Abstract
Histone deacetylase (HDAC) 10, a class II family, has been implicated in various tumors and non-tumor diseases, which makes the discovery of biological functions and novel inhibitors a fundamental endeavor. In cancers, HDAC10 plays crucial roles in regulating various cellular processes through its epigenetic functions or targeting some decisive molecular or signaling pathways. It also has potential clinical utility for targeting tumors and non-tumor diseases, such as renal cell carcinoma, prostate cancer, immunoglobulin A nephropathy (IgAN), intracerebral hemorrhage, human immunodeficiency virus (HIV) infection and schizophrenia. To date, relatively few studies have investigated HDAC10-specific inhibitors. Therefore, it is important to study the biological functions of HDAC10 for the future development of specific HDAC10 inhibitors. In this review, we analyzed the biological functions, mechanisms and inhibitors of HDAC10, which makes HDAC10 an appealing therapeutic target.
Keywords: cancer, disease, epigenetics, HDAC10, histone deacetylases
Introduction
Nuclear DNA in eukaryotic cells always combines with histones before packaging into the chromatin. However, a fundamental question is how the chromatin is regulated when and where it is necessary. The existing evidence presented that ATP-dependent chromatin remodeling as well as covalent chromatin modification may contribute to this regulation [1]. Covalent chromatin modification includes acetylation, phosphorylation, methylation, ubiquitination and ADP-ribosylation [2,3]. Acetylation, as the first discovered modification, is the most studied and best characterized among all modifications. There is compelling evidence that acetylation of lysine residues at amino acid termini greatly affects transcriptional regulations [2].
Histone deacetylases (HDACs) serve as an ‘eraser’, which remove acetate from acetylated histone as well as other non-histone proteins [4,5]. Generally, HDACs are also known as lysine acetyltransferases as well as lysine deacetylases [6]. Additionally, HDACs exert effects on post-translational modifications, such as ubiquitination and methylation, and can influence gene transcription by increasing the interaction between DNA and histone [7–9]. In eukaryotic cells, HDACs may come from prokaryotic enzymes, similar to acetylpolyamine amidohydrolases [7,10]. Currently, HDACs are divided into four classes [11]. In 2002, Fischer et al. [12], Tong et al. [5], Kao et al. [13] and Guardiola and Yao [14] reported a novel enzyme that shares functional characterizations with other class II members, and named it HDAC10.
In this review, we focus our attention on the progress of studies on the role of HDAC10 in various tumors and non-tumor diseases. Particular emphasis is given to its functions and mechanisms.
Structures and catalytic mechanisms of different HDACs
Structures of class II HDACs
Class II HDACs are subclassified as IIa and IIb. Class IIa HDACs (HDAC4, 5, 7, 9) are defined by a functionally essential N-terminal domain, which can regulate nuclear–cytoplasmic shuttling as well as specific DNA-binding. The cellular trafficking of class IIa HDACs is determined by intrinsic nuclear import or export signals as well as specific binding sites for 14-3-3 proteins [15]. Through binding with the 14-3-3 proteins, Class IIa HDACs can stimulate cytoplasmatic retention and nuclear export in a phosphorylation-dependent manner. These processes in turn affect the activity of transcription factors, such as myocyte enhancing factor-2 [16,17].
Class IIb HDACs include HDAC6 and HDAC10. HDAC6 contains two tandem deacetylase domains and a C-terminal zinc finger [18]. HDAC10 is structurally related to HDAC6. The HDAC10 gene localizes to chromosome 22 [14]. Consisting of 20 exons, HDAC10 contains two spliced transcripts [5,12] and also contains N-terminal catalytic domain and C-terminal leucine-rich domain. Specifically, the N-terminal catalytic domain of HDAC10 is similar to the deacetylase domain of other known class II HDACs, but the C-terminal catalytic domain does not contain residues that are necessary for enzymatic functions. The existence of both domains may confer resistance to trapoxin B and sodium butyrate [5,14]. HDAC10 is member of the arginase/deacetylase superfamily and is expressed differentially in the cytoplasm and nucleus [10,12]. It may also repress transcription except for the deacetylation activity in the nucleus [14]. Moreover, HDAC10 expressed in most human tissues, including the kidney, pancreas, liver, spleen, heart, testis, brain and placenta [5]. Emerging evidence has established that HDAC10 is responsible for the regulation of polyamine. Using X-ray crystallography, researchers have observed that HDAC10 has a unique conserved glutamate gatekeeper that may promote N8-acetylspermidine hydrolysis, serving as a polyamine deacetylase (PDAC) [6,19].
Noticeably, class II HDACs show 23–81% amino acid similarity with conserved deacetylase domains. Moreover, they all have cytoplasmic localization, which implies that class II HDACs play significant role in cytoplasmic functions [13,20,21].
Catalytic mechanisms of HDACs
Class I, II and IV HDACs require zinc ion to exert enzyme activity. The X-ray crystallography has determined catalytic domain structures of Class I, II and IV HDACs, which contain open α/β-family of folds and stranded parallel β-sheets [22]. These structures make contributions to the binding of HDACs active sites and HDAC inhibitors (HDACIs). A model of action proposed by Wu et al. [23] suggested that H143, a histidine residue, could function as a base to accept protons from zinc-bound molecules in a rate-determining step. Then the accepted proton will be shuttled to the amide nitrogen atom to facilitate the cleavage of amide bond [24].
The class III HDACs has a common model of action that requires cofactor NAD+ [25]. A structural study found that the catalytic domain of class III HDACs locates in a cleft, which could constitute a protein tunnel where the substrate could combine with NAD+. Mechanically, based on the catalytic structure of class III HDACs, cleavable nicotinamide from NAD+ and ADP-ribose could transfer to acetylated lysine [26].
Biological characterization and mechanism of HDAC10 in tumors
In 2000 and 2011, Hanahan and Weinberg proposed six and additional four hallmarks of tumors, respectively [27,28]. These conceptions promoted the understanding of the diversity of tumors. Here, we focus mostly on HDACs that affect the biology of tumors and tumorigenesis, and summarize the mechanistic underpinnings concerning cell proliferation, cell apoptosis, cell invasion, migration, metastasis, angiogenesis and other tumor-related biological functions (Figure 1 and Table 1).
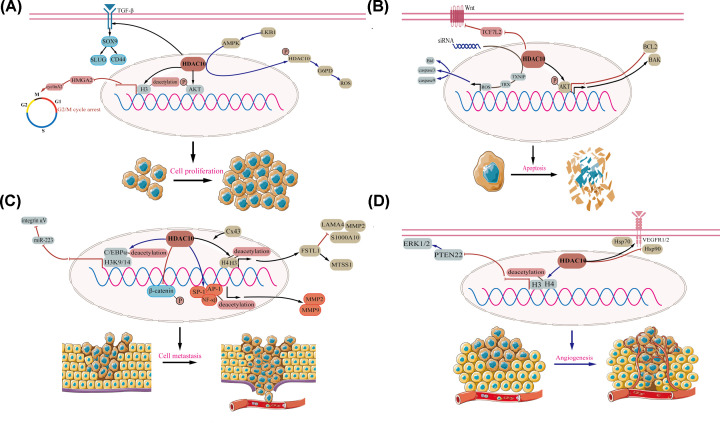
Figure 1. HDAC10 regulate cell proliferation, cell apoptosis, cell metastasis and angiogenesis in cancer cells.
(A) Roles of HDAC10 in cell proliferation. (B) Roles of HDAC10 in cell apoptosis. (C) Roles of HDAC10 in cell metastasis. (D) Roles of HDAC10 in angiogenesis.
Table 1. Overview of biological roles of HDAC10 in several malignant tumors.
| Tumor | Expression level | Function | Target/Signaling pathway | Cell line/Tissue | Reference |
|---|---|---|---|---|---|
| NSCLC | Elevated | Cell proliferation, Cell apoptosis | AKT, BCL2, BAK | A549, H358 and H460 cell | [18] |
| Lung adenocarcinoma | Elevated | Cell proliferation, cancer stemness, Tumor immune | TGF-β pathway, SOX9, CD44, SLUG | HDAC10 KO mice, HDAC10 WT mice | [19] |
| Lung cancer | Elevated | Cell proliferation | LKB1–AMPK signaling, G6PD, ROS | H1299, H157, H1944, H460 and A549 cell | [23] |
| Lung cancer | Elevated | Cell proliferation, Cell cycle | H3, HMGA2 | H1299, H441, H23, H157, H2122, H358, A549, PC9, H1975, H322, H292, H460, H522, H661 and ADLC-5M2 cell | [25] |
| Lung cancer | Decreased | Cell metastasis | Cx43, FSTL1, H3, H4, S100A10, MMP-2, LAMA4, MTSS1 | PG cell | [41] |
| RCC | Decreased | Cell proliferation, cell invasion | β-catenin | ACHN, Caki-2 cell, 45 primary RCC tissues and adjacent normal tissues | [20] |
| Gastric cancer | / | Cell apoptosis | Caspase-3, Caspase-9, Bid, TXNIP, ROS | SNU-620 cell | [35] |
| Gastric cancer | / | Angiogenesis | Hsp70, VEGFR1, VEGFR2 | SNU620 cell | [51] |
| Colorectal cancer | / | Cell apoptosis | Wnt pathway, TCF7L2 | SW480, HCT116 cell | [32] |
| Colon cancer | / | Angiogenesis | Hsp70, VEGFR1, VEGFR2 | HCT116 cell | [51] |
| Colon cancer | Elevated | Mismatch repair | MSH2, MLH1, MSH6 | 100 colon cancer tissues | [81] |
| Cervical cancer | / | Cell metastasis | MMP-2, MMP-9 | HeLa-S3 cell, 60 cancer tissue and normal tissue | [42] |
| Cervical cancer | / | Autophagy | LAMP2A-positive lysosome | HeLa cell | [75] |
| Cervical cancer | / | Mismatch repair | MSH2 | HeLa cell | [80] |
| Cervical cancer and Osteosarcoma | / | Drug resistance | hSSB1, p300 | HeLa and U2OS cells | [62] |
| HCC | / | Cell metastasis | H3K9/14, C/EBPα, miR-223, integrin αV subunit | SMMC-7721 cell | [43] |
| HCC | / | HBV-infected HCC | Promoter polymorphism | 1095 HBV infection patient tissues | [82] |
| Neuroblastoma | / | Drug resistance | Lysosomal exocytosis, DNA damage | SK-N-BE (2)-C, IMR-32 and SK-N-AS cell | [55] |
| Neuroblastoma | / | Drug resistance | Hsp70, Hsc70 | BE [2]-C, Kelly and IMR32 cell | [57,58] |
| Ovarian | Decreased | Drug resistance | DNA repair | UWB1.289 cell | [59] |
| Prostate cancer | / | Drug resistance | AR-V7, flAR, BRD4, NCOR2, DUB3, BRD4 | C4-2, PC-3, 22Rv1, DU145, LNCaP, VCaP and LAPC4 cells | [66,67] |
| Adrenocortical cancer | / | Chromatin modulator | Methylation | 44 adrenocortical cancer and 6 normal tissues | [83] |
| Melanoma | Decreased | Gene expression | HDAC | Melanoma tissue and normal samples | [84] |
Abbreviations: AKT, protein kinase B; AMPK, AMP-activated protein kinase; AR-V7, androgen receptor-V7; BAK, BCL2 antagonist/killer; BCL2, B-cell lymphoma-2; Bid, BH3 interacting domain death agonist; BRD4, bromodomain-containing protein 4; Cx43, connexin 43; C/EBPα, CCAAT enhancer-binding protein; DUB3, ubiquitin-specific peptidase 17 like family member 2; flAR, full length androgen receptor; FSTL1, follistatin-like 1; G6PD, glucose-6-phosphate dehydrogenase; HBV, hepatitis B virus; HCC, hepatic cell carcinoma; HMGA2, high mobility group A 2; Hsp70, heat shock protein 70; hSSB1, human single-stranded DNA binding protein; LAMA4, laminin subunit α 4; LAMP2A, lysosomal associated protein 2 A; LKB1, liver kinase B1; MLH1, mutL homolog 1; MMP-2, matrix metalloproteinase-2; MMP-9, matrix metalloproteinase-9; MSH2, mutS homolog 2; MSH6, mutS homolog 6; MTSS1, MTSS I-BAR domain containing 1; NCOR2, nuclear receptor co-repressor 2; NSCLC, non-small-cell lung cancer; RCC, renal cell carcinoma; ROS, reactive oxygen species; SOX9, sex-determining region Y box protein 9; TCF7L2, transcription factor 7 like 2; TGF-β, transforming growth factor-β; TXNIP, thioredoxin interacting protein; VEGFR1, vascular endothelial growth factor receptor 1; VEGFR2, vascular endothelial growth factor receptor 2.
HDAC10 and cell proliferation
HDAC10 plays an essential role in the regulation of cancer cell growth. It has been demonstrated that it can prompt cell proliferation in non-small-cell lung cancer (NSCLC) [29]. In contrast, HDAC10 suppresses cancer cell growth in other malignant tumors, including lung adenocarcinoma [30] and renal cell carcinoma (RCC) [31]. Akt is a key molecule in the PI3K-AKT pathway, and is involved in cell proliferation, survival and other key functions [32]. Analogously, HDAC10 overexpression significantly facilitates NSCLC cell growth by regulating the Ser473 phosphorylation of AKT [29]. In lung adenocarcinoma, deletion of HDAC10 accelerates the progression of KRAS-driven cancer both in vivo and in vitro. Specifically, by activating the transforming growth-factor β (TGF-β) pathway, deletion of HDAC10 promotes the expression of sex-determining region Y box protein 9 (SOX9), which subsequently up-regulates the expression of SLUG, as well as CD44. These processes contributes to the growth of lung cancer spheres via SOX9-mediated stem-like properties, suggesting that HDAC10-TGF-β-SOX9-SLUG/CD44 axis plays an essential role in lung adenocarcinoma [30].
AMP-activated protein kinase (AMPK) regulates biological functions in tumors that are mediated by liver kinase B1 (LKB1), such as cell survival and transcription, via the mTOR pathway [33]. Induced by LKB1–AMPK signaling, phosphorylated HDAC10 is transported from the the nucleus to the cytoplasm and further enhances the expression of glucose-6-phosphate dehydrogenase (G6PD). This decreases the level of reactive oxygen species (ROS) and promotes lung cancer cell proliferation [34]. The cases described above suggest that the LKB1–AMPK-HDAC10-G6PD-ROS pathway might be important to tumor cell proliferation. However, in RCC cells, suppressed expression of HDAC10 significantly promotes the phosphorylation of β-catenin and thus plays a part in anti-proliferation [31] (Figure 1A).
Perturbed cell cycle is also a growth-regulation way in cancer cell [35]. Previous studies have shown that by inhibiting histone H3 deacetylation around the let-7f-2/miR-98 promoter, HDAC10 suppresses HMGA2 expression which target to cyclin A2 promoter, and further inhibits the transcription of cyclin A2 [30,36]. The signaling pathways: HDAC10-let-7f-2/miR-98-HMGA2-cyclin A2 arrests the G2/M transition and finally inhibits lung cancer cell proliferation. Evidence suggests that both cell cycle inhibitors (such as P21 and P27) and promoters (such as cyclins E1 and D1) play a vital role in cancer progression [37–39]. HDAC10 plays an oncogenic role by inhibiting the expression of P27, P21 and enhancing that of cyclins D1 and E1 [29] (Figure 1A).
HDAC10 and cell apoptosis
Cancer cells will not die in a scenario that too little apoptosis happens [40]. Numbers of proteins and signaling are involved in cell apoptosis. It has been established that overexpressed anti-apoptotic proteins (such as those in the Bcl-2 family) as well as down-regulated proteins (such as Bid, BIK and BAK) may disrupt the balance between apoptosis and anti-apoptosis [41,42]. By targeting AKT, HDAC10 affects the expression of B-cell lymphoma-2 (BCL2) as well as BCL2 antagonist/killer (BAK), which induces apoptosis in lung carcinoma [29]. In colorectal cancer, inhibited HDAC10 expression promotes cell apoptosis by depleting transcription factor 7 like 2 (TCF7L2), which attenuates the Wnt pathway [43]. ROS, generated from mitochondrial damage or oxidative stressors, may promote caspases and induce apoptosis [44,45]. Lee et al. found that a low level of HDAC10 in gastric cancer may activate proapoptotic molecules including caspase-3, caspase-9, and Bid through the thioredoxin interacting protein (TXNIP)-induced ROS signaling pathway [46] (Figure 1B). Although previous studies have determined limited mechanisms in lung, colorectal and gastric tumors, including the HDAC10-AKT-BCL2-BAK pathway, the HDAC10-TCF7L2-Wnt pathway and the HDAC10-TXNIP-ROS-caspase-3/caspase-9/Bid pathway, it is not yet clear whether these mechanisms exist in other tumors [29,43,46]. Therefore, advances in research on the mechanisms of HDAC10 will be key to unraveling its potential importance in tumors.
HDAC10 and cell metastasis
Tumor cell metastasis is a multistep process including cell adhesion, invasion, migration and dissemination at distant organs [47,48]. Invasion- and migration-related molecules, such as matrix metalloproteinases (MMPs), and S100A10 are dysregulated in various cancer cell lines [49–51]. Zhao et al. [52] reported that HDAC10 shows low expression levels in pulmonary giant cell carcinoma cells and is subject to regulation by connexin 43 (Cx43). As Cx43 is overexpressed, the expression of follistatin-like 1 (FSTL1) is elevated, via the enhanced binding between HDAC10-mediated acetylation of H3 and H4 and the promoter of FSTL1. The Cx43-HDAC10-FSTL1 axis not only contributes to the low expression levels of S100A10, MMP-2 and laminin subunit α 4 (LAMA4), but also enhances the expression of MTSS I-BAR domain containing 1 (MTSS1), which plays a pivotal role in the suppression of both invasion and metastasis [52]. In metastatic cervical squamous cancer cells, HDAC10 serves a tumor suppressor role through the regulation of MMPs. Mechanistically, due to the combination of HDAC10 and the promoters of MMP-2 (especially the AP1-binding site) and MMP-9 (especially the NF-κB- and sp1-binding sites) the decreased level of histone acetylation impedes the binding function of polymerase II, weakens the expression levels of MMP-2 and MMP-9 and restrains cancer cell invasion and migration [53]. HDAC10 also deacetylates acetyl-H3K9/14 and acetyl-C/EBPα, preventing their recruitment to the promoter of miR-223 in sulfatide-treated hepatocellular cancer (HCC) cells. The low expression of miR-223 further facilitates the expression of integrin αV subunit and then attenuates HCC cell migration as well as metastasis [54]. The β-catenin pathway is altered in various tumors and also plays important roles in carcinogenesis [55,56]. In renal cell carcinoma (RCC), overexpressed HDAC10 restrains RCC cell invasion by inhibiting phosphorylated nuclear β-catenin expression [31] (Figure 1C). Although these studies are interesting, they were mainly based on overexpression experiments. Therefore, the mechanism of HDAC10 in tumor metastasis must be characterized in more detail.
HDAC10 and angiogenesis
Angiogenesis is an indispensable process in the early stage of tumor growth [57]. Angiogenesis involves several pro-angiogenic proteins, including vascular endothelial growth factor (VEGF), TGF-β, extracellular regulated protein kinases (ERKs) and others [58–60]. Duan et al. [61] reported that overexpressed HDAC10 deacetylates H3 and H4 in promoter of phosphatase and tensin homolog 22 (PTEN22) and consequently inhibits polymerase II from binding to PTEN22 promoter. The suppressed expression of PTEN22 subsequently increases ERK1/2 activation and ultimately facilitates the formation of tubes in vivo and in vitro. Because of this, it was speculated that the HDAC10-PTEN22-ERK1/2 signaling pathway may contribute to tumor angiogenesis. HDAC10 also regulates angiogenesis via depletion of VEGFR in gastric and colon cancer cells. Mechanistically, HDAC10 obstructs heat shock protein (Hsp) 90 bound to VEGF receptor 1 (VEGFR1)/VEGFR2, but promotes the combination between Hsp70 and VEGFR1/VEGFR2 [62]. This imbalanced binding leads to proteasomal-dependent degradation of VEGFRs, which may regulate angiogenesis (Figure 1D).
HDAC10 and drug resistance
Drug resistance is a common phenomenon during chemotherapy. It not only limits the therapeutic effect but can even lead to treatment failure [63]. The altered drug metabolism, mutated drug targets and elevated drug efflux rate contribute to drug resistance [64,65]. Down-regulation of HDAC10 may restore the sensitivity of doxorubicin in neuroblastoma. Ridinger et al. [66] reported that suppressing HDAC10 expression promotes intracellular accumulation of doxorubicin by inhibiting lysosomal exocytosis, which results in cell death with enhanced double-strand breaks (DSBs) as well as DNA damage. Additionally, autophagy, activated via therapeutic stress, participates in drug resistance and multidrug resistance (MDR) [67]. Consistent with this point, Oehme et al. [68,69] found that HDAC10 deletion acetylated Hsp70/heat shock cognate 70 (Hsc70), and impairs the autophagic flux by thwarting the blend between autophagosomes and lysosomes, which allows drug-resistant neuroblastoma cell to recover sensitivity to doxorubicin. A study that combined bioinformatics analyses and cell experiments found that low HDAC10 expression both intensifies the cytotoxicity of ovarian cancer cells to cisplatin and constrains DNA repair [70].
Many cytotoxic events can break DNA double strands and result in cell death [71]. Encountering DNA damage, various proteins, such as human single-stranded DNA binding protein 1 (hSSB1), will repair the flawed sites [72]. Wu et al. [73] found that p300 acetylates hSSB1 Lys94 when it localizes to impaired sites, and recovers DNA stability. However, up-regulated HDAC10 reverses this process and makes tumor cells more responsive to chemotherapy. Castration-resistant prostate cancer (CRPC), arising from resistance to androgen deprivation therapy (ADT), is a tricky problem [74]. Previous studies have reported that bromodomain and extra-terminal protein inhibitor (BETi) can deteriorate the androgen receptor (AR) signaling pathway. Moreover, in prostate cancer cells, elevated expression of bromodomain-containing protein 4 (BRD4, a member of BET family), induced by BETi, confers resistance to these inhibitors [75,76]. Jin et al. [77] found that low expressed nuclear receptor co-repressor 2 (NCOR2)–HDAC10 complex is negatively associated with deubiquitinase ubiquitin-specific peptidase 17 like family member 2 (DUB3), which may interact with BRD4 and further inhibit sensitivity to BETi in prostate cancer cells and in vitro. Although the roles of HDAC10 in drug resistance have not been explored completely, experiments suggest that the NCOR2-HDAC10-DUB3-BRD4 signaling pathway might be useful as a regulator of drug resistance in tumors [77]. AR is a significant target in treating CRPC, and silencing HDAC10 also reduces the transcriptional activity of AR-V7 and full-length AR (flAR) in prostate cells [78], which implies that HDAC10 has potential therapeutic value.
Other biological functions
Cancer stem cells (CSCs) are cells hidden in tumors that are capable of promoting tumor growth, progression and resistance treatments [79,80]. Many molecules, such as SOX9, SLUG and CD44, may help determine stem cell state [81,82]. In lung adenocarcinoma, SOX9, SLUG and CD44 are activated by the down-regulation of HDAC10 and stimulation of TGF-β, which mediates CSC-related cancer progression [30]. HDAC10 impacts immune and inflammation function as well. HDAC10-deleted Foxp3+ Treg cells have immunosuppressive functions and may play roles in alleviating immune-related colitis and prolonging cardiac allograft survival in MHC-mismatched mice [83]. HDAC10 deletion also recruits massive macrophages (especially M2 macrophages) into the lung cancer microenvironment. Considering that M2 macrophages may secrete anti-inflammatory molecules and promote tumor activity, HDAC10 may accelerate tumor progression by increasing tumor inflammation [30,84]. An additional effect of deletion of HDAC10 is chaperone-mediated autophagy (CMA), a form of autophagy that occurs via selective cytosolic protein transfer [85]. In knockout HDAC10 cervical cancer cells, Obayashi et al. [86] identified that increased expression levels of lysosomal associated protein 2 A (LAMP2A) and increased accumulation of LAMP2A-positive lysosomes near the nucleus raised, which may be a sign of activated CMA [87,88]. This characteristic may be effective for cancer treatment.
The mismatch repair system (MMR) is important in correcting flaws occurring during the stage of DNA replication [89]. It has been shown that mutL homolog 1 (MLH1), mutS homolog 2 (MSH2) and MSH6 commonly participated in MMR [90]. HDAC10 can trigger MSH2 activity initiated by MSH2 deacetylation at Lys73 in HeLa cells, following improved MMR activation [91]. A study that compared colon cancer cells and adjacent cells also revealed that low HDAC expression level in normal cells were negatively correlated with MSH2, MLH1 and MSH6 [92]. This may account for the different prognoses between normal and tumor cells.
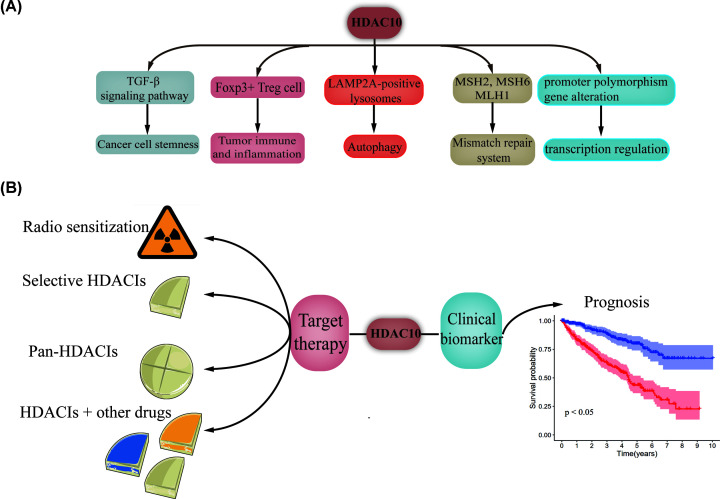
HDAC10 can perform regulatory roles through other epigenetic modifications, i.e. transcription regulation, methylation and promoter polymorphism [93–95]. In adrenocortical cancer cells and adenoma cells, HDAC10 is significantly methylated and its expression level dramatically decreases with hypermethylation [94]. Uzdensky et al. [95] observed down-regulated HDAC10 during transcription deregulation of melanoma progression. HDAC10 also participates in influencing the occurrence and onset age of HBV infection in HCC patients due to the promoter polymorphism, notably HDAC10-589C>T. The ‘T’ allele in HDAC10-589C>T enhances transcription activity, and might spur HCC progression by increasing HDAC10 expression [93]. Other study identified that frameshift deletions, one type of repeated alterations, occurred in HDAC10 among anaplastic thyroid cancer specimen [96]. According to Lourdusamy et al. [97], a large number of genes, including BRD1, HIRA and HDAC10 are expressed at low levels in spinal ependymoma. However, the detailed mechanisms underlying the activity of HDAC10 in spinal ependymoma still need to be elucidated. These studies may manifest an alternate form of tumor regulation (Figure 2A). It is worth noting that the specific mechanisms of the involvement of HDAC10 in other functions also remain unresolved, and further studies are needed. Nonetheless, the studies reviewed herein demonstrate the promise of HDAC10 as a molecular target for cancer.
Figure 2. Role of HDAC10 in non-tumor diseases and its clinical applications.
(A) Role of HDAC10 in non-tumor diseases. (B) Clinical applications of HDAC10.
Prognostic roles of HDAC10
The potential prognostic roles of HDAC10 have been explored. Up-regulated HDAC10 is associated with favorable overall survival (OS) in colon cancer, gastric cancer, RCC and NSCLC patients [31,92,98,99]. However, it should be noted that Liu et al. [100] studied the OS data of 180 NSCLC patients and found that up-regulated HDAC10 predicted poor OS. Moreover, HDAC10 is also an independent predictor of treatment-free survival (TFS) as well. HDAC10 level is associated with poor TFS in chronic lymphocytic leukemia (CLL) [101]. In this disease, high expression levels of HDAC are indirectly associated with a poor prognosis [102–104]. PD-L1 is expressed in various cancers and its clinical therapeutic value has been validated [105]. Due to enhanced PD-L1 expression in NSCLC patients with high HDAC10 expression, HDAC10 could function as a biomarker for the evaluation of PD-L1 treatment [100]. The elevated radiosensitization caused by inhibiting HDAC10 involves down-regulation of Rad51a, a component of DNA repair pathways [106]. In addition, positive HDAC10 expression is correlated with adverse clinical features, such as larger tumor size, poor tumor stage and more lymph node metastasis [99] (Figure 2B).
HDAC10 in other non-tumor diseases and its pathophysiological functions
Respiratory system diseases
An important role of HDAC10 has been found in asthma-induced eosinophilic airway inflammation. Zhang et al. [107] demonstrated that glucocorticoid (GC) and anacardic acid (AA) have the ability to alleviate IL-4, IL-5 and IL-13 and ameliorate airway inflammation in vivo, which depends on the low histone acetylation induced by elevated HDAC10 in memory T cells. A study that examined titanium dioxide (TiO2), a latent occupational carcinogen [108] in lung cells, reported that TiO2 particles provoked ROS (especially superoxide) and HDAC10 expression after lung cell are treated at 100 and 160 μg/cm2 for 24 h, respectively [109]. This implies that heritable epigenetic changes may serve as a reply to TiO2 particle exposure. Moreover, HDAC10 is also a potential pathogenic gene in emphysema [110].
Urinary system diseases
HDAC10 is also a latent biomarker in immunoglobulin A nephropathy (IgAN) and renal fibrosis as well [111,112]. HDAC10 protein levels are significantly higher in fibrotic kidneys. However, piceatannol treatment does not diminish HDAC10 expression but the results hint that HDAC10 may regulate renal fibrosis via other signaling pathways [111]. To determine a biomarker for IgAN, Guo et al. [112] utilized samples from the Gene Expression Omnibus (GEO) database and found that HDAC10 existed in monocytes of IgAN samples, which enriches in both alcoholism and chromatin modifying enzymes via construction of functional interaction (FI) network.
Nervous system diseases
HDAC10 is associated with many nervous system diseases. Kebir et al. [113] indicated that one single nucleotide polymorphism (SNP) (rs7634112) located in HDAC10 is significantly associated with schizophrenia. They devised a model that incorporates HDAC10, HDAC3 and HDAC9 to divide schizophrenia patients with high accuracy. A Korean study confirmed these results and selected a new SNP (rs1555048) for the Korean population [114]. Wang et al. [115] reported that decreased HDAC10 expression may aggravate post-intracerebral hemorrhage inflammation by elevating downstream PTPN22 and activating NLRP3 inflammasomes in rats. HDAC10 can modulate drug addiction as well. González et al. [116] reported that single-dose methamphetamine (METH) enhances HDAC10 expression in the medial prefrontal cortex of rat and that such up-regulation influences H4ac enrichment, which may account for the modified cognitive functions observed after using psychostimulants. The translocation of Olig1 protein involves brain development as well as multiple sclerosis [117,118]. In oligodendrocytes, HDAC10 represses Olig1 Lys150 acetylation, which leads to Olig1 protein being retained in cytoplasm [119]. In mesial temporal lobe epilepsy with hippocampal sclerosis (MTLE-HS), HDAC10 mRNA is significantly up-regulated. Meanwhile, increased HDAC10 expression has also been found in cytoplasm instead of the nucleus [120]. This subcellular distribution hints at potential role of HDAC10 in regulating MTLE-HS pathophysiology.
Hormone regulation
Many studies have opened new doors into the potential functions of HDAC10 in hormone regulation. Xu et al. [121] found that the alteration of decreased luteinizing hormone (LH) surge stimulated by estradiol (E2) in middle age female rats may mediated by HDAC10. Specifically, in the anterior hypothalamus, the attenuated HDAC10 expression and enhanced H3 acetylation induced by E2 were not observed in middle-aged female rats. A study from China illustrated the important role of HDAC10 in melanogenesis [122]. HDAC10 restores melanogenesis by deacetylating paired box protein 3 (Pax3) and KRAB-associated protein 1 (KAP1), which relieves the repressed promoters of microphthalmia-associated transcription factor (MITF) as well as tyrosinase-related protein 1 (TRP-1) and TRP-2 in melanoma cells [122]. These processes ultimately prove that melanogenesis is mainly achieved through HDAC10/Pax3/KAP1 pathway.
Lipid metabolism
The role of HDAC10 in regulating lipid metabolism has also been investigated. Qian et al. [123] found that HDAC10 expression decreases in obese humans and other animals (including mice and monkeys). In one study, the level of HDAC10 was markedly reduced in the medial hypothalamus of fasting mice, but researchers did not investigate its impact on AcH3 and AcH4 expression [124]. A study of benzyl butyl phthalate (BBP) found that it reduces HDACs (including HDAC10) and further contributes to acetylation of H3K9, which drives adipogenesis in mesenchymal stem cells (MSCs) [125].
Human immunodeficiency virus infection
The pathogenetic mechanisms of human immunodeficiency virus (HIV) are still being explored, which is crucial to develop blockbuster drugs [126]. Recent research suggests that HDAC10 takes part in HIV infectivity and thus can be used for its treatment [127–129]. Ran et al. identified that a virus-associated envelope glycoprotein (vEnv), one type of defective viral particle, mediates histone deacetylation via down-regulation of HDAC10 in Jurkat T cell, which ultimately elevates the infectivity of the virus and leads to HIV infection [127]. In CD4+ T cells, down-regulating HDAC10 expression promotes HIV-1 replication by increasing its interaction with DNA. Meanwhile, HDAC10 also combines with HIV-1 integrase (IN), lessening its reciprocal action with cellular protein lens epithelium-derived growth factor (LEDGF/p75), and ultimately deteriorating HIV-1 integration, which makes HDAC10 an inhibitor of HIV-1 replication [128]. Currently, antiretroviral drugs can prolong the life expectancy of HIV patients since it was introducted [130]. Rodriguez et al. reported that both raltegravir alone and in combination with morphine facilitate the level of HDAC10 [129]. However, further investigation is necessary to illustrate the underlying therapeutic mechanism.
Other diseases
HDAC10 orchestrates various biological functions. Pinto et al. [131] found that HDAC10 may mediate nutrition-associated growth. After restricting food in vivo or starving serum in vitro, HDAC10 expression apparently increased which is induced by inhibiting mTOR, and elevated levels of HDAC10 activated autophagy via Hsp70 deacetylation, thereby repressing cell viability [131]. HDAC10 also affects inflammation in the temporomandibular joint. Down-regulated HDAC10 suppresses the IL-1β/NF-κB pathway and its related inflammatory response in vitro [132]. In addition, in embryonic as well as induced pluripotent stem cells, HDAC10 enhances the level of chromatin remodelers [133]. In retinal ganglion cells, excitotoxicity alters HDAC10 localization to the cytoplasm [134]. In patients with vasospastic angina, apparently DNA gain was observed in HDAC10 [135]. However, it remains to be explored the detailed HDAC10-related mechanism or therapeutic value (Table 2).
Table 2. Overview of selected biological roles of HDAC10 in non-tumor diseases.
| Disease | Expression level | Functions | Targets/Signaling pathways | Cell lines/Tissues/Animal model | Reference |
|---|---|---|---|---|---|
| Eosinophilic airway inflammation | Elevated | Inhibit airway inflammation | IL-4, IL-5, IL-13 | CD4+ CD45RBlow cell | [96] |
| TiO2-associated lung disease | Elevated | Cellular response | Reactive oxygen species | A549 cell | [97] |
| Emphysema | / | Pathogenic gene | Copy number gains | 32 emphysema blood samples | [99] |
| IgAN | / | Chromatin modifying enzymes | / | 8 IgAN blood samples and 9 normal blood samples | [101] |
| Renal fibrosis | Elevated | Regulate renal fibrosis | / | C57BL/6 male mice | [100] |
| Schizophrenia | / | Diagnostic value | / | 278 Korean schizophrenia patients, 626 Caucasian schizophrenia parents | [102,103] |
| Post-ICH inflammation | Decreased | Aggravate inflammation response | PTPN22/NLRP3 | 6 ICH samples and S-D rat | [104] |
| Drug addiction | Elevated | Affect cognitive function | H4ac enrichment | C57BL/6 mice | [105] |
| Multiple sclerosis | Decreased | Affect oligodendrocytes | Oligo1 lysine 150 acetylation | Mouse neural progenitor cells, HEK293T cell | [108] |
| MTLE-HS | Elevated | Transcription regulation | / | 28 MTLE-HS samples | [109] |
| LH secretion | Decreased | Reduce LH secretion | H3 acetylation | S-D rat | [110] |
| Melanogenesis | / | Promote melanogenesis | Pax3/KAP1 | HEK293 cell and B16F10 cell | [111] |
| Adipogenesis | Decreased | Promote adipogenesis | H3K9 acetylation | MSCs | [114] |
| HIV infection | Decreased | Promote HIV infection | IN/LEDGF/p75, histone deacetylation | CD4+ T cell | [116,117] |
| Nutrition-associated growth | Elevated | Inhibit cell viability | mTOR, Hsp70, autophagy | S-D rat, Huh7 cell | [120] |
| TMJ inflammation | Decreased | Inhibit inflammatory response | IL-1β/NF-κB | 13 TMJ samples | [121] |
Abbreviations: ICH, intracerebral hemorrhage; IL, interleukin; IN, HIV-1 integrase; mTOR, mammalian target of rapamycin; NF-κB, nuclear factor κB; NLRP3, NOD-like receptors 3; PTPN22, protein tyrosine phosphatase nonreceptor type 22; TMJ, temporomandibular joint.
HDAC10 inhibitors
After constructing the homology model structure of HDAC10, Uba et al. further validated its reliability [136,137]. A group, focused on the tunnel behavior of HDAC10, suggested that the amino acids in tunnel-like sites of HDAC10 are subtly different from other HDACs (such as HDAC8 and HDAC11) identified a non-conserved amino acid in the wall of these sites [138]. Researchers have also found that HDAC10 contains Zn2+-binding groups and has additional interactions with inhibitors when accommodating with them [139]. Moreover, the X-ray crystal structure of zebrafish HDAC10 indicates that the gatekeeper E274 and humanized PEACE motif helix are the structural bases of highly selective HDAC10 inhibitors [140]. These results conduct a useful model and support the development of HDACIs with reliable information.
Selective and non-selective HDAC10 inhibitors
Nowadays, many HDACIs exhibit potent broad-spectrum repression ability to HDACs (including HDAC10), examples in broad-spectrum HDACIs include suberoylanilide hydroxamic acid [141], CRA-024781 [142], CRA-026440 [143], AR42 [144], trichostatin A (TSA) [145], valproic acid (VPA) [146] and so on. Both CRA-024781 and CRA-026440 are novel hydroxamic acid-based HDACIs that show potency against a series of HDACs, including HDAC1, 2, 3, 6, 8 and 10. CRA-024781 and CRA-026440 have in vitro efficacy in suppressing tumor cell proliferation as well as angiogenesis, inducing apoptosis and inhibiting tumor growth in vivo [142,143]. Another group reported pan-HDACIs AR42 and sodium valproate, which also have great selectivity over HDAC1, 3, 8 and 10 and obvious anti-tumor properties were observed in vivo and in vitro following pre-treatment with these HDACIs. Notably, AR42 and sodium valproate diminish PD-L1 and PD-L2 expression, and consequently elevate the efficacy of pazopanib, which increases multiple immune cells, for instance M1 macrophage, neutrophils, NK cells and T cells [144]. In addition, TSA can inhibit HDAC10 and 6, and the selectivity over HDAC10 arises from its N-terminal domain [147]. In colorectal cancer cells, TSA induces the inhibition of HDAC10 as well as HDAC6, and subsequently attenuates the Wnt pathway by the deletion of the Wnt signaling pathway-associated factor TCF7L2 in a proteasome-dependent way, which suppresses cell proliferation in vitro [43]. Bufexamac, an anti-inflammatory drug screened out via a new protocol, is also selective for HDAC10 and HDAC6 and its cellular potency may rely on interferon-α secreted by mononuclear cells [148] (Figure 3).
Figure 3. Mechanisms of action of selective and non-selective HDAC10 inhibitors.
However, due to some severe adverse effects of multitarget HDACIs [149], Géraldy et al. [150] investigated HDAC10-specific inhibitors and found that tubastatin A tightly bound to HDAC10 in HeLa cells with its basic amine in the cap group. Based on an HDAC10 homology model, they revealed that a hydrogen bond falls in between a cap group nitrogen, and a gatekeeper residue Glu272 contributes to the tight binding. Unfortunately, however, the development of a highly selective HDAC10 inhibitor is still far off because of the inability to abrogate HDAC6 activity.
Design or synthesis of HDAC10 inhibitors
We also examined previous works on the design or synthesis of HDAC10 inhibitors. By changing the condition of building block reaction, Villadsen et al. [151] synthesized azumamides A–E. In addition, they examined the inhibitory properties of HDAC10 under different drug concentrations (5 and 50 μM), which indicates both azumamides C and azumamides E markedly inhibit HDAC10. The HDACIs tubastatin A and PCI-34051 have the same N-benzylindole core [150,152]. After hybridizing these two inhibitors, Morgen et al. [153] synthesized dihydroxamic acids, which significantly suppressed the viability of neuroblastoma cells and played the role of potent inhibitor of HDAC10. Uba et al. [137] constructed M0017, the best model of human HDAC10, and filtered out ZINC19749069, the highest rank compound from the ZINC database, which displayed high stability when docking it into the catalytic channel of an HDAC10 model with quisinostat. Thus, M0017 and ZINC19749069 present considerable potential as an optimal HDACI and scaffold.
Although more and more studies could be reliably used for the HDACI designation, further exploration of the HDAC molecule structure and its biological characterization remain fundamental to shed light on the selection and development of the site-specific HDACIs.
Conclusion
Like other HDAC isoforms, HDAC10 exhibits various complex functions and clinic values [4,154,155]. As mentioned above, its functions involve in DNA damage repair, gene transcription and autophagy, and HDAC10-mediated tumor cell proliferation, apoptosis, invasion, migration, angiogenesis, metastasis and immune regulation, laying the theoretical foundation for clinical application. However, there are still challenges to be met. First, more details on the mechanisms of aberrant HDAC10 regulatory are essential to provide a more integrative understanding of HDAC10 and other HDACs. Besides, it is yet to be determined how to treat patients with HDACI resistance as well as the precise dose of HDACI. Furthermore, it remains unclear whether extending HDACI to other solid tumors would lead to satisfactory outcomes. With further in-depth study, HDAC10 may serve as an essential anti-tumor target and contribute to clinical applications in the future.
Acknowledgements
The authors gratefully thank the academic editor and the anonymous reviewers for their insightful comments and suggestions to improve this manuscript.
Abbreviations
- AMPK
AMP-activated protein kinase
- AR
androgen receptor
- BAK
BCL2 antagonist/killer
- BCL2
B-cell lymphoma-2
- BETi
bromodomain and extra-terminal protein inhibitor
- BRD4
bromodomain-containing protein 4
- CMA
chaperone-mediated autophagy
- CRPC
castration-resistant prostate cancer
- CSC
cancer stem cell
- Cx43
connexin 43
- DUB3
deubiquitinase ubiquitin-specific peptidase 17 like family member 2
- ERK
extracellular regulated protein kinase
- E2
estradiol
- FSTL1
follistatin-like 1
- G6PD
glucose-6-phosphate dehydrogenase
- HCC
hepatocellular cancer
- HDAC
histone deacetylase
- HDACI
HDAC inhibitor
- HIV
human immunodeficiency virus
- Hsp
heat shock protein
- hSSB1
human single-stranded DNA binding protein 1
- IgAN
immunoglobulin A nephropathy
- KAP1
KRAB-associated protein 1
- KRAB
kruppel-associated box domain
- LH
luteinizing hormone
- LKB1
liver kinase B1
- MLH1
mutL homolog 1
- MMP
matrix metalloproteinase
- MMR
mismatch repair system
- MSH2
mutS homolog 2
- MTLE-HS
mesial temporal lobe epilepsy with hippocampal sclerosis
- NCOR2
nuclear receptor co-repressor 2
- NSCLC
non-small-cell lung cancer
- OS
overall survival
- Pax3
paired box protein 3
- PD-L1
programmed cell death 1 ligand 1
- PD-L2
programmed cell death 1 ligand 2
- PTEN22
phosphatase and tensin homolog 22
- SNP
single nucleotide polymorphism
- SOX9
sex-determining region Y box protein 9
- TCF7L2
transcription factor 7 like 2
- TGF-β
transforming growth-factor β
- TiO2
titanium dioxide
- Treg
regulatory T cell
- TSA
trichostatin A
- TXNIP
thioredoxin interacting protein
- VEGF
vascular endothelial growth factor
- VEGFR1
vascular endothelial growth factor receptor 1/2
Contributor Information
Zhihong Niu, Email: nzh1789@163.com.
Wei He, Email: Hewei@bjmu.edu.cn.
Data Availability
The present study includes no data deposited in external repositories.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Author Contribution
B.Z. and F.j.C. drafted the manuscript. J.w.W., G.t.Z., Z.s.Y., Z.h.N. and W.H. revised the manuscript. All authors approved the final manuscript.
References
- 1.Workman J.L. and Kingston R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67, 545–579 10.1146/annurev.biochem.67.1.545 [DOI] [PubMed] [Google Scholar]
- 2.Strahl B.D. and Allis C.D. (2000) The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 3.Berger S.L. (2001) An embarrassment of niches: the many covalent modifications of histones in transcriptional regulation. Oncogene 20, 3007–3013 10.1038/sj.onc.1204324 [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti A., Oehme I., Witt O., Oliveira G., Sippl W., Romier C.et al. (2015) HDAC8: a multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 36, 481–492 10.1016/j.tips.2015.04.013 [DOI] [PubMed] [Google Scholar]
- 5.Tong J.J., Liu J., Bertos N.R. and Yang X.J. (2002) Identification of HDAC10, a novel class II human histone deacetylase containing a leucine-rich domain. Nucleic Acids Res. 30, 1114–1123 10.1093/nar/30.5.1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hai Y., Shinsky S.A., Porter N.J. and Christianson D.W. (2017) Histone deacetylase 10 structure and molecular function as a polyamine deacetylase. Nat. Commun. 8, 15368 10.1038/ncomms15368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seto E. and Yoshida M. (2014) Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caron C., Boyault C. and Khochbin S. (2005) Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. BioEssays 27, 408–415 10.1002/bies.20210 [DOI] [PubMed] [Google Scholar]
- 9.Fischle W., Wang Y. and Allis C.D. (2003) Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 15, 172–183 10.1016/S0955-0674(03)00013-9 [DOI] [PubMed] [Google Scholar]
- 10.Leipe D.D. and Landsman D. (1997) Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic Acids Res. 25, 3693–3697 10.1093/nar/25.18.3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang P., Wang Z. and Liu J. (2020) Role of HDACs in normal and malignant hematopoiesis. Mol. Can. 19, 5 10.1186/s12943-019-1127-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer D.D., Cai R., Bhatia U., Asselbergs F.A., Song C., Terry R.et al. (2002) Isolation and characterization of a novel class II histone deacetylase, HDAC10. J. Biol. Chem. 277, 6656–6666 10.1074/jbc.M108055200 [DOI] [PubMed] [Google Scholar]
- 13.Kao H.Y., Lee C.H., Komarov A., Han C.C. and Evans R.M. (2002) Isolation and characterization of mammalian HDAC10, a novel histone deacetylase. J. Biol. Chem. 277, 187–193 10.1074/jbc.M108931200 [DOI] [PubMed] [Google Scholar]
- 14.Guardiola A.R. and Yao T.P. (2002) Molecular cloning and characterization of a novel histone deacetylase HDAC10. J. Biol. Chem. 277, 3350–3356 10.1074/jbc.M109861200 [DOI] [PubMed] [Google Scholar]
- 15.Grozinger C.M. and Schreiber S.L. (2000) Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc. Natl. Acad. Sci. U.S.A. 97, 7835–7840 10.1073/pnas.140199597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKinsey T.A., Zhang C.L., Lu J. and Olson E.N. (2000) Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408, 106–111 10.1038/35040593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verdin E., Dequiedt F. and Kasler H.G. (2003) Class II histone deacetylases: versatile regulators. Trends Genet. 19, 286–293 10.1016/S0168-9525(03)00073-8 [DOI] [PubMed] [Google Scholar]
- 18.Hubbert C., Guardiola A., Shao R., Kawaguchi Y., Ito A., Nixon A.et al. (2002) HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 10.1038/417455a [DOI] [PubMed] [Google Scholar]
- 19.Shinsky S.A. and Christianson D.W. (2018) Polyamine deacetylase structure and catalysis: prokaryotic acetylpolyamine amidohydrolase and eukaryotic HDAC10. Biochemistry 57, 3105–3114 10.1021/acs.biochem.8b00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao H.Y., Downes M., Ordentlich P. and Evans R.M. (2000) Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes Dev. 14, 55–66 [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X., Richon V.M., Rifkind R.A. and Marks P.A. (2000) Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc. Natl. Acad. Sci. U.S.A. 97, 1056–1061 10.1073/pnas.97.3.1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finnin M.S., Donigian J.R., Cohen A., Richon V.M., Rifkind R.A., Marks P.A.et al. (1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193 10.1038/43710 [DOI] [PubMed] [Google Scholar]
- 23.Wu R., Lu Z., Cao Z. and Zhang Y. (2011) Zinc chelation with hydroxamate in histone deacetylases modulated by water access to the linker binding channel. J. Am. Chem. Soc. 133, 6110–6113 10.1021/ja111104p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gantt S.L., Joseph C.G. and Fierke C.A. (2010) Activation and inhibition of histone deacetylase 8 by monovalent cations. J. Biol. Chem. 285, 6036–6043 10.1074/jbc.M109.033399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai S., Armstrong C.M., Kaeberlein M. and Guarente L. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 10.1038/35001622 [DOI] [PubMed] [Google Scholar]
- 26.Avalos J.L., Boeke J.D. and Wolberger C. (2004) Structural basis for the mechanism and regulation of Sir2 enzymes. Mol. Cell 13, 639–648 10.1016/S1097-2765(04)00082-6 [DOI] [PubMed] [Google Scholar]
- 27.Hanahan D. and Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 28.Hanahan D. and Weinberg R.A. (2000) The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- 29.Yang Y., Huang Y., Wang Z., Wang H.T., Duan B., Ye D.et al. (2016) HDAC10 promotes lung cancer proliferation via AKT phosphorylation. Oncotarget 7, 59388–59401 10.18632/oncotarget.10673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y., Zhang X., Zhu S., Dejene E.A., Peng W., Sepulveda A.et al. (2020) HDAC10 regulates cancer stem-like cell properties in KRAS-driven lung adenocarcinoma. Cancer Res. 80, 3265–3278 10.1158/0008-5472.CAN-19-3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan W., Huang J. and Xiao H. (2015) Histone deacetylase 10 suppresses proliferation and invasion by inhibiting the phosphorylation of β-catenin and serves as an independent prognostic factor for human clear cell renal cell carcinoma. Int. J. Clin. Exp. Med. 8, 3734–3742 [PMC free article] [PubMed] [Google Scholar]
- 32.Revathidevi S. and Munirajan A.K. (2019) Akt in cancer: mediator and more. Semin. Cancer Biol. 59, 80–91 10.1016/j.semcancer.2019.06.002 [DOI] [PubMed] [Google Scholar]
- 33.Guertin D.A. and Sabatini D.M. (2007) Defining the role of mTOR in cancer. Cancer Cell. 12, 9–22 10.1016/j.ccr.2007.05.008 [DOI] [PubMed] [Google Scholar]
- 34.Shan C., Lu Z., Li Z., Sheng H., Fan J., Qi Q.et al. (2019) 4-hydroxyphenylpyruvate dioxygenase promotes lung cancer growth via pentose phosphate pathway (PPP) flux mediated by LKB1-AMPK/HDAC10/G6PD axis. Cell Death Dis. 10, 525 10.1038/s41419-019-1756-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evan G.I. and Vousden K.H. (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348 10.1038/35077213 [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Peng L. and Seto E. (2015) Histone deacetylase 10 regulates the cell cycle G2/M phase transition via a novel Let-7-HMGA2-cyclin A2 pathway. Mol. Cell. Biol. 35, 3547–3565 10.1128/MCB.00400-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coqueret O. (2003) New roles for p21 and p27 cell-cycle inhibitors: a function for each cell compartment? Trends Cell Biol. 13, 65–70 10.1016/S0962-8924(02)00043-0 [DOI] [PubMed] [Google Scholar]
- 38.Qie S. and Diehl J.A. (2016) Cyclin D1, cancer progression, and opportunities in cancer treatment. J. Mol. Med. 94, 1313–1326 10.1007/s00109-016-1475-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutherland R.L. and Musgrove E.A. (2004) Cyclins and breast cancer. J. Mamm. Gland Biol. Neoplasia 9, 95–104 10.1023/B:JOMG.0000023591.45568.77 [DOI] [PubMed] [Google Scholar]
- 40.Wong R.S. (2011) Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30, 87 10.1186/1756-9966-30-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gross A., McDonnell J.M. and Korsmeyer S.J. (1999) BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 13, 1899–1911 10.1101/gad.13.15.1899 [DOI] [PubMed] [Google Scholar]
- 42.Reed J.C. (1997) Bcl-2 family proteins: regulators of apoptosis and chemoresistance in hematologic malignancies. Semin. Hematol. 34, 9–19 [PubMed] [Google Scholar]
- 43.Götze S., Coersmeyer M., Müller O. and Sievers S. (2014) Histone deacetylase inhibitors induce attenuation of Wnt signaling and TCF7L2 depletion in colorectal carcinoma cells. Int. J. Oncol. 45, 1715–1723 10.3892/ijo.2014.2550 [DOI] [PubMed] [Google Scholar]
- 44.Circu M.L. and Aw T.Y. (2010) Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 48, 749–762 10.1016/j.freeradbiomed.2009.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zorov D.B., Juhaszova M. and Sollott S.J. (2014) Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 94, 909–950 10.1152/physrev.00026.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.H., Jeong E.G., Choi M.C., Kim S.H., Park J.H., Song S.H.et al. (2010) Inhibition of histone deacetylase 10 induces thioredoxin-interacting protein and causes accumulation of reactive oxygen species in SNU-620 human gastric cancer cells. Mol. Cells 30, 107–112 10.1007/s10059-010-0094-z [DOI] [PubMed] [Google Scholar]
- 47.Polacheck W.J., Zervantonakis I.K. and Kamm R.D. (2013) Tumor cell migration in complex microenvironments. Cell. Mol. Life Sci. 70, 1335–1356 10.1007/s00018-012-1115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamidi H. and Ivaska J. (2018) Every step of the way: integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 10.1038/s41568-018-0038-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saiki Y. and Horii A. (2019) Multiple functions of S100A10, an important cancer promoter. Pathol. Int. 69, 629–636 10.1111/pin.12861 [DOI] [PubMed] [Google Scholar]
- 50.Castro-Castro A., Marchesin V., Monteiro P., Lodillinsky C., Rossé C. and Chavrier P. (2016) Cellular and molecular mechanisms of MT1-MMP-dependent cancer cell invasion. Annu. Rev. Cell Dev. Biol. 32, 555–576 10.1146/annurev-cellbio-111315-125227 [DOI] [PubMed] [Google Scholar]
- 51.Cui N., Hu M. and Khalil R.A. (2017) Biochemical and biological attributes of matrix metalloproteinases. Prog. Mol. Biol. Transl. Sci. 147, 1–73 10.1016/bs.pmbts.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao W., Han H.B. and Zhang Z.Q. (2011) Suppression of lung cancer cell invasion and metastasis by connexin43 involves the secretion of follistatin-like 1 mediated via histone acetylation. Int. J. Biochem. Cell Biol. 43, 1459–1468 10.1016/j.biocel.2011.06.009 [DOI] [PubMed] [Google Scholar]
- 53.Song C., Zhu S., Wu C. and Kang J. (2013) Histone deacetylase (HDAC) 10 suppresses cervical cancer metastasis through inhibition of matrix metalloproteinase (MMP) 2 and 9 expression. J. Biol. Chem. 288, 28021–28033 10.1074/jbc.M113.498758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong Y.W., Wang R., Cai Q.Q., Qi B., Wu W., Zhang Y.H.et al. (2014) Sulfatide epigenetically regulates miR-223 and promotes the migration of human hepatocellular carcinoma cells. J. Hepatol. 60, 792–801 10.1016/j.jhep.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 55.VON Schulz-Hausmann S.A., Schmeel L.C., Schmeel F.C. and Schmidt-Wolf I.G. (2014) Targeting the Wnt/beta-catenin pathway in renal cell carcinoma. Anticancer Res. 34, 4101–4108 [PubMed] [Google Scholar]
- 56.Shang S., Hua F. and Hu Z.W. (2017) The regulation of β-catenin activity and function in cancer: therapeutic opportunities. Oncotarget 8, 33972–33989 10.18632/oncotarget.15687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerbel R. and Folkman J. (2002) Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer 2, 727–739 10.1038/nrc905 [DOI] [PubMed] [Google Scholar]
- 58.Arias H.R., Richards V.E., Ng D., Ghafoori M.E., Le V. and Mousa S.A. (2009) Role of non-neuronal nicotinic acetylcholine receptors in angiogenesis. Int. J. Biochem. Cell Biol. 41, 1441–1451 10.1016/j.biocel.2009.01.013 [DOI] [PubMed] [Google Scholar]
- 59.Rak J., Yu J.L., Klement G. and Kerbel R.S. (2000) Oncogenes and angiogenesis: signaling three-dimensional tumor growth. J. Investig. Dermatol. Symp. Proc. 5, 24–33 10.1046/j.1087-0024.2000.00012.x [DOI] [PubMed] [Google Scholar]
- 60.Relf M., LeJeune S., Scott P.A., Fox S., Smith K., Leek R.et al. (1997) Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor beta-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer Res. 57, 963–969 [PubMed] [Google Scholar]
- 61.Duan B., Ye D., Zhu S., Jia W., Lu C., Wang G.et al. (2017) HDAC10 promotes angiogenesis in endothelial cells through the PTPN22/ERK axis. Oncotarget 8, 61338–61349 10.18632/oncotarget.18130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park J.H., Kim S.H., Choi M.C., Lee J., Oh D.Y., Im S.A.et al. (2008) Class II histone deacetylases play pivotal roles in heat shock protein 90-mediated proteasomal degradation of vascular endothelial growth factor receptors. Biochem. Biophys. Res. Commun. 368, 318–322 10.1016/j.bbrc.2008.01.056 [DOI] [PubMed] [Google Scholar]
- 63.Holohan C., Van Schaeybroeck S., Longley D.B. and Johnston P.G. (2013) Cancer drug resistance: an evolving paradigm. Nat. Rev. Cancer 13, 714–726 10.1038/nrc3599 [DOI] [PubMed] [Google Scholar]
- 64.Longley D.B. and Johnston P.G. (2005) Molecular mechanisms of drug resistance. J. Pathol. 205, 275–292 10.1002/path.1706 [DOI] [PubMed] [Google Scholar]
- 65.Gottesman M.M., Fojo T. and Bates S.E. (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat. Rev. Cancer 2, 48–58 10.1038/nrc706 [DOI] [PubMed] [Google Scholar]
- 66.Ridinger J., Koeneke E., Kolbinger F.R., Koerholz K., Mahboobi S., Hellweg L.et al. (2018) Dual role of HDAC10 in lysosomal exocytosis and DNA repair promotes neuroblastoma chemoresistance. Sci. Rep. 8, 10039 10.1038/s41598-018-28265-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Y.J., Lei Y.H., Yao N., Wang C.R., Hu N., Ye W.C.et al. (2017) Autophagy and multidrug resistance in cancer. Chin. J. Cancer 36, 52 10.1186/s40880-017-0219-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oehme I., Linke J.P., Böck B.C., Milde T., Lodrini M., Hartenstein B.et al. (2013) Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc. Natl. Acad. Sci. U.S.A. 110, E2592–E2601 10.1073/pnas.1300113110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oehme I., Lodrini M., Brady N.R. and Witt O. (2013) Histone deacetylase 10-promoted autophagy as a druggable point of interference to improve the treatment response of advanced neuroblastomas. Autophagy 9, 2163–2165 10.4161/auto.26450 [DOI] [PubMed] [Google Scholar]
- 70.Islam M.M., Banerjee T., Packard C.Z., Kotian S., Selvendiran K., Cohn D.E.et al. (2017) HDAC10 as a potential therapeutic target in ovarian cancer. Gynecol. Oncol. 144, 613–620 10.1016/j.ygyno.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K.et al. (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 10.1038/nature03482 [DOI] [PubMed] [Google Scholar]
- 72.Richard D.J., Savage K., Bolderson E., Cubeddu L., So S., Ghita M.et al. (2011) hSSB1 rapidly binds at the sites of DNA double-strand breaks and is required for the efficient recruitment of the MRN complex. Nucleic Acids Res. 39, 1692–1702 10.1093/nar/gkq1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu Y., Chen H., Lu J., Zhang M., Zhang R., Duan T.et al. (2015) Acetylation-dependent function of human single-stranded DNA binding protein 1. Nucleic Acids Res. 43, 7878–7887 10.1093/nar/gkv707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shafi A.A., Yen A.E. and Weigel N.L. (2013) Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Ther. 140, 223–238 10.1016/j.pharmthera.2013.07.003 [DOI] [PubMed] [Google Scholar]
- 75.Welti J., Sharp A., Yuan W., Dolling D., Nava Rodrigues D., Figueiredo I.et al. (2018) Targeting bromodomain and extra-terminal (BET) family proteins in castration-resistant prostate cancer (CRPC). Clin. Cancer Res. 24, 3149–3162 10.1158/1078-0432.CCR-17-3571 [DOI] [PubMed] [Google Scholar]
- 76.Dai X., Gan W., Li X., Wang S., Zhang W., Huang L.et al. (2017) Prostate cancer-associated SPOP mutations confer resistance to BET inhibitors through stabilization of BRD4. Nat. Med. 23, 1063–1071 10.1038/nm.4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin X., Yan Y., Wang D., Ding D., Ma T., Ye Z.et al. (2018) DUB3 Promotes BET Inhibitor Resistance and Cancer Progression by Deubiquitinating BRD4. Mol. Cell 71, 592.e4–605.e4 10.1016/j.molcel.2018.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun H., Mediwala S.N., Szafran A.T., Mancini M.A. and Marcelli M. (2016) CUDC-101, a novel inhibitor of full-length androgen receptor (flAR) and androgen receptor variant 7 (AR-V7) activity: mechanism of action and in vivo efficacy. Hormones Cancer 7, 196–210 10.1007/s12672-016-0257-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saygin C., Matei D., Majeti R., Reizes O. and Lathia J.D. (2019) Targeting cancer stemness in the clinic: from hype to hope. Cell Stem Cell 24, 25–40 10.1016/j.stem.2018.11.017 [DOI] [PubMed] [Google Scholar]
- 80.Batlle E. and Clevers H. (2017) Cancer stem cells revisited. Nat. Med. 23, 1124–1134 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 81.Guo W., Keckesova Z., Donaher J.L., Shibue T., Tischler V., Reinhardt F.et al. (2012) Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 148, 1015–1028 10.1016/j.cell.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ye X., Tam W.L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E.et al. (2015) Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dahiya S., Beier U.H., Wang L., Han R., Jiao J., Akimova T.et al. (2020) HDAC10 deletion promotes Foxp3 (+) T-regulatory cell function. Sci. Rep. 10, 424 10.1038/s41598-019-57294-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen Y., Song Y., Du W., Gong L., Chang H. and Zou Z. (2019) Tumor-associated macrophages: an accomplice in solid tumor progression. J. Biomed. Sci. 26, 78 10.1186/s12929-019-0568-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kaushik S. and Cuervo A.M. (2018) The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 10.1038/s41580-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Obayashi H., Nagano Y., Takahashi T., Seki T., Tanaka S., Sakai N.et al. (2020) Histone deacetylase 10 knockout activates chaperone-mediated autophagy and accelerates the decomposition of its substrate. Biochem. Biophys. Res. Commun. 523, 246–252 10.1016/j.bbrc.2019.12.048 [DOI] [PubMed] [Google Scholar]
- 87.Arias E. (2017) Methods to study chaperone-mediated autophagy. Methods Enzymol. 588, 283–305 10.1016/bs.mie.2016.10.009 [DOI] [PubMed] [Google Scholar]
- 88.Kaushik S. and Cuervo A.M. (2009) Methods to monitor chaperone-mediated autophagy. Methods Enzymol. 452, 297–324 10.1016/S0076-6879(08)03619-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baretti M. and Le D.T. (2018) DNA mismatch repair in cancer. Pharmacol. Ther. 189, 45–62 10.1016/j.pharmthera.2018.04.004 [DOI] [PubMed] [Google Scholar]
- 90.Li G.M. (2008) Mechanisms and functions of DNA mismatch repair. Cell Res. 18, 85–98 10.1038/cr.2007.115 [DOI] [PubMed] [Google Scholar]
- 91.Radhakrishnan R., Li Y., Xiang S., Yuan F., Yuan Z., Telles E.et al. (2015) Histone deacetylase 10 regulates DNA mismatch repair and may involve the deacetylation of MutS homolog 2. J. Biol. Chem. 290, 22795–22804 10.1074/jbc.M114.612945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tao X., Yan Y., Lu L. and Chen B. (2017) HDAC10 expression is associated with DNA mismatch repair gene and is a predictor of good prognosis in colon carcinoma. Oncol. Lett. 14, 4923–4929 10.3892/ol.2017.6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park B.L., Kim Y.J., Cheong H.S., Lee S.O., Han C.S., Yoon J.H.et al. (2007) HDAC10 promoter polymorphism associated with development of HCC among chronic HBV patients. Biochem. Biophys. Res. Commun. 363, 776–781 10.1016/j.bbrc.2007.09.026 [DOI] [PubMed] [Google Scholar]
- 94.Fonseca A.L., Kugelberg J., Starker L.F., Scholl U., Choi M., Hellman P.et al. (2012) Comprehensive DNA methylation analysis of benign and malignant adrenocortical tumors. Genes Chromosomes Cancer 51, 949–960 10.1002/gcc.21978 [DOI] [PubMed] [Google Scholar]
- 95.Uzdensky A., Demyanenko S., Bibov M., Sharifulina S., Kit O., Przhedetski Y.et al. (2014) Expression of proteins involved in epigenetic regulation in human cutaneous melanoma and peritumoral skin. Tumour Biol. 35, 8225–8233 10.1007/s13277-014-2098-3 [DOI] [PubMed] [Google Scholar]
- 96.Kasaian K., Wiseman S.M., Walker B.A., Schein J.E., Zhao Y., Hirst M.et al. (2015) The genomic and transcriptomic landscape of anaplastic thyroid cancer: implications for therapy. BMC Cancer 15, 984 10.1186/s12885-015-1955-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lourdusamy A., Rahman R. and Grundy R.G. (2015) Expression alterations define unique molecular characteristics of spinal ependymomas. Oncotarget 6, 19780–19791 10.18632/oncotarget.3715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Osada H., Tatematsu Y., Saito H., Yatabe Y., Mitsudomi T. and Takahashi T. (2004) Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int. J. Cancer 112, 26–32 10.1002/ijc.20395 [DOI] [PubMed] [Google Scholar]
- 99.Jin Z., Jiang W., Jiao F., Guo Z., Hu H., Wang L.et al. (2014) Decreased expression of histone deacetylase 10 predicts poor prognosis of gastric cancer patients. Int. J. Clin. Exp. Pathol. 7, 5872–5879 [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X., Wang Y., Zhang R., Jin T., Qu L., Jin Q.et al. (2020) HDAC10 is positively associated with PD-L1 expression and poor prognosis in patients with NSCLC. Front. Oncol. 10, 485 10.3389/fonc.2020.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Van Damme M., Crompot E., Meuleman N., Mineur P., Bron D., Lagneaux L.et al. (2012) HDAC isoenzyme expression is deregulated in chronic lymphocytic leukemia B-cells and has a complex prognostic significance. Epigenetics 7, 1403–1412 10.4161/epi.22674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J.C., Kafeel M.I., Avezbakiyev B., Chen C., Sun Y., Rathnasabapathy C.et al. (2011) Histone deacetylase in chronic lymphocytic leukemia. Oncology 81, 325–329 10.1159/000334577 [DOI] [PubMed] [Google Scholar]
- 103.De Rossi G., Marroni P., Paganuzzi M., Mauro F.R., Tenca C., Zarcone D.et al. (1997) Increased serum levels of soluble CD44 standard, but not of variant isoforms v5 and v6, in B cell chronic lymphocytic leukemia. Leukemia 11, 134–141 10.1038/sj.leu.2400525 [DOI] [PubMed] [Google Scholar]
- 104.De Rossi G., Zarcone D., Mauro F., Cerruti G., Tenca C., Puccetti A.et al. (1993) Adhesion molecule expression on B-cell chronic lymphocytic leukemia cells: malignant cell phenotypes define distinct disease subsets. Blood 81, 2679–2687 10.1182/blood.V81.10.2679.2679 [DOI] [PubMed] [Google Scholar]
- 105.Dermani F.K., Samadi P., Rahmani G., Kohlan A.K. and Najafi R. (2019) PD-1/PD-L1 immune checkpoint: Potential target for cancer therapy. J. Cell. Physiol. 234, 1313–1325 10.1002/jcp.27172 [DOI] [PubMed] [Google Scholar]
- 106.Kim J.H., Moon S.H., No M., Kim J.J., Choi E.J., Cho B.J.et al. (2016) Isotype-specific inhibition of histone deacetylases: identification of optimal targets for radiosensitization. Cancer Res. Treat. 48, 1130–1140 10.4143/crt.2015.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang H.P., Wang L., Fu J.J., Fan T., Wang Z.L. and Wang G. (2016) Association between histone hyperacetylation status in memory T lymphocytes and allergen-induced eosinophilic airway inflammation. Respirology 21, 850–857 10.1111/resp.12774 [DOI] [PubMed] [Google Scholar]
- 108.Rocco L., Santonastaso M., Mottola F., Costagliola D., Suero T., Pacifico S.et al. (2015) Genotoxicity assessment of TiO2 nanoparticles in the teleost Danio rerio. Ecotoxicol. Environ. Saf. 113, 223–230 10.1016/j.ecoenv.2014.12.012 [DOI] [PubMed] [Google Scholar]
- 109.Jayaram D.T. and Payne C.K. (2020) Food-grade TiO (2) particles generate intracellular superoxide and alter epigenetic modifiers in human lung cells. Chem. Res. Toxicol. 33, 2872–2879 10.1021/acs.chemrestox.0c00331 [DOI] [PubMed] [Google Scholar]
- 110.Choi J.S., Lee W.J., Baik S.H., Yoon H.K., Lee K.H., Kim Y.H.et al. (2009) Array CGH reveals genomic aberrations in human emphysema. Lung 187, 165–172 10.1007/s00408-009-9142-x [DOI] [PubMed] [Google Scholar]
- 111.Choi S.Y., Piao Z.H., Jin L., Kim J.H., Kim G.R., Ryu Y.et al. (2016) Piceatannol attenuates renal fibrosis induced by unilateral ureteral obstruction via downregulation of histone deacetylase 4/5 or p38-MAPK signaling. PLoS ONE 11, e0167340 10.1371/journal.pone.0167340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo Y., Gao W., Wang D., Liu W. and Liu Z. (2018) Gene alterations in monocytes are pathogenic factors for immunoglobulin a nephropathy by bioinformatics analysis of microarray data. BMC Nephrol. 19, 184 10.1186/s12882-018-0944-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kebir O., Chaumette B., Fatjó-Vilas M., Ambalavanan A., Ramoz N., Xiong L.et al. (2014) Family-based association study of common variants, rare mutation study and epistatic interaction detection in HDAC genes in schizophrenia. Schizophr. Res. 160, 97–103 10.1016/j.schres.2014.09.029 [DOI] [PubMed] [Google Scholar]
- 114.Kim T., Park J.K., Kim H.J., Chung J.H. and Kim J.W. (2010) Association of histone deacetylase genes with schizophrenia in Korean population. Psychiatry Res. 178, 266–269 10.1016/j.psychres.2009.05.007 [DOI] [PubMed] [Google Scholar]
- 115.Wang L., Zheng S., Zhang L., Xiao H., Gan H., Chen H.et al. (2020) Histone deacetylation 10 alleviates inflammation after intracerebral hemorrhage via the PTPN22/NLRP3 pathway in rats. Neuroscience 432, 247–259 10.1016/j.neuroscience.2020.02.027 [DOI] [PubMed] [Google Scholar]
- 116.González B., Bernardi A., Torres O.V., Jayanthi S., Gomez N., Sosa M.H.et al. (2020) HDAC superfamily promoters acetylation is differentially regulated by modafinil and methamphetamine in the mouse medial prefrontal cortex. Addict. Biol. 25, e12737 10.1111/adb.12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xin M., Yue T., Ma Z., Wu F.F., Gow A. and Lu Q.R. (2005) Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1-null mice. J. Neurosci. 25, 1354–1365 10.1523/JNEUROSCI.3034-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu Q.R., Sun T., Zhu Z., Ma N., Garcia M., Stiles C.D.et al. (2002) Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 109, 75–86 10.1016/S0092-8674(02)00678-5 [DOI] [PubMed] [Google Scholar]
- 119.Dai J., Bercury K.K., Jin W. and Macklin W.B. (2015) Olig1 acetylation and nuclear export mediate oligodendrocyte development. J. Neurosci. 35, 15875–15893 10.1523/JNEUROSCI.0882-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Srivastava A., Banerjee J., Dubey V., Tripathi M., Chandra P.S., Sharma M.C.et al. (2020) Role of altered expression, activity and sub-cellular distribution of various histone deacetylases (HDACs) in mesial temporal lobe epilepsy with hippocampal sclerosis. Cell. Mol. Neurobiol. 10.1007/s10571-020-00994-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu W., An X., Zhang N., Li L., Zhang X., Wang Y.et al. (2019) Middle-aged female rats lack changes in histone H3 acetylation in the anterior hypothalamus observed in young females on the day of a luteinizing hormone surge. Biosci. Trends 13, 334–341 10.5582/bst.2019.01162 [DOI] [PubMed] [Google Scholar]
- 122.Lai I.L., Lin T.P., Yao Y.L., Lin C.Y., Hsieh M.J. and Yang W.M. (2010) Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J. Biol. Chem. 285, 7187–7196 10.1074/jbc.M109.061861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qian H., Chen Y., Nian Z., Su L., Yu H., Chen F.J.et al. (2017) HDAC6-mediated acetylation of lipid droplet-binding protein CIDEC regulates fat-induced lipid storage. J. Clin. Invest. 127, 1353–1369 10.1172/JCI85963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Funato H., Oda S., Yokofujita J., Igarashi H. and Kuroda M. (2011) Fasting and high-fat diet alter histone deacetylase expression in the medial hypothalamus. PLoS ONE 6, e18950 10.1371/journal.pone.0018950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sonkar R., Powell C.A. and Choudhury M. (2016) Benzyl butyl phthalate induces epigenetic stress to enhance adipogenesis in mesenchymal stem cells. Mol. Cell. Endocrinol. 431, 109–122 10.1016/j.mce.2016.04.025 [DOI] [PubMed] [Google Scholar]
- 126.Fanales-Belasio E., Raimondo M., Suligoi B. and Buttò S. (2010) HIV virology and pathogenetic mechanisms of infection: a brief overview. Annali Dell’Istituto Superiore Di Sanita 46, 5–14 10.1590/S0021-25712010000100002 [DOI] [PubMed] [Google Scholar]
- 127.Ran X., Ao Z., Trajtman A., Xu W., Kobinger G., Keynan Y.et al. (2017) HIV-1 envelope glycoprotein stimulates viral transcription and increases the infectivity of the progeny virus through the manipulation of cellular machinery. Sci. Rep. 7, 9487 10.1038/s41598-017-10272-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ran X., Ao Z., Olukitibi T. and Yao X. (2019) Characterization of the role of host cellular factor histone deacetylase 10 during HIV-1 replication. Viruses 12, 28 10.3390/v12010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rodriguez M., Lapierre J., Ojha C.R., Pawitwar S., Karuppan M.K.M., Kashanchi F.et al. (2019) Morphine counteracts the antiviral effect of antiretroviral drugs and causes upregulation of p62/SQSTM1 and histone-modifying enzymes in HIV-infected astrocytes. J. Neurovirol. 25, 263–274 10.1007/s13365-018-0715-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Samji H., Cescon A., Hogg R.S., Modur S.P., Althoff K.N., Buchacz K.et al. (2013) Closing the gap: increases in life expectancy among treated HIV-positive individuals in the United States and Canada. PLoS ONE 8, e81355 10.1371/journal.pone.0081355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pinto G., Shtaif B., Phillip M. and Gat-Yablonski G. (2016) Growth attenuation is associated with histone deacetylase 10-induced autophagy in the liver. J. Nutr. Biochem. 27, 171–180 10.1016/j.jnutbio.2015.08.031 [DOI] [PubMed] [Google Scholar]
- 132.Liao W., Sun J., Liu W., Li W., Jia J., Ou F.et al. (2019) HDAC10 upregulation contributes to interleukin 1β-mediated inflammatory activation of synovium-derived mesenchymal stem cells in temporomandibular joint. J. Cell. Physiol. 234, 12646–12662 10.1002/jcp.27873 [DOI] [PubMed] [Google Scholar]
- 133.Luzzani C., Solari C., Losino N., Ariel W., Romorini L., Bluguermann C.et al. (2011) Modulation of chromatin modifying factors’ gene expression in embryonic and induced pluripotent stem cells. Biochem. Biophys. Res. Commun. 410, 816–822 10.1016/j.bbrc.2011.06.070 [DOI] [PubMed] [Google Scholar]
- 134.Schlüter A., Aksan B., Fioravanti R., Valente S., Mai A. and Mauceri D. (2019) Histone deacetylases contribute to excitotoxicity-triggered degeneration of retinal ganglion cells in vivo. Mol. Neurobiol. 56, 8018–8034 10.1007/s12035-019-01658-x [DOI] [PubMed] [Google Scholar]
- 135.Seo S.M., Koh Y.S., Jung H.O., Choi J.S., Kim P.J., Baek S.H.et al. (2011) Deoxyribonucleic acid copy number aberrations in vasospastic angina patients using an array comparative genomic hybridization. Korean Circ. J. 41, 385–393 10.4070/kcj.2011.41.7.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Uba A.I. and Yelekçi K. (2020) Crystallographic structure versus homology model: a case study of molecular dynamics simulation of human and zebrafish histone deacetylase 10. J. Biomol. Struct. Dyn. 38, 4397–4406 10.1080/07391102.2019.1691658 [DOI] [PubMed] [Google Scholar]
- 137.Ibrahim Uba A. and Yelekçi K. (2019) Homology modeling of human histone deacetylase 10 and design of potential selective inhibitors. J. Biomol. Struct. Dyn. 37, 3627–3636 10.1080/07391102.2018.1521747 [DOI] [PubMed] [Google Scholar]
- 138.Thangapandian S., John S., Lee Y., Arulalapperumal V. and Lee K.W. (2012) Molecular modeling study on tunnel behavior in different histone deacetylase isoforms. PLoS ONE 7, e49327 10.1371/journal.pone.0049327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Herbst-Gervasoni C.J. and Christianson D.W. (2019) Binding of N (8)-acetylspermidine analogues to histone deacetylase 10 reveals molecular strategies for blocking polyamine deacetylation. Biochemistry 58, 4957–4969 10.1021/acs.biochem.9b00906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Herbst-Gervasoni C.J., Steimbach R.R., Morgen M., Miller A.K. and Christianson D.W. (2020) Structural basis for the selective inhibition of HDAC10, the cytosolic polyamine deacetylase. ACS Chem. Biol. 15, 2154–2163 10.1021/acschembio.0c00362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Codd R., Braich N., Liu J., Soe C.Z. and Pakchung A.A. (2009) Zn (II)-dependent histone deacetylase inhibitors: suberoylanilide hydroxamic acid and trichostatin A. Int. J. Biochem. Cell Biol. 41, 736–739 10.1016/j.biocel.2008.05.026 [DOI] [PubMed] [Google Scholar]
- 142.Buggy J.J., Cao Z.A., Bass K.E., Verner E., Balasubramanian S., Liu L.et al. (2006) CRA-024781: a novel synthetic inhibitor of histone deacetylase enzymes with antitumor activity in vitro and in vivo. Mol. Cancer Ther. 5, 1309–1317 10.1158/1535-7163.MCT-05-0442 [DOI] [PubMed] [Google Scholar]
- 143.Cao Z.A., Bass K.E., Balasubramanian S., Liu L., Schultz B., Verner E.et al. (2006) CRA-026440: a potent, broad-spectrum, hydroxamic histone deacetylase inhibitor with antiproliferative and antiangiogenic activity in vitro and in vivo. Mol. Cancer Ther. 5, 1693–1701 10.1158/1535-7163.MCT-06-0042 [DOI] [PubMed] [Google Scholar]
- 144.Booth L., Roberts J.L., Poklepovic A., Kirkwood J. and Dent P. (2017) HDAC inhibitors enhance the immunotherapy response of melanoma cells. Oncotarget 8, 83155–83170 10.18632/oncotarget.17950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.De Souza C. and Chatterji B.P. (2015) HDAC inhibitors as novel anti-cancer therapeutics. Recent Patents on Anti-cancer Drug Discovery 10, 145–162 10.2174/1574892810666150317144511 [DOI] [PubMed] [Google Scholar]
- 146.Gurvich N., Tsygankova O.M., Meinkoth J.L. and Klein P.S. (2004) Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 64, 1079–1086 10.1158/0008-5472.CAN-03-0799 [DOI] [PubMed] [Google Scholar]
- 147.Brush M.H., Guardiola A., Connor J.H., Yao T.P. and Shenolikar S. (2004) Deactylase inhibitors disrupt cellular complexes containing protein phosphatases and deacetylases. J. Biol. Chem. 279, 7685–7691 10.1074/jbc.M310997200 [DOI] [PubMed] [Google Scholar]
- 148.Bantscheff M., Hopf C., Savitski M.M., Dittmann A., Grandi P., Michon A.M.et al. (2011) Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 29, 255–265 10.1038/nbt.1759 [DOI] [PubMed] [Google Scholar]
- 149.Roche J. and Bertrand P. (2016) Inside HDACs with more selective HDAC inhibitors. Eur. J. Med. Chem. 121, 451–483 10.1016/j.ejmech.2016.05.047 [DOI] [PubMed] [Google Scholar]
- 150.Géraldy M., Morgen M., Sehr P., Steimbach R.R., Moi D., Ridinger J.et al. (2019) Selective inhibition of histone deacetylase 10: hydrogen bonding to the gatekeeper residue is implicated. J. Med. Chem. 62, 4426–4443 10.1021/acs.jmedchem.8b01936 [DOI] [PubMed] [Google Scholar]
- 151.Villadsen J.S., Stephansen H.M., Maolanon A.R., Harris P. and Olsen C.A. (2013) Total synthesis and full histone deacetylase inhibitory profiling of Azumamides A-E as well as β²- epi-Azumamide E and β³-epi-Azumamide E. J. Med. Chem. 56, 6512–6520 10.1021/jm4008449 [DOI] [PubMed] [Google Scholar]
- 152.Balasubramanian S., Ramos J., Luo W., Sirisawad M., Verner E. and Buggy J.J. (2008) A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 22, 1026–1034 10.1038/leu.2008.9 [DOI] [PubMed] [Google Scholar]
- 153.Morgen M., Steimbach R.R., Géraldy M., Hellweg L., Sehr P., Ridinger J.et al. (2020) Design and synthesis of dihydroxamic acids as HDAC6/8/10 inhibitors. Chem. Med. Chem. 15, 1163–1174 10.1002/cmdc.202000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Li T., Zhang C., Hassan S., Liu X., Song F., Chen K.et al. (2018) Histone deacetylase 6 in cancer. J. Hematol. Oncol. 11, 111 10.1186/s13045-018-0654-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu S.S., Wu F., Jin Y.M., Chang W.Q. and Xu T.M. (2020) HDAC11: a rising star in epigenetics. Biomed. Pharmacother. 131, 110607 10.1016/j.biopha.2020.110607 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The present study includes no data deposited in external repositories.