SUMMARY
New methods for investigating human astrocytes are urgently needed, given their critical role in the central nervous system. Here we show that CD49f is a novel marker for human astrocytes, expressed in fetal and adult brains from healthy and diseased individuals. CD49f can be used to purify fetal astrocytes and human induced pluripotent stem cell (hiPSC)-derived astrocytes. We provide single-cell and bulk transcriptome analyses of CD49f+ hiPSC-astrocytes, and demonstrate that they perform key astrocytic functions in vitro, including trophic support of neurons, glutamate uptake, and phagocytosis. Notably, CD49f+ hiPSC-astrocytes respond to inflammatory stimuli, acquiring an A1-like reactive state, in which they display impaired phagocytosis and glutamate uptake and fail to support neuronal maturation. Most importantly, we show that conditioned medium from human reactive A1-like astrocytes is toxic to human and rodent neurons. CD49f+ hiPSC-astrocytes are thus a valuable resource for investigating human astrocyte function and dysfunction in health and disease.
Keywords: induced pluripotent stem cells, astrocytes, FACS purification, neurodegeneration, A1 reactive astrocytes
Graphical Abstract
eTOC blurb
Barbar et al. identify CD49f as a novel surface marker expressed by human astrocytes, that can purify hiPSC-astrocytes and primary fetal astrocytes. CD49f+ hiPSC-astrocytes respond to proinflammatory stimuli and become A1 reactive astrocytes, which are dysfunctional and secrete neurotoxic factors that induce apoptosis in human and rodent neurons.
INTRODUCTION
Astrocytes play a critical role in the central nervous system (CNS) by maintaining brain homeostasis, providing metabolic support to neurons, regulating connectivity of neural circuits, and controlling blood flow as an integral part of the blood-brain barrier (reviewed in (Sofroniew and Vinters 2010)). They also undergo a pronounced transformation called reactive astrogliosis following injury and in disease (Zamanian et al. 2012). Accumulating evidence has implicated astrocytes, such as A1 reactive astrocytes (Liddelow et al. 2017), in the onset and progression of many neurological diseases (Phatnani and Maniatis 2015; Sloan and Barres 2014), prompting increased efforts to identify novel astrocyte targets for therapeutic intervention.
Studies of astrocyte biology have often relied on reductionist cell culture models, of which many have been produced since the 1970s. The first method to purify primary astrocytes from rodent brains was based on selective adhesion to tissue culture plates, which eliminates most non-astrocytes, but still retains contaminating cells — largely microglia and oligodendrocyte lineage cells (K. D. McCarthy and de Vellis 1980). Furthermore, astrocytes selected with this method are primarily immature, proliferating cells, and require serum to grow in vitro, which induces a reactive pathological state (Lynette C. Foo et al. 2011). While these methods have been extremely powerful for understanding important astrocyte functions, due to inducing a baseline pathological state, they have had limited success in investigating disease states of astrocytes.
More recently, immunopanning methods that take advantage of the cell surface antigens Integrin Beta-5 (encoded by Itgb5) to purify rodent astrocytes (Lynette C. Foo et al. 2011) and GlialCAM (or HepaCAM) adhesion protein to purify human astrocytes (Zhang et al. 2016) (in rodents, HepaCAM is also highly expressed by oligodendrocyte progenitor cells) have become more commonly used. Commercial magnetic-activated cell sorting based on GLAST (Slc1a3) and ASCA-2 has also been widely adopted (Jungblut et al. 2012). All these methods allow isolation of post-mitotic astrocytes, and in the case of immunopanning, allow maintaining cells in serum-free conditions. Nonetheless, these methods have been largely focused on rodent cells, whereas access to human primary CNS cells has been largely limited by the availability of brain specimens. Therefore, our knowledge of astrocyte biology has been mainly built from rodent models, either in vivo or in vitro. Recently, mounting evidence has revealed critical astrocyte contributions to the development and progression of neurological diseases, such as Alzheimer’s disease (AD) (Long and Holtzman 2019), Parkinson’s disease (PD) (di Domenico et al. 2019) and progressive multiple sclerosis (MS) (Ponath et al. 2018), for which optimal astrocyte-specific animal models are lacking. Moreover, it is now clear that human astrocytes have several distinct features from their rodent counterparts (Zhang et al. 2016; Oberheim et al. 2006), which could well be driving pathogenic mechanisms of human diseases. Taken together, there is an urgent need for better human in vitro modeling of astrocyte (dys)function.
Human induced pluripotent stem cell (hiPSC) technology has emerged as a powerful tool to generate human astrocytes and other CNS cells in vitro, starting from skin fibroblasts or peripheral blood mononuclear cells of patients and healthy individuals (Shi et al. 2017). Protocols to differentiate hiPSCs into astrocytes use either a specific gradient of patterning agents to mimic embryonic development (Krencik and Zhang 2011; Santos et al. 2017; Roybon et al. 2013; Jiang et al. 2013; Palm et al. 2015; Perriot et al. 2018; Tcw et al. 2017) or overexpression of critical transcription factors (Tchieu et al. 2019; Canals et al. 2018). Many of these protocols require a few consecutive passages to eliminate neuronal cells and achieve a mature state. Alternative 3D cultures of CNS organoids generate neural progenitor cells, neurons, oligodendrocyte lineage cells, astrocytes, and can incorporate microglia (Lancaster et al. 2013; Quadrato et al. 2017; Madhavan et al. 2018; Sloan et al. 2017). With organoids being increasingly utilized to model CNS diseases, methods for purifying specific cell types are becoming highly desirable for downstream analyses. To our knowledge, the GlialCAM marker used for purifying adult primary astrocytes via immunopanning is not expressed in hiPSC-derived astrocytes until 100 days in culture (Sloan et al. 2017), prioritizing the need for a novel method to isolate astrocytes at earlier time points.
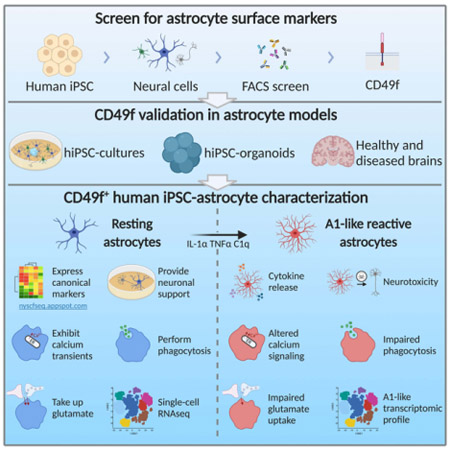
Here we set out to advance the tools available to purify and study astrocytes derived from hiPSCs. We used iPSC lines generated by the NYSCF Global Stem Cell Array®, our fully automated reprogramming platform that has been demonstrated to reduce line-to-line variability(Paull et al. 2015). We leveraged a differentiation protocol that we previously developed to generate oligodendrocytes (Douvaras et al. 2014) within mixed cultures, which also include neurons and astrocytes in serum-free conditions. Our unbiased screen for surface molecules in these cultures identified CD49f as a novel marker for purifying hiPSC-astrocytes from neurons and oligodendrocyte lineage cells through fluorescence-activated cell sorting (FACS). CD49f, encoded by the gene ITGA6, is a member of the integrin alpha chain family of proteins and interacts with extracellular matrices, including laminin. The Brain RNA-Seq database (https://www.brainrnaseq.org/) confirms that ITGA6 expression is higher in human fetal and mature astrocytes compared to neurons, oligodendrocytes and microglia. ITGA6 is also expressed in endothelial cells, but hiPSC differentiations toward the ectodermal lineage produce neural populations with no mesodermal/endothelial cell contribution. In this study, we show that CD49f can be used to sort astrocytes from monolayer cultures and 3D cortical organoids containing oligodendrocyte lineage cells (i.e., oligodendrocyte progenitor cells, immature and mature oligodendrocytes), neural progenitors, and neurons. CD49f+ astrocytes express typical markers, display similar gene expression profiles to human primary astrocytes and perform critical astrocyte functions in vitro. Specifically, they support neuronal growth and synaptogenesis, generate spontaneous Ca2+ transients, respond to ATP, perform glutamate uptake, and secrete inflammatory cytokines in response to inflammatory stimuli. Moreover, as previously reported in rodents (Liddelow et al. 2017), CD49f+ hiPSC-astrocytes acquire an A1-like reactive phenotype upon stimulation with TNFα, IL-1α, and C1q, while maintaining CD49f expression. Transcriptome analysis revealed a conserved A1 signature with similar losses of function as reported in rodent cells, including impaired glutamate uptake, phagocytosis, and support of neuronal maturation. Most importantly, rodent and hiPSC-derived neurons treated with A1 conditioned medium show significant increases in apoptosis, providing a human in vitro model for A1-driven neurotoxicity. Single-cell transcriptome analysis revealed slightly different A1 profiles in astrocytes at different maturation stages – suggesting that disease responses may change at different stages of development and during aging. Taken together, our findings establish a novel human-based platform to model astrocytes in vitro, in which CD49f+ hiPSC-derived astrocytes from early stage organoids and monolayer cultures can be used to study reactive states and interrogate their role in neurodevelopmental and neurodegenerative diseases.
RESULTS
A screen for astrocyte-specific surface markers identifies CD49f
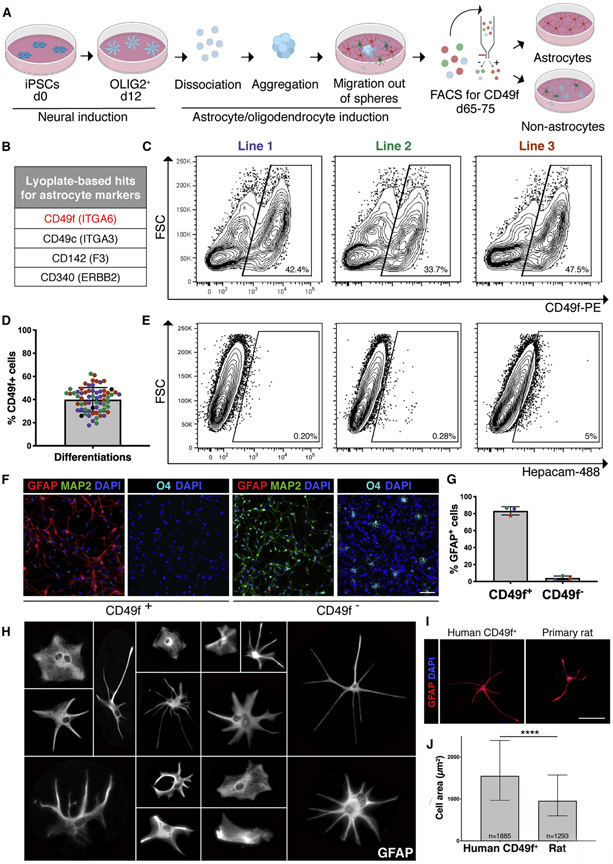
To enable the isolation of pure hiPSC-astrocytes, we performed a screen of surface antigens on a mix of neural cells derived from an oligodendrocyte protocol that we previously developed. This protocol generates OLIG2+ progenitors through retinoic acid and sonic hedgehog signaling, mimicking embryonic development in the spinal cord (Douvaras and Fossati 2015). When OLIG2+-enriched neural spheres are plated down, neurons, astrocytes and oligodendrocyte progenitor cells migrate out in order, and after day 65 immature oligodendrocytes can be purified using the O4 sulfate glycolipid antigen. We sought to develop an analogous sorting strategy to isolate astrocytes for functional studies (Fig. 1a). To identify astrocyte-specific markers, we digested day 78 cultures into single cells and tested a panel of 242 antibodies to surface antigens within the O4+ and O4− populations. Sorted cells were re-plated, fixed and stained for glial fibrillary acidic protein (GFAP), an astrocyte-specific cytoplasm intermediate filament. Among the candidates that showed an enrichment of GFAP+ cells, we identified 4 antigens whose mRNA expression was reported to be higher in astrocytes than in other CNS cell types (Fig. 1 b). We then selected CD49f for further validation using three independent iPSC lines from healthy subjects, from which we isolated an average of 40% CD49f+ cells (Fig. 1c, 1d, S1a, S1b). Of note, HepaCAM was minimally expressed by hiPSC-derived cells at this stage (Fig.le, S1c, S1d), emphasizing the need for an alternative marker. GFAP quantification in the FACS-sorted fractions revealed an average of 83% GFAP+ cells in the CD49f+ versus 3% in the CD49f− population, which was enriched in MAP2+ cells and O4+ oligodendrocytes (Fig.1 f,1g). Interestingly, CD49f+ astrocytes were highly heterogeneous in morphology, echoing the complexity and diversity of astrocytes reported in vivo (Oberheim et al. 2006) (Fig. 1h). Consistent with previous studies of human astrocytes (Oberheim et al. 2006), CD49f+ astrocytes were also larger than primary rat astrocytes (Fig. 1i, 1j).
Figure 1: CD49f surface antigen purifies hiPSC-derived astrocytes.
See also Figures S1 and S2.
a) Schematic of hiPSC-astrocyte differentiation protocol depicting the major steps leading to the CD49f sort.
b) Top four hits for astrocyte markers identified from the Lyoplate screen.
c) Representative flow-cytometry contour plots (with outliers shown) of the CD49f sort of hiPSC-cells generated from 3 hiPSC lines from three individuals using the astrocyte differentiation protocol in a).
d) Bar chart with individual data points plotted as circles, where colors represent 3 hiPSC lines, displays the consistent proportion of CD49f+ cells obtained from independent differentiations for each line. Error bars show mean ± standard deviation (n=65). At least 5 independent differentiations per line were performed.
e) Representative contour plots (with outliers shown) of the EpCAM sort of hiPSC-cells generated from 3 lines.
f) Representative immunofluorescence images of CD49f+ and CD49f− cells 24 hours post-sort showing GFAP+ astrocytes (red), MAP2+ neurons (green), O4+ immature oligodendrocytes (cyan), and total DAPI cells (blue). Scale bar, 100μm.
g) Vast majority of sorted CD49f+ cells are also GFAP+, while almost no CD49f− cells are GFAP+ cells. Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=3 independent lines).
h) Representative images of individual magnified CD49f+ astrocytes cultured at low density and stained with GFAP, showing their morphological heterogeneity. Each cell was cropped and placed in the image.
i) Representative immunofluorescence images of hiPSC-derived CD49f+ astrocytes and primary rat astrocytes stained with GFAP (red) and DAPI (blue) show human cells are larger in size. Scale bar, 100μm.
j) Bar graph showing the median of the cell area in μm2 of GFAP+ hiPSC-derived CD49f+ astrocytes (n= 1885 cells from 3 lines with 1-2 replicates each) is significantly greater than that of primary rat astrocytes (n=1293 cells from 6 replicates). Error bars represent the interquartile range. p-value was calculated using a two-tailed, unpaired t-test.
As CD49f is a laminin receptor, and our differentiation protocol uses laminin coating, we analyzed astrocytes from 3D cortical organoids (Madhavan et al. 2018) cultured in the absence of laminin. The organoid protocol generates iPSC-derived cortical astrocytes, whereas our astrocyte protocol is patterned towards spinal cord. Nevertheless, we found that in sorted cells from digested cortical organoids, CD49f was still co-expressed with AQP4 and GFAP (Fig. S2a), and astrocytes were enriched in the CD49f+ fraction, with an average of 31% GFAP+ cells and 77% AQP4+ cells, as opposed to 1% GFAP+ cells and 8% AQP4+ cells in the CD49f− fraction (Fig. S2b-g). These findings confirm that CD49f expression is neither dependent on laminin coating nor spinal cord–specific.
Transcriptome profile of CD49f+ astrocytes reveals typical astrocyte markers
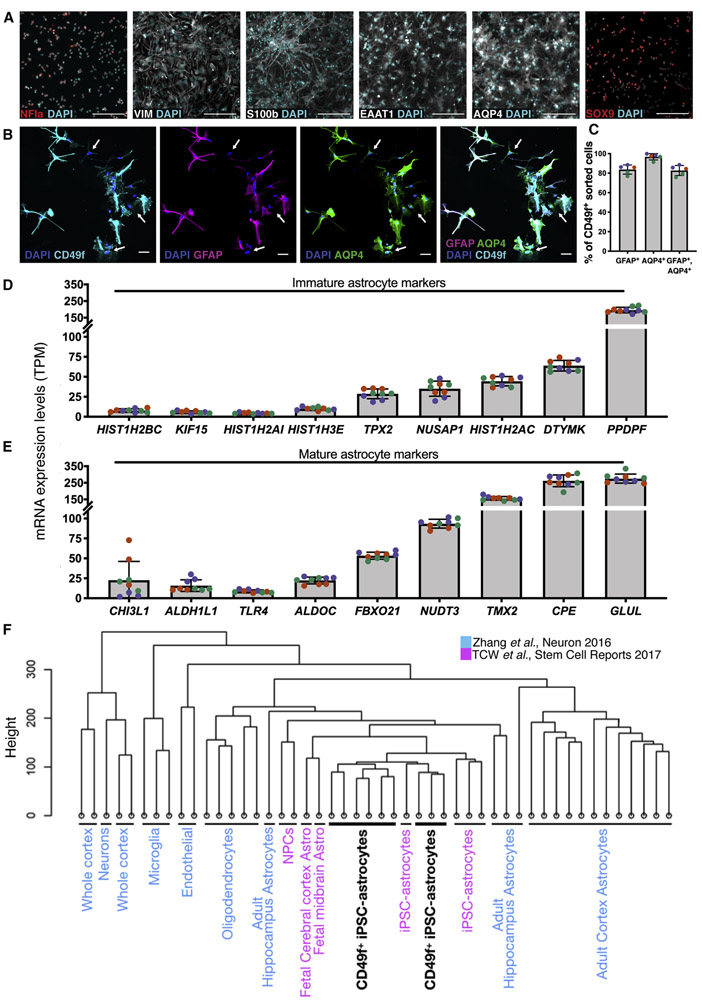
To verify the identity of CD49f+ cells, we assessed the presence of additional astrocyte makers. CD49f+ astrocytes stained positive for AQP4, SOX9, EAAT1, NFIa, VIM, and S100β (Fig. 2a). Interestingly, about 97% of the cells are AQP4+, but only 84% are GFAP+ (Fig. 2b, 2c), in line with findings that GFAP does not identify all CNS astrocytes (Liu et al. 2010; Roessmann et al. 1980) and with the reported heterogeneity of GFAP levels in different brain regions (Cahoy et al. 2008). Transcriptomic analysis via RNA-Seq of CD49f+ cells revealed expression of several mature and immature astrocyte genes, with low inter-line variability (Fig. 2d, 2e). Hierarchical clustering of RNA-Seq data confirmed that hiPSC-astrocytes cluster close to fetal primary human astrocytes, and to hiPSC-astrocytes generated with an alternative differentiation protocol (Tcw et al. 2017), but are distinct from neurons, oligodendrocytes, microglia, and endothelial cells (Zhang et al. 2016) (Fig. 2f). We also confirmed the spinal cord identity of the CD49f+ cells generated using our protocol, by performing hierarchical clustering of RNA-Seq data with a recent study of regionally specified hiPSC-astrocytes (Bradley et al. 2019) (Fig. S3a-b). We have made our gene expression data available in a user-friendly, searchable online database (https://nyscfseq.appspot.com/).
Figure 2: Immunofluorescence and transcription profiling of CD49f+ hiPSC-derived astrocytes confirm expression of canonical markers.
See also Figure S3.
a) Immunofluorescence images of CD49f+ astrocytes showing expression of astrocyte markers in red or white, and total DAPI cells (cyan). Scale bar, 200μm.
b) Immunofluorescence images of CD49f+ astrocytes showing CD49f+ (cyan), GFAP+ (red), AQP4+ (green) and total DAPI cells (blue). White arrows indicate cells that are CD49f+/AQP4+/GFAP−. Scale bar, 50μm.
c) Percentages of CD49f+ cells across different CD49f+ hiPSC-astrocyte lines that are also GFAP+, AQP4+, and triple positive for GFAP, AQP4, and CD49f. Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=5, 1-2 technical replicates per line).
d) RNA-Seq expression levels of immature astrocyte markers in human CD49f+ astrocytes, expressed in transcripts per million (TPM). Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=9, 3 replicates per line).
e) mRNA expression levels of mature astrocyte markers in human CD49f+ astrocytes, expressed in transcripts per million (TPM). Colors and error bars are as in d).
f) Hierarchical clustering of RNA-Seq data shows that CD49f+ hiPSC-astrocytes (black) closely resemble primary and iPSC-derived astrocytes from independent studies and are distinct from other brain cell types (GEO: GSE73721 in blue; GEO: GSE97904 in pink). Analysis is based on transcriptome-wide expression. CD49f+ samples consist of 3 different lines in 3 replicates each.
Single-cell RNA transcriptome analysis demonstrates that CD49f+ enriches for mature hiPSC-derived astrocytes
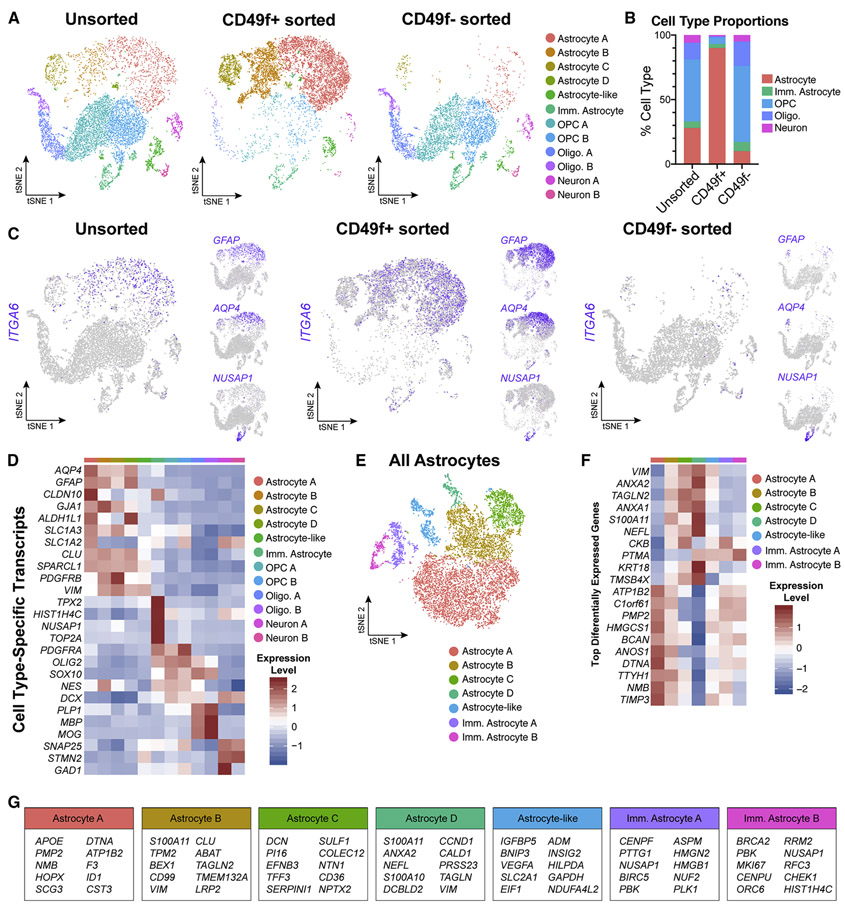
The expression of both mature and immature astrocyte markers in our bulk transcriptome analysis of CD49f+ iPSC-astrocytes could reflect either the cells being in an intermediate state of maturation, or a mixture of immature and mature astrocytes. To resolve this question, we performed single-cell RNA sequencing of unsorted cultures following differentiation as well as sorted CD49f+ astrocytes and CD49f− cells. We found that unsorted cultures contained the following subpopulations: mature astrocytes (28%; defined as GFAP+), immature astrocytes (5%; defined as NUSAP1+), oligodendrocyte progenitor cells (OPC, 48%), oligodendrocytes (13%), and neurons (6%). The sorted CD49f+ fraction was enriched for mature astrocytes (90%), while the sorted CD49f− cells were depleted of mature astrocytes (10%) and enriched for all other cell types (Fig. 3a, 3b). ITGA6 expression primarily overlaps with that of mature markers GFAP and AQP4, but not immature marker NUSAP1 or OPC markers PDGFRA and CSPG4, confirming that sorting based on CD49f enriched for mature astrocytes (Fig. 3c, 3d, S4). Additionally, CD49f+ hiPSC-astrocytes expressed the human-specific astrocyte markers LRRC3B, HSSD17B6, FAM198B, RYR3, STOX1, and MRVI1 identified by Zhang et al. (Fig. S5).
Figure 3: Single-cell transcriptome data confirms that CD49f+ sorting strategy from heterogeneous hiPSC-derived cultures enriches for mature astrocytes.
All data from one control line (line 3, n=1). See also Figures S4, S5, S6, and Supplemental Table 1.
a) tSNE plots of single-cell RNA-Seq data from unsorted (left, n = 7,744), CD49f+ (middle, n = 9,047), and CD49f sorted (right, n = 5,057) cells. In total, 12 clusters were identified.
b) Quantification of cell type proportions from unsorted, CD49f+, and CD49f− sorted cells based on tSNE analysis. CD49f+ sorted cells are mostly astrocytes.
c) tSNE feature plots of CD49f (ITGA6), mature astrocyte (GFAP, AQP4), and immature astrocyte (NUSAP1) transcripts from unsorted, CD49f+ sorted, and CD49f− sorted cells, showing that CD49f+ cells express primarily mature astrocyte markers.
d) Heatmap of cell type–specific transcript expression across identified clusters.
e) tSNE plot of all astrocytes from unsorted, CD49f+, and CD49f− sorted cells (n = 12,061 astrocytes). After subsetting and reintegrating only astrocytes from the initial clustering scheme, we identified 2 immature and 4 mature astrocyte clusters, and 1 astrocyte-like cluster.
f) Heatmap of top differentially expressed genes identified across all astrocyte-related clusters.
g) Top 10 differentially expressed genes for each astrocyte-related cluster.
Abbreviations: Imm.=immature; Oligo=oligodendrocyte; OPC=oligodendrocyte progenitor cell.
To further evaluate potential astrocyte heterogeneity indicated by the scRNA-seq analysis, we subsetted and reintegrated only astrocytes defined by the initial clustering scheme, and subsequently identified two immature astrocyte clusters, four mature astrocyte clusters, and one astrocyte-like cluster (Fig. 3e, 3f). Immature astrocyte clusters, which are enriched in the CD49f− fraction, have higher expression of early pseudotime transcripts from Sloan et al. (Sloan et al. 2017), while mature astrocyte clusters, enriched in the CD49f+ fraction, have higher expression of middle and late pseudotime transcripts (Fig. S6).
CD49f+ astrocytes can be isolated from human fetal brains and are present in human adult brains
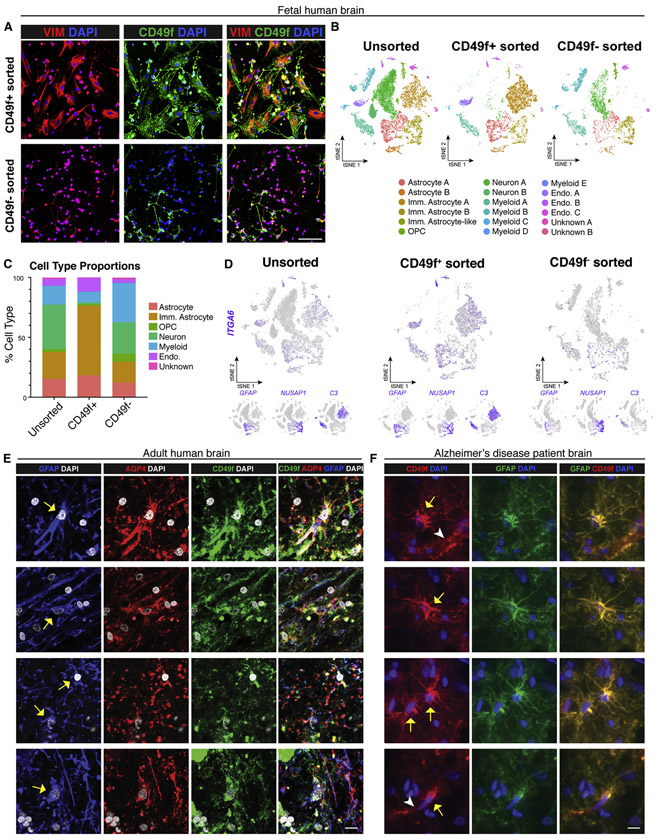
To assess whether CD49f can be used to isolate astrocytes from primary human tissues, we dissociated and sorted cells from human fetal brain, and found that the CD49f+ fraction was highly enriched with vimentin+ astrocytes (Fig. 4a). Single-cell transcriptomic analysis showed that total digested fetal brain cells consisted of mature astrocytes, immature astrocytes, OPCs, neurons, myeloid cells, and endothelial cells (Fig. 4b). Sorting for CD49f resulted in an enrichment of astrocytes (from 37% to 76%) and endothelial cells (from 6% to 11%) (Fig. 4c). ITGA6 expression overlapped with expression of mature astrocyte marker GFAP and immature astrocyte marker C3 (Holst et al. 2019) (Fig. 4d). This indicates potential variation in ITGA6 expression across immature astrocyte populations, as C3+ immature astrocytes appear to have higher ITGA6 expression than NUSAP1+ immature astrocytes, which make up a large proportion of the immature astrocytes in the CD49f fraction of the sort (Fig. 4b, 4d).
Figure 4: CD49f-sort from human fetal brain enriches for astrocytes, and CD49f is localized in astrocytes in slides from human adult brains.
See also Figure S7 and Supplemental Table 1.
a) Representative immunofluorescence images showing vimentin (red), CD49f (green), and DAPI (blue) in CD49f+ and CD49f− sorted cells from human fetal brain tissue. Scale bar, 100μm.
b) tSNE plots of single-cell RNA-Seq data from unsorted (left, n = 11,817), CD49f+ (middle, n = 6,069), and CD49f sorted (right, n = 4,409) cells from fetal brain tissue. In total, 18 clusters were identified. All data are from an 18-week-old human fetus (n=1).
c) Quantification of cell type proportions from unsorted, CD49f+, and CD49f− sorted cells from fetal brain tissue. CD49f+ cells are highly enriched in astrocytes and immature astrocytes.
d) tSNE feature plots highlighting in purple cells that express CD49f (ITGA6), mature astrocyte (GFAP), and immature astrocyte (NUSAP1, C3) transcripts from unsorted, CD49f+, and CD49f− sorted cells from fetal brain tissue.
e) Representative immunofluorescence images showing GFAP+ (blue), AQP4+ (red), and CD49f+ (green) cells with DAPI nuclei (grey) in cryosections from the subventricular zone of an adult brain from healthy individual. Yellow arrows indicate cells that are CD49f+, AQP4+, and GFAP+. Scale bar, 10μm.
f) Representative immunofluorescence images showing CD49f+ (red) and GFAP+ (green) cells with DAPI nuclei (blue) in cryosections from the prefrontal cortex of an Alzheimer’s disease patient. Yellow arrows indicate cells that are CD49f+ and GFAP+. White arrowheads indicate CD49f+ endothelial cells. Scale bar, 10μm.
Abbreviations: Endo.=endothelial cell; Imm.=immature; OPC=oligodendrocyte progenitor cell.
We next investigated whether CD49f is maintained in astrocytes from adult human brains. Staining showed that CD49f co-localizes with AQP4 and GFAP in adult brain tissue sections from a healthy donor (Fig. 4e) as well as in an AD patient (Fig. 4f), corroborating CD49f as a novel marker for human astrocytes in vivo as well as in hiPSC cultures. Brain sections also showed CD49f+ endothelial cells that were not GFAP+ (Fig. 4f).
Interestingly, when we tested CD49f isolation from whole mouse brain of Aldh1l1eGFP mice, no CD49f+ astrocytes were ALDH1L1+ (Fig. S7), suggesting that CD49f could be human-specific.
CD49f+ astrocytes perform physiological astrocyte functions in vitro
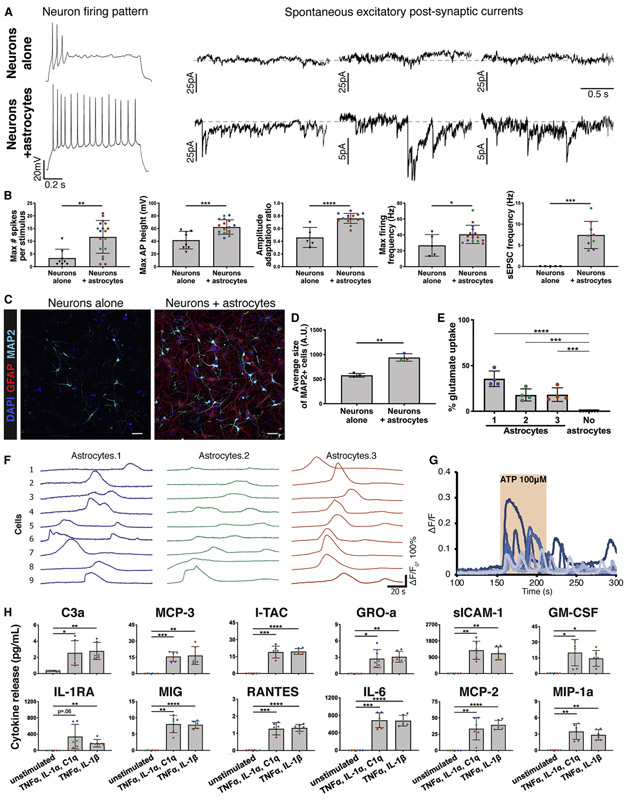
To assess the capacity of astrocytes to support neuronal function in vitro, we set up co-cultures of CD49f+ hiPSC-astrocytes with hiPSC-derived neurons. Neurons that were co-cultured with astrocytes for two weeks displayed more developed electrophysiological properties than neurons alone, including increases in number of action potentials per 1s depolarizing stimulus, maximum firing frequency, maximum height of action potential, and amplitude adaptation ratio (Fig. 5a, 5b). Furthermore, only neurons co-cultured with astrocytes exhibited spontaneous excitatory post synaptic currents (sEPSC) indicating advanced synaptogenesis (Fig. 5a, 5b), as previously reported for adult human (Zhang et al. 2016) and rodent astrocytes (Khakh and North 2012). We also found that neurons co-cultured with CD49f+ astrocytes had a larger MAP2 area than those cultured without astrocytes (Fig. 5c, d), highlighting an increase in neurite length in these cells that also corroborates previous observations (Banker 1980). Together, these data demonstrate that CD49f+ hiPSC-astrocytes are functional in supporting neurite outgrowth, neuronal function, and synapse formation.
Figure 5: CD49f+ hiPSC-derived astrocytes provide neuronal support and exhibit other astrocytic functions in vitro.
See also Figure S8.
a) Representative recordings of firing patterns and spontaneous excitatory post-synaptic currents (sEPSCs) measured in hiPSC-neurons at day 50 when cultured alone, or with CD49f+ hiPSC-astrocytes during days 33-50. Neurons co-cultured with astrocytes exhibited more mature firing patterns and a greater number of sEPSCs.
b) CD49f+ hiPSC-astrocytes enhance electrophysiological properties of hiPSC-neurons in co-culture. Bar graphs show the maximum number of evoked spikes per 1 second stimulus (n=8/18), the maximum firing frequency in hertz (Hz) (n=5/15), the amplitude adaptation ratio between first and last action potential (n=5/15), the maximum action potential height (mV) (n=8/17), and the frequency of spontaneous excitatory post-synaptic currents (Hz) (n=5/9) in hiPSC-neurons at day 50 when cultured alone vs. with astrocytes during days 33-50. Colored dots correspond to 3 different lines. Each dot represents an independent cell. n=neurons alone/neurons co-cultured with astrocytes. Error bars show mean ± standard deviation. p-values were calculated using a two-tailed, unpaired t-test.
c) Representative immunofluorescence images showing MAP2+ neurons (cyan), GFAP+ astrocytes (red), and DAPI nuclei in hiPSC-derived neurons at 40 days in vitro cultured alone or with CD49f+ hiPSC-astrocytes for one week. Scale bar, 50μm.
d) Average size of MAP2+ cells. Colored dots correspond to 3 different CD49f+ hiPSC-astrocyte lines. Error bars show mean ± standard deviation (n=3 technical replicates). p-values were calculated using a two-tailed, unpaired t-test.
e) CD49f+ astrocytes take up glutamate. Bar graphs show percent of glutamate taken up by CD49f+ hiPSC-astrocytes after incubation with 100 μM glutamate for 3 hours, compared to wells without cells (media only). Colored dots correspond to 3 different astrocyte lines. Error bars show mean ± standard deviation (n=4 technical replicates). p-values were calculated using a one-way ANOVA with Dunnett’s correction for multiple comparisons.
f) hiPSC-astrocytes show spontaneous Ca2+ transients. Nine representative traces of spontaneous [Ca2+]i transients from 3 independent CD49f+ hiPSC-astrocyte lines loaded with the Ca2+ indicator Rhod-3/AM are shown. Each line is represented by a different color (lines 1,2,3).
g) CD49f+ hiPSC-astrocytes respond to ATP. Representative traces of [Ca2+]i transients from nine astrocytes from one iPSC line loaded with the Ca2+ indicator Rhod-3/AM following 100 μM ATP application.
h) CD49f+ hiPSC-astrocytes secrete proinflammatory cytokines when stimulated for 24 hours with TNFα, IL-1α, and C1q, or TNFα and IL-1β. Bar charts with individual data points plotted as dots show concentration of cytokines secreted in the supernatant of CD49f+ astrocytes with and without stimulation. Concentrations are expressed in pg/ml and normalized to 1,000 cells. Colored dots correspond to 3 lines (n=6, 2 technical replicates per line). Error bars show mean ± standard deviation. p-values were calculated using a one-way ANOVA with Dunnett’s correction for multiple comparisons.
We next tested other important astrocyte physiological functions in vitro. CD49f+ hiPSC-astrocyte cultures efficiently take up glutamate, mimicking a crucial function in vivo for preventing neuronal glutamate excitotoxicity (Fig. 5e). We also investigated if they would exhibit calcium transients, as it is known that astrocytes have both spontaneous and inducible calcium signals that can be detected in vivo in different brain regions (Srinivasan et al. 2016; Tanaka 1997) and in vitro (Khakh and North 2012). We monitored cytosolic calcium levels and found that—like primary purified human astrocytes in vitro and mouse astrocytes visualized in vivo—spontaneous calcium transients were present in CD49f+ astrocytes from all lines (Fig. 5f). Furthermore, we found that CD49f+ astrocytes robustly responded to 60s application of extracellular ATP at 100 μM (Fig. 5g).
Astrocytes are immunocompetent cells, able to respond to inflammatory stimuli by releasing additional pro-inflammatory molecules (reviewed in (Sofroniew 2014)). To test this in vitro, we stimulated CD49f+ astrocytes either with IL-1β and TNFα (Mayo et al. 2014) or with TNFα, IL-1α, and C1q, which drives the neurotoxic A1 state reported in rodent cells (Liddelow and Barres 2017). Pro-inflammatory cytokine secretion was significantly increased following both types of stimulation. In particular, IL-6 and soluble ICAM-1 showed the greatest fold changes compared to unstimulated cells, and notably, IL-1α was secreted upon IL-1β and TNFα stimulation (Fig. 5h, S8).
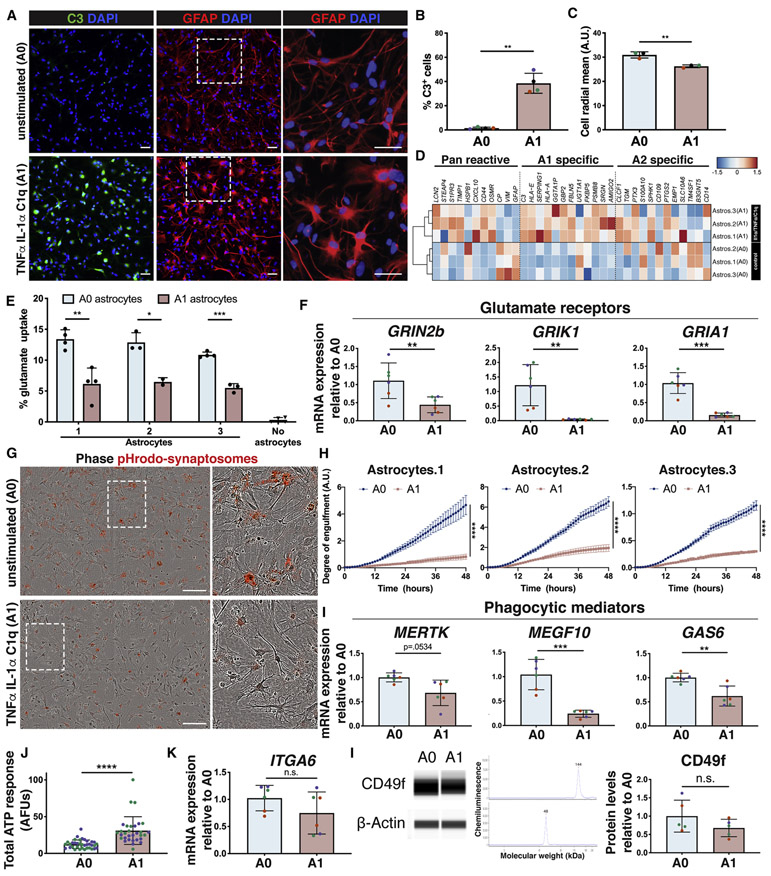
Human A1-like reactive astrocytes lose functional capacity for phagocytosis and glutamate uptake
Previous studies have modeled inflammation-stimulated reactivity in hiPSC-derived astrocytes in vitro (Santos et al. 2017; Roybon et al. 2013; Perriot et al. 2018; Tcw et al. 2017; Tchieu et al. 2019; Zhou et al. 2019) and we expanded on these findings by characterizing the transcriptomic profile and functionality of CD49f+ hiPSC-astrocytes stimulated with TNFα, IL-1α, and C1q to drive an A1 reactive state. Human iPSC-derived A1-like astrocytes were C3-positive like their rodent counterparts (Liddelow et al. 2017) (Fig. 6a, 6b), and they also exhibited the morphological changes observed in reactive astrocytes in vivo (reviewed in (Sun and Jakobs 2012)), losing finer processes and becoming hypertrophied (Fig. 6a, 6c). To evaluate conservation of the defined rodent A1 signature in human cells, we performed RNA-Seq on human stimulated (A1) vs. unstimulated (A0) astrocytes (Fig. 6d), and assessed which transcripts were upregulated in reactive astrocytes in rodents, classified as pan-reactive, specifically induced by neuroinflammation (A1) or induced by ischemia (A2) (Liddelow et al. 2017). We found the human A1-like cells also upregulated most rodent A1-specific genes (Fig. 6d, S9a). SLC1A3, encoding the glutamate transporter GLAST, was downregulated in A1-like astrocytes, while SLC1A2, encoding the glutamate transporter EAAT2 (GLT-1) was unchanged at both the RNA and protein level (Fig. S9b-c). Most importantly, downregulation of glutamate receptors corresponded to impaired glutamate uptake in A1-like reactive cells (Fig 6e-f). Moreover, the decrease in mRNA levels of phagocytic receptors MERTK and MEGF10 and bridging molecule GAS6 paralleled a significant decrease in phagocytosis of synaptosomes in hiPSC-derived A1-like reactive astrocytes (Fig. 6g, 6h, 6i). This was accompanied by a decrease in expression of lysosomal markers LAMP1, LAMP2, and RAB7A (Fig. S9d). When assessing ATP response, we found that hiPSC-derived A1-like astrocytes had a stronger response than A0 astrocytes (Fig. 6j), as opposed to rodent astrocytes, where ATP response does not change in the A1 state (Fig. S9e). Of note, hiPSC-derived A1-like astrocytes retained both ITGA6 expression and CD49f protein levels, verifying CD49f as a reactivity-independent marker (Fig. 6k-l). The full gene expression dataset from iPSC-astrocytes is available in the online database mentioned above. Taken together, our characterization of human A1-like astrocytes, enabled by our technique for astrocyte isolation, establishes hiPSC-derived astrocytes as a powerful in vitro model to investigate molecular mechanisms linked to A1 pathogenesis.
Figure 6: CD49f+ hiPSC-derived astrocytes astrocytes can be stimulated in vitro to take on an A1-like reactive state that loses key astrocytic functions.
See also Figure S9.
a) Representative immunofluorescence images showing the reactive marker C3 (green) in CD49f+ astrocytes upon stimulation with TNFα, IL-1α, and C1q. Cells are also stained for GFAP (red) and DAPI (blue). White dashed boxes indicate the areas of the magnified images on the right to highlight changes in morphology. Scale bar, 50μm.
b) Percentage of C3+ cells in unstimulated vs. stimulated CD49f+ astrocytes as in a). Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=4 independent lines). p-values were calculated using a two-tailed, paired t-test.
c) Cell radial mean, in arbitrary unit (A.U.) across different lines in unstimulated vs. stimulated CD49f+ astrocytes, depicting the shift in morphology between the two conditions. Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=3 independent lines). p-values were calculated using a two-tailed, paired t-test.
d) CD49f+ hiPSC-derived astrocytes upregulate the A1 reactive transcripts previously identified in mouse astrocytes. Heat map shows expression levels of reactive transcripts (pan-reactive, A1 astrocytes or A2 astrocytes) in A1-like vs. unstimulated (A0) CD49f+ hiPSC-astrocytes.
e) Glutamate uptake is reduced in CD49f+ A1-like astrocytes. Percent of glutamate taken up by A0 vs. A1 astrocytes and compared to wells without cells (media only). Error bars show mean ± standard deviation (n=2-4 technical replicates). p-values were calculated using multiple t-tests with Holm-Sidak’s correction for multiple comparisons.
f) Relative mRNA expression of genes encoding glutamate receptors (GRIN2b, GRIK1, GRIA1), quantified via qPCR analysis in two independent experiments, is decreased in CD49f+ stimulated astrocytes (A1) vs. unstimulated (A0). Colored dots correspond to 3 lines. Error bars show mean ± standard deviation (n=6, 2 replicates per line). p-values were calculated using a two-tailed, paired t-test.
g) Representative images of A0 vs. A1 astrocytes engulfing pHrodo-synaptosomes (red), showing reduced phagocytic capacity in A1 astrocytes. White dashed boxes indicate the areas of the magnified images on the right. Scale bar, 200μm.
h) Time course analysis and quantification of A0 vs. reactive A1 astrocytes engulfing pHrodo-synaptosomes. The average degree of engulfment (normalized to confluence) with standard error of the mean (n=9-12 replicates per line), indicates a reduced phagocytic capacity in A1 astrocytes. p-values were calculated using a two-way ANOVA.
i) Relative mRNA expression of genes encoding phagocytic receptors (MERTK, MEGF10) and bridging molecule GAS6, quantified via qPCR analysis, is decreased in CD49f+ A1 vs. A0. Colored dots correspond to 3 lines. Error bars show mean ± standard deviation (n=6, 2 replicates per line). p-values were calculated using a two-tailed, paired t-test.
j) Human A1-like astrocytes have a stronger ATP response than unstimulated astrocytes. Area under the curve of ΔF/F traces for one minute following 100μM ATP application of hiPSC-derived CD49f+ A0 (n=36 cells) and A1 (n=31 cells) astrocytes loaded with the Ca2+ indicator Rhod-3/AM. Unit is in arbitrary fluorescence units (AFUs). Colored dots correspond to 2 lines. Error bars show mean ± standard deviation. p-values were calculated using a two-tailed, unpaired t-test.
k) Relative mRNA expression of ITGA6, quantified via qPCR analysis does not significantly differ between A0 vs. A1 astrocytes in two independent experiments. Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=6, 2 replicates per line). p-values were calculated using a two-tailed, paired t-test.
l) CD49f protein levels do not significantly differ between A0 vs. A1 astrocytes. Representative western bands and electropherograms for CD49f and b-actin, and quantification of protein levels for CD49f in A0 and A1 astrocytes, normalized to b-actin. Error bars show mean ± standard deviation (n=5, 1-2 replicates per line). Colored dots correspond to 3 different lines. p-values were calculated using a two-tailed, paired t-test.
Human A1 reactive astrocytes are neurotoxic
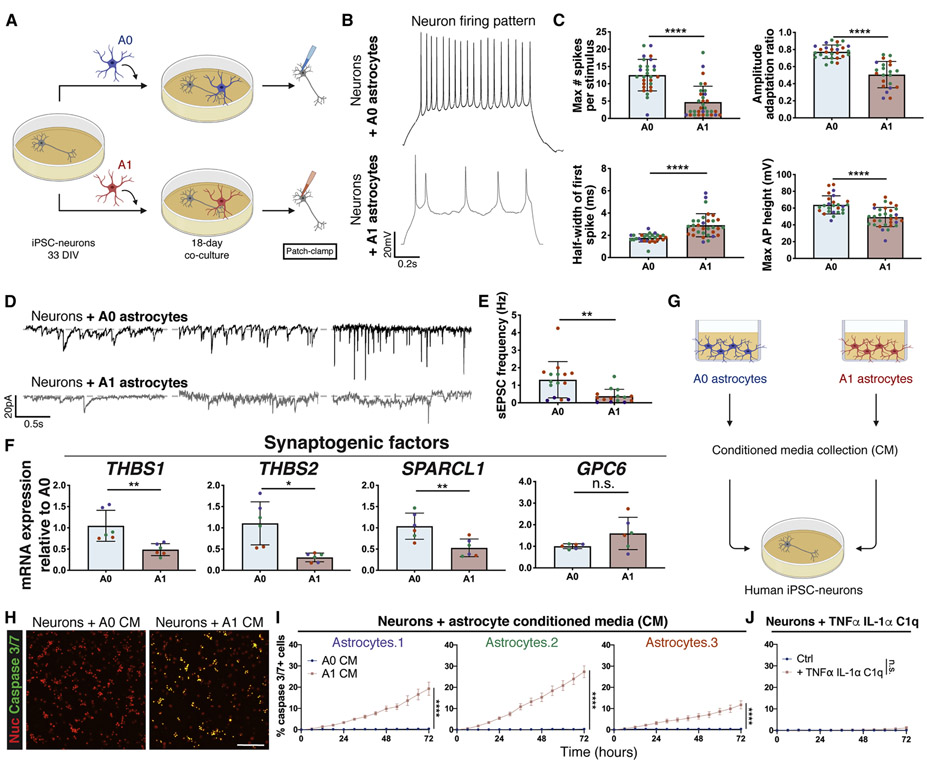
To evaluate the effects of hiPSC-derived A1-like astrocytes on neurons, we co-cultured unstimulated (A0) or A1-like astrocytes with hiPSC-neurons for 18 days and performed electrophysiological analysis (Fig. 7a). Neurons co-cultured with A1-like astrocytes had a less mature firing pattern than neurons cultured with unstimulated astrocytes, including a lower number of spikes per stimulus, a larger spike half-width, a smaller amplitude adaptation ratio, and a smaller spike height (Fig. 7b, 7c). Neurons co-cultured with A1-like astrocytes also had a smaller sEPSC frequency than neurons cultured with unstimulated astrocytes (Fig. 7d, 7e). This is consistent with the significant downregulation of synaptogenic factors seen in A1 astrocytes, which could lead to dysfunctional astrocytes that are unable to support neuronal function and synapse formation, as previously shown in rodents (Liddelow et al. 2017) (Fig. 7f). Direct treatment of hiPSC-neurons with TNFα, IL-1α, and C1q had no effect on neuronal maturation (Fig. S10a-b), supporting the hypothesis that the impaired maturation observed in neurons was astrocyte-mediated. To assess apoptosis, we treated neurons with astrocyte conditioned media collected from A0 and A1 astrocytes and measured caspase 3/7 levels. We found that both hiPSC-neurons and primary mouse neurons exhibited increased apoptosis following treatment with A1-like conditioned media, but not in response to direct treatment with TNFα, IL-1α, and C1q (Fig. 7g-j, S10c-e). These findings demonstrate the specific neurotoxicity of A1 hiPSC-derived astrocytes.
Figure 7: A1-like reactive CD49f+ hiPSC-derived astrocytes impair neuronal maturation and connectivity and are neurotoxic.
See also Figure S10.
a) Schematic of neuron co-culture experiment with A0 and A1 hiPSC-astrocytes.
b) Representative recordings of firing patterns measured in hiPSC-neurons at day 51 when cultured with A0 or A1 astrocytes during days 33-51. Neurons co-cultured with A1 astrocytes exhibited less mature firing patterns.
c) A1 astrocytes provide markedly lower enhancements of neuronal electrophysiological properties in co-culture than A0 astrocytes. Bar graphs with individual data points plotted show the maximum number of evoked spikes per 1s stimulus (n=28/32), the half-width of the first spike (ms) (n=28/32), the amplitude adaptation ratio between first and last action potential (n=27/22), and the maximum action potential height (mV) (n=26/32) in hiPSC-neurons at day 51 when cultured with A0 or A1 astrocytes during days 33-51. Colored dots correspond to 3 different lines. Each dot represents an independent cell from which we recorded. n=neurons co-cultured with A0 astrocytes/neurons co-cultured with A1 astrocytes. Error bars show mean ± standard deviation. p-values were calculated using a two-tailed, unpaired t-test.
d) Representative recordings of spontaneous excitatory post-synaptic currents (sEPSCs) measured in hiPSC-neurons at day 51 when cultured with A0 or A1 astrocytes during days 33-51. Neurons co-cultured with A1 astrocytes exhibited a smaller number of sEPSCs. Representative traces of sEPSCs are each from a different cell.
e) Frequency of spontaneous excitatory post-synaptic currents (Hz) (n=15 co-cultured with A0 astrocytes;15 co-cultured with A1 astrocytes) in hiPSC-neurons at day 51 when cultured with A0 or A1-like astrocytes during days 33-51. Colored dots correspond to 3 different lines. Each dot represents an independent cell from which we recorded. Error bars show mean ± standard deviation. p-values were calculated using a two-tailed, unpaired t-test.
f) Relative mRNA expression of genes encoding synaptogenic factors, quantified via qPCR analysis, is decreased in A1 vs. A0 astrocytes. Colored dots correspond to 3 different lines. Error bars show mean ± standard deviation (n=6, 2 replicates per line). p-values were calculated using a two-tailed, paired t-test.
g) Schematic of A0 vs. A1 astrocyte conditioned media experiment on hiPSC-neurons.
h) Representative images of neurons treated with A0 or A1 astrocyte conditioned media (CM) for 3 days, subjected to a caspase 3/7 apoptosis assay (green) and cell nuclei (red); neurons exposed to A1 CM show increased apoptosis. Scale bar, 100μm.
i) Time course apoptosis analysis and quantification of the percentage of caspase 3/7+ neurons during treatment with A0 or A1 astrocyte conditioned media (CM). Error bars represent the standard error of the mean (n=12-24 replicates per line), indicating a neurotoxic effect of A1 CM on neurons. p-values were calculated using a two-way ANOVA.
j) Time course apoptosis analysis and quantification of the percentage of caspase 3/7+ apoptotic neurons during stimulation with TNFα, IL-1α, and C1q, demonstrating no direct effects of the cytokine cocktail on neuronal apoptosis. Error bars represent the standard error of the mean (n=12 replicates). p-values were calculated using a two-way ANOVA.
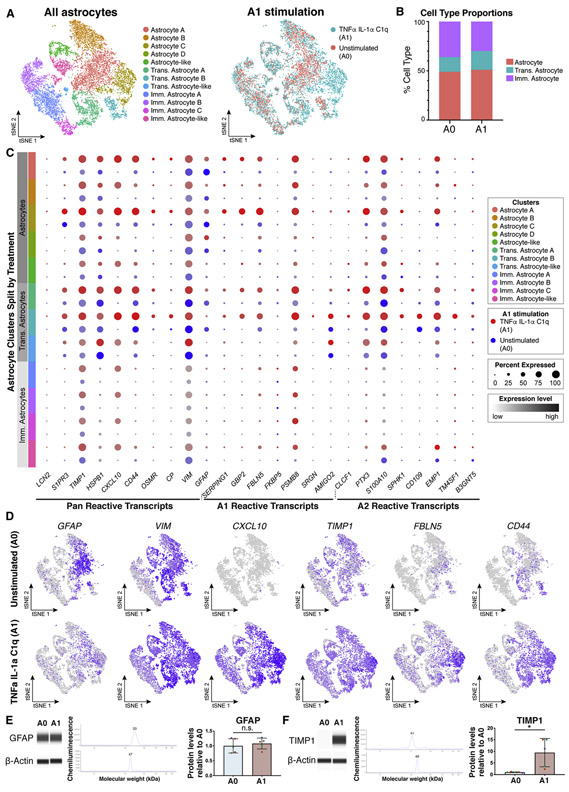
Astrocyte maturity influences response to A1 stimulation
Given that our unsorted cultures contain astrocytes at sequential stages of maturation (Fig. 3a), we assessed their response to TNFα, IL-1α, and C1q stimulation by performing single-cell RNA-Seq 24 hours after treatment with this inflammatory cocktail. After subsetting and reintegrating only astrocytes at all levels of maturation, we identified 5 mature astrocyte clusters, 3 transitioning astrocyte clusters, and 4 immature astrocyte clusters (Fig. 8a-b). Intriguingly, comparing single-cell data from A0 versus A1 indicated a maturation-dependent response to inflammatory stimuli, with mature astrocytes showing a greater response then immature astrocyte (Fig. 8c). Our expression data also revealed CXCL10, TIMP1, FBLN5 and CD44 as better markers of reactivity than GFAP and VIM (Fig. 8d). We confirmed this at the protein level: GFAP levels were similar between A0 and A1 astrocytes (Fig. 8e), while TIMP1 levels were upregulated 9.5-fold in A1 astrocytes (Fig. 8f).
Figure 8: Maturation state affects CD49f+ hiPSC-derived astrocytes response to stimulation with TNFα, IL1α, and C1q.
See also Supplemental Table 1.
a) tSNE plots of single-cell RNA-Seq data from unstimulated (A0, n = 5,881) and TNFα, IL-1α, and C1q stimulated (A1, n = 6,701) astrocytes, shown by cluster (left) and by treatment type (right). Data from two iPSC lines (lines 1 and 3, n = 2).
b) Quantification of cell type proportions from unstimulated (A0) and A1 astrocytes.
c) Dot plot of pan, A1-specific, and A2-specific astrocyte transcripts in A0 (blue) and A1 astrocytes (red), highlighting a stronger gene expression response in mature astrocytes. Dot size represents the percentage of cells that express a transcript, and color intensity represents the expression level of a transcript.
d) tSNE feature plots of reactive astrocyte transcripts.
e) GFAP protein levels are stable in A0 and A1 CD49f+ hiPSC-astrocytes. Western blots for GFAP and b-actin, and quantification of protein levels, normalized to b-actin. Error bars show mean ± standard deviation (n=5, 1-2 replicates per line). Colored dots correspond to 3 different lines. p-values were calculated using a two-tailed, paired t-test.
f) TIMP1 level increases in CD49f+ hiPSC-astrocytes stimulated to A1. Western blots for TIMP1 and b-actin, and protein quantification, normalized to b-actin. Error bars show mean ± standard deviation (n=5, 1-2 replicates per line). Colored dots correspond to 3 different lines. p-values were calculated using a two-tailed, paired t-test.
Abbreviations: Imm.=immature; Trans.=transitioning.
DISCUSSION
A wealth of evidence implicates astrocytes in CNS disease pathology, and efforts to identify novel therapeutic targets have increasingly focused on astrocytes (Cai, Wan, and Liu 2017; Booth, Hirst, and Wade-Martins 2017; Khakh et al. 2017). Given that many of today’s incurable diseases, such as AD, PD, and MS, are specific to humans and critical interspecies differences are evident (Zhang et al. 2016; Han et al. 2013; Eidsvaag et al. 2017), human patient-specific models are a necessary tool to complement traditional animal models for elucidating pathogenic mechanisms and developing effective treatments. Here, we demonstrate that hiPSC-astrocytes purified using the surface marker CD49f are a compelling tool for modeling primary human astrocytes and for studying astrocytic function and dysfunction in vitro. CD49f is a laminin receptor and has been previously reported as a marker for stem cells, including glioblastoma and other cancer stem cells (Lathia et al. 2010). We show that CD49f can also be used for enriching primary fetal human astrocytes and is present in astrocytes from adult human brains in both healthy individuals and neurological disease patients. Importantly, CD49f is a reactivity-independent marker (expressed in both unstimulated and reactive astrocytes), making it ideal for purification strategies in the study of neurodegenerative disease. Established markers for isolating rodent cells, such as HepaCAM, are ineffective for hiPSC-astrocytes, except following prolonged culture to enable complete maturation — just as many available differentiation protocols do not achieve a maturation state equivalent to in vivo adult cells (Parr, Yamanaka, and Saito 2017). Indeed, our transcriptomic analysis of hiPSC-astrocytes showed expression of both immature and mature markers, but single-cell analysis determined that the differentiation yields distinct populations of both immature and mature astrocytes. It should be noted that, due to the patterning during the initial differentiation step with the caudalizing and ventralizing agents retinoic acid and sonic hedgehog (Douvaras and Fossati 2015), the transcriptomic profile of our hiPSC-astrocytes is very similar to that of ventral spinal cord astrocytes generated by the Zhang lab (Tcw et al. 2017). In line with recent findings about regionally specified astrocytes (Bradley et al. 2019), our data suggest that regional heterogeneity exists, stemming from cell-intrinsic developmental differences (Bayraktar et al. 2014) and can be recapitulated in vitro, further emphasizing the usefulness of this resource. Nonetheless, it is important to point out that CD49f is not a spinal cord-specific marker. We successfully isolated astrocytes from cortical organoids and from fetal brain and revealed CD49f+ astrocytes in sections of the subventricular zone and pre-frontal cortex of adult brains. Additionally, independent transcriptomic analyses showed that ITGA6 expression is higher in human forebrain astrocytes than in spinal cord astrocytes (Bradley et al. 2019).
It is important to highlight that CD49f+astrocytes generated through our differentiation protocol achieve within 75 days a maturation stage comparable to that of astrocytes derived from organoids at much later time points, making our strategy optimal for functional studies in vitro. However, it is difficult to directly compare these two protocols, as they use different media formulations and patterning agents, and because the cells are grown in a 2D vs. 3D format (Sloan et al. 2017). Interestingly, scRNA-seq analysis also revealed that our CD49f+ astrocytes consist of multiple astrocyte subclusters with varying expression of genes involved in lipid biosynthesis, neurotransmitter uptake, gliogenesis, antigen presentation, neural development, cell motility and many other important biological processes. Further investigations into these subclusters may provide valuable insights into the functional heterogeneity of astrocytes.
We place the greatest emphasis on our findings from functional assays, establishing CD49f+ astrocyte cultures as a strong platform for disease modeling and for investigating neuronal support, engulfment of debris, glutamate uptake, response to inflammatory stimuli, and neurotoxicity. In the context of neuroinflammation, we showed that CD49f+ astrocytes respond to pro-inflammatory stimuli by secreting typical chemokines and cytokines. Interestingly, we did not observe major differences between stimulation with TNFα, IL-1α, C1q (driving the A1 phenotype (Liddelow et al. 2017)) vs. TNFα and IL-1β, which are typically released by microglia in neurodegenerative diseases. The similarity in cytokine release after stimulation with either cocktail is likely explained by the dominant effect of TNFα, present in both, and by the fact that IL-1α is released by astrocytes upon stimulation with TNFα and IL-1β, triggering autocrine signaling. Our transcriptomic profiles of hiPSC-derived A0 and A1-like reactive astrocytes are available as a resource through a searchable online database (https://nyscfseq.appspot.com/). This transcriptomic analysis reveals that human A1-like reactive astrocytes largely conserve the A1 signature identified in rodent cells and indicates loss of function related to phagocytosis and glutamate uptake. This underscores the value of our methods for isolating and generating patient-specific astrocytes to better understand the pathogenic mechanisms linked to human A1 neurotoxicity in neurological diseases. Furthermore, single-cell RNA-Seq analysis highlighted differences in response to TNFα , IL-1α, C1q treatment based on the developmental stages of hiPSC-astrocytes (immature, transitioning, mature). This likely reflects a difference in the response of astrocytes to infection/injury/disease across different stages of development, which can now be further investigated thanks to our hiPSC-based platform.
In conclusion, we show that CD49f is a reactivity-independent, astrocyte-specific cell surface antigen that is present at all stages of astrocyte development in hiPSC-derived cultures. Astrocytes isolated with this marker recapitulate in vitro critical physiological functions, and following inflammatory stimulation become reactive, dysfunctional, and toxic, triggering neuronal death — opening a window for the study of their role in neurodegenerative diseases. Importantly, hiPSC-derived CD49f+ astrocytes can be used to advance a “clinical trials in a dish” approach to drug discovery (Haston and Finkbeiner 2016; Elitt, Barbar, and Tesar 2018; Gorshkov et al. 2018), and we anticipate that incorporating hiPSC-based models in the preclinical phase of drug development will improve the success of drug discovery for neurological diseases with a high unmet need (Yang et al. 2013; Gribkoff and Kaczmarek 2017). Our strategy to purify hiPSC-derived astrocytes using CD49f will facilitate disease modeling with patient-specific to better understand pathogenic mechanisms of astrocyte reactivity in infection, injury, and developmental and degenerative diseases.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Valentina Fossati (vfossati@nyscf.org).
Materials Availability
Human induced pluripotent stem cell lines utilized in this study can be requested through the NYSCF Repository (https://nyscf.org/research-institute/repository-stem-cell-search/) upon Material Transfer Agreement.
Data and Code Availability
The RNA sequencing datasets generated during this study are available in a user-friendly searchable online database (https://nyscfseq.appspot.com/).
Data are also available from the Synapse open source platform: https://www.synapse.org/#!Synapse:syn21861229.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human induced pluripotent stem cell lines
All iPSC lines were derived from skin biopsies of healthy donors. The participants were enrolled in a study approved by the Western Institutional Review Board (WIRB). This IRB-approved protocol includes the collection of biological samples, research use of these samples, and biobanking of samples. A broad consent form is utilized. iPSC lines 050743-01-MR-023 (51 y.o. male; line 1; represented in purple), 051106-01-MR-046 (57 y.o. female; line 2; represented in green), 051121-01-MR-017 (52 y.o. female; line 3; represented in orange), 051104-01-MR-040 (56 y.o. female), 050659-01-MR-013 (65 y.o. female) were reprogrammed using the NYSCF Global Stem Cell Array® with the mRNA/miRNA method (StemGent), where line-to-line variability has been minimized due to the fully automated reprogramming process (Paull et al. 2015). iPSC lines were cultured and expanded onto Matrigel-coated dishes in mTeSR1 medium (StemCell Technologies) or StemFlex medium (ThermoFisher). Lines were passaged every 3-4 days using enzymatic detachment with Stempro Accutase (ThermoFisher; A1110501) for 5 minutes and re-plated in mTeSR1 medium with 10μM ROCK Inhibitor (Y27632, Stemgent) for 24 hours. All five lines were used for CD49f+ astrocyte isolation and lines 1, 2, and 3 were then used for subsequent functional studies. All iPSC lines made though the NYSCF Global Stem Cell Array® undergo a rigorous quality check including a sterility check, mycoplasma testing, karyotyping, and a pluripotency check. A certificate of analysis (CoA) is provided upon delivery of the first cryovial. A representative CoA (from line 051121-01-MR-017) is shown in Figure S11.
Human brain samples
For healthy brain immunohistochemistry, a fresh sample from the subventricular zone of a 94-year-old male brain was obtained from Advanced Tissue Services. For Alzheimer’s disease brain immunohistochemistry, a fresh frozen sample of prefrontal cortex from an 80-year-old male patient diagnosed with Alzheimer’s disease (Braak score VI/VI) was obtained from Rhode Island Hospital’s Brain Tissue Resource Center (Title 45 CRF Part 46.102(f)). For sorting from fetal brain tissue, de-identified fetal cortical tissues from gestational week 18 (no abnormalities) were obtained under approval from the Albert Einstein College of Medicine Institutional Review Board (IRB; Study protocol 2019-10439). Due to the nature of the tissue collection procedure, we were unable to determine the precise location of the brain where tissue originated.
Animals
All animal procedures were conducted in accordance with guidelines from the National Institute of Health and Stanford University’s Administrative Panel on Laboratory Animal Care (#10726) and NYU School of Medicine’s Institutional Animal Care and Use Committee (#IA18-00249). All rodents were housed with food and water available ad libitum in a 12-h light/dark environment. For experiments using mice, adult female Aldh1l1eGFP transgenic mice (postnatal day, P30) on a C57BL/6J background (GENSAT, MMRRC 036071-UCD) were used. For experiments using rats, Sprague Dawley dams with 4-6-day old postnatal pups (P4-6) were purchased from Charles River (Strain code: 400). All astrocyte purification was completed in rat pups before P7.
METHOD DETAILS
Human healthy brain immunohistochemistry
Chunks were drop fixed in 4% paraformaldehyde overnight at 4°C, washed 3X in PBS, then stored in 30% sucrose in PBS at 4°C overnight. These were then embedded in O.C.T. (Fisher Scientific; 50-363-579), cryosectioned at 20μm thickness and stained using the immunofluorescence protocol described below. Tissue was incubated at 40°C for 10 mins and blocked with PBS containing 0.1% saponin and 2.5% donkey serum for 1 hour. Primary antibodies (see Antibody Table) were applied overnight at 4°C. The next day, slides were washed 3X in PBS, incubated with secondary antibodies (Alexa Fluor) and HOECHST for 1 hour at room temperature, washed 3X for 10 min in PBS. Secondary antibodies were used at 1:500 dilution (all Alexa Fluor from ThermoFisher). Slides were mounted and imaged on a Zeiss Confocal Microscope.
Human AD brain immunohistochemistry
Tissue was drop fixed in 4% PFA (overnight, 4°C), incubated with 30% sucrose (24 hrs, 4°C), embedded in OCT, and cryosectioned onto slides (20 μm thickness). Tissue was incubated at 40°C for 10 mins, blocked with 10% normal goat serum, 0.1% Tween-20 for 1 hour, then stained for CD49f (Biolegend 313602, 1:1000) and GFAP (Sigma G3893, 1:1000), and DAPI (overnight, 4C). After secondary antibodies were applied (Abcam ab150160, 1:5000; Abcam ab150113, 1:5000), TrueBlack (Biotium 23007) staining was conducted according to manufacturer’s protocol. Slides were imaged on a Keyence BZ-X fluorescence microscope with a 60x oil-emersion objective. Images were taken at z-stack, then full-focus merged by channel in FIJI software. Secondary-only controls were performed, showing no observable non-specific staining.
Primary rat astrocyte purification, culture, and staining
Astrocytes were purified by immunopanning and cultured in serum-free conditions as previously described (L. C. Foo 2013). Briefly cortices from 5-6 postnatal day 4-6 Sprague Dawley rat pups (Charles River) were dissected out and meninges and choroid plexus removed. The cortices were minced with a scalpel and digested in Papain for 40 min at 34°C under constant CO2/O2 gas equilibration. The digested brain pieces were washed with CO2/O2-equibiriated Ovomucoid inhibitor solution, triturated, and spun down through a cushion gradient containing low and high Ovomucoid inhibitor layers. The resulting cell pellet was passed through a 20 μm nylon mesh to create a single cell suspension. The cells were then incubated in a 34°C water bath for 30-45 mins to allow cell-specific antigens to return to the cell surface. Negative selection was performed using Goat anti-mouse IgG + IgM (H + L), Griffonia (Bandeiraea) simplicifolia lectin 1 (BSL-1), Rat anti-mouse CD45, and O4 hybridoma supernatant mouse IgM(Bansal et al. 1989), followed by positive selection for astrocytes using mouse anti-human integrin β5 (ITGB5). Purified astrocytes were detached from the panning plate with trypsin at 37°C for 3-4 min, neutralized by 30% fetal calf serum, counted, pelleted, and resuspended in 0.02% BSA in DPBS. All isolation and immunopanning steps occurred at room temperature, except for the heated digestion, incubation, and trypsinization steps. Cells were plated at 70,000 cells per well in 6 well plates containing 2 mL/well of serum-free Astrocyte Growth Medium (50% Neurobasal Medium, 50% Dulbecco’s Modified Eagle Medium (DMEM), 100 U/mL Penicillin & 100 μg/mL Streptomycin, 1 mM sodium pyruvate, 292 μg/mL L-glutamine, 5 μg/mL N-acetyl-L-cysteine (NAC), 100 μg/mL BSA, 100 μg/ml Transferrin, 16 μg/mL putrescine dihydrochloride, 60 ng/mL (0.2 μM) progesterone, and 40 ng/mL sodium selenite. Immediately before plating, the astrocyte trophic factor Heparin-binding EGF-like growth factor (HBEGF) was added (5 ng/mL) and media equilibrated to 37°C in a 10% CO2 incubator). Cells were incubated at 37°C in 10% CO2 and grown for 1 week. Cells were washed with room-temperature DPBS 3x, fixed with ice-cold methanol for 20 mins, and washed 3x with DPBS. Cells were incubated for one hour in blocking solution (PBS containing 5% donkey serum). Primary GFAP antibody (Dako; Z0334) was applied overnight at 4°C at 1:1000. The next day, cells were washed 3X in PBS, incubated with secondary antibodies (Alexa Fluor) and HOECHST for 1 hour at room temperature, and washed 3X for 10 min in PBS. Secondary antibodies were used at 1:500 dilution (all Alexa Fluor from ThermoFisher). Fluorescent imaging was performed on the Opera Phenix High-Content Screening System (PerkinElmer) using Harmony analysis software.
Primary mouse astrocyte sort
Using Aldh1l1eGFP transgenic mice on a C57BL/6J background (GENSAT, MMRRC 036071-UCD), a single cell suspension from the brain was created as previously described (L. C. Foo 2013), with modifications. Briefly, following CO2 euthanasia, brains of four adult female (P30) mice were dissected out and then the hindbrain was removed. The remaining brains were minced with a scalpel and enzymatically digested in a CO2-equilibrated papain solution for 40 mins in a 34°C water bath, one brain per tube in a sealed glass bottle. The digested brain pieces were washed with CO2-equilibrated ice cold ovomucoid inhibitor solution, triturated, and spun down through a cushion gradient containing low and high ovomucoid inhibitor layers. The resulting cell pellet was passed through a 20 μm nylon mesh to create a single cell suspension. The cells from each brain were then pooled, split into 3 conditions, and stained for 30 mins on ice with either CD49f antibody (BD 555736), its isotype control (BD 555844), or mock stained with the working buffer. Resulting cells were then washed three times and underwent fluorescence-activated cell sorting on a Sony SH800Z. Red blood cells, doublets, debris, and DAPI+ events were excluded, and gates were drawn around the Aldh1l1+, Aldh1l1−CD49f+, and Aldh1l1−CD49f− populations and were sorted into either 100 μl working buffer and imaged or directly into 350 μl Buffer RLT (Qiagen). The cells were imaged on a Keyence BZ-X fluorescence microscope. RNA was extracted from the cells sorted into Buffer RLT using the RNeasy kit (Qiagen). In order to determine the identity of the Aldh1l1− populations, reverse transcription PCR (RT-PCR) was performed using previously verified primers targeting Aldh1l1, Gfap, Snap25, Mog, Tmem119, and Cd31 (PECAM-1). Samples containing lysed unsorted brain cells and sorted red blood cells (RBCs) were also run, along with negative controls.
Differentiation of hiPSCs into astrocytes
Cells were cultured in a 37°C incubator, at 5% CO2. hiPSCs were induced along the neural lineage and differentiated using our previously published protocol (Douvaras and Fossati 2015). hiPSCs were plated at 1-2 × 105 cells per well on a matrigel-coated six-well plate in hPSC maintenance media with 10 μM Y27632 (Stemgent; 04-0012) for 24 hours. Cells were then fed daily with hPSC maintenance media. Once colonies were ~100–250 μm in diameter (day 0), differentiation was induced by adding neural induction medium (see table below). Cells were fed daily until day 8. On day 8, medium was switched to N2 medium (see table below) and cells were fed daily until day 12. On day 12, cells were mechanically dissociated using the StemPro™ EZPassage™ Disposable Stem Cell Passaging Tool (ThermoFisher; 23181010). Cells from each well were split into two wells of an ultra-low attachment 6-well plate and plated in N2B27 medium (see table below). From day 12 onwards, two-third media changes were performed every other day. On day 20, cells were switched to PDGF medium using a two-third media change (see table below). On the same day, aggregates that are round, with a diameter between 300 and 800μm, and with a brown center were picked. Picked spheres were plated (20 spheres per well of a 6-well plate) onto Nunclon-Δ plates coated with 0.1 mg mL−1 poly-L-ornithine (Sigma) followed by 10 μg mL−1 laminin (PO/Lam coating, ThermoFisher; 23017015). Spheres were allowed to attach for 24 hours and were gently fed with PDGF medium every other day (2/3 media change). At day 60-80, spheres and the cells migrating out of the spheres were dissociated with StemPro Accutase (ThermoFisher; A1110501) for 30 minutes and passed through a 70μm strainer. The resulting single-cell suspension was sorted for CD49f-positive cells. After the sort, cells were frozen in Synth-a-Freeze (ThermoFisher; A1254201) or plated onto PO/Lam coated 96-well plates for functional analyses. 24 hours after plating, medium was switched to glial medium (see table below) and cells were fed with two-third media changes every other day. Spheres remaining on the strainer at the time of the sort were plated back onto a PO/Lam Nunclon-Δ plates for up to three times (named first, second and third round) to maximize astrocyte yield.
| Neural induction medium (d0 - d7) | ||
|---|---|---|
| mTesr Custom | StemCell Technologies; | |
| PenStrep (100x) | 1x | Life Technologies; 15070063 |
| SB431542 | 10 μM | Stemgent; 04-0010 |
| LDN193189 | 250nM | Stemgent; 04-0074 |
| Retinoic acid | 100nM | Sigma-Aldrich; R2625 |
| Basal medium | ||
| DMEM/F12, GlutaMAX | ThermoFisher; 10565018 | |
| PenStrep (100x) | 1x | Life Technologies; 15070063 |
| 2-Mercaptoethanol (1000x) | 1x | Life Technologies; 21985023 |
| MEM non-essential amino acids (NEAA) solution (100x) | 1x | Life Technologies; 11140-050 |
| N2 medium (d8 - 11) | ||
| Basal medium | ||
| N2 supplement (100x) | 1x | Life Technologies; 15070063 |
| Retinoic acid | 100nM | Sigma-Aldrich; R2625 |
| Smoothened agonist (SAG) | 1μM | EMD Millipore; 566660 |
| N2B27 medium (d12 - 19) | ||
| Basal medium | ||
| N2 supplement (100x) | 1x | Life Technologies; 15070063 |
| B27 Supplement without VitA (50x) | 1x | Life Technologies; 12587-010 |
| Insulin solution, human | 25μg/mL | Sigma-Aldrich; 19278 |
| Retinoic acid | 100nM | Sigma-Aldrich; R2625 |
| Smoothened agonist (SAG) | 1μM | EMD Millipore; 566660 |
| PDGF medium (d20 – sort) | ||
| Basal medium | ||
| N2 supplement (100x) | 1x | Life Technologies; 15070063 |
| B27 Supplement without VitA (50x) | 1x | Life Technologies; 12587-010 |
| Insulin solution, human | 25μ/mL | Sigma-Aldrich; 19278 |
| PDGFaa | 10ng/mL | R&D Systems; 221-AA-050 |
| IGF-1 | 10ng/mL | R&D Systems; 291-G1-200 |
| HGF | 5ng/mL | R&D Systems; 294-HG-025 |
| NT3 | 10ng/mL | EMD Millipore; GF031 |
| T3 | 60ng/mL | Sigma-Aldrich; T2877 |
| Biotin | 100ng/mL | Sigma-Aldrich; 4639 |
| cAMP | 1μM | Sigma-Aldrich; D0260 |
| Glial medium (post-sort) | ||
| Basal medium | ||
| N2 supplement (100x) | 1x | Life Technologies; 15070063 |
| B27 Supplement without VitA (50x) | 1x | Life Technologies; 12587-010 |
| Insulin solution, human | 25μg/mL | Sigma-Aldrich; 19278 |
| T3 | 60ng/mL | Sigma-Aldrich T2877 |
| Biotin | 100ng/mL | Sigma-Aldrich 4639 |
| cAMP | 1μM | Sigma-Aldrich D0260 |
| HEPES | 10mM | Sigma-Aldrich H4034 |
| Ascorbic acid | 20μg/mL | Sigma-Aldrich A4403 |
Fluorescence-activated cell sorting (FACS) for CD49f+ astrocyte isolation
Cells were lifted by incubation with Stempro Accutase for 30 minutes. Cell suspension was triturated 8-10 times and passed through a 70μm cell strainer (Sigma; CLS431751) then diluted >7x with DMEM/F12 medium. Cells were spun in a 15mL conical tube at 300g for 5 minutes at room temperature. The cell pellet was resuspended in 200μL of FACS buffer (PBS, 0.5% BSA, 2mM EDTA, 20mM Glucose) with 1:50 PE Rat Anti-Human CD49f antibody (BD Biosciences; 555736) and incubated on ice for 20 minutes. Cells were then washed in FACS buffer, pelleted at 300g for 5 minutes and resuspended in FACS buffer containing propidium iodide for dead cell exclusion. The respective unstained, CD49f-only stained, and propidium iodide-only stained controls were run in parallel. CD49f+ cells were isolated via FACS on an ARIA-IIu™ Cell Sorter (BD Biosciences) using the 100μm ceramic nozzle, at 20 or 23 psi. Data were analyzed using FlowJo v9.
For initial screening, BD Lyoplate-Human cell surface marker screening panel (BD Biosciences; 560747) and O4 antibody (gift from Dr. Jim Goldman’s lab) were used on day 78 cultures digested as described above.
For A1 stimulation, cells were treated with TNFα (30ng/mL), IL-1α (3ng/mL), and C1q (400ng/mL) for 24-48 hours.
Differentiation into oligocortical organoids and single-cell digestion
hiPSC line 051121-01-MR-017 was induced along the neural lineage and differentiated into oligocortical organoids using our previously published protocol (Madhavan et al. 2018). At days 117 and 169, organoids were digested into a single-cell suspension for FACS sorting. For digestion, 4 organoids were pooled and incubated in papain (Worthington; LK003150) for 30 minutes at 37°C on a shaker. The cell suspension was triturated 10 times and placed back at 37°C for 10 more minutes. The ovomucoid protease inhibitor was added and the cell suspension was spun down at 300g for 4 minutes, resuspended in FACS buffer, and filtered through a 45μm filter. The cell suspension was then stained for PE Rat Anti-Human CD49f antibody with appropriate controls and CD49f positive and negative fractions were isolated as described above. CD49f+ and CD49f− cell fractions were plated down on poly-ornithine and laminin-coated plates and fixed in 4% PFA for immunofluorescence analysis.
Oligocortical organoids were also fixed in 4% paraformaldehyde 1 hour at R.T., washed 3X in PBS, then stored in 30% sucrose in PBS at 4°C overnight. Organoids were then embedded in O.C.T. compound and cryosectioned at 20μm thickness.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 10 minutes, washed 3X in PBS, and incubated for one hour in blocking solution (PBS containing 0.1% Triton-X100 and 5% donkey serum). Primary antibodies (see Antibody Table) were applied overnight at 4°C. The next day, cells were washed 3X in PBS, incubated with secondary antibodies (Alexa Fluor) and HOECHST for 1 hour at room temperature, and washed 3X for 10 min in PBS. Secondary antibodies were used at 1:500 dilution (all Alexa Fluor from ThermoFisher).
For fluorescent image analysis and quantification of GFAP, AQP4, C3, and cell radial mean plates were imaged on the Opera Phenix High-Content Screening System (PerkinElmer) using Harmony analysis software. For MAP2 quantification we imaged plates on the ArrayScan XTI live high content platform (Thermo Fisher) and used CellProfiler software.
Antibodies used in this study are as follows:
| Name | Dilution | Vendor | Cat. No. (RRID) |
|---|---|---|---|
| GFAP | 1:1000 | EMD Millipore | MAB360 (AB_11212597) |
| S100B | 1:1000 | Sigma | S2532 (AB_477499) |
| Vimentin | 1:250 | Abcam | ab8978 (AB_306907) |
| C3D | 1:1000 | DAKO | A0063 (AB_578478) |
| NFIa | 1:250 | Active Motif | 39397 (AB_2314931) |
| AQP4 | 1:500 | Sigma | HPA014784 (AB_1844967) |
| CD49f | 1:1000 | BioLegend | 313602 (AB_345296) |
| SOX9 | 1:250 | Cell Signaling | 82630T (AB_2665492) |
| EAAT1 | 1:250 | Abcam | ab416 (AB_304334) |
| MAP2 | 1:1000 | Abcam | ab5392 (AB_2138153) |
| O4 | 1:50 | Gift from Dr. J. Goldman | |
| CD49f | 1:50 (FACS) | BD Biosciences | 555736 (AB_396079) |
Immunofluorescence on slides
Slides with cryosectioned organoids were incubated for one hour in blocking solution (PBS containing 0.1% saponin and 2.5% donkey serum). Primary antibodies (see Antibody Table) were applied overnight at 4°C. The next day, slides were washed 3X in PBS, incubated with secondary antibodies (Alexa Fluor) and HOECHST for 1 hour at room temperature, washed 3X for 10 min in PBS. Secondary antibodies were used at 1:500 dilution (all Alexa Fluor from ThermoFisher). Slides were mounted and imaged on a Zeiss Confocal Microscope.
Glutamate uptake assay
CD49f+ astrocytes were incubated for 30 minutes in Hank’s balanced salt solution (HBSS) buffer without calcium and magnesium (Gibco), then for 3 hours in HBSS with calcium and magnesium (Gibco) containing 100 μM glutamate. At the same time, identical volumes of HBSS with calcium and magnesium (Gibco) containing 100 μM glutamate were incubated in empty cell-free wells for determining the percentage of glutamate uptake. Samples of medium were collected after 3h and analyzed with a colorimetric glutamate assay kit (Sigma-Aldrich; MAK004-1KT), according to the manufacturer’s instructions. Samples of HBSS with calcium and magnesium (Gibco) without glutamate were also run as negative controls. For A1 astrocytes, cells were treated with 3 ng/mL IL-1α (Sigma; I3901), 30 ng/mL tumor necrosis factor alpha (R&D Systems; 210-TA-020) and 400 ng/mL C1q (MyBioSource; MBS143105) for 24h prior to the experiment. Three iPSC lines and two to four technical replicates per lines were used. p-values were calculated using a one-way ANOVA with Dunnett’s correction for multiple comparisons for comparing astrocytic glutamate uptake to the no astrocyte control or using multiple t-tests with Holm-Sidak’s correction for multiple comparisons for comparing A1 astrocytes to A0 astrocytes.
Intracellular Ca2+ imaging on hiPSC-astrocytes
CD49f+ astrocytes from three iPSC lines were cultured on glass coverslips coated with 0.1 mg/ml poly-L-ornithine followed by 10 μg/ml laminin. For Ca2+ dye loading, cells were treated with Rhod-3/AM (ThermoFisher; R10145) for 30 minutes at 37 °C, washed twice with glial medium and imaged 30-60 minutes later. Live fluorescence imaging of spontaneous Ca2+ activity was done with an ArrayScan XTi high-content imager (ThermoFisher) equipped with live cell module maintaining 37 °C, 5% CO2 and >90% relative humidity environment. Whole field of view images at 20x magnification were acquired with Photometrics X1 cooled CCD camera (ThermoFisher) at 4Hz for 2 minutes. For Ca2+ imaging experiments involving drug application cells were grown on 1.5x PO/Lam coated plastic coverslips (Nunc Thermanox) and then transferred to a heated (31°C) recording chamber mounted onto an upright Olympus BX61 microscope. Fluorescence was recorded at 2Hz by a cooled CCD camera (Hamamatsu Orca R2). Images were taken 2 minutes before and 3 minutes after the addition of ATP (100 μM), and drug application was done via whole chamber perfusion for a period of 60s. For quantification of the change in intensity over time, astrocytes were outlined as regions of interest (ROIs) and analyzed with ImageJ software. [Ca2+]i transients are expressed in the form of ΔF(t)/F0, where F0 is a baseline fluorescence of a given region of interest and ΔF is the difference between current level of fluorescence F(t) and F0. Fluctuations of ΔF(t)/F0 of less than 0.05 were considered non-responses.
Intracellular Ca2+ imaging on rodent astrocytes
Postnatal rat astrocytes were purified by immunpanning (see above), plated at a density of 5,000 cells/35mm glass-bottom dish (MatTek, No. 1.5) coated with poly-D-lysine, and maintained in serum-free culture conditions for 5-7 days. For imaging, astrocytes were pre-incubated for 15 minutes with 2 μM Fluo-4 AM (Invitrogen, F-14201) and washed with 1x PBS and replaced with normal astrocyte growth medium. Fluorescent image stacks were taken at 0.7 s intervals and intensity analyzed for 10-30 randomly selected cells per stack in ImageJ. Data was collected from three separate preparations of astrocyte cultures from at least three different plates of cells per preparation.
Cytokine detection and measurement
CD49f+ astrocytes were plated on PO/Lam coated 96 well plates and treated with 3 ng/mL IL-1α (Sigma; 3901), 30 ng/mL tumor necrosis factor alpha (TNFα R&D Systems; 210-TA-020) and 400 ng/mL C1q (MyBioSource; MBS143105) for 24h or with 10 ng/mL tumor necrosis factor alpha and 10ng/ml IL-1β (R&D Systems; 401-ML-005) for 24h. The medium was collected and spun down at 1,500 rpm for 5 minutes to remove debris, and frozen at −80°C. Samples were thawed on ice and a ProcartaPlex Custom Panel of cytokines (ThermoFisher) was quantified using the Luminex instrument, as per the manufacturer’s instructions. Three different iPSC lines and two replicates per line were used. p-values were calculated using a one-way ANOVA with Dunnett’s correction for multiple comparisons.
Neuronal differentiation and co-culture with astrocytes
For neuronal differentiation, hiPSCs (line 3) were plated in a 12-well plate in hPSC maintenance media with 10μM ROCK inhibitor (Y2732, Stemgent). The next day, the cells were induced and fed daily with neural induction media (DMEM/F12 (ThermoFisher; 11320033) 1:1 Neurobasal (ThermoFisher; 21103049) with 1x Glutamax, 1x N2 supplement, 1x B27 supplement without Vitamin A) with SB431542 (20μM), LDN193189 (100nM), XAV939 (1μM). On day 10, the media was switched to neural induction media with XAV939 (1μM), and the daily media changes continued. On day 15, cells were dissociated using Accutase and either frozen in Synth-a-freeze, or plated in neuronal media (Brainphys (StemCell Technologies; 05790) with 1x B27 supplement (ThermoFisher; 17504001), and 10μM ROCK inhibitor) at 50k/well in a PO/Lam coated 96-well plate (Corning; 353376). On day 16, the media was switched to neuronal media with BDNF (40ng/mL), GDNF (40ng/mL), Laminin (1μg/mL), dbcAMP (250μM), ascorbic acid (200μM), PD0325901 (10μM), SU5402 (10μM), DAPT (10μM)(Qi et al. 2017). Cells were fed every other day. PD0325901, SU5402, and DAPT were taken out of the media after two weeks. CD49f+ astrocytes were plated on top of neurons on day 33 of neuronal differentiation and cells were fed every other day with neuronal media until day 50.
To study the effect of A1 astrocytes on neuronal maturation, CD49f+ astrocytes (15k/well of a 96wp) were plated on top of neurons on day 33 of neuronal differentiation and cells were fed twice per week with neuronal media with or without TNFα, IL-1α, and C1q until day 51-53. To evaluate the potential direct effect of cytokines on neuronal maturation, neurons cultured alone were fed twice per week, starting at day 34 with neuronal media with or without TNFα, IL-1α, and C1q until day 53.
Electrophysiology
For whole-cell recordings, hiPSC-derived neurons from one iPSC line (line 3) were visualized using an upright Olympus BX61 microscope equipped with a 40x objective and differential interference contrast optics. Neurons were constantly perfused with BrainPhys® medium (STEMCELL Technologies, Catalog #05790) preheated to 30-31°C. Patch electrodes were filled with internal solutions containing 130 mM K+gluconate, 6 mM KCl, 4 mM NaCl, 10mM Na+HEPES, 0.2 mM K+EGTA; 0.3mM GTP, 2mM Mg2+ATP, 0.2 mM cAMP, 10mM D-glucose. The pH and osmolarity of the internal solution were adjusted to resemble physiological conditions (pH 7.3, 290–300 mOsmol). Current- and voltage-clamp recordings were carried out using a Multiclamp 700B amplifier (Molecular Devices), digitized with Digidata 1440A digitizer and fed to pClamp 10.0 software package (Molecular Devices). For spontaneous EPSC recordings, neurons were held at chloride reversal potential of −75 mV. Data processing and analysis were performed using ClampFit 10.0 (Molecular Devices) and Prism software. CD49f+ astrocytes for co-cultures were from three iPSC lines. p-values to compare neurons alone to neurons with astrocytes or neurons with A0 astrocytes to neurons with A1 astrocytes were calculated using a two-tailed, unpaired t-test.
Neurotoxicity
Primary mouse cortical neurons (ThermoFisher; A15586) were thawed and plated into PO/Lam coated 96-well plates at a density of 20k/well in Neurobasal media (ThermoFisher; 21103049) with 1x Glutamax, 1x B27 supplement (ThermoFisher; 17504001), and 1x PenStrep (Life Technologies; 15070063). Cells were fed the next day, then every other day until the addition of astrocyte conditioned media ten days later. CD49f+ astrocytes were plated into PO/Lam coated 24-well plates at a density of 200k/well and kept in PDGF medium for 24 hours. The next day, the medium was switched to Neurobasal medium with 1x Glutamax, 1x PenStrep and 1x B27 supplement, and half-media changes were performed every other day, for one week. At day 8, a full media change was performed, with 600μL of Neurobasal medium, 1x Glutamax, 1x PenStrep and 1x B27 supplement, minus antioxidants (ThermoFisher; 10889038) per well, with or without TNFα, IL-1α, and C1q. Astrocyte conditioned media (ACM) from CD49f+ A0 astrocytes (unstimulated) and A1 astrocytes (cultured with TNFα, IL-1α, and C1q) were collected 48 hours later and added to the mouse neuronal cultures without concentration. Mouse neurons were treated with 70% ACM with 5μM IncuCyte Caspase-3/7 Green Apoptosis Assay Reagent (Sartorius; 4440) and imaged every 6 hours for 60 hours on an Incucyte S3 epifluorescence time lapse microscope (Sartorius). Mouse neurons were also treated with fresh media controls, with or without TNFα, IL-1α, and C1q. Two to four wells were analyzed per condition. For image analysis, we took 3 images per well using a 10× objective lens from random areas of the 96-well plate and plotted the total integrated intensity, known as the total sum of the objects’ fluorescent intensity in the image. Data was normalized to the confluence per image. Data analysis was done using the Incucyte Analysis Software (Sartorius). Graphpad Prism software was used to perform a two-way ANOVA to determine statistical significance per line across conditions.
Human iPSC-neurons from one iPSC line (line 3) were differentiated as previously described until the addition of astrocyte conditioned media from three iPSC lines at day 66 of the differentiation. CD49f+ astrocytes were plated into PO/Lam coated 24-well plates at a density of 200k/well in PDGF medium. The next day, the medium was switched to Brainphys medium with 1x PenStrep and 1x B27 supplement, then half-media changes were performed every other day. At day 8, a full media change was performed, with 600μL of Brainphys, 1x PenStrep, and 1x B27 supplement minus antioxidants per well, with or without TNFα, IL-1α, and C1q. Astrocyte conditioned media (ACM) from CD49f+ A0 astrocytes (unstimulated) and A1 astrocytes (cultured with TNFα, IL-1α, and C1q) was collected 48 hours later and applied to the hiPSC-neuron cultures without any concentration. hiPSC-neurons were treated with 67% ACM with 5μM IncuCyte Caspase-3/7 Green Apoptosis Assay Reagent (Sartorius; 4440) and 1:2000 IncuCyte NucLight Rapid Red Reagent for nuclear labeling (Sartorius; 4717) and imaged every 6 hours for 72 hours on an Incucyte S3 epifluorescence time lapse microscope (Sartorius). hiPSC-neurons were also treated with fresh media controls, with or without TNFα, IL-1α, and C1q. Four to eight wells were analyzed per condition. For image analysis, we took 3 images per well using a 10× objective lens from random areas of the 96-well plate and plotted the percentage of caspase-3/7 positive nuclei. Data analysis was done using the Incucyte Analysis Software (Sartorius). Graphpad Prism software was used to perform a two-way ANOVA to determine statistical significance per line across conditions.
Synaptosome engulfment assay
Live CD49f+ astrocytes were plated on PO/Lam coated 96 well plates and treated with TNFα, IL-1α, and C1q for 48 hours. Cells were then incubated with 2μL/mL pHrodo-conjugated synaptosomes in glial medium with or without TNFα, IL-1α, and C1q and imaged every hour with an Incucyte S3 epifluorescence time lapse microscope (Sartorius) for 2 days. Three iPSC lines and three or four wells/line were analyzed per condition. For image analysis, we took 3 images per well using a 10× objective lens from random areas of the 96-well plate and plotted the total integrated intensity, known as the total sum of the objects’ fluorescent intensity in the image. Data was normalized to the confluence per image. Data analysis was done using the Incucyte Analysis Software (Sartorius). Graphpad Prism software was used to perform a two-way ANOVA to determine statistical significance per line across conditions.
Bulk RNA sequencing and analysis
RNA isolation was performed using the RNeasy Plus Micro Kit (Qiagen; 74034). Media was aspirated off CD49f+ cells in culture, and cells were lysed in Buffer RLT Plus with 1:100 β-mercaptoethanol. Samples were then stored at −80°C until processed further according to manufacturer’s instructions. RNA was eluted in 17μl RNase free ddH2O and quantified with a Qubit 4 Fluorometer (ThermoFisher; Q33227). Paired-end RNAseq data were generated with the Illumina HiSeq 4000 platform following the Illumina protocol. The raw sequencing reads were aligned to human hg19 genome using star aligner (Dobin et al. 2013)(version 2.4.0g1). Following read alignment, featureCounts (Liao, Smyth, and Shi 2014) (v1.6.3) was used to quantify the gene expression at the gene level based on Ensembl gene model GRCh37.70. For re-analysis of human primary astrocyte RNAseq data from Zhang et al. (Zhang et al. 2016), we downloaded the raw RNAseq data from gene expression omnibus (GEO: accession GSE73721). Similarly, for comparison with a recently published hiPSC-derived astrocyte datasets, we downloaded three bulk RNAseq raw data from TCW et al. (Tcw et al. 2017)(GSE97904), Bradley et al.(Bradley et al. 2019) (GSE133489), and Sloan et al. (Sloan et al. 2017)(GSE99951). The RNAseq data from each of these published studies were processed using the same star/featureCounts pipeline as described above and then the gene level read counts were combined with the gene count data of our samples. Genes with at least 1 count per million (CPM) in more than 2 samples in the merged data were considered expressed and hence retained for further analysis, otherwise removed. Then the read count data were normalized using trimmed mean of M-values normalization (TMM) method (Robinson and Oshlack 2010) to adjust for sequencing library size difference and then corrected for batch using linear regression. To examine similarities among samples, hierarchical cluster analysis was performed based on the respective transcriptome-wide gene expression data using R programming language. Meanwhile, a separate expression abundance, transcript per million (TPM), was also calculated using salmon (Patro et al. 2017) (v0.14.1) with 50 bootstraps and fragment-level GC biases correction enabled for optimizing the abundance estimation.
Single-cell RNA sequencing.
For A1-like astrocyte analysis, day 73 (line 3) or day 80 (line 1) unsorted hiPSC-derived cultures (to include astrocytes at different stages of development) were left untreated or were treated for 24 hours with TNFα, IL-1α, and C1q, then harvested in parallel using papain (Worthington; LK003153) and processed using the 10X Single Cell 3’ v2 or v3.1 protocols. For CD49f sorting experiment, day 73 (line 3) unsorted cultures were harvested using papain, sorted for CD49f, and the unsorted, CD49f+, and CD49f− fractions were processed using the 10X Single Cell 3’ v3.1 protocol. For fetal samples, unsorted, sorted CD49f+, and sorted CD49f cells were processed using the 10X Single Cell 3’ v3.1. All samples were filtered through 40μm Flowmi Cell Strainers (Scienceware; H13680-0040) to obtain a single cell suspension. In brief, the Chromium Single Cell A Chip (10X Genomics; PN-1000009) or the Chromium Next GEM Chip G Single Cell Kit (10X Genomics; PN-1000120) was loaded with ~7,000 cells/sample and library preparation was performed as per the Chromium Single Cell 3’ Library & Gel Bead Kit manufacturer’s recommendations (10X Genomics; PN-120237 and PN-1000121). The Chromium i7 Multiplex Kit (10X Genomics; PN-120262) was used. Quality control was performed using the Qubit 4 Fluorometer (ThermoFisher; Q33227) and the Agilent 4200 TapeStation system. The resulting prepared cDNA library was sequenced on a NovaSeq/HiSeq 2x150 bp, and ~50,000 reads per cell were obtained.
Single-cell data analysis.
Sequenced samples were initially processed using Cell Ranger software version 3.0.2 (10x Genomics) and were aligned to the GRCh38 (hg38) human reference genome. Through the Cell Ranger pipeline, digital gene expression matrices (DGE) were generated containing the raw unique molecular identifier (UMI) counts for each sample. In order to compare between samples, DGEs were merged using the muscat R package (Crowell et al. 2019) Doublets were removed using the hybrid method of the scds package, therefore excluding the predicted 1% per thousand cells with the highest doublet scores (Bais and Kostka 2019). Quality control and filtering was completed using the scater R package (D. J. McCarthy et al. 2017). Cells were removed if their feature count, number of expressed features, and/or percentage of mitochondrial genes was outside of the median value ± 2.5 median absolute deviations. Genes were removed if they were undetected across all cells or if they were expressed by fewer than 20 cells. For hiPSC samples, of the 53,599 cells and 28,158 genes originally identified, 43,127 cells (81%) and 13,710 genes (49%) met these criteria and were included in the following analyses. For fetal samples, of the 29,585 cells and 28,161 genes originally identified, 22,281 cells (75%) and 14,146 genes (50%) met these criteria and were included in the following analyses.
Next, samples were integrated, clustered, and dimensionally reduced using Seurat version 3.1.0 (Stuart et al. 2019). The top 2000 variable genes, identified using FindVariableFeatures function, were used to integrate and cluster samples. Integration was performed using the first 30 dimensions of the Canonical Correlation Analysis (CCA) cell embeddings. Clusters and tSNE plots were generated using the first 20-30 principle components, and a resolution of 0.1-0.3 was used to cluster all cells (please see Table below for specifics depending on round of analysis). Clusters were then identified manually based on a known set of neural cell-specific markers (Cahoy et al. 2008; Darmanis et al. 2015; Campanelli et al. 2008). New subset objects were made in order to analyze specific astrocyte-related clusters and/or samples (please see Table below for subsetting strategy). This process was repeated separately for fetal samples.
Principle components and resolution used for analyses.
Object key: so_astro_plated, data from only astrocyte-related clusters from only A0/A1-like plated hiPSC-derived samples; so_sorted, data from only sorted hiPSC-derived samples; so_sorted_astro, data from only astrocyte-related clusters from only sorted hiPSC-derived samples; so fetal data from all fetal samples.
| Purpose | Object name | Object from which subset was generated |
Principle Components |
Resolution | Fig. |
|---|---|---|---|---|---|
| Analyze hiPSC astrocyte- Related clusters from plated samples | so_astro_plated | so_astro | 20 | 0.3 | 8a-d |
| Analyze only | so_sorted | so | 30 | 0.3 | 3a-d |
| sorted | S4 | ||||
| samples | S5 | ||||
| Analyze astrocyte-related clusters from only sorted samples | so_sorted_astro | so_sorted | 20 | 0.2 | 3e-g S6 |
| Analyze all fetal samples | so_fetal | N/A | 30 | 0.1 | 4b-d |
Through these analyses, in all hiPSC samples, we identified 13 subpopulations, including mature astrocytes, transitioning astrocytes, immature astrocytes, neural progenitor cells, oligodendrocytes, and neurons. In fetal samples, we identified 18 subpopulations, including mature astrocytes, immature astrocytes, oligodendrocyte precursor cells, myeloid cells, endothelial cells, and cells of unknown origin. tSNE plots, feature plots, and dot plots were generated using Seurat standard functions. Additionally, differential gene expression supplementary tables were generated using the FindAllMarkers function. Additionally, differential gene expression supplemental tables (Supplemental Table 1) were generated using the FindAllMarkers function.
qRT-PCR
Reverse transcription was performed using the iScript cDNA Synthesis Kit (Biorad; 1708891) with 500ng of RNA per reaction. Real-Time PCR was then performed on an Applied Biosystems 7300 Real-time PCR system with 5ng cDNA per sample in triplicate using Taqman gene expression master mix (ThermoFisher; 4369514) and the following pre-designed Taqman gene expression assays (ThermoFisher; 4331182): ITGA6 (Hs01041011_m1), MEGF10 (Hs01002798_m1), MERTK (Hs00179024_m1), GAS6 (Hs01090305_m1), GRIN2b (Hs01002012_m1), GRIA1 (Hs00181348_m1), GRIK1 (Hs00543710_m1), THBS1 (Hs00962908_m1), THBS2 (Hs01568063_m1), SPARCL1 (Hs00949886_m1), GPC6 (Hs00170677_m1), and ACTB (Hs01060665_g1). StepOnePlus Software (ThermoFisher) was used to determine Ct values. Expression values were normalized to ACTB and to A0 samples. CD49f+ astrocytes from three lines were untreated or treated with TNFα, IL-1α, and C1q for 24 hours before mRNA was collected. Two independent experiments were performed per line. Three technical replicates were run per sample. Graphpad Prism software was used to perform a paired t-test analysis to determine statistical significance across conditions.
Fetal brain digestion for single cell suspension
Tissues were chopped and incubated in papain (Worthington; LK003150) for 30 minutes at 37°C on a shaker. The cell suspension was triturated 10 times and placed back at 37°C for 10 more minutes. The ovomucoid protease inhibitor was added and the cell suspension was spun down at 300g for 4 minutes, resuspended in distilled water for 30 seconds for red blood cell lysis, diluted in FACS buffer, spun down at 300g for 4 minutes, resuspended in FACS buffer, and filtered through a 45μm filter. The cell suspension was then stained for PE Rat Anti-Human CD49f antibody with appropriate controls and CD49f positive and negative fractions were FACS-isolated as described above, except using a 130μm nozzle, which is recommended for larger and adherent cells to reduce the likelihood of clogging; however, it may reduce the purity of the sorted populations. Unsorted, CD49f+, and CD49f− cell fractions were processed using the 10X Single Cell 3’ v3.1 protocol as described above or plated down on poly-ornithine and laminin-coated plates, fixed in 4% PFA in PBS 3 days later for 10 minutes, then washed 3X and stored in PBS.
Protein quantification
Unstimulated astrocytes (A0) or astrocytes stimulated for 24 hours with TNFα, IL-1α, and C1q were lysed with protein lysis buffer consisting of RIPA buffer (Sigma; R0278), cOmpleteTM Mini EDTA-free Protease Inhibitor 534 Cocktail (Sigma; 11836170001), Phosphatase Inhibitor Cocktail 3 (Sigma; P0044), and Phosphatase 535 Inhibitor Cocktail 2 (Sigma; P5726). Protein concentration for lysate samples was determined using a Pierce BCA Protein Assay Kit (Thermo Scientific; 23225). To quantify protein levels, equal amounts of protein per sample were run on the Wes™ (ProteinSimple). The following primary antibodies were used at a 1 to 50 dilution: ITGA6 antibody (Novus Biologicals; NBP1-85747), EAAT2 antibody (Santa Cruz Biotechnology; sc-365634), GFAP antibody (Dako; Z0334), TIMP-1 antibody (R&D Systems; AF970), beta-actin antibody (Santa Cruz Biotechnology; sc-47778). ProteinSimple Detection Modules were used for as secondary antibodies. Protein was collected from three cell lines and two independent experiments. Two technical replicates were run per sample. Protein levels were determined using Compass software (ProteinSimple) as the area under the curve and were normalized to beta-actin. Graphpad Prism software was used to perform a paired t-test to determine statistical significance between conditions.
QUANTIFICATION AND STATISTICAL ANALYSES
The software used for quantification is specified for each assay. Briefly, Graphpad Prism software was used for all statistical analyses. The statistical test used, value of n, and meaning of n are indicated in the figure legends and the corresponding methods section. The definition of center and dispersion and precision measures are indicated in the figure legends. Statistical significance is defined as p<.05 (* = p<.05 ; ** = p<.01, *** = p<.001, **** = p<.0001).
ADDITIONAL RESOURCES
We have made RNA sequencing datasets from this study available in a user-friendly searchable online database (https://nyscfseq.appspot.com/).
Supplementary Material
Supplemental Table 1: Cell counts and differentially expressed genes per scRNA-seq cluster (Related to Fig. 3, 4, 8, S4, S5, S6)
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Purified anti-human/mouse CD49f Antibody | BioLegend | Cat#313602; RRID: AB_345296 |
| Monoclonal Anti-Glial Fibrillary Acidic Protein (GFAP) antibody produced in mouse | Sigma-Aldrich | Cat#G3893; RRID: AB_477010 |
| Glial Fibrillary Acidic Protein, Polyclonal (Concentrate) | Dako | Cat#Z0334; RRID: AB_10013382 |
| Anti-Glial Fibrillary Acidic Protein Antibody, clone GA5 | EMD Millipore | Cat#MAB360; RRID: AB_11212597 |
| PE Rat Anti-Human CD49f | BD Biosciences | Cat#555736; RRID: AB_396079 |
| Monoclonal Anti-S-100 (β-Subunit) antibody produced in mouse | Sigma | Cat#S2532; RRID: AB_477499 |
| Anti-Vimentin antibody | Abcam | Cat#ab8978; RRID: AB_306907 |
| C3d Complement (Multipurpose) | Dako | Cat#A0063; RRID: AB_578478 |
| NF-1A antibody (pAb) | Active Motif | Cat#39397; RRID: AB_2314931 |
| Anti-AQP4 antibody produced in rabbit | Sigma | Cat#HPA014784; RRID: AB_1844967 |
| Sox9 (D8G8H) Rabbit mAb | Cell Signaling | Cat#82630T; RRID: AB_2665492 |
| Anti-EAAT1 antibody | Abcam | Cat#ab416; RRID: AB_304334 |
| Anti-MAP2 antibody | Abcam | Cat#ab5392; RRID: AB_2138153 |
| Integrin alpha 6/CD49f Antibody | Novus Biologicals | Cat#NBP1-85747; RRID: AB_11039415 |
| EAAT2 Antibody (E-1) | Santa Cruz Biotechnology | Cat#sc-365634; RRID: AB_10844832 |
| Human TIMP-1 Antibody | R&D Systems | Cat#AF970; RRID: AB_355751 |
| β-Actin Antibody (C4) | Santa Cruz Biotechnology | Cat#sc-47778; RRID: AB_2714189 |
| Biological Samples | ||
| Human healthy brain tissue | Advanced Tissue Services | http://advancedtissueservices.com/ |
| Human Azheimer’s disease patient brain tissue | Rhode Island Hospital Brain Tissue Resource Center | https://www.brown.edu/research/facilities/brain-tissue-resource-center/ |
| Primary mouse cortical neurons | ThermoFisher | Cat#A15586 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Stemolecule SB431542 | Stemgent | Cat#04-0010 |
| Stemolecule LDN-193189 | Stemgent | Cat#04-0074 |
| Retinoic acid | Sigma-Aldrich | Cat#R2625 |
| Smoothened Agonist, SAG | EMD Millipore | Cat#566660 |
| Recombinant Human PDGF-AA Protein, CF | R&D Systems | Cat#221-AA-050 |
| Recombinant Human IGF-I/IGF-1 Protein, CF | R&D Systems | Cat#291-G1-200 |
| Recombinant Human HGF Protein | R&D Systems | Cat#294-HG-025 |
| Neurotrophin 3 | EMD Millipore | Cat#GF031 |
| 3,3’,5-Triiodo-L-thyronine | Sigma-Aldrich | Cat#T2877 |
| Biotin | Sigma-Aldrich | Cat#4639 |
| N6,2’-O-Dibutyryladenosine 3’,5’-cyclic monophosphate sodium salt | Sigma-Aldrich | Cat#D0260 |
| L-Ascorbic acid | Sigma-Aldrich | Cat#A4403 |
| Recombinant Human TNF-alpha Protein | R&D Systems | Cat#210-TA-020 |
| Interleukin-1α from rat | Sigma | Cat#I3901 |
| Complement Component C1q Native Protein | MyBioSource | Cat#MBS143105 |
| Recombinant Mouse IL-1 beta/IL-1F2 Protein | R&D Systems | Cat#401-ML-005 |
| B27 supplement, minus antioxidants | ThermoFisher | 10889038 |
| Critical Commercial Assays | ||
| Human Cell Surface Marker Screening Panel | BD Biosciences | Cat#560747; RRID: AB_1953343 |
| Glutamate Assay Kit | Sigma-Aldrich | Cat#MAK004 |
| Rhod-3 Calcium Imaging Kit | ThermoFisher | Cat#R10145 |
| Fluo-4, AM, cell permeant | Invitrogen | Cat#F14201 |
| ProcartaPlex Custom Panel of cytokines | ThermoFisher | https://www.thermofisher.com/us/en/home/life-science/antibodies/immunoassays/procartaplex-assays-luminex/procartaplex-immunoassays/procartaplex-custom-panels.html |
| IncuCyte® Caspase-3/7 Green Apoptosis Assay Reagent | Sartorius | Cat#4440 |
| IncuCyte® NucLight Rapid Red Reagent for nuclear labeling | Sartorius | Cat#4717 |
| Deposited Data | ||
| Bulk RNA sequencing data | This paper | AD Knowledge Portal (insert accession number) |
| Single-cell RNA sequencing data | This paper | AD Knowledge Portal (insert accession number) |
| Primary astrocyte RNA sequencing data | Zhang et al., 2016 | GEO: GSE73721 |
| hiPSC-derived astrocyte RNA sequencing data | TCW et al., 2017 | GEO: GSE97904 |
| Regional hiPSC-astrocyte RNA sequencing data | Bradley et al., 2019 | GEO: GSE133489 |
| hiPSC-astrocyte RNAseq data at different maturation stages | Sloan et al. 2017 | GEO: GSE99951 |
| Experimental Models: Cell Lines | ||
| Human induced pluripotent stem cell lines | The NYSCF Repository | https://nyscf.org/research-institute/repository-stem-cell-search/ |
| Experimental Models: Organisms/Strains | ||
| Mouse: Aldh1l1eGFP transgenic mice on a C57BL/6J background | GENSAT | RRID: MMRRC 036071-UCD |
| Rat: Sprague Dawley dams | Charles River | Strain code: 400 |
| Oligonucleotides | ||
| Taqman gene expression assays | ThermoFisher | Cat#4331182 |
| Software and Algorithms | ||
| Harmony High-Content Imaging and Analysis Software | PerkinElmer | https://www.perkinelmer.com/product/harmony-4-9-office-license-hh17000010 |
| FlowJo v9 | BD Biosciences | RRID: SCR_008520; https://www.flowjo.com/ |
| CellProfiler Cell Image Analysis Software | Broad Institute | RRID: SCR_007358 https://cellprofiler.org/ |
| Prism Software | GraphPad | RRID: SCR_002798 https://www.graphpad.com/scientific-software/prism/ |
| ImageJ | National Institutes of Health | RRID: SCR_003070 https://imagej.nih.gov/ij/ |
| xPONENT Software | Luminex Corporation | https://www.luminexcorp.com/xponent/ |
| pCLAMP Electrophysiology Data Acquisition & Analysis Software | Molecular Devices | RRID: SCR_011323 https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite |
| IncuCyte® Analysis Software | Sartorius | https://www.essenbioscience.com/en/products/software/incucyte-base-software/ |
| Compass software for Simple Western | ProteinSimple | https://www.proteinsimple.com/software_compass_simplewestern.html |
| STAR aligner | Dobin et al., 2013 | RRID: SCR_015899 https://github.com/alexdobin/STAR |
| featureCounts | Liao et al., 2014 | RRID: SCR_012919 http://bioinf.wehi.edu.au/featureCounts/ |
| salmon | Patro et al., 2017 | RRID: SCR_017036 https://combine-lab.github.io/salmon/ |
| muscat | Crowell et al., 2019 | https://bioconductor.org/packages/3.11/bioc/html/muscat.html |
| scds | Bais et al., 2019 | https://bioconductor.org/packages/release/bioc/html/scds.html |
| scater | McCarthy et al., 2017 | RRID: SCR_015954 https://bioconductor.org/packages/release/bioc/html/scater.html |
| Seurat v3.1.0 | Stuart et al., 2019 | RRID: SCR_016341 https://satijalab.org/seurat/ |
| Other | ||
| User-friendly resource website with all RNA sequencing data | This paper | https://nyscfseq.appspot.com/ |
| The bulk and single cell RNA-sequencing data | This paper | The Synapse open source platform (syn21861229) https://www.synapse.org/#!Synapse:syn21861229 |
Highlights.
CD49f is a novel, reactivity independent marker for human astrocytes
CD49f can be used to purify human fetal astrocytes and iPSC-derided astrocytes
CD49f+ hiPSC-astrocytes acquire an A1-like reactive state upon cytokine stimulation
CD49f+ A1-like reactive astrocytes are dysfunctional and toxic to neurons in vitro
ACKNOWLEDGMENTS
This work was supported by the National Stem Cell Foundation (NSCF), the New York Stem Cell Foundation Research Institute (NYSCF) (including all experiments involving fetal tissues), NINDS grant 1R21NS111186 and NYSTEM grant DOH01-STEM5-2016-00114 (to V.F.), NIA grants U01AG046170 and RF1AG057440 (to B.Z.), NYU School of Medicine, Anonymous Donors and Cure AD Fund (to S.A.L.). Schematics were created created with BioRender.com. We thank Jim Goldman for the O4 antibody, Davide Marotta and Anahita Sharma, for assistance with FACS analysis and organoid sectioning respectively, Daniel Paull and the NYSCF Global Stem Cell Array® team for iPSC line derivation, Edward Stopa, Terra D. Velilla, and the Brain Tissue Resource Center for providing primary human AD brain samples, Raeka Aiyar for manuscript editing, Frederick Monsma, Maria Sapar and all members of the lab for critical feedback.
The bulk and single cell RNA-sequencing data are available from the Synapse open source platform (https://www.synapse.org/#!Synapse:syn21861229).
Footnotes
DECLARATION OF INTERESTS
-SAL is an academic founder of AstronauTx Ltd.
-NYSCF. U.S. Patent Pending.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bais Abha S, and Kostka Dennis. 2019. “Scds: Computational Annotation of Doublets in Single Cell RNA Sequencing Data.” Preprint. Bioinformatics. 10.1101/564021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banker GA 1980. “Trophic Interactions between Astroglial Cells and Hippocampal Neurons in Culture.” Science (New York, N.Y.) 209 (4458): 809–10. 10.1126/science.7403847. [DOI] [PubMed] [Google Scholar]
- Bansal R, Warrington AE, Gard AL, Ranscht B, and Pfeiffer SE. 1989. “Multiple and Novel Specificities of Monoclonal Antibodies O1, O4, and R-MAb Used in the Analysis of Oligodendrocyte Development.” Journal of Neuroscience Research 24 (4): 548–57. 10.1002/jnr.490240413. [DOI] [PubMed] [Google Scholar]
- Bayraktar Omer Ali, Fuentealba Luis C., Alvarez-Buylla Arturo, and Rowitch David H.. 2014. “Astrocyte Development and Heterogeneity.” Cold Spring Harbor Perspectives in Biology 7 (1): a020362. 10.1101/cshperspect.a020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth Heather D. E., Hirst Warren D., and Wade-Martins Richard. 2017. “The Role of Astrocyte Dysfunction in Parkinson’s Disease Pathogenesis.” Trends in Neurosciences 40 (6): 358–70. 10.1016/j.tins.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley Robert A., Shireman Jack, McFalls Caya, Choi Jeea, Canfield Scott G., Dong Yi, Liu Katie, et al. 2019. “Regionally Specified Human Pluripotent Stem Cell-Derived Astrocytes Exhibit Different Molecular Signatures and Functional Properties.” Development (Cambridge, England) 146 (13). 10.1242/dev.170910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy John D., Emery Ben, Kaushal Amit, Foo Lynette C., Zamanian Jennifer L., Christopherson Karen S., Xing Yi, et al. 2008. “A Transcriptome Database for Astrocytes, Neurons, and Oligodendrocytes: A New Resource for Understanding Brain Development and Function.” The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 28 (1): 264–78. 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Zhiyou, Wan Cheng-Qun, and Liu Zhou. 2017. “Astrocyte and Alzheimer’s Disease.” Journal of Neurology 264 (10): 2068–74. 10.1007/s00415-017-8593-x. [DOI] [PubMed] [Google Scholar]
- Campanelli James T., Sandrock Robert W., Wheatley Will, Xue Haipeng, Zheng Jianhua, Liang Feng, Chesnut Jonathan D., Zhan Ming, Rao Mahendra S., and Liu Ying. 2008. “Expression Profiling of Human Glial Precursors.” BMC Developmental Biology 8 (October): 102. 10.1186/1471-213X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canals Isaac, Ginisty Aurélie, Quist Ella, Timmerman Raissa, Fritze Jonas, Miskinyte Giedre, Monni Emanuela, et al. 2018. “Rapid and Efficient Induction of Functional Astrocytes from Human Pluripotent Stem Cells.” Nature Methods 15 (9): 693–96. 10.1038/s41592-018-0103-2. [DOI] [PubMed] [Google Scholar]
- Crowell Helena L., Soneson Charlotte, Germain Pierre-Luc, Calini Daniela, Collin Ludovic, Raposo Catarina, Malhotra Dheeraj, and Robinson Mark D.. 2019. “On the Discovery of Population-Specific State Transitions from Multi-Sample Multi-Condition Single-Cell RNA Sequencing Data.” Preprint. Bioinformatics. 10.1101/713412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis Spyros, Sloan Steven A., Zhang Ye, Enge Martin, Caneda Christine, Shuer Lawrence M., Gephart Melanie G. Hayden, Barres Ben A., and Quake Stephen R.. 2015. “A Survey of Human Brain Transcriptome Diversity at the Single Cell Level.” Proceedings of the National Academy of Sciences of the United States of America 112 (23): 7285–90. 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin Alexander, Davis Carrie A., Schlesinger Felix, Drenkow Jorg, Zaleski Chris, Jha Sonali, Batut Philippe, Chaisson Mark, and Gingeras Thomas R.. 2013. “STAR: Ultrafast Universal RNA-Seq Aligner.” Bioinformatics (Oxford, England) 29 (1): 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- di Domenico Angelique, Carola Giulia, Calatayud Carles, Pons-Espinal Meritxell, Muñoz Juan Pablo, Richaud-Patin Yvonne, Fernandez-Carasa Irene, et al. 2019. “Patient-Specific IPSC-Derived Astrocytes Contribute to Non-Cell-Autonomous Neurodegeneration in Parkinson’s Disease.” Stem Cell Reports 12 (2): 213–29. 10.1016/j.stemcr.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras Panagiotis, and Fossati Valentina. 2015. “Generation and Isolation of Oligodendrocyte Progenitor Cells from Human Pluripotent Stem Cells.” Nature Protocols 10 (8): 1143–54. 10.1038/nprot.2015.075. [DOI] [PubMed] [Google Scholar]
- Douvaras Panagiotis, Wang Jing, Zimmer Matthew, Hanchuk Stephanie, O’Bara Melanie A., Sadiq Saud, Sim Fraser J., Goldman James, and Fossati Valentina. 2014. “Efficient Generation of Myelinating Oligodendrocytes from Primary Progressive Multiple Sclerosis Patients by Induced Pluripotent Stem Cells.” Stem Cell Reports 3 (2): 250–59. 10.1016/j.stemcr.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidsvaag Vigdis Andersen, Enger Rune, Hansson Hans-Arne, Eide Per Kristian, and Nagelhus Erlend A.. 2017. “Human and Mouse Cortical Astrocytes Differ in Aquaporin-4 Polarization toward Microvessels: EIDSVAAG et Al.” Glia 65 (6): 964–73. 10.1002/glia.23138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elitt Matthew S, Barbar Lilianne, and Tesar Paul J. 2018. “Drug Screening for Human Genetic Diseases Using IPSC Models.” Human Molecular Genetics 27 (R2): R89–98. 10.1093/hmg/ddy186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo LC 2013. “Purification of Rat and Mouse Astrocytes by Immunopanning.” Cold Spring Harbor Protocols 2013 (5): pdb.prot074211-pdb.prot074211. 10.1101/pdb.prot074211. [DOI] [PubMed] [Google Scholar]
- Foo Lynette C., Allen Nicola J., Bushong Eric A., Ventura P. Britten, Chung Won-Suk, Zhou Lu, Cahoy John D., et al. 2011. “Development of a Method for the Purification and Culture of Rodent Astrocytes.” Neuron 71 (5): 799–811. 10.1016/j.neuron.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorshkov Kirill, Aguisanda Francis, Thorne Natasha, and Zheng Wei. 2018. “Astrocytes as Targets for Drug Discovery.” Drug Discovery Today 23 (3): 673–80. 10.1016/j.drudis.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribkoff Valentin K., and Kaczmarek Leonard K.. 2017. “The Need for New Approaches in CNS Drug Discovery: Why Drugs Have Failed, and What Can Be Done to Improve Outcomes.” Neuropharmacology 120 (July): 11–19. 10.1016/j.neuropharm.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Xiaoning, Chen Michael, Wang Fushun, Windrem Martha, Wang Su, Shanz Steven, Xu Qiwu, et al. 2013. “Forebrain Engraftment by Human Glial Progenitor Cells Enhances Synaptic Plasticity and Learning in Adult Mice.” Cell Stem Cell 12 (3): 342–53. 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haston Kelly M., and Finkbeiner Steven. 2016. “Clinical Trials in a Dish: The Potential of Pluripotent Stem Cells to Develop Therapies for Neurodegenerative Diseases.” Annual Review of Pharmacology and Toxicology 56: 489–510. 10.1146/annurev-pharmtox-010715-103548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst Camilla Bjørnbak, Brøchner Christian Beltoft, Vitting-Seerup Kristoffer, and Møllgård Kjeld. 2019. “Astrogliogenesis in Human Fetal Brain: Complex Spatiotemporal Immunoreactivity Patterns of GFAP , S100, AQP 4 and YKL -40.” Journal of Anatomy 235 (3): 590–615. 10.1111/joa.12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Peng, Chen Chen, Wang Ruimin, Chechneva Olga V., Chung Seung-Hyuk, Rao Mahendra S., Pleasure David E., Liu Ying, Zhang Quanguang, and Deng Wenbin. 2013. “HESC-Derived Olig2+ Progenitors Generate a Subtype of Astroglia with Protective Effects against Ischaemic Brain Injury.” Nature Communications 4 (1). 10.1038/ncomms3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungblut Melanie, Tiveron Marie Catherine, Barral Serena, Abrahamsen Bjarke, Knöbel Sebastian, Pennartz Sandra, Schmitz Jürgen, et al. 2012. “Isolation and Characterization of Living Primary Astroglial Cells Using the New GLAST-Specific Monoclonal Antibody ACSA-1.” Glia 60 (6): 894–907. 10.1002/glia.22322. [DOI] [PubMed] [Google Scholar]
- Khakh Baljit S., Beaumont Vahri, Cachope Roger, Munoz-Sanjuan Ignacio, Goldman Steven A., and Grantyn Rosemarie. 2017. “Unravelling and Exploiting Astrocyte Dysfunction in Huntington’s Disease.” Trends in Neurosciences 40 (7): 422–37. 10.1016/j.tins.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh Baljit S., and North R. Alan. 2012. “Neuromodulation by Extracellular ATP and P2X Receptors in the CNS.” Neuron 76 (1): 51–69. 10.1016/j.neuron.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik Robert, and Zhang Su-Chun. 2011. “Directed Differentiation of Functional Astroglial Subtypes from Human Pluripotent Stem Cells.” Nature Protocols 6 (11): 1710–17. 10.1038/nprot.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster Madeline A., Renner Magdalena, Martin Carol-Anne, Wenzel Daniel, Bicknell Louise S., Hurles Matthew E., Homfray Tessa, Penninger Josef M., Jackson Andrew P., and Knoblich Juergen A.. 2013. “Cerebral Organoids Model Human Brain Development and Microcephaly.” Nature 501 (7467): 373–79. 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia Justin D., Gallagher Joseph, Heddleston John M., Wang Jialiang, Eyler Christine E., MacSwords Jennifer, Wu Qiulian, et al. 2010. “Integrin Alpha 6 Regulates Glioblastoma Stem Cells.” Cell Stem Cell 6 (5): 421–32. 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Yang, Smyth Gordon K., and Shi Wei. 2014. “FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features.” Bioinformatics (Oxford, England) 30 (7): 923–30. 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liddelow Shane A., and Barres Ben A.. 2017. “Reactive Astrocytes: Production, Function, and Therapeutic Potential.” Immunity 46 (6): 957–67. 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Liddelow Shane A., Guttenplan Kevin A., Clarke Laura E., Bennett Frederick C., Bohlen Christopher J., Schirmer Lucas, Bennett Mariko L., et al. 2017. “Neurotoxic Reactive Astrocytes Are Induced by Activated Microglia.” Nature 541 (7638): 481–87. 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Namba T, Liu J, Suzuki R, Shioda S, and Seki T. 2010. “Glial Fibrillary Acidic Protein-Expressing Neural Progenitors Give Rise to Immature Neurons via Early Intermediate Progenitors Expressing Both Glial Fibrillary Acidic Protein and Neuronal Markers in the Adult Hippocampus.” Neuroscience 166 (1): 241–51. 10.1016/j.neuroscience.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Long Justin M., and Holtzman David M.. 2019. “Alzheimer Disease: An Update on Pathobiology and Treatment Strategies.” Cell 179 (2): 312–39. 10.1016/j.cell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhavan Mayur, Nevin Zachary S., Shick H. Elizabeth, Garrison Eric, Clarkson-Paredes Cheryl, Karl Molly, Clayton Benjamin L. L., et al. 2018. “Induction of Myelinating Oligodendrocytes in Human Cortical Spheroids.” Nature Methods 15 (9): 700–706. 10.1038/s41592-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo Lior, Trauger Sunia A., Blain Manon, Nadeau Meghan, Patel Bonny, Alvarez Jorge I., Mascanfroni Ivan D., et al. 2014. “Regulation of Astrocyte Activation by Glycolipids Drives Chronic CNS Inflammation.” Nature Medicine 20 (10): 1147–56. 10.1038/nm.3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy Davis J., Campbell Kieran R., Lun Aaron T. L., and Wills Quin F.. 2017. “Scater: Pre-Processing, Quality Control, Normalization and Visualization of Single-Cell RNA-Seq Data in R.” Bioinformatics (Oxford, England) 33 (8): 1179–86. 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KD, and de Vellis J. 1980. “Preparation of Separate Astroglial and Oligodendroglial Cell Cultures from Rat Cerebral Tissue.” The Journal of Cell Biology 85 (3): 890–902. 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim Nancy Ann, Wang Xiaohai, Goldman Steven, and Nedergaard Maiken. 2006. “Astrocytic Complexity Distinguishes the Human Brain.” Trends in Neurosciences 29 (10): 547–53. 10.1016/j.tins.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Palm Thomas, Bolognin Silvia, Meiser Johannes, Nickels Sarah, Träger Claudia, Meilenbrock Ralf-Leslie, Brockhaus Johannes, Schreitmüller Miriam, Missler Markus, and Schwamborn Jens Christian. 2015. “Rapid and Robust Generation of Long-Term Self-Renewing Human Neural Stem Cells with the Ability to Generate Mature Astroglia.” Scientific Reports 5 (1). 10.1038/srep16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parr CJC, Yamanaka S, and Saito H. 2017. “An Update on Stem Cell Biology and Engineering for Brain Development.” Molecular Psychiatry 22 (6): 808–19. 10.1038/mp.2017.66. [DOI] [PubMed] [Google Scholar]
- Patro Rob, Duggal Geet, Love Michael I., Irizarry Rafael A., and Kingsford Carl. 2017. “Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression.” Nature Methods 14 (4): 417–19. 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull Daniel, Sevilla Ana, Zhou Hongyan, Hahn Aana Kim, Kim Hesed, Napolitano Christopher, Tsankov Alexander, et al. 2015. “Automated, High-Throughput Derivation, Characterization and Differentiation of Induced Pluripotent Stem Cells.” Nature Methods 12 (9): 885–92. 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- Perriot Sylvain, Mathias Amandine, Perriard Guillaume, Canales Mathieu, Jonkmans Nils, Merienne Nicolas, Meunier Cécile, et al. 2018. “Human Induced Pluripotent Stem Cell-Derived Astrocytes Are Differentially Activated by Multiple Sclerosis-Associated Cytokines.” Stem Cell Reports 11 (5): 1199–1210. https://doi.Org/10.1016/j.stemcr.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phatnani Hemali, and Maniatis Tom. 2015. “Astrocytes in Neurodegenerative Disease.” Cold Spring Harbor Perspectives in Biology 7 (6). 10.1101/cshperspect.a020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponath Gerald, Lincoln Matthew R., Levine-Ritterman Maya, Park Calvin, Dahlawi Somiah, Mubarak Mayyan, Sumida Tomokazu, et al. 2018. “Enhanced Astrocyte Responses Are Driven by a Genetic Risk Allele Associated with Multiple Sclerosis.” Nature Communications 9 (1): 5337. 10.1038/s41467-018-07785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Yuchen, Zhang Xin-Jun, Renier Nicolas, Wu Zhuhao, Atkin Talia, Sun Ziyi, Ozair M. Zeeshan, et al. 2017. “Combined Small-Molecule Inhibition Accelerates the Derivation of Functional Cortical Neurons from Human Pluripotent Stem Cells.” Nature Biotechnology 35 (2): 154–63. 10.1038/nbt.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato Giorgia, Nguyen Tuan, Macosko Evan Z., Sherwood John L., Yang Sung Min, Berger Daniel R., Maria Natalie, et al. 2017. “Cell Diversity and Network Dynamics in Photosensitive Human Brain Organoids.” Nature 545 (7652): 48–53. 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson Mark D., and Oshlack Alicia. 2010. “A Scaling Normalization Method for Differential Expression Analysis of RNA-Seq Data.” Genome Biology 11 (3): R25. 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessmann U, Velasco ME, Sindely SD, and Gambetti P. 1980. “Glial Fibrillary Acidic Protein (GFAP) in Ependymal Cells during Development. An Immunocytochemical Study.” Brain Research 200 (1): 13–21. 10.1016/0006-8993(80)91090-2. [DOI] [PubMed] [Google Scholar]
- Roybon Laurent, Lamas Nuno J., Garcia Alejandro D., Yang Eun Ju, Sattler Rita, Lewis Vernice J., Kim Yoon A., et al. 2013. “Human Stem Cell-Derived Spinal Cord Astrocytes with Defined Mature or Reactive Phenotypes.” Cell Reports 4 (5): 1035–48. 10.1016/j.celrep.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Renata, Vadodaria Krishna C., Jaeger Baptiste N., Mei Arianna, Lefcochilos-Fogelquist Sabrina, Mendes Ana P. D., Erikson Galina, et al. 2017. “Differentiation of Inflammation-Responsive Astrocytes from Glial Progenitors Generated from Human Induced Pluripotent Stem Cells.” Stem Cell Reports 8 (6): 1757–69. 10.1016/j.stemcr.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Yanhong, Inoue Haruhisa, Wu Joseph C., and Yamanaka Shinya. 2017. “Induced Pluripotent Stem Cell Technology: A Decade of Progress.” Nature Reviews. Drug Discovery 16 (2): 115–30. 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan Steven A., and Barres Ben A.. 2014. “Mechanisms of Astrocyte Development and Their Contributions to Neurodevelopmental Disorders.” Current Opinion in Neurobiology 27 (August): 75–81. 10.1016/j.conb.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan Steven A., Darmanis Spyros, Huber Nina, Khan Themasap A., Birey Fikri, Caneda Christine, Reimer Richard, Quake Stephen R., Barres Ben A., and Paşca Sergiu P.. 2017. “Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells.” Neuron 95 (4): 779–790.e6. 10.1016/j.neuron.2017.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew Michael V. 2014. “Multiple Roles for Astrocytes as Effectors of Cytokines and Inflammatory Mediators.” The Neuroscientist: A Review Journal Bringing Neurobiology, Neurology and Psychiatry 20 (2): 160–72. 10.1177/1073858413504466. [DOI] [PubMed] [Google Scholar]
- Sofroniew Michael V., and Vinters Harry V.. 2010. “Astrocytes: Biology and Pathology.” Acta Neuropathologica 119 (1): 7–35. 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan Rahul, Lu Tsai-Yi, Chai Hua, Xu Ji, Huang Ben S., Golshani Peyman, Coppola Giovanni, and Khakh Baljit S.. 2016. “New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo.” Neuron 92 (6): 1181–95. 10.1016/j.neuron.2016.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart Tim, Butler Andrew, Hoffman Paul, Hafemeister Christoph, Papalexi Efthymia, Mauck William M., Hao Yuhan, Stoeckius Marlon, Smibert Peter, and Satija Rahul. 2019. “Comprehensive Integration of Single-Cell Data.” Cell 177 (7): 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Daniel, and Jakobs Tatjana C.. 2012. “Structural Remodeling of Astrocytes in the Injured CNS.” The Neuroscientist: A Review Journal Bringing Neurobiology, Neurology and Psychiatry 18 (6): 567–88. 10.1177/1073858411423441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K 1997. “Epilepsy and Exacerbation of Brain Injury in Mice Lacking the Glutamate Transporter GLT-1.” Science 276 (5319): 1699–1702. 10.1126/science.276.5319.1699. [DOI] [PubMed] [Google Scholar]
- Tchieu Jason, Calder Elizabeth L., Guttikonda Sudha R., Gutzwiller Eveline M., Aromolaran Kelly A., Steinbeck Julius A., Goldstein Peter A., and Studer Lorenz. 2019. “NFIA Is a Gliogenic Switch Enabling Rapid Derivation of Functional Human Astrocytes from Pluripotent Stem Cells.” Nature Biotechnology 37 (3): 267–75. 10.1038/s41587-019-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcw Julia, Wang Minghui, Pimenova Anna A., Bowles Kathryn R., Hartley Brigham J., Lacin Emre, Machlovi Saima I., et al. 2017. “An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells.” Stem Cell Reports 9 (2): 600–614. 10.1016/j.stemcr.2017.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Yin M., Gupta Shailesh K., Kim Kevin J., Powers Berit E., Cerqueira Antonio, Wainger Brian J., Ngo Hien D., et al. 2013. “A Small Molecule Screen in Stem-Cell-Derived Motor Neurons Identifies a Kinase Inhibitor as a Candidate Therapeutic for ALS.” Cell Stem Cell 12 (6): 713–26. 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian Jennifer L., Xu Lijun, Foo Lynette C., Nouri Navid, Zhou Lu, Giffard Rona G., and Barres Ben A.. 2012. “Genomic Analysis of Reactive Astrogliosis.” The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 32 (18): 6391–6410. 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Ye, Sloan Steven A., Clarke Laura E., Caneda Christine, Plaza Colton A., Blumenthal Paul D., Vogel Hannes, et al. 2016. “Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse.” Neuron 89 (1): 37–53. 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Qiong, Viollet Coralie, Efthymiou Anastasia, Khayrullina Guzal, Moritz Kasey E., Wilkerson Matthew D., Sukumar Gauthaman, Dalgard Clifton L., and Doughty Martin L.. 2019. “Neuroinflammatory Astrocytes Generated from Cord Blood-Derived Human Induced Pluripotent Stem Cells.” Journal of Neuroinflammation 16 (1): 164. 10.1186/s12974-019-1553-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Cell counts and differentially expressed genes per scRNA-seq cluster (Related to Fig. 3, 4, 8, S4, S5, S6)
Data Availability Statement
The RNA sequencing datasets generated during this study are available in a user-friendly searchable online database (https://nyscfseq.appspot.com/).
Data are also available from the Synapse open source platform: https://www.synapse.org/#!Synapse:syn21861229.