Significance
Immune memory causes the earliest infection by influenza virus to impart a lasting imprint on an individual’s response to later influenza infections or vaccinations. Until recently, that first exposure was always an infection. In the United States and Europe, many infants and toddlers now receive an influenza vaccine before they experience an infection. Because we have little information about immune imprinting by initial exposure to a vaccine, we have studied, in a nonhuman-primate model, how the mode of initial exposure affects primary and secondary antibody responses. Vaccination with influenza hemaglutinnin (HA) protein and infection with a strain bearing the same HA appear to leave distinct imprints—a difference, if confirmed in humans, relevant for next-generation influenza vaccine design.
Keywords: influenza virus hemagglutinin, immune memory, antibodies, vaccines
Abstract
Immune memory of a first infection with influenza virus establishes a lasting imprint. Recall of that memory dominates the response to later infections or vaccinations by antigenically drifted strains. Early childhood immunization before infection may leave an imprint with different characteristics. We report here a comparison of imprinting by vaccination and infection in a small cohort of nonhuman primates (NHPs). We assayed serum antibody responses for binding with hemaglutinnins (HAs) both from the infecting or immunizing strain (H3 A/Aichi 02/1968) and from strains representing later H3 antigenic clusters (“forward breadth”) and examined the effects of defined HA mutations on serum titers. Initial exposure by infection elicited strong HA-binding and neutralizing serum antibody responses but with little forward breadth; initial vaccination with HA from the same strain elicited a weaker response with little neutralizing activity but considerable breadth of binding, not only for later H3 HAs but also for HA of the 2009 H1 new pandemic virus. Memory imprinted by infection, reflected in the response to two immunizing boosts, was largely restricted (as in humans) to the outward-facing HA surface, the principal region of historical variation. Memory imprinted by immunization showed exposure to more widely distributed epitopes, including sites that have not varied during evolution of the H3 HA but that yield nonneutralizing responses. The mode of initial exposure thus affects both the strength of the response and the breadth of the imprint; design of next-generation vaccines will need to take the differences into account.
Antigenic exposures determine the acquisition and development of adaptive immunity. The humoral response in a naive individual yields both antibody-secreting plasma cells that recognize the new antigen and memory B cells that can respond to future encounters with related antigens. The combination of these two components can confer long-lasting protection against antigenically stable pathogens. For antigenically diverse pathogens and those that evolve to evade immune pressure (e.g., influenza virus and HIV), serum responses often confer incomplete immunity to future variants (1, 2).
The hemagglutinin (HA) and neuraminidase (NA) define the serotype of an influenza virus isolate (3). Antigenic shifts occur when novel animal influenza viruses can transmit to humans, spread rapidly, and initiate pandemics, owing to absence of any preexisting immunity (4, 5). Historically, the descendants of pandemic viruses have become endemic seasonal variants that undergo antigenic drift as they evolve over time to evade dominant human herd immunity (6, 7). For most adults, both processes have shaped human immunity to influenza.
Immune memory causes a primary infection to impart an enduring imprint (8–11). Despite a lifetime of repeated exposures to divergent influenza viruses, the relative strength of an individual’s immune response to vaccination or infection correlates with the antigenic similarity of the vaccine or infecting strain to that person’s initial exposure. Until recently, the first encounter was invariably an infection. Because of recent changes in vaccine policy in the United States and Europe, infants and toddlers are now encouraged to receive influenza vaccines before they experience an influenza infection (12, 13). We have little information, however, about the immunological memory to influenza virus established when the primary exposure is vaccination rather than infection.
Using nonhuman primates (rhesus macaques) as a model, we have examined how the mode of influenza exposure affects both primary and secondary antibody responses. We found that an initial exposure by infection elicited strong, strain-specific, HA-binding, and neutralizing serum antibody responses. Initial exposure by immunization with the HA protein from the strain used in the infection arm of the study elicited relatively weaker HA-binding responses that lacked neutralization potency but had greater interseasonal forward breadth. Subsequent exposures, by immunization, generated antibodies with increased interseasonal breadth in infected animals and neutralizing activity in the initially immunized monkeys. Initially infected macaques maintained responses that were strongly imprinted by the infecting strain, while those initially immunized with protein retained a serum repertoire that cross-reacted with heterologous HAs. Moreover, the distribution of epitopes bound by serum IgG was different in the two cases. These data suggest that the mode of HA exposure influences its immune imprint and that next-generation vaccine design will need to take this influence into account.
Results
Infection and Immunization Protocol.
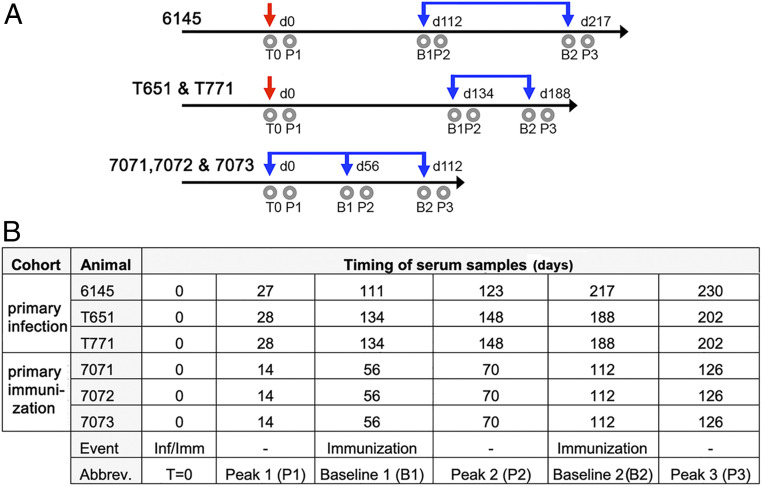
Six adult, rhesus macaques were divided into two cohorts of three animals each (Fig. 1 and SI Appendix, Table S1). We infected one cohort (the “infection cohort”) with A/Aichi/02/1968(H3N2) (X31) (H3-HK-1968), with one animal 6145 infected before T651 and T671 to ensure the safety and efficacy of the infection protocol. The other, “immunization” cohort received recombinant HA protein (rHA) ectodomain from H3-HK-1968 mixed with an oil-in-water adjuvant containing TLR7/8 agonist R848 and TLR9 agonist CpG oligonucleotides (14) (animals 7071, 7072, and 7073). This HA trimer is similar to those delivered in recombinant flu vaccines and differs from classical “split” vaccines that are extracted from virions and retain a membrane spanning region. After periods of rest, the animals in both cohorts were twice boosted with this same adjuvanted rHA (Fig. 1). We obtained blood samples at specific time points throughout the study.
Fig. 1.
Study timeline and overview. (A) The timing of infections (red downward arrows), immunizations (blue downward arrows), and blood draws (gray circles) are shown along a timeline. The days, denoted with a “d,” beginning on the day of immunization or infection (d0), are indicated above each line. The animals on each schedule are indicated. The time points to which each blood draw corresponds in subsequent analyses are indicated and abbreviated as follows: time 0 (T0), peak 1 (p1), baseline 1 (b1), peak 2 (p2), baseline 2 (b2), and peak 3 (p3). (B) Tabulated version of the timeline in A.
We used the immune sera to follow the development and dynamics of the HA-directed humoral immune response. We chose HA of the H3-HK-1968 isolate as the antigen in order to assay the resulting immune sera against HA variants that had subsequently arisen during more than 35 y of antigenic drift in humans (SI Appendix, Fig. S1). From 1968 to 2005, 73 amino acid substitutions were acquired in the HA ectodomain, altering 14% of its amino acid sequence. We produced a panel of 11 H3N2 rHAs that span the defined antigenic clusters of this interval (SI Appendix, Fig. S1) (15). Our panel also included H1, H18, and influenza B rHAs and matched H1 and influenza B viruses.
Primary Infection and Immunization Elicit Distinct Serum Responses.
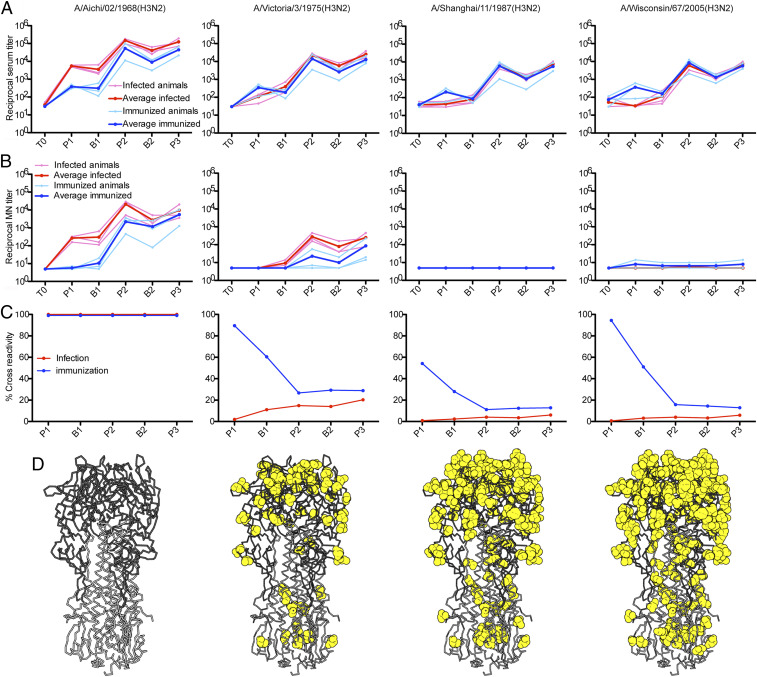
Intracohort variability was low, and the immune signatures imparted by infection and immunization were distinct. In the primary response, all three animals in the infection cohort produced strong, HA-binding antibody responses to H3-HK-1968, with a mean enzyme-linked immunosorbent assay (ELISA) titer at the peak of the primary serum response (Fig. 2A, peak 1) more than one log higher than that of the animals in the immunization cohort. Animals in the infection cohort also had measurable neutralizing titers to the 1968 strain (Fig. 2B, peak 1), while animals in the immunization cohort had neutralizing titers at or near the limit of detection. For neither group could we detect measurable neutralizing titers to H3-VI-1975 and later isolates, indicating that the substitutions acquired over a period of 7 y of antigenic drift were sufficient to evade the neutralizing antibodies elicited by either route (Fig. 1B and SI Appendix, Fig. S3).
Fig. 2.
Serologic profiles of infected and immunized cohorts. (A) Reactivities of the macaque sera to the indicated HA proteins were measured by ELISA and the reciprocal serum titers are graphed. All ELISA data are in SI Appendix, Fig. S2. (B) Neutralization of selected influenza viruses. Macaque sera were assayed for neutralizing activity by microneutralization assay, and the reciprocal neutralizing titers graphed. All microneutralization data are in SI Appendix, Fig. S3. (C) Percent cross-reactivity of macaque serum. Using the averaged ELISA titers for each cohort, we calculated the percentage of the HA directed serum response that reacted with heterologous HAs [(heterologous titer ÷ H3-HK-1968 titer)*100]. All percent cross-reactivity data are in SI Appendix, Fig. S4. For each panel, the time points on the x axis correspond to Fig. 1 and are as follows: time 0 (T0), peak 1 (p1), baseline 1 (b1), peak 2 (p2), baseline 2 (b2), and peak 3 (p3). (D) Differences from A/Aichi/02/1968(H3N2), mapped to the HA structure (Protein Data Bank [PDB] 2VIU), are shown as yellow spheres. The HA head domain is in dark gray; the stem, in light gray.
Despite lower HA-binding titers to HA-HK-1968, animals in the immunization cohort mounted responses with greater forward, interseasonal breadth (Fig. 2A and SI Appendix, Fig. S2, peak 1). Serum titers for drifted H3 isolates, covering 37 y of antigenic drift, were similar to those for H3-HK-1968 and were higher than the corresponding titers from the animals in the infection cohort. Serum reactivity from infected animals declined rapidly with increased drift (Fig. 2A and SI Appendix, Fig. S2). This sensitivity to antigenic variation in the infection cohort was evident even in the mean titer for the least drifted isolate, H3-PC-1973, which was 17-fold lower than the titer for H3-HK-1968 (SI Appendix, Fig. S2). At the peak of the primary response, HA-binding titers for HAs from viruses isolated after 1982 were at or near the limit of detection.
Further Immunizations Elicit Strong Recall Responses.
We subsequently immunized both cohorts with rHA H3-HK-1968, after a period of rest, to examine recall from immunologic memory. We used the homologous HA to maximize recall response. For both cohorts, peak serum HA-binding titers (peak 2) to H3-HK-1968 exceeded those from the primary exposure (Fig. 2 and SI Appendix, Figs. S2 and S3). The boost was stronger in animals of the immunization cohort, with secondary peak titers that were over 100 times greater than the primary peak titers (Fig. 2A). Serum titers of the infection cohort animals were boosted by about 25-fold. The secondary immunization also increased neutralizing titers in both groups, particularly in the immunization cohort, with neutralizing titers increasing from near the limit of detection at the primary peak by over 200-fold at the secondary peak, exceeding the peak titers in the infection cohort during the primary response (Fig. 2B). The latter cohort also mounted a strong boost in neutralizing titers above the primary peak, reaching secondary titers 10-fold higher than those for the immunization cohort.
For all six monkeys, the secondary immunization increased the overall interseasonal breadth and the magnitude of ELISA titers to heterologous HAs (Fig. 2A and SI Appendix, Fig. S2). The relative broadening of serum reactivity was greater in the infection cohort, which had shown little forward breadth in the primary response. For example, these monkeys had detectable HA-binding titers to the most drifted variant, H3-WI-2005. Animals in the infection cohort also gained modest serum reactivity to H1-CA-09 (SI Appendix, Fig. S2). These animals developed neutralizing titers to H3-VI-1975 that were similar to the primary HA-HK-1968 peak neutralizing titers and had weak neutralizing activity against H3-PH-1982 (SI Appendix, Fig. S3). In the immunization cohort, peak reactivity to H3-WI-2005 increased by about 25-fold, and reactivity to H1-CA-09 also increased modestly. Only one of the three animals developed neutralizing titers to H3-VI-1975 (Fig. 2 and SI Appendix, Fig. S3)
Because the time interval between primary and secondary exposures varied between groups, we boosted the animals again with rHA from H3-HK-1968 a second time, roughly 7 wk after their secondary exposure. The response to the tertiary exposure was similar to that of the secondary response (Fig. 2 and SI Appendix, Figs. S2 and S3). The timing of immunization does not appear to influence the characteristics of resulting recall response.
Infection Imparts a Strain-Biased Immune Imprint.
We plotted, for both cohorts, the percentage of the serum titer that cross-reacts with heterologous HAs (heterologous HA titer divided by the corresponding H3-HK-1968 titer and multiplied by 100). In the infection cohort, the serum repertoire had little cross-reactivity with heterologous HAs (Fig. 2C and SI Appendix, Fig. S4). Boosting immunizations led to marginal increases. The immunization cohort produced and maintained a higher proportion of cross-reactive serum antibodies than the infection cohort. Repeated boosting immunizations in the immunization cohort reduced the percentage of cross-reactive serum antibodies, but cross-reactivity still remained severalfold higher than the infection cohort (Fig. 2C and SI Appendix, Fig. S4). Maintenance, for the duration of the study, of strain specificity in the infection cohort and cross-reactivity in the immunization cohort suggests that a primary exposure may impart an enduring immune imprint with distinct characteristics that depend on its mode.
Infection Focuses Antibody Responses to the Variable Periphery of the Receptor Binding Site (RBS).
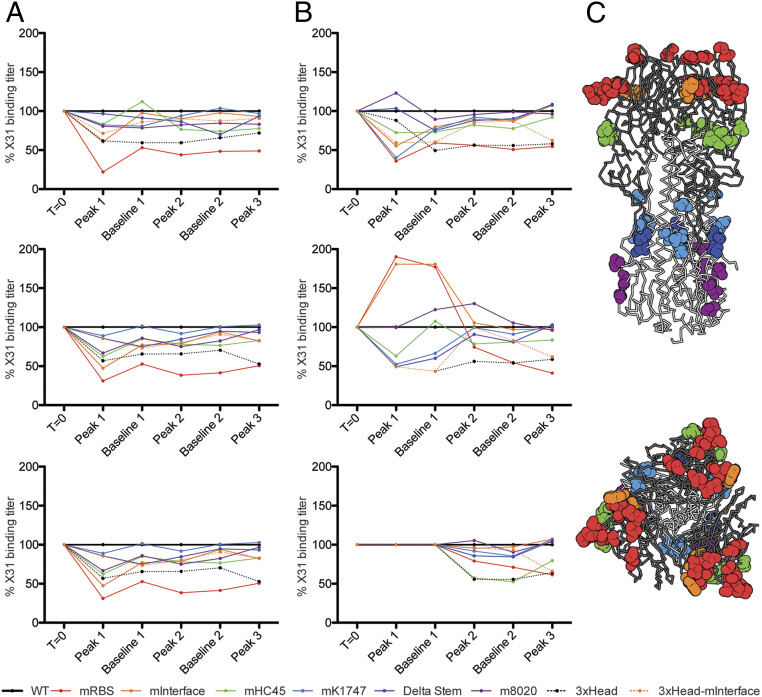
Sera from the two cohorts differ in neutralization potency, strain specificity, and interseasonal breadth of HA binding. Does the route of influenza exposure also influence the distribution of targeted HA epitopes, as we might expect from their differential exposure on virions and free HA trimer? We produced a panel of six variants of the 1968 Hong Kong (X31) HA, each with a set of mutations designed to disrupt the epitope of a particular, well-characterized antibody (16, 17–23) (SI Appendix, Fig. S5), and validated the designs by assessing binding with the corresponding set of antibodies (SI Appendix, Table S2). We then used this panel, augmented by trimeric, head-only constructs, to probe serum reactivity for each of the animals and time points in the study.
Sera from all three animals in the infection arm yielded similar reactivity patterns. The variant with mutations encircling the RBS (X31-mRBS) showed the greatest loss of binding with respect to the wild-type HA; the head-interface variant (X31-mInterface) showed a smaller effect (Fig. 3 and SI Appendix, Fig. S6). By contrast, the three animals in the immunization arm appeared to have mounted more varied responses with respect to epitope distribution. We found reduced binding to X31-mRBS only for sera drawn after the secondary and ternary immunizations—i.e., the time points at which neutralizing titers became comparable to those following infection and at which strain specificity became more pronounced (Figs. 2B and 3 and SI Appendix, Fig. S6). For all six animals, higher neutralizing titers and strain specificity correlated with greater loss of affinity for the X31-mRBS HA. We conclude that primary infection in these animals focused the B cell immune response on epitopes in and around the RBS, leaving an imprint that persisted even after repeated subsequent vaccinations, while primary immunization with HA trimer elicited antibodies with a wider epitope distribution. Correlation of neutralization and X31-mRBS sensitivity is consistent with the low neutralizing activity of stem- and head interface-directed antibodies.
Fig. 3.
Route of exposure influences the distribution of epitopes bound by serum antibodies. We produced a panel of seven HA variants each with mutations that disrupt a single epitopic region, and trimeric HA head domains (3xHead) (SI Appendix, Fig. S5). Using ELISA ED50s (SI Appendix, Fig. S6), we calculated and graphed the percent serum binding compared to wild-type H3-HK-1968. (A) The infected cohort. (B) The immunized cohort. For each graph panel, the time points on the x axis correspond to Fig. 1 and are as follows: time 0 (T0), peak 1 (p1), baseline 1 (b1), peak 2 (p2), baseline 2 (b2), and peak 3 (p3). (C) Locations of mutations mapped to the structure of the HA trimer (PDB 2VIU). Sites that were mutated are shown in spheres that are colored to match the key for the graphs. The HA head domain is shown in dark gray, and the stem domain is shown in light gray. (Top) A side view of the molecule. (Bottom) A top view of HA looking down upon the apex of the molecule and the receptor binding domain.
Discussion
Influenza infection presents the classic example of immune imprinting, the basis of a phenomenon that T. Francis called “original antigenic sin” (11). Recall of B cell memory established during an initial exposure—and in some cases further affinity maturation—appears to dominate any fresh, naive response to subsequent exposure to a drifted strain. Many studies document the immune imprints left by early influenza exposures in adolescent and adult humans, all of whom were likely to have first experienced influenza by infection (1, 2, 24). Because childhood vaccination did not become widely encouraged in the United States until around the year 2000, we lack comparable molecular analyses of the immune imprint left by immunization (12, 13). The results reported here are an attempt to compare imprinting by infection and vaccination in a nonhuman primate model. In this initial study, we used a small cohort of rhesus macaques; larger studies will be necessary to add statistical significance to the trends we describe.
After a first infection, the serum binding titer for the homologous HA rose more than two logs from the naive, preinfection titer in all three animals, declined slightly over the ensuing 3–4 mo, and rose to almost four logs over preinfection after just a single boosting immunization. A second boost merely compensated for the waning titer during the interval after the first. Vaccination produced a much more modest initial response than infection, as expected for vaccinations in general, but a single boost brought the binding titer to within about one log of the infection-induced level. Neutralization titers for the homologous virus followed similar patterns, but there was essentially no measurable neutralizing response to vaccination until the first boost, and only after the second boost did the mean titer for the three animals in the initial-vaccination arm approach the mean for the three initially infected ones.
We tested the forward breadth of the response to antigenically drifted H3 variants with a panel of strains that sampled the successive antigenic clusters since the H3 HA entered human circulation in 1968. The pattern of serum titers following infection in these macaques recapitulates the observed evolution, in response to herd immunity, of the H3 HA. The A/Port Chalmers/1/1973 strain represents the first new antigenic cluster to appear in the years following 1968, and the A/Victoria/3/1975 represents the second (15). Sera from all three infected animals bound weakly to HA from the former isolate and barely at all to HA from the latter. Binding to the 1987 isolate in our panel was essentially undetectable. Therefore, the mutations in HA that defined (historically) the antigenic cluster containing A/Port Chalmers/1/1973 and that presumably were selected because they resisted neutralization by prevalent antibodies, would likewise have produced resistance to the initial response of the rhesus macaques in our study. That is, the B cell arm of the rhesus macaque immune system appears to have “seen” the 1968 HA in the same way as did the B cell arm of the population-averaged human immune system from 1968 to 2005.
The interseasonal breadth of response to monovalent vaccination with recombinant HA followed a very different pattern. Binding to all heterologous HAs in our panel of antibodies from animals in the initially vaccinated group was nearly the same as it was for the homologous antigen; that breadth even included HA from the 2009 H1 new pandemic strain. Boosting the initially infected animals by vaccination with recombinant protein brought the breadth of response in that arm into concordance with its level in the initially vaccinated arm. Thus, previous exposure by infection and consequent circulating antibody levels did not appear to have any effect measurable by this assay on the outcome of the boost. Childhood imprinting by infection in humans likewise appears not to affect the strength of the serum response to a first vaccination later (25)
Unlike the binding titer, the neutralizing response in both arms was quite narrow. Despite their substantial neutralizing response to the homologous, 1968 strain, the infected animals had essentially no neutralizing titer even for HA of A/Victoria/3/1975, only two antigenic clusters removed from HA of the 1968 virus (15). Boosting with protein produced a modest neutralizing titer for the A/Victoria HA in the infected animals and in one of the vaccinated animals, but almost no detectable interseasonal breadth beyond the 7-y interval separating the 1975 isolate from A/Aichi/02/1968.
Can differential response to particular epitopes account for the difference between binding and neutralization for the drifted strains? The most likely explanation for the disparity, consistent with our data on binding to mutated HAs, is that during infection, when virions are the principal immunogens, selective exposure of the outward-facing surface of the HA head focuses the response on the well-studied epitopes, targets of neutralizing antibodies, close to the RBS (26). Indeed, previous studies of the contributions of specific mutations to the appearance of a new antigenic cluster suggest that just one or a few key changes, especially in the periphery of the RBS, can account for the drift (15, 27, 28). The dense packing of HA on a virion or at sites of viral budding occludes many of the HA surfaces. The rHA immunogen we administered in this study was a soluble trimer and similar to the immunogen in recombinant vaccines. Classical, “split” flu vaccines deliver virion-extracted HA in heterogenous, membrane-like contexts, including micellar “rosettes,” all of which dilute the HA and probably create much greater access to stem and interface epitopes than on virions (29). Unlike virion-bound HA, vaccination exposes epitopes distributed across the protein, including conserved surfaces on the stem and possibly at the head interface. Antibodies directed at these epitopes do not neutralize as defined by our single infectious cycle assay, although they protect mice by Fc-mediated mechanisms (19, 20, 30, 31).
If confirmed by more extensive studies in humans, our observations have implications for the design, deployment, and administration policy for improved influenza vaccines. The more potently neutralizing sera following infection probably focused on the antigenically variable, RBS periphery; the broader but nonneutralizing responses following vaccination focused on more conserved epitopes elsewhere on HA. Although the latter will not impart sterilizing immunity, because they rely for protection on Fc-mediated mechanisms that clear already infected cells (19, 20, 30, 31), they could reduce the severity of the infection and thus its pathogenic consequences. Judicious choices, for improved influenza immunogens, of timing and route of exposure might imprint on naive recipients a favorable balance of these qualities and boost them in experienced vaccinees.
Methods
Animal Studies.
Six adult rhesus macaques (Macaca mulatta) were housed at BioQual and maintained in accordance with the Association for Accreditation of Laboratory Animal Care guidelines at the NIH. Three animals were infected with H3N2 A/Aichi/2/1968 influenza virus at 2.36 × 106 plaque-forming units per animal in a divided dose; for each animal, half of the infectious dose was given in 1 mL intranasally and the other half as 1 mL intratracheally. Animals were monitored for signs of infection and no intervention was needed. For all immunizations, animals were immunized intramuscularly with 100 μg of H3 A/Aichi/2/1968 recombinant protein per animal per time point given as 500-μL total injection volume divided into two sites—250 μL each at right and left quadriceps. The final immunization mixture contained 15% STS+R848+oCpG adjuvant (same reference as above) (14) with the remaining volume being sterile saline. Blood was collected in EDTA tubes per the schedule shown in Fig. 1 and processed for plasma and cells, which were cryopreserved.
Serological Assays.
ELISAs used recombinant HA proteins as described (32) modified for detecting rhesus antibodies. Briefly, high-binding 384-well microtiter plates were coated with recombinant HA protein at a final concentration of 2 µg/mL diluted in 0.1 M NaHCO3 and incubated at 4 °C overnight. The plates were washed with 1× phosphate-buffered saline (PBS) containing 0.1% Tween 20 and blocked for 1 h using block buffer (40 g of whey protein, 150 mL of goat serum, 5 mL of Tween 20, 0.5 g of NaN3, 40 mL of 25× PBS, brought up to 1 L with water). Plates were washed, and 10 µL of diluted plasma (starting at 1:30 and serially diluted in block buffer) was directly added to each well and incubated for 1.5 h. Plates were washed and 10 µL of horseradish peroxidase (HRP)-conjugated secondary antibody goat anti-monkey IgG heavy- and light-chain specific antibody (Jackson Immunoresearch) diluted in blocking buffer without NaN3 at a 1:12,000 dilution was added and incubated for 1 h. The plates were washed and developed with 3′,5-tetramethylbenzidine substrate (KPL), plates for 15 min. Development was stopped using 1% HCl (Fisher Scientific). Plates were read on a plate reader (Molecular Devices) at 450 nm. The background for each analyte was determined based on nonimmune plasma. Midpoint (ED50) titers were calculated by applying four-parameter logistic regression to the binding data using the drc package in R (33).
Virus neutralization endpoint titers were determined using the influenza microneutralization assay as described (34–38). Serum samples were heat inactivated for 60 min at 56 °C and diluted 1:10 in virus diluent. Each sample was then diluted twofold in virus diluent yielding a range of 1:10 to 1:1,280 in a flat-bottomed 96-well tissue culture plate. Samples were then mixed equimolar with virus diluent containing 100 TCID50 of each influenza virus of interest. Control samples included known antisera and naive sera treated in exactly the same manner as experimental samples. After virus addition, samples are incubated for 60 min at 37 °C, 5% CO2. The 1.5e4 MDCK cells (London strain; IRR FR-58) were added to each well. Plates were incubated overnight at 37 °C, 5% CO2. Each well was aspirated, and cells were washed one time with PBS. The PBS was aspirated. Then, 250 µL of −20 °C, 80% acetone was added to each well, and plates were incubated at room temperature for 10 min. The acetone was removed and plates air-dried. Each well was washed three times with wash buffer. Primary antibody (Mouse Anti-Influenza A NP, Millipore MAB8251; or Mouse Anti-Influenza B NP, Millipore MAB8661) was diluted 1:4,000 in antibody diluent, and 50 µL was added to every well. Plates were incubated for 60 min at room temperature. Each well was washed three times with wash buffer. Secondary antibody (goat anti-mouse + horseradish peroxidase; KPL 474-1802) was diluted 1:4,000 in antibody diluent, and 50 µL was added to every well. Plates were incubated for 60 min at room temperature. Each well was washed five times with wash buffer. One hundred microliters of substrate was added to every well and incubated at room temperature. The reaction was stopped with 100 µL of 0.5 N sulfuric acid after apparent color change was observed in virus-only control wells. Absorbance was read at 490 nM in an Synergy H1 automated microplate reader (BioTek Instruments). Wells with absorbance values less than or equal to 50% of virus-only control wells were scored as neutralization positive. Data were expressed as the geometric mean of the reciprocal of the final dilution factor that was positive for neutralization. All samples were assayed in at least duplicates.
Influenza viruses were propagated in embryonated specific-pathogen–free chicken hen eggs or MDCK(CCL-34) cells as described (38). Reagents obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), NIH, include the following: influenza A viruses A/Aichi/2/1968 (H3N2) NR-3177; Kilbourne F123: A/Victoria/3/1975 (HA, NA) × A/Puerto Rico/8/1934 (H3N2), Reassortant X-47 NR-3663; Kilbourne F118: A/Port Chalmers/1/1973 (H3N2) × A/Puerto Rico/8/1934, Reassortant X-41 NR-3575; Kilbourne F148: A/Texas/1/1977 × A/Puerto Rico/8/1934, Reassortant X-61 NR-3633; Kilbourne F156: A/Bangkok/1/1979 (H3N3) × A/Puerto Rico/8/1934, Reassortant X-73 NR-3515; Kilbourne F73: A/Beijing/353/1989 (H3N2) × A/Puerto Rico/8/1934, Reassortant X-109 NR-3508; A/Philippines/2/1982 (H3N2) NR-28649; Kilbourne F178: A/Shanghai/11/1987 (HA, NA) × A/Puerto Rico/8/1934 (H3N2), High Yield, Reassortant X-99a NR-3505; Kilbourne F86: A/Johannesburg/33/1994 (HA, NA) × A/Puerto Rico/8/1934 (H3N2), Reassortant X-123a NR-3580; Kilbourne F97: A/Moscow/10/1999 (HA, NA) × A/Puerto Rico/8/1934 (H3N2), Reassortant X-137 NR-3587; A/California/04/2009 (H1N1)pdm09, cell isolate (produced in eggs) NR-13659; polyclonal influenza virus, A/Aichi/2/1968 (H3N2) serum (guinea pig), NR-3126; NR-4282. Influenza A virus A/Wisconsin/67/2005 (H3N2), FR-397; influenza B virus B/Phuket/3073/2013 FR-1364, and MDCK London cells (FR-58) were obtained through the International Reagent Resource (formerly the Influenza Reagent Resource), Influenza Division, World Health Organization Collaborating Center for Surveillance, Epidemiology and Control of Influenza, Centers for Disease Control and Prevention.
Cell Lines.
Human 293F cells were maintained at 37 °C with 5% CO2 in FreeStyle 293 Expression Medium (Thermo Fisher) supplemented with penicillin and streptomycin. High Five Cells (BTI-TN-5B1-4) (Trichoplusia ni) were maintained at 28 °C in EX-CELL 405 medium (Sigma) supplemented with penicillin and streptomycin.
Recombinant HA Expression and Purification.
All wild-type recombinant (HA) constructs were expressed by infection of insect cells with recombinant baculovirus as previously described. In brief, synthetic DNA corresponding to the full-length ectodomain were subcloned into a pFastBac vector modified to encode a C-terminal thrombin cleavage site, a T4 fibritin (foldon) trimerization tag, and a 6×His tag (19, 39). The resulting baculoviruses produce HA-trimers. Supernatant from recombinant baculovirus infected High Five Cells (Trichoplusia ni) was harvested 72 h postinfection and clarified by centrifugation. Proteins were purified by adsorption to cobalt-nitrilotriacetic acid (Co-NTA) agarose resin (Clontech), followed by a wash in 10 mM Tris⋅HCl, 150 mM NaCl at pH 7.5 (buffer A), elution in buffer A plus 350 mM imidazole (pH 8) and gel filtration chromatography on a Superdex 200 column (GE Healthcare) in buffer A.
HA proteins for immunization were prepared in buffers made with endotoxin-free water (HyClone HyPure Cell Culture Grade Water; SH30539.03). Following gel filtration, the tags were removed using thrombin protease (Thrombin CleanCleave Kit; Sigma; catalog #RECOMT-1KT) and the protein repurified on Co-NTA agarose to remove the protease, tag, and uncleaved protein. The protein was concentrated and buffer exchanged into Endotoxin-Free Dulbecco’s PBS (EMD Milipore; TMS-012-A) and further purified using gel filtration chromatography on Superdex 200 (GE Healthcare) in PBS to further remove impurities. Endotoxins were removed using Pierce High Capacity Endotoxin Removal Spin Columns (catalog #88274). Proteins were then concentrated and sterile filtered.
All mutant HAs were produced from synthetic DNAs that corresponded to the to the full-length ectodomain (FLsE) or the globular HA-head. These were cloned into a pVRC vector that has been modified to encode a C-terminal thrombin cleavage site, a T4 fibritin (foldon) trimerization tag, and a 6×His tag. The resulting proteins are rHA trimers and trimeric HA heads. All were produced by polyethylenimine-facilitated, transient transfection of 293F cells that were maintained in FreeStyle 293 Expression Medium. Transfection complexes were prepared in Opti-MEM and added to cells. Supernatants were harvested 4–5 d posttransfection and clarified by low-speed centrifugation. Subsequent purification was then identical to those for wild-type insect cell-produced HAs.
Recombinant IgG Expression and Purification.
The heavy chain variable domains of selected antibodies were cloned into a modified pVRC8400 expression vector to produce a full-length human IgG1 heavy chain (39). IgGs were produced by transient transfection of 293F cells as specified above. Five days posttransfection, supernatants were harvested, clarified by low-speed centrifugation, and incubated overnight with Protein A Agarose Resin (GoldBio). The resin was collected in a chromatography column, washed with a column volume buffer A, and eluted in 0.1 M glycine (pH 2.5), which was immediately neutralized by 1 M Tris(hydroxymethyl)aminomethane (pH 8). Antibodies were then dialyzed against PBS, pH 7.4.
Supplementary Material
Acknowledgments
We acknowledge Tori Duback for assistance in recombinant protein production and purification and Lindsey Robinson-McCarthy for contributions in drafting this manuscript. Influenza virus neutralization assays were performed in the Duke Regional Biocontainment Laboratory, which received partial support for construction from the NIH/NIAID (Grant UC6AI058607 to G.D.S.). The research was supported by NIH/NIAID Grant P01 AI089618 (to S.C.H.). S.C.H. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: L.L.S. and S.H. are co-authors of a research paper published in 2018.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026752118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Monto A. S., Malosh R. E., Petrie J. G., Martin E. T., The doctrine of original antigenic sin: Separating good from evil. J. Infect. Dis. 215, 1782–1788 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobey S., Hensley S. E., Immune history and influenza virus susceptibility. Curr. Opin. Virol. 22, 105–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Air G. M., Influenza virus antigenicity and broadly neutralizing epitopes. Curr. Opin. Virol. 11, 113–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon S. W., Webby R. J., Webster R. G., Evolution and ecology of influenza A viruses. Curr. Top. Microbiol. Immunol. 385, 359–375 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Kim H., Webster R. G., Webby R. J., Influenza virus: Dealing with a drifting and shifting pathogen. Viral Immunol. 31, 174–183 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Kilbourne E. D., Influenza pandemics of the 20th century. Emerg. Infect. Dis. 12, 9–14 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knossow M., Skehel J. J., Variation and infectivity neutralization in influenza. Immunology 119, 1–7 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gostic K. M., Ambrose M., Worobey M., Lloyd-Smith J. O., Potent protection against H5N1 and H7N9 influenza via childhood hemagglutinin imprinting. Science 354, 722–726 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viboud C., Epstein S. L., First flu is forever. Science 354, 706–707 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Schmidt A. G., et al., Immunogenic stimulus for germline precursors of antibodies that engage the influenza hemagglutinin receptor-binding site. Cell Rep. 13, 2842–2850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francis T., On the doctrine of original antigenic sin. Proc. Am. Philos. Soc. 104, 572–578 (1960). [Google Scholar]
- 12.Centers for Disease Control and Prevention , Childhood influenza-vaccination coverage—United States, 2002–03 influenza season. MMWR Morb. Mortal. Wkly. Rep. 53, 863–866 (2004). [PubMed] [Google Scholar]
- 13.Harper S. A., Fukuda K., Uyeki T. M., Cox N. J., Bridges C. B.; Centers for Disease Control and Prevention Advisory Committee on Immunization Practices , Prevention and control of influenza: Recommendations of the advisory committee on immunization practices (ACIP). MMWR Recomm. Rep. 53, 1–40 (2004). [PubMed] [Google Scholar]
- 14.Moody M. A., et al., Toll-like receptor 7/8 (TLR7/8) and TLR9 agonists cooperate to enhance HIV-1 envelope antibody responses in rhesus macaques. J. Virol. 88, 3329–3339 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bedford T., et al., Integrating influenza antigenic dynamics with molecular evolution. eLife 3, e01914 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bizebard T., et al., Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376, 92–94 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Ekiert D. C., et al., Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fleury D., Wharton S. A., Skehel J. J., Knossow M., Bizebard T., Antigen distortion allows influenza virus to escape neutralization. Nat. Struct. Biol. 5, 119–123 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Watanabe A., et al., Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177, 1124–1135.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bajic G., et al., Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 25, 827–835.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moody M. A., et al., H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 6, e25797 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corti D., et al., A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333, 850–856 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Ekiert D. C., et al., A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333, 843–850 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arevalo P., McLean H. Q., Belongia E. A., Cobey S., Earliest infections predict the age distribution of seasonal influenza A cases. eLife 9, e50060 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurwitz E. S., et al., Studies of the 1996–1997 inactivated influenza vaccine among children attending day care: Immunologic response, protection against infection, and clinical effectiveness. J. Infect. Dis. 182, 1218–1221 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Wu N. C., Wilson I. A., Influenza hemagglutinin structures and antibody recognition. Cold Spring Harb. Perspect. Med. 10, a038778 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonville J. M., et al., Antibody landscapes after influenza virus infection or vaccination. Science 346, 996–1000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith D. J., et al., Mapping the antigenic and genetic evolution of influenza virus. Science 305, 371–376 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Renfrey S., Watts A., Morphological and biochemical characterization of influenza vaccines commercially available in the United Kingdom. Vaccine 12, 747–752 (1994). [DOI] [PubMed] [Google Scholar]
- 30.DiLillo D. J., Tan G. S., Palese P., Ravetch J. V., Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 20, 143–151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bangaru S., et al., A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177, 1136–1152.e18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alam S. M., et al., Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: Antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82, 115–125 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritz C., Streibig J. C., Bioassay analysis using R. J. Stat. Softw. 12, 22 (2005). [Google Scholar]
- 34.CDC , Influenza virus microneutralization assay (Centers for Disease Control, Atlanta, GA, 2007). [Google Scholar]
- 35.CDC , Influenza virus microneutralization assay H1N1 pandemic response (Centers for Disease Control, US Department of Health and Human Services, Atlanta, GA, 2009). [Google Scholar]
- 36.World Health Organization , Serological diagnosis of influenza by microneutralization assay (World Health Organization, Geneva, Switzerland, 2010). [Google Scholar]
- 37.World Health Organization , Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza (World Health Organization, Geneva, 2011). [Google Scholar]
- 38.McCarthy K. R., et al., Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 48, 174–184.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt A. G., et al., Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc. Natl. Acad. Sci. U.S.A. 110, 264–269 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.