The interplay of photosynthesis with carbon acquisition and fixation in Chlamydomonas, regulating its inducible carbon concentration mechanism that is dependent on cellular machineries of transcription, protein folding, and retrograde signalling, is reviewed here.
Keywords: Carbon concentration mechanism (CCM), chaperones, Chlamydomonas, CIA5, photorespiration, photosynthesis, pyrenoid, retrograde signalling
Abstract
The inducible carbon concentration mechanism (CCM) in Chlamydomonas reinhardtii has been well defined from a molecular and ultrastructural perspective. Inorganic carbon transport proteins, and strategically located carbonic anhydrases deliver CO2 within the chloroplast pyrenoid matrix where Rubisco is packaged. However, there is little understanding of the fundamental signalling and sensing processes leading to CCM induction. While external CO2 limitation has been believed to be the primary cue, the coupling between energetic supply and inorganic carbon demand through regulatory feedback from light harvesting and photorespiration signals could provide the original CCM trigger. Key questions regarding the integration of these processes are addressed in this review. We consider how the chloroplast functions as a crucible for photosynthesis, importing and integrating nuclear-encoded components from the cytoplasm, and sending retrograde signals to the nucleus to regulate CCM induction. We hypothesize that induction of the CCM is associated with retrograde signals associated with photorespiration and/or light stress. We have also examined the significance of common evolutionary pressures for origins of two co-regulated processes, namely the CCM and photorespiration, in addition to identifying genes of interest involved in transcription, protein folding, and regulatory processes which are needed to fully understand the processes leading to CCM induction.
Introduction
The carbon concentration mechanism (CCM) traits found in algae (and cyanobacteria) have evolved to improve the operating efficiency of Rubisco, which is normally packaged within a specific microcompartment: in algae, this is the chloroplast pyrenoid. Inorganic carbon, in the form of bicarbonate, is delivered to the chloroplast stroma using a series of membrane transporters. Saturating internal CO2 concentrations (Ci), ~40× above ambient (Badger et al., 1980), are generated within the pyrenoid by strategically placed transporters of inorganic carbon and carbonic anhydrases (CAs) (Moroney and Ynalvez, 2007; Meyer and Griffiths, 2013; Hennacy and Jonikas, 2020). The availability of a sequenced genome (Merchant et al., 2007), transcriptomic studies for synchronized cells across 24 h light/dark cycles (Zones et al., 2015; Strenkert et al., 2019), and extensive mutant libraries (Li et al., 2016, 2019; Vilarrasa-Blasi et al., 2020, Preprint) for Chlamydomonas have provided additional opportunities for CCM characterization.
Substantial molecular and mechanistic advances in our understanding of the algal CCM have been recently reviewed (Meyer and Griffiths, 2013; Meyer et al., 2017; Goudet et al., 2020; Hennacy and Jonikas, 2020). CCM induction is associated with enhancement of aggregation of Rubisco with specific linker proteins (Mackinder et al., 2016; He et al., 2020; Meyer et al., 2020) in the pyrenoid surrounded by a starch sheath, with an existing network of knotted tubules making connections with thylakoid stacks (Engel et al., 2015; Meyer et al., 2017). Establishment of the CCM must be co-ordinated between the chloroplast and nucleus in sensing induction stimuli, triggering CCM gene expression, translation, and intracellular transport and assembly of CCM proteins.
The aim of this review is to characterize the various novel aspects of molecular mechanisms leading to mechanisms of CCM induction and establishment. We explore the regulatory interplay between environmental sensing, photosynthesis, and CCM induction that is critical for Chlamydomonas and identify future avenues for investigation. First, we consider the control of nuclear gene expression and the need to identify transcription factors (TFs) associated with sensing the different environmental stimuli—light and CO2; second, regulation of export of translated proteins and folding within the chloroplast, and associated chaperone systems; third, formation of the pyrenoid matrix, starch sheath, and intrapyrenoidal tubule network; fourth, the role of retrograde signalling in delivering signals to alter nuclear gene expression; and, finally, we discuss the potential evolution of CCM induction from existing photorespiration regulatory mechanisms, through the master regulator CIA5.
CCM induction and control of gene expression
Changes in [CO2] in the external medium have traditionally been thought to be sensed through Ci of the photosynthesizing algal cell and conveyed to the nucleus to change the expression of genes that turn on/off the CCM. Studies carried out with asynchronous cells grown under continuous light revealed [CO2]-dependent expression for >5000 genes at the transcription level (Brueggeman et al., 2012; Fang et al., 2012). Over 600 of these differentially expressed genes identified in these genome-wide studies have been implicated in the CCM (Mackinder et al., 2017). With several hundred genes orchestrating the CCM, the following questions come to the fore: (i) what are the regulators in the nucleus that respond to [CO2]/Ci changes, and (ii) how do they bring about changes at the transcriptional level? In this section, we discuss regulatory mechanisms operating at the transcriptional level to modulate the inducible CCM in Chlamydomonas.
CIA5: the ‘master-regulator’ of the CCM
One of the first interesting candidates to be identified as a ‘CCM master regulator’ was CIA5/CCM1. CIA5 was identified in 1989 as essential for growth in limiting CO2 conditions through studies on a UV-generated mutant, cia5 (Moroney et al., 1989). Studies in the early 2000s established CIA5 as being essential for induction of expression of several CCM genes encoding inorganic carbon transporters, HLA3 and LCI1; CAs, CAH3 and CAH1; alanine α-ketoglutarate aminotransferase, ATT1; pyrenoid protein, EPYC1; a TF, LCR1; and mitochondrial membrane proteins, CCP1 and CCP2 (Fukuzawa et al., 2001; Xiang et al., 2001; Miura et al., 2004). It must be noted that genes denoted as CCM genes in this review are based on previous findings (Mackinder et al., 2017; Strenkert et al., 2019). Genome-wide studies (Fang et al., 2012) showed CIA5-dependent expression for 15% genes, but only around half of these CIA5-dependent genes responded to changes in [CO2]. Furthermore, the mechanism of CIA5 in helping cells acclimate to [CO2] changes is not understood. CIA5 is proposed to be a TF based on the presence of two Zn-finger domains (Fukuzawa et al., 2001; Xiang et al., 2001). The DNA-activating region (Chen, 2016) and the [CO2]-dependent domain that triggers the expression of CIA5-dependent CCM genes (Xiang et al., 2001) lie in the C-terminal end sequences of 130 and 54 residues, respectively. Primarily, the expression of CIA5 seems to be [CO2] independent (Fang et al., 2012), and associated [CO2]-dependent expression changes in the genome are believed to be mediated by post-translational modifications of CIA5 (Chen, 2016). This is supported by CIA5 having several putative sites for phosphorylation, glycosylation, and myristoylation (Fukuzawa et al., 2001), and anomalous electrophoretic mobility (Chen, 2016).
Absence of evidence for DNA–CIA5 complexes suggests that CIA5 might act indirectly through other proteins, although no such proteins have been identified (Kohinata et al., 2008). Recombinantly expressed full-length CIA5 showed very weak affinity in vitro for the 9 bp sequence (GGGGCGGGG), identified from analysis of upstream sequences of select CIA5-dependent genes (Chen, 2016). However, no motif-dependent binding for CIA5 could be established in vivo when genes with upstream mutated motifs showed similar expression patterns to those of non-mutated motifs (Chen, 2016).
Understanding the CIA5 mechanism will require identification of the different post-translationally modified forms of CIA5 and the corresponding cis-regulatory elements of CCM genes. The roles played by CIA5 are revisited in the context of chaperone expression in ‘Chaperones and the import and assembly of chloroplastic CCM proteins’, and the implications of strikingly similar CIA5-dependent expression profiles of photorespiratory and CCM genes are explored below.
Other transcriptional and post-transcriptional regulators of the CCM
A search for other CCM TFs and transcription regulators (TRs) has been made over the years. LCR1 (Yoshioka et al., 2004) is a Myb TF that plays a crucial role in the CCM by regulating expression of CAH1, LCI1, and LCI6. LCR1 expression is [CO2] and CIA5 dependent. Absence of LCR1 leads to a reduction in affinity for Ci (Yoshioka et al., 2004).
Sequence analysis identified 234 genes as potential TFs and TRs in Chlamydomonas (Riano-Pachon et al., 2008), and they need to be checked for their activity in CCM regulation. A recent proteomic analysis (Arias et al., 2020) on nuclei obtained from Chlamydomonas grown in 5% CO2/0.04% CO2 revealed the presence of 117 proteins which were potential TFs/TRs. Of these 117 nuclear proteins, 35 were of differential abundance dependent on [CO2], and these are listed in Table 1. It is worth noting that neither CIA5 nor LCR1 was detected in the nuclear proteome in this study. However, the candidates identified in this study might act as a good starting point for investigating other TFs and TRs regulating the CCM.
Table 1.
Transcription factors and regulators occurring with different relative abundances in Chlamydomonas grown in low (0.04%) and high (5%) CO2 conditions (Arias et al., 2020)
| Protein | Description | TF familya | Fold change |
|---|---|---|---|
| Transcription factors | |||
| Cre01.g000050.t1.1 | RWP-RK Transcription Factor | RWP-RK | 5.5 |
| Cre10.g444450.t1.1 | Predicted Protein | C3H | 3.1 |
| Cre14.g625802.t1.1 | Ring Finger Protein-Related | FHA | 2.9 |
| Cre16.g656250.t1.1 | U1 Small Nuclear Ribonucleoprotein | CSD | 2.5 |
| Cre17.g714500.t1.2 | Histone H2A | CCAAT | 2.5 |
| Cre06.g288750.t1.2 | Nuclear Rna Cap-Binding Protein | CSD | 2.4 |
| Cre06.g254650.t1.2 | Zinc Finger Protein 183 | C3H | 1.9 |
| Cre14.g632050.t1.2 | RPGR-Interacting Protein 1 Related | VARL | 1.9 |
| Cre01.g035150.t1.1 | Zinc Finger (CCCH-Type) Family Protein | C3H | 1.9 |
| Cre10.g446900.t1.2 | WD40 Repeat Proteinprl1/PRLl2-Related | Orphans | 1.7 |
| Cre02.g115250.t1.1 | Centriole Proteome Protein | Orphans | 1.6 |
| Cre12.g523200.t1.1 | Nucleosome Remodeling Factor | Orphans | 1.6 |
| Cre09.g389550.t1.1 | Dnaj-Like Protein | MYB-related | 1.6 |
| Cre03.g197350.t1.2 | Cell Division Cycle 5-Like Protein | MYB-related | 1.5 |
| Cre17.g713900.t1.2 | Tor Kinase Binding Protein | Orphans | – b |
| Cre01.g020400.t1.2 | WD40 Repeat Protein | Orphans | 2.3 |
| Cre09.g392350.t2.1 | Rna Recognition Motif (Rnp Domain) (Rrm_1) | CSD | 1.9 |
| Cre01.g035000.t1.2 | Wd Repeat Protein | Orphans | 1.8 |
| Cre17.g729150.t1.2 | Rna Recognition Motif. (Rnp Domain) (Rrm_1) | CSD | 1.7 |
| Cre16.g662800.t1.2 | Splicing Factor, Component Of The U4/U6-U5 Snrnp Complex | Orphans | 1.6 |
| Cre06.g275100.t1.2 | Splicing Factor 3B, Subunit 4 | CSD | 1.5 |
| Cre06.g274200.t1.2 | Histone H2A | CCAAT | – b |
| Cre12.g507650.t2.1 | Chloroplast Dnaj-Like Protein | MYB-related | – b |
| Transcription regulators | |||
| Cre16.g668200.t1.1 | Chromatin Remodeling Protein, Contains Phd Zn-Finger | ARID | 3.9 |
| Cre16.g672300.t1.2 | Swi/Snf-Related Chromatin Binding Protein | HMG | 2.8 |
| Cre01.g015050.t1.1 | Unknown | SNF2 | 2.2 |
| Cre07.g322450.t1.1 | Pwwp Domain (Pwwp)//Set Domain (Set) | PHD | 1.7 |
| Cre08.g380151.t1.1 | Phd-Finger (Phd)//Wstf, Hb1, Itc1P, Mbd9 Motif | PHD | 1.7 |
| Cre07.g334200.t1.2 | Atp-Dependent Rna Helicase Ddx41-Related | SNF2 | 1.6 |
| Cre02.g078700.t1.1 | Lysine-Specific Demethylase 4A-Related | JUMONJI | 1.5 |
| Cre06.g261450.t1.2 | Swi/Snf-Related Chromatin Binding Protein | HMG | – b |
| Cre17.g709550.t1.2 | Lysine-36 Demethylase/Jmjc Domain-Containing Histone Demethylase 1A | JUMONJI | 1.8 |
| Cre01.g029450.t1.1 | Non-Histone Protein 10 | HMG | 1.5 |
| Cre08.g367300.t1.1 | Bromodomain Extra-Terminal - Bet | DDT | – b |
| Cre08.g358532.t1.1 | Gata Zinc Finger (Gata) // Bah Domain (Bah) | PHD | – b |
Only proteins with fold change ≥1.5 are shown in the table. Proteins more abundant in low CO2 conditions are indicated in bold.
a The transcription factor (TF) family has been determined from the Plant transcription factor database.
b Proteins have been detected only in one of the two conditions—low/high CO2.
What also remains to be determined are the cis-DNA elements that respond to these TFs and TRs. Chlamydomonas CAH1 is the only CCM gene for which regulatory elements have been systematically investigated and identified. The 5′ upstream 543 bp region of the CAH1 gene was shown to contain a silencer region and an enhancer region with enhancer elements EE-1 (AGATTTTCACCGGTTGGAAGGAGGT) and EE-2 (CGACTTACGAA) (Kucho et al., 1999, 2003). The upstream regions of duplicated genes, CAH4 and CAH5, which confer CO2 dependence in the presence of light was narrowed to 194 bp. No similarities were seen between the upstream regions of CAH4/5 and CAH1, and no shorter segments in this 194 bp have been identified as the elements responsible for the [CO2]-dependent transcription (Villand et al., 1997). Potential genome-wide cis-DNA regulatory elements in Chlamydomonas that had been shifted from high to low [CO2] were identified by FAIRE-seq (Winck et al., 2013). The potential regulatory regions in these genes require further validation.
While the above studies help identify specific transcription regulatory elements, Chlamydomonas also carries a post-transcriptional regulatory machinery of an extensive system of small RNAs (sRNAs), with three Argonaute and three Dicer-like proteins (Molnár et al., 2007; Zhao et al., 2007; Valli et al., 2016; Chung et al., 2019). This gene expression modulatory system with 6164 loci predicted to give rise to sRNAs (Muller et al., 2020) requires identification of its potential targets. Whether this extensive system of sRNAs regulates the CCM requires investigation.
Mechanisms of regulation of CCM induction: [CO2] is not the only cue
The CCM genes that appear to be responsive to [CO2] changes may be responding to photosynthetic activity or carbohydrate metabolism as an indicator of Ci, and not directly to [CO2] changes. A process allied to photosynthetic carbon reduction that is of particular interest is photorespiration (PR). PR in photosynthetic organisms removes the toxic metabolite 2-phosphoglycolate (2-PG) arising from Rubisco’s oxygenase activity (Fig. 1; Table 2). A series of PR reactions occurring in chloroplasts and mitochondria regenerates the Calvin–Benson–Bassham (CBB) cycle intermediate 3-phosphoglycerate (3-PGA) from 2-PG, with the associated loss of 25% of fixed carbon as CO2. It must be noted that the Chlamydomonas PR differs from that in higher plants in two aspects: (i) glycolate is converted to glyoxylate in the mitochondria rather than the peroxisomes; and (ii) glyoxylate formation is catalysed by algal glycolate dehydrogenase (GDH), as opposed to glycolate oxidase. PR also acts as a sink for energy and reducing power from photosynthetic electron transfer (PET) when CBB cycle activity is limiting (Kozaki and Takeba, 1996; Moroney et al., 2013). PR helps prevent blockades in reduced PET chains, which otherwise would result in formation of reactive oxygen species (ROS), with deleterious effects. ROS resulting from over-reduction of PET include 1O2 (singlet oxygen), H2O2 (hydrogen peroxide), O−2 (superoxide), and ·OH (hydroxyl radical). While 1O2 is generated predominantly at the reaction centre of PSII and is the main ROS responsible for photo-oxidative damage, the other three are formed at the acceptor side of PSI (Erickson et al., 2015). 1O2 is formed primarily by energy transfer from the triplet state of photosensitizers such as chlorophyll, tetrapyrroles, and flavins. Chlamydomonas exhibits acclimation to 1O2 (Ledford et al., 2007), and this acclimation response is an indicator of the 1O2 signalling mechanism (Erickson et al., 2015). PR is thus intricately connected with photosynthesis and the CCM, and could help resolve imbalances in PET and the CBB cycle (Fig. 2) (Caspari et al., 2017), by altering ROS levels. This interconnectedness suggests that metabolites resulting from PR and the CBB cycle, or ROS from light absorption by saturated PET chains, could be signals leading to CCM induction.
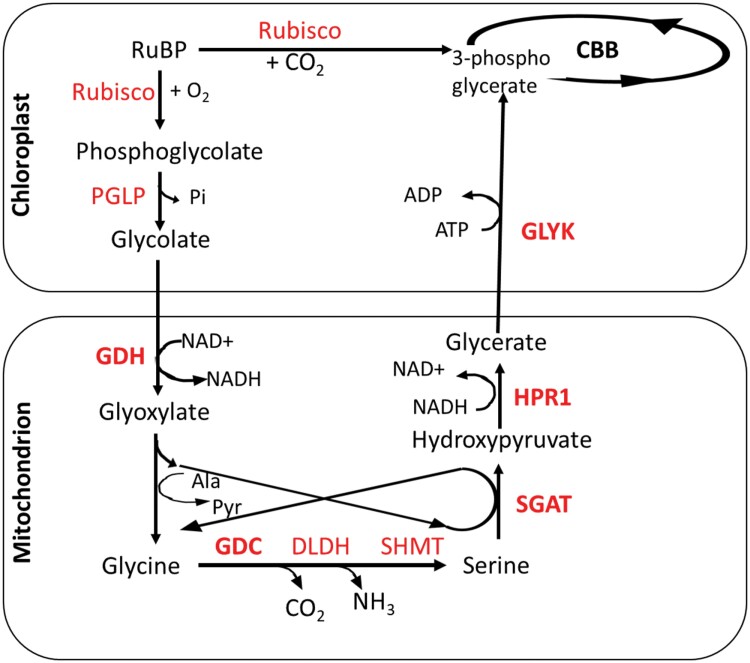
Fig. 1.
Photorespiratory cycle in Chlamydomonas. The enzymes Rubisco, PGLP (phosphoglycolate phosphatase), GDH (glycolate dehydrogenase), GGT (glutamate glyoxalate aminotransferase), GDC (glycine decarboxylase complex), SHMT (serine hydroxymethyl transferase), SGAT (serine/alanine glyoxalate aminotransferase), HPR1 (hydroxypyruvate reductase), and GLYK (glycerate kinase) are in red. Other abbreviations used: 2-OG, 2-oxoglutarate; Pyr, pyruvate. The enzymes highlighted in bold have expression dependent on both [CO2] and CIA5, similar to several CCM genes (Fang et al., 2012).
Table 2.
List of photorespiration genes in Chlamydomonas
| Gene ID | Short name | Brief description |
|---|---|---|
| Cre03.g168700 | PGLP1 | Phosphoglycolate phosphatase/4-nitrophenylphosphatase |
| Cre10.g438100 | PGLP2 | Phosphoglycolate phosphatase/4-nitrophenylphosphatase |
| Cre03.g162601 | PGLP3 | CDP-diacylglycerol-glycerol-3-phosphate 3-phosphatidyltransferase |
| Cre06.g288700 | GDH | Glycolate dehydrogenase |
| Cre10.g451950 | AAT | Alanine aminotransferase |
| Cre06.g294650 | AGT1 | Alanine-glyoxylate transaminase |
| Cre03.g182800 | AGT2 | Alanine-glyoxylate transaminase |
| Cre12.g534800 | GDC-P | Glycine cleavage system, P protein |
| Cre06.g253350 | GDC-H | Glycine cleavage system, H-protein |
| Cre03.g193750 | GDC-T | Glycine cleavage system, T protein |
| Cre18.g749847 | DLDH | Dihydrolipoyl dehydrogenase |
| Cre16.g664550 | SHMT1 | Serine hydroxymethyltransferase |
| Cre06.g293950 | SHMT2 | Serine hydroxymethyltransferase 2 |
| Cre09.g411900 | SHMT3 | Serine hydroxymethyltransferase 3 |
| Cre01.g005150 | SGAT | Serine glyoxylate aminotransferase |
| Cre06.g295450 | HPR1 | Hydroxypyruvate reductase |
| Cre12.g542300 | GLYK | Glycerate kinase |
The genes highlighted in bold were identified to be regulated by both CIA5 and CO2, and were classified as having expression patterns similar to CCM clusters (Fang et al., 2012)
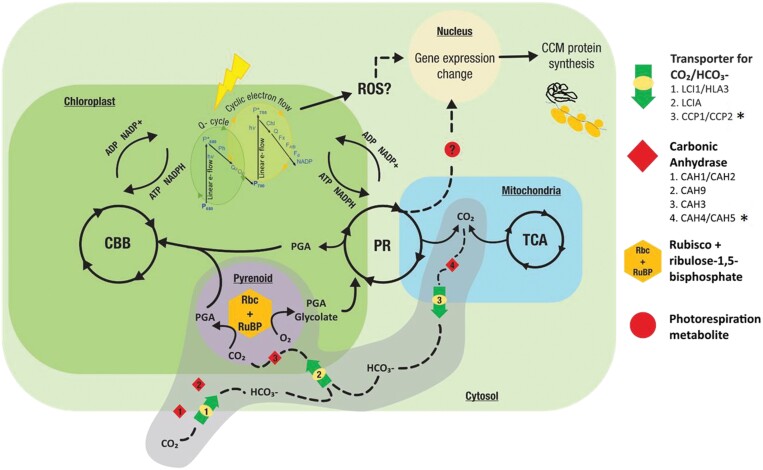
Fig. 2.
Schematic representation of crosstalk between photosynthetic electron transport (PET), the Calvin–Benson–Basham (CBB) cycle, photorespiration (PR), and the carbon concentration mechanism (CCM) in Chlamydomonas. CCM components: inorganic carbon transporters and carbonic anhydrases, occurring in various parts of the cell are highlighted in grey. *The role of mitochondrial proteins CCP1, CCP2, CAH4, and CAH5 is hypothesized, and remains to be explored. Reactive oxygen species (ROS) generated during PET, and PR metabolites are hypothesized to act as signalling molecules for the CCM.
The impact of PET rates was demonstrated by down-regulation of CCM genes CAH1, HLA3, HLA1, HLA2, and HLA4 by 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU)—an inhibitor of PSII (Im and Grossman, 2002). DCMU also affects the intrachloroplastic localization of a retrograde signalling CCM protein called CAS, which in turn affects expression of 13 CCM genes including HLA3 and LCI1 (as discussed later). The use of DCMU leads to 1O2 accumulation (Fufezan et al., 2002). Whether it is the downstream effects of 1O2 or the reduced photosynthesis that impacts gene expression requires further systematic investigations using modulators of photosynthesis and intracellular ROS. This study also showed that the above CCM genes require high-intensity light, in addition to low [CO2], to up-regulate their expression, again hinting towards PET or allied CBB cycle activity as potential influencers of CCM induction.
Like the CCM, PR activity is observed due to low [CO2]/[O2] in actively photosynthesizing organisms. Organisms with mutations of both PR and CCM genes have reduced ability to survive, unless in a high [CO2] environment (Moroney et al., 2013). This led researchers to suggest a metabolite of PR as the signalling molecule for CCM induction in Chlamydomonas over three decades ago (Spalding et al., 1985). A Chlamydomonas PR mutant lacking the phosphoglycolate phosphatase1 (PGP1) that converts 2-PG to glycolate, had affinity for inorganic carbon comparable with the wild type (WT) with an induced CCM (Suzuki et al., 1990), leading the authors to suggest 2-PG as a CCM signalling molecule.
Further evidence for CCM–PR crosstalk comes from differential processing of glycolate in Chlamydomonas with or without a CCM (Moroney et al., 1986). While glycolate is excreted under high CO2 conditions (CCM uninduced and PR down-regulated), glycolate excretion is minimal (1/80th of that in high CO2 conditions) in low CO2, CCM-induced conditions. Experiments with labelled CO2, labelled glycolate, and inhibitors of PR showed that this is not owing to lowered Rubisco oxygenase activity, but rather due to high rates of processing of glycolate by PR enzymes. This crosstalk between the algal CCM and PR (Fig. 1) is further strengthened by co-regulation of several CCM and PR genes by similar environmental stimuli of CO2 and the master regulator CIA5 (Fang et al., 2012). The PR genes are shown in Table 2, with those highlighted being co-expressed with CCM genes. Co-regulation of expression of PR and CCM genes is discussed further later in this review where we propose a hypothesis for evolution of CCM regulatory mechanisms. In addition to the transfer of metabolites between chloroplasts and mitochondria in Chlamydomonas during PR, light and CO2 induce changes in mitochondrial arrangement within the cells (Geraghty and Spalding, 1996; Polukhina et al., 2016). The mitochondria lying between the chloroplast and cell membrane in CCM-active cells (Geraghty and Spalding, 1996) probably capture the glycolate exiting the chloroplast, and the implications for mitochondrial CAs and inorganic carbon transporters for the CCM are discussed in the last section. Whether the PR metabolites moving between organelles with light- and CO2-dependent intracellular location impact expression of nuclear CCM genes needs further investigation.
There have only been a few studies to explore the flux between PR, the CCM, and photosynthesis. Caspari et al. (2017) studied a pyrenoid-less CCM-defective mutant expressing a higher plant version of the Rubisco small subunit. This mutant exhibited a lower PET rate without affecting the intrapyrenoid thylakoid morphologies to compensate for the limited CO2 supply arising from lack of the CCM. The rate of PET was restored to levels comparable with those of WT cells when exposed to high CO2 and the mutant also had increased non-photochemical quenching (NPQ) to dissipate the energy reaching the photosystems.
A large-scale experiment studying metabolite flux and gene expression in photoautotrophic Chlamydomonas cells grown under low CO2 (0.04%) and high CO2 (10%) showed that the mitochondrial processes of glycolysis, gluconeogenesis, the glyoxylate pathway, dicarboxylate, and metabolism rates of amino acids in PR were responsive to [CO2] (Winck et al., 2016). A more systematic analysis of PET, PR, and CBB cycle reactions and inorganic carbon uptake under different conditions of [CO2] is needed. Engineering of the biophysical (algal/cyanobacterial) CCM into higher plants might require tinkering with PR processes, in addition to introducing various CCM components (Atkinson et al., 2016, 2020), for optimizing fluxes between light and dark reactions of photosynthesis.
While the CCM is active in the presence of light and the transcription of several of the CCM genes is light and [CO2] dependent (Brueggeman et al., 2012; Fang et al., 2012; Tirumani et al., 2014), there have been observations of transcription of CAH1, CAH3, CAH6, and LCIB in low [CO2] conditions even in the dark (Rawat and Moroney, 1995; Mitchell et al., 2014; Tirumani et al., 2014, 2019). However, the increase of protein levels of these dark–low [CO2] transcribed genes, and localization of CAH3 to the pyrenoid, does not happen until the cells are exposed to light (Mitchell et al., 2014; Tirumani et al., 2014). These observations point towards regulation of expression not just during transcription, but also during translation and transport.
In conclusion, much ground remains to be covered for identification of key regulatory elements of the inducible CCM transcription machinery. Efforts are needed to utilize the extensive temporal transcription data from Chlamydomonas (Zones et al., 2015; Strenkert et al., 2019) to identify potential CCM TFs and TRs by developing gene regulatory networks (Emmert-Streib et al., 2014). Experiments are needed to understand the CIA5 mechanism, and validate and characterize potential CCM TFs and TRs identified computationally or via organism-wide experiments discussed in this section. Understanding of CCM regulation requires a general comparison with other physiological mechanisms associated with sensing light and signalling photosynthetic, photorespiratory, and photoinhibitory responses. This interconnectedness of PR and CCM led us to consider their evolutionary origins in the section ‘Insights for evolution of CCM regulation’.
Chaperones and the import and assembly of chloroplastic CCM proteins
Fewer than 100 genes encoding proteins are found in the chloroplast, with the remaining plastidic proteins, including those involved in the CCM, encoded in the nucleus (Martin et al., 2002). The spatial segregation between nuclear gene expression and localization of the photosynthetic apparatus in the chloroplast means that many of the photosynthetic and CCM components need to be translated on cytoplasmic ribosomes and transported to/across the chloroplast membranes. This implies the need for chaperones for transport and folding of CCM proteins. In this section, we evaluate experimental evidence for chaperones and cellular transport machinery having roles in the assembly and functioning of the CCM.
Folding and transport of CCM proteins
Based on existing data (Brueggeman et al., 2012; Fang et al., 2012; Wang et al., 2015; Mackinder et al., 2016) and proteomics studies, Mackinder et al. (2017) collated 624 nuclear genes involved in the CCM. Analysis of the encoded protein sequences suggests that a significant proportion are significantly disordered (140 sequences >70% disorder, 201 sequences >50% disorder, and 291 sequences >30% disorder). These included 76 chloroplast-localized CCM proteins of which 29, 21, and 16 sequences have disorderliness >30, 50, and 70%, respectively (Mackinder et al., 2017). The occurrence of disorderliness is often an indicator of the presence of scaffolding modules for interaction with other proteins. Such disordered regions also destabilize proteins, and very often chaperones are required to prevent their irreversible misfolding (Pechmann and Frydman, 2014). Analysis of the 624 sequences also showed that 142 nuclear and 21 chloroplastic sequences carry at least one transmembrane (TM) domain (Krogh et al., 2001). The insertion of TM regions into membranes requires the assistance of chaperones within the cell (Jarvis and Kessler, 2014; Guna and Hegde, 2018). The disorderliness and the presence of TM domains suggest a requirement for assisted folding in several of the CCM proteins that are translated on cytosolic ribosomes, as the proteins need to be transported across the chloroplast membranes (envelope and/or thylakoid) or embedded within them. Chloroplast TM transport is achieved through channels formed by TOC and TIC (translocon on the outer/inner chloroplast membrane) complexes. This transport is a complex process involving recognition of a transit peptide, protein unfolding and threading through TOC–TIC complexes, cleavage of the transit peptide in the stroma, and finally refolding of the protein. The chaperones Hsp70 (cytosolic and stromal), cytosolic Hsp90, stromal Hsp93, and Cpn60 are involved in chloroplast import of proteins (Flores-Pérez and Jarvis, 2013).
Chaperones involved in the CCM
The roles of chaperones in organellar transport and folding mean that they could be vital for establishing the CCM, although little work has been carried out in this area. Here, using previously published protein interaction data, we have identified the CCM proteins that interact with chaperones HSP90, HSP70, and sHSPs (22E and 22F) (Table 3) (Mackinder et al., 2017; Rütgers et al., 2017). The list indicates that several key CCM proteins including EPYC1, Rubisco small subunits, Cas, and transporters might need chaperone assistance.
Table 3.
CCM proteins that are found to interact with chaperones from proteomics studies (Mackinder et al., 2017; Rutgers et al., 2017)
| Protein ID | Description | Protein ID | Description |
|---|---|---|---|
| Interactors of HSP22C | Interactors of HSP70A (continued) | ||
| Cre06.g295450 | HRP1, hydroxypyruvate reductase | Cre03.g162800 | LCI, low-CO2-inducible membrane protein |
| Cre05.g248450 | CAH5, mitochondrial carbonic anhydrase | Cre16.g652800 | Unannotated |
| Interactors of HSP22E | Cre04.g229300 | RCA1, Rubisco activase | |
| Cre10.g444700 | SBE3, starch-binding enzyme | Cre12.g509050 | PSBP3,OEE2-like protein of thylakoid lumen |
| Cre03.g151650 | Unannotated | Cre13.g577100 | ACP2, acyl-carrier protein |
| Cre16.g651050 | CYC6, cytochrome c6 | Cre16.g651050 | CYC6, cytochrome c6 |
| Interactors of HSP22F | Cre09.g394473 | LCI9, low-CO2-inducible protein | |
| Cre17.g724300 | PSAK, PSI reaction centre subunit | Cre12.g560950 | PSAG, PSI reaction centre subunit V |
| Cre16.g651050 | CYC6, cytochrome c6 | Cre10.g436550 | EPYC1/LCI5, low-CO2-inducible protein |
| Cre12.g509050 | PSBP3, OEE2-like protein of thylakoid lumen | Cre02.g120150 | Unannotated |
| Cre10.g444700 | SBE3, starch-binding enzyme | Cre02.g120100 | RBCS1, Rubisco small subunit 1 |
| Cre03.g179800 | LCI24, low-CO2-inducible membrane protein | Cre07.g330250 | PSAH, subunit H of PSI |
| Cre16.g663450 | LCI11, low-CO2-inducible membrane protein | Cre14.g626700 | PETF, apoferredoxin |
| Interactors of HSP70A | Cre12.g507300 | LCI30, low-CO2-inducible protein | |
| Cre16.g662600 | Unannotated | Cre12.g519300 | TEF9, unannotated |
| Cre10.g444700 | SBE3, starch-binding enzyme | Interactors of HSP70B | |
| Cre06.g307500 | LCIC, low-CO2 inducible protein | Cre16.g662600 | Unannotated |
| Cre01.g054850 | Unannotated | Cre16.g663450 | LCI11, low-CO2-inducible membrane protein |
| Cre16.g663450 | LCI11, low-CO2-inducible membrane protein | Cre06.g283750 | HST1, homogentisate solanesyltransferase |
| Cre08.g372450 | PSBQ, oxygen-evolving enhancer protein 3 | Cre10.g444700 | SBE3, starch-binding enzyme |
| Cre02.g097800 | HLA3, ABC transporter | Cre03.g151650 | Unannotated |
| Cre17.g724300 | PSAK, PSI reaction centre subunit | Cre16.g652800 | Unannotated |
| Cre09.g415700 | CAH3, carbonic anhydrase 3 | Cre17.g724300 | PSAK, PSI reaction centre subunit |
| Cre04.g223300 | CCP1, low-CO2-inducible chloroplast envelope protein | Cre09.g394473 | LCI9, low-CO2-inducible protein |
| Cre10.g452800 | LCIB, low-CO2-inducible protein | Cre06.g307500 | LCIC, low-CO2-inducible protein |
| Cre05.g248450 | CAH5, mitochondrial carbonic anhydrase | Cre06.g309000 | NAR1.2, anion transporter |
| Cre06.g309000 | NAR1.2, anion transporter | Cre12.g519300 | TEF9, unannotated |
| Cre06.g295450 | HRP1, putative hydroxypyruvate reductase | Cre03.g191250 | LCI34, low-CO2-inducible protein |
| Cre08.g362900 | PSBP4, lumenal PsbP-like protein | Cre01.g051500 | ULP1, uncharacterized lumenal polypeptide |
| Cre03.g151650 | Unannotated | Cre09.g415700 | CAH3, carbonic anhydrase 3 |
| Cre12.g485050 | CAH6, carbonic anhydrase 6 | Cre02.g097800 | HLA3, ABC transporter |
| Cre01.g051500 | ULP1, uncharacterized lumenal polypeptide | Cre10.g452800 | LCIB, low-CO2-inducible protein |
| Cre03.g179800 | LCI24, low-CO2-inducible membrane protein | Cre04.g229300 | RCA1, Rubisco activase |
| Cre06.g283750 | HST1, homogentisate solanesyltransferase | Cre12.g509050 | PSBP3, OEE2-like protein of thylakoid lumen |
| Cre04.g223050 | CAH2, carbonic anhydrase, alpha type, periplasmic | Cre02.g120100 | RBCS1, Rubisco small subunit 1 |
| Cre03.g191250 | LCI34, low-CO2-inducible protein | Cre12.g560950 | PSAG, PSI reaction centre subunit V |
| Interactors of HSP70B (continued) | Interactors of HSP90A | ||
| Cre01.g054850 | Unannotated | Cre02.g097800 | HLA3, ABC transporter |
| Cre02.g120150 | Unannotated | Cre04.g229300 | RCA1, Rubisco activase |
| Cre03.g179800 | LCI24, low-CO2-inducible membrane protein | Cre04.g223300 | CCP1, low-CO2-inducible mitochondrial envelope protein |
| Cre04.g223300 | CCP1, low-CO2-inducible chloroplast envelope protein | Cre07.g330250 | PSAH, subunit H of PSI |
| Cre16.g651050 | CYC6, cytochrome c6 | Cre17.g724300 | PSAK, PSI reaction centre subunit |
| Cre08.g372450 | PSBQ, oxygen-evolving enhancer protein 3 | Cre03.g151650 | Unannotated |
| Cre10.g436550 | EPYC1/LCI5, low-CO2-inducible protein | Cre12.g485050 | CAH6, carbonic anhydrase 6 |
| Cre08.g362900 | PSBP4, lumenal PsbP-like protein | Cre16.g651050 | CYC6, cytochrome c6 |
| Cre04.g223050 | CAH2, carbonic anhydrase, alpha type, periplasmic | Cre04.g223050 | CAH2, carbonic anhydrase, alpha type, periplasmic |
| Cre12.g485050 | CAH6, carbonic anhydrase 6 | Cre09.g415700 | CAH3, carbonic anhydrase 3 |
| Cre03.g162800 | LCI1, low-CO2-inducible membrane protein | Cre16.g662600 | Unannotated |
| Cre05.g248450 | CAH5, mitochondrial carbonic anhydrase | Cre01.g054850 | Unannotated |
| Cre07.g330250 | PSAH, subunit H of PSI | Cre16.g663450 | LCI11, low-CO2-inducible membrane protein |
| Cre12.g507300 | LCI30, low-CO2-inducible protein | Cre09.g394473 | LCI9, low-CO2-inducible protein |
| Cre06.g295450 | HRP1, putative hydroxypyruvate reductase | Cre10.g452800 | LCIB, low-CO2-inducible protein |
| Cre14.g626700 | PETF, apoferredoxin | Cre12.g507300 | LCI30, low-CO2-inducible protein |
| Cre13.g577100 | ACP2, acyl-carrier protein | Cre16.g652800 | Unannotated |
| Cre17.g721500 | STA2, granule-bound starch synthase I | Cre08.g362900 | PSBP4, lumenal PsbP-like protein |
| Interactors of HSP70C | Interactors of HSP90B | ||
| Cre05.g248450 | CAH5, mitochondrial carbonic anhydrase | Cre07.g330250 | PSAH, subunit H of PSI |
| Cre16.g662600 | Unannotated | Cre03.g191250 | LCI34, low-CO2-inducible protein |
| Cre10.g436550 | EPYC1/LCI5, low-CO2-inducible protein | Cre12.g509050 | PSBP3, OEE2-like protein of thylakoid lumen |
| Cre09.g394473 | LCI9, low-CO2-inducible protein | Interactors of HSP90C | |
| Cre06.g295450 | HRP1, putative hydroxypyruvate reductase | Cre10.g444700 | SBE3, starch-binding enzyme |
| Cre06.g307500 | LCIC, low-CO2-inducible protein | Cre17.g724300 | PSAK, PSI reaction centre subunit |
| Cre16.g663450 | LCI11, low-CO2-inducible membrane protein | Cre01.g054850 | Unannotated |
| Cre03.g151650 | Unannotated | Cre03.g151650 | Unannotated |
| Cre06.g283750 | HST1, homogentisate solanesyltransferase | Cre04.g229300 | RCA1, Rubisco activase |
| Cre04.g229300 | RCA1, Rubisco activase | Cre16.g663450 | LCI11, low-CO2-inducible membrane protein |
| Cre16.g652800 | Unannotated | Cre09.g415700 | CAH3, carbonic anhydrase 3 |
| Cre10.g444700 | SBE3, starch-binding enzyme | Cre16.g652800 | Unannotated |
| Cre06.g309000 | NAR1.2m anion transporter | ||
| Cre02.g097800 | HLA3, ABC transporter | ||
| Cre17.g724300 | PSAK, PSI reaction centre subunit | ||
| Cre16.g651050 | CYC6, cytochrome c6 | ||
| Cre09.g415700 | CAH3- carbonic anhydrase 3 | ||
| Cre04.g223300 | CCP1, low-CO2-inducible mitochondrial envelope protein |
Despite the expectation that chaperones are essential for the CCM, the only chaperone which has been suggested to be essential for photosynthesis in a large-scale mutant screen is CDJ2, a chloroplastic DnaJ protein (Li et al., 2019). Previously, expression of DNJ12 (DnaJ protein) was shown to be dependent on both CO2 and CIA5 (Fang et al., 2012). Two other DnaJ chaperones, DNJ15 and DNJ31, have CIA5-dependent expression (Fang et al., 2012), and hence feature as CCM proteins in a list compiled recently (Mackinder et al., 2017). The expression of DNJ31 is also dependent on the retrograde signalling mediated by the CCM protein CAS (Wang et al., 2016) (the role of CAS is discussed in ‘Retrograde signalling in CCM regulation’), further hinting at a role in the CCM. DnaJ or Hsp40 proteins are chaperones that work in conjunction with Hsp70 to help fold nascent proteins. Certain DnaJ proteins are known to confer substrate specificity. Whether the algal DnaJ proteins are indeed chaperones and are specifically interacting with CCM proteins needs further characterization. An up-regulation of HSF1 in response to low CO2 conditions in Chlamydomonas has also been shown (Winck et al., 2013). HSF1 is a TF that is known to bind to promoter elements of HSP22F and HSP70A (Strenkert et al., 2011), suggesting that CCM responses include an up-regulation of at least two chaperones. Overexpression of chaperones important for CCM expression might be worth considering in a strategy for engineering the CCM.
The limited evidence for chaperones associated with gene expression and protein import essential for the CCM should not negate their importance. Chaperones which are CCM specific might be few, and there might be several, such as Hsp70 and Hsp90 members, which cater for varied substrates, including CCM proteins. The varied nature of substrates for several chaperones makes it difficult to identify CCM-associated chaperones. The chaperones discussed in this section which have featured in genome-wide studies, and in interactomes, are good candidates to further explore this area.
Dynamics of the starch sheath, pyrenoid matrix, and thylakoid tubules
While the response to extracellular environment changes rests primarily with the nuclear genes, the vital process of carbon capture occurs in the pyrenoids. The discovery of the role of EPYC1 as a linker protein (Mackinder et al., 2016; Wunder et al., 2018), the visualization of Rubisco–EPYC1 dynamics during pyrenoid division (Freeman Rosenzweig et al., 2017), proteomics studies (Mackinder et al., 2016, 2017; Zhan et al., 2018), and the identification of a motif linking key elements (Meyer et al., 2020) have given the CCM community much needed information about pyrenoidal composition and dynamics. In this section, we focus on specific aspects of the pyrenoid matrix, the extra-pyrenoidal starch sheath, the intra-pyrenoidal tubule network, as well as the linkages and interactions that promote their assembly.
Role of the extra-pyrenoidal starch sheath
An extra-pyrenoidal starch sheath in Chlamydomonas was first clearly defined in 1957 (Sager and Palade, 1957). While the formation of a starch sheath under low [CO2] suggested that starch prevents CO2 leakage (Ramazanov et al., 1994), there have been contradictory findings showing CCM induction in starchless algal mutants with a >10-fold increase in affinity for inorganic carbon (Plumed et al., 1996; Villarejo et al., 1996). However, recent studies in Chlamydomonas are indicative of a more fundamental role for the starch sheath in the CCM.
An important CCM protein associated with the starch sheath is LCIB that has low [CO2]- and light-dependent localization around the pyrenoid in a complex with LCIC. The localization of the LCIB–LCIC complex close to thylakoid tubule emergence and starch plate convergence (Yamano et al., 2010, 2014), together with a preferential role in uptake of CO2 over HCO3– during pH-dependent photosynthetic activity measurements (Wang and Spalding, 2014), are consistent with a role for the complex in capturing CO2 retro-diffusing from the pyrenoid. Whether the LCIB–LCIC complex acts as a physical barrier or functions as an inducible CA needs examination. Structures of both LCIB and LCIB–LCIC have attributes of CAs, but no detectable CA activity (Jin et al., 2016). LCIB localization in extra-pyrenoidal starch affects pyrenoid size and number (Yamano et al., 2014), and inorganic carbon affinity (Toyokawa et al., 2020). LCID and LCIE are two more members of the same predicted CA family as LCIB occurring in Chlamydomonas, needing characterization (Wang and Spalding, 2014; Jin et al., 2016).
The importance of starch in affecting pyrenoid number and orientation around the tubule network was further supported by studies with Chlamydomonas saga1 mutants with lowered photosynthetic efficiency, and containing many pyrenoid-like structures, in contrast to the single pyrenoid in the WT (Itakura et al., 2019). SAGA1 is believed to link starch and Rubisco. Pyrenoids harbour a thylakoid tubule network that acts as a conduit between thylakoid lumen and pyrenoid. In saga1 mutants, while the number of pyrenoid-like structures is increased, there was only one tubule network which was often displaced to the periphery of a pyrenoid. Both formation of an entire starch sheath enclosing a single pyrenoid in the canonical position and a central thylakoid tubule network appear important for maintaining photosynthetic efficiency of the saga1 mutants. Recent findings show that SAGA2, 30% similar in sequence to SAGA1 with a starch-binding domain, localizes to the interface of the pyrenoid and starch sheath (Meyer et al., 2020). Both SAGA1 and SAGA2 also contain a Rubisco-binding motif (RbM), the role of which is discussed later in this section.
Thylakoid membrane proteins involved in the CCM
The function of the thylakoid tubule network is also thought to be critical for CCM induction. An important protein in the thylakoid lumen is the carbonic anhydrase CAH3, that becomes phosphorylated and localizes to the tubule network. Though interactions of CAH3 with thylakoid-associated kinases (Depège et al., 2003; Lemeille et al., 2010; Mackinder et al., 2017) have been observed, the exact kinase responsible for its phosphorylation remains unknown. CAH3 is responsible for converting HCO3– to CO2 for release into the heart of the pyrenoid (Blanco-Rivero et al., 2012). Such a mechanism will require a HCO3– transporter to be located in the thylakoid membrane, and Chlamydomonas cells lacking CAH3 grow more slowly compared with WT cells, particularly in low [CO2], highlighting its role in the upkeep of the CCM (Sinetova et al., 2012). Recently, three genes coding for bestrophin-like proteins in Chlamydomonas called BST1–BST3 were identified (Mukherjee et al., 2019). These genes are under the regulation of the CCM master TF CIA5 (Fang et al., 2012), and interact with CCM components such as LCIB (Mackinder et al., 2017). BST1–BST3 localize to the thylakoid membrane and are thought to transport HCO3– ions to CAH3, with down-regulation of these proteins hampering cell growth at low [CO2] (Mukherjee et al., 2019).
Linkages involved in the formation of the pyrenoid matrix and starch sheath
Previous studies have shown that EPYC1, an intrinsically disordered repeat linker protein, is necessary to form a Rubisco–EPYC1 pyrenoidal matrix (Mackinder et al., 2016). Recently, five Rubisco-binding regions in EPYC1 were identified using structural data for complexes of Rubisco and peptides representing different regions of EPYC1 (He et al., 2020). EPYC1 acts as a ‘molecular glue’ that tethers multiple Rubisco molecules together to give rise to the pyrenoid matrix.
The presence of an RbM similar to that found in EPYC1 was identified in other pyrenoid-localized Rubisco-interacting proteins (Meyer et al., 2020). Identified first in EPYC1, an RbM [D/N]W[R/K]XX[L/I/V/A] has been found in a putative chloroplast epimerase CSP41A, SAGA1, SAGA2, and in the thylakoid-localizing proteins RBMP1 and RBMP2. Disruption of the motif caused these pyrenoid-targeted proteins to diffuse homogeneously across the chloroplast, while introduction of the motif into a non-pyrenoidal protein led to their accumulation in the pyrenoid matrix (Meyer et al., 2020). It must be noted that the presence of this motif does not always lead to pyrenoidal localization. As discussed previously, the presence of the motif in EPYC1 allows for its interaction with Rubisco to give rise to the EPYC1–Rubisco condensate forming the pyrenoid matrix (Wunder et al., 2018; Atkinson et al., 2020). Similarly, the presence of the RbM along with the starch-binding regions in SAGA2 might help the starch sheath envelop the pyrenoid. Two newly identified thylakoid tubule network-localizing proteins RBMP1(Cre06.g261750) and RBMP2 (Cre09.g416850) might help tether the pyrenoid to the tubules (Fig. 3). Among them, RBMP1 is predicted to be a member of the bestrophin family and may play a role in transporting HCO3– to the pyrenoid directly through the tubule network, in contrast to the other bestrophin-like proteins which are found outside the pyrenoid. How these tubule network proteins are localized to the tubule network, which is formed even in cases where there is no pyrenoid matrix formation (Caspari et al., 2017), needs investigation.
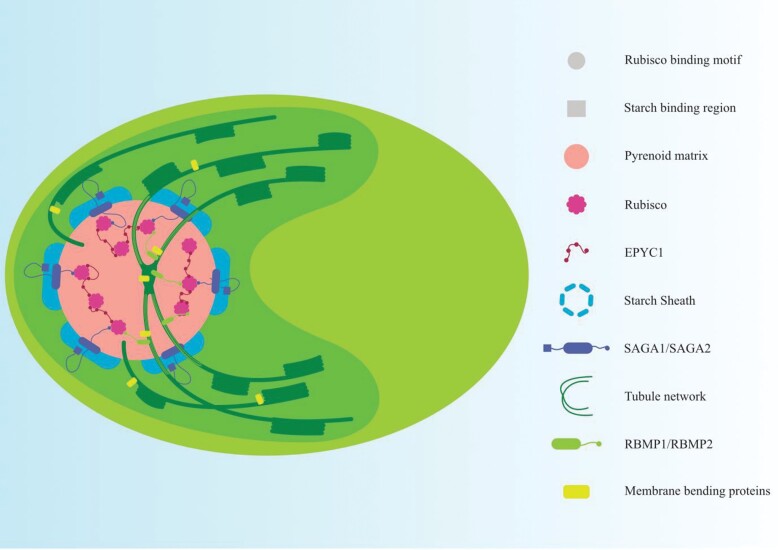
Fig. 3.
Assembly of the pyrenoid. The Rubisco-binding motif (RbM) mediates the formation of three regions of the pyrenoid. The RbM-bearing protein EPYC1 binds to multiple Rubisco holoenzymes and creates a Rubisco–EPYC1 condensate that forms the pyrenoid matrix. The interaction of RbM-bearing thylakoid-anchored proteins RBMP1 and RBMP2 with Rubisco tethers the pyrenoid matrix to the tubule network. The starch sheath is moulded around the pyrenoid matrix through the action of SAGA1 and SAGA2, which bind to Rubisco through their RbM domain and bind to the starch sheath through their starch-binding domain.
Membrane bending and plasticity of the tubule network for CCM maintenance
A key area requiring study is the mechanism regulating the formation of the thylakoid tubule network for effective CCM operation. Insights for these processes may arise from studies of cell division in Chlamydomonas, when each of the four daughter cells normally contains a nascent pyrenoid as identified from fluorophore-tagged components (Freeman Rosenzweig et al., 2017). Although a small proportion of cells synthesize a pyrenoid de novo, the extent to which the thylakoid tubule network is partitioned is not known. The intricate tubule network is formed as thylakoid membranes coalesce near entry points into the pyrenoid, but an added complexity arises from the internal mini-tubules which provide connectivity and allow CBB cycle intermediates to exchange between the pyrenoid matrix and the chloroplast stroma (Engel et al., 2015). How this complex array of thylakoid membrane remodelling is regulated needs examination.
Membrane-remodelling proteins have been discovered in cyanobacteria and chloroplasts of algae and plants. However, there is limited understanding of their interactions during thylakoid tubule network formation and their potential to mediate CCM development. A set of proteins called CURT1 (Curvature Thylakoid 1) have emerged as important modulators of thylakoid membrane bending and plasticity. First identified in Arabidopsis thaliana (CURT1A, B, C, and D), these proteins are conserved across photosynthetic organisms, including three homologues in Chlamydomonas (Armbruster et al., 2013). Arabidopsis CURT1 proteins concentrate around granal margins and oligomerize to induce membrane tubulation, with their inhibition negatively affecting photosynthetic efficiency (Armbruster et al., 2013; Pribil et al., 2018). The role of CURT in Chlamydomonas is pending investigation, but it probably contributes to similar membrane dynamics, and potentially plays a role in coordinating the assembly of CCM components, and the possible exclusion of PSII from thylakoid tubules within the pyrenoid matrix (McKay and Gibbs, 1991).
Another protein implicated in membrane remodelling is VIPP1, a member of the ESCRT-III family found in eukaryotes (Liu et al., 2020, Preprint). Disrupting VIPP1 in vascular plants altered thylakoid structure and decreased the volume of thylakoid membranes, suggesting roles in thylakoid membrane upkeep, biogenesis, and remodelling (Kroll et al., 2001; Westphal et al., 2001; Hennig et al., 2015, 2017; Heidrich et al., 2017; Gutu et al., 2018). VIPP1 localizes to the Chlamydomonas pyrenoid (Zhan et al., 2018). The vipp1 mutant is sensitive to high-light and heat stress, and shows altered thylakoid membrane structures close to the pyrenoid, suggesting a role in tubule biogenesis (Nordhues et al., 2012). Chlamydomonas also harbours a paralogue of VIPP1, called VIPP2, that is not found in many land plants. VIPP1 and VIPP2 are both up-regulated under high-light- or H2O2-induced stress. Both oligomerize to form rod-like structures (Theis et al., 2020). VIPP2 is expressed only in high-light conditions, in contrast to the constitutive expression of VIPP1, and forms a complex with VIPP1 and HSP22E/F. The lack of up-regulation of HSP22E/F in a vipp2 mutant led the authors to suggest a role for VIPP2 in conveying chloroplastic stress to the nucleus. This recurring pattern of thylakoid membrane-remodelling proteins being up-regulated during high-light stress in Chlamydomonas is consistent with CCM-inductive stimuli (Im and Grossman, 2002). Notably, HSP22E/F interact with various starch synthesis proteins (Table 1), with the role of remobilization and starch plate formation being important for pyrenoid assembly and CCM induction (Ramazanov et al., 1994; Itakura et al., 2019). Another candidate involved in shaping thylakoid morphology is Fzl, which is a dynamin-like protein involved in grana organization in Arabidopsis (Gao et al., 2006). A GTP-binding, oligomerizing homologue of this protein in Chlamydomonas called crFZL was recently found to be important for coping with high-light stress (Findinier et al., 2019).
While the recent identifications of EPYC1 as a linker protein and of an RbM (Meyer et al., 2020) are major developments, many intriguing questions related to thylakoid organization during CCM induction, whether during transfer from high to low CO2, or in synchronized cells during cell division and development, remain unanswered. Structural and mechanistic insights about the thylakoid membrane organizational proteins discussed in this section, in conjunction with life cycle and environmental regulation of the pyrenoid starch and tubule network, are needed to further our understanding of the CCM.
Retrograde signalling in CCM regulation
In previous sections, we have navigated from the nucleus to the pyrenoid examining different algal mechanisms to establish and regulate the CCM. The chloroplast, effectively operating as the hub orchestrating photosynthetic reactions, must be sensitive to environmental factors that affect photosynthesis and/or the CCM. In this section, we see how changes in the chloroplast are communicated to the nucleus via signalling molecules (Rea et al., 2018) for CCM regulation. In Chlamydomonas, molecular transducers such as tetrapyrrole intermediates, ROS, as well as Ca2+ ions help relay signals to the nucleus, with potential changes to the nuclear transcriptome. This section describes how changes within the chloroplast affect nuclear gene expression with implications for photosynthesis and the CCM.
Redox status signalling in photosynthesis
Photosynthesis is a redox-centred metabolic process, subject to fluctuations in environmental conditions (Dietz et al., 2016). Maintaining a functional PET requires balanced excitation of the two photosystems (Rea et al., 2018) without which over-reduced PET components might generate harmful ROS. As discussed earlier, PET rates need to be tuned for the CCM, the CBB cycle, and PR (Caspari et al., 2017). The influence of changes in PET affecting CCM gene expression (Im and Grossman, 2002), discussed above, suggests that the redox status of PET components acts as a regulator of CCM gene expression. It is therefore essential to have inter- and intraorganellar redox status communication (Pfannschmidt et al., 2020) so that both the CCM and photosynthesis rates are optimal. Here, we explore the gun4 mutant, which demonstrates a link between ROS generation and retrograde signalling.
Retrograde signalling by GUN4
The biosynthesis of tetrapyrroles for chlorophyll generation is a process that is sensitive to oxidative stress and needs to be tightly regulated. Tetrapyrroles, as well as many of their biosynthetic intermediates, interact with oxygen in their triplet state to generate ROS, mainly singlet oxygen (Tanaka and Tanaka, 2007; Rea et al., 2018). Tetrapyrrole metabolism occurs in the chloroplast, making use of nuclear-encoded enzymes, necessitating communication between the chloroplast and the nucleus. The role of tetrapyrrole intermediates in ‘retrograde signalling’ to modulate exyhpression of certain nuclear genes has been studied using ‘GUN mutants’ in plants and algae (Larkin, 2016). In WT cells, disrupted chlorophyll biosynthesis is known to activate a specific retrograde signal, which results in down-regulation of photosynthetic nuclear genes, such as Lhcb2. The gun mutant alleles are defective in this particular signal, instead elevating levels of these usually suppressed gene transcripts (Larkin, 2016). One such mutant—gun4—has been identified in Chlamydomonas with half as much chlorophyll as WT cells (Formighieri et al., 2012). While GUN4 is not essential for chlorophyll synthesis, gun4 mutants in Arabidopsis and Chlamydomonas exhibit impaired chlorophyll accumulation, suggesting a role for GUN4 in regulation of the enzyme Mg2+ chelatase (MgCh) (Fig. 4). RNA-seq analysis has shown that the expression of 803 nuclear-encoded genes in Chlamydomonas is altered in the gun4 mutans as compared with the WT (Formighieri et al., 2012).
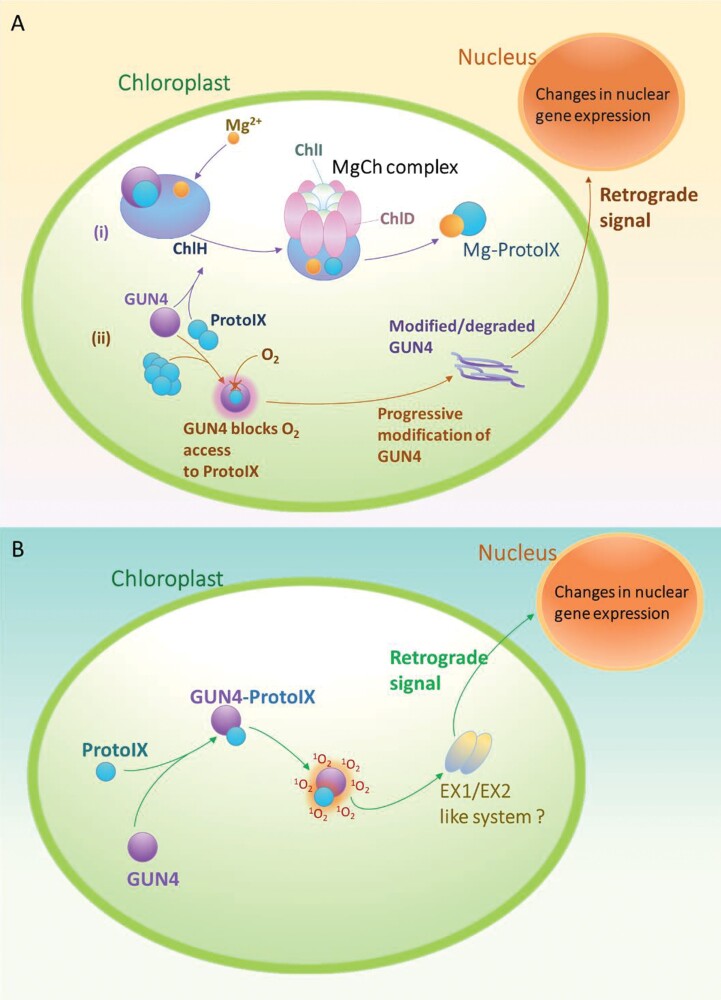
Fig. 4.
(A) GUN4 retrograde signalling model (after Brzezowski et al., 2014). (i) GUN4 is proposed to be an activator of MgCh activity, interacting with the chlorophyll H subunit to promote the catalytic integration of Mg2+ with ProtoIX to form the chlorophyll biosynthesis pathway intermediate Mg-ProtoIX. (ii) The accumulation of excess tetrapyrrole intermediates, such as ProtoIX, in the chloroplast can lead to generation of ROS. GUN4 is proposed to bind ProtoIX, shielding its reaction with ROS. In shielding ProtoIX, GUN4 may be progressively modified or degraded, with degradation products hypothesized to act as the retrograde signals. (B) A contrasting model for GUN4 (after Tahari Tabrizi et al., 2016). Instead of having a ‘shielding’ effect when bound to ProtoIX, the GUN4–ProtoIX complex appeared to escalate 1O2 generation. The elevated 1O2 produced by GUN4–ProtoIX may be sensed by an 1O2-sensing system (like the Arabidopsis EXECUTER1/EXECUTER2 or EX1/EX2 system) yet to be discovered, that relays a signal to the nucleus.
While the role of retrograde signalling in regulating photosynthesis is accepted, we checked if it also plays a part in modulating the CCM. We compared the 803 differentially expressed genes in the gun4 mutant (Formighieri et al., 2012) and those identified as being important for the CCM (Mackinder et al., 2017; Strenkert et al., 2019). Our examination of these datasets shows that expression of nuclear genes encoding 52 CCM genes is affected >8-fold by GUN4 and, by extension, retrograde signalling (Table 4). This list of 52 genes includes a redox protein (peroxiredoxin), a PR pathway protein (serine hydroxymethyl transferase2), three photosystem components (PSAK, PSAH, and PSIIPbs27), bestrophin-3, three CAs (mitochondrial CAH4 and CAH5, and periplasmic CAH8), and the chaperone DNJ15. That PR, mitochondrial CAs, and PET proteins are influenced by GUN4 further strengthens the crosstalk between these processes which has been highlighted in the section- ‘CCM induction and control of gene expression’. GUN4-influenced CCM genes also include DNJ15, implying protein homeostasis regulation by retrograde signalling. Seven of the CCM genes in Table 4 are uncharacterized. These proteins of unknown function which are influenced by GUN4 make a case for further exploration of the connections between the CCM and retrograde signalling.
Table 4.
Nuclear-encoded CCM genesa with >8-fold change in expression in the gun4 mutant with respect to WT Chlamydomonas
| Gene ID | Short name | Short description | Gene ID | Short name | Short description |
|---|---|---|---|---|---|
| Cre01.g014350 | PRX5 | Type II peroxiredoxin | Cre08.g360200 | DUR3 | Urea active transporter |
| Cre01.g015350 | Light-dependent protochlorophyllide reductase | Cre09.g405750 | CAH8 | Carbonic anhydrase | |
| Cre01.g029250 | Amino acid hydroxylase-like protein | Cre10.g426050 | CTPA1 | C-terminal processing peptidase | |
| Cre01.g036950 | Cobalamin-5′-phosphate synthase | Cre10.g455700 | Non-canonical poly(A) polymerase | ||
| Cre01.g053950 | MOX | Monooxygenase | Cre12.g485150 | GAP1 | Glyceraldehyde 3-phosphate dehydrogenase |
| Cre02.g078507 | PSII Pbs27 | PSII Pbs27 | Cre12.g519300 | TEF9 | Predicted protein |
| Cre02.g085500 | Putative transposase DNA-binding domain | Cre12.g535250 | RNA polymerase s factor | ||
| Cre02.g107000 | Cyclin dependent kinase-2 | Cre12.g541550 | Unknown function | ||
| Cre02.g143450 | Unknown function | Cre12.g555700 | DNJ15 | DnaJ-like protein | |
| Cre02.g144800 | LCI8 | Acetylglutamate kinase | Cre13.g569600 | Antibiotic biosynthesis monooxygenase | |
| Cre03.g149050 | Cyt b561 | Cytochrome b561 | Cre14.g625450 | Methyltransferase | |
| Cre03.g158000 | Glu-1-semialdehyde aminotransferase | Cre14.g630350 | Unknown function | ||
| Cre03.g171350 | SEC61A | SEC61-α subunit | Cre16.g652800 | Unknown function | |
| Cre03.g200350 | Methyltransferase | Cre16.g658400 | FDX2 | Ferredoxin | |
| Cre04.g223250 | LCIB-like gene | Cre16.g659800 | Unknown function | ||
| Cre05.g236650 | CYG63 | Guanylate cyclase | Cre16.g663450 | BST-3 | Bestrophin-3 |
| Cre05.g248400 | CAH4 | Mitochondrial carbonic anhydrase | Cre16.g685100 | Cobalamin synthesis protein | |
| Cre05.g248450 | CAH5 | Mitochondrial carbonic anhydrase | Cre17.g700950 | FDX5 | Apoferredoxin |
| Cre06.g258850 | Stage V sporulation protein S | Cre17.g713700 | Tryptophan pyrrolase | ||
| Cre06.g266450 | Protein kinase (MEC-15) | Cre17.g720900 | Unknown function | ||
| Cre06.g284150 | RHP2 | Ammonium transport protein | Cre01.g038400 | Calreticulin 2 | |
| Cre06.g303050 | Nitrate reductase | Cre02.g111450 | Rhodanese-like protein | ||
| Cre06.g310950 | Sarcosine dehydrogenase | Cre03.g198950 | PSBP domain carrying protein | ||
| Cre07.g315050 | Gamma-glutamyl hydrolase | Cre06.g293950 | SHMT2 | Serine hydroxymethyltransferase 2 | |
| Cre07.g330250 | PSAH | PSI subunit H | Cre07.g321400 | FAP113 | Flagellar associated protein |
| Cre07.g337100 | Unknown function | Cre17.g724300 | PSAK | PSI subunit PsaK |
All genes up-regulated in the gun4 mutant are highlighted in bold.
a CCM genes were compiled from Mackinder et al. (2017) and Strenkert et al. (2019).
The mechanistic action of GUN4 in relaying a signal to the nucleus remains undetermined. GUN4 orthologues are present only in species that carry out oxygenic photosynthesis (Formighieri et al., 2012), suggesting a role in photo-oxidative acclimation strategies. High resolution structures of Synechocystis GUN4 in the unliganded (Verdecia et al., 2005) and protoporphyrin IX (ProtoIX) bound form (Chen et al., 2015) have been solved. The binding pocket of GUN4 was shown to be amphiphilic and partially-open (Chen et al., 2015). GUN4 that senses and binds the excess ProtoIX not entering chlorophyll synthesis, could be susceptible to modification by singlet oxygen (1O2). Modified and degraded GUN4 products are hypothesized to initiate the retrograde signalling to nucleus (Brzezowski et al., 2014) (Fig 4a). Tahari Tabrizi et al., suggest that the partially open binding pocket of GUN4 makes bound ProtoIX susceptible to photosensitization releasing 1O2. This 1O2 is hypothesized to relay signals through a system such as EXECUTER1 and EXECUTER2 (Tahari Tabrizi et al., 2016) (Fig 4b), as seen in Arabidopsis (Singh et al., 2015). It is proposed that a similar sensing system is required in Chlamydomonas. However, no corresponding homologue has been found; the most similarity shared with EXECUTER 1 and 2 was 10.2% and 11.2% by the Chlamydomonas protein Cre03.g163500. Mechanistic regulation of CCM gene expression by retrograde signalling in Chlamydomonas, as discussed in above for nuclear regulators, needs to be explored in detail with systematic analysis for signalling molecules, TFs, and cis-elements.
Another CCM protein that has been characterized in the last 5 years is CAS. It is the only CCM protein that has been studied in a systematic manner for its potential role in retrograde signalling independent of GUN4, and is described in the following paragraphs.
Retrograde signalling by CAS
Chlamydomonas Ca2+-binding protein (CAS) is a chloroplastic thylakoid membrane protein that mediates signalling, as part of acclimation to high-light and low-carbon (LC) conditions (Wang et al., 2016). CAS was initially studied in Arabidopsis, where it acts as a Ca2+-binding protein, regulating stomatal closure (Nomura and Shiina, 2014). Although no catalytic activity has been demonstrated for CAS, A. thaliana and Chlamydomonas CAS have Ca2+ binding ability in their N-terminus, and a rhodanese domain of unknown function (Wang, 2017). A comparison of the transcriptomes of a cas mutant strain with the WT and complemented strains demonstrated that absence of CAS leads to a >4-fold decrease in transcript levels of 13 genes (Wang et al., 2016; Wang, 2017) (Table 5). All 13 genes except one coding for a predicted phosphatase have previously been identified as CCM genes. Of particular interest was reduced gene transcription and accumulation of transporters HLA3 and LCIA for the uptake of inorganic carbon into the cell (Yamano et al., 2015) in the cas mutant. Three of the genes (LCID, Cre12.g541550, and Cre26.g756747) in Table 5 have been identified as CCM genes based on previous large-scale expression studies, but their cellular functions are unknown. The expression of CIA5-dependent DNJ31, encoding a DnaJ chaperone discussed above, is also influenced by CAS. These expression features of DNJ31 make it an interesting candidate requiring its functional characterization and identification of its substrates. Two mitochondrial carbonic anhydrases (CAH4 and CAH5, also regulated by GUN4) and two mitochondrial envelope proteins (CCP1 and CCP2) of unknown function (Atkinson et al., 2016; Mackinder et al., 2017) are among those encoded by the 13 genes with CAS-dependent expression. The chloroplastic CCM protein CAS influencing the expression of four mitochondrial CCM proteins represents another feature of the mitochondria–chloroplast crosstalk needed to establish the CCM. Considering the importance of PR–CCM connections as described above, it is probable that CAH4 and CAH5 help recapture photorespired and respired CO2 in mitochondria as bicarbonate ions prior to export by unidentified transporters. CAS also influences PET by affecting expression of genes encoding two proteins involved in NPQ, namely LHCSR2 and LHCSR3. The over-reduction of the PET chain caused by high light intensity could be a potential trigger for CAS activation, and its modulation of NPQ (Caspari et al., 2017). These observations further reiterate the need for tuning the rates of PET, PR, the CBB cycle, and inorganic carbon uptake with CCM induction, as discussed in above.
Table 5.
Nuclear-encoded genes with upregulation >4-fold in WT Chlamydomonas with respect to the cas mutant
| Protein id | Short name | Short description |
|---|---|---|
| Cre02.g097800 | HLA3 | ABC transporter |
| Cre06.g309000 | LCIA | Anion transporter |
| Cre03.g204577 | DNJ31 | DnaJ-like protein |
| Cre05.g248400 | CAH4 | Mitochondrial carbonic anhydrase, beta type |
| Cre05.g248450 | CAH5 | Mitochondrial carbonic anhydrase |
| Cre07.g334750 a | PPP30 | Protein phosphatase 2C |
| Cre04.g223300 | CCP1 | Low-CO2-inducible mitochondrial protein |
| Cre04.g222750 | CCP2 | Low-CO2-inducible mitochondrial protein |
| Cre04.g222800 | LCID | Low-CO2-inducible protein |
| Cre08.g367500 | LHCSR3.1 | Stress-related chlorophyll a/b binding protein 2 |
| Cre08.g367400 | LHCSR3.2 | Stress-related chlorophyll a/b binding protein 3 |
| Cre12.g541550 | – | Uncharacterized |
| Cre26.g756747 | – | Uncharacterized |
a PPP30 is an uncharacterized gene, which has not been identified as a CCM gene in any previous study.
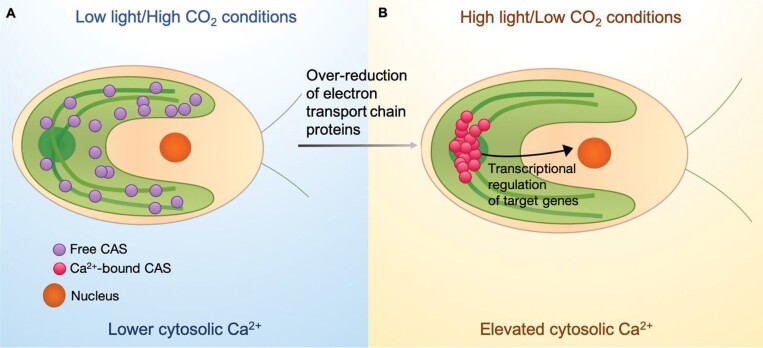
Similar to Rubisco, CAS was also revealed to move into the pyrenoid upon transition from high to low [CO2], with light being a prerequisite for this relocalization to occur (Yamano et al., 2018). The importance of Ca2+ binding is demonstrated by use of the chelator BAPTA that lessens CAS-mediated accumulation of LCIA and HLA3 (Yamano et al., 2018). The Ca2+-rich environment in the pyrenoid leads to Ca2+–CAS binding, which is thought to trigger a conformational change and mediate a signal to the nucleus, regulating CCM gene expression. The precise manner in which this protein signals for LC acclimation in Chlamydomonas is unknown, but a broad model is depicted in Fig. 5. The PET inhibitor DCMU prevents CAS relocalization under low [CO2] (Wang, 2017). Upon activation, CAS appears to relocate to a focal region within the pyrenoid. Though unclear, the signal propagation is affected by intracellular Ca2+ levels, [CO2], and light intensity. While the CAS mode of action is still unknown, these results suggest that the protein mediates a Ca2+-dependent retrograde signal to the nucleus, as part of the Chlamydomonas high light/LC acclimation. High light acclimation and CCM induction appear as important factors in the case of CIA5-mediated CCM gene expression and thylakoid membrane structuring proteins, and also in retrograde signalling in Chlamydomonas. CAS-mediated retrograde signalling further strengthens the ideas presented above, integrating the various physical and biological factors regulating CCM.
Fig. 5.
A schematic of a tentative mechanism for CAS activity in Chlamydomonas retrograde signalling. (A) Under low light/high CO2 conditions, CAS is dispersed throughout the chloroplast. (B) Under high light/low CO2, the ETC proteins become over-reduced, triggering the movement of CAS into the pyrenoid along the pyrenoid tubules. In the Ca2+-rich pyrenoid, CAS binds to Ca2+ and becomes activated. This form of CAS signals back to the nucleus to modulate target genes. It also induces an increase in intracellular Ca2+.
Insights for evolution of CCM regulation
The cues regulating CCM genes, which have been frequently reiterated throughout this review, are not low ambient [CO2] alone, but also light intensity. The response to CO2 and light intensity changes, as discussed above, could be indirectly mediated by metabolites or ROS that result from photosynthesis and/or PR. The physiological connections between PR and the CCM along with the uncanny similarity in gene expression regulation of several CCM and PR genes, led us to hypothesize on the evolutionary origins of CCM regulatory mechanisms.
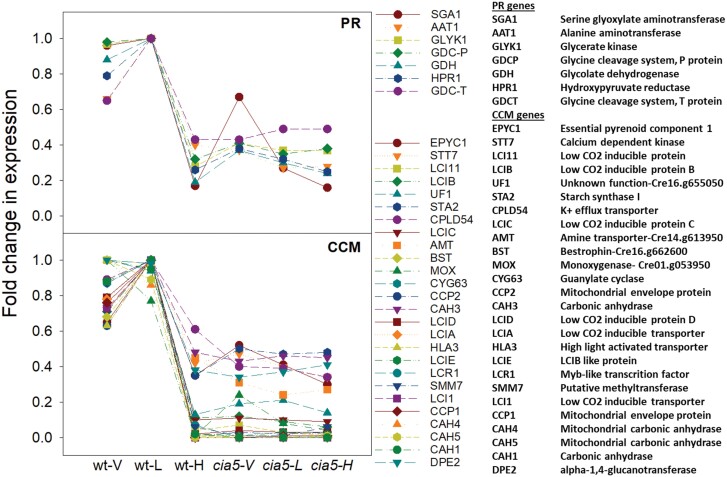
The regulation of PR in Chlamydomonas bears several similarities to that of the CCM. The genome-wide expression study with cia5 showed that several key PR enzymes (Fig. 1; Table 2) [alanine aminotransferase1 (AAT1), glycerate kinase (GLYK), glycolate dehydrogenase (GDH), hydroxypyruvate reductase1(HPR1), serine glyoxylate aminotransferase1 (SGAT1), and glycine decarboxylase (GDC) complex enzymes] were dependent on the master regulator CIA5 and CO2 in a manner similar to several CCM genes (Fang et al., 2012). The expression profiles of the PR and CCM genes that clustered in ‘CCM clusters’ (Fang et al., 2012) are shown in Fig. 6. It is worth noting that not all PR genes display this expression pattern. Notable exceptions are PGP genes, serine hydroxymethyltransferase (SHMT) genes, and the GDC-H gene. GDC-H is in a GDC complex with two other subunits (GDC-T and GDC-P) that are encoded by genes which are part of ‘CCM clusters’. PGP1 has already been discussed above. Although the PGP genes PGP1, PGP2, and PGP3 did not show CIA5-dependent differential expression (Fang et al., 2012), the expression of these PR genes was up-regulated under low [CO2] (Tural and Moroney, 2005). [CO2]-independent expression of PGP genes, in contrast to most genes that encode enzymes downstream of PGP in PR, lends more weight to the PGP substrate (2-PG) or product (glycolate) as CCM induction signalling molecules. 2-PG was suggested as a potential signalling molecule in a pgp1 algal mutant study (Suzuki et al., 1990). SHMT genes, which also have expression profiles different from that of other PR genes, encode proteins that catalyse conversion of glycine to serine. Whether the mitochondrial glycine:serine ratio impacts CCM induction requires investigation.
Fig. 6.
Relative expression of PR (top) and CCM (bottom) genes in wild-type (wt) and cia5 Chlamydomonas grown in different [CO2]: <0.02% (V, very low), 0.03–0.05% (L, low), and 5% (H, high). The expression of only genes classified as being in CCM clusters (Fang et al., 2012) is shown here.
A recent study (Tirumani et al., 2019) showed that genes encoding five of the PR enzymes, namely AAT1, GDH, HPR1, PGP1, and GDC-H, were not only dependent on low [CO2] for enhanced expression, but were also affected by light intensity. These genes also displayed diel regulation of expression without any circadian rhythmicity. These expression characteristics displayed by the five PR genes were also seen for four CCM genes (CAH3, LCIB, LCI1, and CCP1) in the same study. This similarity in expression profiles dependent on [CO2] changes and the presence of a common TF led us to look at the evolutionary origins of these processes. These studies provide compelling evidence that the stimulus for the CCM is likely to be determined by interactions between light intensity and PR activity when external CO2 is limiting.
The origin of oxygenic photosynthesis in cyanobacteria is placed ~2.4 billion years ago (bya) (Kopp et al., 2005). The endosymbiotic event conferring eukaryotic cells with a photosynthetic chloroplast through engulfment of a cyanobacterium is believed to have occurred 1.8 bya, and the origin of chlorophytes (the branch carrying green algae such as Chlamydomonas) from a common ancestor for all green plants and algae is dated ~700 million years ago (mya) (Becker, 2013). The endosymbiotic events of internalizing cyanobacteria and proteobacteria are believed to have not only conveyed the photosynthetic machinery via chloroplast (cyanobacterial endosymbiosis) establishment, but also the PR enzymes from both cyano- and proteobacteria (mitochondrial evolution) (Eisenhut et al., 2008; Bauwe et al., 2012). PR had to develop in response to the oxygenase activity of Rubisco. PR is seen in all photosynthetic organisms, with and without biophysical (cyanobacteria, algae, and hornworts) or biochemical CCMs [C4 and Crassulacean acid metabolism (CAM) plants] (Hagemann et al., 2016). It is an ancillary pathway to photosynthesis that evolved >1.8 bya, before the evolution of the algal CCM, which was perhaps as early as 500 mya, and C4 and CAM in land plants evolved <100 mya. The driver for evolution of PR, the biophysical CCM, C4, and CAM was the changing atmospheric composition with increasing [O2]:[CO2] ratios (Griffiths et al., 2017). This similarity in evolutionary drivers and co-regulation by CIA5 of both PR and CCM led us to hypothesize that the cellular machinery adopted a pre-existing PR regulatory tool to establish the CCM.
The observation that Rubisco occurring in organisms with CCM/C4/CAM have greater affinity for O2 (i.e. a lower specificity factor) than those lacking concentration mechanisms (Griffiths et al., 2017) hints at PR being an essential physiologically linked process for concentration mechanisms to function. Whether there are common TFs between PR and C4/CAM in higher plants, such as CIA5 of Chlamydomonas, requires further analysis. While common TFs such as CIA5 as a co-regulatory tool for concentration mechanisms and PR may not exist due to homoplastic origins of eukaryotic CCM, C4, and CAM pathways (Sage et al., 2011; Sage, 2016; Raven et al., 2017; Edwards, 2019), there is a possibility of evolution of other regulatory modes for coordinating the processes. However, the identification of common TFs regulating PR responses, as well as coordinating expression of the algal CCM and C4/CAM pathways in higher plants, is a promising line of investigation.
Conclusions
We began this review by considering the role of the TF CIA5, and were faced with the confounding observation that this key regulatory molecule not only activates some CCM genes but is also involved in activating PR. Working from the hypothesis that [CO2] changes are sensed through indirect cues from photosynthetic activity or carbohydrate metabolism, we propose that signals associated with the imbalance of PET and CBB cycle rates in response to [CO2] and light intensity changes, such as photorespiratory metabolites, or associated redox signalling, mediated by specific chloroplastic retrograde signalling mediators such as CAS or GUN4, may be effectors in CCM induction. Whilst the CCM is a response to CO2 limitation, the associated signalling has been co-opted secondarily from the requirement to regulate genes processing photorespiratory intermediates, or cope with associated light stress when the PET is overenergized and NPQ is up-regulated (Caspari et al., 2017). From an evolutionary perspective, this signalling would be consistent with the theory that CCM evolved in Chlorophyceae at the point of dissolved [CO2]:[O2] ratios being equivalent, some 400–500 mya (Griffiths et al., 2017), probably building on the evolution of PR machinery that had evolved 1.8 bya (Becker, 2013). The close cooperation between mitochondria and chloroplast metabolite exchanges, PR activity, and inducible CCM components strengthens this notion. We also propose that the interconnections between PET, the CBB cycle, the CCM, and PR might be further strengthened by recapturing photorespired CO2 by the CCM machinery and suggest that optimization of an engineered algal CCM in higher plants requires consideration of the PR apparatus.
Whilst building on the tremendous progress that has been made in recent years in identifying novel CCM components and assembly of a functional pyrenoid (Meyer et al., 2012, 2020; Mackinder et al., 2016, 2017; Atkinson et al., 2020), future studies should seek to identify the key effectors for CCM activation. The promise of identifying additional TFs (Arias et al., 2020) and potential cis-activational motifs (Winck et al., 2013), and post-transcriptional regulation through sRNA systems (Muller et al., 2020) will underpin efforts to understand CCM regulation. The analyses of large transcriptional datasets (Zones et al., 2015; Strenkert et al., 2019) using gene regulatory networks (Emmert-Streib et al., 2014), in conjunction with cis-element analysis (Winck et al., 2013), can help identify key TFs and TRs for future focus. Equally, by revisiting earlier studies which associated photorespiratory activity and CCM induction (Spalding et al., 1985; Moroney et al., 1986; Im and Grossman, 2002), we have highlighted additional regulatory processes which perhaps lead to CCM induction. Subsequent investigations with relevant knockout mutants from the large repositories generated (Li et al., 2016, 2019; Vilarrasa-Blasi et al., 2020, Preprint) will help test the above hypotheses and obtain further insights about regulation.
Major questions still remain, such as the origins and regulatory processes leading to the knotted thylakoid tubule network at the heart of the pyrenoid, the formation of associated connective minitubules, and spatial segregation between PSI and PSII. The energetic balance between CO2 reduction in the pyrenoid matrix, associated metabolite exchange, and regulation of the CBB cycle operating in the stroma may also account for the regulatory signalling complexities outlined above. The answers to questions about the complex regulatory processes leading to CCM induction will be revealed by molecular and physiological analyses and will require critical decisions on appropriate growth conditions (light intensity, photoperiods, synchronization of cells, and [CO2]) to identify the signalling mechanisms which orchestrate the biophysical CCM.
Acknowledgements
The authors thank Dr Gitanjali Yadav for helpful discussions, and Dr Jessica Royles for meticulous proofreading of this document. We are grateful for financial support from the GCRF Collective Call: BBSRC BB/P027970/1 (TIGR2ESS) programme.
Contributor Information
Indu Santhanagopalan, Department of Plant Sciences, Downing Street, University of Cambridge, Cambridge, UK.
Rachel Wong, Department of Plant Sciences, Downing Street, University of Cambridge, Cambridge, UK.
Tanya Mathur, Department of Plant Sciences, Downing Street, University of Cambridge, Cambridge, UK.
Howard Griffiths, Department of Plant Sciences, Downing Street, University of Cambridge, Cambridge, UK.
Donald Ort, University of Illinois, USA.
References
- Arias C, Obudulub O, Zhaoa X, et al. 2020. Nuclear proteome analysis of Chlamydomonas with response to CO2 limitation. Algal Research 46, 101765. [Google Scholar]
- Armbruster U, Labs M, Pribil M, et al. 2013. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. The Plant Cell 25, 2661–2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LC, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ. 2016. Introducing an algal carbon-concentrating mechanism into higher plants: location and incorporation of key components. Plant Biotechnology Journal 14, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N, Mao Y, Chan KX, McCormick AJ. 2020. Condensation of Rubisco into a proto-pyrenoid in higher plant chloroplasts. Nature Communications 11, 6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Kaplan A, Berry JA. 1980. Internal inorganic carbon pool of Chlamydomonas reinhardtii: evidence for a carbon dioxide-concentrating mechanism. Plant Physiology 66, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Kern R, Timm S. 2012. Photorespiration has a dual origin and manifold links to central metabolism. Current Opinion in Plant Biology 15, 269–275. [DOI] [PubMed] [Google Scholar]
- Becker B. 2013. Snow ball earth and the split of Streptophyta and Chlorophyta. Trends in Plant Science 18, 180–183. [DOI] [PubMed] [Google Scholar]
- Blanco-Rivero A, Shutova T, Román MJ, Villarejo A, Martinez F. 2012. Phosphorylation controls the localization and activation of the lumenal carbonic anhydrase in Chlamydomonas reinhardtii. PLoS One 7, e49063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueggeman AJ, Gangadharaiah DS, Cserhati MF, Casero D, Weeks DP, Ladunga I. 2012. Activation of the carbon concentrating mechanism by CO2 deprivation coincides with massive transcriptional restructuring in Chlamydomonas reinhardtii. The Plant Cell 24, 1860–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezowski P, Schlicke H, Richter A, Dent RM, Niyogi KK, Grimm B. 2014. The GUN4 protein plays a regulatory role in tetrapyrrole biosynthesis and chloroplast-to-nucleus signalling in Chlamydomonas reinhardtii. The Plant Journal 79, 285–298. [DOI] [PubMed] [Google Scholar]
- Caspari OD, Meyer MT, Tolleter D, Wittkopp TM, Cunniffe NJ, Lawson T, Grossman AR, Griffiths H. 2017. Pyrenoid loss in Chlamydomonas reinhardtii causes limitations in CO2 supply, but not thylakoid operating efficiency. Journal of Experimental Botany 68, 3903–3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B. 2016. The function and regulation of CIA5/CCM1 in Chlamydomonas reinhardtii. PhD thesis, Iowa State University.
- Chen X, Pu H, Fang Y, Wang X, Zhao S, Lin Y, Zhang M, Dai HE, Gong W, Liu L. 2015. Crystal structure of the catalytic subunit of magnesium chelatase. Nature Plants 1, 15125. [DOI] [PubMed] [Google Scholar]
- Chung BY, Valli A, Deery MJ, Navarro FJ, Brown K, Hnatova S, Howard J, Molnar A, Baulcombe DC. 2019. Distinct roles of Argonaute in the green alga Chlamydomonas reveal evolutionary conserved mode of miRNA-mediated gene expression. Scientific Reports 9, 11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depège N, Bellafiore S, Rochaix JD. 2003. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science 299, 1572–1575. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Turkan I, Krieger-Liszkay A. 2016. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiology 171, 1541–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ. 2019. Evolutionary trajectories, accessibility and other metaphors: the case of C4 and CAM photosynthesis. New Phytologist 223, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. 2008. The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proceedings of the National Academy of Sciences, USA 105, 17199–17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmert-Streib F, Dehmer M, Haibe-Kains B. 2014. Gene regulatory networks and their applications: understanding biological and medical problems in terms of networks. Frontiers in Cell and Developmental Biology 2, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel BD, Schaffer M, Kuhn Cuellar L, Villa E, Plitzko JM, Baumeister W. 2015. Native architecture of the Chlamydomonas chloroplast revealed by in situ cryo-electron tomography. eLife 4, e04889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson E, Wakao S, Niyogi KK. 2015. Light stress and photoprotection in Chlamydomonas reinhardtii. The Plant Journal 82, 449–465. [DOI] [PubMed] [Google Scholar]
- Fang W, Si Y, Douglass S, Casero D, Merchant SS, Pellegrini M, Ladunga I, Liu P, Spalding MH. 2012. Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. The Plant Cell 24, 1876–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findinier J, Delevoye C, Cohen MM. 2019. The dynamin-like protein Fzl promotes thylakoid fusion and resistance to light stress in Chlamydomonas reinhardtii. PLoS Genetics 15, e1008047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Pérez Ú, Jarvis P. 2013. Molecular chaperone involvement in chloroplast protein import. Biochimica et Biophysica Acta 1833, 332–340. [DOI] [PubMed] [Google Scholar]
- Formighieri C, Ceol M, Bonente G, Rochaix JD, Bassi R. 2012. Retrograde signaling and photoprotection in a gun4 mutant of Chlamydomonas reinhardtii. Molecular Plant 5, 1242–1262. [DOI] [PubMed] [Google Scholar]
- Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, et al. 2017. The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171, 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fufezan C, Rutherford AW, Krieger-Liszkay A. 2002. Singlet oxygen production in herbicide-treated photosystem II. FEBS Letters 532, 407–410. [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho KI, Saito T, Kohinata T, Ohyama K. 2001. Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proceedings of the National Academy of Sciences, USA 98, 5347–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Sage TL, Osteryoung KW. 2006. FZL, an FZO-like protein in plants, is a determinant of thylakoid and chloroplast morphology. Proceedings of the National Academy of Sciences, USA 103, 6759–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty AM, Spalding MH. 1996. Molecular and structural changes in Chlamydomonas under limiting CO2 (a possible mitochondrial role in adaptation). Plant Physiology 111, 1339–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet MMM, Orr DJ, Melkonian M, Müller KH, Meyer MT, Carmo-Silva E, Griffiths H. 2020. Rubisco and carbon-concentrating mechanism co-evolution across chlorophyte and streptophyte green algae. New Phytologist 227, 810–823. [DOI] [PubMed] [Google Scholar]
- Griffiths H, Meyer MT, Rickaby REM. 2017. Overcoming adversity through diversity: aquatic carbon concentrating mechanisms. Journal of Experimental Botany 68, 3689–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna A, Hegde RS. 2018. Transmembrane domain recognition during membrane protein biogenesis and quality control. Current Biology 28, R498–R511. [DOI] [PubMed] [Google Scholar]
- Gutu A, Chang F, O’Shea EK. 2018. Dynamical localization of a thylakoid membrane binding protein is required for acquisition of photosynthetic competency. Molecular Microbiology 108, 16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann M, Kern R, Maurino VG, Hanson DT, Weber AP, Sage RF, Bauwe H. 2016. Evolution of photorespiration from cyanobacteria to land plants, considering protein phylogenies and acquisition of carbon concentrating mechanisms. Journal of Experimental Botany 67, 2963–2976. [DOI] [PubMed] [Google Scholar]
- He S, Chou HT, Matthies D, et al. 2020. The structural basis of Rubisco phase separation in the pyrenoid. Nature Plants 6, 1480–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich J, Thurotte A, Schneider D. 2017. Specific interaction of IM30/Vipp1 with cyanobacterial and chloroplast membranes results in membrane remodeling and eventually in membrane fusion. Biochimica et Biophysica Acta 1859, 537–549. [DOI] [PubMed] [Google Scholar]
- Hennacy JH, Jonikas MC. 2020. Prospects for engineering biophysical CO2 concentrating mechanisms into land plants to enhance yields. Annual Review of Plant Biology 71, 461–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig R, Heidrich J, Saur M, et al. 2015. IM30 triggers membrane fusion in cyanobacteria and chloroplasts. Nature Communications 6, 7018. [DOI] [PubMed] [Google Scholar]
- Hennig R, West A, Debus M, Saur M, Markl J, Sachs JN, Schneider D. 2017. The IM30/Vipp1 C-terminus associates with the lipid bilayer and modulates membrane fusion. Biochimica et Biophysica Acta 1858, 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im CS, Grossman AR. 2002. Identification and regulation of high light-induced genes in Chlamydomonas reinhardtii. The Plant Journal 30, 301–313. [DOI] [PubMed] [Google Scholar]
- Itakura AK, Chan KX, Atkinson N, et al. 2019. A Rubisco-binding protein is required for normal pyrenoid number and starch sheath morphology in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 116, 18445–18454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Kessler F. 2014. Mechanisms of chloroplast protein import in plants. In: Theg S., Wollman FA, eds. Plastid biology. Advances in Plant Biology, vol. 5. New York: Springer, 241–270. [Google Scholar]
- Jin S, Sun J, Wunder T, Tang D, Cousins AB, Sze SK, Mueller-Cajar O, Gao YG. 2016. Structural insights into the LCIB protein family reveals a new group of beta-carbonic anhydrases. Proceedings of the National Academy of Sciences, USA 113, 14716–14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohinata T, Nishino H, Fukuzawa H. 2008. Significance of zinc in a regulatory protein, CCM1, which regulates the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant & Cell Physiology 49, 273–283. [DOI] [PubMed] [Google Scholar]
- Kopp RE, Kirschvink JL, Hilburn IA, Nash CZ. 2005. The Paleoproterozoic snowball Earth: a climate disaster triggered by the evolution of oxygenic photosynthesis. Proceedings of the National Academy of Sciences, USA 102, 11131–11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki A, Takeba G. 1996. Photorespiration protects C3 plants from photooxidation. Nature 384, 557–560. [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of Molecular Biology 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. 2001. VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proceedings of the National Academy of Sciences, USA 98, 4238–4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Ohyama K, Fukuzawa H. 1999. CO2-responsive transcriptional regulation of CAH1 encoding carbonic anhydrase is mediated by enhancer and silencer regions in Chlamydomonas reinhardtii. Plant Physiology 121, 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Yoshioka S, Taniguchi F, Ohyama K, Fukuzawa H. 2003. Cis-acting elements and DNA-binding proteins involved in CO2-responsive transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiology 133, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin RM. 2016. Tetrapyrrole signaling in plants. Frontiers in Plant Science 7, 1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford HK, Chin BL, Niyogi KK. 2007. Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryotic Cell 6, 919–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemeille S, Turkina MV, Vener AV, Rochaix JD. 2010. Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Molecular & Cellular Proteomics 9, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Patena W, Fauser F, et al. 2019. A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nature Genetics 51, 627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Patena W, et al. 2016. An indexed, mapped mutant library enables reverse genetics studies of biological processes in Chlamydomonas reinhardtii. The Plant Cell 28, 367–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Tassinari M, Souza DP, Naskar S, Noel JK, Bohuszewicz O, Buck M, Williams TA, Baum B, Low HH. 2020. Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodelling superfamily. BioRxiv doi: 10.1101/2020.08.13.249979. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LCM, Chen C, Leib RD, Patena W, Blum SR, Rodman M, Ramundo S, Adams CM, Jonikas MC. 2017. A spatial interactome reveals the protein organization of the algal CO2-concentrating mechanism. Cell 171, 133–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinder LC, Meyer MT, Mettler-Altmann T, et al. 2016. A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proceedings of the National Academy of Sciences, USA 113, 5958–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Rujan T, Richly E, Hansen A, Cornelsen S, Lins T, Leister D, Stoebe B, Hasegawa M, Penny D. 2002. Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proceedings of the National Academy of Sciences, USA 99, 12246–12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RML, Gibbs SP. 1991. Composition and function of pyrenoids: cytochemical and immunocytochemical approaches. Canadian Journal of Botany 69, 1040–1052. [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, Genkov T, Skepper JN, Jouhet J, Mitchell MC, Spreitzer RJ, Griffiths H. 2012. Rubisco small-subunit α-helices control pyrenoid formation in Chlamydomonas. Proceedings of the National Academy of Sciences, USA 109, 19474–19479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Griffiths H. 2013. Origins and diversity of eukaryotic CO2-concentrating mechanisms: lessons for the future. Journal of Experimental Botany 64, 769–786. [DOI] [PubMed] [Google Scholar]
- Meyer MT, Itakura AK, Patena W, Wang L, He S, Emrich-Mills T, Lau CS, Yates G, Mackinder LCM, Jonikas MC. 2020. Assembly of the algal CO2-fixing organelle, the pyrenoid, is guided by a Rubisco-binding motif. Science Advances 6, eabd2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MT, Whittaker C, Griffiths H. 2017. The algal pyrenoid: key unanswered questions. Journal of Experimental Botany 68, 3739–3749. [DOI] [PubMed] [Google Scholar]
- Mitchell MC, Meyer MT, Griffiths H. 2014. Dynamics of carbon-concentrating mechanism induction and protein relocalization during the dark-to-light transition in synchronized Chlamydomonas reinhardtii. Plant Physiology 166, 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Yamano T, Yoshioka S, et al. 2004. Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiology 135, 1595–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. 2007. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447, 1126–1129. [DOI] [PubMed] [Google Scholar]
- Moroney JV, Husic HD, Tolbert NE, Kitayama M, Manuel LJ, Togasaki RK. 1989. Isolation and characterization of a mutant of Chlamydomonas reinhardtii deficient in the CO2 concentrating mechanism. Plant Physiology 89, 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Jungnick N, Dimario RJ, Longstreth DJ. 2013. Photorespiration and carbon concentrating mechanisms: two adaptations to high O2, low CO2 conditions. Photosynthesis Research 117, 121–131. [DOI] [PubMed] [Google Scholar]
- Moroney JV, Wilson BJ, Tolbert NE. 1986. Glycolate metabolism and excretion by Chlamydomonas reinhardtii. Plant Physiology 82, 821–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroney JV, Ynalvez RA. 2007. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryotic Cell 6, 1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Lau CS, Walker CE, et al. 2019. Thylakoid localized bestrophin-like proteins are essential for the CO2 concentrating mechanism of Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 116, 16915–16920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller SY, Matthews NE, Valli AA, Baulcombe DC. 2020. The small RNA locus map for Chlamydomonas reinhardtii. PLoS One 15, e0242516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura H, Shiina T. 2014. Calcium signaling in plant endosymbiotic organelles: mechanism and role in physiology. Molecular Plant 7, 1094–1104. [DOI] [PubMed] [Google Scholar]
- Nordhues A, Schöttler MA, Unger AK, et al. 2012. Evidence for a role of VIPP1 in the structural organization of the photosynthetic apparatus in Chlamydomonas. The Plant Cell 24, 637–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pechmann S, Frydman J. 2014. Interplay between chaperones and protein disorder promotes the evolution of protein networks. PLoS Computational Biology 10, e1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Terry MJ, Van Aken O, Quiros PM. 2020. Retrograde signals from endosymbiotic organelles: a common control principle in eukaryotic cells. Philosophical Transactions of the Royal Society B: Biological Sciences 375, 20190396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumed MP, Villarejo A, Roos A, Garcia-Reina G, Ramazanov Z. 1996. The CO2-concentrating mechanism in a starchless mutant of the green unicellular alga Chlorella pyrenoidosa. Planta 200, 28–31. [Google Scholar]
- Polukhina I, Fristedt R, Dinc E, Cardol P, Croce R. 2016. Carbon supply and photoacclimation cross talk in the green alga Chlamydomonas reinhardtii. Plant Physiology 172, 1494–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribil M, Sandoval-Ibáñez O, Xu W, et al. 2018. Fine-tuning of photosynthesis requires CURVATURE THYLAKOID1-mediated thylakoid plasticity. Plant Physiology 176, 2351–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramazanov Z, Rawat M, Henk MC, Mason CB, Matthews SW, Moroney JV. 1994. The induction of the CO2-concentrating mechanism is correlated with the formation of the starch sheath around the pyrenoid of Chlamydomonas reinhardtii. Planta 195, 210–216. [Google Scholar]
- Raven JA, Beardall J, Sánchez-Baracaldo P. 2017. The possible evolution and future of CO2-concentrating mechanisms. Journal of Experimental Botany 68, 3701–3716. [DOI] [PubMed] [Google Scholar]
- Rawat M, Moroney JV. 1995. The regulation of carbonic anhydrase and ribulose-1,5-bisphosphate carboxylase/oxygenase activase by light and CO2 in Chlamydomonas reinhardtii. Plant Physiology 109, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea G, Antonacci A, Lambreva MD, Mattoo AK. 2018. Features of cues and processes during chloroplast-mediated retrograde signaling in the alga Chlamydomonas. Plant Science 272, 193–206. [DOI] [PubMed] [Google Scholar]
- Riaño-Pachón DM, Corrêa LG, Trejos-Espinosa R, Mueller-Roeber B. 2008. Green transcription factors: a Chlamydomonas overview. Genetics 179, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rütgers M, Muranaka LS, Mühlhaus T, Sommer F, Thoms S, Schurig J, Willmund F, Schulz-Raffelt M, Schroda M. 2017. Substrates of the chloroplast small heat shock proteins 22E/F point to thermolability as a regulative switch for heat acclimation in Chlamydomonas reinhardtii. Plant Molecular Biology 95, 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2016. A portrait of the C4 photosynthetic family on the 50th anniversary of its discovery: species number, evolutionary lineages, and Hall of Fame. Journal of Experimental Botany 67, 4039–4056. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin PA, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62, 3155–3169. [DOI] [PubMed] [Google Scholar]
- Sager R, Palade GE. 1957. Structure and development of the chloroplast in Chlamydomonas. I. The normal green cell. Journal of Biophysical and Biochemical Cytology 3, 463–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinetova MA, Kupriyanova EV, Markelova AG, Allakhverdiev SI, Pronina NA. 2012. Identification and functional role of the carbonic anhydrase Cah3 in thylakoid membranes of pyrenoid of Chlamydomonas reinhardtii. Biochimica et Biophysica Acta 1817, 1248–1255. [DOI] [PubMed] [Google Scholar]
- Singh R, Singh S, Parihar P, Singh VP, Prasad SM. 2015. Retrograde signaling between plastid and nucleus: a review. Journal of Plant Physiology 181, 55–66. [DOI] [PubMed] [Google Scholar]
- Spalding MH, Spritzer RJ, Ogren WL. 1985. Use of mutants in analysis of the CO2-concentrating pathway of Chlamydomonas reinhardtii. Botany Publication and Papers 72, http://lib.dr.iastate.edu/bot_pubs/72. [Google Scholar]
- Strenkert D, Schmollinger S, Gallaher SD, et al. 2019. Multiomics resolution of molecular events during a day in the life of Chlamydomonas. Proceedings of the National Academy of Sciences, USA 116, 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M. 2011. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. The Plant Cell 23, 2285–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Marek LF, Spalding MH. 1990. A photorespiratory mutant of Chlamydomonas reinhardtii. Plant Physiology 93, 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka R, Tanaka A. 2007. Tetrapyrrole biosynthesis in higher plants. Annual Review of Plant Biology 58, 321–346. [DOI] [PubMed] [Google Scholar]
- Tarahi Tabrizi S, Sawicki A, Zhou S, Luo M, Willows RD. 2016. GUN4–protoporphyrin IX is a singlet oxygen generator with consequences for plastid retrograde signaling. Journal of Biological Chemistry 291, 8978–8984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis J, Niemeyer J, Schmollinger S, et al. 2020. VIPP2 interacts with VIPP1 and HSP22E/F at chloroplast membranes and modulates a retrograde signal for HSP22E/F gene expression. Plant, Cell & Environment 43, 1212–1229. [DOI] [PubMed] [Google Scholar]
- Tirumani S, Gothandam KM, J Rao B. 2019. Coordination between photorespiration and carbon concentrating mechanism in Chlamydomonas reinhardtii: transcript and protein changes during light–dark diurnal cycles and mixotrophy conditions. Protoplasma 256, 117–130. [DOI] [PubMed] [Google Scholar]
- Tirumani S, Kokkanti M, Chaudhari V, Shukla M, Rao BJ. 2014. Regulation of CCM genes in Chlamydomonas reinhardtii during conditions of light–dark cycles in synchronous cultures. Plant Molecular Biology 85, 277–286. [DOI] [PubMed] [Google Scholar]
- Toyokawa C, Yamano T, Fukuzawa H. 2020. Pyrenoid starch sheath is required for LCIB localization and the CO2-concentrating mechanism in green algae. Plant Physiology 182, 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tural B, Moroney JV. 2005. Regulation of the expression of photorespiratory genes in Chlamydomonas reinhardtii. Canadian Journal of Botany 83, 810–819. [Google Scholar]
- Valli AA, Santos BA, Hnatova S, Bassett AR, Molnar A, Chung BY, Baulcombe DC. 2016. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Research 26, 519–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdecia MA, Larkin RM, Ferrer JL, Riek R, Chory J, Noel JP. 2005. Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biology 3, e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilarrasa-Blasi J, Fauser F, Onishi M, et al. 2020. Systematic characterization of gene function in a photosynthetic organism. BioRxiv doi: 10.1101/2020.12.11.420950 [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villand P, Eriksson M, Samuelsson G. 1997. Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. The Biochemical Journal 327, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A, Martınez F, Plumed MP, Ramazanov Z. 1996. The induction of the CO2 concentrating mechanism in a starch-less mutant of Chlamydomonas reinhardtii. Physiologia Plantarum 98 [Google Scholar]
- Wang L. 2017. A calcium-binding protein CAS regulates the CO2-concentrating mechanism in the green alga Chlamydomonas reinhardtii. PhD thesis, Kyoto University.
- Wang L, Yamano T, Takane S, et al. 2016. Chloroplast-mediated regulation of CO2-concentrating mechanism by Ca2+-binding protein CAS in the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 113, 12586–12591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Spalding MH. 2014. Acclimation to very low CO2: contribution of limiting CO2 inducible proteins, LCIB and LCIA, to inorganic carbon uptake in Chlamydomonas reinhardtii. Plant Physiology 166, 2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Stessman DJ, Spalding MH. 2015. The CO2 concentrating mechanism and photosynthetic carbon assimilation in limiting CO2: how Chlamydomonas works against the gradient. The Plant Journal 82, 429–448. [DOI] [PubMed] [Google Scholar]
- Westphal S, Heins L, Soll J, Vothknecht UC. 2001. Vipp1 deletion mutant of Synechocystis: a connection between bacterial phage shock and thylakoid biogenesis? Proceedings of the National Academy of Sciences, USA 98, 4243–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winck FV, Melo DO, Riaño-Pachón DM, Martins MC, Caldana C, Barrios AF. 2016. Analysis of sensitive CO2 pathways and genes related to carbon uptake and accumulation in Chlamydomonas reinhardtii through genomic scale modeling and experimental validation. Frontiers in Plant Science 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winck FV, Vischi Winck F, Arvidsson S, Riaño-Pachón DM, Hempel S, Koseska A, Nikoloski Z, Urbina Gomez DA, Rupprecht J, Mueller-Roeber B. 2013. Genome-wide identification of regulatory elements and reconstruction of gene regulatory networks of the green alga Chlamydomonas reinhardtii under carbon deprivation. PLoS One 8, e79909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunder T, Cheng SLH, Lai SK, Li HY, Mueller-Cajar O. 2018. The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nature Communications 9, 5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhang J, Weeks DP. 2001. The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 98, 5341–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Asada A, Sato E, Fukuzawa H. 2014. Isolation and characterization of mutants defective in the localization of LCIB, an essential factor for the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Photosynthesis Research 121, 193–200. [DOI] [PubMed] [Google Scholar]
- Yamano T, Sato E, Iguchi H, Fukuda Y, Fukuzawa H. 2015. Characterization of cooperative bicarbonate uptake into chloroplast stroma in the green alga Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences, USA 112, 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Toyokawa C, Fukuzawa H. 2018. High-resolution suborganellar localization of Ca2+-binding protein CAS, a novel regulator of CO2-concentrating mechanism. Protoplasma 255, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Yamano T, Tsujikawa T, Hatano K, Ozawa S, Takahashi Y, Fukuzawa H. 2010. Light and low-CO2-dependent LCIB–LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant & Cell Physiology 51, 1453–1468. [DOI] [PubMed] [Google Scholar]
- Yoshioka S, Taniguchi F, Miura K, Inoue T, Yamano T, Fukuzawa H. 2004. The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. The Plant Cell 16, 1466–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Marchand CH, Maes A, et al. 2018. Pyrenoid functions revealed by proteomics in Chlamydomonas reinhardtii. PLoS One 13, e0185039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. 2007. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes & Development 21, 1190–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zones JM, Blaby IK, Merchant SS, Umen JG. 2015. High-resolution profiling of a synchronized diurnal transcriptome from Chlamydomonas reinhardtii reveals continuous cell and metabolic differentiation. The Plant Cell 27, 2743–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]