Bacterial adhesins are key virulence factors that are essential for the pathogen-host interaction and biofilm formation that cause most infections. Many of the adhesin-driven cell-cell interactions are mediated by lectins.
KEYWORDS: repeats-in-toxin adhesins, bacterial colonization and infection, lectin-carbohydrate interactions, multispecies biofilms, structural biology
ABSTRACT
Carbohydrate recognition by lectins governs critical host-microbe interactions. MpPA14 (Marinomonas primoryensis PA14 domain) lectin is a domain of a 1.5-MDa adhesin responsible for a symbiotic bacterium-diatom interaction in Antarctica. Here, we show that MpPA14 binds various monosaccharides, with l-fucose and N-acetylglucosamine being the strongest ligands (dissociation constant [Kd], ∼150 μM). High-resolution structures of MpPA14 with 15 different sugars bound elucidated the molecular basis for the lectin’s apparent binding promiscuity but underlying selectivity. MpPA14 mediates strong Ca2+-dependent interactions with the 3,4-diols of l-fucopyranose and glucopyranoses, and it binds other sugars via their specific minor isomers. Thus, MpPA14 only binds polysaccharides like branched glucans and fucoidans with these free end groups. Consistent with our findings, adhesion of MpPA14 to diatom cells was selectively blocked by l-fucose, but not by N-acetyl galactosamine. The MpPA14 lectin homolog present in a Vibrio cholerae adhesin was produced and was shown to have the same sugar binding preferences as MpPA14. The pathogen’s lectin was unable to effectively bind the diatom in the presence of fucose, thus demonstrating the antiadhesion strategy of blocking infection via ligand-based antagonists.
INTRODUCTION
Carbohydrate-based polymers (glycans) are abundant on the exterior of cells (1, 2). The recognition of glycans by carbohydrate-binding proteins, or lectins, underlies many essential biological events such as fertilization, immunological responses, cell-to-cell communications, and host-pathogen interactions (3–9). Some lectins are components of larger adhesion proteins (adhesins), which are key virulence factors that mediate the attachment of bacteria to host cells and other surfaces (6, 10–12). Adhesins further mediate the colonization of bacteria in their favorable niches by helping develop biofilms, which cause over 80% of chronic infections in humans. Once formed, biofilms are resistant to various bactericidal treatments, including the use of antibiotics. With a current shortage of methods to treat biofilm-related diseases and the emerging prevalence of antibiotic-resistant bacteria, there is an urgent need to develop new strategies to treat bacterial infections. Since adhesins play a critical role in the initial stages of biofilm formation, the development of adhesin lectin antagonists holds great promise for treating various infections by blocking bacterial adhesion to human cells. To date, this antiadhesion strategy has led to the development of some promising treatments against diseases. For example, binding of uropathogenic Escherichia coli (UPEC) to mannose-containing glycoproteins of human uroepithelium via the adhesin lectin FimH is an enabling step toward most urinary tract infections (13, 14). Mannoside-based FimH antagonists developed through structure-guided design can effectively block UPEC from binding to the human uroepithelium (15, 16). These compounds have demonstrated fast-acting efficacy against chronic urinary tract infections and can prevent the disease when used as prophylactics (17–19). Despite these successes, widespread application of this antiadhesion approach to treat other bacterial infections is hampered by a lack of knowledge at the molecular level of ligand recognition by other adhesin lectin modules.
Marinomonas primoryensis ice-binding protein (MpIBP) is a large (∼1.5 MDa) repeats-in-toxin (RTX) adhesin found on the surface of its Antarctic Gram-negative bacterium (11, 20–22). While the N terminus of MpIBP anchors the giant protein to the bacterial outer membrane, the ligand-binding modules near the C terminus bind the bacterium to both ice and photosynthetic diatoms to form symbiotic biofilms on the underside of sea ice. MpIBP was initially extracted from the cell lysate of its native bacterium by an ice-affinity purification step (20). Intriguingly, the protein failed to elute from a Superdex S200 size exclusion column intended for further purification. This suggested that the adhesin interacts with the Superdex matrix, which is based on porous agarose particles covalently linked to dextran, a complex branched polymer of α-d-1,6-glucose. Bioinformatic analyses indicated the presence of an ∼20-kDa domain near the C terminus of MpIBP that is a member of the PA14 family, which is a carbohydrate-binding lectin module widely distributed across several kingdoms of life (23, 24). PA14 homologues are found in human proteins like fibrocystin (25) and in fungal and bacterial proteins such as β-glucosidases (26, 27), proteases (23), and adhesins (10, 21, 28, 29). PA14 domains share a β-sandwich fold and the presence of two consecutive aspartate residues in a cis peptide linkage (DcisD). The DcisD motif coordinates a Ca2+ ion that is directly involved in binding polar vicinal hydroxyl groups of various carbohydrates (10, 12, 29). Despite these conserved features in their ligand-binding sites, PA14 lectins in microbial adhesins have a broad specificity profile for a range of carbohydrates. In this regard, how PA14 lectins of bacterial adhesins recognize their ligands remains unclear. Yet, this highly conserved module is widespread in adhesins of many different bacteria, including those of human pathogens. These important attributes justify the pursuit of structural studies to elucidate the molecular basis of ligand recognition by MpPA14 (Marinomonas primoryensis PA14 domain), which may set the stage for the subsequent development of antagonists to block harmful adhesion.
In this report, we use various types of binding assays and glycan microarrays to show that MpPA14 is a lectin with an unusual binding promiscuity to monosaccharides but is specific in binding certain polysaccharides. X-ray crystal structures of MpPA14 in complex with 15 different simple sugars at atomic resolution reveal the molecular basis for the uncommon ligand selectivity by the lectin. We further show that the adhesion of MpPA14 to its host diatom cells can be fully abolished by a micromolar concentration of l-fucose. Since bioinformatic analyses indicate that lectins highly similar to MpPA14 are present in many bacterial adhesins, including those from human pathogens such as Vibrio cholerae and Vibrio vulnificus (12), there is an opportunity to use a structure-based approach to devise high-affinity lectin antagonists to block harmful biofilm formation or to develop molecular probes to detect these microbes.
RESULTS AND DISCUSSION
MpPA14 interacts strongly with fucose and N-acetylglucosamine.
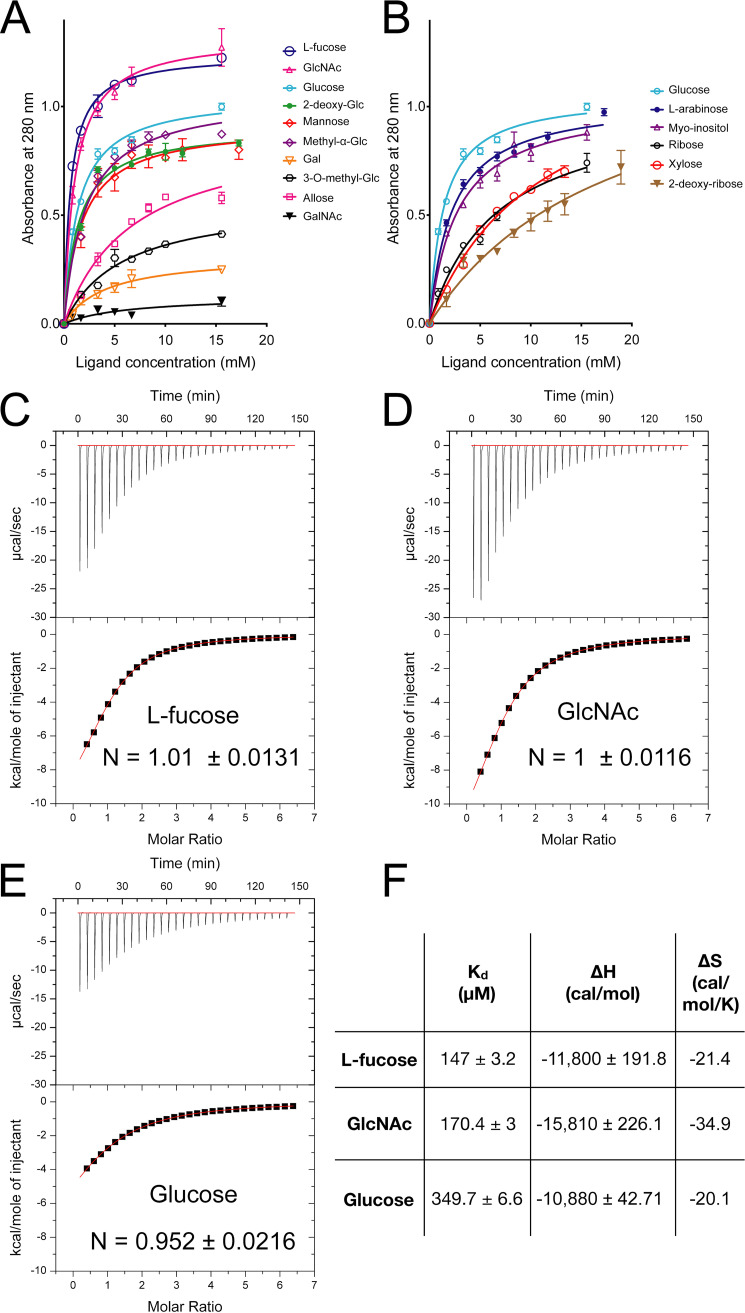
To gain insight into the binding specificity of MpPA14, we investigated the relative affinity of various monosaccharides for the lectin by a comparative competition assay (12). MpPA14 bound to Superdex resin was competitively released into solution by the progressive addition of free sugars. The released protein concentrations measured by absorbance at 280 nm were plotted as a function of free sugar concentration to produce semiquantitative binding curves (Fig. 1A and B). The apparent dissociation constant (Kdapp) calculated from these binding curves was used as a relative measure of affinity for each assayed sugar (Table 1). MpPA14 lectin bound l-fucose most strongly, with a Kdapp of 0.65 mM, followed by N-acetylglucosamine (GlcNAc) (Kdapp = 1.07 mM; Fig. 1A). Glucose and 2-deoxy-glucose bound the lectin with similar affinity, giving Kdapp values of 1.36 and 1.37 mM, respectively. d-Mannose and methyl-α-d-glucose bound MpPA14 with slightly weaker affinity (Kdapp = 1.7 mM and 2.1 mM, respectively), and the binding of d-allose, 3-O-methyl-d-glucose and d-galactose was significantly diminished to Kdapp values between 4.1 and 6.8 mM. There was no measurable interaction between MpPA14 and N-acetyl-galactosamine (GalNAc). d-Ribose bound dextran more strongly than its derivative 2-deoxy-d-ribose (Kdapp of 6.8 mM as opposed to 18 mM; Fig. 1B and Table 1). l-Arabinose exhibited higher affinity than the other pentoses for MpPA14, with a Kdapp of 2.2 mM.
FIG 1.
Binding assays to determine relative affinity of MpPA14 for simple carbohydrates. (A) Dextran-based competition binding assay to determine relative affinity of MpPA14 for various hexoses and their derivatives. Each experiment was done in triplicate. (B) Dextran-based competition binding assay to determine relative affinity of MpPA14 for selected pentoses and myo-inositol. Isothermal titration calorimetry of MpPA14 binding to fucose (C), GlcNAc (D), and glucose (E), respectively. The binding stoichiometry (N value) is indicated on each graph. (F) Table of the calculated thermodynamic parameters, including the dissociation constant (Kd), enthalpy (ΔH), and entropy (ΔS), for each of the three sugars.
TABLE 1.
Apparent dissociation constants for the binding of different sugars
| Sugar | Kdapp (mM) |
|---|---|
| l-Fucose | 0.65 |
| N-acetyl-glucosamine | 1.07 |
| Glucose | 1.4 |
| 2-Deoxy-glucose | 1.4 |
| Mannose | 1.7 |
| Methyl-α-glucose | 2.1 |
| l-Arabinose | 2.2 |
| Inositol | 2.7 |
| Galactose | 4.1 |
| 3-O-methyl-glucose | 5.7 |
| Allose | 6.8 |
| Ribose | 6.8 |
| Xylose | 11 |
| 2-Deoxy-ribose | 18 |
| N-acetyl-galactosamine | –a |
No detectable binding was observed for N-acetyl-galactosamine.
To further assess the binding thermodynamic parameters of MpPA14 ligands, we performed isothermal titration calorimetry (ITC) with l-fucose, N-acetylglucosamine (GlcNAc) and glucose (Fig. 1C to E). The ITC measurements produced rectangular hyperbolic curves for all three simple saccharides, and fitted well to a one-binding-site model with calculated binding stoichiometry (N) values being close to 1 (12, 21). ITC ranked the affinity of these three ligands in the same order as that shown by the competition binding assay, with l-fucose as the strongest ligand, followed by GlcNAc and then glucose. The dissociation constant (Kd) values calculated from the ITC measurements were significantly lower than the Kdapp values obtained from the competition assay (Fig. 1F and Table 1). However, this is to be expected, because the dextran beads used in the competition assay have multiple binding sites nearby that attract the lectin, whereas the calorimetry was done with free sugars in solution. With Kd values of 147 μM and 170 μM for l-fucose and GlcNAc, these two ligands had a greater than 2-fold higher affinity for MpPA14 than did glucose (Kd = 350 μM). In general, the affinity of lectins for monosaccharides (Kd values) lies within the high micromolar to millimolar range (8, 30, 31). Thus, our results showed the MpPA14 domain from its bacterial adhesin had relatively high affinity for the three strongest ligands in comparison to other lectins. Furthermore, negative enthalpy (ΔH) and entropy (ΔS) contributions were calculated for all three carbohydrates when they bound to MpPA14, which indicated the binding was driven primarily by polar interaction such as the formation of hydrogen and ionic bonds rather than by hydrophobic interactions. This was consistent with the observation that the Ca2+-dependent ligand-binding site of MpPA14 consisted of mainly polar and charged amino acids without any residues with large hydrophobic side chains. We therefore acquired detailed structural information to study MpPA14 ligand recognition.
Structural basis of MpPA14 selectivity for glucopyranoses.
To examine the molecular basis of carbohydrate recognition by MpPA14, we determined the X-ray crystal structures of the lectin in complex with 14 new saccharides to a resolution of 1 to 1.3 Å (see Table S1 to S3 in the supplemental material). The lectin fold is a β-sandwich domain that binds four to seven Ca2+ ions (Ca1 to Ca7) on its surface (21) (see Fig. S1A in the supplemental material. Ca1 is coordinated by the DcisD motif (Asp110 and Asp111) on the periphery of the protein, which is directly involved in binding carbohydrate with help from amino acid residues in loops 9 and 11 (L9 and L11; Fig. S1B). While Ca2 to Ca4 likely have a role in lectin folding (12, 21) (Fig. S1C to E), Ca5-7 have few ligands from the lectin and are probably an artifact from the crystallization condition that contained over 100 mM CaCl2. The highly electronegative surface of MpPA14 is consistent with its capacity to bind a high number of Ca2+ ions (Fig. S1F). There were no substantial conformational changes to the overall lectin fold when it was complexed with different sugars (root mean square deviation [RMSD] < 0.1 Å).
Structural features of MpPA14. (A) Overview of the MpPA14 structure in complex with glucose. (Right) Interface from a view that is rotated approximately 180° around a vertical axis from the left. Carbon atoms are colored grey, oxygen atoms are red, and nitrogen atoms are blue. The glucose-coordinating Ca1 is shown as a dark blue sphere, while the other Ca2+ ions are shown as green spheres. (B) Zoomed-in view of the ligand-binding site of MpPA14. The DcisD motif is indicated by an arrow. (C to E) Ca2+ coordination sites for Ca2 to Ca4. (F) Surface electrostatic potential of MpPA14. Surfaces that are electronegative are colored in red, while those that are neutral are colored white, and the electropositive surfaces are colored blue. (G) Amino acid alignment of PA14 domains from Marinomonas primoryensis (MpPA14), the oil-eating bacterium Marinobacter hydrocarbonoclasticus (MhPA14), and the cholera-causing human pathogen Vibrio cholerae (VcPA14). Amino acid residue number and secondary structure components for MpPA14 are indicated. Conserved residues that constitute the carbohydrate-binding site are highlighted in magenta. Red arrowhead points to the DcisD motif. Loops (L9 and L11) involved in carbohydrate binding are indicated in red. Download FIG S1, PDF file, 0.3 MB (324.8KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallographic statistics for MpPA14 in complex with l-fucose, mannose, α-methyl-glucose, inositol, GlcNAc, allose, and 3-O-methyl glucose. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
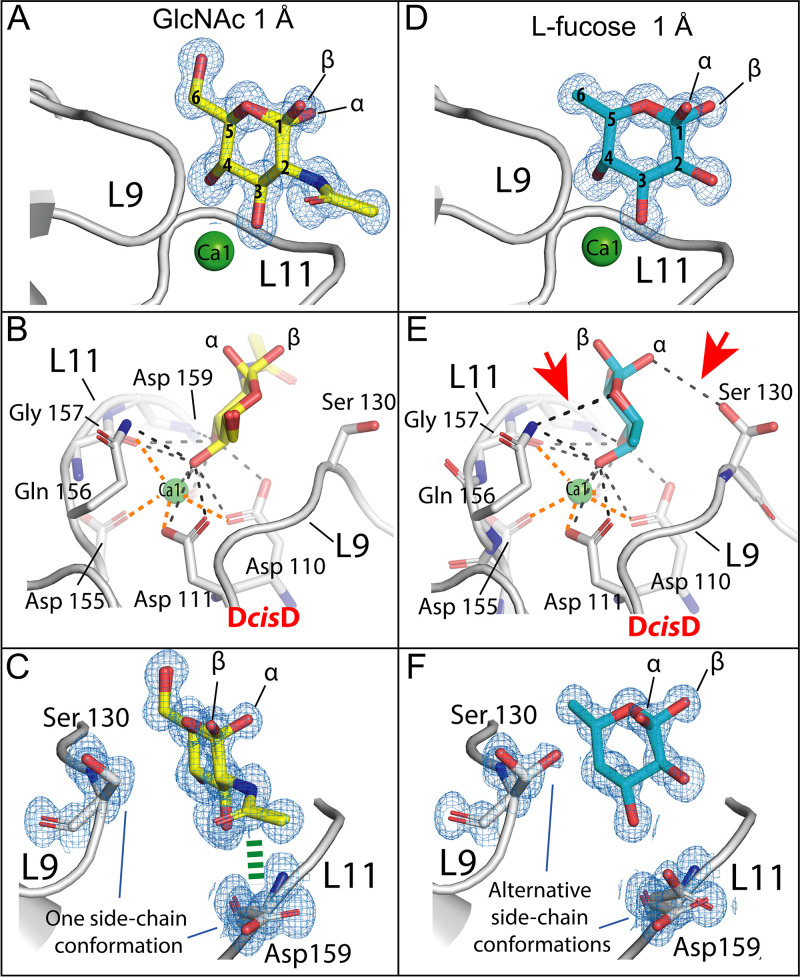
As suggested by ITC, sugar recognition by MpPA14 is primarily driven by polar interactions in a Ca2+-dependent manner. Glucose, GlcNAc, and other glucopyranose-containing carbohydrates, including 2-deoxy-glucose, methyl-α-glucose, and two disaccharides (sucrose and trehalose), all bound to the Mp-PA14-Ca1 via their trans vicinal 3,4-diols (ionic bond length, 2.5 Å) in gauche configuration with a dihedral angle of ∼60° (Fig. 2A; see also Fig. S2 and Fig. S3A and B in the supplemental material). This interaction is further enhanced with the diol being coordinated by the side chain carboxyl and hydroxyl oxygens of the DcisD motif, and main-chain and side chain protein ligands from L11 (Gln156, Gly157, and Asp159; Fig. 2B). The acetyl group on the C-2 position of GlcNAc interacts with the side chain atoms of Asp159 on L11 (Fig. 2C), holding the aspartate side chain down in one stable conformation. This additional interaction likely accounts for the higher affinity of GlcNAc to the lectin than that of the other glucopyranoses.
FIG 2.
Ligand-binding site of MpPA14 in complex with GlcNAc and l-fucose. Structural features of the MpPA14 ligand-binding sites with GlcNAc (A) and l-fucose (D). Detailed carbohydrate-protein interactions between MpPA14 and GlcNAc (B), and l-fucose (E), respectively. Ionic bonds (∼2.5 Å) between Ca1 and MpPA14 are indicated by thicker orange dashed lines, while polar interactions between the lectin and carbohydrates are indicated with black dashed lines (shorter than 3.5 Å). Red arrowheads in panel E indicate the additional polar interactions between l-fucose and MpPA14 compared to those in panel B. (C, F) Angled view comparing the conformations of the side chains of L9-Ser130, and L11-Asp159 in the ligand-binding sites of MpPA14-GlcNAc (C) and MpPA14-fucose (F) complexes. Carbon atoms of GlcNAc are colored yellow, while those of l-fucose and MpPA14 are colored cyan and light gray, respectively. The interaction between the acetyl group of GlcNAc and side chain of Asp159 is indicated by a green dashed line (∼4 Å). Oxygens are colored red, nitrogens are colored blue, and Ca2+ ions are colored green. (A, C, D, and F) The 2 Fo − Fc maps shown for sugars and ligand-binding site residues are shown as blue meshes (contoured at σ = 1).
Structures of MpPA14 in complex with glucopyranose-containing sugars. The color scheme and labeling are the same as those in Fig. 2. Download FIG S2, PDF file, 0.2 MB (251.5KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structures of strongly binding (A), moderately binding (B), and weakly binding (C) sugars of MpPA14. The specific isomer shown for each monosaccharide was revealed by X-ray crystallography as described in Fig. 2 and 3 and Fig. S2 and S5. Double arrowheads point to the diol groups used by the sugar for interaction with Ca1 in the ligand-binding site of MpPA14. Alternative binding configurations are indicated with double arrowheads in blue or green instead of red. Download FIG S3, PDF file, 0.1 MB (148KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structural data shown here for MpPA14 contrast with the previously reported structure of MhPA14 in complex with glucose (12). MhPA14 is an MpPA14 homolog from an RTX adhesin of the oil-degrading bacterium Marinobacter hydrocarbonoclasticus. With a similar ligand-binding site to that of MpPA14 (12), MhPA14 also had a strong preference for binding l-fucose and glucopyranoses over other monosaccharides. However, X-ray crystallography showed MhPA14 complexing glucopyranose via its 1,2-diol. Given that the C-2 position of GlcNAc lacks the hydroxyl group required for interacting with MpPA14 via the 1,2-diol, the binding mode shown by the MhPA14-glucose complex could not explain the lectin’s high affinity for this acetylated sugar. Close inspection of the MhPA14-glucose complex structure revealed that the monosaccharide in the carbohydrate-binding site is in direct contact with a neighboring symmetry-related molecule (see Fig. S4A and B in the supplemental material), indicating the tight packing of the unit cell caused the sugar to bind in a less favorable configuration. The observed crystal-packing artifacts explained why cocrystallization of MhPA14 with various other sugars, and direct soaking experiments with the apo-MhPA14 crystal, were futile. In contrast, symmetry-related molecules in the unit cells of MpPA14 are far apart from the ligand-binding site and thus do not impact the binding of monosaccharides to MpPA14 (Fig. S4C and D).
Crystal packing of MhPA14 and MpPA14. (A) Crystal unit cell of MhPA14 in complex with glucose. The box with black dashed lines indicates the interface between one molecule of MhPA14-glucose (grey) and its neighboring symmetry-related molecules (magenta) in the crystal unit cell. (B) The glucose molecule bound by MhPA14 is directly involved in crystal contact with a symmetry-related molecule. Lengths for the protein-sugar interaction bonds (black dashed lines) are indicated. (C) Crystal unit cell of MpPA14 in complex with GlcNAc. The box with black dashed lines indicates the interface between one molecule of MpPA14-GlcNAc (grey) and its neighboring symmetry-related molecules (magenta) in the crystal unit cell. (D) The GlcNAc bound by MpPA14 does not interact with a symmetry-related molecule in the crystal unit cell. The distances between the GlcNAc and the nearest atoms from a neighboring symmetry-related molecule are indicated by green dashed lines. Download FIG S4, PDF file, 0.3 MB (338.3KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Why l-fucose is a better ligand than glucopyranoses.
Based on results from docking experiments, it was proposed that l-fucose binds MhPA14 via its 2,3-diol (12). However, the well-resolved 1-Å electron density map in this study unambiguously showed that fucose bound MpPA14 with its cis 3,4-diol in the gauche conformation with a dihedral angle of 47° (Fig. 2D). In contrast to the MpPA14 hexose ligands, which are all in the d-configuration, fucose is in the l-configuration, with hydroxyl groups of the fucopyranose ring pointed in opposite directions. This helps the endocyclic oxygen atom of fucose point toward L11 and hydrogen bond with the sidechain of Gln156 (Fig. 2E). Additionally, the hydroxyl on the α-anomeric carbon may hydrogen bond with the sidechain of Ser130 on L9, clamping the pyranose ring tightly into the binding site (Fig. 2E and F). The l-fucose–MpPA14 interaction is distinct from that shown by the bacterial C-type lectin, LecB, from Pseudomonas aeruginosa, which uses two side-by-side Ca2+ ions to directly coordinate the 2,3,4-triol of l-fucose (32, 33). The additional ionic interaction between LecB and l-fucose can explain its enhanced affinity (Kd = 58 μM) for the sugar compared to that of MpPA14 (Kd = 147 μM).
Promiscuity of MpPA14 in monosaccharide recognition.
To investigate the molecular basis for the plastic nature of MpPA14 in binding various monosaccharides, we investigated MpPA14 structures in complex with various glucose epimers and derivatives.
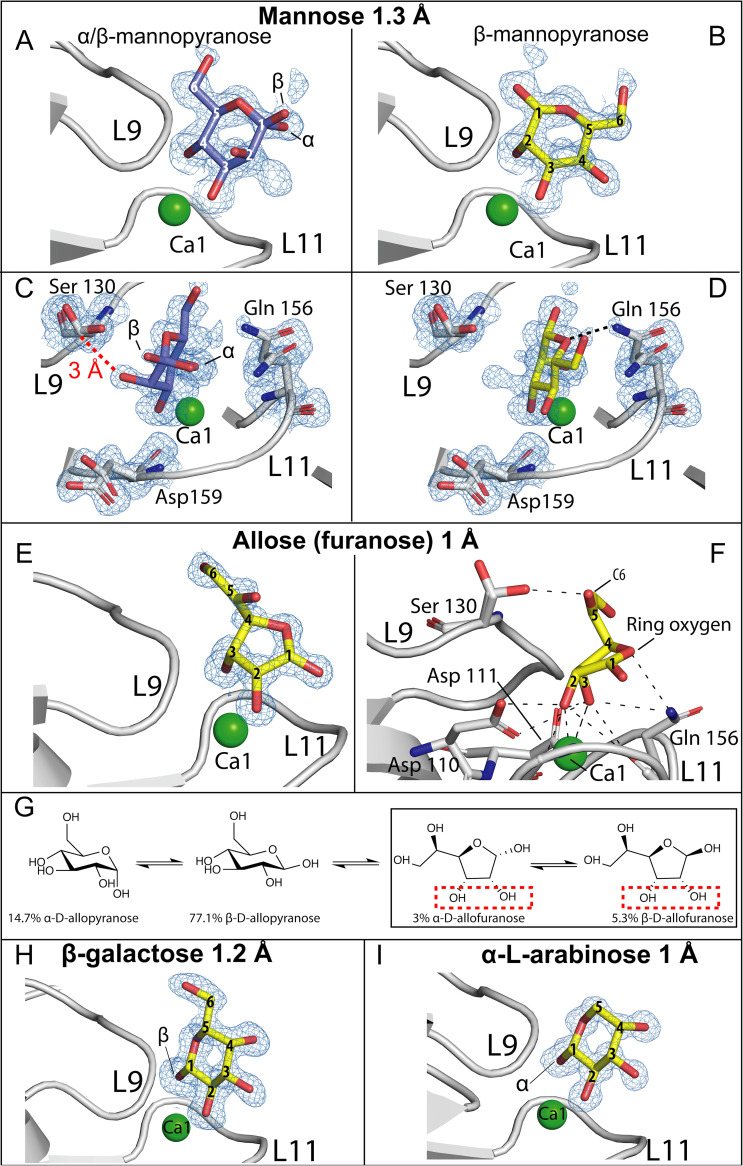
Mannose bound MpPA14 slightly more weakly than glucose (Fig. 1A and Table 1). The electron density map for the mannose-MpPA14 complex indicated that the sugar bound in two distinct conformations (Fig. 3A and B; see also Fig. S3B). Like glucose, d-mannose bound MpPA14 via the 3,4-diol. However, since the mannose C-2 hydroxyl moiety is in an axial position, its oxygen atom may clash with the β-carbon of the Ser130, as they are only 3 Å away from each other (Fig. 3C). In addition, as the C-2 and C-3 hydroxyl groups of mannose are positioned in cis, they may form an intramolecular hydrogen bond that further weakens the 3,4-diol from binding Ca1. Alternatively, β-mannopyranose can bind MpPA14 using a second configuration where its 2,3-diol anchors the saccharide ring in an inverted fashion, allowing the ring oxygen to hydrogen bond with the side chain amide group of the Gln156 (Fig. 3D). However, α-mannopyranose failed to fit into the electron density via this binding mode, which indicates that MpPA14 can only recognize the less prevalent β-anomer in the equilibrium via its 2,3-diol (33% β-mannopyranose as opposed to 62% α-mannopyranose at 30°C) (34). This apparent lack of one distinct stable binding configuration explains the relatively inferior affinity of mannose compared to glucose.
FIG 3.
Ligand-binding site of MpPA14 in complex with mannose, allose, galactose, and l-arabinose. Side (A) and top views (C) of α- and β-mannopyranose rings anchored to MpPA14 via their 3,4-diols. Electron density for only β-mannopyranose was seen for the binding mode via the 2,3-diol. (B) Side view; (D) top-down view. The distance between the β-carbon of serine on L9 and C-2 hydroxyl oxygen of α/β-mannopyranose is indicated by a red dashed line. (E) Side-view of β-allofuranose in the MpPA14 ligand-binding site. (F) Detailed polar interactions (black dashed lines) between β-allofuranose and MpPA14. Amino acid residues involved in allose-MpPA14 interaction are labeled. The color scheme is the same as that in Fig. 3. (G) Equilibria of allose anomers in aqueous solution at 30°C. The diol of allofuranose responsible for binding MpPA14 is indicated by a red dashed box. Side-view of β-galactose (H) and α-l-arabinose (I) in the MpPA14 ligand-binding site.
With the C-3 hydroxyl in the axial position, allopyranose cannot bind MpPA14 via its cis vicinal 3,4-diol, as this would cause the saccharide ring to clash with residues of L11 (e.g., Gln156). Unexpectedly, the binding site of the lectin contained the furanose form of allose, with its 2,3-diol interacting with Ca1 (Fig. 3E and Fig. S3C). The furanose rings lean toward L11, with their endocyclic oxygen interacting with the side chain amide group of Gln156, and the C-5 hydroxyl group extends out to hydrogen bond with the side chain hydroxyl of Ser130 (Fig. 3F). At 30°C, approximately 92% of allose exists as pyranoses in solution (Fig. 3G), yet MpPA14 binding appears to be dependent on the rare presence of the allofuranose, which only makes up ∼8% of allose in solution, explaining the feeble affinity of this sugar for the lectin (Table 1). Similarly, 3-O-methyl-glucose cannot bind MpPA14 via the 3,4-diol because of the substituted methyl group on its C-3. This sugar compensates by binding using the 1,2-diol of the β-anomer (Fig. S3C), resulting in its considerably weaker binding to the lectin compared to that of glucopyranoses and mannose.
Galactose has its C-4 hydroxyl in the axial position instead of being equatorial as in glucose. Docking of the galactopyranose ring to MpPA14 via the 3,4-diol is not possible due to steric hinderance against Gln129 and Ser130 on L-9. Instead, galactose can only interact with MpPA14 via the 1,2-diol of its rare β-anomer (Fig. 3H). These limitations explain the relatively weak interaction between galactose and MpPA14 (Table 1). Results from the binding analysis of GalNAc verify this assessment. Having just shown the 3,4-diol of galactopyranose cannot complex MpPA14, binding of GalNAc is completely abolished because its 1,2-diol is unavailable due to the C-2 hydroxyl being substituted with an acetyl group.
As a comparison to MpPA14 and its close homologs, C-type lectins also bind monosaccharide vicinal hydroxyl groups via Ca2+. In addition, sugar selectivity by C-type lectins typically involves interactions through amino acid residues with aromatic sidechains. For instance, the C-type lectins that selectively bind galactose require the presence of a tryptophan that aligns with the open face of the pyranose ring akin to a hydrophobic stacking interaction (or histidine that selects for mannose [35, 36]). This interaction with hydrophobic amino acids is common not only in lectins but also in other carbohydrate-binding modules (8) and in carbohydrate-active enzymes (37), where it often imparts stringent selectivity for the sugar type bound. Sometimes multiple aromatic side chains situated along the glycan binding site contribute to the recognition of multiple monomers in a glycan, further increasing selectivity not only for the monomers but also for how they are linked. MpPA14 lacks these hydrophobic residues in its interactions with only the terminal sugars. As a result, the relatively more flexible ligand-binding site of MpPA14 may contribute to its higher degree of plasticity to engage sugar monomers that vary in stereochemistry.
We further analyzed the crystal structures of MpPA14 in complex with three pentoses, l-arabinose, ribose, and 2-deoxy-ribose, as well as inositol, which has an unusual 6-carbon saccharide ring without an endocyclic oxygen (see Fig. S3B and C and Fig. S5 in the supplemental material). These four carbohydrates bound MpPA14 more weakly than glucose and mannose (Table 1). Crystal structures showed that MpPA14 selects the pyranose form of pentoses for binding. As pentoses have a higher percentage of furanose present in the conformational equilibria than do hexoses, selectivity for the more thermodynamically stable pyranoses might contribute to the weaker affinity of pentoses toward the lectin. For instance, despite l-arabinose existing in solution predominantly in the pyranose form at 25°C (57% α-arabinopyranose versus 30.5% β-arabinopyranose) (38), only the α-anomer complexes MpPA14 via its 1,2-diol (Fig. 3I). The same rationale can be used to explain the even weaker affinity of ribose and its derivative 2-deoxy ribose for MpPA14 (se Fig. S5B and C in the supplemental material) (39). In the case of inositol, the composite electron density map indicates it binds MpPA14 in several different conformations (Fig. S5D to F). This promiscuous binding mode is indicative of a lack of one stable binding conformation, which might explain inositol’s moderate affinity for MpPA14.
Structures of MpPA14 in complex with its moderately and weakly binding ligands. (A) β-3-O-Methyl-glucose. (B) Ribose. (C) 2-deoxy-ribose. (D to F) Three binding conformations of Inositol. The color scheme and labeling are the same as those in in Fig. S2. Download FIG S5, PDF file, 0.2 MB (202.4KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In summary, X-ray crystallography has elucidated the molecular basis of MpPA14’s promiscuity in binding a range of monosaccharides. Remarkably, the lectin can discern favorable conformations of these monosaccharides from their nonbinding anomers, even when the latter are much more prevalent in the equilibria (e.g., allofuranose as opposed to allopyranose). Nevertheless, monovalent or simple carbohydrate oligomers are typically not the physiological targets of lectins (30). In the context of MpPA14, the lectin likely binds complex glycans or glycoproteins present on the surfaces of microbes, where the proximity effect of having numerous identical or similar end groups increases the avidity of the lectin interaction. In this way the sugar-binding activity of MpPA14 can help to form biofilms. This prompted us to survey the lectin-sugar interactions from a broad spectrum of complex carbohydrates using glycan microarray technology.
MpPA14 binds glucopyranose and fucose moieties of complex glycans.
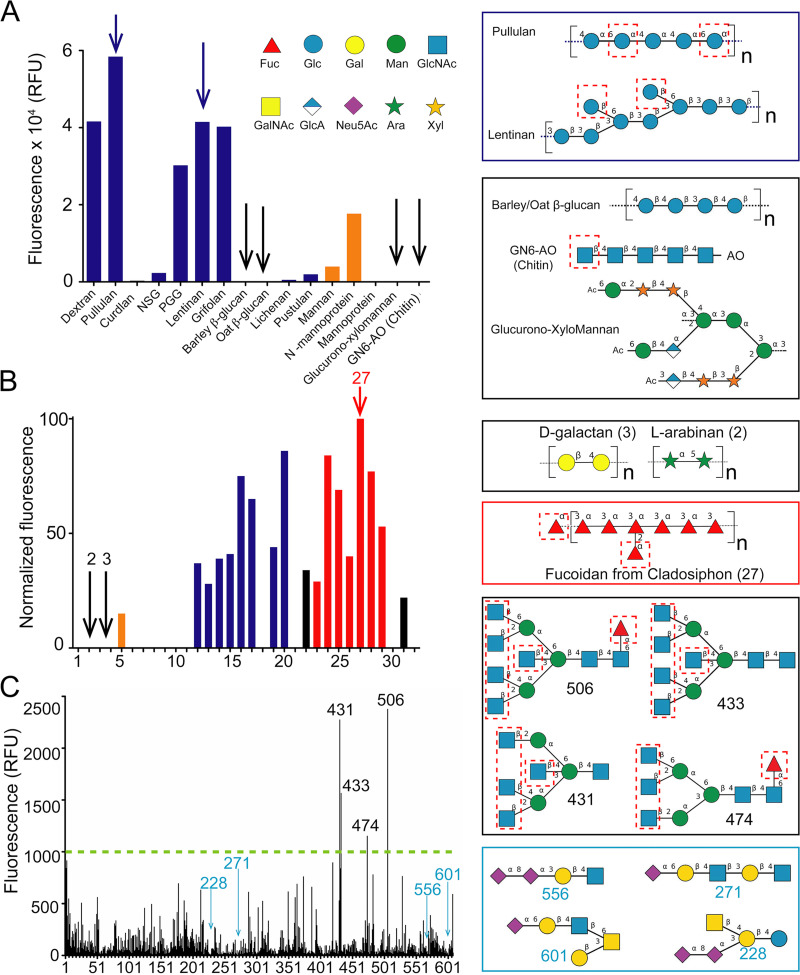
To dissect MpPA14’s role in the formation of mixed-species biofilms, we probed two different microbial glycan microarrays for lectin-binding partners (40). We first analyzed the binding of GFP-MpPA14 to 16 different polysaccharides consisting primarily of glucans and mannans from bacteria and fungi (Imperial College Glycosciences Laboratory). Four glucans, namely pullulan, lentinan, dextran, and grifolan, bound most avidly to MpPA14 (Fig. 4A). To emphasize the specificity of this interaction, six of the other polysaccharides showed negligible or no detectable binding; these included glucans such as curdlan and those purified from oat and barley, as well as mannoprotein, glucurono-xylomannan, and GN6-AO, which is a hexasaccharide of 1,4-linked GlcNAc (Chitin) with an aminooxy group (AO; see Table S4 in the supplemental material. Consistent with findings from our structural and binding analyses (Fig. 1 and 3), the four strongest binders contain multiple glucopyranoses with unoccupied 3,4-diols either as internal or terminal moieties in their linear backbone (pullulan) as well as in their branches (lentinan, dextran, and grifolan; Fig. 4A). In contrast, weak or negligible binding to MpPA14 was demonstrated for linear glucans formed through 1,3 and 1,4 linkages, such as β-glucans of oat and barley. Each of these linear polysaccharides has only one 3,4-diol set from its terminal glucopyranose accessible for MpPA14 binding, while all potential 3,4-diols in the backbone are involved in the formation of glycosidic bonds (Fig. 4A). Similarly, GN6-AO interacted poorly with MpPA14 because it too has only one free 3,4-diol in the terminal GlcNAc. Furthermore, moderate lectin-glycan interactions were observed for the highly branched N-mannoprotein from the fungus Candida albicans (Fig. 4A, orange bar; see also Table S4), while binding to glucurono-xylomannan and mannoprotein from the fungus Aspergillus fumigatus was negligible, as these glycans lack the more favorable structural epitopes of 3,4-diols on either glucopyranose or l-fucose.
FIG 4.
Binding of MpPA14 to sugar oligomers identified by three different glycan microarrays. (A) Microbial glycan array analysis conducted at the Imperial College Glycosciences Laboratory. Data bars showing binding intensity for glucans and mannans are colored blue and orange, respectively. Examples of strong binders are drawn in the blue box (top right, blue arrows in the graph), while those for weak binders are drawn below (black arrows in the graph). The putative sites for MpPA14 binding to the glycans are indicated by dashed red boxes. Glycan nomenclature symbols are included. Abbreviations are as follows: Fuc, l-fucose; Glc, glucose; Gal, galactose; Man, mannose; GlcNAc, N-acetylglucosamine; GalNAc, N-acetyl-galactosamine; GlcA, glucuronic acid; Neu5Ac, N-acetyl-neuraminic acid; Ara, l-arabinose; Xyl, xylose. (B) Microarray data of glycans from marine algae and land plants. Glucans (blue), mannans (orange) fucoidans (red), and other types of glycans (black). Examples of nonbinders (right, black arrows in the graph), while the structure for Cladosiphon fucoidan is shown below (red arrow in the graph). (C) Mammalian glycan array analyzed at the Consortium for Functional Glycomics. Representative structures for the strong MpPA14 binders (black box) and nonbinders (blue box) are drawn on the right.
X-ray crystallographic statistics for MpPA14 in complex with 2-deoxy-glucose, galactose, l-arabinose, ribose, 2-deoxy-ribose, sucrose, and trehalose. Download Table S2, DOCX file, 0.02 MB (19.2KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polysaccharides used in the first glycan array. The first column on the left presents the relative lectin-binding activity detected. Download Table S3, DOCX file, 0.02 MB (16.4KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polysaccharides contained in the second glycan array. The first column on the left presents the lectin-binding activity detected (normalized GFP-MpPA14 signal intensity). Polysaccharide structures are listed in the same order as in the x axis from Fig. 4B (order of appearance shown in second column). Download Table S4, DOCX file, 0.02 MB (20.3KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To expand the repertoire of glycans beyond glucans and mannans, we performed a second focused microarray composed of 32 polysaccharides found in microbes such as fungi and bacteria, as well as in macroalgae and plants (Fig. 4B). Similarly to the first microarray, MpPA14 showed significant binding to some glucans that present multiple sets of unoccupied 3,4-diols, which included pustulan, pachyman, and scleroglucan (Fig. 4B, dark blue bars, glycans 20, 16 and 17, respectively; see also Table S4 in the supplemental material). In addition, MpPA14 interacted avidly with fucoidans from macroalgae. The strongest MpPA14-binding fucoidans were to Cladosiphon (41), Sargassum, and Ascophyllum nodosum (glycans 27, 24, and 28, red bars). Since fucoidans are algal polysaccharides primarily consisting of a linear backbone of sulfated α-1,3- or α-1,4-linked l-fucose (42, 43), MpPA14 can interact by binding the terminal moieties on the backbone and branches with unoccupied 3,4-diols. In contrast, MpPA14 did not bind to polysaccharides such as arabinan, galactan, galactomannan, xylan, xyloglucan, and porphyran (glycans 2, 3, 6, 11, 21, and 32, respectively; Fig. 4B), as these lack the structural epitopes required for favorable interactions with MpPA14. For instance, while arabinan is a polymer of arabinose, a sugar that MpPA14 binds, X-ray crystallography showed that MpPA14 is selective in only forming a complex with α-l-arabinopyranose (Fig. 3I). This conformer is not present in the arabinan polymer composed of 1,5-linked α-l-arabino-furanoses, which are then not free to transition to the pyranose form. Similarly, since the MpPA14 binds galactose via the 1,2-diol (β-anomer) (Fig. 3H), the lectin cannot interact with galactans, as they are polymers of β-1,4-d-galactose (Fig. 4B and Table S5).
Some differences were observed in the binding results, with several polysaccharides present in both the first and second glycan microarrays. Examples include mannan from Saccharomyces cerevisiae, which showed weak binding to MpPA14 in the first array (Fig. 4A, glycan number 12) but had no detectable interaction with the lectin in the second array (Fig. 4B; glycan number 8; pullulan bound MpPA14 more strongly than pustulan in the first array (Fig. 4A; glycan numbers 2 and 11, respectively), whereas their relative affinities were reversed in the second array (Fig. 4B; glycan numbers 12 and 20, respectively. These minor discrepancies in the binding results could be due to differences in the polysaccharide sources or the different methods used to immobilize them onto the microarrays (44, 45). Nevertheless, both arrays pointed to the key result that MpPA14 selectively binds glucans with multiple unoccupied 3,4-diols, while the lectin does not recognize glycans such as arabinans, galactans, and xylans. Furthermore, the finding that MpPA14 interacted strongly with fucoidans is consistent with the MpPA14-fucose interaction demonstrated by the binding and structural data. This result may have physiological relevance, as l-fucose-containing polysaccharides are highly prevalent in the exudates of diatoms (46–49), at least one of which is a natural host of M. primoryensis.
Since the PA14 domain is widespread in bacteria, including some that are human commensals and others that are pathogens, we reasoned that MpPA14 and its homologs might interact with mammalian glycans. Therefore, we tested the lectin on a microarray consisting of 609 complex mammalian glycans (Consortium for Functional Glycomics, version 5.2). Four glycans (506, 431, 433, and 474) stood out among the strongest binders (Fig. 4C). They share an architecture as moderately branched mannose-containing oligomers with a bisecting GlcNAc motif. Each of the four glycans has three to five terminal GlcNAc moieties with 3,4-diols available for complexing MpPA14. Glycans 506 and 474 also have one α-l-fucose moiety linked to the surface-immobilized GlcNAc. Interestingly, the strongest binder, glycan 506, differs from glycan 433 only by the addition of the α-l-fucose, suggesting that this monosaccharide might contribute to the higher affinity of glycan 506 by presenting an extra binding site for the lectin. Glycans with fewer binding epitopes of GlcNAc and fucose bound more weakly in general. Out of the 61 glycans that did not bind MpPA14 (relative fluorescence units [RFUs] of 20 or below), 33 have N-acetyl-neuraminic acid as their terminal sugar, while another 25 of these nonbinders end with either galactose or GalNAc (Fig. 4C). Indeed, as shown by our structural analyses above, these sugars lack the 3,4-diol motifs of glucopyranoses and l-fucose preferred for MpPA14 recognition.
l-Fucose blocks PA14-diatom interactions.
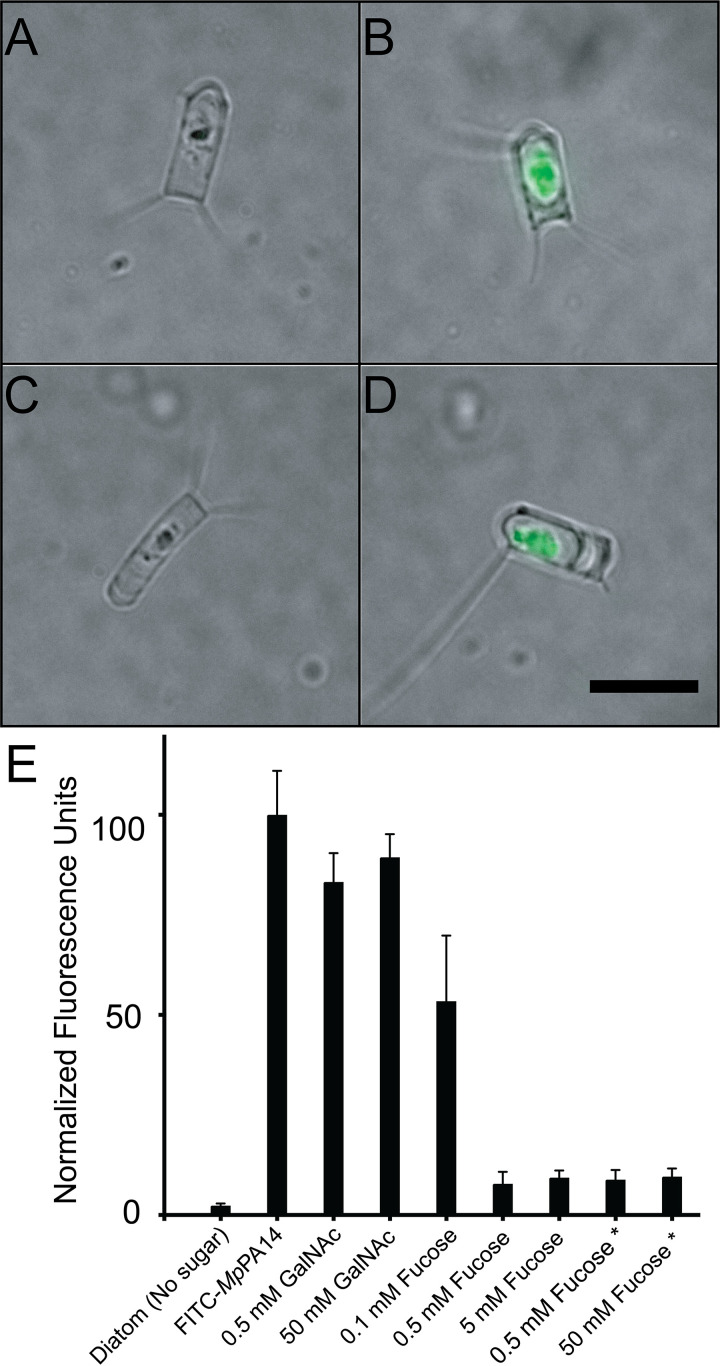
Having identified l-fucose as the strongest monosaccharide ligand for MpPA14, we set out to validate its potential as an inhibitor for the lectin-dependent bacteria-diatom interaction that led to the discovery and characterization of this protein (21). Here, we tested if l-fucose can block fluorescently labeled MpPA14 from binding to the diatom Chaetoceros neogracile.
C. neogracile is a psychrophilic marine diatom found in Antarctic waters (50). As shown in Fig. 5A and D, C. neogracile diatoms are roughly 10 μm in length with a width of 3 to 4 μm. Each diatom cell has 1 to 4 projections protruding from the corners. Given its photosynthetic capability, C. neogracile contains chlorophyll that is intrinsically fluorescent. However, the binding of fluorescein isothiocyanate (FITC)-labeled MpPA14 to C. neogracile resulted in a 40-fold increase of fluorescence over the basal autofluorescence of the diatom (Fig. 5A and B; see also Fig. S6 in the supplemental material). The addition of 0.5 mM l-fucose was extremely effective at blocking accumulation of lectin on the diatom (Fig. 5C), as the free sugar outcompeted the cell surface glycans for the binding MpPA14 and displaced 95% of the fluorescent signal (Fig. 5E). This competitive effect fell off to ∼40% as the l-fucose concentration was reduced to 0.1 mM (Fig. 5E). The effective concentration of l-fucose needed to block association is significantly higher than the Kd of 147 μM for MpPA14-fucose interaction determined by ITC. We reason that glycans coating the diatom cell membrane present numerous end group binding sites in close proximity that can serve as a “molecular Velcro” for MpPA14 binding (30).
FIG 5.
Inhibition of MpPA14 binding to the diatom C. neogracile. Representative images showing (A) an untreated C. neogracile diatom with basal autofluorescence in the center from its cell; (B) a C. neogracile cell treated with 0.2 mg/ml fluorescein isothiocyanate (FITC)-labeled MpPA14; (C) a C. neogracile cell treated with 0.5 mM l-fucose and 0.2 mg/ml FITC-labeled MpPA14; and (D) a C. neogracile cell treated with 0.2 mg/ml FITC-labeled MpPA14 and 50 mM GalNAc. All four images (A to D) are at the same scale, with the black scale bar in panel D indicating 10 μm. (E) Fluorescence levels shown by the C. neogracile cells alone and those treated with various ligands. Each bar represents the quantification of average fluorescence from 30 individual diatoms. Data bars with an asterisk underneath represent experiments where diatoms were incubated with FITC-MpPA14 before l-fucose was added.
Representative images of diatoms with various fluorescein isothiocyanate (FITC)-MpPA14 and sugar treatments shown in Fig. 5. Asterisks (*) represent experiments where diatoms were incubated with FITC-MpPA14 before l-fucose was added. Download FIG S6, PDF file, 0.1 MB (107.9KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In contrast to the inhibitory effect of l-fucose, the nonbinder of MpPA14, GalNAc, was unable to prevent the lectin from binding the diatom even at 50 mM (Fig. 5D and E and Fig. S6), validating the results from the binding and structural studies. Importantly, adding 0.5 mM l-fucose to diatoms precoated with FITC-labeled MpPA14 resulted in the dissociation of the lectin from the cells. These results suggest that l-fucose can disrupt preexisting associations between bacteria and diatoms.
MpPA14 lectin homologs are found in pathogens.
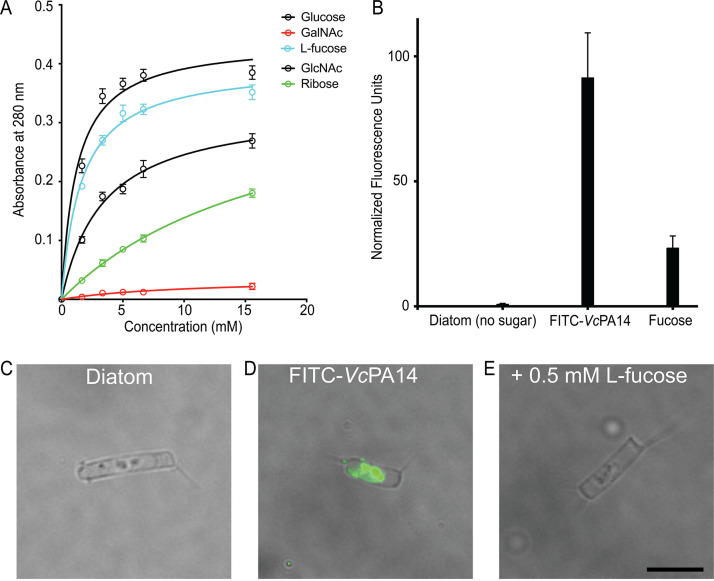
The binding conformations of various monosaccharides identified in the structural analyses can lay the foundation for the structure-guided design of glycan-based probes for detecting microbes or for making inhibitors to disrupt bacterial adhesion. MpPA14 homologs are widespread in the adhesins of Gram-negative bacteria such as the previously reported MhPA14 from the oil-degrading M. hydrocarbonoclasticus, as well as those adhesins that help pathogenic bacteria infect specific niches. For example, a large RTX adhesin from the cholera-causing human pathogen, Vibrio cholerae, contains a MpPA14 homolog (VcPA14) with 44% identity at the protein level. Moreover, the amino acid residues involved in coordinating Ca1 and recognizing glycans are conserved between the MpPA14 and VcPA14 (see Fig. S1G in the supplemental material). Despite small deviations in amino acid sequence between MpPA14 and MhPA14 from M. hydrocarbonoclasticus, these lectins have the same monosaccharide ligands, and there is insignificant variation in their complex glycan recognition (12). With its ligand-binding site even more like MpPA14 than that of MhPA14, VcPA14 probably binds to the same simple sugars. It is therefore of interest to test the inhibitory effect of strong PA14 binders identified in this study to set the stage for developing novel strategies for modulating bacterial adhesion.
As proof of concept for the antiadhesion strategy that targets microbial pathogens, we studied the VcPA14 lectin domain from a large 6,938-amino-acid RTX adhesin (GenBank accession number WP_154597608) of the cholera-causing bacterium, V. cholerae. As discussed, VcPA14 has a similar ligand-binding site as that of MpPA14 (Fig. S1), with key sugar-binding residues conserved (Fig. S1G). Consistent with our structural and functional studies on MpPA14, VcPA14 interacts with GlcNAc, and fucose more strongly than glucose, while the lectin cannot bind GalNAc (Fig. 6A). Moreover, FITC-labeled VcPA14 bound to the cell membrane of C. neogracile within the frustule in the same way as MpPA14 did, and this binding was largely blocked by addition of 0.5 mM fucose (Fig. 6B to E).
FIG 6.
Sugar selectivity of VcPA14 and its interactions with C. neogracile. (A) Dextran-based competition binding assay to determine relative affinity of MpPA14 for glucose, GlcNAc, l-fucose, ribose, and GalNAc. (B) Fluorescence levels shown by the C. neogracile cells alone and those treated with FITC-VcPA14 in the absence or presence of 0.5 mM l-fucose. Each bar represents the quantification of normalized average fluorescence from 30 individual diatoms. (C to E) Representative images of diatoms alone (C) and those incubated with FITC-VcPA14 in the absence (D) and presence of 0.5 mM l-fucose (E).
Conclusions and outlook.
In this study, we elucidated the molecular basis for ligand recognition by a lectin module widespread in bacterial adhesins. The atomic details revealed by X-ray crystallography not only helped clarify the plasticity of MpPA14 in binding various monosaccharide ligands but also revealed how the lectin recognizes complex polysaccharides in a more specific manner. We further show that a low millimolar amount of l-fucose can be used to disrupt binding of the lectin to diatom cells. The atomic details of the lectin-carbohydrate interactions elucidated here serve as the starting points for the development of adhesin antagonists via ligand-based design. For instance, the fact that GlcNAc gains a 2-fold higher affinity for MpPA14 than glucose simply from the replacement of the C-2 hydroxyl with an acetyl group suggests that appending designed substituents on various positions of avid binders such as the C-2 and C-5 of l-fucose might further enhance their potency (Fig. 2C and F).
Given the high similarity between MpPA14 and lectin folds in the adhesins of pathogenic bacteria, this work gives insight into how harmful bacterium-host interactions might be controlled through modulation of the lectin-glycan interactions. This antiadhesion approach holds promise as an alternative or additive approach to treat infections without the excessive use of antibiotics and may thus help mitigate problems with multidrug-resistant bacteria (17, 51, 52).
MATERIALS AND METHODS
Dextran-based comparative competition assay.
The dextran resin competition assay was performed as previously described for MhPA14 (12). Briefly, Superdex 200 (S200) resin was washed twice with 50 mM Tris-HCl (pH 9), 150 mM NaCl, and 5 mM CaCl2. One ml of 1 mg/ml MpPA14 with green fluorescent protein (GFP) fused to its N terminus (GFP-MpPA14) or VcPA14 labeled with FITC was suspended with 300 μl of equilibrated Superdex 200 (S200) resin. Following an incubation period with gentle mixing, the S200 resin bound with GFP-MpPA14 was pelleted by centrifugation. The pellet was washed twice with 50 mM Tris-HCl (pH 9), 150 mM NaCl, and 5 mM CaCl2, and the A280 of the supernatant from the second wash was used as the baseline reading. Next, after resuspension in the same buffer, aliquots of 1.67 μmol saccharide were sequentially added to the solution six or seven times, with the A280 of the supernatant being measured after each addition to quantify the release of lectin. The final addition of saccharide was 5 μM. Data from the dextran affinity assay were plotted using GraphPad Prism after subtracting the background. Next, the data were fitted to a nonlinear regression of one-site-specific binding, which follows the model y/Bmax = x/(Kd + x), with Bmax as the maximum specific binding and Kd as the equilibrium binding constant.
Isothermal titration calorimetry.
Isothermal titration calorimetry (ITC) measurements were performed at 30°C with a MicroCal VP-ITC instrument (Malvern). MpPA14 (400 μM) was mixed with serial 5-μl aliquots of 8 mM sugar solution (l-fucose, GlcNAc, or glucose). Sugars were automatically added by a rotating syringe (400 rpm) at 5-min intervals into the MpPA14 solution for a total of 50 injections. The data were analyzed with Origin software version 5.0 (MicroCal).
Cocrystallization, X-ray diffraction, and structure solutions of MpPA14 with various sugars.
Details for the cloning, expression, purification, and crystallization of MpPA14 were previously reported (12, 21). Cocrystallization of MpPA14 with various sugars was performed using the “microbatch-under-oil” method by mixing equal volumes of ∼20 mg/ml protein with a precipitant solution composed of 0.2 M calcium chloride, 0.1 M HEPES (pH 7), 20% (vol/vol) polyethylene glycol 3350, and 0.5 to 1 M different sugars. In addition to the previously reported MpPA14-glucose structure, the 14 different new sugars that were complexed with MpPA14 were l-fucose, GlcNAc, galactose, allose, mannose, 3-O-methyl-glucose, 2-deoxy-glucose, α-methyl-glucose, myo-inositol, sucrose, trehalose, l-arabinose, ribose, and 2-deoxy-ribose.
X-ray crystallographic data were collected at either the 08ID-1 beamline of the Canadian Light Source synchrotron facility or at the 23-ID-B beamline of the Advanced Photon Source via remote access. Data were indexed and integrated with X-ray Detector Software (XDS) (53) and CCP4-Aimless (54) or the DIALS/xia2 in the CCP4i2 software suite (55). The structure solutions for all complexes were obtained by molecular replacement using the MpPA14 glucose-bound structure as the search model (21). The structures were refined using CCP4-Refmac5 (56) or Phenix (57).
Glycan arrays.
Three different glycan arrays were probed with MpPA14. Two of the arrays focused on fungal, bacterial, algal, and plant polysaccharides, and the other on mammalian glycans. The first glycan array was done at the Carbohydrate Microarray Facility (Glycosciences Laboratory, Imperial College). GFP-MpPA14 (50 μg/ml) was exposed to the “Fungal, bacterial and plant polysaccharide array set 2,” which contained duplicates of 20 saccharide probes from a variety of organisms. An Alexa Fluor 647-tagged anti-GFP antibody was used for detecting the lectin, and the duplicates were averaged to produce the final relative fluorescence unit (RFU) values. In a negative-control experiment, where anti-GFP antibody was directly reacted to the saccharide probes, four glycans, namely lipomannan and lipoarabinomannan from Mycobacterium tuberculosis, lipoarabinomannan from Mycobacterium smegmatis, and native O-glycoprotein from M. tuberculosis, showed significant binding to the anti-GFP antibody, as they produced RFUs of greater than 1,000. This indicated that these four glycan samples yielded false-positive results. Therefore, these four polysaccharides were discarded from the analyses shown in Results of our paper.
The second glycan array was performed at the Max Planck Institute for Marine Microbiology (Bremen, Germany). The array contained duplicates of 32 polysaccharides, including those from macroalgae, bacteria, fungi, and land plants (see details in Table S5 in the supplemental material). N-terminally His-tagged MpPA14 was incubated with the array, and binding of the lectin to the polysaccharides was detected by an anti-His tag secondary antibody conjugated to alkaline phosphatase (Sigma-Aldrich). Microarray probing and quantification were performed as previously described (45). Maximal mean (average of the duplicates) signal intensity was set to 100, and the rest of values were normalized accordingly. A cutoff of 5 was applied (58).
The third glycan array screening was done by the Consortium for Functional Glycomics (Harvard Medical School) using version 5.2 of a printed mammalian glycan array, which contained 609 glycans (59). Tetramethyl rhodamine isocyanate (TRITC)-labeled MpPA14 was incubated with the surface-immobilized glycans, and the array was scanned at an excitation wavelength of 532 nm. The resulting RFUs were used as a measure of the bound protein. Each glycan was present in six replicates on the array, and the highest and lowest value from each set was omitted to avoid outlying values. The RFU values from the remaining four replicates were averaged.
Diatom binding experiments.
The Antarctic diatom, Chaetoceros neogracile, was cultured as previously described (21, 50). FITC-labeled MpPA14 or VcPA14 (FITC-MpPA14 or FITC-VcPA14, 0.2 mg/ml) in the presence or absence of sugars was incubated with diatoms in buffer (50 mM Tris-HCl [pH 9], 300 mM NaCl, and 5 mM CaCl2) with gentle mixing for 2 h. Next, diatoms were pelleted by centrifugation for 3 min at 7,000 rpm, and the resulting supernatant was discarded. This procedure was repeated three times to wash away unbound FITC-MpPA14 before the diatom pellet was finally resuspended in 20 μl buffer, which was then used to make slides for fluorescence microscopy. In a separate experiment to test if fucose could compete off the MpPA14 that was already bound to diatoms, FITC-MpPA14 was incubated with diatom for 1.5 h before fucose was added. The rest of the experiment followed the same procedure as described above.
Images were obtained using an Olympus IX83 inverted fluorescence microscope equipped with an Andor Zyla 4.2 Plus camera. Quantification of the fluorescence intensity was done using Fiji ImageJ. The corrected total cell fluorescence (CTCF) was calculated using the following formula: CTCF = integrated density − (area of selected cell × mean fluorescence of the background) (60). Quantification of 30 individual diatom cells was done for each treatment. Graphs were made using GraphPad Prism.
Data availability.
X-ray crystal structure coordinates solved in this study have been deposited in the Protein Data Bank under accession codes 6X7J (MpPA14-fucose), 6X7X (MpPA14-mannose), 6XAQ (MpPA14-α-methyl-glucose), 6X7Z (MpPA14-inositol), 6X7Y (MpPA14-GlcNAc), 6X7T (MpPA14-allose), 6X9M (MpPA14-3-O-methyl-glucose), 6X95 (MpPA14-2-deoxy-glucose), 6XAC (MpPA14-galactose), 6X8D (MpPA14-arabinose), 6X8Y (MpPA14-ribose), 6X9P (MpPA14-2-deoxy-ribose), 6X8A (MpPA14-sucrose), and 6XA5 (MpPA14-trehalose). The data that support the findings of this study are available from the corresponding author, P. L. Davies, upon reasonable request.
ACKNOWLEDGMENTS
We are grateful to John Allingham for the use of his home source X-ray diffractometer at Queen’s University and to staff members at the Canadian Light Source in Saskatoon and the Advanced Photon Source in Chicago for access to data collection at these synchrotrons. We thank EonSeon Jin, Hanyang University, Seoul, for the gift of the diatom Chaetoceros neogracile and Saeed Rismani Yazdi for assistance with the diatom cultures. We thank Xu Deng from Central South University (China) for fruitful discussions and for his thoughtful comments on the initial draft of the manuscript; we also thank Chantelle Capicciotti and Inka Brockhausen for comments. We thank Kim Munro for acquiring and interpreting ITC data. We appreciate Sherry Gauthier’s assistance with molecular cloning. In addition, we acknowledge the glycan array facilities at the Consortium for Functional Glycomics (CFG), Harvard Medical School, and the Glycosciences Laboratory at Imperial College for their help.
This project was funded by a Natural Science and Engineering Research Council (NSERC; http://www.nserc-crsng.gc.ca/index_eng.asp) Discovery Grant (RGPIN-2016-04810). P.L.D. holds the Canadian Research Chair in Protein Engineering, and T.D.R.V. held a Canadian Graduate Scholarship from NSERC.
S.G. and P.L.D. conceived the study, designed the experiments, and wrote the manuscript. S.G. performed cocrystallization, data collection, and structure determination of the X-ray crystal structures. R.E. and T.D.R.V. performed the comparative binding assays. H.Z. and C.S. performed the diatom binding experiments and analyzed data. S.V.-M. and J.-H.H. performed one of the microbial glycan microarrays and analyzed data. All authors contributed to the revision of drafts of the manuscript.
We declare that we have no competing interests.
Footnotes
Citation Guo S, Vance TDR, Zahiri H, Eves R, Stevens C, Hehemann J-H, Vidal-Melgosa S, Davies PL. 2021. Structural basis of ligand selectivity by a bacterial adhesin lectin involved in multispecies biofilm formation. mBio 12:e00130-21. https://doi.org/10.1128/mBio.00130-21.
REFERENCES
- 1.Varki A. 2011. Evolutionary forces shaping the Golgi glycosylation machinery: why cell surface glycans are universal to living cells. Cold Spring Harb Perspect Biol 3:a005462. doi: 10.1101/cshperspect.a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki A. 2017. Biological roles of glycans. Glycobiology 27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark GF. 2013. The role of carbohydrate recognition during human sperm-egg binding. Hum Reprod 28:566–577. doi: 10.1093/humrep/des447. [DOI] [PubMed] [Google Scholar]
- 4.Lepenies B, Lang R. 2019. Editorial: Lectins and their ligands in shaping immune responses. Front Immunol 10:2379. doi: 10.3389/fimmu.2019.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hebert E. 2000. Endogenous lectins as cell surface transducers. Bioscience Rep 20:213–237. doi: 10.1023/A:1026484722248. [DOI] [PubMed] [Google Scholar]
- 6.Ielasi FS, Alioscha-Perez M, Donohue D, Claes S, Sahli H, Schols D, Willaert RG. 2016. Lectin-glycan interaction network-based identification of host receptors of microbial pathogenic adhesins. mBio 7:e00584-16. doi: 10.1128/mBio.00584-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wesener DA, Wangkanont K, McBride R, Song XZ, Kraft MB, Hodges HL, Zarling LC, Splain RA, Smith DF, Cummings RD, Paulson JC, Forest KT, Kiessling LL. 2015. Recognition of microbial glycans by human intelectin-1. Nat Struct Mol Biol 22:603–610. doi: 10.1038/nsmb.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Kooyk Y, Rabinovich GA. 2008. Protein-glycan interactions in the control of innate and adaptive immune responses. Nat Immunol 9:593–601. doi: 10.1038/ni.f.203. [DOI] [PubMed] [Google Scholar]
- 10.Maestre-Reyna M, Diderrich R, Veelders MS, Eulenburg G, Kalugin V, Bruckner S, Keller P, Rupp S, Mosch HU, Essen LO. 2012. Structural basis for promiscuity and specificity during Candida glabrata invasion of host epithelia. Proc Natl Acad Sci U S A 109:16864–16869. doi: 10.1073/pnas.1207653109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo S, Vance TDR, Stevens CA, Voets IK, Davies PL. 2019. RTX adhesins are key bacterial surface megaproteins in the formation of biofilms. Trends Microbiol 27:470. doi: 10.1016/j.tim.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Vance TDR, Guo SQ, Assaie-Ardakany S, Conroy B, Davies PL. 2019. Structure and functional analysis of a bacterial adhesin sugar-binding domain. PLoS One 14:e0220045. doi: 10.1371/journal.pone.0220045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krogfelt KA, Bergmans H, Klemm P. 1990. Direct evidence that the FimH protein is the mannose-specific adhesin of Escherichia coli type 1 fimbriae. Infect Immun 58:1995–1998. doi: 10.1128/IAI.58.6.1995-1998.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wellens A, Garofalo C, Nguyen H, Van Gerven N, Slattegard R, Hernalsteens JP, Wyns L, Oscarson S, De Greve H, Hultgren S, Bouckaert J. 2008. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS One 3:e2040. doi: 10.1371/journal.pone.0002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mydock-McGrane LK, Hannan TJ, Janetka JW. 2017. Rational design strategies for FimH antagonists: new drugs on the horizon for urinary tract infection and Crohn’s disease. Expert Opin Drug Discov 12:711–731. doi: 10.1080/17460441.2017.1331216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sauer MM, Jakob RP, Luber T, Canonica F, Navarra G, Ernst B, Unverzagt C, Maier T, Glockshuber R. 2019. Binding of the bacterial adhesin FimH to its natural, multivalent high-mannose type glycan targets. J Am Chem Soc 141:936–944. doi: 10.1021/jacs.8b10736. [DOI] [PubMed] [Google Scholar]
- 17.Cusumano CK, Pinkner JS, Han Z, Greene SE, Ford BA, Crowley JR, Henderson JP, Janetka JW, Hultgren SJ. 2011. Treatment and prevention of urinary tract infection with orally active FimH inhibitors. Sci Transl Med 3:109–115. doi: 10.1126/scitranslmed.3003021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Totsika M, Kostakioti M, Hannan TJ, Upton M, Beatson SA, Janetka JW, Hultgren SJ, Schembri MA. 2013. A FimH inhibitor prevents acute bladder infection and treats chronic cystitis caused by multidrug-resistant uropathogenic Escherichia coli ST131. J Infect Dis 208:921–928. doi: 10.1093/infdis/jit245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spaulding CN, Klein RD, Ruer S, Kau AL, Schreiber HL, Cusumano ZT, Dodson KW, Pinkner JS, Fremont DH, Janetka JW, Remaut H, Gordon JI, Hultgren SJ. 2017. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 546:528–532. doi: 10.1038/nature22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo S, Garnham CP, Whitney JC, Graham LA, Davies PL. 2012. Re-evaluation of a bacterial antifreeze protein as an adhesin with ice-binding activity. PLoS One 7:e48805. doi: 10.1371/journal.pone.0048805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo S, Stevens CA, Vance TDR, Olijve LLC, Graham LA, Campbell RL, Yazdi SR, Escobedo C, Bar-Dolev M, Yashunsky V, Braslavsky I, Langelaan DN, Smith SP, Allingham JS, Voets IK, Davies PL. 2017. Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice. Sci Adv 3:e1701440. doi: 10.1126/sciadv.1701440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo S, Langelaan DN, Phippen SW, Smith SP, Voets IK, Davies PL. 2018. Conserved structural features anchor biofilm-associated RTX-adhesins to the outer membrane of bacteria. FEBS J 285:1812–1826. doi: 10.1111/febs.14441. [DOI] [PubMed] [Google Scholar]
- 23.Rigden DJ, Mello LV, Galperin MY. 2004. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem Sci 29:335–339. doi: 10.1016/j.tibs.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Taylor ME, Drickamer K. 2014. Convergent and divergent mechanisms of sugar recognition across kingdoms. Curr Opin Struct Biol 28:14–22. doi: 10.1016/j.sbi.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergmann C, Senderek J, Sedlacek B, Pegiazoglou I, Puglia P, Eggermann T, Rudnik-Schoneborn S, Furu L, Onuchic LF, De Baca M, Germino GG, Guay-Woodford L, Somlo S, Moser M, Buttner R, Zerres K. 2003. Spectrum of mutations in the gene for autosomal recessive polycystic kidney disease (ARPKD/PKHD1). J Am Soc Nephrol 14:76–89. doi: 10.1097/01.asn.0000039578.55705.6e. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida E, Hidaka M, Fushinobu S, Koyanagi T, Minami H, Tamaki H, Kitaoka M, Katayama T, Kumagai H. 2010. Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus. Biochem J 431:39–49. doi: 10.1042/BJ20100351. [DOI] [PubMed] [Google Scholar]
- 27.Zverlov VV, Volkov IY, Velikodvorskaya TV, Schwarz WH. 1997. Thermotoga neapolitana bglB gene, upstream of lamA, encodes a highly thermostable beta-glucosidase that is a laminaribiase. Microbiology 143:3537–3542. doi: 10.1099/00221287-143-11-3537. [DOI] [PubMed] [Google Scholar]
- 28.Cormack BP, Ghori N, Falkow S. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285:578–582. doi: 10.1126/science.285.5427.578. [DOI] [PubMed] [Google Scholar]
- 29.Veelders M, Bruckner S, Ott D, Unverzagt C, Mosch HU, Essen LO. 2010. Structural basis of flocculin-mediated social behavior in yeast. Proc Natl Acad Sci U S A 107:22511–22516. doi: 10.1073/pnas.1013210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiessling LL. 2018. Chemistry-driven glycoscience. Bioorg Med Chem 26:5229–5238. doi: 10.1016/j.bmc.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 31.Ambrosi M, Cameron NR, Davis BG. 2005. Lectins: tools for the molecular understanding of the glycocode. Org Biomol Chem 3:1593–1608. doi: 10.1039/b414350g. [DOI] [PubMed] [Google Scholar]
- 32.Garber N, Guempel U, Gilboa-Garber N, Royle RJ. 1987. Specificity of the fucose-binding lectin of Pseudomonas aeruginosa. FEMS Microbiology Lett 48:331–334. doi: 10.1111/j.1574-6968.1987.tb02619.x. [DOI] [Google Scholar]
- 33.Loris R, Tielker D, Jaeger KE, Wyns L. 2003. Structural basis of carbohydrate recognition by the lectin LecB from Pseudomonas aeruginosa. J Mol Biol 331:861–870. doi: 10.1016/s0022-2836(03)00754-x. [DOI] [PubMed] [Google Scholar]
- 34.Wilbur DJ, Williams C, Allerhand A. 1977. Detection of the furanose anomers of d-mannose in aqueous solution: application of carbon-13 nuclear magnetic resonance spectroscopy at 68 MHz. J Am Chem Soc 99:5450–5452. doi: 10.1021/ja00458a036. [DOI] [Google Scholar]
- 35.Kolatkar AR, Weis WI. 1996. Structural basis of galactose recognition by C-type animal lectins. J Biol Chem 271:6679–6685. doi: 10.1074/jbc.271.12.6679. [DOI] [PubMed] [Google Scholar]
- 36.Zelensky AN, Gready JE. 2005. The C-type lectin-like domain superfamily. FEBS J 272:6179–6217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 37.Gilbert HJ, Stalbrand H, Brumer H. 2008. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr Opin Plant Biol 11:338–348. doi: 10.1016/j.pbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Ueberschar KH, Blachnitzky EO, Kurz G. 1974. Reaction mechanism of d-galactose dehydrogenases from Pseudomonas saccharophila and Pseudomonas fluorescens. Formation and rearrangement of aldono-1,5-lactones. Eur J Biochem 48:389–405. doi: 10.1111/j.1432-1033.1974.tb03780.x. [DOI] [PubMed] [Google Scholar]
- 39.Angyal SJ. 1969. The composition and conformation of sugars in solution. Angew Chem Int Ed Engl 8:157–226. doi: 10.1002/anie.196901571. [DOI] [Google Scholar]
- 40.Rillahan CD, Paulson JC. 2011. Glycan microarrays for decoding the glycome. Annu Rev Biochem 80:797–823. doi: 10.1146/annurev-biochem-061809-152236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim SJ, Wan Aida WM, Schiehser S, Rosenau T, Bohmdorfer S. 2019. Structural elucidation of fucoidan from Cladosiphon okamuranus (Okinawa mozuku). Food Chem 272:222–226. doi: 10.1016/j.foodchem.2018.08.034. [DOI] [PubMed] [Google Scholar]
- 42.Ale MT, Meyer AS. 2013. Fucoidans from brown seaweeds: an update on structures, extraction techniques and use of enzymes as tools for structural elucidation. RSC Advances 3:8131–8141. doi: 10.1039/C3RA23373A. [DOI] [Google Scholar]
- 43.Vidal-Melgosa S, Sichert A, Francis TB, Bartosik D, Niggemann J, Wichels A, Willats WGT, Fuchs BM, Teeling H, Becher D, Schweder T, Amann R, Hehemann JH. 2021. Diatom fucan polysaccharide precipitates carbon during algal blooms. Nat Commun 12:1150. doi: 10.1038/s41467-021-21009-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Feizi T. 2018. The neoglycolipid (NGL) technology-based microarrays and future prospects. FEBS Lett 592:3976–3991. doi: 10.1002/1873-3468.13217. [DOI] [PubMed] [Google Scholar]
- 45.Vidal-Melgosa S, Pedersen HL, Schuckel J, Arnal G, Dumon C, Amby DB, Monrad RN, Westereng B, Willats WG. 2015. A new versatile microarray-based method for high throughput screening of carbohydrate-active enzymes. J Biol Chem 290:9020–9036. doi: 10.1074/jbc.M114.630673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gugi B, Le Costaouec T, Burel C, Lerouge P, Helbert W, Bardor M. 2015. Diatom-specific oligosaccharide and polysaccharide structures help to unravel biosynthetic capabilities in diatoms. Mar Drugs 13:5993–6018. doi: 10.3390/md13095993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith DC, Steward GF, Long RA, Azam F. 1995. Bacterial mediation of carbon fluxes during a diatom bloom in a mesocosm. Deep Sea Res Part II Top Stud Oceanogr 42:75–97. doi: 10.1016/0967-0645(95)00005-B. [DOI] [Google Scholar]
- 48.Haug A, Myklestad S. 1976. Polysaccharides of marine diatoms with special reference to Chaetoceros species. Marine Biology 34:217–222. doi: 10.1007/BF00388798. [DOI] [Google Scholar]
- 49.Torode TA, Marcus SE, Jam M, Tonon T, Blackburn RS, Herve C, Knox JP. 2015. Monoclonal antibodies directed to fucoidan preparations from brown algae. PLoS One 10:e0118366. doi: 10.1371/journal.pone.0118366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gwak IG, Jung WS, Kim HJ, Kang SH, Jin E. 2010. Antifreeze protein in Antarctic marine diatom, Chaetoceros neogracile. Mar Biotechnol (NY) 12:630–639. doi: 10.1007/s10126-009-9250-x. [DOI] [PubMed] [Google Scholar]
- 51.Krachler AM, Orth K. 2013. Targeting the bacteria-host interface: strategies in anti-adhesion therapy. Virulence 4:284–294. doi: 10.4161/viru.24606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalas V, Hibbing ME, Maddirala AR, Chugani R, Pinkner JS, Mydock-McGrane LK, Conover MS, Janetka JW, Hultgren SJ. 2018. Structure-based discovery of glycomimetic FmlH ligands as inhibitors of bacterial adhesion during urinary tract infection. Proc Natl Acad Sci U S A 115:E2819–E2828. doi: 10.1073/pnas.1720140115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W. 2010. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans P. 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 55.Beilsten-Edmands J, Winter G, Gildea R, Parkhurst J, Waterman D, Evans G. 2020. Scaling diffraction data in the DIALS software package: algorithms and new approaches for multi-crystal scaling. Acta Crystallogr D Struct Biol 76:385–399. doi: 10.1107/S2059798320003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 57.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moller I, Sorensen I, Bernal AJ, Blaukopf C, Lee K, Obro J, Pettolino F, Roberts A, Mikkelsen JD, Knox JP, Bacic A, Willats WGT. 2007. High-throughput mapping of cell-wall polymers within and between plants using novel microarrays. Plant J 50:1118–1128. doi: 10.1111/j.1365-313X.2007.03114.x. [DOI] [PubMed] [Google Scholar]
- 59.Heimburg-Molinaro J, Song X, Smith DF, Cummings RD. 2011. Preparation and analysis of glycan microarrays. Curr Protoc Protein Sci Chapter 12:Unit12.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A 107:12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural features of MpPA14. (A) Overview of the MpPA14 structure in complex with glucose. (Right) Interface from a view that is rotated approximately 180° around a vertical axis from the left. Carbon atoms are colored grey, oxygen atoms are red, and nitrogen atoms are blue. The glucose-coordinating Ca1 is shown as a dark blue sphere, while the other Ca2+ ions are shown as green spheres. (B) Zoomed-in view of the ligand-binding site of MpPA14. The DcisD motif is indicated by an arrow. (C to E) Ca2+ coordination sites for Ca2 to Ca4. (F) Surface electrostatic potential of MpPA14. Surfaces that are electronegative are colored in red, while those that are neutral are colored white, and the electropositive surfaces are colored blue. (G) Amino acid alignment of PA14 domains from Marinomonas primoryensis (MpPA14), the oil-eating bacterium Marinobacter hydrocarbonoclasticus (MhPA14), and the cholera-causing human pathogen Vibrio cholerae (VcPA14). Amino acid residue number and secondary structure components for MpPA14 are indicated. Conserved residues that constitute the carbohydrate-binding site are highlighted in magenta. Red arrowhead points to the DcisD motif. Loops (L9 and L11) involved in carbohydrate binding are indicated in red. Download FIG S1, PDF file, 0.3 MB (324.8KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallographic statistics for MpPA14 in complex with l-fucose, mannose, α-methyl-glucose, inositol, GlcNAc, allose, and 3-O-methyl glucose. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structures of MpPA14 in complex with glucopyranose-containing sugars. The color scheme and labeling are the same as those in Fig. 2. Download FIG S2, PDF file, 0.2 MB (251.5KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structures of strongly binding (A), moderately binding (B), and weakly binding (C) sugars of MpPA14. The specific isomer shown for each monosaccharide was revealed by X-ray crystallography as described in Fig. 2 and 3 and Fig. S2 and S5. Double arrowheads point to the diol groups used by the sugar for interaction with Ca1 in the ligand-binding site of MpPA14. Alternative binding configurations are indicated with double arrowheads in blue or green instead of red. Download FIG S3, PDF file, 0.1 MB (148KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Crystal packing of MhPA14 and MpPA14. (A) Crystal unit cell of MhPA14 in complex with glucose. The box with black dashed lines indicates the interface between one molecule of MhPA14-glucose (grey) and its neighboring symmetry-related molecules (magenta) in the crystal unit cell. (B) The glucose molecule bound by MhPA14 is directly involved in crystal contact with a symmetry-related molecule. Lengths for the protein-sugar interaction bonds (black dashed lines) are indicated. (C) Crystal unit cell of MpPA14 in complex with GlcNAc. The box with black dashed lines indicates the interface between one molecule of MpPA14-GlcNAc (grey) and its neighboring symmetry-related molecules (magenta) in the crystal unit cell. (D) The GlcNAc bound by MpPA14 does not interact with a symmetry-related molecule in the crystal unit cell. The distances between the GlcNAc and the nearest atoms from a neighboring symmetry-related molecule are indicated by green dashed lines. Download FIG S4, PDF file, 0.3 MB (338.3KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Structures of MpPA14 in complex with its moderately and weakly binding ligands. (A) β-3-O-Methyl-glucose. (B) Ribose. (C) 2-deoxy-ribose. (D to F) Three binding conformations of Inositol. The color scheme and labeling are the same as those in in Fig. S2. Download FIG S5, PDF file, 0.2 MB (202.4KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
X-ray crystallographic statistics for MpPA14 in complex with 2-deoxy-glucose, galactose, l-arabinose, ribose, 2-deoxy-ribose, sucrose, and trehalose. Download Table S2, DOCX file, 0.02 MB (19.2KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polysaccharides used in the first glycan array. The first column on the left presents the relative lectin-binding activity detected. Download Table S3, DOCX file, 0.02 MB (16.4KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Polysaccharides contained in the second glycan array. The first column on the left presents the lectin-binding activity detected (normalized GFP-MpPA14 signal intensity). Polysaccharide structures are listed in the same order as in the x axis from Fig. 4B (order of appearance shown in second column). Download Table S4, DOCX file, 0.02 MB (20.3KB, docx) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Representative images of diatoms with various fluorescein isothiocyanate (FITC)-MpPA14 and sugar treatments shown in Fig. 5. Asterisks (*) represent experiments where diatoms were incubated with FITC-MpPA14 before l-fucose was added. Download FIG S6, PDF file, 0.1 MB (107.9KB, pdf) .
Copyright © 2021 Guo et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
X-ray crystal structure coordinates solved in this study have been deposited in the Protein Data Bank under accession codes 6X7J (MpPA14-fucose), 6X7X (MpPA14-mannose), 6XAQ (MpPA14-α-methyl-glucose), 6X7Z (MpPA14-inositol), 6X7Y (MpPA14-GlcNAc), 6X7T (MpPA14-allose), 6X9M (MpPA14-3-O-methyl-glucose), 6X95 (MpPA14-2-deoxy-glucose), 6XAC (MpPA14-galactose), 6X8D (MpPA14-arabinose), 6X8Y (MpPA14-ribose), 6X9P (MpPA14-2-deoxy-ribose), 6X8A (MpPA14-sucrose), and 6XA5 (MpPA14-trehalose). The data that support the findings of this study are available from the corresponding author, P. L. Davies, upon reasonable request.