Abstract
Fibroblasts are a critical component of tumor microenvironments and associate with cancer cells physically and biochemically during different stages of the disease. Existing cell culture models to study interactions between fibroblasts and cancer cells lack native tumor architecture or scalability. We developed a scalable organotypic model by robotically encapsulating a triple negative breast cancer (TNBC) cell spheroid within a natural extracellular matrix containing dispersed fibroblasts. We utilized an established CXCL12 – CXCR4 chemokine-receptor signaling in breast tumors to validate our model. Using imaging techniques and molecular analyses, we demonstrated that CXCL12-secreting fibroblasts have elevated activity of RhoA/ROCK/myosin light chain-2 pathway and rapidly and significantly contract collagen matrices. Signaling between TNBC cells and CXCL12-producing fibroblasts promoted matrix invasion of cancer cells by activating oncogenic mitogen-activated protein kinase signaling, whereas normal fibroblasts significantly diminished TNBC cell invasiveness. We demonstrated that disrupting CXCL12 – CXCR4 signaling using a molecular inhibitor significantly inhibited invasiveness of cancer cells, suggesting blocking of tumor-stromal interactions as a therapeutic strategy especially for cancers such as TNBC that lack targeted therapies. Our organotypic tumor model mimics native solid tumors, enables modular addition of different stromal cells and extracellular matrix proteins, and allows high throughput compound screening against tumor-stromal interactions to identify novel therapeutics.
Keywords: breast cancer, tumor microenvironment, organotypic tumor model, invasion, contractility
1. Introduction
The tumor microenvironment is a complex tissue containing cancer cells and various stromal cells such as fibroblasts, immune cells, and endothelial cells embedded in an extracellular matrix (ECM) network [1]. Among different stromal cells, fibroblasts play pivotal roles in initiation and progression of epithelial tumors [2,3]. During tumor progression, cancer cells secrete signaling molecules to recruit resident tissue fibroblasts and activate them. These cells, known as cancer-associated fibroblasts (CAFs), are the most abundant stromal cells in solid tumors and associate with cancer cells at all stages of tumor progression [2,4]. Histopathological analysis of human tumors shows that advanced tumors with poor prognosis are abundant in CAFs [5]. In breast cancer, CAFs and their ECM cause a tumor to be palpable [6]. When epithelial cancer cells breach the basement membrane and invade the ECM, they come in direct contact with CAFs [7]. Additionally, CAFs secrete various pro-invasive signaling molecules into the tumor microenvironment and deposit and remodel the ECM [8]. Physical and biochemical interactions with CAFs and their associated ECM promote proliferation of cancer cells and alter their morphology to an invasive mesenchymal-like shape [9]. Therefore, CAFs are a critical component of solid tumors and a key regulator of tumor-stromal interactions.
Cell cultures and animal models are typically used to study tumor-stromal interactions. Culture of cancer cells and fibroblasts in an intermixed monolayer or in separated sheets provides a convenient method to study interactions between the two cell types. However, monolayer cultures do not reproduce complexities of tumors that are three-dimensional (3D), including oxygen and nutrients distribution profiles, cell-cell and cell-ECM interactions, diffusive barrier to drug delivery, etc. [10,11]. Mouse models provide physiological systems for cancer research but lack human tumor stroma and need complex biological assays and sophisticated imaging to study effects of stroma on tumor cells [12]. To address the technological need for human tumor models, 3D cultures of cancer cells as spheroids have been developed. Spheroids are compact clusters of cells that reproduce properties of solid tumors such as diffusive transport of oxygen and nutrients, hypoxia and necrosis, drug delivery barriers, and deposition and degradation of ECM proteins [13–15]. Co-culture spheroids of cancer cells with various stromal cells have been used to study tumor-stromal interactions that promote proliferation and drug resistance of breast cancer cells [16], and their epithelial-mesenchymal transition (EMT) [17]. Despite their benefits, co-culture spheroids do not mimic the architecture of native solid tumors in terms of spatial distribution of cancer and stromal cells and cell-ECM interactions. Matrigel has been used to develop spheroid cultures to study cell-cell and cell-ECM interactions and ECM invasion of cancer cells [18,19]. However, Matrigel is an animal-derived protein cocktail and not amenable to adjusting its composition and physicochemical properties [20]. To gain control over spatial distribution of cells and ECM properties, microfluidics technology has been widely used to develop 3D tumor models. Recently, microfluidic co-culture systems containing hydrogel-based 3D matrices were used to study tumor-stromal interactions, cancer cell invasion of ECM, and anti-metastatic drug testing [21–24]. Major drawbacks of microfluidic cell culture devices are their complexity and need for user expertise to develop and maintain them especially for long-term cultures, incompatibility with industrial-scale high throughput drug screening applications, and adsorption of drugs and proteins on the surface of polydimethylsiloxane (PDMS) microfluidic devices [25,26].
To overcome these limitations of existing cultures, we developed a 3D organotypic breast tumor model that resembles the architecture of avascular solid tumors and consists of three major components of the tumor microenvironment: mass of cancer cells, dispersed fibroblasts, and ECM. We also adapted this technology to a microwell plate format to enable convenient, reproducible formation of cultures, high throughput drug screening, and adaption by other users. To establish the validity of our model, we used triple negative breast cancer (TNBC) with CXCL12 – CXCR4 signaling as a disease model. CXCL12, or stromal-derived factor-1α (SDF-1α), is a major paracrine signaling molecule produced by CAFs and signals through its cognate CXCR4 receptor often overexpressed on the surface of TNBC cells [27,28]. Therefore, we use engineered CXCL12-secreting fibroblasts and CXCR4-expressing breast cancer cells in our microtissues to recapitulate paracrine stromal-cancer cells signaling. Using different molecular analyses and imaging techniques, we demonstrated that CXCL12-secreting fibroblasts significantly contract the ECM. Additionally, signaling between the fibroblasts and TNBC cells promoted ECM invasion of cancer cells by activating oncogenic mitogen-activated protein kinase (MAPK) pathway. We also demonstrated that blocking tumor-stromal interactions inhibits cancer cell invasiveness as a potential treatment strategy. This organotypic model is a convenient-to-adapt tool broadly useful to study different cancers. It offers straightforward implementation to facilitate dissemination to academic and industrial settings, allows adjusting added components including single to multiple cell types and ECM, and its compatibility with microwell plates uniquely enables high throughput drug testing against tumor-stromal interactions to identify novel therapeutics.
2. Materials and Methods
2.1. Cell culture
Two different fibroblast cells were used: normal human mammary fibroblasts (gift of Dr. Daniel Hayes, University of Michigan) stably transduced with mCherry protein (labeled HMFs) and human mammary fibroblasts stably transduced to secrete 0.75 ng/ml/h of CXCL12α fused with mCherry (labeled CXCL12+HMFs). MDA-MB-231 TNBC cells (purchased from the ATCC and labeled TNBC) overexpressing CXCR4 receptors (labeled CXCR4+TNBC) were transduced to stably express eGFP [29]. These cells were generated in Dr. Luker’s laboratory in University of Michigan. The authenticity of cells after stable transduction was verified by short tandem repeat profiling. Cells were tested for mycoplasma and maintained in Plasmocin prophylactic (Invivogen) according to the manufacturer’s directions. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Sigma) supplemented with 10% fetal bovine serum (FBS, Sigma), 1% glutamine (Life Technologies), and 1% antibiotic (Life Technologies). Cells were cultured in a humidified incubator at 37°C and 5% CO2 in a T75 flask (Thermo Fisher Scientific). When the cells were about 80% confluent, they were rinsed with phosphate buffered saline (PBS, Sigma), dislodged using 0.25% trypsin (Life Technologies), and sub-cultured.
2.2. Culture of fibroblasts in collagen
Fibroblast cells were harvested from a confluent monolayer and mixed with ice-cold collagen solution. Collagen solutions of desired concentrations were prepared from stock solutions of rat tail type I collagen (Corning) using the manufacturer’s protocol [30]. All the reagents were kept on ice during collagen preparation to prevent premature gelation of collagen. The pH of solutions was measured with a pH meter (Mettler Toledo) and maintained at 7.5 by adding 1 N NaOH or 1 N HCL dropwise. Of the stock collagen solution, 1 mg/ml and 4 mg/ml solutions were prepared and stored at 4°C for up to 1 h before use. The 1 mg/ml and 4 mg/ml collagen gels are referred to as softer and stiffer gels, respectively, for convenience. Different densities (number of cells per well) of HMFs and CXCL12+HMFs were used to study collagen contractility. For example, to form stiffer collagen gels with 1.5 × 104 cells in each well of a 384-well plate, 7.5 × 105 fibroblasts were mixed with 1 ml of 4 mg/ml collagen solution, and 20 μl of the resulting suspension solution was dispensed into each well. Incubation at 37°C for 30 min resulted in hydrogel formation. Cultures were maintained in DMEM supplemented with only 1% FBS. Phase images were captured using an inverted fluorescence microscope (Axio Observer A1, Zeiss). Contraction of collagen gels was quantified as a matrix contractility = (1 – ratio of projected area of a culture before and after contraction). Prior to this experiment, nanoindentation of collagen gels prepared from various concentration of 1.0, 2.5, 4.0, 5.5, 7.0, and 8.5 mg/ml was conducted using an atomic force microscope (AFM, Digital Instruments). Plasma cleaning and UV radiation were performed on indentation probe and tip-holder to remove organic contaminants before each experiment. PBS buffer was used as a rehydrating medium for gels during indentation. We used 300 nm radius silicon-oxide probe with a cantilever spring constant of 0.06 N/m. Velocity of extension and recession of cantilever during indentation was controlled at 500 nm/s, slow enough to avoid hydrodynamic force from flow of buffer solution. A Poisson’s ratio of 0.5 was used. At least 30 indentations were made on each sample to generate force versus indentation curves that were analyzed using the Hertz model to generate elastic modulus values for collagen gels of various protein concentrations.
2.3. Spheroid formation
CXCR4+TNBC spheroids were formed using our established protocol with an aqueous two-phase system [31]. Polyethylene glycol (PEG) (Mw: 35 kDa, Sigma) and dextran (DEX) (Mw: 500 kDa, Pharmacosmos) were used to form 5.0% (w/v) aqueous PEG phase and 6.4% (w/v) aqueous DEX phase solutions in a complete growth medium [32,33]. The PEG phase solution was supplemented with 0.24% of methylcellulose powder. CXCR4+TNBC cells were mixed thoroughly with the DEX phase solution to form a cell suspension with a density of 2.5×104 cells/μl. Next, 30 μl of the PEG solution was loaded into a round-bottom, ultralow attachment 384-well plate (Corning) labeled as the destination plate. A robotic liquid handler (SRT Bravo, Agilent Technologies) was used to dispense a 0.3 μl drop of the DEX phase containing 7.5×103 cells into each well of the destination plate [34]. This process was done column-by-column to reduce the need for large volumes of high-density cell suspensions. The plate was incubated at 37°C for 24 h to allow formation of a spheroid in the DEX phase drop within each microwell.
2.4. Immunofluorescence of 3D cultures of dispersed fibroblasts
After 5 days of incubating collagen gels containing only fibroblasts (1.5 ×104 cells), cultures were transferred from the microwells into a Petri dish using a micro-spatula (Fine Science Tools). The gels were fixed using 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature and washed three times with PBS, each time for 10 min. The gels were blocked for 1 h at room temperature with 5% goat serum. Cells were permeabilized using 0.3% Triton X-100 in PBS. Samples were incubated overnight with a mouse anti-β-actin antibody (1:200 dilution, Abcam, Cat. No. ab6276) prepared in 1% BSA and 0.3% Triton X-100 in PBS. Gels were washed and then incubated with a goat anti-mouse FITC-conjugated secondary antibody (Jackson ImmunoResearch) for 1 h followed by nuclear staining with Hoechst 33342 (Thermo Fisher Scientific) and imaged at 60x with a confocal microscope (Nikon A1). FITC was excited by a 488 nm laser and the emission isolated with a 500–550 nm filter, while Hoechst was excited by 405 nm and isolated with a 425–475 nm emission filter.
2.5. Metabolic activity of fibroblast cells in 3D cultures
HMFs and CXCL12+HMFs were included toin separate collagen solutions in a 384-well plate at a density of 1.5 ×104 cells per well. The collagen solutions were gelled and then medium with 1% FBS was added. The growth of the fibroblasts in collagen was evaluated for 2 weeks using a standard Prestoblue metabolic activity assay (Invitrogen). After 15 min of incubation, the fluorescent signal was measured with a standard plate reader (Synergy H1M, BioTek Instruments) at 560 nm excitation and 590 nm emission wavelengths. Collagen gels without fibroblasts were used as a negative control.
2.6. Conditioned medium cultures
CXCL12+HMFs cells were seeded in a flask containing medium with 1% FBS. Conditioned medium was collected from a confluent monolayer of the CXCL12+HMFs and added to a culture of HMFs in 3D collagen gels every other day by replacing 60 μl of existing medium. Contraction of ECM was quantified using phase contrast images.
2.7. Wound healing assay
Both HMFs and CXCL12+HMFs were seeded as a monolayer culture in 35 mm Petri dishes (Corning) and cultured until confluent. Using a pipette tip, a straight scratch was made in the cell layer. Cells were imaged for 48 h and the migration of cells in the cultures was quantified as gap closure=, where A1 was the initial gap area and A2 was the final gap area.
2.8. Organotypic Microtissue formation, ECM invasion, and AMD3100 treatment
After formation of compact spheroids, medium was robotically removed from the wells. The protocol was optimized to ensure that spheroids remained in the wells during aspiration. This was done by adjusting the height of the pipette tip from the bottom of each well to 0.2 mm, maintaining a low aspiration rate of 0.6 μl/sec, and offsetting the position of the pipette tip 30% from the center of the well where spheroids were located. Cold collagen solution containing 1.5×104 fibroblasts (HMFs or CXCL12+HMFs) was dispensed into each well containing a spheroid. The well plate was incubated at 37°C for 30 min to form a collagen gel and then 60 μl of medium containing only 1% FBS was added (Fig. 1A). For convenience, microtissues containing HMFs or CXCL12+HMFs are labelled +HMFs and +CXCL12+HMFs, respectively. As a negative control, microtissues without fibroblasts were generated by adding the collagen solution to wells containing a cancer cell spheroid. Medium was refreshed every 72 hrs. Both aspirating and dispensing of collagen solution and medium were carried out robotically for accuracy and speed of sample preparation. Confocal images were taken with a 10x objective on days 1, 3, and 5 to assess matrix invasion of cancer cells. A 488 nm laser was used to excite the fluorophore, while a 500–550 nm filter was used to capture eGFP fluorescence from CXCR4+TNBC cells. Z-stacks were constructed from images of samples acquired with a z-spacing of 20 μm. NIS software was used for image acquisition and Fiji (ImageJ, NIH) was used for analysis and 3D reconstruction. CXCR4+TNBC cell invasion of the ECM was quantified by analyzing images from both z-projected images and from the individual images of each z-stack.
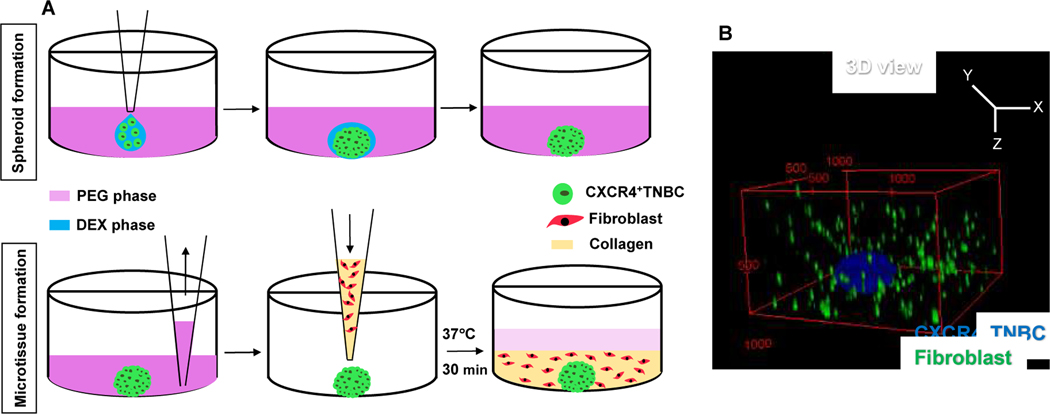
Fig. 1.
(A) Schematic process of formation of a microtissue containing a CXCR4+TNBC spheroid and fibroblasts dispersed in collagen. (B) 3D confocal reconstruction of a microtissue containing CXCR4+TNBC spheroids (blue) and fibroblasts (green). Collagen was not stained. The numbers in the red box represent distances in micrometers.
ECM invasion of cells was analyzed in two different ways. First, all the confocal images from each sample were collapsed to form a z-projected image. This image was opened in ImageJ and the scale was removed to set the scale to pixels and measure area in pixels. Then, color thresholding was used to select the area covered by the cells, and pixel areas were measured to quantify invasion area. Because this commonly-used method masks vertical displacement of cells, the vertical scattering of cancer cells was also analyzed. Briefly, each image from a stack of the confocal images was analyzed to measure the pixel area occupied by the CXCR4+TNBC cells. The area of pixels (labeled as invasion area) was then plotted against the z-distance of the images from each stack to obtain a histogram. Each microtissue resulted in one histogram and each data point in the histogram represented pixels area from one image. Area under the curve (AUC) was measured from each histogram in MATLAB (MathWorks). AUC values were averaged to express as 3D invasion cell area.
Because dispersed fibroblasts contracted the collagen matrix, the contractility of the matrix in each microtissue containing a cancer cell spheroid and fibroblasts was expressed between 0 – 1, with 1 being the highest matrix contraction. Then, ECM invasion of the CXCR4+TNBC cells was normalized with contractility and expressed as normalized invasion cell area=* invasion cell area. To block signaling from CXCL12+HMFs to CXCR4+TNBC cells, microtissues were treated with 1 μM of AMD3100. Medium change was done every other day using 40 μl of fresh medium containing AMD3100. ECM invasion of CXCR4+TNBC cells was imaged using a confocal microscope and analyzed as above.
2.9. Western blotting
Microtissues were treated with collagenase I (Sigma) for about 10 min with added mechanical agitation by pipetting to dissociate and dissolve the matrix. Collagenase activity was neutralized by adding triple amounts of complete growth medium using the manufacturer’s protocol. The resulting suspension was centrifuged to obtain a cell pellet. The cells were washed with PBS and lysed with 500 μl of complete RIPA buffer containing 1% protease inhibitors and 1% phosphatase inhibitors. The suspension was further sonicated twice at a 20% amplitude for 5 sec (Vibra-Cell, Sonics). A BCA protein quantification assay (Life Technologies) was used to measure total protein concentration from the samples, of which 25 μl was loaded onto a 4–15% gel (Bio-rad) for electrophoresis. The gel was transferred onto a nitrocellulose membrane by electroblotting, blocked in 5% non-fat dried milk prepared in wash buffer for 1 h, and incubated overnight with a primary antibody. Primary antibodies used were rabbit anti-phospho-Myosin Light Chain 2 (Thr18/Ser19) antibody (Cat. No. 3674), rabbit anti-Myosin Light Chain 2 antibody (Cat. No. 3672), rabbit anti-phospho-PI3K antibody (Cat. No. 4228), rabbit anti-phospho-p44/42 MAPK (p-Erk1/2) antibody (Cat. No. 9101), rabbit anti-p44/42 MAPK (Erk1/2) antibody (Cat. No. 9102), rabbit anti-phospho-AKT (Ser473) antibody (Cat. No. 4060) and GAPDH antibody (Cat. No. 5174), all purchased from Cell Signaling Technology. The membranes were washed and then incubated with anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody (Cell Signaling Technology, Cat. No. 7074) for 1 h. Detection was carried out using ECL chemiluminescence detection kit (GE Healthcare) with FluorChem E imaging system (ProteinSimple).
2.10. Statistical Analysis
Data were first checked for normality using Anderson-Darling method in MINITAB. For normally distributed data, one-way ANOVA with Tukey’s pairwise comparisons were used to compare means among three or more samples. Two-tailed, unpaired t-test was used to compare two experimental groups. Normally distributed data from experiments were expressed as mean ± standard error. Non-Gaussian distributed data were analyzed using two-tailed Mann-Whitney test. Values of p < 0.05 denoted statistical significance.
3. Results
3.1. Fibroblasts contract the ECM
To reproduce solid breast tumors, we generated organotypic tumor models containing a CXCR4+TNBC cell mass within a collagen matrix with dispersed fibroblasts (HMFs or CXCL12+HMFs). After forming a compact CXCR4+TNBC spheroid using our ATPS technology, we aspirated the medium robotically, dispensed collagen solution containing fibroblasts to embed the spheroid in each well and incubated the plate to allow collagen to gel (Fig. 1A). Prior to this, we also performed nanoindentation of collagen gels to determine the elastic modulus value of the collagen gel to match that of breast tumor tissues (Fig. S1A-B). Fig. 1B shows a 3D reconstructed volume view of the microtissue where a spheroid of CXCR4+TNBC cells (MDA-MB-231; blue) is surrounded by fibroblasts (green) dispersed in the collagen gel.
We observed that the cultures detached from walls of microwells and shrank over time. To understand this phenomenon, we created microtissues of collagen-embedded CXCR4+TNBC spheroids only or collagen-embedded dispersed fibroblasts only. Cultures containing only CXCR4+TNBC spheroids in collagen did not shrink the gel even after two weeks of culture, whereas both HMFs and CXCL12+HMFs contracted the collagen gel time-dependently. To quantitatively study contraction of the ECM by fibroblasts, we used known numbers of fibroblast cells in collagen gels of two different protein concentrations. We first examined the spatial distribution of fibroblasts in collagen and consistency of preparation. Fibroblasts were uniformity distributed in the 3D space and the number of cells included closely matched the number of cells quantified from the confocal images (Fig. 2A-B). Next, we captured phase images of the cultures and quantified ECM contraction over time. This study considered effects of collagen gel elastic modulus, fibroblast cell density, and fibroblast type (HMFs or CXCL12+HMFs) (Fig. 2C-G).
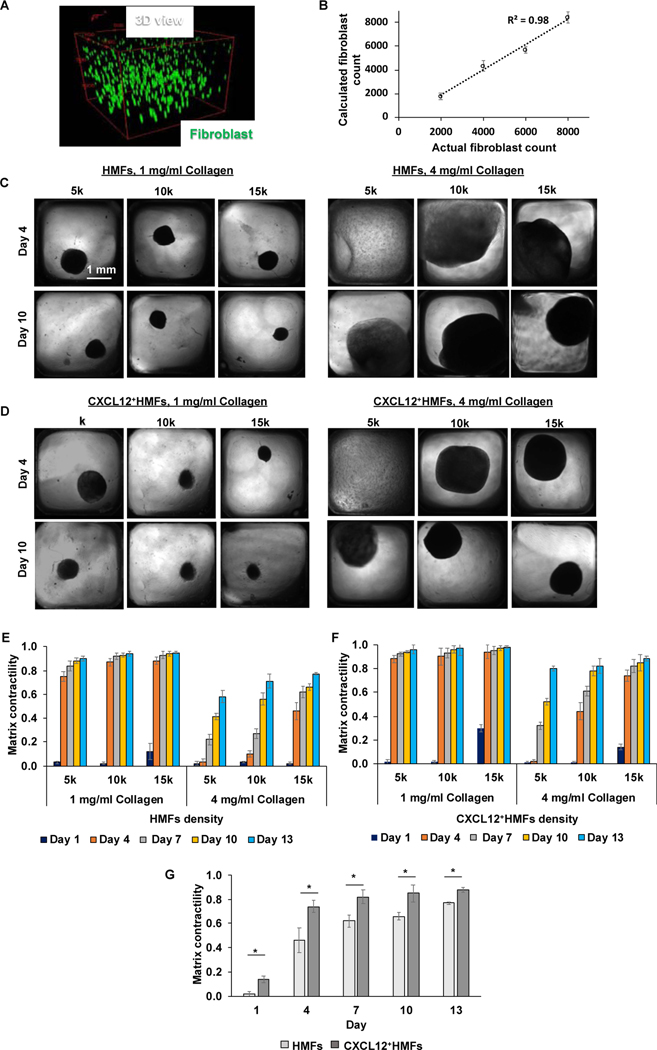
Fig. 2.
(A) 3D view of spatial distribution of fibroblasts (green) in collagen. (B) Fibroblasts were dispersed in collagen at a density of 2×103, 4×103, 6×103, or 8×103 cells per well. Each well contained 20 μl of collagen solution. The number of cells in each well was estimated by analyzing each stack of confocal images in MATLAB. Briefly, images from each stack were opened in ImageJ and XY coordinates of the cells were determined in an excel sheet. The coordinates were imported in MATLAB to count the total number of cells. The cells that had similar XY coordinates in four consecutive images were considered redundant and hence counted only once. Each cell XY coordinates usually repeated in two or three consecutive images. (C,D) Phase contrast images show contraction of collagen gels, 1 mg/ml and 4 mg/ml, by fibroblasts at different densities of 5×103 (5k), 10×103 (10k), and 1.5×104 (15k) on day 4 and day 10. Each density indicates the number of cells per well of 384-well plates. (E,F) Time-dependent contractility of collagen gel, 1 mg/ml and 4 mg/ml, by fibroblasts of different densities (5k,10k, and 15k). (G) Comparison of matrix contractility of 4 mg/ml collagen gels by 15k density of HMFs and CXCL12+HMFs. *p<0.05, n=16. Two-tailed unpaired t-test was used to compare the two experimental groups.
We found that with the softer ECM, the effect of fibroblast cell density was minimal and by day 4, the ECM shrank by 75–88% with HMFs and 88–94% with CXCL12+HMFs (Fig. 2E-F). The stiffer ECM showed significantly less contraction than the softer ECM for a given cell density. For example, on day 4 and with 1×104 HMFs, the stiffer ECM showed only 10% contraction, whereas the softer ECM had 87% contraction. With both fibroblasts, there was a cell density-dependent contractility effect, and larger cell densities caused a greater ECM contraction. For example, with the stiffer ECM on day 10, increasing the density of HMFs from 5×103 to 1.5×104 cells increased ECM contraction from 41% to 66%. Increasing density of CXCL12+HMFs in this range increased ECM contraction from 52% to 85%. CXCL12+HMFs more significantly contracted the ECM than HMFs at all cell densities used, and this effect was more pronounced with the stiffer ECM. For example, with a similar 1×104 cell density and on day 10, CXCL12+HMFs caused 78% ECM contraction compared to 56% with HMFs. Fig. 2G statistically compares ECM contraction by CXCL12+HMFs and HMFs at the largest cell density used. This analysis used for different cell densities showed that CXCL12+HMFs always generate significantly greater matrix contraction than HMFs (p<0.05).
3.2. Mechanism of ECM Contraction by Fibroblasts
Consistent with our results, fibroblasts in human tumor stroma show ECM remodeling ability and increased contractility [35,36]. To determine whether potential morphological differences between HMFs and CXCL12+HMFs contribute to their ability to generate force and contract the ECM, we conducted whole gel immunofluorescence of fibroblasts stained with β-actin and Hoechst. Confocal imaging showed that unlike HMFs, CXCL12+HMFs had a spindle-like, elongated morphology (Fig. 3A). CXCL12+HMFs had a five-fold higher cell aspect ratio than HMFs (Fig. 3B).
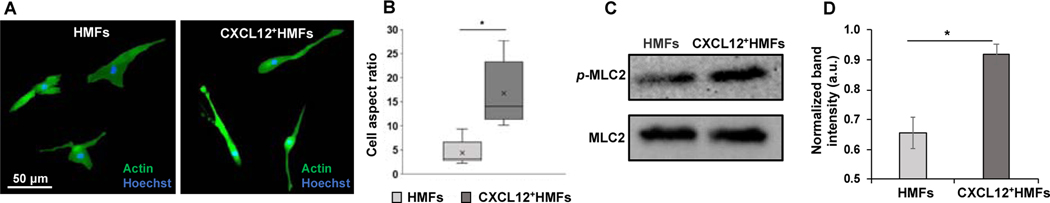
Fig. 3.
Collagen contraction by fibroblasts. (A) Immunofluorescence staining of β-actin (green) and Hoechst (blue) in HMFs and CXCL12+HMFs dispersed in collagen on day 5 of culture. (B) Box plot of cell aspect ratio () for HMFs and CXCL12+HMFs dispersed in collagen. The boxes represent the 25th and 75th percentiles with the median shown with a horizontal line inside each box. The mean is shown by a cross symbol inside each box. The whiskers represent the 10th and 90th percentiles of the data. Two-tailed Mann-Whitney test was used to calculate p-value. *p<0.01, n=10. (C) Western blot analysis of expression levels of phosphorylated and total MLC2 in HMFs and CXCL12+HMFs dispersed in collagen. (D) MLC2 activity quantified as p-MLC2/MLC2 from three separate experiments. Data were normally distributed and two-tailed, unpaired t-test was used to calculate p-value. *p<0.01.
To understand the underlying molecular mechanism, we evaluated activity of myosin light chain 2 (MLC2) protein that regulates actomyosin contractility and cell polarity. CXCL12+HMFs had 1.40-fold higher p-MLC2/MLC2 than HMFs (Fig. 3C-D). This was also consistent with the significantly higher motility (Fig. S2A-B) and RhoA and ROCK I activity (Fig. S2C) in CXCL12+HMFs than HMFs. Using a ROCK inhibitor, Y27632, fully blocked collagen contraction by both fibroblasts (Fig. S2D), establishing the RhoA/ROCK/MLC2 signaling as a major mechanism of collagen contraction by fibroblasts. Both fibroblasts dispersed in collagen had a similar level of metabolic activity, indicating that the difference in their contractility was not due to potential differences in their proliferation in culture (Fig. S3). Additionally, supplementing CXCL12-conditioned medium to HMFs did not make these cells more contractile (Fig. S4), negating a role for this chemokine in fibroblast cell contractility. Overall, activities of the signaling molecules that regulate cell-ECM mechnaotransduction support the greater contractility of CXCL12+HMFs than HMFs in the organotypic tumor model.
3.3. The role of fibroblasts in matrix invasion of cancer cells
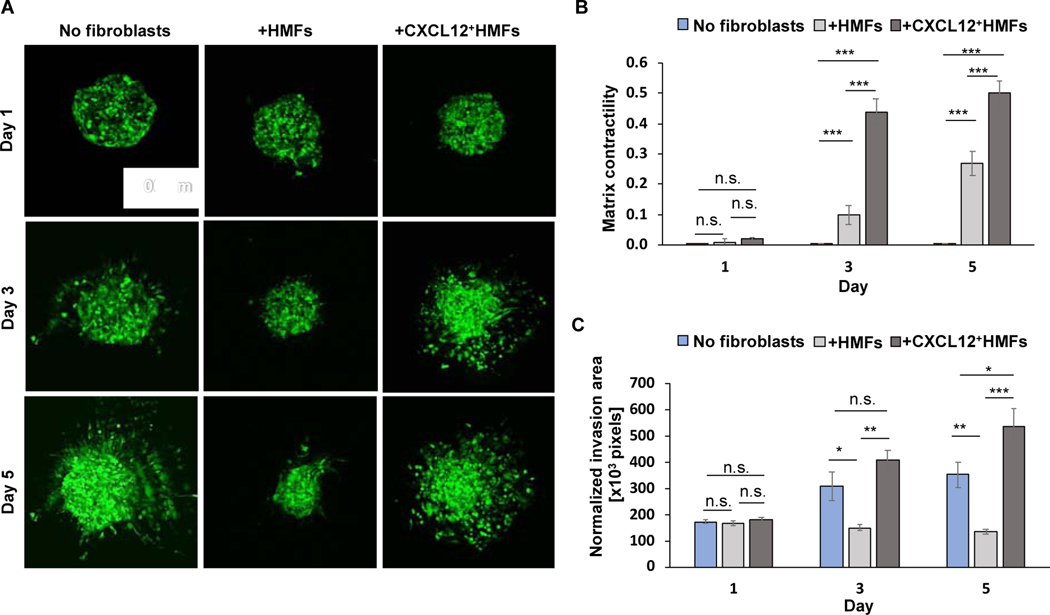
Next, we studied the role of fibroblasts on CXCR4+TNBC cell invasion of ECM in the organotypic cultures. We used collagen-embedded CXCR4+TNBC cell mass as a negative control to evaluate how HMFs and CXCL12+HMFs regulate breast cancer cell invasiveness. In the absence of fibroblasts, CXCR4+TNBC cells invaded the ECM and showed astral-like protrusions from the spheroid (Fig. 4A). CXCR4+TNBC cells showed contrasting behaviors when cultured with different fibroblasts. While CXCL12+HMFs promoted a significant ECM invasion of CXCR4+TNBC cells, HMFs suppressed CXCR4+TNBC cell invasiveness (Fig. 4A). To quantitatively compare cancer cell invasion of ECM among the three models, we normalized the invasion of CXCR4+TNBC cells of each model on a measurement day by its matrix contractility (Fig. 4B). This compensated the effect of ECM contraction on masking the invasion distance of cancer cells. Cancer cell spheroids in the three models showed a similar morphology after 24 h of incubation. However, significant differences emerged after 3 days. CXCR4+TNBC cells in the model containing CXCL12+HMFs showed a significantly more invasion than the other two models containing HMFs or without any fibroblasts. With CXCL12+HMFs, invasion of CXCR4+TNBC cells was 2.70-fold higher than that with HMFs, and 1.30-fold higher than that without fibroblasts (Fig. 4C). Maintaining the cultures for 5 days increased these differences to 4.0-fold and 1.5-fold, respectively.
Fig. 4.
Influence of fibroblasts on ECM invasion of cancer cell. (A) Confocal images of CXCR4+TNBC cell mass in three different microtissues over a 5-day culture. (B) Matrix contractility for CXCR4+TNBC-containing microtissues with and without 1.5×104 fibroblasts. Unpaired t-test was used to calculate the p-values. *p<0.05, **p<0.001, ***p<0.0001, n=10. (C) Normalized invasion of CXCR4+TNBC cells in microtissues with and without fibroblasts. Invasion area values were normally distributed and two-tailed, unpaired t-test was used to calculate the p-values. *p<0.05, **p<0.001, ***p<0.0001, n=8.
To develop a holistic view of 3D distribution of invading CXCR4+TNBC cells in the three models, we quantified CXCR4+TNBC cell numbers in different imaging planes and constructed histograms of position of the cells along the z-axis (Fig. 5A, Fig. S5). On day 3, CXCR4+TNBC cells in models including CXCL12+HMFs or without fibroblasts showed similar invasion profiles and a larger number of cells that invaded a greater distance than the model with HMFs (Fig. 5A). Results from day 5 samples showed a striking difference in the distribution of invading cells between these models, which could not be captured from the commonly-used analysis of a collapsed z-stack of images. In the presence of CXCL12+HMFs, CXCR4+TNBC cells showed the most scattering along the z-direction, whereas HMFs significantly inhibited invasion of CXCR4+TNBC cells. In the absence of fibroblasts, there was no significant difference in the travel distance of cancer cells in the z-direction from day 3 to day 5. To enable statistical comparison, we averaged the invasion results from each model that confirmed a significant increase in the ECM invasion of CXCR4+TNBC cells from day 3 to day 5 only in the presence of CXCL12+HMFs (Fig. 5B).
Fig. 5.
Analysis of ECM invasion of cancer cells along the z-axis. (A) Histograms of invasion of CXCR4+TNBC cells in three different microtissues on day 3 and day 5. Each colored curve in a graph represents pixels area of CXCR4+TNBC cells from one microtissue sample and each data point is obtained from one image of the confocal stack of images of that microtissue. Histograms have non-Gaussian distribution. (B) Normalized 3D spreading in three different tumor models. Normalized invasion area and AUC values were normally distributed. *p<0.05, **p<0.001, and ***p<0.0001 were calculated using two-tailed, unpaired t-test with n=8.
To identify potential molecular mechanisms underlying invasion of CXCR4+TNBC cells due to signaling with CXCL12+HMFs, we evaluated activities of several prominent protein kinases associated with tumor cell invasiveness. Results showed that ERK1/2 activity was significantly higher by 1.4-fold in microtissues containing CXCL12+HMFs than those with HMFs (Fig. 6A-B). In addition, we determined ERK1/2 phosphorylation in mono-cultures of only fibroblasts or TNBC spheroids in collagen gel. The active levels of ERK1/2 in both fibroblasts were comparable and even lower in TNBC spheroids. These results suggest that CXCL12 signaling with CXCR4+TNBC cells in the organotypic culture accounts for the increased ERK1/2 activity. We did not observe any significant difference in activities of p-AKT or p-PI3K between the two organotypic models (data not shown).
Fig. 6.
(A) Western blot analysis of ERK1/2 phosphorylation in different microtissues. (B) Quantified p-ERK1/2 normalized with t-ERK1/2 in five different microtissues. Data represent three separate experiments. +HMFs and +CXCL12+HMFs indicate organotypic cultures containing a TNBC spheroid and fibroblasts dispersed in collagen. For statistics, t-test was used to compare +HMFs and +CXCL12+HMFs, whereas one-way ANOVA with Tukey’s pairwise comparisons were used to compare CXCR4+TNBC, HMFs, and CXCL12+HMFs. *p<0.05 and ***p<0.0001.
3.4. Inhibition of ECM invasion of CXCR4+TNBC cells
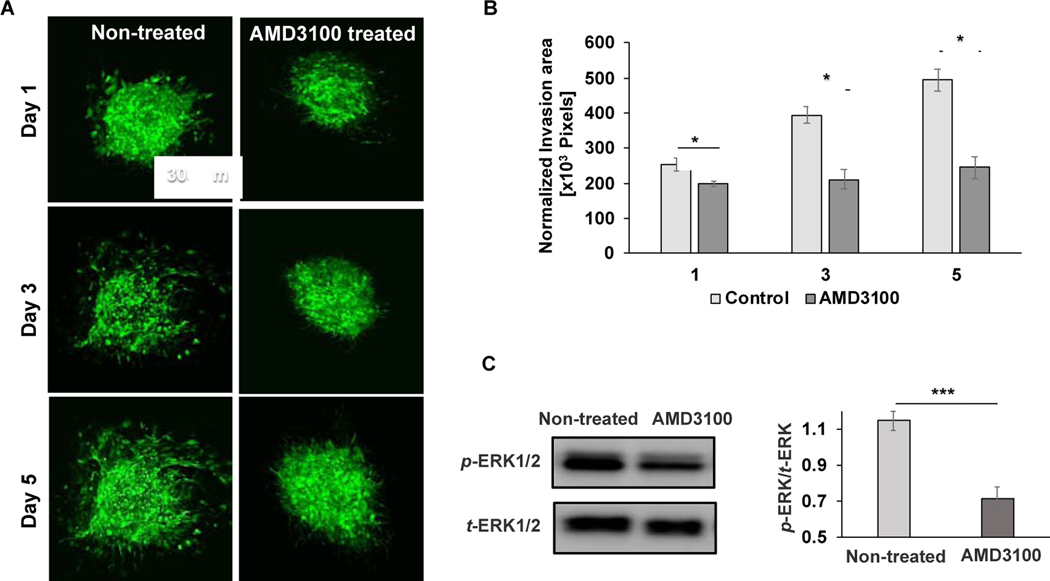
We treated the CXCL12+HMFs-containing microtissues with 1 μM of AMD3100, which is a potent antagonist of CXCR4. We imaged the microtissues on days 1, 3, and 5 and used the z-projected images to investigate the effect of blocking CXCL12 – CXCR4 signaling on ECM invasion of CXCR4+TNBC cells. Only after 24 h of incubation, the treatment significantly reduced ECM invasion of cancer cells (Fig. 7A-B). While cancer cell invasion of the matrix increased during culture without treatment, blocking CXCR4 signaling maintained the cancer cells as a minimally-invasive spheroid (Fig. 7A-B). Our molecular analysis showed that blocking CXCR4 signaling significantly reduced p-ERK1/2 levels in the CXCR4+TNBC:CXCL12+HMFs microtissue (Fig. 7C). These results further established the role of MAPK pathway in TNBC cell invasiveness and the potential of inhibiting tumor-stromal signaling as a therapeutic approach.
Fig. 7.
Inhibition of tumor-stromal interactions. (A) Confocal z-projected images of the CXCR4+TNBC cells in CXCL12+HMFs-containing microtissues without and with AMD3100 treatment. (B) Normalized invasion area for AMD3100-treated and non-treated microtissues. *p<0.01 was calculated using two-tailed, unpaired t-test with n=8. (C) Western blot analysis of effect of AMD3100 treatment on ERK1/2 activity. The graph shows quantified phospho-protein levels after normalizing with t-ERK1/2. Data represent three separate experiments. *p<0.05 and ***p<0.0001 were calculated using two-tailed, unpaired t-test.
4. Discussion
Unlike approaches using co-cultures or Matrigel to form 3D cultures, our high throughput organotypic tumor model both mimics the architecture of solid tumors and reproduces physical and biochemical properties of the stroma. As a proof of concept in this study, we used breast tissue fibroblasts as stromal cells and type I collagen as the primary component of ECM in breast tissue. To demonstrate feasibility of adjusting properties of the system, we developed tumor models that replicated elastic modulus of breast tumors. The 1 mg/ml and 4 mg/ml collagen matrices had elastic moduli of 0.96 and 2.53 kPa to mimic bimodal stiffness of human breast tumors [37,38]. We also adjusted the cellular composition of the model to a 2:1 ratio of fibroblast:cancer cells to mimic advanced breast tumors that have a larger stromal content, which correlates with tumor progression and poorer prognosis [39,40].
We observed significant contraction of the collagen matrix as early as 48 h after forming the models. This ECM contraction was solely due to the dispersed fibroblasts as a spheroid of CXCR4+TNBC cells did not shrink the matrix even after 2 weeks of culture. Additionally, there was no significant difference in the matrix contraction of fibroblasts when the culture contained or lacked tumor spheroids (data not shown). This is consistent with the known capability of fibroblasts to remodel the ECM in normal and pathological conditions such as wound healing, fibrosis, and cancer [41,42]. We characterized ECM contraction as a function of density and type of the fibroblasts (HMFs and CXCL12+HMFs), and elastic modulus of collagen gel. Consistent with results from 3D contraction assays [42,43], our finding suggests that fibroblasts contract softer collagen hydrogels faster than stiffer ones, and that a greater density of fibroblasts expedites the process. More importantly, we found that CXCL12+HMFs contract the collagen matrix more significantly than their normal counterparts, regardless of cell density or elastic modulus of the matrix. We verified that the two fibroblasts in the organotypic model have similar proliferation rates (Fig. S3), excluding the possibility of potential differences in their proliferative activities causing the difference in their contractility.
Previous studies showed that activated fibroblasts, known as myofibroblasts, enhance contraction of collagen lattices and feature elongated morphology with stress fibers [44–46]. In addition, myofibroblastic stromal cells derived from diseased fibrotic tissue have enhanced collagen contraction than stromal cells derived from normal tissue [47]. We hypothesized that CXCL12+HMFs may exhibit myofibroblastic morphology responsible for their higher ECM contraction than HMFs. Indeed, CXCL12+HMFs dispersed in collagen hydrogels had an elongated morphology compared to HMFs that showed a branching morphology (Fig. 3A). Seeding the fibroblasts on the surface of collagen gels also reproduced this morphological difference (Fig. S6A). Additionally, the clear difference in the morphology of these two fibroblasts persisted at the same contractile time point. That is, CXCL12+HMFs on day 13 and HMFs on day 4 had a similar matrix contractility of about 0.75 (Fig. 2G) but displayed drastically different morphologies (Fig. S6B). Consistent with this finding, CXCL12+HMFs were more migratory than HMFs in a wound healing assay (Fig. S2A-B). Our molecular analysis showed that CXCL12+HMFs dispersed in collagen gels have a significantly greater cell-matrix mechanotransduction activity than HMFs, i.e., higher levels of contractility-associated proteins in the RhoA/ROCK I/MLC2 pathway (Fig. 3C-D & Fig. S2C). This is consistent with studies that show CAFs generate actomyosin-mediated contraction and remodel ECM through this pathway [48–51]. These results suggest that the different morphologies of HMFs and CXCL12+HMFs are inherent properties of the cells.
To validate the biological relevance of our organotypic tumor model, we investigated the effect of CXCL12-producing fibroblasts on matrix invasion of breast cancer cells by focusing on the CXCL12 – CXCR4 signaling axis. Among different soluble signaling molecules of CAFs, CXCL12 is a prominent chemokine promoting invasion of cancer cells [52,53]. CXCL12 enhanced invasiveness of CXCR4+TNBC cells significantly and by four-fold compared to HMFs that lacked this chemokine. It is important to note that we normalized invasion of CXCR4+TNBC cells with matrix contractility that was quantified from 2D projected area of collagen gels. However, gels contract volumetrically. Considering volumetric contraction of the matrix would increase invasion of cancer cells in the model containing CXCL12+HMFs even more. For example, approximating the shapes of hydrogels and quantifying invasion of cancer cells in day 5 cultures using the volumetric approach resulted in eight-fold increase in the invasion of cancer cells in the model containing CXCL12+HMFs than that with HMFs. Overall, this is consistent with studies that showed increased cancer cell invasion in microenvironments enriched with CAFs [50,54], and that CAFs promote invasion and metastasis of tumor cells in vitro and in mouse models [55–57]. We found this signaling converges on MAPK pathway and amplifies ERK1/2 phosphorylation in CXCR4+TNBC cells to mediate enhanced invasion of cancer cells, consistent with the established role of ERK in tumor cell migration and invasiveness [58–60]. As a therapy strategy, we demonstrated that blocking CXCR4 leads to a significant reduction in ECM invasion of CXCR4+TNBC cells. This is a promising treatment strategy for TNBC disease because the lack of hormone receptors and low HER2 expression renders endocrine and targeted therapies not feasible.
To further delineate the importance of CXCL12 – CXCR4 signaling in cancer cell invasion, we generated organotypic models containing parental TNBC cells and CXCL12+HMFs fibroblasts. Results showed minimal matrix invasion of parental TNBC cells on day 5 of cultures (Fig. S7). This is because parental TNBC cells do not express detectable levels of CXCR4 receptors [29], highlighting that invasiveness of breast cancer cells depends on both components of this signaling axis. It is important to recognize that in tumor microenvironments, there are temporally dynamic changes in the expression levels of signaling molecules as the disease progresses. That is, the expression of CXCR4 receptor may be induced due to the persistent presence of its chemokine ligand in tumor microenvironments. These changes often occur over several months to years [61], and capturing them with in vitro systems may not be readily feasible. In addition, in the organotypic CXCR4+TNBC:HMFs model that lacked CXCL12 to initiate CXCR4 signaling, invasion of cancer cells was significantly reduced and cancer cells remained confined to a spheroid (Fig. 4). This is consistent with studies that showed normal mammary fibroblasts limit and even reverse malignant transformation of breast epithelial cells, which restore their baso-apical polarity, secretion of a tissue-specific glycoprotein into the acinar lumens, and deposition of basement membrane [62]. Normal fibroblasts were also shown to inhibit proliferation and invasion of malignant cells through processes including changes in ECM architecture and pro-inflammatory molecules [63]. Elucidating inhibitory effects of normal fibroblasts on invasiveness of TNBC cells in our model requires further studies.
Because co-culture spheroids are routinely used in cancer research, we aimed to compare the ECM invasion of cancer cells in our organotypic model with intermixed co-cultures of fibroblasts and cancer cells embedded in collagen. Results from intermixed co-cultures showed that cancer cells from both CXCR4+TNBC:HMFs and CXCR4+TNBC:CXCL12+HMFs co-cultures invaded the ECM without any significant difference (Fig. S8A-B) [64]. Consistent with incompatibility of cell adhesion molecules of fibroblasts and cancer cells [65,66], our previous study showed that the fibroblasts and CXCR4+TNBC cells segregate from an intermixed spheroid, and that this effect was greater with HMFs than with CXCL12+HMFs [16]. Considering that intermixed co-cultures do not mimic the architecture of solid tumors in terms of spatial distribution of cancer and stromal cells, our results suggest that their use for cancer cell invasion studies can lead to misleading information.
Overall, this study underscores the value of our organotypic model that reproduces spatial positioning of cells in solid tumors where cancer cells are bordered by stroma containing dispersed CAFs [67,68]. The modularity of our model allows convenient compositional adjustments, including addition of other stromal cells such as immune and endothelial cells and adjusting physicochemical properties of the ECM by supplementing various ECM elements of tumor microenvironment such as hyaluronan and fibronectin or using functionalized synthetic matrices. The compatibility of our model with microwell plates enables high throughput testing of chemical compounds using off-the-shelf robotic liquid handling tools for cancer drug discovery applications.
5. Conclusions
We developed high throughput organotypic tumor microtissues that mimic the structure and composition of solid breast tumors and reproduce dynamic interactions between breast cancer and stromal cells in presence of a natural ECM. This model allowed us to mechanistically study the role of fibroblasts in contraction of collagen matrix and invasiveness of TNBC cells. We demonstrated that both components of the CXCR4 – CXCL12 tumor-stromal signaling are required to promote ECM invasion of TNBC cells through oncogenic MAPK pathway. We showed that blocking tumor-stromal signaling using a targeted therapeutic molecule may be used as a therapy strategy to inhibit matrix invasion of breast cancer cells. This convenient-to-use and scalable tumor model offers a great potential to study tumor-stromal interactions and as a drug discovery tool in compound screening applications.
Supplementary Material
Acknowledgements
This work was supported by grants 1801591 from National Science Foundation and CA225549 from National Institutes of Health. The authors are grateful to Dr. Yuanzhong Zhang and Dr. Younjin Min for helping with AFM measurements.
Footnotes
Data Availability
The data required to reproduce these findings, both raw and processed, is available upon request.
Conflicts of Interest
Authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Balkwill FR, Capasso M, Hagemann T, The tumor microenvironment at a glance, Journal of Cell Science. 125 (2012) 5591–5596. doi: 10.1242/jcs.116392. [DOI] [PubMed] [Google Scholar]
- [2].Kalluri R, Zeisberg M, Fibroblasts in cancer, Nature Reviews Cancer. 6 (2006) 392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- [3].Kalluri R, The biology and function of exosomes in cancer, Journal of Clinical Investigation. (2016) 1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].De Kruijf EM, Van Nes JGH, Van De Velde CJH, Putter H, Smit VTHBM, Liefers GJ, Kuppen PJK, Tollenaar RAEM, Mesker WE, Tumor-stroma ratio in the primary tumor is a prognostic factor in early breast cancer patients, especially in triple-negative carcinoma patients, Breast Cancer Research and Treatment. 125 (2011) 687–696. doi: 10.1007/s10549-010-0855-6. [DOI] [PubMed] [Google Scholar]
- [5].Amornsupak K, Jamjuntra P, Warnnissorn M, O Charoenrat P, Sanguanraksa D, Thuwajit P, Eccles SA, Thuwajit C, High ASMA + Fibroblasts and Low Cytoplasmic HMGB1 + Breast Cancer Cells Predict Poor Prognosis, Clinical Breast Cancer. 17 (2017) 441–452. doi: 10.1016/j.clbc.2017.04.007. [DOI] [PubMed] [Google Scholar]
- [6].Marsh T, Pietras K, McAllister SS, Fibroblasts as architects of cancer pathogenesis, Biochimica et Biophysica Acta - Molecular Basis of Disease. 1832 (2013) 1070–1078. doi: 10.1016/j.bbadis2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ronnov-Jessen L, A function for filamentous alpha-smooth muscle actin: retardation of motility in fibroblasts, The Journal of Cell Biology. 134 (2004) 67–80. doi: 10.1083/jcb.134.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Attieh Y, Vignjevic DM, The hallmarks of CAFs in cancer invasion, European Journal of Cell Biology. 95 (2016) 493–502. doi: 10.1016/j.ejcb.2016.07.004. [DOI] [PubMed] [Google Scholar]
- [9].Ungefroren H, Sebens S, Seidl D, Lehnert H, Hass R, Interaction of tumor cells with the microenvironment, Cell Communication and Signaling. 9 (2011) 18. doi: 10.1186/1478-811X-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lee GY, Kenny PA, Lee EH, Bissell MJ, Three-dimensional culture models of normal and malignant breast epithelial cells, Nature Methods. 4 (2007) 359–365. doi: 10.1038/nmeth1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Weigelt B, Lo AT, Park CC, Gray JW, Bissell MJ, HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment, Breast Cancer Research and Treatment. 122 (2010) 35–43. doi: 10.1007/s10549-009-0502-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bradford JR, Wappett M, Beran G, Logie A, Delpuech O, Brown H, Boros J, Camp NJ, McEwen R, Mazzola AM, D’Cruz C, Barry ST, Whole transcriptome profiling of patient-derived xenograft models as a tool to identify both tumor and stromal specific biomarkers, Oncotarget. 7 (2016) 20773. doi: 10.18632/oncotarget.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zanoni M, Piccinini F, Arienti C, Zamagni A, Santi S, Polico R, Bevilacqua A, Tesei A, 3D tumor spheroid models for in vitro therapeutic screening: a systematic approach to enhance the biological relevance of data obtained., Scientific Reports. 6 (2016) 19103. doi: 10.1038/srep19103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lee GH, Lee JS, Wang X, Lee SH, Bottom-Up Engineering of Well-Defined 3D Microtissues Using Microplatforms and Biomedical Applications, Advanced Healthcare Materials. 5 (2016) 56–74. doi: 10.1002/adhm.201500107. [DOI] [PubMed] [Google Scholar]
- [15].Andrew IM, Ian FT, Drug penetration in solid tumours, Nature Reviews Cancer. 6 (2006) 583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- [16].Ham SL, Thakuri PS, Plaster M, Li J, Luker KE, Luker GD, Tavana H, Three-dimensional tumor model mimics stromal-breast cancer cells signaling, Oncotarget. 9 (2018) 249. doi: 10.18632/oncotarget.22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kim SA, Lee EK, Kuh HJ, Co-culture of Tumor Spheroids with Fibroblasts as a Model for EMT, Experimental Cell Research. (2015). doi: 10.1016/j.yexcr.2015.05.016. [DOI] [PubMed] [Google Scholar]
- [18].Yamaguchi H, Yoshida N, Takanashi M, Ito Y, Fukami K, Yanagihara K, Yashiro M, Sakai R, Stromal fibroblasts mediate extracellular matrix remodeling and invasion of scirrhous gastric carcinoma cells, PLoS ONE. 9 (2014) e85485. doi: 10.1371/journal.pone.0085485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Izumi D, Ishimoto T, Miyake K, Sugihara H, Eto K, Sawayama H, Yasuda T, Kiyozumi Y, Kaida T, Kurashige J, Imamura Y, Hiyoshi Y, Iwatsuki M, Iwagami S, Baba Y, Sakamoto Y, Miyamoto Y, Yoshida N, Watanabe M, Takamori H, Araki N, Tan P, Baba H, CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin β1 clustering and invasiveness in gastric cancer, International Journal of Cancer. 138 (2016) 1207–1219. doi: 10.1002/ijc.29864. [DOI] [PubMed] [Google Scholar]
- [20].Graeser R, Arundkar SC, Chong Y, Gjerde K, Stock K, Oren M, Rudisch A, Stein Y, Santo VE, Boghaert E, Brito C, de Hoogt R, van Weerden W, Vidic S, Rotter V, Gruenewald S, Barbier M, Osswald A, Sommergruber W, Blom S, Selvam I, Estrada MF, Capturing tumor complexity in vitro: Comparative analysis of 2D and 3D tumor models for drug discovery, Scientific Reports. 6 (2016) 1–15. doi: 10.1038/srep28951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Truong DD, Kratz A, Park JG, Barrientos ES, Saini H, Nguyen T, Pockaj B, Mouneimne G, LaBaer J, Nikkhah M, A human organotypic microfluidic tumor model permits investigation of the interplay between patient-derived fibroblasts and breast cancer cells, Cancer Research. (2019) canres.2293.2018. doi: 10.1158/0008-5472.can-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee JH, Kim SK, Khawar IA, Jeong SY, Chung S, Kuh HJ, Microfluidic co-culture of pancreatic tumor spheroids with stellate cells as a novel 3D model for investigation of stroma-mediated cell motility and drug resistance, Journal of Experimental and Clinical Cancer Research. 37 (2018) 4. doi: 10.1186/s13046-017-0654-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Xu Y, Mi S, Wu Z, Qian X, Du Z, Zhang M, Sun W, Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening, Scientific Reports. 6 (2016) 1–11. doi: 10.1038/srep35544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Menon NV, Chuah YJ, Cao B, Lim M, Kang Y, A microfluidic co-culture system to monitor tumor-stromal interactions on a chip, Biomicrofluidics. 8 (2014) 064118. doi: 10.1063/1.4903762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang JD, Douville NJ, Takayama S, Elsayed M, Quantitative analysis of molecular absorption into PDMS microfluidic channels, Annals of Biomedical Engineering. 40 (2012) 1862–1873. doi: 10.1007/s10439-012-0562-z. [DOI] [PubMed] [Google Scholar]
- [26].McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu H, Schueller OJA, Whitesides GM, Fabrication of microfluidic systems in poly(dimethylsiloxane), An International Journal. 21 (2000) 27–40. doi:. [DOI] [PubMed] [Google Scholar]
- [27].Luker KE, Luker GD, Functions of CXCL12 and CXCR4 in breast cancer, Cancer Letters. 238 (2006) 30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- [28].Smith MCP, Luker KE, Garbow JR, Prior JL, Jackson E, Piwnica-Worms D, Luker GD, CXCR4 regulates growth of both primary and metastatic breast cancer, Cancer Research. 64 (2004) 8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- [29].Song JW, Cavnar SP, Walker AC, Luker KE, Gupta M, Tung YC, Luker GD, Takayama S, Microfluidic endothelium for studying the intravascular adhesion of metastatic breast cancer cells, PLoS ONE. 4 (2009) e5756. doi: 10.1371/journal.pone.0005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Singh S, Tavana H, Collagen Partition in Polymeric Aqueous Two-Phase Systems for Tissue Engineering, Frontiers in Chemistry. 6 (2018). doi: 10.3389/fchem.2018.00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Tavana H, Jovic A, Mosadegh B, Lee QY, Liu X, Luker KE, Luker GD, Weiss SJ, Takayama S, Nanolitre liquid patterning in aqueous environments for spatially defined reagent delivery to mammalian cells, Nature Materials. 8 (2009) 736–741. doi: 10.1038/nmat2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Atefi E, Lemmo S, Fyffe D, Luker GD, Tavana H, High throughput, polymeric aqueous two-phase printing of tumor spheroids, Advanced Functional Materials. 24 (2014) 6509–6515. doi: 10.1002/adfm.201401302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lemmo S, Atefi E, Luker GD, Tavana H, Optimization of aqueous biphasic tumor spheroid microtechnology for anti-cancer drug testing in 3D culture, Cellular and Molecular Bioengineering. 7 (2014) 344–354. doi: 10.1007/s12195-014-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ham SL, Atefi E, Fyffe D, Tavana H, Robotic production of cancer cell spheroids with an aqueous two-phase system for drug testing, Journal of Visualized Experiments. (2015) e52754. doi: 10.3791/52754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ronnov-Jessen L, Petersen OW, Bissell MJ, Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction, Physiological Reviews. 76 (2017) 69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- [36].Xing F, Saidou J, Watabe K, Cancer associated fibroblasts (CAFs) in tumor microenvironment., Frontiers in Bioscience. 15 (2010) 166–79. doi: 10.2741/3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Marija P, Marko L, Christophe AM, Ellen CO, Rosanna Z-D, Philipp O, Janne TH, Ueli A, Mohamed B-A, Roderick YHL, Cora-Ann S, The nanomechanical signature of breast cancer, Nature Nanotechnology. 7 (2012) 757. doi: 10.1038/nnano.2012.167. [DOI] [PubMed] [Google Scholar]
- [38].Boettiger D, Hammer DA, Weaver VM, Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Tensional homeostasis and the malignant phenotype, Cancer Cell. 8 (2005) 241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [39].Wu J, Liang C, Chen M, Su W, Association between tumor-stroma ratio and prognosis in solid tumor patients: a systematic review and meta-analysis, Oncotarget. 7 (2016). doi: 10.18632/oncotarget.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kramer CJH, Vangangelt KMH, van Pelt GW, Dekker TJA, Tollenaar RAEM, Mesker WE, The prognostic value of tumour–stroma ratio in primary breast cancer with special attention to triple-negative tumours: a review, Breast Cancer Research and Treatment. 173 (2019) 55–64. doi: 10.1007/s10549-018-4987-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Grinnell F, Fibroblast biology in 3D collagen matrices, Trends in Cell Biology. 13 (2003) 264–269. doi: 10.1016/S0962-8924(03)00057-6. [DOI] [PubMed] [Google Scholar]
- [42].Bell E, Ivarsson B, Merrill C, Production of a tissue-like structure by contraction of collagen lattices by human fibroblasts of different proliferative potential in vitro., Proceedings of the National Academy of Sciences of the United States of America. 76 (1979) 1274–8. doi: 10.1073/pnas.76.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhu YK, Umino T, Liu XD, Wang HJ, Romberger DJ, Spurzem JR, Rennard SI, Contraction of fibroblast-containing collagen gels: Initial collagen concentration regulates the degree of contraction and cell survival, In Vitro Cellular & Developmental Biology - Animal. 37 (2002) 10. doi:. [DOI] [PubMed] [Google Scholar]
- [44].Tomasek JJ, Haaksma CJ, Eddy RJ, Vaughan MB, Fibroblast contraction occurs on release of tension in attached collagen lattices: Dependency on an organized actin cytoskeleton and serum, The Anatomical Record. 232 (1992) 359–368. doi: 10.1002/ar.1092320305. [DOI] [PubMed] [Google Scholar]
- [45].Burridge K, Are stress fibres contractile?, Nature. 294 (1981) 691–692. doi: 10.1038/294691a0. [DOI] [PubMed] [Google Scholar]
- [46].Katoh K, Kano Y, Masuda M, Onishi H, Fujiwara K, Isolation and Contraction of the Stress Fiber, Molecular Biology of the Cell. 9 (1998) 1919–1938. doi: 10.1091/mbc.9.7.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yuge A, Nasu K, Matsumoto H, Nishida M, Narahara H, Collagen gel contractility is enhanced in human endometriotic stromal cells: A possible mechanism underlying the pathogenesis of endometriosis- associated fibrosis, Human Reproduction. 22 (2007) 938–944. doi: 10.1093/humrep/del485. [DOI] [PubMed] [Google Scholar]
- [48].Tomasek JJ, Vaughan MB, Kropp BP, Gabbiani G, Martin MD, Haaksma CJ, Hinz B, Contraction of myofibroblasts in granulation tissue is dependent on Rho/Rho kinase/myosin light chain phosphatase activity, Wound Repair and Regeneration. 14 (2006) 313–320. doi: 10.1111/j.1743-6109.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- [49].Kolodney M, Elson E, Correlation of myosin light chain phosphorylation with isometric contraction of fibroblasts., Journal of Biological Chemistry. 32 (1993) 23850–23855. [PubMed] [Google Scholar]
- [50].Hooper S, Hidalgo-Carcedo C, Sahai E, Gaggioli C, Harrington K, Grosse R, Marshall JF, Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells, Nature Cell Biology. 9 (2007) 1392–1400. doi: 10.1038/ncb1658. [DOI] [PubMed] [Google Scholar]
- [51].Olson MF, Sahai E, The actin cytoskeleton in cancer cell motility, Clinical & Experimental Metastasis. 26 (2009) 273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- [52].De Wever O, Nguyen QD, Van Hoorde L, Bracke M, Bruyneel E, Gespach C, Mareel M, Tenascin-C and SF/HGF produced by myofibroblasts in vitro provide convergent pro-invasive signals to human colon cancer cells through RhoA and Rac, The FASEB Journal. 18 (2004) 1016–1018. doi: 10.1096/fj.03-1110fje. [DOI] [PubMed] [Google Scholar]
- [53].Weinberg RA, Carey VJ, Naeem R, Delaunay T, Orimo A, Arenzana-Seisdedos F, Sgroi DC, Richardson AL, Gupta PB, Stromal Fibroblasts Present in Invasive Human Breast Carcinomas Promote Tumor Growth and Angiogenesis through Elevated SDF-1/CXCL12 Secretion, Cell. 121 (2005) 335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- [54].Fiering S, Ang L-H, Lacoste J, Smith TD, Griner E, Registered report: Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis, ELife. 4 (2015). doi: 10.7554/elife.04796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Goetz J, Minguet S, Navarro-Lérida I, Lazcano J, Samaniego R, Calvo E, Tello M, Osteso-Ibáñez T, Pellinen T, Echarri A, Cerezo A, Szanto AJP, Garcia R, Keely PJ, Mateos P, Cukierman E, Miguel A, Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis, Cell. 146 (2011) 148–163. doi: 10.1016/j.cell.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Karagiannis GS, Poutahidis T, Erdman SE, Kirsch R, Riddell RH, Diamandis EP, Cancer-Associated Fibroblasts Drive the Progression of Metastasis through both Paracrine and Mechanical Pressure on Cancer Tissue, Molecular Cancer Research. 10 (2012) 1403–1418. doi: 10.1158/1541-7786.MCR-12-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liao D, Luo Y, Markowitz D, Xiang R, Reisfeld RA, Cancer associated fibroblasts promote tumor growth and metastasis by modulating the tumor immune microenvironment in a 4T1 murine breast cancer model, PLoS ONE. 4 (2009) e7965. doi: 10.1371/journal.pone.0007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ham SL, Nasrollahi S, Shah KN, Soltisz A, Paruchuri S, Yun YH, Luker GD, Bishayee A, Tavana H, Phytochemicals potently inhibit migration of metastatic breast cancer cells, Integrative Biology. 7 (2015) 792–800. doi: 10.1039/c5ib00121h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Neophytou C, Boutsikos P, Papageorgis P, Molecular Mechanisms and Emerging Therapeutic Targets of Triple-Negative Breast Cancer Metastasis, Frontiers in Oncology. 8 (2018). doi: 10.3389/fonc.2018.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Masuda T, Endo M, Yamamoto Y, Odagiri H, Kadomatsu T, Nakamura T, Tanoue H, Ito H, Yugami M, Miyata K, Morinaga J, Horiguchi H, Motokawa I, Terada K, Suimye Morioka M, Manabe I, Iwase H, Mizuta H, Oike Y, ANGPTL2 increases bone metastasis of breast cancer cells through enhancing CXCR4 signaling, Scientific Reports. 5 (2015) 9170. doi: 10.1038/srep09170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Papatheodorou H, Papanastasiou A, Sirinian C, Scopa C, Kalofonos HP, Leotsinidis M, Papadaki H, Expression patterns of SDF1/CXCR4 in human invasive breast carcinoma and adjacent normal stroma: correlation with tumor clinicopathological parameters, Pathology - Research and Practice. 10 (2014) 662–667. doi: 10.1016/j.prp.2014.06.015. [DOI] [PubMed] [Google Scholar]
- [62].Roemer A, Luehr I, Klein A Friedl, Sebens S, Roesel F, Bauer M, Normal mammary fibroblasts induce reversion of the malignant phenotype in human primary breast cancer, Anticancer Research. 4 (2013) 1525–1536. [PubMed] [Google Scholar]
- [63].Alkasalias T, Flaberg E, Kashuba V, Alexeyenko A, Pavlova T, Savchenko A, Szekely L, Klein G, Guven H, Inhibition of tumor cell proliferation and motility by fibroblasts is both contact and soluble factor dependent, Proceedings of the National Academy of Sciences. 111 (2014) 17188–17193. doi: 10.1073/pnas.1419554111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Plaster M, Singh S, Tavana H, Fibroblasts Promote Proliferation and Matrix Invasion of Breast Cancer Cells in Co‐Culture Models, Advanced Therapeutics. 2 (2019) 1900121. doi: 10.1002/adtp.201900121. [DOI] [Google Scholar]
- [65].Krens SFG, Veldhuis JH, Barone V, Capek D, Maître JL, Brodland GW, Heisenberg CP, Interstitial fluid osmolarity modulates the action of differential tissue surface tension in progenitor cell segregation during gastrulation, Development. 144 (2017) 1798–1806. doi: 10.1242/dev.144964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Foty RA, Steinberg MS, The differential adhesion hypothesis: A direct evaluation, Developmental Biology. 278 (2005) 255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- [67].Ishii G, Ochiai A, Neri S, Phenotypic and functional heterogeneity of cancer-associated fibroblast, Advanced Drug Delivery Reviews. 99 (2016) 186–196. doi: 10.1016/j.addr.2015.07.007. [DOI] [PubMed] [Google Scholar]
- [68].Banerjee S, Yuan Y, Lan C, Huang X, Heindl A, Xi S, Liu J, Quantitative histology analysis of the ovarian tumour microenvironment, Scientific Reports. 5 (2015) 1–12. doi: 10.1038/srep16317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.