Abstract
The kinase complex mTORC2 is widely accepted as controlling phosphorylation of the hydrophobic motif, a key regulatory switch in the C-terminal tail of protein kinase C (PKC), Akt, and other AGC kinases. Yet the biochemical mechanism by which it controls this site and whether mTOR is the direct hydrophobic motif kinase remain controversial. Here we identify a distinct mTOR-mediated phosphorylation site we term the TOR-Interaction Motif (TIM; F-x3-F-pT), which controls hydrophobic motif phosphorylation and activity of PKC and Akt. The TIM is invariant in all mTOR-dependent kinases, is evolutionarily conserved, and co-evolved with mTORC2 components. Mutation of this motif alone in Akt1 (Thr443) or together with the turn motif in PKCβII (Thr634/Thr641) abolishes cellular kinase activity by impairing activation loop and hydrophobic motif phosphorylation. mTORC2 directly phosphorylates the PKC TIM in vitro, and its phosphorylation is detected in mouse brain by mass spectrometry. Overexpression of PDK1 in cells lacking mTORC2 rescues hydrophobic motif phosphorylation of PKC and Akt by a mechanism that depends on their intrinsic catalytic, revealing that mTORC2 facilitates the PDK1 phosphorylation step, which in turn permits autophosphorylation. Analysis of a previously reported PKCβII crystal structure reveals a PKC homodimer driven by a helix containing the TIM. Biophysical proximity assays show that unphosphorylated PKC, but not phosphorylated PKC, dynamically dimerizes in cells. Furthermore, disruption of the dimer interface by stapled peptides promotes hydrophobic motif phosphorylation. Our data support a model in which mTORC2 relieves nascent PKC dimerization through TIM phosphorylation, recruiting PDK1 to phosphorylate the activation loop, and triggering intramolecular hydrophobic motif autophosphorylation. Identification of TIM phosphorylation and its role in the regulation of PKC provides the basis for AGC kinase regulation by mTORC2.
One Sentence Summary:
mTORC2 binds and phosphorylates a conserved TOR-Interaction Motif in the C-tail of PKC and Akt to regulate hydrophobic motif autophosphorylation and kinase activation.
Main Text:
Activation of nearly all eukaryotic protein kinases involves the structuring of an activation loop segment in the kinase domain, most commonly achieved by phosphorylation (1). The activation loop of AGC kinase family members (2, 3), including protein kinase C (PKC) and Akt, is phosphorylated directly by the phosphoinositide-dependent kinase-1, PDK1 (4-6). AGC kinases additionally possess a C-terminal extension of the kinase domain (C-tail) that harbors a conserved phosphorylation site, the hydrophobic motif (7, 8), which regulates their activity (9, 10). The kinase complex mTORC2 regulates the phosphorylation of the C-tail sites for a subset of AGC kinases, including most (but not all) PKC isozymes and Akt, by mechanism that has remained elusive. Given the prevalence of mTOR inhibitor development for a variety of diseases (11), understanding the biochemical mechanisms of how mTORC2 controls AGC kinase function is of critical importance.
For PKC to become catalytically competent, newly-synthesized enzyme is matured by constitutive phosphorylations at three conserved sites (7): the activation loop phosphorylated by PDK1 (6, 12) and two phosphorylations in the C-tail, the turn motif and hydrophobic motif, which are mTORC2-sensitive for all but 3 of the 9 PKC isozymes (13, 14). The activation loop and hydrophobic motif phosphorylations are required for cellular PKC activity (10, 15); whereas, turn motif phosphorylation is not necessary for catalysis, and instead promotes protein stability (16). For Akt, turn motif phosphorylation occurs co-translationally at the ribosome by direct mTORC2 phosphorylation (17). In contrast, phosphorylation of the activation loop and hydrophobic motif sites is agonist-evoked (18), dependent upon the generation of phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2) or phosphatidylinositol (3,4,5)-trisphosphate (PIP3) at cellular membranes (19-22). The Akt hydrophobic motif phosphorylation, distinct from that of PKC, enhances Akt activity (23), but is not required for catalysis (24); rather, activation loop phosphorylation is necessary and sufficient for Akt activity (25). Thus, hydrophobic motif phosphorylation of PKC is constitutive and controls the steady-state levels of PKC, whereas phosphorylation of the corresponding site on Akt is agonist dependent and controls the acute activity, but not stability, of Akt. Phosphorylation of the hydrophobic motif is opposed by the PH domain Leucine rich repeat Protein Phosphatase (PHLPP), with dephosphorylation promoting the down-regulation of PKC and the inactivation of Akt (26). The dysregulation of the hydrophobic motif phosphorylation on both kinases in cancer highlights the importance of understanding the mechanism of phosphorylation.
The current dogma proposes that the atypical kinase complex mTORC2 is the long-sought-after “hydrophobic motif kinase” (27) that directly activates several AGC kinases by phosphorylating the turn motif and hydrophobic motif sites (28, 29). However, cellular and biochemical studies point to autophosphorylation controlling this site. First, the catalytic competence of both PKC and Akt is required for hydrophobic motif phosphorylation in cells (30, 31). Second, autophosphorylation at this site on the purified kinases is triggered in vitro by PDK1 phosphorylation of the activation loop (6, 32). Third, pure PKC selectively dephosphorylated at the hydrophobic motif in vitro re-autophosphorylates at this site by an intramolecular mechanism (6). Additionally, we have previously reported that hydrophobic motif phosphorylation of Akt, but not kinase-inactive Akt, is induced in the absence of mTORC2 by any signal that disengages the PH domain of Akt from its kinase domain, as occurs upon binding to the plasma membrane (33). These results suggest that phosphate incorporation at the hydrophobic motif is governed by a kinase-intrinsic mechanism. The related AGC kinases SGK, PKN, and RSK also depend upon mTORC2, (34-37), which is evolutionarily conserved in yeast (38). Hydrophobic motif phosphorylation of the AGC kinase member S6K depends, instead, upon mTORC1 (39-41). Contributing to the puzzle of understanding mTORC2 regulation, three novel PKCs (δ, θ, and η) are mTOR-independent and do not require mTOR kinase activity for phosphorylation (13). Thus, in the absence of a mechanism for hydrophobic motif phosphorylation, the function of mTORC2 in regulating AGC kinases remains unclear.
Here we identify a distinct AGC kinase phosphorylation site regulated by mTORC2, which we term the TOR-Interaction Motif (TIM), that controls the activation of PKC and Akt by facilitating PDK1 phosphorylation of the activation loop. We find that phosphorylation of the PKC and Akt hydrophobic motif is governed by autophosphorylation in an mTORC2- and PDK1-dependent manner. We detect phosphorylation of the PKCβII TOR-interaction motif (Thr634) by mass spectrometry in mouse brain and show that mTORC2 phosphorylates the TIM site in vitro. TIM phosphorylation is sensitive to mTOR inhibition and mTORC2-deficiency in cells, and mutation of the TIM site abolishes PKC and Akt cellular kinase activity by impairing activation loop and hydrophobic motif phosphorylation. Accordingly, we show that the role of TIM phosphorylation in PKC activation involves conformational rearrangements that promote PDK1 docking to the C-tail. Reanalysis of a PKCβII crystal structure reveals an unappreciated PKC homodimer that is regulated by TIM phosphorylation and mTOR activity. Targeting the TOR-interaction motif dimer interface with stapled peptides is sufficient to enhance PKC phosphorylation in cells. Thus, these data are consistent with a model whereby mTORC2 phosphorylates nascent PKC at the TOR-interaction motif and turn motif sites to relieve dimerization and expose the PDK1 binding site in the C-tail, leading to transphosphorylation of the activation loop and intramolecular autophosphorylation at the hydrophobic motif. Additionally, we show that the TOR-interaction motif consensus sequence (F-x3-F-T) is evolutionarily conserved in AGC kinases from major eukaryotic clades and co-evolved with TOR complex components, suggesting ancient origins for mTORC2-dependent regulation of AGC kinases. In summary, our study reveals the long-elusive role of mTORC2 in PKC regulation: this kinase complex binds and phosphorylates a previously unidentified TOR-interaction motif (TIM) to facilitate the critical binding of PDK1 to the C-terminal tail of newly-synthesized PKC, thus initiating the maturation of the enzyme to the fully-phosphorylated and catalytically competent state.
Results
Defect in PKC Maturation Upon Loss of mTORC2
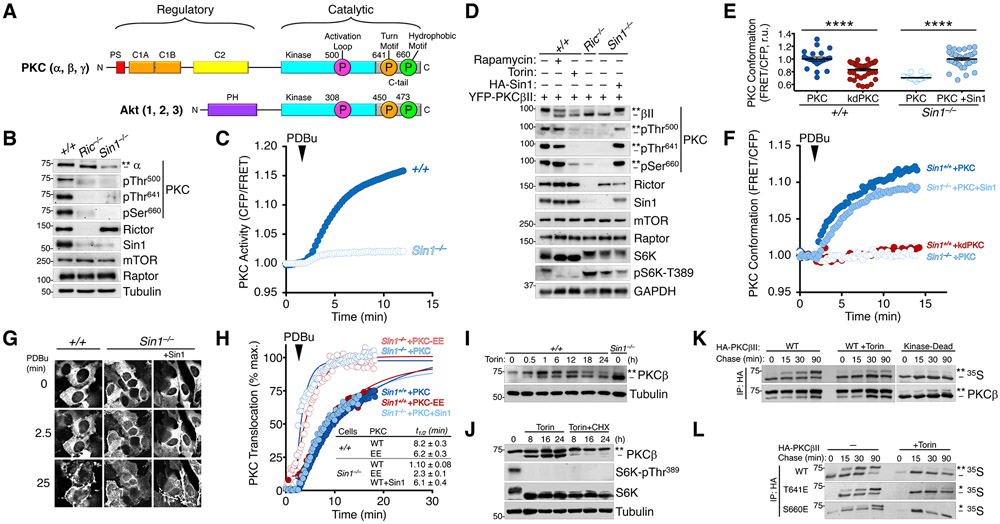
The phosphorylation of most PKC isozymes, including the conventional PKCs (α, β, γ), and all Akt isozymes (Akt 1, 2, 3) (Fig. 1A) depends on mTORC2, which ultimately controls the enzymatic activity and cellular stability of these kinases (13, 14). To assess the nature of the mTORC2 requirement, we characterized PKC function in Sin1 KO (Sin1−/−) and Rictor KO (Ric−/−) mouse-embryonic fibroblasts (MEFs), which lack critical components of the mTORC2 complex and abolish mTORC2 function (42-45). Endogenous PKC phosphorylation at the activation loop, turn motif, and hydrophobic motif sites was ablated in Sin1 KO and Rictor KO MEFs (Fig. 1B). Additionally, the steady-state levels of PKC were reduced, consistent with the unphosphorylated species of PKC being unstable (46). Accompanying the reduced PKC levels, the cellular activity of PKC was reduced in Sin1 KO MEFs compared to WT MEFs as assessed using the C Kinase Activity Reporter (CKAR (47, 48)) (Fig. 1C, S1A). Similarly, mTOR inhibition with the mTORC1/2 inhibitor Torin (49), but not the mTORC1-specific inhibitor Rapamycin, also impaired PKC phosphorylation (Fig. 1D), which could not be rescued by agonist stimulation, targeting to the plasma membrane, or protecting PKC from dephosphorylation with active-site inhibitors (50) (Fig. S1B-E). Thus, mTOR activity and mTORC2 complex integrity are necessary for PKC function, the loss of which results in an unphosphorylated, inactive and unstable enzyme.
Fig. 1. Defect in PKC Maturation Upon Loss of mTORC2.
(A) Schematic of conventional PKC (cPKC) and Akt domain structures. cPKCs (α, β, γ) have an autoinhibitory pseudosubstrate (PS, red), tandem diacylglycerol sensing C1 domains (orange), and a Ca2+-dependent plasma membrane sensing C2 domain (yellow) in their N-terminal regulatory moiety and a kinase domain (cyan) and C-terminal tail (C-tail, grey) in the catalytic moiety. Akt has a PIP3-sensing PH domain and and a kinase domain (cyan) and C-terminal tail (C-tail, grey) in the catalytic moiety. Both kinases have three conserved phosphorylations at the activation loop (magenta) of the kinase domain and the turn motif (orange) and hydrophobic motif (green) in the C-tail, indicated with circles (PKCβII and Akt1 numbering). For PKC, these phosphorylations are constitutive whereas for Akt only the turn motif is constitutive, with the activation loop and hydrophobic motif phosphorylated in an agonist-dependent manner. The C-tail sites are mTORC2-sensitive.
(B) Western blot of Triton-solubilized lysates from WT (+/+), Rictor KO (Ric−/−), or Sin1 KO (Sin1−/−) MEFs probed with the indicated total and phospho-specific antibodies. The double asterisk (**) denotes the position of mature, fully-phosphorylated PKC and the dash (−) indicates the position of unphosphorylated PKC. Note the activation loop does not cause a mobility shift and is modified in C-terminally phosphorylated species. Blots are representative of three independent experiments.
(C) PKC activity in WT (+/+) or Sin1 KO (Sin1−/−) cells expressing the PKC activity reporter, CKAR, and treated with PDBu (200 nM) to maximally activate PKC. Data represent the normalized FRET ratio changes (mean ± SEM) from three independent experiments.
(D) Western blot of Triton-solubilized lysates from WT (+/+), Rictor KO (Ric−/−), or Sin1 KO (Sin1−/−) MEFs, expressing YFP-PKCβII and HA-Sin1, treated with Rapamycin (10 nM; 24 h) or Torin (200 nM; 24 h), and probed with the indicated antibodies. Mobility shifts as described in B. Blots are representative of three independent experiments.
(E) Basal conformation of PKC analyzed using the conformational reporter Kinameleon which comprises a donor:acceptor pair flanking the N and C termini of PKC. Autoinhibited PKC has a low FRET ratio and the open conformation has high FRET ratio. Indicated are the FRET ratio (mean ± SEM) of PKCβII Kinameleon wild-type (WT) or kinase-dead K371R (KD) expressed without or with HA-Sin1 in WT (+/+) or Sin1 KO (Sin1−/−) MEFs. Each data point represents the FRET ratio from an individual cell relative to the average maximum signal in three independent experiments. ****p < 0.0001 by Student’s t-test.
(F) Agonist-induced conformational changes of PKCβII assessed using the reporter Kinameleon. Wild-type (WT) or kinase-dead K371R (KD) Kinameleon was expressed without or with HA-Sin1 in WT (Sin1+/+) or Sin1 KO (Sin1−/−) MEFs and treated with PDBu (200 nM) at the indicated time. Data represent the normalized FRET ratio changes (mean ± SEM) from three independent experiments.
(G) Fluorescence images of WT (+/+) or Sin1 KO (Sin1−/−) MEFs expressing PKCβII Kinameleon alone or with coexpression of HA-Sin1, before (0) or after treatment with PDBu (200 nM) for the indicated timepoints. Images are representative of three independent experiments.
(H) Analysis of plasma membrane translocation of mYFP-PKCβII WT or T641E/S660E (EE) in WT (+/+) or Sin1 KO (−/−) MEFs co-expressing myristoylated-palmitoylated mCFP with or without HA-Sin1, and treated with PDBu (100 nM). Data represent the FRET ratio signal from the CFP to YFP and are normalized to the maximum FRET ratio signal determined by fitting the data to a single phase logarithmic nonlinear regression (solid lines). Data represent the normalized FRET ratio changes (mean ± SEM) from three independent experiments.
(I) Western blot of Triton-solubilized lysates from WT (+/+) MEFs overexpressing PKCβII and treated with Torin (250 nM) for 24 h prior to lysis. Mobility shifts as described in B. Blots are representative of three independent experiments.
(J) Western blot of Triton-solubilized lysates from COS7 cells transfected with cDNA for PKCβII for 24 h prior to treatment with Torin (250 nM) and Cycloheximide (250 μM) for the indicated times. Mobility shifts as described in B. Blots are representative of three independent experiments.
(K) Autoradiogram (detecting 35S-labeled newly-synthesized PKC) and Western blot (detecting total pool of PKC) of HA immunoprecipitates from a pulse-chase analysis of COS7 cells expressing HA-PKCβII WT or K371R (Kinase-Dead) and treated with Torin (250 nM) during the chase. Mobility shifts as described in B. Blots are representative of three independent experiments.
(L) Autoradiogram (detecting 35S-labeled newly-synthesized PKC) and Western blot (detecting total pool of PKC) HA immunoprecipitates from a pulse-chase analysis of WT MEFs expressing the indicated HA-PKCβII constructs and treated with Torin (250 nM) during the chase. The double asterisk (**) denotes the position of mature, fully-phosphorylated PKC; the single asterisk (*) denotes the position of PKC phosphorylated at either the turn motif or hydrophobic motif; and the dash (−) indicates the position of unphosphorylated PKC. Blots are representative of three independent experiments.
To further characterize the PKC defect in the absence of mTORC2, we used a live-cell PKC conformation reporter, Kinameleon, which detects intramolecular rearrangements of functional PKC by increases in the FRET ratio between CFP and YFP flanking the N- and C-termini of the enzyme (51). The PKCβII-Kinameleon reporter exhibits increasing FRET ratios in each of the conformation transitions: 1) unprimed (unphosphorylated) has the lowest FRET ratio, 2) primed (phosphorylated and autoinhibited) has an intermediate FRET ratio, and 3) active (membrane-associated) has the highest FRET ratio. Consistent with a role for mTORC2 in PKC priming, PKCβII-Kinameleon expressed in Sin1 KO MEFs displayed a reduced FRET ratio compared to that in Sin1 KO MEFs reconstituted with Sin1; this low FRET ratio was similar to that of kinase-dead PKCβII (K371R (52), kdPKC), which is incapable of priming because it cannot autophosphorylate (Fig. 1E). PKCβII-Kinameleon expressed in Sin1 KO MEFs also did not undergo a FRET change upon activation in response to PDBu stimulation; likewise, unprimed kinase-dead PKC did not undergo a conformational change because it is incapable of transition to the primed or active conformations, which are predicated on autophosphorylation (46) (Fig. 1F). However, reconstitution of Sin1 into Sin1 KO MEFs rescued the phenotype and allowed PKC to display a FRET change characteristic of primed PKC activation (Fig. 1F). These data reveal that mTORC2 promotes the mature, autoinhibited conformation of PKC.
As an additional measure of the PKC folding deficiency in the absence of mTORC2, we used a FRET-based translocation assay to assess PKC ligand sensitivity in living cells. We have previously shown that unprimed PKC translocates to plasma membrane more rapidly than primed PKC upon agonist stimulation, owing to exposed ligand-binding C1 domains; these domains become masked in primed (phosphorylated) and autoinhibited PKC. Consistent with the characteristics of unprimed enzyme, WT PKCβII translocated more rapidly from the cytosol to plasma membrane (Fig. 1G) in Sin1 KO MEFs (Sin1−/− +PKC) compared with WT MEFs (Sin1+/+ +PKC) (Fig. 1H). Reconstitution of Sin1 KO MEFs with Sin1 (Sin1−/− +PKC+Sin1) slowed the translocation rate to that observed in WT MEFs. The enhanced rate of PKC translocation in mTORC2-deficient cells was attributable to an exposed C1A domain: impairing ligand binding of the domain by introduction of a W58A mutation (53) reduced the rate of PKCβII translocation in Rictor KO MEFs (Ric−/− +PKC-W58A) (Fig. S1F). Furthermore, PKC with phosphomimetic Glu substitutions at the turn and hydrophobic motif phospho-acceptor sites (T641E/S660E) translocated rapidly in Sin1 KO MEFs (Sin1−/− +PKC-EE), indicative of the unprimed conformation of PKC. This suggests that mTORC2 regulates PKC conformation by a mechanism independent of phosphorylation at the C-tail turn motif and hydrophobic motif sites (Fig. 1H). This rapid translocation was not a result of the phosphomimetics not allowing the primed, autoinhibited conformation because this same construct translocated slowly in WT MEFs, indicating normal autoinhibition. Thus, mTORC2 promotes the autoinhibited and ‘primed’ conformation of PKC by a mechanism that is independent of phosphorylation of the known C-tail sites.
We next addressed whether mTOR activity regulates the initial phosphorylation of newly-synthesized PKC or the steady-state phosphorylation of mature PKC. The stoichiometry of PKC phosphorylation can be assessed by monitoring the electrophoretic mobility shift that accompanies phosphorylation of the two C-terminal sites (7) :the slower mobility species (asterisk, upper band) is phosphorylated at both the turn motif and hydrophobic motif sites and the faster mobility species (dash, lower band) is unphosphorylated. Using electrophoretic mobility to assess the phosphorylation state of PKC, we observed that Torin treatment of WT MEFs overexpressing PKCβII resulted in a relatively slow accumulation of unphosphorylated PKCβII (faster electrophoretic mobility). This slow rate of appearance of unphosphorylated PKC contrasted with the rapid loss of phosphate on the mTORC1-regulated S6K1 (Fig. 1I, Fig. S1G). Treatment with the protein synthesis inhibitor cycloheximide (CHX) prevented the Torin-dependent accumulation of unphosphorylated PKC without affecting the amount of pre-existing phosphorylated PKC, revealing that mTORC2 exclusively controlled PKC maturation (Fig. 1J). Thus, mTORC2 controls the initial phosphorylation in the maturation of PKC and does not affect the phosphorylation of primed PKC.
To examine the mechanism by which mTORC2 controls the maturation of PKC, we took advantage of pulse-chase experiments to interrogate the pool of newly-synthesized enzyme. Cells were metabolically labeled with 35S-Cys/Met for 7 min, and then chased with unlabeled amino acids for up to 90 min. PKC was immunoprecipitated and the phosphorylation state of newly-synthesized PKC monitored by the electrophoretic mobility shift accompanying C-tail phosphorylations. As reported previously, PKC matured with a half-time of approximately 30 min (Fig. 1K) (54).The presence of the mTOR inhibitor during the chase prevented PKC progression to the slower-mobility phosphorylated species, similar to the inability of a kinase-deficient PKC mutant to progress to the phosphorylated species (Fig. 1K). Phosphomimetic Glu substitutions at the turn (T641E) or hydrophobic (S660E) motifs did not rescue PKC phosphorylation upon mTOR inhibition, revealing that neither site can bypass the rate-limiting mTORC2 requirement in PKC processing (Fig. 1L, S1H). Taken together, mTORC2 regulates PKC activity, conformation, and the rate-limiting step of PKC phosphorylation independently of the turn motif and hydrophobic motif sites.
The Hydrophobic Motif of PKC and Akt is Regulated by Autophosphorylation
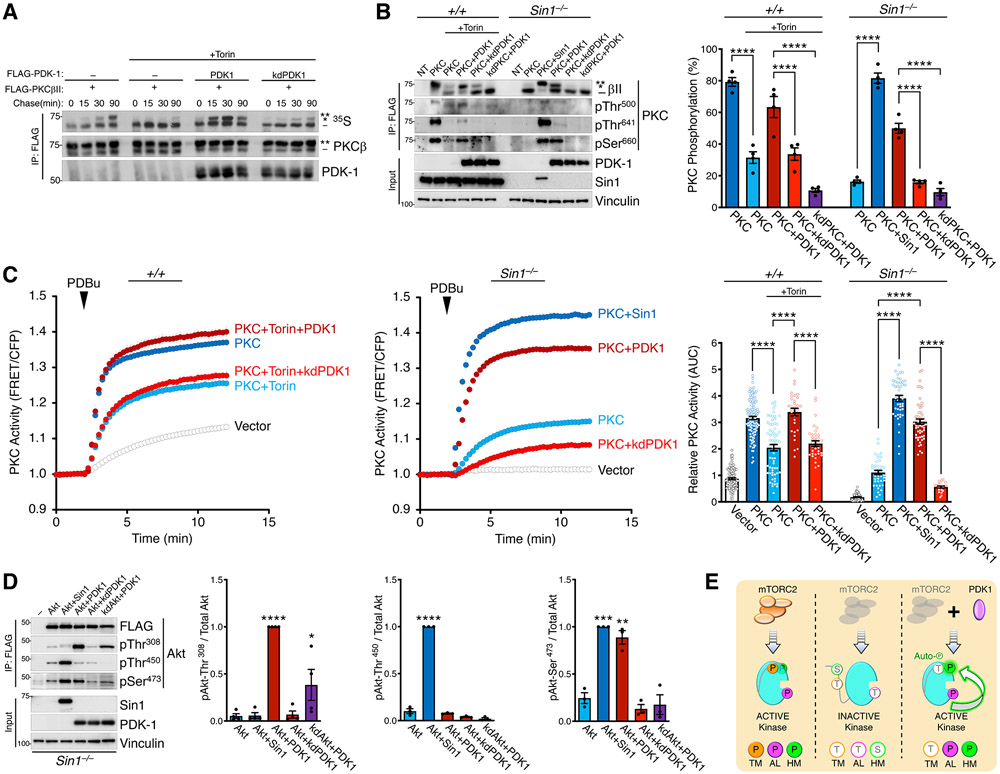
Given that mTORC2 is the proposed hydrophobic motif kinase (55, 56), regulates PKC independently of this site, we investigated the mechanism by which mTORC2 regulates the hydrophobic motif phosphorylation. Accordingly, we examined whether mTORC2 facilitates the PDK1-catalyzed phosphorylation of the activation loop, which is necessary for subsequent phosphorylation at the hydrophobic motif (6, 12). Pulse-chase analyses in the presence of mTOR inhibitors revealed that co-expression of WT PDK1, but not kinase-dead PDK1 (K110N; kdPDK1), rescued the phosphorylation of newly-synthesized PKC as assessed by electrophoretic mobility shift (Fig. 2A). Further analysis with phospho-specific antibodies indicated that PDK1 expression restored phosphorylation at the activation loop and hydrophobic motif, but not the turn motif, both in WT MEFs treated with Torin and in Sin1 KO MEFs (Fig. 2B). Consistent with previous reports that phosphorylation of the PKC hydrophobic motif is governed by autophosphorylation, PDK1 did not rescue hydrophobic motif phosphorylation of kinase-dead PKCβII (K371R; kdPKC) in either Torin-treated WT MEFs or Sin1 KO MEFs (Fig. 2B). Similar rescue of hydrophobic motif phosphorylation by PDK1 overexpression was also observed in Torin-treated COS7 cells and Rictor KO MEFs (Fig. S8A, S8B). Correlating with the rescue of hydrophobic motif phosphorylation, overexpression of WT, but not kinase-dead (kd), PDK1 restored cellular PKC activity in Torin-treated WT MEFs or Sin1 KO MEFs (Fig. 2C). These results reveal that PDK1 can bypass the requirement for mTORC2 by a mechanism that depends upon the intrinsic catalytic activity of both PDK1 and PKC.
Fig. 2. The Hydrophobic Motif of PKC and Akt is Regulated by Autophosphorylation.
(A) Pulse-chase experiment monitoring newly-synthesized FLAG-PKCβII (35S; autoradiograph) immunoprecipitated (IP) from COS7 cells co-expressing FLAG-PDK1 WT or kinase-dead K110N (kd) and treated without or with Torin (250 nM) during the chase. Blots are representative of three independent experiments.
(B) Western blot analysis of Triton-solubilized lysates (Input) or FLAG immunoprecipitates (IP:FLAG) from WT (+/+) or Sin1 KO (Sin1−/−) MEFs expressing FLAG-PKCβII WT or kinase-dead K371R (kd), and FLAG-PDK1 WT, kinase-dead K110N (kd), or HA-Sin1. Non-transfected cells (NT) also shown. Cells were treated with or without Torin (200 nM) during transfection and lysed 24 h later. (right) Quantification of % PKC phosphorylation obtained from the ratio of the slower mobility species (phosphorylated on hydrophobic motif, indicated by asterisks) over total PKC from 4 independent experiments.
(C) PKC activity in WT (+/+) or Sin1 KO (Sin1−/−) MEFs expressing CKAR2 (48) and mCherry-PKCβII, and treated with PDBu (200 nM). Data represent the normalized FRET ratio changes (mean ± SEM) from three independent experiments. (right) Quantification of PKC activity reflects the normalized area under the curve from baseline of 1.0 (AUC; mean ± SEM) 10 min after PDBu treatment. Each point represents data from one cell.
(D) Western blot analysis of Triton-solubilized lysates (Input) or FLAG immunoprecipitates (IP:FLAG) from Sin1 KO (Sin1−/−) MEFs expressing FLAG-mAkt1 catalytic domain (141-480) WT or kinase-dead K179M (kd), and FLAG-PDK1 WT, kinase-dead K110N (kd), or HA-Sin1. (right) Quantification of Akt phosphorylation reflects the normalized phospho-signal relative to total Akt for the activation loop (pThr308), turn motif (pThr450), or hydrophobic motif (pSer473) from 4 independent experiments.
(E) The hydrophobic motif of PKC and Akt is regulated by autophosphorylation in a PDK1-dependent and mTORC2-sensitive manner. Schematic of mTORC2 and PDK1 function in the phosphorylation of PKC and Akt: (left) mTORC2-containing cells produce kinase that is phosphorylated at the activation loop (AL), Turn Motif (TM), and Hydrophobic Motif (HM); (middle) mTORC2-deficient cells produce kinase with impaired phosphorylation at all three sites; (right) PDK1 overexpression in mTORC2-deficient cells rescues phosphorylation at the AL and HM, but not the TM. This rescue depends on the intrinsic catalytic activity of PKC and Akt, indicating that the hydrophobic motif of both kinases is regulated by autophosphorylation (green arrow) in a reaction that is promoted by PDK1 phosphorylation of the activation loop. mTORC2 facilitates the PDK1 step and can by bypassed with PDK1 overexpression.
In Western blots, the double asterisk (**) denotes the position of mature, phosphorylated PKC; the single asterisk (*) denotes the position of PKC phosphorylated at the hydrophobic motif, and the dash (−) indicates the position of unphosphorylated PKC.
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by One-way ANOVA and Tukey HSD Test. Western blot quantifications represent the mean ± SEM from at least three independent experiments.
Given that Akt is also regulated by PDK1 and mTORC2 (5, 27), we addressed whether the Akt hydrophobic motif in cells lacking mTORC2 could also be rescued by PDK1 by a mechanisms depending on Akt autophosphorylation. The isolated catalytic domain of Akt1 expressed in Sin1 KO cells was not appreciably phosphorylated at the activation loop (Thr308), turn motif (Thr450), or hydrophobic motif (Ser473), as reported previously (33). Similar to our result above with PKC, co-expression of PDK1 rescued Akt phosphorylation at the activation loop and hydrophobic motif, but not turn motif, sites (Fig. 2D). This restoration of Akt hydrophobic motif phosphorylation required the catalytic activity of both PDK1 and Akt: kinase-dead PDK1 (K110N; kdPDK1) or kinase-dead Akt1 (K179M; kdAkt) were ineffective in promoting hydrophobic motif phosphorylation in the absence of mTORC2 (Fig. 2D). It has previously been shown that kinase-dead Akt becomes phosphorylated at the turn motif in an mTORC2-dependent manner (14). Thus, mTORC2 is only necessary for phosphorylation of the turn motif for both PKC and Akt. PDK1 can effectively restore hydrophobic motif phosphorylation in cells lacking mTORC2 by a mechanism that depends on the intrinsic catalytic activity of both PDK1 and the recipient kinase (Fig. 2E). Taken together, these data establish that the hydrophobic motif is not a direct mTORC2 target, but rather is modified by autophosphorylation following PDK1 phosphorylation of the activation loop. It is the latter event that is controlled by mTORC2. Therefore, for both PKC and Akt, mTORC2 facilitates, but is not required for, phosphorylations that control the activity of each kinase.
mTORC2 Binds and Phosphorylates a Novel TOR-Interaction Motif
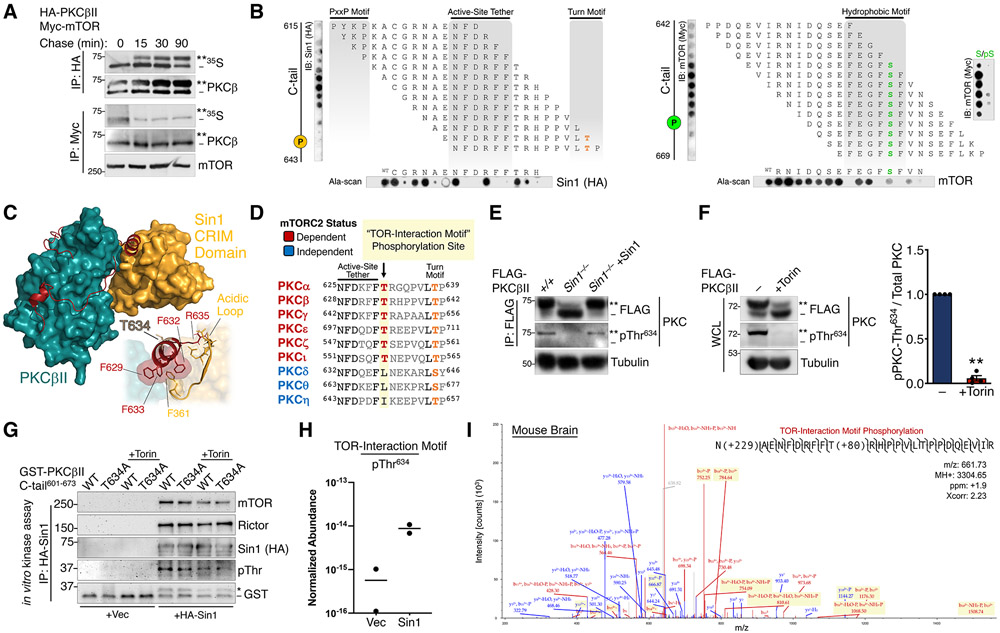
Having established that mTOR is not the physiological hydrophobic motif kinase, we explored the mechanism by which mTORC2 facilitates hydrophobic motif phosphorylation and kinase activation. To identify critical regions for mTOR-mediated PKC and Akt phosphorylation, we pursued a structural approach to map the mTORC2 binding site on PKC. Using pulse-chase analysis to label newly-synthesized PKC, we found that mTOR and Sin1 co-immunoprecipitated exclusively the unphosphorylated, newly-synthesized PKC (Fig. 3A). This association with newly-synthesized PKC was mediated by determinants primarily in the catalytic domain (Fig. S2A, S2B). In order to identify the interaction regions, we performed a peptide array of the PKCβII catalytic domain sequence and asked which segments bound mTORC2 components. Specifically, 15-mer synthesized peptides derived from the kinase + C-tail (a.a.296-673), at one residue intervals, were spotted onto a membrane which was overlaid with lysate from cells expressing Sin1, Raptor, or the mTOR kinase. Both components of mTORC2 (mTOR and Sin1), but not the mTORC1 subunit Raptor, bound to peptides containing the active-site tether (57) and hydrophobic motif regions in the C-tail (Fig. 3B, S2C). Alanine-scanning of the peptide displaying the greatest binding to Sin1 or mTOR identified Phe residues in the PKC active-site tether (PKCβII Phe629, Phe632, Phe633) and hydrophobic motif (PKCβII Phe656, Phe659, Phe661) as critical for binding to Sin1 and mTOR, respectively (Fig. 3B). Furthermore, substitution of the hydrophobic motif phospho-acceptor Ser660 with phospho-Ser (pS) abolished mTOR binding (Fig. 3B), consistent with mTORC2 specifically associating with unphosphorylated PKC. To gain insight into the structural determinants mediating this interaction, we modeled the mTORC2:PKC complex by docking the kinase domain of PKCβII (58) with the Sin1 CRIM domain (59), the known direct interaction partner that recruits AGC kinases to mTORC2 (60). Corroborating our peptide arrays, unbiased docking positioned the CRIM domain in contact with the PKC active-site tether (Fig. 3C, S4A). It has been previously shown that an acidic loop region of the Sin1 CRIM domain is required for mTORC2-mediated phosphorylation of PKC and Akt (60). Consistent with this finding, our model revealed that the acidic loop was anchored to the C-tail by hydrophobic interactions between Phe residues in the active-site tether determined in our peptide arrays to be critical for Sin1 binding (Fig. 3C).
Fig. 3. mTORC2 Binds and Phosphorylates a Novel TOR-Interaction Motif.
(A) Autoradiograph (35S) and Western blot of HA or Myc immunoprecipitates from a pulse-chase experiment of COS7 cells co-expressing HA-PKCβII and Myc-mTOR. The double asterisk (**) denotes the position of mature, fully phosphorylated PKC and the dash (−) indicates the position of unphosphorylated PKC. Blots are representative of three independent experiments.
(B) (top) Immunoblot analysis of 1-step, 15-mer peptide arrays spanning residues 615-643 (left) and residues 642-669 (right) of the PKCβII C-tail overlaid with Triton-solubilized lysate from WT MEFs expressing HA-Sin1 or Myc-mTOR and probed with antibodies for Sin1 (HA) or mTOR (Myc). The positions of the turn motif Thr641 and hydrophobic motif Ser660 are indicated. (far right) Peptide arrays of the indicated hydrophobic motif sequences were generated with phospho-Ser (pS) or unphosphorylated Ser (S) at the hydrophobic motif site and probed for mTOR as in panels on left. (bottom) Ala-scans of the indicated C-tail peptides were probed for Sin1 (HA) and mTOR; first spot is the wild-type peptide (WT). Blots are representative of three independent experiments.
(C) Docking of the Sin1 CRIM domain (NMR structure, PDB ID: 2RVK) to the PKCβII catalytic domain (X-ray structure, PDB ID: 2I0E). (inset) Interactions of the CRIM domain acidic loop with the PKCβII TOR-interaction motif helix are shown.
(D) Sequence alignment of the active-site tether and turn motif regions in the PKC C-tail, indicating the novel TOR-interaction motif Thr conserved in mTORC2-dependent PKC isozymes.
(E) Western blot of FLAG immunoprecipitates (IP:FLAG) from WT (+/+) or Sin1 KO (Sin1−/−) MEFs expressing FLAG-PKCβII alone or with HA-Sin1 and probed with an antibody to pThr634 or FLAG. Tubulin blot represents 10% whole-cell lysate input prior to immunoprecipitation.
(F) Western blot of whole-cell lysates (WCL) from HEK-293t cells expressing FLAG-PKCβII, treated with Torin (250 nM) for 36 h co-transfection, and probed with the indicated antibodies. (right) Quantification reflects the TIM phospho-signal (pThr634) relative to total PKC.
(G) Western blot from in vitro mTORC2 kinase assay, performed by incubation of HA-Sin1 or HA empty vector control (Vec) immunoprecipitated from HEK-293t cells transfected with these HA constructs, and GST-tagged PKCβII C-tail (a.a.601-673) WT or T634A purified by GST pulldown, in the presence or absence of Torin (200 nM) and probed with antibodies to mTORC2 components, GST, or an antibody to phospho-Thr. The asterisk (*) represents phosphorylated GST-PKCβII C-tail peptide and the dash (−) represents unphosphorylated peptide.
(H) Relative abundance of PKCβII C-tail peptide or phosphorylation at Thr634 for the in vitro kinase assay shown in (H) as determined by LC-MS. Data were obtained from two independent kinase assays.
(I) Representative spectrum of in vivo PKCβII Thr634 phosphorylation from analysis of a mouse brain phospho-proteomic dataset. Blue color denotes y ions and red color denotes b ions. (ppm-parts per million, Xcorr- spectral match correlation score).
**p < 0.01 by One-way ANOVA and Tukey HSD Test or Student’s t-test. Error bars represent SEM from at least three independent experiments.
To gain insight into how the binding of mTORC2 could affect phosphorylation of the C-terminal sites, we took advantage of the fact that the novel PKCδ is not regulated by mTORC2 yet also has a turn motif and hydrophobic motif that is phosphorylated during maturation. We engineered PKCδ/β chimeras containing the mTORC2-insensitive PKCδ kinase domain fused to PKCβII C-tail fragments (Fig. S3A). Substitution of the PKCδ C-terminus with 45 amino acids of PKCβII sequence containing the turn motif and hydrophobic motif phosphorylation sites (PKCδ/β TM; PKCδ1-631/βII628-673) produced a kinase that required mTORC2 for phosphorylation of the turn motif, but not the hydrophobic motif in cells (Fig. S3B). However, extending the junction eight residues further to include the PKCβII active-site tether (PKCδ/β AST; PKCδ1-622/βII620-673) resulted in a chimera that was dependent upon mTORC2 for turn motif and hydrophobic motif phosphorylation (Fig. S3B). Thus, turn motif phosphorylation is controlled by the specific sequences of the mTORC2-dependent and -independent PKCs, respectively, while phosphorylation of the hydrophobic motif is regulated by the region preceding the turn motif in the active-site tether. Comparative structural analysis of several AGC kinase C-tails revealed that the critical active-site tether region that confers mTORC2-independence diverges in PKCβII to form the “novel α-helix” (58) (Fig. S3C, S3D), perhaps providing a structural basis for the differential regulation of PKCs by mTORC2. Therefore, the active-site tether functions as a molecular switch to confer mTORC2-dependent hydrophobic motif phosphorylation, independently of turn motif phosphorylation.
We next hypothesized that a distinct C-tail phosphorylation site in the active-site tether region may account for the requirement of mTORC2 activity in the regulation of a subset of PKCs. Sequence alignment of the PKC active-site tether revealed a Thr preceding the turn motif site (TM-7), which we have termed the “TOR-Interaction Motif” (TIM; PKCβII Thr634), that is conserved, and exclusively present, in all mTORC2-sensitive PKCs (Fig. 3D). Cellular phosphorylation of the TOR-interaction motif was effectively suppressed in Sin1 KO MEFs (Fig. 3E) or upon mTOR kinase inhibition (Fig. 3F), as detected by a pThr634 antiserum (61). Incubation of PKCβII C-tail peptide (a.a.601-673) with immunoprecipitated mTORC2 (Fig. S5A) resulted in phosphorylation of the TIM site in vitro (Fig. 3G, 3H). Furthermore, phosphorylation of this site in mouse brain was detected by tandem mass spectrometry (Fig. 3I, S5B), validating its presence in vivo. Thus, the TOR-interaction motif is a direct mTORC2 phosphorylation site that specifies the mTORC2-dependence of PKC isozymes.
The TOR-interaction Motif Is Evolutionarily Conserved in Eukaryotes
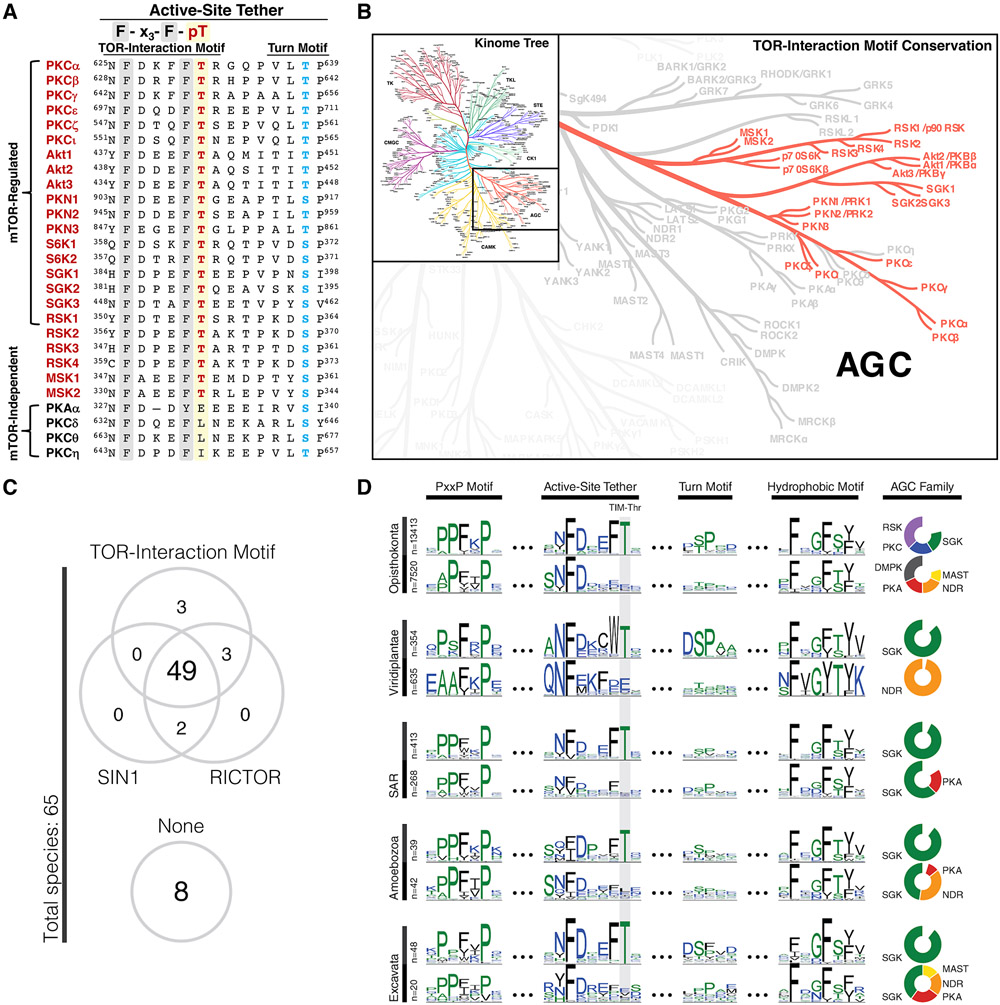
Next, we investigated the conservation of the TOR-interaction motif phosphorylation site in other AGC kinases. Human kinome analysis revealed that the TIM-Thr, located in an invariant F-x3-F-T motif, was conserved in all known mTORC2 regulated kinases (Fig. 4A). The TOR-interaction motif was also found in the mTORC1-regulated p70S6K family, as well as in p90RSK and MSK family kinases, which are not known to be regulated by mTOR (Fig. 4A, 4B). We note that while conservation of the TIM-Thr does not explicitly signify a phosphorylation site, TIM phosphorylation is detected in several unbiased proteomics studies (62), although to a lesser degree than either the turn motif or hydrophobic motif sites (Fig. S6A).
Fig. 4. The TOR-Interaction Motif Is Evolutionarily Conserved in Eukaryotes.
(A) Sequence alignment of the active-site tether region of AGC kinases indicating TOR-interaction motif (red) and turn motif (blue) phosphorylation sites for selected AGC kinases.
(B) AGC kinase branch of the human Kinome tree (111) indicating conservation of the TOR-interaction motif Thr in the highlighted kinases.
(C) Co-conservation of the TOR-interaction motif with TORC2 components SIN1 and RICTOR in various species (detailed in Fig. S7). Conservation of the TOR-interaction motif was defined by species that showed at least one AGC kinase conserving the TIM Thr.
(D) Conservation of the TOR-interaction motif Thr (TIM-Thr) in the AGC C-terminal tail across 5 major taxonomic groups. Sequence logos are shown for four distinct regions of the AGC tail: PxxP motif (124), active-site tether (57), turn motif, and hydrophobic motif. Sequences are stratified by taxonomic clades across the Y-axis: opisthokonta (animals, fungi, yeast), viridiplantae (terrestrial & aquatic plants), SAR (protozoa), amoebozoa (protozoa), and excavata (protozoa). Within each clade, sequences are further stratified by conservation of the TIM-Thr (top row). Pie charts (right) show the composition of AGC families within strata. AGC families accounting for less than 10% of each strata are not shown.
In order to assess the evolutionary association of the TOR-interaction motif and the presence of TORC2 components, we analyzed diverse species for the presence of kinases harboring the TOR-interaction motif and for the TORC2 components Rictor and Sin1 (Fig. 4C). The vast majority of species possessing kinases with the TOR-interaction motif (52 of 55) also contained TORC2 components (Fig. 4C, S7), supporting the role of the TOR-interaction motif as a TORC2-regulated phosphorylation site throughout evolution. Comparative sequence analyses show that the AGC C-terminal tail is well conserved across eukaryotes, and the TIM-Thr consistently appeared in major eukaryotic clades within the F-x3-F-T motif of the active-site tether (Fig. 4D, S6A). Patterns of conservation and variation in the C-terminal motifs reveal that constraints on the TIM-Thr were lost within some families and species. For example, the NDR family, which is present in most eukaryotes, consistently lack the TIM-Thr. . Likewise, SGK family members in multicellular eukaryotes (opisthokonta and viridiplantae) conserve the TIM-Thr, whereas SGKs in unicellular eukaryotes (SAR, amoebozoa, and excavata) lack the TIM-Thr. (Fig. 4D, S6B). Conserved within all five major eukaryotic clades, the TOR-interaction motif was likely present in the last eukaryotic common ancestor (LECA). An evolutionary study characterizing the widespread conservation of TOR signaling components in eukaryotes suggests that TOR signaling was also conserved in the LECA and has remained vital in almost all eukaryotes (63). While phylogenetic studies have traced the appearance of the TOR signaling pathways to the earliest eukaryotes, TORC2 is not found in plants. Our data indicate a plant-specific variation in the TOR-interaction motif in which the Phe preceding the TIM-Thr is replaced by a Trp (Fig. 4D, S6A, S6B). Thus, the F-x3-W-T motif in plants may reflect a unique variation on TOR signaling compared to eukaryotes which have retained the canonical F-x3-F-T motif. Taken together, these observations suggest that the TOR-interaction motif is evolutionarily conserved in eukaryotes and may have coevolved with mTORC2 to regulate a subset of AGC kinases.
TOR-Interaction Motif Phosphorylation is Critical for PKC and Akt Activity
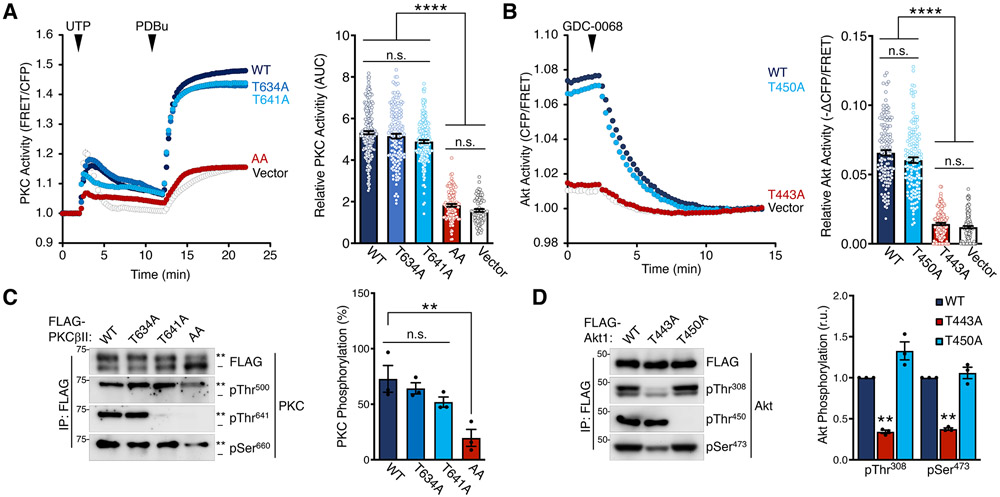
Next, we assessed the effect of TOR-interaction motif phosphorylation on PKC and Akt activity in cells using biosensors for each kinase (48, 64). Although mutation of the TIM (T634A) or turn motif (T641A) alone had minimal effect on PKC activity stimulated by either the natural agonist UTP (to transiently elevate diacylglycerol and Ca2+) or PDBu (for long-term activation), the double mutation (AA; T634A/T641A) abolished PKC cellular activity (Fig. 5A). Akt1 TIM mutation (T443A) alone, however, abolished the constitutive activity of the isolated catalytic domain: the drop of FRET ratio in cells expressing the WT Akt1 catalytic domain and treated with the Akt inhibitor treatment (GDC-0068) represent its high constitutive activity, which was abrogated with the T443A mutation (Fig. 5B). In contrast, mutation of the turn motif (T450A) had no effect on the inhibitor-sensitive activity of Akt1 (Fig. 5B). Western blot analysis revealed that mutation of both the TIM and turn motif in PKC, and only the TIM in Akt, resulted in reduced activation loop and hydrophobic motif phosphorylation of the inactive PKC and Akt TIM mutants (Fig. 5C, 5D). The related AGC kinase PKN2, in contrast, showed a greater dependence upon the turn motif than the TIM for activation loop phosphorylation, and TIM mutation in PKN1 actually enhanced activation loop phosphorylation (Fig. S8A).
Fig. 5. TOR-Interaction Motif Phosphorylation is Critical for PKC and Akt Activity.
(A) PKC activity in COS7 cells expressing CKAR2 (48) alone or with mCherry-PKCβII WT, T634A, T641A, or T634A/T641A (AA) and treated with UTP (100 μM) and then PDBu (200 nM) at the times noted. Data represent the normalized FRET ratio changes (mean ± SEM) from three independent experiments. (right) Quantification of PKC activity represents the normalized area under the curve from baseline of 1.0 (AUC; mean ± SEM) for the 20 min following UTP addition. Each data point reflects AUC of single cells from three independent experiments.
(B) Akt activity in COS7 cells expressing BKAR and mCherry-mAkt1 kinase domain (a.a.141-480) WT, T450A, or T443A and treated with the Akt inhibitor GDC-0068 (20 μM). The drop in FRET upon inhibitor addition indicates the degree of basal activity of the isolated kinase domain. Data represent the normalized FRET ratio changes (mean ± SEM) from three independent experiments. (right) Quantification of basal Akt activity measured by magnitude of the FRET change (mean ± SEM) 12 min after inhibitor addition. Each data point reflects AUC of a single cells from three independent experiments.
(C) Western blot of FLAG immunoprecipitates from Triton-solubilized lysates of HEK-293t cells expressing FLAG-PKCβII WT, T634A, T641A, or T634A/T641A (AA) and probed with the indicated antibodies. (right) Quantification of PKC phosphorylation (mean ± SEM) from three independent experiments represents the percent of the slower-mobility, phosphorylated species (**) over total PKC. Blots are representative of three independent experiments.
(D) Western blot of FLAG immunoprecipitates from Triton-solubilized lysates of HEK-293t cells expressing FLAG-Akt1 catalytic domain (a.a.141-480) WT, T443A, or T450A constructs and probed with the indicated antibodies. (right) Quantification of Akt phosphorylation (mean ± SEM) from three independent experiments reflects the normalized phospho-signal relative to total Akt for the activation loop (pThr308) or hydrophobic motif (pSer473). Blots are representative of three independent experiments.
**p < 0.01; ****p < 0.0001; n.s., not significant by One-way ANOVA and Tukey HSD Test or Student’s t-test. Western blot quantifications represent the mean ± SEM from at least three independent experiments.
Supporting a role for mTORC2 in promoting PKC activation loop phosphorylation, PDK1 bound the nascent, unphosphorylated PKC species two-fold better than the phosphorylated form; this preference for unphosphorylated PKC was lost upon mTOR inhibition (Fig. S9C). PDK1 binds AGC kinases through a polyvalent interaction with the active-site tether and hydrophobic motif in the C-tail (65); however, the ability of PDK1 to bind PKC C-tail peptides was independent of phosphorylation or phosphomimetics at the TIM (T634E) or turn motif (T641E) sites as assessed by peptide array (Fig. S9D-F). Competition with PDK1-interacting fragment (PIF) peptide, which binds the PDK1 hydrophobic docking site (PIF pocket) with high affinity (66, 67), suppressed the fraction of phosphorylated PKC in the presence or absence of mTOR inhibition as indicated by electrophoretic mobility (Fig. S9G). This finding suggests that mTOR regulation of PDK1 binding is mediated through the hydrophobic motif binding site and is regulated at the level of C-tail accessibility, rather than by altered binding affinity upon phosphorylation. This result supports our previous finding which shows that although PDK1 exhibits higher affinity for phosphorylated hydrophobic motif peptide, PDK1 preferentially associates with unphosphorylated PKC in cells due to increased accessibility of the C-tail in the unphosphorylated state (68).
We next assessed the role of TIM and turn motif phosphorylation on kinase domain conformational dynamics with the live-cell reporter KinCon (Fig. S10A) (69, 70). This bioluminescence-based reporter utilizes the protein-fragment complementation luciferase to characterize kinase conformational states in response to various stimuli. TIM/turn motif mutation in PKCβII (T634A/T641A) was sufficient to alter kinase dynamics (Fig. S10B), as assessed by reduced complementation of the split luciferase at N- and C-termini of the kinase. Mutation of the corresponding residues in S6K1 (T387A/T394A), an mTORC1 substrate, also resulted in a reduction in bioluminescence (Fig. S10C), indicating that these residues may serve similar functions in each kinase despite regulation by different TOR complexes. The reduced bioluminescence observed upon mutation of the C-tail phosphorylation sites reflects reduced interaction between the unstructured PKC N- and C-termini, suggesting that docking of the N-term pseudosubstrate in the active site cleft and packing of the C-tail against the kinase domain, respectively, may be impaired. Acidic residues (T387DT394D or T387E/T394E), however, restored complementation to that of WT for S6K1 (Fig. S10C), indicating their ability to serve as phosphomimetics for this kinase. Glu or Asp substitutions in PKC (T634D/T641D, T634E/T641E), conversely, did not rescue bioluminescence to WT levels, reflecting the inability for acidic residues to effectively recapitulate phosphorylations in some proteins and positions. For example, Glu at the PKC activation loop phosphorylation site (T500E) can produce an active PKC; whereas, Asp (T500D) cannot (15). Thus, these findings provide evidence that phosphorylation of the segment comprising the TOR-interaction motif and turn motif dominantly controls PKC activity by altering accessibility of the C-tail to facilitate activation loop phosphorylation by PDK1.
The TOR-interaction Motif Coordinates PKC Dimerization
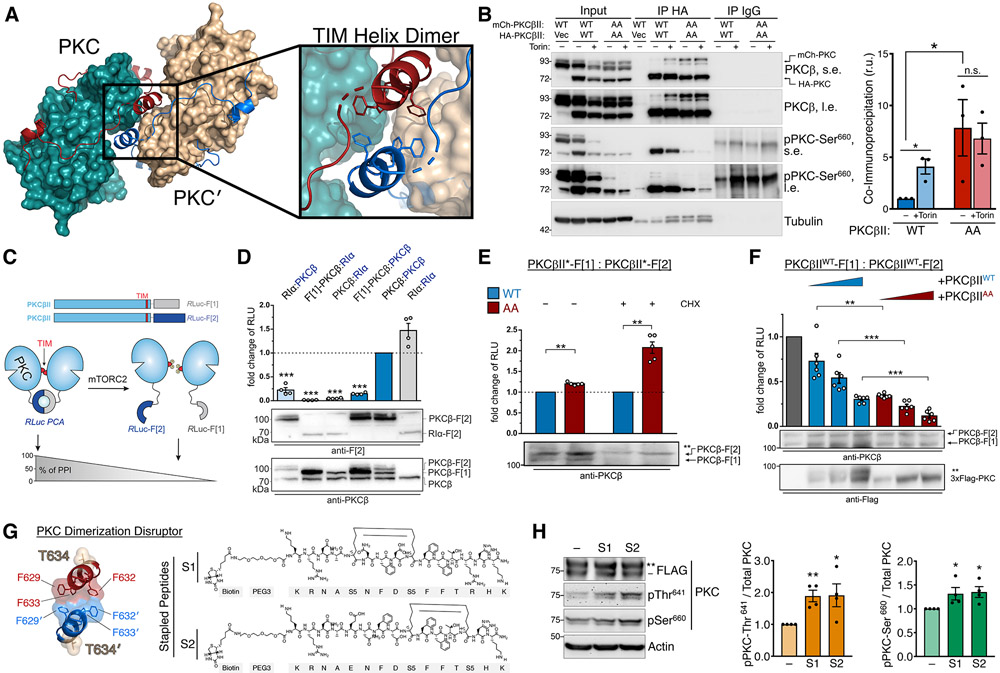
To explore how TIM phosphorylation by mTORC2 alters C-tail accessibility, we re-analyzed a reported PKCβII structure of the isolated catalytic domain, removed of any intramolecular contacts with the regulatory domain, in which the active-site tether is resolved (55). Examination of the crystal packing revealed a symmetrical, head-to-head PKC homodimer, forming a dimerization interface through hydrophobic interactions in the “novel α helix” (58) that contains the TOR-interaction motif (TIM Helix) (Fig. 6A). To determine whether mTOR regulates PKC dimerization in a cellular context, we performed co-immunoprecipitation assays between differentially tagged PKC proteins. Whereas little association of WT PKCβII proteins was observed in untreated cells in which the majority of PKC is in the mature form, mTOR inhibition or TIM/turn mutation (PKCβII AA) greatly enhanced PKC self-association, consistent with dimerization (Fig. 6B). The co-immunoprecipitated PKC was devoid of phosphorylation at the hydrophobic motif (Fig. 6B), indicating that association occurred only between unphosphorylated PKC molecules. Note that Torin treatment resulted in decreased steady-state levels of WT PKC, but not PKCβII AA, because the unphosphorylated species is unstable. Furthermore, the isolated PKC catalytic domain showed enhanced self-association compared to full-length PKC (Fig. S11), suggesting that the PKC regulatory domain impairs dimerization, perhaps by occluding the dimerization interface in the mature form.
Fig. 6. The TOR-Interaction Motif Coordinates PKC Dimerization.
(A) PKC dimer from PKCβII X-ray structure (PDB ID: 2I0E) showing the PKC kinase (teal) and C-tail (red) with dimer partner (PKC′) kinase (tan) and C-tail (blue). The dimerization interface at the TOR-interaction helix (TIM Helix) is shown with interacting Phe residues.
(B) Western blot of Triton-solubilized lysates (Input) and HA or IgG control immunoprecipitates from HEK-293t cells expressing HA- and mCherry-PKCβII WT or T634A/T641A (AA), and probed with the indicated antibodies. s.e., short exposure; l.e., long exposure. (right) Quantification of immunoprecipitated mCherry-PKC normalized to the input mCherry-PKC (mean ± SEM) from three independent experiments.
(C) Domain organization of PKCβII constructs tagged with the Renilla luciferase (Rluc) based protein-fragment complementation assay (PCA) fragments Rluc-F[1] and Rluc-F[2]. Scheme illustrates an involvement of the PKC TOR-interaction motif (TIM) in PKC dimer formation analyzed using the PCA system. PKC dimerization induces the complementation of Rluc fragments and bioluminescent signal reflects the indicated protein-protein interactions (PPI).
(D) Indicated C-terminally tagged Rluc PCA reporter constructs were transiently co-expressed in HEK293 cells (the exception is the N terminally tagged F[1]-PKCβ). Data are presented as the fold change of bioluminescent signals (relative light units (RLU)) relative to PKCβII homodimer formation. Each point represents the mean value of an individual experiment performed at least in triplicate; bars indicate the mean ± SEM from four independent experiments. The PKA subunit RIα was used as a dimerization positive control.
(E) PKC dimer formation of PKCβII WT and T634A/T641A (AA), measured in the presence or absence of cycloheximide (CHX; 250 μM for 6 h), assessed by the RLuc PCA reporter assay as in (D). Data are presented as the fold change of the PPI signal relative to the WT PKCβII dimer signal. Each point represents the mean value of an individual experiment performed at least in triplicate; bars indicate the mean ± SEM from five independent experiments. *p< 0.05; **p<0.01; ***p<0.001; n.s., not significant by paired Student's t-test.
(F) Effect of increasing expression of FLAG-PKCβII WT or T634A/T641A (AA) on dimer formation of PKCbII-Rluc-F[1] and PKCbII-Rluc-F[2]. Data are presented as the fold change of the PPI signal in relation to the mock control. Each point represents the mean value of an individual experiment performed at least in triplicate; bars indicate the mean ± SEM from three independent experiments. *p< 0.05; **p<0.01; ***p<0.001; n.s., not significant by One-way ANOVA.
(G) Dimerization interface of the TIM helix showing hydrophobic interactions and TIM phosphorylation site (T634). Kekulé structures of two PKC Dimerization Disruptor (PKC-DD) stapled peptides targeting the TOR-interaction motif.
(H) Western blot of HEK293t cells expressing FLAG-PKCβII, treated with 10 μM of the indicated PKC-DD stapled peptides for 24 h prior to lysis, and probed with the indicated antibodies. (right) Quantification represents the amount of PKC phosphorylated at Thr641 or Ser660 normalized to total PKC from three independent experiments.
*p < 0.05; **p<0.01; ***p<0.001; n.s., not significant by Student’s t-test or One-way ANOVA. Error bars represent SEM from at least n=3 experiments.
To further interrogate PKC self-association in a cellular system, we employed a luciferase-based protein-fragment complementation assay (PCA) that has been successfully used to detect protein dimerization in intact cells (71) (Fig. 6C). Using this system, we observed a robust association of PKC molecules by PCA indicative of steady-state PKC dimerization (Fig. 6D), which was modestly enhanced by TIM/turn motif mutation (AA) (Fig 6E). Dimerization of TIM/turn motif mutant PKC was more pronounced when protein synthesis was inhibited for 6 h prior to the assay (Fig 6E, +CHX). When overexpressed, some unphosphorylated WT PKC accumulates in cells, which may result in higher baseline dimerization of unprocessed PKC. Thus, inhibiting protein synthesis presumably increases the proportion of phosphorylated PKC in the WT condition by allowing sufficient time for the unphosphorylated and unstable protein (46) to become phosphorylated or degraded. Since mature (phosphorylated) PKC is monomeric as demonstrated by co-immunoprecipitation (Fig 6B), reducing the amount of unprocessed WT PKC made the impact of PKC AA mutation more pronounced. Therefore, the TIM/turn motif mutant PKC (AA), which remains unphosphorylated, results in relatively enhanced dimerization compared to phosphorylated WT PKC (Fig 6E). We next assessed whether PKC dimers could dynamically assemble in cells. In a competition assay, increasing amounts of FLAG-PKCβII was sufficient to displace the PCA protein-protein interaction in a dose-dependent manner (Fig. 6F), indicating that PKC dimerization is a dynamic association. Furthermore, the ability to dissociate the interaction by competition is consistent with protein dimerization rather than aggregation accounting for the complementation of the split luciferase probes (72). The TIM/turn motif mutant PKCβII (AA) more effectively competed with PCA dimerization than did WT PKCβII, supporting the preferential dimerization of unprocessed PKC. Furthermore, this finding shows that TIM/turn motif phosphorylation controls the formation of PKC dimers. In summary, these data are consistent with nascent, unphosphorylated PKC forming a dimer which is disrupted by phosphorylation the TOR-interaction motif and turn motif.
Since mTORC2 phosphorylation of the TOR-interaction motif promotes hydrophobic motif phosphorylation and also dissociates PKC dimers, we next addressed whether disrupting PKC dimerization would be sufficient to enhance PKC phosphorylation at the hydrophobic motif. To test this possibility, we designed cell-permeable α-helical stapled peptides (73, 74) targeting the TOR-interaction motif to relieve dimerization (Fig. 6G). Treatment of cells overexpressing PKCβII with either of two different PKC dimerization disruptor stapled peptides (S1, S2) resulted in increased levels of PKC phosphorylation compared to DMSO-treated controls (Fig. 6H). Thus, peptides targeting the TOR-interaction motif designed to disrupt the dimer interface effectively promoted PKC processing. These data are consistent with a model in which the TOR-interaction motif helix coordinates an immature PKC homodimer, which is dissociated by mTORC2 phosphorylation to promote PKC maturation. Moreover, the TIM dimerization interface, previously identified as a “novel α-helix” in the original crystallization, is therapeutically actionable and can be targeted with stapled peptides in cells to induce PKC processing.
Discussion
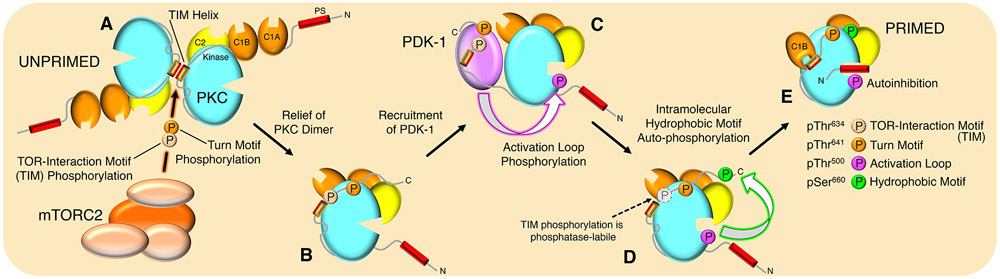
The requirement of mTORC2 for both PKC and Akt hydrophobic motif phosphorylation has been firmly established, yet the mechanism of this regulation has remained elusive. In this study, we present evidence that mTORC2 performs the first and rate-limiting step of PKC maturation by phosphorylating the newly identified TOR-interaction motif (TIM) (Fig. 7A), which ultimately facilitates hydrophobic motif autophosphorylation. This regulation by mTORC2 is accomplished by recognition of determinants in the C-tail active-site tether and hydrophobic motif that confer binding to the immature, unphosphorylated, and dimerized PKC species (UNPRIMED). Dissociation of the PKC dimer upon TIM phosphorylation (Fig. 7B) recruits PDK1 to the unphosphorylated hydrophobic motif to promote activation loop phosphorylation (Fig. 7C). For Akt, activation loop phosphorylation is sufficient to activate the kinase; whereas, for PKC, this site alone does not enable phosphorylation of downstream substrates but may provide the low rate of catalytic activity required to autophosphorylate at the hydrophobic motif (Fig. 7D). Following hydrophobic motif phosphorylation, the final step of PKC maturation is autoinhibition by the pseudosubstrate to yield the catalytically-competent and stable PKC species (46) (PRIMED) (Fig. 7E). This process serves the interdependent functions of stabilizing the catalytically-competent and phosphorylated PKC, as well as preventing self-association of the mature form. Moreover, this study identifies the TOR-interaction motif as the physiological target of the mTOR kinase and provides a mechanism for AGC kinase regulation by mTOR. Thus, our findings establish that the PKC and Akt hydrophobic motif sites, similar to autoregulatory sites in the C-terminal tail of receptor tyrosine kinases (75), are regulated by autophosphorylation (Fig. 8).
Fig. 7. Model of PKC maturation by phosphorylation.
(A) Newly-synthesized PKC exists as a homodimer mediated by the TOR-interaction motif (TIM) helix, with membrane targeting domains exposed, and is neither phosphorylated nor catalytically active (UNPRIMED).
(B) mTORC2 binds to disrupt the dimer interface, phosphorylates TIM and turn motif phosphorylation to relieve the PKC dimer, exposing the C-terminal tail to recruit PDK1.
(C) Bound PDK1 phosphorylates PKC at the activation loop.
(D) Activation loop phosphorylation triggers intramolecular autophosphorylation at the hydrophobic motif.
(E) Phosphorylation at the hydrophobic motif triggers binding of the pseudosubstrate to the substrate binding cavity to effect autoinhibition (PRIMED). This species of PKC, which is stable and catalytically competent, is maintained in an inactive state by the pseudosubstrate, poised for activation by the second-messengers diacylglycerol and Ca2+.
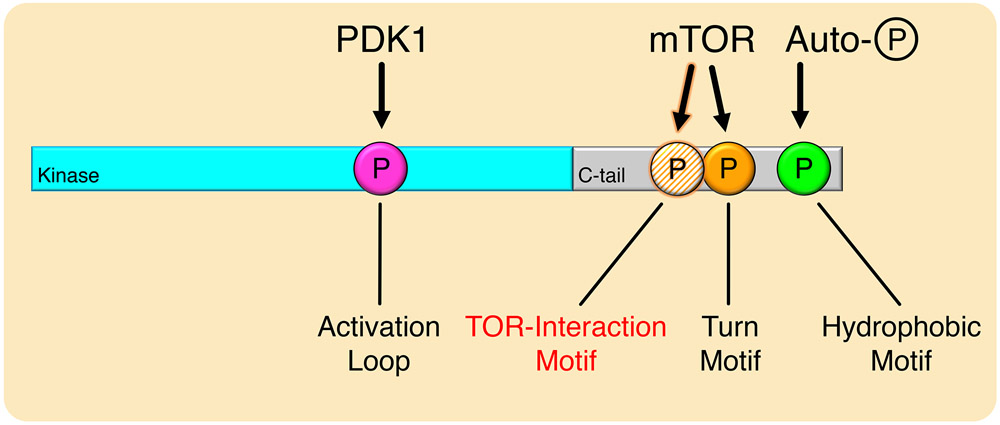
Fig. 8. Model for Phosphorylation of mTOR-Regulated AGC Kinases.
mTOR-regulated kinases are phosphorylated at the activation loop, turn motif, hydrophobic motif, and the newly-identified TOR-interaction motif. The TOR-interaction motif and turn motif are phosphorylated by mTOR, which facilitates activation loop phosphorylation by PDK1, and triggers hydrophobic motif autophosphorylation to activate the kinase.
Characterization of TOR-interaction motif phosphorylation may have been hindered by the potentially transient nature of phosphorylation at this site, as evidenced by the low rate of identification in proteomic studies (62). First detected by mass spectrometry as a PKC phosphorylation in insect cell expression (76), TIM phosphorylation may be more sensitive to phosphatases because of its outward orientation on an exposed segment of the C-tail. TIM phosphorylation was previously detected as a “protected” PKC phosphorylation in neurons during long-term potentiation. In this study, we detected PKC TIM phosphorylation by mass spectrometry in mouse brain, suggesting that stable phosphorylation at the TIM site may exist in certain phosphatase contexts. The critical role for TIM phosphorylation specifically during PKC processing and compensation by the turn motif phosphorylation suggests that this site may only be required in the recruitment of PDK1 and is dispensable thereafter. Whether this phosphorylation plays a more dynamic role in kinases such as Akt that are transiently activated by PDK1 merits further investigation.
The turn motif phosphorylation, which is the constitutive and stable mTOR phosphorylation, may additionally confer the ability to autophosphorylate at the hydrophobic motif. In support of this, mature PKC that is dephosphorylated at the activation loop and hydrophobic motif in vitro, but retains the turn motif phosphorylation, autophosphorylates at the hydrophobic motif (7). Dephosphorylation of the turn motif site, however, abolishes PKC re-autophosphorylation, which is rescued by incubation with catalytically-competent PDK1 (6). Therefore, turn motif phosphorylation may lock into place the conformational changes conferred by activation loop phosphorylation, facilitating PDK1-independent re-autophosphorylation that extends the signaling lifetime of PKC (77). Furthermore, we have shown here that TIM phosphorylation and turn motif phosphorylation can compensate for one another in PKC, but the TIM site is dominant in Akt. Indeed, the turn motif does not directly regulate the activity of PKC, Akt, or PKA in vitro, but instead anchors the C-tail to the kinase domain to stabilize AGC kinases (65). Curiously, even in the case of PKA, which has a divergent turn motif site that is mTORC2-independent, phosphorylation of the turn motif is a prerequisite for activation loop phosphorylation in cells (78). Thus, phosphorylation of the TIM and/or turn motif sites may coordinately facilitate the initial activation by PDK1 in a variety of AGC kinases and sustain a regulatory “memory” of activation. That the TOR-interaction motif is more highly conserved than the turn motif in eukaryotic kinases across evolution substantiates our findings that the TIM is the primary target of mTOR and the basis of mTORC2 regulation for a subset of AGC kinases.
Elucidating the hydrophobic motif as an mTORC2-facilitated and PDK1-dependent autophosphorylation highlights the intricate regulation of AGC kinase activation. PKC hydrophobic motif phosphorylation, which occurs during priming, is essential for its catalytic activity in cells, whereas this acutely-regulated site in Akt tunes its activity. For some AGC kinases, however, hydrophobic motif phosphorylation occurs first to create a docking site for PDK1 (2). In all cases, activation loop phosphorylation is the critical event for activation, and this process appears to be regulated principally by PDK1 recruitment, which is achieved by a multitude of mechanisms (79). That PDK1-dependent activation is regulated at the level of binding is not surprising, as PDK1 exists at low nanomolar concentrations, while its substrates may outnumber that by as many as two orders of magnitude in the cell (80). Additionally, PDK1 lacks a canonical AGC kinase C-tail and requires the transinteracting C-tail of the recipient kinase for activation. In fact, the PDK1 C-tail sequence diverges immediately preceding the TOR-interaction motif (57), suggesting that binding this regulatory element, like the hydrophobic motif, directs PDK1 activity and specificity towards its substrates. Thus, recruitment of PDK1 to the C-tail is the rate-determining step of PKC maturation, regulated by mTORC2 phosphorylation of the TIM, and is likely the limiting factor in AGC kinase activation. The TOR-interaction motif is also conserved in S6K1 and S6K2, which are well known to be regulated by mTORC1. A unique C-terminal extension in S6K precludes regulation by mTORC2 (81), and the hydrophobic motif is instead mediated by mTORC1 through a distinct TOR-signaling (TOS) motif (82). Early studies reporting the S6K hydrophobic motif as an autophosphorylation (83) raise the interesting possibility that TIM phosphorylation by mTORC1 may serve a similar function in S6K activation by recruiting PDK1 (84), but is subject to regulation by the multitude of nutrient-sensing inputs that feed into mTORC1 (85). Furthermore, the TOR-interaction motif is present in RSK family kinases, which has recently been identified as an mTORC2-regulated kinases (37), and MSK family kinases, which are not known to be regulated by mTOR (86). Establishing the specific role of TIM phosphorylation in distinct kinase families awaits future studies.
The ability to regulate function by autophosphorylation is a prevalent theme in kinase biology (87). In general, conformational switches convert kinases into a mode “prone-to-autophosphorylation,” which allows stabilizing or activating autophosphorylation. In vitro studies with pure PKC first showed that the enzyme autophosphorylates by an intramolecular reaction at sites later identified to be C-terminal sites including the hydrophobic motif (76, 88). Furthermore, purified enzyme selectively dephosphorylated at this site in vitro could re-incorporate phosphate at that position in a concentration-independent manner (7, 31, 89). The inability of kinase-inactive constructs to incorporate phosphate at the hydrophobic motif in cells supported the physiological regulation of this site as a cis-autophosphorylation (31, 46). This finding was refuted in a study showing that active site inhibitors allowed phosphate accumulation of the hydrophobic motif on kinase-inactive constructs (90). However, occupancy of the active site by inhibitors or peptide substrates prevents PKC dephosphorylation, and mutants that have weak activity accumulate phosphate at this site (46, 50). In the case of Akt, multiple mechanisms have been proposed to control the phosphorylation of the hydrophobic motif (91). But, as for PKC, Akt activation loop phosphorylation by PDK1 was shown to trigger hydrophobic motif phosphorylation of active, but not inactive, Akt (30). Furthermore, several studies have reported that phosphorylation of the hydrophobic motif is impaired in kinase-dead Akt (14, 30, 33). The discovery that the mTOR kinase promotes hydrophobic motif phosphorylation for a number of AGC kinases has confounded a consensus on the mechanism of regulation at this site. Our current study resolves this question by showing that mTORC2 “primes” PKC for activation loop phosphorylation by the critical kinase, PDK1, with the immediate consequence being autophosphorylation of the hydrophobic motif. Thus, mTORC2 facilitates the PDK1 step which converts PKC into a “autophosphorylation-prone” conformation.
An unexpected finding from this study is the ability of immature PKC to homodimerize. Dimerization of mature PKC has previously been proposed in vitro in the presence of Ca2+ and phosphatidylserine, or in cells upon activation (92, 93); however, these studies do not take into account the reduction in dimensionality upon PKC engagement on a lipid membrane or vesicle, and may have limited physiological basis. Instead, biophysical studies support mature (phosphorylated) PKC functioning as a monomer in cells (94). Additionally, biochemical studies reveal that the purified mature PKC monomeric even when bound to activating lipid surfaces (95), where it autophosphorylates in a concentration-independent manner, indicative of monomeric function (88). Consistent with the activated form being unlikely to dimerize, the C2-mediated translocation of PKCβII is a diffusion-driven process (96) and relies upon intramolecular, rather than intermolecular, contacts with the kinase domain (97). Thus, our finding in this study that newly-synthesized PKC exists as an immature homodimer regulated by mTORC2-mediated TIM phosphorylation is likely to be the only physiologically relevant dimer that is consistent with PKC biochemistry. Akt has been proposed to dimerize (98, 99); however, whether mTORC2-mediated TIM phosphorylation also regulates Akt dimerization or relieves PH domain autoinhibition to facilitate its activation remains to be determined.
Strategies to manipulate hydrophobic motif phosphorylation may be applicable in cancer therapies, where this site is frequently dysregulated. The AGC kinase hydrophobic motif is a hotspot for cancer-associated mutations in sites of post-translational modifications (100). Additionally, PKC quality control, which involves PHLPP-mediated hydrophobic motif dephosphorylation and degradation, is exploited in cancer to suppress PKC protein levels (46, 101). Thus, maintaining the integrity of the PKC processing machinery that permits hydrophobic motif phosphorylation is critical for PKC’s tumor-suppressive function (102, 103). Akt hydrophobic motif phosphorylation, conversely, generally serves an oncogenic role, and constitutively active or amplified Akt is prevalent in the wide variety of cancers that frequently harbor PI3K pathway alterations (104). In these cancers, the mTOR kinase is an attractive therapeutic target (105); however, this strategy should be approached with caution as mTORC1/2 inhibitors would have the unwanted consequence of inhibiting PKC processing, depleting the levels of an important tumor-suppressor. Therefore, utilizing 3rd generation mTOR inhibitors that selectively target mTORC1 (106), or exploring methods to promote mTOR-independent PKC processing by targeting PKC dimerization with stapled peptides as we have done here, may potentiate the efficacy of mTOR therapies.
Our work detailing the molecular basis of PKC and Akt regulation by mTORC2 provides new lines of investigation into the mechanisms of AGC kinase activation. Understanding the determinants for signal propagation through these kinases will facilitate the identification of new therapeutic strategies to modulate diverse cellular processes and disease states.
Materials and Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Alexandra Newton (anewton@health.ucsd.edu).
Experimental Model and Subject Details
Cell Culture and Transfection
+/+, Ric−/−, and Sin1−/− MEFs were previously described (40) MEFs, COS7, and HEK-293t cells were cultured in DMEM (Corning) containing 10% fetal bovine serum (Atlanta Biologicals) and 1% penicillin/streptomycin (Gibco) at 37°C in 5% CO2. Transient transfection was carried out using the Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific). Cells used were periodically tested for Mycoplasma contamination using a PCR-based protocol (107) and showed no evidence of contamination.
Method Details
Comparative Sequence Analysis:
An alignment profile of the human AGC C-terminal tail was created ranging from the PxxP motif to the hydrophobic motif. The profile was expanded with iterative BLASTp (108) searches on UniprotKB until convergence, then aligned using MAFFT v7.310 (109). Members of the AGC kinases group were identified from Uniprot Proteomes (retrieved Oct 9, 2019) using MAPGAPS (110). From this filtered set, we identified and aligned AGC tails with the aforementioned profile using MAPGAPS (110). Kinase domains residing N-terminal to the AGC tails were classified according to AGC families defined by the Manning classification (111) using MAPGAPS (110). Sequence taxonomy was determined by mapping Uniprot OX values to NCBI taxdump (retrieved March 23, 2019). Phosphorylation sites were mapped using PhosphoSitePlus (62). Sequence logos were created using WebLogo3 (112).
FRET Imaging and Analysis
Cells were imaged as described previously (113). For activity experiments MEFs and COS7 cells were co-transfected with the indicated mCherry-tagged PKC construct and CKAR or CKAR2. For Kinameleon experiments, the indicated Kinameleon construct containing mYFP and mCFP was transfected alone. For translocation experiments, MEFs cells were co-transfected with the indicated mYFP-tagged construct and plasma-membrane targeted mCFP. Baseline images were acquired every 15 s for 2 min prior to drug addition. Förster resonance energy transfer (FRET) ratios represent the mean ± SEM from at least three independent experiments. FRET ratios (FRET/CFP or CFP/FRET) were standardized such that greater values correspond to increased phosphorylation of the reporter, indicative of increased kinase activity. All data were normalized to the baseline FRET ratio of each individual cell unless noted that absolute FRET ratio was plotted or traces were normalized to levels post-inhibitor addition. Area under the curve (AUC) represents mean ± SEM of individual cells from the time of the first drug addition (t = 2 min) until the indicated time point, integrated over the normalized initial value y = 1. When comparing translocation kinetics, data were also normalized to the maximal amplitude of translocation for each cell, as previously described (51), in order to compare translocation rates. This normalization was performed because the maximal amplitude of translocation of the mutants varied, possibly because of changes in the orientation or distance of the fluorophores caused by differential folding of the mutant kinases.
Immunoblotting and antibodies
Cells were lysed in PPHB: 50 mM NaPO4 (pH 7.5), 1% Triton X-100, 20 mM NaF, 1 mM Na4P2O7, 100 mM NaCl, 2 mM EDTA, 2mM EGTA, 1 mM Na3VO4, 1 mM PMSF, 40 mg/ml leupeptin, 1mM DTT, and 1 mM microcystin. Whole-cell lysates were sonicated and Triton-soluble lysates were centrifuged at 13,000g x g for three minutes and supernatant was isolated. Lysates were analyzed by SDS-PAGE, transferred to PVDF membrane (Biorad), blocked in 5% non-fat dry milk for 1h at room temperature, incubated with primary antibodies at 1:1000 dilutions overnight at 4°C, incubated with HRP-conjugated secondary antibody at 1:10,000 dilution for 1h at room temperature, and detected using chemiluminescence SuperSignal West reagent (Thermo Fisher) on a FluorChem Q imaging system (ProteinSimple). Blots were washed three times with 1X TBS-T between incubations. The pan anti-phospho-PKC activation loop antibody (PKC pT500) (114) and TOR-interaction motif antibody (PKC pT634) (61) were described previously. The anti-phospho-PKCα/βII turn motif (pT638/641; 9375S), anti-PKCδ/θ turn motif (9376S), anti-Myc (D84C12), anti-mTOR (7C10), anti-Rictor (53A2), anti-Raptor (24C12), anti-S6K (9202S), anti-pS6K T389 (9205), anti-PDK1 (3062S), anti-phospho-PKN1/PKN2 activation loop antibody (2611), anti-Vinculin (4650S), anti-phospho-Akt pT308 (244F9) / pT450 (D5G4) / pS473 (D9E), pan anti-phospho-PKC hydrophobic motif (βII pS660; 9371S), and anti-phospho-Thr (clone 42H4; 9386S) antibodies were purchased from Cell Signaling Technologies. The anti-Sin1 antibody was from Abcam (ab71152). Anti-PKCβ antibody was purchased from BD Transduction Laboratories (610128). The anti-dsRED antibody was purchased from Clonetech. The anti-GST (clone B-14; sc-138) and rabbit (sc-2491) and mouse (H-270; sc-66931) IgG antibodies were purchased from Santa Cruz Biotechnology. The anti-HA antibody for immunoblot was purchased from Roche. The anti-HA (clone 16B12; 901515) and anti-FLAG (Clone L5; 637301) antibodies used for immunoprecipitation were purchased from BioLegend. The anti-α-tubulin (T6074) antibody, anti-FLAG antibody for immunoblot (clone M2, F1804), and the anti-β-actin antibody (A228) were from Sigma-Aldrich. Protein-fragment complementation assay lysates were probed with mouse anti-Rluc antibody (Chemi-Con, #MAB4400) against Rluc-F[2]. HRP-conjugated anti-mouse (7076), anti-rabbit (7074), and anti-rat (7077) secondary antibodies were from Cell Signaling Technologies. Gö6976 (115), Gö6983 (116), PDBu, UTP, CHX, Torin1, and Calyculin A were purchased from Calbiochem. GDC-0068 (117) (S2808) was purchased from Selleck Chemicals.
In Vitro Kinase Assays
In vitro kinase assays were performed similarly to mTOR kinase assays described previously (27, 118). Briefly, for the mTORC2 kinase assays experiments, due to poor stability of mTORC2 complex, kinase assays were performed on the same day of the isolation of the complex. After 48 h transfection of HEK293t cells with the HA-Sin1 plasmid, cells were lysed in CHAPS buffer (40 mM HEPES, pH 7.4, 120 mM NaCl, 1 mM EDTA, 0.3% CHAPS, 50 mM NaF, 5 mM sodium pyrophosphate (Na4P2O7). 10 mM sodium glycerol phosphate (C3H7Na2O6P), 1 mM DTT, 1 mM Na3VO4, 1 mM PMSF, 50 μg/mL leupeptin, 1 μM microcystin, and 2 mM benzamidine). Lysates were incubated for 30 min on ice and centrifuged at 13000 x g for 15 min at 4 °C. Supernatants were collected and quantified by Bradford assay, and 5 mg of lysate were used per condition. 5 mg of lysate were incubated with 15 μl of HA beads (Thermo Scientific, Cat. Number #26181), previously washed three times with PBS, for 1h at 4 °C in a nutator. Samples were centrifuged at 500 x g for 5 min at 4 °C, and pellets washed 6 times with CHAPS buffer. Then, pellets were washed twice with kinase assay buffer (25 mM HEPES, pH 7.4, 50 mM KCl). Beads were incubated with 0.5 ug of the GST-tagged C-tails of PKC and kinase assay was performed for 30 min at 30 °C. The conditions of the kinase assay were: 25 mM HEPES, pH 7.4, 50 mM KCl, 0.5 μg substrate protein, 10 mM MgCl and 250 μM ATP in a final reaction volume of 25 μl. After the 30 min incubation at 30 °C, reactions were stopped by adding 5 μl of sample buffer 4X and boiled at 100 °C for 5 min (for western blot analysis) or by 500 x g for 5 min to separate the mTORC2 complex from the PKC C-tails (for mass spectrometry analysis). Reactions were analyzed by SDS-PAGE and immunoblotting as described.
Liquid Chromatography/Mass Spectrometry (LC/MS) Analysis
In vitro kinase assays were analyzed by LC/MS as follows. Samples were dried then resuspended in digest buffer (1 M urea, 50 mM HEPES, pH 8.5) for digestion with sequencing grade trypsin (Promega – 6h, 37 °C). Digested peptides were desalted with Sep-Paks (Waters), dried, then resuspended in 5% formic acid/5% acetonitrile for LC/MS analysis. LC/MS analysis was conducted as previously described (119) with the following modifications. Peptides were eluted on a gradient of 85 mins and only MS1 and MS2 scans were taken. Raw files were searched against the human Uniprot database (downloaded: 03/2019) using the Sequest algorithm with a decoy search conducted on sequences in reverse order to filter matches to an FDR < 1%. Mass tolerances were set to 50 ppm for MS1 and 0.6 Da for MS2. Oxidation of methionine and phosphorylation of serine, threonine and tyrosine were specified as variable modifications. Quantification of phospho-peptides and proteins was based on MS1 area under the curve. The representative spectrum of PKCβII T634 phosphorylation from mouse brains was derived from an unpublished phospho-proteomic dataset. All procedures involving animals were approved by The Scripps Research Institute’s Institutional Animal Care and Usage Committee (IACUC) and met the guidelines of the National Institute of Health detailed in the Guide for the Care and Use of Laboratory Animals (120).
Luciferase PCA Analyses
HEK293 cells were grown in DMEM supplemented with 10% FBS. We transiently overexpressed indicated versions of the Rluc-PCA–based reporter in 24-well plate formats. Transient transfections were performed with TransFectin reagent (Bio-Rad, #1703352) according to manufacturer’s instructions. Experiments were performed 48 h post transfection. For the luciferase-PCA measurements, the growth medium was carefully removed and the cells were washed with phosphate-buffered saline (PBS). Cell suspensions were transferred to 96-well plates and subjected to luminescence analysis using the PHERAstar FSX (BMG Labtech). Luciferase luminescence signals were integrated for 10s following addition of the Rluc substrate benzyl-coelenterazine (NanoLight, #301).
Molecular Modeling
Molecular docking of the Sin1 CRIM domain (PDB ID: 2RVK) (59) and PKCβII (PDB ID: 2I0E) (58) was performed using ClusPro (121) without any restrictions and using the default settings. Structure diagrams were generated with PyMOL (122). Kinome tree (111) plots were generated using CORAL (123).
Peptide Arrays
Peptide arrays were generated by the INTAVIS MultiPep RS peptide synthesizer (INTAVIS Bioanalytical Instruments AG, Koeln, Germany) using standard Fmoc (9-fluorenylmethoxycarbonyl) protection-based solid phase peptide synthesis to produce short, overlapping 15-mer or 18-mer peptides directly conjugated onto amino-PEG modified cellulose membranes (ACS01, Intavis AG). Each peptide spot sequence corresponds to a segment of the rat PKCβII kinase domain and C-terminal tail (296-673) by shifting in increments of one or three amino acids. Arrays were vacuum dried, sealed in plastic, frozen at −20 °C, rehydrated in 100% ethanol for 30 min, and washed three times with TBS-T before use. Array strips were blocked with 5% non-fat dry milk in TBS-T (0.0005% Tween 20) for 1 h at room temperature, overlaid with cell lysate from WT MEFs or HEK293t cells over-expressing Sin1, mTOR, Raptor, or PDK1 for 1 h, incubated with 1:1000 HA, Myc, or mTOR primary antibody for 1 h at room temperature, incubated with 1:5000 HRP-conjugated secondary antibody for 1h at room temperature, and developed by immunoblot protocol described above. Arrays were washed three times for 5 min each with TBS-T between each incubation.
Plasmids and Constructs
The C Kinase Activity Reporter (CKAR) (47) and CKAR2 (48), and B Kinase Activity Reporter (BKAR) (64) were previously described. mCherry-tagged constructs were cloned into pcDNA3 with mCherry at the N-terminus at the BamHI and XbaI sites. mYFP-tagged constructs were cloned into pcDNA3 with mYFP at the N-terminus at the XhoI and XbaI sites. HA-tagged constructs were cloned into pcDNA3 with HA at the N-terminus at the NotI and XbaI sites. FLAG-tagged constructs were cloned into pCMV at the N-terminus at the NotI and XbaI sites. Kinameleon was cloned into pcDNA3 as mYFP-PKCβII-mCFP. Point mutations were generated by site-directed mutagenesis. GST constructs of the C-terminal tail of PKCβII (601-673) with specified mutations (T634A, T641A, S660A, T634E, T634E/S660A), and GST-PIF, were generated as described previously (68). MyrPalm constructs contain N-terminal Lyn kinase (MGCIKSK) sequence. Myr constructs contain the N-terminal src sequence (MGSSKSKPK). The Rluc-PCA–based hybrid proteins were designed as previously described (72). In brief, the coding regions of RIα (NP_037313.1) and PKCβII (NP_002729.2); cDNA template was kindly provided by Gottfried Baier) were PCR amplified and cloned into an eukaryotic expression vector and fused either N- or C-terminally with -F[1] or -F[2] of the Rluc-PCA. PKC chimera fusion proteins were generated by cloning the N-terminus of mTORC2-independent PKCδ, including the regulatory domains, kinase domain, and various fragments of the C-tail as indicated, to C-terminal fragments of the PKCβII C-tail to create a full-length chimera at various C-tail junctions. Chimeras were cloned pcDNA3 vectors with mCherry tags. An identical approach was used for the PKCθ/β chimera. A site-directed mutagenesis approach was used to generate the T634A/T641A double-mutant of PKCβII. The Rluc-PCA–based hybrid proteins F[1]-PKCβII-F[2] and F[1]-S6Kinase-1-F[2] (NP_003152.1) were generated using an identical cloning approach as described previously (70). A site-directed mutagenesis approach was used to generate the T634A/T641A, T634D/T641D, and T634E/T641E double-mutants of PKCβII as well as the T387A/S394A, T387D/S394D, and T387E/S394E double-mutants of S6Kinase. Catalytic domains of murine PKN2 (UniProtID Q8BWW9, 645-983) and PKN1 (UniProtID P70268, 607-946) were cloned into pcDNA™3.1/Zeo vector (Thermo Fisher) with N-terminal FLAG tag. Cloning and mutagenesis were performed using In-Fusion HD Cloning Kit (Clontech) and verified by sequencing.
Pulse-chase & Immunoprecipitation Experiments
For Pulse-Chase experiments, COS7 cells were incubated with Met/Cys-deficient DMEM (Corning) with 10% FBS for 30 min at 37 °C. The cells were then pulse-labeled with 0.5 mCi/ml [35S]Met/Cys (Perkin Elmer) in Met/Cys-deficient DMEM for 7 min at 37 °C, media was removed, cells were washed with dPBS (Corning), and cells were chased with DMEM culture media (Corning) containing 200 mM unlabeled methionine and 200 mM unlabeled cysteine and 10% FBS. At the indicated times, cells were lysed in PPHB and centrifuged at 13,000 x g for three min at 22 °C, supernatants were pre-cleared for 30 min at 4 °C with Protein A/G Beads (Santa Cruz), and protein complexes were immunoprecipitated from the supernatant with 1:100 dilution of either an anti-HA or anti-FLAG monoclonal antibody (BioLegend, 16B12; BioLegend L5) overnight at 4°C. The immune complexes were collected with Protein A/G Beads (Santa Cruz) for 2 h at 4 °C, washed 4 times with PPHB, separated by SDS-PAGE, transferred to PVDF membrane (Biorad), and analyzed by autoradiography and western blot. GST pull-down experiments utilized a similar approach with Glutathione Resin (Pierce; 16101) for purification.
Purification of GST-tagged C-tails of PKC for in Vitro Kinase Assays
After 48 h transfection of HEK293t cells with effectene with the GST-PKCβ C-tail (601-673) WT or T634A plasmids, cells were lysed in lysis buffer (20 mM Tri, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate (Na4P2O7), 1 mM β-glycerol phosphate, 1 mM DTT, 1 mM Na3VO4, 1 mM PMSF, 50 μg/mL leupeptin, 1 μM microcystin, and 2 mM benzamidine). Lysates were incubated for 10 min on ice and centrifuged at 13000 x g for 10 min at 4 °C. Supernatants were incubated with 65 μl of GST beads (Pierce; 16101) previously washed three times with PBS, for 1 h at 4 °C in a nutator. Samples were centrifuged at 500 x g for 2 min at 4 °C, and pellets washed 2 times with lysis buffer and 2 times with elution buffer (50 mM Hepes, pH 7.5, 100 mM NaCl, 5 mM β-glycerol phosphate, 1 mM DTT, 0.01% NP-40 and 10% glycerol). Beads were incubated with glutathione 10 mM in elution times at 4 °C for 15 min, centrifuged at 500 x g for 2 min at 4 °C and supernatants collected. This step was repeated three more times to increase the yield. Purified protein was exchanged into 20 mM Hepes, pH 7.5, 1 mM EDTA, 1 mM EGTA, and 1 mM DTT using 10-kDa Amicon centrifugal filter unit (EMD Millipore). Protein concentration was determined using BSA standards on an SDS/PAGE gel stained with Coomassie Brilliant Blue stain.
Stapled Peptide Experiments
Peptide synthesis: peptides used in this study were synthesized using Fmoc (fluorenylmethoxycarbonyl)-protected solid phase peptide synthesis on Rink Amide MBHA resin. Synthesis was performed in NMP (1-methyl-2-pyrrolidinone). Deprotections were performed using 25% v/v solution of piperidine in NMP for30 minutes. Amino acid coupling was carried out by adding 10 equivalents of Fmoc-protected amino acid (0.25 M final concentration) along with HCTU [O-(1H-6-Chlorobenzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate] in NMP (0.24 M final concentration) followed by 8% v/v DIEA (N, N-Diisopropylethylamine) for 45 min. The olefinic amino acid S 5 [Fmoc-(S)-2-(4-pentenyl)alanine] was incorporated as denoted in the sequences below using standard coupling conditions at i, i+4 positions. Ring closing metathesis was carried out on protected and resin bound peptides using 0.4 equivalents of Grubb’s 1st Generation catalyst [Benzylidene-bis(tricyclohexylphosphine)dichlororuthenium] in 1,2-dichloroethane for 1 hour. Olefin metathesis was performed twice to ensure completion of staple formation. N-terminal addition of PEG three (Fmoc-11-amino-3,6,9-trioxaundecanoic acid, denoted as asterisks in sequences below) was conducted with 4 molar equivalents under standard coupling conditions. Peptides were biotin labeled at the N-terminus using 10 equivalents of D-biotin, 0.14 M HCTU and 4% v/v DIEA in a 1:1 mixture of DMF and DMSO (dimethyl sulfoxide) overnight at room temperature. Peptides were cleaved from resin using 95% TFA (trifluoroacetic acid), 2.5% water and 2.5% TIS (triisopropylsilane) for 4 h. Products were then precipitated in ice cold MTBE (methyl-tert-butyl ether) and air dried. Crude products were dissolved in methanol and purified using high performance liquid chromatography over a 10-100% acetonitrile gradient. Products were verified by mass spectroscopy and quantified by measuring diminished absorbance of HABA[2-(4’-hydroxybenzeneazo)benzoic acid]-avidin complex at 500 nm. Molecular weights of the purified products are as follows: S1: Biotin-PEG3-KRNA*NFD*FFTRHK, actual mass 2345.4 (expected mass: 2345.8); and S2: Biotin-PEG3-KRNAENFD*FFT*HK; actual mass 2318.2 (expected mass= 2318.7).
Transient transfection of 3XFlag-rPKCβII was carried out using Effectene® transfection reagent (Qiagen) for 24 h. At the same time of transfection, HEK293t cells were treated with vehicle (DMSO) or one of the two stapled peptides (S1 or S2) (10 μM) designed to bind PKC and disrupt its dimerization. Cells were incubated at 37°C in 5% CO2 for 24 h and then lysed in 50 mM Tris, pH 7.4, 1% Triton X-100, 50 mM NaF, 10 mM Na4P2O7, 100mM NaCl, 5mM EDTA, 1 mM Na3VO4, 1 mM PMSF, 50 μg/mL leupeptin, 1 μM microcystin, and 2 mM benzamidine. Homogenates were sonicated and protein was quantified using a BCA protein assay kit (Thermo Fisher Scientific). Thirty micrograms of protein were separated by standard SDS/PAGE and transferred to PVDF membranes (BioRad). Membranes were blocked with 5% BSA or 5% milk for 1 h at room temperature and analyzed by immunoblotting with specific antibodies. Detection of immunoreactive bands was performed using chemiluminescence on a FluorChemQ imaging system (Alpha Innotech).
Quantification and Statistical Analysis
Statistical significance was determined using Repeated Measures One-Way ANOVA and Brown-Forsythe Test or Student’s t-test performed in GraphPad Prism 6.0a (GraphPad Software). The half-time of translocation was calculated by fitting the data to a non-linear regression using a one-phase exponential association equation with Graph Pad Prism 6.0a (GraphPad Software). Area under the curve (AUC) calculations were performed in GraphPad Prism 6.0a (GraphPad Software). Western blots and Autoradiographs were quantified by densitometry using the AlphaView (Protein Simple) and ImageJ software, respectively. PKC phosphorylation (%) was determined by measuring the proportion of phosphorylated PKC (slower mobility band) over total PKC by densitometry.
Supplementary Material
Fig. S1. Characterization of PKC Defect in TORC2-Deficient Cells
Fig. S2. Determinants for mTORC2 Association with PKC
Fig. S3. Active-Site Tether Region of PKC C-tail Confers mTORC2 Dependence
Fig. S4. Molecular Modeling of the Sin1:PKC Interaction
Fig. S5. Mass Spectrometry Analysis of mTORC2 Kinase Assays
Fig. S6. TOR-Interaction Motif Phosphorylation and Evolutionary Conservation
Fig. S7. Conservation of the TOR-Interaction Motif with TOR Pathway Components
Fig. S8. Protein Kinase N Dependence on TOR-Interaction Motif and Turn Motif Sites
Fig. S9. mTORC2 Recruits PDK1 to the PKC C-tail
Fig. S10. TOR-Interaction Motif Mutations Affect Kinase Conformations
Fig. S11. Regulatory Domains Attenuate PKC Dimerization
Acknowledgments:
We would like to thank Andrea Raffeiner for technical assistance (cloning & initial PCA experiments).
Funding:
This work was supported by NIH R35 GM122523 and NIH GM43154 to A.C.N.; NIH GM134168 to EJK; and NIH R01GM114409 and U01CA239106 to N. K. T.R.B. was supported by the PhRMA Foundation Pre Doctoral Fellowship in Pharmacology Toxicology (#20183844) and the UCSD Graduate Training Program in Cellular and Molecular Pharmacology (T32 GM007752). J.M.W. was supported by the UCSD Graduate Training Programs in Cellular and Molecular Pharmacology (T32 GM007752) and Rheumatic Diseases Research (T32 AR064194). E.S. was supported by grants from the Austrian Science Fund (P30441, P32960).
Footnotes
Competing interests: The authors declare they have no competing interests.
Data and materials availability: The phosphoproteomics data presented in Figure 3I has been deposited to ProteomeXchange (PXD022233) and MassIVE (MSV000086380). All other data needed to evaluate the conclusions in the paper are present in the Main Text or the Supplementary Materials.
References and Notes:
- 1.Nolen B, Taylor S, Ghosh G, Regulation of protein kinases: Controlling activity through activation segment conformation. Mol. Cell 15 (2004), pp. 661–675. [DOI] [PubMed] [Google Scholar]
- 2.Pearce LR, Komander D, Alessi DR, The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. cell Biol 11 (2010), doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 3.Leroux AE, Schulze JO, Biondi RM, AGC kinases, mechanisms of regulation and innovative drug development. Semin. Cancer Biol 48, 1–17 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Mora A, Komander D, Van Aalten DMF, Alessi DR, PDK1, the master regulator of AGC kinase signal transduction. Semin. Cell Dev. Biol 15, 161–170 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PRJ, Reese CB, Cohen P, Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Bα. Curr. Biol 7, 261–269 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Dutil EM, Toker A, Newton AC, Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr. Biol 8, 1366–1375. [DOI] [PubMed] [Google Scholar]
- 7.Keranen LM, Dutil EM, Newton AC, Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr. Biol 5, 1394–1403 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Pearson RB, Dennis PB, Han JW, Williamson NA, Kozma SC, Wettenhall RE, Thomas G, The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14, 5279–5287 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA, Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 15, 6541–51 (1996). [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards AS, Newton AC, Phosphorylation at conserved carboxyl-terminal hydrophobic motif regulates the catalytic and regulatory domains of protein kinase C. J. Biol Chem 272, 18382–18390(1997). [DOI] [PubMed] [Google Scholar]
- 11.Liu GY, Sabatini DM, mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol 21 (2020), pp. 183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ, Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science (80-. ). 281, 2042–2045 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan K-L, Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J. 27, 1919–1931 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, Sessa WC, Qin J, Zhang P, Su B, Jacinto E, The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J. 27, 1932–1943 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orr JW, Newton AC, Requirement for negative charge on “activation loop” of protein kinase C. J Biol Chem. 269, 27715–27718 (1994). [PubMed] [Google Scholar]
- 16.Bornancin F, Parker PJ, Phosphorylation of threonine 638 critically controls the dephosphorylation and inactivation of protein kinase Calpha. Curr Biol. 6, 1114–1123 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Oh WJ, Wu C, Kim SJ, Facchinetti V, Julien L-A, Finlan M, Roux PP, Su B, Jacinto E, mTORC2 can associate with ribosomes to promote cotranslational phosphorylation and stability of nascent Akt polypeptide. EMBO J. 29, 3939–3951 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chan TO, Rittenhouse SE, Tsichlis PN, AKT/PKB and Other D3 Phosphoinositide-Regulated Kinases: Kinase Activation by Phosphoinositide-Dependent Phosphorylation. Annu. Rev. Biochem 68, 965–1014 (1999). [DOI] [PubMed] [Google Scholar]
- 19.Ebner M, Lučić I, Leonard TA, Yudushkin I, PI(3,4,5)P 3 Engagement Restricts Akt Activity to Cellular Membranes. Mol. Cell 65, 416–431.e6 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Klippel A, Kavanaugh WM, Pot D, Williams LT, A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol. Cell. Biol 17, 338–344 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freeh M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA, High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J. Biol. Chem 272, 8474–8481 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Franke TF, Kaplan DR, Cantley LC, Toker A, Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science (80-. ). 275, 665–668 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Chu N, Salguero AL, Liu AZ, Chen Z, Dempsey DR, Ficarro SB, Alexander WM, Marto JA, Li Y, Amzel LM, Gabelli SB, Cole PA, Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis. Cell. 174, 897–907.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Cron P, Thompson V, Good VM, Hess D, Hemmings BA, Barford D, Molecular Mechanism for the Regulation of Protein Kinase B/Akt by Hydrophobic Motif Phosphorylation. Mol. Cell 9, 1227–1240 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Balasuriya N, Kunkel MT, Liu X, Biggar KK, Li SSC, Newton AC, O’Donoghue P, Genetic code expansion and live cell imaging reveal that Thr-308 phosphorylation is irreplaceable and sufficient for Akt1 activity. J. Biol. Chem 293, 10744–10756(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baffi TR, Katsenelson KC, Newton AC, Annu. Rev. Pharmacol. Toxicol, in press, doi: 10.1146/annurev-pharmtox-031820-122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM, Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Alessi DR, Pearce LR, García-Martínez JM, New insights into mTOR signaling: mTORC2 and beyond. Sci. Signal 2 (2009), pp. pe27–pe27. [DOI] [PubMed] [Google Scholar]
- 29.Su B, Jacinto E, Mammalian TOR signaling to the AGC kinases. Crit. Rev. Biochem. Mol. Biol 46 (2011), pp. 527–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toker A, Newton AC, Akt/protein kinase B is regulated by autophosphorylation at the hypothetical PDK-2 site. J. Biol. Chem 275, 8271–8274 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Behn-Krappa A, Newton AC, The hydrophobic phosphorylation motif of conventional protein kinase C is regulated by autophosphorylation. Curr. Biol 9, 728–737 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Balendran A, Casamayor A, Deak M, Paterson A, Gaffney P, Currie R, Downes CP, Alessi DR, PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol 9, 393–404 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Warfel NA, Niederst M, Newton AC, Disruption of the interface between the pleckstrin homology (PH) and kinase domains of Akt protein is sufficient for hydrophobic motif site phosphorylation in the absence of mTORC2. J Biol Chem. 286, 39122–39129 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Martinez JM, Alessi DR, mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J. 416, 375–385 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Yang C-S, Melhuish TA, Spencer A, Ni L, Hao Y, Jividen K, Harris TE, Snow C, Frierson HF, Wotton D, Paschal BM, The protein kinase C super-family member PKN is regulated by mTOR and influences differentiation during prostate cancer progression. Prostate. 77, 1452–1467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, Hall MN, Ohsumi Y, Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol. Cell. Biol 25, 7239–48 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou P-C, Rajput S, Zhao X, Patel C, Albaciete D, Oh WJ, Daguplo HQ, Patel N, Su B, Werlen G, Jacinto E, mTORC2 Is Involved in the Induction of RSK Phosphorylation by Serum or Nutrient Starvation. Cells. 9, 1567 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roelants FM, Leskoske KL, Martinez Marshall MN, Locke MN, Thorner J, The TORC2-Dependent Signaling Network in the Yeast Saccharomyces cerevisiae. Biomolecules. 7 (2017), doi: 10.3390/biom7030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isotani S, Hara K, Tokunaga C, Inoue H, Avruch J, Yonezawa K, Immunopurified mammalian target of rapamycin phosphorylates and activates p70 S6 kinase a in vitro. J. Biol Chem 21A, 34493–34498 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM, Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell. 11, 859–871 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM, RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci 95, 1432–1437 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM, Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 14, 1296–1302 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B, SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell. 127, 125–137 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Yang Q, Inoki K, Ikenoue T, Guan KL, Identification of Sin1 as an essential TORC2 component required for complex formation and kinase activity. Genes Dev. 20, 2820–2832 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frias MA, Thoreen CC, Jaffe JD, Schroder W, Sculley T, Carr SA, Sabatini DM, mSin1 Is Necessary for Akt/PKB Phosphorylation, and Its Isoforms Define Three Distinct mTORC2s. Curr. Biol 16, 1865–1870 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Baffi TR, Van A-AN, Zhao W, Mills GB, Newton AC, Protein Kinase C Quality Control by Phosphatase PHLPP1 Unveils Loss-of-Function Mechanism in Cancer. Mol. Cell 74, 378–392.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Violin JD, Zhang J, Tsien RY, Newton AC, A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J. Cell Biol 161, 899–909 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ross BL, Tenner B, Markwardt ML, Zviman A, Shi G, Kerr JP, Snell NE, McFarland JJ, Mauban JR, Ward CW, Rizzo MA, Zhang J, Single-color, ratiometric biosensors for detecting signaling activities in live cells. Elife. 7 (2018), doi: 10.7554/eLife.35458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS, An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gould CM, Antal CE, Reyes G, Kunkel MT, Adams RA, Ziyar A, Riveros T, Newton AC, Active site inhibitors protect protein kinase C from dephosphorylation and stabilize its mature form. J Biol Chem. 286, 28922–28930 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antal CE, Violin JD, Kunkel MT, Skovso S, Newton AC, Intramolecular conformational changes optimize protein kinase C signaling. Chem Biol. 21, 459–469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iyer GH, Moore MJ, Taylor SS, Consequences of lysine 72 mutation on the phosphorylation and activation state of cAMP-dependent kinase. J. Biol. Chem 280, 8800–7 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Dries DR, Gallegos LL, Newton AC, A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J Biol Chem. 282, 826–830 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Dutil EM, Dual Role of Pseudosubstrate in the Coordinated Regulation of Protein Kinase C by Phosphorylation and Diacylglycerol. J. Biol. Chem 275, 10697–10701 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Sarbassov DD, Ali SM, Sabatini DM, Growing roles for the mTOR pathway. Curr. Opin. Cell Biol 17 (2005), pp. 596–603. [DOI] [PubMed] [Google Scholar]
- 56.Yan L, Mieulet V, Lamb RF, mTORC2 is the hydrophobic motif kinase for SGK1. Biochem. J 416 (2008), pp. e19–e21. [DOI] [PubMed] [Google Scholar]
- 57.Kannan N, Haste N, Taylor SS, Neuwald AF, The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc. Natl. Acad. Sci. U. S. A 104, 1272–1277 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grodsky N, Li Y, Bouzida D, Love R, Jensen J, Nodes B, Nonomiya J, Grant S, Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry. 45, 13970–13981 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Tatebe H, Murayama S, Yonekura T, Hatano T, Richter D, Furuya T, Kataoka S, Furuita K, Kojima C, Shiozaki K, Substrate specificity of TOR complex 2 is determined by a ubiquitin-fold domain of the Sin1 subunit. Elife. 6 (2017), doi: 10.7554/eLife.19594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cameron AJM, Linch MD, Saurin AT, Escribano C, Parker PJ, mTORC2 targets AGC kinases through Sin1-dependent recruitment. Biochem. J 439, 287–297 (2011). [DOI] [PubMed] [Google Scholar]
- 61.Sweatt JD, Atkins CM, Johnson J, English JD, Roberson ED, Chen S-J, Newton A, Klann E, Protected-Site Phosphorylation of Protein Kinase C in Hippocampal Long-Term Potentiation. J. Neurochem 71, 1075–1085 (2002). [DOI] [PubMed] [Google Scholar]
- 62.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E, PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tatebe H, Shiozaki K, Evolutionary Conservation of the Components in the TOR Signaling Pathways. Biomolecules. 7 (2017), doi: 10.3390/biom7040077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kunkel MT, Ni Q, Tsien RY, Zhang J, Newton AC, Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem 280, 5581–5587 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Romano RA, Kannan N, Kornev AP, Allison CJ, Taylor SS, A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 18, 1486–97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frödin M, Antal TL, Dümmler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM, A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 21, 5396–5407 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biondi RM, Cheung PC, Casamayor A, Deak M, Currie RA, Alessi DR, Identification of a pocket in the PDK1 kinase domain that interacts with PIF and the C-terminal residues of PKA. EMBO J. 19, 979–88 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gao T, Toker A, Newton AC, The Carboxyl Terminus of Protein Kinase C Provides a Switch to Regulate Its Interaction with the Phosphoinositide-dependent Kinase, PDK-1. J. Biol. Chem 276, 19588–19596 (2001). [DOI] [PubMed] [Google Scholar]
- 69.Enzler F, Tschaikner P, Schneider R, Stefan E, KinCon: Cell-based recording of full-length kinase conformations. IUBMB Life. 72, 1168–1174 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Röck R, Mayrhofer JE, Torres-Quesada O, Enzler F, Raffeiner A, Raffeiner P, Feichtner A, Huber RG, Koide S, Taylor SS, Troppmair J, Stefan E, BRAF inhibitors promote intermediate BRAF(V600E) conformations and binary interactions with activated RAS. Sci. Adv 5, eaav8463 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stefan E, Aquin S, Berger N, Landry CR, Nyfeler B, Bouvier M, Michnick SW, Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl. Acad. Sci. U. S. A 104, 16916–16921 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Röck R, Bachmann V, Bhang HEC, Malleshaiah M, Raffeiner P, Mayrhofer JE, Tschaikner PM, Bister K, Aanstad P, Pomper MG, Michnick SW, Stefan E, In-vivo detection of binary PKA network interactions upon activation of endogenous GPCRs. Sci. Rep 5, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schafmeister CE, Po J, Verdine GL, An all-hydrocarbon cross-linking system for enhancing the helicity and metabolic stability of peptides [8]. J. Am. Chem. Soc 122 (2000), pp. 5891–5892. [Google Scholar]
- 74.Verdine GL, Hilinski GJ, in Methods in Enzymology (Academic Press Inc., 2012), vol. 503, pp. 3–33. [DOI] [PubMed] [Google Scholar]
- 75.Lemmon MA, Schlessinger J, Cell signaling by receptor tyrosine kinases. Cell. 141 (2010), pp. 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Flint AJ, Paladini RD, Koshland DE, Autophosphorylation of protein kinase C at three separated regions of its primary sequence. Science. 249, 408–11 (1990). [DOI] [PubMed] [Google Scholar]
- 77.Keshwani MM, Klammt C, von Daake S, Ma Y, Kornev AP, Choe S, Insel PA, Taylor SS, Cotranslational cis-phosphorylation of the COOH-terminal tail is a key priming step in the maturation of cAMP-dependent protein kinase. Proc Natl Acad Sci U S A. 109, E1221–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hauge C, Antal TL, Hirschberg D, Doehn U, Thorup K, Idrissova L, Hansen K, Jensen ON, Jorgensen TJ, Biondi RM, Frodin M, Mechanism for activation of the growth factor-activated AGC kinases by turn motif phosphorylation. Embo j. 26, 2251–2261 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Toker A, Newton AC, Cellular signaling: Pivoting around PDK-1. Cell 103 (2000), pp. 185–188. [DOI] [PubMed] [Google Scholar]
- 80.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M, A Human Interactome in Three Quantitative Dimensions Organized by Stoichiometries and Abundances. Cell. 163, 712–723 (2015). [DOI] [PubMed] [Google Scholar]
- 81.Ali SM, Sabatini DM, Structure of S6 kinase 1 determines whether raptor-mTOR or rictor-mTOR phosphorylates its hydrophobic motif site. J. Biol Chem 280, 19445–19448 (2005). [DOI] [PubMed] [Google Scholar]
- 82.Schalm SS, Blenis J, Identification of a conserved motif required for mTOR signaling. Curr. Biol 12, 632–9 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Romanelli A, Dreisbach VC, Blenis J, Characterization of phosphatidylinositol 3-kinase-dependent phosphorylation of the hydrophobic motif site Thr(389) in p70 S6 kinase 1. J. Biol Chem 277, 40281–9 (2002). [DOI] [PubMed] [Google Scholar]
- 84.Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G, Phosphorylation and activation of p70(s6k) by PDK1. Science (80-. ). 279, 707–710 (1998). [DOI] [PubMed] [Google Scholar]
- 85.Sabatini DM, Twenty-five years of mTOR: Uncovering the link from nutrients to growth. Proc. Natl. Acad. Sci. U. S. A 114 (2017), pp. 11818–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hauge C, Frödin M, RSK aand MSK in MAP kinase signalling. J. Cell Sci 119, 3021–3023 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Beenstock J, Mooshayef N, Engelberg D, How Do Protein Kinases Take a Selfie (Autophosphorylate)? Trends Biochem. Sci 41 (2016), pp. 938–953. [DOI] [PubMed] [Google Scholar]
- 88.Newton AC, Koshland DE, Protein kinase C autophosphorylates by an intrapeptide reaction. J. Biol. Chem 262, 10185–10188 (1987). [PubMed] [Google Scholar]
- 89.Dutil EM, Keranen LM, DePaoli-Roach AA, Newton AC, In vivo regulation of protein kinase C by trans-phosphorylation followed by autophosphorylation. J. Biol. Chem 269, 29359–62 (1994). [PubMed] [Google Scholar]
- 90.Cameron AJM, Escribano C, Saurin AT, Kostelecky B, Parker PJ, PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat. Struct. Mol. Biol 16, 624–630 (2009). [DOI] [PubMed] [Google Scholar]
- 91.Cole PA, Chu N, Salguero AL, Bae H, AKTivation mechanisms. Curr. Opin. Struct. Biol 59 (2019), pp. 47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Swanson CJ, Ritt M, Wang W, Lang MJ, Narayan A, Tesmer JJ, Westfall M, Sivaramakrishnan S, Conserved Modular Domains Team up to Latch-open Active Protein Kinase Cα. J. Biol. Chem 289, 17812–17829 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang S-M, Leventhal PS, Wiepz GJ, Bertics PJ, Calcium and Phosphatidylserine Stimulate the Self-Association of Conventional Protein Kinase C Isoforms †. Biochemistry. 38, 12020–12027 (1999). [DOI] [PubMed] [Google Scholar]
- 94.Ziemba BP, Li J, Landgraf KE, Knight JD, Voth GA, Falke JJ, Single-molecule studies reveal a hidden key step in the activation mechanism of membrane-bound protein kinase C-α. Biochemistry. 53, 1697–1713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hannun YA, Bell RM, Phorbol ester binding and activation of protein kinase C on triton X-100 mixed micelles containing phosphatidylserine. J. Biol. Chem . 261, 9341–9347 (1986). [PubMed] [Google Scholar]
- 96.Hui X, Sauer B, Kaestner L, Kruse K, Lipp P, PKCα diffusion and translocation are independent of an intact cytoskeleton. Sci. Rep 7 (2017), doi: 10.1038/S41598-017-00560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Antal CE, Callender JA, Kornev AP, Taylor SS, Newton AC, Intramolecular C2 Domain-Mediated Autoinhibition of Protein Kinase C βII. CellReports. 12, 1252–1260 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME, Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 275, 661–5 (1997). [DOI] [PubMed] [Google Scholar]
- 99.Datta K, Franke TF, Chan TO, Makris A, Yang SI, Kaplan DR, Morrison DK, Golemis EA, Tsichlis PN, AH/PH domain-mediated interaction between Akt molecules and its potential role in Akt regulation. Mol. Cell. Biol 15, 2304–10 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang L-C, Ross KE, Baffi TR, Drabkin H, Kochut KJ, Ruan Z, D’Eustachio P, McSkimming D, Arighi C, Chen C, Natale DA, Smith C, Gaudet P, Newton AC, Wu C, Kannan N, Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci. Rep 8 (2018), doi: 10.1038/s41598-018-24457-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baffi TR, Katsenelson KC, Newton AC, Annu. Rev. Pharmacol. Toxicol, in press, doi: 10.1146/annurev-pharmtox-031820-122108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Antal CE, Hudson AM, Kang E, Zanca C, Wirth C, Stephenson NL, Trotter EW, Gallegos LL, Miller CJ, Furnari FB, Hunter T, Brognard J, Newton AC, Cancer-associated protein kinase C mutations reveal kinase’s role as tumor suppressor. Cell. 160, 489–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Newton AC, Brognard J, Reversing the Paradigm: Protein Kinase C as a Tumor Suppressor. Trends Pharmacol Sci. 38, 438–447 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuan TL, Cantley LC, PI3K pathway alterations in cancer: variations on a theme. Oncogene. 27, 5497–510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tian T, Li X, Zhang J, mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int. J. Mol. Sci 20, 755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rodrik-Outmezguine VS, Okaniwa M, Yao Z, Novotny CJ, McWhirter C, Banaji A, Won H, Wong W, Berger M, De Stanchina E, Barratt DG, Cosulich S, Klinowska T, Rosen N, Shokat KM, Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature. 534, 272–276 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Uphoff CC, Drexler HG, in Cancer Cell Culture: Methods and Protocols, Cree IA, Ed. (Humana Press, Totowa, NJ, 2011; 10.1007/978-1-61779-080-5_8), pp. 93–103. [DOI] [Google Scholar]
- 108.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, Basic local alignment search tool. J. Mol. Biol 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 109.Katoh K, MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neuwald AF, Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics. 25, 1869–75 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, The protein kinase complement of the human genome. Science (80-. ). 298 (2002), pp. 1912–1934. [DOI] [PubMed] [Google Scholar]
- 112.Crooks GE, Hon G, Chandonia J-M, Brenner SE, WebLogo: A Sequence Logo Generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gallegos LL, Kunkel MT, Newton AC, Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem. 281, 30947–30956 (2006). [DOI] [PubMed] [Google Scholar]
- 114.Dutil EM, Toker A, Newton AC, Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr Biol. 8, 1366–1375 (1998). [DOI] [PubMed] [Google Scholar]
- 115.Martiny-Baron G, Kazanietz MG, Mischak H, Blumberg PM, Kochs G, Hug H, Marme D, Schachtele C, Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem 268, 9194–9197 (1993). [PubMed] [Google Scholar]
- 116.Gschwendt M, Dieterich S, Rennecke J, Kittstein W, Mueller HJ, Johannes FJ, Inhibition of protein kinase C μ by various inhibitors. Differentiation from protein kinase c isoenzymes. FEBS Lett. 392, 77–80 (1996). [DOI] [PubMed] [Google Scholar]
- 117.Lin J, Sampath D, Nannini MA, Lee BB, Degtyarev M, Oeh J, Savage H, Guan Z, Hong R, Kassees R, Lee LB, Risom T, Gross S, Liederer BM, Koeppen H, Skelton NJ, Wallin JJ, Belvin M, Punnoose E, Friedman LS, Lin K, Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin. Cancer Res 19, 1760–1772 (2013). [DOI] [PubMed] [Google Scholar]
- 118.Pearce LR, Sommer EM, Sakamoto K, Wullschleger S, Alessi DR, Protor-1 is required for efticient mT0RC2-mediated activation of SGK1 in the kidney. Biochem. J 436, 169–179 (2011). [DOI] [PubMed] [Google Scholar]
- 119.Lapek JD, Lewinski MK, Wozniak JM, Guatelli J, Gonzalez DJ, Quantitative temporal viromics of an inducible HIV-1 model yields insight to global host targets and phospho-dynamics associated with protein Vpr. Mol. Cell. Proteomics 16, 1447–1461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.N. R. C. (US) C. for the U. of the G. for the C. and U. of L. Animals, Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). [PubMed] [Google Scholar]
- 121.Kozakov D, Hall DR, Xia B, Porter KA, Padhorny D, Yueh C, Beglov D, Vajda S, The ClusPro web server for protein-protein docking. Nat. Protoc 12, 255–278 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schrodinger LLC, The PyMOL Molecular Graphics System, Version 1.8 (2015). [Google Scholar]
- 123.Metz KS, Deoudes EM, Berginski ME, Jimenez-Ruiz I, Aksoy BA, Hammerbacher J, Gomez SM, Phanstiel DH, Coral: Clear and Customizable Visualization of Human Kinome Data. Cell Syst. 7, 347–350.e1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gould CM, Kannan N, Taylor SS, Newton AC, The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem. 284, 4921–4935 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.McSkimming DI, Dastgheib S, Baffi TR, Byrne DP, Ferries S, Scott ST, Newton AC, Eyers CE, Kochut KJ, Eyers PA, Kannan N, KinView: a visual comparative sequence analysis tool for integrated kinome research. Mol. Biosyst 12 (2016), doi: 10.1039/c6mb00466k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.§,∥ Neil Grodsky, §,⊥ Ying Li, ∥ Djamal Bouzida, ∥ Robert Love, ⊥ Jordan Jensen, ⊥ Beverly Nodes, ⊥ and Jim Nonomiya, ⊥ Stephan Grant*, Structure of the Catalytic Domain of Human Protein Kinase C β II Complexed with a Bisindolylmaleimide Inhibitor‡ (2006), doi: 10.1021/BI061128H. [DOI] [PubMed] [Google Scholar]
- 127.Xu Z-B, Chaudhary D, Olland S, Wolfrom S, Czerwinski R, Malakian K, Lin L, Stahl ML, Joseph-McCarthy D, Benander C, Fitz L, Greco R, Somers WS, Mosyak L, Catalytic Domain Crystal Structure of Protein Kinase C-θ (PKCθ). J. Biol. Chem 279, 50401–50409 (2004). [DOI] [PubMed] [Google Scholar]
- 128.Van Eis MJ, Evenou JP, Floersheim P, Gaul C, Cowan-Jacob SW, Monovich L, Rummel G, Schuler W, Stark W, Strauss A, Von Matt A, Vangrevelinghe E, Wagner J, Soldermann N, 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes. Bioorganic Med. Chem. Lett 21, 7367–7372 (2011). [DOI] [PubMed] [Google Scholar]
- 129.Lin K, Lin J, Wu W-I, Ballard J, Lee BB, Gloor SL, Vigers GPA, Morales TH, Friedman LS, Skelton N, Brandhuber BJ, An ATP-site on-off switch that restricts phosphatase accessibility of Akt. Sci. Signal 5, ra37 (2012). [DOI] [PubMed] [Google Scholar]
- 130.Knighton D, Zheng J, Ten Eyck L, Ashford V, Xuong N, Taylor S, Sowadski J, Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science (80-. ). 253, 407–414 (1991). [DOI] [PubMed] [Google Scholar]
- 131.Leonard TA, Różycki B, Saidi LF, Hummer G, Hurley JH, Crystal Structure and Allosteric Activation of Protein Kinase C βII. Cell. 144, 55–66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Katoh T, Takai T, Yukawa T, Tsukamoto T, Watanabe E, Mototani H, Arita T, Hayashi H, Nakagawa H, Klein MG, Zou H, Sang BC, Snell G, Nakada Y, Discovery and optimization of 1,7-disubstituted-2,2-dimethyl-2,3-dihydroquinazolin-4(1H)-ones as potent and selective PKCθ inhibitors. Bioorganic Med. Chem 24, 2466–2475 (2016). [DOI] [PubMed] [Google Scholar]
- 133.Gibbs CS, Zoller MJ, Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J. Biol. Chem 266, 8923–31 (1991). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Characterization of PKC Defect in TORC2-Deficient Cells
Fig. S2. Determinants for mTORC2 Association with PKC
Fig. S3. Active-Site Tether Region of PKC C-tail Confers mTORC2 Dependence
Fig. S4. Molecular Modeling of the Sin1:PKC Interaction
Fig. S5. Mass Spectrometry Analysis of mTORC2 Kinase Assays
Fig. S6. TOR-Interaction Motif Phosphorylation and Evolutionary Conservation
Fig. S7. Conservation of the TOR-Interaction Motif with TOR Pathway Components
Fig. S8. Protein Kinase N Dependence on TOR-Interaction Motif and Turn Motif Sites
Fig. S9. mTORC2 Recruits PDK1 to the PKC C-tail
Fig. S10. TOR-Interaction Motif Mutations Affect Kinase Conformations
Fig. S11. Regulatory Domains Attenuate PKC Dimerization