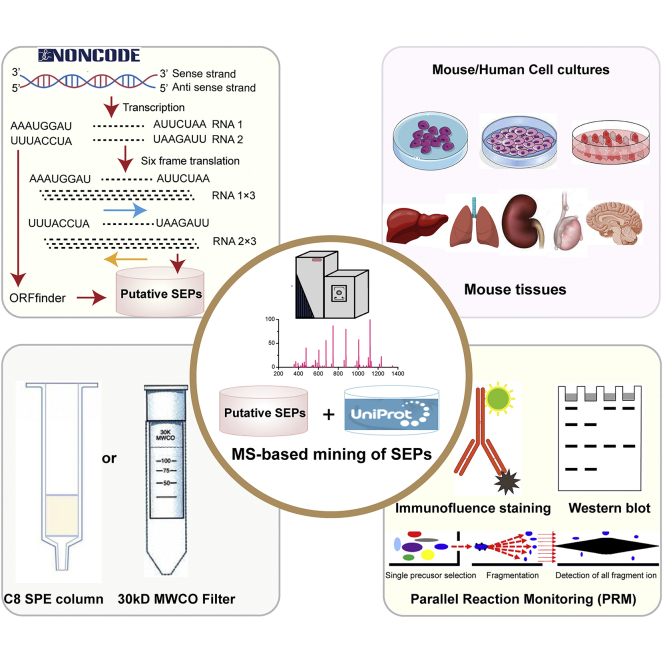
Abstract
Many small ORFs embedded in long noncoding RNA (lncRNA) transcripts have been shown to encode biologically functional polypeptides (small ORF-encoded polypeptides [SEPs]) in different organisms. Despite some novel SEPs have been found, the identification is still hampered by their poor predictability, diminutive size, and low relative abundance. Here, we take advantage of NONCODE, a repository containing the most complete collection and annotation of lncRNA transcripts from different species, to build a novel database that attempts to maximize a collection of SEPs from human and mouse lncRNA transcripts. In order to further improve SEP discovery, we implemented two effective and complementary polypeptide enrichment strategies using 30-kDa molecular weight cutoff filter and C8 solid-phase extraction column. These combined strategies enabled us to discover 353 SEPs from eight human cell lines and 409 SEPs from three mouse cell lines and eight mouse tissues. Importantly, 19 of them were then verified through in vitro expression, immunoblotting, parallel reaction monitoring, and synthetic peptides. Subsequent bioinformatics analysis revealed that some of the physical and chemical properties of these novel SEPs, including amino acid composition and codon usage, are different from those commonly found in canonical proteins. Intriguingly, nearly 65% of the identified SEPs were found to be initiated with non-AUG start codons. The 762 novel SEPs probably represent the largest number of SEPs detected by MS reported to date. These novel SEPs might not only provide new clues for the annotation of noncoding elements in the genome but also serve as a valuable resource for functional study.
Keywords: long noncoding RNA, smORF-encoded polypeptides, NONCODE database, enrichment, MS
Abbreviations: ACN, acetonitrile; DMEM, Dulbecco's modified Eagle's medium; EGFP, enhanced GFP; FA, formic acid; HEK293T, human embryonic kidney 293T; LC–MS/MS, LC–tandem MS; lincRNA, long intergenic noncoding RNA; lncRNA, long noncoding RNA; MEF, mouse embryonic fibroblast; mESC, mouse embryonic stem cell; MWCO, molecular weight cutoff; PRM, parallel reaction monitoring; SEP, small ORF-encoded polypeptide; smORF, short or small ORF; SPE, solid-phase extraction
Graphical abstract
Highlights
-
•
Complementary enrichment strategies combined with membrane filtration and C8 SPE.
-
•
A combined database with the comprehensive putative SEPs and canonical proteins used.
-
•
Seven hundred sixty-two novel SEPs identified from human cell lines, mouse cell lines, and mouse tissues.
-
•
Nineteen SEPs have been validated by fusion expression or synthetic peptides.
In Brief
This study proposed a new and effective strategy for the improved discovery and identification of novel SEPs, including the construction of databases maximally collecting all putative small ORFs from human and mouse lncRNA transcripts in NONCODE and the effective enrichment of polypeptides based on 30-kDa molecular weight cutoff (MWCO) membrane and C8 solid-phase extraction column. This effort led to the discovery of 762 novel lncRNA-encoded SEPs from multiple cell lines and tissues.
Long noncoding RNAs (lncRNAs), a family of noncoding RNAs that are greater than 200 nucleotides in length and lack long or conserved ORFs, were formerly regarded as “junk RNAs.” Recently, however, a growing amount of evidence has demonstrated that many short or small ORFs (smORFs) embedded in lncRNA transcripts are able to encode functional polypeptides (smORFs-encoded polypeptides [SEPs]). These SEPs contain less than 100 amino acids in eukaryotes (50 amino acids in prokaryotes) and play vital regulatory roles in diverse physiological processes, including cancer growth (1), mucosal immunity (2), and fatty acid β-oxidation (3). These findings have subverted our understanding of lncRNAs and expanded our knowledge of the coding potential of the genome. Moreover, the development of genomics and bioinformatics, in particular the advent of high-throughput sequencing technology, accelerated the discovery of thousands of additional lncRNA transcripts with smORFs. Considering such large numbers of lncRNAs and smORFs, it is expected that SEPs may represent a large albeit neglected portion of nonannotated peptides involved in diverse physiological process. Therefore, large-scale discovery and functional characterization of unknown SEPs might provide new clues for the annotation and functional analysis of noncoding elements in the genome and their effects on biological evolution.
A variety of different methodologies, such as smORF predictions by computational sequence analysis, deep sequencing–based ribosome profiling, and MS-based proteomics, have been developed for the identification and characterization of SEPs across different biological samples. However, each of these strategies presents caveats. First of all, while bioinformatics analysis of lncRNA transcript sequences is a typical first step to predict the existence of smORFs, achieving high prediction sensitivity and specificity remains a significant challenge (4, 5). Furthermore, despite the power of deep sequencing–based ribosome profiling for the identification of the region of active translation in lncRNA transcripts, it nevertheless only provides indirect evidence of translation (6, 7, 8). Finally, while MS-based proteomics directly identifies SEPs by detecting the peptides generated from smORFs embedded in lncRNA transcripts (9, 10, 11), the number of SEPs identified by MS from different biological samples is still small (12).
The relatively low number of SEPs detected by MS is largely attributed to the fact that this type of detection is still analytically challenging. First of all, because of the actual low concentration and small size of SEPs, an accurate, consistent, and comprehensive measurement can be quite challenging and significantly affected by sample preparation. Even though multiple methods are available to enrich SEPs from different biological samples by fractionating or removing highly abundant and large proteins to reduce sample complexity, the various physical and chemical properties of different SEPs are often overlooked by different methods, which may negatively bias their discovery. Second, the identification of SEPs using MS is achieved by matching them against the theoretical spectra of all candidate peptides present in a reference protein sequence database. Crucially, this implies that the strategy behind the generation of a reference database can dramatically impact the identification of novel SEPs. For example, the most straightforward approach is six-frame translation of the entire genome. Unfortunately, such a dataset is difficult to use because of its extremely large size and the abundant presence of unknown protein sequences. While it is possible to, alternatively, create a smaller database by utilizing RNA transcripts from RNA-Seq or Ribo-Seq data, this strategy only captures actively translated RNA transcripts and mainly relies on sequencing depth.
In the present study, we address the challenges presented previously by developing an effective SEP enrichment workflow through the integration of two complementary enrichment methods based on 30-kDa MWCO filter and C8 solid-phase extraction (SPE) column. This approach allowed us to build a robust SEP database containing all putative smORFs from lncRNA transcripts deposited in the NONCODE database, a strategy that significantly improves the discovery of SEPs from different cell lines and tissues. We subsequently employed multiple technologies to experimentally validate the existence of these SEPs.
Experimental Procedures
Cell Lines and Cell Cultures
Human HeLa, human embryonic kidney 293T (HEK293T), 22Rv1, Du145, LNCap, PC3, and A375 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% (v/v) fetal bovine serum (Gibco) and 1% (v/v) penicillin/streptomycin (Gibco). Human U251 cells were cultured in DMEM/Nutrient Mixture F-12 (Gibco) supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin. Mouse 4T1 cells were cultured in RPMI1640 (Gibco) supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin. Mouse embryonic fibroblast (MEF) and mouse embryonic stem cell (mESC) D3 cells were obtained from the stem cell core facility at the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). mESCs were grown in MEFs treated with mitomycin C. mESCs were cultured in DMEM supplemented with 15% (v/v) fetal bovine serum and 1% (v/v) penicillin/streptomycin, plus 2 Mm l-glutamine, 0.1 mM 2-mercaptoethanol, 0.1 mM nonessential amino acids, and 103 units/ml mouse leukemia inhibitory factor.
Animals and Tissue Collection
Twelve-week-old male and female mice (C57BL/6J) were obtained from the Animal Core Facility at the Institute of Biophysics, Chinese Academy of Sciences. All animal protocols were approved by the Animal Care and Use Committee of the Institute of Biophysics, Chinese Academy of Sciences.
Protein Extraction and SEP Enrichment
For total cell protein extraction, ~1 × 106 cells were resuspended with 100 μl extraction buffer (8 M urea/100 mM NH4HCO3) containing Protease Inhibitor Cocktail Tablets (Roche), followed by sonication for 24 bursts with a 50% duty cycle (Scientz-IID), and then the supernatant was carefully collected after centrifugation at 20,000g at 4 °C for 20 min.
For whole tissue protein extraction, ~20 mg tissues were cut into small pieces and homogenized with 500 μl extraction buffer (8 M urea/100 mM NH4HCO3) containing Protease Inhibitor Cocktail Tablets. The lysate was sonicated for 24 bursts with a 50% duty cycle, and the remaining debris was removed by centrifugation at 20,000g for 20 min at 4 °C.
SEP enrichment from cell samples was performed with 30-kDa MWCO filters by resuspending ~1 × 107 cells in 500 μl ice-cold water containing Protease Inhibitor Cocktail Tablets. After three bursts of sonication with a 50% duty cycle, the mixture was heated at 95 °C for 5 min and then cooled down on ice for a few more minutes. Subsequently, 0.1 N ice-cold HCl was added to the sample to a final concentration of 10 mM and incubated on ice for 10 min. After centrifugation at 20,000g for 20 min at 4 °C in a bench-top centrifuge, the supernatant was filtered through a 30-kDa MWCO filter (Millipore), and the flow through was collected and evaporated to dryness by vacuum centrifugation at 4 °C. The pellet was then dissolved in 50 μl 8 M urea/100 mM NH4HCO3.
We performed SEP enrichment from tissue with 30-kDa MWCO filters by cutting ~200 mg frozen tissue into small pieces and then homogenizing in 500 μl ice-cold water containing Protease Inhibitor Cocktail Tablets. The subsequent steps were the same as described previously for SEP enrichment from cells.
In order to perform SEP extraction and enrichment from cell samples using C8 SPE columns, we used acidic lysis buffer containing detergent and C8 SPE columns. We slightly modified the C8 SPE method following previously described protocols (13, 14). Specifically, ~1 × 107 cells were lysed in 1 ml lysis buffer (50 mM HCl, 0.1% β-mercaptoethanol, and 0.05% Triton X-100) containing Protease Inhibitor Cocktail Tablets for 30 min at room temperature. After centrifugation at 20,000g for 20 min at 4 °C, the supernatant was collected. Subsequently, Bond Elute C8 silica cartridges (Agilent Technologies) were prepared with one-column volume of methanol and two-column volumes of triethylammonium formate buffer (pH 3.0) before the lysate was applied. Enriched SEPs were eluted, in turn, with 400 μl of 25%, 50%, and 75% acetonitrile (ACN) in triethylammonium formate buffer. The eluted fractions were then combined and concentrated to less than 100 μl at 4 °C by vacuum concentrator. Finally, enriched SEPs were precipitated with chloroform/methanol to remove residual detergent, and the precipitate was dissolved in 50 μl 8 M urea/100 mM NH4HCO3.
For SEP enrichment from tissue samples using C8 SPE column, ~200 mg of frozen tissue was initially cut into small pieces and homogenized in 1.5 ml lysis buffer (50 mM HCl, 0.1% β-mercaptoethanol, and 0.05% Triton X-100) containing Protease Inhibitor Cocktail Tablets. The subsequent steps were the same as described previously for SEP extraction from cell samples.
Tricine Gel Analysis of Enriched SEP Samples
An aliquot of 20 μg proteins was dissolved with loading buffer (50 mmol/l Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 0.1% bromophenol blue, and 1% β-mercaptoethanol). After denaturation for 5 min at 95 °C, the protein samples were loaded onto homemade 10% tricine SDS-PAGE gels (15) and ran at 120 V for 80 min. The gel was stained with One-Step Blue Protein Gel Stain (BIOTIUM) and then washed with distilled water.
Protein Reduction, Alkylation, and Tryptic Digestion
Proteins/SEPs were reduced with 10 mM dithiothreitol (37 °C, 1 h), alkylated with 20 mM iodoacetamide (at room temperature, in the dark, for 45 min), after which they were digested overnight with trypsin (Promega) at a ratio of 1:50 (enzyme/protein, w/w) at 37 °C in less than 2 M urea/100 mM NH4HCO3. Formic acid (FA) was added to the digested samples with 0.1% final concentration to stop the reaction. The tryptic peptide sample was then desalted using Pierce C18 Tips (Thermo Fisher Scientific) with 0.1% FA. The peptides were eluted with 50 μl of 20% ACN/0.1% FA, 50 μl of 40% ACN/0.1% FA, and 50 μl of 60% ACN/0.1% FA. The eluted peptide solutions were combined and evaporated to dryness by vacuum concentrator.
LC–MS/MS Analysis
Digested peptides were analyzed by LC–tandem MS (LC–MS/MS) by combining an Easy-nLC1000 (Thermo Fisher Scientific) with a Q Exactive Mass Spectrometer (Thermo Fisher Scientific). A 100 μm × 2 cm trap column packed with Reprosil-Pur C18 5 μm particles (Dr Maisch GmbH) and a 75 μm × 25 cm analytical column packed with Reprosil-Pur C18 3 μm particles (Dr Maisch GmbH) were used to separate the peptides with mobile phase A (0.1% FA in water) and mobile phase B (0.1% FA in ACN) at a 78 min gradient: 5 to 8% B in 8 min, 8 to 22% B in 50 min, 22 to 32% B in 12 min, 32 to 95% B in 1 min, and then kept B at 95% for 7 min. The flow rate was set as 300 nl/min.
The Q Exactive Mass Spectrometer was operated in a data-dependent acquisition mode with a spray voltage of 2 kV and a heated capillary temperature of 320 °C. MS1 data were collected at a high resolution of 70,000 (m/z 200) with a mass range of 300 to 1600 m/z, a target value of 3e6 and a maximum injection time of 60 ms. For each full MS scan, the 20 most abundant precursor ions were selected for MS2 with an isolation window of 2 m/z and the higher energy collision dissociation with normalized collision energy of 27. MS2 spectrums were collected at a resolution of 17,500 (m/z 200). The target value was 5e4 with a maximum fill time of 80 ms and a dynamic exclusion time of 40 s.
Construction of Putative SEP Database
We downloaded lncRNA transcripts for human (NONCODE V4) and mouse (NONCODE 2016) from the NONCODE database (http://www.noncode.org/). The ORFfinder and six-frame translation were employed to ensure we could detect all possible smORFs, which were then considered putative SEPs. We built SEP databases for human and mouse by collecting all putative SEPs with a length of 5 to 100 amino acids.
Identification of Annotated Proteins and SEPs
The LC–MS/MS raw data were analyzed with Thermo Scientific Proteome Discoverer (version 1.4) using the SEQUEST HT search engine. Four different protein databases were used in this study. The details of these databases are as follows: (1) Homo sapiens canonical protein database, downloaded from the Uniprot Web site on February 2, 2018 and consisting of 93,637 entries; (2) Mus musculus canonical protein database, downloaded from the Uniprot Web site on February 2, 2018 and consisting of 61,314 entries; (3) in-house putative human SEP database, including 3,969,981 entries; and (4) in-house putative mouse SEP database, including 8,710,195 entries.
For identification of candidate novel peptides from the digests, data were searched against the merged database of corresponding species described previously, which included canonical protein database and in-house putative SEP database. The search space included all fully tryptic and semitryptic peptides. Other common searching parameters were set as follows: peptides with a maximum of two missed cleavages were considered; the mass tolerance of precursor and product ions was set as 10 ppm and 0.02 Da, respectively; Carbamidomethylation on cysteine was considered as static modification; Oxidation on methionine was selected as dynamic modification; For protein identification, we set a significance threshold of p < 0.05 (with 95% confidence) and a false discovery rate <1%, which was estimated using a target-decoy search strategy.
For data derived from nondigested samples, no enzyme was chosen. For identification of canonical proteins, data were searched against the Uniprot protein database of corresponding species. Other common searching parameters were set as mentioned previously.
Validation of Novel SEPs with Nonsynthetic Peptide-Based Parallel Reaction Monitoring
All parallel reaction monitoring (PRM) experiments were performed on the same LC–MS/MS system as aforementioned. In this study, 21 SEPs were randomly selected from 196 SEPs identified in HEK293T cells for PRM analyses. The theoretically predicted and identified tryptic peptides in the selected endogenous SEPs were chosen for PRM analyses with a semitargeted data acquisition approach in order to verify the identified SEPs. Briefly, using high-resolution data-dependent scanning, an extensive MS1 fragmentation inclusion list of the theoretically predicted and identified tryptic peptides in the selected endogenous SEPs was generated to confirm the identified peptides and discover novel peptides in the selected endogenous SEPs. The peptides, which are identical to annotated proteins or nonunique in the putative SEP database, were excluded from the list. A total of 53 peptide targets (corresponding to 21 SEPs) were generated in the inclusion list.
Validation of Novel SEPs with Synthetic Standard Peptides
To further validate the identification of novel SEPs, 15 standard peptides from 14 SEPs were synthesized by GenScript Biotech Corporation and analyzed on the same LC–MS/MS instruments as mentioned previously.
Plasmid Constructs
In order to generate fusion protein constructs for the SEP ORF and enhanced GFP (EGFP), we amplified SEP ORF sequences without the endogenous 5′ UTR using RT-PCR and cloned them into a pEGFPmut-N1 vector in which the GFP start codon (ATGGTG) was mutated to ATTGTT (pEGFPmut). A list of the primers used in this study is available in supplemental Table S1.
Cell Transfection
HEK293T cells were transfected with the plasmid constructs using Lipofectamine TM 2000 (Invitrogen; 11668-019) according to manufacturers' instructions.
RT-PCR
Total RNA was extracted from cells using the Trizol Total RNA Isolation Reagent (Invitrogen). RNA levels of GFP, GFPmut, and SEP ORF-EGFPmut were detected by RT-PCR. A list containing all the primers used in this study is available in supplemental Table S2.
Western Blotting
Western blotting was performed according to standard protocols. The primary antibodies used in this study were obtained as follows: anti-GFP (ABclonal Technology; AE012), anti-β-tubulin (Yeasen Tech; 30303ES50), anti-NONHSAT130014+unORF+2+peptide9, and anti-NONHSAT077882+1+orf4 were customized and raised by GeneScript Biotech Corporation.
Immunofluorescence Staining
HEK293T cells were transfected with SEP ORF-EGFPmut, EGFPwt, and EGFPmut vectors for 24 h, and GFP fluorescence was directly visualized and recorded. HEK293T cells were plated on glass coverslips and then fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, incubated with anti-NONHSAT077882+1+orf4 antibodies and, subsequently, incubated with Goat Anti-Rabbit IgG H&L (Alexa Fluor 488). Cellular nuclei were stained with 4′,6-diamidino-2-phenylindole.
Experimental Design and Statistical Rationale
To test the two different SEP enrichment methods implemented, we performed and analyzed three technical replicates per method using the same cell or tissue samples. Data were analyzed by a two-tailed unpaired Student's t test (unless otherwise indicated), and p < 0.05 was chosen as the statistical limit of significance. We chose a notation of ∗, ∗∗, and ∗∗∗ for p < 0.05, p < 0.01, and p < 0.001, respectively. Unless otherwise indicated, all the data in the figures were represented as arithmetic means ± the standard deviations from at least three independent experiments.
Results
Design Rationale and Optimized MS-Based Workflow for Improved SEP Discovery
As discussed previously, the inherent low abundance and small sizes of SEPs contribute to their poor detectability, whereby it is critical to carefully consider sample preparation and build putative SEP reference databases from MS-based analysis in order to improve SEP discovery from different biological samples.
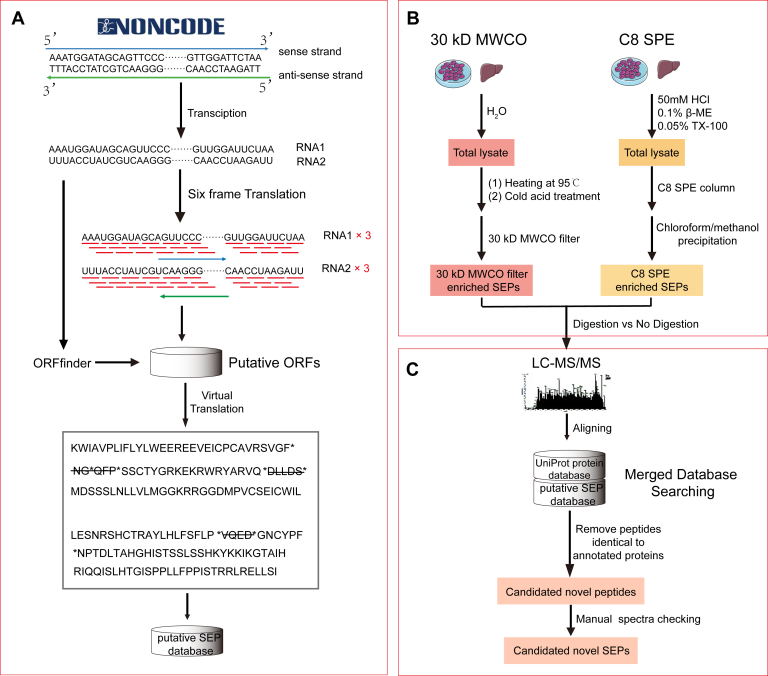
MS-based database searching is the key step for MS-based SEP identification. In order to build a SEP database that could maximally cover all the putative SEPs in human and mouse, we scanned lncRNA transcripts deposited in the NONCODE database (http://www.noncode.org/), an interactive repository that currently represents the most complete collection and annotation of noncoding RNAs, especially lncRNAs. Specifically, lncRNA transcripts were scanned by ORFfinder and six-frame translation mode to make it possible to obtain all possible smORFs, which were then assumed to represent putative SEPs (Fig. 1A). This resulted in 3,969,981 and 8,710,195 polypeptides in the newly constructed human and mouse putative SEP databases, respectively. To verify the quality of these two databases, we chose recently reported functional SEPs, including myoregulin (16), myomixer (17), minion (18), SPAR (19), HOXB-AS3 (1), NoBody (20), and LINC-PINT (21) and BLAST-ed them against our newly assembled database. All these SEPs could be found within our putative SEP database, which attests the high quality, accuracy, and comprehensiveness of our database.
Fig. 1.
Schematic illustration of the workflow for MS-based discovery of lncRNA-encoded SEPs.A, construction of putative lncRNA-encoded SEPs. The lncRNA transcripts of human and mouse deposited in the NONCODE database were screened by ORFfinder and six-frame translation to find all possible ORFs and then assumed to represent putative SEPs. All SEPs with a length of 5 to 100 amino acids were collected into the human and mouse putative SEPs database. B, enrichment of lncRNA-encoded SEPs based on the combination of 30-kDa MWCO filter and C8 SPE column. C, MS-based identification of lncRNA-encoded SEPs. lncRNA, long noncoding RNA; MWCO, molecular weight cutoff; SEP, small ORF-encoded polypeptide; SPE, solid-phase extraction.
The isolation and enrichment of SEPs from biological samples is another critical step for their characterization. Accordingly, various methodologies have been applied for this purpose, including 30-kDa MWCO filter, C8 SPE, and organic solvent–based or inorganic salt–based precipitation. Among these, the 30-kDa MWCO filter and C8 SPE are commonly used albeit based on different principles. In the case of 30-kDa MWCO filter, SEPs are separated and enriched based on their molecular size and/or weight. On the contrary, selective adsorption and selective elution are utilized to enrich, separate, and purify SEPs using C8 SPE. Therefore, we hypothesized these may represent two complimentary strategies for SEP enrichment, and that their combined use could significantly improve SEP discovery. We tested our hypothesis by enriching SEPs from equal amounts of HEK293T cell lysates using both 30-kDa MWCO filter and C8 SPE and then performing SDS-PAGE and LC–MS/MS analysis based on our in-house database (Fig. 1, B and C).
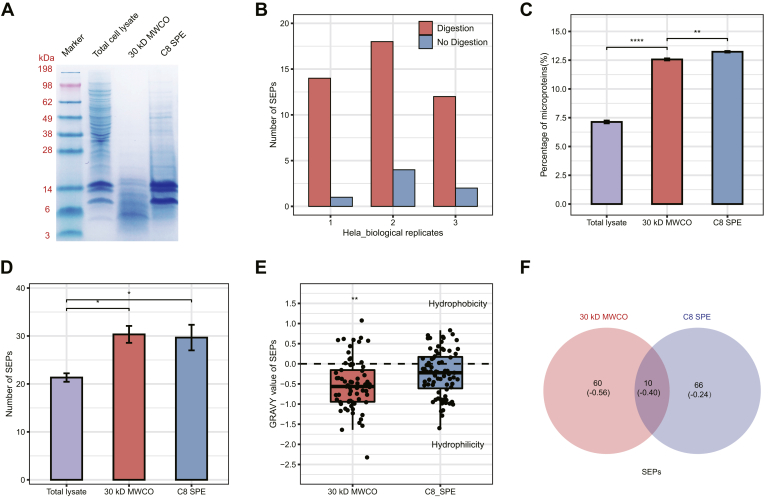
Tricine SDS-PAGE showed that both 30-kDa MWCO filter and C8 SPE are very effective in enriching for proteins/peptides in the molecular weight range between 5 and 15 kDa (Fig. 2A). LC–MS/MS data further confirmed these results by showing that 12.6% and 13.2% of the total identified annotated proteins in the 30-kDa MWCO filter and C8 SPE approaches, respectively, were low molecular weight proteins/peptides (≤100 aa), in comparison to only 7.1% in total lysates without enrichment (Fig. 2C). Importantly, an average of 30 and 29 candidate SEPs were identified from the 30-kDa MWCO filter and C8 SPE, respectively, which are both significantly higher than the 21 candidates identified using total lysates without enrichment (Fig. 2D). Interestingly, and as expected, given the complimentary nature of the two approaches, there are only a few overlaps between SEP candidates identified with 30-kDa MWCO filter and C8 SPE (Fig. 2F), despite the observed comparable enrichment efficiency. Similar results showing low overlap between the two methods were obtained in mouse kidney lysate, HeLa, and MEF cell lysate (supplemental Fig. S1, A and C–E). This is likely the result of differences in enrichment efficiency according to SEP hydrophobicity between the two methods, since protein hydrophobicity analysis showed that hydrophobic SEPs accounted for 18.2% and 32.7% of the total SEPs identified in HEK293T cells enriched with 30-kDa MWCO filter and C8 SPE, respectively (supplemental Fig. S1B). In fact, the grand average of hydropathicity value (22) of all SEPs identified in C8 SPE was −0.26, compared with only −0.52 for those detected using the 30-kDa MWCO filter (Fig. 2E). Moreover, the grand average of hydropathicity values obtained for SEPs uniquely identified by each method were −0.24 and −0.56 for C8 SPE and 30-kDa MWCO filter, respectively. Among the possible explanations for these differences are the low extraction efficiency of hydrophobic proteins in the 30-kDa MWCO filter because of the lack of detergent in the solvent, and the fact that hydrophilic proteins tend to be lost in the processes of C8 SEP, such as methanol/chloroform precipitation (23).
Fig. 2.
Comparison of different strategies for the identification of lncRNA-encoded SEPs.A, tricine SDS-PAGE analysis of total cell lysates, SEP fractions enriched by 30-kDa MWCO membrane and C8 SPE column from HEK293T cells are shown from left to right, respectively. B, the number of SEPs identified in HeLa cells from three biological replicates at a single MS run with (red) and without (blue) tryptic digestion. C, percentage of annotated microproteins identified in the total cell lysate and enriched fractions from HEK293T cells with three technical replicates. D, the number of SEPs identified in total cell lysate and enriched fractions from HEK293T cells with three technical replicates. E, GRAVY values of the total SEPs identified in the fractions enriched by 30-kDa MWCO membrane and C8 SPE column from HEK293T cells. F, Venn diagram of the total SEPs identified in the fractions enriched by 30-kDa membrane and C8 SPE column from HEK293T cells. (Values in parentheses show the average GRAVY values of SEPs in each region). GRAVY, grand average of hydropathicity; HEK293T, human embryonic kidney 293T; lncRNA, long noncoding RNA; MWCO, molecular weight cutoff; SEP, small ORF-encoded polypeptide; SPE, solid-phase extraction.
For the MS-based analysis, we employed trypsin-based digestion for the identification of SEPs, since most of putative SEPs deposited in our newly generated in-house database were longer than 25 amino acids, in accordance with previous studies showing that only 27% of SEPs identified in human cell lines or tissues are less than 25 amino acids long (10). Moreover, our results also demonstrate that a higher number of SEPs can be detected in HeLa samples treated with tryptic digestion compared with those with no digestion (Fig. 2B).
Comprehensive SEP Discovery from Multiple Cell Lines and Tissues
In order to allow for a diverse and comprehensive detection of novel SEPs, the optimized MS-based workflow was applied to eight human cell lines, three mouse cell lines, and eight mouse tissues. In addition, and to ensure high-confidence SEP identification, MS-detected SEPs were strictly filtered through manual sequence and spectrum checking (9, 24). The peptides satisfying any of the following filtering criteria were removed from the list: (1) peptide sequences identical to annotated proteins in the UniProt database (treating isoleuvine and leucine as equivalent); (2) peptides with less than eight amino acids in length; (3) spectra containing less than four continuous b- or y-ions and many impure peaks with high intensity; and (4) spectra containing less than 40% b- and y-ions coverage. After filtering, the remaining peptides were considered as novel SEP-derived peptides. This resulted in the confident identification of 353 SEPs from eight human cell lines and 409 SEPs from three mouse cell lines and eight mouse tissues (Table 1), which should represent, to the best of our knowledge, the largest number of SEPs identified by MS reported to date.
Table 1.
The number of lncRNA-encoded SEPs discovered in different cell lines and tissues from human and mouse
| Samples | Number of identified SEPs | Total identified SEPs | |
|---|---|---|---|
| Human cell lines | 293T | 249 | 353 |
| HeLa | 126 | ||
| 22Rv1 | 46 | ||
| Du145 | 30 | ||
| LNCap | 44 | ||
| PC3 | 24 | ||
| A375 | 33 | ||
| U251 | 19 | ||
| Mouse cell lines | MEF | 75 | 409 |
| mESC | 47 | ||
| 4T1 | 60 | ||
| Mouse tissues | Kidney | 213 | |
| Liver | 85 | ||
| Heart | 36 | ||
| Brain | 46 | ||
| Cerebellum | 64 | ||
| Testicle | 14 | ||
| Lung | 6 | ||
| Spleen | 9 | ||
Specifically, we identified 373 novel peptides derived from 353 human SEPs, with 131 (35.1%) being detected at least twice from eight human cell lines. Similarly, we discovered 425 novel peptides derived from 409 mouse SEPs, with 103 (24.2%) being detected twice or more across cell lines and tissues. In addition, we have also been able to identify the previously reported functional peptide NoBody. Additional information regarding the 762 identified SEPs, including SEP sequences, unique peptide sequences, and genomic location, can be found in supplemental Tables S3 and S4. Representative mass spectra of several identified lncRNA-SEP peptides are listed in supplemental Figure S2. The raw data files, including search result files, are available at ProteomeXchange with the identifier PXD019486.
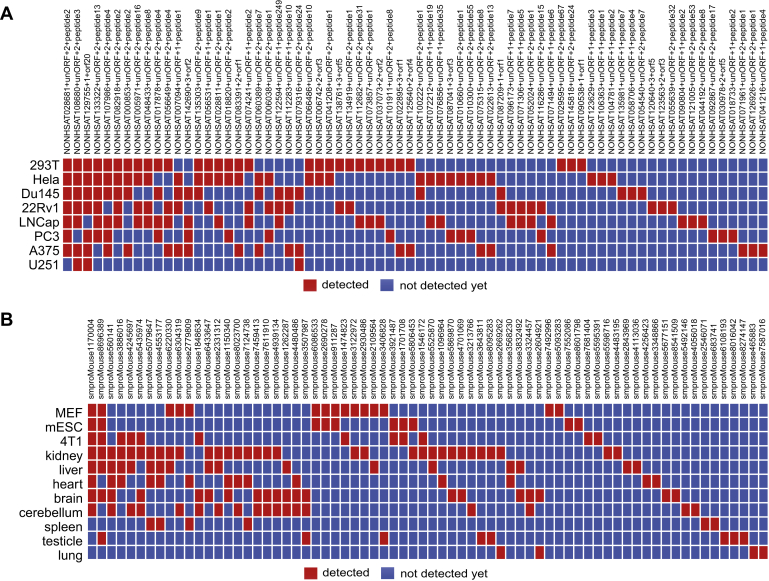
Importantly, we have identified a handful of SEPs that are found in most human cell lines used in our study, even though many SEPs are cell line specific (Fig. 3A). A total of 26 of the 353 (7.4%) identified human SEPs were present in at least three different human cell lines, with similar results found in mouse cell lines (Fig. 3B). These results suggest that SEPs are widely present in different cell lines and tissues in different species. Moreover, it is worth considering the fact that biological and technical replicates might significantly increase the number of SEPs discovered. Hence, we performed LC–MS/MS analysis on 18 technical replicates of SEPs extracted from mouse kidney samples in order to test how many technical replicates are necessary to achieve a relative saturation level for SEP identification from a biological sample. Our results show that an average of 23 SEPs was detected per run with a range between 18 and 34 SEPs in each sample (supplemental Fig. S3), which brings the total number of novel SEPs detected from the mouse kidney to 169. Similar results were found for the HEK293T cell line, in which 196 SEPs were identified in total across the different biological and technical replicates. These findings are consistent with previous studies (10) and support the idea that lncRNA-SEP detection is variable.
Fig. 3.
Distribution of representative lncRNA-encoded SEPs. In different human cell lines (A) and mouse tissues (B). Nearly 20% of the identified human lncRNA-encoded SEPs were present in more than two different human cell lines, similar to results found in mouse tissues. lncRNA, long noncoding RNA; SEP, small ORF-encoded polypeptide.
Computational and Experimental Validation of the Novel MS-Detected SEPs
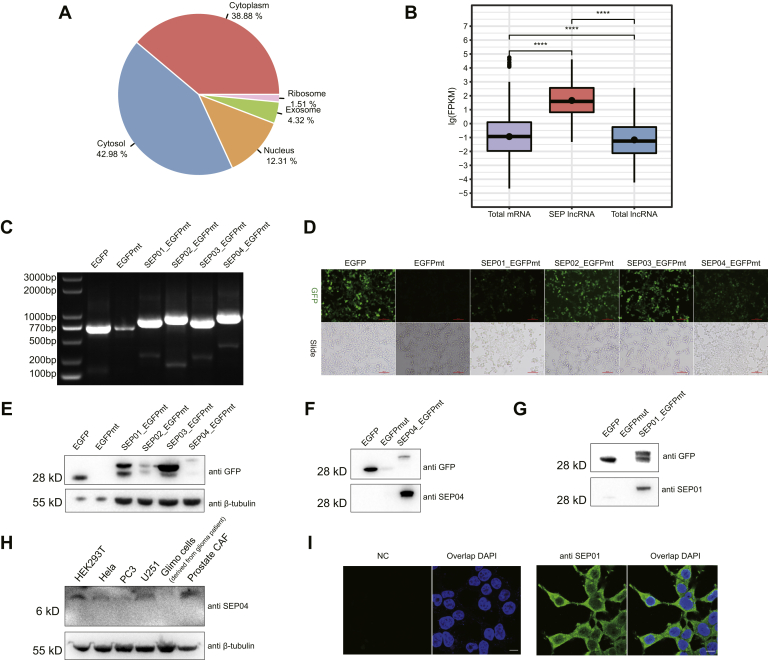
To confirm the reliability of our data, several bioinformatics and experimental approaches were implemented to validate our findings. Computing-based methods included prediction of cellular location and expression analysis for SEP-coding lncRNA transcripts. Experimental-based methods included expression of lncRNAs, identification of additional tryptic peptides based on a PRM strategy, and synthesis of peptide standards. First, cellular location is an important factor to understand the functional roles of lncRNAs. For SEP-coding lncRNAs, we expected that they tend to reside in the cytoplasm rather than the nucleus to enable ribosomal translation. We tested this hypothesis by collecting and analyzing the lncRNA transcripts corresponding to the identified SEPs using LncLocator, a subcellular localization predictor for lncRNAs based on a stacked ensemble classifier (25). As expected, we found that more than 80% of SEP-coding lncRNAs are predicted to locate in the cytoplasm, whereas less than 13% are predicted to locate in the nucleus (Fig. 4A). Similar results were observed when analyzing mouse SEPs (supplemental Fig. S4A). Our observations are different from those found in a previous study (26), which detected that 17% of lncRNAs are enriched in the nucleus, 4% in the cytoplasm, whereas 15% of mRNAs are enriched in the nucleus, 26% in the cytoplasm, it suggested that these SEP-coding lncRNAs are highly likely to bind to ribosomes for active translation.
Fig. 4.
Computational and experimental validation of lncRNA-encoded SEPs.A, prediction of the subcellular localization of human SEP-coding lncRNA transcripts. More than 80% of SEP-coding lncRNAs were predicted to locate in the cytoplasm, whereas less than 13% were found in the nucleus. B, comparison of the expression levels of SEP-coding lncRNAs, whole cell mRNAs, and lncRNAs in HEK293T cells. C, RT-PCR–based validation of the transcription of SEP–GFP fusion coding sequences. The RNA transcripts corresponding to EGFPwt-, EGFPmt-, SEP01-EGFPmt-, SEP02-EGFPmt-, SEP03-EGFPmt-, and SEP04-EGFPmt-constructs were subjected to RT-PCR analysis. D, detection of GFP fluorescence for different GFP fusion constructs after transfection into HEK293T cells for 24 h. The scale bar represents 100 μm. E, Western blot of the SEP–EGFP fusion proteins by anti-GFP antibody. About 2 μg of the total proteins from the cell lysates of HEK293T cells transfected with EGFP-n1 and EGFPmt-n1 and 15 μg of that from HEK293T cells transfected with different SEP–GFP fusion constructs were subjected to gel separation and detection by both GFP and β-tubulin antibodies. F, Western blot of the SEP04–EGFP fusion protein by the customized anti-SEP04 polyclonal antibody. G, Western blot of the SEP01–EGFP fusion protein by customized anti-SEP01. H, Western blot of the endogenous SEP04 in different human cell lines. I, immune fluorescence of SEP01 in HEK293T cells using anti-SEP01 polyclonal antibody. The scale bar represents 10 μm. HEK293T, human embryonic kidney 293T; lncRNA, long noncoding RNA; SEP, small ORF-encoded polypeptide.
Second, the levels of protein-coding RNA transcripts generally reflect the levels of expression of their corresponding proteins (27). This directly affects MS-based SEP detection, as higher levels of SEP-coding lncRNA expression may make it easier for the corresponding SEPs to be detected. To investigate this, we retrieved the levels of expression of the 196 SEP-coding lncRNAs identified and the whole cell mRNAs in HEK293T cells from an RNA-Seq dataset (Gene Expression Omnibus: GSE122633). Interestingly, we observed that the average levels of expression of the identified 196 SEP-coding lncRNAs were significantly higher than those of the whole cell expressed mRNAs in HEK293T cells (Fig. 4B). In addition, we found similar results when analyzing mouse kidney SEPs (supplemental Fig. S4B). While these results are not in agreement with previous studies showing that the levels of expression of lncRNAs are comparable to those of mRNAs, the higher levels of expression found here could partially explain why these SEPs can be readily detected in our study.
Third, considering that most SEPs were only detected with a single peptide, likely because of the relatively low abundance of SEPs in cells, we implemented a PRM strategy based on the predicted or identified tryptic peptides and the data-independent acquisition MS method in order to present additional evidence to support our detection claims. Specifically, we performed LC–PRM–MS analysis on 32 of the 196 identified SEPs in HEK293T cells and identified eight additional peptides for eight SEPs (supplemental Table S5), increasing SEP sequence coverage.
Fourth, to further increase the reliability of SEPs identified in this study, we selected 15 peptides from 14 SEPs identified in HEK2932T cell lines more than twice to synthesize as peptide standards. We then analyzed the mixture of 15 synthetic peptide standards by LC–MS/MS and compared the MS/MS spectra of both synthetic and identified SEP peptides. Our results show that 15 previously identified peptides from 14 SEPs were successfully matched with the synthetic peptides (supplemental Fig. S5). The raw data files, including search result files, are available in ProteomeXchange with the identifier PXD019486.
In addition, we selected four human SEPs (defined here as SEP01, SEP02, SEP03, and SEP04, shorted for NONHSAT077882+1+orf4, NONHSAT096173+unORF+1+peptide7, NONHSAT126926+unORF+2+peptide1, NONHSAT130014+unORF+2+peptide9, respectively) that were found in most of the human cell lines and cloned the corresponding genes into pEGFPmut-N1 plasmids (supplemental Fig. S6). The RNA transcripts corresponding to these four SEP–GFP fusion coding sequences were successfully identified by RT-PCR 24 h after being transfected into HEK293T cells from EGFPwt-transfected, SEP01-EGFPmut-transfected, SEP02-EGFPmut-transfected, SEP03-EGFPmut-transfected, and SEP04-EGFPmut-transfected cells (Fig. 4C). We also observed substantial expression of GFP or SEP ORF–GFP fusion proteins (Fig. 4D), whereas no expression of GFP was found in cells transfected with the EGFPmt plasmid, in which the start codon ATGGTG of GFP was mutated to ATTGTT to eliminate translation initiation. The results were further confirmed by Western blotting analysis using an anti-GFP antibody (Fig. 4E and supplemental Fig. S7).
We next attempted to provide direct evidence of SEP expression by detecting endogenous SEPs from HEK293T cells. We raised polyclonal antibodies specifically against the SEP01 and SEP04 using their specific peptides that contained antigen epitopes. Immunoblotting results showed clear and specific bands in SEP04-EGFPmut-transfected and SEP01-EGFPmut-transfected cells at their respective expected molecular weights (Fig. 4, F and G), suggesting high specificity of these two antibodies. Importantly, endogenous SEP04 also exhibited the predicted relative molecular weight in multiple human cell lysates (Fig. 4H), suggesting it exists in full length and stable forms in vivo. We verified the existence of endogenous SEP01 polypeptides in HEK293T cells by immunofluorescence (Fig. 4I).
Taken together, our analysis validated the existence of multiple SEPs through different strategies including MS, expression of lncRNAs, and antibody evidence.
Characterization of Total Discovered SEPs
In order to obtain a clear picture and draw deeper insights into the properties of the identified SEPs, we characterized them from multiple aspects, including type, codon usage, length distribution, amino acid composition, and protein stability.
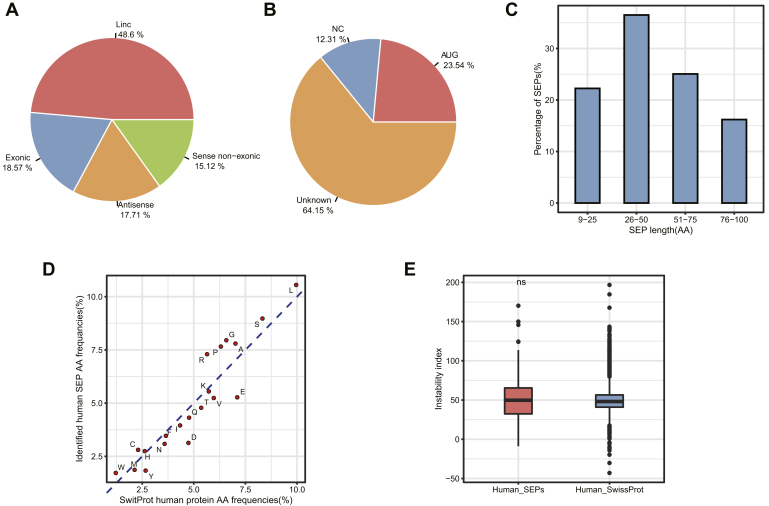
LncRNAs were mainly categorized into four subclasses based on their genomic location and context: long intergenic noncoding RNAs (lincRNAs), antisense, exonic, and sense nonexonic (28). Interestingly, it has been found that lincRNAs are transcriptionally activated in a similar fashion to mRNAs, as they are more conserved than introns and antisense transcripts. This observation is consistent with our results, which showed that lincRNA-encoded SEPs accounted for 48.6% of the total identified human SEPs, whereas antisense, exonic, and sense nonexonic lncRNAs accounted for 17.71%, 18.57%, and 15.12%, respectively (Fig. 5A). Similarly, lincRNA-encoded SEPs accounted for 53.38% of the total identified mouse SEPs, compared with 12.68%, 5.39%, and 28.54% for antisense, exonic, and sense nonexonic lncRNA, respectively (supplemental Fig. S8A).
Fig. 5.
Characterization of the identified SEPs and their corresponding lncRNAs.A, classification of human SEP-coding lncRNAs based on their location on the genome with respect to protein-coding genes. B, usage of start codon in human SEPs. Nearly 77% of the identified lncRNA-SEPs were found to be initiated with non-AUG start codons. AUG, smORF initiates with AUG; NC, near cognate start codon, containing only one nucleotide different from AUG; unknown, smORF not initiated with AUG and not an NC. C, length distribution of human SEPs. D, the amino acid usage of canonical proteins and identified human SEPs. The human SEPs identified in this study tend to utilize more positively charged amino acids (such as K) and less negatively charged amino acids (such as D and E), while using a similar amount of uncharged amino acids, as observed in canonical proteins. E, stability of the identified human SEPs. The instability index was calculated by ProtParam and used to characterize the protein stability of human SEPs. There is only a minor deviation between the instability index distributions of the identified SEPs and canonical proteins. lncRNA, long noncoding RNA; SEP, small ORF-encoded polypeptide.
Codon usage bias is an important evolutionary feature in a genome and provides important information for studying gene function and gene expression. The codon usage bias of the identified SEPs was analyzed by predicting the start and stop codons, as previously reported (14). Briefly, any in-frame ATG or near cognate codon in a Kozak sequence was predicted to be a start codon. In all other cases, SEPs were predicted to have an unknown start codon. Of the 357 identified human SEPs, 23.54% and 12.31% were initiated with AUG or a near cognate codon, respectively, whereas the majority (64%) had an unknown start codon (Fig. 5B). Similarly, 28.44% and 11.1% of the 409 identified mouse SEPs started with AUG or a near cognate codon, respectively, with 60% exhibiting an unknown start codon (supplemental Fig. S8B). By contrast, almost all canonical proteins were initiated with an AUG start codon, in accordance with previous reports (10). The fact that a majority of SEPs is initiated with an unknown start codon makes their novel detection less likely.
We determined the length of SEPs with a known start codon by the predicted ORF length. In contrast, for SEPs with an unknown start codon, length was defined as the interval between two contiguous stop codons. As such, a majority of the identified human (Fig. 5C) and mouse (supplemental Fig. S8C) SEPs were predicted to have a length between 26 and 50 aa, with the shortest one being only nine amino acids long. Importantly, considering that nearly 75% of SEPs were predicted to be larger than 25 amino acids in length, enzyme-based digestion seems to be a good choice for discovering SEPs using the MS-based approach.
Although it is not yet possible to fully explain protein function from its amino acid sequence, it is nevertheless feasible to establish correlations between protein structure and function by studying the properties of the amino acids that compose it (29). Our amino acid composition analysis revealed that both identified human and mouse SEPs tend to utilize more positively charged amino acids (e.g., K) and less negatively charged amino acids (e.g., D and E) than canonical proteins, while using a similar amount of uncharged amino acids (Fig. 5D and supplemental Fig. S8D), an observation in line with previous studies (30, 31). Furthermore, it has been proposed that proteins containing more positively charged amino acids and a hydrophobic region were commonly found across the cell membrane and organelles (6, 32, 33, 34). This opens the possibility for SEPs to act as transmembrane peptides. It is also possible that SEPs containing a higher proportion of positively charged amino acids may play a role in binding to negatively charged DNA or RNA (35, 36, 37). Finally, the higher proportion of K and R amino acids in SEPs suggests that trypsin digestion might not be the best choice for SEP discovery, since it can generate very small peptides that are not suitable for MS detection. Instead, a combination of multiple proteases would likely improve SEP discovery and sequence coverage.
Finally, we calculated instability indexes using ExPASy ProtParam (38, 39) and found only minor differences between the distributions of the identified SEPs and canonical proteins (Fig. 5E and supplemental Fig. S8E), which is in accordance with previous results (40). This suggests that SEPs might be as stable as canonical proteins.
Discussion
Technological advances over the past few years have led to the discovery of numerous biologically relevant SEPs from different species. While it is expected that many more are yet to be discovered, SEP detection is technically challenging because of their relatively low abundance and small size. In the current study, we implemented a comprehensive strategy for SEP discovery and characterization from multiple human and mouse cell lines and mouse tissues through an optimized MS-based workflow by combining two effective and complementary polypeptide enrichment methods with the de novo construction of a high-quality SEP database. Our strategy enabled the discovery of 762 novel SEPs from different human and murine cell lines and tissues, which, to our knowledge, represent the largest number of MS-detected SEPs ever to be reported.
The improved SEP discovery rate reported here can be attributed to several reasons. First, our in-house SEP reference database collects a maximum number of putative smORFs from lncRNA transcripts deposited in the NONCODE database by combining the six-frame translation mode with ORFfinder. Even though the strategy employed in building the database may result in an elevated false discovery rate, it allowed us to investigate a higher number of SEPs across cell lines and tissues and to subsequently validate this identification with stringent criteria. Furthermore, the combination of two effective and complementary polypeptide enrichment strategies, 30-kDa MWCO filter and C8 SPE, helps circumventing SEP detection biases of different approaches and thus greatly improves SEP discovery. Moreover, the implementation of trypsin-based digestion and multiple biological and technical replicates further improved MS-based SEP identification.
Importantly, the increased number of discovered SEPs allowed us to gain deeper insights into their physical and chemical properties, some of which might partially explain why previous studies have failed to identify them. For example, the identified SEPs were derived from lncRNA transcripts with a higher average level of expression than mRNA, which means the translation efficiency of SEPs might be lower than mRNA, and therefore reducing the sensitivity of SEP identification. In fact, more than 60% of the identified SEPs from both human or mouse are initiated by unknown start codons (i.e., non-AUG), which typically have reduced efficiency when compared with AUG codons (41, 42). Moreover, non-AUG initiation may itself make it less likely for SEPs to be discovered and included in a reference database for downstream MS-based identification. Furthermore, SEPs are commonly shorter in amino acid length and enriched with more basic residues, which are typical features of known coding genes lacking MS evidence (43). Together, these observations suggest that the SEPs identified in this study might just represent the tip of the iceberg, and that many more low abundance SEPs remain undiscovered.
Importantly, nearly 20% of identified human SEPs were present in more than two different human cell lines. Experimental evidence acquired from in vitro translation, MS, immunoblotting, and bioinformatics also confirmed the existence of 19 novel SEPs in HEK293T cells. Moreover, these newly discovered SEPs are predicted to be as stable as canonical proteins and in their full-length form in human cell lines, as demonstrated by Western blotting. These results strongly suggest that the identified SEPs do not result from random noise but instead are of biological significance, despite individual SEPs still lacking a complete functional characterization.
In summary, we demonstrate that an optimized MS-based workflow allows for comprehensive discovery of hundreds of novel SEPs from different human and mouse cell lines and tissues, which can not only provide new clues for the annotation of noncoding elements in the genome but might also serve as a valuable resource for the functional characterization of individual SEPs.
Data availability
The MS raw data and research results are publicly available in ProteomeXchange (http://proteomecentral.proteomexchange.org) with the identifiers PXD019486. The SEP databases have been uploaded to the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, with an accession number OMIX266 (http://bigd.big.ac.cn/omix/release/OMIX266).
Supplemental data
This article contains supplemental data.
Conflict of interest
The authors declare no competing interests.
Acknowledgments
We thank Junjie Hou, from the Institute of Biophysics, Chinese Academy of Sciences, for his assistance on MS data analysis. We thank Xiang Ding, Zhensheng Xie, Lili Niu, Peng Xue, Nali Zhu, and Mengmeng Zhang from the Laboratory of Proteomics, Institute of Biophysics, Chinese Academy of Sciences, for their assistance with the LC–MS/MS experiment. Data analysis and computational resources were supported by the Center for Big Data Research in Health (http://bigdata.ibp.ac.cn), Institute of Biophysics, Chinese Academy of Sciences. This work was supported by the National Natural Science Foundation of China (91640112 and 31670185) and the National Key Research and Development Program of the Ministry of Science and Technology of China (2018YFA0106901 and 2017YFC0907503).
Author contributions
F. Y., R. C., T. C., and J. L. supervision; F. Y., R. C., T. C., and J. L. writing-review and editing; Q. Z., Y. T., L. Z., Y. H., J. W., Y. Z., and X. G. investigation; Q. Z. writing-original draft; E. W. and B. Z. validation.
Contributor Information
Jianjun Luo, Email: luojianj@ibp.ac.cn.
Runsheng Chen, Email: rschen@ibp.ac.cn.
Fuquan Yang, Email: fqyang@ibp.ac.cn.
Supplemental data
References
- 1.Huang J.Z., Chen M., Chen fnm, Gao X.C., Zhu S., Huang H., Hu M., Zhu H., Yan G.R. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell. 2017;68:171–171.e6. doi: 10.1016/j.molcel.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Jackson R., Kroehling L., Khitun A., Bailis W., Jarret A., York A.G., Khan O.M., Brewer J.R., Skadow M.H., Duizer C., Harman C.C.D., Chang L., Bielecki P., Solis A.G., Steach H.R. The translation of non-canonical open reading frames controls mucosal immunity. Nature. 2018;564:434–438. doi: 10.1038/s41586-018-0794-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Makarewich C.A., Baskin K.K., Munir A.Z., Bezprozvannaya S., Sharma G., Khemtong C., Shah A.M., McAnally J.R., Malloy C.R., Szweda L.I., Bassel-Duby R., Olson E.N. MOXI is a mitochondrial micropeptide that enhances fatty acid β-oxidation. Cell Rep. 2018;23:3701–3709. doi: 10.1016/j.celrep.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauli A., Valen E., Schier A.F. Identifying (non-)coding RNAs and small peptides: challenges and opportunities. Bioessays. 2015;37:103–112. doi: 10.1002/bies.201400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen S.M. Everything old is new again: (linc)RNAs make proteins! EMBO J. 2014;33:937–938. doi: 10.1002/embj.201488303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P. Extensive translation of small open reading frames revealed by Poly-Ribo-seq. Elife. 2014;3 doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013;154:240–251. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chew G.L., Pauli A., Rinn J.L., Regev A., Schier A.F., Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 2013;140:2828–2834. doi: 10.1242/dev.098343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013;9:59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J., Ward C.C., Jungreis I., Slavoff S.A., Schwaid A.G., Neveu J., Budnik B.A., Kellis M., Saghatelian A. Discovery of human sORF-encoded polypeptides (SEPs) in cell lines and tissue. J. Proteome Res. 2014;13:1757–1765. doi: 10.1021/pr401280w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budamgunta H., Olexiouk V., Luyten W., Schildermans K., Maes E., Boonen K., Menschaert G., Baggerman G. Comprehensive peptide analysis of mouse brain Striatum identifies novel sORF-encoded polypeptides. Proteomics. 2018;18 doi: 10.1002/pmic.201700218. [DOI] [PubMed] [Google Scholar]
- 12.Pueyo J.I., Magny E.G., Couso J.P. New peptides under the s(ORF)ace of the genome. Trends Biochem. Sci. 2016;41:665–678. doi: 10.1016/j.tibs.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Vale W., Vaughan J., Jolley D., Yamamoto G., Bruhn T., Seifert H., Perrin M., Thorner M., Rivier J. Assay of growth hormone-releasing factor. Methods Enzymol. 1986;124:389–401. doi: 10.1016/0076-6879(86)24030-6. [DOI] [PubMed] [Google Scholar]
- 14.Ma J., Diedrich J.K., Jungreis I., Donaldson C., Vaughan J., Kellis M., Yates J.R., Saghatelian A. Improved identification and analysis of small open reading frame encoded polypeptides. Anal. Chem. 2016;88:3967–3975. doi: 10.1021/acs.analchem.6b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schagger H. Tricine-SDS-PAGE. Nat. Protoc. 2006;1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 16.Anderson D.M., Anderson K.M., Chang C.L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi P., Ramirez-Martinez A., Li H., Cannavino J., McAnally J.R., Shelton J.M., Sánchez-Ortiz E., Bassel-Duby R., Olson E.N. Control of muscle formation by the fusogenic micropeptide myomixer. Science. 2017;356:323–327. doi: 10.1126/science.aam9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q., Vashisht A.A., O'Rourke J., Corbel S.Y., Moran R., Romero A., Miraglia L., Zhang J., Durrant E., Schmedt C., Sampath S.C., Sampath S.C. The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 2017;8:15664. doi: 10.1038/ncomms15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K.I., Clohessy J.G., Pandolfi P.P. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017;541:228–232. doi: 10.1038/nature21034. [DOI] [PubMed] [Google Scholar]
- 20.D'Lima N.G., Ma J., Winkler L., Chu Q., Loh K.H., Corpuz E.O., Budnik B.A., Lykke-Andersen J., Saghatelian A., Slavoff S.A. A human microprotein that interacts with the mRNA decapping complex. Nat. Chem. Biol. 2017;13:174–180. doi: 10.1038/nchembio.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H., Yang X., Huang N., Liu J., He K., Xie K. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 23.Liu J., Wang F., Mao J., Zhang Z., Liu Z., Huang G., Cheng K., Zou H. High-sensitivity N-glycoproteomic analysis of mouse brain tissue by protein extraction with a mild detergent of N-dodecyl β-D-maltoside. Anal. Chem. 2015;87:2054–2057. doi: 10.1021/ac504700t. [DOI] [PubMed] [Google Scholar]
- 24.He C., Jia C., Zhang Y., Xu P. Enrichment-based proteogenomics identifies microproteins, missing proteins, and novel smORFs in Saccharomyces cerevisiae. J. Proteome Res. 2018;17:2335–2344. doi: 10.1021/acs.jproteome.8b00032. [DOI] [PubMed] [Google Scholar]
- 25.Cao Z., Pan X., Yang Y., Huang Y., Shen H.B. The lncLocator: A subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics. 2018;34:2185–2194. doi: 10.1093/bioinformatics/bty085. [DOI] [PubMed] [Google Scholar]
- 26.Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G., Lagarde J., Veeravalli L., Ruan X., Ruan Y., Lassmann T. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H., Mathieson T., Lemeer S., Schnatbaum K., Reimer U., Wenschuh H. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 28.Xie C., Yuan J., Li H., Li M., Zhao G., Bu D., Zhu W., Wu W., Chen R., Zhao Y. NONCODEv4: Exploring the world of long non-coding RNA genes. Nucleic Acids Res. 2014;42:D98–D103. doi: 10.1093/nar/gkt1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couso J.P., Patraquim P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 2017;18:575–589. doi: 10.1038/nrm.2017.58. [DOI] [PubMed] [Google Scholar]
- 30.Aspden J.L., Eyre-Walker Y.C., Phillips R.J., Amin U., Mumtaz M.A., Brocard M., Couso J.P. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. Elife. 2014;3:e03528. doi: 10.7554/eLife.03528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu S., Zhang J., Lian X., Sun L., Meng K., Chen Y., Sun Z., Yin X., Li Y., Zhao J., Wang T., Zhang G., He Q.Y. A hidden human proteome encoded by 'non-coding' genes. Nucleic Acids Res. 2019;47:8111–8125. doi: 10.1093/nar/gkz646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones S.W., Christison R., Bundell K., Voyce C.J., Brockbank S.M., Newham P., Lindsay M.A. Characterisation of cell-penetrating peptide-mediated peptide delivery. Br. J. Pharmacol. 2005;145:1093–1102. doi: 10.1038/sj.bjp.0706279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy M.P. Targeting lipophilic cations to mitochondria. Biochim. Biophys. Acta. 2008;1777:1028–1031. doi: 10.1016/j.bbabio.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 34.Saghatelian A., Couso J.P. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 2015;11:909–916. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanyu-Nakamura K., Sonobe-Nojima H., Tanigawa A., Lasko P., Nakamura A. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 2008;451:730–733. doi: 10.1038/nature06498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauressergues D., Couzigou J.M., Clemente H.S., Martinez Y., Dunand C., Bécard G., Combier J.P. Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015;520:90–93. doi: 10.1038/nature14346. [DOI] [PubMed] [Google Scholar]
- 37.Slavoff S.A., Heo J., Budnik B.A., Hanakahi L.A., Saghatelian A. A human short open reading frame (sORF)-encoded polypeptide that stimulates DNA end joining. J. Biol. Chem. 2014;289:10950–10957. doi: 10.1074/jbc.C113.533968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guruprasad K., Reddy B.V., Pandit M.W. Correlation between stability of a protein and its dipeptide composition: A novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 39.Wilkins M.R., Gasteiger E., Bairoch A., Sanchez J.C., Williams K.L., Appel R.D., Hochstrasser D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 40.Verheggen K., Volders P.J., Mestdagh P., Menschaert G., Van Damme P., Gevaert K., Martens L., Vandesompele J. Noncoding after all: Biases in proteomics data do not explain observed absence of lncRNA translation products. J. Proteome Res. 2017;16:2508–2515. doi: 10.1021/acs.jproteome.7b00085. [DOI] [PubMed] [Google Scholar]
- 41.Clements J.M., Laz T.M., Sherman F. Efficiency of translation initiation by non-AUG codons in Saccharomyces cerevisiae. Mol. Cell. Biol. 1988;8:4533–4536. doi: 10.1128/mcb.8.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kearse M.G., Wilusz J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017;31:1717–1731. doi: 10.1101/gad.305250.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omenn G.S., Lane L., Overall C.M., Corrales F.J., Schwenk J.M., Paik Y.K., Van Eyk J.E., Liu S., Snyder M., Baker M.S., Deutsch E.W. Progress on identifying and characterizing the human proteome: 2018 metrics from the HUPO human proteome project. J. Proteome Res. 2018;17:4031–4041. doi: 10.1021/acs.jproteome.8b00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS raw data and research results are publicly available in ProteomeXchange (http://proteomecentral.proteomexchange.org) with the identifiers PXD019486. The SEP databases have been uploaded to the OMIX, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, with an accession number OMIX266 (http://bigd.big.ac.cn/omix/release/OMIX266).