Summary
The extra-embryonic yolk sac contains the first definitive multipotent hematopoietic cells, denominated erythromyeloid progenitors. They originate in situ prior to the emergence of hematopoietic stem cells and give rise to erythroid, monocytes, granulocytes, mast cells and macrophages, the latter in a Myb transcription factor-independent manner. We uncovered here the heterogeneity of yolk sac erythromyeloid progenitors, at the single cell level, and discriminated multipotent from committed progenitors, prior to fetal liver colonization. We identified two temporally distinct megakaryocyte differentiation pathways. The first occurs in the yolk sac, bypasses intermediate bipotent megakaryocyte-erythroid progenitors and, similar to the differentiation of macrophages, is Myb-independent. By contrast, the second originates later, from Myb-dependent bipotent progenitors expressing Csf2rb and colonize the fetal liver, where they give rise to megakaryocytes and to large numbers of erythrocytes. Understanding megakaryocyte development is crucial as they play key functions during vascular development, in particular in separating blood and lymphatic networks.
Keywords: megakaryopoiesis, erythropoiesis, embryo, yolk sac, fetal liver, hematopoiesis, resident macrophages, mast cells, fate mapping, scRNAseq
Graphical abstract
Highlights
-
•
EMP produce locally committed progenitors prior to fetal liver colonization
-
•
Two sequential waves of EMP differentiation revealed at the single cell level
-
•
Yolk sac Mk develop mainly from EMPs in a Myb-independent manner
-
•
CD131 identifies Myb-dependent EMP-derived MEP progenitors in the yolk sac
The yolk sac produces the first blood cells during embryonic development. Iturri et al. now show the extent of local definitive hematopoietic differentiation occurring in this organ before colonization of the fetal liver and demonstrate the existence of a direct pathway of megakaryopoiesis produced by EMPs, independently of hematopoietic stem cells.
Introduction
Erythromyeloid progenitors (EMPs) are developmentally restricted hematopoietic progenitors that emerge from the hemogenic endothelium of the yolk sac and have the potential to give rise to definitive hematopoietic cells from both the erythroid and myeloid lineages (Gomez Perdiguero et al., 2015; McGrath et al., 2015; Frame et al., 2016; Kasaai et al., 2017). While the extent of their contribution to fetal and adult hematopoietic systems is still under investigation, EMP-derived hematopoiesis is required for embryo survival through erythrocyte production until birth (Chen et al., 2011; Soares-da-Silva et al., 2021) and for the generation of certain adult resident macrophage populations, such as brain microglia, liver Kupffer cells and epidermal Langerhans cells (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015), and adult resident mast cells (Gentek et al., 2018; Li et al., 2018).
EMPs emerge from the yolk sac vasculature from E8.25 to E11.5, where they are observed as intravascular cell clusters (Frame et al., 2016). In contrast to hematopoietic stem cells (HSCs), EMP emergence is regulated by canonical Wnt signaling but does not require Notch signaling (Hadland et al., 2004) or blood flow (Frame et al., 2016; Kasaai et al., 2017). Importantly, while HSCs need to colonize the fetal liver in order to differentiate (Godin et al., 1999; Kieusseian et al., 2012), EMPs can also differentiate in situ in the yolk sac at least into macrophages, but other lineages have not been formally investigated. EMPs then enter the bloodstream at the beginning of circulation and colonize the fetal liver, where they expand and differentiate into erythrocytes, megakaryocytes, macrophages, monocytes, granulocytes and mast cells (Gomez Perdiguero et al., 2015; Hoeffel et al., 2015; McGrath et al., 2015; Gentek et al., 2018; Li et al., 2018; Stremmel et al., 2018; Soares-da-Silva et al., 2021).
Another key difference between EMPs and HSCs is their dependence on the transcription factor Myb. While both progenitors express Myb (Mukouyama et al., 1999; Hoeffel et al., 2015), only HSCs require Myb for their survival and self-renewal (Mukouyama et al., 1999; Sumner et al., 2000). Consequently, Myb-deficient embryos lack HSC and HSC-derived cells (Mucenski et al., 1991) but still possess normal EMP-derived resident macrophages (Schulz et al., 2012).
EMPs have been characterized by different groups through the expression of Kit, CD41 and CD16/32 (McGrath et al., 2015) or Kit, AA4.1 and low expression of CD45 (Bertrand et al., 2005; Gomez Perdiguero et al., 2015), which can all be expressed by HSC themselves or HSC-derived progenitors. Indeed, from E9.5–10.5 onward, HSCs emerge from the dorsal aorta hemogenic endothelium and contribute to circulating and fetal liver progenitor pools. As such, the study of EMP-derived hematopoiesis requires control of progenitor labeling in a short temporal window. Unfortunately, most tamoxifen-inducible fate mapping strategies based on targeting of hemogenic endothelium (Runx1MericreMer; Cdh5creERT2; Tie2MercreMer) lead to labeling of both EMPs and HSCs, even when tamoxifen or 4-hydroxytamoxifen (OHT) is delivered very early in development (E6.5–E7.5) (Samokhvalov, Samokhvalova, and Nishikawa, 2007; Gomez Perdiguero et al., 2015; Gentek et al., 2018). We thus decided to employ the Csf1rMericreMer fate mapping strategy, as it has been demonstrated that it labels EMPs without any HSC labeling when OHT is administered at E8.5 (Gomez Perdiguero et al., 2015).
Erythro-myeloid commitment and differentiation from HSCs is a process relatively well understood. In the adult bone marrow, HSCs give rise to common myeloid progenitors (CMPs) after sequentially losing self-renewal (multipotent progenitor, MPP) and lymphoid potential (Debili et al., 1996; Akashi et al., 2000). CMPs hold the potential to form myeloid and erythroid lineages, and they could be thus considered as a functional equivalent to EMPs. CMPs then differentiate into megakaryocyte-erythrocyte progenitors (MEPs) and granulocyte-monocyte progenitors (GMPs). Lineage-determining transcription factors orchestrate the activation of specific gene expression programs that lead to commitment and differentiation (Orkin and Zon, 2008), such as Pu.1 for myeloid fate (Scott et al., 1994) and Gata1 for erythroid lineages (Pevny et al., 1991), and their interplay is essential for myeloid versus erythroid lineage choice (Nerlov et al., 2000; Zhang et al., 2000; Hoppe et al., 2016).
Nevertheless, the dynamics and differentiation steps by which EMPs give rise to diverse blood and immune cell types in development are currently unclear. The paucity of progenitor cells, the coexistence with HSCs, and the limited expression of specific cell surface markers at these stages have to date hampered the faithful characterization of definitive yolk sac progenitors. We thus embarked in an unbiased characterization of yolk sac progenitor heterogeneity by combining high-parameter flow cytometry, single cell RNA-sequencing (scRNA-seq), and functional potency assays together with fate mapping analysis of wild type and Myb mutant embryos. We uncovered the heterogeneity and progressive commitment of yolk sac HSC-independent EMP progenitors into myeloid, megakaryocyte, and erythroid fates in their niche of emergence. We further demonstrated that EMPs committed and differentiated within the yolk sac not only into macrophages but also into megakaryocytes (Mks). EMPs first commit toward Mks in a Myb-independent manner that did not require a shared Mk-Erythroid common progenitor (MEP). Such bipotent MEP was produced later by EMPs in the yolk sac and was absent in Myb-deficient embryos. We identified CD131 (Csf2rb) as a marker of these EMP-derived Myb-dependent MEPs in the yolk sac that colonize the fetal liver and differentiate there into Mk and mostly erythrocytes.
Results
Yolk sac progenitor heterogeneity over time corresponds to progressive commitment to erythroid lineages
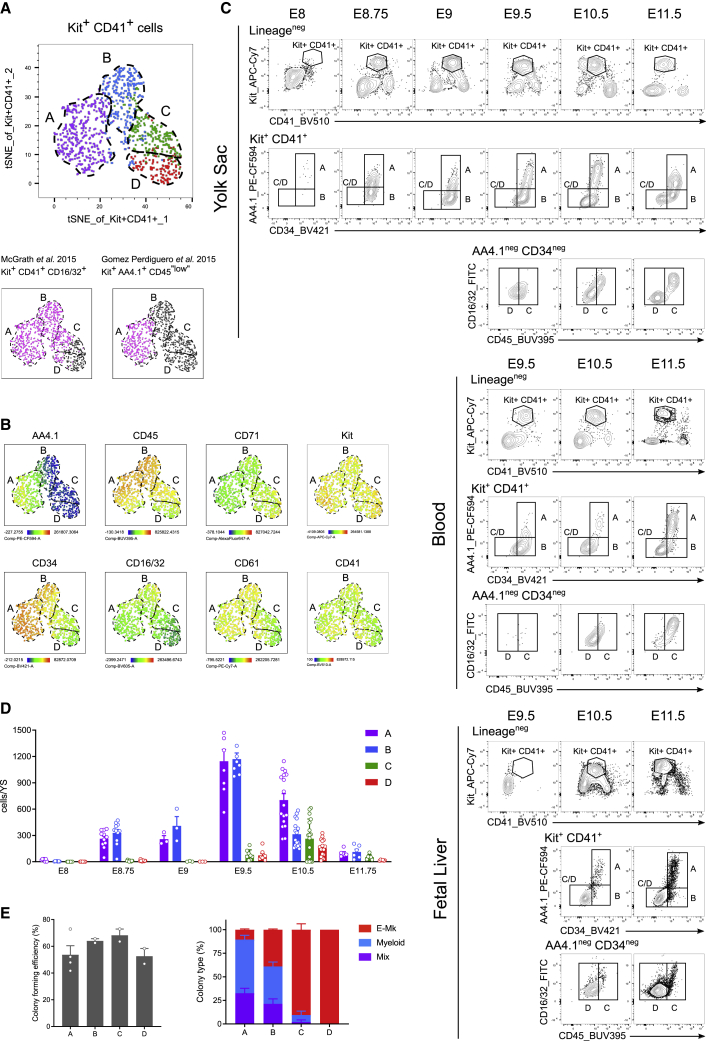
We sought to characterize the heterogeneity among yolk sac (YS) definitive progenitors using high parameter flow cytometry with fluorescence-conjugated antibodies against well-known hematopoietic and progenitor markers. We probed expression of CD45 (pan-hematopoietic marker), AA4.1 (also known as CD93, C-type lectin like type I transmembrane protein; endothelial and progenitor marker), CD16/32 (Fc receptor, myeloid progenitor marker), CD34 (transmembrane phosphoglycoprotein, hematopoietic progenitor marker), CD61 (Integrin b3, binding partner to the Mk marker CD41), and CD71 (transferrin receptor, erythroid precursor marker) at the surface of Kit+ CD41+ progenitor cells in the YS (gating strategy in Figure S1A).
We selected AA4.1, CD34, and CD45 as they were the most discriminating markers and used them to further subdivide YS progenitor cells into four major subsets (A, B, C, and D). The four subsets of cells could be visualized in two dimensions using a t-distributed stochastic neighbor embedding (t-SNE) plot (Figures 1A, 1B, and S1B). We next performed a time-course analysis of the emergence and dynamics of these populations in the three main developmental locations of EMPs: the yolk sac, blood and fetal liver (FL) (Figure 1C).
Figure 1.
Spatiotemporal dynamics of yolk sac progenitor subsets endowed with progressive commitment to erythroid lineages
(A) Yolk sac E10.5 CD41+ Kit+ cells can be subdivided in a tSNE representation (upper panel) into 4 populations A, B, C, and D (dashed lines). (Lower panel) Overlay of previous phenotypic definitions of EMPs (pink) on the t-SNE space (gray).
(B) t-SNE representation of the fluorophore intensity of AA4.1, CD45, CD71, Kit, CD34, CD16/32, CD61, and CD41 in the four subsets. Please also see Figure S1B.
(C) Representative contour plots of Kit and CD41 expression among lineage negative cells, of AA4.1 and CD34 among Kit+ CD41+ progenitor cells and of CD16/32 and CD45 among AA4.1neg CD34neg cells from E8 to E11.5 in the yolk sac (upper panel), blood (middle panel) and fetal liver (bottom panel). At least two independent litters analyzed per time point, except for E8 and E9. Number of analyzed embryos per time point indicated in (D).
(D) Quantification of the number of cells per yolk sac in each population from E8.5 to E11.5. n = 3 at E8 (6–8 somite pairs [sp]), 11 at E8.75 (16–28 sp); 3 at E9 (20–21 sp); 7 at E9.5 (27–29 sp); 17 at E10.5 (34–38 sp); and 5 at E11.75 (51–53 sp). Bars represent mean SEM.
(E) Colony forming units (CFU-C) assays of sorted single cells in liquid culture analyzed 12 days after. Cloning efficiency (left) and frequency of colony types (E-Mk, erythroid and/or Mk; Myeloid and Mix colonies) (right); mean SEM. Two or four technical replicates of one pooled litter. Please also see Figure S1.
Population A, defined as AA4.1+ CD34+ CD45+/lo, was detected in the yolk sac from E8 (6–8 sp stage), albeit at very low numbers (21.21 ± 5.35 cells per yolk sac) that increased by 54-fold by E9.5 (1146.6 ± 133.94 cells per yolk sac) (Figure 1C and quantification in 1D). Population A could be detected from E9.5 onward in blood while it was only detected in the FL from E10.5, suggesting that A was generated in the YS and reached and seeded the FL through circulation. Although A was CD45neg at E8, it gained expression of CD45 at its cell surface from E8.75 (Figure S1C). Population B, defined as AA4.1neg CD34+ CD45+, followed a similar trend to A in dynamics and cell numbers, but delayed, as no B population could be detected yet at E8.
Population C, defined as AA4.1neg CD34neg CD45+, and D, AA4.1neg CD34neg CD45neg, only appeared in the YS at E9.5 and expressed higher amounts of the erythroid lineage marker CD71 (Figures 1B and S1B). In blood and FL, they were only detected after E10.5 and represented the most abundant subsets in the FL (Figure 1C).
These population dynamics combined with the enrichment of CD71 in C-D suggested that A gave rise to B and then to erythroid committed progenitors (C and D). Indeed, colony forming unit (CFU) assays confirmed that, while all populations had a similar cloning efficiency, multipotent progenitors were enriched in A and B, while C and D contained almost strictly erythroid and/or megakaryocyte potential (Figure 1E). Thus bona fide multipotential EMPs were found in A and B, while C and D correspond to already committed progenitors toward E-Mk lineages.
EMP-derived Erythroid and Megakaryocyte-committed Progenitors (MEP) give rise to Megakaryocytes in the yolk sac
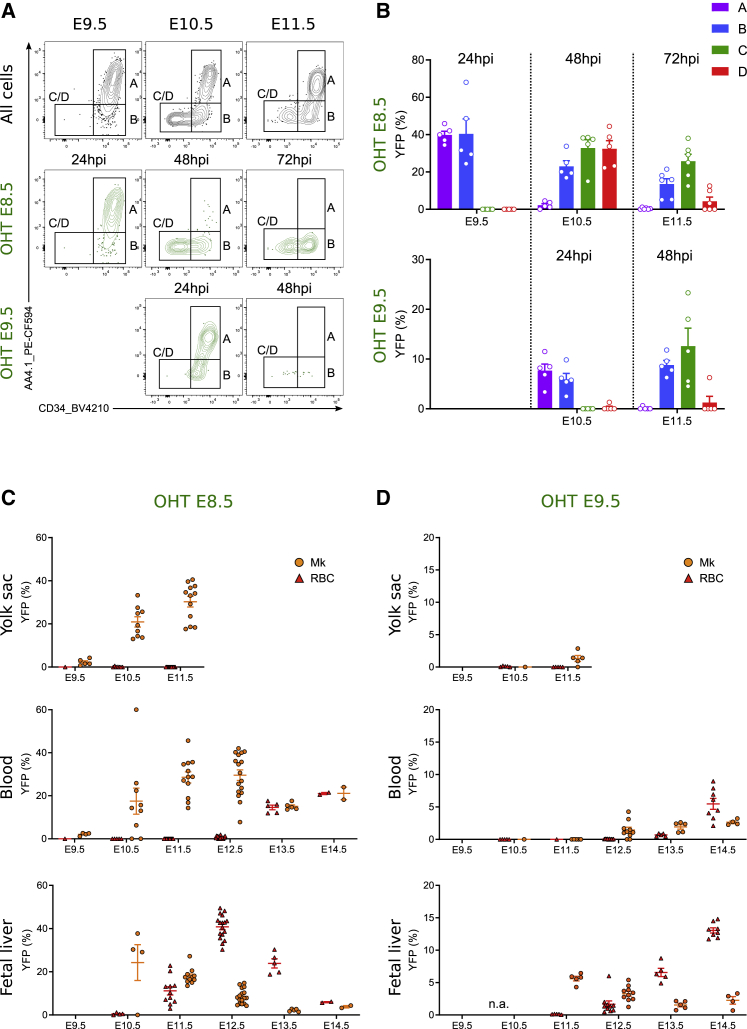
To confirm the lineage relationship suggested above, we performed pulse-and-chase experiments in the transgenic fate-mapping mouse line Csf1rMericreMer Rosayfp by administering 4-hydroxytamoxifen (OHT) to pregnant females at E8.5 or E9.5 (Figure 2A) and analyzing the labeling efficiency of the different subsets of yolk sac progenitors 24, 48, and 72 h after pulse (Figure 2B). Populations A and B were equally labeled 24 h after pulse, albeit at different frequencies depending on the timing of the pulse (40% in OHT E8.5 and 10% in OHT E9.5). Importantly, A was barely labeled 48 h after pulse, whereas B labeling efficiency was maintained. These results showed that YFP-labeled A cells differentiated into B progenitors within 24 h, regardless of the timing of their labeling (“early” at E8.5 versus “late” at E9.5). Erythroid-committed progenitors (C and D) were only labeled 48 h after pulse and labeling efficiency of C equilibrated with B, indicating that C and D correspond to EMP-derived E-Mk-committed progenitors and that transiting from A/B into C also took 24 h or less.
Figure 2.
Lineage relationship between yolk sac progenitor subpopulations
(A) Flow cytometric analysis of Kit+ CD41+ cells from Csf1rMericreMerRosayfp embryos pulsed at E8.5 (middle panel) and E9.5 (bottom panel) and analyzed 24, 48, and 72 h after. hpi, hours post-induction.
(B) Labeling efficiency (% of YFP+ cells) of progenitor subsets after E8.5 (upper) or E9.5 pulse (bottom). OHT at E8.5: n = 5 at E9.5; 9 at E10.5; and 12 at E11.5. OHT at E9.5: n = 6 at E10.5 and 5 at E11.5. Bars represent mean SEM. hpi, hours post-induction.
(C) Labeling efficiency of megakaryocytes (Mk) and red blood cells (RBC) in the yolk sac, blood, and fetal liver after an E8.5 pulse. n = 5 at E9.5; 9 at E10.5; 12 at E11.5; 17 at E12.5; 5 at E13.5; and 2 at E14.5. Error bar respresent SEM.
(D) Labeling efficiency of Mk and RBC in the yolk sac, blood and fetal liver after an E9.5 pulse. n = 6 at E10.5; 5 at E11.5; 10 at E12.5; 5 at E13.5; and 4–8 at E14.5. Error bar respresent SEM. Please also see Figure S2.
We observed differences in E-Mk-committed progenitor labeling between E8.5 and E9.5 OHT pulses and this was reflected in the YFP labeling of mature Mks (Figures 2C and 2D). Early E8.5 pulse labeling led to equal labeling of C and D at E10.5 (48 h post-pulse) and labeling peaking at 30% of mature Mks in YS and blood until E11.5 and E14.5, respectively (Figure 2C, gating strategy in the FL in Figure S2). However, the E9.5 pulse barely labeled mature cells in the yolk sac and blood, and YFP+ Mks in the FL preceded circulating YFP+ Mks, suggesting that the YFP+ Mks found in FL were locally produced (Figure 2D). In conclusion, early-pulsed EMPs produced mature circulating Mks within 24–48 h in the yolk sac, while late E9.5 pulsed EMPs required the fetal liver to produce Mks. Thus, it appeared there were two megakaryopoiesis pathways from EMPs: a YS pathway, in which Mk differentiation occurred rapidly from early-pulsed EMPs in situ in the yolk sac with Mks released immediately into circulation, and a second FL pathway, in which late-pulsed EMPs differentiated more slowly into Mk and released them later into circulation.
No erythrocytes were labeled in the yolk sac, as previously published (Gomez Perdiguero et al., 2015), and the first EMP-derived (YFP+) erythrocytes were found in the FL three days after pulse (Figures 2C–2D, gating strategy in Figure S2). EMP-derived erythrocytes were consistently released into the blood in high numbers 5 days after pulse in both pulses, demonstrating it takes 5 days for yolk sac EMPs to commit, differentiate, and release erythrocytes to the circulation, regardless of time of the pulse. Altogether, these results showed that the in vivo output of EMPs changed over time, while their cell intrinsic potential was unchanged: first EMPs differentiate rapidly into mature Mks in the yolk sac, while red blood cell production is gradual and becomes predominant in the fetal liver.
Single cell expression analysis reveals lineage commitment and differentiation among fate-mapped progenitors
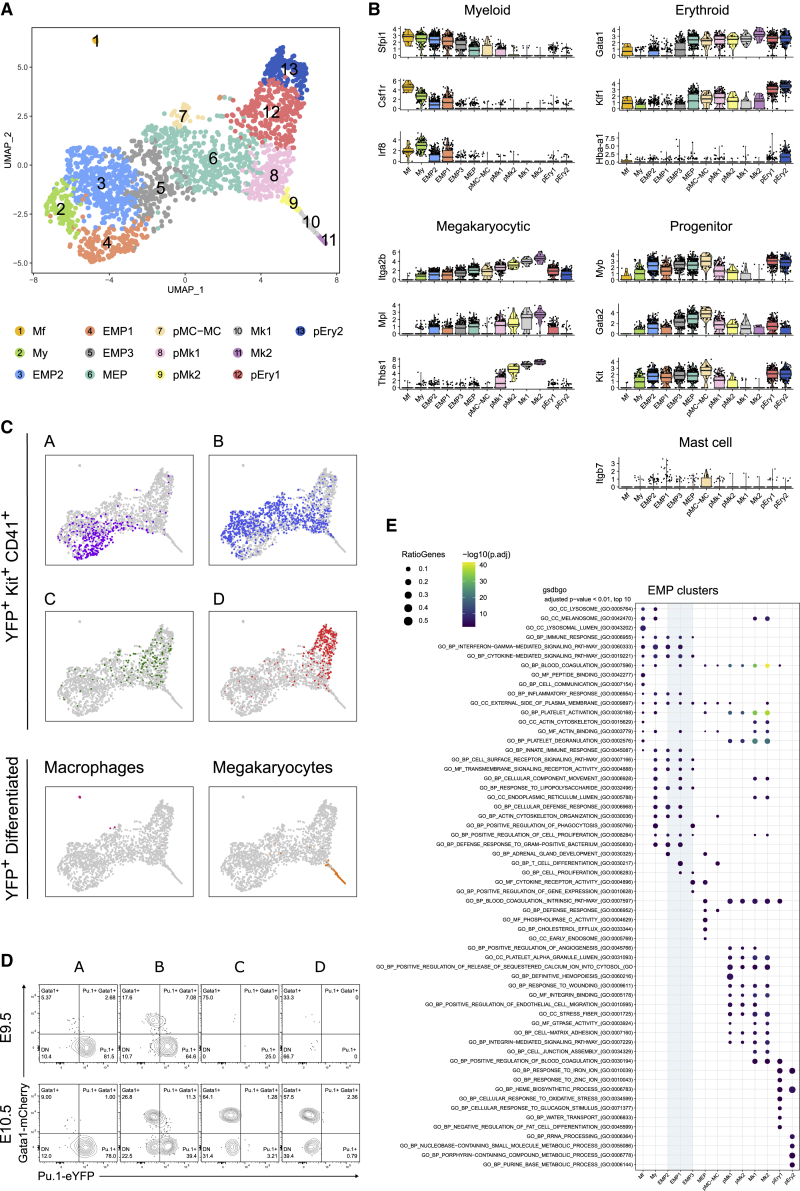
In order to further unravel the relationship between the different progenitor populations, we performed gene expression analysis of single EMP-derived progenitor cells in the yolk sac. We index-sorted yolk sac YFP+ single cells from E9.5 Csf1ricre Rosayfp and E10.5 Csf1rMericreMer Rosayfp embryos pulsed at E8.5 in order to analyze the different populations at key stages. The cells were stained with fluorescent antibodies for the previously described populations and processed following the MARS-Seq pipeline (Jaitin et al., 2014). After quality control and doublet exclusion, we analyzed a total of 781 E9.5 cells and 1,234 E10.5 cells (Figures S3A and S3B). Among those, 1,952 cells were Kit+ CD41+ progenitors, 13 were macrophages, and 64 were megakaryocytes. Normalization and data merging minimized batch effects generated during plate processing and sequencing runs, and dimensionality reduction was conducted using uniform manifold approximation and projection (UMAP) (see Figures S3C–S3E). Random walk clustering (Pons and Latapy, 2006) allowed to identify 13 well-defined cell states or clusters, which were annotated as the following: macrophages (Mf), myeloid progenitors (My), undifferentiated erythro-myeloid clusters EMP1, EMP2, and EMP3, megakaryocyte-erythrocyte progenitors (MEPs), mast cell and/or mast cell progenitors (MCp-MC), Mk progenitors (pMk1 and pMk2), megakaryocytes (Mk1, Mk2), and erythroid progenitors (pEry1 and pEry2) (Figure 3A). Clusters were annotated on the basis of literature-established lineage genes (Figure 3B), top differentially expressed genes of each pairwise comparison (Figure S3F), and index sorting information (Figure 3C). Finally, gene set enrichment analysis (GSEA) uncovered in specific clusters innate immunity (clusters My and Mf), platelet activation, degranulation and coagulation (clusters pMk1 to Mk2), and heme synthesis (clusters pEry1 and pEry2) pathways, thus further confirming the cluster annotations (Figure 3E).
Figure 3.
Single cell expression profiling reveals different transcriptional states and commitment stages in fate mapped progenitors
(A) UMAP representation of 1965 yolk sac YFP+ cells from E9.5 Csf1ricreRosayfp and E10.5 Csf1rMericreMerRosayfp pulsed at E8.5, index-sorted and processed with the MARS-Seq pipeline. Clustering was performed using shared nearest-neighbor graph (SNN) and random walks method (FAST-MNN). Mf, macrophage; My, myeloid progenitor; MEP, megakaryocyte-erythroid progenitor; pMC-MC, mast cell progenitor and/or mast cell; pMK, Mk progenitor; pEry, erythroid progenitor.
(B) Violin plot representation of key lineage-associated gene expression by the hematopoietic clusters represented as log-normalized UMI counts.
(C) UMAP representation of the cells colored by index sorting. A (purple), B (blue), C (green), D (red), macrophages (Kitneg CD16/32hi, pink) and megakaryocytes (Kitneg CD41hi, orange).
(D) Representative contour plots showing expression of Gata1-mCherry and Pu.1-YFP among E9.5 (upper) and E10.5 (bottom) yolk sac progenitor subsets. Two independent litters analyzed; n = 9 and 3 at E9.5 and E10.5, respectively.
(E) Gene set enrichment analysis (GSEA) of the hematopoietic clusters. Dot color indicates adjusted p value and dot size indicates the percentage of genes that are expressed from the pathway. Only the top 10 gene ontology (GO) pathways with Benjamini and Hochberg adjusted p value < 0.01 in at least one cluster are represented. The three GO categories were used simultaneously (BP [biological process], CC [cellular component], MF [molecular function]). Please also see Figure S3.
Surface marker annotations allowed us to correlate the scRNAseq clusters with the progenitor subsets identified above and they showed that population A was mainly restricted to EMP1 and part of EMP2 and EMP3 clusters, while population B was a very heterogeneous population that corresponded to EMP2 and EMP3, myeloid progenitors (My), MEP, and megakaryocyte progenitors (pMk1 and pMk2) (Figure 3C). As expected, C and D were restricted to the E-Mk clusters (MEP, pEry1 and pMk1), although D corresponded principally to the most committed erythroid clusters (pEry1 and pEry2). The expression of two key hematopoietic master regulators, the transcription factors Sfpi1 (Pu.1) and Gata1 for myeloid and erythroid lineages, respectively, confirmed the lineage-commitment of the clusters and immunophenotypic populations. At the gene expression level, EMP1 expressed Pu.1, but Gata1 expression was not detected. In accordance with progressive erythroid commitment, Pu.1 expression was downregulated from EMP1 to MEP, while Gata1 expression was upregulated from EMP3 to fully committed progenitors (pEry2 and pMk2). Concurrently, Pu.1 expression was upregulated along myeloid-macrophage differentiation (Figure 3B). To confirm these findings at the protein level, we analyzed the expression of Pu.1 and Gata1 proteins tagged with fluorescent proteins in Pu.1eyfp;Gata1mcherry embryos (Hoppe et al., 2016) (Figure 3D). As expected, population A (EMP1) expressed only Pu.1-YFP at both E9.5 and E10.5 while population B contained cells expressing Pu.1, Gata1, or coexpressing both markers, consistent with the transcriptional heterogeneity observed in the scRNASeq dataset. Pu.1-YFP cells were not detected in populations C and D. Thus, in accordance with the pulse-and-chase experiments (Figure 2), A was mostly composed of multipotent EMPs, B was a mixed population of committed or lineage-primed progenitors, C contained E-Mk-committed progenitors, and D was mostly composed of fully committed erythroid progenitors.
Importantly, there was a difference in the contribution of the two analyzed time points to the committed clusters (Figure S3E). Myeloid (My) and Mk (pMk1 and pMk2) progenitor clusters were composed of cells from both E9.5 and E10.5. However, erythroid clusters (pEry1 and pEry2) were almost exclusively composed of E10.5 cells. Altogether, the results confirmed at the single cell level that the in vivo output of EMPs changed over time in regards to the Mk-E balance, with erythroid differentiation only starting after E9.5.
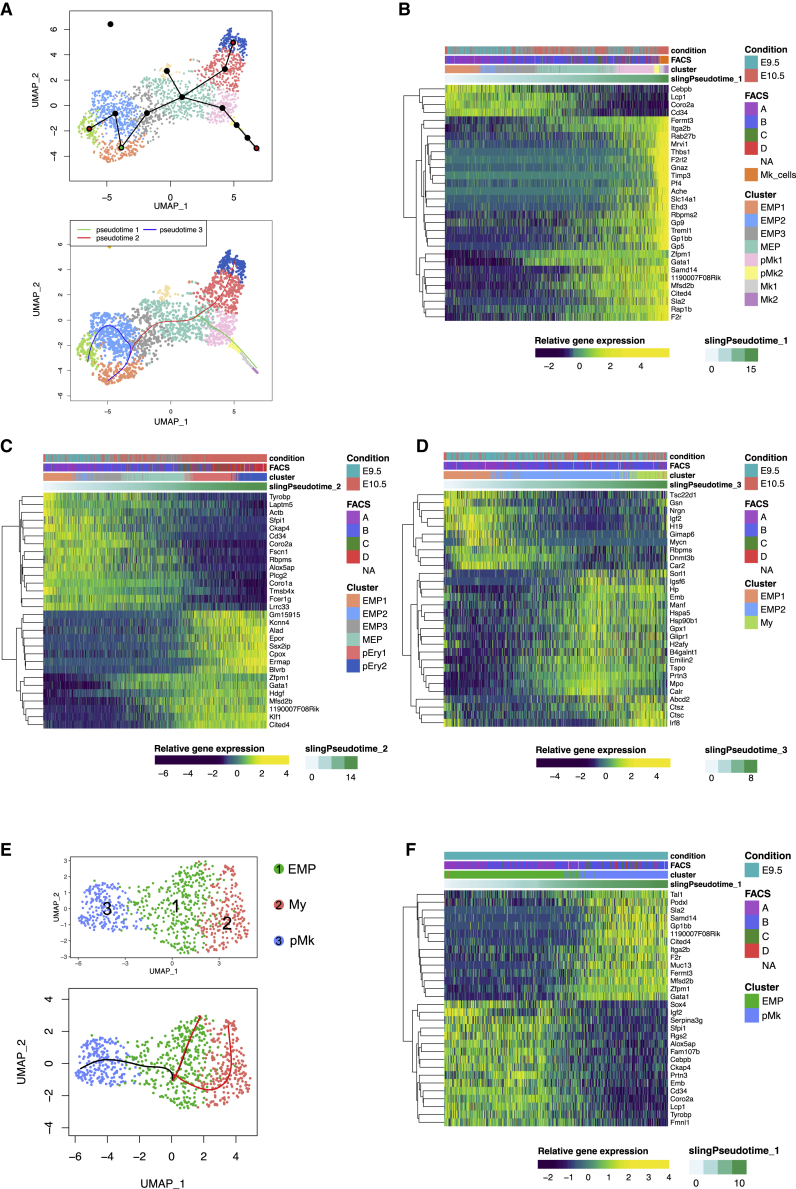
Trajectory analysis of yolk sac progenitors uncovers an E9.5-specific Mk lineage pathway
We then performed trajectory analysis of the lineages that were annotated in the dataset with the Slingshot package (Street et al., 2018). Starting from cluster EMP1, three trajectories were generated: (1) a megakaryocyte lineage trajectory (Mk trajectory, green); (2) an erythroid trajectory (E trajectory, red); and (3) a shorter myeloid trajectory (My trajectory, blue) (Figure 4A). 1,468, 2,246, and 431 genes out of 10,188 were associated with the megakaryocyte, erythrocyte and myeloid trajectories, respectively (adjusted p value < 0.01). The trajectory analysis suggested that, among EMP clusters, EMP2 corresponded to myeloid-primed EMPs and EMP3 to erythroid-primed EMPs. This was supported by higher expression of myeloid genes in EMP2 (Pu.1, Csf1r, Irf8) and lower expression of E-Mk genes (Gata1, Klf1) (Figures 3B and S3F). Conversely, EMP3 had higher expression of E-Mk genes (Gata1, Klf1, and Gata2).
Figure 4.
Lineage trajectories of EMP commitment and differentiation in the yolk sac
(A) (Upper panel) Green node represents the starting point (EMP1) while red nodes represent the finishing points indicated to Slingshot to create the lineage trajectories. (Lower panel) Three lineage trajectories inferred by Slingshot toward Mk2 (green), pEry2 (red), and My clusters (blue).
(B) Heatmap of key genes of the trajectory from EMP1 to Mk2. Heatmap representing the differentially expressed genes along the trajectory.
(C) Heatmap of key genes of the trajectory from EMP1 toward Erythroid progenitors (pEry2).
(D) Heatmap of key genes of the trajectory from EMP1 to Myeloid progenitor cluster (My).
(E) Upper panel, UMAP representation of YS E9.5 Csf1ricreRosayfp YFP+ Kit+ cells colored by cluster: EMP (green), Myeloid (red) and Mk progenitor (blue) cluster. Lower panel, Slingshot infers two trajectories starting from EMP toward pMk or My. See also Figure S4.
(F) Heatmap of key genes of the trajectory of E9.5 EMP to Mk progenitor cluster (pMk). Top 30 genes (30 smallest p values) for the trajectories are represented in all the heatmaps. Please also see Figure S4.
In the E and Mk shared trajectory, leading from EMP1 to MEP, Cebpb, Lcp1, Coro2a, and Cd34 were downregulated while Gata1, Zfpm1, Cited4, and Mfsd2b were upregulated at the MEP stage (Figures 4B and 4C). In the Mk trajectory, the first genes specific for megakaryopoiesis to be upregulated in MEP were Fermt3, Itga2b (CD41), F2r, and Rap1b. A large number of genes were upregulated in more differentiated progenitors (pMk1 and pMk2) and in mature Mks, such as Thbs1, Pf4, Treml1, F2rl2, and components of the platelet von Willebrand factor (vWF) receptor (Gp1bb, Gp5, and Gp9). Few cells with a C and D phenotype were part of the trajectory, but cells from population B, especially from E9.5, were found in the committed pMk1 and pMk2 clusters (Figures 4B and S4C). The Erythroid trajectory had two sets of genes that clustered depending on the timing of upregulation. The first set of genes that was upregulated early corresponded to MEP differentiation genes (Gata1, Zfpm1, Cited4, and Mfsd2b). The second set of genes was upregulated in more mature progenitors (pEry1 and pEry2) at E10.5 and included genes linked to erythrocyte function such as Alad, Epor, Cpox and Ermap (Figure 4C). The Myeloid trajectory was characterized by a first large gene set upregulated in EMP2 that included innate immune and myeloid genes (Emb, Tspo, and Mpo) and then a second set of upregulated genes in the My cluster, containing a macrophage specific transcription factor (Irf8), genes involved in peroxisomal or lysosomal activity (Abcd2) and cathepsin-encoding genes (Ctsz and Ctsc) (Figure 4D).
Since E9.5 progenitors seemed particularly enriched in megakaryocyte progenitor but not erythroid progenitor clusters (Figure S3E and Figures 4B and 4C) and as the erythroid trajectory was mostly composed of E10.5 progenitors, we analyzed separately the E9.5 progenitors and annotated three cell states or clusters on the basis of gene expression (Figure 4E). A distinct megakaryocyte progenitor cluster was observed at E9.5 (Figure 4B, S4A, and S4B). We performed trajectory analysis starting from the EMP cluster at E9.5 and observed a rapid Mk-differentiation trajectory that involved the significant regulation of 586 genes (adjusted p value < 0.01) (Figures 4F and S4D). These included the downregulation of multiple myeloid genes such as Sfpi1, Cebpb, Emb and Cd34, the upregulation of Tal1 starting in the EMP cluster and a later upregulation of Mk lineage genes such as Gata1, Podxl, Gp1bb, F2r and Itga2b (CD41) in the pMk cluster. This gene set upregulation was also closely correlated with the loss of AA4.1 at the surface (phenotype A to B) (Figure S4E) and suggested the existence of a rapid yolk sac megakaryopoiesis that does not involve a MEP intermediate.
CD131 is a marker of MEP commitment in the yolk sac
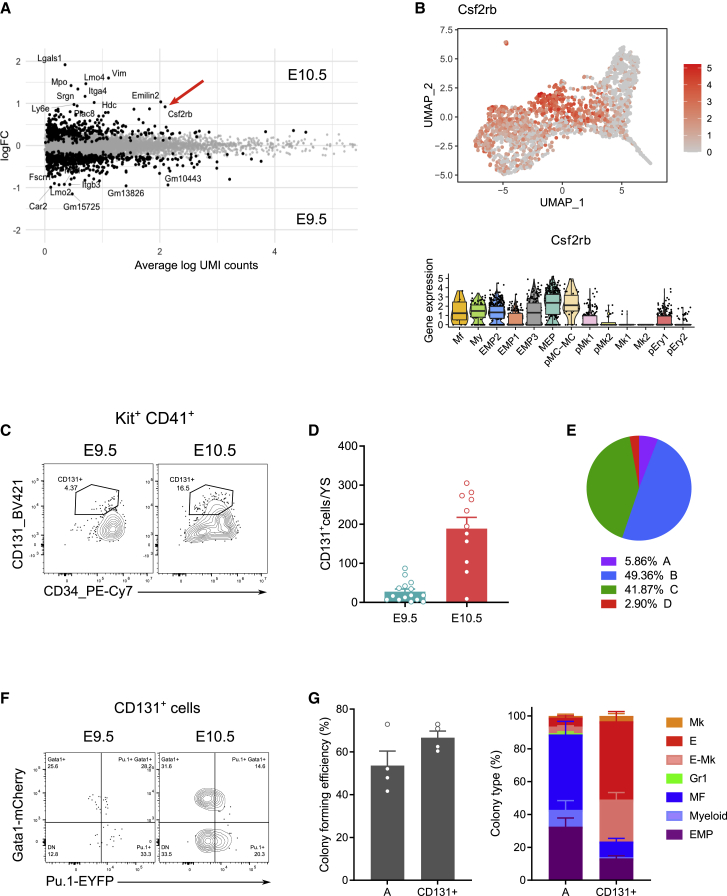
In order to resolve the erythroid and megakaryocyte commitment steps in the yolk sac, we performed pairwise comparison of the genes that were differentially expressed by E9.5 and E10.5 cells from the MEP cluster. We observed that E10.5 MEPs distinctly expressed Csf2rb (the common β chain of GM-CSF, IL-3, and IL-5 receptors) (Figure 5A). Importantly, while cells in EMP and myeloid clusters also expressed Csf2rb at a lower amount, the highest Cs2rb-expressing cells were those of the MEP cluster. Further, Cs2rb-expressing cells were effectively located in the UMAP region where E10.5 cells were most abundant (Figures 5B and S3E).
Figure 5.
CD131 is a marker of Megakaryocyte and Erythroid Progenitors (MEP) in the yolk sac
(A) MA-plot of the MEP cluster comparing E9.5 to E10.5 cells. The genes that are differentially expressed with FDR <1% are shown as black dots, and the names of the genes with FDR <1% and logFC >0.9 are shown. Arrow points to Csf2rb.
(B) Upper panel, UMAP visualization of the expression of Csf2rb. Lower panel, violin plot representation with boxplots of Csf2rb expression in all hematopoietic clusters.
(C) Representative contour plots showing cytometric analysis of CD131+ cells among YS Kit+ CD41+ cells at E9.5 and E10.5. At least two independent litters analyzed per time point.
(D) Quantification of the number of CD131+ cells per YS at E9.5 and E10.5. n = 15 and 11 at E9.5 and E10.5, respectively. Bars represent mean SEM.
(E) Pie chart indicating the proportion of CD131+ cells among A–D progenitor subsets.
(F) Expression of Gata1-mCherry and Pu.1-YFP among E9.5 and E10.5 YS CD131+ progenitors. n = 9 and 3 at E9.5 and E10.5, respectively.
(G) Colony forming units (CFU-C) assays of sorted CD131+ progenitors compared to population A and analyzed 12 days after. Cloning efficiency (left) and frequency of colony types (Mk, megakaryocyte; E, erythroid; E-Mk, E and Mk; Gr1, granulocytes; MF, macrophages; Myeloid, macrophages and granulocytes; EMP, mixed colonies) (right); mean SEM Four technical replicates of one pooled litter. Please also see Figure S5.
We proceeded to verify the expression of CD131 at the surface of YS progenitor populations, and we observed few CD131+ cells at E9.5 (27.51 ± 6.69 SEM) but considerably more at E10.5 (188.8 ± 28.6 SEM) (Figures 5C and 5D). CD131+ cells were CD45+ AA4.1neg and expressed less CD34 than B at their cell surface and as such corresponded to phenotypic intermediates between B and C populations (Figure S5). Since most CD131+ cells were found in B (49.36%) and C (41.87%) (Figure 5E) and expressed Gata1 protein (46%) (Figure 5F), we hypothesized they corresponded to an erythroid and Mk- committed progenitor. We therefore sorted E10.5 CD131+ yolk sac cells for colony forming assays to investigate their differentiation potential. The enrichment in highly proliferative erythroid colonies and mixed erythroid and megakaryocyte colonies suggested an early E-Mk committed progenitor (Figure 5G). Altogether, the results identified CD131 as a marker of yolk sac MEPs produced at E10.5.
Deletion of the transcription factor Myb blocks production of yolk sac MEP but allows direct megakaryocyte differentiation
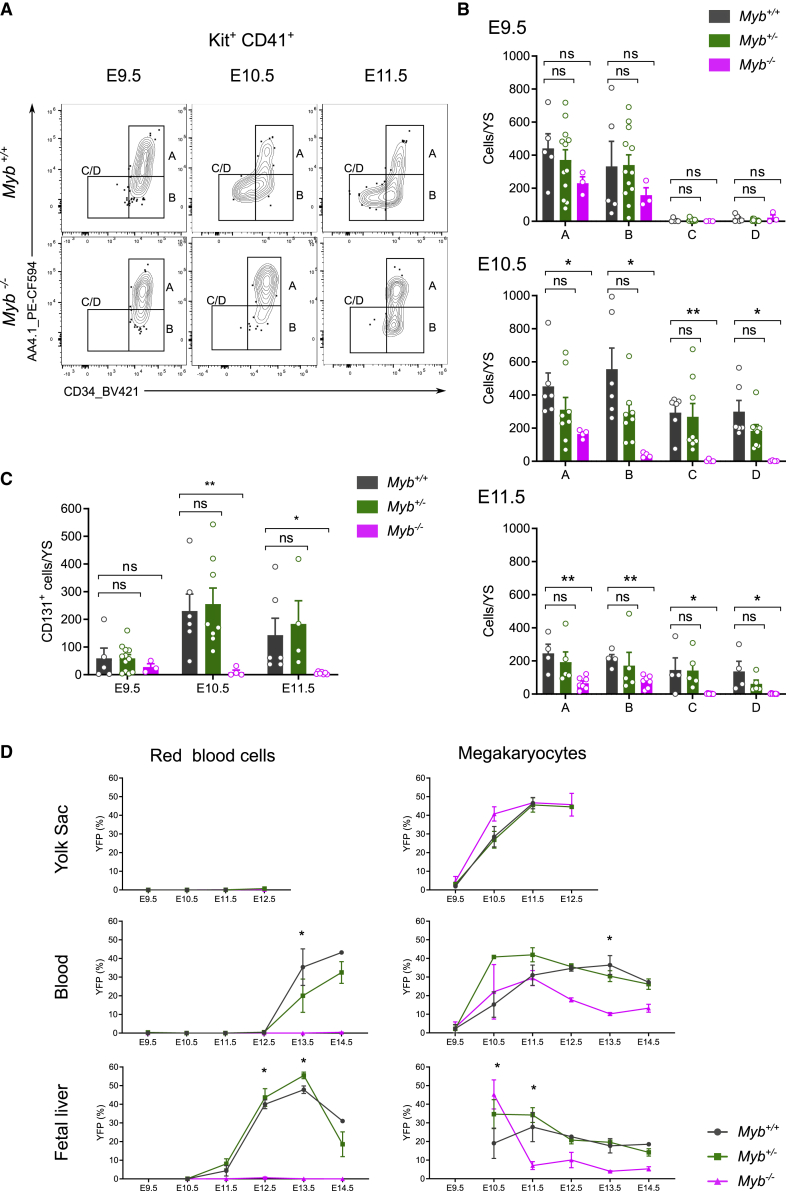
We next analyzed Csf1rMericreMer Rosayfp Myb-deficient embryos pulsed at E8.5 in order to follow the fate of EMP and EMP-derived cells in the absence of the hematopoietic transcription factor Myb. Erythroid-committed progenitors C and D were completely absent in Myb-deficient embryos at all stages analyzed (Figures 6A and 6B). While there were no significant differences in A and B numbers at E9.5, A and B were reduced from E10.5 onward in Myb-deficient yolk sacs and no gene dosage effects were observed (Figure 6B). Pulse-labeled YFP+ Myb-deficient progenitors followed the same trend as all cells, therefore showing no difference between YFP+ and YFPneg progenitors with regard to Myb-dependency (Figure S6A). By E11.5, only a few YFP+ cells were still present in the yolk sac and had a B phenotype (Figure S6A). Further, CD131+ cells were also absent both at E10.5 and E11.5 in Myb-deficient yolk sacs, suggesting an early blockage at the MEP level during YS hematopoiesis (Figure 6C). This complete lack of E-Mk-committed progenitors in Myb-deficient embryos should result in the lack of definitive YFP+ Erythroid and Mk mature cells. It is indeed the case for erythrocytes (Figure 6D), as no YFP+ red blood cells were observed in the fetal liver and blood of Myb-deficient embryos. However, yolk sac EMP-derived megakaryopoiesis was unaffected until E12.5 (Figures 6D and S6B). Moreover, Mk labeling was normal in Myb-deficient Csf1rMericreMer Rosayfp embryos pulsed at E8.5, in the blood until E11.5 and in the fetal liver at E10.5, but strongly decreased thereafter (Figure 6D). These results suggested that EMP-derived Mks found in blood and fetal liver at these early stages bypassed the MEP intermediate and were produced in the yolk sac until E12.5. No EMP-derived Mk production was observed in the FL after E11.5, coinciding with the absence of MEPs in Myb-deficient embryos and the lack of production of mature red blood cells (Figures 6B–6D).
Figure 6.
Deletion of the transcription factor Myb does not affect megakaryocyte differentiation in the yolk sac
(A) Flow cytometric analysis of Kit+ CD41+ cells from Myb-deficient embryos and wild-type littermates from E9.5 to E11.5.
(B) Number of cells per yolk sac in each population in Myb+/+,Myb+/− and Myb−/− at E9.5, E10.5, and E11.5. Results from 3 independent litters per stage. Bars represent mean SEM ns, not significant; ∗p < 0.1; ∗∗p < 0.01. See also Figure S6.
(C) Number of CD131+ cells per yolk in Myb+/+, Myb+/− and Myb−/− at E9.5, E10.5, and E11.5. Results from 3 independent litters per stage. Bars represent mean SEM. ns, not significant; ∗p < 0.1; ∗∗p < 0.01.
(D) Labeling efficiency (% of YFP+ cells) of red blood cells (RBC) and megakaryocytes (Mk) in the yolk sac, blood and fetal liver of Csf1rMericreMerRosayfp embryos pulsed at E8.5 with Myb+/+,Myb+/− or Myb−/− genotype. Results from 3 independent litters per stage. Bars represent mean SEM Statistical analysis compared Myb+/+ and Myb−/− groups: ∗p < 0.1. Please also see Figure S6.
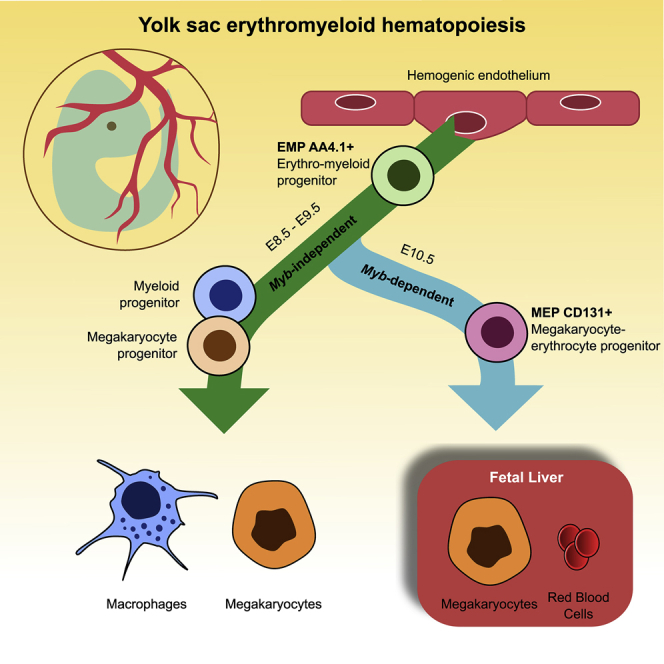
Altogether, these results demonstrate the existence of two successive and independent waves of Mk production from EMPs. In the yolk sac, EMPs were able to provide the first burst of definitive megakaryocytes in circulation in a Myb-independent manner from E9.5 until at least E12.5, which were later replaced by a wave of MEP-produced Mk. EMP-derived MEPs were produced in the yolk sac after E9.5 in a Myb-dependent manner and they terminally differentiated in the fetal liver before Mk were released into circulation.
Discussion
Kit and CD41 coexpression in the yolk sac is considered a marker of definitive hematopoiesis and both are expressed in the hemogenic clusters in this niche (Frame et al., 2016). While it is known that EMPs in the yolk sac locally differentiate into macrophages and that the other erythromyeloid lineages are found in the fetal liver (Gomez Perdiguero et al., 2015; McGrath et al., 2015), less is known about the spatiotemporal dynamics of EMP hematopoiesis. The presence of cells able to produce BFU-E and CFU-E type colonies from E9.5 in the yolk sac also led to the hypothesis that some maturation occurs in this niche before liver colonization (Palis et al., 1999) and implied that intermediate progenitors can be present in the yolk sac. We therefore decided to decipher the EMP-derived hematopoiesis in the yolk sac focusing on the progenitor heterogeneity among Kit and CD41 expressing cells.
While McGrath and colleagues proposed a yolk sac EMP phenotype based on CD16/32 expression, Gomez Perdiguero and colleagues used AA4.1 and CD45 low expression (Gomez Perdiguero et al., 2015), as initially defined by Bertrand and colleagues (Bertrand et al., 2005). We here showed that at least four populations of EMP-derived progenitors are present in the yolk sac before fetal liver colonization. These four populations (hereby named A, B, C, and D) can be distinguished by the surface expression of AA4.1, CD34, and CD45, and they are endowed with different differentiation potential. In particular, the sequential loss of CD34 and CD45 observed in populations C and D enriched for yolk sac definitive megakaryocyte and erythroid progenitors (MEP). We did not select CD16/32 expression for further analysis, as it also labeled many already committed erythroid progenitors (found in populations C and, to a minor extent, D) and it did not help us to unravel the progenitor heterogeneity at E10.5.
Pulse-and-chase experiments in Csf1rMericreMer Rosayfp embryos showed that A (AA4.1+ CD34+ CD45+) gave rise to the other subsets and that these progenitors could only be traced for 24 h before downregulating AA4.1 and pursuing differentiation. Single-cell transcriptomic analysis further revealed the heterogeneity of yolk sac progenitors and confirmed that A (AA4.1+ CD34+ CD45+) was the most undifferentiated multipotent population, which expressed Pu.1 but not Gata1. We showed that population B (AA4.1neg CD34+ CD45+) was very heterogeneous and contained mostly committed or lineage-primed myeloid, megakaryocytic and early erythroid progenitors. Finally, C was enriched in E-Mk-committed progenitors, and D was mostly composed of highly committed erythroid progenitors. The notion of differentiation from A toward myeloid, megakaryocyte, and erythroid committed progenitors was further supported by trajectory inference. Since E9.5 and E10.5 cells segregated inside the MEP (megakaryocyte and erythroid progenitors) cluster and contributed differently to the trajectories of megakaryocyte and erythrocytes, we proceeded to investigate in depth E-Mk pathways of differentiation in the yolk sac.
We were able to further refine the identification of the intermediate steps in the E-Mk pathway thanks to a cell surface marker identified with the scRNA-seq analysis by pairwise statistical comparison of E9.5 and E10.5 cells from the MEP cluster. Csf2rb was found to be expressed 2.4-fold more in E10.5 cells, and it was also expressed higher in E10.5 MEP when compared to other clusters. This gene encodes for the high affinity receptor subunit CD131 or βc, the common β chain shared among granulocyte-macrophage colony-stimulating factor (GM-CSF or CSF2), interleukin-3 (IL-3) and interleukin 5 (IL-5) receptors. CD131 is involved in the differentiation and survival of progenitor cells and the growth and differentiation of myeloid cells and megakaryocytes (Scott and Begley, 1999). The expression of CD131 was recently described in the literature to enrich in E-Mk biased progenitors of the human bone marrow (Drissen et al., 2019), and the cytokine GM-CSF is used in culture to promote the initial proliferation of progenitors of the erythroid and megakaryocyte lineage, although it is not required for the terminal cell divisions in these pathways (Metcalf, Johnson, and Burgess, 1980). In this study, we showed that, at E10.5, EMPs had differentiated in the yolk sac into functional and transcriptomic MEPs, characterized first by the expression of CD131 and defined by in vitro clonal differentiation potential. CD131 is thus a marker of MEP commitment in the yolk sac that is situated upstream of CD34 loss at the cell surface.
We also showed that this differentiation pathway was dependent on the expression of Myb. Embryos lacking the hematopoietic transcription factor Myb have normal number of tissue resident macrophages (Schulz et al., 2012) but fetal liver erythropoiesis is blocked (Mucenski et al., 1991). While AA4.1+ EMPs emerge normally in Myb−/− embryos, their AA4.1neg CD34+ CD45+ progeny is rapidly reduced from E10.5 onward and all the erythroid and Mk intermediates (including CD131+ MEP) are completely absent at all stages. No EMP-derived erythrocyte production was consequently found in Myb−/− embryos; however, EMP-derived megakaryopoiesis was unaffected in the yolk sac at all stages. After E10.5, EMP-derived Mk were severely reduced in the fetal liver, suggesting that Mk production through a Myb-dependent MEP intermediate occurred primarily in the fetal liver niche.
It was proposed that normal Mk numbers in the Myb-deficient embryos derive from primitive hematopoiesis (Tober, McGrath and Palis, 2008). However, by combining Myb deficiency with EMP pulse labeling, we could clearly demonstrate that EMPs emerged normally in Myb−/− embryos and differentiated directly into megakaryocytes bypassing yolk sac CD131+ MEP progenitors. Importantly, this direct MEP-independent megakaryopoiesis uncovered in Myb-deficient embryos was also found in Myb-competent embryos, as evidenced during EMP fate mapping and scRNaseq studies. Yolk sac MEP-independent megakaryopoiesis started rapidly after EMP emergence in a limited time window, as evidenced by the fact that yolk sac Mks are only labeled when EMP are pulsed at E8.5 and not at E9.5. E9.5 pulse did not label HSC or HSC-derived cells as previously shown ((Elsaid et al., 2020) and unpublished data). This yolk-sac-specific direct megakaryopoiesis was faster than the MEP-derived counterpart, as mature Mks were found in the yolk sac 24–48 h after EMP pulse labeling, and Mks were released into circulation as soon as they were produced. Further scRNaseq analysis of yolk sac progenitors confirmed the existence of a megakaryocyte differentiation pathway at E9.5 that differed from the adult-like MEP-dependent differentiation, which was predominant at E10.5. Collectively, our results demonstrate that Myb-deficient Mk progenitors are not just a restricted form of definitive megakaryopoiesis (Tober, McGrath and Palis, 2008), in which Myb−/− EMP retain the capability to give rise to Mks as they fail to differentiate into MEP in the absence of Myb. Rather, megakaryopoiesis in Myb-deficient embryos corresponds to the first wave of rapid and local Mk production from EMPs before the production of CD131 MEPs, that later mature into a second EMP-derived wave of Mk in the fetal liver. This is reminiscent of local macrophage differentiation in the yolk sac that is proposed to bypass monocytic intermediates (Takahashi, Yamamura and Naito, 1989; Naito et al., 1996).
Altogether, these results demonstrate that EMPs can differentiate into megakaryocytes in the yolk sac very rapidly (in 24–48 h) and that they do not depend on a MEP intermediate to do so. These results uncover two pathways to produce definitive megakaryocytes: a direct yolk sac-specific pathway that starts at E9.5 directly from EMP progenitors in a Myb-independent manner, and a second adult-like pathway, where EMP produce first MEP in the yolk sac in a Myb-dependent manner and that terminally differentiate into both erythrocytes and megakaryocytes in the fetal liver. These MEP progenitors seed the fetal liver starting from E10.5 and are responsible for the rapid production of millions of definitive red blood cells that are released into circulation from E12.5. Commitment of erythroid progenitors occurs at E10.5, and it is characterized by the acquisition of CD131 at the surface and a sequential loss of CD34 and CD45.
Limitations of the study
This study has been performed in mouse embryos, and it would be important therefore to assess if this process is also conserved in other species, and humans in particular. In regards to the differences in EMP output depending on the timing of pulse-labeling, the production of red blood cells also displayed different dynamics. The differences in EMP output between pulses that we report here in erythroid lineages could be explained by a temporal specialization of progenitors in the yolk sac in response to changes in the different signals provided by the hematopoietic niches over time (cell extrinsic hypothesis) or by the emergence of two waves of EMP progenitors (cell intrinsic hypothesis). In regards to the latter, comparison of E9.5 and E10.5 yolk sac progenitors did not reveal any major differences at the scRNA-seq level in the EMP clusters or in the overall number of CFU-E and Mac-CFC from E9.5 and E10.5 YS (Palis et al., 1999) but futures studies should investigate this further. Finally, our study also showed that cells with a mast cell progenitor signature are present as early as E10.5 in the yolk sac, in accordance with results from in vitro potency assays (Palis et al., 1999). In the dimensionality reduction projection (UMAP), mast cell progenitors were found adjacent to the MEP cluster, as previously shown in human fetal liver and adult bone marrow hematopoietic scRNA-seq datasets (Franco et al., 2010; Drissen et al., 2019; Popescu et al., 2019). However, we did not pursue this lineage further due to the very low number of mast cell progenitors found in the YS, limiting our ability to draw conclusions, and because this was not the scope of the present study. Future studies focusing on resolving the developmental trajectory of early EMP-derived mast cells could use the dataset created here to pursue the question of mast cell commitment.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AA4.1/CD93 PE-CF594 (Clone AA4.1) | BD Biosciences | Cat# 563805; RRID:AB_2738431 |
| CD131 BV421 (Clone JORO50.2) | BD Biosciences | Cat# 740050; RRID:AB_2739817 |
| CD131 PE (Clone JORO50) | BD Biosciences | Cat# 559920; RRID:AB_397374 |
| CD16/32 BV605 (Clone 2.4G2) | BD Biosciences | Cat# 563006; RRID:AB_2737947 |
| CD16/32 FITC (Clone 2.4G2) | BD Biosciences | Cat# 553144; RRID:AB_394659 |
| CD16/32 Fc Block™ (Clone 2.4G2) | BD Biosciences | Cat# 553142; RRID:AB_394657 |
| CD19 Biotin (Clone 1D3) | BD Biosciences | Cat# 553784; RRID:AB_395048 |
| CD3 Biotin (Clone 145-2C11) | BD Biosciences | Cat# 553060; RRID:AB_394593 |
| CD34 Biotin (Clone RAM34) | eBioscience | Cat# 13-0341-82; RRID:AB_466425 |
| CD34 BV421 (Clone RAM34) | BD Biosciences | Cat# 562608; RRID:AB_11154576 |
| CD4 Biotin (Clone H129) | BD Biosciences | Cat# 553649; RRID:AB_394969 |
| CD41 BV510 (Clone MWReg30) | Biolegend | Cat# 133923; RRID:AB_2564013 |
| CD41 PE (Clone MWReg30) | SONY Biotechnology | Cat# 1269530; RRID: AB_2265179 |
| CD45 BUV395 (Clone 30-F11) | BD Biosciences | Cat# 564279; RRID:AB_2651134 |
| CD45.2 APC-Cy7 (Clone 104) | SONY Biotechnology | Cat# 1149120; RRID: AB_830788 |
| CD61 PE-Cy7 (Clone 2C9.G2) | Biolegend | Cat# 104318; RRID:AB_2687361 |
| CD71 AF647 (Clone C2) | BD Biosciences | Cat# 563504; RRID:AB_2738245 |
| CD8 Biotin (Clone 53-6.7) | Biolegend | Cat# 100704; RRID: AB_312743 |
| ChromPure Mouse IgG | Jackson ImmunoResearch | Cat# 015-000-003; RRID:AB_2337188 |
| F4/80 Biotin (Clone BM8) | Biolegend | Cat# 123106; RRID:AB_893501 |
| F4/80 BV421 (Clone BM8) | SONY Biotechnology | Cat# 1215660; RRID: AB_11203717 |
| Gr1 Biotin (Clone RB6-8C5) | BD Biosciences | Cat# 553124; RRID:AB_394640 |
| Gr1 APC(Clone RB6-8C5) | BD Biosciences | Cat# 553129; RRID:AB_398532 |
| Kit APC-Cy7 (Clone 2B8) | SONY Biotechnology | Cat# 1129130; RRID: AB_1626278 |
| Kit PE (Clone 2B8) | BD Biosciences | Cat# 553355; RRID:AB_394806 |
| Streptavidin BUV737 | BD Biosciences | Cat# 564293; RRID:AB_2869560 |
| Streptavidin BV785 | Biolegend | Cat# 405249 |
| Streptavidin PE-Cy7 | Biolegend | Cat# 405206 |
| Ter119 Biotin (Clone TER-119) | Biolegend | Cat# 116204; RRID: AB_313705 |
| Ter119 PerCP-Cy5.5 (Clone TER-119) | BD Biosciences | Cat# 560512; RRID:AB_10561844 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Csf1rMeriCreMer / tg(Csf1r-Mer-iCre-Mer)1Jwp | Jeffrey W. Pollard | MGI:5433683 |
| Mouse: RosaYFP / B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J) | Jackson Laboratory | MGI:2449038 |
| Mouse: Myb / c-myb | Michael Mucenski | MGI:97249 |
| Mouse: Pu.1eYFP Gata1mCherry | Timm Schroeder | N/A |
| Oligonucleotides | ||
| iCre | Sigma | F 5'- GTC TCC AAC CTG CTG ACT GTG C −3' |
| R 5'- CCA ATG CTG TGT CCC TGG TGA TG −3' | ||
| Myb | Sigma | Common F 5'- CCA TGC GTC GCA AGG TGG AAC −3' |
| WT R 5'- GTG CTT CGG CGA TGT GGT AA −3' | ||
| KO R 5'- TGG CCG CTT TTC TGG ATT CAT C −3' | ||
| Rosa-YFP | Sigma | Common F 5'- AAG TCG CTC TGA GTT GTT AT −3' |
| WT R 5'- GGA GCG GGA GAA ATG GAT ATG 3' | ||
| YFP R 5'- GCG AAG AGT TTG TCC TCA ACC −3' | ||
| mCherry | Sigma | Common R 5'- GCA GGA GAA TGG GAA ATG TG −3' |
| WT F 5'- AGG TAC TGC CCA CCT CTA TC −3' | ||
| mCherry F 5'- GCA TGG ACG AGC TGT ACA AG −3' | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase D from C. histolyticum | Roche | 11088882001 |
| Bovine Serum albumine (BSA) Heat Shock Fraction | Sigma | A7906-500G |
| DNaseI | Sigma | DN25-100MG |
| Stem Cell Factor (Kit ligand) | N/A | N/A |
| Mouse Granulocyte Macrophage Colony Stimulating Factor (mGM-CSF) | OZYME | 5191SC |
| Mouse Macrophage Colony Stimulating Factor (mM-CSF) | OZYME | 5228SC |
| rmEpo (Recombinant Mouse Erythropoietin) | R&D Systems | 959-ME-010 |
| rmTpo (Recombinant Mouse Thrombopoietin) | R&D Systems | 488-TO-025 |
| 4-Hydroxytamoxifen (OHT) | Sigma | H7904-25MG |
| Kolliphor EL | Sigma | C5135-500G |
| Progesterone | Sigma | P3972-5G |
| Sunflower Oil | Sigma | S5007-250ML |
| OptiMEM-I W/Glutamax | Fisher | 51985026 |
| 2-Mercaptoethanol | Fisher | 31350010 |
| Penicillin Streptomycin Solution | Fisher | 15070063 |
| Critical Commercial Assays | ||
| NextSeq® 500/550 High Output Kit v2 (75 cycles) | Illumina | FC-404-2005 |
| Deposited Data | ||
| Single-cell sequencing of yolk sac and fetal liver erytho-myeloid progenitors | Gomez Perdiguero Lab | GEO: GSE166223 |
| Code | Gomez Perdiguero Lab | https://gitlab.pasteur.fr/abiton/scmce_publi |
| Software and Algorithms | ||
| FlowJo Software V 10.6 | FlowJo Inc. | https://www.flowjo.com/solutions/flowjo/downloads |
| R version 3.6.3 (2020-02-29)–“Holding the Windsock” | © The R Foundation | https://cran.r-project.org/mirrors.html |
| Rstudio version 1.2.5033 “Orange Blossom” (330255dd, 2019-12-03) | © 2009-2019 RStudio, Inc. | https://rstudio.com/products/rstudio/download/ |
| Prism 7 for Mac OS X | GraphPad Software Inc. | https://www.graphpad.com/scientific-software/prism/ |
| Inkscape for Mac v1.0 | Free Software Foundation, Inc. | https://inkscape.org/release/inkscape-1.0/ |
Resource availability
Lead contact
Further information and requests for resources and reagents may be directed to and will be fulfilled by the Lead Contact Elisa Gómez Perdiguero (elisa.gomez-perdiguero@pasteur.fr).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The RNA-seq datasets and scripts generated during this study are available on https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166223 (GEO: GSE166223) and https://gitlab.pasteur.fr/abiton/scmce_publi.
Experimental model and subject details
Mice
Csf1ricre(Deng et al., 2010), Csf1rMericreMer (Qian et al., 2011), Pu.1eyfp Gata1mcherry (Hoppe et al., 2016), c-Myb mutant (Mucenski et al., 1991) and Rosayfp(Srinivas et al., 2001) reporter mice have been previously described. Csf1ricre and Csf1rMericreMer mice were on FVB background, other strains were on C57BL/6 background. Csf1ricre and Csf1rMericreMer mice were generated by J. W. Pollard. Myb−/− mice were generated by M. L. Mucenski and were a kind gift from F. Geissmann, Centre for Molecular and Cellular Biology of Inflammation (CMCBI), King’s College London, London SE1 1UL, UK. Rosayfp (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J) reporter mice were purchased from The Jackson Laboratory. Embryonic development was estimated considering the day of vaginal plug formation as 0.5 day embryonic day (E0.5), and staged by developmental criteria. All experiments included littermate controls. No randomization method was used and the investigators were blinded to the genotype of the animals during the experimental procedure. Animal procedures were performed in accordance with the Care and Use Committee of the Institut Pasteur (CETEA) guidelines and with their approval (dap160091).
Heterozygous Csf1ricre or Csf1rMericreMer females were crossed to homozygous Rosayfp reporter males in Figures 2, 3, 4, and 5. Homozygous PU.1eyfp and heterozygous GATA1mcherry females were crossed with homozygous PU.1eyfpGATA1mcherry males in Figures 3 and 6. Heterozygous Myb+/− females were crossed with males homozygous for Csf1rMericreMer; Rosayfp and heterozygous for Myb+/− in Figure 6.
Method details
Embryo genotyping
PCR genotyping of Csf1ricre and Csf1rMericreMer (Qian et al., 2011), PU.1eYFPGATA1mCherry (Hoppe et al., 2016) and c-myb mutant (Mucenski et al., 1991) embryos was performed according to protocols described previously.
In utero pulse labeling of Csf1r+ hematopoietic progenitors
Recombination was induced by single injection at E8.5 or E9.5 of 75 μg/g (body weight) of 4-hydroxytamoxifen (Sigma) into pregnant females. The 4-hydroxytamoxifen was supplemented with 37.5 μg per g (body weight) progesterone (Sigma).
Processing of tissues for flow cytometry
Pregnant females were euthanized by cervical dislocation and embryos ranging from embryonic day (E) 8 to E14.5 were dissected out from the uterus and washed in 4°C phosphate buffered saline (PBS, Invitrogen). Harvest of embryonic blood was performed by severing the umbilical and vitelline vessels and immediately transferring the embryo into a 12-well tissue culture plate filled with 2 mM 4°C EDTA, embryos smaller than E11.5 were decapitated in the well. Organs were dissected in PBS at 4°C and digested for 30 min at 37°C in with 0.5 mL digestion solution (PBS containing 1mg/mL Collagenase D (Roche), 100 U/mL DNase I (Sigma) and 3% fetal calf serum (FCS) (Invitrogen)). Tissues were then mechanically disrupted with a 2ml-syringe piston on top of 100 μm filters to obtain a single cell suspension in FACS buffer containing PBS with 0.5% bovine serum albumin (BSA) and 2mM ethylene-diamine-tetra-acetic acid (EDTA).
Flow cytometric analysis of embryonic tissues and cell sorting
Cells were centrifuged at 320 g for 7 min, resuspended in 4°C FACS buffer, plated in multi-well round-bottom plates and immunolabelled for FACS analysis. After 15 min incubation with purified anti-CD16/32 (FcgRIII/II) diluted 1/50 or ChromPure Mouse IgG diluted 1/20, antibody mixes were added and incubated for 30 min. Where appropriate, cells were further incubated with streptavidin conjugates for 20 min.
Samples were acquired using a Beckman Coulter CytoFLEX LX or a BD Symphony A5 cell analyzer. Cells were sorted using a BD FACSAria III cell sorter. All data was analyzed using FlowJo 10.7. t-Distributed Stochastic Neighbor Embedding (t-SNE) representation was obtained using FlowJo 10.7 default parameters. Cell density was calculated using the following formula: Cells/tissue = Count x Vsample/Vacq x Vblock/Vplated; with Count, quantification by FlowJo; Vsample, volume of the analyzed sample; Vacq, volume acquired by cytometer; Vblock, volume in antibody blocking buffer after first centrifugation; Vplated, volume of Vblock plated with antibody mix. List of antibodies in Key resources table.
Colony forming assays
Colony-forming-unit-culture (CFU-C) assays were performed after sorting single cells into 96-well Nunc UpCell Microplates (ThermoFisher) in complete medium OPTI-MEM with 10% FCS, penicillin (50 units/mL), streptomycin (50 mg/mL) and β-mercaptoethanol (50 mM) supplemented with a saturating amount of the following cytokines: M-CSF (5 ng/mL), GM-CSF (5 ng/mL), SCF in saturation, EPO (2ng/mL) and TPO (5ng/mL) for myeloid and erythroid differentiation. SCF was obtained from the supernatant of myeloma cell lines (provided by F. Melchers) transfected with cDNA encoding this cytokine.
Cultures were grown at 37°C with 5% CO2 and colonies scored after 12 days. Plates were incubated at 4°C for 20 min and colonies were transferred into multi-well round-bottom plates and washed once with FACS buffer. Wells were stained following same protocol with antibodies against Ter119, CD41, CD11b, F4/80 and Gr-1 and analyzed by flow cytometry using a Beckman Coulter CytoFLEX LX.
Gene expression analysis
Experimental design for scRNA-Seq
Yolk sac YFP+ single cells from E9.5 Csf1ricre Rosayfp (n = 3 litters, 27-29sp) and E10.5 Csf1rMericreMer Rosayfp (n = 6 litters, 34-38sp) embryos pulsed at E8.5 were stained with fluorescent antibodies for the populations A, B, C D, Mf and Mk. Cells were index-sorted into 384-well plates with a lysis mix containing UPW, 10% Triton X-100 and RNasin plus 40U/mL (Promega N2611) with MARS-seq barcodes (prepared by Baptiste Saudemont (Keren-Shaul et al., 2019)) and immediately placed in dry ice after sorting. Plates were stored until processing at −80°C.
The plates were processed following the MARS-Seq pipeline (Jaitin et al., 2014) and sequenced in an Illumina NextSeq 500 sequencer using NextSeq® 500/550 High Output Kit v2 (75 cycles) (Illumina). Nine plates were sequenced in three independent sequencing runs, megakaryocytes and macrophages were sorted in the first run (Figures S3A–S3E)
Cell and gene filtering for scRNA-Seq analysis
Cells for which mitochondrial genes took up more than 5% of the total UMI counts, and that had less than 2,750 detected genes were filtered out. We excluded 38 cells that were detected as doublets by the scDblFinder R package (Germain, 2020) using default parameters and running the analysis for each plate separately. The genes detected in less than 5% of the cells of each condition (E9.5 and E10.5), the ERCC-spike-ins, and all mitochondrial genes were excluded. In total, 10,188 genes were kept and 781 and 1,234 cells were selected in the E9.5 and E10.5 conditions, respectively.
Data normalization
Cells were pre-clustered within each plate with the ‘quickCluster’ function of the scran R package (Lun, McCarthy and Marioni, 2016) using a minimum cluster size of 50 cells. Size factors were calculated using the ‘computeSumFactors’ function from the scran R package. Size factors were then rescaled to adjust for differences in sequencing depth between plates and used to normalize the UMI counts of each cell using the ‘multiBatchNorm’ function of the R batchelor package (Haghverdi et al., 2018). The resulting normalized and log-transformed counts were used for further processing.
Selection of highly variable genes
A mean-variance trend was fitted for each gene using the ‘modelGeneVar’ and ‘combineVar’ functions from the scran R package using plate as a blocking factor. The top 3,000 genes with larger variance than the fitted trend were retained as the highly variable genes (HVGs).
Batch effect correction
Batch effects were removed by hierarchically merging the samples using the ‘reduceMNN’ function of the batchelor R package (Haghverdi et al., 2018) on 50 principal components calculated with the multiBatchPCA function from the HVGs only. The samples were first merged across plates within each sequencing run for each condition (E9.5 and E10.5) separately, then across sequencing runs for each condition, and finally across conditions from the oldest (E10.5) to youngest (E9.5) time point. In the E9.5 dataset alone, plates were merged from the plate with the highest number of cells to the lowest number of cells.
The corrected low-dimensional coordinates of each cell returned by fastMNN were used as input for computation of clustering and UMAP (Uniform Manifold Approximation and Projection for Dimension Reduction) for visualization. UMAPs were calculated using the ‘runUMAP’ function of the Seurat package v3.2.2 (Stuart et al., 2019) with 16 neighboring points (neighbors = 16) and 0.32 as the minimal distance to be considered (min.dist = 0.32).
Cell Clustering
A shared nearest-neighbor graph (SNN) was built using the ‘buildSNNGraph’ function of the scran R package. Communities were searched for in the SNN graph using the method of random walks implemented in the `cluster_walktrap` function of the igraph v1.2.5 package. The number k of nearest neighbors used in the SNN graph differed depending on the dataset under consideration: k = 13 was used for the full yolk sac dataset and k = 9 for the E9.5 yolk sac dataset.
Gene differential expression between clusters
Differential expression analyses were performed by computing pairwise t tests using the ‘findMarkers’ function in the scran R package. The option ‘pval.type = ”some”’ was used to only consider markers differential in at least half of the pairwise comparisons between clusters. Heatmaps were generated using the ‘DoHeatmap’ function of the Seurat package.
Gene set enrichment analysis
Using the genes detected as differentially expressed between each cluster and at least half of the other clusters (FDR < 1%), we performed an over-representation analysis using the MSigDB (http://software.broadinstitute.org/gsea/msigdb) and the GeneSetDB databases (https://www.genesetdb.auckland.ac.nz/haeremai.html).
Pseudotime analysis
Cell lineage trajectories were inferred using the R package slingshot v1.4.0 (Street et al., 2018). The UMAP coordinates and the cell clustering assignments were used as inputs. The EMP1 cluster was used as the root node and the ‘My’, ’Mk2’, and ‘pEry2’ clusters as ending points of the trajectories. We used ‘shrink.method = ”density”’ and the parameter “extend='n'” was used in the ‘getCurves’ function in order to ask for the trajectories to not extend beyond the center of the endpoint clusters. In order to find genes associated with each trajectory, a general additive model (GAM) was fitted to each gene as recommended in the slingshot vignette, p values were corrected using the Bonferroni method. Heatmaps of the top associated genes were generated using the pheatmap package in R. Hierarchical clustering of centered and scaled gene expression (represented as normalized-log UMI read count) was performed using complete linkage and Euclidean distance.
Quantification and statistical analysis
Mann-Whitney test was used for pairwise comparisons. Statistical analysis was performed with Graphpad Prism 7.0 (GraphPad Software). The number of subjects used in each experiment is defined in figure legends. Results are displayed as mean ± SEM The following symbols were used in figures to indicate statistical significance: p < 0.1 (∗); p < 0.01 (∗∗); p < 0.001 (∗∗∗).
Acknowledgments
We are thankful for insightful discussions, technical support, and critical reading of the manuscript by Ana Cumano. We gratefully acknowledge the Plateforme de Cytometrie (Sandrine Schmutz and Sophie Novault) at the Center for Innovation and Technological Research of the Institut Pasteur for support in conducting this study. We thank Yann Loe Mie and Baptiste Saudemont for their help with the MARS-Seq pipeline. This work was supported by recurrent funding from the Institut Pasteur, the CNRS, and Revive (Investissement d’Avenir; ANR-10-LABX-73), and by an ERC investigator award (2016-StG-715320) from the European Research Council to E.G.P. E.G.P. also acknowledges financial support from the Fondation Schlumberger (FRM FSER 2017) and the Emergence(s) program from Ville de Paris (2016 DAE 190). L.I. was supported by a fellowship from the Revive Labex and L.F. by a Florence Gould-Pasteur Foundation fellowship.
Author contributions
Conceptualization: L.I. and E.G.P.; Methodology and data collection: L.I., L.F., P.D., Y.L., and E.G.P.; scRNA-seq data analysis: L.I., L.F., and A.B.; Writing - Original Draft: L.I. and E.G.P.; Writing - Review & Editing: L.I., L.F., A.B., Y.L., and E.G.P.; Funding Acquisition: E.G.P.; Supervision: E.G.P.
Declaration of interests
The authors declare no competing interests.
Published: May 31, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.immuni.2021.04.026.
Supplemental information
References
- Akashi K., Traver D., Miyamoto T., Weissman I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Bertrand J.Y. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:134–139. doi: 10.1073/pnas.0402270102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Li Y., De Obaldia M.E., Yang Q., Yzaguirre A.D., Yamada-Inagawa T., Vink C.S., Bhandoola A., Dzierzak E., Speck N.A. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debili N., Coulombel L., Croisille L., Katz A., Guichard J., Breton-Gorius J., Vainchenker W. Characterization of a bipotent erythro-megakaryocytic progenitor in human bone marrow. Blood. 1996;88:1284–1296. doi: 10.1182/blood.v88.4.1284. bloodjournal8841284. [DOI] [PubMed] [Google Scholar]
- Deng L., Zhou J.F., Sellers R.S., Li J.F., Nguyen A.V., Wang Y., Orlofsky A., Liu Q., Hume D.A., Pollard J.W. A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. Am. J. Pathol. 2010;176:952–967. doi: 10.2353/ajpath.2010.090622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissen R., Thongjuea S., Theilgaard-Mönch K., Nerlov C. Identification of two distinct pathways of human myelopoiesis. Sci. Immunol. 2019;4:7148. doi: 10.1126/sciimmunol.aau7148. [DOI] [PubMed] [Google Scholar]
- Elsaid R. A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner. Blood. 2020;137:1024–1036. doi: 10.1182/blood.2020006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame J.M. Definitive Hematopoiesis in the Yolk Sac Emerges from Wnt-Responsive Hemogenic Endothelium Independently of Circulation and Arterial Identity. STEM CELLS. 2016;34:431–444. doi: 10.1002/stem.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco C.B., Chen C.C., Drukker M., Weissman I.L., Galli S.J. Distinguishing mast cell and granulocyte differentiation at the single-cell level. Cell Stem Cell. 2010;6:361–368. doi: 10.1016/j.stem.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentek R., Ghigo C., Hoeffel G., Bulle M.J., Msallam R., Gautier G., Launay P., Chen J., Ginhoux F., Bajénoff M. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity. 2018;48:1160–1171.e5. doi: 10.1016/j.immuni.2018.04.025. [DOI] [PubMed] [Google Scholar]
- Germain P.-L. 2020. scDblFinder: scDblFinder. R package version 1.1.8. Available at.https://github.com/plger/scDblFinder [Google Scholar]
- Godin I., Garcia-Porrero J.A., Dieterlen-Lièvre F., Cumano A. Stem cell emergence and hemopoietic activity are incompatible in mouse intraembryonic sites. J. Exp. Med. 1999;190:43–52. doi: 10.1084/jem.190.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M.F., Geissmann F., Rodewald H.R. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B.K., Huppert S.S., Kanungo J., Xue Y., Jiang R., Gridley T., Conlon R.A., Cheng A.M., Kopan R., Longmore G.D. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 2004;104:3097–3105. doi: 10.1182/blood-2004-03-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghverdi L., Lun A.T.L., Morgan M.D., Marioni J.C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Chen J., Lavin Y., Low D., Almeida F.F., See P., Beaudin A.E., Lum J., Low I., Forsberg E.C. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe P.S., Schwarzfischer M., Loeffler D., Kokkaliaris K.D., Hilsenbeck O., Moritz N., Endele M., Filipczyk A., Gambardella A., Ahmed N. Early myeloid lineage choice is not initiated by random PU.1 to GATA1 protein ratios. Nature. 2016;535:299–302. doi: 10.1038/nature18320. [DOI] [PubMed] [Google Scholar]
- Jaitin D.A. Massively Parallel Single-Cell RNA-Seq for Marker-Free Decomposition of Tissues into Cell Types. Science. 2014;343:776 LP–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasaai B., Caolo V., Peacock H.M., Lehoux S., Gomez-Perdiguero E., Luttun A., Jones E.A. Erythro-myeloid progenitors can differentiate from endothelial cells and modulate embryonic vascular remodeling. Sci. Rep. 2017;7:43817. doi: 10.1038/srep43817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H., Kenigsberg E., Jaitin D.A., David E., Paul F., Tanay A., Amit I. MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 2019;14:1841–1862. doi: 10.1038/s41596-019-0164-4. [DOI] [PubMed] [Google Scholar]
- Kieusseian A. Immature hematopoietic stem cells undergo maturation in the fetal liver. Development. 2012;139:3521–3530. doi: 10.1242/dev.079210. [DOI] [PubMed] [Google Scholar]
- Li Z., Liu S., Xu J., Zhang X., Han D., Liu J., Xia M., Yi L., Shen Q., Xu S. Adult Connective Tissue-Resident Mast Cells Originate from Late Erythro-Myeloid Progenitors. Immunity. 2018;49:640–653.e5. doi: 10.1016/j.immuni.2018.09.023. [DOI] [PubMed] [Google Scholar]
- Lun A.T.L., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath K.E., Frame J.M., Fegan K.H., Bowen J.R., Conway S.J., Catherman S.C., Kingsley P.D., Koniski A.D., Palis J. Distinct Sources of Hematopoietic Progenitors Emerge before HSCs and Provide Functional Blood Cells in the Mammalian Embryo. Cell Rep. 2015;11:1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D., Johnson G.R., Burgess A.W. Direct stimulation by purified GM-CSF of the proliferation of multipotential and erythroid precursor cells. Blood. 1980;55:138–147. doi: 10.1182/blood.v55.1.138.138. [DOI] [PubMed] [Google Scholar]
- Mucenski M.L., McLain K., Kier A.B., Swerdlow S.H., Schreiner C.M., Miller T.A., Pietryga D.W., Scott W.J., Jr., Potter S.S. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-K. [DOI] [PubMed] [Google Scholar]
- Mukouyama Ys., Chiba N., Mucenski M.L., Satake M., Miyajima A., Hara T., Watanabe T. Hematopoietic cells in cultures of the murine embryonic aorta-gonad-mesonephros region are induced by c-Myb. Curr. Biol. 1999;9:833–836. doi: 10.1016/S0960-9822(99)80368-6. [DOI] [PubMed] [Google Scholar]
- Naito M., Umeda S., Yamamoto T., Moriyama H., Umezu H., Hasegawa G., Usuda H., Shultz L.D., Takahashi K. Development, differentiation, and phenotypic heterogeneity of murine tissue macrophages. J. Leukoc. Biol. 1996;59:133–138. doi: 10.1002/jlb.59.2.133. [DOI] [PubMed] [Google Scholar]
- Nerlov C., Querfurth E., Kulessa H., Graf T. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. doi: 10.1182/blood.v95.8.2543. [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. Hematopoiesis: An Evolving Paradigm for Stem Cell Biology. Cell. 2008:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.F., D’Agati V., Orkin S.H., Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Pons P., Latapy M. ‘Computing communities in large networks using random walks’, Journal of Graph Algorithms and Applications. Brown University. 2006;10:191–218. doi: 10.7155/jgaa.00124. [DOI] [Google Scholar]
- Popescu D.M., Botting R.A., Stephenson E., Green K., Webb S., Jardine L., Calderbank E.F., Polanski K., Goh I., Efremova M. Decoding human fetal liver haematopoiesis. Nature. 2019;574:365–371. doi: 10.1038/s41586-019-1652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian B.Z., Li J., Zhang H., Kitamura T., Zhang J., Campion L.R., Kaiser E.A., Snyder L.A., Pollard J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samokhvalov I.M., Samokhvalova N.I., Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- Schulz C. A Lineage of Myeloid Cells Independent of Myb and Hematopoietic Stem Cells. Science. 2012;336:86 LP–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- Scott C.L., Begley C.G. The beta common chain (bc) of the granulocyte macrophage-colony stimulating factor, interleukin-3 and interleukin-5 receptors. International Journal of Biochemistry and Cell Biology. 1999;31:1011–1015. doi: 10.1016/S1357-2725(99)00077-1. [DOI] [PubMed] [Google Scholar]
- Scott E.W., Simon M.C., Anastasi J., Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Soares-da-Silva F., Freyer L., Elsaid R., Burlen-Defranoux O., Iturri L., Sismeiro O., Pinto-do-Ó P., Gomez-Perdiguero E., Cumano A. Yolk sac, but not hematopoietic stem cell-derived progenitors, sustain erythropoiesis throughout murine embryonic life. J. Exp. Med. 2021;218:e20201729. doi: 10.1084/jem.20201729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas S., Watanabe T., Lin C.S., William C.M., Tanabe Y., Jessell T.M., Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremmel C., Schuchert R., Wagner F., Thaler R., Weinberger T., Pick R., Mass E., Ishikawa-Ankerhold H.C., Margraf A., Hutter S. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat. Commun. 2018;9:75. doi: 10.1038/s41467-017-02492-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner R., Crawford A., Mucenski M., Frampton J. Initiation of adult myelopoiesis can occur in the absence of c-Myb whereas subsequent development is strictly dependent on the transcription factor. Oncogene. 2000;19:3335–3342. doi: 10.1038/sj.onc.1203660. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Yamamura F., Naito M. Differentiation, maturation, and proliferation of macrophages in the mouse yolk sac: a light-microscopic, enzyme-cytochemical, immunohistochemical, and ultrastructural study. J. Leukoc. Biol. 1989;45:87–96. doi: 10.1002/jlb.45.2.87. [DOI] [PubMed] [Google Scholar]
- Tober J., McGrath K.E., Palis J. Primitive erythropoiesis and megakaryopoiesis in the yolk sac are independent of c-myb. Blood. 2008;111:2636–2639. doi: 10.1182/blood-2007-11-124685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Zhang X., Iwama A., Yu C., Smith K.A., Mueller B.U., Narravula S., Torbett B.E., Orkin S.H., Tenen D.G. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648. doi: 10.1182/blood.v96.8.2641.h8002641_2641_2648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq datasets and scripts generated during this study are available on https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE166223 (GEO: GSE166223) and https://gitlab.pasteur.fr/abiton/scmce_publi.