Abstract
Objective
To develop machine learning models employing administrative health data that can estimate risk of adverse outcomes within 30 days of an opioid dispensation for use by health departments or prescription monitoring programmes.
Design, setting and participants
This prognostic study was conducted in Alberta, Canada between 2017 and 2018. Participants included all patients 18 years of age and older who received at least one opioid dispensation. Pregnant and cancer patients were excluded.
Exposure
Each opioid dispensation served as an exposure.
Main outcomes/measures
Opioid-related adverse outcomes were identified from linked administrative health data. Machine learning algorithms were trained using 2017 data to predict risk of hospitalisation, emergency department visit and mortality within 30 days of an opioid dispensation. Two validation sets, using 2017 and 2018 data, were used to evaluate model performance. Model discrimination and calibration performance were assessed for all patients and those at higher risk. Machine learning discrimination was compared with current opioid guidelines.
Results
Participants in the 2017 training set (n=275 150) and validation set (n=117 829) had similar baseline characteristics. In the 2017 validation set, c-statistics for the XGBoost, logistic regression and neural network classifiers were 0.87, 0.87 and 0.80, respectively. In the 2018 validation set (n=393 023), the corresponding c-statistics were 0.88, 0.88 and 0.82. C-statistics from the Canadian guidelines ranged from 0.54 to 0.69 while the US guidelines ranged from 0.50 to 0.62. The top five percentile of predicted risk for the XGBoost and logistic regression classifiers captured 42% of all events and translated into post-test probabilities of 13.38% and 13.45%, respectively, up from the pretest probability of 1.6%.
Conclusion
Machine learning classifiers, especially incorporating hospitalisation/physician claims data, have better predictive performance compared with guideline or prescription history only approaches when predicting 30-day risk of adverse outcomes. Prescription monitoring programmes and health departments with access to administrative data can use machine learning classifiers to effectively identify those at higher risk compared with current guideline-based approaches.
Keywords: public health, epidemiology, adverse events, health & safety, clinical governance
Strengths and limitations of this study.
This study incorporated near complete capture of opioid dispensations from community pharmacies and used validated administrative health data.
This study used commonly available algorithms to train machine learning models using data which is available to government health departments in all provinces in Canada and other single payer jurisdictions; machine learning classifiers were evaluated with informative prognostic metrics not usually seen in other studies.
Our predictive models used dispense events and not medication utilisation, which is difficult to capture in administrative data.
Our training dataset does not account for non-prescription opioids, opioids administered in hospitals, and other risks associated with non-prescription use.
Introduction
Canada is among the countries with the highest rates of opioid prescribing in the world, making prescription opioid use a key driver of the current opioid crisis1; a major part of the policy response to the opioid crisis focuses on endorsing safe, appropriate opioid prescribing.2–4 In order to minimise high-risk opioid prescribing and to identify patients at high risk of opioid-related adverse outcomes, numerous health regulatory bodies have released clinical practice recommendations for health providers regarding appropriate opioid prescribing.3 5 6
Prescription monitoring programmes (PMPs) have been implemented around the world, like Alberta’s provincial Triplicate Prescription Programme7 in Canada, and are mandated to monitor the utilisation and appropriate use of opioids to reduce adverse outcomes. In most jurisdictions, both population-level monitoring metrics and clinical decision aids are used to identify patients at risk of hospitalisation or death and are most often based on prescribing guidelines. However, a comprehensive infrastructure of administrative data containing patient level International Statistical Classification of Diseases and Related Health Problems (ICD)8 codes and prescription drug histories exists in Alberta and other provinces in Canada which could be further integrated to predict opioid-related risk. Furthermore, current guidelines addressing high risk prescribing and utilisation of opioids were derived from studies that used traditional statistical methods to identify population level risk factors for overdose rather than an individual’s absolute risk3 9 10; these population estimates may not be generalisable to different populations.11 Thus, a functional gap exists in many health jurisdictions where much of the available administrative health data is not being leveraged for opioid prescription monitoring.
Supervised machine learning (ML)12 13 is an approach that uses computer algorithms to build predictive models in the clinical setting that can make use of the large amounts of available administrative data,14 15 all within a well-defined process.16 Supervised ML trains on labelled data to develop prediction models that are specific to different populations and, in many cases, can provide better predictive performance than traditional, population-based statistical models.10 15 17 We identified one study10 that applied ML techniques to predict overdose risk in opioid patients pursuant to a prescription. In their validation sample, they found that the deep neural network (DNN) and gradient boosting machines algorithms carried the best discrimination performance based on estimated c-statistics and that the ML approach out-performed the guideline approach in terms of risk prediction; neural networks have little interpretability and are not necessarily better at predicting outcomes when trained on structured data.18 This study relied on c-statistics to evaluate their ML models and did not emphasise other performance metrics (eg, positive likelihood ratios (PLR), pre and post-test probabilities) required to assess clinical utility that are recommended by medical reporting guidelines.11 13 19 20 It also did not address the important issue of ML model interpretability.21 Reporting informative prognostic metrics is needed to better understand the capabilities of ML classifiers if health departments and PMPs are to incorporate them into their decision-making processes.
The objective of our study was to further develop and validate ML algorithms (beyond just DNN) to predict the 30-day risk of hospitalisation, emergency visit and mortality for a patient in Alberta, Canada at the time of an opioid dispensation using administrative data routinely available to health departments and PMPs and evaluate them using the above referenced reporting guidelines. We also analysed feature importance to provide meaningful interpretations of the ML models. Comparing discrimination performance (area under the receiver operating characteristics curves (AUROC)), we hypothesised that the ML process would perform better than the current guideline approach for predicting risk of adverse outcomes related to opioid prescribing.
Methods
Study design and participants
This prognostic study used a supervised ML scheme. All patients in Alberta, Canada who received a dispensation for an opioid, were 18 years of age and older between 1 January 2017 and 31 December 2018 were eligible. Patients were excluded from all analyses if they had any previous diagnosis of cancer, received palliative interventions or were pregnant during the study period (online supplemental eTable 1) as use of opioids in these contexts is clinically different.
bmjopen-2020-043964supp001.pdf (665.6KB, pdf)
Government health departments and payers in many jurisdictions have systems to capture prescription histories and ICD diagnostic codes. As such, we linked various administrative health data sets available in Alberta, Canada using unique patient identifiers in order to establish a complete description of patient demographics, drug exposures and health outcomes. These databases include (1) Pharmaceutical Information Network (PIN): PIN data include all dispensing records from community pharmacies from all prescriber types occurring in the province outside of the hospital setting. PIN collects all drug dispensations irrespective of age or insurance status in Alberta; Anatomical Therapeutic Chemical classification (ATC) codes22 were used to identify opioid dispensations and their respective opioid molecules (online supplemental eTable 5), (2) Population and Vital Statistics Data (vs, Alberta Services): sex, age, date of birth, death date, immigration and emigration data, and underlying cause of death according to the WHO algorithm using ICD codes,8 (3) Hospitalisations and Emergency Department (ED) Visits (National Ambulatory Care Reporting System (NACRS), Discharge Abstract Database (DAD)): all services, length of stay, diagnosis (up to 25 ICD-108 based diagnoses). Data and coding accuracy are routinely validated both provincially and centrally via the Canadian Institute for Health Information, and (4) Physician Visits/Claims (Alberta Health): all claims from all settings (eg, outpatient, office visits, EDs, inpatient) with associated date of service, ICD code, procedure and billing information.
This study followed the Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD) and Standards for Reporting of Diagnostic Accuracy Studies (STARD) reporting guidelines.23–25
Measures and outcome
ML models were trained on a labelled dataset in which the observation/analysis unit was an opioid dispensation. Every opioid dispensation, not just the incident one, was used as a potential instance to predict the risk of our outcome. The primary outcome was a composite of a drug-related hospitalisation, ED visit or mortality within 30 days of an opioid dispensation based on ICD-10 codes used by others and identified from DAD, NACRS and Vital Statistics (T40, F55, F10–19; online supplemental eTable 2).2 10 26
We anticipated that our defined outcome would be a rare event, leading to a class imbalanced dataset.27 To address this, we relied on specifying balanced class weightage for supporting algorithms; other approaches were deemed not suitable (eg, oversampling using randomly repeating minority class); undersampling (subsampling within the majority class) resulted in changes in outcome prevalence. Class weightage is a commonly used method28 to address class imbalance along with over and undersampling approaches. However, oversampling, which involves generating new opioid dispensations from the original data distribution and is prone to introducing bias, is difficult due to the categorical nature of the data and beyond the scope of this study. With undersampling, which takes samples from the majority class (in this case, no 30-day event after dispensation), we would not be able to use all of the information provided by the data in instances with no outcome. Hence, we decided to use the class weightage method which does not alter the data distribution. Instead, the learning process is adjusted in a way that increases the importance of the positive class (instances that led to a 30-day event).29
Predictor candidates for ML models
Predictor variables in our ML models included those that were informed by the literature3 4 10 and those directly obtained from the data sets. These included features based on demographics (age, sex, income using Forward Sortation index from postal codes,30 comorbidity history using ICD-based Elixhauser score categories,31 healthcare utilisation (number of unique providers, number of hospital and ED visits) and drug utilisation (level 3 ATC codes,22 oral morphine equivalents,32 concurrent use with benzodiazepines, number of opioid and benzodiazepine dispensations, number of unique opioid and benzodiazepine molecules). Depending on the potential predictor and data availability, we used data from 30 days to 5 years before the opioid dispensation to generate model features (online supplemental eFigure 1); 30 days was used to reflect the immediate nature of the risk and 5 years to fully capture comorbidities. This approach aligns with how health providers would assess patients using the entire history of comorbidities and then the more immediate factors in deciding on the need for a therapeutic as well as risk in patients. We performed experiments to identify the features and data sets that contributed most to predicting the outcomes with a view to minimising the potential future data requirements for health departments and PMPs.
Statistical analyses and ML prediction evaluation
We randomly divided the patients in the 2017 portion of our study cohort into training (70%) and validation (30%) sets13 by patients and opioid dispensations such that no patients in the training set were in the validation set. Baseline characteristics and event rates were compared in the training vs validation group, and between those who experienced the outcome and those who did not using χ2 tests of independence. As well, we used all the 2018 data as another independent validation set.
We trained commonly used13 33 ML algorithms (online supplemental eAppendix) and further tuned out-of-box models using fivefold cross-validation on the training data to address model overfitting.13 34 As is common in ML validation studies,10 13 we reported model discrimination performance (ie, how well a model differentiates those at higher risk from those at lower risk)11 using AUROC (c-statistic). We then stratified the two ML models with the highest c-statistics into percentile categories (deciles) according to absolute risk of our outcome, as was done in previous studies.10 35 We also plotted AUROC11 and precision-recall curves (PRCs).36
Because discrimination alone is insufficient to assess ML model prediction capability, we assessed a second necessary property, namely, calibration (ie, how similar the predicted absolute risk is to the observed risk across different risk strata).11 37 Using the two ML models with the highest discrimination performance, we assessed calibration performance on the 2018 data by plotting observed (fraction of positives) vs predicted risk (mean predicted value). Using these same two ML classifiers, we analysed the top 0.1, 1, 5 and 10 percentiles of predicted risk by the number of true and false positives, PLR,20 positive predictive values (PPV), post-test probabilities and number needed to screen. We also performed a simulation of daily data uploads for 2018 quarter 1 to view the predictive capabilities if an ML risk predictor were to be deployed into a monitoring workflow.
For the XGBoost and logistic regression classifiers, we reported feature importance33 and plotted PRCs that compared all dispenses to those within the top 10 percentiles of estimated risk. As well, for the XGBoost classifier, we described feature importance on model outcome using Shapley Additive Explanations (SHAP) values38 39 to add an additional layer of interpretability.
Finally, we compared ML risk prediction (the two ML models with highest discrimination performance) to current guideline approaches as others have,10 using the 2019 Centers for Medicare & Medicaid Services opioid safety measures40 and the 2017 Canadian Opioid Prescribing Guideline.3 This was done by using the guidelines as ‘rules’ when coding for the 30-day risk of event at the time of each opioid dispensation on the entire 2018 validation set. We also compared the discrimination performance of different logistic regression classifier models using various combinations of features derived from their respective databases: (1) demographic and drug/health utilisation features from PIN and (2) comorbidity features derived from DAD, NACRS and Claims.
All analyses were done using Python (V.3.6.8,), SciKit Learn41 (V.0.23.2) SHAP39 (V.0.35), XGBoost (V.0.90),42 Pandas (V.1.0.5)43 and H20 Driverless AI (V.1.9).
Patient and public involvement
This research was done without patient involvement. Patients were not invited to comment on the study design and were not consulted to develop patient-relevant outcomes or interpret the results. Patients were not invited to contribute to the writing or editing of this document for readability or accuracy. There are no plans to disseminate the results of the research to study participants.
Results
Patient characteristics and predictors
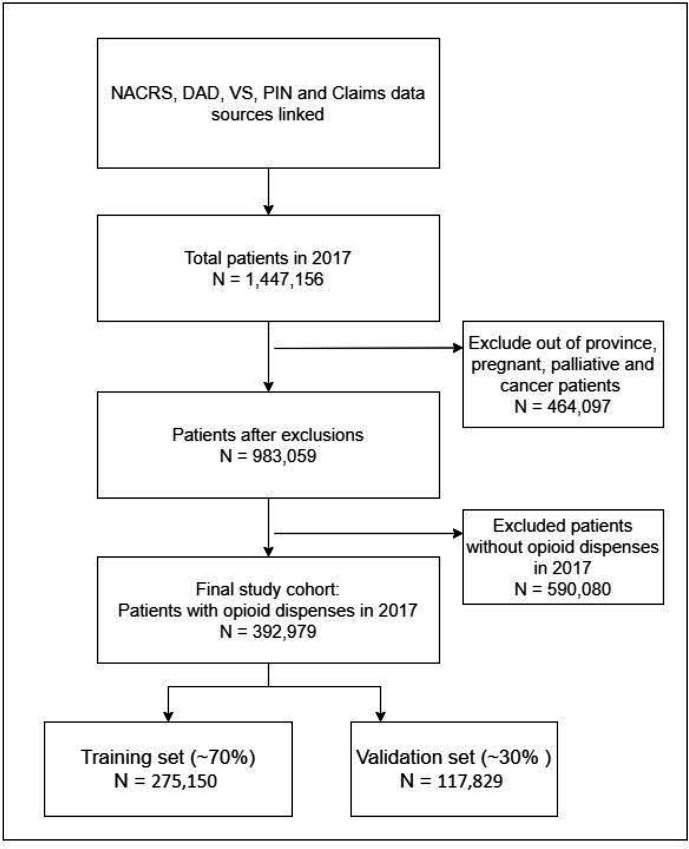
We identified 392 979 patients with at least one opioid dispensation in 2017 (figure 1). This cohort was used to train (n=275 150, 70%) and validate (n=117 829, 30%) ML models. In 2017 and 2018, 6608 and 5423 patients experienced the defined outcome, respectively. Baseline characteristics were different between those who experienced the outcome and those who did not (online supplemental eTable 3) while characteristics were similar between the training and validation sets (online supplemental eTable 4). There were 2 283 075 opioid dispensations in 2017 and 1 977 389 in 2018. Overall, in 2017, 2.03% (n=45 757) of opioid dispensations were associated with the outcome; in 2018, the estimate was 1.6% (n=31 392).
Figure 1.
Patientflow diagram of study participants used for training and validating ML models. DAD, Discharge Abstract Database; ML, machine learning; PIN, Pharmaceutical Information Network; NACR, National Ambulatory Care Reporting System; VS, vital statistics.
As described above, we categorised our candidate features into four groups (online supplemental eTable 5). When using all of the databases, the total number of features was 283 and 34 when considering only co-morbidities.
ML prediction performance
Using the 2017 validation set, AUROCs for the XGBoost and logistic regression classifiers had the highest discrimination performance at 0.87, while the neural network classifier had lower performance at 0.80 (online supplemental eTable 6).
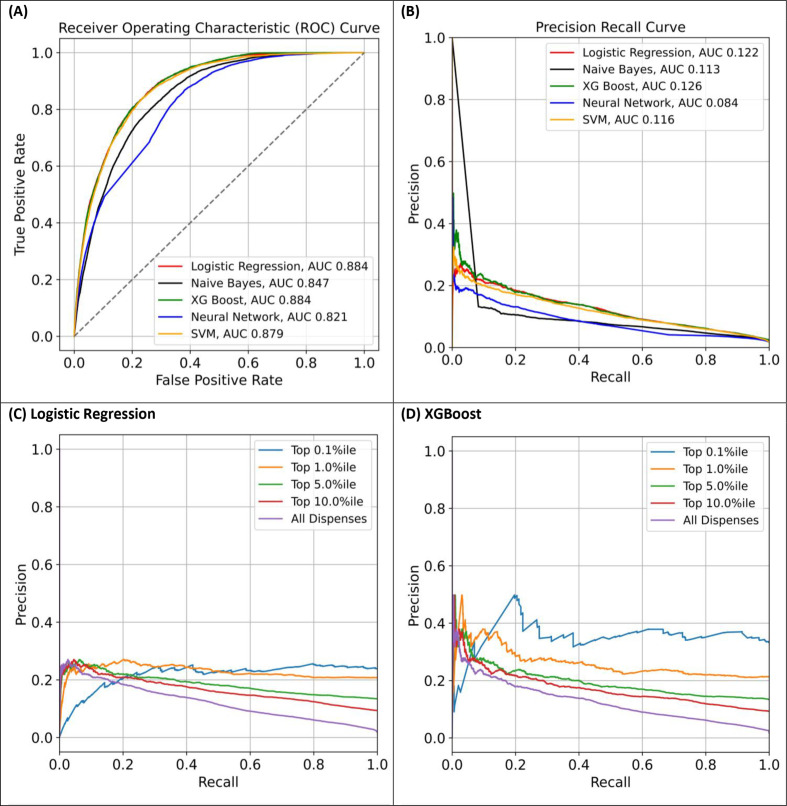
Discrimination performance was similar for the 2018 validation set (n=393 023; online supplemental eTable 6). XGBoost and logistic regression had the highest estimated AUROCs and area under PRCs while the neural network classifier was lower (figure 2A, B). As expected, PRCs indicate stronger predictive performance in opioid dispensations at higher predicted risk percentiles (figure 2C, D).
Figure 2.
Area under the receiver operating characteristic curve (AUROC) (A) and precision-recall curves (B) for all dispensations using logistic regression (L1), neural network, support vector machine (SVM), XGBoost and Naïve-Bayes; precision-recall curves for higher risk dispensations according to predicted risk percentile categories for logistic regression (C) and XGBoost (D) using the 2018 validation set.
In the 2018 validation set, although discrimination performance was similar (0.88), individual feature importance was different between the logistic regression and XGBoost classifiers, with logistic regression feature importance more reliant on co-morbidity data from DAD, NACRS and Claims while XGBoost relied more on drug utilisation data from PIN (online supplemental eFigure 2). With the XGBoost classifier, history of drug abuse, alcoholism and prior hospitalisation/emergency visit carried the highest importance for predicting the study outcome (online supplemental eFigure 3A) where the presence of these features in a patient suggested a strong prediction towards having the defined outcome (online supplemental eFigure 3B, C).
Calibration
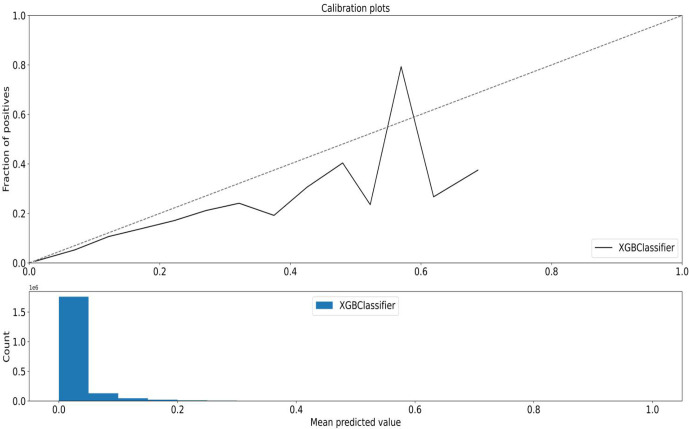
When considering dispensations predicted to be in the highest percentiles of risk, the top five percentile captured 42% of all outcomes using the XGBoost and logistic regression classifiers (table 1). Also, as the predicted risk percentiles get higher (top 10 percentile to top 0.1 percentile), so too do the corresponding PPVs with the top 0.1 percentile associated with a PPV of 33% for the XGBoost classifier. As well, lower categories of risk percentiles were associated with lower outcomes (figure 3, online supplemental eFigure 4). When we simulated a monitoring workflow scenario with daily data uploads, a similar pattern was illustrated where the dispensations predicted to be higher risk had higher event rates (figure 4).
Table 1.
Highest percentiles of estimated risk and predictive performance using the XGBoost and logistic regression classifiers for the 2018 validation dataset (n=393 023)
| Metric | Top 0.1%ile | Top 1%ile | Top 5%ile | Top 10%ile | ||||
| XGBoost | Logistic regression | XGBoost | Logistic regression | XGBoost | Logistic regression | XGBoost | Logistic regression | |
| No of dispenses | 1977 | 1977 | 19 774 | 19 774 | 98 869 | 98 869 | 197 739 | 197 739 |
| TP captured | 655 | 472 | 4204 | 4100 | 13 224 | 13 293 | 18 404 | 18 409 |
| Per cent of TP | 2.09 | 1.50 | 13.39 | 13.06 | 42.13 | 42.35 | 58.63 | 58.64 |
| FP captured | 1322 | 1505 | 15 570 | 15 674 | 85 645 | 85 576 | 179 335 | 179 330 |
| PPV | 33.13 | 23.87 | 21.26 | 20.73 | 13.38 | 13.45 | 9.31 | 9.31 |
| PLR | 30.71 | 19.44 | 16.74 | 16.22 | 9.57 | 9.63 | 6.36 | 6.36 |
| Post-test Probability* | 33.13 | 23.87 | 21.26 | 20.73 | 13.38 | 13.45 | 9.31 | 9.31 |
| NNS | 3.17 | 4.49 | 5.08 | 5.22 | 8.48 | 8.43 | 12.95 | 12.95 |
Logistic regression used L1 (lasso) parameter regularisation.
Total number of dispenses=1 977 389; total number of outcomes=31 392.
*Pretest probability estimated at 1.6% using prevalence.
FP, false positives; NNS, number needed to screen; PLR, positive likelihood ratio; PPV, positive predictive value; TP, true positives.
Figure 3.
Calibration curve plotting observed vs quantiles (deciles) of estimated risk for the XGBoost classifier using the 2018 validation dataset. The majority of counts (dispensations) were predicted to be lower risk.
Figure 4.
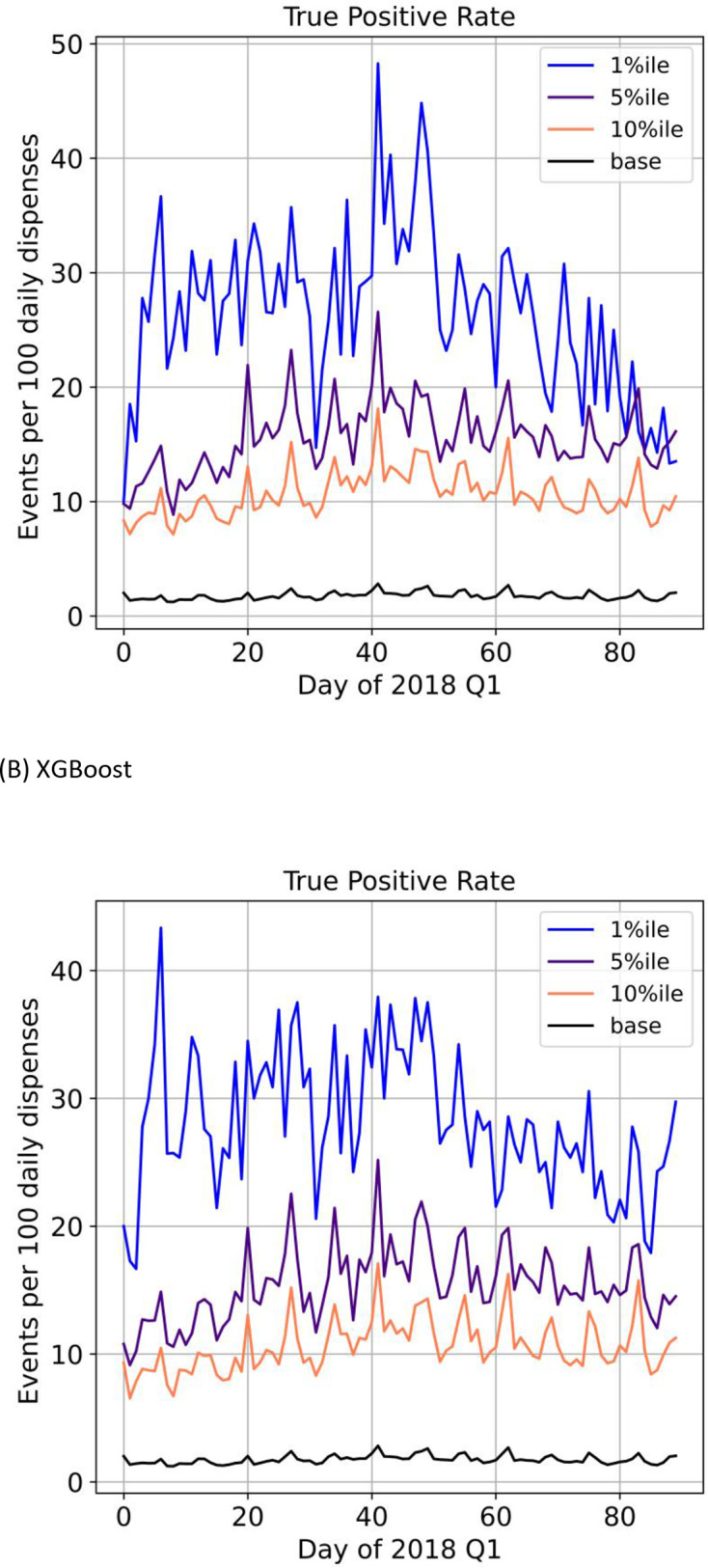
Simulation of a clinical workflow with daily uploads and events per 100 daily dispenses by risk percentiles using 2018 quarter 1 (Q1) data for logistic regression (A) and XGBoost (B) classifiers.
After using the XGBoost and logistic regression classifiers to identify the dispensations in the highest predicted risk percentiles, the pretest probability of the outcome (1.6%) was transformed into higher post-test probabilities, with higher probabilities in the riskier percentiles (table 1). The number needed to screen also decreased as predicted risk increased (table 1).
Comparing discrimination performance, ML risk prediction outperformed the current guideline approaches when using various combinations of guideline recommendations (table 2). In many of the guideline scenarios, the estimated AUROCs were close to the 0.5 mark. When we estimated the discrimination performance of the logistic regression classifier based on database source, using all databases produced an AUROC of 0.88. Reducing the database source to only DAD, NACRS, Claims (comorbidities only) resulted in an AUROC of 0.85, while PIN (prescription history) only was 0.78 (table 3).
Table 2.
Discrimination performance of guideline approach using the 2018 validation set
| Canadian guidelines* | AUROC | Sensitivity | Specificity |
| History of mental disorder only | 0.620 | 0.90 | 0.34 |
| Substance abuse only | 0.686 | 0.99 | 0.37 |
| OME/day >90 only | 0.539 | 0.22 | 0.85 |
| (Mental disorder and substance abuse) or OME/day>90 | 0.690 | 0.91 | 0.47 |
| Mental disorder and substance abuse and OME/day>90 | 0.560 | 0.20 | 0.91 |
| Mental disorder or substance abuse or OME/day>90 | 0.589 | 0.99 | 0.18 |
| CMS guidelines† | |||
| High opioid dose (>120 OME/day for 90+days) | 0.507 | 0.081 | 0.933 |
| Concurrency (Opioid and BZRA for 30+days) | 0.575 | 0.423 | 0.727 |
| Multiple doctors (>4) | 0.591 | 0.294 | 0.888 |
| Multiple pharmacies (>4) | 0.537 | 0.120 | 0.959 |
| All conditions | 0.50 | 0.001 | 0.999 |
| Any condition | 0.622 | 0.62 | 0.625 |
Guideline approaches were adapted from the 2017 Canadian opioid prescribing guideline and 2019 CMS opioid safety measures and compared with logistic regression and XGBoost classifiers (each with an estimated area under the receiver operating characteristic curve of 0.88). These guidelines were used as rules to predict the 30-day risk of event at the time of opioid dispensation.
*The Canadian guidelines do not specify timelines. >90 OME was determined by taking the average daily OME over the 30 days prior to dispensation.
†The CMS guidelines specify 90 or more days at >120 OME and concurrent use of opioids and benzodiazepines for 30 days or more within an assessment period of 180 days.
AUROC, area under the receiver operating characteristic curve; BZRA, benzodiazepine receptor agonist; CMS, Centers for Medicare & Medicaid services; OME, daily oral morphine equivalents.
Table 3.
Discrimination performance based on database source using AUROC for the logistic regression classifier on the 2018 validation set
| Database source | Predictor variables formed from database | AUROC | No of features |
| PIN only | Drug utilisation+prescription history | 0.78 | 248* |
| DAD, NACRS, Claims | Co-morbidities | 0.85 | 34 |
| PIN, DAD NACRS, Claims (all databases used in study) | Demographic+drug utilisation+healthcare utilisation+comorbidities | 0.88 | 283 |
Drug utilisation includes features describing oral morphine equivalents,32 concurrent use with benzodiazepines, number of opioid and benzodiazepine dispensations, number of unique opioid and benzodiazepine molecules; healthcare utilisation includes features describing number of unique health providers visited, number of hospital/emergency department visits; logistic regression used L1 (lasso) parameter regularisation.
*excludes mean income
AUROC, area under the receiver operating characteristic curve; DAD, Discharge Abstract Database; NACRS, National Ambulatory Care Reporting System; PIN, Pharmaceutical Information Network.
Discussion
This study showed that ML techniques using available administrative data (prescription histories and ICD codes) may provide enough discriminatory performance to predict adverse outcomes associated with opioid prescribing. Indeed, our ML analyses showed very high discrimination performance at 0.88. The linear model (logistic regression) and XGBoost carried higher discrimination and calibration performance, while the neural network classifier did not perform as well. By identifying the predicted top 5–10 percentile of absolute risk pursuant to an opioid dispensation, we were able to capture approximately half of all outcomes using ML methods. All ML models we trained had higher discrimination performance using the validation sets compared with the clinical guideline approach.
Since the prevalence of our defined outcome is relatively low in the general population, PPVs would also be expectedly low. However, estimated PPVs increased when we considered higher risk dispensations, as is expected since PPV is related to event prevalence. This is important because different users of a risk predictor will require different predictive capabilities. Similarly, our estimates of PLRs and associated post-test probabilities also increased in dispensations with higher predicted risk indicating the strong predictive capabilities of the XGBoost and logistic regression classifiers; likelihood ratios >10 generate conclusive changes from pretest to post-test probabilities.20
The current guideline approach to assess absolute opioid prescribing risk produced c-statistic estimates closer to 0.5 indicating that discrimination was not much better than chance alone. ML models with higher predictive performance can better support health departments and PMPs with monitoring mandates to identify and intervene on those at high risk and their associated prescribers. We also found that adding co-morbidity features from administrative databases increased prediction performance compared with prescription history alone, thus making the case for the use of this data by PMPs and health departments. However, if only prescription history is available, our trained XGBoost classifier still had strong discrimination performance.
We found only one study that used ML approaches to quantify the absolute risk of an event pursuant to an opioid dispensation.10 Their methodology used rolling 3-month windows for estimating risk and ML model training while we used historic records to estimate 30-day risk. Differences in study population and feature selection may explain why their highest performing ML model was deep learning (neural network classifier) and ours was not. Nevertheless, we were able to replicate their predictive performance using our ML approach as we both showed that ML approaches have higher predictive capabilities than guideline approaches. Both of our studies used predicted percentile risk estimates to identify high-risk dispensations and were able to do so with strong discrimination and calibration performance. Furthermore, we emphasised prognostic metrics which are more informative to assess the clinical utility of ML classifiers using pretest and post-test probabilities, something not done in other studies and recommended in medical guidelines.20 This major aspect of our study, not done previously, is important because any ML classifier that does not increase prognostic information compared with baseline cannot be incorporated into decision making for the purpose of intervening on higher-risk instead of lower-risk patients. Indeed, another study we found describes how identifying cases in higher predicted risk percentiles using ML methods can be deployed in hospital settings for the purpose of targeted interventions35 on discharge, however, the effect on outcomes is still to be determined.
The limitations of our study are similar to other ML studies10 and need to be addressed when considering deployment of ML risk predictors. Our training dataset was not able to account for non-prescription opioid consumption and the risk associated with non-prescription use, both of which are substantial contributors to overall risk.2 Regarding our analysis, we assumed that all dispensations were independent events; future research in this area should focus on employing ML methods using correlated data. As with all ML projects, our models were trained using Alberta data and might not be generalisable to other populations, or to specific populations within Alberta. However, one of the benefits of the ML process is that models can be retrained or similar methods could be used to develop new models to accommodate different populations.
This study suggests that ML risk prediction can support PMPs, especially if readily available administrative health data is used. PMPs currently use population-based guidelines which we, and others, have shown cannot predict absolute individual risk. The ML process allows for flexibility in model training, validation and deployment to specific settings in which, for the case of PMPs, high-risk patients can be identified and targeted for intervention either at the patient or provider level. For example, an ML classifier can be trained on accessible data to create an aggregated list of ‘high-risk’ patients at regular time intervals to identify points of intervention. Moreover, ML classifiers can be retrained over time as changes in populations and trends in prescribing occur and are therefore specific to the population unlike broadly based guidelines. Further research can assess whether implementation of an ML-based monitoring system by PMPs leads to improved clinical outcomes within their own jurisdictions and whether other available features or feature reduction can yield sufficiently valid results for their own intended purposes.
Supplementary Material
Acknowledgments
This study is based on data provided by The Alberta Strategy for Patient Orientated Research (AbSPORU) SUPPORT unit and Alberta Health.
Footnotes
Contributors: VS, VK, LK, SS and DTE were involved in the conception and design of the study. VS, VK, LK, SS and DTE analysed the data. VS, VK and LK drafted the article. VS, VK, LK, DTE and SS revised the article. All authors gave final approval of the version to be published. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted. DTE is the guarantor.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: SS has received grants from the College of Physicians and Surgeons of Alberta.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data may be obtained from a third party and are not publicly available. No data are available. The data used in this study is not available for external analysis. However, administrative health data can be accessed from Alberta Health by following defined research protocols and confidentiality agreements.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
Ethical approval: This study was approved by the Health Research Ethics board at the University of Alberta (#Pro00083807_AME1)
References
- 1.Belzak L, Halverson J. Evidence synthesis - The opioid crisis in Canada: a national perspective. Health Promotion and Chronic Disease Prevention in Canada 2018;38:224–33. 10.24095/hpcdp.38.6.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gomes T, Khuu W, Martins D, et al. Contributions of prescribed and non-prescribed opioids to opioid related deaths: population based cohort study in Ontario, Canada. BMJ 2018;362:k3207. 10.1136/bmj.k3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busse JW, Craigie S, Juurlink DN, et al. Guideline for opioid therapy and chronic noncancer pain. CMAJ 2017;189:E659–66. 10.1503/cmaj.170363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowell D. CDC guideline for prescribing opioids for chronic pain, 2016. [Google Scholar]
- 5.ismp Canada . Essential clinical skills for opioid prescribers, 2017. Available: https://www.ismp-canada.org/download/OpioidStewardship/Opioid-Prescribing-Skills.pdf [Accessed Nov 2018].
- 6.Centre for Effective Practice . Management of chronic non cancer pain, 2017. Available: thewellhealth.ca/cncp
- 7.College of Physicians and Surgeons of Alberta . Tpp Alberta – OME and DDD conversion factors, 2020. Available: http://www.cpsa.ca/tpp/ [Accessed Jun 2020].
- 8.World health Organization . Classification of diseases (ICD), 2019. Available: https://www.who.int/classifications/icd/icdonlineversions/en/ [Accessed Jun 2020].
- 9.Gomes T, Mamdani MM, Dhalla IA. Opioid dose and drug-related mortality in patients with nonmalignant PainOpioid dose and drug-related mortality. JAMA Internal Medicine 2011;171:686–91. [DOI] [PubMed] [Google Scholar]
- 10.Lo-Ciganic W-H, Huang JL, Zhang HH, et al. Evaluation of Machine-Learning algorithms for predicting opioid overdose risk among Medicare beneficiaries with opioid prescriptions. JAMA Netw Open 2019;2:e190968. 10.1001/jamanetworkopen.2019.0968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alba AC, Agoritsas T, Walsh M, et al. Discrimination and calibration of clinical prediction models: users' guides to the medical literature. JAMA 2017;318:1377–84. 10.1001/jama.2017.12126 [DOI] [PubMed] [Google Scholar]
- 12.Shah NH, Milstein A, Bagley PhD SC. Making machine learning models clinically useful. JAMA 2019;322:1351–2. 10.1001/jama.2019.10306 [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Chen P-HC, Krause J, et al. How to read articles that use machine learning: users' guides to the medical literature. JAMA 2019;322:1806–16. 10.1001/jama.2019.16489 [DOI] [PubMed] [Google Scholar]
- 14.Baştanlar Y, Ozuysal M. Introduction to machine learning. Methods Mol Biol 2014;1107:105–28. 10.1007/978-1-62703-748-8_7 [DOI] [PubMed] [Google Scholar]
- 15.Thottakkara P, Ozrazgat-Baslanti T, Hupf BB, et al. Application of machine learning techniques to high-dimensional clinical data to forecast postoperative complications. PLoS One 2016;11:e0155705. 10.1371/journal.pone.0155705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alberta Machine Intelligence Institute . Machine learning process lifecycle, 2019. [Google Scholar]
- 17.Hsich E, Gorodeski EZ, Blackstone EH, et al. Identifying important risk factors for survival in patient with systolic heart failure using random survival forests. Circ Cardiovasc Qual Outcomes 2011;4:39–45. 10.1161/CIRCOUTCOMES.110.939371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caruana R, Lou Y, Gehrke J. Intelligible models for healthcare: predicting pneumonia risk and hospital 30-day readmission. Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, 2015. [Google Scholar]
- 19.Yusuf M, Atal I, Li J, et al. Reporting quality of studies using machine learning models for medical diagnosis: a systematic review. BMJ Open 2020;10:e034568. 10.1136/bmjopen-2019-034568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeschke R, Guyatt GH, Sackett DL. Users' guides to the medical literature. III. How to use an article about a diagnostic test. B. what are the results and will they help me in caring for my patients? the evidence-based medicine Working group. JAMA 1994;271:703–7. 10.1001/jama.271.9.703 [DOI] [PubMed] [Google Scholar]
- 21.Shortliffe EH, Sepúlveda MJ. Clinical decision support in the era of artificial intelligence. JAMA 2018;320:2199–200. 10.1001/jama.2018.17163 [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization . International language for drug utilization research, ATC/DDD, 2020. Available: https://www.whocc.no/ [Accessed Jun 2020].
- 23.Moons KGM, Altman DG, Reitsma JB, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1–73. 10.7326/M14-0698 [DOI] [PubMed] [Google Scholar]
- 24.equator network . Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement, 2020. Available: https://www.equator-network.org/reporting-guidelines/tripod-statement/ [Accessed Feb 2020]. [DOI] [PMC free article] [PubMed]
- 25.Cohen JF, Korevaar DA, Altman DG, et al. Stard 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016;6:e012799. 10.1136/bmjopen-2016-012799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou H, Della PR, Roberts P, et al. Utility of models to predict 28-day or 30-day unplanned Hospital readmissions: an updated systematic review. BMJ Open 2016;6:e011060. 10.1136/bmjopen-2016-011060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownlee J . A gentle introduction to imbalanced classification, 2020. Available: https://machinelearningmastery.com/what-is-imbalanced-classification/ [Accessed Jan 2021].
- 28.King G, Zeng L. Logistic regression in rare events data. Political Analysis 2001;9:137–63. 10.1093/oxfordjournals.pan.a004868 [DOI] [Google Scholar]
- 29.Johnson JM, Khoshgoftaar TM. Survey on deep learning with class imbalance. J Big Data 2019;6:1–54. 10.1186/s40537-019-0192-5 [DOI] [Google Scholar]
- 30.Government of Canada . Forward Sortation Area—Definition, 2015. Available: https://www.ic.gc.ca/eic/site/bsf-osb.nsf/eng/br03396.html [Accessed April 2020].
- 31.Quan H, Sundararajan V, Halfon P, et al. Coding algorithms for defining comorbidities in ICD-9-CM and ICD-10 administrative data. Med Care 2005;43:1130–9. 10.1097/01.mlr.0000182534.19832.83 [DOI] [PubMed] [Google Scholar]
- 32.College of Physicians and Surgeons of Alberta . Ome and DDD conversion factors. Available: http://www.cpsa.ca/wp-content/uploads/2017/06/OME-and-DDD-Conversion-Factors.pdf
- 33.Goldstein BA, Navar AM, Carter RE. Moving beyond regression techniques in cardiovascular risk prediction: applying machine learning to address analytic challenges. Eur Heart J 2017;38:1805–14. 10.1093/eurheartj/ehw302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose S. Machine learning for prediction in electronic health data. JAMA Netw Open 2018;1:e181404. 10.1001/jamanetworkopen.2018.1404 [DOI] [PubMed] [Google Scholar]
- 35.Morgan DJ, Bame B, Zimand P, et al. Assessment of machine learning vs standard prediction rules for predicting Hospital readmissions. JAMA Netw Open 2019;2:e190348. 10.1001/jamanetworkopen.2019.0348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10:e0118432. 10.1371/journal.pone.0118432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shah ND, Steyerberg EW, Kent DM. Big data and predictive analytics: recalibrating expectations. JAMA 2018;320:27–8. 10.1001/jama.2018.5602 [DOI] [PubMed] [Google Scholar]
- 38.Molnar C. Interpretable machine learning. A Guide for Making Black Box Models Explainable, 2019. [Google Scholar]
- 39.Lundberg SM, Lee S-I. A unified approach to interpreting model predictions. Paper presented at: Advances in neural information processing systems, 2017. [Google Scholar]
- 40.Centers for Medicare & Medicaid Services (CMS) . Announcement of calendar year (cy) 2019 Medicare advantage capitation rates and Medicare advantage and part D payment policies and final call letter.
- 41.Buitinck L, Louppe G, Blondel M. Api design for machine learning software: experiences from the scikit-learn project. arXiv preprint arXiv 2013:13090238. [Google Scholar]
- 42.Chen T, Guestrin C. Xgboost: a scalable tree boosting system. Paper presented at: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, 2016. [Google Scholar]
- 43.The PANDAS development team. pandas-dev/pandas: PANDAS 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-043964supp001.pdf (665.6KB, pdf)
Data Availability Statement
Data may be obtained from a third party and are not publicly available. No data are available. The data used in this study is not available for external analysis. However, administrative health data can be accessed from Alberta Health by following defined research protocols and confidentiality agreements.