Abstract
Background:
The germline serves as a conduit for transmission of genetic and epigenetic information from one generation to the next. In males, spermatozoa are the final carriers of inheritance and their continual production is supported by a foundational population of spermatogonial stem cells (SSCs) that forms from prospermatogonial precursors during the early stages of neonatal development. In mammals, the timing for which SSCs are specified and the underlying mechanisms guiding this process remain to be completely understood.
Objectives:
To propose an evolving concept for how the foundational SSC population is established.
Materials and methods:
This review summarizes recent and historical findings from peer-reviewed publications made primarily with mouse models while incorporating limited studies from humans and livestock.
Results and conclusion:
Establishment of the SSC population appears to follow a biphasic pattern involving a period of fate programming followed by an establishment phase that culminates in formation of the SSC population. This model for establishment of the foundational SSC population from precursors is anticipated to extend across mammalian species and include humans and livestock, albeit on different timescales.
Keywords: establishment, germline, gonocyte, prospermatogonia, specification, spermatogonial stem cel
1 |. INTRODUCTION
1.1 |. Overview
Germ cells are the cellular link between generations, and although studies of their development in higher order mammals such as livestock and humans are dwarfed by the number using rodent models, accumulating knowledge suggests that the broad milestones in germline developmental are generally conserved. Between mammalian species, however, the timescales vary dramatically, but are generally equivalent to the total length of gestation. For example, developmental events that occur on the order of days in mice with a gestational period of 19–21 days take place on the order of weeks in humans and livestock with significantly longer gestational windows. While male germline development among mammalian species will be the primary focus of this review due to recent insights from mouse studies, readers are encouraged to explore the unique developmental events among other model organisms (reviewed in1).
In addition to species-specific timescales, events in germline development are largely asynchronous in nature. Rather than a series of switches or steps, changes occur progressively over the course of days or weeks. At any given time in development, germ cells are generally heterogeneous with respect to their developmental status. Thus, defining stages of development is often difficult. However, with the advent of technologies that allow for studying biological processes at single-cell resolution, the asynchronous and heterogenous nature of germline development that is otherwise masked by bulk methods of analysis can be accurately assessed.
1.2 |. Germline development
Primordial germ cells (PGCs) mark the most ancestral cell type of the spermatogenic lineage. PGCs are specified from the epiblast in response to inductive signaling around E6.5-E7.25 in mice, E11.5-E15.5 in pigs, and 2–3 weeks of gestation in humans.2–4 Nascent PGCs retain a latent pluripotency program while the remaining post-implantation epiblast cells destined for somatic tissues experience increased DNA methylation, H3K9me2, and X inactivation.5 Following specification, PGCs proliferate, undergo extensive epigenetic remodeling, and migrate to the developing gonad.6 Prior to gonadal colonization, PGCs have both spermatogenic and teratoma-forming potential 7,8 and can form pluripotent cell lines in vitro,9,10 likely owing to a latent pluripotency network.5,9,11,12 After gonadal colonization, however, PGCs eventually become unipotent 13 and differentiate through their respective sex-specific pathways in concert with the bipotential soma.14
Following sex determination, PGCs committed to male germ cell fate transition to form precursors to the entire spermatogenic lineage, commonly known as prospermatogonia (Prospg). Initially, Prospg remain mitotically active (referred to as fetal mitotic-Prospg or ProspgM) before entering a period of sustained quiescence (referred to as transitional 1-Prospg or ProspgT1). After birth, germ cells progressively re-enter the cell cycle (referred to as transitional 2-Prospg or ProspgT2) before transitioning to form SSCs and the remaining spermatogenic lineage. Postnatally, germ cells also migrate from the center of seminiferous cords to the basement membrane concurrent with re-activation of proliferation. Importantly, previous studies indicate that proliferation and migration are temporally independent events that are likely regulated by different mechanisms.15,16 Thus, subsets of Prospg are best defined based on their respective proliferative activities. While Prospg are also referred to as gonocytes, prespermatogonia, PGCs, or primitive germ cells depending on the study,17,18 this review will follow the established nomenclature for male germ cell types through development outlined in.17 Within these defined subtypes of Prospg, only a portion of the germline will ultimately form SSCs tasked with maintaining the entire spermatogenic lineage long into adulthood.
Pioneering studies by Kluin and de Rooij in 1981 first described evidence that a division of fate occurs in the male germline such that one subset of Prospg directly enters terminal differentiation, while another forms the undifferentiated population containing presumptive SSCs.19 Using histological examination of neonatal mouse testes, the authors described two populations of Prospg present shortly after birth that resembled either differentiating spermatogonia (denoted “Type II”) or undifferentiated spermatogonia (denoted “Type I”), representing ~ 70% or the remaining ~ 30% of the germline, respectively.19 Importantly, these populations were readily discernable at P1-P2, prior to the onset of retinoic acid signaling and differentiation at P3.20,21 These findings were later validated by studies demonstrating that germ cells negative for the transcription factor neurogenin-3 (Ngn3) form a population of initial differentiating spermatogonia that contribute to a unique first round or cohort of spermatogenesis, while successive and continuous sperm production is derived from Ngn3-positive progenitors originating from the SSC population.22 Collectively, these studies demonstrated that only a subset of Prospg forms SSCs during development and that SSC fate, along with other fates, may be specified prior to formation of the SSC population.
Consistent with the notion of fate programming, recent findings by our laboratory suggest that formation of the SSC population follows a biphasic scheme, comprising an initial period of specification and fate programming among ProspgT1 followed by a window of SSC establishment in ProspgT2 that culminates in formation of the foundational SSC population that will support continuous sperm production and inheritance through the male germline (depicted in Figure 1).
FIGURE 1.
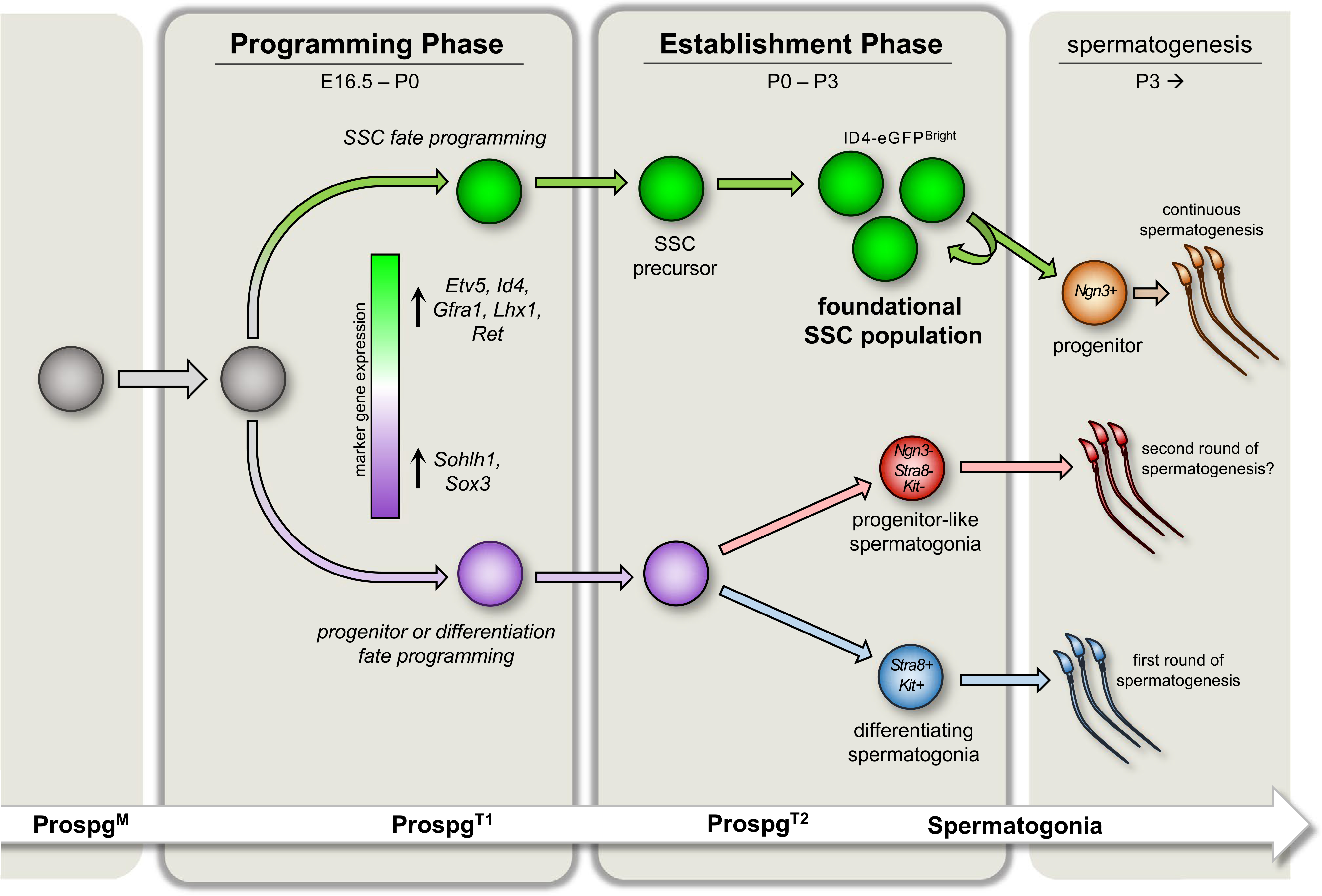
Evolving biphasic model underlying formation of the foundational SSC population in mice. Mitotically active fetal prospermatogonia (ProspM) progressively enter quiescence leading up to E16.5 to form transitional Prospg (ProspgT1). During the Programming phase, genes necessary for postnatal SSC function are upregulated in a subset of quiescent ProspgT1, including Etv5, Id4, Gfra1, Lhx1, and Ret, while a different subset upregulates genes associated with progenitors or differentiation, including Sohlh1 and Sox3. Based on the outcomes from transplantation analyses, stem cell potential within the germline becomes confined to those ProsgT1 that upregulate SSC-associated transcripts, indicating that postnatal fate is programmed during this time. After birth, the germline enters an Establishment phase. ProspgT1asynchronously re-enter the cell cycle, which marks formation of ProspgT2. A dramatic shift in gene expression also occurs, including upregulation of genes necessary for postnatal SSC function. Conclusion of the Establishment phase is marked by a brief period of self-renewal that builds the foundational SSC population from which continuous spermatogenesis will arise throughout adulthood. By contrast, Prospg programmed for non-SSC fate either enter the differentiating spermatogonial path directly to generate the first round of spermatogenesis or possibly form a population of initial progenitor-like germ cells that will be the source of a second round of spermatogenesis that does not emanate from the foundational SSC pool
2 |. PROGRAMMING PHASE
Proliferating fetal ProspgM progressively enter G1/G0 arrest to become ProspgT1 from around E12.5-E16.5 in mice, E80-E105 in cattle, and 20–25 weeks of gestation in humans.23–28 This transition represents not only a shift in mitotic potential, but also bookmarks the start of several hallmark and essential milestones in germline development.
2.1 |. Programming of spermatogenic fate
Recent studies by our laboratory and others indicate that during mitotic arrest, ProspgT1 progressively acquire distinct transcriptional characteristics that align with postnatal functional fates.29,30 Using single-cell RNA-sequencing (scRNA-seq), ProspgT1 were found to be transcriptionally heterogeneous. In fact, opposing transcriptional signatures divide the perinatal germline. Genes that are necessary for postnatal SSC self-renewal and maintenance, including Etv5, Id4, Lhx1, and Ret,31–34 are upregulated in a subset of the germline at E16.5. By contrast, transcripts associated with differentiation, including Sohlh1 and Sox3,35,36 are upregulated among the opposite portion of the Prospg population. Evidence of these opposing transcriptional signatures is not present prior to E16.5 (Law and Oatley, unpublished). Assessment of ID4 expression in vivo using an Id4e-Gfp transgenic mouse line confirmed that only a subset of Prospg express ID4 prior to birth. Importantly, this heterogeneity is not simply a byproduct of asynchronous development because the opposing transcriptomic signatures are present at P0, P3, and P6 as well, suggesting a division of fate between the SSC lineage and a lineage primed for differentiation.
To begin exploring the functional significance of this transcriptional heterogeneity, we performed transplantation analysis of isolated fetal ProspgT1 at E18.5. ProspgT1 were subdivided based on ID4-eGFP expression and transplanted into the testes of germ cell–depleted recipients using well-established methods.37 Outcomes of transplantation analysis revealed that only ProspgT1 that are ID4+ possess the latent capacity to generate spermatogenic colonies, thus demonstrating that regenerative capacity within the perinatal germline is restricted to a subset of the mitotically arrested population. Strikingly, colonization was never observed among ProspgT1 that are ID4-. Together, these findings suggest that SSC fate is ingrained within a subset of Prospg prior to birth, far earlier in development than previously suspected.
As noted above, multiple studies have demonstrated that germ cells prior to E18.5 are capable of regenerating spermatogenesis upon transplantation.7,8,38 Although one study described a lack of complete colonization among germ cells isolated prior to P4,39 the consensus from several other studies indicates that the capacity to regenerate complete spermatogenesis is present prior to P4.7,8,29,38,40 Strikingly, donor cells isolated as early as E6.5, at the time of PGC specification, are capable to generating colonies of spermatogenesis upon transplantation.7 Therefore, stem cell potential is not necessarily acquired during the Programming phase. Rather, regenerative capacity is restricted to a subset of germline given that no colonization was observed among ID4- Prospg. Therefore, stem cell potential is retained within those ProspgT1 that express the machinery necessary for postnatal SSC function prior to behaving as an SSC.
The ability of Prospg and PGCs to regenerate spermatogenesis when transplanted into adult recipients is anomalous given that neither are normally present in adult testes. The onset of gene expression for factors necessary for postnatal SSC function within ProspgT1 indicates that germ cells prior to E16.5 lack certain critical components to function as SSCs postnatally, but that these attributes are likely acquired upon transplantation. For example, Id4 expression is not detected prior to E16.529; yet, ID4 is important for long-term maintenance of the SSC population in postnatal life.41 Thus, one could envision that upon transplantation of donor cells from prenatal fetuses, germ cells likely transition through intermediate states analogous to developmental stages in vivo before forming SSCs that support continuous spermatogenesis. To date, no studies have evaluated the dynamics or characteristics of prenatal germ cells following transplantation into adult recipients. Interestingly, as donor cell suspensions are obtained from progressively more developmentally advanced tissues, regenerative capacity from germ cells within donor cell suspensions appears to increase in an age-dependent manner.7,38,40 However, these studies utilized donor cell suspensions generated from the entire tissue. Given that the distribution of germ cells within these whole-tissue suspensions changes with time, quantitative studies are necessary to fully appreciate changes in regenerative capacity or distribution of regenerative capacity within the germline beyond our recent studies.
2.2 |. Epigenetic modifications
Following sex determination, the germline of both sexes undergoes dynamic epigenetic shifts in a manner necessary to gain the developmental competence to ultimately form either spermatozoa or oocytes.42 Studies utilizing cloning by nuclear transfer eloquently demonstrated that a dramatic shift in developmental potential occurs in the days preceding and during prospermatogonial mitotic arrest. Germ cell nuclei from E8.5-E10.5 fetuses are fully competent to support viable offspring, while nuclei from E11.5 and later progressively lose this potential, particularly following mitotic arrest.43–46 This loss of competence is associated with the loss of parental-specific imprinting and biallelic resetting of the genome through DNA methylation.
DNA methylation levels in the germline are low following erasure that occurs within migratory PGCs, thereby leaving germ cells at a ground state for sex-specific re-methylation.47–49 Most de novo re-methylation in the male germline occurs during mitotic arrest, with a lesser fraction occurring postnatally.50,51 By contrast, DNA methylation is predominantly catalyzed postnatally in females.52 Members of the DNA methyltransferase (DNMT) family facilitate germline de novo methylation. DNMT3A and DNMT3B possess direct catalytic activity for methylation and appear to generate methylation signatures within male germ cells that are both unique and overlapping.53,54 DNMT3L acts as a non-catalytic co-factor that is also essential for germline methylation.55,56 Finally, DNMT1 passively maintains DNA methylation levels in a replication-dependent manner.57,58 PIWI-interacting RNAs (piRNAs) derived from repeat sequences direct de novo DNA methylation in the male germline.59 DNA methylation not only re-establishes genomic imprints, but also silences transposable elements which is key to maintaining genomic integrity within the germline.42 Additionally, resetting the germline epigenome is critical for early embryo totipotency. Core regulators of the pluripotency network are progressively silenced leading up to birth but must remain poised for activation following fertilization 5,60; aberrant expression or disrupted downregulation of these pluripotency factors are thought to underlie the formation of some testicular germ cell tumors.60
In addition to DNA methylation, post-translational modifications of histones are dynamically altered during prospermatogonial mitotic arrest. Limited studies have evaluated histone modifications during germline quiescence. However, studies utilizing immunostaining have evaluated global changes in histone marks within Prospg. For example, H3K4me3, H3K9me3, H3K27me3, and H3K79me2/3 increase in the days preceding or during mitotic arrest through temporally distinct patterns.61,62 While the functional significance of these histone modifications is not currently understood, studies suggest that histone marks may target DNA methylation to specific genomic loci.50 Furthermore, these marks may underlie broader shifts in gene expression coincident with developmental milestones that occur during mitotic arrest. H3K4me3 is generally activating in nature and typically localizes to gene promoter regions (reviewed in63). By contrast, H3K9me3 and H3K27me3 are generally repressive, with H3K9me3 associating with permanent developmental repression and H3K27me3 linking to transcriptional silencing (reviewed in63). Future studies are needed to better understand the developmental significance of histone modification during mitotic arrest and whether epigenetic modifications may underlie differences in germline fate during perinatal development.
3 |. ESTABLISHMENT PHASE
After birth, formation of the foundational SSC population enters a second phase following fate programming. Several developmental milestones occur following birth, thus, demarcating the Programming and Establishment Phases of SSC pool formation.
3.1 |. SSC establishment
In contrast to the limited studies of ProspgT1 heterogeneity, numerous studies have reported considerable diversity within the neonatal male germline (eg P0-P3 in mice) in terms of cellular morphology and marker gene expression.19,22,64–72 In vivo protein expression of markers associated with SSC function, including RET, PAX7, GFRA1, and ID4, are heterogenous among P0-P3 germ cells.71,73–75 Likewise, expression of markers associated with differentiation, including SOHLH1, is also heterogenous within the germline at this time in development.76 The distribution of these markers among the germline is variable. For example, among P0 Prospg, ~55% of Prospg are RET+,73 ~47% are ID4+,29 and ~28% are PAX7+.71 However, the relationship between these markers has not been explored in detail. Therefore, to gain a better perspective on this heterogeneity in the germline while accounting for asynchrony during development, scRNA-seq analysis was conducted on isolated germ cells at P0 and P3 in our recent studies.29 Outcomes revealed that opposing transcriptome signatures, either SSC- or differentiation/progenitorassociated, that are present during prenatal development (E16.5) are maintained at P0 and P3. Taken together, these results suggest that two distinct lineages can be identified shortly after birth based on gene expression. The pressing question is whether this heterogeneity represents differences in function and fate or merely temporal variances in gene expression?
Fortunately, past functional studies employing lineage tracing have begun addressing this question. Lineage tracing in animal models utilizes reporter transgenes that are both inducible and inherited.77 The most common form of lineage tracing utilizes tamoxifen-inducible Cre systems (CreERT2) that recombine floxed expression cassettes to drive constitutive expression of either fluorescent or biochemical reporters, thereby “marking” cells that express the Cre and any daughter cells derived from the labeled parent cell. Using this technology, a few studies have demonstrated that stem cell fate lies within neonatal Prospg that express Id4 and Pax7.
With a CreERT2 expression cassette inserted within the endogenous Id4 locus, Sun et al (2015) demonstrated that injection of P0 mice with tamoxifen generated labeled clones of complete spermatogenesis that persisted long into adulthood.78 Likewise, sustained clones of spermatogenesis were observed with tamoxifen administration between P1 and P3 in transgenic animals with CreERT2 within the Pax7 locus.71 Given that continuous spermatogenesis is derived from SSCs, these studies demonstrate that Prospg expressing Id4 or Pax7 between P0 and P3 are fated to the SSC pool. Furthermore, given the heterogenous expression of both ID4 and PAX7, these studies support the notion that a subset of the P0-P3 germline is fated to the SSC lineage. However, the dynamics and timing for which the SSC population is established was not determined in previous studies.
Prior studies utilizing an Id4-eGfp transgenic mouse line have shown that regenerative capacity in the germline from P6-P8 is highly concentrated within those germ cells expressing the highest levels of Id4, identified as the ID4-eGFPBright fraction.32 Therefore, to better understand the kinetics underlying formation of the SSC population while accounting for temporal shifts in marker gene expression, both ID4-eGFP fluorescence and the distribution of ID4-eGFP+ germ cells were quantified using flow cytometric analysis.29 Outcomes revealed that ID4-eGFP expression is progressively upregulated from P0-P3, such that the germline can be subdivided into Bright, Mid, Dim, and Negative fractions based on eGFP fluorescence. The ID4-eGFPBright fraction is a subset of the germ cell population from P0-P3. Strikingly, the size of the ID4-eGFPBright population reaches an upper maximum of ~12 500 cells per testis (or ~25 000 per animal) at P3 that remains constant long into adulthood. Thus, based on tracking ID4-eGFP expression and distribution within the heterogenous germ cell population, the SSC population is fully established at P3. Furthermore, while the adoption of different fates within the germline occurs during the Programming phase of SSC formation, those fates appear distinct by birth and the foundational SSC population is thus established from P0 to P3.
In addition to lineage tracing, transplantation analyses have indirectly demonstrated that the SSC population is established at P3. Studies of Shinohara et al (2001) found that the germ cell population from neonatal mice (P0-P2) generated 4-fold less spermatogenic colonies following transplantation into an adult recipient testis compared to the population in pre-pubertal pups (P6).40 Unfortunately, developmental ages between P2 and P6 were not assessed, including P3. While further transplantation studies are necessary to evaluate regenerative content during neonatal development surrounding P3, these data align with the conclusion that the SSC population is established at P3. Beyond P3, little is understood regarding the maturation and dynamics of the SSC population postnatally. Transplantation analyses by Shinohara et al (2001) reported that SSC number is higher in adult cryptorchid testes compared to P6 pups 40; however, cryptorchid testes are enriched for SSCs compared to wild-type adults.79 Studies by Nagano (2003) using transplantation analyses concluded that 3000–6000 SSCs are present in an adult mouse testis.80 This value subtly contrasts with the reported 12 500 ID4-eGFPBright germ cells in our recent studies, which may reflect technical limitations underlying transplantation or potential shifts in the SSC population during postnatal development. Indeed, transcriptomic studies have demonstrated significant differences in the gene expression signatures of spermatogonial populations that are seemingly enriched for SSCs in pre-pubertal versus adult mice69,81 that may underlie functional differences between pup and adult SSCs, including proliferative potential.29 However, considering that the spermatogonial populations studied from adult and pre-pubertal mice have appreciably different functional capacities to behave as SSCs in a transplantation assay (eg the ID4-eGFBright population in pre-pubertal testes produces ~ 2.5-fold more colonies of donor-derived spermatogenesis compared to the TertHi-GFRα1+ population in adult testes), further studies are necessary to enumerate SSCs during development and understand how differences in transcriptome profiles align with biological functions throughout postnatal development.
Interestingly, studies by Aloisio et al (2014) reported that expression of Pax7, which labels rapidly dividing SSCs in adult testes, is suppressed in Prospg during fetal development from E11.5-E18.5.71 Outcomes of recent scRNA-seq analysis of isolated mouse germ cells at E16.5, P0, P3, and P6 not only confirmed that Pax7 expression initiates after birth, but also revealed that numerous SSC-associated genes are either dramatically upregulated or initiated at this time as well,29 including Bcl6b, Bmi1, Eomes, Gfra1, T, and Shisa6.68,72,82–84 Furthermore, mapping of temporal shifts in gene expression within the SSC lineage using trajectory inference modeling revealed a massive shift in gene expression around the time of birth and prior to SSC establishment such that > 4500 transcripts are significantly upregulated during this time.29 Gene ontology terms associated with these transcripts include numerous transcription factors, receptors, and metabolic regulators. Thus, P0 marks a dramatic shift in the expression of genes underlying SSC function.
3.2 |. Proliferative and population dynamics
In mammals, quiescent Prospg initiate re-entry to the cell cycle after birth. Studies utilizing radioactive thymidine, EdU, or BrdU incorporation, which label cells following S-phase DNA replication, determined that active cell divisions are first detectable at P1 in mice with a small percentage of labeled Prospg, but proliferation markedly increases between P2 and P3.15,25,85 Similar results have been reported with immunolabeling for the mitotic marker Ki67.86 As with other aspects of germline development, postnatal re-entry into the cell cycle occurs on different timescales depending on the species. Outside the mouse, Prospg begin to resume proliferation a day later (P2) in rats,87 sometime prior to P30 in pigs,88 4 weeks postnatally in cattle,28 and approximately 8 weeks of age in humans (reviewed in18,89).
Interestingly, our recent studies showed that activation of proliferation differs among Prospg based on predicted postnatal fate.29 The change in proliferation that occurs between P2 and P3 in mice is not uniform across the entire germ cell population. Specifically, the ID4-eGFPBright population that presumably labels the SSC lineage appears to re-enter the cell cycle with greatest proliferative index among the germline and rapidly expands by > 4-fold between P2 and P3, which parallels transplantation studies performed by Shinohara et al (2001) that demonstrated an equivalent increase regenerative capacity between P0-P2 and P6.40 In contrast to the ID4-eGFPBright population, postnatal germ cells with lower ID4-eGFP expression, which presumably marks germ cells transitioning out of the SSC pool, possess significantly lower proliferative activity between P2 and P3 and expansion of this population does not initiate until after the ID4-eGFPBright population reaches maximal size at P3. Thus, while all Prospg in the testes of mice re-enter the cell cycle postnatally over the course of approximately 2–3 developmental days, proliferative potential may be unique or enhanced within the SSC lineage.
Unexpectedly, while the total number of ID4-eGFPBright cells reaches steady-state at P3, the proliferative index of this population does not significantly decline until after P7, with > 50% of the population in S, G2, or M phases of the cell cycle.29 Thus, why are ID4-eGFPBright actively cycling when the population is fully established? Based on population kinetics, growth of the ID4-eGFPMid population trails establishment of the ID4-eGFPBright pool. Therefore, the combination of population dynamics and proliferative indices suggest a shift in the symmetry of cell divisions among ID4-eGFPBright spermatogonia, from symmetric to asymmetric, in order to build a progenitor pool that will serve as a source for successive rounds of spermatogenesis. Likewise, the ID4-eGFPDim population expands after formation of the ID4-eGFPMid population until reaching an upper maximum around P9. This layering effect indicates that once the SSC population is established at P3, subsequent layers of spermatogonia assemble in a top-down fashion, starting with progenitors derived from the SSC pool via asymmetric division. Collectively, further experimentation is needed to confirm these predictions.
3.3 |. Migration
In addition to re-entering the cell cycle, Prospg transit from the center of seminiferous cords to the basement membrane during the first days postnatally.15,16,90,91 Neonatal migration is associated with an increased formation of pseudopodia among germ cells and intimate cell-to-cell contacts with adjacent Sertoli cells.90,91 Approximately half of neonatal Prospg form pseudopodia, and as opposed to rounded germ cells, pseudopod-shaped Prospg are capable of recolonizing the testes of recipients upon transplantation, indicating that migration is a critical milestone in formation of the SSC population.64 Platelet-derived growth factor and Notch signaling have been implicated in the regulation of Prospg migration92,93; however, the precise mechanisms that guide colonization of the basement are not completely understood. Once germ cells adhere to the basement membrane, a distinct shift in morphology occurs to resemble spermatogonia in the mature testis. Some consider this change in appearance sufficient to identify the transition from Prospg to spermatogonia; however, our recent findings suggest that active and functional changes in proliferation better define this event (discussed above). The purpose of migration is not completely understood. However, a portion of the germ cell population is cleared from the testis during neonatal development, particularly those that fail to migrate and are round in appearance.64,94 Thus, fitness for migration to the basement membrane and therefore proper placement within the seminiferous epithelium may select for germ cells that will ultimately form the postnatal spermatogonial lineage and serve as the source for continual spermatogenesis throughout adulthood.
Recent studies have postulated that another subtype of Prospg is present in the neonatal male germline, referred to as “intermediate” or “I-Prospg,” that possesses unique migratory characteristics but lacks the proliferative phenotype of ProspgT2.30 While this is a tempting speculation, it assumes a clear distinction between ProspgT1 and this intermediate population, which is difficult to pinpoint. First, some ProspgT1 initiate the luminal to peripheral migration prior to birth,15,19 indicating that ProspgT1 also functionally possess migratory capacity. Second, there is clear evidence that proliferation and migration are independent events in germline development because proliferative activation, which marks the transition to ProspgT2, occurs in both luminal and peripheral Prospg during neonatal development 15; thus, ProspgT2 also possess migratory capacity. Interestingly, Kun et al (2020) reported that this intermediate population possesses a distinct transcriptional signature from ProspgT1 based on scRNA-seq analysis.30 However, the intermediate population was only detected at P2, and other studies,29,95–97 indicate that there are major differences in gene expression between developmental ages such that ages segregate based on global transcriptional signature after dimensional reduction strategies such as PCA, graph-based clustering, tSNE, and/or UMAP. Therefore, a unique gene expression signature present within a single developmental age seems insufficient for defining a unique population. Collectively, neither migratory behavior nor transcriptome signature appear to define a functionally unique, intermediate subset of Prospg whose characteristics more closely align with postnatal ProspgT1. Ultimately, defining prospermatogonial subtypes based on proliferation alone is not only logical, but also prevents over-complicating what is already a heavily debated nomenclature system.17,18
3.4 |. Nesting of germ cells
PGCs that colonize the genital ridge undergo successive rounds of mitosis and form aggregates or clusters of germ cells (reviewed in98). In the developing fetal ovary, aggregates of germ cells form nests with somatic cells, and within these nests, oogonia are fated to either form primary follicles or succumb for atresia (reviewed in99). Interestingly, Prospg with similar levels of ID4-eGFP expression cluster together within distinct regions along the length of seminiferous cords, forming nests of germ cells akin to oogonia in the developing ovary.29 Quantitative assessment of this clustering and associated dynamics through developmental time revealed that > 97% ID4-eGFPBright Prospg from P0 to P2 localize to nests with an average size of ~15 cells per nest. At P2, ~210 ID4eGFPBright nests were present throughout the entire testis. During expansion of the ID4-eGFPBright population from P2 to P3, the average ID4-eGFPBright nest size does not change but the number of total nests dramatically increases, indicating the start of nest breakdown. In subsequent postnatal days, nests continue to disperse throughout the tissue mass. Similar to the ID4-eGFPBright population, ~75% of ID4-eGFPMid Prospg are present in nests. However, breakdown of ID4-eGFPMid nests initiates around P1-P2. Given that nests of ID4-eGFPBright germ cells were sustained leading up to establishment of the SSC population at P3, these findings suggest that all cells within a nest adopt a common fate. By contrast, fate determination between primary and apoptotic follicles among oogonia occurs within nests via a nursing mechanism (reviewed in100). Collectively, it appears as though nesting behavior during development, albeit in different forms, may be a conserved feature of both male and female germline establishment.
4 |. ADDITIONAL FATES IN THE NEONATAL GERMLINE
While establishment of the foundational SSC population has been the foremost focus of recent studies on male germline development, interesting patterns of fate during neonatal development outside the SSC lineage have also been made.29,69,70,101 Given that a subset of the germline forms the SSC pool (ID4-eGFPBright germ cells), what is the fate of the remaining populations? Evaluating marker gene expression and population distribution reveals insights into the complexity within the neonatal germline. First, at P3 ~67% of the germline constitutes ID4-eGFPBright SSCs.29 Approximately 17% of germ cells are ID4-eGFPDim/Negative, ~93% of which becomes KIT+ in response to RA signaling that initiates at P3 and forms an initial differentiating spermatogonial pool.21,29 If SSC-derived progenitors, classically labeled by NGN3 expression, emerge around P4 and after establishment of the SSC population,22,65 what is the identity of the remaining ~16% of the germline at P3? Using scRNA-seq, SSC, progenitor, and differentiating spermatogonial populations can be identified based on their distribution within the germline and/or gene expression signatures. Interestingly, a fourth population is present around P3 that does not express genes necessary for SSC function at the same level as established SSCs while also lacking Ngn3, Stra8, and Kit expression.29 Therefore, this population may represent a pool of initial progenitor-like cells present at P3 derived from Prospg that neither form SSCs nor initially respond to RA and enter differentiation; rather, these cells may be poised to differentiate following the first round of spermatogenesis, akin to a second round of spermatogenesis. Although further experimentation is necessary to validate the presence of this population, these results underscore the complexity of cell subtype development in the germline during neonatal life.
5 |. CONCLUSION
Discoveries in the mouse over the last few years have greatly advanced our knowledge of the dynamics and mechanisms that guide formation of the foundational SSC population. Fortunately, the kinetics underlying SSC formation are accelerated in the mouse relative to higher order mammalian species, thus providing an experimentally tractable model for stepwise sampling and analysis of the germline during late fetal and neonatal development that enables stitching together of maps that explain the process on a basic level. By contrast, this can also make interpreting findings complicated as developmental milestones are often concurrent. For example, establishment of the SSC population overlaps with the onset of RA signaling and the formation of the first differentiating spermatogonia in mice which may not be the case in other mammals in which the timeline of development is extended. Ultimately, how the dynamics of SSC establishment translate from rodents to humans and livestock remains an ongoing area of interest. Histomorphological studies of pre-pubertal human tissues describe the formation of differentiating spermatogonia prior to the onset of puberty as well as heterogeneity in the appearance of newborn germ cells102; both observations are comparable to those reported in rodents and further suggest the potential for conserved mechanisms of fate determination. Additionally, recent studies utilizing scRNA-seq analysis of human germ cells from neonatal and infant tissues provide key resources to begin understanding formation of the foundational SSC population in humans.103,104 By contrast, studies in pre-pubertal domestic animal species are even more limited and represent an area ripe for generation of new knowledge.
ACKNOWLEDGEMENTS
This work was funded by the National Institute of Childhood Health and Disease grant HD061665 awarded to JMO
Funding information
Eunice Kennedy Shriver National Institute of Child Health and Human Development, Grant/Award Number: HD061665
REFERENCES
- 1.Santos AC, Lehmann R. Germ cell specification and migration in Drosophila and beyond. Curr Biol. 2004;14(14):R578–R589. [DOI] [PubMed] [Google Scholar]
- 2.Ohinata Y, Payer B, O’Carroll D, et al. Blimp1 is a critical determinant of the germ cell lineage in mice. Nature. 2005;436(7048):207–213. [DOI] [PubMed] [Google Scholar]
- 3.Kobayashi T, Zhang H, Tang WWC, et al. Principles of early human development and germ cell program from conserved model systems. Nature. 2017;546(7658):416–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leitch HG, Tang WW, Surani MA. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr Top Dev Biol. 2013;104:149–187. [DOI] [PubMed] [Google Scholar]
- 5.Magnusdottir E, Surani MA. How to make a primordial germ cell. Development. 2014;141(2):245–252. [DOI] [PubMed] [Google Scholar]
- 6.Richardson BE, Lehmann R. Mechanisms guiding primordial germ cell migration: strategies from different organisms. Nat Rev Mol Cell Biol. 2010;11(1):37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuma S, Kanatsu-Shinohara M, Inoue K, et al. Spermatogenesis from epiblast and primordial germ cells following transplantation into postnatal mouse testis. Development. 2005;132(1):117–122. [DOI] [PubMed] [Google Scholar]
- 8.Stevens LC. Experimental production of testicular teratomas in mice. Proc Natl Acad Sci USA. 1964;52:654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70(5):841–847. [DOI] [PubMed] [Google Scholar]
- 10.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359(6395):550–551. [DOI] [PubMed] [Google Scholar]
- 11.Hargan-Calvopina J, Taylor S, Cook H, et al. Stage-specific demethylation in primordial germ cells safeguards against precocious differentiation. Dev Cell. 2016;39(1):75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cinalli RM, Rangan P, Lehmann R. Germ cells are forever. Cell. 2008;132(4):559–562. [DOI] [PubMed] [Google Scholar]
- 13.Nicholls PK, Schorle H, Naqvi S, et al. Mammalian germ cells are determined after PGC colonization of the nascent gonad. Proc Natl Acad Sci USA. 2019;116(51):25677–25687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowles J, Koopman P. Sex determination in mammalian germ cells: extrinsic versus intrinsic factors. Reproduction. 2010;139(6):943–958. [DOI] [PubMed] [Google Scholar]
- 15.Nagano R, Tabata S, Nakanishi Y, Ohsako S, Kurohmaru M, Hayashi Y. Reproliferation and relocation of mouse male germ cells (gonocytes) during prespermatogenesis. Anat Rec. 2000;258(2):210–220. [DOI] [PubMed] [Google Scholar]
- 16.McGuinness MP, Orth JM. Reinitiation of gonocyte mitosis and movement of gonocytes to the basement membrane in testes of newborn rats in vivo and in vitro. Anat Rec. 1992;233(4):527–537. [DOI] [PubMed] [Google Scholar]
- 17.McCarrey JR. Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biol Reprod. 2013;89(2):47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonocytes Culty M., from the fifties to the present: is there a reason to change the name? Biol Reprod. 2013;89(2):46. [DOI] [PubMed] [Google Scholar]
- 19.Kluin PM, de Rooij DG. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int J Androl. 1981;4(4):475–493. [DOI] [PubMed] [Google Scholar]
- 20.Geyer CB. Setting the Stage: The First Round of Spermatogenesis. New York, NY: Springer; 2017. [Google Scholar]
- 21.Griswold MD. Spermatogenesis: the commitment to meiosis. Physiol Rev. 2016;96(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshida S, Sukeno M, Nakagawa T, et al. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133(8):1495–1505. [DOI] [PubMed] [Google Scholar]
- 23.Western PS, Miles DC, van den Bergen JA, Burton M, Sinclair AH. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells. 2008;26(2):339–347. [DOI] [PubMed] [Google Scholar]
- 24.Hilscher B, Engemann A. Histological and morphometric studies on the kinetics of germ cells and immature Sertoli cells during human prespermatogenesis. Andrologia. 1992;24(1):7–10. [DOI] [PubMed] [Google Scholar]
- 25.Vergouwen RP, Jacobs SG, Huiskamp R, Davids JA, de Rooij DG. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J Reprod Fertil. 1991;93(1):233–243. [DOI] [PubMed] [Google Scholar]
- 26.Franke FE, Pauls K, Rey R, Marks A, Bergmann M, Steger K. Differentiation markers of Sertoli cells and germ cells in fetal and early postnatal human testis. Anat Embryol (Berl). 2004;209(2):169–177. [DOI] [PubMed] [Google Scholar]
- 27.Ketola I, Toppari J, Vaskivuo T, Herva R, Tapanainen JS, Heikinheimo M. Transcription factor GATA-6, cell proliferation, apoptosis, and apoptosis-related proteins Bcl-2 and Bax in human fetal testis. J Clin Endocrinol Metab. 2003;88(4):1858–1865. [DOI] [PubMed] [Google Scholar]
- 28.Wrobel KH. Prespermatogenesis and spermatogoniogenesis in the bovine testis. Anat Embryol (Berl). 2000;202(3):209–222. [DOI] [PubMed] [Google Scholar]
- 29.Law NC, Oatley MJ, Oatley JM. Developmental kinetics and transcriptome dynamics of stem cell specification in the spermatogenic lineage. Nat Commun. 2019;10(1):2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan K, Song HW, Wilkinson MF. Single-cell RNAseq analysis of testicular germ and somatic cell development during the perinatal period. Development. 2020;147(3):dev183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen C, Ouyang W, Grigura V, et al. ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature. 2005;436(7053):1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Helsel AR, Yang QE, Oatley MJ, Lord T, Sablitzky F, Oatley JM. ID4 levels dictate the stem cell state in mouse spermatogonia. Development. 2017;144(4):624–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oatley JM, Avarbock MR, Brinster RL. Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J Biol Chem. 2007;282(35):25842–25851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jain S, Naughton CK, Yang M, et al. Mice expressing a dominant-negative Ret mutation phenocopy human Hirschsprung disease and delineate a direct role of Ret in spermatogenesis. Development. 2004;131(21):5503–5513. [DOI] [PubMed] [Google Scholar]
- 35.Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294(1):161–167. [DOI] [PubMed] [Google Scholar]
- 36.Laronda MM, Jameson JL. Sox3 functions in a cell-autonomous manner to regulate spermatogonial differentiation in mice. Endocrinology. 2011;152(4):1606–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helsel AR, Oatley JM. Transplantation as a quantitative assay to study mammalian male germline stem cells. Methods Mol Biol. 2017;1463:155–172. [DOI] [PubMed] [Google Scholar]
- 38.Ohta H, Wakayama T, Nishimune Y. Commitment of fetal male germ cells to spermatogonial stem cells during mouse embryonic development. Biol Reprod. 2004;70(5):1286–1291. [DOI] [PubMed] [Google Scholar]
- 39.McLean DJ, Friel PJ, Johnston DS, Griswold MD. Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol Reprod. 2003;69(6):2085–2091. [DOI] [PubMed] [Google Scholar]
- 40.Shinohara T, Orwig KE, Avarbock MR, Brinster RL. Remodeling of the postnatal mouse testis is accompanied by dramatic changes in stem cell number and niche accessibility. Proc Natl Acad Sci USA. 2001;98(11):6186–6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oatley MJ, Kaucher AV, Racicot KE, Oatley JM. Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol Reprod. 2011;85(2):347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9(2):129–140. [DOI] [PubMed] [Google Scholar]
- 43.Kato Y, Rideout WM 3rd, Hilton K, Barton SC, Tsunoda Y, Surani MA. Developmental potential of mouse primordial germ cells. Development. 1999;126(9):1823–1832. [DOI] [PubMed] [Google Scholar]
- 44.Lee J, Inoue K, Ono R, et al. Erasing genomic imprinting memory in mouse clone embryos produced from day 11.5 primordial germ cells. Development. 2002;129(8):1807–1817. [DOI] [PubMed] [Google Scholar]
- 45.Yamazaki Y, Mann MRW, Lee SS, et al. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci USA. 2003;100(21):12207–12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miki H, Inoue K, Kohda T, et al. Birth of mice produced by germ cell nuclear transfer. Genesis. 2005;41(2):81–86. [DOI] [PubMed] [Google Scholar]
- 47.Lane N, Dean W, Erhardt S, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35(2):88–93. [DOI] [PubMed] [Google Scholar]
- 48.Seisenberger S, Andrews S, Krueger F, et al. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48(6):849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 2005;278(2):440–458. [DOI] [PubMed] [Google Scholar]
- 50.Morselli M, Pastor WA, Montanini B, et al. In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. Elife. 2015;4:e06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Molaro A, Falciatori I, Hodges E, et al. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 2014;28(14):1544–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smallwood SA, Tomizawa S-I, Krueger F, et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet. 2011;43(8):811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato Y, Kaneda M, Hata K, et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet. 2007;16(19):2272–2280. [DOI] [PubMed] [Google Scholar]
- 54.Kaneda M, Okano M, Hata K, et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429(6994):900–903. [DOI] [PubMed] [Google Scholar]
- 55.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotrans-poson reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431(7004):96–99. [DOI] [PubMed] [Google Scholar]
- 56.La Salle S, Oakes CC, Neaga OR, Bourc’his D, Bestor TH, Trasler JM. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev Biol. 2007;7:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ohno R, Nakayama M, Naruse C, et al. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development. 2013;140(14):2892–2903. [DOI] [PubMed] [Google Scholar]
- 58.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe T, Tomizawa S-I, Mitsuya K, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332(6031):848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Clark AT. The stem cell identity of testicular cancer. Stem Cell Rev. 2007;3(1):49–59. [DOI] [PubMed] [Google Scholar]
- 61.Abe M, Tsai SY, Jin SG, Pfeifer GP, Szabo PE. Sex-specific dynamics of global chromatin changes in fetal mouse germ cells. PLoS ONE. 2011;6(8):e23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshioka H, McCarrey JR, Yamazaki Y. Dynamic nuclear organization of constitutive heterochromatin during fetal male germ cell development in mice. Biol Reprod. 2009;80(4):804–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lawrence M, Daujat S, Schneider R. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. [DOI] [PubMed] [Google Scholar]
- 64.Orwig KE, Ryu BY, Avarbock MR, Brinster RL. Male germ-line stem cell potential is predicted by morphology of cells in neonatal rat testes. Proc Natl Acad Sci USA. 2002;99(18):11706–11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida S, Takakura A, Ohbo K, et al. Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev Biol. 2004;269(2):447–458. [DOI] [PubMed] [Google Scholar]
- 66.Yoshida S, Nabeshima Y, Nakagawa T. Stem cell heterogeneity: actual and potential stem cell compartments in mouse spermatogenesis. Ann N Y Acad Sci. 2007;1120:47–58. [DOI] [PubMed] [Google Scholar]
- 67.Suzuki H, Sada A, Yoshida S, Saga Y. The heterogeneity of spermatogonia is revealed by their topology and expression of marker proteins including the germ cell-specific proteins Nanos2 and Nanos3. Dev Biol. 2009;336(2):222–231. [DOI] [PubMed] [Google Scholar]
- 68.Nakagawa T, Sharma M, Nabeshima Y, Braun RE, Yoshida S. Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science. 2010;328(5974):62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hermann BP, Mutoji KN, Velte EK, et al. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod. 2015;92(2):54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction. 2015;149(4):329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aloisio GM, Nakada Y, Saatcioglu HD, et al. PAX7 expression defines germline stem cells in the adult testis. J Clin Invest. 2014;124(9):3929–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Komai Y, Tanaka T, Tokuyama Y, et al. Bmi1 expression in long-term germ stem cells. Sci Rep. 2014;4:6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naughton CK, Jain S, Strickland AM, Gupta A, Milbrandt J. Glial cell-line derived neurotrophic factor-mediated RET signaling regulates spermatogonial stem cell fate. Biol Reprod. 2006;74(2):314–321. [DOI] [PubMed] [Google Scholar]
- 74.Chan F, Oatley MJ, Kaucher AV, et al. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28(12):1351–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pui HP, Saga Y. Gonocytes-to-spermatogonia transition initiates prior to birth in murine testes and it requires FGF signaling. Mech Dev. 2017;144(Pt B):125–139. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki H, Dann CT, Rajkovic A. Generation of a germ cell-specific mouse transgenic CHERRY reporter, Sohlh1-mCherryFlag. Genesis. 2013;51(1):50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148(1–2):33–45. [DOI] [PubMed] [Google Scholar]
- 78.Sun F, Xu Q, Zhao D, Degui CC. Id4 marks spermatogonial stem cells in the mouse testis. Sci Rep. 2015;5:17594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shinohara T, Avarbock MR, Brinster RL. Functional analysis of spermatogonial stem cells in Steel and cryptorchid infertile mouse models. Dev Biol. 2000;220(2):401–411. [DOI] [PubMed] [Google Scholar]
- 80.Nagano MC. Homing efficiency and proliferation kinetics of male germ line stem cells following transplantation in mice. Biol Reprod. 2003;69(2):701–707. [DOI] [PubMed] [Google Scholar]
- 81.Garbuzov A, Pech MF, Hasegawa K, et al. Purification of GFRalpha1+ and GFRalpha1- spermatogonial stem cells reveals a niche-dependent mechanism for fate determination. Stem Cell Reports. 2018;10(2):553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sharma M, Srivastava A, Fairfield HE, Bergstrom D, Flynn WF, Braun RE. Identification of EOMES-expressing spermatogonial stem cells and their regulation by PLZF. eLife. 2019;8:pii:e43352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tokue M, Ikami K, Mizuno S, et al. SHISA6 confers resistance to differentiation-promoting Wnt/beta-catenin signaling in mouse spermatogenic stem cells. Stem Cell Reports. 2017;8(3):561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA. 2006;103(25):9524–9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang QE, Gwost I, Oatley MJ, Oatley JM. Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol Reprod. 2013;89(5):113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lei N, Hornbaker KI, Rice DA, Karpova T, Agbor VA, Heckert LL. Sex-specific differences in mouse DMRT1 expression are both cell type- and stage-dependent during gonad development. Biol Reprod. 2007;77(3):466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Boulogne B, Olaso R, Levacher C, Durand P, Habert R. Apoptosis and mitosis in gonocytes of the rat testis during foetal and neonatal development. Int J Androl. 1999;22(6):356–365. [DOI] [PubMed] [Google Scholar]
- 88.Franca LR, Silva VA Jr, Chiarini-Garcia H, Garcia SK, Debeljuk L. Cell proliferation and hormonal changes during postnatal development of the testis in the pig. Biol Reprod. 2000;63(6):1629–1636. [DOI] [PubMed] [Google Scholar]
- 89.Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res C Embryo Today. 2009;87(1):1–26. [DOI] [PubMed] [Google Scholar]
- 90.Orth JM, Qiu J, Jester WF Jr, Pilder S. Expression of the c-kit gene is critical for migration of neonatal rat gonocytes in vitro. Biol Reprod. 1997;57(3):676–683. [DOI] [PubMed] [Google Scholar]
- 91.Clermont Y, Perey B. Quantitative study of the cell population of the seminiferous tubules in immature rats. Am J Anat. 1957;100(2):241–267. [DOI] [PubMed] [Google Scholar]
- 92.Garcia TX, DeFalco T, Capel B, Hofmann MC. Constitutive activation of NOTCH1 signaling in Sertoli cells causes gonocyte exit from quiescence. Dev Biol. 2013;377(1):188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Basciani S, De Luca G, Dolci S, et al. Platelet-derived growth factor receptor beta-subtype regulates proliferation and migration of gonocytes. Endocrinology. 2008;149(12):6226–6235. [DOI] [PubMed] [Google Scholar]
- 94.Rodriguez I, Ody C, Araki K, Garcia I, Vassalli P. An early and massive wave of germinal cell apoptosis is required for the development of functional spermatogenesis. EMBO J. 1997;16(9):2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song H-W, Bettegowda A, Lake B, et al. The homeobox transcription factor RHOX10 drives mouse spermatogonial stem cell establishment. Cell Rep. 2016;17(1):149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grive KJ, Hu Y, Shu E, et al. Dynamic transcriptome profiles within spermatogonial and spermatocyte populations during postnatal testis maturation revealed by single-cell sequencing. PLoS Genet. 2019;15(3):e1007810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Makino Y, Jensen NH, Yokota N, et al. Single cell RNA-sequencing identified Dec2 as a suppressive factor for spermatogonial differentiation by inhibiting Sohlh1 expression. Sci Rep. 2019;9(1):6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mork L, Tang H, Batchvarov I, Capel B. Mouse germ cell clusters form by aggregation as well as clonal divisions. Mech Dev. 2012;128(11–12):591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tingen C, Kim A, Woodruff TK. The primordial pool of follicles and nest breakdown in mammalian ovaries. Mol Hum Reprod. 2009;15(12):795–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu K, Jensen L, Lei L, Yamashita YM. Stay connected: a germ cell strategy. Trends Genet. 2017;33(12):971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Velte EK, Niedenberger BA, Serra ND, et al. Differential RA responsiveness directs formation of functionally distinct spermatogonial populations at the initiation of spermatogenesis in the mouse. Development. 2019;146(12):dev173088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Paniagua R, Nistal M. Morphological and histometric study of human spermatogonia from birth to the onset of puberty. J Anat. 1984;139(Pt 3):535–552. [PMC free article] [PubMed] [Google Scholar]
- 103.Sohni A, Tan K, Song H-W, et al. The Neonatal and Adult Human Testis Defined at the Single-Cell Level. Cell Rep. 2019;26(6):1501–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Guo J, Grow EJ, Mlcochova H, et al. The adult human testis transcriptional cell atlas. Cell Res. 2018;28(12):1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]


