Abstract
A reporter assay system is an essential tool for investigating gene expression mechanisms. In the case of bifidobacteria, several convenient and sensitive reporter systems have been developed. Here, we developed a new reporter system for bifidobacteria using the chloramphenicol acetyltransferase gene (cat) from Staphylococcus aureus. This enzyme stoichiometrically produced free CoA-SH, which was analyzed quantitatively with Ellman’s test using 2-nitrobenzoic acid (DTNB). The 2-nitro-5-thiobenzoate (TNB2-) produced showed a strong yellowish color with maximum absorbance at 412 nm. We also constructed a new pBCMAT plasmid series for CAT assays in bifidobacteria to evaluate promoters and terminators. Analyses using promoters from Bifidobacterium longum NCC2705 indicated that the CAT assay using these promoters is quantitative, has a wide measurement range, and is stable. In addition, this assay was useful for several bifidobacterial species, including B. longum, Bifidobacterium breve, and Bifidobacterium adolescentis. Compared with evoglow-Bs2, a fluorescent protein used under anaerobic conditions, the CAT assay showed about 0.25% background activity. In analyses using this CAT assay, we identified 11 promoters and 12 terminators of B. longum NCC2705. The genes encoding ribosomal proteins, elongation factors, and transfer RNAs possessed strong promoters, and terminators that include strong stem-loops and poly-U tails structures tended to show high activities. Although the abovementioned promoters made stronger contributions to expression activities than the terminators, the maximum fold difference in the activities among the tested terminators was approximately 17-fold. Modification of the -10 box and 5’-UTR in the promoters and the structure around the stem-loop in the terminators affected expression levels. These results suggest that the CAT assay is useful for various analyses of bifidobacterial gene expression.
Keywords: Bifidobacterium, promoter, chloramphenicol acetyltransferase, terminator, reporter assay
INTRODUCTION
Over the last several decades, numerous bacterial promoters have been elucidated in model organisms [1]. Bifidobacterial promoters have also been investigated in recent studies by gene expression profiling [2, 3]. These recent studies indicated that the core structure of bifidobacterial promoters was similar to that of a typical bacterial promoter, and that the consensus sequences of the -35 and -10 boxes were TTGACA and TATAAT, respectively [2, 3]. However, there was a difference in the optimal spacer length between the -35 and -10 boxes. Bifidobacterial promoters possess an 11-bp spacer as well as a 17-bp spacer, which were thought to be the lengths of bacterial promoters using the primary sigma-factor [2]. Since bacterial transcription is likely to be regulated by organism-specific rules, we should reevaluate these rules in bifidobacteria. Therefore, it is essential to establish a tool to evaluate promoter activities in these organisms.
Green fluorescent protein (GFP), first isolated from the jellyfish Aequorea victoria (avGFP), is one of the most commonly used reporters, and the system does not require specific reagents or cell disruption procedures. However, in an anaerobic organism, avGFP is not useful because activation of GFP requires oxidation at Ser65-Tyr66-Gly67 with molecular oxygen [4]. Although some anaerobic fluorescent proteins, such as evoglow [5], have been developed using flavoproteins, their sensitivity in bifidobacteria remains unclear.
Several studies have described reporter systems for bifidobacteria using β-glucuronidase, α-galactosidase, arabinofuranosidase, β-glucosidase, and luciferase as reporter genes [6,7,8,9,10,11]. However, bifidobacteria possess various gene sets for sugar metabolism, with each strain having different gene sets, most of which are still unknown. Thus, they may compete with reporter genes, except for luciferase, and increase the background signal in the assay.
In this study, we employed the cat gene, which encodes chloramphenicol acetyltransferase (CAT), in a new reporter system for bifidobacteria. CAT confers chloramphenicol resistance to the host bacterium. It catalyzes the transfer of an acetyl group from acetyl-CoA to chloramphenicol, producing an inactive product, chloramphenicol 3-acetate [5], which causes it to lose its bactericidal activity.
Researchers have frequently used this reaction, designated the “CAT assay”, to evaluate eukaryotic promoter activity [12]. In these assays, the cat gene is introduced downstream of the target promoter and then transferred into target cells. Cell lysates are prepared, and an enzymatic reaction using a 14C-labeled radioisotopic substrate is measured. Thus, thin-layer chromatography (TLC) separation and autoradiography are required to assess its activity [13]. This method requires these complicated techniques (TLC and autoradiography), and it has a limited dynamic range. In the 1990s, more convenient reporter genes, such as luciferase and GFP, were introduced, and they have become more widely used because of their simpler and safer procedures, wide quantitative range, and applicability in living cells [14, 15]. Thus, the CAT assay has become a forgotten method for reporter assay experiments.
Ellman’s test is a well-known method for quantifying free SH-groups using 5,5’-Dithiobis (2-nitrobenzoic acid, DTNB). This reaction stoichiometrically produces TNB2−, which shows strong absorbance (ε = 13,700 M−1 cm−1) in the yellow range (412 nm). CAT catalyzes a reaction in which acetyl-CoA donates an acetyl group to chloramphenicol and produces free CoA-SH, which can be quantified by Ellman’s test [16]. This should be useful to assay CAT activity with a convenient and low-cost procedure.
In this study, we applied this reporter assay system to bifidobacteria using the cat gene. As target factors for the analysis, we focused on the terminators and promoters. Terminators generally affect both transcription and the half-life of mRNA. Bifidobacterial terminators have not been well studied with wet assays and need to be more thoroughly characterized.
MATERIALS AND METHODS
Bacterial strains and plasmid construction
The strains and plasmids used in this study are listed in Supplementary Table 1. PCR for cloning promoters and terminators was performed with KOD-Plus-Neo (TOYOBO, Osaka, Japan) and specific oligo-DNA primers, as listed in Supplementary Table 2, using the Bifidobacterium longum NCC2705 genome as a template. The plasmid backbone of the pBCMAT series and pBIFGLOW series (Supplementary Fig. 1, Supplementary Table 1) is a derivative based on the pKKT427 vector [17]. The reporter genes evoglow-Bs2 and cat were cloned from pGLOW-Bs2 [5] and pKO403-TPCTcon [2]. The complete plasmids were constructed through the Golden Gate method [18] with several DNA fragments (vector backbone, terminators, promoters, and reporter genes). Modifications of promoters, TrpsO, and chimeric promoters were constructed using the strategy shown in Supplementary Fig. 2. PCR was performed using the primers listed in Supplementary Table 3. For improved promoters and TrpsO, 5′-phosphate labeled primers were used for amplification, and the products were self-ligated. In the case of chimeric promoters, the ligation reaction was performed using the Golden Gate method. All plasmids were constructed in Escherichia coli TOP10 cells. The cells were lysed, and the constructs were extracted and purified to confirm their sequences.
Bacterial culture
Bifidobacterium strains were grown in MRS medium at 37°C under anaerobic conditions. E. coli TOP10 strains were grown in LB medium (1% tryptone, 0.5% yeast extract, and 1% NaCl) at 37°C with shaking. For solid media, agar was added at 1.5% (w/v). For cultivation of transformants, spectinomycin was added to the medium at 75–300 µg/mL.
Transformation into bifidobacterial strains
The plasmids were introduced into B. longum NCC2705 [19], Bifidobacterium breve JCM 1192 [20], and Bifidobacterium adolescentis ATCC15703 [21]. Cultivated cells (600 µL) reaching an OD660 of 0.55 were centrifuged, and the pellets were suspended in 600 µL 0.05 M buffered sucrose (99:1 ratio of 0.05 M sucrose and 100 mM tri-ammonium citrate, pH 6.0). After centrifugation, the pellets were suspended in 50 µL of 0.05 M buffered sucrose and used as the electro-competent cells. Plasmid samples (100 ng) were added to the cells, and the mixtures were incubated for 5 min on ice. After incubation, electroporation was performed using a MicroPulser (2.10 kV, Bio-Rad, Hercules, CA, USA), and the cells were cultivated in MRS media for 3 hr at 37°C. After incubation, the cells were transferred into MRS agar plates containing 75 or 300 µg/mL spectinomycin (300 µg/mL) and were used for B. adolescentis ATCC 15703.
CAT assay
The CAT assays were performed using extracts from three independent bacterial cultures. Bifidobacterial strains were cultured in MRS medium until the OD660 reached 0.37 to 0.40. The cells were washed with PBS and sonicated at level 5 for 45 sec (XL-2000, Misonix, Farmingdale, NY, USA). The sonicated samples were centrifuged (15,000 × g, 3 min, 4°C), and the supernatants were transferred to new tubes. A 15 µL aliquot of the supernatant was then mixed with 650 µL of CAT assay solution (100 mM Tris-HCl [pH 8.0], 2.5 mM DTNB, 5.0 mM acetyl-CoA, and 0.3% [w/v] chloramphenicol) in a 1.5 mL tube, incubated at 37°C for 10 min, and then transferred to a 96-well plate (200 µL × 3 wells). Finally, the absorbance was measured at 412 nm using a microplate reader (SpectraMax M5, Molecular Devices Japan, Tokyo, Japan). The interval time between when incubation at 37°C was halted and measurement began was exactly 30 seconds. The CAT activity was calculated as follows:
The total amount of protein in each sample was measured using the Bradford method, with a Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad) and a standard protein gamma globulin. The total amount of protein for the CAT assay ranged from 20 to 25 µg, except for the assay shown in Fig. 1A.
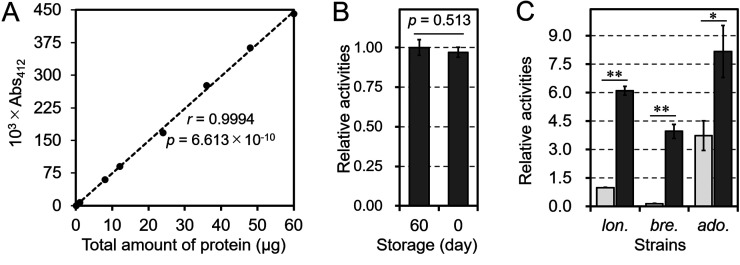
Fig. 1.
Evaluation of the chloramphenicol acetyltransferase (CAT) assay for bifidobacterial strains.
A: Analysis of the quantitativeness of the CAT assay. The pBCMAT-Pgap-TdppA2 plasmid was transformed into B. longum NCC2705. Different dilutions of the crude cell extract were prepared and assayed. B: Comparison of samples stored for different time periods. The results are shown relative to the results obtained using Pgap stored at −28°C for 60 days. C: Comparison of the assays in different bifidobacterial strains. The plasmids pBCMAT-Pgap-TdppA2 and pBCMAT-PrpmB-TdppA2 were transformed into B. longum NCC2705 (lon.), B. breve JCM 1192 (bre.), and B. adolescentis ATCC 15703 (ado.). The results are relative to those obtained using Pgap in B. longum NCC2705. The light-gray and dark-gray bars show the CAT activities with Pgap and PrpmB, respectively. Values are presented as mean values ± SD. CAT assays were performed on extracts prepared from three independent bacterial cultures. Statistically distinguishable differences in B and C between two samples were assessed by Student’s t-test (*p<0.05; **p<0.01). r is the Pearson’s correlation coefficient between the total protein of the sample and absorbance at 412 nm. The p value in A was calculated using a test of no correlation based on the null hypothesis that there is no correlation between the 103 × A412 and the total amount of protein.
Measurement of the fluorescence from evoglow-Bs2
Bifidobacterial strains carrying evoglow-Bs2 were cultivated at 37°C until they reached an OD660 of about 0.37 to 0.40. Then, the cells were washed with PBS, and fluorescence was measured at the excitation and emission wavelengths of 450 nm and 495 nm (λex 450 nm and λem 495 nm), respectively, with a microplate reader (SpectraMax M5).
Selection of test promoters and terminators
The selected test promoters were positioned in the regions of the upstream genes and were classified in the high expression group, as in a previous study [2]. Except for Phup and Ptuf, the promoters contained the motif TTGNNN-N17–18-TANNNT. In contrast, the tested terminators were selected from the WebGeSTer DB terminator database [22].
RESULTS
Expression analysis in Bifidobacterium using the developed CAT assay
B. longum NCC2705 carrying pBCMAT-Pgap-TdppA2 was used for the CAT assay. Several samples of different dilutions showed linear signals (Abs412 = 0.001–0.450), with a significant correlation (r=0.994, p=6.613 × 10−10). In addition, a crude extract stored at −28°C for 60 days showed almost the same activity as a fresh sample (Fig. 1B). These data indicate that this method is reliable for routine reporter assay experiments.
Next, the two generated plasmids, pBCMAT-Pgap-TdppA2 and pBCMAT-PrpmB-TdppA2, were transformed into three strains, B. longum NCC2705, B. breve JCM 1192, and B. adolescentis ATCC 15703, and CAT assays were performed. As shown in Fig. 1C, all transformants showed CAT activity, and expression levels, measured as CAT activity, tended to be higher in strains carrying PrpmB than in strains carrying Pgap. In addition, the highest to lowest activity levels were in the order of B. adolescentis, B. longum, and B. breve.
Comparison of the performance of the CAT and evoglow-Bs2 assays
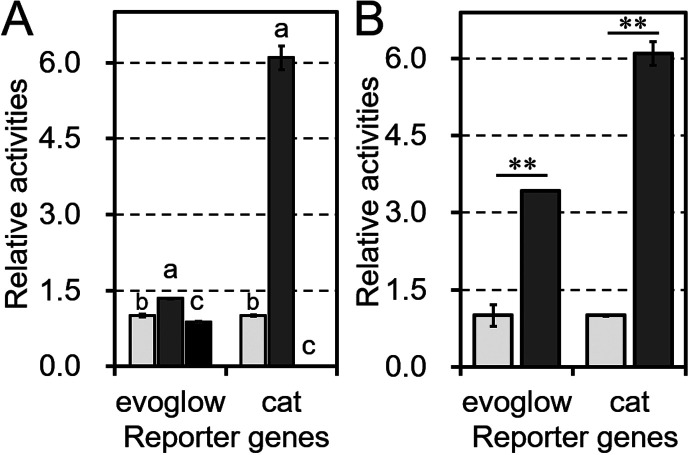
To objectively evaluate the performance of the CAT assay, we tested another reporter system using a fluorescent flavoprotein designed for anaerobic bacteria, evoglow-Bs2. Two plasmids, pBIFGLOW-Bs2-Pgap-TdppA2 and pBIFGLOW-Bs2-PrpmB-TdppA2, were constructed by replacing the respective CAT coding sequences in pBCMAT-Pgap-TdppA2 and pBCMAT-PrpmB-TdppA2 with the evoglow-Bs2 coding sequence. As shown in Fig. 2A, although evoglow-Bs2 fluorescence could be detected in the transformants, the background signal from the transformants carrying the empty vector pKKT427 was 86% of the fluorescence of the Pgap plasmid. In contrast, the CAT assay empty vector showed only 0.2% of the activity of the Pgap plasmid, which was about 0.25% of the background level in the evoglow-Bs2 assay. After subtracting the background levels, PrpmB showed significantly higher activity levels than Pgap in both the CAT and evoglow-Bs2 assays (Fig. 2B).
Fig. 2.
Comparison of the performance of the chloramphenicol acetyltransferase (CAT) and evoglow-Bs2 assays.
A: Relative activity levels in the CAT and evoglow-Bs2 assays. Tested transformants carried a cat reporter gene driven by either Pgap (light-gray bars), PrpmB (dark-gray bars), or the promoter-less pBCMAT or pBIFGLOW vectors (black bars), which were used to measure background levels. All plasmids were transformed into B. longum NCC2705. B: Relative activities after subtracting the background. The results are shown relative to those obtained with Pgap, which was set as 1. Values are presented as means ± SD. The assays were performed on the extracts of three independent bacterial cultures. Statistically distinguishable differences between two samples were evaluated by Student’s t-test (**p<0.01). Different letters indicate statistically distinguishable groups (p<0.05; Tukey’s multiple-comparison test).
Effect of promoters on bifidobacterial transcription
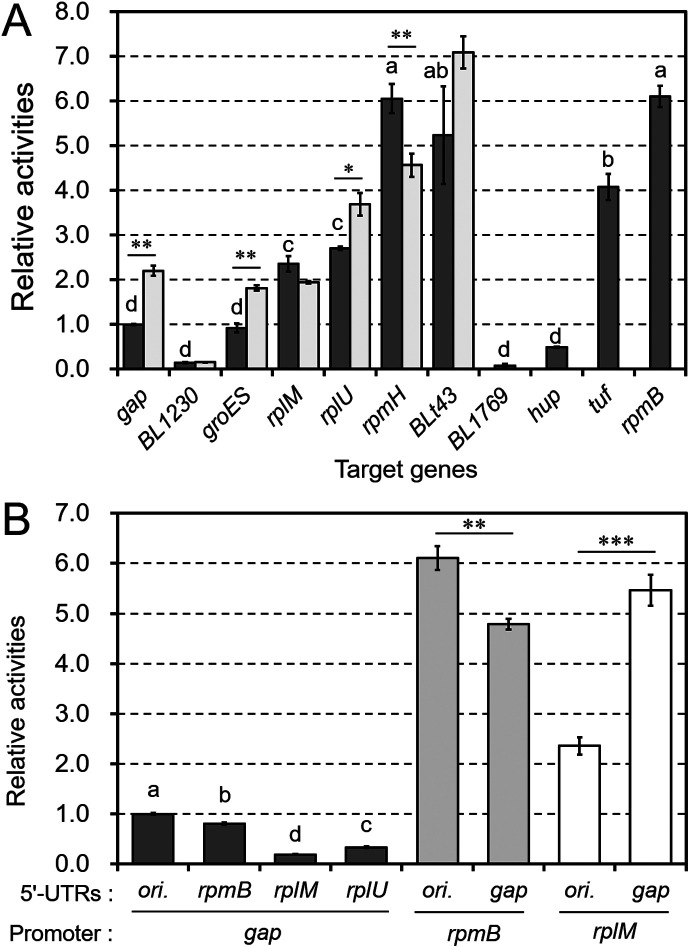
We used this assay to assess the transcriptional activity of several promoters (Fig. 3). We selected and cloned the promoters from B. longum NCC2705 listed in Table 1. These promoters contain the core motifs of the -35 and -10 boxes, TTGNNN and TANNNT, respectively, as described in previous study [2]. In the transformed B. longum NCC2705, there were significant differences in the CAT activities of these promoters (Fig. 3A). Several promoters, such as PrpmB, PrpmH, PBLt43, Ptuf, PrplU, and PrplM, showed higher activities than Pgap, which is commonly used as a constitutive control promoter.
Fig. 3.
The impact of promoters on chloramphenicol acetyltransferase (CAT) activity.
A: Eleven promoter regions were inserted into the pBCMAT construct. The genes driven by these promoters are shown on the horizontal axis. The relative activities of the promoters are shown as dark-gray bars, and the modified Pgap, PgroES, PrpmH, PBLt43, PrplU, PrplM, and PBL1230 are shown as light-gray bars. B: Chimeric promoters generated using different core regions and 5’-UTRs were also tested in the CAT assay. All constructed plasmids were transformed into B. longum NCC2705. The results are shown relative to the activity of Pgap. Values are presented as means ± SD. CAT assays were performed on extracts from three independent bacterial cultures. Different letters indicate statistically distinguishable groups (p<0.05; Tukey’s multiple-comparison test). Statistically distinguishable differences between two samples were assessed using Student’s t-test (*p<0.05; **p<0.01; ***p<0.001).
Table 1. List and structures of the promoters tested with the chloramphenicol acetyltransferase (CAT) assay.
| Genes | -35 box | Spacer (length [bp]) | -10 box |
|---|---|---|---|
| groES (BL1558) | TTGGCA | GAGTGCTAATAATCGCGC (18) | TACGAT |
| gap (BL1363) | TTGCCA | TGTGTACAGAGTCGGCAT (18) | TACAGT |
| rpmB (BL0330) | TTGCGG | TTGTCGGTATTGGGCTA (17) | TATATT |
| rpmH (BL0642) | TTGACT | TGAGGGTAGGGGTGAGAG (18) | TACTTT |
| BLt43 | TTGCGA | TAATCCACCGGAGCCGT (17) | TACTAT |
| rplU (BL1282) | TTGATT | TAGTCCGCATGGTGGTG (17) | TAGATT |
| tuf (BL1097) | GTGGCA | CATAACCCAGAAACCCAG (18) | TAGAAT |
| rplM (BL1571) | TTGCCC | TATGTCAAGCCCGGCGA (17) | TATACT |
| BL1230 | TTGTGA | TTCAAATTATCATTCAT (17) | TACAAT |
| hup (BL1798) | TTCGCA | GAAACATGCGC (11) | TAGTAT |
| BL1769 | TTGACA | ACATTGGTAAAAGGTTA (17) | TATCAT |
The target promoters of the CAT assay were the upstream regions of the listed genes. We also showed the promoter core motifs, termed -35 and -10 boxes, and the sequences of the spacers.
Next, we elucidated the effect of modifying the core motifs on promoter activity. Promoters rarely possess consensus -35 and -10 boxes (TTGACA and TATAAT, respectively); however, the -10 box is thought to be an essential element for transcriptional initiation, as it is involved in the dissociation of the double-stranded structure of DNA [23]. Therefore, we replaced the -10 box of several promoters (Pgap, PgroES, PrpmH, PBLt43, PrplU, PrplM, and PBL1230) with the consensus motif TATAAT and measured their CAT activities (Fig. 3A, Supplementary Fig. 2). Modified Pgap, PgroES, and PrplU showed significantly higher activities than the corresponding original promoters. These results indicate that the degree of matching within the -10 box consensus motif influences transcription efficiency. To elucidate the impact of the 5’-UTR on gene expression, we created several chimeric promoters. The original promoters were divided into two regions, the core and 5’-UTR regions, which were upstream and downstream of the -10 box, respectively (Supplementary Fig. 2). As shown in Fig. 3B, exchanging the 5’-UTRs significantly altered expression levels, indicating that the 5’-UTR is an important factor affecting expression levels.
Effect of terminators on bifidobacterial expression
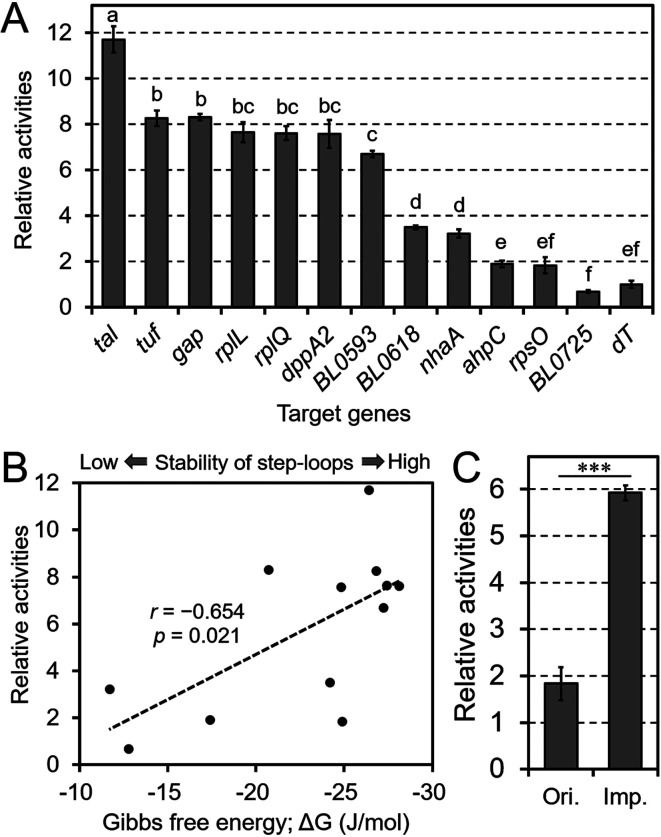
To investigate the impact of terminator sequences on gene expression, we performed a CAT assay with 11 different terminators (Table 2). B. longum NCC2705 was transformed with the pBCMAT-Pgap-Txxx series plasmids containing a terminator following the cat gene (Supplementary Fig. 1). All terminators were presumed to be of the rho-independent type because of the existence of U-rich regions following strong stem-loops (Table 2). The results of the CAT assays showed that the absence of a terminator significantly reduced CAT activity (Fig. 4A). However, the results also showed differences in CAT activities among the terminators, with the largest difference observed between the Ttal and TBL0725 terminators.
Table 2. List and structures of the terminators tested in the chloramphenicol acetyltransferase (CAT) assay.
| Genes | Sequences of terminators | dG |
|---|---|---|
| rplQ (BL1607) | UAGCUGAAAGGGCUGGAAUCCGUUUGGGUUCCAGCCCUUUUGCGUUGUUUU | -28.1 |
| rplL (BL1550) | UUUUCG(A)GGAACCCCGCACCUGUGUUUCAGG(C)UGCGGGGUUUCCGCAUACU | -27.4 |
| BL0593 | UUAUGAGAAAGGCCGACCUCC(C)GUAUGCGGAGGUCGGCCUUUUUCGUGCCUU | -27.2 |
| tuf (BL1097) | GUUAUAGGAAAUCCCCUUCGAGGAAACUCGGAGGGGAUUUUCUAUAAC | -26.8 |
| tal (BL0715) | ACAGAAGCCCCGACCCCAACGGUCGGGGCUUCUUUCGUUUGUU | -26.4 |
| rpsO (BL1545a) | AAGACUUCCGCCCGAGUACCUAGGAUGGUGCUCGGGCGUUUUACAUAU | -24.9 |
| [imp.] | (AAGAAAAACGCCCGAGUACCUAGGAUGGUGCUCGGGCGUUUUACAUAU ) | |
| dppA2 (BL1386) | AAAUAAAGGGCG(A)GGAUCGCAAGAUCC(C)CGCCCUUUUUGUUGUGU | -24.8 |
| BL0618 | AGUAUUCCGUGGCUCCCCUCAAAGAGGGGAGCCACUUUCGUCU | -24.2 |
| gap (BL1363) | AGAACUAAGCUGUGAGACCCCCGCCUUGUGCGGGGGUUUCUUCGUAGU | -20.7 |
| ahpC (BL0615) | GUAAUCUUGGC(C)CCCCUUUCUGAGGGG(A)GCUGUCGCUGAAA | -17.4 |
| BL0725 | ACCCGCGC(A)ACCGGUUUUAUCGGU(AC)GCGCAAUAAGACAAU | -12.8 |
| nhaA (BL0942) | GUGAAAACCGUACG(C)UGAGACGCGCUCA(C)CGUACGUUCUUCACG | -11.7 |
The targeted terminators evaluated in the CAT assay were downstream of the listed genes. The sequences around the stem-loops of the terminators and the Gibbs free energy (ΔG, kcal/mol) of the stem-loops are shown. Underlined and boldfaced sequences correspond to stem-loops and U-rich regions, respectively. The bases inside parentheses do not contribute to the stem-loops.
Fig. 4.
The impact of terminators on chloramphenicol acetyltransferase (CAT) activities.
A, C: The regions downstream of the target genes were inserted into pBCMAT. All constructed plasmids were transformed into B. longum NCC2705. The results are shown relative to the activity levels obtained with a terminator-less vector (dT). The terminators are shown on the horizontal axis. The original and improved TrpsO constructs in C are shown as “Ori.” and “Imp.,” respectively. Values are presented as means ± SD. CAT assays were performed on extracts from three independent bacterial cultures. Different letters indicate statistically distinguishable groups (p<0.05; Tukey’s multiple-comparison test). Statistically distinguishable differences between two samples were evaluated using Student’s t-test (***p<0.001). B: Correlation between CAT activity and Gibbs free energy (ΔG, kcal/mol) for each stem-loop. Each black dot represents each terminator shown in A and Table 2. r is the Pearson’s correlation coefficient between the ΔG and CAT activities of the tested terminators. The p value in B was calculated using a test of no correlation based on the null hypothesis that there is no correlation between CAT activities and the Gibbs free energy.
To clarify the factors causing these differences, we assessed the correlation between CAT activity and the stability of the stem-loops of the terminators, as predicted by their Gibbs free energy (Fig. 4B). The analysis showed a correlation between these factors (r=−0.654, p=0.021). In addition, TnhaA and TahpC, which possess weaker stem-loops, showed comparatively low activities. In contrast, although TrpsO possesses an ideal terminator structure, with a stable stem-loop (ΔG =−24.9 kcal/mol) followed by a poly-U tail, the CAT activities of constructs containing this terminator were low. Further analysis of the structure of the terminators showed that poly-A tails upstream of the stem-loops were conserved in six terminators: TdppA2, TrplQ, TBL0593, Ttuf, Ttal, and TnhaA (Table 2). To confirm whether the poly-A tail affects gene expression, we modified TrpsO, as shown in Fig. 4C. The results of the assay showed that the modified TrpsO had 3.2-fold higher activity than the original TrpsO, indicating the strong impact of the poly-A tail on gene expression.
DISCUSSION
In a previous study [2], we constructed a pKO403-TPCT series for a CAT assay in B. longum NCC2705. Plasmids based on pKO403 have a temperature-sensitive replication origin (ori Ts). Due to this specific origin, the previous plasmids were not available for the assay under heat-shock stress. Moreover, they have three hup terminators, as shown in Supplementary Fig. 1A. Such a structure may cause the loss of the CAT gene by inter-plasmid homologous recombination after transformation into bifidobacteria. To overcome these disadvantages, we constructed a pBCMAT series (Supplementary Fig. 1B) for the CAT assay. Its members are based on a pKKT427 vector and have TdppA2 or other terminators in the downstream region of the cat gene instead of Thup.
In this study, we developed a CAT assay to measure promoter or terminator activities in some species of the genus Bifidobacterium. The results shown in Fig. 1A and 1C indicate that this CAT assay is useful for quantitative analyses of expression with a wide measurement range in B. breve, B. adolescentis, and B. longum. Remarkably, B. adolescentis showed higher activities than B. longum despite using Pgap and PrpmB from B. longum. This is likely due to the difference in incubation time during cultivation. It took longer for B. adolescentis to grow to a certain level (OD660 = 0.37–0.40) in culture compared with B. longum (data not shown). This difference could depend on strain-specific factors, such as oxygen tolerance. Such a delay in growth was likely caused by the accumulation of CAT protein. Another possible reason for the difference in the activities is that the difference in the activities could depend on the plasmid copy number in each strain; therefore, the activities of different strains were not comparable in this study. Moreover, the crude extracts used for the CAT assay showed high stability at −28°C for at least 60 days (Fig. 1B). This is a strong advantage in that samples can be utilized repeatedly for later experiments.
Another reporter gene, evoglow-Bs2, was also useful. However, the background level from control bacterial cells was obviously higher than that in the CAT assay (Fig. 2A). This indicates that the evoglow-Bs2 assay was unlikely to be useful for weak promoters. The background activity of the CAT reporter system (about 0.2% of Pgap) is comparable to those of other reporter genes, α-galactosidase (about 0.4% of Pgap) and β-glucuronidase (0% of Pgap) [6, 7]. Existing reporter genes for bifidobacteria are associated with sugar metabolism and may compete with intrinsic genes. In contrast, bifidobacteria generally do not possess the cat gene, indicating the versatility of this gene compared with existing reporter genes. Moreover, another advantage for constructing plasmids easily is that the sequence coding the cat gene is 651 bp long, which is shorter than the length of existing reporter genes (Supplementary Table 4). However, cat is also used as a selection marker for the chloramphenicol resistance gene (cmr). Therefore, the cat reporter gene is unavailable for plasmids with cmr as a selection marker.
Promoters driving the expression of major essential genes encoding tRNAs, ribosomal proteins, and elongation factors, such as PBLt43, PrpmH, PrpmB, PrplM, PrplU, and Ptuf, showed higher activities. Modification of the -10 box (Fig. 3A) and replacement of the 5’-UTR (Fig. 3B) affected transcription levels. Therefore, modifications focused on the -10 box and 5’-UTR should be targeted strategies for improvement of transcriptional efficiency and analysis of transcriptional regulation involving the 5’-UTR structure, such as riboswitches.
Our results showed that terminators contribute to gene expression because most constructs with terminators showed significantly higher expression levels than constructs without terminators (Fig. 4A). However, the degree of enhancement differed among terminators, and this may be controlled by some physicochemical factors. Two such factors may be the stability of the stem-loop and the presence of poly-U tails. TahpC and TBL0725, which possess more unstable stem-loops than the other terminators as well as unclear poly-U tails, showed low activity. As another factor, we focused on the poly-A tails upstream of the stem-loops because bidirectional terminators tended to show higher activities (Fig. 4A, Table 2). A modified TrpsO (Fig. 4C) showed increased gene expression. This structure is well-conserved in terminators in other bacteria, such as E. coli [24]. In addition, terminator modification affects the efficiency of termination in E. coli [25]. One likely role of the poly-A tail is to stop transcription from the opposite strand. Stopping this transcription likely prevents collisions between transcriptional complexes on both strands.
The results shown in Figs. 3A and 4A indicate that the maximum fold difference in activities among the tested promoters was approximately 73-fold (PrpmB/PBL1769), while that among terminators was approximately 17-fold (Ttal/TBL0725). This result indicated that promoters have a greater impact on transcription levels than terminators, which is in agreement with findings in other organisms [26].
In conclusion, our data suggest that this CAT assay can be a good reporter system for use in bifidobacterial strains because of its quantitativeness, wide measurement range, and stability. Moreover, this assay is useful for the analysis of terminators, 5’-UTRs, and promoters. In the future, we believe that this CAT assay will aid in the elucidation of the bifidobacterial gene expression mechanisms.
Supplementary Material
Acknowledgments
This work was supported by JSPS KAKENHI Grant Number 16H04896.
REFERENCES
- 1.Harley CB, Reynolds RP. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res 15: 2343–2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozakai T, Izumi A, Horigome A, Odamaki T, Xiao JZ, Nomura I, Suzuki T. 2020. Structure of a core promoter in Bifidobacterium longum NCC2705. J Bacteriol 202: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bottacini F, Zomer A, Milani C, Ferrario C, Lugli GA, Egan M, Ventura M, van Sinderen D. 2017. Global transcriptional landscape and promoter mapping of the gut commensal Bifidobacterium breve UCC2003. BMC Genomics 18: 991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormö M, Remington SJ. 1997. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci USA 94: 2306–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drepper T, Eggert T, Circolone F, Heck A, Krauss U, Guterl JK, Wendorff M, Losi A, Gärtner W, Jaeger KE. 2007. Reporter proteins for in vivo fluorescence without oxygen. Nat Biotechnol 25: 443–445. [DOI] [PubMed] [Google Scholar]
- 6.Sakanaka M, Tamai S, Hirayama Y, Onodera A, Koguchi H, Kano Y, Yokota A, Fukiya S. 2014. Functional analysis of bifidobacterial promoters in Bifidobacterium longum and Escherichia coli using the α-galactosidase gene as a reporter. J Biosci Bioeng 118: 489–495. [DOI] [PubMed] [Google Scholar]
- 7.Klijn A, Moine D, Delley M, Mercenier A, Arigoni F, Pridmore RD. 2006. Construction of a reporter vector for the analysis of Bifidobacterium longum promoters. Appl Environ Microbiol 72: 7401–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz L, Álvarez-Martín P, Mayo B, de los Reyes-Gavilán CG, Gueimonde M, Margolles A. 2012. Controlled gene expression in bifidobacteria by use of a bile-responsive element. Appl Environ Microbiol 78: 581–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Kim JY, Park MS, Ji GE. 2012. Novel Bifidobacterium promoters selected through microarray analysis lead to constitutive high-level gene expression. J Microbiol 50: 638–643. [DOI] [PubMed] [Google Scholar]
- 10.Cronin M, Sleator RD, Hill C, Fitzgerald GF, van Sinderen D. 2008. Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol 8: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Z, Westermann C, Yuan J, Riedel CU. 2014. Experimental determination and characterization of the gap promoter of Bifidobacterium bifidum S17. Bioengineered 5: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman CM, Moffat LF, Howard BH. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol 2: 1044–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordeen SK, Green PP, 3rd, Fowlkes DM. 1987. A rapid, sensitive, and inexpensive assay for chloramphenicol acetyltransferase. DNA 6: 173–178. [DOI] [PubMed] [Google Scholar]
- 14.Chiu W, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J. 1996. Engineered GFP as a vital reporter in plants. Curr Biol 6: 325–330. [DOI] [PubMed] [Google Scholar]
- 15.Grentzmann G, Ingram JA, Kelly PJ, Gesteland RF, Atkins JF. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4: 479–486. [PMC free article] [PubMed] [Google Scholar]
- 16.Ellman GL. 1959. Tissue sulfhydryl groups. Arch Biochem Biophys 82: 70–77. [DOI] [PubMed] [Google Scholar]
- 17.Yasui K, Kano Y, Tanaka K, Watanabe K, Shimizu-Kadota M, Yoshikawa H, Suzuki T. 2009. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic Acids Res 37: e3–e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altaib H, Ozaki Y, Kozakai T, Badr Y, Nomura I, Suzuki T. 2019. A new Escherichia coli entry vector series (pIIS18) for seamless gene cloning using type IIS restriction enzymes. Microbiol Resour Announc 8: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci USA 99: 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita H, Toh H, Oshima K, Nakano A, Omori E, Hattori Y, Arakawa K, Suda W, Honda K, Hattori M. 2015. Complete genome sequence of Bifidobacterium breve JCM 1192(T) isolated from infant feces. J Biotechnol 210: 81–82. [DOI] [PubMed] [Google Scholar]
- 21.Yasui K, Tabata M, Yamada S, Abe T, Ikemura T, Osawa R, Suzuki T. 2009. Intra-species diversity between seven Bifidobacterium adolescentis strains identified by genome-wide tiling array analysis. Biosci Biotechnol Biochem 73: 1422–1424. [DOI] [PubMed] [Google Scholar]
- 22.Mitra A, Kesarwani AK, Pal D, Nagaraja V. 2011. WebGeSTer DB—a transcription terminator database. Nucleic Acids Res 39: D129–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feklistov A, Darst SA. 2011. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell 147: 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reynolds R, Bermúdez-Cruz RM, Chamberlin MJ. 1992. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol 224: 31–51. [DOI] [PubMed] [Google Scholar]
- 25.Reynolds R, Chamberlin MJ. 1992. Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J Mol Biol 224: 53–63. [DOI] [PubMed] [Google Scholar]
- 26.Yamanishi M, Ito Y, Kintaka R, Imamura C, Katahira S, Ikeuchi A, Moriya H, Matsuyama T. 2013. A genome-wide activity assessment of terminator regions in Saccharomyces cerevisiae provides a “terminatome” toolbox. ACS Synth Biol 2: 337–347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.