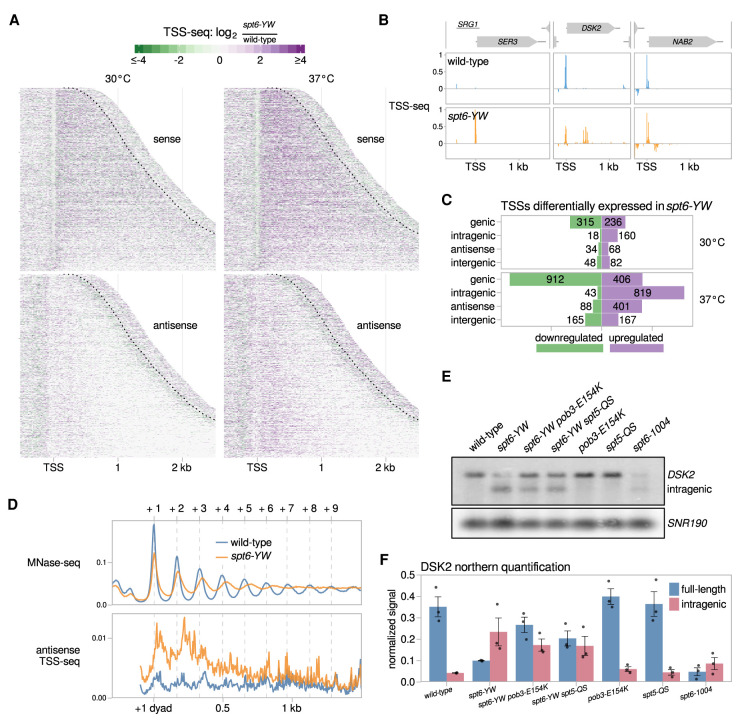
Our results showed that there are extensive changes in spt6-YW compared with wild type after the shift to 37°C (Fig. 3A,B; Supplemental Fig. S3B). Under these conditions, >1300 genic TSSs were misregulated and several hundred intragenic TSSs on both sense and antisense strands of genes were induced in the spt6-YW mutant (Fig. 3C). The intragenic TSSs induced in spt6-YW were mostly a subset of those induced in an spt6-1004 mutant (Supplemental Fig. S3C,D). In addition, the antisense TSSs induced in both mutants showed preferences for the 5′ ends of genes (Supplemental Fig. S3E), similar to antisense transcription observed in other cases (Kim et al. 2012; Smolle et al. 2012; Mayer et al. 2015; Lavender et al. 2016; Shetty et al. 2017). Interestingly, the mean antisense TSS-seq signal in spt6-YW was greatest in the regions between the average wild-type positions of the +1, +2, and +3 nucleosomes (Fig. 3D), suggesting a potential connection between these nucleosomes and antisense transcription.
Figure 3.
The spt6-YW mutation causes altered sense and antisense transcription. (A) Heat maps of the ratio of TSS-seq signal in the spt6-YW (FY3223) strain over wild type (FY87), grown at 30°C or with an 80 min shift to 37°C. Data are shown for the sense and antisense strands of 3087 nonoverlapping verified coding genes aligned by wild-type genic TSS and sorted by length. The region shown for each gene extends up to 300 nt 3′ from the cleavage and polyadenylation site (CPS), which is indicated by the dotted line. (B) Examples of altered mRNA level (SER3), intragenic initiation (DSK2), and antisense initiation (NAB2) in spt6-YW. Relative TSS-seq signal in wild-type and spt6-YW strains shifted to 37°C is shown for each region, with sense and antisense signals plotted above and below the X-axis, respectively. The signal is independently scaled for each region shown. (C) Bar plots showing the number of TSS-seq peaks differentially expressed in spt6-YW versus wild type. “Intragenic” and “antisense” refer to sense strand and antisense strand intragenic TSSs, respectively. (D, top panel) The average positions of the +1 through +9 nucleosome dyads in wild type and spt6-YW as determined from MNase-seq are indicated with vertical dashed lines. (Bottom panel) The median antisense TSS-seq signal in wild type and spt6-YW at 37°C, over 3086 nonoverlapping verified coding genes aligned by wild type +1 nucleosome dyad. (E) Northern analysis of the DSK2 gene after a shift to 37°C, using a probe from the 3′ region of DSK2, to assay DSK2 full-length and intragenic transcripts. SNR190 served as the loading control. (F) Quantification of the full-length and intragenic DSK2 transcript levels from three Northerns. Error bars indicate the mean ± standard error of the Northern signal for DSK2 normalized to the SNR190 signal.
As TSS-seq measures steady-state transcript levels, the intragenic and antisense transcripts in spt6-YW could result from increased synthesis and/or reduced degradation of these transcripts. To differentiate between the two, we performed native elongating transcript sequencing (NET-seq) to quantify elongating RNAPII in wild-type and spt6-YW strains. We then examined the NET-seq signal corresponding to antisense TSSs, as antisense transcription is not obscured by overlapping genic transcription. We observed increased NET-seq signal in spt6-YW versus wild type, consistent with increased synthesis contributing to the intragenic transcripts observed in spt6-YW (Supplemental Fig. S3F).
To test whether either pob3-E154K or spt5-QS suppresses the spt6-YW transcriptional changes, we used Northern blots to measure the transcript levels for two genes with altered RNA levels in spt6-YW: SER3, which is elevated, and DSK2, which is decreased. In both cases we observed suppression, although the effects were modest (Fig. 3E,F; Supplemental Fig. S3G,H). A more comprehensive approach will be required to discern the global degree of suppression. Overall, we can conclude that the Spt6-Spn1 interaction is required for normal transcription, possibly in coordination with FACT and Spt5.
Altered FACT bypasses the requirement for the Spt6-Spn1 interaction and for Spn1 itself
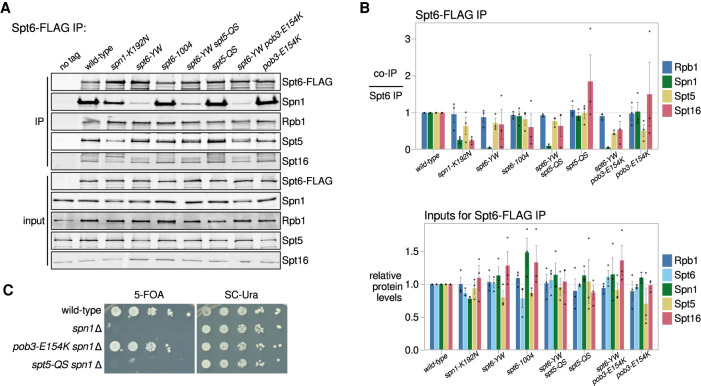
One obvious possible mechanism by which changes in FACT and Spt5 suppress spt6-YW would be by restoring the Spt6-Spn1 interaction. To test this, we conducted extensive co-IP experiments to assay interactions among five factors: Spt6, Spn1, FACT (Spt16), Spt5, and RNAPII (Rpb1 or Rpb3). In a wild-type strain, when we immunoprecipitated Spt6-FLAG, we observed co-IP of the other proteins, Spn1, Spt16, Spt5, and Rpb1, as expected (Fig. 4A,B; Lindstrom and Hartzog 2001; Krogan et al. 2002; Lindstrom et al. 2003). In spt6-YW extracts, while there was a 20-fold decrease in Spt6-Spn1 co-IP, the co-IPs of Spt6 with Spt16, Spt5, and Rpb1 were not significantly altered (Figs. 1A, 4A,B), supporting that spt6-YW specifically impairs binding of Spt6 with Spn1 without affecting other interactions of Spt6. Reciprocal co-IPs revealed that the association of Spn1 with RNAPII, Spt5, and Spt16 is also impaired in spt6-YW (Supplemental Fig. S4), suggesting that the Spt6-Spn1 interaction is necessary to recruit Spn1 to the elongation complex. Surprisingly, neither the pob3-E154K nor spt5-QS suppressor had an effect on the Spn1 co-IP profile in an spt6-YW background (Fig. 4A,B; Supplemental Fig. S4), suggesting that both suppressors circumvent the requirement for the Spt6-Spn1 interaction.
Figure 4.
The pob3-E154K and spt5-QS suppressors do not restore the Spt6-Spn1 interaction. (A) Western blots for Spt6-FLAG co-IP analysis, as in Figure 1A. Spt5 and Spt16 were detected using their respective polyclonal antibodies. (B) Quantification of Spt6-FLAG co-IP experiments (top) and inputs (bottom). Error bars indicate the mean ± standard error of the relative Western blot signal from the replicates shown. The co-IP signal was normalized to the Spt6-FLAG pull-down signal. (C) Assay for the ability of pob3-E154K or spt5-QS to suppress spn1Δ inviability. Growth in the presence of 5-fluoroorotic acid (5-FOA) indicates viability after the loss of a SPN1-URA3 plasmid as the sole source of Spn1. Strains were grown to saturation in YPD, serially diluted 10-fold, and spotted for growth on the indicated media.
Given our co-IP results, we asked whether the pob3-E154K or spt5-QS mutations allowed bypass of Spn1 genetically by testing whether either mutation could suppress the inviability caused by a complete deletion of SPN1 (spn1Δ). Our results showed that a spn1Δ spt5-QS double mutant was inviable (Fig. 4C; Supplemental Table S2), indicating that suppression of spt6-YW by spt5-QS is dependent on Spn1. However, a spn1Δ pob3-E154K double mutant was viable and grew comparably with a wild-type strain (Fig. 4C; Supplemental Table S2). Thus, the pob3-E154K mutation bypasses the requirement for Spn1, an essential histone chaperone.
Suppression by pob3-E154K does not restore the loss of Spn1 recruitment in an spt6-YW mutant
To examine the bypass of Spn1 by an independent approach, we studied Spn1 recruitment to chromatin, using chromatin immunoprecipitation sequencing (ChIP-seq) in wild-type and spt6-YW strains, with and without the pob3-E154K suppressor. To enable detection of global changes in Spn1 occupancy, we used exogenously added S. pombe chromatin for spike-in normalization, and to account for differences in Spn1 occupancy resulting from altered levels of transcription, we performed Rpb1 ChIP-seq from the same chromatin samples used for Spn1 ChIP-seq.
In wild type, Spn1 was distributed over coding genes at levels highly correlated to levels of Rbp1, as seen in previous studies (Supplemental Fig. S5A; Mayer et al. 2010; Reim et al. 2020). In contrast, in spt6-YW, Spn1 occupancy over coding genes was decreased to ∼18% of wild-type levels. This decrease occurred uniformly over the length of RNAPII-transcribed genes, including virtually all protein-coding genes (Fig. 5A; Supplemental Fig. S5B), snRNA genes, and snoRNA genes. Furthermore, the levels of Spn1 occupancy in spt6-YW were not rescued by pob3-E154K, consistent with the bypass of Spn1 (Fig. 5A; Supplemental Fig. S5B). Together, these results show that the Spt6-Spn1 interaction is required to recruit Spn1 to transcribed genes and provide additional evidence that pob3-E154K bypasses the requirement for Spn1.
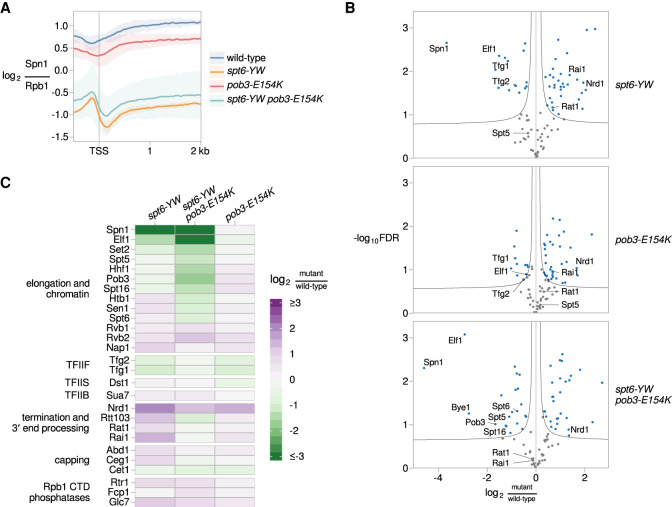
Figure 5.
Suppression by pob3-E154K does not restore the loss of Spn1 recruitment in an spt6-YW mutant. (A) The average Rpb1-normalized Spn1 ChIP enrichment over 3087 nonoverlapping verified coding genes aligned by TSS in wild type (FY3292), spt6-YW (FY3289), pob3-E154K (FY3294), and spt6-YW pob3-E154K (FY3293). The solid line and shading are the mean and 95% confidence interval of the mean ratio over the genes considered from two replicates. (B) Volcano plots comparing the Rpb3-FLAG interactome in spt6-YW, pob3-E154K, and spt6-YW pob3-E154K versus wild type, as measured by mass spectrometry. Fold changes and significance values are calculated from significance analysis of microarrays (Tusher et al. 2001), using either two (wild type, pob3-E154K, and spt6-YW pob3-E154K) or three (spt6-YW) replicates. Black lines indicate significance cutoffs at an FDR of 0.1 and s0 of 0.1. Each point is an Rpb3-interacting protein enriched in Rpb3-FLAG IP samples over untagged mock IP samples, with blue points indicating proteins significantly changed between strains. (C) A heat map of the ratio of mass spectrometry signal in spt6-YW, spt6-YW pob3-E154K, and pob3-E154K versus wild type, for selected RNAPII-interacting factors.
The RNAPII interactome is changed by the spt6-YW and the pob3-E154K mutations
To better understand the bypass of Spn1, we investigated how the spt6-YW and pob3-E154K mutations affect the RNAPII elongation complex. We immunopurified RNAPII complexes from wild-type, spt6-YW, pob3-E154K, and spt6-YW pob3-E154K strains and identified the copurified proteins by quantitative mass spectrometry (Materials and Methods). Similar to previous studies (Tardiff et al. 2007; Mosley et al. 2013; Harlen and Churchman 2017), we identified 89 Rpb3-interacting proteins that were significantly enriched in at least one strain when comparing an Rpb3 immunopurification with a mock immunopurification (Supplemental Table S3).
The mass spectrometry data recapitulated our co-IP results: Spn1 association was reduced 13-fold in spt6-YW compared with wild type, and this decrease was not rescued in the spt6-YW pob3-E154K double mutant (Fig. 5B,C). In addition to the depletion of Spn1, there were other smaller-scale changes in the spt6-YW strains (Fig. 5B,C), including reductions in the association of Elf1 and the TFIIF subunits Tfg1 and Tfg2, as well as increases in the association of the termination factors Rai1, Rat1, and Nrd1.
Interestingly, while some changes in the RNAPII interactome for spt6-YW were rescued by pob3-E154K, other changes were found specifically in the spt6-YW pob3-E154K double mutant and not in either single mutant. These changes included significantly reduced association of Spt6, Spt5, and both FACT subunits in the double mutant (Fig. 5B,C). This suggests that decreased transcription elongation in spt6-YW pob3-E154K may be one mechanism by which this double mutant overcomes loss of the Spt6-Spn1 interaction. Overall, our mass spectrometry results support the model that the major effect of spt6-YW is decreased interaction with Spn1 and that the pob3-E154K mutation bypasses the requirement for Spn1, possibly due to additional changes in the RNAPII interactome.
The Pob3 and Spt16 suppressors are clustered in the FACT domain that interfaces with nucleosomal DNA
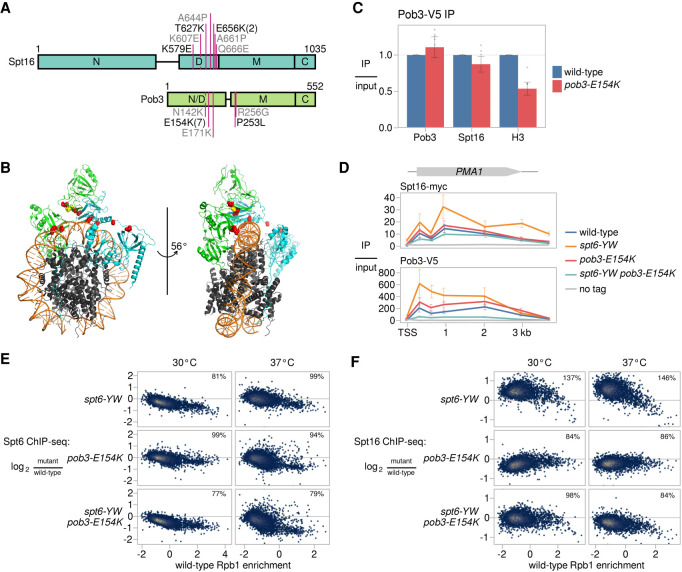
To gain insight into how changes in FACT might bypass the need for Spn1, we examined the locations of all five different pob3 and spt16 suppressors of spt6-YW, as well as eight additional pob3 and spt16 mutations that were isolated as suppressors of spn1Δ inviability, to be described elsewhere (F López-Rivera, J Chuang, R Gopalakrishnan, et al., unpubl.). Most of the amino acid changes caused by these mutations are charge changes of conserved residues and are clustered within the dimerization domains of Pob3 and Spt16 (Fig. 6A). The suppressor mutants are distinct in location from previously characterized pob3 and spt16 mutants that do not suppress spt6-YW, including pob3-272 (I282K) and spt16-197 (G132D) (Malone et al. 1991; Rowley et al. 1991; Lycan et al. 1994; Costa and Arndt 2000; Jamai et al. 2009; Feng et al. 2016). Remarkably, the conserved Spt16 and Pob3 residues changed by the suppressor mutations map to the inner surface of the “saddle” module of FACT (Fig. 6B), based on the cryogenic electron microscopy structure of human FACT bound to a subnucleosome (Liu et al. 2020). Since this region of FACT interfaces with nucleosomal DNA, structural analysis predicts that pob3-E154K would affect FACT-nucleosome interactions. We tested this prediction by immunoprecipitating Pob3 and assaying co-IP of histone H3 and Spt16. Our results show that pob3-E154K caused a twofold decrease in co-IP with histone H3, while the co-IP with Spt16 was unaffected (Fig. 6C). These results provide support for the recently proposed FACT-nucleosome structure (Liu et al. 2020) and suggest that pob3-E154K bypasses the requirement for Spn1 by weakening the interaction between FACT and nucleosomes.
Figure 6.
Mutant FACT suppresses the Spt6-Spn1 defect by restoring the balance between FACT and Spt6 on chromatin. (A) A schematic of the FACT subunits Spt16 and Pob3, depicting the amino acid changes caused by mutations suppressing spt6-YW (black) and spn1Δ (gray). The numbers in parentheses indicate the number of times a mutation was isolated if it was more than once. The labeled rectangles represent the domains of FACT. (N) N-terminal, (D) dimerization, (M) middle,(C) C-terminal. (B) The structure of human FACT bound to a subnucleosomal particle (PDB: 6UPL) (Liu et al. 2020), highlighting conserved residues corresponding to the locations of the yeast suppressor changes shown in A. Human Spt16 (cyan), SSRP1 (green), and histones (gray) are shown as ribbon diagrams. The suppressor residues are shown as red spheres, except for SSRP1-E149, which corresponds to yeast Pob3-E154 and is shown as yellow spheres. Nucleosomal DNA is shown in orange. (C) Quantification of Pob3-V5 co-IP experiments. Error bars indicate the mean ± standard error of relative co-IP signal normalized to Pob3-V5 pull-down signal in the replicates shown. (D) ChIP analysis of Spt16 and Pob3 over the PMA1 gene. The diagram shows Spt16 and Pob3 ChIP enrichment over input based on ChIP-qPCR measurements at the PMA1 gene in wild-type, spt6-YW, pob3-E154K, and spt6-YW pob3-E154K strains. Error bars indicate the mean ± standard error of two replicates for each qPCR amplicon. (E) Scatter plots showing change in Spt6 ChIP enrichment in mutants (FY3277, FY3281, FY3282) over wild type (FY3276) versus wild-type Rpb1 ChIP enrichment for 5091 verified coding genes. Rpb1 enrichment values are the relative log2 enrichment of IP over input. (F) As in E, but for Spt16 ChIP enrichment, using strains FY3299-3302.
Evidence that the Spt6:FACT ratio on chromatin is critical for their functions
Our co-IP results suggest that the pob3-E154K mutation causes weakened FACT-chromatin interactions. To test this possibility and to see whether it might be connected to suppression of spt6-YW, we performed ChIP-seq of Spt6 and Spt16 in our mutants. First, our results showed that, in most cases, the median recruitment of the mutant Spt6-YW protein was reduced to ∼80% of wild-type Spt6 (Fig. 6E; Supplemental Fig. S6A). The reduction in Spt6-YW occupancy was detected in both the POB3 and the pob3-E154K backgrounds. In the one case where a reduction was not observed, the spt6-YW single mutant after a shift to 37°C, this was likely caused by the increased occupancy of RNAPII over gene bodies, particularly near 5′ ends (Supplemental Fig. S6B). Overall, these data suggest that spt6-YW caused a moderate decrease in Spt6 occupancy. This decrease is likely the result of the lack of Spn1 recruitment in this mutant (Reim et al. 2020).
In contrast to the decreased level of recruitment of Spt6-YW, Spt16 recruitment was broadly elevated in the spt6-YW mutant (Fig. 6D,F), revealing a major imbalance of the levels of these three essential histone chaperones: Spt6 (decreased), Spn1 (barely detectable), and FACT (increased). In the spt6-YW pob3-E154K double mutant, the recruitment of Spt16 was either restored to wild-type levels (at 30°C) or modestly decreased (at 37°C), suggesting that the elevated FACT recruitment caused by spt6-YW was suppressed by pob3-E154K. In the pob3-E154K single mutant, there was a moderate decrease in Spt16 occupancy over most genes, particularly after a shift to 37°C (Fig. 6F; Supplemental Fig. S6A), consistent with the structural prediction and co-IP results that pob3-E154K causes decreased association of FACT with nucleosomes.
We note that when we normalized Spt16 occupancy by Rpb1 ChIP-seq levels, we observed that the relative Spt16 occupancies are equivalent between wild type and the spt6-YW mutant, suggesting that the altered recruitment of Spt16 in spt6-YW is coupled to changes in transcription (Supplemental Fig. S6A). From our data, we cannot distinguish whether increased FACT recruitment alters transcription or vice versa. However, regardless of the causal direction of this relationship, our data show that spt6-YW causes increased recruitment of Spt16 and that this increase is suppressed by pob3-E154K. From these results, we propose a model in which the levels of Spt6 and FACT on chromatin must be properly balanced for optimal function and that Spt6 and/or Spn1 modulate the level of FACT association with chromatin.
Discussion
In this work, we have discovered several functional interactions between three essential and conserved histone chaperones, Spt6, Spn1, and FACT, during transcription in S. cerevisiae. First, we showed that disruption of the Spt6-Spn1 physical interaction by spt6-YW impairs recruitment of Spn1 to the elongation complex, resulting in widespread changes in both transcription and chromatin structure. Second, we identified suppressors of spt6-YW, revealing functional interactions with several regulators of chromatin and transcription. Third, focusing on two novel sets of suppressor mutations, in FACT (SPT16 and POB3) and in SPT5, we showed that they suppress spt6-YW by distinct mechanisms. Fourth, while our understanding of suppression by spt5-QS remains unclear, we demonstrated that a change in FACT bypasses the need for Spn1, likely by restoring a balanced level of FACT and Spt6 associated with chromatin. Taken together, our studies have revealed previously unknown and surprising connections between FACT activity and the functions of Spt6 and Spn1.
One striking result from our studies was the elevated level of FACT that was recruited to chromatin in an spt6-YW mutant, altering the normal ratio of chromatin-associated Spt6 and FACT. We propose a model for this increased level based on recent evidence that FACT associates with an altered nucleosome structure (Martin et al. 2018). In our model, Spt6 and Spn1 normally prevent accumulation of such altered nucleosomes during transcription. However, in an spt6-YW mutant, there is a disruption of normal chromatin structure, possibly due to loss of Spn1 recruitment, causing an increased level of altered nucleosomes, leading to increased recruitment of FACT. Interestingly, FACT levels and chromatin association are increased in cancer cells, consistent with the possibility that higher levels of FACT induce alterations in growth and transcription (Chang et al. 2018).
Suppression of spt6-YW by pob3-E154K likely occurs by a weakened FACT-nucleosome interaction, resulting in a balanced level of FACT and Spt6 on chromatin. We note that, during growth at 37°C, the levels of chromatin-bound Spt6 and FACT are both reduced in the spt6-YW pob3-E154K double mutant (Fig. 6E,F; Supplemental Figure S6A), yet the strain grows almost as well as wild type (Fig. 1C). This supports the idea that it is the Spt6:FACT ratio, rather than the absolute levels, that are critical for function. There is precedent for this among proteins that form structures, such as bacteriophage heads (Floor 1970; Sternberg 1976) and histone proteins (Meeks-Wagner and Hartwell 1986; Clark-Adams et al. 1988). As the functions of Spt6, FACT, and Spt5 are all sensitive to altered levels (Clark-Adams and Winston 1987; Malone et al. 1991; Swanson et al. 1991), it seems likely that these proteins function by a mechanism that requires a specific stoichiometry. Our results have provided insight into how this stoichiometry might be maintained, as Spt6 directly recruits Spn1, and Spt6-Spn1 modulates the chromatin association of FACT. This network may be important in processes in addition to transcription, as FACT is also required for DNA replication (Formosa and Winston 2020) and all three chaperones have been suggested to be required for genome stability (Herrera-Moyano et al. 2014; Nojima et al. 2018; Thurston et al. 2018).
An unexpected result from our studies was the discovery that the spt6-YW and spn1-K192N mutations cause an increase in internucleosome distances. Although we do not know the exact impact of this change on chromatin in our mutants, it might affect higher-order chromatin folding, chromatin compaction, and DNA accessibility (Correll et al. 2012; Li et al. 2016), possibly leading to the transcriptional changes observed in spt6-YW. An increase in internucleosome distances has been observed in mutants that impair several other factors (Lombardi et al. 2011; Vasseur et al. 2016; McCullough et al. 2019; Prajapati et al. 2020). Interestingly, we show that inactivation of one of these factors, Chd1, suppresses spt6-YW and spn1-K192N mutant phenotypes (Supplemental Table S2). We cannot distinguish whether Spt6 and Spn1 directly control spacing as part of their interactions with histones or whether they function with other proteins. Whatever the mechanism, it also requires FACT and Spt5, as the change in the internucleosome distances observed in spt6-YW is strongly suppressed by both the spt5-QS and pob3-E154K suppressors. We note that our results provide the first in vivo demonstration that Spt5 controls nucleosome organization, complementing recent structural and in vitro studies that implicated Spt5 in facilitating transcription past nucleosomes (Crickard et al. 2017; Ehara et al. 2019).
In conclusion, our studies have shed light on a network of interactions between histone chaperones that controls transcription and chromatin structure. Another recent study has shown that this network extends beyond Spt6, Spn1, and FACT to additional histone chaperones (Jeronimo et al. 2019). Given the essential and conserved nature of the three chaperones we studied, Spt6, Spn1, and FACT, it is surprising that complete loss of Spn1 can be strongly compensated by a single amino acid change in FACT. In spite of this, there must be strong selection to maintain this network of factors over evolutionary time, something that will be understood in greater depth as we learn the full range of the functions of these factors.
Materials and methods
Yeast strains, media, and growth conditions
All S. cerevisiae strains (Supplemental Table S4) are in the S288C background (Winston et al. 1995) and were constructed by either yeast transformation or crosses. Oligonucleotides and plasmids are listed in Supplemental Table S5. For TSS-seq, NET-seq, ChIP-seq, and MNase-seq, yeast cultures were grown in YPD at 30°C or were shifted to growth for 80 min at 37°C (Cheung et al. 2008). The shift for 80 min to 37°C did not greatly affect the viability of the spt6-YW and spn1-K192N mutants, resulting in 0.83 ± 0.17 and 0.85 ± 0.07 cell survival, respectively, compared with 30°C. YPD plates were supplemented with hydroxyurea at a final concentration of 150 mM or phleomycin at 13 μg/mL (Diebold et al. 2010).
Coimmunoprecipitation
Coimmunoprecipitations for Spt6, Spn1, and Rpb3 were performed with antibody-conjugated beads: either the anti-FLAG M2-FLAG affinity gel (20 µL per IP; Sigma) or anti-V5-conjugated magnetic beads (30 µL per IP; MBL International Corporation) as previously described (Reim et al. 2020). For FACT-histone co-IP experiments, the cell extracts were prepared using buffer B for lysis and coimmunoprecipitation (100 mM HEPES at pH 7.9, 20% glycerol, 1 mM EDTA, 25 mM magnesium acetate, 0.4% NP-40 [IGEPAL, Sigma], 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail [Sigma]) followed by the pulldowns as described. The eluates were analyzed using Western blotting. The antibodies are listed in Supplemental Table S5.
Isolation and analysis of spt6-YW suppressors
Yeast strains FY3019, FY3297, and FY3298 were used to isolate spontaneous and UV-induced suppressors of the spt6-YW temperature-sensitive (Ts−) phenotype as follows. Independent cultures were inoculated from single colonies and grown overnight to saturation in liquid YPD medium. Then, 200 µL of each culture was plated on duplicate YPD plates, with one of them UV-irradiated (Winston 2008). The plates were incubated at 37°C and colonies were purified after either the third or fifth day of incubation, yielding 52 independent suppressor candidates. Three purified colonies of each candidate were retested for suppression of the Ts− phenotype. Genetic analysis was performed for the confirmed suppressor strains, excluding those that did not sporulate. The remaining 38 strains were crossed to an spt6-YW strain to test for single-gene segregation of the suppressor phenotype. Three strains had suppressors tightly linked to SPT6. Sanger sequencing of the SPT6 open reading frame for SPT6 from these independent suppressors uncovered the same nucleotide change resulting in a P231L substitution. The genetic analysis resulted in the identification of 25 strains with suppressor mutations (Supplemental Table S1).
Identification of suppressor mutations
To identify the causative suppressor mutations, we performed pooled linkage analysis, followed by whole-genome sequencing (WGS) (Birkeland et al. 2010). For 20 of the suppressors, we crossed a suppressor strain (spt6-YW sup) with a parental spt6-YW strain, pooled an equal number of suppressor and nonsuppressor progeny (12–50 segregants per pool), and sequenced the two pools. DNA from each pool was extracted and used to generate genomic libraries (Gopalakrishnan and Winston 2019). Single-read sequencing was performed using an Illumina MiSeq according to the manufacturer's instructions, or on an Illumina HiSeq 2500 by the Harvard Bauer Core facility. The WGS data were processed using a custom pipeline (Gopalakrishnan and Winston 2019) to identify point mutations unique to the suppressor pools, and to monitor ploidy in the suppressor strains. Additional analysis for identification of polymorphisms was performed using Geneious Prime version 2019.0.3 (Kearse et al. 2012). The identified candidate mutations were verified by Sanger sequencing, and the ability of each mutation to confer suppression was verified by genetic tests (Supplemental Table S1), including (1) complementation using plasmids expressing the wild-type candidate genes, (2) reconstitution of the suppressor phenotype by null alleles of nonessential genes, and (3) allele replacement of essential genes. In parallel, while the WGS results for a subset of suppressors were emerging, we used linkage analysis and plasmid complementation to screen the remaining suppressors for mutations in already identified genes. This screen revealed that five suppressors likely contained mutations in POB3, which was further validated by Sanger sequencing identifying a pob3-E154K allele in each of these independent suppressors.
Micrococcal nuclease sequencing
Cultures (500 mL) for strains FY87, FY3125, FY3223, FY3205, FY3206, and FY3272-3274 were grown in YPD at 30°C and also after a shift to 37°C for 80 min. The cells were processed and MNase-seq libraries constructed and sequenced as previously described (Doris et al. 2018).
Transcription start site sequencing (TSS-seq), native elongating transcript sequencing (NET-seq), and Northern blotting
TSS-seq was performed as previously described (Doris et al. 2018). Analysis was done on yeast strains FY87, FY3223, and FY3125. TSS-seq libraries were single-read-sequenced on an Illumina HiSeq 2500 at the Harvard Bauer Core Facility. NET-seq was done as previously described (Churchman and Weissman 2011). Northern blotting was performed as previously described (Gopalakrishnan et al. 2019). The primer pairs used to generate Northern probes are listed in Supplemental Table S5.
Mass spectrometry analysis of the RNA polymerase II (RNAPII) interactome
Yeast strains bearing a C-terminal triple FLAG tag on Rpb3 were grown as 1 L of cultures to OD600 ∼0.8 in duplicate (FY2912, FY3287, FY3288) or triplicate (FY3019). Cells were collected by filtration and flash-frozen in liquid nitrogen. The cells were lysed in a mixer-mill using eight cycles at 15 Hz using buffer A (20 mM HEPES at pH 7.6, 20% glycerol, 1 mM DTT, 1 mM EDTA, 125 mM potassium acetate, 1% NP-40, 1× protease inhibitors [Sigma], 1× phosphatase inhibitors [Sigma]). The lysates were diluted to have equal total protein concentration, and Rpb3 was immunoprecipitated using 200 μL of FLAG M2 beads (Sigma). Samples were then incubated for 2 h at 4°C on a roller, washed three times with buffer A, and eluted with FLAG peptide (0.25 mg/mL, 10 mM Tris at pH 7.4, 150 mM NaCl, 10% glycerol). The eluates were submitted to the Thermo Fisher Center for Multiplexed Proteomics (Harvard Medical School) for tandem mass tag (TMT)-based mass spectrometry analysis (Zhang and Elias 2017) according to the standard workflow. The resulting peptide spectra were searched using the SEQUEST algorithm against a Uniprot composite database for the S. cerevisiae proteome and known contaminants. Peptide spectral matches were filtered to a 1% false discovery rate (FDR) using the target-decoy strategy combined with linear discriminant analysis. Proteins were quantified only from peptides with a summed SN threshold of ≥100 and MS2 isolation specificity of 0.5. Differential protein abundance analysis was performed for proteins with two or more identified peptides, using the Perseus software platform (Hubner et al. 2010; Hubner and Mann 2011) as previously described (Harlen and Churchman 2017).
Chromatin immunoprecipitation
For Spt6 and Spt16 ChIP-seq studies, yeast strains containing Spt6 fused with the triple FLAG epitope tag (FY3276, FY3277, FY3281, and FY3282) or Myc-tagged Spt16 fusions (FY3299-3302) were grown in YPD at 30°C or shifted to 37°C as described above. For Spn1 ChIP-seq and for Pob3 ChIP-qPCR, strains containing an N-terminal V5 epitope tag on Spn1 (FY3289 and FY3292-3294) or C-terminal triple V5 tag on Pob3 (FY3303-3306) were grown in YPD at 30°C. The cultures were processed for cross-linking and collection as previously described (Doris et al. 2018). Chromatin was prepared using standard methods (Gopalakrishnan et al. 2019). Each chromatin sample used for ChIP-seq was mixed with S. pombe chromatin (strain FWP570) at 10% level by protein mass for spike-in normalization and split into aliquots of 500 µg of chromatin for immunoprecipitation. ChIP-qPCR assays for Spt16 and Pob3, depicted on Figure 6D, were performed without spike-in normalization. Chromatin precipitations were performed using either 50 µL of anti-FLAG M2 affinity gel (Sigma) for Spt6, 30 µL of anti-Myc antibodies (Santa Cruz Biotechnology 9E10), 8 µL of 8WG16 antibodies (Millipore Sigma) for Rpb1, or 5 µL of anti-V5 antibodies (Invitrogen) per 500 µg of chromatin and the DNA library generated as previously described (Gopalakrishnan et al. 2019). ChIP-qPCR was performed using Brilliant III Ultra-Fast SYBR Green qPCR mix (Agilent). The antibodies, oligonucleotides, and main reagents are listed in Supplemental Table S5.
Data analysis and management
Processing and analysis of the TSS-seq, NET-seq, and MNase-seq data sets were largely performed as previously described (Doris et al. 2018). Analysis of the ChIP-seq data sets was done as in Reim et al. (2020). The details on these analyses and modifications are summarized in the Supplemental Material. All data analyses except for mass spectrometry analyses and polymorphism identification using Geneious were managed using the Snakemake workflow management system (Koster and Rahmann 2012).
Data and code availability
All high-throughput sequencing data except for whole-genome sequencing data are available on GEO under accession number GSE160821. An archive containing code and raw data for reproducing all analyses except for whole-genome sequencing and mass spectrometry analyses is available at Zenodo (https://doi.org/10.5281/zenodo.4174464). Additionally, updated versions of the Snakemake pipelines used are available at http://github.com/winston-lab.
Supplementary Material
Acknowledgments
We thank Laurie Stargell for Spn1 antisera; Karen Arndt for H3 antisera; Tim Formosa for sharing yeast strains, plasmids, and Spt6 and Spt16 antisera; Karolin Luger for helpful discussions; and Sarah Boswell and Mike Springer for use of their MiSeq. We also thank Karen Arndt and Catherine Weiner for helpful comments on the manuscript, and Isabelle Washkurak for help with some of the experiments. Part of this research was conducted on the O2 High-Performance Computing Cluster supported by the Research Computing Group at Harvard Medical School. This work was supported by National Institutes of Health (NIH) Fellowship F32GM119291 to O.V., a Ford Foundation Predoctoral Fellowship to F.L.-R., NIH Fellowship F31GM112370 to N.I.R., NIH grant R01HG007173 to L.S.C., and NIH grants R01GM032967 and R01GM120038 to F.W.
Author contributions: O.V. and F.W. designed most of the experiments. O.V. performed the TSS-seq, ChIP-seq, MNase-seq, and co-IP experiments, and isolated and performed the genetic analysis of the spt6-YW suppressors. J.C. and D.J. performed and interpreted the bioinformatic analyses, with equal contributions, and D.J. working under the supervision of P.J.P. F.L.-R. isolated and analyzed suppressors of spn1Δ. N.I.R. performed the Western blots to measure protein levels in wild-type and mutant backgrounds and contributed to the genetic studies. M.M., F.W., and L.S.C. designed the NET-seq experiments and M.M. performed them. D.S. performed the single-gene ChIP experiments and Northern blots. O.V., J.C., and F.W. wrote the manuscript with feedback from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348431.121.
Competing interest statement
The authors declare no competing interests.
References
- Arribere JA, Gilbert WV. 2013. Roles for transcript leaders in translation and mRNA decay revealed by transcript leader sequencing. Genome Res 23: 977–987. 10.1101/gr.150342.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland SR, Jin N, Ozdemir AC, Lyons RH Jr., Weisman LS, Wilson TE. 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186: 1127–1137. 10.1534/genetics.110.123232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Dutta-Biswas R, Stillman DJ. 2007. Chd1 and yFACT act in opposition in regulating transcription. Mol Cell Biol 27: 6279–6287. 10.1128/MCB.00978-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Winston F. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272: 1473–1476. 10.1126/science.272.5267.1473 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- Chang HW, Valieva ME, Safina A, Chereji RV, Wang J, Kulaeva OI, Morozov AV, Kirpichnikov MP, Feofanov AV, Gurova KV, et al. 2018. Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci Adv 4: eaav2131. 10.1126/sciadv.aav2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277. 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Sutton A, Sternglanz R, Prelich G. 2006. The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol Cell Biol 26: 3029–3038. 10.1128/MCB.26.8.3029-3038.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. 10.1038/nature09652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams CD, Winston F. 1987. The SPT6 gene is essential for growth and is required for δ-mediated transcription in Saccharomyces cerevisiae. Mol Cell Biol 7: 679–686. 10.1128/MCB.7.2.679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F. 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev 2: 150–159. 10.1101/gad.2.2.150 [DOI] [PubMed] [Google Scholar]
- Correll SJ, Schubert MH, Grigoryev SA. 2012. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J 31: 2416–2426. 10.1038/emboj.2012.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156: 535–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickard JB, Lee J, Lee TH, Reese JC. 2017. The elongation factor Spt4/5 regulates RNA polymerase II transcription through the nucleosome. Nucleic Acids Res 45: 6362–6374. 10.1093/nar/gkx220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta CE, Hildreth AE, McShane BM, Shirra MK, Arndt KM. 2019. The nucleosome acidic patch directly interacts with subunits of the Paf1 and FACT complexes and controls chromatin architecture in vivo. Nucleic Acids Res 47: 8410–8423. 10.1093/nar/gkz549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold ML, Koch M, Loeliger E, Cura V, Winston F, Cavarelli J, Romier C. 2010. The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26. EMBO J 29: 3979-3991. 10.1038/emboj.2010.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doris SM, Chuang J, Viktorovskaya O, Murawska M, Spatt D, Churchman LS, Winston F. 2018. Spt6 is required for the fidelity of promoter selection. Mol Cell 72: 687–699.e6. 10.1016/j.molcel.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dronamraju R, Hepperla AJ, Shibata Y, Adams AT, Magnuson T, Davis IJ, Strahl BD. 2018. Spt6 association with RNA polymerase II directs mRNA turnover during transcription. Mol Cell 70: 1054–1066.e4. 10.1016/j.molcel.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duina AA. 2011. Histone chaperones Spt6 and FACT: similarities and differences in modes of action at transcribed genes. Genet Res Int 2011: 625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara H, Kujirai T, Fujino Y, Shirouzu M, Kurumizaka H, Sekine SI. 2019. Structural insight into nucleosome transcription by RNA polymerase II with elongation factors. Science 363: 744–747. 10.1126/science.aav8912 [DOI] [PubMed] [Google Scholar]
- English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. 2006. Structural basis for the histone chaperone activity of Asf1. Cell 127: 495–508. 10.1016/j.cell.2006.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Gan H, Eaton ML, Zhou H, Li S, Belsky JA, MacAlpine DM, Zhang Z, Li Q. 2016. Noncoding transcription is a driving force for nucleosome instability in spt16 mutant cells. Mol Cell Biol 36: 1856–1867. 10.1128/MCB.00152-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. 2008. H2b ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31: 57–66. 10.1016/j.molcel.2008.04.025 [DOI] [PubMed] [Google Scholar]
- Floor E. 1970. Interaction of morphogenetic genes of bacteriophage T4. J Mol Biol 47: 293–306. 10.1016/0022-2836(70)90303-7 [DOI] [PubMed] [Google Scholar]
- Formosa T, Winston F. 2020. The role of FACT in managing chromatin: disruption, assembly, or repair? Nucleic Acids Res 48: 11929–11941. 10.1093/nar/gkaa912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan R, Winston F. 2019. Whole-genome sequencing of yeast cells. Curr Protoc Mol Biol 128: e103. 10.1002/cpmb.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan R, Marr SK, Kingston RE, Winston F. 2019. A conserved genetic interaction between Spt6 and Set2 regulates H3K36 methylation. Nucleic Acids Res 47: 3888–3903. 10.1093/nar/gkz119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurova K, Chang HW, Valieva ME, Sandlesh P, Studitsky VM. 2018. Structure and function of the histone chaperone FACT—resolving FACTual issues. Biochim Biophys Acta Gene Regul Mech 1861: 892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A. 2017. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 18: 141–158. 10.1038/nrm.2016.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlen KM, Churchman LS. 2017. Subgenic Pol II interactomes identify region-specific transcription elongation regulators. Mol Syst Biol 13: 900. 10.15252/msb.20167279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Fu J. 2013. The Spt4-Spt5 complex: a multi-faceted regulator of transcription elongation. Biochim Biophys Acta 1829: 105–115. 10.1016/j.bbagrm.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Moyano E, Mergui X, Garcia-Rubio ML, Barroso S, Aguilera A. 2014. The yeast and human FACT chromatin-reorganizing complexes solve R-loop-mediated transcription-replication conflicts. Genes Dev 28: 735–748. 10.1101/gad.234070.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges AJ, Gloss LM, Wyrick JJ. 2017. Residues in the nucleosome acidic patch regulate histone occupancy and are important for FACT binding in Saccharomyces cerevisiae. Genetics 206: 1339–1348. 10.1534/genetics.117.201939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Mann M. 2011. Extracting gene function from protein-protein interactions using Quantitative BAC InteraCtomics (QUBIC). Methods 53: 453–459. 10.1016/j.ymeth.2010.12.016 [DOI] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754. 10.1083/jcb.200911091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I, Jacques PE, Rando OJ, Robert F, Winston F. 2011. Control of chromatin structure by Spt6: different consequences in coding and regulatory regions. Mol Cell Biol 31: 531–541. 10.1128/MCB.01068-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Puglisi A, Strubin M. 2009. Histone chaperone Spt16 promotes redeposition of the original H3-H4 histones evicted by elongating RNA polymerase. Mol Cell 35: 377–383. 10.1016/j.molcel.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Jeronimo C, Watanabe S, Kaplan CD, Peterson CL, Robert F. 2015. The histone chaperones FACT and Spt6 restrict H2A.Z from intragenic locations. Mol Cell 58: 1113–1123. 10.1016/j.molcel.2015.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Poitras C, Robert F. 2019. Histone recycling by FACT and Spt6 during transcription prevents the scrambling of histone modifications. Cell Rep 28: 1206–1218.e8. 10.1016/j.celrep.2019.06.097 [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. 10.1126/science.1087374 [DOI] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. 2005. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123: 593–605. 10.1016/j.cell.2005.10.025 [DOI] [PubMed] [Google Scholar]
- Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. 2012. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150: 1158–1169. 10.1016/j.cell.2012.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster J, Rahmann S. 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522. 10.1093/bioinformatics/bts480 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992. 10.1128/MCB.22.20.6979-6992.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavender CA, Cannady KR, Hoffman JA, Trotter KW, Gilchrist DA, Bennett BD, Burkholder AB, Burd CJ, Fargo DC, Archer TK. 2016. Downstream antisense transcription predicts genomic features that define the specific chromatin environment at mammalian promoters. PLoS Genet 12: e1006224. 10.1371/journal.pgen.1006224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Ranger M, Meneghini MD. 2018. Combinatorial genetic control of Rpd3S through histone H3K4 and H3K36 methylation in budding yeast. G3 8: 3411–3420. 10.1534/g3.118.200589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Pattenden SG, Seidel C, Workman JL. 2007. Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev 21: 1422–1430. 10.1101/gad.1539307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen P, Yu J, Dong L, Liang D, Feng J, Yan J, Wang PY, Li Q, Zhang Z, et al. 2016. FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol Cell 64: 120–133. 10.1016/j.molcel.2016.08.024 [DOI] [PubMed] [Google Scholar]
- Li S, Almeida AR, Radebaugh CA, Zhang L, Chen X, Huang L, Thurston AK, Kalashnikova AA, Hansen JC, Luger K, et al. 2018. The elongation factor Spn1 is a multi-functional chromatin binding protein. Nucleic Acids Res 46: 2321–2334. 10.1093/nar/gkx1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Hartzog GA. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR 3rd, Hartzog GA. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol Cell Biol 23: 1368–1378. 10.1128/MCB.23.4.1368-1378.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, Zhang Z, Carragher B, Potter CS, D'Arcy S, Luger K. 2020. FACT caught in the act of manipulating the nucleosome. Nature 577: 426–431. 10.1038/s41586-019-1820-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi LM, Ellahi A, Rine J. 2011. Direct regulation of nucleosome density by the conserved AAA-ATPase Yta7. Proc Natl Acad Sci 108: E1302–E1311. 10.1073/pnas.1116819108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycan D, Mikesell G, Bunger M, Breeden L. 1994. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol Cell Biol 14: 7455–7465. 10.1128/MCB.14.11.7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabat C, Feuerbach F, Ma L, Saveanu C, Jacquier A. 2015. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. Elife 4: e06722. 10.7554/eLife.06722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone EA, Clark CD, Chiang A, Winston F. 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol Cell Biol 11: 5710–5717. 10.1128/MCB.11.11.5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin BJE, Chruscicki AT, Howe LJ. 2018. Transcription promotes the interaction of the FAcilitates Chromatin Transactions (FACT) complex with nucleosomes in Saccharomyces cerevisiae. Genetics 210: 869–881. 10.1534/genetics.118.301349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. 2010. Uniform transitions of the general RNA polymerase II transcription complex. Nat Struct Mol Biol 17: 1272–1278. 10.1038/nsmb.1903 [DOI] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. 2015. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough L, Connell Z, Petersen C, Formosa T. 2015. The abundant histone chaperones Spt6 and FACT collaborate to assemble, inspect, and maintain chromatin structure in Saccharomyces cerevisiae. Genetics 201: 1031–1045. 10.1534/genetics.115.180794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough LL, Pham TH, Parnell TJ, Connell Z, Chandrasekharan MB, Stillman DJ, Formosa T. 2019. Establishment and maintenance of chromatin architecture are promoted independently of transcription by the histone chaperone FACT and H3-K56 acetylation in Saccharomyces cerevisiae. Genetics 211: 877–892. 10.1534/genetics.118.301853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald SM, Close D, Xin H, Formosa T, Hill CP. 2010. Structure and biological importance of the Spn1-Spt6 interaction, and its regulatory role in nucleosome binding. Mol Cell 40: 725–735. 10.1016/j.molcel.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D, Hartwell LH. 1986. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell 44: 43–52. 10.1016/0092-8674(86)90483-6 [DOI] [PubMed] [Google Scholar]
- Mosley AL, Hunter GO, Sardiu ME, Smolle M, Workman JL, Florens L, Washburn MP. 2013. Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcriptional elongation. Mol Cell Proteomics 12: 1530–1538. 10.1074/mcp.M112.024034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawska M, Schauer T, Matsuda A, Wilson MD, Pysik T, Wojcik F, Muir TW, Hiraoka Y, Straub T, Ladurner AG. 2020. The chaperone FACT and histone H2B ubiquitination maintain S. pombe genome architecture through genic and subtelomeric functions. Mol Cell 77: 501–513 e507. 10.1016/j.molcel.2019.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Tellier M, Foxwell J, Ribeiro de Almeida C, Tan-Wong SM, Dhir S, Dujardin G, Dhir A, Murphy S, Proudfoot NJ. 2018. Deregulated expression of mammalian lncRNA through loss of SPT6 induces R-loop formation, replication stress, and cellular senescence. Mol Cell 72: 970–984.e7. 10.1016/j.molcel.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R, Singh P, Ananthakrishnan S, Adamczyk S, Schimmel O, Govind CK. 2018. Acetylation-dependent recruitment of the FACT complex and its role in regulating Pol II occupancy genome-wide in Saccharomyces cerevisiae. Genetics 209: 743–756. 10.1534/genetics.118.300943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales R, Erickson B, Zhang L, Kim H, Valiquett E, Bentley D. 2013. Gene promoters dictate histone occupancy within genes. EMBO J 32: 2645–2656. 10.1038/emboj.2013.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prajapati HK, Ocampo J, Clark DJ. 2020. Interplay among ATP-dependent chromatin remodelers determines chromatin organisation in yeast. Biology 9: 190. 10.3390/biology9080190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan TK, Hartzog GA. 2010. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4-Spt5 in transcription. Genetics 184: 321–334. 10.1534/genetics.109.111526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim NI, Chuang J, Jain D, Alver BH, Park PJ, Winston F. 2020. The conserved elongation factor Spn1 is required for normal transcription, histone modifications, and splicing in Saccharomyces cerevisiae. Nucleic Acids Res 48: 10241–10258. 10.1093/nar/gkaa745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Singer RA, Johnston GC. 1991. CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol Cell Biol 11: 5718–5726. 10.1128/MCB.11.11.5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier AC, Taatjes DJ. 2020. Structure and mechanism of the RNA polymerase II transcription machinery. Genes Dev 34: 465–488. 10.1101/gad.335679.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sdano MA, Fulcher JM, Palani S, Chandrasekharan MB, Parnell TJ, Whitby FG, Formosa T, Hill CP. 2017. A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription. Elife 6: e28723. 10.7554/eLife.28723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty A, Kallgren SP, Demel C, Maier KC, Spatt D, Alver BH, Cramer P, Park PJ, Winston F. 2017. Spt5 plays vital roles in the control of sense and antisense transcription elongation. Mol Cell 66: 77–88 e75. 10.1016/j.molcel.2017.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856. 10.1093/emboj/cdg179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol 19: 884–892. 10.1038/nsmb.2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N. 1976. A genetic analysis of bacteriophage λ head assembly. Virology 71: 568–582. 10.1016/0042-6822(76)90382-2 [DOI] [PubMed] [Google Scholar]
- Swanson MS, Winston F. 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132: 325–336. 10.1093/genetics/132.2.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Malone EA, Winston F. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol Cell Biol 11: 3009–3019. 10.1128/MCB.11.6.3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff DF, Abruzzi KC, Rosbash M. 2007. Protein characterization of Saccharomyces cerevisiae RNA polymerase II after in vivo cross-linking. Proc Natl Acad Sci 104: 19948–19953. 10.1073/pnas.0710179104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston AK, Radebaugh CA, Almeida AR, Argueso JL, Stargell LA. 2018. Genome instability is promoted by the chromatin-binding protein Spn1 in Saccharomyces cerevisiae. Genetics 210: 1227–1237. 10.1534/genetics.118.301600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwimana N, Collin P, Jeronimo C, Haibe-Kains B, Robert F. 2017. Bidirectional terminators in Saccharomyces cerevisiae prevent cryptic transcription from invading neighboring genes. Nucleic Acids Res 45: 6417–6426. 10.1093/nar/gkx242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Tsui K, Gebbia M, Mnaimneh S, Hughes TR, Nislow C. 2013. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet 9: e1003479. 10.1371/journal.pgen.1003479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasseur P, Tonazzini S, Ziane R, Camasses A, Rando OJ, Radman-Livaja M. 2016. Dynamics of nucleosome positioning maturation following genomic replication. Cell Rep 16: 2651–2665. 10.1016/j.celrep.2016.07.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos SM, Farnung L, Boehning M, Wigge C, Linden A, Urlaub H, Cramer P. 2018. Structure of activated transcription complex Pol II-DSIF-PAF-SPT6. Nature 560: 607–612. 10.1038/s41586-018-0440-4 [DOI] [PubMed] [Google Scholar]
- Warren C, Shechter D. 2017. Fly fishing for histones: catch and release by histone chaperone intrinsically disordered regions and acidic stretches. J Mol Biol 429: 2401–2426. 10.1016/j.jmb.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. 2008. EMS and UV mutagenesis in yeast. Curr Protoc Mol Biol 82: 13.3B.1–13.3B.5. 10.1002/0471142727.mb1303bs82 [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. 10.1002/yea.320110107 [DOI] [PubMed] [Google Scholar]
- Yoh SM, Lucas JS, Jones KA. 2008. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev 22: 3422–3434. 10.1101/gad.1720008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youdell ML, Kizer KO, Kisseleva-Romanova E, Fuchs SM, Duro E, Strahl BD, Mellor J. 2008. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol Cell Biol 28: 4915–4926. 10.1128/MCB.00001-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Elias JE. 2017. Relative protein quantification using tandem mass tag mass spectrometry. Methods Mol Biol 1550: 185–198. 10.1007/978-1-4939-6747-6_14 [DOI] [PubMed] [Google Scholar]
- Zhang L, Fletcher AG, Cheung V, Winston F, Stargell LA. 2008. Spn1 regulates the recruitment of Spt6 and the Swi/Snf complex during transcriptional activation by RNA polymerase II. Mol Cell Biol 28: 1393–1403. 10.1128/MCB.01733-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All high-throughput sequencing data except for whole-genome sequencing data are available on GEO under accession number GSE160821. An archive containing code and raw data for reproducing all analyses except for whole-genome sequencing and mass spectrometry analyses is available at Zenodo (https://doi.org/10.5281/zenodo.4174464). Additionally, updated versions of the Snakemake pipelines used are available at http://github.com/winston-lab.