SUMMARY
The family Enterobacteriaceae has undergone significant morphogenetic changes in its more than 85-year history, particularly during the past 2 decades (2000 to 2020). The development and introduction of new and novel molecular methods coupled with innovative laboratory techniques have led to many advances. We now know that the global range of enterobacteria is much more expansive than previously recognized, as they play important roles in the environment in vegetative processes and through widespread environmental distribution through insect vectors. In humans, many new species have been described, some associated with specific disease processes. Some established species are now observed in new infectious disease settings and syndromes. The results of molecular taxonomic and phylogenetics studies suggest that the current family Enterobacteriaceae should possibly be divided into seven or more separate families. The logarithmic explosion in the number of enterobacterial species described brings into question the relevancy, need, and mechanisms to potentially identify these taxa. This review covers the progression, transformation, and morphogenesis of the family from the seminal Centers for Disease Control and Prevention publication (J. J. Farmer III, B. R. Davis, F. W. Hickman-Brenner, A. McWhorter, et al., J Clin Microbiol 21:46–76, 1985, https://doi.org/10.1128/JCM.21.1.46-76.1985) to the present.
KEYWORDS: Enterobacteriaceae, enterobacteria, insect vectors, liver abscess, meninigitis, phylogeny, plant diseases, taxonomy, transmissible gastroenteritis virus
INTRODUCTION
Historical Aspects
No collective group of currently defined prokaryotic bacteria has had a greater medical, public health, and veterinary impact on the global community than the family Enterobacteriaceae (1–3). Not only are the enterobacteria associated with a wide range of clinical syndromes, but the family is also a major causative agent of foodborne enteritis and zoonotic infections, which include sporadic to pandemic outbreaks of human plague (1).
The impact of the family does not stop there. Widely dispersed in nature in many naturally occurring ecosystems, members of the family are increasingly being implicated as pathogens of piscine species (natural, aquaculture) (4) as well as the etiologic agent of a variety of plant diseases (5). Finally, the family has been recognized for its impact on molecular and cell biology, gene structure and function, and microbial pathogenicity. An excellent list of notable discoveries associated with the Enterobacteriaceae can be found in the chapter by Farmer et al. (1) in the 10th edition of Topley & Wilson’s Microbiology & Microbial Infections.
While current members of this family have long been recognized for well over 100 years, the formalized origins of this family can be traced back to the 1937 publication of Rahn (6). The family Enterobacteriaceae was subsequently created with a single type genus (Enterobacter) to house a collection of 112 species previously referred to as the “colon-typhoid group” (2, 7). This group was originally defined by a set of unifying phenotypic traits, including Gram stain reaction (negative), good growth on artificial media, acid formation from d-glucose (often with gas), and production of nitrites from nitrate. Many genera with current taxonomic standing were represented within the genus “Enterobacter” when the family was first proposed, including Escherichia, Klebsiella, Shigella, and Proteus (2).
Over the next 20+ years, considerable controversy occurred regarding the exact number and names for delineated taxa that existed within the family Enterobacteriaceae (8). Multiple issues caused these nomenclature problems, including taxonomic proposals based upon minimal phenotypic data, a limited array of biochemical features available, the use of nonstandardized test methodologies, and no Bacteriological Code to govern the legitimacy or correctness of proposed taxa. By 1944, considerable difficulty was reported on defining criteria for inclusion of members in this family, since the fifth edition of Bergey’s Manual of Determinative Bacteriology already contained such names as Paracolobactrum, Colobactrum, Proshigella, and Shigella ambigua (8). Excellent historical perspectives on this topic can be found elsewhere (1, 2).
During the early and mid-1960s, a more formalized system was developed by the U.S. Centers for Disease Control and Prevention (CDC), which included a standardized set of 50 or more biochemical and phenotypic properties and computer-based software programs employing numerical taxonomy that compared phenotypes of two groups by matching similarities (Ssm) (9). By the mid-1970s, Don Brenner and his colleagues (10) at the CDC were defining existing and previously unrecognized taxa of enteric bacteria by using a polyphasic approach. This approach coupled 50 to 200 morphologic, cultural, and biochemical features to genetic studies of DNA relatedness by DNA-DNA hybridization (DDH) as well as mol% G+C content (10). Such pioneering efforts led to classification changes, such as Enterobacter sakazakii (cloacae), Hafnia alvei (Enterobacter hafniae), and Morganella (Proteus) morganii as examples. By 1985, the CDC published a 30-page landmark study in the Journal of Clinical Microbiology describing new species and biogroups within the family Enterobacteriaceae isolated from clinical specimens (11).
Both the family name (Enterobacteriaceae versus Enterobacteraceae) and type genus (Escherichia versus Enterobacter) have been unsuccessfully challenged over the past several decades (7). For the purposes of this review, the family name Enterobacteriaceae is used in the traditional or classic sense (pre-2016). Proposed classification changes for members of the family are discussed below (see Nomenclature and Taxonomy).
Traditional Phenotypic and Molecular Markers of the Family
From the family’s infancy in the late 1930s until the late 1980s, the primary means of initially identifying potential taxa (genera, species) residing in the enterobacteria was a series of collective properties almost exclusively associated with this family. These taxonomic markers were key elements in recognizing unusual strains, new biotypes, or potentially unnamed genomospecies prior to determining genetic relatedness using DDH. The traits utilized for this purpose were gradually expanded and refined over time and helped define the family from a taxonomic point of view.
The defining general reactions that have been used to identify true members of the Enterobacteriaceae for decades are listed in Table 1. These biochemical markers have provided the phenotypic definition for the family and have been used by many commercial companies in developing semiautomated and automated biochemical platforms for their recognition. DDH, which came into vogue in the 1970s, further helped to refine this family on a genetic basis prior to modern-day phylogenetic studies and full genome sequencing.
TABLE 1.
Traditional markers associated with inclusion in the family Enterobacteriaceae
| Characteristic | Trait | Typical reaction or value | Exceptions |
|---|---|---|---|
| Gram stain | Structural | Negative | |
| Flagella (polar) | Structural | Negative | Plesiomonas, Tatumella |
| ECAa | Structural | Positive | Dickeya chrysanthemi |
| Spore formation | Structural | Negative | Serratia marcescens subsp. sakuensisb |
| Oxidase | Biochemical | Negative | Alterococcusc, Franconibacterd, Plesiomonas |
| Catalase | Biochemical | Positive | Chania, Shigella dysenteriae 1, Xenorhabdus |
| Nitrate reductase | Biochemical | Positive | Photorhabdus, Erwinia, Lonsdalea, Rosenbergiella, Yersiniae |
| O/129 susceptibility | Biochemical | Positive | Plesiomonas |
| d-Glucose | Biochemical | Positivef | |
| d-Mannitol | Biochemical | Positive | Edwardsiella tarda, Providencia rustigianii, Rosenbergiella, Shigella dysenteriae type 1, among others |
| d-Xylose | Biochemical | Positive | Cedeceae, Cosenzea, Edwardsiella, Izhakiella, Lonsdalea, Morganella, Plesiomonas shigelloides, Yersinia ruckeri, among others |
| G+C (mol%) | Genetic | 38–60% | |
| DNA relatednessg | |||
| Core | Genetic | 40–50% | Salmonella, Klebsiella, Citrobacter, Enterobacter |
| Periphery | Genetic | 5–20% | Edwardsiella, Morganella, Proteus, Providencia, Yersinia |
ECA, enterobacterial common antigen.
The legitimacy of this subspecies and its ability to produce spores have been questioned (275).
This genus and species is listed in the family Enterobacteriaceae by the Editorial Board in the 2nd edition of Bergey’s Manual of Systematic Bacteriology (276).
Weak reaction.
Some isolates.
Rare strain exceptions.
DNA relatedness to the type species Escherichia coli. Core and periphery members are based upon phylogenetic analysis.
Several of the properties listed in Table 1 bear special mention. A key marker almost exclusively associated with this family is the enterobacterial common antigen or ECA (12). ECA is a carbohydrate moiety whose exact function is presently unknown but which is thought to play a role in maintaining the outer membrane permeability barrier (12, 13). ECA is almost exclusively associated with members of the Enterobacteriaceae and is one taxonomic criterion for the transfer of Plesiomonas shigelloides, which is ECA positive, to this family in 2005 (14). Only two nonenteric species, Actinobacillus equuli and Actinobacillus suis, have been found to possess this antigen (15). Another point of demarcation used from the 1960s onward is G+C content. G+C content for bacteria varies from 25 to 75 mol% (16). In the case of the enterobacteria, the G+C content has a wide range (38 to 60 mol%), which is not typically found for phylogenetically “tight” families (Table 1). The family Moraxellaceae, as an example, has a much narrower G+C content range of 38 to 50 mol% (17). However, most genera in the family Enterobacteriaceae have a G+C content of 49 to 59 mol%, which is more in line with other families containing phylogenetically related genera (1).
When the characteristics listed in Table 1 were first established, few exceptions to the metrics were noted. However, with the expansion in the family in both the number of genera and species and the transfer of other taxa to this family (e.g., P. shigelloides), more exceptions have been observed. Most of these exceptions have been detected in more recently described taxa since 2010 which are rarely associated with clinical infections or public health issues (Table 1, exceptions).
Major Clinical and Public Health Aspects of the Family
Foodborne disease and outbreaks.
Infectious diarrhea is a leading cause of morbidity and mortality on a worldwide basis. The most recent figures released by the Global Disease Burden (GBD) Diarrheal Diseases Collaborators (18) estimate that there were 2.39 billion episodes of diarrheal disease in 2015, almost 1 billion of which occurred in children under 5 years of age. The World Health Organization (WHO) further estimates that from 550 to 600 million of these cases of diarrhea are foodborne infections (19, 20). Projections that 70% of all diarrheal disease are food related (21) translates to between 350 to 420 million episodes of gastroenteritis being of bacterial etiology (19, 20). The figures are staggering.
Among major foodborne bacterial pathogens, the family Enterobacteriaceae is well represented by several groups, including Salmonella, Escherichia coli (O157, non-O157), Shigella, and Yersinia enterocolitica (19, 21–23). The CDC Foodborne Diseases Active Surveillance Network (FoodNet) provides active surveillance and epidemiologic studies in conjunction with 10 state health departments regarding the above-listed agents (21). FoodNet covers approximately 15% of the U.S. population. Not included in this list are other members of the family Enterobacteriaceae such as Cronobacter sakazakii (23). In addition to its association with neonatal meningitis and powdered infant formula (24, 25), C. sakazakii has been implicated in a large-scale foodborne outbreak of gastroenteritis in high school students and school employees (26).
The latest data from FoodNet for years 2016 to 2019 reported over 25,000 cases of laboratory-diagnosed infections for 9 major bacterial and parasitic pathogens surveyed by 10 state health departments. These cumulative infections resulted in over 6,000 hospitalizations and 122 deaths (27). High incidence rates (per 100,000 population) were recorded for Salmonella (17.1 cases), Shiga toxin-producing E. coli (STEC) (6.3 cases), and Shigella (4.8 cases), which ranked these three pathogens, after Campylobacter, as the second, third, and fourth most common foodborne pathogens in incidence. For all members of this family (including Yersinia), the incidence numbers remained unchanged or increased in comparison to 2006-2017 figures, suggesting that progress in controlling such infections has stalled (27, 28). Prior CDC investigations have found nontyphoidal Salmonella to be the leading cause of hospitalizations and death from foodborne infections (29). Comparable numbers for 26 member states in the European Union (EU) for 2015 included over 4,000 foodborne and waterborne outbreaks and 45,874 cases of illness (23). The most common bacterial pathogen responsible for foodborne outbreaks in the EU was Salmonella (∼34% of all bacterial outbreaks).
Several alarming trends appear to be occurring with foodborne pathogens in the family Enterobacteriaceae. First, in addition to traditional sources of foodborne outbreaks associated with enterobacteria (dairy, poultry, beef, pork, melons, sprouts), an increasing number of other consumable products or condiments are being linked to outbreaks. These include basil (Shigella), bagged salad (Y. enterocolitica), cookie dough and sprouted seeds (E. coli), and peanut butter and jalapeno and serrano peppers (Salmonella) (22). C. sakazakii has also been isolated from various retail foods, including legumes, nuts, dried flour, and spices (23). A second issue is the recent appearance of enterobacterial strains with “hybrid” virulence characteristics causing foodborne outbreaks. In 2011, a major outbreak of food-related illness associated with E. coli O104:H4 occurred in Germany (22). This outbreak was linked to the consumption of sprouts (30). Of the more than 3,186 infections, approximately 22% of patients developed hemolytic-uremic syndrome and 54 persons (1.7%) died (22). Subsequent molecular analysis of the infecting strain revealed that it contained properties of enteroaggregative E. coli and also had the capacity to produce Shiga toxin 2 (22). Two more recent California outbreaks involving 56 patients were found to be caused by a Shiga toxin 1-producing strain of Shigella sonnei (31). This genotype is rarely found in shigellae other than Shigella dysenteriae type 1. Finally, in addition to CDC data, in Europe (GBD collaborators) Shigella has become the most common bacterial foodborne pathogen associated with mortality in children under the age of 5, with an estimated 54,900 deaths annually (18). The wide distribution of enterobacteria in foods coupled with hygiene and sanitation issues poses serious problems for developing nations (19).
HAI.
The WHO estimates that there are over 4.5 million episodes of health care-associated infections (HAI) in Europe each year, with a projected 37,000 deaths annually (https://www.who.int/gpsc/country_work/gpsc_ccisc_fact_sheet_en.pdf). Comparable figures last reported for the United States include 99,000 deaths and $6.5 billion in costs associated with extra days of hospital stay.
Genera in the family Enterobacteriaceae are important pathogens for three of the four major HAI categories according to the CDC, namely, central line-associated bloodstream infections (CLABSI), catheter-associated urinary tract infections (CAUTI), and surgical site infections (SSI) (https://www.cdc.gov/hai/index.html). One investigation studied the relative frequency of HAI pathogens within the University of North Carolina Health Care database over a 28-year period (1980 to 2008) (32). Based upon overall frequency, E. coli, Klebsiella, and Enterobacter ranked 2nd, 7th, and 8th, respectively, in number of infections reported, with the genera Proteus, Serratia, and Citrobacter ranking between 11th and 16th (32). A recent CDC study summarizing data collected from 2011 to 2017 on CLABSI found the Enterobacteriaceae to be causing 23% to 31% of HAI in adult, pediatric, and oncology wards (33).
To further complicate the health care setting with enterobacterial infections is the fact that over the last 10 to 15 years, antimicrobial resistance to carbapenem compounds has emerged, dramatically restricting treatment options for serious life-threatening infections (34). A variety of Ambler class A, D, and B enzymes inactivate carbapenems, including Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo-β-lactamase (NDM), and Verona integron-encoded metallo-β-lactamase (VIM), among others (34). Enteric species most commonly associated with these drug resistance patterns are the most common HAI-associated species, such as E. coli, K. pneumoniae, and Enterobacter cloacae complex. Increasing numbers of carbapenemase-resistant Enterobacteriaceae (CRE) have been thought to be due to expansion of clonal groups and horizontal gene transfer. However, a phylogenetic analysis of many CRE strains at three hospitals in the Boston area suggest not only remarkable genetic diversity but also limited clonal expansion (35). This suggests that CRE transmission is occurring at multiple unsampled transmission points throughout the health care process. This rapid rise has been recently documented in a National Healthcare Safety Network (NHSN) summary report of 5,626 acute care facilities from 2015 to 2017 (36). In that epidemiologic survey of antimicrobial-resistant bacteria, E. coli and K. pneumoniae were the most common and the third most frequently reported HAI pathogens, respectively (36).
THE FAMILY ENTEROBACTERIACEAE
Post-1980 Era: The Family That Wasn’t
Modern bacterial nomenclature and taxonomy was born on 1 January 1980 with the publication of the Approved Lists of Bacterial Names (AL) in the International Journal of Systematic Bacteriology (IJSB) (37). The AL was created based upon currently valid names of bacteria that were cultivatable and those published in the International Journal of Systematic Bacteriology prior to 1 January 1978 (37). What many microbiologists and scientists do not recognize is the fact that with the publication of the AL, the family name Enterobacteriaceae automatically became invalid and technically could not be used in subsequent publications because the name had been omitted under the rank of “family.” Subsequently, the matter was considered sub judice (under judicial review) because of a challenge to the legitimacy of the name by Lapage (38, 39), who recommended changing the name to Enterobacteraceae. However, it was immediately recognized that the AL had serious exclusions to it and that validly published legitimate names had been arbitrarily omitted (40). In companion articles, the CDC opposed rejection of the family name (7) and Ewing and coauthors (40) validly published and proposed reviving the name Enterobacteriaceae under rules defined by the Bacteriological Code. The Judicial Commission subsequently determined that the family name Enterobacteriaceae Rahn 1937 with Escherichia Castellani and Chalmers 1919 as the type genus did have standing and belonged on the AL (39). While other proposals such as changing the family name to Escherichiaceae have appeared, they have almost always been uniformly opposed by subcommittees on the enterobacteria (41).
Genus and Species Expansion (1980 to 2020)
The original AL contained 2,366 valid prokaryotic names, of which 2,213 were at the rank of genus (n = 290), species (n = 1,792), and subspecies (n = 131) (42). While these numbers are sizable, they pale in comparison to the computational prokaryotic diversity in the global biosphere, estimated to range between 2 and 4 million species (43). This figure may actually be much higher (43). Thus, like other families in the prokaryotes, given the above projected numbers, it is not surprising that membership in the family Enterobacteriaceae has also increased dramatically over the past 40 years.
The history of taxon expansion in the family Enterobacteriaceae post-1980 has for the most part mirrored that of other large prokaryotic families. Those increases again paralleled technical developments in the characterization of prokaryotes which enabled taxonomists to more easily assess both the uniqueness (new species) and relatedness (phylogeny) of sets of strains to other named groups. From the early 1980s to the mid-1990s, the main systematic approach involved the use of polyphasic taxonomy, that is, a combination of phenotypic (culture, biochemical, serology, cellular fatty acids) and genetic (DDH, G+C mol%) characteristics. Polyphasic taxonomy is still used today but is less in vogue. Groups spearheading the description of new species in the family Enterobacteriaceae during this period included the CDC in Atlanta, the Institute of Pasteur (IOP) in France, and the National Institutes of Health in Japan (NIH Japan) (44–47). Virtually all of the named species during this period were accompanied by extensive biochemical profiles, which allowed laboratories to differentiate on a phenotypic basis a new taxon from previously named species. When a significant number of strains were not available to clearly define an unnamed taxon, they were typically given a generic name, such as CDC enteric group 58 (11).
The “gold standard” for defining these new species during this period was DDH (48, 49). However, DDH has a number of limitations. The traditional technique was expensive, labor-intensive, time-consuming, and subject to technical errors (temperature, ionic strength) (48–50). Only a few large international laboratories (CDC, IOP, NIH Japan) had the personnel and resources to employ such technologies on a routine basis. Additionally, DNA studies indicated that many existing species in the family Enterobacteriaceae were polyphyletic, that is, composed of genetically distinct species that could not easily be resolved by simple biochemical tests, the principal mechanism most clinical microbiology laboratories used for final species identification. Thus, such organisms as Enterobacter cloacae, Pantoea (Enterobacter) agglomerans, and Hafnia alvei (Enterobacter hafniae) were known by DDH to be composed of multiple hybridization groups (HGs), each representing a unique genomospecies although they all exhibited common characteristics and could not be separated phenotypically from each other (51). Such species in actuality were at that time “phenospecies” or a “complex” of species rather than, for instance, E. cloacae sensu stricto.
Family expansion due to newer technology.
In the 1990s, sequencing of housekeeping genes, such as the 16S rRNA gene, was thought at that time to be a major breakthrough in bacterial systematics (48, 50, 52). The 16S gene could be sequenced with relative ease on different platforms and could provide informatics more quickly than DDH (53). Threshold values were determined for comparing a proposed new taxon to closely related neighbors, with <98.7% sequence similarity strongly suggesting that the unnamed group represented a new species (50, 54). Full-length 16S rRNA gene sequences (∼1,500 bp) quickly became a standard requirement for the publication of new species proposals, while16S similarity values above 98.7% still required DDH to determine relatedness. While 16S gene sequencing clearly provided quicker resolution of taxonomic issues, problems still remained (49, 55). This was especially true for the Enterobacteriaceae, where in a number of instances 16S gene sequencing did not have particularly good resolving power at the species level (48). This was partially due to the highly conserved nature of the ribosomal gene (55). Species in genera clearly resolvable by DDH (Edwardsiella, Enterobacter) yielded 16S rRNA gene sequence results that were either identical or showed very little variation (48).
The advent of 16S rRNA gene sequencing as both a taxonomic tool and a routine platform for bacterial identification in the clinical laboratory was rapidly followed by the introduction of a wide range of molecular techniques with which to assess genetic diversity and similarities of named or proposed species. These techniques included matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF), multilocus sequence analysis (MLSA), average nucleotide identity (ANI), percentage of conserved proteins (POCP), digital DDH (dDDH), genome-to-genome distance calculator (GGDC), and whole-genome sequencing (WGS), among others (49, 54, 56). This avalanche of new molecular technologies and techniques were less expensive and labor-intensive and available to both clinical and research laboratories, which resulted in an explosion in the number of new genera and species descriptions.
While the number of described species was relatively steady through 1994 (50), the numbers have dramatically risen since then. By 2007, over 8,000 species had been described, and this was more than a 400% increase since the AL in 1980 (42, 48). The family Enterobacteriaceae experienced a similar increase in the numbers of genera and species, with the greatest increase in percentages after 2005, which is a reflection of technologic developments related to the description and phylogenetic classifications of new taxa (Table 2). The single technology with the greatest impact has been WGS.
TABLE 2.
Genus and species expansion in the family Enterobacteriaceae, 1974–2020
| Source | Yr | No. of genera | No. of species | % Increasea | Reference |
|---|---|---|---|---|---|
| Bergey’s Manual of Determinative Bacteriology (8th ed.) | 1974 | 12 | 36 | −32b | 1 |
| Approved List | 1980 | 18 | 80 | 122 | 37 |
| Bergey’s Manual of Systematic Bacteriology | 1984 | 20 | 76 | −5 | 1 |
| Bergey’s Manual of Determinative Bacteriology (9th ed.) | 1994 | 30 | 107 | 29 | 1 |
| Topley & Wilson’s Microbiology & Microbial Infections (10th ed.) | 2005 | 40 | 150 | 41 | 1 |
| LPSNc | 2020 | 68 | 355 | 136 |
Based on number of species from the previous source or edition.
From Rahn, 1937 (6).
LPSN, List of Prokaryotic Names with Standing in Nomenclature (https://www.bacterio.net/).
Because of the rapidly evolving landscape of microbial taxonomy, the family has radically changed over the past 4 decades (1980 to 2020). Table 3 shows a snapshot of that change, comparing a few selected genera that Farmer et al. (11) described in their landmark publication of 1985 with their present status in 2020. The 2020 reference point is the List of Prokaryotic Names with Standing in Nomenclature (LPSN) website (https://www.bacterio.net/) curated by Aidan C. Parte, Leibniz Institute DSMZ.
TABLE 3.
Comparison of new genera and species proposed by the CDC in 1985 and present status in 2020a
| Genusb | 1985 |
Classification change | 2020 |
Classification change | Comments | ||||
|---|---|---|---|---|---|---|---|---|---|
| Species | Human sources | Disease associated | Species | Human sources | Disease associated | ||||
| Buttiauxella (1)/(6) | agrestis | − | − | No | brennerae | − | − | No | |
| ferragutiae | − | − | No | ||||||
| gaviniae | − | − | No | ||||||
| izardii | − | − | No | ||||||
| noackiae | + | − | No | ||||||
| warmboldiae | − | − | No | ||||||
| Cedecea (3)/(0) | davisae | + | − | No | Unnamed HGs | ||||
| lapagei | − | − | No | ||||||
| neteri | + | + | No | ||||||
| Citrobacter (3)/(12) | amalonaticus | + | + | No | braakii | + | + | No | |
| diversusc | + | + | Yes | cronae | + | − | No | ||
| freundiid | + | + | No | europaeus | + | − | No | ||
| farmeri | + | + | No | C. amalonaticus biogroup 1 | |||||
| gillenii | + | + | No | ||||||
| murliniae | + | + | No | ||||||
| pasteurii | + | − | No | ||||||
| portucalensis | − | − | No | ||||||
| rodentium | − | − | No | Mouse pathogen | |||||
| sedlakii | + | + | No | ||||||
| werkmanii | + | + | No | ||||||
| youngae | + | + | No | ||||||
| Edwardsiella (3)/(2) | hoshinae | − | − | No | anguillarum | − | − | No | Fish/eel pathogen |
| ictaluri | − | − | No | piscidia | − | − | No | Fish pathogen | |
| tarda | + | + | No | ||||||
| Enterobacter (8)/(34) | aerogenes | + | + | Yes | asburiae | + | + | No | |
| agglomerans | + | + | Yes | arachidis | − | − | Yes | ||
| amnigenus | + | − | Yes | bugandensis | + | + | No | ||
| cloacaed | + | − | No | chengduensise | + | + | No | ||
| gergoviae | + | + | Yes | chuandensise | + | + | No | ||
| intermedium | − | − | Yes | cowanii | + | + | Yes | ||
| sakazakii | + | + | Yes | dissolvens | + | + | Yes | ||
| tayloraef | + | + | Yes | helveticus | − | − | Yes | ||
| hormaechei | + | + | No | ||||||
| huaxensise | + | + | No | ||||||
| kobei | + | + | No | ||||||
| ludwigii | + | + | No | ||||||
| massiliensis | + | − | Yes | ||||||
| mori | − | − | No | ||||||
| muelleri | + | + | Yes | ||||||
| nimipressuralis | + | − | Yes | ||||||
| oligotrophicus | − | − | Yes | ||||||
| oryzae | − | − | Yes | ||||||
| oryzendophyticus | − | − | Yes | ||||||
| oriziphilus | − | − | Yes | ||||||
| pulveris | − | − | Yes | ||||||
| pyrinus | − | − | Yes | ||||||
| quasihormaechei | + | − | No | ||||||
| radicincitans | + | + | Yes | ||||||
| roggenkampii | + | − | No | ||||||
| sacchari | − | − | Yes | ||||||
| siamensis | − | − | No | ||||||
| sichuanensis | + | − | No | ||||||
| soli | − | − | No | ||||||
| tabachi | − | − | Yes | ||||||
| taylorae | + | + | Yes | ||||||
| turicensis | − | − | Yes | ||||||
| wuhouensis | + | − | No | ||||||
| xiangfangensis | − | − | Yes | ||||||
| Escherichia (5)/(2) | coli | + | + | Yes | albertii | + | + | No | |
| fergusonii | + | + | Yes | marmotae | − | − | No | ||
| hermannii | + | + | Yes | ||||||
| vulneris | + | + | No | ||||||
| blattae | − | − | No | ||||||
| Ewingella (1)/(0) | americana | + | + | No | |||||
| Klebsiella (6)/(13) | pneumoniae | + | + | No | aerogenes | + | + | No | E. aerogenes |
| oxytoca | + | No | africana | + | − | No | |||
| planticola | + | Yes | alba | − | − | Yes | |||
| ozaenaeg | + | Yes | grimontii | + | + | No | |||
| rhinoscleromatisg | + | Yes | huaxiensis | + | − | No | |||
| terrigena | Yes | indica | − | − | No | ||||
| michiganensis | + | + | No | ||||||
| ornithinolytica | + | + | Yes | ||||||
| pasteurii | + | − | No | ||||||
| quasipneumoniae | + | + | No | ||||||
| singaporensis | − | − | Yes | K. variicola | |||||
| spallanzanii | + | − | No | ||||||
| variicola | + | + | No | ||||||
| Kluyvera (2)/(2) | ascorbata | + | + | No | “cochleae” | − | − | Yes | K. intermedia |
| cryocrescens | + | + | No | georgiana | + | − | No | ||
| intermedia | + | + | No | ||||||
| Moellerella (1)/(0) | wisconsensis | No | |||||||
| Proteus (4)/(7) | mirabilis | + | + | No | alimentorum | − | − | No | |
| vulgaris | + | + | No | cibarius | − | − | No | ||
| penneri | + | No | cibi | − | − | No | |||
| myxofaciens | Yes | columbae | − | − | No | ||||
| faecis | + | − | No | ||||||
| hauseri | − | − | No | ||||||
| terrae | − | − | No | ||||||
| Providencia (4)/(6) | alcalifaciens | + | No | burhodogranariea | − | − | No | ||
| rettgeri | + | No | heimbachae | + | − | No | |||
| rustigianii | + | No | huaxiensis | + | − | No | |||
| stuartii | + | No | sneebia | − | − | No | |||
| thailandensis | − | − | No | ||||||
| vermicola | − | − | No | ||||||
| Serratia (7)/(12) | ficaria | + | No | aquatilis | − | − | No | ||
| fonticola | + | No | entomophila | − | − | No | |||
| liquefaciens | + | No | “glossinae” | + | + | Yes | S. fonticola | ||
| marcescens | + | No | grimesii | − | − | No | |||
| oderifera | + | No | inhibens | − | − | No | |||
| plymuthica | + | No | microhaemolytica | − | − | No | |||
| rubidaea | + | No | myotis | − | − | No | |||
| nematodiphila | − | − | No | ||||||
| oryzae | − | − | No | ||||||
| “quinivora” | + | + | Yes | S. quinovorans | |||||
| quinivorans | + | + | No | ||||||
| symbiotica | − | − | No | ||||||
| ureilytica | − | − | No | ||||||
| vespertilionis | − | − | No | ||||||
| Tatumella (1)/(5) | ptyseos | + | No | citrea | − | − | No | ||
| morbirosei | − | − | No | ||||||
| punctata | − | − | No | ||||||
| saanichensis | + | − | No | ||||||
| terrea | − | − | No | ||||||
1985 data are from Table 1 (11) regarding genera with addition of new species (post-1980); 2020 data are from the LPSN website (https://www.bacterio.net/) and include all new species validly published since the data of Farmer et al. (11). +, found in clinical samples and/or infections; −, found in nonclinical samples.
Numbers in parentheses are number of species in 1985/number of new species (2020).
Rejected name; identical to C. koseri.
Existed as a phenospecies (genetically heterogeneous).
Recovered from blood.
Correct name, Enterobacter cancerogenus.
Subspecies of K. pneumoniae.
The 1985 CDC data revealed 23 new species (∼40% of which were associated with one of six new genera, namely, Buttiauxella. Cedecea, Ewingella, Kluyvera, Moellerella, and Tatumella). Most of these species were recovered from clinical sources or infections (80%) and could be biochemically separated from all other named taxa in the family at that time. Furthermore, phenotypic properties for these species were based on the characterization of multiple strains of each taxon. The 49 total species listed in Table 3 for 1985 mushroomed to a total of 152 species in 2020 (one species, aerogenes, remained the same but was changed to a different genus). By 2020, 16 of the 49 species (33%) listed in 1985 have undergone taxonomic/classification revisions. Two genera, one that has always presented taxonomic issues (Enterobacter) and another thought to be a relatively uncomplicated genus (Klebsiella), had 87.5% and 67% of their respective species reclassified post-1985. In the case of Enterobacter, all but two species listed in 1985 now phylogenetically reside in other genera, including E. agglomerans (Pantoea), E. amnigenus (Lelliottia), E. intermedius (Kluyvera), E. gergoviae (Pluralibacter), E. aerogenes (Klebsiella), and E. sakazakii (Cronobacter) (57–61).
As of 2020, of the 14 genera listed, all but three (Cedecea, Ewingella, Moellerella) have had new members added to each genus, with the greatest number involving Enterobacter (n = 33). A noted difference in the 2020 proposed species is the increasing percentage of taxa (51/103, 49.5%) described from strictly environmental sources. This is a reflection not only of the general availability of WGS and newer systematic methods but also the fact that natural ecosystems (water, soil, plants) have not been extensively mined or explored for potential enterobacterial species. Additionally, many of these more recently described nomenspecies can no longer be identified by simple phenotypic characteristics. Additional classification changes (23.5%) between 1985 and 2020 have been due to phylogenetic investigations that resulted in the transfer of established taxa to new genera (homotypic synonyms). Prime examples include Kosakonia (Enterobacter spp.), Raoultella (Klebsiella spp.), and Tatumella (Pantoea spp.) (58, 62, 63).
Modern taxonomy and family expansion issues.
The changing panorama of the family Enterobacteriaceae in the modern molecular taxonomy era has resulted in a number of advantages along with some limitations as well as issues (55, 56, 64). A central issue for clinical microbiologists is the way new species are described. Already an observable trend dating back to the late 1990s, the vast majority of new species are described analyzing only a single strain (type). Today, >90% of new taxa are described on this basis (50, 65). Although some taxonomists have questioned whether or not a single strain should be sufficient to propose a new species (66) and ad hoc committees have encouraged microbiologists to describe new species on the basis of more than one strain (67), this has not happened. Already many of the recently described new genera and species in the family Enterobacteriaceae have been proposed with only a type strain. Recent examples in the enterobacteria include Mangrovibacter (68), Chania (69), Limnobaculum (70), and Scandinavium (71). While such reports are of general academic interest, the limited amount of information that can be provided, including genetic data, is troubling and whether the type strain is actually a centrist isolate (center of properties for the species) is unknown.
A second issue involves phenotypic properties. With phenotype long considered one of the cardinal features in the description of new species, its importance in regard to classification is diminishing (55). Many taxonomic proposals, whether involving a single isolate or a small number of strains, utilize primarily API miniaturized systems (20E, 32E, 50CH, ZYM) or carbon source utilization panels such as the Biolog Gen III MicroPlate for comprehensive biochemical characterizations of newly proposed groups (68–72). While these systems are generally accurate in bacterial identifications, miniaturized systems do not always parallel phenotypic properties generated by traditional methods (11) and some tests are subject to considerable variation. An additional quandary is the fact that when new species are proposed and compared to nearest neighbors, they are not tested in-house; instead, data are pulled from previous publications which may not have used the same test methodology. Such factors bring into question how reliable biochemical data may be, an important issue for diagnostic laboratories.
The general availability of WGS has led to a significant increase in laboratories attempting to become systematists overnight without understanding the fundamental rules of nomenclature and taxonomy. This has led to species proposal submissions without any recognition of technical requirements to formally propose a new taxon (67). A considerable number of recently proposed species have also been found to represent heterotypic synonyms, that is, different names and types that in the opinion of taxonomists represent the same taxon. Table 3 has a number of such examples, including the publication of Enterobacter muelleri in 2015 (73) that by computational analysis of sequenced Enterobacter genomes (74) is identical to Enterobacter asburiae, which was described in 1986 (75). Furthermore, many proposed genera for the family Enterobacteriaceae although effectively published (in a public journal easily accessible) have not been validly published (see Nomenclature and Taxonomy). Without validation, these genera and species have no standing in the medical and scientific literature. A prime example of this situation is the 2005 publication of the taxon Averyella dalhousiensis (76). This report details over 20 isolates, including one from a case of septicemia, but as of 2020 has no standing in the literature according to the LPSN website for one or more reasons. Many such genera proposed for the family are in the same situation and, if validated, would considerably increase the size of the family listed in Table 2.
NOMENCLATURE AND TAXONOMY
Taxonomy
Nomenclature and the ICNP.
The correct name and publication of new taxa within an ordered system of prokaryotes, including the family Enterobacteriaceae, are governed by the International Code of Nomenclature of Prokaryotes (ICNP), with the ranks of genus and species being the cornerstone for such a categorical classification system. In 2019, an update of the ICNP (2008 revision) was published for the first time in more than 25 years (77). General Consideration 2 essentially defines the purpose of the ICNP, which is “To achieve order in nomenclature, it is essential that scientific names be regulated by internationally accepted Rules” (77). The ICNP covers not only the correct naming of a genus and/or species but also the requirements to publish and validate a taxon, which constitutes an official form of registering or indexing a name through a centralized system (55). This includes such things as Enterobacter oligotrophicus (correct name) rather than Enterobacter oligotrophica (misspelling) and various homotypic (more than one name associated with the same types that belong to the same taxon) or heterotypic synonyms of previously published genera and species (Table 3).
Validation.
Rule 27 of the ICNP requires the publication of a new taxon either in the International Journal of Systematic and Evolutionary Microbiology (IJSEM) or effective publication in another journal with subsequent publication on a Validation List in IJSEM (78). Along with this are additional requirements concerning the deposition of type strains, derivation (etymology) of a new name, and others. A problematic issue related to this concerns the fact that almost 50% of all taxa between the ranks of subspecies and class that are effectively published in journals other than IJSEM are never validated (78). Reasons for this trend are not clear. For the family Enterobacteriaceae, in addition to Averyella (76), this includes the genera Atlantibacter, Edaphovirga, Jejubacter, Nissabacter, and Oceanomonas [sic] (https://lpsn.dsmz.de/family/enterobacteriaceae). Failure to validate creates confusion in the scientific and medical literature. As an example, the genus Atlantibacter was created in 2016 to reclassify two species (Escherichia hermannii and Salmonella subterranean, the latter of which is not a salmonella but an organism closely related to E. hermannii) to the new genus (79). Because of lack of validation, it is unclear whether the authors simply failed to submit the publication for validation to IJSEM or lacked the required components for validation in their publication or whether there were technical flaws in their analyses. Since E. hermannii is a known although uncommon human pathogen, this has clinical significance.
Nomenclature anomalies in the family Enterobacteriaceae.
With the emergence of modern-day metrics, including DDH and ANI, with which to assess or reassess relatedness among taxa in the family Enterobacteriaceae, a number of nomenclature issues which are of great importance to the medical community have arisen. As early as 1968, Brenner et al. (80) determined that E. coli and Shigella flexneri were more than 75% related by DDH when tested at 75°C, which is well above the threshold level of species identity of 70% reassociation with ≤5°C change in melting temperature (ΔTm) (81). These studies were subsequently confirmed for all Shigella species, which were 80% to 89% related to E. coli strains (51). Shigella spp. have evolved over time through a number of gene deletion or plasmid acquisition events but are still, on a genetic basis, the same species (82, 83). In a similar fashion, Yersinia pestis and Yersinia pseudotuberculosis are highly related by DDH and whole genome-to-genome sequence analysis (84, 85). Together they constitute a single species on a genetic basis. According to the rules of the ICPN (Principle 6), the earliest published name has priority over synonyms (77). In the case of Yersinia, Y. pseudotuberculosis (1889) has priority over Y. pestis (1896). Similarly, for Shigella, E. coli (1895) has precedence over Shigella dysenteriae (1897). However, while genetically a single species, the disease manifestations of shigellosis (versus E. coli) and plague (versus Y. pseudotuberculosis) have immense clinical, medical, and epidemiologic ramifications that would result from the merging of each group. However, Principle 1 of the ICPN requires the maintenance of the stability of names, particularly when the use of a name could cause error or confusion. So, these species remain as originally published because of their medical importance, although both constitute “artificial species” at the DNA level.
Classification.
Bacterial classification is the orderly arrangement of taxonomically defined entities (species) on a genetic basis (55). The present system is data driven and theoretically attempts to order such groupings on an evolutionary or phylogenetic basis mimicking nature (86). Like bacterial nomenclature, prokaryotic classification has relied on similar traits or characteristics to produce the present organizational structure consisting of a series of artificial hierarchical ranks above species (class, order, family, genus) (55). These traits or characteristics have, as in the case of bacterial nomenclature, evolved over time from phenotypic (morphology, physiology, chemotaxonomy) to genetic (DDH, MLSA, ANI) markers. An excellent minireview on the subject is by Schleifer (86). Unlike nomenclature, however, the 16S ribosomal gene provides the cornerstone to such a classification system and is viewed as the gold standard for bacterial phylogeny (55, 86). The 16S ribosomal gene and a limited number of other housekeeping genes found in essentially all prokaryotic species (recA, rpoB, EF-Tu, gyrA) together form the foundation for modern-day bacterial phylogeny (55, 86).
Unlike bacterial nomenclature, there is no formal body or international organization that is universally approved to oversee classification changes (1, 55, 86). Rather, the acceptance or rejection of classification changes is dependent upon the formal data presented and whether or not the classification proposal is widely accepted or rejected by the scientific community. This latter point means that it is basically decided by usage in the scientific and medical literature, which usually takes at least 2 or 3 years from date of publication (1). These collective facts mean that unlike the proposed name for a new taxon, a classification change never becomes “official” (1). A second relevant point is the fact that because there are no governing rules, any researcher may propose a classification change without any focus on taxonomy or without having been trained in the classification or identification of bacteria (87). This can lead to additional taxonomic confusion in an already confusing field. Finally, by its very nature, bacterial classification is always in a state of flux and never relatively stable. Classification changes can occur rapidly and can be based on the use of new or different technology, bacterial populations analyzed, and analytical or computational methods chosen. The family Enterobacteriaceae is an excellent example of this. The genus Metakosakonia was proposed by Alnajar and Gupta (88) in 2017 to house the taxon Enterobacter massiliensis based upon comparative genomic studies. Recently, a proposal has been made to unify the genus Metakosakonia with the genus Phytobacter, since Phytobacter is based upon digital DDH, average amino acid sequence identity, and conserved signature insertions/deletions (indels) (89). Other proposed taxonomic classification changes involve the genus Izhakiella (90), two subspecies of Klebsiella pneumoniae (91), and reassignment of Proteus cibarius as a later heterotypic synonym of Proteus terrae (92).
The Family Enterobacteriaceae—Current Status
Present definition for the family.
The vast majority of established genera and species presently included in the family Enterobacteriaceae, order “Enterobacterales,” have been recognized for over 50 years (15). Early taxonomic classifications at the rank of family relied on a number of common traits in addition to phenotypes and genetic characteristics. A perfect example of the use of this classification system was the family Vibrionaceae proposed by Véron in 1965 (93). This family was initially composed of three genera (Vibrio, Aeromonas, Plesiomonas) that had several features in common, including ecologic habitats (freshwater, marine), similar disease syndromes (gastroenteritis, wound infections), and phenotypic features (oxidase positivity, facultatively anaerobic). It was not until years later that phylogenetic investigations clearly demonstrated that aeromonads belonged in their own family (94) while Plesiomonas should be transferred to the family Enterobacteriaceae (14). Despite these technologic advances, clinical microbiologists continue to think of these genera as a single cohort because of clinical histories (water exposure) and common cultural and biochemical properties.
The family Enterobacteriaceae can be thought of in a similar fashion. Members added to this group over the years share a number of common features, including phenotypes (oxidase negative, ECA positive, nitrate reductase), habitats (gastrointestinal tract of vertebrates, including humans), and disease patterns (diarrhea, sepsis, urinary tract infections) (1–3, 15). They are also recovered from clinical samples on a variety of common selective and differential agars used in the laboratory for decades. Some of these groupings within the family have stood the test of time. For instance, the tribe Proteeae, which was named by Castellani and Chalmers in 1918 and subsequently contained the genera Proteus, Providencia, and Morganella (https://lpsn.dsmz.de/tribe/proteeae), still group together today on a phylogenetic level. By 2005, the inclusion of Plesiomonas (oxidase positive) in the family along with endosymbionts of insects and other fastidious species has made a literal description of the Enterobacteriaceae difficult (15). Furthermore, while many new enteric genera with more unusual properties have been described over the past 20 years, microbiologists again still think of the main members of this family in a fashion similar to those originally included in the Vibrionaceae (95).
Core and peripheral members.
The present definition for the family became more uncertain with the introduction of DDH (51). Using DDH as the criterion, DNA from members of the family exhibited at least 20% relatedness to the type species, E. coli (15). However, in the 2005 edition of Bergey’s Manual of Systematic Bacteriology, noted exceptions to this rule included the Proteeae, Hafnia, and Edwardsiella, which were only 5% to 20% related (15). Core members were defined as enterobacteria that were 40% to 50% related to E. coli. This included such genera as Enterobacter, Klebsiella, Citrobacter, and Salmonella (96). Some other genera were intermediate in their DNA relatedness values (25% to 30%) compared to core and peripheral groups. Taxa including Cedecea, Kluyvera, and Serratia showed this intermediate level of relatedness (96).
Phylogenetic studies.
There have been surprisingly few phylogenetic investigations in which a large collection of enterobacterial genera and species have been analyzed in any great detail. Most of these studies have involved the analysis of one or more housekeeping genes such as the 16S gene. Unfortunately, in regard to the Enterobacteriaceae, 16S rRNA gene sequencing has low discriminatory power, particularly in reference to closely related species (48, 55, 97, 98). Housekeeping genes used to assess phylogenetic relatedness within the family with higher resolving power than the 16S gene include elongation factor Tu (tuf), the F-ATPase β-subunit gene (atpD), the DNase gyrase subunit B gene (gyrB), and the chromosomal replication origin (oriC), among others (97, 99–102).
Notwithstanding the fact that different genes and taxa have been analyzed and assessed in various publications, several basic conclusions can be drawn from these collective phylogenetic studies of the family. First, in general, neighbor-joining phylogenetic trees constructed from housekeeping gene sequence analysis support previous DDH studies on the position of genera relative to the deep-rooted core members of the family, including E. coli. The genera least related to core members and those located at the periphery of dendritic trees include Plesiomonas, Xenorhabdus/Photorhabdus, the Proteeae, Yersinia, and Serratia (97, 99, 102). Second, the family displays polyphyletic branching (98). While some of the cladistic discrepancies previously noted in the family can be resolved by recent classification changes proposed for certain species, others cannot. Thus, the present family appears to contain a number of taxa whose lineage is distinct from core members.
Species or taxa of uncertain status.
There are a number of nomenspecies or taxonomic groups at the genus and/or species rank that have nomenclature or classification issues at present.
(i) Salmonella species.
The genus Salmonella is extremely complex on an evolutionary basis (103). Currently, three Salmonella spp. are validly published as correct names (https://lpsn.dsmz.de/genus/salmonella). One of these species, Salmonella subterranea, was proposed in 2004 for an acid-resistant bacterium recovered from subsurface sediment (104). A recent study including 16S rRNA gene sequencing, MLSA of four housekeeping genes, and concatenated hypervariable sequences of 10 housekeeping proteins indicate that this species aligns with Escherichia hermannii into a separate clade distinct from both Salmonella and Escherichia (79). The authors have proposed to transfer both species to a new genus, “Atlantibacter” (79). However, at the time of writing, this genus still has not been validated, so both species remain in their originally assigned genera. Other phylogenetic investigations suggest that two subspecies of Salmonella enterica (salamae, houtenae) are polyphyletic in nature, being composed of at least two distinct phylogroups (103). This suggests that more taxa are yet to be described.
(ii) Enterobacter and Enterobacter hormaechei.
Enterobacter hormaechei, previously referred to as enteric group 75, was originally part of the E. cloacae complex (105). Population studies suggest that E. hormaechei may be the predominant species within the E. cloacae complex causing HAI (74). The LPSN website presently lists five validated subspecies (https://lpsn.dsmz.de/species/enterobacter-hormaechei). E. hormaechei is presently in “taxonomic chaos” and disarray. Using WGS, Sutton and others (74) have proposed that two subspecies of E. hormaechei (subsp. hoffmannii and subsp. xiangfangensis) are heterotypic synonyms of a separate species, E. asburiae. An entirely different conclusion using similar techniques has just been drawn by Wu et al. (106). In their investigations, the authors propose the following: (i) that E. hormaechei subsp. hoffmannii should be elevated to species status and not assigned to E. asburiae (74); (ii) that all present subspecies assignments in the genus Enterobacter are incorrect; (iii) that many Enterobacter genome sequences in GenBank need to be corrected and curated; and (iv) at least 14 tentative novel species have been detected in the genus (106). Other investigators have also identified new lineages representing novel species within E. hormaechei (107). If the majority of findings from Wu et al. (106) are confirmed, it throws the genus Enterobacter into total confusion from a clinical and medical perspective in regard to terminology, nomenclature, epidemiology, and disease pathogenesis.
(iii) Yersinia ruckeri.
Yersinia ruckeri has a very checkered taxonomic history. This bacterium is one infectious cause of redmouth disease in rainbow trout (108) and has also been recovered from a human wound infection (109). The taxon was initially placed in the genus Yersinia based upon mol% G+C content and biochemical similarities; however, considerable data suggest that it is not a true yersinia. DNA relatedness investigations indicate that it is 30% related to both Yersinia and Serratia, while early studies suggest that it biochemically resembles Serratia and Salmonella (“Arizona”) (108, 110). Phylogenetic studies constructing a multilocus enzyme electrophoresis dendrogram (110) and a 16S RNA maximum-likelihood tree (111) place this taxon at the extreme periphery (distantly related) of Yersinia, with the cumulative data implying that Y. ruckeri should not be classified within the genus.
(iv) Edwardsiella tarda biogroup 1.
The vernacular name Edwardsiella tarda biogroup 1 was coined by the CDC in 1985 for a biochemically atypical group of E. tarda isolates (11). In contrast to classic strains, the atypical isolates were sucrose positive and H2S negative. All of the original isolates identified by the Japanese were from snakes (112). Since this report, a biogroup 1 strain has been associated with human cases of cholelithiasis (113) and sepsis (114). DDH indicates that the biogroup 1 strains are 69% to 74% related to the type strain of E. tarda (112). Phylogenetic investigations have not been conducted to date. It may be that these unusual isolates are simply a biotype of E. tarda or that they may represent a new subspecies unless phylogenetic investigations indicate otherwise.
(v) Enteric groups, HGs, and related taxa.
There are many apparently distinct taxa residing in the family Enterobacteriaceae that have yet to be named but have been identified on the basis of both phenotypic and genotypic properties. The CDC has given a number of these groups the general designation of “enteric group,” followed by a corresponding number, such as enteric group 17 (1, 11). Table 4 lists some of these groups that have been described in the literature with their present standing.
TABLE 4.
Present status of CDC enteric groupsa
| Enteric group | Phenotype | Human source(s)b | Current designation | Reference(s) |
|---|---|---|---|---|
| 17 | Atypical Citrobacter or Enterobacter | bld, wd, rt, urn | Enterobacter asburiae | 75 |
| 45 | Atypical “Hafnia-like” | bld, wd, stl | Yokenella regensburgei (“Koserella trabulsii”) | 45, 277 |
| 57 | H2S+ bacteria | stl | Leminorella | 278 |
| 58 | wd | Averyella dalhouensis | 77 | |
| 59 | ADH+ “E. agglomerans-like” | rt | Buttiauxella noackiae | 279 |
| 60 | Inactive group | urn, rt | Unnamed | 1, 11 |
| 63 | Buttiauxella | None | Unnamed | 1, 11 |
| 64 | Buttiauxella | None | Unnamed | 1, 11 |
| 68 | DNase+ | urn | Unnamed | 1, 11 |
| 69 | “Enterobacter sakazakii-like” | bld | Unnamed | 1, 11, 280 |
| 137 | “Citrobacter farmeri” or “Citrobacter amalonaticus-like) | rt, wd, urn | Unnamed | 1, 281 |
In addition to numbered enteric groups, other nomenspecies are known to contain HGs that have not been characterized to date. This includes Cedecea (genomospecies 3 and 5), Ewingella (strain 0679-79), and Kluyvera (genomospecies 3), among others (11, 115).
Cladistic Analysis, and Phylogenetic Investigations
A polyphyletic family.
Bacterial taxonomy involves three important components, including identification, nomenclature, and classification (97). Classification involves the orderly arrangement of genetically similar organisms into a related evolutionary group or clade (cladistic analysis). Past classification of members of the family Enterobacteriaceae were primarily restricted to phenotypic properties, DNA relatedness studies, and 16S rRNA gene sequence studies.
Past phylogenetic findings have augmented earlier DNA relatedness reports, indicating that there is extensive evolutionary divergence within the family as presently constructed and defined (see “Phylogenetic studies” above). These phylogenetic investigations, however, were limited by the number of housekeeping genes investigated and the number of taxa (genera and species) analyzed. In a landmark 2016 publication by Adeolu and associates (98), the authors constructed three phylogenetic trees based upon 1,548 core proteins, 53 ribosomal proteins, and 4 MLSAs, respectively, for 179 whole genomes of members representing 49 validated taxa of the order “Enterobacterales.” These collective trees indicated that the order could be broken down into seven monophyletic groups at the rank of family. Only 3 of the 49 genera evaluated gave atypical results (Buchnera, Plesiomonas, Wigglesworthia). Furthermore, a series of indel signatures were detected such that five conserved signature indels (CSIs) were order specific, while an additional 66 CSIs were family or clade specific. A follow-up investigation by Alnajar and Gupta (88) investigating only members of a potentially redefined family Enterobacteriaceae found that 78 genome-sequenced species could be subdivided into six well-circumscribed subfamilies, with a seventh termed “Enterobacteriaceae incertae sedis clade” containing Mangrovibacter and Shimwellia (88). Again, in this study, a number of CSIs that were specific to one or more groups were identified.
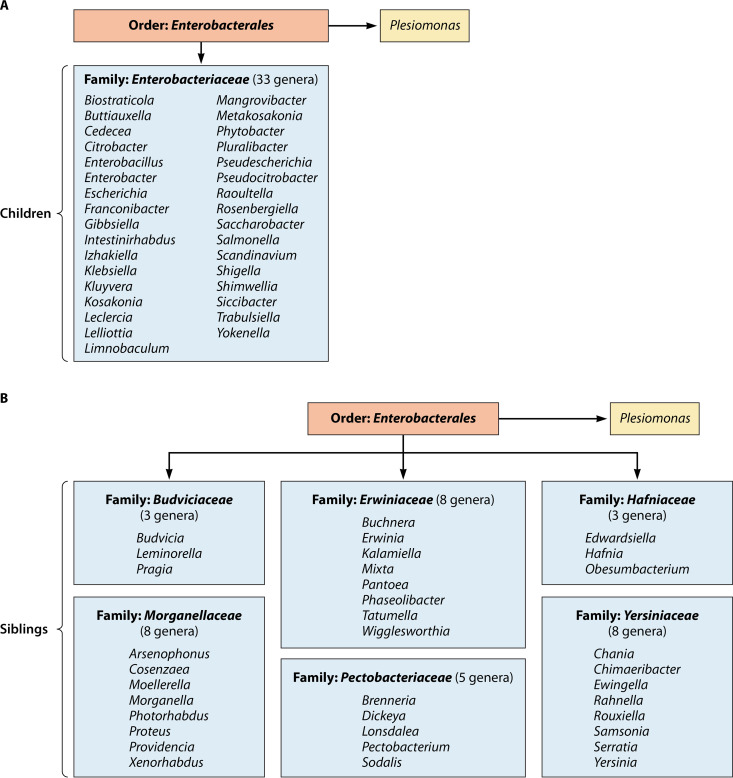
Proposal to reclassify members of the order “Enterobacterales.”
Based upon a significant amount of phylogenetic information in the literature, Adeolu et al. (98) have made a number of classification proposals for members previously assigned to the family Enterobacteriaceae. In addition to proposing the name “Enterobacterales” to replace the previous order name “Enterobacteriales,” which has never been validated, the authors propose placing some current family members into six newly created families while leaving other genera in the family Enterobacteriaceae. These six new families include the Erwiniaceae, Pectobacteriaceae, Yersiniaceae, Hafniaceae, Morganellaceae, and Budiviciaceae, which exhibit >60% genome-to-genome relatedness (98). Figure 1 illustrates how genera would be assigned and reclassified within the order “Enterobacterales” if formally accepted and approved. The LPSN website has already adopted these classification changes (Aidan C. Parte, Curator, Leibniz Institute DSMZ). Figure 1A illustrates what genera would still remain within the family Enterobacteriaceae, excluding nonvalidated genera. The family Enterobacteriaceae with 33 genera would still be the largest family within the order “Enterobacterales,” although many genera with long-standing associations with this family, such as Edwardsiella, Hafnia, Morganella, Proteus, Providencia, Serratia, and Yersinia, would be transferred into one of six new families (Fig. 1B). Plesiomonas, located in the top right of Fig. 1A, appears there because it belongs to the order but has not been assigned to a specific family as of this date.
FIG 1.
Proposed classification of current members of the family Enterobacteriaceae according to Adeolu et al. (98). (A) Revised family Enterobacteriaceae; (B) six newly proposed families for inclusion in the order Enterobacterales.
Figure 1B depicts the remaining six proposed families, all of which are much smaller in size than the Enterobacteriaceae. A number of new terms are used in addressing the phylogenetic relationships of members of this order at the LPSN website. “Children” refers to a taxon belonging to a hierarchical classification above it, such as the genus Salmonella being one of a number of “children” of the family Enterobacteriaceae. “Siblings” refers to other genera in the same family (Fig. 1B), such as Proteus and Providencia being siblings (along with others) in the proposed family Morganellaceae (Aidan C. Parte, personal communication). The family Thorselliaceae, which currently contains two genera, has been listed as a member of the “Enterobacterales” (Enterobacteriales). However, although a member of the Gammaproteobacteria, it has not as of this date been assigned to an order (95, 116).
Clinical and public health implications of proposed taxonomic changes.
The combined phylogenetic studies of both Adeolu et al. (98) and Alnajar and Gupta (88) clearly highlight numerous taxonomic and classification flaws within the Enterobacteriaceae on an evolutionary basis. Whether these proposed classification changes will be generally accepted by the medical and scientific communities remains to be seen. The same group has previously proposed sweeping changes to the genus Mycobacterium (117) and creation of a new order, families, and genera currently containing Ureaplasma and other genera (118). Both of these proposals have met with considerable resistance from both august subcommittees of the ICSP and an international group of medical and scientific professionals and researchers (119, 120). Reasons for the proposed rejections include violation of one or more rules of the ICNP and confusion for health care and harm for patients.
In a similar fashion, there is concern regarding how such proposals might affect the family Enterobacteriaceae, in particular in regard to the medical community and clinical microbiologists. If accepted, archival data accumulated for decades on the family would not be directly comparable to future research studies of the Enterobacteriaceae as redefined. This could conceivably impact reports on virulence factors, pathogenicity, and clinical studies involving isolation, identification, and susceptibility profiles of organisms such as carbapenem-resistant Enterobacteriaceae (34). Epidemiologic investigations might also be impacted. As an example, FoodNet foodborne disease surveillance data currently include four species within the family (Salmonella, Shigella, Shiga toxin-producing E. coli, Yersinia). If the proposal is subsequently approved, Yersinia would be removed from the Enterobacteriaceae, even though the four foodborne disease agents exhibit many properties in common. A similar impact could be seen for HAI that currently include Proteus and Serratia as enterobacteria (https://www.cdc.gov/nhsn/datastat/index.html). While most of the changes proposed above do not involve genus or species designations, classic references and texts would need to be revised. More importantly, continued upheaval in nomenclature and taxonomy involving the fields of medicine and clinical microbiology leads to suggestions to completely disregard new taxonomy (118), a viewpoint that is counterproductive to both the medical and scientific communities.
ENTEROBACTERIACEAE—ENVIRONMENTAL DISTRIBUTION
Extension in Ecohabitats of Enterobacteria
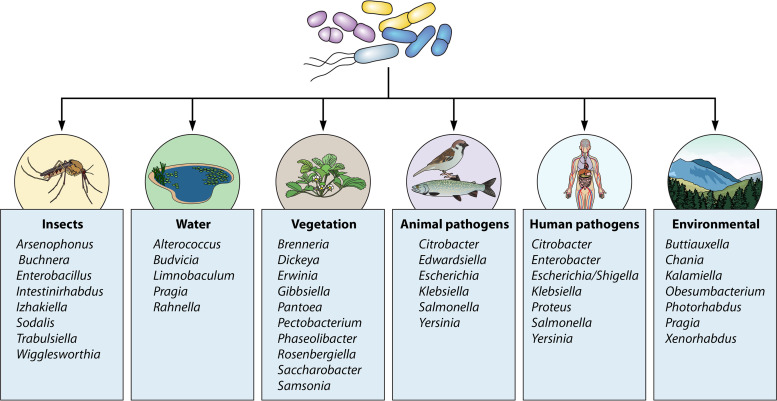
One of the less well appreciated aspects of the family is the increasing detection of members in an expanding panorama of environmental niches and ecosystems. The majority of these ecohabitats can be broken down into several major categories, with the predominant genera depicted in Fig. 2. These include the gastrointestinal tracts of vertebrates (humans, animals), vegetation (plants, trees, fruit, flowering ornamentals, grains), insects (including endosymbionts), and aquatic habitats such as freshwater, thermal springs, and marine or saline sources (1, 2, 15). A catch-all category for lack of a better term has been coined “miscellaneous,” which include such reservoirs as soil (landfill), shellfish, nematodes, and the International Space Station (69, 121). None of these categories occur as “silos”; rather, these ecohabitats coexist together in various symbiotic relationships, such as the rhizosphere and submerged aquatic vegetation as two examples (122, 123).
FIG 2.
Categorical representation of key enterobacterial genera associated with various ecosystems. (Adapted from reference 2.)
Selected Ecosystems
Vegetation.
Depending upon the genus and species, plant-associated members can exist in a number of different states associated with various vegetations (fig. 2). These include intimate relationships with plant species (epiphyte), within the internal structures of a plant (endophyte), or as a symbiont, a saprophyte, or a pathogen (124). Of these states, the one of cardinal importance and concern is the role of enterobacterial species as pathogens of agricultural produce, ornamental flowers, and trees. There are basically four types of plant diseases or infections associated with enterobacteria. These include (i) rapid necrosis, (ii) progressive tissue maceration (soft rot), (iii) occlusion of vascular vessels (wilt), and (iv) hypertrophy leading to gall, tumor, or canker formation (125).
By far, the plant disease that has received the greatest attention due to its impact is soft rot, which is most frequently associated with species of two genera, Pectobacterium and Dickeya (5, 126–128). Both genera secrete a number of extracellular enzymes, including pectinases, which degrade pectin-containing structures in plant cell walls, leading to wet rot of storage organs such as tubers and bulbs (128). Data from the early 1980s indicate that at that time, the economic loss from soft rot ranges from $50 to $100 million per annum (5, 15). The disease can be sporadic in nature or devastating, attacking either growing fields or storage units containing beets, corn, and lettuce. Soft rot affects only vegetables and ornamental plant production.
Another important soft rot infection is blackleg disease of potatoes. This infection, caused by both Dickeya and Pectobacterium species, but in particular P. atrosepticum, occurs worldwide, including New York and the northeastern United States. (129). Blackleg primarily manifests itself by attacking the stems of potatoes after initial growth in seed tubers. In its more severe form, the stems become darkened and decay, causing the “blackleg” appearance (128, 130). One study suggests that Dickeya alone can be responsible for a 20% to 25% reduction in potato yields (5).
Fire blight is a devastating necrogenic disease of pome fruit trees, particularly pears and apples (126, 130, 131). The etiologic agent is Erwinia amylovora. Fire blight occurs worldwide but is more associated with young trees (<2 years) growing in temperate climates (126). In 2003 alone, fire blight was estimated to cause more than $100 million annual loss to the fruit industry in the United States (131), and massive national pandemics involving pear trees have occurred multiple times in Israel (132). The disease progresses from the epiphytic state associated with cankers, where it can subsequently be released, multiply, and attack various parts of the tree, including flowers, leaves, branches, and roots (131, 132). Fire blight most often manifests itself in a burnt-like appearance on infected tissues along with wilting and watermark (132). Currently there is no cure for this as well as other enterobacterial plant-associated diseases.
Several other enterobacterial plant-associated diseases bear special mention (133). Acute oak decline (AOD) is a relatively new disease affecting oaks in southeast England and the midlands bordering Wales. AOD is characterized by weeping lesions on oak trees, dark fluid seeping from cracks in the outer bark, and irregularly shaped lesions in the inner bark (133, 134). While the microbial flora of AOD is complex, two predominant bacteria are thought to play important roles in the disease process, namely, Brenneria goodwinii and Gibbsiella quercinecans (134). Severely affected trees can die within 4 to 5 years. Both Pantoea agglomerans and Pantoea ananatis cause a variety of diseases in fruit, vegetables, and grains (124, 135). P. agglomerans, which causes boll rot in cotton, has been reported to be responsible for 10% to 15% of annual crop losses.
Table 5 lists some of the key plant diseases linked to enterobacterial species. There are a number of excellent reviews that cover various aspects of plant diseases concerning the etiologic agents, taxonomy, susceptible hosts, disease manifestations, pathogenesis, and control and remediation processes (128, 130, 135–138).
TABLE 5.
Key enterobacterial species associated with plant diseases
| Pathogen | Disease | Type of infectionb | Hostc | Reference(s) |
|---|---|---|---|---|
| Brenneria alni | Canker | 1 | Alder (Alnus spp.) | 125, 136 |
| Brenneria nigrifluens | Canker | 1 | Walnut (Juglans regia) | 125 |
| Brenneria populi | Canker | 1 | Poplar (Populus euramericana) | 282 |
| Brenneria roseae | Acute oak disease | 1 | Oak (Quercus robur, Quercus petraea) | 283 |
| Brenneria rubrifaciens | Canker | 1 | Walnut (Juglans regia) | 125 |
| Brenneria salicis | Wilt, watermark | 3 | Willows (Salix spp.) | 125, 284, 285 |
| Dickeya chrysanthemi | Wilt, soft rot | 2, 3 | Bananas (Musa spp.), maize (Zea mays), chrysanthemum (Chrysanthemum spp.), orchids (Vanda spp.) | 128, 136, 286, 287 |
| Dickeya dadantiia | Blackleg | 2 | Potato (Solanum tuberosum), African violets (Saintpaulia ionantha) | 126 |
| Dickeya dianthicola | Blackleg | 2 | Potato (Solanum tuberosum) | 129 |
| Dickeya paradisciaca | Root rot | 1 | Bananas (Musa spp.) | 125, 136, 288 |
| Dickeya solania | Blackleg | 2 | Potato (Solanum tuberosum) | 126, 128 |
| Dickeya zeae | Bacterial stalk rot, rice foot rot, soft rot | 2 | Bananas (Musa spp.), maize (Zea mays), rice (Oryza sativa) | 128, 289, 290 |
| Erwinia amylovoraa | Fire blight, wilt | 1 | Apples (Malus domestica), pears (Pyrus communis), other fruit | 125, 126, 131 |
| Erwinia mallotivora | Dieback disease, black leaf spot | 1 | Papaya (Carica papaya) | 125, 291, 292 |
| Erwinia tracheiphila | Cucumber wilt | 3 | Cucumber (Cucumis sativus), squash (Cucurbita spp.), pumpkins (Cucurbita spp.), gourds (Cucurbita pepo) | 125, 293 |
| Lonsdalea quercina | Drippy blight | 2 | Coast live oak (Quercus agrifolia) | 125, 155 |
| Pantoea agglomerans | Blight, boll rot, center rot, gall formation, wilt | 1, 2, 3, 4 | Beet (Beta vulgans), cotton (Gossypium hirsutum), maize (Zea mays), onion (Allium cepa), wisteria (Wisteria) | 135, 137 |
| Pantoea ananatis | Brown spot, brown stalk rot, center rot | 1, 2 | Cantaloupe (Cucumis melo), honeydew melons (Cucumis melo), maize (Zea mays), onion (Allium cepa), rice (Oryza sativa) | 124, 135, 137 |
| Pantoea stewartii | Leaf blight, Stewart’s wilt | 3 | Cotton (Gossypium hirsutum), eucalyptus, maize (Zea mays) | 135 |
| Pectobacterium atrosepticuma | Blackleg | 2 | Potato (Solanum tuberosum) | 125, 128, 129 |
| Pectobacterium carotovoruma | Soft rot | 2 | African violets (Saintpaulia ionantha), cucumber (Cucumis sativus), lettuce (Lactuca sativa), okra (Abelmoschus esculentus), potato (Solanum tuberosum), sugar beets (Beta vulgans), watermelon (Citrellus lanatus) | 125, 126, 128 |
One of the top 10 plant pathogens according to Mansfield et al. (126).
Types: 1, necrosis; 2, maceration; 3, vascular wilt; 4, gall or tumor formation.
Selected hosts.
Insects.
Insects collectively comprise the largest number of genera and species on planet Earth and therefore exhibit the greatest diversity phylogenetically as well as in total biomass and environmental distribution (139). While insects provide a number of beneficial effects globally, including pollination, pest control, bioremediation, and saprophytic “recycling,” they can also have deleterious effects, such as the destruction of agricultural crops and deforestation (see “Vegetation” above). Very few studies to date have looked at other roles insects may play in the biosphere.
Enterobacteria are carried by a wide range of insects, including flies, moths, and cockroaches. The two most extensively investigated species are the house fly (Musca domestica) and blow flies (Chrysomya megacephala, Protophormia terraenovae) (140–142). Studies of insects collected from various sites (farms, dairies, kennels, fresh food markets, restaurants, garbage piles) all show a number of common findings. These include the following: (i) regardless of the geographic region (Indian subcontinent, Southeast Asia, Africa), all studies exhibit high enterobacterial positivity rates from samples analyzed, typically ranging between 20% and 70% (142); (ii) the predominant species identified in most studies are Escherichia coli and Klebsiella pneumoniae (140, 142); (iii) isolates from a variety of genera have been recovered, including Citrobacter, Enterobacter, and the Proteeae, among others (140, 142); and (iv) many recognized human pathogens have been identified, including Shiga toxin-producing E. coli, Salmonella species including Salmonella enterica serovar Typhi, Shigella, and Y. enterocolitica (140, 142–144). Members of the family Enterobacteriaceae have also been found to be predominant flora in flying insects of the order Diptera recovered from British hospitals (145). Together, these data suggest that these insects might be important vectors for foodborne or health care-associated infections.
Cockroaches are an omnipresent insect group that inhabit many different niches, including the hospital environment (146, 147). Some historical data suggest a role for cockroaches in both human and health care-associated infections. However, much of these data predate modern taxonomy, laboratory techniques, and epidemiologic methods (147). Cockroaches have also been implicated as potential pathogens of consumable products due to the recovery of many foodborne pathogens from this potential vector, including Salmonella, Shigella, and E. coli O157:H7 (148, 149). The German cockroach (Blattella germanica) has been studied by several investigators, and results are surprisingly similar to those from studies on house and blow flies (146, 149). These cumulative results show that enterobacterial genera and species are commonly found in B. germanica, including food-associated pathogens, and as with flies, the principal species isolated are E. coli and K. pneumoniae.
(i) Insects as mechanical vectors.
Insects play an important role in the perpetuation of vegetative diseases by transmission of many viral agents from infected to uninfected plants (150). It is likely that insects serve as vectors of bacterium-associated plant diseases in a fashion similar to that of viruses, although to what extent is not completely understood. Phytopathogens and their associations with insects can have narrow or broad host specificity, may exist in one of several symbiotic states, and can vary geographically based upon environmental factors like climate, moisture, and vegetation (151). The pea aphid (Acyrthosiphon pisum) can serve as host to several phytopathogens, including Dickeya dadantii, Pantoea stewartii, and Erwinia aphidicola, which cause disease in potatoes, maize, peas, and beans (151). P. atrosepticum has been isolated from trapped insects and symptomatic potato plants exhibiting soft rot, as have turnip root flies, cabbage moths, and green lacewing larvae (152). In contrast, P. stewartii has a very restricted association with the corn flea beetle (Chaetocnema pulicaria) in causing Stewart’s wilt (leaf blight) of maize (151, 152).
Many other insect vectors have been involved in the transmission of plant diseases. The green stink bug (Nezara viridula) can transmit P. agglomerans to cotton, precipitating boll rot (135, 153). Center rot of sweet onions, which produces bacterial stalk and leaf necrosis, is caused by P. agglomerans (135). The probable vector of this disease is onion thrips (Thrips tabaci) (135, 154). Drippy blight disease of red oaks caused by Lonsdalea quercina is associated with a kermes scale insect, Allokermes galliformis (155).
An excellent article on a variety of alternative and direct insect hosts of bacterial phytopathogens including the Enterobacteriaceae is the review by Nadarasah and Stavrinides (151).
(ii) Edible insects.
Since 2013, when the Food and Agricultural Organization of the United Nations issued an edible insects document, a concerted international effort has been under way to investigate the usefulness of insects as a potential edible and nutritional food source (156, 157). The impetus for this directive concerns the increasing global population (estimated at 9 billion), the decreasing acreage of agricultural lands due to climate change, limited availability of water resources, and the projected need to double food production by 2050 (157). Edible insects are being strongly looked at as a “novel food” potentially rich in protein, lipids, fiber, and micronutrients to help fill the expected void (158). Of the myriad of insect species on the planet, those eliciting the most general interest include the mealworn (Tenebrio molitor), grasshoppers (Locusta migrans), cockroaches (Blattodea), and house crickets (Acheta domesticus) (158, 159).
Entomophagy, or the practice of consuming insects, has been practiced by many nations throughout Africa, Asia, Latin America, and Oceania for countless years. It is estimated that 113 countries consume 2,000 distinct species of insects as food sources (157). The consumption of edible insects in the future, if expanded, raises a number of questions and issues, including acceptability of such food sources by western cultures, technical production, and regulations that include food safety. Part of the food safety risk assessment concerns potential human illnesses related to biologic and chemical agents (158). Many different microbial species, including bacteria and parasites, have been isolated from most insect species studied (158–160). These include the family Enterobacteriaceae and many common genera, such as Escherichia, Klebsiella, Enterobacter, Salmonella, Proteus, and Pantoea (158, 160). Concentrations of enterobacteria vary by family or genus but range from <1 to >7 log CFU/g (158). Some studies have focused on the detection of specific human pathogens, such as Cronobacter (161), in ready-to-eat edible insects, although C. sakazakii species identification is questionable.
An outstanding and comprehensive publication by Garofalo et al. (158) reviews studies conducted between 2000 and 2019 on the isolation of microbial pathogens from different insect species. Data tabulated include their relative load (microbial burden), sources (market, wild), and the insect form analyzed (fresh, processed).
Animals.
Comparable to plants and insects, animals have an intimate association with genera in the family Enterobacteriaceae. These associations can be as commensals or occasionally as pathogens for a number of vertebrate species (2). Salmonella as an example is a natural commensal of reptiles and amphibians but can also be introduced into poultry stock, where it can live asymptomatically or on occasion cause disease. Likewise, E. coli (pathogenic and nonpathogenic types) is another example, living symbiotically in cattle, deer, and other animals but can be introduced through fecal contamination into consumable products such as ground beef, fruit, melons, and sprouts (2). Surprisingly, there have been few comprehensive studies looking at the distribution of Enterobacteriaceae in mammals, for example, with the exception of the study by Gordon and FitzGibbon in 1999 (162). In that investigation, the authors isolated enterobacteria from 642 mammalian hosts in Australia and calculated the relative abundance and diversity of genera and species.
An increasing but less-well-documented aspect of the enterobacterial group is the potentially increasing importance of family members in causing animal diseases. Many traditional taxa within the family produce sporadic or episodic intestinal or extraintestinal infections in numerous different hosts (1, 2, 15). However, new agent-disease associations are consistently being described, such as Morganella morganii causing fatal infections in chickens (163) and hypermucoid K. pneumoniae producing sepsis and meningitis in sea lion pups (164). What is less well appreciated is the increasing association of various taxa with specific infectious disease syndromes, as shown in Table 6. While some disease syndromes have a very specific host range (C. rodentium and murine colonic hyperplasia), others like bovine mastitis are caused by many different microbes in addition to E. coli and K. pneumoniae. Collectively, the group listed in Table 6 can cause large outbreaks of disease, the listed syndromes are recognized worldwide, and infections result in huge financial losses, particularly in the food industry such as aquaculture systems.
TABLE 6.
Selected examples of enterobacteria-associated disease syndromes in animals
| Pathogen | Host | Disease | Symptoms | Reference(s) |
|---|---|---|---|---|
| Citrobacter rodentium | Mouse | Transmissible murine colonic hyperplasia | Thickened colon, rectal prolapse, diarrhea | 294 |
| Edwardsiella ictaluria | Channel catfish (farmed, tilapia) | Enteric septicemia | Petechial hemorrhages (mouth), pale gills, exophthalmia, multiple epithelial lesions | 295 |
| Edwardsiella piscicida | Fish (farmed, aquaculture) | Edwardsiellosis | Loss of pigmentation, external hemorrhage, septicemia | 4, 296 |
| Escherichia coli | Cow | Bovine mastitis | Inflammation of mammary gland; abnormal milk from a mammary quarter | 297 |
| Escherichia colia | Broiler chicken | Colibacillosis | Respiratory tract syndrome; swollen head syndrome | 298 |
| Escherichia colib | Pigs | Porcine diarrhea, postweaning diarrhea | Profuse diarrhea, dehydration, death | 299, 300 |
| Klebsiella pneumoniae | Cow | Bovine mastitis | Inflammation of mammary gland; abnormal milk from a mammary quarter | 297, 301 |
| Yersinia ruckeri | Fish (salmonid) | Enteric redmouth disease | Darkening of skin, subcutaneous hemorrhages (mouth, throat), internal petechial hemorrhages | 302 |
Avian-pathogenic E. coli strains.
Enterotoxigenic and enteropathogenic E. coli.
Soil and water.
The microbial ecology of soil and water is extremely complex. Sewage, sewage sludges, animal excreta, manure, contaminated agricultural products, and runoff can result in the contamination of soil and freshwater sources with enteric bacilli (165). Direct studies linking contaminated soil such as farm soil to human or animal infections is lacking (165). Compounding this problem is the fact that pathogenic enterobacteria released into soil may not simply colonize resident plants transiently but may develop a longer-term relationship with them as alternative hosts (166). Plants could then act as hosts for enteric species when existing as aquatic vegetation in freshwater habitats, including lakes (123).
ENTEROBACTERIACEAE—OLD AND NEW AGENTS IN NEW DISEASE SETTINGS
Overview
Advances in molecular techniques and phylogenetic methods over the past 30 years have resulted in a dramatic transformation in the size and scope of the family Enterobacteriaceae in terms of the number and type of taxa identified, environmental distribution, and disease associations. These two events, modernization of bacterial taxonomy and phylogenetic classification, coupled with better laboratory methods to achieve a final bacterial identification (16S gene sequencing, MALDI-TOF, WGS) have revolutionized the field of clinical microbiology.
The ability to generate a more exacting and definitive identification of pathogenic bacteria, in particular regarding less common or rare species, has led to a better understanding of their occurrence, pathogenicity, and disease associations in the clinical environment. This, in turn, has led to significant advances in our knowledge of these microbial agents with respect to clinical microbiology. Klebsiella variicola is an emerging human pathogen (167). In the past, this species has constantly been misidentified in the microbiology laboratory as K. pneumoniae by conventional methods prior to the introduction of molecular techniques (168–170). K. variicola is now known to cause serious bloodstream infections, produce hypervirulent strains, and exhibit multidrug resistance (167). The misidentification of many isolates for years prior to 16S gene sequencing, MALDI-TOF, and WGS leaves a number of unanswered questions regarding this taxon, including clinical frequency, environmental distribution, relative pathogenicity, and composite disease associations. Other taxa with recently identified and expanded syndromic presentations include Raoultella ornithinolytica with osteoarticular and intrathoracic respiratory infections (171), Leclercia adecarboxylata disease in pediatric patients (172), and the epidemiology, clinical presentation, and antimicrobial resistance of Escherichia (“Atlantibacter”) hermannii illnesses (173).
Additional changes witnessed within the family Enterobacteriaceae that have been precipitated by the “molecular revolution” also extend to traditional infectious disease syndromes and etiologic agents. The family’s natural habitat is the gastrointestinal tract of vertebrates, and members have long been recognized as a major cause of various gastrointestinal infections, including enteritis, colitis (including hemorrhagic), and dysentery. As late as 1985, only three enteric groups, Escherichia coli-Shigella, Salmonella, and Yersinia enterocolitica, were universally recognized as gastrointestinal pathogens from a limited list of documented prokaryotic species. However, this list has continued to expand over time, with more than 40 different agents or pathotypes now recognized as definitive, probable, or possible etiologic agents of gastrointestinal disturbances based upon reported outbreaks, case histories, other clinical features (serologic responses, histopathology), and identification of enteropathogenic virulence-associated factors (174, 175). For the enterobacteria, over 50% of these 40 agents currently reside within the family Enterobacteriaceae (175) and several presumptive pathogens are now recognized as emerging agents of diarrheal infections (176, 177).
Recently Identified Gastrointestinal Pathogens
Providencia alcalifaciens.
In 1989, Haynes and Hawkey (178) found a significant association between the presence of P. alcalifaciens and travelers’ diarrhea in persons between the ages of 15 and 64 who were primarily returning from travel to Mediterranean countries. Shortly thereafter, Albert and coinvestigators (179) studied three cases of diarrhea, one in a deceased Bangladeshi child and two in adults. P. alcalifaciens was present in all three stools in either pure culture (child) or as predominant enteric flora, and these strains were later shown to produce potential in vitro and in vivo enteropathogenic mechanisms, including invasion of HEp-2 cells and diarrhea in the removable intestinal tie adult rabbit model (179). Together, these two studies formed the cornerstone for interest in P. alcalifaciens as a possible gastrointestinal pathogen. Further evidence supporting a causal role for P. alcalifaciens in diarrheal disease stems from studies showing a statistically significant association of this species with symptomatic persons versus controls (178, 180, 181), elevated immune responses in infected persons (182), and identification of multiple virulence-associated factors potentially operative in the gut. Virulence-associated characteristics included invasion of cell monolayers (178, 182), production of a cytolethal distending toxin (183), and barrier dysfunction and apoptosis in tissue culture cells (184).
However, the most compelling evidence to date supporting a role for P. alcalifaciens in bacterial gastroenteritis comes from several reports on outbreaks of diarrheal disease attributed to this bacterium (Table 7). These outbreaks ranged in size from 2 (father and son) to 270 involving three schools in Fukui City, Japan, in 1996. In that later outbreak, 7 of 18 patients were culture positive for a clonal strain of P. alcalifaciens and 7 of 8 symptomatic persons displayed elevated serologic titers against the outbreak strain (182). Furthermore, the outbreak strains were demonstrated to be invasive in Caco-2 cells and to illicit moderate fluid accumulation in rabbit ileal loops (RIL) (182). In three of the four outbreaks listed in Table 7, the outbreak strains were found to be clonal in nature by a combination of techniques including serogrouping, plasmid analysis, pulsed-field gel electrophoresis (PFGE), and randomly amplified polymorphic DNA (RAPD) (182, 185, 186). A food source was implicated in three instances, two associated with potato dishes, one of which appeared contaminated by a culture-positive cook (181). The largest outbreak in Japan appeared to originate from a catering company (182). Travel-related infections are suspected in two outbreaks (181, 185).
TABLE 7.
Reported outbreaks of gastroenteritis caused by P. alcalifaciens
| Cohort | No. of cases | % Attack rate | Location | Food | Clonala | Reference |
|---|---|---|---|---|---|---|
| Father and son | 2 | Unknown | Mexico | Unknown | + | 185 |
| Elementary and high schools | 270 | 27.6–54.7 | Japan | Teriyaki chicken burger (warm bread) | + | 182 |
| Army hospital personnel | 27 | 41.5 | Turkey | Potato salad | ND | 181 |
| Church social event | 11 | Unknown | Kenya | Mashed potatoes (mukimo) | + | 186 |
ND, not determined.
Shah et al. (187) recently reviewed the literature on P. alcalifaciens infections; however, many questions remain unanswered in regard to the role of P. alcalifaciens in diarrheal disease. Since most laboratories do not look for this organism routinely, its relative frequency as an enteropathogen in select patient populations is presently unknown, although a selective medium, polymyxin-mannitol-xylitol medium-Providencia or PMXMP, has been developed to aid in its recovery and identification (188). While several studies have found an association between providenciae and gastroenteritis, P. alcalifaciens has also been found to be present with other copathogens (188) and in very high percentages in one study (180). In an outbreak in Turkey, at least 44% of infected military personnel who were now healthy were still culture positive for P. alcalifaciens 10 days later (181). Finally, we currently do not know which of several reported pathogenic mechanisms may be operative in all or select strains involved in gastroenteritis.
Escherichia albertii.
In 2003, a new Escherichia species, E. albertii, was proposed for five fecal isolates recovered from diarrheal stools of children <5 years old residing in Bangladesh (189). These strains had been isolated more than a decade earlier and had been thought to belong to the genus Hafnia or to be “Hafnia-like” in phenotypic characteristics. However, they differed markedly in a number of ways from true Hafnia strains by being Voges-Proskauer negative, acetate positive, and susceptible to cephalothin, by their failure to be lysed by the Hafnia-specific phage 1672, and by possessing the E. coli/Shigella outer membrane protein gene phoE (190). DNA relatedness studies by Huys et al. (189) indicated that these isolates represent a new species residing in the genus Escherichia.
E. albertii appears to be fairly widely dispersed in the animal kingdom, although its exact range, distribution, and prevalence in vertebrate hosts still remain to be determined. Two major animal reservoirs of E. albertii have been identified. Wild and domestic avian species in both healthy and diseased states have yielded E. albertii (191, 192). Birds found positive for this taxon include ducks, finches, magpies, wrens, woodpeckers, and chickens (191–193). This list even extends to the isolation of this species from a penguin in Patagonia (194). A second major reservoir that has just been discovered is raccoons. A study of over 400 wild raccoons in Osaka, Japan, found that 57.7% of rectal swabs obtained were positive for E. albertii by PCR; in this study, 143 viable E. albertii isolates were isolated from 62 PCR-positive samples (195). In addition to these two sources, E. albertii has been recovered from a cat, seals, and processed meats, including pork and mutton (193, 194, 196).
E. albertii causes diarrheal syndromes in humans, although the actual prevalence of the disease worldwide is presently unknown. Essentially all E. albertii strains carry two virulence determinants, the intimin gene (eae) and a cytolethal distending toxin gene (cdt) (193, 197). The intimin gene appears to be important in disease pathogenesis, while much less is known regarding what role the cytolethal distending toxin might play (193). Sporadically, strains of E. albertii also carry variants of Shiga toxin 2 (stx2) (193), but again, there are less than a handful of case reports describing such infections in the literature (196, 198, 199). Most of our knowledge regarding E. albertii-associated diarrhea comes from epidemiologic data on Japanese outbreaks of gastroenteritis associated with this organism. Six of these outbreaks between 2003 and 2015 were recently summarized by Masuda et al. (200). The chief symptoms of E. albertii-associated diarrhea compiled from these outbreaks are watery diarrhea (80% to 100%), abdominal pain (50% to 84%), and fever (26% to 100%) (200). Less common complications include headaches, nausea, and vomiting. The overall recorded attack rate was 457/741 or 61.2% (200). The largest of these outbreaks involved 409 students and teachers from a high school staying at a campground. The suspected cause of the outbreak was sump water that was unchlorinated and used as a potable water source (200). In most of these outbreaks, the vehicle of transmission was not definitively identified, but consumables including restaurant items (salad, other foods, water used) and purchased lunch box meals were suspected. Two cases of gastroenteritis linked to Shiga toxin-producing E. albertii have been published (198, 199). In one instance, bloody diarrhea resulted (198), while in the other case, the child experienced only watery diarrhea (199). There has been one reported case of extraintestinal infection caused by E. albertii, that occurring in a 76-year-old woman with multiple comorbid conditions who developed bacteremia (201). She recovered uneventfully after appropriate chemotherapy.
The present lack of both laboratory and epidemiologic data concerning E. albertii stems from general difficulties in the recognition of this Escherichia species. E. albertii has been dubbed “the evasive enemy,” no doubt in large part due to the difficulty in isolation and identification of this pathogen in the clinical laboratory (193). This taxon was originally proposed and described as an indole-negative, lysine decarboxylase (LDC)-positive species that could be separated from other members of this genus on the basis of these properties and several others, such as d-xylose (189). However, it quickly became apparent that variability in phenotypic expression among E. albertii strains was significantly greater than originally thought. Sequence analysis of multiple housekeeping genes coupled to eae and cdt gene sequencing by Hyma and colleagues (197) indicated that strains previously classified as Shigella boydii serotype 13 (known not to be a true shigella) were actually E. albertii on a genetic basis. These strains differed biochemically from those described by Huys and others (189) in being indole positive and LDC negative. Two biogroups were subsequently proposed for these strains (202). Biogroup 1 represented the former group described by Huys et al. (2018), while biogroup 2 strains were identical to those previously referred to as Shigella boydii 13. Ongoing research studies have identified many field isolates of E. albertii that do not fit into either of the two previously recognized biogroups, so a biogroup 3 has been proposed (203). Table 8 lists distinctive phenotypic features of all E. albertii biogroups and their differentiation from normal reactive E. coli.
TABLE 8.
Distinguishing features of Escherichia albertii biogroupsa
| Test | % Positive |
|||
|---|---|---|---|---|
|
E. albertii |
E. colib | |||
| Biogroup 1 | Biogroup 2 | Biogroup 3 | ||
| Motility | 0 | 0 | 0 | 95 |
| Indole | 0 | 100 | 100 | 98 |
| LDC | 100 | 0 | 100 | 90 |
| Lactose, acid | 0 | 0 | 0 | 95 |
| l-Rhamnose, acid | 0 | 0 | 0 | 80 |
| d-Sorbitol, acid | 0 | 100 | 0 | 94 |
| d-Xylose | 0 | 0 | 0 | 95 |
E. albertii has been misidentified frequently. Biochemical methods have often reported E. albertii isolates as “Hafnia-like” or Hafnia alvei, while molecular methods assessing both the eae and cdt genes have reported strains as belonging to either the enteropathogenic E. coli (EPEC) or enterohemorrhagic E. coli (EHEC) groups. Ooka and others (196) retrospectively analyzed 278 eae-positive E. coli strains and found that 26 (9.3%) were in fact E. albertii. In 2019, a similar study found 17 (4.5%) E. albertii isolates from a collection of 373 strains to be misclassified as either EPEC or EHEC (204). Conventional biochemical tests can help to distinguish E. albertii from phenotypically similar bacteria (Table 8); however, inactive E. coli bacteria, although uncommon, still present problems (202). Other tests not mentioned in Table 8 can also be helpful (193, 196). Bhatt and coinvestigators (193) have published a composite table on the main biochemical or phenotypic properties of E. albertii, E. coli, and H. alvei that is also quite useful.
Two recent developments may significantly help in the accurate identification of E. albertii in the diagnostic laboratory. A recent report found that enhancing the Bruker MALDI-TOF database with additional E. albertii spectra not present from the manufacturer was extremely productive (205). Of 58 E. albertii stains originally tested on the Bruker MALDI-TOF with a database library of 7,311, only 4 (6.8%) were correctly identified. However, when enhanced with additional spectra, MALDI-TOF correctly identified all 58 strains (205). This same group has also developed a selective agar for the recovery of E. albertii. Named XRM-MacConkey (xylose-rhamnose-melibiose), this medium was found superior to several other selective media for Enterobacteriaceae, producing colorless colonies that invariably were identified as E. albertii (206). The detection limit of XRM-MacConkey was 105 CFU/g of stool. These two innovations together should significantly improve the ability of laboratories to detect and identify this enteric pathogen (205, 206).
Klebsiella oxytoca and colitis.
Acute colitis is typically a transient inflammation of the colon accompanied by one or more symptoms such as bloody diarrhea and abdominal pain or distension. The syndrome can be precipitated by a number of different drugs, including antibiotics or other irritants. For over half a century, the genus Klebsiella has been recognized as a possible cause of acute enterocolitis (207). In the late 1980s, Japanese researchers identified a unique cytotoxin produced by Klebsiella oxytoca strains associated with cases of hemorrhagic enterocolitis (208). The toxin had a molecular mass of approximately 271 Da, was protease resistant, and produced rounding and/or cell death in HEp-2, HeLa, CHO, and Vero cell lines (208, 209). Subsequent investigations by the same group found that the cytotoxin elicited fluid accumulation in RIL (210). Histologic examination of these infected tissues revealed intense mucosal hemorrhaging with erosion of the ileum (210).
Following these discoveries, a series of prospective studies and case reports provided further documentation linking certain strains of K. oxytoca to antibiotic-associated hemorrhagic colitis (AAHC) (209). All of these studies had a number of common features, which included patients on penicillin derivative therapy of short durations, some persons additionally on nonsteroidal anti-inflammatory drugs (NSAID), and stools that were negative for Clostridium difficile with no other enteric pathogens detected. A French study of 20 suspected cases of antibiotic-associated diarrhea found 11 patients with AAHC (right-sided hemorrhagic to diffuse ulcerative colitis) primarily consisting of bloody and mucousy diarrhea (211). Of 11 such persons, 8 were positive for K. oxytoca. In contrast, none of 36 colonic biopsy specimens of control patients contained K. oxytoca (211). The definitive study on K. oxytoca AAHC was published by Högenauer et al. (212) in 2006. In that investigation, 5 of 6 persons with AAHC as determined by colonoscopy were positive for cytotoxigenic K. oxytoca on HEp-2 cells, as opposed to only 1.6% of healthy subjects. The age range of persons with K. oxytoca-associated AAHC was 28 to 65 years, younger than the typical age range for Clostridium difficile-associated colitis. In all five positive patients with K. oxytoca, amoxicillin-clavulanate appeared to be the triggering antimicrobial, and two persons were also noted to be on an NSAID (212). Furthermore, the authors fulfilled Koch’s postulates by inducing right-sided hemorrhagic colitis in Sprague-Dawley rats given cytotoxigenic K. oxytoca and amoxicillin-clavulanate but not in either of two control groups that did not contain this combination (212). Patient’s conditions resolved completely after discontinuation of antimicrobial therapy.
The exact frequency of K. oxytoca in gastrointestinal contents is unknown and has varied in studies from different geographical locales from 1.6% to >4.0% (212–214). Cytotoxigenic strains are found in higher percentages in patients with C. difficile-negative AAHC than in those with acute colitis or in healthy carriers (215). The disease can develop rapidly, with AAHC manifesting itself within 2 to 4 days of the initiation of antimicrobial chemotherapy (215). A limited study of four patients with AAHC found that K. oxytoca could be recovered in relatively high concentrations (>106 CFU/ml) from patient stools (209, 213). A selective medium, Simmons citrate-inositol-tryptophan-bile salts (SCITB), has been used in a Hong Kong survey to appreciably increase the recovery rate of K. oxytoca from diarrheal stools (214).
Whether cytotoxigenic K. oxytoca plays a role in other gastrointestinal syndromes is not known. One large study of 371 patients broken down into four different groups based upon the presence of diarrhea and whether or not individuals received antibiotics found no association between the presence of K. oxytoca and nonhemorrhagic antibiotic-associated gastroenteritis (213). However, Paveglio and colleagues (216) recovered cytotoxigenic K. oxytoca from 6 of 10 infants with necrotizing enterocolitis characterized by bloody diarrhea, pneumatosis, and abdominal distension. K. oxytoca has also been linked to a fatal case of antibiotic-associated pseudomembranous colitis, but the pathogenic characteristics of the infecting strain were not investigated (217).
Potential enteropathogens with inconclusive data.
A number of enterobacterial species or biotypes have been occasionally implicated in gastroenteritis based upon clinical disease associations, case reports, and possession of virulence-associated factors (2, 174, 175). While the data in many instances are very limited, several bear mentioning, as many recognized enteric pathogens (see above) started under similar circumstances.
A recent review has suggested that Proteus species might be putative gastrointestinal pathogens, as has already been demonstrated conclusively for other Proteeae such as P. alcalifaciens (218). The primary evidence supporting a possible role for protei in gastroenteritis involves their possession of some potential pathogenic characteristics (hemolysin, intracellular invasion) and a “guilt by association” recovery of P. mirabilis from one foodborne outbreak of gastroenteritis in Beijing, China (218). A more recent publication has characterized potential pathogenic mechanisms in one food poisoning-associated strain when it was compared to two reference cultures (219). However, there are presently no clinical or epidemiologic data linking isolations of Proteus to case-controlled studies of gastroenteritis, detection of common virulence-associated factors in outbreak-related isolates, animal models of infection, or demonstration of clonality among isolates associated with instances of food poisoning (219). Furthermore, there are only a couple of studies with contradictory conclusions on whether or not the frequencies of Proteus spp. in diarrheal versus healthy stools are different (218, 220).
Over the last 4 decades, there have been sporadic reports describing the association of Citrobacter freundii with episodes and outbreaks of gastroenteritis (2). In 1995, Tschäpe and others (221) described a nursery school outbreak of diarrhea associated with hemolytic-uremic syndrome (HUS) and thrombocytopenic purpura with anemia. At least 14 children between the ages of 2 and 6 years were infected, 9 of which required hospitalization due to renal failure. All C. freundii strains recovered from these children produced a Shiga toxin (stx2) (221). Sandwiches prepared with green butter containing contaminated parsley appeared to the likely vehicle of infection. A C. freundii strain producing an aggregative adherence pattern on HeLa cells has also been isolated from a child with severe diarrhea (222). Cytotoxigenic and adherent C. freundii isolates have been identified in the stools of persons with diarrhea (223). One strain, recovered from a goat, was not only cytotoxic but produced an aggregative adherence pattern on Hep-2 cells. These publications may be a reflection of the horizontal transmission of virulence factors from a traditional pathogen (E. coli) to citrobacteria as an isolated event, or they could signal that C. freundii has been overlooked as a potential enteropathogen.
Extraintestinal Diseases and Infectious Syndromes
Extraintestinal community-associated infections linked to current members of the family Enterobacteriaceae appear to be on the rise. Some of these infections are the result of the translocation of pathogenic strains from the gastrointestinal lumen into the circulatory system or lymph nodes, producing systemic disease, including bloodstream infections (BSI). Conditions in the community that exacerbate such situations include an increasingly elderly population and more residents with underlying diseases or immunocompromised conditions. Community-acquired extraintestinal infections can also result from the consumption of contaminated foods, contact with infected animals, or penetrating traumas (2). Some examples are listed below.
Klebsiella pneumoniae and PLA.
A pyogenic liver abscess (PLA) is a solitary mass or collection of pus-filled masses due to bacterial infection, often located within the right lobe of the liver. PLAs are typically polymicrobic in nature, with Gram-negative pathogens predominating, including certain enterobacterial species such as E. coli and Klebsiella spp. (224). PLA can develop from a number of sources, including trauma, the circulatory system, due to biliary tract infections, or peritonitis subsequent to bacterial translocation from the gastrointestinal tract (225). The incidence of PLA has been found to be higher in Southeast Asia (11.99 to 17.59/100,000) than in various western countries, including the United States (2.7 to 4.1/100,000), although reasons for these geographic differences are not well appreciated (226).
Beginning in the mid-1980s, a new variant of K. pneumoniae associated with PLA and metastatic disease first appeared in Taiwan (227). The seven infections reported in this clinical series differed from classical K. pneumoniae (cKp) illnesses in that they were associated with septic endophthalmitis and other metastatic complications, including meningitis and pulmonary embolism; all seven patients recovered but completely lost their vision (n = 6) or visual acuity (n = 1). By 1998, a Taiwanese review of 182 cases of liver abscesses caused by K. pneumoniae occurring between 1990 and 1996 found 160 (88%) to be monomicrobic (228). Differences noted between monomicrobic and polymicrobic abscess infections included a higher frequency of diabetes or glucose intolerance and metastatic disease in the former group (228). Today, while this disease is still centered in Southeast Asia, and in particular in Taiwan and South Korea, it has now been reported worldwide from India, Europe, Australia, and the United States (229, 230).
This new variant of K. pneumoniae is referred to as hypervirulent K. pneumoniae, or hvKp (230). Another term used less frequently for this group is hypermucoviscous. It can be distinguished from cKp strains clinically because illnesses associated with the variant pathotype are normally community acquired and PLA occurs in the absence of biliary disease, often presents as metastatic infections at multiple anatomic sites, and is typically monomicrobic in composition (230). Genetically, these strains harbor a number of virulence factors that are either chromosomally encoded or on virulence plasmids such as pK2044 and pLVPK (229, 230). These virulence factors include genes encoding capsule formation (cps), mucoid regulators (rmpA, rmpA2), K1 and K2 antigenic capsule types, and several siderophores, including yersiniabactin (229, 230). The predominant phenotype in all of these strains is hypermucoviscocity (HMV), which is easily detectable on common media such a blood agar by a “string test” (231). For K1 strains, this hyperviscous phenotype is linked to the gene magA (mucoviscocity-associated gene A). Other pathogenicity factors have been described but are less well characterized (230).
Most cases of PLA are preceded by gastrointestinal tract colonization by hvKp prior to invasion, although initial sources of environmental acquisition are poorly defined. Risk factors associated with developing hvKp disease include Asian ancestry and diabetes mellitus (225, 229, 230). In its infancy as a recognized emerging pathogen, hvKp infections were initially defined by their association with cryptogenic PLA. Symptoms associated with hvKp PLA are not variant specific but rather typical for the syndrome and include fever, chills, abdominal pain, and leukocytosis (225, 230). Common metastatic sites of hvKp PLA infection identified early in their history included the eye, central nervous system (CNS), and pulmonary tree in addition to bacteremia (229, 230); in one large study of over 800 persons, 12% of patients with hepatic abscesses developed metastatic disease (229). Approximately 5% of patients presenting with hvKp bacteremia develop endogenous endophthalmitis, which carries with it a devastating prognosis of loss of visual acuity for 89% of patients (230). The mortality rate associated with PLA is roughly 5% but increases to 10% to 16% for metastatic disease (225). Some studies report an even higher range (3% to 31%) with mortality rates as high as 35% (229).
Unfortunately, over the last few years, a number of new and emergent trends concerning hvKp disease have been detected. Various studies have described extrahepatic illnesses associated with hvKp, including bacteremia, pneumonia, and musculoskeletal and soft tissue infections along with a variety of less frequently encountered complications like Bartholin’s abscess (229, 230). Another alarming trend is the increasing incidence of hvKp causing health care problems, including ventilator-associated infections (229). A 2020 study from China found that a retrospective analysis of 79 hvKp strains determined that 53 (67.1%) of these isolates were health care associated and 19 (24.1%) of these caused true HAI. Only 8.8% of the hvKp isolates in this survey were community acquired (232). Probably the most disturbing and evolving tendency of this variant is increasing acquisition of resistance to antibiotics. Historically, the frequency of drug resistance in this variant was low, with <2% of isolates resistant to cephalosporins (229). However, detection of extended-spectrum β-lactamases (ESBLs) in this group has risen from <5% to 35% or more in some studies (232). An outbreak of pneumonia in surgical patients who were subsequently placed on mechanical ventilation was caused by hvKp (233). The infecting strain belonged to sequence type 11 (ST11) and was carbapenem resistant (233). All five patients in that study died. This increasing trend in drug resistance in hvKp will clearly impact disease frequency and associated mortality rates in hospitalized persons in the future if this trend continues to increase and disseminate globally.
PLA requires a clinical diagnosis via abdominal imaging (225). Suspicion of hvKp playing a role in PLA is presently challenging, requiring an astute review of clinical and demographic information that might suggest the presence of K. pneumoniae (230). The clinical laboratory, however, can play a key role in the diagnosis of such infections by the identification of hvKp from suspected (blood) or occult (eye) clinical samples. The defining phenotype for hvKp strains is HMV. The HMV phenotype is detected by a “string test” test, which is defined as the extension of a mucous string by an inoculating loop >5 mm from a bacterial colony grown overnight on a blood agar plate (231). In the initial studies, the string test had a 98% sensitivity in recognizing the HMV phenotype (231). More recent studies suggest that this value is somewhat lower, with an accuracy of 0.90 (234). Regardless of this difference, it is a simple test than can be performed in any microbiology laboratory. A number of genotypes have been found to have a diagnostic accuracy of >0.95. These include rmpA, rmpA2, peg-344 (putative transporter), and iroB (salmochelin siderophore) (234). In addition to the HMV phenotype, capsule typing using commercial-grade antisera can be useful, as the majority of hvKp strains are either K1 or K2 (229, 230). However, other capsular types can be hvKp with K1 > K2 > K5 > K57 in decreasing order of frequency (229).
One of several problematic issues regarding the laboratory diagnosis of this group is that no single test encompasses or detects all hvKp isolates. The current gold standard for identifying these strains is rmpA+ rmpA2+ magA+ HMV+ (229). However, again variants that deviate from the ideal phenotype but are still hvKp have been detected. Due to their increasing significance worldwide, there has been a call for a consensus definition for hvKp (235). This will no doubt be a challenging task, as the extent of genetic and phenotypic variation in the group continues to grow.
A number of excellent reviews on various topical issues of hvKp are available (225, 229, 230).
Cronobacter sakazakii and neonatal meningitis.
In 1980, the CDC proposed a new species for inclusion in the genus Enterobacter, now Cronobacter (“Enterobacter”) sakazakii (24, 236). This collection of 57 strains came from diverse clinical sources. The authors commented that strains isolated from sputum, wounds, and species were probably not clinically significant, although rare cases of neonatal meningitis were identified. Just 3 years later, the CDC in partnership with international collaborators in The Netherlands reported 8 additional cases of C. (Enterobacter) sakazakii neonatal meningitis and septicemia (237). For two of these neonates, their episodes of meningitis were accompanied by necrotizing enterocolitis. Six of these eight infants subsequently died (75% fatality). Over the last 15 years, the incidence and breadth of C. sakazakii invasive disease in neonates and infants have mushroomed on a global basis. An excellent historical timeline of the progression of C. sakazakii infections and associated diseases from their initial retrospective recognition (1950, prior to naming) to 2015 can be found in the review by Farmer (24).
While C. sakazakii can cause illnesses on a sporadic basis in adults and older persons, its paramount clinical and public health importance is as a causative agent of two types of invasive neonatal disease, bacteremia and meningitis (238). In up to 40% of cases of neonatal or infantile meningitis, such infections are accompanied by brain lesions, typically abscess formation. This makes C. sakazakii one of only three enterobacterial species (E. coli K1, Citrobacter koseri) intimately linked to causing brain abscesses in young infants and children (2). The major risk factors in this population setting for developing invasive C. sakazakii include neonatality (<28 days), low birth weights (<2,500 g), and consumption of powdered infant formula (PIF) (238–240). Case fatality rates have been reported as high as 40% to 80% in those presenting with bacteremia, meningitis, and necrotizing enterocolitis (238). Henry and Fouladkhah (25) have recently summarized a number of sporadic cases and outbreaks of invasive C. sakazakii disease in infants. The vast majority of these cases were associated with infant formula, including PIF.
As in the case of K. pneumoniae and hvKp, infections involving C. sakazakii appear to be on the rise and changing with respect to demographics in the United States. A report reviewing the published literature (1961 to 2018) plus all systemic isolates of C. sakazakii referred to the CDC identified 183 cases of infant infections (240). Of these illnesses, 63% involved episodes of meningitis, while the remainder were bloodborne infections. Neonates were the predominant age group impacted, accounting for 67% of all infections. A review of published studies and information accompanying isolates forwarded to the CDC indicated that 79% of infants were reported as having consumed PIF (240). The observed mortality rate was 38%. One of the interesting findings of this CDC epidemiologic investigation was the changing demographics of disease presentation. Prior to 2004, only 44% of infections were found in nonhospitalized children. Between 2004 and 2018, this figure had risen to 78%. Similarly, the percentage of full-term neonates infected with C. sakazakii rose from 22% before 2004 to 50% during the last quarter of the study (240). The incidence in cases also rose from 1.2 cases/year before 2004 to 8.7 cases/year between 2004 and 2018. The reasons behind these changing demographics in the United States are not apparent. Cronobacter invasive disease also appears to be emerging outside the United States. An Egyptian study of 100 cases of neonatal sepsis found that 12% of cases were due to C. sakazakii (241). Of these 12 cases, two infants died and two others suffered marked physical and mental impairment. The authors report these dozen infections as the first cases of this emergent disease in Egypt (241).
Presently, C. sakazakii, like the other above-mentioned agents, is not a reportable disease in the United States or elsewhere (240). This severely limits our understanding of the magnitude of neonatal infections caused by this pathogen worldwide. Two other confounding problems involve their environmental distribution and laboratory identification. Cronobacter spp., and C. sakazakii in particular, are ubiquitous in the environment, being found in a variety of consumable products and water. While PIF is clearly the primary implicated vehicle for most infant infections (242), C. sakazakii illnesses have also been connected to expressed breast milk on several occasions (240). A key to decreasing the incidence of infections is to find substitutes for PIF, and the WHO has established recommendations for hospitals and home use of PIF and its preparation for the first 2 months of life (25, 242). In addition to PIF, C. sakazakii has been recovered from flour-based products, flour, cereal kernels, herbs and spices, insects, meats, and other commodities (25, 161, 238, 241, 243, 244). Their extensive environmental distribution is in no doubt partially related to the fact that they are resistant to desiccation and dry and acid growth conditions in comparison to other enterobacteria (238).
For many years, C. (Enterobacter) sakazakii was considered to be a fairly tight genetic species composed of 15 biotypes (25). However, that opinion changed with the creation and publication of a new genus, Cronobacter, in 2007 (25). Today the genus has seven validated species, including C. sakazakii (https://lpsn.dsmz.de/species?page=C#Crabtreella). For the clinical laboratory, this presents problems, because although most cronobacters have been associated with human disease, definitive identification of C. sakazakii is of immense importance because of its paramount role as the preeminent pathogen of the genus. The Vitek GN identification system lists most species under the phenospecies designation “Cronobacter sakazakii group” (245). Identification matches based on various commercial products (Vitek GN, API 20E, ID32E) ranged from 82.3% to 90% (245). A multicenter European study involving 11 countries found that only 59 of 77 (76.7%) isolates submitted as C. sakazakii were, in fact, this genetic species, as determined by MALDI-TOF and WGS (245). Some misidentified isolates did not even belong to the genus Cronobacter but rather belonged to a variety of genera, including Enterobacter, Klebsiella, Kluyvera, Kosakonia, and Siccibacter (246). These results suggest that better methods for identifying this important clinical and public health pathogen need to be developed.
Chronic conditions and potential long-term sequelae.
The vast majority of medical personnel and allied scientists associate the family Enterobacteriaceae with foodborne disease, health care-associated illnesses, and a variety of syndromic diseases, including gastroenteritis, urinary tract infections, and soft tissue infections subsequent to traumatic events. However, less well appreciated is the fact that many of these species as resident commensal gastrointestinal flora may be associated in some aspect with much longer-term chronic or persistent conditions.
Inflammatory bowel diseases, which include Crohn’s disease and ulcerative colitis, are thought to be due to host genetic factors, environmental triggers, and endogenous microbial communities. E. coli is one of the leading candidates to play some role in these two disease processes. Studies of the microbiome have demonstrated that the relative abundance of E. coli increases in Crohn’s patients (as does Serratia marcescens) when compared to healthy individuals and familial controls while beneficial bacteria decrease (247, 248). The particular pathotype implicated in these inflammatory processes is adherent-invasive E. coli (AIEC) (249, 250). The data seem clear that adherent-invasive E. coli bacteria are associated with both conditions, but whether they are opportunistic bystanders or are intimately involved in the entire immune-mediated process remains to be determined. Proteus spp. have also been proposed to have a possible role in Crohn’s disease (218). This proposal is based on two pediatric studies where there was an overabundance of protei in Crohn’s patients compared to individuals with other gastrointestinal conditions.
E. coli strains have also been linked to irritable bowel syndrome, a condition affecting 10% to 20% of the population (251). The two pathotypes associated with this condition are AIEC and enteroaggregative E. coli (251, 252). Increased numbers of E. coli correlate with disease symptomatology, and patients respond to antimicrobial therapy directed against this microbe (251).
Enterobacteria are increasingly being implicated as potential infectious agents associated with immune and nonimmune inflammatory processes of bones, joints, and associated tissues. Rheumatoid arthritis (RA) is a chronic inflammatory arthritic condition most often involving symmetrical joints. It primarily affects middle-aged women. RA is thought to involve genetic, environmental, and microbial factors (253). Among enterobacteria, several different species have been implicated in RA, but by far the greatest amount of data linking a group to the etiology of RA is that concerning the genus Proteus, and in particular P. mirabilis (2, 254). A number of studies have found significantly elevated antibodies (IgM, IgG, IgA) against P. mirabilis antigens (hemolysin, urease) in RA patients in comparison to controls (255). These antigens share common motifs with HLA subtypes found in RA patients. The six-amino-acid sequence ESRRAL (glutamic acid-serine-arginine-arginine-alanine-leucine) in P. mirabilis hemolysins shares homology with the EQRRAA (glutamic acid-glutamine-arginine-arginine-alanine-alanine) motif found in RA-associated HLA-DR (subtypes HALA-DR1 and HLA-DR4) (2, 254). The prevailing theory is that P. mirabilis urinary tract infections elicit significant immune responses against this uropathogen such that, through molecular mimicry, cross-reacting antibodies that target self-antigens in the joints are elicited.
A variety of other secondary complications have been associated with gastrointestinal enterobacterial infections. Reactive arthritis, a sterile nonpurulent inflammation of limbs, can be triggered by the acquisition of or infection with various enterobacteria, including diarrheagenic E. coli (DEC) and Yersinia enterocolitica (256, 257). Some cases of reactive arthritis are associated with human leukocyte antigens, part of the inflammatory process, such as HLA-B27 (257). In one investigation, 17% of persons developed musculoskeletal symptoms posttravel (256). Multivariate analysis found a significant association between this illness only with the exclusive isolation of DEC. Other less frequently encountered infection-associated chronic conditions include Reiter’s syndrome, ankylosing spondylitis, and prostatitis (2).
Colorectal cancer and related malignancies.
An area of potential research that has received little attention to date concerns the possible association of members of the Enterobacteriaceae with malignancies. Dysbiosis is a clinical condition where there is an imbalance in the normal microflora of a person related to ill health. For enterobacteria, this mainly involves their natural habitat, the gastrointestinal tract. The handful of published studies to date have primarily shown an abnormal rate or abundance of various enterobacteria in disease states associated with cancer. Higher rates of colorectal cancer have been detected in patients with pyogenic liver abscesses (230, 258). In one 11-year study, the rate of colon cancer was 2.68 times greater for those with PLA due to K. pneumoniae than for those patients without this enteric pathogen. A 2019 study has found both Escherichia-Shigella and Enterobacter in higher numbers in patients with primary liver cancer (PLC) than in healthy controls or those with liver cirrhosis (259). The relative abundance of Enterobacter ludwigii was 100 times greater in the PLC group than in controls. E. coli has also been implicated as possibly associated with colorectal cancer (259, 260).
What role some or any enterobacteria play in cancer is unknown. It could be that the amplification of these species in dysbiotic states is a simple reflection of them as possible indicators of occult malignancies in a fashion similar to that recorded for Streptococcus bovis and Clostridium septicum (230). Another possibility is that these bacteria are the direct causative agents of certain cancers. However, this hypothesis seems unlikely in that host factors (immunologic, physiologic) as well as environmental triggers are thought to play important key roles. Finally, enterobacteria might act as important cofactors in dysplasia formation by one of several mechanisms, including activation or enhancement of inflammatory processes or release of virulence factors causing DNA damage or genetic instability (260). Further research in this area seems important.
THE FUTURE
Issue: The Family Enterobacteriaceae—Expansion or Contraction?
The landscape of clinical and diagnostic microbiology has forever been changed by the introduction of the molecular taxonomic revolution brought about by WGS and related phylogenetic techniques. These “technologic wonders” have generated a number of important and positive changes in the field of bacterial taxonomy, including (i) the ability to detect and identify new genetic species of potential medical, public health, or veterinary importance (261), (ii) more precise identifications in the laboratory, especially for difficult-to-identify pathogens, (iii) less reliance on federal, state, or reference laboratories for definitive identifications, which could take days to weeks, and (iv) advances in tracing, fingerprinting, and identifying the origins, transmission, and spread of infectious disease outbreaks associated with the family Enterobacteriaceae (262).
Intimately linked to these novel and constructive taxonomic achievements are classification issues which have no rules or boundaries regarding their acceptance or rejection. The work of Gupta and colleagues (98, 117, 118) on various groups of medical importance is impressive, but the bottom line of whether these changes should be accepted into practice in medicine and related allied disciplines is questionable. There has already been negative feedback on proposed classification (division) changes by this group of researchers for long-standing taxa, including Borrelia (263), Mycobacterium (119), and Mycoplasma (120), as well as the Enterobacteriaceae. The reasons for rejecting such changes include inadequate data or evidence, avoidance of unnecessary changes, importance of the stability in nomenclature, and potential impact on adverse medical outcomes. Many of these reasons are listed in the rules of the ICNP. Classification changes can also be a double-edged sword. As an example, Enterobacter aerogenes has been recently transferred to the genus Klebsiella as K. aerogenes (60). Wesevich et al. (264) have found that the presence of K. aerogenes in the bloodstream of patients is associated with a poor clinical outcome (death before discharge, recurrent BSI) in comparison to other Enterobacter species. In contrast to these findings, there is a concern that the transfer of this species from the genus Enterobacter to Klebsiella may influence suboptimal therapy (265). Munson (265) in a recent newsletter has done an outstanding job of providing a detailed and comprehensive overview summarizing clinical issues concerning proposed taxonomic revisions.
Possible classification models.
One of the important aspects of bacterial taxonomy for clinical microbiologists, physicians, epidemiologists, and related groups is that nomenclature and taxonomy need to be practical, useful, and workable with the above-mentioned groups in regard to daily communications concerning diagnosis, prognosis, treatment, and the frequency and distribution of infectious diseases on a global basis. While the ICNP establishes the rules for nomenclature, many exceptions have been granted so that certain species that are genetically identical on the DNA level can remain separate and distinct because of their enormous medical importance. Several have previously been mentioned, including E. coli/Shigella and Yersinia pestis/Yersinia pseudotuberculosis. While taxonomic progress is important, it needs to be balanced by a needs assessment and general utility. Two obvious options are that the Enterobacteriaceae could remain “status quo” or the proposals of Adeolu et al. (98) could be adopted as published. Some other possible proposals are listed below.
(i) Proposal 1. Classification of genera into tribes.
The classification of genera into tribes would leave the family Enterobacteriaceae intact at the family level but create 7 to 9 tribes based upon the evolutionary work of Adeolu et al. (98). These tribes could include the following: tribe 1, Escherichieae (revised definition); tribe 2, Erwinieae (revised definition); tribe 3, Pectobacterieae (tribe nov.); tribe 4, Yersinieae (tribe nov.); tribe 5, Hafneae (tribe nov.); tribe 6, Proteeae (revised definition); tribe 7, Budviceae (tribe nov.); tribe 8, Plesiomonadeae (tribe nov.); and tribe 9, Thorselleae (tribe nov.).
(ii) Proposal 2. Classification of genera into subfamilies.
The classification of genera into subfamilies is similar to that of proposal 1 but uses the taxonomic rank of subfamily instead of tribes to group all genera.
(iii) Proposal 3. Classification of Enterobacteriaceae into “groups.”
The classification of Enterobacteriaceae into “groups” would use nonscientific names to group genera that are close together in an evolutionary sense. Such a system could include the following: group 1, core genera (Escherichia-Shigella-Salmonella-Citrobacter-Enterobacter-Klebsiella-Cronobacter and others; group 2, Erwinia-Pantoea group; group 3, Pectobacterium group; group 4, Yersinia-Serratia group; group 5, Hafnia-Edwardsiella group; group 6, Proteus-Providencia-Morganella group; group 7, Budvicia-Leminorella-Pragia group; group 8, Plesiomonas group; group 9, Thorsellia-Coetzeea group.
(iv) Proposal 4. Nonclassification system.
The nonclassification system would simply be an alphabetical listing of genera in the family but with the type genus listed first, similar to what was published in Bergey’s Manual of Systematic Bacteriology, 2nd edition.
Issue: Species Identification in the Modern Era for Clinical Laboratories
From conventional to commercial to molecular approaches.
Despite all the technological advances over the past 20 years, phenotypic identification currently remains the gold standard for identification of microorganisms (266). Both semiautomated and automated commercial testing platforms that provide both an identification and a susceptibility test result still carry the major workload in volume in the United States and elsewhere in diagnostic settings. Many molecular approaches are available to aid microbiologists in achieving final identifications when initial test results provide correct identification with low probabilities or no identification at all (267, 268). Advances continue to be made in this arena. One investigation has proposed that broad-range dnaJ PCR and sequencing can produce a higher resolution of species identification within the family than 16S rRNA gene sequencing (269). The only major impediment is the Escherichia-Shigella group, which is one species at the genomic level.
Techniques versus a plethora of taxa: problematic issues.
The relative explosion in the number of new enterobacterial species isolated from human samples or clinical infections over the past decade brings with it a number of clinical decisions for microbiologists regarding how far and to what extent an isolate should be characterized prior to generating a final identification. The relevancy of modern taxonomy for clinical laboratories has also been the subject of some questions and controversy (270). One of the difficulties microbiologists face with the molecular taxonomic revolution in the current era is that newly described species are typically “phenotypically light” in regard to biochemical characteristics which are based upon gene expression and not gene structure (271). Phenotypic methods remain the mainstay for most laboratories in regard to bacterial identifications as of 2020 (266). Thus, as modern taxonomy drives us forward, how do laboratories deal with the onslaught of new genera and species whether phenotypic or molecular methods are used?
The first issue microbiologists face is keeping abreast of all the new enterobacterial species, including classification changes. A few years ago, one had several options in this area. Three journals, International Journal of Systematic and Evolutionary Microbiology, Systematic and Applied Microbiology, and Antonie van Leeuwenhoek, published the vast majority of new species. Today, while these journals still publish the majority of new proposals, many other journals now routinely publish articles on new taxa. This makes the time and effort needed to review all of these periodicals more demanding. Microbiologists can also search PubMed (https://pubmed.ncbi.nlm.nih.gov/) for new groups using key words or terms, but this can also be fairly complicated and articles can be missed. The website LPSN—List of Prokaryotic Names with Standing in Nomenclature (https://www.bacterio.net/) is useful, but there are no easy methods to search for new taxa of clinical relevancy for specific ranks such as families. A second issue with the LPSN is that only validated species are published, and currently up to 40% of recently published species have not been validated according to ICNP rules. Finally, two journals routinely publish updates on newly proposed species, Diagnostic Microbiology and Infectious Diseases (since 2014) and Journal of Clinical Microbiology (since 2017). While the articles published in both journals differ slightly in scope, length, and structure, these are excellent sources for microbiologists to review for important updates (65, 272).
A second issue is identification, phenotypic versus molecular. For the vast majority of common infections such as those originating from the urinary tract (E. coli, P. mirabilis), simple and inexpensive phenotypic tests should provide a definitive identification. In some instances, even rapid spot tests may be applicable. For more-difficult-to-identify or unusual microorganisms not often seen in the laboratory, such approaches will not be sufficient. In these cases, molecular approaches are needed, such as 16S ribosomal gene sequencing, MALDI-TOF, and WGS. Use of molecular approaches for a final identification must be counterbalanced with need, as these techniques are more expensive and require skilled personnel and sophisticated instrumentation with maintenance contracts that are more costly (262). For newly published groups or species, the majority of publications are generated on the basis of a single type strain (65). This has several ramifications. Commercial panels or platforms will typically not include new taxa without a data matrix of 25 or more strains. For MALDI-TOF users, this will require laboratories to obtain the type strain from an international type culture collection (not inexpensive) and then add the spectral analysis to the manufacturer’s database. However, single species strains may not be enough to recognize all isolates or variants of a recently described taxon. For the present, it seems unwise to immediately attempt to identify new infectious agents without recognizable biochemical markers adaptable to multiple formats for phenotypic identifications (270).
Molecular identifications should be restricted to situations where the data generated are both relevant and required. Examples of situations where such analyses are needed include the following: (i) where specific species identification is important in a patient prognosis, diagnosis, or treatment, (ii) in outbreak disease investigations, (iii) for description of new species, infectious agents, or pathotypes (such as STEC or PLA-associated K. pneumonia), (iv) in epidemiologic or environmental surveillance surveys, and (v) in publications defining unique infectious disease syndromes, pathogenic characteristics, or unusual biochemical or metabolic properties.
CONCLUSIONS
Despite the voluminous number of publications, periodicals, and textbooks on members of the family Enterobacteriaceae, we are just in our infancy in understanding the scope, magnitude, and impact of this group on the world around us. Molecular technology, taxonomy, and phylogenetic analysis have ushered in a new era such that 28 new validated genera and more than 200 new species have been described since 2005 (Table 2). This trend will continue to increase and evolve for the foreseeable future, as witnessed by the recent description of Intestinirhabdus alba isolated from the gut of plastic-eating larvae (273). Some species such as E. albertii (189) have already been found to be important gastrointestinal pathogens in the Far East, while reclassified taxa such as K. aerogenes are being redefined by new epidemiologic criteria as important pathogens (264). Determination of the taxon frequency, clinical significance, and defining markers of these recently described species seems daunting, and our success in these endeavors remains to be determined.
What might eventually be of greater consequence is increasing our understanding of how enterobacteria affect the biosphere outside of human hosts. Climate changes will bring on a specter of new infectious diseases (274). Global warming will extend the geographic range of many insects carrying enterobacterial species with them. As vectors, insects could promote new and devastating diseases to trees, shrubs, flowers, and consumable fruits and vegetables (Table 5). Whether we will be able to detect and treat such an onslaught of vegetative pathogens remains to be seen.
The family or divisional constructs will continue to challenge a variety of scientific professionals for both their positive benefits and their negative impact for decades to come. The hope is that technologic developments and scientific advances will occur at speeds able to adapt to our changing and evolving world.
ACKNOWLEDGMENT
This review is dedicated to the memory of John James Farmer III (Jim), a dedicated public health professional who worked at the Centers for Disease Control and Prevention (CDC) for 28 years. Jim served as Chief of the Enterics Reference Laboratory for most of this time. Along with Don Brenner and many other researchers at the CDC, he isolated, identified, and named many of the genera and species listed in the present review, including Cronobacter (Enterobacter) sakazakii, Proteus penneri, Ewingella americana, Moellerella wisconsensis, Escherichia fergusonii, Enterobacter asburiae, Enterobacter hormaechei, and many others. In 1985, along with numerous CDC colleagues, Jim published a 30-page article describing the biochemical characteristics, clinical features, and diagnostic characteristics of new members of the family Enterobacteriaceae that became a cornerstone reference for many microbiologists over many years.
Biographies

J. Michael Janda received his undergraduate degree from Loyola University (Westchester), his M.S. in Microbiology from California State University-Los Angeles, and his Ph.D. in Microbiology and Immunology from UCLA. From 1979 to 1986, he was first a postdoctoral fellow (ABMM) at The Mount Sinai Medical Center and then Assistant/Associate Professor of Clinical Microbiology there, where he was Assistant Director of the Microbiology Laboratory and Coordinator of the ABMM program. From 1986 to 2011, he was Section Chief and then Chief of the Microbial Diseases Laboratory Branch, California Department of Public Health. Since 2015, he has been the Laboratory Director for Kern County. He is an Associate Editor of Diagnostic Microbiology and Infectious Diseases and is on the editorial board of Clinical Microbiology Reviews. Past activities include membership in Bergey’s Trust and on two subcommittees of the International Committee on the Systematics of Prokaryotes (ICSP). Over his career of more than 35 years, Dr. Janda has focused on the study of bacterial gastroenteritis, enteric bacteriology, aquatic pathogens, and microbial taxonomy, with particular emphasis on the genera Aeromonas, Plesiomonas, and Vibrio and the family Enterobacteriaceae.

Sharon L. Abbott received her Bachelor of Arts degree from California State University at Chico. For over 35 years she worked in the Microbial Diseases Laboratory Branch (MDL) of the California Department of Public Health, primarily in enteric bacteriology. In 1990, she became Supervisor of the Enterics Unit and served in that position for almost 15 years. After retiring from the State, she was employed by the University of California at Berkeley as a Training Coordinator for a Postdoctoral Fellowship program at MDL. Her research focus has centered on the identification of enteric bacteria, particularly Aeromonas species, and the development of in vitro pathogenicity assays.
REFERENCES
- 1.Farmer JJ, III, Farmer MK, Holmes B. 2005. The Enterobacteriaceae: general characters, p 1317–1359. In Borriello SP, Murray PR, Funke G (ed), Topley & Wilson’s microbiology & microbial infections, 10th ed, vol 2. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 2.Janda JM, Abbott SL. 2006. The enterobacteria, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 3.Janda JM, Abbott SL. 2015. The family Enterobacteriaceae, p 307–319. In Goldman E, Green LH (ed), Practical handbook of microbiology, 3rd ed. CRC Press, Boca Raton, FL. [Google Scholar]
- 4.Buján N, Toranzo AE, Magariños B. 2018. Edwardsiella piscicida: a significant bacterial pathogen of cultured fish. Dis Aquat Organ 131:59–71. doi: 10.3354/dao03281. [DOI] [PubMed] [Google Scholar]
- 5.Motyka A, Zoledowska S, Sledz W, Lojkowska E. 2017. Molecular methods as tools to control plant diseases caused by Dickeya and Pectobacterium spp.: a minireview. N Biotechnol 39:181–189. doi: 10.1016/j.nbt.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Rahn O. 1937. New principles for the classification of bacteria. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt 96:273–286. [Google Scholar]
- 7.Farmer JJ, III, Brenner DJ, Ewing WH. 1980. Opposition to recent proposals which would reject the family name Enterobacteriaceae and Escherichia as its type genus. Int J Syst Bacteriol 30:660–673. doi: 10.1099/00207713-30-4-660. [DOI] [Google Scholar]
- 8.Borman EK, Stuart CA, Wheeler KM. 1944. Taxonomy of the family Enterobacteriaceae. J Bacteriol 48:351–367. doi: 10.1128/JB.48.3.351-367.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewing WH. 1986. Taxonomy and nomenclature, p 1–16. In Edwards PR, Ewing WH (ed), Edwards and Ewing’s identification of Enterobacteriaceae, 4th ed. Elsevier, New York, NY. [Google Scholar]
- 10.Brenner DJ, Farmer JJ, III, Hickman FW, Asbury MA, Steigerwalt AG. 1979. Taxonomic and nomenclature changes in Enterobacteriaceae. HEW publication no. (CDC) 79-8356. CDC, Atlanta, GA. [Google Scholar]
- 11.Farmer JJ, III, Davis BR, Hickman-Brenner FW, McWhorter A, Huntley-Carter GF, Ashbury MA, Riddle C, Wathen-Grady HG, Elias C, Fanning GR, Steigerwalt AG, O’Hara CM, Morris GK, Smith PB, Brenner DJ. 1985. Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J Clin Microbiol 21:46–76. doi: 10.1128/JCM.21.1.46-76.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holst O. 2007. The structure of core regions from enterobacterial lipopolysaccharides—an update. FEMS Microbiol Lett 271:3–11. doi: 10.1111/j.1574-6968.2007.00708.x. [DOI] [PubMed] [Google Scholar]
- 13.Westfall CS. 2018. An “uncommon” role for cyclic enterobacterial common antigen in maintaining outer membrane integrity. mBio 9:e02162-18. doi: 10.1128/mBio.02162-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janda JM, Abbott SL, McIver CJ. 2016. Plesiomonas shigelloides revisited. Clin Microbiol Rev 29:349–374. doi: 10.1128/CMR.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brenner DJ, Farmer JJ, III, 2005. Family 1. Enterobacteriaceae Rahn 1937, Nom. Fam. Cons. Opin. 15, Jud. Comm. 1958a, 73; Ewing, Farmer, and Brenner 1980, 674; Judicial Commission 1981, 104, p 587–607. In Brenner DJ, Krieg NR, Staley JT (ed), Bergey’s manual of systematic bacteriology, 2nd ed, vol 2. Springer, New York, NY. [Google Scholar]
- 16.Lightfield J, Fram NR, Ely B. 2011. Across bacterial phyla, distantly-related genomes with similar genomic GC content have similar patterns of amino acid usage. PLoS One 6:e17677. doi: 10.1371/journal.pone.0017677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossau R, Van Landschoot A, de Gillis M, Ley J. 1991. Taxonomy of Moraxellaceae fam. nov., a new bacterial family to accommodate the genera Moraxella, Acinetobacter, and Psychrobacter and related organisms. Int J Syst Bacteriol 41:310–319. doi: 10.1099/00207713-41-2-310. [DOI] [Google Scholar]
- 18.GBD Diarrhoeal Diseases Collaborators. 2017. Estimates of global, regional, and national morbidity, mortality, and aetiologies of diarrhoeal diseases: a systematic analysis for the Global Burden Disease Study 2015. Lancet Infect Dis 17:909–948. doi: 10.1016/S1473-3099(17)30276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chlebicz A, Śliżewska K. 2018. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: a review. Int J Environ Res Public Health 15:863. doi: 10.3390/ijerph15050863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland D, Thomson L, Mahmoudzadeh N, Khaled A. 2020. Estimating deaths from foodborne diseases in the UK for 11 key pathogens. BMJ Open Gastroenterol 7:e000377. doi: 10.1136/bmjgast-2020-000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buzby JC, Roberts T. 2009. The economics of enteric infections: human foodborne disease costs. Gastroenterology 136:1851–1862. doi: 10.1053/j.gastro.2009.01.074. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien SJ. 2012. The public health impact of food-related illness. Curr Opin Infect Dis 25:537–545. doi: 10.1097/QCO.0b013e328356aeba. [DOI] [PubMed] [Google Scholar]
- 23.Bintsis T. 2017. Foodborne pathogens. AIMS Microbiol 3:529–563. doi: 10.3934/microbiol.2017.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer JJ, III. 2015. My 40-year history with Cronobacter/Enterobacter sakazakii—lessons learned, myths debunked, and recommendations. Front Pediatr 3:84. doi: 10.3389/fped.2015.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henry M, Fouladkhah A. 2019. Outbreak history, biofilm formation, and preventive measures for control of Cronobacter sakazakii in infant formula and infant care settings. Microorganisms 7:77. doi: 10.3390/microorganisms7030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yong W, Guo B, Shi X, Cheng T, Chen M, Jiang X, Ye Y, Wang J, Xie G, Ding J. 2018. An investigation of an acute gastroenteritis outbreak: Cronobacter sakazakii, a potential cause of food-borne illness. Front Microbiol 9:2549. doi: 10.3389/fmicb.2018.02549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tack DM, Ray L, Griffin PM, Cieslak PR, Dunn J, Rissman T, Jervis R, Lathrop S, Muse A, Duwell M, Smith K, Tobin-D'Angelo M, Vugia DJ, Zablotsky Kufel J, Wolpert BJ, Tauxe R, Payne DC. 2020. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 2016–2019. MMWR Morb Mortal Wkly Rep 69:509–514. doi: 10.15585/mmwr.mm6917a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Jervis R, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2018. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—foodborne diseases active surveillance network, 10 U.S. sites, 2006–2017. Morb Mortal Wkly Rep 67:324–328. doi: 10.15585/mmwr.mm6711a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buchholz U, Bernard H, Werber D, Bohmer MM, Remschmidt C, Wilking H, Delere Y, An der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Höhle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kühne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365:1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 31.Lamba K, Nelson JA, Kimura AC, Poe A, Collins J, Kao AS, Cruz L, Inami G, Vaishampayan J, Garza A, Chaturvedi V, Vugia D. 2016. Shiga toxin 1-producing Shigella sonnei infections, California, United States, 2014–2015. Emerg Infect Dis 22:679–686. doi: 10.3201/eid2204.151825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kang J, Sickbert-Bennett EE, Brown VM, Weber DJ, Rutala WA. 2012. Relative frequency of health care-associated pathogens by infection site at a university hospital from 1980 to 2008. Am J Infect Control 40:416–420. doi: 10.1016/j.ajic.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Novosad SA, Fike L, Dudeck MA, Allen-Bridson K, Edwards JR, Edens C, Sinkowitz-Cochran R, Powell K, Kuhar D. 2020. Pathogens causing central-line-associated bloodstream infections in acute-care hospitals—United States, 2011–2017. Infect Control Hosp Epidemiol 41:313–319. doi: 10.1017/ice.2019.303. [DOI] [PubMed] [Google Scholar]
- 34.Potter RF, D’Souza AW, Dantas G. 2016. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat 29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerqueira GC, Earl AM, Ernst CM, Grad YH, Dekker JP, Feldgarden M, Chapman SB, Reis-Cunha JL, Shea TP, Young S, Zeng Q, Delaney ML, Kim D, Peterson EM, O'Brien TF, Ferraro MJ, Hooper DC, Huang SS, Kirby JE, Onderdonk AB, Birren BW, Hung DT, Cosimi LA, Wortman JR, Murphy CI, Hanage WP. 2017. Multi-institute analysis of carbapenem resistance reveals remarkable diversity, unexplained mechanisms, and limited clonal outbreaks. Proc Natl Acad Sci U S A 114:1135–1140. doi: 10.1073/pnas.1616248114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiner-Lastinger LM, Abner S, Edwards JR, Kallen AJ, Karlsson M, Magill SS, Pollock D, See I, Soe MM, Walters MS, Dudeck MA. 2020. Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect Control Hosp Epidemiol 41:1–18. doi: 10.1017/ice.2019.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skerman VBD, McGowan V, Sneath PHA. 1980. Approved lists of bacterial names. Int J Syst Bacteriol 30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- 38.Lapage SP. 1979. Proposal of Enterobacteraceae nom. nov. as a substitute for the illegitimate but conserved name Enterobacteriaceae Rahn 1937. Request for an opinion. Int J Syst Bacteriol 29:265–266. doi: 10.1099/00207713-29-3-265. [DOI] [Google Scholar]
- 39.Judicial Commission of the International Committee on Systematic Bacteriology. 1981. Present standing of the family name Enterobacteriaceae Rahn 1937. Int J Syst Bacteriol 31:104. doi: 10.1099/00207713-31-1-104. [DOI] [Google Scholar]
- 40.Ewing WH, Farmer JJ, III, Brenner DJ. 1980. Proposal of Enterobacteriaceae fam. nov., nom. rev. to replace Enterobacteriaceae Rahn 1937, nom. fam. cons. (Opin. 15, Jud. Comm. 1958), which lost standing in nomenclature on 1 January 1980. Int J Syst Bacteriol 30:674–675. doi: 10.1099/00207713-30-4-674. [DOI] [Google Scholar]
- 41.Brenner DJ. 1983. Opposition to the proposal to replace the family name Enterobacteriaceae. Int J Syst Bacteriol 33:892–895. doi: 10.1099/00207713-33-4-892. [DOI] [Google Scholar]
- 42.Parte AC. 2018. LPSN—List of Prokaryotic Names with Standing in Nomenclature (bacterio.net), 20 years on. Int J Syst Evol Microbiol 68:1825–1829. doi: 10.1099/ijsem.0.002786. [DOI] [PubMed] [Google Scholar]
- 43.Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci U S A 99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimont PAD, Grimont F, Starr MP. 1979. Serratia ficaria sp. nov., a bacterial species associated with Smyrna figs and the fig wasp Blastophaga psenes. Curr Microbiol 2:277–280. doi: 10.1007/BF02602859. [DOI] [Google Scholar]
- 45.Kosako Y, Sakazaki R, Yoshizaki E. 1984. Yokenella regensburgei gen. nov., sp. nov.: a new genus and species in the family Enterobacteriaceae. Jpn J Med Sci Biol 37:117–124. doi: 10.7883/yoken1952.37.117. [DOI] [PubMed] [Google Scholar]
- 46.O'Hara CM, Steigerwalt AG, Hill BC, Farmer JJ, Fanning GR, Brenner DJ. 1989. Enterobacter hormaechei, a new species of the family Enterobacteriaceae formerly known as enteric group 75. J Clin Microbiol 27:2046–2049. doi: 10.1128/JCM.27.9.2046-2049.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McWhorter AC, Haddock RL, Nocon FA, Steigerwalt AG, Brenner DJ, Aleksic S, Bockemühl J, Farmer JJ, III. 1991. Trabusiella guamensis, a new genus and species of the family Enterobacteriaceae that resembles Salmonella subgroups 4 and 5. J Clin Microbiol 29:1480–1485. doi: 10.1128/JCM.29.7.1480-1485.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore ERB, Mihaylova SA, Vandamme P, Krichevsky MI, Dijkshoorn L. 2010. Microbial systematics and taxonomy: relevance for a microbial commons. Res Microbiol 161:430–438. doi: 10.1016/j.resmic.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 50.Oren A, Garrity GM. 2014. Then and now: a systematic review of the systematics of prokaryotes in the last 80 years. Antonie Van Leeuwenhoek 106:43–56. doi: 10.1007/s10482-013-0084-1. [DOI] [PubMed] [Google Scholar]
- 51.Brenner DJ. 1978. Characterization and clinical identification of Enterobacteriaceae by DNA hybridization. Prog Clin Pathol 7:71–117. [PubMed] [Google Scholar]
- 52.Woo PCY, Lau SKP, Teng JLL, Tse H, Yuen KY. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 14:908–934. doi: 10.1111/j.1469-0691.2008.02070.x. [DOI] [PubMed] [Google Scholar]
- 53.Lau SKP, Ng KHL, Woo PCY, Yip K-T, Fung AMY, Woo GKS, Chan K-M, Que T-L, Yuen K-Y. 2006. Usefulness of the MicroSeq 500 16S rDNA bacterial identification system for identification of anaerobic Gram positive bacilli isolated from blood cultures. J Clin Pathol 59:219–222. doi: 10.1136/jcp.2004.025247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayashi Sant'Anna F, Bach E, Porto RZ, Guella F, Hayashi Sant'Anna E, Passaglia LMP. 2019. Genomic metrics made easy: what to do and where to go in the new era of bacterial taxonomy. Crit Rev Microbiol 45:182–200. doi: 10.1080/1040841X.2019.1569587. [DOI] [PubMed] [Google Scholar]
- 55.Kämpfer P. 2012. Systematics of prokaryotes: the state of the art. Antonie Van Leeuwenhoek 101:3–11. doi: 10.1007/s10482-011-9660-4. [DOI] [PubMed] [Google Scholar]
- 56.Mahato NK, Gupta V, Singh P, Kumari R, Verma H, Tripathi C, Rani P, Sharma A, Singhvi N, Sood U, Hira P, Kohli P, Nayyar N, Puri A, Bajaj A, Kumar R, Negi V, Talwar C, Khurana H, Nagar S, Sharma M, Mishra H, Singh AK, Dhingra G, Negi RK, Shakarad M, Singh Y, Lal R. 2017. Microbial taxonomy in the era of OMICS: application of DNA sequences, computational tools and techniques. Antonie Van Leeuwenhoek 110:1357–1371. doi: 10.1007/s10482-017-0928-1. [DOI] [PubMed] [Google Scholar]
- 57.Gavini F, Mergaert J, Beji A, Mielcarek C, Izard D, Kersters K, De Ley J. 1989. Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea disoersa sp. nov. Int J Syst Bacteriol 39:337–345. doi: 10.1099/00207713-39-3-337. [DOI] [Google Scholar]
- 58.Brady C, Cleenwerck I, Venter S, Coutinho T, De Los P. 2013. Taxonomic evaluation of the genus Enterobacter based on multilocus-sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amingenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 59.Pavan ME, Franco RJ, Rodriguez JM, Gadaleta P, Abbott SL, Janda JM, Zorzópulos J. 2005. Phylogenetic relationships of the genus Kluyvera: transfer of Enterobacter intermedius Izard et al. 1980 to the genus Kluyvera as Kluyvera intermedia comb. nov. and reclassification of Kluyvera cochleae as a later synonym of K. intermedia. Int J Syst Evol Microbiol 55:437–442. doi: 10.1099/ijs.0.63071-0. [DOI] [PubMed] [Google Scholar]
- 60.Tindall BJ, Sutton G, Garrity GM. 2017. Enterobacter aerogenes Hormaeche and Edwards 1960 (Approved Lists 1980) and Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980) share the same nomenclatural type (ATCC 13408) on the Approved Lists and are homotypic synonyms, with consequences for the name Klebsiella mobilis Bascomb et al. 1971 (Approved Lists 1980). Int J Syst Evol Microbiol 67:502–504. doi: 10.1099/ijsem.0.001572. [DOI] [PubMed] [Google Scholar]
- 61.Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, Stephan R, Joosten H. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactariid subsp. nov. Int J Syst Evol Microbiol 58:1442–1447. doi: 10.1099/ijs.0.65577-0. [DOI] [PubMed] [Google Scholar]
- 62.Drancourt M, Bollet C, Carta A, Rousselier P. 2001. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov, Raolutella terrigena comb. nov., and Raoultella planticola comb. nov. Int J Syst Evol Microbiol 51:925–932. doi: 10.1099/00207713-51-3-925. [DOI] [PubMed] [Google Scholar]
- 63.Brady CL, Venter SN, Cleenwerck I, Vandemeulebroecke K, De Vos P, Coutinho TA. 2010. Transfer of Pantoea cirea, Pantoea punctate and Pantoea terrea to the genus Tatumella emend. as Tatumella citrea comb. nov., Tatumella punctate comb. nov. and Tatumella terrea comb. nov. and description of Tatumella morbirosei sp. nov. Int J Syst Evol Microbiol 60:484–494. doi: 10.1099/ijs.0.012070-0. [DOI] [PubMed] [Google Scholar]
- 64.Sentausa E, Fournier P-E. 2013. Advantages and limitations of genomics in prokaryotic taxonomy. Clin Microbiol Infect 19:790–795. doi: 10.1111/1469-0691.12181. [DOI] [PubMed] [Google Scholar]
- 65.Janda JM. 2017. Taxonomic update on proposed nomenclature and classification changes for bacteria of medical importance, 2016. Diagn Microbiol Infect Dis 88:100–105. doi: 10.1016/j.diagmicrobio.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 66.Christensen H, Bisgaard M, Frederiksen W, Mutters R, Kuhnert P, Olsen E. 2001. Is characterization of a single isolate sufficient for valid publication of a new genus or species? Proposal to modify Recommendation 30b of the Bacteriological Code (1990 Revision). Int J Syst Evol Microbiol 51:2221–2225. doi: 10.1099/00207713-51-6-2221. [DOI] [PubMed] [Google Scholar]
- 67.Stackebrandt E, Frederiksen W, Garrity GM, Grimont PAD, Kämpfer P, Maiden MCJ, Nesme X, Rosselló-Mora R, Swings J, Trüper HG, Vauterin L, Ward AC, Whitman WB. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int J Syst Evol Microbiol 52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- 68.Rameshkumar N, Lang E, Nair S. 2010. Mangrovibacter plantisponsor gen. nov., a nitrogen-fixing bacterium isolated from a mangrove-associated wild rice (Porteresia coarctata Tateoka). Int J Syst Evol Microbiol 60:179–186. doi: 10.1099/ijs.0.008292-0. [DOI] [PubMed] [Google Scholar]
- 69.Ee R, Madhaiyan M, Ji L, Lim Y-L, Nor NM, Tee K-K, Chen J-W, Yin W-F. 2016. Chania multitudinisentens gen. nov., sp. nov., an N-acyl-homoserine-lactone-producing bacterium in the family Enterobacteriaceae isolated from landfill site soil. Int J Syst Evol Microbiol 66:2297–2304. doi: 10.1099/ijsem.0.001025. [DOI] [PubMed] [Google Scholar]
- 70.Baek C, Shin S-K, Yi H. 2019. Limnobaculum parvum gen. nov., sp. nov., isolated from a freshwater lake. Int J Syst Evol Microbiol 69:1826–1830. doi: 10.1099/ijsem.0.003401. [DOI] [PubMed] [Google Scholar]
- 71.Marathe NP, Salvà-Serra F, Karlsson R, Larsson DGJ, Moore ERB, Svensson-Stadler L, Jakobsson HE. 2019. Scandinavium goeteborgense gen. nov., sp. nov., a new member of the family Enterobacteriaceae isolated from a wound infection, carries a novel quinolone resistance gene variant. Front Microbiol 10:2511. doi: 10.3389/fmicd.2019.02511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang L, Wang D, Lee J-S, Kim D-H, Jeong JC, Kim CY, Kim SW, Lee J. 2020. Jejubacter calystegiae gen. nov., sp. nov., moderately halophilic, a new member of the family Enterobacteriaceae, isolated from beach morning glory. J Microbiol 58:357–366. doi: 10.1007/s12275-020-9294-1. [DOI] [PubMed] [Google Scholar]
- 73.Kämpfer P, McInroy JA, Glaeser SP. 2015. Enterobacter muelleri sp. nov., isolated from the rhizosphere of Zea mays. Int J Syst Evol Microbiol 65:4093–4099. doi: 10.1099/ijsem.0.000547. [DOI] [PubMed] [Google Scholar]
- 74.Sutton GG, Brinkac LM, Clarke TH, Fouts DE. 2018. Enterobacter hormaechei subsp. hoffmannii subsp. nov., Enterobacter hormaechei subsp. Xianfangensis comb. nov., Enterobacter roggenkampii sp. nov, and Enterobacter muelleri is a later heterotypic synonym of Enterobacter asburiae based upon computational analysis of sequenced Enterobacter genomes. F1000Res 7:521. doi: 10.12688/f1000research.14566.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brenner DJ, McWhorter AC, Kai A, Steigerwalt AG, Farmer JJ, III. 1986. Enterobacter asburiae sp. nov., a new species found in clinical specimens, and reassignment of Erwinia dissolvens and Erwinia nimipressuralis to the genus Enterobacter as Enterobacter dissolvens comb. nov. and Enterobacter nimipressuralis comb. nov. J Clin Microbiol 23:1114–1120. doi: 10.1128/JCM.23.6.1114-1120.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Johnson AS, Tarr CL, Brown BH, Jr, Birkhead KM, Farmer JJ, III. 2005. First case of septicemia due to a strain belonging to enteric group 58 (Enterobacteriaceae) and its designation as Averyella dalhousiensis gen. nov., based on analysis of strains from 20 additional cases. J Clin Microbiol 43:5195–5201. doi: 10.1128/JCM.43.10.5195-5201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Parker CT, Tindall BJ, Garrity GM. 2019. International Code of Nomenclature of Prokaryotes: Prokaryotic Code (2008 Revision). Int J Syst Evol Microbiol 69:S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 78.Oren A, Garrity GM, Parte AC. 2018. Why are so many effectively published names of prokaryotic taxa never validated? Int J Syst Evol Microbiol 68:2125–2129. doi: 10.1099/ijsem.0.002851. [DOI] [PubMed] [Google Scholar]
- 79.Hata H, Natori T, Mizuno T, Kanazawa I, Eldesouky I, Hayashi M, Miyata M, Fukunaga H, Ohji S, Hosoyama A, Aono E, Yamazoe A, Tsuchikane K, Fujita N, Ezaki T. 2016. Phylogenetics of the family Enterobacteriaceae and proposal to reclassify Escherichia hermannii and Salmonella subterranean as Atlantibacter hermannii and Atlantibacter subterranea gen. nov, comb. nov. Microbiol Immunol 60:303–313. doi: 10.1111/1348-0421.12374. [DOI] [PubMed] [Google Scholar]
- 80.Brenner DJ, Fanning GR, Johnson KE, Citarella RV, Falkow S. 1969. Polynucleotide sequence relationships among members of Enterobacteriaceae. J Bacteriol 98:637–650. doi: 10.1128/JB.98.2.637-650.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Trüper HG. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464. doi: 10.1099/00207713-37-4-463. [DOI] [Google Scholar]
- 82.Lan R, Reeves PR. 2002. Escherichia coli in disguise: molecular origins of Shigella. Microbes Infect 4:1125–1132. doi: 10.1016/s1286-4579(02)01637-4. [DOI] [PubMed] [Google Scholar]
- 83.Peng J, Yang J, Jin Q. 2009. The molecular evolutionary history of Shigella spp. Infect Genet Evol 9:147–152. doi: 10.1016/j.meegid.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 84.Bercovier H, Mollaret HH, Alonso JM, Brault J, Fanning GR, Steigerwalt AG, Brenner DJ. 1980. Intra- and interspecies relatedness of Yersinia pestis by DNA hybridization and its relationship to Yersinia pseudotuberculosis. Curr Microbiol 4:225–229. doi: 10.1007/BF02605861. [DOI] [Google Scholar]
- 85.Chain PSG, Carniel E, Larimer FW, Lamerdin J, Stoutland PO, Regala WM, Georgescu AM, Vergez LM, Land ML, Motin VL, Brubaker RR, Fowler J, Hinnebusch J, Marceau M, Medigue C, Simonet M, Chenal-Francisque V, Souza B, Dacheux D, Elliott JM, Derbise A, Hauser LJ, Garcia E. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A 101:13826–13831. doi: 10.1073/pnas.0404012101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schleifer KH. 2009. Classification of Bacteria and Archaea: past, present and future. Syst Appl Microbiol 32:533–542. doi: 10.1016/j.syapm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 87.Baltrus DA. 2016. Divorcing strain classification from species names. Trends Microbiol 24:431–439. doi: 10.1016/j.tim.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 88.Alnajar S, Gupta RS. 2017. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect Genet Evol 54:108–127. doi: 10.1016/j.meegid.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 89.Ma Y, Yao R, Li Y, Wu X, Li S, An Q. 2020. Proposal for unification of the genus Metakosakonia and the genus Phytobacter to a single genus Phytobacter and reclassification of Metakosakonia massiliensis as Phytobacter massiliensis comb. nov. Curr Microbiol 77:1945–1954. doi: 10.1007/s00284-020-02004-4. [DOI] [PubMed] [Google Scholar]
- 90.Jiang L, Wang D, Kim J-S, Lee JH, Kim D-H, Kim SW, Lee J. 2020. Reclassification of genus Izhakiella into the family Erwiniaceae based on phylogenetic and genomic analyses. Int J Syst Evol Microbiol 70:3541–3546. doi: 10.1099/ijsem.0.004192. [DOI] [PubMed] [Google Scholar]
- 91.Caputo A, Merhej V, Georgiades K, Fournier P-E, Croce O, Robert C, Raoult D. 2015. Pan-genomic analysis to redefine species and subspecies based upon quantum discontinuous variation: the Klebsiella paradigm. Biol Direct 10:55. doi: 10.1186/s13062-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dai H, Lu B, Li Z, Huang Z, Cai H, Yu K, Wang D. 2020. Multilocus sequence analysis for the taxonomic updating and identification of the genus Proteus and reclassification of Proteus genospecies 5 O’Hara et al. 2000, Proteus cibarius Hyun et al. 2016 as later heterotypic synonyms of Proteus terrae Behrendt et al. 2015. BMC Microbiol 20:152. doi: 10.1186/s12866-020-01844-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Véron M. 1965. La position taxonomique des Vibrio et de certaines bacteries comparables. C R Hebd Seances Acad Sci 261:5243–5246. [PubMed] [Google Scholar]
- 94.Colwell RR, MacDonell MT, De Ley J. 1986. Proposal to recognize the family Aeromonadaceae fam. nov. Int J Syst Bacteriol 36:473–477. doi: 10.1099/00207713-36-3-473. [DOI] [Google Scholar]
- 95.Morales-López S, Yepes JA, Prada-Herrera JC, Torres-Jiménez A. 2019. Enterobacteria in the 21st century: a review focused on taxonomic changes. J Infect Dev Ctries 13:265–273. doi: 10.3855/jidc.11216. [DOI] [PubMed] [Google Scholar]
- 96.Ewing WH. 1986. Deoxyribonucleic acid (DNA) hybridization. p 17–27. In Edwards PR, Ewing WH (ed), Edwards and Ewing’s identification of Enterobacteriaceae, 4th ed. Elsevier, New York, NY. [Google Scholar]
- 97.Paradis S, Boissinot M, Paquette N, Bélanger SD, Martel EA, Boudreau DK, Picard FJ, Ouellette M, Roy PH, Bergeron MG. 2005. Phylogeny of the Enterobacteriaceae based on genes encoding elongation factor Tu and F-ATPase β-subunit. Int J Syst Evol Microbiol 55:2013–2025. doi: 10.1099/ijs.0.63539-0. [DOI] [PubMed] [Google Scholar]
- 98.Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 99.Dauga C. 2002. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int J Syst Evol Microbiol 52:531–547. doi: 10.1099/00207713-52-2-531. [DOI] [PubMed] [Google Scholar]
- 100.Roggenkamp A. 2007. Phylogenetic analysis of enteric species of the family Enterobacteriaceae using the oriC-locus. Syst Appl Microbiol 30:180–188. doi: 10.1016/j.syapm.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 101.Wertz JE, Goldstone C, Gordon DM, Riley MA. 2003. A molecular phylogeny of enteric bacteria and implications for a bacterial species concept. J Evol Biol 16:1236–1248. doi: 10.1046/j.1420-9101.2003.00612.x. [DOI] [PubMed] [Google Scholar]
- 102.Spröer C, Mendrock U, Swiderski J, Lang E, Stackebrandt E. 1999. The phylogenetic position of Serratia, Buttiauxella and some other genera of the family Enterobacteriaceae. Int J Syst Bacteriol 49:1433–1438. doi: 10.1099/00207713-49-4-1433. [DOI] [PubMed] [Google Scholar]
- 103.Criscuolo A, Issenhuth-Jeanjean S, Didelot X, Thoreil K, Hale J, Parkhill J, Thomson NR, Weill F-X, Falush D, Brisse S. 2019. The speciation and hybridization history of the genus Salmonella. Microb Genom 5:e000284. doi: 10.1099/mgen.0.000284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shelobolina ES, Sullivan SA, O'Neill KR, Nevin KP, Lovley DR. 2004. Isolation, characterization, and U(VI)-reducing potential of a facultatively anaerobic, acid-resistant bacterium from low-pH, nitrate- and U(VI)-contaminated subsurface sediment and description of Salmonella subterranean sp. nov. Appl Environ Microbiol 70:2959–2965. doi: 10.1128/aem.70.5.2959-2965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Davin-Regli A, Lavigne J-P, Pagès J-M. 2019. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev 32:e00002-19. doi: 10.1128/CMR.00002-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu W, Feng Y, Zong Z. 2020. Precise species identification for Enterobacter: a genome sequence-based study with reporting of two novel species, Enterobacter quasiroggenkampii sp. nov. and Enterobacter quasimori sp. nov. mSystems 5:e00527-20. doi: 10.1128/mSystems.00527-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beyrouthy R, Barets M, Marion E, Dananché C, Dauwalder O, Robin F, Gauthier L, Jousset A, Dortet L, Guérin F, Bénet T, Cassier P, Vanhems P, Bonnet R. 2018. Novel Enterobacter lineage as leading cause of nosocomial outbreak involving carbapenemase-producing strains. Emerg Infect Dis 24:1505–1515. doi: 10.3201/eid2408.180151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ewing WH, Ross AJ, Brenner DJ, Fanning GR. 1978. Yersinina ruckeri sp. nov., the redmouth (RM) bacterium. Int J Syst Bacteriol 28:37–44. doi: 10.1099/00207713-28-1-37. [DOI] [Google Scholar]
- 109.De Keukeleire S, De Bel A, Jansen Y, Janssens M, Wauters G, Piérard D. 2014. Yersinia ruckeri, an unusual microorganism isolated from a human wound infection. New Microbes New Infect 2:134–135. doi: 10.1002/nmi2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sulakvelidze A. 2000. Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect 2:497–513. doi: 10.1016/S1286-4579(00)00311-7. [DOI] [PubMed] [Google Scholar]
- 111.Kotetishvili M, Kreger A, Wauters G, Morris JG, Jr, Sulakvelidze A, Stine OC. 2005. Multilocus sequence typing for studying genetic relationships among Yersinia species. J Clin Microbiol 43:2674–2684. doi: 10.1128/JCM.43.6.2674-2684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Grimont PAD, Grimont F, Richard C, Sakazaki R. 1980. Edwardsiella hoshinae, a new species of Enterobacteriaceae. Curr Microbiol 4:347–351. doi: 10.1007/BF02605375. [DOI] [Google Scholar]
- 113.Walton DT, Abbott SL, Janda JM. 1993. Sucrose-positive Edwardsiella tarda mimicking a biogroup 1 strain isolated from a patient with cholelithiasis. J Clin Microbiol 31:155–156. doi: 10.1128/JCM.31.1.155-156.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Leung MJ. 1996. Plesiomonas shigelloides and sucrose-positive Edwardsiella tarda bacteremia in a man with obstructive jaundice. Pathology 28:68–69. doi: 10.1080/00313029600169563. [DOI] [PubMed] [Google Scholar]
- 115.Grimont PAD, Farmer JJ, III, Grimont F, Asbury MA, Brenner DJ, Deval C. 1983. Ewingella americana gen. nov., sp. nov., a new Enterobacteriaceae isolated from clinical specimens. Ann Microbiol (Inst Pasteur) 134:39–52. doi: 10.1016/0769-2609(83)90102-3. [DOI] [PubMed] [Google Scholar]
- 116.Kämpfer P, Glaeser SP, Nilsson KJ, Eberhard T, Håkansson S, Guy L, Roos S, Busse H-J, Terenius O. 2015. Proposal of Thorsellia kenyensis sp. nov., and Thorsellia kandunguensis sp. nov., isolated from larvae of Anopheles arabiensis, as members of the family Thorselliaceae fam. nov. Int J Syst Evol Microbiol 65:444–451. doi: 10.1099/ijs.0.070292-0. [DOI] [PubMed] [Google Scholar]
- 117.Gupta RS, Lo B, Son J. 2018. Phylogenomics and comparative genomic studies robustly support division of the genus Mycobacterium into an emended genus Mycobacterium and four novel genera. Front Microbiol 9:67. doi: 10.3389/fmicb.2018.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gupta RS, Sawnani S, Adeolu M, Alnajar S, Oren A. 2018. Phylogenetic framework for the phylum Tenericutes based on genome sequence data: proposal for the creation of a new order Mycoplasmoidales ord. nov., containing two new families Mycoplasmoidaceae fam. nov and Metamycoplasmataceae fam. nov. harboring Eperythrozoon, Ureaplasma and five novel genera. Antonie Van Leeuwenhoek 111:1583–1630. doi: 10.1007/s10482-018-1047-3. [DOI] [PubMed] [Google Scholar]
- 119.Tortoli E, Brown-Elliott BA, Chalmers JD, Cirillo DM, Daley CL, Emler S, Floto RA, Garcia MJ, Hoefsloot W, Koh W-J, Lange C, Loebinger M, Maurer FP, Morimoto K, Niemann S, Richter E, Turenne CY, Vasireddy R, Vasireddy S, Wagner D, Wallace RJ, Jr, Wengenack N, van Ingen J. 2019. Same meat, different gravy: ignore the new names of mycobacteria. Eur Respir J 54:1900795. doi: 10.1183/13993003.00795-2019. [DOI] [PubMed] [Google Scholar]
- 120.Balish M, Bertaccini A, Blanchard A, Brown D, Browning G, Chalker V, Frey J, Gasparich G, Hoelzle L, Knight T, Jr, Knox C, Kuo C-H, Manso-Silván L, May M, Pollack JD, Ramirez AS, Spergser J, Taylor-Robinson D, Voloknov D, Zhao Y. 2019. Recommended rejection of the names Malacoplasma gen. nov., Mesomycoplasma gen. nov., Metamycoplasma gen. nov., Metamycoplasmataceae fam. nov. Mycoplasmoidaceae fam. nov., Mycoplasmoidales ord. nov., Mycoplasmoides gen. nov., Mycoplasmopsis gen. nov. [Gupta, Sawnani, Adeolu, Alnajar, and Oren 2018] and all proposed species comb. nov. placed therein. Int J Syst Evol Microbiol 69:3650–3653. doi: 10.1099/ijsem.0.003632. [DOI] [PubMed] [Google Scholar]
- 121.Singh NK, Wood JM, Mhatre SS, Venkateswaran K. 2019. Metagenome to phenome approach enables isolation and genomics characterization of Kalamiella piersonii gen. nov. from the International Space Station. Appl Microbiol Biotechnol 103:4483–4497. doi: 10.1007/s00253-019-09813-z. [DOI] [PubMed] [Google Scholar]
- 122.Zhang Y, Jewett C, Gilley J, Bartelt-Hunt SL, Snow DD, Hodges L, Li X. 2018. Microbial communities in the rhizosphere and the root of lettuce as affected by Salmonella-contaminated irrigation water. FEMS Microbiol Ecol 94:fiy135. doi: 10.1093/femsec/fiy135. [DOI] [PubMed] [Google Scholar]
- 123.Mathai PP, Dunn HM, Magnone P, Zhang Q, Ishii S, Chun CL, Sadowsky MJ. 2019. Association between submerged aquatic vegetation and elevated levels of Escherichia coli and potential bacterial pathogens in freshwater lakes. Sci Total Environ 657:319–324. doi: 10.1016/j.scitotenv.2018.11.484. [DOI] [PubMed] [Google Scholar]
- 124.Coutinho TA, Venter SN. 2009. Pantoea ananastis: an unconventional pathogen. Mol Plant Pathol 10:325–335. doi: 10.1111/j.1364-3703.2009.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kado CI. 2006. Erwinia and related genera, p 443–451. In Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (ed), The prokaryotes: a handbook, 3rd ed, vol 6. Springer, New York, NY. [Google Scholar]
- 126.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow M, Verdier V, Beer SV, Machado MA, Toth I, Salmond G, Foster GD. 2012. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang Y, Fan Q, Loria R. 2016. A re-evaluation of the taxonomy of phytopathogenic genera Dickeya and Pectobacterium using whole-genome sequencing data. Syst Appl Microbiol 39:252–259. doi: 10.1016/j.syapm.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 128.Charkowski AO. 2018. The changing face of bacterial soft-rot diseases. Annu Rev Phytopathol 56:269–288. doi: 10.1146/annurev-phyto-080417-045906. [DOI] [PubMed] [Google Scholar]
- 129.Ma X, Schloop A, Swingle B, Perry KL. 2018. Pectobacterium and Dickeya responsible for potato backleg disease in New York State in 2016. Plant Dis 102:1834–1840. doi: 10.1094/PDIS-10-17-1595-RE. [DOI] [PubMed] [Google Scholar]
- 130.Pérombelon MCM. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol 51:1–12. doi: 10.1046/j.0032-0862.2001.Shorttitle.doc.x. [DOI] [Google Scholar]
- 131.Khan MA, Zhao Y, Korban SS. 2012. Molecular mechanisms of pathogenesis and resistance to the bacterial pathogen Erwinia amylovora, causal agent of fire blight disease in Rosaceae. Plant Mol Biol Rep 30:247–260. doi: 10.1007/s11105-011-0334-1. [DOI] [Google Scholar]
- 132.Shtienberg D, Manulis-Sasson S, Zilberstaine M, Oppenheim D, Shwartz H. 2015. The incessant battle against fire blight in pears: 30 years of challenges and successes in managing the disease in Israel. Plant Dis 99:1048–1058. doi: 10.1094/PDIS-01-15-0101-FE. [DOI] [PubMed] [Google Scholar]
- 133.Brady C, Arnold D, McDonald J, Denman S. 2017. Taxonomy and identification of bacteria associated with acute oak decline. World J Microbiol Biotechnol 33:143. doi: 10.1007/s11274-017-2296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brady C, Allainguillaume J, Denman S, Arnold D. 2016. Rapid identification of bacteria associated with Acute Oak Decline by high-resolution melt analysis. Lett Appl Microbiol 63:89–95. doi: 10.1111/lam.12593. [DOI] [PubMed] [Google Scholar]
- 135.Dutkiewicz J, Mackiewicz B, Lemieszek MK, Golec M, Milanowski J. 2016. Pantoea agglomerans: a mysterious bacterium of evil and good. Part III. Deleterious effects: infections of humans, animals, and plants. Ann Agric Environ Med 23:197–205. doi: 10.5604/12321966.1203878. [DOI] [PubMed] [Google Scholar]
- 136.Hauben L, Moore ERB, Vauterin L, Steenackers M, Mergaert J, Verdonck L, Swings J. 1998. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst Appl Microbiol 21:384–397. doi: 10.1016/S0723-2020(98)80048-9. [DOI] [PubMed] [Google Scholar]
- 137.Walterson AM, Stavrinides J. 2015. Pantoea insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol Rev 39:968–984. doi: 10.1093/femsre/fuv027. [DOI] [PubMed] [Google Scholar]
- 138.Aremu BR, Babalola OO. 2015. Classification and taxonomy of vegetable macergens. Front Microbiol 6:1361. doi: 10.3389/fmicb.2015.01361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 140.Chaiwong T, Srivoramas T, Sueabsamran P, Sukontason K, Sanford MR, Sukontason KL. 2014. The blow fly, Chrysomya megacephala, and the house fly, Musca domestica, as mechanical vectors of pathogenic bacteria in northeast Thailand. Trop Biomed 31:336–346. [PubMed] [Google Scholar]
- 141.Poudel A, Hathcock T, Butaye P, Kang Y, Price S, Macklin K, Walz P, Cattley R, Katalah A, Adekanmbi F, Wang C. 2019. Multidrug-resistant Escherichia coli, Klebsiella pneumoniae, and Staphylococcus spp. in houseflies and blowflies from farms and their environment settings. J Environ Res Public Health 16:3583. doi: 10.3390/ijerph16193583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Khamesipour F, Lankarani KB, Honarvar TE, Kwenti TE. 2018. A systematic review of human pathogens carried by the housefly (Musca domestica L.). BMC Public Health 18:1049. doi: 10.1186/s12889-018-5934-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Puri-Giri R, Ghosh A, Thomson JL, Zurek L. 2017. House flies in the confined cattle environment carry non-O157 Shiga toxin-producing Escherichia coli. J Med Entomol 54:726–732. doi: 10.1093/jme/tjw240. [DOI] [PubMed] [Google Scholar]
- 144.Burrus RG, Hogsette JA, Kaufman PE, Maruniak JE, Simonne AH, Mai V. 2017. Prevalence of Escherichia coli O157:H7 from house flies (Diptera: Muscidae) and dairy samples in north central Florida. J Med Entomol 54:733–741. doi: 10.1093/jme/tjw205. [DOI] [PubMed] [Google Scholar]
- 145.Boiocchi F, Davies MP, Hilton AC. 2019. An examination of flying insects in seven hospitals in the United Kingdom and carriage of bacteria by true flies (Diptera: Calliphoridae, Dolichopodidae, Fanniidae, Muscidae, Phoridae, Psychodidae, Sphaeroceridae). J Med Entomol 56:1684–1697. doi: 10.1093/jme/tjz086. [DOI] [PubMed] [Google Scholar]
- 146.Oliva GR, Diaz C, Fuentes González O, Martinez MD, Fernández C, Cordoví R, Lago RM, Herrera N. 2010. Blatella germanica as a possible cockroach vector of micro-organisms in a hospital. J Hosp Infect 74:93–95. doi: 10.1016/j.jhin.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 147.Donkor ES. 2019. Nosocomial pathogens: an in-depth analysis of the vectorial potential of cockroaches. Trop Med 4:14. doi: 10.3390/tropicalmed4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Donkor ES. 2020. Cockroaches and food-borne pathogens. Environ Health Insights 14:1178630220913365. doi: 10.1177/1178630220913365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Solomon F, Belayneh F, Kibru G, Ali S. 2016. Vector potential of Blatella germanica (L.) (Dictyoptera: Blattidae) for medically important bacteria at food handling establishments in Jimma Town, Southwest Ethiopia. BioMed Res Int 2016:1–6. doi: 10.1155/2016/3490906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Islam W, Noman A, Naveed H, Alamri SA, Hashem M, Huang Z, Chen HYH. 2020. Plant-insect vector-virus interactions under environmental change. Sci Total Environ 701:135044. doi: 10.1016/j.scitotenv.2019.135044. [DOI] [PubMed] [Google Scholar]
- 151.Nadarasah G, Stavrinides J. 2011. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev 35:555–575. doi: 10.1111/j.1574-6976.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- 152.Rossmann S, Dees MW, Perminow J, Meadow R, Brurberg MB. 2018. Soft rot Enterobacteriaceae are carried by a large range of insect species in potato fields. Appl Environ Microbiol 84:e00281-18. doi: 10.1128/AEM.00281-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Medrano EG, Esquivel J, Bell A, Greene J, Roberts P, Bacheler J, Marois J, Wright D, Nichols R, Lopez J. 2009. Potential for Nezara viridula (Hemiptera: Pentatomidae) to transmit bacterial and fungal pathogens in cotton bolls. Curr Microbiol 59:405–412. doi: 10.1007/s00284-009-9452-5. [DOI] [PubMed] [Google Scholar]
- 154.Grode AS, Brisco-McCann E, Wiriyajitsonboom P, Hausbeck MK, Szendrei Z. 2019. Managing onion thrips can limit bacterial stalk and leaf necrosis in Michigan onion fields. Plant Dis 103:938–943. doi: 10.1094/PDIS-07-18-1271-RE. [DOI] [PubMed] [Google Scholar]
- 155.Sitz RA, Aquino VM, Tisserat NA, Cranshaw WS, Stewart JE. 2019. Insects visiting drippy blight diseased red oak trees are contaminated with the pathogenic bacterium Lonsdalea quercina. Plant Dis 103:1940–1946. doi: 10.1094/PDIS-12-18-2248-RE. [DOI] [PubMed] [Google Scholar]
- 156.Da Silva Lucas AJ, Menegon de Oliveira L, de Rocha M, Prentice C. 2020. Edible insects: an alternative of nutritional, functional and bioactive compounds. Food Chem 311:126022. doi: 10.1016/j.foodchem.2019.126022. [DOI] [PubMed] [Google Scholar]
- 157.Kim T-K, Yong HI, Kim Y-B, Kim H-W, Choi Y-S. 2019. Edible insects as a protein source: a review of public health perception, processing technology, and research trends. Food Sci Anim Resour 39:521–540. doi: 10.5851/kosfa.2019.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Garofalo C, Milanović V, Cardinali F, Aquilanti L, Clementi F, Osimani A. 2019. Current knowledge on the microbiota of edible insects intended for human consumption: a state-of-the art review. Food Res Int 125:108527. doi: 10.1016/j.foodres.2019.108527. [DOI] [PubMed] [Google Scholar]
- 159.Galęcki R, Sokól R. 2019. A parasitological evaluation of edible insects and their role in the transmission of parasitic diseases to humans and animals. PLoS One 14:e0219303. doi: 10.1371/journal.pone.0219303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Grabowski NT, Tchibozo S, Abdulmawjood A, Acheuk F, M’Saad Guerfali M, Sayed WAA, Plötz M. 2020. Edible insects in Africa in terms of food, wildlife resource, and pest management legislation. Foods 9:502. doi: 10.3390/foods9040502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Greenhalgh JP, Amund D. 2019. Examining the presence of Cronobacter spp. in ready-to-eat edible insects. Food Saf (Tokyo) 7:74–78. doi: 10.14252/foodsafetyfscj.D-19-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gordon DM, FitzGibbon F. 1999. The distribution of enteric bacteria from Australian mammals: host and geographical effects. Microbiology 145:2663–2671. doi: 10.1099/00221287-145-10-2663. [DOI] [PubMed] [Google Scholar]
- 163.Zhao C, Tang N, Wu Y, Zhang Y, Wu Z, Li W, Qin X, Zhao J, Zhang G. 2012. First reported fatal Morganella morganii infections in chickens. Vet Microbiol 156:452–455. doi: 10.1016/j.vetmic.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 164.Roe WD, Rogers L, Pinpimai K, Dittmer K, Marshall J, Chilvers BL. 2015. Septicaemia and meningitis caused by infection of New Zealand sea lion pups with a hypermucoviscous strain of Klebsiella pneumoniae. Vet Microbiol 176:301–308. doi: 10.1016/j.vetmic.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 165.Santamaria J, Toranzos GA. 2003. Enteric pathogens and soil: a short review. Int Microbiol 6:5–9. doi: 10.1007/s10123-003-0096-1. [DOI] [PubMed] [Google Scholar]
- 166.Holden N, Pritchard L, Toth I. 2009. Colonization outwith the colon: plants as an alternative environmental reservoir for human pathogenic enterobacteria. FEMS Microbiol Rev 33:689–703. doi: 10.1111/j.1574-6976.2008.00153.x. [DOI] [PubMed] [Google Scholar]
- 167.Rodriguez-Medina N, Barrios-Camacho H, Duran-Bedolla J, Garza-Ramos U. 2019. Klebsiella variicola: an emerging pathogen in humans. Emerg Microbes Infect 8:973–988. doi: 10.1080/22221751.2019.1634981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Seki M, Gotoh K, Nakamura S, Akeda Y, Yoshii T, Miyaguchi S, Inohara H, Horii T, Oishi K, Iida T, Tomono K. 2013. Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. J Med Microbiol 62:801–803. doi: 10.1099/jmm.0.051334-0. [DOI] [PubMed] [Google Scholar]
- 169.Berry GJ, Loeffelholz MJ, Williams-Bouyer N. 2015. An investigation into laboratory misidentification of a bloodstream Klebsiella variicola infection. J Clin Microbiol 53:2793–2794. doi: 10.1128/JCM.00841-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fontana L, Bonura E, Lyski Z, Messer W. 2019. The brief case: Klebsiella variicola—identifying the misidentified. J Clin Microbiol 57:e00826-18. doi: 10.1128/JCM.00825-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hajjar R, Ambaraghassi G, Sebajang H, Schwenter F, Su S-H. 2020. Raoultella ornithinolytica: emergence and resistance. Infect Drug Resist 13:1091–1104. doi: 10.2147/IDR.S191387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Keyes J, Johnson EP, Epelman M, Cadilla A, Ali S. 2020. Leclercia adecarboxylata: an emerging pathogen among pediatric infections. Cureus 12:e8049. doi: 10.7759/cureus.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ioannou P. 2019. Escherichia hermannii infections in humans: a systematic review. Trop Med Infect Dis 4:17. doi: 10.3390/tropicalmed4010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Janda JM, Abbott SL. 2006. New gram-negative enteropathogens: fact or fancy? Rev Med Microbiol 17:27–37. doi: 10.1097/01.revmedmi.0000237166.02265.1e. [DOI] [Google Scholar]
- 175.Janda JM, Abbott SL. 2011. Revisiting bacterial gastroenteritis, part I: issues, possible approaches, and an ever-expanding list of etiologic agents. Clin Microbiol Newslett 33:71–76. doi: 10.1016/j.clinmicnews.2011.04.002. [DOI] [Google Scholar]
- 176.Humphries RM, Linscott AJ. 2015. Laboratory diagnosis of bacterial gastroenteritis. Clin Microbiol Rev 28:3–31. doi: 10.1128/CMR.00073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schuetz AN. 2019. Emerging agents of gastroenteritis: Aeromonas, Plesiomonas, and the diarrheagenic pathotypes of Escherichia coli. Semin Diagn Pathol 36:187–192. doi: 10.1053/j.semdp.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 178.Haynes J, Hawkey PM. 1989. Providencia alcalifaciens and travellers’ diarrhoea. BMJ 299:94–95. doi: 10.1136/bmj.299.6691.94-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Albert MJ, Alam K, Ansaruzzaman M, Islam MM, Rahman ASMH, Haider K, Bhuiyan NA, Nahar S, Ryan N, Montanaro J, Mathan MM. 1992. Pathogenesis of Providencia alcalifaciens-induced diarrhea. Infect Immun 60:5017–5024. doi: 10.1128/IAI.60.12.5017-5024.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Albert MJ, Faruque ASG, Mahalanabis D. 1998. Association of Providencia alcalifaciens with diarrhea in children. J Clin Microbiol 36:1433–1435. doi: 10.1128/JCM.36.5.1433-1435.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chlibek R, Jirous J, Beran J. 2002. Diarrhea outbreak among Czech Army Field Hospital personnel caused by Providencia alcalifaciens. J Travel Med 9:151–152. doi: 10.2310/7060.2002.23190. [DOI] [PubMed] [Google Scholar]
- 182.Murata T, Iida T, Shiomi Y, Tagomori K, Akeda Y, Yanagihara I, Mushiake S, Ishiguro F, Honda T. 2001. A large outbreak of foodborne infection attributed to Providencia alcalifaciens. J Infect Dis 184:1050–1055. doi: 10.1086/323458. [DOI] [PubMed] [Google Scholar]
- 183.Shima A, Hinenoya A, Asakura M, Sugimoto N, Tsukamoto T, Ito H, Nagita A, Faruque SM, Yamasaki S. 2012. Molecular characterizations of cytolethal distending toxin produced by Providencia alcalifaciens strains isolated from patients with diarrhea. Infect Immun 80:1323–1332. doi: 10.1128/IAI.05831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Asakura H, Momose Y, Ryu C-H, Kasuga F, Yamamoto S, Kumagai S, Igimi S. 2013. Providencia alcalifaciens causes barrier dysfunction and apoptosis in tissue cell culture: potent role of lipopolysaccharides on diarrheagenicity. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 30:1459–1466. doi: 10.1080/19440049.2013.790086. [DOI] [PubMed] [Google Scholar]
- 185.Janda JM, Abbott SL, Woodward D, Khashe S. 1998. Invasion of HEp-2 and other eukaryotic cell lines by providenciae: further evidence supporting the role of Providencia alcalifaciens in bacterial gastroenteritis. Curr Microbiol 37:159–165. doi: 10.1007/s002849900357. [DOI] [PubMed] [Google Scholar]
- 186.Shah M, Odoyo E, Larson PS, Apondi E, Kathiiko C, Miringu G, Nakashima M, Ichinose Y. 2015. First report of a foodborne Providencia alcalifaciens outbreak in Kenya. Am J Trop Med Hyg 93:497–500. doi: 10.4269/ajtmh.15-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Shah MM, Odoyo E, Ichinose Y. 2019. Epidemiology and pathogenesis of Providencia alcalifaciens infections. Am J Trop Med Hyg 101:290–293. doi: 10.4269/ajtmh.18-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yoh M, Matsuyama J, Ohnishi M, Takagi K, Miyagi H, Mori K, Park K-S, Ono T, Honda T. 2005. Importance of Providencia species as a major cause of travellers’ diarrhea. J Med Microbiol 54:1077–1082. doi: 10.1099/jmm.0.45846-0. [DOI] [PubMed] [Google Scholar]
- 189.Huys G, Cnockaert M, Janda JM, Swings J. 2003. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int J Syst Evol Microbiol 53:807–810. doi: 10.1099/ijs.0.02475-0. [DOI] [PubMed] [Google Scholar]
- 190.Janda JM, Abbott SL, Albert MJ. 1999. Prototypal diarrheagenic strains of Hafnia alvei are actually members of the genus Escherichia. J Clin Microbiol 37:2399–2401. doi: 10.1128/JCM.37.8.2399-2401.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Oh J-Y, Kang M-S, Hwang H-T, An B-K, Kwon J-H, Kwon Y-K. 2011. Epidemiological investigation of eae-positive Escherichia coli and Escherichia albertii strains isolated from healthy wild birds. J Microbiol 49:747–752. doi: 10.1007/s12275-011-1133-y. [DOI] [PubMed] [Google Scholar]
- 192.Oaks JL, Besser TE, Walk ST, Gordon DM, Beckmen KB, Burek KA, Haldorson GJ, Bradway DS, Ouellette L, Rurangirwa FR, Davis MA, Dobbin G, Whittam TS. 2010. Escherichia albertii in wild and domestic birds. Emerg Infect Dis 16:638–646. doi: 10.3201/eid1604.090695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bhatt S, Egan M, Critelli B, Kouse A, Kalman D, Upreti C. 2018. The evasive enemy: insights into the virulence and epidemiology of the emerging attaching and effacing pathogen Escherichia albertii. Infect Immun 87:e00254-18. doi: 10.1128/IAI.00254-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Grillová L, Sedláček I, Páchníková G, Staňková E, Švec P, Holochová P, Micenková L, Bosák J, Slaninová I, Šmajs D. 2018. Characterization of four Escherichia albertii isolates collected from animals living in Antarctica and Patagonia. J Vet Med Sci 80:138–146. doi: 10.1292/jvms.17-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hinenoya A, Nagano K, Awasthi SP, Hatanaka N, Yamasaki S. 2020. Prevalence of Escherichia albertii in raccoons (Procyon lotor), Japan. Emerg Infect Dis 26:1304–1307. doi: 10.3201/eid2606.191436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, Ichihara S, Kaneko A, Isobe J, Yamaguchi K, Horikawa K, Gomes TAT, Linden A, Bardiau M, Mainil JG, Beutin L, Ogura Y, Hayashi T. 2012. Clinical significance of Escherichia albertii. Emerg Infect Dis 18:488–492. doi: 10.3201/eid1803.111401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hyma KE, Lacher DW, Nelson AM, Bumbaugh AC, Janda JM, Strockbine NA, Young VB, Whittam TS. 2005. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J Bacteriol 187:619–628. doi: 10.1128/JB.187.2.619-628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Brandal LT, Tunsjø HS, Ranheim TE, Løbersli I, Lange H, Wester AL. 2015. Shiga toxin 2a in Escherichia albertii. J Clin Microbiol 53:1454–1455. doi: 10.1128/JCM.03378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hinenoya A, Yasuda N, Hibino T, Shima A, Nagita A, Tsukamoto T, Yamasaki S. 2017. Isolation and characterization of an Escherichia albertii strain producing three different toxins from a child with diarrhea. Jpn J Infect Dis 70:252–257. doi: 10.7883/yoken.JJID.2016.186. [DOI] [PubMed] [Google Scholar]
- 200.Masuda K, Ooka T, Akita H, Hiratsuka T, Takao S, Fukada M, Inoue K, Honda M, Toda J, Sugitani W, Narimatsu H, Ishioka T, Hirai S, Sekizuka T, Kuroda M, Morita Y, Hayashi T, Kimura H, Oishi K, Ohnishi M, Fujimoto S, Murakami K. 2020. Epidemiological aspects of Escherichia albertii outbreaks in Japan and genetic characteristics of the causative pathogen. Foodborne Pathog Dis 17:144–150. doi: 10.1089/fpd.2019.2654. [DOI] [PubMed] [Google Scholar]
- 201.Inglis TJJ, Merritt AJ, Bzdyl N, Lansley S, Urosevic MN. 2015. First bacteraemic human infection with Escherichia albertii. New Microbes New Infect 8:171–173. doi: 10.1016/j.nmni.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Strockbine NA, Bopp CA, Fields PI, Kaper JB, Nataro JP. 2015. Escherichia, Shigella, and Salmonella, p 685–713. In Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, Warnock DW (ed), Manual of clinical microbiology, 11th ed, vol 1. ASM Press, Washington, DC. [Google Scholar]
- 203.Murakami K, Maeda-Mitani E, Kimura H, Honda M, Ikeda T, Sugitani W, Konno T, Kawano K, Etoh Y, Sera N, Mizukoshi F, Saitoh T, Kawamura Y, Ishioka T, Ohnishi M, Oishi K, Fujimoto S. 2019. Non-biogroup 1 or 2 strains of the emerging zoonotic pathogen Escherichia albertii, their proposed assignment to biogroup 3, and their commonly detected characteristics. Front Microbiol 10:1543. doi: 10.3389/fmicb.2019.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hinenoya A, Ichimura H, Awasthi SP, Yasuda N, Yatsuyanagi J, Yamasaki S. 2019. Phenotypic and molecular characterization of Escherichia albertii: further surrogates to avoid potential laboratory misidentification. Int J Med Microbiol 309:108–115. doi: 10.1016/j.ijmm.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 205.Hatanaka N, Awasthi SP, Hinenoya A, Ueda O, Yamasaki S. 2020. Accurate identification of Escherichia albertii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Microbiol Methods 173:105916. doi: 10.1016/j.mimet.2020.105916. [DOI] [PubMed] [Google Scholar]
- 206.Hinenoya A, Nagano K, Okuno K, Nagita A, Hatanaka N, Awasthi SP, Yamasaki S. 2020. Development of XRM-MacConkey agar selective medium for the isolation of Escherichia albertii. Diagn Microbiol Infect Dis 97:115006. doi: 10.1016/j.diagmicrobio.2020.115006. [DOI] [PubMed] [Google Scholar]
- 207.Rosen E. 1954. Acute ulcerative colitis due to Klebsiella. Calif Med 80:322–323. [PMC free article] [PubMed] [Google Scholar]
- 208.Minami J, Okabe A, Shiode J, Hayashi H. 1989. Production of a unique cytotoxin by Klebsiella oxytoca. Microb Pathog 7:203–211. doi: 10.1016/0882-4010(89)90056-9. [DOI] [PubMed] [Google Scholar]
- 209.Gorkiewicz G. 2009. Nosocomial and antibiotic-associated diarrhoea caused by organisms other than Clostridium difficile. Int J Antimicrob Agents 33:S37–S41. doi: 10.1016/S0924-8579(09)70015-9. [DOI] [PubMed] [Google Scholar]
- 210.Minami J, Katayama S-I, Matsushita O, Sakamoto H, Okabe A. 1994. Enterotoxic activity of Klebsiella oxytoca cytotoxin in rabbit intestinal loops. Infect Immun 62:172–177. doi: 10.1128/IAI.62.1.172-177.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Benoit R, Danquechin Dorval E, Loulergue J, Bacq Y, Oliver J-M, Audurier A, Metman EH. 1992. Diarrhée post-antibiotique: role de Klebsiella oxytoca. Gastroenterol Clin Biol 16:860–864. [PubMed] [Google Scholar]
- 212.Högenauer C, Langner C, Beubler E, Lippe IT, Schicho R, Gorkiewicz G, Krause R, Gerstgrasser N, Krejs GJ, Hinterleitner TA. 2006. Klebsiella oxytoca as a causative organism of antibiotic-associated hemorrhagic colitis. N Engl J Med 355:2418–2426. doi: 10.1056/NEJMoa054765. [DOI] [PubMed] [Google Scholar]
- 213.Zollner-Schwetz I, Högenauer C, Joainig M, Weberhofer P, Gorkiewicz G, Valentin T, Hinterleitner TA, Krause R. 2008. Role of Klebsiella oxytoca in antibiotic-associated diarrhea. Clin Infect Dis 47:e74–e78. doi: 10.1086/592074. [DOI] [PubMed] [Google Scholar]
- 214.Cheng VCC, Yam W-C, Tsang L-L, Yau MCY, Siu GKH, Wong SCY, Chan JFW, To KKW, Tse H, Hung IFN, Tai JWM, Ho P-L, Yuen K-Y. 2012. Epidemiology of Klebsiella oxytoca-associated diarrhea detected by Simmons citrate agar supplemented with inositol, tryptophan, and bile salts. J Clin Microbiol 50:1571–1579. doi: 10.1128/JCM.00163-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Beaugerie L, Metz M, Barbut F, Bellaiche G, Bouhnik Y, Raskine L, Nicolas J-C, Chatelet F-P, Lehn N, Petit J-C, The Infectious Colitis Study Group . 2003. Klebsiella oxytoca as an agent of antibiotic-associated hemorrhagic colitis. Clin Gastroenterol Hepatol 1:370–376. doi: 10.1053/S1542-3565(03)00183-6. [DOI] [PubMed] [Google Scholar]
- 216.Paveglio S, Ledala N, Rezaul K, Lin Q, Zhou Y, Provatas AA, Bennett E, Lindberg T, Camano M, Matson AP. 2020. Cytotoxin-producing Klebsiella oxytoca in the preterm gut and its association with necrotizing enterocolitis. Emerg Microbes Infect 9:1321–1329. doi: 10.1080/22221751.2020.1773743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nagamura T, Tanaka Y, Terayama T, Higashiyama D, Seno S, Isoi N, Katsurada Y, Matsubara A, Yoshimura Y, Sekine Y, Akitomi S, Sato K, Tsuda H, Saitoh D, Ikeuchi H. 2019. Fulminant pseudomembranous enterocolitis caused by Klebsiella oxytoca: an autopsy case report. Acute Med Surg 6:78–82. doi: 10.1002/ams2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hamilton AL, Kamm MA, Ng SC, Morrison M. 2019. Proteus spp. as putative gastrointestinal pathogens. Clin Microbiol Rev 31:e00085-17. doi: 10.1128/CMR.00085-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gong Z, Shi X, Bai F, He X, Zhang H, Li Y, Wan Y, Lin Y, Qiu Y, Chen Q, Hu Q, Cao H. 2019. Characterization of a novel diarrheagenic strain of Proteus mirabilis associated with food poisoning in China. Front Microbiol 10:2810. doi: 10.3389/fmicb.2019.02810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Drzewiecka D. 2016. Significance and roles of Proteus spp. bacteria in natural environments. Microb Ecol 72:741–758. doi: 10.1007/s00248-015-0720-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tschäpe H, Prager R, Streckel W, Fruth A, Tietze E, Böhme G. 1995. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infectious source. Epidemiol Infect 114:441–450. doi: 10.1017/s0950268800052158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pereira AL, Silva TN, Gomes ACMM, Araújo ACG, Giugliano LG. 2010. Diarrhea-associated biofilm formed by enteroaggregative Escherichia coli and aggregative Citrobacter freundii: a consortium mediated by putative F pili. BMC Microbiol 10:57. doi: 10.1186/1471-2180-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Bai L, Xia S, Lan R, Liu L, Ye C, Wang Y, Jin D, Cui Z, Jing H, Xiong Y, Bai X, Sun H, Zhang J, Wang L, Xu J. 2012. Isolation and characterization of cytotoxic, aggregative Citrobacter freundii. PLoS One 7:e33054. doi: 10.1371/journal.pone.0033054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Greenstein AJ, Lowenthal D, Hammer GS, Schaffner F, Aufses AH. 1984. Continuing changing patterns of disease in pyogenic liver abscess: a study of 38 patients. Am J Gastroenterol 79:217–226. [PubMed] [Google Scholar]
- 225.Jun J-B. 2018. Klebsiella pneumoniae liver abscess. Infect Chemother 50:210–218. doi: 10.3947/ic.2018.50.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Song H, Wang X, Lian Y, Wan T. 2020. Analysis of the clinical characteristics of 202 patients with liver abscess associated with diabetes mellitus and biliary tract disease. J Int Med Res 48:300060520949404. doi: 10.1177/0300060520949404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu Y-C, Cheng DL, Lin CL. 1986. Klebsiella pneumoniae liver abscess associated with septic arthritis. Arch Intern Med 146:1913–1916. doi: 10.1001/archinte.1986.00360220057011. [DOI] [PubMed] [Google Scholar]
- 228.Wang J-H, Liu Y-C, Lee SS-J, Yen M-Y, Chen Y-S, Wang J-H, Wann S-R, Lin H-H. 1998. Primary liver abscess due to Klebsiella pneumoniae in Taiwan. Clin Infect Dis 26:1434–1438. doi: 10.1086/516369. [DOI] [PubMed] [Google Scholar]
- 229.Choby JE, Howard-Anderson J, Weiss DS. 2020. Hypervirulent Klebsiella pneumoniae – clinical and molecular perspectives. J Intern Med 287:283–300. doi: 10.1111/joim.13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Russo TA, Marr CM. 2019. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev 32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Fang C-T, Chuang Y-P, Shun C-T, Chang S-C, Wang J-T. 2004. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 199:697–705. doi: 10.1084/jem.20030857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. 2020. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence 11:1215–1224. doi: 10.1080/21505594.2020.1809322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 234.Russo TA, Olson R, Fang C-T, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR, Backer M, Bajwa R, Catanzaro AT, Crook D, de Almeda K, Fierer J, Greenberg DE, Klevay M, Patel P, Ratner A, Wang J-T, Zola J. 2018. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol 56:e00776-18. doi: 10.1128/JCM.00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Harada S, Doi Y. 2018. Hypervirulent Klebsiella pneumoniae: a call for consensus definition and international collaboration. J Clin Microbiol 56:e00959-18. doi: 10.1128/JCM.00959-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Farmer JJ, III, Asbury MA, Hickman FW, Brenner DJ, The Enterobacteriaceae Study Group . 1980. Enterobacter sakazakii: a new species of "Enterobacteriaceae" isolated from clinical specimens. Int J Syst Bacteriol 30:569–584. doi: 10.1099/00207713-30-3-569. [DOI] [Google Scholar]
- 237.Muytjens HL, Zanen HC, Sonderkamp HJ, Kollée LA, Wachsmuth K, Farmer JJ, III. 1983. Analysis of eight cases of neonatal meningitis and sepsis due to Enterobacter sakazakii. J Clin Microbiol 18:115–120. doi: 10.1128/JCM.18.1.115-120.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Jaradat ZW, Al Mousa W, Elbetieha A, Al Nabulsi A, Tall BD. 2014. Cronobacter spp.—opportunistic food-borne pathogens. A review of their virulence and environmental-adaptive traits. J Med Microbiol 63:1023–1037. doi: 10.1099/jmm.0.073742-0. [DOI] [PubMed] [Google Scholar]
- 239.Tall B, Chen YC, Yan Q, Gopinath GR, Grim CJ, Jarvis KG, Fanning JS, Lampel KA. 2014. Cronobacter: an emergent pathogen causing meningitis to neonates through their feeds. Sci Prog 97:154–172. doi: 10.3184/003685014X13994743930498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Strysko J, Cope JR, Martin H, Tarr C, Hise K, Collier S, Bowen A. 2020. Food safety and invasive Cronobacter infections during early infancy, 1961–2018. Emerg Infect Dis 26:857–865. doi: 10.3201/eid2605.190858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Elkhawaga AA, Hetta HF, Osman NS, Hosni A, El-Mokhtar MA. 2020. Emergence of Cronobacter sakazakii in cases of neonatal sepsis in upper Egypt: first report in North Africa. Front Microbiol 11:215. doi: 10.3389/fmicb.2020.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kalyantanda G, Shumyak L, Archibald LK. 2015. Cronobacter species contamination of powdered infant formula and implications for neonatal health. Front Pediatr 3:56. doi: 10.3389/fped.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lou X, Liu T, Zhang W, Yu H, Wang H, Song S, Chen Q, Fang Z. 2019. The occurrence and distribution characteristics of Cronobacter in diverse cereal kernels, flour, and flour-based products. Food Microbiol 84:103269. doi: 10.1016/j.fm.2019.103269. [DOI] [PubMed] [Google Scholar]
- 244.Forsythe SJ. 2018. Updates on the Cronobacter genus. Annu Rev Food Sci Technol 9:23–44. doi: 10.1146/annurev-food-030117-012246. [DOI] [PubMed] [Google Scholar]
- 245.Jackson EE, Forsythe SJ. 2016. Comparative study of Cronobacter identification according to phenotyping methods. BMC Microbiol 16:146. doi: 10.1186/s12866-016-0768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Lepuschitz S, Ruppitsch W, Pekard-Amenitsch S, Forsythe SJ, Cormican M, Mach RL, Piérard D, Allerberger F, The EUCRONI Study Group . 2019. Multicenter study of Cronobacter sakazakii infections in humans, Europe, 2017. Emerg Infect Dis 25:515–522. doi: 10.3201/eid2503.181652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7:e01250-16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Glassner KL, Abraham BP, Quigley EMM. 2020. The microbiome and inflammatory bowel disease. J Allergy Clin Immunol 145:16–27. doi: 10.1016/j.jaci.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 249.Strober W. 2011. Adherent-invasive E. coli in Crohn disease: bacterial “agent provocateur.” J Clin Invest 121:841–844. doi: 10.1172/JCI46333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chervy M, Barnich N, Denizot J. 2020. Adherent-invasive E. coli: update on the lifestyle of a troublemaker in Crohn’s disease. Int J Mol Sci 21:3734. doi: 10.3390/ijms21103734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Dogan B, Belcher-Timme HF, Dogan EI, Jiang Z-D, DuPont HL, Snyder N, Yangf S, Chandler B, Scherl EJ, Simpson KW. 2018. Evaluation of Escherichia coli pathotypes associated with irritable bowel syndrome. FEMS Microbiol Lett 365:fny249. doi: 10.1093/femsle/fny249. [DOI] [PubMed] [Google Scholar]
- 252.Sobieszczańska BM, Osek J, Waśko-Czopnik D, Dworniczek E, Jermakow K. 2007. Association of enteroaggregative Escherichia coli with irritable bowel syndrome. Clin Microbiol Rev 13:404–407. doi: 10.1111/j.1469-0691.2006.01669.x. [DOI] [PubMed] [Google Scholar]
- 253.Rashid T, Ebringer A, Wilson C. 2017. The link between Proteus mirabilis, environmental factors and autoantibodies in rheumatoid arthritis. Clin Exp Rheumatol 35:865–871. [PubMed] [Google Scholar]
- 254.Rashid T, Ebringer A. 2007. Rheumatoid arthritis is linked to Proteus—the evidence. Clin Rheumatol 26:1036–1043. doi: 10.1007/s10067-006-0491-z. [DOI] [PubMed] [Google Scholar]
- 255.Christopoulos G, Christopoulou V, Routsias JG, Babionitakis A, Antoniadis C, Vaiopoulos G. 2017. Greek rheumatoid arthritis patients have elevated levels of antibodies against antigens of P mirabilis. Clin Rheumatol 36:527–535. doi: 10.1007/s10067-016-3441-4. [DOI] [PubMed] [Google Scholar]
- 256.Tuompo R, Lääveri T, Hannu T, Pakkanen SH, Kirveskari J, Leirisalo-Repo M, Kantele A. 2020. Reactive arthritis and other muscoskeletal symptoms associated with acquisition of diarrhoeagenic Escherichia coli (DEC). Ann Rheum Dis 79:605–611. doi: 10.1136/annrheumdis-2019-216736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Honda K, Iwanaga N, Izumi Y, Tsuji Y, Kawahara C, Michitsuji T, Higashi S, Kawakami A, Migita K. 2017. Reactive arthritis caused by Yersinia enterocolitica enteritis. Intern Med 56:1239–1242. doi: 10.2169/internalmedicine.56.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Huang W-K, Chang W-C, See L-C, Tu H-T, Chen J-S, Liaw C-C, Lin Y-C, Yang T-S. 2012. Higher rate of colorectal cancer among patients with pyogenic liver abscess with Klebsiella pneumoniae than those without: an 11-year follow-up study. Colorectal Dis 14:e794–e801. doi: 10.1111/j.1463-1318.2012.03174.x. [DOI] [PubMed] [Google Scholar]
- 259.Zhang L, Wu Y-N, Chen T, Ren C-H, Li X, Liu G-X. 2019. Relationship between intestinal microbial dysbiosis and primary liver cancer. Hepatobiliary Pancreat Dis Int 18:149–157. doi: 10.1016/j.hbpd.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 260.Allen-Vercoe E, Jobin C. 2014. Fusobacterium and Enterobacteriaceae: important players for CRC? Immunol Lett 162:54–61. doi: 10.1016/j.imlet.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zhang Z, Li D, Shi X, Zhai Y, Guo Y, Zheng Y, Zhao L, He Y, Chen Y, Wang Z, Su J, Kang Y, Gao Z. 2020. Genomic characterization of an emerging Enterobacteriaceae species: the first case of co-infection with a typical pathogen in a human patient. BMC Genomics 21:297. doi: 10.1186/s12864-020-6720-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ciotti M, Angeletti S, Ciccozzi M. 2020. Bringing phylogeny and clinical microbiology together. Future Microbiol 15:5–7. doi: 10.2217/fmb-2019-0264. [DOI] [PubMed] [Google Scholar]
- 263.Margos G, Marosevic D, Cutler S, Derdakova M, Diuk-Wasser M, Emler S, Fish D, Gray J, Hunfeldt K-P, Jaulhac B, Kahl O, Kovalev S, Kraiczy P, Lane RS, Lienhard R, Lindgren PE, Ogden N, Ornstein K, Rupprecht T, Schwartz I, Sing A, Straubinger RK, Strle F, Voordouw M, Rizzoli A, Stevenson B, Fingerle V. 2017. There is inadequate evidence to support the division of the genus Borrelia. Int J Syst Evol Microbiol 67:1081–1084. doi: 10.1099/ijsem.0.001717. [DOI] [PubMed] [Google Scholar]
- 264.Wesevich A, Sutton G, Ruffin F, Park LP, Fouts DE, Fowler VG, Jr, Thaden JT. 2020. Newly named Klebsiella aerogenes (formerly Enterobacter aerogenes) is associated with poor clinical outcomes relative to other Enterobacter species in patients with bloodstream infection. J Clin Microbiol 58:e00582-20. doi: 10.1128/JCM.00582-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Munson E. 2020. Moving targets of bacterial taxonomy revision: what are they and why should we care? Clin Microbiol Newslett 42:111–118. doi: 10.1016/j.clinmicnews.2020.06.002. [DOI] [Google Scholar]
- 266.Chakraborty T, Doijad S. 2018. Reviewer report for “Enterobacter hormaechei subsp. hoffmannii subsp. nov., Enterobacter hormaechei subsp. Xianfangensis comb. nov., Enterobacter roggenkampii sp. nov, and Enterobacter muelleri is a later heterotypic synonym of Enterobacter asburiae based upon computational analysis of sequenced Enterobacter genomes.” F1000Res. doi: 10.5256/f1000research.15853.r34159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wolk DM, Dunne WM. 2011. New technologies in clinical microbiology. J Clin Microbiol 49:S62–S67. doi: 10.1128/JCM.00834-11. [DOI] [Google Scholar]
- 268.Laupland KB, Valiquette L. 2013. The changing culture of the microbiology laboratory. Can J Infect Dis Med Microbiol 24:125–128. doi: 10.1155/2013/101630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.McLean K, Rosenthal CA, Sengupta D, Owens J, Cookson BT, Hoffman NG, Salipante SJ. 2019. Improved species-level clinical identification of Enterobacteriaceae through broad-range dnaJ PCR and sequencing. J Clin Microbiol 57:e00986-19. doi: 10.1128/JCM.00986-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Janda JM. 2018. Clinical decisions: how relevant is modern bacterial taxonomy for the clinical microbiologist? Clin Microbiol Newslett 40:51–57. doi: 10.1016/j.clinmicnews.2018.03.005. [DOI] [Google Scholar]
- 271.Tabssum F, Ahmad Q-U-A, Qazi JI. 2018. DNA sequenced based bacterial taxonomy should entail decisive phenotypic remarks: towards a balanced approach. J Basic Microbiol 58:918–927. doi: 10.1002/jobm.201800319. [DOI] [PubMed] [Google Scholar]
- 272.Munson E, Carroll KC. 2018. An update on the novel genera and species and revised taxonomic status of bacterial organisms described in 2016 and 2017. J Clin Microbiol 57:e01181-18. doi: 10.1128/JCM.01181-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Xu Z, Xia M, Hua Y-X, Yang Y. 2020. Intestinirhabdus alba gen. nov., a novel genus of the family Enterobacteriaceae, isolated from the gut of plastic-eating larvae of the Coleoptera insect Zophobus atratus. Int J Syst Evol Microbiol 70:4951–4959. doi: 10.1099/ijsem.0.004364. [DOI] [PubMed] [Google Scholar]
- 274.Casadevall A. 2020. Climate change brings the specter of new infectious diseases. J Clin Invest 130:553–555. doi: 10.1172/JCI135003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cho G-S, Stein M, Brinks E, Rathje J, Lee W, Suh SH, Franz CMAP. 2020. Serratia nevei sp. nov. and Serratia backelmqnnii sp. nov. isolated from fresh produce in Germany and reclassification of Serratia marcescens subsp. Sakuensis Ajithkumar et al. 2003 as a later heterotypic synonym of Serratia marcescens subsp. Marcescens. Syst Appl Microbiol 43:126055. doi: 10.1016/j.syapm.2020.126055. [DOI] [PubMed] [Google Scholar]
- 276.The Editorial Board. 2005. Genus II. Alterococcus Shieh and Jean 1999, 341VP (effective publication: Shieh and Jean 1998, 644), p 625. In Brenner DJ, Krieg NR, Staley JT (ed), Bergey’s manual of systematic bacteriology, 2nd ed, vol 2. Springer, New York, NY. [Google Scholar]
- 277.Hickman-Brenner FW, Huntley-Carter GP, Fanning GR, Brenner DJ, Farmer JJ, III.1985. Koserella trabulsii, a new genus and species of Enterobacteriaceae formerly known as enteric group 45. J Clin Microbiol 21:39–42. doi: 10.1128/JCM.21.1.39-42.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Hickman-Brenner FW, Vohra MP, Huntley-Carter GP, Fanning GR, Lowery VA, III, Brenner DJ, Farmer JJ, III. 1985. Leminorella, a new genus of Enterobacteriaceae: identification of Leminorella grimontii sp. nov. and Leminorella richardii sp. nov. found in clinical specimens. J Clin Microbiol 23:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Müller HE, Brenner DJ, Fanning GR, Grimont PAD, Kämpfer P. 1996. Emended description of Buttiauxella agrestis with recognition of six new species of Buttiauxella and two new species of Kluyvera: Buttiauxella ferragutiae sp. nov., Buttiauxella gaviniae sp. nov., Buttiauxella brennerae sp. nov., Buttiauxella izardii sp. nov., Buttiauxella noackiae sp. nov., Buttiauxella warmboldiae sp. nov., Kluyvera cochleae sp. nov., and Kluyvera georgiana sp. nov. Int J Syst Bacteriol 46:50–63. doi: 10.1099/00207713-46-1-50. [DOI] [PubMed] [Google Scholar]
- 280.Søgaard P, Kjaeldgaard P. 1986. Two isolations of enteric group 69 from human clinical specimens. Acta Pathol Microbiol Immunol Scand B 94:365–367. doi: 10.1111/j.1699-0463.1986.tb03068.x. [DOI] [PubMed] [Google Scholar]
- 281.Warren JR, Farmer JJ, III, Dewhirst FE, Birkhead K, Zembower T, Peterson LR, Sims L, Bhattacharya M. 2000. Outbreak of nosocomial infections due to extended-spectrum β-lactamase-producing strains of enteric group 137, a new member of the family Enterobacteriaceae closely related to Citrobacter farmer and Citrobacter amalonaticus. J Clin Microbiol 38:3946–3952. doi: 10.1128/JCM.38.11.3946-3952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Li Y, Fang W, Xue H, Liang W-x, Wang L-f, Tian G-z, Wang X-z, Lin C-l, Li X, Piao C-g. 2015. Brenneria populi sp. nov., isolated from symptomatic bark of Populus×euramericana canker. Int J Syst Evol Microbiol 65:432–437. doi: 10.1099/ijs.0.066068-0. [DOI] [PubMed] [Google Scholar]
- 283.Brady C, Hunter G, Kirk S, Arnold D, Denman S. 2014. Description of Brenneria rosae sp. nov. and two subspecies, Brenneria rosae subspecies rosae ssp. nov. and Brenneria rosae subspecies americana ssp. nov. isolated from symptomatic oak. Syst Appl Microbiol 37:396–401. doi: 10.1016/j.syapm.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 284.Maes M, Baeyen S, De Croo H, De Smet K, Steenackers M. 2017. Monitoring of endophytic Brenneria salicis in willow and its relations to watermark disease. Plant Prot Sci 38:528–530. doi: 10.17221/10545-PPS. [DOI] [Google Scholar]
- 285.Grosso S, Mason G, Ortalda E, Scortichini M. 2011. Brenneria salicis associated with watermark disease symptoms on Salix alba in Italy. Plant Dis 95:772. doi: 10.1094/PDIS-11-10-0781. [DOI] [PubMed] [Google Scholar]
- 286.Cating RA, Hong JC, Palmateer AJ, Stiles CM, Dickstein ER. 2008. First report of bacterial soft rot on Vanda orchids caused by Dickeya chrysanthemi (Erwinia chrysanthemi) in the United States. Plant Dis 92:977. doi: 10.1094/PDIS-92-6-0977A. [DOI] [PubMed] [Google Scholar]
- 287.Végh A, Némethy Z, Salamon P, Mándoki Z, Palkovics L. 2014. First report of bacterial wilt on chrysanthemum caused by Dickeya chrysanthemi (syn: Erwinia chrysanthemi) in Hungary. Plant Dis 98:988. doi: 10.1094/PDIS-09-13-0948-PDN. [DOI] [PubMed] [Google Scholar]
- 288.Blomme G, Dita M, Jacobsen KS, Vicente LP, Molina A, Ocimati W, Poussier S, Prior P. 2017. Bacterial diseases of bananas and enset: current state of knowledge and integrated approaches toward sustainable management. Front Plant Sci 8:1290. doi: 10.3389/fpls.2017.01290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Martinez-Cisneros BA, Juarez-Lopez G, Valencia-Torres N, Duran-Peralta E, Mezzalama M. 2014. First report of bacterial stalk rot of maize caused by Dickeya zeae in Mexico. Plant Dis 98:1267. doi: 10.1094/PDIS-02-14-0198-PDN. [DOI] [PubMed] [Google Scholar]
- 290.Hu M, Li J, Chen R, Li W, Feng L, Shi L, Xue Y, Feng X, Zhang L, Zhou J. 2018. Dickeya zeae strains isolated from rice, banana and clivia rot plants show great virulence differentials. BMC Microbiol 18:136. doi: 10.1186/s12866-018-1300-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Amin NM, Bunawan H, Redzuan A, Jaganath IBS. 2010. Erwinia mallotivora sp., a new pathogen of papaya (Carica papaya) in peninsular Malaysia. Int J Mol Sci 12:39–45. doi: 10.3390/ijms12010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Taha MD, Jaini MFM, Saldi NB, Rahim RA, Shah UKM, Hashim AM. 2019. Biological control of Erwinia mallotivora, the causal agent of papaya dieback disease by indigenous seed-borne endophytic lactic acid bacteria consortium. PLoS One 14:e0224431. doi: 10.1371/journal.pone.0224431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Nazareno ES, Dumenyo CK. 2015. Modified inoculation and disease assessment methods reveal host specificity in Erwinia tracheiphila-Cucurbitaceae interactions. Microb Pathog 89:184–187. doi: 10.1016/j.micpath.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 294.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. 2005. Citrobacter rodentium of mice and man. Cell Microbiol 7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 295.Akgul A, Akgul A, Lawrence ML, Karsi A. 2018. Stress-related genes promote Edwardsiella ictaluri pathogenesis. PLoS One 13:e019669. doi: 10.1371/journal.pone.0194669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Leung KY, Wang Q, Yang Z, Siame BA. 2019. Edwardsiella piscicida: a versatile emerging pathogen of fish. Virulence 10:555–567. doi: 10.1080/21505594.2019.1621648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Gao J, Barkema HW, Zhang L, Liu G, Deng Z, Cai L, Shan R, Zhang S, Zou J, Kastelic JP, Han B. 2017. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci 100:4797–4806. doi: 10.3168/jds.2016-12334. [DOI] [PubMed] [Google Scholar]
- 298.Kim YB, Yoon MY, Ha JS, Seo KW, Noh EB, Son SH, Lee YJ. 2020. Molecular characterization of avian pathogenic Escherichia coli from broiler chickens with colibacillosis. Poult Sci 99:1088–1095. doi: 10.1016/j.psj.2019.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Rhouma M, Fairbrother JM, Beaudry F, Letellier A. 2017. Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet Scand 59:31. doi: 10.1186/s13028-017-0299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Fairbrother JM, Nadeau E, Gyles CL. 2005. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev 6:17–39. doi: 10.1079/ahr2005105. [DOI] [PubMed] [Google Scholar]
- 301.Massé J, Dufour S, Archambault M. 2020. Characterization of Klebsiella isolates obtained from clinical mastitis cases in dairy cattle. J Dairy Sci 103:3392–3400. doi: 10.3168/jds.2019-17324. [DOI] [PubMed] [Google Scholar]
- 302.Kumar G, Menanteau-Ledouble S, Saleh M, El-Matbouli M. 2015. Yersinia ruckeri, the causative agent of enteric redmouth disease in fish. Vet Res 46:103. doi: 10.1186/s13567-015-0238-4. [DOI] [PMC free article] [PubMed] [Google Scholar]