Abstract
The circadian rhythm has a strong influence on both cardiac physiology and disease in humans. Preclinical studies primarily using tissue specific transgenic mouse models have contributed to our understanding of the molecular mechanism of the circadian clock in the cardiovascular system. The core clock driven by CLOCK:BMAL1 complex functions as a universal timing machinery that primarily sets the pace in all mammalian cell types. In one specific cell or tissue type, core clock may control a secondary transcriptional oscillator, conceptualized as slave clock, which confers the oscillatory expression of tissue-specific effectors. Here, we discuss a core clock-slave clock-effectors network, which links the molecular clock to cardiac function.
Keywords: Circadian rhythm, cardiac function, BMAL1, KLF15, ion channel, NAD+, H2O2, AMPK, PI3K, AKT, PKA, Ca2+, calcineurin
Graphical abstract
1. Introduction
Circadian rhythm is a cell-autonomous timing system evolved to optimize biological processes in anticipation of recurring external cues [1,2]. In the cardiovascular system physiological parameters such as heart rate, blood pressure and endothelial function show diurnal rhythmicity [3,4]. Whilst disruption of circadian rhythm is associated with cardiovascular disease (CVD) such as myocardial infarction, strokes and sudden cardiac death [5**,6,7,8**,9,10*].
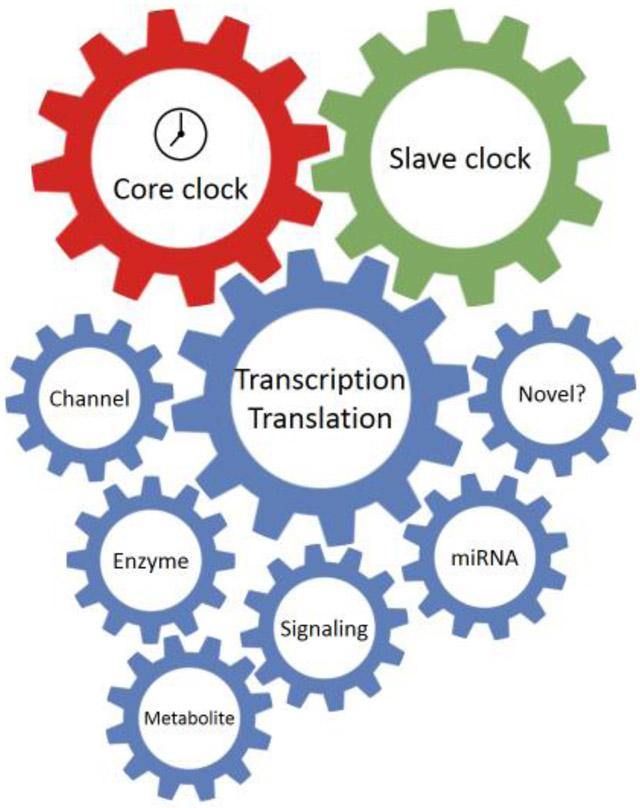
Here we will discuss a core clock-slave clock-effector network which governs the cardiac function (Figure 1). The primary molecular motor for circadian rhythm is the core clock. It consists of both transcriptional activators and repressors, which rhythmically express within an autoregulatory transcription-translation feedback circuit. The core clock presides over the slave clock, a secondary oscillator that governs the gene transcription of essential downstream output effectors and enables rhythmic tissue-specific functions. The core clock may also directly regulate certain effectors. Finally, dual regulation from the core clock and slave clock is also possible.
Figure 1. The core clock-slave clock-effectors network.
This schematic diagram illustrates the proposed model of the molecular mechanism of circadian regulation in the heart. The core clock generates the primary oscillation, which is either transduced by a tissue specific slave clock or directly to a variety of downstream effectors. The slave clock allows tissue specific circadian regulation as well as integration of additional environmental inputs, such as metabolic or disease status. The effectors ultimately affe ct cardiac physiology or pathophysiology.
In this review, we will first discuss the function and regulation of the core clock and the slave clock in the heart. Kruppel-like factor 15 (KLF15) will be highlighted as an essential slave clock in heart. In the latter part, we will focus on downstream oscillatory effectors which are critical in cardiac function and have shown significant cardiac phenotypes in animal studies when their circadian regulation is disrupted. We will discuss the circadian regulation of these effectors by core clock and slave clock as well as their impact on cardiac pathophysiology.
2. The core clock
2.1. The molecular core clock
The molecular circuit of the core clock is identical across all tissues and cell types. The CLOCK:BMAL1 complex activates Cry and Per gene expression. Accumulation of cytoplasmic CRY:PER complex leads to their nuclear translocation and switches off the CLOCK:BMAL1-driven gene transcription of Cry and Per [11,12]. Moreover, CLOCK:BMAL1 complex also drives the transcription of Nr1d1/2 and Nr1f1 which encodes nuclear receptors REV-ERBα/β and RORα. REV-ERB competes with RORα at the ROR binding elements (ROREs), which suppresses and actives the transcription of Bmal1, respectively [13,14,15].
The critical function of the peripheral core clock in the heart has been directly demonstrated by the significant cardiac phenotypes in mouse models where core clock function was genetically ablated in a tissue specific manner. Mice bearing cardiomyocyte-specific Clock mutant (CCM) [16,17] and Bmal1 knockout (CBK) [18,19,20] display significant disturbance of circadian rhythm in blood pressure and heart rate accompanied with abnormal cardiac electrophysiology and function. Additionally, PER2 regulates the rhythmicity of heart rate [21] and susceptibility to ischemia-reperfusion injury (I/R injury) in mice [22,23,24,25]. Further, REV-ERBα agonists and antagonists have been shown to provide cardioprotection against heart failure in mouse models and I/R injury in both mice and humans [26,27,28,29**].
2.2. Regulation of the core clock by post-translational modification
The core clock components undergo extensive and diverse post-translational modifications, including acetylation, phosphorylation, methylation, ubiquitination, SUMOylation, O-GlcNAcylation, ADP-ribosylation and S-nitrosylation [8**,30,31,32,33,34]. Particularly, acetylation, phosphorylation, ubiquitination and O-GlcNAcylation of core clock are known to modulate circadian rhythm in heart. Acetylation of BMAL1 directly regulates the CLOCK:BMAL1 activity. In fact, CLOCK itself is an acetyltransferase and acetylates its partner BMAL1 on K536 [35], which is required for gene transactivation [31]. In contrast, deacetylation of BMAL1 by sirtuin 1 (SIRT1) suppresses the core clock-activated transcription [31,36,37]. SIRT1 also deacetylates PER2 and promotes its degradation [38]. Either loss or overexpression of SIRT1 in heart can lead to cardiac dysfunction and heart failure [39,40]. Phosphorylation mediated by casein kinase 1 delta/epsilon (CK1δ/ε) promotes nuclear translocation and degradation of PER1/2 [12,41], which is critical for the clock period [8**,42*,43]. Mutation in CK1ε (tau mutant) that impairs the kinase activity reduces the amplitude of Bmal1 circadian expression and induces a phase shift for Per1 and Dbp in hamster heart [43]. In addition, Durgan et al. showed that Bmal1 is O-GlcNAcylated in mouse heart with the peak O-GlcNAcylation level in the middle of the active phase [44**]. O-GlcNAcylation of BMAL1/CLOCK inhibits their ubiquitination and promotes the transcriptional activity [45,46]. As the O-GlcNacylation modification is sensitive to glucose level, future study may investigate whether O-GlcNacylation can serve as a metabolic sensor to regulate circadian clock in heart [47*].
2.3. The core clock affects cardiac function through both transcription and translation
2.3.1. The core clock regulates circadian transcription
Approximately 10% of the gene transcription oscillates in the heart similar to other organs in the body [16,17,20,48,49]. In addition to acetylating BMAL1, CLOCK also functions as a histone acetyltransferase (HAT) for histone H3 [50]. BMAL1 enhances the HAT function and serves as pioneer factor for driving the circadian transcription due to its accessibility to closed chromatin [51,52]. A comprehensive interrogation of the cistrome profile uncovered the circadian rhythmic recruitments of BMAL1 and RNA polymerase II (RNAPII) that occur on a genome-wide scale [53,54]. This landmark finding suggests that global mobilization of BMAL1 and RNAPII is the foundation for circadian oscillation in gene transcription and cardiac function.
2.3.2. The core clock regulates circadian translation
The activity of protein synthesis also oscillates in a circadian fashion in the heart [55]. CBK mouse hearts have increased translation level [55]. Detailed biochemical study showed that BMAL1 serves as a negative regulator for AKT signaling in the heart [55] which activates the mammalian target of rapamycin (mTOR)/ribosomal protein S6/eukaryotic translation initiation factor 4E-binding protein 1 signaling axis, a classic mechanism that promotes protein synthesis [56]. Importantly, loss of BMAL1 persistently activates mTOR signaling leading to increased protein synthesis level as well as attenuated autophagy, which causes cardiac hypertrophy [55].
3. The slave clock in heart
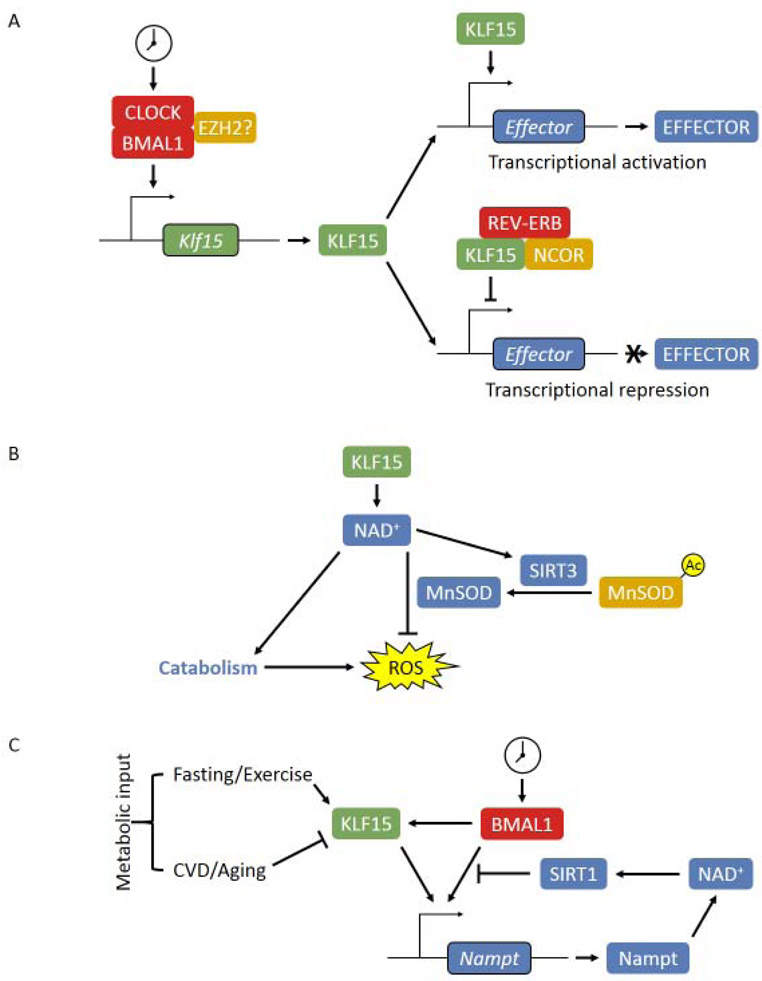
While each tissue has about 5-10% of the genes oscillating, the overlap between different tissues is exceedingly small, suggesting a local factor interfacing with the core clock and determines the tissue specificity of transcript oscillation [48,57]. The concept of “slave clocks” has long been proposed to explain this phenomenon, however, there is very few examples of well described slave clocks. Our work demonstrated that the Kruppel-like factor 15 (KLF15) is an essential slave clock in the heart (Figure 2A) [58]. KLF15 is a transcription factor that oscillates in a circadian fashion under the control of BMAL1 [59]. Additionally, a recent study identified enhancer of zeste homolog 2 (EZH2) as a novel suppressor for Klf15 transcription in the heart in the setting of ischemic heart failure [60**]. EZH2 may also modulates the circadian function of KLF15 as it directly associates with CLOCK:BMAL1 [61], the details of which, remain to be studied.
Figure 2. Cardiac slave clock KLF15.
(A) Circadian regulation of KLF15 and its slave clock function in both gene activation and repression. (B) KLF15 coordinates catabolism and ROS clearance in the active phase by regulation of myocardial NAD+ level. (C) The dual regulation of BMAL1 and KLF15 on cardiac NAD+ biosynthesis. Both BMAL1 and KLF15 activates Nampt transcription through direct binding on the promoter/enhancer. The increased NAD+ level activates SIRT1 activity, which deacetylates BMAL1 and histone H3 at the cis-acting site and suppresses the transactivation of BMAL1, forming a feedback inhibition loop. Additionally, KLF15 is subjected to the regulation of various metabolic inputs, which fine-tunes the circadian oscillatory expression of Nampt to achieve precise control of cardiac NAD+ level.
Circadian RNA-seq revealed that KLF15 governs 75% of oscillating genes in the heart without affecting the oscillation of the core clock, including direct regulation of catabolic genes in the active phase [58]. Additionally, KLF15 allows the recruitment of REV-ERB and nuclear receptor corepressor which suppresses hundreds of genes from aberrant oscillation [58]. KLF15 deficiency leads to cardiac lipid utilization defect and cardiomyopathy [58,62*]. Further, our recent work demonstrated that KLF15 also regulates MnSOD activity through acetylation on lysine 122 in a SIRT3-dependent fashion and provides circadian cardioprotection against oxidative stress induced by I/R injury [63**]. Thus, KLF15 coordinates the catabolic activity and clearance of the accompanied oxidative stresses in a circadian fashion, thereby exemplifying one of the proposed evolutionary benefit of circadian rhythm (Figure 2B). In addition to the core clock, KLF15 is also regulated by other physiological and pathological inputs such as diet and exercise, disease status and aging. For instance, Prosdocimo et al. showed that KLF15 expression in the mouse heart may be induced by fasting, and activates lipid metabolic genes and promotes myocardial lipid utilization [62*]. Thus in contrast to what’s implied by their name, “slave clocks” are not completely subordinate to the core clock, but integrate circadian rhythmic gene expression with local lineage specific information as well as metabolic signals (Figure 2C).
4. Effectors for circadian rhythm in cardiac function
Ultimately, the diverse rhythmic activity of the heart is carried out by various effectors, such as ion channels, metabolic enzymes, and signaling molecules. Here, we will review the major categories of effectors with implications in cardiac function (Table 1). The effectors that have exhibited circadian rhythm in in vivo studies will be discussed here in more details.
Table 1.
Effectors for circadian rhythm in cardiac function.
| Category | Effector | Circadian regulation on cardiac physiology & disease susceptibility |
|---|---|---|
| Ion channel | SCN5A, CACNA1C, KChIP2 | Heart rate and cardiac electrophysiology (repolarization); arrhythmia |
| Metabolic enzyme | NAMPT | NAD+ biosynthesis |
| DGCT2 | Triacylglycerol biosynthesis | |
| BDH1 | Ketone body catabolism | |
| Metabolite | NAD+ | Cardiac metabolism and SIRT1/3-dependent regulation; I/R injury |
| H2O2 | Redox cycling of Prx III and mitochondrial function | |
| Intracellular signaling | AMPK | Cardiac metabolism (NAMPT and hormone-sensitive lipase) |
| PI3K/AKT (mTOR & GSK3β) | Translation, autophagy, hormone (insulin) response and Cav1.2 expression; hypertrophy and I/R injury | |
| PKA | β-adrenergic signaling, SERCA2 activity and contractility | |
| CaN | β-adrenergic signaling and CaN signaling; I/R injury and heart failure | |
| MicroRNA | miR-21 | Glycolysis and cardiomyocyte survival; I/R injury |
| Novel mechanism | ? | Mitochondrial dynamics (mitophagy, fission and fusion), alternative splicing, long non-coding RNA and BMAL1-independent clock |
4.1. Ion channels
Ion channels are the molecular basis for cardiac electrophysiology [64]. Comparing to neurological tissues, cardiac ion channels have shorter half-lives (hours) and undergo more rapid turnover [65,66]. Therefore, by modulating the expression of cardiac ion channel genes (e.g. Scn5a, Kcnip2 and Cacna1c) the circadian clock readily regulates cardiac electrophysiology in a rhythmic fashion [19,59,67**,68]. For instance, BMAL1 and KLF15 directly regulates the rhythmic expression of Nav1.5 channel (SCN5A) [19,59] and Kv channel interacting protein 2 (KChIP2) [59], respectively. Studies from CCM, CBK and KLF15-null models indicate that loss of circadian clock can cause abnormal cardiac electrophysiology through effector channels and result in slowed heart rate, prolonged QRS intervals [19], reduced rhythmic amplitude of heart rate [16] and absence of oscillation in QT variation [59], which increases the risk for arrhythmia.
4.2. Metabolic enzymes
The heart demands continuous energy to support its contractile function [69,70]. Cardiac metabolism is tightly controlled by metabolic enzymes where dysregulation may lead to energy failure, metabolic remodeling and heart failure [71,72,73]. Early expression analysis of CBK mice revealed 19 putative direct BMAL1 target genes including three genes encoding essential metabolic enzymes in the heart [20], which are Nampt for nicotinamide adenine dinucleotide (NAD+) biosynthesis [63**,74,75,76**,77], Dgat2 for triacylglycerol biosynthesis [78,79,80,81*], and Bdh1 for ketone body catabolism [82].
Circadian rhythm in metabolic enzyme highly correlates with the metabolic status in the heart. For instance, nicotinamide phosphoribosyltransferase (NAMPT) level is increased prior to the sleep-to-wake transition in the heart in anticipation of the upcoming active phase when the demand of NAD+ will be elevated for fuel catabolism [20,63**,77,83]. Mechanistically, circadian transcription of Nampt is controlled by the dual regulation of BMAL1 (core clock) [36,37,84,85,86,87] and KLF15 (slave clock) [63**], in which KLF15 integrates external metabolic inputs [88,89**,90,91] and fine-tunes NAD+ biosynthesis besides the clock control (Figure 2C). KLF15 also transcriptionally activates numerous other effector enzymes in the fatty acid and multiple amino acid metabolic pathways [58]. Dysregulation of these effectors has been associated with I/R injury [63**,74,80] and heart failure [62*,74,75,76**,78,92].
4.3. Metabolites
In addition to providing building blocks and energy, metabolites serve important signaling functions and connects cardiac metabolism with contractile function and cell survival in the heart [93,94].
4.3.1. NAD+
The NAD+ level in the heart is higher than most of the other organs [95]. Besides interacting with over 500 enzymes, NAD+ is involved in almost every key biological process in the mammalian cells [96,97**]. Dysregulated NAD+ homeostasis has been associated with various CVD and NAD+-boosting is emerging as a novel strategy for treating and preventing CVD [97**, 98*,99,100**].
The oscillatory NAD+ level in heart is primarily governed by the circadian NAMPT levels [20,97**,77,83]. This is likely further driving the rhythmic activity of certain NAD+-dependent enzymes that have Km values within the physiological range of NAD+(300-700 μM) [97**,101], such as SIRT1 (94-888 μM) [102,103,104,105] and SIRT3 (280-880 μM) [106,107]. Indeed, we found that KLF15 regulates circadian susceptibility to I/R injury through the KLF15-Nampt-NAD+-SIRT3-MnSOD axis [63**]. Further, we found administration of an NAD+ precursor, nicotinamide mononucleotide (NMN) provided cardioprotection against I/R injury when the NAD+ level was at nadir but not when the NAD+ level was at peak, supporting the use of chronotherapy in NAD+-boosting strategies [63**].
4.3.2. H2O2
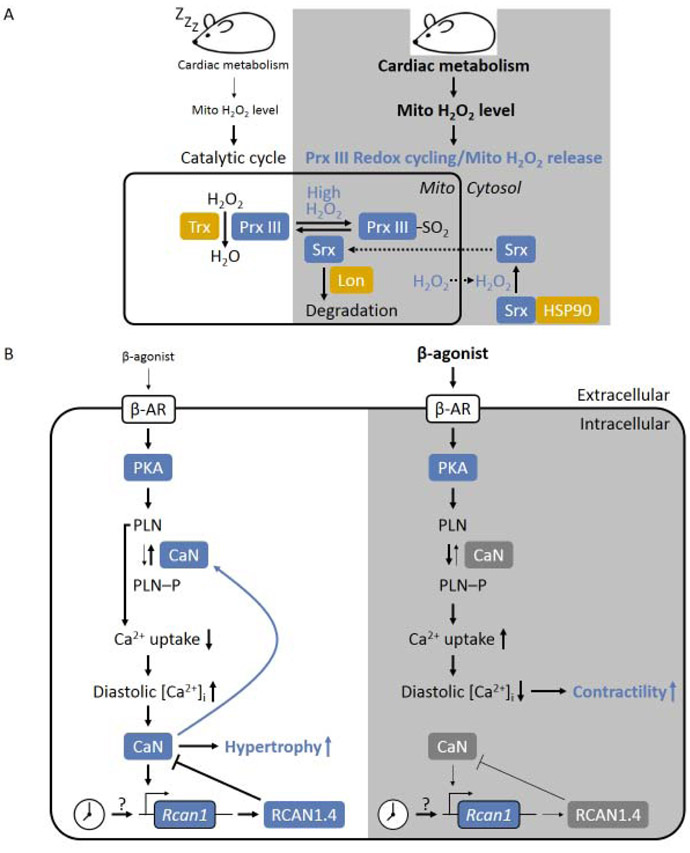
In mammalian cells, most of the H2O2 is produced in the mitochondria [108]. Excessive production of H2O2 can be detrimental to cardiac function [109**] which underlies a number of CVD [110,111,112,113]. Peroxiredoxin III (Prx III) is the major H2O2-eliminating enzyme in the mitochondria where it reduces H2O2 into H2O in the presence of thioredoxin (Trx) through the catalytic cycle [114,115,116,117]. However, when H2O2 production rate is boosted due to increased energy demand during the active phase, Prx III can undergo hyperoxidization cycle, through which it turns into its inactive form (PrxIII-SO2) [114,117]. A redox cycling between Prx III and PrxIII-SO2 is then established based on H2O2 release from the mitochondria and mitochondrial translocation of sulfiredoxin (Srx) that reactivates Prx III (Figure 3A) [117,118]. This mechanism was found to exhibit a circadian rhythm in many types of cells/tissues including the heart [118,119,120,121,122,123]. H2O2 could potentially damage the mitochondrial [124] and the cardiac function [109**], however, a direct investigation of mitochondrial function correlating with oscillating H2O2 level in a circadian fashion has not be performed.
Figure 3. Effectors of cardiac circadian clock.
(A) Circadian H2O2 release and Prx III redox cycling. During the resting phase, mitochondrial H2O2 level is low, Prx III is able to scavenge all the mitochondrial H2O2 through catalytic cycle. While in the active phase, the H2O2 production is increased due to increased cardiac metabolism, which lead to hyperoxidization of Prx III and the formation of the inactive Prx III-SO2. Excessive H2O2 released into cytosol oxidizes Srx leading to the formation of the Srx:HSP30 complex, which is required to reactivate Prx III.
(B) Circadian regulation of PKA/β-adrenergic and CaN signaling, resulting in low diastolic intracellular Ca2+ concentration ([Ca2+]i) in the active phase which favors increased contractility, and high diastolic [Ca2+]I in the resting phase which favors hypertrophy.
4.4. Intracellular signaling pathways
4.4.1. AMPK signaling
AMPK is a metabolic sensor that integrates the energy status with circadian regulation on key metabolic pathways [125,126]. For example, AMPK regulates the rhythmic activity of NAMPT [77] and hormone-sensitive lipase [127] which affects the NAD+ metabolism and the triglyceride metabolism in heart, respectively. The core clock regulation of AMPK is evidenced by the CCM model where oscillation in AMPK activity is lost [127]. Notably, CCM only disrupts the rhythm without affecting the baseline expression [127]. On the other hand, AMPK can also influence the core clock activity by phosphorylation on core clock suppressor CRY1/2 [128] and PER2 [129] which promotes their degradation.
4.4.2. PI3K/AKT (mTOR and GSK3β) signaling
PI3K/AKT is involved in diverse biological processes such as cell proliferation, survival and migration [130,131]. PI3K/AKT signaling activates mTOR but inhibits GSK3β [131,132]. The core clock controls the circadian rhythm in PI3K/AKT signaling as well as mTOR and GSK3β. Expression analysis of CBK mice revealed that BMAL1 directly regulates Pik3r1 which encodes p85α, the regulatory subunit of PI3K [20]. Consistently, CCM or CBK heart exhibits disrupted circadian rhythm in p85α expression and also the phosphorylation states of AKT, mTOR and GSK3β [55,133]. The importance of circadian PI3K/AKT signaling in cardiac function has been demonstrated in vivo. McGinnis et al. showed that loss of circadian PI3K signaling in CBK mouse disrupted circadian mTOR signaling, which led to increased protein synthesis rate, suppressed autophagy and cardiac hypertrophy [55]. Additionally, PI3K/AKT signaling has been shown to contribute to the oscillatory physiological response to insulin [55] and the circadian rhythmic expression of Cav1.2 channel (CACNA1C) [68].
4.4.3. PKA signaling
PKA signaling transduces β-adrenergic signaling and phosphorylates several proteins that are essential for Ca2+ transient such as L-type calcium channel (Cav1.2 [134] and β2a subunits [135]), ryanodine receptor 2 (RYR2) [136], and phospholamban (PLN) [137], which ultimately increases the contractile function in heart (inotropic) (Figure 3B) [138,139,140,141]. One study found modest transcriptional oscillation of Prkar1a, a gene encodes the type 1a regulatory subunit of PKA [142], which was disrupted in the CCM heart [16]. Both of the Prkar1a transcriptional level [16] and the circulating β-agonist level exhibit in-phase rhythm with peak during the active phase [143]. Therefore, although lacking direct experimental data, it is likely that the actual activity of the PKA signaling in the heart also oscillates in a similar pattern, which may contribute to circadian Ca2+ transient, sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 2 (SECRCA2) activity and contractility in heart [8**,144,145].
4.4.4. Ca2+ and calcineurin signaling
The calcium-activated protein phosphatase or CaN links the Ca2+ signaling and protein phosphorylation, which is particularly important for excitatory cells such as the cardiomyocytes [146]. CaN also regulates mitochondrial dynamics [146,147,148,149,150], which is associated with I/R injury [151**], cardiomyocyte hypertrophy [152] and heart failure [147]. Activity of CaN exhibits circadian rhythm in the heart [144]. The oscillatory transcription of an important CaN-targeted gene, Rcan1 (regulator of CaN 1) is diminished in the CCM heart, suggesting that circadian CaN activity requires the core clock [16]. Circadian rhythm in CaN is influenced by two interconnected mechanisms (Figure 3B) [144,153]. First, CaN responses to cytosolic Ca2+ level change [146] which is influenced by the aforementioned circadian β-adrenergic/PKA signaling cascade [144]. Second, the expression of RCAN1 isoform 4 (RCAN1.4 with exon 4 being the first exon [154,155]), a CaN inhibitor [156,157], is circadian in the heart [144]. The rhythmic RCAN1.4 level is partially regulated by CaN-triggered nuclear translocation of nuclear factor of activated T cells (NFAT) [144,158,159]. However, CaN-independent mechanism also exists as inhibition of CaN activity only reduces the peak transcription of Rcan1.4 but does not abolish its rhythmicity in the heart [144].
Circadian CaN activity modulates the functional consequence of β-agonist/PKA signaling in a daily fashion (Figure 3B). CaN activity reaches trough at the sleep-to-wake transition so that key modulators for β-adrenergic signaling such as PLN [160,161] could maintain in the phosphorylated state (inactive). This helps maximize the stimulation of contractile function by β adrenergic signaling and helps anticipate the elevation of cardiac workload in the upcoming active phase [144]. Indeed, epinephrine induces a greater increase in cardiac power ex vivo during the active phase than inactive phase [16]. On the other hand, isoproterenol induces a more pronounced hypertrophic growth at the wake-to-sleep transition, [162] likely due to high CaN activity that promotes cardiac hypertrophy [163]. Moreover, high CaN activity increases dephosphorylation of PLN and impairs Ca2+ uptake by SERCA2 which elevates diastolic Ca2+ level [140,164,165,166] and may further enhance the CaN-induced hypertrophy.
4.5. MicroRNAs
Several oscillatory miRNAs have been implicated for cardiac function, however, the molecular mechanism remains sparse [167,168]. Recently, Bartman et al. demonstrated that miR-21 is a downstream target of PER2 that exhibits rhythmic expression in the heart. They showed that miR-21 plays a crucial role in glycolysis, metabolic adaptation and cardioprotection in response to I/R injury [22,169]. Intense light exposure induces PER2 expression [170,171] and upregulates miR-21 level in mouse heart and human plasma [169], suggesting a potential therapeutic benefit for intense light therapy through this pathway [167]. Future study is required to elucidate the detailed mechanisms of circadian miRNAs in various diseases settings.
5. Novel mechanisms
Recent evidence suggests that circadian rhythm also exist in mitochondrial dynamics (mitophagy, fission and fusion) [8**,172,173,174,175], alternative splicing [176,177**] and long non-coding RNA [178] in the heart. In addition, BMAL1-independent clock that has been found in other types of cells/tissues [119,120,121,122,179**,180,181,182,183**] may also play a role in the heart. Future studies exploring these novel mechanisms will elucidate exciting new avenues in the circadian regulation of the cardiac function.
6. Conclusion
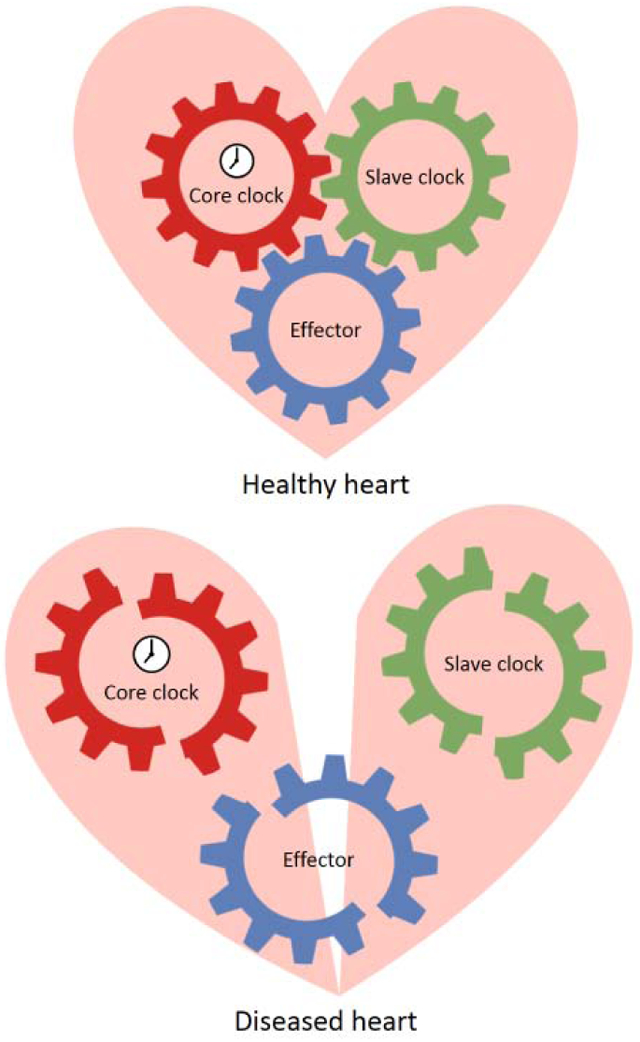
In the modern society exposures to artificial lighting and temperature control disrupt our intrinsic circadian rhythm. Approximately 15%-20% of industrial workers are shift workers who have increased risks for hypertension and coronary artery disease [184,185,186,187,188,189]. The contribution of circadian disruption to CVD inevitably increases with continued modernization. Thus, understanding the molecular mechanisms and designing novel therapeutic strategies targeting the core clock or the slave clocks as well as chronotherapies targeting the effectors are in urgent need for the treatment and prevention of CVD.
The core clock-slave clock-effectors forms an expansive and intricate system to optimize the physiological function of the heart. We have just begun to understand this sophisticated system. The discovery of KLF15 as an essential slave clock that coordinate s intrinsic core clock and metabolic output has open up a new dimension in the cardiac circadian rhythm research. Although KLF15 regulates about 75% of the oscillatory gene expression in heart, it still remains an open question whether there’s other slave clock in the heart. In addition, it is not well understood how external metabolic signals regulate KLF15. Future study may also explore the roles of novel effectors in regulating circadian rhythm in heart (Table 5), particularly through non-transcriptional mechanisms, such as microRNA, long non-coding RNA and alternative splicing, all of which can potentially lead to broad regulation of gene program and cellular function.
Highlights.
A network of core clock-slave clock-effectors links the molecular clock to cardiac function
The core clock machinery BMAL1:CLOCK regulates circadian rhythm through both transcription and translation in the heart
KLF15 is an essential slave clock in the heart that coordinates cardiac catabolism and clearance of the accompanied oxidative stresses in a circadian fashion
Circadian rhythm in cardiac function is carried out by various circadian effectors including ion channel, metabolic enzyme, metabolite, intracellular signaling, microRNA as well as other novel mechanisms.
Disruption of intrinsic circadian rhythm is highly associated with cardiovascular disease
Acknowledgements
This work was supported by National Institutes of Health Grants K08HL123551, R01HL143067 (L.Z.)and R01HL119195 (M.K.J.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
** of special interest
** of outstanding interest
- 1.Gerhart-Hines Z, Lazar MA: Circadian metabolism in the light of evolution. Endocr Rev 2015, 36:289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhadra U, Thakkar N, Das P, Pal Bhadra M: Evolution of circadian rhythms: from bacteria to human. Sleep Med 2017, 35:49–61. [DOI] [PubMed] [Google Scholar]
- 3.Kollias GE, Stamatelopoulos KS, Papaioannou TG, Zakopoulos NA, Alevizaki M, Alexopoulos GP, Kontoyannis DA, Karga H, Koroboki E, Lekakis JP, et al. : Diurnal variation of endothelial function and arterial stiffness in hypertension. J Hum Hypertens 2009, 23:597–604. [DOI] [PubMed] [Google Scholar]
- 4.Degaute JP, van de Borne P, Linkowski P, Van Cauter E: Quantitative analysis of the 24-hour blood pressure and heart rate patterns in young men. Hypertension 1991, 18:199–210. [DOI] [PubMed] [Google Scholar]
- 5.Crnko S, Du Pre BC, Sluijter JPG, Van Laake LW: Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat Rev Cardiol 2019, 16:437–447.** This is an excellent review on circadian rhythm in cardiac biology and diease.
- 6.Abbott SM, Malkani RG, Zee PC: Circadian disruption and human health: A bidirectional relationship. Eur J Neurosci 2020, 51:567–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jain M, Zhang L, Sabeh M: Circadian rhythm and cardiovascular disorders. ChronoPhysiology and Therapy 2014, 2014:27. [Google Scholar]
- 8.Zhang J, Chatham JC, Young ME: Circadian Regulation of Cardiac Physiology: Rhythms That Keep the Heart Beating. Annu Rev Physiol 2020, 82:79–101.** This is an excellent review on circadian regulation of cardiac physiology.
- 9.Roenneberg T, Merrow M: The Circadian Clock and Human Health. Curr Biol 2016, 26:R432–443. [DOI] [PubMed] [Google Scholar]
- 10.Walker WH, 2nd, Walton JC, DeVries AC, Nelson RJ: Circadian rhythm disruption and mental health. Transl Psychiatry 2020, 10:28.* This is an excellent review on circadian rhythm and mental health.
- 11.Buhr ED, Takahashi JS: Molecular components of the Mammalian circadian clock. Handb Exp Pharmacol 2013:3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM: Posttranslational mechanisms regulate the mammalian circadian clock. Cell 2001, 107:855–867. [DOI] [PubMed] [Google Scholar]
- 13.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U: The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 2002, 110:251–260. [DOI] [PubMed] [Google Scholar]
- 14.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB: A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 2004, 43:527–537. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, Armour SM, Remsberg JR, Jager J, Soccio RE, et al. : GENE REGULATION. Discrete functions of nuclear receptor Rev-erbalpha couple metabolism to the clock. Science 2015, 348:1488–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bray MS, Shaw CA, Moore MW, Garcia RA, Zanquetta MM, Durgan DJ, Jeong WJ, Tsai JY, Bugger H, Zhang D, et al. : Disruption of the circadian clock within the cardiomyocyte influences myocardial contractile function, metabolism, and gene expression. Am J Physiol Heart Circ Physiol 2008, 294:H1036–1047. [DOI] [PubMed] [Google Scholar]
- 17.Podobed P, Pyle WG, Ackloo S, Alibhai FJ, Tsimakouridze EV, Ratcliffe WF, Mackay A, Simpson J, Wright DC, Kirby GM, et al. : The day/night proteome in the murine heart. Am J Physiol Regul Integr Comp Physiol 2014, 307:R121–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis AM, Cheng Y, Kapoor S, Reilly D, Price TS, Fitzgerald GA: Circadian variation of blood pressure and the vascular response to asynchronous stress. Proc Natl Acad Sci U S A 2007, 104:3450–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroder EA, Lefta M, Zhang X, Bartos DC, Feng HZ, Zhao Y, Patwardhan A, Jin JP, Esser KA, Delisle BP: The cardiomyocyte molecular clock, regulation of Scn5a, and arrhythmia susceptibility. Am J Physiol Cell Physiol 2013, 304:C954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young ME, Brewer RA, Peliciari-Garcia RA, Collins HE, He L, Birky TL, Peden BW, Thompson EG, Ammons BJ, Bray MS, et al. : Cardiomyocyte-specific BMAL1 plays critical roles in metabolism, signaling, and maintenance of contractile function of the heart. J Biol Rhythms 2014, 29:257–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vukolic A, Antic V, Van Vliet BN, Yang Z, Albrecht U, Montani JP: Role of mutation of the circadian clock gene Per2 in cardiovascular circadian rhythms. Am J Physiol Regul Integr Comp Physiol 2010, 298:R627–634. [DOI] [PubMed] [Google Scholar]
- 22.Eckle T, Hartmann K, Bonney S, Reithel S, Mittelbronn M, Walker LA, Lowes BD, Han J, Borchers CH, Buttrick PM, et al. : Adora2b-elicited Per2 stabilization promotes a HIF-dependent metabolic switch crucial for myocardial adaptation to ischemia. Nat Med 2012, 18:774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonney S, Kominsky D, Brodsky K, Eltzschig H, Walker L, Eckle T: Cardiac Per2 functions as novel link between fatty acid metabolism and myocardial inflammation during ischemia and reperfusion injury of the heart. PLoS One 2013, 8:e71493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virag JA, Anderson EJ, Kent SD, Blanton HD, Johnson TL, Moukdar F, DeAntonio JH, Thayne K, Ding JM, Lust RM: Cardioprotection via preserved mitochondrial structure and function in the mPer2-mutant mouse myocardium. Am J Physiol Heart Circ Physiol 2013, 305:H477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun YY, Bai WW, Wang B, Lu XT, Xing YF, Cheng W, Liu XQ, Zhao YX: Period 2 is essential to maintain early endothelial progenitor cell function in vitro and angiogenesis after myocardial infarction in mice. J Cell Mol Med 2014, 18:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solt LA, Wang Y, Banerjee S, Hughes T, Kojetin DJ, Lundasen T, Shin Y, Liu J, Cameron MD, Noel R, et al. : Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012, 485:62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Zhang R, Tien CL, Chan RE, Sugi K, Fu C, Griffin AC, Shen Y, Burris TP, Liao X, et al. : REV-ERBalpha ameliorates heart failure through transcription repression. JCI Insight 2017, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reitz CJ, Alibhai FJ, Khatua TN, Rasouli M, Bridle BW, Burris TP, Martino TA: SR9009 administered for one day after myocardial ischemia-reperfusion prevents heart failure in mice by targeting the cardiac inflammasome. Commun Biol 2019, 2:353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Montaigne D, Marechal X, Modine T, Coisne A, Mouton S, Fayad G, Ninni S, Klein C, Ortmans S, Seunes C, et al. : Daytime variation of perioperative myocardial injury in cardiac surgery and its prevention by Rev-Erbalpha antagonism: a single-centre propensity-matched cohort study and a randomised study. Lancet 2018, 391:59–69.** A comprehensive randomised clinical study combined with transcriptomic and ex-vivo analysis of human myocardium samples. The results showed circadian hypoxia-reoxygenation tolerance and highest gene expression of Nr1d1 in the morning. Genetic ablation of Rev-Erb alpha gene or phamacological inhibition offered cardioprotection at sleep-to-wake transition in vivo.
- 30.Gallego M, Virshup DM: Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 2007, 8:139–148. [DOI] [PubMed] [Google Scholar]
- 31.Hirano A, Fu YH, Ptacek LJ: The intricate dance of post-translational modifications in the rhythm of life. Nat Struct Mol Biol 2016, 23:1053–1060. [DOI] [PubMed] [Google Scholar]
- 32.Tamaru T, Takamatsu K: Circadian modification network of a core clock driver BMAL1 to harmonize physiology from brain to peripheral tissues. Neurochem Int 2018, 119:11–16. [DOI] [PubMed] [Google Scholar]
- 33.Papazyan R, Zhang Y, Lazar MA: Genetic and epigenomic mechanisms of mammalian circadian transcription. Nat Struct Mol Biol 2016, 23:1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stojkovic K, Wing SS, Cermakian N: A central role for ubiquitination within a circadian clock protein modification code. Front Mol Neurosci 2014, 7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirayama J, Sahar S, Grimaldi B, Tamaru T, Takamatsu K, Nakahata Y, Sassone-Corsi P: CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature 2007, 450:1086–1090. [DOI] [PubMed] [Google Scholar]
- 36.Bellet MM, Sassone-Corsi P: Mammalian circadian clock and metabolism - the epigenetic link. J Cell Sci 2010, 123:3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P: Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol 2009, 41:81–86. [DOI] [PubMed] [Google Scholar]
- 38.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U: SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 2008, 134:317–328. [DOI] [PubMed] [Google Scholar]
- 39.Sanz MN, Grimbert L, Moulin M, Gressette M, Rucker-Martin C, Lemaire C, Mericskay M, Veksler V, Ventura-Clapier R, Garnier A, et al. : Inducible Cardiac-Specific Deletion of Sirt1 in Male Mice Reveals Progressive Cardiac Dysfunction and Sensitization of the Heart to Pressure Overload. Int J Mol Sci 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcendor RR, Gao S, Zhai P, Zablocki D, Holle E, Yu X, Tian B, Wagner T, Vatner SF, Sadoshima J: Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res 2007, 100:1512–1521. [DOI] [PubMed] [Google Scholar]
- 41.Etchegaray JP, Machida KK, Noton E, Constance CM, Dallmann R, Di Napoli MN, DeBruyne JP, Lambert CM, Yu EA, Reppert SM, et al. : Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol Cell Biol 2009, 29:3853–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narasimamurthy R, Hunt SR, Lu Y, Fustin JM, Okamura H, Partch CL, Forger DB, Kim JK, Virshup DM: CK1delta/epsilon protein kinase primes the PER2 circadian phosphoswitch. Proc Natl Acad Sci USA 2018, 115:5986–5991.* This study unraveled the regulation of CK1δ/ε on the essential core clock element PER2.
- 43.Dey J, Carr AJ, Cagampang FR, Semikhodskii AS, Loudon AS, Hastings MH, Maywood ES: The tau mutation in the Syrian hamster differentially reprograms the circadian clock in the SCN and peripheral tissues. J Biol Rhythms 2005, 20:99–110. [DOI] [PubMed] [Google Scholar]
- 44.Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, et al. : O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 2011, 286:44606–44619.** This is the first publication demonstrated the direct link between O-GlcNAcylation and circadian clock in heart.
- 45.Li MD, Ruan HB, Hughes ME, Lee JS, Singh JP, Jones SP, Nitabach MN, Yang X: O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab 2013, 17:303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma YT, Luo H, Guan WJ, Zhang H, Chen C, Wang Z, Li JD: O-GlcNAcylation of BMAL1 regulates circadian rhythms in NIH3T3 fibroblasts. Biochem Biophys Res Commun 2013, 431:382–387. [DOI] [PubMed] [Google Scholar]
- 47.Kaasik K, Kivimae S, Allen JJ, Chalkley RJ, Huang Y, Baer K, Kissel H, Burlingame AL, Shokat KM, Ptacek LJ, et al. : Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab 2013, 17:291–302.* This study demonstrates the regulation of O-GlcNAc transferase-mediated O-GlcNacylation on GSKb and PER2 activity. Importantly, O-GlcNAcylation of PER2 is partially mediated by glucose level, indicating that O-GlcNAcylation serves as a metabolic sensor for clock regulation and works coordinately with phosphorylation to fine-tune circadian clock.
- 48.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB: Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 2002, 109:307–320. [DOI] [PubMed] [Google Scholar]
- 49.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ: Extensive and divergent circadian gene expression in liver and heart. Nature 2002, 417:78–83. [DOI] [PubMed] [Google Scholar]
- 50.Doi M, Hirayama J, Sassone-Corsi P: Circadian regulator CLOCK is a histone acetyltransferase. Cell 2006, 125:497–508. [DOI] [PubMed] [Google Scholar]
- 51.Menet JS, Pescatore S, Rosbash M: CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev 2014, 28:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O'Malley BW: Coactivator-Dependent Oscillation of Chromatin Accessibility Dictates Circadian Gene Amplitude via REV-ERB Loading. Mol Cell 2015, 60:769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS: Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338:349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Partch CL, Green CB, Takahashi JS: Molecular architecture of the mammalian circadian clock. Trends Cell Biol 2014, 24:90–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGinnis GR, Tang Y, Brewer RA, Brahma MK, Stanley HL, Shanmugam G, Rajasekaran NS, Rowe GC, Frank SJ, Wende AR, et al. : Genetic disruption of the cardiomyocyte circadian clock differentially influences insulin-mediated processes in the heart. J Mol Cell Cardiol 2017, 110:80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma XM, Blenis J: Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009, 10:307–318. [DOI] [PubMed] [Google Scholar]
- 57.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB: A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A 2014, 111:16219–16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang L, Prosdocimo DA, Bai X, Fu C, Zhang R, Campbell F, Liao X, Coller J, Jain MK: KLF15 Establishes the Landscape of Diurnal Expression in the Heart. Cell Rep 2015, 13:2368–2375. [DOI] [PubMed] [Google Scholar]
- 59.Jeyaraj D, Haldar SM, Wan X, McCauley MD, Ripperger JA, Hu K, Lu Y, Eapen BL, Sharma N, Ficker E, et al. : Circadian rhythms govern cardiac repolarization and arrhythmogenesis. Nature 2012, 483:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pepin ME, Ha CM, Crossman DK, Litovsky SH, Varambally S, Barchue JP, Pamboukian SV, Diakos NA, Drakos SG, Pogwizd SM, et al. : Genome-wide DNA methylation encodes cardiac transcriptional reprogramming in human ischemic heart failure. Lab Invest 2019, 99:371–386.** A study using ischemic left ventricular tissue demonstrated a genome-wide alteration in DNA methylation that is associated with ischemic heart failure. Remarkably, EZH2 and KLF15 were identified to be the essential master regulators.
- 61.Etchegaray JP, Yang X, DeBruyne JP, Peters AH, Weaver DR, Jenuwein T, Reppert SM: The polycomb group protein EZH2 is required for mammalian circadian clock function. J Biol Chem 2006, 281:21209–21215. [DOI] [PubMed] [Google Scholar]
- 62.Prosdocimo DA, Anand P, Liao X, Zhu H, Shelkay S, Artero-Calderon P, Zhang L, Kirsh J, Moore D, Rosca MG, et al. : Kruppel-like factor 15 is a critical regulator of cardiac lipid metabolism. J Biol Chem 2014, 289:5914–5924.* This study established KLF15 as a key regulator of myocardial lipid utilization.
- 63.Li L, Li H, Tien CL, Jain MK, Zhang L: Kruppel-Like Factor 15 Regulates the Circadian Susceptibility to Ischemia Reperfusion Injury in the Heart. Circulation 2020, 141:1427–1429.** This is the first study that demonstrated KLF15 regulates circadian susceptibility to I/R injury through rhythmic NAD+ biosynthesis by Nampt, Sirt3 and MnSOD activities in vivo. Importantly, this paper also showed that cardioprotection from NAD+ repletion is most prominent when endogenous NAD+ level is low, which implicates NAD+-boosting therapy should coordinate with intrinsic NAD+ level to acheive maxiumal clinical outcome.
- 64.Bartos DC, Grandi E, Ripplinger CM: Ion Channels in the Heart. Compr Physiol 2015, 5:1423–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conrad R, Stolting G, Hendriks J, Ruello G, Kortzak D, Jordan N, Gensch T, Hidalgo P: Rapid Turnover of the Cardiac L-Type CaV1.2 Channel by Endocytic Recycling Regulates Its Cell Surface Availability. iScience 2018, 7:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Basheer WA, Shaw RM: Connexin 43 and CaV1.2 Ion Channel Trafficking in Healthy and Diseased Myocardium. Circ Arrhythm Electrophysiol 2016, 9:e001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Black N, D'Souza A, Wang Y, Piggins H, Dobrzynski H, Morris G, Boyett MR: Circadian rhythm of cardiac electrophysiology, arrhythmogenesis, and the underlying mechanisms. Heart Rhythm 2019, 16:298–307.** This is an excellent review on circadian rhythm in cardiac electrophysiology.
- 68.Chen Y, Zhu D, Yuan J, Han Z, Wang Y, Qian Z, Hou X, Wu T, Zou J: CLOCK-BMAL1 regulate the cardiac L-type calcium channel subunit CACNA1C through PI3K-Akt signaling pathway. Can J Physiol Pharmacol 2016, 94:1023–1032. [DOI] [PubMed] [Google Scholar]
- 69.Bing RJ: The metabolism of the heart. Harvey Lect 1954, 50:27–70. [PubMed] [Google Scholar]
- 70.Taegtmeyer H, Young ME, Lopaschuk GD, Abel ED, Brunengraber H, Darley-Usmar V, Des Rosiers C, Gerszten R, Glatz JF, Griffin JL, et al. : Assessing Cardiac Metabolism: A Scientific Statement From the American Heart Association. Circ Res 2016, 118:1659–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doenst T, Nguyen TD, Abel ED: Cardiac metabolism in heart failure: implications beyond ATP production. Circ Res 2013, 113:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosano GM, Vitale C: Metabolic Modulation of Cardiac Metabolism in Heart Failure. Card Fail Rev 2018, 4:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parihar P, Parihar MS: Metabolic enzymes dysregulation in heart failure: the prospective therapy. Heart Fail Rev 2017, 22:109–121. [DOI] [PubMed] [Google Scholar]
- 74.Hsu CP, Oka S, Shao D, Hariharan N, Sadoshima J: Nicotinamide phosphoribosyltransferase regulates cell survival through NAD+ synthesis in cardiac myocytes. Circ Res 2009, 105:481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee CF, Chavez JD, Garcia-Menendez L, Choi Y, Roe ND, Chiao YA, Edgar JS, Goo YA, Goodlett DR, Bruce JE, et al. : Normalization of NAD+ Redox Balance as a Therapy for Heart Failure. Circulation 2016, 134:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Byun J, Oka SI, Imai N, Huang CY, Ralda G, Zhai P, Ikeda Y, Ikeda S, Sadoshima J: Both gain and loss of Nampt function promote pressure overload-induced heart failure. Am J Physiol Heart Circ Physiol 2019, 317:H711–H725.** This paper provided direct evidance showing that the expression level of Nampt siginificantly influences the functional outcome on cardiac function in vitro and in vivo.
- 77.Um JH, Pendergast JS, Springer DA, Foretz M, Viollet B, Brown A, Kim MK, Yamazaki S, Chung JH: AMPK regulates circadian rhythms in a tissue- and isoform-specific manner. PLoS One 2011, 6:e18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu L, Trent CM, Fang X, Son NH, Jiang H, Blaner WS, Hu Y, Yin YX, Farese RV Jr., Homma S, et al. : Cardiomyocyte-specific loss of diacylglycerol acyltransferase 1 (DGAT1) reproduces the abnormalities in lipids found in severe heart failure. J Biol Chem 2014, 289:29881–29891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu L, Shi X, Bharadwaj KG, Ikeda S, Yamashita H, Yagyu H, Schaffer JE, Yu YH, Goldberg IJ: DGAT1 expression increases heart triglyceride content but ameliorates lipotoxicity. J Biol Chem 2009, 284:36312–36323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolwicz SC Jr., Liu L, Goldberg IJ, Tian R: Enhancing Cardiac Triacylglycerol Metabolism Improves Recovery From Ischemic Stress. Diabetes 2015, 64:2817–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Roe ND, Handzlik MK, Li T, Tian R: The Role of Diacylglycerol Acyltransferase (DGAT) 1 and 2 in Cardiac Metabolism and Function. Sci Rep 2018, 8:4983.* This study performed a comprehensive analysis on two important lipid metabolic enzymes and their functional impacts on cardiac metabolism and funciton in vivo.
- 82.Uchihashi M, Hoshino A, Okawa Y, Ariyoshi M, Kaimoto S, Tateishi S, Ono K, Yamanaka R, Hato D, Fushimura Y, et al. : Cardiac-Specific Bdh1 Overexpression Ameliorates Oxidative Stress and Cardiac Remodeling in Pressure Overload-Induced Heart Failure. Circ Heart Fail 2017, 10. [DOI] [PubMed] [Google Scholar]
- 83.Peliciari-Garcia RA, Goel M, Aristorenas JA, Shah K, He L, Yang Q, Shalev A, Bailey SM, Prabhu SD, Chatham JC, et al. : Altered myocardial metabolic adaptation to increased fatty acid availability in cardiomyocyte-specific CLOCK mutant mice. Biochim Biophys Acta 2016, 1861:1579–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P: The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 2008, 134:329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, et al. : Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 2009, 324:651–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P: Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 2009, 324:654–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wijnen H: Circadian rhythms. A circadian loop asSIRTs itself. Science 2009, 324:598–599. [DOI] [PubMed] [Google Scholar]
- 88.Sugi K, Hsieh PN, Ilkayeva O, Shelkay S, Moroney B, Baadh P, Haynes B, Pophal M, Fan L, Newgard CB, et al. : Kruppel-like factor 15 is required for the cardiac adaptive response to fasting. PLoS One 2018, 13:e0192376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao Y, Song W, Wang L, Rane MJ, Han F, Cai L: Multiple roles of KLF15 in the heart: Underlying mechanisms and therapeutic implications. J Mol Cell Cardiol 2019, 129:193–196.** This is an excellent review on the physiological and pathophysiological roles of KLF15 in heart.
- 90.Haldar SM, Lu Y, Jeyaraj D, Kawanami D, Cui Y, Eapen SJ, Hao C, Li Y, Doughman YQ, Watanabe M, et al. : Klf15 deficiency is a molecular link between heart failure and aortic aneurysm formation. Sci Transl Med 2010, 2:26ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hsieh PN, Sweet DR, Fan L, Jain MK: Aging and the Kruppel-like factors. Trends Cell Mol Biol 2017, 12:1–15. [PMC free article] [PubMed] [Google Scholar]
- 92.Sun H, Olson KC, Gao C, Prosdocimo DA, Zhou M, Wang Z, Jeyaraj D, Youn JY, Ren S, Liu Y, et al. : Catabolic Defect of Branched-Chain Amino Acids Promotes Heart Failure. Circulation 2016, 133:2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kolwicz SC Jr., Purohit S, Tian R: Cardiac metabolism and its interactions with contraction, growth, and survival of cardiomyocytes. Circ Res 2013, 113:603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chanda D, Luiken JJ, Glatz JF: Signaling pathways involved in cardiac energy metabolism. FEBS Lett 2016, 590:2364–2374. [DOI] [PubMed] [Google Scholar]
- 95.Mori V, Amici A, Mazzola F, Di Stefano M, Conforti L, Magni G, Ruggieri S, Raffaelli N, Orsomando G: Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS One 2014, 9:e113939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ansari HR, Raghava GP: Identification of NAD interacting residues in proteins. BMC Bioinformatics 2010, 11:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katsyuba E, Romani M, Hofer D, Auwerx J: NAD+ homeostasis in health and disease. Nature Metabolism 2020, 2:9–31.** This is an excellent review on NAD+ biology, NAD+-associated diseases and therapies in animal and human studies.
- 98.Matasic DS, Brenner C, London B: Emerging potential benefits of modulating NAD(+) metabolism in cardiovascular disease. Am J Physiol Heart Circ Physiol 2018, 314:H839–H852.* This is an excellent review on NAD+ metabolism and cardiovascular disease.
- 99.Hershberger KA, Martin AS, Hirschey MD: Role of NAD(+) and mitochondrial sirtuins in cardiac and renal diseases. Nat Rev Nephrol 2017, 13:213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rajman L, Chwalek K, Sinclair DA: Therapeutic Potential of NAD-Boosting Molecules: The In Vivo Evidence. Cell Metab 2018, 27:529–547.** This is an excellent review that summaried in vivo studies on NAD+-boosting intervention in various disease models.
- 101.Anderson KA, Madsen AS, Olsen CA, Hirschey MD: Metabolic control by sirtuins and other enzymes that sense NAD(+), NADH, or their ratio. Biochim Biophys Acta Bioenerg 2017, 1858:991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pacholec M, Bleasdale JE, Chrunyk B, Cunningham D, Flynn D, Garofalo RS, Griffith D, Griffor M, Loulakis P, Pabst B, et al. : SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem 2010, 285:8340–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smith BC, Hallows WC, Denu JM: A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal Biochem 2009, 394:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Madsen AS, Andersen C, Daoud M, Anderson KA, Laursen JS, Chakladar S, Huynh FK, Colaco AR, Backos DS, Fristrup P, et al. : Investigating the Sensitivity of NAD+-dependent Sirtuin Deacylation Activities to NADH. J Biol Chem 2016, 291:7128–7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gerhart-Hines Z, Dominy JE Jr., Blattler SM, Jedrychowski MP, Banks AS, Lim JH, Chim H, Gygi SP, Puigserver P: The cAMP/PKA pathway rapidly activates SIRT1 to promote fatty acid oxidation independently of changes in NAD(+). Mol Cell 2011, 44:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, et al. : Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci 2009, 18:514–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, et al. : SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 2011, 44:177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Munro D, Treberg JR: A radical shift in perspective: mitochondria as regulators of reactive oxygen species. J Exp Biol 2017, 220:1170–1180. [DOI] [PubMed] [Google Scholar]
- 109.Steinhorn B, Sorrentino A, Badole S, Bogdanova Y, Belousov V, Michel T: Chemogenetic generation of hydrogen peroxide in the heart induces severe cardiac dysfunction. Nat Commun 2018, 9:4044.** This study developed a novel fusion protein tool that allows for simultaneous production (DAAO) and measurement (HyPer) of H2O2.
- 110.Panth N, Paudel KR, Parajuli K: Reactive Oxygen Species: A Key Hallmark of Cardiovascular Disease. Adv Med 2016, 2016:9152732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moris D, Spartalis M, Spartalis E, Karachaliou GS, Karaolanis GI, Tsourouflis G, Tsilimigras DI, Tzatzaki E, Theocharis S: The role of reactive oxygen species in the pathophysiology of cardiovascular diseases and the clinical significance of myocardial redox. Ann Transl Med 2017, 5:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bagheri F, Khori V, Alizadeh AM, Khalighfard S, Khodayari S, Khodayari H: Reactive oxygen species-mediated cardiac-reperfusion injury: Mechanisms and therapies. Life Sci 2016, 165:43–55. [DOI] [PubMed] [Google Scholar]
- 113.Tsutsui H, Kinugawa S, Matsushima S: Oxidative stress and heart failure. Am J Physiol Heart Circ Physiol 2011, 301:H2181–2190. [DOI] [PubMed] [Google Scholar]
- 114.Rhee SG, Woo HA: Multiple functions of peroxiredoxins: peroxidases, sensors and regulators of the intracellular messenger H(2)O(2), and protein chaperones. Antioxid Redox Signal 2011, 15:781–794. [DOI] [PubMed] [Google Scholar]
- 115.Watabe S, Kohno H, Kouyama H, Hiroi T, Yago N, Nakazawa T: Purification and characterization of a substrate protein for mitochondrial ATP-dependent protease in bovine adrenal cortex. J Biochem 1994, 115:648–654. [DOI] [PubMed] [Google Scholar]
- 116.Yamamoto T, Matsui Y, Natori S, Obinata M: Cloning of a housekeeping-type gene (MER5) preferentially expressed in murine erythroleukemia cells. Gene 1989, 80:337–343. [DOI] [PubMed] [Google Scholar]
- 117.Rhee SG, Kil IS: Mitochondrial H2O2 signaling is controlled by the concerted action of peroxiredoxin III and sulfiredoxin: Linking mitochondrial function to circadian rhythm. Free Radic Biol Med 2016, 100:73–80. [DOI] [PubMed] [Google Scholar]
- 118.Kil IS, Ryu KW, Lee SK, Kim JY, Chu SY, Kim JH, Park S, Rhee SG: Circadian Oscillation of Sulfiredoxin in the Mitochondria. Mol Cell 2015, 59:651–663. [DOI] [PubMed] [Google Scholar]
- 119.Saez L, Young MW: Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron 1996, 17:911–920. [DOI] [PubMed] [Google Scholar]
- 120.Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TD, Weitz CJ, Takahashi JS, Kay SA: Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 1998, 280:1599–1603. [DOI] [PubMed] [Google Scholar]
- 121.Rey G, Milev NB, Valekunja UK, Ch R, Ray S, Silva Dos Santos M, Nagy AD, Antrobus R, MacRae JI, Reddy AB: Metabolic oscillations on the circadian time scale in Drosophila cells lacking clock genes. Mol Syst Biol 2018, 14:e8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ode KL, Ueda HR: Lost in clocks: non-canonical circadian oscillation discovered in Drosophila cells. Mol Syst Biol 2018, 14:e8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, et al. : Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485:459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nulton-Persson AC, Szweda LI: Modulation of mitochondrial function by hydrogen peroxide. J Biol Chem 2001, 276:23357–23361. [DOI] [PubMed] [Google Scholar]
- 125.Lee Y, Kim EK: AMP-activated protein kinase as a key molecular link between metabolism and clockwork. Exp Mol Med 2013, 45:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jordan SD, Lamia KA: AMPK at the crossroads of circadian clocks and metabolism. Mol Cell Endocrinol 2013, 366:163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsai JY, Kienesberger PC, Pulinilkunnil T, Sailors MH, Durgan DJ, Villegas-Montoya C, Jahoor A, Gonzalez R, Garvey ME, Boland B, et al. : Direct regulation of myocardial triglyceride metabolism by the cardiomyocyte circadian clock. J Biol Chem 2010, 285:2918–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, et al. : AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 2009, 326:437–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Um JH, Yang S, Yamazaki S, Kang H, Viollet B, Foretz M, Chung JH: Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem 2007, 282:20794–20798. [DOI] [PubMed] [Google Scholar]
- 130.Ghigo A, Li M: Phosphoinositide 3-kinase: friend and foe in cardiovascular disease. Front Pharmacol 2015, 6:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Manning BD, Toker A: AKT/PKB Signaling: Navigating the Network. Cell 2017, 169:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jere SW, Houreld NN, Abrahamse H: Role of the PI3K/AKT (mTOR and GSK3beta) signalling pathway and photobiomodulation in diabetic wound healing. Cytokine Growth Factor Rev 2019, 50:52–59. [DOI] [PubMed] [Google Scholar]
- 133.Ko ML, Shi L, Tsai JY, Young ME, Neuendorff N, Earnest DJ, Ko GY: Cardiac-specific mutation of Clock alters the quantitative measurements of physical activities without changing behavioral circadian rhythms. J Biol Rhythms 2011, 26:412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gao T, Yatani A, Dell'Acqua ML, Sako H, Green SA, Dascal N, Scott JD, Hosey MM: cAMP-dependent regulation of cardiac L-type Ca2+ channels requires membrane targeting of PKA and phosphorylation of channel subunits. Neuron 1997, 19:185–196. [DOI] [PubMed] [Google Scholar]
- 135.Gerhardstein BL, Puri TS, Chien AJ, Hosey MM: Identification of the sites phosphorylated by cyclic AMP-dependent protein kinase on the beta 2 subunit of L-type voltage-dependent calcium channels. Biochemistry 1999, 38:10361–10370. [DOI] [PubMed] [Google Scholar]
- 136.Marx SO, Reiken S, Hisamatsu Y, Jayaraman T, Burkhoff D, Rosemblit N, Marks AR: PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell 2000, 101:365–376. [DOI] [PubMed] [Google Scholar]
- 137.Simmerman HK, Collins JH, Theibert JL, Wegener AD, Jones LR: Sequence analysis of phospholamban. Identification of phosphorylation sites and two major structural domains. J Biol Chem 1986, 261:13333–13341. [PubMed] [Google Scholar]
- 138.Najafi A, Sequeira V, Kuster DW, van der Velden J: beta-adrenergic receptor signalling and its functional consequences in the diseased heart. Eur J Clin Invest 2016, 46:362–374. [DOI] [PubMed] [Google Scholar]
- 139.Lohse MJ, Engelhardt S, Eschenhagen T: What is the role of beta-adrenergic signaling in heart failure? Circ Res 2003, 93:896–906. [DOI] [PubMed] [Google Scholar]
- 140.Marks AR: Calcium cycling proteins and heart failure: mechanisms and therapeutics. J Clin Invest 2013, 123:46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Grimm M, Brown JH: Beta-adrenergic receptor signaling in the heart: role of CaMKII. J Mol Cell Cardiol 2010, 48:322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bossis I, Stratakis CA: Minireview: PRKAR1A: normal and abnormal functions. Endocrinology 2004, 145:5452–5458. [DOI] [PubMed] [Google Scholar]
- 143.Richards AM, Nicholls MG, Espiner EA, Ikram H, Cullens M, Hinton D: Diurnal patterns of blood pressure, heart rate and vasoactive hormones in normal man. Clin Exp Hypertens A 1986, 8:153–166. [DOI] [PubMed] [Google Scholar]
- 144.Sachan N, Dey A, Rotter D, Grinsfelder DB, Battiprolu PK, Sikder D, Copeland V, Oh M, Bush E, Shelton JM, et al. : Sustained hemodynamic stress disrupts normal circadian rhythms in calcineurin-dependent signaling and protein phosphorylation in the heart. Circ Res 2011, 108:437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Collins HE, Rodrigo GC: Inotropic response of cardiac ventricular myocytes to beta-adrenergic stimulation with isoproterenol exhibits diurnal variation: involvement of nitric oxide. Circ Res 2010, 106:1244–1252. [DOI] [PubMed] [Google Scholar]
- 146.Parra V, Rothermel BA: Calcineurin signaling in the heart: The importance of time and place. J Mol Cell Cardiol 2017, 103:121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marin-Garcia J, Akhmedov AT: Mitochondrial dynamics and cell death in heart failure. Heart Fail Rev 2016, 21:123–136. [DOI] [PubMed] [Google Scholar]
- 148.Vasquez-Trincado C, Garcia-Carvajal I, Pennanen C, Parra V, Hill JA, Rothermel BA, Lavandero S: Mitochondrial dynamics, mitophagy and cardiovascular disease. J Physiol 2016, 594:509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nan J, Zhu W, Rahman MS, Liu M, Li D, Su S, Zhang N, Hu X, Yu H, Gupta MP, et al. : Molecular regulation of mitochondrial dynamics in cardiac disease. Biochim Biophys Acta Mol Cell Res 2017, 1864:1260–1273. [DOI] [PubMed] [Google Scholar]
- 150.Saad NS, Elnakish MT, Ahmed AAE, Janssen PML: Protein Kinase A as a Promising Target for Heart Failure Drug Development. Arch Med Res 2018, 49:530–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Parra V, Altamirano F, Hernandez-Fuentes CP, Tong D, Kyrychenko V, Rotter D, Pedrozo Z, Hill JA, Eisner V, Lavandero S, et al. : Down Syndrome Critical Region 1 Gene, Rcan1, Helps Maintain a More Fused Mitochondrial Network. Circ Res 2018, 122:e20–e33.** This paper demonstrated that the RCAN1-calcineurin axis impinges on mitochondrial fussion through DRP1 which protects the heart from I/R injury. iPSC-derived cardiomyocyte was applied in this study.
- 152.Pennanen C, Parra V, Lopez-Crisosto C, Morales PE, Del Campo A, Gutierrez T, Rivera-Mejias P, Kuzmicic J, Chiong M, Zorzano A, et al. : Mitochondrial fission is required for cardiomyocyte hypertrophy mediated by a Ca2+-calcineurin signaling pathway. J Cell Sci 2014, 127:2659–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rana S, Prabhu SD, Young ME: Chronobiological Influence Over Cardiovascular Function: The Good, the Bad, and the Ugly. Circ Res 2020, 126:258–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fuentes JJ, Pritchard MA, Planas AM, Bosch A, Ferrer I, Estivill X: A new human gene from the Down syndrome critical region encodes a proline-rich protein highly expressed in fetal brain and heart. Hum Mol Genet 1995, 4:1935–1944. [DOI] [PubMed] [Google Scholar]
- 155.Fuentes JJ, Pritchard MA, Estivill X: Genomic organization, alternative splicing, and expression patterns of the DSCR1 (Down syndrome candidate region 1) gene. Genomics 1997, 44:358–361. [DOI] [PubMed] [Google Scholar]
- 156.Rothermel BA, Vega RB, Williams RS: The role of modulatory calcineurin-interacting proteins in calcineurin signaling. Trends Cardiovasc Med 2003, 13:15–21. [DOI] [PubMed] [Google Scholar]
- 157.Yang J, Rothermel B, Vega RB, Frey N, McKinsey TA, Olson EN, Bassel-Duby R, Williams RS: Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ Res 2000, 87:E61–68. [DOI] [PubMed] [Google Scholar]
- 158.Lange AW, Molkentin JD, Yutzey KE: DSCR1 gene expression is dependent on NFATc1 during cardiac valve formation and colocalizes with anomalous organ development in trisomy 16 mice. Dev Biol 2004, 266:346–360. [DOI] [PubMed] [Google Scholar]
- 159.Panther F, Williams T, Ritter O: Inhibition of the calcineurin-NFAT signalling cascade in the treatment of heart failure. Recent Pat Cardiovasc Drug Discov 2009, 4:180–186. [DOI] [PubMed] [Google Scholar]
- 160.Mattiazzi A, Kranias EG: The role of CaMKII regulation of phospholamban activity in heart disease. Front Pharmacol 2014, 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Munch G, Bolck B, Karczewski P, Schwinger RH: Evidence for calcineurin-mediated regulation of SERCA 2a activity in human myocardium. J Mol Cell Cardiol 2002, 34:321–334. [DOI] [PubMed] [Google Scholar]
- 162.Durgan DJ, Tsai JY, Grenett MH, Pat BM, Ratcliffe WF, Villegas-Montoya C, Garvey ME, Nagendran J, Dyck JR, Bray MS, et al. : Evidence suggesting that the cardiomyocyte circadian clock modulates responsiveness of the heart to hypertrophic stimuli in mice. Chronobiol Int 2011, 28:187–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wilkins BJ, Molkentin JD: Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem Biophys Res Commun 2004, 322:1178–1191. [DOI] [PubMed] [Google Scholar]
- 164.Negretti N, O'Neill SC, Eisner DA: The effects of inhibitors of sarcoplasmic reticulum function on the systolic Ca2+ transient in rat ventricular myocytes. J Physiol 1993, 468:35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sankaranarayanan R, Kistamas K, Greensmith DJ, Venetucci LA, Eisner DA: Systolic [Ca(2+)]i regulates diastolic levels in rat ventricular myocytes. J Physiol 2017, 595:5545–5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lompre AM, Hajjar RJ, Harding SE, Kranias EG, Lohse MJ, Marks AR: Ca2+ cycling and new therapeutic approaches for heart failure. Circulation 2010, 121:822–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Oyama Y, Bartman CM, Gile J, Eckle T: Circadian MicroRNAs in Cardioprotection. Curr Pharm Des 2017, 23:3723–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Noyan H, El-Mounayri O, Isserlin R, Arab S, Momen A, Cheng HS, Wu J, Afroze T, Li RK, Fish JE, et al. : Cardioprotective Signature of Short-Term Caloric Restriction. PLoS One 2015, 10:e0130658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bartman CM, Oyama Y, Brodsky K, Khailova L, Walker L, Koeppen M, Eckle T: Intense light-elicited upregulation of miR-21 facilitates glycolysis and cardioprotection through Per2-dependent mechanisms. PLoS One 2017, 12:e0176243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bendova Z, Sumova A: Photoperiodic regulation of PER1 and PER2 protein expression in rat peripheral tissues. Physiol Res 2006, 55:623–632. [DOI] [PubMed] [Google Scholar]
- 171.Reppert SM, Weaver DR: Coordination of circadian timing in mammals. Nature 2002, 418:935–941. [DOI] [PubMed] [Google Scholar]
- 172.Young ME, Razeghi P, Cedars AM, Guthrie PH, Taegtmeyer H: Intrinsic diurnal variations in cardiac metabolism and contractile function. Circ Res 2001, 89:1199–1208. [DOI] [PubMed] [Google Scholar]
- 173.Kohsaka A, Das P, Hashimoto I, Nakao T, Deguchi Y, Gouraud SS, Waki H, Muragaki Y, Maeda M: The circadian clock maintains cardiac function by regulating mitochondrial metabolism in mice. PLoS One 2014, 9:e112811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Sardon Puig L, Valera-Alberni M, Canto C, Pillon NJ: Circadian Rhythms and Mitochondria: Connecting the Dots. Front Genet 2018, 9:452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Manella G, Asher G: The Circadian Nature of Mitochondrial Biology. Front Endocrinol (Lausanne) 2016, 7:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Li BJ, Zhu ZX, Qin H, Meng ZN, Lin HR, Xia JH: Genome-Wide Characterization of Alternative Splicing Events and Their Responses to Cold Stress in Tilapia. Front Genet 2020, 11:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.El-Athman R, Knezevic D, Fuhr L, Relogio A: A Computational Analysis of Alternative Splicing across Mammalian Tissues Reveals Circadian and Ultradian Rhythms in Splicing Events. Int J Mol Sci 2019, 20.** A comprehensive computational analysis of alternative splicing which revealed that circadian oscillation in alternative splicing events is universal across mammalian tissues including heart. Interestingly, 97% of the circadian alternative splicing events are 12-hr rhythmic despite that only few genes are 12-hr rhythmic in heart.
- 178.Fan Z, Zhao M, Joshi PD, Li P, Zhang Y, Guo W, Xu Y, Wang H, Zhao Z, Yan J: A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res 2017, 45:5720–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Millius A, Ode KL, Ueda HR: A period without PER: understanding 24-hour rhythms without classic transcription and translation feedback loops. F1000Res 2019, 8:499.** This is an excellent review on non-canonical circadian clock.
- 180.Lakin-Thomas PL: Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms 2006, 21:83–92. [DOI] [PubMed] [Google Scholar]
- 181.Ashkenazi IE, Hartman H, Strulovitz B, Dar O: Activity rhythms of enzymes in human red blood cell suspensions. Journal of Interdisciplinary Cycle Research 1975, 6:291–301. [Google Scholar]
- 182.O'Neill JS, Reddy AB: Circadian clocks in human red blood cells. Nature 2011, 469:498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ray S, Valekunja UK, Stangherlin A, Howell SA, Snijders AP, Damodaran G, Reddy AB: Circadian rhythms in the absence of the clock gene Bmal1. Science 2020, 367:800–806.** This is the first study that provided experimental evidence showing the presence of BMAL-independent non-canonical circadian mechanism throughout transcriptional, translational and post-translational levels in mammalian organs, which is primarily driven by E26 transformation-specific factors.
- 184.Kawachi I, Colditz GA, Stampfer MJ, Willett WC, Manson JE, Speizer FE, Hennekens CH: Prospective study of shift work and risk of coronary heart disease in women. Circulation 1995, 92:3178–3182. [DOI] [PubMed] [Google Scholar]
- 185.Esquirol Y, Perret B, Ruidavets JB, Marquie JC, Dienne E, Niezborala M, Ferrieres J: Shift work and cardiovascular risk factors: new knowledge from the past decade. Arch Cardiovasc Dis 2011, 104:636–668. [DOI] [PubMed] [Google Scholar]
- 186.Vyas MV, Garg AX, Iansavichus AV, Costella J, Donner A, Laugsand LE, Janszky I, Mrkobrada M, Parraga G, Hackam DG: Shift work and vascular events: systematic review and meta-analysis. BMJ 2012, 345:e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lo SH, Lin LY, Hwang JS, Chang YY, Liau CS, Wang JD: Working the night shift causes increased vascular stress and delayed recovery in young women. Chronobiol Int 2010, 27:1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yeom JH, Sim CS, Lee J, Yun SH, Park SJ, Yoo CI, Sung JH: Effect of shift work on hypertension: cross sectional study. Ann Occup Environ Med 2017, 29:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Puttonen S, Harma M, Hublin C: Shift work and cardiovascular disease - pathways from circadian stress to morbidity. Scand J Work Environ Health 2010, 36:96–108. [DOI] [PubMed] [Google Scholar]