Abstract
The airborne fungus Aspergillus fumigatus poses a serious health threat to humans by causing numerous invasive infections and a notable mortality in humans, especially in immunocompromised patients. Mould-active azoles are the frontline therapeutics employed to treat aspergillosis. The global emergence of azole-resistant A. fumigatus isolates in clinic and environment, however, notoriously limits the therapeutic options of mould-active antifungals and potentially can be attributed to a mortality rate reaching up to 100 %. Although specific mutations in CYP51A are the main cause of azole resistance, there is a new wave of azole-resistant isolates with wild-type CYP51A genotype challenging the efficacy of the current diagnostic tools. Therefore, applications of whole-genome sequencing are increasingly gaining popularity to overcome such challenges. Prominent echinocandin tolerance, as well as liver and kidney toxicity posed by amphotericin B, necessitate a continuous quest for novel antifungal drugs to combat emerging azole-resistant A. fumigatus isolates. Animal models and the tools used for genetic engineering require further refinement to facilitate a better understanding about the resistance mechanisms, virulence, and immune reactions orchestrated against A. fumigatus. This review paper comprehensively discusses the current clinical challenges caused by A. fumigatus and provides insights on how to address them.
Key words: Aspergillus fumigatus, Azole-resistance, Drug-resistance mechanism, Invasive aspergillosis
Introduction
Species within the genus Aspergillus have been long exploited as an invaluable biotechnological resource to produce pharmaceuticals, food and food ingredients, and enzymes (reviewed in Meyer et al. 2011). Among species within this genus, Aspergillus fumigatus is the most ubiquitous fungal species in the environment (reviewed in Kwon-Chung & Sugui 2013). Aspergillus fumigatus can withstand and survive in a wide range of pH and temperature and its hydrophobic cell wall allows this species to be efficiently dispersed by even slight air currents. Similarly, a number of features allow this species to be the most predominant mould species causing infections in humans (Kwon-Chung & Sugui 2013). Small conidia size allows penetration to the lower respiratory tract system and escaping clearance by mucociliary forces, presence of melanin in the cell wall enables withstanding reactive oxygen species and phagocytosis, and abundance of negatively charged sialic acid on the surface permits A. fumigatus to effectively bind to the basal lamina proteins once inside the host lung (reviewed in Kwon-Chung & Sugui 2013). Although conidia can be easily cleared by counteracting host mechanisms in the lung, A. fumigatus can cause a wide range of infections in both immunocompromised and immunocompetent individuals (Denning & Chakrabarti 2017), including an estimated annual number of 16 million pulmonary-infections (Denning et al. 2013, 2016) with fatal outcomes in many hundred thousand patients annually (Brown et al. 2012, Lowes et al. 2017).
Triazoles are the first-line antifungals used to treat patients suffering from aspergillosis and have brought down mortality rates to 30 % or lower in invasive aspergillosis, which is almost always fatal if untreated (Neofytos et al. 2009). Extensive use of azoles in various sectors ranging from agriculture and industry to clinics, however, promotes selective pressure allowing emergence of azole-resistant A. fumigatus (ARAF) isolates in numerous niches (Verweij et al. 2015). Subsequently, deposition of such ARAF spores in the lung of immunocompromised patients may cause azole-resistant invasive aspergillosis (IA), often in patients that have never been prescribed any azoles, resulting in treatment guidelines favouring initial treatment with liposomal amphotericin B in settings with high prevalence of azole resistance (i.e. > 10 %) (Thom & Church 1926). Therefore, the high mortality rate and wide range of infections together with the emergence of ARAF isolates severely complicates the management of patients suffering from aspergillosis. In this review, we discuss the current paradigm and challenges of aspergillosis, and subsequently provide suggestions to more effectively tackle these challenges utilising worldwide initiatives across multiple disciplines.
Taxonomy and prevalence of clinically important Aspergillus species
History
The genus Aspergillus has a long history which dates back to Micheli’s “Nova Plantarum Genera” of 1729. Micheli, being a priest, noted that the fungus he described resembled the shape of an aspergillum (sprinkler of holy water), hence the name Aspergillus. The genus gained more attention from 1850 onwards, because it was recognised as a causal agent of spoilage, human disease and producer of useful metabolites. Due to its economic significance, the taxonomy of the genus was studied various times in history. In 1926, Thom and Church brought all available material on Aspergillus together and published the first major monograph on the genus (Thom & Church 1926). This monograph was revised in 1945 (Thom & Raper 1945) and subsequently in 1965 (Raper & Fennell 1965). Their taxonomic schemes were based on macroscopic (e.g., conidial colour and growth rates) and microscopic characters (vesicle shape, presence/absence of metulae). These monographs were the standard until the introduction of the molecular techniques in the 1990’s. Due to DNA sequence analysis, and to a lesser extent extrolite analysis, morphologically well-defined species turn out to consist of multiple species. Nowadays, a polyphasic approach, integrating different kinds of data and information (phenotypic, genotypic and phylogenetic), is the standard for describing new species in Aspergillus. To date, an updated Aspergillus monograph is lacking; however, there are some more recent taxonomic overviews on various Aspergillus sections, e.g., sect. Aenei (Varga et al. 2010), Aspergillus (Chen et al. 2017), Cervini (Chen et al. 2016a, 2016c), Circumdati (Visagie et al. 2014), Clavati (Varga et al. 2007), Flavi (Frisvad et al. 2019), Flavipedes (Hubka et al. 2015), Fumigati (Samson et al. 2007a), Nidulantes (Chen et al. 2016a, 2016c), Nigri (Samson et al. 2007b, Varga et al. 2011), Polypaecilum (Tanney et al. 2017), Restricti (Sklenář et al. 2017), Terrei (Samson et al. 2011) and Usti (Houbraken et al. 2007).
Nomenclature and Aspergillus
The International Code of Nomenclature for Algae, Fungi and Plants (ICN) governs the naming of fungi (McNeill et al. 2012). For a long time, dual nomenclature was used and asexually reproducing fungi got separate names from their sexual states. When strictly following these old rules, the name of the sexual morph had priority over the asexual morph name. For example, Neosartorya fumigata and Petromyces flavus should be used instead of the more well-known names A. fumigatus and A. flavus. The separate naming of these morphs was debated for many years and the principle “One fungus, One name” was introduced on January 1, 2013. In practice this means after that date, a fungus can only have one name. Nowadays, the name Aspergillus is used in a broad sense. Species producing (different) sexual morphs and previously described in teleomorph genera (e.g., Emericella, Eurotium, Neosartorya, Petromyces) are treated as synonyms (Kocsubé et al. 2016). The single name nomenclature led to various name changes. Many of the clinically relevant species were already known under its current Aspergillus name and therefore these changes did not have a big impact in the field of medical mycology. For example, in medical mycology Aspergillus nidulans was already a well-known name, while Emericella nidulans was more commonly used in food and indoor mycology. In some cases, the species epithet already indicates the connection between the old and current name (e.g., A. chevalieri/Eurotium chevalieri, A. fischeri/Neosartorya fischeri, A. udagawae/Neosartorya udagawae), but in other cases this is less obvious (e.g., A. glaucus/Eurotium herbariorum, A. montevidensis/Eurotium amstelodami and A. thermomutatus/Neosartorya pseudofischeri). In order to help the users with these changes in nomenclature, a list of all accepted species was prepared for Aspergillus (and related genera) (Houbraken et al. 2020).
Classification of Aspergillus
The genus Aspergillus is classified in the family Aspergillaceae, order Eurotiales (Houbraken & Samson 2011). There is a long tradition of using an infrageneric classification in Aspergillus; these are names of taxa between the ranks of genus and species (e.g., subgenera, sections, series). Using morphological characters, Raper & Fennell divided Aspergillus in 18 groups; however, these groups do not have any standing nomenclature and should not be used anymore (Raper & Fennell 1965). To avoid confusion and to promote taxonomic stability, a formal infrageneric classification system was introduced by Gams et al. (1985) and they replaced the “group” structure by a subgeneric and sectional structure. Nowadays, the genus is subdivided in six subgenera, 27 sections and 87 series (Houbraken et al. 2020). These formal infrageneric ranks are not commonly used in medical mycology. Articles often refer to various other informal ranks, for example “species complexes” (e.g., “A. fumigatus species complex”) (Salsé et al. 2019, Dos Santos et al. 2020), “cryptic species”, “cryptic A. fumigatus” (Wiederhold et al. 2018, Rivero-Menendez et al. 2019a) “species clades” (e.g., A. fumigatus-clade) (Balajee et al. 2005, 2007a), “sensu lato” (e.g., A. fumigatus sensu lato) (Li et al. 2014, Hagiwara et al. 2019) and “sensu stricto” (e.g., A. fumigatus sensu stricto) (Li et al. 2014, Monteiro et al. 2019)). The main disadvantage of using these informal ranks is lack of consensus. For example, it is not clear whether “A. fumigatus sensu lato”, “A. fumigatus species complex”, “cryptic A. fumigatus” and the “A. fumigatus-clade” are actually representing the same (group of) species. It is therefore recommended to use, when possible, a formal classification system of subgenera, sections and series. In the case of A. fumigatus, it is recommended to refer to A. fumigatus (the species), series Fumigati (for A. fumigatus and the related species Aspergillus fischeri, A. fumigatiaffinis, A. fumigatus, A. fumisynnematus, A. laciniosus, A. lentulus, A. novofumigatus, A. oerlinghausensis, A. spinosus, A. takakii) or section Fumigati (59 species) (Houbraken et al. 2020).
Identification of Aspergillus species from pure culture
In the last decade, there is steep increase of the number of accepted Aspergillus species (Houbraken et al. 2020). The driving forces behind this steep increase are twofold: firstly, there is a large diversity and high interest in this genus and secondly, phenotypically well-known species are turn out to be species complexes that are genetically and evolutionary distinct (Chen et al. 2016a, 2016c, 2017, Houbraken et al. 2016a, Sklenář et al. 2017). Morphology was for a long time the mainstay in Aspergillus identification (Raper & Fennell 1965). As morphologically well-defined species turned out to be species complexes, accurate phenotype-based identification became more difficult and unreliable. There are often only small differences between species within a complex and sometimes they need to be grown on special agar media to observe those differences. Identification based on phenotypic characters is therefore challenging, even for experienced mycologists and (well-trained) staff of routine labs. These phenotypically similar species are also referred to as “cryptic species”; however, they can be identified using a molecular based approach. These phenotypically closely related (cryptic) species can have strikingly different patterns of antifungal susceptibility patterns against the most important antifungals, including triazoles and amphotericin B (AMB), and some of these species are intrinsically resistant or have acquired resistance against these antifungals (Alastruey-Izquierdo et al. 2013, 2014, Escribano et al. 2013, Negri et al. 2014, Iatta et al. 2016, Heo et al. 2017, Talbot & Barrs 2018, Zoran et al. 2018, Salah et al. 2019, Mendoza et al. 2020, Glampedakis et al. 2021). For correct identification of Aspergillus species, calmodulin gene sequencing is recommended (Samson et al. 2019), and partial β-tubulin gene sequencing can be used as an alternative. ITS sequencing lacks resolution and is therefore not suitable. The public databases are well-stocked with calmodulin gene sequences and 96.9 % of the 446 accepted Aspergillus taxa are represented with a calmodulin gene sequence in GenBank (Houbraken et al. 2020). Actually, the taxonomic position of the species lacking a calmodulin sequence needs to be determined (Houbraken et al. 2020) and are unlikely to be relevant in medical mycology. Besides an overview of the accepted species, also calmodulin and β-tubulin references sequences are given.
Since its introduction into clinical microbiology diagnostics, MALDI-TOF MS has become the standard workhorse system for the identification of bacteria and yeasts (Kostrzewa 2018). Because of their rigid cell wall and the phenotypic variability by sporulation, the identification of moulds has been shown to be more challenging. Different approaches for sample preparation have been developed to overcome these hurdles, e.g., tube extraction of proteins before spotting on the MALDI-TOF MS target or liquid culturing to avoid sporulation. A prerequisite of successful identification of moulds are extensive libraries containing high-quality reference spectra of well characterised strains. This has led to a number of dedicated, user-specific databases and database supplements besides the libraries supplied by manufacturers (Sanguinetti & Posteraro 2017, Patel 2019). A study of US academic centres using a database established at the NIH, a solid media extraction method and a challenge set of 80 clinical mould isolates demonstrated the requirement of instrument optimisation and high standardisation for mould identification across different laboratories (Lau et al. 2019). MALDI-TOF MS has been successfully applied to Aspergillus spp. identification in several studies. Thereby, it could be demonstrated that many but not all rare and cryptic species can be correctly identified if they are represented well in the according database (Vidal-Acuña et al. 2018, Imbert et al. 2019, Américo et al. 2020). Closely related Aspergillus species are sometimes difficult to differentiate by MALDI-TOF MS because of their similar spectral pattern, today, but this can be improved by further extension of databases and utilisation of alternative identification algorithms.
Taxonomic notes on A. fumigatus and other clinically relevant Aspergilli
Aspergillus section Fumigati
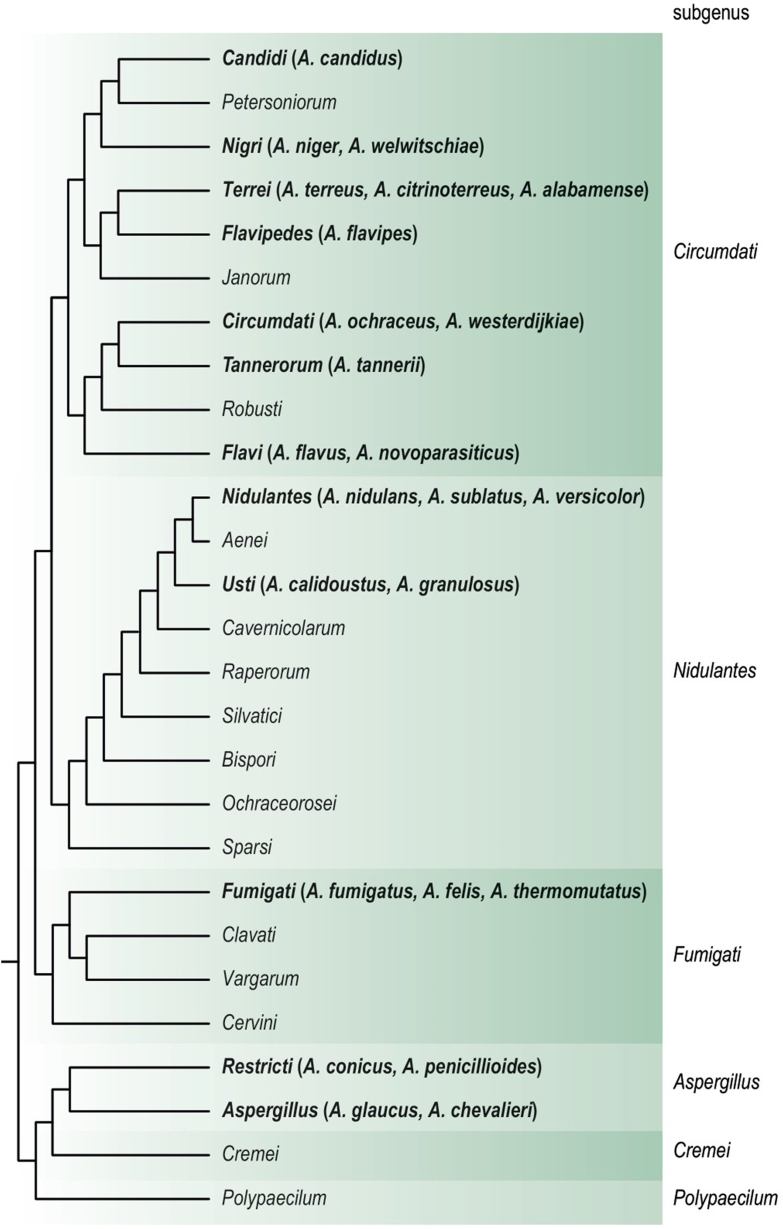
The species in this section produce uniseriate, columnar conidial heads in shades of green and flask shaped vesicles (Raper & Fennell 1965). Traditionally, the identification of these related species is performed using the colony patterns and the morphology of the conidiogenous structures, conidia, ascomata and ascsopores (Raper & Fennell 1965). However, clinical isolates can be markedly abnormal being more floccose with fewer conidia and more recent taxonomic studies showed that section Fumigati species can have a highly similar morphology. For accurate identification, a sequence-based approach is therefore recommended (Samson et al. 2007a). Aspergillus section Fumigati includes 63 species and the species of the section are thermotolerant. Aspergillus fischeri, A. fumigatus and A. oerlinghausenensis are able to grow at 50 °C (Houbraken et al. 2016b), while other species of the section have lower maximum growth temperatures (e.g., 45 °C: A. fumigatiaffinis, A. fumisynnematus, A. lentulus, A. novofumigatus; 42 °C: A. felis) (Balajee et al. 2007a). The sexual morph is of the neosartorya-type (Samson et al. 2007a). The most well-known member of this section is A. fumigatus, though other species in the section are also clinically relevant: A. felis, A. fischeri, A. fumigatiaffinis, A. fumisynnematus, A. hiratsukae, A. laciniosus, A. lentulus, A. novofumigatus, A. pseudoviridinutans, A. spinosus, A. thermomutatus, A. udagawae and A. viridinutans (Sugui et al. 2014, Frisvad & Larsen 2016). Taxonomic evaluation of A. pseudofelis and A. parafelis (also known as A. felis) showed that these species are included in the genetically diverse A. felis (Hubka et al. 2018) (Fig. 1).
Fig. 1.
Cladogram of the genus Aspergillus and the relationship between sections and subgenera. A selection of the species mentioned in the text are given in brackets in bold font after the section name. Adopted from Houbraken et al. (2020) with permission.
Aspergillus fumigatus is reported as the most prevalent species from this section in different countries (Binder & Lass-Flörl 2013). Other species of section Fumigati, the so-called cryptic A. fumigatus, have been increasingly identified in the clinical setting in the last two decades, which is because of the increasing use of the polyphasic approach for the discrimination and identification of Aspergillus species in clinical samples. Based on multilocus comparative sequence analysis, other species from this section are recovered from 3 to 6 % of patients with IA. Among cryptic species, A. felis, A. lentulus, A. thermomutatus and series Viridinutantes members (= A. viridinutans complex) are the most common isolates from clinical disease in human (Balajee et al. 2009, Alastruey-Izquierdo et al. 2013, 2014, Escribano et al. 2013, Negri et al. 2014, Sabino et al. 2014, Frisvad & Larsen 2016, Talbot et al. 2018, Paccoud et al. 2019, Yagi et al. 2019).
Aspergillus fumigatus biosynthesizes a variety of secondary metabolites such as fumagillin, fumitoxins, fumigaclavines A & C, fumitremorgins, gliotoxin, trypacidin, pseurotins, helvolic acid, pyripyropens, methyl-sulochrin, verruculogen, fumiquinazolines. Several of these metabolites may cause serious health hazard (Frisvad & Samson 1990, Fujimoto et al. 1993, Larsen et al. 2007, Frisvad & Larsen 2016), though none of them are actually regulated mycotoxins. Some of these metabolites are involved in impairing the host immune system (Steenwyk et al. 2020) e.g., gliotoxin has been shown to inhibit the host immune response (Sugui et al. 2007, Spikes et al. 2008).
Other Aspergillus sections with clinically relevant species
Besides the section Fumigati species, a wide variety of other Aspergilli are clinically relevant. Most of them belong to the species-rich sections Flavi, Nidulantes, Nigri, Terrei and Usti. The taxonomy of these sections is well-studied and correct identification using sequence data should therefore not be problematic (Houbraken et al. 2020).
The taxonomy of section Flavi was recently updated and contains 35 species (Visagie et al. 2014, Frisvad et al. 2019, Houbraken et al. 2020), of which nine species are known to cause infection in humans (Hedayati et al. 2007, Frisvad et al. 2019, Rudramurthy et al. 2019, Alshehri & Palanisamy 2020). Aspergillus flavus is the main and most commonly occurring species of the section. Most of the reports of A. oryzae in clinical settings are likely to be erroneous. Aspergillus oryzae is the domesticated form of A. flavus and they can be regarded as conspecific. It is impossible to reliably differentiate A. oryzae and A. flavus using morphology and calmodulin or β-tubulin gene sequencing. The differentiation between both species is mainly driven by an applied aspect: A. oryzae is extensively used in food fermentations (e.g., soy sauce, sake) and for the production of enzymes, and these industries do not want to use the name A. flavus, which has a strong association with aflatoxin production. As a consensus, strains that do not produce aflatoxin and have a food fermentation or biotechnological background can be identified as A. oryzae; wild-type strains are A. flavus.
The clinically most important species of section Terrei is A. terreus, which is the second or third most common cause of IA in immunocompromised patients (Lass-Flörl et al. 2005, Blum et al. 2008). Of the 17 accepted species in section Terrei (Houbraken et al. 2020), five are human pathogens (A. alabamensis, A. citrinoterreus, A. floccosus, A. hortae (= A. hortai) and A. neoafricanus) (Zoran et al. 2018, Lackner et al. 2019). According to a recent report, infections caused by the A. terreus species complex were identified in 21 countries and 38 centres, and account for 5.2 % of all mould infections (Risslegger et al. 2017). However, a high incidence of A. terreus infections was reported in Innsbruck (Austria) and Houston (USA) (Lass-Flörl et al. 2007).
Species belonging to section Nigri (“the black Aspergilli”) are phenotypically very similar. Eight (A. brasiliensis, A. carbonarius, A. japonicus, A. luchuensis (= A. acidus), A. niger (= A. foetidus), A. tubingensis, A. uvarum, A. welwitschiae (= A. awamori) of the 30 accepted species in section Nigri are reported to cause infections in humans. The identified pathogenic species from this section are generally reported as the third leading causative agents of IA (Samson et al. 2014, Huang et al. 2017). Within this section, A. niger sensu stricto is the most prevalent clinical isolate (68.4 % cases vs. A. tubingensis, 31.6 % cases) (Balajee et al. 2009). However, recent analyses based on β-tubulin and calmodulin gene sequencing revealed a shift toward other cryptic species, including A. tubingensis, and A. welwitschiae, in different countries (Iatta et al. 2016, Hedayati et al. 2019, Alshehri & Palanisamy 2020, Carrara et al. 2020, Takeda et al. 2020).
The majority of the 74 accepted species in section Nidulantes are isolated from the soil, plant material, or the indoor environment (Sklenář et al. 2020). Eleven species have been isolated from patients with Aspergillus infections, of which A. nidulans was reported as the main agent of IA in different countries (Gabrielli et al. 2014, Chrenkova et al. 2018, Seyedmousavi et al. 2018, Tavakoli et al. 2020). Aspergillus nidulans was also reported as the second most frequently encountered mould in patients with chronic granulomatous disease characterised by sudden invasive features (Blumental et al. 2011, King et al. 2016b, Khalid & Ali 2018). Section Versicolores is a synonym of section Nidulantes, and series Versicolores is nowadays used. The clinically relevant species A. sydowii and A. creber belong to series Versicolores (Borgohain et al. 2019, Alshehri & Palanisamy 2020); of all 25 members in the section Usti, A. calidoustus is most often reported as the causal agent of invasive infections. Prior to the description of A. calidoustus, clinical strains were attributed to A. ustus and A. calidoustus are easy to differentiate, since the latter grows rapidly at 37 °C, while the former does not (Balajee et al. 2007a, Varga et al. 2008). Other members of the section isolated from proven/probable IA cases include A. granulosus, A. pseudodeflectus and A. ustus; A. insuetus, A. keveii, A. puniceus, A. pseudodeflectus and A. ustus were reported from respiratory samples (Fakih et al. 1995, Glampedakis et al. 2021).
In addition to the species mentioned above, other taxa can also cause infections in humans. Others include A. chevalieri, A. costiformis, A. glaucus, A. montevidensis, A. proliferans and A. pseudoglaucus (sect. Aspergillus) (Aznar et al. 1989, Naidu & Singh 1994, Traboulsi et al. 2007, Hubka et al. 2012, Alshehri & Palanisamy 2020); A. insulicola, A. melleus, A. ochraceopetaliformis, A. ochraceus, A. persii, A. sclerotiorum, A. subramanianii and A. westerdijkiae (sect. Circumdati) (Novey & Wells 1978, García-Martos et al. 2001, Harima et al. 2004, Brasch et al. 2009, Zotti et al. 2010, 2015, Hubka et al. 2012, Babamahmoodi et al. 2015, Bongomin et al. 2018, Seyedmousavi et al. 2018, Amri et al. 2020); A. flavipes (sect. Flavipides) (Seyedmousavi et al. 2018); A. tanneri (sect. Tannerorum) (Seyedmousavi et al. 2018); A. candidus (sect. Candidi) (Bongomin et al. 2018); and A. penicillioides and A. conicus (sect. Restricti) (Sklenář et al. 2017).
Virulence, immunology and pathogenesis of Aspergillus
In its natural environment, Aspergillus behaves as a saprobe that survives under different stress conditions. Likewise, during human infection, it has evolved adaptive mechanisms that allow it to withstand the unfavourable conditions in the lungs and to counter environmental changes in temperature, pH, water and nutrient balance, oxidative stress, and host molecules with antifungal properties.
Among the many virulence traits exhibited by Aspergillus, its plasticity in nutrient acquisition and metabolism confers a major advantage for growth during fungal infection under conditions of limited nutrient availability (Brock 2009, Blatzer & Latgé 2017). In addition, in experimental models of A. fumigatus infection, sites of hypoxia are commonly observed in the lungs, highlighting a remarkable ability to survive and thrive in conditions of low oxygen (Grahl et al. 2011, Kowalski et al. 2019). Besides these and several other relevant traits elicited in the context of infection (reviewed in Latgé & Chamilos 2019), the cell wall is a unique virulence factor, since it protects A. fumigatus from external aggression, while at the same time, it plays an active role in infection by influencing and modulating the immune response of the host (Latgé et al. 2017, van de Veerdonk et al. 2017). Owing to its dynamic structural properties according to morphotype, growth stage, and environmental conditions, the fungal cell wall is the main source of fungal ligands that activate the immune system (Latgé 2010). The physical barrier of the respiratory tract affords the first line of defence against inhaled conidia of Aspergillus, after which the respiratory epithelium is the initial point of contact with inhaled conidia (Filler & Sheppard 2006). Indeed, an increasing body of evidence has revealed a critical role of the airway’s epithelium in fungal clearance (Amich et al. 2020, Seidel et al. 2020) and production of cytokines and antimicrobial peptides (Bellanger et al. 2009, Sharon et al. 2011, Richard et al. 2018). Under certain conditions conidia escape the respiratory epithelium and are then challenged by cells of the innate immune system, including resident alveolar macrophages and dendritic cells (van de Veerdonk et al. 2017). Germinating conidia that escape macrophages are eliminated by recruited neutrophils and monocytes. Neutrophil extracellular traps (NETs) contribute to the innate host defence in vivo and neutrophils exert a considerable variety of antifungal effector functions, which include recognition, phagocytosis, intracellular clearance mediated by both oxidative and non-oxidative mechanisms, secretion of antimicrobial molecules and the release of neutrophil extracellular traps (NETs) (Urban & Backman 2020). Failure to prevent conidial germination results in hyphal growth, tissue invasion, and marks the initiation of fungal disease. Innate immune cells express a vast repertoire of pattern recognition receptors (PRRs) that recognise pathogen-associated molecular patterns in the fungus and activate effector functions, including phagocytosis and the production of proinflammatory cytokines and chemokines that orchestrate innate and adaptive immune responses (Patin et al. 2019). IL-8, also known as neutrophil chemotactic factor, is produced by macrophages and epithelial cells as an important chemoattractant for neutrophils, also during early phases of IA, where conidia are killed by local alveolar macrophages, and has been extensively used as biomarker for invasive aspergillosis (Winn et al. 2003, Camargo & Husain 2014, Gonçalves et al. 2017, Heldt et al. 2017, 2018, Jenks et al. 2019d). Up-regulation of gene transcription by A. fumigatus proteases has been suggested as cause of increased release of IL-8 by A549 pulmonary epithelial cells and primary epithelial cells (Borger et al. 1999). Other studies have shown that in vitro opsonization of A. fumigatus conidia with H-ficolin, L-ficolin and M-ficolin, which play essential roles in pathogen recognition and complement activation through the lectin pathway, potentiate IL-8 secretion of A549 lung epithelial cells (Houser et al. 2013, Bidula et al. 2015, Ghufran et al. 2017).
The family of C-type lectin receptors (CLRs) is the best-studied with regard to antifungal immunity (Brown et al. 2018). For example, the importance of dectin-1 in the recognition of β-1,3-glucan and activation of downstream immune responses has been confirmed in patients with recurrent fungal infections carrying an early stop codon polymorphism (Ferwerda et al. 2009). This polymorphism results in a truncated form of dectin-1 lacking several amino acids, with a detrimental effect on recognition of β-1,3-glucan and cytokine production after fungal stimulation (Ferwerda et al. 2009, Cunha et al. 2010). As a result, this polymorphism was found to predispose hematopoietic stem-cell transplant (HSCT) recipients to the development of IA in different patient cohorts (Cunha et al. 2010, Chai et al. 2011, Fisher et al. 2017). More recently, another CLR named MelLec was identified as the receptor for fungal melanin (Stappers et al. 2018). Macrophages from carriers of a polymorphism in the cytoplasmic tail of MelLec displayed a generalised defect in the production of cytokines after fungal stimulation. Likely owing to this defect, HSCT recipients receiving grafts from affected donors displayed a markedly increased risk for invasive pulmonary aspergillosis (IPA) after transplantation (Bassetti et al. 2020, Donnelly et al. 2020).
The efficiency of fungal recognition also relies largely on the opsonization by different soluble pattern recognition molecules, including collectins, pentraxins, ficolins and components of the complement pathway (Bidula & Schelenz 2016). One molecule that has received a great deal of recent attention in the field of aspergillosis is the long pentraxin-3 (PTX3) (Foo et al. 2015). This molecule binds microbial moieties from a wide range of microorganisms, including A. fumigatus (Garlanda et al. 2002). Accordingly, genetic variation in PTX3 was identified as a major risk factor for IPA after HSCT (Cunha et al. 2014), an association that was validated in independent cohorts of recipients of HSCT (Fisher et al. 2017) and solid organ transplant (Cunha et al. 2015, Wojtowicz et al. 2015), as well as in patients with chronic obstructive pulmonary disease (Cunha & Carvalho 2018, He et al. 2018). Mechanistically, genetic variants in PTX3 compromised the normal expression of the protein in the lungs and, at a cellular level, the antifungal effector mechanisms of neutrophils (Cunha et al. 2014). The impact of PTX3 deficiency on neutrophil function was confirmed in a recent study describing a similar association in patients with acute myeloid leukemia undergoing chemotherapy courses without pre-existing neutropenia (White et al. 2018).
The interaction of Aspergillus with the immune system is being harnessed to propose novel and improved fungal diagnostics, but also the implementation of clinical models aimed at the effective prediction of infection in high-risk patients. A recent study evaluating the combination of multiple genetic and clinical factors into a predictive model has demonstrated that such information could be used to successfully guide pre-emptive therapy in haematological patients (White et al. 2018). Besides improved diagnostics, functional analyses of genetic variation influencing susceptibility to aspergillosis may assist in the design of innovative and personalised immunotherapeutic approaches. This is illustrated by the preclinical evidence showing that genetic PTX3 deficiency can be rescued by the administration of the recombinant protein, a finding that supports its personalised use in specific patients at high-risk of infection. In conclusion, the success of novel diagnostic and immunotherapeutic approaches for aspergillosis may benefit from personalisation based on the interindividual variability in antifungal immune function.
Clinical significance of Aspergillus
Infection in humans
Aspergillus is the most common cause of mould infections in humans and can cause a variety of serious diseases in both immunocompetent and immunocompromised patients (Lass-Flörl 2019). The most clinically relevant Aspergillus species is A. fumigatus, followed by A. flavus, A. terreus and A. niger. Non-invasive infections in immunocompetent patients (e.g. with cystic fibrosis or post-tuberculosis) are allergic sinusitis or allergic bronchopulmonary aspergillosis (ABPA), fungal balls in the sinus or lung, chronic pulmonary aspergillosis, otitis externa or onychomycosis (Denning et al. 2018). For invasive infections, the respiratory tract is the most common primary site of IA –due to inhalation of conidia– but any organ might be involved as a single organ infection or as part of dissemination. Sino-nasal and cerebral aspergillosis may occur particularly in immunocompromised patients (Reischies & Hoenigl 2014). Aspergillus endocarditis is rare and risk factors include prior valve replacement, indwelling central venous catheters or broad-spectrum antibiotic treatment (Aldosari et al. 2020). Hematogenous spread to the spleen leads to either infarction or abscesses (Smolovic et al. 2018). Aspergillosis of the kidney is rare and derives from hematogenous dissemination or ascending from pan-urothelial aspergillosis or secondary to obstructive uropathy. Gastrointestinal tract IA may occur when the mucosal barrier is impaired (Chasan et al. 2013). Other rare manifestations of IA are (vertebral) osteomyelitis, arthritis, or subacute thyroiditis. Endophthalmitis may be consequent to intraocular surgery or trauma of the eye or after hematogenous spread and is associated with poor ocular prognosis (Dave et al. 2020).
Cutaneous aspergillosis may be caused by inoculation into disrupted skin or secondary to hematogenous dissemination (Lass-Flörl 2019). Primary extrapulmonary invasive aspergillosis often requires surgery in addition to systemic antifungal therapy (Reischies & Hoenigl 2014, Dave et al. 2020). Depending on the type of immunosuppression of the host, invasive pulmonary aspergillosis may present primarily angio-invasive in those with neutropenia, or primarily airway-invasive in those with corticosteroid associated immunosuppression, resulting in distinct radiological and clinical manifestations (Bergeron et al. 2012, Jenks et al. 2019b). The most common site of hematogenic spread is the central nervous system leading to brain abscess, stroke or less frequent to meningitis, and associated with devastating mortality rates (Hoenigl & Krause 2013).
Emergent susceptible population to acquire pulmonary aspergillosis
Mould-active prophylaxis has shown some success in reducing IA in patients with traditional risk factors for IA, such as those with underlying hematologic malignancy and prolonged neutropenia (Cornillet et al. 2006, Duarte et al. 2014). However, the prevalence of IA continues to increase in non-neutropenic patients with severe underlying diseases, including those in intensive care units (Pappas et al. 2010, Eigl et al. 2015, Bassetti et al. 2018, Schauwvlieghe et al. 2018b), solid organ transplant recipients (Lewis & Kontoyiannis 2009), those receiving systemic glucocorticoids (Chamilos et al. 2018), those with underlying structural lung damage (Prattes et al. 2014), those with advanced liver cirrhosis and liver failure (Prattes et al. 2017), those receiving tyrosine kinase inhibitors such as ibrutinib (Lenczuk et al. 2018), those with solid cancers (Yan et al. 2009, Bassetti et al. 2018), and others (Guinea et al. 2010, Prattes et al. 2014, Bassetti et al. 2018, Ghez et al. 2018).
Aspergillus species, especially A. fumigatus, can cause co-infection with viruses, including cytomegalovirus, and –importantly– influenza virus leading to complication of management of patients inflicted (Schauwvlieghe et al. 2018b). In one multicentre study from the Netherlands and Belgium, invasive pulmonary aspergillosis was diagnosed in 83 (19 %) of 432 patients admitted with influenza to the ICU, a median of 3 days after admission to the ICU (Schauwvlieghe et al. 2018b), and independently associated with mortality. In another study from Canada, IA complicated only 7.2 % of influenza associated respiratory failure ICU admissions, although the rate varied between 0 and 23 % between influenza seasons (Schwartz et al. 2020).
In November 2019, a novel virus termed severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was first identified as the causative agent of pneumonia from a cluster of individuals with pneumonia in Wuhan City, Hubei province, China (WHO 2020). Since the first reported case, SARS-CoV-2, which causes the disease now called coronavirus disease 2019 (COVID-19), has spread throughout China and to almost every country in the world as of May 2020. Although the vast majority of COVID-19 cases are mild to moderate, up to 20 % of patients with symptomatic COVID-19 may develop acute respiratory distress syndrome (ARDS) (Wang et al. 2020, Wu & McGoogan 2020, Xu et al. 2020). There is increasing evidence that COVID-19 patients are at risk of developing COVID-19 associated invasive pulmonary aspergillosis (CAPA) co-infection, with more than 100 cases reported to date (Blaize et al. 2020, Dupont et al. 2020, Gangneux et al. 2020, Hoenigl 2021, Koehler et al. 2020b, Lescure et al. 2020, Mitaka et al. 2020, van Arkel et al. 2020, Verweij et al. 2020, Bartoletti et al. 2021, Prattes et al. 2021, White et al. 2021). The pathophysiology of COVID-19 is not well understood, but leukopenia, lymphopenia and T-cell perturbations, including immune dysregulation impacting Th2 as well as Th1 responses in severe COVID19, are frequently observed among symptomatic patients (Chen et al. 2020a, 2020b, Huang et al. 2020, Zheng et al. 2020), and may predispose patients to fungal diseases. Furthermore, the utilisation of – often high dose – systemic and inhaled glucocorticoids, which may further predispose to opportunistic infections such as CAPA, has been described in close to 50 % of patients with COVID-19-associated ARDS (Wang et al. 2020). This immune dysregulation together with epithelial lung damage stemming from COVID-19 immunopathology may facilitate Aspergillus superinfection.
From Wuhan, epidemiological studies indicate that invasive fungal infections may occur in 4–5 % of COVID-19 episodes requiring ICU admission (Baxter et al. 2011, Yang et al. 2020). In a cohort from China, fungal infections were diagnosed in seven (3 %) out of 221 COVID-19 patients, all of whom were admitted to the ICU (Zhang et al. 2020). Given that in Wuhan, galactomannan (GM) testing is rarely available and fungal diagnostics are sparse (Chindamporn et al. 2018), this is likely an underestimate of the true burden of IA in patients with SARS-CoV-2 requiring ICU admission. A study from the United Kingdom reported a 14.1 % incidence of pulmonary aspergillosis among 135 ICU patients (White et al. 2021). Higher rates of CAPA were recently reported in a single centre study from the Netherlands, where a high incidence (19.4 %) of putative aspergillosis was observed in a cohort of 31 mechanically ventilated ICU patients with COVID-19 (van Arkel et al. 2020). In another study from Germany, five of 19 (26 %) consecutive critically ill COVID-19 patients with ARDS were diagnosed with putative CAPA, highlighting that rates may be comparable to those observed in association with severe influenza (Koehler et al. 2020b). In a study from Italy, probable CAPA was diagnosed in 30 (27.7 %) patients after a median of 4 (2–8) days from intensive care unit (ICU) admission (Bartoletti et al. 2021). In a study from France, putative CAPA was reported in 33 % of 27 critically ill COVID-19 patients in an enriched population undergoing bronchoscopy (Alanio et al. 2020). Finally, the highest rate of CAPA has been noted in Belgium reaching 35 % (7/20) of the COVID-19 patients presented with ARDS (Rutsaert et al. 2020). The vast majority of reported CAPA cases lacked EORTC/MSGERC host factors, highlighting the need for improved criteria for defining IPA in non-neutropenic patients, as reported elsewhere (Jenks et al. 2019b). Additional cases of fatal CAPA were reported from Argentina, Australia, Austria, Brazil, France, Ireland, Italy, Pakistan, Switzerland, United States and many other countries (Blaize et al. 2020, Mitaka et al. 2020, Prattes et al. 2021). Importantly, three cases of azole resistance have been reported, indicating that ARAF is emerging also among the ICU population at risk for IPA (Meijer et al. 2020, Ghelfenstein-Ferreira et al. 2021, Mohamed et al. 2021).
As clinical presentation and imaging findings of COVID-19 and IPA may overlap (fever, shortness of breath, cough, unspecific infiltrates and consolidations, halo sign), biomarker and culture based diagnostic work-up is essential. Serum GM may have imperfect sensitivity of 20 % and below (Alanio et al. 2020, Koehler et al. 2020b). While reasons for the lower sensitivity in CAPA versus influenza associated pulmonary aspergillosis are unknown, treatment with chloroquine, which exhibits in vitro activity against A. fumigatus (Henriet et al. 2013), may have explained the lower sensitivity in some of the earlier studies, given that exposure to antifungals is a well-known factor that decreases the sensitivity of GM-testing and may explain the lower sensitivity compared to influenza associated IPA (Eigl et al. 2015).
Future studies are needed to evaluate other blood tests for CAPA, including Aspergillus PCR (Egger et al. 2020), β-D-glucan (Heldt et al. 2018), and the two newly CE-marked point of care tests, the Aspergillus GM lateral flow assay (LFA) and the Aspergillus-specific lateral flow device test (Eigl et al. 2015, Jenks et al. 2019c, 2019e, Mercier et al. 2020, Wahidi et al. 2020). Further complicating diagnosis of CAPA is the extremely limited role of bronchoscopy in COVID-19 as this aerosol generating procedure increases the risk of exposure for patients and personnel (Jenks et al. 2020), although detailed instructions on how to safely perform bronchoscopy have been published (Koehler et al. 2020c). In some centres, however, collection of tracheal aspirates remains the preferred method for diagnosis. Although Aspergillus can be detected in sputum and tracheal aspirates in CAPA-patients, its presence might reflect oral pharyngeal colonisation as Aspergillus is considered a core component of the basal oral mycobiome (Krüger et al. 2019). Importantly, GM-testing, presented in detail below, which is an important tool for IPA diagnosis in BALF specimens and represents active growth (Eigl et al. 2017), is not validated for upper respiratory tract specimens.
These early findings suggest IA may be an important and underrecognised complication of SARS-CoV-2 infection, due to the absence of typical host factors. Since bacterial and fungal superinfections are difficult to distinguish from severe COVID-19 based on clinical or imaging findings, histopathology has a central role in determining prevalence and also outcomes of CAPA (reviewed in Arastehfar et al. 2020a). However, autopsies of COVID-19 patients are rarely performed due to the risk of aerosol transmission. Criteria for defining COVID-19 associated aspergillosis have been developed and will help classifying this important disease (Koehler et al. 2020a). The frequency of post-COVID-19 aspergillosis is likely to differ significantly between hospitals and geographic sites, and environmental factors may also play a large role in increasing exposure beyond what would normally be encountered within hospitals and ICUs. The rapid spread of COVID-19 to a non-immune population has been seen in temporary facilities/hospitals rapidly assembled that do not adhere to the rigorous ventilation requirements that are present within permanent hospitals. These temporary sites are essential to increase healthcare capacity; however, dust and construction-related increases in ambient air spore counts will very likely increase patient colonisation with Aspergillus and other fungal species predisposing to infection. Finally, drug-drug interactions may limit the use of voriconazole (Jenks et al. 2019a), the gold-standard treatment for IA in the ICU. Future studies need to evaluate effectiveness of isavuconazole (Jenks et al. 2018), and new antifungals currently under development like fosmanogepix or olorofim (Kupferschmidt 2019), which may have comparable efficacy without the same burden of drug-drug interactions, while other drugs such as rezafungin or ibrexafungerp may offer options for combination therapy or even prophylaxis.
Diagnosis of aspergillosis in clinic – Serology to PCR
For the diagnosis of IA, culture and microscopy are essential, but show limited sensitivity. Detection of the fungal cell wall component galactomannan (GM) has therefore become the imperfect gold standard (Hoenigl et al. 2012, 2013b, Duettmann et al. 2014, Eigl et al. 2017, Rawlings et al. 2019) as it is more sensitive than culture. Galactomannan is a polysaccharide that exists primarily in the cell walls of Aspergillus species (Zhou et al. 2017) and a commercially-available double sandwich enzyme immunoassay (EIA) utilises the monoclonal antibody EB-A2 (Platelia™, Bio-Rad, Marnes-la-Coquette, France) to detect the GM antigen. It is approved by the U.S. Food and Drug Administration (FDA) for testing of GM from serum and bronchoalveolar fluid (BALF). This antibody detects multiple epitopes on galactofuranose side chains that link to the mannan backbone (Kudoh et al. 2015), although galactofuranose is not specific to Aspergillus and is present in other fungi such as Fusarium species (Tortorano et al. 2012), Penicillium species (Huang et al. 2007), and Histoplasma species (Wheat et al. 2007). The current sensitivity and specificity of BALF GM is 82 % and 81 %, respectively (Rawlings et al. 2019). The optimal optical density index (ODI) threshold is debatable, although the FDA considers an ODI of ≥ 0.5 to be positive for GM in both serum and BALF.
Galactomannan from BALF has shown better diagnostic performance for IA than GM from blood, particularly in patients on mould-active antifungal prophylaxis (Heldt et al. 2018). In a systematic review investigating the accuracy of GM from BALF for the diagnosis of IA in immunocompromised patients, the sensitivity and specificity of GM was 0.88 and 0.81, respectively, at an ODI of 0.5; at an ODI of 1.0 the sensitivity was 0.78 and specificity 0.93 (de Heer et al. 2019). Particularly in non-neutropenic patients a higher cut-off of 1.0 ODI in BALF may be preferable (Prattes et al. 2014) as false-positive results may occur with the lower cut-off (Martinelli et al. 2019). Given the airway-invasive growth, BALF GM is always the preferable option for IA diagnosis in non-neutropenic patients, and GM from BALF has found to be superior to GM from serum (Bassetti et al. 2020).
Molecular tests such as PCR (Heldt et al. 2018, Prattes et al. 2018, Jenks et al. 2019f, Rawlings et al. 2019) have emerged as alternative options to diagnose IA and are widely used (Hoenigl et al. 2014b, Buchheidt et al. 2017), although there is a lack of standardisation (White et al. 2010) and a large variation in diagnostic performance across studies and settings (White et al. 2015b, Springer et al. 2016). PCR is most useful in high-risk groups such as neutropenic patients who are not receiving mould-active prophylaxis, where a negative result is reassuring in ruling out IA. In other settings including non-neutropenic patients and patients at low risk for IA, like GM the utility of PCR testing is limited, particularly from blood (Egger et al. 2020), and overall it suffers from poor precision, with a specificity of 76 % (Arvanitis et al. 2014). Performance of blood PCR is particularly poor in patients on mould-active prophylaxis (Egger et al. 2020).
Two point-of-care tests are now available for the diagnosis of IA. The Aspergillus-specific Lateral Flow Device (LFD) test (OLM Diagnostics, Newcastle Upon Tyne, United Kingdom) detects an extracellular mannoprotein antigen secreted exclusively during active growth of Aspergillus species via the JF5 monoclonal antibody (Hoenigl et al. 2014b, 2018, Prattes et al. 2015, Orasch et al. 2017). Another new test, the sōna Aspergillus GM-LFA (IMMY, Norman, OK, United States) detects GM but has a shorter turnaround time compared to the conventional GM ELISA test. In patients with hematologic malignancy, both the LFD and LFA have a comparable sensitivity and specificity to GM from BALF for diagnosing IA (Heldt et al. 2018, de Heer et al. 2019, Jenks et al. 2019c, 2021, Mercier et al. 2020), and sensitivity increased further when combined with inflammatory markers, triacetylfusarinine C, or PCR (Hoenigl et al. 2019, Jenks et al. 2019d, 2019e, Rawlings et al. 2019). In non-neutropenic patients, the LFA and LFD have demonstrated a sensitivity and specificity between 60–70 % when used alone and up to 80 % when used in combination (Jenks et al. 2019b), with tendencies towards better performance of the LFA (Jenks et al. 2020). Particularly the LFA, but also the LFD have also shown promise for diagnosing IA in serum samples from patients with haematological malignancies (Jenks & Hoenigl 2020). Unmet needs for the diagnosis of IA include a true point-of-care test that can be done at the bedside or in the clinic in the matter of minutes. In addition, improved diagnostic algorithms to diagnose IA in non-neutropenic patients are needed as well.
The detection of Aspergillus serum precipitin antibodies (i.e., subsets of IgG and IgM antibodies) is useful for the diagnosis of ABPA (Agarwal et al. 2013). The detection of Aspergillus IgG antibodies via commercial ELISAs or POC tests (Richardson & Page 2018) is the mainstay diagnostic test for aspergilloma (Hope et al. 2005) and chronic pulmonary aspergillosis (CPA) (Denning et al. 2016), when used in conjunction with pulmonary imaging, but Aspergillus IgG lacks the specificity in the diagnosis of IA (Richardson & Page 2017), and GM and also the LFD have limited sensitivity in those with CPA (Salzer et al. 2018). Aspergillus IgG are also present in Aspergillus bronchitis, Aspergillus nodule and chronic rhinosinusitis, and can be used for treatment stratification in CPA (Denning et al. 2018).
Clinical treatment of aspergillosis
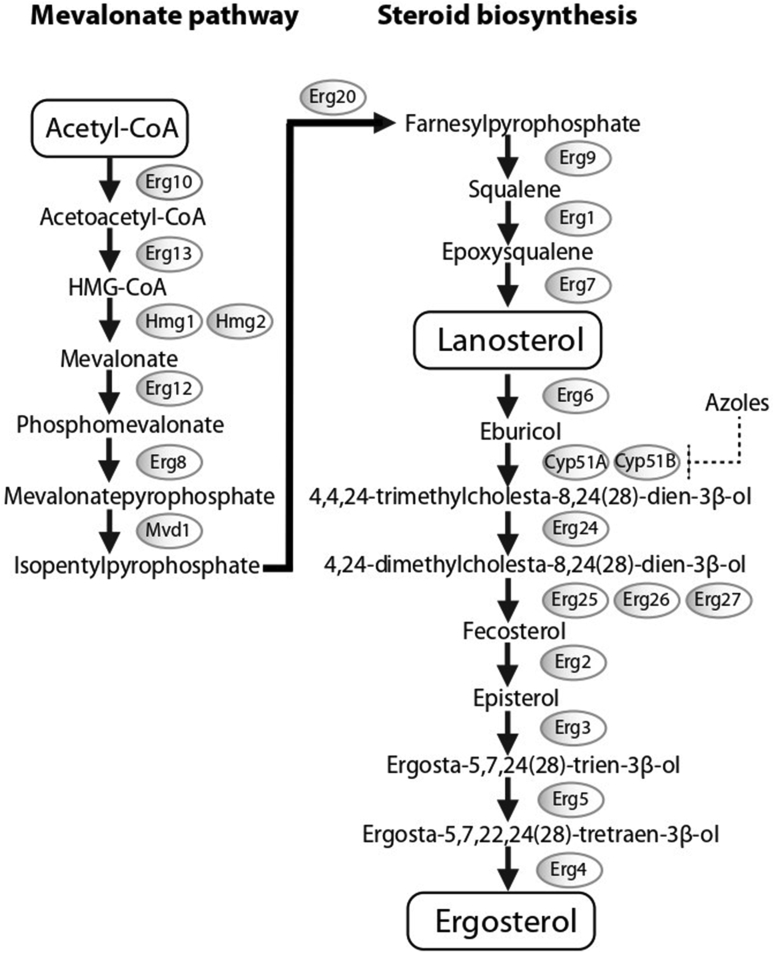
Triazoles, particularly voriconazole, isavuconazole and posaconazole for invasive infections, and voriconazole or itraconazole for chronic infections, are the first line antifungal agents used to treat aspergillosis (Denning et al. 2016, Cornely et al. 2019a, Jenks et al. 2019a). Voriconazole and isavuconazole exert fungicidal activity by inhibiting ergosterol biosynthesis. Ergosterol is one of the main structural components of the fungal cell membrane, allowing membrane fluidity, proper positioning and function of membrane-integrated proteins, and the cell cycle. Azoles bind and inhibit at the active site of the cytochrome P450 enzyme lanosterol 14-α-demethylase, which contains a heme cofactor and is encoded by two homologous genes, CYP51A and the CYP51B (a third homolog CYP51C has been found in A. flavus) (Dudakova et al. 2017). This induces the accumulation of 14-methyl sterols, such as lanosterol, which alters functions of cell membrane leading to fungal growth arrest. Furthermore, inhibition of ergosterol biosynthesis results in both accumulation of toxic sterol intermediates and creation of cell wall carbohydrate patches that extend to the plasma membrane (Geiβel et al. 2018). Drug-drug interactions may limit the use of voriconazole and other triazoles not only in the ICU setting, but also in patients with haematological malignancies where some newer drugs, including ibrutinib, venetoclax, and midostaurin, may complicate the use of triazoles (Groll et al. 2017, Tapaninen et al. 2020).
The therapeutic approach of IA has been changed with the development of the second-generation mould-active triazoles, voriconazole, posaconazole and isavuconazole in addition to the first-generation azole itraconazole. The chemical structure of voriconazole closely resembles fluconazole and shares a similarity with isavuconazole, while posaconazole more closely resembles itraconazole. Voriconazole was derived from fluconazole by replacing one triazole moiety in fluconazole with a 4-fluoropyrimidine group and adding an α-methyl group (Bellmann & Smuszkiewicz 2017). Posaconazole derives from itraconazole through the replacement of the chlorine substituents with flourine in the phenyl ring, as well as hydroxylation of the triazolone side chain (https://pubchem.ncbi.nlm.nih.gov/). Unlike the other second-generation triazoles, isavuconazole is administered as a prodrug; the isavuconazoilum sulfate ester which is hydrolysed rapidly by serum esterases, is highly water soluble and does not require the addition of a beta-cyclodextrin to facilitate solubility (Jenks et al. 2018).
The availability of both intravenous and different oral formulations of triazoles increases the therapeutic options and improves their pharmacokinetics (Table 1) (Cornely et al. 2019b). The variable pharmacokinetics (80–100 %) of voriconazole (oral solution, tablet and intravenous [IV] formulation) and the oral solution of posaconazole due to erratic hepatic metabolism and absorption, respectively, have been improved with the new formulations of posaconazole i.v./tablet and isavuconazole i.v./capsules (∼50 % variation) (Jović et al. 2019). The bioavailability of oral formulations ranges from poor with the “old” posaconazole oral solution to high with isavuconazole, and intake with fatty food is most important for the posaconazole oral solution (Hoenigl et al. 2014a). All four azoles exhibit non-linear pharmacokinetics because of saturable absorption or metabolism except isavuconazole (Bellmann & Smuszkiewicz 2017). They are highly protein bound (> 98 %) except voriconazole (58 %). All azoles are characterised by a large volume of distributions (3–11 lt/kg) and they are extensively metabolised with minimal amounts of parent drug excreted renally or hepatobiliarily. Their half-lives and total drug exposure area under curve (AUC) varies from short with voriconazole (6–12 h and 13–16 mg∗h/l) to long with isavuconazole (110–130 h and 98–121 mg∗h/l, respectively) (Table 1). As will be discussed in detail later in this review, the emergence of ARAF isolates threatens the efficacy of azoles, and lipid formulations of amphotericin B, as well as echinocandins (which should be used in combination with another antifungal drug) are alternative treatment options for IA (Patterson et al. 2016). Surgery in addition to systemic antifungal therapy plays an important role in the treatment of primary extrapulmonary invasive aspergillosis (Aldosari et al. 2020).
Table 1.
Pharmacokinetic and pharmacodynamic properties of anti-Aspergillus azoles.
| Parameters | Voriconazole | Posaconazole | Isavuconazole | Itraconazole |
|---|---|---|---|---|
| Chemical structure | 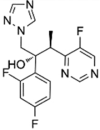 |
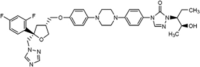 |
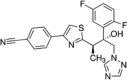 |
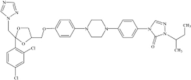 |
| Molecular weight | 349.3 | 700.8 | 717.77 | 705.64 |
| Water solubility (mg/mL) | 0.0978 | 0.012 | 0.0162 | 0.00964 |
| Log D (pH 7.4) | 1.8 | 2.15 | 4.14 | > 5 |
| Formulations | Oral solution, tablet, iv | Oral solution, tablet, iv | Capsules, iv | Oral solution, tablet, capsules, iv |
| Standard dose | LD: 6 mg/kg q12h, MD: 4 mg/kg q12h or 200 mg PO | Oral = 200 mg q6h/q8h or 400 mg q12h Tablet / iv = 300 mg q24h |
LD: 200 mg q6h for 2 d; MD: 200 mg q24h | Oral = LD: 800 mg q12h/g24h for 2 d, MD: 200 mg q12h IV = LD: 200 mg q12h for 2d, MD: 200 mg q24h |
| Dose adjustment | < 40 kg = 50 % MD dose, RI = avoid iv1, HI = 50 % MD, IR = 300mg q12h | RI = avoid iv1 | No | RI = avoid iv1 |
| Tmax | 1.43–1.81 | Oral = 3, tablet = 2.2, iv = 1.5 | Oral = 2–3, iv = 0.75–1 | Oral = 5, Iv = 1 |
| Bioavailability | 90 % (healthy) / 83 % (patients) | Oral = 8–47 % | 98 % | Capsules = 22 %; oral solution = 55–92 % |
| Effect of food | 22 % reduction | Oral = ↑ with high fat meal/low pH, tablet = ↓ mucositis, ↑ with high fat meal | No | Capsules = ↑ with high fat meal/low pH, oral solution = ↑ empty stomach |
| Protein binding | 42–58 % | 98–99 % | 99 % | 99.8 % |
| Vd (Lt/kg) | 2–4.6 | Oral = 3.7, tablet = 5, iv = 20 | 6.42 | 11 |
| Tissue penetration | Brain, ELF | Alveolar cells, liver kidney, lung, myocardium | Muscle, fat, liver, brain | Skin, fat, liver, lung, kidney, spleen, bone, muscle |
| CL (mL/min/kg) | 3–8.3 | 9.39–16.4 L/h | 1.9–5 | 5.1 |
| Hepatic/Urinary (metabolites and drug) excretion % | 20 (M)+< 1(D) / 80 (M)+< 2(D) | 77 (M)+< 5(D) / 14 (M)+< 5(D) | 46 (M) / 46 (M)+0.38(D) | 54 (M)+< 0.03(D) / 35 (M)+3–18(D) |
| Metabolizing enzymes | CYP2C19 > CYP2C9,CYP3A4 | UGT1A4 | CYP3A4, CYP3A5, UGT | CYP3A4, CYP2C9, CYP2C19 |
| T1/2 (h) | 6–12 | 27–35 | 110–130 | 24 |
| Cmax (mg/L) | 4.84 (300 mg), 5.27 (400 mg) | Oral = 0.851, tablet = 1.96, iv = 3.28 | 2.6 | 0.5–2.3 |
| AUC0–24 (mg∗h/mL) | 13.21–16.38 | Oral = 17.24, tablet / iv = 34.3 | 98–121 | 29.2 |
| Variation in AUC0–24 | 82 % | Oral = 82 %, tablet / iv = 35–40 % | 10–43 % | 30–60 % |
| Pharmacokinetics | Non-linear (saturable metabolism) | Oral = non-linear (saturable absorption), tablet / iv = linear | Linear | Non-linear (saturable absorption and metabolism) |
| tAUC/MIC (50 % survival in animals) | 17.61–21.96 | 167–178 | 25 | NA |
| TDM targets (mg/L) | Prophylaxis: Cmin > 0.5; therapy: Cmin > 1–2; toxicity: Cmin < 4–6 | prophylaxis: Cmin > 0.5–0.7; therapy: Cmin > 1–1.25 | NA | Prophylaxis: Cmin > 0.5; therapy: Cmin > 0.5–1; toxicity: Cavg < 17 (bioassay), < 3.5 (HPLC) |
NA, not applicable.
Unless an assessment of the benefit/risk to the patient justifies the use of intravenous formulations.
Antifungal combination therapy with voriconazole or amphotericin B and an echinocandin is often employed as primary or salvage therapy for management particularly of refractory aspergillosis (Elefanti et al. 2013). Resistance to first line triazole antifungal agents among Aspergillus species has resulted in the increased use of second-line monotherapy with echinocandin drugs (caspofungin, micafungin or anidulafungin) (Aruanno et al. 2019). Echinocandin class drugs inhibit the cell wall biosynthetic enzyme β-(1,3)-d-glucan synthase (Perlin 2015), and were initially approved by the FDA for the treatment of invasive aspergillosis refractory to conventional therapy (Johnson & Perfect 2003). Given a strong 20-year history of safety and efficacy, it is being used increasingly in patients being treated for chronic pulmonary aspergillosis. A recent meta-analysis of 12 studies covering 380 patients who received IV antifungals, either amphotericin B (n = 143) or an echinocandin (n = 237) reported a response of 58 % for amphotericin B and 62 % for echinocandins (micafungin). Echinocandins, especially micafungin are well-tolerated and effective prophylactic antifungal agents used in patients with hematologic diseases at high risk for invasive mould infections (Ziakas et al. 2014, Park et al. 2019).
Worldwide emergence of azole-resistant A. fumigatus: environmental and clinical routes
Azole drugs play a major role in prevention and treatment of infections caused by Aspergillus species. Azole drugs belong to a wider group of compounds called demethylation inhibitors (DMIs) whose common target is the 14-α sterol demethylase. Demethylation inhibitors are widely used in the clinical setting, both as treatment and prophylaxis, but also in the agriculture setting due to their high-efficiency and broad-spectrum activity (Price et al. 2015, Hollomon 2017).
Several azole-based fungicides used in agriculture to protect cereals and wine from phytopathogenic moulds have a similar chemical structure to medical triazoles and the development of cross-resistance between them has been proved (Snelders et al. 2012, Zhang et al. 2017). A large number of demethylation inhibitor fungicides have been used intensively in agriculture and medicine (human and veterinary) since 1970 (Rochette et al. 2003). Though azole fungicides are not used to target A. fumigatus, it transpires that many of these fungicides are active against A. fumigatus, a condition that led to the emergence of resistance. More than 30 azole fungicides have been studied for their in vitro activity against A. fumigatus, including propiconazole, bromuconazole, epoxiconazole, difenconazole, and tebuconazole. In this context, one of the presumed routes for triazole-resistance emergence is through selection pressure from the DMIs used as fungicides in the environment (Verweij et al. 2007, 2009). The resulting azole resistant isolates are associated with a particular resistance mechanism constituted by a variable number of tandem repeat (TR) integrations in the CYP51A promoter followed or not by point mutations in the coding gene (Snelders et al. 2008). There is a lot of evidence that supports the idea of an environmental driven mechanism, such as the fact that these TR azole resistant strains (TR34/L98H, TR34/L98H/S297T/F495I, TR46/Y121F/T289A, and TR53) have been detected throughout the world (Garcia-Rubio et al. 2017) but showed shorter genetic distances among them than with other azole-susceptible and non TR-resistant isolates, which suggests that they could have developed from a reduced set of clonally related strains (Snelders et al. 2008, Camps et al. 2012b, Garcia-Rubio et al. 2018b). Besides, the fact that TR azole resistant strains have been isolated from azole-naïve patients with IA also endorses the existence of this environmental resistance route, indicating that infections would have been caused by the inhalation of already ARAF spores harbouring aforementioned mutations in CYP51A (Snelders et al. 2009, Verweij et al. 2009). However, other single mutations occurring in CYP51A arise during the course of azole therapy (Howard et al. 2006, 2009, Albarrag et al. 2011, Camps et al. 2012c, Wiederhold et al. 2016).
Nevertheless, there are also some findings that support that TR azole-resistance mechanisms do not seem to be restricted to the environment. A clinical case of fatal aspergillosis caused by an A. fumigatus strain that developed a TR120 insertion in the CYP51A promoter during long-term azole treatment has been recently reported using both STRAf typing and whole-genome sequencing (Hare et al. 2019). This challenges the existence of a link between resistance mechanisms and specific routes of resistance selection and may fade the presumed boundaries between the environmental and clinical routes of resistance. In line with these facts, strains carrying G432S and G432A mutations have been isolated from azole-naïve patients (Howard et al. 2006, 2009, Alanio et al. 2011, Albarrag et al. 2011), while strains carrying TR53 (and also TR120) mutations have been isolated from patients exposed to azole antifungals in prior treatments (Hodiamont et al. 2009, Hare et al. 2019). Furthermore, studies about how these supposedly environmental resistance mechanisms originated are still scarce, although it has been hypothesised that environmental conditions or even a more complex reproductive method, such as sexual reproduction, could play a role. In addition, the dispersion of A. fumigatus spores from the human lung into the environment has been suggested lately as a possible transmission path in cystic fibrosis patients as an alternative transmission route from patient to environment (Engel et al. 2019). Although azole resistance may predominantly originate from environmental sources, further research is warranted in order to gain a deeper knowledge about how azole resistance emerges and is transmitted, which has implications for patient management.
The worldwide clinical burden of ARAF
The increasing burden of azole resistance on a global scale notoriously limits the therapeutic options to treat aspergillosis (Denning & Perlin 2011). Over the last two decades, a rapid local and global emergence of triazole resistance has been observed. The first ARAF isolates were obtained from two patients treated with itraconazole, one of whom died early 1990, in California, in a case that dates back to the late 1980s (Denning et al. 1997b). A Dutch study later reported three ARAF isolates recovered after long-term itraconazole treatment from a lung transplant recipient in 1997 (Verweij et al. 2002). Moreover, a study in France found four itraconazole-resistant isolates with high itraconazole minimum inhibitory concentrations (MICs) values, > 16 mg/L, in 1999 (Dannaoui et al. 1999b). Later, in 2007, a comprehensive study of nine cases of azole-resistant IA found that four out of nine patients had never previously been treated with azole antifungals (Hussain et al. 2007).
Studies have investigated the distribution of ARAF in relation to the TR34/L98H mutation, which was first found in the Netherlands in 1998 (Snelders et al. 2008). Indeed, surveillance studies and case series recently reported the global presence of ARAF harbouring TR34/L98H, with reports from Europe, the Middle East, including Australia, Tanzania, Kuwait and Iran, North and South Asia, including China and Japan, Australia and Tanzania; Africa, and North and South America, including Brazil and Columbia (Mellado et al. 2007, Verweij et al. 2007, Snelders et al. 2008, Howard et al. 2009, Lockhart et al. 2011, van der Linden et al. 2011, 2013, Chowdhary et al. 2012a, 2015, Rath et al. 2012, Seyedmousavi et al. 2013b, Astvad et al. 2014, Ahmad et al. 2015, Choukri et al. 2015, Fuhren et al. 2015, Liu et al. 2015, Steinmann et al. 2015, Vermeulen et al. 2015, Wu et al. 2015, 2020, Chen et al. 2016b, Jensen et al. 2016, Lazzarini et al. 2016, Mushi et al. 2016, Nabili et al. 2016, Perveen et al. 2016, Wiederhold et al. 2016, Koehler et al. 2017, Montesinos et al. 2017, Prigitano et al. 2017, Toyotome et al. 2017, Abdolrasouli et al. 2018a, 2018b, Berkow et al. 2018, Denardi et al. 2018, Pinto et al. 2018, Riat et al. 2018, Seufert et al. 2018, Talbot et al. 2018, Gonzalez-Lara et al. 2019, Rivero-Menendez et al. 2019b, Tsuchido et al. 2019, Li et al. 2020, Pontes et al. 2020) (Table 2). These studies describe the most recent discoveries of the TR46/Y121F/T289A resistance mechanism involving environmental mutations, which the Netherlands first reported in 2009 (van der Linden et al. 2013); another report revealed that a US patient was recovering from TR46/Y121F/T289A A. fumigatus infection in 2008 (Wiederhold et al. 2015).
Table 2.
Diversity and prevalence of CYP51A mutations causing azole resistance in clinical A. fumigatus isolates.
| MIC values (μg/mL) |
Azole exposure prior resistance |
Azole therapeutic failure |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Country (year; number of isolates) | Amino acid substitutions (total number) (references) | VRZ | PSZ | ITZ | Present | Naïve | VRZ | PSZ | ITZ |
| the Netherlands (1994–2013; 186), Germany (2002–2018; 111), Italy (1995–2006, 2013–2015; 28), China (2008–2016; 19), Belgium (2011–2016; 18), Taiwan (2011–2018; 16), Denmark (2010–2014; 12), India (2008–2014; 11), UK (1992–2017; 11), USA (2001–2017; 7), Iran (2003–2016; 5), Tanzania (2016; 5), France (2012; 3), Spain (2014–2018; 3), Brazil (2014–2018; 3), Pakistan (2016; 3), Japan (2016–2018; 3), Switzerland (2016; 2), Portugal (2010–2016; 2), Kuwait (2015; 2), Mexico (2014–2017; 2), Australia (2015–2017; 1) | TR34 / L98H (T297S / F495I) (453) (Mellado et al. 2007, Verweij et al. 2007, Snelders et al. 2008, Howard et al. 2009, Lockhart et al. 2011, van der Linden et al. 2011, 2013, Rath et al. 2012, Seyedmousavi et al. 2013b, Ahmad et al. 2015, Chowdhary et al. 2015, Liu et al. 2015, Vermeulen et al. 2015, Wu et al. 2015, 2020, Chen et al. 2016b, Jensen et al. 2016, Lazzarini et al. 2016, Mushi et al. 2016, Nabili et al. 2016, Perveen et al. 2016, Wiederhold et al. 2016, Koehler et al. 2017, Montesinos et al. 2017, Prigitano et al. 2017, Abdolrasouli et al. 2018b, Berkow et al. 2018, Denardi et al. 2018, Pinto et al. 2018, Riat et al. 2018, Seufert et al. 2018, Talbot et al. 2018, Gonzalez-Lara et al. 2019, Rivero-Menendez et al. 2019b, Tsuchido et al. 2019, Pontes et al. 2020) | 0.5–>16 | 0.125–>8 | >4–32 | Yes FLZ VRZ ITZ PSZ |
Yes | Yes | Yes | Yes |
| the Netherlands (2009–2013; 29), Belgium (2011–2016; 11), Denmark (2010–2014; 6), Germany (2015–2018; 3), USA (2001–2016; 3), China (2010–2016; 3), France (2014; 2), Spain (2014–2018; 2), UK (2016; 1), Portugal (2010–2016; 1), Argentina (2009; 1), Japan (2013; 1), Taiwan (2011–2018; 1) | TR46 / Y121F / T289A (64) (Verweij et al. 1998, Kuipers et al. 2011, Lockhart et al. 2011, Vermeulen et al. 2012, 2015, Montesinos et al. 2013, van der Linden et al. 2011, 2013, Astvad et al. 2014, Chen et al. 2015, 2016b, Peláez et al. 2015, Steinmann et al. 2015, Jensen et al. 2016, Vazquez & Manavathu 2016, Wiederhold et al. 2016, Lavergne et al. 2017, Moore et al. 2017, Isla et al. 2018, Pinto et al. 2018, Seufert et al. 2018, Rivero-Menendez et al. 2019b, Li et al. 2020, Wu et al. 2020) | >8–>32 | 0.125–2 | 1–>16 | ITZ, VRZ PSZ FLZ |
Yes | Yes | Yes | Yes |
| The Netherlands (2009; 1) | TR53 (1) (Hodiamont et al. 2009) | 16 | 0.5 | 16 | ITZ | Yes/No | No | Yes | |
| Denmark (2016; 1) | TR120 (1) (Hare et al. 2019) | 4 | 0.5 | 16–>16 | Yes VRZ PSZ |
Yes | Yes | ND | |
| Germany (2018; 8), Japan (2012; 8), Italy (1995–2006; 7), UK (1992–2009; 6), USA (2001–2014; 5), China (2001; 3), Australia (2015–2017; 2), the Netherlands (2007–2009; 2), India (2011–2014; 2), Spain (2014–2018; 2), Denmark (2010–2014; 1) | G54R/E/W (46) (Chen et al. 2005, Bueid et al. 2010, van der Linden et al. 2011, Camps et al. 2012c, Tashiro et al. 2012, Chowdhary et al. 2015, Wiederhold et al. 2015, 2016, Jensen et al. 2016, Lazzarini et al. 2016, Talbot et al. 2018, Rivero-Menendez et al. 2019b) | 0.125–4 | 1–>16 | 1–>16 | Yes ITZ PSZ VRZ |
ND | Yes | ND | Yes |
| UK (1992–2007; 16), USA (2001–2014; 2) | G138C/S (18) (Howard et al. 2006, 2009, Albarrag et al. 2011, Wiederhold et al. 2016) | 16–8 | 1–4 | >8–16 | Yes ITZ VRZ |
ND | Yes | ND | Yes |
| The Netherlands (2007–2009; 3), India (2012–2016; 3), Denmark (2010–2014; 2), UK (1992–2009; 2), USA (2015–2017; 1), Japan (2009–2011; 1) | P216L (12) (Howard et al. 2009, Bueid et al. 2010, Camps et al. 2012c, Hagiwara et al. 2014, Ahmad et al. 2015, Jensen et al. 2016, Berkow et al. 2018, Dabas et al. 2018) | 0.25–2 | 0.06–2 | 2–>16 | ND | ND | ND | ND | ND |
| The Netherlands (2007–2009; 8), USA (2001–2014; 1), Italy (2013–2015; 1) | F219I/S/L (10) (van der Linden et al. 2011, Wiederhold et al. 2016, Prigitano et al. 2017) | 0.25–8 | 0.25–>16 | >16 | Yes ITZ |
ND | Yes | Yes | Yes |
| UK (1992–2009; 10), USA (2001–2014; 4), the Netherlands (1994–2007; 1), Denmark (2010–2014; 1), Sweden (1997–1998; 1), Japan (2017–2018; 1), China (2001; 1) | M220V/K/I/R/W (19) (Chen et al. 2005, Snelders et al. 2008, Howard et al. 2009, Bueid et al. 2010, Jensen et al. 2016, Wiederhold et al. 2016, Dabas et al. 2018, Tsuchido et al. 2019) | 0.5–4 | 0.5–>4 | >4–>16 | Yes ITZ |
ITZ | |||
| Japan (2000; 1) | F332K (Asano et al. 2011) | 0.25 | 0.5 | 32 | ND | ND | ND | ND | ND |
| USA (2001–2014; 4), UK (1992–2009; 3), Belgium (2015–2016; 2), Spain (2011; 1), Japan (2017–2018; 1), France (2006; 1) | G448S (12) (Howard et al. 2009, Bellete et al. 2010, Bueid et al. 2010, Peláez et al. 2012, Wiederhold et al. 2016, Montesinos et al. 2017) | 0.5–>16 | 0.25–4 | 0.5–>16 | Yes ITZ VRZ |
ND | Yes | ND | Yes |
| UK (1992–2007; 2), India (2012–2016; 2) | Y431C (4) (Howard et al. 2006, 2009, Albarrag et al. 2011, Dabas et al. 2018) | 0.5–4 | 0.06–2 | >8–16 | ND | ND | ND | ND | ND |
| China (2011–2014; 1), France (2006–2009; 1) | G432A/S (2) (Alanio et al. 2011, Liu et al. 2015) | 0.25–2 | 0.25–0.5 | 4–16 | ND | ND | ND | ND | ND |
| UK (1992–2007; 2) | G434C (2) (Howard et al. 2006, 2009, Albarrag et al. 2011) | 4 | 1 | >8 | No | Yes | ND | ND | ND |
| UK (1992–2017; 70),# China (2008–2009; 16), Spain (2014–2018; 17), Japan (2009–2018; 8), Denmark (2010–2014;7), USA (2015–2017; 7), Italy (1995–2006, 2013–2015; 6), Taiwan (2011–2018; 6), the Netherlands (1994–2013; 4), Belgium (2011–2016; 4), Portugal (2008–2016; 4), Sweden (1997–1998; 2), South Korea (2012–2013; 2), Iran (2003–2016; 2), Brazil (2008–2009; 1), Czech Republic (2008–2009; 1), Germany (2015; 1) | WT (158) (Chryssanthou 1997, Snelders et al. 2008, Bueid et al. 2010, Lockhart et al. 2011, Tashiro et al. 2012, Hagiwara et al. 2014, Fuhren et al. 2015, Steinmann et al. 2015, Vermeulen et al. 2015, Wiederhold et al. 2015, 2016, Jensen et al. 2016, Lazzarini et al. 2016, Nabili et al. 2016, Lavergne et al. 2017, Montesinos et al. 2017, Prigitano et al. 2017, Abdolrasouli et al. 2018b, Berkow et al. 2018, Isla et al. 2018, Pinto et al. 2018, Seufert et al. 2018, Tsuchido et al. 2019, Won et al. 2020) | 0.25–16 | 0.06–16 | 0.5–>16 | ITZ | ND | ND | ND | Yes |
MIC, minimum inhibitory concentration; FLZ, fluconazole; ITZ, itraconazole; PSZ, posaconazole; VRZ, voriconazole.
In the following years, many more studies have been reported, from Argentina, China, Europe (Portugal, Spain, UK), Japan, Taiwan and US (Table 2) (Lockhart et al. 2011, van der Linden et al. 2013, Astvad et al. 2014, Steinmann et al. 2015, Vermeulen et al. 2015, Chen et al. 2016b, Jensen et al. 2016, Wiederhold et al. 2016, Lavergne et al. 2017, Montesinos et al. 2017, Isla et al. 2018, Pinto et al. 2018, Seufert et al. 2018, Li et al. 2020, Wu et al. 2020). Wild-type isolates have developed resistance to itraconazole in many countries and continents, such as the UK and the USA and in Europe and South Asia (Chryssanthou 1997, Snelders et al. 2008, Bueid et al. 2010, Lockhart et al. 2011, Tashiro et al. 2012, van der Linden et al. 2013, Hagiwara et al. 2014, Fuhren et al. 2015, Steinmann et al. 2015, Vermeulen et al. 2015, Jensen et al. 2016, Lazzarini et al. 2016, Nabili et al. 2016, Wiederhold et al. 2016, Prigitano et al. 2017, Abdolrasouli et al. 2018b, Berkow et al. 2018, Seufert et al. 2018). Other single nucleotide polymorphisms, in gene positions G54, M220, and G448 of the CYP51A gene, have been reported to be frequently observed in patients with chronic pulmonary aspergillosis, Invasive aspergillosis bronchitis, aspergilloma, and chronic cavitary pulmonary aspergillosis (CCPA) treated long term with azole antifungals, as well as several clinical treatment failures (Chryssanthou 1997, Chen et al. 2005, 2016b, Snelders et al. 2008, Howard et al. 2009, Bellete et al. 2010, van der Linden et al. 2011, Camps et al. 2012c, Tashiro et al. 2012, Chowdhary et al. 2015, Wiederhold et al. 2016, Montesinos et al. 2017, Dabas et al. 2018, Denardi et al. 2018, Riat et al. 2018, Seufert et al. 2018, Talbot et al. 2018, Tsuchido et al. 2019). Moreover, other single point mutations associated with resistance to azole antifungals have also been reported: G138C, F219I, P216L, G432S, and G432A (Howard et al. 2006, 2009, Bueid et al. 2010, Albarrag et al. 2011, van der Linden et al. 2011, Camps et al. 2012c, Hagiwara et al. 2014, Jensen et al. 2016, Wiederhold et al. 2016, Berkow et al. 2018, Dabas et al. 2018) (Table 2).
While matched control studies involving patients infected with azole-resistant and/or azole-susceptible isolates have not been conducted, patients with azole-resistant Aspergillus infections are at high risk for therapeutic failure. In the Netherlands surveillance study, a high mortality rate was reported among culture-positive ICU patients with ARAF; 14 patients, 10 of whom died, were identified with azole-resistant IA and several underlying conditions, such as autoimmune hepatitis, allogeneic stem cell transplant, hematologic malignancy non-small cell lung cancer, and chronic obstructive pulmonary diseases (COPD) (van Paassen et al. 2016). Two patients died in Belgium of IA that progressed to cerebrospinal aspergillosis (Vermeulen et al. 2012). Other cases support these findings, including a recent German study in which seven of eight azole-resistant IA patients experienced failed therapeutic treatment and died (Salsé et al. 2019). It has also been shown that CPA patients have failed azole treatment due to azole resistance (Howard et al. 2009, Steinmann et al. 2015), while several other case series reported mortality rates of 50–100 % in patients with triazole-resistant IA. Resistance rates as high as 29 % have been observed in specific patient populations, such as critically ill patients (van Paassen et al. 2016, Verweij et al. 2016a).
Worldwide burden of ARAF in the environment
In recent years, an increasing proportion of A. fumigatus isolates has been observed to be resistant in patients (Table 2) and environments (Table 3) due to the presence of mutations in the CYP51A gene (Snelders et al. 2008). Numerous fungicides were able to inhibit wild-type strains, and some azole fungicides were active against wild-type strains but ineffective against isolates with the TR34/L98H mutations, which have high MICs. In the Netherlands, these fungicides were introduced between 1990 and 1996, just before the first clinical TR34/L98H strain was found in 1998 (Snelders et al. 2008). Indeed, evidence of an ARAF environmental route of acquisition was first found in the Netherlands (Zhang et al. 2017). As a result, it has become evident that azole resistance has a potentially global distribution and is therefore a global problem (Mortensen et al. 2010, Verweij et al. 2016a).
Table 3.
Diversity and prevalence of CYP51A mutations causing azole resistance in environmental A. fumigatus isolates.
| MIC1 values (mg/L) |
Azole exposure prior resistance |
Source of isolate | |||||
|---|---|---|---|---|---|---|---|
| Country (year of isolation; number of isolation) | Amino acid substitutions (total number) (references) | VRZ | PSZ | ITZ | Present | Naive | |
| The Netherlands (2002–2019; 203); Denmark (2009; 4); India (2011–2014; 56); Iran (2013; 5); China (200–2015; 4); Thailand (2014–2015; 8); Italy (2011–2018; 61); Romania (2013–2014; 16);UK (2009–2018; 43); France (2010–2017; 62); Taiwan (2014–2018; 33); Tanzania (2013–2014; 11); Australia (2013–2017; 1); Kuwait (2013–2015; 9); USA (2015; 38); Germany (2012–2013; 45); Colombia (2015; 2) | TR34/L98H (S297T/F495I/Q141H/S52T) (601) (Verweij et al. 2009, Mortensen et al. 2010, Chowdhary et al. 2012b, 2014a, 2014b, Badali et al. 2013, Ahmad et al. 2014, 2015, Bromley et al. 2014, Prigitano et al. 2014, Wang et al. 2014, Bader et al. 2015, Sharma et al. 2015, Chen et al. 2016b, 2019b, Nabili et al. 2016, van der Linden et al. 2016, Álvarez-Moreno et al. 2017, 2019, Hurst et al. 2017, Jeanvoine et al. 2017, Lavergne et al. 2017, Tangwattanachuleeporn et al. 2017, Rocchi et al. 2018, Talbot et al. 2018, Trovato et al. 2018, Tsitsopoulou et al. 2018, Sewell et al. 2019, Ahangarkani et al. 2020) | 0.5–>32 | 0.25–3 | >4–>16 | Yes Bromuconazole, cyproconazole, difenoconazole, epoxiconazole, hexaconazole, metconazole, penconazole, propiconazole, tebuconazole, tricyclazole, triticonazole |
No/Yes | Air, patient room, water filter, soil, corps, seeds, rose pot compost, compost. Hospital garden, cucurbit fields |
| Iran (2017–2018; 3); The Netherlands (2009–2011; 14); UK (2018; 6); Tanzania (2013–2014; 4); France (2014–2017; 31); Colombia (2015–2018; 38); India (2012–2013; 6); Germany (2012–2013; 6); Taiwan (2016–2018; 3); Greece (2016–2017; 1) | TR46 / Y121F / T289A (112) (van der Linden et al. 2013, 2016, Chowdhary et al. 2014a, 2014b, Bader et al. 2015, Sharma et al. 2015, Álvarez-Moreno et al. 2017, 2019, Lavergne et al. 2017, Rocchi et al. 2018, Chen et al. 2019b, Sewell et al. 2019, Siopi et al. 2020) | >2–≥16 | 0.25–2 | >8–>16 | Yes | No/Yes | Air, patient room, water filter, soil, corps, seeds, rose pot compost, compost, raisins, grapes. Hospital garden, cucurbit fields, farmland |
| India (2013–2014; 1); Romania (2013–2014; 7); Tanzania (2013–2014; 13); Australia (2013–2017; 2); Germany (2012–2013; 1); Thailand (2014–2015; 2) | G54R/E/W/A (26) (Bader et al. 2015, Sharma et al. 2015, Tangwattanachuleeporn et al. 2017, Talbot et al. 2018) | 0.25–>32 | 0.25–2 | 16–>32 | Yes Bromuconazole, cyproconazole, difenoconazole, epoxiconazole, hexaconazole, metconazole, penconazole, tebuconazole, tricyclazole, triticonazole |
ND | Hospital soil, woody debris of trunk hollows, garden soil |
| Australia (2013–2017; 41); Germany (2012–2013; 1); Taiwan (2014–2016; 5); Colombia (2015–2018; 5) | WT(52) (Bader et al. 2015, Álvarez-Moreno et al. 2017, 2019, Talbot et al. 2018, Wang et al. 2018b) | 1–>32 | 0.125–1 | 0.25–>32 | Yes | No/Yes | Azole-naïve and azole exposed soil, air |
| Italy (2011-2012; 1) Australia (2013-2017; 1); Germany (2012–2013; 1) | F46Y; M172I/V; N248K/T; D255E (3) (Prigitano et al. 2014, Bader et al. 2015, Talbot et al. 2018) | 1–>32 | 0.5–2 | >16–>32 | Yes | ND | Cucurbit fields |
| France (2014–2016; 20) | P216L (Lazzarini et al. 2016, Jeanvoine et al. 2017) | 0.5 | 0.5 | >16 | Yes | ND | Sawmills and air |
| Germany (2012–2013; 1) | M220I (Bader et al. 2015) | 1–>32 | 0.125–1 | 0.125–>32 | ND | ND | Soils |
| Colombia (2015–2018; 4) | TR53 (van der Linden et al. 2016, Álvarez-Moreno et al. 2017, 2019) | 4 | ND | 4–>8 | Yes | No | Flower fields, soil and greenhouse soil |
MIC, minimum inhibitory concentration; ITZ, itraconazole; PSZ, posaconazole; VRZ, voriconazole.
Such environmental route-related mutations are present in many geographically diverse countries and continents (Table 3). Many countries have reported environmental ARAF isolates harbouring TR34/L98H, including Australia, China, Colombia, Denmark, France, Germany, India, Iran, Italy, Kuwait, the Netherlands, Romania, Taiwan, Tanzania, Thailand, UK and the US (Verweij et al. 2009, Mortensen et al. 2010, Chowdhary et al. 2012b, 2014a, Badali et al. 2013, van der Linden et al. 2013, Ahmad et al. 2014, 2015, Bromley et al. 2014, Prigitano et al. 2014, Wang et al. 2014, Bader et al. 2015, Sharma et al. 2015, Chen et al. 2016b, Nabili et al. 2016, van der Linden et al. 2016, Álvarez-Moreno et al. 2017, Hurst et al. 2017, Jeanvoine et al. 2017, Tangwattanachuleeporn et al. 2017, Rocchi et al. 2018, Talbot et al. 2018, Trovato et al. 2018, Tsitsopoulou et al. 2018, Chen et al. 2019b, Sewell et al. 2019, Ahangarkani et al. 2020) (Table 3). Additionally, several studies have reported ARAF isolates harbouring TR46/Y121F/T289A in Iran, the Netherlands, the UK, Tanzania, France, Colombia, India, Germany, and Taiwan (Chowdhary et al. 2014a, 2014b, Bader et al. 2015, Sharma et al. 2015, Nabili et al. 2016, van der Linden et al. 2016, Álvarez-Moreno et al. 2017, Lavergne et al. 2017, Rocchi et al. 2018, Chen et al. 2019b, Sewell et al. 2019). Moreover, wild-type strains have been reported in several countries, including Australia, Taiwan, Germany, and Colombia, for which the MICs values of itraconazole, voriconazole, and posaconazole ranged from 0.125 to > 32 (Bader et al. 2015, Álvarez-Moreno et al. 2017, 2019, Wang et al. 2018b).
Increasingly, other point mutations in azole-resistant strains have been reported in studies in Europe, North America, and Asia (Prigitano et al. 2014, Bader et al. 2015, Sharma et al. 2015, van der Linden et al. 2016, Álvarez-Moreno et al. 2017, 2019, Jeanvoine et al. 2017, Tangwattanachuleeporn et al. 2017, Talbot et al. 2018). Therefore, it is crucial to change processing practices to reduce the use and spread of azole fungicides in the environment that result in cross-resistance with medical azoles. Nonetheless, while TR34/L98H and TR46/Y121F/T289A mutations are currently the most prevalent mutations associated with the environmental route, ARAF strains without the CYP51A gene mutations may also emerge from the environment (Table 3). Indeed, the evidence demonstrates that environmental azole-resistance is increasing due to azole fungicide drugs in the environment; further study is needed.
Antifungal tolerance in A. fumigatus
Antifungal tolerance is different from antifungal resistance and a relatively new concept for medical mycology. Among the human pathogenic fungi, antifungal tolerance has been mainly studied in Candida albicans and Candida glabrata. Tolerance is defined as the ability of a subpopulation of cells (> 1 %) in a drug-susceptible strain to persist or grow in presence of drug concentrations above the minimal inhibitory concentration (MIC), often resulting in “trailing growth” in MIC assays. This is in contrast to heteroresistance or persistence, where very rare cells (< 1 %) grow above the MIC (Delarze & Sanglard 2015, Berman & Krysan 2020). Candida albicans tolerance is most commonly seen with azole antifungals and is quantified by disk diffusion or broth microdilution assays; in C. glabrata tolerance occurs readily with echinocandins. Tolerance is affected by strain genetics, especially chromosomal rearrangements or mutations in genes that participate in core stress pathways; it is also influenced by differences in growth conditions such as pH, temperature and nutrient availability and in physiological differences between genetically identical cells (Berman & Krysan 2020). The clinical relevance of antifungal tolerance in Candida species is still unresolved, with some studies failing to find a connection (Rueda et al. 2017), whereas others show a positive correlation between tolerance and disease persistence (Astvad et al. 2018, Healey & Perlin 2018, Rosenberg et al. 2018).
In A. fumigatus, tolerance has been best characterised for the echinocandin antifungals: micafungin, anidulafungin and especially caspofungin, which inhibit fungal β-1,3-d-glucan synthase activity, thereby disrupting cell wall integrity (Patil & Majumdar 2017). Echinocandin tolerance is defined by partial growth inhibition or trailing, under increasing drug concentrations, followed by an unusual phenomenon called the “paradoxical effect”, during which hyphal growth intensifies despite increasing drug concentration (Wagener & Loiko 2017) until complete inhibition (MIC) at very high drug levels. Growth inhibition is characterised by the formation of stubby, highly budded compact colonies in which the growing hyphae undergo tip lysis followed by regenerative intrahyphal growth initiating from viable internal compartments (Moreno-Velásquez et al. 2017), with the minimal effective concentration (MEC) defined as the lowest drug concentration that induces compact colony formation. The mechanism(s) underlying A. fumigatus tolerance and paradoxical growth are not fully understood. A factor in overcoming echinocandin stress is the upregulation of chitin synthesis and a large increase in cell wall chitin levels (Walker et al. 2015). Three signalling pathways, the high osmolarity glycerol (HOG), HSP90/calcineurin and cell wall integrity pathway, upregulate chitin synthesis in response to echinocandin treatment (Wagener & Loiko 2017). Genetic or pharmacological targeting of these pathways blocked the upregulation of chitin synthesis and abolished A. fumigatus tolerance and paradoxical growth. Another important event that occurs after sustained echinocandin exposure is the relocalisation of glucan synthase from the vacuoles to the hyphal tips (Moreno-Velásquez et al. 2017). This is accompanied by increased cell wall β-1,3-d-glucan, and a reduction in chitin (Loiko & Wagener 2017). In animal models, increased drug levels elevate fungal burden but have no effect on overall survival (Wagener & Loiko 2017). Compact fragmented colonies are seen by histology, which may increase CFU counts while decreasing tissue penetration, organ injury and mortality (Petraitiene et al. 2002). Thus, the clinical relevance of echinocandin tolerance and the paradoxical effect is not yet clear.
In contrast to the literature on Candida spp., very few reports describe azole tolerance in aspergilli. Trailing reported for some clinical isolates of A. flavus and A. niger was dependent on inoculum size and growth medium, respectively (Mosquera et al. 2001, Wang et al. 2018a). Trailing also was described for A. fumigatus exposed to the allylamine antifungal terbinafine (Moore et al. 2001). No trailing growth has been reported with amphotericin B exposure. Recently, an important new mechanism for tolerance and clinical resistance to echinocandins emerged in A. fumigatus in which it was reported that caspofungin induces a change in the membrane sphingolipid content rendering glucan synthase insensitive to the drug (Satish et al. 2019).
In summary, for A. fumigatus, antifungal tolerance to echinocandins is associated with physiological changes in cell wall and membrane composition; tolerance to antifungal azoles remains to be investigated.
Azole resistance mechanisms
Azole resistance in A. fumigatus is mainly associated with the acquisition of mutations in and overexpression of CYP51A, and overexpression of efflux pumps, as discussed in detail below. Adaptation to a new environment before acquisition of stable mutations in the azole drug target, e.g., CYP51A, requires orchestration of a rapid, robust, and coordinated response that allows the cell to thrive despite the presence of drugs. It has been shown that mounting an appropriate physiological response and phenotypic plasticity to itraconazole may take 60 min (Hokken et al. 2019). Among the most highly expressed genes 30 and 60 min after itraconazole exposure are efflux pump genes mdr1 and mdr4; hmg1, encoding a 3-hydroxyl-3-methylglutaryl-CoA 1 reductase from the mevalonate pathway; and ERG6, the most upregulated gene at all time points studied, involved in ergosterol biosynthesis (Hokken et al. 2019) (Fig. 2). Further, increased expression of hmg1 may lead to an increased production of mevalonate, a precursor in sterol biosynthesis, which can positively regulate the overexpression of ERG6, resulting in increased eburicol levels (Yasmin et al. 2012, Hokken et al. 2019). The higher activity of ERG6 also results in the higher quantity of eburicol, the substrate for CYP51A, which also results in higher expression of CYP51A to keep up with the substrate elevation. The detailed mechanisms of azole resistance are discussed below (Fig. 2).
Fig. 2.
The ergosterol pathway involves multiple enzymes and is tightly regulated by the key rate-limiting enzyme Hmg1 producing mevalonate. Lanosterol produced by ERG6 is the substrate catalysed by CYP51A and CYP51B, while azoles competitively occupy the catalytic site and hence reduce the ergosterol synthesis. Adopted from Moreno-Velásquez et al. (2017), with permission.
Role of CYP51A and CYP51B in azole resistance
In Candida species, azole drugs target ERG11 catalyses demethylation of 14-α-lanosterol. In A. fumigatus, ERG6 catalyses this reaction and acts upstream of the triazole drug target cytochrome P450 51 (Fig. 2). Cytochrome P450 51 is encoded by two isoforms, CYP51A and CYP51B (Alcazar-Fuoli et al. 2008, Hokken et al. 2019), which share 59–63 % similarity (Warrilow et al. 2010, Hargrove et al. 2015). A gene essentiality study in A. fumigatus based on the gene expression levels revealed that CYP51A encodes the major enzyme required for mycelial growth; the biological function of CYP51B remains elusive (Hu et al. 2007). Further, deletion of CYP51A abrogates fluconazole resistance, but this effect is not observed when CYP51B is deleted (Mellado et al. 2005). In addition, CYP51A weakly binds to triazoles, which is in contrast with tighter binding observed for CYP51B (Warrilow et al. 2010). Collectively, these observations indicate that CYP51A is the major enzyme in the ergosterol biosynthesis pathway and azole resistance. The presence of both CYP51A and CYP51B is vital to cell survival, and deletion of either isoform is compensated by the presence of other, without major apparent abnormalities in cell morphology (Hu et al. 2007). Further, the intrinsic resistance of Aspergillus to fluconazole is thought to be mediated by I301T substitution in CYP51A and a higher expression of CYP51A than CYP51B upon fluconazole exposure (Blosser & Cramer 2012, Leonardelli et al. 2016).
Mutations in CYP51A and CYP51B
Mutations in the CYP51A gene identified in clinical and environmental ARAF isolates are listed in Table 2, Table 3. Most of these mutations within the CYP51A coding sequence are accompanied by tandem repeats (TRs) in the promoter region, such as TR34/L98H or TR46/Y121F/T289A (TR-mediated azole resistance mechanism is detailed in the following section) (Fig. 3). The effect of the mutations on MIC values and CYP51A structure requires high-resolution structure and/or simulation analysis. Although the crystal structure of CYP51B has been determined, that of CYP51A has not yet been defined (Hargrove et al. 2015). Therefore, the association of such mutations with azole resistance is mainly derived from heterologous expression experiments and simulation studies, in which CYP51A from human, Mycobacterium tuberculosis, and Saccharomyces cerevisiae, and CYP51B from A. fumigatus are used as models to evaluate the effect of specific amino acid substitutions on protein structure (Liu et al. 2016, Nash & Rhodes 2018). According to in silico modelling, the impact of an amino acid substitution on CYP51A structure depends on the position and the substituted residue (Liu et al. 2016). For instance, substitutions of G54, L98, M220, and Y431 decrease the binding affinity of CYP51A to azoles, while substitutions of G432 and also L98 reduce the stability of CYP51A, which can lead to conformational changes in the substrate and/or inhibitor binding pocket by causing dramatic changes in specific loop structures close to these sites (Liu et al. 2016, Nash & Rhodes 2018). Furthermore, L98H substitution reduces hydrogen bond formation between the residue at site 98 and polar side chains of adjacent residues, which could prevent docking of triazoles in the binding pocket (Nash & Rhodes 2018). Typically, L98 changes are accompanied by amino acid substitutions of S297 and F495, which are both adjacent to the binding pocket and, hence, may synergistically confer azole resistance (Liu et al. 2016). Conversely, G54, G138, and M220 are close to the opening channels 1 and 2, which are close to the ligand access channel. Based on the same analysis, substitutions of amino acids located on the periphery of the protein, such as E130D, L252L, S400I, F46Y, M172V, N248T, D255E, L358L, E427K, and C454C, do not cause pronounced conformational changes as they are far from the critical residues and, therefore, do not play a role in azole resistance (Liu et al. 2016).
Fig. 3.
Molecular mechanisms contributing to triazole resistance observed in A. fumigatus. The scheme represents an A. fumigatus cell - with particular focus on the nucleus (N), the endoplasmic reticulum (ER), and the plasma membrane (PM), which depicts the most relevant mechanisms of triazole resistance in this fungus. In A. fumigatus, a major role in ergosterol biosynthesis is played by the sterol demethylase CYP51A (a1). The CYP51A gene is regulated positively by the SrbA protein, which activates its expression by binding to two Sterol Regulatory Elements (SRE) in the promoter region. When ergosterol biosynthesis is repressed, the access of SrbA to SREs is prevented by both the CBC complex and the HapX transcription factor binding to regulatory elements located downstream of SREs, resulting in negative regulation of CYP51A expression. The sterol demethylase CYP51A, whose native substrate is eburicol, an intermediate of ergosterol biosynthesis, is the target of azole drugs (a2). As a result, changes in CYP51A sequence or expression are associated with increased MIC to triazoles. Amino acid substitutions in either the ligand binding site or the catalytic site (b1) modulates triazole binding affinity to CYP51A (b2). A different mutation that can be found in combination with SNPs in the CYP51A gene is the presence of tandem repeats (TRs) in the promoter region, resulting in an expansion of the SREs, unimpeded SrbA binding, and ultimately hyper-activation of CYP51A expression (c1). The same outcome was observed in the case of the P88L mutation in the HapE subunit of the CCAAT-binding complex (CBC) complex, which diminishes CBC binding affinity and its negative regulation of CYP51A expression, although this genotype had only been observed in the clinical isolates in which it was first described (d1). In both cases, the increased amount of the CYP51A enzyme prevents saturation by triazoles and sustains ergosterol biosynthesis (c2 and d2). As for other pathogenic fungi, overexpression of either ATP-binding cassettes (ABC) or Major Facilitator Superfamily (MFS) type drug efflux pumps had been observed among triazole-resistant clinical isolates, which prevents the accumulation of active concentration of drug in the cell. In particular, the transcription factor AtrR positively regulates the expression of the ABC transporter CDR1B (d1 and d2). Notably, AtrR is also involved in the positive regulation of CYP51A. A clinically relevant mutation of a different kind is the one affecting the Hmg-CoA reductase encoded by hmg1, which takes part in ergosterol biosynthesis by converting Hmg-CoA into Mevalonate. Hmg1 has a conserved Sterol Sensing Domain (SSD) involved in regulation of sterol biosynthesis. Mutations in the SSD result in a dysregulation of the sterol pathway that eventually translates to an increased cellular ergosterol production and triazole resistance (f1 and f2).
Resistance mechanisms that do not involve CYP51A mutations
Upregulation of CYP51A
Upregulation of CYP51A expression is an important mechanism of azole resistance. It is partly mediated by a steroid regulatory element-binding protein (SREBP), SrbA. Apart from CYP51A, SrbA also controls many other genes involved in sterol biosynthesis, adaptation to hypoxic conditions, virulence, normal cell polarity and hyphal morphogenesis, iron uptake, nitrate assimilation, and heme biosynthesis (Willger et al. 2008, Chung et al. 2014, Dhingra & Cramer 2017). Following complex processing in the endoplasmic reticulum and Golgi apparatus, the N-terminal DNA-binding domain of SrbA, a basic helix-loop-helix leucine zipper transcription factor, is liberated and translocated to the nucleus, where it binds to steroid regulatory elements (SRE) and activates the transcription of target genes (Fig. 3) (Willger et al. 2008, Chung et al. 2014, Dhingra & Cramer 2017). SRE elements typically contain two SrbA recognition sites, SRE1 and SRE2 (Willger et al. 2008, Gsaller et al. 2016). Hence, TRs identified in ARAF isolates (TR34, TR48, TR53, and TR120) act as additional SrbA-binding motifs, leading to the recruitment of additional SrbA molecules, and increased expression of srbA and CYP51A, and, to a lesser extent, CYP51B (Willger et al. 2008, Gsaller et al. 2016). Indeed, the activity of SrbA is similar to that of Upc2 in C. albicans, which upregulates the expression of ERG11 (Willger et al. 2008, Gsaller et al. 2016). It should be noted that CYP51A is not exclusively regulated by SrbA, since CYP51A expression is not completely inhibited in mutants lacking srbA (Blosser & Cramer 2012). Of note, a 1822-bp insertion (type II transposon Aft1) was identified upstream of the start codon of CYP51A in an ARAF isolate overexpressing CYP51A; however, its role in azole resistance remains to be determined (Albarrag et al. 2011). Finally, although upregulation of CYP51B in ARAF isolates appears to be rare, a baseline and/or induced overexpression of CYP51B has been observed in a limited number of clinical isolates lacking mutations in CYP51A (Buied et al. 2013).
CCAAT-binding complex-mediated azole resistance
Whole-genome sequencing of an ARAF strain with wild-type (WT) CYP51A isolated from a Dutch patient identified HapE, a new factor involved in the regulation of sterol synthesis (Camps et al. 2012a). HapE is a subunit of the CCAAT-binding complex (CBC); it harboured the amino acid substitution P88L in the clinical isolate (Camps et al. 2012a).
CBC is a trimeric transcription factor complex (HapB, HapC, and HapE), which together with the monomeric transcription factor HapX regulates sterol synthesis by binding at positions –293 to –289 and –275 to –269 upstream of the CYP51A start codon, respectively (Fig. 3) (Gsaller et al. 2016). Knock-out analysis of CBC subunit genes and hapX, and heterologous expression of HapEP88L result in increased triazole MIC values and overexpression of CYP51A, HMG-CoA synthase (paralog of erg13A and erg13B), and HMG-CoA reductase (paralog of hmg1), indicating that these transcription factors act as repressors of genes involved in sterol biosynthesis (Gsaller et al. 2016). Further studies revealed that the N-terminal DNA-binding domains of CBC and HapX physically interact with one another (Hortschansky et al. 2015). Further, experiments under iron-limiting conditions demonstrated that the initial binding of CBC to a CBC motif allows the recruitment of HapX to HapX motif (5′-GAT-3′) located 11–12 bp downstream of the CBC motif (Hortschansky et al. 2015). Interestingly, although both SrbA and CBC competitively bind to the same SRE site, position –293, the binding affinity of SrbA is 8-fold higher than that of CBC (Gsaller et al. 2016). Further, SrbA shows a higher binding affinity for the original motif located in position –293 than to the additional motifs in isolates with TRs, located in position –327 (Gsaller et al. 2016). These studies implicated other determinants involved in azole resistance, which negatively regulate the expression of azole drug target and other genes involved in ergosterol biosynthesis.
The role of efflux pumps
Identification of a relatively large number of clinical ARAF isolates lacking CYP51A mutations and comparative genomics studies in yeasts prompted the investigation of alternative mechanisms of azole resistance. This led to the discovery of the role of efflux pumps in azole resistance. Efflux pumps are categorised into two main classes, the major-facilitator superfamily (MFS), encoded by 278 genes, and ATP-binding cassette (ABC) proteins that require ATP for activity, encoded by 49 genes (Loiko & Wagener 2017).
An initial comparative genomics analysis revealed that the two paralogs CDR1A and CDR1B (also known as abcC) are orthologous to C. albicans CDR1, and abcA the same as Afumdr1 (Fraczek et al. 2013). The efflux pump genes atrI and mdrA partially contribute to azole resistance in clinical isolates, while atrF appeared to be upregulated in environmental ARAF isolates (Meneau et al. 2016). Studies focused on generating AFAR strains in vitro (UV-irradiated and itraconazole-treated strains) and those exploring the response of A. fumigatus to itraconazole and voriconazole revealed a pronounced overexpression of several efflux genes, namely, Afumdr1, Afumdr3, Afumdr4, and atrF (Nascimento et al. 2003, da Silva Ferreira et al. 2004, 2006b, Hokken et al. 2019). Of note, the triazole MIC values of isolates lacking Afumdr1 and mfs56 showed a slight change, which raises the question whether these proteins are the main efflux pumps involved in triazole resistance (Fraczek et al. 2013). Several studies dissecting the molecular mechanisms underpinning triazole resistance in clinical ARAF isolates, however, showed that atrF, mfsC, and CDR1B (orthologous to CaCDR1), especially CDR1B and atrF, are greatly upregulated in these isolates (Slaven et al. 2002, Fraczek et al. 2013, Meneau et al. 2016, Paul et al. 2017, Sharma et al. 2019, Wu et al. 2020). This observation is further supported by the observation that heterologous expression of CDR1B in S. cerevisiae isolate lacking Pdr5, or deleting this gene from an ARAF isolate carrying TR34/L98H, TR34, or L98H, results in a profound decrease of voriconazole MIC values (Paul & Moye-Rowley 2013, Paul et al. 2017). Furthermore, these efflux pumps have different substrate specificity, i.e., a narrow or broad-spectrum and functionality, with CDR1B showing the broadest substrate specificity (Esquivel et al. 2020).
Attempts to identify the regulator of CDR1B expression resulted in the identification of AtrR, (ABC transporter–regulating transcription factor). AtrR is a Gal-4 type Zn2-Cys6 cluster-containing transcription factor, which shares homology with CgPdr1 (Fig. 3) (Hagiwara et al. 2017). Interestingly, AtrR is responsible for the upregulation of CYP51A and also CYP51B, as predicted by experiments that suggested that proteins other than SrbA also control CYP51A expression (Blosser & Cramer 2012, Paul et al. 2019). Intriguingly, the AtrR response element (ATRE) is located within the 34-bp repeat element and, hence, both AtrR and SrbA share overlapping binding sites (Fig. 3) (Paul et al. 2019). Therefore, it is plausible that both SrbA and AtrR cooperatively mediate the upregulation of CYP51A expression, as well as the expression of several genes that control the ergosterol biosynthesis pathway (Hagiwara et al. 2017, Paul et al. 2019). Importantly, deletion of atrR resulted in azole hypersensitivity of a strain with CYP51A with G54E substitution, which indicates that AtrR contributes to azole resistance even in isolates with mutated CYP51A (Hagiwara et al. 2017). Strains lacking both atrR and srbA are viable (Hagiwara et al. 2017), which may indicate the presence of other transcription factors that control the expression of other efflux pumps.
Although the regulator of atrF expression remains to be identified in A. fumigatus, a recent study using whole genome sequencing of a laboratory-derived voriconazole-resistant A. flavus isolate revealed that yap1 controls the expression of atrF (Ukai et al. 2018). Yap1 is a bZIP master-regulator transcription factor involved in the oxidative stress response (Ukai et al. 2018). Interestingly, the voriconazole-resistant A. flavus isolates harboured the mutation Yap1L558T, which resulted in atrF overexpression, and reverting Yap1L558T to WT form and deletion of atrF significantly decreased the voriconazole MIC values (8- to 16-fold) (Ukai et al. 2018). The gain-of-function mutation resulted in changes in the C-terminus of Yap1, leading to a constitutive localisation of this transcription factor in the nucleus, binding to a putative Yap1 response element (YRE) at positions –462 to –456 relative to the start codon of the target genes, and overexpression of the target genes (Ukai et al. 2018). Of note, YRE is also present upstream of CYP51A, but this gene was not greatly upregulated in the respective azole-resistant isolate, which suggests that Yap1 does not control the CYP51A. This finding encourages evaluation of the yap1 sequence in ARAF isolates overexpressing atrF.
Hmg1 and azole resistance
Losada et al. (2015) identified Hmg1 as another player in azole resistance, in an experiment involving successive in vitro exposure of A. fumigatus to various azole compounds. Hmg1 is bound to the endoplasmic reticulum membrane via an N-terminal anchor domain linked to the catalytic site via a linker (Fig. 3) (Sever et al. 2003, Friesen & Rodwell 2004). In the experiment of Losada et al. (2015), all voriconazole-resistant progenies had specific amino acid substitutions in the sterol-sensing domain of Hmg1. Sterol negatively regulates the activity of Hmg1, and in the presence of high sterol levels, the membrane-bound domain of Hmg1 is targeted to proteasome-mediated proteolysis (Sever et al. 2003). Therefore, it is plausible to associate mutations in the sterol-sensing domain with an increased enzyme stability, which would lead to sterol overproduction and, potentially, azole resistance (Sever et al. 2003, Friesen & Rodwell 2004, Losada et al. 2015, Jiang et al. 2018). Indeed, this was the case with ARAF clinical isolates from Japan and the US (Hagiwara et al. 2018, Rybak et al. 2019). These isolates, lacking CYP51A mutations but harbouring mutations in hmg1, produced more ergosterol and were more susceptible to polyenes, such as AMB, than isolates without mutations in hmg1. Other studies conducted in India and Taiwan identified similar mutations in hmg1 in clinical ARAF isolates (Sharma et al. 2019, Wu et al. 2020). Intriguingly, a study conducted in the US revealed that a high proportion of isolates with WT CYP51A (11/21; 52 %) harbour mutations in the hmg1 portion encoding the sterol-sensing domain (Siopi et al. 2017). Although initial ectopic expression experiments failed to associate the discovered hmg1 mutations with azole resistance, a recent study using CRISPR-Cas9 methodology revealed that F262_del, S305P, and I412S dramatically increase the triazole MIC values (Rybak et al. 2019). Similarly, other genes involved in ergosterol biosynthesis, including ERG6, are mutated and may potentially contribute to triazole resistance, although these mutations are not as prevalent as those of hmg1 (Hagiwara et al. 2018).
Master regulators of azole resistance
It is still unclear how genes involving ergosterol biosynthesis and SrbA, AtrR, CDR1B, and Hap complex genes are regulated on a larger scale, and which master regulators control their expression. In a recent study, potential master regulators that could simultaneously be involved in azole resistance and pathogenicity were systematically analysed (Furukawa et al. 2020). The authors showed that negative cofactor two (Nct2), consisting of the NctA and NctB subunits, regulates ergosterol biosynthesis and iron-responsive pathways by co-localising and interacting with the TATA-box located upstream of the target genes (an estimated nearly 30 % of coding genes in A. fumigatus). Interestingly, nctA and nctB mutants are not only pan-azole and AMB resistant, but they present no fitness cost as their pathogenicity is comparable with that of the WT (Furukawa et al. 2020). The controversial AMB resistance despite a modest increase of ergosterol content could be explained by an upregulation of oxidative stress-reducing enzymes and the notion that altered cell wall morphology may act as a barrier to AMB penetration in these mutants (Furukawa et al. 2020). These findings warrant future studies to assess the role of loss-of-function mutations in Nct2 complex genes in clinical ARAF isolates.
Additional mechanisms of azole resistance
In addition to the already mentioned major mechanisms of azole resistance, some other, relatively rare, mechanisms are also implicated in azole resistance. Damage resistance protein 1 (Dap1) is a cytochrome b5-like heme-binding protein that regulates the function of CYP51A and ERG5. It is located at the endoplasmic reticulum membrane and is composed of three subunits, DapA, DapB, and DapC. DapA stabilises CYP51A and ERG5, allowing electron transfer, while DapB and DapC suppress electron transfer and prevent the activity of target proteins through depletion of heme (Song et al. 2016). Of note, although DAPA and DAPC co-localise at the endoplasmic reticulum membrane, and form complexes with CYP51A and ERG5, DAPB is located in the nucleus (Song et al. 2016, 2017). Gene deletion analysis revealed that ΔdapA was susceptible and ΔdapC was more resistant against itraconazole, while the itraconazole susceptibility of ΔdapB was indistinguishable from the parental strain (Song et al. 2016). The observation that even upon azole stress Dap1 family proteins remain at the endoplasmic reticulum membrane indicate that other transcription factors that translocate to the nucleus may regulate the expression of these genes (Song et al. 2017). In keeping with this anticipation, it was revealed that SrbA is required for the overexpression of dapA and dapC, and Dap1 family proteins per se do not sense ergosterol depletion, indicating that the expression of Dap1 protein genes is controlled by SRE (Song et al. 2017).
According to a recent study, mutation in a gene encoding farnesyl transferase (Afcox10R243Q) and loss of algA, a component of the calcium signalling pathway, leads to itraconazole resistance (Wei et al. 2017). Interestingly, a collection of ARAF clinical isolates with WT CYP51A harbour several amino acid substitutions in Afcox10 but their effect on azole resistance was not evaluated (Sharma et al. 2019). A mismatch repair gene (MMR, also known as MSH2) plays an important role in facilitating the acquisition of drug resistance in C. glabrata (Healey et al. 2016). Unlike C. glabrata, however, the clinical and environmental Aspergillus isolates do not harbour many nonsense mutations in MSH2. Nonetheless, deleting msh2 profoundly impacts genetic stability, antifungal resistance, and virulence in A. fumigatus (Dos Reis et al. 2019). Finally, some studies have implicated OrmA, the rate-limiting enzyme of the sphingolipid biosynthesis pathway (Zhai et al. 2019), b5 CybE (Misslinger et al. 2017), mitochondrial dynamics (Neubauer et al. 2015), and oxidoreductase HorA (Kroll et al. 2016), in azole resistance.
It should be noted that triazole resistance is a multifactorial phenomenon, involving alteration of the drug target, and upregulation of the drug target and efflux pumps (Nascimento et al. 2003, Fraczek et al. 2013). In Candida species belonging to the CTG clade, such as C. albicans, gain-of-function mutations in UPC2, TAC1, and MRR1 result in upregulation of ERG11 and efflux pump genes (Gsaller et al. 2016). Since genes with similar function exist in A. fumigatus, e.g., srbA and atrR, it would be interesting to explore the presence of such gain-of-function mutations in ARAF isolates, and their association with CYP51A and CDR1B overexpression. Further, gain-of-function mutations in CgPDR1 lead to increased virulence and immune evasion and, hence, their implications for A. fumigatus represent an interesting area for future investigations (Vale-Silva et al. 2013). The most important factor that would facilitate gene expression analysis in A. fumigatus is gene characterisation, since most genes remain uncharacterised, which makes the interpretation of transcriptomic studies challenging (Hokken et al. 2019). Nonetheless, the current understanding of azole resistance in A. fumigatus offers a wide range of potential targets that can inspire the development of novel and potent antifungal drugs.
Azole resistance and in-host fitness cost of A. fumigatus
Aspergillus fumigatus exhibits tremendous phenotypic, physiologic, and genomic plasticity, which allows it to adapt to and survive azole exposure. However, bona fide azole resistance requires the acquisition of permanent mutations in azole resistance-conferring genes and/or rewiring of the transcriptomic landscape to enable fungal persistence and survival in the presence of antifungal drugs within the host. Occurrence of such changes, in turn, results in therapeutic failure and dramatically increases the mortality rates, to over 80 % and even up to 100 % in real-life clinical settings (van der Linden et al. 2011, van Paassen et al. 2016). However, since resistance-conferring mutations negatively affect the catalytic activity of key enzymes, such as CYP51A, the mutated isolates may show fitness defects in the absence of azoles compared with the WT population. Indeed, the growth rate and conidia production by isolates harbouring various mutations and with large chromosomal deletions are markedly lower than those of isogenic susceptible isolates (Hagiwara et al. 2014). Further, as shown in in vivo studies, ARAF isolates harbouring HapE with P88L are less virulent than azole-susceptible isogenic ancestors and WT isolates, and exhibit a 4-h growth delay relative to susceptible and WT isolates (Arendrup et al. 2010). In keeping with these observations, ARAF isolates are not detected following discontinuation of the azole therapy, while resistant isolates reappear following itraconazole treatment in the clinical setting (Chen et al. 2005). By contrast, ARAF isolates carrying mutations in CYP51A, especially those with TRs, may harbour additional mutations in the genome, acting as a compensatory mechanism, which allow them to thrive within the host and/or the environment (Verweij et al. 2016b). Indeed, no significant differences in the sterol (and ergosterol) content of several of azole susceptible and ARAF isolates were detected in one study (Alcazar-Fuoli et al. 2008). These observations highlight the notion that while mutations might affect the docking of an antifungal at the enzyme active site, they might not affect the binding of the sterol substrate, as also suggested in simulation studies (Nash & Rhodes 2018). Further, the presence of two copies of the CYP51 gene might enable rapid ergosterol biosynthesis and increased azole resistance (Hu et al. 2007). The presence of multiple paralogs impacts the fitness cost, which is unexpected, based on what is known about yeasts species, such as Candida. Consistent with this notion, in one study, mutant ARAF isolates were persistently isolated from clinical samples of a patient following discontinuation of azoles (Tashiro et al. 2012), and isolates carrying TR46/Y121F/T289A exhibit the same growth rate and conidia production on PDA medium as WT isolates (Hagiwara et al. 2016).
There are multiple possible explanations for such contradictory observations. First, since the fitness cost is an outcome of accumulation of multiple mutations in the genome (Verweij et al. 2016b), assessment of the effect of various mutations requires a study of isogenic isolates, which only differ with respect to the presence of mutations of interest in a locus of interest. Accordingly, WT status should not be assigned solely on the sequencing of CYP51A or few genes. Indeed, to obtain reliable data, studies focusing on fitness-cost evaluation should utilise a well-defined WT isolate whose entire genome has been sequenced. Second, the fitness cost may vary depending on the gene and mutation studied, e.g., the virulence attenuation observed in HapEP88L strains (Arendrup et al. 2010) vs. lack of pronounced virulence attenuation in isolates lacking nct2 (Furukawa et al. 2020). Third, pronounced growth defects in vitro do not always mirror isolate behaviour in vivo, since, as shown in some studies, the in vivo virulence of mutant strains with a growth defect in vitro does not significantly differ from that of WT isolates (Furukawa et al. 2020), which reflects the complex nature of growth in the host. Further complicating matters is simultaneous recovery of both, azole-susceptible and -resistant isolates from clinical samples, which undermines the notion of predominance of a single genotype in a specific ecological niche. Indeed, a remarkable 20 % rate of isolation of mixed susceptible-resistant colonies from clinical samples was reported in one study (Fuhren et al. 2015), which reinforced the analysis of multiple isolates per clinical sample to verify the concomitant presence of isolates with different susceptibility profiles (Camps et al. 2012c).
Resistance to other classes of antifungals
As explained above, the continuous increase in the prevalence of triazole-resistant A. fumigatus in the clinic promote physicians to use other antifungal drugs, most notably AMB and echinocandins. Although the degree of resistance may vary depending on the fungicidal and fungistatic nature of these antifungals, as is the case with azoles, it is rational to assume that the selective pressure exerted by these antifungals will allow the selection of drug-resistant A. fumigatus isolates. Overall, the AMB resistance is a rare phenomenon among patients with IPA and although not generalising the case, some studies have shown that AMB MIC did not differ among IPA patients with/ without AMB exposure (Moosa et al. 2002). The AMB resistance rarity also may explain the lack of knowledge on underlying resistance mechanisms involved in A. fumigatus, which appears to be associated with less drug uptake and a higher catalase activity in A. terreus (Blum et al. 2013). Although counterintuitive, the ergosterol level does not seem to differ among AMB-resistant and AMB-susceptible A. terreus isolates (Blum et al. 2013). To close this knowledge gap, developing in vitro AMB-resistant A. fumigatus isolates followed by unravelling the subcellular mechanism may provide some insights on this context.
Similar to AMB, echinocandin resistance is rarely reported among clinical isolates of A. fumigatus (Arastehfar et al. 2020b) and few studies conducted in this regard have shown that acquisition of mutations in a hotspot of the FKS gene, which encodes the catalytic subunit of β-1,3-D-glucan synthase (Jiménez-Ortigosa et al. 2017), along with the changes in the lipid profile surrounding β-1,3-D-glucan synthase (Satish et al. 2019) may serve as the possible cellular factors underlying echinocandin resistance.
Antifungal drug resistance detection in A. fumigatus: from phenotypic assays to MALDI-TOF MS
As discussed, mortality rates are notably high in patients infected with A. fumigatus azole resistant strains (van der Linden et al. 2011, Lestrade et al. 2019), evincing the importance of making an early resistance detection in order to start an appropriate therapy. Antifungal susceptibility testing has the ultimate goal of helping clinicians to anticipate the chance of treatment success or failure. This generally follows the “90/60” rule in which approximately 90 % of the infections caused by susceptible isolates and 60 % of those due to resistant isolates respond to therapy (Rex & Pfaller 2002). Antifungal resistance can be in vitro detected by performing broth microdilution assays such as those developed and standardised by the Clinical and Laboratory Standard Institute (CLSI) (CLSI 2008) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (Arendrup et al. 2017a), considered as the reference for yeasts and moulds. These methods determined the MIC, defined as the lowest drug concentration required to inhibit fungal growth. However, some antifungals are only able to partially inhibit certain species, causing changes in their hyphal growth. Such is the case of echinocandins on moulds, for which the two methodologies propose the determination of the minimum effective concentration (MEC), the lowest concentration of antifungal resulting in hyphal morphological changes (Kurtz et al. 1994). MEC assessment has the difficulty of being subjective and requiring expertise in microscopic observation in order to determine what is considered as aberrant growth. Even though there are some methodological differences between CLSI and EUCAST procedures, their results have been proved to be comparable (Espinel-Ingroff et al. 2013) and allow the categorisation of the strains as susceptible or resistant by applying the established clinical breakpoints (CBPs). Although EUCAST has defined drug-related and species-related CBPs for A. flavus, A. fumigatus, A. nidulans, A. niger and A. terreus (EUCAST 2020), CLSI has recently adopted CBPs for voriconazole only for A. fumigatus species and uses epidemiological cut-offs (ECVs) to discriminate between wild-type susceptible strains and others with acquired resistance to other antifungal drugs (https://clsi.org/media/3682/m61ed2_sample.pdf, Espinel-Ingroff & Turnidge 2016).
In addition, several complementary methods have been commercialised and are easily carried out in the daily routine of a clinical microbiology laboratory, which include colorimetric endpoint methodologies such as Sensititre YeastOne, Micronaut-AM and the XTT assay, or agar-based methods using strips with a gradient of antifungal concentrations (Etest or MIC Test Strips) or four-well plates for azole (VIPCheck) and/or echinocandin susceptibility testing. The performance of these methods has been compared with the reference standard with generally positive results. Sensititre YeastOne yielded high essential agreement rates with CLSI for itraconazole, voriconazole, posaconazole and amphotericin B (Castro et al. 2004, Guinea et al. 2006, Mello et al. 2017), although its concordance for echinocandins was lower so its use is not recommended in this case (Siopi et al. 2017). Micronaut-AM showed good categorical agreements (≥96 %) with CLSI for anidulafungin, amphotericin B, voriconazole and itraconazole, being able to detect azole resistance in A. fumigatus (Nuh et al. 2020). As echinocandin susceptibility testing for Aspergillus spp. using reference methods is not easy, the XTT-based assay proved to be a feasible alternative and showed promising results when compared with EUCAST MEC values (Meletiadis et al. 2020). Etest and MIC Test strips have been confirmed to be reliable alternatives for antifungal susceptibility testing for Aspergillus species, as well as for detecting A. fumigatus azole resistance, after obtaining categorical and essential agreements of ≥90 % when correlating their results with those yielded by CLSI (Guinea et al. 2008, Martos et al. 2010, Lamoth & Alexander 2015) and EUCAST (Arendrup et al. 2017b, Idelevich et al. 2018). Azole and echinocandin resistance in A. fumigatus is successfully detected using four-well agar plates, showing comparable results to those obtained with the reference methodologies (Arendrup et al. 2017c, Buil et al. 2017b, Tsitsopoulou et al. 2018, Meletiadis et al. 2019). EUCAST has proposed several recommendations for their use as a screening procedure for the detection of azole resistance (Guinea et al. 2019).
Nevertheless, these methodologies have important limitations, especially for moulds: while EUCAST and CLSI are time-consuming and laborious, colorimetric and agar-based methods are easier to perform. Besides, all of them require a fungal pure culture. This can be a critical issue due to the usual low recovery rate of Aspergillus species in culture from clinical samples, which also leads to the underestimation of azole resistance rates (van der Linden et al. 2016). Although the potential of culturing high volume sputum samples yielded positive results for patients with chronic and pulmonary aspergillosis (Vergidis et al. 2020), the development of alternative non-culture-based techniques is essential.
In this context, molecular tools detecting resistance mutations directly from clinical samples are proving to be complementary to phenotypic assays by reducing turnaround times for the initiation of an effective therapy (Jenks et al. 2019f). Several in-house and commercial PCR-based methodologies to directly detect mutations in the A. fumigatus azole-resistance related gene CYP51A and its promoter, including the most frequent point mutations (G54, G138, M220, G448, L98H, Y121F, T289A) and tandem repeat insertions (TR34 and TR46), have been developed (Table 4). While some consist in conventional PCR assays (Spiess et al. 2012, 2014, Postina et al. 2018), the vast majority are real-time PCR based methods that avoid the delay associated with sequencing, something also successfully achieved with loop-mediated isothermal amplification (LAMP) assays (van der Linden et al. 2010, Denning et al. 2011, Zhao et al. 2013, 2016, Chong et al. 2015, 2016, White et al. 2015a, 2017, Dannaoui et al. 2017, Montesinos et al. 2017, Schauwvlieghe et al. 2017, de Groot et al. 2018, Denis et al. 2018, Guegan et al. 2018, Morio et al. 2018, Mikulska et al. 2019, Yu et al. 2019, Pelzer et al. 2020). Pyrosequencing also shows promising results, as it has the advantage of being adaptable to other genes of interest (Trama et al. 2005, van der Torre et al. 2020). However, one of their main limitations is that they need to be very sensitive and specific to detect the low concentration of Aspergillus DNA and prevent cross-reactivity with human DNA. Besides, CYP51A is a single-copy gene, which impairs its amplification, although this can be improved by the use of a nested PCR assay in order to achieve a higher sensitivity (Denning et al. 2011, Zhao et al. 2013, 2016). Although other in-house and commercial molecular methods for detecting CYP51A mutations in A. fumigatus have been successfully developed, they have not been evaluated in clinical samples yet (García-Effron et al. 2008, Klaassen et al. 2010, Bernal-Martínez et al. 2017, Wang et al. 2019, Fungiplex® 2020). The emergence of CYP51A-WT strains increasingly being identified in clinical settings in addition to a diverse range of mutations occurring in the CYP51A challenge the applicability of such tools. Despite the limited nature of resistance-associated mutations, there may be a role for whole-genome sequencing as an alternative strategy (see section 13).
Table 4.
Studies using in-house and commercial methodologies for the direct detection of azole resistance in A. fumigatus from clinical samples.
| Method | Type of assay | CYP51A mutations detected | Clinical samples (n) | CYP51A amplification (false positives) | Additional results | References |
|---|---|---|---|---|---|---|
| Conventional PCR + sequencing | In-house | TR34, L98H, M220 | BAL fluids (6); tissue (2) | 100 % | 75 % WT; 12.5 % TR34/L98H, 12.5 % L98H | Spiess et al. 2012 |
| In-house | TR34, L98H, M220, TR46 | Blood (25); BAL fluids (120); CSF (19); tissue (17) | 100 % (39.8 %) | 58.6 % WT; 1.1 % TR34/L98H; 0.5 % L98H | Spiess et al. 2014 | |
| In-house | TR34, L98H, M220, TR46, Y121F, T289A | BAL fluids (22) | 74.2 % (65.1 %) | 7.6 % not sequenced; 1.5 % TR34/L98H | Postina et al. 2018 | |
| Tissue (15) | 68.9 % (54.5 %) | 7.8 % not sequenced; 1.1 % L98H; 3.3 % TR46/Y121F/T289A; 2.2 % TR34/L98H | ||||
| CSF (15) | 39 % (28.7 %) | 10.3 % not sequenced | ||||
| Pyrosequencing | In-house | G54 | Blood (56) | 3.6 % | 3.6 % WT | Trama et al. 2005 |
| In-house | All of those described | Respiratory samples | van der Torre et al. 2020 | |||
| Real-time PCR | In-house | TR34, L98H | Sputum (1); tissue (4) | 60 % | 60 % TR34/L98H | van der Linden et al. 2010 |
| AsperGenius® | TR34, L98H, Y121F, T289A | BAL fluids (77) | 18.2 % | 15.6 % WT; 1.3 % TR34/L98H; 1.3 % TR46/Y121F/T289A | Chong et al. 2015 | |
| AsperGenius® | TR34, L98H, Y121F, T289A | Serum (72) | 16.7 % | 6.9 % TR46/Y121F/T289A; 5.6 % L98H; 2.8 % TR34/L98H; 1.4 % Y121F | White et al. 2015a | |
| AsperGenius® | TR34, L98H, Y121F, T289A | BAL fluids (201) | 33.8 % | 28.3 % WT; 3.5 % TR34/L98H; 1.5 % TR34/L98H+WT; 0.5 % TR46/Y121F/T289A | Chong et al. 2016 | |
| MycoGENIE® | TR34, L98H | Respiratory samples (88); serum (69) | 0 % TR34/L98H | Dannaoui et al. 2017 | ||
| AsperGenius® | TR34, L98H, Y121F, T289A | BAL fluids (100) | 20 % | 17 % WT; 3 % unspecified mutations | Montesinos et al. 2017 | |
| AsperGenius® | TR34, L98H, Y121F, T289A | BAL fluids (91) | 49.5 % | 34.1 % WT; 8.8 % TR34/L98H; 3.3 % TR34/L98H +WT; 3.3 % TR46/Y121F/T289A | Schauwvlieghe et al. 2017 | |
| AsperGenius® | TR34, L98H, Y121F, T289A | Plasma (86) | 100 % WT | White et al. 2017 | ||
| AsperGenius® | TR34, L98H, Y121F, T289A | Serum (9); tissue (8) | 76.5 % | 76.5 % WT | de Groot et al. 2018 | |
| MycoGENIE® | TR34, L98H | BAL fluids (31) | 0 % TR34/L98H | Denis et al. 2018 | ||
| AsperGenius® | TR34, L98H, Y121F, T289A | Sputum (119) | 47.9 % | 47.9 % WT | Guegan et al. 2018 | |
| MycoGENIE® | TR34, L98H | 0 % TR34/L98H | Guegan et al. 2018 | |||
| MycoGENIE® | TR34, L98H | Respiratory samples (147) | 0 % TR34/L98H | Burckhardt & Zimmermann 2018 | ||
| AsperGenius® | TR34, L98H, Y121F, T289A | BAL fluids (22) | 59.1 % | 56.8 % WT; 2.3 % TR34/L98H | Postina et al. 2018 | |
| Tissue (15) | 46.7 % | 43.4 % WT; 3.3 % TR46/Y121F/T289A | ||||
| CSF (15) | 41.7 % | 41.7 % WT | ||||
| MycoGENIE® | TR34, L98H | BAL fluids (123) | 0.8 % TR34/L98H | Mikulska et al. 2019 | ||
| AsperGenius® | TR34, L98H, Y121F, T289A | BAL fluids (23) | 65.2 % | 52.2 % WT; 8.7 % TR34/L98H; 4.3 % TR46/Y121F/T289A | Pelzer et al. 2020 | |
| Nested PCR + real-time PCR | In-house | TR34, L98H, G54, G138, M220 | Sputum (29) | 100 % | 48.3 % TR34/L98H; 31 % L98H; 6.9 % TR34; 6.9 % TR34/L98H+M220; 6.9 % M220 | Denning et al. 2011 |
| In-house | TR34, L98H, G54, G138, M220, G448 | BAL fluids (94) | 64.9 % | Zhao et al. 2013 | ||
| In-house | TR34, L98H, G54, G138, M220, G448 | Respiratory samples (97) | 39.2 % | Zhao et al. 2016 | ||
| LAMP | In-house | TR34 | Clinical samples (11) | 100 % TR34/L98H | Yu et al. 2019 |
LAMP, loop-mediated isothermal amplification; BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid; WT, wild-type.
Taking all of these into account, new molecular assays should intend to cover a broader range of azole resistance-related mutations and mechanisms, as not all of the CYP51A reported alterations are detected by the available methodologies, while increasing their sensitivity in order to become a feasible option to detect azole resistance in more clinical settings. Besides, there are no molecular options for the direct detection of azole resistance in Aspergillus species other than A. fumigatus, which should be further studied and developed.
Since MALDI-TOF MS has been introduced as a routine identification tool, laboratories are interested to explore this rapid technology as an alternative methodology potentially accelerating antifungal susceptibility testing (Burckhardt & Zimmermann 2018). A simple approach using specific marker peaks in mass spectra of resistant microorganisms could be applied to a number of specific resistances in certain bacteria, but this approach was yet not successful for fungi. Several functional assays have been proposed for antibiotic susceptibility testing in bacteria, e.g., a test for the hydrolytic degradation of β-lactam antibiotics, the observation of incorporation of stable isotope-labelled metabolites, or the utilisation of MALDI-TOF MS as semi-quantitative read out for microbial growth in the presence of an antibiotic drug. Less work has been performed until today to apply such MALDI-TOF MS based assays to antifungal resistance. Most of the studies in this regard have focused on the detection of resistance in yeasts by MALDI-TOF MS. Detection of peak pattern changes after incubation of cells in presence of an antifungal has been demonstrated to determine antifungal resistance in Candida species (Delavy et al. 2019). In two studies, this method was also applied to test susceptibility in Aspergillus species. One study described the detection of caspofungin resistance in A. fumigatus and A. flavus strains (De Carolis et al. 2012). The same approach was applied to strains of Aspergillus species and voriconazole (Gitman et al. 2017). Although the results were in good agreement with reference methods, there was no obvious advantage over traditional methods, in particular because of the similar time to result. A promising method which has been shown to detect antifungal resistance in Candida by semi-quantitative detection of fungal growth in the presence of the drug after only several hours of incubation (Vatanshenassan et al. 2018, 2019) has not yet been applied to Aspergillus yet, according studies should be performed.
Application of typing techniques to identify infection and resistance routes
Strain genotyping is considered one of the most basic tools in the clinical setting since it fulfils many needs, among which the establishment of epidemiological relationships between isolates stands out. As in many other settings, typing methodologies have had an important impact in the aspergillosis field, since they have been used, among many others, for outbreak analysis (Menotti et al. 2005, Doll et al. 2017), environmental monitoring of the isolates that constitute a specific population (Deng et al. 2017, Fan et al. 2020), also for patient monitoring in order to study how clinical strains evolve under drug pressure within the antifungal therapy (Escribano et al. 2017) or to assess local and global Aspergillus spp. epidemiology (Garcia-Rubio et al. 2018a, 2018c, Choi et al. 2019). Thus, the molecular analysis of the genetic and epidemiological relationship between environmental and clinical strains could potentially assess strain origin and route of transmission. Besides all these applications at the subspecies level, molecular typing methods have also been used at the genus level for discriminating between species and also for the definition and recognition of new fungal species (Klaassen & Osherov 2007).
Different methodologies have been developed to genotype Aspergillus species strains (Table 5). However, due to its clinical significance, most of them have been implemented for A. fumigatus strains (Latgé & Chamilos 2019). Classically, genotyping techniques can be grouped in two different categories; methods either based on PCR amplification and sequencing, which are described in detail below, or based on non-coding repetitive sequences paired with restriction fragment length polymorphisms, such as random amplified polymorphic DNA (RAPD) (Loudon et al. 1993) amplified fragment length polymorphism analysis (AFLP) (Warris et al. 2003), and restriction fragment length polymorphism analysis (RFLP) (Neuveglise et al. 1996). The latter ones show a poor inter-laboratory reproducibility which is the reason why they have been replaced by other techniques. Thus, the selection of the most appropriate method in each context will highly depend on the technical resources of a particular setting (Klaassen & Osherov 2007).
Table 5.
Advantages and disadvantages of technologies used for typing clinical and environmental A. fumigatus isolates.
| Assay | Methodology | D∗ | R∗∗ | References |
|---|---|---|---|---|
| RAPD | Random amplified polymorphic DNA | Low | Low | Bertout et al. 2001 |
| SSDP | PCR typing method combining RAPD and specific DNA primers | Low | Low | Mondon et al. 1997 |
| MLEE | Multilocus enzyme electrophoresis | Low | Low | Rodriguez et al. 1996, Bertout et al. 2000 |
| AFLP | Amplified fragment length polymorphism analysis | Moderate | Low | Warris et al. 2003 |
| RFLP | Restriction fragment length polymorphism analysis | Low | Low | Neuveglise et al. 1996 |
| MLP | Microsatellite length polymorphism | High | Moderate | Bart-Delabesse et al. 1998, Bertout et al. 2001 |
| RISC | Retrotransposon insertion-site PCR amplification | Moderate | Moderate | de Ruiter et al. 2007 |
| MLST | Multilocus sequence typing | Moderate | High | Bain et al. 2007 |
| STRAf | Short coding tandem repeats | High | Moderate | de Valk et al. 2005, Guinea et al. 2011, Escribano et al. 2015, Fan et al. 2020 |
| CSP | Cell-surface protein sequencing | Moderate | High | Balajee et al. 2007b, Levdansky et al. 2007 |
| TRESPERG | Sequencing of tandem repeats surface protein coding genes | High | High | Garcia-Rubio et al. 2018a, Fan et al. 2020 |
| WGS | Whole Genome Sequencing | High | High | Hagiwara et al. 2014, Garcia-Rubio et al. 2018b, Puértolas-Balint et al. 2019 |
D∗, discriminatory power; R∗∗ reproducibility considering stability and availability.
Although not a bona fide gold standard technique, the microsatellite analysis assay called STRAf is the most popular and widely used technique used to type A. fumigatus, which stands for short tandem repeats of A. fumigatus (de Valk et al. 2005). This assay, developed more than a decade ago, is based on a panel of microsatellites divided into three multicolour multiplex PCRs. Each multiplex reaction amplifies three di-, tri-, or tetra-nucleotide repeats, respectively. One of the biggest advantages of this technique is the multicolour multiplex approach which allows large numbers of markers to be tested in a short period of time, which is why this assay is a very suitable tool for large-scale epidemiological studies. Moreover, it can even be used to genotype A. fumigatus isolates directly from clinical samples, such as formalin-fixed paraffin-embedded tissues or serum samples (de Groot et al. 2018). The Simpson’s diversity index of this assay (Hunter & Gaston 1988), which shows the discriminatory power of the methodology, is really high –0.9994 (de Valk et al. 2005), 0.988–0.995 (Escribano et al. 2015), 0.984 (Guinea et al. 2011) and 0.9993 (Garcia-Rubio et al. 2018a, 2018c). From a methodological point of view, the STRAf assay presents some major difficulties associated with sizing of the obtained PCR products; high-resolution equipment such as capillary-based or acrylamide-based electrophoresis platforms is required to translate the fragment electrophoretic mobility to their repeat number. However, this mobility is dependent on many critical factors such as the presence, or not, of denaturing compounds, the matrix, the run temperature, the sequence of the fragment, the fluorescent labels, the sizing marker, etc. (Klaassen & Osherov 2007). In order to get exchangeable typing results between laboratories, it is necessary to run allelic ladders for calibrating every platform (de Valk et al. 2009). Also, the low-level instability of two of the markers (de Groot & Meis 2019), together with the availability of the required laboratory technology, dedicated software and personnel specifically trained for its performance comprise some of the main disadvantages of this assay (Klaassen & Osherov 2007, Garcia-Rubio et al. 2016).
As a result, the development of novel and more accessible typing methods has been encouraged. One simple and rapid single-locus typing method was developed based on sequencing the coding tandem repeats present on the cell surface protein (CSP) gene (Balajee et al. 2007b, Levdansky et al. 2007). This method has a lower discriminatory power than microsatellite-based typing – 0.78 (Klaassen et al. 2009), 0.83 (Gao et al. 2013) – and that is why a new typing method, called TRESPERG assay, was described combining four coding tandem repeat markers (Garcia-Rubio et al. 2018a, 2018c). This assay has a sufficiently high discriminatory power to compete with STRAf, and its main advantage is that it does not require trained personnel, specific equipment, or software for analysis, as it only consists of conventional PCR amplification and Sanger sequencing (Garcia-Rubio et al. 2018a, 2018c). Moreover, TRESPERG assay clustered tandem-repeat (TR) azole resistant strains in a better manner than STRAf assay did compared to whole genome sequencing (WGS) studies. Many authors have described these TR azole resistant strains as genetically more closely related than other A. fumigatus isolates (Camps et al. 2012b, Abdolrasouli et al. 2015, Garcia-Rubio et al. 2018b, Wang et al. 2018b). This fact is supported by TRESPERG results in which every A. fumigatus TR isolate tested grouped in only one cluster, endorsing their genetic closeness (Garcia-Rubio et al. 2018a, 2018c).
Finally, whole genome sequencing (WGS) has recently emerged as an invaluable tool for the analysis of genetic differences between A. fumigatus strains and has turned into the typing technique with the highest resolution, becoming increasingly affordable and widely available (Hagiwara et al. 2014, Garcia-Rubio et al. 2018b, Puértolas-Balint et al. 2019). The details of this technology is presented below.
Whole-genome sequencing applications in the clinic
The phenotypic traits of an Aspergillus strain, including its virulence potential and its ability to survive drug exposure, are ultimately encoded in its genome. Reference genome sequences for A. fumigatus, and A. nidulans are available since 2005 (Galagan et al. 2005, Nierman et al. 2005), and have undoubtedly served to sustain major advances in the field. However, as many other fungi, Aspergillus have very plastic genomes and phenotypes, with clinically relevant traits varying widely across isolates. The lowering costs and continuing developments in high-throughput sequencing have recently allowed zooming into the specific genomes of particular strains, revealing how genomic and phenotypic plasticity are connected. In addition, our ability to access full genomic sequences in a timely and cost-effective manner are opening new avenues for clinical applications such as species identification and diagnosis of resistance potential (Consortium & Gabaldón 2019). Currently, three major sequencing platforms exist that differ in their functionalities and suitability for different purposes (Table 6). Their combined use has enabled deciphering genome variation in the Aspergillus genus with increasing level of resolution. For instance, reference genomes for virtually all major species in the genus have been produced in the last few years (Kjærbølling et al. 2018, 2020, Vesth et al. 2018). More recently, re-sequencing of evolved isolates has served to estimate mutation rates in the three major species (Álvarez-Escribano et al. 2019) – A. flavus (4.2 × 10-11 mutations per site and mitotic division), A. fumigatus (1.1. × 10-11) and A. nidulans (4.1 × 10-11), which can be used, for instance, to more precisely date the divergence between two strains or the origin of clinical outbreaks. Other sequencing studies have shown that relationships between genome and phenotypic plasticity can be mapped by comparing genomes and phenotypes from different isolates (Bastos et al. 2020, Drott et al. 2020). The sequencing of serially collected isolates from the same patients with aspergillosis can reveal mutations acquired during the infection process (Hagiwara et al. 2014) and whole genome sequencing can identify mutations leading to azole resistance, particularly in resistant isolates not bearing mutations in CYP51A (Ukai et al. 2018, Sharma et al. 2019). Finally, another promising application of high-throughput sequencing is the direct detection of pathogenic species through targeted barcode sequencing or whole genome shotgun sequencing of complex samples, without the need of isolation, allowed by the high sensitivity of high-throughput sequencing approaches coupled to the increasing resolution power of comparisons with sequence databases (McTaggart et al. 2019).
Table 6.
Comparison of the main features of the three most widely used high-throughput sequencing platforms. Data has been compiled from vendors information and the literature mentioned in the text.
| Platform | Advantages | Disadvantages | Comments |
|---|---|---|---|
| Illumina |
|
|
|
| Oxford Nanopore |
|
|
|
| PacBio |
|
|
|
Although these developments are very promising, the implementation of whole genome sequencing in the clinic faces important challenges which limits its implementation (Greninger 2018, Consortium & Gabaldón 2019, Kidd et al. 2019). Three major clinical applications of genome sequencing are considered: (i) genome-wide profiling of resistance conferring mutations, (ii) genome-wide molecular epidemiology for the study of outbreaks and, (iii) detection and identification of pathogens from complex, patient samples. For the three applications, analytical methodologies are ready, as we have seen in the examples above. However, the translation of these methodologies from an academic study to a clinical context faces many issues, of which we will discuss the more relevant. The first issue is cost. Although the prices of high-throughput sequencing equipment continue to drop, they are still far away from those of other routine analyses in the clinical mycology lab, particularly when one considers the combined costs of reagents, equipment purchase and maintenance, computational infrastructure, and necessary personnel (Greninger 2018). Another important aspect is time, particularly for diagnostic purposes, less so for epidemiological studies. Most sequencing approaches require multiple steps before sequencing can be run (isolation, culture, DNA extraction, library preparation, etc.). These factors coupled with the required bioinformatics analyses that are often not fully automatised, require high-level expertise, which delay results over the limits that are reasonable in the context of clinical needs. This problem can be aggravated if the sequencing and bioinformatics resources are not on site, which still is currently the case for most hospitals. Finally, despite many developments in computational tools and databases, they are still not mature for routine infection control and outbreak investigations. For instance, many public genome repositories are not curated, leading to wrong annotations that can lead to errors in species identification (Stavrou et al. 2018). In our view, full integration of such methodologies into the clinics requires the following developments: (i) equipment should evolve into smaller, more robust and easier to handle devices minimising expert dedication and maintenance cost; (ii) these should be coupled to a computer storage and artificial intelligence-driven computation system that will automatically process data into clinically meaningful results, this system could be on-site or securely accessed remotely; (iii) curated databases and pipelines should be developed that are directed to specific needs in the clinics; and (iv) expert personnel with bioinformatics and microbial genomics expertise should be incorporated into the clinical system. Some of these developments, particularly (i) and (ii), are progressing significantly thanks to the push of personalised medicine applications based on the human genome. However, (iii) and (iv), only partially overlap with other medical applications of genomics, and require a specific microbial genomics focus, and expertise. We envision that joint international efforts with the participation of regulatory authorities, researchers, and clinicians will help to make progress through pilot proof of concept studies and the standardisation of methodologies.
Genetic toolbox used to identify antifungal resistance and virulence determinants
Inducing mating in A. fumigatus under laboratory conditions is a time-consuming process (O’Gorman et al. 2009), and studies of gene function have therefore heavily relied on site-directed mutagenesis (O’Gorman et al. 2009). However, the effect of the cell wall on the uptake of exogenous DNA and the low rate of homology driven repair (HDR) which ranges from 1–10 % in different strains makes gene manipulation of A. fumigatus an uphill road (Krappmann et al. 2006). Different techniques have been used in the attempt to find a balance between increasing transformation efficiency and limiting ectopic genome integration (Sánchez & Aguirre 1996, Sugui et al. 2005, Szewczyk et al. 2006); usually, polyethylene glycol-mediated transformation of protoplasts is preferred for site-directed mutagenesis (Brakhage & Langfelder 2002). Deleting essential players of the non-homology end-joining (NHEJ) pathway such as Ku70 (Krappmann et al. 2006) and Ku80 (da Silva Ferreira et al. 2006a) dramatically increases HDR frequency, so that 0.5–1 kb homology arms can efficiently mediate gene deletion (Krappmann et al. 2006). Although metabolic markers are available (Weidner et al. 1998, Xue et al. 2004) dominant selection markers are preferred, especially when using animal models, because position effects due to the marker integration site may ultimately affect fungal survival within the host (Liebmann et al. 2004, Greenstein et al. 2006). Commonly used markers (resistance towards hygromycin, phleomycin, or pyrithiamine) have been combined with site-specific recombinase systems to allow for marker recycling and targeting of multiple genes in the same background (Punt & van den Hondel 1992, Kubodera et al. 2002, Krappmann et al. 2005, Hartmann et al. 2010). Using a split-marker approach and a cloning-free fusion PCR strategy to assemble the transformation cassette further streamlined the process (Gravelat et al. 2012, Furukawa et al. 2020). Recently, the generation of a library of 484 transcription factor null mutants in a Δku80 A. fumigatus strain offered insights into the complex transcriptional regulation of azole response in this species (Szewczyk et al. 2006). Comprehensive libraries like this are a powerful resource to dissect pathways involved in antifungal resistance, virulence, and nutritional versatility, which all contribute to the multifaceted pathogenicity of A. fumigatus (Ries et al. 2018, Pérez-Cantero et al. 2020). Nonetheless, using NHEJ-deficient strains in animal models calls for caution due to possible genome instability/sensitivity to abiotic stress, and reconstitution of a functional ku80 gene is not feasible in large-scale efforts (Cairns et al. 2016). Moreover, dissecting the - still largely unclear - molecular basis of azole resistance in clinical isolates requires the ability to manipulate wild-type strains (van der Linden et al. 2015). Functional redundancy further muddies the water, and the sequential deletion of gene family members, albeit possible, is a painstaking process. These drawbacks can be overcome with the implementation of CRISPR-Cas9 editing technology (Fig. 4) (Morio et al. 2020). In 2016, microhomology-mediated end joining (MMEJ) was exploited to efficiently replace a Cas9-targeted gene with an hph cassette flanked by 28-bp homology arms in a NHEJ competent strain (Zhang et al. 2016). The same year, the expression of a ribozyme-flanked sgRNA from a RNA pol II promoter was combined with a split-marker approach to induce a single-nucleotide deletion in a Δku80 strain containing an integrated tetracycline-inducible CAS9 (Weber et al. 2017). Alternatively, Cas9-sgRNA ribonucleoproteins (RNP) can be used in conjunction with MMEJ to induce gene replacement with a hph marker in NHEJ competent strains (Al Abdallah et al. 2017). Multiplexing and protein-tagging have also been accomplished (Zhang et al. 2016). Recently, two gene-free intergenic safe haven regions were discovered in A. fumigatus, to which CAS9/sgRNA expressing constructs or selection markers may be directed, thus resolving potential position effects resulting from random ectopic integration (Pham et al. 2020). Notably, CRISPR-Cas9 finally allows high efficiency site-directed mutagenesis of wild-type strains. However, marker integration in the genome was originally required, either for replacing the target gene (Zhang et al. 2016, Al Abdallah et al. 2017) or for selecting the CRISPR elements (Weber et al. 2017). Last year, Ballard and colleagues (2019) tweaked the systems developed by Nødvig et al. (2015) and Weber et al. (2017) into a two-plasmid system for introducing SNPs into a clinical isolate without marker integration. Single-base CRISPR-Cas9 editing streamlined the association of amino acid substitutions with azole resistance in A. fumigatus, and opens up the way to the discovery of new clinically relevant and CYP51A-independent resistance mechanisms (Umeyama et al. 2018, Ballard et al. 2019).
Fig. 4.
CRISPR-Cas9 technology in A. fumigatus. The vignettes illustrate representative methods for CRISPR–Cas9 genetic manipulation of A. fumigatus. (a) MMEJ can be used to integrate an HPH cassette into a desired locus by using short homology arms. The strain is first transformed with a plasmid expressing Cas9 and containing a PYR4 marker and then with an in vitro transcribed sgRNA and the HPH cassette. (b) The need to set up a suitable system to express the CRISPR elements can be circumnavigated by using CRISPR RNPs, in which two different crRNAs and tracrRNA are assembled in vitro with Cas9 and then transformed into the cells to target the upstream and downstream regions of YFG. MMEJ results in the integration of a HPH cassette into the targeted gene. (c) Ballard and colleagues tweaked the systems developed by into a two-plasmid system for introducing SNPs into a clinical isolate without marker integration. The AMA1 sequence supports the replication of the plasmid harbouring CAS9 in A. fumigatus, which confers resistance to hygromycin. A different plasmid carries a cassette for the expression of a ribozyme-flanked sgRNA from a A. nidulans RNA pol II promoter; after expression in A. fumigatus, the self-splicing activity of the rybozymes releases the mature sgRNA. This plasmid contains a PTRA split marker, interrupted by the same protospacer sequence that is being targeted on the gene of interest. After transformation of both plasmids and a RT containing the SNP to introduce in YFG, Cas9 targets both the protospacer on the desired locus in the genome and the twin protospacer interrupting the split marker. HDR then simultaneously mediates the insertion of the SNP into YFG and the reconstitution of the PTRA marker, thus allowing for selection of the transformants on pyrithiamine without marker integration. (d) The RNP system can also be exploited to affect gene expression, and it was recently used to replace a native promoter with a constitutive hspA promoter by transforming the cells with the RNP particle and a repair template carrying HPH, hspA, and homology arms flanking the insertion site. Af, A. fumigatus; CRISPR, clustered regularly interspaced short palindromic repeats; crRNA, CRISPR-RNA; HDR, homology-directed repair; HDV, human hepatitis delta virus; HH, hammerhead; MMEJ, microhomology-mediated end joining; RNA–Cas9 protein complex; sgRNA, single- guide RNA; tracrRNA, trans-activating RNA; YFG, your favourite gene. HA, Homology Arms; RT, Repair Template. Panels a and b were adopted from Morio et al. (2020) with permission.
Recently, the versatility of the CRISPR-Cas9 technology was exploited to dissect the molecular players of triazole resistance in a collection of A. fumigatus clinical isolates (Rybak et al. 2019). Cas9-sgRNA complexes were used to induce site directed mutagenesis and promoter replacement and to show that – surprisingly – mutations in CYP51A and overexpression of either CYP51A/CYP51B or the efflux pump abcC could not recapitulate the triazole MIC observed. Instead, the combination of CRISPR-Cas9 technology with a split hygromycin B marker approach confirmed that clinically occurring mutations in the sterol-sensing domain of the 3-hydroxy-3-methyl-glutaryl-coenzyme A (HMG-CoA) – observed in more than half of the clinical isolates examined – were indeed responsible for the triazole resistance observed (Rybak et al. 2019).
Non-editing applications developed in other fungi may also be adapted in the future to A. fumigatus, including the use of a catalytically dead Cas9 fused to transcriptional regulators to either activate (CRISPRa) or repress (CRISPRi) gene expression, which could address some of the shortcomings of RNA interference in this species (Cairns et al. 2016, Schultz et al. 2018). Disease outcome in patients at high risk of IA also depends on a timely diagnosis (Latgé & Chamilos 2019). The CRISPR technology may revolutionise the field of diagnostics with regards to nucleic acid detection and antimicrobial resistance profiling directly from clinical samples (Myhrvold et al. 2018, Quan et al. 2019). Overall, we may be at a turning point in the study of virulence and antifungal resistance, as the full potential of CRISPR-Cas9 removes long-standing roadblocks and re-shapes the A. fumigatus genetic toolbox.
Application of murine and insect models to identify virulence determinants and antifungal resistance
In vivo infection models are essential for understanding pathogenesis, dissecting host-pathogen interactions, and identification of pathogen and host traits that contribute to susceptibility and disease development. For Aspergillus species, and A. fumigatus in particular, infection models for a variety of mammalian species, especially mice, rats, and rabbits have been used to model different types of disease caused by Aspergillus (Table 7). Technical aspects and the need for standardisation have been extensively reviewed recently by several authors (Paulussen et al. 2014, Desoubeaux & Cray 2017, 2018, Banfalvi 2018, Mirkov et al. 2019). Therefore, we will focus here on (i) differences between leukopenic and non-leukopenic mouse models of IA, as well as murine models of systemic aspergillosis, regarding the contribution of distinct fungal factors to virulence, (ii) the use of in vivo models, including insect models, to determine the impact of antifungal resistance, and (iii) the role of comorbidities.
Table 7.
Examples of mammalian models for different types of disease caused by Aspergillus.
| Type of disease | Host species | Route of infection or exposure | Immunosuppression | References1 |
|---|---|---|---|---|
| Invasive aspergillosis, pulmonary | Mouse | Intranasal | Cortisone acetate; cyclophosphamide; cyclophosphamide and cortisone acetate | Sarfati et al. 2002, Ibrahim-Granet et al. 2010, Wong et al. 2017, Morio et al. 2020 |
| Oropharyngeal | Triamcinolone acetate | Shepardson et al. 2014 | ||
| Intratracheal | Anti-Ly-6G / anti-Ly6C antibody | Shibata et al. 2014, Rolle et al. 2016 | ||
| Inhalation chamber | Cyclophosphamide and cortisone acetate | Sheppard et al. 2004, Steinbach et al. 2004 | ||
| Rabbit | Intratracheal | Hydrocortisone and cyclophosphamide; cyclosporin A and methylprednisolone | Chilvers et al. 1989, Berenguer et al. 1995 | |
| Rat | Intratracheal | Cyclophosphamide and cortisone acetate | Schmitt et al. 1988, Chandenier et al. 2009 | |
| Intrabronchial | Cyclophosphamide | Leenders et al. 1996 | ||
| Intrapulmonal | Cyclophosphamide | Habicht et al. 2002, Becker et al. 2006 | ||
| Invasive aspergillosis, systemic | Guinea pig | Intravenous | Cyclophosphamide and triamcinolone acetate | Kirkpatrick et al. 2000 |
| Mouse | Intravenous | None; cyclophosphamide | Johnson et al. 2000, Sarfati et al. 2002 | |
| Rabbit | Intravenous | Cyclophosphamide; cyclophosphamide and triamcinolone acetate | Patterson et al. 1988, 1991 | |
| Invasive aspergillosis, cerebral | Mouse | Intracranial, Intracerebral | Cyclophosphamide | Chiller et al. 2002, Clemons et al. 2012 |
| Rat | Intracisternal | None | Zimmerli et al. 2007 | |
| ABPA | Mouse | Intranasal, intratracheal or inhalation | None | Hogaboam et al. 2000, Kurup et al. 2001, Ramaprakash et al. 2009, Hoselton et al. 2010, Fei et al. 2011, Moretti et al. 2014 |
| Aspergilloma | Rabbit | Intrapulmonal | None, but surgically induced artificial stenosis of the bronchus and the ligature of pulmonary artery | Sawasaki et al. 1967 |
| Keratitis | Mouse | Intracorneal | None | Clark et al. 2016 |
| Rabbit | Intrastromal | Triamcinolone acetate (locally) | Komadina et al. 1985 | |
| Endocarditis | Guinea pig | Intravenous | None | Martin et al. 1997 |
Selected references with detailed description of materials and methods.
Respiratory infection of leukopenic mice or mice receiving high-dose corticosteroids represent the most commonly used models to study IA (Desoubeaux & Cray 2017). These two models represent the two main groups of human patients at risk for IA, oncological patients undergoing stem cell transplantation or aggressive chemotherapy and patients after solid organ transplantation, respectively. Both models differ fundamentally in the role of the host response in pathogenesis: Leukopenia prevents substantial influx of immune cells into the lung and results in fungal growth unrestricted by the host responses, but driven by the fungal ability to germinate and acquire nutrients from the host tissue for growth (Stephens-Romero et al. 2005, Ibrahim-Granet et al. 2010, Kalleda et al. 2016). In contrast, large numbers of immune cells are recruited to the lungs of corticosteroid-treated mice, and while partially restricting fungal growth, immune cell recruitment contributes to pathogenesis in this model (Ibrahim-Granet et al. 2010, Kalleda et al. 2016). Thus, fungal factors required for growth in the lung generally impact virulence in both models, whereas determinants involved in the interaction with immune cells might only affect virulence in corticosteroid models. An example for this dichotomy is the secondary metabolite gliotoxin; while reduced gliotoxin production attenuates virulence in corticosteroid-treated mice, no significant effect was observed in the leukopenic model (Kupfahl et al. 2006, Sugui et al. 2007, Spikes et al. 2008). Another example is the transcription factor DvrA that negatively regulates virulence in cell culture and the corticosteroid model but not in leukopenic mice (Ejzykowicz et al. 2010). IA furthermore differs from intravenous, systemic infection of mice that targets the liver, spleen and kidney (Jouvion et al. 2012, Paulussen et al. 2014). Organ-specific differences in nutrient supply are the likely reason why deletion of hcsA, involved in lysine biosynthesis, renders the mutant attenuated in IA but not systemic infection models (Schobel et al. 2010). These examples demonstrate the relevance of using different models for aspergillosis to fully dissect the contribution of fungal factors to host-pathogen-interaction.
While animal models have been and continue to be essential for studying pathogenesis of aspergillosis and identification of fungal virulence determinants, ethical concerns and practical considerations have led to the development and increasing use of invertebrate models, especially insects like adult Drosophila melanogaster and Galleria mellonella larvae (reviewed in Lionakis & Kontoyiannis (2012), Binder et al. (2016)). Important aspects of the innate immune response are conserved between insects and mammals (Medzhitov 2001, Müller et al. 2008, Sheehan et al. 2018), but insects lack lymphocytes and an adaptive immune system. While different levels of immunosuppression can be achieved by genetic manipulation in Drosophila (Neyen et al. 2014) or application of drugs (Chamilos et al. 2008), Galleria mutants are not yet available and non-treated insect hosts are commonly used. Furthermore, insect anatomy is dramatically different from mammals, and while both Drosophila and Galleria can be infected via different routes (Lionakis & Kontoyiannis 2012, Kavanagh & Sheehan 2018), it is not possible to mimic pulmonary infection. Thus, taking into account that the mode of immunosuppression and the route of infection determine the relevance of some fungal factors in mammalian aspergillosis models, results obtained in insect models cannot be directly translated to humans without confirmation in mammalian hosts. In fact, it yet remains to be determined which murine aspergillosis model the insect hosts resemble best, although infection of insects, which share several aspects with systemic infection of mice (Kavanagh & Sheehan 2018). Other limitations include the difficulties of infecting Drosophila with an accurate dose, and the necessity to maintain Drosophila at temperatures below 30 °C. Despite this, insect models have not only been used successfully to identify fungal virulence traits, but also in testing antifungal therapy.
Due to the lack of ethical restrictions, the short generation time, and limited space needed for insect models, they are ideally suited for comparison of a large number of fungal strains, semi-high throughput screening of novel antifungal compounds, and evaluation of combination therapies (reviewed in Kavanagh & Sheehan 2018 and Jemel et al. 2020). One limitation in many studies is however the lack of information on compound concentration at the site of infection. Although this has been addressed for Galleria in several studies (Forastiero et al. 2015, Maurer et al. 2015, 2019, Astvad et al. 2017, Kloezen et al. 2018), systematic analyses of pharmacokinetics in infected larvae (Jemel et al. 2020) and a comparison of larvae from different vendors are lacking. In addition, it remains unclear to which extent the metabolisation of antifungal compounds is comparable in insects and mammals. This is important for azoles, which target fungal cytochrome P450 oxidase and are also metabolised by the host (Sugar & Liu 2000, MacCallum & Odds 2002, Mavridou et al. 2010).
Animal models are essential for the development of therapies for aspergillosis, and especially murine models have been widely applied (reviewed in Paulussen et al. (2014), Lewis & Verweij (2017)). Both murine (reviewed in Lewis & Verweij (2017)) and insects (reviewed in Jemel et al. (2020)) models have more recently also been used to address consequences of antifungal resistance for treatment outcome (Table 8). Important developments are the use of coinfection with susceptible and resistant strains or species (Alcazar-Fuoli et al. 2015), the analysis of the consequences of resistance on fungal fitness in vivo (Lackner et al. 2018), and characterisation of strains evolved during chronic infection (Ballard et al. 2018).
Table 8.
Consequences of antifungal resistance of Aspergillus species in murine models and Galleria.
| Species/mutation | Antifungal | Main finding | References |
|---|---|---|---|
| Murine models | |||
| A. fumigatus CYP51A (M220K, G54W) | Itraconazole, posaconazole | Systemic infection: resistance did not impact fitness and might increase virulence | Lackner et al. 2018 |
| A. fumigatus CYP51A (M220K, G54W) | Itraconazole, posaconazole | IPA: resistance did not impact fitness | Valsecchi et al. 2015 |
| A. fumigatus CYP51A (G448S) | Voriconazole | IPA: resistance was associated with reduced treatment efficacy | Krishnan-Natesan et al. 2012 |
| A. fumigatus CYP51A (M220I, G54W, TR34/L98H) | Isavuconazole | Systemic infection: resistance was associated with reduced treatment efficacy in a dose-dependent manner | Seyedmousavi et al. 2015a |
| A. fumigatus CYP51A (M220K) and unknown | Caspofungin, posaconazole | Systemic infection: resistance was associated with reduced treatment efficacy in a dose-dependent manner | Arendrup et al. 2008 |
| A. fumigatus | Posaconazole | IPA: resistance was associated with reduced prophylaxis efficacy | Seyedmousavi et al. 2015b |
| A. fumigatus | Itraconazole | Systemic infection: resistance was associated with reduced treatment efficacy | Denning et al. 1997a, 1997b, Dannaoui et al. 1999a, 2001 |
| A. fumigatus CYP51A and Fks1 | Posaconazole | IPA: resistance was associated with reduced treatment efficacy in a dose-dependent manner | Mavridou et al. 2010, Lepak et al. 2013 |
| A. fumigatus | Anidulafungin, voriconazole | Systemic infection: resistance was associated with reduced treatment efficacy in a dose-dependent manner | Seyedmousavi et al. 2013a |
| A. fumigatus sequential isolates from CGD patient | anidulafungin, Posaconazole | Systemic infection: resistance was associated with reduced treatment efficacy | Arendrup et al. 2010 |
| A. terreus | Voriconazole | Systemic infection: resistance was associated with reduced treatment efficacy | Salas et al. 2013 |
| A. terreus | Amphotericin B, Itraconazole | Systemic infection: only itraconazole was effective | Dannaoui et al. 2000 |
| A. flavus CYP51C | Voriconazole | Systemic infection: efficacy of voriconazole depended on drug exposure but correlated inversely with MIC | Rudramurthy et al. 2017 |
| Insect models | |||
| A. fumigatus CYP51A (M220K, G54E, G54W, TR/L98H) | Posaconazole, voriconazole | In vitro resistance associated with reduced treatment efficacy | Forastiero et al. 2015 |
| A. fumigatus CYP51A (M172V, N248T, D255E, E427K, F46Y) | Voriconazole | Resistance did not impact virulence but was associated with reduced treatment efficacy | Garcia-Rubio et al. 2018a |
| A. fumigatus CYP51A (N248K/V436A, Y433N substitution) | Itraconazole, posaconazole, 8voriconazole | Resistance did not impact virulence but was associated with reduced treatment efficacy | Chen et al. 2019a |
| A. fumigatus CYP51A (G54, M220, TR/L98) | N/A | Resistance did not impact virulence | Gomez-Lopez et al. 2014 |
| A. fumigatus sequential isolates from CGD patient | N/A | No association between virulence and resistance profile | Ballard et al. 2018 |
| A. lentulus vs A. fumigatus | Voriconazole | Aspergillus lentulus could not be eradicated by voriconazole treatment in single and mixed infections | Alcazar-Fuoli et al. 2015 |
| A. terreus; resistance mechanism not defined | Amphotericin B | In vitro susceptibility to L-AMB correlated with in vivo outcome | Maurer et al. 2015 |
One aspect that has not been extensively addressed in animal models for aspergillosis is the presence of comorbidities. Due to differences in anatomy and physiology, insect models are innately limited regarding options to model underlying diseases that affect humans at risk for aspergillosis. However, although murine models for various oncological and metabolic disorders, and infections that might occur in association with aspergillosis exist, they have not been applied to aspergillosis research. One example is aspergillosis in COVID-19 patients, discussed above. Murine models of influenza infection have been described (Thangavel & Bouvier 2014), as well as Mycobacterium tuberculosis, but a combination with aspergillosis has not yet been published. In fact, healthy young mice that are rendered immunocompromised shortly before infection might not adequately represent elderly patients with complex comorbidities receiving a variety of medications that might possibly interfere with antifungal therapy. They furthermore do not reflect the group of “non-immunocompromised” patients that develop IA (Stevens & Melikian 2011). The challenge in addressing comorbidities in animal models is that it makes the models significantly more complex and likely more difficult to standardise. Addressing this requires a combination of expertise for aspergillosis models and animal models for the respective comorbidity. This can likely be only realised by collaboration of groups across disciplines and might require a shift from the predominant use of murine models to other mammalian species like rabbits, which more closely reflect human immune response. Although laboratory mice are convenient to use and their genetic tractability allowed identification of distinct genetic polymorphisms associated with an increased risk for aspergillosis (Garlanda et al. 2002, Gresnigt et al. 2018), their small size limits repeated sampling and the course of the standard models for IA in mice is acute. For chronic types of aspergillosis, several models for ABPA have been developed (summarised in Takazono & Sheppard (2017)). In contrast, only a single model for chronic airway colonisation of immunocompetent mice has been described (Nawada et al. 1996, Urb et al. 2015, Wang et al. 2017) that has not yet been widely applied to study the chronic forms of aspergillosis. This model could be useful to address the possible role of Aspergillus in chronic pulmonary diseases such as cystic fibrosis (King et al. 2016a) and chronic pulmonary obstructive disease (COPD) (Gago et al. 2019). As mouse models for COPD and cystic fibrosis exist (Vlahos & Bozinovski 2014, Semaniakou et al. 2018), these could be combined with models of aspergillosis to gain more insight into pathogenesis.
In summary, a variety of in vivo models for aspergillosis using different host species have been described. Insect models are valuable tools for questions that require a large number of strains or compounds to be screened and can be used to generate hypotheses later to be tested in mammalian models. Mice are the most widely used mammal to study aspergillosis, with practical advantages, but also some limitations. A variety of murine models have been established, that allow addressing different types of aspergillosis. For some aspects of aspergillosis, especially in the context of different comorbidities and also co-infections, it will however be necessary to refine existing models or to develop new high order models to adequately mimic human disease physiology and pathogenesis.
Therapeutic drug monitoring and its role in adjusting optimal azole drug dosage in the clinics
The pharmacokinetic/pharmacodynamic index that describes azoles activity against Aspergillus is the time/AUC (Lepak & Andes 2014) with an fAUC/MIC 0.33–25 corresponding to 50 % survival in mice or net stasis in fungal burden in lung (Lepak & Andes 2014). Preclinical and clinical studies have demonstrated exposure-effect and -toxicity relationships for voriconazole, posaconazole and itraconazole and targets for therapeutic drug monitoring (TDM) for treatment, prophylaxis and toxicity have been determined (Table 1), although most of them were based on low quality evidence (Ashbee et al. 2014). However, the benefit of TDM in treatment response as well as in adverse events of voriconazole have been demonstrated in a randomised controlled clinical trial (Park et al. 2012). Furthermore, TDM could prevent the development of resistance which usually occurs after 4 m (range 3 to 23 mo) of azole therapy with itraconazole, posaconazole (oral solution) and voriconazole (Arendrup et al. 2010, Camps et al. 2012c) as subtherapeutic levels may be associated with emergence of azole resistance (Howard et al. 2010, Moazam et al. 2020).
Given the large number of patients (40–60 %) having unpredictably subtherapeutic/undetectable levels of voriconazole, itraconazole and posaconazole (oral solution) (Park et al. 2012, Hoenigl et al. 2013a, 2014a, Prattes et al. 2016, Yi et al. 2017), TDM is recommended for most patients even for susceptible isolates (Arendrup et al. 2020). Patients for whom TDM is particularly recommended are those with erratic absorption (e.g., due to non-compliance, mucositis, diarrhoea), distribution and elimination (e.g., due to altered pathophysiology, genetic predisposition, insufficiencies, extracorporeal devices), potential drug interactions, poor response, difficult-to-treat infections (difficult sites of infection, resistant isolates) and those belonging to special patient population (e.g., neonates, obese, elderly) (Hoenigl et al. 2014a, Lenczuk et al. 2018). Although the tablet/iv formulation of posaconazole provides sufficient exposure in most patients, the steady state with the current dosing regimens is reached after 7 d of therapy (Dekkers et al. 2016), although posaconazole levels (solution) obtained on day 3 to 5 showed high correlation with day 7 levels (Prattes et al. 2016). Considering the extra delay for TDM and dose adjustment, it may take > 2 wk until the new steady states are reached and verify that target levels are attained particularly for drugs with long half-lives like isavuconazole (Cornely et al. 2019b). Such a delay may be detrimental for invasive infections particularly in neutropenic patients since mortality increases from 41 to 90 % if effective antifungal therapy is delayed by 10 d (von Eiff et al. 1995). Population pharmacokinetic models (Hennig et al. 2006, van Iersel et al. 2018, Shi et al. 2019) and algorithms for early TDM (after 48 h) can be used to identify patients with subtherapeutic levels and speed up the dose adjustment process (Dekkers et al. 2016, Prattes et al. 2016). TDM can be coupled with monitoring fungal biomarkers like galactomannan (von Eiff et al. 1995) and PCR (Moazam et al. 2020) in order to optimise efficacy of azole therapy.
Azoles’ efficacy is reduced by resistance which is often associated with high therapeutic failure rates (∼90 %) (Howard et al. 2009, van der Linden et al. 2011). Most resistance strains have mutations in the CYP51A gene either tandem repeats in the promoter region (TR34, TR46, and TR53) and/or single point mutations (mainly in codons 54, 98, 138, 220 and 448) whereas several other non-CYP51A gene mutations have been described (CYP51B overexpression, overexpression/modification of efflux pumps, mutations in other genes) (Dudakova et al. 2017). TDM could be used to optimise azole exposure against azole-resistant isolates. An exposure-MIC relationship has been demonstrated in experimental pharmacodynamic models (Siopi et al. 2014) and in a retrospective study (Troke et al. 2011) with a recommended TDM target for voriconazole of Cmin/MIC 2 for CLSI and 1 for EUCAST. The probability of a standard voriconazole dosing regimen to attain the pharmacokinetic/pharmacodynamic (PK/PD) target for an isolate with EUCAST MIC 2 mg/L is ∼40 % requiring trough levels > 2 mg/L, which are feasible, whereas for isolates an MIC of 4 mg/L the probability drops to < 5 % requiring trough levels > 4 mg/L, which are usually associated with increased toxicity (Siopi et al. 2014). Similarly, the probability attaining the corresponding PK/PD targets of posaconazole and isavuconazole is very low for isolates with MICs > 0.5 mg/L (Seyedmousavi et al. 2014) > 2 mg/L (Espinel-Ingroff & Turnidge 2016), respectively. However, even if a clinically relevant PK/PD target has been determined, MIC-guided TDM approaches should consider the variation in MIC and in the PK/PD target. Isolates with voriconazole MIC 2 mg/L may harbour either the TR34/L98H or the M220K/R/V CYP51A mutations with MICs 2–16 mg/L and 1–4 mg/L, respectively (Seyedmousavi et al. 2014, Arendrup et al. 2017c). Thus, an isolate with MIC of 2 mg/L may have a real MIC of 4 mg/L or higher depending on the underlying resistance mechanism and the corresponding MIC distribution of mutants leaving no space of optimisation via TDM. Given that resistant isolates have usually high modal MICs to itraconazole (> 16 mg/L), voriconazole (4 mg/L for TR34/L98H and other mechanisms, > 16 mg/L for TR46/Y121F/T289A) and isavuconazole (8 mg/L for TR34/L98H, > 16 mg/L for TR46/Y121F/T289A and other mechanisms though with wide variation) but lower to posaconazole (0.5 mg/L) (Meletiadis et al. 2012, Buil et al. 2018a, 2018b), posaconazole could be a good candidate for dose optimisation against azole-resistant aspergillosis. High dose posaconazole has been used to treat IA by azole-resistant isolates with posaconazole MIC 0.25–2 mg/L in patients (4/7 patients survived and 3/7 died from underlying diseases) (Mouton et al. 2018) and in dolphin after TDM (Bunskoek et al. 2017). A high dose regimen of isavuconazole (400 mg o.d.) has also been proposed for wildtype isolates with MIC 2 mg/L (Espinel-Ingroff & Turnidge 2016) previously considered resistant but now belonging to the new category of area of technical uncertainty as isolates with posaconazole MICs 0.25 mg/L previously considered intermediate susceptible (Arendrup et al. 2020). An MIC-guided TDM has been proposed using the epidemiological cut-off value for the wild-type isolates and 4 × MIC for the non-wild type isolates (Mouton et al. 2018) limiting thus the role of TDM for azole dose optimisation against even the low-level azole-resistant isolates.
In addition to reduced efficacy, subtherapeutic levels may also be associated with emergence of azole resistance particularly in the setting of chronic aspergillosis or HSCT prophylaxis where long azole therapy is required (Steinmann et al. 2015, Moazam et al. 2020). Azole induced resistance has been described in vitro for isolates with non-CYP51A mutants and it has been associated with the overexpression of CYP51B gene and CDR1B efflux pump after exposure to itraconazole (Buied et al. 2013, Fraczek et al. 2013). Thus, it was hypothesised that azole-induced resistance which increases slightly the azole’s MICs allows fungal survival until the development of stable resistance via mutation in target genes. Slight increases in MICs of posaconazole and itraconazole with ultimately development of resistance due to CYP51A overexpression has been reported in isogenic isolates recovered from patients after prolonged azole therapy (Arendrup et al. 2010). Those isogenic resistant isolates are unusually, harbour single point mutations (Camps et al. 2012c) or other non-CYP51A mutations (Howard et al. 2010). Subtherapeutic levels have been associated with the emergence of isogenic resistant isolates with non-CYP51A mutations and of non-isogenic resistant isolates harbouring the TR34/L98H mutation (Howard et al. 2010). The PK/PD relationships and the clinical relevance of these phenomena should be further explored in order to estimate the potential role of TDM in preventing in situ development and de novo emergence of resistance.
Several methods have been used for TDM of azoles. Although chromatographic techniques (HPLC, LC-MS) are sensitive and specific, they are expensive, they have slow runtimes, they are not widely available and usually performed in central laboratories (Ashbee et al. 2014). Bioassays are cheap and simple to perform by each laboratory and can be adapted in different clinical settings including combination therapy (Siopi et al. 2016) but they lack specificity and measure total drug activity including any active metabolites. An immunoassay for quantitative measurement of voriconazole levels in serum samples has been developed and can be used in non-specialised centres (van der Elst et al. 2013). A dry spot blood technique has been developed for TDM of posaconazole, voriconazole and fluconazole (van der Elst et al. 2013). The DBS technique is simple, can be performed at home with finger prick blood, requires small sampling volumes, is less invasive and samples can be shipped at room temperature to the laboratory for analysis. A more innovative technique for TDM is the biosensor technology which provides real-time monitoring and dose adjustment of antimicrobials in a minimally invasive fashion (Rawson et al. 2018). An aptamer for sensing azole antifungal drugs has been developed moving forward this technology (Wiedman et al. 2017).
In conclusion, TDM of azoles is important in order to identify and optimise drug exposure in patients with subtherapeutic levels which are often observed in a large subset of patients treated particularly with voriconazole, itraconazole and the oral solution of posaconazole. Optimising azole exposure against low-level azole-resistant isolates in clinical settings is challenged by the precise and rapid determination of pathogen’s MIC and the underlying resistance mechanism, the turnaround time of drug levels and the dose optimisation techniques for attaining steady target levels. Azole-induced resistance is an area that requires further research to understand the PK/PD relationships and define target levels. The development of bedside point-of-care tests for measuring drug levels is crucial for efficient TDM strategies. Because of azoles’ complex pharmacology and non-linear pharmacokinetics, dose adjustment should be coupled with TDM early on therapy and frequent monitoring of drug levels depending on the disease and patient status.
Treating azole-resistant aspergillosis in the clinic
As discussed, the triazoles including voriconazole, posaconazole, and isavuconazole are recommended first-line antifungal therapy for IA, with itraconazole an option for the treatment of chronic pulmonary aspergillosis. Unfortunately, azole resistance has been an emerging problem over the last two decades, and treatment relies on either monotherapy with lipid formulations of amphotericin B or an echinocandin or combination therapy with an echinocandin or triazole plus lipid formulations of amphotericin B. Recent recommendations from the Netherlands (Schauwvlieghe et al. 2018a) outline the importance of primary treatment with lipid formulations of amphotericin B in settings where azole resistance is above 10 %, with a later switch to an azole if the isolate turns out to be susceptible to azoles. Several promising new antifungal agents are on the horizon that may serve an important role in the treatment of azole-resistant aspergillosis.
Amphotericin B appears inferior to the triazoles in the treatment of aspergillosis. In a large randomised trial comparing amphotericin B deoxycholate to voriconazole in patients with hematologic malignancy, the group receiving amphotericin B had lower rates of treatment response and survival at 12 wk compared to those who received voriconazole (Herbrecht et al. 2002). When treating azole-resistant aspergillosis with amphotericin B, lipid formulations are preferable to amphotericin B deoxycholate due to a decreased risk of toxicity and fewer infusion-related side effects (Hiemenz & Walsh 1996). In addition, a dose of 3 mg/kg of liposomal amphotericin B daily is preferable to 10 mg/kg daily given that no benefit has been seen and higher rates of nephrotoxicity associated with higher doses (Cornely et al. 2007). While the Infectious Diseases Society of America (IDSA) and European Conference on Infections in Leukemia (ECIL-6) do not specifically give recommendations on the treatment of azole-resistant aspergillosis, the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) gives liposomal amphotericin B a strong recommendation for the treatment of voriconazole-resistant IA (MIC = 2 mg/mL), a moderate recommendation for the treatment of chronic pulmonary aspergillosis and a strong recommendation for the treatment of aspergillosis with an MIC to voriconazole > 2 mg/mL (Hiemenz & Walsh 1996, Ullmann et al. 2018).
The echinocandins are not preferable for first-line treatment of IA, although they can be used in salvage therapy or combination therapy as discussed in more detail below. While caspofungin is approved by the FDA for the treatment of IA, other echinocandins such as micafungin and anidulafungin are not FDA approved, although they have activity against Aspergillus species and appear equally efficacious to caspofungin. In one study of patients with IA who were intolerant to or refractory to other preferable agents, caspofungin showed utility as salvage therapy (Maertens et al. 2004). The ESCMID gives monotherapy with caspofungin or micafungin a marginal recommendation for the treatment of voriconazole-resistant aspergillosis (MIC > 2 mg/mL) (Ullmann et al. 2018) and a marginal recommendation (caspofungin) and a moderate recommendation (micafungin) for the treatment of progressive chronic pulmonary aspergillosis with triazole resistance (Hiemenz & Walsh 1996).
Combination therapy with liposomal amphotericin B and caspofungin has shown some benefit compared to monotherapy with liposomal amphotericin B for the treatment of IA. In a small prospective study of patients with hematologic malignancy, this antifungal combination was associated with favourable overall response and increased survival at 12 wk, although the survival difference was not statistically significant (Caillot et al. 2007). This combination has also shown promise in retrospective studies as salvage therapy (Aliff et al. 2003, Marr et al. 2004), although conclusions were limited due to the study design and small sample sizes.
Combination therapy with voriconazole plus an echinocandin has shown promise for the treatment of IA in some studies (Marr et al. 2004, 2015, Viscoli 2004, Singh et al. 2006), although again conclusions for this approach are limited based on study design and small sample sizes. In addition, none of these studies specifically looked at the treatment of azole-resistant aspergillosis. Still, the ESCMID gives combination therapy with voriconazole plus an echinocandin strong recommendation for voriconazole-resistant aspergillosis (MIC > 2 mg/mL) and voriconazole plus anidulafungin a moderate recommendation and posaconazole plus caspofungin a marginal recommendation for voriconazole-resistant aspergillosis with an MIC > 2 mg/mL (Ullmann et al. 2018).
New horizon
There are several new antifungal agents with promise for the treatment of azole-resistant aspergillosis. Fosmanogepix targets the highly conserved enzyme Gwt1 which catalyses an early step in glycosylphosphatidylinositol anchor biosynthesis, compromising cell wall integrity and fungal growth. This compound has been evaluated in vivo (Zhao et al. 2019) and in the murine model of IA (Gebremariam et al. 2019) and a phase II study is underway (ClinicalTrials.gov 2020b). Olorofim (previously F901318) is a novel member of the orotomide class that targets dihydroorotate dehydrogenase (DHODH), an important enzyme for pyrimidine biosynthesis. It has been evaluated in vitro (Buil et al. 2017a, du Pré et al. 2018), in the murine model of IA (Hope et al. 2017), and is currently being evaluated in a phase II clinical trial (ClinicalTrials.gov 2020a). Ibrexafungerp and rezafungin are other novel antifungals that show synergy when used in combination with azoles. Ibrexafungerp has shown to improve azole susceptibility for azole-resistant strains of A. calidoustus and A. terreus when used in combination. Rezafungin has shown some activity against azole resistant Aspergillus in a mouse model (Miesel et al. 2019). Further clinical data on these compounds is needed before determining their place in the treatment of azole-resistant aspergillosis.
Future perspectives
In the near future, there is a need for antifungal susceptibility testing of Aspergillus species to become the standard of care around the world. To accomplish this goal, more rapid and sensitive methods to detect ARAF need to be developed. Optimally, these methods would be applied as rapid tests and detect a broader spectrum of resistance markers directly in the clinical sample. Knowledge about the epidemiology of Aspergillus susceptibility patterns will represent a cornerstone for guiding the appropriate selection of antifungal prophylaxis and treatment. Given that more areas may be burdened with high rates of environmental triazole resistance, triazoles may not be universally recommended as primary antifungal treatment, but instead, treatment choice may depend on local epidemiology of ARAF. New antifungal agents, such as ibrexafungerp, that are currently in clinical stage evaluation with novel mechanisms of action may have central roles in treating these azole resistant infections, as well as species like A. terreus which are often less-susceptible to amphotericin B.
Funding disclosure
AA, RGR, and DSP were supported by NIH AI 109025. MH was supported by NIH UL1TR001442. AC was supported by the Fundação para a Ciência e a Tecnologia (FCT) (CEECIND/03628/2017 and PTDC/MED GEN/28778/2017). Additional support was provided by FCT (UIDB/50026/2020 and UIDP/50026/2020), the Northern Portugal Regional Operational Programme (NORTE 2020), under the Portugal 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) (NORTE-01-0145-FEDER-000013 and NORTE-01-0145-FEDER-000023), the European Union's Horizon 2020 Research and Innovation programme under grant agreement no. 847507, and the “la Caixa” Foundation (ID 100010434) and FCT under the agreement LCF/PR/HP17/52190003. DJA was supported by CF Trust Strategic Research Centre TrIFIC (SRC015), Wellcome Trust Collaborative Award 219551/Z/19/Z and the NIHR Centre for Antimicrobial Optimisation.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Contributor Information
D.S. Perlin, Email: david.perlin@hmh-cdi.org.
M. Hoenigl, Email: hoeniglmartin@gmail.com.
References
- Abdolrasouli A., Petrou M.A., Park H. Surveillance for azole-resistant Aspergillus fumigatus in a centralized diagnostic mycology service, London, United Kingdom, 1998–2017. Frontiers in Microbiology. 2018;9:2234. doi: 10.3389/fmicb.2018.02234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolrasouli A., Rhodes J., Beale M.A. Genomic context of azole resistance mutations in Aspergillus fumigatus determined using whole-genome sequencing. mBio. 2015;6 doi: 10.1128/mBio.00536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdolrasouli A., Scourfield A., Rhodes J. High prevalence of triazole resistance in clinical Aspergillus fumigatus isolates in a specialist cardiothoracic centre. International Journal of Antimicrobial Agents. 2018;52:637–642. doi: 10.1016/j.ijantimicag.2018.08.004. [DOI] [PubMed] [Google Scholar]
- Agarwal R., Chakrabarti A., Shah A. Allergic bronchopulmonary aspergillosis: review of literature and proposal of new diagnostic and classification criteria. Clinical and Experimental Allergy. 2013;43:850–873. doi: 10.1111/cea.12141. [DOI] [PubMed] [Google Scholar]
- Ahangarkani F., Puts Y., Nabili M. First azole-resistant Aspergillus fumigatus isolates with the environmental TR46/Y121F/T289A mutation in Iran. Mycoses. 2020;63:430–436. doi: 10.1111/myc.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad S., Joseph L., Hagen F. Concomitant occurrence of itraconazole-resistant and -susceptible strains of Aspergillus fumigatus in routine cultures. Journal of Antimicrobial Chemotherapy. 2015;70:412–415. doi: 10.1093/jac/dku410. [DOI] [PubMed] [Google Scholar]
- Ahmad S., Khan Z., Hagen F. Occurrence of triazole-resistant Aspergillus fumigatus with TR34/L98H mutations in outdoor and hospital environment in Kuwait. Environmental Research. 2014;133:20–26. doi: 10.1016/j.envres.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Al Abdallah Q., Ge W., Fortwendel J.R. A simple and universal system for gene manipulation in Aspergillus fumigatus: In vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere. 2017;2 doi: 10.1128/mSphere.00446-17. e00446–00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanio A., Delliére S., Fodil S. Prevalence of putative invasive pulmonary aspergillosis in critically ill patients with COVID-19. Lancet Respiratory Medicine. 2020;8:e48–e49. doi: 10.1016/S2213-2600(20)30237-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alanio A., Sitterle E., Liance M. Low prevalence of resistance to azoles in Aspergillus fumigatus in a French cohort of patients treated for haematological malignancies. Journal of Antimicrobial Chemotherapy. 2011;66:371–374. doi: 10.1093/jac/dkq450. [DOI] [PubMed] [Google Scholar]
- Alastruey-Izquierdo A., Alcazar-Fuoli L., Cuenca-Estrella M. Antifungal susceptibility profile of cryptic species of Aspergillus. Mycopathologia. 2014;178:427–433. doi: 10.1007/s11046-014-9775-z. [DOI] [PubMed] [Google Scholar]
- Alastruey-Izquierdo A., Mellado E., Peláez T. Population-based survey of filamentous fungi and antifungal resistance in Spain (FILPOP Study) Antimicrobial Agents and Chemotherapy. 2013;57:4604. doi: 10.1128/AAC.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albarrag A.M., Anderson M.J., Howard S.J. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrobial Agents and Chemotherapy. 2011;55:5113–5121. doi: 10.1128/AAC.00517-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Buitrago M., Gomez-Lopez A. An alternative host model of a mixed fungal infection by azole susceptible and resistant Aspergillus spp strains. Virulence. 2015;6:376–384. doi: 10.1080/21505594.2015.1025192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcazar-Fuoli L., Mellado E., García-Effron G. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids. 2008;73:339–347. doi: 10.1016/j.steroids.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Aldosari M.A., Alghamdi M.H., Alhamdan A.A. Native valve fungal endocarditis caused by Aspergillus fumigatus: management dilemma. Oxford Medical Case Reports. 2020;2020 doi: 10.1093/omcr/omz147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliff T.B., Maslak P.G., Jurcic J.G. Refractory Aspergillus pneumonia in patients with acute leukemia: successful therapy with combination caspofungin and liposomal amphotericin. Cancer. 2003;97:1025–1032. doi: 10.1002/cncr.11115. [DOI] [PubMed] [Google Scholar]
- Alshehri B., Palanisamy M. Evaluation of molecular identification of Aspergillus species causing fungal keratitis. Saudi Journal of Biological Sciences. 2020;27:751–756. doi: 10.1016/j.sjbs.2019.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Escribano I., Sasse C., Bok J.W. Genome sequencing of evolved aspergilli populations reveals robust genomes, transversions in A. flavus, and sexual aberrancy in non-homologous end-joining mutants. BMC Biology. 2019;17:88. doi: 10.1186/s12915-019-0702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Moreno C., Lavergne R.A., Hagen F. Azole-resistant Aspergillus fumigatus harboring TR34/L98H, TR46/Y121F/T289A and TR53 mutations related to flower fields in Colombia. Scientific Reports. 2017;7:45631. doi: 10.1038/srep45631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Moreno C., Lavergne R.A., Hagen F. Fungicide-driven alterations in azole-resistant Aspergillus fumigatus are related to vegetable crops in Colombia, South America. Mycologia. 2019;111:217–224. doi: 10.1080/00275514.2018.1557796. [DOI] [PubMed] [Google Scholar]
- Américo F.M., Machado Siqueira L.P., Del Negro G.M.B. Evaluating VITEK MS for the identification of clinically relevant Aspergillus species. Medical Mycology. 2020;58:322–327. doi: 10.1093/mmy/myz066. [DOI] [PubMed] [Google Scholar]
- Amich J., Mokhtari Z., Strobel M. Three-dimensional light sheet fluorescence microscopy of lungs to dissect local host immune-Aspergillus fumigatus interactions. mBio. 2020;11 doi: 10.1128/mBio.02752-19. e02752–e02719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amri M., Gorcii M., Essabbah N. Aspergillus sclerotiorum: à propos d’un cas d’onychomycose en Tunisie. Journal of Medical Mycology. 2020;20:128–132. [Google Scholar]
- Arastehfar A., Carvalho A., van de Veerdonk F.L. COVID-19 associated pulmonary aspergillosis (CAPA) –From immunology to treatment. Journal of Fungi. 2020;6:91. doi: 10.3390/jof6020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arastehfar A., Lass-Flörl C., Garcia-Rubio R. The quiet and underappreciated rise of drug-resistant invasive fungal pathogens. Journal of Fungi. 2020;6:E138. doi: 10.3390/jof6030138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M.C., Friberg N., Mares M. How to interpret MICs of antifungal compounds according to the revised clinical breakpoints v. 10.0 European committee on antimicrobial susceptibility testing (EUCAST) Clinical Microbiology and Infection. 2020;26:1464–1472. doi: 10.1016/j.cmi.2020.06.007. [DOI] [PubMed] [Google Scholar]
- Arendrup M.C., Mavridou E., Mortensen K.L. Development of azole resistance in Aspergillus fumigatus during azole therapy associated with change in virulence. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M.C., Meletiadis J., Mouton J.W. 2017. EUCAST Definitive Document E.DEF 9.3.1: Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia forming moulds.http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_9_3_1_Mould_testing__definitive.pdf [DOI] [PubMed] [Google Scholar]
- Arendrup M.C., Perkhofer S., Howard S.J. Establishing in vitro-in vivo correlations for Aspergillus fumigatus: the challenge of azoles versus echinocandins. Antimicrobial Agents and Chemotherapy. 2008;52:3504–3511. doi: 10.1128/AAC.00190-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M.C., Verweij P., Nielsen H.V. Evaluation of MIC strip isavuconazole test for susceptibility testing of wild-type and non-wild-type Aspergillus fumigatus isolates. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.01659-16. e01659–e01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendrup M.C., Verweij P.E., Mouton J.W. Multicentre validation of 4-well azole agar plates as a screening method for detection of clinically relevant azole-resistant Aspergillus fumigatus. Journal of Antimicrobial Chemotherapy. 2017;72:3325–3333. doi: 10.1093/jac/dkx319. [DOI] [PubMed] [Google Scholar]
- Aruanno M., Glampedakis E., Lamoth F. Echinocandins for the treatment of invasive aspergillosis: from laboratory to bedside. Antimicrobial Agents and Chemotherapy. 2019;63:e00399–e00419. doi: 10.1128/AAC.00399-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis M., Ziakas P.D., Zacharioudakis I.M. PCR in diagnosis of invasive aspergillosis: a meta-analysis of diagnostic performance. Journal of Clinical Microbiology. 2014;52:3731–3742. doi: 10.1128/JCM.01365-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano M., Kano R., Makimura K. Molecular typing and in-vitro activity of azoles against clinical isolates of Aspergillus fumigatus and A. niger in Japan. Journal of Infection and Chemotherapy. 2011;17:483–486. doi: 10.1007/s10156-010-0202-1. [DOI] [PubMed] [Google Scholar]
- Ashbee H.R., Barnes R.A., Johnson E.M. Therapeutic drug monitoring (TDM) of antifungal agents: guidelines from the British Society for Medical Mycology. Journal of Antimicrobial Chemotherapy. 2014;69:1162–1176. doi: 10.1093/jac/dkt508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astvad K.M., Jensen R.H., Hassan T.M. First detection of TR46/Y121F/T289A and TR34/L98H alterations in Aspergillus fumigatus isolates from azole-naïve patients in Denmark despite negative findings in the environment. Antimicrobial Agents and Chemotherapy. 2014;58:5096–5101. doi: 10.1128/AAC.02855-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astvad K.M.T., Meletiadis J., Whalley S. Fluconazole pharmacokinetics in Galleria mellonella larvae and performance evaluation of a bioassay compared to liquid chromatography-tandem mass spectrometry for hemolymph specimens. Antimicrobial Agents and Chemotherapy. 2017;61:e00895–e00917. doi: 10.1128/AAC.00895-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astvad K.M.T., Sanglard D., Delarze E. Implications of the EUCAST trailing phenomenon in Candida tropicalis for the in vivo susceptibility in invertebrate and murine models. Antimicrobial Agents and Chemotherapy. 2018;62 doi: 10.1128/AAC.01624-18. e01624–01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aznar C., de Bievre C., Guiguen C. Maxillary sinusitis from Microascus cinereus and Aspergillus repens. Mycopathologia. 1989;105:93–97. doi: 10.1007/BF00444031. [DOI] [PubMed] [Google Scholar]
- Babamahmoodi F., Shokohi T., Ahangarkani F. Rare case of Aspergillus ochraceus osteomyelitis of calcaneus bone in a patient with diabetic foot ulcers. Case Reports in Medicine. 2015;2015:509827. doi: 10.1155/2015/509827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badali H., Vaezi A., Haghani I. Environmental study of azole-resistant Aspergillus fumigatus with TR34/L98H mutations in the cyp51A gene in Iran. Mycoses. 2013;56:659–663. doi: 10.1111/myc.12089. [DOI] [PubMed] [Google Scholar]
- Bader O., Tünnermann J., Dudakova A. Environmental isolates of azole-resistant Aspergillus fumigatus in Germany. Antimicrobial Agents and Chemotherapy. 2015;59:4356–4359. doi: 10.1128/AAC.00100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J.M., Tavanti A., Davidson A.D. Multilocus sequence typing of the pathogenic fungus Aspergillus fumigatus. Journal of Clinical Microbiology. 2007;45:1469–1477. doi: 10.1128/JCM.00064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee S.A., Gribskov J.L., Hanley E. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryotic Cell. 2005;4:625–632. doi: 10.1128/EC.4.3.625-632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee S.A., Houbraken J., Verweij P.E. Aspergillus species identification in the clinical setting. Studies in Mycology. 2007;59:39–46. doi: 10.3114/sim.2007.59.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee S.A., Kano R., Baddley J.W. Molecular identification of Aspergillus species collected for the Transplant-Associated Infection Surveillance Network. Journal of Clinical Microbiology. 2009;47:3138–3141. doi: 10.1128/JCM.01070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balajee S.A., Tay S.T., Lasker B.A. Characterization of a novel gene for strain typing reveals substructuring of Aspergillus fumigatus across North America. Eukaryotic Cell. 2007;6:1392–1399. doi: 10.1128/EC.00164-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard E., Melchers W.J.G., Zoll J. In-host microevolution of Aspergillus fumigatus: A phenotypic and genotypic analysis. Fungal Genetics and Biology. 2018;113:1–13. doi: 10.1016/j.fgb.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballard E., Weber J., Melchers W.J.G. Recreation of in-host acquired single nucleotide polymorphisms by CRISPR-Cas9 reveals an uncharacterised gene playing a role in Aspergillus fumigatus azole resistance via a non-cyp51A mediated resistance mechanism. Fungal Genetics and Biology. 2019;130:98–106. doi: 10.1016/j.fgb.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfalvi G. Improved and adopted murine models to combat pulmonary aspergillosis. Applied Microbiology and Biotechnology. 2018;102:6865–6875. doi: 10.1007/s00253-018-9161-8. [DOI] [PubMed] [Google Scholar]
- Bart-Delabesse E., Humbert J.F., Delabesse E. Microsatellite markers for typing Aspergillus fumigatus isolates. Journal of Clinical Microbiology. 1998;36:2413–2418. doi: 10.1128/jcm.36.9.2413-2418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoletti M., Pascale R., Cricca M. Epidemiology of invasive pulmonary aspergillosis among COVID-19 intubated patients: a prospective study. Clinical Infectious Diseases. 2021;28 doi: 10.1093/cid/ciaa1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassetti M., Giacobbe D.R., Grecchi C. Performance of existing definitions and tests for the diagnosis of invasive aspergillosis in critically ill, adult patients: A systematic review with qualitative evidence synthesis. Journal of Infection. 2020;81:131–146. doi: 10.1016/j.jinf.2020.03.065. [DOI] [PubMed] [Google Scholar]
- Bassetti M., Peghin M., Vena A. Challenges and solution of invasive aspergillosis in non-neutropenic patients: A review. Infectious Diseases and Therapy. 2018;7:17–27. doi: 10.1007/s40121-017-0183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R.W., Valero C., Silva L.P. Functional characterization of clinical isolates of the opportunistic fungal pathogen Aspergillus nidulans. mSphere. 2020;5 doi: 10.1128/mSphere.00153-20. e00153–00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter C.G., Bishop P., Low S.E. Pulmonary aspergillosis: an alternative diagnosis to lung cancer after positive [18F]FDG positron emission tomography. Thorax. 2011;66:638–640. doi: 10.1136/thx.2010.155515. [DOI] [PubMed] [Google Scholar]
- Becker M.J., De Marie S., Fens M.H. Pathophysiology of unilateral pulmonary aspergillosis in an experimental rat model. Medical Mycology. 2006;44:133–139. doi: 10.1080/13693780500271749. [DOI] [PubMed] [Google Scholar]
- Bellanger A.P., Millon L., Khoufache K. Aspergillus fumigatus germ tube growth and not conidia ingestion induces expression of inflammatory mediator genes in the human lung epithelial cell line A549. Journal of Medical Microbiology. 2009;58:174–179. doi: 10.1099/jmm.0.005488-0. [DOI] [PubMed] [Google Scholar]
- Bellete B., Raberin H., Morel J. Acquired resistance to voriconazole and itraconazole in a patient with pulmonary aspergilloma. Medical Mycology. 2010;48:197–200. doi: 10.3109/13693780902717018. [DOI] [PubMed] [Google Scholar]
- Bellmann R., Smuszkiewicz P. Pharmacokinetics of antifungal drugs: practical implications for optimized treatment of patients. Infection. 2017;45:737–779. doi: 10.1007/s15010-017-1042-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenguer J., Allende M.C., Lee J.W. Pathogenesis of pulmonary aspergillosis. Granulocytopenia versus cyclosporine and methylprednisolone-induced immunosuppression. American Journal of Respiratory and Critical Care Medicine. 1995;152:1079–1086. doi: 10.1164/ajrccm.152.3.7663787. [DOI] [PubMed] [Google Scholar]
- Bergeron A., Porcher R., Sulahian A. The strategy for the diagnosis of invasive pulmonary aspergillosis should depend on both the underlying condition and the leukocyte count of patients with hematologic malignancies. Blood. 2012;119:1831–1837. doi: 10.1182/blood-2011-04-351601. quiz 1956. [DOI] [PubMed] [Google Scholar]
- Berkow E.L., Nunnally N.S., Bandea A. Detection of TR34/L98H CYP51A mutation through passive surveillance for azole-resistant Aspergillus fumigatus in the United States from 2015 to 2017. Antimicrobial Agents and Chemotherapy. 2018;62 doi: 10.1128/AAC.02240-17. e02240–02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J., Krysan D.J. Drug resistance and tolerance in fungi. Nature Reviews Microbiology. 2020;18:319–331. doi: 10.1038/s41579-019-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Martínez L., Gil H., Rivero-Menéndez O. Development and validation of a high-resolution melting assay to detect azole resistance in Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 2017;61:e01083–17. doi: 10.1128/AAC.01083-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertout S., Renaud F., Barton R. Genetic polymorphism of Aspergillus fumigatus in clinical samples from patients with invasive aspergillosis: investigation using multiple typing methods. Journal of Clinical Microbiology. 2001;39:1731–1737. doi: 10.1128/JCM.39.5.1731-1737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertout S., Renaud F., De Meeüs T., The Ebga Network Multilocus enzyme electrophoresis analysis of Aspergillus fumigatus strains isolated from the first clinical sample from patients with invasive aspergillosis. EBGA Network. European Research Group on Biotype and Genotype of Aspergillus. Journal of Medical Microbiology. 2000;49:375–381. doi: 10.1099/0022-1317-49-4-375. [DOI] [PubMed] [Google Scholar]
- Bidula S., Schelenz S. A sweet response to a sour situation: The role of soluble pattern pecognition receptors in the innate immune response to invasive Aspergillus fumigatus infections. PLoS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidula S., Sexton D.W., Abdolrasouli A. The serum opsonin L-ficolin is detected in lungs of human transplant recipients following fungal infections and modulates inflammation and killing of Aspergillus fumigatus. Journal of Infectious Diseases. 2015;212:234–246. doi: 10.1093/infdis/jiv027. [DOI] [PubMed] [Google Scholar]
- Binder U., Lass-Flörl C. New insights into invasive aspergillosis – from the pathogen to the disease. Current Pharmaceutical Design. 2013;19:3679–3688. doi: 10.2174/13816128113199990366. [DOI] [PubMed] [Google Scholar]
- Binder U., Maurer E., Lass-Flörl C. Galleria mellonella: An invertebrate model to study pathogenicity in correctly defined fungal species. Fungal Biology. 2016;120:288–295. doi: 10.1016/j.funbio.2015.06.002. [DOI] [PubMed] [Google Scholar]
- Blaize M., Mayaux J., Nabet C. Fatal invasive aspergillosis and coronavirus disease in an immunocompetent patient. Emerging Infectious Diseases. 2020;26:1636–1637. doi: 10.3201/eid2607.201603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatzer M., Latgé J.P. Metal-homeostasis in the pathobiology of the opportunistic human fungal pathogen Aspergillus fumigatus. Current Opinion in Microbiology. 2017;40:152–159. doi: 10.1016/j.mib.2017.11.015. [DOI] [PubMed] [Google Scholar]
- Blosser S.J., Cramer R.A. SREBP-dependent triazole susceptibility in Aspergillus fumigatus is mediated through direct transcriptional regulation of erg11A (cyp51A) Antimicrobial Agents and Chemotherapy. 2012;56:248–257. doi: 10.1128/AAC.05027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G., Hörtnagl C., Jukic E. New insight into amphotericin B resistance in Aspergillus terreus. Antimicrobial Agents and Chemotherapy. 2013;57:1583–1588. doi: 10.1128/AAC.01283-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G., Perkhofer S., Grif K. A 1-year Aspergillus terreus surveillance study at the University Hospital of Innsbruck: molecular typing of environmental and clinical isolates. Clinical Microbiology and Infection. 2008;14:1146–1151. doi: 10.1111/j.1469-0691.2008.02099.x. [DOI] [PubMed] [Google Scholar]
- Blumental S., Mouy R., Mahlaoui N. Invasive mold infections in chronic granulomatous disease: a 25-year retrospective survey. Clinical Infectious Diseases. 2011;53:e159–e169. doi: 10.1093/cid/cir731. [DOI] [PubMed] [Google Scholar]
- Bongomin F., Batac C.R., Richardson M.D. A review of onychomycosis due to Aspergillus species. Mycopathologia. 2018;183:485–493. doi: 10.1007/s11046-017-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger P., Koëter G.H., Timmerman J.A. Proteases from Aspergillus fumigatus induce interleukin (IL)-6 and IL-8 production in airway epithelial cell lines by transcriptional mechanisms. Journal of Infectious Diseases. 1999;180:1267–1274. doi: 10.1086/315027. [DOI] [PubMed] [Google Scholar]
- Borgohain P., Barua P., Dutta P.J. Onychomycosis associated with superficial skin infection due to Aspergillus sydowii in an immunocompromised patient. Mycopathologia. 2019;184:683–689. doi: 10.1007/s11046-019-00383-2. [DOI] [PubMed] [Google Scholar]
- Brakhage A.A., Langfelder K. Menacing mold: the molecular biology of Aspergillus fumigatus. Annual Review of Microbiology. 2002;56:433–455. doi: 10.1146/annurev.micro.56.012302.160625. [DOI] [PubMed] [Google Scholar]
- Brasch J., Varga J., Jensen J.M. Nail infection by Aspergillus ochraceopetaliformis. Medical Mycology. 2009;47:658–662. doi: 10.1080/13693780902803032. [DOI] [PubMed] [Google Scholar]
- Brock M. Fungal metabolism in host niches. Current Opinion in Microbiology. 2009;12:371–376. doi: 10.1016/j.mib.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Bromley M.J., van Muijlwijk G., Fraczek M.G. Occurrence of azole-resistant species of Aspergillus in the UK environment. Journal of Global Antimicrobial Resistance. 2014;2:276–279. doi: 10.1016/j.jgar.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Brown G.D., Denning D.W., Gow N.A. Hidden killers: human fungal infections. Science Translational Medicine. 2012;4:165rv113. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- Brown G.D., Willment J.A., Whitehead L. C-type lectins in immunity and homeostasis. Nature Reviews Immunology. 2018;18:374–389. doi: 10.1038/s41577-018-0004-8. [DOI] [PubMed] [Google Scholar]
- Buchheidt D., Reinwald M., Hoenigl M. The evolving landscape of new diagnostic tests for invasive aspergillosis in hematology patients: strengths and weaknesses. Current Opinion in Infectious Diseases. 2017;30:539–544. doi: 10.1097/QCO.0000000000000408. [DOI] [PubMed] [Google Scholar]
- Bueid A., Howard S.J., Moore C.B. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. Journal of Antimicrobial Chemotherapy. 2010;65:2116–2118. doi: 10.1093/jac/dkq279. [DOI] [PubMed] [Google Scholar]
- Buied A., Moore C.B., Denning D.W. High-level expression of cyp51B in azole-resistant clinical Aspergillus fumigatus isolates. Journal of Antimicrobial Chemotherapy. 2013;68:512–514. doi: 10.1093/jac/dks451. [DOI] [PubMed] [Google Scholar]
- Buil J.B., Bruggemann R.J.M., Wasmann R.E. Isavuconazole susceptibility of clinical Aspergillus fumigatus isolates and feasibility of isavuconazole dose escalation to treat isolates with elevated MICs. Journal of Antimicrobial Chemotherapy. 2018;73:134–142. doi: 10.1093/jac/dkx354. [DOI] [PubMed] [Google Scholar]
- Buil J.B., Hagen F., Chowdhary A. Itraconazole, voriconazole, and posaconazole CLSI MIC distributions for wild-type and azole-resistant Aspergillus fumigatus isolates. Journal of Fungi. 2018;4:103. doi: 10.3390/jof4030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buil J.B., Rijs A., Meis J.F. In vitro activity of the novel antifungal compound F901318 against difficult-to-treat Aspergillus isolates. Journal of Antimicrobial Chemotherapy. 2017;72:2548–2552. doi: 10.1093/jac/dkx177. [DOI] [PubMed] [Google Scholar]
- Buil J.B., van der Lee H.A.L., Rijs A. Single-center evaluation of an agar-based screening for azole resistance in Aspergillus fumigatus by using VIPcheck. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.01250-17. e01250–01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunskoek P.E., Seyedmousavi S., Gans S.J. Successful treatment of azole-resistant invasive aspergillosis in a bottlenose dolphin with high-dose posaconazole. Medical Mycology Case Reports. 2017;16:16–19. doi: 10.1016/j.mmcr.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt I., Zimmermann S. Susceptibility testing of bacteria using MALDI-ToF mass spectrometry. Frontiers in Microbiology. 2018;9:1744. doi: 10.3389/fmicb.2018.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillot D., Thiebaut A., Herbrecht R. Liposomal amphotericin B in combination with caspofungin for invasive aspergillosis in patients with hematologic malignancies: a randomized pilot study (Combistrat trial) Cancer. 2007;110:2740–2746. doi: 10.1002/cncr.23109. [DOI] [PubMed] [Google Scholar]
- Cairns T.C., Studholme D.J., Talbot N.J. New and improved techniques for the study of pathogenic fungi. Trends in Microbiology. 2016;24:35–50. doi: 10.1016/j.tim.2015.09.008. [DOI] [PubMed] [Google Scholar]
- Camargo J.F., Husain S. Immune correlates of protection in human invasive aspergillosis. Clinical Infectious Diseases. 2014;59:569–577. doi: 10.1093/cid/ciu337. [DOI] [PubMed] [Google Scholar]
- Camps S.M., Dutilh B.E., Arendrup M.C. Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps S.M., Rijs A.J., Klaassen C.H. Molecular epidemiology of Aspergillus fumigatus isolates harboring the TR34/L98H azole resistance mechanism. Journal of Clinical Microbiology. 2012;50:2674–2680. doi: 10.1128/JCM.00335-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps S.M., van der Linden J.W., Li Y. Rapid induction of multiple resistance mechanisms in Aspergillus fumigatus during azole therapy: a case study and review of the literature. Antimicrobial Agents and Chemotherapy. 2012;56:10–16. doi: 10.1128/AAC.05088-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara B., Richards R., Imbert S. Species distribution and comparison between EUCAST and gradient concentration strips methods for antifungal susceptibility testing of 112 Aspergillus section Nigri isolates. Antimicrobial Agents and Chemotherapy. 2020;64 doi: 10.1128/AAC.02510-19. e02510–02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro C., Serrano M.C., Flores B. Comparison of the Sensititre YeastOne colorimetric antifungal panel with a modified NCCLS M38-A method to determine the activity of voriconazole against clinical isolates of Aspergillus spp. Journal of Clinical Microbiology. 2004;42:4358–4360. doi: 10.1128/JCM.42.9.4358-4360.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai L.Y., de Boer M.G., van der Velden W.J. The Y238X stop codon polymorphism in the human beta-glucan receptor dectin-1 and susceptibility to invasive aspergillosis. Journal of Infectious Diseases. 2011;203:736–743. doi: 10.1093/infdis/jiq102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos G., Lewis R.E., Hu J. Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9367–9372. doi: 10.1073/pnas.0709578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos G., Lionakis M.S., Kontoyiannis D.P. Call for action: Invasive fungal infections associated with ibrutinib and other small molecule kinase inhibitors targeting immune signaling pathways. Clinical Infectious Diseases. 2018;66:140–148. doi: 10.1093/cid/cix687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandenier J., Bernard S., Montharu J. The utility of a nebulised intra-tracheal rat model of invasive pulmonary aspergillosis. Mycoses. 2009;52:239–245. doi: 10.1111/j.1439-0507.2009.01695.x. [DOI] [PubMed] [Google Scholar]
- Chasan R., Patel G., Malone A. Primary hepatic aspergillosis following induction chemotherapy for acute leukemia. Transplant Infectious Disease. 2013;15:E201–E205. doi: 10.1111/tid.12127. [DOI] [PubMed] [Google Scholar]
- Chen A.J., Frisvad J.C., Sun B.D. Aspergillus section Nidulantes (formerly Emericella): Polyphasic taxonomy, chemistry and biology. Studies in Mycology. 2016;84:1–118. doi: 10.1016/j.simyco.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.J., Hubka V., Frisvad J.C. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology. 2017;88:37–135. doi: 10.1016/j.simyco.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A.J., Varga J., Frisvad J.C. Polyphasic taxonomy of Aspergillus section Cervini. Studies in Mycology. 2016;85:65–89. doi: 10.1016/j.simyco.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Wu D., Guo W. Clinical and immunological features of severe and moderate coronavirus disease 2019. Journal of Clinical Investigation. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li H., Li R. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. Journal of Antimicrobial Chemotherapy. 2005;55:31–37. doi: 10.1093/jac/dkh507. [DOI] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Liu M., Zeng Q. Uncovering new mutations conferring azole resistance in the Aspergillus fumigatus cyp51A gene. Frontiers in Microbiology. 2019;10:3127. doi: 10.3389/fmicb.2019.03127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Lu Z., Zhao J. Epidemiology and molecular characterizations of azole resistance in clinical and environmental Aspergillus fumigatus isolates from China. Antimicrobial Agents and Chemotherapy. 2016;60:5878–5884. doi: 10.1128/AAC.01005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Wang H., Lu Z. Emergence of TR46/Y121F/T289A in an Aspergillus fumigatus isolate from a Chinese patient. Antimicrobial Agents and Chemotherapy. 2015;59:7148–7150. doi: 10.1128/AAC.00887-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.C., Kuo S.F., Wang H.C. Azole resistance in Aspergillus species in Southern Taiwan: An epidemiological surveillance study. Mycoses. 2019;62:1174–1181. doi: 10.1111/myc.13008. [DOI] [PubMed] [Google Scholar]
- Chiller T.M., Luque J.C., Sobel R.A. Development of a murine model of cerebral aspergillosis. Journal of Infectious Diseases. 2002;186:574–577. doi: 10.1086/341567. [DOI] [PubMed] [Google Scholar]
- Chilvers E.R., Spreadbury C.L., Cohen J. Bronchoalveolar lavage in an immunosuppressed rabbit model of invasive pulmonary aspergillosis. Mycopathologia. 1989;108:163–171. doi: 10.1007/BF00436221. [DOI] [PubMed] [Google Scholar]
- Chindamporn A., Chakrabarti A., Li R. Survey of laboratory practices for diagnosis of fungal infection in seven Asian countries: An Asia Fungal Working Group (AFWG) initiative. Medical Mycology. 2018;56:416–425. doi: 10.1093/mmy/myx066. [DOI] [PubMed] [Google Scholar]
- Choi M.J., Won E.J., Joo M.Y. Microsatellite typing and resistance mechanism analysis of voriconazole-resistant Aspergillus flavus isolates in South Korean hospitals. Antimicrobial Agents and Chemotherapy. 2019;63:e01610–e01618. doi: 10.1128/AAC.01610-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong G.L., van de Sande W.W., Dingemans G.J. Validation of a new Aspergillus real-time PCR assay for direct detection of Aspergillus and azole resistance of Aspergillus fumigatus on bronchoalveolar lavage fluid. Journal of Clinical Microbiology. 2015;53:868–874. doi: 10.1128/JCM.03216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong G.M., van der Beek M.T., von dem Borne P.A. PCR-based detection of Aspergillus fumigatus Cyp51A mutations on bronchoalveolar lavage: a multicentre validation of the AsperGenius assay® in 201 patients with haematological disease suspected for invasive aspergillosis. Journal of Antimicrobial Chemotherapy. 2016;71:3528–3535. doi: 10.1093/jac/dkw323. [DOI] [PubMed] [Google Scholar]
- Choukri F., Botterel F., Sitterle E. Prospective evaluation of azole resistance in Aspergillus fumigatus clinical isolates in France. Medical Mycology. 2015;53:593–596. doi: 10.1093/mmy/myv029. [DOI] [PubMed] [Google Scholar]
- Chowdhary A., Kathuria S., Randhawa H.S. Isolation of multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR/L98H mutations in the cyp51A gene in India. Journal of Antimicrobial Chemotherapy. 2012;67:362–366. doi: 10.1093/jac/dkr443. [DOI] [PubMed] [Google Scholar]
- Chowdhary A., Kathuria S., Xu J. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the TR34/L98H mutations in the cyp51A gene in India. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A., Sharma C., Kathuria S. Azole-resistant Aspergillus fumigatus with the environmental TR46/Y121F/T289A mutation in India. Journal of Antimicrobial Chemotherapy. 2014;69:555–557. doi: 10.1093/jac/dkt397. [DOI] [PubMed] [Google Scholar]
- Chowdhary A., Sharma C., Kathuria S. Prevalence and mechanism of triazole resistance in Aspergillus fumigatus in a referral chest hospital in Delhi, India and an update of the situation in Asia. Frontiers in Microbiology. 2015;6:428. doi: 10.3389/fmicb.2015.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary A., Sharma C., van den Boom M. Multi-azole-resistant Aspergillus fumigatus in the environment in Tanzania. Journal of Antimicrobial Chemotherapy. 2014;69:2979–2983. doi: 10.1093/jac/dku259. [DOI] [PubMed] [Google Scholar]
- Chrenkova V., Hubka V., Cetkovsky P. Proven invasive pulmonary aspergillosis in stem cell transplant recipient due to Aspergillus sublatus, a cryptic species of A. nidulans. Mycopathologia. 2018;183:423–429. doi: 10.1007/s11046-017-0223-8. [DOI] [PubMed] [Google Scholar]
- Chryssanthou E. In vitro susceptibility of respiratory isolates of Aspergillus species to itraconazole and amphotericin B. Acquired resistance to itraconazole. Scandinavian Journal of Infectious Diseases. 1997;29:509–512. doi: 10.3109/00365549709011864. [DOI] [PubMed] [Google Scholar]
- Chung D., Barker B.M., Carey C.C. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark H.L., Jhingran A., Sun Y. Zinc and manganese chelation by neutrophil S100A8/A9 (Calprotectin) limits extracellular Aspergillus fumigatus hyphal growth and corneal infection. Journal of Immunology. 2016;196:336–344. doi: 10.4049/jimmunol.1502037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons K.V., Schwartz J.A., Stevens D.A. Experimental central nervous system aspergillosis therapy: efficacy, drug levels and localization, immunohistopathology, and toxicity. Antimicrobial Agents and Chemotherapy. 2012;56:4439–4449. doi: 10.1128/AAC.06015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinicalTrials.gov . 2020. Evaluate F901318 treatment of invasive fungal infections in patients lacking treatment options (FORMULA-OLS) [Google Scholar]
- ClinicalTrials.gov . 2020. An open-label study of APX001 for treatment of patients with invasive mold infections caused by Aspergillus species or rare molds (AEGIS) [Google Scholar]
- CLSI . 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard-second edition (CLSI Document M38-A2) [Google Scholar]
- Consortium OPATHY, Gabaldón T. Recent trends in molecular diagnostics of yeast infections: from PCR to NGS. FEMS Microbiology Reviews. 2019;43:517–547. doi: 10.1093/femsre/fuz015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornely O.A., Alastruey-Izquierdo A., Arenz D. Global guideline for the diagnosis and management of mucormycosis: an initiative of the European Confederation of Medical Mycology in cooperation with the Mycoses Study Group Education and Research Consortium. Lancet Infectious Diseases. 2019;19:e405–e421. doi: 10.1016/S1473-3099(19)30312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornely O.A., Hoenigl M., Lass-Flörl C. Defining breakthrough invasive fungal infection–Position paper of the mycoses study group education and research consortium and the European Confederation of Medical Mycology. Mycoses. 2019;62:716–729. doi: 10.1111/myc.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornely O.A., Maertens J., Bresnik M. Liposomal amphotericin B as initial therapy for invasive mold infection: a randomized trial comparing a high-loading dose regimen with standard dosing (AmBiLoad trial) Clinical Infectious Diseases. 2007;44:1289–1297. doi: 10.1086/514341. [DOI] [PubMed] [Google Scholar]
- Cornillet A., Camus C., Nimubona S. Comparison of epidemiological, clinical, and biological features of invasive aspergillosis in neutropenic and nonneutropenic patients: a 6-year survey. Clinical Infectious Diseases. 2006;43:577–584. doi: 10.1086/505870. [DOI] [PubMed] [Google Scholar]
- Cunha C., Aversa F., Lacerda J.F. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. New England Journal of Medicine. 2014;370:421–432. doi: 10.1056/NEJMoa1211161. [DOI] [PubMed] [Google Scholar]
- Cunha C., Carvalho A. Toward the identification of a genetic risk signature for pulmonary aspergillosis in chronic obstructive pulmonary disease. Clinical Infectious Diseases. 2018;66:1153–1154. doi: 10.1093/cid/cix944. [DOI] [PubMed] [Google Scholar]
- Cunha C., Di Ianni M., Bozza S. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood. 2010;116:5394–5402. doi: 10.1182/blood-2010-04-279307. [DOI] [PubMed] [Google Scholar]
- Cunha C., Monteiro A.A., Oliveira-Coelho A. PTX3-based genetic testing for risk of aspergillosis after lung transplant. Clinical Infectious Diseases. 2015;61:1893–1894. doi: 10.1093/cid/civ679. [DOI] [PubMed] [Google Scholar]
- da Silva Ferreira M.E., Capellaro J.L., dos Reis Marques E. In vitro evolution of itraconazole resistance in Aspergillus fumigatus involves multiple mechanisms of resistance. Antimicrobial Agents and Chemotherapy. 2004;48:4405–4413. doi: 10.1128/AAC.48.11.4405-4413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Ferreira M.E., Kress M.R., Savoldi M. The akuB(KU80) mutant deficient for nonhomologous end joining is a powerful tool for analyzing pathogenicity in Aspergillus fumigatus. Eukaryotic Cell. 2006;5:207–211. doi: 10.1128/EC.5.1.207-211.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva Ferreira M.E., Malavazi I., Savoldi M. Transcriptome analysis of Aspergillus fumigatus exposed to voriconazole. Current Genetics. 2006;50:32–44. doi: 10.1007/s00294-006-0073-2. [DOI] [PubMed] [Google Scholar]
- Dabas Y., Xess I., Bakshi S. Emergence of azole-resistant Aspergillus fumigatus from immunocompromised hosts in India. Antimicrobial Agents and Chemotherapy. 2018;62 doi: 10.1128/AAC.02264-17. e02264–e02217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannaoui E., Borel E., Monier M.F. Acquired itraconazole resistance in Aspergillus fumigatus. Journal of Antimicrobial Chemotherapy. 2001;47:333–340. doi: 10.1093/jac/47.3.333. [DOI] [PubMed] [Google Scholar]
- Dannaoui E., Borel E., Persat F., Monier M.F., Piens M.A., Ebga Network In-vivo itraconazole resistance of Aspergillus fumigatus in systemic murine aspergillosis. EBGA Network. European research group on biotypes and genotypes of Aspergillus fumigatus. Journal of Medical Microbiology. 1999;48:1087–1093. doi: 10.1099/00222615-48-12-1087. [DOI] [PubMed] [Google Scholar]
- Dannaoui E., Borel E., Persat F. Amphotericin B resistance of Aspergillus terreus in a murine model of disseminated aspergillosis. Journal of Medical Microbiology. 2000;49:601–606. doi: 10.1099/0022-1317-49-7-601. [DOI] [PubMed] [Google Scholar]
- Dannaoui E., Gabriel F., Gaboyard M. Molecular diagnosis of invasive aspergillosis and detection of azole resistance by a newly commercialized PCR kit. Journal of Clinical Microbiology. 2017;55:3210–3218. doi: 10.1128/JCM.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannaoui E., Persat F., Monier M.F. In-vitro susceptibility of Aspergillus spp. isolates to amphotericin B and itraconazole. Journal of Antimicrobial Chemotherapy. 1999;44:553–555. doi: 10.1093/jac/44.4.553. [DOI] [PubMed] [Google Scholar]
- Dave V.P., Pappuru R.R., Pathengay A. Aspergillus endophthalmitis: Clinical presentations and factors determining outcomes. Asia-Pacific Journal of Ophthalmology. 2020;9:9–13. doi: 10.1097/01.APO.0000617928.43993.7e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carolis E., Vella A., Florio A.R. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry for caspofungin susceptibility testing of Candida and Aspergillus species. Journal of Clinical Microbiology. 2012;50:2479–2483. doi: 10.1128/JCM.00224-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot T., Hagen F., Vreuls W. Genotyping of Aspergillus fumigatus in formalin-fixed paraffin-embedded tissues and serum samples from patients with invasive aspergillosis. Frontiers in Cellular and Infection Microbiology. 2018;8:377. doi: 10.3389/fcimb.2018.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot T., Meis J.F. Microsatellite stability in STR analysis Aspergillus fumigatus depends on number of repeat units. Frontiers in Cellular and Infection Microbiology. 2019;9:82. doi: 10.3389/fcimb.2019.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Heer K., Gerritsen M.G., Visser C.E. Galactomannan detection in broncho-alveolar lavage fluid for invasive aspergillosis in immunocompromised patients. The Cochrane Database of Systematic Reviews. 2019;5 doi: 10.1002/14651858.CD012399.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Ruiter M.T., de Valk H.A., Meis J.F. Retrotransposon insertion-site context (RISC) typing: a novel typing method for Aspergillus fumigatus and a convenient PCR alternative to restriction fragment length polymorphism analysis. Journal of Microbiological Methods. 2007;70:528–534. doi: 10.1016/j.mimet.2007.06.009. [DOI] [PubMed] [Google Scholar]
- de Valk H.A., Meis J.F., Bretagne S. Interlaboratory reproducibility of a microsatellite-based typing assay for Aspergillus fumigatus through the use of allelic ladders: proof of concept. Clinical Microbiology and Infection. 2009;15:180–187. doi: 10.1111/j.1469-0691.2008.02656.x. [DOI] [PubMed] [Google Scholar]
- de Valk H.A., Meis J.F., Curfs I.M. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. Journal of Clinical Microbiology. 2005;43:4112–4120. doi: 10.1128/JCM.43.8.4112-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekkers B.G.J., Bakker M., van der Elst K.C.M. Therapeutic drug monitoring of posaconazole: an update. Current Fungal Infection Reports. 2016;10:51–61. doi: 10.1007/s12281-016-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarze E., Sanglard D. Defining the frontiers between antifungal resistance, tolerance and the concept of persistence. Drug Resistance Updates. 2015;23:12–19. doi: 10.1016/j.drup.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Delavy M., Dos Santos A.R., Heiman C.M. Investigating antifungal susceptibility in Candida species with MALDI-TOF MS-based assays. Frontiers in Cellular and Infection Microbiology. 2019;9:19. doi: 10.3389/fcimb.2019.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denardi L.B., Melchers W.J.G., Zoll J. 20th Congress of the International Society for Human and Animal Mycology, Amsterdam, The Netherlands: Abstract no: S6.6c. 2018. First report of azole-resistant Aspergillus fumigatus harboring TR34/L98H and M220R in Brazil. [Google Scholar]
- Deng S., Zhang L., Ji Y. Triazole phenotypes and genotypic characterization of clinical Aspergillus fumigatus isolates in China. Emerging Microbes & Infections. 2017;6:e109. doi: 10.1038/emi.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis J., Forouzanfar F., Herbrecht R. Evaluation of two commercial real-time PCR kits for Aspergillus DNA detection in bronchoalveolar lavage fluid in patients with invasive pulmonary aspergillosis. Journal of Molecular Diagnostics. 2018;20:298–306. doi: 10.1016/j.jmoldx.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.W., Cadranel J., Beigelman-Aubry C. Chronic pulmonary aspergillosis: rationale and clinical guidelines for diagnosis and management. European Respiratory Journal. 2016;47:45–68. doi: 10.1183/13993003.00583-2015. [DOI] [PubMed] [Google Scholar]
- Denning D.W., Chakrabarti A. Pulmonary and sinus fungal diseases in non-immunocompromised patients. Lancet Infectious Diseases. 2017;17:e357–e366. doi: 10.1016/S1473-3099(17)30309-2. [DOI] [PubMed] [Google Scholar]
- Denning D.W., Page I.D., Chakaya J. Case definition of chronic pulmonary aspergillosis in resource-constrained settings. Emerging Infectious Diseases. 2018;24 doi: 10.3201/eid2408.171312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.W., Park S., Lass-Flörl C. High-frequency triazole resistance found in nonculturable Aspergillus fumigatus from lungs of patients with chronic fungal disease. Clinical Infectious Diseases. 2011;52:1123–1129. doi: 10.1093/cid/cir179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D.W., Perlin D.S. Azole resistance in Aspergillus: a growing public health menace. Future Microbiology. 2011;6:1229–1232. doi: 10.2217/fmb.11.118. [DOI] [PubMed] [Google Scholar]
- Denning D.W., Pleuvry A., Cole D.C. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Medical Mycology. 2013;51:361–370. doi: 10.3109/13693786.2012.738312. [DOI] [PubMed] [Google Scholar]
- Denning D.W., Radford S.A., Oakley K.L. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. Journal of Antimicrobial Chemotherapy. 1997;40:401–414. doi: 10.1093/jac/40.3.401. [DOI] [PubMed] [Google Scholar]
- Denning D.W., Venkateswarlu K., Oakley K.L. Itraconazole resistance in Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 1997;41:1364–1368. doi: 10.1128/aac.41.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoubeaux G., Cray C. Rodent models of invasive aspergillosis due to Aspergillus fumigatus: Still a long path toward standardization. Frontiers in Microbiology. 2017;8:841. doi: 10.3389/fmicb.2017.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoubeaux G., Cray C. Animal models of aspergillosis. Comparative Medicine. 2018;68:109–123. [PMC free article] [PubMed] [Google Scholar]
- Dhingra S., Cramer R.A. Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: Opportunities for therapeutic development. Frontiers in Microbiology. 2017;8:92. doi: 10.3389/fmicb.2017.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll M., Preas M.A., Johnson J.K. A pseudo-outbreak of aspergillosis at a tertiary care hospital: Thinking beyond the infection control risk assessment. Infection Control and Hospital Epidemiology. 2017;38:115–118. doi: 10.1017/ice.2016.220. [DOI] [PubMed] [Google Scholar]
- Donnelly J.P., Chen S.C., Kauffman C.A. Revision and Update of the Consensus Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clinical Infectious Diseases. 2020;71:1367–1376. doi: 10.1093/cid/ciz1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Reis T.F., Silva L.P., de Castro P.A. The Aspergillus fumigatus mismatch repair MSH2 homolog is important for virulence and azole resistance. mSphere. 2019;4:e00416–e00419. doi: 10.1128/mSphere.00416-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos R.A.C., Steenwyk J.L., Rivero-Menendez O. Genomic and phenotypic heterogeneity of clinical isolates of the human pathogens Aspergillus fumigatus, Aspergillus lentulus, and Aspergillus fumigatiaffinis. Frontiers in Genetics. 2020;11:459. doi: 10.3389/fgene.2020.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drott M.T., Bastos R.W., Rokas A. Diversity of secondary metabolism in Aspergillus nidulans clinical isolates. mSphere. 2020;5:e00156–20. doi: 10.1128/mSphere.00156-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- du Pré S., Beckmann N., Almeida M.C. Effect of the novel antifungal drug F901318 (Olorofim) on growth and viability of Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 2018;62:e00231–18. doi: 10.1128/AAC.00231-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte R.F., Sánchez-Ortega I., Cuesta I. Serum galactomannan-based early detection of invasive aspergillosis in hematology patients receiving effective antimold prophylaxis. Clinical Infectious Diseases. 2014;59:1696–1702. doi: 10.1093/cid/ciu673. [DOI] [PubMed] [Google Scholar]
- Dudakova A., Spiess B., Tangwattanachuleeporn M. Molecular tools for the detection and deduction of azole antifungal drug resistance phenotypes in Aspergillus species. Clinical Microbiology Reviews. 2017;30:1065–1091. doi: 10.1128/CMR.00095-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duettmann W., Koidl C., Troppan K. Serum and urine galactomannan testing for screening in patients with hematological malignancies. Medical Mycology. 2014;52:647–652. doi: 10.1093/mmy/myu019. [DOI] [PubMed] [Google Scholar]
- Dupont D., Menotti J., Turc J. Pulmonary aspergillosis in critically ill patients with Coronavirus Disease 2019 (COVID-19) Medical Mycology. 2020;59:110–114. doi: 10.1093/mmy/myaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M., Jenks J.D., Hoenigl M. Blood Aspergillus PCR: The Good, the Bad, and the Ugly. Journal of Fungi. 2020;6:18. doi: 10.3390/jof6010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigl S., Hoenigl M., Spiess B. Galactomannan testing and Aspergillus PCR in same-day bronchoalveolar lavage and blood samples for diagnosis of invasive aspergillosis. Medical Mycology. 2017;55:528–534. doi: 10.1093/mmy/myw102. [DOI] [PubMed] [Google Scholar]
- Eigl S., Prattes J., Lackner M. Multicenter evaluation of a lateral-flow device test for diagnosing invasive pulmonary aspergillosis in ICU patients. Critical Care. 2015;19:178. doi: 10.1186/s13054-015-0905-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejzykowicz D.E., Solis N.V., Gravelat F.N. Role of Aspergillus fumigatus DvrA in host cell interactions and virulence. Eukaryotic Cell. 2010;9:1432–1440. doi: 10.1128/EC.00055-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefanti A., Mouton J.W., Verweij P.E. Amphotericin B- and voriconazole-echinocandin combinations against Aspergillus spp.: Effect of serum on inhibitory and fungicidal interactions. Antimicrobial Agents and Chemotherapy. 2013;57:4656–4663. doi: 10.1128/AAC.00597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel T.G.P., Slabbers L., de Jong C. Prevalence and diversity of filamentous fungi in the airways of cystic fibrosis patients – A Dutch, multicentre study. Journal of Cystic Fibrosis. 2019;18:221–226. doi: 10.1016/j.jcf.2018.11.012. [DOI] [PubMed] [Google Scholar]
- Escribano P., Muñoz P., Montilla P. Genotyping of Aspergillus fumigatus reveals compartmentalization of genotypes in disseminated disease after invasive pulmonary aspergillosis. Journal of Clinical Microbiology. 2017;55:331–333. doi: 10.1128/JCM.02037-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano P., Peláez T., Bouza E. Microsatellite (STRAf) genotyping cannot differentiate between invasive and colonizing Aspergillus fumigatus isolates. Journal of Clinical Microbiology. 2015;53:667–670. doi: 10.1128/JCM.02636-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano P., Peláez T., Muñoz P. Is azole resistance in Aspergillus fumigatus a problem in Spain? Antimicrobial Agents and Chemotherapy. 2013;57:2815–2820. doi: 10.1128/AAC.02487-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinel-Ingroff A., Cuenca-Estrella M., Cantón E. EUCAST and CLSI: Working together towards a harmonized method for antifungal susceptibility testing. Current Fungal Infection Reports. 2013;7:59–67. [Google Scholar]
- Espinel-Ingroff A., Turnidge J. The role of epidemiological cutoff values (ECVs/ECOFFs) in antifungal susceptibility testing and interpretation for uncommon yeasts and moulds. Revista Iberoamericana de Micologia. 2016;33:63–75. doi: 10.1016/j.riam.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Esquivel B.D., Rybak J.M., Barker K.S. Characterization of the efflux capability and substrate specificity of Aspergillus fumigatus PDR5-like ABC transporters expressed in Saccharomyces cerevisiae. mBio. 2020;11 doi: 10.1128/mBio.00338-20. e00338–e00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST . 2020. European Committee for Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs for antifungal agents. [Google Scholar]
- Fakih M.G., Barden G.E., Oakes C.A. First reported case of Aspergillus granulosus infection in a cardiac transplant patient. Journal of Clinical Microbiology. 1995;33:471–473. doi: 10.1128/jcm.33.2.471-473.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Chen Y., Duan L. Comparison of two typing methods for characterization of azole resistance in Aspergillus fumigatus from potting soil samples in a Chinese Hospital. Antimicrobial Agents and Chemotherapy. 2020;64:e01578–e01619. doi: 10.1128/AAC.01578-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei M., Bhatia S., Oriss T.B. TNF-alpha from inflammatory dendritic cells (DCs) regulates lung IL-17A/IL-5 levels and neutrophilia versus eosinophilia during persistent fungal infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5360–5365. doi: 10.1073/pnas.1015476108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferwerda B., Ferwerda G., Plantinga T.S. Human dectin-1 deficiency and mucocutaneous fungal infections. New England Journal of Medicine. 2009;361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filler S.G., Sheppard D.C. Fungal invasion of normally non-phagocytic host cells. PLoS Pathogens. 2006;2:e129. doi: 10.1371/journal.ppat.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C.E., Hohl T.M., Fan W. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood. 2017;129:2693–2701. doi: 10.1182/blood-2016-10-743294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo S.S., Reading P.C., Jaillon S. Pentraxins and collectins: Friend or foe during pathogen invasion? Trends in Microbiology. 2015;23:799–811. doi: 10.1016/j.tim.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forastiero A., Bernal-Martinez L., Mellado E. In vivo efficacy of voriconazole and posaconazole therapy in a novel invertebrate model of Aspergillus fumigatus infection. International Journal of Antimicrobial Agents. 2015;46:511–517. doi: 10.1016/j.ijantimicag.2015.07.007. [DOI] [PubMed] [Google Scholar]
- Fraczek M.G., Bromley M., Buied A. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. Journal of Antimicrobial Chemotherapy. 2013;68:1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- Friesen J.A., Rodwell V.W. The 3-hydroxy-3-methylglutaryl coenzyme-A (HMG-CoA) reductases. Genome Biology. 2004;5:248. doi: 10.1186/gb-2004-5-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Hubka V., Ezekiel C.N. Taxonomy of Aspergillus section Flavi and their production of aflatoxins, ochratoxins and other mycotoxins. Studies in Mycology. 2019;93:1–63. doi: 10.1016/j.simyco.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Larsen T.O. Extrolites of Aspergillus fumigatus and other pathogenic species in Aspergillus section Fumigati. Frontiers in Microbiology. 2016;6:1485. doi: 10.3389/fmicb.2015.01485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisvad J.C., Samson R.A. Chemotaxonomy and morphology of Aspergillus fumigatus and related taxa. In: Samson R.A., Pitt J.I., editors. Modern concepts in Penicillium and Aspergillus classification. Plenum Press; New York: 1990. pp. 201–208. [Google Scholar]
- Fuhren J., Voskuil W.S., Boel C.H. High prevalence of azole resistance in Aspergillus fumigatus isolates from high-risk patients. Journal of Antimicrobial Chemotherapy. 2015;70:2894–2898. doi: 10.1093/jac/dkv177. [DOI] [PubMed] [Google Scholar]
- Fujimoto H., Ikeda M., Yamamoto K. Structure of fischerin, a new toxic metabolite from an ascomycete, Neosartorya fischeri var. fischeri. Journal of Natural Products. 1993;56:1268–1275. doi: 10.1021/np50098a010. [DOI] [PubMed] [Google Scholar]
- Fungiplex . 2020. Fungiplex® Aspergillus Azole-R IVD PCR. [Google Scholar]
- Furukawa T., van Rhijn N., Fraczek M. The negative cofactor 2 complex is a key regulator of drug resistance in Aspergillus fumigatus. Nature Communications. 2020;11:427. doi: 10.1038/s41467-019-14191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli E., Fothergill A.W., Brescini L. Osteomyelitis caused by Aspergillus species: a review of 310 reported cases. Clinical Microbiology and Infection. 2014;20:559–565. doi: 10.1111/1469-0691.12389. [DOI] [PubMed] [Google Scholar]
- Gago S., Denning D.W., Bowyer P. Pathophysiological aspects of Aspergillus colonization in disease. Medical Mycology. 2019;57:S219–S227. doi: 10.1093/mmy/myy076. [DOI] [PubMed] [Google Scholar]
- Galagan J.E., Calvo S.E., Cuomo C. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gams W., Christensen M., Onions A.H. Infrageneric taxa of Aspergillus. In: Samson R.A., Pitt J.I., editors. Advances in Penicillium and Aspergillus Systematics. Plenum Press; New York: 1985. pp. 55–62. [Google Scholar]
- Gangneux J.P., Bougnoux M.E., Dannaoui E. Invasive fungal diseases during COVID-19: We should be prepared. Journal de Mycologie Medicale. 2020;30:100971. doi: 10.1016/j.mycmed.2020.100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L.J., Sun Y., Wan Z. CSP typing of Chinese Aspergillus fumigatus isolates: identification of additional CSP types. Medical Mycology. 2013;51:683–687. doi: 10.3109/13693786.2013.770609. [DOI] [PubMed] [Google Scholar]
- García-Effron G., Dilger A., Alcazar-Fuoli L. Rapid detection of triazole antifungal resistance in Aspergillus fumigatus. Journal of Clinical Microbiology. 2008;46:1200–1206. doi: 10.1128/JCM.02330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Martos P., Guarro J., Gené J. Onychomycosis caused by Aspergillus sclerotiorum. Journal de Mycologie Medicale. 2001;11:222–224. [Google Scholar]
- Garcia-Rubio R., Alcazar-Fuoli L., Monteiro M.C. Insight into the significance of Aspergillus fumigatus cyp51A polymorphisms. Antimicrobial Agents and Chemotherapy. 2018;62:e00241–18. doi: 10.1128/AAC.00241-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio R., Cuenca-Estrella M., Mellado E. Triazole resistance in Aspergillus species: An emerging problem. Drugs. 2017;77:599–613. doi: 10.1007/s40265-017-0714-4. [DOI] [PubMed] [Google Scholar]
- Garcia-Rubio R., Escribano P., Gomez A. Comparison of two highly discriminatory typing methods to analyze Aspergillus fumigatus azole resistance. Frontiers in Microbiology. 2018;9:1626. doi: 10.3389/fmicb.2018.01626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio R., Gil H., Monteiro M.C. A new Aspergillus fumigatus typing method based on hypervariable Tandem Repeats located within Exons of Surface Protein coding genes (TRESP) PLoS One. 2016;11 doi: 10.1371/journal.pone.0163869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rubio R., Monzon S., Alcazar-Fuoli L. Genome-wide comparative analysis of Aspergillus fumigatus strains: The reference genome as a matter of concern. Genes (Basel) 2018;9:363. doi: 10.3390/genes9070363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlanda C., Hirsch E., Bozza S. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature. 2002;420:182–186. doi: 10.1038/nature01195. [DOI] [PubMed] [Google Scholar]
- Gebremariam T., Alkhazraji S., Alqarihi A. APX001 is effective in the treatment of murine invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 2019;63:e01713–e01718. doi: 10.1128/AAC.01713-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiβel B., Loiko V., Klugherz I. Azole-induced cell wall carbohydrate patches kill Aspergillus fumigatus. Nature Communications. 2018;9:3098. doi: 10.1038/s41467-018-05497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghelfenstein-Ferreira T., Saade A., Alanio A. Recovery of a triazole-resistant Aspergillus fumigatus in respiratory specimen of COVID-19 patient in ICU – A case report. Medical Mycology Case Reports. 2021;31:15–18. doi: 10.1016/j.mmcr.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghez D., Calleja A., Protin C. Early-onset invasive aspergillosis and other fungal infections in patients treated with ibrutinib. Blood. 2018;131:1955–1959. doi: 10.1182/blood-2017-11-818286. [DOI] [PubMed] [Google Scholar]
- Ghufran M.S., Ghosh K., Kanade S.R. A fucose specific lectin from Aspergillus flavus induced interleukin-8 expression is mediated by mitogen activated protein kinase p38. Medical Mycology. 2017;55:323–333. doi: 10.1093/mmy/myw066. [DOI] [PubMed] [Google Scholar]
- Gitman M.R., McTaggart L., Spinato J. Antifungal susceptibility testing of Aspergillus spp. by using a Composite Correlation Index (CCI)-based Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry method appears to not offer benefit over traditional broth microdilution testing. Journal of Clinical Microbiology. 2017;55:2030–2034. doi: 10.1128/JCM.00254-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glampedakis E., Cassaing S., Fekkar A. Invasive aspergillosis due to Aspergillus section Usti: a multicenter retrospective study. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciaa230. [DOI] [PubMed] [Google Scholar]
- Gomez-Lopez A., Forastiero A., Cendejas-Bueno E. An invertebrate model to evaluate virulence in Aspergillus fumigatus: the role of azole resistance. Medical Mycology. 2014;52:311–319. doi: 10.1093/mmy/myt022. [DOI] [PubMed] [Google Scholar]
- Gonçalves S.M., Lagrou K., Rodrigues C.S. Evaluation of bronchoalveolar lavage fluid cytokines as biomarkers for invasive pulmonary aspergillosis in at-risk patients. Frontiers in Microbiology. 2017;8:2362. doi: 10.3389/fmicb.2017.02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lara M.F., Roman-Montes C.M., Diaz-Lomeli P. Azole resistance and cyp51A mutation screening in Aspergillus fumigatus in Mexico. Journal of Antimicrobial Chemotherapy. 2019;74:2047–2050. doi: 10.1093/jac/dkz121. [DOI] [PubMed] [Google Scholar]
- Grahl N., Puttikamonkul S., Macdonald J.M. In vivo hypoxia and a fungal alcohol dehydrogenase influence the pathogenesis of invasive pulmonary aspergillosis. PLoS Pathogens. 2011;7 doi: 10.1371/journal.ppat.1002145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat F.N., Askew D.S., Sheppard D.C. Targeted gene deletion in Aspergillus fumigatus using the hygromycin-resistance split-marker approach. Methods in Molecular Biology. 2012;845:119–130. doi: 10.1007/978-1-61779-539-8_8. [DOI] [PubMed] [Google Scholar]
- Greenstein S., Shadkchan Y., Jadoun J. Analysis of the Aspergillus nidulans thaumatin-like cetA gene and evidence for transcriptional repression of pyr4 expression in the cetA-disrupted strain. Fungal Genetics and Biology. 2006;43:42–53. doi: 10.1016/j.fgb.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Greninger A.L. The challenge of diagnostic metagenomics. Expert Review of Molecular Diagnostics. 2018;18:605–615. doi: 10.1080/14737159.2018.1487292. [DOI] [PubMed] [Google Scholar]
- Gresnigt M.S., Cunha C., Jaeger M. Genetic deficiency of NOD2 confers resistance to invasive aspergillosis. Nature Communications. 2018;9:2636. doi: 10.1038/s41467-018-04912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll A.H., Townsend R., Desai A. Drug-drug interactions between triazole antifungal agents used to treat invasive aspergillosis and immunosuppressants metabolized by cytochrome P450 3A4. Transplant Infectious Disease. 2017;19 doi: 10.1111/tid.12751. [DOI] [PubMed] [Google Scholar]
- Gsaller F., Hortschansky P., Furukawa T. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guegan H., Chevrier S., Belleguic C. Performance of molecular approaches for Aspergillus detection and azole resistance surveillance in cystic fibrosis. Frontiers in Microbiology. 2018;9:531. doi: 10.3389/fmicb.2018.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea J., Garcia de Viedma D., Peláez T. Molecular epidemiology of Aspergillus fumigatus: an in-depth genotypic analysis of isolates involved in an outbreak of invasive aspergillosis. Journal of Clinical Microbiology. 2011;49:3498–3503. doi: 10.1128/JCM.01159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea J., Peláez T., Alcalá L. Comparison of Sensititre YeastOne with the NCCLS M38-A microdilution method to determine the activity of amphotericin B, voriconazole, and itraconazole against clinical isolates of Aspergillus fumigatus. Diagnostic Microbiology and Infectious Disease. 2006;56:53–55. doi: 10.1016/j.diagmicrobio.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Guinea J., Peláez T., Recio S. In vitro antifungal activities of isavuconazole (BAL 4815), voriconazole, and fluconazole against 1,007 isolates of zygomycete, Candida, Aspergillus, Fusarium, and Scedosporium species. Antimicrobial Agents and Chemotherapy. 2008;52:1396–1400. doi: 10.1128/AAC.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinea J., Torres-Narbona M., Gijón P. Pulmonary aspergillosis in patients with chronic obstructive pulmonary disease: incidence, risk factors, and outcome. Clinical Microbiology and Infection. 2010;16:870–877. doi: 10.1111/j.1469-0691.2009.03015.x. [DOI] [PubMed] [Google Scholar]
- Guinea J., Verweij P.E., Meletiadis J. How to: EUCAST recommendations on the screening procedure E.Def 10.1 for the detection of azole resistance in Aspergillus fumigatus isolates using four-well azole-containing agar plates. Clinical Microbiology and Infection. 2019;25:681–687. doi: 10.1016/j.cmi.2018.09.008. [DOI] [PubMed] [Google Scholar]
- Habicht J.M., Preiss M., Passweg J. Invasive pulmonary aspergillosis: effects of early resection in a neutropenic rat model. European Journal of Cardio-Thoracic Surgery. 2002;22:728–732. doi: 10.1016/s1010-7940(02)00467-0. [DOI] [PubMed] [Google Scholar]
- Hagiwara D., Arai T., Takahashi H. Non-cyp51A azole-resistant Aspergillus fumigatus isolates with mutation in HMG-CoA reductase. Emerging Infectious Diseases. 2018;24:1889–1897. doi: 10.3201/eid2410.180730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Miura D., Shimizu K. A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by co-regulating cyp51A and cdr1B expressions. PLoS Pathogens. 2017;13 doi: 10.1371/journal.ppat.1006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara D., Takahashi H., Fujimoto M. Multi-azole resistant Aspergillus fumigatus harboring Cyp51A TR46/Y121F/T289A isolated in Japan. Journal of Infection and Chemotherapy. 2016;22:577–579. doi: 10.1016/j.jiac.2016.01.015. [DOI] [PubMed] [Google Scholar]
- Hagiwara D., Takahashi H., Watanabe A. Whole-genome comparison of Aspergillus fumigatus strains serially isolated from patients with aspergillosis. Journal of Clinical Microbiology. 2014;52:4202–4209. doi: 10.1128/JCM.01105-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagiwara S., Tamura T., Satoh K. The molecular identification and antifungal susceptibilities of Aspergillus species causing otomycosis in Tochigi, Japan. Mycopathologia. 2019;184:13–21. doi: 10.1007/s11046-018-0299-9. [DOI] [PubMed] [Google Scholar]
- Hare R.K., Gertsen J.B., Astvad K.M.T. In vivo selection of a unique tandem repeat mediated azole resistance mechanism (TR120) in Aspergillus fumigatus cyp51A, Denmark. Emerging Infectious Diseases. 2019;25:577–580. doi: 10.3201/eid2503.180297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrove T.Y., Wawrzak Z., Lamb D.C. Structure-functional characterization of cytochrome P450 sterol 14 alpha-Demethylase (CYP51B) from Aspergillus fumigatus and molecular basis for the development of antifungal drugs. Journal of Biological Chemistry. 2015;290:23916–23934. doi: 10.1074/jbc.M115.677310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harima N., Inoue T., Kubota T. A case of otomycosis caused by Aspergillus sclerotiorum. Journal of Dermatology. 2004;31:949–950. doi: 10.1111/j.1346-8138.2004.tb00635.x. [DOI] [PubMed] [Google Scholar]
- Hartmann T., Dumig M., Jaber B.M. Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the beta-rec/six site-specific recombination system. Applied and Environmental Microbiology. 2010;76:6313–6317. doi: 10.1128/AEM.00882-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q., Li H., Rui Y. Pentraxin 3 gene polymorphisms and pulmonary aspergillosis in chronic obstructive pulmonary disease patients. Clinical Infectious Diseases. 2018;66:261–267. doi: 10.1093/cid/cix749. [DOI] [PubMed] [Google Scholar]
- Healey K.R., Perlin D.S. Fungal resistance to echinocandins and the MDR phenomenon in Candida glabrata. Journal of Fungi. 2018;4:105. doi: 10.3390/jof4030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey K.R., Zhao Y., Perez W.B. Prevalent mutator genotype identified in fungal pathogen Candida glabrata promotes multi-drug resistance. Nature Communications. 2016;7:11128. doi: 10.1038/ncomms11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedayati M.T., Pasqualotto A.C., Warn P.A. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology. 2007;153:1677–1692. doi: 10.1099/mic.0.2007/007641-0. [DOI] [PubMed] [Google Scholar]
- Hedayati M.T., Tavakoli M., Maleki M. Fungal epidemiology in cystic fibrosis patients with a special focus on Scedosporium species complex. Microbial Pathogenesis. 2019;129:168–175. doi: 10.1016/j.micpath.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Heldt S., Eigl S., Prattes J. Levels of interleukin (IL)-6 and IL-8 are elevated in serum and bronchoalveolar lavage fluid of haematological patients with invasive pulmonary aspergillosis. Mycoses. 2017;60:818–825. doi: 10.1111/myc.12679. [DOI] [PubMed] [Google Scholar]
- Heldt S., Prattes J., Eigl S. Diagnosis of invasive aspergillosis in hematological malignancy patients: Performance of cytokines, Asp LFD, and Aspergillus PCR in same day blood and bronchoalveolar lavage samples. Journal of Infection. 2018;77:235–241. doi: 10.1016/j.jinf.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S., Wainwright C.E., Bell S.C. Population pharmacokinetics of itraconazole and its active metabolite hydroxy-itraconazole in paediatric cystic fibrosis and bone marrow transplant patients. Clinical Pharmacokinetics. 2006;45:1099–1114. doi: 10.2165/00003088-200645110-00004. [DOI] [PubMed] [Google Scholar]
- Henriet S.S., Jans J., Simonetti E. Chloroquine modulates the fungal immune response in phagocytic cells from patients with chronic granulomatous disease. Journal of Infectious Diseases. 2013;207:1932–1939. doi: 10.1093/infdis/jit103. [DOI] [PubMed] [Google Scholar]
- Heo S.T., Tatara A.M., Jiménez-Ortigosa C. Changes in in vitro susceptibility patterns of Aspergillus to triazoles and correlation with aspergillosis outcome in a tertiary care cancer center, 1999–2015. Clinical Infectious Diseases. 2017;65:216–225. doi: 10.1093/cid/cix297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrecht R., Denning D.W., Patterson T.F. Voriconazole versus amphotericin B for primary therapy of invasive aspergillosis. New England Journal of Medicine. 2002;347:408–415. doi: 10.1056/NEJMoa020191. [DOI] [PubMed] [Google Scholar]
- Hiemenz J.W., Walsh T.J. Lipid formulations of amphotericin B: recent progress and future directions. Clinical Infectious Diseases. 1996;22:S133–S144. doi: 10.1093/clinids/22.supplement_2.s133. [DOI] [PubMed] [Google Scholar]
- Hodiamont C.J., Dolman K.M., Ten Berge I.J. Multiple-azole-resistant Aspergillus fumigatus osteomyelitis in a patient with chronic granulomatous disease successfully treated with long-term oral posaconazole and surgery. Medical Mycology. 2009;47:217–220. doi: 10.1080/13693780802545600. [DOI] [PubMed] [Google Scholar]
- Hoenigl M. Invasive fungal disease complicating COVID-19: when it rains it pours. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciaa1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenigl M., Duettmann W., Raggam R.B. Impact of structured personal on-site patient education on low posaconazole plasma concentrations in patients with haematological malignancies. International Journal of Antimicrobial Agents. 2014;44:140–144. doi: 10.1016/j.ijantimicag.2014.03.013. [DOI] [PubMed] [Google Scholar]
- Hoenigl M., Duettmann W., Raggam R.B. Potential factors for inadequate voriconazole plasma concentrations in intensive care unit patients and patients with hematological malignancies. Antimicrobial Agents and Chemotherapy. 2013;57:3262–3267. doi: 10.1128/AAC.00251-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenigl M., Eigl S., Heldt S. Clinical evaluation of the newly formatted lateral-flow device for invasive pulmonary aspergillosis. Mycoses. 2018;61:40–43. doi: 10.1111/myc.12704. [DOI] [PubMed] [Google Scholar]
- Hoenigl M., Krause R. Antifungal therapy of aspergillosis of the central nervous system and Aspergillus endophthalmitis. Current Pharmaceutical Design. 2013;19:3648–3668. doi: 10.2174/13816128113199990342. [DOI] [PubMed] [Google Scholar]
- Hoenigl M., Orasch T., Faserl K. Triacetylfusarinine C: A urine biomarker for diagnosis of invasive aspergillosis. Journal of Infection. 2019;78:150–157. doi: 10.1016/j.jinf.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenigl M., Prattes J., Spiess B. Performance of galactomannan, beta-d-glucan, Aspergillus lateral-flow device, conventional culture, and PCR tests with bronchoalveolar lavage fluid for diagnosis of invasive pulmonary aspergillosis. Journal of Clinical Microbiology. 2014;52:2039–2045. doi: 10.1128/JCM.00467-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenigl M., Salzer H.J., Raggam R.B. Impact of galactomannan testing on the prevalence of invasive aspergillosis in patients with hematological malignancies. Medical Mycology. 2012;50:266–269. doi: 10.3109/13693786.2011.603102. [DOI] [PubMed] [Google Scholar]
- Hoenigl M., Seeber K., Koidl C. Sensitivity of galactomannan enzyme immunoassay for diagnosing breakthrough invasive aspergillosis under antifungal prophylaxis and empirical therapy. Mycoses. 2013;56:471–476. doi: 10.1111/myc.12060. [DOI] [PubMed] [Google Scholar]
- Hogaboam C.M., Blease K., Mehrad B. Chronic airway hyperreactivity, goblet cell hyperplasia, and peribronchial fibrosis during allergic airway disease induced by Aspergillus fumigatus. American Journal of Pathology. 2000;156:723–732. doi: 10.1016/S0002-9440(10)64775-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hokken M.W.J., Zoll J., Coolen J.P.M. Phenotypic plasticity and the evolution of azole resistance in Aspergillus fumigatus; an expression profile of clinical isolates upon exposure to itraconazole. BMC Genomics. 2019;20:28. doi: 10.1186/s12864-018-5255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollomon D. Does agricultural use of azole fungicides contribute to resistance in the human pathogen Aspergillus fumigatus? Pest Management Science. 2017;73:1987–1993. doi: 10.1002/ps.4607. [DOI] [PubMed] [Google Scholar]
- Hope W.W., McEntee L., Livermore J. Pharmacodynamics of the orotomides against Aspergillus fumigatus: New opportunities for treatment of multidrug-resistant fungal disease. mBio. 2017;8 doi: 10.1128/mBio.01157-17. e01157–e01117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope W.W., Walsh T.J., Denning D.W. The invasive and saprophytic syndromes due to Aspergillus spp. Medical Mycology. 2005;43:S207–S238. doi: 10.1080/13693780400025179. [DOI] [PubMed] [Google Scholar]
- Hortschansky P., Ando E., Tuppatsch K. Deciphering the combinatorial DNA-binding code of the CCAAT-binding complex and the iron-regulatory basic region leucine zipper (bZIP) transcription factor HapX. Journal of Biological Chemistry. 2015;290:6058–6070. doi: 10.1074/jbc.M114.628677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoselton S.A., Samarasinghe A.E., Seydel J.M. An inhalation model of airway allergic response to inhalation of environmental Aspergillus fumigatus conidia in sensitized BALB/c mice. Medical Mycology. 2010;48:1056–1065. doi: 10.3109/13693786.2010.485582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Due M., Varga J. Polyphasic taxonomy of Aspergillus section Usti. Studies in Mycology. 2007;59:107–128. doi: 10.3114/sim.2007.59.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Kocsubé S., Visagie C.M. Classification of Aspergillus, Penicillium, Talaromyces and related genera (Eurotiales): An overview of families, genera, subgenera, sections, series and species. Studies in Mycology. 2020;95:5–169. doi: 10.1016/j.simyco.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Studies in Mycology. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Samson R.A., Yılmaz N. Taxonomy of Aspergillus, Penicillium and Talaromyces and its significance for biotechnology. In: de Vries R.P., Benoit-Gelber I., Rørdam-Andersen M., editors. Aspergillus and Penicillium in the post-genomic era. Caister Academic Press; Poole, UK: 2016. pp. 1–16. [Google Scholar]
- Houbraken J., Weig M., Groβ U. Aspergillus oerlinghausenensis, a new mould species closely related to A. fumigatus. FEMS Microbiology Letters. 2016;363:fnv236. doi: 10.1093/femsle/fnv236. [DOI] [PubMed] [Google Scholar]
- Houser J., Komarek J., Kostlanova N. A soluble fucose-specific lectin from Aspergillus fumigatus conidia--structure, specificity and possible role in fungal pathogenicity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0083077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S.J., Cerar D., Anderson M.J. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerging Infectious Diseases. 2009;15:1068–1076. doi: 10.3201/eid1507.090043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard S.J., Pasqualotto A.C., Denning D.W. Azole resistance in allergic bronchopulmonary aspergillosis and Aspergillus bronchitis. Clinical Microbiology and Infection. 2010;16:683–688. doi: 10.1111/j.1469-0691.2009.02911.x. [DOI] [PubMed] [Google Scholar]
- Howard S.J., Webster I., Moore C.B. Multi-azole resistance in Aspergillus fumigatus. International Journal of Antimicrobial Agents. 2006;28:450–453. doi: 10.1016/j.ijantimicag.2006.08.017. [DOI] [PubMed] [Google Scholar]
- https://clsi.org/media/3682/m61ed2_sample.pdf Accessed on 08 January 2021.
- Hu W., Sillaots S., Lemieux S. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathogens. 2007;3:e24. doi: 10.1371/journal.ppat.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang J., Zhang M. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry for rapid identification of fungal rhinosinusitis pathogens. Journal of Medical Microbiology. 2017;66:328–333. doi: 10.1099/jmm.0.000435. [DOI] [PubMed] [Google Scholar]
- Huang Y.T., Hung C.C., Hsueh P.R. Aspergillus galactomannan antigenemia in penicilliosis marneffei. AIDS. 2007;21:1990–1991. doi: 10.1097/QAD.0b013e3282eeb413. [DOI] [PubMed] [Google Scholar]
- Hubka V., Barrs V., Dudova Z. Unravelling species boundaries in the Aspergillus viridinutans complex (section Fumigati): opportunistic human and animal pathogens capable of interspecific hybridization. Persoonia. 2018;41:142–174. doi: 10.3767/persoonia.2018.41.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubka V., Kubátová A., Mallatova N. Rare and new etiological agents revealed among 178 clinical Aspergillus strains obtained from Czech patients and characterized by molecular sequencing. Medical Mycology. 2012;50:601–610. doi: 10.3109/13693786.2012.667578. [DOI] [PubMed] [Google Scholar]
- Hubka V., Nováková A., Kolařík M. Revision of Aspergillus section Flavipedes: seven new species and proposal of section Jani sect. nov. Mycologia. 2015;107:169–208. doi: 10.3852/14-059. [DOI] [PubMed] [Google Scholar]
- Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. Journal of Clinical Microbiology. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst S.F., Berkow E.L., Stevenson K.L. Isolation of azole-resistant Aspergillus fumigatus from the environment in the south-eastern USA. Journal of Antimicrobial Chemotherapy. 2017;72:2443–2446. doi: 10.1093/jac/dkx168. [DOI] [PubMed] [Google Scholar]
- Hussain H.M., Hotopf M., Oyebode F. Atypical antipsychotic drugs and Alzheimer’s disease (retracted) New England Journal of Medicine. 2007;356:416. doi: 10.1056/NEJMc063094. author reply 417–418. [DOI] [PubMed] [Google Scholar]
- Iatta R., Nuccio F., Immediato D. Species distribution and in vitro azole susceptibility of Aspergillus section Nigri isolates from clinical and environmental settings. Journal of Clinical Microbiology. 2016;54:2365–2372. doi: 10.1128/JCM.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim-Granet O., Jouvion G., Hohl T.M. In vivo bioluminescence imaging and histopathopathologic analysis reveal distinct roles for resident and recruited immune effector cells in defense against invasive aspergillosis. BMC Microbiology. 2010;10:105. doi: 10.1186/1471-2180-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idelevich E.A., Groβ U., Becker K. Comparative evaluation of different gradient diffusion tests for detection of azole resistance in Aspergillus fumigatus. Diagnostic Microbiology and Infectious Disease. 2018;91:52–54. doi: 10.1016/j.diagmicrobio.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Imbert S., Normand A.C., Gabriel F. Multi-centric evaluation of the online MSI platform for the identification of cryptic and rare species of Aspergillus by MALDI-TOF. Medical Mycology. 2019;57:962–968. doi: 10.1093/mmy/myz004. [DOI] [PubMed] [Google Scholar]
- Isla G., Leonardelli F., Tiraboschi I.N. First clinical isolation of an azole-resistant Aspergillus fumigatus isolate harboring a TR46/Y121F/T289A mutation in South America. Antimicrobial Agents and Chemotherapy. 2018;62:e00872–e00918. doi: 10.1128/AAC.00872-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanvoine A., Rocchi S., Reboux G. Azole-resistant Aspergillus fumigatus in sawmills of Eastern France. Journal of Applied Microbiology. 2017;123:172–184. doi: 10.1111/jam.13488. [DOI] [PubMed] [Google Scholar]
- Jemel S., Guillot J., Kallel K. Galleria mellonella for the evaluation of antifungal efficacy against medically important fungi, a narrative review. Microorganisms. 2020;8:390. doi: 10.3390/microorganisms8030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks J.D., Hoenigl M. Point-of-care diagnostics for invasive aspergillosis: nearing the finish line. Expert Review of Molecular Diagnostics. 2020;20:1009–1017. doi: 10.1080/14737159.2020.1820864. [DOI] [PubMed] [Google Scholar]
- Jenks J.D., Mehta S.R., Hoenigl M. Broad spectrum triazoles for invasive mould infections in adults: Which drug and when? Medical Mycology. 2019;57:S168–S178. doi: 10.1093/mmy/myy052. [DOI] [PubMed] [Google Scholar]
- Jenks J.D., Mehta S.R., Taplitz R. Point-of-care diagnosis of invasive aspergillosis in non-neutropenic patients: Aspergillus galactomannan lateral flow assay versus Aspergillus-specific lateral flow device test in bronchoalveolar lavage. Mycoses. 2019;62:230–236. doi: 10.1111/myc.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks J.D., Mehta S.R., Taplitz R. Bronchoalveolar lavage Aspergillus galactomannan lateral flow assay versus Aspergillus-specific lateral flow device test for diagnosis of invasive pulmonary aspergillosis in patients with hematological malignancies. Journal of Infection. 2019;78:249–259. doi: 10.1016/j.jinf.2018.10.014. [DOI] [PubMed] [Google Scholar]
- Jenks J.D., Prattes J., Frank J. Performance of the bronchoalveolar lavage fluid Aspergillus galactomannan lateral flow assay with Cube Reader for diagnosis of invasive pulmonary aspergillosis: a multicenter cohort study. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciaa1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks J.D., Rawlings S.A., Garcia-Vidal C. Immune parameters for diagnosis and treatment monitoring in invasive mold infection. Journal of Fungi. 2019;5:116. doi: 10.3390/jof5040116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks J.D., Salzer H.J., Prattes J. Spotlight on isavuconazole in the treatment of invasive aspergillosis and mucormycosis: design, development, and place in therapy. Drug Design, Development and Therapy. 2018;12:1033–1044. doi: 10.2147/DDDT.S145545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks J.D., Salzer H.J.F., Hoenigl M. Improving the rates of Aspergillus detection: an update on current diagnostic strategies. Expert Review of Anti-Infective Therapy. 2019;17:39–50. doi: 10.1080/14787210.2018.1558054. [DOI] [PubMed] [Google Scholar]
- Jenks J.D., Spiess B., Buchheidt D. New) methods for detection of Aspergillus fumigatus resistance in clinical samples. Current Fungal Infection Reports. 2019;13:129–136. doi: 10.1007/s12281-019-00342-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.H., Hagen F., Astvad K.M. Azole-resistant Aspergillus fumigatus in Denmark: a laboratory-based study on resistance mechanisms and genotypes. Clinical Microbiology and Infection. 2016;22 doi: 10.1016/j.cmi.2016.04.001. 570.e1–9. [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Li H., Tang J.J. Discovery of a potent HMG-CoA reductase degrader that eliminates statin-induced reductase accumulation and lowers cholesterol. Nature Communications. 2018;9:5138. doi: 10.1038/s41467-018-07590-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Ortigosa C., Moore C., Denning D.W. Emergence of echinocandin resistance due to a point mutation in the fks1 gene of Aspergillus fumigatus in a patient with chronic pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.01277-17. e01277–01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.M., Oakley K.L., Radford S.A. Lack of correlation of in vitro amphotericin B susceptibility testing with outcome in a murine model of Aspergillus infection. Journal of Antimicrobial Chemotherapy. 2000;45:85–93. doi: 10.1093/jac/45.1.85. [DOI] [PubMed] [Google Scholar]
- Johnson M.D., Perfect J.R. Caspofungin: first approved agent in a new class of antifungals. Expert Opinion on Pharmacotherapy. 2003;4:807–823. doi: 10.1517/14656566.4.5.807. [DOI] [PubMed] [Google Scholar]
- Jouvion G., Brock M., Droin-Bergere S. Duality of liver and kidney lesions after systemic infection of immunosuppressed and immunocompetent mice with Aspergillus fumigatus. Virulence. 2012;3:43–50. doi: 10.4161/viru.3.1.18654. [DOI] [PubMed] [Google Scholar]
- Jović Z., Janković S.M., Ružić-Zečević D. Clinical pharmacokinetics of second-generation triazoles for the treatment of invasive aspergillosis and candidiasis. European Journal of Drug Metabolism and Pharmacokinetics. 2019;44:139–157. doi: 10.1007/s13318-018-0513-7. [DOI] [PubMed] [Google Scholar]
- Kalleda N., Amich J., Arslan B. Dynamic immune cell recruitment after murine pulmonary Aspergillus fumigatus infection under different immunosuppressive regimens. Frontiers in Microbiology. 2016;7:1107. doi: 10.3389/fmicb.2016.01107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanagh K., Sheehan G. The use of Galleria mellonella larvae to identify novel antimicrobial agents against fungal species of medical interest. Journal of Fungi. 2018;4:113. doi: 10.3390/jof4030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid M., Ali S.A. Fungal osteomyelitis in a patient with chronic granulomatous disease: Case report and review of the literature. Journal of the Pakistan Medical Association. 2018;68:1387–1390. [PubMed] [Google Scholar]
- Kidd S.E., Chen S.C., Meyer W. A new age in molecular diagnostics for invasive fungal disease: Are we ready? Frontiers in Microbiology. 2019;10:2903. doi: 10.3389/fmicb.2019.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J., Brunel S.F., Warris A. Aspergillus infections in cystic fibrosis. Journal of Infection. 2016;72:S50–S55. doi: 10.1016/j.jinf.2016.04.022. [DOI] [PubMed] [Google Scholar]
- King J., Henriet S.S.V., Warris A. Aspergillosis in chronic granulomatous disease. Journal of Fungi. 2016;2:15. doi: 10.3390/jof2020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick W.R., McAtee R.K., Fothergill A.W. Efficacy of voriconazole in a guinea pig model of disseminated invasive aspergillosis. Antimicrobial Agents and Chemotherapy. 2000;44:2865–2868. doi: 10.1128/aac.44.10.2865-2868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærbølling I., Vesth T., Frisvad J.C. A comparative genomics study of 23 Aspergillus species from section Flavi. Nature Communications. 2020;11:1106. doi: 10.1038/s41467-019-14051-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærbølling I., Vesth T.C., Frisvad J.C. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E753–E761. doi: 10.1073/pnas.1715954115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen C.H., de Valk H.A., Balajee S.A. Utility of CSP typing to sub-type clinical Aspergillus fumigatus isolates and proposal for a new CSP type nomenclature. Journal of Microbiological Methods. 2009;77:292–296. doi: 10.1016/j.mimet.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Klaassen C.H., de Valk H.A., Curfs-Breuker I.M. Novel mixed-format real-time PCR assay to detect mutations conferring resistance to triazoles in Aspergillus fumigatus and prevalence of multi-triazole resistance among clinical isolates in the Netherlands. Journal of Antimicrobial Chemotherapy. 2010;65:901–905. doi: 10.1093/jac/dkq041. [DOI] [PubMed] [Google Scholar]
- Klaassen C.H., Osherov N. Aspergillus strain typing in the genomics era. Studies in Mycology. 2007;59:47–51. doi: 10.3114/sim.2007.59.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloezen W., Parel F., Bruggemann R. Amphotericin B and terbinafine but not the azoles prolong survival in Galleria mellonella larvae infected with Madurella mycetomatis. Medical Mycology. 2018;56:469–478. doi: 10.1093/mmy/myx064. [DOI] [PubMed] [Google Scholar]
- Kocsubé S., Perrone G., Magistà D. Aspergillus is monophyletic: Evidence from multiple gene phylogenies and extrolites profiles. Studies in Mycology. 2016;85:199–213. doi: 10.1016/j.simyco.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler P., Bassetti M., Chakrabarti A. Defining and managing COVID-19 associated pulmonary aspergillosis: The 2020 ECMM/ISHAM consensus criteria for research and clinical guidance. Lancet Infectious Diseases. 2020 doi: 10.1016/S1473-3099(20)30847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler P., Cornely O.A., Bottiger B.W. COVID–19 associated pulmonary aspergillosis. Mycoses. 2020;63:528–534. doi: 10.1111/myc.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler P., Cornely O.A., Kochanek M. Bronchoscopy safety precautions for diagnosing COVID-19 associated pulmonary aspergillosis – A simulation study. Mycoses. 2020;64:55–59. doi: 10.1111/myc.13183. [DOI] [PubMed] [Google Scholar]
- Koehler P., Hamprecht A., Bader O. Epidemiology of invasive aspergillosis and azole resistance in patients with acute leukaemia: the SEPIA Study. International Journal of Antimicrobial Agents. 2017;49:218–223. doi: 10.1016/j.ijantimicag.2016.10.019. [DOI] [PubMed] [Google Scholar]
- Komadina T.G., Wilkes T.D., Shock J.P. Treatment of Aspergillus fumigatus keratitis in rabbits with oral and topical ketoconazole. American Journal of Ophthalmology. 1985;99:476–479. doi: 10.1016/0002-9394(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Kostrzewa M. Application of the MALDI Biotyper to clinical microbiology: progress and potential. Expert Review of Proteomics. 2018;15:193–202. doi: 10.1080/14789450.2018.1438193. [DOI] [PubMed] [Google Scholar]
- Kowalski C.H., Kerkaert J.D., Liu K.W. Fungal biofilm morphology impacts hypoxia fitness and disease progression. Nature Microbiology. 2019;4:2430–2441. doi: 10.1038/s41564-019-0558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S., Bayram O., Braus G.H. Deletion and allelic exchange of the Aspergillus fumigatus veA locus via a novel recyclable marker module. Eukaryotic Cell. 2005;4:1298–1307. doi: 10.1128/EC.4.7.1298-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krappmann S., Sasse C., Braus G.H. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryotic Cell. 2006;5:212–215. doi: 10.1128/EC.5.1.212-215.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan-Natesan S., Wu W., Cutright J.L., Chandrasekar P.H. In vitro-in vivo correlation of voriconazole resistance due to G448S mutation (cyp51A gene) in Aspergillus fumigatus. Diagnostic Microbiology and Infectious Disease. 2012;74:272–277. doi: 10.1016/j.diagmicrobio.2012.06.030. [DOI] [PubMed] [Google Scholar]
- Kroll K., Shekhova E., Mattern D.J. The hypoxia-induced dehydrogenase HorA is required for coenzyme Q10 biosynthesis, azole sensitivity and virulence of Aspergillus fumigatus. Molecular Microbiology. 2016;101:92–108. doi: 10.1111/mmi.13377. [DOI] [PubMed] [Google Scholar]
- Krüger W., Vielreicher S., Kapitan M. Fungal-bacterial interactions in ealth and disease. Pathogens. 2019;8:70. doi: 10.3390/pathogens8020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T., Yamashita N., Nishimura A. Transformation of Aspergillus sp. and Trichoderma reesei using the pyrithiamine resistance gene (ptrA) of Aspergillus oryzae. Bioscience, Biotechnology, and Biochemistry. 2002;66:404–406. doi: 10.1271/bbb.66.404. [DOI] [PubMed] [Google Scholar]
- Kudoh A., Okawa Y., Shibata N. Significant structural change in both O- and N-linked carbohydrate moieties of the antigenic galactomannan from Aspergillus fumigatus grown under different culture conditions. Glycobiology. 2015;25:74–87. doi: 10.1093/glycob/cwu091. [DOI] [PubMed] [Google Scholar]
- Kuipers S., Bruggemann R.J., de Sevaux R.G. Failure of posaconazole therapy in a renal transplant patient with invasive aspergillosis due to Aspergillus fumigatus with attenuated susceptibility to posaconazole. Antimicrobial Agents and Chemotherapy. 2011;55:3564–3566. doi: 10.1128/AAC.01544-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupfahl C., Heinekamp T., Geginat G. Deletion of the gliP gene of Aspergillus fumigatus results in loss of gliotoxin production but has no effect on virulence of the fungus in a low-dose mouse infection model. Molecular Microbiology. 2006;62:292–302. doi: 10.1111/j.1365-2958.2006.05373.x. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. New drugs target growing threat of fatal fungi. Science. 2019;366:407. doi: 10.1126/science.366.6464.407. [DOI] [PubMed] [Google Scholar]
- Kurtz M.B., Heath I.B., Marrinan J. Morphological effects of lipopeptides against Aspergillus fumigatus correlate with activities against (1,3)-beta-D-glucan synthase. Antimicrobial Agents and Chemotherapy. 1994;38:1480–1489. doi: 10.1128/aac.38.7.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurup V.P., Xia J.Q., Crameri R. Purified recombinant A. fumigatus allergens induce different responses in mice. Clinical Immunology. 2001;98:327–336. doi: 10.1006/clim.2000.4993. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung K.J., Sugui J.A. Aspergillus fumigatus – what makes the species a ubiquitous human fungal pathogen? PLoS Pathogens. 2013;9 doi: 10.1371/journal.ppat.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M., Obermair J., Naschberger V. Cryptic species of Aspergillus section Terrei display essential physiological features to cause infection and are similar in their virulence potential in Galleria mellonella. Virulence. 2019;10:542–554. doi: 10.1080/21505594.2019.1614382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner M., Rambach G., Jukic E. Azole-resistant and -susceptible Aspergillus fumigatus isolates show comparable fitness and azole treatment outcome in immunocompetent mice. Medical Mycology. 2018;56:703–710. doi: 10.1093/mmy/myx109. [DOI] [PubMed] [Google Scholar]
- Lamoth F., Alexander B.D. Comparing Etest and broth microdilution for antifungal susceptibility testing of the most–relevant pathogenic molds. Journal of Clinical Microbiology. 2015;53:3176–3181. doi: 10.1128/JCM.00925-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen T.O., Smedsgaard J., Nielsen K.F. Production of mycotoxins by Aspergillus lentulus and other medically important and closely related species in section Fumigati. Medical Mycology. 2007;45:225–232. doi: 10.1080/13693780601185939. [DOI] [PubMed] [Google Scholar]
- Lass-Flörl C. How to make a fast diagnosis in invasive aspergillosis. Medical Mycology. 2019;57:S155–S160. doi: 10.1093/mmy/myy103. [DOI] [PubMed] [Google Scholar]
- Lass-Flörl C., Grif K., Kontoyiannis D.P. Molecular typing of Aspergillus terreus isolates collected in Houston, Texas, and Innsbruck, Austria: evidence of great genetic diversity. Journal of Clinical Microbiology. 2007;45:2686–2690. doi: 10.1128/JCM.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass-Flörl C., Griff K., Mayr A. Epidemiology and outcome of infections due to Aspergillus terreus: 10–year single centre experience. British Journal of Haematology. 2005;131:201–207. doi: 10.1111/j.1365-2141.2005.05763.x. [DOI] [PubMed] [Google Scholar]
- Latgé J.P. Tasting the fungal cell wall. Cellular Microbiology. 2010;12:863–872. doi: 10.1111/j.1462-5822.2010.01474.x. [DOI] [PubMed] [Google Scholar]
- Latgé J.P., Beauvais A., Chamilos G. The cell wall of the human fungal pathogen Aspergillus fumigatus: Biosynthesis, organization, immune response, and virulence. Annual Review of Microbiology. 2017;71:99–116. doi: 10.1146/annurev-micro-030117-020406. [DOI] [PubMed] [Google Scholar]
- Latgé J.P., Chamilos G. Aspergillus fumigatus and Aspergillosis in 2019. Clinical Microbiology Reviews. 2019;33 doi: 10.1128/CMR.00140-18. e00140–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A.F., Walchak R.C., Miller H.B. Multicenter study demonstrates standardization requirements for mold identification by MALDI-TOF MS. Frontiers in Microbiology. 2019;10:2098. doi: 10.3389/fmicb.2019.02098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavergne R.A., Chouaki T., Hagen F. Home environment as a source of life-threatening azole-resistant Aspergillus fumigatus in immunocompromised patients. Clinical Infectious Diseases. 2017;64:76–78. doi: 10.1093/cid/ciw664. [DOI] [PubMed] [Google Scholar]
- Lazzarini C., Esposto M.C., Prigitano A. Azole resistance in Aspergillus fumigatus clinical isolates from an Italian Culture Collection. Antimicrobial Agents and Chemotherapy. 2016;60:682–685. doi: 10.1128/AAC.02234-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenders A.C., de Marie S., ten Kate M.T. Liposomal amphotericin B (AmBisome) reduces dissemination of infection as compared with amphotericin B deoxycholate (Fungizone) in a rate model of pulmonary aspergillosis. Journal of Antimicrobial Chemotherapy. 1996;38:215–225. doi: 10.1093/jac/38.2.215. [DOI] [PubMed] [Google Scholar]
- Lenczuk D., Zinke-Cerwenka W., Greinix H. Antifungal prophylaxis with posaconazole delayed-release tablet and oral suspension in a real-life setting: Plasma levels, efficacy, and tolerability. Antimicrobial Agents and Chemotherapy. 2018;62:e02655–17. doi: 10.1128/AAC.02655-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardelli F., Macedo D., Dudiuk C. Aspergillus fumigatus intrinsic fluconazole resistance is due to the naturally occurring T301I substitution in Cyp51Ap. Antimicrobial Agents and Chemotherapy. 2016;60:5420–5426. doi: 10.1128/AAC.00905-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepak A.J., Andes D.R. Antifungal pharmacokinetics and pharmacodynamics. Cold Spring Harbor Perspectives in Medicine. 2014;5 doi: 10.1101/cshperspect.a019653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepak A.J., Marchillo K., Vanhecker J. Posaconazole pharmacodynamic target determination against wild-type and Cyp51 mutant isolates of Aspergillus fumigatus in an in vivo model of invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 2013;57:579–585. doi: 10.1128/AAC.01279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D. Clinical and virological data of the first cases of COVID-19 in Europe: A case series. Lancet Infectious Diseases. 2020;20:697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lestrade P.P., Bentvelsen R.G., Schauwvlieghe A. Voriconazole resistance and mortality in invasive aspergillosis: A multicenter retrospective cohort study. Clinical Infectious Diseases. 2019;68:1463–1471. doi: 10.1093/cid/ciy859. [DOI] [PubMed] [Google Scholar]
- Levdansky E., Romano J., Shadkchan Y. Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryotic Cell. 2007;6:1380–1391. doi: 10.1128/EC.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R.E., Kontoyiannis D.P. Invasive aspergillosis in glucocorticoid-treated patients. Medical Mycology. 2009;47:S271–S281. doi: 10.1080/13693780802227159. [DOI] [PubMed] [Google Scholar]
- Lewis R.E., Verweij P.E. Animal models for studying triazole resistance in Aspergillus fumigatus. Journal of Infectious Diseases. 2017;216:S466–S473. doi: 10.1093/infdis/jix222. [DOI] [PubMed] [Google Scholar]
- Li F., Wang B., Wang L. Phylogenetic analyses on the diversity of Aspergillus fumigatus sensu lato based on five orthologous loci. Mycopathologia. 2014;178:163–176. doi: 10.1007/s11046-014-9790-0. [DOI] [PubMed] [Google Scholar]
- Li Y., Wang H., Zhao Y.P. Antifungal susceptibility of clinical isolates of 25 genetically confirmed Aspergillus species collected from Taiwan and Mainland China. Journal of Microbiology, Immunology, and Infection. 2020;53:125–132. doi: 10.1016/j.jmii.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Liebmann B., Muhleisen T.W., Müller M. Deletion of the Aspergillus fumigatus lysine biosynthesis gene lysF encoding homoaconitase leads to attenuated virulence in a low-dose mouse infection model of invasive aspergillosis. Archives of Microbiology. 2004;181:378–383. doi: 10.1007/s00203-004-0667-3. [DOI] [PubMed] [Google Scholar]
- Lionakis M.S., Kontoyiannis D.P. Drosophila melanogaster as a model organism for invasive aspergillosis. Methods in Molecular Biology. 2012;845:455–468. doi: 10.1007/978-1-61779-539-8_32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zeng R., Zhang L. Multiple cyp51A-based mechanisms identified in azole-resistant isolates of Aspergillus fumigatus from China. Antimicrobial Agents and Chemotherapy. 2015;59:4321–4325. doi: 10.1128/AAC.00003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Zheng N., Li D. Cyp51A-based mechanism of azole resistance in Aspergillus fumigatus: Illustration by a new 3D structural model of Aspergillus fumigatus CYP51A protein. Medical Mycology. 2016;54:400–408. doi: 10.1093/mmy/myv102. [DOI] [PubMed] [Google Scholar]
- Lockhart S.R., Frade J.P., Etienne K.A. Azole resistance in Aspergillus fumigatus isolates from the ARTEMIS global surveillance study is primarily due to the TR/L98H mutation in the cyp51A gene. Antimicrobial Agents and Chemotherapy. 2011;55:4465–4468. doi: 10.1128/AAC.00185-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiko V., Wagener J. The paradoxical effect of echinocandins in Aspergillus fumigatus relies on recovery of the β-1,3-glucan synthase Fks1. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.01690-16. e01690–01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada L., Sugui J.A., Eckhaus M.A. Genetic analysis using an isogenic mating pair of Aspergillus fumigatus identifies azole resistance genes and lack of MAT locus’s role in virulence. PLoS Pathogens. 2015;11 doi: 10.1371/journal.ppat.1004834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon K.W., Burnie J.P., Coke A.P. Application of polymerase chain reaction to fingerprinting Aspergillus fumigatus by random amplification of polymorphic DNA. Journal of Clinical Microbiology. 1993;31:1117–1121. doi: 10.1128/jcm.31.5.1117-1121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowes D., Al-Shair K., Newton P.J. Predictors of mortality in chronic pulmonary aspergillosis. European Respiratory Journal. 2017;49:1601062. doi: 10.1183/13993003.01062-2016. [DOI] [PubMed] [Google Scholar]
- MacCallum D.M., Odds F.C. Influence of grapefruit juice on itraconazole plasma levels in mice and guinea pigs. Journal of Antimicrobial Chemotherapy. 2002;50:219–224. doi: 10.1093/jac/dkf103. [DOI] [PubMed] [Google Scholar]
- Maertens J., Raad I., Petrikkos G. Efficacy and safety of caspofungin for treatment of invasive aspergillosis in patients refractory to or intolerant of conventional antifungal therapy. Clinical Infectious Diseases. 2004;39:1563–1571. doi: 10.1086/423381. [DOI] [PubMed] [Google Scholar]
- Marr K.A., Boeckh M., Carter R.A. Combination antifungal therapy for invasive aspergillosis. Clinical Infectious Diseases. 2004;39:797–802. doi: 10.1086/423380. [DOI] [PubMed] [Google Scholar]
- Marr K.A., Schlamm H.T., Herbrecht R. Combination antifungal therapy for invasive aspergillosis: a randomized trial. Annals of Internal Medicine. 2015;162:81–89. doi: 10.7326/M13-2508. [DOI] [PubMed] [Google Scholar]
- Martin M.V., Yates J., Hitchcock C.A. Comparison of voriconazole (UK-109, 496) and itraconazole in prevention and treatment of Aspergillus fumigatus endocarditis in guinea pigs. Antimicrobial Agents and Chemotherapy. 1997;41:13–16. doi: 10.1128/aac.41.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli A.W., Patil P., Wong V.K. A positive BAL galactomannan in non-haemato-oncology patients risks harmful overtreatment. Journal of Medical Microbiology. 2019;68:1766–1770. doi: 10.1099/jmm.0.001109. [DOI] [PubMed] [Google Scholar]
- Martos A.I., Romero A., Gonzalez M.T. Evaluation of the Etest method for susceptibility testing of Aspergillus spp. and Fusarium spp. to three echinocandins. Medical Mycology. 2010;48:858–861. doi: 10.3109/13693781003586943. [DOI] [PubMed] [Google Scholar]
- Maurer E., Browne N., Surlis C. Galleria mellonella as a host model to study Aspergillus terreus virulence and amphotericin B resistance. Virulence. 2015;6:591–598. doi: 10.1080/21505594.2015.1045183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer E., Hörtnagl C., Lackner M. Galleria mellonella as a model system to study virulence potential of mucormycetes and evaluation of antifungal treatment. Medical Mycology. 2019;57:351–362. doi: 10.1093/mmy/myy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavridou E., Bruggemann R.J., Melchers W.J. Impact of cyp51A mutations on the pharmacokinetic and pharmacodynamic properties of voriconazole in a murine model of disseminated aspergillosis. Antimicrobial Agents and Chemotherapy. 2010;54:4758–4764. doi: 10.1128/AAC.00606-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. Koeltz Scientific Books; Königstein: 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code) adopted by the Eighteenth International Botanical Congress Melbourne, Australia, July 2011. [Regnum Vegetabile No. 154.] [Google Scholar]
- McTaggart L.R., Copeland J.K., Surendra A. Mycobiome sequencing and analysis applied to fungal community profiling of the lower respiratory tract during fungal pathogenesis. Frontiers in Microbiology. 2019;10:512. doi: 10.3389/fmicb.2019.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nature Reviews Immunology. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Meijer E.F.J., Dofferhoff A.S.M., Hoiting O. Azole-resistant COVID-19-associated pulmonary aspergillosis in an immunocompetent host: A case report. Journal of Fungi. 2020;6:79. doi: 10.3390/jof6020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J., Mavridou E., Melchers W.J. Epidemiological cutoff values for azoles and Aspergillus fumigatus based on a novel mathematical approach incorporating cyp51A sequence analysis. Antimicrobial Agents and Chemotherapy. 2012;56:2524–2529. doi: 10.1128/AAC.05959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J., Siopi M., Kanioura L. Development and multicentre validation of an agar-based screening method for echinocandin susceptibility testing of Aspergillus species. Journal of Antimicrobial Chemotherapy. 2019;74:2247–2254. doi: 10.1093/jac/dkz154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meletiadis J., Siopi M., Kanioura L. A multicentre study to optimize echinocandin susceptibility testing of Aspergillus species with the EUCAST methodology and a broth microdilution colorimetric method. Journal of Antimicrobial Chemotherapy. 2020;75:1799–1806. doi: 10.1093/jac/dkaa102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., García-Effron G., Alcazar-Fuoli L. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrobial Agents and Chemotherapy. 2007;51:1897–1904. doi: 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., García-Effron G., Buitrago M.J. Targeted gene disruption of the 14-alpha sterol demethylase (cyp51A) in Aspergillus fumigatus and its role in azole drug susceptibility. Antimicrobial Agents and Chemotherapy. 2005;49:2536–2538. doi: 10.1128/AAC.49.6.2536-2538.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello E., Posteraro B., Vella A. Susceptibility testing of common and uncommon Aspergillus species against posaconazole and other mold-active antifungal azoles using the Sensititre method. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.00168-17. e00168–00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.A., Anderson A., Morris M.I. Successful treatment of invasive fungal infection due to highly resistant Aspergillus calidoustus in an allogeneic hematopoietic cell transplant recipient. Mycopathologia. 2020;185:399–403. doi: 10.1007/s11046-019-00423-x. [DOI] [PubMed] [Google Scholar]
- Meneau I., Coste A.T., Sanglard D. Identification of Aspergillus fumigatus multidrug transporter genes and their potential involvement in antifungal resistance. Medical Mycology. 2016;54:616–627. doi: 10.1093/mmy/myw005. [DOI] [PubMed] [Google Scholar]
- Menotti J., Waller J., Meunier O. Epidemiological study of invasive pulmonary aspergillosis in a haematology unit by molecular typing of environmental and patient isolates of Aspergillus fumigatus. Journal of Hospital Infection. 2005;60:61–68. doi: 10.1016/j.jhin.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Mercier T., Dunbar A., de Kort E. Lateral flow assays for diagnosing invasive pulmonary aspergillosis in adult hematology patients: A comparative multicenter study. Medical Mycology. 2020;58:444–452. doi: 10.1093/mmy/myz079. [DOI] [PubMed] [Google Scholar]
- Meyer V., Wu B., Ram A.F. Aspergillus as a multi-purpose cell factory: current status and perspectives. Biotechnology Letters. 2011;33:469–476. doi: 10.1007/s10529-010-0473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel L., Lin K.Y., Ong V. Rezafungin treatment in mouse models of invasive candidiasis and aspergillosis: Insights on the PK/PD pharmacometrics of rezafungin efficacy. Pharmacology Research & Perspectives. 2019;7 doi: 10.1002/prp2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulska M., Furfaro E., De Carolis E. Use of Aspergillus fumigatus real-time PCR in bronchoalveolar lavage samples (BAL) for diagnosis of invasive aspergillosis, including azole-resistant cases, in high risk haematology patients: the need for a combined use with galactomannan. Medical Mycology. 2019;57:987–996. doi: 10.1093/mmy/myz002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkov I., Popov Aleksandrov A., Lazovic B. Usefulness of animal models of aspergillosis in studying immunity against Aspergillus infections. Journal de Mycologie Medicale. 2019;29:84–96. doi: 10.1016/j.mycmed.2019.01.006. [DOI] [PubMed] [Google Scholar]
- Misslinger M., Gsaller F., Hortschansky P. The cytochrome b5 CybE is regulated by iron availability and is crucial for azole resistance in A. fumigatus. Metallomics. 2017;9:1655–1665. doi: 10.1039/c7mt00110j. [DOI] [PubMed] [Google Scholar]
- Mitaka H., Perlman D.C., Javaid W. Putative invasive pulmonary aspergillosis in critically ill patients with COVID-19: An observational study from New York City. Mycoses. 2020;63:1368–1372. doi: 10.1111/myc.13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazam S., Eades C.P., Muldoon E.G. Positive Aspergillus PCR as a marker of azole resistance or sub–therapeutic antifungal therapy in patients with chronic pulmonary aspergillosis. Mycoses. 2020;63:376–381. doi: 10.1111/myc.13052. [DOI] [PubMed] [Google Scholar]
- Mohamed A., Hassan T., Trzos-Grzybowska M. Multi-triazole-resistant Aspergillus fumigatus and SARS-CoV-2 co-infection: A lethal combination. Medical Mycology Case Reports. 2021;31:11–14. doi: 10.1016/j.mmcr.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondon P., Brenier M.P., Symoens F. Molecular typing of Aspergillus fumigatus strains by sequence-specific DNA primer (SSDP) analysis. FEMS Immunology and Medical Microbiology. 1997;17:95–102. doi: 10.1111/j.1574-695X.1997.tb01001.x. [DOI] [PubMed] [Google Scholar]
- Monteiro C., Pinheiro D., Maia M. Aspergillus species collected from environmental air samples in Portugal-molecular identification, antifungal susceptibility and sequencing of cyp51A gene on A. fumigatus sensu stricto itraconazole resistant. Journal of Applied Microbiology. 2019;126:1140–1148. doi: 10.1111/jam.14217. [DOI] [PubMed] [Google Scholar]
- Montesinos I., Argudin M.A., Hites M. Culture-based methods and molecular tools for azole-resistant Aspergillus fumigatus detection in a Belgian University Hospital. Journal of Clinical Microbiology. 2017;55:2391–2399. doi: 10.1128/JCM.00520-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesinos I., Dodemont M., Lagrou K. New case of azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation in Belgium, July 2012. Journal of Antimicrobial Chemotherapy. 2013;69:3439–3440. doi: 10.1093/jac/dku289. [DOI] [PubMed] [Google Scholar]
- Moore C.B., Novak-Frazer L., Muldoon E. First isolation of the pan-azole-resistant Aspergillus fumigatus cyp51A TR46/Y121F/T289A mutant in a UK patient. International Journal of Antimicrobial Agents. 2017;49:512–514. doi: 10.1016/j.ijantimicag.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Moore C.B., Walls C.M., Denning D.W. In vitro activities of terbinafine against Aspergillus species in comparison with those of itraconazole and amphotericin B. Antimicrobial Agents and Chemotherapy. 2001;45:1882–1885. doi: 10.1128/AAC.45.6.1882-1885.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa M.Y., Alangaden G.J., Manavathu E. Resistance to amphotericin B does not emerge during treatment for invasive aspergillosis. Journal of Antimicrobial Chemotherapy. 2002;49:209–213. doi: 10.1093/jac/49.1.209. [DOI] [PubMed] [Google Scholar]
- Moreno-Velásquez S.D., Seidel C., Juvvadi P.R. Caspofungin-mediated growth inhibition and paradoxical growth in Aspergillus fumigatus involve fungicidal hyphal tip lysis coupled with regenerative intrahyphal growth and dynamic changes in β-1,3-glucan synthase localization. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.00710-17. e00710–00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S., Bozza S., Oikonomou V. IL-37 inhibits inflammasome activation and disease severity in murine aspergillosis. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morio F., Dannaoui E., Chouaki T. PCR-based detection of Aspergillus fumigatus and absence of azole resistance due to TR34/L98H in a french multicenter cohort of 137 patients with fungal rhinosinusitis. Mycoses. 2018;61:30–34. doi: 10.1111/myc.12702. [DOI] [PubMed] [Google Scholar]
- Morio F., Lombardi L., Butler G. The CRISPR toolbox in medical mycology: State of the art and perspectives. PLoS Pathogens. 2020;16 doi: 10.1371/journal.ppat.1008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen K.L., Mellado E., Lass-Flörl C. Environmental study of azole-resistant Aspergillus fumigatus and other aspergilli in Austria, Denmark, and Spain. Antimicrobial Agents and Chemotherapy. 2010;54:4545–4549. doi: 10.1128/AAC.00692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosquera J., Warn P.A., Morrissey J. Susceptibility testing of Aspergillus flavus: inoculum dependence with itraconazole and lack of correlation between susceptibility to amphotericin B in vitro and outcome in vivo. Antimicrobial Agents and Chemotherapy. 2001;45:1456–1462. doi: 10.1128/AAC.45.5.1456-1462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouton J.W., Muller A.E., Canton R. MIC-based dose adjustment: facts and fables. Journal of Antimicrobial Chemotherapy. 2018;73:564–568. doi: 10.1093/jac/dkx427. [DOI] [PubMed] [Google Scholar]
- Mushi M.F., Buname G., Bader O. Aspergillus fumigatus carrying TR34/L98H resistance allele causing complicated suppurative otitis media in Tanzania: Call for improved diagnosis of fungi in sub-Saharan Africa. BMC Infectious Diseases. 2016;16:464. doi: 10.1186/s12879-016-1796-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U., Vogel P., Alber G. The innate immune system of mammals and insects. Contributions to Microbiology. 2008;15:21–44. doi: 10.1159/000135684. [DOI] [PubMed] [Google Scholar]
- Myhrvold C., Freije C.A., Gootenberg J.S. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabili M., Shokohi T., Moazeni M. High prevalence of clinical and environmental triazole-resistant Aspergillus fumigatus in Iran: is it a challenging issue? Journal of Medical Microbiology. 2016;65:468–475. doi: 10.1099/jmm.0.000255. [DOI] [PubMed] [Google Scholar]
- Naidu J., Singh S.M. Aspergillus chevalieri (Mangin) Thom and Church: a new opportunistic pathogen of human cutaneous aspergillosis. Mycoses. 1994;37:271–274. doi: 10.1111/j.1439-0507.1994.tb00425.x. [DOI] [PubMed] [Google Scholar]
- Nascimento A.M., Goldman G.H., Park S. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to itraconazole. Antimicrobial Agents and Chemotherapy. 2003;47:1719–1726. doi: 10.1128/AAC.47.5.1719-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash A., Rhodes J. Simulations of CYP51A from Aspergillus fumigatus in a model bilayer provide insights into triazole drug resistance. Medical Mycology. 2018;56:361–373. doi: 10.1093/mmy/myx056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawada R., Amitani R., Tanaka E. Murine model of invasive pulmonary aspergillosis following an earlier stage, noninvasive Aspergillus infection. Journal of Clinical Microbiology. 1996;34:1433–1439. doi: 10.1128/jcm.34.6.1433-1439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negri C.E., Goncalves S.S., Xafranski H. Cryptic and rare Aspergillus species in Brazil: prevalence in clinical samples and in vitro susceptibility to triazoles. Journal of Clinical Microbiology. 2014;52:3633–3640. doi: 10.1128/JCM.01582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neofytos D., Horn D., Anaissie E. Epidemiology and outcome of invasive fungal infection in adult hematopoietic stem cell transplant recipients: analysis of Multicenter Prospective Antifungal Therapy (PATH) Alliance registry. Clinical Infectious Diseases. 2009;48:265–273. doi: 10.1086/595846. [DOI] [PubMed] [Google Scholar]
- Neubauer M., Zhu Z., Penka M. Mitochondrial dynamics in the pathogenic mold Aspergillus fumigatus: therapeutic and evolutionary implications. Molecular Microbiology. 2015;98:930–945. doi: 10.1111/mmi.13167. [DOI] [PubMed] [Google Scholar]
- Neuveglise C., Sarfati J., Latgé J.P. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Research. 1996;24:1428–1434. doi: 10.1093/nar/24.8.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyen C., Bretscher A.J., Binggeli O. Methods to study Drosophila immunity. Methods. 2014;68:116–128. doi: 10.1016/j.ymeth.2014.02.023. [DOI] [PubMed] [Google Scholar]
- Nierman W.C., Pain A., Anderson M.J. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- Nødvig C.S., Nielsen J.B., Kogle M.E. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novey H.S., Wells I.D. Allergic bronchopulmonary aspergillosis caused by Aspergillus ochraceus. American Journal of Clinical Pathology. 1978;70:840–843. doi: 10.1093/ajcp/70.5.840. [DOI] [PubMed] [Google Scholar]
- Nuh A., Ramadan N., Schelenz S., Armstrong-James D. Comparative evaluation of MIRONAUT-AM and CLSI broth microdilution method for antifungal susceptibility testing of Aspergillus species against four commonly used antifungals. Medical Mycology. 2020;58:1085–1090. doi: 10.1093/mmy/myaa020. [DOI] [PubMed] [Google Scholar]
- O’Gorman C.M., Fuller H., Dyer P.S. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Orasch T., Prattes J., Faserl K. Bronchoalveolar lavage triacetylfusarinine C (TAFC) determination for diagnosis of invasive pulmonary aspergillosis in patients with hematological malignancies. Journal of Infection. 2017;75:370–373. doi: 10.1016/j.jinf.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccoud O., Guery R., Poirée S. Aspergillus felis in patient with chronic granulomatous disease. Emerging Infectious Diseases. 2019;25:2319–2321. doi: 10.3201/eid2512.191020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas P.G., Alexander B.D., Andes D.R. Invasive fungal infections among organ transplant recipients: results of the Transplant-Associated Infection Surveillance Network (TRANSNET) Clinical Infectious Diseases. 2010;50:1101–1111. doi: 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- Park H., Youk J., Shin D.Y. Micafungin prophylaxis for acute leukemia patients undergoing induction chemotherapy. BMC Cancer. 2019;19:358. doi: 10.1186/s12885-019-5557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park W.B., Kim N.H., Kim K.H. The effect of therapeutic drug monitoring on safety and efficacy of voriconazole in invasive fungal infections: a randomized controlled trial. Clinical Infectious Diseases. 2012;55:1080–1087. doi: 10.1093/cid/cis599. [DOI] [PubMed] [Google Scholar]
- Patel R. A moldy application of MALDI: MALDI-ToF mass spectrometry for fungal identification. Journal of Fungi. 2019;5:4. doi: 10.3390/jof5010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil A., Majumdar S. Echinocandins in antifungal pharmacotherapy. Journal of Pharmacy and Pharmacology. 2017;69:1635–1660. doi: 10.1111/jphp.12780. [DOI] [PubMed] [Google Scholar]
- Patin E.C., Thompson A., Orr S.J. Pattern recognition receptors in fungal immunity. Seminars in Cell & Developmental Biology. 2019;89:24–33. doi: 10.1016/j.semcdb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson T.F., George D., Ingersoll R. Efficacy of SCH 39304 in treatment of experimental invasive aspergillosis. Antimicrobial Agents and Chemotherapy. 1991;35:1985–1988. doi: 10.1128/aac.35.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson T.F., Miniter P., Ryan J.L. Effect of immunosuppression and amphotericin B on Aspergillus antigenemia in an experimental model. Journal of Infectious Diseases. 1988;158:415–422. doi: 10.1093/infdis/158.2.415. [DOI] [PubMed] [Google Scholar]
- Patterson T.F., Thompson G.R., 3rd, Denning D.W. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clinical Infectious Diseases. 2016;63:e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Diekema D., Moye-Rowley W.S. Contributions of both ATP-binding cassette transporter and Cyp51A proteins are essential for azole resistance in Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 2017;61:e02748–16. doi: 10.1128/AAC.02748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Moye-Rowley W.S. Functional analysis of an ATP-binding cassette transporter protein from Aspergillus fumigatus by heterologous expression in Saccharomyces cerevisiae. Fungal Genetics and Biology. 2013;57:85–91. doi: 10.1016/j.fgb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S., Stamnes M., Thomas G.H. AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. mBio. 2019;10:e02563–18. doi: 10.1128/mBio.02563-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulussen C., Boulet G.A., Cos P. Animal models of invasive aspergillosis for drug discovery. Drug Discovery Today. 2014;19:1380–1386. doi: 10.1016/j.drudis.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Peláez T., Gijon P., Bunsow E. Resistance to voriconazole due to a G448S substitution in Aspergillus fumigatus in a patient with cerebral aspergillosis. Journal of Clinical Microbiology. 2012;50:2531–2534. doi: 10.1128/JCM.00329-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez T., Monteiro M.C., Garcia-Rubio R. First detection of Aspergillus fumigatus azole-resistant strain due to Cyp51A TR46/Y121F/T289A in an azole-naïve patient in Spain. New Microbes and New Infections. 2015;6:33–34. doi: 10.1016/j.nmni.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelzer B.W., Seufert R., Koldehoff M. Performance of the AsperGenius(R) PCR assay for detecting azole resistant Aspergillus fumigatus in BAL fluids from allogeneic HSCT recipients: A prospective cohort study from Essen, West Germany. Medical Mycology. 2020;58:268–271. doi: 10.1093/mmy/myz050. [DOI] [PubMed] [Google Scholar]
- Pérez-Cantero A., López-Fernandez L., Guarro J., Capilla J. Azole resistance mechanisms in Aspergillus: update and recent advances. International Journal of Antimicrobial Agents. 2020;55:105807. doi: 10.1016/j.ijantimicag.2019.09.011. [DOI] [PubMed] [Google Scholar]
- Perlin D.S. Echinocandin resistance in Candida. Clinical Infectious Diseases. 2015;61:S612–S617. doi: 10.1093/cid/civ791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perveen I., Sehar S., Naz I. 7th. Advances Against Aspergillosis conference, March 3–5, 2016. Manchester, UK. 2016. Prospective evaluation of azole resistance in Aspergillus fumigatus clinical isolates in Pakistan. Abstract no: 1. [Google Scholar]
- Petraitiene R., Petraitis V., Groll A.H. Antifungal efficacy of caspofungin (MK-0991) in experimental pulmonary aspergillosis in persistently neutropenic rabbits: pharmacokinetics, drug disposition, and relationship to galactomannan antigenemia. Antimicrobial Agents and Chemotherapy. 2002;46:12–23. doi: 10.1128/AAC.46.1.12-23.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T., Xie X., Lin X. An intergenic “safe haven” region in Aspergillus fumigatus. Medical Mycology. 2020;58:1178–1186. doi: 10.1093/mmy/myaa009. [DOI] [PubMed] [Google Scholar]
- Pinto E., Monteiro C., Maia M. Aspergillus species and antifungals susceptibility in clinical setting in the North of Portugal: Cryptic species and emerging azoles resistance in A. fumigatus. Frontiers in Microbiology. 2018;9:1656. doi: 10.3389/fmicb.2018.01656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes L., Beraquet C.A.G., Arai T. Aspergillus fumigatus clinical isolates carrying CYP51A with TR34/L98H/S297T/F495I substitutions detected after four–year retrospective azole resistance screening in Brazil. Antimicrobial Agents and Chemotherapy. 2020;64:e02059–19. doi: 10.1128/AAC.02059-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postina P., Skladny J., Boch T. Comparison of two molecular assays for detection and characterization of Aspergillus fumigatus triazole resistance and Cyp51A mutations in clinical isolates and primary clinical samples of immunocompromised patients. Frontiers in Microbiology. 2018;9:555. doi: 10.3389/fmicb.2018.00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattes J., Duettmann W., Hoenigl M. Posaconazole plasma concentrations on days three to five predict steady–state levels. Antimicrobial Agents and Chemotherapy. 2016;60:5595–5599. doi: 10.1128/AAC.00389-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattes J., Flick H., Pruller F. Novel tests for diagnosis of invasive aspergillosis in patients with underlying respiratory diseases. American Journal of Respiratory and Critical Care Medicine. 2014;190:922–929. doi: 10.1164/rccm.201407-1275OC. [DOI] [PubMed] [Google Scholar]
- Prattes J., Hoenigl M., Krause R. Invasive aspergillosis in patients with underlying liver cirrhosis: a prospective cohort study. Medical Mycology. 2017;55:803–812. doi: 10.1093/mmy/myx011. [DOI] [PubMed] [Google Scholar]
- Prattes J., Hoenigl M., Zinke S.E. Evaluation of the new AspID polymerase chain reaction assay for detection of Aspergillus species: A pilot study. Mycoses. 2018;61:355–359. doi: 10.1111/myc.12757. [DOI] [PubMed] [Google Scholar]
- Prattes J., Lackner M., Eigl S. Diagnostic accuracy of the Aspergillus-specific bronchoalveolar lavage lateral-flow assay in haematological malignancy patients. Mycoses. 2015;58:461–469. doi: 10.1111/myc.12343. [DOI] [PubMed] [Google Scholar]
- Prattes J., Valentin T., Hoenigl M., Talakic E., Reisinger A.C., Eller P. Invasive pulmonary aspergillosis complicating COVID-19 in the ICU – A case report. Medical Mycology Case Reports. 2021;31:2–5. doi: 10.1016/j.mmcr.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C.L., Parker J.E., Warrilow A.G. Azole fungicides – understanding resistance mechanisms in agricultural fungal pathogens. Pest Management Science. 2015;71:1054–1058. doi: 10.1002/ps.4029. [DOI] [PubMed] [Google Scholar]
- Prigitano A., Esposto M.C., Biffi A. Triazole resistance in Aspergillus fumigatus isolates from patients with cystic fibrosis in Italy. Journal of Cystic Fibrosis. 2017;16:64–69. doi: 10.1016/j.jcf.2016.06.006. [DOI] [PubMed] [Google Scholar]
- Prigitano A., Venier V., Cogliati M. Azole-resistant Aspergillus fumigatus in the environment of northern Italy, May 2011 to June 2012. Euro Surveillance. 2014;19:20747. doi: 10.2807/1560-7917.es2014.19.12.20747. [DOI] [PubMed] [Google Scholar]
- Puértolas-Balint F., Rossen J.W.A., Oliveira Dos Santos C. Revealing the virulence potential of clinical and environmental Aspergillus fumigatus isolates using whole-genome sequencing. Frontiers in Microbiology. 2019;10:1970. doi: 10.3389/fmicb.2019.01970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt P.J., van den Hondel C.A. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods in Enzymology. 1992;216:447–457. doi: 10.1016/0076-6879(92)16041-h. [DOI] [PubMed] [Google Scholar]
- Quan J., Langelier C., Kuchta A. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Research. 2019;47:e83. doi: 10.1093/nar/gkz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaprakash H., Ito T., Standiford T.J. Toll-like receptor 9 modulates immune responses to Aspergillus fumigatus conidia in immunodeficient and allergic mice. Infection and Immunity. 2009;77:108–119. doi: 10.1128/IAI.00998-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper K.B., Fennell D.I. Williams and Wilkins; Baltimore, Maryland: 1965. The Genus Aspergillus. [Google Scholar]
- Rath P.M., Buchheidt D., Spiess B. First reported case of azole-resistant Aspergillus fumigatus due to the TR/L98H mutation in Germany. Antimicrobial Agents and Chemotherapy. 2012;56:6060–6061. doi: 10.1128/AAC.01017-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings S.A., Heldt S., Prattes J. Using interleukin 6 and 8 in blood and bronchoalveolar lavage fluid to predict survival in hematological malignancy patients with suspected pulmonary mold infection. Frontiers in Immunology. 2019;10:1798. doi: 10.3389/fimmu.2019.01798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson T.M., O’Hare D., Herrero P. Delivering precision antimicrobial therapy through closed-loop control systems. Journal of Antimicrobial Chemotherapy. 2018;73:835–843. doi: 10.1093/jac/dkx458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reischies F., Hoenigl M. The role of surgical debridement in different clinical manifestations of invasive aspergillosis. Mycoses. 2014;57(suppl. 2):1–14. doi: 10.1111/myc.12224. [DOI] [PubMed] [Google Scholar]
- Rex J.H., Pfaller M.A. Has antifungal susceptibility testing come of age? Clinical Infectious Diseases. 2002;35:982–989. doi: 10.1086/342384. [DOI] [PubMed] [Google Scholar]
- Riat A., Plojoux J., Gindro K. Azole resistance of environmental and clinical Aspergillus fumigatus isolates from Switzerland. Antimicrobial Agents and Chemotherapy. 2018;62:e02088–17. doi: 10.1128/AAC.02088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard N., Marti L., Varrot A. Human bronchial epithelial cells inhibit Aspergillus fumigatus germination of extracellular conidia via FleA recognition. Scientific Reports. 2018;8:15699. doi: 10.1038/s41598-018-33902-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M., Page I. Role of serological tests in the diagnosis of mold infections. Current Fungal Infection Reports. 2018;12:127–136. doi: 10.1007/s12281-018-0321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M.D., Page I.D. Aspergillus serology: Have we arrived yet? Medical Mycology. 2017;55:48–55. doi: 10.1093/mmy/myw116. [DOI] [PubMed] [Google Scholar]
- Ries L.N.A., Beattie S., Cramer R.A. Overview of carbon and nitrogen catabolite metabolism in the virulence of human pathogenic fungi. Molecular Microbiology. 2018;107:277–297. doi: 10.1111/mmi.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risslegger B., Zoran T., Lackner M. A prospective international Aspergillus terreus survey: an EFISG, ISHAM and ECMM joint study. Clinical Microbiology and Infection. 2017;23 doi: 10.1016/j.cmi.2017.04.012. 776.e1–776.e5. [DOI] [PubMed] [Google Scholar]
- Rivero-Menendez O., Cuenca-Estrella M., Alastruey-Izquierdo A. In vitro activity of olorofim (F901318) against clinical isolates of cryptic species of Aspergillus by EUCAST and CLSI methodologies. Journal of Antimicrobial Chemotherapy. 2019;74:1586–1590. doi: 10.1093/jac/dkz078. [DOI] [PubMed] [Google Scholar]
- Rivero-Menendez O., Soto-Debran J.C., Medina N. Molecular identification, antifungal susceptibility testing, and mechanisms of azole resistance in Aspergillus species received within a surveillance program on antifungal resistance in Spain. Antimicrobial Agents and Chemotherapy. 2019;63:e00865–e00919. doi: 10.1128/AAC.00865-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchi S., Ponçot M., Morin-Crini N. Determination of azole fungal residues in soils and detection of Aspergillus fumigatus-resistant strains in market gardens of Eastern France. Environmental Science and Pollution Research International. 2018;25:32015–32023. doi: 10.1007/s11356-018-3177-6. [DOI] [PubMed] [Google Scholar]
- Rochette F., Engelen M., Vanden Bossche H. Antifungal agents of use in animal health – practical applications. Journal of Veterinary Pharmacology and Therapeutics. 2003;26:31–53. doi: 10.1046/j.1365-2885.2003.00457.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez E., De Meeûs T., Mallie M. Multicentric epidemiological study of Aspergillus fumigatus isolates by multilocus enzyme electrophoresis. Journal of Clinical Microbiology. 1996;34:2559–2568. doi: 10.1128/jcm.34.10.2559-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolle A.M., Hasenberg M., Thornton C.R. ImmunoPET/MR imaging allows specific detection of Aspergillus fumigatus lung infection in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E1026–E1033. doi: 10.1073/pnas.1518836113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A., Ene I.V., Bibi M. Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nature Communications. 2018;9:2470. doi: 10.1038/s41467-018-04926-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudramurthy S.M., Paul R.A., Chakrabarti A. Invasive aspergillosis by Aspergillus flavus: Epidemiology, diagnosis, antifungal resistance, and management. Journal of Fungi. 2019;5:55. doi: 10.3390/jof5030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudramurthy S.M., Seyedmousavi S., Dhaliwal M. Pharmacodynamics of voriconazole against wild-type and azole-resistant Aspergillus flavus isolates in a nonneutropenic murine model of disseminated aspergillosis. Antimicrobial Agents and Chemotherapy. 2017;61 doi: 10.1128/AAC.01491-16. e01491–01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda C., Puig-Asensio M., Guinea J. Evaluation of the possible influence of trailing and paradoxical effects on the clinical outcome of patients with candidemia. Clinical Microbiology and Infection. 2017;23:49. doi: 10.1016/j.cmi.2016.09.016. e1–49.e8. [DOI] [PubMed] [Google Scholar]
- Rutsaert L., Steinfort N., Van Hunsel T. COVID-19-associated invasive pulmonary aspergillosis. Annals of Intensive Care. 2020;10:71. doi: 10.1186/s13613-020-00686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak J.M., Ge W., Wiederhold N.P. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in Aspergillus fumigatus. mBio. 2019;10:e00437–e00519. doi: 10.1128/mBio.00437-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino R., Veríssimo C., Parada H. Molecular screening of 246 Portuguese Aspergillus isolates among different clinical and environmental sources. Medical Mycology. 2014;52:519–529. doi: 10.1093/mmy/myu006. [DOI] [PubMed] [Google Scholar]
- Salah H., Lackner M., Houbraken J. The emergence of rare clinical Aspergillus species in Qatar: Molecular characterization and antifungal susceptibility profiles. Frontiers in Microbiology. 2019;10:1677. doi: 10.3389/fmicb.2019.01677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas V., Pastor F.J., Sutton D.A. MIC values of voriconazole are predictive of treatment results in murine infections by Aspergillus terreus species complex. Antimicrobial Agents and Chemotherapy. 2013;57:1532–1534. doi: 10.1128/AAC.01436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsé M., Gangneux J.P., Cassaing S. Multicentre study to determine the Etest epidemiological cut-off values of antifungal drugs in Candida spp. and Aspergillus fumigatus species complex. Clinical Microbiology and Infection. 2019;25:1546–1552. doi: 10.1016/j.cmi.2019.04.027. [DOI] [PubMed] [Google Scholar]
- Salzer H.J.F., Prattes J., Flick H. Evaluation of galactomannan testing, the Aspergillus-specific lateral-flow device test and levels of cytokines in bronchoalveolar lavage fluid for diagnosis of chronic pulmonary aspergillosis. Frontiers in Microbiology. 2018;9:2223. doi: 10.3389/fmicb.2018.02223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Hong S., Peterson S.W. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Studies in Mycology. 2007;59:147–203. doi: 10.3114/sim.2007.59.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Houbraken J., Thrane U. CBS-KNAW Fungal Biodiversity Centre; Utrecht, The Netherlands: 2019. Food and Indoor Fungi. [Google Scholar]
- Samson R.A., Noonim P., Meijer M. Diagnostic tools to identify black aspergilli. Studies in Mycology. 2007;59:129–145. doi: 10.3114/sim.2007.59.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Peterson S.W., Frisvad J.C. New species in Aspergillus section Terrei. Studies in Mycology. 2011;69:39–55. doi: 10.3114/sim.2011.69.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Visagie C.M., Houbraken J. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez O., Aguirre J. Efficient transformation of Aspergillus nidulans by electroporation of germinated conidia. Fungal Genetics Reports. 1996;43:21. [Google Scholar]
- Sanguinetti M., Posteraro B. Identification of molds by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Journal of Clinical Microbiology. 2017;55:369–379. doi: 10.1128/JCM.01640-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarfati J., Diaquin M., Debeaupuis J.P. A new experimental murine aspergillosis model to identify strains of Aspergillus fumigatus with reduced virulence. Nihon Ishinkin Gakkai Zasshi. 2002;43:203–213. doi: 10.3314/jjmm.43.203. [DOI] [PubMed] [Google Scholar]
- Satish S., Jiménez-Ortigosa C., Zhao Y. Stress-induced changes in the lipid microenvironment of β-(1,3)-d-glucan synthase cause clinically important echinocandin resistance in Aspergillus fumigatus. mBio. 2019;10:e00779–e00819. doi: 10.1128/mBio.00779-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasaki H., Horie K., Naito Y. Experimental pulmonary aspergilloma. Mycopathologia et Mycologia Applicata. 1967;32:265–274. doi: 10.1007/BF02202064. [DOI] [PubMed] [Google Scholar]
- Schauwvlieghe A., de Jonge N., van Dijk K. The diagnosis and treatment of invasive aspergillosis in Dutch haematology units facing a rapidly increasing prevalence of azole-resistance. A nationwide survey and rationale for the DB–MSG 002 study protocol. Mycoses. 2018;61:656–664. doi: 10.1111/myc.12788. [DOI] [PubMed] [Google Scholar]
- Schauwvlieghe A., Rijnders B.J.A., Philips N. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. Lancet Respiratory Medicine. 2018;6:782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- Schauwvlieghe A., Vonk A.G., Buddingh E.P. Detection of azole-susceptible and azole-resistant Aspergillus coinfection by cyp51A PCR amplicon melting curve analysis. Journal of Antimicrobial Chemotherapy. 2017;72:3047–3050. doi: 10.1093/jac/dkx262. [DOI] [PubMed] [Google Scholar]
- Schmitt H.J., Bernard E.M., Hauser M. Aerosol amphotericin B is effective for prophylaxis and therapy in a rat model of pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 1988;32:1676–1679. doi: 10.1128/aac.32.11.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobel F., Jacobsen I.D., Brock M. Evaluation of lysine biosynthesis as an antifungal drug target: biochemical characterization of Aspergillus fumigatus homocitrate synthase and virulence studies. Eukaryotic Cell. 2010;9:878–893. doi: 10.1128/EC.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz C., Lian J., Zhao H. Metabolic engineering of Saccharomyces cerevisiae using a trifunctional CRISPR/Cas system for simultaneous gene activation, interference, and deletion. Methods in Enzymology. 2018;608:265–276. doi: 10.1016/bs.mie.2018.04.010. [DOI] [PubMed] [Google Scholar]
- Schwartz I.S., Friedman D.Z.P., Zapernick L. High rates of influenza-associated invasive pulmonary aspergillosis may not be universal: A retrospective cohort study from Alberta, Canada. Clinical Infectious Diseases. 2020;71:1760–1763. doi: 10.1093/cid/ciaa007. [DOI] [PubMed] [Google Scholar]
- Seidel C., Moreno-Velásquez S.D., Ben-Ghazzi N. Phagolysosomal survival enables non-lytic hyphal escape and ramification through lung epithelium during Aspergillus fumigatus infection. Frontiers in Microbiology. 2020;11:1955. doi: 10.3389/fmicb.2020.01955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaniakou A., Croll R.P., Chappe V. Animal models in the pathophysiology of cystic fibrosis. Frontiers in pharmacology. 2018;9:1475. doi: 10.3389/fphar.2018.01475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert R., Sedlacek L., Kahl B. Prevalence and characterization of azole-resistant Aspergillus fumigatus in patients with cystic fibrosis: a prospective multicentre study in Germany. Journal of Antimicrobial Chemotherapy. 2018;73:2047–2053. doi: 10.1093/jac/dky147. [DOI] [PubMed] [Google Scholar]
- Sever N., Yang T., Brown M.S. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Molecular Cell. 2003;11:25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- Sewell T.R., Zhang Y., Brackin A.P. Elevated prevalence of azole-resistant Aspergillus fumigatus in urban versus rural environments in the United Kingdom. Antimicrobial Agents and Chemotherapy. 2019;63:e00548–e00619. doi: 10.1128/AAC.00548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S., Bruggemann R.J., Meis J.F. Pharmacodynamics of isavuconazole in an Aspergillus fumigatus mouse infection model. Antimicrobial Agents and Chemotherapy. 2015;59:2855–2866. doi: 10.1128/AAC.04907-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S., Bruggemann R.J., Melchers W.J. Efficacy and pharmacodynamics of voriconazole combined with anidulafungin in azole-resistant invasive aspergillosis. Journal of Antimicrobial Chemotherapy. 2013;68:385–393. doi: 10.1093/jac/dks402. [DOI] [PubMed] [Google Scholar]
- Seyedmousavi S., Hashemi S.J., Zibafar E. Azole-resistant Aspergillus fumigatus, Iran. Emerging Infectious Diseases. 2013;19:832–834. doi: 10.3201/eid1905.130075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S., Lionakis M.S., Parta M. Emerging Aspergillus species almost exclusively associated with primary immunodeficiencies. Open Forum Infectious Diseases. 2018;5:ofy213. doi: 10.1093/ofid/ofy213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedmousavi S., Mouton J.W., Melchers W.J. The role of azoles in the management of azole-resistant aspergillosis: from the bench to the bedside. Drug Resistance Updates. 2014;17:37–50. doi: 10.1016/j.drup.2014.06.001. [DOI] [PubMed] [Google Scholar]
- Seyedmousavi S., Mouton J.W., Melchers W.J. Posaconazole prophylaxis in experimental azole-resistant invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 2015;59:1487–1494. doi: 10.1128/AAC.03850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C., Hagen F., Moroti R. Triazole-resistant Aspergillus fumigatus harbouring G54 mutation: Is it de novo or environmentally acquired? Journal of Global Antimicrobial Resistance. 2015;3:69–74. doi: 10.1016/j.jgar.2015.01.005. [DOI] [PubMed] [Google Scholar]
- Sharma C., Nelson-Sathi S., Singh A. Genomic perspective of triazole resistance in clinical and environmental Aspergillus fumigatus isolates without cyp51A mutations. Fungal Genetics and Biology. 2019;132:103265. doi: 10.1016/j.fgb.2019.103265. [DOI] [PubMed] [Google Scholar]
- Sharon H., Amar D., Levdansky E. PrtT-regulated proteins secreted by Aspergillus fumigatus activate MAPK signaling in exposed A549 lung cells leading to necrotic cell death. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan G., Garvey A., Croke M. Innate humoral immune defences in mammals and insects: The same, with differences? Virulence. 2018;9:1625–1639. doi: 10.1080/21505594.2018.1526531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepardson K.M., Jhingran A., Caffrey A. Myeloid derived hypoxia inducible factor 1-alpha is required for protection against pulmonary Aspergillus fumigatus infection. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard D.C., Rieg G., Chiang L.Y. Novel inhalational murine model of invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 2004;48:1908–1911. doi: 10.1128/AAC.48.5.1908-1911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Xiao Y., Mao Y. Voriconazole: A review of population pharmacokinetic analyses. Clinical Pharmacokinetics. 2019;58:687–703. doi: 10.1007/s40262-019-00735-7. [DOI] [PubMed] [Google Scholar]
- Shibata T., Habiel D.M., Coelho A.L. Axl receptor blockade protects from invasive pulmonary aspergillosis in mice. Journal of Immunology. 2014;193:3559–3565. doi: 10.4049/jimmunol.1401258. [DOI] [PubMed] [Google Scholar]
- Singh N., Limaye A.P., Forrest G. Combination of voriconazole and caspofungin as primary therapy for invasive aspergillosis in solid organ transplant recipients: a prospective, multicenter, observational study. Transplantation. 2006;81:320–326. doi: 10.1097/01.tp.0000202421.94822.f7. [DOI] [PubMed] [Google Scholar]
- Siopi M., Mavridou E., Mouton J.W. Susceptibility breakpoints and target values for therapeutic drug monitoring of voriconazole and Aspergillus fumigatus in an in vitro pharmacokinetic/pharmacodynamic model. Journal of Antimicrobial Chemotherapy. 2014;69:1611–1619. doi: 10.1093/jac/dku023. [DOI] [PubMed] [Google Scholar]
- Siopi M., Neroutsos E., Zisaki K. Bioassay for determining voriconazole serum levels in patients receiving combination therapy with echinocandins. Antimicrobial Agents and Chemotherapy. 2016;60:632–636. doi: 10.1128/AAC.01688-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siopi M., Pournaras S., Meletiadis J. Comparative evaluation of Sensititre YeastOne and CLSI M38-A2 reference method for antifungal susceptibility testing of Aspergillus spp. against echinocandins. Journal of Clinical Microbiology. 2017;55:1714–1719. doi: 10.1128/JCM.00044-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siopi M., Rivero-Menendez O., Gkotsis G. Nationwide surveillance of azole-resistant Aspergillus fumigatus environmental isolates in Greece: detection of pan-azole resistance associated with the TR46/Y121F/T289A cyp51A mutation. Journal of Antimicrobial Chemotherapy. 2020;75:3181–3188. doi: 10.1093/jac/dkaa316. [DOI] [PubMed] [Google Scholar]
- Sklenář F., Ž Jurjević, Peterson S.W. Increasing the species diversity in the Aspergillus section Nidulantes: Six novel species mainly from the indoor environment. Mycologia. 2020;112:342–370. doi: 10.1080/00275514.2019.1698923. [DOI] [PubMed] [Google Scholar]
- Sklenář F., Ž Jurjević, Zalar P. Phylogeny of xerophilic aspergilli (subgenus Aspergillus) and taxonomic revision of section Restricti. Studies in Mycology. 2017;88:161–236. doi: 10.1016/j.simyco.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaven J.W., Anderson M.J., Sanglard D. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genetics and Biology. 2002;36:199–206. doi: 10.1016/s1087-1845(02)00016-6. [DOI] [PubMed] [Google Scholar]
- Smolovic B., Vukcevic B., Muhovic D. Renal aspergillosis in a liver transplant patient: A case report and review of literature. World Journal of Clinical Cases. 2018;6:1155–1159. doi: 10.12998/wjcc.v6.i16.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E., Camps S.M., Karawajczyk A. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E., Huis In’t Veld R.A., Rijs A.J. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Applied and Environmental Microbiology. 2009;75:4053–4057. doi: 10.1128/AEM.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders E., van der Lee H.A., Kuijpers J. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Medicine. 2008;5:e219. doi: 10.1371/journal.pmed.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Zhai P., Lu L. Damage resistance protein (Dap) contributes to azole resistance in a sterol-regulatory-element-binding protein SrbA-dependent way. Applied Microbiology and Biotechnology. 2017;101:3729–3741. doi: 10.1007/s00253-016-8072-9. [DOI] [PubMed] [Google Scholar]
- Song J., Zhai P., Zhang Y. The Aspergillus fumigatus damage resistance protein family coordinately regulates ergosterol biosynthesis and azole susceptibility. mBio. 2016;7 doi: 10.1128/mBio.01919-15. e01919–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess B., Postina P., Reinwald M. Incidence of Cyp51 A key mutations in Aspergillus fumigatus – a study on primary clinical samples of immunocompromised patients in the period of 1995–2013. PLoS One. 2014;9 doi: 10.1371/journal.pone.0103113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess B., Seifarth W., Merker N. Development of novel PCR assays to detect azole resistance-mediating mutations of the Aspergillus fumigatus cyp51A gene in primary clinical samples from neutropenic patients. Antimicrobial Agents and Chemotherapy. 2012;56:3905–3910. doi: 10.1128/AAC.05902-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spikes S., Xu R., Nguyen C.K. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. Journal of Infectious Diseases. 2008;197:479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- Springer J., Lackner M., Nachbaur D. Prospective multicentre PCR-based Aspergillus DNA screening in high-risk patients with and without primary antifungal mould prophylaxis. Clinical Microbiology and Infection. 2016;22:80–86. doi: 10.1016/j.cmi.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Stappers M.H.T., Clark A.E., Aimanianda V. Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus. Nature. 2018;555:382–386. doi: 10.1038/nature25974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrou A.A., Mixao V., Boekhout T. Misidentification of genome assemblies in public databases: The case of Naumovozyma dairenensis and proposal of a protocol to correct misidentifications. Yeast. 2018;35:425–429. doi: 10.1002/yea.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk J.L., Mead M.E., Knowles S.L. Variation among biosynthetic gene clusters, secondary metabolite profiles, and cards of virulence across Aspergillus species. Genetics. 2020;216:481–497. doi: 10.1534/genetics.120.303549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach W.J., Benjamin D.K., Jr., Trasi S.A. Value of an inhalational model of invasive aspergillosis. Medical Mycology. 2004;42:417–425. doi: 10.1080/13693780410001712034. [DOI] [PubMed] [Google Scholar]
- Steinmann J., Hamprecht A., Vehreschild M.J. Emergence of azole-resistant invasive aspergillosis in HSCT recipients in Germany. Journal of Antimicrobial Chemotherapy. 2015;70:1522–1526. doi: 10.1093/jac/dku566. [DOI] [PubMed] [Google Scholar]
- Stephens-Romero S.D., Mednick A.J., Feldmesser M. The pathogenesis of fatal outcome in murine pulmonary aspergillosis depends on the neutrophil depletion strategy. Infection and Immunity. 2005;73:114–125. doi: 10.1128/IAI.73.1.114-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.A., Melikian G.L. Aspergillosis in the “nonimmunocompromised” host. Immunological Investigations. 2011;40:751–766. doi: 10.3109/08820139.2011.614307. [DOI] [PubMed] [Google Scholar]
- Sugar A.M., Liu X.P. Effect of grapefruit juice on serum voriconazole concentrations in the mouse. Medical Mycology. 2000;38:209–212. doi: 10.1080/mmy.38.3.209.212. [DOI] [PubMed] [Google Scholar]
- Sugui J.A., Chang Y.C., Kwon-Chung K.J. Agrobacterium tumefaciens-mediated transformation of Aspergillus fumigatus: an efficient tool for insertional mutagenesis and targeted gene disruption. Applied and Environmental Microbiology. 2005;71:1798–1802. doi: 10.1128/AEM.71.4.1798-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui J.A., Pardo J., Chang Y.C. Gliotoxin is a virulence factor of Aspergillus fumigatus: gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryotic Cell. 2007;6:1562–1569. doi: 10.1128/EC.00141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui J.A., Peterson S.W., Figat A. Genetic relatedness versus biological compatibility between Aspergillus fumigatus and related species. Journal of Clinical Microbiology. 2014;52:3707–3721. doi: 10.1128/JCM.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szewczyk E., Nayak T., Oakley C.E. Fusion PCR and gene targeting in Aspergillus nidulans. Nature Protocols. 2006;1:3111–3120. doi: 10.1038/nprot.2006.405. [DOI] [PubMed] [Google Scholar]
- Takazono T., Sheppard D.C. Aspergillus in chronic lung disease: Modeling what goes on in the airways. Medical Mycology. 2017;55:39–47. doi: 10.1093/mmy/myw117. [DOI] [PubMed] [Google Scholar]
- Takeda K., Suzuki J., Watanabe A. Species identification, antifungal susceptibility, and clinical feature association of Aspergillus section Nigri isolates from the lower respiratory tract. Medical Mycology. 2020;58:310–314. doi: 10.1093/mmy/myz072. [DOI] [PubMed] [Google Scholar]
- Talbot J.J., Barrs V.R. One-health pathogens in the Aspergillus viridinutans complex. Medical Mycology. 2018;56:1–12. doi: 10.1093/mmy/myx016. [DOI] [PubMed] [Google Scholar]
- Talbot J.J., Subedi S., Halliday C.L. Surveillance for azole resistance in clinical and environmental isolates of Aspergillus fumigatus in Australia and cyp51A homology modelling of azole-resistant isolates. Journal of Antimicrobial Chemotherapy. 2018;73:2347–2351. doi: 10.1093/jac/dky187. [DOI] [PubMed] [Google Scholar]
- Tangwattanachuleeporn M., Minarin N., Saichan S. Prevalence of azole-resistant Aspergillus fumigatus in the environment of Thailand. Medical Mycology. 2017;55:429–435. doi: 10.1093/mmy/myw090. [DOI] [PubMed] [Google Scholar]
- Tanney J.B., Visagie C.M., Yilmaz N. Aspergillus subgenus Polypaecilum from the built environment. Studies in Mycology. 2017;88:237–267. doi: 10.1016/j.simyco.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapaninen T., Olkkola A.M., Tornio A. Itraconazole increases ibrutinib exposure 10-fold and reduces interindividual variation – A potentially beneficial drug-drug interaction. Clinical and Translational Science. 2020;13:345–351. doi: 10.1111/cts.12716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro M., Izumikawa K., Hirano K. Correlation between triazole treatment history and susceptibility in clinically isolated Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 2012;56:4870–4875. doi: 10.1128/AAC.00514-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli M., Rivero-Menendez O., Abastabar M. Genetic diversity and antifungal susceptibility patterns of Aspergillus nidulans complex obtained from clinical and environmental sources. Mycoses. 2020;63:78–88. doi: 10.1111/myc.13019. [DOI] [PubMed] [Google Scholar]
- Thangavel R.R., Bouvier N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. Journal of Immunological Methods. 2014;410:60–79. doi: 10.1016/j.jim.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom C., Church M. Williams and Wilkins; Baltimore, Maryland: 1926. The Aspergilli. [Google Scholar]
- Thom C., Raper K.B. Williams and Wilkins; Baltimore, Maryland: 1945. Manual of the Aspergilli. [Google Scholar]
- Tortorano A.M., Esposto M.C., Prigitano A. Cross-reactivity of Fusarium spp. in the Aspergillus galactomannan enzyme-linked immunosorbent assay. Journal of Clinical Microbiology. 2012;50:1051–1053. doi: 10.1128/JCM.05946-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyotome T., Hagiwara D., Kida H. First clinical isolation report of azole-resistant Aspergillus fumigatus with TR34/L98H-type mutation in Japan. Journal of Infection and Chemotherapy. 2017;23:579–581. doi: 10.1016/j.jiac.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Traboulsi R.S., Kattar M.M., Dbouni O. Fatal brain infection caused by Aspergillus glaucus in an immunocompetent patient identified by sequencing of the ribosomal 18S–28S internal transcribed spacer. European Journal of Clinical Microbiology & Infectious Diseases. 2007;26:747–750. doi: 10.1007/s10096-007-0361-x. [DOI] [PubMed] [Google Scholar]
- Trama J.P., Mordechai E., Adelson M.E. Detection of Aspergillus fumigatus and a mutation that confers reduced susceptibility to itraconazole and posaconazole by real-time PCR and pyrosequencing. Journal of Clinical Microbiology. 2005;43:906–908. doi: 10.1128/JCM.43.2.906-908.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troke P.F., Hockey H.P., Hope W.W. Observational study of the clinical efficacy of voriconazole and its relationship to plasma concentrations in patients. Antimicrobial Agents and Chemotherapy. 2011;55:4782–4788. doi: 10.1128/AAC.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trovato L., Scalia G., Domina M. Environmental isolates of multi-azole-resistant Aspergillus spp. in Southern Italy. Journal of Fungi. 2018;4:131. doi: 10.3390/jof4040131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsopoulou A., Posso R., Vale L. Determination of the prevalence of triazole resistance in environmental Aspergillus fumigatus strains isolated in South Wales, UK. Frontiers in Microbiology. 2018;9:1395. doi: 10.3389/fmicb.2018.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchido Y., Tanaka M., Nakano S. Prospective multicenter surveillance of clinically isolated Aspergillus species revealed azole-resistant Aspergillus fumigatus isolates with TR34/L98H mutation in the Kyoto and Shiga regions of Japan. Medical Mycology. 2019;57:997–1003. doi: 10.1093/mmy/myz003. [DOI] [PubMed] [Google Scholar]
- Ukai Y., Kuroiwa M., Kurihara N. Contributions of yap1 mutation and subsequent atrF upregulation to voriconazole resistance in Aspergillus flavus. Antimicrobial Agents and Chemotherapy. 2018;62:e01216–e01218. doi: 10.1128/AAC.01216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullmann A.J., Aguado J.M., Arikan-Akdagli S. Diagnosis and management of Aspergillus diseases: executive summary of the 2017 ESCMID-ECMM-ERS guideline. Clinical Microbiology and Infection. 2018;24:e1–e38. doi: 10.1016/j.cmi.2018.01.002. [DOI] [PubMed] [Google Scholar]
- Umeyama T., Hayashi Y., Shimosaka H. CRISPR/Cas9 genome editing to demonstrate the contribution of Cyp51A Gly138Ser to azole resistance in Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 2018;62:e00894–18. doi: 10.1128/AAC.00894-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urb M., Snarr B.D., Wojewodka G. Evolution of the immune response to chronic airway colonization with Aspergillus fumigatus hyphae. Infection and Immunity. 2015;83:3590–3600. doi: 10.1128/IAI.00359-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban C.F., Backman E. Eradicating, retaining, balancing, swarming, shuttling and dumping: a myriad of tasks for neutrophils during fungal infection. Current Opinion in Microbiology. 2020;58:106–115. doi: 10.1016/j.mib.2020.09.011. [DOI] [PubMed] [Google Scholar]
- Vale-Silva L., Ischer F., Leibundgut-Landmann S. Gain-of-function mutations in PDR1, a regulator of antifungal drug resistance in Candida glabrata, control adherence to host cells. Infection and Immunity. 2013;81:1709–1720. doi: 10.1128/IAI.00074-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsecchi I., Mellado E., Beau R. Fitness studies of azole-resistant strains of Aspergillus fumigatus. Antimicrobial Agents and Chemotherapy. 2015;59:7866–7869. doi: 10.1128/AAC.01594-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arkel A.L.E., Rijpstra T.A., Belderbos H.N.A. COVID-19-associated pulmonary aspergillosis. American Journal of Respiratory and Critical Care Medicine. 2020;202:132–135. doi: 10.1164/rccm.202004-1038LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Veerdonk F.L., Gresnigt M.S., Romani L. Aspergillus fumigatus morphology and dynamic host interactions. Nature Reviews Microbiology. 2017;15:661–674. doi: 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- van der Elst K.C., Span L.F., van Hateren K. Dried blood spot analysis suitable for therapeutic drug monitoring of voriconazole, fluconazole, and posaconazole. Antimicrobial Agents and Chemotherapy. 2013;57:4999–5004. doi: 10.1128/AAC.00707-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden J.W., Arendrup M.C., Melchers W.J. Azole resistance of Aspergillus fumigatus in immunocompromised patients with invasive aspergillosis. Emerging Infectious Diseases. 2016;22:158–159. doi: 10.3201/eid2201.151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden J.W., Arendrup M.C., Warris A. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerging Infectious Diseases. 2015;21:1041–1044. doi: 10.3201/eid2106.140717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden J.W., Camps S.M., Kampinga G.A. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clinical Infectious Diseases. 2013;57:513–520. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- van der Linden J.W., Snelders E., Arends J.P. Rapid diagnosis of azole-resistant aspergillosis by direct PCR using tissue specimens. Journal of Clinical Microbiology. 2010;48:1478–1480. doi: 10.1128/JCM.02221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden J.W., Snelders E., Kampinga G.A. Clinical implications of azole resistance in Aspergillus fumigatus, The Netherlands, 2007–2009. Emerging Infectious Diseases. 2011;17:1846–1854. doi: 10.3201/eid1710.110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Torre M.H., Novak-Frazer L., Rautemaa-Richardson R. Detecting azole-antifungal resistance in Aspergillus fumigatus by pyrosequencing. Journal of Fungi. 2020;6:12. doi: 10.3390/jof6010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iersel M.L.P.S., Rossenu S., de Greef R. A population pharmacokinetic model for a solid oral tablet formulation of posaconazole. Antimicrobial Agents and Chemotherapy. 2018;62 doi: 10.1128/AAC.02465-17. e02465–02417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Paassen J., Russcher A., In’t Veld-van Wingerden A.W., Verweij P., Kuijper E.J. Emerging aspergillosis by azole-resistant Aspergillus fumigatus at an intensive care unit in the Netherlands, 2010 to 2013. Euro Surveillance. 2016;21 doi: 10.2807/1560-7917.ES.2016.21.30.30300. [DOI] [PubMed] [Google Scholar]
- Varga J., Due M., Frisvad J.C. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Studies in Mycology. 2007;59:89–106. doi: 10.3114/sim.2007.59.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J.C., Kocsubé S. New and revisited species in Aspergillus section Nigri. Studies in Mycology. 2011;69:1–17. doi: 10.3114/sim.2011.69.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Frisvad J.C., Samson R.A. Aspergillus sect. Aeni sect. nov., a new section of the genus for A. karnatakaensis sp. nov. and some allied fungi. IMA Fungus. 2010;1:197–205. doi: 10.5598/imafungus.2010.01.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Houbraken J., van der Lee H.A. Aspergillus calidoustus sp. nov., causative agent of human infections previously assigned to Aspergillus ustus. Eukaryotic Cell. 2008;7:630–638. doi: 10.1128/EC.00425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanshenassan M., Boekhout T., Lass-Flörl C. Proof of concept for MBT ASTRA, a rapid matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS)-based method to detect caspofungin resistance in Candida albicans and Candida glabrata. Journal of Clinical Microbiology. 2018;56:e00420–18. doi: 10.1128/JCM.00420-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanshenassan M., Boekhout T., Meis J.F. Candida auris identification and rapid antifungal susceptibility testing against echinocandins by MALDI-TOF MS. Frontiers in Cellular and Infection Microbiology. 2019;9:20. doi: 10.3389/fcimb.2019.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez J.A., Manavathu E.K. Molecular characterization of a voriconazole-resistant, posaconazole-susceptible Aspergillus fumigatus isolate in a lung transplant recipient in the United States. Antimicrobial Agents and Chemotherapy. 2016;60:1129–1133. doi: 10.1128/AAC.01130-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergidis P., Moore C.B., Novak-Frazer L. High-volume culture and quantitative real-time PCR for the detection of Aspergillus in sputum. Clinical Microbiology and Infection. 2020;26:935–940. doi: 10.1016/j.cmi.2019.11.019. [DOI] [PubMed] [Google Scholar]
- Vermeulen E., Maertens J., De Bel A. Nationwide surveillance of azole resistance in Aspergillus diseases. Antimicrobial Agents and Chemotherapy. 2015;59:4569–4576. doi: 10.1128/AAC.00233-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen E., Maertens J., Schoemans H., Lagrou K. Azole-resistant Aspergillus fumigatus due to TR46/Y121F/T289A mutation emerging in Belgium, July 2012. Euro Surveillance. 2012;17:20326. [PubMed] [Google Scholar]
- Verweij P.E., Ananda-Rajah M., Andes D. International expert opinion on the management of infection caused by azole-resistant Aspergillus fumigatus. Drug Resistance Updates. 2015;21–22:30–40. doi: 10.1016/j.drup.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Verweij P.E., Chowdhary A., Melchers W.J. Azole resistance in Aspergillus fumigatus: Can we retain the clinical use of mold–active antifungal azoles? Clinical Infectious Diseases. 2016;62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij P.E., Gangneux J.P., Bassetti M. Diagnosing COVID-19-associated pulmonary aspergillosis. Lancet Microbe. 2020;1:e53–e55. doi: 10.1016/S2666-5247(20)30027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij P.E., Mellado E., Melchers W.J. Multiple-triazole-resistant aspergillosis. New England Journal of Medicine. 2007;356:1481–1483. doi: 10.1056/NEJMc061720. [DOI] [PubMed] [Google Scholar]
- Verweij P.E., Mensink M., Rijs A.J. In-vitro activities of amphotericin B, itraconazole and voriconazole against 150 clinical and environmental Aspergillus fumigatus isolates. Journal of Antimicrobial Chemotherapy. 1998;42:389–392. doi: 10.1093/jac/42.3.389. [DOI] [PubMed] [Google Scholar]
- Verweij P.E., Snelders E., Kema G.H. Azole resistance in Aspergillus fumigatus: a side-effect of environmental fungicide use? Lancet Infectious Diseases. 2009;9:789–795. doi: 10.1016/S1473-3099(09)70265-8. [DOI] [PubMed] [Google Scholar]
- Verweij P.E., Te Dorsthorst D.T., Rijs A.J. Nationwide survey of in vitro activities of itraconazole and voriconazole against clinical Aspergillus fumigatus isolates cultured between 1945 and 1998. Journal of Clinical Microbiology. 2002;40:2648–2650. doi: 10.1128/JCM.40.7.2648-2650.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij P.E., Zhang J., Debets A.J.M. In-host adaptation and acquired triazole resistance in Aspergillus fumigatus: a dilemma for clinical management. Lancet Infectious Diseases. 2016;16:e251–e260. doi: 10.1016/S1473-3099(16)30138-4. [DOI] [PubMed] [Google Scholar]
- Vesth T.C., Nybo J.L., Theobald S. Investigation of inter- and intra-species variation through genome sequencing of Aspergillus section Nigri. Nature Genetics. 2018;50:1688–1695. doi: 10.1038/s41588-018-0246-1. [DOI] [PubMed] [Google Scholar]
- Vidal-Acuña M.R., Ruiz-Pérez de Pipaón M., Torres-Sánchez M.J. Identification of clinical isolates of Aspergillus, including cryptic species, by matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI–TOF MS) Medical Mycology. 2018;56:838–846. doi: 10.1093/mmy/myx115. [DOI] [PubMed] [Google Scholar]
- Visagie C.M., Varga J., Houbraken J. Ochratoxin production and taxonomy of the yellow aspergilli (Aspergillus section Circumdati) Studies in Mycology. 2014;78:1–61. doi: 10.1016/j.simyco.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viscoli C. Combination therapy for invasive aspergillosis. Clinical Infectious Diseases. 2004;39:803–805. doi: 10.1086/423389. [DOI] [PubMed] [Google Scholar]
- Vlahos R., Bozinovski S. Recent advances in pre-clinical mouse models of COPD. Clinical Science. 2014;126:253–265. doi: 10.1042/CS20130182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Eiff M., Roos N., Schulten R. Pulmonary aspergillosis: early diagnosis improves survival. Respiration. 1995;62:341–347. doi: 10.1159/000196477. [DOI] [PubMed] [Google Scholar]
- Wagener J., Loiko V. Recent insights into the paradoxical efffect of echinocandins. Journal of Fungi. 2017;4:5. doi: 10.3390/jof4010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahidi M.M., Lamb C., Murgu S. American Association for Bronchology and Interventional Pulmonology (AABIP) statement on the use of bronchoscopy and respiratory specimen collection in patients with suspected or confirmed COVID-19 infection. Journal of Bronchology & Interventional Pulmonology. 2020;27:e52–e54. doi: 10.1097/LBR.0000000000000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker L.A., Lee K.K., Munro C.A. Caspofungin treatment of Aspergillus fumigatus results in ChsG–dependent upregulation of chitin synthesis and the formation of chitin-rich microcolonies. Antimicrobial Agents and Chemotherapy. 2015;59:5932–5941. doi: 10.1128/AAC.00862-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Hu B., Hu C. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.Y., Gricourt M., Arne P. Mutations in the Cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus isolated from avian farms in France and China. Poultry Science. 2014;93:12–15. doi: 10.3382/ps.2013-03541. [DOI] [PubMed] [Google Scholar]
- Wang F., Zhang C., Jiang Y. Innate and adaptive immune response to chronic pulmonary infection of hyphae of Aspergillus fumigatus in a new murine model. Journal of Medical Microbiology. 2017;66:1400–1408. doi: 10.1099/jmm.0.000590. [DOI] [PubMed] [Google Scholar]
- Wang H.C., Hsieh M.I., Choi P.C. Comparison of the sensititre YeastOne and CLSI M38-A2 microdilution methods in determining the activity of amphotericin B, itraconazole, voriconazole, and posaconazole against Aspergillus species. Journal of Clinical Microbiology. 2018;56:e00780–18. doi: 10.1128/JCM.00780-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.C., Huang J.C., Lin Y.H. Prevalence, mechanisms and genetic relatedness of the human pathogenic fungus Aspergillus fumigatus exhibiting resistance to medical azoles in the environment of Taiwan. Environmental Microbiology. 2018;20:270–280. doi: 10.1111/1462-2920.13988. [DOI] [PubMed] [Google Scholar]
- Wang Q., Kontoyiannis D.P., Li R. A novel broad allele-specific TaqMan real-time PCR method to detect triazole-resistant strains of Aspergillus fumigatus, even with a very low percentage of triazole-resistant cells mixed with triazole-susceptible cells. Journal of Clinical Microbiology. 2019;57:e00604–e00619. doi: 10.1128/JCM.00604-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrilow A.G., Melo N., Martel C.M. Expression, purification, and characterization of Aspergillus fumigatus sterol 14-alpha demethylase (CYP51) isoenzymes A and B. Antimicrobial Agents and Chemotherapy. 2010;54:4225–4234. doi: 10.1128/AAC.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warris A., Klaassen C.H., Meis J.F. Molecular epidemiology of Aspergillus fumigatus isolates recovered from water, air, and patients shows two clusters of genetically distinct strains. Journal of Clinical Microbiology. 2003;41:4101–4106. doi: 10.1128/JCM.41.9.4101-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Valiante V., Nødvig C.S. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synthetic Biology. 2017;6:62–68. doi: 10.1021/acssynbio.6b00203. [DOI] [PubMed] [Google Scholar]
- Wei X., Chen P., Gao R. Screening and characterization of a non-cyp51A mutation in an Aspergillus fumigatus cox10 strain conferring azole resistance. Antimicrobial Agents and Chemotherapy. 2017;61:e02101–e02116. doi: 10.1128/AAC.02101-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner G., d’Enfert C., Koch A. Development of a homologous transformation system for the human pathogenic fungus Aspergillus fumigatus based on the pyrG gene encoding orotidine 5'-monophosphate decarboxylase. Current Genetics. 1998;33:378–385. doi: 10.1007/s002940050350. [DOI] [PubMed] [Google Scholar]
- Wheat L.J., Hackett E., Durkin M. Histoplasmosis-associated cross-reactivity in the BioRad Platelia Aspergillus enzyme immunoassay. Clinical and Vaccine Immunology. 2007;14:638–640. doi: 10.1128/CVI.00479-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.L., Bretagne S., Klingspor L. Aspergillus PCR: one step closer to standardization. Journal of Clinical Microbiology. 2010;48:1231–1240. doi: 10.1128/JCM.01767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.L., Dhillon R., Cordey A. A national strategy to diagnose COVID-19 associated invasive fungal disease in the ICU. Clinical Infectious Diseases. 2021 doi: 10.1093/cid/ciaa1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.L., Parr C., Barnes R.A. Predicting invasive aspergillosis in hematology patients by combining clinical and genetic risk factors with early diagnostic biomarkers. Journal of Clinical Microbiology. 2018;56 doi: 10.1128/JCM.01122-17. e01122–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.L., Posso R.B., Barnes R.A. Analytical and clinical evaluation of the PathoNostics AsperGenius Assay for detection of invasive aspergillosis and resistance to azole antifungal drugs during testing of serum samples. Journal of Clinical Microbiology. 2015;53:2115–2121. doi: 10.1128/JCM.00667-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.L., Posso R.B., Barnes R.A. Analytical and clinical evaluation of the PathoNostics AsperGenius Assay for detection of invasive aspergillosis and resistance to azole antifungal drugs directly from plasma samples. Journal of Clinical Microbiology. 2017;55:2356–2366. doi: 10.1128/JCM.00411-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.L., Wingard J.R., Bretagne S. Aspergillus polymerase chain reaction: Systematic review of evidence for clinical use in comparison with antigen testing. Clinical Infectious Diseases. 2015;61:1293–1303. doi: 10.1093/cid/civ507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . World Health Organization; 2020. Coronavirus disease (COVID-19) outbreak. [Google Scholar]
- Wiederhold N., Gil V., Lindner J. Evaluation of Cyp51A mechanisms of azole resistance in Aspergillus fumigatus isolates from the United States. In 7th Trends in Medical Mycology, 9–12 October 2015, Lisbon, Portugal (poster no. P009) Mycoses. 2015;58(suppl. 4):55. [Google Scholar]
- Wiederhold N.P., Gil V.G., Gutierrez F. First detection of TR34L98H and TR46Y121FT289A Cyp51 mutations in Aspergillus fumigatus isolates in the United States. Journal of Clinical Microbiology. 2016;54(suppl. 1):168–171. doi: 10.1128/JCM.02478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold N.P., Locke J.B., Daruwala P. Rezafungin (CD101) demonstrates potent in vitro activity against Aspergillus, including azole-resistant Aspergillus fumigatus isolates and cryptic species. Journal of Antimicrobial Chemotherapy. 2018;73:3063–3067. doi: 10.1093/jac/dky280. [DOI] [PubMed] [Google Scholar]
- Wiedman G.R., Zhao Y., Mustaev A. An aptamer-based biosensor for the azole class of antifungal drugs. mSphere. 2017;2 doi: 10.1128/mSphere.00274-17. e00274–00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willger S.D., Puttikamonkul S., Kim K.H. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathogens. 2008;4 doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn R.M., Gil-Lamaignere C., Roilides E. Selective effects of interleukin (IL)-15 on antifungal activity and IL-8 release by polymorphonuclear leukocytes in response to hyphae of Aspergillus species. Journal of Infectious Diseases. 2003;188:585–590. doi: 10.1086/377099. [DOI] [PubMed] [Google Scholar]
- Wojtowicz A., Lecompte T.D., Bibert S. PTX3 polymorphisms and invasive mold infections after solid organ transplant. Clinical Infectious Diseases. 2015;61:619–622. doi: 10.1093/cid/civ386. [DOI] [PubMed] [Google Scholar]
- Won E.J., Joo M.Y., Lee D. Antifungal susceptibility tests and the cyp51 mutant strains among clinical Aspergillus fumigatus isolates from Korean multicenters. Mycobiology. 2020;48:148–152. doi: 10.1080/12298093.2020.1744955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S.S.W., Rasid O., Laskaris P. Treatment of cyclosporin A retains host defense against invasive pulmonary aspergillosis in a non-immunosuppressive murine model by preserving the myeloid cell population. Virulence. 2017;8:1744–1752. doi: 10.1080/21505594.2017.1339007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Liu W.L., Lai C.C. Multicenter study of azole-resistant Aspergillus fumigatus clinical isolates, Taiwan. Emerging Infectious Diseases. 2020;26:804–806. doi: 10.3201/eid2604.190840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.J., Wang H.C., Lee J.C. Azole-resistant Aspergillus fumigatus isolates carrying TR34/L98H mutations in Taiwan. Mycoses. 2015;58:544–549. doi: 10.1111/myc.12354. [DOI] [PubMed] [Google Scholar]
- Wu Z., McGoogan J.M. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: Summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- Xu Z., Li S., Tian S. Full spectrum of COVID-19 severity still being depicted. Lancet. 2020;395:947–948. doi: 10.1016/S0140-6736(20)30308-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T., Nguyen C.K., Romans A. Isogenic auxotrophic mutant strains in the Aspergillus fumigatus genome reference strain AF293. Archives of Microbiology. 2004;182:346–353. doi: 10.1007/s00203-004-0707-z. [DOI] [PubMed] [Google Scholar]
- Yagi K., Ushikubo M., Maeshima A. Invasive pulmonary aspergillosis due to Aspergillus lentulus in an adult patient: A case report and literature review. Journal of Infection and Chemotherapy. 2019;25:547–551. doi: 10.1016/j.jiac.2019.02.003. [DOI] [PubMed] [Google Scholar]
- Yan X., Li M., Jiang M. Clinical characteristics of 45 patients with invasive pulmonary aspergillosis: retrospective analysis of 1711 lung cancer cases. Cancer. 2009;115:5018–5025. doi: 10.1002/cncr.24559. [DOI] [PubMed] [Google Scholar]
- Yang X., Yu Y., Xu J. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respiratory Medicine. 2020;8:475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmin S., Alcazar-Fuoli L., Gründlinger M. Mevalonate governs interdependency of ergosterol and siderophore biosyntheses in the fungal pathogen Aspergillus fumigatus. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E497–E504. doi: 10.1073/pnas.1106399108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W.M., Schoeppler K.E., Jaeger J. Voriconazole and posaconazole therapeutic drug monitoring: a retrospective study. Annals of Clinical Microbiology and Antimicrobials. 2017;16:60. doi: 10.1186/s12941-017-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L.S., Rodriguez-Manzano J., Malpartida-Cardenas K. Rapid and sensitive detection of azole-resistant Aspergillus fumigatus by tandem repeat loop-mediated isothermal amplification. Journal of Molecular Diagnostics. 2019;21:286–295. doi: 10.1016/j.jmoldx.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai P., Song J., Gao L. A sphingolipid synthesis-related protein OrmA in Aspergillus fumigatus is responsible for azole susceptibility and virulence. Cellular Microbiology. 2019;21 doi: 10.1111/cmi.13092. [DOI] [PubMed] [Google Scholar]
- Zhang C., Meng X., Wei X. Highly efficient CRISPR mutagenesis by microhomology-mediated end joining in Aspergillus fumigatus. Fungal Genetics and Biology. 2016;86:47–57. doi: 10.1016/j.fgb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- Zhang G., Hu C., Luo L. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. Journal of Clinical Virology. 2020;127:104364. doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., van den Heuvel J., Debets A.J.M. Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proceedings Biological Sciences. 2017;284:20170635. doi: 10.1098/rspb.2017.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Lepak A.J., Marchillo K. APX001 pharmacokinetic/pharmacodynamic target determination against Aspergillus fumigatus in an in vivo model of invasive pulmonary aspergillosis. Antimicrobial Agents and Chemotherapy. 2019;63:e02372–18. doi: 10.1128/AAC.02372-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Garnaud C., Brenier-Pinchart M.P. Direct molecular diagnosis of aspergillosis and CYP51A profiling from respiratory samples of French patients. Frontiers in Microbiology. 2016;7:1164. doi: 10.3389/fmicb.2016.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Stensvold C.R., Perlin D.S. Azole resistance in Aspergillus fumigatus from bronchoalveolar lavage fluid samples of patients with chronic diseases. Journal of Antimicrobial Chemotherapy. 2013;68:1497–1504. doi: 10.1093/jac/dkt071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.Y., Zhang M., Yang C.X. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cellular & Molecular Immunology. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Li H., Zhang Y. Diagnostic value of galactomannan antigen test in serum and bronchoalveolar lavage fluid samples from patients with nonneutropenic invasive pulmonary aspergillosis. Journal of Clinical Microbiology. 2017;55:2153–2161. doi: 10.1128/JCM.00345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziakas P.D., Kourbeti I.S., Mylonakis E. Systemic antifungal prophylaxis after hematopoietic stem cell transplantation: a meta-analysis. Clinical Therapeutics. 2014;36:292–306.e1. doi: 10.1016/j.clinthera.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Zimmerli S., Knecht U., Leib S.L. A model of cerebral aspergillosis in non-immunosuppressed nursing rats. Acta Neuropathologica. 2007;114:411–418. doi: 10.1007/s00401-007-0255-0. [DOI] [PubMed] [Google Scholar]
- Zoran T., Sartori B., Sappl L. Azole-resistance in Aspergillus terreus and related species: An emerging problem or a rare phenomenon? Frontiers in Microbiology. 2018;9:516. doi: 10.3389/fmicb.2018.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotti M., Agnoletti A.F., Vizzini A. Onychomycosis from Aspergillus melleus, a novel pathogen for humans. Fungal identification and in vitro drug susceptibility. Experimental Dermatology. 2015;24:966–968. doi: 10.1111/exd.12807. [DOI] [PubMed] [Google Scholar]
- Zotti M., Machetti M., Perotti M. A new species, Aspergillus persii, as an agent of onychomycosis. Medical Mycology. 2010;48:656–660. doi: 10.3109/13693780903420641. [DOI] [PubMed] [Google Scholar]