Abstract
WWP1 is an E3 ubiquitin ligase that has been reported to target the tumor suppressor lipid phosphatase PTEN. K740N and N745S are recently identified germline variants of WWP1 that have been linked to PTEN-associated cancers (Y.R. Lee et al. New England Journal of Medicine, 2020). These WWP1 variants have been suggested to release WWP1 from its native autoinhibited state, thereby promoting enhanced PTEN ubiquitination as a mechanism for driving cancer. Using purified proteins and in vitro enzymatic assays, we investigate the possibility that K740N and N745S WWP1 possess enhanced ubiquitin ligase activity and demonstrate that these variants are similar to WT in both autoubiquitination and PTEN ubiquitination. Furthermore, K740N and N745S WWP1 show similar dependencies to WT in terms of allosteric activation by an engineered ubiquitin variant, upstream E2 concentration, and substrate ubiquitin concentration. Transfected WWP1 WT and mutants demonstrate comparable effects on cellular PTEN levels. These findings challenge the idea that K740N and N745S WWP1 variants promote cancer by enhanced PTEN ubiquitination.
Introduction
PTEN is a phosphatidyl 3,4,5-triphosphate (PIP3) lipid phosphatase that regulates cell growth and metabolism1–5. Loss of PTEN function by mutation, epigenetically, or through post-translational modifications is a common feature of human cancer4, 6, 7. It has been proposed that the ubiquitination of PTEN can lead to its inactivation or destruction8, 9. Several ubiquitin E3 ligases in the NEDD4 family including WWP1, WWP2, and NEDD4-1 have been reported to be PTEN ubiquitin ligases10–13. NEDD4-1, WWP1, and WWP2 are comprised of an N-terminal C2 domain that can bind membranes followed by four WW domains that interact with Pro-rich peptide motifs and culminating in a C-terminal catalytic (HECT) domain14. Recently, Y.R. Lee et al. reported that two germline variants in the catalytic domain of WWP1, K740N and N745S, are associated with increased PTEN-like cancers15. They proposed that these WWP1 mutations increase its enzymatic activity and PTEN ubiquitination by relieving the intramolecular or intermolecular autoinhibition conferred by the 2,3-linker. Prior studies have shown that WWP1 and several NEDD4 E3 ligases are autoinhibited by an internal ~30 aa alpha-helical peptide linker positioned between the WW domains, the 2,3-linker (connecting the WW2 and WW3 domains) in WWP1 and WWP2, and the 1,2-linker (connecting the WW1 and WW2 domains) in NEDD4-15, 16–18 (Figure 1A). Displacing these linkers and activation of these ubiquitin ligases can occur through several physiological mechanisms including allosteric protein activators and Tyr/Thr phosphorylation16, 19, 20 (Figure 1B). Among protein activators, ubiquitin itself can engage the so-called exosite of the NEDD4 family catalytic domain, a pocket distinct from the ubiquitin substrate-binding site21, 22. Engineered ubiquitin variants (UbV) such as UbV for WWP1 and WWP2 (UbV P2.3 reported by W. Zhang et al.23) that have high affinity and selectivity for particular NEDD4 family member exosites, and lack the ability to serve as E3 ubiquitin ligase substrates, have been developed23. UbV can efficiently compete away the 2,3-linker of WWP1 and WWP2 and stimulate the catalytic activities of these enzymes in both autoubiquitination and protein substrate ubiquitination18, 23. Beyond physiological activation mechanisms of NEDD4 family members, a mutation found in renal cell carcinoma (M752T) in the WWP2 HECT domain that lies at the linker/HECT domain interface has been shown to enhance ubiquitination ligase activity18, 24.
Figure 1.
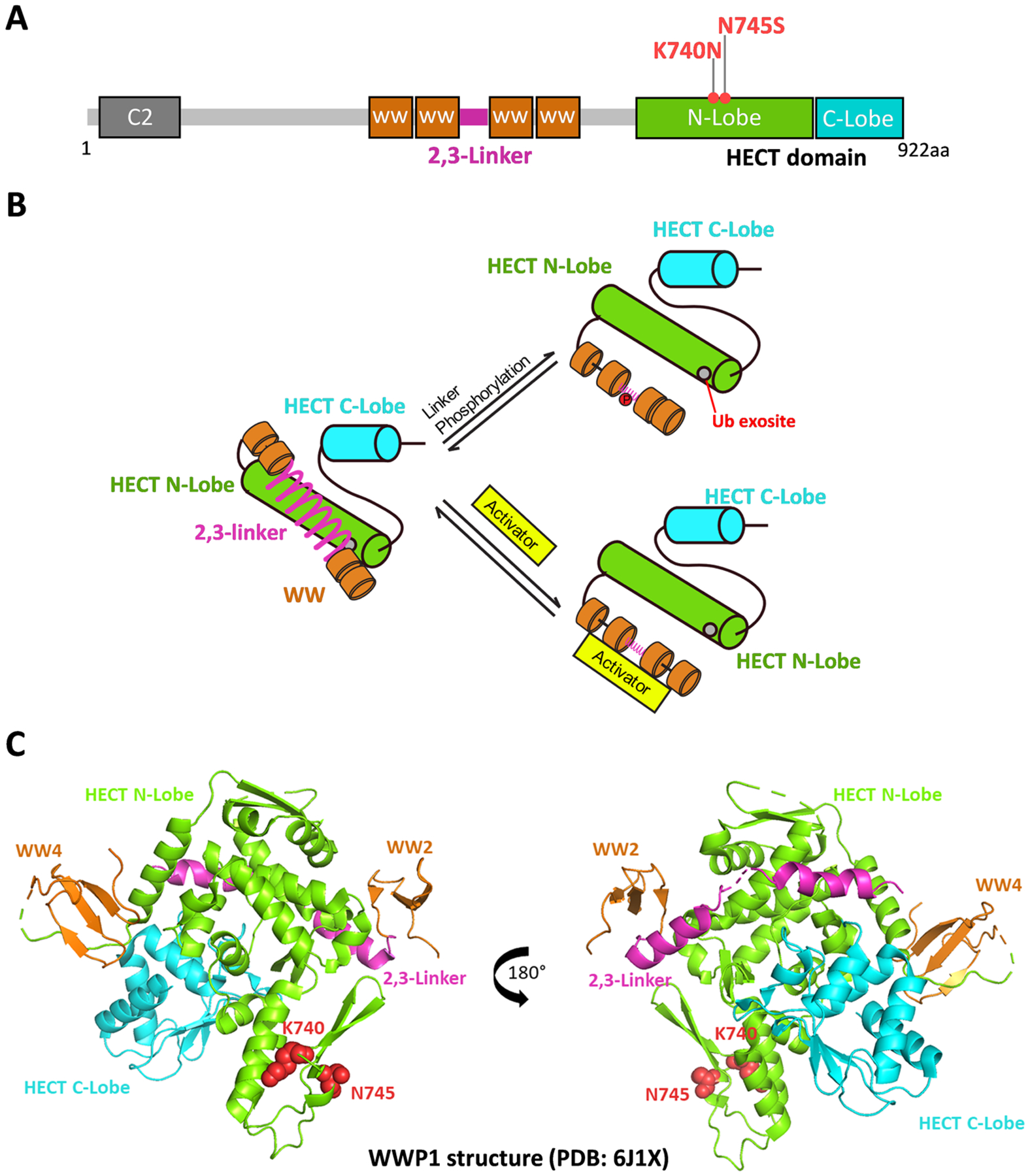
WWP1 and its mechanisms of regulation. (A) WWP1 protein overview. WWP1 has an N-terminal C2 domain, four WW domain, and a C-terminal catalytic HECT domain. The autoinhibitory link (2,3-Linker) lies between WW2 and WW3 domains. The HECT domain consists of N-Lobe and C-Lobe. The two germline mutants K740N and N745S locate on the N-Lobe of HECT. (B) A scheme shows linker activation mechanisms. The WWP1 linker can be displaced by either phosphorylation of Tyr in the linker or binding to allosteric protein activators. (C) Crystal structure of WWP1 with autoinhibitory 2,3-Linker (PDB: 6J1X). The HECT N-Lobe, C-Lobe, linker, and WW domains are colored in cyan, green, purple, and brown, accordingly. K740 and N745 are highlighted in red spheres.
An X-ray crystal structure of WWP15 shows that both K740 and N745 are rather remote from the 2,3-linker-catalytic domain interaction surface in WWP1 (Figure 1C). Based on our analysis of the X-ray structures of the various NEDD4 family members, it was difficult to rationalize that K740N and N745S replacements in WWP1 would destabilize linker/HECT interaction or otherwise stimulate catalysis. In this study, we investigate these variants in the context of purified WWP1 proteins. The enzymatic experiments on these WWP1 variants performed here do not support a role for these germline variants as activating WWP1 ubiquitin ligase catalysis.
Experimental Procedures
Reagents
Human WWP1 cDNA (aa2-922) was purchased from Genscript. The ubiquitin variant gene DNA was synthesized by IDT. The WWP1 and ubiquitin variant gene cDNAs were subcloned into a pGEX6p-2 vector for E. coli expression. The WWP1 K740N and N745S mutations were prepared by QuikChange mutagenesis and the sequence of the entire open reading frames were confirmed by DNA sequencing. Wild-type ubiquitin, ubiquitin-activating enzyme UBA1 (E1), and conjugating enzyme UbcH5b (E2) were prepared as previously described25. The Colloidal Blue staining kit was purchased from Thermo Fisher. The anti-PTEN antibody (N-19 and A2B1) was purchased from Santa Cruz Biotechnology. The anti-GAPDH antibody (14C10) was purchased from Cell signaling Technology. The transfection reagent Lipofectamine 3000 was purchased from Thermo Fisher Scientific. All other reagents were of the highest quality and obtained commercially from either MilliporeSigma or Thermo Fisher Scientific.
Expression and Purification of PTEN Protein
PTEN was prepared as previously described by Bolduc et al26. Briefly, PTEN (amino acids 1-378) fused to Mycobacterium xenopi GyrA intein and the chitin-binding domain (CBD) was subcloned into the pFastBac-1 vector and transformed into DH10Bac competent cells for production of a bacmid containing the PTEN sequence. The bacmid containing PTEN-intein-CBD was then transfected into Sf-21 insect cells to generate the corresponding baculovirus needed for protein expression. After successful virus production, High Five insect cells were infected at a multiplicity of infection (MOI) of 1 with the baculovirus at a cell density of 1 million/ml and cultured for 48 hr at 27 °C for the protein expression. The final culture was then harvested by centrifugation, and the insect cells were lysed by a Dounce homogenizer by 40 stokes in 40 ml of 50 mM HEPES pH 7.5, 250 mM NaCl, 1 mM EDTA, 10% glycerol and one table of complete EDTA-free Roche cocktail protease inhibitor (Roche). The soluble lysate was then incubated with fibrous cellulose (MilliporeSigma) for 30 min at 4°C to remove chitinase. The fusion protein then was immobilized on chitin resin and incubated with 50 mM HEPES pH 7.3, 250 mM NaCl, 400 mM MESNa, and 50 mM cysteine for 24 hr at room temperature. After incubation, PTEN was eluted from resin followed by dialysis with the same buffer to remove unreacted cysteine and salt from the reaction. The PTEN protein was then further purified by FPLC anion-exchange chromatography (MonoQ column, GE healthcare). using a gradient of 0–50% buffer B where buffer A is 50 mM TRIS pH 8.0, 5 mM NaCl, 10 mM DTT and buffer B is 50 mM TRIS pH 8.0, 1 M NaCl, 10 mM DTT. Pure fractions determine by 10% SDS-PAGE were concentrated, flash frozen, and stored at −80°C.
Expression and Purification of WT and Mutant WWP1 and Ubiquitin Variant (UbV) Proteins
The pGEX6p-2 plasmids were transformed into E.coli BL21 Codon plus competent cells for expression. The expression and purification procedures were performed as previously described16. In brief, the E.coli cells were cultured at 37°C to OD600= 0.6. 0.5 mM IPTG was added to induce protein expression at 16°C for 20 hr. The cells were harvested and lysed with a french press in buffer containing 25 mM HEPES pH 7.8, 250 mM NaCl, 1 mM TCEP, and a 1X mixture of protease inhibitor cocktail (Roche Applied Science). The cell lysate was loaded onto GSH-agarose and washed with the same buffer containing 0.1% Triton-X. The desired GST-tagged protein was eluted with 50 mM reduced GSH and treated with Prescission protease (GE Healthcare) at 4°C overnight while being dialyzed against a buffer containing 25 mM HEPES pH 7.8, 250 mM NaCl, and 1 mM TCEP. After cleavage, the protein was loaded to GSH resin again to remove the free GST. The protein was then concentrated and further purified by Superdex 200 increased 10/300 GL (GE Healthcare). The purified fractions were combined, concentrated, and stored at −80°C.
In vitro Ubiquitination Assays
The in vitro ubiquitination assays were conducted using conditions similar to those previously described for WWP2 and NEDD4-1.13 Reactions were carried out at 30°C (or 37°C as indicated) in a total volume of 20 μl. The reactions contained 5 mM ATP, 50 μM wt ubiquitin, 50 nM E1 protein, 1 μM E2 protein (unless indicated otherwise), 1 μM WWP1 with 40 mM Tris-HCl, pH 7.5, 50 mM NaCl, 0.5 mM TCEP and 5 mM MgCl2 as reaction buffer. Substrate PTEN at 5 μM was preincubated with everything except E1 in the reaction at 30°C for 20 min before E1 was added to the system to initiate the reactions. In the UbV activation assays, the addition of UbV at 5 μM was included in the requisite reactions. In the mutant/wild type mixing experiments, wild type at 0.5 μM and WWP1 mutants K740N or N745S at 0.5 μM were added in the indicated group. In the E2 titration assays, the E2 concentration was varied at 0.1, 0.3, 1, and 5 μM. In the ubiquitin concentration titration assays, the ubiquitin concentration used in the assays were at 10, 30, 100 and 300 μM. Reactions were quenched at the indicated time points by adding SDS loading buffer containing reducing agent β-mercaptoethanol. The reaction samples were then resolved on SDS-PAGE gels and analyzed by either Colloidal Coomassie Blue staining or Western Blots. All assays were repeated on at least three independent occasions with replicates revealing similar results to the data presented in the figures.
Cell Culture and Cellular Transfection Experiments
HCT 116 colon cancer cells were obtained from ATCC and cultured in McCoy’s 5A medium with L-Glutaminer supplemented with 10% FBS and antibiotics-penicillin/streptomycin in a 37°C incubator with 5% CO2. HCT116 cells were seeded in 6-well plate. At around 90% confluence, the cells were transfected with 0.6 μg pcDNA3.1 PTEN C124S (catalytic defective PTEN is used as wt PTEN was toxic to cells under these conditions), 0.5 μg pRK HA-wt ubiquitin and 1.2 μg pcDNA3.1 Myc-WWP1 (WT, K740N, or N745S) using Lipofectamine 3000 reagent(Invitrogen). 48 hours after the transfection, the cells were lysed with RIPA buffer (Cell signaling) containing 0.5 mM PMSF and 1x Pierce cocktail protease inhibitor (Thermo Fisher Scientific). The cell lysate was mixed with SDS loading dye and boiled for 5 min. 30 μg of total protein (determined by BCA assay) was resolved on SDS-PAGE gels, and transferred to a nitrocellulose membranes using an iBlot dry blotting system (Thermo Fisher Scientific). All assays were repeated on four independent occasions with replicates showing similar results.
Western Blotting
After SDS-PAGE, the proteins were transferred to nitrocellulose membranes using an iBlot dry-blotting system (Thermo Fisher Scientific). The membranes were then blocked with 5% BSA in PBST buffer for 1 hr and then incubated with anti-PTEN antibody (1:500 for in vitro assays, 1:100 for cell lysate) or anti-GAPDH (1:1000) at 4°C overnight. After this, the membranes were washed with PBST and probed with HRP-conjugated secondary antibody. The bands were detected by chemiluminescence using an ECL Western Blot detection kit (Bio-Rad).
Data Analysis
To quantify the rate of the ubiquitination reactions, the WWP1 bands were quantified by densitometric analysis using ImageJ software, and the decrease of unmodified WWP1 was calculated as a percentage of the baseline WWP1 band. All the ubiquitination assays were repeated at least three times and the average values and standard deviations were calculated and stated in the Figure legends. The Western blots for cell transfection studies were repeated at lease three times. The bands were quantified using ImageJ and the error bars represent the standard error of the mean (SEM). The statistical significance and p values between groups were calculated using Graphpad Prism software using paired T-tests and reported in the figures.
Results
We produced full-length recombinant human wild-type (WT), K740N, and N745S WWP1 proteins from E. coli as N-terminal GST fusion proteins and after glutathione affinity chromatography used Prescission protease to remove the GST. These WWP1 proteins were further subjected to size exclusion chromatography which revealed that the final purified proteins were monomeric and were >70% pure using SDS-PAGE (Supplemental Figure S1). The ubiquitin ligase activities of these WWP1 proteins were measured by mixing with purified recombinant E1 and E2 enzymes as well as recombinant PTEN protein as a substrate. The depletion of WWP1 and formation of autoubiquitinated WWP1 forms were measured as a function of time by SDS-PAGE stained with Colloidal Coomassie Blue whereas PTEN ubiquitination was determined by Western blot. These experiments (Figure 2A and 2B) revealed that neither K740N nor N745S WWP1 showed enhanced autoubiquitination or PTEN ubiquitination relative to WT and K740N showed slightly reduced activities in these assay conditions. These ubiquitin ligase results were similar whether performed at the routine temperature of 30°C18 (Figure 2A) or physiological temperature of 37°C (Figure 2B). As the germline variants are generally heterozygous15, we also evaluated the K740N and N745S WWP1 ubiquitin ligase kinetics in the context of WT WWP1 as 1:1 mixture (Figure 2C). These assays showed no apparent activation of WT WWP1 conferred by the presence of K740N or N745S WWP1.
Figure 2.
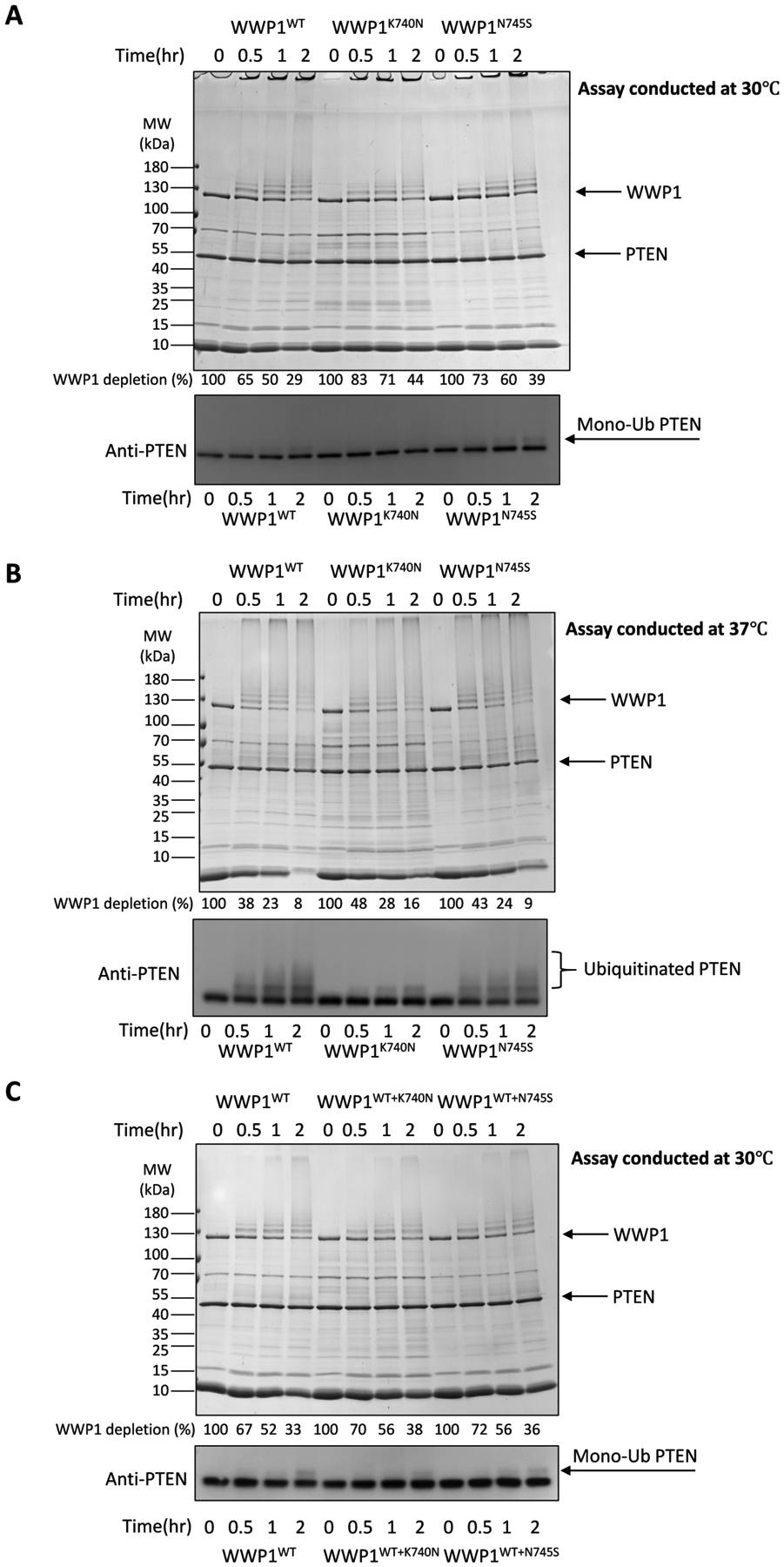
WWP1 K740N and N745S forms show similar E3 ubiquitin ligase activity compared to WT WWP1. (A) Auto- and PTEN ubiquitination assays for WWP1 wild-type, or K740N, N740S germline mutants at 30°C (n=3). The activity of WWP1 autoubiquitination was determined by the time-dependent depletion of the unmodified WWP1 band and the appearance of higher MW bands presumed to represent mono- or poly-ubiquitination. The depletion of WWP1 was quantified by densitometry of the Colloidal stained SDS-PAGE and the average percentages are listed below each lane corresponding to the indicated time points. The average ± S.D. from 3 repeats are as follows (in %): 100, 65±8, 50±7, 29±3; 100, 83±4, 71±6, 44±3; 100, 73±2, 60±4, 39±5. The same samples were analyzed by PTEN western blot. Monoubiquitinated PTEN bands are noted by the mono-Ub arrow (ubiquitination reactions were conducted at 30°C, with 50 nM E1, 1 μM E2, 1 μM WWP1, 5 μM PTEN, and 100 μM WT-Ub). (B) Auto- and PTEN ubiquitination assays for WWP1 wild-type, or K740N, N740S germline mutants at physiological temperature 37°C (n=3). The depletion of WWP1 was quantified and the average percentages are listed below each lane of the Colloidal Blue stained SDS-PAGE corresponding to the indicated reaction conditions. The average ± S.D. from 3 repeats are as follows (in %): 100, 38±6, 23±4, 8±1; 100, 48±2, 28±1, 16±2; 100, 43±3, 24±2, 9±1. The same samples were analyzed by PTEN Western blot. Ubiquitinated PTEN bands are noted. (C) Auto- and PTEN ubiquitination assays for WWP1 wild-type, wild type and K740N 1:1 mixture, or wild type and N740S 1:1 mixture (n=3). In the 1:1 mixture, wild type at 0.5 μM and the mutant form at 0.5 μM were added in the assays. The depletion of WWP1 was quantified and the average percentages are listed below each lane of the Colloidal Blue stained SDS-PAGE corresponding to the indicated reaction conditions. The average ± S.D. from 3 repeats are as follows (in %): 100, 67±1, 52±2, 33±2; 100, 70±1, 56±1, 38±1; 100, 72±1, 56±2, 36±3. The same samples were analyzed by PTEN Western blot. Monoubiquitinated PTEN bands are noted by the mono-Ub arrow.
To address whether the WWP1 mutants might show distinct sensitivity to allosteric activation, we performed similar ubiquitin ligase measurements in the presence of the exosite ligand UbV protein23 (Figure 3A). UbV addition to the various WWP1 forms sharply accelerated autoubiquitination and also stimulated PTEN ubiquitination (Figure 3B). The degree of catalytic enhancement was similar when comparing WT and mutant enzymes. These results demonstrate that K740N and N745S mutations are still sensitive to allosteric activation and are therefore likely autoinhibited by the 2,3-linker in a manner that resembles the WT WWP1.
Figure 3.
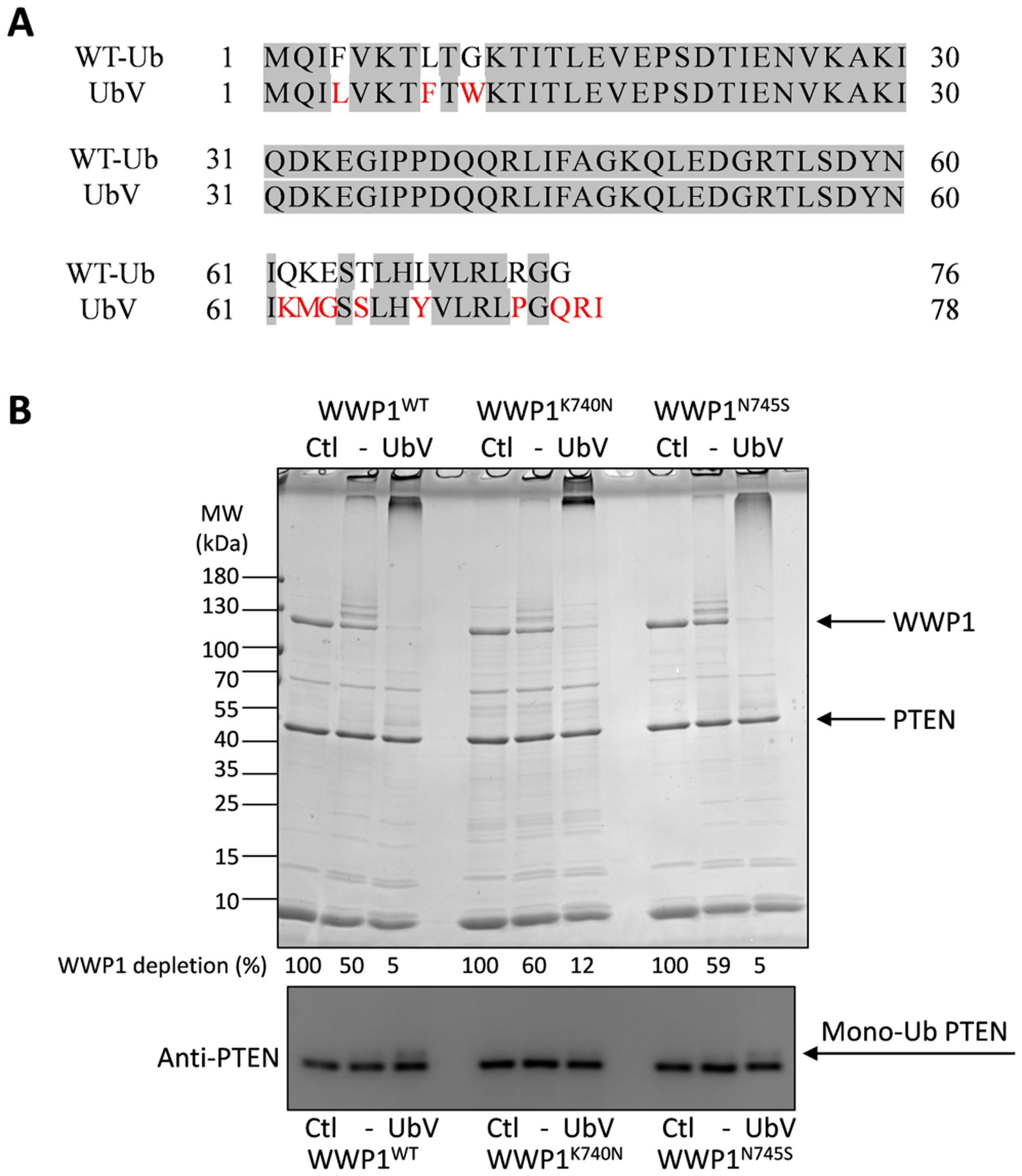
WWP1 K740N and N745S forms can be allosterically activated by UbV. (A) Protein sequence alignment of wild-type ubiquitin (WT-Ub) and allosteric activator ubiquitin variant (Ubv). The mutation includes: F4L, L8F, G10W, Q62K, K63M, E64G, T66S, H68Y, R74P, G76Q, and two extra residues R77 and I78 at the C-terminus. (B) Auto- and PTEN ubiquitination by WWP1 WT, or K740N, N745S, in response to allosteric activator UbV. The ubiquitination reactions were conducted under the same conditions as in Figure 2 for 60 min (n=3). The depletion of WWP1 was quantified and the average percentages are listed below each lane of the Colloidal Blue stained SDS-PAGE corresponding to the indicated reaction conditions. The average ± S.D. from 3 repeats are as follows (in %): 100, 50±1, 5±1; 100, 60±2, 12±1; 100, 59±4, 5±1. The same samples were analyzed by PTEN Western blot. Monoubiquitinated PTEN bands are noted by the mono-Ub arrow.
We also considered the possibility that K740N and N745S might convey a differential reactivity with the E2 enzyme that loads ubiquitin onto WWP1 or ubiquitin concentration. To test these possibilities, we performed ubiquitin ligase experiments with varying concentrations of E2 ranging from 0.1 to 5 μM (Figure 4A) or varying concentrations of ubiquitin from 10 to 300 μM (Figure 4B). As expected, the rate of WWP1 autoubiquitination and PTEN ubiquitination increased with higher concentrations of E2 and ubiquitin. However, the catalytic activities of WT, K740N, and N745S displayed similar dependencies on the E2 ubiquitin levels, revealing that the WWP1 mutations do not promote the E2 loading of the E3s or the lowering of apparent affinity of ubiquitin to the E3s.
Figure 4.
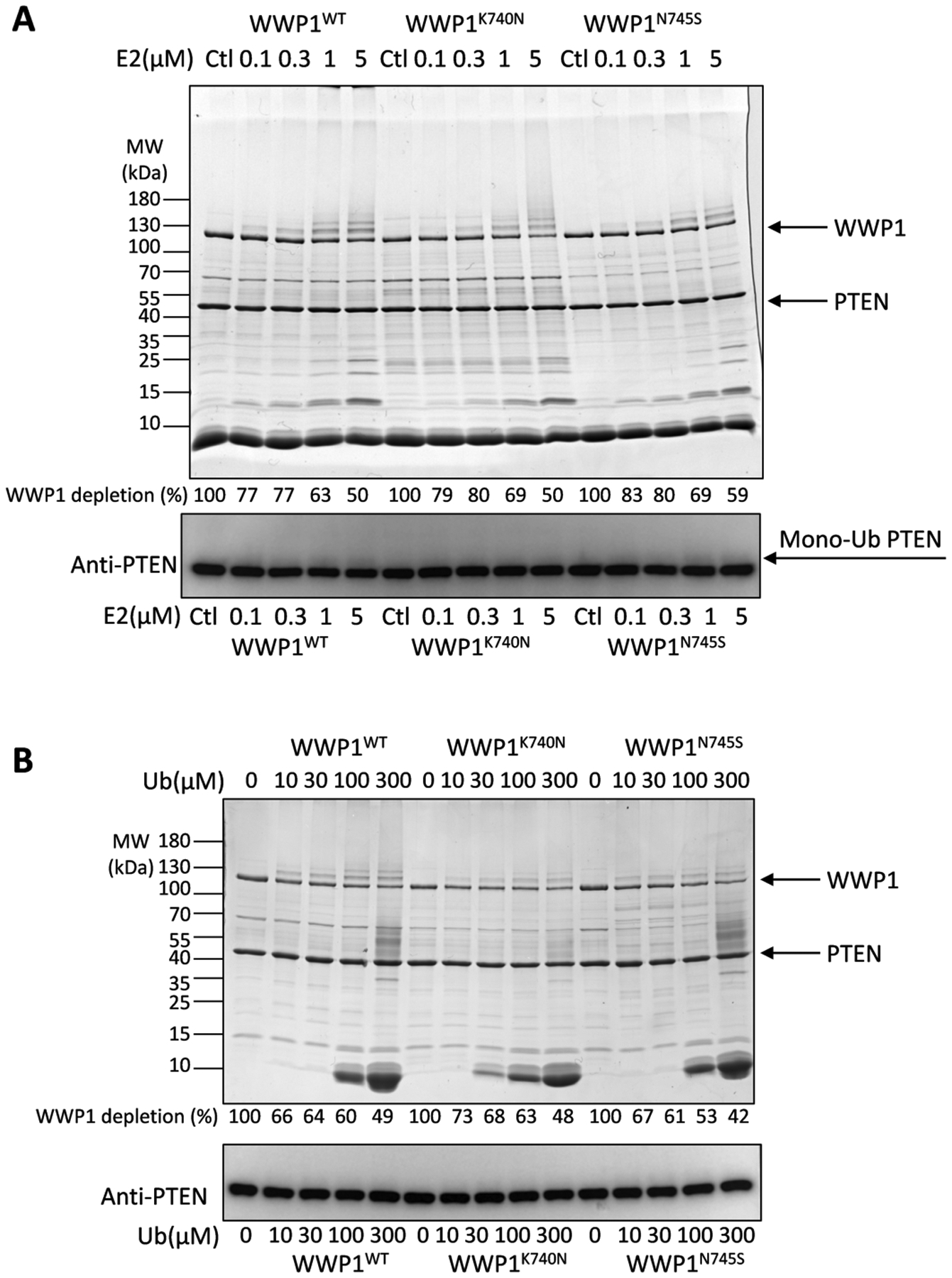
Analysis of the ubiquitin ligase activities of the WWP1 K740N and N745S forms as a function of E2 concentration or ubiquitin concentration. (A) The E2 concentrations employed were 0.1, 0.3, 1, and 5 μM. The ubiquitination reactions were conducted under the same conditions as in Figure 2 for 60 min (n=3). The depletion of WWP1 was quantified by densitometry of the Colloidal stained SDS-PAGE and the average percentages listed below the lanes corresponding to indicated reaction conditions. The average ± S.D. from 3 repeats are as follows (in %): 100, 77±1, 77±2, 63±3, 50±2; 100, 79±4, 80±3, 69±3, 50±6; 100, 83±1, 80±2, 69±1, 59±5. The same samples were analyzed by PTEN Western blot. Monoubiquitinated PTEN bands are noted by the mono-Ub arrow. (B) The ubiquitin concentrations employed were 10, 30, 100, and 300 μM. The ubiquitination reactions were conducted under the same conditions as in Figure 2 for 60 min (n=3). The depletion of WWP1 was quantified by densitometry of the Colloidal stained SDS-PAGE and the average percentages listed below the lanes corresponding to the indicated reaction conditions. The average ± S.D. from 3 repeats are as follows (in %): 100, 66±6, 64±2, 60±6, 49±6; 100, 73±1, 68±2, 63±3, 48±1; 100, 67±3, 61±2, 53±4, 42±2. The same samples were analyzed by PTEN western blot.
To assess the impact of K740N and N745S mutations on WWP1 in a cellular environment, we adapted an assay previously used to analyze the paralog WWP2’s E3 ligase actions on PTEN18. HCT116 colon cancer cells were co-transfected with the WWP1 forms along with PTEN and ubiquitin. Transfected WT WWP1 led to a modest reduction in PTEN protein as assessed by Western blot, similar to what has been observed for WWP2, previously18(Figure 5A). As observed for the in vitro ubiquitination assays, transfected K740N and N745S WWP1 showed similar behavior to WT WWP1 with regard to PTEN protein levels (Figure 5B).
Figure 5.
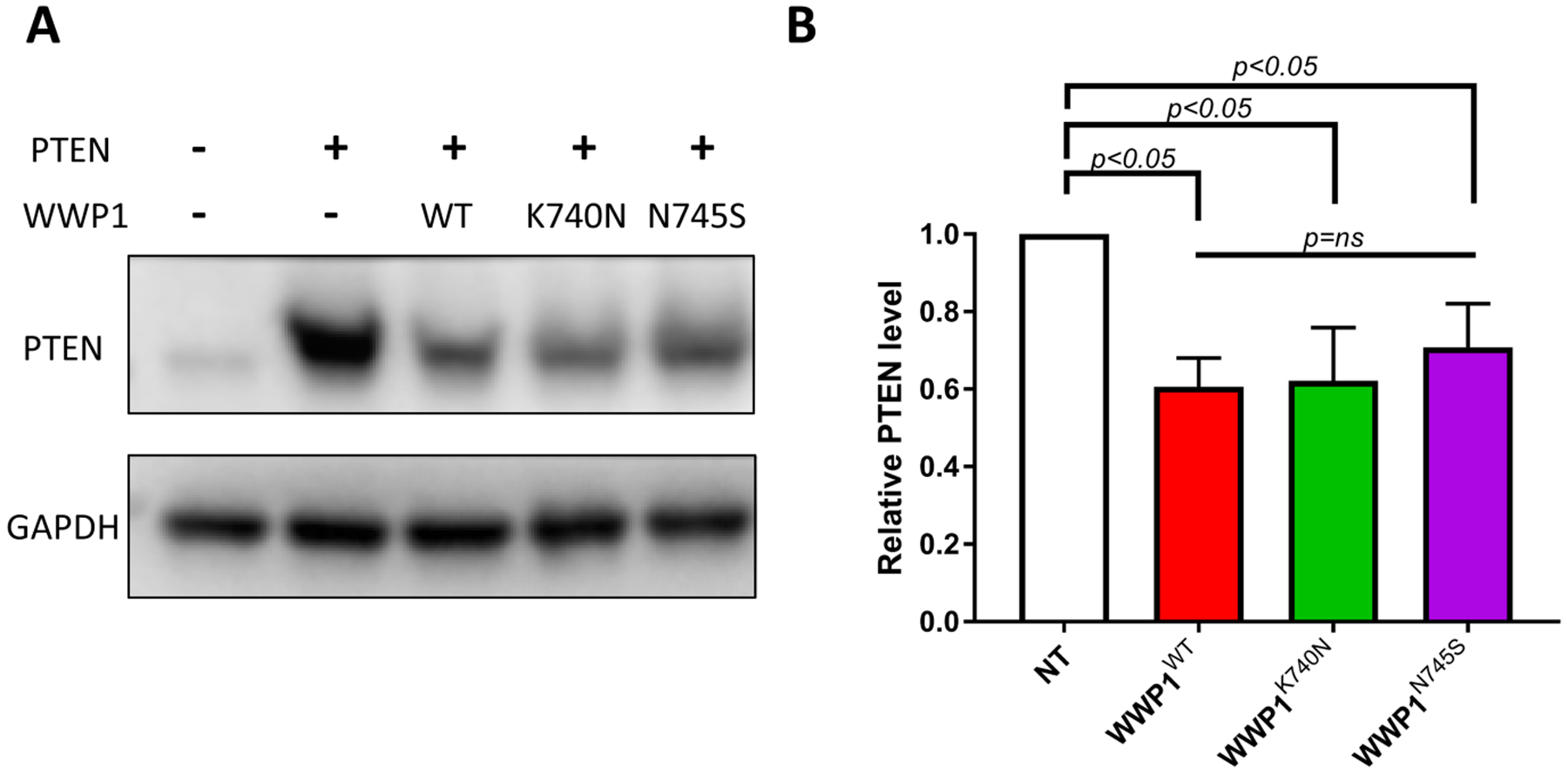
Analysis of the WWP1 mutants K740N and N745S on cellular PTEN level in cell transfection assays. (A) Western blot analysis of WWP1 wild type, K740N or N745S in HCT116 colon cancer cells. The HCT116 cells were transfected with plasmids expressing N-Myc-tagged full length WWP1: WT, K740N, and N745s, and co-transfected with PTEN (C124S) and wild type ubiquitin for 48 hr. Cells were lysed and analyzed by Western blot using anti-PTEN and anti-GAPDH antibody (n=4) (B) Quantification of PTEN expression level. PTEN and GAPDH bands were quantified by densitometric analysis using ImageJ software. The relative PTEN protein level was calculated by normalization with GAPDH as loading control (n=4, SEM shown as error bar). The statistical significance (p values) were detemined by Graphpad Prism and labeled as indicated in the graph (ns=non significant).
Discussion
A genomics analysis suggested an association between K740N and N745S WWP1 forms and cancer15. Cellular studies suggested that these WWP1 mutations could promote Akt activation which was attributed to the ubiquitination of PTEN as reported for K740N WWP115. Y.R. Lee et al. also reported that the K740N HECT domain of WWP1 interacted less with the N-terminal segment C2-WW1-WW2-WW3-WW4 in trans relative to the corresponding binding with WT HECT15. Based on these findings, the authors proposed that K740N and N745S can stimulate WWP1 catalysis by destabilizing 2,3-linker/HECT domain binding. Our data here suggest that there is no intrinsic or extrinsic capacity for enhanced ubiquitin ligase activity conferred by these mutations in WWP1. It is possible that these WWP1 mutations can have functional impacts beyond altered ubiquitin ligase activity or that WWP1 regulation is more complicated in cancer. Our transfection experiments, however, did not reveal any differences in impacts on PTEN levels conferred by the germline variants. Overall, our findings challenge the concept that K740N and N745S WWP1 drive cancer solely through enhancing PTEN ubiquitination.
Supplementary Material
Acknowledgments
This work was supported by funds from the NIH. We thank the Cole lab members for helpful advice.
Footnotes
Accession Codes
Accession codes for proteins in this study from the UniProt database are listed as follows:
WWP1: Q9H0M0-1
UbcH5b (E2): P62837-1
UBA1 (E1): P22314-1
PTEN: P60484-1
Supporting Information. Purified proteins that were used for ubiquitination assay of WWP1 (Figure S1)
References
- [1].Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner SH, Giovanella BC, Ittmann M, Tycko B, Hibshoosh H, Wigler MH, and Parsons R (1997) PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer, Science 275, 1943–1947. [DOI] [PubMed] [Google Scholar]
- [2].Maehama T, and Dixon JE (1998) The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate, J Biol Chem 273, 13375–13378. [DOI] [PubMed] [Google Scholar]
- [3].Li J, Simpson L, Takahashi M, Miliaresis C, Myers MP, Tonks N, and Parsons R (1998) The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene, Cancer Res 58, 5667–5672. [PubMed] [Google Scholar]
- [4].Sun H, Lesche R, Li DM, Liliental J, Zhang H, Gao J, Gavrilova N, Mueller B, Liu X, and Wu H (1999) PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway, Proc Natl Acad Sci U S A 96, 6199–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Z, Liu Z, Chen X, Li J, Yao W, Huang S, Gu A, Lei QY, Mao Y, and Wen W (2019) A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases, Nat Commun 10, 3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Perren A, Weng LP, Boag AH, Ziebold U, Thakore K, Dahia PL, Komminoth P, Lees JA, Mulligan LM, Mutter GL, and Eng C (1999) Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast, Am J Pathol 155, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yao YJ, Ping XL, Zhang H, Chen FF, Lee PK, Ahsan H, Chen CJ, Lee PH, Peacocke M, Santella RM, and Tsou HC (1999) PTEN/MMAC1 mutations in hepatocellular carcinomas, Oncogene 18, 3181–3185. [DOI] [PubMed] [Google Scholar]
- [8].Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, and Pandolfi PP (2007) Ubiquitination regulates PTEN nuclear import and tumor suppression, Cell 128, 141–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lee MS, Jeong MH, Lee HW, Han HJ, Ko A, Hewitt SM, Kim JH, Chun KH, Chung JY, Lee C, Cho H, and Song J (2015) PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis, Nat Commun 6, 7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, and Jiang X (2007) NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN,Cell 128, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Maddika S, Kavela S, Rani N, Palicharla VR, Pokorny JL, Sarkaria JN, and Chen J (2011) WWP2 is an E3 ubiquitin ligase for PTEN, Nat Cell Biol 13, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lee Y-R, Chen M, Lee JD, Zhang J, Lin S-Y, Fu T-M, Chen H, Ishikawa T, Chiang S-Y, Katon J, Zhang Y, Shulga YV, Bester AC, Fung J, Monteleone E, Wan L, Shen C, Hsu C-H, Papa A, Clohessy JG, Teruya-Feldstein J, Jain S, Wu H, Matesic L, Chen R-H, Wei W, and Pandolfi PP (2019) Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway, Science 364, eaau0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen Z, Thomas SN, Bolduc DM, Jiang X, Zhang X, Wolberger C, and Cole PA (2016) Enzymatic Analysis of PTEN Ubiquitylation by WWP2 and NEDD4-1 E3 Ligases, Biochemistry 55, 3658–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ingham RJ, Gish G, and Pawson T (2004) The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture, Oncogene 23, 1972–1984. [DOI] [PubMed] [Google Scholar]
- [15].Lee YR, Yehia L, Kishikawa T, Ni Y, Leach B, Zhang J, Panch N, Liu J, Wei W, Eng C, and Pandolfi PP (2020) WWP1 Gain-of-Function Inactivation of PTEN in Cancer Predisposition, N Engl J Med 382, 2103–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jiang H, Thomas SN, Chen Z, Chiang CY, and Cole PA (2019) Comparative analysis of the catalytic regulation of NEDD4-1 and WWP2 ubiquitin ligases, J Biol Chem 294, 17421–17436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhu K, Shan Z, Chen X, Cai Y, Cui L, Yao W, Wang Z, Shi P, Tian C, Lou J, Xie Y, and Wen W (2017) Allosteric auto-inhibition and activation of the Nedd4 family E3 ligase Itch, EMBO Rep 18, 1618–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, Devreotes P, Wolberger C, Amzel LM, Gabelli SB, and Cole PA (2017) A Tunable Brake for HECT Ubiquitin Ligases, Mol Cell 66, 345–357 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Grimsey NJ, Narala R, Rada CC, Mehta S, Stephens BS, Kufareva I, Lapek J, Gonzalez DJ, Handel TM, Zhang J, and Trejo J (2018) A Tyrosine Switch on NEDD4-2 E3 Ligase Transmits GPCR Inflammatory Signaling, Cell Rep 24, 3312–3323 e3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Riling C, Kamadurai H, Kumar S, O’Leary CE, Wu KP, Manion EE, Ying M, Schulman BA, and Oliver PM (2015) Itch WW Domains Inhibit Its E3 Ubiquitin Ligase Activity by Blocking E2-E3 Ligase Trans-thiolation, J Biol Chem 290, 23875–23887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim HC, Steffen AM, Oldham ML, Chen J, and Huibregtse JM (2011) Structure and function of a HECT domain ubiquitin-binding site, EMBO Rep 12, 334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Maspero E, Mari S, Valentini E, Musacchio A, Fish A, Pasqualato S, and Polo S (2011) Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation, EMBO Rep 12, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang W, Wu KP, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, Mercredi PY, Murchie R, Hu J, Persaud A, Mukherjee M, Li N, Doye A, Walker JR, Sheng Y, Hao Z, Li Y, Brown KR, Lemichez E, Chen J, Tong Y, Harper JW, Moffat J, Rotin D, Schulman BA, and Sidhu SS (2016) System-Wide Modulation of HECT E3 Ligases with Selective Ubiquitin Variant Probes, Mol Cell 62, 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, Nagata Y, Yoshida K, Kon A, Suzuki Y, Chiba K, Tanaka H, Niida A, Fujimoto A, Tsunoda T, Morikawa T, Maeda D, Kume H, Sugano S, Fukayama M, Aburatani H, Sanada M, Miyano S, Homma Y, and Ogawa S (2013) Integrated molecular analysis of clear-cell renal cell carcinoma, Nat Genet 45, 860–867. [DOI] [PubMed] [Google Scholar]
- [25].Wiener R, DiBello AT, Lombardi PM, Guzzo CM, Zhang X, Matunis MJ, and Wolberger C (2013) E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1, Nat Struct Mol Biol 20, 1033–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bolduc D, Rahdar M, Tu-Sekine B, Sivakumaren SC, Raben D, Amzel LM, Devreotes P, Gabelli SB, and Cole P (2013) Phosphorylation-mediated PTEN conformational closure and deactivation revealed with protein semisynthesis, Elife 2, e00691. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


