Significance
Disease-relevant targets of infection by obligatory intracellular pathogens are beyond the reach of conventional antibodies. The neutralizing targets of intracellular pathogens in host cells are mostly unknown. The present study attempted to overcome these limitations with a combination of nanobodies, transfection, and a cell-permeable peptide delivery method to inhibit infection and determine inhibitory mechanisms. Ehrlichia chaffeensis uses the type IV secretion system to deliver bacterial effector Etf-1 to establish intracellular infection. Our study demonstrates that a nanobody can specifically block Etf-1 functions and thereby block E. chaffeensis infection in cell culture and in a mouse model, suggesting that this approach can be developed as a therapeutic intervention for human monocytic ehrlichiosis and other diseases caused by intracellular infections.
Keywords: Ehrlichia chaffeensis, type IV secretion system, Etf-1, nanobody, cell-permeable peptide
Abstract
Infection with obligatory intracellular bacteria is difficult to treat, as intracellular targets and delivery methods of therapeutics are not well known. Ehrlichia translocated factor-1 (Etf-1), a type IV secretion system (T4SS) effector, is a primary virulence factor for an obligatory intracellular bacterium, Ehrlichia chaffeensis. In this study, we developed Etf-1–specific nanobodies (Nbs) by immunizing a llama to determine if intracellular Nbs block Etf-1 functions and Ehrlichia infection. Of 24 distinct anti–Etf-1 Nbs, NbD7 blocked mitochondrial localization of Etf-1–GFP in cotransfected cells. NbD7 and control Nb (NbD3) bound to different regions of Etf-1. Size-exclusion chromatography showed that the NbD7 and Etf-1 complex was more stable than the NbD3 and Etf-1 complex. Intracellular expression of NbD7 inhibited three activities of Etf-1 and E. chaffeensis: up-regulation of mitochondrial manganese superoxide dismutase, reduction of intracellular reactive oxygen species, and inhibition of cellular apoptosis. Consequently, intracellular NbD7 inhibited Ehrlichia infection, whereas NbD3 did not. To safely and effectively deliver Nbs into the host cell cytoplasm, NbD7 was conjugated to cyclized cell-permeable peptide 12 (CPP12-NbD7). CPP12-NbD7 effectively entered mammalian cells and abrogated the blockade of cellular apoptosis caused by E. chaffeensis and inhibited infection by E. chaffeensis in cell culture and in a severe combined-immunodeficiency mouse model. Our results demonstrate the development of an Nb that interferes with T4SS effector functions and intracellular pathogen infection, along with an intracellular delivery method for this Nb. This strategy should overcome current barriers to advance mechanistic research and develop therapies complementary or alternative to the current broad-spectrum antibiotic.
Human monocytic ehrlichiosis (HME), one of the most prevalent, life-threatening, and emerging tick-borne diseases in the United States (1, 2) is caused by infection with Ehrlichia chaffeensis, an obligatory intracellular bacterium in the order Rickettsiales. E. chaffeensis replicates within human monocytes-macrophages and causes severe flu-like symptoms accompanied by hematologic abnormalities and hepatitis. Currently, the only HME therapy is the broad-spectrum antibiotic doxycycline, which is effective only if initiated early because delayed initiation (e.g., because of misdiagnosis can lead to severe complications or death). In addition, doxycycline is contraindicated for pregnant women and children or those with drug allergies. The presence of underlying illness or injury, immunosuppression, and coinfection with other tick-borne pathogens can similarly lead to severe complications or death (3). No vaccine exists for HME. Tick-borne diseases have risen dramatically in the past 20 y and continue to rise, underscoring the importance of developing new therapeutic approaches and preventive measures (4).
The type IV secretion system (T4SS) is conserved among all rickettsial organisms. The recent elucidation of critical roles of T4SS for E. chaffeensis and Anaplasma phagocytophilum infection (5) may provide potential targets for new approaches against rickettsial diseases. For example, the T4SS effectors Ehrlichial translocated factors 1 and 2 (Etf-1 and Etf-2) are critical E. chaffeensis proteins secreted via T4SS into the host cell cytoplasm, as knockdown of Etf-1 or Etf-2 by transfection of E. chaffeensis with specific antisense peptide nucleic acids significantly inhibits E. chaffeensis infection (6, 7). Secreted Etf-1 localizes to mitochondria and blocks mitochondria-mediated host cell apoptosis to keep the infected host cell alive for bacterial intracellular replication (8). A subpopulation of Etf-1 molecules that are not localized to mitochondria interacts with Beclin 1 (ATG6) and active Rab5 (Rab5-GTP), and induces Rab5-regulated autophagy for E. chaffeensis to acquire catabolites as nutrients (9). Etf-2 directly binds Rab5-GTP on Ehrlichia-containing inclusion membranes and blocks Rab5 GTPase activating protein (RabGAP-5) engagement with Rab5-GTP to prevent Ehrlichia-containing inclusions from maturing into late endosomes and fusing with lysosomes (7).
Camelidae produce two types of antibodies: conventional antibodies and heavy-chain–only antibodies (10). The variable domain of the heavy chain of heavy-chain–only antibodies (VHHs) of camelids is the smallest (11 to 15 kDa) antigen-binding fragment relative to conventional antibodies. VHHs are soluble and display long surface loops, which are often larger than those of conventional murine and human antibodies (11, 12). The VHHs can be cloned into bacterial or mammalian expression plasmids (13) to produce a nanobody (Nb), a monomeric variable antibody. VHHs cloned into mammalian expression vectors can produce intracellular Nbs within mammalian cells that are superior to conventional antibodies for modulating intracellular functions because they can operate in the reducing intracellular environment, are proteolytically stable, can target subcellular sites, can penetrate cavities in target antigens, and can bind efficiently to antigens, such as enzyme catalytic sites (13–16). Although the therapeutic potential of Nbs has been investigated for several infectious diseases (14, 17–19), the use of Nbs as a therapeutic agent against intracellular bacteria such as E. chaffeensis has not been reported. In the present study, we developed an intracellular Nb approach to block T4SS effectors within mammalian cells, thereby inhibiting intracellular pathogen infection.
Progress in developing effective therapy and investigative approach for obligatory intracellular pathogens has been hindered by many factors, not the least of which is the lack of safe and efficient intracellular delivery methods of macromolecules. Although cyclic peptides are generally unable to cross the cell membrane, some naturally occurring cyclic peptides (e.g., cyclosporine A) possess the unusual ability of crossing the cell membrane by passive diffusion and are orally bioavailable (20). Cyclized Arg-rich cell-permeable peptides (CPPs)—such as cyclo(FΦRRRRQ) or cFΦR4, where Φ is l-2-naphthylalanine—or newer and more effective CPPs, such as CPP9 and CPP12, that include d-arginine or d-phenylalanine, provide rapid and efficient cytosolic delivery of their linked cargo proteins into >95% of cells (21–23). They are not cytotoxic at effective concentrations and have oral and intravenous bioavailability based on preliminary pharmacokinetics in mice (22). The cyclic CPPs (and the CPP-cargo conjugates) bind directly to plasma membrane phospholipids and enter cells by endocytosis (22). They then efficiently escape from the early endosome into the cytosol unlike Tat, which escapes only from late endosomes (22–24).
In the present study, we have developed anti–Etf-1 Nbs. We obtained a Nb that blocks Etf-1 functions and demonstrated its effectiveness in combination with a cyclic CPP for inhibition of E. chaffeensis infection in cell culture and in a mouse model. These findings represent a significant advance in developing therapeutic and investigative strategy of obligatory intracellular pathogens.
Results
Generation and Characterization of anti–Etf-1 Nbs.
To generate Etf-1–specific Nbs, a llama was immunized with soluble full-length recombinant Etf-1 protein (rEtf-1) immediately after purification by fast protein liquid chromatography (FPLC) (SI Appendix, Fig. S1A). A strong antisera titer of 4 × 1011 against rEtf-1 was detected by ELISA after the third immunization (SI Appendix, Fig. S1B). The serum from the immunized llama recognized both rEtf-1 and native Etf-1 from E. chaffeensis-infected THP-1 cells, as demonstrated by Western blotting (SI Appendix, Fig. S1C). For Nb library construction, cDNA was synthesized from total RNA extracted from peripheral blood leukocytes isolated from the llama after the third rEtf-1 immunization (SI Appendix, Fig. S2A), and was used as a template for amplification of the Nb-encoding VHH regions by two-step PCR (SI Appendix, Fig. S2B), as described previously with slight modification (25). A phagemid Nb library comprising 2.32 × 1010 individual transformants was generated (SI Appendix, Fig. S2C), of which 83% had the proper insert (SI Appendix, Fig. S2D). The native Etf-1 protein synthesized by E. chaffeensis is assumed to be linearized when it passes through the two membranes of E. chaffeensis and one membrane of the E. chaffeensis–containing inclusion via the T4SS transmembrane channel, since the diameter of the inner channel is only 10 Å at the cytoplasm side (26).
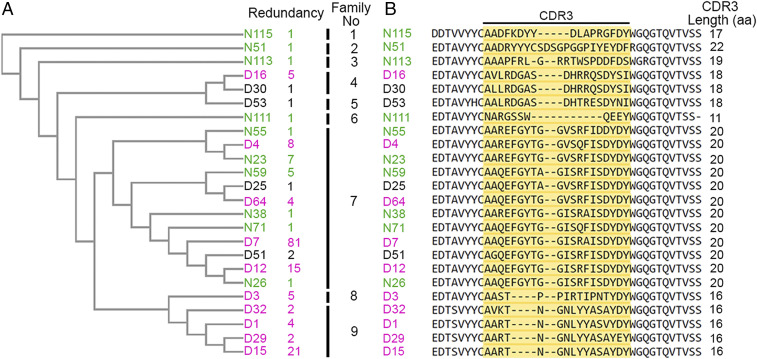
After the native Etf-1 protein is secreted into the cytoplasm of the host cell, it will be refolded back into its tertiary structure, as demonstrated in the type III secretion system (27). Thus, we enriched for phages encoding anti–Etf-1 Nbs by panning using nondenatured (folded) and denatured (linearized) rEtf-1 as antigen. After panning, we screened 368 individual Nbs expressed by TG1—an amber codon suppression (supE) strain of Escherichia coli (25)—by ELISA. A total of 172 rEtf-1–binding phagemid clones (107 and 65 positive clones against nondenatured and denatured Etf-1, respectively) was obtained and used for DNA sequencing of the VHHs. Complementarity-determining region 3 (CDR3), which is the most variable region within VHHs, was used for phylogenetic analysis. Alignment of the deduced CDR3 amino acid sequences of the 172 clones revealed clones encoding 24 Nbs with distinct CDR3 sequences (Fig. 1). Ten Nbs bound to both denatured and nondenatured rEtf-1, 10 Nbs bound only to nondenatured rEtf-1, and 4 Nbs bound only to denatured rEtf-1 (Fig. 1A). Nbs with a high similarity in their CDR3 sequences (identical length and >80% amino acid sequence identity) are derived from the same B cell clonal lineage and can be grouped into a single family (25). Based on this, 24 Nbs were grouped into nine different families (Fig. 1A).
Fig. 1.
Phylogenetic analysis and CDR3 alignment of anti–Etf-1 Nbs. (A) Phylogenetic analysis of 24 distinct anti–Etf-1 Nbs based on their CDR3 amino acid sequences. Redundancy and family classification are shown on the right. Font colors indicate Nbs that bind denatured Etf-1 (black), nondenatured Etf-1 (green), or both (magenta). (B) Amino acid sequence alignment of anti–Etf-1 Nbs. CDR3 amino acid sequences are highlighted in yellow, and the number of amino acids in each CDR3 is shown on the right.
NbD7 Blocks Mitochondrial Localization of Etf-1.
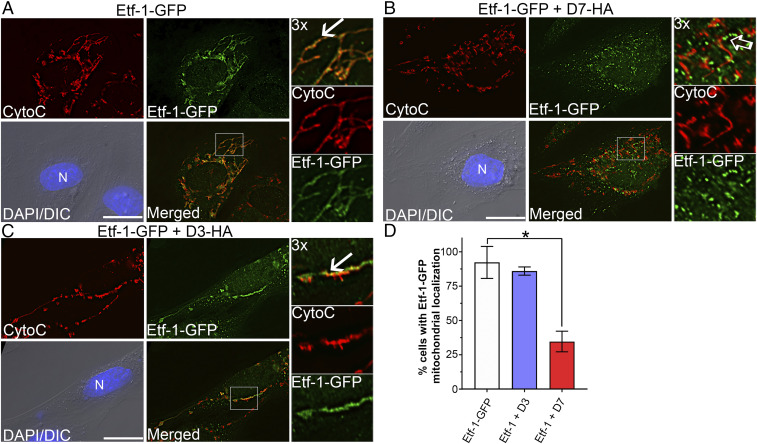
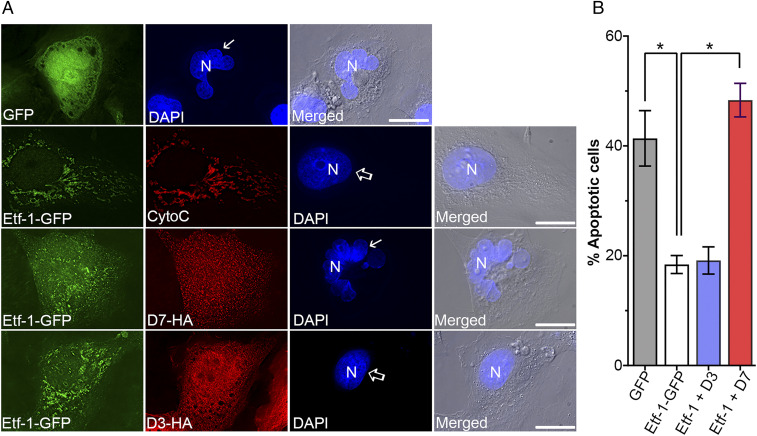
To determine whether anti–Etf-1 Nbs can block the mitochondrial localization of Etf-1, we recloned 24 Nbs into the mammalian expression vector pEGFP-C1 by replacing EGFP with a C-terminal hemagglutinin (HA) tag for intracellular Nb expression. Rhesus monkey RF/6A endothelial cells, which are thinly spread adherent cells and are thus suitable for cellular localization studies, were cotransfected with each Nb and Etf-1–GFP. Ectopically expressed Etf-1–GFP in RF/6A cells localizes to the mitochondria, which are visible as filamentous structures and colocalize with cytochrome c, a marker of mitochondria (8) (Fig. 2A). Among 24 Nbs, only NbD7 (referred to hereafter as D7) unequivocally blocked Etf-1–GFP localization to mitochondria (Fig. 2 B and D), and NbD3 (referred to hereafter as D3) unequivocally did not block Etf-1–GFP localization to mitochondria (Fig. 2 C and D). Therefore, D7 and D3 were selected for detailed studies of intracellular Nb activities and mechanisms.
Fig. 2.
NbD7, but not D3, blocks localization of Etf-1–GFP to mitochondria. (A−C) RF/6A cells were transfected with plasmids expressing Etf-1–GFP (A) or cotransfected with Etf-1–GFP and HA-tagged D7 (B) or D3 (C). At 2 dpt, cells were fixed, and mitochondria were labeled with mouse anti-cytochrome c (CytoC, red). Etf-1–GFP was labeled with rabbit anti–Etf-1 (green). DAPI/DIC, the image stained with DAPI was merged with the DIC image. N, nucleus. Each boxed area in the merged image was enlarged 3× on the Right. White arrows: colocalization; open arrow: lack of colocalization. (Scale bars, 10 μm.) (D) Quantification of effects of Nbs on Etf-1–GFP localization with mitochondria. Data were obtained by counting ∼100 RF/6A cells from three independent experiments showing expression of Etf-1–GFP and D7-HA or D3-HA and represented as the mean ± SD (n = 3). *P < 0.05 by one-way ANOVA.
The N-Terminal 24 aa of Etf-1 Are Critical for Mitochondrial Localization.
Most eukaryotic mitochondria-targeting proteins are synthesized as precursor proteins in the cytosol and then are directed into mitochondria by one or more signal sequences (28). The position of the signal sequence in each precursor protein determines its destination in mitochondria. An N-terminal presequence is required for many proteins entering the matrix and is rapidly removed by a signal peptidase in the mitochondrial matrix after import (28). In contrast, an internal signal sequence that is present on all proteins that target the mitochondrial outer membrane and several sequences that target the inner membrane or intermembrane space are required for their import and proper localization; these sequences are not removed (28). As D7 blocked Etf-1–GFP localization to mitochondria, we first examined the mitochondria localization signal sequence of Etf-1.
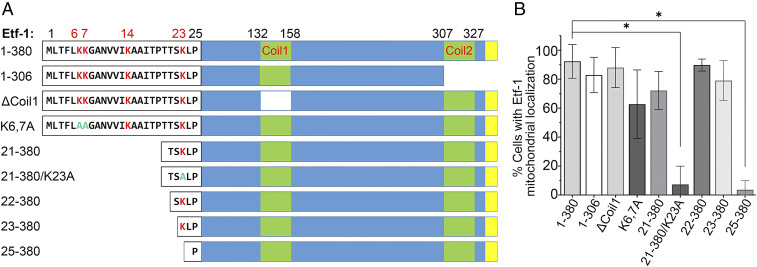
To determine whether the N terminus of Etf-1 is essential for mitochondrial localization, sequential truncation of the N-terminal 20 to 24 aa of Etf-1, in conjunction with internal deletions or point mutations, was carried out (Fig. 3A), and the resulting constructs were ectopically expressed in RF/6A cells to examine their mitochondrial localization by double immunofluorescence labeling. Etf-1–GFP (full-length Etf-1, 1 to 380 aa) (8) was used as a positive control. C-terminal deletion (1 to 306 aa) or deletion of the first coiled-coil domain motif (ΔCoil1) of Etf-1 did not affect the mitochondrial localization as compared with Etf-1 (Fig. 3B and SI Appendix, Fig. S3). Etf-1 with the deletion of the N-terminal 24 aa (Δ1–24, 25 to 380 aa) resulted in the complete loss of mitochondrial localization (Fig. 3B and SI Appendix, Fig. S3), whereas Etf-1 Δ1−20 (21 to 380 aa), Δ1−21 (22 to 380 aa), and Δ1−22 (23 to 380 aa) were targeted to mitochondria (Fig. 3B), indicating that the N-terminal amino acids 23 and 24 of Etf-1 are critical for Etf-1 targeting to mitochondria. N-terminal basic amino acids are known to be critical for mitochondrial targeting of proteins (28). As four lysine (K) residues are present at the N terminus of Etf-1, we determined whether these lysine residues are critical for mitochondrial localization of Etf-1. Lysines at positions 6, 7, and 23 of Etf-1 were individually point-mutated to alanine (A) and cloned into plasmid pGFP-N1 (Fig. 3A). The K6A, K7A double mutant of Etf-1 was constructed based on full-length Etf-1, whereas the K23A mutant was based on Etf-1 Δ1−20 (residues 21 to 380) (Fig. 3A). Ectopic expression of the K/A mutants of Etf-1 in RF/6A cells showed that the mitochondrial localization of Etf-1 was only slightly reduced by the K6A/K7A double mutant but was completely abolished by Etf-1 Δ1−20 K23A (Fig. 3B and SI Appendix, Fig. S3). These results indicated that the N-terminal 24 aa of Etf-1, especially the positively charged residue K23, plays a critical role in mitochondrial targeting of Etf-1, as both deletion and mutation of this residue significantly decreased the percentage of cells with Etf-1 that was targeted to mitochondria (Fig. 3B and SI Appendix, Fig. S3).
Fig. 3.
K23 within the N-terminal 24 aa of Etf-1 is critical for mitochondrial targeting. (A) Domains and mutations of Etf-1 plasmid constructs showing truncations, internal deletions, and point mutations of Etf-1. The N-terminal 25 amino acids are indicated, with K (lysine) residues shown in red and point mutations of K to alanine (A) in aqua. ΔCoil1, internal deletions of the first coiled-coil domain (amino acids 132 to 158); K6,7A, double mutations of K at residues 6 and 7 to A of full-length Etf-1; 21–380/K23A, deletion of the N-terminal 20 aa with point mutation of K at residue 23 to A. Yellow box, putative T4SS secretion motif. Protein lengths are not drawn to scale. (B) Percentage of cells with Etf-1–GFP localized to mitochondria. RF/6A cells were transfected with plasmids expressing full-length (1 to 380 aa) or mutant Etf-1–GFP, and mitochondria were labeled with anti-cytochrome c at 2 dpt. Etf-1–GFP localization with mitochondria in ∼100 cells each from three independent experiments was scored. Data are represented as the mean ± SD (n = 3). *P < 0.05 by one-way ANOVA. Representative images are shown in SI Appendix, Fig. S3.
Characterization of D7 and D3 Binding to Etf-1.
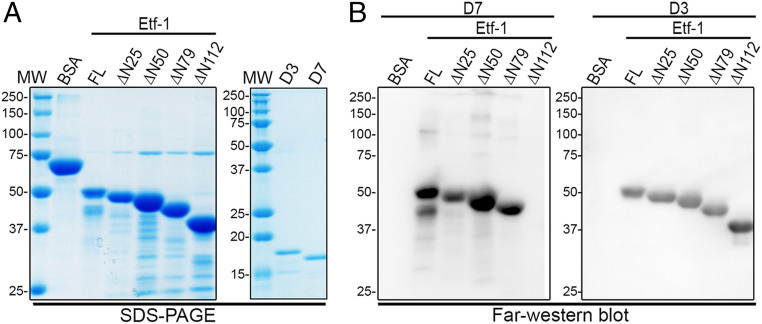
Using recombinant D7 and D3 and progressively truncated rEtf-1, Nb binding epitopes were determined by far-Western blot analysis (Fig. 4). D7 bound to full-length and Etf-1 ΔN25, ΔN50, and ΔN79 (residues 26 to 380, 51 to 380, and 80 to 380, respectively) but not to Etf-1 Δ112 (residues 113 to 380) of Etf-1 protein (Fig. 4B). D3 bound to full-length and all four N-terminal–truncated Etf-1 proteins (Fig. 4B), indicating that the binding epitope of Etf-1 resides between residues 81 and 112 for D7 and between residues 113 and 380 for D3. Thus, inhibition of Etf-1 localization to mitochondria by D7 was not due to masking or blockade of the N-terminal Etf-1 mitochondria localization signal.
Fig. 4.
D7 and D3 bind different regions of Etf-1. (A) Purified full-length (FL) and various N-terminal truncations of rEtf-1 (ΔN25, residues 26 to 380; ΔN50, residues 51 to 380; ΔN79, residues 80 to 380; and ΔN112, residues 113 to 380) were subjected to SDS/PAGE, and proteins were stained by Coomassie blue. BSA was used as a negative control, and D3 and D7 were also stained to verify their purity. MW, molecular mass marker. (B) Far-Western blotting. Proteins separated by SDS-PAGE as in A were transferred onto PVDF membranes. Proteins were denatured, renatured, and incubated with purified D7-HA or D3-HA. The binding of D7 or D3 to Etf-1 was detected with anti-HA. Molecular mass is indicated in kilodaltons (kDa).
Next, size-exclusion chromatography was used to determine the stability of the rEtf-1 and D7 complex. The analysis showed that D7 formed a more stable complex with rEtf-1 in solution, as indicated by the comigration of the two proteins, but not D3 (Fig. 5 A and B). By using open surface plasmon resonance (OpenSPR), we determined that the dissociation constant of rEtf-1 and D7 was 3.15 ± 0.07 µM, whereas that of rEtf-1 and D3 was 11.60 ± 0.87 µM (Fig. 5C), consistent with the analytical size-exclusion chromatography result. Thus, the stability of D7 binding to rEtf-1 likely results in steric misfolding or steric hindrance of Etf-1 that reduces its localization to mitochondria. In contrast, binding of D3 to Etf-1 is so unstable that it did not alter Etf-1 functions.
Fig. 5.
NbD7 forms a more stable complex with Etf-1 than does NbD3. (A) To obtain the elution profile of individual protein, recombinant protein Etf-1 (blue), NbD7 (red), or NbD3 (green), was individually separated by size exclusion chromatography. Fractions (numbered and shown in red at the bottom) corresponding to the elution peaks indicated by the absorbance at 280 nm were collected, and proteins in each fraction were subjected to SDS-PAGE and Coomassie blue staining. (B) To determine stability of binding complexes of recombinant Etf-1 + NbD7 (red) or NbD3 (green) in solution, each mixture was subjected to size exclusion chromatography. Coomassie blue staining shows the coelution of the stable complex of Etf-1 and NbD7 (red box), but coelution of Etf-1 and NbD3 was undetectable (green box). MW, molecular mass marker (indicated in kDa; A and B). (C) Binding affinity of D7 and D3 to Etf-1 was determined by OpenSPR. A series of five dilutions of Nbs was used to test their binding against biotinylated Etf-1 immobilized on a sensor chip. Colored lines are signals detected by OpenSPR, thin black lines are fitted models generated by the TraceDrawer software.
D7 Abrogates Etf-1 Inhibition of Host Cell Apoptosis Induced by Etoposide.
Etoposide, a topoisomerase II inhibitor, can induce DNA double-strand breaks that lead to the activation of caspase 2 and subsequent induction of Bax translocation to mitochondria and cytochrome c release, which results in apoptosis (29). Ectopically expressed Etf-1 localizes to mitochondria and inhibits mitochondria-mediated cellular apoptosis induced by etoposide, including Bax translocation to mitochondria, cytochrome c release, loss of mitochondrial membrane potential, nuclear fragmentation, as well as human Bax-induced yeast apoptosis (8). Since Etf-1(25–380) truncation and Etf-1(21–380/K23A) mutant completely abolished their mitochondrial localization, we examined whether these two Etf-1 mutants affect mitochondria-mediated cellular apoptosis induced by etoposide. The results showed that both Etf-1(25–380) and Etf-1(21–380/K23A) mutants could not inhibit etoposide-induced apoptosis (SI Appendix, Fig. S4), indicating that the mitochondrial localization of Etf-1 is essential for its inhibition of mitochondria-mediated cellular apoptosis.
Because D7 blocked Etf-1 targeting to mitochondria (Fig. 2), we examined whether D7 abrogates Etf-1−mediated inhibition of apoptosis. RF/6A cells cotransfected with Nb and Etf-1−GFP or GFP (as a negative control) were treated with 100 µM etoposide at 24 h posttransfection (hpt), and apoptotic cells were scored based on characteristic condensed or fragmented nuclei at 41 h (65 hpt) after etoposide treatment (Fig. 6). D7-HA transfection abrogated apoptosis inhibition induced by Etf-1–GFP, resulting in a level similar to that of GFP-transfected control cells (Fig. 6), whereas D3-HA transfection did not alter apoptosis inhibition induced by Etf-1−GFP (Fig. 6).
Fig. 6.
D7 abrogates Etf-1 inhibition of etoposide-induced apoptosis. RF/6A cells were transfected with GFP or Etf-1–GFP, or cotransfected with Etf-1–GFP and D7 or D3, followed by treatment with 100 µM etoposide at 24 hpt for 41 h. Cells were labeled with mouse monoclonal anti-cytochrome c (CytoC) and rabbit polyclonal anti–Etf-1, or mouse monoclonal anti-GFP and rabbit monoclonal anti-HA. (A) Images show the localization of Etf-1 and Nbs, and nuclear morphology was stained by DAPI (arrows, apoptotic nuclei; open arrows, nonapoptotic nuclei). Merged, the fluorescent image of DAPI channel merged with the DIC image. (Scale bars, 10μm.) (B) Quantification of apoptosis (nuclear fragmentation) in 100 cells expressing transfected genes from three independent experiments. Data are represented as the mean ± SD (n = 3). *P < 0.05, by one-way ANOVA.
D7 Abrogates the Etf-1−Induced Increase in Mn-Superoxide Dismutase and Decrease in Cellular Reactive Oxygen Species.
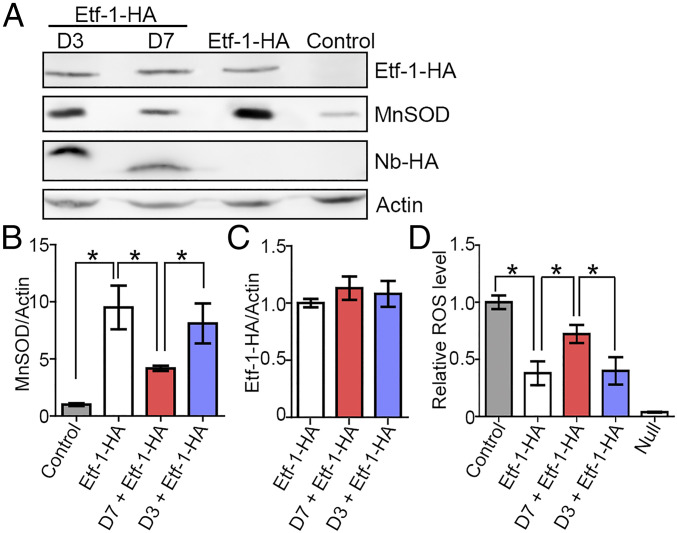
During aerobic respiration to produce ATP from oxygen in mitochondria, toxic reactive oxygen species (ROS) are produced as 1 to 3% of electrons leak from the electron transport chain and interact with oxygen directly to yield superoxide (30). This leads to DNA damage and is a critical factor for inducing host cell apoptosis (31, 32). To prevent oxidative damage to mitochondria and the cell, Mn-superoxide dismutase (MnSOD) and the glutathione/glutathione peroxidase-1 system in the mitochondrial matrix act cooperatively to reduce the levels of ROS produced during aerobic metabolism (33, 34). Our previous study showed that ectopically expressed Etf-1–GFP up-regulates mitochondrial MnSOD at the protein level, but not at the mRNA level, and reduces intracellular ROS levels, consequently inhibiting cellular apoptosis induced by etoposide (8). Based on Western blotting, D7, but not D3, lowered the level of MnSOD in cells transfected with Etf-1–HA (Fig. 7 A–C). By using 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA), which can be oxidized by ROS to yield DCF, a fluorescent compound detectable by fluorescence spectrophotometry (35), we measured whether D7 affects intracellular ROS levels. ROS levels in E. chaffeensis-infected or Etf-1–transfected cells are lower than those in uninfected or untransfected cells, respectively, suggesting that Etf-1 in mitochondria reduces the ROS level in the host cells (8). D7, but not D3, abrogated ROS reduction induced by Etf-1–HA transfection (Fig. 7D). Thus, these results are in agreement with our previous prediction that not simply the intracellular presence of Etf-1 but specifically its mitochondrial localization (8) is required for MnSOD up-regulation and cellular ROS reduction.
Fig. 7.
D7, but not D3, abrogates the increase in MnSOD and attenuation of intracellular ROS generation by Etf-1. HEK293 cells were transfected with Etf-1–HA, or cotransfected with Etf-1–HA and D7 or D3. Control, untransfected cells. (A) At 2 dpt, cell lysates were subjected to Western blotting using antibodies against HA, MnSOD, and human actin. (B and C) Quantification of relative band densities of MnSOD (B) and Etf-1 (C) normalized against actin. (D) ROS production at 2 dpt was analyzed by the fluorescent indicator H2DCFDA. Null, buffer control without H2DCFDA. (B−D) Data are presented as the mean ± SD from three independent experiments with triplicates per sample. *P < 0.05, by one-way ANOVA.
D7 Abrogates E. chaffeensis-Induced Increase in MnSOD and Reduction in ROS and Inhibits Infection.
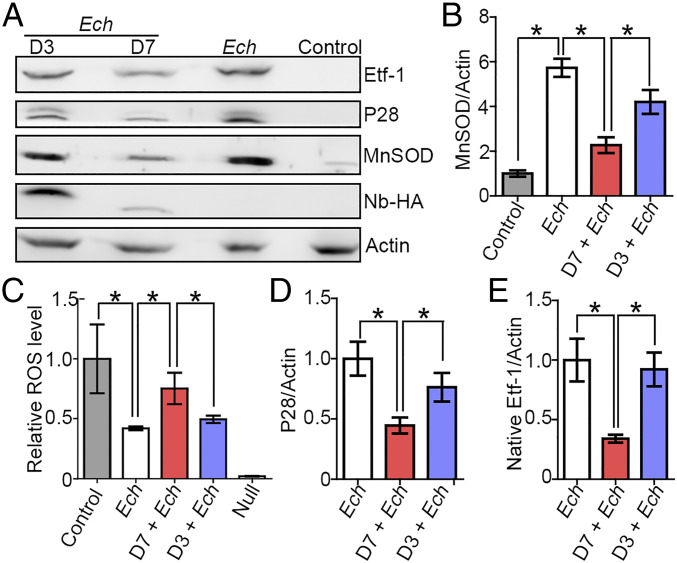
The MnSOD protein level is greater and ROS levels are lower in E. chaffeensis-infected cells than are those in uninfected cells (8). Ectopically expressed D7, but not D3, abrogated the increase in MnSOD in E. chaffeensis-infected cells (Fig. 8 A and B) and abrogated the reduction in intracellular ROS levels in E. chaffeensis-infected cells (Fig. 8C). Most importantly, D7 (but not D3) also inhibited E. chaffeensis infection of host cells based on the expression of E. chaffeensis P28/OMP-1F (major outer membrane proteins) (36, 37) as determined by Western blot analysis (Fig. 8 A and D). Etf-1 protein itself was decreased (Fig. 8E), most likely as a result of the inhibition of E. chaffeensis infection.
Fig. 8.
D7, but not D3, abrogates E. chaffeensis-induced increase in MnSOD and reduction in ROS and inhibits infection. (A) HEK293 cells were transfected with HA-tagged Nbs and infected with E. chaffeensis (Ech) at 1 dpt. Native E. chaffeensis Etf-1, E. chaffeensis outer membrane proteins P28/OMP-1F, Nbs, MnSOD, and human actin were detected at 2 dpi by Western blotting using their respective antibodies. (B, D, and E) Quantification of relative densities of MnSOD (B), P28 (D), and Etf-1 (E) normalized against actin. (C) ROS production at 2 dpi was analyzed by the fluorescent indicator H2DCFDA. Null, buffer control without H2DCFDA. (B−E) Data are presented as the mean ± SD from three independent experiments with triplicates per sample. *P < 0.05, by one-way ANOVA.
Intracellular Delivery of CPP-D7 Abrogates Etf-1 Inhibition of Host Cell Apoptosis and Inhibits E. chaffeensis Infection.
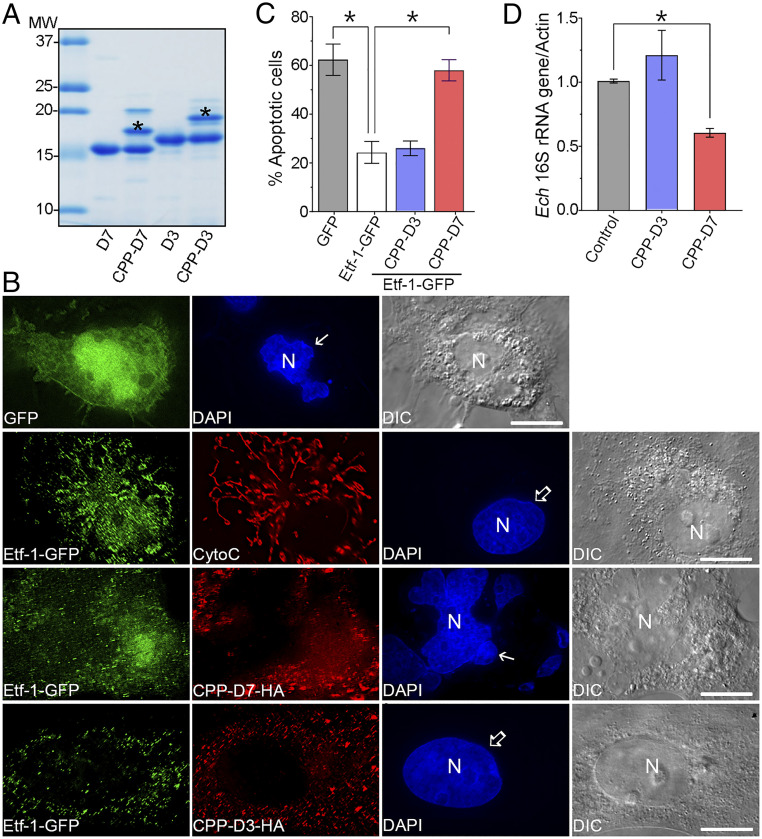
Whereas transfection with plasmids encoding Nb is useful for in vitro analyses, in vivo application of Nb requires a different intracellular Nb delivery method. We therefore used the cyclic CPP12—cyclo(FfΦRrRrQ), where Φ is l-2-naphthylalanine, f is d-phenylalanine, and r is d-arginine (22)—for D7 and D3 conjugation. D7 and D3 were purified to >95% purity for CPP conjugation (Fig. 9A). D7 and D3 were specifically labeled with CPP at their N-terminal amine with ∼40% labeling efficiency (Fig. 9A). The CPP-Nbs were effectively taken up by almost 100% of RF/6A cells based on flow cytometry (SI Appendix, Fig. S5A). There was no cytotoxicity of CPP-Nb−treated cells as determined by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide] assay (SI Appendix, Fig. S5B). CPP-D7 abrogated apoptosis inhibition induced by Etf-1–GFP, to a level similar to that of GFP-transfected control cells (Fig. 9 B and C), but CPP-D3 did not. When E. chaffeensis-infected THP-1 cells were treated with CPP-Nbs, bacterial infection was significantly reduced by CPP-D7 but not by CPP-D3 (Fig. 9D), similar to the effects of ectopically expressed Nbs (Fig. 8 A and D).
Fig. 9.
Intracellular delivery of CPP-D7 abrogates Etf-1 inhibition of host cell apoptosis and inhibits E. chaffeensis infection. (A) SDS/PAGE and Coomassie blue staining show that D7 and D3 were purified with >95% purity and that ∼40% of Nbs were specifically conjugated with CPP12, as indicated by the band labeled with an asterisk. (B) RF/6A cells were transfected with GFP or Etf-1–GFP, and treated with HA-labeled CPP-D7 or CPP-D3 for 12 h at 12 hpt. Cells were treated with 100 µM etoposide at 24 hpt for an additional 41 h and then were double-labeled with mouse anti-cytochrome c (CytoC) and rabbit anti–Etf-1 or with mouse anti-GFP and rabbit anti-HA. N, nucleus by DAPI staining (solid arrows, apoptotic nuclei; open arrows, nonapoptotic nuclei). (Scale bars, 10 μm.) (C) Quantification of the percentage of apoptotic cells (i.e., those showing nuclear fragmentation) among cells expressing Etf-1–GFP in the absence and presence of intracellularly delivered CPP-Nbs; 100 cells were analyzed for each condition in three independent experiments. (D) THP-1 cells were infected with host cell-free E. chaffeensis for 2 h and then were incubated with 10 μM of CPP-D7 or CPP-D3 for 2 d. DNA was extracted from samples and qPCR was performed to amplify E. chaffeensis 16S rRNA gene normalized against with human actin gene. (C and D) Data are represented as the mean ± SD (n = 3); *P < 0.05, by one-way ANOVA.
CPP-D7 Reduces E. chaffeensis Infection in Severe-Combined Immunodeficiency Mice.
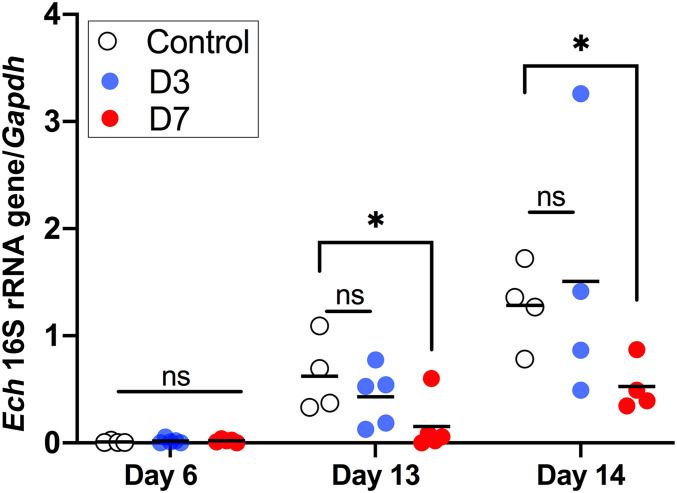
Immunocompetent mice clear E. chaffeensis Arkansas infection within 2 wk and do not develop clinical signs (38). Consequently, severe-combined immunodeficiency (SCID) mice have been used to investigate E. chaffeensis infection and pathogenesis. When SCID mice are inoculated with one of three distinct E. chaffeensis strains, the order of the severity of clinical signs and the bacterial burden detected in the mice is Wakulla > Liberty > Arkansas, and the Wakulla strain rapidly multiplies in the blood >2,000-fold from days 5 to 15 and kills mice in <15 d (39). Etf-1 amino acid sequences are identical between the E. chaffeensis Arkansas and Wakulla strains (9). Thus, analyzing the Wakulla strain in a SCID mouse infection model allowed us to investigate the effects of CPP-D7 on Ehrlichia infection and pathogenesis in the absence of adaptive immune responses. Mice were intraperitoneally inoculated with E. chaffeensis Wakulla-infected DH82 cells preincubated with CPP-D7, CPP-D3, or PBS for 12 h, and mice were intraperitoneally inoculated with CPP-D7, CPP-D3, or PBS once per day on 1 and 2 d postinfection (dpi). Whereas there was no significant difference in E. chaffeensis in the blood at 6 dpi among three groups of mice, subsequent bacterial proliferation at 13 and 14 dpi was significantly inhibited in CPP-D7–inoculated SCID mice as compared with mice inoculated with CPP-D3 or PBS (Fig. 10). The result indicates inhibitory effects of CPP-D7 on the virulent Wakulla strain in culture and subsequent intraperitoneal inoculation of mice was sustained almost 2 wk. These results further support in vitro results presented in Fig. 9, which concludes CPP-mediated intracellular Nb delivery is effective in inhibiting E. chaffeensis infection.
Fig. 10.
CPP-conjugated D7, but not D3, reduces E. chaffeensis Wakulla infection in SCID mice. Three groups of five SCID mice were ip inoculated with E. chaffeensis Wakulla-infected DH82 cells that were preincubated with CPP-D7, CPP-D3, or PBS, respectively, for 12 h. For the following 1 ∼2 dpi, mice were intraperitoneally inoculated with CPP-D7 or CPP-D3 at 20 μg/g body weight per day or with PBS. Infection with Ehrlichia was determined by qPCR of the blood samples collected on day 6, 13, and 14 using the E. chaffeensis (Ech) 16S rRNA gene and normalized with mouse Gapdh. The scatter plot shows the normalized Ehrlichia levels in individual mice, with the horizontal bar representing the mean value. *P < 0.05, by one-way ANOVA; ns, not significant.
Discussion
In the present study, we developed anti–Etf-1 Nbs and demonstrated that Nbs can be delivered intracellularly and that a particular Nb, D7, blocks mitochondrial localization of Etf-1. The mechanism by which intracellular D7 blocks Etf-1 localization to mitochondria is currently unknown. Although we determined the critical mitochondrial localization signal in the N terminus of Etf-1, this was not the D7 binding site. Gel-filtration chromatography revealed the stability of the Etf-1–D7 complex in solution, suggesting that D7 binding caused steric hindrance and misfolding of Etf-1 to prevent Etf-1 from being targeted to mitochondria. Although D3 binds Etf-1 with a similar affinity based on OpenSPR, the greatly reduced stability of the Etf-1–D3 complex in solution may not be able to prevent Etf-1 from being targeted to mitochondria. Furthermore, Etf-1(25–380) truncation and the Etf-1(21–380/K23A) mutant of Etf-1 completely abolished mitochondrial localization, and failed to inhibit etoposide-induced apoptosis, indicating that the mitochondrial localization of Etf-1 is essential for its inhibition of mitochondria-mediated apoptosis. Future analysis of the structure of the Etf-1–D7 complex would facilitate refining the D7 structure for stronger affinity and inhibition by mutating the binding site amino acids.
Etf-1 is critical for E. chaffeensis infection, because knockdown of Etf-1 by Etf-1 antisense peptide nucleic acid transfection of E. chaffeensis inhibits its ability to infect cells, and this inhibition is transcomplemented by intracellular expression of Etf-1 (6). Ectopically expressed Etf-1 in mammalian cells inhibits cellular apoptosis, which allows sufficient time for E. chaffeensis to replicate (8). However, it is unclear whether mitochondrial localization of Etf-1 is required for this inhibition. By using NbD7, the present study demonstrated that mitochondrial localization of Etf-1 is indeed required for host cell apoptosis inhibition and effective E. chaffeensis infection. Both E. chaffeensis infection and Etf-1 ectopic expression up-regulate mitochondrial MnSOD at the protein level and reduce intracellular ROS derived from mitochondrial and E. chaffeensis metabolism (8), which likely benefits E. chaffeensis intracellular survival as well as the survival of the infected host cell. Based on our results with D7, this ROS inhibition requires mitochondrial localization of Etf-1. Molecular mechanisms of up-regulation of MnSOD by Etf-1 localized in the mitochondria remain to be studied. Nonetheless, intracellularly delivered Nbs are an effective research tool for dissecting complex mechanisms of the pathogen–host interaction that occur inside host cells.
Intracellular antibodies could be developed that block bacterial virulence factors/mechanisms, host cell receptors/partners, and signaling pathways. Indeed, we have shown that intracellular delivery of conventional antibodies against Etf-1 by the Chariot protein transfection system (8), or transfection of Nbs against human heterogeneous nuclear ribonucleoprotein K (hnRNP-K) can block E. chaffeensis infection of human cells (40). However, one of the critical steps for therapeutic application of intracellular Nbs is effective and safe in vivo delivery of Nbs. To overcome this universal difficulty, we used a new approach that relies on CPP, which was never tried for Nbs. Furthermore, we took advantage of the research progress on a newer synthetic cyclic CPP, CPP12, which is over six times more effective than the prototype cyclic CPP cFΦR4 (22). By covalently linking individual Nb molecules with CPP12, we demonstrated the effective penetration of CPP12-Nb into almost 100% of cells by simple coincubation for 12 h at 37 °C, without any cytotoxic effects. Inhibition of apoptosis and E. chaffeensis infection were more effective with CPP delivery of the Nb than with transfection, indicating the feasibility of this mode of intracellular Nb delivery. Indeed, CPP-D7 showed significant protection of SCID mice from the highly virulent strain Wakulla without the help of the adaptive immune system, supporting the potential therapeutic potential of CPP-Nbs for intracellular infections and other cellular ailments.
Another important function of Etf-1 is to induce Rab5-regulated autophagy and recruit Etf-1–positive early autophagosomes (amphisomes) to Ehrlichia-replicating inclusions, which provides the necessary nutrients for intracellular growth of E. chaffeensis (9). It will be important to develop and identify anti–Etf-1 Nbs that block Rab5-regulated autophagy in the future, so that we can further define Etf-1–induced autophagy and molecular mechanisms. Furthermore, if such Nbs can be found and combined with D7, they would most likely be more effective in inhibiting E. chaffeensis than D7 alone.
Etf-1 homologs are found in every sequenced member of the genera Ehrlichia and Anaplasma, all of which are human or animal pathogens (SI Appendix, Table S1). In fact, Anaplasma translocated substrate 1 (Ats-1) is an A. phagocytophilum T4SS substrate that targets host cell (neutrophil) mitochondria in an N-terminal sequence-dependent manner and prevents apoptosis in A. phagocytophilum-infected human neutrophils (41). Ats-1 also induces host cell autophagy to deliver host cytosolic catabolites to Anaplasma-containing inclusions for bacterial growth (42, 43). Ats-1 and Etf-1 have 21% identity at the amino acid level. Thus, this approach of developing T4SS-blocking Nbs is likely to be applicable to broader members of the Ehrlichia and Anaplasma genera. Taken together, our findings support the feasibility of therapeutic use of intracellular Nbs for blocking intracellular infection.
Materials and Methods
Ethics Statement.
All animal experiments were performed in accordance with the Ohio State University Institutional Animal Care and Use Committee guidelines and approved e-protocol. The university program has full continued accreditation by the Association for Assessment and Accreditation of Laboratory Animal Care International under 000028, dated 9 June 2000, and has Public Health Services assurance renewal A3261-01, dated 6 February 2019 through 28 February 2023. The program is licensed by the US Department of Agriculture, number 31-R-014, and is in full compliance with Animal Welfare Regulations.
Antibodies.
Antibodies used were affinity-purified rabbit IgG against the C-terminal 250 aa of Etf-1 (residues 152 to 264) (8); rabbit anti-E. chaffeensis recombinant major outer membrane proteins P28 (36); mouse monoclonal anti-HA (BioLegend); mouse monoclonal anti-GFP, anti-MnSOD, and anti-cytochrome c (Santa Cruz Biotechnology); rabbit monoclonal anti-HA (Cell Signaling Technology); rabbit anti-actin (Sigma-Aldrich); Alexa Fluor (AF)488- and AF555-conjugated goat anti-rabbit IgG and anti-mouse IgG (Life Technologies); horseradish peroxidase (HRP)-conjugated goat anti-llama IgG (Bethyl Laboratories); and HRP-conjugated goat anti-mouse IgG and HRP-conjugated goat anti-rabbit IgG (KPL).
Plasmid Construction.
For screening of anti–Etf-1 Nbs and OpenSPR analysis, full-length Etf-1 with an N-terminal 6× His-tag followed by a tobacco etch virus (TEV) protease cleavage site and C-terminal Avi-tag was ligated into pET33b(+) vector to create His–TEV–Etf-1–Avi fusion protein. For far-Western blotting, truncated Etf-1 plasmids were constructed based on His–TEV–Etf-1–Avi. For mitochondrial localization analysis, truncated and/or mutated Etf-1–GFP was constructed from codon-optimized full-length Etf-1–GFP (8). For Nbs expression in mammalian cells, each full-length Nb was cloned into pEGFP-C1 vector by replacing EGFP with a C-terminal HA-tag. The primers used for cloning are listed in SI Appendix, Table S2.
Recombinant Protein Expression, Purification, and Biotinylation.
Avi-tagged full-length recombinant Etf-1 protein (rEtf-1) and truncated rEtf-1 proteins (primers shown in SI Appendix, Table S2) were expressed in Escherichia coli BL21(DE3) (New England Biolabs) and purified by affinity chromatography using HisPur Cobalt resin (Thermo Scientific) as described previously (7). The proteins were further purified by size-exclusion chromatography on an AKTA express (GE Healthcare) with a Superdex 200 Increase 10/300 GL column (GE Healthcare). For biotinylation, rEtf-1 was separated in Tris buffer (10 mM Tris, pH 8.0; 10 mM NaCl; 0.05% CHAPS; 0.05% sodium deoxycholate [NaDoc]) and concentrated to 2.2 mg/mL and was then biotinylated at the C-terminal Avi-tag using BirA Biotin-Protein Ligase Kit (Avidity). The reaction mixture was subjected to size-exclusion chromatography with a Superdex 200 Increase 10/300 GL column to remove BirA and d-biotin and then underwent a buffer exchange to PBS (8 mM Na2HPO4, 1.47 mM KH2PO4, 2.67 mM KCl, 137.9 mM NaCl, pH 7.4) containing 0.05% CHAPS and 0.05% NaDoc. The biotinylation of rEtf-1 was verified by Western blotting, and the labeled protein was then used for ELISA, panning, and OpenSPR analyses.
Llama Immunization and Analysis of Serum Conversion.
For llama immunization, full-length Etf-1 with an N-terminal 6× His-tag (8) was purified by affinity chromatography using HisPur Cobalt resin followed by size-exclusion chromatography on an AKTA express with a Superdex 200 Increase 10/300 GL column in Hepes buffer (20 mM Hepes, pH 7.2; 150 mM NaCl). Freshly prepared Etf-1 protein (1 mg Etf-1 in 2 mL buffer) was mixed gently with 2 mL of GERBU adjuvant (GERBU Biotechnik) to form an emulsion, and then polymyxin B (Sigma-Aldrich) was added to a final concentration of 20 µg/mL. A 3-y-old castrated male llama was injected subcutaneously with the emulsion at two bilaterally symmetrical sites at the base of the neck near the lymph node; this was repeated two additional times with 2-wk intervals between injections. Prior to the first immunization, 20 mL of blood was collected in a Venosafe serum gel tube (Becton Dickinson) and clotted for 2 h at room temperature; the supernatant was then recovered after centrifugation (preimmune serum). This process was repeated at 3 d after the third immunization (postimmune serum).
An ELISA was performed as described previously (25). Briefly, a MaxiSorp 96-well plate (Thermo Fisher Scientific) was coated with 4 µg/mL of NeutrAvidin (Thermo Fisher Scientific) followed by 3 µg/mL of biotinylated rEtf-1 in PBST (PBS containing 0.05% Tween 20). BSA was biotin-labeled with EZ-Link Sulfo-NHS-Biotin kit (Thermo Fisher Scientific) and used as a negative control. The wells were first blocked with 5% skim milk (Difco) in PBST and then were incubated with fourfold serially diluted serum samples starting from a 1:64 dilution in PBST containing 0.1% skim milk for 1 h and then with 1: 10,000 HRP-conjugated goat anti-llama IgG. The absorbance was measured by a SpectraMax PLUS384 spectrophotometer (Molecular Devices) following the addition of H2O2 and TMB substrate (Thermo Fisher Scientific). To verify antibody recognition of rEtf-1 and native E. chaffeensis Etf-1, Western blotting analyses were performed as described previously (7, 8).
Nb Library Construction and Isolation of Etf-1–Binding Nbs.
The Nb library was constructed as described with slight modifications (25). Briefly, 5 d after the third immunization, 400 mL of blood was collected from the jugular vein in a Venosafe hematology citrate phosphate dextrose-coated blood collection bag (Fenwal), and lymphocytes were separated immediately using Ficoll-Paque Plus (GE Healthcare). Total RNA was extracted using RNeasy Plus Mini kit (Qiagen), and the first-strand cDNA was synthesized from 50 µg of total RNA using SuperScript III Reverse Transcriptase (Invitrogen). The variable domains of all immunoglobulin heavy chains were PCR-amplified using two gene-specific primers, CALL001 and CALL002 (25), with KOD Hot Start DNA Polymerase (Toyobo). The 700-bp PCR products were gel purified and further PCR-amplified using Nb-For and Nb-Back primers with Eco91I and PstI sites (25). The final 400-bp PCR products encoding Nb sequences were ligated into pMECS phagemid vector (received from Serge Muyldermans, Vlaams Instituut voor Biotechnologie, Brussels, Belgium) following digestion with PstI/Eco91I restriction enzymes (with the addition of XbaI restriction enzyme to reduce vector self-ligation) and transformed into the E. coli TG1 strain (Lucigen) using a Gene Pulser Xcell electroporation system (Bio-Rad). Colonies were titrated and analyzed by colony PCR using MP57 and GIII primers (25) to determine the total and functional library sizes.
Nbs were expressed on the surface of rescued recombinant VCSM13 phages after the TG1 cells of the immune library were infected with VCSM13 helper phage (Agilent). Phages with specificity for Etf-1 were enriched by one round of panning: Phages (1011 phages per well) were added to either denatured (by boiling for 10 min) or nondenatured biotinylated rEtf-1 that had been coated on microtiter plates (405 ng per well) as described previously (25) and bound phages were eluted with trypsin (Thermo Fisher Scientific). The eluate was transferred to separate microcentrifuge tubes prefilled with 5 μL of a 4 mg/mL AEBSF (Thermo Fisher Scientific) solution to inhibit protease activity and was used to infect freshly cultured exponentially growing TG1 cells. The enrichment of phage particles carrying the Etf-1–specific Nbs was calculated by comparing the number of phages eluted from rEtf-1–coated wells vs. negative control wells. After panning, 368 individual clones eluted either from denatured rEtf-1 or nondenatured rEtf-1 were screened by standard ELISA procedures using mouse anti-HA at a 1:1,000 dilution and HRP-conjugated goat anti-mouse IgG at a 1:7,500 dilution.
Plasmids of the positive clones were purified from TG1 cells, transformed into DH5α cells, and sequenced as described previously (25). The CDR3 amino acid sequences of 107 and 65 Nbs bound to nondenatured and denatured rEtf-1, respectively, were aligned using the MegAlign program with the ClustalW algorithm of Lasergene DNASTAR software (DNASTAR). A phylogenetic tree based on the alignment of 24 distinct CDR3 amino acid sequences was obtained using the MegAlign program.
Expression and Purification of anti–Etf-1 Nbs.
Plasmids encoding anti–Etf-1 Nbs were purified from TG1 cells and transformed into WK6 cells, a nonsuppressor strain (supE−) of E. coli (ATCC). Nbs, which contain the periplasmic localization sequence pelB signal peptide at their N terminus, were expressed and purified from the periplasm as described previously (25). The supernatant containing the Nbs, which contain a C-terminal HA-tag followed by a 6× His-tag, was affinity-purified with the cobalt resin followed by size-exclusion chromatography on an AKTA express using a Superdex 75 Increase 10/300 GL column (GE Healthcare) for OpenSPR and far-Western blot analysis.
Characterization of D7 and D3 Binding to rEtf-1 by FPLC and OpenSPR.
Binding of D7 and D3 with rEtf-1 was qualitatively assessed by first determining elution profiles of each of three proteins, then mixing the purified rEtf-1 with molar excess D7 or D3 proteins, separating the mixture with a Superdex 200 Increase 10/300 GL gel filtration column in PBS running buffer (PBS containing 0.05% CHAPS and 0.05% NaDoc), and analyzing each fraction for the presence rEtf-1 and D7 or D3 by Western blotting. The affinity binding constant was measured by OpenSPR (Nicoya, Kitchener, ON, Canada). After immobilization of biotinylated rEtf-1 to a streptavidin sensor chip (Nicoya), dilutions of D7 and D3 in PBST running buffer were slowly flowed over the sensor chip at a rate of 20 µL/min with a contact time of 270 s and dissociation time of 330 s. Binding kinetics were obtained using TraceDrawer software (Nicoya).
CPP Conjugation of D7 and D3.
CPP12 was synthesized and purified by high-performance liquid chromatography (HPLC), as described previously (22). FPLC-purified D7 and D3 with >95% purity in PBS at 3 mg/mL were adjusted to pH 6.2 with 1 M MES buffer (pH 5.5). Proteins were incubated with 8-M equivalents of CPP12 for 1 h at room temperature on an end-to-end rotator, and then freshly prepared 10 mM NaBH3CN solution was added to the mixture. Reactions were continued at 4 °C for 48 h and 72 h for D7 and D3, respectively. Buffer containing the CPP12 and D7 or D3 conjugates was exchanged to PBS, and excess CPP12 was removed by FPLC through a HiTrap desalting column (GE Healthcare). CPP-D7 and CPP-D3 were concentrated to 3.6 mg/mL (0.22 mM) and 3.7 mg/mL (0.24 mM), respectively, for subsequent experiments.
Analysis of Mitochondrial Localization of Etf-1 and Apoptosis Assay.
For transfection of mammalian cells, plasmids were transformed into E. coli strain DH5α (Invitrogen) and purified using the Endo-Free Plasmid Purification kit (Qiagen, or Omega).
To examine the cellular distribution of truncated and mutated Etf-1–GFP, plasmids were transfected into RF/6A cells using Fugene HD (Promega). For apoptosis assays, RF/6A cells were transfected with full-length, truncated, or mutated Etf-1–GFP, or cotransfected with Etf-1–GFP and D7-HA or D3-HA plasmids by electroporation at 100 V and 1,000 µF using the Gene Pulser Xcell System and were then seeded onto coverslips at 1.3 × 105 cells per well in a 12-well plate. To determine the ability of CPP-Nbs to inhibit Etf-1–induced apoptosis, RF/6A cells transfected with Etf-1–GFP at 12 hpt were treated with 10 µM CPP-D7 or CPP-D3 in AMEM with 1% FBS for 12 h. At 24 hpt, cells were washed to remove uninternalized CPP-Nbs and changed to AMEM with 10% FBS. The cells were then treated with 100 µM etoposide (Sigma-Aldrich) for 4 h.
At 2 d posttransfection (dpt), cells were fixed with 4% paraformaldehyde in PBS at room temperature for 20 min. The cells were then labeled with primary antibodies (mouse anti-cytochrome c, rabbit anti–Etf-1, mouse anti-GFP, and/or rabbit anti-HA) diluted 1:50 in PGS (PBS with 0.1% gelatin and 0.1% saponin) for 2 h at room temperature, followed by AF488- or AF555-conjugated goat anti-mouse IgG and/or anti-rabbit IgG diluted 1:100 in PGS at room temperature for 1 h. Nuclei were stained with 300 nM DAPI, and coverslips were mounted and sealed with nail polish. Fluorescence images and differential interference contrast (DIC) images were captured with a DeltaVision deconvolution microscope (Applied Precision). Mitochondria exist as distinct filaments in RF/6A cells; therefore, when GFP-tagged Etf-1 or Etf-1 mutants showed filamentous distribution and colocalized with cytochrome c, these cells were scored as positively localized to mitochondria. When GFP-tagged Etf-1 or Etf-1 mutants were expressed in punctate or diffuse patterns, and did not colocalize with cytochrome c, these cells were scored as no colocalization with mitochondria. To determine the effects of Nbs on the localization of Etf-1 to mitochondria, only RF/6A cells expressing both Etf-1–GFP and D7- or D3-HA were selected.
Far-Western Blot Analysis.
Purified full-length rEtf-1 and various truncated rEtf-1 proteins and BSA were run on SDS-polyacrylamide gels and transferred to PVDF membranes using a semidry blotter (WEP). Proteins on the membranes were denatured with 6 M guanidine-HCl in a basic buffer (20 mM Tris, pH 7.6; 100 mM NaCl; 0.5 mM EDTA; 10% glycerol; 0.1% Tween-20; 2% skim milk; 1 mM DTT), followed by renaturing in the basic buffer containing serially diluted guanidine-HCl as described previously (44). The membrane was blocked with 5% skim milk in PBST at room temperature for 1 h and then was incubated with 5 mL of 2 µg/mL NbD7-HA-His or NbD3-HA-His overnight at 4 °C. Membranes were probed with mouse anti-HA (1:2,000 dilution) followed by HRP-conjugated goat anti−mouse IgG (1:1,000 dilution). ECL Western blotting substrate (Thermo Scientific) was used for chemiluminescence detection, and images were captured by an Amersham Imager 680 (GE Healthcare).
E. chaffeensis and Cell Culture.
E. chaffeensis Arkansas strain (45) was cultured in THP-1 cells (ATCC) in RPMI medium 1640 (Corning) supplemented with 8% fetal bovine serum (FBS; Atlanta Biologicals) at 37 °C under 5% CO2 in a humidified atmosphere. E. chaffeensis Wakulla strain was propagated in DH82 cells (46) in DMEM (Corning) with 8% FBS, as described previously (39, 47). RF/6A cells (ATCC) were cultured in Advanced Minimum Essential Medium (AMEM; Gibco) with 10% FBS. HEK293 cells (ATCC) were cultured in DMEM with 5% FBS. HEK293T cells (ATCC) were cultured in DMEM with 10% FBS. All culture media were supplemented with an additional 2 mM l-glutamine (Gibco).
ROS Assay with H2DCFDA and Western Blot Analysis.
HEK293 cells were seeded in a 96-well flat- and clear-bottom black plate (Tecan) at 1 × 104 cells per well. HEK293 cells were transfected with D7-HA or D3-HA using Fugene HD and then were infected by E. chaffeensis (∼50 multiplicity of infection [MOI]) at 1 dpt or were cotransfected with plasmids encoding Etf-1–HA and D7- or D3-HA. At 2 dpi (3 dpt) or at 2 dpt for the cotransfection experiments, the amount of ROS in whole cells was detected by using the fluorescent dye H2DCFDA (Invitrogen). Briefly, cells were washed with PBS and then incubated with 200 μL of 10 μM H2DCFDA in prewarmed PBS for 30 min at 37 °C under 5% CO2. The cells were washed once with PBS, and the fluorescence intensity of DCF (corresponding to the ROS level) was measured with an Infinite 200 PRO Microplate Reader (Tecan) at excitation and emission wavelengths of 492 nm and 520 nm, respectively.
For Western blot analysis, 2 × 106 HEK293 cells were transfected and infected with E. chaffeensis as described above for 2 d. Cell lysates were subjected to Western blotting using primary antibodies, including rabbit antirecombinant P28 (diluted 1:2,000), rabbit anti–Etf-1 (1:2,000), mouse anti-MnSOD (1:1,000), rabbit anti-actin (1:2,000), and mouse anti-HA (1:1,000) to detect Nbs, followed by HRP-conjugated secondary antibodies (1:2,000). Images were captured by the Amersham Imager 680, and band densities were quantitated by ImageQuantTL (GE Healthcare). As the rabbit antirecombinant P28 antisera (36) recognized both native P28 and OMP-1F expressed by E. chaffeensis (37), band densities of both proteins were used in the quantitation to determine E. chaffeensis infection levels.
Analysis of CPP12-Nbs on E. chaffeensis Infection and qPCR.
THP-1 cells were seeded at 2 × 105 cells per well in a 12-well plate and infected with host cell-free E. chaffeensis at 20 MOI. At 2 hpi, cells were treated with 10 μM CPP-D3/D7 or with an equal volume of PBS (control) in 1 mL RPMI medium 1640 with 1% FBS. After a 12-h incubation, cells were washed to remove uninternalized CPP-Nbs and cultured in fresh RPMI medium 1640 with 8% FBS for 2 d. DNA was extracted from each culture, and qPCR analysis was performed in an MX3000P qPCR instrument (Stratagene) using SYBR Green Real-Time PCR Master Mix (Thermo Fisher Scientific), as described previously (48). Fivefold serial dilutions of the control group was used to generate a standard curve to calculate the relative copy numbers of E. chaffeensis 16S rRNA and human actin (ACTB) genes using the MX3000P software from Stratagene.
Infection of SCID Mice with E. chaffeensis Wakulla and Their Treatment with CPP-Conjugated Nbs.
E. chaffeensis Wakulla-infected DH82 cells at ∼30% infectivity were centrifuged and resuspended at 1 × 105 cells in 0.5 mL of DMEM containing 1% FBS. The cells were incubated with CPP12-conjugated Nbs (∼50% conjugation efficiency, 80 μg CPP12-D3 or -D7 in 30 μL PBS) or a PBS only control at 37 °C for 12 h. Cells were then mixed with 0.47 mg of CPP-Nbs or an equal volume of PBS and injected with a 26-gauge needle into the peritoneal cavity of a 4-wk-old SCID ICR mouse (obtained from Taconic). Five mice were used for each group. At 1 and 2 dpi, each mouse was inoculated intraperitoneally with ∼0.47 mg of CPP-Nb diluted in 500 μL of PBS (equivalent to 0.02 mg CPP-Nb per gram body weight with an average mouse body weight of 23 g) or with PBS alone. Mice were monitored daily for body weight and clinical signs (squinty eyes, anorexia, and inactivity) of infection with E. chaffeensis. In addition, blood samples were collected from the sublingual sinus on days 0, 6, and 13 postinfection. One mouse in each group died at 13 dpi during blood collection. All remaining mice were killed at 14 dpi. DNA was isolated from the blood samples using Chelex 100 Resin (Bio-Rad) as described previously (49) or QIAamp DNA Blood Mini kit (Qiagen), and qPCR analysis was performed by using primers targeting the E. chaffeensis 16S rRNA gene and mouse Gapdh (39).
Flow Cytometry.
HEK293T cells were cultured in 24-well plates (2.5 × 105 cells per well) for 24 h, and incubated for 12 h with 10 μM CPP-D7, CPP-D3, D7, D3, or an equivalent volume of PBS in DMEM with 1% FBS. Cells were washed to remove unbound CPP-Nbs or Nbs, fixed with 4% paraformaldehyde for 10 min. Internalized HA-tagged Nbs were labeled with monoclonal rabbit anti-HA (1:1,600 dilution in PGS), followed by AF555-conjugated goat anti-rabbit IgG (1:200 dilution in PGS) at room temperature for 1 h each. Cells were analyzed on an Attune NxT acoustic focusing cytometer (Life Technologies) using the YL1 channel (excitation, 561 nm; emission, 585 nm). Data were analyzed by FlowJo software (Becton Dickinson) to visualize differences in relative percentages of cell populations among groups.
MTT Assay.
RF/6A cells were cultured in 96-well plates at 5,000 cells per well for 6 h, and incubated for 12 h with 10 μM CPP-D7, CPP-D3, D7, D3, or an equivalent volume of PBS in AMEM medium containing 1% FBS and 2 mM l-glutamine. Cells were washed with PBS to remove unbound CPP-Nbs or Nbs, and cultured in AMEM medium with 10% FBS and 2 mM l-glutamine for an additional 24 h. MTT (Roche) solution was added to each well (10 μL per well) and incubated for 4 h at 37 °C. The formazan crystals were solubilized by the addition of 100 μL of solubilization buffer (Roche) to each well, and the plates were incubated overnight at 37 °C. Absorbance at 565 nm was determined using a SpectraMax PLUS384 spectrophotometer (Molecular Devices).
Statistical Analysis.
All statistical analyses were performed with a one-way ANOVA using Prism 8 software (GraphPad). A post hoc test was then used to determine significant differences between different treatments. P < 0.05 was considered to reflect a statistically significant difference.
Supplementary Material
Acknowledgments
This study was funded by NIH Grant R21 AI146736 (to Y.R.).
Footnotes
Competing interest statement: D.P. is a cofounder and shareholder of Entrada Therapeutics.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2024102118/-/DCSupplemental.
Data Availability
All experimental data described in this study are included in the main text and SI Appendix.
References
- 1.Adams D. A.et al.; Nationally Notifiable Infectious Conditions Group , Summary of notifiable infectious diseases and conditions—United States, 2015. MMWR Morb. Mortal. Wkly. Rep. 64, 1–143 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Kuriakose K., Pettit A. C., Schmitz J., Moncayo A., Bloch K. C., Assessment of risk factors and outcomes of severe Ehrlichiosis infection. JAMA Netw. Open 3, e2025577 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paddock C. D., Childs J. E., Ehrlichia chaffeensis: A prototypical emerging pathogen. Clin. Microbiol. Rev. 16, 37–64 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Institute of Medicine , Critical Needs and Gaps in Understanding Prevention, Amelioration, and Resolution of Lyme and Other Tick-Borne Diseases: The Short-Term and Long-Term Outcomes, Workshop Report, (National Academies Press, Washington, DC, 2011). [PubMed] [Google Scholar]
- 5.Rikihisa Y., Role and function of type IV secretion systems in Anaplasma and Ehrlichia species. Curr. Top. Microbiol. Immunol. 413, 297–321 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Sharma P., Teymournejad O., Rikihisa Y., Peptide nucleic acid knockdown and Intra-host cell complementation of Ehrlichia type IV secretion system effector. Front. Cell. Infect. Microbiol. 7, 228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan Q., et al., Ehrlichia type IV secretion system effector Etf-2 binds to active RAB5 and delays endosome maturation. Proc. Natl. Acad. Sci. U.S.A. 115, E8977–E8986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H., Bao W., Lin M., Niu H., Rikihisa Y., Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell. Microbiol. 14, 1037–1050 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin M., et al., Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12, 2145–2166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamers-Casterman C., et al., Naturally occurring antibodies devoid of light chains. Nature 363, 446–448 (1993). [DOI] [PubMed] [Google Scholar]
- 11.Desmyter A., Decanniere K., Muyldermans S., Wyns L., Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J. Biol. Chem. 276, 26285–26290 (2001). [DOI] [PubMed] [Google Scholar]
- 12.De Genst E., Saerens D., Muyldermans S., Conrath K., Antibody repertoire development in camelids. Dev. Comp. Immunol. 30, 187–198 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Dumoulin M., et al., Single-domain antibody fragments with high conformational stability. Protein Sci. 11, 500–515 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y., et al., Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. Int. J. Nanomedicine 11, 3287–3303 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser P. D., Maier J., Traenkle B., Emele F., Rothbauer U., Recent progress in generating intracellular functional antibody fragments to target and trace cellular components in living cells. Biochim. Biophys. Acta 1844, 1933–1942 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Jobling S. A., et al., Immunomodulation of enzyme function in plants by single-domain antibody fragments. Nat. Biotechnol. 21, 77–80 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Mejías M. P., et al., Development of camelid single chain antibodies against Shiga toxin type 2 (Stx2) with therapeutic potential against Hemolytic Uremic Syndrome (HUS). Sci. Rep. 6, 24913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudolph M. J., et al., Intracellular neutralization of ricin toxin by single-domain antibodies targeting the active site. J. Mol. Biol. 432, 1109–1125 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrapp D.et al.; VIB-CMB COVID-19 Response Team , Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell 181, 1004–1015.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothbard J. B., et al., Conjugation of arginine oligomers to cyclosporin A facilitates topical delivery and inhibition of inflammation. Nat. Med. 6, 1253–1257 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Trinh T. B., Upadhyaya P., Qian Z., Pei D., Discovery of a direct Ras inhibitor by screening a combinatorial library of cell-permeable bicyclic peptides. ACS Comb. Sci. 18, 75–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian Z., et al., Discovery and mechanism of highly efficient cyclic cell-penetrating peptides. Biochemistry 55, 2601–2612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian Z., et al., Early endosomal escape of a cyclic cell-penetrating peptide allows effective cytosolic cargo delivery. Biochemistry 53, 4034–4046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brugnano J. L., Chan B. K., Seal B. L., Panitch A., Cell-penetrating peptides can confer biological function: Regulation of inflammatory cytokines in human monocytes by MK2 inhibitor peptides. J. Control. Release 155, 128–133 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Pardon E., et al., A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 9, 674–693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fronzes R., Christie P. J., Waksman G., The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7, 703–714 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metcalf K. J., et al., Proteins adopt functionally active conformations after type III secretion. Microb. Cell Fact. 15, 213 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Truscott K. N., Brandner K., Pfanner N., Mechanisms of protein import into mitochondria. Curr. Biol. 13, R326–R337 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Karpinich N. O., Tafani M., Rothman R. J., Russo M. A., Farber J. L., The course of etoposide-induced apoptosis from damage to DNA and p53 activation to mitochondrial release of cytochrome c. J. Biol. Chem. 277, 16547–16552 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Halliwell B., Gutteridge J. M. C., Free Radicals in Biology and Medicine (Oxford University Press, New York, ed. 3, 1999). [Google Scholar]
- 31.Cai J., Jones D. P., Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J. Biol. Chem. 273, 11401–11404 (1998). [DOI] [PubMed] [Google Scholar]
- 32.Parone P., Priault M., James D., Nothwehr S. F., Martinou J. C., Apoptosis: Bombarding the mitochondria. Essays Biochem. 39, 41–51 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Oberley L. W., Oberley T. D., Role of antioxidant enzymes in cell immortalization and transformation. Mol. Cell. Biochem. 84, 147–153 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Zelko I. N., Mariani T. J., Folz R. J., Superoxide dismutase multigene family: A comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic. Biol. Med. 33, 337–349 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Armstrong J. S., Whiteman M., Measurement of reactive oxygen species in cells and mitochondria. Methods Cell Biol. 80, 355–377 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Ohashi N., Zhi N., Zhang Y., Rikihisa Y., Immunodominant major outer membrane proteins of Ehrlichia chaffeensis are encoded by a polymorphic multigene family. Infect. Immun. 66, 132–139 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumagai Y., Huang H., Rikihisa Y., Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. J. Bacteriol. 190, 3597–3605 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winslow G. M., Yager E., Shilo K., Collins D. N., Chu F. K., Infection of the laboratory mouse with the intracellular pathogen Ehrlichia chaffeensis. Infect. Immun. 66, 3892–3899 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura K., Rikihisa Y., Virulence potential of Ehrlichia chaffeensis strains of distinct genome sequences. Infect. Immun. 75, 3604–3613 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohan Kumar D., et al., EtpE binding to DNase X induces Ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. MBio 6, e01541–e15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu H., Kozjak-Pavlovic V., Rudel T., Rikihisa Y., Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog. 6, e1000774 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu H., Rikihisa Y., Ats-1: A novel bacterial molecule that links autophagy to bacterial nutrition. Autophagy 9, 787–788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu H., Xiong Q., Yamamoto A., Hayashi-Nishino M., Rikihisa Y., Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc. Natl. Acad. Sci. U.S.A. 109, 20800–20807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu Y., Li Q., Chen X. Z., Detecting protein-protein interactions by far Western blotting. Nat. Protoc. 2, 3278–3284 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Dawson J. E., et al., Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29, 2741–2745 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellman M. L., Krakowka S., Jacobs R. M., Kociba G. J., A macrophage-monocyte cell line from a dog with malignant histiocytosis. In Vitro Cell. Dev. Biol. 24, 223–229 (1988). [DOI] [PubMed] [Google Scholar]
- 47.Sumner J. W., Childs J. E., Paddock C. D., Molecular cloning and characterization of the Ehrlichia chaffeensis variable-length PCR target: An antigen-expressing gene that exhibits interstrain variation. J. Clin. Microbiol. 37, 1447–1453 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng Z., Miura K., Popov V. L., Kumagai Y., Rikihisa Y., Insights into the CtrA regulon in development of stress resistance in obligatory intracellular pathogen Ehrlichia chaffeensis. Mol. Microbiol. 82, 1217–1234 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rikihisa Y., Zhang C., Christensen B. M., Molecular characterization of Aegyptianella pullorum (Rickettsiales, Anaplasmataceae). J. Clin. Microbiol. 41, 5294–5297 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All experimental data described in this study are included in the main text and SI Appendix.