Significance
The crystal structures of photosynthetic reaction centers from purple bacteria (PbRCs) and photosystem II show large structural similarity. However, the proposed mechanisms of proton transfer toward the terminal electron acceptor quinone (QB) are not consistent. In particular, not His-L190, which is an H-bond partner of QB, but rather Glu-L212, which is ∼6 Å away from QB, was assumed to be the direct proton donor for QB. We demonstrate that the H-bond between His-L190 and QB is a low-barrier H-bond, which facilitates proton transfer from singly protonated His-L190 to QB. Furthermore, Glu-L212 is not a direct H-bond donor for QB. However, it facilitates proton transfer toward deprotonated His-L190 via water molecules after QBH2 forms and leaves the PbRC.
Keywords: proton-coupled electron transfer, low-barrier hydrogen bond, photosystem II, conformational gating, artificial photosynthesis
Abstract
In photosynthetic reaction centers from purple bacteria (PbRCs) from Rhodobacter sphaeroides, the secondary quinone QB accepts two electrons and two protons via electron-coupled proton transfer (PT). Here, we identify PT pathways that proceed toward the QB binding site, using a quantum mechanical/molecular mechanical approach. As the first electron is transferred to QB, the formation of the Grotthuss-like pre-PT H-bond network is observed along Asp-L213, Ser-L223, and the distal QB carbonyl O site. As the second electron is transferred, the formation of a low-barrier H-bond is observed between His-L190 at Fe and the proximal QB carbonyl O site, which facilitates the second PT. As QBH2 leaves PbRC, a chain of water molecules connects protonated Glu-L212 and deprotonated His-L190 forms, which serves as a pathway for the His-L190 reprotonation. The findings of the second pathway, which does not involve Glu-L212, and the third pathway, which proceeds from Glu-L212 to His-L190, provide a mechanism for PT commonly used among PbRCs.
Purple bacterial photosynthetic reaction centers (PbRCs) have special pair bacteriochlorophylls (PL/PM), accessory bacteriochlorophylls (BL/BM), bacteriopheophytins (HL/HM), and ubiquinones (QA/QB) in the heterodimeric L/M protein subunit pair. PL and PM form the electronically coupled special pair [PLPM]. The electronic excitation of [PLPM] leads to the formation of the charge-separated state, [PLPM]•+BL•−, and subsequent electron transfer occurs to QB via HL and QA (1–3). QB accepts two electrons via QA and two protons via the proton transfer (PT) pathways, forming QBH2, and leaves the PbRC.
The first and second electron transfers from QA to QB occur with rates kAB(1) (104 s−1) and kAB(2) (103 s−1), respectively (4). The first PT leads to the protonation of the distal and carbonyl O site of QB (with respect to the nonheme Fe). The proton donor of the distal QB O site is Ser-L223, for which the H-bond donor is Asp-L213. The Ser-L223 side chain, which donates an H-bond with Asp-L213 due to the highly polarized carboxyl O site, reorients toward the distal QB O site in response to QB•− formation (5, 6). Notably, in photosystem II (PSII), D1-Ser264 and D1-His252, which correspond to Ser-L223 and Asp-L213 in the PbRC, respectively (7), serve as a PT pathway toward the distal QB O site (8). As D(L213)N (9) and S(L223)A (10, 11) mutations decrease the kAB(2) significantly, these residues are likely involved in the PT pathway toward the distal QB O site in the PbRC. Although Asp-H124, His-H126, and His-H128 are likely to form the entry point of the major PT pathway (12), it also seems plausible that the PT pathway is delocalized toward the protein bulk surface (13–15). The delocalization of the PT pathway is also observed in PSII; specifically, the PT pathway is branched as it proceeds from the oxygen-evolving complex via D1-Asp61 toward the protein bulk surface (16).
Glu-L212 is the titratable residue that is nearest to and a candidate for the proton donor to the proximal QB O site. Indeed, the uptake of 0.3 to 0.8 H+ by Glu-L212 has been reported upon QB•− formation (17–21). The protonation of Glu-L212 plays a role in electron transfer, increasing the redox potential Em(QB) with respect to Em(QA) (22). However, the PT pathway from Glu-L212 to QBH− is unclear (4), because the crystal structure shows that Glu-L212 cannot form an H-bond with the proximal QB O site (Glu-L212…QB = 5.7 Å) (23). PT occurs most efficiently along H-bonds (24). Okamura et al. (4) proposed that the movement of QBH− toward Glu-L212 and the formation of an H-bond might be required for PT. Alternatively, His-L190, which forms an H-bond with the proximal QB O site (His-L190…QB = 2.81 Å) (23), might serve as a proton donor for QBH−, as proposed by Wraight (25). However, Wraight also argued that the pKa value for the deprotonation of singly protonated His-L190 might be too high even in the presence of the cationic nonheme Fe.
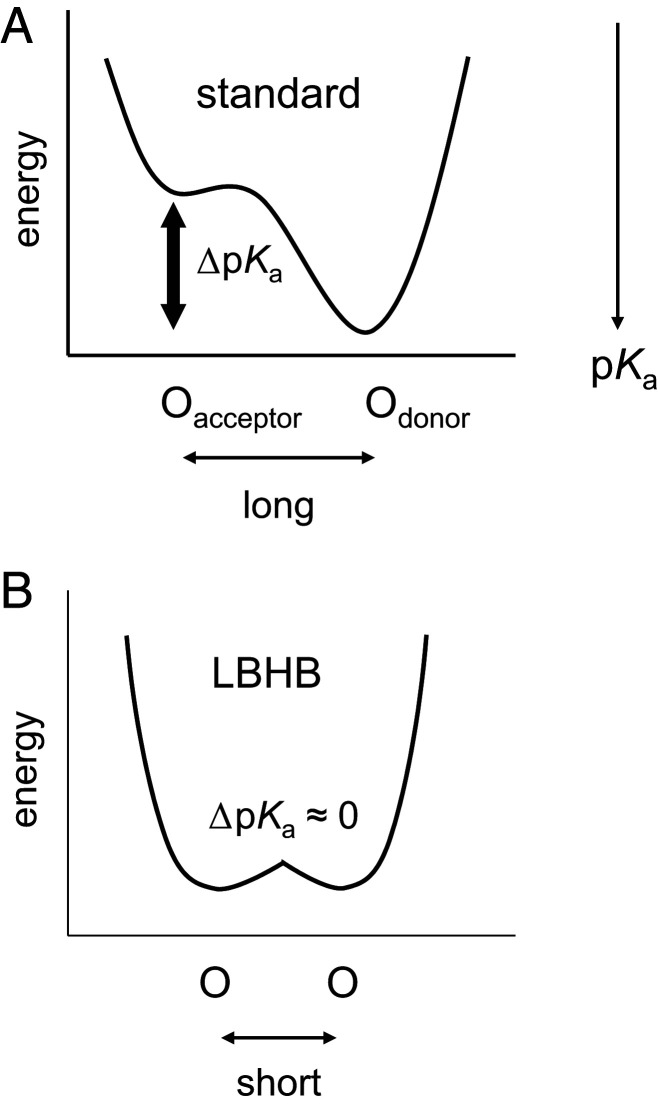
Notably, the crystal structures of PSII and PbRC show large structural similarity (7). His-L190 is conserved as D1-His215 at the nonheme Fe complex in PSII (7). In PSII, D1-His215 can form a low-barrier H-bond with QBH−, which facilitates QBH2 formation (8, 26, 27). Low-barrier H-bonds can form when the pKa values of the H-bond donor and acceptor moieties are nearly equal (28, 29). The shape of the potential energy curve of a low-barrier H-bond is symmetric, while that of a standard H-bond is asymmetric because pKa(donor) > pKa(acceptor) (30) (Fig. 1). In addition, Glu-L212 in PbRCs is not conserved as a titratable residue in PSII. These findings for PSII might provide an opportunity to revisit the mechanism of PT toward the proximal QB O site in PbRCs. Here, we report how PT pathways form in response to electron transfer in the PbRC protein environment, by adopting a large-scale quantum mechanical/molecular mechanical (QM/MM) approach based on the PbRC crystal structure (23).
Fig. 1.
Typical potential-energy profiles of H-bonds. (A) Standard H-bond. (B) Low-barrier H-bond (LBHB). In low-barrier H-bonds, the H-bond acceptor (Oacceptor) and donor (Odonor) cannot be distinguishable due to the same pKa values.
Results
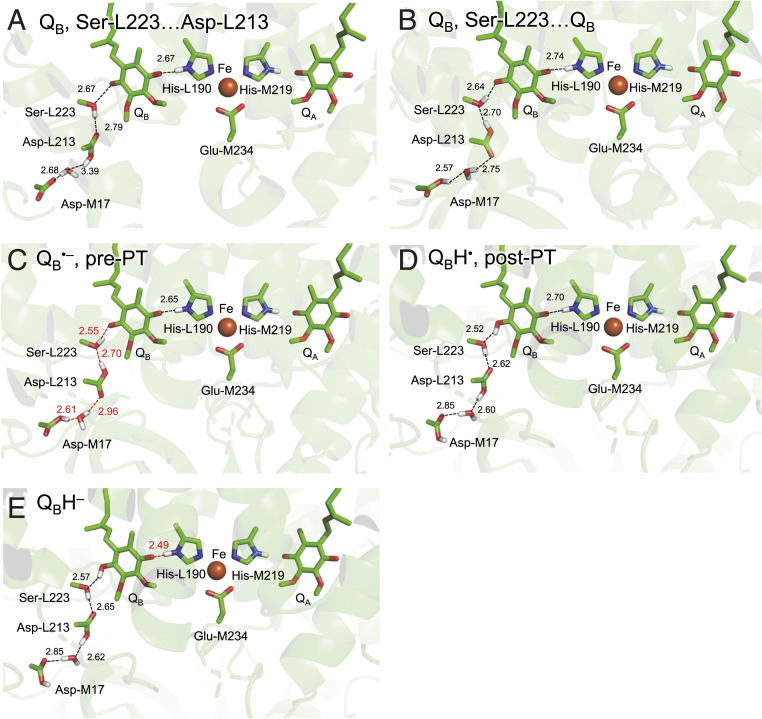
The PbRC crystal structure shows that the H-bond network proceeds from the distal QB carbonyl O site via Ser-L223, Asp-L213, a water molecule, and Asp-M17 (23) (i.e., QM region) (Fig. 2). Note that Asp-L210 cannot form an H-bond with the residues in the H-bond network, since it is more than 5.0 Å away from the nearest residue, Asp-M17 (23). When the first electron transfer occurs to QB, Glu-L212 protonates and the Ser-L223 hydroxyl group, which initially forms an H-bond with Asp-L213, reorients toward QB•− (6, 31, 32). The H-bond distances along the QB…Ser-L223…Asp-L213…H2O…Asp-M17 decrease in response to the proton uptake of Glu-L212 and the reorientation of the Ser-L223 hydroxyl group (Table 1). Specifically, the H-bond between the distal QB O site and Ser-L223 is shortened from 2.65 to 2.55 Å in response to QB•− formation (Table 1), which implies that the first PT proceeds toward the distal QB O site. The potential energy profile indicates that PT is energetically downhill from Ser-L223 to the distal QB O site in the presence of QB•− (Fig. 3A).
Fig. 2.
H-bond network of QB. (A) QB (before the first electron transfer): Ser-L223 donates an H-bond to Asp-L213. (B) QB: Ser-L223 donates an H-bond to QB. (C) QB•− (after the first electron transfer) in the pre-PT H-bond pattern. (D) QBH• in the post-PT H-bond pattern. (E) QBH− (after the second electron transfer). Black dotted lines indicate H-bonds. Red dotted lines indicate proton-conducting H-bonds. H atoms in the H-bond network are depicted as white sticks. Note that Glu-L212 is deprotonated in neutral unprotonated QB and protonated in other states.
Table 1.
H-bond distances near QB (Å)
| PT pattern | L190…QB | QB…L223 | L223…L213 | L213…H2O | H2O…M17 | |
| PDB | ||||||
| 1AIG* | 2.81 | 3.21 | 3.07 | — | — | |
| 3I4D† | 2.57 | 2.60 | 2.51 | 2.73 | 3.53 | |
| QM/MM | ||||||
| [deprotonated Glu-L212]‡ | ||||||
| QB | L223…L213§ | 2.67 | 2.67 | 2.79 | 3.39 | 2.68 |
| QB | L223…QB¶ | 2.74 | 2.64 | 2.70 | 2.75 | 2.57 |
| [protonated Glu-L212]‡ | ||||||
| QB•– | pre-PT# | 2.65 | 2.55 | 2.70 | 2.98 | 2.61 |
| QBH• | post-PT# | 2.70 | 2.52 | 2.62 | 2.60 | 2.85 |
| QBH– | pre-PT# | 2.49 | 2.57 | 2.65 | 2.62 | 2.85 |
| QBH2 | post-PT | 2.60 | 2.57 | 2.64 | 2.62 | 2.86 |
—, not applicable. Distances of low-barrier H-bonds are in bold.
Reported as the light-exposed charge-separated QB•− structure (23). The distances in the dark-adapted structure (PDB ID code 1AIJ) (23) are not shown: the QB binding site in the dark-adapted structure is not consistent with that in the light-exposed structure (23), whereas the difference in the QB binding site is not observed in a FTIR difference spectroscopy (67).
The redox state is not reported (34).
Glu-L212 is deprotonated for neutral unprotonated QB and protonated for QB•− in the present calculation.
Ser-L223 donates an H-bond to Asp-L213 (Fig. 2A).
Ser-L223 donates an H-bond to QB (Fig. 2B).
See Fig. 2.
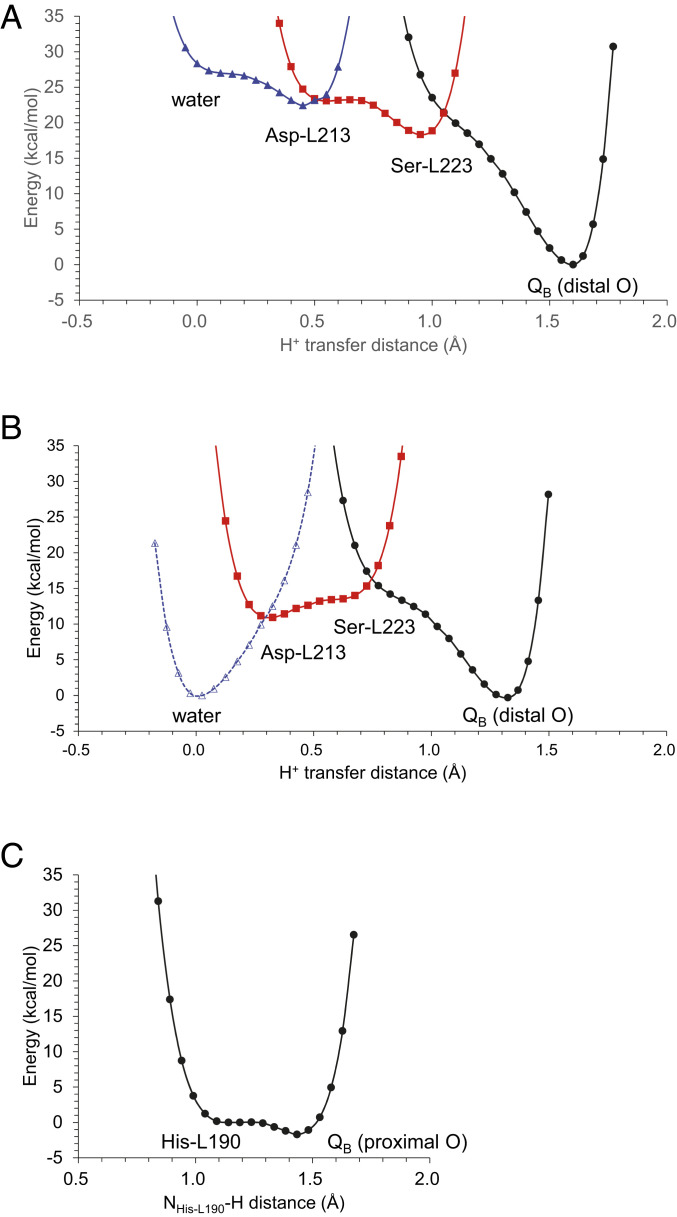
Fig. 3.
Potential-energy profiles for the first and second PT processes. (A) PT toward the distal QB O site in the presence of (A) protonated Asp-M17 and (B) ionized Asp-M17. The potential-energy profile was calculated, modeling a water molecule at the Asp-M17 and Asp-L213 moieties, which is observed in the crystal structure reported by Fujii and colleagues (34) (PDB ID code 3I4D). (C) PT toward the proximal QB O site.
Asp-M17 has been reported to be involved in the PT pathway (33). When it accepts a proton from the bulk region, the entire H-bond network that proceeds from Asp-M17 via a water molecule, Asp-L213, and Ser-L223 forms the pre-PT H-bond pattern (Fig. 2B), which serves as a downhill pathway (Fig. 3A): the proton at Asp-M17 is ultimately transferred to the distal QB O site concertedly via the Grotthuss-like mechanism.
The water molecule that connects Asp-M17 and Asp-L213 (W-H267) is present in the 2.01-Å crystal structure reported by Fujii and colleagues (34) (PDB ID code 3I4D), whereas it is absent in the light-exposed QB•− structure reported by Stowell et al. (23) at 2.60-Å resolution (PDB ID code 1AIG). It should also be noted that the distribution pattern of water molecules shows that the Asp-L213 and Asp-M17 moieties are highly accessible from the protein bulk surface (Fig. 4A). Thus, Asp-L213…Ser-L223…QB is likely to be the essential component for the first PT, as the corresponding H-bond network, D1-His252…D1-Ser264…QB is conserved in PSII (8). Consistently, the potential energy profile indicates that the proton at Asp-L213 can be transferred via Ser-L223 to the distal QB O site along a downhill pathway even if Asp-M17 is ionized (Fig. 3B).
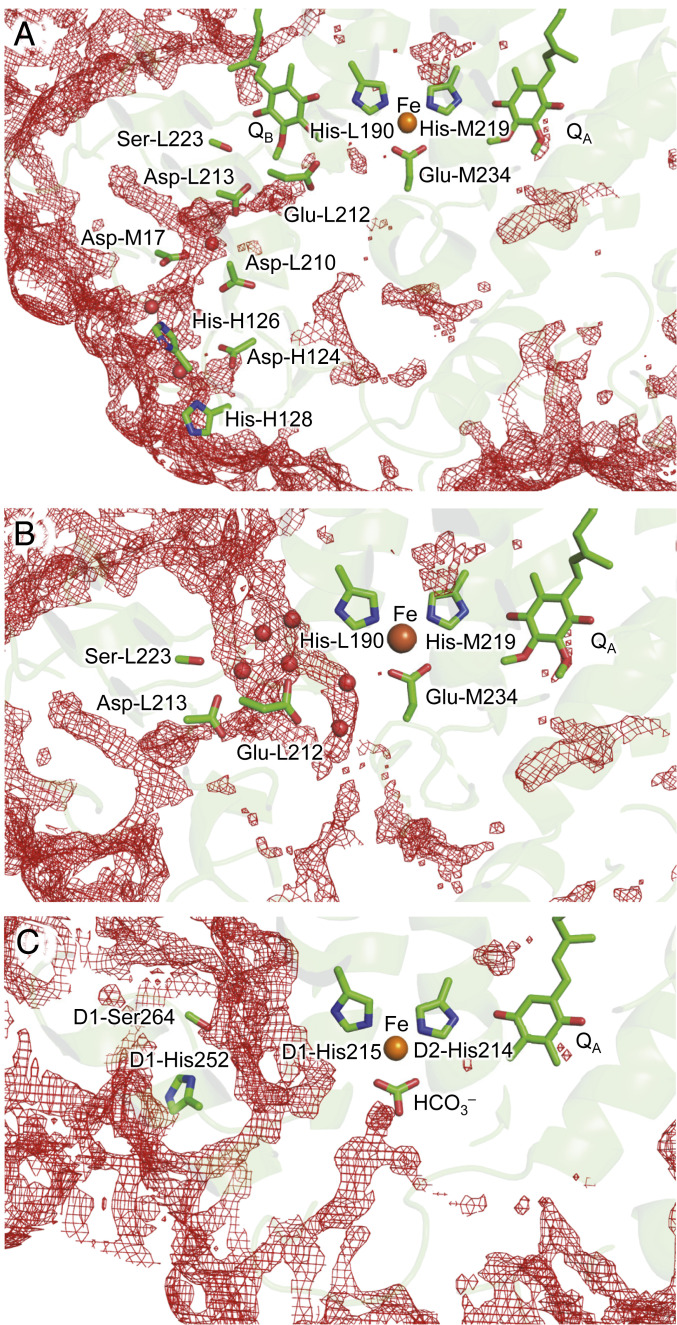
Fig. 4.
Distribution patterns of water molecules (red mesh). (A) In the presence of QB in PbRC (PDB ID code 1AIG) (23). (B) In the absence of QB in the PbRC. QB was removed from the PbRC crystal structure. (C) In the absence of QB in PSII (PDB ID code 3JCU) (66). The threshold of 3D distribution function is 2.0.
As the QBH− forms (via the first PT and the second electron transfer), the H-bond between His-L190 and the proximal QB O site is significantly shortened to 2.49 Å (Table 1). The potential-energy profile for the H-bond between His-L190 and the proximal QB O site resembles that for a typical low-barrier H-bond (Fig. 3C), as observed for the H-bond between D1-His215 and the proximal QB O site in PSII (8, 26, 27). Thus, the second PT can proceed along the barrierless potential, leading to the release of QBH2 from the PbRC.
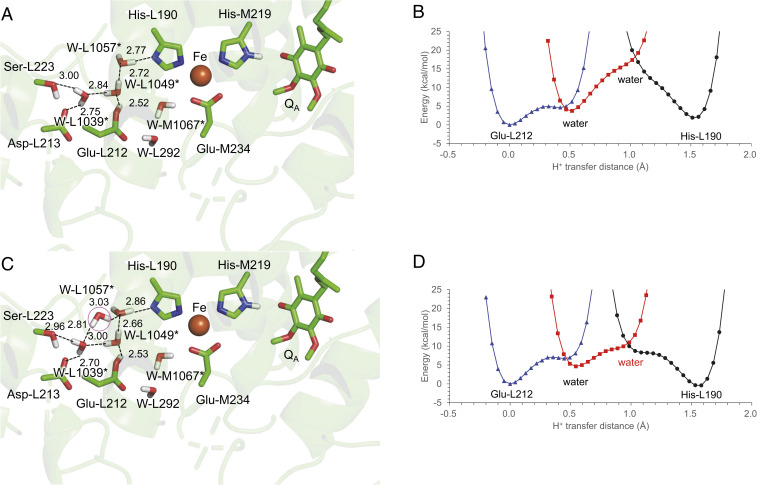
Not only the crystal structure (23) but also the calculated distribution pattern of water molecules shows that no H-bond network exists between Glu-L212 and His-L190 in the presence of QB (Fig. 4A). In contrast, the QB-lacking crystal structure shows that water molecules form an H-bond network that connects Glu-L212 and His-L190 (e.g., PDB ID code 1L9B) (35) (Fig. 4B), which may also be the case with the QBH2-released PbRC. QM/MM calculations also indicate that an H-bond network forms among protonated Glu-L212, water molecules, and deprotonated His-L190 (Fig. 5A). The potential-energy profile along the H-bond network suggests that pKa(Glu-L212) ∼ pKa(His-L190) (Fig. 5B).
Fig. 5.
(A) H-bond network that connects protonated Glu-L212 and ionized His-L190 in the absence of QB and (B) the potential-energy profile. The potential-energy profile was calculated adding four water molecules (*: L1039, L1049, L1057, and M1067), which are visible in the QB-lacking crystal structure (PDB ID code 1L9B) (35). (C) Addition of a water molecule (pink circle) to the H-bond network and (D) the potential-energy profile.
The distribution pattern of water molecules suggests that there are more water molecules than are identified at the QB binding moiety in the crystal structure (Fig. 4B). As the addition of a water molecule to the H-bond network (Fig. 5C) decreases the energy barrier for the PT (Fig. 5D), the existence of water molecules, which are not visible in the reported crystal structure, may facilitate PT from Glu-L212 to His-L190 in the QBH2-released PbRC.
Discussion
The initial protonation of QB occurs at the distal O site, as the H-bond between Ser-L223 and the distal QB O site is specifically shortened in response to the formation of QB•− (Fig. 2 A and B). The present results indicate that Ser-L223, Asp-L213, a water molecule, and Asp-M17 form a Grotthuss-like PT pathway toward the distal QB O site (Figs. 2B and 3A). Among them, protonated Asp-L213 (5, 22, 31, 36–38) [pKa(Asp-L213) = 8.9 (38)] and Ser-L223 are conserved as D1-His252 and D1-Ser264 in PSII, respectively (39). Because D1-His252 and D1-Ser264 serve as a PT pathway toward the distal QB O site in PSII (8), Ser-L223 and Asp-L213 are the primary components for PT toward the distal QB O site in the PbRC. Consistently, the D(L213)N (9) and S(L223)A (10, 11) mutations affected kAB(2) by factors of 6,000 and 400, respectively, whereas the D(M17)N mutation affected kAB(2) only by a factor of 2 (4). The extension of the H-bond network [e.g., Asp-M17, Asp-L210 (33, 40), Asp-H124, His-H126, His-H128 (12, 41), and water molecules] may increase the surface of the proton entry point. It seems plausible that the proton can enter either directly from Asp-L213 or the extension of the H-bond network.
Glu-L212 was assumed to serve as a proton donor to the proximal QB O site (e.g., refs. 4, 42, and 43). However, Glu-L212 cannot form an H-bond with QB and is unlikely to serve as a direct proton donor for QB, because PT occurs most efficiently along H-bonds (24). The findings of His-L190 forming a low-barrier H-bond with the proximal QB O site and being able to serve as a direct proton donor to QBH− (Fig. 3C) suggest that pKa(His-L190) ∼ pKa(QBH−/QBH2). The involvement of His-L190 in QBH2 formation is consistent with the mechanism proposed by Wraight (25) and the mechanism suggested for PSII (8, 26, 27). In PSII, QM/MM calculations showed that D1-His215 forms a low barrier H-bond with QBH−, facilitating PT (8, 26). Recent Fourier transform infrared (FTIR) studies suggested that D1-His215 can serve as a proton donor to anionic QBH−, as D1-His215 releases the proton upon the oxidation of the nonheme Fe complex (27). His-L190 is conserved as D1-His215 at the nonheme Fe cluster in PSII, whereas Glu-L212 is not conserved. These features suggest that His-L190 is the proton donor for the proximal O site of QB in the PbRC.
The QB binding pocket has a large cavity. The QBH2 channel is oriented toward the protein bulk surface in the transmembrane region (44) (Fig. 4B). Although the energy barrier for PT from Glu-L212 to His-L190 is not significantly low with respect to PT to the distal (Fig. 3 A and B) and proximal QB O (Fig. 3C) sites, the PT profile suggests that pKa(His-L190) ∼ pKa(Glu-L212) (= 9.4) (38) (Fig. 5 B and D), which would make the protonation of His-L190 by Glu-L212 possible before an unprotonated quinone occupies the QB binding site. The H-bond network presented here is likely to represent a minimum component of the reprotonation pathway for ionized His-L190, which is consistent with the mechanism proposed by Wraight (25), specifically a water molecule serving as a direct proton donor to reprotonate ionized His-L190. Cherepanov et al. (45) proposed that the H-bond network that connects Glu-L212 and His-L190 (water bridge) forms in response to the release of QBH2 and prevents the QBH2 to QBH− reversion. The estimated QB exchange time is ∼1 ms (46), which is sufficiently long for a few water molecules to approach the QB binding site and form the H-bond network: for example, among 103 water molecules in the protein interior near the Mn4CaO5 cluster of the PSII crystal structure, 90 water molecules are incorporated into the binding positions in the 50-ns molecular dynamics simulation (47). In addition, some PbRC crystal structures show that the corresponding water molecule also exists near Glu-L212 and His-L190 even when QB is only slightly displaced from His-L190 in the binding pocket (e.g., PDB ID code 1AIJ) (23).
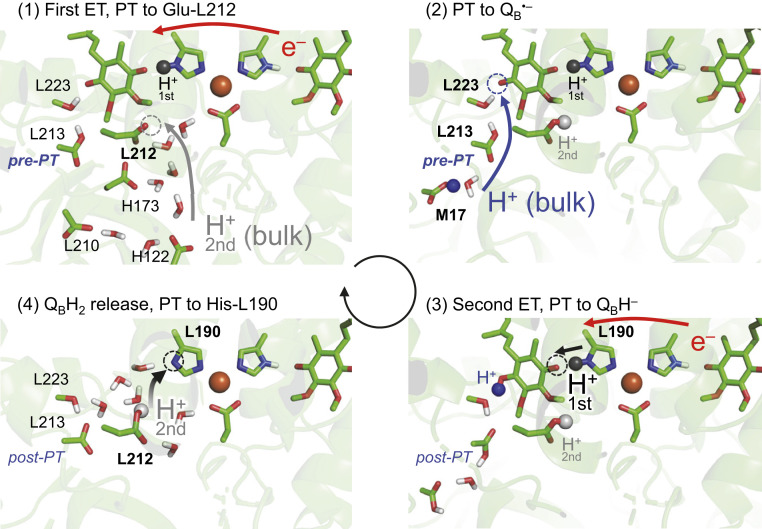
The PbRC crystal structure shows that Glu-L212 has an H-bond network that proceeds toward the protein bulk surface [e.g., the P1a and P1b water chains (48)], which may serve as a pathway for the uptake of 0.3 to 0.8 H+ (17, 18) for ionized Glu-L212 (19–21) upon QB•− formation (Fig. 6, panel 1). The protonation of Glu-L212 increases Em(QB) with respect to Em(QA), leading to an energetically downhill electron transfer to QB (22). As QB•− forms, the -OH group of Ser-L223 is oriented away from Asp-L213 toward the distal QB O site, further increasing Em(QB) (6), transforming the post-PT (Fig. 6, panel 4) into pre-PT patterns (Fig. 6, panel 1), and facilitating QBH• formation (6) (Fig. 6, panel 2). Thus, Glu-L212 plays a role in facilitating the first electron transfer to QB, donating a proton to ionized His-L190 in the absence of QB (Fig. 6, panel 4), and eventually providing the second proton for transfer to QBH− via His-L190 (Fig. 6, panel 3).
Fig. 6.
Overview of PT (blue and gray arrows) and electron transfer (ET, red arrows). PT events toward the distal/proximal QB O site are gray/blue colored, respectively. Proton acceptor sites are indicated by dotted circles. H+-first already exists at His-L190 (panel 1) and is transferred to the proximal QB O site (panel 3). H+-second is transferred from the bulk water region to deprotonated Glu-L212 (panels 1 to 2) and is further transferred to deprotonated His-L190 (panel 4), being H+-first in the next turnover.
Remarkably, Glu-L212 is not conserved in PSII. The absence of a subunit H-like protein in PSII renders D1-His215 (i.e., Asp-L213 in PbRC) exposed to the protein bulk surface (Fig. 4C), which may explain why PSII has no corresponding protonatable residue. Because subunit H is likely to restrict the access of water molecules to the QB binding pocket in the PbRC, having protonated Glu-L212 in the QB binding pocket could be advantageous for the immediate reprotonation of ionized His-L190 and binding of unprotonated QB at protonated His-L190.
In summary, the pathway to the distal QB O site is a Grotthuss-like H-bond network (e.g., Ser-L223 and Asp-L213), which facilitates PT by transforming the pre-PT to post-PT H-bond patterns (Fig. 2 B and C). In contrast, the pathway to the proximal QB O site is a single, low-barrier H-bond between QBH− and His-L190 (Fig. 2D), as observed in PSII (8, 26, 27). His-L190 is the proton donor for QBH− and no movement of QBH− toward Glu-L212 (e.g., ref. 4) is required for the QBH− protonation. However, Glu-L212 is ultimately involved in PT to QBH−, because protonated Glu-L212 is likely to serve as a proton donor to ionized His-L190 via water molecules in response to the release of QBH2 (Fig. 5). Moreover, the proton delivered to Glu-L212 during the initial photo-cycle is delivered to QBH− during the next photo-cycle (Fig. 6). The mechanism presented here can also explain why Glu-L212 is crucial for PT (e.g., refs. 5 and 17–22) irrespective of the absence of an H-bond with QB (see discussions in ref. 4) and how the protonation of QBH− to QBH2 proceeds via His-L190 (25).
Methods
Coordinates and Atomic Partial Charges.
The atomic coordinates were taken from the X-ray structure of PbRC from Rhodobacter sphaeroides (PDB ID code 1AIG) (23). The H atom positions were optimized with CHARMM (49), whereas the heavy atom positions were fixed. During the procedure, all titratable groups (e.g., acidic and basic groups) were ionized. Atomic partial charges of the amino acids were obtained from the CHARMM22 (50) parameter set, whereas those of cofactors were obtained from the previous studies (51). Additional counter ions were added to neutralize the entire system in QM/MM calculations.
Protonation Pattern.
The computation was based on the electrostatic continuum model, solving the linear Poisson–Boltzmann equation with the MEAD program (52). The difference in electrostatic energy between the two protonation states, protonated and deprotonated, in a reference model system was calculated using a known experimentally measured pKa value [e.g., 4.0 for Asp (53)]. The difference in the pKa value of the protein relative to the reference system was added to the known reference pKa value. The experimentally measured pKa values employed as references were 12.0 for Arg, 4.0 for Asp, 9.5 for Cys, 4.4 for Glu, 10.4 for Lys, 9.6 for Tyr (53), and 7.0 and 6.6 for the Nε and Nδ atoms of His, respectively (54–56). All other titratable sites were fully equilibrated to the protonation state of the target site during titration. The dielectric constants were set to 4 inside the protein and 80 for water. All computations were performed at 300 K, pH 7.0, and with an ionic strength of 100 mM. The linear Poisson–Boltzmann equation was solved using a three-step grid-focusing procedure at resolutions of 2.5, 1.0, and 0.3 Å. The ensemble of the protonation patterns was sampled by the Monte Carlo method with the Karlsberg program (57). The Monte Carlo sampling yielded the probabilities [protonated] and [deprotonated] of the two protonation states of the molecule.
QM/MM Calculations.
The unrestricted density functional theory method was employed with the B3LYP functional and LACVP* basis sets, using the QSite (58) program. In the QM region, all the atomic coordinates were fully relaxed. In the MM region, the positions of H atoms were optimized using the OPLS2005 force field, while the positions of heavy atoms were fixed. The initial-guess wavefunctions were obtained using the ligand field theory (59) implemented in the QSite program. To analyze the potential-energy profiles for PT to QB, the QM region was defined as QB, the nonheme Fe complex (Fe, the side-chains of His-L190, His-L230, His-M219, His-M266, and Glu-M234), the side-chains of Asp-M17, Asp-L213, and Ser-L223, and a water molecule at the Asp-M17 and Asp-L213 moiety, which is visible in the 2.01-Å crystal structure (34) (PDB ID code 3I4D). To analyze the potential-energy profiles for PT from protonated Glu-L212 to ionzed His-L190 in the absence of QB, QB was removed and the QM region was defined as the nonheme Fe complex (Fe, the side-chains of His-L190, His-L230, His-M219, His-M266, and Glu-M234), the side-chains of Glu-L212, Asp-L213, and Ser-L223, and five water molecules (W-L292, L1039, L1049, L1057, and M1067), which are visible in the QB-lacking crystal structure (PDB ID code 1L9B) (35) if not otherwise specified. All other protein units and cofactors were approximated by the MM force field (i.e., electrostatic influences are sufficiently considered in the MM region). Note that the residues in the PT pathways must be included in the QM region to consider the formation/breakage of the covalent (H-)bonds during PT. See Dataset S1 for the atomic coordinates of the resulting QM region.
The potential-energy profiles of H-bonds were obtained as follows. First, we prepared the QM/MM optimized geometry without constraints and used the resulting geometry as the initial geometry. The H atom under investigation was then moved from the H-bond donor atom (O/Ndonor) toward the acceptor atom (O/Nacceptor) by 0.05 Å, after which the geometry was optimized by constraining the O/Ndonor–H and H–O/Nacceptor distances. The energy of the resulting geometry was calculated. This procedure was repeated until the H atom reached the O/Nacceptor atom.
Analysis of Water Molecule Distribution in the Protein.
To analyze the distribution of water molecules in the protein environment, we used a three-dimensional reference interaction site model (3D-RISM) with Placevent analysis (60–64). It should be noted that the distribution pattern of water molecules obtained from the 3D-RISM with Placevent analysis was consistent with the positions of the water molecules (65).
Supplementary Material
Acknowledgments
This research was supported by Japan Science and Technology Core Research for Evolutionary Science and Technology (JST CREST) Grant JPMJCR1656 (to H.I.); Japan Society for the Promotion of Science (JSPS) Grants KAKENHI JP18H01937, JP18H05155, JP20H03217, and JP20H05090 (to H.I.) and JP16H06560 and JP18H01186 (to K.S.); and the Interdisciplinary Computational Science Program in Computational Sciences, University of Tsukuba.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2103203118/-/DCSupplemental.
Data Availability
All study data are included in the article and Dataset S1.
References
- 1.Zankel K. L., Reed D. W., Clayton R. K., Fluorescence and photochemical quenching in photosynthetic reaction centers. Proc. Natl. Acad. Sci. U.S.A. 61, 1243–1249 (1968). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finkele U., Lauterwasser C., Zinth W., Gray K. A., Oesterhelt D., Role of tyrosine M210 in the initial charge separation of reaction centers of Rhodobacter sphaeroides. Biochemistry 29, 8517–8521 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Tamura H., Saito K., Ishikita H., Acquirement of water-splitting ability and alteration of the charge-separation mechanism in photosynthetic reaction centers. Proc. Natl. Acad. Sci. U.S.A. 117, 16373–16382 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamura M. Y., Paddock M. L., Graige M. S., Feher G., Proton and electron transfer in bacterial reaction centers. Biochim. Biophys. Acta 1458, 148–163 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Alexov E. G., Gunner M. R., Calculated protein and proton motions coupled to electron transfer: Electron transfer from QA− to QB in bacterial photosynthetic reaction centers. Biochemistry 38, 8253–8270 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Ishikita H., Knapp E.-W., Variation of Ser-L223 hydrogen bonding with the QB redox state in reaction centers from Rhodobacter sphaeroides. J. Am. Chem. Soc. 126, 8059–8064 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Michel H., Deisenhofer J., Relevance of the photosynthetic reaction center from purple bacteria to the structure of photosystem II. Biochemistry 27, 1–7 (1988). [Google Scholar]
- 8.Saito K., Rutherford A. W., Ishikita H., Mechanism of proton-coupled quinone reduction in Photosystem II. Proc. Natl. Acad. Sci. U.S.A. 110, 954–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi E., Wraight C. A., A crucial role for AspL213 in the proton transfer pathway to the secondary quinone of reaction centers from Rhodobacter sphaeroides. Biochim. Biophys. Acta 1020, 107–111 (1990). [Google Scholar]
- 10.Paddock M. L., McPherson P. H., Feher G., Okamura M. Y., Pathway of proton transfer in bacterial reaction centers: Replacement of serine-L223 by alanine inhibits electron and proton transfers associated with reduction of quinone to dihydroquinone. Proc. Natl. Acad. Sci. U.S.A. 87, 6803–6807 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paddock M. L., et al., Pathway of proton transfer in bacterial reaction centers: Role of aspartate-L213 in proton transfers associated with reduction of quinone to dihydroquinone. Biochemistry 33, 734–745 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Axelrod H. L., Abresch E. C., Paddock M. L., Okamura M. Y., Feher G., Determination of the binding sites of the proton transfer inhibitors Cd2+ and Zn2+ in bacterial reaction centers. Proc. Natl. Acad. Sci. U.S.A. 97, 1542–1547 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maróti P., Hanson D. K., Baciou L., Schiffer M., Sebban P., Proton conduction within the reaction centers of Rhodobacter capsulatus: The electrostatic role of the protein. Proc. Natl. Acad. Sci. U.S.A. 91, 5617–5621 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerencsér L., Maróti P., Retardation of proton transfer caused by binding of the transition metal ion to the bacterial reaction center is due to pKa shifts of key protonatable residues. Biochemistry 40, 1850–1860 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Gerencser L., Taly A., Baciou L., Maroti P., Sebban P., Effect of binding of Cd2+ on bacterial reaction center mutants: Proton-transfer uses interdependent pathways. Biochemistry 41, 9132–9138 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Kuroda H., et al., Proton transfer pathway from the oxygen-evolving complex in photosystem II substantiated by extensive mutagenesis. Biochim. Biophys. Acta Bioenerg. 1862, 148329 (2021). [DOI] [PubMed] [Google Scholar]
- 17.Maróti P., Wraight C. W., Flash-induced H+ binding by bacterial photosynthetic reaction centers: Influences of the redox states of the acceptor quinones and primary donor. Biochim. Biophys. Acta 934, 329–347 (1988). [Google Scholar]
- 18.McPherson P. H., Nagarajan V., Parson W. W., Okamura M. Y., Feher G., pH-dependence of the free energy gap between DQA and D+Q−A determined from delayed fluorescence in reaction centers from Rhodobacter sphaeroides R-26. Biochim. Biophys. Acta 1019, 91–94 (1990). [Google Scholar]
- 19.Hienerwadel R., et al., Protonation of Glu L212 following QB− formation in the photosynthetic reaction center of Rhodobacter sphaeroides: Evidence from time-resolved infrared spectroscopy. Biochemistry 34, 2832–2843 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Nabedryk E., et al., Fourier transforms infrared difference spectroscopy of secondary quinone acceptor photoreduction in proton transfer mutants of Rhodobacter sphaeroides. Biochemistry 34, 14722–14732 (1995). [DOI] [PubMed] [Google Scholar]
- 21.Miksovska J., et al., Distant electrostatic interactions modulate the free energy level of QA− in the photosynthetic reaction center. Biochemistry 35, 15411–15417 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Ishikita H., Morra G., Knapp E.-W., Redox potential of quinones in photosynthetic reaction centers from Rhodobacter sphaeroides: Dependence on protonation of Glu-L212 and Asp-L213. Biochemistry 42, 3882–3892 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Stowell M. H. B., et al., Light-induced structural changes in photosynthetic reaction center: Implications for mechanism of electron-proton transfer. Science 276, 812–816 (1997). [DOI] [PubMed] [Google Scholar]
- 24.Ishikita H., Saito K., Proton transfer reactions and hydrogen-bond networks in protein environments. J. R. Soc. Interface 11, 20130518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wraight C. A., Proton and electron transfer in the acceptor quinone complex of photosynthetic reaction centers from Rhodobacter sphaeroides. Front. Biosci. 9, 309–337 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Saito K., Mandal M., Ishikita H., Redox potentials along the redox-active low-barrier H-bonds in electron transfer pathways. Phys. Chem. Chem. Phys. 22, 25467–25473 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Kimura M., Kato Y., Noguchi T., Protonation state of a key histidine ligand in the iron–quinone complex of photosystem II as revealed by light-induced ATR-FTIR spectroscopy. Biochemistry 59, 4336–4343 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Warshel A., Papazyan A., Kollman P. A., On low-barrier hydrogen bonds and enzyme catalysis. Science 269, 102–106 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Schutz C. N., Warshel A., The low barrier hydrogen bond (LBHB) proposal revisited: The case of the Asp... His pair in serine proteases. Proteins 55, 711–723 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Perrin C. L., Nielson J. B., “Strong” hydrogen bonds in chemistry and biology. Annu. Rev. Phys. Chem. 48, 511–544 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Zhu Z., Gunner M. R., Energetics of quinone-dependent electron and proton transfers in Rhodobacter sphaeroides photosynthetic reaction centers. Biochemistry 44, 82–96 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Paddock M. L., et al., ENDOR spectroscopy reveals light induced movement of the H-bond from Ser-L223 upon forming the semiquinone (QB−•) in reaction centers from Rhodobacter sphaeroides. Biochemistry 46, 8234–8243 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paddock M. L., Feher G., Okamura M. Y., Identification of the proton pathway in bacterial reaction centers: Replacement of Asp-M17 and Asp-L210 with Asn reduces the proton transfer rate in the presence of Cd2+. Proc. Natl. Acad. Sci. U.S.A. 97, 1548–1553 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roszak A. W., et al., New insights into the structure of the reaction centre from Blastochloris viridis: Evolution in the laboratory. Biochem. J. 442, 27–37 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Axelrod H. L., et al., X-ray structure determination of the cytochrome c2: Reaction center electron transfer complex from Rhodobacter sphaeroides. J. Mol. Biol. 319, 501–515 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Grafton A. K., Wheeler R. A., Amino acid protonation states determine binding sites of the secondary ubiquinone and its anion in the Rhodobacter sphaeroides photosynthetic reaction center. J. Phys. Chem. B 103, 5380–5387 (1999). [Google Scholar]
- 37.Rabenstein B., Ullmann G. M., Knapp E.-W., Electron transfer between the quinones in the photosynthetic reaction center and its coupling to conformational changes. Biochemistry 39, 10487–10496 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Ishikita H., Knapp E.-W., Energetics of proton transfer pathways in reaction centers from Rhodobacter sphaeroides. The Glu-H173 activated mutants. J. Biol. Chem. 280, 12446–12450 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Ishikita H., Knapp E.-W., Control of quinone redox potentials in photosystem II: Electron transfer and photoprotection. J. Am. Chem. Soc. 127, 14714–14720 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Paddock M. L., et al., Identification of the proton pathway in bacterial reaction centers: Cooperation between Asp-M17 and Asp-L210 facilitates proton transfer to the secondary quinone (QB). Biochemistry 40, 6893–6902 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Paddock M. L., Graige M. S., Feher G., Okamura M. Y., Identification of the proton pathway in bacterial reaction centers: Inhibition of proton transfer by binding of Zn2+ or Cd2+. Proc. Natl. Acad. Sci. U.S.A. 96, 6183–6188 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ädelroth P., et al., Identification of the proton pathway in bacterial reaction centers: Decrease of proton transfer rate by mutation of surface histidines at H126 and H128 and chemical rescue by imidazole identifies the initial proton donors. Biochemistry 40, 14538–14546 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Paddock M. L., Feher G., Okamura M. Y., Proton transfer pathways and mechanism in bacterial reaction centers. FEBS Lett. 555, 45–50 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Niwa S., et al., Structure of the LH1-RC complex from Thermochromatium tepidum at 3.0 Å. Nature 508, 228–232 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Cherepanov D. A., et al., Reduction and protonation of the secondary quinone acceptor of Rhodobacter sphaeroides photosynthetic reaction center: Kinetic model based on a comparison of wild-type chromatophores with mutants carrying Arg→Ile substitution at sites 207 and 217 in the L-subunit. Biochim. Biophys. Acta 1459, 10–34 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Osváth S., Maróti P., Coupling of cytochrome and quinone turnovers in the photocycle of reaction centers from the photosynthetic bacterium Rhodobacter sphaeroides. Biophys. J. 73, 972–982 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakashita N., Watanabe H. C., Ikeda T., Saito K., Ishikita H., Origins of water molecules in the photosystem II crystal structure. Biochemistry 56, 3049–3057 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Abresch E. C., et al., Identification of proton transfer pathways in the X-ray crystal structure of the bacterial reaction center from Rhodobacter sphaeroides. Photosynth. Res. 55, 119–125 (1998). [Google Scholar]
- 49.Brooks B. R., et al., CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983). [Google Scholar]
- 50.MacKerell A. D. Jr, et al., All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- 51.Kawashima K., Ishikita H., Energetic insights into two electron transfer pathways in light-driven energy-converting enzymes. Chem. Sci. (Camb.) 9, 4083–4092 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bashford D., Karplus M., pKa’s of ionizable groups in proteins: Atomic detail from a continuum electrostatic model. Biochemistry 29, 10219–10225 (1990). [DOI] [PubMed] [Google Scholar]
- 53.Nozaki Y., Tanford C., Acid-base titrations in concentrated guanidine hydrochloride. Dissociation constants of the guamidinium ion and of some amino acids. J. Am. Chem. Soc. 89, 736–742 (1967). [DOI] [PubMed] [Google Scholar]
- 54.Tanokura M., 1H nuclear magnetic resonance titration curves and microenvironments of aromatic residues in bovine pancreatic ribonuclease A. J. Biochem. 94, 51–62 (1983). [DOI] [PubMed] [Google Scholar]
- 55.Tanokura M., 1H-NMR study on the tautomerism of the imidazole ring of histidine residues. I. Microscopic pK values and molar ratios of tautomers in histidine-containing peptides. Biochim. Biophys. Acta 742, 576–585 (1983). [DOI] [PubMed] [Google Scholar]
- 56.Tanokura M., 1H-NMR study on the tautomerism of the imidazole ring of histidine residues. II. Microenvironments of histidine-12 and histidine-119 of bovine pancreatic ribonuclease A. Biochim. Biophys. Acta 742, 586–596 (1983). [DOI] [PubMed] [Google Scholar]
- 57.Rabenstein B., Knapp E.-W., Calculated pH-dependent population and protonation of carbon-monoxy-myoglobin conformers. Biophys. J. 80, 1141–1150 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.QSite, Version 5.8 (Schrödinger, New York, NY, 2012).
- 59.Vacek G., Perry J. K., Langlois J. M., Advanced initial-guess algorithm for self-consistent-field calculations on organometallic systems. Chem. Phys. Lett. 310, 189–194 (1999). [Google Scholar]
- 60.Beglov D., Roux B., An integral equation to describe the solvation of polar molecules in liquid water. J. Phys. Chem. B 101, 7821–7826 (1997). [Google Scholar]
- 61.Kovalenko A., Hirata F., Potential of mean force between two molecular ions in a polar molecular solvent: A study by the three-dimensional reference interaction site model. J. Phys. Chem. B 103, 7942–7957 (1999). [Google Scholar]
- 62.Luchko T., et al., Three-dimensional molecular theory of solvation coupled with molecular dynamics in Amber. J. Chem. Theory Comput. 6, 607–624 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Case D. A., et al., AMBER 12 (University of California, San Francisco, 2012). [Google Scholar]
- 64.Sindhikara D. J., Yoshida N., Hirata F., Placevent: An algorithm for prediction of explicit solvent atom distribution-application to HIV-1 protease and F-ATP synthase. J. Comput. Chem. 33, 1536–1543 (2012). [DOI] [PubMed] [Google Scholar]
- 65.Takaoka T., Sakashita N., Saito K., Ishikita H., pKa of a proton-conducting water chain in photosystem II. J. Phys. Chem. Lett. 7, 1925–1932 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Wei X., et al., Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534, 69–74 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Breton J., Absence of large-scale displacement of quinone QB in bacterial photosynthetic reaction centers. Biochemistry 43, 3318–3326 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and Dataset S1.