Abstract
The hypocotyls of germinating seedlings elongate in a search for light to enable autotrophic sugar production. Upon exposure to light, photoreceptors that are activated by blue and red light halt elongation by preventing the degradation of the hypocotyl-elongation inhibitor HY5 and by inhibiting the activity of the elongation-promoting transcription factors PIFs. The question of how sugar affects hypocotyl elongation and which cell types stimulate and stop that elongation remains unresolved. We found that overexpression of a sugar sensor, Arabidopsis hexokinase 1 (HXK1), in guard cells promotes hypocotyl elongation under white and blue light through PIF4. Furthermore, expression of PIF4 in guard cells is sufficient to promote hypocotyl elongation in the light, while expression of HY5 in guard cells is sufficient to inhibit the elongation of the hy5 mutant and the elongation stimulated by HXK1. HY5 exits the guard cells and inhibits hypocotyl elongation, but is degraded in the dark. We also show that the inhibition of hypocotyl elongation by guard cells’ HY5 involves auto-activation of HY5 expression in other tissues. It appears that guard cells are capable of coordinating hypocotyl elongation and that sugar and HXK1 have the opposite effect of light on hypocotyl elongation, converging at PIF4.
Subject terms: Plant physiology, Light responses, Stomata, Plant molecular biology, Plant morphogenesis
Kelly et al. show that Arabidopsis hexokinase1 (HXK1) expressed in guard-cells is sufficient to drive hypocotyl elongation through increasing the activity of PIF4 and auxin level, and competing with the effects of HY5. This study provides insights into how light and sucrose antagonistically coordinate the effort to achieve the height necessary for efficient photosynthetic, autotrophic sugar production.
Introduction
The growth of seedlings from germinating seeds is dependent on the conversion of seed carbon reserves to sugar, usually sucrose, and the movement of that sugar to the developing hypocotyl and cotyledons1–3. The hypocotyls of germinating seeds elongate in the dark, in a search for light that will allow the cotyledons to switch from relying on reserve generated sugars to autotrophic photosynthetic sugar production. Stomata, composed of two guard cells, appear on the cotyledons soon after germination, to allow gas exchange for photosynthesis and sugar production, once the seedlings reach the light4,5. In dark-grown Arabidopsis seedlings, CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1, an E3 ubiquitin ligase)/SUPPRESSOR OF PHYA (SPA) and DE-ETIOLATED 1 (DET1) complexes act as master suppressors of light signaling6–8. These complexes target elongation suppressors such as ELONGATED HYPOCOTYL5 (HY5) for degradation, thereby enabling hypocotyl elongation9–11. The hypocotyl elongation is mediated by PHYTOCHROME INTERACTING TRANSCRIPTION FACTORs (PIFs), which stimulate the production of the auxin that is necessary to promote hypocotyl elongation11,12. Light halts hypocotyl elongation through the activation of blue-light photoreceptors called cryptochromes (CRY) and red-light photoreceptors called phytochromes (PHY)13. Light-activated photoreceptors interfere with the activity of the COP1/SPA and DET1 complexes, preventing the degradation of the hypocotyl elongation inhibitor HY5 and blocking the transcriptional activity of PIFs, thereby inhibiting hypocotyl elongation6,10,12,14–19.
Shade (a low ratio of red/far red light) and low levels of blue light also promote hypocotyl elongation via PIFs20,21. Those conditions are sensed by the photoreceptors in the cotyledons, which drive the synthesis of auxin, which is transported to the hypocotyl to induce hypocotyl elongation22–26. In the hypocotyl, auxin stimulates the synthesis of brassinosteroids (BR) and the responses that are required for hypocotyl elongation27,28. Evidence suggests that the epidermis is a central player in the regulation of hypocotyl elongation. The hypocotyls of photochrome B mutants (phyB) elongate in red light and expression of PHYB in the epidermis of phyB mutant is sufficient to prevent elongation in red light, suggesting that red-light perception occurs in the epidermis and prevents both auxin production and the transport of auxin out of the cotyledon29. Suppression of BR biosynthesis in the epidermis restricted shoot growth; whereas restoring BR biosynthesis or BR receptors in the epidermis of BR mutants rescued the dwarf phenotype28. High temperatures also cause hypocotyl elongation and the epidermis coordinates thermo-responsive growth through the PHYB-PIF4-auxin pathway30. These studies suggest that the epidermis plays a role in hypocotyl elongation. However, the epidermis is composed of epidermal pavement and guard cells and it is not clear which of those two types of cells is involved in the elongation signals.
In addition to light, the role of sugars in hypocotyl elongation has also been studied. Sucrose and glucose have been shown to stimulate auxin biosynthesis in Arabidopsis and several studies have observed that external sugars such as sucrose or glucose promote hypocotyl elongation, which is mediated by PIF transcription factors and auxin31–37. Sucrose is a disaccharide that must be cleaved, yielding the hexose monomers glucose and fructose, which must be phosphorylated before they can be metabolized38. There are only two distinct groups of enzymes that can phosphorylate the glucose and fructose: hexokinases (HXK) and fructokinases (FRK)38. HXKs phosphorylate both glucose and fructose; whereas FRKs are specific to fructose39. HXK is an important sugar sensor that monitors sugar levels in various tissues, in addition to its enzymatic activity40–43. For example, the Arabidopsis HXK1 inhibits the expression of photosynthetic genes in mesophyll cells in response to high sugar levels40–42, and HXK1 within guard cells controls guard-cell behavior and reduces stomatal apertures in response to high sugar levels, thereby coordinating sugar production with transpiration43–46. Previous studies have examined the behavior of the HXK1 mutant (gin2) in terms of hypocotyl elongation and auxin levels in response to sugar treatments and reached ambiguous conclusions, perhaps due to redundant activity with the other HXKs, HXK2, and HXK334–38.
In this study, we took a different approach in which we overexpressed HXK1. We found that expression of HXK1, either globally or only in guard cells, stimulates hypocotyl elongation under long-day conditions. This HXK1-induced elongation is mediated by COP1, PIF4 and auxin signals, and competes with the effects of light and HY5. Furthermore, we show that increased expression of PIF4 or HY5 only in guard cells is sufficient to promote or inhibit hypocotyl elongation, respectively. Thus, HXK1 mediates the effects of sugar on hypocotyl elongation and guard cells alone are capable of controlling hypocotyl elongation.
Results
Hexokinase promotes hypocotyl elongation
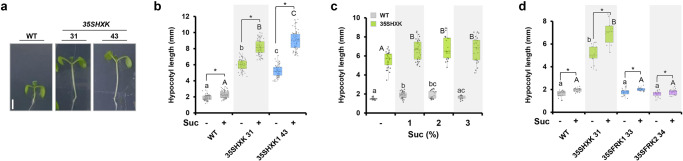
Sucrose promotes hypocotyl elongation in the presence of light32,33,35–37. Indeed, the hypocotyls of Arabidopsis seedlings grown under long-day conditions (16 h light/8 h dark, 40 μE) on plates containing 1% sucrose elongated about 20% further than the hypocotyls of similar seedlings grown on control plates without sucrose (Fig. 1b–d). To explore the role of the known sugar sensor HXK1 in sucrose-induced hypocotyl elongation, we measured the length of the hypocotyls of 7-day-old seedlings of independent lines overexpressing HXK1 under the 35S global promoter (35SHXK plants). Under long-day conditions, two previously described independent lines, 35SHXK31 and 35SHXK43, with high levels of HXK1 expression47, had hypocotyls that were about 3 times longer than those of WT seedlings (Fig. 1). Supplementation of the medium with 1% sucrose promoted the elongation of the 35SHXK lines about 35% further, suggesting that the sucrose-induced elongation effect is mediated by HXK1 (Fig. 1b–d). Higher concentrations (2–3%) of sucrose affected elongation much like 1% sucrose (Fig. 1c). Hypocotyl elongation was not observed in Arabidopsis plants overexpressing fructokinases, either FRK1 or FRK2, which are fructose-specific phosphorylating enzymes distinct from HXK (Fig. 1d)38,48. This suggests that the elongation observed in 35SHXK is specific to HXK and is not a metabolic or a general hexose-phosphorylation effect.
Fig. 1. Hexokinase promotes hypocotyl elongation.
a–d Seedlings were grown under long-day (16 h light/8 h dark), white-light (40 µmolm−2 s−1) conditions for 7 days. a Representative images of 7-day-old 35SHXK seedlings. Bar = 2 mm. b Hypocotyl lengths of 35SHXK seedlings grown with and without 1% sucrose. Data from two independent lines (#31, #43) are shown. c Hypocotyl lengths of 35SHXK seedlings treated with increasing concentrations of sucrose. d Hypocotyl lengths of 35SHXK, 35SFRK1 and 35SFRK2 seedlings grown with and without 1% sucrose. b–d The box plots extend from the first to third quartiles and the whiskers extend from the minimum to the maximum levels. Lines within the boxes signify median values and light gray dots represent individual data points (n = 55–70 for b; n = 20–30 for c; n = 15–25 for d). Different lower-case and capital letters indicate significant differences (Tukey’s HSD test, P < 0.05). Asterisks indicate significant differences between the compared treatments (t test, P < 0.05).
Hexokinase in guard cells is sufficient to promote hypocotyl growth
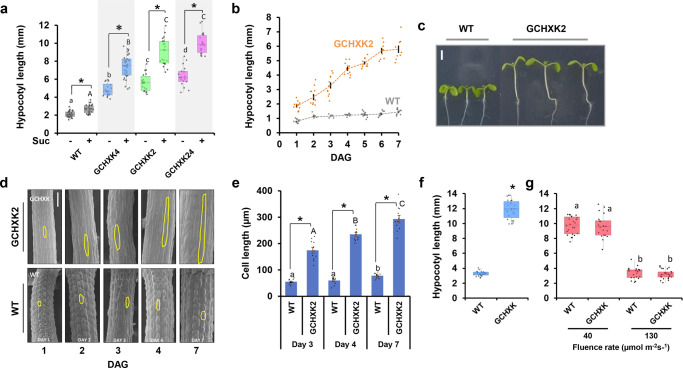
While a few studies have suggested that the epidermis affects hypocotyl elongation by generating hormonal cues22,23,27,28,30, the contribution of guard cells per se to hypocotyl elongation is not known. Stomata, each composed of two guard cells, appear on the epidermis of the cotyledons immediately after the emergence of the cotyledons from the seed coat5. To examine the involvement of guard cells in hypocotyl elongation, we used seedlings that express HXK1 specifically in their guard cells (named GCHXK plants for guard-cell HXK)44. Similar to 35SHXK seedlings, the hypocotyls of independent GCHXK lines (GCHXK2 and GCHXK4) were about three times longer than those of WT seedlings under long-day conditions (Fig. 2a–c), indicating that guard cells are sufficient to trigger HXK-mediated hypocotyl elongation. The hypocotyls of GCHXK2 seedlings were longer than those of GCHXK4 seedlings and the hypocotyls of the offspring of the cross between the two lines (referred to as GCHXK24) were slightly longer than those of the GCHXK2 seedlings, indicating a near maximal elongation effect of GCHXK (Fig. 2a). Like 35SHXK, the presence of 1% sucrose caused the hypocotyls to elongate significantly further (Fig. 2a). A day-by-day analysis showed that elongation of WT seedlings ceased almost entirely 2 days after germination, while that of GCHXK seedlings continued up to 6 days after germination (Fig. 2b). These results suggest that HXK stimulates elongation by extending the hypocotyl growth period, in line with an earlier study showing that sucrose extends the growth period of WT seedlings32. Scanning electron microscopy (SEM) has indicated that the hypocotyl elongation of GCHXK is due to cell elongation, as is the case in regular hypocotyl elongation (Fig. 2d, e)32.
Fig. 2. Guard-cell expression of hexokinase is sufficient to promote hypocotyl elongation.
a–g Seedlings were grown under long-day (16 h light/8 h dark, 40 μE) conditions for 7 days on 0.5× MS agar medium. a Hypocotyl length of GCHXK seedlings grown with and without 1% sucrose. Two independent lines (GCHXK2, GCHXK4) and homozygote offspring of the cross between those two lines (GCHXK24) are shown. b Hypocotyl lengths of GCHXK2 and WT seedlings measured for 7 days after germination. Data are means ± SE (n ≥ 7). Orange and gray dots represent individual data points for GCHXK2 and WT, respectively. c Representative images of 7-day-old WT and GCHXK2 seedlings; bar = 2 mm. d SEM images of WT and GCHXK hypocotyls taken at Days 1–4 and 7 following germination. Representative cell borders are highlighted, Bar = 100 µm. e Average cell length of WT and GCHXK epidermal cells at days 3, 4, and 7 following germination. Data are means ± SE (n ≥ 12). Gray dots represent individual data points. f, g Hypocotyl lengths of WT and GCHXK seedlings under (f) blue and (g) red light conditions with 1% sucrose. Light intensities were kept at 14 μE for blue light and 40 or 130 μE for red light. a, e, g Different lower-case and capital letters indicate significant differences (Tukey’s HSD test, P < 0.05). Asterisks indicate a significant difference relative to the control (a) or relative to WT (e, f, t test, P < 0.05). a, f, g The box plots extend from the first to third quartiles and the whiskers extend from the minimum to the maximum levels. Lines within the boxes signify median values and light gray dots represent individual data points (n = 20–35).
To explore the molecular response triggered by GCHXK, we performed a transcriptomic analysis of seedlings grown under long-day conditions (16 h light/8 h dark, 40 μE), 4 days after germination (Supplementary Data 1, 2). We identified 1011 differentially expressed genes (DEGs) that were downregulated (843 genes) or upregulated (168 genes) by GCHXK (Supplementary Figs. 1, 2, 3, 4). The pathways containing the highest number of DEGs are primarily associated with metabolic activity, the biosynthesis of secondary metabolites and carbon metabolism (Supplementary Fig. 2). Among the metabolic processes affected by GCHXK, a downregulation of photosynthesis-related pathways stood out (Supplementary Fig. 1). The transcript levels of genes involved in the light reaction step, as well as those of genes associated with the Calvin–Benson cycle pathway and carbonic anhydrases, were significantly lower, as also seen in the functional overview (Supplementary Fig. 1, 1-photosynthesis). Inhibition of photosynthesis during hypocotyl elongation was reported previously9,17 and it appears that the hypocotyl elongation promoted by GCHXK has a similar effect.
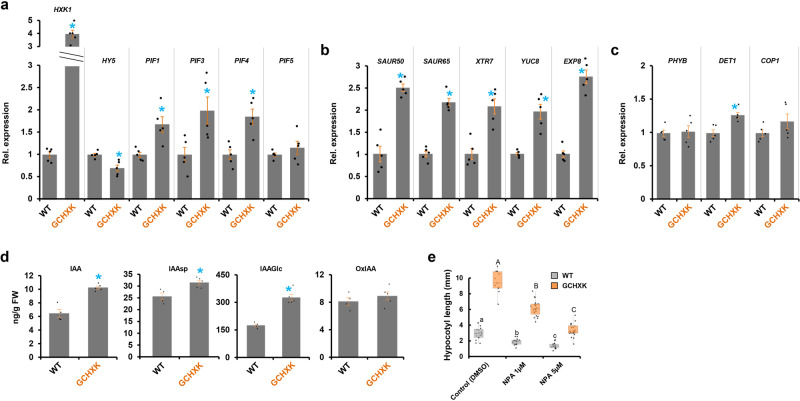
A qPCR analysis of hypocotyl elongation-related genes verified that the expression of HY5, a suppressor of hypocotyl elongation in the light, was lower in GCHXK plants. The expression of COP1 and PHYB remained unchanged, but the expression of DET1 (a repressor of HY5) and of PIF1,3,4, which promote hypocotyl elongation in the dark12, were higher (Fig. 3a, c). Along with the upregulation of PIFs, the expression of the auxin-responsive genes SMALL AUXIN UP RNA 50, 65 (SAUR50, SAUR65) and genes involved in cell elongation [XYLOGLUCAN ENDOTRANSGLUCOSYLASE-RELATED PROTEIN7 (XTR7) and EXPANSIN8 (EXP8)], whose expression is stimulated by PIF49,50, was also upregulated (Fig. 3b). The auxin biosynthesis gene YUCCA8 (YUC8) that promotes elongation in the dark49 was upregulated as well (Fig. 3b). Accordingly, auxin levels in GCHXK seedlings were higher than those in WT seedlings (Fig. 3d) and inhibiting auxin transport via the polar auxin transport inhibitor N-1-naphthylphthalamic acid (NPA) prevented the elongation of the hypocotyls of GCHXK seedlings (Fig. 3e). These results support the notion that GCHXK promotes hypocotyl elongation under light conditions via the known elongation-related molecular pathways that stimulate auxin production and signals.
Fig. 3. Relative expression of genes related to hypocotyl elongation, auxin levels, and auxin transport activity in GCHXK seedlings.
a–c RT-PCR expression analysis of genes related to the hypocotyl elongation of WT and GCHXK seedlings grown in 0.5 MS agar media. TUB2 (β-tubulin) was used for normalization and the expression level in the WT was set to one. Data points are mean ± SE (n = 5). d GCHXK promotes the accumulation of auxin. IAA, IAAsp, IAGlu, and OxIAA quantification in 5-day-old WT and GCHXK seedlings. Data points for are means ± SE (n = 4 and 5 for WT and GCHXK, respectively). a–d Black dots represent individual data points, and blue asterisks indicate a significant difference relative to the WT (t test, P < 0.05). e Auxin transport activity is required for hypocotyl growth of GCHXK. WT and GCHXK seedlings grown in 0.5× MS agar media containing 1 or 5 µM of NPA for 9 days. The box plots extend from the first to third quartiles and the whiskers extend from the minimum to the maximum levels. Lines within the boxes signify median values and light gray dots represent individual data points (n = 12–25). Different lower-case and capital letters indicate significant differences (Tukey’s HSD test, P < 0.05).
GCHXK stimulates elongation under blue light
Since either blue or red light can inhibit hypocotyl elongation, we tested whether GCHXK elongation under white light overcomes the inhibition caused by blue or red light. The hypocotyls of GCHXK seedlings grown in blue light (16 h light/8 h dark, 14 µE) were 4 times longer than those of WT seedlings, indicating that sucrose and GCHXK can overcome the blue-light inhibition of hypocotyl elongation (Fig. 2f). However, GCHXK failed to stimulate hypocotyl elongation when seedlings were grown in red light (16 h light/8 h dark). Under a low red-light intensity of 40 µE, the hypocotyls of both WT and GCHXK were similarly elongated; whereas at 130 µE, the hypocotyls of both lines were relatively short (Fig. 2g). We concluded that GCHXK overcomes the elongation–inhibition effect of blue light, but not that of red light. Below, we discuss the biological meaning of the different effects of blue and the red light on GCHXK elongation.
Sucrose and hexokinase in guard cells act upstream of PIF4 and compete with HY5
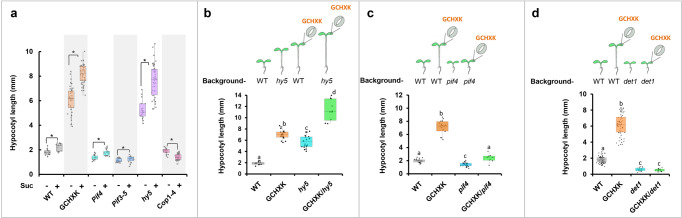
To explore the interplay between sucrose, HXK and various genes involved in hypocotyl elongation, we analyzed the effect of sucrose on elongation-related mutants. The hypocotyls of hy5 mutant elongate in the presence of light and elongate even further in the presence of sucrose (Fig. 4a). These results suggest that the additive elongation promoted by sucrose does not require the suppression of HY5 by sucrose, as this gene was already mutated. Rather, it suggests that the promotion of hypocotyl elongation by sucrose occurs via an independent pathway, which probably competes with the inhibitory effects of HY5. Unlike hy5 mutant, cop1-4 mutants, which do not elongate under dark or light conditions, do not elongate any further in presence of sucrose; whereas the pif4 mutant and the triple pif3-5 mutant exhibit slightly more elongation in the presence of sucrose (Fig. 4a). These results support the notion that sucrose enhances elongation via COP1. The fact that sucrose affects elongation independent of HY5, but fails to promote hypocotyl elongation in the cop1 mutant indicates that COP1 activates hypocotyl elongation not only by targeting HY5 for degradation, but also via additional effects on other transcription factors, a notion that has already been proposed previously6.
Fig. 4. Hypocotyl lengths of mutants treated with sucrose and the combined effects of GCHXK with hy5, pif4, and det1 mutants.
a Hypocotyl lengths of 7-day-old WT, GCHXK, pif4, pif3-5, hy5, and cop1-4 seedlings grown under long-day conditions with or without 1% sucrose. b–d Hypocotyl lengths of (b) GCHXK/hy5, (c) GCHXK/pif4, and (d) GCHXK/det1 seedlings grown on 0.5× MS agar media with 1% sucrose. a–d The box plots extend from the first to third quartiles and the whiskers extend from the minimum to the maximum levels. Lines within the boxes signify median values and gray dots represent individual data points (n ≥ 10 for a–c; n = 20–40 for d). a Asterisks indicate a significant difference relative to the control (t test, P < 0.05). b–d Different letters indicate a significant difference (Tukey’s HSD test, P < 0.05). The illustration above each figure indicates the gene expressed in guard cells, the genomic background (WT, hy5, pif4, or det1) and the hypocotyl-growth response.
To further explore the interplay of GCHXK with HY5 and PIF4, we studied hy5 and pif4 mutants expressing GCHXK: the GCHXK/hy5 and GCHXK/pif4 lines, which were obtained by crossing the GCHXK2 line with hy5 and pif4, respectively. In the light, GCHXK/hy5 elongated further than GCHXK or hy5 single mutants did (Fig. 4b). The additive elongation effect of GCHXK over that of the hy5 mutant further suggests that GCHXK stimulates hypocotyl elongation via a pathway that is independent of HY5 (i.e., it is not exerted through the suppression of HY5, but rather competes with the inhibitory effects of HY5). Unlike GCHXK/hy5, the hypocotyls of GCHXK/pif4 failed to elongate, indicating that PIF4 is central for GCHXK elongation and that GCHXK acts upstream of PIF4 (Fig. 4c).
The DET1 complex is needed to target HY5 for degradation and the hypocotyls of the det1 mutant did not elongate, even under dark conditions6. We wondered whether the elongation observed in GCHXK (under light) operates through DET1. To explore that issue, we constructed plants that expressed GCHXK in the background of det1 (GCHXK/det1) by crossing GCHXK2 with the det1 mutant. While the hypocotyl elongation of GCHXK seedlings increased about threefold, it was abolished entirely in GCHXK/det1, where it was similar to that observed for det1 alone (Fig. 4d). These results suggest that DET1 is essential for the hypocotyl elongation of GCHXK seedlings. The dominant inhibitory effects of det1 and pif4 on GCHXK hypocotyl elongation demonstrate that the hypocotyl elongation observed among the GCHXK seedlings involved primarily DET1 and PIF4. The fact that GCHXK had an elongation effect even in the presence of HY5 (Fig. 4b), but failed to promote elongation of the det1 mutant indicates that DET1 activates elongation not only by assisting in targeting HY5 for degradation, but perhaps also by other previously proposed roles of DET1, such as chromatin regulation that affects gene expression6.
Expression of PIF4 and HY5 in guard cells affects hypocotyl elongation
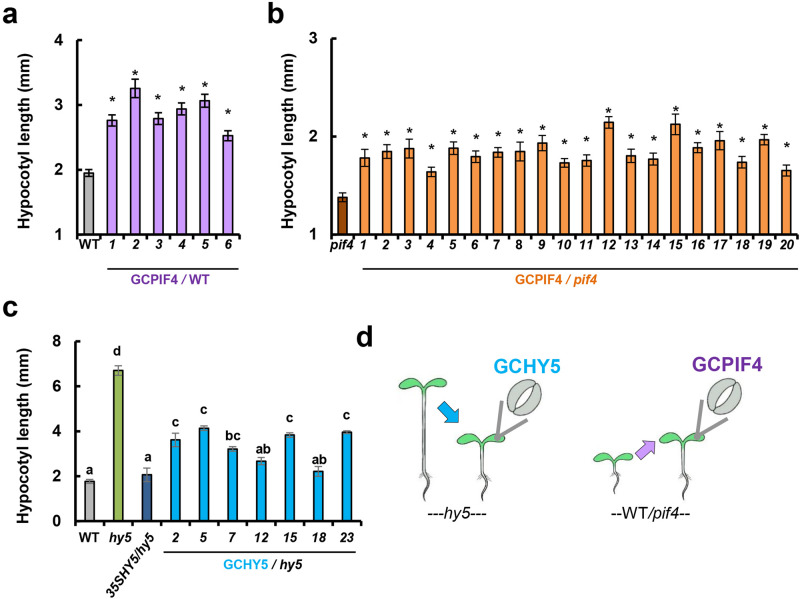
The results so far suggest that HXK1 promotes hypocotyl elongation and that expression of HXK1 specifically in guard cells is sufficient to stimulate that elongation. These results raise the possibility that signals generated exclusively within guard cells can control hypocotyl elongation. To examine this possibility, we expressed the transcription factors PIF4 and HY5 specifically in guard cells (GCPIF4 and GCHY5 lines, respectively) and examined their effects on hypocotyl elongation (Fig. 5). Exclusive expression of PIF4 in guard cells of WT plants (GCPIF4 plants) promoted hypocotyl elongation in the light (increase of 30 to 67%) relative to the WT (Fig. 5a). Similar behavior was observed when GCPIF4 was expressed in the background of the pif4 mutant (GCPIF4/pif4), in which the short hypocotyls of the pif4 mutants (1.4 mm) were elongated by GCPIF4 (GCPIF4/pif4) to about 2 mm (Fig. 5b). Elongation was observed in all of the 20 independent lines tested, with the increases in length ranging from 18 to 55% (Fig. 5b, d). We concluded that the expression of PIF4 in guard cells promotes hypocotyl elongation under light conditions.
Fig. 5. Expression of PIF4 (GCPIF4) and HY5 (GCHY5) in guard cells affects hypocotyl elongation.
a GCPIF4 in the WT background (purple columns) stimulates hypocotyl elongation of independent GCPIF4/WT lines relative to the WT. b GCPIF4 in the pif4-mutant background (orange columns) stimulates hypocotyl elongation of independent GCPIF4/pif4 lines. c Hypocotyl lengths of independent lines expressing HY5 in the guard cells (GCHY5/hy5) or globally under the 35S promoter (35SHY5/hy5) in the hy5-mutant background (light-blue columns). Data points for (a–c) are means ± SE (n = 20–25 for a; n = 15–25 for b; n = 13–25 for c). a, b Asterisks indicate significant differences relative to the WT (Dunnett’s test, P < 0.05). c Different letters indicate a significant differences (Tukey’s HSD test, P < 0.05). d The illustration indicates the gene expressed in guard cells (GCHY5 or GCPIF4) and the genomic background (hy5, WT/pif4). The blue and purple arrows indicate the hypocotyl-growth response stimulated by GCHY5 and GCPIF4, respectively.
With regard to HY5, hypocotyls of hy5 mutants were 3 times longer than WT hypocotyls (6.7 mm vs. 1.76 mm) and global overexpression of HY5 under the 35S promoter (35SHY5) completely abolished the long hypocotyls of hy5 (Fig. 5c). Similarly, exclusive expression of HY5 in the guard cells of the hy5 mutant (GCHY5/hy5 plants) partially to fully inhibited the hy5 elongated phenotype, reducing the hypocotyl lengths of independent GCHY5/hy5 lines (Fig. 5c, d). These results from GCPIF4 and GCHY5 plants demonstrate that guard cells alone are capable of influencing hypocotyl elongation and that signals generated within the guard cells are exported and affect other tissues involved in the elongation process.
GCHY5 counteracts the effect of GCHXK in an AtHY5-dependent manner
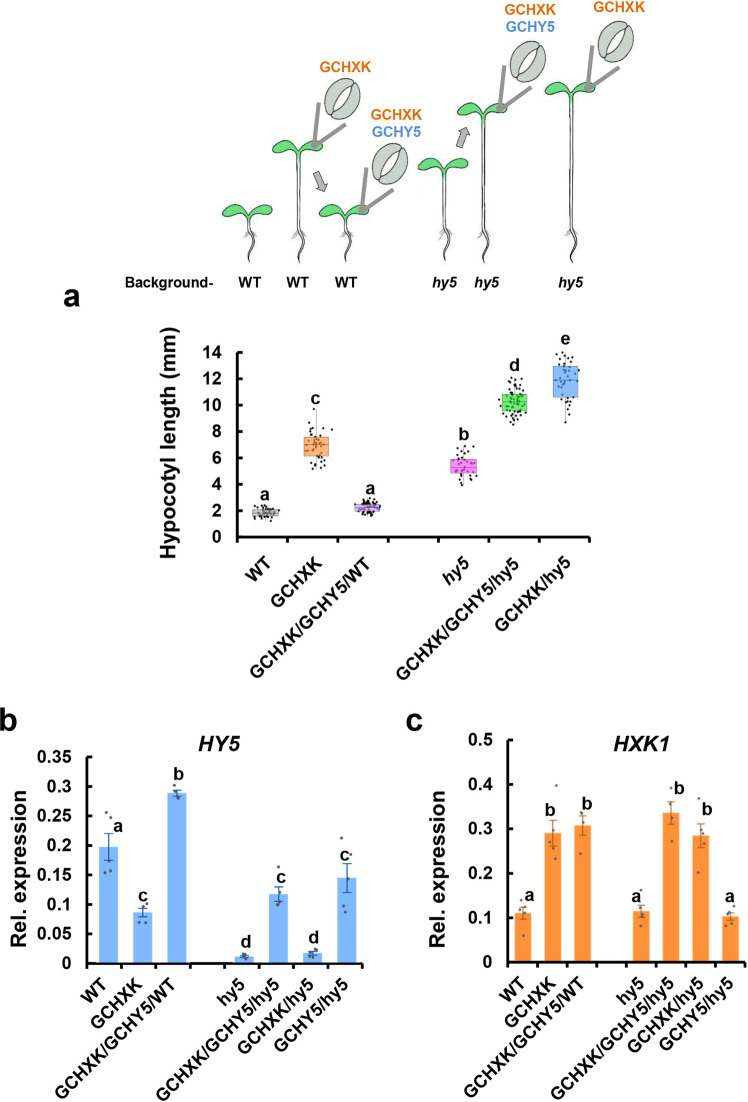
As shown above, expression of HXK1 in guard cells (GCHXK plants) in a WT background is dominant over the inhibitory effect of the endogenous HY5 and stimulates elongation in the light (Fig. 2); whereas the expression of HY5 in guard cells in the background of the hy5 mutant partially inhibits elongation (Fig. 5c). To better understand the interplay between HXK1 and HY5 in guard cells, we generated plants that co-expressed HXK1 and HY5 in their guard cells (GCHXK/GCHY5 plants). These plants were created by crossing GCHXK2 with GCHY5/WT (WT background) and with GCHY5/hy5 (hy5-mutant background). The hypocotyls of GCHXK/GCHY5 with the WT background (GCHXK/GCHY5/WT) were shortened from about 7 mm in GCHXK to only 2 mm, indicating that expression of HY5 in guard cells is dominant over the effect of GCHXK (Fig. 6a). However, the hypocotyls of GCHXK/GCHY5 with the hy5-mutant background (GCHXK/GCHY5/hy5) were long (Fig. 6a), indicating that having HY5 only in the guard cells is insufficient to inhibit the elongation induced by GCHXK, and that the inhibition of elongation by GCHY5 probably requires the presence of HY5 in tissues other than guard cells.
Fig. 6. The ability of GCHY5 to reduce the hypocotyl elongation of GCHXK is dependent on the endogenous HY5.
a Hypocotyl lengths of GCHXK/GCHY5/WT (WT background) and GCHXK/GCHY5/hy5 (hy5 background) seedlings grown with 1% sucrose. The box plots extend from the first to third quartiles and the whiskers extend from the minimum to the maximum levels. Lines within the boxes signify median values and gray dots represent individual data points (n = 30–60). The illustration at the top of the figure indicates the genes expressed in guard cells (GCHXK, GCHY5), the genomic background (WT or hy5). Arrows indicate the hypocotyl-elongation response. b, c RT-PCR expression analysis of HY5 (b) and HXK1 (c) in GCHXK/GCHY5/WT and GCHXK/GCHY5/hy5 seedlings. TUB2 (β-tubulin) was used for normalization. Data points are means ± SE (n = 5). Gray dots represent individual data points. a–c Different letters indicate a significant difference (Tukey’s HSD test, P < 0.05).
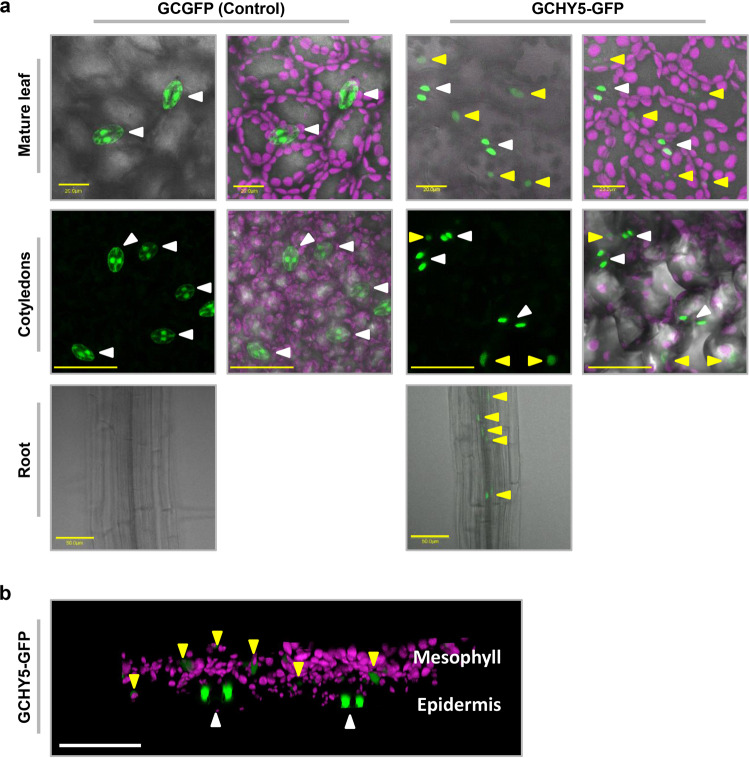
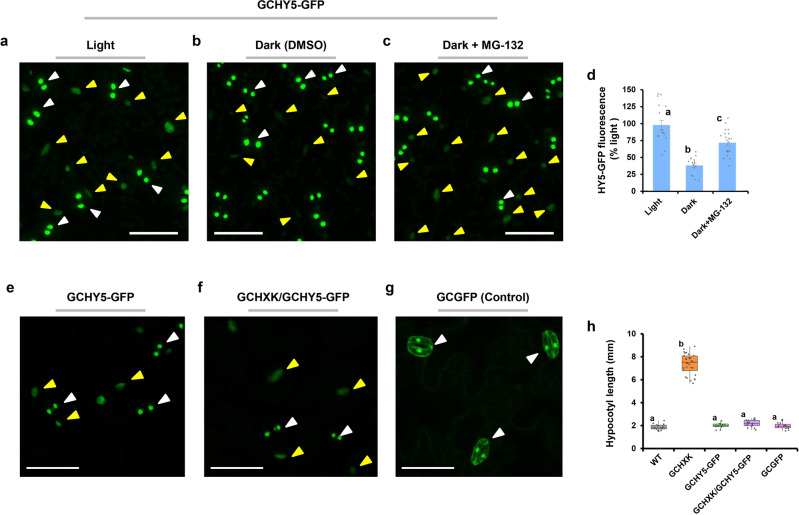
HY5 is a shoot-to-root mobile transcription factor and HY5-GFP expressed under mesophyll- and phloem-specific promoters accumulates in the roots51. It has also been shown that HY5 protein binds to the HY5 promoter and auto-activates its expression51,52. We, therefore, assumed that HY5 exits the guard cells of seedlings expressing GCHY5 and auto-activates the expression of HY5 in other tissues. However, guard cells do not have plasmodesmata and are symplastically isolated53–55. We, therefore, decided to take a closer look at the question of whether HY5 is capable of exiting the guard cells. We generated transgenic plants expressing GFP fused to the HY5 protein under the control of the guard cell-specific promoter (GCHY5-GFP plants) and searched for GFP signal in tissues other than guard cells (Fig. 7). In control plants in which GFP was expressed in guard cells (GCGFP), the GFP-fluorescence signal was restricted to the guard cells and was not detected anywhere else (Fig. 7a)5. However, in the GCHY5-GFP plants (in which HY5 was fused to GFP), GFP signal was also detected in the nuclei of mesophyll cells and in phloem cells in the root, indicating that HY5-GFP produced within guard cells is transported outside those cells and enters the nuclei of cells in other tissues (Fig. 7a, b). The presence of HY5-GFP outside the guard cells was observed in seedlings grown under light (long-day) conditions and in seedlings grown in the dark (Fig. 8b). Yet, the HY5-GFP signal in the seedlings grown in the dark was half as strong as that of the seedlings grown in the light (Fig. 8a–d). This result suggests that HY5-GFP exits the guard cells even in the dark, but is probably targeted for degradation by the proteasome. To examine whether HY5-GFP is degraded by the proteasome, we applied the proteosome inhibitor MG-132 to dark-grown seedlings and found that the level of HY5-GFP signal was indeed higher in the presence of MG-132 (Fig. 8c, d). We concluded that HY5 exists in the guard cells in the dark, but is then degraded and, therefore, fails to prevent elongation. These results suggest that HY5 produced in guard cells moves out of the guard cells and inhibits hypocotyl elongation under light conditions by auto-activating the endogenous expression of HY5 in other tissues51,52. To examine this assumption, we analyzed the expression levels of the endogenous HY5 in 7-day-old seedlings. The expression level of HY5 in GCHXK/GCHY5/WT (WT background) was twice as high as that observed in the GCHXK/GCHY5/hy5 or GCHY5/hy5 lines, in which the expression of HY5 was solely derived from GCHY5 (Fig. 6b). These results support the notion that HY5 expressed in guard cells activates the expression of the endogenous HY5. Unlike HY5, the expression of HXK1 derived from GCHXK was not affected in any of the crosses (Fig. 6c).
Fig. 7. HY5 produced within guard cells is translocated to mesophyll and root cells.
a Distribution of GCGFP (control) or GCHY5-GFP in mature leaves, cotyledons and roots. All panels are merged images of white-light, chlorophyll-autofluorescence (stained magenta), and GFP-fluorescence (stained green). Scale bars (yellow) are 20 µm for mature leaves and 50 µm for cotyledons and roots. b 3D simulation providing side views of GCHY5-GFP cotyledons, composed of epidermis and mesophyll cell layers. Image is a merge of chlorophyll-autofluorescence (stained magenta), and GFP-fluorescence (stained green). Bar = 50 µm. a, b White arrows indicate the location of GFP in guard cells and yellow arrows indicate the location of GFP in mesophyll cells of mature leaves and the cotyledons, and within the phloem of the roots.
Fig. 8. Dark and GCHXK do not block the export of HY5 from guard cells.
a–d HY5-GFP-fluorescence intensity is light-dependent. Ten-day-old GCHY5-GFP seedlings treated with 15 µM proteasome inhibitor; MG-132 c, kept in the dark for 16 h prior to image acquisition. b Dark-grown seedlings treated with 0.1% DMSO served as a control, and (a) light-grown seedlings served as an additional control. d Relative fluorescence intensity of HY5-GFP. The fluorescence intensity in the light was set to 100%. Data points are means ± SE (n > 15). Light gray dots represent individual data points. Different letters indicate a significant difference (Tukey’s HSD test, P < 0.05). e–h GCHXK did not block the export of HY5 from guard cells. e–g Distribution of GFP signal in GCHY5-GFP (e), GCHXK/GFHY5-GFP (f), and GCGFP (g, control) in cotyledons of developing seedlings. h Hypocotyl lengths of GCHXK/GFHY5-GFP seedlings grown with 1% sucrose. The box plots extend from the first to third quartiles and the whiskers extend from the minimum to the maximum levels. Lines within the boxes signify median values and dots represent individual data points (n > 15). Different letters indicate a significant difference (Tukey’s HSD test, P < 0.05). a–c, e–g All panels are GFP-fluorescence (stained green) images. White arrows indicate the location of GFP in guard cells and yellow arrows indicate the location of GFP in mesophyll cells. Bar = 50 µm (a–c), 25 µm (e–g).
Since GCHXK per se promotes hypocotyl elongation against the WT background (Fig. 2), it could be that GCHXK suppresses the export of the endogenous HY5 from guard cells and, as a result, promotes hypocotyl elongation. To examine this possibility, we made plants that co-expressed HXK1 and HY5-GFP in their guard cells (GCHXK/GCHY5-GFP plants) by crossing GCHXK2 with GCHY5-GFP and monitored GFP fluorescence (Fig. 8e–h). GFP fluorescence appeared in both the guard cells and mesophyll cells of the GCHXK/GCHY5-GFP, demonstrating that GCHXK does not block the export of HY5 from guard cells (Fig. 8f). Furthermore, the hypocotyls of GCHXK/GCHY5-GFP were short (Fig. 8h), similar to those of GCHXK/GCHY5/WT (Fig. 6a). This indicates that the fusion HY5-GFP retains its natural activity and inhibits the elongation imposed by GCHXK, most likely by auto-activating the expression of the endogenous HY5.
Discussion
Germinating seedlings elongate in a search for light, which is perceived by photoreceptors and halts hypocotyl elongation. The perception of light prevents the degradation of HY5 and targets PIFs for degradation, thereby blocking hypocotyl elongation. Researchers have long wondered in which tissues and cell types the light signal is perceived. In the current study, we found that modulated expression of the sugar sensor HXK1, HY5, and PIF4 in guard cells is sufficient to promote or stop hypocotyl elongation under light conditions, uncovering a role for guard cells in hypocotyl elongation. A recent study reported that expression of PHYB in the epidermis under the ML1 promoter is sufficient for the perception of a red-light signal and the inhibition of hypocotyl elongation in the phyB mutant29. Yet, the epidermis is composed of epidermal pavement cells and guard cells, with the latter appearing on the cotyledons immediately after germination4,5. Since the ML1 promoter is also active in guard cells (56; http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), it is possible that the light perception and the effects of PHYB that have been attributed to the epidermis are generated in the guard cells.
Many of the genes involved in hypocotyl elongation, including PHYB, CRY1, CRY2, COP1, SPA, DET1, HY5, PIFs, YUC8, and HXK, are expressed in guard cells57 (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi). We propose that light-activated photoreceptors within guard cells prevent the degradation of guard cells’ HY5. HY5 then interferes with PIF4 activity to prevent the production of auxin within the guard cells9–11. In addition, HY5 exits the guard cells and apparently auto-activates HY5 expression in other tissues, thereby interfering with the activity of PIF4 (and perhaps other PIFs, as well) in those tissues, and blocking hypocotyl elongation. Under dark conditions, HY5 that exits the guard cells is targeted for degradation by the proteosome, as the GCHY5-GFP signal in mesophyll cells of seedlings germinating in the dark was less than half of that of seedlings germinating in the light and the degradation was attenuated by a proteasome inhibitor (Fig. 8d).
Expression of HY5 in guard cells is sufficient to partially inhibit hypocotyl elongation of the hy5 mutant (Fig. 5c). It was previously shown that a chloroplast retrograde signal (sent from the chloroplast to the nucleus) is required for HY5-mediated repression of hypocotyl elongation17. It was also reported that the absence of functional chloroplasts prevents normal light perception and signal transduction by phytochromes58. While guard cells contain active chloroplasts59, the presence of chloroplasts in the epidermal pavement cells has been debated60. In the course of this study, we failed to spot any chloroplast fluorescence (autofluorescence of chlorophyll) in the pavement cells of epidermal peels, while chloroplast fluorescence was easily spotted in guard cells (Supplementary Fig. 5). It is likely that epidermal pavement cells in Arabidopsis cotyledons lack active chloroplasts and that guard cells rather than epidermal pavement cells mediate the de-elongation signal. Whether epidermal cells are also involved in the de-elongation signal should be examined using epidermal-specific promoters that do not drive expression in guard cells.
It has previously been shown that shade (a low ratio of red/far red light) is perceived in the cotyledons, where PIF4 drives the synthesis of the growth hormone auxin. Auxin is then transported to the hypocotyl to induce hypocotyl elongation22,23,25. Since hypocotyl elongation driven by HXK1 expressed in guard cells (GCHXK) requires PIF4 (Fig. 4c) and since increased expression of PIF4 in guard cells (GCPIF4) is sufficient to promote hypocotyl elongation in WT and pif4 backgrounds even under long-day conditions (Fig. 5a, b), it is likely that GCHXK and GCPIF4 stimulate auxin production in the guard cells. The auxin is then transported to the epidermis of the hypocotyl, in which BR and gibberellin (GA) signals stimulate an elongation response22,23,25. Indeed, a higher level of auxin was measured in GCHXK seedlings (Fig. 3d), and the expression of PIF and auxin-induced genes were upregulated (Fig. 3a, b). In addition, inhibitor of auxin transport prevented GCHXK-mediated hypocotyl elongation (Fig. 3e), supporting our assumption that GCHXK stimulates PIF4-mediated auxin production in guard cells.
The levels of hypocotyl elongation in the light of GCPIF4 seedlings with the pif4-mutant or WT backgrounds seem to be similar to one another and moderate (~30%; Fig. 5a, b). Unlike the expression of PIF4 in guard cells, global overexpression of PIF4 under 35S in the WT background stimulated much greater (doubled) hypocotyl elongation61,62, indicating an additive elongation effect when PIF4 expression is not limited to the guard cells. This conclusion is in line with our results; the more pronounced hypocotyl elongation of GCHXK (about 3 times greater than the WT) as compared to the moderate hypocotyl elongation of GCPIF4 (in which PIF4 was overexpressed only in guard cells) is probably due to the expression of PIF4 in GCHXK tissues other than guard cells. That is, GCHXK stimulates PIF4 activity not only in guard cells, but probably in other tissues as well. This assumption is further supported by the increased expression of PIF1, 3 and 4 in GCHXK seedlings (Fig. 3a). That may suggest that expression of PIF4 in the hypocotyl elongating tissues is also important for hypocotyl elongation63. The question of whether expression of any of the other PIFs, aside from PIF4, in guard cells and in the hypocotyl tissues is also required for elongation awaits further examination. Yet, the use of PIF4 in this study shows that GCHXK stimulates hypocotyl elongation via the known COP1/SPA, DET1, and PIF pathways.
Unlike PIF4, the results from GCHY5 suggest that HY5 exits the guard cells and not only activates its expression in other tissues, but also prevents PIF-stimulated elongation effects in those tissues. This is based on the marked difference between GCHY5 expressed concomitantly with GCHXK in the WT background, as compared to the hy5-mutant background (Fig. 6). Expression of GCHXK and GCHY5 in the WT background completely abolished the elongation induced by GCHXK; whereas hypocotyl elongation was not abolished when GCHXK and GCHY5 were expressed in the hy5-mutant background (Fig. 6a). Since HY5 antagonizes the effect of PIFs, which are essential for hypocotyl elongation12, these results indicate that auto-activated expression of HY5 in locations other than the guard cells helps to eliminate PIF-stimulated elongation effect.
Our finding that HY5 moves from guard cells to other tissues (Fig. 7) is in line with the results reported by Chen51, who demonstrated shoot-to-root HY5 movement. It has recently been shown that local expression of N-terminal GFP-tagged HY5 in epidermal, mesophyll or vascular tissues is confined to the specific tissue, with no movement of the tagged-HY5 out of the specific tissue64. Yet, this local expression was sufficient to partially (but not completely) shorten the long hypocotyls of the hy5 mutant64. The authors of that work suggested that these results support the notion that HY5 acts in various tissues. Indeed, both that study and the current study suggest that various tissues are likely targets for autoactivation of HY5 expression and participate in the inhibition of hypocotyl elongation by HY5. It is worth noting that in Burko’s et al.64 study, epidermal expression of the confined tagged-HY5 was driven by the CER6 promoter that also drives expression in the guard cells65,66. Yet, in our study, HY5 expressed in guard cells of GCHY5 was not confined and moved out of the guard cells, which may explain why in some of the GCHY5/hy5 lines (background of hy5 mutant), the suppression of hypocotyl elongation was as complete as that observed in the WT (Fig. 5c).
The very fact that sugars promote hypocotyl elongation of WT Arabidopsis seedlings under short-32 and long-day conditions (this study) suggests that light is not the only signal that controls hypocotyl elongation and that sugars can overcome the effects of light on this process. Furthermore, expression of HXK1, an established sugar sensor, under the global 35S or guard-cell promoter (35SHXK and GCHXK, respectively) stimulates hypocotyl elongation in the light, which also supports the notion that sugars constitute an additional pathway that affects hypocotyl elongation.
The observed effect of sucrose on the hypocotyl elongation of hy5 mutants also supports the hypothesis that sugars constitute an independent regulatory pathway for hypocotyl elongation. The hypocotyls of hy5 mutants elongate in the light, but the addition of sucrose causes it to elongate significantly further (Fig. 4a). That indicates an additive effect of sucrose over the absence of hy5, suggesting that sucrose does not require suppression of HY5 to promote hypocotyl elongation. Unlike the effect of sucrose on hy5 mutants, sucrose added to cop1 mutants does not promote hypocotyl elongation, suggesting that the effect of sucrose and HXK1 is dependent on COP1. COP1 is an E3 ubiquitin ligase which in darkness targets various transcription factors that inhibit hypocotyl elongation for ubiquitination and degradation, including HY5, HY5 HOMOLOG (HYH), LONG AFTER FAR RED LIGHT (LAF1), LONG HYPOCOTYL IN FAR RED (HFR1), B-box zinc finger proteins (BBXs), and a BR-regulated GATA transcriptional factor (GATA2)6,49. The fact that sucrose stimulates elongation beyond that of hy5 mutant, but requires COP1 to stimulate elongation might indicate that sucrose stimulates elongation via the other targets of COP1, independent of HY5. Yet, it is still possible that sucrose also targets HY5 for degradation via COP1. The application of sugar represses the transcription of HY5 and its homolog HYH, as seen in a database search (Supplementary Fig. 6). In line with those results, we found that GCHXK reduced the expression of HY5 (Figs. 3a, 6b). Therefore, it is possible that sucrose and HXK1 promote hypocotyl elongation via several mechanisms: inducing expression of DET1 (Fig. 3c) helping DET1 and COP1 to target various hypocotyl-elongation inhibitors for degradation (including HY5), repressing the expression of HY5 (perhaps by reducing autoactivation of HY5 expression) and possibly inducing the expression of PIF genes independent of HY5 (Fig. 3a).
The results of the current study suggest that the effect of guard cells on hypocotyl elongation involves two opposite signals converging at PIF433. The first is sugar-sensing by HXK1, which overcomes the effect of the endogenous HY5 to promote hypocotyl elongation. This effect of HXK1 is dependent on COP1, DET1, and PIF4 (and perhaps other PIFs) and involves the generation of an auxin signal that exits the guard cells and activates elongation effects in the target tissues. The second signal is perceived by photoreceptors, probably within guard and mesophyll cells29,67, prevents degradation of HY5 and blocks PIF activity. It is likely that there is some balance between HXK1 and HY5 signals within guard cells. HXK1 tilts the balance toward hypocotyl elongation, while HY5 tilts the balance towards movement and autoactivation of HY5 expression in other tissues, which eventually inhibit hypocotyl elongation.
We have shown that GCHXK overcomes the inhibition of elongation caused by blue light, but not that caused by red light (Fig. 2f, g). We suggest the following possible explanation for this difference: At dawn, the primary light is blue light, which is photosynthetically inefficient and, therefore, the primary carbon source at that time of day is sucrose generated from seed storage. As the sun rises, the blue light is followed by photosynthetically efficient red light, with which the immediate products of photosynthesis are triose phosphates. While triose phosphates are phosphorylated independently of HXK activity, sucrose metabolism requires hexose phosphorylation of the sucrose-cleavage products, glucose and fructose, by HXK. We, therefore, suggest that HXK activity acts as a sensor for photosynthetic efficiency. That is, HXK indicates how much of the available carbon is derived from storage reserves versus photosynthesis-derived triose phosphates. Under blue-light conditions, a high level of HXK activity, which is needed for sucrose metabolism, indicates that the carbon is coming from storage reserves, which promotes hypocotyl elongation in a search for photosynthetically efficient red light. However, under red-light conditions associated with the production of triose phosphates, HXK activity is dispensable, and therefore, no further elongation is promoted.
In summary, the current study reveals that guard cells are central players in hypocotyl elongation. It also reveals the role of HXK1 in that elongation and indicates that light and sucrose antagonistically coordinate the effort to achieve the height necessary for efficient photosynthetic, autotrophic sugar production.
Methods
Plant material
All of the plants used in this study were of the Arabidopsis thaliana Col-0 ecotype. The hy5 mutant was obtained from the Arabidopsis Information Resource (ABRC) stock (Salk_096651, https://abrc.osu.edu). The pif4-101, cop1-4, pif3-568, det169, GCHXK, GCGFP44,46, 35SHXK47, and 35SFRK148 plants have been described previously. Cloning, transformation and characterization of additional transgenic lines, as well as crosses and T-DNA mutants used in this study, are described below.
Plasmid construction
To generate the KSTppro::AtPIF4 (GCPIF4), KSTppro::AtHY5 (GCHY5), 35Spro::AtHY5 (35SHY5) and KSTppro::AtHY5-GFP (GCHY5-GFP) transgenic plants, segments for the coding regions of AtHY5 (AT5G11260), AtHY5 fused to GFP (HY5-GFP) and AtPIF4 (AT2G43010), as well as a segment of the KST1 partial promoter (KSTppro,5) were synthesized by GENEWIZ (https://www.genewiz.com). Specific restriction sites (NotI, XhoI, XmaI and XbaI) were added to the sequence. The binary vector pGreen containing an insertion of “NotI-35Spro-XhoI-XmaI-GFP-XbaI” that was kindly provided by the lab of Dr. Arthur Schaffer (ARO, Israel) was used to clone the GCPIF4, GCHY5, 35SHY5, and GCHY5-GFP constructs. For the generation of the 35SFRK2 construct, the Solanum tuberosum Fructokinase2 (GeneBank accession number AF106068) was inserted under the 35S promoter in the binary vector pBI12170.
Generation of transgenic plants
Electrocompetent Agrobacterium tumefaciens (GV3101 strain) was transformed using 100 ng plasmid by electroporation. Arabidopsis WT, hy5, and pif4 plants were transformed using the floral-dip method71. Screening was performed on 0.5× MS selection media containing 3% sucrose (Suc, Duchefa) and 50 mg l−1 kanamycin (Kan; Duchefa). The primers used to identify positive transgenic events are listed in Supplementary Table 3. T-DNA single mutant lines were genotyped by primers designed using the SIGnAL primer design tool (http://signal.salk.edu/tdnaprimers.2.html) powered by the Genome Express Browser Server (GEBD). Homozygous mutants were identified using the primers listed in Supplementary Table 2. GCHXK crosses conducted in this study were all carried out using the same GCHXK line (GCHXK2). For the GCHXK/GCHY5, GCHXK/GCHY5/hy5 crosses, we used lines GCHY5/hy5 #12 (described in Fig. 5c) together with GCHXK2. The primers used for the identification of the crosses, GCHXK/pif4, GCHXK/hy5, GCHXK/det1, GCHXK/GCHY5, GCHXK/GCHY5/hy5 and GCHXK/GCHY5-GFP, are listed in Supplementary Tables 2, 3.
Hypocotyl-growth assay
For the hypocotyl-growth assay, seeds were sterilized and sown onto Petri plates (9 mm diameter) containing half Murashige and Skoog (0.5× MS; Duchefa Biochemie, The Netherlands) medium as a control (pH 5.8, 0.8% plant agar) or 0.5× MS with varying sugar concentrations; 1, 2, or 3% sucrose (Suc; Duchefa), as indicated for each experiment. Seeds were cold-treated at 4 °C in darkness for 3 days before they were transferred to a growth chamber (16 h light/8 h dark photoperiod at 22–23 °C, 40 µmol m−2 s−1 light intensity). For plants grown in soil, a potting mix containing (w/w) 70% peat, 30% perlite, supplemented with slow-release fertilizer (Even-Ari, Israel) was used. For seedlings grown under blue and red light conditions, we used the adjustable led lighting system, pro 325 (Lumigrow, CA, USA). Seedlings were imaged 7 days after the transfer to the growth room, unless mentioned otherwise. To determine hypocotyl length, images were analyzed using the ImageJ software (http://rsb.info.nih.gov/ij/) fit-line tool, following size calibration.
Scanning electron microscopy
For the scanning electron microscopy (SEM), seedlings were fixed in 3.7% formaldehyde, 50% ethanol, and 5% acetic acid by vacuum infiltration for 30 min. Seedlings were later kept in the fixative for 8 h, followed by a slow dehydration through a series of ethanol concentrations: 50%, 70% and 90%, 100%, 100%, 60 min each. Seedlings were critical-point dried in liquid CO2 in a Quorum K850 critical-point dryer (Quorum Technologies, East Sussex, UK), and sputter-coated with gold palladium using a Quorum SC7620 mini sputter coater (Quorum Technologies, East Sussex, UK). Images were taken with a JEOL JCM6000 benchtop SEM (Jeol, Japan).
Microarray database processing and analysis
Expression data for AtHY5 and AtHYH were obtained from the NASCarrays microarray database72, Experiment No. 593. A heat-map diagram was computed using Expander 7 software73 based on the expression data obtained from the NASCArray database. The accession number and the Affimetrix probe ID number for each gene are listed in Supplementary Table 4. Gene accession numbers were assigned according to the information in the TAIR database (http://www.arabidopsis.org/).
RNA extraction and cDNA preparation
Samples were collected from seedlings grown on 0.5× MS agar plates, 4 h after lights were switched on. Each sample included at least 40 seedlings. Samples were snap-frozen in liquid nitrogen and total RNA was extracted using the Logspin method74. Samples were ground using a Geno/grinder (SPEX SamplePrep, Metuchen, NJ, USA) and RNA was extracted in 8M guanidine hydrochloride buffer (Duchefa Biochemie) and then transferred to tubes containing 96% EtOH (Bio Lab, Jerusalem, Israel). Then, samples were transferred through a plasmid DNA extraction column (RBC Bioscience, New Taipei City, Taiwan), washed twice with 3M Na-acetate (BDH Chemicals, Mumbai, India) and twice in 75% EtOH and eluted with DEPC (diethylpyrocarbonate) water (Biological Industries, Co., Beit Haemek, Israel) that had been preheated to 65 °C. The RNA was treated with RQ1-DNase (ProMega, Madison, WI, USA) according to the manufacturer’s instructions, to degrade any residual DNA. The purity of all RNA samples was assessed by 260/280 and 260/230 nm absorbance ratios. For the preparation of cDNA, total RNA (1 µg) was taken for reverse-transcription PCR using qscriptTM cDNA Synthesis Kit (Quanta BioSciences, Gaithersburg, MD, USA) following the manufacturer’s instructions.
Quantitative real-time PCR
For qPCR, cDNA samples were diluted 1:7 in DEPC water. Quantitative real-time PCR reactions were performed using SYBR Green mix (Thermo-Scientific, Waltham, MA, USA) and reactions were run in a RotorGene 6000 cycler (Corbett, Mortlake, New South Wales, Australia). Following an initial pre-heating step at 95 °C for 15 min, there were 40 cycles of amplification consisting of 10 s at 95 °C, 15 s at 55 °C, 10 s at 60 °C, and 20 s at 72 °C. The melting point was determined for each sample to validate the specificity of the primers. Results were analyzed using the RotorGene software. The Arabidopsis AtTUB2 (accession no. At5g62690) was used as a reference for the normalization of cDNA amounts. The primers used for the amplification procedure are listed in Supplementary Table 1.
Library preparation and sequencing
Library construction and sequencing were performed by the Genomics Unit at the Grand Israel National Center for Personalized Medicine (https://g-incpm.weizmann.ac.il), Weizmann Institute of Science (Rehovot, Israel). Briefly, the poly(A) fraction (mRNA) was purified from 500 ng of total RNA, followed by fragmentation and generation of double-stranded cDNA. Then end repair, A-base addition, adaptor ligation and PCR amplification were carried out. Sequencing libraries were constructed with barcodes to allow the multiplexing of eight samples in one lane. An Illumina HiSeq 2500 V4 instrument was used to sequence single-end, non-stranded 60-bp reads. The number of reads was similar for all samples.
Filtering and mapping of reads to the reference genome
Raw reads were subjected to a filtering and cleaning procedure. Adaptors were removed using Trimmomatic software, version 0.3275. Then, the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13.2) was used to (i) trim read-end nucleotides with quality scores <30, with the Fastq quality_trimmer, and (ii) remove reads with <70% of base pairs with a quality score ≤30 using the Fastq quality filter. Transcript quantification (the number of reads per gene) from the RNA-Seq data was performed using the Bowtie2 aligner76, the Arabidopsis reference genome (TAIR database) and the expectation-maximization method (RSEM) to estimate maximum-likelihood expression levels77. The expression level was calculated as trimmed mean of M values (TMM)-normalized counts78.
Differential-expression analysis, GO analysis, and pathway enrichment
Differential-expression (DE) analysis was performed with the DESeq2 package79 of the R software. Transcripts that were more than 2-fold differentially expressed with an adjusted P value of no more than 0.05 were considered differentially expressed. GO and pathway-enrichment analyses were performed using KOBAS program80, to determine significant annotated processes of the main biological functions. The PANTHER (http://www.pantherdb.org), KEGG PATHWAY (www.genome.jp/kegg/) and BioCyc (https://biocyc.org) Gene Ontology databases were used with multiple testing correction of false discovery rate (FDR)81. The threshold was set to a FDR with a corrected P value of <0.05. The REVIGO web server82 was used for removing redundant GO terms. DE transcript diagrams were displayed using MapMan 3.6.0RC1 (https://mapman.gabipd.org/mapman-version-3.6.0) and TAIR10 mapping (Ath_AGI_TAIR9_Jan2010).
Confocal microscopy imaging
Image acquisition was done using a Leica SP8 laser scanning microscope (Leica, Wetzlar, Germany) equipped with a solid-state laser with 488 nm light, a HC PL APO CS 63×/1.2 water immersion objective (Leica) and Leica Application Suite X software (LASX, Leica). Images of GFP signal were acquired using the 488-nm laser light and the emission was detected with HyD (hybrid) detector in a range of 500–525 nm. Autofluorescence of the chloroplasts was detected in a range of 650–750 nm with a PMT detector.
Auxin quantification using LC–MS
Hormone extraction was performed using standard protocols83 with slight modifications. Five-day-old seedlings grown on control agar plates (0.5× MS, 0.8% plant agar, pH 5.8) were collected and frozen in liquid nitrogen. Each sample collected, included at least 80 seedlings. The frozen tissue was ground to a fine powder using a Geno/grinder (SPEX SamplePrep, Metuchen, NJ, USA). Two hundred mg of the powder were transferred to a 1.5-ml tube containing 1 ml of an extraction solvent (ES) mixture [79% isopropanol (Bio Lab, Israel): 20% MeOH (Bio Lab): 1% acetic acid, (Gadot, Israel)], supplemented with 20 ng of each deuterium-labeled internal standards (IS, Olomouc, Czech Republic). The tubes were incubated for 60 min at 4 °C with rapid shaking and centrifuged at 14,000 g for 15 min at 4 °C. The supernatant was collected and transferred to a 2 ml tube. ES (0.5 ml) was added to the pellet and the extraction steps were repeated twice. The combined extracts were evaporated using a speed-vac (Hetovac VR-1, Denmark) at room temperature. Dried samples were dissolved in 200 μl of 50% methanol and filtered through a 0.22 μm PVDF syringe filter (Agela Technologies, Torrance, CA, USA). Five μl were injected for each analysis.
LC–MS-MS analyses were conducted using UPLC-Triple Quadrupole-MS (Waters Xevo TQ MS). Separation was performed on a Waters Acquity UPLC BEH C18 1.7 μm 2.1 × 100 mm column with a VanGuard pre-column (BEH C18 1.7 μm 2.1 × 5 mm). Chromatographic parameters were as follows: The mobile phase consisted of water (phase A) and acetonitrile (phase B), both containing 0.1% formic acid in the gradient-elution mode. The solvent gradient program for auxins is presented in Supplementary Table 6. The flow rate was 0.3 ml/min and the column temperature was kept at 35 °C. The retention times and MS-MS parameters for each plant hormone and its internal standard are listed in Supplementary Table 5. Acquisition of LC–MS data was performed using MassLynx V4.1 software (Waters). Quantification was done using isotope-labeled IS, except for OxIAA, which was quantified using calibration curves.
Auxin transport activity assay
For the auxin transport activity assay, the auxin transport inhibitor N-1-naphthylphthalamic acid (NPA, Sigma-Aldrich, Israel) dissolved in dimethyl sulfide (DMSO, Sigma-Aldrich, Israel) was added to the 0.5× MS agar media, containing 1% Suc. A 0.5× MS + 1% Suc media, supplemented with 0.1% DMSO without NPA, served as the control. Hypocotyl length was measured at 9 days after germination.
Proteasome inhibitor assay
For the proteasome inhibitor treatment, we used carbobenzoxy-Leu-Leu-leucinal (MG-132, Sigma-Aldrich, Israel) dissolved in DMSO. Ten-day-old GCHY5-GFP seedlings grown on 0.5x MS agar media containing 1% Suc (grown under long-day conditions) were treated with MG-132 to a final concentration of 15 µM. Following the application of MG-132, seedlings were moved to the dark for 16 h. DMSO (0.1%) served as a control. Following MG-132/dark treatment, cotyledons were taken for confocal imaging. Images were analyzed using the ImageJ software histogram tool to evaluate fluorescence intensity.
Calculated lengths of hypocotyl epidermal cells
The average cell length of 4-day-old seedlings was calculated by dividing hypocotyl length by the average number of cells counted following the SEM analysis.
Statistical analysis
Statistical analysis was performed using the JMP 14 software program. Box plots were prepared using the Graph Builder tool. Means were compared using Student’s t test, Tukey’s HSD test or Dunnett’s method, as described for each experiment. Means were considered to be significantly different at P < 0.05.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We would like to thank the reviewers for their valuable comments that have improved the paper. We thank Prof. Jorge Casal and Manuel Pacín for providing the pif4-101, cop1-4, pif3-5seeds, Prof. Meng Chen for the det1 seeds and Prof. Danny Chamovitz for the phyB seeds. We thank Dr. Mira Carmeli-Weissberg (Volcani Center, ARO) for assisting us with the LC–MS-MS analysis. We would also like to thank Dr. Joshua Klein for providing the LED lighting systems.
Author contributions
G.K., N.C., and D.G. planned and designed the research. G.K. carried out plasmid construction and generation of transgenic plants. G.K., O.S., N.L., and A.E. generated, genotyped, and analysed the crosses presented in this study. G.K., A.E., N.S., and D.B. performed the experiments, analysed the data, and interpreted the results. A.D.-F. carried out bioinformatics analysis and interpretation. E.B. carried out confocal microscopy imaging, F.S. and G.K. conducted auxin quantification, and H.Z., D.B., and N.L. performed SEM experiments. G.K., N.S., and D.G. wrote the paper.
Data availability
The sequencing data from this study have been deposited in the The National Center for Biotechnology Information BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under the accession number PRJNA687355. The raw data from the Illumina sequencing have been deposited in NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR13316969-SRR13316975. The raw data referring to the plots shown in the figures are provided in Supplementary Data 1, 2. All relevant data are available from the authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Yuan Qin and Luke R. Grinham. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-021-02283-y.
References
- 1.Kornberg HL, Beevers H. A mechanism of conversion of fat to carbohydrate in castor beans. Nature. 1957;180:35–36. doi: 10.1038/180035a0. [DOI] [PubMed] [Google Scholar]
- 2.Penfield S, et al. Reserve mobilization in the Arabidopsis endosperm fuels hypocotyl elongation in the dark, is independent of abscisic acid, and requires PHOSPHOENOLPYRUVATE CARBOXYKINASE1. Plant Cell. 2004;16:2705–2718. doi: 10.1105/tpc.104.024711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eastmond PJ, et al. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 2015;6:6659. doi: 10.1038/ncomms7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geisler MJ, Sack FD. Variable timing of developmental progression in the stomatal pathway in Arabidopsis cotyledons. N. Phytol. 2002;153:469–476. doi: 10.1046/j.0028-646X.2001.00332.x. [DOI] [PubMed] [Google Scholar]
- 5.Kelly G, et al. The Solanum tuberosum KST1 partial promoter as a tool for guard cell expression in multiple plant species. J. Exp. Bot. 2017;68:2885–2897. doi: 10.1093/jxb/erx159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trend Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Delker C, et al. The DET1-COP1-HY5 pathway constitutes a multipurpose signaling module regulating plant photomorphogenesis and thermomorphogenesis. Cell Rep. 2014;9:1983–1989. doi: 10.1016/j.celrep.2014.11.043. [DOI] [PubMed] [Google Scholar]
- 8.Legris M, Nieto C, Sellaro R, Prat S, Casal JJ. Perception and signalling of light and temperature cues in plants. Plant J. 2017;90:683–697. doi: 10.1111/tpj.13467. [DOI] [PubMed] [Google Scholar]
- 9.Toledo-Ortiz G, et al. The HY5-PIF regulatory module coordinates light and temperature control of photosynthetic gene transcription. PLoS Genet. 2014;10:e1004416. doi: 10.1371/journal.pgen.1004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gangappa SN, Botto JF. The multifaceted roles of HY5 in plant growth and development. Mol. Plant. 2016;9:1353–1365. doi: 10.1016/j.molp.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Legris M, Ince YÇ, Fankhauser C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat. Commun. 2019;10:5219. doi: 10.1038/s41467-019-13045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leivar P, Monte E. PIFs: systems integrators in plant development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H. Signaling mechanisms of higher plant photoreceptors: a structure‐function perspective. Curr. Top. Dev. Biol. 2005;68:227–261. doi: 10.1016/S0070-2153(05)68008-8. [DOI] [PubMed] [Google Scholar]
- 14.Li QH, Yang HQ. Cryptochrome signaling in plants. Photochem. Photobiol. 2007;83:94–101. doi: 10.1562/2006-02-28-IR-826. [DOI] [PubMed] [Google Scholar]
- 15.Lian H-L, et al. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Gene Dev. 2011;25:1023–1028. doi: 10.1101/gad.2025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu X-D, et al. Red-light-dependent interaction of phyB with SPA1 promotes COP1–SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol. Plant. 2015;8:467–478. doi: 10.1016/j.molp.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Xu X, et al. Convergence of light and chloroplast signals for de-etiolation through ABI4–HY5 and COP1. Nat. Plant. 2016;2:16066. doi: 10.1038/nplants.2016.66. [DOI] [PubMed] [Google Scholar]
- 18.Pedmale UV, et al. Cryptochromes interact directly with PIFs to control plant growth in limiting blue light. Cell. 2016;164:233–245. doi: 10.1016/j.cell.2015.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Podolec R, Ulm R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018;45:18–25. doi: 10.1016/j.pbi.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 20.Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome‐mediated inhibition of shade avoidance involves degradation of growth‐promoting bHLH transcription factors. Plant J. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- 21.Boccaccini A, et al. Low blue light enhances phototropism by releasing cryptochrome1-mediated inhibition of PIF4 expression. Plant Physiol. 2020;183:1780–1793. doi: 10.1104/pp.20.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao Y, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Procko C, Crenshaw CM, Ljung K, Noel JP, Chory J. Cotyledon-generated auxin is required for shade-induced hypocotyl growth in Brassica rapa. Plant Physiol. 2014;165:1285–1301. doi: 10.1104/pp.114.241844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Wit M, Martınez-Cero NC, Fankhauser C, Pierik R. Integration of phytochrome and cryptochrome signals determines plant growth during competition for light. Curr. Biol. 2016;26:3320–3326. doi: 10.1016/j.cub.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 25.Küpers JJ, van Gelderen K, Pierik R. Location matters: canopy light responses over spatial scales. Trends Plant Sci. 2018;23:865–873. doi: 10.1016/j.tplants.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 26.Kupers, J.J., Oskam, L. & Pierik, R. Photoreceptors regulate plant developmental plasticity through auxin. Plants9, 940, 10.3390/plants9080940 (2020). [DOI] [PMC free article] [PubMed]
- 27.Procko C, et al. The epidermis coordinates auxin-induced stem growth in response to shade. Gene Dev. 2016;30:1529–1541. doi: 10.1101/gad.283234.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savaldi-Goldstein S, Peto C, Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199–202. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, et al. Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell autonomously. Plant Cell. 2016;28:2770–2785. doi: 10.1105/tpc.16.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, et al. The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 2020;11:1053. doi: 10.1038/s41467-020-14905-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, et al. Phytochrome interacting factors (PIFs) are essential regulators for sucrose-induced hypocotyl elongation in Arabidopsis. J. Plant Physiol. 2011;168:1771–1779. doi: 10.1016/j.jplph.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Stewart JL, Maloof JN, Nemhauser JL. PIF genes mediate the effect of sucrose on seedling growth dynamics. PloS ONE. 2011;6:e19894. doi: 10.1371/journal.pone.0019894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart Lilley JL, Gee CW, Sairanen I, Ljung K, Nemhauser JL. An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol. 2012;160:2261–2270. doi: 10.1104/pp.112.205575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sairanen I, et al. Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell. 2012;24:4907–4916. doi: 10.1105/tpc.112.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh M, Gupta A, Singh D, Khurana JP, Laxmi A. Arabidopsis RSS1 mediates cross-talk between glucose and light signaling during hypocotyl elongation growth. Sci. Rep. 2017;7:16101. doi: 10.1038/s41598-017-16239-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simon NM, et al. The energy-signalling hub SnRK1 is important for sucrose-induced hypocotyl elongation. Plant Physiol. 2017;176:1299–1310. doi: 10.1104/pp.17.01395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, et al. TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 2016;26:1854–1860. doi: 10.1016/j.cub.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Granot D, David-Schwartz R, Kelly G. Hexose kinases and their role in sugar-sensing and plant development. Front. Plant Sci. 2013;4:44. doi: 10.3389/fpls.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Granot D. Putting plant hexokinases in their proper place. Phytochemistry. 2008;69:2649–2654. doi: 10.1016/j.phytochem.2008.08.026. [DOI] [PubMed] [Google Scholar]
- 40.Jang JC, Leon P, Zhou L, Sheen J. Hexokinase as a sugar sensor in higher plants. Plant Cell. 1997;9:5–19. doi: 10.1105/tpc.9.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai N, et al. Overexpression of Arabidopsis hexokinase in tomato plants inhibits growth, reduces photosynthesis, and induces rapid senescence. Plant Cell. 1999;11:1253–1266. doi: 10.1105/tpc.11.7.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore B, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–336. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- 43.Granot D, Kelly G. Evolution of guard-cell theories: the story of sugars. Trends Plant Sci. 2019;24:507–518. doi: 10.1016/j.tplants.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 44.Kelly G, et al. Hexokinase mediates stomatal closure. Plant J. 2013;75:977–988. doi: 10.1111/tpj.12258. [DOI] [PubMed] [Google Scholar]
- 45.Lugassi N, et al. Expression of Arabidopsis hexokinase in tobacco guard cells increases water-use efficiency and confers tolerance to drought and salt stress. Plants. 2019;8:613. doi: 10.3390/plants8120613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelly G, et al. Guard-cell hexokinase increases water-use efficiency under normal and drought conditions. Front. Plant Sci. 2019;10:1499. doi: 10.3389/fpls.2019.01499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly G, et al. The pitfalls of transgenic selection and new roles of AtHXK1: a high level of AtHXK1 expression uncouples hexokinase1-dependent sugar signaling from exogenous sugar. Plant Physiol. 2012;159:47–51. doi: 10.1104/pp.112.196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stein O, et al. Arabidopsis fructokinases are important for seed oil accumulation and vascular development. Front. Plant Sci. 2017;7:2047. doi: 10.3389/fpls.2016.02047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gangappa SN, Kumar SV. DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 2017;18:344–351. doi: 10.1016/j.celrep.2016.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun N, et al. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc. Nat. Acad. Sci. U.S.A. 2016;113:6071–6076. doi: 10.1073/pnas.1604782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen X, et al. Shoot-to-root mobile transcription factor HY5 coordinates plant carbon and nitrogen acquisition. Curr. Biol. 2016;26:640–646. doi: 10.1016/j.cub.2015.12.066. [DOI] [PubMed] [Google Scholar]
- 52.Abbas N, Maurya JP, Senapati D, Gangappa SN, Chattopadhyay S. Arabidopsis CAM7 and HY5 physically interact and directly bind to the HY5 promoter to regulate its expression and thereby promote photomorphogenesis. Plant Cell. 2014;26:1036–1052. doi: 10.1105/tpc.113.122515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faulkner C, Oparka KJ. Plasmodesmata. eLS. 2001;3:1–7. [Google Scholar]
- 54.Wille AC, Lucas WJ. Ultrastructural and histochemical studies on guard cells. Planta. 1984;160:129–142. doi: 10.1007/BF00392861. [DOI] [PubMed] [Google Scholar]
- 55.Fitzgibbon J, et al. A developmental framework for complex plasmodesmata formation revealed by large-scale imaging of the Arabidopsis leaf epidermis. Plant Cell. 2013;25:57–70. doi: 10.1105/tpc.112.105890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu J, et al. Regulation of stomatal development by stomatal lineage miRNAs. Proc. Nat. Acad. Sci. U.S.A. 2020;117:6237–6245. doi: 10.1073/pnas.1919722117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bates GW, et al. A comparative study of the Arabidopsis thaliana guard-cell transcriptome and its modulation by sucrose. PLoS ONE. 2012;7:e49641. doi: 10.1371/journal.pone.0049641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ortiz-Alcaide M, et al. Chloroplasts modulate elongation responses to canopy shade by retrograde pathways Involving HY5 and abscisic acid. Plant Cell. 2019;31:384. doi: 10.1105/tpc.18.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lawson T. Guard cell photosynthesis and stomatal function. N. Phytol. 2009;181:13–34. doi: 10.1111/j.1469-8137.2008.02685.x. [DOI] [PubMed] [Google Scholar]
- 60.Barton KA, et al. Epidermal pavement cells of Arabidopsis have chloroplasts. Plant Physiol. 2016;171:723–726. doi: 10.1104/pp.16.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594–e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hwang G, et al. Trehalose-6-phosphate signaling regulates thermoresponsive hypocotyl growth in Arabidopsis thaliana. EMBO Rep. 2019;20:e47828. doi: 10.15252/embr.201947828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh E, Zhu J-Y, Wang Z-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burko Y, Gaillochet C, Seluzicki A, Chory J, Busch W. Local HY5 activity mediates hypocotyl growth and shoot-to-root communication. Plant Commun. 2020;1:100078. doi: 10.1016/j.xplc.2020.100078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsuzuki T, Takahashi K, Tomiyama M, Inoue S, Kinoshita T. Overexpression of the Mg-chelatase H subunit in guard cells confers drought tolerance via promotion of stomatal closure in Arabidopsis thaliana. Front. Plant Sci. 2013;4:440. doi: 10.3389/fpls.2013.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kinoshita T, et al. FLOWERING LOCUS T regulates stomatal opening. Curr. Biol. 2011;21:1232–1238. doi: 10.1016/j.cub.2011.06.025. [DOI] [PubMed] [Google Scholar]
- 67.Endo M, Nakamura S, Araki T, Mochizuki N, Nagatani A. Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles. Plant Cell. 2005;17:1941–1952. doi: 10.1105/tpc.105.032342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pacín M, Semmoloni M, Legris M, Finlayson SA, Casal JJ. Convergence of CONSTITUTIVE PHOTOMORPHOGENESIS 1 and PHYTOCHROME INTERACTING FACTOR signalling during shade avoidance. N. Phytol. 2016;211:967–979. doi: 10.1111/nph.13965. [DOI] [PubMed] [Google Scholar]
- 69.Feng CM, Qiu Y, Van Buskirk EK, Yang EJ, Chen M. Light-regulated gene repositioning in Arabidopsis. Nat. Commun. 2014;5:3027. doi: 10.1038/ncomms4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Odanaka S, Bennett AB, Kanayama Y. Distinct physiological roles of fructokinase isozymes revealed by gene-specific suppression of frk1 and frk2 expression in tomato. Plant Physiol. 2002;129:1119–1126. doi: 10.1104/pp.000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 72.Craigon DJ, et al. NASCArrays: a repository for microarray data generated by NASC’s transcriptomics service. Nucl. Acid Res. 2004;32:D575–D577. doi: 10.1093/nar/gkh133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ulitsky I, et al. Expander: from expression microarrays to networks and functions. Nat. Protoc. 2010;5:303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 74.Yaffe H, et al. LogSpin: a simple, economical and fast method for RNA isolation from infected or healthy plants and other eukaryotic tissues. BMC Res. Notes. 2012;5:45. doi: 10.1186/1756-0500-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat. Method. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genom. Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom. Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie C, et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucl. Acid Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B (Method.) 1995;57:289–300. [Google Scholar]
- 82.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PloS ONe. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muller D, Leyser O. Auxin, cytokinin and the control of shoot branching. Ann. Bot. 2011;107:1203–1212. doi: 10.1093/aob/mcr069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The sequencing data from this study have been deposited in the The National Center for Biotechnology Information BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under the accession number PRJNA687355. The raw data from the Illumina sequencing have been deposited in NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR13316969-SRR13316975. The raw data referring to the plots shown in the figures are provided in Supplementary Data 1, 2. All relevant data are available from the authors upon request.