Abstract
Key processes characterizing human aging are immunosenescence and inflammaging. The capacity of the immune system to adequately respond to external perturbations (e.g., pathogens, injuries, and biochemical irritants) and to repair somatic mutations that may cause cancers or cellular senescence declines. An important goal remains to identify genetic or biochemical, predictive biomarkers for healthy aging. We recruited two cohorts in the age range 70 to 82, one afflicted by chronic illnesses (non-healthy aging, NHA) and the other in good health (healthy aging, HA). NHA criteria included major cardiovascular, neurodegenerative, and chronic pulmonary diseases, diabetes, and cancers. Quantitative analysis of forty proinflammatory cytokines in blood plasma and more than 500 proteins in urine was performed to identify candidate biomarkers for and biological pathway implications of healthy aging. Nine cytokines revealed lower quantities in blood plasma for the NHA compared with the HA groups (fold change > 1.5; p value < 0.025) including IL-12p40 and IL-12p70. We note that, sampling at two timepoints, intra-individual cytokine abundance patterns clustered in 86% of all 60 cases, indicative of person-specific, highly controlled multi-cytokine signatures in blood plasma. Twenty-three urinary proteins were differentially abundant (HA versus NHA; fold change > 1.5; p value < 0.01). Among the proteins increased in abundance in the HA cohort were glycoprotein MUC18, ephrin type-B receptor 4, matrix remodeling–associated protein 8, angiopoietin-related protein 2, K-cadherin, and plasma protease C1 inhibitor. These proteins have been linked to the extracellular matrix, cell adhesion, and vascular remodeling and repair processes. In silico network analysis identified the regulation of coagulation, antimicrobial humoral immune responses, and the IL-12 signaling pathway as enriched GO terms. To validate links of these preliminary biomarkers and IL-12 signaling with healthy aging, clinical studies using larger cohorts and functional characterization of the genes/proteins in cellular models of aging need to be conducted.
Electronic supplementary material
The online version of this article (10.1007/s11357-020-00269-y) contains supplementary material, which is available to authorized users.
Keywords: Urinary proteome, Proinflammatory cytokines, Healthy aging, Chronic disease, Proteomics, Vasculature, Metabolic syndrome, Senescence, Inflammaging, Cathepsin D, Interleukin 12, Afamin, IL-12 signaling
Introduction
Environmental/lifestyle factors such as nutrition, the gastrointestinal microbiome, smoking, physical activity, and mental stress as well as genetic attributes influence human aging. Declining physiological and immunological functions can trigger acute or chronic diseases including organ failure. Aging-associated immunological changes (immunosenescence) involve changes in the adaptive immune system, such as lymphocyte migration, maturation, and function, but also the innate immune system [22]. Chronic low-grade inflammation (inflammaging) is considered a contributing factor to increased disease risks when people age [70]. It is unclear to what extent immunosenescence and inflammaging involve similar molecular and cellular changes, and if they entail health and longevity benefits [19]. Immunosenescence is thought to transition through stages of initiation, early, and late senescence. Cellular traits are mitochondrial dysfunction, chromatin remodeling, and altered lysosomal capacity for autophagy. Chronic inflammation mediated by the release of various inflammatory molecules results, which is termed the senescence-associated secretory phenotype (SASP) [25]. Cytokines are components of the SASP. A myriad of cytokines contribute to the maturation of and modulate our immune function, thus shaping the morphology and functional capacity of lymphoid organs and the thymus where hematopoiesis and T cell development, respectively, primarily originate [22, 65]. The cytokine network is pleiotropic; it has redundant and overlapping activities related to the development, proliferation, activation, chemotaxis, and migration of distinct cell types, including vascular endothelial cells [20, 21]. Thymic atrophy appears to be a distinct morphological feature of immunosenescence and leads to decline and/or dysregulation of the human T cell immune repertoire [54, 55]. A key cytokine stimulating thymopoiesis and peripheral T cell expansion is IL-7 [7, 22].
Cellular manifestations of the aging process are particularly evident in postmitotic cells such as cardiac myocytes and neurons [10, 61]. Subcellular organelles such as mitochondria and lysosomes have important roles in cellular waste product removal and recycle their molecular building blocks. Reduced functions of lysosomes [12] and mitochondria [9] can lead to waste accumulation which, in turn, triggers the inflammation embodied by the SASP phenotype. SASP proteins are released into body fluids (blood plasma and urine) and may become relevant as surrogate biomarkers for senescence and chronic inflammation-associated aging. One commonly used model system for research on aging in multicellular organisms with a digestive tract is the nematode Caenorhabditis elegans. Studies using this model established the links between mitochondrial function and aging [49] and identified genes of potential importance to aging. One such longevity gene, encoding a NAD-dependent deacetylase, is sir2. Other genes code for the insulin receptor Daf2 and forkhead transcription factor Daf16 [35, 62]. The human Sir2 ortholog SIRT3 has a potential mitochondrial NAD-dependent deacetylase function, supporting the interplay between mitochondrial functional decline and aging. Furthermore, genotype variability in SIRT3 has been statistically linked to longevity [44]. SIRT3 is involved in reactive oxygen species (ROS) suppression and mitochondrial biogenesis [29] and a tumor suppressor protein [13]. The insulin/IGF-1 signaling (IIS) pathway may be an important, evolutionarily conserved pathway with a role in NF-kB signaling and the aging process. SIRT1, a paralog of SIRT3, and the class O Forkhead box protein (FoxO), the human ortholog of the C. elegans protein Daf16, are NF-kB signaling inhibitors [45]. In another study, genetic variability in the gene encoding FOXO3A was found to be strongly associated with longevity [68].
It is of medical interest to identify surrogate biomarkers predictive of healthy aging (i.e., the aging of human subjects who are not afflicted with major diseases such as cancer, chronic and acute cardiovascular diseases, chronic pulmonary diseases, major neurological diseases and stroke, diabetes and diabetic complications in their lifetimes compared with individuals who do). No biomarkers are currently in clinical use as specific tests to predict healthy aging although age-independent biomarkers of deteriorating health are monitored to diagnose and predict onset of the most common chronic diseases [33]. Those include C-reactive protein (CRP), low-density lipoprotein (LDL), glucose, and HbA1C concentrations (all measured in blood), hypertension, and proteinuria. These tests are often included during a patient’s physical examination while the readout focuses on specific pathologies: CRP and LDL to identify cardiovascular risk [64] and HbA1C, glucose, and proteinuria to determine onset or progression of diabetes [24]. Specifically relevant to aging, the NIH funded a study to measure phosphorylated Tau-181 as a potential blood diagnostic for Alzheimer’s dementia [26]. Predictive aging biomarkers may be genetic (longevity-associated genes) or biochemical, the latter of which may represent changes in abundance, post-transcriptional or post-translational modifications (RNAs, proteins, and small molecules). Inflammaging-associated markers may be cytokines, lysosomal enzymes, ROS-modified macromolecules, or mitochondrial DNA mutations [10, 19]. Studies to identify protein and metabolite biomarkers in body fluids for the elderly and for healthy aging (HA) have been performed [1, 27, 51, 53]. To our knowledge, no study has used a cohort design with elderly individuals discordant for the incidence of major chronic diseases such as cardiovascular, pulmonary, neurodegenerative, metabolic, and cancer in their lifetimes. We recruited 65 human subjects in the age range 70 to 82 years, ~ 50% each in the HA and non-healthy aging (NHA) groups. Blood plasma and urine were collected at two timepoints, 4 to 6 months apart, for each study participant. Stool and saliva samples from this cohort were analyzed for differences in the microbiome, with outcomes that we previously reported [50]. We conducted studies to quantitatively analyze the urinary proteome (untargeted proteomics) and forty proinflammatory cytokines and chemokines in blood (via cytokine antibody arrays) reported in this paper.
Materials and methods
Study population, recruitment, and specimen collections
This case-control, prospective study consisted of patients who visited a physician at Danbury Hospital, Danbury, CT, in the context of routine or disease follow-up health care. If in the age range 70 to 82, they were asked about their interest in volunteering for a study intended to identify biomarkers of healthy aging. Internal review boards (IRB) of Danbury Hospital and the J. Craig Venter Institute (JCVI), Rockville, MD approved a consent form and human subject protocol outlining risks and benefits of participation in 2013. Data on medical histories were collected. Sixty-five human subjects (both genders) provided saliva, stool, blood, and urine specimens twice during the active enrollment period. Enrolment and specimen collections (two samples were obtained from sixty and one from five participants) were completed in 2015. Medical histories were used to divide the subjects into healthy aging (HA) and non-healthy aging (NHA) groups, with 33 and 32 subjects, respectively. The NHA group included those diagnosed with (1) cancer, (2) acute or chronic cardiovascular diseases, (3) acute or chronic pulmonary diseases, (4) chronic liver disease, (5) diabetes, and/or (6) stroke or neurodegenerative disorders at any point in their lifetimes. Subjects part of the HA group did not report diagnosis of any of the illnesses. Blood was collected via venipuncture by a phlebotomist. Clean-catch urine specimens were collected on-site with instructions by professional medical staff.
Sample processing for cytokine arrays
Blood samples were collected in BD Vacutainer® trace element plastic blood collection tubes with K2-EDTA as the anticoagulant on the day of the study participant’s visit. Blood plasma was separated by centrifugation at 1500×g to remove cells. Plasma supernatants were frozen at − 80 °C, shipped, and stored frozen until thawed to remove aliquots for the cytokine abundance measurements. We used a commercial product, the G-series HIAA 3 cytokine antibody microarray (Ray Biotech, Inc., Norcross, GA). Following the manufacturer’s instructions, fluorescence signals for 40 cytokines were measured. The Ray Biotech Q Analyzer program was used for data analysis. On the day the HIAA array was used, plasma aliquots were vortexed and centrifuged for 10 min at 10,000×g to remove particulate matter. Following 30 min of exposure to a sample diluent, the product’s glass plates were washed, wells were overlaid with 70 μL sample, covered, and incubated at 20 °C for 2 h. Wash steps with buffers I and II were followed by covering the sub-arrays with an equal volume (70 μL) of biotin-conjugated anticytokine antibodies, incubation at 20 °C for 2 h, a 2nd round of buffers I/II wash steps, and incubation at 4 °C overnight. The Alexa Fluor 647-conjugated streptavidin reagent was added and incubated at 20 °C for 2 h. Fluorescence signals (Cy5 range; 655 nm emission) were scanned and extracted using a GenePix 4000B laser scanner (Axon Instruments, Foster City, CA). Normalization steps consisted of (a) subtracting background signals, (b) normalizing signals to positive controls, and (c) comparing the signal intensities of antigen-specific array spots among the images from different array slides. Relative normalized abundance values for the detected cytokine analytes were computed. We chose 1.5- and 0.65-fold-changes cutoff values to designate an increase or decrease in protein abundance, respectively, in the data comparison of the HA and NHA groups. The “A” and “B” samples for a single individual (two timepoints) were not placed in sub-arrays on the same array (slide) to avoid bias in data interpretation. The methods were analogous to those used previously for HIAA-3 arrays [60]. The quantified and normalized cytokine datasets are provided in Supplemental File S1, Dataset.
Sample processing for urinary shotgun proteomics
Urine samples collected in 50 mL Falcon tubes on the day of the study participant’s visit were centrifuged for 10 min at 3000×g, and sediments were removed. Urine supernatant samples were frozen at − 80 °C, shipped to JCVI, and stored frozen until thawed for ultrafiltration-based protein concentration with a 10 kDa MWCO membrane filter. The procedures all urine samples and protein concentrates underwent, including protein visualization by staining in SDS-PAGE gels, were previously described in detail [59]. Aliquots of approximately 100 μg total protein were subjected to filter-aided sample preparation (FASP) [69] to digest proteins, separate, and enrich the resulting tryptic peptides [75]. Peptide mixtures were lyophilized and stored frozen until resuspended for liquid chromatography tandem mass spectrometry (LC-MS/MS) analyses. The “A” and “B” samples from a given patient were not processed together in batches, thus avoiding biases in the data interpretation.
Shotgun proteomics on an accurate mass/high mass resolution Q-Exactive system
Peptide concentrates were subjected to LC-MS/MS on an Ultimate 3000 nano-LC coupled to a Q-Exactive MS system via a FLEX nano-electrospray ion source (Thermo Scientific). The peptide samples were resuspended in 60 μL HPLC solvent A (0.1% formic acid in water), and 10 μL were loaded onto a trap column (C18 PepMap100, 300 μm × 5 mm, 5 μm, 100 Å, Thermo Scientific). The analytical column used was a PicoFrit column (75 μm × 10 cm, 5 μm BetaBasic C18, 150 Å, New Objective, MA). A 130 min LC gradient with a flow rate of 300 nL/min started from 2% solvent B to 35% solvent B (0.1% formic acid in acetonitrile) for 110 min, then followed by a steeper gradient to 80% solvent B over 15 min. The column was re-equilibrated with solvent A for 5 more minutes. The eluting peptides were sprayed at a voltage of 2.1 kV and acquired in a MS data-dependent mode using Xcalibur software (version 2.2, Thermo Scientific). Full scan MS spectra were acquired at a resolution of 70,000 over a mass range of m/z 350 to 1800 with an automatic gain control (AGC) target of 106. Up to ten of the most abundant ions were subjected to fragmentation by higher energy collisional dissociation (HCD) with a normalized collision energy of 27. The peptide ion fragments from the MS/MS scans were acquired at a resolution of 17,500 with an AGC target of 5 × 104. Dynamic exclusion was set to 20 s. Unassigned ions were rejected and only those with a charge ≥ 2 were subjected to HCD fragmentation.
Analysis of urinary proteomes and methods for quantification with the MaxQuant software tool
LC-MS/MS raw data were processed using the MaxQuant-Andromeda software (version 1.5.1.0). The analysis methods were previously described in detail [75]. Only reviewed protein sequence entries in the non-redundant Human UniProt database (release 2015–2016; 20,195 sequences) were used for database searches. Search parameters were set to allow two missed tryptic cleavages with oxidation (M), N-terminal acetylation, and deamidation (N, Q) as variable modifications and carbamidomethylation (C) as a fixed modification. The peptide identifications (IDs) were limited to top-ranked peptides with a length of at least seven amino acids. The MS and MS/MS ion tolerances were set at 10 ppm and 0.02 Da, respectively. False discovery rates (FDR) were estimated using the integrated decoy database search tool Percolator. Protein hits identified with an FDR threshold set at 1% were accepted in the final protein list.
A total of 124 datasets (124 samples, all but 5 study participants were represented by two samples) were available for quantitative analysis using the MaxQuant software suite. Default settings for MS1 peak integration and normalization among datasets provided in the MaxQuant-Andromeda software suite were accepted. The label-free quantitation (LFQ) algorithm was enabled. Only proteins that were quantified by at least two unique peptides were used for further analysis. When all peptides of a protein were shared by multiple proteins, they were combined and reported as a single protein group (this was a rare event). The proteins’ calculated abundances were log (base 2) transformed. Proteins absent in at least 30% of the samples (separately for the HA or NHA group) were removed for the statistical analyses. The datasets for which quantitative information was available for less than 25% of the human proteins that were represented in the MaxQuant analyses were also eliminated. Missing values were imputed using the LSimpute algorithm with the LSimpute_adaptive option [36]. Histograms were displayed for Log2 transformed MaxLFQ values prior to and after imputation. The data (Supplemental File S2, Dataset) showed improved normalization after imputation. Thus, data including missing value imputations were subjected to statistical analyses. The mass spectrometry proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012477 (reviewer account details: username: reviewer44087@ebi.ac.uk; password: qAT4NwUo).
Statistical analysis of proteomic and cytokine data
Differential analyses of cytokine and urinary proteome data were performed using the Limma software package, which is part of the R package and used for the analysis of expression data using a linear model which is fit to gene and protein expression data [43]. Application of the linear model allowed for analysis of the entire experiment as an integrated whole, sharing information between samples. Limma accepts log-ratios or log-intensity values of the expressed proteins as input. The p value cutoffs for significant proteins were set at 0.05; most of those discussed in the “Results and discussion” had lower p values (for urinary proteins 0.01, for cytokines 0.025). Multiple testing adjustments were performed using the Benjamini-Hochberg correction method. Only nine urinary proteins remained statistically significant after multiple testing corrections (adjusted p value ≤ 0.05).
Gene ontology, clustering, and classification analyses
Gene ontology (GO) information of proteins was obtained using the ClueGO version 2.2.5 and CluePedia version 1.2.5 plugin in Cytoscape version 3.3.0 [4, 5, 48]. Biological process, cellular component, molecular function, and immune system process ontology data were from a database update 31.03.2016. KEGG pathway data were from a 10.02.2016 update. For GO enrichment analysis, p values of less than 0.05 were considered, selecting all experimental evidence codes (EXP_IDA_IPI_IMP_IGI_IEP). The GO terms were connected by the proteins that shared those GO terms. Differentially expressed proteins and samples were clustered using the dist function in R, which calculates the Euclidean distance matrix between different proteins and samples. The distance matrix was used to generate hierarchical clusters using the hclust function in R and allowed conversion into a heatmap with the heatmap function in R. The Pearson correlation was used to cluster the urinary proteome datasets to generate hierarchical clusters for the datasets. Principle component analysis (PCA) of proteins was performed in the R software package. The volcano plots were generated using the Enhanced Volcano package in R.
Results and discussion
Cohort description and study design
The studies described here were performed along with those of oral and gastrointestinal microbiomes for 65 human subjects aged 70 to 82, recruited at a single site and composed of 31 male and 34 female human subjects. Half of the subjects had a history of serious, mostly chronic diseases (the NHA group), and the other half reached 70 or more years of age without being diagnosed with (1) cancer, (2) acute or chronic cardiovascular diseases, (3) acute or chronic pulmonary diseases, (4) chronic liver disease, (5) diabetes, and/or (6) stroke/neurodegenerative disorders (the HA group). Study participants were also asked questions about chronic pain, memory loss, hospitalizations, their mental state, and dietary habits, but these factors were considered less to place an individual in the NHA group. Details of the medical records are found in Supplemental File S3, Dataset. Eleven patients had cancer, 8 patients had diabetes, and 15 and 7 patients reported suffering from chronic cardiovascular and pulmonary diseases, respectively. One study participant had a neurodegenerative disorder. Overall, 125 blood plasma and urine specimens were obtained during recruitment over a 15-month time frame. For all but five participants, two timepoints (and datapoints) were available to determine proteins differentially abundant in HA vs. NHA cohorts. To facilitate participant consent and retention in the study and ensure absence of health risks, no instructions to change diet or alter therapeutic drug intake were provided during an individual’s participation and visits of the clinic. Based on self-reported medical data re-evaluated at the 2nd timepoint’s visit, none of the HA subjects needed to be recategorized as NHA.
Quantitative differences for urinary proteins (HA vs. NHA) many of which have known or putative roles in cell adhesion, blood vessel formation/repair, and inflammation
Collectively, we quantified nearly 2000 urinary proteins from the surveyed sample set. The urinary proteome span over seven orders of magnitude, suggesting excellent depth of coverage (Fig. 1). On average, 752 proteins were identified from each specimen (± 157, n = 125, FDR at 1%). Based on the frequency of detection in the 125 datasets (filtered for proteins to be present in at least 30% of all samples), 505 proteins were retained for quantitative analysis. Principal component analysis (PCA) did not reveal separate clusters for the HA and NHA proteome datasets (Fig. 2). Pearson correlation analysis revealed that the timepoints per subject clustered pairwise in 16 out of 58 cases (Supplemental File S4, Figure). Intra-individual variability in the urinary proteome was previously reported [39]. Without separating for gender, statistical differences in abundance (HA vs. NHA cohorts) were determined via unequal variance non-parametric t tests and yielded 23 proteins at a fold change > 1.5 (p value < 0.01). Nine proteins were statistically significant following Benjamini-Hochberg corrections (adj. p value < 0.05). These and five additional functionally interesting proteins with low p values are listed in Table 1 and included in the dynamic range plot of Fig. 1. The entire set of 23 proteins is displayed in the volcano plot of Fig. 3.
Fig. 1.
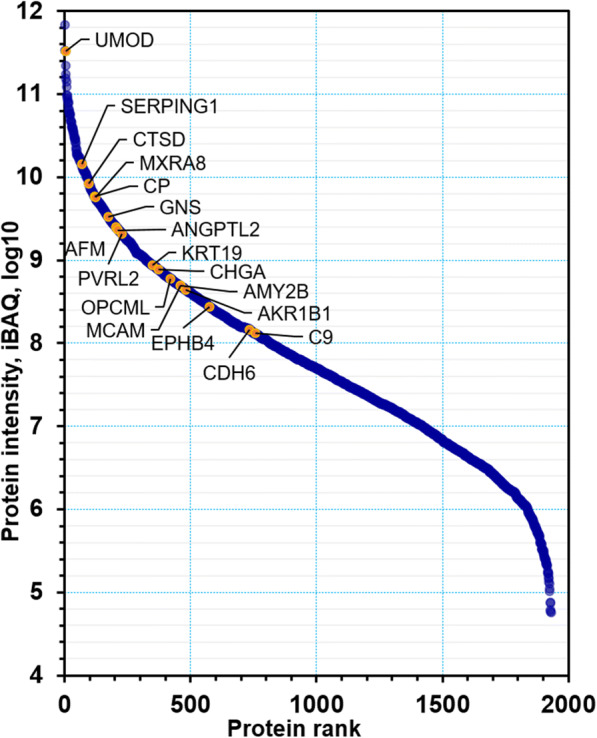
Dynamic range of urinary proteins. A total of 1959 proteins were ranked by the iBAQ intensity derived from the MaxQuant analysis. Some representative significant proteins as it pertains to quantitative differences (HA vs. NHA datasets) are marked in the plot
Fig. 2.
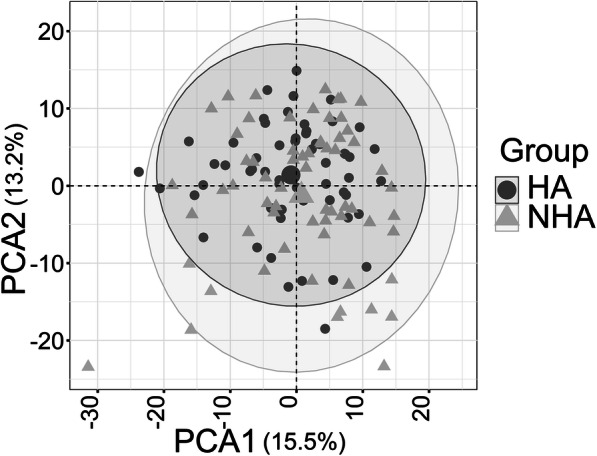
PCA plot comparing healthy (HA) and non-healthy (NHA) group using 505 urinary proteins as input data. The PCA plot includes all timepoints (two for most subjects). They are not averaged for a given subject. There is no separate cluster formation for the NHA (triangles) and HA (circles) datasets
Table 1.
Subset of differentially abundant proteins (healthy aging vs. non-healthy aging) with functional roles
| Protein acc. no. | Protein name | Fold change | t | p value | Adj. p value | A selection of putative or known protein functional roles |
|---|---|---|---|---|---|---|
| P07911 | Uromodulin (UMOD) | ↑ 2.12 | − 4.04 | 9.08E−05 | 0.032 | Leukocyte cell-cell adhesion and migration, water/electrolyte balance in urinary tract |
| P07339 | Cathepsin D (CTSD) | ↓ 1.69 | 3.96 | 0.00013 | 0.032 | Lysosomal enzyme with proteolytic functions mediating ECM degradation, proliferation and migration of microvascular endothelial cells, part of PI3K/AKT and FBF pathways |
| Q9UKU9 | Angiopoietin-related protein 2 (ANGPTL2) | ↑ 1.66 | − 3.79 | 0.00023 | 0.038 | Induces angiogenesis via paracrine/autocrine actions, promotes leukocyte attachment to vascular endothelial cells, integrin signaling and PI3K/AKT pathways, inflammation of vasculature |
| Q9BRK3 | Matrix remodeling–associated protein 8 (MXRA8), limitrin | ↑ 1.82 | − 3.66 | 0.00037 | 0.038 | Cell adhesion protein suppressing migration and promoting apoptosis of vascular endothelial cells, active in their cell junctions (angiogenesis), involved in integrin signaling and VGEF pathways, blood-brain barrier |
| Q14982 | Opioid binding and cell adhesion protein (OPCML) | ↑ 1.78 | − 3.63 | 0.00041 | 0.038 | Glycosylphosphatidylinositol-anchored cell adhesion molecule, opioid receptors are present in microvascular endothelial cells |
| P55285 | Cadherin-6 (CDH6), K-cadherin | ↑ 1.64 | − 3.60 | 0.00046 | 0.038 | Calcium-dependent cell adhesion protein (in adherence cell junctions), E-cadherin is known to be active in β-catenin, fibroblast growth factor and PI3K/AKT pathways |
| P43121 | Cell surface glycoprotein MUC18 (MCAM) | ↑ 1.62 | − 3.47 | 0.00071 | 0.050 | Receptor on vascular endothelial cells triggering intracellular signaling pathways via E-cadherin and galectin-3, promotes tumor metastasis, vascular wound healing, cell adhesion |
| P54760 | Ephrin type-B receptor 4 (EPHB4) | ↑ 1.66 | − 3.41 | 0.00086 | 0.050 | Receptor tyrosine kinase in VGEF signaling pathway, involved in regulation of cell adhesion and migration, vascular remodeling, and wound healing (angiogenesis), blood-brain barrier |
| P43652 | Afamin (AFM) | ↓ 2.15 | 3.41 | 0.00089 | 0.050 | Carrier for hydrophobic molecules in body fluids (vitamin E, palmitoleic acid), ass. with development of metabolic syndrome and Wnt signaling, proinflammatory, blood-brain barrier |
| P02748 | Complement component C9 | ↓ 1.74 | 3.33 | 0.00114 | 0.053 | Acute phase response, MAC complex of complement system, innate immunity |
| P19961 | Alpha-amylase 2B (AMY2B) | ↑ 1.97 | − 3.30 | 0.00125 | 0.053 | Carbohydrate metabolism |
| P05155 | Plasma protease C1 inhibitor (SERPING1) | ↑ 1.60 | − 3.30 | 0.00125 | 0.053 | Activates complement system, fibrinolysis and kinins, regulates vascular permeability and suppression of inflammation |
| P00450 | Ceruloplasmin (CP) | ↓ 1.77 | 3.18 | 0.00180 | 0.062 | Acute phase response, iron ion homeostasis |
| P15121 | Aldo-keto reductase family 1 member B1 AKR1B1 | ↑ 1.73 | − 3.14 | 0.00210 | 0.066 | Key enzyme in the polyol pathway, plays a role in detoxifying dietary and lipid-derived unsaturated carbonyls, renal water homeostasis |
Proteins are listed with UniProt accession numbers (acc. no.), names, differential abundance data, and selected functional roles with a focus on cell adhesion, angiogenesis, vascular repair and remodeling, coagulation and fibrinolysis, and inflammation and acute phase response. With two groups of 33 HA and 32 NHA subjects, urinary proteome datasets were analyzed quantitatively. Fold change ↑, ↓: up and down in the HA compared with the NHA group, respectively. Adj. p value: adjusted p value via Benjamini-Hochberg corrections. PI3K/AKT phosphatidylinositol-3-kinase/Akt signal transduction pathway, ECM extracellular matrix
Fig. 3.
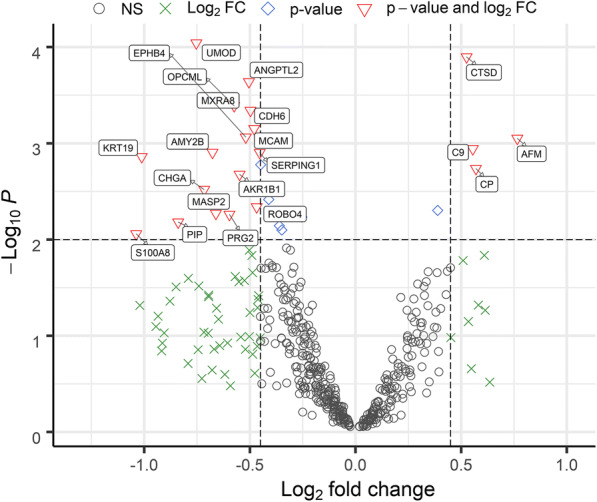
Differentially abundant urinary proteins in volcano plot. Urinary proteins displaying quantitative differences with statistical significance are depicted in red triangles. Non-significant proteins are shown in black circles. The proteins meeting the log2 fold change (FC) cutoff and with a -log10 p value lower than 2 are shown as green crosses. Proteins not meeting the log2 FC cutoff and with a -log10 p value higher than 2 are shown as blue diamonds, respectively. The p value threshold is 0.01. A log FC cutoff of 0.45 was used to generate the plot
Four proteins were decreased in abundance in the HA cohort: cathepsin D, ceruloplasmin (CP), afamin (AFM), and complement factor C9 (C9). These proteins are associated with acute phase and inflammatory processes. Afamin has been linked to metabolic syndrome and type 2 diabetes [28, 30, 47]. Metabolic syndrome, defined as a cluster of cardiovascular risk factors associated with obesity and insulin resistance [8], is linked to excess ROS production and mitochondrial oxidoreductive dysfunction. The aging process is in part attributed to these pathologies. Lysosomal cathepsins have altered proteolytic activities in the aging brain. This is relevant to neurodegenerative diseases [56]. Cathepsin D is a quantitative marker of cellular senescence [11]. However, cathepsin D was also found to be critical for protection against alpha-synuclein aggregation and toxicity [41]. CP and C9 are acute phase reactants. The abundance changes of these four proteins in urine likely reflect systemic differences in abundance and support the notion that the HA cohort is less perturbed by low-grade inflammation compared with the NHA cohort. None of the proteins has been specifically brought into context with healthy aging [15, 71].
Nineteen proteins were increased in abundance in the HA cohort. Interpretation of data on partial or extensive biochemical characterization revealed protein associations with angiogenesis, vascular remodeling and repair, and signaling pathways and adhesion involving the ECM, endothelial and epithelial cells, and leukocytes. Two proteins altered in abundance in the NHA vs. HA groups, matrix remodeling–associated protein 8 (MXRA8) and ephrin type-B receptor 4 (EPHB4), contribute to maintaining the blood-brain barrier and neurovasculature [37, 74]. The prominent role of aldo-keto reductases including AKR1B1 in the detoxification of drugs and xenobiotics is relevant to the response to cellular oxidative stress, a hallmark of the aging process [3]. Nine proteins have been implicated in endothelial function, adhesion, and repair in the context of the vasculature (seven were increased in the HA cohort, Table 1). Vascular dysfunction is a prominent factor in chronic disease–associated non-healthy aging. Endothelial vascular function was defined as a surrogate of vascular risk and aging in women [14]. Atherosclerosis is a major peripheral vascular pathology and also strongly associated with cytokine-mediated chronic inflammation and senescence [67]. Impaired angiogenesis and induced endothelial cell senescence factor into major aging-associated diseases such as diabetes. This is mediated by phosphatidylinositol 3-kinase (PI3K)/AKT signal transduction and VGEF signaling pathways [6]. There were links between candidate biomarkers and the PI3K/Akt pathway. Cathepsin D induces proliferation of omental microvascular endothelial cells by activating the PI3K/pAKT pathway [40]. E-cadherin (like K-cadherin a cadherin superfamily member) slows tumor cell growth by suppressing the PI3K/Akt signaling [34]. Two angiopoietin-related proteins (Angptl1 and Angptl2) have antiapoptotic activities via the phosphatidylinositol 3-kinase/Akt pathway using a zebrafish model [31]. MXRA8 influences the activation of AKT and p38 MAP kinase via VGEF signaling in endothelial cells [23]. Data for proteins changed with statistical significance (p value < 0.05) are provided in Supplemental File S5, Table. Most conventional healthy aging biomarker discovery studies target specific molecules based on assay availability. This urinary proteome survey is untargeted and thus allowed identifying several barely characterized proteins as novel potential aging biomarkers.
Proinflammatory cytokines in blood plasma are only modestly changed in abundance comparing HA and NHA cohorts and display strikingly consistent person-specific patterns assessed at two timepoints
The low-grade proinflammatory phenotype SASP, human aging, and age-related chronic diseases are linked to perturbations in circulating cytokine levels (e.g., IL-6 and TNF-α) and acute phase proteins (e.g., CRP) [18, 38, 42]. We measured differences for 40 proinflammatory cytokines and chemokines. Nine cytokines and chemokines were modestly altered in abundance, all of them increased in the HA cohort (p value < 0.025). None were statistically significant adjusting for multiple testing (p value < 0.05). These proteins are displayed in the volcano plot of Fig. 4. The changes were more than 2-fold for IL-12p40 (the IL-12 subunit p40 or β), IL-12p70 (the dimer of the α (p35) and β subunits), IL-16, and MIP-1α. The pleiotropic cytokine IL-12 can have pro- and antiinflammatory activities within the cardiovascular system [2], but more is known about its proinflammatory functions influencing both innate and adaptive immunity [63]. IL-12 has been attributed a major role as an antiangiogenic factor [66], which is of interest given that angiogenesis and vascular function were highly represented terms in the context of HA vs. NHA urinary proteome surveys. We elaborate more on the role of IL-12 in the protein network analysis section. Both IL-16 and MIP-1α are chemoattractants, IL-16 for lymphocytes, monocytes, and eosinophils [16] and MIP-1α for diverse cell types. Chemoattractant properties are relevant to the pathology of atherosclerosis and vascular remodeling/repair. Macrophage-secreted protein MIP-1α was reported to contribute to atherosclerotic plaque formation [46] (PMID: 23288165). In line with the absence of multiple testing–corrected statistically significant changes, PCA did not result in separate clusters for the two cohorts (Fig. 5). Strikingly, Euclidian distance correlation analysis revealed that the timepoints for a given subject clustered pairwise in 47 of 60 cases and, together with a 3rd unrelated cytokine profile, in three cases (Supplemental File S6, Figure). Cytokine and chemokine concentrations in blood appear to be tightly controlled in a “normal” range. The baseline abundance range differs among individuals, regardless of good health or affliction with a chronic disease. We are unaware of other studies demonstrating that cytokine/chemokine abundance patterns are person-specific signatures. We observed that the cytokines with low p values strongly correlated with each other in abundance. Only IL-7 and IL-8 did not have R2 correlation values > 0.9. Master regulators for the expression of proinflammatory cytokines and their secretion into plasma may have strong roles in the observed high correlations (Supplemental File S7, Figure). In summary, this data corroborates the notion that the measurement of cytokine quantities requires a longitudinal approach to monitor a subject’s baseline cytokine expression level and its perturbations in the context of any inflammatory pathology. Cross-sectional cytokine biomarker discovery and validation studies are less likely to yield conclusive results.
Fig. 4.
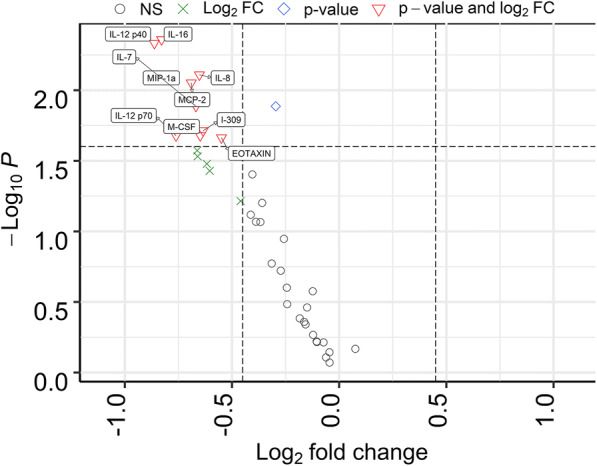
Differentially abundant cytokines/chemokines in volcano plot. The cytokines displaying quantitative differences with statistical significance are depicted in red triangles. Non-significant proteins are shown in black circles, and proteins not meeting either p value or log2 FC cutoffs are depicted as green crosses and blue diamonds, respectively. The p value threshold is 0.05. A log FC cutoff of 0.45 was used to generate the plot
Fig. 5.
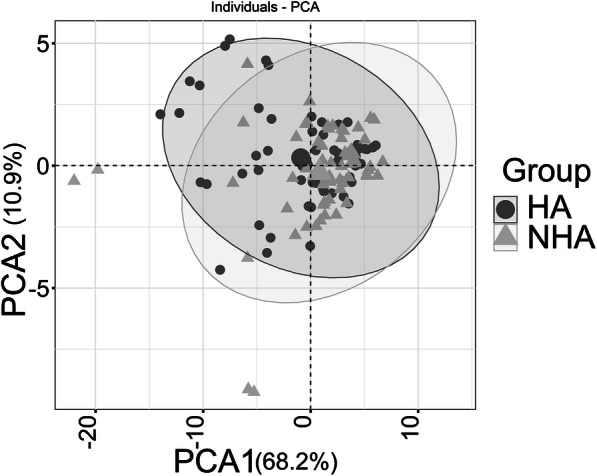
The PCA plot between the healthy and the non-healthy group using 40 cytokines and chemokines from blood plasma. The PCA plot includes all timepoints (two for most subjects). They are not averaged for a given subject. There is no separate cluster formation for the NHA (triangles) and HA (circles) datasets
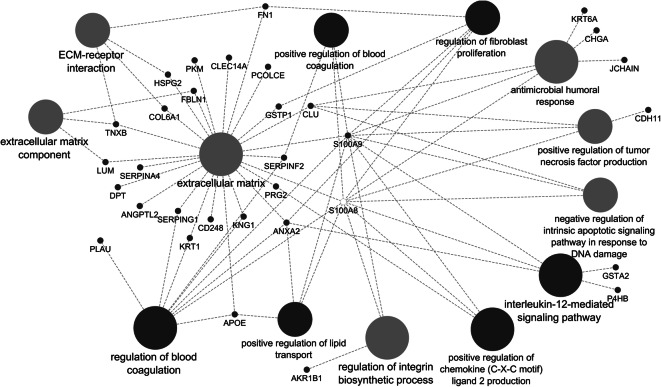
Protein network analysis applied to differentially abundant urinary proteins
Combined GO term and KEGG pathway enrichment analyses for 35 differentially abundant urinary proteins (HA vs. NHA; p value < 0.05) resulted in eight terms decreased in the HA vs. NHA dataset, most of which pointed towards lysosomal hydrolytic, proteolytic, and glucosaminoglycan-degrading functions, as shown in Supplemental File S9 (Figure). Twenty-five biologically diverse terms were increased in the HA dataset vs. the NHA dataset (Supplemental File S8, Dataset). As depicted in the respective protein network analysis of Fig. 6, among the highly connected terms were extracellular matrix (ECM), ECM-receptor interactions, and regulation of blood coagulation. Other terms were antimicrobial humoral immune response and the regulation of fibroblast proliferation and integrin biosynthesis. All of these terms are implicated in vascular remodeling/repair and inflammation, with cell adhesion as a major facilitating cellular process. Given the IL-12 subunit abundance differences in the cytokine dataset, it is interesting that the urinary protein network analysis identified the IL-12 signaling pathway as enriched. ANXA2, GSTA2, P4HB, S100A8, and S100A9 (p value < 0.05 comparing the HA and the NHA datasets) contribute to this pathway. IL-12 signaling activity reduces VGEF receptor 3 expression in the tumor vasculature [52]. IL-12p35 has a major role in regulating the inflammatory response during hypertension [72, 73]. IL-12 is also part of an antiangiogenic program mediated by IFN-γ-inducible genes, inhibits endothelial cell functions, and affects the lymphocyte-endothelial cell crosstalk [17, 58]. The fact that the abundance of IL-12 in plasma, along with proteins detected in urine that are able to modulate the vasculature and endothelial function (for instance, cathepsin D, ANGPTL2, MXRA8, cell surface glycoprotein MUC18, EPHB4, and SERPING1) are changed supports an association between healthy aging and balanced vascular function.
Fig. 6.
Protein network displaying selected enriched GO terms based on increased urinary protein abundances in the HA versus the NHA cohort. This data is included in tabular form in Supplemental File S8, Dataset
Conclusions
In a study comparing a cohort aging healthily with one afflicted by chronic diseases, we identified candidate protein biomarkers and their associated biological processes, such as cell adhesion, angiogenesis, vascular repair/remodeling, and IL-12 signaling. These outcomes set the stage for studies to validate their biomarker potential. Predictive biomarkers and those measuring responses to therapeutic intervention to chronic diseases are increasingly recognized as important for patient stratification, specifically in the context of cancer and neurodegenerative diseases [32, 57]. We propose that quantitatively validated biomarkers can predict healthy aging (i.e., low risk of chronic diseases), individually or as a panel, and allow development of blood plasma or urine clinical tests to monitor the health of the population above 50 years of age, not unlike current medical practice of colonoscopies (imaging data) to diagnose pre-neoplastic and colorectal cancer lesions. A panel of biomarkers may also have utility to assess the efficacy of investigational new drugs (INDs) as companion biomarkers in clinical trials with the purpose to ameliorate or reverse chronic diseases. Further molecular and physiological investigations of the candidate biomarkers and their pathways, especially with respect to an inflamed or otherwise malfunctional vasculature, may reveal new rationales for therapeutic intervention to chronic diseases.
Electronic supplementary material
Dataset: Relative abundances of Blood Plasma Cytokines and Chemokines from 124 Specimens (65 Human Subjects) for "Healthy Aging" and "Non-Healthy Aging" Cohorts (XLSX 74 kb)
Dataset: Urinary Proteome Datasets from 125 Specimens (65 Human Subjects) for "Healthy Aging" and "Non-Healthy Aging" Cohorts (XLSX 473 kb)
Dataset: Medical Histories for the 65 Study Participants and their Differentiation into "Healthy Aging" and "Non-Healthy Aging" Cohorts (XLSX 28 kb)
Figure: Pearson correlation analysis generates hierarchical clusters of urinary proteome datasets (PNG 1691 kb)
Table: Differentially Abundant Proteins Comparing Healthy Aging (HA) and Non-Healthy Aging (NHA) Cohorts Based on Urinary Proteome Analysis. (DOCX 33 kb)
Figure: Blood Plasma Cytokine Abundance Profiles from 65 Human Subjects Assigned to Healthy Aging and Non-Healthy Aging Cohorts Analyzed Clustered Using the Euclidian Clustering Algorithm. (PNG 2074 kb)
Figure: Correlation Analysis for Differentially Abundant Cytokines and Chemokines (Healthy Aging vs. Non-Healthy Aging Cohorts) (PNG 1695 kb)
Dataset: Pathway Enrichment Analysis Using GO and KEGG Terms Based on Statistically Significant Proteins in the Urinary Proteome Survey (p-value < 0.05) Comparing the Healthy Aging (HA) and Non-Healthy Aging (NHA) Cohorts. (XLSX 56 kb)
Figure: GO term and KEGG pathway enrichment with associated protein network based on proteins decreased in abundance in the HA versus the NHA cohort (HA vs. NHA; p-value < 0.05). (PNG 677 kb)
Acknowledgments
We thank Mr. Zachary Gortler for contributions to human subject recruitment and specimen collections, and Ms. Lauren DiLello for medical data collection and discussions on biomarker analysis strategies.
Authors’ contributions
YY, responsible for LC-MS/MS experiments and LC-MS/MS data analysis, participation in writing manuscript; HS, responsible for biostatistical data and network analyses, graphic art work, participation in writing manuscript; KK, responsible for cytokine array experiments and data analysis; TT, sample preparation to analyze urinary proteomes; JP, design, organization, and implementation of human subject recruitment and medical data collection; KEN, conceptualization of the study and manuscript review; RP, conceptualization and implementation of the study, biological data analysis, wrote manuscript.
Funding
This work was generously funded by the Ruggles Family Foundation, Moline, IL and Mr. and Mrs. Rudy Ruggles.
Data availability
All detailed datasets (medical data, quantitative cytokine and urinary proteome surveys) and methods used for analysis are included in the “Materials and methods” section and in the Supplemental Files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012477 (reviewer account details: username: reviewer44087@ebi.ac.uk; password: qAT4NwUo).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethics approval (include appropriate approvals or waivers)
Internal review boards (IRB) of Danbury Hospital and the J. Craig Venter Institute (JCVI), Rockville, MD approved a consent form and human subject protocol outlining risks and benefits of participation in 2013. JP and RP wrote the consent form and human subject protocol.
Consent to participate (include appropriate statements)
All human subjects consented to participate and enable specimen and medical data collections.
Consent for publication (include appropriate statements)
N/A
Code availability
For all analyses that involved coding, appropriate literature references are provided.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yanbao Yu and Harinder Singh contributed equally to this work.
References
- 1.Bakun M, Senatorski G, Rubel T, Lukasik A, Zielenkiewicz P, Dadlez M, Paczek L. Urine proteomes of healthy aging humans reveal extracellular matrix (ECM) alterations and immune system dysfunction. Age (Dordr) 2014;36(1):299–311. doi: 10.1007/s11357-013-9562-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasubbramanian D, Goodlett BL, Mitchell BM. Is IL-12 pro-inflammatory or anti-inflammatory? Depends on the blood pressure. Cardiovasc Res. 2019;115(6):998–999. doi: 10.1093/cvr/cvz028. [DOI] [PubMed] [Google Scholar]
- 3.Barski OA, Tipparaju SM, Bhatnagar A. The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev. 2008;40(4):553–624. doi: 10.1080/03602530802431439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bitar MS. Diabetes impairs angiogenesis and induces endothelial cell senescence by up-regulating thrombospondin-CD47-dependent signaling. Int J Mol Sci. 2019;20(3). 10.3390/ijms20030673. [DOI] [PMC free article] [PubMed]
- 7.Bolotin E, Smogorzewska M, Smith S, Widmer M, Weinberg K. Enhancement of thymopoiesis after bone marrow transplant by in vivo interleukin-7. Blood. 1996;88(5):1887–1894. doi: 10.1182/blood.V88.5.1887.1887. [DOI] [PubMed] [Google Scholar]
- 8.Bonomini F, Rodella LF, Rezzani R. Metabolic syndrome, aging and involvement of oxidative stress. Aging Dis. 2015;6(2):109–120. doi: 10.14336/AD.2014.0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bratic A, Larsson NG. The role of mitochondria in aging. J Clin Invest. 2013;123(3):951–957. doi: 10.1172/JCI64125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunk UT, Terman A. The mitochondrial-lysosomal axis theory of aging: accumulation of damaged mitochondria as a result of imperfect autophagocytosis. Eur J Biochem. 2002;269(8):1996–2002. doi: 10.1046/j.1432-1033.2002.02869.x. [DOI] [PubMed] [Google Scholar]
- 11.Byun HO, Han NK, Lee HJ, Kim KB, Ko YG, Yoon G, Lee YS, Hong SI, Lee JS. Cathepsin D and eukaryotic translation elongation factor 1 as promising markers of cellular senescence. Cancer Res. 2009;69(11):4638–4647. doi: 10.1158/0008-5472.CAN-08-4042. [DOI] [PubMed] [Google Scholar]
- 12.Carmona-Gutierrez D, Hughes AL, Madeo F, Ruckenstuhl C. The crucial impact of lysosomes in aging and longevity. Ageing Res Rev. 2016;32:2–12. doi: 10.1016/j.arr.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Fu LL, Wen X, Wang XY, Liu J, Cheng Y, Huang J. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014;5:e1047. doi: 10.1038/cddis.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins P, Maas A, Prasad M, Schierbeck L, Lerman A. Endothelial vascular function as a surrogate of vascular risk and aging in women. Mayo Clin Proc. 2020;95(3):541–553. doi: 10.1016/j.mayocp.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Crimmins E, Vasunilashorn S, Kim JK, Alley D. Biomarkers related to aging in human populations. Adv Clin Chem. 2008;46:161–216. doi: 10.1016/s0065-2423(08)00405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruikshank W, Little F. lnterleukin-16: the ins and outs of regulating T-cell activation. Crit Rev Immunol. 2008;28(6):467–483. doi: 10.1615/critrevimmunol.v28.i6.10. [DOI] [PubMed] [Google Scholar]
- 17.Del Vecchio M, Bajetta E, Canova S, Lotze MT, Wesa A, Parmiani G, et al. Interleukin-12: biological properties and clinical application. Clin Cancer Res. 2007;13(16):4677–4685. doi: 10.1158/1078-0432.CCR-07-0776. [DOI] [PubMed] [Google Scholar]
- 18.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol A Biol Sci Med Sci. 2014;69(Suppl 1):S4–S9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 19.Fulop T, Larbi A, Dupuis G, Le Page A, Frost EH, Cohen AA, et al. Immunosenescence and inflammaging as two sides of the same coin: friends or foes? Front Immunol. 2017;8:1960. doi: 10.3389/fimmu.2017.01960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gimbrone MA, Jr, Garcia-Cardena G. Endothelial cell dysfunction and the pathobiology of atherosclerosis. Circ Res. 2016;118(4):620–636. doi: 10.1161/CIRCRESAHA.115.306301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granger JP. Inflammatory cytokines, vascular function, and hypertension. Am J Physiol Regul Integr Comp Physiol. 2004;286(6):R989–R990. doi: 10.1152/ajpregu.00157.2004. [DOI] [PubMed] [Google Scholar]
- 22.Gruver AL, Hudson LL, Sempowski GD. Immunosenescence of ageing. J Pathol. 2007;211(2):144–156. doi: 10.1002/path.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han SW, Jung YK, Lee EJ, Park HR, Kim GW, Jeong JH, Han MS, Choi JY. DICAM inhibits angiogenesis via suppression of AKT and p38 MAP kinase signalling. Cardiovasc Res. 2013;98(1):73–82. doi: 10.1093/cvr/cvt019. [DOI] [PubMed] [Google Scholar]
- 24.Herman WH, Cohen RM. Racial and ethnic differences in the relationship between HbA1c and blood glucose: implications for the diagnosis of diabetes. J Clin Endocrinol Metab. 2012;97(4):1067–1072. doi: 10.1210/jc.2011-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. 2018;128(4):1238–1246. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janelidze S, Mattsson N, Palmqvist S, Smith R, Beach TG, Serrano GE, Chai X, Proctor NK, Eichenlaub U, Zetterberg H, Blennow K, Reiman EM, Stomrud E, Dage JL, Hansson O. Plasma P-tau181 in Alzheimer’s disease: relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nat Med. 2020;26(3):379–386. doi: 10.1038/s41591-020-0755-1. [DOI] [PubMed] [Google Scholar]
- 27.Kim HO, Kim HS, Youn JC, Shin EC, Park S. Serum cytokine profiles in healthy young and elderly population assessed using multiplexed bead-based immunoassays. J Transl Med. 2011;9:113. doi: 10.1186/1479-5876-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kollerits B, Lamina C, Huth C, Marques-Vidal P, Kiechl S, Seppala I, et al. Plasma concentrations of Afamin are associated with prevalent and incident type 2 diabetes: a pooled analysis in more than 20,000 individuals. Diabetes Care. 2017;40(10):1386–1393. doi: 10.2337/dc17-0201. [DOI] [PubMed] [Google Scholar]
- 29.Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5(7):e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kronenberg F, Kollerits B, Kiechl S, Lamina C, Kedenko L, Meisinger C, Willeit J, Huth C, Wietzorrek G, Altmann ME, Thorand B, Melmer A, Dähnhardt D, Santer P, Rathmann W, Paulweber B, Koenig W, Peters A, Adham IM, Dieplinger H. Plasma concentrations of afamin are associated with the prevalence and development of metabolic syndrome. Circ Cardiovasc Genet. 2014;7(6):822–829. doi: 10.1161/CIRCGENETICS.113.000654. [DOI] [PubMed] [Google Scholar]
- 31.Kubota Y, Oike Y, Satoh S, Tabata Y, Niikura Y, Morisada T, Akao M, Urano T, Ito Y, Miyamoto T, Nagai N, Koh GY, Watanabe S, Suda T. Cooperative interaction of angiopoietin-like proteins 1 and 2 in zebrafish vascular development. Proc Natl Acad Sci U S A. 2005;102(38):13502–13507. doi: 10.1073/pnas.0501902102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Thangue NB, Kerr DJ. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat Rev Clin Oncol. 2011;8(10):587–596. doi: 10.1038/nrclinonc.2011.121. [DOI] [PubMed] [Google Scholar]
- 33.Lara J, Cooper R, Nissan J, Ginty AT, Khaw KT, Deary IJ, Lord JM, Kuh D, Mathers JC. A proposed panel of biomarkers of healthy ageing. BMC Med. 2015;13:222. doi: 10.1186/s12916-015-0470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lau MT, Klausen C, Leung PC. E-cadherin inhibits tumor cell growth by suppressing PI3K/Akt signaling via beta-catenin-Egr1-mediated PTEN expression. Oncogene. 2011;30(24):2753–2766. doi: 10.1038/onc.2011.6. [DOI] [PubMed] [Google Scholar]
- 35.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: an HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278(5341):1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 36.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malik VA, Di Benedetto B. The blood-brain barrier and the EphR/Ephrin system: perspectives on a link between neurovascular and neuropsychiatric disorders. Front Mol Neurosci. 2018;11:127. doi: 10.3389/fnmol.2018.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minciullo PL, Catalano A, Mandraffino G, Casciaro M, Crucitti A, Maltese G, Morabito N, Lasco A, Gangemi S, Basile G. Inflammaging and anti-inflammaging: the role of cytokines in extreme longevity. Arch Immunol Ther Exp. 2016;64(2):111–126. doi: 10.1007/s00005-015-0377-3. [DOI] [PubMed] [Google Scholar]
- 39.Nagaraj N, Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10(2):637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 40.Pranjol MZI, Gutowski NJ, Hannemann M, Whatmore JL. Cathepsin D non-proteolytically induces proliferation and migration in human omental microvascular endothelial cells via activation of the ERK1/2 and PI3K/AKT pathways. Biochim Biophys Acta Mol Cell Res. 2018;1865(1):25–33. doi: 10.1016/j.bbamcr.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 41.Qiao L, Hamamichi S, Caldwell KA, Caldwell GA, Yacoubian TA, Wilson S, Xie ZL, Speake LD, Parks R, Crabtree D, Liang Q, Crimmins S, Schneider L, Uchiyama Y, Iwatsubo T, Zhou Y, Peng L, Lu YM, Standaert DG, Walls KC, Shacka JJ, Roth KA, Zhang J. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol Brain. 2008;1:17. doi: 10.1186/1756-6606-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rea IM, Gibson DS, McGilligan V, McNerlan SE, Alexander HD, Ross OA. Age and age-related diseases: role of inflammation triggers and cytokines. Front Immunol. 2018;9:586. doi: 10.3389/fimmu.2018.00586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, de Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38(10):1065–1070. doi: 10.1016/S0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 45.Salminen A, Kaarniranta K. Insulin/IGF-1 paradox of aging: regulation via AKT/IKK/NF-kappaB signaling. Cell Signal. 2010;22(4):573–577. doi: 10.1016/j.cellsig.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 46.de Jager SCA, Bot I, Kraaijeveld AO, Korporaal SJA, Bot M, van Santbrink PJ, et al. Leukocyte-specific CCL3 deficiency inhibits atherosclerotic lesion development by affecting neutrophil accumulation. Arterioscler Thromb Vasc Biol. 2013;33:e75–e83. [DOI] [PubMed]
- 47.Seeber B, Morandell E, Lunger F, Wildt L, Dieplinger H. Afamin serum concentrations are associated with insulin resistance and metabolic syndrome in polycystic ovary syndrome. Reprod Biol Endocrinol. 2014;12:88. doi: 10.1186/1477-7827-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen EZ, Song CQ, Lin Y, Zhang WH, Su PF, Liu WY, Zhang P, Xu J, Lin N, Zhan C, Wang X, Shyr Y, Cheng H, Dong MQ. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature. 2014;508(7494):128–132. doi: 10.1038/nature13012. [DOI] [PubMed] [Google Scholar]
- 50.Singh H, Torralba MG, Moncera KJ, DiLello L, Petrini J, Nelson KE, Pieper R. Gastro-intestinal and oral microbiome signatures associated with healthy aging. Geroscience. 2019;41(6):907–921. doi: 10.1007/s11357-019-00098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sood S, Gallagher IJ, Lunnon K, Rullman E, Keohane A, Crossland H, Phillips BE, Cederholm T, Jensen T, van Loon LJC, Lannfelt L, Kraus WE, Atherton PJ, Howard R, Gustafsson T, Hodges A, Timmons JA. A novel multi-tissue RNA diagnostic of healthy ageing relates to cognitive health status. Genome Biol. 2015;16:185. doi: 10.1186/s13059-015-0750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sorensen EW, Gerber SA, Frelinger JG, Lord EM. IL-12 suppresses vascular endothelial growth factor receptor 3 expression on tumor vessels by two distinct IFN-gamma-dependent mechanisms. J Immunol. 2010;184(4):1858–1866. doi: 10.4049/jimmunol.0903210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Starr ME, Saito M, Evers BM, Saito H. Age-associated increase in cytokine production during systemic inflammation-II: the role of IL-1beta in age-dependent IL-6 upregulation in adipose tissue. J Gerontol A Biol Sci Med Sci. 2015;70(12):1508–1515. doi: 10.1093/gerona/glu197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinmann GG. Changes in the human thymus during aging. Curr Top Pathol. 1986;75:43–88. doi: 10.1007/978-3-642-82480-7_2. [DOI] [PubMed] [Google Scholar]
- 55.Steinmann GG, Klaus B, Muller-Hermelink HK. The involution of the ageing human thymic epithelium is independent of puberty. A morphometric study. Scand J Immunol. 1985;22(5):563–575. doi: 10.1111/j.1365-3083.1985.tb01916.x. [DOI] [PubMed] [Google Scholar]
- 56.Stoka V, Turk V, Turk B. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res Rev. 2016;32:22–37. doi: 10.1016/j.arr.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 57.Strafella C, Caputo V, Galota MR, Zampatti S, Marella G, Mauriello S, Cascella R, Giardina E. Application of precision medicine in neurodegenerative diseases. Front Neurol. 2018;9:701. doi: 10.3389/fneur.2018.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strasly M, Cavallo F, Geuna M, Mitola S, Colombo MP, Forni G, Bussolino F. IL-12 inhibition of endothelial cell functions and angiogenesis depends on lymphocyte-endothelial cell cross-talk. J Immunol. 2001;166(6):3890–3899. doi: 10.4049/jimmunol.166.6.3890. [DOI] [PubMed] [Google Scholar]
- 59.Suh MJ, Tovchigrechko A, Thovarai V, Rolfe MA, Torralba MG, Wang J, Adkins JN, Webb-Robertson BJM, Osborne W, Cogen FR, Kaplowitz PB, Metz TO, Nelson KE, Madupu R, Pieper R. Quantitative differences in the urinary proteome of siblings discordant for type 1 diabetes include lysosomal enzymes. J Proteome Res. 2015;14(8):3123–3135. doi: 10.1021/acs.jproteome.5b00052. [DOI] [PubMed] [Google Scholar]
- 60.Teklu T, Kwon K, Wondale B, HaileMariam M, Zewude A, Medhin G, et al. Potential immunological biomarkers for detection of mycobacterium tuberculosis infection in a setting where M. tuberculosis is endemic, Ethiopia. Infect Immun. 2018;86(4). 10.1128/IAI.00759-17. [DOI] [PMC free article] [PubMed]
- 61.Terman A, Kurz T, Navratil M, Arriaga EA, Brunk UT. Mitochondrial turnover and aging of long-lived postmitotic cells: the mitochondrial-lysosomal axis theory of aging. Antioxid Redox Signal. 2010;12(4):503–535. doi: 10.1089/ars.2009.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410(6825):227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 63.Trinchieri G. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu Rev Immunol. 1995;13:251–276. doi: 10.1146/annurev.iy.13.040195.001343. [DOI] [PubMed] [Google Scholar]
- 64.Tsimikas S, Willerson JT, Ridker PM. C-reactive protein and other emerging blood biomarkers to optimize risk stratification of vulnerable patients. J Am Coll Cardiol. 2006;47(8 Suppl):C19–C31. doi: 10.1016/j.jacc.2005.10.066. [DOI] [PubMed] [Google Scholar]
- 65.Vacchelli E, Galluzzi L, Eggermont A, Galon J, Tartour E, Zitvogel L, Kroemer G. Trial watch: immunostimulatory cytokines. Oncoimmunology. 2012;1(4):493–506. doi: 10.4161/onci.20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Voest EE, Kenyon BM, O'Reilly MS, Truitt G, D'Amato RJ, Folkman J. Inhibition of angiogenesis in vivo by interleukin 12. J Natl Cancer Inst. 1995;87(8):581–586. doi: 10.1093/jnci/87.8.581. [DOI] [PubMed] [Google Scholar]
- 67.Wang JC, Bennett M. Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ Res. 2012;111(2):245–259. doi: 10.1161/CIRCRESAHA.111.261388. [DOI] [PubMed] [Google Scholar]
- 68.Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105(37):13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 70.Woods JA, Wilund KR, Martin SA, Kistler BM. Exercise, inflammation and aging. Aging Dis. 2012;3(1):130–140. [PMC free article] [PubMed] [Google Scholar]
- 71.Xia X, Chen W, McDermott J, Han JJ. Molecular and phenotypic biomarkers of aging. F1000Res. 2017;6:860. doi: 10.12688/f1000research.10692.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye J, Huang Y, Que B, Chang C, Liu W, Hu H, Liu L, Shi Y, Wang Y, Wang M, Zeng T, Zhen W, Xu Y, Shi L, Liu J, Jiang H, Ye D, Lin Y, Wan J, Ji Q. Interleukin-12p35 knock out aggravates doxorubicin-induced cardiac injury and dysfunction by aggravating the inflammatory response, oxidative stress, apoptosis and autophagy in mice. EBioMedicine. 2018;35:29–39. doi: 10.1016/j.ebiom.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ye J, Que B, Huang Y, Lin Y, Chen J, Liu L, Shi Y, Wang Y, Wang M, Zeng T, Wang Z, Hu H, Xu Y, Shi L, Ye D, Liu J, Jiang H, Wan J, Ji Q. Interleukin-12p35 knockout promotes macrophage differentiation, aggravates vascular dysfunction, and elevates blood pressure in angiotensin II-infused mice. Cardiovasc Res. 2019;115(6):1102–1113. doi: 10.1093/cvr/cvy263. [DOI] [PubMed] [Google Scholar]
- 74.Yonezawa T, Ohtsuka A, Yoshitaka T, Hirano S, Nomoto H, Yamamoto K, Ninomiya Y. Limitrin, a novel immunoglobulin superfamily protein localized to glia limitans formed by astrocyte endfeet. Glia. 2003;44(3):190–204. doi: 10.1002/glia.10279. [DOI] [PubMed] [Google Scholar]
- 75.Yu Y, Sikorski P, Smith M, Bowman-Gholston C, Cacciabeve N, Nelson KE, Pieper R. Comprehensive metaproteomic analyses of urine in the presence and absence of neutrophil-associated inflammation in the urinary tract. Theranostics. 2017;7(2):238–252. doi: 10.7150/thno.16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset: Relative abundances of Blood Plasma Cytokines and Chemokines from 124 Specimens (65 Human Subjects) for "Healthy Aging" and "Non-Healthy Aging" Cohorts (XLSX 74 kb)
Dataset: Urinary Proteome Datasets from 125 Specimens (65 Human Subjects) for "Healthy Aging" and "Non-Healthy Aging" Cohorts (XLSX 473 kb)
Dataset: Medical Histories for the 65 Study Participants and their Differentiation into "Healthy Aging" and "Non-Healthy Aging" Cohorts (XLSX 28 kb)
Figure: Pearson correlation analysis generates hierarchical clusters of urinary proteome datasets (PNG 1691 kb)
Table: Differentially Abundant Proteins Comparing Healthy Aging (HA) and Non-Healthy Aging (NHA) Cohorts Based on Urinary Proteome Analysis. (DOCX 33 kb)
Figure: Blood Plasma Cytokine Abundance Profiles from 65 Human Subjects Assigned to Healthy Aging and Non-Healthy Aging Cohorts Analyzed Clustered Using the Euclidian Clustering Algorithm. (PNG 2074 kb)
Figure: Correlation Analysis for Differentially Abundant Cytokines and Chemokines (Healthy Aging vs. Non-Healthy Aging Cohorts) (PNG 1695 kb)
Dataset: Pathway Enrichment Analysis Using GO and KEGG Terms Based on Statistically Significant Proteins in the Urinary Proteome Survey (p-value < 0.05) Comparing the Healthy Aging (HA) and Non-Healthy Aging (NHA) Cohorts. (XLSX 56 kb)
Figure: GO term and KEGG pathway enrichment with associated protein network based on proteins decreased in abundance in the HA versus the NHA cohort (HA vs. NHA; p-value < 0.05). (PNG 677 kb)
Data Availability Statement
All detailed datasets (medical data, quantitative cytokine and urinary proteome surveys) and methods used for analysis are included in the “Materials and methods” section and in the Supplemental Files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012477 (reviewer account details: username: reviewer44087@ebi.ac.uk; password: qAT4NwUo).