Abstract
Banana is one of the most important fruit crops consumed globally owing to its high nutritional value. Previously, we demonstrated that the ripe pulp of the banana cultivar (cv.) Nendran (AAB) contained a high amount of pro-vitamin A carotenoids. However, the molecular factors involved in the ripening process in Nendran fruit are unexplored. Hence, we commenced a transcriptome study by using the Illumina HiSeq 2500 at two stages i.e. unripe and ripe fruit-pulp of Nendran. Overall, 3474 up and 4727 down-regulated genes were obtained. A large number of identified transcripts were related to genes involved in ripening, cell wall degradation and aroma formation. Gene ontology analysis highlighted differentially expressed genes that play a key role in various pathways. These pathways were mainly linked to cellular, molecular and biological processes. The present transcriptome study also reveals a crucial role of up-regulated carotenoid biosynthesis pathway genes namely, lycopene beta cyclase and geranylgeranyl pyrophosphate synthase at the ripening stage. Genes related to the ripening and other processes like aroma and flavor were highly expressed in the ripe pulp. Expression of numerous transcription factor family genes was also identified. This study lays a path towards understanding the ripening, carotenoid accumulation and other related processes in banana.
Introduction
Banana is amongst one of the most essential staple food cultivated in both tropical and subtropical countries and consumed worldwide [1]. The banana plant is a flowering monocot belonging to the family Musaceae and mainly originated from intra- and inter-cross among Musa acuminata (A genome) and Musa balbisiana (B genome) [2]. This resulted in several genome groups viz. AA, AB, AAA, AAB, ABB, AABB, AAAB and ABBB [3]. Ripening process in banana leads to various changes in gene expression that results in changes in flavor, texture and color of these fruits [4, 5]. These irrevocable biological and physiological changes due to ripening often result in shortening of the shelf life of a banana, causing losses at its postharvest level [6] Previously, some chemical treatments were extensively employed to minimize postharvest losses but due to economical and health concerns, these were not favored [7]. Previous studies have reported that ripening in banana involves various gene families that are associated mainly with cell wall degradation and few genes have also been identified which are associated with transcription factors (TFs), signal transduction and ethylene biosynthesis [8–10]. However, the role of molecular factors related to carotenoid accumulation during the ripening process is not explored much in banana. Carotenoids play a significant role during fruit ripening and banana represent a low to moderate amount of their content. Screening of banana germplasm is found to have a significant variation in carotenoid content in their pulp tissue [11]. Banana cultivar (cv.) Nendran (AAB) is identified with high pro-vitamin A content in ripe fruit-pulp [12]. Hence, it will be alluring to study the molecular mechanism associated with ripening and pro-vitamin A accumulation in Nendran.
DNA sequencing has grown as an inescapable medium for studies related to molecular biology. The availability of the draft sequence of the banana genome (523 megabase) from Musa acuminata a double haploid provided imperative information for genetic improvement of the banana plant [13]. Being a quick and economical method, next-generation sequencing (NGS) tools provide high throughput transcriptome analysis with techniques like RNA-Seq [14]. In comparison to whole-genome sequencing, transcriptome profiling is favored as it is confined to study only a subset of the genome (transcribed portions of the genome) [15]. This analysis helps to understand the expression of genes in varying biological environments like in cells and tissues [15]. Further, factors related to stresses and different metabolic pathways can be well studied using genome-scale NGS based technologies. In banana (Dwarf Cavendish), transcriptome analysis has been done to understand the molecular mechanisms involved during the ripening process [16]. Transcriptome analysis of different varieties of Musa spp. has been performed on leaf, root, pulp, rhizome in response to fungal infection, to analyze the role of TF in ripening and to study metabolic processes under low potassium stress [17]. These analyses could further enhance our understanding of the molecular mechanism underlying biosynthesis and defense, and can also contribute to elucidate evolutionary aspects of its genes, and genomes. Databases such as Arabidopsis Next-gen sequences DBs and prediction algorithms have been used to provide information on genes associated with fruit ripening [18]. Further, transcriptome analysis of fruits like kiwi [19] blueberry [20], Cucumis melo [21], orange [22, 23] watermelon [24] and tomato [25] have led to identifying pathways and genes associated with fruit ripening and development. Recent developments in genomics comprising molecular markers (simple sequence repeats) have also enhanced our current knowledge in understanding various functional characteristics of the plant genome, which can help to improve banana by breeding approaches [16]. In-silico databases also use to harbor information of novel molecular markers that can be utilized for genetic improvement programs in banana.
The present study is commenced to get the global expression profile about the key molecular factors involved in ripening, carotenoid accumulation and other related processes in the economically and nutritionally important banana cv. Nendran. To elucidate the role of various up- and down-regulated genes in Nendran, we have generated and analysed transcriptomic data at two fruit developmental stages i.e. unripe and ripe using NGS technology hosted on the Illumina platform.
Materials and methods
Plant material
Banana cv. Nendran was used for experimental purposes. The fruit samples from unripe (6 weeks/w) and ripe (15 weeks/w) stages of Nendran were collected from the banana germplasm plot at National Agri-Food Biotechnology Institute (NABI), Mohali, Punjab. Sampling was performed during the summer and winter seasons. The tissues were then kept in liquid nitrogen and stored at 80°C till further use.
RNA extraction, cDNA library preparation and illumina sequencing
Total RNA was isolated from fruit pulp using the RNA extraction kit (Sigma-Aldrich, USA). In total three biological replicates were taken for each sample and used further for the isolation of RNA. Isolated RNA was treated with DNase I kit (Ambion Thermo Scientific, USA) to eliminate DNA contamination. Total RNA was analyzed by agarose gel electrophoresis for size and integrity. The quantification of total RNA was done with a NanoQuant (Infinite 200 PRO NanoQuant, Austria). The sample for RNA sequencing was derived from the pooling of the RNA samples in two groups i.e. replicates isolated from the fruit-pulp of 6w (unripe) and 15w (ripe) stages. DNA-free RNA was used for cDNA first-strand synthesis by using revert aid first-strand cDNA synthesis kit (Thermo Scientific, USA) as per manufacturer’s protocol. Oligo dT primer was used for cDNA preparation. Consequently, the integrity of RNA used for library preparations was checked with a value of ≥ 8.5 using Bioanalyzer (Agilent, USA). The quality control (QC) passed RNA samples were then processed for library preparation. The paired-end libraries were prepared from the total RNA using Illumina TruSeq stranded mRNA library prep kit as per the instructions (Illumina Inc., USA). The generated libraries were sequenced on Illumina HiSeq 2500 platform.
Transcriptome assembly and RNA seq analysis
Raw reads obtained from sequencing were processed to obtain high-quality reads. Moreover, all reads were trimmed by using the Trimmomatic 0.35 tool [26] to remove low-quality reads and any adapter sequences if present. The resultant high-quality reads of each sample were used for mapping on Musa acuminata DH-Pahang v2 on banana genome hub [27] (https://banana-genome-hub.southgreen.fr/download). The reads were mapped using STAR 2.6 [28]. Cufflinks v2.2.1 [29] program was used to assemble the STAR aligned transcripts to quantify their expression. Cufflinks, Cuffmerge and Cuffdiff were then used for further mapping and expression analysis of differentially expressed genes (between unripe and ripe samples). Cuffdiff software was also used to quantify the abundance of transcripts in the form of Fragment Per Kilobase of transcript per Million mapped reads (FPKMs). The genes were additionally categorized as differentially expressed by considering statistical significance (p<0.05, p<0.01) and false discovery rate for significant expression.
Functional annotation of Differentially Expressed Genes (DEG) and pathways
For functional annotation of DEG and to identify putative pathways associated with them, we annotated identified DEG’s with banana genome hub, NCBI protein database and GO databases. Significant GO IDs were extracted from the banana genome hub ontology browser. The g:Profiler web server was employed for functional enrichment analysis [30]. Further, the WEGO tool [31] was used to calculate the statistical enrichment of DEGs in various pathways using FDR values of < 0.05 (threshold).
Quantitative real-time PCR (qRT-PCR) validation
Total RNA was isolated from ripe and unripe fruit pulp samples and cDNA was prepared as discussed above. The qRT-PCR study was implemented with ABI 7500 Sequence Detector (Applied Biosystems, USA). Housekeeping gene Actin1 (GenBank Accession No. AF246288) was used to normalize the variant expression of chosen genes [32, 33]. The primers were firstly tested for single-band amplification using conventional end-point PCR. The expression of each gene was tested in unripe and ripe conditions of Nendran fruit samples. A melting curve study was carried out using qRT-PCR. The total volume of each reaction was adjusted to 10 μl and contained 1X SYBR Green Master Mix (Applied Biosystems, USA); 5 pmol of each primer (forward and reverse); 0.5 μl cDNA template and sterile distilled H2O. PCR conditions followed during real-time PCR experiment were: step (1) 50°C 2 min, step (2) 95°C 10 min, step (3) (95°C 0.15 min, 60°C 1 min) x 40 cycles, followed by the thermal dissociation curve. The relative expression level was analyzed using the 2-ΔΔCt method [34]. Primer details along with corresponding gene IDs are mentioned in the S1 Table. All the primers used in the qRT-PCR analysis were unique to each gene and were designed using Primer3 software [35]. All experiments were executed in biological triplicates and each experiment entailed three technical replicates. Statistical significance was determined by using the Student’s paired t-test.
Results
Transcriptome sequencing, alignment and analysis of banana fruit samples
The whole transcriptome sequencing i.e. RNA-seq (paired-end) of fruit (ripe and unripe) samples of cv. Nendran was performed using Illumina HiSeq 2500 platform. On average for each sample, 96,013,558 reads in NEN-Ripe and 107,849,342 in NEN-Unripe samples were recovered from two cDNA libraries. Approximately 89.30% of total reads have a Phred quality score > 30 (a measure of the quality of nucleobases). After exclusion of low-quality reads, 95,320,622 and 107,351,986 reads were obtained from NEN-Ripe and NEN-Unripe samples, respectively. Clean reads were then selected for aligning to the banana genome. By mapping the selected transcripts, 94% (Ripe) and 92% (Unripe) reads matched with the banana genome (Table 1).
Table 1. RNA sequencing statistics of ripe and unripe banana cv. Nendran samples.
| Sample Name | Total Read Count | Read Count after rRNA removal | QC Pass % | Aligned Read Count | Aligned % | Unaligned % |
|---|---|---|---|---|---|---|
| NEN-Ripe | 96,013,558 | 95,320,622 | 99.28 | 8,99,28,387 | 94.34 | 5.66 |
| NEN-Unripe | 107,849,342 | 107,351,986 | 99.54 | 9,91,05,193 | 92.32 | 7.68 |
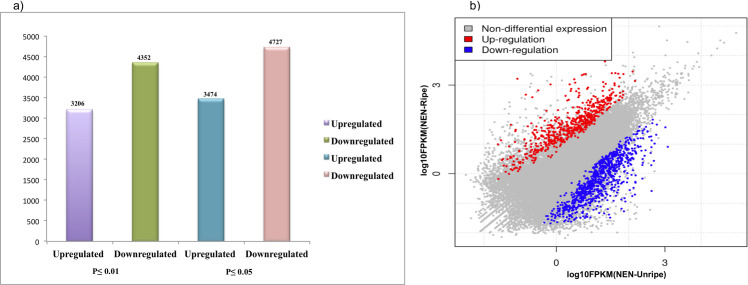
Further, the expression is evaluated in FPKMs using the Cufflink software package [29]. Based on log2 fold change parameter and p-value ≤ 0.01, we obtained 3206 up-regulated (≥ 2 fold) and 4352 (≤ -2 fold) down-regulated genes in unripe vs. ripe samples (Fig 1A). Similarly, with p-value ≤ 0.05, a total of 3474 up- and 4727 down-regulated genes were obtained (Fig 1A). The scatter plot of the expressed genes at unripe and ripe stages of fruit-pulp is presented in Fig 1B. Different colors were used in scatter plot to specify up-regulated genes, down-regulated genes and genes in which expression was not affected. We evaluated unripe and ripe samples and created scatter plot of expressed genes where specific colors were used to exemplify down-regulated, up-regulated and non-regulated genes. The DEGs were examined between control (Unripe) and test (Ripe) samples using the FPKM method log2 fold change ≥ 2 as a threshold. The signifying log2 values of gene expression and screening conditions are represented in Fig 1B.
Fig 1. General features of Nendran banana transcriptome.
a) Total number of DEGs with p-value ≤ 0.01and ≤0.05. b) Scatter plot of all differentially expressed genes. Significant genes with log fold change >2 are indicated by red color and those having log fold change <2 are indicated by blue color, while genes without significant difference are depicted by grey color.
Functional enrichment of differentially expressed genes
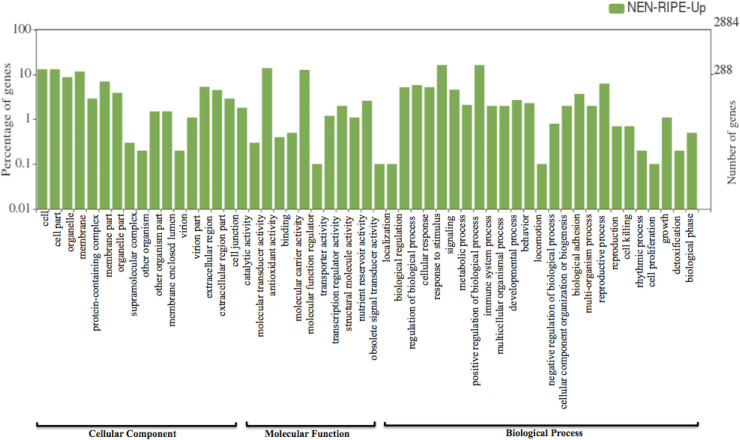
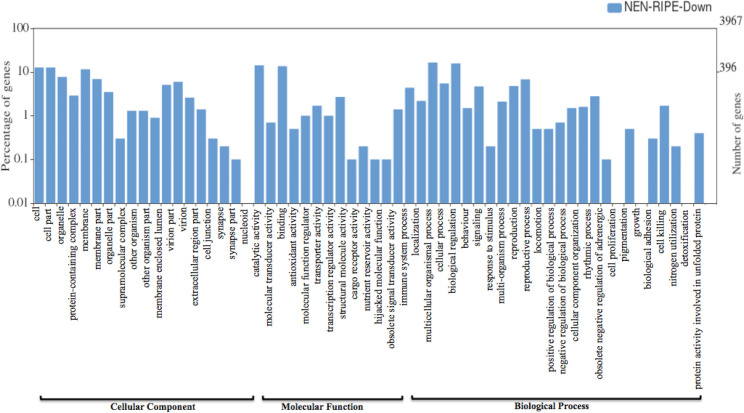
The significantly differentially expressed genes were then mapped to the banana genome hub database (https://banana-genome-hub.southgreen.fr/). Further, the study on gene ontology (GO) and classification of DEGs was performed to get the information of genes involved in cellular, molecular and biological processes in ripened fruit pulp of Nendran. All the DEGs with annotated GO terms were visualized using the WEGO tool (Figs 2 and 3). In the cellular component, most of the genes were classified into the extracellular region, cell part and membrane part, while in the molecular function, most of the genes were involved in catalytic activity, binding and molecular function regulators. In biological processes, genes were mainly involved in response to stimuli, biogenesis or cellular component organization, biological regulation, etc. Overall, cellular component organization, localization, developmental process and response to stimuli were the most considerably enriched processes in DEGs (S1 Fig).
Fig 2. WEGO plot representing annotation and classification of differentially expressed genes in Nendran ripe fruit-pulp.
X-axis (right) displaying selected GO terms of up-regulated genes and y-axis (left) displaying the percentage of genes.
Fig 3. WEGO plot representing annotation and classification of differentially expressed genes in Nendran unripe fruit-pulp.
X-axis (right) displaying selected GO terms of down-regulated genes and y-axis (left) displaying the percentage of genes.
Identification of differentially expressed genes
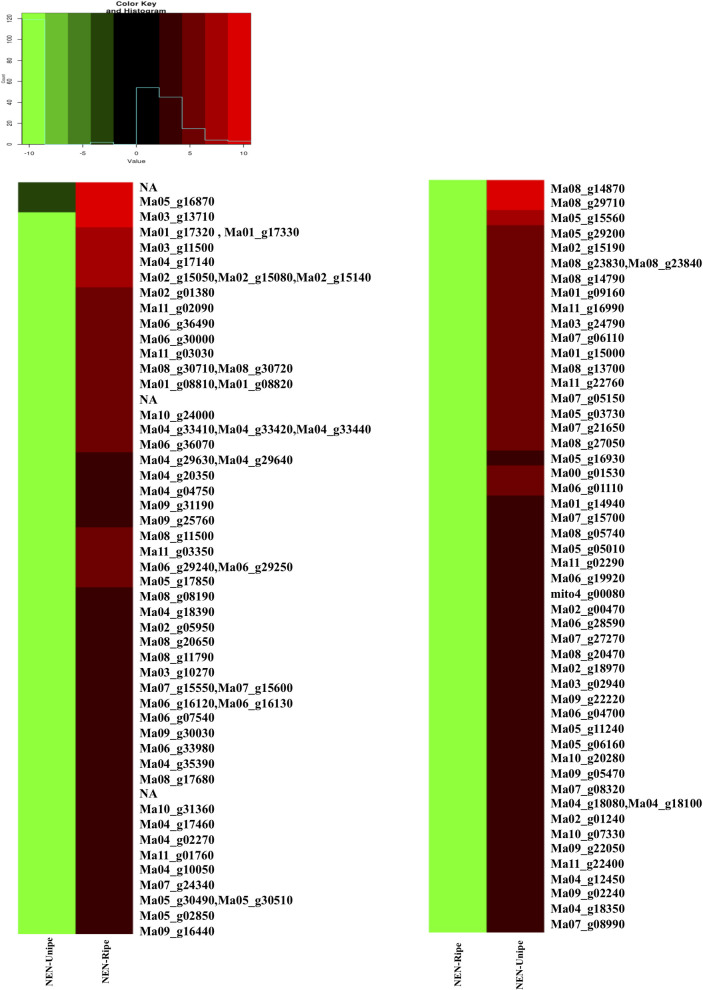
To identify transcripts that are expressed differentially in response to ripening, the top 50 up-regulated and down-regulated genes were selected for further analysis. Gene expression of the most up-regulated transcripts varied from 14 to 6.6 folds (Table 2). The acyl carrier protein and cytochrome P450 encoding genes are shown to be up-regulated significantly. Categorically, other genes encoding for stress and pathogenesis-related proteins were also up-regulated in ripen fruit-pulp samples. Similarly, the genes that were down-regulated are mostly linked with TF, hydrolase, and cellulose related genes. A detailed list of top up- and down-regulated genes is depicted by a heat map (Fig 4). The top 50 up- and down-regulated genes are listed in Tables 2 and 3, respectively.
Table 2. List of top 50 up-regulated genes in Nendran ripe fruit samples.
| Musa acuminata IDs | Unripe FPKM | Ripe FPKM | Log2 (fold change) | P-value |
|---|---|---|---|---|
| NA | 0.084 | 1631.93 | 14.2455 | 0.00435 |
| Ma05_g16870 | 0.2272 | 2196.7 | 13.2392 | 0.22265 |
| Ma03_g13710 | 0.3616 | 2292.69 | 12.6306 | 0.22785 |
| Ma01_g17320, Ma01_g17330 | 0.0881 | 368.309 | 12.0301 | 0.17305 |
| Ma03_g11500 | 0.1433 | 473.92 | 11.881 | 0.00435 |
| Ma04_g17140 | 0.3154 | 663.35 | 11.0384 | 0.0315 |
| Ma02_g15050,Ma02_g15080, Ma02_g15140 | 0.2177 | 387.42 | 10.7974 | 0.04925 |
| Ma02_g01380 | 0.691 | 1051.4 | 10.5714 | 0.03525 |
| Ma11_g02090 | 0.0557 | 45.48 | 9.674 | 0.01545 |
| Ma06_g36490 | 0 | 676.19 | 9.4034 | 5.00E-05 |
| Ma06_g30000 | 3.1304 | 1572.4 | 8.9724 | 0.0368 |
| Ma11_g03030 | 0.0297 | 13.42 | 8.8177 | 0.0439 |
| Ma08_g30710, Ma08_g30720 | 2.3397 | 1055.65 | 8.8176 | 0.0006 |
| Ma01_g08810, Ma01_g08820 | 5.11 | 2165.23 | 8.727 | 5.00E-05 |
| NA | 0.0754 | 30.64 | 8.6664 | 0.00435 |
| Ma10_g24000 | 0.0244 | 9.9 | 8.666 | 0.04935 |
| Ma04_g33410, Ma04_g33420, Ma04_g33440 | 5.7266 | 2293.89 | 8.6459 | 5.00E-05 |
| Ma06_g36070 | 0.0738 | 27.39 | 8.5367 | 0.0104 |
| Ma04_g29630, Ma04_g29640 | 6.4843 | 2380.04 | 8.5198 | 0.00375 |
| Ma04_g20350 | 3.3082 | 1206.63 | 8.5107 | 0.00055 |
| Ma04_g04750 | 21.9975 | 6325.5 | 8.1677 | 0.03775 |
| Ma09_g31190 | 2.5786 | 741.18 | 8.1671 | 0.00955 |
| Ma09_g25760 | 0.5324 | 135.71 | 7.9938 | 0.00875 |
| Ma08_g11500 | 2.4939 | 631.55 | 7.9844 | 0.00035 |
| Ma11_g03350 | 0.2043 | 50.78 | 7.9573 | 0.037 |
| Ma06_g29240, Ma06_g29250 | 0.1632 | 40.08 | 7.9398 | 0.01065 |
| Ma05_g17850 | 1.8005 | 431.87 | 7.9061 | 0.0191 |
| Ma08_g08190 | 0.3605 | 78.63 | 7.7691 | 0.01995 |
| Ma04_g18390 | 4.12 | 756.6 | 7.5207 | 0.00025 |
| Ma02_g05950 | 3.5512 | 617.78 | 7.4427 | 0.0025 |
| Ma08_g20650 | 0.0493 | 8.37 | 7.4071 | 0.01545 |
| Ma08_g11790 | 0.072 | 11.7 | 7.3435 | 0.00455 |
| Ma03_g10270 | 0 | 159.22 | 7.3239 | 5.00E-05 |
| Ma07_g15550, Ma07_g15600 | 2.0088 | 300.7 | 7.2259 | 0.0034 |
| Ma06_g16120, Ma06_g16130 | 2.204 | 318.29 | 7.1741 | 0.00385 |
| Ma06_g07540 | 0.282 | 39.53 | 7.1312 | 0.00865 |
| Ma09_g30030 | 0.4778 | 62.16 | 7.0235 | 0.02725 |
| Ma06_g33980 | 4.2437 | 547.44 | 7.0112 | 0.00185 |
| Ma04_g35390 | 1.1122 | 141.47 | 6.9909 | 0.00045 |
| Ma08_g17680 | 0.2461 | 30.13 | 6.9358 | 0.03105 |
| NA | 0 | 120.5 | 6.9248 | 0.00535 |
| Ma10_g31360 | 0.2208 | 24.87 | 6.8157 | 0.0103 |
| Ma04_g17460 | 0.1326 | 14.71 | 6.7938 | 0.00435 |
| Ma04_g02270 | 7.4929 | 825.12 | 6.7829 | 0.0047 |
| Ma11_g01760 | 0.1312 | 14.42 | 6.7803 | 0.01715 |
| Ma04_g10050 | 0.2748 | 30.15 | 6.7779 | 0.0362 |
| Ma07_g24340 | 7.0938 | 740.83 | 6.7064 | 0.005 |
| Ma05_g30490, Ma05_g30510 | 4.3015 | 446.84 | 6.6988 | 0.00115 |
| Ma05_g02850 | 0 | 98.94 | 6.6429 | 0.01335 |
| Ma09_g16440 | 4.0667 | 387.78 | 6.5753 | 0.0065 |
Fig 4.
Clustered heatmap depicting top 50 up-regulated (left panel) and down-regulated genes (right panel) in ripe fruit pulp. Red color indicates up-regulated genes and green color indicates down-regulated genes in Nendran fruit samples.
Table 3. List of top 50 down-regulated genes in Nendran ripe fruit samples.
| Musa acuminata IDs | Unripe FPKM | Ripe FPKM | Log2 (fold change) | P-value |
|---|---|---|---|---|
| Ma08_g14870 | 57.9970 | 12.3140 | -2.2357 | 0.0422 |
| Ma08_g29710 | 18.6179 | 3.6973 | -2.3321 | 0.0459 |
| Ma05_g15560 | 198.1540 | 38.7050 | -2.3560 | 0.04935 |
| Ma05_g29200 | 82.2009 | 15.9326 | -2.3672 | 0.0323 |
| Ma02_g15190 | 6.4337 | 1.2186 | -2.4005 | 0.04805 |
| Ma08_g23830, Ma08_g23840 | 53.1387 | 9.9826 | -2.4123 | 0.03 |
| Ma08_g14790 | 90.0315 | 16.7500 | -2.4263 | 0.04685 |
| Ma01_g09160 | 2.6453 | 0.4839 | -2.4508 | 0.0317 |
| Ma11_g16990 | 12.0605 | 2.0735 | -2.5402 | 0.04245 |
| Ma03_g24790 | 11.7536 | 2.0114 | -2.5468 | 0.03935 |
| Ma07_g06110 | 15.2059 | 2.5859 | -2.5559 | 0.0428 |
| Ma01_g15000 | 16.7579 | 2.8328 | -2.5646 | 0.038 |
| Ma08_g13700 | 55.7171 | 9.4159 | -2.5649 | 0.04005 |
| Ma11_g22760 | 24.0255 | 4.0311 | -2.5753 | 0.0439 |
| Ma07_g05150 | 13.5948 | 2.2438 | -2.5990 | 0.04545 |
| Ma05_g03730 | 18.2312 | 2.9992 | -2.6038 | 0.0464 |
| Ma07_g21650 | 31.7084 | 5.1810 | -2.6136 | 0.02875 |
| Ma08_g27050 | 23.9941 | 3.9084 | -2.6180 | 0.037 |
| Ma05_g16930 | 23.3441 | 3.7838 | -2.6252 | 0.0362 |
| Ma00_g01530 | 20.2872 | 3.2693 | -2.6335 | 0.0239 |
| Ma06_g01110 | 30.6584 | 4.9257 | -2.6379 | 0.04155 |
| Ma01_g14940 | 136.9260 | 21.7356 | -2.6553 | 0.03425 |
| Ma07_g15700 | 179.5230 | 28.0934 | -2.6759 | 0.0445 |
| Ma08_g05740 | 29.6195 | 4.6249 | -2.6791 | 0.04335 |
| Ma05_g05010 | 105.5100 | 16.4617 | -2.6802 | 0.0486 |
| Ma11_g02290 | 12.5888 | 1.9597 | -2.6835 | 0.0328 |
| Ma06_g19920 | 10.2797 | 1.5929 | -2.6901 | 0.02795 |
| mito4_g00080 | 10.7582 | 1.6646 | -2.6922 | 0.0327 |
| Ma02_g00470 | 175.1800 | 27.0540 | -2.6949 | 0.02425 |
| Ma06_g28590 | 32.6655 | 5.0367 | -2.6972 | 0.0341 |
| Ma07_g27270 | 54.7385 | 8.3345 | -2.7154 | 0.02555 |
| Ma08_g20470 | 54.1184 | 8.2262 | -2.7178 | 0.04435 |
| Ma02_g18970 | 29.7441 | 4.4848 | -2.7295 | 0.04965 |
| Ma03_g02940 | 23.3713 | 3.5238 | -2.7296 | 0.0315 |
| Ma09_g22220 | 45.9947 | 6.9188 | -2.7329 | 0.03515 |
| Ma06_g04700 | 16.4684 | 2.4648 | -2.7402 | 0.0328 |
| Ma05_g11240 | 186.9100 | 27.9561 | -2.7411 | 0.02375 |
| Ma05_g06160 | 84.3011 | 12.5721 | -2.7453 | 0.0389 |
| Ma10_g20280 | 15.5856 | 2.3120 | -2.7530 | 0.04475 |
| Ma09_g05470 | 20.7883 | 3.0835 | -2.7531 | 0.04635 |
| Ma07_g08320 | 193.6760 | 28.4678 | -2.7662 | 0.04545 |
| Ma04_g18080, Ma04_g18100 | 18.5819 | 2.7309 | -2.7665 | 0.01615 |
| Ma02_g01240 | 460.2760 | 67.5250 | -2.7690 | 0.02775 |
| Ma10_g07330 | 34.0326 | 4.9770 | -2.7736 | 0.02735 |
| Ma09_g22050 | 38.0633 | 5.5625 | -2.7746 | 0.0358 |
| Ma11_g22400 | 20.6719 | 2.9902 | -2.7894 | 0.03615 |
| Ma04_g12450 | 95.9664 | 13.8414 | -2.7935 | 0.04895 |
| Ma09_g02240 | 26.2597 | 3.7843 | -2.7947 | 0.03605 |
| Ma04_g18350 | 20.4148 | 2.9223 | -2.8045 | 0.02455 |
Differential expression pattern of genes involved in the carotenoid biosynthesis pathway
The fruit ripening response and the expression of carotenoid biosynthesis pathway genes in cv. Nendran was correlated and presented in S2 Fig. It was observed that expression of isopentenyl-diphosphate delta isomerase 1-like (IPP) (1 fold), lycopene beta cyclase (LCYβ) (1.29 fold), geranylgeranyl pyrophosphate synthase (GGPS) (6 fold), prolycopene isomerase (CRTISO) (1.5 fold), cytokinin dehydrogenase (5 fold), 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (HDS) (4 fold) and phytoene desaturase (PDS) (1.44 fold) was significantly enhanced at the ripe stage of fruit compared to the unripe stage. While the gene expression of 9-cis-epoxycarotenoid dehydrogenase (NCED), carotenoid 9%2C10(9%2C10’)-cleavage dioxygenase 1-like (CCD1), lycopene epsilon cyclase (LCYε), β-carotene 3-hydroxylase 2 (BCH2) and phytoene synthase 2 (PSY2) was highly down-regulated at ripened stage (ranging from -1 to -9 folds) (S2 Fig).
Genes involved in ripening, aroma and flavor
The banana ripening process is known to be involved in softening of fruit tissues that lead to the formation of aromatic compounds. The softening is mainly governed by the degradation of cell wall components [36]. This process is associated with a repertoire of genes, which are differentially expressed to regulate these events. Therefore, we have also analyzed the expression of genes that are linked with the ripening, flavor and aroma formation.
The expression pattern of methyltransferase, expansins, pectin lyase (PL), xyloglucan endotransglucosylase/hydrolase protein 32 (XTH), polygalactouronase (PG) which are ripening associated genes has been evaluated in the transcriptome data generated at the ripe stage of Nendran. The expression of the XTH gene family was highest (13 fold) followed by PL (12 fold) and trans-resveratrol di-O-methyltransferase-like (11 fold) in ripe fruit-pulp tissue in comparison to the unripe stage of fruit-pulp.
From the putative XTH gene family, 15 genes were up-regulated with the highest fold change (13 fold) and 17 genes were down-regulated (-7 fold). Similarly, six genes from the PL family were highly expressed during ripening stage of the banana. Nine expansin genes were identified and their expression was increased up to 9 fold. Few members of the PG gene family were highest in expression (up to 6 fold), while other gene families like glucosidases (GSMUA_Achr11G06230_001) (7 fold) are also expressed in the fruit-pulp of cv. Nendran. However, the expression of some of the members of these gene families like pectinesterase (5 fold) was considerably enhanced but still on the lower side when compared to the expression of other ripening associated families such as XTH. The expression details of ripening associated genes are provided in the S2 Table. Genes involved in softening of the cell wall were amongst the highly up-regulated genes (PL, PE, XTH) indicating that softening is the main event during the ripening of banana.
The presence of various volatiles viz. butyl acetate, isoamyl alcohol, and isoamyl acetate attributes to the aroma of banana fruit. Fatty acid biosynthesis and other pathways like the phenylpropanoid pathway mainly produce these volatile compounds. In this study, the expression pattern of genes involved in the biosynthesis pathways of fatty acid, unsaturated fatty acid and amino acid formation was analyzed. We have checked the expression of alcohol dehydrogenase (ADH) that mediates the conversion of alcohol from sugars. ADH genes are usually expressed during the fruit ripening process and reported to play a major role in the development of flavor. In total 5 transcripts annotated for ADH (ADH1—ADH5) were identified and among them ADH1 (GSMUA_Achr2G08040_001) has shown one-fold increased expression, while others have not indicated any significant change in expression levels. Likewise, lipoxygenases (LOX) genes are also involved in aroma development and the expression of one gene belonging to LOX (GSMUA_Achr9G12470_001) was up-regulated (5 fold) in ripened banana. Further analysis of transcriptome data revealed that various transferases like benzoyltransferases, methyltransferases and acyltransferases were significantly up-regulated in ripe banana indicating their potential role in the aroma. The highest expression was observed in putative 3-N-debenzoyl-2-deoxytaxol N-benzoyltransferase that was increased maximum by 12 fold in a ripe banana. Similarly, 3-oxoacyl-[acyl-carrier-protein] reductase and acyltransferase genes also exhibited 11 and 2 fold increased expression, respectively (S3 Table).
Ethylene exposure also accelerates ripening in banana. ACC synthase (ACS) and ACC oxidase (ACO) are the main regulators that govern ethylene biosynthesis in fruits [37]. ACO is also considered to be the rate-limiting step in ethylene production [38]. In the current study, expression change in ACS and ACO genes to understand their role in fruit ripening of cv. Nendran is explored. It has been observed that the expression of ACS (3 fold) and ACO (1 fold) was up-regulated in a ripe fruit-pulp than that of unripe fruit-pulp (S4 Table).
Identification of Transcription Factors (TFs)
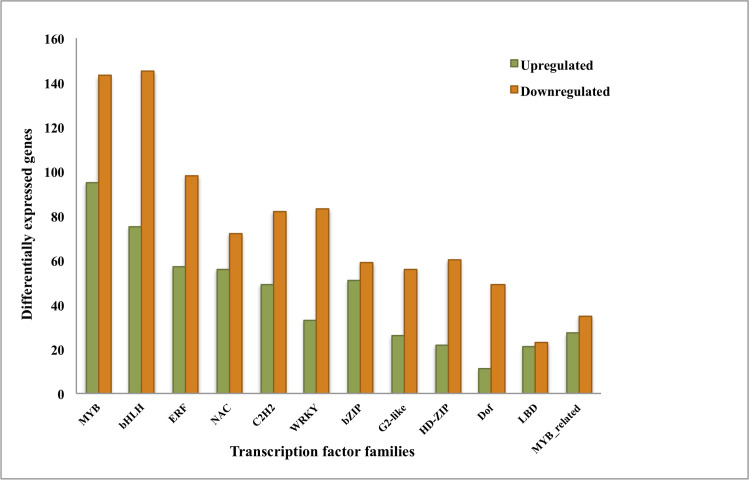
TFs regulate the expression of genes. Hence, we have downloaded TFs from the PlantTFDB database [39]. This database harbors 2896 TFs from Musa acuminata which are classified into 57 families. It was examined that most of the TFs belong to the multigene family and mainly the TFs were related to MYB, bHLH, ERF, NAC and C2H2 gene family. These gene families showed varied expression at the ripening stage (up and down). A list of the top 12 TF family members is given in Fig 5 and a detailed list of all transcription factors family members along with fold change expression is provided in the S5 Table.
Fig 5. Transcription factors expressively correlated with cv. Nendran ripe fruit-pulp.
Orange bars represent down-regulated genes while green bars represent up-regulated genes.
Validation of differential gene expression by qRT-PCR
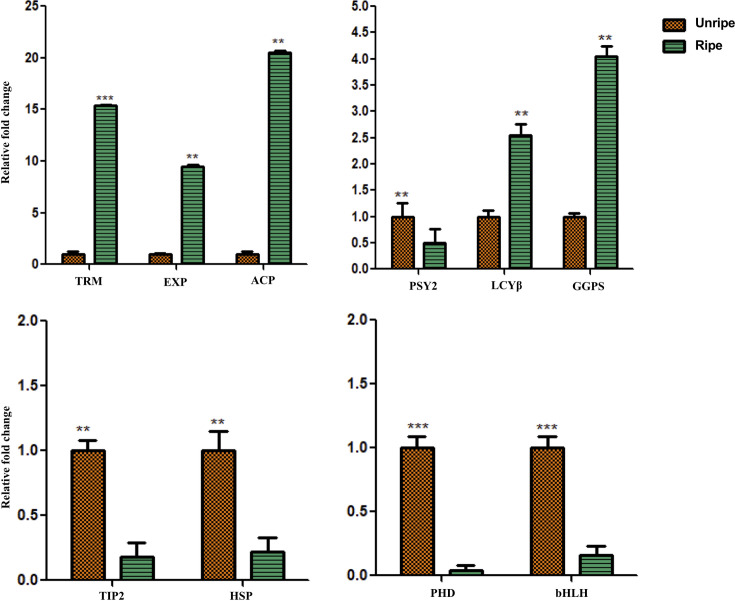
The validation of differential expression of selected genes belonging to different pathways was checked by qRT-PCR assay. Total ten genes were selected based on their significant differential expression pattern and potential role in acting as TFs stress response and carotenoid pathway genes. All the genes exhibited a comparable trend of expression in unripe and ripe stages as attained by transcriptomic data. It was observed that expression of putative 3-oxoacyl-[acyl-carrier-protein] reductase, chloroplastic (ACP), trans-resveratrol di-O-methyltransferase (TRM) and expansin-A2 (EXP) was up-regulated which is in accordance with the transcriptome data. Similarly, the expression of genes involved in the carotenoid pathway LCYβ and GGPS was highly up-regulated in ripe fruit-pulp of cv. Nendran as compared to unripe sample (Fig 6). Further, the expression of Actin 1 (LOC103992183) was also analysed and no change in expression pattern was observed in both ripe and unripe conditions.
Fig 6. Quantitative real-time PCR of selected genes.
Expression profile of selected genes in ripe Nendran banana. Gene expression was normalized using Actin1 as an internal control. Orange bars represent the expression of unripe while green bars depict the expression of ripe genes. Statistical analysis was executed using Student’s paired t-test and statistical significance was checked at **P ≤ 0.001; ***P ≤ 0.0001. TRM (Trans-resveratrol di-O-methyltransferase); EXP (Expansin-A2); ACP (Putative 3-oxoacyl-[acyl-carrier-protein] reductase, chloroplastic); PSY2 (phytoene synthase 2); LCYβ (lycopene beta cyclase); GGPS (geranylgeranyl pyrophosphate synthase); TIP2 (Probable aquaporin TIP2); HSP (Heat shock 70 kDa protein 8); PHD (PHD-type domain-containing protein); bHLH (transcription factor bHLH62).(Corresponding gene IDs given in S1 Table).
Discussion
Banana is a staple fruit crop worldwide. Therefore, the understanding of molecular mechanisms that are associated with various traits is crucial. Reports showed that ripening in various fruit crops including banana is initiated by a set of genes that brings physico-chemical changes in the quality of fruit [40]. These changes mainly alter cell wall degradation, synthesis of volatile compounds and alteration in phenolic constituents [40]. Sequencing technologies now provide ways to identify ripening and associated process-related genes and subsequently those can be used for genetic manipulation for the improvement of banana. Henceforth, insights into the genes responsible for ripening are necessary to understand the molecular basis of these genes that can also be implemented to improve post-harvest losses of this highly perishable crop.
In the present study, cv. Nendran of banana has been selected as it is having a high content of carotenoid at the ripening stage of fruit-pulp [12]. Further, early ripening (15w of bunch emergence) of fruits in this nutritional rich cultivar has also influenced us to understand the molecular basis of this observation. Using transcriptome analysis hosted on Illumina (HiSeq 2500) 3474 up- and 4727 down-regulated genes were obtained in the Nendran ripe fruit-pulp samples. Functional enrichment of the genes revealed their probable role in various metabolic processes.
Nendran has been reported as an excellent source of β-carotene and its activity was correlated with enhanced antioxidant activity [12]. Ripening plays a crucial role in the biosynthesis of carotenoids in the fruit-pulp tissue of banana [41]. In the current study, the expression pattern of carotenoid genes indicated that LCYβ, GGPS, and PDS were up-regulated in ripe fruit pulp. Moreover, the role of LCYβ in β-carotene biosynthesis has already been demonstrated by the regulation of lycopene flux [42]. Similarly, GGPS serves as a precursor for important compounds like tocopherols, carotenoids, and chlorophyll [43]. Expression of GGPS was up-regulated in ripe pulp samples. PDS is one of the first enzymes in the carotenoid biosynthesis pathway. It has also been reported as a positive regulator for ripening in tomato fruit [44]. Mutation in PDS gene by CRISPR/Cas technology resulted in decreased chlorophyll and carotenoid content in banana cv. Rasthali [32]. The transcriptome data have shown a ~2 fold increase in expression of the PDS gene (GSMUA_Achr3G21400_001) in the ripe fruit. It indicates the possible role of PDS in early fruit ripening and high carotenoid deposition in cv. Nendran. BCH gene is reported to involve in the biosynthesis of zeaxanthin which is a precursor of abscisic acid [45]. In our analysis expression of gene annotated as β-carotene 3-hydroxylase 2 (BCH2) (GSMUA_Achr11G02930_001) in Musa was down-regulated by ~ -6 fold. Other carotenoid pathway genes like CCD, NCED are considered carotenoid degrading enzymes [46]. In this study expression of these genes was down-regulated in ripened banana, signifying their less role in the ripening and carotenoid degradation process in Nendran.
Softening is mainly initiated with the inception of ripening [47], which leads to cell wall degradation. Cell wall hydrolysis plays a crucial role in plant growth and development, stress response and ripening process. Utmost of the genes involved in this process are mainly members of multigene families and are associated with specified functions like cell wall metabolism [48]. The significant cell wall degrading proteins are pectin methyl esterase, polygalacturonase (PG), XTH, expansins, PL, galactosidases and endoglucanases [49]. XTH gene family members have been reported to play important role in the ripening of fruits like tomato and apple [50, 51]. Studies in fruit crops such as mango and banana have reported the role of these genes in cell wall loosening [52, 53]. The expression pattern of most of the genes belonging to XTH and expansins gene families was reported to be highly up-regulated during ripening conditions. Amongst them, some members of these gene families were also down-regulated indicating that they might not be playing any significant function in the ripening process.
Ethylene is considered one of the major plant hormones that control many aspects of fruit ripening [54]. The initial step in ethylene biosynthesis is the conversion of S-adenosyl methionine to 1-aminocyclopropane-1-carboxylic-acid catalyzed by ACS [55]. The ACS and ACO genes are reported to regulate ethylene biosynthesis in the tomato and apple during the fruit ripening stage [56, 57]. Transcriptome data in this study revealed up-regulation of both genes in ripe fruit-pulp tissue specifying their potential role in ethylene synthesis and ripening.
We also analysed the expression of transcription gene families in fruit-pulp of cv. Nendran and found that most of the members of these multigene families of TFs were down-regulated at the ripening stage. These TFs may not be required at this stage hence, their expression is declined during the ripening process.
Conclusion
The comparative analysis of transcriptome at the unripe and ripen stages of fruit-pulp of cv. Nendran provides a comprehensive landscape of differentially expressed genes that are mostly associated with ripening, carotenoid biosynthesis, aroma and other related processes. The expression data acquired by RNA-seq were validated by qRT-PCR analysis. The results of this study suggested that many differentially expressed genes in the unripe and ripe banana are associated with aroma and ripening processes. Gene families in ripening like PL, expansins, XTH etc. were showed differential expression patterns. Genes like acyltransferases known to be responsible for cell wall hydrolysis and production of aromatic volatiles and flavor have shown higher expression at the ripening stage. The expression pattern of the carotenoid synthesis pathway genes indicated that LCYβ and GGPS were highly up-regulated during the ripening stage while CCDs and NCEDs were downregulated. Overall, the present study has provided information about the promising role of genes such as acyltransferases, LCYβ and GGPS to develop a better understanding of the ripening process and their link with carotenoid synthesis, aroma and flavor formation in banana fruit-pulp.
Supporting information
X-axis represents significance of gene ontology term enrichment and y-axis represents the log P-values.
(PDF)
Red color represents up-regulated genes and green color represents down-regulated genes.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
The authors express their gratitude to the National Agri-Food Biotechnology Institute (NABI) for research facilities and the Department of Biotechnology (DBT), Ministry of Science & Technology, Government of India for the research facilities and support. Authors are also thankful to the Biotechnology Industry Research Assistance Council (BIRAC) for the banana biofortification project support. The authors would like to acknowledge the DBT-eLibrary Consortium (Del-CON) for providing access to online journals.
Data Availability
Relevant datasets (transcriptomic) supporting results of this work have been deposited in NCBI SRA repository under BioProject ID: PRJNA699099.
Funding Statement
The authors express their gratitude to the National Agri-Food Biotechnology Institute (NABI) for research facilities and the Department of Biotechnology (DBT), Government of India for the grant (BT/PR25789/GET/119/97/2017) under the scheme of Genome Engineering Technology and Their Applications. The present research was also supported by the Biotechnology Industry Research Assistance Council (BIRAC) for a banana biofortification project grant (BIRAC/Tech Transfer/08/I2/QUT-BBF dated 04.09.2012).
References
- 1.Heslop-Harrison JS, Schwarzacher T. Domestication, genomics and the future for banana. Annals of botany. 2007;100(5):1073–84. doi: 10.1093/aob/mcm191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Miao H, Liu J, Xu B, Yao X, Xu C, et al. Musa balbisiana genome reveals subgenome evolution and functional divergence. Nature plants. 2019;5(8):810–21. doi: 10.1038/s41477-019-0452-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jesus ON, Silva Sde O, Amorim EP, Ferreira CF, de Campos JM, Silva Gde G, et al. Genetic diversity and population structure of Musa accessions in ex situ conservation. BMC plant biology. 2013;13:41. doi: 10.1186/1471-2229-13-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning K, Tör M, Poole M, Hong Y, Thompson AJ, King GJ, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nature genetics. 2006;38(8):948–52. doi: 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- 5.Clendennen SK, May GD. Differential gene expression in ripening banana fruit. Plant physiology. 1997;115(2):463–9. doi: 10.1104/pp.115.2.463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchita Biswas Murmu HNM. Post-harvest shelf-life of banana and guava: Mechanisms of common degradation problems and emerging counteracting strategies. Innovative Food Science & Emerging Technologies. 20 July 2018;49:20–30. [Google Scholar]
- 7.Maqbool M AA, Ramachandran S, Smith DR, Alderson PG. Control of postharvest anthracnose of banana using a new edible composite coating. International Society for Horticultural Science. 2010:639–44. [Google Scholar]
- 8.Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H. The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. Journal of experimental botany. 2010;61(5):1523–35. doi: 10.1093/jxb/erq017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams-Phillips L, Barry C, Giovannoni J. Signal transduction systems regulating fruit ripening. Trends in plant science. 2004;9(7):331–8. doi: 10.1016/j.tplants.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 10.Yan SC, Chen JY, Yu WM, Kuang JF, Chen WX, Li XP, et al. Expression of genes associated with ethylene-signalling pathway in harvested banana fruit in response to temperature and 1-MCP treatment. Journal of the science of food and agriculture. 2011;91(4):650–7. doi: 10.1002/jsfa.4226 [DOI] [PubMed] [Google Scholar]
- 11.Englberger L, Wills RB, Blades B, Dufficy L, Daniells JW, Coyne T. Carotenoid content and flesh color of selected banana cultivars growing in Australia. Food and nutrition bulletin. 2006;27(4):281–91. doi: 10.1177/156482650602700401 [DOI] [PubMed] [Google Scholar]
- 12.Kaur N, Pandey A, Shivani, Kumar P, Pandey P, Kesarwani AK, et al. Regulation of Banana Phytoene Synthase (MaPSY) Expression, Characterization and Their Modulation under Various Abiotic Stress Conditions. Frontiers in plant science. 2017;8:462. doi: 10.3389/fpls.2017.00462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–7. doi: 10.1038/nature11241 [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nature reviews Genetics. 2009;10(1):57–63. doi: 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hrdlickova R, Toloue M, Tian B. RNA-Seq methods for transcriptome analysis. Wiley interdisciplinary reviews RNA. 2017;8(1). doi: 10.1002/wrna.1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asif MH, Lakhwani D, Pathak S, Gupta P, Bag SK, Nath P, et al. Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC plant biology. 2014;14:316. doi: 10.1186/s12870-014-0316-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun J, Zhang J, Fang H, Peng L, Wei S, Li C, et al. Comparative transcriptome analysis reveals resistance-related genes and pathways in Musa acuminata banana ’Guijiao 9’ in response to Fusarium wilt. Plant physiology and biochemistry: PPB. 2019;141:83–94. doi: 10.1016/j.plaphy.2019.05.022 [DOI] [PubMed] [Google Scholar]
- 18.Nakano M, McCormick K, Demirci C, Demirci F, Gurazada SGR, Ramachandruni D, et al. Next-Generation Sequence Databases: RNA and Genomic Informatics Resources for Plants. Plant physiology. 2020;182(1):136–46. doi: 10.1104/pp.19.00957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang A, Zhang Q, Li J, Gong H, Fan X, Yang Y, et al. Transcriptome co-expression network analysis identifies key genes and regulators of ripening kiwifruit ester biosynthesis. BMC plant biology. 2020;20(1):103. doi: 10.1186/s12870-020-2314-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi X, Ogden EL, Die JV, Ehlenfeldt MK, Polashock JJ, Darwish O, et al. Transcriptome analysis identifies genes related to the waxy coating on blueberry fruit in two northern-adapted rabbiteye breeding populations. BMC plant biology. 2019;19(1):460. doi: 10.1186/s12870-019-2073-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanca J, Esteras C, Ziarsolo P, Pérez D, Fernã Ndez-Pedrosa V, Collado C, et al. Transcriptome sequencing for SNP discovery across Cucumis melo. BMC genomics. 2012;13:280. doi: 10.1186/1471-2164-13-280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu K, Xu Q, Da X, Guo F, Ding Y, Deng X. Transcriptome changes during fruit development and ripening of sweet orange (Citrus sinensis). BMC genomics. 2012;13:10. doi: 10.1186/1471-2164-13-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JH, Liu JJ, Chen KL, Li HW, He J, Guan B, et al. Comparative transcriptome and proteome profiling of two Citrus sinensis cultivars during fruit development and ripening. BMC genomics. 2017;18(1):984. doi: 10.1186/s12864-017-4366-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Fan M, He Y. Transcriptome Analysis of Watermelon Leaves Reveals Candidate Genes Responsive to Cucumber green mottle mosaic virus Infection. International journal of molecular sciences. 2019;20(3). doi: 10.3390/ijms20030610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan Y, Qu Y, Zhu L, Shen C, Feng X, Yu C. Transcriptome analysis of tomato (Solanum lycopersicum L.) shoots reveals a crosstalk between auxin and strigolactone. PloS one. 2018;13(7):e0201124. doi: 10.1371/journal.pone.0201124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–20. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Droc G, Larivière D, Guignon V, Yahiaoui N, This D, Garsmeur O, et al. The banana genome hub. Database: the journal of biological databases and curation. 2013;2013:bat035. doi: 10.1093/database/bat035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England). 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols. 2012;7(3):562–78. doi: 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic acids research. 2019;47(W1):W191–w8. doi: 10.1093/nar/gkz369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye J, Zhang Y, Cui H, Liu J, Wu Y, Cheng Y, et al. WEGO 2.0: a web tool for analyzing and plotting GO annotations, 2018 update. Nucleic acids research. 2018;46(W1):W71–w5. doi: 10.1093/nar/gky400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur N, Alok A, Shivani, Kaur N, Pandey P, Awasthi P, et al. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Functional & integrative genomics. 2018;18(1):89–99. doi: 10.1007/s10142-017-0577-5 [DOI] [PubMed] [Google Scholar]
- 33.Kaur N, Alok A, Shivani, Kumar P, Kaur N, Awasthi P, et al. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for β-carotene biosynthesis in banana fruit. Metabolic engineering. 2020;59:76–86. doi: 10.1016/j.ymben.2020.01.008 [DOI] [PubMed] [Google Scholar]
- 34.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3(6):1101–8. doi: 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 35.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3—new capabilities and interfaces. Nucleic acids research. 2012;40(15):e115. doi: 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiao C, Anderson CT. Roles of pectin in biomass yield and processing for biofuels. Frontiers in plant science. 2013;4:67. doi: 10.3389/fpls.2013.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bleecker AB, Schaller GE. The Mechanism of Ethylene Perception. Plant physiology. 1996;111(3):653–60. doi: 10.1104/pp.111.3.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van de Poel B, Bulens I, Markoula A, Hertog ML, Dreesen R, Wirtz M, et al. Targeted systems biology profiling of tomato fruit reveals coordination of the Yang cycle and a distinct regulation of ethylene biosynthesis during postclimacteric ripening. Plant physiology. 2012;160(3):1498–514. doi: 10.1104/pp.112.206086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin J, Zhang H, Kong L, Gao G, Luo J. PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic acids research. 2014;42(Database issue):D1182–7. doi: 10.1093/nar/gkt1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulkarni SG, Kudachikar VB, Keshava Prakash MN. Studies on physico-chemical changes during artificial ripening of banana (Musa sp) variety ’Robusta’. Journal of food science and technology. 2011;48(6):730–4. doi: 10.1007/s13197-010-0133-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su L, Diretto G, Purgatto E, Danoun S, Zouine M, Li Z, et al. Carotenoid accumulation during tomato fruit ripening is modulated by the auxin-ethylene balance. BMC plant biology. 2015;15:114. doi: 10.1186/s12870-015-0495-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeng J, Wang C, Chen X, Zang M, Yuan C, Wang X, et al. The lycopene β-cyclase plays a significant role in provitamin A biosynthesis in wheat endosperm. BMC plant biology. 2015;15:112. doi: 10.1186/s12870-015-0514-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beck G, Coman D, Herren E, Ruiz-Sola MA, Rodríguez-Concepción M, Gruissem W, et al. Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant molecular biology. 2013;82(4–5):393–416. doi: 10.1007/s11103-013-0070-z [DOI] [PubMed] [Google Scholar]
- 44.Naing AH, Kyu SY, Pe PPW, Park KI, Lee JM, Lim KB, et al. Silencing of the phytoene desaturase (PDS) gene affects the expression of fruit-ripening genes in tomatoes. Plant methods. 2019;15:110. doi: 10.1186/s13007-019-0491-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du H, Wang N, Cui F, Li X, Xiao J, Xiong L. Characterization of the beta-carotene hydroxylase gene DSM2 conferring drought and oxidative stress resistance by increasing xanthophylls and abscisic acid synthesis in rice. Plant physiology. 2010;154(3):1304–18. doi: 10.1104/pp.110.163741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vallabhaneni R, Bradbury LM, Wurtzel ET. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Archives of biochemistry and biophysics. 2010;504(1):104–11. doi: 10.1016/j.abb.2010.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Payasi A, Mishra NN, Chaves AL, Singh R. Biochemistry of fruit softening: an overview. Physiology and molecular biology of plants: an international journal of functional plant biology. 2009;15(2):103–13. doi: 10.1007/s12298-009-0012-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brummell DA, Harpster MH. Cell wall metabolism in fruit softening and quality and its manipulation in transgenic plants. Plant molecular biology. 2001;47(1–2):311–40. [PubMed] [Google Scholar]
- 49.Paull RE, Chen NJ. Postharvest Variation in Cell Wall-Degrading Enzymes of Papaya (Carica papaya L.) during Fruit Ripening. Plant physiology. 1983;72(2):382–5. doi: 10.1104/pp.72.2.382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saladié M, Rose JK, Cosgrove DJ, Catalá C. Characterization of a new xyloglucan endotransglucosylase/hydrolase (XTH) from ripening tomato fruit and implications for the diverse modes of enzymic action. The Plant journal: for cell and molecular biology. 2006;47(2):282–95. doi: 10.1111/j.1365-313X.2006.02784.x [DOI] [PubMed] [Google Scholar]
- 51.Muñoz-Bertomeu J, Miedes E, Lorences EP. Expression of xyloglucan endotransglucosylase/hydrolase (XTH) genes and XET activity in ethylene treated apple and tomato fruits. Journal of plant physiology. 2013;170(13):1194–201. doi: 10.1016/j.jplph.2013.03.015 [DOI] [PubMed] [Google Scholar]
- 52.Srivastava S, Singh RK, Pathak G, Goel R, Asif MH, Sane AP, et al. Comparative transcriptome analysis of unripe and mid-ripe fruit of Mangifera indica (var. "Dashehari") unravels ripening associated genes. Scientific reports. 2016;6:32557. doi: 10.1038/srep32557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asif MH, Nath P. Expression of multiple forms of polygalacturonase gene during ripening in banana fruit. Plant physiology and biochemistry: PPB. 2005;43(2):177–84. doi: 10.1016/j.plaphy.2005.01.011 [DOI] [PubMed] [Google Scholar]
- 54.Liu M, Pirrello J, Chervin C, Roustan JP, Bouzayen M. Ethylene Control of Fruit Ripening: Revisiting the Complex Network of Transcriptional Regulation. Plant physiology. 2015;169(4):2380–90. doi: 10.1104/pp.15.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iqbal N, Khan NA, Ferrante A, Trivellini A, Francini A, Khan MIR. Ethylene Role in Plant Growth, Development and Senescence: Interaction with Other Phytohormones. Frontiers in plant science. 2017;8:475. doi: 10.3389/fpls.2017.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nakatsuka A, Murachi S, Okunishi H, Shiomi S, Nakano R, Kubo Y, et al. Differential expression and internal feedback regulation of 1-aminocyclopropane-1-carboxylate synthase, 1-aminocyclopropane-1-carboxylate oxidase, and ethylene receptor genes in tomato fruit during development and ripening. Plant physiology. 1998;118(4):1295–305. doi: 10.1104/pp.118.4.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong Li DT, Xuyuan Yang, Aide Wang. Exploring the apple genome reveals six ACC synthase genes expressed during fruit ripening Author links open overlay panel. Scientia Horticulturae. 2013;157:119–23. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
X-axis represents significance of gene ontology term enrichment and y-axis represents the log P-values.
(PDF)
Red color represents up-regulated genes and green color represents down-regulated genes.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
Relevant datasets (transcriptomic) supporting results of this work have been deposited in NCBI SRA repository under BioProject ID: PRJNA699099.