Significance
The microbiome plays key roles in human health, but little is known about its evolution. We investigate the evolutionary history of the African hominid oral microbiome by analyzing dental biofilms of humans and Neanderthals spanning the past 100,000 years and comparing them with those of chimpanzees, gorillas, and howler monkeys. We identify 10 core bacterial genera that have been maintained within the human lineage and play key biofilm structural roles. However, many remain understudied and unnamed. We find major taxonomic and functional differences between the oral microbiomes of Homo and chimpanzees but a high degree of similarity between Neanderthals and modern humans, including an apparent Homo-specific acquisition of starch digestion capability in oral streptococci, suggesting microbial coadaptation with host diet.
Keywords: dental calculus, microbiome, Neanderthal, primate, salivary amylase
Abstract
The oral microbiome plays key roles in human biology, health, and disease, but little is known about the global diversity, variation, or evolution of this microbial community. To better understand the evolution and changing ecology of the human oral microbiome, we analyzed 124 dental biofilm metagenomes from humans, including Neanderthals and Late Pleistocene to present-day modern humans, chimpanzees, and gorillas, as well as New World howler monkeys for comparison. We find that a core microbiome of primarily biofilm structural taxa has been maintained throughout African hominid evolution, and these microbial groups are also shared with howler monkeys, suggesting that they have been important oral members since before the catarrhine–platyrrhine split ca. 40 Mya. However, community structure and individual microbial phylogenies do not closely reflect host relationships, and the dental biofilms of Homo and chimpanzees are distinguished by major taxonomic and functional differences. Reconstructing oral metagenomes from up to 100 thousand years ago, we show that the microbial profiles of both Neanderthals and modern humans are highly similar, sharing functional adaptations in nutrient metabolism. These include an apparent Homo-specific acquisition of salivary amylase-binding capability by oral streptococci, suggesting microbial coadaptation with host diet. We additionally find evidence of shared genetic diversity in the oral bacteria of Neanderthal and Upper Paleolithic modern humans that is not observed in later modern human populations. Differences in the oral microbiomes of African hominids provide insights into human evolution, the ancestral state of the human microbiome, and a temporal framework for understanding microbial health and disease.
The oral cavity is colonized by one of the most diverse sets of microbial communities of the human body, currently estimated at over 600 prevalent taxa (1). Dental diseases, such as caries and periodontitis, remain health burdens in all human populations despite hygiene interventions (2, 3), and oral microbes are often implicated in extraoral inflammatory diseases (4, 5). To date, most oral microbiome research has focused on clinical samples obtained from industrialized populations that have daily oral hygiene routines and access to antibiotics (1, 6), but far less is known about the global diversity of the oral microbiome, especially from diverse past and present nonindustrialized societies (7). The oral cavity contains at least six distinct habitats, but dental biofilms, including both supra- and subgingival dental plaque, are among the most diverse and clinically important (1, 6, 8). During life, these dental biofilms naturally and repeatedly calcify, forming dental calculus (tooth tartar) (9), a robust, long-term record of the oral microbiome (10). Archaeological dental calculus has been shown to preserve authentic oral bacterial metagenomes in a wide range of historic and prehistoric populations and up to 50 thousand years ago (ka) (10–13). As such, dental calculus presents an opportunity to directly investigate the evolution of the hominid microbiome and to reconstruct ancestral states of the modern human oral microbiome. In addition, because research has shown that evolutionary traits, diet, and cultural behaviors shape modern human microbiome structure and function at other body sites, such as the gut and skin microbiomes (14–18), investigating ancient oral metagenomes has the potential to reveal valuable information about major events in modern human evolution and prehistory, such as predicted dietary changes during the speciation of Homo (19–21) and the direct interaction of Neanderthals and modern humans during the Late Pleistocene (22).
To better understand the evolutionary ecology of the African hominid microbiome, we generated and analyzed 109 dental calculus metagenomes from present-day modern humans (n = 8), gorillas (Gorilla, n = 29), chimpanzees (Pan, n = 20), Neanderthals (n = 13), and two groups of archaeological modern humans associated with major lifestyle transitions (preagricultural, n = 20; preantibiotic, n = 14), as well as New World howler monkeys (n = 5) for comparison (SI Appendix, Fig. S1). To account for potential sampling biases, we analyzed multiple subspecies and populations of each African great ape genus, which were obtained from C20th or C21st-collected museum collections, and for modern humans we sampled multiple populations from both Africa and Europe. To this, we added previously published microbiome data from chimpanzees (n = 1) (13), Neanderthals (n = 4) (13), and present-day modern humans (n = 10) (23), for a total dataset of 124 individuals (Fig. 1A, SI Appendix, Table S1, and Dataset S1). We also generated eight new radiocarbon dates for archaeological individuals, for a total of 44 directly or indirectly dated ancient individuals in this study (Dataset S1).
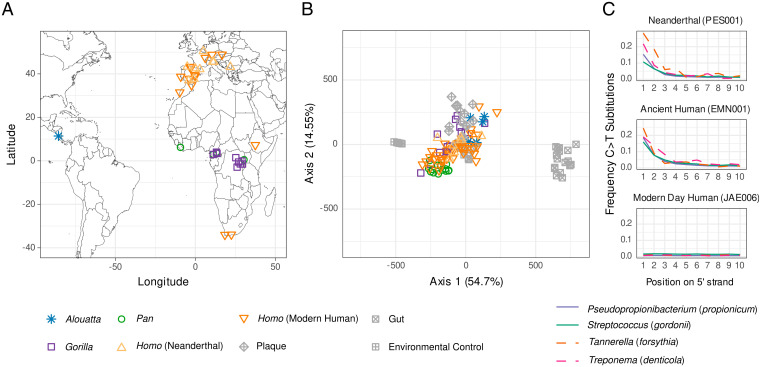
Fig. 1.
Sample locations and oral microbiome authentication of ancient dental calculus. (A) Sample locations. (B) PCoA comparing euclidean distances of microbial genera of well-preserved ancient and present-day dental calculus to environmental proxy controls (degraded archaeological bone) and present-day dental plaque and feces. Ancient dental calculus is distinct from gut and archaeological bone but overlaps with present-day dental plaque. (C) Representative DNA damage patterns for Neanderthals and ancient and present-day modern humans for four oral-specific bacterial species. The Neanderthal and upper Paleolithic modern human individuals show expected damage patterns consistent with authentic aDNA, whereas the present-day individual does not. See also SI Appendix, Fig. S4.
Here, we investigate the structure, function, and core microbial members of the human oral microbiome within an evolutionary framework, seeking to determine whether a core microbiome can be defined for each African hominid group, whether the core is phylogenetically coherent, and whether some members of the core are specific to certain host groups. We test whether the oral microbiome of hominids reflects host phylogeny, finding that African hominid oral microbiota are distinguished by major taxonomic and functional differences that only weakly reflect host relationships and are likely influenced by other physiological, dietary, or behavioral factors. We compare the microbial profiles of Neanderthals and modern humans and, contrary to expectations (12, 13), find a high consistency of oral microbiome structure within Homo, regardless of geography, time period, or diet/lifestyle. We detect the persistence of shared genetic diversity in core taxa between Neanderthals and Upper Paleolithic humans prior to 14 ka, supporting a growing body of evidence for earlier admixture and interaction in Ice Age Europe (24, 25). Finally, we explore possible implications of our findings on Homo-associated encephalization (19, 26) and the role of dietary starch in human evolution (20, 21) by investigating the evolutionary history of amylase-binding capability by oral streptococci. We find that amylase binding is an apparent Homo-specific trait, suggestive of microbial coadaptation to starch-rich diets early in human evolution.
Results
Preservation of Oral Microbiota in Dental Calculus.
Authenticating ancient DNA (aDNA) preservation is a necessary and essential step for all paleogenomic studies. However, these methods have been underdeveloped for ancient microbiomes. Here, we apply a multistep procedure of both conventional and new methods to evaluate and validate oral microbiome preservation in our dataset (SI Appendix, Fig. S2). First, we applied a reference-based metagenomic binning of reads to the National Center for Biotechnology Information (NCBI) nucleotide (nt) database (27) (Dataset S2) and then developed and applied a method to assess the decay of the cumulative percentage of known oral taxa in samples compared to a panel of oral and nonoral reference metagenomes (SI Appendix, Fig. S3A and section S3.4.1). This allowed us to remove samples that did not exhibit a taxonomic composition consistent with an oral origin. We then cross validated these results using SourceTracker (28) (SI Appendix, Fig. S3B) and inspection by principal coordinate analysis (PCoA, SI Appendix, Fig. S5 A–D). To samples exhibiting good oral microbiome preservation (Fig. 1B), we then applied the R package decontam (29) to detect and remove putative laboratory and environmental contaminants prior to downstream analysis (SI Appendix, section S3.6). Next, we examined each dataset and confirmed the presence of DNA damage characteristics of ancient samples, including short fragment lengths and elevated levels of cytosine to thymine deamination (Fig. 1C and SI Appendix, Fig. S4) (30). Finally, to reduce potentially spurious assignments for compositional analysis, we removed low-abundance taxa using thresholds optimized at different taxonomic levels (SI Appendix, Figs. S7 and S8 and sections S3.6 and S5.2). The resulting 89 well-preserved dental calculus datasets consist of samples ranging from the present day up to 100 ka.
The Core African Hominid Oral Microbiome.
We performed PCoA on our dataset of well-preserved samples and found considerable overlap in the microbial composition of African hominid dental calculus, as well as howler monkeys (Fig. 1B), suggesting the existence of a core microbiome that has been maintained for more than 8 My, based on fossil and molecular evidence of host divergence among African hominids (31, 32), and possibly since before the catarrhine–platyrrhine split ca. 40 Mya (33, 34). At the same time, small but significant differences were indicated by Permutational Multivariate Analysis of Variance (PERMANOVA) (35) at both the microbial genus and species levels between each hominid genus (100 bootstrap replicates, ɑ = 0.05; genus: F = 5.22 ± 1.42, df = 3, R2 = 0.27 ± 0.05, P = 0.001; species: F = 6.67 ± 2.52, df = 3, R2 = 0.32 ± 0.07, P = 0.001; SI Appendix, Fig. S5), and this pattern remained robust after controlling for unequal sample sizes (SI Appendix, section S4.2).
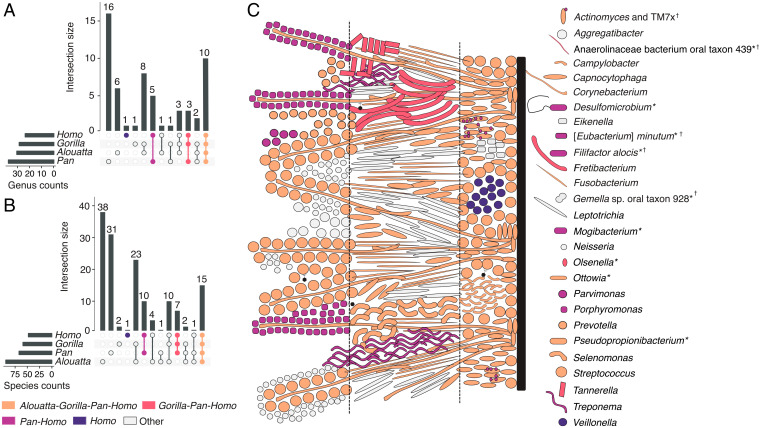
Dental plaque biofilms in humans form by the microbial succession of early, bridging, and late colonizers (36), and in contrast to the gut, which has high interindividual variability at the microbial phylum level (37) and is sensitive to subsistence changes over short and long timescales (15, 38, 39), oral microbial communities have been found to be more stable and consistent, particularly at the genus level (40–42), and even when challenged by antibiotics (43). Because of this, we sought to begin to define the African hominid core oral microbiome as a group and for each genus separately. For a microbial taxon to be considered “core” (44, 45), we required it to be present in at least two-thirds of the populations making up a given host genus, counting as present only those populations in which it is found in at least half of individuals to account for variation in preservation (SI Appendix, Fig. S9A and section S5.2). We then calculated the intersection of each core microbial genus (Fig. 2A) and species (Fig. 2B) across all host taxa (Dataset S3). Most “core” taxa are shared across all three African hominid genera (Gorilla, Pan, and Homo) and howler monkeys, whereas fewer are “core” only to African hominids (Gorilla, Pan, and Homo), Pan and Homo, or Homo (Fig. 2 and SI Appendix, Fig. S1). Despite smaller sample sizes than studies of present-day microbiomes, bootstrapping analysis to assess consistency of calculations supported most core microbiome assignments, with lower values possibly indicating taxa influenced by factors such as biofilm maturity (SI Appendix, section S5.3). This suggests a high degree of genus-level microbial taxonomic conservation during African hominid, and possibly broader primate, host evolution and speciation.
Fig. 2.
Core oral microbiome of African hominids shows a deep evolutionary conservation of biofilm structure. UpSet plots showing the number of microbial genera (A) and species (B) core to host groups and group combinations. (C) Core taxa of the human oral microbiome (inclusive of all African hominid and howler monkey ranks). Human biofilm spatial organization based on refs. 8 and 100. Taxa are colored by the broadest host group for which they are core. “Other” taxa are those that fall into paraphyletic host groupings (e.g., Alouatta:Homo). Dashed lines separate the biofilm into basal, intermediate, and peripheral regions (100). Taxa with unknown spatial location are marked with an asterisk (*); taxa core to Homo with any combination of other host genera at the species level but not at the genus level are marked with a dagger (†). Reference Dataset S3 for additional information.
Core taxa at both the genus and species levels include well-known members of each stage of plaque biofilm formation (8, 36), including the early colonizers Streptococcus and Actinomyces, the bridging taxa Fusobacterium and Corynebacterium, and the late colonizers Porphyromonas and Treponema, although the latter two are “core” to only chimpanzees and Homo (Fig. 2C). Major periopathogens, bacteria associated with periodontal disease, are found among the different host core combinations, and, focusing on Porphyromonas and Tannerella specifically because of their clinical significance today, we find that their major virulence factors are shared across multiple primates and thus are not specific to modern humans (SI Appendix, Fig. S9 B and C). The presence of periopathogens within the core microbiome supports the hypothesis that they are not pathogens in a conventional sense but rather that their pathogenic character in present-day humans may be related to an imbalance between the biofilm and the host, as has been suggested by recent ecological studies (46). Although some of the African hominid “core” taxa are periopathogens or their close relatives, most core members are known today to play important structural and functional roles in the formation and maturation of plaque, implying deep coevolutionary relationships between these taxa and their hosts.
African Hominid Oral Microbiome Structure Shows a Weak Relationship with Host Phylogeny.
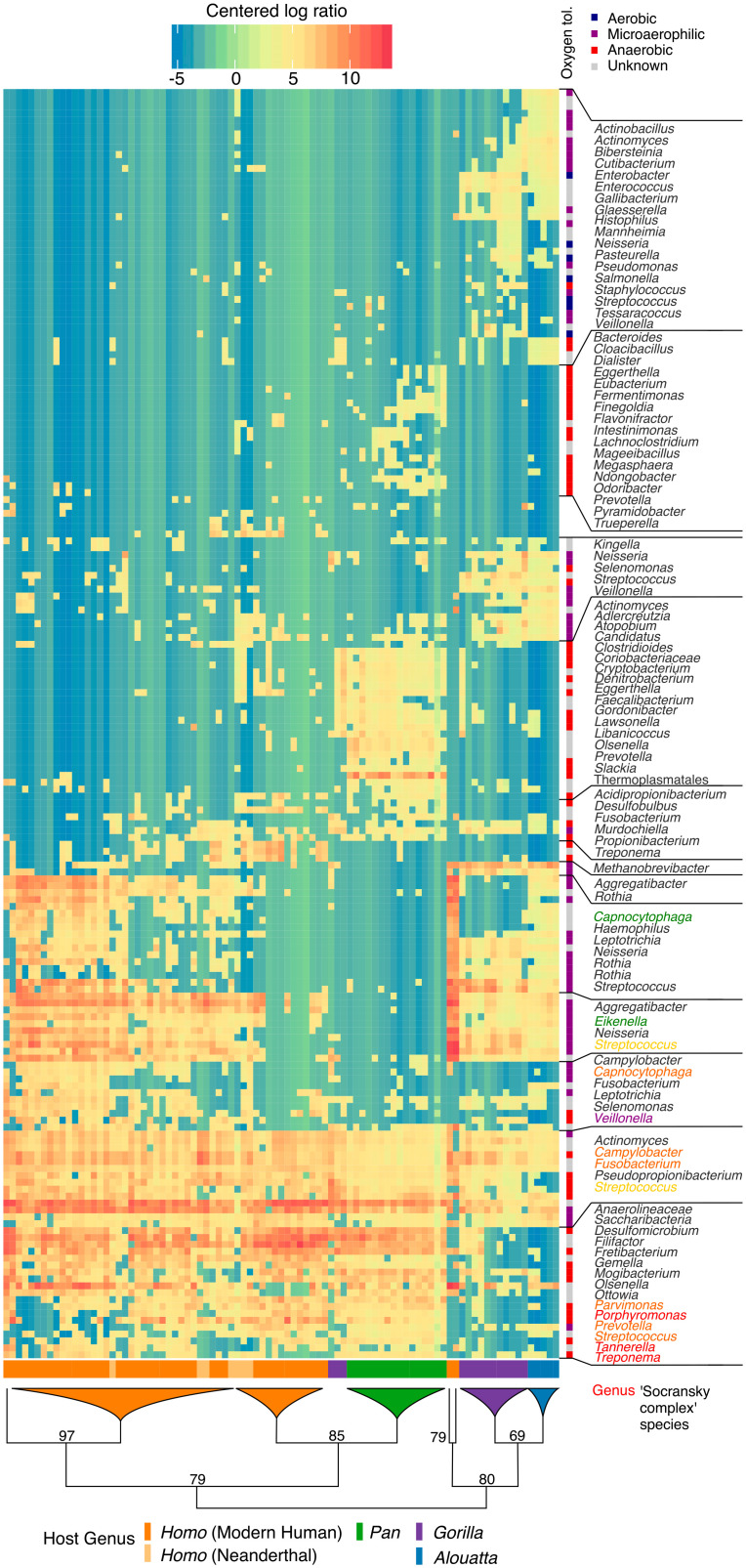
Hierarchical clustering shows that calculus metagenomes tend to cluster by host genus, confirming intragroup similarity, but these relationships exhibit differences from host phylogeny (Fig. 3). We find, for example, that howler monkeys and gorillas fall together in a single clade and a subset of Homo clusters with chimpanzees. With respect to the latter, available metadata do not provide any clear associations with factors such as geography, time period, or disease to explain this pattern (SI Appendix, section S4.3). Overall, gorillas and howler monkeys are characterized by a wide diversity of aerobic and facultatively anaerobic taxa, while chimpanzees have higher levels of obligately anaerobic taxa, including many putative periopathogens (e.g., Porphyromonas gingivalis, Treponema denticola, Tannerella forsythia, Filifactor alocis, and Fretibacterium fastidosum). Neanderthals consistently fall within the diversity of modern humans. Homo is notable for its high abundance of Streptococcus spp., while this genus is found at substantially lower levels in Pan.
Fig. 3.
African hominid dental calculus microbiomes cluster by host genus and other factors. Hierarchical clustering of howler monkeys, chimpanzees, gorillas, Neanderthals, and ancient and present-day modern humans based on species-level prokaryotic taxonomic assignments. Bacterial oxygen tolerance is associated with biofilm maturation stage in modern humans, and colored names indicate species corresponding to Socransky complexes (111) (reference SI Appendix section S5.1.1 for a summary). Microaerophilic is defined based on the BacDive database and is roughly synonymous to facultative anaerobe. The tree is schematic, and bifurcations are shown until all host genera are represented. Microbial species names are collapsed to genus level. Species and sample names can be located in SI Appendix, section S4.3.
Many of the taxa identified in human and nonhuman primate dental calculus are poorly characterized, making further exploration difficult. Indeed, several species within the human core genera remain unnamed (Ottowia sp. oral taxon 894, Olsenella sp. oral taxon 807) or understudied (Pseudopropionibacterium propionicum, F. fastidiosum) and some even lack genus designations ([Eubacterium] minutum, TM7x, Anaerolinaceae bacterium oral taxon 439). Their absence from most discussions of the modern human oral microbiome points to a major gap in current oral microbiology research, and targeted investigation of these species is needed to identify their functional and structural roles within plaque biofilms (47–49). Host genus patterns in community structure may be influenced by differences in salivary flow or composition (50), as well as differences in diet texture, quality, and nutrient content (51) (SI Appendix, sections S1, S5.1, and S5.6). We also investigated microbial community structure within modern humans, but in contrast to previous studies (13), we found no difference among broad dietary patterns or time periods (SI Appendix, Fig. S6 and section S4.5). These findings accord with the results of modern oral microbiome studies, which also show minimal, if any, broad and sustained compositional changes in response to diet (e.g., refs. 41, 52, and 53). The relative stability of the oral microbiome may be due in part to the extensive community interdependencies (54, 55) that have developed within these biofilms to metabolize complex host salivary glycoproteins, which are the major nutrient source for most members of the oral microbiota (56). This is in contrast to studies demonstrating strong associations between diet and taxonomic/functional composition in modern gut microbiomes (57, 58).
Evolutionary Histories of Oral Microbial Species Reflect Homo Interactions.
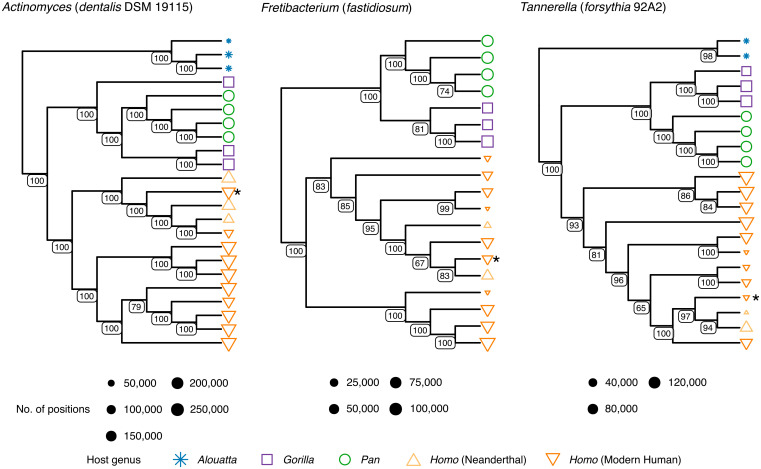
We next examined the phylogenies of individual microbial taxa to determine if host evolutionary relationships are reflected at the microbial genome level. To improve genome coverage and reduce potential noise from DNA damage, we selected a representative subset of well-preserved calculus samples across all host genera (n = 19; SI Appendix, Table S1 and Dataset S1) and constructed uracil-DNA glycosylase-treated (UDG) libraries to remove deaminated cytosines (59), which we then deeply sequenced and analyzed together with a subset of four of the present-day modern humans. Genome-level sequence reconstruction from diversity-rich ancient microbiomes is challenging due to both the highly fragmented nature of aDNA and the low relative abundance of each species, which makes strain separation difficult (SI Appendix, section S6). Furthermore, a lack of sufficient reference genomes for many commensal oral microbes increases mismapping and noise artifacts when identifying single-nucleotide polymorphisms (SNPs) (SI Appendix, Fig. S10). Nevertheless, despite these challenges and using representative genomes from core taxa, we were able to reconstruct phylogenetic trees with high bootstrap support on internal nodes for eight oral bacteria (Fig. 4 and SI Appendix, Fig. S11).
Fig. 4.
African hominid oral taxa cluster phylogenetically by host genus. Selected neighbor-joining SNP-based phylogenetic cladograms of representative core oral microbiome genomes from deep-sequenced calculus metagenomes (SI Appendix, section S6.6). Actinomyces and Tannerella trees are rooted on the branch leading to howler monkeys (Alouatta, blue), Fretibacterium tree is midpoint rooted. Positions refer to non-N nucleotide calls in the alignment. Node values represent node support out of 100 bootstrap replicates. Asterisk (*) represents the Upper Paleolithic individual from El Mirón (EMN001), which consistently falls near Neanderthal individuals. The remaining eight trees, with tip labels, are provided in SI Appendix, Fig. S11.
As with compositional analysis, reconstructed genome-level sequences tend to cluster with those from the same host genus but do not closely reflect host phylogeny (Fig. 4 and SI Appendix, Figs. S11 and S12). Overall, genome-level sequences reconstructed from gorillas and chimpanzees fall closer to each other than do those of chimpanzees and Homo. Biases from the use of modern human-derived microbial reference genomes may in part contribute to this pattern, but microbial exchange due to overlapping territorial ranges of gorillas and chimpanzees throughout their evolution may also be a contributing factor. Within Homo, Neanderthals consistently group together, indicating shared within-species microbial diversity. However, we also note that the Upper Paleolithic individual from El Mirón in Iberia (18.6 ka) clusters in all trees with Neanderthals, rather than with other Pleistocene hunter-gatherers of the African Later Stone Age or more recent Holocene-era European or African populations. Recently published human genomic data including this individual has revealed that its associated genetic ancestry component was largely displaced across Europe after 14 ka (24, 60) during postglacial warming. Turning to our low-coverage metagenomic datasets, we assessed additional European Upper Paleolithic and Mesolithic groups (SI Appendix, section S6.6) and found that they show a similar pattern (albeit at lower resolution), with the oral taxa of individuals dated to before 14 ka mostly falling with Neanderthals and those after 14 ka mostly clustering with present-day modern humans (24, 60). This pattern suggests that the reconstructed oral bacterial genomes from El Mirón reflect a standing microbial diversity in Homo that was present in Europe during the Middle and Upper Paleolithic, but which was later replaced following subsequent migrations of modern human populations from elsewhere. Because oral microbiota are primarily inherited through caregivers (61, 62), additional sampling and ultradeep sequencing of Paleolithic European and Asian dental calculus may prove informative about the poorly understood interaction dynamics between archaic and modern humans.
Homo-Specific Shifts in Oral Biofilm Are Linked to Dietary Starch Availability.
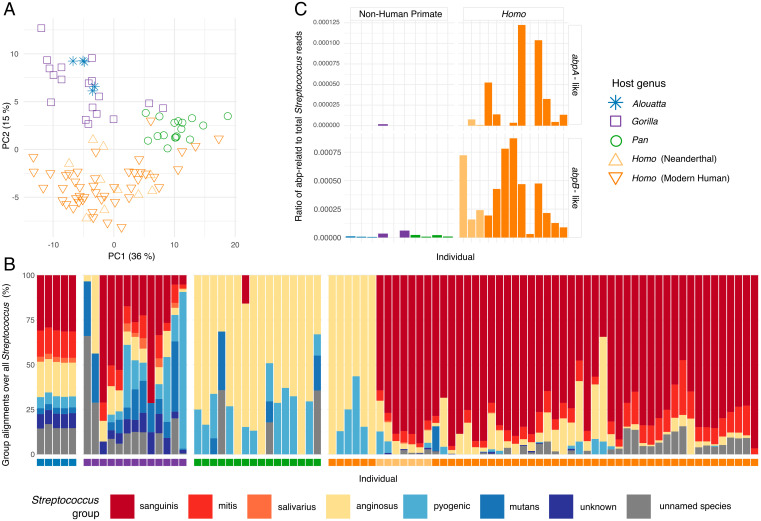
The metabolic potential of a microbial community, which is inferred from its total gene content, can offer insights into biofilm ecology and function that cannot be understood from taxonomy alone. To better characterize the metabolic and functional differences among hominid oral biofilms, we compared the gene content of dental calculus metagenomes from well-preserved samples of the larger sequencing dataset using two different methods of functional classification, HUMAnN2 (63) and AADDER (64), and found moderate concordance in overall results. Principal Components Analysis (PCA) of the protein-level functional assignments cluster host genera distinctly with a high degree of separation between hosts and functional content (Fig. 5A and SI Appendix, Figs. S13 and S14), whereas we observe only a moderate degree of separation in the taxonomic PCoA (Fig. 1B), suggesting that gene content of the taxa shared by hominids is more host-specific than taxonomic assignments, a pattern that has also been seen for other microbial systems (65). The genes that drive separation of Homo from nonhuman primates consistently relate to carbohydrate processing (SI Appendix, Fig. S14), are much more abundant in Homo, and largely derive from Streptococcus (SI Appendix, Fig. S13), something also observed in primate gut microbiomes (66). We therefore investigated the distribution of Streptococcus across our samples (Fig. 5B) using a classification system based on biochemical characteristics and genetic relatedness (67). We find that Streptococcus species belonging to the Mitis, Sanguinis, and Salivarius groups are dominant in Homo, while these same groups are effectively absent in chimpanzees, and nonhuman primates in general are characterized by much higher proportions of Streptococcus species in the Anginosus, Mutans, and Pyogenic groups (Fig. 5B).
Fig. 5.
Metabolic function and Streptococcus amylase-binding gene content is distinct between African hominid oral microbiomes. (A) PCA of microbial gene functions (SEED classification) clusters well-preserved samples by host genus (PERMANOVA R2 = 0.345). Homo is functionally distinct from nonhuman African hominids and howler monkeys, particularly with respect to carbohydrate metabolism (SI Appendix, Fig. S14). (B) Bar plot of proportion of alignments to different Streptococcus groups show differences between host genera. Color of squares below bars corresponds to legend in C. Amylase-binding activity has been observed among members of the Sanguinis, Mitis, and Salivarius groups (68). (C) Ratios of reads aligning to amylase-binding-protein annotated sequences versus a genus-wide Streptococcus “superreference” show higher values in Homo than nonhuman primates, based on a deep-sequenced subset of samples and four present-day modern humans. Note the ratio on the y-axes of abpA and abpB are scaled differently.
The Mitis, Sanguinis, and Salivarius groups are notable for their ability to express amylase-binding proteins to capture salivary ɑ-amylase (68, 69), which they use for their own nutrient acquisition from dietary starch, as well as dental adhesion (70, 71). Amylase-binding protein genes (e.g., abpA and abpB) share no homology but rather confer a similar phenotype through convergent evolution, and they are found almost exclusively in oral Streptococcus species (68). Alpha-amylase is the most abundant enzyme in modern human saliva and modern humans express it at higher levels than any other hominid (50, 72). In contrast to most other nonhuman primates, modern humans exhibit high salivary ɑ-amylase (AMY1) copy number variation, with a reported range of up to 30 diploid copies (16, 73, 74). This copy number expansion is estimated to have occurred along the modern human lineage after the divergence from Neanderthals in the Middle Pleistocene (75, 76). It has been argued this increase relates to dietary shifts during the evolutionary history of modern humans and specifically to an increased reliance on starch-rich foods (20, 21).
We next calculated the ratio of reads aligning to abpA and abpB sequences compared to all Streptococcus reads in the deep-sequenced dataset. We find that abpA and abpB reads are nearly absent in the nonhuman groups but are prevalent and significantly more abundant in Homo (Mann–Whitney U test Homo versus non-Homo: abpB, α = 0.05, U = 128, P = < 0.001, 95% CI = 0.686 to 0.851; abpA, α = 0.05, U = 112, P = < 0.001, 95% CI = 0.398 to 0.861). In particular, abpB is present in all deeply sequenced Homo individuals, and abpA is especially prevalent in modern humans (Fig. 5C). This suggests that oral streptococci evolved in association with changes in host diet and supports an early importance of starch-rich foods in Homo evolution.
Discussion
Commensal microbes of the oral microbiome represent an underutilized and independent source of information about host evolutionary and ecological differences (15, 77). With generation times orders-of-magnitude shorter than their hosts and the ability to acquire new functions through horizontal gene transfer across distantly related groups, microbes are a particularly dynamic and temporally resolved system for understanding human evolution. After applying a rigorous strategy to identify, decontaminate, and authenticate well-preserved dental calculus specimens up to 100 ka, we identify a core group of 10 bacterial genera within the African hominid primate oral microbiome that are also shared with howler monkeys, suggesting that these microbial groups have played a key role in oral biofilms since before the catarrhine–platyrrhine split ca. 40 Mya (33, 34). Today, these core taxa are primarily involved in providing structural support within the dental plaque biofilm, and their study holds promise for understanding biofilm growth and maturation in the ancestral human microbiome (78, 79). Identifying the role of such taxa is critical for the successful long-term treatment, prevention, and control of dysbiotic biofilms, such as those found in dental and periodontal diseases (80).
Further, we identify 27 genus-level members of the Homo core oral microbiome, and these include many well-known and clinically relevant taxa, such as Streptococcus and the periopathogens P. gingivalis, T. forsythia, and T. denticola; however, nearly all of these are also core microbiome members of other African hominids. Only Veillonella parvula, a commensal species known to have a synergistic relationship with the cariopathogen Streptococcus mutans (81), is primarily found in humans. Surprisingly, not all members of the core Homo oral microbiome are well-known—three have no genus designation and several lack species names, revealing a major gap in oral microbiology research that in part relates to the difficulties in growing and propagating these microbes.
Focusing on oral microbiome evolution within Homo, we reconstruct authentic oral metagenomes of Neanderthals dating up to 100 ka and modern humans dating up to 30 ka, finding a high degree of similarity in microbial community structure, while also documenting indications of strain-level differences within core taxa. Interestingly, we find that Neanderthal-associated strain-level sequence variants are consistently present in Upper Paleolithic Europeans but not afterward, which accords with a described modern human genomic turnover around 14 ka (24, 60). Comparing human and nonhuman primates, we show that within Streptococcus, amylase-binding groups play a central role in the oral biofilms of Homo, likely aided by both their enhanced ability to colonize the dentition and their exclusive access to dietary starches. These Streptococcus groups and abpB are a general feature of Homo, suggesting that starch-rich foods, possibly modified by cooking (20) (SI Appendix, section S5.8), first became important early in Homo evolution prior to the split between Neanderthal and modern human lineages more than 600 ka (82, 83), a finding with potential implications for the energetics of Homo-associated encephalization (19–21, 26). Subsequent copy number expansion of AMY1 in the modern human genome and the rise of abpA in oral streptococci may signal an even greater reliance on starch-rich foods by modern humans.
Further research on the evolution of abpA, abpB, and other amylase-binding proteins, including phylogenetic reconstruction and demographic modeling, promises to refine questions regarding biofilm formation and the nature and timing of dietary change in Homo. In addition, future research on non-African hominids (orangutans) and additional catarrhines, in particular cercopithecenes with high or unusual salivary amylase expression, such as gelada and hamadryas baboons (73, 84, 85), may yield further insights into the diverse evolutionary trajectories of primate oral microbiomes in response to habitat and dietary change. In addition, it is clear that more research on core genera is urgently needed, as many of the highly conserved and potentially key structural taxa in hominid oral biofilms are understudied and even lack formal names. Furthermore, future sequencing projects focusing on within-species genomic diversity will be critical to understanding microbiome evolution and coadaptation within the human lineage. This study demonstrates that integrating evolutionary studies of the modern human microbiome with wild primate and ancient Homo metagenomic data provides valuable insights into the ancestral states of the human oral microbiome, the nature of microbial–host relationships, and major events in the evolution of modern humans and Neanderthals.
Materials and Methods
Materials.
Our sampling strategy aimed to collect dental calculus from a minimum of two independent populations, each consisting of at least five individuals, for each host genus and modern human lifestyle group (excepting Alouatta) (SI Appendix, Table S1 and Dataset S1). Dental calculus was sampled from twentieth-century skeletal remains of wild Alouatta (A. palliata), Gorilla (G. berengei beringei; G. berengei graueri; G. gorilla gorilla), and Pan (P. troglodytes schweinfurthii; P. troglodytes ellioti; P. troglodytes verus) and from archaeological Neanderthals and modern humans using established protocols (DOIs: 10.17504/protocols.io.7vrhn56 and 10.17504/protocols.io.7hphj5n). Although many present-day human dental plaque datasets are publicly available, they have been shown to not be directly comparable to dental calculus (23), and consequently we generated dental calculus data for present-day humans. The study of deidentified present-day dental calculus was approved by the Institutional Review Board for Human Research Participant Protection at the University of Oklahoma (IRB no. 4543). All samples were collected under informed consent during routine dental cleaning procedures by practicing dental odontologists. For additional sample context descriptions and additional ethical approval information, reference SI Appendix, section S2.1.
Laboratory Methods.
For all museum and field station samples, we performed DNA extraction in dedicated cleanroom facilities using a protocol optimized for the recovery of degraded and fragmentary DNA (86). Present-day calculus was extracted as previously described (23). For all samples, DNA was built into dual-indexed Illumina libraries (87) and shotgun sequenced. In addition, a subset of samples were separately subjected to UDG treatment (88), followed by deep sequencing. Negative controls were included in all extraction and library construction batches. Sequencing was performed on either Illumina NextSeq. 500 or HiSeq. 4000 platforms. For details, reference SI Appendix, section S2.2–S2.4 and protocols.io under DOI: 10.17504/protocols.io.bq7wmzpe.
Data Processing and Quality Filtering.
For detailed descriptions of preprocessing and analysis procedures, including code, reference SI Appendix and external data repository (GitHub repository: https://github.com/jfy133/Hominid_Calculus_Microbiome_Evolution; Archive DOI: 10.5281/zenodo.3740493). Additional ancient (13) and present-day dental calculus (23) data from previous studies were downloaded from the Online Ancient Genome Repository (OAGR) (https://www.oagr.org.au/) and the European Bioinformatics Institute (EBI) European Nucleotide Archive (ENA) (https://www.ebi.ac.uk/ena/) databases, respectively. Comparative metagenomes from present-day modern human microbiome and environmental sources were additionally downloaded from the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/). Accession numbers and download instructions for all FASTQ files are provided in SI Appendix, section S3.1. The EAGER pipeline (89) was used to perform initial preprocessing of sequencing data to remove possible modern human DNA sequences that can interfere with taxonomic profiling (due to present-day modern human DNA contamination in microbial reference genomes). We used relaxed bwa aln (90) mapping parameters for aDNA (−n 0.01), and nonhuman reads from replicate samples and libraries were then concatenated per individual. Human-mapped sequences were then poly-G clipped prior to reporting of mapping statistics. Processing statistics are provided in SI Appendix, section S3.2.
Taxonomic Binning and Preservation Assessment.
For taxonomic binning, we used the aDNA-optimized high-throughput aligner MALT (27, 91) together with the NCBI nt database (October 2017; uploaded to Zenodo under DOI: 10.5281/zenodo.4382154) and a custom NCBI RefSeq database (containing bacteria, archaea, and Homo sapiens, October 2018, SI Appendix, section S3.3) and employed a relaxed percent identity parameter of 85% and a base tail cut off (“minimum support”) of 0.01%. Resulting RMA6 files were loaded into MEGAN6 CE (64) and prokaryotic Operational Taxonomic Unit (OTU) tables were exported (Dataset S2). A comparison of the two databases is provided in SI Appendix, section S3.3. Given the challenges of low preservation and contamination in ancient microbiome studies, we performed a multistep procedure to screen for and remove poorly preserved samples and contaminant OTUs from the non-UDG-treated dataset (SI Appendix, Fig. S2). We developed a visualization for the identification of calculus samples with weak oral microbiome signatures (SI Appendix, Fig. S3). This procedure involves comparing identified taxa to their previously reported isolation source(s), ranking these taxa from most to least abundant, and tracking the cumulative percentage of oral taxa along this rank (termed here as “decay”). Samples with a low percentage of oral taxa after an initial “burn-in” based on stabilization of curve fluctuation were removed from downstream analysis. Reference SI Appendix, section S3.4 for details. We compared this method to results obtained using SourceTracker (28)—which was performed on 16S-mapped reads filtered from shotgun data using EAGER (with comparative present-day modern human and environmental metagenomes as sources), followed by closed-reference clustering using QIIME (92)—and found concordance between the two methods (SI Appendix, Fig. S3). We next used the R package decontam (29) to statistically detect putative laboratory and environmental contaminants (as present in negative controls and a set of archaeological bone samples—SI Appendix, section S3.6), which were then removed prior to downstream analysis. To authenticate the remaining OTUs, we utilized the output of MaltExtract (93) in the MaltExtract-Interactive Plotting App (MEx-IPA) tool (DOI: 10.5281/zenodo.3380011), which we developed for rapid visualization of characteristic aDNA patterns, such as cytosine to thymine deamination, short fragment lengths, and edit distance from reference (SI Appendix, Fig. S4 and section S3.5). After mapping with EAGER to well-known oral taxa, we also validated DNA damage patterns using DamageProfiler (Fig. 1C) (94).
Microbial Compositional Analysis.
To remove low-abundance environmental contaminants or spurious hits, we selected a minimum abundance cutoff of 0.07% of alignments for genus-level and 0.04% of alignments for species-level identifications (SI Appendix, Figs. S7 and S8 and section 5.2). We normalized profiles through phylogenetic isometric-log-ratio transformation (95) of the abundance-filtered OTU tables and then performed PCoA on the resulting euclidean distances (SI Appendix Fig. S5 C and D and SI Appendix, section S4.1). To statistically verify host genus clusters, we used the adonis function from the R package vegan to perform PERMANOVA (35) analysis after controlling from unequal sample sizes (SI Appendix, section S4.2). After removal of poorly preserved samples, oral communities show distinct centroids for each host genus (bootstrapped PERMANOVA, ɑ = 0.05, P = 0.001, pseudo-F = 5.23, R2 = 0.28); Alouatta was excluded due to small sample size. We performed bootstrapped hierarchical clustering (96) on the euclidean distances of centered log ratio–transformed OTU tables and visualized the results in the form of a heatmap (Fig. 3 and SI Appendix section S4.3). Sample and taxon clustering was performed with the McQuitty hierarchical clustering algorithm, and taxon blocks within the heatmap were selected by visual inspection. Bootstrap values of sample clusters were estimated through the R package pvclust (96). Species oxygen-tolerance metadata was obtained from the BacDive database (97) via the BacDiveR R package (DOI: 10.5281/zenodo.1308060). For validation of the observations made on the heatmaps, we also performed grouped indicator analysis (98) (SI Appendix, section S4.4). Clustering of human oral microbiomes by variables such as time, geography, and dietary subsistence was assessed using PCoA, PERMANOVA, and hierarchical clustering (SI Appendix, section S4).
Core Microbiome Analysis.
Using the contaminant-filtered OTU tables of well-preserved samples, we converted all taxa above the minimum support threshold to a presence/absence profile. Taxa were required to be present in at least half (50%) of the members of a population for it to be considered core to the population and to be present in at least two-thirds (66%) of populations to be considered core to a host group (SI Appendix, Fig. S9; reference SI Appendix, section S5.2 for parameter experimentation details). We then generated UpSet plots (99) to visualize the microbial intersections of each host group at both the species and genus levels (Fig. 2 A and B), and we also compared the results between both databases. Further discussion on the exclusion of the common soil genus Mycobacterium from core genera is provided in SI Appendix, section S5.2. Validation of results through smaller sample sizes was carried out by bootstrapping analysis, which was performed by randomly subsampling (with replacement) individuals from each host genus and rerunning the core calculation procedure to 1,000 replicates (SI Appendix, section S5.3). We created a diagram of the core human oral microbiome (Fig. 2C) based on published fluorescence in situ hybridization (FISH) images of human dental plaque (8, 100). For species/genera that were not analyzed in these publications, literature searches were performed to find evidence of their localization within plaque based on immunohistochemistry, immunofluorescence, or FISH (SI Appendix, sections S5.1 and S5.4). All members of the human core microbiome are shown, including those also shared with other African hominids and howler monkeys. For further details, reference SI Appendix, section S5.3.
Genomic Analysis.
We used EAGER to map (see below for more details) the deep-sequenced UDG-treated dataset and four samples from present-day individuals (Alouatta, 3; Gorilla, 3; Pan, 4; Neanderthal, 3; ancient modern human, 6; present-day modern human, 4; total: 23) against the reference genomes of Tannerella forsythia and Porphyromonas gingivalis (SI Appendix, section S5.5). We used bedtools (101) to calculate the breadth and depth coverage of a set of known virulence factors for these two taxa. To reduce the risk of spurious alignments (e.g., from cross mapping of conserved sequences), we filtered out genes that had a breadth of coverage less than 70% and/or that appeared to have strongly different coverage depths compared to the rest of the genome (reference SI Appendix, section S5.5 for more details). The resulting genes were visualized as a heatmap for comparison (SI Appendix, Fig. S9). We selected all species-level Streptococcus alignments from the shallow sequenced dataset minimum support filtered NCBI nt–MALT OTU tables and assigned them to one of eight species groups based on the literature (67) (reference SI Appendix, section S5.6 for group definitions). We then calculated the fraction of alignments for each species group over all taxonomic alignments for each sample (Fig. 5B). To further validate the results, we calculated a similar ratio but based on the mapping of the deep-sequenced dataset against a superreference of 166 Streptococcus genomes (see below). We identified abpA- and abpB-like gene coordinates from the superreference using panX (102), then extracted the number of reads mapping to these annotations and calculated the fraction of these reads over all Streptococcus superreference mapped reads. We then applied a Mann–Whitney U test to test the null hypothesis of no difference between the distributions of ratios of Homo and nonhuman primates, as well as compared these results to a distribution of P values of 100 randomly shuffled group assignments (reference SI Appendix, section S5.7 for more details). Reference sequences of abpA and abpB were extracted from Streptococcus genomes in RefSeq and indexed for mapping. All shallow sequencing dataset samples were mapped against all reference strains. For samples with a gene coverage of at least 40% at 1×, a consensus sequence was exported from the Integrative Genome Viewer (IGV) (103). An input file of the consensus sequences and references was generated in BEAUTi and used to run BEAST2 (104) for Bayesian skyline plot analysis. For details, reference SI Appendix, section S5.9.
Microbial Phylogenetics.
We first attempted a competitive-mapping strategy against genus-wide superreferences of identified core taxa (reference SI Appendix, sections S6.1 and S6.2), but this approach yielded only limited results (SI Appendix Fig. S10 and section S6.3). We then instead performed phylogenetic reconstruction by mapping the same dataset to a single representative genome for each genus, considered as representing a population of related taxa. To account for challenges with low-coverage ancient data, we called SNPs using MultiVCFAnalyzer and required each SNP call to have a minimum of 2× coverage and a support of ≥70% of reads (SI Appendix, section S6.5). The resulting FASTA alignments were loaded into R. Samples with fewer than 1,000 SNPs were removed, and pairwise distances were calculated based on the JC69 model (105). A bootstrapped neighbor-joining algorithm from the R package ape (106) was applied to the distance matrices with 100 replicates (SI Appendix, section S6.6). Trees were visualized with ggtree (107). Finally, we retained trees where the basal internal nodes had bootstrap supports of ≥70% (SI Appendix, Fig. S11). The same procedure was then applied to the shallow sequencing dataset with the additional samples described above in the main text (SI Appendix, Fig. S12). To test whether pre-14 ka individuals clustered with Neanderthals due to reference bias, we calculated the median number of positions that were shared between EMN001 and Neanderthals to a histogram of median pairwise comparisons between all modern human individuals (SI Appendix, section S6.6).
Functional and Metabolic Pathway Analysis.
We took two approaches to characterizing the functional profiles of the calculus metagenomes. First, we used HUMANn2 (63) [with MetaPhlAn2 (108) generated taxonomic profiles] to generate functional profiles based on the UniRef90 (109) and ChocoPhlAn (July 2018) (63) databases. Preservation was independently assessed for pathway abundance and Kyoto Encyclopedia of Genes and Genomes (KEGG) ortholog functional profiles, SI Appendix, section S7.1. We compared the functional profiles of well-preserved calculus between host groups using pathway abundance (n = 94) and gene families converted to KEGG orthologs (n = 109) using PCA (SI Appendix, Fig. S13). Orthologs with the strongest loadings were visualized with biplots (SI Appendix, Fig. S13 A–C), and the species from which these orthologs were derived were determined (SI Appendix, Fig. S13 B–D). The clustering of host genera in PCAs using only orthologs in specific pathways (carbohydrates, amino acids, lipids) was also explored (SI Appendix, Fig. S10 A–C). For details, reference SI Appendix, section S7.1.4. Second, we used AADDER (included within MEGAN6 CE) (64) to profile the number of alignments to annotations present in the custom RefSeq database as aligned by MALT (see above). We then used MEGAN6 to export SEED category (110) profiles. Preservation was independently assessed for SEED protein functional profiles, reference SI Appendix, section S7.2. We compared the functional profiles of well-preserved calculus (n = 95) between host groups using proteins but not higher-level pathways (SI Appendix, Fig. S13). The proteins with the strongest loadings were visualized using biplots (SI Appendix, Fig. S13 E–G), and the species from which these proteins were derived were determined (SI Appendix, Fig. S13 F–H). The clustering of host genera in PCAs using only proteins in specific pathways (carbohydrates, amino acids, lipids) was also explored (SI Appendix, Fig. S14 D–F). For details, reference SI Appendix, section S7.2.3.
Supplementary Material
Acknowledgments
We thank Ethiopia's Authority for Research and Conservation of Cultural Heritage (ARCCH), the Museu de Prehistòria de València, Marta Negra at the Burgos Museum, Ottmar Kullmer at the Naturmuseum Senckenberg, Lyman Jellema at the Cleveland Museum of Natural History, Reneé Molina of the Maderas Rainforest Conservancy at the Ometepe Biological Field Station, Daniela C. Kalthoff at the Swedish Museum of Natural History, Emmanuel Gilissen at the Royal Museum for Central Africa, Tom Geerinckx and Patrick Semal at the Royal Belgian Institute of Natural Sciences, and Abdeljalil Bouzouggar at the Institut National des Sciences de l'Archéologie et du Patrimoine (INSAP) for archaeological and museum collection assistance. We thank the Uganda Wildlife Authority for giving permission to carry out research on chimpanzees from Kibale National Park. We thank Miriam Carbo Toran, Alicia Hernández Fuster, Juan Bautista Rodriguez Martinez, and Eros Chaves for present-day calculus sampling assistance. We thank Dr. Dominique Henry-Gambier (University of Bordeaux I) for her initial examination of the mandible of Rigney 1. We thank Dr. Dawn Mulhern and Jaelle Brealey for consultation on nonhuman primate samples. We thank Rachel Carmody for suggestions on early drafts. We thank Alex Hübner for advice on data analysis. We thank Zandra Fagernäs, Richard Hagan, Maria Spyrou, and Antje Wissgott for their additional laboratory assistance. Research at the De Nadale Cave is coordinated by the University of Ferrara within the framework of a project supported by the Ministry of Culture–Western Veneto Archaeological Superintendence, the Soprintendenza Archeologia, belle Arti e Paesaggio per le Provincie di Verona, Rovigo e Vicenza (SABAP), and the Zovencedo Municipality, financed by the H. Obermaier Society, local private companies (RAASM and Saf), and local sponsors. The Calleva Foundation supported the excavation and research during which sampling of the Taforalt calculus was carried out. This project was funded by grants from the US National Science Foundation (NSF) (BSC-1516633 to C.W. and C.M.L.; BSC-1027607 to K.W.A, M.C.C., and J.W.A.; SBR-0416125 to R.W.W.), the US National Institutes of Health (NIH) (2R01 GM089886 to C.M.L., C.W., and K.S.; R37DE016937 and R01DE024468 to F.E.D.), the European Research Council (ERC) (ERC-STG 677576 “HARVEST” to A.G.H.; ERC-CG 617627 “ADaPt” to J.T.S.), the Deutsche Forschungsgemeinschaft (DFG FOR 2237 to K.H.; EXC 2051-390713860 to C.W.), the National Research Foundation of South Africa (NRF 115257 and 12081 to V.E.G.), the Natural Sciences and Engineering Research Council of Canada (RGPIN-2017-04702 and RGPIN-2019-04113 to M.R.), Czech National Institutional Support (RVO 68081758 to S.S.), the Ministry of Culture and Information and the Ministry of Education, Science and Technological Development of the Republic of Serbia (177023 to D.M.), Junta de Castilla y León (BU028A09 to J.C.D.F-L.), the Swedish Research Council Formas (2016-00835 and 2019-00275 to K.G.), the University of South Florida, the University of Oklahoma, the Werner Siemens Foundation (Paleobiotechnology to C.W.), and the Max Planck Society. Any opinions, findings, and conclusions expressed in this study are those of the authors and do not necessarily reflect the views of the granting agencies.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.R.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021655118/-/DCSupplemental.
Data Availability
All newly generated sequencing data have been deposited in the ENA repository (https://www.ebi.ac.uk/ena/browser/home) under project accession ID PRJEB34569. R notebooks, bioinformatic scripts, additional supporting figures, and intermediate analysis files are provided in an external data repository hosted on GitHub (http://github.com/jfy133/Hominid_Calculus_Microbiome_Evolution) and archived with Zenodo under DOI: 10.5281/zenodo.3740493.
Change History
May 12, 2021: The author line has been updated.
References
- 1.Dewhirst F. E., et al., The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kassebaum N. J., et al., Global burden of untreated caries: A systematic review and metaregression. J. Dent. Res. 94, 650–658 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Eke P. I., et al., Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J. Periodontol. 86, 611–622 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scannapieco F. A., Bush R. B., Paju S., Associations between periodontal disease and risk for nosocomial bacterial pneumonia and chronic obstructive pulmonary disease. A systematic review. Ann. Periodontol. 8, 54–69 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Lockhart P. B.et al.; American Heart Association Rheumatic Fever, Endocarditis, and Kawasaki Disease Committee of the Council on Cardiovascular Disease in the Young, Council on Epidemiology and Prevention, Council on Peripheral Vascular Disease, and Council on Clinical Cardiology , Periodontal disease and atherosclerotic vascular disease: Does the evidence support an independent association?: A scientific statement from the American heart association. Circulation 125, 2520–2544 (2012). [DOI] [PubMed] [Google Scholar]
- 6.The Human Microbiome Project Consortium , Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemente J. C., et al., The microbiome of uncontacted Amerindians. Sci. Adv. 1, e1500183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mark Welch J. L., Rossetti B. J., Rieken C. W., Dewhirst F. E., Borisy G. G., Biogeography of a human oral microbiome at the micron scale. Proc. Natl. Acad. Sci. U.S.A. 113, E791–E800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akcalı A., Lang N. P., Dental calculus: The calcified biofilm and its role in disease development. Periodontol. 2000 76, 109–115 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Warinner C., et al., Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 46, 336–344 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de La Fuente C., Flores S., Moraga M., DNA from human ancient bacteria: A novel source of genetic evidence from archaeological dental calculus. Archaeometry 55, 767–778 (2013). [Google Scholar]
- 12.Adler C. J., et al., Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and industrial revolutions. Nat. Genet. 45, 450–455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weyrich L. S., et al., Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 544, 357–361 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Carmody R. N., et al., Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol. 4, 2052–2063 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schnorr S. L., Sankaranarayanan K., Lewis C. M. Jr, Warinner C., Insights into human evolution from ancient and contemporary microbiome studies. Curr. Opin. Genet. Dev. 41, 14–26 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poole A. C., et al., Human salivary amylase gene copy number impacts oral and gut microbiomes. Cell Host Microbe 25, 553–564.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Council S. E., et al., Diversity and evolution of the primate skin microbiome. Proc. Biol. Sci. 283, 20152586 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ross A. A., Rodrigues Hoffmann A., Neufeld J. D., The skin microbiome of vertebrates. Microbiome 7, 79 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiello L. C., Wheeler P., The expensive-tissue hypothesis: The brain and the digestive system in human and primate evolution. Curr. Anthropol. 36, 199–221 (1995). [Google Scholar]
- 20.Carmody R. N., Wrangham R. W., The energetic significance of cooking. J. Hum. Evol. 57, 379–391 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Hardy K., Brand-Miller J., Brown K. D., Thomas M. G., Copeland L., The importance of dietary carbohydrate in human evolution. Q. Rev. Biol. 90, 251–268 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Villanea F. A., Schraiber J. G., Multiple episodes of interbreeding between Neanderthal and modern humans. Nat. Ecol. Evol. 3, 39–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Velsko I. M., et al., Microbial differences between dental plaque and historic dental calculus are related to oral biofilm maturation stage. Microbiome 7, 102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu Q., et al., The genetic history of Ice Age Europe. Nature 534, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Q., et al., An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Navarrete A., van Schaik C. P., Isler K., Energetics and the evolution of human brain size. Nature 480, 91–93 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Vågene Å. J., et al., Salmonella enterica genomes from victims of a major sixteenth-century epidemic in Mexico. Nat. Ecol. Evol. 2, 520–528 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Knights D., et al., Bayesian community-wide culture-independent microbial source tracking. Nat. Methods 8, 761–763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J., Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briggs A. W., et al., Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl. Acad. Sci. U.S.A. 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wood B., Grabowski M., “Macroevolution in and around the hominin clade” in Macroevolution: Explanation, Interpretation and Evidence, Serrelli E., Gontier N., Eds. (Springer International Publishing, 2015), pp. 345–376. [Google Scholar]
- 32.Prado-Martinez J., et al., Great ape genetic diversity and population history. Nature 499, 471–475 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bond M., et al., Corrigendum: Eocene primates of South America and the African origins of New World monkeys. Nature 525, 552 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Schrago C. G., On the time scale of New World primate diversification. Am. J. Phys. Anthropol. 132, 344–354 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Anderson M. J., A new method for non-parametric multivariate analysis of variance: Non-parametric manova for ecology. Austral Ecol. 26, 32–46 (2001). [Google Scholar]
- 36.Kolenbrander P. E., Palmer R. J. Jr, Periasamy S., Jakubovics N. S., Oral multispecies biofilm development and the key role of cell-cell distance. Nat. Rev. Microbiol. 8, 471–480 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Turnbaugh P. J., et al., A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.David L. A., et al., Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonnenburg E. D., et al., Diet-induced extinctions in the gut microbiota compound over generations. Nature 529, 212–215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Utter D. R., Mark Welch J. L., Borisy G. G., Individuality, stability, and variability of the plaque microbiome. Front. Microbiol. 7, 564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato I., et al., Nutritional correlates of human oral microbiome. J. Am. Coll. Nutr. 36, 88–98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y., et al., Biogeography of the ecosystems of the healthy human body. Genome Biol. 14, R1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaura E., et al., Same exposure but two radically different responses to antibiotics: Resilience of the salivary microbiome versus long-term microbial shifts in feces. mBio 6, e01693-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shade A., Handelsman J., Beyond the venn diagram: The hunt for a core microbiome. Environ. Microbiol. 14, 4–12 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Risely A., Applying the core microbiome to understand host-microbe systems. J. Anim. Ecol. 89, 1549–1558 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Hajishengallis G., Lamont R. J., Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27, 409–419 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasolli E., et al., Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aleti G., et al., Identification of the bacterial biosynthetic gene clusters of the oral microbiome illuminates the unexplored social language of bacteria during health and disease. mBio 10, e00321-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edlund A., et al., Metabolic fingerprints from the human oral microbiome reveal a vast knowledge gap of secreted small peptidic molecules. mSystems 2, e00058-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thamadilok S., et al., Human and nonhuman primate lineage-specific footprints in the salivary proteome. Mol. Biol. Evol. 37, 395–405 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott R. S., Teaford M. F., Ungar P. S., Dental microwear texture and anthropoid diets. Am. J. Phys. Anthropol. 147, 551–579 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Anderson A. C., et al., Long-term fluctuation of oral biofilm microbiota following different dietary phases. Appl. Environ. Microbiol. 86, e01421-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Filippis F., et al., The same microbiota and a potentially discriminant metabolome in the saliva of omnivore, ovo-lacto-vegetarian and vegan individuals. PLoS One 9, e112373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mark Welch J. L., Dewhirst F. E., Borisy G. G., Biogeography of the oral microbiome: The site-specialist hypothesis. Annu. Rev. Microbiol. 73, 335–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mark Welch J. L., Ramírez-Puebla S. T., Borisy G. G., Oral microbiome geography: Micron-scale habitat and niche. Cell Host Microbe 28, 160–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marsh P. D., Do T., Beighton D., Devine D. A., Influence of saliva on the oral microbiota. Periodontol. 2000 70, 80–92 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Schnorr S. L., et al., Gut microbiome of the Hadza hunter-gatherers. Nat. Commun. 5, 3654 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Angelis M., et al., Diet influences the functions of the human intestinal microbiome. Sci. Rep. 10, 4247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Briggs A. W., et al., Removal of deaminated cytosines and detection of in vivo methylation in ancient DNA. Nucleic Acids Res. 38, e87 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Posth C., et al., Pleistocene mitochondrial genomes suggest a single major dispersal of non-Africans and a late glacial population turnover in Europe. Curr. Biol. 26, 827–833 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Tanner A. C. R., et al., Similarity of the oral microbiota of pre-school children with that of their caregivers in a population-based study. Oral Microbiol. Immunol. 17, 379–387 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Shaw L., et al., The human salivary microbiome is shaped by shared environment rather than genetics: Evidence from a large family of closely related individuals. mBio 8, e01237-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Franzosa E. A., et al., Species-level functional profiling of metagenomes and metatranscriptomes. Nat. Methods 15, 962–968 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huson D. H., et al., MEGAN community edition–Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Louca S., et al., Function and functional redundancy in microbial systems. Nat. Ecol. Evol. 2, 936–943 (2018). [DOI] [PubMed] [Google Scholar]
- 66.Amato K. R., et al., Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 20, 201 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richards V. P., et al., Phylogenomics and the dynamic genome evolution of the genus Streptococcus. Genome Biol. Evol. 6, 741–753 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haase E. M., et al., Comparative genomics and evolution of the amylase-binding proteins of oral streptococci. BMC Microbiol. 17, 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nikitkova A. E., Haase E. M., Scannapieco F. A., Taking the starch out of oral biofilm formation: Molecular basis and functional significance of salivary α-amylase binding to oral streptococci. Appl. Environ. Microbiol. 79, 416–423 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Deimling D., et al., Electron microscopic detection of salivary α-amylase in the pellicle formed in situ. Eur. J. Oral Sci. 112, 503–509 (2004). [DOI] [PubMed] [Google Scholar]
- 71.Rogers J. D., Palmer R. J. Jr, Kolenbrander P. E., Scannapieco F. A., Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69, 7046–7056 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Behringer V., et al., Measurements of salivary alpha amylase and salivary cortisol in hominoid primates reveal within-species consistency and between-species differences. PLoS One 8, e60773 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pajic P., et al., Independent amylase gene copy number bursts correlate with dietary preferences in mammals. eLife 8, e44628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernández C. I., Wiley A. S., Rethinking the starch digestion hypothesis for AMY1 copy number variation in humans. Am. J. Phys. Anthropol. 163, 645–657 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Lazaridis I., et al., Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inchley C. E., et al., Selective sweep on human amylase genes postdates the split with Neanderthals. Sci. Rep. 6, 37198 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Warinner C., Speller C., Collins M. J., Lewis C. M. Jr, Ancient human microbiomes. J. Hum. Evol. 79, 125–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Warinner C., Dental calculus and the evolution of the human oral microbiome. J. Calif. Dent. Assoc. 44, 411–420 (2016). [PubMed] [Google Scholar]
- 79.Warinner C., Lewis C. M., Microbiome and health in past and present human populations. Am. Anthropol. 117, 740–741 (2015). [Google Scholar]
- 80.Kilian M., et al., The oral microbiome–An update for oral healthcare professionals. Br. Dent. J. 221, 657–666 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Liu S., et al., Effect of Veillonella parvula on the physiological activity of Streptococcus mutans. Arch. Oral Biol. 109, 104578 (2020). [DOI] [PubMed] [Google Scholar]
- 82.Prüfer K., et al., A high-coverage Veandertal genome from Vindija Cave in Croatia. Science 358, 655–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gómez-Robles A., Dental evolutionary rates and its implications for the Neanderthal-modern human divergence. Sci. Adv. 5, eaaw1268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mau M., Südekum K.-H., Johann A., Sliwa A., Kaiser T. M., Indication of higher salivary alpha-amylase expression in hamadryas baboons and geladas compared to chimpanzees and humans. J. Med. Primatol. 39, 187–190 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Mau M., Südekum K.-H., Johann A., Sliwa A., Kaiser T. M., Saliva of the graminivorous Theropithecus gelada lacks proline-rich proteins and tannin-binding capacity. Am. J. Primatol. 71, 663–669 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Dabney J., et al., Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. U.S.A. 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyer M., Kircher M., Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb. Protoc. 2010, pdb.prot5448 (2010). [DOI] [PubMed] [Google Scholar]
- 88.Briggs A. W., Heyn P., Preparation of next-generation sequencing libraries from damaged DNA. Methods Mol. Biol. 840, 143–154 (2012). [DOI] [PubMed] [Google Scholar]
- 89.Peltzer A., et al., EAGER: Efficient ancient genome reconstruction. Genome Biol. 17, 60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Li H., Durbin R., Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herbig A., et al., MALT: Fast alignment and analysis of metagenomic DNA sequence data applied to the Tyrolean Iceman 10.1101/050559 (Accessed 28 April 2016). [DOI]
- 92.Caporaso J. G., et al., QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hübler R., et al., HOPS: Automated detection and authentication of pathogen DNA in archaeological remains. Genome Biol. 20, 280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Neukamm J., Peltzer A., Nieselt K., DamageProfiler: Fast damage pattern calculation for ancient DNA. Bioinformatics, btab190, 10.1093/bioinformatics/btab190 (2021). [DOI] [PubMed] [Google Scholar]
- 95.Silverman J. D., Washburne A. D., Mukherjee S., David L. A., A phylogenetic transform enhances analysis of compositional microbiota data. eLife 6, e21887 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Suzuki R., Shimodaira H., Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 (2006). [DOI] [PubMed] [Google Scholar]
- 97.Reimer L. C.et al., BacDive in 2019: bacterial phenotypic data for High-throughput biodiversity analysis. Nucleic Acids Res. 47, D631–D636 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.De Cáceres M., Legendre P., Moretti M., Improving indicator species analysis by combining groups of sites. Oikos 119, 1674–1684 (2010). [Google Scholar]
- 99.Conway J. R., Lex A., Gehlenborg N., UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zijnge V., et al., Oral biofilm architecture on natural teeth. PLoS One 5, e9321 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ding W., Baumdicker F., Neher R. A., panX: Pan-genome analysis and exploration. Nucleic Acids Res. 46, e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Thorvaldsdóttir H.et al., Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bouckaert R., et al., BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e100650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jukes T. H., Cantor C. R., “Evolution of protein molecules” in Mammalian Protein Metabolism, Munro H. N., Ed. (Academic Pres, New York, USA, 1969), III, pp. 21–135. [Google Scholar]
- 106.Paradis E., Schliep K., Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35, 526–528 (2019). [DOI] [PubMed] [Google Scholar]
- 107.Yu G., Smith D. K., Zhu H., Guan Y., Lam T. T.-Y., ggtree: An R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017). [Google Scholar]
- 108.Truong D. T., et al., MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat. Methods 12, 902–903 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Suzek B. E., Wang Y., Huang H., McGarvey P. B., Wu C. H.; UniProt Consortium , UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Overbeek R., et al., The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Socransky S. S., Haffajee A. D., Cugini M. A., Smith C., Kent R. L. Jr, Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25, 134–144 (1998). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All newly generated sequencing data have been deposited in the ENA repository (https://www.ebi.ac.uk/ena/browser/home) under project accession ID PRJEB34569. R notebooks, bioinformatic scripts, additional supporting figures, and intermediate analysis files are provided in an external data repository hosted on GitHub (http://github.com/jfy133/Hominid_Calculus_Microbiome_Evolution) and archived with Zenodo under DOI: 10.5281/zenodo.3740493.