Abstract
Because it is the precursor for various essential cellular components, the amino acid serine is indispensable for every living organism. In plants, serine is synthesized by two major pathways: photorespiration and the phosphorylated pathway of serine biosynthesis (PPSB). However, the importance of these pathways in providing serine for plant development is not fully understood. In this study, we examine the relative contributions of photorespiration and PPSB to providing serine for growth and metabolism in the C3 model plant Arabidopsis thaliana. Our analyses of cell proliferation and elongation reveal that PPSB-derived serine is indispensable for plant growth and its loss cannot be compensated by photorespiratory serine biosynthesis. Using isotope labeling, we show that PPSB-deficiency impairs the synthesis of proteins and purine nucleotides in plants. Furthermore, deficiency in PPSB-mediated serine biosynthesis leads to a strong accumulation of metabolites related to nitrogen metabolism. This result corroborates 15N-isotope labeling in which we observed an increased enrichment in labeled amino acids in PPSB-deficient plants. Expression studies indicate that elevated ammonium uptake and higher glutamine synthetase/glutamine oxoglutarate aminotransferase (GS/GOGAT) activity causes this phenotype. Metabolic analyses further show that elevated nitrogen assimilation and reduced amino acid turnover into proteins and nucleotides are the most likely driving forces for changes in respiratory metabolism and amino acid catabolism in PPSB-deficient plants. Accordingly, we conclude that even though photorespiration generates high amounts of serine in plants, PPSB-derived serine is more important for plant growth and its deficiency triggers the induction of nitrogen assimilation, most likely as an amino acid starvation response.
The phosphorylated pathway of serine biosynthesis is required to synthesize serine for plant growth; and its deficiency triggers an amino acid starvation response by inducing nitrogen assimilation.
Introduction
The availability of serine is crucial for every living organism, as it is a constituent of proteins and indispensable for several cellular processes, such as the metabolism of folate and the biosynthesis of nucleotides, amino acids, the antioxidant glutathione, phospholipids, and sphingolipids (Kalhan and Hanson, 2012; Ros et al., 2014). In most organisms, serine is synthesized via the phosphorylated pathway of serine biosynthesis (PPSB; Supplemental Figure S1), starting from the glycolytic intermediate 3-phosphoglycerate (3-PGA; Pizer, 1963; Snell, 1984; Achouri et al., 1997; Dey et al., 2005; Tabatabaie et al., 2010; Locasale et al., 2011). The first and rate-limiting reaction of the PPSB is catalyzed by 3-PGA dehydrogenase (PGDH), which oxidizes 3-PGA into 3-phosphohydroxypyruvate. The latter is converted to 3-phosphoserine by 3-phosphoserine aminotransferase (PSAT) via a transamination reaction using glutamate as an amino-group donor. The final reaction of the pathway, the conversion of 3-phosphoserine into serine, is catalyzed by 3-phosphoserine phosphatase (PSP). This pathway is well conserved across prokaryotic and eukaryotic species (Wulfert and Krueger, 2018), indicating its essential role for cellular metabolism.
In heterotrophic organisms, PPSB-mediated serine biosynthesis acts as a central node in the metabolic network crucial for cell proliferation (Locasale et al., 2011; Yang and Vousden, 2016). It has been previously shown that the PPSB represents an important metabolic flux control point balancing the glycolytic carbon flow between the biosynthesis of serine and the tricarboxylic acid (TCA) cycle in fast proliferating cancer cells (Locasale et al., 2011; Chaneton et al., 2012; Labuschagne et al., 2014; Kottakis et al., 2016; Mattaini et al., 2016; Gao et al., 2018). Consequently, the PPSB has become of major interest in cancer research (Possemato et al., 2011; Amelio et al., 2014).
In contrast to heterotrophic organisms, serine metabolism in plants is still poorly understood. Plants possess three possible routes for the formation of serine (Supplemental Figure S1): the PPSB in heterotrophic tissue, the glycolate pathway associated with photorespiration in autotrophic tissue, and the glycerate pathway (Kleczkowski and Givan, 1988; Kleczkowski et al., 1988; Ho and Saito, 2001; Ros et al., 2014). Photorespiration is considered to be the main source of serine in plants (Bauwe et al., 2012), whereas the importance of the glycerate pathway for serine biosynthesis is still unknown due to the lack of clear genetic evidence for the rate limiting enzyme 3-PGA phosphatase (Robinson, 1982).
Photorespiration is required in photoautotrophic organisms due to the dual nature of RuBisCO (Andrews et al., 1971; Bowes et al., 1971). Although the carboxylase activity of RuBisCo yields two molecules 3-PGA, the oxygenase activity produces one molecule 3-PGA and one molecule 2-phosphoglycolate, a toxic intermediate. The photorespiratory pathway converts two molecules 2-phosphoglycolate to CO2 and 3-PGA via a series of complex reactions. Although photorespiration is often seen as a wasteful pathway due to the loss of up to 25% of the photosynthetic fixed carbon, it is also considered to be an important source for the amino acids glycine and serine in plants (Bauwe et al., 2010).
The presence of the PPSB in plants was initially described by Handford and Davies (1958) and the first genetic evidence of its importance was previously obtained by studying mutants deficient in the major enzymes of the pathway (Benstein et al., 2013; Cascales-Minana et al., 2013). The Arabidopsis thaliana genome encodes three genes for PGDH (At4g34200, PGDH1; At1g17745, PGDH2; At3g19480, PGDH3), two for PSAT (At4g35630, PSAT1; At2g17630, PSAT2), and one gene for PSP (At1g18640, PSP1). The in vitro activity of the respective enzymes has been demonstrated (Ho et al., 1998, 1999a, 1999b; Ho and Saito, 2001; Benstein et al., 2013). In Arabidopsis, all enzymes of the pathway are localized in plastids and the genes are highly expressed in heterotrophic tissue, except for PGDH3, which is expressed at low levels in autotrophic tissues (Benstein et al., 2013). Although mutations in PGDH2 or PGDH3 revealed no obvious phenotype, loss of either PGDH1 or PSP causes embryo lethality and impairs pollen development (Benstein et al., 2013; Cascales-Minana et al., 2013). In addition, preliminary characterization of conditional PSP mutants and PGDH1- and PSAT1-silenced plants revealed that PPSB-deficiency also impairs vegetative plant growth (Benstein et al., 2013; Cascales-Minana et al., 2013). However, it is still unknown which serine-requiring cellular functions are disrupted in these plants to cause growth inhibition. Furthermore, inhibition of PPSB-mediated serine biosynthesis strongly alters amino acid levels in plants, but the reason for this phenotype is also unclear (Benstein et al., 2013; Cascales-Minana et al., 2013). Considering the generally accepted assumption that photorespiration represents the major source of serine in plants (Douce et al., 2001; Ros et al., 2014), the cause of the growth and metabolic phenotypes in PPSB-deficient plants is even more puzzling.
The aim of our study was to carry out an in-depth analysis to further clarify the function of the PPSB in plants, and to better understand the relative contributions of the PPSB and photorespiration to serine biosynthesis and subsequently to plant metabolism and growth. For this purpose, a combination of techniques including growth analyses, isotope labeling experiments, and targeted metabolomics were performed under carefully chosen growth conditions. The results of our study demonstrate that the PPSB is more important than photorespiration in providing serine for plant growth. This finding was confirmed by performing isotope labeling experiments to determine the metabolic fate of serine produced by the PPSB. In addition, the analysis of transcripts, metabolites and 15N-isotope enrichment reveals that PPSB-deficient plants have an unusual metabolic phenotype caused by enhanced nitrogen assimilation and amino acid catabolism in response to serine starvation. Altogether, these results imply that even in the presence of photorespiration, PPSB-derived serine is indispensable for the plant and its deficiency triggers the induction of nitrogen assimilation, most likely as an amino acid starvation response.
Results
Growth analyses indicate that deficiency in PPSB-mediated serine synthesis cannot be compensated by photorespiration
To dissect the individual contributions that photorespiration and PPSB have on plant serine metabolism, we analyzed changes in the growth rate and biomass production of empty vector (EV) control plants, PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2) and psp1-1 mutants at photorespiratory CO2 (400 µL L−1 CO2) conditions and at elevated CO2 (4,000 µL L−1 CO2) conditions where photorespiration is severely repressed (Figure 1; Supplemental Figure S2). As incubation at elevated CO2 strongly inhibits seedling growth of PGDH1-silenced lines, plants were germinated and precultured for 8 d at photorespiratory conditions before half of them were transferred for additional 8 d to elevated CO2 conditions.
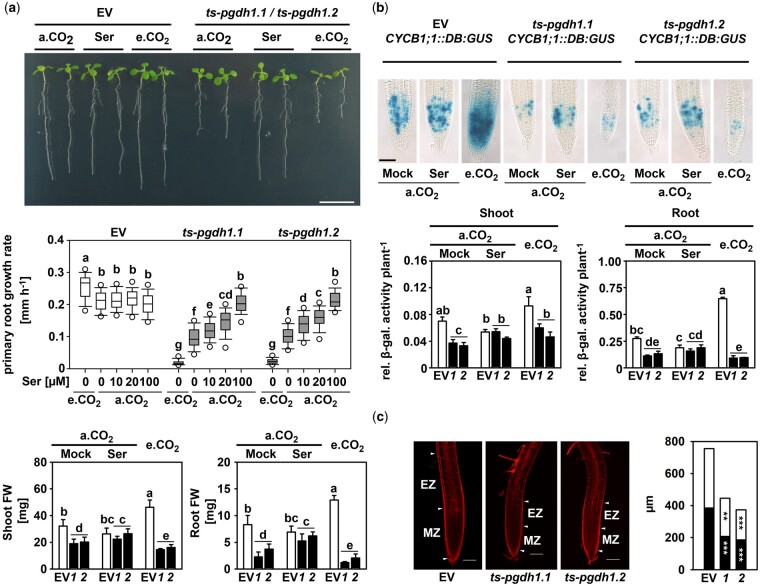
Figure 1.
PGDH1-deficiency inhibits cell division and elongation. A, PRGR and plant biomass of EV control plants and PGDH1-silenced plants (ts-pgdh1.1 [1] or ts-pgdh1.2 [2]) were determined under different growth conditions: a.CO2 (ambient CO2, 400 µL L−1), a.CO2 supplemented with 0.1-mM serine (Ser) and e.CO2 (elevated CO2, 4,000 µL L−1). Scale bar = 1 cm. Primary root growth rate was determined at a.CO2, e.CO2 and at a.CO2 with increasing serine concentration. Data (n > 20) are shown as box plot; the line represents median; the whiskers represent 1.5 interquartile range (IQR) and dots show 5th/95th percentiles. FW of shoots and roots are the mean ± sd of n > 5. B, CYCB1;1::DB:GUS expressing plants were transformed with the PGDH1 silencing construct and cultivated under different conditions. GUS activity was visualized by histological staining with X-Gluc for 12 h at 37°C (Scale bar = 50µm) or quantified by a fluorescent β-galactosidase assay. Values are the mean and se of n = 5. For (A) and (B), one-way analysis of variance (ANOVA) followed by Fisher’s least significant difference test (P < 0.05) were performed and columns with the same letter are not significantly different. C, Roots were stained with propidium iodine and cell size and number were analyzed by confocal laser scanning microscopy. Black bars represent the size of the meristem zone (MZ) and white bars represent the size of the first 11 cells above the transition zone, also called elongation zone (EZ). White arrowheads mark the boundaries of the MZ and EZ. Scale bars indicate 100 µm. The cell area and meristem size were determined using the ImageJ software. Data presented are means ± se of n > 11. Asterisks indicate significantly different values between EV control plants and PGDH1-silenced lines by the Student’s t test (**P < 0.01; ***P < 0.001).
Cultivation of control plants at elevated CO2 conditions enhanced the primary root growth rate (PRGR) from 0.21 to 0.26 mm h−1 (Figure 1A). In accordance, the shoot biomass increased by around 10 mg and root biomass by around 4 mg compared with plants grown at ambient CO2 conditions (Figure 1A). The increase in growth and biomass was accompanied by a higher abundance of the B-type cyclin B1 (Figure 1B), a commonly used marker for cell proliferation (Weingartner et al., 2004; Rodriguez et al., 2010). Therefore, suppression of photorespiration by elevated CO2 has no negative impact on plant growth. Interestingly, expression analysis showed that the transcript levels of major PPSB genes were significantly increased in plants after the suppression of photorespiration (Supplemental Figure S3), suggesting that enhanced growth of plants at elevated CO2 requires the activation of the PPSB.
In contrast to plants with repressed photorespiration, plants deficient in the PPSB were strongly impaired in growth (Figure 1A;Supplemental Figure S2). Compared to control plants, the PRGR of PGDH1-silenced lines and psp1-1 mutants was reduced to 45% and 15%, respectively (Figure 1A;Supplemental Figure S2). The shoot and root biomasses of PGDH1-silenced lines decreased to 55% and 35% of control plants, and the biomasses of psp1-1 mutants to 47% and 22% of control plants (Figure 1A;Supplemental Figure S2). In accordance with the strong growth phenotype of PPSB-deficient plants, the abundance of the B-type cyclin B1 was significantly reduced in shoot and root tissues of PGDH1-silenced lines (Figure 1B).
The PRGR, as well as the shoot and root biomasses, of PGDH1-silenced lines and psp1-1 mutants improved substantially when these plants were cultivated in the presence of 0.1-mM serine in the growth medium (Figure 1A;Supplemental Figure S2). In line with this finding, the cyclin B1 abundance in the shoot and root tissues of PGDH1-silenced lines increased after external application of serine (Figure 1B). Thus, the growth defects of PPSB-deficient plants were mainly caused by serine starvation. To support this conclusion, plants were germinated and continuously cultivated on growth medium supplemented with 0.1-mM serine before transfer to serine-free medium (Supplemental Figure S4). While withdrawal of external serine has no significant impact on the PRGR of control plants, the PRGR of PGDH1-silenced lines decreased constantly from 0.22 mm h−1 at Day 1 to 0.08 mm h−1 at Day 5 (Supplemental Figure S4). Thus, plants without a functional PPSB are dependent on continuous application of external serine to maintain normal plant growth.
In contrast to the control plants, the growth rate of roots of PGDH1-silenced lines and psp1-1 mutants deteriorated significantly when these plants were grown under low-photorespiratory conditions (Figure 1A;Supplemental Figure S2). Furthermore, the shoot and root biomasses of PGDH1-silenced lines were also exacerbated, and decreased to around 35% and 12% of those of the EV control, respectively (Figure 1A). Accordingly, the cyclin B1 abundance was reduced in shoot and root tissues of PGDH1-silenced lines compared to control plants (Figure 1B). Thus, simultaneous inhibition of PPSB-mediated and photorespiratory serine biosynthesis by growing PGDH1-silenced plants and psp1-1 mutants at high CO2 exacerbated the growth phenotypes of these plants, which was most likely the consequence of enhanced serine deficiency. This assumption was supported by the finding that the deterioration of the growth phenotype of PGDH1-silenced lines at elevated CO2 could be significantly mitigated by serine supplementation (Supplemental Figure S5).
Plant growth is a combination of cell division and elongation (Gonzalez et al., 2012). To test whether deficiency of PGDH1 or PSP also alters cell elongation, we determined the size of cells in the elongation zone above the root apical meristem (Figure 1C;Supplemental Figure S6). The size of the root apical meristem and the size of the cells above the transition zone were both significantly reduced in PGDH1-silenced lines and psp1-1 mutants compared to control plants (Figure 1C;Supplemental Figure S6).
Collectively, our study showed that plants lacking a functional PPSB are impaired in growth due to reduced cell division and elongation, while inhibition of photorespiration alone has no negative impact on plant growth. However, simultaneous inhibition of both pathways aggravated the growth defects of PPSB-deficient plants, indicating that photorespiratory serine partially contributes to plant growth.
Tracer studies reveal elevated incorporation of carbon derived from external applied [3-14C] serine into major cellular components in shoot and root tissues of PGDH1-silenced lines
PGDH1-silenced lines and psp1.1 mutants require a continuous supply of external serine to maintain normal plant growth (Figure 1A;Supplemental Figures S2 and S4). This finding suggests that external serine is incorporated into cellular components, which are insufficiently supplied with serine in PPSB-deficient plants. Amino acid feeding has been widely used to rescue amino acid auxotrophic mutants (Last and Fink, 1988; Muralla et al., 2007), indicating that external supplied amino acids are metabolized in the same way as plant’s own.
To follow the fate of externally applied serine throughout the plant, a feeding experiment using 14C-labeled serine [3-14C] was conducted (Figure 2). For this purpose, plants were constantly grown on medium supplemented with serine before incubating them for 2 d on medium additionally supplemented with 50-µCi 14C-labeled serine. Under such conditions PGDH1-silenced lines were phenotypically indistinguishable from control plants. Shoot and root tissue of the traced plants were harvested separately, fractionated into four fractions containing DNA, proteins, polar metabolites, and lipophilic metabolites according to Valledor et al. (2014), and the amount of incorporated radioactivity was analyzed by scintillation counting (Figure 2).
Figure 2.
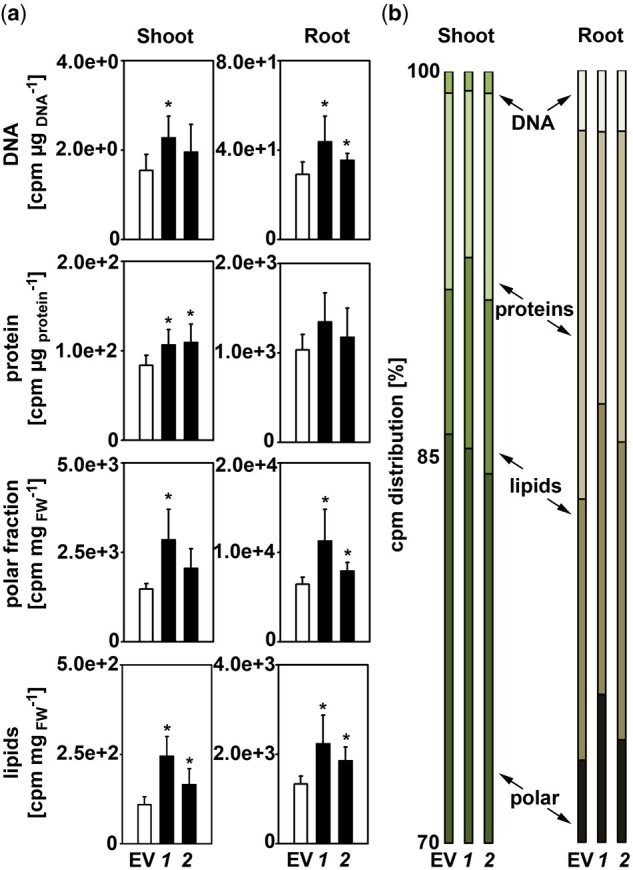
PGDH1-silenced lines incorporate more [14C-3] serine-derived radioactivity. Plants were germinated and pre-cultivated on plates containing half-strength MS medium supplemented with 0.1-mM serine. Subsequently, plants were transferred to liquid half-strength MS medium supplemented with 0.1-mM serine and traced with 50 µCi [14C-3] serine for 48 h. After incubation, shoot and root materials were harvested separately and fractionated into DNA, proteins, polar, and lipophilic compounds. A, The incorporation of radioactive carbon derived from 14C-labeled serine into DNA, proteins, polar, and lipophilic compounds in shoot and root tissues of EV and PGDH1-silenced lines (ts-pgdh1.1 [1], ts-pgdh1.1 [2]) was determined as counts per minute using a scintillation counter. B, Based on these data, the percent distribution of radioactivity in the individual compounds was calculated. For (A) and (B), data presented are means ± sd of n = 5. Asterisks indicate significantly different values between EV and PGDH1-silenced lines by the Student’s t test (*P < 0.05).
The feeding experiment revealed that serine was efficiently taken up by the plant, allocated into shoot and root tissues and rapidly incorporated into cellular components, such as lipids, DNA, polar metabolites, and proteins (Figure 2A). Generally, PGDH1-silenced lines accumulated significantly more radioactivity in shoots and roots than control plants (Figure 2A). The analysis of the radioactivity enrichment in different cellular components revealed a significantly increased incorporation of radioactivity into lipids and proteins in shoots of both PGDH1-silenced lines, and there was also a clear trend for a higher amount of label incorporation into the DNA and polar fractions. In roots, the amount of radioactivity in the polar fraction as well as in DNA and lipids was significantly higher in both PGDH1-silenced lines compared to control plants, while incorporation into proteins was more variable.
Assessing the proportional distribution of radioactivity between the analyzed compounds revealed no clear indications that any of the analyzed compounds was preferentially labeled in PGDH1-silenced lines (Figure 2B).
Altogether, our findings revealed that radioactivity derived from externally applied 14C labeled serine [3-14C] is more efficiently incorporated into major cellular components in shoots and roots of PGDH1-silenced lines, indicating that the lack of PGDH1-mediated serine biosynthesis causes a general shortage of serine for the synthesis of these components in plants.
Protein and purine biosynthesis are impaired in PGDH1-silenced lines
PGDH1-silenced lines and psp1-1 mutants are substantially impaired in plant growth, indicating that serine produced by the PPSB is required for the formation of essential cellular components, such as proteins and nucleotides (Ros et al., 2014). In order to elucidate the role of PPSB-derived serine for the synthesis of these components, we analyzed protein and adenosine monophosphate (AMP) biosynthesis in shoot and root tissues of PGDH1-silenced lines (Figure 3).
Figure 3.
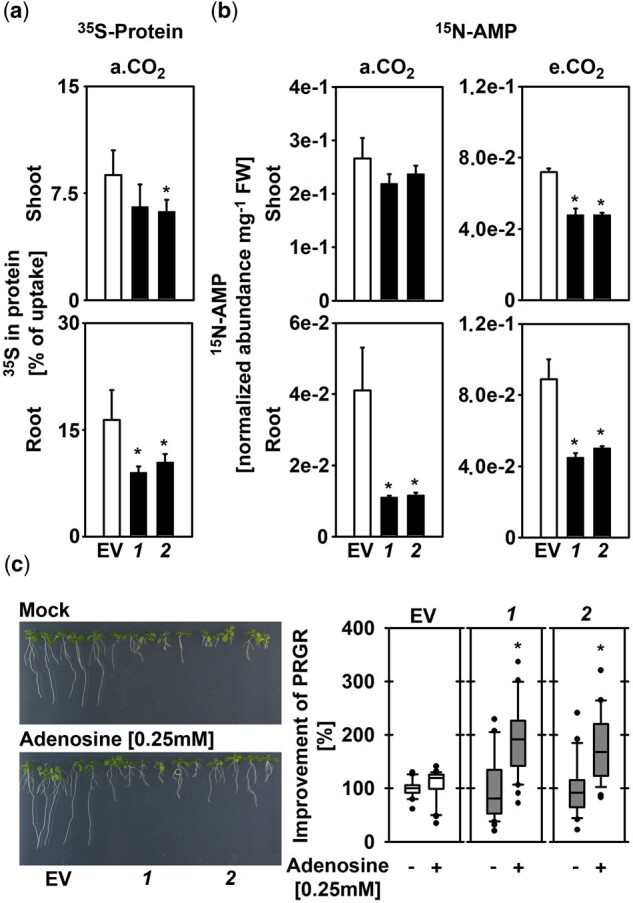
PGDH1-deficiency impairs protein and purine biosynthesis. A, Protein synthesis in shoot and root tissues of EV and PGDH1-silenced lines (1 and 2) was determined by analyzing 35S-methionine incorporation into proteins. Data presented are means ± sd of n = 5. B, AMP synthesis was examined by quantifying fully labeled 15N-AMP in shoots and roots of plants grown at a.CO2 (ambient CO2, 400 µL L−1) and at e.CO2 (elevated CO2, 4,000 µL L−1) on medium supplemented with 15NO3. For (A) and (B), data presented are means ± sd of n = 5. Asterisks indicate significantly different values between EV and PGDH1-silenced lines by the Student’s t test (*P < 0.05). C, PRGR of EV control plants and PGDH1-silenced lines in the absence (Mock; −) or the presence (Adenosine [0.25 mM]; +) of 0.25-mM adenosine is shown. Data (n > 20) are shown as box plot (white, EV control; gray, PGDH1-silenced lines 1 and 2); the line represents median; the whiskers represent 1.5 IQR and dots show outliers. Asterisks indicate significantly different values between Mock and adenosine-treated EV plants and PGDH1-silenced lines by the Student’s t test (*P < 0.05).
Protein biosynthesis was quantified by analyzing the incorporation rate of 35S-labeled methionine into proteins in shoot and root tissues (Figure 3A). Therefore, plants were incubated with 35S-labeled methionine for 6 h during the light period, and total proteins were isolated from shoot and root tissues according to Koprivova et al. (2000). The amount of newly synthesized proteins was determined by scintillation counting and finally normalized to the total amount of radioactivity taken up by the plants. By this method, we found that around 8.7% of the absorbed radioactivity was integrated into shoot proteins of control plants, while 6.5% and 6.1% were incorporated into shoot proteins of ts-pgdh1.1 and ts-pgdh1.2 lines (Figure 3A). However, this slight reduction in the biosynthesis of shoot proteins was significant only for the ts-pgdh1.2 line. In agreement with the strong inhibition of root growth of PGDH1-silenced lines, only 9% and 10% of the absorbed radioactivity was incorporated into root proteins of ts-pgdh1.1 and ts-pgdh1.2 lines, respectively, while 17% was incorporated into root proteins extracted from control plants (Figure 3A).
To investigate purine biosynthesis, we quantified the amount of 15N-labeled AMP extracted from plants grown in the presence of 15N-labeled ammonium nitrate (Figure 3B). Similarly, as observed for protein biosynthesis, the synthesis of AMP was significantly reduced in roots, but not substantially altered in shoots of PGDH1-silenced lines grown at ambient CO2 (Figure 3B). However, when plants were transferred to elevated CO2 conditions, the amount of labeled 15N-AMP was significantly lower in shoot and root tissues compared to control plants (Figure 3B). Thus, our data indicate that in PGDH1-silenced lines the biosynthesis of AMP in shoots is preferentially supplied with serine produced by photorespiration, while the roots are starved for serine for the synthesis of purine nucleotides.
To further elucidate the importance of the PPSB for purine biosynthesis, PGDH1-silenced lines were cultivated on medium supplemented with 0.25-mM adenosine (Figure 3C). Externally applied adenosine is efficiently taken up by the plants via nucleoside carriers (Möhlmann et al., 2001) and enters purine metabolism through the salvage pathway (Witte and Herde, 2020). Previously, it has been reported that adenosine feeding is sufficient to rescue the growth phenotype of AMP-deficient plants lacking the plastid localized AMP exporter (Kirchberger et al., 2008).
Interestingly, external application of 0.25-mM adenosine significantly improves the PRGR of PGDH1-silenced plants (Figure 3C). Although the effect of adenosine supplementation on the PRGR of control plants was weak, the PRGR of PGDH1-silenced lines improved significantly by 190% (ts-pgdh1.1) and 175% (ts-pgdh1.2), respectively (Figure 3C).
Conclusively, our tracer studies and feeding experiments revealed that protein and nucleotide biosynthesis were impaired in PGDH1-silenced lines, which very likely contributed to the strong growth phenotype of these plants.
Photorespiration and PPSB contribute to the steady-state serine content in plants
To elucidate the relative contribution of photorespiration and PPSB to the steady-state serine content in plants, we examined the serine content in shoot and root tissues of PGDH1-silenced lines and control plants, before and after repression of photorespiration (Figure 4).
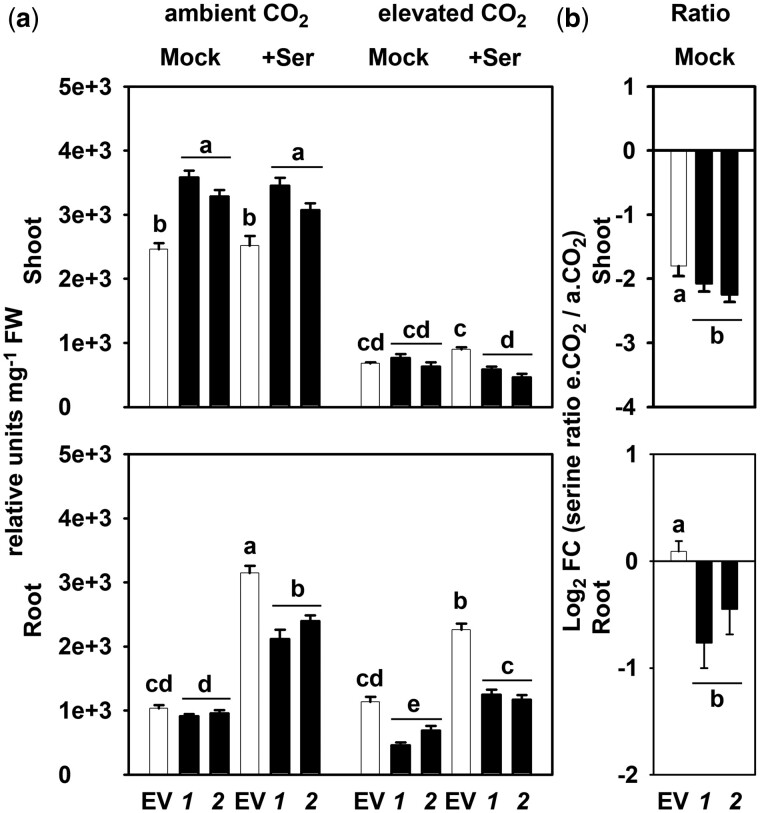
Figure 4.
Contribution by photorespiration and PPSB to the steady-state serine content in plants. A, Total serine content in shoot and root tissues of EV and PGDH1-silenced lines (1 and 2), grown at different conditions, a.CO2 (ambient CO2, 400 µL L−1), +Ser (supplemented with 0.1-mM serine), and e.CO2 (elevated CO2, 4,000 µL L−1). B, Calculated ratio between serine levels determined at ambient and elevated CO2. Values are shown as Log2 fold change (FC). For (A) and (B), data are presented as mean ± se of n = 8; one-way ANOVA followed by Fisher’s least significant difference test (P < 0.05) were performed and columns with the same letter are not significantly different.
We found that the serine content was not reduced in PGDH1-silenced lines when grown at ambient CO2 (Figure 4). These plants actually contain higher serine levels in shoots than controls, while the serine content in roots was not significantly altered (Figure 4A).
External application of serine at a concentration of 0.1 mM, which rescued the shoot and root growth phenotypes of PGDH1 and PSP deficient plants (Figure 1A;Supplemental Figures S2 and S4), had no effect on the internal serine content in shoots of both control and PGDH1-silenced lines. However, this serine feeding caused a significant accumulation of internal serine in roots, although to a lesser extent in PGDH1-silenced lines (Figure 4A). These data support the fast metabolization of externally applied serine in plants, and confirmed that PGDH1-silenced lines are starved of serine. In addition, our results indicate that PGDH1-silenced lines adjust their growth rate to the reduced availability of serine, most likely to maintain a certain cellular concentration.
Repression of photorespiration strongly reduced the shoot serine content in control plants, while the root serine content was not altered (Figure 4A). In PGDH1-silenced lines, repression of photorespiration reduced the serine content in shoots more drastically than observed in control plants (Figure 4A). The analysis of the serine ratio at elevated and ambient CO2 (e.CO2/a.CO2) confirmed this observation (Figure 4B).
The root serine content was significantly decreased in PGDH1-silenced lines after repression of photorespiration (Figure 4). This finding was also reflected by the significantly lower e.CO2/a.CO2 ratio for serine in roots of the silenced lines compared to control plants (Figure 4B). Therefore, our data indicate that PPSB and photorespiration both contribute to the serine pool in roots.
External application of serine to plants grown at elevated CO2 caused a moderate serine accumulation in shoots of control plants, but had no significant effect on the internal serine content in shoots of PGDH1-silenced lines (Figure 4). In roots, serine feeding caused a significant increase in the internal serine content in both control and PGDH1-silenced lines. However, this increase was significantly lower in the silenced lines (Figure 4). Furthermore, the accumulation of serine in roots after serine feeding was substantially lower in plants grown at elevated CO2 compared with plants grown at ambient CO2 (Figure 4). Thus, the consumption of externally applied serine is higher in PGDH1-silenced lines and in EV control plants after repression of photorespiration.
Overall, our results show that both photorespiration and the PPSB contribute to the steady-state serine content in shoot and root tissues of plants, although only inhibition of the PPSB has a negative impact on plant growth linked to serine deficiency.
Photorespiration and PPSB have different impacts on plant metabolism
To gather more information about the role of the PPSB for plant metabolism, we analyzed the composition of primary metabolites in shoot and root tissues of plants impaired in either photorespiration, the PPSB or in both pathways (Figure 5). To test whether the observed metabolic phenotypes originate specifically from serine deficiency, we examined the effect of external serine application on the metabolic changes in these plants.
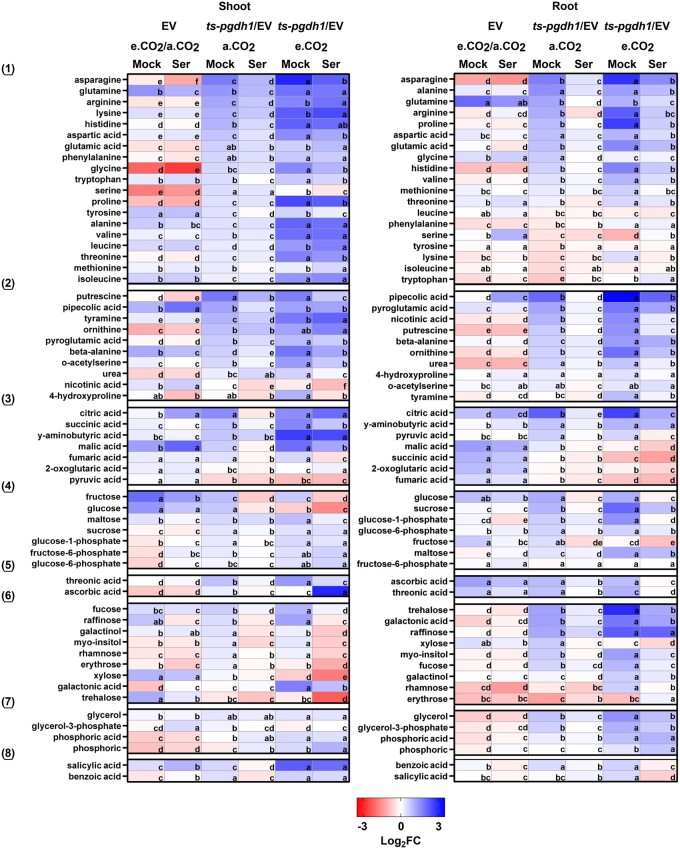
Figure 5.
Changes in metabolite composition in response to the inhibition of photorespiration or PPSB. Changes in the content of metabolites extracted from shoot and root tissues of EV control plants and PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2) grown at a.CO2 (ambient CO2, 400 µL L−1) and e.CO2 (elevated CO2, 4,000 µL L−1) with 0.1-mM serine (Ser) or without (Mock) in the growth medium. Data are presented as heatmap. Blue indicates elevated metabolite content and red indicates decreased metabolite content. Data are presented as Log2 FC. To simplify the data presentation, the average between the values obtained for ts-pgdh1.1 and ts-pgdh1.2 (i.e. “ts-pgdh1” in figure) was used for all calculations. For EV, the ratio (Log2 FC) of measured analytes in plants grown under a.CO2 and e.CO2 (a.CO2/e.CO2) is presented. Metabolites were grouped into (1) amino acids, (2) nitrogen-containing compounds, (3) organic acids related to respiratory metabolism, (4) carbohydrates and glycolytic intermediates, (5) organic acids, (6) carbohydrates, (7) sugar alcohols and phosphoric ester, and (8) aromatic compounds. Data are means of n = 8. One-way ANOVA followed by Fisher’s least significant difference test (P < 0.05) were performed and fields within one raw with the same letter are not significantly different.
Repression of photorespiration in control plants resulted in distinct changes in the content of many primary metabolites in shoot and root tissues (Figure 5). The most obvious change was the strong decrease in glycine and serine content in the shoot. Apart from minor differences in the content of some compounds, serine feeding had no substantial impact on the metabolite profile of these plants (Figure 5). Thus, the vast majority of the observed metabolic changes in these plants were not related to serine starvation, and were most likely caused by changes in the C/N balance due to alterations in carbon and nitrogen assimilation after repression of photorespiration by elevated CO2 (Rachmilevitch et al., 2004).
The analysis of the metabolite composition in PGDH1-silenced lines revealed only a very small overlap with the metabolic changes observed after repression of photorespiration (Figure 5). Although the majority of analyzed metabolites were significantly increased in shoot and root tissues of PGDH1-silenced lines, the most important changes were observed for amino acids, nitrogen-containing compounds, and TCA cycle-related organic acids (Figure 5).
In shoots the contents of most amino acids, nitrogen-containing compounds (except for nicotinic acid and 4-hydroxyproline) and TCA cycle-related organic acids (except for 2-oxoglutarate and pyruvate) were elevated. In roots, many of the amino acids, nitrogen-containing compounds and TCA cycle-related organic acids were also increased. However, several amino acids were not significantly altered or were even decreased (leucine, phenylalanine, serine, tyrosine, lysine, isoleucine, tryptophan; Figure 5). In addition, the content of the N-containing metabolite tyramine and the organic acids succinic acid, 2-oxoglutarate and fumaric acids were not altered in roots.
To test whether the alteration of TCA cycle intermediates is caused by reduced or enhanced activity of the pathway, we measured the activity of the citrate synthase enzyme (Supplemental Figure S7). Citrate synthase represents a key enzyme of this pathway, as it catalyzes the first committed step in the TCA cycle (Schmidtmann et al., 2014). The citrate synthase activity was significantly increased in shoot and root tissues of PGDH1-silenced lines, indicating that the alteration in TCA cycle intermediates was caused by increased activity of the respiratory metabolism. This finding is supported by the elevated respiration in roots of the silenced lines (Supplemental Figure S7).
Apart from the abovementioned metabolites, the contents of various glycolytic intermediates, carbohydrates, sugar alcohols, and secondary metabolites were also elevated in shoot and root tissues of PGDH1-silenced lines (Figure 5).
Most of the metabolic changes observed in PGDH1-silenced lines were significantly attenuated by serine feeding (Figure 5). Thus, the metabolic alterations in these plants clearly correspond to the availability of serine. This assumption is further supported by the finding that simultaneous inhibition of photorespiration and PPSB, by growing PGDH1-silenced lines at elevated CO2, enhanced the metabolic phenotypes of these plants, and most of the changes were also attenuated by serine feeding, at least in root tissue.
In summary, our comprehensive metabolic analysis revealed that repression of photorespiration causes substantially different changes in plant metabolism than inhibition of the PPSB. In addition, the metabolic phenotype of PGDH1-silenced lines was clearly serine dependent, while the vast majority of metabolic changes after repression of photorespiration were not related to serine deficiency. However, simultaneous inhibition of photorespiration and PPSB enhanced the metabolic phenotype of PGDH1-silenced lines, confirming that both pathways interact in providing serine to the plant.
Shortage in PPSB-mediated serine biosynthesis alters nitrogen metabolism in plants
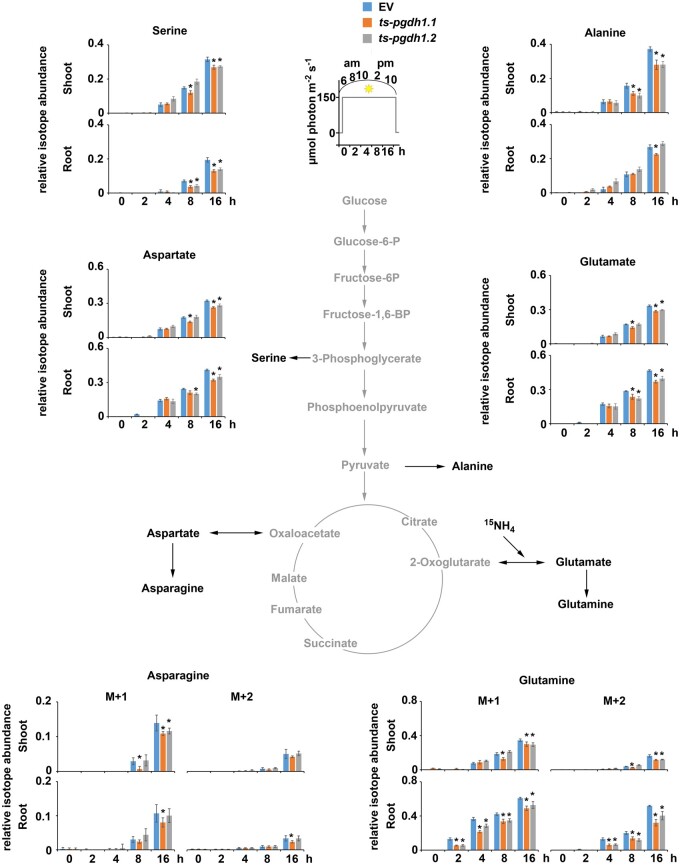
Based on the metabolic characterization of PGDH1-silenced lines, we became interested in whether the high content of amino acids in shoot and root tissues of these plants originates from elevated nitrogen assimilation or impaired amino acid turnover. To answer this question, we measured the enrichment of 15N in amino acids in shoot and root tissues of PGDH1-silenced lines after labeling the plants with 15N-labeled ammonium nitrate (Figure 6; Supplemental Figures S8 and S9). Labeling started right after the end of the dark phase, and samples for the first time point (0 h) were immediately harvested (see insets in Figures 6 and 7; Supplemental Figures S8 and S9). Further samples were progressively taken during the light period at 2, 4, 8, and 16 h after labeling began, corresponding to 8 and 10 am, and 2 and 10 pm, respectively. The contents of amino acids were determined and the extent of labeling was evaluated by quantification of the heavy isotope peak for the respective amino acid by liquid chromatography–mass spectrometry (LC–MS; Figure 6; Supplemental Figures S8 and S9). Changes in the abundance of the 15N-labeled isotopes of major amino acids are shown in Figure 7 and changes in the total amino acid content (sum of 14N and 15N isotopes) are depicted in Supplemental Figures S8 and S9.
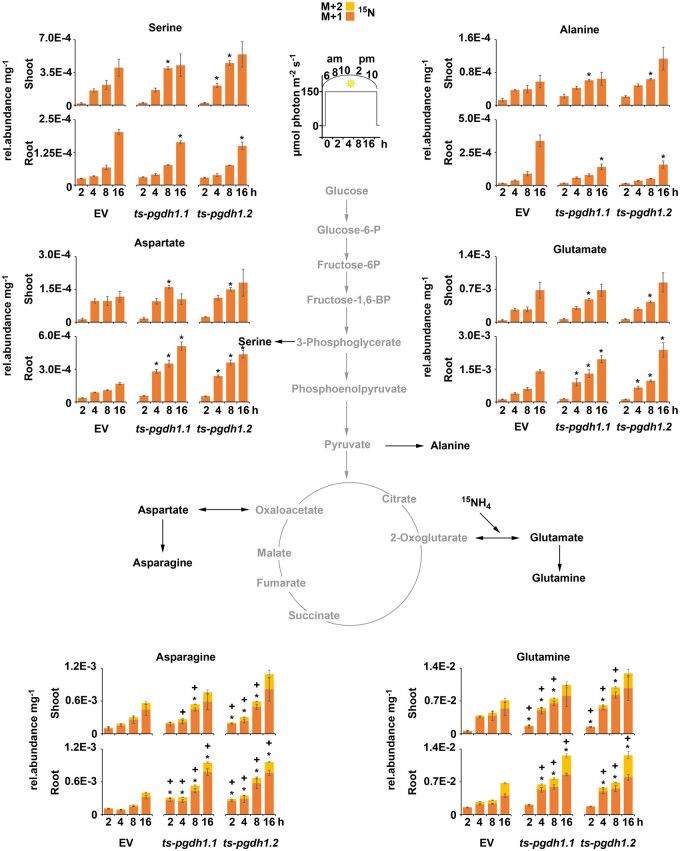
Figure 6.
Quantification of 15N-labeled amino acids in PGDH1-silenced lines. 15N-isotopes (M + 1 orange; M + 2 yellow) of major amino acids are shown in shoot and root tissues of EV control plants and PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2) in a 16-h time course (see inset). Data presented as relative (rel.) abundance per mg FW are means ± se of n = 5. Asterisks (M + 1) and cross (M + 2) indicate significantly different values between EV and PGDH1-silenced lines by the Student’s t test (*P < 0.05).
Figure 7.
Changes in the RIA in PGDH1-silenced lines. Based on the 15N-enrichment in major amino acids, the RIA was calculated for the individual amino acids in shoot and root tissues of EV control plants and PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2) grown in a 16-h time course (see inset). Data presented are means ± se of n = 5. Asterisks indicate significantly different values between EV and PGDH1-silenced lines by the Student’s t test (*P < 0.05).
The total amino acid content in shoots of PGDH1-silenced lines and control plants varied over the day along the diurnal cycle, which is in agreement with previous observations (Gibon et al., 2006; Usadel et al., 2008). However, we observed some differences between the lines. Although in shoots of control plants the amino acid content was highest at 10 am (4 h after labeling) and decreased significantly until 2 pm (8 h after labeling), the amino acid content in shoots of PGDH1-silenced lines remained at high levels until 2 pm (Supplemental Figures S8 and S9). In roots, the levels of all amino acids were highest at 6 am (0 h) and either decreased continuously over the day (aspartate, asparagine, threonine, leucine, valines, isoleucine, lysine, tyrosine), kept constant (phenylalanine) or increased again at the end of the light period (serine, alanine, glutamate, glutamine, glycine, methionine; Supplemental Figures S8 and S9). Except for alanine, which accumulated in control plants at the end of the day, no differences in the diurnal variation of total amino acid accumulation in roots were observed between the control and the PGDH1-silenced lines. (Supplemental Figures S8 and S9).
Interestingly, the intensity of 15N labeling of several amino acids was significantly different in PGDH1-silenced lines compared to control plants. The flux of 15N-labeled nitrogen into aspartate, asparagine, glutamate, glutamine, and threonine was significantly elevated in root tissue and to some extent also in shoot tissue of PGDH1-silenced lines (Figure 6; Supplemental Figures S8 and S9). However, some amino acids revealed tissue specific differences. While labeling of serine and alanine was elevated in shoots, the flux of 15N into both amino acids was significantly reduced in roots (Figure 6; Supplemental Figure S8). Similarly, the flux of 15N into glycine and methionine was also significantly lower in roots of PGDH1-silenced lines, but not altered in shoots (Supplemental Figure S9). This finding was in agreement with the reduced biosynthesis of serine in roots of PGDH1-silenced lines, and supports the intimate association of serine metabolism with the biosynthesis of glycine, methionine, and alanine (Hesse and Hoefgen, 2003; Engel et al., 2011; Zhang et al., 2013). In addition, slightly elevated labeling was also observed for the minor-abundant amino acids leucine, valine, and phenylalanine (Supplemental Figure S9). Although labeling of leucine was marginally enhanced in shoots, labeling of valine and phenylalanine was slightly higher in roots.
Next, we investigated whether the elevated amino acid levels in PGDH1-silenced lines are caused only by enhanced synthesis. Therefore, we calculated the relative isotope abundance (RIA) for the respective amino acids based on the 15N labeling data (Figure 7). The RIA indicates to what extent the pool of an individual amino acid is labeled, and therefore represents a useful measure for determining the turnover rate of each individual amino acid pool. Our evaluation of the RIA revealed lower values for most of the amino acids in the shoot and root tissues of PGDH1-silenced lines, indicating that the increase in the steady-state content was not proportional to the increase in the content of 15N-labeled amino acids. Thus, the accumulation of amino acids in shoot and root tissues of PGDH1-silenced plants cannot be explained only by enhanced synthesis and might be additionally caused by reduced turnover or increased remobilization of older nitrogen storages.
To further test whether the elevated flux of nitrogen in PGDH1-silenced lines was accompanied by an increased content of ammonium, we quantified the total ammonium content in shoot and root tissues of these plants (Supplemental Figure S10). As expected from the higher incorporation of 15N into amino acids, the absolute amount of ammonium was significantly higher in shoot and root tissues of PGDH1-silenced lines (Supplemental Figure S10). Thus, the elevated levels of ammonium in PGDH1-silenced lines support our finding that nitrogen is more efficiently assimilated in these plants.
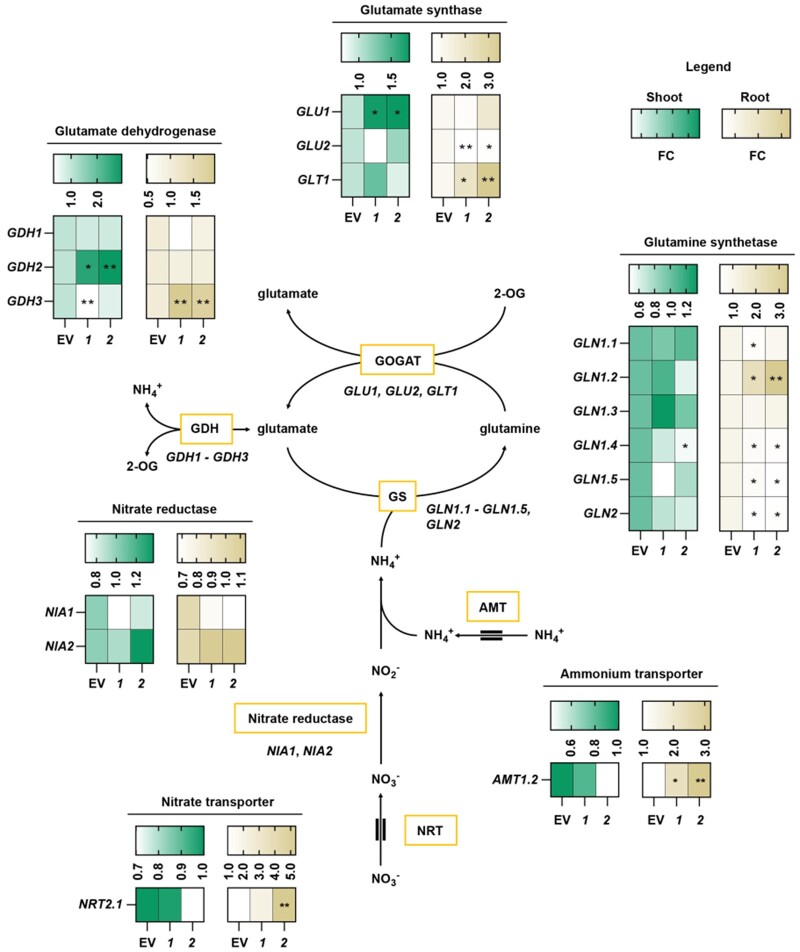
To elucidate whether the elevated 15N labeling of amino acids in PGDH1-silenced lines was accompanied by an activation of the nitrogen assimilation pathway, we determined the transcript levels of genes encoding for major components involved in nitrogen uptake and assimilation (Figure 8). These genes included nitrate transporter (NRT2.1), ammonium transporter (AMT1.2), nitrate reductase (NIA1, NIA2), glutamine synthetase (GLN1.1-GLN1.5, GLN2), glutamate synthase (GLU1, GLU2, GLT1), and glutamate dehydrogenase (GDH1-GDH3).
Figure 8.
PGDH1-deficiency triggers the induction of genes involved in nitrogen assimilation. The expression of the genes encoding for the glutamate synthases GLU1, GLU2, and GLT1, the glutamine synthetase GLN1.1-GLN1.5 and GLN2, the glutamate dehydrogenase GDH1-GDH3, the nitrate reductases NIA1 and NIA2, the ammonium transporter AMT1.2 and the nitrate transporter NRT2.1 is shown in shoots and roots of EV and PGDH1-silenced lines (1 and 2) as FC of n = 5. Lower values are displayed whiter and higher values are displayed darker. Yellow boxes indicate proteins and double lines around arrows for AMT and NRT indicate membrane transport processes. Asterisks indicate significantly different values between EV and PGDH1-silenced lines by the Student’s t test (*P < 0.05; **P < 0.01).
Note: Figure Replacement Requested.
Although the expression of NIA genes was not altered in PGDH1-silenced lines, the expression of the cytosolic glutamine synthetase GLN1.2, a marker for an oversupply of ammonium in roots (Lothier et al., 2011), was significantly induced in roots of these plants (Figure 8).
Furthermore, the expression of the glutamate synthase isoform GLU1 was increased in shoots, while that of GLT1 and GLU2 was higher in roots of the silenced lines (Figure 8). In addition to the genes belonging to the GS/GOGAT pathway, the expression of the ammonium transporter AMT1.2 was also elevated in roots of PGDH1-silenced lines (Figure 8). Furthermore, we found that the expression of GDH2 and GDH3 were also significantly elevated in PGDH1-silenced lines (Figure 8). The GDH enzymes play a central role in amino acid catabolism as they catalyze the deamination of glutamate into 2-oxoglutarate and ammonium (Glevarec et al., 2004; Masclaux-Daubresse et al., 2006; Skopelitis et al., 2007; Labboun et al., 2009; Fontaine et al., 2012). Therefore, the expression data indicate that elevated nitrogen assimilation is driven by an enhanced ammonium uptake and assimilation, mainly in the roots of PGDH1-silenced lines. In addition, the induced expression of GDHs indicates that not only nitrogen assimilation and uptake, but also amino acid recycling, are enhanced in these plants.
Conclusively, the higher content of 15N-labeled amino acids, the elevated content of ammonium and the enhanced expression of N-assimilatory genes hint at an increased nitrogen assimilation in the absence of PGDH1 function.
Discussion
PPSB-derived serine is the driving force for plant growth
Plants are continuously growing by a combination of cell division and cell elongation (Gonzalez et al., 2012). Both processes require a constant supply of serine for the biosynthesis of proteins, purine bases for nucleic acid synthesis, phospho- and sphingolipids for the synthesis of cellular membranes, glutathione, and also the phytohormone auxin, which regulates cell proliferation (Ros et al., 2014). Thus, it is reasonable to assume that serine deficiency would lead to a strong developmental phenotype in plants.
Photorespiration is assumed to represent the main producer of serine in plants since photorespiratory serine biosynthesis can exceed 1,200 nmol serine mg−1 protein min−1 (Douce et al., 2001; Ros et al., 2014). However, repression of photorespiration by growing plants at elevated CO2 has no negative impact on plant growth (Figure 1A;Supplemental Figure S2). In addition, it has been previously reported that bypassing photorespiration even improves plant growth by preventing energy-intensive photorespiratory 2-phosphoglycolate recycling (Kebeish et al., 2007; Maier et al., 2012; South et al., 2019). Thus, plant growth cannot rely only on serine produced by photorespiration.
In this study, we dissect the relative contributions made by photorespiration and the PPSB towards plant serine biosynthesis. We present multiple lines of evidence demonstrating that the PPSB makes an irreplaceable contribution to whole-plant serine supply. Previously, the essential role of PGDH1 and PSP for pollen and embryo development has been demonstrated (Benstein et al., 2013; Cascales-Minana et al., 2013). However, the function of the PPSB during vegetative plant growth was still enigmatic. It has been proposed that the PPSB is mainly active during the night (Modde et al., 2017), but the genes of the PPSB are highly expressed during the day and required for light-dependent growth promotion (Wulfert and Krueger, 2018), indicating that the PPSB is also functional during the day time.
To elucidate the role of the PPSB during vegetative plant growth, we performed a number of experiments analyzing growth of PGDH1-silenced lines and psp1-1 mutants under various conditions. These data revealed that plants deficient in either PGDH1 or PSP were significantly impaired in growth due to reduced cell division and elongation, which consequently also resulted in lower plant biomass (Figure 1A;Supplemental Figures S2 and S6). We found that this phenotype was directly linked to the availability of serine since growth of PGDH1-silenced lines and psp1-1 mutants could be completely rescued by serine feeding (Figure 1A;Supplemental Figure S2). A continuous supply of external serine was required to maintain normal growth of PGDH1-silenced lines and withdrawal immediately lowered the growth rate (Supplemental Figure S4). The requirement of externally supplied serine for plant growth was further supported by the observation that radioactivity derived from externally supplied 14C-labeled serine was significantly enriched in major cellular components, such as DNA, lipids, and proteins in shoot and root tissues of PGDH1-silenced lines (Figure 2). This result is in agreement with the reduced synthesis of proteins and AMP in the silenced lines, although this phenotype was more pronounced in root tissue (Figure 3). The latter finding indicates that photorespiration partially compensates for the lack of PPSB-derived serine in shoots, which was supported by the observation that AMP synthesis was additionally reduced in shoot tissue after simultaneous inhibition of PPSB and photorespiration (Figure 3B).
A relevant question arising from our observations was whether photorespiratory serine contributes to plant growth. As mentioned before, repression of photorespiration has no negative impact on plant growth. However, expression analysis revealed that plants compensate for repression of photorespiratory serine biosynthesis by activating the PPSB (Supplemental Figure S3). In addition, we found, by transferring PGDH1-silenced lines to elevated CO2, that simultaneous inhibition of photorespiration and PPSB resulted in a stronger growth phenotype of these plants (Figure 1), which could be substantially mitigated by serine feeding (Supplemental Figure S5). These findings indicate that serine deficiency, and not secondary effects induced by elevated CO2 assimilation, causes the deterioration of the growth phenotype of PGDH1-silenced lines. Furthermore, it supports our assumption that photorespiration and PPSB partially cooperate to provide serine to the plant. Accordingly, the steady-state serine content was more rapidly reduced in shoot and root tissues of PGDH1-silenced lines than in control plants after transfer to elevated CO2. Furthermore, PGDH1-silenced lines accumulated less serine after serine feeding when grown at nonphotorespiratory CO2 conditions (Figure 4). Taking into consideration that approximately 30% of the photorespiratory serine constantly leaves the photorespiratory cycle (Abadie et al., 2016; Busch et al., 2018), it is very likely that part of the released serine contributes to plant growth even though it is not sufficient to compensate for the lack of PPSB-derived serine.
Altogether, from our analysis, it became evident that PPSB-derived serine is required for the synthesis of major cellular components in shoot and root tissues, which finally explains the strong growth phenotype of PPSB-deficient plants. In addition, our data indicate that photorespiratory serine is insufficient to compensate for PPSB-mediated serine biosynthesis and its contribution to plant growth is limited.
Deficiency in PPSB-mediated serine biosynthesis alters plant primary metabolism, possibly triggered by the induction of nitrogen assimilation
Previous studies revealed that plants deficient in one of the major enzymes of the PPSB extensively accumulate amino acids (Benstein et al., 2013; Cascales-Minana et al., 2013; Wulfert and Krueger, 2018). However, the reason for this unexpected metabolic phenotype was unknown.
To gather more information about the metabolic consequences of PPSB deficiency, we conducted a comprehensive metabolic analysis of PGDH1-silenced lines and control plants (Figure 5).
Metabolite profiling confirmed that accumulation of amino acids represented the most intense metabolic alterations in PGDH1-silenced lines (Figure 5). However, a variety of different nitrogen-containing compounds, organic acids, and carbohydrates were also altered in these plants (Figure 5). We found that this metabolic phenotype clearly corresponds to the availability of serine, as it was attenuated by serine feeding and enhanced after simultaneous repression of PPSB and photorespiration (Figure 5). In contrast, repression of photorespiration alone also altered plant metabolism, but these changes exhibited a substantially different pattern and only a few were connected to serine starvation (Figure 5).
Detailed analysis of nitrogen-containing compounds altered in PGDH1-silenced lines revealed that most of them, namely β-alanine, pipecolic acid, ornithine, putrescine, and γ-aminobutyric acid, are intermediates of amino acid anabolic as well as catabolic pathways (Alcázar et al., 2006; Hildebrandt et al., 2015; Parthasarathy et al., 2019). Thus, amino acid catabolism seemed to also be elevated in PGDH1-silenced lines, most likely to attenuate the excessive accumulation of amino acids caused by repression of protein and nucleotide biosynthesis in these plants (Figure 3). This conclusion is supported by two additional findings: first, ammonium, which is released during amino acid degradation, accumulates to high concentrations in shoot and root tissues of these plants (Supplemental Figure S10). Second, respiratory metabolism, required to respire the carbon skeleton released by amino acid degradation (Hildebrandt et al., 2015), was also enhanced, indicated by an increased content of TCA cycle intermediates and an elevated activity of the citrate synthase (Supplemental Figure S7A). However, a closer inspection revealed specific differences in the accumulation of amino acids and organic acids between shoot and root tissues of PGDH1-silenced lines (Figure 5).
In contrast to shoots, the contents of the amino acids isoleucine, leucine, phenylalanine, tryptophan, and tyrosine were not increased and even decreased in roots (Figure 5). In addition, the contents of pyruvate and citrate were increased, while the contents of 2-oxoglutarate, succinic acid, fumaric acid and malic acid were not altered at ambient CO2 and even decreased at elevated CO2 (Figure 5). The pattern of changes in the content of organic acids (Figure 5) in combination with a high activity of the citrate synthase (Figure S7A) suggests that the drain of carbon from the TCA cycle was elevated in roots of PGDH1-silenced lines. This is in agreement with the higher content of glycolytic carbohydrates and amino acid breakdown products (Figure 5), indicating that anaplerotic reactions might counteract the enhanced drain of carbon from the TCA cycle in roots of these plants (Sweetlove et al., 2010; Hildebrandt et al., 2015). The low content of leucine, isoleucine, and the aromatic amino acids (Figure 5) support this assumption, because their oxidative degradations provide carbon for the TCA cycle and function as alternative substrates to feed electrons into the mitochondrial electron transport chain (Däschner et al., 2001; Araújo et al., 2010; Engqvist et al., 2011). In agreement with this hypothesis, we found that respiration was elevated in roots of PGDH1-silenced lines (Supplemental Figure S7B). A question arising from our observation was which metabolic pathway consumes more TCA cycle intermediates in these plants. It is well established that nitrogen assimilation represents a main consumer of organic acids derived from the TCA cycle (Araújo et al., 2014). Accordingly, serine deficiency in PGDH1-silenced plants could activate nitrogen assimilation as an amino acid starvation response, a mechanism well established in heterotrophic organisms (Gaba et al., 2001; Natarajan et al., 2001).
To more precisely answer the question whether nitrogen assimilation was altered in PGDH1-silenced lines, stable isotope labeling was performed by using 15N-labeled ammonium nitrate. Using this approach, we found that the flux of 15N-labeled nitrogen into the major abundant amino acids was significantly elevated in PGDH1-silenced lines (Figure 6; Supplemental Figures S8 and S9). Although labeling of glutamate, glutamine, aspartate, and asparagine was slightly higher in shoots, it was substantially increased in roots of PGDH1-silenced lines. This finding supported our assumption that nitrogen assimilation is elevated in PGDH1-silenced lines and that elevated nitrogen assimilation might be the reason for the observed changes in the content of TCA cycle intermediates in these plants.
To further elucidate the mechanism behind the elevated nitrogen assimilation phenotype of PGDH1-silenced plants, we analyzed the expression of a set of central genes involved in nitrogen uptake and assimilation as well as in amino acid biosynthesis and degradation (Lancien et al., 2002; Ishiyama et al., 2004; Guan et al., 2016; Konishi et al., 2017; Moison et al., 2018). These data revealed that the expressions of the ammonium transporter AMT1.2 and the GS/GOGAT genes GLN1.2, GLU1, and GLT1 were significantly upregulated in PGDH1-silenced lines (Figure 8), indicating that elevated nitrogen assimilation in these plants is driven by a higher capacity of ammonium uptake and increased activity of the GS/GOGAT pathway. This result also explained the high concentrations of ammonium in shoot and root tissues of PGDH1-silenced lines (Supplemental Figure S10). However, we also found elevated expression of GDH2 and GDH3; both enzymes are involved in deamination of glutamate to provide 2-oxoglutarate to the TCA-cycle (Fontaine et al., 2012). This finding further indicates that amino acid catabolism was also elevated in PGDH1-silenced lines, most likely to prevent their excessive accumulation.
To gather more information about the origin of the amino acids accumulating in PGDH1-silenced lines, we utilized the 15N labeling data to calculate the RIA for the major abundant amino acids. The evaluation of the RIA revealed that the increase in the steady-state amino acid content in shoot and root tissues of the silenced lines was not proportional to the increase of the content of 15N-labeled amino acids (Figure 7). Thus, the accumulation of amino acids in PGDH1-silenced lines cannot be explained only by enhanced de novo synthesis, and is most likely the consequence of two effects: enhanced de novo synthesis and reduced turnover. This conclusion is in agreement with our finding that the biosynthesis of proteins and purine nucleotides, two major consumers of amino acids, were significantly impaired in PGDH1-silenced lines (Figure 3).
Taken together, our data elucidate a serine-dependent metabolic phenotype, where inhibition of the PPSB leads to a strong increase in ammonium uptake, followed by its assimilation into amino acids. This remarkable phenotype indicates that plants sense the cellular status of serine and its deficiency triggers the activation of nitrogen assimilation. Although this phenotype is accompanied by enhanced expression of genes encoding ammonium transporter and GS/GOGAT enzymes, the signaling cascade starting from serine starvation and followed by the activation of ammonium uptake and assimilation still remains elusive. Its mechanistic elucidation will provide us information about the metabolic regulation of nitrogen assimilation in plants.
Conclusion
Our study aimed to better understand serine metabolism in plants. Therefore, we dissected the individual contributions made by photorespiration and the PPSB to provide serine for plant growth. We found that in contrast to photorespiration, the PPSB is indispensable for maintaining cell division and elongation and, therefore, plant growth. Accordingly, we found that the PPSB provides serine for the synthesis of major cellular components and its deficiency triggers changes in plant primary metabolism driven by alterations in nitrogen metabolism. Alongside slower rates of amino acid turnover into proteins and purine nucleotides, we found that PPSB-deficient plants exhibit enhanced levels of nitrogen assimilation, which most likely represent a global plant response to amino acid starvation. Conclusively, our data show that the PPSB plays a crucial role in plant performance and constitutes an important future target for crop improvement, especially in the context of plant growth and nitrogen use efficiency.
Materials and methods
Plant material
Arabidopsis thaliana plants of the ecotype Columbia (Col-0) were used. Empty vector control plants (EV), PGDH1-silenced lines (ts-pgdh1.1 and ts-pgdh1.2), and conditional psp1-1 mutant plants have been described previously (Benstein et al., 2013; Cascales-Minana et al., 2013; Krueger et al., 2017). The annotation of PGDH1-silenced lines was changed from #133.7 and #133.15 (Benstein et al., 2013) to ts-pgdh1.1 and ts-pgdh1.2. The efficiency of PGDH1 silencing under the current growth conditions was analyzed by quantitative RT-qPCR (Supplemental Figure S11).
For the analysis of cell proliferation, CYCB1;1::DB:GUS plants were transformed with the PGDH1-silencing construct previously generated by Benstein et al. (2013). In CYCB1;1::DB:GUS plants, the cyclin-destruction-box (DB) of CYCB1;1 is fused to the GUS gene, causing the degradation of the GUS protein at the end of mitosis, allowing visualization and quantification of cell-cycle progression by staining or measuring the GUS activity.
For all experiments, Arabidopsis seeds were surface-sterilized by incubation with ethanol (2 × 10 min in 70% [v/v], 1 × 10 min in 100% [v/v]) and sown on plates containing half-strength Murashige and Skoog (MS) basic salt medium (Duchefa). After 2–4 d of stratification at 4°C, plates were transferred to longday conditions (16 h photoperiod) for germination. Except where stated otherwise, all experimental plants were grown under controlled conditions (16 h photoperiod, 20°C /18°C, 150 μmol·m−2·s−1 irradiance, 60% humidity) in growth chambers.
For all experiments, each biological replicate was collected from plants grown in different plates. Unless otherwise stated, all experiments were performed independently at least twice with similar results.
Analysis of CYCB1;1::DB:GUS activity
For the analysis of cell proliferation, CYCB1;1::DB:GUS ts-pgdh1 plants were transferred 4 d after germination to vertical plates containing half-strength MS medium supplemented with or without 0.1-mM serine. The plates were further incubated for 2 d at 16 h photoperiod, 20/18°C, 150 μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2. After 2 d, half of the plates were transferred to elevated (4,000 µL L−1) CO2. Eight days after transfer to elevated CO2, plant material was harvested and the GUS activity was determined. For histochemical staining, the GUS signal was visualized by staining seedlings with X-Gluc for 12 h at 37°C, followed by 3-h destaining of the chlorophyll in 75% ethanol. GUS-stained seedlings were cleared by incubation in Hoyer’s solution (Anderson, 1954; mixture of chloral hydrate:water:glycerol 3.0:0.8:0.2 [w/v/v]), placed on a glass slide and photographed either using a Leica S8AP0 binocular microscope and the respective LAS-EZ (2.1.0) software package or using differential interference contrast microscopy (Nikon Eclipse E800).
To quantify alterations in CYCLIN B1;1 accumulation, total protein was extracted from seedlings of CYCB1;1::DB:GUS and CYCB1;1::DB:GUS ts-pgdh1 plants and used for a fluorescent β-galactosidase assay (MUG). Product formation was determined fluorometrically (365/455 nm) every 90 s. The rate of product formation corresponds to the amount of β-galactosidase activity, which can be equated with the abundance of CYCB1;1:DB:GUS fusion protein and is an indirect measure of cell proliferation.
Analysis of cell elongation
For the analysis of cell elongation, PGDH1-silenced lines and EV control plants were transferred 4 d after germination to vertical plates containing half-strength MS medium. The plates were further incubated for 8 d at 16-h photoperiod, 20°C/18°C, 150 μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2.
Eight days after transfer to vertical plates, seedlings were harvested and incubated in the dark for ten minutes in 10 μg mL−1 propidium iodide (PI) solution. Seedlings were washed twice in distilled water, placed on a glass slide and PI fluorescence (Ex488/Em503) was monitored using a Zeiss LSM 700 confocal laser-scanning microscope. Images were collected with a 25× lens (Plan-Apochromat 25×/.8 NA water immersion), a laser intensity of 10% and with 543 nm excitation. Fluorescence was measured at 610–700 nm. All measurements were performed with an averaging of eight repetitive scans per line for improved signal-to-noise ratios. The cell size was determined by analyzing the confocal images using ImageJ software (https://imagej.nih.gov/ij/index.html).
14C-labeled serine tracer experiments
For the tracer experiment using 14C-labeled serine [3-14C] (Hartmann Analytic), plants were germinated on half-strength MS medium supplemented with 0.1-mM serine for 4 d before transfer to vertical plates containing the same medium. The plates were incubated for 6 d at 16-h photoperiod, 20/18°C, 150 μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2 prior to transfer of the plants to freshly prepared plates to avoid nutrient starvation effects. After additional 6 d, the plants were transferred from vertical plates to hydroponic solutions containing liquid half-strength MS medium supplemented with 0.1-mM serine, and incubated for 24 h. For the labeling experiment, the liquid medium was exchanged to freshly prepared half-strength MS medium containing 0.1-mM serine traced by 5-µCi 14C-labeled serine [3-14C]. Plant material was harvested after 48 h of incubation in the middle of the light period and immediately frozen in liquid nitrogen.
For the analysis of tracer incorporation into DNA, proteins, and polar and apolar metabolites, a fractionation procedure was adapted from Valledor et al. (2014) using the MACHEREY-NAGEL DNA purification kit for total DNA isolation. Shoot and root material was homogenized using a ball mill (18 Hz, 30 s) and solubilized in 400-µL PL buffer provided in the MACHEREY-NAGEL purification kit. Ultra-pure DNA was isolated from the plant material following the instruction manual. Solubilized plant material was incubated for 10 min at 65°C, continuously shaking at 900 rpm. After centrifugation for 5 min at 11,000g, the supernatant was transferred into a new tube and mixed with 400-µL PC DNA binding buffer by pipetting. The mixture was loaded onto a silica column and centrifuged for 1 min at 11,000g for DNA binding. The flow through was transferred into a new tube, while the bound DNA was washed and eluted according the instruction manual. The flow through was further processed by adding 1,200 µL of a mix containing methanol, water, and chloroform (1:1:1.25 [v/v/v]). The mixture was vortexed and incubated for 5 min at room temperature by continuously shaking at 900 rpm. For phase separation, the mixture was centrifuged for 5 min at 11,000g. The polar phase was completely transferred into a new tube, contamination with the apolar/lipophilic and interphase was avoided. Similarly, the apolar phase was also transferred into a new tube. The remaining protein disc was washed twice with 1 mL 100% MeOH (v/v). The protein pellet was resuspended in 100 µL 0.1M NaOH by incubation for 5 min at 65°C. The total volumes of the polar phase, the lipid/apolar phase, the DNA eluate and the protein suspension were mixed with 4 mL of the scintillation cocktail (Rotiszint eco plus; Roth) and counted in a liquid scintillation counter (Beckman Coulter LS6500).
Analysis of protein translation using 35S-labeled methionine
For the tracer experiment using 35S-labeled methionine, plants were transferred 4 d after germination to vertical plates containing half-strength MS medium. The plates were incubated for 6 d at 16-h photoperiod, 20/18°C, 150 μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2 prior to transfer of the plants to freshly prepared plates to avoid nutrient starvation effects. After additional 6 d, the plants were transferred from vertical plates to hydroponic solutions containing liquid half-strength MS medium supplemented with 0.1-mM methionine and incubated for 24 h. For the labeling experiment, the liquid medium was exchanged with freshly prepared half-strength MS medium containing 0.2-mM methionine traced with 1 µCi 35S-labeled methionine (Hartmann Analytic). Plant material was harvested after 6 h of incubation and immediately frozen in liquid nitrogen.
For the analysis of tracer incorporation into proteins, plant material was homogenized and extracted with 500-µL 0.1M HCl for shoot and 250-µL 0.1M HCl for root tissue in Eppendorf tubes. Ten microliters of the extract was transferred into a scintillation vial, 1-mL scintillation cocktail was added and the radioactivity was determined in the scintillation counter as a measure of total methionine uptake. One-hundred microliters of the residual extract were mixed with 25-μL 100% TCA solution and kept on ice for 15 min to precipitate the proteins. The mixture was centrifuged at 11,000g for 10 min and supernatant was discarded. The remaining protein pellet was washed once in 200-μL 1% TCA and once in 400-μL EtOH, and finally dissolved in 100-μL 0.1M NaOH. The total volume of the protein suspension was mixed with 2 mL of the scintillation cocktail (Rotiszint eco plus; Roth) and counted in a liquid scintillation counter (Beckman Coulter LS6500). To calculate the protein translation rate, radioactivity incorporated into proteins was normalized to the incubation time and total methionine uptake.
Adenosine feeding
For adenosine feeding, plants were transferred 4 d after germination to vertical plates containing half-strength MS medium supplemented with or without 0.25-mM adenosine. The plates were incubated at 16 h photoperiod, 20/18°C, 150-μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2 and the PRGR was continuously monitored for 10 d.
Metabolite analysis
For metabolite profiling, plants were transferred 4 d after germination to vertical plates containing half-strength MS medium supplemented with or without 0.1-mM serine. The plates were incubated for 6 d at 16-h photoperiod, 20/18°C, 150-μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2 prior to transfer of the plants to freshly prepared plates to avoid nutrient starvation effects. After an additional 4 d, half of the plates were transferred to elevated (4,000 µL L−1) CO2 and further incubated for 4 d. Plant material was harvested in the middle of the light period.
Primary metabolites were analyzed either by high-performance liquid chromatography (HPLC) or by gas chromatography coupled mass spectrometry (GC–MS). HPLC analysis was conducted to quantify the amino acid content in plants, as described previously (Krueger et al., 2017). For GC–MS based metabolite profiling, the plant material was freeze-dried and homogenized using a ball mill. Metabolite extraction, derivatization, and analysis was conducted as described previously by Lisec et al. (2006). Metabolites were identified in comparison to database entries of authentic standards (Kopka et al., 2005). Chromatograms and mass spectra were evaluated with the Chroma TOF 1.0 (LECO) and TagFinder 4.0 software (Luedemann et al., 2008).
LC–MS analysis of 15N-labeled compounds
For the time-resolved analysis of 15N incorporation into amino acids, plants were transferred 4 d after germination to vertical plates containing half-strength MS medium. The plates were incubated for 6 d at 16-h photoperiod, 20/18°C, 150-μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2 prior to transfer of the plants to freshly prepared plates to avoid nutrient starvation effects. After additional 6 d, half of the plates were transferred to elevated (4,000 µL L−1) CO2. After additional 4 d, the plants were transferred from vertical plates to hydroponic solutions containing liquid half-strength MS medium and incubated for 24 h. For the labeling experiment, the liquid medium was exchanged to either half-strength MS medium containing 10-mM 14NO3, or to medium in which 14NO3 was replaced by 15NO3. Plant material was harvested during the light period after 0, 2, 4, 8, and 16 h for plants grown at ambient CO2.
To determine the amount of 15N incorporated into amino acids and AMP, LC–MS analysis was conducted. Shortly before extraction, frozen plant material was freeze-dried and homogenized in a ball mill. The dry plant powder was dissolved in 80% MeOH (v/v) including 20-µM methyl-tryptophan (MeOH/MeTrp) as an internal standard, and metabolites were extracted by shaking for 30 min at 600 rpm. To minimize the influence of differences between shoot and root weight on the extraction efficiency, shoot and root materials were dissolved in 500 and 150 µL 80% MeOH/MeTrp (v/v), respectively, corresponding to approximately 3 µL mg−1 fresh weight. Various dilutions of metabolite extracts were tested to avoid ion suppression effects: a 1:50 dilution of shoot extracts and 1:10 of root extracts were found to be most suitable. The diluted samples (5 µL) were injected for LC-MS analysis. Metabolites were separated on a Dionex HPG 3200 HPLC system (Thermo Scientific) equipped with a 150 × 3 mm2, 2.7 μm, XP Xselect HSS T3 C18 column (Waters) with a binary gradient system. Mobile phase A consisted of water + 0.1% formic acid (FA; v/v) and mobile phase B consisted of MeOH + 0.1% FA (v/v). The mobile phase gradients were: 0–1 min, 1% B; 1–10 min, 40% B; 10–15 min, 99% B; 15–16 min, 99% B; and 16–20 min, 1% B. The conditions were kept at 1% mobile phase B for another 3 min for equilibration prior to injection of the next sample. The flow rate was 0.5 mL min−1. Metabolites were analyzed by Q-TOF MS on a maXis 4G instrument (Bruker Daltonics) equipped with an electrospray ionization source. The instrument was operated in positive ion mode. The operating conditions were: dry gas (nitrogen): 8.0 L min−1, dry heater: 220°C, nebulizer pressure: 1.8 bar, capillary voltage: 4,500 V. Collision RF voltage was optimized to 250 V to allow analysis of small metabolites. Mass spectrometry data were evaluated using Compass Quant Analysis software (Bruker). For identification, standard substances for all metabolites of interest were analyzed. Except where stated otherwise, all compounds were purchased from Sigma.
The RIA was calculated as follows:
M represents the abundance of the light isotope, M1 the abundance of the heavy (M + 1) isotope and cf is a factor to correct for the naturally occurring M1 peaks mainly derived from 13C atoms. For each molecule analyzed, the appropriate cf was calculated using the molecular mass calculator (http://www.lfd.uci.edu/∼gohlke/molmass/), a publicly available software tool.
For the analysis of AMP synthesis, plants were grown at ambient CO2 and elevated CO2 as described above. The 15N incorporation into AMP was determined by LC–MS similarly as described for amino acids. For quantification of 15N-labeled AMP, the abundances of M + 1, M + 2, M + 3, M + 4, and M + 5 were summarized and normalized to the amount of 15N-labeled glutamate. Glutamate represents the unique nitrogen source for all amino acid precursors of AMP synthesis. Thus, normalization to glutamate was used to avoid artificial effects coming from differences in nitrogen uptake and assimilation.
Quantification of ammonium
The amount of ammonium in plant tissue was determined according to the modified method previously described by Bräutigam et al. (2007). Therefore, frozen plant tissue was homogenized by using a ball mill. Fifty microliters of 0.1M HCl per 10 mg fresh weight (FW) was added to the frozen powder and ammonium was extracted by incubation of the sample for 5 min at 80° C. Insoluble plant material was removed by centrifugation of the extract at 10,000g for 20 min. The supernatant was transferred into a fresh tube and ammonium was determined based on the Berthelot method. The absorbance at 635 nm was determined after sequential addition of 150-µL sodium phenoxide trihydrate (0.3 M), 300-µL sodium nitroprusside (0.02%), and 150-µL sodium hypochlorite (1:50) to 150 µL of the extract.
Determination of citrate synthase activity and cellular respiration
To measure citrate synthase activity, proteins were extracted with a buffer containing 50-mM Hepes/KOH, pH 7.4, 1-mM EDTA, 1-mM EGTA, 2-mM benzamidine, 2-mM E-aminocaproic acid, 0.5-mM PMSF, 10% glycerol, and 0.1% Triton x-100. Enzyme activity was assayed as described by Stitt (1989). Cellular respiration in plant roots was analyzed according to Jacoby et al. (2015) using a Clark-type oxygen electrode (Hansatech). Entire root systems (∼40 mg fresh weight) were excised, and oxygen consumption rate was measured in respiration buffer (half-strength MS, 50-mM MES, pH 5.8) over 10 min at 25°C.
Expression analysis by reverse transcription quantitative PCR
For the analysis of transcripts by reverse transcription quantitative polymerase chain reaction (RT-qPCR), plants were transferred 4 d after germination to vertical plates containing half-strength MS medium. The plates were further incubated for 10 d at 16-h photoperiod, 20/18°C, 150-μmol·m−2·s−1 irradiance, 60% humidity at ambient (400 µL L−1) CO2. RT-qPCR was conducted according to a previously published protocol (Czechowski et al., 2005). Therefore, total RNA was extracted from 20 to 50 mg of shoot and root material by hot phenol extraction, followed by a LiCl precipitation. Isolated RNA was digested with the TURBO DNA-free kit (Ambion) and 5-μg digested RNA was used for first-strand cDNA synthesis using Bioscript Reverse Transcriptase (Bioline), according to the manufacturer’s instructions. To quantify the expression of the respective genes, quantitative PCR was performed using the 7300 Real-Time PCR System and SYBR Green (Applied Biosystems), according to the manufacturer’s instructions. Sequences of the primers used for the PCR reactions are listed in Supplemental Table 1.
Statistical analysis
Bar plots and box plots were generated in SigmaPlot 10 software. The Shapiro–Wilk test of normality was used before the one-way ANOVA followed by Fisher’s least significant difference test was conducted. P-values were adjusted with the Benjamini–Hochberg method. Statistical analyses were performed in RStudio Version 1.1.456, except for Student’s t test, which was performed in Excel (Microsoft). All Figures were finalized in ADOBE Photoshop CS3 EXTENDED Version 10 software.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers_At4g34200; At1g17745; At3g19480; At4g35630; At2g17630; At1g18640.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Pathways for serine biosynthesis in plants.
Supplemental Figure S2. Growth of psp1-1 mutants.
Supplemental Figure S3. Expression analysis of PPSB genes at ambient and elevated CO2.
Supplemental Figure S4. PGDH1-silenced lines require continuous supply of external serine to maintain normal plant growth.
Supplemental Figure S5. Growth of PGDH1-silenced lines at elevated CO2 is significantly improved by external serine.
Supplemental Figure S6. Size of the meristem and elongation zone in wild type and psp1-1 mutant plants.
Supplemental Figure S7. Citrate synthase activity in PGDH1-silenced lines.
Supplemental Figure S8. Quantification of 15N-labeled amino acids in PGDH1-silenced lines.
Supplemental Figure S9. Quantification of 15N-labeled amino acids in PGDH1-silenced plants.
Supplemental Figure S10. Changes in the ammonium content in PGDH1-silenced lines.
Supplemental Figure S11. Expression analysis of PGDH genes in PGDH1-silenced lines.
Supplemental Table S1. Oligonucleotide sequences.
Funding
The research of S.K. was supported by the Deutsche Forschungsgemeinschaft (Grants Kr4245/1-1 and Kr4245/2-1). The research of S.E.Z., V.W., S.Kop., and S.C.G. was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy (EXC-1028 and EXC-2048/1 project ID 390686111). The research of R.R. was supported by the Spanish Government and the European Union (FEDER/BFU2015-64204R), the Valencian Regional Government: PROMETEO II/2014/052; and the University of Valencia: V segles fellowship to M.F.-T.
Conflict of interest statement: The author declare that they have no conflict of interest.
Supplementary Material
S.E.Z., R.M.B., M.F.-T., A.D.A., S.A., A.R.F., S.R.T., S.C.G., S.B., M.A.S., P.G., V.W., S.Kop., and S.K. performed research. R.R., R.P.J., U.-I.F., and S.K. wrote the article. R.R. and S.K. designed the research.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Stephan Krueger (stephan.krueger@uni-koeln.de).
References
- Abadie C, Boex-Fontvieille ER, Carroll AJ, Tcherkez G (2016) In vivo stoichiometry of photorespiratory metabolism. Nat Plants 2: 15220. [DOI] [PubMed] [Google Scholar]
- Achouri Y, Rider MH, Schaftingen EV, Robbi M (1997) Cloning, sequencing and expression of rat liver 3-phosphoglycerate dehydrogenase. Biochem J 323: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcázar R, Marco F, Cuevas JC, Patron M, Ferrando A, Carrasco P, Tiburcio AF, Altabella T (2006) Involvement of polyamines in plant response to abiotic stress. Biotechnol Lett 28: 1867–1876 [DOI] [PubMed] [Google Scholar]
- Amelio I, Cutruzzola F, Antonov A, Agostini M, Melino G (2014) Serine and glycine metabolism in cancer. Trends Biochem Sci 39: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LE (1954) Hoyer’s solution as a rapid permanent mounting medium for bryophytes. Bryologist 57: 242–244 [Google Scholar]
- Andrews TJ, Lorimer GH, Tolbert NE (1971) Incorporation of molecular oxygen into glycine and serine during photorespiration in spinach leaves. Biochemistry 10: 4777–4782 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Martins AO, Fernie AR, Tohge T (2014) 2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front Plant Sci 5: 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo WL, Ishizaki K, Nunes-Nesi A, Larson TR, Tohge T, Krahnert I, Witt S, Obata T, Schauer N, Graham IA, et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Kern R, Timm S (2012) Photorespiration has a dual origin and manifold links to central metabolism. Curr Opin Plant Biol 15: 269–275 [DOI] [PubMed] [Google Scholar]
- Benstein RM, Ludewig K, Wulfert S, Wittek S, Gigolashvili T, Frerigmann H, Gierth M, Flugge UI, Krueger S (2013) Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25: 5011–5029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowes G, Ogren WL, Hageman RH (1971). Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem Biophys Res Commun 45: 716–722 [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Gagneul D, Weber AP (2007) High-throughput colorimetric method for the parallel assay of glyoxylic acid and ammonium in a single extract. Anal Biochem 362: 151–153 [DOI] [PubMed] [Google Scholar]
- Busch FA, Sage RF, Farquhar GD (2018) Plants increase CO2 uptake by assimilating nitrogen via the photorespiratory pathway. Nat Plants 4: 46–54 [DOI] [PubMed] [Google Scholar]
- Cascales-Minana B, Munoz-Bertomeu J, Flores-Tornero M, Anoman AD, Pertusa J, Alaiz M, Osorio S, Fernie AR, Segura J, Ros R (2013) The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25: 2084–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin ACL, Maddocks ODK, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, et al. (2012). Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Däschner K, Couée I, Binder S (2001). The mitochondrial isovaleryl-coenzyme a dehydrogenase of arabidopsis oxidizes intermediates of leucine and valine catabolism. Plant Physiol 126: 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Hu Z, Xu XL, Sacchettini JC, Grant GA (2005) D-3-phosphoglycerate dehydrogenase from mycobacterium tuberculosis is a link between the Escherichia coli and mammalian enzymes. J Biol Chem 280: 14884–14891 [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Engel N, Ewald R, Gupta KJ, Zrenner R, Hagemann M, Bauwe H (2011). The presequence of Arabidopsis serine hydroxymethyltransferase SHM2 selectively prevents import into mesophyll mitochondria. Plant Physiol 157: 1711–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist MK, Kuhn A, Wienstroer J, Weber K, Jansen EE, Jakobs C, Weber AP, Maurino VG (2011) Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J Biol Chem 286: 11382–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine JX, Tercé-Laforgue T, Armengaud P, Clément G, Renou JP, Pelletier S, Catterou M, Azzopardi M, Gibon Y, Lea PJ, et al. (2012) Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism. Plant Cell 24: 4044–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaba A, Wang Z, Krishnamoorthy T, Hinnebusch AG, Sachs MS (2001) Physical evidence for distinct mechanisms of translational control by upstream open reading frames. EMBO J 20: 6453–6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Lee K, Reid MA, Sanderson SM, Qiu C, Li S, Liu J, Locasale JW (2018) Serine availability influences mitochondrial dynamics and function through lipid metabolism. Cell Rep 22: 3507–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibon Y, Usadel B, Blaesing OE, Kamlage B, Hoehne M, Trethewey R, Stitt M (2006) Integration of metabolite with transcript and enzyme activity profiling during diurnal cycles in Arabidopsis rosettes. Genome Biol 7: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glevarec G, Bouton S, Jaspard E, Riou MT, Cliquet JB, Suzuki A, Limami AM (2004) Respective roles of the glutamine synthetase/glutamate synthase cycle and glutamate dehydrogenase in ammonium and amino acid metabolism during germination and post-germinative growth in the model legume Medicago truncatula. Planta 219: 286–297 [DOI] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inze D (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Guan M, de Bang TC, Pedersen C, Schjoerring JK (2016) Cytosolic glutamine synthetase Gln1;2 is the main isozyme contributing to GS1 activity and can be up-regulated to relieve ammonium toxicity. Plant Physiol 171: 1921–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handford J, Davies DD (1958) Formation of phosphoserine from 3-phopho-glycerate in higher plants. Nature 182: 532–533 [Google Scholar]
- Hesse H, Hoefgen R (2003) Molecular aspects of methionine biosynthesis. Trends Plant Sci 8: 259–262 [DOI] [PubMed] [Google Scholar]
- Hildebrandt TM, Nunes Nesi A, Araujo WL, Braun HP (2015) Amino acid catabolism in plants. Mol Plant 8: 1563–1579 [DOI] [PubMed] [Google Scholar]
- Ho CL, Saito K (2001) Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20: 243–259 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito K (1999a) Plastidic pathway of serine biosynthesis. Molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. J Biol Chem 274: 11007–11012 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Saito K (1999b) Regulation of serine biosynthesis in Arabidopsis. Crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J Biol Chem 274: 397–402 [DOI] [PubMed] [Google Scholar]
- Ho CL, Noji M, Saito M, Yamazaki M, Saito K (1998) Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J 16: 443–452 [DOI] [PubMed] [Google Scholar]
- Ishiyama K, Inoue E, Watanabe-Takahashi A, Obara M, Yamaya T, Takahashi H (2004) Kinetic properties and ammonium-dependent regulation of cytosolic isoenzymes of glutamine synthetase in Arabidopsis. J Biol Chem 279: 16598–16605 [DOI] [PubMed] [Google Scholar]
- Jacoby RP, Millar AH, Taylor NL (2015) Assessment of respiration in isolated plant mitochondria using Clark-type electrodes. Methods Mol Biol 1305: 165–185 [DOI] [PubMed] [Google Scholar]
- Kalhan SC, Hanson RW (2012) Resurgence of serine: an often neglected but indispensable amino acid. J Biol Chem 287: 19786–19791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebeish R, Niessen M, Thiruveedhi K, Bari R, Hirsch HJ, Rosenkranz R, Stabler N, Schonfeld B, Kreuzaler F, Peterhansel C (2007) Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nat Biotechnol 25: 593–599 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Tjaden J, Neuhaus HE (2008) Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J 56: 51–63 [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA, Givan CV (1988) Serine formation in leaves by mechanisms other than the glycolate pathway. J Plant Physiol 132: 641–652 [Google Scholar]
- Kleczkowski LA, Givan CV, Hodgson JM, Randall DD (1988) Subcellular location of NADPH-dependent hydroxypyruvate reductase activity in leaf protoplasts of Pisum sativum L. and its role in photorespiratory metabolism. Plant Physiol 88: 1182–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi N, Ishiyama K, Beier MP, Inoue E, Kanno K, Yamaya T, Takahashi H, Kojima S (2017) Contributions of two cytosolic glutamine synthetase isozymes to ammonium assimilation in Arabidopsis roots. J Exp Bot 68: 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dormann P, Weckwerth W, Gibon Y, Stitt M, et al. (2005) GMD@CSB.DB: the Golm metabolome database. Bioinformatics 21: 1635–1638 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Suter M, den Camp RO, Brunold C, Kopriva S (2000). Regulation of sulfate assimilation by nitrogen in Arabidopsis. Plant Physiol 122: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, et al. (2016) LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539: 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger S, Benstein RM, Wulfert S, Anoman AD, Flores-Tornero M, Ros R (2017) Studying the function of the phosphorylated pathway of serine biosynthesis in Arabidopsis thaliana. Methods Mol Biol 1653: 227–242 [DOI] [PubMed] [Google Scholar]
- Labboun S, Terce-Laforgue T, Roscher A, Bedu M, Restivo FM, Velanis CN, Skopelitis DS, Moschou PN, Roubelakis-Angelakis KA, Suzuki A, et al. (2009) Resolving the role of plant glutamate dehydrogenase. I. In vivo real time nuclear magnetic resonance spectroscopy experiments. Plant Cell Physiol 50: 1761–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD (2014) Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep 7: 1248–1258 [DOI] [PubMed] [Google Scholar]
- Lancien M, Martin M, Hsieh MH, Leustek T, Goodman H, Coruzzi GM (2002) Arabidopsis glt1-T mutant defines a role for NADH-GOGAT in the non-photorespiratory ammonium assimilatory pathway. Plant J 29: 347–358 [DOI] [PubMed] [Google Scholar]
- Last RL, Fink GR (1988) Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240: 305–310 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, et al. (2011) Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lothier J, Gaufichon L, Sormani R, Lemaître T, Azzopardi M, Morin H, Chardon F, Reisdorf-Cren M, Avice JC, Masclaux-Daubresse C (2011) The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J Exp Bot 62: 1375–1390 [DOI] [PubMed] [Google Scholar]
- Luedemann A, Strassburg K, Erban A, Kopka J (2008) TagFinder for the quantitative analysis of gas chromatography–mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 24: 732–737 [DOI] [PubMed] [Google Scholar]
- Maier A, Fahnenstich H, von Caemmerer S, Engqvist MK, Weber AP, Flugge UI, Maurino VG (2012) Transgenic introduction of a glycolate oxidative cycle into A. thaliana chloroplasts leads to growth improvement. Front Plant Sci 3: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Reisdorf-Cren M, Pageau K, Lelandais M, Grandjean O, Kronenberger J, Valadier MH, Feraud M, Jouglet T, Suzuki A (2006) Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaini KR, Sullivan MR, Vander Heiden MG (2016) The importance of serine metabolism in cancer. J Cell Biol 214: 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modde K, Timm S, Florian A, Michl K, Fernie AR, Bauwe H (2017) High serine:glyoxylate aminotransferase activity lowers leaf daytime serine levels, inducing the phosphoserine pathway in Arabidopsis. J Exp Bot 68: 643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möhlmann T, Mezher Z, Schwerdtfeger G, Neuhaus HE (2001) Characterisation of a concentrative type of adenosine transporter from Arabidopsis thaliana (ENT1,At). FEBS Lett 509: 370–374 [DOI] [PubMed] [Google Scholar]
- Moison M, Marmagne A, Dinant S, Soulay F, Azzopardi M, Lothier J, Citerne S, Morin H, Legay N, Chardon F, et al. (2018) Three cytosolic glutamine synthetase isoforms localized in different-order veins act together for N remobilization and seed filling in Arabidopsis. J Exp Bot 69: 4379–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralla R, Sweeney C, Stepansky A, Leustek T, Meinke D (2007) Genetic dissection of histidine biosynthesis in Arabidopsis. Plant Physiol 144: 890–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ (2001) Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol 21: 4347–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy A, Savka MA, Hudson AO (2019) The synthesis and role of β-alanine in plants. Front Plant Sci 10: 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer LI (1963) The pathway and control of serine biosynthesis in Escherichia coli. J Biol Chem 238: 3934–3944 [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et al. (2011). Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476: 346–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilevitch S, Cousins AB, Bloom AJ (2004). Nitrate assimilation in plant shoots depends on photorespiration. Proc Natl Acad Sci USA 101: 11506–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SP (1982) 3-phosphoglycerate phosphatase activity in chloroplast preparations as a result of contamination by acid phosphatase. Plant Physiol 70: 645–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RE, Mecchia MA, Debernardi JM, Schommer C, Weigel D, Palatnik JF (2010) Control of cell proliferation in Arabidopsis thaliana by microRNA miR396. Development 137: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros R, Munoz-Bertomeu J, Krueger S (2014) Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci 19: 564–569 [DOI] [PubMed] [Google Scholar]
- Schmidtmann E, Konig AC, Orwat A, Leister D, Hartl M, Finkemeier I (2014) Redox regulation of Arabidopsis mitochondrial citrate synthase. Mol Plant 7: 156–169 [DOI] [PubMed] [Google Scholar]
- Skopelitis DS, Paranychianakis NV, Kouvarakis A, Spyros A, Stephanou EG, Roubelakis-Angelakis KA (2007) The isoenzyme 7 of tobacco NAD(H)-dependent glutamate dehydrogenase exhibits high deaminating and low aminating activities in vivo. Plant Physiol 145: 1726–1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K (1984) Enzymes of serine metabolism in normal, developing and neoplastic rat tissues. Adv Enzyme Regul 22: 325–400 [DOI] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR (2019) Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M (1989) Product inhibition of potato tuber pyrophosphate:fructose-6-phosphate phosphotransferase by phosphate and pyrophosphate. Plant Physiol 89: 628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabatabaie L, Klomp LW, Berger R, de Koning TJ (2010) L-serine synthesis in the central nervous system: a review on serine deficiency disorders. Mol Genet Metab 99: 256–262 [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valledor L, Escandon M, Meijon M, Nukarinen E, Canal MJ, Weckwerth W (2014) A universal protocol for the combined isolation of metabolites, DNA, long RNAs, small RNAs, and proteins from plants and microorganisms. Plant J 79: 173–180 [DOI] [PubMed] [Google Scholar]
- Weingartner M, Criqui MC, Meszaros T, Binarova P, Schmit AC, Helfer A, Derevier A, Erhardt M, Bogre L, Genschik P (2004) Expression of a nondegradable cyclin B1 affects plant development and leads to endomitosis by inhibiting the formation of a phragmoplast. Plant Cell 16: 643–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte CP, Herde M (2020) Nucleotide metabolism in plants. Plant Physiol 182: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulfert S, Krueger S (2018) Phosphoserine aminotransferase1 is part of the phosphorylated pathways for serine biosynthesis and essential for light and sugar-dependent growth promotion. Front Plant Sci 9: 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Vousden KH (2016) Serine and one-carbon metabolism in cancer. Nat Rev Cancer 16: 650–662 [DOI] [PubMed] [Google Scholar]
- Zhang Q, Lee J, Pandurangan S, Clarke M, Pajak A, Marsolais F (2013). Characterization of Arabidopsis serine:glyoxylate aminotransferase, AGT1, as an asparagine aminotransferase. Phytochemistry 85: 30–35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.