Summary:
N6-methyladenosine (m6A) on mRNAs mediates different biological processes and its dysregulation contributes to tumorigenesis. How m6A dictates its diverse molecular and cellular effects in leukemias remains unknown. We found that YTHDC1 is the essential m6A reader in myeloid leukemia from a genome-wide CRISPR screen and m6A is required for YTHDC1 to undergo liquid-liquid phase separation and form nuclear YTHDC1-m6A Condensates (nYACs). The number of nYACs increases in acute myeloid leukemia (AML) cells compared to normal hematopoietic stem and progenitor cells. AML cells require the nYACs to maintain cell survival and the undifferentiated state that is critical for leukemia maintenance. Furthermore, nYACs enable YTHDC1 to protect m6A-mRNAs from the PAXT-complex and exosome-associated RNA degradation. Collectively, m6A is required for the formation of a nuclear body mediated by phase separation that maintains mRNA stability and control cancer cell survival and differentiation.
Graphical Abstract
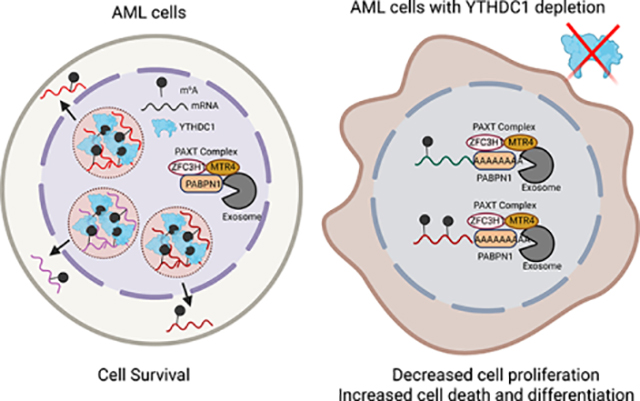
eTOC blurb
Using AML cell lines and patient samples Cheng et al. identify a requirement for YTHDC1 in myeloid leukemogenesis. YTHDC1 undergoes liquid-liquid phase separation by binding to m6A to form dynamic nuclear condensates. These nuclear bodies are increased in myeloid leukemia cells and protect mRNAs from the PAXT-exosome complex.
INTRODUCTION
The most prevalent chemical modification on mRNAs is N6-methyladenosine (m6A). The METTL3 writer complex co-transcriptionally methylates mRNAs. A set of “reader” proteins in the nucleus (YTHDC1 etc.) and the cytoplasm (YTHDF1, 2 and 3, etc.) bind directly or indirectly to m6A-mRNAs and thus alter their fate. This regulatory process is tightly controlled, as its dysregulation has been implicated in tumorigenesis(Chang et al., 2020; Huang et al., 2020; Zhang et al., 2017).
A number of studies have highlighted the requirement of m6A-methylation program for myeloid leukemogenesis (Bansal et al., 2014; Barbieri et al., 2017; Cheng et al., 2019; Lee et al., 2019; Li et al., 2017b; Shen et al., 2020; Vu et al., 2019; Vu et al., 2017a; Wang et al., 2020; Weng et al., 2018). Both m6A writers (METTL3, METTL14 and WTAP) and erasers (FTO and ALKBH5) have been reported to contribute to myeloid leukemogenesis, suggesting m6A regulators as potential therapeutic targets in AML. Several inhibitors targeting FTO and METTL3 are currently being developed (Bedi et al., 2020; Huang et al., 2019; Su et al., 2020). Additionally, various readers have been also implicated in leukemia including YTHDF2 and IGFBPs (Elcheva et al., 2020; He et al., 2018; Paris et al., 2019). However, it remains unclear how m6A directly determines the fate of m6A-mRNAs to control leukemia cell growth, differentiation state and cell survival remains largely unknown.
RESULTS
YTHDC1 is highly expressed in AML
To explore how m6A mediates its effect in myeloid leukemia, we examined the rankings of all the known m6A readers in a genome-wide CRISPR-based screen for essential genes in 14 acute myeloid leukemia cell lines(Wang et al., 2017) and nuclear reader YTHDC1 scored as the top essential reader (Figure 1A; Figure S1A). YTHDF2 was not found to be as essential even though recent studies have implicated YTHDF2 as a critical regulator in leukemia stem cells and not essential for normal hematopoietic stem and progenitors cells (HSPC)(Li et al., 2018; Paris et al., 2019). This could be explained by the fact that the other YTHDF paralogs could compensate for YTHDF2’s function in normal HSPCs(Lasman et al., 2020; Zaccara and Jaffrey, 2020). The CRISPR data suggests a role for nuclear m6A through YTHDC1 in leukemia cell survival.
Figure 1: YTHDC1 is required for leukemia cell survival.
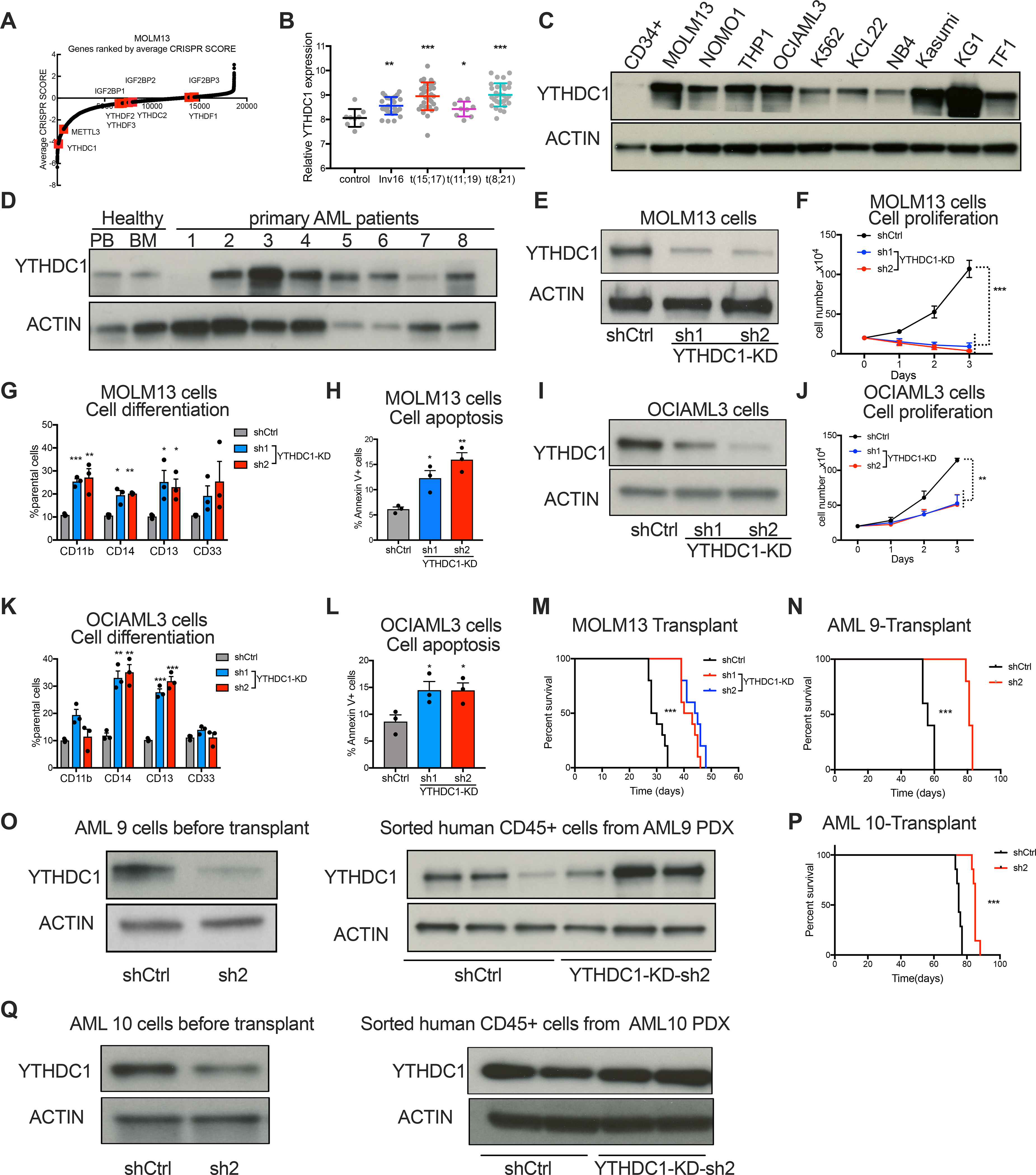
(A) CRISPR score rank of the m6A “readers” in MOLM13 cells. m6A readers are highlighted in red and METTL3 was a control. (B) YTHDC1 expression in human primary AML cases with inv(16), t(8;21) t(15;17), t(11;19) or normal controls (NC). Data is from GSE34184 and GSE30285. (C) Immunoblot analysis YTHDC1 protein expression in AML cell lines compared to normal HSPCs (CD34+ cells). (D) Immunoblot analysis of YTHDC1 protein abundance in primary AML patient cells and peripheral blood and bone marrow from healthy donors. (E-H) MOLM13 cells were transduced with lentiviruses expressing control shRNA(shCtrl) or two independent shRNAs targeting YTHDC1 (sh1 and sh2; YTHDC1-knockdown). n=3 independent replicants. (E) Representative immunoblot for control and YTHDC1 depleted MOLM13 cells. (F) Cell proliferation of control versus YTHDC1 depleted MOLM13 cells. (G) Myeloid differentiation of cells was quantified by flow cytometry. (H) Apoptotic cells were determined by flow cytometry analysis of Annexin V and 7-AAD staining. (I-L) OCIAML3 cells were transduced with lentiviruses expressing control shRNA(shCtrl) or two independent shRNAs targeting YTHDC1 (sh1 and sh2; YTHDC1-knockdown). n=3 independent replicants. (I) Immunoblot of YTHDC1 expression in OCIAML3 cells. (J) Cell proliferation of OCIAML3 cells upon YTHDC1 depletion. (K) Myeloid differentiation of OCIAML3 cells was determined by flow cytometry. (L) Apoptotic cells were determined by flow cytometry analysis of Annexin V and 7-AAD staining. (M) Survival curve of NSG mice transplanted with control or YTHDC1 depleted MOLM13 cells by shRNA. n=10 for each group. (N) Survival curves of NSG mice xeno-transplanted with human primary AML deprived cells (AML 9 cells) that were transduced with control or YTHDC1-targeting shRNA. n=5 for each group. (O) Immunoblot of PDX cells before injection to NSG mice or after xenograft in (N). (P) Survival curves in of NSG mice xeno-transplanted with AML 10 cells that were transduced with control or YTHDC1-targeting shRNA. n=10 for each group. (Q) Immunoblot of PDX cells before injection to NSG mice or after xenograft in (P). Error bars, s.e.m. *p<0.05, **p<0.01, ***p<0.001, two-tailed t test.
We next assessed YTHDC1 expression in human acute myeloid leukemia (AML) and found significantly higher expression than the majority of other cancer types (Figure S1B). We found that YTHDC1 expression is increased in AML patients with different genetic backgrounds compared to normal controls (Figure 1B). Moreover, YTHDC1 protein is more abundant in 10 myeloid leukemia cell lines compared to cord blood derived CD34+ (CB-CD34+) HSPCs (Figure 1C). Furthermore, in 8 primary acute myeloid leukemia patient samples, we found higher YTHDC1 abundance in 6/8 samples in comparison to healthy human peripheral blood and bone marrow or CB-CD34+ cells (Figure 1D; Figure S1C; Table S1). YTHDC1 expression is comparable across different AML subtypes and genetic mutations do not correlate with YTHDC1 expression (Figures S1D–F). These data suggest that YTHDC1 expression is dysregulated across different subtypes and diverse oncogenic drivers of myeloid leukemia. However, we found that YTHDC1 is expressed at a significantly higher level in M0 (undifferentiated acute myeloblastic leukemia; French-American-British, FAB classification) compared to other FAB groups of AML patients (Figure S1G). These data suggest a role for YTHDC1 in controlling myeloid differentiation. Interestingly, YTHDC1 expression was not enriched in the leukemic stem cell (LSC) fraction in AML (Figure S1H). Overall, our data suggest that YTHDC1 is dysregulated in leukemia compared to normal cells and is enriched in the most histologically immature leukemias.
YTHDC1 is essential for maintaining myeloid cell state
With the increased expression of YTHDC1, we next depleted YTHDC1 by shRNAs and observed a marked reduction in cell growth (from 3–100-fold), increased differentiation (3–4-fold depending on the myeloid marker and cell line) and apoptosis (2–3-fold) in multiple AML cell lines: MOLM13 (MLL-AF9, FLT3-ITD) (Figures 1E–H; Figures S2A and B), OCIAML3 (NPM1 mut) (Figures 1I–L; Figures S2C and D), NOMO1 (MLL-AF9) (Figures S2E–J), THP1 (MLL-AF9) (Figures S2K–P) and HL60 (MYC amplification) (Figures S2Q–V). Similar to the results with shRNAs, we found decreased cell proliferation (>2-fold), increased cell differentiation (3-fold with CD11b and CD13 markers) and apoptosis (2–3-fold) using CRISPR-Cas9 mediated YTHDC1 knockout in MOLM13 cells (Figures S3A–F). All these data suggest that the nuclear m6A reader YTHDC1 is essential for maintaining the undifferentiated state and survival of myeloid leukemia cells.
YTHDC1 is essential for leukemogenesis
To assess if YTHDC1 is required for leukemogenesis, MOLM13 cells were transduced with YTHDC1-shRNAs and transplanted into immunodeficient recipient mice. These cells exhibited significantly delayed leukemia development compared to MOLM13 cells transduced with the control shRNA (Figure 1M; Figures S3G and H). Sorted engrafted human YTHDC1-shRNA-transduced leukemic cells from mice showed equivalent abundance of YTHDC1 compared to the controls, indicating a negative selection of YTHDC1 depleted cells in vivo (Figure S3I). Additionally, in two patient-derived xenotransplantation (PDX) AML models, we found that shRNA-mediated depletion of YTHDC1 dramatically delayed leukemogenesis (Figures 1N–Q; Table S1). We examined the leukemia cells that successfully engrafted in the YTHDC1 knockdown group and found wild-type (WT) levels of YTHDC1 expression. These data indicate that the few cells that engraft were those that evaded successful knockdown (Figures 1N–Q). Furthermore, in another PDX model, we observed significantly reduced engraftment of leukemia cells upon YTHDC1 depletion (Figures S3J and K). Thus, these data support the requirement of YTHDC1 for leukemogenesis.
YTHDC1 forms more condensates in AML than HSPCs
To understand how YTHDC1 mediates its functional requirement in leukemia, we first examined the amino acid sequence of YTHDC1. Similar to the cytosolic m6A readers YTHDFs, YTHDC1 comprise a YTH domain that recognizes m6A and intrinsically disordered regions (IDR) (Figure S4A). Different from YTHDFs, YTHDC1 contains a Glu-rich N-terminal IDR and an Arg/Pro-rich C-terminal IDR (Figure S4B). These IDRs share features with the IDRs of several proteins known to facilitate condensate formation(Alberti and Dormann, 2019; Banani et al., 2017; Shin and Brangwynne, 2017).
Previous studies have found that YTHDFs direct m6A-mRNAs to stress granules through condensates mediated by liquid-liquid phase separation (LLPS) in the cytoplasm(Fu and Zhuang, 2020; Liu et al., 2020b; Ries et al., 2019). LLPS is a fundamental process in cells that drives the formation of many biomolecular condensates such as nucleoli, nuclear speckles, stress granules, P bodies and others (Feric et al., 2016; Molliex et al., 2015; Shin and Brangwynne, 2017) and it is required for regulation of heterochromatin formation, gene expression, and nucleocytoplasmic transport(Larson et al., 2017; Lu et al., 2018; Sabari et al., 2018; Schmidt and Gorlich, 2016). Dysregulation of LLPS is corelated with development of many diseases including cancer(Alberti and Dormann, 2019; Boija et al., 2021).
It was found that the m6A reader YTHDC1 forms nuclear YT bodies(Hartmann et al., 1999; Nayler et al., 2000). Thus, we postulated that YTHDC1 has intrinsic biophysical properties that facilitate LLPS and this property contributes to its requirement for leukemia survival. We observed that YTHDC1 resembled a puncta-like staining pattern in normal and malignant cell types including in both suspension cells (cord blood CD34+, MOLM13 and OCIAML3 cells) and adherent cells (Hela, 293T and MCF7 cells) (Figure 2A; Figure S4C). Surprisingly, we found an increase number of YTHDC1 puncta and overall increase in staining intensity in human leukemia cell lines (MOLM13 and OCIAML3) compared to CB-CD34+ cells (Figures 2B and C; Figure S4C). These data suggest that increased YTHDC1 expression also correlates with aberrant nuclear YTHDC1-puncta.
Figure 2: YTHDC1 proteins undergo liquid-liquid phase separation in vitro and in vivo.
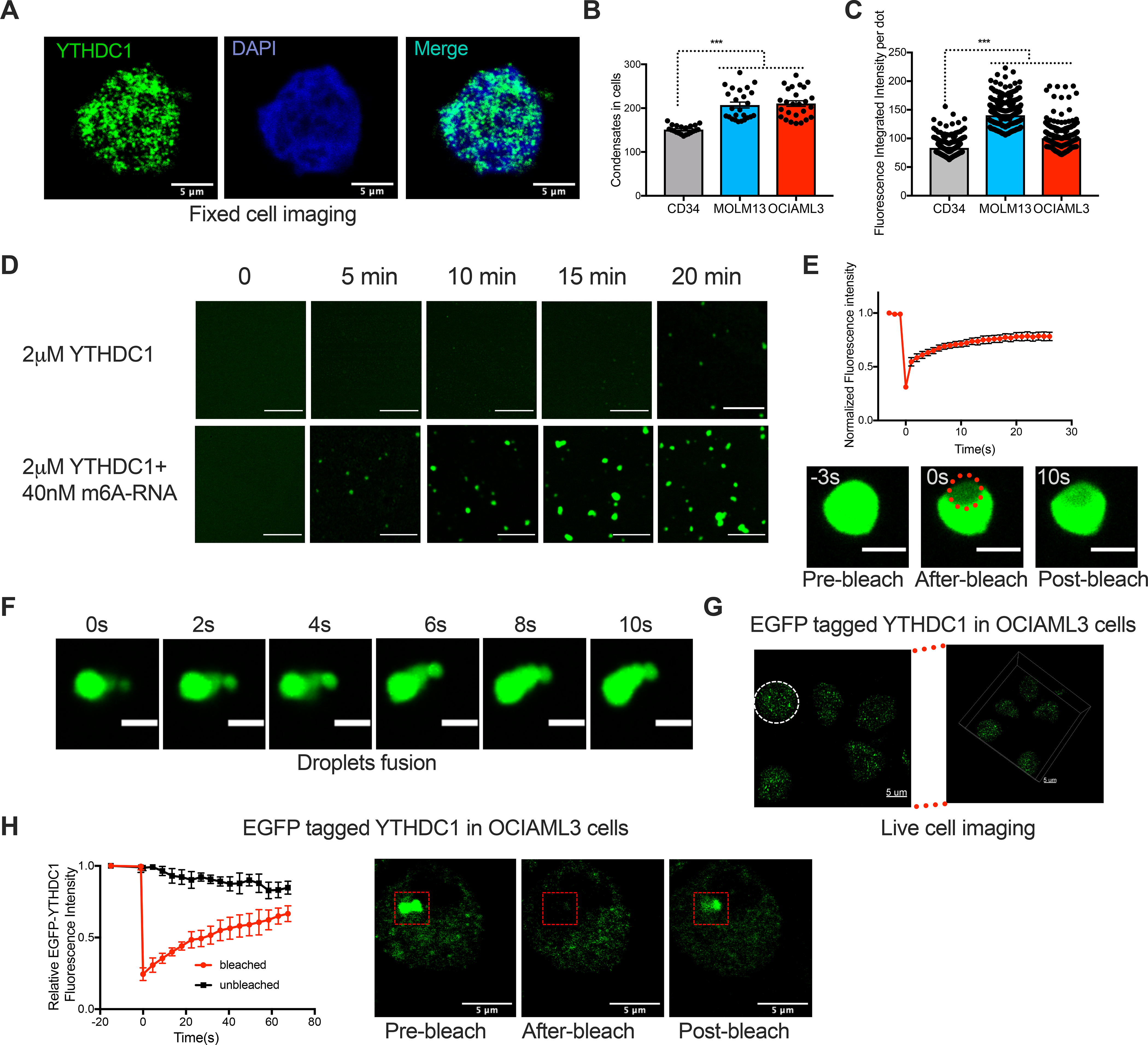
(A) Immunofluorescence (IF) imaging of YTHDC1 in OCIAML3 cells shows nuclear YTHDC1 puncta. YTHDC1: green, DAPI: blue. Scale bars, 5μm. (B) Quantitative summary of YTHDC1 condensates in CB-CD34+, MOLM13 and OCIAML3 cells by IF. Mean+s.e.m, n=21,25,25 from 3 independent experiments. (C) Quantitative summary of fluorescence intensity per condensate in CB-CD34+, MOLM13 and OCIAML3 cells by IF. Mean+s.e.m, n=304,333,365. (D) Time-lapse images to show phase separation of YTHDC1 protein. Scale bars, 5μm. (E) Top: Quantification of FRAP data for YTHDC1-m6A RNA droplets. The bleaching event occurs at t = 0 s. Mean+s.e.m, n= 5; Bottom: representative images of fluorescence recovery. Scale bars, 2μm. (F) Time-lapse images of YTHDC1-m6A RNA droplets showing a droplet fusion event at indicated time. Scale bars, 2μm. (G) Live imaging of endogenously tagged EGFP-YTHDC1 in OCIAML3 cells. The white line highlights the nuclear periphery. Left: 2D image. Right:3D image. Scale bars, 5μm. (H) Left: Quantification of FRAP. Mean+ s.e.m, n=3. Right: representative images of fluorescence recovery. Scale bars, 5μm.
We next sought to determine if these YTHDC1 nuclear puncta are part of any known previously characterized nuclear bodies. We found that YTHDC1-interacting proteins from the BioGRID database were enriched in gene functional categories associated with splicing, RNA surveillance and RNA transport (Figure S4D, Table S2). Additionally, these factors were also known to be a part of nuclear bodies, speckles and RNA polymerase II complex (Figure S4D). These data support that YTHDC1 puncta is involved in multiple steps of RNA metabolism. We next quantified the relative association of YTHDC1 with known nuclear bodies including nuclear speckles (SRSF2), Cajal bodies (COILIN), PML bodies (PML), super-enhancer condensates (BRD4) and NPM1 bodies (NPM1) in leukemia cells (Figure S4E). YTHDC1 puncta were rarely (~1–2%) colocalized with Cajal bodies, PML bodies and NPM1 bodies. However, around 40% of YTHDC1 is colocalized with nuclear speckles and 35% of YTHDC1 is colocalized with super-enhancer condensates, indicating that YTHDC1 puncta could function in regulating gene transcription and mRNA processing. Thus, nuclear YTHDC1 condensates only partially overlap with these well-characterized nuclear structures.
YTHDC1 undergoes LLPS
We then asked if the YTHDC1 condensates could be associated with a phase transition. We purified full-length recombinant EGFP (enhanced green fluorescent protein)-YTHDC1 and observed droplet formation suggesting a phase separation of YTHDC1 (Figure 2D, Figure S5A). We next tested whether the binding of m6A-RNA regulates phase separation of YTHDC1 protein. Addition of a m6A containing RNA triggered droplets formation in minutes with increased number, size and intensity, showing much stronger phase separation than YTHDC1 protein alone (Figure 2D). Compared to methylated RNA, the non-m6A-containing nucleotides induced YTHDC1 droplets to a lesser extent with slower kinetics and made up of smaller sized droplets (Figure S5B). Both protein concentrations and methylated RNA concentrations affected YTHDC1 phase separation (Figure S5C). Compared to solid-like condensates, liquid-like condensates are more dynamic with rapid fluorescence recovery after photobleaching (FRAP). We found that these condensates have liquid-liquid like properties since photobleaching of the region of EGFP-YTHDC1 droplets containing methylated RNA resulted in a rapid fluorescence recovery (Figure 2E; Video S1). Additionally, time lapse imaging revealed droplets fusing to form larger droplets dynamically (Figure 2F; Video S2). These data suggest that m6A enhances YTHDC1 to form liquid-liquid droplets through phase separation in vitro at low concentrations that are likely to be physiologically relevant.
We then wanted to determine if endogenous YTHDC1 puncta exhibits liquid-liquid like properties by CRISPR-Cas9 mediated tagging endogenous YTHDC1 with EGFP in OCIAML3 and 293T cells (Figures S5D and E). Live-cell imaging revealed discrete nuclear puncta (Figure 2G; Figure S5F; Video S3) that resembled staining of endogenous YTHDC1 in unmodified cells. These EGFP-YTHDC1 puncta recovered fluorescence rapidly and exhibited liquid-liquid like properties (Figures 2H). These data suggested that YTHDC1 can from dynamic liquid-liquid phased condensates in cells.
IDR domains and m6A binding are essential for the function of nYACs in AML
We next characterized the domains in YTHDC1 that are essential for LLPS. We generated different YTHDC1 mutants including the N-terminal IDR-deletion (Δpoly E) and the C-terminal IDR-deletion (ΔR+P) (Figure 3A). EGFP-YTHDC1 formed distinct nuclear dynamic condensates with rapid fluorescence recovery in a FRAP assay (Figure 3B, Figure S5G, Video S4 and 5). However, disruption of either of the YTHDC1 IDRs affected LLPS resulted in more irregular-shaped and decreased number of condensates indicating impaired puncta formation capacity (Figures 3B and C). These results demonstrate that the IDR domains are critical for proper YTHDC1 condensates in cells.
Figure 3: m6A dependent nYACs are essential for leukemia cell survival and differentiation.
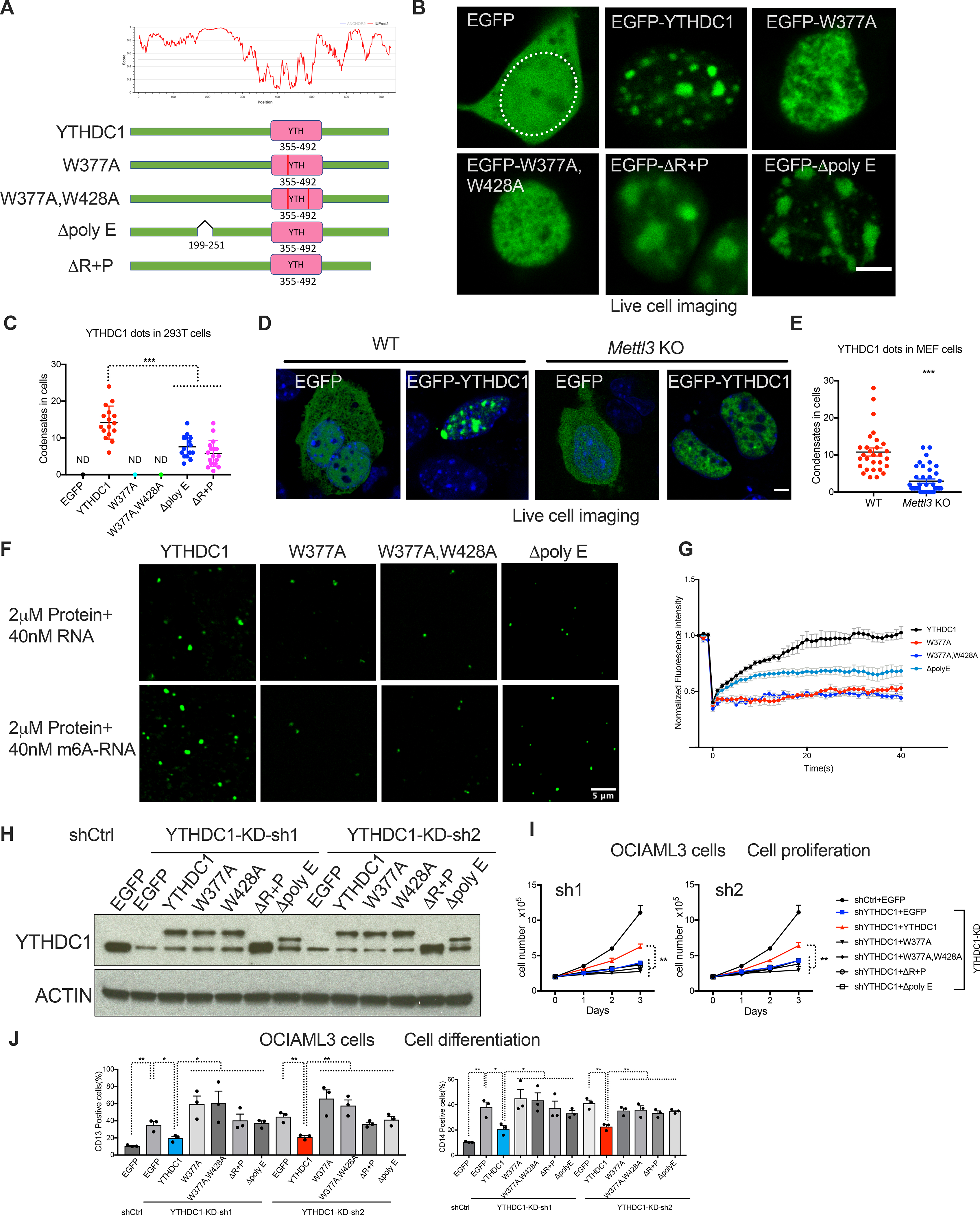
(A) Top: Graphs plotting intrinsic disorder for YTHDC1. PONDR (Predictor of Natural Disordered Regions) VSL2 scores are shown on the y axis, and amino acid positions are shown on the x axis. Bottom: Schematic of EGFP fused WT YTHDC1 and different YTHDC1 mutants used in this study. Pink boxes indicate the YTH domain. Green boxes indicate predicted IDRs. (B) Live imaging of 293T cells expressing EGFP fused WT YTHDC1 and different YTHDC1 mutants as indicated. The white line highlights the cell nuclear. Scale bars, 5μm. (C) Quantitative summary of YTHDC1 condensates in 293T cells related to (B). ND: not detected. Mean+s.e.m, n=16,15,17 from 3 independent experiments. (D) Live imaging of WT and Mettl3 KO MEF cells transfected with EGFP–YTHDC1 or EGFP as control. Scale bars, 5μm. (E) Quantitative summary of YTHDC1 condensates in WT and Mettl3 KO cells related to (D). Mean+s.e.m, n=29,39 from 2 independent experiments. (F) In vitro phase separation of YTHDC1 and its mutants proteins. Top: 2μM YTHDC1 protein plus 40nM 65-nucleotide non-m6A RNA. Bottom: 2μM YTHDC1 protein plus 40nM 65-nucleotide RNA containing 10 m6A nucleotides. Scale bars, 5μm. (G) Quantification of FRAP data for droplets of YTHDC1 and its mutants plus m6A RNA as indicated. The bleaching event occurs at t = 0 s. For YTHDC1 and Δpoly E, n=9; for W377A, W428A, n=6. (H-J) OCIAML3 cells overexpressed with EGFP (control), WT YTHDC1 or different YTHDC1 mutants as indicated were followed endogenous YTHDC1 knockdown by viral transduction. n=3 independent experiments. (H) Representative immunoblot probed with indicated antibodies. (I) Cell numbers measured over time of OCIAML3 cells. (J) Quantitative summary of myeloid differentiation determined by flow cytometry using CD13 (left) and CD14 (right) in OCIAML3 cells.
Error bars, s.e.m. *p<0.05, **p<0.01, ***p<0.001, two-tailed t test.
To determine if these nuclear condensates were regulated by m6A, we generated two YTH domain-mutants (W377A, W428A) to disrupt the tryptophan cage required for m6A binding(Xu et al., 2014) (Figure 3A). Consistent with our in vitro data that m6A-containing mRNAs enhance LLPS of YTHDC1, we observed that disruption of the YTH domain resulted in the disappearance of puncta, which we have named as nuclear YTHDC1-m6A condensates (nYACs) (Figures 3B and C, Figure S5G). To further examine whether m6A binding is required for YTHDC1-mediated LLPS, we utilized m6A-deficient MEF cells generated by tamoxifen-inducing expression of ERT-Cre in Mettl3 flox/flox MEFs (Figures S5H). Overexpression of YTHDC1 in Mettl3 KO MEF cells rarely form puncta (Figures 3D and E).
We next purified YTHDC1 protein containing mutants in YTH domain or IDR domain and tested the effects of these mutants on YTHDC1 phase separation in vitro (Figure 3F). Compared to the WT YTHDC1 protein, both YTH and IDR mutants exhibited attenuated capacity to form droplets in vitro. Consistent with our in vivo data that m6A binding is essential for nYACs formation, m6A-RNA cannot enhance phase separation of YTHDC1 containing mutants in YTH domain compared to unmodified RNA (Figure 3F). In contrast, m6A RNA can still promote droplets formation of IDR mutant that contains a WT YTH domain. However, fluorescence recovery of YTHDC1 proteins was reduced in IDR mutants and almost lost in YTH mutants (Figure 3G). Overall, these data further support that both IDR domain and YTH domain are essential for YTHDC1 LLPS.
We then tested if the ability of YTHDC1 to form nYACs is required for AML survival. Thus, we performed a set of rescue experiments with shRNA-resistant YTHDC1 constructs. Importantly, we were able to partially rescue both proliferation and the increase of differentiation by overexpressing shRNA-resistant YTHDC1 (Figures 3H–J; Figure S5I) indicating that our shRNA-mediated depletion was an on-target effect. In contrast, all of YTHDC1 mutants (YTH domain mutant and IDR deleted), that were defective in forming nYACs, failed to reverse the reduction in proliferation and increase in differentiation induced by YTHDC1 depletion despite their equivalent abundance to the WT (Figures 3H–J; Figure S5I). These data suggest that nYACs abundance is essential for leukemia growth and differentiation control.
YTHDC1 prevents myeloid differentiation of human HSPCs
We next sought to address the role for YTHDC1 in normal hematopoietic cells to determine if there is differential dependency for YTHDC1 in normal blood cells compared to leukemia cells. Thus, we first depleted YTHDC1 using shRNAs in human CB-CD34+ HSPCs (Figure 4A). Loss of YTHDC1 resulted in modest inhibition of cell growth (Figure 4B) and colony-forming units (CFUs) in all lineages with no effect on apoptosis (Figures 4C and D). Moreover, we observed a significant but modest increase in myeloid differentiation ((Figures 4E–G) and no effect on erythroid differentiation (Figures 4H and I) after YTHDC1 depletion. These data indicate that leukemia cell survival and differentiation state is more dependent on YTHDC1 than normal HSPCs.
Figure 4: YTHDC1 contributes to human HSPC myeloid differentiation.
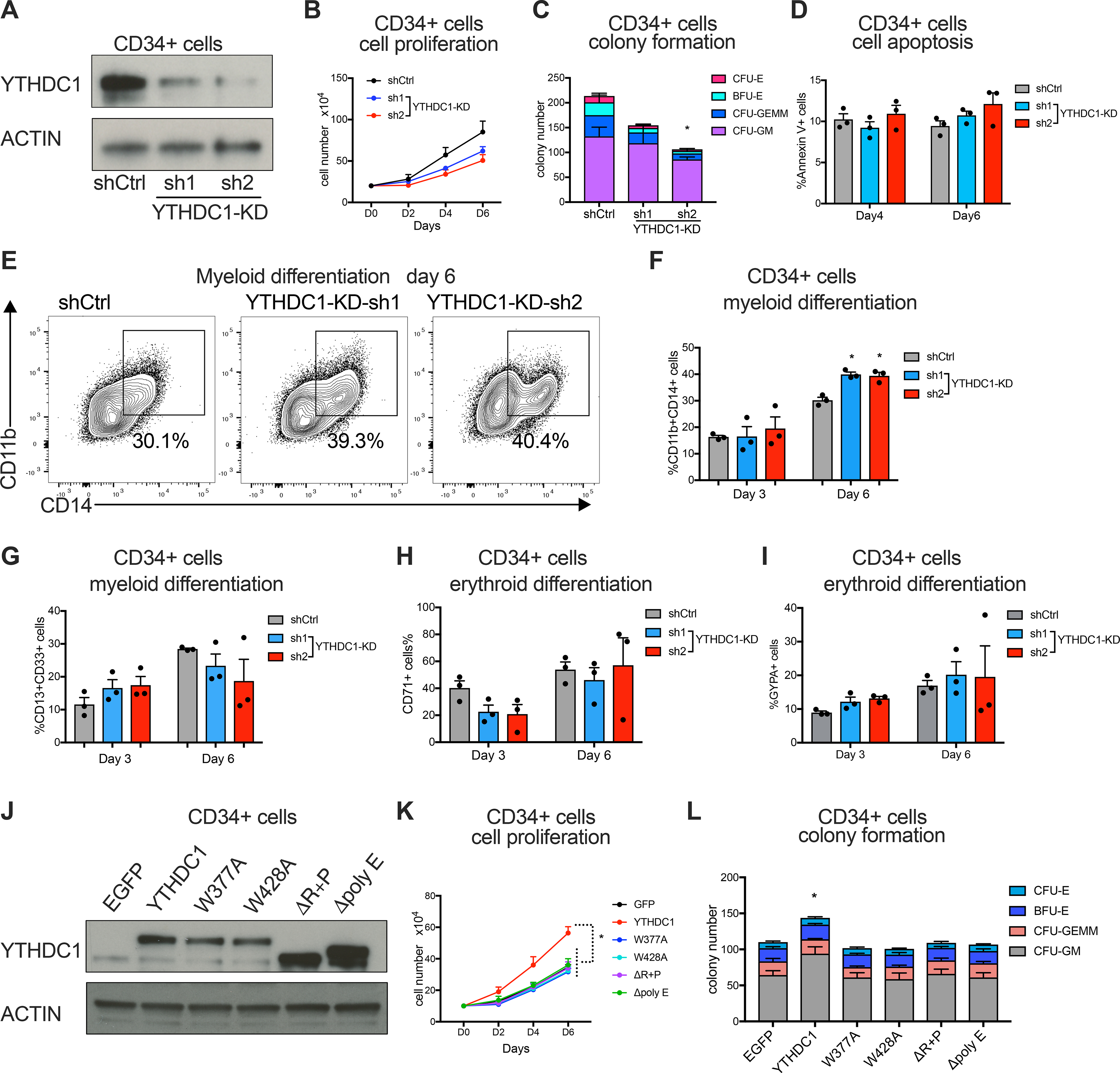
(A-I) Human CB-CD34+ cells were transduced with lentiviruses expressing control shRNA or two independent shRNAs targeting YTHDC1. Cells were used for following experiments after puromycin selection. n=3 independent experiments. (A) Immunoblot of YTHDC1 expression in CB-CD34+ cells. (B) Cell proliferation of control and YTHDC1 depleted CB-CD34+ cells were determined. (C) Colony forming assay of control and YTHDC1 depleted HSPCs. The total number of colony forming units (CFUs) was scored two weeks after plating. (D) Apoptotic cells were determined by flow cytometry at day four and six post-transduction. (E-G) Myeloid differentiation of CB-CD34+ cells was measured by flow cytometry using CD11b, CD13, CD14 and CD33 as markers at indicated timepoint. Representative flow plot was shown in (E). (H and I) Erythroid differentiation of CB-CD34+ cells was measured by flow cytometry using CD71 and glycophorin A(GYPA) as markers at indicated timepoint. (J-L) CB-CD34+ cells were transduced with lentiviruses expressing control, YTHDC1 and its different mutants as indicated. Sorted cells were used for following experiments. n=3 independent experiments. (J) Representative immunoblot of YTHDC1 expression in indicated CB-CD34+ cells. (K) Cell proliferation of CB-CD34+ cells were determined. (L) Cells in (K) were plated on methylcellulose (5000 cells for each replicate). The total number of colony forming units (CFUs) was scored two weeks after plating.
Error bars, s.e.m. * p<0.05, **p<0.01, ***p<0.001, two-tailed t test.
Since YTHDC1 expression is lower in human HSPCs than AML cells, we next overexpressed WT YTHDC1 and mutants that fail to form nYACs in CB-CD34+ cells (Figure 4J). Forced expression of YTHDC1 but not YTHDC1 mutants resulted in an increase in cell proliferation and colony formation of CB-CD34+ cells (Figures 4K and L). These data indicate that ectopic YTHDC1 expression is sufficient to induce dysregulated cell growth of HSPCs and this effect depends on nYACs formation.
YTHDC1 and nYACs promote abundance of target mRNAs
To understand the molecular basis for nYACs on leukemia survival, we performed RNA-seq on MOLM13 cells after YTHDC1 depletion. We found large transcriptional changes that included 1320 downregulated genes and 520 upregulated genes (Figure 5A; Table S3). Among the downregulated genes, we found genes enriched in DNA replication and cell cycle pathway and related transcription factors including E2F1, MYC, TP53, FOXM1 and SP1 (Figures S6A–C). In support of our cellular phenotypes, we also observed enrichment for myeloid differentiation and a loss of the HOXA9-MEIS1 gene signatures (Figures S6D and E). These data suggest that YTHDC1 maintains a cell growth and undifferentiated gene expression program.
Figure 5: YTHDC1 and nYACs are essential for maintaining mRNA abundance of its targets.
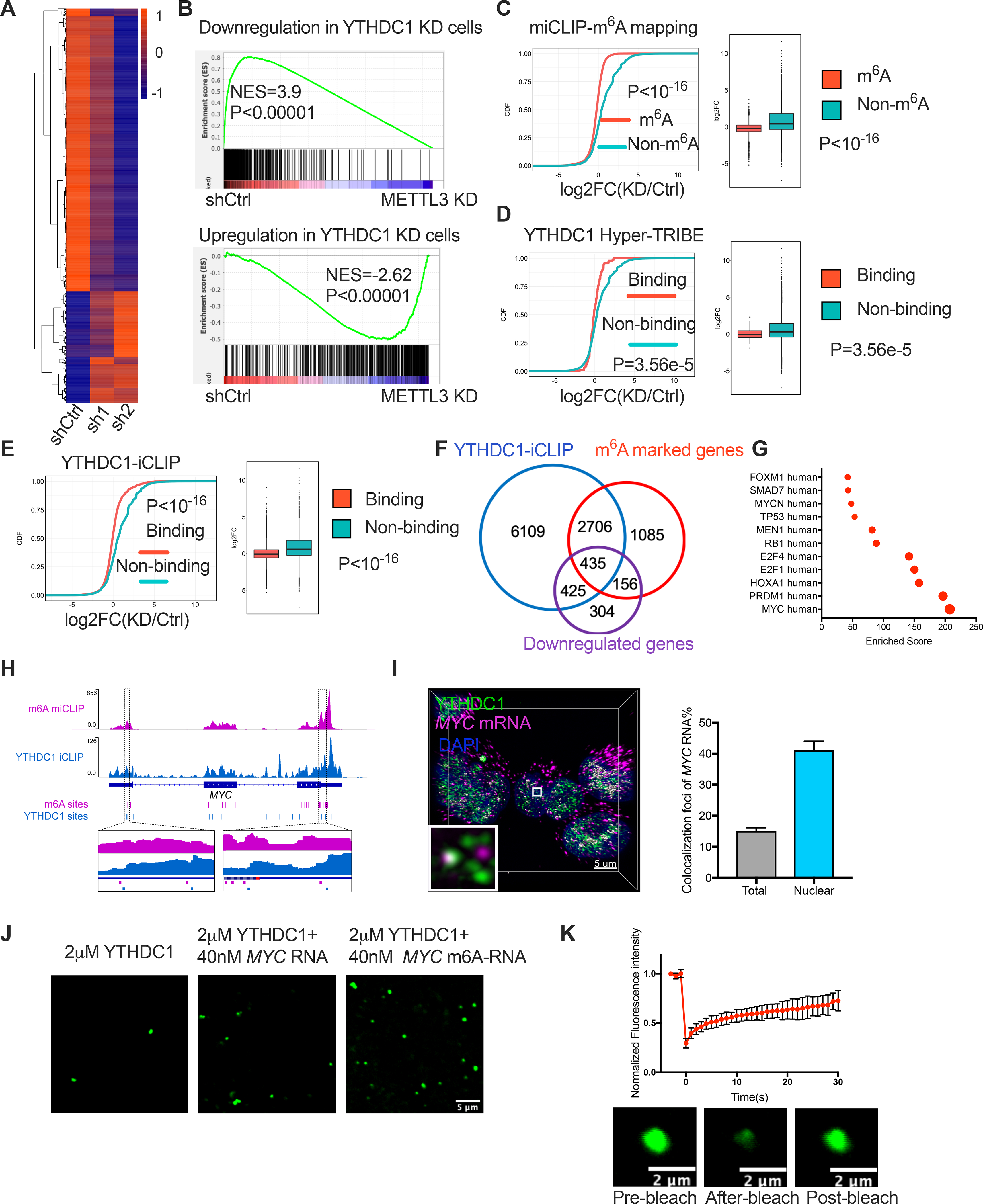
(A) Significant differentially expressed genes upon YTHDC1 depletion in MOLM13 cells by RNA-seq were shown as heatmap. n=3 independent replicants. (B) Gene set enrichment analysis of differentially expressed genes upon YTHDC1 depletion with differentially expressed genes upon METTL3 knockdown in MOLM13 cells. (C) Cumulative distribution (left) and boxplots (right) to show the abundance of m6A methylated and non-methylated transcripts upon YTHDC1 knockdown. (D-E) Cumulative distribution (left) and boxplots (right) to show the abundance of YTHDC1 binding and non-binding transcripts upon YTHDC1 depletion. These binding targets were identified by Hyper-TRIBE of YTHDC1 in (D) and by iCLIP in (E). (F) Venn diagram shows overlapped genes 1: genes containing YTHDC1 binding sits identified by iCLIP; 2: genes containing at least 1 m6A site mapped by miCLIP; 3: downregulated genes upon YTHDC1 depletion. (G) Transcription factors that enriched with overlapped 435 genes from (F). (H) Gene tracks displaying the iCLIP(YTHDC1 binding sites) and miCLIP(m6A sites) read coverage at the MYC locus. (I) Left: Representative 3D image of of MYC mRNAs (magenta) by FISH and YTHDC1 (green) by IF and DAPI (blue). Scale bars, 5μm. Right: Quantitative summary of colocalization of total or nuclear MYC mRNAs with YTHDC1 protein. (J) In vitro phase separation of YTHDC1 protein as well as YTHDC1 protein plus 200nt MYC RNA with no m6A sites or 4 m6A sites. Scale bars, 5μm. (K) Top: Quantification of FRAP data for YTHDC1-m6A MYC RNA droplets in (J). The bleaching event occurs at t = 0 s. Mean+s.e.m, n= 7. Bottom: Representative images showing FRAP. Scale bars, 2μm.
Given that nYACs are m6A dependent, we next examined how YTHDC1 regulates the expression of m6A-containing transcripts. Previously we found that METTL3 depletion resulted in reduced translation of m6A-marked mRNAs with a concomitant of increased mRNA abundance of these targets in the cytoplasmic fraction in AML cells(Vu et al., 2017a). Increased cytoplasmic mRNAs could be due to the block of cytoplasmic m6A reader YTHDF2 mediated m6A-mRNA decay upon METTL3 depletion. In contrast to METTL3 or YTHDFs depletion, we found that YTHDC1 depletion has an opposite effect on the expression of METTL3-regulated transcripts as demonstrated by a negative enrichment with gene set in METTL3-depleted cells(Vu et al., 2017a) (Figure 5B). Furthermore, by overlapping YTHDC1 shRNA-RNA-seq dataset with our m6A profiling dataset by miCLIP (m6A individual-nucleotide-resolution cross-linking and immunoprecipitation)(Vu et al., 2017a), we observed that even one m6A site was sufficient for reduced expression of these m6A mRNAs (~45%) (Figure 5C; Figure S6F).These data indicated that YTHDC1 depletion reduces the expression m6A marked transcripts, which is opposite to the regulation of METTL3/YTHDFs on transcripts, suggesting that the fate of m6A on mRNA transcripts in the nucleus differs from the YTHDF-mediated effects of m6A in the cytosol.
To identify the direct targets of YTHDC1, we then mapped its transcriptome-wide RNA-binding sites using several approaches. Recently, we identified targets of RNA-binding proteins (RBPs) in mammalian cells using Hyper-TRIBE (Nguyen et al., 2020). Thus, we performed YTHDC1 Hyper-TRIBE and identified ~300 significant sites representing ~200 genes after filter (Figure S6G and H, Table S4). We then compared data from YTHDC1 Hyper-TRIBE to YTHDC1-iCLIP (individual-nucleotide-resolution cross-linking and immunoprecipitation) generated previously that identified over 9000 YTHDC1 potential binding genes (Patil et al., 2016). Although these targets from Hyper-TRIBE were representing only a subset of YTHDC1 target genes identified by iCLIP (Table S5), de novo motif finder identified a similar m6A binding motif (DRACH) and an enrichment for m6A sites from the miCLIP dataset (Figures S6I and J). Furthermore, both the YTHDC1-binding targets from our Hyper-TRIBE and the YTHDC1-iCLIP enriched for YTHDC1-downregulated genes by RNA-seq (Figures 5D and E). Thus, these data suggest that YTHDC1 directly binds to m6A-marked mRNAs and this is critical to maintain the mRNA expression of these targets.
We next overlapped YTHDC1 binding genes from iCLIP with m6A modified genes by miCLIP and downregulated genes upon YTHDC1 depletion by RNA-seq to identify the YTHDC1 direct regulated gene signature (Figure 5F). High expression of the YTHDC1 regulated gene signature predicted a poor prognosis in AML patients (Figure S6K). In contrast, expression of the upregulated gene signature upon YTHDC1 depletion that is indirectly controlled by YTHDC1 was not associated with patient survival (Figure S6K). We then identified the minimal set of YTHDC1 direct regulated genes (12-gene set) that could have predictive value using three different AML cohorts (Figure S6L–N). Overall, these data indicate that YTHDC1 promotes expression of target genes, which is associated with clinical survival outcome of AML patients.
YTHDC1 indirectly regulates splicing in AML
As previous study reported that YTHDC1 regulates mRNA splicing (Xiao et al., 2016), we next assessed the impact of YTHDC1 on global splicing in myeloid leukemia. We observed over 4000 different types of alternative splicing events representing 2486 genes and around 54% spliced exons are more included upon YTHDC1 depletion (Figures S6O and P; Table S6). About 43% of the genes with altered splicing upon YTHDC1 depletion contain m6A sites (Figure S6Q). However, the location of m6A sites on the vast majority of these transcripts are far from both donor and acceptor of spliced exons (>1kb), suggesting these splicing events may be not directly regulated by YTHDC1 binding to m6A (Figure S6R). More importantly, only 5% of these genes were downregulated and direct YTHDC1-m6A targets (Figure S6Q). Therefore, these data demonstrate that although YTHDC1 is associated with m6A-related splicing alterations, the YTHDC1 does not directly maintain the expression of these genes through a splicing mechanism.
MYC is a functional direct target of YTHDC1 in AML
Given that splicing does not explain how YTHDC1 controls m6A target expression, we next sought to further understand how YTHDC1 depletion reduces expression by focusing on specific m6A-YTHDC1 direct targets. The downregulated YTHDC1-m6A-direct targets were strongly enriched in the MYC signaling pathway and the MYC transcript itself contains at least 20 m6A sites in MOLM13 cells(Vu et al., 2017a) (Figures 5G and H). Based on this data and the known role for MYC in the RNA methylation pathway, we examined how MYC is affected by YTHDC1. Co-localization of MYC mRNA (fluorescence in situ hybridization, FISH) and the YTHDC1 puncta (Immunofluorescence, IF) in the nucleus suggests that nYACs directly bind mRNA and functionally affect the mRNA fate of specific targets (Figure 5I). Additionally, m6A modified MYC RNA can promote droplet formation of YTHDC1 protein in vitro and rapid FRAP indicated the liquid-liquid like phase transition (Figures 5J and K). Overall, the data associate YTHDC1 binding to its targets with condensate formation and expression control.
To determine if nYACs directly regulate YTHDC1 targets in AML cells, we next validated by qPCR the down-regulation of several critical cell survival targets including MYC, GINS1 and FOXM1 in MOLM13 cells (Figure 6A). Corresponding with MYC being an important target of YTHDC1 in AML patients, we found that high YTHDC1 expression correlated with an increased MYC expression (Figure S7A). Consistent with the effects on the mRNA, MYC protein was reduced upon YTHDC1 depletion in multiple AML cell lines (Figure S7B). Most importantly, MYC was required for YTHDC1’s function as MYC overexpression could partially rescue the proliferation and differentiation effects after YTHDC1 depletion (Figures 6B–G). These data suggest that MYC functionally contributes to the YTHDC1’s effects in AML.
Figure 6: MYC is a functional target of YTHDC1 in AML.
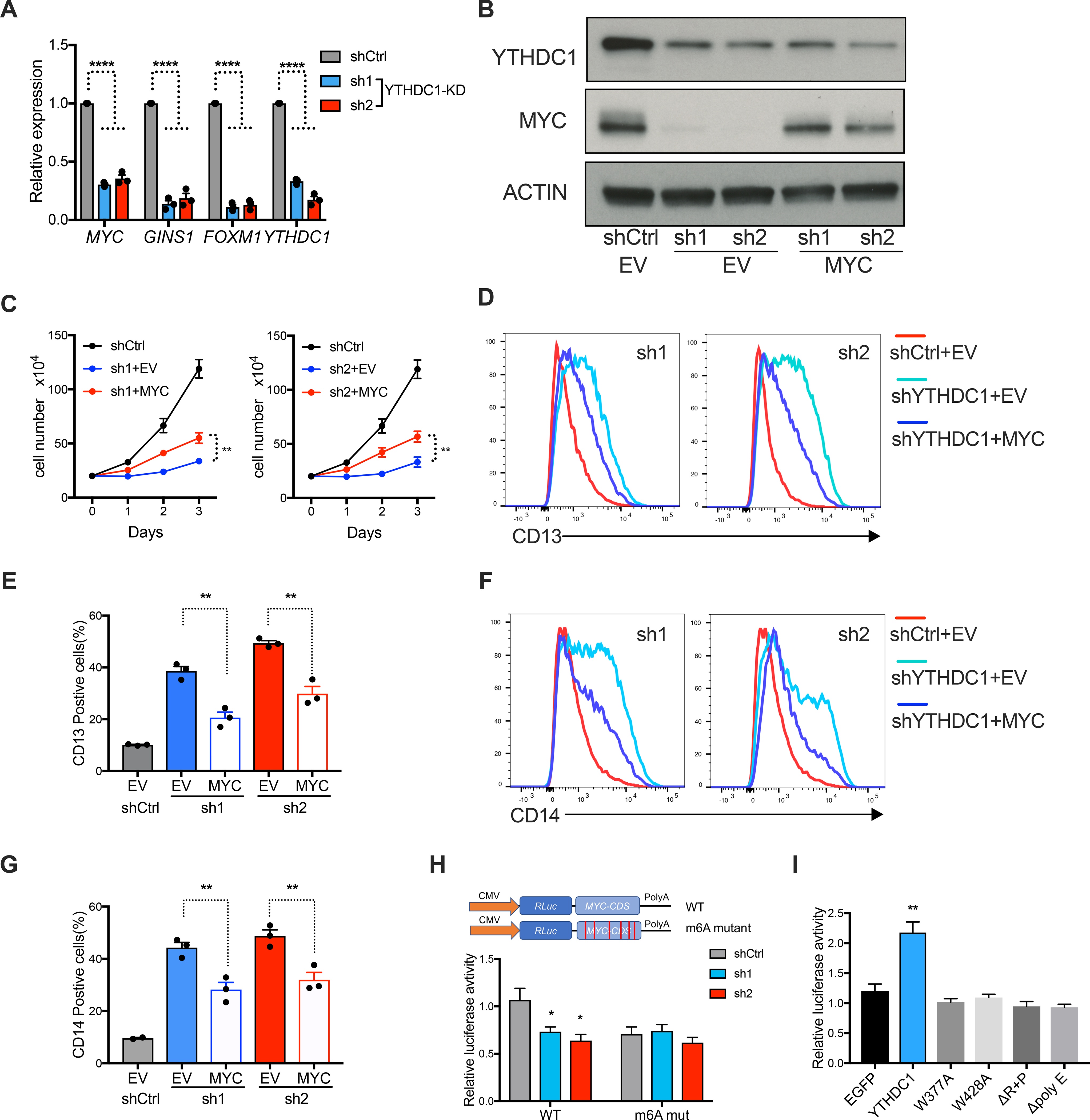
(A) qPCR to measure mRNA expression of YTHDC1 targets in MOLM13 cells upon YTHDC1 depletion. n=3 independent experiments. (B-G) OCIAML3 cells overexpressing empty vector (EV) or MYC as indicated were followed with endogenous YTHDC1 knockdown by viral transduction. EV is used as control for overexpression. shCtrl is used as control for shRNAs. n=3 independent experiments. (B) Representative immunoblot in OCIAML3 cells probed with indicated antibodies. (C) Cell proliferation of OCIAML3 cells. (D-G) Myeloid differentiation of OCIAML3 cells. (D) and (F): Representative flow plot to show expression of myeloid marks CD13 and CD14. (E) and (G): Quantitative summary of myeloid differentiation using CD13 (E) and CD14 (G) determined by flow cytometry. (H) Up: Diagram of vector used in luciferase reporter assay. Bottom: Luciferase reporter assay using the original MYC CDS or the m6A sites mutated MYC CDS in 293T cells. 293T cells were transfected with control or YTHDC1 shRNA constructs. Normalized luciferase activity was calculated. n=4 independent experiments. (I) Luciferase constructs are the same as (H). 293T cells were transfected with control vector (EGFP), YTHDC1, or indicated YTHDC1 mutants. Normalized luciferase activity was calculated. n=4 independent experiments. Mean and s.e.m are shown (*, P < 0.05; **, P < 0.01; ***, P < 0.001, ****, P < 0.0001). two-tailed t test.
We next sought to assess if m6A binding is required for YTHDC1 mediated gene regulation. YTHDC1 knockdown reduced MYC luciferase activity and this effect was mostly abrogated if the m6A sites were mutated (Figure 6H; Figures S7C and D). Furthermore, only WT YTHDC1 but not the YTHDC1 mutants increased MYC luciferase activity (Figure 6I; Figure S7E). These data demonstrate that YTHDC1 can regulate MYC expression in a m6A dependent manner and LLPS of YTHDC1 is essential for this regulation.
YTHDC1 and nYACs protect target mRNAs from degradation
Recent studies demonstrated that YTHDC1 can affect transcriptional activity by regulating chromatin states (Li et al., 2020; Liu et al., 2020a). We find that nascent transcription of YTHDC1 targets (MYC, GINS1 and FOXM1) in leukemia cells were not affected by YTHDC1 depletion (Figure 7A; Figure S7F). These data suggest that nYACs can maintain transcript abundance through a post-transcriptional mechanism rather than altering transcriptional activity of these targets. We then asked whether YTHDC1’s direct targets were being downregulated through reduced mRNA stability and nuclear mRNA degradation mechanisms. Decreased half-life of MYC and GINS1 mRNAs suggested that YTHDC1 stabilizes its targets (Figure 7B). We next tested the colocalization of MYC mRNAs to nuclear degradation pathways, as assessed by RNA-FISH co-staining with nuclear exosome component RRP6. We observed increased co-localization of MYC mRNA with RRP6 (from ~13 to 30%) upon YTHDC1 depletion (Figure 7C). These data suggest that loss of nYACs results in an increase in degradation of MYC mRNA matching with the reduced mRNA half-life. As reported previously, the nuclear exosome is essential for RNA elimination of aberrant transcripts (i.e. intron-containing splice-defective mRNAs, export-defective or improperly processed, etc.)(Singh et al., 2018). Our data raise the possibility that nYACs protect m6A-mRNAs from the degradation machinery.
Figure 7: nYACs protect mRNAs from the PAXT complex and exosome mediated degradation.
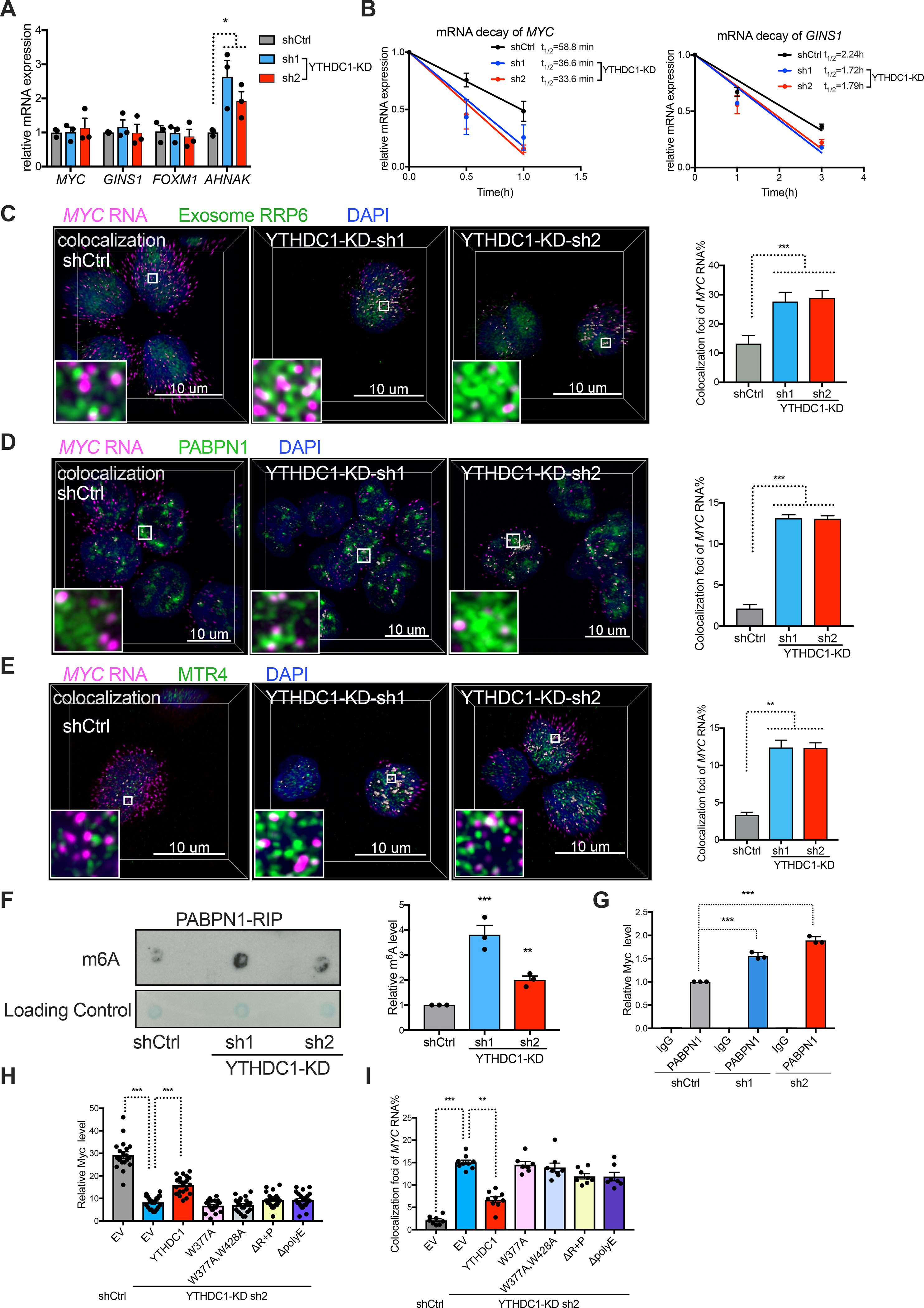
(A) Analysis of nascent RNA synthesis of specific genes in control or YTHDC1 depleted MOLM13 cells. n=3 independent experiments. (B) The mRNA half-life (t1/2) of MYC and GINS1 transcripts in control and YTHDC1 depleted cells. n=3 independent experiments. (C-E) Left: Representative 3D images of MYC mRNA (magenta) by FISH con-staining with exosome protein RRP6 (C) or PAXT protein PABPN1 (D) or PAXT protein MTR4 (E) (green) by IF and DAPI (blue) in control and YTHDC1 depleted cells. White dots indicate co-localization between MYC mRNA and protein as indicated. Scale bars, 5μm. Right: Quantitative summary of co-localization of MYC mRNA with indicated protein (n=10). (F) Left: m6A dot blot assays of PABPN1 binding RNAs by RNA-Immunoprecipitation (RIP) from control and YTHDC1 depleted MOLM13 cells. Right: Quantitative summary of m6A level of PABPN1 binding RNAs by RIP. n=3 independent experiments. (G) qPCR of PABPN1-RIP recovered RNA at MYC locus in control and YTHDC1 depleted MOLM13 cells. IgG served as a non-specific binding control. n=3 independent experiments. (H and I) OCIAML3 cells overexpressed with control, WT YTHDC1 or different YTHDC1 mutants as indicated were followed endogenous YTHDC1 knockdown by viral transduction. n=3 independent experiments. (H) Quantitative summary of MYC mRNA by RNA-FISH. (I) Quantitative summary of co-localization of MYC mRNA and PABPN1 protein.
Error bars, s.e.m. *p<0.05, **p<0.01, ***p<0.001, two-tailed t test.
Depletion of YTHDC1 resulted in increased PAXT mediated nuclear m6A mRNA decay
There are two major known complexes NEXT (nuclear exosome targeting complex) and PAXT (polyA tail exosome targeting complex) in the nucleus targeting RNAs to degradation(Wolin and Maquat, 2019). The NEXT complex mediates decay of non-polyA ncRNAs whereas the PAXT targets polyadenylated RNAs for degradation(Meola et al., 2016; Silla et al., 2018). Thus, we tested if nYACs protect MYC mRNA from the PAXT complex mediated degradation. YTHDC1 depletion resulted in significantly increased colocalization of MYC mRNA with PAXT components PABPN1 and MTR4 and this increase is not due to their elevated protein abundance (Figures 7D and E; Figure S7G). We also observed that PABPN1 binds (PABPN1-RIP) to more m6A modified RNAs upon YTHDC1 depletion including MYC (Figure 7F–G). These data support the hypothesis that nYACs protect m6A-RNAs from the PAXT complex mediated RNA degradation. Furthermore, this mechanism was also associated with a reduction of nuclear MYC mRNAs, a more pronounced reduction in cytoplasmic MYC mRNAs and an increase in the nuclear to cytoplasmic ratio of MYC mRNAs (Figures S7H–J). As YTHDC1 was reported as an important factor in facilitating mRNA export(Roundtree et al., 2017), our data with reduced nuclear MYC mRNA suggests that nYACs play a dominant role in mRNA surveillance.
To determine whether phase transition of YTHDC1 into nYACs is essential for protecting or to ensure proper processing of target mRNAs from degradation, we performed rescue experiments. We observed that only WT YTHDC1 but not nYACs mutants could partially rescue MYC mRNA reduction and increased colocalization of MYC mRNA with PABPN1 after YTHDC1 depletion (Figures 7H and I; Figure S7K). Overall, these findings indicate that nYACs either maintain proper mRNA surveillance and/or protect mRNAs from the PAXT complex and the exosome degradation machinery.
DISCUSSION
In this study, we identified a nuclear condensate formed by YTHDC1 binding to m6A. This suggest that m6A could mediate its effects on transcripts in two compartments, via YTHDC1 in the nucleus first, and then in the cytoplasm via YTHDF2 or other YTHDFs. We observed that YTHDC1 can be found in nuclear speckles, super enhancer condensates and other nuclear bodies. However, the specific functional roles for nYACs in these specific complexes remain to be determined. Based on the lack of known functional domains beside the YTH domain, YTHDC1 is made of N or C-terminal IDRs. These stretches of repetitive sequences are required for LLPS and for YTHDC1’s function in leukemia. However, it remains a possibility that the inability to form nYACs abrogates a specific protein-protein interaction resulting in defect of YTHDC1 function. Future studies will help to determine how disruption of LLPS alters the composition and function of these protein complexes.
YTHDC1 has been linked to disparate effects on m6A mRNAs in the nucleus, including the regulation of transcription, heterochromatin, retrotransposons, splicing, RNA export and homologous recombination-mediated repair of DSBs(Li et al., 2020; Liu et al., 2020a; Liu et al., 2021; Roundtree et al., 2017; Xiao et al., 2016; Xu et al., 2021; Zhang et al., 2020). We find that the changes in m6A-mRNAs after YTHDC1 depletion are not mainly due to changes in gene transcription, splicing or export but from reduced mRNA stability of its targets in AML cells. Mechanistically, our data demonstrates that nYACs prevent m6A-mRNAs from being degraded by reducing PAXT-nuclear-exosome mediated degradation. This may reflect the ability of nYACs to promote nuclear export of m6A-mRNAs by evading the mRNA degradation machinery. Our data provides a framework to understand how YTHDC1 impacts mRNA metabolism through nYACs and thus explaining the disparate previously reported effects of YTHDC1. Our data support the idea that YTHDC1 could be a therapeutic target in myeloid leukemia. However, it is important to expand our study on nYACs and their contribution in primary AML patient samples. Also, future work will determine the requirement of LLPS and its clinical relevance. Importantly, similar to the m6A writers, YTHDC1 could contribute to normal hematopoiesis and blood stem cell function. Development of YTHDC1 selective inhibitors and in vivo models would clarify the therapeutic potential (Li et al., 2019). We propose that nYACs phase separation provides a regulatory mechanism for controlling m6A-mRNA fate in the nucleus and we link this to the functional requirements in maintaining cell growth and differentiation state.
STAR Methods:
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for reagents should be directed to and will be fulfilled by the corresponding author Michael G. Kharas (kharasm@mskcc.org).
Materials Availability
There are no restrictions to the availability of all materials mentioned in the manuscript.
Data and Code Availability
The raw and processed RNA-seq and Hyper-TRIBE data described in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) -- Accession number: GSE168565 and GSE168568.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture.
MOLM13, OCIAML3, THP1, NOMO1 and HL60 cells were cultured in RPMI1640 medium containing 10% FBS and 1%L-Glutamine PenStrep. 293T cells were cultured in DMEM medium containing10% FBS and 1%L-Glutamine PenStrep. All leukemia cells and 293T cells were from ATCC and tested negative for mycoplasma contamination. Mettl3 flox/flox MEF cells were derived from Mettl3 flox/flox embryo and immortalized by continuous passage(Cheng et al., 2019). Mettl3 flox/flox MEF cells were cultured in DMEM medium containing10% FBS and 1%L-Glutamine PenStrep. For Mettl3 knockout, Mettl3 flox/flox MEF cells were transduced with puro-CreER viruses and selected with puromycin. After selection, these cells were treated with 100nM tamoxifen for 24h to induce CRE expression.
Purification and culture of cord blood (CB) derived HSPC-CD34+ cells
CD34+ HSPCs were purified from at least 10 mixed CB units (each unit from one healthy donor) in each purification. After Hespan and Ficoll-Hypaque Plus density centrifugation, mononuclear cells from CB units were performed positive selection using the Auto MACS Pro Seperator and isolation Kit (Miltenyi). CD34+ cells were cultured in basic medium containing Iscove’s modified Dulbecco’s medium (IMDM, Cellgro) 20% BIT 9500 medium (Stem Cell Technologies) supplemented with SCF (100 ng/ml), FLT-3 ligand (10 ng/ml), IL-6 (20 ng/ml) and TPO (100 ng/ml). To differentiate HSPCs, cells were cultured under the myeloid-promoting conditions: SCF (100 ng/ml), FLT-3 ligands (10 ng/ml), IL-3 (20 ng/ml), IL-6 (20 ng/ml), GM-CSF (20 ng/ml) and G-CSF (20 ng/ml) or the erythroid-promoting conditions: Epo (6 IU/ml) and SCF (100 ng/ml). Cytokines were purchased from Peprotech, NJ.
Primary Patient samples
All primary patient samples were collected under the Biospecimen collection/banking study 09–141, and the use of the samples for research purposes was covered under the Biospecimen research protocol 16–354. Briefly, frozen AML patient cells were thawed, and cell lysis was used for immunoblotting.
Primary AML patient cells are cultured in IMDM medium containing 10% dialyzed FBS, beta-mercaptoethanol, 100 ng/ml SCF, 10 ng/ml IL-3, 10 ng/ml TPO, 10 ng/ml FLT-3, 10 ng/ml GM-CSF, L-Glutamine, Penicillin and Streptomycin. Cells were transduced with concentrated virus expressing scramble or YTHDC1 shRNA together with GFP and 10 ug/ml polybrene. Cells were incubated for 6 hrs and new media was added. Cells were then sorted by GFP 24h after transduction for following transplantation.
In vivo transplantation of human leukemia cells
MOLM13 human leukemia cells were transduced with lentiviruses expressing puromycin resistance gene and shRNAs against YTHDC1or a control scramble shRNA. Transduced cells were selected by 3μg/ml puromycin for 2 days. 500,000 selected cells were injected via tail vein into female NSG (6–8 week old, Jackson Laboratories) recipient mice that had been sublethally irradiated with 200 cGy one day before transplantation.
For patient-derived xenograft, the cells from primary patient were first transplanted in irradiated (200 rad) NSG mice at a dose of 3×106 cells/mouse in order to expand them. For secondary transplant, bone marrow cells from primary mouse was sacrificed and transduced with lentiviruses expressing shRNAs against YTHDC1or a control scramble shRNA. 50K or 100K sorted PDX cells were injected into irradiated (200 rad) NSG mice. Survival curve of mice was monitored. All animal studies were performed on animal protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Memorial Sloan Kettering Cancer Center.
Plasmid constructs and transfection
For YTHDC1 knockdown experiment, all shRNAs including shCtrl(Scramble), sh1 and sh2 were cloned in PLKO.1 backbone with puromycin selection or GFP. The sequences were: sh1: TGGATTTGCAGGCGTGAATT; sh2: TGCCTCCAGAGAACCTTATA. sgRNAs were cloned in PLKO.5 backbone with GFP. The sequences were: sg4: GGAAGGAGTGGAAGAAGATG; sg7: GGATCCACGATACCAGGAAG.
For YTHDC1 overexpression, EGFP, EGFP-YTHDC1 and EGFP-YTHDC1 mutants including W377A, W428A, ΔpolyE and ΔR+P, were cloned in lentiviral backbone with blasticidin selection.
For plasmid transfection, 293T cells or MEF cells were transfected with certain constructs using lipofectamine 2000 or lipofectamine 3000 (Invitrogen) following instruction.
METHOD DETAILS
Lentiviral production and transductions
Lentiviral packaging of shRNAs or overexpression constructs was performed in 293T cells as previously prescribed(Vu et al., 2017a; Vu et al., 2017b). Lentivirus was kept at 4°C and used within 2 weeks of production. For transduction, cells were transduced with high-titer lentiviral supernatant and 10μg/ml polybrene followed with spin infection for 1.5 hr. Infected cells were changed with fresh medium and cultured in incubator before downstream experiments.
Proliferation assay
Human leukemia cells or CD34+ cells were transduced with viruses by spin-infection. After 48 hours of infection, cells transduced with shRNAs against YTHDC1 or control scramble shRNA were treated with 3μg/ml puromycin for leukemia cells and 2μg/ml puromycin for CD34+ cells. Two days after puromycin selection, cells were plated at 200,000 cells/ml for proliferation assay.
Colony forming unit (CFU) assay
Transduced CD34+ cells were plated in methylcellulose (MethoCult TMH4434 Classic – Stem Cell Technologies). CFU colonies: erythroid progenitor cells (BFU-E and CFU-E), granulocyte-macrophage progenitor cells (CFU-GM, CFU-G and CFU-M), and multipotent granulocyte, erythroid, macrophage and megakaryocyte progenitor cells (CFU-GEMM) were scored 14 days after seeding.
Flow cytometry
To monitor the differentiation status, cells were stained with the following antibodies: APC-CD11b (ThermoFisher, CD11b 05), APC-CD13 (ThermoFisher, MHCD1305), PE-CD14 (BD Pharmingen, 555398), PE-CD33 (BD Pharmingen, 555450). To measure apoptosis, cells were washed with PBS and incubated with anti– PE-Annexin V (BD Pharmingen, 556421) and 7-AAD in the ANNEXIN-V binding buffer in a reaction volume for 15 minutes based on manufacturer’s instruction. Cells were analyzed on a BD FACS LSR or Fortessa instrument.
To monitor human leukemia cell engraftment, bone marrow cells from recipient mice were stained with PB-mouse CD45.1 (Invitrogen 110722) and PerCP-Cyanine 5.5-human CD45 (eBioscience, 45–9459-42). Immunoblot analysis of engrafted human cells was performed using sorted human CD45 positive cells.
Immunoblot analysis
Cells were counted and washed twice with cold PBS prior to collection. ~ 250,000 were resuspended and lysed in 40μl 1X Lamine protein running buffer and boiled for 5 minutes. Whole cell lysates were run on 4%–15% gradient SDS-PAGE and transferred to nitrocellulose membrane. Membranes were blocked in 5% milk PBST for 30 min at room temperature (RT), incubated in a diluted primary antibody solution at 4 °C overnight, washed and incubated in a dilution of secondary antibody conjugated to HRP for 1h at RT. Antibodies used are shown in the resource table.
RNA isolation and qPCR
Total RNA was extracted from cells using TRIZOL (Life Technologies) following the standard manual. Equal amount of RNA from samples was reverse transcribed into cDNA with Verso cDNA Synthesis Kit (Thermo Fisher), and qPCR was performed using an ABI 7500 sequence detection system using primers together with SYBR green master mix (ABI systems). Primers are listed below:
MYC-F: TTCGGGTAGTGGAAAACCAG
MYC-R: CAGCAGCTCGAATTTCTTCC
GINS1-F: GGTCACTGGGAGGAGATGAA
GINS1-R: GCTCACATTTCCATCGAGGT
FOXM1-F: ACCCAAACCAGCTATGATGC
FOXM1-R: GAAGCCACTGGATGTTGGAT
YTHDC1-F: AAGGAGGGCCAAATCTCCTA
YTHDC1-R: CAGTGTTGTTCCCTTGCTCA
AHNAK-F: CAGGAGGTGACGCAGAACTC
AHNAK-R: GACTTCACGGGTCCAGGTCT
ACTIN-F: GGACTTCGAGCAAGAGATGG
ACTIN-R: AGCACTGTGTTGGCGTACAG
RNA-FISH in conjugation with fluorescent immunostaining
Control and YTHDC1 depleted cells were fixed with 4% paraformaldehyde and permeabilized with cold methanol. Fixed cells were then cytospun onto glass slides. RNA in situ hybridization was performed using RNAscope multiplex fluorescent detection kit according to the manufacturer’s instructions (Advanced Cell Diagnostics). RNAscope probes targeting human MYC was designed and produced by ACDbio. After the in stiu hybridization was completed, slides were washed twice and subjected to immunostaining.
For immunostaining, cells were fixed with 4% paraformaldehyde 15 mins at room temperature and permeabilized with cold methanol. Fixed cells were then cytospun onto glass slides and were blocked (with buffer PBST+0.5%BSA) for 1h followed with staining on slides with primary antibody and secondary Ab (Goat anti–rabbit Alexa flour 488, Invitrogen) with DAPI counterstaining. Antibodies used are shown in the resource table.
mRNA stability assay
mRNA stability analysis is performed as previously described. Briefly, control and YTHDC1 depleted cells were treated with 5 μg/ml actinomycin D (Sigma) for inhibition of mRNA transcription. Cells were harvested at indicated time points and total RNA was extracted and used for qRT-PCR. Relative mRNA levels are normalized to the starting point of treatment.
YTHDC1 protein expression and purification
Full-length human YTHDC1 sequence with a non-cleavable C-terminal EGFP-His6 tag and an N-terminal MBP tag followed by a tobacco etch virus (TEV) protease cleavage site was cloned into plasmid pVL1393 (AB vector). The pVL1393 containing MBP-YTHDC1-EGFP sequence was co-transfected with ProGreen (AB vector) into Sf9 insect cells for protein expression, following the manufacturer’s instructions. After 72h, 1L cells were collected and lysed in the buffer (50 mM Tris-HCl pH 8.0, 500 mM NaCl, 5% (v/v) glycerol, 5 mM β-mercaptoethanol, and protease inhibitors cocktail (Roche)). After Ni2+ affinity chromatography, the proteins were eluted with the buffer (50 mM Tris-HCl pH 8.0, 250 mM NaCl, 300 mM Imidazole, 5 mM β-mercaptoethanol), and directly loaded on the Heparin column (GE healthcare). The target proteins were eluted with a linear gradient from 250 mM NaCl to 1 M NaCl monitoring with UV280/260/486. The target proteins were detected using SDS-PAGE, concentrated in the buffer (20 mM HEPES 7.3, 500 mM NaCl, 1mM DTT), flash-frozen in liquid nitrogen, and stored at −80°C. The expressions and purifications of YTHDC1 mutants were the same as those for the wild-type proteins.
In vitro RNA transcription
The DNA sequences containing ten tandem GGACU consensus motifs or wild-type and m6A sites mutated MYC CDS C-terminal regions (sequences in resource table) were synthesized by Integrated DNA Technologies (IDT) and cloned into the pUT-7 vector with a T7 promoter. The plasmid was amplified in DH5α cells and linearized with the HindIII restriction endonuclease to obtain the DNA templates. Transcription of RNAs was performed by mixing the DNA templates with bacteriophage T7 RNA polymerase (lab stock) as previously described(Pikovskaya et al., 2009). We added m6 ATP (TriLink) instead of ATP into the reaction to obtain the m6A version of the RNA. All transcribed RNAs were digested with DNase for 30 min at 37°C, then purified with NucleoSpin RNA set for NucleoZOL (MACHEREY-NAGEL) and then dissolved in DEPC water and stored at −20 °C.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| YTHDC1 Ab | Abcam | Cat#Ab122340 RRID: AB_11128253 |
| Human CD11b-APC | Thermo Fisher Scientific | Cat# CD11B05 RRID:AB_1464195 |
| Human CD13-APC | Thermo Fisher Scientific | Cat# MHCD1305 RRID:AB_10372037 |
| Human CD14-PE | BD Biosciences | Cat# 555398 RRID:AB_395799 |
| Human CD33-PE | BD Biosciences | Cat# 555450 RRID:AB_395843 |
| Human CD45 clone 2D1 | Invitrogen | Cat#45-9459-42 RRID:AB_1548697 |
| Mouse CD45.1 clone A20 | BioLegend | Cat#110722 RRID:AB_492866 |
| Annexin V-PE | BD Biosciences | Cat#556421 RRID:AB_2869071 |
| RRP6 Ab | Abcam | Cat#Ab50558 RRID:AB_869937 |
| PABPN1 Ab | Abcam | Cat#Ab75855 RRID:AB_1310538 |
| MTR4 Ab | Abcam | Cat#Ab70551 RRID:AB_1270701 |
| GFP Ab | Cell signaling Technology | Cat#2956 RRID:AB_1196615 |
| METTL3 Ab | Proteintech | Cat# 150731-1-AP; RRID: AB_2142033 |
| METTL14 Ab | Sigma | Cat#HPA038002RRID:AB_10672401 |
| MYC Ab | Cell signaling Technology | Cat# 5605S; RRID:AB_1903938 |
| Actin Ab | Sigma Aldrich | Cat# A3854; RRID:AB_262011 |
| m6A Ab | Synaptic Systems | Cat# 202003 RRID:AB_2279214 |
| BRD4 Ab | Atlas Antibodies | Cat# AMAb90841 RRID:AB_2665685 |
| NPM1 Ab | Thermo Fisher Scientific | Cat# MA512508 RRID:AB_10981922 |
| SRSF2 Ab | Abcam | Cat# Ab11826 RRID:AB_298608 |
| Coilin Ab | Abcam | Cat# Ab87913 RRID:AB_10860831 |
| PML Ab | Abcam | Cat# Ab96051 RRID:AB_10679887 |
| Biological Samples | ||
| Cord Blood | National Cord Blood Program, NY Blood Center | N/A |
| AML patient cells | Hematologic Oncology Tissue Bank at Memorial Sloan Kettering Cancer Center | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Puromycin | Thermo Fisher Scientific | A11138-031 |
| Polybrene | Millipore | TR-1003-G |
| 7AAD | eBioscience | 00-6993-50 |
| 4-Hydroxytamoxifen | Sigma Aldrich | Cat# H7904 |
| Human SCF | Peprotech | Cat# 300-07 |
| Human IL-3 | Peprotech | Cat# 200-03 |
| Human IL-7 | Peprotech | Cat# 200-07 |
| Human GSCF | Peprotech | Cat# 300-23 |
| Human FLT-3 Ligand | Peprotech | Cat# 300-19 |
| Human TPO | Peprotech | Cat# 300-18 |
| Human IL-6 | Peprotech | Cat# 200-06 |
| Human GM-CSF | Peprotech | Cat# 300-03 |
| MethoCult H3434 | Stem Cell Technologies | Cat# 04434 |
| Actinomycin D | Sigma | Cat# 50-76-0 |
| Critical commercial assays | ||
| RNeasy Plus Micro Kit | QIAGEN | Cat# 74034 |
| CD34 MicroBeads Kit | Miltenyi Biotec | Cat# 130-046-702 |
| Verso cDNA Synthesis Kit | Thermo Fisher Scientific | Cat# AB1453-B |
| Power SYBR Green PCR Master Mix | Thermo Fisher Scientific | Cat# 4367659 |
| Click-iT Nascent RNA Capture Kit | Thermo Fisher Scientific | Cat# C10365 |
| Dual-Luciferase Reporter | Promega | Cat# E1910 |
| Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit | Millipore | Cat# 17-700 |
| RNAscope multiplex fluorescent detection kit | Advanced Cell Diagnostics | Cat# 320851 |
| Deposited Data | ||
| RNA-sequencing data | National Center for Biotechnolongy Information (NCBI) Gene Expression Omnibus (GEO) | GSE168565 |
| YTHDC1-HyperTRIBE data | National Center for Biotechnolongy Information (NCBI) Gene Expression Omnibus (GEO) | GSE168568 |
| Experimental Models: Strains | ||
| Mouse: NOD.Cg-Prkdcscid //2rgtm1Wjl/SzJ (NSG) | The Jackson Laboratory | Stock No: 005557 |
| Oligonucleotides | ||
| MYC- Forward | TTCGGGTAGTGGAAAACCAG | N/A |
| MYC-Reverse | CAGCAGCTCGAATTTCTTCC | N/A |
| GINS1- Forward | GGTCACTGGGAGGAGATGAA | N/A |
| GINS1-Reverse | GCTCACATTTCCATCGAGGT | N/A |
| FOXM1- Forward | ACCCAAACCAGCTATGATGC | N/A |
| FOXM1-Reverse | GAAGCCACTGGATGTTGGAT | N/A |
| AHNAK- Forward | CAGGAGGTGACGCAGAACTC | N/A |
| AHNAK-Reverse | GACTTCACGGGTCCAGGTCT | N/A |
| YTHDC1- Forward | AAGGAGGGCCAAATCTCCTA | N/A |
| YTHDC1-Reverse | CAGTGTTGTTCCCTTGCTCA | N/A |
| ACTIN- Forward | GGACTTCGAGCAAGAGATGG | N/A |
| ACTIN-Reverse | AGCACTGTGTTGGCGTACAG | N/A |
| Ten tandem GGACU consensus motifs | GGACTCGGACTTGGACTCTGGACTTTGGACTTGGACTTGGACTTCGGACTCGGACTTTGGACT | N/A |
| Wild-type MYC CDS | GAGGACTTGTTGCGGAAACGACGAGAACAGTTGAAACACAAACTTGAACAGCTACGGAACTCTTGTGCG | N/A |
| m6A mutated MYC CDS | GtGGACTTGTTGCGGtttCGtCGtGAACtGTTGtttCtCtAACTTGAACtGCTtCGGAACTCTTGTGCG | N/A |
| shRNAs and sgRNA sequence | ||
| human YTHDC1 sh1 | TGGATTTGCAGGCGTGAATT | TRCN0000243987 |
| human YTHDC1 sh2 | TGCCTCCAGAGAACCTTATA | TRCN0000243989 |
| human YTHDC1 sg4 | GGAAGGAGTGGAAGAAGATG | N/A |
| human YTHDC1 sg7 | GGATCCACGATACCAGGAAG | N/A |
| EGFP knockin sg1 | CTCCCGACTGTCAGCCGCCA | N/A |
| EGFP knockin sg2 | GAGCCATGGCGGCTGACAGT | N/A |
| Recombinant DNA | ||
| MSCV-MYC-IRES-GFP | This paper | N/A |
| Lenti-EGFP-Blast | This paper | N/A |
| Lenti-EGFP-YTHDC1-Blast | This paper | N/A |
| Lenti-EGFP-YTHDC1-W377A-Blast | This paper | N/A |
| Lenti-EGFP-YTHDC1-W377A, W428A-Blast | This paper | N/A |
| Lenti-EGFP-YTHDC1- ΔR+P -Blast | This paper | N/A |
| Lenti-EGFP-YTHDC1- ΔE -Blast | This paper | N/A |
| Lenti-ADAR-YTHDC1-Blast | This paper | N/A |
| pRL-CMV-Rluc-MYC-CDS-WT | This paper | N/A |
| pRL-CMV-Rluc-MYC-CDS-m6A mutant | This paper | N/A |
| pRL-CMV-Rluc-MYC-3’UTR-WT | This paper | N/A |
| pRL-CMV-Rluc-MYC-3’UTR-m6A mutant | This paper | N/A |
| PGL3-Fluc-MYC-5’UTR-WT | This paper | N/A |
| PGL3-Fluc-MYC-5’UTR-m6A mutant | This paper | N/A |
| pUT7-10GGACUmotifs | This paper | N/A |
| pUT7-mycCDS-wild-type | This paper | N/A |
| pUT7-mycCDS-mutant | This paper | N/A |
| Software and Algorithms | ||
| Image J version 2.1.0 | Image J | N/A |
| FlowJo software (version10.2) | FlowJo | N/A |
| GraphPad Prism 7 | GraphPad Software | N/A |
| R version 3.5.2 | R development core team | N/A |
| Imaris | BITPLANE | N/A |
In vitro phase separation assay
Before droplet formation experiments, purified MBP-YTHDC1-EGFP protein stock was incubated with TEV protease (lab stock at 200:1 molar ratio) for 2 hours on ice to cleave off the MBP tag. All phase separation assays were performed in the phase separation buffer (20 mM HEPES 7.3, 300 mM NaCl, 1mM MgCl2, 1mM DTT, 5% Dextran T500 (Pharmacosmos).
The YTHDC-EGFP1 proteins were centrifuged at 13,000 g for 5 min to remove small protein pellets and diluted into the desired concentrations with phase separation buffer. The RNA samples were also diluted into the desired concentrations in phase separation buffer. To start liquid droplet formation, 5 μL YTHDC-EGFP1 protein sample was mixed with 5 μL RNA sample into a 384-well glass-bottom microplate (Greiner bio-one) pre-coated with 1 mg/ml BSA (Sigma). The final concentration of proteins and RNAs are indicated in the figures. The droplets were visualized with confocal microscopy (ZEISS LSM 880). The time-lapse data were processed using FIJI/ImageJ.
CRISPR knockin of EGFP into the endogenous YTHDC1 locus.
EGFP was inserted into the endogenous locus at N’ terminal of YTHDC1 in 293T and OCIAML3 cells using CRISPR. Knock-in by CRISPR was performed as described previously(Li et al., 2017a). ssDNA containing 500- nucleotide-long homology arms flanking an EGFP coding sequence was synthesized as template. The sequence of the guide RNAs used is gRNA1 (CTCCCGACTGTCAGCCGCCA) and gRNA2 (GAGCCATGGCGGCTGACAGT). 293T cells were transfected with RNP complex (Cas9 protein and gRNAs) plus templates with lipofectamine 2000. OCIAML3 cells were transfected with RNP complex (Cas9 protein and gRNAs) plus templates through electroporation using Neon transfection system (Thermo Fisher Scientific) under a condition:1375V, 25ms, 1 pulse. Successful incorporation was validated by western blotting using YTHDC1-specific antibody and GFP antibody for EGFP–YTHDC1, which exhibited the expected mobility shift relative to YTHDC1.
Confocal microscopy
For immunostaining, confocal imaging was performed using Leica SP5 upright with 63x/1.4 Oil objective. For live cell imaging, cells were plated on 4-well glass bottom chamber and transfected with constructs described above. Cells were incubated with a LiveCell imaging chamber at 37°C and 5% CO2 and imaged in cell culture medium using Leica SP5 invert with 63x/1.4 Oil objective. Z stack images were captured with the interval size of 130 nm. Excitations were performed sequentially using 405, 488, 594 or 633 nm laser wavelength and imaging conditions were experimentally optimized to minimize bleed-through. Images were prepared with the Leica software and three-dimensional reconstruction was carried out using volume rendering with Imaris software (BITPLANE). Co-localization was also analyzed with Imaris software (BITPLANE).For in vitro YTHDC1 droplets imaging, ZEISS LSM 880 was used with Airyscan super-resolution mode. Images were analyzed by FIJI software. Time lapse movies were captured using the same method as described above. The movie is rendered at 10 frames per second.
Fluorescence Recovery After Photobleaching (FRAP)
For in vitro FRAP analysis, the droplet was photobleached by ZEISS LSM 880 in three regions ROIs that were defined for these experiments. ROI-1 was the indicated circular region in the droplet, and ROI-2 was a similarly sized circular region in the same droplet but in an area that was not photobleached. ROI-3 was defined as background and drawn outside the droplet and its signal was subtracted from both ROI-1 and ROI-2. Fluorescence intensity was measured using FIJI and plotted using Prism software.
For FRAP experiments in living cells, an area of diameter 1μm of YTHDC1 puncta was bleached with a 405 nm laser using Leica SP5 invert confocal. GFP fluorescence signal was collected over time. Each data point is representative of the mean and standard deviation of fluorescence intensities in three unbleached (control) or three bleached (experimental) granules. The prebleached fluorescence intensity was normalized to 1 and the signal after bleach was normalized to the prebleach level.
Nascent RNA transcription of specific gene measurement by qPCR
Control and YTHDC1 depleted cells were incubated with EU (5-ethynyl uridine) for 1hr and harvested. Total RNA was purified by Trizol and nascent RNA was captured by using Click-iT Nascent RNA Capture Kit (Invitrogen) following the manual instruction. Nascent RNA was determined by RT-qPCR using primers targeting specific gens as indicated.
Luciferase reporter assay
For CDS and 3’UTR reporter, original CDS, 3’UTR near stop codon(200nt) or m6A sites mutated CDS and 3’UTR (A to T) of human MYC were cloned downstream of Renilla luciferase reporter gene in pRL-CMV vector (Promega AF025843). For 5’UTR reporter, 5’UTR near initiation codon(200nt) or m6A sites mutated 5’UTR of human MYC was cloned upstream of Firefly luciferase reporter gene in in PGL3 basic vector. Reporters or m6A mutant constructs was co-transfected with Firefly luciferase control, as well as YTHDC1 knockdown constructs (shCtrl, sh1 and sh2) or the indicated YTHDC1 overexpression constructs (YTHDC1 and mutants). 48 hours post transfection, expression of Renilla and Firefly luciferase was determined by Dual-Luciferase Reporter Assay System (Promega) following the manufacturer instructions.
RNA-Immunoprecipitation (RIP)
Control and YTHDC1 depleted MOLM13 cells by shRNAs were collected (20 × 106 cells were used per IP reaction) and washed twice with ice-cold PBS. Cells were lysed in ice-cold IP lysis buffer (50mM Tris-HCL pH 7.5; 300mM NaCl and 0.5% NP40) for 30 minutes on ice and frozen down at −80°C immediately to aid the lysis. On the IP day, lysate was thawed out and spin down at max speed to precipitate the debris at 4°C. Supernatant was collected and incubated with 5ug anti-PABPN1 antibody (Abcam, ab75855) or equivalent amount of Rabbit IgG (Millipore) by rotating overnight at 4°C. RNA-PABPN1-antibody complexes were pulled down using Dynabeads Protein A/G (Millipore) and wash 5 times in 100% IP lysis buffer, 70% IP lysis buffer and 30% PBS, 50% IP lysis buffer and 50% PBS, 30% IP lysis buffer and 70% PBS, 100% PBS. RNA was extracted using phenol-chroloroform method and quantified for qRT-PCR.
Dot blot assay
PABPN1 binding RNAs from control and YTHDC1 depleted cells were purified by RIP as described above. RNA samples were quantified using Nanodrop. The m6A-dot-blot was performed on Amersham Hybond-N+ membrane (GE Healthcare, catalog number: RPN203B). After RNA loading, the RNA was crosslinked to the membrane using a UV Stratalinker 1800 by running the auto-crosslink program twice. The membrane was washed twice with 1XPBST and then block in PBST + 5% milk for 1h. After blocking, the membrane was incubated with m6A primary antibody (Synaptic Systems, Cat. #202 003, 1:1000) overnight at 4°C. Next day, the membrane was washed 3 times in PBST, and incubated with the secondary anti rabbit antibody (1:1000 dilution) for 1h at RT. The membrane was washed again in PBST and exposed on an auto radiographic film using enhanced chemiluminescence substrate. RNA levels were normalized with methylene blue staining.
RNA-seq
RNA from control and YTHDC1 depleted cells (sh1 and sh2) in 3 replicates was extracted with chloroform. Isopropanol and linear acrylamide were added, and the RNA was precipitated with 75% ethanol. Samples were resuspended in RNase-free water. High purity mRNAs were enriched from total RNAs using Dynabeads mRNA purification kit (Thermo Fisher). After PicoGreen quantification and quality control by Agilent BioAnalyzer, mRNA input was used for library preparation (TrueSeq Stranded mRNA LT Sample Prep Kit). Libraries were run on a HiSeq 4000 in a 100bp/100bp paired end run, using the HiSeq 3000/4000 SBS Kit (Illumina). The average number of read pairs per sample was 100 million. Sequence data were aligned using STAR aligner to human reference genome (version hg19). Fragments Per Kilobase of transcript per Million mapped reads(FPKM) were calculated and differential expression analysis was conducted using the DESeq software package. Differentially expressed genes were identified as those with FPKM greater than 1 showing differential expression greater than twofold (up or down) with an adjustment P value less than 0.05. Alternative splicing analysis was performed as previously reported(Xiao et al., 2016). Briefly, RNA-seq reads were mapped to the EEJ (exon-exon junction) libraries with BWA, and PSI (percentage spliced in) level of the exon was quantified.
YTHDC1 Hyper-TRIBE
YTHDC1 Hyper-TRIBE was performed as previously described(Xu et al., 2018). ADAR-YTHDC1 fusion was constructed by fusing the A-I deaminase domain of the Drosophila enzyme ADAR containing a hyperactive mutant E488Q to the human YTHDC1 CDS, with a linker. The construct was codon-optimized for expression in human cells before gene synthesis and cloning into lentiviral-blast vector. MOLM-13 cells were infected with virus expressing ADAR-YTHDC1 or vector controls. After incubation and selection, cells were harvested and used for RNA extraction and sequencing. The expression of ADAR-YTHDC1 was confirmed by western-blot. The RNA editing events identification was followed the workflow as previously reported(Nguyen et al., 2020). Briefly, paired-end RNA-seq reads were aligned to human (hg19) genome using STAR aligner. Next we followed the GATK workflow for calling variants in RNA-seq (https://software.broadinstitute.org/gatk/documentation/article?id=3891) to identify all the mutations in each RNA-seq library. We then restricted to the mutations within annotated mRNA transcripts, as well as restricting to A-to-G mutations in transcripts encoded by the forward strand and T-to-C mutations in transcripts encoded by the reverse strand. We filtered out mutations found in the dbSNP database. We then combined the filtered sets of RNA editing events from all RNA-seq libraries of the same experiment and counted the number of reads containing reference (A/T) and alternative (G/C) alleles from each library at each site. To identify difference of edit frequencies between control and ADAR-YTHDC1, beta-binomial distribution was employed as we did before. Significant sites were determined by filtering for FDR-adjusted (Benjamin-Hochberg correction), using FDR < 0.05.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were processed using GraphPad Prism v.7 and the R statistical environment. All analyses were performed using two-tailed Student’s t tests, except where stated otherwise. P values less than 0.05 were considered to be significant. Graphs and error bars reflect means ± s.e.m., except where stated otherwise. For animal studies, survival probabilities were estimated using the Kaplan-Meier method and compared with the log-rank test. Image analysis was processed using FIJI and Imaris software.
Supplementary Material
Video S1: FRAP of YTHDC1-m6A-droplets, Related to Figure 2.
Video S2: YTHDC1-droplets in solution, Related to Figure 2.
Video S3: 3D image of YTHDC1 condensates, Related to Figure 2.
Video S4: FRAP of nYACs in cells, Related to Figure 3.
Video S5: Dynamics of nYACs in cells, Related to Figure 3.
Highlights.
YTHDC1 is required for AML cell survival, differentiation state and leukemogenesis
YTHDC1 binds to m6A and forms nuclear condensates (nYACs) mediated by LLPS
nYACs are more abundant in AML cells compared to normal blood cells
nYACs protect mRNAs (i.e MYC and others) from degradation by the PAXT-complex
Acknowledgements
We would like to thank members of the Kharas, Jaffrey laboratories for helpful advice and suggestions. We thank Integrated Genomics Operation (IGO) core in MSKCC for their help with our RNA-sequence. We thank Molecular Cytology core in MSKCC for their help with imaging and data analysis. M.G.K. is a Scholar of the Leukemia and Lymphoma Society and supported by NIDDK NIH R01-DK101989–01A1, NCI 1R01CA193842–01, NCI 1R01CA193842–06A1, 5R01CA186702–07, 1R01DK1010989–06A1, R01HL135564, and R01CA225231–01; NYSTEM 0266-A121–4609, the Kimmel Scholar Award; the V-Scholar Award; the Geoffrey Beene Award; the Starr Cancer Consortium; the Alex’s Lemonade Stand A Award; the LLS Translation Research Program; the Susan and Peter Solomon Fund; and the Tri-Institutional Stem Cell Initiative 2016–014. MSK core facilities are supported by P30CA008748.
Declaration of Interests:
S.R.J. is a scientific founder of Gotham Therapeutics and has equity in this company. M.G.K. is a consultant for Accent Therapeutics and M.G.K.’s laboratory receives some financial support from 28–7. These disclosures are not directly related to these studies. There is a patent pending.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberti S, and Dormann D (2019). Liquid-Liquid Phase Separation in Disease. Annu Rev Genet 53, 171–194. [DOI] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, and Rosen MK (2017). Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18, 285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal H, Yihua Q, Iyer SP, Ganapathy S, Proia DA, Penalva LO, Uren PJ, Suresh U, Carew JS, Karnad AB, et al. (2014). WTAP is a novel oncogenic protein in acute myeloid leukemia. Leukemia 28, 1171–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi RK, Huang D, Eberle SA, Wiedmer L, Sledz P, and Caflisch A (2020). Small-Molecule Inhibitors of METTL3, the Major Human Epitranscriptomic Writer. ChemMedChem 15, 744–748. [DOI] [PubMed] [Google Scholar]
- Boija A, Klein IA, and Young RA (2021). Biomolecular Condensates and Cancer. Cancer Cell 39, 174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, Huse JT, Huo L, Ma L, Ma Y, et al. (2020). YTHDF3 Induces the Translation of m(6)A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 38, 857–871 e857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Luo H, Izzo F, Pickering BF, Nguyen D, Myers R, Schurer A, Gourkanti S, Bruning JC, Vu LP, et al. (2019). m(6)A RNA Methylation Maintains Hematopoietic Stem Cell Identity and Symmetric Commitment. Cell Rep 28, 1703–1716 e1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva IA, Wood T, Chiarolanzio K, Chim B, Wong M, Singh V, Gowda CP, Lu Q, Hafner M, Dovat S, et al. (2020). RNA-binding protein IGF2BP1 maintains leukemia stem cell properties by regulating HOXB4, MYB, and ALDH1A1. Leukemia 34, 1354–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, and Brangwynne CP (2016). Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 165, 1686–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, and Zhuang X (2020). m(6)A-binding YTHDF proteins promote stress granule formation. Nat Chem Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann AM, Nayler O, Schwaiger FW, Obermeier A, and Stamm S (1999). The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn). Mol Biol Cell 10, 3909–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Li W, Liang X, Zhu X, Zhang L, Huang Y, Yu T, Li S, and Chen Z (2018). IGF2BP2 Overexpression Indicates Poor Survival in Patients with Acute Myelocytic Leukemia. Cell Physiol Biochem 51, 1945–1956. [DOI] [PubMed] [Google Scholar]
- Huang H, Weng H, and Chen J (2020). m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 37, 270–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, Ni T, Zhang ZS, Zhang T, Li C, et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35, 677–691 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S, and Narlikar GJ (2017). Liquid droplet formation by HP1alpha suggests a role for phase separation in heterochromatin. Nature 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasman L, Krupalnik V, Geula S, Zerbib M, Viukov S, Mor N, Aguilera Castrejon A, Mizrahi O, Shashank S, Nachshon A, et al. (2020). Context-dependent functional compensation between Ythdf m6A readers. bioRxiv, 2020.2006.2003.131441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Bao S, Qian Y, Geula S, Leslie J, Zhang C, Hanna JH, and Ding L (2019). Stage-specific requirement for Mettl3-dependent m(6)A mRNA methylation during haematopoietic stem cell differentiation. Nat Cell Biol 21, 700–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Beckman KA, Pessino V, Huang B, Weissman JS, and Leonetti MD (2017a). Design and specificity of long ssDNA donors for CRISPR-based knock-in. bioRxiv, 178905. [Google Scholar]
- Li Y, Bedi RK, Wiedmer L, Huang D, Sledz P, and Caflisch A (2019). Flexible Binding of m(6)A Reader Protein YTHDC1 to Its Preferred RNA Motif. J Chem Theory Comput 15, 7004–7014. [DOI] [PubMed] [Google Scholar]
- Li Y, Xia L, Tan K, Ye X, Zuo Z, Li M, Xiao R, Wang Z, Liu X, Deng M, et al. (2020). N(6)-Methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2. Nat Genet. [DOI] [PubMed] [Google Scholar]
- Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, Yu Z, Wang Y, Qi M, Zhu Y, et al. (2018). Suppression of m(6)A reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res 28, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. (2017b). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dou X, Chen C, Chen C, Liu C, Xu MM, Zhao S, Shen B, Gao Y, Han D, et al. (2020a). N (6)-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gao M, He J, Wu K, Lin S, Jin L, Chen Y, Liu H, Shi J, Wang X, et al. (2021). The RNA m(6)A reader YTHDC1 silences retrotransposons and guards ES cell identity. Nature. [DOI] [PubMed] [Google Scholar]
- Liu SY, Feng Y, Wu JJ, Zou ML, Sun ZL, Li X, and Yuan FL (2020b). m(6) A facilitates YTHDF-independent phase separation. J Cell Mol Med 24, 2070–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Yu D, Hansen AS, Ganguly S, Liu R, Heckert A, Darzacq X, and Zhou Q (2018). Phase-separation mechanism for C-terminal hyperphosphorylation of RNA polymerase II. Nature 558, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola N, Domanski M, Karadoulama E, Chen Y, Gentil C, Pultz D, Vitting-Seerup K, Lykke-Andersen S, Andersen JS, Sandelin A, et al. (2016). Identification of a Nuclear Exosome Decay Pathway for Processed Transcripts. Mol Cell 64, 520–533. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayler O, Hartmann AM, and Stamm S (2000). The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J Cell Biol 150, 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DTT, Lu Y, Chu KL, Yang X, Park SM, Choo ZN, Chin CR, Prieto C, Schurer A, Barin E, et al. (2020). HyperTRIBE uncovers increased MUSASHI-2 RNA binding activity and differential regulation in leukemic stem cells. Nat Commun 11, 2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, Mapperley C, Lawson H, Wotherspoon DA, Sepulveda C, et al. (2019). Targeting the RNA m(6)A Reader YTHDF2 Selectively Compromises Cancer Stem Cells in Acute Myeloid Leukemia. Cell Stem Cell 25, 137–148 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, and Jaffrey SR (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikovskaya O, Serganov AA, Polonskaia A, Serganov A, and Patel DJ (2009). Preparation and crystallization of riboswitch-ligand complexes. Methods Mol Biol 540, 115–128. [DOI] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, and Jaffrey SR (2019). m(6)A enhances the phase separation potential of mRNA. Nature 571, 424–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari BR, Dall’Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, et al. (2018). Coactivator condensation at super-enhancers links phase separation and gene control. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, and Gorlich D (2016). Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci 41, 46–61. [DOI] [PubMed] [Google Scholar]
- Shen C, Sheng Y, Zhu AC, Robinson S, Jiang X, Dong L, Chen H, Su R, Yin Z, Li W, et al. (2020). RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin Y, and Brangwynne CP (2017). Liquid phase condensation in cell physiology and disease. Science 357. [DOI] [PubMed] [Google Scholar]
- Silla T, Karadoulama E, Makosa D, Lubas M, and Jensen TH (2018). The RNA Exosome Adaptor ZFC3H1 Functionally Competes with Nuclear Export Activity to Retain Target Transcripts. Cell Rep 23, 2199–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Saha U, Paira S, and Das B (2018). Nuclear mRNA Surveillance Mechanisms: Function and Links to Human Disease. J Mol Biol 430, 1993–2013. [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li Y, Gao M, Han L, Wunderlich M, Deng X, Li H, Huang Y, Gao L, et al. (2020). Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 38, 79–96 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LP, Cheng Y, and Kharas MG (2019). The Biology of m(6)A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer Discov 9, 25–33. [DOI] [PubMed] [Google Scholar]
- Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. (2017a). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 23, 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu LP, Prieto C, Amin EM, Chhangawala S, Krivtsov A, Calvo-Vidal MN, Chou T, Chow A, Minuesa G, Park SM, et al. (2017b). Functional screen of MSI2 interactors identifies an essential role for SYNCRIP in myeloid leukemia stem cells. Nat Genet 49, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li Y, Wang P, Han G, Zhang T, Chang J, Yin R, Shan Y, Wen J, Xie X, et al. (2020). Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell. [DOI] [PubMed] [Google Scholar]
- Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, Chen WW, Lander ES, and Sabatini DM (2017). Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell 168, 890–903 e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 22, 191–205 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, and Maquat LE (2019). Cellular RNA surveillance in health and disease. Science 366, 822–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell 61, 507–519. [DOI] [PubMed] [Google Scholar]
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, and Min J (2014). Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 10, 927–929. [DOI] [PubMed] [Google Scholar]
- Xu W, Li J, He C, Wen J, Ma H, Rong B, Diao J, Wang L, Wang J, Wu F, et al. (2021). METTL3 regulates heterochromatin in mouse embryonic stem cells. Nature. [DOI] [PubMed] [Google Scholar]
- Xu W, Rahman R, and Rosbash M (2018). Mechanistic implications of enhanced editing by a HyperTRIBE RNA-binding protein. RNA 24, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, and Jaffrey SR (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 181, 1582–1595 e1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Chen L, Peng D, Jiang A, He Y, Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al. (2020). METTL3 and N6-Methyladenosine Promote Homologous Recombination-Mediated Repair of DSBs by Modulating DNA-RNA Hybrid Accumulation. Mol Cell. [DOI] [PubMed] [Google Scholar]
- Zhang S, Zhao BS, Zhou A, Lin K, Zheng S, Lu Z, Chen Y, Sulman EP, Xie K, Bogler O, et al. (2017). m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 31, 591–606 e596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1: FRAP of YTHDC1-m6A-droplets, Related to Figure 2.
Video S2: YTHDC1-droplets in solution, Related to Figure 2.
Video S3: 3D image of YTHDC1 condensates, Related to Figure 2.
Video S4: FRAP of nYACs in cells, Related to Figure 3.
Video S5: Dynamics of nYACs in cells, Related to Figure 3.
Data Availability Statement
The raw and processed RNA-seq and Hyper-TRIBE data described in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) -- Accession number: GSE168565 and GSE168568.


