Abstract
In order to increase the hit rate of discovering diverse, beneficial protein variants via high-throughput screening, we have developed a computational method to optimize combinatorial mutagenesis libraries for overall enrichment in two distinct properties of interest. Given scoring functions for evaluating individual variants, POCoM (Pareto Optimal Combinatorial Mutagenesis) scores entire libraries in terms of averages over their constituent members, and designs optimal libraries as sets of mutations whose combinations make the best trade-offs between average scores. This represents the first general-purpose method to directly design combinatorial libraries for multiple objectives characterizing their constituent members. Despite being rigorous in mapping out the Pareto frontier, it is also very fast even for very large libraries (e.g., designing 30 mutation, billion-member libraries in only hours). We here instantiate POCoM with scores based on a target’s protein structure and its homologs’ sequences, enabling the design of libraries containing variants balancing these two important yet quite different types of information. We demonstrate POCoM’s generality and power in case study applications to green fluorescent protein, cytochrome P450, and β-lactamase. Analysis of the POCoM library designs provides insights into the trade-offs between structure- and sequence-based scores, as well as the impacts of experimental constraints on library designs. POCoM libraries incorporate mutations that have previously been found favorable experimentally, while diversifying the contexts in which these mutations are situated and maintaining overall variant quality.
Keywords: Combinatorial mutagenesis, protein library design, sequence-based protein design, structure-based protein design, Pareto optimization, high-throughput screening
1. Introduction
COMBINATORIAL mutagenesis enables facile construction and characterization of a targeted set of related protein variants that incorporate at each of a set of residue positions one of a position-specific set of amino acids (Fig. 1(a)). All combinations of the alternative amino acids are represented in a library, and experimental selection or screening methods are employed to identify variants with beneficial properties. In contrast to other methods for generating mutational diversity for experimental discovery, such as error-prone PCR [1] and site-saturation mutagenesis [2], combinatorial mutagenesis allows finer control over the variants being experimentally tested and thus the opportunity to better focus effort on variants likely to be favorable.
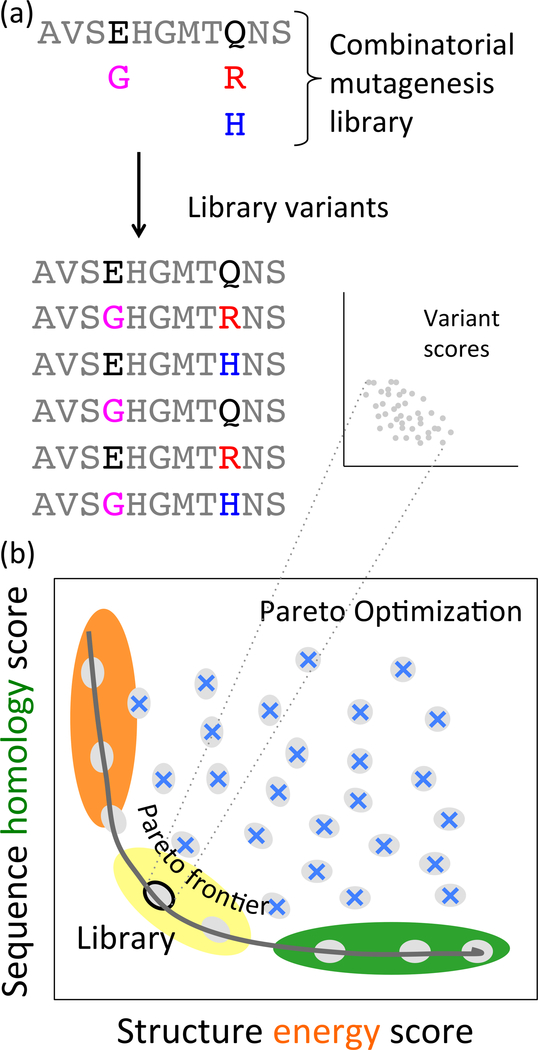
Fig. 1:
Combinatorial mutagenesis libraries, (a) A tiny example combinatorial mutagenesis library, with two possible amino acids at one position and three possible amino acids at another position, yielding six variants representing all combinations of these choices, (b) POCoM optimizes libraries to balance two objectives, here sequence homology and structure energy, each to be minimized. It evaluates a library in terms of the average score of its variants. Pareto optimal designs, comprising the Pareto frontier (on the black curve), make the best trade-offs between the two objectives, while dominated designs (blue Xes) are worse on both.
Computational methods have guided the development of combinatorial mutagenesis libraries yielding improved proteins for a variety of goals, including altered specificity, increased thermostability, improved spectroscopic properties, reduced immunogenicity, and improved catalytic activity [3]-[8]. Different metrics have been used for computationally assessing the quality of a library and thereby establishing a design objective. Sequence-based library design methods [9], [10], [11] leverage information from the evolutionary record regarding which mutations and combinations of mutations have been accepted. For example, our OCoM algorithm [9] optimizes libraries based on a one- and two-body statistical sequence potential derived from a multiple sequence alignment (MSA) of the target protein [12]. Significantly, while OCoM evaluates a library in terms of the average sequence potential of its constituent variants, it is able to do that efficiently, without having to explicitly enumerate the variants. Structure-based library design methods [6], [13], [14], [15] evaluate biophysical properties of library members, e.g., assessing potential impacts of mutations on stability. While some structure-based methods require explicit evaluation of each library member and can therefore only handle small libraries [6], [16], our SOCoM method [14] and the independently developed SHARPEN method [15] overcome that limitation by employing a Cluster Expansion (CE) technique [17], [18] to convert structure-based biophysical evaluations into protein-specific functions of amino acid sequence alone. That essentially reduces structure-based library design to sequence-based library design, rendering it amenable to much more efficient evaluation and design of libraries in terms of average scores over their constituents. We leverage here these key insights from both OCoM and SOCoM to enable large-scale but fast library optimization.
In order to utilize the complementary valuable information provided by sequence- and structure-based scores, we have developed a library design method called POCoM (Pareto Optimal Combinatorial Mutagenesis) that integrates two objectives, optimizing designs for the best tradeoffs between them (Fig. 1(b)). POCoM is thus the first general-purpose design method to directly optimize, in “library space”, combinatorial mutagenesis libraries for multiple objectives characterizing the constituent variants that they will generate. While POCoM is generic to the scoring functions, we focus here on structure-based assessment of energy, ensuring overall stability of variants, combined with sequence-based assessment of evolutionary constraints, accounting for additional properties (maintaining functional sites, folding pathways, etc.). A Pareto optimal design is undominated, in that no other design is as good for one objective without being worse for the other objective; such designs provide the best balance between objectives and are thus a natural set to consider for experimental evaluation. Pareto optimization techniques have been developed for and proved valuable in other protein engineering contexts [3], [15], [19]–[22], and POCoM demonstrates, for the first time, how to leverage this principle in library design for an arbitrary pair of sequence- and structure-based scoring functions. It is important to note that, as discussed above, POCoM is efficient in its evaluation of a library, using averages to avoid enumeration of individual members. Furthermore, POCoM is efficient in its optimization of designs, identifying all and only the Pareto optimal ones without explicitly considering all those that are dominated by them. By defining the Pareto frontier of undominated designs, POCoM provides an unbiased characterization of the two-objective design space.
We demonstrate the generality, power, and efficiency of POCoM in case study applications to green fluorescent protein (GFP), cytochrome P450, and β-lactamase. Libraries were designed for a variety of experimental contexts: allowing mutations throughout a protein or focused in an active site region, planning for library construction using specified mutations or degenerate oligonucleotides, and allowing from 10 to 20 mutation sites and correspondingly different library sizes. These case studies demonstrate how POCoM libraries incorporate mutations balancing evolutionary favorability with energetic favorability. The designs include a number of mutations previously experimentally determined to be beneficial, along with diverse new combinations manifesting different balances of sequence and structural scores. POCoM provides a clear characterization of designs to consider for experimental evaluation, along with insights into how best to target a study according to its particular goals and constraints.
2. Methods
Fig. 2 overviews the main steps of POCoM. Each panel in the figure corresponds to one of the following subsections detailing the computational steps.
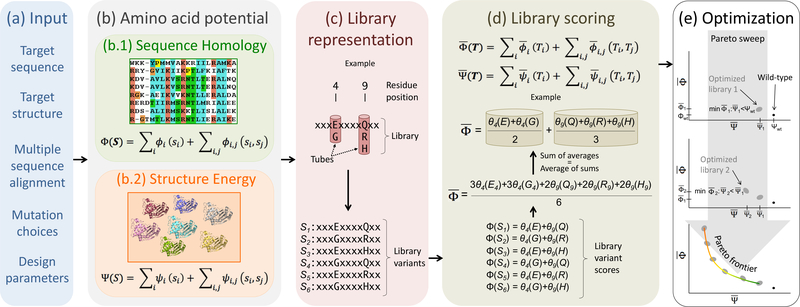
Fig. 2:
POCoM overview, (a) Input data are collected and preprocessed, (b) Amino acid potentials are extracted from the target’s sequence and structure, (c) Possible sets of amino acids are determined at each position, thereby defining choices representing a library, (d) Scores for each position-specific amino acid set, or “tube”, are averaged in order to assign contributions to the sequence homology and structure energy library scores, (e) Pareto optimal libraries are optimized with a sweeping technique that repeatedly finds the next undominated design as that with the best while constrained to have reduced .
2.1. Input (step a)
We denote the sequence of the target as S = {s1, s2, …, sn}, where si, represents the amino acid at position i and N is the total number of amino acids. Depending on usage, in addition to S, POCoM also requires the structure of the target and an MSA of related proteins (Fig. 2(a)). Mutation choices to consider at each position can be prespecified, or can be computed directly from these inputs, as is common and we have detailed elsewhere [23], e.g., amino acids with sufficient frequency at that position in the MSA or suitable in the local environment at that position in the structure. Details are provided in the Supplementary Material for the case studies in this paper. Additional parameters control the design process, in terms of the number of sites to mutate, the maximum number of mutations at each site, the total library size, and whether the library will be constructed with point mutations or degenerate oligos.
2.2. Amino Acid Potential (step b)
POCoM optimizes libraries for two complementary scores (Fig. 2(b). It is generic to the details of the scoring functions being used, but we focus here on optimizing a score based on evolutionary information (“sequence homology” score) along with one based on molecular modeling (“structure energy” score). We further restrict the elaboration here to pairwise-decomposable scores (i.e., one- and two-body terms), as commonly used in practice and sufficient to capture the key contributions needed for library design.
We employ as the sequence homology score Φ (Fig. 2(b.1)) a straightforward model also used in our earlier work [9], [24] and similar to many other sequence potentials. These potentials have been used in a wide range of practically useful applications; e.g., we (among others) have engineered stable, active enzyme variants based on optimizing this potential [25], [26], and others have used similar potentials in additional applications such as predicting residue contacts [27]–[29], predicting 3D structures of proteins and protein complexes [30], [31], and assessing allosteric mechanisms [32]. Given an MSA, the one-body terms ϕi(si) capture conservation for individual positions (i.e., how common amino acid si is at position i), while the two-body terms ϕij(si, sj) capture correlated/compensating mutations for pairs of positions (i.e., how common it is for a sequence to have both amino acid si at position i while also having sj at j). Which terms to include in the potential and their position and amino acid specific contributions are derived from a statistical analysis of the MSA, computing log-frequencies so as to yield an additive model, and keeping only those that are statistically significant. We note that higher-order terms are possible [24], [33], and the method here generalizes to incorporate them, but in practice a pairwise deomposition often suffices.
We derive the structure energy score Ψ (Fig. 2(b.2)) from the target protein’s structure by employing a Cluster Expansion (CE) technique [18] to accurately represent structural properties (here energy, as a surrogate for stability) as a protein-specific function of amino acid sequence. CE derives one-body ψi(si) and two-body ψi,j(si, sj) potentials from a large training set of <sequence, energy> pairs, determining which possible “clusters” (positions i for terms ψi and position pairs i,j for terms ψi,j) to include in the model. We employ Rosetta [34] to predict structures and calculate energies for a large set of randomly sampled sequences in the training set. Values for the potential terms are then optimized to construct a protein-specific model as previously described [35], enabling rapid and accurate prediction of the structure energy of any variant of the target based on only its sequence. The predictive quality of the trained potential model is assessed with a unique set of test sequences, typically reaching ~80% in our studies here, and previously demonstrated to be suitable for experimentally successful design of individual proteins [17], [18], [36] and SOCoM-based libraries [3].
2.3. Library Representation with Tubes (step c)
A combinatorial mutagenesis library is constructed from all combinations of specified amino acids (including wild-type) at specified mutation sites (Fig. 2(c)). We refer to an amino acid set Ti at position i as a “tube” [9], [14]. For the example in Fig. 2(c), with two amino acids possible at position 4 (T4 ={E4, G4}) and three at position 9 (T9 ={Q9, R9, H9}), there are six possible variants derived from the cross product of these two tubes: T4×T9 = {{E4,Q9}, {E4,R9}, {E4,H9}, {G4,Q9}, {G4,R9}, {G4,H9}}.
In library design, each position has one or more different tubes to choose from; selecting exactly one tube at each site specifies a library design. The tube choices at a position are precomputed from the combinations of allowed amino acid choices at the position (an input from (a)). Only tube choices including the wild-type amino acid are considered (so the wild-type is represented in the library); a position with no additional amino acid choices is assigned a single tube choice containing just the wild-type. Different library construction contexts give rise to different possibilities for tube choices. If the library is to be constructed by an arbitrary set of point mutations at each position, then the tube choices include all possible combinations of allowed amino acids up to a user-specified size of the amino acid set. If, as is common to save expense, the library is to be constructed using degenerate oligonucleotides (i.e., a mixture of nucleotides at each of the three positions in a codon), then tube choices are determined from degenerate oligos that encode combinations of allowed amino acids. Since nucleotide mixtures in a degenerate oligo may translate to extra amino acids beyond those desired, tubes are filtered to those with at most a user-specified amount of extras. We find that in practice, due to the typically small number of allowed amino acids at each position, along with constraints on overall library size, the potential combinatorial explosion of tube choices remains under control.
2.4. Library Scoring (step d)
In order to optimize a library, we need an efficient way to evaluate its overall quality for the two objectives. Each variant can be assessed by the given amino acid potentials. However, a library includes a combinatorial set of variants, so explicit scoring of each would be extremely inefficient for evaluating, much less optimizing, large libraries. Instead, we take as our objective the average quality over the library. This allows us to “lift” scoring functions for individual variants to average scoring functions for combinatorial libraries, as illustrated in Fig. 2(d). The key is that each amino acid in a tube occurs the same number of times in the library. Thus the contribution from a tube at a position to the average library score can be precomputed as a tube-averaged score, and similarly for pairs of positions and pairs of tubes [9], [14], [24], [37]. Then the library average score is simply the sum of these tube and tube-pair average scores. We note that this technique works for the objective of optimizing average library quality, assuming that if the average is good enough, experimental screening/selection will identify good candidates. It remains future work to optimize libraries for other overall metrics (e.g., score of a top percentile).
2.5. Library Optimization (step e)
Protein library design is an NP-hard problem [38], [9]; it follows that Pareto optimal design is also NP-hard, with the additional practical difficulty of needing to identify just the undominated designs. POCoM is based on an integer linear programming technique that has proven effective in practice for solving the core protein design problem in a number of related protein engineering contexts [9], [14], [21], [39]–[41].
Let us start by considering just one of the scores, say the library-average sequence homology score . Let xi,t, be a single-position binary variable indicating whether or not tube t is present at position i. Let yi,j,t,u be a pairwise binary variable derived from the corresponding single-position binary variables, indicating whether or not both tube t is at i and u is at j. Then the overall library score is simply the sum of these binary variables, weighted by the corresponding coefficients and .
| (1) |
A similar formula gives in terms of these same variables and the tube-averaged coefficients and .
The variable values are constrained to allow exactly one tube at a position (Equation 2) and to enforce consistency between corresponding single and pairwise variables (Equations 3 and 4). Furthermore, the number of mutated positions and total library size are constrained to user-specified ranges. Equation 5 ensures that between μ and M positions have a selected tube that is not just the wild-type residue, whereas Equation 6 ensures that the product of the number of amino acids in the selected tubes (expressed as the sum of the logs, so as to be linear) is between λ and Λ. An additional constraint, not shown due to lack of a simple expression, ensures that for each previous solution, at least one tube in the next solution is different.
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
In order to move from optimizing a single objective to mapping the Pareto frontier (all and only the undominated designs; see again Fig. 1), we follow a “sweep” approach inspired by our previous work on the design of deimmunized enzymes [3], [21], [39], [40] and protein-protein interactions [17]. This optimization technique produces the Pareto frontier in the two-objective design space, without explicitly considering the dominated designs. As illustrated in Fig. 2(e), the algorithm starts from the homology optimal region (lower right) and traverses towards the energy optimal region (upper left). At each step, it optimizes only the sequence homology score, as described in the above integer linear program, but at each step it imposes successively tighter constraints to force better successively better structure energy scores. It first identifies the optimal design for the sequence homology score. It then constrains the structure energy score to be better than that of the first design, and reoptimizes, finding a design with a somewhat worse sequence homology score (but best of all those with better structure energy scores, so undominated). Note that in order to implement a “better than” (<) constraint using the “at least as good as” (<=) constraint provided by the integer program solver, we use “at least as good as” epsilon better, where the epsilon is a small value precomputed from the library size to ensure that no solution is missed. The process is repeated to obtain additional designs with iteratively better structure energy scores but iteratively worse sequence homology scores. Thus the full Pareto frontier is generated, invoking the optimizer once per design.
POCoM is implemented in Python, specifying and solving the integer linear program by invoking CPLEX via the Python API. The source code is freely available for academic use by contacting the authors.
3. Results and discussion
We employed POCoM to optimize combinatorial mutagenesis libraries for three different proteins for different experimental contexts (point mutations vs. degenerate oligos, considering mutations anywhere vs. only near active sites, 10 vs. 15 vs. 20 mutational sites). After preprocessing and construction of scoring potentials, detailed in the supplementary material, POCoM was able to generate each set of Pareto optimal library designs in less than an hour on an off-the-shelf 2012 Macbook Pro. By way of calibration, optimizing a single design for just one of the objectives took under a minute; this relationship follows expectations because there may be hundreds of two-objective designs per Pareto curve. To push the limits, we also tried designing a 30-site GFP library and found that even that massive library design task required less than 6 hours on that same machine.
3.1. Green fluorescent protein
GFP and its derivatives provide powerful tools for biological research, with their unique fluorescent properties enabling researchers to track, visualize, and map subcellular structures and interactions [42]–[48]. The fluorescent protein field is constantly seeking additional proteins with different properties, and GFP is ideally suited for high-throughput characterization of large libraries that can produce such candidates. Here we design libraries for GFP from Aequorea victoria (UniProt ID GFP_AEQVI and PDB ID 1GFL), the same protein used in previous structure-only library studies [14], [6] seeking to modulate fluorescent properties by allowing mutations anywhere including the core of the protein. POCoM builds upon that work by complementing structural information with sequence homology information and thereby enabling libraries to balance these criteria. Moreover, the resulting GFP libraries are much larger and more diverse than those produced by other library design methods [6].
POCoM elucidates sequence-structure design tradeoffs
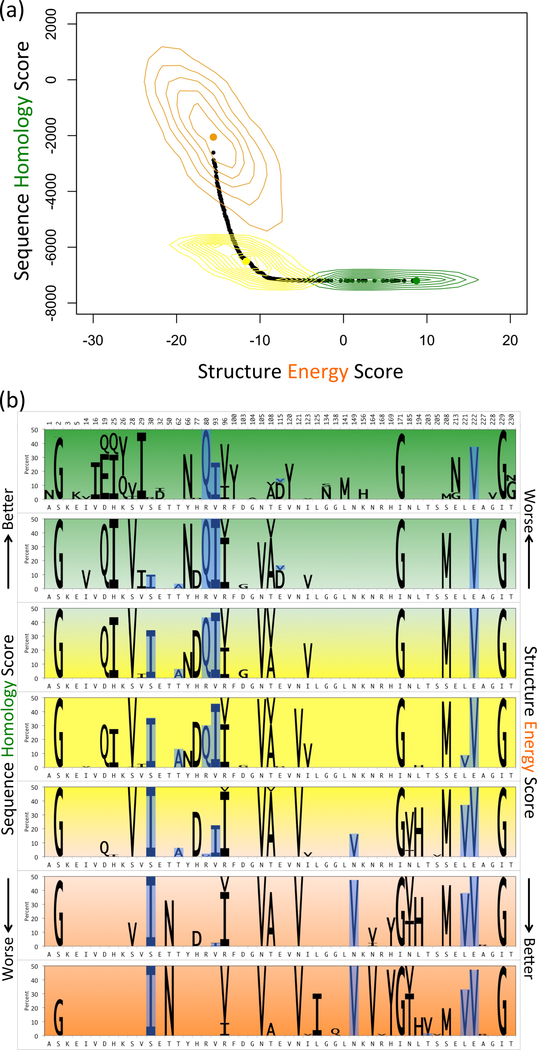
We first designed 15-site libraries for construction by point mutation. The shape of the resulting Pareto frontier (Fig. 3(a)) highlights the complementarity between the sequence homology and structure energy scores. Plans toward the lower-right corner are optimized primarily for sequence homology, whereas plans toward the upper-left corner are more structurally optimized. Moving from the lower-right incurs only a small penalty to the sequence homology score for a substantial improvement in structure energy, suggesting that there are many possibilities for variants to achieve the same degree of sequence acceptability with substantial stability implications. This also suggests that optimizing for the homology dimension alone could result in libraries with poor energies, illustrated by the distribution of enumerated variants in the homology score optimized library (green contours in Fig. 3(a)). Similarly, when optimizing a library for energy score alone, the variant distribution suggests that a significant portion of the library is wasted in unproductive portions according to the homology score (orange contours in Fig. 3(a)). But POCoM also designs libraries with more balanced criteria, as illustrated with the “elbow” library plan (i.e., that closest to the origin when scores are normalized to 0–1). Variants enumerated from the elbow plan (yellow contours in Fig. 3(a)) manifest a strong anti-correlation between the two objectives, suggesting that here the sequence record contains information complementary to energy minimization in producing folded and functional proteins.
Fig. 3:
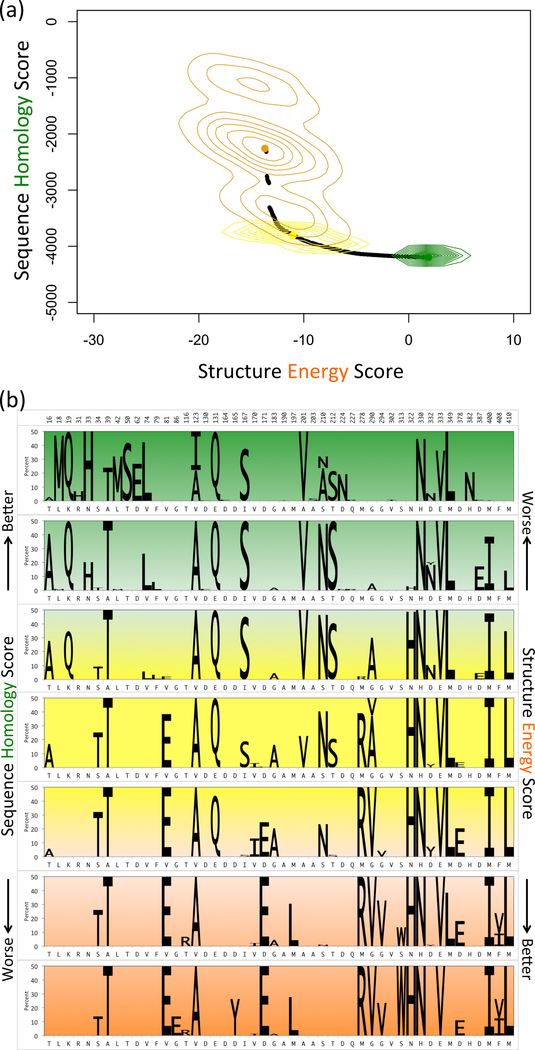
(a) Pareto frontier of 15-site point mutation GFP libraries. Selected library plans in sequence-optimized (green), structure-optimized (orange) and sequence-structure-optimized (yellow) regions are enumerated (random 1000 variants in each) and displayed as contours, (b) Amino acid composition of Pareto optimal 15-site point mutation GFP libraries. The seven panels represent different regions of the Pareto frontier, green for sequence-optimized, orange for structure-optimized and yellow for sequence-structure-optimized. The height of sequence logos corresponds to the abundance of mutations in the plans (wild-type not shown). Experimentally observed mutations are highlighted in blue.
Differences in plan composition reflect sequence-structure implications of mutational combinations
For exploration of plan details, the Pareto frontier was divided into seven intervals and mutation frequencies computed separately for the plans in each interval (Fig. 3(b)). While some mutations (e.g., S208M) contribute to plans in all intervals, others are more common in the sequence homology intervals (e.g., H25I) or structure energy intervals (e.g., N121V), and still others are predominant in intervals around the balanced elbow (e.g., T62A). Some positions employ different choices in different portions of the curve, for example, V or I at position 96. These differences in library composition elucidate the power of POCoM to uncover a diverse set of alternative designs for consideration.
A number of mutations – S30I, T62A, R80Q, V93I, L221V, and E222V – have been experimentally found in GFP variants, accounting for their improved or different color fluorescent properties [6], [49]–[53]. Strikingly (blue boxes in Fig. 3(b)), these mutations tend to be in designs near the balanced elbow plan, though some permeate all plans and others also span one side or the other of the sequence-structure trade-off.
POCoM explicates impacts of experimental constraints
We varied two input parameters, the number of mutational sites to employ (10, 15, or 20) and how the library is to be constructed (point mutations or degenerate oligos); for the sake of comparison purposes the tube size was limited to 2 (wild-type plus one alternative). Fig. S1 illustrates that, as would be expected, relaxed constraints (point mutations, larger library) yield better-scoring libraries. Tubes in degenerate oligo libraries are constrained by the genetic code, and thus can’t readily pair disparate amino acid types that might have better scores. Likewise, a higher mutational load enables libraries to use more conservative mutations to achieve the same energy score. While not required by the optimization approach, we found that, regardless of construction method, the positions targeted in 10-mutation site libraries are a subset of those targeted in 15-mutation site libraries which are in turn a subset of positions targeted in 20-mutation site libraries. This suggests that POCoM targets more productive sites at lower mutational loads and augments them with additional beneficial ones at higher mutational loads. There are differences at the level of amino acid selections, though; e.g., T62A occurs only in the 15- and 20-site libraries.
Potential contributions govern amino acid selections
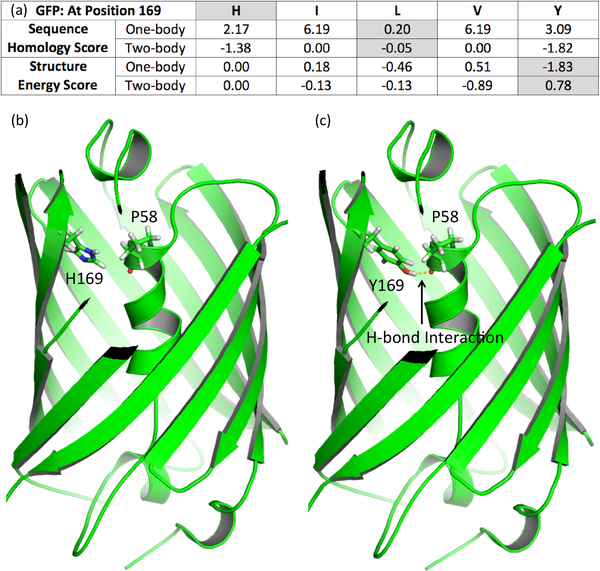
Each mutation in a specific library plan on the Pareto curve can be characterized by energy and homology score contributions. We selected the structurally most optimized plan, with mutations S30IS, T49NT, R96VR, N105VN, N121NV, L125IL, N149VN, N164VN, R168VR, H169YH, I171GI, N185TN, T203VT, L221VL and I229GI, and compared its side-chain placements with those in the original structure. Fig. 4 illustrates the impact of POCoM’s optimization at position 169, whose side chain interacts with the chromophore core region, a location that can affect GFP’s spectroscopic properties. The figure also illustrates the contributions to homology and energy scores for each of the allowed mutations from the MSA. Homology scores suggest that L is highly conserved (lower scores are better) and should be favored for library construction. However, energetically more favorable Y is chosen over L for this plan because it can form a strong hydrogen bond interaction with P58 in the GFP core, which is also indicated by the best averaged two-body homology score. Such two-body interactions are reflective of coevolution, which in addition to physical proximity [28] in a structure may also capture indirect relationships, e.g., for allosteric propagation [54]. The example presented here clearly demonstrates a strong interplay between sequence homology and structure energy, suggesting that both potentials may be important for developing favorable variants.
Fig. 4:
(a) Comparison of sequence homology and structure energy scores of available choices at position 169 that lead to the selection of tyrosine in GFP. Lower scores correspond to more favorable contributions. Comparison of (b) wild-type GFP and (c) structure energy optimized 15-point mutation plan variant illustrating formation of a favorable hydrogen bond with P58 upon H169Y mutation.
3.2. Cytochrome P450
Cytochrome P450 enzymes play major roles in the synthesis and breakdown of molecules in cells. Some P450s synthesize molecules including fatty acids (including cholesterol) [55], vitamins, and steroid hormones, while others are involved in the breakdown of external (medication) and internal (cellular toxins) substances [56]. Reengineered variants of P450 are widely used in the biotechnology industry, medicine, and bioremediation [57]. Constructing diverse P450 libraries can yield variants with new properties facilitating synthesis and breakdown of additional molecules.
We designed degenerate oligo libraries of Bacillus subtilis cytochrome P450 (UniProt ID CYPC_BACSU and PDB ID 2ZQJ), which catalyzes hydroxylation of long-alkyl-chain fatty acids. Previous library studies for this P450 have focused on sequence information [58], whereas POCoM integrates sequence and structure to construct diverse libraries allowing mutations anywhere on the protein that have the potential to modify its catalytic and thermostability properties.
POCoM elucidates sequence-structure design tradeoffs and impacts of experimental constraints
The Pareto frontier of 15-site P450 libraries (Fig. 5(a)) is less curved than for 15-site GFP libraries (Fig. 3(a)), suggesting a stronger sequence homology and structure energy relationship. The trends in variant distribution remain the same, however, with the homology-optimized library (green contours) tightly distributed, the energy-optimized library (orange contours) exploring a large region of non-conserved mutations, and the elbow library (yellow contours) maintaining a balance of both objectives.
Fig. 5:
(a) Pareto frontier of 15-site degenerate oligo P450 libraries. Selected library plans in sequence-optimized (green), structure-optimized (orange) and sequence-structure-optimized (yellow) regions are enumerated (random 1000 variants in each) and displayed as contours, (b) Amino acid composition of Pareto optimal 15-site degenerate oligo P450 libraries. The seven panels represent different regions of the Pareto frontier, green for sequence-optimized, orange for structure-optimized and yellow for sequence-structure-optimized. The height of sequence logos corresponds to the abundance of mutations in the plans (wild-type not shown).
We constructed additional P450 libraries targeting 10 or 20 sites, and also explored the differences between point mutation and degenerate oligo library construction methods. As illustrated in Fig. S2, the curves for P450 are less sparse and more distinct than those for GFP (Fig. S1). The larger size of P450 provides relatively more positions for mutations, leading to more possible plans with slightly different scores. As observed for GFP, higher mutational loads can explore a larger design space and thus achieve better scores, and degenerate oligos are restricted by the genetic code in possible mutation combinations and thus obtain worse scores. Also as observed for GFP, smaller libraries select highly productive sites that remain in use and are augmented in larger libraries.
Differences in plan composition reflect sequence-structure implications of mutational combinations
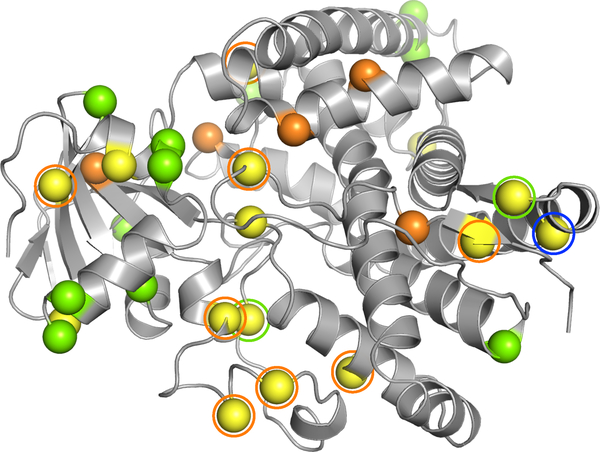
Fig. 5(b) illustrates the 15-mutation Pareto optimal degenerate oligo plans, binned into intervals as for GFP. Out of 48 targeted positions, 43 positions have only one alternative amino acid type and the remaining 5 having two alternatives. 44 mutated positions are on the surface, including 25 hydrophilic and 19 hydrophobic. Interestingly, none of the buried residues were targeted in the homology biased library plans, whereas 4 core hydrophobic residues were targeted for mutation in the structure energy biased library plans. Considering the mutations of all plans in the elbow region, we see that they are enriched with mutations from homology- and energy-biased potentials as well as the potentials balanced on both the criteria: out of 22 targeted sites, 7 mutations are homology-biased, 8 are energy-biased, and the remaining 7 mutations are balanced between the criteria. Fig. 6 further elaborates this trend, highlighting mutations selected for the sequence-only optimized plan vs. the structure-only optimized plan vs. the elbow plan. Only position 123 is shared among all three plans, while two positions are shared between sequence-only and elbow, and eight between structure-only and elbow, with the elbow plan having four unique sites. Pareto optimization thus includes many important mutations that would be missed by sequence-only or structure-only design, either because they derive from the other scoring function, or because they balance and complement both.
Fig. 6:
Comparison of sites targeted by sequence-only optimization (green spheres), structure-only optimization (orange spheres) and sequence-structure optimization (yellow spheres) for P450 15-site degenerate oligo library plans. Balanced sequence-structure optimization (elbow plan) sites that overlap with sequence-only and structure-only sites are shown by green and orange circles, respectively. Position 123, targeted by all three plans, is shown by a yellow sphere with a blue circle.
3.3. β-lactamase
The β-lactamase family of enzymes provide bacteria with antibiotic resistance by hydrolyzing the β-lactam ring of penicillin-like drugs [59]. The inexpensive screening of β-lactamase coupled with its diversity in extended spectrum recognition [60] has allowed researchers to characterize functional and stability contributions of active site residues [61], and provided insights regarding the hydrolysis activity of β-lactam derivatives. Constructing diverse active site libraries can generate variants with highly modified substrate specificities that can elucidate structure-activity relationships and antibiotic drug resistance properties [62], [63].
Previous library design methods have targeted TEM-1 β-lactamase from Escheridia coli for sequence-only [9] and structure-only [14], [64] optimized libraries, allowing mutations in the active site or throughout the protein. Here, in contrast, we integrate sequence and structure information (from UniProt BLAT_ECOLX and PDB 1BT5) [65] and explore the impact of targeting the active site region vs. allowing mutations anywhere.
Active site residues are more sensitive to mutations
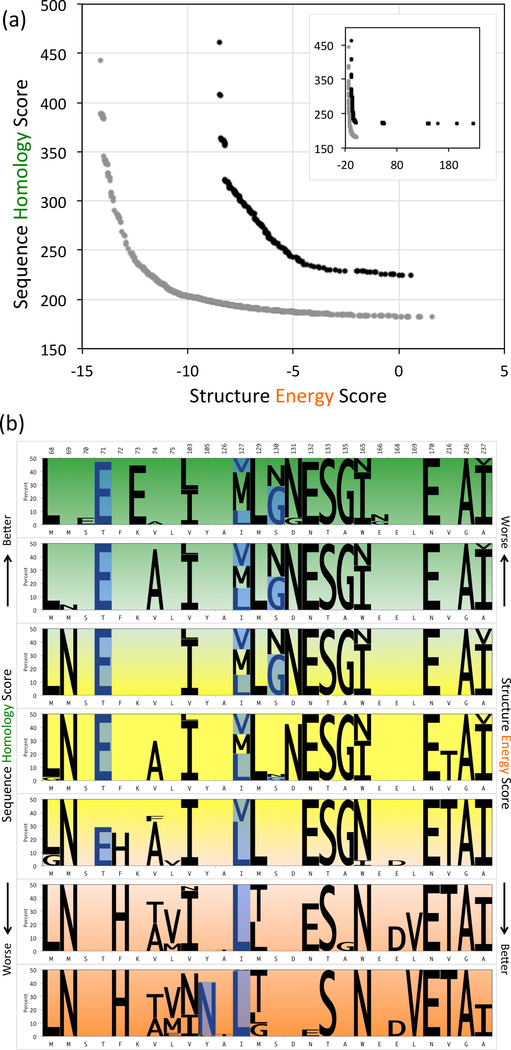
Libraries were designed to target any of 32 residues within CB-CB 8Å of catalytic residues S70, K73, S130, and E166. Fig. 7(a) illustrates the difference between Pareto frontiers for 15-site libraries focused on the active site vs. considering the whole protein. With limited choices available for focused design, large energy and homology penalties are incurred at both ends of the Pareto frontier (see inset in Fig. 7(a)), also suggesting that a very small portion of the curve offers libraries with balanced sequence and structure objectives. In comparison to the Pareto frontier for full protein libraries, that for focused libraries is less curved in the elbow region, suggesting strong sequence-structure complementarity around the active site. Comparing their elbow plans, 4 mutation sites (68, 69, 165, and 236) are shared between full and focused libraries of which 3 have identical mutation selections (M69N, W165N, and G236A). The difference in mutation at site 68 is due to the existence of residue-residue interaction between positions 68 and 71 in the focused elbow library. Overall, the focused elbow plan exhibits 8 residue-residue mutation site interactions in comparison to just 2 in the full-protein elbow plan, which is freer to spread out and decouple mutations.
Fig. 7:
(a) Pareto frontiers comparing 15-site point mutation (β-lactamase active-site (black curve) and complete (grey curve) libraries. (b) Amino acid composition of Pareto optimal 15-site point mutation β-lactamase active-site libraries. The seven panels represent different regions of the Pareto frontier, green for sequence-optimized, orange for structure-optimized and yellow for sequence-structure-optimized. The height of sequence logos corresponds to the abundance of mutations in the plans (wild-type not shown). Experimentally observed mutations are highlighted in blue.
Mutations are highly constrained at catalytic sites
The seven representative regions in Fig. 7(b) illustrate mutation choices used for the variable positions in targeted library designs. One additional choice at catalytic sites S70 and K73, and two additional choices at catalytic sites SI30 and E166 were allowed for mutation. In spite of these additional choices, three of the catalytic sites remained unmutated across the full Pareto frontier. The only mutation chosen at a catalytic site, S130G, captures a mutation that has been experimentally verified to be functional [66]. Also, we note that focused elbow plans capture mutations at active site residues T71E, I127V, and I127L, which previous experimental studies have revealed to maintain stability and functional activity [61], [64], [66], [67].
Other experimental evidence
We found an experimentally validated mutation W165Y[68] that POCoM did not select, opting instead for W165I and W165N instead (Fig. 7(b)). While tyrosine was considered as a choice, both isoleucine and asparagine had better sequence potential scores and asparagine had the best structure energy score. This analysis suggests that the selection of choices for library construction is a function of Pareto optimality and the interplay between the potentials.
4. Conclusions
We have introduced a novel method called POCoM that aims to improve the hit rate of discovering beneficial variants in combinatorial mutagenesis studies by enriching the libraries with complementary information assessing the quality of constituent variants. POCoM designs directly in “library space,” choosing sets of mutations whose combinations make the best library-averaged trade offs between two criteria. This approach enables POCoM to quite quickly design massive libraries expected to have good variants. By generating the entire Pareto frontier of libraries, POCoM elucidates, in an unbiased fashion, the trade offs between the complementary criteria.
In case study applications to GFP, cytochrome P450, and β-lactamase, we instantiate POCoM to integrate sequence homology and structure energy scores and explore the trade-offs between these scores as well as the impacts of experimental constraints. GFP and cytochrome P450 designs reveal that libraries constructed from point mutations achieve better potential scores in comparison to libraries constructed from degenerate oligonucleotides which are constrained by the genetic code. Subsequent analyses of specific designs provide insights into energy and homology contributions that lead to selection of specific amino acids for mutation. β-lactamase active-site focused libraries suggest that POCoM appropriately accounts for functional site mutation sensitivity during the design process by simultaneously considering evolutionary and structure energy information.
Overall, the modular nature of POCoM, its ability to quickly optimize large libraries enriched for complementary protein objectives, and the insights it provides into the library design space promise a high success rate of discovering beneficial variants in high throughput protein engineering experiments.
Supplementary Material
5. Acknowledgments
This work was supported in part by NIH grant R01-GM-098977 to CBK. We also gratefully acknowledge computational resources provided by NSF grant CNS-1205521. Correspondence should be addressed to Chris Bailey-Kellogg: cbk@cs.dartmouth.edu; 6211 Sudikoff Laboratory, Hanover, NH 03755.
Biography
 Deeptak Verma is a postdoctoral researcher in the Department of Computer Science at Dartmouth College, NH. He received his PhD degree in Bioinformatics and Computational Biology from the University of North Carolina at Charlotte. His PhD work elucidated the changes in proteins’ biophysical and physiochemical properties due to mutations and evolutionary changes. Currently, he is working in the area of combinatorial protein design targeting specific properties and computational vaccine design. His other research interests are in the areas of protein therapeutics, computer-aided drug design, biological data visualization and protein sequence-structure-function relationships.
Deeptak Verma is a postdoctoral researcher in the Department of Computer Science at Dartmouth College, NH. He received his PhD degree in Bioinformatics and Computational Biology from the University of North Carolina at Charlotte. His PhD work elucidated the changes in proteins’ biophysical and physiochemical properties due to mutations and evolutionary changes. Currently, he is working in the area of combinatorial protein design targeting specific properties and computational vaccine design. His other research interests are in the areas of protein therapeutics, computer-aided drug design, biological data visualization and protein sequence-structure-function relationships.
 Gevorg Grigoryan, Ph.D., is an Assistant Professor in the Department of Computer Science, an Adjunct Assistant Professor in the Departments of Biological Sciences, and an Adjunct Assistant Professor in the Department of Chemistry at Dartmouth College. He received two Bachelor’s degrees, in Computer Science and Biochemistry, from the University of Maryland Baltimore County, and a PhD from the Massachusetts Institute of Technology, where he studied computational protein design and modeling of protein-protein interactions. As a postdoctoral fellow at the University of Pennsylvania, in the laboratory of Dr. William F. DeGrado, he continued to work on designing proteins with novel functions. In 2011, Dr. Grigoryan joined the faculty of Dartmouth College, establishing a laboratory towards continued innovation in protein structural modeling and computational protein design. Dr. Grigoryan has authored over thirty peer-reviewed papers and the work in his laboratory has been funded by the National Institutes of Health, the National Science Foundation, and other funding organizations.
Gevorg Grigoryan, Ph.D., is an Assistant Professor in the Department of Computer Science, an Adjunct Assistant Professor in the Departments of Biological Sciences, and an Adjunct Assistant Professor in the Department of Chemistry at Dartmouth College. He received two Bachelor’s degrees, in Computer Science and Biochemistry, from the University of Maryland Baltimore County, and a PhD from the Massachusetts Institute of Technology, where he studied computational protein design and modeling of protein-protein interactions. As a postdoctoral fellow at the University of Pennsylvania, in the laboratory of Dr. William F. DeGrado, he continued to work on designing proteins with novel functions. In 2011, Dr. Grigoryan joined the faculty of Dartmouth College, establishing a laboratory towards continued innovation in protein structural modeling and computational protein design. Dr. Grigoryan has authored over thirty peer-reviewed papers and the work in his laboratory has been funded by the National Institutes of Health, the National Science Foundation, and other funding organizations.
 Chris Bailey-Kellogg is Professor of Computer Science at Dartmouth. He earned a BS/MS with Sandy Pentland at MIT and a PhD with Feng Zhao at Ohio State and Xerox PARC, conducted postdoctoral research with Bruce Donald at Dartmouth, and started his faculty career at Purdue. Research in his lab focuses on the development and application of computational methods to enable studies of protein sequence, structure, and function. He is seeking to engineer the immune response to foreign proteins, both desired responses to infection and vaccination and undesired responses to therapeutics. He has received an NSF Career award and an Alfred P. Sloan Foundation fellowship, and is currently funded by the NIH, NSF, and as part of a Gates Foundation Collaboration for AIDS Vaccine Discovery.
Chris Bailey-Kellogg is Professor of Computer Science at Dartmouth. He earned a BS/MS with Sandy Pentland at MIT and a PhD with Feng Zhao at Ohio State and Xerox PARC, conducted postdoctoral research with Bruce Donald at Dartmouth, and started his faculty career at Purdue. Research in his lab focuses on the development and application of computational methods to enable studies of protein sequence, structure, and function. He is seeking to engineer the immune response to foreign proteins, both desired responses to infection and vaccination and undesired responses to therapeutics. He has received an NSF Career award and an Alfred P. Sloan Foundation fellowship, and is currently funded by the NIH, NSF, and as part of a Gates Foundation Collaboration for AIDS Vaccine Discovery.
Contributor Information
Deeptak Verma, Department of Computer Science at Dartmouth College, Hanover, NH 03755..
Gevorg Grigoryan, Department of Computer Science and the Department of Biological Sciences at Dartmouth College, Hanover, NH 03755..
Chris Bailey-Kellogg, Department of Computer Science at Dartmouth College, Hanover, NH 03755..
6 References
- [1].McCullum EO, Williams BAR, Zhang J, and Chaput JC, “Random mutagenesis by error-prone PCR.,” Methods in molecular biology (Clifton, N.J.), vol. 634, pp. 103–9, January. 2010. [DOI] [PubMed] [Google Scholar]
- [2].Siloto RMP and Weselake RJ, “Site saturation mutagenesis: Methods and applications in protein engineering,” Biocatalysis and Agricultural Biotechnology, vol. 1, no. 3, pp. 181–189, July. 2012. [Google Scholar]
- [3].Zhao H, Verma D, Li W, Choi Y, Ndong C, Fiering SNSN, Bailey-Kellogg C, and Griswold KEKE, “Depletion of T cell epitopes in lysostaphin mitigates anti-drug antibody response and enhances antibacterial efficacy in vivo.,” Chemistry & biology, vol. 22, no. 5, pp. 629–39, May 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chica RA, Moore MM, Allen BD, and Mayo SL, “Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 107, no. 47, pp. 20257–62, November. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chica RA, Doucet N, and Pelletier JN, “Semi-rational approaches to engineering enzyme activity: Combining the benefits of directed evolution and rational design,” Current Opinion in Biotechnology, vol. 16, no. 4. pp. 378–384, 2005. [DOI] [PubMed] [Google Scholar]
- [6].Treynor TP, Vizcarra CL, Nedelcu D, and Mayo SL, “Computationally designed libraries of fluorescent proteins evaluated by preservation and diversity of function.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 104, no. 1, pp. 48–53, January. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kang S. gu and Saven JG, “Computational protein design: structure, function and combinatorial diversity,” Current Opinion in Chemical Biology, vol. 11, no. 3. pp. 329–334, 2007. [DOI] [PubMed] [Google Scholar]
- [8].Shivange AV, Marienhagen J, Mundhada H, Schenk A, and Schwaneberg U, “Advances in generating functional diversity for directed protein evolution,” Current Opinion in Chemical Biology, vol. 13, no. 1. pp. 19–25, 2009. [DOI] [PubMed] [Google Scholar]
- [9].Parker AS, Griswold KE, and Bailey-Kellogg C, “Optimization of combinatorial mutagenesis.,” Journal of computational biology: a journal of computational molecular cell biology, vol. 18, no. 11, pp. 1743–56, November. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Mena MA and Daugherty PS, “Automated design of degenerate codon libraries.,” Protein engineering, design & selection : PEDS, vol. 18, no. 12, pp. 559–61, December. 2005. [DOI] [PubMed] [Google Scholar]
- [11].Bendl J, Stourac J, Sebestova E, Vavra O, Musil M, Brezovsky J, and Damborsky J, “HotSpot Wizard 2.0: automated design of site-specific mutations and smart libraries in protein engineering.,” Nucleic acids research, p. gkw416-, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zheng W, Griswold KE, and Bailey-Kellogg C, “Protein fragment swapping: a method for asymmetric, selective site-directed recombination.,” Journal of computational biology : a journal of computational molecular cell biology, vol. 17, no. 3, pp. 459–75, March. 2010. [DOI] [PubMed] [Google Scholar]
- [13].Voigt CA, Mayo SL, Arnold FH, and Wang ZG, “Computational method to reduce the search space for directed protein evolution.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 98, no. 7, pp. 3778–83, March. 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Verma D, Grigoryan G, and Bailey-Kellogg C, “Structure-based design of combinatorial mutagenesis libraries.,” Protein science : a publication of the Protein Society, vol. 24, no. 5, pp. 895–908, May 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lunt MW and Snow CD, “A Structure-Based Design Protocol for Optimizing Combinatorial Protein Libraries.,” Methods in molecular biology (Clifton, N.J.), vol. 1414, pp. 99–138, January. 2016. [DOI] [PubMed] [Google Scholar]
- [16].Saraf MC, Moore GL, Goodey NM, Cao VY, Benkovic SJ, and Marañas CD, “IPRO: an iterative computational protein library redesign and optimization procedure.,” Biophysical journal, vol. 90, no. 11, pp. 4167–4180, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Grigoryan G, Reinke AW, and Keating AE, “Design of protein-interaction specificity gives selective bZIP-binding peptides.,” Nature, vol. 458, no. 7240, pp. 859–64, April. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Grigoryan G, Zhou F, Lustig SR, Ceder G, Morgan D, and Keating AE, “Ultra-fast evaluation of protein energies directly from sequence.,” PLoS computational biology, vol. 2, no. 6, p. e63, June. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].He L, Friedman AM, and Bailey-Kellogg C, “A divide-and-conquer approach to determine the Pareto frontier for optimization of protein engineering experiments.,” Proteins, vol. 80, no. 3, pp. 790–806, March. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Suarez M, Tortosa P, Garcia-Mira MM, Rodríguez-Larrea D, Godoy-Ruiz R, Ibarra-Molero B, Sanchez-Ruiz JM, and Jaramillo A, “Using multi-objective computational design to extend protein promiscuity.,” Biophysical chemistry, vol. 147, no. 1–2, pp. 13–9, March. 2010. [DOI] [PubMed] [Google Scholar]
- [21].Salvat RS, Choi Y, Bishop A, Bailey-Kellogg C, and Griswold KE, “Protein deimmunization via structure-based design enables efficient epitope deletion at high mutational loads.,” Biotechnology and bioengineering, vol. 112, no. 7, pp. 1306–18, July. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Salvat RS, Parker AS, Guilliams A, Choi Y, Bailey-Kellogg C, and Griswold KE, “Computationally driven deletion of broadly distributed T cell epitopes in a biotherapeutic candidate.,” Cellular and molecular life sciences : CMLS, June. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Choi Y, Verma D, Griswold KE, and Bailey-Kellogg C, “EpiSweep: Computationally Driven Reengineering of Therapeutic Proteins to Reduce Immunogenicity While Maintaining Function,” in Methods in molecular biology (Clifton, N.J.), vol. 1529, 2017, pp. 375–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ye X, Friedman AM, and Bailey-Kellogg C, “Hypergraph model of multi-residue interactions in proteins: sequentially-constrained partitioning algorithms for optimization of site-directed protein recombination.,” Journal of computational biology : a journal of computational molecular cell biology, vol. 14, no. 6, pp. 777–90, January. 2007. [DOI] [PubMed] [Google Scholar]
- [25].Salvat RS, Parker AS, Choi Y, Bailey-Kellogg C, and Griswold KE, “Mapping the Pareto Optimal Design Space for a Functionally Deimmunized Biotherapeutic Candidate,” PLoS Computational Biology, vol. 11, no. 1, p. e1003988, January. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Salvat RS, Verma D, Parker AS, Kirsch JR, Brooks SA, Bailey-Kellogg C, and Griswold KE, “Computationally optimized deimmunization libraries yield highly mutated enzymes with low immunogenicity and enhanced activity.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 114, no. 26, pp. E5085–E5093, June. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kaján L, Hopf TA, Kalaš M, Marks DS, and Rost B, “FreeContact: fast and free software for protein contact prediction from residue co-evolution.,” BMC bioinformatics, vol. 15, p. 85, March. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Morcos F, Pagnani A, Lunt B, Bertolino A, Marks DS, Sander C, Zecchina R, Onuchic JN, Hwa T, and Weigt M, “Direct-coupling analysis of residue coevolution captures native contacts across many protein families.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 108, no. 49, pp. E1293–301, December. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kamisetty H, Ovchinnikov S, and Baker D, “Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era,” Proceedings of the National Academy of Sciences, vol. 110, no. 39, pp. 15674–15679, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, and Sander C, “Protein 3D Structure Computed from Evolutionary Sequence Variation,” PLoS ONE, vol. 6, no. 12, p. e28766, December. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hopf TA, Schärfe CPI, Rodrigues JPGLM, Green AG, Kohlbacher O, Sander C, Bonvin AMJJ, and Marks DS, “Sequence co-evolution gives 3D contacts and structures of protein complexes,” eLife, vol. 3, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Livesay DR, Kreth KE, and Fodor AA, “A Critical Evaluation of Correlated Mutation Algorithms and Coevolution Within Allosteric Mechanisms,” in Methods in molecular biology (Clifton, N.J.), vol. 796, 2012, pp. 385–398. [DOI] [PubMed] [Google Scholar]
- [33].Cocco S, Monasson R, and Weigt M, “From Principal Component to Direct Coupling Analysis of Coevolution in Proteins: Low-Eigenvalue Modes are Needed for Structure Prediction,” PLoS Computational Biology, vol. 9, no. 8, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rohl CA, Strauss CEMM, Misura KMSS, and Baker D, “Protein Structure Prediction Using Rosetta,” Methods in Enzymology, vol. 383, pp. 66–93, January. 2004. [DOI] [PubMed] [Google Scholar]
- [35].Hahn S, Ashenberg O, Grigoryan G, and Keating AE, “Identifying and reducing error in clusterexpansion approximations of protein energies.,” Journal of computational chemistry, vol. 31, no. 16, pp. 2900–14, December. 2010. [DOI] [PubMed] [Google Scholar]
- [36].Zhou F, Grigoryan G, Lustig SR, Keating AE, Ceder G, and Morgan D, “Coarse-graining protein energetics in sequence variables.,” Physical review letters, vol. 95, no. 14, p. 148103, September. 2005. [DOI] [PubMed] [Google Scholar]
- [37].Zheng W, Friedman AM, and Bailey-Kellogg C, “Algorithms for joint optimization of stability and diversity in planning combinatorial libraries of chimeric proteins.,” Journal of computational biology : a journal of computational molecular cell biology, vol. 16, no. 8, pp. 1151–68, August. 2009. [DOI] [PubMed] [Google Scholar]
- [38].Pierce NA and Winfree E, “Protein Design is NP-hard,” Protein Engineering Design and Selection, vol. 15, no. 10, pp. 779–782, October. 2002. [DOI] [PubMed] [Google Scholar]
- [39].Parker AS, Griswold KE, and Bailey-Kellogg C, “Optimization of therapeutic proteins to delete T-cell epitopes while maintaining beneficial residue interactions.,” Journal of bioinformatics and computational biology, vol. 9, no. 2, pp. 207–29, April. 2011. [DOI] [PubMed] [Google Scholar]
- [40].Parker AS, Choi Y, Griswold KE, and Bailey-Kellogg C, “Structure-guided deimmunization of therapeutic proteins.,” Journal of computational biology: a journal of computational molecular cell biology, vol. 20, no. 2, pp. 152–65, February. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Salvat RS, Parker AS, Choi Y, Bailey-Kellogg C, and Griswold KE, “Mapping the Pareto Optimal Design Space for a Functionally Deimmunized Biotherapeutic Candidate,” PLoS Computational Biology, vol. 11, no. 1, p. e1003988, January. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, and O’Shea EK, “Global analysis of protein localization in budding yeast.,” Nature, vol. 425, no. 6959, pp. 686–91, October. 2003. [DOI] [PubMed] [Google Scholar]
- [43].Heim R, Prasher DC, and Tsien RY, “Wavelength mutations and posttranslational autoxidation of green fluorescent protein.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 91, no. 26, pp. 12501–4, December. 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Soboleski MR, Oaks J, and Halford WP, “Green fluorescent protein is a quantitative reporter of gene expression in individual eukaryotic cells.,” FASEB journal : official publication of the Federation of American Societies for Experimental Biology, vol. 19, no. 3, pp. 440–2, March. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang J, Campbell RE, Ting AY, and Tsien RY, “Creating new fluorescent probes for cell biology.,” Nature reviews. Molecular cell biology, vol. 3, no. 12, pp. 906–18, December. 2002. [DOI] [PubMed] [Google Scholar]
- [46].Rolls MM, Stein PA, Taylor SS, Ha E, McKeon F, and Rapoport TA, “A visual screen of a GFP-fusion library identifies a new type of nuclear envelope membrane protein.,” The Journal of cell biology, vol. 146, no. 1, pp. 29–44, July. 1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cutler SR, Ehrhardt DW, Griffitts JS, and Somerville CR, “Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 97, no. 7, pp. 3718–23, March. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jackrel ME, Cortajarena AL, Liu TY, and Regan L, “Screening libraries to identify proteins with desired binding activities using a split-GFP reassembly assay.,” ACS chemical biology, vol. 5, no. 6, pp. 553–62, June. 2010. [DOI] [PubMed] [Google Scholar]
- [49].Saeger J, Hytonen VP, Klotzsch E, and Vogel V, “GFP’s mechanical intermediate states.,” PloS one, vol. 7, no. 10, p. e46962, January. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tallini YN, Ohkura M, Choi B-R, Ji G, Imoto K, Doran R, Lee J, Plan P, Wilson J, Xin H-B, Sanbe A, Gulick J, Mathai J, Robbins J, Salama G, Nakai J, and Kotlikoff MI, “Imaging cellular signals in the heart in vivo: Cardiac expression of the high-signal Ca2+ indicator GCaMP2.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 103, no. 12, pp. 4753–8, March. 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Pavoor TV, Cho YK, and Shusta EV, “Development of GFP-based biosensors possessing the binding properties of antibodies.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 106, no. 29, pp. 11895–900, July. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bjorn SP, Pagliaro L, and Thastrup O, “Fluorescent proteins.” Google Patents, 21-February-2006. [Google Scholar]
- [53].Paschke M, Tiede C, and Höhne W, “Engineering a circularly permuted GFP scaffold for peptide presentation.,” Journal of molecular recognition : JMR, vol. 20, no. 5, pp. 367–78, January. [DOI] [PubMed] [Google Scholar]
- [54].Süel GM, Lockless SW, Wall MA, and Ranganathan R, “Evolutionarily conserved networks of residues mediate allosteric communication in proteins.,” Nature structural biology, vol. 10, no. 1, pp. 59–69, January. 2003. [DOI] [PubMed] [Google Scholar]
- [55].Pikuleva I. a, “Cytochrome P450s and cholesterol homeostasis.,” Pharmacology & therapeutics, vol. 112, pp. 761–773, 2006. [DOI] [PubMed] [Google Scholar]
- [56].Guengerich FP, “Cytochrome P450 and chemical toxicology,” Chemical Research in Toxicology, vol. 21, no. 1. pp. 70–83, 2008. [DOI] [PubMed] [Google Scholar]
- [57].Kumar S, “Engineering cytochrome P450 biocatalysts for biotechnology, medicine and bioremediation.,” Expert opinion on drug metabolism & toxicology, vol. 6, no. 2, pp. 115–31, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Pantazes RJ, Saraf MC, and Maranas CD, “Optimal protein library design using recombination or point mutations based on sequence-based scoring functions.,” Protein engineering, design & selection : PEDS, vol. 20, no. 8, pp. 361–73, August. 2007. [DOI] [PubMed] [Google Scholar]
- [59].WALLMARK G, “The production of penicillinase in Staphylococcus aureus pyogenes and its relation to penicillin resistance.,” Acta pathologica et microbiologica Scandinavica, vol. 34, no. 2, pp. 182–90, January. 1954. [DOI] [PubMed] [Google Scholar]
- [60].De Wals P-Y, Doucet N, and Pelletier JN, “High tolerance to simultaneous active-site mutations in TEM-1 beta-lactamase: Distinct mutational paths provide more generalized beta-lactam recognition.,” Protein science : a publication of the Protein Society, vol. 18, no. 1, pp. 147–60, January. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schultz SC and Richards JH, “Site-saturation studies of beta-lactamase: production and characterization of mutant beta-lactamases with all possible amino acid substitutions at residue 71.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 83, no. 6, pp. 1588–92, March. 1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Palzkill T and Botstein D, “Identification of amino acid substitutions that alter the substrate specificity of TEM-1 beta-lactamase.,” Journal of bacteriology, vol. 174, no. 16, pp. 5237–43, August. 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Matagne A, Lamotte-Brasseur J, and Frere JM, “Catalytic properties of class A beta-lactamases: efficiency and diversity.,” The Biochemical journal, vol. 330 ( Pt 2, pp. 581–98, March. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hayes RJ, Bentzien J, Ary ML, Hwang MY, Jacinto JM, Vielmetter J, Kundu A, and Dahiyat BI, “Combining computational and experimental screening for rapid optimization of protein properties.,” Proceedings of the National Academy of Sciences of the United States of America, vol. 99, no. 25, pp. 15926–31, December. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Maveyraud L, Mourey L, Kotra LP, Pedelacq J-D, Guillet V, Mobashery S, and Samama J-R, “Structural Basis for Clinical Longevity of Carbapenem Antibiotics in the Face of Challenge by the Common Class A β-lactamases from the Antibiotic-Resistant Bacteria,” Journal of the American Chemical Society, vol. 120, no. 38, pp. 9748–9752, September. 1998. [Google Scholar]
- [66].Thomas VL, Golemi-Kotra D, Kim C, Vakulenko SB, Mobashery S, and Shoichet BK, “Structural consequences of the inhibitor-resistant Ser130Gly substitution in TEM beta-lactamase.,” Biochemistry, vol. 44, no. 26, pp. 9330–8, July. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Arpin C, Labia R, Dubois V, Noury P, Souquet M, and Quentin C, “TEM-80, a novel inhibitor-resistant beta-lactamase in a clinical isolate of Enterobacter cloacae.,” Antimicrobial agents and chemotherapy, vol. 46, no. 5, pp. 1183–9, May 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Stojanoski V, Chow DC, Hu L, Sankaran B, Gilbert HF, Prasad BVV, and Palzkill T, “A triple mutant in the Ω-loop of TEM-1 β-lactamase changes the substrate profile via a large conformational change and an altered general base for catalysis,” Journal of Biological Chemistry, vol. 290, no. 16, pp. 10382–10394, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.