Abstract
Bacterial blight (BB) is a globally devastating rice disease caused by Xanthomonas oryzae pv. oryzae (Xoo). The use of disease resistance (R) genes in rice breeding is an effective and economical strategy for the control of this disease. Nevertheless, a majority of R genes lack durable resistance for long-term use under global warming conditions. Here, we report the isolation of a novel executor R gene, Xa7, that confers extremely durable, broad-spectrum, and heat-tolerant resistance to Xoo. The expression of Xa7 was induced by incompatible Xoo strains that secreted the transcription activator-like effector (TALE) AvrXa7 or PthXo3, which recognized effector binding elements (EBEs) in the Xa7 promoter. Furthermore, Xa7 induction was faster and stronger under high temperatures. Overexpression of Xa7 or co-transformation of Xa7 with avrXa7 triggered a hypersensitive response in plants. Constitutive expression of Xa7 activated a defense response in the absence of Xoo but inhibited the growth of transgenic rice plants. In addition, analysis of over 3000 rice varieties showed that the Xa7 locus was found primarily in the indica and aus subgroups. A variation consisting of an 11-bp insertion and a base substitution (G to T) was found in EBEAvrXa7 in the tested varieties, resulting in a loss of Xa7 BB resistance. Through a decade of effort, we have identified an important BB resistance gene and characterized its distinctive interaction with Xoo strains; these findings will greatly facilitate research on the molecular mechanism of Xa7-mediated resistance and promote the use of this valuable gene in breeding.
Keywords: Xa7, executor R gene, durable resistance, TALE, bacterial blight, rice
This study isolated Xa7, a novel executor R gene that confers durable and broad-spectrum resistance to bacterial blight disease, by map-based cloning from rice. Xa7 expression can be induced by incompatible Xoo strains whose secreted TALEs recognize EBEs in the Xa7 promoter. Variation in the EBE sequence of Xa7 destroys its associated disease resistance.
Introduction
Plants are frequently attacked by pathogenic microbes in the ecosystem (Chisholm et al., 2006; Boller and He, 2009). In response to pathogen invasion, plants have evolved mechanisms that rapidly recognize pathogen-associated molecular patterns (PAMPs) via receptors on the cell surface and activate PAMP-triggered immunity (PTI) to defend against pathogen attack (Jones and Dangl, 2006; Tang et al., 2017). In turn, pathogens have successfully deployed a mechanism to secrete so-called effectors, which can interfere with PTI in host cells, resulting in effector-triggered plant susceptibility (Jones and Dangl, 2006; Antony et al., 2010). Evidence from crop research has shown that pathogen effectors can target the so-called susceptibility (S) genes in host cells, and genetic variability in S genes may cause a recessive resistance to pathogens (Liu et al., 2009; White and Yang, 2009; Antony et al., 2010). Interestingly, plants have also co-evolved dominant resistance (R) genes that can specifically recognize pathogen effectors to activate effector-triggered immunity (Jones and Dangl, 2006; Zhang et al., 2020a, 2020b). Therefore, plants undergo endless attacks by pathogens and in turn evolve novel R genes for survival. More than 300 R genes have been cloned from various plants, but the molecular mechanisms underlying their functions are understood for only a small fraction of R genes (Kourelis and van der Hoorn, 2018).
Rice is an economically important staple food crop for an enormous number of people worldwide (Ainsworth, 2008). Rice production is always affected by various diseases in the field, among the most devastating of which is bacterial blight (BB), which is caused by Xanthomonas oryzae pv. oryzae (Xoo; Mew, 1987). BB can affect rice growth, development, or reproduction, and it causes severe yield losses of up to 50% (Liu et al., 2014). Breeding rice varieties with genetic resistance is an effective, economical, and environmentally friendly strategy for the control of BB disease in rice production (1; Jiang et al., 2020). To date, at least 46 genes that confer dominant or recessive host resistance to Xoo have been identified (Chen et al., 2020). Among them, 15 genes (or alleles) have been cloned, including Xa1 (Yoshimura et al., 1998) and its alleles Xa2, Xa14, Xa31(t), and Xa45(t) (Ji et al., 2020; Zhang et al., 2020a, 2020b); Xa3/Xa26 (Sun et al., 2004; Xiang et al., 2006); Xa4 (Hu et al., 2017); xa5 (Iyer and McCouch, 2004; Jiang et al., 2006); Xa10 (Tian et al., 2014); xa13 (Yang et al., 2006); Xa21(Song et al., 1995); Xa23 (Wang et al., 2015); xa25 (Liu et al., 2011); Xa27 (Gu et al., 2005); and xa41(t) (Hutin et al., 2016). Based on the functions of their encoded proteins, these R genes or S genes (dominant alleles of the recessive resistance genes) can be classified into four categories: nucleotide-binding leucine-rich repeat receptor genes (Xa21, Xa3/Xa26, and Xa4), sugars will eventually be exported transporter (SWEET) genes (Xa13/OsSWEET11, Xa25/OsSWEET13, and Xa41/OsSWEET14), executor R genes (Xa10, Xa23, and Xa27), and others (Xa1 and Xa5). Interestingly, eight of the above genes (Xa1, Xa5, Xa10, Xa13, Xa23, Xa25, Xa27, and Xa41) have been shown to interact with a type of pathogen effector called transcription activator-like effectors (TALEs) during the defense response of host cells (Jiang et al., 2020).
TALEs, the virulent or avirulent proteins secreted by Xoo, are translocated into host cells by the bacterial Type III secretion system (Bogdanove et al., 2010). TALE protein sequences are highly conserved, with the exception of the repetitive central region that consists of 33- to 35-amino-acid repeats. In the host cell, TALEs are located in the nucleus where they bind to specific DNA sequences called effector binding elements (EBEs) in order to regulate the expression of host genes. The recognition between TALE and EBE is determined by the repeat variable di-residues (RVDs) at the 12th and 13th positions of each repeat in the central TALE region (Boch et al., 2009; (1)Moscou and Bogdanove, 2009). Binding of pathogen TALEs to EBEs transcriptionally activates target genes in the host cell, resulting in susceptibility or resistance of the host plant (Bogdanove et al., 2010; Bogdanove and Voytas 2011). For instance, Xoo TALEs can directly target corresponding EBEs in the promoters of the OsSWEET genes Xa13/OsSWEET11 (Antony et al., 2010), Xa25/OsSWEET13 (Zhou et al., 2015), and Xa41/OsSWEET14 (Hutin et al., 2016), upregulating their expression in rice and causing BB disease. The SWEET proteins are cellular sugar transporters, and induction of OsSWEET genes may provide sufficient sugars for the nutrition of the pathogen (Chen, 2014; Bezrutczyk et al., 2017). Therefore, natural variations or CRISPR-Cas9-mediated genome editing in EBE regions of the OsSWEET genes abolished cognate TALE binding and produced recessive resistance to Xoo (Yang et al., 2006; Liu et al., 2011; Hutin et al., 2016; Oliva et al., 2019; Xu et al., 2019). Alternatively, the expression of the rice executor R genes Xa10, Xa23, and Xa27 can be transcriptionally activated by the TALEs AvrXa10, AvrXa23, and AvrXa27, respectively, triggering a hypersensitive response (HR) to restrict Xoo growth in rice, resulting in dominant resistance to BB disease (Gu et al., 2005; Tian et al., 2014; Wang et al., 2015). However, all the known executor R genes encode small proteins that lack conserved domains or display little homology with one another (Wang et al., 2014). The biological functions and molecular mechanisms of the executor R genes are still unclear.
Xa7 is a dominant R gene that provides broad-spectrum and extremely durable resistance to Xoo (Vera Cruz et al., 2000; (1)White et al., 2009; Zhang et al., 2015). Over the past decades, Xa7 has been the subject of ongoing research since its original identification from the Bangladeshi rice cultivar DV85 in the 1970s (Sidhu et al., 1978). Field tests over continuous years confirmed that the BB resistance of Xa7 was more durable than that of Xa4 and Xa10 (Vera Cruz et al., 2000). Furthermore, evidence from different studies has shown that Xa7 is more effective against BB at high temperatures, a characteristic that differentiates it from most other R genes (Webb et al., 2010; Cohen et al., 2017; Dossa et al., 2020). Likewise, the TALE AvrXa7, encoded by the avirulent gene avrXa7, triggers the resistance of Xa7 in rice and is found in various Xoo strains (Hopkins et al., 1992; Yang et al., 2000). AvrXa7 has also been shown to be an important virulence factor for Xoo because isolated strains that lost the ability to induce Xa7-associated resistance were weakly virulent, presumably due to mutations in AvrXa7 (Vera-Cruzetal., 2000). In addition, AvrXa7 can induce the expression of Xa41/OsSWEET14 by binding to a specific EBE in its promoter (Antony et al., 2010; (1)). Because the BB resistance conferred by Xa7 is particularly persistent and tolerant of high temperature, and because its cognate TALE, AvrXa7, plays double roles as both an avirulent and virulent factor in Xoo strains, the cloning and functional identification of Xa7 may provide insight into a novel molecular mechanism of plant–pathogen interaction.
In 1995, Xa7 was initially located at 107.5 cM on the current Rice Genome Research Project map (Kaji and Ogawa, 1995). It was later finely mapped to a 2.7-cM region by Porter et al. (2003) using the BB-resistant rice variety IRBB7, and it was ultimately mapped to a 118.5-kb region on chromosome 6 by Chen et al. (2008) based on the Nipponbare reference genome. However, the molecular nature of Xa7 remains to be revealed. We began work on Xa7 in 2005 and finely mapped it to a 200-kb physical region in 2009 using the Chinese BB-resistant rice variety Zhen-hui 084, which was bred from DV85 (Zhang et al., 2009). After another 11 years of effort, we now report the molecular cloning and characterization of the mysterious Xa7 gene. Our results show that Xa7 is a new executor R gene that encodes a novel protein significantly different from the known executor R proteins XA10, XA27, and XA23, and it has no significant homolog in any other plants. We identified a putative EBE targeted by AvrXa7 in the Xa7 promoter and documented Xa7 expression patterns in response to various Xoo strains and at different temperatures. These results help to explain why Xa7 confers a broad-spectrum and durable BB resistance in rice and provide a solid foundation for future investigations of its molecular mechanisms during host defense.
Results
Fine mapping of Xa7
To narrow down the Xa7 mapping region, an F2 population with 11 285 individual plants was derived from a cross between Zhen-hui 084 (which contains Xa7) and the BB-susceptible variety Cheng-hui 448. The BB-resistant or -susceptible phenotypes of the F2 plants were observed after inoculation with the Xoo strain PXO86 (which contains avrXa7) (Supplemental Figure 1). Statistical results showed that 8490 plants were highly resistant, and 2795 plants were highly susceptible, with a significant segregation ratio of 3:1 (χ2 = 0.326, P < 0.05; Supplemental Figure 1). All the susceptible F2 plants were used for genetic analysis by DNA markers, and Xa7 was placed in a 51-kb interval flanked by the markers M5 and M10 on the Nipponbare rice genome (https://rapdb.dna.affrc.go.jp; Figure 1A). To develop new polymorphism markers for fine mapping, PCR primers inside the 51-kb region were designed based on Nipponbare, and DNA fragments were successfully amplified from Cheng-hui 448 and other varieties, but not from Xa7-containing varieties such as Zhen-hui 084, IRBB7, and DV85 (data not shown). This result indicated that the sequences in the Xa7-mapping region might be specific for Xa7-containing varieties.
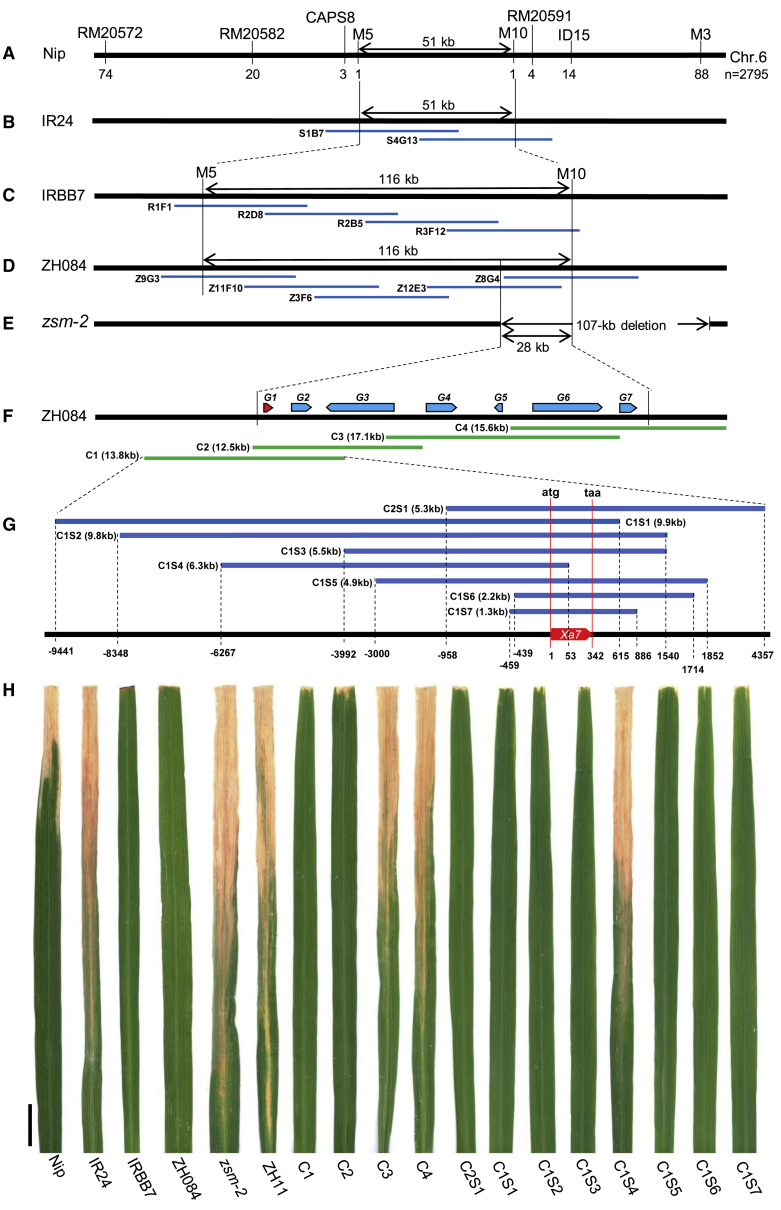
Figure 1.
Map-based cloning of Xa7.
(A) Physical map of Xa7 on rice chromosome 6 based on the reference genome of Nipponbare (Nip); n refers to the total number of BB-susceptible F2 plants used for fine mapping; the number under each marker represents the number of recombinants detected by the corresponding marker.
(B–D) Physical maps of Xa7 based on the rice varieties (B) IR24, (C) IRBB7, and (D) Zhen-hui 084 (ZH084). Maps were built using contigs of clones isolated from the genomic libraries of the corresponding varieties.
(E) The 28-kb overlapping region between the 116-kb Xa7 mapping region and the 107-kb deletion in a BB-susceptible mutant, zsm-2, mutagenized from ZH084.
(F) Predicted genes (G1 to G7) in the 28-kb overlapping region and relative positions and sizes of the four fragments (C1 to C4) used for complementation transformation in Zhonghua 11 (ZH11).
(G) Contigs and sizes of subclones (C2S1, C1S1 to C1S7) used for the transgenic complementation test in ZH11. Numbers under the line represent positions relative to the start codon (atg) of the Xa7 gene. Negative numbers refer to positions before atg, and positive numbers refer to positions starting at atg. taa refers to the stop codon of Xa7, and the sequence from atg to taa is 342 bp in length.
(H) Leaves inoculated with Xoo strain PXO86. Photographs were taken 2 weeks after inoculation. Nip, IR24, and ZH11 are BB-susceptible rice varieties. IRBB7 and ZH084 are Xa7-containing rice varieties. zsm-2 refers to a BB-susceptible mutant mutagenized from ZH084. C1 to C4, C1S1 to C1S7, and C2S1 are the transgenic complementation lines of ZH11. Bar, 2 cm.
To identify the Xa7 sequence, a Zhen-hui 084 genomic library was constructed using a fosmid strategy and screened by PCR using markers M5 and M10. The positive clones were subjected to paired-end sequencing using the fosmid vector's primers to obtain the insertion sequences, which were in turn used to develop PCR primers to screen new overlapping clones. Finally, five clones (Z9G3, Z11F10, Z3F6, Z12E3, and Z8G4) were screened out, constituting a contig that covered the whole Xa7 mapping region (Figure 1D), and the assembled sequences were obtained using the Illumina HiSeq 2500 platform. Surprisingly, the physical distance from M5 to M10 was 116 kb in Zhen-hui 084 (Supplemental Data 2), very different from the 51 kb distance in Nipponbare (Supplemental Figure 2). To confirm this result, genomic libraries of IRBB7 and its near-isogenic line IR24, a BB-susceptible variety, were constructed and screened. Among the positive clones sequenced, two (S1B7 and S4G13) from IR24 covered the mapped 51-kb region in IR24 (Figure 1B), and four (R1F1, R2D8, R2B5, and R3F12) from IRBB7 covered the 116-kb region in IRBB7 (Figure 1C). Remarkably, the 116-kb sequence of IRBB7 was identical to that of Zhen-hui 084 (Supplemental Figure 2), which was completely different from that of IR24 (Supplemental Figure 2). By contrast, the sequence between markers M5 and M10 in IR24 was almost identical to that in Nipponbare (Supplemental Figure 2).
Bioinformatic analysis of the 116-kb sequence was performed using online databases, but it was not helpful in identifying the Xa7 candidate gene. Therefore, we began to screen BB-susceptible mutants in our Zhen-hui 084 radiation-mutagenesis library in 2015. More than 20 000 M1 lines were phenotyped by inoculation with the Xoo strain PXO86, and nine highly susceptible mutant lines were identified. All mutant lines were assayed by PCR using primers located in the 116-kb region to find any possible variation. Fortunately, we found one mutant line, zsm-2 (Zhen-hui 084 susceptible mutant 2), in which the expected DNA fragments could not be amplified by several primers consecutively distributed in the 116-kb region (data not shown). A 107-kb deletion on chromosome 6 was then detected in the zsm-2 mutant by high-throughput sequencing (Supplemental Figure 3) and further verified by PCR amplification and sequencing. We compared the 107-kb fragment with the 116-kb region and found a 28-kb sequence overlap (Figure 1E) in which Xa7 may reside.
Functional complementation of Xa7 by genetic transformation
Seven putative genes were predicted in the 28-kb sequence (Figure 1F). However, none were homologous or related to known R genes. Therefore, four overlapping fragments (designated C1 to C4) that contained different putative genes were used for a transgenic complementation test (Figure 1F; Supplemental Data 2). The fragments were amplified from fosmid clone Z12E3 or Z8G4 by high-fidelity proofreading PCR and then cloned separately into the pCAMBIA1300 vector. The correct constructs were transformed into Zhong-hua 11, a japonica rice variety that is highly susceptible to the Xoo strain PXO86. Among 261 hygromycin phosphotransferase-resistant plants in the T0 generation, 15 T0 plants were highly resistant to PXO86 (Supplemental Table 1). All the resistant plants were derived from transformation with the C1 or C2 constructs, whereas all the T0 plants that contained C3 or C4 constructs were highly susceptible to PXO86 (Supplemental Table 1; Figure 1F and 1H). The T1 resistant plants from line C1 were then inoculated with 10 Xoo strains from the Philippines that represented different types of Xoo physiological races identified by the International Rice Research Institute and have been widely used to evaluate BB-resistance genes. The results showed that the resistance spectrum of transgenic line C1 was identical to that of Zhen-hui 084 (Supplemental Figure 4).
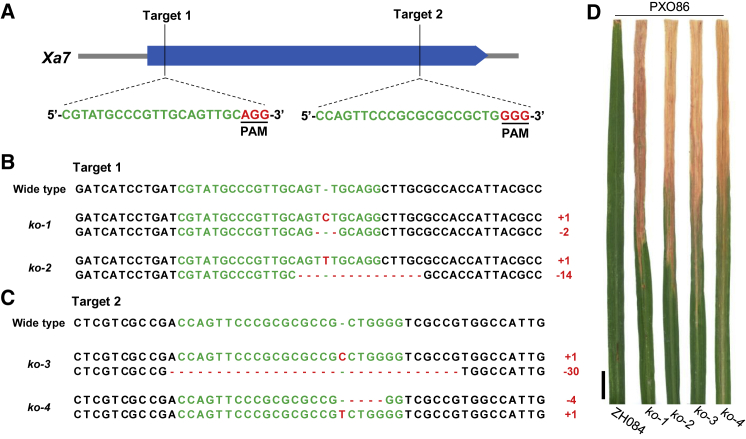
To isolate the Xa7 sequence, eight constructs containing different-sized fragments amplified from the C1 clone (Figure 1H; Supplemental Data 2) were constructed and used to transform Zhong-hua 11. In total, 68 of 465 T0 transgenic plants showed resistance to PXO86 (Supplemental Table 2). All the BB-resistant plants were derived from transformation with constructs that contained the predicted gene G1, with the exception of construct C1S4, which had partially lost the G1 sequence (Figure 1G and 1H). In addition, CRISPR-Cas9-mediated gene editing was used to knock out G1 in Zhen-hui 084. To increase the efficiency of gene editing, two sgRNAs targeting different sites in the G1 coding sequence (CDS) were designed and cloned into CRISPR-Cas9 vectors for the transformation of Zhen-hui 084 (Figure 2A). In total, 49 T0 transgenic plants were characterized by PCR-based sequencing of G1 and inoculation with PXO86. Twelve G1-knockout plants with different editing types were highly susceptible to PXO86 (Figure 2B–2D; Supplemental Tables 3 and 4). By contrast, the other 41 transgenic plants in which G1 was not edited or only one allele was edited maintained the BB resistance of wild-type Zhen-hui 084 (Supplemental Table 3 and 4). All these results indicated that G1 was the BB-resistant gene Xa7.
Figure 2.
CRISPR-Cas9-mediated knockout of Xa7.
(A) The CRISPR-Cas9 editing sites of targets 1 and 2 in Xa7. The blue box represents the exon of Xa7. The sgRNA sequences of targets 1 and 2 are shown in green, and the protospacer adjacent motif (PAM) is shown in red.
(B and C) Sequences of targets 1 (B) and 2 (C) in the CRISPR-Cas9 transgenic lines of rice variety Zhen-hui 084 (ZH084). Wild type refers to the sequence of ZH084. ko-1 to ko-4 refer to the different types of knockout mutants. The sgRNA sequence regions are shown in green, and mutations are shown in red. Numbers in red are the base-mutation changes in the single chromosome, and base deletions and insertions are indicated with minus and plus, respectively.
(D) Leaves of the ko1 to ko4 mutants of Xa7 and the wild-type control ZH084 inoculated with Xoo strain PXO86. Photographs were taken 2 weeks after inoculation. Bar, 2 cm.
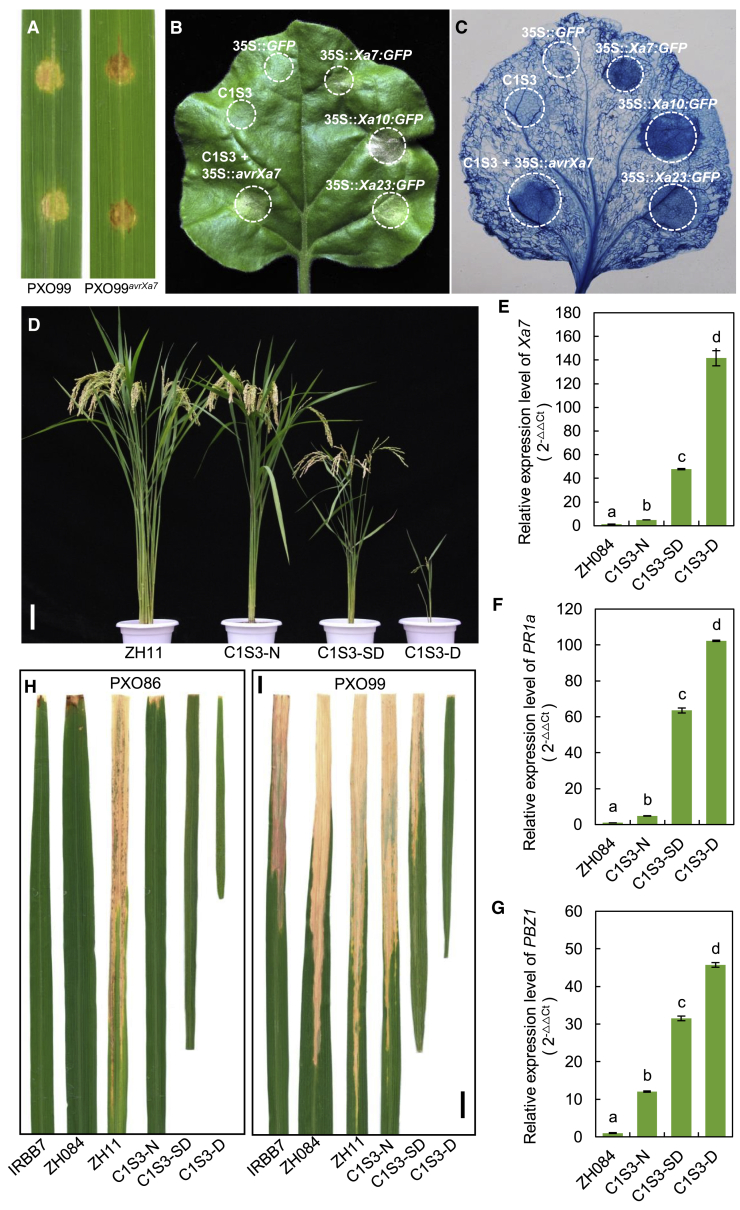
Figure 4.
Phenotypes of hypersensitive response (HR) and defense response triggered by Xa7.
(A) IRBB7 in response to inoculation with different Xoo strains. HR resistant (dark brown inoculation spots) or susceptible (pale inoculation spots and spreading) symptoms were scored 3 days after inoculation. PXO99 is an Xa7-compatible strain used as a control, and PXO99avrXa7 is the modified strain transformed with the avrXa7 gene in PXO99.
(B) HR assays in tobacco leaves. Agrobacterium tumefaciens strains harboring different constructs were injected individually or co-infiltrated with avrXa7-overexpressing A. tumefaciens into leaves using a needle-less syringe. 35S::Xa7:GFP, 35S::Xa10:GFP, and 35S::Xa23:GFP refer to constructs that overexpressed Xa7, Xa10, and Xa23 fused with eGFP, respectively, and 35S::GFP refers to the negative control. C1S3 refers to the C1S3 construct that contained Xa7 with its native promoter. The infiltrated areas are shown by dashed circles. The picture was taken at 4 days after infiltration.
(C) Cell death histochemical staining assay. The corresponding leaf from (B) was stained with trypan blue and cleared in ethanol to visualize cell death. The infiltrated areas are shown in dashed circles.
(D) Phenotypes of transgenic plants transformed with the construct C1S3. Zhong-hua 11 (ZH11) is the wild-type rice variety selected as the receptor for genetic transformation. C1S3-N, C1S3-SD, and C1S3-D refer to normal, semi-dwarf, or dwarf plants, respectively, from the C1S3 transgenic lines. Bar, 10 cm.
(E–G) Expression of Xa7(E), PR1a(F), and PBZ1(G) analyzed by qRT–PCR in the transgenic plants in the absence of Xoo. Zheng-hui 084 (ZH084) was used as the wild-type control. Data are presented as the means of three independent replicates ± SD and were statistically analyzed by one-way ANOVA. Bars with different letters are significantly different from each other (P < 0.01).
(H and I) Leaves of the transgenic plants and the wild-type control inoculated with Xoo strain PXO86 (H) and PXO99 (I). Photographs were taken 2 weeks after inoculation. Bar, 2 cm.
Xa7 is an executor R gene whose TALE-induced transcription increases at high temperature
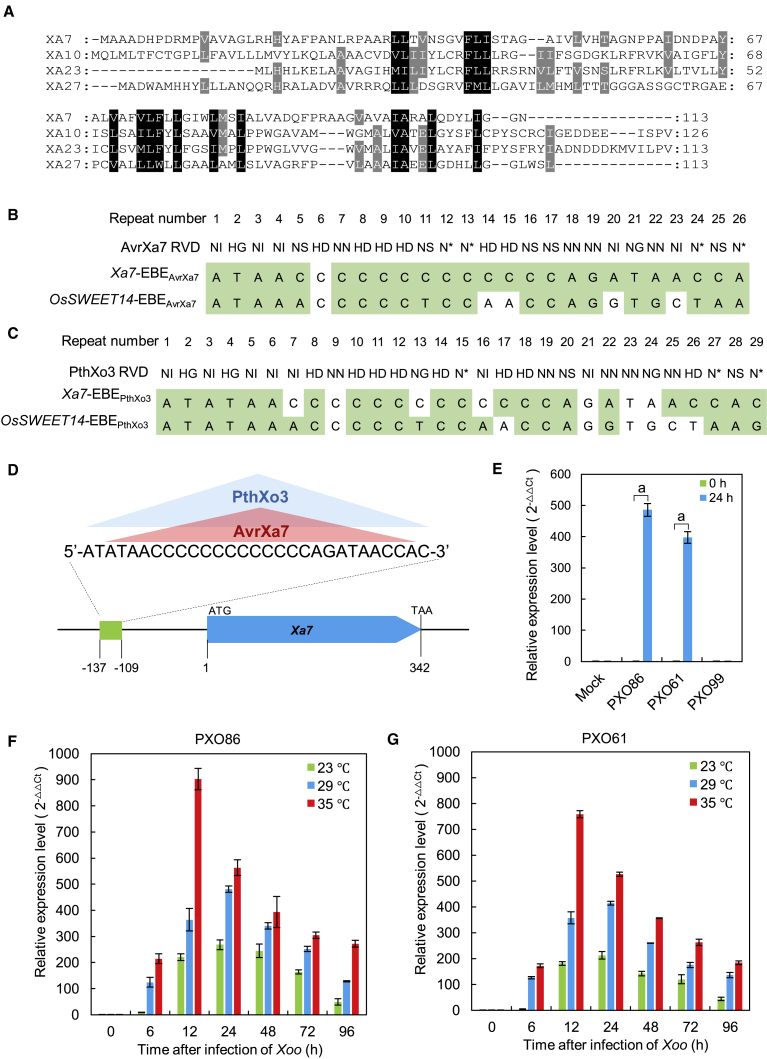
PCR amplification and high-throughput sequencing showed that the open reading frame (ORF) of Xa7 and its promoter (>3000 bp) were completely identical in IRBB7 and Zhen-hui 084 but did not exist in PXO86-susceptible varieties Nipponbare, IR24, and Zhong-hua 11. The Xa7 gene has only one exon, with a 342-bp ORF that encodes a 113-aa unknown protein (Supplemental Figure 5). Coincidentally, the amino acid number of the deduced XA7 protein is equal to that of executor R proteins XA23 (Wang et al., 2015) and XA27 (Gu et al., 2005), and XA7 contains putative transmembrane domains similar to those of XA10, XA23, and XA27 (Zhang et al., 2015). Nevertheless, XA7 shares few sequence similarities with known executor R proteins (Figure 3A) and has no homologs in current plant databases. These results suggest that Xa7 may encode a novel executor R protein.
Figure 3.
Sequence characteristics and induced expression of Xa.
(A) Deduced amino acid sequence alignment of XA7 with other executor R proteins, XA10, XA23, and XA27. Conserved residues are boxed in black or highlighted in gray based on the degree of conservation. Dashes indicate missing sequences or gaps generated by the alignment.
(B) Pairing recognition relationship between AvrXa7 and the putative Xa7-EBEAvrXa7 compared with OsSWEET14-EBEAvrXa7.
(C) Pairing recognition relationship between PthXo3 and the putative Xa7-EBEPthXo3 compared with OsSWEET14-EBEPthXo3. Repeat number refers to the specific repeat in the central region of the AvrXa7 or PthXo3 protein. RVD represents the repeat variable di-residues at the 12th and 13th positions of each repeat in the central region of AvrXa7 or PthXo3. Perfect recognition between the bases of EBE and the RVD of the TALE is highlighted in green.
(D) Overlapping EBEs corresponding to AvrXa7 and PthXo3 in the promoter of Xa7. The blue box represents the Xa7 gene from ATG to TAA. The green box represents the EBEs. Numbers indicate the sites of EBEs. Red and blue triangles show the EBEs recognized by AvrXa7 and PthXo3, respectively.
(E) Expression of Xa7 in rice variety IRBB7 in response to different Xoo strains: PXO86 (AvrXa7), PXO61 (PthXo3), PXO99 (neither AvrXa7 nor PthXo3), and mock inoculation (H2O). Leaves were sampled at 0 and 24 h after inoculation. Data are presented as the mean of three independent replicates ± SD. Statistical analysis was performed by t-test, and significant differences (P < 0.01) between 0 and 24 h inoculation with each strain or the mock control are indicated by “a” on the bars.
(F and G) Expression of Xa7 in IRBB7 in response to strains PXO86 (F) and PXO61 (G) under different temperatures (23°C, 29°C, and 35°C). Leaves were sampled at 0, 6, 12, 24, 48, 72, and 96 h after inoculation. Gene expression levels were measured by qRT–PCR, and data are presented as the means of three independent replicates ± SD.
Executor R genes are reported to be transcriptionally induced by TALEs that recognize and bind to EBEs in R gene promoters (Zhang et al., 2015). To search for a possible EBE in the Xa7 promoter, the 2000-bp sequence before the start codon was scanned using TALE-NT with a 3.0 cutoff (Doyle et al., 2012). A 26-bp putative EBE (Figure 3B) for AvrXa7 was detected from −135 to −110 bp with a high TALE-NT score (Table 1) (Figure 3D). Smaller scores indicate greater binding capacities of EBEs to TALEs, and the EBE in the Xa7 promoter (score: 19.65) was therefore predicted to have a greater binding capacity than that in the OsSWEET14 promoter (score: 26.72) identified by Antony et al. (2010; Table 1). In addition, based on the principle of recognition between TALEs and EBEs (Boch et al., 2009), the 25.5 RVDs of AvrXa7 could be perfectly manually aligned with the 26-bp EBE in the Xa7 promoter, and this alignment was better than that of the OsSWEET14 promoter (Figure 3B). To distinguish between these two EBEs, we designated that in the Xa7 promoter Xa7-EBEAvrXA7 and that in the OsSWEET14 promoter OsSWEET14-EBEAvrXA7. We also found an EBE in the Xa7 promoter that may be recognized by the TALE PthXo3 (designated Xa7-EBEPthXo3; Table 1; Figure 3C). Interestingly, the Xa7-EBEAvrXA7 and Xa7-EBEPthXo3 sequences overlap (Figure 3D), just like the EBEs in the OsSWEET14 promoter (Antony et al., 2010).
Table 1.
EBE prediction scores for Xa7 and OsSWEET14 genes by TALE-NT 2.0 (cutoff: 3.0).
| Gene | TALE | a EBE | b Score |
|---|---|---|---|
| Xa7 | AvrXa7 | Xa7-EBEAvrXa7 | 19.65 |
| PthXo3 | Xa7-EBEPthXo3 | 29.95 | |
| OsSWEET14 | AvrXa7 | OsSWEET14-EBEAvrXa7 | 26.72 |
| PthXo3 | OsSWEET14-EBEPthXo3 | 31.87 |
Effector binding element in the promoter of Xa7 or SWEET14.
EBE prediction scores for Xa7 and OsSWEET14.
To demonstrate that Xa7 could be transcriptionally induced by AvrXa7 and PthXo3, the Xoo strains PXO86 (AvrXa7), PXO61 (PthXo3), and PXO99 (neither AvrXa7 nor PthXo3; Oliva et al., 2019) were used to infect the leaves of IRBB7, and Xa7 expression was analyzed by qRT–PCR 24 h after inoculation. Compared with the control, Xa7 was highly induced by PXO86 and PXO61 but not by PXO99 (Figure 3E). Because high temperature can increase the BB resistance conferred by Xa7 (Webb et al., 2010; Supplemental Figure 6), the Xoo induction of Xa7 was compared under different temperatures (23°C, 29°C, and 35°C) by qRT–PCR. After inoculations with PXO86 and PXO61, Xa7 was highly induced at 6 h under 29°C and 35°C, but its induction was delayed at 12 h under 23°C (Figure 3F and 3G). Xoo-induced expression of Xa7 was therefore proportional to temperature and gradually increased from 23°C to 35°C (Figure 3F and 3G).
Xa7 activates plant defense response
The executor R genes Xa10 and Xa23 are reported to activate defense responses such as HR in plants other than rice (Tian et al., 2014; Wang et al., 2015). We transferred an avrXa7 overexpression construct into a Xa7-compatible strain, PXO99, which was designated PXO99avrXa7. This strain and the control PXO99 were infiltrated into the leaves of IRBB7. Three days after injection, the leaves with PXO99avrXa7 displayed a strong HR and resistance phenotype, whereas those infiltrated with PXO99 did not (Figure 4A). Moreover, a vector overexpressing Xa7 driven by the CaMV35S promoter was constructed and infiltrated into tobacco leaves by the Agrobacterium-mediated method. Vectors that overexpressed Xa10 or Xa23 were used as positive controls. The overexpression of Xa7 triggered a strong HR, just like Xa10 and Xa23 (Figure 4B and 4C). A construct of Xa7 driven by its native promoter (C1S3) also induced HR in tobacco leaves (Figure 4B and 4C), and co-infiltration of C1S3 with the avrXa7-expressing vector triggered a stronger HR (Figure 4B and 4C; Supplemental Figure 7).
The plants of some Xa7 transgenic lines displayed abnormal phenotypes such as dwarfism, short leaves, and small panicles. We speculated that Xa7 might be constitutively expressed and activate a defense reaction in these lines, resulting in the inhibition of plant growth or development. Three types of Xa7-transgenic plants, normal (C1S3-N), semi-dwarf (C1S3-SD), and dwarf (C1S3-D), were selected from the transgenic line C1S3 (Figure 4D) for Xa7 expression assay by qRT–PCR. In the absence of Xoo, the expression levels of Xa7 were 5-, 48-, and 142-fold higher in the C1S3-N, C1S3-SD, and C1S3-D plants than in the Zhen-hui 084 control (Figure 4E). The higher the expression level, the lower the plant height. Expression of the defense response marker genes PR1a and PBZ1 ((1)Peng et al., 2008) was measured by qRT–PCR. Both were induced in the three plant types, and expression was particularly high in the C1S3-SD and C1S3-D plants (Figure 4F and 4G). These results indicated that the induction of Xa7 could activate a defense response. Furthermore, the BB resistance of the three plant types was tested using the Xa7-incompatible strain PXO86 and the compatible strain PXO99. The IRBB7 and Zhen-hui 084 controls were resistant to PXO86 and susceptible to PXO99 (Figure 4H and 4I). As expected, all three types of Xa7 transgenic plants were resistant to PXO86 (Figure 4H). However, they exhibited different responses to PXO99: the C1S3-N plants were highly susceptible, the C1S3-SD plants were moderately susceptible, and the C1S3-D plants were resistant (Figure 4I).
Genetic diversity of Xa7 in rice germplasm
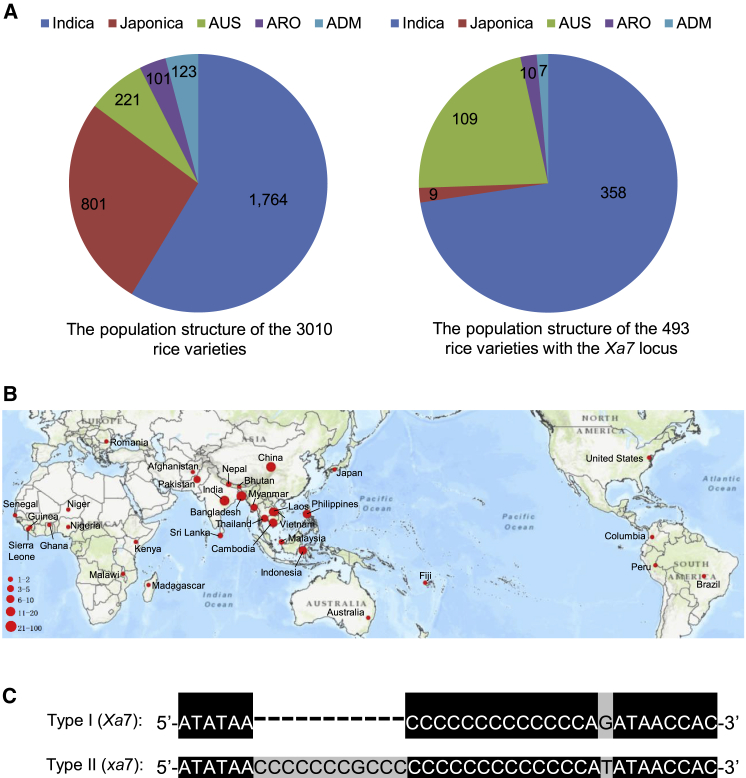
Xa7 was initially identified from DV85, an indica rice variety from Bangladesh (Sidhu et al., 1978). The Xa7 sequence was used for the analysis of genetic diversity in a database of 3010 cultivated rice varieties (Wang et al., 2018), and 493 varieties were found to contain the Xa7 CDS (Supplemental Data 2). These varieties were mainly from the indica and aus subgroups (Figure 5A), and their origins were geographically distributed in South Asia (India and Bangladesh), Southeast Asia (Laos, Vietnam, Thailand, Philippines, and Indonesia), and the south of China. These are the exact cultivation areas of the indica and aus subgroups (Figure 5B). Compared with the subgroup distribution of the 3010 varieties, the Xa7 locus showed an obvious expansion in the aus subgroup (Figure 5A).
Figure 5.
Geographic distribution and genetic variation of the Xa7 locus.
(A) Comparison of the population structure of 3010 rice accessions and 493 rice varieties that contain the Xa7 locus. Indica, Japonica, AUS, ARO, and ADM are different subgroups of rice. Numbers refer to the number of varieties in each subgroup.
(B) Geographic distribution of the Xa7 locus among the 3010 cultivated rice accessions. The size of the red dots represents the number of varieties distributed in the indicated countries.
(C) Comparison of EBEs in the Xa7 locus based on the sequencing results of the 12 varieties (shown in Supplemental Figure 8). Type I (Xa7) belongs to the PXO86-resistant varieties, and Type II (xa7) belongs to the PXO86-susceptible varieties.
Sequence alignments showed that most of the 493 varieties had an Xa7 CDS identical to that of Zhen-hui 084 (Supplemental Data 2). However, we were unable to determine whether the EBE sequences of the 493 varieties were consistent with that of Xa7-EBEAvrXa7 in Zhen-hui 084, particularly at the site of polycytosine (C) bases (Supplemental Figure 5). There are 13 C bases in the Xa7-EBEAvrXa7 sequence (Figure 3D), and they are difficult to sequence completely by high throughput methods and then assemble correctly. To confirm the genetic diversity of the Xa7 gene in rice germplasm, 27 of the 493 varieties from our lab were used for identification (Table 2). PCR primers were designed based on the sequences of Zhen-hui 084, and amplification was performed using genomic DNA from the 27 varieties as templates. Xa7 CDS sequences were amplified from all 27 varieties (Table 2), and all the amplified sequences were identical to that of Zhen-hui 084 (data not shown). However, the promoter sequences of Xa7 could be amplified from only 12 varieties, and two types of sequence diversity were found in the EBE region (Supplemental Figure 8). The Type I sequences from five varieties (IRBB7, DV85, AUS 242, AUS 299, and AUS 308) were identical to that of Zhen-hui 084, and the Type II sequences from the other seven varieties had an 11-bp (5′-CCCCCCCGCCC-3′) insertion and a G-to-T base substitution (Figure 5C; Supplemental Figure 8). This insertion directly broke the EBE sequence and may result in nonrecognition of AvrXa7. The promoter sequences of the remaining 15 varieties were unknown, as they could not be amplified; there may be other types of EBE in their promoters. The 27 varieties were then inoculated with PXO86 to test their BB resistance. Consistent with the PCR results, the Type I varieties were highly resistant, whereas the Type II varieties and the remaining 15 varieties were highly susceptible (Table 2). These results indicated that the EBEAvrXa7 in the Xa7 promoter was essential for the BB resistance in rice.
Table 2.
Information on 27 rice varieties that contain the Xa7 CDS.
| a Accession ID | Variety name | Origin | b Group | c EBE | d Lesion length | e Phenotype |
|---|---|---|---|---|---|---|
| IRGC 117725 | DV85 | Bangladesh | AUS | Type I | 0.4 ± 0.154 | R |
| IRIS_313-11049 | AUS 219 | Bangladesh | AUS | – | 30.4 ± 2.08 | S |
| IRIS_313-11051 | AUS 242 | Bangladesh | AUS | Type I | 0.3 ± 0.06 | R |
| IRIS_313-11055 | AUS 299 | Bangladesh | AUS | Type I | 0.4 ± 0.08 | R |
| IRIS_313-11057 | AUS 308 | Bangladesh | AUS | Type I | 0.4 ± 0.07 | R |
| B120 | HONG WAN 1 | China | IND | – | 27.8 ± 3.15 | S |
| B207 | AI HE CHI | China | IND | – | 24.8 ± 2.33 | S |
| B246 | LAO ZAO GU | China | IND | Type II | 29.2 ± 2.50 | S |
| IRIS_313-9555 | CHUA DAU | China | IND | – | 32.2 ± 1.96 | S |
| IRIS_313-10875 | ARC 12021 | India | AUS | – | 30.3 ± 1.85 | S |
| IRIS_313-11175 | SONA AUS | India | AUS | – | 31.1 ± 1.97 | S |
| IRIS_313-11374 | W 398 | India | AUS | – | 31.8 ± 2.65 | S |
| IRIS_313-11454 | BARI SUTAR | India | AUS | Type II | 33.2 ± 5.78 | S |
| IRIS_313-11456 | KOLAMBA | India | AUS | – | 27.2 ± 3.00 | S |
| IRIS_313-9137 | ARC 10100 | India | AUS | Type II | 37.1 ± 2.37 | S |
| IRIS_313-9610 | DANGAR | India | AUS | Type II | 26.6 ± 1.99 | S |
| IRIS_313-11809 | MHARAKA | Kenya | AUS | Type II | 34.4 ± 2.22 | S |
| IRIS_313-11071 | CHAO HAI | Laos | IND | – | 30.4 ± 2.47 | S |
| IRIS_313-12139 | MANSARA DHAN | Nepal | AUS | Type II | 31.3 ± 0.92 | S |
| IRIS_313-11020 | BAMLA SUFFAID 320 | Pakistan | AUS | – | 37.2 ± 1.25 | S |
| IRIS_313-11028 | MOTIA | Pakistan | AUS | Type II | 28.0 ± 3.67 | S |
| IRGC 135948 | IRBB7 | Philippines | IND | Type I | 0.6 ± 0.287 | R |
| IRIS_313-7809 | WAS 174-B-3-5 | Senegal | IND | – | 28.6 ± 2.50 | S |
| IRIS_313-11510 | A 69-1 | Sri Lanka | IND | – | 29.7 ± 2.46 | S |
| IRIS_313-10928 | KHAO SIM | Thailand | IND | – | 28.0 ± 1.54 | S |
| IRIS_313-8073 | DAWN CI 9534 | United States | AUS | – | 29.1 ± 1.05 | S |
| IRIS_313-8341 | BAT DO | Vietnam | IND | – | 21.2 ± 4.70 | S |
Accession ID of variety in the 3K Rice database or IRGC.
SNP-based subgroups of varieties.
EBE types in the Xa7 promoter. Type I is identical to Zhen-hui 084, Type II is a variation shown in Figure 6, and ‘–’ indicates that the type is unknown.
Lesion length (mean ± SD) of leaves measured at 2 weeks after inoculation with Xoo strain PXO86. More than 15 individual leaves were measured for each variety.
Resistant (R) or susceptible (S) phenotype of the variety against PXO86.
Discussion
Map-based cloning remains a classical and effective method for the mapping and isolation of important genes from crops (Song et al., 1995; Tian et al., 2019; Wang et al., 2020). Nevertheless, we encountered great difficulties in map-based cloning of the BB resistance gene Xa7 in rice. It took us many years to overcome these challenges and finally isolate the gene. We had successfully fine mapped Xa7 to a 51-kb interval using available DNA markers (Figure 1A). However, no products could be amplified using primers located in the 51-kb region based on the rice reference genome, indicating that there must be a large structural variation in this region of Xa7-containing varieties such as IRBB7 and Zhen-hui84. In that situation, map-based cloning of a target gene could not continue until the sequence had been determined. By constructing and sequencing BAC libraries, we identified a 116-kb sequence from Zhen-hui84 and IRBB7 that corresponded to the 51-kb region in IR24 (Figure 1D). Moreover, there was almost no homology between the 116-kb and 51-kb sequences (Supplemental Figure 2). Therefore, it was likely that chromosome recombination of the Xa7 mapping region was inhibited in our F2 population, thereby blocking the fine mapping of Xa7. Because no candidate genes were predicted in the interval, we subsequently turned to screening for Xa7 mutants in a Zhen-hui84 radiation-mutagenesis library. After 5 years of effort, we screened out one BB-susceptible mutant from 20 000 lines; it contained a 107-kb deletion overlapping the Xa7 mapping region (Supplemental Figure 3). Combining the regions identified by fine mapping and the 107-kb deletion, we finally placed the target gene in a 28-kb candidate region and identified Xa7 through the analysis of a large number of transgenic plants. Our experiences in this study showed that if difficulties are encountered in the map-based cloning of a gene of interest, other approaches such as mutant screening can be carried out simultaneously.
Just like Xa23 (Wang et al., 2015) and Xa27 (Gu et al., 2005), Xa7 encodes an executor R protein that confers broad-spectrum resistance to Xoo strains. However, Xa7 has some unique characteristics, including extremely durable and heat-tolerant resistance (Vera Cruz et al., 2000; Webb et al., 2010). First, the durable resistance of Xa7 may be partly explained by the distinctive functions of its cognate avirulence gene avrXa7 in Xoo. The cognate avirulence genes of the R genes Xa23 and Xa27 are avrXa23 and avrXa27, respectively. The AvrXa23 and AvrXa27 proteins are not major virulent TALEs in Xoo and function only as avirulence factors whose mutation or loss of function does not affect Xoo survival (Oliva et al., 2019). Under the pressure of long-term, continuous use of a single R gene in crop breeding and production, pathogens accumulating gene mutations may overcome this R gene in host plants (Quibod et al., 2019). However, avrXa7 plays dual roles in Xoo, not only encoding an avirulent TALE that induces the R gene expression of host plants, but also acting as a virulence gene that maintains the toxicity of Xoo for survival. Thus, an Xoo strain that contains avrXa7 is subject to defense by the cognate R gene in plants, but the loss of avrXa7 causes a risk of being eliminated in nature (Vera Cruz et al., 2000). This places the Xoo strain in a dilemma but enables Xa7 to become a durable R gene in plants. Our results suggested that the expression of Xa7 could be induced by a Xoo strain that contained avrxa7 (Figure 3E and 3F), thereby activating a defense response (like HR) to inhibit Xoo (Figure 4). Xa7 induction appeared to result from the binding of AvrXa7 to EBEAvrXa7 in the Xa7 promoter (Figure 3B). We provided a preliminary demonstration of an interaction between Xa7 and AvrXa7 and shed light on the durable resistance of Xa7.
Second, Xa7 provides broad-spectrum resistance to BB, suggesting that avrXa7 is generally widespread in Xoo strains. We searched for avrXa7 in the genomes of hundreds of Xoo strains using Blast but found that it was not widely present in Xoo (data not shown). It has been reported that some Xoo strains such as PXO61 that lack a credible avrXa7 gene are still incompatible with Xa7 (Oliva et al., 2019). Naturally, a question arises: how does Xa7 provide broad-spectrum resistance to Xoo? In fact, PXO61 contains the virulent gene pthXo3, which encodes a major TALE, PthXo3 (Yang and White, 2004), and is highly homologous to avrXa7 in genomic DNA (data not shown). We speculated that avrXa7 and pthXo3 may have originated from the same ancestor. Interestingly, a sequence containing two overlapping EBEs in the OsSWEET14 promoter could be recognized by both AvrXa7 and PthXo3 (Antony et al., 2010). We also found that two putative EBEs for AvrXa7 and PthXo3 overlapped in the Xa7 promoter (Figure 3D), and Xa7 expression was significantly induced by both PXO86 (avrXa7) and PXO61 (pthXo3) with similar expression patterns (Figure 3F and 3G). This means that pthXo3 may be another cognate avirulent gene for Xa7 in host plants. Therefore, two major TALEs of Xoo, AvrXa7 and PthXo3, target Xa7 in host plants, enabling this R gene to confer broad-spectrum disease resistance. More evidence should certainly be collected to confirm this speculation.
Third, global warming is affecting the survival of human beings and plants. Studies have confirmed that heat stress reduces the effectiveness of plant resistance against pathogens (De Jong et al., 2002; Zhu et al., 2010). At present, the underlying mechanisms by which R genes lose effectiveness in disease resistance at high temperatures are poorly understood. Fascinatingly, Xa7 acts in the completely opposite manner, enhancing the BB resistance of rice to a greater extent at higher temperatures (Cohen et al., 2017; Dossa et al., 2020; Webb et al., 2010; Supplemental Figure 6). Here, we showed that Xa7 induction by Xoo was faster and stronger at higher temperatures (Figure 3F and 3G), which may promptly activate and enhance the defense response to inhibit Xoo in rice. This trait is very valuable for rice breeding in the face of global warming.
The isolation of Xa7 provides a unique model with which to investigate the co-evolution between host plants and pathogens. Although the molecular mechanism of Xa7-mediated defense response remains unclear, our results provide valuable clues for uncovering it in the future and will greatly facilitate the use of this gene in breeding. This study also sheds light on breeding varieties with broad-spectrum and durable disease resistance, for example by mimicking the recognition of specific TALEs (like AvrXa7) by R genes or by pyramiding different EBEs of the major TALEs into the promoter of a single R gene. These strategies may provide an effective means of breeding durable disease-resistant varieties to control BB disease.
Methods
Plant materials and growth conditions
The rice (Oryza sativa L.) varieties Zhen-hui 084 and IRBB7 both contain Xa7 with broad-spectrum resistance to Xoo, and the varieties Nipponbare, IR24, Cheng-hui 448, and Zhong-hua 11 are highly susceptible to Xoo. For fine mapping of the Xa7 gene, Cheng-hui 448 was selected as the pollen accepter and crossed with Zhen-hui 084; the heterozygous F1 plants were self-crossed to develop the F2 population. Rice plants were grown in the field. For temperature treatment, rice seedlings at the tillering stage were transferred into a growth chamber (14 h light/10 h dark) with different temperatures. Tobacco (Nicotiana benthamiana) plants were cultured in a growth chamber at 25°C (16 h light/8 h dark).
Xoo strains and inoculation
The Xoo strains used in this study included ten races from the Philippines. Strains were cultured on agar medium that contained 20 g sucrose, 5 g peptone, 0.5 g Ca(NO3)2, 0.43 g Na2HPO4, and 0.05 g FeSO4 per liter and were allowed to grow at 28°C for 2–3 days. The bacterial colony was suspended in sterile distilled water at an optical density of OD600 = 1.0 and immediately used for plant inoculation. The leaf-tip clipping method (Kauffman et al., 1973) was used for Xoo inoculation. From the inoculated leaves, 1 cm of leaf tissue below the cut edge was collected for gene expression analysis. Lesions on the inoculated leaves were measured for the evaluation of BB resistance 2 weeks after inoculation (Yin et al., 2000). Based on the genomic sequence of PXO86 (NCBI: CP031463.1), the coding sequence of AvrXa7 was cloned by PCR amplification and inserted into the pHM1 vector by homologous recombination. The constructed vector was transformed into strain PXO99 by electroporation. The resulting PXO99AvrXa7 strain was injected into tobacco or rice leaves through a needle-less syringe, and photos were taken 3 days after injection.
Genomic library construction and sequencing
Seeds of rice were germinated and grown in the dark, and the etiolated seedlings were sent to the Takara Biomedical Technology (Beijing) Company in China for the construction of genomic fosmid libraries. The library from each variety provided greater than 10-fold coverage of the rice genome, and the inserted chromosome fragments of each clone were 35 kb on average. Positive clones were screened by normal PCR from the constructed libraries and sequenced on the Illumina HiSeq 2500 platform. The clean reads were assembled using SPAdes 3.5.0.
Complementary vector construction and rice transformation
Complementary fragments C1 to C4, C1S1 to C1S7, and C2S1 (Supplemental Data 2) were amplified from the plasmid of fosmid clones by proofreading PCR using a PrimeSTAR HS DNA Polymerase Kit (TaKaRa) and cloned into the pCAMBIA1300 vector using an In-Fusion HD Cloning Kit (TaKaRa) according to the manufacturer's instructions. CRISPR-Cas9 technology was used to knock out the Xa7 gene. Two gRNAs targeting the CDS of Xa7 were designed by the CRISPR Design program (http://crispr.mit.edu). The sequences of Target-1 and Target-2 were 5′-CGTATGCCCGTTGCAGTTGCAGG-3′ and 5′-CCAGTTCCCGCGCGCCGCTGGGG-3′, respectively. The underlined bases represent the protospacer adjacent motif sequences. The pCAMBIA1300-pYAO-cas9 vector was used to make the two CRISPR/Cas9 constructs. All of the vectors were transformed into rice by the Agrobacterium-mediated method described previously (Nishimura et al., 2006).
RNA extraction and qRT–PCR analysis
Total RNA was extracted from leaves using the TRIzol reagent (Life Technologies) and purified using an RNeasy mini kit (QIAGEN) and RNase-Free DNase Set (QIAGEN) following the manufacturer's instructions. Synthesis of first-strand cDNAs from RNA was performed using the M-MLV Reverse Transcriptase kit (Promega) according to the manufacturer's instructions. Fast SYBR Green Master Mix reagent (Applied Biosystems) was used for the real-time PCR experiment. The thermal cycle was performed on a StepOne Real-Time PCR system (Applied Biosystems) with the following program: 95°C for 20 s, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. A rice housekeeping gene, Actin, was used as the standardization control, and the relative expression level of each gene was analyzed by the 2−ΔΔCT method (Livak and Schmittgen, 2001). Standard errors were calculated based on a minimum of three biological replicates. Primers are listed in Supplemental Table 5.
HR analysis in tobacco leaves
CDSs of Xa7, Xa10, and Xa23 were amplified and inserted into the pCAMBIA1300-GFP vector by homologous recombination. The CDS of avrXa7 was amplified from Xoo strain PXO86 and inserted into the pCAMBIA1300 vector driven by the CaM35S promoter. All constructs were transformed into Agrobacterium tumefaciens strain EHA105. The bacteria were cultured in 5 ml of LB liquid medium with kanamycin (50 mg/ml) and grown at 28°C for 2 days. A single clone was picked to inoculate in 5 ml LB liquid medium with kanamycin (50 mg/ml) until the density reached OD600 = 0.8 and was then subcultured in 20 ml of LB liquid medium until the density reached OD600 = 0.6. Qualified bacterial cells were collected by centrifugation at 4°C for 10 min at 3000 rpm, resuspended in 5 ml buffer (10 mM MES, 10 nM MgCl2, 200 μM AS, [pH 5.7]), and activated at 28°C for 4 h. Leaves of 4-week-old tobacco plants were used for infiltration as described by Kay et al. (2007). The infected leaves were photographed and then stained with trypan blue 2–3 days after infiltration (Wilson and Coffey, 1980).
Bioinformatic analysis
Gene prediction in the Xa7 mapping region was performed with Fgenesh (http://www.softberry.com/). Sequence alignment was performed with ClustalX 2.0, and the output was colored using GENEDOC 2.1. TALE-NT 2.0 (https://tale-nt.cac.cornell.edu/) was used with a stringency cutoff of 3.0 to search for putative EBEs recognized by AvrXa7 and PthXo3 (Doyle et al., 2012). The RFGB database (http://www.rmbreeding.cn/Blast; Wang et al., 2018) was used to investigate the genetic evolution of Xa7 in different rice varieties. The geographic distribution of rice varieties was visualized with ArcGIS (version 10.5) based on longitude and latitude of the varieties obtained from the MBKbase database (http://www.mbkbase.org/rice; Peng et al., 2020).
Funding
This work was supported by the Ministry of Agriculture and Rural Affairs of China (2016ZX08009003-001), the National Natural Science Foundation of China (32071987, 31871605), and the Natural Science Foundation of Zhejiang Province (LD19C130001).
Author contributions
B.M. and Q.Q. designed and supervised the research. X.C., P.L., L.M., and X.H. performed the pivotal experiments. L.C., H.L., S.S., Z.J., X.Z., and Y.Z. contributed the preparatory work. Z.G. and D.Z. provided the rice varieties. X.C., P.L., and L.M. wrote the manuscript with input from all other co-authors.
Acknowledgments
We are grateful to Professor Hongsheng Zhang and Professor Jianfei Wang (Nanjing Agricultural University) for kindly providing help and rice materials and to Dr. Lihuang Zhu and Dr. Wenxue Zhai (Institute of Genetics and Developmental Biology, Chinese Academy of Science) for guidance. We also thank Dr. Kaijun Zhao and Zhiyuan Ji (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for their kind help and for donating strains. No conflict of interest declared.
Published: January 9, 2021
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Ainsworth E.A. Rice production in a changing climate: a meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration. Glob. Change Biol. 2008;14:1642–1650. [Google Scholar]
- Antony G., Zhou J., Huang S., Li T., Liu B., White F., Yang B. Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 2010;22:3864–3876. doi: 10.1105/tpc.110.078964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezrutczyk M., Yang J., Eom J.S., Prior M., Sosso D., Hartwig T., Szurek B., Oliva R., Vera-Cruz C., White F.F. Sugar flux and signaling in plant-microbe interactions. Plant J. 2017;93:675–685. doi: 10.1111/tpj.13775. [DOI] [PubMed] [Google Scholar]
- Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove A.J., Voytas D.F. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Bogdanove A.J., Schornack S., Lahaye T. TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Boller T., He S.Y. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324:742–744. doi: 10.1126/science.1171647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol. 2014;201:1150–1155. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- Chen S., Huang Z., Zeng L., Yang J., Liu Q., Zhu X. High-resolution mapping and gene prediction of Xanthomonas oryzae pv. oryzae resistance gene Xa7. Mol. Breed. 2008;22:433–441. [Google Scholar]
- Chen S., Wang C., Yang J., Chen B., Wang W., Su J., Feng A., Zeng L., Zhu X. Identification of the novel bacterial blight resistance gene Xa46(t) by mapping and expression analysis of the rice mutant H120. Sci. Rep. 2020;10:12642. doi: 10.1038/s41598-020-69639-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Cohen S.P., Liu H., Argueso C.T., Pereira A., Vera Cruz C., Verdier V., Leach J.E. RNA-Seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS One. 2017;12:e0187625. doi: 10.1371/journal.pone.0187625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong C.F., Takken F.L.W., Cai X., De Wit P.J.G.M., Joosten M.H.A.J. Attenuation of Cf-mediated defense responses at elevated temperatures correlates with a decrease in elicitor-binding sites. Mol. Plant Microbe Interact. 2002;15:1040–1049. doi: 10.1094/MPMI.2002.15.10.1040. [DOI] [PubMed] [Google Scholar]
- Dossa G.S., Quibod I., Atienza-Grande G., Oliva R., Maiss E., Vera Cruz C., Wydra K. Rice pyramided line IRBB67 (Xa4/Xa7) homeostasis under combined stress of high temperature and bacterial blight. Sci. Rep. 2020;10:683. doi: 10.1038/s41598-020-57499-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle E., Booher N., Standage D., Voytas D., Brendel V., Vandyk J., Bogdanove A. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu K., Yang B., Tian D., Wu L., Wang D., Sreekala C., Yang F., Chu Z., Wang G.L., White F.F. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- Hopkins C.M., White F.F., Choi S.H., Guo A., Leach J.E. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- Hu K., Cao J., Zhang J., Xia F., Ke Y., Zhang H., Xie W., Liu H., Cui Y., Cao Y. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants. 2017;3:17009. doi: 10.1038/nplants.2017.9. [DOI] [PubMed] [Google Scholar]
- Hutin M., Sabot F.O., Ghesquière A., Koebnik R., Szurek B. A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J. 2016;84:694–703. doi: 10.1111/tpj.13042. [DOI] [PubMed] [Google Scholar]
- Iyer A.S., McCouch S.R. The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 2004;17:1348–1354. doi: 10.1094/MPMI.2004.17.12.1348. [DOI] [PubMed] [Google Scholar]
- Ji C., Ji Z., Liu B., Cheng H., Liu H., Liu S., Yang B., Chen G. Xa1 allelic R genes activate rice blight resistance suppressed by interfering TAL effectors. Plant Commun. 2020;1:100087. doi: 10.1016/j.xplc.2020.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Xia Z., Zhou Y., Wan J., Li D., Chen R., Zhai W., Zhu L. Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAγ1. Mol. Genet. Genomics. 2006;275:354–366. doi: 10.1007/s00438-005-0091-7. [DOI] [PubMed] [Google Scholar]
- Jiang N., Yan J., Liang Y., Shi Y., He Z., Wu Y., Zeng Q., Liu X., Peng J. Resistance genes and their interactions with bacterial blight/leaf streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)-an Updated Review. Rice. 2020;13:3. doi: 10.1186/s12284-019-0358-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Kaji R., Ogawa T. Identification of the located chromosome of the resistance gene, Xa7, to bacterial leaf blight in rice. Breed. Sci. 1995;45:79. [Google Scholar]
- Kauffman H.E., Reddy A.P., Hsieh S.P., Merca S.D. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis. 1973;57:537–541. [Google Scholar]
- Kay S., Hahn S., Marois E., Hause G., Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- Khush G.S., Mackill D.J., Sidhu G.S. Breeding rice for resistance to bacterial blight. International Rice Research Institute; ManilaPhilippines: 1989. pp. 207–217. [Google Scholar]
- Kourelis J., van der Hoorn R. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 2018;30:285–299. doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Yuan M., Zhou Y., Li X., Xiao J., Wang S. A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 2011;34:1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x. [DOI] [PubMed] [Google Scholar]
- Liu W., Liu J., Triplett L., Leach J.E., Wang G. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- Liu Z., Faris J.D., Oliver R.P., Tan K.C., Solomon P.S., McDonald M.C., McDonald B.A., Nunez A., Lu S., Rasmussen J.B. SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog. 2009;5:e1000581. doi: 10.1371/journal.ppat.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mew T.W. Current status and future prospects of research on bacterial blight of rice. Annu. Rev. Phytopathol. 1987;25:359–382. [Google Scholar]
- Nishimura A., Aichi I., Matsuoka M. A protocol for Agrobacterium-mediated transformation in rice. Nat. Protoc. 2006;1:2796–2802. doi: 10.1038/nprot.2006.469. [DOI] [PubMed] [Google Scholar]
- Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.S., Li C., Nguyen H., Liu Bo. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019;37:1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Wang K., Chen Z., Cao Y., Gao Q., Li Y., Li X., Lu H., Du H., Lu M. MBKbase for rice: an integrated omics knowledgebase for molecular breeding in rice. Nucleic Acids Res. 2020;48:D1085–D1092. doi: 10.1093/nar/gkz921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter B.W., Chittoor J.M., Yano M., Sasaki T., White F.F. Development and mapping of markers linked to the rice bacterial blight resistance gene Xa7. Crop Sci. 2003;43:1484–1492. [Google Scholar]
- Quibod I.L., Atieza-Grande G., Oreiro E.G., Palmos D., Oliva R. The green revolution shaped the population structure of the rice pathogen Xanthomonas oryzae pv. oryzae. ISME J. 2019;14:492–505. doi: 10.1038/s41396-019-0545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu G.S., Khush G.S., Mew T.W. Genetic analysis of bacterial blight resistance in seventy-four cultivars of rice, Oryza sativa L. Theor. Appl. Genet. 1978;53:105–111. doi: 10.1007/BF00272687. [DOI] [PubMed] [Google Scholar]
- Song W., Wang G., Chen L., Kim H.S., Pi L., Holsten T., Gardner J., Wang B., Zhai W., Zhu L. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- Sun X., Cao Y., Yang Z., Xu C., Li X., Wang S., Zhang Q. Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 2004;37:517–527. doi: 10.1046/j.1365-313x.2003.01976.x. [DOI] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J. Receptor kinases in plant pathogen interactions: more than pattern recognition. Plant Cell. 2017;29:618–637. doi: 10.1105/tpc.16.00891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D., Wang J., Zeng X., Gu K., Qiu C., Yang X., Zhou Z., Goh M., Luo Y., Murata-Hori M. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell. 2014;26:497–515. doi: 10.1105/tpc.113.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J., Wang C., Xia J., Wu L., Xu G., Wu W., Li D., Qin W., Han X., Chen Q. Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science. 2019;365:658–664. doi: 10.1126/science.aax5482. [DOI] [PubMed] [Google Scholar]
- Vera Cruz C.M., Bai J., Ona I., Leung H., Nelson R.J., Mew T.W., Leach J.E. Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. U S A. 2000;97:13500–13505. doi: 10.1073/pnas.250271997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Zhang X., Fan Y., Gao Y., Zhu Q., Zheng C., Qin T., Li Y., Che J., Zhang M. XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant. 2015;8:290–302. doi: 10.1016/j.molp.2014.10.010. [DOI] [PubMed] [Google Scholar]
- Wang H., Sun S., Ge W., Zhao L., Hou B., Wang K., Lyu Z., Chen L., Xu S., Guo J. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science. 2020;368:eaba5435. doi: 10.1126/science.aba5435. [DOI] [PubMed] [Google Scholar]
- Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., Li M., Zheng T., Fuentes R.R., Zhang F. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018;557:43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb K.M., Oña I., Bai J., Garrett A.K., Mew T., Vera Cruz C.M., Leach J.E. A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 2010;185:568–576. doi: 10.1111/j.1469-8137.2009.03076.x. [DOI] [PubMed] [Google Scholar]
- White F.F., Yang B. Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol. 2009;150:1677–1686. doi: 10.1104/pp.109.139360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson U.E., Coffey M.D. Cytological evaluation of general resistance to phytophthora infestans in potato foliage. Ann. Bot. 1980;45:81–90. [Google Scholar]
- Xiang Y., Cao Y., Xu C., Li X., Wang S. Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor. Appl. Genet. 2006;113:1347–1355. doi: 10.1007/s00122-006-0388-x. [DOI] [PubMed] [Google Scholar]
- Xu Z., Xu X., Gong Q., Li Z., Li Y., Wang S., Yang Y., Ma W., Liu L., Zhu B. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant. 2019;12:1434–1446. doi: 10.1016/j.molp.2019.08.006. [DOI] [PubMed] [Google Scholar]
- Yang B., Sugio A., White F.F. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U S A. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., White F.F. Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant Microbe Interact. 2004;17:1192–1200. doi: 10.1094/MPMI.2004.17.11.1192. [DOI] [PubMed] [Google Scholar]
- Yang B., Zhu W., Johnson L.B., White F.F. The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U S A. 2000;97:9807–9812. doi: 10.1073/pnas.170286897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Chen J., Zeng L., Goh M., Leung H., Khush G.S. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant Microbe Interact. 2000;13:869–876. doi: 10.1094/MPMI.2000.13.8.869. [DOI] [PubMed] [Google Scholar]
- Yoshimura S., Yamanouchi U., Katayose Y., Toki S., Wang Z., Kono I., Kurata N., Yano M., Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U S A. 1998;95:1663–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zhang H., Li F., Ouyang Y., Yuan M., Li X., Xiao J., Wang S. Multiple alleles encoding atypical NLRs with unique central tandem repeats in rice confer resistance to Xanthomonas oryzae pv. oryzae. Plant Commun. 2020;1:100088. doi: 10.1016/j.xplc.2020.100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Coaker G., Zhou J., Dong X. Plant immune mechanisms: from reductionistic to holistic points of view. Mol. Plant. 2020;13:1358–1378. doi: 10.1016/j.molp.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yin Z., White F.F. TAL effectors and the executor R genes. Front. Plant Sci. 2015;6:641. doi: 10.3389/fpls.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang J., Pan J., Gu Z., Chen X., Jin Y., Liu F., Zhang H., Ma B. Identification and molecular mapping of the rice bacterial blight resistance gene allelic to Xa7 from an elite restorer line Zhenhui 084. Eur. J. Plant Pathol. 2009;125:235–244. [Google Scholar]
- Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J.S., Huang S., Liu S., Vera Cruz C., Frommer W.B. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015;82:632–643. doi: 10.1111/tpj.12838. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qian W., Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLos Pathog. 2010;6:e1000844. doi: 10.1371/journal.ppat.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Hou Bi-Huei., Lalonde Sylvie., Takanaga Hitomi., Hartung Mara L., Qu Xiao-Qing., Guo Woei-Jiun., Kim Jung-Gun., Underwood William., Chaudhuri Bhavna., Chermak Diane., Antony Ginny., White Frank F., Somerville Shauna C., Mudgett Mary Beth., Frommer Wolf B. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M.J., Bogdanove A.J A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- White F.F., Yang B Host and pathogen factors controlling the rice-Xanthomonas oryzae interaction. Plant Physiol. 2009;150:1677–1686. doi: 10.1104/pp.109.139360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Bartley L.E., Chen X., Dardick C., Chern M., Ruan R., Canlas P.E., Ronald P.C OsWRKY62 is a negative regulator of basal and Xa21-mediated defense against Xanthomonas oryzae pv. oryzae in rice. Mol Plant. 2008;1:446–458. doi: 10.1093/mp/ssn024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.