ABSTRACT
Comparative genomics of bacterial pathogens has been useful for revealing potential virulence factors. Escherichia coli is a significant cause of human morbidity and mortality worldwide but can also exist as a commensal in the human gastrointestinal tract. With many sequenced genomes, it has served as a model organism for comparative genomic studies to understand the link between genetic content and potential for virulence. To date, however, no comprehensive analysis of its complete “virulome” has been performed for the purpose of identifying universal or pathotype-specific targets for vaccine development. Here, we describe the construction of a pathotype database of 107 well-characterized completely sequenced pathogenic and nonpathogenic E. coli strains, which we annotated for major virulence factors (VFs). The data are cross referenced for patterns against pathotype, phylogroup, and sequence type, and the results were verified against all 1,348 complete E. coli chromosomes in the NCBI RefSeq database. Our results demonstrate that phylogroup drives many of the “pathotype-associated” VFs, and ExPEC-associated VFs are found predominantly within the B2/D/F/G phylogenetic clade, suggesting that these phylogroups are better adapted to infect human hosts. Finally, we used this information to propose polyvalent vaccine targets with specificity toward extraintestinal strains, targeting key invasive strategies, including immune evasion (group 2 capsule), iron acquisition (FyuA, IutA, and Sit), adherence (SinH, Afa, Pap, Sfa, and Iha), and toxins (Usp, Sat, Vat, Cdt, Cnf1, and HlyA). While many of these targets have been proposed before, this work is the first to examine their pathotype and phylogroup distribution and how they may be targeted together to prevent disease.
KEYWORDS: Escherichia coli, vaccine development, comparative genomics, pathogenomics, ExPEC, InPEC, enteric pathogens, genomics, pathogenesis, vaccines, virulence factors
INTRODUCTION
Escherichia coli has long been a significant cause of human morbidity and mortality. It is a common inhabitant of the gastrointestinal tract, readily exchanges elements associated with antibiotic resistance, and has an extensive pangenome of at least 13,000 genes (and possibly over 100,000) which include numerous virulence factors (VFs) that complicate an understanding of disease etiology (1–12). Currently, there is no vaccine available, and attempts to generate one have been limited by the lack of universal protection against all invasive strains or types of infection this organism is capable of causing (13–15). Unlike the causative agent of diphtheria and pertussis, which are largely viewed as toxigenic diseases caused by secreted toxins from Corynebacterium diph theriae and Bordetella pertussis, respectively, E. coli infections largely resist being pigeon-holed by a single VF, instead being multifactorial—a composite of virulence factors contributing to each step in the diverse range of diseases this organism causes (11, 16–20). This and the fact that the organism borders on being an opportunistic pathogen—with commensal strains often apparently possessing the same virulence factors—makes the development of vaccines challenging (21). Along these lines, strains are organized into so-called pathological types or “pathotypes” (also known as pathovars). Pathotypes are groups of pathogenic strains that share the same phenotype of disease. In a broad sense, these can be broken up into two types based on whether the strain causes disease outside (extraintestinal pathogenic E. coli [ExPEC]) or inside (intestinal pathogenic E. coli [InPEC]) the intestines (12, 22). These two pathotypes are broken down further based on the specifics of the disease they cause. For example, neonatal meningitis E. coli (NMEC) and uropathogenic E. coli (UPEC) are considered ExPECs, while enteroaggregative E. coli (EAEC) and enterohemorrhagic E. coli (EHEC) are InPECs. A full list of the pathotypes discussed here, along with their acronyms and characteristics, can be found in Table 1.
TABLE 1.
E. coli pathotypes, acronyms, disease presentations, and associated VFs
| Pathotype | Acronym | Disease presentation(s) | Associated VFs (references) |
|---|---|---|---|
| Extraintestinal Pathogenic E. coli | ExPEC | Disease outside the intestines | See below |
| Uropathogenic E. coli | UPEC | Urinary tract infections (23, 24) | fim, hlyA, csg, pap, sfa, afa, cdtAB, iha, iutA, iroN, fyuA, sitA, chuA, hma, kpsMT, agn43/flu, pic, sat, vat, usp (12, 24, 25) |
| Neonatal Meningitis E. coli | NMEC | Bacterial meningitis | fim, sfa, mat, ibeA, irp, iroN, kpsMT, K1 (25) |
| Avian Pathogenic E. coli | APEC | Multiple ExPEC diseases in avian species (26–28) | Colicin, ibeA, iutA, iroN, sitA, tsh, fim, fyuA, pap, vat (12, 25, 28) |
| Intestinal Pathogenic E. coli | InPEC | Disease of the intestines | See below |
| Adherent-Invasive E. coli | AIEC | Associated with intestinal inflammation (29, 30) | fimH, lpf, pap, sfa, afa, vat, hlyA, cnf I, cdtAB, ibeA (31–33) |
| Enterohemorrhagic E. coli | EHEC | Bloody diarrhea, hemorrhagic colitis, hemolytic-uremic syndrome (32) | LEE pathogenicity island; stx, espP, lpf, efa, toxB, eibG, ehaA, ompA, iha, paa (32, 34) |
| Enteroaggregative E. coli | EAEC | Acute and chronic watery diarrhea (32, 35) | set, agg, aaf, agg3, astA, pet, sat, aap, aagR, shf, pic, irp2, hly, tia (32, 36) |
| Enteroaggregative Hemorrhagic E. coli | EAHEC | Similar to EHEC, with increased adherence and antibiotic resistance (32, 37) | iha, pic, pet, stx (32) |
| Enterotoxigenic E. coli | ETEC | Mild to severe watery diarrhea (32) | CFA fimbriae, astA, eltAB, estIa, clyA, eatA (32, 38) |
| Enteropathogenic E. coli | EPEC | Severe acute watery diarrhea (32, 39, 40) | LEE pathogenicity island, set, paa, lpf, iha, ehaA (32, 41) |
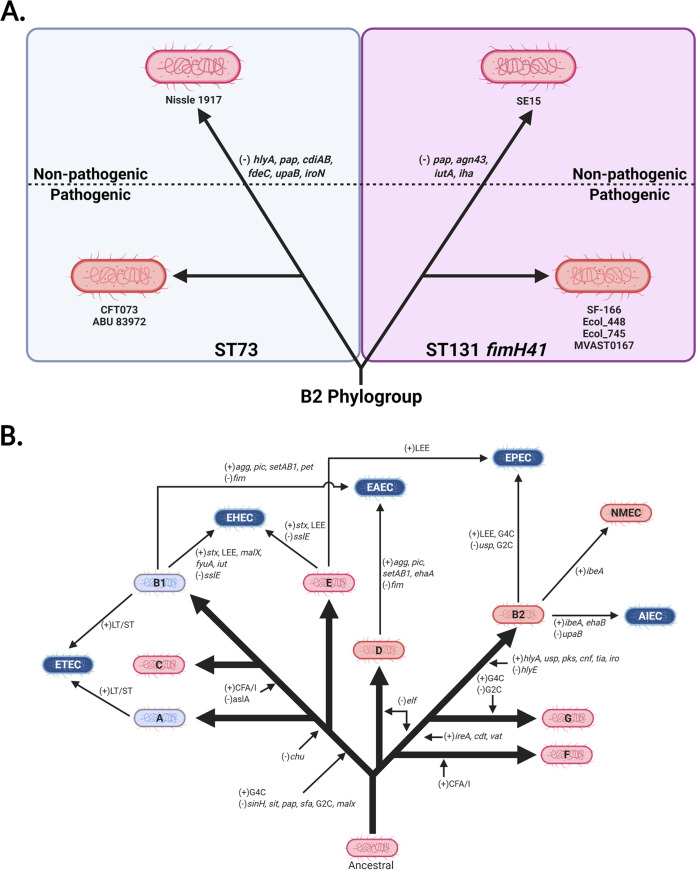
Strains are also characterized by the phylogenetic group (phylogroup) to which they belong. In contrast to pathotype, which are based largely on disease phenotype, phylogroup assignments are based on genetic lineage. There are four major (A, B1, B2, and D) and five minor (C, E, F, G, and cryptic clade I) phylogroups in E. coli (42). The four major phylogroups were first observed by multilocus enzyme electrophoresis and subsequently found to be resolved by grouping core E. coli genomes (43, 44). One main, if underappreciated, difference between phylogroups is the presence of the chu system responsible for heme importation. This system is found in phylogroup B2, D, E, F, and G strains but is absent in phylogroup A, B1, and C strains. Iron is an essential, possibly growth rate-limiting, nutrient that is found predominantly in heme of red blood cells in mammalian hosts (45–47). Phylogroups A and B1 appear to be the source of most human commensals (44, 48). Strains of the B1 phylogroup also may be more prevalent in domesticated animal isolates (44). In contrast to the A and B1 phylogroups, strains from the B2 and D phylogroups are more commonly found in ExPEC strains (48–50). Less is known about the minor phylogroups, but ST23 and ST88 of phylogroup C are often found to be avian pathogenic E. coli (APEC) strains (51–53), while phylogroup E contains most of the known EHEC isolates, such as the well-known O157:H7 serotype. Phylogroup F is very similar to phylogroup B2, but with the exception of certain sequence types, they generally lack many of the ExPEC-associated virulence factors (11, 54). Lastly, phylogroup G has only recently been resolved, and little is known about it other than being found between phylogroups F and B2 phylogenetically (55).
Here, we use a comparative genomics approach to comprehensively organize every major virulence factor in E. coli into a holistic picture that allows comparison across pathotype, phylogroup, and VF category. This organization reveals vaccine targets that speciate by phylogroup specifically but also highlight some unexpected entry points that may disarm this pathogen from multiple angles. Many of the vaccine targets proposed here have been proposed before as monovalent targets. However, to our knowledge, there has been no work looking at their pathotype and phylogroup distribution or how this could be leveraged to identify novel vaccine targets or polyvalent strategies. Furthermore, we reveal here several novel findings that provide insight into E. coli evolution, pathogen-versus-commensal delineation, and diagnostic classification.
RESULTS AND DISCUSSION
Curation of the E. coli virulome and visualization of results.
The first step in our analysis was to curate a database of E. coli virulence factors by known strains. This included retrieving known virulence factors from the Virulence Factor Database (VFDB), VICTORs, and PATRIC, cross-referencing and confirming their function from hundreds of literature sources, and using a tiered approach to analyze strains (56–58). On the first tier, we used a strain database of 107 strains which had complete chromosome sequences (sequences of high quality) and had published evidence for their pathotype assignment. Two incomplete strains were grandfathered in from preliminary analyses (NC101 and REL606), but these nonetheless had published pathotype evidence. These strains were organized by their pathotype and visualized in detail. This is referred to here as our “pathotype database.” The next tier database contained 1,348 complete E. coli chromosomes that were organized into phylogroups using an in silico method based on Clermont phylotyping developed in-house (see Materials and Methods; see also Appendix S1 in the supplemental material) (59–61). This is referred to as our “phylogroup database.” Genetic insights gleaned from the first tier (pathotype database) were tested against this larger phylogroup database, mainly in the form of gene distribution. Using both methods, we found many apparently novel associations between virulence factors, phylogroups, and pathotypes. Figure S1 details the pipeline for separation of the E. coli genomes into phylogroups.
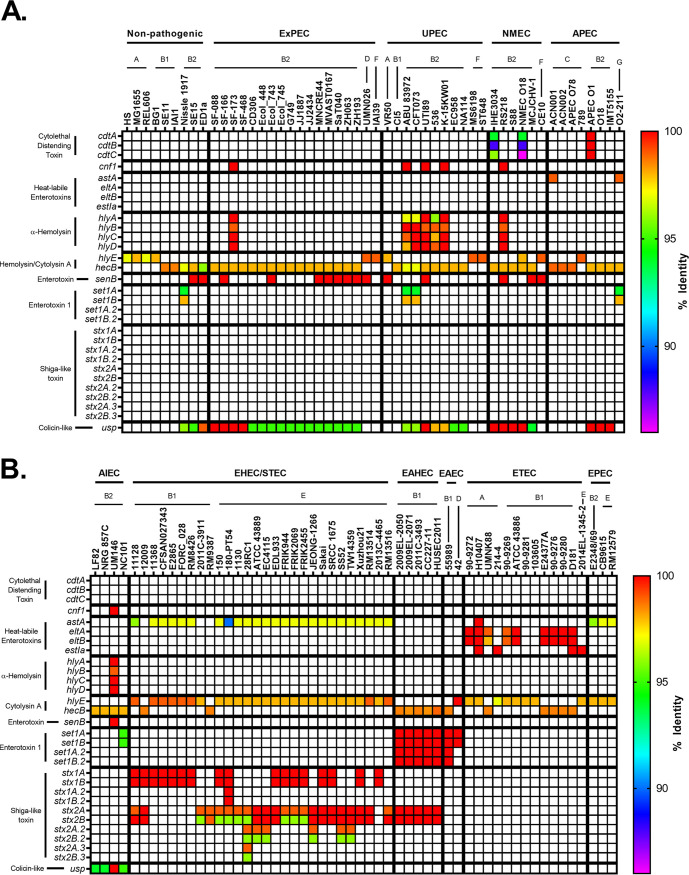
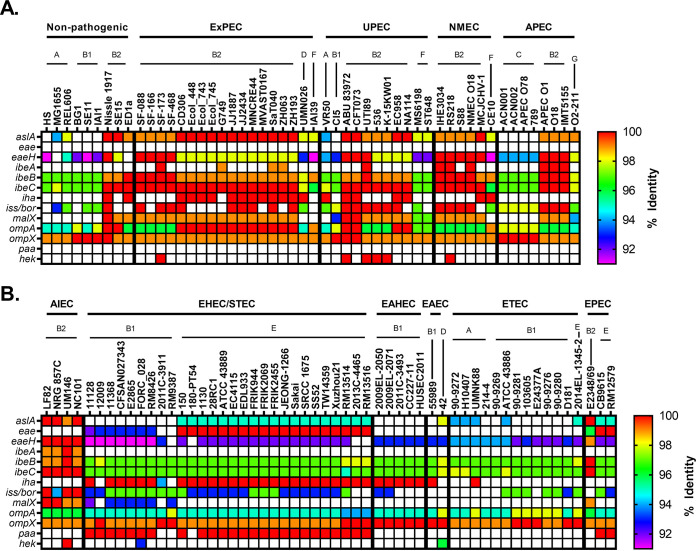
To visualize the relationship between pathotype, phylogroup, type of virulence factor, and any polymorphisms in genes associated with virulence factors, a heatmap template was developed. These heatmaps are divided into two panels: nonpathogenic strains and ExPECs (general ExPECs, UPECs, NMECs, and APECs) (see, for example, Fig. 1A) and InPECs (AIECs, EHECs/STECs, EAHECs, EAECs, ETECs, and EPECs [Table 1]) (see, for example, Fig. 1B). Each column represents a single strain that is listed at the top of the heatmap. These strains are organized first by pathotype, then by phylogroup, and finally by sequence type. Rows represent a single gene, which is listed to the left of the heatmap. Genes are generally organized by class, operon, or otherwise related function.
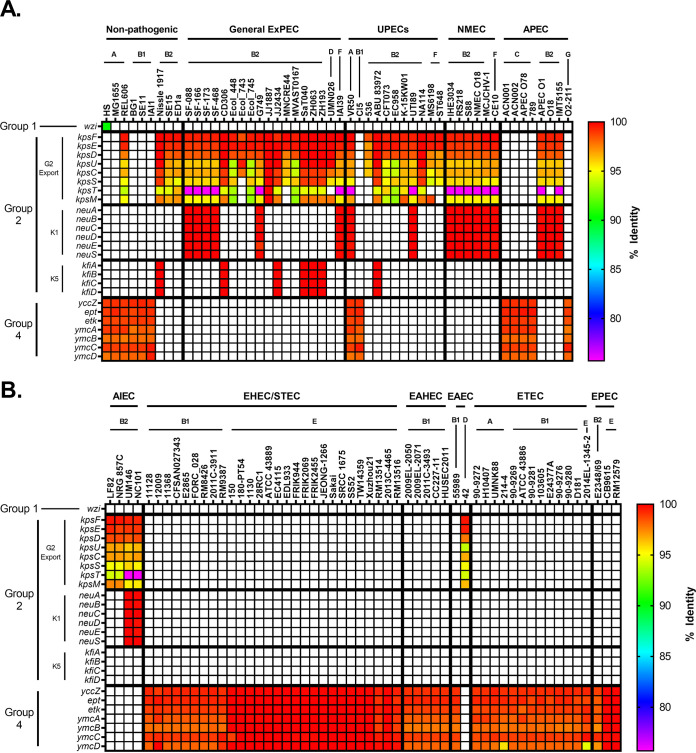
FIG 1.
Pathotype distribution of E. coli capsule. (A and B) Heatmap showing nonpathogenic E. coli and ExPECs (A) and InPECs (B). Columns are organized first by pathotype, then by phylogroup, and finally by sequence type (sequence type not shown). The wzi gene used as a reference is specific for G1C. G2C and G3C both use the kpsFEDUCS and kpsTM operons for export. The biosynthetic operons for the most widely distributed and studied K-types, K1 (neu) and K5 (kfi), are also shown. G4C is synthesized by yccZ, ept, etk, and ymcABCD genes. The percent identity was determined using megaBLAST with reference genes found in Data Set S1 in the supplemental material.
Each cell in the heatmap is colored based on percent nucleotide identity compared to reference used to generate the alignments. The range of colors for each figure are based on the lowest value—which indicates divergence from the reference gene—and so varies from figure to figure. Coloring cells this way allows the user to use color as a proxy for both conservation and allelic distribution, and our organization allows the reader to investigate this distribution by pathotype, phylogroup, or sequence type. It is important to note that matching colors do not necessarily mean identical alleles. Instead, it indicates that the gene in those strains have the same number of mutations relative to the reference gene. However, we did not come across a case where matching colors were from separate alleles.
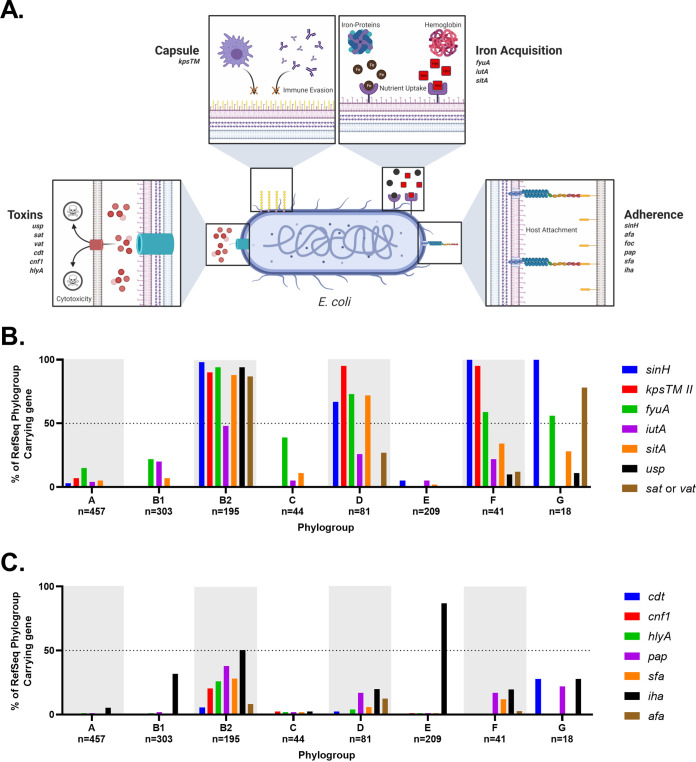
K-capsule group types are strongly associated with phylogroup.
Surface sugars are an important virulence factor in many pathogenic bacteria as they generally act as protectins, hiding the bacterium from the host’s immune system (62). Three U.S. Food and Drug Administration-approved vaccines (for N. meningitidis, H. influenzae, and S. pneumoniae) have capsule as their component and thus may guide E. coli vaccine development since E. coli also produces capsule. In E. coli there are three major types of surface polysaccharides: lipopolysaccharide (LPS), capsular polysaccharide (CPS or K-antigen), and exopolysaccharide (EPS) (63–66). Of these, LPS and CPS are serospecific surface polysaccharides (64, 66). E. coli K-antigens are characterized into groups 1, 2, 3, and 4 (64). K-antigens that belong to groups 1 (G1C) and 4 (G4C) are related to LPS O-antigens and use similar biosynthetic machinery. G1C and G4C are found in intestinal pathogenic E. coli, including EPECs, ETECs, and EHECs (64). Group 2 capsule (G2C) and 3 (G3C) are found in extraintestinal pathogenic E. coli and are the group of interest here (64). K-antigens from these groups utilize a separate assembly and transport system (the kps operon) to those of the group 1 and 4 K-types. The structure of G2C and G3C are similar to those found in N. meningitidis and H. influenzae bacteria, which strengthens the case for using them as vaccine targets. G2C often appear similar to polysaccharides found on the surface of eukaryotic cells, and these include two of the most extensively studied K-antigens, K1 and K5 (64).
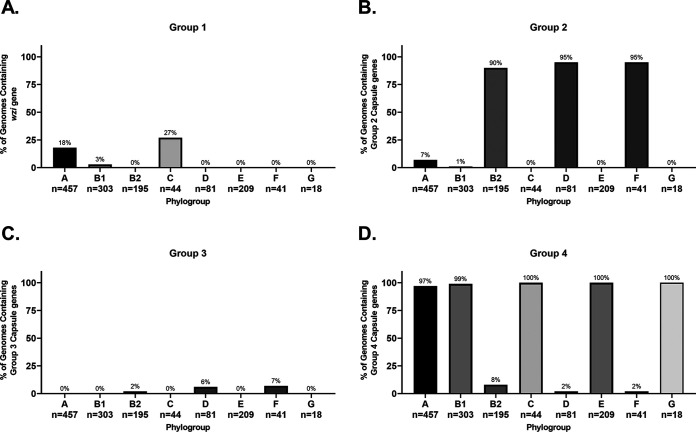
G1C was found only in one of the strains examined in our pathotype database, the commensal strain HS (nonpathogenic; A) (Fig. 1). G3C was found in none of the strains. While initially puzzling, the G1C and G3C distribution in our RefSeq phylogroup databases indicate that G1C and G3C are not widespread and found predominantly in phylogroups C and D, respectively. These two phylogroups are underrepresented in our pathotype database (Fig. 1). Given the rarity of G1C and G3C in both databases, they are not recommended candidates for vaccine development.
G2C and G4C were more common, being found mostly in ExPECs and InPECs of our pathotype database, respectively (Fig. 1). These results suggested that G2C are by far the most common capsule type found in ExPECs and are almost completely absent from InPECs, with the exception of E. coli 042 (EAEC), a member of the D phylogroup. This trend agreed with the literature that G2C are virulence factors of ExPECs, and it further strengthens the notion that capsular polysaccharide from group 2 should be considered a vaccine target. The G2C distribution in our RefSeq phylogroup database showed that G2C is associated with the B2, D, and F phylogroups, rather than with the ExPEC pathotype, which explains G2C in the 042 (EAEC; D) strain (Fig. 1B). However, this strong phylogroup association does not exclude a pathotype-based selection. Instead, this may partly explain why ExPEC strains tend to be from these three phylogroups. This is highlighted by exceptions to the rule in strain VR50, an asymptomatic bacteriuria (ABU) strain of the A phylogroup, which contains both G2C and G4C, and the E2348/69 strain, which is an EPEC strain and the only member of the B2 phylogroup in our pathotype database that lacks G2C and contains G4C capsule (Fig. 1). In our phylogroup database, only 8% (15/195) of B2 strains carried the genes to produce G4C (Fig. 2). Of these, 87% (13/15) also carried the locus of enterocyte effacement (LEE) which is a pathogenicity island associated with diarrheal E. coli (namely, EHECs and EPECs) (67). Of B2 strains not carrying genes to encode the G4C, none of these 180 strains carry the LEE pathogenicity island. This, as well as the overall pathotype distribution of G2C and G4C, suggests that G4C may be important for survival within the intestines, while G2C is important for survival in other parts of the host. Thus, it would seem that targeting G2C capsule might be protective against strains that disseminate from the gastrointestinal tract into extraintestinal tissues. This has interesting implications since the phylogroup distribution of these capsule groups suggests that the B2, D, and F phylogroups are adapted to infections outside the intestines and opens up the range of potential vaccine targets from known virulence factors and surface antigens to those that are associated specifically with these phylogroups.
FIG 2.
Phylogroup distribution of E. coli capsule. megaBLAST was used to bin strains of each phylogroup based on hit versus no hit. (A) Distribution of wzi from strain HS, which is specific for G1C. (B) Distribution of kpsMTII. (C) Distribution of kpsMIII (using AAC38078.1 as a reference). (D) Distribution of ymcDCBA, yccZ, ept, and etk, which are specific for G4C. Distributions were determined using megaBLAST to bin strains from each phylogroup into hit versus no hit.
Interestingly, all AIEC strains, which are found in the intestines, also carried G2C (Fig. 1B) (68). This appears to be because the AIEC strains in this data set are from the B2 phylogroup; however, the majority of AIEC strains isolated thus far belong to the B2 phylogroup, so that does not necessarily mean our strains are not representative. This deviation from the phylogroups of other InPECs may be explained by the association of AIECs with inflammatory bowel disease. In such a dysregulated and immune-factor-heavy environment, a capsule type that is associated with avoidance of the immune system provides obvious benefits. This dovetails with our findings above: if G2C acts as a protectin against the immune system outside the intestines and G2C is predominantly found in B2 strains, this could explain why most AIEC strains belong to the B2 phylogroup—they simply are more resistant to the immune system and possibly survive better intracellularly (69–72).
Lastly, another interesting finding is that strains from the recently described phylogroup G, which is a sister phylogroup to the B2 phylogroup, apparently carry G4C (Fig. 2) (55). This may suggest that phylogroup G, like B2 EPEC strains, diverged from phylogroups B2, D, and F and is more adapted to an intestinal niche. This also has interesting implications for the evolution and acquisition of capsular genes, since it suggests that these divergent G and B2 strains may have exchanged G2C for G4C, which has been an important step in their evolutionary development.
kpsT is predictive for group 2 capsule K-types.
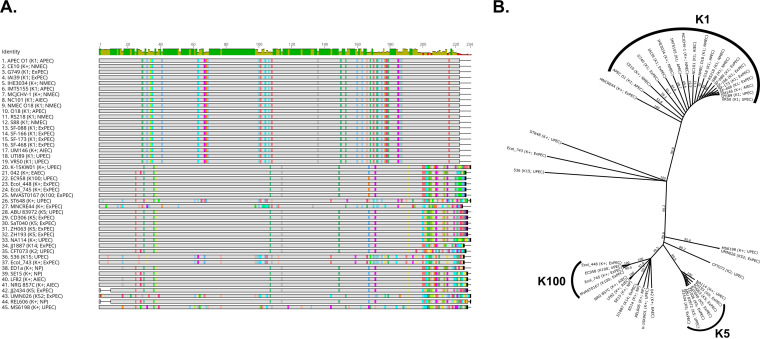
One of the most exciting things about the work presented here is the novel patterns that emerge from this layout. A good example of this was a striking result from the capsule comparisons where there was an apparent 1:1 association of the K-type with specific alleles of kpsT, a gene which encodes a ATP-binding protein member of the KpsTM ABC transport complex (Fig. 1; see also Fig. S2 in the supplemental material). While other genes in the kps operon showed either significant differences within each K-type or high conservation across K-types, kpsT showed a high degree of difference between K-types, but little-to-no change within a given K-type (Fig. 3; see also Fig. S3 in the supplemental material). This can clearly be seen in the amino acid alignment of all the KpsT sequences in our pathotype database (Fig. 3A). To test whether this trend holds up in a larger data set, we took advantage of the fact that the biosynthetic operons for two well-known G2C K-types are known: neu for K1 and kfi for K5. We then used megaBLAST to search our phylogroup database for 100% identical matches for the kpsT allele found in strains known to have K1 (kpsTK1) or K5 (kpsTK5) and cross referenced them against strains that contained the neu or kfi operons, respectively. Overall, our results showed that 89% (58/65) of all strains carrying the neu operon also contain kpsTK1, while 94% of all strains carrying the kfi operon contain kpsTK5. However, if we lower the stringency and include kpsT alleles that were >99% identical (<5 single nucleotide polymorphisms) to kpsTK1 or kpsTK5, 99% (64/65) of strains carrying the neu operon carry a kpsTK1-like allele, and 100% (50/50) of strains carrying the kfi operon carry a kpsTK5-like allele. These results show that kpsT is tightly linked to K-type and suggest that it may interact directly with variable regions of the exported polysaccharide. If true, it is likely that this kpsT-to-K-type association will hold for other K-types. Due to its location inside the cell, KpsT is unlikely to be a candidate antigen for vaccine consideration (see Fig. S2). However, these results do have important implications outside vaccine development. For one, K-typing can be achieved by simply sequencing this gene, thereby allowing for K-types to be reported alongside phylogroup and virulence factors in clinical studies. This will be especially important for rarer K-types. It could also allow for rapid bacteriophage therapeutic selection, since several types of phages specifically target capsule (73, 74). Finally, our results suggest that KpsT could be an attractive drug target, since disruption of this gene should be sufficient to prevent capsule export, thus targeting can be focused on K-types that are associated with extraintestinal pathogenesis while sparing commensals.
FIG 3.
Alignment of KpsT shows strong association between kpsT allele and K-antigen. (A) Geneious alignment of the amino acid sequence of KpsT. Sequences were sorted by differences to compared to K1 strain KpsT. An identity histogram is shown at the top, and colors represent amino acid differences from the majority consensus. (B) Unrooted phylogenetic tree built from using Geneious TreeMaker and the alignment shown in panel A, with bootstrap support with 1,000 replicates. Branch labels indicate percent consensus support. Branches were transformed proportionally to allow for clearer comparison.
Fimbriae and adhesins are associated with phylogroup rather than pathotype.
Fimbriae and adhesins are proteins found on the surface of all E. coli. Many are thought to be associated with adhesion to certain molecules, environments, or cell types (75). They are highly immunogenic and, if their distribution and mechanisms can be understood, may make excellent vaccine targets (76). Our results showed that fimbriae tend to be associated with phylogroup rather than pathotype (Fig. 4).
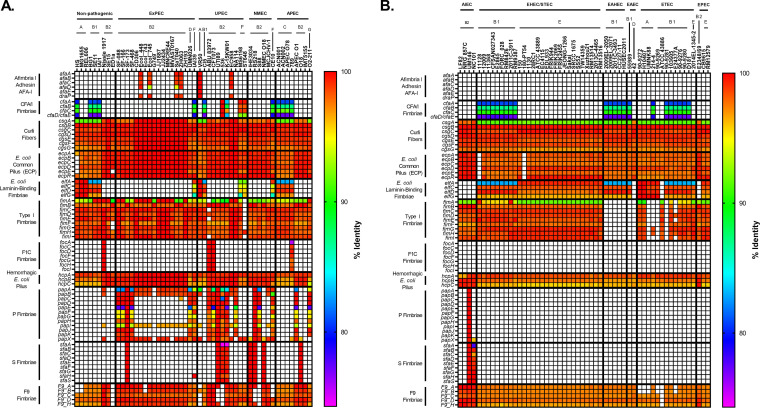
FIG 4.
Pathotype distribution of adherence fimbriae. (A) megaBLAST percent identity results for nonpathogenic and ExPEC strains. (B) megaBLAST percent identity results for InPEC strains. The percent identity was determined using megaBLAST with the reference genes in Data Set S1 in the supplemental material.
Type I fimbriae are encoded by the fim operon, target α-mannose, and are known to be in both commensals and pathogenic E. coli (77–81). It has been reported to bind several other molecules and cell types, including collagen, fibronectin, laminin, and macrophages (82–85). AIEC adherence to intestinal epithelial cells of Crohn’s disease (CD) patients is dependent on type I fimbriae, apparently because of the overexpression of CEACAM6 in CD patients (86). These fimbriae have been shown to promote urinary tract colonization and persistence, as well as cellular invasion (87, 88). However, there are some conflicting results where the ABU strain 83972 (which lacks the complete fim operon) did not promote adherence in a murine urinary tract when fim was complemented (89). Our results show that type I fimbriae are indeed widely distributed and well conserved in nonpathogenic, ExPEC, and InPEC strains (Fig. 4). Surprisingly, EAEC and EAHEC appear to completely lack the fim operon, and this absence does not correlate with phylogroup, sequence type, or serotype (Fig. 4B). This is despite the fact that the fim operon of prototypical EAEC 042 strain has been studied (90). It is possible that the F9 or sfm fimbriae that are found in 042 were mistaken for the type I fimbriae given their similarity and the fact that they are often mistakenly annotated as fim. In each case for EAEC and EAHEC, the only hit found for the fim locus is a partial 171 bp hit for fimH. This partial hit is found in the correct location on the chromosome (between the gntP and nanC genes), but it is interrupted by an IS1-family transposable element. This supports the notion that the B1 EAEC and EAHEC strains studied here may share an ancestor. Interestingly, of the eight non-EAEC/EAHEC strains lacking the full fimH operon, this partial 171-bp hit and transposable element was also found in the ST648 (ExPEC; F phylogroup) strain and 90-9281 (ETEC; B1 phylogroup). It should be noted that there are two E. coli 042 strains found in the NCBI database. The one analyzed here has a 5,241,977-bp chromosome with accession number FN554766 (RefSeq NC_017626) and has had its analysis as an EAEC strain published (91). The other has a 4,692,707 bp chromosome with accession number CP042934. This entry does not have an accompanying article, and the title gives no indication that it is an EAEC or EAHEC strain.
The afimbrial adhesin Afa is encoded by the afa genes and is generally associated with ExPEC, UPEC, and diffusely adhering E. coli (DAEC) strains (81, 92). The vast majority of strains containing afa genes belonged to the ST131 clonal group (Fig. 4). The full set of genes to make this adhesin protein were only found in the VR50 strain (A; ABU/UPEC), which is an asymptomatic bacteriuria strain (Fig. 4A) (93). But this is because there are multiple divergent alleles of afaE, and a single reference (afaE-I was used here) will not cover them all using our method (94). The limited distribution of afa genes make it a poor vaccine target by itself, but combining it with other antigens in a polyvalent vaccine is a viable option.
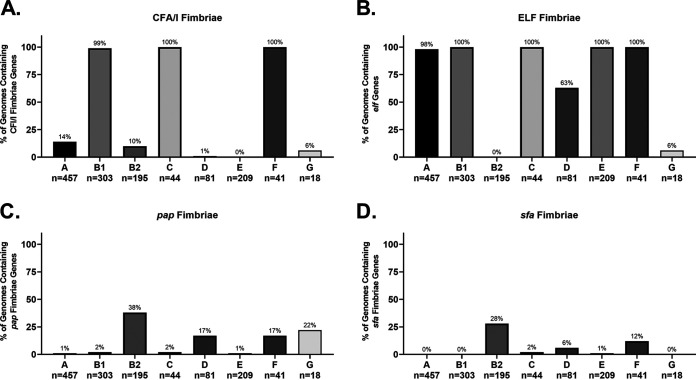
Colonization factor antigen I (CFA/I) fimbria is a class 5 fimbria generally considered associated with human colonization of ETEC strains (95). The CFA/I fimbriae are found in ETEC strains are encoded by a plasmid, but there is apparently a divergent form (∼30% identical to the ETEC plasmid version) on the chromosome that was widely found in strains belonging to the B1, C, and F phylogroups (Fig. 4 and 5). This trend is verified by our phylogroup database, where 100% of B1, C, and F strains contain hits for CFA/I (Fig. 5A). The most surprising result here is that all strains from the F phylogroup in both our databases contained this type of fimbria because the F phylogroup is more closely related to B2 strains (Fig. 4 and 5). This finding may be a clue to the lifestyle of the understudied F phylogroup. Interestingly, the only B2 strains that contained CFA/I were those from ST127 (Fig. 4A). Given that B1 strains are most often found as commensals in domesticated animals, it is possible that this fimbria promotes colonization in nonhuman hosts. Our finding also suggests that CFA/I as a vaccine target against ETECs will need to be carefully studied, since there is a possibility of cross-reactivity with nonpathogenic strains.
FIG 5.
Phylogroup distribution of select fimbriae. (A) Distribution of CFA/I fimbriae, which has until now been associated with ETEC strains. (B) Distribution of the ELF fimbrial genes. (C and D) Distributions of UPEC-associated P and S fimbriae, respectively. Note that only papCDEFH genes were used to differentiate strains that contained a full array of pap genes and those that contain a disrupted pap operon. Distributions were determined using megaBLAST to bin strains from each phylogroup into hit versus no hit.
E. coli laminin-binding fimbriae (ELF) were first described in EDL933, an O157:H7 EHEC/STEC strain, where is was found to contribute to the ability to bind Hep-2 cells (96). That group also found that antibodies against ELF were able to partially block adherence of EDL933. Our results suggested that ELF is more generally found among strains that do not belong to the B2 or D phylogroup (Fig. 4 and 5). In fact, it appears that no strain from the B2 phylogroup (0/39; 0%) in either our pathotype database or phylogroup database (0/195; 0%) harbor ELF on their chromosome, but it is found in all other phylogroups in our pathotype database (Fig. 4 and 5). It was also found in more than 97% of strains from phylogroups A, B1, C, E, and, surprisingly, F in our phylogroup database (Fig. 5B). It is interesting that, like CFA/I fimbriae, ELF is found in a phylogroup F that is more closely related to phylogroups B2 and D, which are generally considered to be more like ExPECs. This all makes ELF unlikely to be a viable vaccine target for either ExPECs or InPECs, but again may offer clues to the niche of phylogroup F strains.
Fimbriae of serotype 1C (F1C) is encoded by the focA, focC, focD, focF, focG, focH, and focI genes (97). These fimbriae are associated with uropathogenic strains and selectively bind to glycosphingolipids found on bladder, urethra, and kidney cells (98–101). It has also been shown to play a role in intestinal colonization of Nissle 1917 (nonpathogenic) (102). Despite the association with uropathogenicity, our search located true F1C fimbria hits in only three strains—Nissle 1917 (nonpathogenic), ABU 83972 (UPEC), and CFT073 (UPEC)—but that several cross hits with sfa genes did occur. The three strains with F1C were all members of the ST73 sequence type, which may indicate that this F1C is specific to certain sequence types.
F1C fimbriae are closely related to S-fimbrial adhesins, which are encoded by sfa genes and found on PAI III536 (103). However, they have distinct receptors, with S-fimbria-binding sialyl galactosides (104, 105). This matches our results where the foc reference genes from CFT073 (UPEC) showed significant homology for sfa reference genes from the UTI89 (UPEC) strain. The major differences were found in the major subunits focA and sfaA, which only shared 75.7% identity, and the adhesins focH and sfaH, which shared 84.8% identity. The other genes of each loci could be considered alleles of the same gene: the first minor subunits (focI and sfaD) shared 98.7% identity, the second minor subunits (focF and sfaG) shared 99.2% identity, the periplasmic chaperones (focC and sfeE) shared 98.7% identity, and the outer membrane ushers (focD and sfaF) shared 99.6% identity. The second gene downstream of the sfa locus is a previously uncharacterized bona fide papX regulator (100% coverage, 96% identical to reference papX). The sfa locus also contains two other regulatory genes that are like pap genes: sfaB, which shares high similarity with papB, and sfaC, which shares high similarity with papI. This all suggests a connected evolutionary history between these three fimbriae and may link them with uropathogenicity. On a practical level, this relatedness made it difficult to determine whether to score hits as sfa or foc. In the present study, we considered the hits to be hits for the F1C fimbriae if a full-length focG was present and S fimbriae if sfaS was present. In general, this method agreed with deciding which fimbriae were present by determining whether the adhesin was a better hit for focH or sfaH. However, strain 789 (APEC) presents an interesting problem for either way: this strain contains what appears to be a focH adhesin in an operon that is predominantly made up of S-fimbrial genes (>99% identical), with the exception of the minor subunit sfaS, which diverged and only had a minor hit (Fig. 4A). This may be an uncharacterized hybrid F1C/S fimbria. For strain 789, hits were included under both the F1C and S fimbria subsection to highlight the relatedness. The divergences between the traditional targets of fimbria vaccines, major subunits focA and sfaA and adhesins focH and sfaH, signifies a high probability that F1C and S fimbriae could only be targeted together with a polyvalent vaccine. However, the relatedness of the minor subunits means that they should be explored as potential targets.
P fimbriae are encoded by pap (pyelonephritis-associated pili) genes and are associated with uropathogenic strains and target glycosphingolipids (101, 106). It is part of both PAI ICFT073 and PAI IICFT073 pathogenicity islands (107). Some studies have found that P fimbriae are associated with 90% of acute pyelonephritis but less than 20% of ABU strains (80). Many ExPEC and NMEC strains contained only a few of the genes required for P-fimbria production, including the major repeating subunit, papA (Fig. 4). In some cases, such as in strains belonging to the ST131 group, full true hits for papA, papB, papI, and papX were found with nearby mobile elements that may explain the absence of the rest of the loci. In others, there were hits for papB and papI that appeared to be cross-hits with some S-fimbria genes (sfaB and sfaC, respectively). There were papX hits returned in any strain with the P or S fimbriae due to the papX homologue found near the sfa operon and in fact was found in over 70% of B2, D, and F strains in our phylogroup database, indicating that it may be ancestral to this lineage. It is important to note that it is possible that the bias toward certain sequence types (i.e., ST131 and ST95) in our pathotype database may skew these percentages, but the papB/sfaB hit does cover the majority of sequence types found in the ExPEC and NMEC category (Fig. 4). Interestingly, the only ST131 with a full complement of pap genes was NA114, a UPEC strain isolated in India (Fig. 4) (108). On the other hand, 70% (7/10) of ST95 strains, 67% (2/3) of ST73 strains, and 100% (3/3) of ST127 strains carried the full complement of pap genes (Fig. 4). P-fimbrial genes are only rarely found outside the B2/D/F/G clade in our phylogroup database (Fig. 5).
The afa, foc, pap, and sfa genes are considered markers for ExPEC potential. However, the results from our pathotype and phylogroup databases suggest that these genes are found almost exclusively in the B2, D, F, and G cluster and are found in less than 2% of strains from the A, B1, C, and E cluster (Fig. 5). Its presence in the latter cluster could be explained by horizontal gene transfer (HGT), but its presence in the former could be more complicated. The leading hypothesis is that these genes give a competitive advantage in intestinal colonization (2, 49, 50). This may be related to the fact that elf and CFA/I fimbriae are well conserved in phylogroups associated with commensalism or intestinal pathogenesis but lacking in B2 and D strains. This again makes the F phylogroup intriguing because it contains both of those fimbriae but can also carry the ExPEC-associated fimbriae. No matter which proposed evolutionary route for phylogroup diversification turns out to be correct, the presence of ELF and CFA/I in F strains indicates these fimbriae were acquired or lost in the population multiple times in other phylogroups.
A polyvalent fimbria vaccine targeting afa, foc, pap, and sfa could potentially target most strains responsible for ExPEC infections. Although these fimbriae are mostly found in the B2, D, F, and G clade, this clade is responsible for the majority of extraintestinal infections. Importantly, such a vaccine could also conceivably prevent long-term colonization by ExPEC, since these adhesins appear to contribute to colonization of the intestines, and many B2 and D strains seem to lack fimbriae found in other intestinal E. coli, such as CFA/I and ELF. This vaccine would also target ExPEC strains from other phylogroups that have acquired them through HGT, such as VR50 (A; ExPEC) and 789 (C; APEC). The most logical targets of such a vaccine would be the adhesin or major subunit, but the overlap between minor subunits of F1C and S fimbriae should be investigated as well.
One major question remains with P fimbriae: which protein to target. Of the highly conserved genes, papB, papI, and papX encode regulatory proteins, excluding them as potential targets (109–111). The only other highly conserved gene is papA, which encodes the major subunit. However, some studies have shown it to be dispensable for binding when papE is present, unlike papF and papG, which are required for binding (75, 112). This presents more questions than answers: conservation of papB, papI, or papX can be explained by trans-regulatory functions (109–111). The conservation of papA is, at first glance, a mystery. However, upon closer inspection, it appears that papA may be susceptible to transposon insertion, since it appears to have been disrupted multiple separate times based on alignments of papA (see Fig. S4 in the supplemental material). Given this, papH appears to be the best target for vaccine intervention.
Strains from the B2 phylogroup are enriched for iron acquisition genes.
Iron acquisition proteins have long been known to be associated with virulence because iron is a limiting essential nutrient for pathogenic bacteria in the host (24, 113–116). Hypothesizing that the pathogenic pathotypes may harbor a preponderance of such genes, we compared genes encoding siderophores, hemophores, and iron transporters between pathogenic and commensal strains. Our results show that pathogenic strains are enriched for iron acquisition genes (Fig. 6). The three exceptions to this were the nonpathogenic phylogroup B2 strains ED1a, Nissle 1917, and SE15 (Fig. 6A). This general trend of increased iron acquisition may be another explanation for why members of the B2 and D phylogroups are overrepresented in ExPECs, since recent work has shown that iron acquisition gene increase intrinsic virulence (117). Because several pathogenic bacteria lacked these virulence-linked iron acquisition genes on their chromosomes, we also searched their plasmids (Fig. 7).
FIG 6.
Pathotype distribution of iron acquisition genes. (A) Nonpathogenic and ExPEC strains. (B) InPEC strains. The chu operon is responsible for heme uptake. Yersiniabactin, aerobactin, and salmochelin are virulence-associated iron-binding siderophores, whereas enterobactin is a ubiquitous iron-binding siderophore. The percent identity was determined using megaBLAST with the reference genes in Data Set S1 in the supplemental material.
FIG 7.
Distribution of plasmid-encoded iron acquisition genes. (A) Nonpathogenic and ExPEC strains. (B) InPEC strains. Plasmids for each strain in the pathotype database were obtained and analyzed using megaBLAST against the iron acquisition data set. The percent identity was determined using megaBLAST with the reference genes in Data Set S1 in the supplemental material.
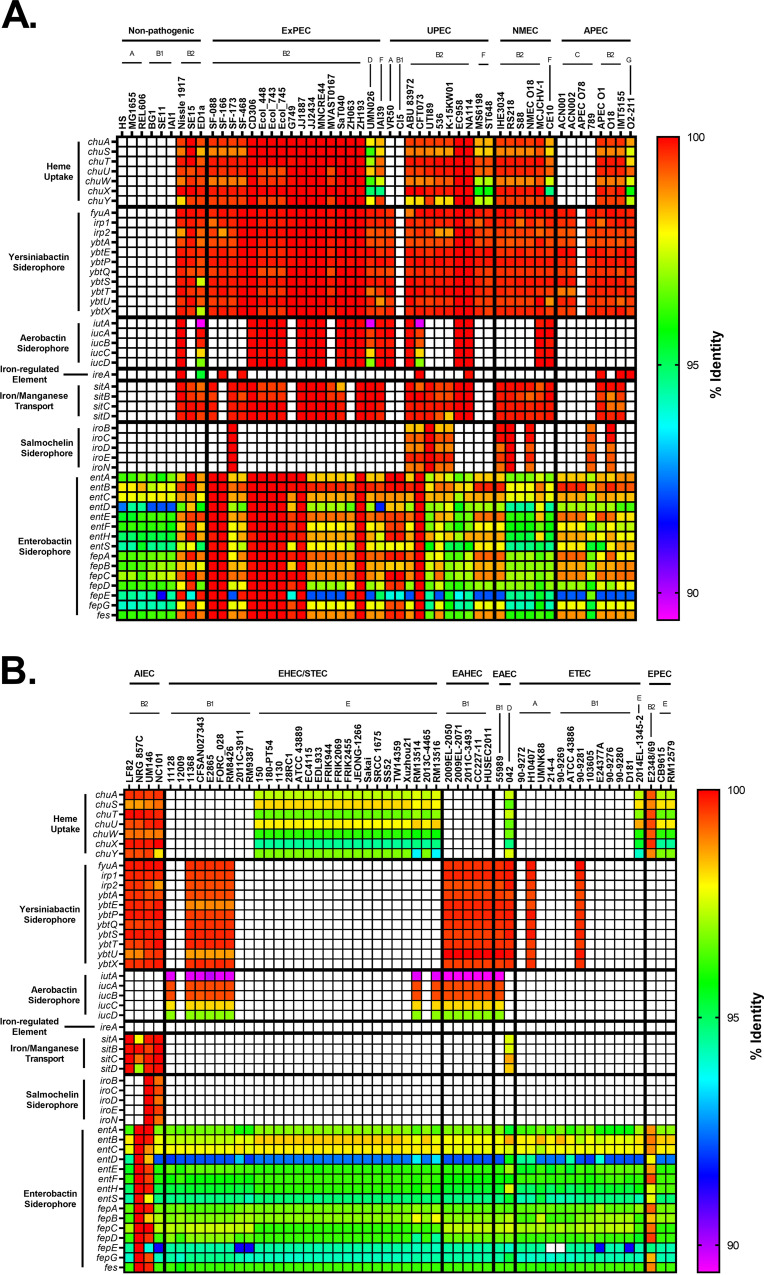
ExPECs tended to have more iron acquisition than InPECs (Fig. 6). With non-APEC ExPECs, only 5 (G749, CI5, MS6198, ST648, and VR50) of 35 (14.3%) strains carry fewer than three of these iron-uptake loci (Fig. 6A). InPECs tended to have few iron acquisition types, with only AIECs and strain 042 (EAEC; D) having more than two of the loci examined (Fig. 6B). All of these trends can be explained by phylogeny: of the five ExPECs deficient in virulence-associated iron acquisition, only one was a B2 strain (Fig. 6A). EHECs/STECs tended to have either the chu heme operon or the yersiniabactin siderophore system, depending on whether they are from the E phylogroup (chu) or the B1 phylogroup (yersiniabactin) (Fig. 6B). This may suggest that EHEC/STEC strains from different phylogroups have different primary sources of iron during infection, since the chu operon is responsible for the uptake of heme, whereas yersiniabactin targets ferric iron (Fe3+) with an affinity that would allow it to steal iron from host iron-bound proteins, including transferrin and lactoferrin (118, 119). The A, B1, and C phylogroups lacked chu because, by definition of the phylogroups, they lack chuA. Interestingly, the ETEC pathotype seems to be the most devoid of iron acquisition genes, possibility indicating a close relationship with commensals, which has been noted for some ETEC strains (120).
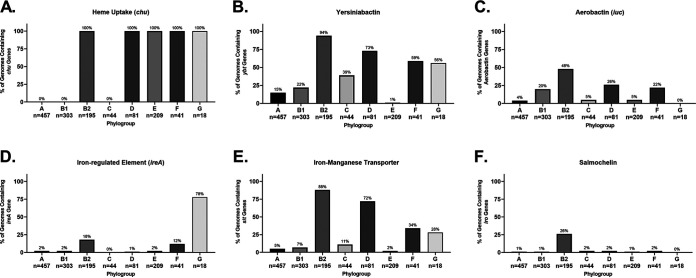
In the phylogroup database, our results for chu show that it is found in 100% of B2, D, E, F, and G strains (Fig. 8A). This is an important control for our method since these phylogroups are characterized by the presence of chuA. Still, the chu system has been shown to play an important role in uropathogenesis (121, 122). The distribution of yersiniabactin, encoded by fyuA, also agrees with the results from our pathotype database and highlights the trends more clearly. Over 90% of B2 strains carry yersiniabactin (fyuA), a siderophore found on PAI IV536 (Fig. 8B) (117). It is surprising to find that such an overwhelming majority of B2 strains carried fyuA because it is a well-known ExPEC-associated gene. In fact, it has been proposed as a gene to differentiate UPEC strains from commensals and other pathotypes (123). While fyuA is only found in 22% (68/303) of B1 strains, 42 of those strains carry the stx toxin, making them EHEC or STEC strains (Fig. 8B). Finding yersiniabactin in many B1 EHEC/STEC strains does make sense, however, since unlike members of the E phylogroup (including the well-known O157 serotype), B1 strains do not have the chu system to extract iron from blood. This could also explain why less than 1% of E-phylogroup strains contain yersiniabactin: the majority of those strains are members of the EHEC/STEC pathotype, and they all have the chu system to extract heme-iron after inducing bloody diarrhea (Fig. 8A and B). This does lead to some obvious questions about selection in cattle, where EHEC/STEC strains are found as asymptomatic intestinal residents. Yersiniabactin does not appear to be highly conserved, but it is enriched for in both the B2/D/F/G clade and the C phylogroup. This pattern could be explained by extensive horizontal transfer coupled with the lack of strong positive selection during intestinal colonization and commensalism. The presence of yersiniabactin in the C phylogroup is probably related to APEC strains from that phylogroup: three of our four strains matching that criteria in our pathotype database contained the siderophore (Fig. 6A).
FIG 8.
Phylogroup distribution of iron acquisition genes. (A) Distribution of the heme uptake genes: chuASTUWXY. This is an important control for our phylogroup database and shows that the B2/D/F/G clade and E phylogroup are correctly distinguished from the A/B1/C clade. (B) Distribution of strains carrying any of the yersiniabactin genes: fyuA, irp1, irp2, and ybtAEPQSTUX. (C) Distribution of strains carrying any of the aerobactin genes: iucABCD or iutA. (D) Distribution of strains carrying the iron-regulated gene ireA. (E) Distribution of strains carrying any of the iron/manganese transporter genes: sitABCD. (F) Distribution of strains carrying any of the salmochelin genes: iroBCDEN. Distributions were determined using megaBLAST to bin strains from each phylogroup into hit versus no hit.
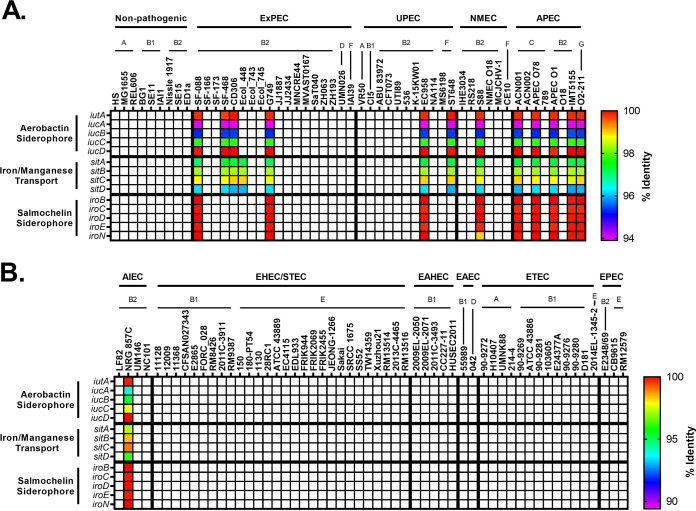
Aerobactin is a siderophore found on PAI ICFT073 that has been shown to be important for uropathogenesis (124, 125). The aerobactin locus is found throughout pathotypes and phylogroups, which is not surprising considering it is found on a mobile element (Fig. 6 and 8). There do appear to be two distinct forms of the operon, with one form being found predominantly in InPEC strains of the B1 phylogroup and the other being found mostly in ExPEC strains of the B2 phylogroup, which raises questions about how or whether this locus is transferred between phylogroups or just within them (Fig. 6). Between these forms of aerobactin, the biggest divergence was found in iutA (89.6% identical to ExPEC ST131 reference), the gene encoding the aerobactin receptor, which may make it difficult to target with a vaccine. This divergent iutA is predominantly found in strains from the B1 and E phylogroups, but there are instances of it being found in the B2 phylogroup, such as with CFT073 (Fig. 6). This is interesting because CFT073 belongs to the ST73, which also contains the ABU 83972 (UPEC) strain and the Nissle 1917 (Nonpathogenic) strain, two strains that carry the less-divergent iutA gene (Fig. 6A). There is less divergence in the chromosomal versions of iucC and iucD, which are aerobactin biosynthesis genes (Fig. 6). Of the 61 B1 strains that carry iutA, 89% (54/61) belong to the EHEC/STEC pathotype (stx+). Outside these EHEC/STEC strains, iutA is again found predominantly in the B2, D, and F cluster (Fig. 8C).
Genes encoding aerobactin were also found on numerous plasmids from the B2, F, and C phylogroups and was in 63% (5/8) of the APEC strains (Fig. 7). The plasmid version of aerobactin appeared to be nearly identical across plasmids, regardless of pathotype or phylogroup, indicating that this plasmid is probably widespread and contributes to virulence (Fig. 7). The plasmid-encoded version was different from the chromosomally encoded version (Fig. 6 and 7). They all contained the ST131-like iutA gene but differed significantly from the chromosomal version in their biosynthetic genes. This indicates that they probably diverged long ago, but the actual receptor was conserved. The plasmid carrying the aerobactin operon also contained nearly identical operons for the sit (iron-manganese transporter) and iro (salmochelin) iron acquisition genes (Fig. 7). This plasmid is the pS88/pColV plasmid associated with APEC virulence and NMEC strains (126, 127).
Iron-regulated element A (ireA) is a siderophore-receptor like protein that is associated with ExPEC strains, implicated in adherence, and found on PAICFT073 II (128). It appears to be present only sporadically in members of the B2 phylogroup and strain O2-211 (APEC), which is a member of the recently characterized G phylogroup (Fig. 6). This gene is relatively rare in our phylogroup database, being found in less than 3% of strains in phylogroups A, B1, C, D, and E (Fig. 8D). However, it is still only found in 20% of phylogroup B2 strains and 12% of phylogroup F strains. Surprisingly, it is found in 78% (14/18) of phylogroup G strains in our database (Fig. 8D).
The iron-manganese transporter system encoded by the sit locus was found almost exclusively in the chromosome of the B2/D/F/G cluster and follows a pattern similar to yersiniabactin, except for being less common in the C phylogroup (Fig. 6). Only 4 of the 39 strains (10%) belonging to the B2 phylogroup in our pathotype database lacked this transporter: G749, SF-166, and ZH063 (all ExPECs) and E2348-69 (EPEC), though G749 carried it on a plasmid (Fig. 6 and 7). Distribution of sit in our phylogroup database agrees with this, with sit being present in 88% of B2 strains, 72% of D strains, and 34% of F strains (Fig. 8E).
The salmochelin siderophore encoded by the iro genes on the PAI III536 and has been implicated in the adherence and invasion of urothelial cells and virulence in an animal model and is upregulated in the presence of human urine (103, 129–133). This locus is found only on the chromosome of a few strains from the B2 phylogroup, mainly from the UPEC (46%; 5/11), NMEC (50%; 3/6), and AIEC (50%; 2/4) pathotypes, from the ST73, ST95, and ST127 sequence types (Fig. 6). Our phylogroup database saw a similar distribution: iroN was found in only 26% of B2 strains and less than 2% of strains from other phylogroups (Fig. 8F).
Iron acquisition genes have a well-known and extensively studied association with uropathogenesis (116, 121, 122, 124, 134–137). Indeed, vaccines targeting siderophores have been proven as a concept in animal models, including mouse models of both E. coli urinary tract infection (UTI) and intestinal colonization by Salmonella (138–145). While some of these studies showed that protective antibodies against siderophores can be generated, the results are not as efficacious as one would hope. This is potentially due to the high level of redundancy in iron acquisition genes seen in uropathogenic strains (135). Still, our work supports previous results suggesting that iron acquisition genes are good targets for an ExPEC vaccine. One obvious bonus to targeting iron acquisition genes is that the risk of targeting commensals may be lower with such a vaccine. This is also supported by evidence that different iron acquisition mechanisms have different levels of importance depending on the type and location of the infection (121, 122, 129, 135). Of course, there is also the risk of redundancy previously mentioned that could make it easy for E. coli to evolve resistance. A solution to this is a polyvalent vaccine. Targeting yersiniabactin, aerobactin, and sit would target many members of the B2, D, and F clade, while targeting ireA and salmochelin would target rarer sequence types and apparently UPECs specifically (Fig. 6 and 8). Targeting aerobactin, sit, and salmochelin would also vaccinate against the pColV plasmid that appears to be associated with ExPECs (Fig. 7).
The B2 phylogroup is enriched for ExPEC-associated toxins.
Bacterial protein toxins have long been studied as critical virulence factors driving pathogenesis. In some cases, pathotype—such as EHEC/STEC and ETEC—assignment can be made based solely on the presence of certain toxins. While there are toxins known to contribute to ExPEC virulence, no single toxin can ensure an ExPEC phenotype. Like our iron acquisition gene analysis, plasmids were included in the toxin analysis because they are often carried on plasmids. However, unlike the iron acquisition graphs, there were no overlaps between genes on the chromosome and those on plasmids, so they are incorporated into a single graph (Fig. 9). Our analysis of E. coli for well-known toxins produced some surprising findings.
FIG 9.
Pathotype distribution of toxins. (A and B) Percent identity results from megaBLAST alignments of nonpathogenic and ExPEC strains (A) and InPEC strains (B). The percent identity was determined using megaBLAST with the reference genes in Data Set S1 in the supplemental material.
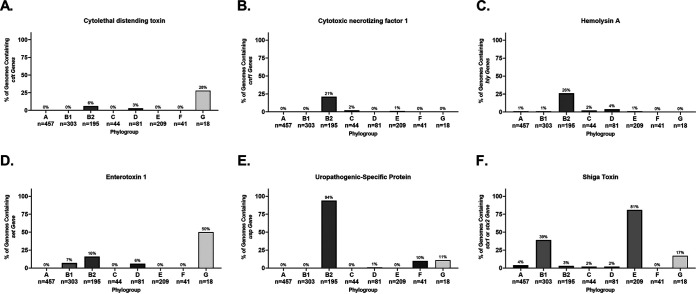
Cytolethal distending toxin (CDT) is a heterotrimeric genotoxin encoded by the cdtABC locus (146). After the CdtABC complex binds to a host cell, CdtB is delivered intracellularly where it causes double-stranded breaks and death of the host cell (146). In our pathotype database, cdtABC is only found in three B2 phylogroup strains, though two of them were from the ST95 sequence type (Fig. 9). This represented only 20% of ST95 strains in our database. In our phylogroup database, cdtA was also rare; only 19 of 1,348 strains carried it on their chromosome (Fig. 10A). The majority (n = 11) of these were strains from the B2 phylogroup, whereas 5 were from the G phylogroup, 2 were from the D phylogroup, and 1 was from the A phylogroup (Fig. 10A). Of these, the G phylogroup had the highest percentage (28%) of strains containing cdtAB, though there are only 18 strains in this phylogroup in our database (Fig. 10A).
FIG 10.
Phylogroup distribution of toxins. (A) Distribution of cytolethal distending toxin (cdt). (B) Distribution of cytotoxic necrotizing factor 1 (cnf1). (C) Distribution of chromosomal α-hemolysin (hlyA). (D) Distribution of chromosomal enterotoxin 1 (setA1). (E) Distribution of uropathogenic-specific protein (usp). (F) Distribution of phage-encoded Shiga toxin (stx1 or stx2). Distributions were determined using megaBLAST to bin strains from each phylogroup into hit versus no hit.
Cytotoxic necrotizing factor 1 (CNF-1) is a chromosome-encoded deamidase toxin that is associated with UPEC and NMEC strains, and a virulence factor that is supposedly found in between 30 and 40% of ExPEC and diarrheal strains (147). The cnf1 locus was not widely spread in our pathotype database, but it does appear to be associated with ExPEC strains and found only in strains from the B2 phylogroup (Fig. 9). These strains were from the ST73, ST95, ST127, and ST643 sequence types, though not all strains from these sequence types contained the cnf1 gene. In our phylogroup database, there were only 43 hits for cnf1, and 40 of these were in the B2 phylogroup, but only in a low percentage of the total B2 strains (21%; 40/195) (Fig. 10B). The other three hits were in phylogroups E (n = 2) and C (n = 1). That cnf1 was found in 30 and 40% of pathogenic strains suggests that an oversized number of ExPEC and diarrheal strains are caused by a relatively small number of strains, particularly from the B2 phylogroup.
Hemolysin E, also known as cytolysin A or silent hemolysin, is a pore-forming toxin encoded by hlyE that under certain conditions can confer hemolytic phenotype to E. coli carrying it (148, 149). It is found on PAI I536 and PAI II536 (103). It belongs to a family of toxins that is also found in Salmonella typhi and Shigella flexneri and reported to be a virulence factor in ETEC strains (32). It appears to have three chromosomal forms, all of which are more than 95% identical to the reference (Fig. 9). In over half of phylogroup B1 (15/27) and C (3/4) strains the hlyE gene is truncated by a frameshift, with both the 5′ and 3′ ends present and possibly translated (Fig. 9). This includes phylogroup B1 strains in the EAEC/EAHEC pathotype. In 100% of B2 strains (39/39), hlyE is truncated as well, but only the C terminus is present and is annotated as a hemolysin-activating protein, HecB, and it is more than 97% identical to reference hlyE. The full hlyE gene is found in the remaining (A, D, F, and G) phylogroups, as well as the other members of the B1 and C phylogroups. This truncation has been published before (148), but to our knowledge, no connection to phylogroup has been made before the present study, and the phylogroup distribution can explain the results showing a pathotype association.
α-hemolysin, also known as hemolysin A, is a pore-forming cytolytic toxin encoded by genes found on the PAI I536 and II536 pathogenicity islands and is only found on the chromosome of a few phylogroup B2 ExPECs and the plasmid of UMNK88 (ETEC; pUMNK88_Hly) (103, 150, 151). The chromosomal version is specifically found in strains from the ST73 (2/3; not found in Nissle 1917 commensal), ST95 (3/10), and ST127 (2/2) (Fig. 9). There is, however, another form that is found on the plasmids of many EHEC/STEC strains. This form is divergent enough to not be hit by megaBLAST alignments except for part of hlyB and is found on a plasmid of 82% of the EHEC/STEC strains examined: 78% (7/9) phylogroup B1 strains and 84% (16/19) of phylogroup E strains. Distribution of chromosomal hlyA in our phylogroup database shows that it occurs almost exclusively in strains from the B2 phylogroup, where it is found in 26% of strains (50/195) (Fig. 10C).
There has been some work on the distribution of hlyA and hlyE, such as by Kerenyi et al. (150), who looked at hundreds of clinical isolates for the presence of hemolytic activity, hlyA, and hlyE (referred to in that study as shaE). These researchers very reasonably concluded that hlyA and hlyE never occur together. Our results suggest an answer: hlyA occurs almost exclusively in phylogroup B2, and no B2 strain in our pathotype database contains the N terminus of hlyE.
Shigella enterotoxin 1 (ShET1) encoded by setA1 and setB1 genes that are found on the antisense strand of the pic gene in the SHI-1 (she) pathogenicity island. In Shigella, it has been found to induce intestinal fluid accumulation (152). It has been associated with EAEC infections but also found in many ExPECs as well (125, 153–155). In our pathotype database, this toxin was found to be highly associated with EAEC strains and found on the chromosome of a few B2 phylogroup members from very diverse pathotypes: Nissle 1917 (nonpathogenic; B2), NC101 (AIEC; B2), ABU 83972 (ABU-UPEC; B2), CFT073 (UPEC; B2), and O2-211 (APEC; G) (Fig. 9). However, three of these five strains (Nissle 1917, ABU 83972, and CFT073) belong to the ST73 sequence type. In B1 EAEC and EAHEC strains, the SHI-1 PAI was duplicated and led to two identical copies of each gene. It is interesting to note that all EAEC and EAHEC strains in our pathotype database contained very similar alleles of setA1 and setB1 (<0.06 and 0% divergence from the reference, respectively), despite being from diverse phylogroups (B1 and D). This is compared to setA1 and setB1 genes in B2 and G strains, which have roughly 5.6 and 1.6% divergence from the reference, respectively. This implicates genes carried on the SHI-1 PAI in EAEC pathogenesis, since it is likely that these divergent phylogroups acquired it independently, but recently. In our phylogroup database, setA and setB were found at low abundance, but predominantly in phylogroup B2 (Fig. 10D). The most surprising result is that 50% (9/18) of strains from the G phylogroup contained set genes (Fig. 10D).
Uropathogenic-specific protein (usp) is a colicin-like bacteriocin toxin that is associated with UPEC strains and increases virulence in a mouse model of UTIs (156). In our pathotype database, this supposedly UPEC-specific protein is actually highly associated with the B2 phylogroup, being found in all but one B2 strain: E2348/69 (B2; EPEC) (Fig. 9). Our pathotype database confirms this association: 94% of B2 strains and 10% of F and G strains contain this protein. It is only found in a single phylogroup D strain and none of the strains from the E phylogroup or the A/B1/C cluster (Fig. 10E). This distribution could explain the propensity for B2 strains to cause UTIs, and being so widespread suggests it entered the B2 phylogroup early in its diversification. Its lack of representation in other phylogroups suggests that it is not very mobile or highly selected against in some contexts. However, if it is not mobile, its presence in 10% (4/41) of the F-phylogroup strains is curious, especially since two of the three sequence types that are known to cause UTIs (ST62 and ST648) are represented in our pathotype database and yet do not contain usp (Fig. 9A) (54).
It is difficult to say whether or not some of these toxins can be used as a vaccine target, since many of them seem to be dispensable for ExPEC virulence. There has been some early success targeting α-hemolysin (hemolysin A) (157). Targeting α-hemolysin and/or CNF-1 could hit a subset of ExPEC strains, particularly some of the more common ExPEC strains from the ST73 and ST127 sequence types (Fig. 9). A vaccine targeting a combination of these proteins has been shown to significantly reduce instances of cystitis in a mouse model and bacterial loads in urine, but not colonization of the kidneys or bladder (158). However, these are both still found in only a small set of B2 strains and appear absent in ST131s. The setAB toxin could potentially be a target against EAECs and some virulent ExPEC sequence types (ST73). Targeting this toxin would also present a unique situation where the protein coded on the opposite strand—pic, an autotransporter—could also be a potential target. The so-called uropathogenic-specific protein (usp) could potentially act as a target for vaccines targeting B2 strains, but our results suggests that commensal B2 strains would also be targeted. Hemolysin E/cytolysin A also may present a B2 target, but it remains to be seen whether the truncated version found in B2 strains—often annotated as hecB—is functional and exported. Of all of these, we believe that α-hemolysin is the more attractive toxin target.
One thing our work does highlight is that the B2 phylogroup is by far the most likely phylogroup to contain these toxins, which does make targeting B2-associated antigens an attractive possibility.
Type Va and Vc secretion systems (autotransporters) are associated with ExPEC strains.
Type V secretion system is composed of the autotransporters (Va or AT-1), two-partner secretion pathway (Vb), and trimeric autotransporter adhesins (Vc or AT-2) (159). This secretion system is made up of secreted and outer membrane proteins involved in adherence and virulence. They consist of N-terminal signal peptide, a passenger domain, an autochaperone domain, and a C-terminal transmembrane β-barrel domain (160). Other domains are possible, including a lectin-like domain found on the end of the invasion protein, Inv (161, 162). Autotransporters are classified based on their domain architecture as AIDA-I, serine protease autotransport of Enterobacteriaceaes (SPATEs), or trimeric autotransporter adhesin (TAA) (160). Overall, autotransporters in our data set were generally associated with ExPEC strains.
Antigen 43 (ang43), an AIDA-I member of the Va pathway, is one of the most abundant phase-varying outer membrane proteins and is encoded on PAI III536, and PAI ICFT073, along with the genes encoding the Vat or Sat autotransporter, respectively (103, 125, 163). This gene is also known as flu for fluffing because it promotes aggregation between cells and colonization of mouse bladders. Some evidence suggests it may be beneficial to UPEC strains (164). Our results suggest that ang43 is widespread, with no apparent phylogroup or pathotype association (Fig. 11). There does appear to be an EAEC- and EAHEC-specific allele. This allele is found in all EAEC and EAHEC strains, regardless of whether they are from the D or the B1 phylogroup (Fig. 11), and appears to be on the same genomic island (GI 3 in strain 042) that carries pic, set1A, and set1B and two type VI secretion systems (see Fig. S4 in the supplemental material).
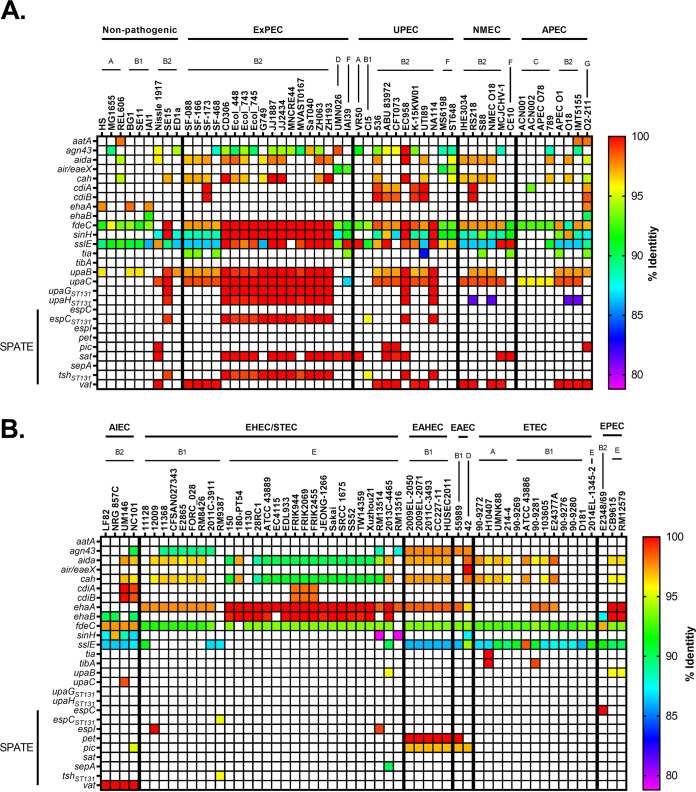
FIG 11.
Pathotype distribution of autotransporters. (A and B) Percent identity results from megaBLAST alignments for nonpathogenic and ExPEC strains (A) and InPEC strains (B). SPATEs, serine protease autotransporters of Enterobacteriaceae. The percent identity was determined using megaBLAST with the reference genes in Data Set S1 in the supplemental material.
EhaA is an autotransporter identified as being associated with the O157:H7 serotype and important for adhesion and biofilm formation (165). However, our results suggest that ehaA is more generally associated with EHEC/STEC, EAEC, and EAHEC (Fig. 11). It is also possibly associated with EPEC strains but appears to be absent in the B2 EPEC strain (E2348/69).
EhaB, also called UpaC, is found in two forms: one (labeled upaC; ST131 is used as a reference) is found predominantly in ExPEC strains and B2 commensal strains (Nissle 1917 and SE15) (Fig. 11). However, it is absent in most AIEC strains (Fig. 11B). The other form, designated ehaB and using the Sakai (EHEC) strain as a reference, is found in 100% of O157 (15/15; all EHEC/STECs) and O55 (3/3; 1 EHEC/STEC and 2 EPECs) serotypes (Fig. 11). It is also found in the B2 phylogroup EPEC strain but in none of the B2 members of the ExPEC phylogroup (Fig. 11). Interestingly, it is also found in three of the four (75%) AIEC strains (Fig. 11B). This means that four of the five InPEC members of the B2 phylogroup carry this allele, while being found in none of the 3 B2 commensals or 32 B2 ExPEC strains. These results suggest that ehaA and ehaB are important virulence factors for intestinal pathogenesis and suggests that ehaB may be an AIEC-associated virulence factor.
Another AIEC- or InPEC-specific finding in our results is the profound changes in upaB, which encodes an autotransporter that binds to fibronectin and glucosaminoglycans (166). It has been shown to promote uropathogenesis and colonization of the bladder in a mouse model (167). Our results suggest upaB is found widely throughout ExPEC and nonpathogenic B2 strains, with only 2 of 35 of these strains missing the gene (Fig. 11). However, the two commensal strains that carry this gene are from the ST73 and ST131 sequence types and appear to have lost virulence factors on their way to becoming nonpathogenic. In contrast to ExPEC B2 strains, all five B2 InPEC strains (four AIEC and one EPEC) lacked a similar upaB (Fig. 11B). In the AIEC strains, the upaB the gene contained either a premature stop codon (LF82 and UM146) or a 189-bp insertion (NC101 or NRG 857C), explaining why this potential connection has not been investigated. The B2 EPEC strain E2348_69 did not elicit even a partial hit.
Several autotransporters appear to be tightly linked in the ST131 group: espCST131, tshST131, upaGST131, and upaHST131 (Fig. 11). Outside ST131 strains, these autotransporters only appear sporadically (Fig. 11). In fact, espCST131 and tshST131 are only found in one strain outside ST131 strains in our pathotype database: CI5 (UPEC; B1) (Fig. 11). However, closer examination shows that this appears to be caused by remarkable diversity in the upaGST131 and upaHST131 genes. UpaH is a Va AIDA-I autotransporter that is involved in biofilm formation and colonization (168, 169). UpaG is a Vc trimeric autotransporter adhesin that has been shown to promote biofilm formation, adherence to host matrix, and abiotic surfaces (170). These genes are highly conserved, but alignments show a large degree of variation from sequence type to sequence type, mainly in the middle of the sequence. This has been noted in both genes (169, 171). To reflect this, upaG and upaH are annotated to indicate they used ST131 references. This could mean that UpaG and UpaH could be useful for quickly determining whether an isolate belongs to virulent sequence types (such as ST131 or ST73) using multiplex PCR. As a vaccine target, UpaG and UpaH may lack specificity, however, for UpaH conserved regions have been identified throughout phylogroups and work on a vaccine is under way (168, 169). The question remains as to whether UpaH is involved in colonization of other areas, since it is found in pathotypes (e.g., EHEC) that do not colonize the bladder.
Two SPATEs--EspCST131 and TshST131--are found almost exclusively in ST131 strains with the interesting exception of also being found in the CI5 strain (B1; UPEC) (172, 173) (Fig. 11A). However, while these were annotated as espC and tsh by the VFDB and some annotation software, it appears that this annotation may be incorrect. Using these references, we saw no hits in any of our EPEC strains or APEC strains, where research on EspC and Tsh has been done, respectively (27, 173–175) (Fig. 11). Closer examination revealed that espC from E2348/69 was only 60% identical to this reference, so a second espC (espCE2348/69) was included, while tsh from APEC strains were only ∼41% identical to tshST131. In fact, tshST131 appears to be related to adcA (CBG90828; 70% identical; 72% coverage) and espCST131 is related to a putative autotransporter gene (CBG91787; 87% identical; 100% coverage) from Citrobacter rodentium (176–178). The espCE2348/69 is found on integrative element 5 (IE5), and the surrounding genes do not match those of espCST131, while tshST131 is found only two genes upstream of espCST131. The only strain carrying espCE2348/69 was the E2348/69 strain (Fig. 11). This is probably because E2348/69 is the only B2 EPEC strain in our database, and this gene appears to be isolated to LEE-containing B2 strains in our phylogroup database. This would explain the results by Mellies et al. (173), who only found these genes in a subset of EPEC strains. The fact that tshST131 and espCST131 are specific for ST131s does make them potential vaccine targets, but more work must be done to determine whether the proteins they encode contribute to virulence and induce an antibody response.
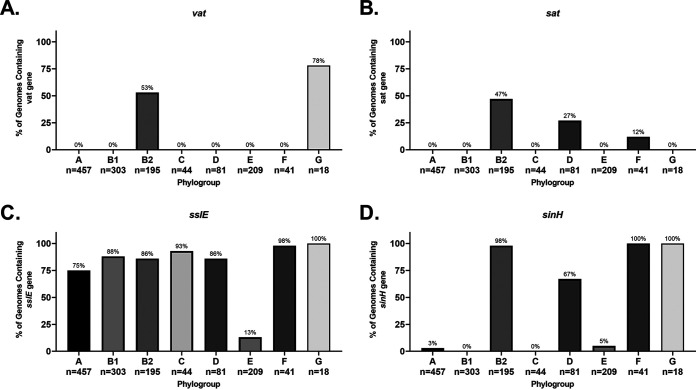
Vacuolating autotransporter toxin (Vat) has been shown to contribute to uropathogenesis (179). Our data show that it is only found in the B2 and G phylogroups. It is found in the well-known ST73 and ST95, as well as lesser-known sequence types, but not ST131 strains. It should also be noted that sometimes this vat gene is mistakenly annotated as temperature-sensitive hemagglutinin, tsh (or the closely related hemoglobin-binding protease, hbp) or sepA, which encodes a Shigella virulence factor (180, 181). Of these genes, only sepA was found on the chromosome of one strain examined in our database, though tsh/hbp was found on the plasmid of 50% (4/8) of APEC strains, and sepA was found on the plasmid of all B1 EAEC and EAHEC strains. To verify each vat hit, we examined each hit for a marA/papX regulator (vatX) immediately downstream and the yag operon roughly 4 kb upstream (179). In our phylogroup database, vat is found exclusively in phylogroups B2 (53%; 103/195) and G (78%; 14/18) (Fig. 12A). Vat is a potential target, as it is found in many of the common ExPEC sequence types (ST73, ST95, and ST127), and it elicits an antibody response in UTI patients infected with vat+ E. coli (179). The largest downside is that targeting Vat would not target ST131, a major cause of ExPEC infections worldwide.
FIG 12.
Phylogroup distribution of autotransporters. (A) Distribution of the gene encoding vacuolating autotransporter toxin (vat). (B) Distribution of the gene encoding secreted autotransporter toxin (sat). (C) Distribution of the gene encoding accessory colonization protein (sslE). (D) Distribution of the gene encoding invasion-like protein (sinH). Distributions were determined using megaBLAST to bin strains from each phylogroup into hit versus no hit.
Secreted autotransporter toxin (Sat) is another vacuolating cytotoxin implicated in uropathogenesis (182, 183). Like Vat, it also elicits a strong antibody response, but unlike vat, it is also found in ST131 and ExPEC strains from the D and F phylogroup in our pathotype database (Fig. 11) (183). It is also more widely distributed in our phylogroup database, where it is found in 47% of B2 strains (92/195), 27% of D strains (22/81), and 12% of F strains (5/41) (Fig. 12B). One interesting note is that sat is found in only one of the 457 strains in our phylogroup A database: VR50. While both sat and vat are only found in only barely 50% of B2 strains, 87% (170/195) of B2 strains carry one or the other (or both), making a polyvalent vaccine targeting both an intriguing prospect (Fig. 12B). Such a hypothetical vaccine would provide protection against 86% (30/35) of the ExPEC, UPEC, and NMEC strains in our pathotype database, while only targeting 11% (1/9) of the nonpathogenic strains.
The invasion-like autotransporter fdeC binds to human epithelial cells and contributes to colonization of kidneys and bladder in an animal model, and it may be protective as a vaccine target (184). In our pathotype database, the gene encoding the FdeC adhesin is found in nearly every strain (Fig. 11). It appears that certain alleles of fdeC may be associated with intestinal lifestyles, although these allelic trends appear to fall along A/B1/C/E and B2/D/F/G clusters (Fig. 11). However, the fact that fdeC genes are found throughout almost all strains makes the protein it encodes a less ideal vaccine target.
The accessory colonization factor encoded by sslE is a zinc-metalloprotease with mucinase activity (185). The sslE gene is found across all phylogroups but is mostly lacking the EHEC/STEC pathotype. This is even seen in EHEC/STEC strains that belong to the B1 phylogroup, despite B1 strains in other phylogroup still having this gene (Fig. 11). This trend is also seen in our phylogroup database (Fig. 12C). Roughly 75% of stx+ B1 strains carry sslE compared to 96% of B1 strains lacking stx (see Fig. S6). In the E phylogroup, only 1% of stx+ strains carry sslE compared to 56% of stx mutant strains (see Fig. S6). This may indicate that sslE is either not required or selected against during EHEC infections.
Interestingly, inoculation with FdeC and SslE have been shown to be protective against UPECs (184, 186). However, our results show that both of these targets could produce significant off-target effects considering how broadly they are conserved in commensals (Fig. 11).
The pet and pic autotransporters are normally associated with EAEC strains (32). In our data set, pet is found almost exclusively on the chromosomes EAEC and EAHEC strain from the B1 phylogroup and, strangely, on the chromosome of ED1a (B2; commensal). However, it is truncated and predicted to be expressed in two parts (Fig. 11). The 042 (D; EAEC) strain lacks pet on its chromosome, but a divergent form annotated as pet and picked up by our reference pet is found on its plasmid (data not shown). The pic gene is associated with EAEC as well, and all six EAEC or EAHEC strains share a near identical allele—EAECs are 97% identical to the reference, while EAHECs are 96.9% identical. There is an allele of pic that is found in B2 strains, specifically those belonging to ST73 (Fig. 11). The only other instance is also the most divergent allele, which is found in the NC101 (AIEC; B2).
The invasin-like protein SinH appears to be strongly associated with ExPEC strains, but the distribution results from our phylogroup database suggests this association is more likely with the B2/D/F/G cluster than specifically with ExPEC strains (Fig. 11 and 12), although this does not rule out a contribution to virulence. Outside of ExPEC strains, B2 commensal strains also carry this gene, but in ED1a the beta-barrel domain that links the protein to the outer membrane is missing due to a premature stop codon (Fig. 11). There also appears to be an ST131-specific allele that is a relatively divergent (ca. 88 to 90% identical) from the sinH found in other ExPECs (see Fig. S7). There are two significant hits in the RM13514 (EHEC/STEC) and RM13516 (EHEC/STEC) strains, but the SinH in RM13514 is missing the beta-barrel domain, and the RM13516 strain is very divergent and aligns poorly with the other SinHs on the protein level (see Fig. S7). Our phylogroup database verifies the association with the B2 phylogroup, where it is found in 98% of strains, and shows that sinH is also strongly associated with the F and G phylogroups (100%) and, to a lesser extent, with the D phylogroup (48%) (Fig. 12).
Other adhesins and miscellaneous virulence genes are either ubiquitous or enriched in B2 strains.
The locus of enterocyte effacement (LEE) carries a type 3 secretion system related to virulence, is known to be associated with EPEC and EHEC/STEC strains, and is responsible for their characteristic attaching-and-effacing (A/E) phenotype (187). The LEE-encoded attachment protein intimin is a product of eae (also called eaeA) and is responsible for EHEC and EPEC characteristic attachment phenotype, and our results are in agreement (Fig. 13). Another adhesin, EaeH, was first identified in ETEC. It is upregulated when ETEC strains interact with epithelial cells and promotes adhesion and toxin delivery (188, 189). However, it seems this adhesin is ubiquitous, since it is found in all but five strains, though there may be some allelic differences in eaeH (Fig. 13). Unlike most molecules that show allelic differences roughly along the A/B1/C/E and B2/D/F/G clades, eaeH appears to have a B2-specific allele.
FIG 13.
Pathotype distribution of other chromosomal virulence factors. (A and B) Percent identity results from megaBLAST alignments for nonpathogenic and ExPEC strains (A) and InPEC strains (B). The percent identity was determined using megaBLAST with the reference genes found in Data Set S1 in the supplemental material.
Porcine A/E-associated protein (encoded by paa) is an LEE-encoded adhesin that is required for EHEC infections (190). It has been found to be more immunogenic than intimin, and it confers a slight protective effect against colonization of EHEC strains in mice when used as a vaccine (191). Our results shows that it is predominantly found in LEE-carrying pathotypes, though it appears to be missing in some B1 EHEC/STEC strains and the B2 EPEC strain (Fig. 13).
The invasion of brain endothelium A (ibeA) gene encodes a protein that has been shown to be important for invasion of the blood-brain barrier (192). It has also been shown to be an important virulence factor in APEC strains (192). Our pathotype results largely support these assertions, but the gene appears to only be found in B2 strains of these pathotypes: 3/5 B2 strains in NMEC and 3/3 B2 strains in APEC (Fig. 13). The F-phylogroup strain (CE10) and two of the B2 strains NMEC strains lacked ibeA, and none of the phylogroup C APEC strains carried the gene (Fig. 13). Interestingly, all four AIEC strains in our database contained ibeA, which supports recent work showing that this gene is important for intestinal colonization, cellular invasion, and macrophage survival of the NRG857 (AIEC) strain (Fig. 13B) (33). In our phylogroup database, ibeA is only found in phylogroups B2 and F, which may further explain why most AIEC strains are from the B2 phylogroup (Fig. 14A).
FIG 14.
Phylogroup distribution of other virulence factors. (A) Distribution of the gene encoding invasion of brain endothelium A (ibeA). (B) Distribution of the gene encoding iron-regulated homologous adhesin (iha). (C) Distribution of the gene encoding the Hek adhesin (hek). (D) Distribution of the genes encoding increased serum survival protein or bor (iss/bor).
The malX gene encodes a phosphotransferase system II enzyme and is associated with ExPEC strains (193–195). It may also increase persistence in the intestines (156). Our results show that it highly conserved in B2 strains and, to a lesser degree, pathogenic B1 strains from the UPEC and EHEC/STEC pathotypes (Fig. 13).
The iron-regulated homologous adhesin encoded by iha gene is part of the PAICFT073 I and PAI536 II pathogenicity islands and, unlike the P fimbriae, appears to be more widespread (107, 196). This suggests that iha has weaker selection against it or greater selection for it. Iha has been shown to be important for colonization of the kidneys and bladder in a mouse model of infection (196). In our work, it is found widely in B2 strains and pathogenic strains in general (Fig. 13). An interesting exception is that it is not found in either ST95 or ST127 B2 strains, two common sources of ExPEC infections. In EHEC/STEC strains, which are all from the B1 and E phylogroups, it appears highly conserved (Fig. 13). However, it is not found in any of the EPEC strains which may suggest that the acquisition of an iha PAI in addition to stx is important for EHEC/STEC pathogenesis and differentiates EHEC/STEC strains from EPEC strains (Fig. 13B). It is also lacking in B2 strains from the AIEC, NMEC, and APEC pathotypes, which may be related to their lifestyle (Fig. 13A). In our phylogroup database, it was found in a relatively diverse set of phylogroups and especially enriched in phylogroup E (Fig. 14B). One intriguing thing about targeting Iha is that, outside EHEC/STEC strains, the gene that encodes it is often found on the same pathogenicity island (e.g., PAI536 II) as sat and iutA (aerobactin), which have already been mentioned as potential vaccine targets. Targeting any of these alone has a serious drawback in that they are not present in the majority of the ST95 strains examined in our pathotype database. However, as with our speculations on combining Sat and Vat to cover the majority of ExPEC strains, Vat combined with either Iha or IutA appears to confer similar or greater coverage.
The Hek adhesin is encoded by a gene that is part of the PAI536 II pathogenicity island and is involved in autoaggregation, hemagglutination, heparin binding, adherence, and invasion of host cells in NMEC strains (103, 197, 198). Despite being found on the same pathogenicity island as iutA, sat, and iha, it has apparently been lost in most of those strains in our pathotype database (Fig. 13). It is only found sporadically in B2 strains, specifically ST127, and occasionally ST73 and ST95 strains. In our phylogroup database, it is found in low abundance in all phylogroups examined (Fig. 14C). The relatively low number of strains carrying this protein makes it less suitable as a vaccine target.
The prophage-encoded increased serum survival gene (iss) and bor genes have been associated with ExPEC infections for at least four decades. The proteins encoded by these genes are known to be surface exposed, have been shown to help E. coli resist the host complement system, and elicit an immune response in avian hosts (199–201). Since the proteins encoded by iss and bor are more than 90% identical, we have not distinguished between the two here (202). Both were first found on plasmids, but more recent work has shown that they are also carried on the chromosome (203). Our results suggest that iss/bor are widespread and found in all pathotypes (Fig. 13). There appears to be no phylogroup or sequence types trends, since these genes are found sporadically through different sequence types and are found often in all phylogroups (Fig. 13 and 14D). This is most likely because they are encoded by prophages and means that they are unlikely to be suitable vaccine targets.
Commensalism and vaccine targets.
Our results show that the majority of virulence factors (VFs) and markers for extraintestinal pathogenesis are associated with the B2 phylogroup or the B2/D/F/G cluster. In many cases, these VFs are found exclusively (or nearly exclusively) in these phylogroups. In other cases, the VFs are found predominantly in the B2/D/F/G cluster but also in a minority of strains from other phylogroups. This suggests that the B2/D/F/G cluster is the dominant source of pathogenic strains, which is known, although estimates of the extent to which they are predominant vary. A similar conclusion with B2 and D strains was reached over 20 years ago in an experimental determination of the lethality of 82 strains from the E. coli reference (ECOR) collection cross-referenced against the presence of seven virulence determinants (204). There, researchers found that these virulence factors and lethality correlated with the B2 phylogroup (this was before phylogroups F and G were described). Our work not only verifies this but expands it to a wider range of virulence factors and, importantly, also shows that the B2/D/F/G cluster is the dominant reservoir for extraintestinal pathogenic virulence factors in general.
A major implication of this is that B2/D/F/G isolates that contain virulence factors should be looked at with suspicion but, without a comprehensive and in-depth overview of virulence factors, it is hard to say whether or not a strain from this cluster can cause disease. This is easy to see with the three nonpathogenic B2 strains in our pathotype database: Nissle 1917, SE15, and ED1a. Nissle 1917 is an ST73 strain, like ABU 83972 (ABU) and CFT073 (UPEC), and SE15 belongs to ST131, of which we have numerous examples of in our pathotype database. In most cases, these commensal strains contain the same virulence factors as pathogenic B2 strains, with some notable exceptions. Compared to other ST73 strains, Nissle 1917 is missing hlyA (toxin), full-length pap (fimbriae), cdiAB (autotransporter), fdeC (autotransporter), upaB (autotransporter), and salmochelin (iron acquisition). With ST131 strains of the fimH41 subclass, SE15 has lost the partial pap (fimbria) genes seen in other ST131 strains, as well as agn43 (autotransporter), sat (autotransporter), aerobactin/iutA (iron acquisition), and iha (adhesion/iron acquisition)—all proteins found on PAICFT073 I (Fig. 15A). The loss of these genes appears to abolish pathogenicity in a strain from a highly pathogenic sequence type. It is important to note that most cursory overviews of these B2 commensals would conclude that they have high pathogenic potential, especially if they were compared to strains outside their phylogroup or sequence type. For example, Nissle 1917, still carries kpsTM, K5 biosynthetic genes, pic, sat, vat, foc fimbriae, papA, fyuA (yersiniabactin), iutA (aerobactin), ireA, iha, malX, and iss. It also still carries the pks genotoxin island (205). It can be reasonably concluded that this is a virulence fingerprint that is from a strain with high probability of causing disease, and yet Nissle 1917 has been used as a probiotic to successfully treat intestinal diseases for a century (206).
FIG 15.
Genetic difference in pathogenicity. (A) Schematic for the loss of pathogenicity in ST73 and ST131 using examples from strains examined in the present study. (B) Gain and loss of virulence factors at various points in E. coli phylogroups. Labels were assigned under the presumption that gaining genetic factors are rare, while loss is common, i.e., the map was built to reflect the lowest gain of genes possible based on the results presented here. ExPEC, extraintestinal pathogenic E. coli; UPEC, uropathogenic E. coli; NMEC, neonatal meningitis-causing E. coli; APEC, avian pathogenic E. coli; InPEC, intestinal pathogenic E. coli; AIEC, adherent-invasive E. coli; EHEC, enterohemorrhagic E. coli; EAEC, enteroaggregative E. coli; ETEC, enterotoxigenic E. coli; EPEC, enteropathogenic E. coli. Light blue, commensals (or those likely to be commensal); dark blue are InPEC strains; red, ExPEC strains; gray, phylogroups of uncertain intrinsic virulence. This illustration was made using BioRender.
Another implication from our work is that most of the ExPEC-associated VFs are found widely and predominantly in B2 and D strains (Fig. 15B). This suggest that strains from the A/B1/C cluster should be viewed with greater suspicion when clinical isolates are shown to contain virulence factors that are generally found in the B2/D/F/G cluster. This is because for these strains to have these genes, they almost certainly had to acquire them through horizontal gene transfer, and then the gene had to most likely be selected for to fix it in the population. With a gene that confers an advantage during infection, increased virulence is the simplest mechanism of fixation.
However, pathotype associations from this work need to be interpreted with care, since several pathotypes we catalogued have few representative strains and are therefore at greater risk of selection bias. There is, of course, the same risk of bias with our phylogroup associations, but we feel it is much less likely because of the much larger sample size and because our phylogroup database comprises all complete E. coli genomes currently available in NCBI’s RefSeq database.
Our work also suggests that vaccines targeting VFs will likely target many strains in the B2/D/F/G cluster, even nonpathogenic ones. Although it has been postulated that an ideal vaccine candidate would only target pathogenic strains, targeting all B2/D/F/G strains is not necessarily a dead end (13). B2 strains are now common commensals in the developed world, but much rarer in older data sets and in the developing world, and there is no indication that the lack of B2 strains in the gut microbiota has been detrimental to health (207, 208). In fact, the opposite may be true since microbiota in developed countries tend to be less diverse (209). Accepting this fact leads to the obvious conclusion that specifically targeting B2 strains may open new avenues for vaccine research because while we have struggled to clearly define the ExPEC pathotypes, phylogroups can be clearly delineated. A successful B2 or B2/D/F/G cluster vaccine could potentially eliminate a large source of both virulent strains and a virulence factor reservoir.
Along these lines, the phylogroup distribution of what we believe are the vaccine targets with the most potential given their prevalence in different pathotypes and phylogroups is shown in Fig. 16. These potential targets are predominantly found on the surface, but it should be noted that the proposed toxin targets are excreted. While many successful vaccines against other organisms have targeted secreted toxins, the multifaceted nature of E. coli virulence means that a monovalent toxin vaccine may reduce the severity of the disease rather than completely prevent an infection. However, a polyvalent vaccine targeting several of these proposed proteins—including toxins—would target multiple mechanisms of virulence such as adherence or immune evasion. This would increase the chances that an infection could be completely prevented rather than simply abated. Such an approach would also make developing resistance while maintaining virulence difficult (Fig. 16A). Targeting uropathogenic-specific protein (Usp), yersiniabactin receptor (FyuA), group 2 capsule (KpsTM), vacuolating toxins (Sat/Vat), and iron/manganese transporter (SitA) would target the vast majority of strains from the B2 and D phylogroups—which are more intrinsically virulent. They would do this while avoiding strains from phylogroups more common among commensal unless those strains have acquired these virulence factors (e.g., strain VR50) (Fig. 16B). Other, less widespread targets such as α-hemolysin (HlyA), afimbrial adhesin (Afa), F1C fimbriae (Foc), P fimbriae (Pap), S fimbriae (Sfa), and aerobactin receptor (IutA) make attractive auxiliary targets because even though they are rare, they have been shown to play a major role in pathogenicity (Fig. 16C).
FIG 16.
Potential polyvalent vaccine targets. Phylogroup distributions of potential targets were determined by using megaBLAST to cross reference all 1,348 complete E. coli chromosomes in the RefSeq database that could be categorized by phylogroup against potential targets. (A) Schematic overview of targets and their virulence mechanisms. Made using Biorender. (B) Targets with high disease impact and wide distributions through ExPEC-associated phylogroups; (C) targets with limited distribution, but still high disease impact. Terms: uropathogenic protein, usp; Salmonella-like invasion H, sinH; secreted autotransporter toxin, sat; vacuolating autotransporter toxin, vat; G2C-specific Kapsel polysaccharide genes M and T, kpsTMII; ferric yersiniabactin uptake receptor, fyuA; aerobactin uptake receptor, iutA; iron/manganese transporter, sitA; cytolethal distending toxin, cdt; cytotoxic necrotizing factor-1, cnf1; α-hemolysin, hlyA; afimbrial adhesin, afa; F1C fimbriae, foc; P fimbriae, pap; S fimbriae, sfa; IrgA homologue adhesin, iha.
This information is being used to guide polyvalent vaccine development in our lab, but these genes and their distribution should also be useful when determining the potential pathogenicity of isolates, as well as in further elucidating the pathogenicity of E. coli.
MATERIALS AND METHODS
Comparative genomics.
Alignments of virulence factors and strains were created using megaBLAST plugin for Geneious Prime 2019.2.3 using a 1/–2 match/mismatch scoring matrix, linear gap score, and a maximum E value of 1E–10. Where necessary, a culling limit of 1 was used to prevent too many off-target hits. Percent identity outputs were exported as a spreadsheet and used to produce heatmaps in GraphPad Prism version 9.0.0 for Windows (GraphPad Software, San Diego, CA).
Virulence factors database was created using the Virulence Factor Database, Victors, and PATRIC virulence databases as a backbone and expanded upon with literature (56–58). Strain and phylogroup databases were constructed as described below. Virulence factors and sequences can be found in Data Set S1 in the supplemental material.
RefSeq phylogroup database construction.
Escherichia coli strains for the phylogroup database were retrieved from NCBI’s RefSeq database (accessed 1 September 2020) with the following query string: Search all[filter] AND “Escherichia coli”[ORGN] AND latest[filter] AND “complete genome”[filter] AND (all[filter] NOT “derived from surveillance project”[filter] AND all[filter] NOT anomalous[filter]) Sort by: SORTORDER. The 4,105 strains were trimmed to 1,351 strains above 3.5 megabases. These strains were classified by phylogroup using in-house methods (see Appendix S1 in the supplemental material). Briefly, megaBLAST was used to align short sequences derived from Clermont phylotyping fragments. Strains were binned based on hits or no hits to each fragment (61). The resulting pattern of “hit/no hit” for each of the main four fragments were used to determine phylogroup (see the Fig. S1 legend for more information). Phylogroups E, C, and G strains were classified next based on the presence of the arpA_gpE, trpA_gpC, and ygbD_G PCR fragments, respectively (55, 61, 210). For more information, see Appendix S1 in the supplemental material. A breakdown of strains in our phylogroup data can be found in Fig. S8. Accession numbers for strains divided by phylogroup can be found in Data Set S2 in the supplemental material.
Strain curation.
Phylogroup and pathotype breakdown of strains in our pathotype database can be found in Fig. S9 in the supplemental material.
The extraintestinal pathogenic E. coli strains (ExPECs) refer to several pathotypes that cause disease outside the intestines; the major pathotypes are uropathogenic E. coli (UPEC), neonatal meningitis E. coli (NMEC), and avian pathogenic E. coli (APEC) (25). There are dozens of virulence factors said to be associated with ExPEC strains, though our results suggest that most of these may actually be found in the majority of strains (25). In the present study, ExPEC strains are labeled as ExPECs unless there is published evidence for the strains to be sorted into the UPEC, NMEC, or APEC pathotypes, even if a strain was collected from the urine of an infected patient (Table 2). This may mostly affect UPEC strains, since the terms ExPEC and UPEC are often used interchangeably. Of the 18 general ExPEC strains, 16 belong to the B2 phylogroup, 1 belongs to the D phylogroup, and 1 belongs to the F phylogroup. Of these strains, 12 belong to the ST131 sequence type. Given the highly virulent nature of ST131 strains, they were all considered ExPECs, with three exceptions: SE15, EC958, and NA114. SE15 was isolated from a healthy adult and shown to lack virulence and also lacks many virulence genes seen in other ExPEC strains, so it is classified as nonpathogenic (Table 1) (211). EC958 and NA114 were classified as UPEC strains because they were isolated in urine and carried UPEC-associated virulence factors not seen in other ExPECs: the selC genomic island in EC958 and the full-length pap fimbriae in NA114 (212, 213).
TABLE 2.
Strains, accession numbers, phylogroups, STs, antigens, and references
| Pathotype | Strain | Accession no. | Isolation source | Phylogroup | ST | O antigen | H antigen | K antigena | FimH typeb | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| Nonpathogenic | HS | CP000802 | Feces, healthy subject | A | ST46 | O9 | 35 | 9 | ||
| MG1655 | CP014225 | Feces, healthy subject | A | ST10 | O16 | H48 | 27 | 227 | ||
| REL606 | CP000819 | Lab strain | A | ST93 | O7 | 427/608 | 228 | |||
| BG1 | MOAH00000000 | Bovine intestines | B1 | ST58 | O159 | H21 | 53 | 229 | ||
| SE11 | AP009240 | Feces, healthy subject | B1 | ST156 | O152 | H28 | 38 | 230 | ||
| IAI1 | CU928160 | Feces, healthy subject | B1 | ST1128 | O8 | H4 | 32 | 231 | ||
| Nissle 1917 | CP007799 | Feces, healthy subject | B2 | ST73 | O6 | H1 | K5 | 30 | 232 | |
| SE15 | AP009378 | Feces, health subject | B2 | ST131 | O150 | H5 | K+ | 41 | 233 | |
| ED1a | CU928162 | Feces, healthy subject | B2 | ST452 | O81 | H27 | K+ | 225/580 | 233, 234 | |
| ExPEC | SF-088 | CP012635 | Bloodstream infection | B2 | ST95 | O1 | H7 | K1 | 30 | 235 |
| SF-166 | CP012633 | Bloodstream infection | B2 | ST95 | O1 | H7 | K1 | 41 | 235 | |
| SF-173 | CP012631 | Bloodstream infection | B2 | ST95 | O18 | H7 | K1 | 18 | 235 | |
| SF-468 | CP012625 | Bloodstream infection | B2 | ST95 | O25 | H4 | K1 | 27 | 235 | |
| CD306 | CP013831 | Feces, cat | B2 | ST131 | O25 | H4 | K5 | 30 | 236 | |
| Ecol_448 | CP015076 | Infected urine | B2 | ST131 | O16 | H5 | K100? | 41 | 237, 238 | |
| Ecol_743 | CP015069 | Infected urine | B2 | ST131 | O16 | H5 | K+ | 1426 | 237, 238 | |
| Ecol_745 | CP015074 | Infected urine | B2 | ST131 | OR | H5 | K100? | 41 | 237, 238 | |
| G749 | CP014488 | B2 | ST131 | O25 | H4 | K1 | 22 | 236 | ||
| JJ1887 | CP014316 | Recurrent cystitis | B2 | ST131 | O25 | H4 | K14 | 30 | 239 | |
| JJ2434 | CP013835 | B2 | ST131 | O25 | H4 | K5 | 30 | 236 | ||
| MNCRE44 | CP010876 | Sputum | B2 | ST131 | O25 | H4 | K+ | 30 | 240 | |
| MVAST0167 | CP014492 | B2 | ST131 | O16 | H5 | K100? | 41 | 236 | ||
| SaT040 | CP014495 | B2 | ST131 | O25 | H4 | K5 | 22 | 236 | ||
| ZH063 | CP014522 | B2 | ST131 | O25 | H4 | K5 | 22 | 236 | ||
| ZH193 | CP014497 | B2 | ST131 | O25 | H4 | K5 | 30 | 236 | ||
| UMN026 | CU928163 | Urine, acute cystitis | D | ST597 | O17 | H18 | K52 | 27 | 241 | |
| IAI39 | CU928164 | Infected urine | F | ST62 | O7 | H45 | K1 | 44 | 233 | |
| UPEC | VR50 | CP011134 | Urine, ABU | A | ST10 | OR | H- | K1 | 27 | 93, 242 |
| CI5 | CP011018 | Pyelonephritis | B1 | ST5082 | O- | H14 | 1155 | 243 | ||
| ABU 83972 | CP001671 | Urine, ABU | B2 | ST73 | O25 | H1 | K5 | 12 | 244 | |
| CFT073 | AE014075 | Blood, Pyelonephritis | B2 | ST73 | O6 | H1 | K2 | 10 | 245, 246 | |
| UTI89 | CP000243 | Urine, cystitis | B2 | ST95 | O18 | H7 | K1 | 18 | 247, 248 | |
| 536 | CP000247 | Pyelonephritis | B2 | ST127 | O6 | H31 | K15 | 1467 | 249 | |
| K-15KW01 | CP016358 | Fecal, ABU | B2 | ST127 | O6 | H100 | K+ | 310 | 250 | |
| EC958 | HG941718 | Infected urine | B2 | ST131 | O25b | H4 | K100 | 30 | 212 | |
| NA114 | CP002797 | urine, UTI | B2 | ST131 | O25 | H4 | K+ | 30 | 213 | |
| MS6198 | CP015834 | Urine | F | ST648 | O1 | H6 | K+ | 27 | 251 | |
| ST648 | CP008697 | Pleural effusion | F | ST648 | O51 | H4 | K+ | 18 | ||
| NMEC | IHE3034 | CP001969 | B2 | ST95 | O18 | H7 | K1 | 18 | 186, 252 | |
| RS218 | CP007149 | Cerebrospinal fluid | B2 | ST95 | O17 | H7 | K1 | 18 | 253–255 | |
| S88 | CU928161 | Cerebrospinal fluid | B2 | ST95 | O45 | H7 | K1 | 54 | 127 | |
| NMEC O18 | CP007275 | Cerebrospinal fluid | B2 | ST416 | O18 | H7 | K1 | 244 | 194, 256, 257 | |
| MCJCHV-1 | CP030111 | Cerebrospinal fluid | B2 | ST1193 | O75 | H5 | K1 | 64 | 258, 259 | |
| CE10 | CP003034 | Cerebrospinal fluid | F | ST62 | O7 | K1 | 44 | 260 | ||
| APEC | ACN001 | CP007442 | Liver, chicken | C | ST23 | O78 | H9 | 35 | 261 | |
| ACN002 | CP007491 | C | ST23 | O79 | H9 | 35 | ||||
| APEC O78 | CP004009 | Lung, turkey | C | ST23 | O79 | 35 | 262 | |||
| 789 | CP010315 | Avian colisepticemia | C | ST88 | O78 | H19 | 27 | 263 | ||
| APEC O1 | CP000468 | Colibacillosis, turkey | B2 | ST95 | O1 | H7 | K1 | 15 | 264 | |
| O18 | CP006830 | Pericardium/lung, chicken | B2 | ST95 | O18 | H7 | K1 | 15 | 265 | |
| O2-211 | CP006834 | Air sack, chicken | G | ST117 | O2 | H4 | 97 | 266 | ||
| IMT5155 | CP005930 | Colisepticemia, chicken | B2 | ST140 | O2 | H5 | K1 | 15 | 267, 268 | |
| AIEC | LF82 | CU651637 | Ileum, Crohn’s disease | B2 | ST135 | O83 | H1 | K+ | 436 | 269 |
| NRG 857C | CP001855 | Ileum, Crohn’s disease | B2 | ST135 | O83 | H1 | K+ | 2 | 270 | |
| UM146 | CP002167 | Ileum, Crohn’s disease | B2 | ST643 | O18 | H7 | K1 | 18 | 271 | |
| NC101 | AEFA01000000 | Feces, mouse | B2 | ST998 | O2 | H6 | K1 | 1477 | 272, 273 | |
| EHEC/STEC | 11128 | AP010960 | Bloody diarrhea | B1 | ST16 | O111 | H- | 86 | 274 | |
| 12009 | AP010958 | Bloody diarrhea | B1 | ST17 | O103 | H2 | 25 | 274 | ||
| 11368 | AP010953 | Bloody diarrhea | B1 | ST21 | O26 | H11 | 440 | 274 | ||
| CFSAN027343 | CP037943 | Stool | B1 | ST21 | O26 | H11 | 440 | 275 | ||
| E2865 | AP018808 | Cattle | B1 | ST21 | O26 | H11 | 440 | |||
| FORC_028 | CP012693 | Stool | B1 | ST21 | O26 | H11 | 440 | |||
| RM8426 | CP028116 | Creek | B1 | ST21 | O26 | H11 | 440 | |||
| 2011C-3911 | CP015240 | Stool | B1 | ST1727 | O79 | H7 | 31 | 276 | ||
| RM9387 | CP009104 | Feces, cattle | B1 | ST2773 | O104 | H7 | 32 | 277 | ||
| 150 | CP028592 | Cattle | E | ST11 | O157 | H7 | 82 | |||
| 180-PT54 | CP015832 | Outbreak isolate | E | ST11 | O157 | H7 | 82 | 278 | ||
| 1130 | CP017434 | Cattle hide | E | ST11 | O157 | 82 | ||||
| 28RC1 | CP015020 | Bovine carcass | E | ST11 | O157 | H7 | 82 | 279, 280 | ||
| ATCC 43889 | CP015854 | Feces, HUS | E | ST11 | O157 | H7 | 82 | 281, 282 | ||
| EC4115 | CP001164 | Spinach outbreak | E | ST11 | O157 | H7 | 82 | 283 | ||
| EDL933 | CP008957 | Ground beef | E | ST11 | O157 | H7 | 36 | 284 | ||
| FRIK944 | CP016625 | Calf feces | E | ST11 | O157 | H7 | 82 | |||
| FRIK2069 | CP015846 | Feces | E | ST11 | O157 | H7 | 82 | |||
| FRIK2455 | CP015844 | Feces, steer | E | ST11 | O157 | H7 | 82 | 285, 286 | ||
| JEONG-1266 | CP014314 | Feces, steer | E | ST11 | O157 | H7 | 82 | 287 | ||
| Sakai | BA000007 | Stool | E | ST11 | O157 | H7 | 36 | 288, 289 | ||
| SRCC 1675 | CP015023 | Apple cider | E | ST11 | O157 | H7 | 36 | 279, 280 | ||
| SS52 | CP010304 | Feces, cattle | E | ST11 | O157 | H7 | 82 | 290 | ||
| TW14359 | CP001368 | Spinach outbreak | E | ST11 | O157 | H7 | 82 | 291 | ||
| Xuzhou21 | CP001925 | Feces | E | ST11 | O157 | H7 | 36 | 292 | ||
| RM13514 | CP006027 | Lettuce outbreak | E | ST32 | O145 | H28 | 331 | 293 | ||
| 2013C-4465 | CP015241 | Stool | E | ST335 | O55 | H7 | 82 | |||
| RM13516 | CP006262 | Ice cream outbreak | E | ST6130 | O145 | H28 | 54 | 293 | ||
| EAHEC | 2009EL-2050 | CP003297 | Bloody diarrhea | B1 | ST678 | O104 | H4 | 294 | ||
| 2009EL-2071 | CP003301 | Bloody diarrhea | B1 | ST678 | O104 | H4 | 294 | |||
| 2011C-3493 | CP003289 | Stool, HUS | B1 | ST678 | O104 | H4 | 295 | |||
| CC227-11 | CP011331 | Bloody diarrhea | B1 | ST678 | O104 | H4 | 296 | |||
| HUSEC2011 | HF572917 | Stool, HUS | B1 | ST678 | O104 | H4 | ||||
| EAEC | 55989 | CU928145 | Watery diarrhea | B1 | ST678 | O104 | H4 | 233 | ||
| 042 | FN554766 | Diarrhea | D | ST414 | O44 | H18 | K+ | 91 | ||
| ETEC | 90-9272 | CP024239 | Diarrhea | A | ST48 | O15 | H11 | 41 | 297 | |
| H10407 | FN649414 | Diarrhea | A | ST48 | O78 | H11 | K80? | 41 | 120, 298 | |
| UMNK88 | CP002729 | Porcine neonatal diarrhea | A | ST100 | O149 | H10 | 299 | |||
| 214-4 | CP025840 | Stool | A | ST398 | Nontypable | 25 | 300 | |||
| 90-9269 | CP024661 | Diarrhea | B1 | ST761* | OUND | H4 | 297 | |||
| ATCC 43886 | CP024256 | Feces | B1 | ST1312 | O25 | H16 | 198 | 301 | ||
| 90-9281 | CP024243 | Diarrhea | B1 | ST58 | O128 | H27 | 297 | |||
| 103605 | CP025920 | Stool | B1 | ST443 | O115 | H5 | 24 | |||
| E24377A | CP000800 | Diarrhea | B1 | ST1132 | O139 | H28 | 54 | 9, 302, 303 | ||
| 90-9276 | CP024299 | Diarrhea | B1 | ST5305 | O114 | H49 | 32 | 297 | ||
| 90-9280 | CP024240 | Diarrhea | B1 | ST5305 | O114 | H49 | 32 | 297 | ||
| D181 | CP024252 | Diarrhea | B1 | ST4493 | O182 | H21 | 24 | 304 | ||
| 2014EL-1345-2 | CP024223 | E | ST182 | O169 | H41 | 30 | 305 | |||
| EPEC | E2348/69 | FM180568 | Diarrhea | B2 | ST15 | O127 | H6 | 57 | 306 | |
| CB9615 | CP001846 | Diarrhea, infant | E | ST335 | O55 | H7 | 82 | 307 | ||
| RM12579 | CP003109 | Urine | E | ST335 | O55 | H7 | 82 | 308 | ||
K+ indicates a strain that contained kps genes without additional information. If the kps genes were identical to those for strains that had experimentally determined K-types or if the K-type was described with some uncertainty, the K-type is followed by a “?” (for example “K100?”).
FimH types were determined using CGE’s FimTyper (226).
UPECs are characterized by their ability to multiply intracellularly in the urinary tract (25, 214). Many of the virulence factors described for this pathotype are represented in this work because UPECs are among the most studied pathogens to date. The loss of some virulence factors leads to asymptomatic bacteriuria (ABU). Our strain collection has 13 strains specifically labeled UPEC strains; two of these are known to cause ABU (83972 and VR50). The UPEC strains represented here belong to five different phylogroups and eight sequence types.
Strains that belong to the NMEC phylogroup can survive in the bloodstream and invade the meninges of infants (25). NMEC strains are generally considered to be difficult to distinguish from commensals (215).
The avian pathogenic E. coli (APEC) causes colibacillosis in poultry and other avian species and has many of the same virulence factors that are found in ExPEC strains (26).
Four strains belong to the adherent-invasive E. coli (AIEC) pathotype. This pathotype was first described in 1999 with the LF82 strain isolated from the ileal mucosa of a Crohn’s disease (CD) patient (216). AIECs have since been associated with inflammatory bowel disease, are characterized by the ability to invade intestinal epithelial cells and replicate in macrophages, and have been found to bind receptors overexpressed in CD patients using type I fimbriae (29, 86, 217). There have been no unique genetic factors described for this pathotype, other than a weak association with certain alleles of chiA chitinase, a gene encoding a protein shown to promote AIEC adherence to IEC (1, 216, 218). The four complete AIEC genomes here all belong to the B2 phylogroup, the phylogroup to which the majority of isolated AIEC strains belong.
Shiga-toxin-producing E. coli (STEC) strains have acquired the phage-associated Stx toxin (34, 39, 219). This pathotype is a foodborne illness that can produce a wide range of symptoms from asymptomatic to life-threatening hemorrhagic colitis and hemolytic-uremic syndrome (HUS). Technically, pathogenic STEC strains that have the ability to cause disease in humans are called enterohemorrhagic E. coli (EHEC) and belong predominantly to seven serotypes, including the well-known O157:H7 serotype. In practice, however, the terms STEC and EHEC are used somewhat interchangeably, along with verotoxigenic E. coli (VTEC). In addition to the Stx toxin, the most prominent virulence factor found in these strains is the LEE pathogenicity island, which encodes intimin among other things (34). Other described virulence genes in the literature include espP, lpf, efa, toxB, eibG, ehaA, ompA, iha, and paa (32).
Enteroaggregative E. coli (EAEC) is an emerging diarrheagenic pathotype that is characterized by its “stacked-brick” adherence pattern on Hep-2 cells. This pathotype is a serious cause of childhood diarrhea in developing countries and traveler’s diarrhea (35). EAEC can be asymptomatic, but during symptomatic infections the most common symptoms are watery diarrhea, abdominal pain, nausea, and vomiting (220).
During an outbreak of bloody diarrhea in Europe in 2011, a new hybrid pathotype called enteroaggregative hemorrhagic E. coli (EAHEC) was described (37). All known members of this pathotype are EAEC strains of the O104:H4 serotype that have acquired the Stx phage (221).
Enterotoxigenic E. coli (ETEC) is a pathotype that gets its name for the plasmid-transmitted heat-labile enterotoxin (LT) and heat-stable enterotoxin (ST). These strains can carry one or both toxins. LT is similar to cholera toxin and has a similar mechanism of action. ST is a much smaller peptide that binds to guanylate cyclase to increase cyclic GMP in the small intestines. ETEC is the most common cause of E. coli diarrheagenic disease and the leading cause of traveler’s diarrhea. It is also a major cause of acute infectious diarrhea, which accounts for an estimated 20% of all childhood deaths (222).
Enteropathogenic E. coli (EPEC) is another major diarrheagenic E. coli. This pathotype is characterized by the formation of A/E lesions. The attachment to intestinal cells is mediated by intimin, encoded by eae found in the LEE pathogenicity island (40).
Of these strains, 6 were nonpathogenic strains, 2 were lab-derived, 4 were AIEC, 29 were ExPEC/UPEC, 6 were NMEC, 8 were APEC, 28 were EHEC/STEC, 5 were EAHEC, 2 were EAEC, 13 were ETEC, and 3 were EPEC strains. For Clermont phylogroups, 8 belonged to the A phylogroup, 27 to the B1 phylogroup, 39 to the B2 phylogroup, 4 to the C phylogroup, 2 to the D phylogroup, 22 to the E phylogroup, 4 to the F phylogroup, and 1 to the G phylogroup. Table 2 contains a complete list of strains in our pathotype database. Figure S5 shows the pathotype and phylogroup distributions.
The sequence type of each strain was determined using Center for Genomic Epidemiology (CGE) MLST 2.0 software (223, 224). For strains where the O and H serotypes were not known, CGE SeroTypeFinder software was used (225). K-types were listed if they had been experimentally determined or if neu (for K1) or kfi (for K5) operons were present. When the strain contained kps genes without additional information, “K+” is listed in Table 2. If the kps genes were identical to strains that had experimentally determined K-types or if the K-type was described with some uncertainty, the K-type was listed followed by a “?” (for example “K100?”). FimH type was determined using CGE’s FimTyper (226).
ACKNOWLEDGMENTS
We thank Richard Sucgang for his advice on analytics and Hannah Carter, Yikun Xing, Nina Poole, and Sabrina Green for recommending and/or directing us toward information on certain virulence factors in this study.
Footnotes
Supplemental material is available online only.
Contributor Information
J. R. Clark, Email: jrclark@bcm.edu.
A. M. Maresso, Email: maresso@bcm.edu.
Denise Monack, Stanford University.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HLT. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Diard M, Garry L, Selva M, Mosser T, Denamur E, Matic I. 2010. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J Bacteriol 192:4885–4893. 10.1128/JB.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abram K, Udaondo Z, Bleker C, Wanchai V, Wassenaar TM, Robeson MS, Ussery DW. 2021. Mash-based analyses of Escherichia coli genomes reveal 14 distinct phylogroups. Commun Biol 4:117. 10.1038/s42003-020-01626-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler DJ, Peplinski RG, Reed JL, Glasner JD, Perna NT. 2011. The evolution of metabolic networks of Escherichia coli. BMC Syst Biol 5:182. 10.1186/1752-0509-5-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 6.Mainil J. 2013. Escherichia coli virulence factors. Vet Immunol Immunopathol 152:2–12. 10.1016/j.vetimm.2012.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Gov Y, Bitler A, Dell’Acqua G, Torres JV, Balaban N. 2001. RNAIII inhibiting peptide (RIP), a global inhibitor of Staphylococcus aureus pathogenesis: structure and function analysis. Peptides 22:1609–1620. 10.1016/S0196-9781(01)00496-X. [DOI] [PubMed] [Google Scholar]
- 8.Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, Svanborg C, Gottschalk G, Karch H, Hacker J. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol 185:1831–1840. 10.1128/jb.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rasko DA, Rosovitz MJ, Myers GSA, Mongodin EF, Fricke WF, Gajer P, Crabtree J, Sebaihia M, Thomson NR, Chaudhuri R, Henderson IR, Sperandio V, Ravel J. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 190:6881–6893. 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohlin J, Brynildsrud OB, Sekse C, Snipen L. 2014. An evolutionary analysis of genome expansion and pathogenicity in Escherichia coli. BMC Genomics 15:882. 10.1186/1471-2164-15-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denamur E, Clermont O, Bonacorsi S, Gordon D. 2021. The population genetics of pathogenic Escherichia coli. Nat Rev Microbiol 19:37–54. 10.1038/s41579-020-0416-x. [DOI] [PubMed] [Google Scholar]
- 12.Pitout JDD. 2012. Extraintestinal pathogenic Escherichia coli: a combination of virulence with antibiotic resistance. Front Microbiol 3. 10.3389/fmicb.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brumbaugh AR, Mobley HL. 2012. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines 11:663–676. 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poolman JT, Wacker M. 2016. Extraintestinal pathogenic Escherichia coli, a common human pathogen: challenges for vaccine development and progress in the field. J Infect Dis 213:6–13. 10.1093/infdis/jiv429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rojas-Lopez M, Monterio R, Pizza M, Desvaux M, Rosini R. 2018. Intestinal pathogenic Escherichia coli: insights for vaccine development. Front Microbiol 9. 10.3389/fmicb.2018.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pappenheimer AM. 1977. Diphtheria toxin. Annu Rev Biochem 46:69–94. 10.1146/annurev.bi.46.070177.000441. [DOI] [PubMed] [Google Scholar]
- 17.Hadfield TL, McEvoy P, Polotsky Y, Tzinserling VA, Yakovlev AA. 2000. The pathology of diphtheria. J Infect Dis 181:S116–S120. 10.1086/315551. [DOI] [PubMed] [Google Scholar]
- 18.Weiss AA, Hewlett EL. 1986. Virulence factors of Bordetella pertussis. Annu Rev Microbiol 40:661–686. 10.1146/annurev.mi.40.100186.003305. [DOI] [PubMed] [Google Scholar]
- 19.Kilgore PE, Salim AM, Zervos MJ, Schmitt H-J. 2016. Pertussis: microbiology, disease, treatment, and prevention. Clin Microbiol Rev 29:449–486. 10.1128/CMR.00083-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plowe CV. 2011. Vaccine design: innovative approaches and novel strategies. Clin Infect Dis 53:318–318. 10.1093/cid/cir415. [DOI] [Google Scholar]
- 21.Leimbach A, Hacker J, Dobrindt U. 2013. Escherichia coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol 858:3–32. [DOI] [PubMed] [Google Scholar]
- 22.Robins-Browne RM, Holt KE, Ingle DJ, Hocking DM, Yang J, Tauschek M. 2016. Are Escherichia coli pathotypes still relevant in the era of whole-genome sequencing? Front Cell Infect Microbiol 6:141. 10.3389/fcimb.2016.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto S. 2007. Molecular epidemiology of uropathogenic Escherichia coli. J Infect Chemother 13:68–73. 10.1007/s10156-007-0506-Y. [DOI] [PubMed] [Google Scholar]
- 24.Wiles TJ, Kulesus RR, Mulvey MA. 2008. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol 85:11–19. 10.1016/j.yexmp.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarowska J, Futoma-Koloch B, Jama-Kmiecik A, Frej-Madrzak M, Ksiazczyk M, Bugla-Ploskonska G, Choroszy-Krol I, Ewers C, Janssen T, Wieler LH. 2019. Virulence factors, prevalence, and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog 11:10. 10.1186/s13099-019-0290-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ewers C, Janssen T, Wieler LH. 2003. Avian pathogenic Escherichia coli (APEC). Berl Munch Tierarztl Wochenschr 116:381–395. [PubMed] [Google Scholar]
- 27.Dho-Moulin M, Fairbrother JM. 1999. Mar-Jun. Avian pathogenic Escherichia coli (APEC). Vet Res 30:299–316. [PubMed] [Google Scholar]
- 28.Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. 2005. Characterizing the APEC pathotype. Vet Res 36:241–256. 10.1051/vetres:2004057. [DOI] [PubMed] [Google Scholar]
- 29.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CHF, Stanners CP, Darfeuille-Michaud A. 2009. Crohn’s disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med 206:2179–2189. 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friswell M, Campbell B, Rhodes J. 2010. The role of bacteria in the pathogenesis of inflammatory bowel disease. Gut Liver 4:295–306. 10.5009/gnl.2010.4.3.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Liao Y, Ma Y, Gong W, Zhu G. 2017. The role of major virulence factors of AIEC involved in inflammatory bowl disease: a mini-review. Appl Microbiol Biotechnol 101:7781–7787. 10.1007/s00253-017-8507-y. [DOI] [PubMed] [Google Scholar]
- 32.Clements A, Young JC, Constantinou N, Frankel G. 2012. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 3:71–87. 10.4161/gmic.19182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cieza RJ, Hu J, Ross BN, Sbrana E, Torres AG. 2015. The IbeA invasin of adherent-invasive Escherichia coli mediates interaction with intestinal epithelia and macrophages. Infect Immun 83:1904–1918. 10.1128/IAI.03003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delannoy S, Beutin L, Fach P. 2013. Discrimination of enterohemorrhagic Escherichia coli (EHEC) from non-EHEC strains based on detection of various combinations of type III effector genes. J Clin Microbiol 51:3257–3262. 10.1128/JCM.01471-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaur P, Chakraborti A, Asea A2. 2010. Enteroaggregative Escherichia coli: an emerging enteric food borne pathogen. Interdiscip Perspect Infect Dis 2010:254159–254110. 10.1155/2010/254159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zamboni A, Fabbricotti SH, Fagundes-Neto U, Scaletsky ICA. 2004. Enteroaggregative Escherichia coli virulence factors are found to be associated with infantile diarrhea in Brazil. J Clin Microbiol 42:1058–1063. 10.1128/jcm.42.3.1058-1063.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beutin L, Martin A. 2012. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J Food Prot 75:408–418. 10.4315/0362-028X.JFP-11-452. [DOI] [PubMed] [Google Scholar]
- 38.Gaastra W, Svennerholm AM. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 4:444–452. 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 39.Blanco M, Schumacher S, Tasara T, Zweifel C, Blanco JE, Dahbi G, Blanco J, Stephan R. 2005. Serotypes, intimin variants, and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-η2). BMC Microbiol 5:23–11. 10.1186/1471-2180-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic Escherichia coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520. 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 41.Peleg A, Shifrin Y, Ilan O, Nadler-Yona C, Nov S, Koby S, Baruch K, Altuvia S, Elgrably-Weiss M, Abe CM, Knutton S, Saper MA, Rosenshine I. 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J Bacteriol 187:5259–5266. 10.1128/JB.187.15.5259-5266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herzer PJ, Inouye S, Inouye M, Whittam TS. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol 172:6175–6181. 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tenaillon O, Skurnik D, Picard B, Denamur E. 2010. The population genetics of commensal Escherichia coli. Nat Rev Microbiol 8:207–217. 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 45.Honsa ES, Maresso AW, Sarah E, Anthony H, Maresso W. 2011. Mechanisms of iron import in anthrax. Biometals 24:533–545. 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 46.Skaar EP. 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6:e1000949. 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Contreras H, Chim N, Credali A, Goulding CW. 2014. Heme uptake in bacterial pathogens. Curr Opin Chem Biol 19:34–41. 10.1016/j.cbpa.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nowrouzian FL, Wold AE, Adlerberth I. 2005. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J Infect Dis 191:1078–1083. 10.1086/427996. [DOI] [PubMed] [Google Scholar]
- 50.Le Gall T, Clermont O, Gouriou S, Picard B, Nassif X, Denamur E, Tenaillon O. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol Biol Evol 24:2373–2384. 10.1093/molbev/msm172. [DOI] [PubMed] [Google Scholar]
- 51.Papouskova A, Masarikova M, Valcek A, Senk D, Cejkova D, Jahodarova E, Cizek A. 2020. Genomic analysis of Escherichia coli strains isolated from diseased chicken in the Czech Republic. BMC Vet Res 16:189. 10.1186/s12917-020-02407-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cordoni G, Woodward MJ, Wu H, Alanazi M, Wallis T, La Ragione RM. 2016. Comparative genomics of European avian pathogenic Escherichia coli (APEC). BMC Genomics 17:960. 10.1186/s12864-016-3289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borzi MM, Cardozo MV, de Oliveira ES, de Pollo AS, Guastalli EAL, dos Santos LF, de Ávila FA. 2018. Characterization of avian pathogenic Escherichia coli isolated from free-range helmeted guineafowl. Brazilian J Microbiol 49:107–112. 10.1016/j.bjm.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vangchhia B, Abraham S, Bell JM, Collignon P, Gibson JS, Ingram PR, Johnson JR, Kennedy K, Trott DJ, Turnidge JD, Gordon DM. 2016. Phylogenetic diversity, antimicrobial susceptibility, and virulence characteristics of phylogroup F Escherichia coli in Australia. Microbiology (Reading) 162:1904–1912. 10.1099/mic.0.000367. [DOI] [PubMed] [Google Scholar]
- 55.Clermont O, Dixit OVA, Vangchhia B, Condamine B, Dion S, Bridier‐Nahmias A, Denamur E, Gordon D. 2019. Characterization and rapid identification of phylogroup G in Escherichia coli, a lineage with high virulence and antibiotic resistance potential. Environ Microbiol 21:3107–3117. 10.1111/1462-2920.14713. [DOI] [PubMed] [Google Scholar]
- 56.Sayers S, Li L, Ong E, Deng S, Fu G, Lin Y, Yang B, Zhang S, Fa Z, Zhao B, Xiang Z, Li Y, Zhao X-M, Olszewski MA, Chen L, He Y. 2019. Victors: a web-based knowledge base of virulence factors in human and animal pathogens. Nucleic Acids Res 47:D693–D700. 10.1093/nar/gky999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 47:D606–D612. 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roubaud Baudron C, Panhard X, Clermont O, Mentre F, Fantin B, Denamur E, Lefort A. 2014. Escherichia coli bacteremia in adults: age-related differences in clinical and bacteriological characteristics, and outcome. Epidemiol Infect 142:2672–2683. 10.1017/S0950268814000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gordon DM, Clermont O, Tolley H, Denamur E. 2008. Assigning Escherichia coli strains to phylogenetic groups: multilocus sequence typing versus the PCR triplex method. Environ Microbiol 10:2484–2496. 10.1111/j.1462-2920.2008.01669.x. [DOI] [PubMed] [Google Scholar]
- 61.Clermont O, Christenson JK, Denamur E, Gordon DM. 2013. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylogroups. Environ Microbiol Rep 5:58–65. 10.1111/1758-2229.12019. [DOI] [PubMed] [Google Scholar]
- 62.Miajlovic H, Smith SG. 2014. Bacterial self-defence: how Escherichia coli evades serum killing. FEMS Microbiol Lett 354:1–9. 10.1111/1574-6968.12419. [DOI] [PubMed] [Google Scholar]
- 63.Sperandeo P, Martorana AM, Polissi A. 2017. Lipopolysaccharide biogenesis and transport at the outer membrane of Gram-negative bacteria. Biochim Biophys Acta Mol Cell Biol Lipids 1862:1451–1460. 10.1016/j.bbalip.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 64.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 65.Whitfield GB, Marmont LS, Howell PL. 2015. Enzymatic modifications of exopolysaccharides enhance bacterial persistence. Front Microbiol 6:471. 10.3389/fmicb.2015.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sachdeva S, Palur RV, Sudhakar KU, Rathinavelan T. 2017. E. coli group 1 capsular polysaccharide exportation nanomachinary as a plausible antivirulence target in the perspective of emerging antimicrobial resistance. Front Microbiol 8:70. 10.3389/fmicb.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Franzin FM, Sircili MP. 2015. Locus of enterocyte effacement: a pathogenicity island involved in the virulence of enteropathogenic and enterohemorragic Escherichia coli subjected to a complex network of gene regulation. Biomed Res Int 2015:534738–534710. 10.1155/2015/534738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. 2007. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 56:669–675. 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buckles EL, Wang X, Lane MC, Lockatell CV, Johnson DE, Rasko DA, Mobley HLT, Donnenberg MS. 2009. Role of the K2 capsule in Escherichia coli urinary tract infection and serum resistance. J Infect Dis 199:1689–1697. 10.1086/598524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Goller CC, Seed PC. 2010. Revisiting the Escherichia coli polysaccharide capsule as a virulence factor during urinary tract infection: contribution to intracellular biofilm development. Virulence 1:333–337. 10.4161/viru.1.4.12388. [DOI] [PubMed] [Google Scholar]
- 71.Hafez M, Hayes K, Goldrick M, Warhurst G, Grencis R, Roberts IS. 2009. The K5 capsule of Escherichia coli strain Nissle 1917 is important in mediating interactions with intestinal epithelial cells and chemokine induction. Infect Immun 77:2995–3003. 10.1128/IAI.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim KJ, Elliott SJ, Di Cello F, Stins MF, Kim KS. 2003. The K1 capsule modulates trafficking of Escherichia coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell Microbiol 5:245–252. 10.1046/j.1462-5822.2003.t01-1-00271.x. [DOI] [PubMed] [Google Scholar]
- 73.Bertozzi Silva J, Storms Z, Sauvageau D. 2016. Host receptors for bacteriophage adsorption. FEMS Microbiol Lett 363:fnw002. 10.1093/femsle/fnw002. [DOI] [PubMed] [Google Scholar]
- 74.Mangalea MR, Duerkop BA. 2020. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect Immun 88:e00926-19. 10.1128/IAI.00926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Antão E-M, Wieler LH, Ewers C. 2009. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog 1:22. 10.1186/1757-4749-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Klemm P. 1985. Fimbrial adhesins of Escherichia coli. Rev Infect Dis 7:321–340. 10.1093/clinids/7.3.321. [DOI] [PubMed] [Google Scholar]
- 77.Brinton CC. 1965. The structure, function, synthesis, and genetic control of bacterial pili and a molecular model for DNA and RNA transport in Gram-negative bacteria. Trans N Y Acad Sci 27:1003–1054. 10.1111/j.2164-0947.1965.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 78.Duguid JP, Smith IW, Dempster G, Edmunds PN. 1955. Non-flagellar filamentous appendages (“fimbriæ”) and hæmagglutinating activity in bacterium coli. J Pathol Bacteriol 70:335–348. 10.1002/path.1700700210. [DOI] [PubMed] [Google Scholar]
- 79.Johnson JR. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin Microbiol Rev 4:80–128. 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bergsten G, Wullt B, Svanborg C. 2005. Escherichia coli, fimbriae, bacterial persistence, and host response induction in the human urinary tract. Int J Med Microbiol 295:487–502. 10.1016/j.ijmm.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 81.Dhakal BK, Kulesus RR, Mulvey MA. 2008. Mechanisms and consequences of bladder cell invasion by uropathogenic Escherichia coli. Eur J Clin Invest 38:2–11. 10.1111/j.1365-2362.2008.01986.x. [DOI] [PubMed] [Google Scholar]
- 82.Pouttu R, Puustinen T, Virkola R, Hacker J, Klemm P, Korhonen TK. 1999. Amino acid residue Ala-62 in the FimH fimbrial adhesin is critical for the adhesiveness of meningitis-associated Escherichia coli to collagens. Mol Microbiol 31:1747–1757. 10.1046/j.1365-2958.1999.01311.x. [DOI] [PubMed] [Google Scholar]
- 83.Sokurenko EV, Courtney HS, Ohman DE, Klemm P, Hasty DL. 1994. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J Bacteriol 176:748–755. 10.1128/jb.176.3.748-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kukkonen M, Raunio T, Virkola R, Lähteenmäki K, Mäkelä PH, Klemm P, Clegg S, Korhonen TK. 1993. Basement membrane carbohydrate as a target for bacterial adhesion: binding of type I fimbriae of Salmonella enterica and Escherichia coli to laminin. Mol Microbiol 7:229–237. 10.1111/j.1365-2958.1993.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 85.Sharon N. 1987. Bacterial lectins, cell-cell recognition and infectious disease. FEBS Lett 217:145–157. 10.1016/0014-5793(87)80654-3. [DOI] [PubMed] [Google Scholar]
- 86.Barnich N, Carvalho FA, Glasser A, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel J-F, Darfeuille-Michaud A. 2007. CEACAM6 acts as a receptor for mucosa colonization in Crohn disease. J Clin Invest 117:1566–1574. 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Connell I, Agace W, Klemm P, Schembri M, Mărild S, Svanborg C. 1996. Type 1 fimbrial expression enhances Escherichia coli virulence for the urinary tract. Proc Natl Acad Sci U S A 93:9827–9832. 10.1073/pnas.93.18.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wright KJ, Seed PC, Hultgren SJ. 2007. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell Microbiol 9:2230–2241. 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 89.Bergsten G, Wullt B, Schembri MA, Leijonhufvud I, Svanborg C. 2007. Do type 1 fimbriae promote inflammation in the human urinary tract? Cell Microbiol 9:1766–1781. 10.1111/j.1462-5822.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- 90.Moreira CG, Carneiro SM, Nataro JP, Trabulsi LR, Elias WP. 2003. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol Lett 226:79–85. 10.1016/S0378-1097(03)00561-5. [DOI] [PubMed] [Google Scholar]
- 91.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. 10.1371/journal.pone.0008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Servin AL. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev 18:264–292. 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beatson SA, Ben Zakour NL, Totsika M, Forde BM, Watts RE, Mabbett AN, Szubert JM, Sarkar S, Phan M-D, Peters KM, Petty NK, Alikhan N-F, Sullivan MJ, Gawthorne JA, Stanton-Cook M, Nhu NTK, Chong TM, Yin W-F, Chan K-G, Hancock V, Ussery DW, Ulett GC, Schembri MA. 2015. Molecular analysis of asymptomatic bacteriuria Escherichia coli strain VR50 reveals adaptation to the urinary tract by gene acquisition. Infect Immun 83:1749–1764. 10.1128/IAI.02810-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Le Bouguenec C, Lalioui L, Du Merle L, Jouve M, Courcoux P, Bouzari S, Selvarangan R, Nowicki BJ, Germani Y, Andremont A, Gounon P, Garcia M-I. 2001. Characterization of AfaE adhesins produced by extraintestinal and intestinal human Escherichia coli isolates: PCR assays for detection of Afa adhesins that do or do not recognize Dr blood group antigens. J Clin Microbiol 39:1738–1745. 10.1128/JCM.39.5.1738-1745.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li Y-F, Poole S, Nishio K, Jang K, Rasulova F, McVeigh A, Savarino SJ, Xia D, Bullitt E. 2009. Structure of CFA/I fimbriae from enterotoxigenic Escherichia coli. Proc Natl Acad Sci U S A 106:10793–10798. 10.1073/pnas.0812843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Samadder P, Xicohtencatl-Cortes J, Saldaña Z, Jordan D, Tarr PI, Kaper JB, Girón JA. 2009. The Escherichia coli ycbQRST operon encodes fimbriae with laminin-binding and epithelial cell adherence properties in Shiga-toxigenic E. coli O157:H7. Environ Microbiol 11:1815–1826. 10.1111/j.1462-2920.2009.01906.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Riegman N, Kusters R, Van Veggel H, Bergmans H, Van Bergen En Henegouwen P, Hacker J, Van Die I. 1990. F1C fimbriae of a uropathogenic Escherichia coli strain: genetic and functional organization of the foc gene cluster and identification of minor subunits. J Bacteriol 172:1114–1120. 10.1128/jb.172.2.1114-1120.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bäckhed F, Alsén B, Roche N, Ångström J, von Euler A, Breimer ME, Westerlund-Wikström B, Teneberg S, Richter-Dahlfors A. 2002. Identification of target tissue glycosphingolipid receptors for uropathogenic, F1C-fimbriated Escherichia coli and its role in mucosal inflammation. J Biol Chem 277:18198–18205. 10.1074/jbc.M111640200. [DOI] [PubMed] [Google Scholar]
- 99.Klemm P, Christiansen G, Kreft B, Marre R, Bergmans H. 1994. Reciprocal exchange of minor components of type 1 and F1C fimbriae results in hybrid organelles with changed receptor specificities. J Bacteriol 176:2227–2234. 10.1128/jb.176.8.2227-2234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Khan AS, Kniep B, Oelschlaeger TA, Van Die I, Korhonen T, Hacker J. 2000. Receptor structure for F1C fimbriae of uropathogenic Escherichia coli. Infect Immun 68:3541–3547. 10.1128/iai.68.6.3541-3547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mulvey MA. 2002. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol 4:257–271. 10.1046/j.1462-5822.2002.00193.x. [DOI] [PubMed] [Google Scholar]
- 102.Lasaro MA, Salinger N, Zhang J, Wang Y, Zhong Z, Goulian M, Zhu J. 2009. F1C fimbriae play an important role in biofilm formation and intestinal colonization by the Escherichia coli commensal strain Nissle 1917. Appl Environ Microbiol 75:246–251. 10.1128/AEM.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, Hacker J. 2002. Genetic structure and distribution of four pathogenicity islands (PAI I536 to PAI IV536) of uropathogenic Escherichia coli strain 536. Infect Immun 70:6365–6372. 10.1128/iai.70.11.6365-6372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ott M, Hoschützky H, Jann K, Van Die I, Hacker J. 1988. Gene clusters for S fimbrial adhesin (sfa) and F1C fimbriae (foc) of Escherichia coli: comparative aspects of structure and function. J Bacteriol 170:3983–3990. 10.1128/jb.170.9.3983-3990.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Adlerberth I, Hanson ÅL, Svanborg C, Svennerholm A, Nordgren S, Wold AE. 1995. Adhesins of Escherichia coli-associated with extra-intestinal pathogenicity confer binding to colonic epithelial cells. Microb Pathog 18:373–385. 10.1006/mpat.1995.0034. [DOI] [PubMed] [Google Scholar]
- 106.Hull RA, Gill RE, Hsu P, Minshew BH, Falkow S. 1981. Construction and expression of recombinant plasmids encoding type 1 or d-mannose-resistant pili from a urinary tract infection Escherichia coli isolate. Infect Immun 33:933–938. 10.1128/IAI.33.3.933-938.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rasko DA, Phillips JA, Li X, Mobley HLT. 2001. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. J Infect Dis 184:1041–1049. 10.1086/323602. [DOI] [PubMed] [Google Scholar]
- 108.Jadhav S, Hussain A, Devi S, Kumar A, Parveen S, Gandham N, Wieler LH, Ewers C, Ahmed N. 2011. Virulence characteristics and genetic affinities of multiple drug-resistant uropathogenic Escherichia coli from a semi-urban locality in India. PLoS One 6:e18063. 10.1371/journal.pone.0018063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lindberg S, Xia Y, Sondén B, Göransson M, Hacker J, Uhlin BE. 2008. Regulatory interactions among adhesin gene systems of uropathogenic Escherichia coli. Infect Immun 76:771–780. 10.1128/IAI.01010-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Simms AN, Mobley HLT. 2008. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect Immun 76:4833–4841. 10.1128/IAI.00630-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kawamura T, Le LUK, Zhou H, Dahlquist FW. 2007. Solution structure of Escherichia coli PapI, a key regulator of the Pap pilus phase variation. J Mol Biol 365:1130–1142. 10.1016/j.jmb.2006.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lund B, Lindberg F, Marklund BI, Normark S. 1987. The PapG protein is the α-d-galactopyranosyl-(1–4)-β-d-galactopyranose-binding adhesin of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 84:5898–5902. 10.1073/pnas.84.16.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilks A, Heinzl G. 2014. Heme oxygenation and the widening paradigm of heme degradation. Arch Biochem Biophys 544:87–95. 10.1016/j.abb.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Weinberg ED. 2009. Iron availability and infection. Biochim Biophys Acta 1790:600–605. 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 115.Skaar EP, Raffatellu M. 2015. Metals in infectious diseases and nutritional immunity. Metallomics 7:926–928. 10.1039/c5mt90021b. [DOI] [PubMed] [Google Scholar]
- 116.Subashchandrabose S, Mobley HLT. 2015. Back to the metal age: battle for metals at the host-pathogen interface during urinary tract infection. Metallomics 7:935–942. 10.1039/c4mt00329b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Galardini M, Clermont O, Baron A, Busby B, Dion S, Schubert S, Beltrao P, Denamur E. 2020. Major role of iron uptake systems in the intrinsic extra-intestinal virulence of the genus Escherichia revealed by a genome-wide association study. PLoS Genet 16:e1009065. 10.1371/journal.pgen.1009065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Perry RD, Fetherston JD. 2011. Yersiniabactin iron uptake: mechanisms and role in Yersinia pestis pathogenesis. Microbes Infect 13:808–817. 10.1016/j.micinf.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Torres AG, Payne SM. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol Microbiol 23:825–833. 10.1046/j.1365-2958.1997.2641628.x. [DOI] [PubMed] [Google Scholar]
- 120.Crossman LC, Chaudhuri RR, Beatson SA, Wells TJ, Desvaux M, Cunningham AF, Petty NK, Mahon V, Brinkley C, Hobman JL, Savarino SJ, Turner SM, Pallen MJ, Penn CW, Parkhill J, Turner AK, Johnson TJ, Thomson NR, Smith SGJ, Henderson IR. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J Bacteriol 192:5822–5831. 10.1128/JB.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Reigstad CS, Hultgren SJ, Gordon JI. 2007. Functional genomic studies of uropathogenic Escherichia coli and host urothelial cells when intracellular bacterial communities are assembled. J Biol Chem 282:21259–21267. 10.1074/jbc.M611502200. [DOI] [PubMed] [Google Scholar]
- 122.Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69:6179–6185. 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Spurbeck RR, Dinh PC, Walk ST, Stapleton AE, Hooton TM, Nolan LK, Kim KS, Johnson JR, Mobley HLT. 2012. Escherichia coli isolates that carry vat, fyuA, chuA, and yfcV efficiently colonize the urinary tract. Infect Immun 80:4115–4122. 10.1128/IAI.00752-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Barnard TJ, Johnson JR. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect Immun 70:7156–7160. 10.1128/iai.70.12.7156-7160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Desvaux M, Dalmasso G, Beyrouthy R, Barnich N, Delmas J, Bonnet R. 2020. Pathogenicity factors of genomic islands in intestinal and extraintestinal Escherichia coli. Front Microbiol 11:2065. 10.3389/fmicb.2020.02065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sabri M, Léveillé S, Dozois CM. 2006. A SitABCD homologue from an avian pathogenic Escherichia coli strain mediates transport of iron and manganese and resistance to hydrogen peroxide. Microbiology (Reading) 152:745–758. 10.1099/mic.0.28682-0. [DOI] [PubMed] [Google Scholar]
- 127.Peigne C, Bidet P, Mahjoub-Messai F, Plainvert C, Barbe V, Médigue C, Frapy E, Nassif X, Denamur E, Bingen E, Bonacorsi S. 2009. The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model. Infect Immun 77:2272–2284. 10.1128/IAI.01333-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Y, Dai J, Zhuge X, Wang H, Hu L, Ren J, Chen L, Li D, Tang F. 2016. Iron-regulated gene ireA in avian pathogenic Escherichia coli participates in adhesion and stress-resistance. BMC Vet Res 12:167. 10.1186/s12917-016-0800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Feldmann F, Sorsa LJ, Hildinger K, Schubert S. 2007. The salmochelin siderophore receptor IroN contributes to invasion of urothelial cells by extraintestinal pathogenic Escherichia coli in vitro. Infect Immun 75:3183–3187. 10.1128/IAI.00656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bauer RJ, Zhang L, Foxman B, Siitonen A, Jantunen ME, Saxen H, Marrs CF. 2002. Molecular epidemiology of 3 putative virulence genes for Escherichia coli urinary tract infection: usp, iha, and iroN E. coli. J Infect Dis 185:1521–1524. 10.1086/340206. [DOI] [PubMed] [Google Scholar]
- 131.Hantke K, Nicholson G, Rabsch W, Winkelmann G. 2003. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A 100:3677–3682. 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Russo TA, Carlino UB, Mong A, Jodush ST. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect Immun 67:5306–5314. 10.1128/IAI.67.10.5306-5314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Caza M, Lépine F, Milot S, Dozois CM. 2008. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect Immun 76:3539–3549. 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HLT. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog 6:e1001187. 10.1371/journal.ppat.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Garcia EC, Brumbaugh AR, Mobley HLT. 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun 79:1225–1235. 10.1128/IAI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HLT. 2004. Transcriptome of uropathogenic Escherichia coli during urinary tract infection. Infect Immun 72:6373–6381. 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hagan EC, Mobley HLT. 2009. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol Microbiol 71:79–91. 10.1111/j.1365-2958.2008.06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mike LA, Smith SN, Sumner CA, Eaton KA, Mobley HLT. 2016. Siderophore vaccine conjugates protect against uropathogenic Escherichia coli urinary tract infection. Proc Natl Acad Sci U S A 113:13468–13473. 10.1073/pnas.1606324113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gorden PJ, Kleinhenz MD, Ydstie JA, Brick TA, Slinden LM, Peterson MP, Straub DE, Burkhardt DT. 2018. Efficacy of vaccination with a Klebsiella pneumoniae siderophore receptor protein vaccine for reduction of Klebsiella mastitis in lactating cattle. J Dairy Sci 101:10398–10408. 10.3168/jds.2017-14267. [DOI] [PubMed] [Google Scholar]
- 140.Fox JT, Thomson DU, Drouillard JS, Thornton AB, Burkhardt DT, Emery DA, Nagaraja TG. 2009. Efficacy of Escherichia coli O157:H7 siderophore receptor/porin protein-based vaccine in feedlot cattle naturally shedding E. coli O157. Foodborne Pathog Dis 6:893–899. 10.1089/fpd.2009.0336. [DOI] [PubMed] [Google Scholar]
- 141.Thornton AB, Thomson DU, Loneragan GH, Fox JT, Burkhardt DT, Emery DA, Nagaraja TG. 2009. Effects of a siderophore receptor and porin protein-based vaccination on fecal shedding of Escherichia coli O157:H7 in experimentally inoculated cattle. J Food Prot 72:866–869. 10.4315/0362-028X-72.4.866. [DOI] [PubMed] [Google Scholar]
- 142.Sassone-Corsi M, Chairatana P, Zheng T, Perez-Lopez A, Edwards RA, George MD, Nolan EM, Raffatellu M. 2016. Siderophore-based immunization strategy to inhibit growth of enteric pathogens. Proc Natl Acad Sci U S A 113:13462–13467. 10.1073/pnas.1606290113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Saha M, Sarkar S, Sarkar B, Sharma BK, Bhattacharjee S, Tribedi P. 2016. Microbial siderophores and their potential applications: a review. Environ Sci Pollut Res Int 23:3984–3999. 10.1007/s11356-015-4294-0. [DOI] [PubMed] [Google Scholar]
- 144.Habibi M, Asadi Karam MR, Bouzari S. 2017. Evaluation of prevalence, immunogenicity and efficacy of FyuA iron receptor in uropathogenic Escherichia coli isolates as a vaccine target against urinary tract infection. Microb Pathog 110:477–483. 10.1016/j.micpath.2017.07.037. [DOI] [PubMed] [Google Scholar]
- 145.Forsyth VS, Himpsl SD, Smith SN, Sarkissian CA, Mike LA, Stocki JA, Sintsova A, Alteri CJ, Mobley HLT. 2020. Optimization of an experimental vaccine to prevent Escherichia coli urinary tract infection. mBio 11:e00555-20. 10.1128/mBio.00555-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. 2011. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology (Reading) 157:1851–1875. 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hofman P, Le Negrate G, Mograbi B, Hofman V, Brest P, Alliana-Schmid A, Flatau G, Boquet P, Rossi B. 2000. Escherichia coli cytotoxic necrotizing factor-1 (CNF-1) increases the adherence to epithelia and the oxidative burst of human polymorphonuclear leukocytes but decreases bacteria phagocytosis. J Leukoc Biol 68:522–528. [PubMed] [Google Scholar]
- 148.Ludwig A, von Rhein C, Bauer S, Hüttinger C, Goebel W. 2004. Molecular analysis of cytolysin A (ClyA) in pathogenic Escherichia coli strains. J Bacteriol 186:5311–5320. 10.1128/JB.186.16.5311-5320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wallace AJ, Stillman TJ, Atkins A, Jamieson SJ, Bullough PA, Green J, Artymiuk PJ. 2000. Escherichia coli hemolysin E (HlyE, ClyA, SheA). Cell 100:265–276. 10.1016/S0092-8674(00)81564-0. [DOI] [PubMed] [Google Scholar]
- 150.Kerenyi M, Allison HE, Batai I, Sonnevend A, Emody L, Plaveczky N, Pal T. 2005. Occurrence of hlyA and sheA genes in extraintestinal Escherichia coli strains. J Clin Microbiol 43:2965–2968. 10.1128/JCM.43.6.2965-2968.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wiles TJ, Mulvey MA. 2013. The RTX pore-forming toxin α-hemolysin of uropathogenic Escherichia coli: progress and perspectives. Future Microbiol 8:73–84. 10.2217/fmb.12.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schroeder GN, Hilbi H. 2008. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev 21:134–156. 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Estrada-Garcia T, Navarro-Garcia F. 2012. Enteroaggregative Escherichia coli pathotype: a genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol Med Microbiol 66:281–298. 10.1111/j.1574-695X.2012.01008.x. [DOI] [PubMed] [Google Scholar]
- 154.Vila J, Vargas M, Henderson IR, Gascón J, Nataro JP. 2000. Enteroaggregative Escherichia coli virulence factors in traveler’s diarrhea strains. J Infect Dis 182:1780–1783. 10.1086/317617. [DOI] [PubMed] [Google Scholar]
- 155.Mirzarazi M, Rezatofighi SE, Pourmahdi M, Mohajeri MR. 2015. Occurrence of genes encoding enterotoxins in uropathogenic Escherichia coli isolates. Braz J Microbiol 46:155–159. 10.1590/S1517-838246120130860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Östblom A, Adlerberth I, Wold AE, Nowrouzian FL. 2011. Pathogenicity island markers, virulence determinants malX and usp, and the capacity of Escherichia coli to persist in infants’ commensal microbiotas. Appl Environ Microbiol 77:2303–2308. 10.1128/AEM.02405-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hanley P, Lalonde G, Ji G. 1991. Alpha-hemolysin contributes to the pathogenicity of piliated digalactoside-binding Escherichia coli in the kidney: efficacy of an alpha-hemolysin vaccine in preventing renal injury in the BALB/c mouse model of pyelonephritis. Infect Immun 59:1153–1161. 10.1128/IAI.59.3.1153-1161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Smith MA, Weingarten RA, Russo LM, Ventura CL, O’Brien AD. 2015. Antibodies against hemolysin and cytotoxic necrotizing factor type 1 (CNF1) reduce bladder inflammation in a mouse model of urinary tract infection with toxigenic uropathogenic Escherichia coli. Infect Immun 83:1661–1673. 10.1128/IAI.02848-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Henderson IR, Navarro-Garcia F, Desvaux M, Fernandez RC, Ala’Aldeen D. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol Mol Biol Rev 68:692–744. 10.1128/MMBR.68.4.692-744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wells TJ, Totsika M, Schembri MA. 2010. Autotransporters of Escherichia coli: a sequence-based characterization. Microbiology (Reading) 156:2459–2469. 10.1099/mic.0.039024-0. [DOI] [PubMed] [Google Scholar]
- 161.Isberg RR, Voorhis DL, Falkow S. 1987. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell 50:769–778. 10.1016/0092-8674(87)90335-7. [DOI] [PubMed] [Google Scholar]
- 162.Haltner E, Easson JH, Lehr C-M. 1997. Lectins and bacterial invasion factors for controlling endo- and transcytosis of bioadhesive drug carrier systems. Eur J Pharm Biopharm 44:3–13. 10.1016/S0939-6411(97)00096-9. [DOI] [Google Scholar]
- 163.Roche AJ, McFadden JP, Owen P. 2001. Antigen 43, the major phase-variable protein of the Escherichia coli outer membrane, can exist as a family of proteins encoded by multiple alleles. Microbiology 147:161–169. 10.1099/00221287-147-1-161. [DOI] [PubMed] [Google Scholar]
- 164.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo J-M, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75:3233–3244. 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol 10:589–604. 10.1111/j.1462-2920.2007.01479.x. [DOI] [PubMed] [Google Scholar]
- 166.Paxman JJ, Lo AW, Sullivan MJ, Panjikar S, Kuiper M, Whitten AE, Wang G, Luan C-H, Moriel DG, Tan L, Peters KM, Phan M-D, Gee CL, Ulett GC, Schembri MA, Heras B. 2019. Unique structural features of a bacterial autotransporter adhesin suggest mechanisms for interaction with host macromolecules. Nat Commun 10:1967. 10.1038/s41467-019-09814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Allsopp LP, Beloin C, Ulett GC, Valle J, Totsika M, Sherlock O, Ghigo J-M, Schembri MA. 2012. Molecular characterization of UpaB and UpaC, two new autotransporter proteins of uropathogenic Escherichia coli CFT073. Infect Immun 80:321–332. 10.1128/IAI.05322-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect Immun 78:1659–1669. 10.1128/IAI.01010-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Allsopp LP, Beloin C, Moriel DG, Totsika M, Ghigo J-M, Schembri MA. 2012. Functional heterogeneity of the UpaH autotransporter protein from uropathogenic Escherichia coli. J Bacteriol 194:5769–5782. 10.1128/JB.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo J-M, Beloin C. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol 190:4147–4161. 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Totsika M, Wells TJ, Beloin C, Valle J, Allsopp LP, King NP, Ghigo J-M, Schembri MA. 2012. Molecular characterization of the EhaG and UpaG trimeric autotransporter proteins from pathogenic Escherichia coli. Appl Environ Microbiol 78:2179–2189. 10.1128/AEM.06680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Stein M, Kenny B, Stein MA, Finlay BB. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J Bacteriol 178:6546–6554. 10.1128/jb.178.22.6546-6554.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mellies JL, Navarro-Garcia F, Okeke I, Frederickson J, Nataro JP, Kaper JB. 2001. espC pathogenicity island of enteropathogenic Escherichia coli encodes an enterotoxin. Infect Immun 69:315–324. 10.1128/IAI.69.1.315-324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Stathopoulos C, Provence DL, Curtiss R. 1999. Characterization of the avian pathogenic Escherichia coli hemagglutinin Tsh, a member of the immunoglobulin A protease-type family of autotransporters. Infect Immun 67:772–781. 10.1128/IAI.67.2.772-781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dozois CM, Dho-Moulin M, Brée A, Fairbrother JM, Desautels C, Curtiss R. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the tsh genetic region. Infect Immun 68:4145–4154. 10.1128/iai.68.7.4145-4154.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Vijayakumar V, Santiago A, Smith R, Smith M, Robins-Browne RM, Nataro JP, Ruiz-Perez F. 2014. Role of class 1 serine protease autotransporter in the pathogenesis of Citrobacter rodentium colitis. Infect Immun 82:2626–2636. 10.1128/IAI.01518-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Celik N, Webb CT, Leyton DL, Holt KE, Heinz E, Gorrell R, Kwok T, Naderer T, Strugnell RA, Speed TP, Teasdale RD, Likić VA, Lithgow T. 2012. A bioinformatic strategy for the detection, classification and analysis of bacterial autotransporters. PLoS One 7:e43245. 10.1371/journal.pone.0043245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hart E, Yang J, Tauschek M, Kelly M, Wakefield MJ, Frankel G, Hartland EL, Robins-Browne RM. 2008. RegA, an AraC-like protein, is a global transcriptional regulator that controls virulence gene expression in Citrobacter rodentium. Infect Immun 76:5247–5256. 10.1128/IAI.00770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nichols KB, Totsika M, Moriel DG, Lo AW, Yang J, Wurpel DJ, Rossiter AE, Strugnell RA, Henderson IR, Ulett GC, Beatson SA, Schembri MA. 2016. Molecular characterization of the vacuolating autotransporter toxin in uropathogenic Escherichia coli. J Bacteriol 198:1487–1498. 10.1128/JB.00791-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, Park S-Y, Tame JRH. 2005. Crystal structure of hemoglobin protease, a heme binding autotransporter protein from pathogenic Escherichia coli. J Biol Chem 280:17339–17345. 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- 181.Benjelloun-Touimi Z, Sansonetti PJ, Parsot C. 1995. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol 17:123–135. 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 182.Guyer DM, Radulovic S, Jones F-E, Mobley HLT. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect Immun 70:4539–4546. 10.1128/IAI.70.8.4539-4546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Guyer DM, Henderson IR, Nataro JP, Mobley HLT. 2000. Identification of Sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol 38:53–66. 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 184.Nesta B, Spraggon G, Alteri C, Gomes Moriel D, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HLT, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010-12. 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Valeri M, Rossi Paccani S, Kasendra M, Nesta B, Serino L, Pizza M, Soriani M. 2015. Pathogenic Escherichia coli exploits SslE mucinase activity to translocate through the mucosal barrier and get access to host cells. PLoS One 10:e0117486. 10.1371/journal.pone.0117486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Moriel DG, Bertoldi I, Spagnuolo A, Marchi S, Rosini R, Nesta B, Pastorello I, Corea VAM, Torricelli G, Cartocci E, Savino S, Scarselli M, Dobrindt U, Hacker J, Tettelin H, Tallon LJ, Sullivan S, Wieler LH, Ewers C, Pickard D, Dougan G, Fontana MR, Rappuoli R, Pizza M, Serino L. 2010. Identification of protective and broadly conserved vaccine antigens from the genome of extraintestinal pathogenic Escherichia coli. Proc Natl Acad Sci U S A 107:9072–9077. 10.1073/pnas.0915077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Slater SL, Frankel G. 2020. Advances and challenges in studying type III secretion effectors of attaching and effacing pathogens. Front Cell Infect Microbiol 10:337. 10.3389/fcimb.2020.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sheikh A, Luo Q, Roy K, Shabaan S, Kumar P, Qadri F, Fleckenstein JM. 2014. Contribution of the highly conserved EaeH surface protein to enterotoxigenic Escherichia coli pathogenesis. Infect Immun 82:3657–3666. 10.1128/IAI.01890-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kansal R, Rasko DA, Sahl JW, Munson GP, Roy K, Luo Q, Sheikh A, Kuhne KJ, Fleckenstein JM. 2013. Transcriptional modulation of enterotoxigenic Escherichia coli virulence genes in response to epithelial cell interactions. Infect Immun 81:259–270. 10.1128/IAI.00919-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Batisson I, Guimond M-P, Girard F, An H, Zhu C, Oswald E, Fairbrother JM, Jacques M, Harel J. 2003. Characterization of the novel factor Paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect Immun 71:4516–4525. 10.1128/iai.71.8.4516-4525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li T, Han R, Wang Q, Wang S, Fang H, Li Z, Tu W, Wang D, Wang H. 2013. Immunogenicity of recombinant porcine attaching and effacing-associated protein compared with intimin fragment in Escherichia coli O157:H7-infected mice. Foodborne Pathog Dis 10:1016–1022. 10.1089/fpd.2013.1496. [DOI] [PubMed] [Google Scholar]
- 192.Germon P, Chen Y-H, He L, Blanco JE, Brée A, Schouler C, Huang S-H, Moulin-Schouleur M. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179–1186. 10.1099/mic.0.27809-0. [DOI] [PubMed] [Google Scholar]
- 193.Reidl J, Boos W. 1991. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J Bacteriol 173:4862–4876. 10.1128/jb.173.15.4862-4876.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Johnson JR, Oswald E, O’Bryan TT, Kuskowski MA, Spanjaard L. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J Infect Dis 185:774–784. 10.1086/339343. [DOI] [PubMed] [Google Scholar]
- 195.Sannes MR, Kuskowski MA, Owens K, Gajewski A, Johnson JR. 2004. Virulence factor profiles and phylogenetic background of Escherichia coli isolates from veterans with bacteremia and uninfected control subjects. J Infect Dis 190:2121–2128. 10.1086/425984. [DOI] [PubMed] [Google Scholar]
- 196.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HLT, Tarr PI. 2005. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. IAI 73:965–971. 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Fagan RP, Lambert MA, Smith SGJ. 2008. The Hek outer membrane protein of Escherichia coli strain RS218 binds to proteoglycan and utilizes a single extracellular loop for adherence, invasion, and autoaggregation. Infect Immun 76:1135–1142. 10.1128/IAI.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Fagan RP, Smith SGJ. 2007. The Hek outer membrane protein of Escherichia coli is an auto-aggregating adhesin and invasin. FEMS Microbiol Lett 269:248–255. 10.1111/j.1574-6968.2006.00628.x. [DOI] [PubMed] [Google Scholar]
- 199.Binns MM, Davies DL, Hardy KG. 1979. Cloned fragments of the plasmid ColV,I-K94 specifying virulence and serum resistance. Nature 279:778–781. 10.1038/279778a0. [DOI] [PubMed] [Google Scholar]
- 200.Binns MM, Mayden J, Levine RP. 1982. Further characterization of complement resistance conferred on Escherichia coli by the plasmid genes traT of R100 and iss of ColV,I-K94. Infect Immun 35:654–659. 10.1128/IAI.35.2.654-659.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lynne AM, Foley SL, Nolan LK. 2006. Immune response to recombinant Escherichia coli Iss protein in poultry. Avian Dis 50:273–276. 10.1637/7441-092105R.1. [DOI] [PubMed] [Google Scholar]
- 202.Lynne AM, Skyberg JA, Logue CM, Nolan LK. 2007. Detection of Iss and Bor on the surface of Escherichia coli. J Appl Microbiol 102:660–666. 10.1111/j.1365-2672.2006.03133.x. [DOI] [PubMed] [Google Scholar]
- 203.Johnson TJ, Wannemuehler YM, Nolan LK. 2008. Evolution of the iss gene in Escherichia coli. Appl Environ Microbiol 74:2360–2369. 10.1128/AEM.02634-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Picard B, Garcia JS, Gouriou S, Duriez P, Brahimi N, Bingen E, Elion J, Denamur E. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect Immun 67:546–553. 10.1128/IAI.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, Bacquié V, Penary M, Gaultier E, Nougayrède J-P, Fioramonti J, Oswald E. 2012. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 3:501–509. 10.4161/gmic.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Schultz M. 2008. Clinical use of Escherichia coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis 14:1012–1018. 10.1002/ibd.20377. [DOI] [PubMed] [Google Scholar]
- 207.Escobar-Páramo P, Grenet K, Le Menac’h A, Rode L, Salgado E, Amorin C, Gouriou S, Picard B, Rahimy MC, Andremont A, Denamur E, Ruimy R. 2004. Large-scale population structure of human commensal Escherichia coli isolates. Appl Environ Microbiol 70:5698–5700. 10.1128/AEM.70.9.5698-5700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventré A, Elion J, Picard B, Denamur E. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology (Reading) 147:1671–1676. 10.1099/00221287-147-6-1671. [DOI] [PubMed] [Google Scholar]
- 209.Gomez A, Petrzelkova KJ, Burns MB, Yeoman CJ, Amato KR, Vlckova K, Modry D, Todd A, Jost Robinson CA, Remis MJ, Torralba MG, Morton E, Umaña JD, Carbonero F, Gaskins HR, Nelson KE, Wilson BA, Stumpf RM, White BA, Leigh SR, Blekhman R. 2016. Gut microbiome of coexisting BaAka pygmies and Bantu reflects gradients of traditional subsistence patterns. Cell Rep 14:2142–2153. 10.1016/j.celrep.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 210.Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, Djossou F, Dupont C, Perroz G, Picard B, Catzeflis F, Andremont A, Denamur E. 2013. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependent population structure. Environ Microbiol Rep 5:49–57. 10.1111/j.1758-2229.2012.00374.x. [DOI] [PubMed] [Google Scholar]
- 211.Toh H, Oshima K, Toyoda A, Ogura Y, Ooka T, Sasamoto H, Park S-H, Iyoda S, Kurokawa K, Morita H, Itoh K, Taylor TD, Hayashi T, Hattori M. 2010. Complete genome sequence of the wild-type commensal Escherichia coli strain SE15, belonging to phylogenetic group B2. J Bacteriol 192:1165–1166. 10.1128/JB.01543-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Forde BM, Ben Zakour NL, Stanton-Cook M, Phan M-D, Totsika M, Peters KM, Chan KG, Schembri MA, Upton M, Beatson SA. 2014. The complete genome sequence of Escherichia coli EC958: a high-quality reference sequence for the globally disseminated multidrug-resistant E. coli O25b:H4-ST131 clone. PLoS One 9:e104400. 10.1371/journal.pone.0104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Avasthi TS, Kumar N, Baddam R, Hussain A, Nandanwar N, Jadhav S, Ahmed N. 2011. Genome of multidrug-resistant uropathogenic Escherichia coli strain NA114 from India. J Bacteriol 193:4272–4273. 10.1128/JB.05413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Lewis AJ, Richards AC, Mulvey MA. 2016. Invasion of host cells and tissues by uropathogenic bacteria. Microbiol Spectr 4:10.1128/microbiolspec.UTI-0026-2016. 10.1128/microbiolspec.UTI-0026-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wijetunge DSS, Gongati S, DebRoy C, Kim KS, Couraud PO, Romero IA, Weksler B, Kariyawasam S. 2015. Characterizing the pathotype of neonatal meningitis causing Escherichia coli (NMEC). BMC Microbiol 15:211. 10.1186/s12866-015-0547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Camprubí-Font C, Ewers C, Lopez-Siles M, Martinez-Medina M. 2019. Genetic and phenotypic features to screen for putative adherent-invasive Escherichia coli. Front Microbiol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Palmela C, Chevarin C, Xu Z, Torres J, Sevrin G, Hirten R, Barnich N, Ng SC, Colombel J-F. 2018. Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67:574–587. 10.1136/gutjnl-2017-314903. [DOI] [PubMed] [Google Scholar]
- 218.Low D, Tran HT, Lee I, Dreux N, Kamba A, Reinecker H, Darfeuille–Michaud A, Barnich N, Mizoguchi E. 2013. Chitin-binding domains of Escherichia coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology 145:602–612.e9. 10.1053/j.gastro.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Nobles CL, Green SI, Maresso AW. 2013. A product of heme catabolism modulates bacterial function and survival. PLoS Pathog 9:e1003507. 10.1371/journal.ppat.1003507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Adachi JA, Ericsson CD, Jiang Z, DuPont MW, Pallegar SR, DuPont HL. 2002. Natural history of enteroaggregative and enterotoxigenic Escherichia coli infection among US travelers to Guadalajara, Mexico. J Infect Dis 185:1681–1683. 10.1086/340419. [DOI] [PubMed] [Google Scholar]
- 221.Grad YH, Godfrey P, Cerquiera GC, Mariani-Kurkdjian P, Gouali M, Bingen E, Shea TP, Haas BJ, Griggs A, Young S, Zeng Q, Lipsitch M, Waldor MK, Weill F-X, Wortman JR, Hanage WP. 2013. Comparative genomics of recent Shiga toxin-producing Escherichia coli O104:H4: short-term evolution of an emerging pathogen. mBio 4 10.1128/mBio.00452-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Qadri F, Svennerholm A-M, Faruque ASG, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18:465–483. 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Roer L, Tchesnokova V, Allesøe R, Muradova M, Chattopadhyay S, Ahrenfeldt J, Thomsen MCF, Lund O, Hansen F, Hammerum AM, Sokurenko E, Hasman H. 2017. Development of a web tool for Escherichia coli subtyping based on fimH alleles. J Clin Microbiol 55:2538–2543. 10.1128/JCM.00737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kurylo CM, Alexander N, Dass RA, Parks MM, Altman RA, Vincent CT, Mason CE, Blanchard SC. 2016. Genome sequence and analysis of Escherichia coli MRE600, a colicinogenic, nonmotile strain that lacks RNase I and the type I methyltransferase, EcoKI. Genome Biol Evol 8:742–752. 10.1093/gbe/evw008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi S-H, Couloux A, Lee S-W, Yoon SH, Cattolico L, Hur C-G, Park H-S, Ségurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. 2009. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394:644–652. 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- 229.Segura A, Auffret P, Klopp C, Bertin Y, Forano E. 2017. Draft genome sequence and characterization of commensal Escherichia coli strain BG1 isolated from bovine gastro-intestinal tract. Stand Genomic Sci 12:61. 10.1186/s40793-017-0272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Oshima K, Toh H, Ogura Y, Sasamoto H, Morita H, Park S-H, Ooka T, Iyoda S, Taylor TD, Hayashi T, Itoh K, Hattori M. 2008. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res 15:375–386. 10.1093/dnares/dsn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, Rose DJ, Darling A, Mau B, Perna NT, Payne SM, Runyen-Janecky LJ, Zhou S, Schwartz DC, Blattner FR. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun 71:2775–2786. 10.1128/iai.71.5.2775-2786.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Reister M, Hoffmeier K, Krezdorn N, Rotter B, Liang C, Rund S, Dandekar T, Sonnenborn U, Oelschlaeger TA. 2014. Complete genome sequence of the Gram-negative probiotic Escherichia coli strain Nissle 1917. J Biotechnol 187:106–107. 10.1016/j.jbiotec.2014.07.442. [DOI] [PubMed] [Google Scholar]
- 233.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, El Karoui M, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Saint Ruf C, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EPC, Denamur E. 2009. Organized genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Clermont O, Lescat M, O’Brien CL, Gordon DM, Tenaillon O, Denamur E. 2008. Evidence for a human-specific Escherichia coli clone. Environ Microbiol 10:1000–1006. 10.1111/j.1462-2920.2007.01520.x. [DOI] [PubMed] [Google Scholar]
- 235.Stephens CM, Skerker JM, Sekhon MS, Arkin AP, Riley LW. 2015. Complete genome sequences of four Escherichia coli ST95 isolates from bloodstream infections. Genome Announc 3:e01241-15. 10.1128/genomeA.01241-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Johnson TJ, Danzeisen JL, Youmans B, Case K, Llop K, Munoz-Aguayo J, Flores-Figueroa C, Aziz M, Stoesser N, Sokurenko E, Price LB, Johnson JR. 2016. Separate F-type plasmids have shaped the evolution of the H30 subclone of Escherichia coli sequence type 131. mSphere 1:e00121-16. 10.1128/mSphere.00121-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stoesser N, Sheppard AE, Peirano G, Sebra R, Lynch T, Anson L, Kasarskis A, Motyl MR, Crook DW, Pitout JD. 2016. Complete sequencing of plasmids containing blaOXA-163 and blaOXA-48 in Escherichia coli sequence type 131. Antimicrob Agents Chemother 60:6948–6951. 10.1128/AAC.01130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, Pitout JDD. 2014. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 20:1928–1931. 10.3201/eid2011.141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Johnson TJ, Aziz M, Liu CM, Sokurenko E, Kisiela DI, Paul S, Andersen P, Johnson JR, Price LB. 2016. Complete genome sequence of a CTX-M-15-producing Escherichia coli strain from the H30Rx subclone of sequence type 131 from a patient with recurrent urinary tract infections, closely related to a lethal urosepsis isolate from the patient’s sister. Genome Announc 4:e00334-16. 10.1128/genomeA.00334-16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Johnson TJ, Hargreaves M, Shaw K, Snippes P, Lynfield R, Aziz M, Price LB. 2015. Complete genome sequence of a carbapenem-resistant extraintestinal pathogenic Escherichia coli strain belonging to the sequence type 131 H30R subclade. Genome Announc 3:e00272-15. 10.1128/genomeA.00272-15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lescat M, Calteau A, Hoede C, Barbe V, Touchon M, Rocha E, Tenaillon O, Médigue C, Johnson JR, Denamur E. 2009. A module located at a chromosomal integration hot spot is responsible for the multidrug resistance of a reference strain from Escherichia coli clonal group A. Antimicrob Agents Chemother 53:2283–2288. 10.1128/AAC.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Roos V, Nielsen EM, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol Lett 262:22–30. 10.1111/j.1574-6968.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 243.Mehershahi KS, Abraham SN, Chen SL. 2015. Complete genome sequence of uropathogenic Escherichia coli strain CI5. Genome Announc 3:e00558-15. 10.1128/genomeA.00558-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Andersson P, Engberg I, Lidin-Janson G, Lincoln K, Hull R, Hull S, Svanborg C. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect Immun 59:2915–2921. 10.1128/IAI.59.9.2915-2921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. 10.1128/IAI.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Welch RA, Burland V, Plunkett G, Redford P, Roesch P, Rasko D, Buckles EL, Liou S-R, Boutin A, Hackett J, Stroud D, Mayhew GF, Rose DJ, Zhou S, Schwartz DC, Perna NT, Mobley HLT, Donnenberg MS, Blattner FR. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 99:17020–17024. 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen SL, Hung C-S, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A 103:5977–5982. 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Mulvey MA, Schilling JD, Hultgren SJ. 2001. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect Immun 69:4572–4579. 10.1128/IAI.69.7.4572-4579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol 61:584–595. 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- 250.Zurfluh K, Tasara T, Stephan R. 2016. Full-genome sequence of Escherichia coli K-15KW01, a uropathogenic E. coli B2 sequence type 127 isolate harboring a chromosomally carried bla CTX-M-15 gene. Genome Announc 4:e00927-16. 10.1128/genomeA.00927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Hancock SJ, Phan M-D, Peters KM, Forde BM, Chong TM, Yin W-F, Chan K-G, Paterson DL, Walsh TR, Beatson SA, Schembri MA. 2017. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob Agents Chemother 61:e01740-16. 10.1128/AAC.01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Achtman M, Mercer A, Kusecek B, Pohl A, Heuzenroeder M, Aaronson W, Sutton A, Silver RP. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect Immun 39:315–335. 10.1128/IAI.39.1.315-335.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Silver RP, Aaronson W, Sutton A, Schneerson R. 1980. Comparative analysis of plasmids and some metabolic characteristics of Escherichia coli K1 from diseased and healthy individuals. Infect Immun 29:200–206. 10.1128/IAI.29.1.200-206.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Day MW, Jackson LA, Akins DR, Dyer DW, Chavez-Bueno S. 2015. Whole-genome sequences of the archetypal K1 Escherichia coli neonatal isolate RS218 and contemporary neonatal bacteremia clinical isolates SCB11, SCB12, and SCB15. Genome Announc 3:e01598-14. 10.1128/genomeA.01598-14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Wijetunge DSS, Katani R, Kapur V, Kariyawasam S. 2015. Complete genome sequence of Escherichia coli strain RS218 (O18:H7:K1), associated with neonatal meningitis. Genome Announc 3:e00804-15. 10.1128/genomeA.00804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nicholson BA, Wannemuehler YM, Logue CM, Li G, Nolan LK. 2016. Complete genome sequence of the neonatal meningitis-causing Escherichia coli strain NMEC O18. Genome Announc 4:e01239-16. 10.1128/genomeA.01239-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Tivendale KA, Logue CM, Kariyawasam S, Jordan D, Hussein A, Li G, Wannemuehler Y, Nolan LK. 2010. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infect Immun 78:3412–3419. 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Iqbal J, Dufendach KR, Wellons JC, Kuba MG, Nickols HH, Gómez-Duarte OG, Wynn JL. 2016. Lethal neonatal meningoencephalitis caused by multidrug-resistant, highly virulent Escherichia coli. Infect Dis (Auckl) 48:461–466. 10.3109/23744235.2016.1144142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Nielsen DW, Ricker N, Barbieri NL, Wynn JL, Gómez-Duarte OG, Iqbal J, Nolan LK, Allen HK, Logue CM. 2018. Complete genome sequence of the multidrug-resistant neonatal meningitis Escherichia coli serotype O75:H5:K1 strain mcjchv-1 (NMEC-O75). Microbiol Resour Announc 7:e01043-18. 10.1128/MRA.01043-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lu S, Zhang X, Zhu Y, Kim KS, Yang J, Jin Q. 2011. Complete genome sequence of the neonatal-meningitis-associated Escherichia coli strain CE10. J Bacteriol 193:7005–7005. 10.1128/JB.06284-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wang X, Wei L, Wang B, Zhang R, Liu C, Bi D, Chen H, Tan C. 2016. Complete genome sequence and characterization of avian pathogenic Escherichia coli field isolate ACN001. Stand Genomic Sci 11:13. 10.1186/s40793-015-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Mangiamele P, Nicholson B, Wannemuehler Y, Seemann T, Logue CM, Li G, Tivendale KA, Nolan LK. 2013. Complete genome sequence of the avian pathogenic Escherichia coli strain APEC O78. Genome Announc 1:e0002613. 10.1128/genomeA.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Huja S, Oren Y, Trost E, Brzuszkiewicz E, Biran D, Blom J, Goesmann A, Gottschalk G, Hacker J, Ron EZ, Dobrindt U. 2015. Genomic avenue to avian colisepticemia. mBio 6:e01681-14. 10.1128/mBio.01681-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol 189:3228–3236. 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mangiamele P. 2014. From sequencing to analysis: building a comparative genomics tool for the biologist end-user. Iowa State University Digital Repository, Ames, IA. [Google Scholar]
- 266.Nielsen DW, Mangiamele P, Ricker N, Barbieri NL, Allen HK, Nolan LK, Logue CM. 2018. Complete genome sequence of avian pathogenic Escherichia coli strain APEC O2-211. Microbiol Resour Announc 7:e01046-18. 10.1128/MRA.01046-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Li G, Laturnus C, Ewers C, Wieler LH. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect Immun 73:2818–2827. 10.1128/IAI.73.5.2818-2827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhu Ge X, Jiang J, Pan Z, Hu L, Wang S, Wang H, Leung FC, Dai J, Fan H. 2014. Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2:K1:H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1:K1 and human ExPEC O18:K1 strains. PLoS One 9:e112048. 10.1371/journal.pone.0112048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, Zineb EH, Segurens B, Barbe V, Sauvanet P, Neut C, Colombel J-F, Medigue C, Mojica FJM, Peyret P, Bonnet R, Darfeuille-Michaud A. 2010. Complete genome sequence of Crohn’s disease-associated adherent-invasive Escherichia coli strain LF82. PLoS One 5:e12714. 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. 2010. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics 11:667. 10.1186/1471-2164-11-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Krause DO, Little AC, Dowd SE, Bernstein CN. 2011. Complete genome sequence of adherent invasive Escherichia coli UM146 isolated from ileal Crohn’s disease biopsy tissue. J Bacteriol 193:583–583. 10.1128/JB.01290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. 2005. Variable phenotypes of enterocolitis in interleukin-10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 128:891–906. 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 273.Ellermann M, Huh EY, Liu B, Carroll IM, Tamayo R, Sartor RB. 2015. Adherent-invasive Escherichia coli production of cellulose influences iron-induced bacterial aggregation, phagocytosis, and induction of colitis. Infect Immun 83:4068–4080. 10.1128/IAI.00904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K, Tobe T, Hattori M, Hayashi T. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci U S A 106:17939–17944. 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.González-Escalona N, Allard MA, Brown EW, Sharma S, Hoffmann M. 2019. Nanopore sequencing for fast determination of plasmids, phages, virulence markers, and antimicrobial resistance genes in Shiga toxin-producing Escherichia coli. PLoS One 14:e0220494. 10.1371/journal.pone.0220494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lindsey RL, Rowe L, Garcia-Toledo L, Loparev V, Knipe K, Stripling D, Martin H, Trees E, Juieng P, Batra D, Strockbine N. 2016. High-quality draft genome sequences for five non-O157 Shiga toxin-producing Escherichia coli strains generated with PacBio sequencing and optical maps. Genome Announc 4:e00626-16. 10.1128/genomeA.00626-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yan X, Fratamico PM, Bono JL, Baranzoni GM, Chen C-Y. 2015. Genome sequencing and comparative genomics provides insights on the evolutionary dynamics and pathogenic potential of different H-serotypes of Shiga toxin-producing Escherichia coli O104. BMC Microbiol 15:83. 10.1186/s12866-015-0413-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Cowley LA, Dallman TJ, Fitzgerald S, Irvine N, Rooney PJ, McAteer SP, Day M, Perry NT, Bono JL, Jenkins C, Gally DL. 2016. Short-term evolution of Shiga toxin-producing Escherichia coli O157:H7 between two food-borne outbreaks. Microb Genom 2:e000084. 10.1099/mgen.0.000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Oh D-H, Pan Y, Berry E, Cooley M, Mandrell R, Breidt F. 2009. Escherichia coli O157:H7 strains isolated from environmental sources differ significantly in acetic acid resistance compared with human outbreak strains. J Food Prot 72:503–509. 10.4315/0362-028X-72.3.503. [DOI] [PubMed] [Google Scholar]
- 280.Baranzoni GM, Fratamico PM, Reichenberger ER, Kim G-H, Breidt F, Kay K, Oh D-H. 2016. Complete genome sequences of Escherichia coli O157:H7 strains SRCC 1675 and 28RC, which vary in acid resistance. Genome Announc 4. 10.1128/genomeA.00743-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Kudva IT, Hatfield PG, Hovde CJ. 1996. Escherichia coli O157:H7 in microbial flora of sheep. J Clin Microbiol 34:431–433. 10.1128/JCM.34.2.431-433.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Marques LRM, Moore MA, Wells JG, Wachsmuth IK, O’brien AD. 1986. Production of Shiga-like toxin by Escherichia coli. J Infect Dis 154:338–341. 10.1093/infdis/154.2.338. [DOI] [PubMed] [Google Scholar]
- 283.Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A 108:20142–20147. 10.1073/pnas.1107176108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 285.Park D, Stanton E, Ciezki K, Parrell D, Bozile M, Pike D, Forst SA, Jeong KC, Ivanek R, Döpfer D, Kaspar CW. 2013. Evolution of the Stx2-encoding prophage in persistent bovine Escherichia coli O157:H7 strains. Appl Environ Microbiol 79:1563–1572. 10.1128/AEM.03158-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Shere JA, Bartlett KJ, Kaspar CW. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl Environ Microbiol 64:1390–1399. 10.1128/AEM.64.4.1390-1399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Teng L, Ginn A, Jeon S, Kang M, Jeong KC. 2016. Complete genome sequence of an Escherichia coli O157:H7 strain isolated from a super-shedder steer. Genome Announc 4:e00258-16. 10.1128/genomeA.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Makino K, Ishii K, Yasunaga T, Hattori M, Yokoyama K, Yutsudo CH, Kubota Y, Yamaichi Y, Iida T, Yamamoto K, Honda T, Han CG, Ohtsubo E, Kasamatsu M, Hayashi T, Kuhara S, Shinagawa H. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res 5:1–9. 10.1093/dnares/5.1.1. [DOI] [PubMed] [Google Scholar]
- 289.Hayashi T. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 290.Katani R, Cote R, Raygoza Garay JA, Li L, Arthur TM, DebRoy C, Mwangi MM, Kapur V. 2015. Complete genome sequence of SS52, a strain of Escherichia coli O157:H7 recovered from supershedder cattle. Genome Announc 3:e01569-14. 10.1128/genomeA.01569-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kulasekara BR, Jacobs M, Zhou Y, Wu Z, Sims E, Saenphimmachak C, Rohmer L, Ritchie JM, Radey M, McKevitt M, Freeman TL, Hayden H, Haugen E, Gillett W, Fong C, Chang J, Beskhlebnaya V, Waldor MK, Samadpour M, Whittam TS, Kaul R, Brittnacher M, Miller SI. 2009. Analysis of the genome of the Escherichia coli O157:H7 2006 spinach-associated outbreak isolate indicates candidate genes that may enhance virulence. Infect Immun 77:3713–3721. 10.1128/IAI.00198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Xiong Y, Wang P, Lan R, Ye C, Wang H, Ren J, Jing H, Wang Y, Zhou Z, Bai X, Cui Z, Luo X, Zhao A, Wang Y, Zhang S, Sun H, Wang L, Xu J. 2012. A novel Escherichia coli O157:H7 clone causing a major hemolytic uremic syndrome outbreak in China. PLoS One 7:e36144. 10.1371/journal.pone.0036144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Cooper KK, Mandrell RE, Louie JW, Korlach J, Clark TA, Parker CT, Huynh S, Chain PS, Ahmed S, Carter M. 2014. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genomics 15:17. 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Scheutz F, Møller Nielsen E, Frimodt-Møller J, Boisen N, Morabito S, Tozzoli R, Nataro JP, Caprioli A. 2011. Characteristics of the enteroaggregative Shiga toxin/verotoxin-producing Escherichia coli O104:H4 strain causing the outbreak of haemolytic uraemic syndrome in Germany, May to June 2011. Eurosurveillance 16:19889. 10.2807/ese.16.24.19889-en. [DOI] [PubMed] [Google Scholar]
- 295.Ahmed SA, Awosika J, Baldwin C, Bishop-Lilly KA, Biswas B, Broomall S, Chain PSG, Chertkov O, Chokoshvili O, Coyne S, Davenport K, Detter JC, Dorman W, Erkkila TH, Folster JP, Frey KG, George M, Gleasner C, Henry M, Hill KK, Hubbard K, Insalaco J, Johnson S, Kitzmiller A, Krepps M, Lo C-C, Luu T, McNew LA, Minogue T, Munk CA, Osborne B, Patel M, Reitenga KG, Rosenzweig CN, Shea A, Shen X, Strockbine N, Tarr C, Teshima H, van Gieson E, Verratti K, Wolcott M, Xie G, Sozhamannan S, Gibbons HS. Threat Characterization Consortium. 2012. Genomic comparison of Escherichia coli O104:H4 isolates from 2009 and 2011 reveals plasmid, and prophage heterogeneity, including Shiga toxin encoding phage stx2. PLoS One 7:e48228. 10.1371/journal.pone.0048228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Beaulaurier J, Zhang X-S, Zhu S, Sebra R, Rosenbluh C, Deikus G, Shen N, Munera D, Waldor MK, Chess A, Blaser MJ, Schadt EE, Fang G. 2015. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nat Commun 6:7438. 10.1038/ncomms8438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Strockbine NA, Faruque SM, Kay BA, Haider K, Alam K, Alam AN, Tzipori S, Wachsmuth IK. 1992. DNA probe analysis of diarrhoeagenic Escherichia coli: detection of EAF-positive isolates of traditional enteropathogenic E. coli serotypes among Bangladeshi paediatric diarrhoea patients. Mol Cell Probes 6:93–99. 10.1016/0890-8508(92)90052-y. [DOI] [PubMed] [Google Scholar]
- 298.Evans DJ, Evans DG. 1973. Three characteristics associated with enterotoxigenic Escherichia coli isolated from man. Infect Immun 8:322–328. 10.1128/IAI.8.3.322-328.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Shepard SM, Danzeisen JL, Isaacson RE, Seemann T, Achtman M, Johnson TJ. 2012. Genome sequences and phylogenetic analysis of K88- and F18-positive porcine enterotoxigenic Escherichia coli. J Bacteriol 194:395–405. 10.1128/JB.06225-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Sack D. 1975. Diarrhea associated with heat-stable enterotoxin-producing strains of Escherichia coli. Lancet 306:239–241. 10.1016/S0140-6736(75)90958-7. [DOI] [PubMed] [Google Scholar]
- 301.Wachsmuth K, Wells J, Shipley P, Ryder R. 1979. Heat-labile enterotoxin production in isolates from a shipboard outbreak of human diarrheal illness. Infect Immun 24:793–797. 10.1128/IAI.24.3.793-797.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sahl JW, Rasko DA. 2012. Analysis of global transcriptional profiles of enterotoxigenic Escherichia coli isolate E24377A. Infect Immun 80:1232–1242. 10.1128/IAI.06138-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Levine MM, Nalin DR, Hoover DL, Bergquist EJ, Hornick RB, Young CR. 1979. Immunity to enterotoxigenic Escherichia coli. Infect Immun 23:729–736. 10.1128/IAI.23.3.729-736.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Youssef M, Shurman A, Bougnoux M-E, Rawashdeh M, Bretagne S, Strockbine N. 2000. Bacterial, viral and parasitic enteric pathogens associated with acute diarrhea in hospitalized children from northern Jordan. FEMS Immunol Med Microbiol 28:257–263. 10.1111/j.1574-695X.2000.tb01485.x. [DOI] [PubMed] [Google Scholar]
- 305.Smith P, Lindsey RL, Rowe LA, Batra D, Stripling D, Garcia-Toledo L, Drapeau D, Knipe K, Strockbine N. 2018. High-quality whole-genome sequences for 21 enterotoxigenic Escherichia coli strains generated with PacBio sequencing. Genome Announc 6:e01311-18. 10.1128/genomeA.01311-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, Henderson IR, Harris D, Asadulghani M, Kurokawa K, Dean P, Kenny B, Quail MA, Thurston S, Dougan G, Hayashi T, Parkhill J, Frankel G. 2009. Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J Bacteriol 191:347–354. 10.1128/JB.01238-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Zhou Z, Li X, Liu B, Beutin L, Xu J, Ren Y, Feng L, Lan R, Reeves PR, Wang L. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One 5:e8700. 10.1371/journal.pone.0008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kyle JL, Cummings CA, Parker CT, Quinones B, Vatta P, Newton E, Huynh S, Swimley M, Degoricija L, Barker M, Fontanoz S, Nguyen K, Patel R, Fang R, Tebbs R, Petrauskene O, Furtado M, Mandrell RE. 2012. Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J Bacteriol 194:1885–1896. 10.1128/JB.00120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download IAI00115-21_Supp_1_seq4.pdf, PDF file, 1.3 MB (1.3MB, pdf)
Supplemental material. Download IAI00115-21_Supp_2_seq1.xlsx, XLSX file, 0.2 MB (157.9KB, xlsx)
Supplemental material. Download IAI00115-21_Supp_3_seq2.xlsx, XLSX file, 0.06 MB (60.4KB, xlsx)
Supplemental material. Download IAI00115-21_Supp_4_seq3.xlsx, XLSX file, 0.02 MB (22.2KB, xlsx)