Abstract
Phenotype-specific omic expression patterns in people with frailty could provide invaluable insight into the underlying multi-systemic pathological processes and targets for intervention. Classical approaches to frailty have not considered the potential for different frailty phenotypes. We characterized associations between frailty (with/without disability) and sets of omic factors (genomic, proteomic, and metabolomic) plus markers measured in routine geriatric care. This study was a prevalent case control using stored biospecimens (urine, whole blood, cells, plasma, and serum) from 1522 individuals (identified as robust (R), pre-frail (P), or frail (F)] from the Toledo Study of Healthy Aging (R=178/P=184/F=109), 3 City Bordeaux (111/269/100), Aging Multidisciplinary Investigation (157/79/54) and InCHIANTI (106/98/77) cohorts. The analysis included over 35,000 omic and routine laboratory variables from robust and frail or pre-frail (with/without disability) individuals using a machine learning framework. We identified three protective biomarkers, vitamin D3 (OR: 0.81 [95% CI: 0.68–0.98]), lutein zeaxanthin (OR: 0.82 [95% CI: 0.70–0.97]), and miRNA125b-5p (OR: 0.73, [95% CI: 0.56–0.97]) and one risk biomarker, cardiac troponin T (OR: 1.25 [95% CI: 1.23–1.27]). Excluding individuals with a disability, one protective biomarker was identified, miR125b-5p (OR: 0.85, [95% CI: 0.81–0.88]). Three risks of frailty biomarkers were detected: pro-BNP (OR: 1.47 [95% CI: 1.27–1.7]), cardiac troponin T (OR: 1.29 [95% CI: 1.21–1.38]), and sRAGE (OR: 1.26 [95% CI: 1.01–1.57]). Three key frailty biomarkers demonstrated a statistical association with frailty (oxidative stress, vitamin D, and cardiovascular system) with relationship patterns differing depending on the presence or absence of a disability.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-021-00334-0.
Keywords: Frailty, Biomarkers, Omics, Clinical phenotype, Disability
Introduction
As a heterogeneous condition [1, 2], frailty relies almost exclusively on expert assessment of clinical and functional parameters for diagnosis [3, 5]. Whereas frailty has been described as a dynamic general impairment of physiological reserves involving multiple organ systems, frailty manifests as progressive vulnerability, impaired ability to address intrinsic and environmental stressors, and impeded the capacity to maintain physiological and psychosocial homeostasis [6–8]. It is associated with an increased risk of geriatric syndromes, dependency, disability, hospitalization, institutional placement, and mortality. Frailty represents a growing burden on societies as populations age [9]. Depending on circumstances, an estimated 11–17% of community-dwelling older persons are frail [10, 11] and 42–49% pre-frail [11, 12].
The underlying pathophysiology of frailty remains largely unknown despite our increasing understanding of the molecular aging processes [13]. Moreover, the usual approach has assessed the relationship between isolated systems, taken one-by-one, in relation to frailty, for example, hormonal dysregulation, vascular function, immune-aging, oxidative stress, pro-coagulation, and pro-inflammatory status [14–17], including several publications from FRAILOMIC initiative [18–20]. Few studies have investigated the complex interrelationships among the different systems underlying frailty [21], which suggest possible multi-system pathogenesis involving several co-existing factors. More than ever, it seems relevant to understand the pathophysiology of frailty since, as recent observations show, these processes are not one directional. There is convincing evidence that they may improve spontaneously [22] or on intervention [23, 24]. Focus is turning to identify factors that influence both negative and positive transitions and how insight could lead to diagnostic biomarkers. To this end, it is required to implement a statistical approach that is able to discriminate the role of several clusters of biomarkers, taking into account their frequent interactions [25, 26]. Our use of a robust machine learning framework to identify biomarkers for frailty and their discriminatory potential is highly appropriate considering the number of pathogenic routes leading to frailty and its consequences. These include metabolic and hormonal factors, inflammation, regulation of cell proliferation, regulation of gene expression, muscle dysfunction, insulin/IGF-1 pathway, stress responses, and cardiovascular homeostasis [27, 28].
Although the classical approach to frailty relies on a single phenotype, emerging evidence suggests different pathways to frailty [2, 29] and different outcomes that patients may experience under various clinical conditions. This suggests several subtypes of frailty [2, 29, 30], such as subtypes with mobility problems [29]. Besides, it is largely understood that frailty and disability are two frequently converging [31–33], but distinct entities need to be dissociated [34]. The expression of different biomarkers is a potential tool to achieve such distinction. Recent studies report biomarker fingerprints in older people with clinical subtypes, such as frailty and sarcopenia [35].
Our aims were, first to identify omic and other lab features, among more than 35,000 potential biomarkers tested, that provide additional information (and possibly mechanistic characterization) on frailty beyond that captured within clinical marker data; second, to identify specific characteristics that are risk or protective factors; and finally, to investigate whether such biomarkers differ according to individual disability status. To this end, the FRAILOMIC initiative assessed both frailty and disability in four large, exploratory, European cohorts by implementing a machine learning framework interrogating clinical and laboratory (genomic, proteomic, and metabolomic (omic)) data where both frailty and disability were assessed.
Methods
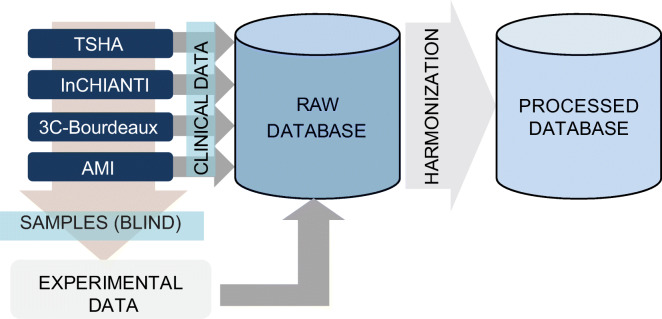
Figure 1 provides an overview of the logistical design for integrating and harmonizing laboratory and clinical data derived from four European cohorts.
Fig. 1.
Graphical description of the data analysis pipeline. The figure depicts the flow initiated at the cohorts to the generation of the FRAILOMIC database. From the cohorts, samples are sent (single-blind) to the experimental labs. A raw database was generated by combining the laboratory and the clinical data for all patients from all cohorts. Harmonization is conducted in order to have the values of all individuals on the same scale for all variables (see eMethods: 1.2 Harmonization)
Study population
This nested case-control study was conducted in the FRAILOMIC Initiative cohorts; data was generated from four population-based European cohorts of older adults: Bordeaux sample data from the Three-City Study (3-C, France) [4], Aging Multidisciplinary Investigation cohort (AMI, Gironde, France) [36], Toledo Study for Healthy Aging (TSHA, Toledo, Spain) [31], and Invecchiare in Chianti Study (InCHIANTI, Chianti geographical area, Tuscany, Italy) [37]. These cohorts and their data harmonization have been described in the online supplement (eMethods, 1.1 to 1.3). An ethical committee approved the original study protocols according to the principles of the Declaration of Helsinki, and all participants signed a written consent.
Clinical and biological data were harmonized using methods described in the online supplement (eMethods 1.2 Harmonization). Harmonized data included gender, age (years), weight (kg), height (cm), body mass index (BMI; kg/m2), and smoking status (current smoker). During the assessment of frailty, information was collected on depressive symptoms, Mini-mental State Examination (MMSE) score corrected by educational status, limitations in basic Activities of Daily Living-BADL (Katz ADL, as a binary variable), Instrumental Activities of Daily Living-IADL (Lawton IADL, as a binary variable), and comorbidity variables (see Supplementary Information (Section A1.1: Cohort Details) for a detailed description of the assessment of covariates and eMethods eTable1).
Frailty classification and participant selection
Frailty was as defined by Fried et al. [38]; subjects were considered to be robust if no criteria of frailty were present, pre-frail when one or two characteristics were present, and frail when three or more items were observed. Participants were drawn from the cohorts to achieve an approximate ‘frail versus pre-frail and robust’ ratio of 1 : 3. Eligibility for study inclusion required data on all five items used to establish frailty status and phenotype.
Biomarker analysis
The study relied on the analysis of stored biospecimens (whole blood, cells, serum/plasma, and urine). Biological samples were identified for specific analyses and shipped to the various laboratories performing the different analyses; laboratories remained blinded to subject phenotype. Candidate biomarkers selected for analysis included muscle function, insulin/type-1 insulin-like growth factors (IGF-1) signaling, stress response, cardiovascular homeostasis, inflammation, cellular senescence, regulation of cell proliferation, and regulation of gene expression. A detailed description of the candidate biomarkers and methods of assessment are provided in the supplementary information (Section B Experimental profiling) [39, 40].
Subpopulations
The analysis was conducted for two overlapping populations. In the first population (F1), we considered all individuals. In the second population (F2), we considered non-disabled individuals (defined by the absence of any restriction in IADL or ADL). The analysis was conducted separately for each population.
Statistics and the machine learning framework
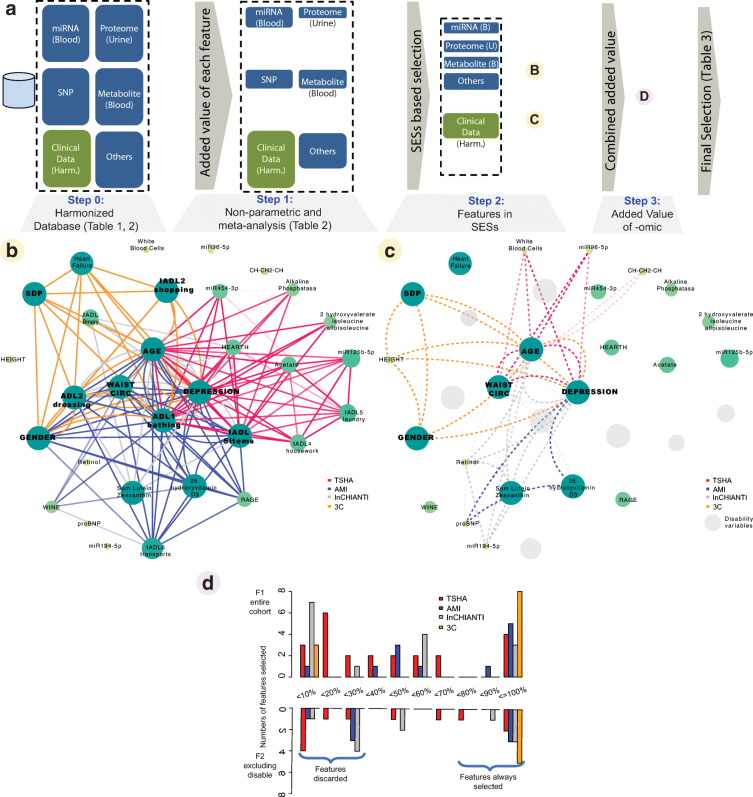
Figure 2a depicts the analytical design of this study aimed to identify frailty omic features providing additional information to that of clinical markers. In brief, following data harmonization (Step 0), a first filtering criteria (Step 1) was applied by defining a frailty classification task using classical clinical biomarkers and one omic feature at a time. Following this, a framework was established to uncover all omic variables that could be combined to provide additional information on the frailty classification problem (Step 2). Finally, all possible combinations of 2 to 10 variables were considered and evaluated (Step 3) while also comparing classification models with only clinical variables or those including also omic features.
Fig. 2.
Robust selection of frailty relevant omic features. a Summary description of the machine learning selection framework (see “Statistics and the machine learning framework”): after harmonization (Step 0), a non-parametric and meta-analysis study selects individual features to be selected (Step 1). Then, a framework is established to uncover all variables that can be combined to provide additional information on frailty using statistically significant signature variables (SESv) (Step 2). Finally, all possible combinations of two to six variables are considered and evaluated (Step 3) while also comparing models with only clinical variables or those, including also omic features. b, c Graphical representation of SESv-derived network. Node size represents the number of times (imputations) a node has been selected in a SESv subset. Edges identify whether nodes have been co-selected in a SESv, where darker shades depict larger number of times the two nodes were connected. F1 and F2 are depicted, respectively, in b and c. In c, gray nodes denote variables associated with disability and excluded in the analysis. d Each feature is shown in one of the x-axis windows depending on the percentage of imputations it was selected in a SESv from the total of 1000 imputations; the y-axis denotes how many features are in each window. Numbers are provided separately per cohort. 3-C 3-City Study; AMI: aging multidisciplinary investigation; TSHA Toledo Study for Healthy Aging; InCHIANTI Invecchiare in Chianti Study
In the analysis, frailty classification models were used to identify omic features that provided information additional to that provided by classical clinical markers. The details for each step are provided below and described in the supplemental at eMethods.
Harmonization (Step 0): variables from all cohorts underwent unified codification, termed harmonization (Fig. 1: described in eMethods: 1.2).
Omic-relevant variables per omic type (Step 1): to limit the number of candidates among the more than 35,000 features (see eTable 2), we aimed to robustly pre-select omic features associated with frailty whose classification power were not influenced by known clinical markers in the first step: age, gender, depression, MMSE corrected by education, disability (as binary), and comorbidity variables. To this end, we implemented a strategy that combined a permutation-based analysis and meta-analysis (see eMethods 1.3: Step 1). We defined the following selection criteria in order to (a) include candidate variables with frailty relevance beyond clinical markers, but also (b) to cover the different data types available:
All variables pertaining to data types (eTable 2) with less than five variables were selected for Step 2.
A variable pertaining to data type with less than 100 variables was selected for the next step (Step 2, eTable 3) if permuted p-value < 0.01 for any cohort or meta-analysis (if possible) derived p-value < 0.001.
A variable pertaining to data type with 100 and more variables was selected for the next step (Step 2, eTable 3) if false discovery rate (FDR) < 0.10 for any cohort or meta-analysis (when possible) derived p-value < 0.0001.
Any variables selected using F1 or F2 populations were included in the Step 2 analysis for both models.
Selection of variables to be considered for the minimal models prediction frailty subtypes (Step 2): to limit the effect of missing data, we first excluded variables with more than 20% of missing values. A thousand imputations were generated for each cohort [41]; to reduce any bias associated with the imputation of missing data, the following analysis (Step 2 and Step 3) was conducted for each imputation.
To select the best combinations of clinical and omic markers for frailty classification for each imputation and each cohort, subsets of variables with maximal or close-to-maximal predictability of frailty [42] were computed; these were denoted as statistically significant signature variables (SESv). The maximum number of independent variables was set to 10 to address over-fitting. The analysis of pairs of variables being co-selected or not in SESv was conducted using network analysis (Fig. 2b and c). Variables that were present in SESvs in more than 50% of the permutations were selected for Step 3 (Fig. 2D).
Using classification models of frailty subtypes for feature selection (Step 3): after selecting the variables derived from SESv analysis (using Fig. 2d), all combinations of variables from two up to 10 were studied as classifiers, and the most accurate combinations as frailty classifiers in each cohort were stored (eTable 4, eTable 5). For each number of variables, and for all possible combinations, we estimated three quality measures for each imputation: the classification error rate using random forests [43], the cross-validation-derived classification error rate of random forests, and the area under the curve (AUC) derived from support vector machine-derived classifiers [44]. We analyzed each frailty subgroup (F1, F2) for each cohort and considering either only clinical variables (clinical, eTable 4, and eTable 5) or all variables (both). A summary of the best classification models is depicted in eFig. 1A and eFig. 1B for F1 and F2, respectively.
We identified clinical markers required to be used as co-variates in any classification model for frailty (ClinS, see detailed in eMethods 3.3) and those omic features with added value in the frailty classification problem as a final outcome from Step 3 (Table 2).
Table 2.
Odds ratios of candidate omics
| TSHA | AMI | InChianti | 3C | Meta–odd | Meta-pvalue | ||
|---|---|---|---|---|---|---|---|
| F1 | 25-Hydroxyvitamin D3 | 0.88 (0.67–1.18) | 0.63 (0.45–0.85) | 0.79 (0.56–1.02) | 0.99 (0.73–1.32) | 0.81 (0.68–0.98) | 0.03 |
| -CHCH2CH- moiety | 0.77 (0.56–1.03) | ||||||
| citrate | 1.49 (1.07–2.09) | ||||||
| Troponin_T | 1.25 (0.92–1.94) | 1.25 (0.86–1.89) | 1.25 (1.23–1.27) | <0.001 | |||
| Lutein/zeaxanthin | 1 (0.76–1.32) | 0.84 (0.62–1.1) | 0.67 (0.51–0.85) | 0.81 (0.55–1.13) | 0.82 (0.7–0.97) | 0.02 | |
| pro-BNP | 1.06 (0.77-1.59) | 1.58 (0.87–4.49) | 1.29 (0.88–1.91) | 0.2 | |||
| Retinol | 1.05 (0.79–1.4) | 1.14 (0.81–1.51) | 0.7 (0.52–0.91) | 1.23 (0.92–1.65) | 1.01 (0.79–1.29) | 0.97 | |
| miR194-5p | 0.96 (0.7–1.34) | 1.24 (0.9–1.73) | 1.09 (0.85–1.39) | 0.5 | |||
| miR125b-5p | 0.64 (0.47–0.86) | 0.85 (0.61–1.13) | 0.73 (0.56–0.97) | 0.03 | |||
| miR454-3p | 0.61 (0.44–0.84) | 1.01 (0.76–1.32) | 0.79 (0.48–1.28) | 0.33 | |||
| MDA | 1.13 (0.81–1.54) | 0.85 (0.64–1.13) | 0.98 (0.74–1.3) | 0.89 | |||
| sRAGE | 0.98 (0.69–1.35) | 1.3 (0.93–1.7) | 1.25 (0.93–1.71) | 1.17 (0.98–1.39) | 0.08 | ||
| Urine peptide 56884 | 0.69 (0.48–0.91) | ||||||
| F2 | 25-Hydroxyvitamin D3 | 0.97 (0.7–1.33) | 0.52 (0.37–0.70) | 0.74 (0.54–0.98) | 1.04 (0.78–1.40) | 0.79 (0.58–1.08) | 0.14 |
| -CHCH2CH- moiety | 0.71 (0.51–0.95) | ||||||
| citrate | 1.19 (0.85–1.68) | ||||||
| Troponin_T | 1.25 (0.97–1.88) | 1.33 (1.01–1.91) | 1.29 (1.21–1.38) | <0.001 | |||
| Lutein/zeaxanthin | 1.22 (0.88–1.67) | 0.87 (0.65–1.16) | 0.61 (0.42–0.82) | 0.92 (0.64–1.23) | 0.88 (0.66–1.16) | 0.36 | |
| pro-BNP | 1.37 (1.02–2.64) | 1.58 (1.20–3.08) | 1.47 (1.27–1.7) | <0.001 | |||
| Retinol | 1.14 (0.85–1.58) | 1.26 (0.94–1.62) | 0.62 (0.45–0.82) | 1.22 (0.91–1.60) | 1.02 (0.74–1.42) | 0.9 | |
| miR194-5p | 0.87 (0.66–1.17) | 1.39 (1.02–1.99) | 1.1 (0.7–1.74) | 0.67 | |||
| miR125b-5p | 0.87 (0.64–1.15) | 0.83 (0.6–1.1) | 0.85 (0.81–0.88) | <0.001 | |||
| miR454-3p | 0.71 (0.49–1.01) | 1.07 (0.82–1.4) | 0.87 (0.58–1.3) | 0.49 | |||
| MDA | 1.38 (1.09–1.78) | 0.85 (0.63–1.16) | 1.08 (0.67–1.75) | 0.74 | |||
| sRAGE | 1.01 (0.70–1.37) | 1.35 (1.02–1.76) | 1.46 (1.08–2.01) | 1.26 (1.01–1.57) | 0.04 | ||
| Urine peptide 56884 | 0.65 (0.47–0.85) |
3C 3-City Study; AMI: Aging Multidisciplinary Investigation; TSHA: Toledo Study for Healthy Aging; InCHIANTI: Invecchiare in Chianti Study; MDA: malondialdehyde; pro-BNP: pro B-type natriuretic peptide; sRAGE: serum soluble receptor for advanced glycation end products
aBoot strapping-based median and 95% confidence interval of the odds ratios derived from the ordinal regression analysis. The columns protective and risk enumerate the number of cohorts where the omic variable was found significant with an odds ratio below 1 and over 1, respectively. In each model, the controlling factors were the clinical variables, including age, gender, and depression as omnipresent variables plus summary measures of ADL and IADL for F1
Characterization of omic selected features
To characterize further the added value of selected omic features, we computed proportional odd ratios (OR) for frailty for each selected feature in F1 and F2 separately. As co-variates in the model, we used ClinS variables. We computed an ordinal logistic regression model using bootstrapping (n=1000) and reporting the median and the 0.025 and 0.975 quantiles to estimate the 95% confidence intervals for each selected feature. In addition, and for those features available in more than one cohort, we computed a meta-analysis-derived OR.
Results
Subject disposition
After excluding subjects with missing values for frailty status (n=114) according to the Frailty Phenotype criteria (see “Frailty classification and participant selection”), we interrogated data from 1522 out of 1636 participants included in the FRAILOMIC database: TSHA (n=471, robust=178/pre-frail=184/frail=109), InCHIANTI (n=281, 106/98/77), 3C-Bordeaux (n=480, 111/269/100), and AMI (n=290, 157/79/54) (see Supplementary Information eMethods 1.1: Cohort Details). Clinical characteristics of study participants by cohort are summarized in Table 1. Mean participant age was 75.3–82.1 years, and, with the exception of the 3-C cohort (where the MMSE score was higher), the general level of education was low. Slowness and low physical activity were the most frequent frailty criteria met by the participants, whereas weight loss and exhaustion were reported least. Available samples per laboratory of analysis, cohort, number of laboratory (including omics) biomarkers per data type, and the features selected from each laboratory in Step 1 are summarized in Supplementary eTable 2.
Table 1.
Sociodemographic and health indicators of the selected participants from each cohort.
| TSHA | AMI | InCHIANTI | 3C | |
|---|---|---|---|---|
| n | 471 | 290 | 281 | 480 |
| Age, years (mean, (SD)) | 75.28 (5.77) | 75.47 (6.44) | 75.53 (6.91) | 82.13 (4.27) |
| Gender, (n, % female) | 280 (59.4) | 110 (37.9) | 163 (58.0) | 300 (62.5) |
| Education (n, % ) | ||||
| Low | 442 (94.4) | 229 (79.0) | 267 (96.0) | 123 (25.6) |
| Intermediate | 13 (2.8) | 55 (19.0) | 11 (4.0) | 242 (50.4) |
| High | 13 (2.8) | 6 (2.1) | 0 (0.0) | 115 (24.0) |
| MMSE, (mean, (SD)) | 23.11 (5.15) | 25.59 (3.51) | 24.93 (3.38) | 27.86 (2.11) |
| MMSE corrected, (n, (%)) | 45 (11.4) | 9 (3.3) | 10 (3.6) | 12 (2.6) |
| Body mass index, kg/m2 (mean, (SD)) | 29.13 (5.24) | 27.79 (4.46) | 27.91 (4.42) | 25.77 (4.13) |
| Smoking | ||||
| Past smoker (n, (%)) | 150 (31.8) | 100 (34.6) | 106 (37.7) | 174 (36.2) |
| Current smoker (n, (%)) | 38 (8.1) | 15 (5.2) | 0 (0.0) | 22 (4.6) |
| Frailty items | ||||
| Sedentarity (n, (%)) | 139 (29.5) | 70 (24.1) | 81 (28.8) | 278 (57.9) |
| Weakness (n, (%)) | 134 (28.5) | 67 (23.1) | 90 (32.0) | 117 (24.4) |
| Shrinking (n, (%)) | 77 (16.3) | 28 (9.7) | 26 (9.3) | 72 (15.0) |
| Slowness (n, (%)) | 160 (34.0) | 67 (23.1) | 104 (37.0) | 149 (31.0) |
| Fatigue (n, (%)) | 90 (19.1) | 42 (14.5) | 78 (27.8) | 74 (15.4) |
| Frailty status | ||||
| Robust | 178 (37.8) | 157 (54.1) | 106 (37.7) | 111 (23.1) |
| Pre-frail | 184 (39.1) | 79 (27.2) | 98 (34.9) | 269 (56.0) |
| Frail | 109 (23.1) | 54 (18.6) | 77 (27.4) | 100 (20.8) |
Assessment of statistically significant signature variables (SESv)
Networks were derived from identified SESv computed for each cohort and each imputation. A network edge characterizes co-occurrence of variables within SESv, whereas a network node characterizes the prevalence of variables within SESvs per imputation. The associations across networks are summarized in Fig. 2b and c for F1 and F2, respectively. As a positive control, age was recognized as being at the core of all SESv for all cohorts. A highly connected set of clinical features common for all cohorts and both frailty subgroups (F1 and F2) was subsequently identified: depression and waist circumference. As expected, IADL and ADL disability-associated features were also at the core for F1-derived networks, where few variables apart from those related to physical functioning were included.
Assessment of biomarkers
Table 2 shows the associated proportional odds ratio (OR) for all identified omic features derived from Step 3 analysis (eTable 4, eTable 5) in relation to the frailty status, differentiating for whether or not disabled individuals were included in the multinomial logistic regression, F1 and F2, respectively, and considering in the models all clinical markers identified in Step 3: ClinS (see eMethods 3.3) and including (F1) or excluding (F2) disability markers. Following meta-analysis in populations including disability (F1), vitamin D3 (OR: 0.81 [95% CI: 0.68, 0.98]), lutein zeaxanthin (OR: 0.82 [95% CI: 0.70, 0.97]), and miRNA125b-5p (OR: 0.73, [95% CI: 0.56, 0.97]) demonstrated significant associations with reduced odds ratios, whereas only one biomarker, cardiac troponin T (OR: 1.25 [95% CI: 1.23, 1.27]), was associated with increased odds of being pre-frail and frail when compared with participants identified as robust. Only miRNA125b-5p (OR: 0.85 [95% CI: 0.81, 0.88]) was associated with reduced odds ratio scores of pre-frailty and frailty in models that excluded disabled individuals (F2), whereas cardiac troponin T (OR: 1.29 [95% CI: 1.21, 1.38]), pro-BNP (OR: 1.47 [95% CI: 1.27, 1.70]), and sRAGE (OR: 1.26 [95% CI: 1.01, 1.57]) were associated with increased odds of pre-frailty and frailty. Four other biomarkers showed an association with frailty in some of the cohorts where they were measured but failed to do in the meta-analysis: retinol (OR: 0.7 [95% CI: 0.52, 0.91]) in the InChianti and miR454-3 (OR: 0.61 [95% CI: 0.44, 0.84]) in the TSHA cohort showed to be protective in older people in the model F1, while retinol (OR: 0.62 [95% CI: 0.45, 0.82]) and miR194-5 (OR: 1.39 [95% CI: 1.02, 1.99]) in the InChianti cohort and malondialdehyde (MDA: OR: 1.38 [95% CI: 1.09, 1.78]) in the TSHA cohort showed an association with frailty in people without disability. Finally, when considering features only available in one cohort, urine peptide 56884 was significantly associated in the InChianti cohort in both F1 (OR: 0.69 [95% CI: 0.48–0.91]) and F2 (OR: 0.65 [95% CI: 0.47–0.85]); citrate in F1 (OR: 1.49 [95% CI: 1.07–2.09]) and -CHCH2CH- moiety in F2 (OR: 0.71 [95% CI: 0.51–0.95]) were associated with frailty in the TSHA cohort.
Discussion
Our study describes the use of a robust machine learning framework to identify biomarkers for frailty and their discriminatory potential in alternative frailty phenotypes (with and without disability). Underlying causes of frailty, as assessed by the frailty phenotype, embrace several potential biological mediators, ranging from hormones to endothelial function [27, 28], whereas other theoretical approaches to frailty do not offer such a wide selection of biological factors possibly associated with the syndrome. The analysis included among 35,312 omic markers selected based on their relevance to aging and included metabolism, inflammation, regulation of cell proliferation, regulation of gene expression, muscle dysfunction, insulin/IGF-1 pathway, stress responses, and cardiovascular homeostasis. We used a frailty classification-oriented analysis to identify omic and non-omic lab features providing added value to that of clinical markers. Thirteen candidate features emerged as having associations with frailty beyond that of classical clinical factors measured in geriatric care, although only six in ten showed to be associated after a meta-analysis, and the other three biomarkers were determined in only one of the cohorts.
It is noteworthy that although some of the features seemed to be involved in deleterious processes, mainly related with cardiovascular biomarkers (Troponin-T, sRAGE, and pro-BNP), others, mainly related to vitamin D and antioxidant systems, seemed to relate to protective roles (miRNA125-5p, vitamin D3, and lutein zeaxanthin). This finding reinforces and expands previous findings [18], obtained in FRAILOMIC using another methodological approach, and provides pathophysiological support to the observation about the dynamic condition of frailty [22–24]. These findings serve as a starting point for identifying both risk and protective factors of frailty in future longitudinal studies. Once again, their inclusion in the classification models is modulated by the presence or absence of a disability.
In terms of gene expression, miRNA 125b-5p demonstrated a robust association with frailty after meta-analysis. The 125b-5p miRNA has previously been linked with several cancers [45] and to the expression of vitamin D receptors [46]. Here, we also confirm a possible role of lower vitamin D3 levels with development of frailty [47]. Vitamin D has been linked to several components involved in the development of frailty and/or its consequences, including sarcopenia and low bone density [48]. The role of bone in frailty is also highlighted by the relationship with citrate in the only cohort where it was determined [49].
Our findings suggest a relevant role for vitamin D3, oxidative stress, and cardiovascular system in older people with pre-frailty and frailty, with potential relationships among them. Inclusion of MDA (an indicator of oxidative stress) along with retinol and lutein zeaxanthin (two well-known antioxidants) underscores the important role of oxidative balance. This fits with our recent demonstration of a decreased expression of genes implicated in the cellular response to stress [20], suggesting that a low defense to oxidative insult, more than increased oxidative stress, is a core aspect of development. The observation that increased levels of sRAGE were also associated with the presence of frailty appears to further support the role of oxidative stress.
Advanced glycation end products (AGE) and the cell-bound receptor called receptor for AGE (RAGE) are implicated in the pathogenesis of numerous diseases. Soluble receptor for AGE (sRAGE) counteracts the adverse effects of AGE-RAGE interaction by competing with RAGE for binding with AGE. Low levels of serum sRAGE are proposed as a biomarker for comorbidities associated with aging and related to increased oxidative stress [50], particularly in people with cardiovascular risk [51–53]. Current research supports the validity of the high ratio AGE/sRAGE as a universal biomarker for such diseases [54]. Interestingly, cardiac troponin T, a regulatory protein integral to muscle contraction, and the propeptide of the brain natriuretic peptide (pro-BNP), both associated with cardiac pathology [55, 56], were increased in participants who expressed the frailty phenotype. Cardiac troponin T is also involved in muscle contraction [57] and pro-BNP in blood pressure regulation [58]. The increase in this highly specific cardiac biomarker fits with the biological and functional impairment of cardiac muscle often associated with frailty.
Our study provides a first large-scale attempt to test these assumptions in the context of aging and frailty. Despite assessing more than 35,000 omic variables in a controlled statistical manner, using carefully harmonized cohorts, it is clear that any emergent predictive power is undermined by participant heterogeneity. Only three of 13 biomarkers (pro-BNP, sRAGE, and vitamin D3) were observed with significant OR in more than one cohort (Table 2). This observation appears to stress the need for further subject stratification as shown from a less clinical point-of-view in a recent study [59], as it is also the case in other clinical syndromes.
Our study has limitations. The observed heterogeneity of our study results is most likely explained by the different characteristics of the populations included in the participant cohorts. The heterogeneity reflects not only differences in cultural and socioeconomic background, but also potentially different frailty subtypes. In addition, while for an omic study our sample size is adequate, it was only powered to identify an odds ratio of 1.7, meaning that only those omic factors with a substantial association with frailty could be identified. Major strengths of this study are the breadth of omic markers—representing (a) inflammation, (b) regulation of cell proliferation, (c) regulation of gene expression, (d) metabolism/muscle dysfunction, insulin—IGF1 signaling pathway/stress response, and cardiovascular homeostasis, measured in highly specialized laboratories, coupled with careful harmonization across well phenotyped, independent European cohorts and the rigorous statistical analysis to minimize the occurrence of false-positive biomarkers. The study’s main contribution is the building of sets of biomarkers (clinical and laboratory), reflecting the alteration in several physiological systems, providing a comprehensive consideration of the multi-systemic nature of frailty. Moreover, our results suggest that the contribution of those biomarkers to the diagnosis of frailty might differ depending upon the presence of a disability.
Concluding this discussion, using a robust methodological approach, we identified six omic and lab biomarkers related to three main pathophysiological pathways (vitamin D, oxidative stress, and cardiovascular system) which are strongly linked to frailty. In addition, we provide evidence about the potential involvement of other seven biomarkers. These biomarkers change according to the disability status and have the potential to provide additional information of frailty and, in the future, may allow improving diagnostic accuracy for frailty and modeling beyond clinical parameters usually assessed in medical practice [60]. Validation of these biomarkers in independent cohorts and in different clinical phenotypes is of paramount importance as well as the evaluation of their biological role in the onset of frailty and its prognosis.
Supplementary Information
(DOCX 8326 kb)
(XLSX 36 kb)
Acknowledgments
On behalf of the FRAILOMIC Initiative the authors would like to thank Perrine André MSc and Hermine Pellay MSc (Univ. Bordeaux, Inserm, Bordeaux Population Health Research Center, UMR 1219, F-33000 Bordeaux, France), Mariam El Assar MSc and Betty Davies MD (Fundación de Investigación Biomedica Hospital Unversitario de Getafe), Eleonora Talluri PhD(USL Centro Toscana, Firenze, Italy), Valeria Orrù MSc and Michele Marongiu MSc (Institute for Genetic and Biomedical Research, Caligari, Italy), Esther García-Esquinas PhD and Esther Lopez-Garcia PhD and Pilar Guallar MD PhD and Fernando Rodriguez Artalejo MD PhD (Faculty of Medicine, Universidad Autonoma de Madrid, Madrid, Spain), Ignacio Ara PhD (GENUD Toledo Research Group, Universidad Castilla-La Mancha, Toledo, Spain), José-María Sánchez-Puelles PhD (Molecular Pharmacology Lab, Centre of Biological Sciences, CSIC, Madrid, Spain), Paloma Moraga MSc (Sistemas Genomicos, Valencia, Spain).
Author contribution
The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal.
Funding
This work was supported by the European Union’s Seventh Framework Programme (FP7/2007-2013) FRAILOMIC Project (grant number 305483). The Three-City Study was conducted under a partnership agreement between the Institut National de la Santé et de la Recherche Médicale, Victor Segalen – Bordeaux2 University and the Sanofi-Synthélabo company. The Fondation pour la Recherche Médicale funded the preparation and beginning of the study. The 3C-Study was also sponsored by the Caisse Nationale Maladie des Travailleurs Salariés, Direction Générale de la Santé, Conseils Régionaux of Aquitaine and Bourgogne, Fondation de France, Ministry of Research-INSERM Program Cohortes et collections de données biologiques, the Fondation Plan Alzheimer (FCS 2009-2012), and the Caisse Nationale pour la Solidarité et l’Autonomie. The InCHIANTI study baseline (1998–2000) was supported as a ‘targeted project’ (ICS110.1/RF97.71) by the Italian Ministry of Health and in part by the U.S. National Institute on Aging (Contracts: 263 MD 9164 and 263 MD 821336) and by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, Baltimore, Maryland.
Data Availability
Data will be made freely available
Declarations
Ethics approval
The original study protocols were approved by ethical committee according to the principles of the Declaration of Helsinki and all participants signed a written consent with participants agree to sample retention/analysis and data publication.
Conflicts of interest
Stefan Walter, Rebeca Miñambres, Lucía Bernard, Lee Butcher, Jorge Erusalimsky, Francisco José García-García, José Antonio Carnicero, Tim Hardman, Mattias Hacki, Johannes Grillari, Edoardo Fiorillo, Francesco Cucca, Matteo Cesari, Isabelle Carrie, Marco Colpo, Stefania Bandinelli, Karine Peres, Jean Francois Dartigues, Catherine Helmer,José Viña, Gloria Olaso, Irene Garcia, Jorge Garcia, Pidder Janssen-Dürr, Tilman Grune, Daniela Weber, Giuseppe Lippi, Chiara Bonaguri, and Alan Sinclair declare no conflicts of interest. David Gomez-Cabrero, Imad Abugesaissa and Jesper Tegner have been paid as consultants by YouHealth SB. David Gomez-Cabrero has received a speaker honorarium from Sanofi Aventis. Harald Mischak is the co-founder and co-owner of Mosaiques Diagnostics. Petra Zürbig is employed by Mosaiques Diagnostics. Catherine Féart received fees for conferences from Danone Institute and Nutricia, and served as consultant for Laboratoire Lescuyer and Cholé'Doc. Leocadio Rodriguez-Mañas has received fees for conferences from Abbott Laboratories and Novartis.
Footnotes
David Gómez-Cabrero and Stefan Walter contributed equally to this work
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Liu L-K, Guo C-Y, Lee W-J, Chen L-Y, Hwang A-C, Lin M-H, Peng LN, Chen LK, Liang KY. Subtypes of physical frailty: Latent class analysis and associations with clinical characteristics and outcomes. Sci Rep. 2017;7(1):46417. doi: 10.1038/srep46417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies B, García F, Ara I, Artalejo FR, Rodriguez-Mañas L, Walter S. Relationship Between sarcopenia and frailty in the toledo study of healthy aging: a population based cross-sectional study. J Am Med Dir Assoc. 2018;19(4):282–286. doi: 10.1016/j.jamda.2017.09.014. [DOI] [PubMed] [Google Scholar]
- 3.Searle SD, Mitnitski A, Gahbauer EA, Gill TM, Rockwood K. A standard procedure for creating a frailty index. BMC Geriatr. 2008;8(1):24. doi: 10.1186/1471-2318-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.3C Study Group Vascular factors and risk of dementia: design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology. 2003;22(6):316–325. doi: 10.1159/000072920. [DOI] [PubMed] [Google Scholar]
- 5.Justice JN, Ferrucci L, Newman AB, Aroda VR, Bahnson JL, Divers J, Espeland MA, Marcovina S, Pollak MN, Kritchevsky SB, Barzilai N, Kuchel GA. A framework for selection of blood-based biomarkers for geroscience-guided clinical trials: report from the TAME Biomarkers Workgroup. GeroScience. 2018;40(5–6):419–436. doi: 10.1007/s11357-018-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Viña J, Tarazona-Santabalbina FJ, Pérez-Ros P, Martínez-Arnau FM, Borras C, Olaso-Gonzalez G, Salvador-Pascual A, Gomez-Cabrera MC. Biology of frailty: Modulation of ageing genes and its importance to prevent age-associated loss of function. Mol Aspects Med. 2016;50:88–108. doi: 10.1016/j.mam.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Cutler RG, Mattson MP. The adversities of aging. Ageing Res Rev. 2006;5(3):221–238. doi: 10.1016/j.arr.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Hayflick L. Biological aging is no longer an unsolved problem. Ann N Y Acad Sci. 2007;1100:1–13. doi: 10.1196/annals.1395.001. [DOI] [PubMed] [Google Scholar]
- 9.Clegg A, Young J, Iliffe S, Rikkert MO, Rockwood K. Frailty in elderly people. Lancet. 2013;381(9868):752–762. doi: 10.1016/S0140-6736(12)62167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collard RM, Boter H, Schoevers RA, Oude Voshaar RC. Prevalence of frailty in community-dwelling older persons: a systematic review. J Am Geriatr Soc. 2012;60(8):1487–1492. doi: 10.1111/j.1532-5415.2012.04054.x. [DOI] [PubMed] [Google Scholar]
- 11.Siriwardhana DD, Hardoon S, Rait G, Weerasinghe MC, Walters KR. Prevalence of frailty and prefrailty among community-dwelling older adults in low-income and middle-income countries: a systematic review and meta-analysis. BMJ Open. 2018;8(3):e018195. doi: 10.1136/bmjopen-2017-018195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liljas AEM, Carvalho LA, Papachristou E, De Oliveira C, Wannamethee SG, Ramsay SE, et al. Self-reported vision impairment and incident prefrailty and frailty in English community-dwelling older adults: findings from a 4-year follow-up study. J Epidemiol Community Health. 2017;71(11):1053–1058. doi: 10.1136/jech-2017-209207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodríguez-Mañas L, Féart C, Mann G, Viña J, Chatterji S, Chodzko-Zajko W, Gonzalez-Colaço Harmand M, Bergman H, Carcaillon L, Nicholson C, Scuteri A, Sinclair A, Pelaez M, van der Cammen T, Beland F, Bickenbach J, Delamarche P, Ferrucci L, Fried LP, Gutiérrez-Robledo LM, Rockwood K, Rodríguez Artalejo F, Serviddio G, Vega E, FOD-CC group (Appendix 1) Searching for an operational definition of frailty: a Delphi method based consensus statement. The Frailty Operative Definition-Consensus Conference Project. Journals Gerontol Ser A Biol Sci Med Sci. 2013;68(1):62–67. doi: 10.1093/gerona/gls119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clegg A, Hassan-Smith Z. Frailty and the endocrine system. Lancet Diabetes Endocrinol. 2018;6(9):743–752. doi: 10.1016/S2213-8587(18)30110-4. [DOI] [PubMed] [Google Scholar]
- 15.Alonso-Bouzón C, Carcaillon L, García-García FJ, Amor-Andrés MS, El Assar M, Rodríguez-Mañas L. Association between endothelial dysfunction and frailty: the Toledo Study for Healthy Aging. Age (Dordr). 2014;36(1):495–505. doi: 10.1007/s11357-013-9576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pansarasa O, Pistono C, Davin A, Bordoni M, Mimmi MC, Guaita A, Cereda C. Altered immune system in frailty: Genetics and diet may influence inflammation. Ageing Res Rev. 2019;54:100935. doi: 10.1016/j.arr.2019.100935. [DOI] [PubMed] [Google Scholar]
- 17.Arauna D, García F, Rodríguez-Mañas L, Marrugat J, Sáez C, Alarcón M, Wehinger S, Espinosa-Parrilla Y, Palomo I, Fuentes E. Older adults with frailty syndrome present an altered platelet function and an increased level of circulating oxidative stress and mitochondrial dysfunction biomarker GDF-15. Free Radic Biol Med. 2020;149:64–71. doi: 10.1016/j.freeradbiomed.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 18.Kochlik B, Stuetz W, Pérès K, Pilleron S, Féart C, García García FJ, Bandinelli S, Gomez-Cabrero D, Rodriguez-Mañas L, Grune T, Weber D. Associations of fat-soluble micronutrients and redox biomarkers with frailty status in the FRAILOMIC initiative. J Cachexia Sarcopenia Muscle. 2019;10(6):1339–1346. doi: 10.1002/jcsm.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butcher L, Carnicero JA, Gomez Cabrero D, Dartigues J-F, Pérès K, Garcia-Garcia FJ, Rodriguez-Mañas L, Erusalimsky JD, FRAILOMIC Consortium Increased levels of soluble receptor for advanced glycation end-products (RAGE) are associated with a higher risk of mortality in frail older adults. Age Ageing. 2019;48(5):696–702. doi: 10.1093/ageing/afz073. [DOI] [PubMed] [Google Scholar]
- 20.El Assar M, Angulo J, Carnicero JA, Walter S, García-García FJ, López-Hernández E, et al. Frailty is associated with lower expression of genes involved in cellular response to stress: results from the Toledo Study for Healthy Aging. J Am Med Dir Assoc. 2017;18(8):734.e1–734.e7. doi: 10.1016/j.jamda.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 21.Fried LP, Xue Q-L, Cappola AR, Ferrucci L, Chaves P, Varadhan R, Guralnik JM, Leng SX, Semba RD, Walston JD, Blaum CS, Bandeen-Roche K. Nonlinear multisystem physiological dysregulation associated with frailty in older women: implications for etiology and treatment. Journals Gerontol Ser A Biol Sci Med Sci. 2009;64A(10):1049–1057. doi: 10.1093/gerona/glp076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trevisan C, Veronese N, Maggi S, Baggio G, Toffanello ED, Zambon S, Sartori L, Musacchio E, Perissinotto E, Crepaldi G, Manzato E, Sergi G. Factors influencing transitions between frailty states in elderly adults: The Progetto Veneto Anziani Longitudinal Study. J Am Geriatr Soc. 2017;65(1):179–184. doi: 10.1111/jgs.14515. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez-Mañas L, Laosa O, Vellas B, Paolisso G, Topinkova E, Oliva-Moreno J, Bourdel-Marchasson I, Izquierdo M, Hood K, Zeyfang A, Gambassi G, Petrovic M, Hardman TC, Kelson MJ, Bautmans I, Abellan G, Barbieri M, Peña-Longobardo LM, Regueme SC, Calvani R, de Buyser S, Sinclair AJ, on behalf of the European MID‐Frail Consortium Effectiveness of a multimodal intervention in functionally impaired older people with type 2 diabetes mellitus. J Cachexia Sarcopenia Muscle. 2019;10(4):721–733. doi: 10.1002/jcsm.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trombetti A, Hars M, Hsu F-C, Reid KF, Church TS, Gill TM, King AC, Liu CK, Manini TM, McDermott MM, Newman AB, Rejeski WJ, Guralnik JM, Pahor M, Fielding RA, for the LIFE Study Investigators Effect of physical activity on frailty. Ann Intern Med. 2018;168(5):309–316. doi: 10.7326/M16-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson R, Björkegren J, Tegnér J. On reliable discovery of molecular signatures. BMC Bioinformatics. 2009;10:38. doi: 10.1186/1471-2105-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsson R, Peña JM, Björkegren J, Tegnér J. Detecting multivariate differentially expressed genes. BMC Bioinformatics. 2007;8:150. doi: 10.1186/1471-2105-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angulo J, El Assar M, Álvarez-Bustos A, Rodríguez-Mañas L. Physical activity and exercise: strategies to manage frailty. Redox Biol. 2020;35:101513. doi: 10.1016/j.redox.2020.101513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angulo J, El Assar M, Rodríguez-Mañas L. Frailty and sarcopenia as the basis for the phenotypic manifestation of chronic diseases in older adults. Mol Aspects Med. 2016;50:1–32. doi: 10.1016/j.mam.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 29.Huang S-T, Tange C, Otsuka R, Nishita Y, Peng L-N, Hsiao F-Y, Tomida M, Shimokata H, Arai H, Chen LK. Subtypes of physical frailty and their long-term outcomes: a longitudinal cohort study. J Cachexia Sarcopenia Muscle. 2020;11(5):1223–1231. doi: 10.1002/jcsm.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pamoukdjian F, Laurent M, Martinez-Tapia C, Rolland Y, Paillaud E, Canoui-Poitrine F. Frailty parameters, morbidity and mortality in older adults with cancer: a structural equation modelling approach based on the fried phenotype. J Clin Med. 2020;9(6):1826. doi: 10.3390/jcm9061826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia-Garcia FJ, Gutierrez Avila G, Alfaro-Acha A, Amor Andres MS, de la Torre Lanza MDLA, Escribano Aparicio MV, et al. The prevalence of frailty syndrome in an older population from Spain. The Toledo study for healthy aging. J Nutr Heal AGING. 2011;15(10):852–856. doi: 10.1007/s12603-011-0075-8. [DOI] [PubMed] [Google Scholar]
- 32.Balzi D, Lauretani F, Barchielli A, Ferrucci L, Bandinelli S, Buiatti E, Milaneschi Y, Guralnik JM. Risk factors for disability in older persons over 3-year follow-up. Age Ageing. 2010;39(1):92–98. doi: 10.1093/ageing/afp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ávila-Funes JA, Helmer C, Amieva H, Barberger-Gateau P, Le Goff M, Ritchie K, et al. Frailty among community-dwelling elderly people in France: the Three-City Study. Journals Gerontol Ser A Biol Sci Med Sci. 2008;63(10):1089–1096. doi: 10.1093/gerona/63.10.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fried LP, Ferrucci L, Darer J, Williamson JD, Anderson G. Untangling the concepts of disability, frailty, and comorbidity: implications for improved targeting and care. Journals Gerontol Ser A Biol Sci Med Sci. 2004;59(3):M255–M263. doi: 10.1093/gerona/59.3.m255. [DOI] [PubMed] [Google Scholar]
- 35.Calvani R, Picca A, Marini F, Biancolillo A, Gervasoni J, Persichilli S, et al. Identification of biomarkers for physical frailty and sarcopenia through a new multi-marker approach: results from the BIOSPHERE study. GeroScience. 2020. 10.1007/s11357-020-00197-x. [DOI] [PMC free article] [PubMed]
- 36.Pérès K, Matharan F, Allard M, Amieva H, Baldi I, Barberger-Gateau P, Bergua V, Bourdel-Marchasson I, Delcourt C, Foubert-Samier A, Fourrier-Réglat A, Gaimard M, Laberon S, Maubaret C, Postal V, Chantal C, Rainfray M, Rascle N, Dartigues JF. Health and aging in elderly farmers: the AMI cohort. BMC Public Health. 2012;12:558. doi: 10.1186/1471-2458-12-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferrucci L, Bandinelli S, Benvenuti E, Di Iorio A, Macchi C, Harris TB, et al. Subsystems contributing to the decline in ability to walk: bridging the gap between epidemiology and geriatric practice in the InCHIANTI study. J Am Geriatr Soc. 2000;48(12):1618–1625. doi: 10.1111/j.1532-5415.2000.tb03873.x. [DOI] [PubMed] [Google Scholar]
- 38.Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, et al. Frailty in older adults: evidence for a phenotype. Journals Gerontol A. 2001;56(3):146–156. doi: 10.1093/gerona/56.3.m146. [DOI] [PubMed] [Google Scholar]
- 39.Erusalimsky JD, Grillari J, Grune T, Jansen-Duerr P, Lippi G, Sinclair AJ, Tegnér J, Viña J, Durrance-Bagale A, Miñambres R, Viegas M, Rodríguez-Mañas L, FRAILOMIC Consortium In search of ‘omics’-based biomarkers to predict risk of frailty and its consequences in older individuals: the FRAILOMIC Initiative. Gerontology. 2015;62(2):182–190. doi: 10.1159/000435853. [DOI] [PubMed] [Google Scholar]
- 40.Lippi G, Jansen-Duerr P, Viña J, Durrance-Bagale A, Abugessaisa I, Gomez-Cabrero D, Tegnér J, Grillari J, Erusalimsky J, Sinclair A, Rodriguez-Manãs L, on behalf of the FRAILOMIC consorti Laboratory biomarkers and frailty: presentation of the FRAILOMIC initiative. Clin Chem Lab Med. 2015;53:e253–e255. doi: 10.1515/cclm-2015-0147. [DOI] [PubMed] [Google Scholar]
- 41.Van Buuren S, Groothuis-Oudshoorn K. Multivariate imputation by chained equations. J Stat Softw. 2011;45(3):1–67. [Google Scholar]
- 42.Lagani V, Athineou G, Farcomeni A, Tsagris M, Tsamardinos I. Feature selection with the R package MXM: discovering statistically equivalent feature subsets. J Stat Software. 2017;1(7) SepAvailable from: https://www.jstatsoft.org/v080/i07.
- 43.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 44.Hand DJ, Till RJ. A simple generalisation of the area under the ROC curve for multiple class classification problems. Mach Learn. 2001;45(2):171–186. [Google Scholar]
- 45.Nagaraj S, Zoltowska KM, Laskowska-Kaszub K, Wojda U. microRNA diagnostic panel for Alzheimer’s disease and epigenetic trade-off between neurodegeneration and cancer. Ageing Res Rev. 2018. 10.1016/j.arr.2018.10.008. [DOI] [PubMed]
- 46.Mohri T, Nakajima M, Takagi S, Komagata S, Yokoi T. MicroRNA regulates human vitamin D receptor. Int J Cancer. 125(6):1328–33. [DOI] [PubMed]
- 47.Semba RD, Varadhan R, Bartali B, Ferrucci L, Ricks MO, Blaum C, Fried LP. Low serum carotenoids and development of severe walking disability among older women living in the community: the Women’s Health and Aging Study I. Age Ageing. 2007;36(1):62–67. doi: 10.1093/ageing/afl122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bruyère O, Cavalier E, Buckinx F, Reginster J-Y. Relevance of vitamin D in the pathogenesis and therapy of frailty. Curr Opin Clin Nutr Metab Care. 2017;20(1) Available from: https://journals.lww.com/co-clinicalnutrition/Fulltext/2017/01000/Relevance_of_vitamin_D_in_the_pathogenesis_and.6.aspx. [DOI] [PubMed]
- 49.Costello LC, Franklin RB. Plasma citrate homeostasis: how it is regulated; and its physiological and clinical implications. an important, but neglected, relationship in medicine. HSOA J Hum Endocrinol. 2016 [cited 2019 Feb 10]. ;1(1). Available from: http://www.ncbi.nlm.nih.gov/pubmed/28286881 [PMC free article] [PubMed]
- 50.Huang M, Que Y, Shen X. Correlation of the plasma levels of soluble RAGE and endogenous secretory RAGE with oxidative stress in pre-diabetic patients. J Diabetes Complications. 2015;29(3):422–426. doi: 10.1016/j.jdiacomp.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura K, Yamagishi S, Adachi H, Kurita-Nakamura Y, Matsui T, Yoshida T, Sato A, Imaizumi T. Elevation of soluble form of receptor for advanced glycation end products (sRAGE) in diabetic subjects with coronary artery disease. Diabetes Metab Res Rev. 2007;23(5):368–371. doi: 10.1002/dmrr.690. [DOI] [PubMed] [Google Scholar]
- 52.Selvin E, Halushka MK, Rawlings AM, Hoogeveen RC, Ballantyne CM, Coresh J, Astor BC. SRAGE and risk of diabetes, cardiovascular disease, and death. Diabetes. 2013;62(6):2116–2121. doi: 10.2337/db12-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colhoun HM, Betteridge DJ, Durrington P, Hitman G, Neil A, Livingstone S, Charlton-Menys V, Bao W, DeMicco DA, Preston GM, Deshmukh H, Tan K, Fuller JH. Total soluble and endogenous secretory receptor for advanced glycation end products as predictive biomarkers of coronary heart disease risk in patients with type 2 diabetes. Diabetes. 2011;60(September):2379–2385. doi: 10.2337/db11-0291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prasad K. Is there any evidence that AGE/sRAGE is a universal biomarker/risk marker for diseases? Mol Cell Biochem. 2019;451(1):139–144. doi: 10.1007/s11010-018-3400-2. [DOI] [PubMed] [Google Scholar]
- 55.Smedsrud MK, Gravning J, Omland T, Eek C, Mørkrid L, Skulstad H, Aaberge L, Bendz B, Kjekshus J, Edvardsen T. Sensitive cardiac troponins and N-terminal pro-B-type natriuretic peptide in stable coronary artery disease: correlation with left ventricular function as assessed by myocardial strain. Int J Cardiovasc Imaging. 2015;31(5):967–973. doi: 10.1007/s10554-015-0646-6. [DOI] [PubMed] [Google Scholar]
- 56.Daniels LB, Clopton P, DeFilippi CR, Sanchez OA, Bahrami H, Lima JAC, et al. Serial measurement of N-terminal pro-B-type natriuretic peptide and cardiac troponin T for cardiovascular disease risk assessment in the Multi-Ethnic Study of Atherosclerosis (MESA) Am Heart J. 2015;170(6):1170–1183. doi: 10.1016/j.ahj.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kajioka S, Takahashi-Yanaga F, Shahab N, Onimaru M, Matsuda M, Takahashi R, et al. Endogenous cardiac troponin T modulates Ca2+-mediated smooth muscle contraction. Sci Rep. 2012;2:1–7. doi: 10.1038/srep00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seidelmann SB, Vardeny O, Claggett B, Yu B, Shah AM, Ballantyne CM, et al. An NPPB promoter polymorphism associated with elevated N-terminal pro-b-type natriuretic peptide and lower blood pressure, hypertension, and mortality. J Am Heart Assoc. 2017;6(4). 10.1161/JAHA.116.005257. [DOI] [PMC free article] [PubMed]
- 59.Rattray NJW, Trivedi DK, Xu Y, Chandola T, Johnson CH, Marshall AD, Mekli K, Rattray Z, Tampubolon G, Vanhoutte B, White IR, Wu FCW, Pendleton N, Nazroo J, Goodacre R. Metabolic dysregulation in vitamin E and carnitine shuttle energy mechanisms associate with human frailty. Nat Commun. 2019;10(1):5027. doi: 10.1038/s41467-019-12716-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tegnér JN, Compte A, Auffray C, An G, Cedersund G, Clermont G, Gutkin B, Oltvai ZN, Stephan KE, Thomas R, Villoslada P. Computational disease modeling - fact or fiction? BMC Syst Biol. 2009;3:56. doi: 10.1186/1752-0509-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 8326 kb)
(XLSX 36 kb)
Data Availability Statement
Data will be made freely available