Abstract
Background
Convalescent plasma containing neutralizing antibody to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is under investigation for coronavirus disease 2019 (COVID-19) treatment. We report diverse virological characteristics of UK intensive care patients enrolled in the Immunoglobulin Domain of the REMAP-CAP randomized controlled trial that potentially influence treatment outcomes.
Methods
SARS-CoV-2 RNA in nasopharyngeal swabs collected pretreatment was quantified by PCR. Antibody status was determined by spike-protein ELISA. B.1.1.7 was differentiated from other SARS-CoV-2 strains using allele-specific probes or restriction site polymorphism (SfcI) targeting D1118H.
Results
Of 1274 subjects, 90% were PCR positive with viral loads 118–1.7 × 1011IU/mL. Median viral loads were 40-fold higher in those IgG seronegative (n = 354; 28%) compared to seropositives (n = 939; 72%). Frequencies of B.1.1.7 increased from <1% in November 2020 to 82% of subjects in January 2021. Seronegative individuals with wild-type SARS-CoV-2 had significantly higher viral loads than seropositives (medians 5.8 × 106 and 2.0 × 105 IU/mL, respectively; P = 2 × 10−15).
Conclusions
High viral loads in seropositive B.1.1.7-infected subjects and resistance to seroconversion indicate less effective clearance by innate and adaptive immune responses. SARS-CoV-2 strain, viral loads, and antibody status define subgroups for analysis of treatment efficacy.
Keywords: clade B.1.1.7, convalescent plasma, coronavirus, COVID-19, ELISA, polymerase chain reaction, randomized clinical trial, SARS-CoV-2, variant of concern, viral load
Patients enrolled in a trial for convalescent plasma treatment were highly heterogeneous in terms of viral loads, infecting SARS-CoV-2 types, and antibody status, each potentially influencing treatment outcomes. B.1.1.7 infections were associated with higher viral loads and reduced clearance postseroconversion.
The catastrophic zoonotic emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the human population in China at the end of 2019, and its subsequent pandemic spread, has caused global devastation [1]. To date, only limited treatment regimens exist for coronavirus disease 2019 (COVID-19) patients [2, 3], and their management is primarily supportive, with case-fatality rates remaining at 1%–2% in Western countries, including the UK [4]. In the search of alternative methods to treat COVID-19, convalescent plasma therapy boosting levels of neutralizing antibody has been considered as a potential means to reduce morbidity and mortality by providing or boosting levels of neutralizing antibody [5, 6], particularly if given in early stages of infection [7]. The Randomised Evaluation of COVID-19 Therapy (RECOVERY) and Randomized, Embedded, Multifactorial Adaptive Platform Trial for Community-Acquired Pneumonia (REMAP-CAP) randomized controlled trials have investigated the efficacy of convalescent plasma therapy conducted in the UK with REMAP-CAP also recruiting participants globally [7, 8]. In the REMAP-CAP trial [8] patients with severe COVID-19, restricted to those in intensive care units (ICUs), are transfused with 1 to 2 plasma units collected from donors with previously documented SARS-CoV-2 infection and confirmed to have high plasma titers of neutralizing antibody [9].
While final statistical analysis is awaited, both trials have been closed as initial analyses did not show a significant benefit of treatment across the overall study population in terms of either COVID-19–associated mortality or number of organ- support–free days [10, 11]. However, the final analyses of the data will need to account for patient variables that may influence treatment efficacy and address patient outcomes other than death. For example, the potential efficacy of convalescent plasma therapy may be influenced by disease stage at trial enrolment. Severe COVID-19 disease may be driven by either the direct effects of virus replication in the respiratory tract or by indirect damage associated with the often intense inflammatory antiviral response that usually leads to clearance of SARS-CoV-2. Treatment with neutralizing antibody may primarily influence outcomes during the early stages of active virus replication-induced disease but could be less relevant in those patients who have already seroconverted for anti-SARS-CoV-2 antibody. Therefore, we have determined the pretreatment viral loads in respiratory samples and serological status of study subjects.
Another variable influencing treatment outcomes may originate from effects of strain variation of SARS-CoV-2. A variant of SARS-CoV-2 with a D614G mutation in the spike protein has been described [12, 13], along with more recent mutants such as B.1.1.7 in the UK, B.1.351 in South Africa [14], and P.1 in Brazil [15], all potentially more transmissible [16–18]; furthermore, B.1.1.7 may result in a higher mortality than the original pandemic strain [19, 20]. As regards B.1.351 (South Africa), clustered amino acid changes in the spike protein gene may crucially render this mutant partially antigenically distinct in the receptor binding domain [21]. These virological differences, including the possibility of antigenic escape from neutralizing antibodies in convalescent plasma collected from individuals with earlier strains of SARS-CoV-2, may individually or collectively affect the efficacy of convalescent plasma therapy. To identify SARS-CoV-2 strains, we developed a real-time polymerase chain reaction (PCR) targeting the D1118H polymorphism in the spike gene that is characteristic of the B.1.1.7 strain. This assay was found to be faster and more effective than high-throughput sequencing for low viral load samples. A simpler agarose gel-based method was also developed and evaluated to enable B.1.1.7 identification in resource-limited settings.
We report substantial variability in pretreatment viral loads, variability in serological status, and an increasing detection of B.1.1.7 during the enrolment period of UK participants in the international REMAP-CAP trial (NCT02735707) [8]. Measurement of these variables will be of considerable value in analyzing effects of convalescent plasma therapy and potentially identifying subgroups of patients for whom this treatment may be effective.
METHODS
Patient Recruitment and Sample Collection
All subjects were enrolled in the UK and had a laboratory-confirmed diagnosis of SARS-CoV-2 infection with concomitant severe pneumonia requiring ICU admission. Patients were not eligible if more than 48 hours had elapsed since their ICU admission, if they had already received treatment with any other nontrial prescribed antibody therapy (monoclonal antibody, hyperimmune immunoglobulin, or convalescent plasma), or if more than 14 days had elapsed since hospital admission. A total of 122 hospitals have recruited patients in this trial in the UK [22]. Nasopharyngeal or oropharyngeal swabs and serum samples for baseline virological testing were collected from patients at enrolment and frozen at −80°C.
Ethics Statement
The study was conducted according to the principles of the latest version of the Declaration of Helsinki (version Fortaleza 2013), and in accordance with regulatory and legal requirements (EudraCT number: 2015-002340-14). The study was approved by London-Surrey Borders Research Ethics Committee London Centre (18/LO/0660). Written or verbal informed consent, in accordance with regional legislation, was obtained from all patients or their surrogates.
Nucleic Acid Extraction
Viral RNA was extracted from patient samples using the QIAamp Viral RNA Mini Kit according to the manufacturer’s protocol (QIAGEN). Respiratory sample, 140 µL, was mixed with 560 µL of Buffer AVL containing 20 µg/mL of linear polyacrylamide (ThermoFisher Scientific) and eluted into 60 µL of buffer AVE. Dry swabs were resuspended in 2 mL of phosphate-buffered saline and incubated at room temperature for 20 minutes prior to extraction. Total nucleic acid was extracted from the NIBSC reagent 19/304 using the QIAamp Viral RNA Mini Kit and serially diluted in RNA storage solution (Thermo Fisher Scientific; 1 mM sodium citrate, pH 6.4) containing herring sperm carrier RNA (50 µg/mL) and RNasin (100 U/mL; New England BioLabs). All samples were subject to a single freeze-thaw cycle between extraction and real time (RT)-PCR quantification.
Real-Time Reverse Transcriptase Polymerase Chain Reaction
Viral RNA was detected and quantified by RT-PCR using the Quantitect Probe RT-PCR kit (QIAGEN) with CDC N1 primers (Supplementary Table 1). RT-PCR was carried out on an Applied Biosystems StepOnePlus Real-Time PCR system (ThermoFisher Scientific) with the following settings: 50°C for 30 minutes, 95°C for 15 minutes, and 40 cycles of 94°C for 15 seconds and 60°C for 1 minute. Intraassay variation was standardized through use of a standard curve of National Institute for Biological Standards and Control (NIBSC) RNA control 19/304 serially diluted from 10 000 copies/reaction to 100 copies/reaction. Ct values were converted to international units/mL (IU/mL) by the conversion rate provided by NIBSC.
Typing Assays to Identify B.1.1.7 SARS-CoV-2 Strains
An RT-PCR using allele-specific probes (ASP) and nested PCR restriction fragment length polymorphism (RFLP) for the G→C polymorphism at position 24814 (D1118H) (Supplementary Table 1) were used to differentiate wild-type (WT) and B.1.1.7 strains (Table 1). Detailed methods are available as Protocols A and B (Supplementary Material).
Table 1.
Specificity and Sensitivity of the 8 Polymorphisms in B.1.1.7 for Strain Identification
| Polymorphisma | Strain | Specificity/Sensitivity, %e | |||
|---|---|---|---|---|---|
| B.1.1.7b | B.1.1.28c | B.1.351d | WTd | ||
| H69/V70 deletion | 98.73/100 | ||||
| WT | 0 | 374 | 834 | 36 577 | |
| Del | 26 328 | 0 | 0 | 470 | |
| Y144 deletion | 99.87/100 | ||||
| WT | 0 | 374 | 834 | 35 708 | |
| Del | 26 328 | 0 | 0 | 46 | |
| N501Y | 99.52/100 | ||||
| A | 0 | 294 | 0 | 36 868 | |
| U | 26 328 | 80 | 834 | 177 | |
| A570D | 99.98/99.99 | ||||
| C | 2 | 374 | 833 | 37 597 | |
| A | 26 359 | 0 | 0 | 0 | |
| P681H | 99.69/99.97 | ||||
| C | 0 | 374 | 832 | 37 442 | |
| A | 26 351 | 0 | 1 | 34 | |
| T716I | 99.94/99.99 | ||||
| C | 2 | 374 | 832 | 37 528 | |
| U | 26 351 | 0 | 1 | 23 | |
| S982A | 100/100 | ||||
| U | 0 | 374 | 831 | 37 563 | |
| G | 26 360 | 0 | 0 | 0 | |
| D1118H | 99.98/99.97 | ||||
| G | 0 | 374 | 834 | 37 579 | |
| C | 26 358 | 0 | 0 | 8 | |
Abbreviations: GISAID, Global Influenza Surveillance and Response System; SARS-CoV-2, severe acute respiratory syndrome coronavirus; WT, wild type.
aSource UK. For each mutation, mutations associated with B.1.1.7 shown in lower row.
bSource all. Available sequences from GISAID Nov, Dec 2020, and Jan 2021.
cSource all. All available sequence worldwide.
dSource UK. Available sequences from GISAID Jan-April, July, Nov, Dec 2020, and Jan 2021.
eSpecificity and sensitivity for differentiation B.1.1.7 from WT SARS-CoV-2 sequences.
Nucleotide Sequencing
Whole-virus genomes were sequenced using the virus-specific PCR-free sequencing method veSEQ. The full method description is provided in Protocol C (Supplementary Material).
Serology
Baseline blood samples were collected from all patients to determine the presence of SARS-CoV-2 immunoglobulin G (IgG) antibodies against spike protein at the time of transfusion. These were determined using a 386-well plate enzyme-linked immunosorbent assay (ELISA) coupled with an automated liquid handler [23].
Statistical Analysis
All analyses of association between demographic, clinical, and virological variables were performed using SPSS version 26. Categorical variables analyzed were patient sex, ethnic background, invasive ventilation, preexisting chronic conditions (asthma, other respiratory disease, liver disease, immunosuppression, cardiovascular disease, and diabetes), and infecting SARS-CoV-2 type (WT or B.1.1.7). Continuous variables were patient age (years), viral load, body mass index (BMI), and Acute Physiology and Chronic Health Evaluation II (APACHE II) score. Analysis of categorial variables associated with viral load differences were performed using the Mann-Whitney or Kruskal-Wallis nonparametric test. Continuous variables were analyzed by linear regression. For multivariate analysis, categorical variables were numerically recoded and analyzed with continuous variables using ANOVA (P IN = .05; P OUT = .1). Type association with patient variables were estimated using Pearson χ 2 (categorial variables), Mann-Whitney U test (continuous variables), and by binary logistic regression.
RESULTS
Trial Enrolment
The study group was derived from patients enrolled in the REMAP-CAP trial in the UK from 25 May 2020 through 7 January 2021, with the main demographic and clinical variables listed in Table 2. Viral loads and anti-SARS-CoV-2 IgG antibody status were determined in respiratory and/or serum samples collected from patients prior to convalescent plasma treatment or standard care.
Table 2.
Baseline Characteristics of the Study Group
| All | WT | B.1.1.7 | |
|---|---|---|---|
| Total No. of patients | 1192 | 485 | 251 |
| Background | |||
| Age, y, median (IQR) | 61 (52–70) | 63 (55–72) | 61 (51–70) |
| Sex, male/total (%) | 804/1192 (67.4) | 345/485 (71.1) | 166/251 (66.1) |
| Ethnicity, nonwhite/total (%) | 86/302 (28.5) | 50/167 (29.9) | 7/21 (33.3) |
| BMI, median (IQR) | 30.8 (26.7–36.0) | 30.1 (26.6–35.4) | 31.8 (27.4–37.4) |
| Preexisting comorbidities, No./total (%) | |||
| Immunocompromised | 54/1190 (4.5) | 28/485 (5.8) | 14/251 (5.6) |
| Diabetes | 361/1190 (30.3) | 163/485 (33.6) | 74/251 (29.5) |
| Respiratory disease | 274/1190 (23.0) | 129/485 (26.6) | 45/251 (17.9) |
| Cardiovascular disease | 98/1162 (8.4) | 44/477 (9.2) | 13/245 (5.3) |
| Severity | |||
| APACHE II score, median (IQR) | 12 (8–19) | 14 (9–20) | 12 (7.5–18) |
| Invasive ventilation, No./total (%) | 415/1192 (34.8) | 192/485 (39.6) | 96/251 (38.2) |
Abbreviations: BMI. Body mass index; IQR, interquartile range; WT, wild type.
Pretreatment Viral Loads
RNA extracted from respiratory samples was amplified by real-time RT-PCR targeting the nucleoprotein (N1) gene (Figure 1A) and viral loads calculated as IU/mL. The assay sensitivity threshold was set at 100 copies/mL. A total of 1141 from 1274 samples (90%) were SARS-CoV-2 RNA positive. Viral loads ranged from 118 IU/mL to 1.7 × 1011 IU/mL, a remarkable >2 billion-fold range in levels of secreted virus.
Figure 1.
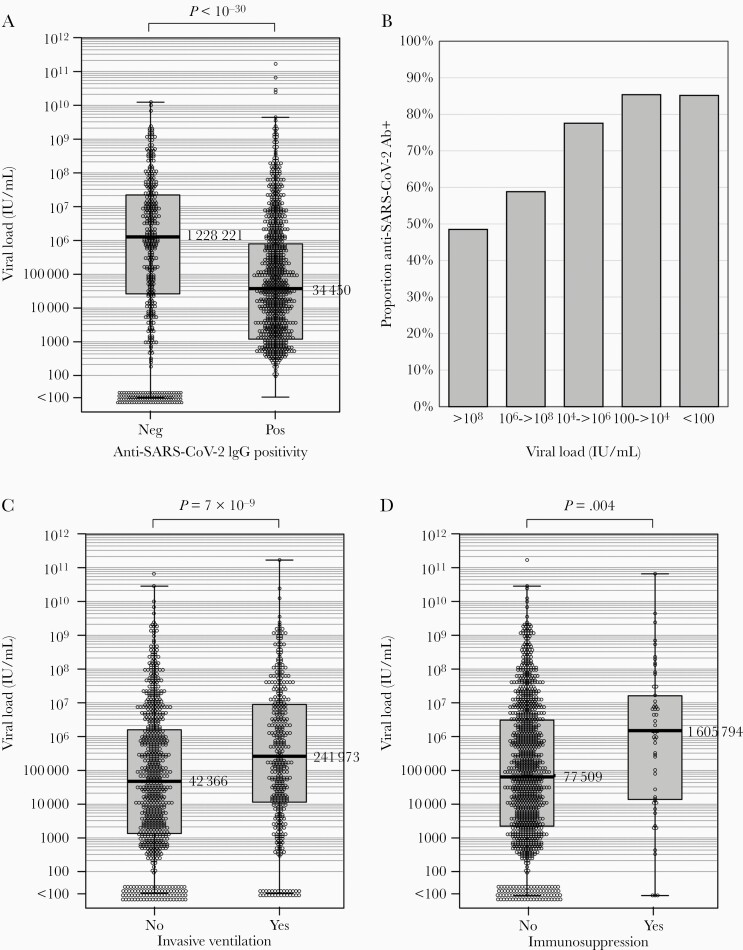
Comparison of viral loads in anti-SARS-CoV-2 seropositive and seronegative subjects. A, C, and D, Associations of anti-SARS-CoV-2 antibody status, invasive ventilation, and immunosuppression on viral load distributions as determined by RT-qPCR of pretreatment respiratory samples. Median values shown to the right of Tukey box plots. Distributions were compared by Mann-Whitney U test. B, Frequency of seropositivity in individuals with different viral loads quantified in respiratory samples. Abbreviations: IgG, immunoglobulin G; RT-qPCR, reverse transcription quantitative polymerase chain reaction; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Serology
The serological status was determined by spike ELISA. Results were expressed as positive and negative although the degree of antibody reactivity could not be quantified by the nature of the assay. Antibody status has been determined in 1293 patients included in this study to date, of whom 804 were determined as seropositive (62%). The median viral load of seronegative individuals was 36 times higher than seropositives with markedly different distributions of viral loads between the 2 groups (Figure 1A). The proportion of seropositive patients varied systematically with viral load ranges, being highest (85%) in those with the lowest viral loads, through to 48% in those with viral loads >108 IU/mL (Figure 1B).
Effects of patient variables and anti-SARS-CoV-2 antibody serostatus on viral loads were investigated (Table 2). Negative SARS-CoV-2 serostatus, increased age, and higher APACHE II score (reflecting disease severity) were each significantly associated with increased viral loads; those with diabetes, immunosuppression, and requiring invasive ventilation additionally showed significantly raised viral loads. Independent variables associated with increased viral loads on multivariate linear regression were age, invasive ventilation (Figure 1C), and immunosuppression (Figure 1D) in addition to antibody status.
SARS-CoV-2 Strain Identification
As infections with the emerging B.1.1.7 strain may respond differently to treatment compared to previously circulating SARS-CoV-2 strains, 2 different typing assays were developed to identify B.1.1.7 in trial patients. We first analyzed the specificity and predictive value of individual polymorphic sites in the spike gene of B.1.1.7 (H69/V70del., Y144del., N501Y, A570D, P681H, T716I, S982A, and D1118H) that differentiated them from other SARS-CoV-2 strains (Supplementary Table 2). All but one of the defined polymorphisms differentiated the B.1.1.7 strain with very high sensitivity and specificity from previously circulating strains in the United Kingdom and from emerging strains in South Africa and Brazil (B.1.351 and P.1). The exception was the H69/V70 del site, which showed increasing frequencies with time in multiple lineages of WT strains (Supplementary Table 2). As the G→C change associated with D1118H also led to the loss of a restriction enzyme site (SfcI), amplicons from this region were used in typing assays with ASP (Supplementary Figure 1A) and nested-PCR RFLP assays (Supplementary Figure 1B).
For baseline genomic characterization, the first 284 respiratory samples collected in November 2020 from the trial patients were sequenced by high-throughput sequencing using SARS-CoV-2–baited target enrichment. Variable coverage of the genome was achieved, associated with viral load (Supplementary Figure 2). All sequences with coverage over codon 164 of the spike gene showed the D614G mutation. Phylogenetic analysis (Supplementary Figure 3) of those samples showing >65% sequence coverage demonstrated that the majority fell into the lineage GV (according to its designation in Global Influenza Surveillance and Response System [GISAID]). Of the sequences obtained, 13 of the 173 sequences with sufficient sequence in the spike gene to enable assignment were identifiable as B.1.1.7. No sequences corresponded to emerging strains B.1.351 (South Africa) or P.1 (Brazil).
A subset of 40 samples sequenced across the spike gene were used for evaluation of the typing assays. These produced highly concordant results with 100% specificity, 97.5% sensitivity for PCR using allele-specific probes, and 97.5% specificity, 93.5% sensitivity for the RFLP assay (Supplementary Table 3). Study samples were typed by a combination of the 3 methods with 808 (527 WT; 281 B.1.1.7) from 1119 tested (72%) successfully typed; >98% of samples with viral loads >106 RNA copies/mL were successfully typed by ASP and RFLP assays (Supplementary Figure 4).
Emergence and Virological Associations of SARS-CoV-2 Strains
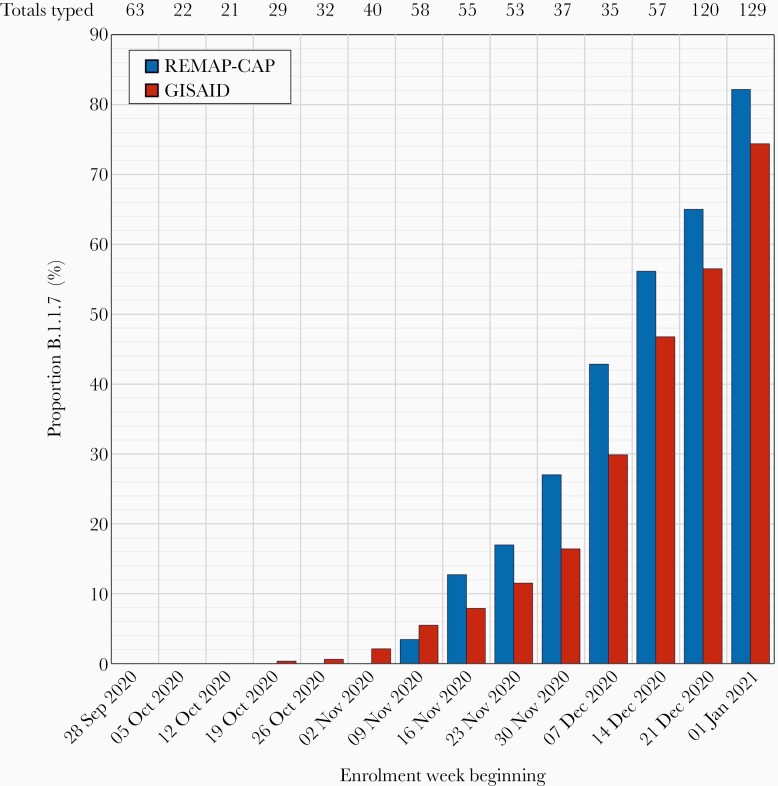
Infections with the B.1.1.7 strain were first detected among enrolled patients in early November 2020 and rapidly increased in frequency through December and early January, representing over 80% of infections in week 1, 2021 (Figure 2). Frequencies of B.1.1.7 infections matched or indeed occurred ahead of their emergence in the general UK population based on analysis of SARS-CoV-2 sequences deposited on GISAID in the corresponding study weeks. Geographically, B.1.1.7 infections occurred at greatest frequency in subjects enrolled in South East England (including London) and in East England (approximately 50%; Supplementary Figure 5). Cohort enrolment from different regions of England closely matched the proportions of COVID-19 diagnoses reported to Public Health England over the study period; B.1.1.7-infected subjects showed comparable demographics (age range, sex distribution, ethnicity, and BMIs) as those infected with WT virus (Table 2).
Figure 2.
Temporal emergence of the SARS-CoV-2 B.1.1.7 clade. Proportion of subjects with the B.1.1.7 clade virus enrolled to the REMAP-CAP trial in different weeks over the study period compared to proportions in the wider UK population from sequences deposited in GISAID. Numbers at the top of the graph indicate total enrolments/week. Abbreviations: GISAID, Global Influenza Surveillance and Response System REMAP-CAP, Randomized, Embedded, Multifactorial Adaptive Platform Trial for Community-Acquired Pneumonia; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
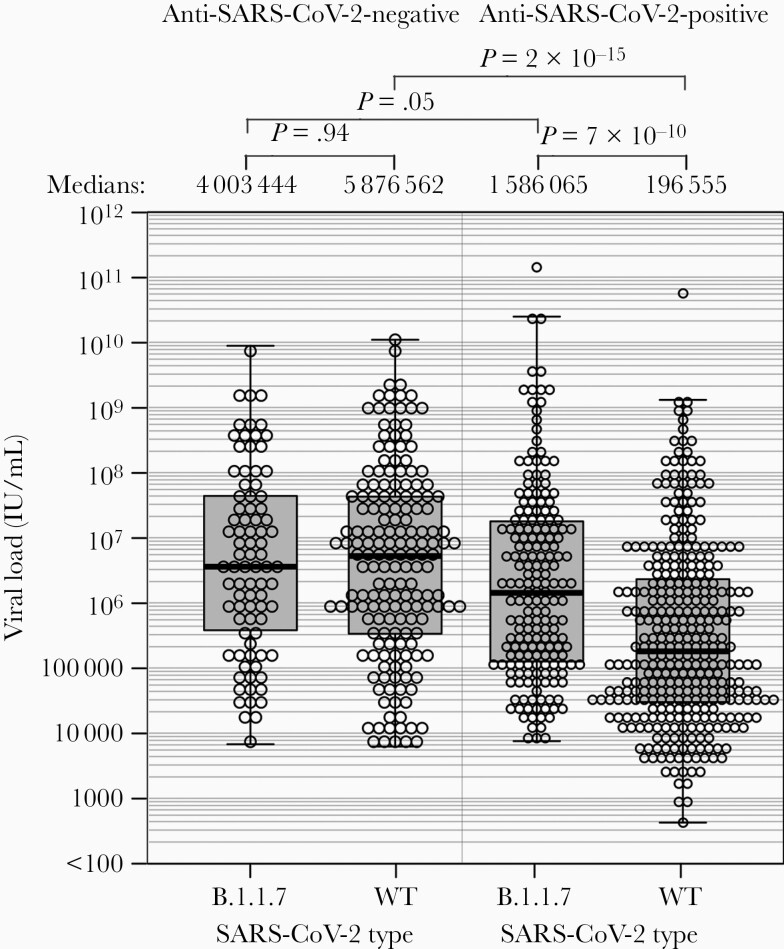
However, viral loads ranges from subjects infected with the B.1.1.7 strain differed substantially from those with WT SARS-CoV-2, with medians of 2 424 129 (n = 273) and 656 801 (n = 512) IU/mL respectively (P = 1.4 × 10–4). Remarkably, significant differences in viral loads were only apparent among the 528 subjects who were seropositive for SARS-CoV-2 antibody (Figure 3), with an approximately 8-fold difference between medians of B.1.1.7 and WT infections. Contrastingly, viral loads varied by less than 1.5-fold between those with B.1.1.7 and WT infections in the 257 seronegative subjects (P = .94). Seropositivity had marginal effect on B.1.1.7 viral loads with viral load distributions elevated in both seronegative (n = 85) and seropositive (n = 188) subjects infected with B.1.1.7 (4.0 × 106 and 1.6 × 106 IU/mL, respectively).
Figure 3.
Viral loads of wild-type and B.1.1.7 strains in seronegative and seropositive subjects. Distributions of viral loads of in samples from patients infected with wild-type and B.1.1.7 strains, subdivided by serostatus. Distributions were compared by Mann-Whitney U test test. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; WT, wild type.
Among the previously listed demographic or clinical variables, only age (medians of 63 and 60 years for WT and B.1.1.7, respectively) and BMI score (31.8 and 30.1) varied significantly between those infected with different SARS-CoV-2 types (Supplementary Table 4), although BMI was not independently associated on multivariate analysis. Such differences were only manifest in those who were anti-SARS-CoV-2 antibody positive. For metrics of infection severity, rates of invasive ventilation were comparable between WT and B.1.1.7 infections (Table 3 and Table 4). Contrastingly, APACHE II scores were higher in the WT infections (median 15 compared to 11; P = 5.6 × 10–5) and remained significantly different on multivariate analysis (Table 4).
Table 3.
Patient and Virological Variables Associated With Virus Load and Virus Type
| Variable | No | Yes | P | MLR Pb |
|---|---|---|---|---|
| Categorical variables | ||||
| Diabetes | 62 570 (2526–3 125 789) | 193 575 (4443–7 863 774) | .015a | .099 |
| Immunosuppression | 77 510 (2790–3 554 804) | 1 605 794 (16 884–18 369 106) | .004a | .013 |
| Invasive ventilation | 42 366 (1661–1 782 723) | 241 973 (13 951–10 369 484) | 7.1 × 10–9a | 5.3 × 10–7 |
| Anti-SARS-CoV Ab | 1 289 633 (39 151–26 681 971) | 36 264 (1543–1 262 019) | <1 × 10–3a | 1.5 × 10–2 |
| Continuous variables | ||||
| Age, R2 | 0.015 | .00003c | .002 | |
| APACHE II, R2 | 0.025 | 5.7 × 10–8c | .15 |
Data listed for categorical variables are viral loads, IU/mL, for continuous variables, correlation coefficients are listed.
Abbreviations: Ab, antibody; APACHE II, Acute Physiology and Chronic Health Evaluation II; MLR, multivariate linear regression; SARS-CoV, severe acute respiratory syndrome coronavirus.
aP values from Mann-Whitney U test.
bP values from MLR with log transformed viral loads.
cP values from simple linear regression with log transformed viral loads.
Table 4.
Associations of Disease Severity With Virus Type
| Characteristic | WT | B.1.1.7 | P | BLR Pa |
|---|---|---|---|---|
| All | ||||
| Viral load, IU/mL | 718 445 (43 785–8 682 234) | 3 536 790 (198 314–32 349 877) | 2.7 × 10–8 | 4.8 × 10–9 |
| APACHE II | 14 (9–20) | 12 (8–18) | .026 | .005 |
| Invasive ventilation, % | 38.2 | 39.6 | .72 | .97 |
| Antibody negative | ||||
| Viral load, IU/mL | 5 746 214 (367 928–48 852 455) | 4 168 803 (335 710–57 126 270) | .57 | .30 |
| APACHE II | 15 (11–21) | 11 (7–17) | 5.6 × 10–5 | .001 |
| Invasive ventilation, % | 29.5 | 42.8 | .048 | .38 |
| Antibody positive | ||||
| Viral load, IU/mL | 196 556 (32 219–2 573 817) | 1 840 314 (137 041–22 308 100) | 1.5 × 10–10 | 2.3 × 10–10 |
| APACHE II | 12 (8–20) | 12 (8–19) | .99 | .27 |
| Invasive ventilation, % | 42.2 | 38.0 | .39 | .50 |
Abbreviations: APACHE II, Acute Physiology and Chronic Health Evaluation II; BLR, binary logistic regression; WT, wild type.
aP values from BLR with log transformed viral loads.
DISCUSSION
The motivation for this study was the need to characterize the virological variables, viral loads, anti-SARS-CoV-2 serology status, and SARS-CoV-2 genotype that may individually or collectively influence the observed treatment efficacy of convalescent plasma. For example, emerging SARS-CoV-2 variants have been associated with reduced susceptibility to neutralization [12, 21, 24] and greater pathogenicity [20, 25], and potentially greater resistance to immunotherapy. Such analyses have been rarely performed in previous or ongoing COVID-19 treatment trials, even though the extensive variability in these parameters is likely to exert potent effects on patient response. In this respect, the calibration of viral loads into international units using the NIBSC standard will be important in enabling better standardization of quantitative data and more effective cross-trial comparisons. The widely reported uncalibrated Ct values provide only an indirect metric of viral load, differing for example through varying efficiencies of primer/probe combinations [26] that do not make quantitative results directly comparable across studies.
Our most striking observation was an approximately billion-fold variation in SARS-CoV-2 viral loads. While the predictive value of viral loads for clinical outcomes or treatment response is unclear, with several studies reporting significant [27–32], minimal [33, 34], or no [35–37] associations with increased severity or mortality from COVID-19, we report significant independent associations of pretreatment variables such as invasive ventilation and immunosuppression with higher viral loads in our cohort, which may subsequently influence their response to treatment (Table 2 and Figure 1C). The range of viral loads we observed is consistent with the kinetics of SARS-CoV-2 secretion in the respiratory tract, where rapid decline following the acute infection stage has been observed [29, 31, 35, 36]. Indeed, the observation that RNA levels were <100 IU/mL in 10% of subjects on PCR screening (Figure 1A) despite severe COVID-19 leading to ICU admission suggests that a substantial element in the disease mechanisms of this latter group is inflammatory or cytokine related, rather than being directly virally induced. The higher frequency of seropositivity for SARS-CoV-2 in subjects who were classed as PCR negative or showed lower viral loads in those infected with the WT genotype (Figure 1B) provides further evidence for differences in the timings of severe disease presentations relative to the evolution of SARS-CoV-2 infection. Overall, 62% of patients included in this study were shown to have SARS-CoV-2 antibodies.
Prospectively, viral load measurements and serological status provides potentially valuable information for patient stratification in treatment selection for COVID-19. Antiviral effect of neutralizing antibodies in convalescent plasma may be most apparent in seronegative patients where respiratory tract pathology is primarily virus driven. Conversely, those presenting after seroconversion where disease mechanisms may primarily derive from inflammation-mediated damage may not be helped by infusion of additional neutralizing antibodies and who may better respond to interventions that modulate host response (corticosteroids, interleukin-6 receptor antagonists [2, 38–40]). The importance of identifying those at early stages of infection is demonstrated by the efficacy of convalescent plasma given within 3 days from disease onset [7] whereas no benefit was seen when plasma was used in later-stage infections (median 8 days from onset [41]).
The effects of mutations in the spike gene on the transmissibility, viral loads, disease severity, and potential antigenic variation is currently an area of substantial concern for COVID-19 pandemic management, public health measures, and immunization programs. The development of practical methods developed here for rapid and large-scale identification of the B.1.1.7 strains is of paramount importance for monitoring its spread worldwide, given its greater propensity to transmit [18]. The RFLP assay furthermore enables typing to be performed in resource-limited diagnostic facilities.
SARS-CoV-2 B.1.1.7 strains became increasingly prevalent in the UK population whilst the REMAP-CAP convalescent plasma trial patients were drawn. The study subjects showed a substantial representation of B.1.1.7 infections, particularly those recruited towards the end of the study period (Figure 2), mirroring the appearance of this variant in the wider UK population over this period among primarily nonhospitalized individuals. In terms of the potential effects of their emergence on trial outcomes, several spike mutations are associated with reductions in susceptibility to neutralization by antibodies elicited by infection or immunization with previously circulating variants of SARS-CoV-2 [24, 42]. However, whether this affects their susceptibility to convalescent plasma treatment is unclear—less than 10-fold reductions in titers are typically observed in in vitro neutralization or pseudotype assays and there was little or no effect on detection of anti-spike binding antibodies by ELISA [24, 43, 44].
In terms of potential pathogenicity differences, recent investigations have shown B.1.1.7 infections to lead to a 1.7-fold higher hazard of death within 28 day of diagnosis [25]. These findings are consistent with some, but not all, ongoing studies (summarized in [20]), including higher rates of hospitalization and intensive care admission [45], higher relative case fatality ratios between B.1.1.7 and WT of 1.29–1.36 (Imperial College London), and mortality hazard ratios of 1.7 (Public Health England) and 1.7 (University of Exeter). The observation that B.1.1.7 patients prior to seroconversion for anti-SARS-CoV-2 antibody were significantly more likely to be ventilated (Table 2B) is consistent with a possible greater severity of B.1.1.7 infections (Table 2). However, contrasting with these conclusions is the recent study of hospitalized COVID-19 patients that found no association between B.1.1.7 infections and disease severity or mortality [46].
The observation that viral loads were higher in B.1.1.7-infected individuals than those infected with WT, but only in those who had already seroconverted for anti-SARS-CoV-2 antibody (40-fold higher; Figure 3), are consistent with replication capacity and/or a less effective cellular or humoral host immune response to contain B.1.1.7 replication than WT. This is supported by the previous observation of higher viral loads in individuals infected with B.1.1.7 [46, 47]. A prolonged duration of respiratory virus shedding in subjects infected with B.1.1.7 compared to other strains (mean values of 13.3 and 8.2 days respectively) but equal peak viral loads [48] supports our evidence for the potentially extended trajectory of B.1.1.7 infections and delayed clearance manifested in Figure 3. This contrasts with the G614 spike mutant that possesses greater in vitro fitness and replicative capacity compared to WT strains with D614 [49, 50].
Virus strain characterization contributes substantially to differentiating patients in the REMAP-CAP cohort and the association of viral genotype with higher viral loads suggests potential differences in treatment response. While current indications from the REMAP-CAP trial demonstrate no overall effect on the treated cohort compared to untreated controls [11], data obtained in the current results will form the basis for further analyses of treatment efficacy in subgroups defined by antibody status, viral load band, and by infecting strain. This post hoc analysis may identify specific patient groups who will benefit from convalescent plasma and related immunotherapies in the future.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We are grateful to the National Institute for Health Research (NIHR) Clinical Research Network, UK for their support of participant recruitment. We are grateful to the UK Blood Services (NHS Blood and Transplant, Northern Ireland Blood Transfusion Service, Scottish National Blood Transfusion Service, and Welsh Blood Service) for the supply of convalescent plasma in the UK.
Disclaimer. The views expressed in this publication are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
Financial support. This work was supported by the National Institute for Health UKRIDHSC COVID-19 Rapid Response Rolling Call (grant number COV19-RECPLAS); the European Commission (SUPPORT-E (grant number 101015756); European Union Platform for European Preparedness Against (Re-) Emerging Epidemics (PREPARE) consortium, FP7-HEALTH-2013-INNOVATION-1 (grant number 602525); and European Union Horizon 2020 Research and Innovation Programme Rapid European COVID-19 Emergency Research response (RECOVER) consortium (grant number 101003589); the UK NIHR; and the NIHR Imperial Biomedical Research Centre. J. R. is supported by the Marshall Scholarship and Clarendon Fund, A. C. G. by an NIHR Research Professorship (grant number RP-2015-06-18), M. S. H. by an NIHR Clinician Scientist Fellowship (grant number CS-2016-16-011), and D. K. M. by the NIHR through the Cambridge NIHR BRC and the Addenbrooke’s Charitable Trust.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World Health Organization. WHO coronavirus (COVID-19) dashboard.https://covid19.who.int. Accessed 12 May 2021.
- 2.Angus DC, Derde L, Al-Beidh F, et al. ; Writing Committee for the REMAP-CAP Investigators . Effect of hydrocortisone on mortality and organ support in patients with severe COVID-19: the REMAP-CAP COVID-19 corticosteroid domain randomized clinical trial. JAMA 2020; 324:1317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sterne JAC, Murthy S, Diaz JV, et al. Association between administration of systemic corticosteroids and mortality among critically ill patients with COVID-19: a meta-analysis. JAMA 2020; 324:1330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harman K, Allen H, Kall M, Dabrera G. Interpretation of COVID-19 case fatality risk measures in England [published online ahead of print 29 January 2021]. J Epidemiol Community Health doi: 10.1136/jech-2020-216140. [DOI] [PubMed] [Google Scholar]
- 5.Duan K, Liu B, Li C, et al. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A 2020; 117:9490–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shankar-Hari M, Estcourt L, Harvala H, Roberts D, Menon DK; United Kingdom SARS-CoV-2 Convalescent Plasma Evaluation (SCoPE) Consortium . Convalescent plasma to treat critically ill patients with COVID-19: framing the need for randomised clinical trials. Crit Care 2020; 24:449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libster R, Pérez Marc G, Wappner D, et al. ; Fundación INFANT–COVID-19 Group . Early high-titer plasma therapy to prevent severe COVID-19 in older adults. N Engl J Med 2021; 384:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angus DC, Berry S, Lewis RJ, et al. The REMAP-CAP (randomized embedded multifactorial adaptive platform for community-acquired pneumonia) study. rationale and design. Ann Am Thorac Soc 2020; 17:879–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvala H, Mehew J, Robb ML, et al. Convalescent plasma treatment for SARS-CoV-2 infection: analysis of the first 436 donors in England, 22 April to 12 May 2020. Euro Surveill 2020; 25:2001260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Randomised Evaluation of COVID-19 Therapy (RECOVERY). RECOVERY trial closes recruitment to convalescent plasma treatment for patients hospitalised with COVID-19. Statement from the RECOVERY trial chief investigators,15. January 2021. https://www.recoverytrial.net/news/statement-from-the-recovery-trial-chief-investigators-15-january-2021-recovery-trial-closes-recruitment-to-convalescent-plasma-treatment-for-patients-hospitalised-with-covid-19. Accessed 12 May 2021.
- 11.O’Hare R. Blood plasma treatment has limited effect for sickest COVID-19 patients,11. January 2021. https://www.imperial.ac.uk/news/211493/blood-plasma-treatment-limited-effect-sickest/. Accessed 12 May 2021.
- 12.Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021; 592:116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021; 184:64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tegally H, Wilkinson E, Giovanetti M, et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021; 592:438–43. [DOI] [PubMed] [Google Scholar]
- 15.Voloch CM, Silva F Rd, de Almeida LGP, et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J Virol 2021; 95:e00119–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung K, Shum MH, Leung GM, Lam TT, Wu JT. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Euro Surveill 2021; 26:2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graham MS, Sudre CH, May A, et al. Changes in symptomatology, re-infection and transmissibility associated with SARS-CoV-2 variant B.1.1.7: an ecological study. Lancet Public Health 2021; 6:e335–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volz E, Mishra S, Chand M, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021; 593:266–9. [DOI] [PubMed] [Google Scholar]
- 19.Iacobucci G. COVID-19: New UK variant may be linked to increased death rate, early data indicate. BMJ 2021; 372:n230. [DOI] [PubMed] [Google Scholar]
- 20.Horby P, Bell I, Breuer J, et al. NERVTAG paper update note on B.1.1.7 severity,11. February 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/961042/S1095_NERVTAG_update_note_on_B.1.1.7_severity_20210211.pdf. Accessed 12 May 2021.
- 21.Wibmer CK, Ayres F, Hermanus T, et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med 2021; 27:622–5. [DOI] [PubMed] [Google Scholar]
- 22.Randomized, Embedded, Multifactorial Adaptive Platform Trial for Community-Acquired Pneumonia (REMAP-CAP). Domain-specific appendix: COVID-19 Immunoglobulin Therapy Domain, 1 June2020. https://static1.squarespace.com/static/5cde3c7d9a69340001d79ffe/t/5f5c842bd7bf523730bdda76/1599898680445/REMAP-CAP+COVID-19+Immunoglobulin+Therapy+Domain-Specific+Appendix+V1.01-+01+June+2020.pdf. Accessed 12 May 2021. [Google Scholar]
- 23.National SARS-CoV-2 Serology Assay Evaluation Group. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020; 20:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muik A, Wallisch AK, Sänger B, et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science 2021; 371:1152–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies NG, Jarvis CI, Edmunds WJ, Jewell NP, Diaz-Ordaz K, Keogh RH. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature 2021; 593:270–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahdouh E, Lázaro-Perona F, Romero-Gómez MP, Mingorance J, García-Rodriguez J. Ct values from SARS-CoV-2 diagnostic PCR assays should not be used as direct estimates of viral load. J Infect 2021; 82:414–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zacharioudakis IM, Zervou FN, Prasad PJ, et al. Association of SARS-CoV-2 genomic load trends with clinical status in COVID-19: a retrospective analysis from an academic hospital center in New York City. PLoS One 2020; 15:e0242399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryan A, Fink SL, Gattuso MA, et al. SARS-CoV-2 viral load on admission is associated with 30-day mortality. Open Forum Infect Dis 2020; 7:ofaa535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fajnzylber J, Regan J, Coxen K, et al. ; Massachusetts Consortium for Pathogen Readiness . SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun 2020; 11:5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faíco-Filho KS, Passarelli VC, Bellei N. Is higher viral load in SARS-CoV-2 associated with death? Am J Trop Med Hyg 2020; 103:2019–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lui G, Ling L, Lai CK, et al. Viral dynamics of SARS-CoV-2 across a spectrum of disease severity in COVID-19. J Infect 2020; 81:318–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X, Sun S, Shi Y, Wang H, Zhao R, Sheng J. SARS-CoV-2 viral load in sputum correlates with risk of COVID-19 progression. Crit Care 2020; 24:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrasquer A, Peiró Ó M, Sanchez-Gimenez R, et al. Lack of association of initial viral load in SARS-CoV-2 patients with in-hospital mortality. Am J Trop Med Hyg 2021; 104: 540–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang Z, Zhang Y, Hang C, Ai J, Li S, Zhang W. Comparisons of viral shedding time of SARS-CoV-2 of different samples in ICU and non-ICU patients. J Infect 2020; 81:147–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yilmaz A, Marklund E, Andersson M, et al. Upper respiratory tract levels of severe acute respiratory syndrome coronavirus 2 RNA and duration of viral RNA shedding do not differ between patients with mild and severe/critical coronavirus disease 2019. J Infect Dis 2021; 223:15–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou L, Ruan F, Huang M, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 2020; 382:1177–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Borgne P, Solis M, Severac F, et al. ; CRICS TRIGGERSEP Group (Clinical Research in Intensive Care and Sepsis Trial Group for Global Evaluation and Research in Sepsis) . SARS-CoV-2 viral load in nasopharyngeal swabs in the emergency department does not predict COVID-19 severity and mortality. Acad Emerg Med 2021; 28:306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.RECOVERY Collaborative Group. Tocilizumab in patients admitted to hospital with COVID-19 (RECOVERY): preliminary results of a randomised, controlled, open-label, platform trial. Lancet 2021; 397:2049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon AC, Mouncey PR, Al-Beidh F, et al. Interleukin-6 receptor antagonists in critically ill patients with COVID-19—preliminary report. N Engl J Med 2021; 384:1491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galani IE, Rovina N, Lampropoulou V, et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat Immunol 2021; 22:32–40. [DOI] [PubMed] [Google Scholar]
- 41.Simonovich VA, Burgos Pratx LD, Scibona P, et al. A randomized trial of convalescent plasma in COVID-19 severe pneumonia. N Engl J Med 2021; 384:619–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes WA, Kamath K, Lucas C, Shon J, Iwasaki A. Impact of B.1.1.7 variant mutations on antibody recognition of linear SARS-CoV-2 epitopes. medRxiv, doi: 10.1101/2021.01.06.20248960, 8. January 2021, preprint: not peer reviewed. [DOI] [Google Scholar]
- 43.Collier D, Meng B, Ferreira I, et al. SARS-CoV-2 B.1.1.7 sensitivity to mRNA vaccine-elicited, convalescent and monoclonal antibodies. medRxiv, doi: 10.1101/2021.01.19.21249840, 15. February 2021, preprint: not peer reviewed. [DOI] [Google Scholar]
- 44.Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 2021; 27:917–24. [DOI] [PubMed] [Google Scholar]
- 45.Funk T, Pharris A, Spiteri G, et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Euro Surveill 2021; 26:2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frampton D, Rampling T, Cross A, et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect Dis 2021; S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kidd M, Richter A, Best A, et al. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J Infect Dis 2021; 223:1666–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kissler SM, Fauver JR, Mack C, et al. Densely sampled viral trajectories suggest longer duration of acute infection with B.1.1.7 variant relative to non-B.1.1.7 SARS-CoV-2. medRxiv, doi: 10.1101/2021.02.16.21251535, 19. February 2021, preprint: not peer reviewed. [DOI] [Google Scholar]
- 49.Zhang L, Jackson CB, Mou H, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun 2020; 11:6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hou YJ, Chiba S, Halfmann P, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020; 370:1464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.