Abstract
Apart from the four canonical nucleobases, DNA molecules carry a number of natural modifications. Substantial evidence shows that DNA modifications can regulate diverse biological processes. Dynamic and reversible modifications of DNA are critical for cell differentiation and development. Dysregulation of DNA modifications is closely related to many human diseases. The research of DNA modifications is a rapidly expanding area and has been significantly stimulated by the innovations of analytical methods. With the recent advances in methods and techniques, a series of new DNA modifications have been discovered in the genomes of prokaryotes and eukaryotes. Deciphering the biological roles of DNA modifications depends on the sensitive detection, accurate quantification, and genome-wide mapping of modifications in genomic DNA. This review provides an overview of the recent advances in analytical methods and techniques for both the quantification and genome-wide mapping of natural DNA modifications. We discuss the principles, advantages, and limitations of these developed methods. It is anticipated that new methods and techniques will resolve the current challenges in this burgeoning research field and expedite the elucidation of the functions of DNA modifications.
Apart from the four canonical nucleobases, DNA molecules carry a number of natural modifications.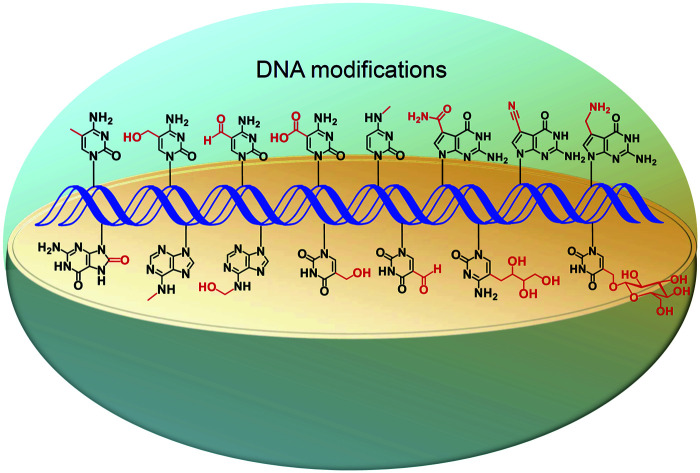
1. Introduction
DNA molecules employ four canonical nucleobases, adenine (A), thymine (T), cytosine (C), and guanine (G), to encode genetic information in living organisms.1 In addition to these four nucleobases, recent years have witnessed the discovery of a variety of modified nucleobases in genomes.2,3 DNA cytosine methylation (5-methylcytosine, 5mC) is the most extensively characterized epigenetic modification, and it plays sophisticated roles in regulating gene expression in mammalian cells.4 The formation of 5mC by DNA methyltransferases (DNMTs) and oxidation by ten-eleven translocation (TET) proteins diversify the genomes and shape the chromatin landscape.5,6 TET proteins catalyze the oxidation of 5mC to 5-hydroxymethylcytosine (5hmC) and 5-formylcytosine (5fC), and finally to 5-carboxycytosine (5caC).7 The 5fC and 5caC undergo base excision repair to restore the unmodified cytosines.8 Apart from being intermediates in the active demethylation pathway, 5hmC, 5fC, and 5caC are also considered to function as epigenetic modifications with potential regulatory roles in gene expression.9
In addition to DNA cytosine modifications, many other types of modifications also exist in the genomes of living organisms.10,11N6-Methyladenosine (6mA) is a natural DNA modification existing in both prokaryotes and eukaryotes.12 6mA functions in the restriction–modification (RM) systems and gene regulatory processes in prokaryotes.13 In 2015, it was reported that 6mA was presented in the genomes of Chlamydomonas reinhardtii,14Caenorhabditis elegans,15 and Drosophila melanogaster.16 Later on, 6mA was detected in a variety of living organisms of vertebrates and mammals.17–21 The most recent studies demonstrated that 6mA mainly existed in the mitochondrial DNA22 and originated from the degraded RNA nucleoside of N6-methyladenosine (m6A), which was further processed through the nucleotide-salvage pathway and incorporated into genomic DNA by DNA polymerases.23,24
5-Hydroxymethyluracil (5hmU) is a thymine modification existing in the genomes of many living organisms, such as leishmania,25 bacteriophages,26 dinoflagellates,27 and eukaryotes.28 TET enzymes can oxidize thymine to form 5hmU and 5-formyluracil (5fU) in mammalian genomic DNA.28 In some parasite genomes, the TET homologue of JBP (J-binding protein) can oxidize thymine to 5hmU, which is further converted to a new nucleobase β-d-glucosyl-5-hydroxymethyluracil (base J) through J-GT (base J glycosyltransferase).29,30 Recently, another TET homologue of the 5mC-modifying enzyme (CMD1) from the alga Chlamydomonas reinhardtii has been reported to catalyze the conversion of 5mC to generate a new modified nucleobase, i.e., 5-glyceryl-methylcytosine (5-glyceryl-mC), which played critical roles in the photoprotective process.31
Reactive oxygen species (ROS) produced by cellular aerobic metabolism can oxidize DNA, and 8-oxo-7,8-dihydroguanine (OG) is the main oxidation product.32 OG could lead to G-to-T transversion mutation.33,34 OG in genomes can be repaired through the base excision repair (BER) mechanism.35 However, OG has recently been considered to have regulatory and epigenetic properties in modulating gene expression.36,37 For example, the increased OG level in genomes was closely related to the increased gene expression through the BER pathway.38 OG could also activate mRNA synthesis by promoting the formation of G-quadruplex in the DNA promoter.39
Phosphorothioate (PT) modification on DNA has been discovered in bacteria and archaea.40 So far, PT modification has not been discovered in the DNA of eukaryotes. In PT modification, the non-bridging oxygen in the phosphate group of DNA strands is replaced by sulfur. For PT modifications, the dndABCDE genes function as part of RM systems.41 The PT modifications in the DNA of bacteria and archaea involve regulating gene expression and cellular stress response, and maintaining cellular redox homeostasis.42 In addition, some 7-deazaguanine modifications, such as 2′-deoxy-7-amido-7-deazaguanosine (dADG), 2′-deoxy-7-cyano-7-deazaguanine (dPreQ0) and 2′-deoxy-7-aminomethyl-7-deazaguanine (dPreQ1), were found to be present in the DNA of bacteria and phage.43,44 These DNA modifications are considered to function in protecting the phage DNA from cleavage by host restriction enzymes.
The innovation of analytical methods has greatly stimulated the research of DNA modifications. In the past few decades, a variety of methods have been developed for the genome-wide quantification and mapping of DNA modifications. With the recent advances in analytical methods and techniques, a series of new endogenous DNA modifications have been discovered. In addition, many previously discovered DNA modifications have been endowed with epigenetic-like properties in regulating biological processes. These methods have enabled the establishment of the correlation between the dynamic changes of DNA modifications and various human diseases, including cancers, imprinting disorders, neurological disorders, cardiovascular diseases, and developmental diseases.4,45 In addition, the interplay between the epigenetic DNA modifications and metabolism emerges as a new research direction by virtue of these sophisticated analytical methods.46,47
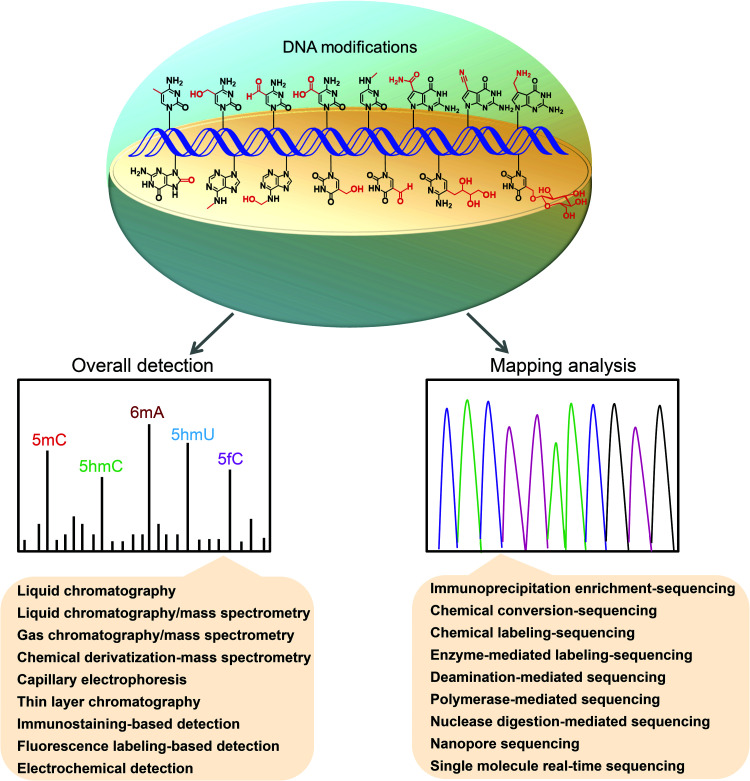
Revealing the functions of DNA modifications depends on the sensitive detection, accurate quantification, and mapping of these modifications in genomes.48–50 Here, we review the recent advances in analytical methods for the quantification and mapping of DNA modifications, including 5mC, 5hmC, 5fC, 5caC, 6mA, 5hmU, 5fU, base J, 5-glyceryl-mC, N4-methylcytosine (4mC), N6-hydroxymethyladenosine (6hmA), OG, PT, and 7-deazaguanine modifications (Fig. 1). We discuss the principles, advantages, and limitations of these established methods.
Fig. 1. Schematic illustration for the overall detection and mapping of DNA modifications. The chemical structures shown in DNA strand represent 5mC, 5hmC, 5fC, 5caC, 6mA, 5hmU, 5fU, base J, 5-glyceryl-mC, 4mC, 6hmA, OG, dADG, dPreQ0 and dPreQ1.
2. Methods for quantification of DNA modifications
The reported methods for the detection of overall DNA modifications mainly include liquid chromatography (LC), liquid chromatography/mass spectrometry (LC/MS), gas chromatography/mass spectrometry (GC/MS), chemical derivatization-mass spectrometry, capillary electrophoresis (CE), thin layer chromatography (TLC), immunostaining-based detection, fluorescence labeling-based detection, and electrochemical detection.
2.1. Liquid chromatography
Determination of DNA modifications by liquid chromatography (LC) is based on the detection of different nucleobases, 2′-deoxynucleosides or 2′-deoxynucleotides, which are obtained from the enzymatic or chemical hydrolysis of DNA. These DNA components are typically separated by reversed-phase LC,51 and then detected using a photometric or electrochemical detector. Thus, the baseline separation of the DNA components is essential since the determination relies on the chromatographic separation to distinguish analytes.
It should be noted that if DNA is hydrolyzed by formic acid to release nucleobases for LC detection, the complete elimination of RNA contamination in isolated DNA is required because the nucleobases released from RNA, such as 5-methylcytosine and N6-methyladenine, will affect the accurate quantification of the corresponding DNA modifications. As for the LC-based detection of DNA modifications, a relatively large amount of genomic DNA (∼1–50 μg) is typically needed owing to the low detection sensitivity of the method. Moreover, the LC-based detection of DNA modifications is less confirmative than the mass spectrometry-based detection.
2.2. Liquid chromatography/mass spectrometry
Mass spectrometry (MS) has been widely used to detect and quantify nucleic acid modifications as MS exhibits high detection sensitivity and good capability to identify compounds.52–58 LC/MS is the preferred method for the sensitive detection and quantification of the global levels of modifications in genomic DNA. Typically, DNA is enzymatically digested into 2′-deoxynucleosides or 2′-deoxynucleotides and then determined by LC/MS. LC/MS now plays a central role in the discovery, identification and quantification of DNA modifications across different cells, tissues and organisms.59,60 LC/MS provides the nucleoside composition but not the sequence context of DNA modifications in genomes.
2.2.1. LC/MS
LC/MS with the multiple reaction monitoring (MRM) detection mode is frequently employed to detect DNA modifications due to the inherent high sensitivity of this detection mode. LC/MS with MRM detection can usually achieve a sub-femtomole level of limit of detection (LOD) towards DNA modifications. We realized the analysis of 5mC in a single cell using liquid chromatography tandem mass spectrometry (LC-MS/MS) with the MRM detection mode.61 The PT modifications in prokaryotic and eukaryotic genomes were quantitatively measured by LC-MS/MS. The results showed that the PT modifications were as low as 1 per 106 nt for d(TPST) and 2 per 108 nt for d(CPST).62,63 The LC-MS/MS analysis showed that dADG, dPreQ0, and dPreQ1 modifications were in the range of 6–790 (per 106 nt).44 Ultra-performance liquid chromatography (UPLC) has also been employed in the LC-MS/MS analysis, thus enabling short chromatographic runs in the analysis of 5mC and 6mA in DNA.18,64,65 Using a stable isotope labeled (SIL) analogue of the nucleoside as an internal standard can calibrate the detection variation caused by the matrix effect and sample pretreatment during the measurements of DNA modifications. Stable isotope labeling coupled with LC/MS methods has been developed for the accurate measurement of DNA modifications, such as 5mC, 5hmC (LOD, 0.056 fmol), 5fC (LOD, 0.098 fmol), 5caC (LOD, 0.14 fmol), 5hmU (LOD, 80 fmol), base J, 6mA (LOQ, 1.6 fmol), PT and 6hmA.66–71 In addition, stable isotope labeling also benefits the qualitative analysis of DNA modifications.
A vitamin-C-derived DNA modification (5-glyceryl-mC) has been recently discovered in the genome of the green alga Chlamydomonas reinhardtii by LC/MS analysis.31 The measured level of 5-glyceryl-mC was about 10 modifications per 106 dC in the genomic DNA of wild-type Chlamydomonas reinhardtii. The discovery of a new modification of 5-glyceryl-mC highlights the structural diversity of DNA associated with environmental adaptation.
2.2.2. High-resolution mass spectrometry analysis
High-resolution mass spectrometry (HRMS) can provide accurate mass measurements and plays an important role in deciphering the structural information of compounds. LC coupled with HRMS (LC-HRMS) has been frequently used in the determination of DNA modifications. For example, LC-HRMS enabled the discovery of 5fC and 5caC in the DNA of mouse embryonic stem cells,72,73 providing confirmatory evidence of the 5mC oxidative demethylation via 5fC and 5caC. Using high-resolution quadrupole TOF MS (qTOF-MS), 5mC and 5hmC in genomic DNA can be simultaneously detected,74 which allowed the sensitive determination of low contents of 5mC and 5hmC with 2 ng of DNA. This study demonstrated that the content of 5hmC in hepatocellular carcinoma (HCC) tumor tissues was decreased compared to that of the tumor adjacent tissues. The dysregulation of 5hmC in cancer tissues has now become a common phenomenon in a variety of cancers.75–78
2.2.3. Inductively coupled plasma mass spectrometry analysis
The use of external elemental tags and inductively coupled plasma-mass spectrometry (ICP-MS) detection has become a new strategy in analytical chemistry.79 This method is based on the exceptional features of elemental mass spectrometry in terms of sensitivity, selectivity, and multi-elemental and isotopic capabilities, which can enhance the quantification of labeled biomolecules. The hyphenation of LC with ICP-MS (LC-ICP-MS) has been developed for evaluating DNA methylation.80 The reversed-phase separation of 5-methyl-2′-deoxycytidine-3′-monophosphate (5mCMP) with specific ICP-MS detection on 31P was established to detect 5mC.80 In addition, potassium acid (K2OsO4) was used to selectively label 5mC in single-stranded DNA (ssDNA) in the presence of strong oxidant K3Fe(CN)6 and N,N,N′,N′-tetramethylethylenediamine (TEMED).80 Then, size-exclusion chromatography-ICP-MS was used to detect 31P (proportional to the total amount of DNA) and 189Os (equivalent to 5mC). These methods were successfully applied in analyzing 5mC from the DNA of salmon testes and commercial oligonucleotides, which provided a proof-of-concept that ICP-MS was a useful and complementary platform to the LC/MS in the detection of DNA modifications.
2.2.4. Metabolic isotope tracing by mass spectrometry
Metabolic isotope tracing is a useful strategy that has been widely used in the detection and confirmation of endogenous biomolecules.81 The in vivo metabolic pathways and dynamics of DNA modifications can be revealed by the combination of isotope tracing and LC-MS/MS analysis. It has been known that ATP and l-methionine could be converted to S-adenosyl-l-methionine (SAM) by methionine adenosyltransferase, and SAM is a universal reagent for the methylation of nucleic acids. Therefore, by feeding cells with SIL l-methionine, the source of methyl groups for DNA methylation and the dynamics of 5mC and 5hmC were uncovered.82 Similarly, various isotopologues of modified bases have been utilized to elucidate the mechanisms of the formation and removal of modifications, such as 5fC,83 5hmU,28 6mA,84 and 5caC.85,86
2.3. Gas chromatography/mass spectrometry
Compared to LC/MS, gas chromatography/mass spectrometry (GC/MS) has the advantage of providing better chromatographic separation. Nevertheless, GC/MS requires the conversion of analytes into volatile derivatives before separation.87 In this regard, nucleobases can only be analyzed after converting them into volatile counterparts through derivatization.
For the GC/MS analysis of 5mC, DNA is typically hydrolyzed using aqueous formic acid (88%), and the resulting nucleobases are then derivatized with appropriate reagents, such as N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide with 1% tert-butyldimethyl-chlorosilane88 and N,O-bis-(trimethylsilyl)-trifluoroacetamide (BSTFA) with 1% cholorotrimethylsilane.89 The results showed that the LOD value of 5mC was as low as 0.8 pg (6.4 fmol).89 GC/MS was also used to detect OG from human breast tissues and lymphocytes.90 However, it was found that the OG content measured by GC/MS was higher than that by other methods. The gas-phase derivatization step could cause the oxidation of guanine, which may lead to increased OG due to the artificially formed OG during sample preparation.
2.4. Chemical derivatization-mass spectrometry
DNA modifications typically present in low abundance in vivo, and it is a challenging task to detect some rare DNA modifications by direct LC/MS analysis. In this respect, the approaches based on chemical labeling in conjugation with LC/MS analysis were developed to analyze the low levels of compounds.91–97 The tagged group that was added to the target modifications from labeling reagents can endow the desired properties of DNA modifications, which can be utilized to achieve the improved ionization efficiencies of DNA modifications during mass spectrometry analysis (summarized in Table 1).
Summary of the detection sensitivities of various DNA modifications analyzed by chemical derivatization-mass spectrometry. LOD, limit of detection.
| Derivatization reagents | Modifications | LOD (fmol) | Ref. |
|---|---|---|---|
| 2-Bromo-1-(4-dimethyl aminophenyl)-ethanone (BDAPE) | 5mC | 0.10 | 98 |
| 5hmC | 0.06 | ||
| 5fC | 0.11 | ||
| 5caC | 0.23 | ||
| 2-Bromo-1-(4-diethylaminophenyl)-ethanone (BDEPE) | 5mC | 0.06 | 99 |
| 5hmC | 0.07 | ||
| 5fC | 0.10 | ||
| 5caC | 0.08 | ||
| 2-Bromo-1-(4-1-pyrrolidinylphenyl)-ethanone (BPPE) | 5mC | 0.43 | |
| 5hmC | 0.69 | ||
| 5fC | 0.79 | ||
| 5caC | 1.21 | ||
| 3-Bromoacetonyltrimethylammonium bromide (BTA) | 5mC | 2.20 | |
| 5hmC | 4.72 | ||
| 5fC | 6.22 | ||
| 5caC | 0.53 | ||
| ω-Bromoacetonylpyridinium bromide (BPB) | 5mC | 5.25 | |
| 5hmC | 2.72 | ||
| 5fC | 16.59 | ||
| 5caC | 4.41 | ||
| 4-(Dimethylamino) benzoic anhydride | 5mC | 2.24 | 100 |
| 5hmC | 2.53 | ||
| 5fC | 2.54 | ||
| 5caC | 1.27 | ||
| Uridine diphosphoglucose (UDP-glucose) | 5hmC | 6.7 | 102 |
| 5gmC | 1.5 | ||
| Girard's T reagent | 5fU | 3.3 | 103 |
| 5fC | 0.09 | 104 | |
| 5caC | 0.77 | ||
| 5fC | 0.08 | 105 | |
| 5fU | 0.14 | ||
| Girard's D reagent | 5fC | 0.15 | 104 |
| 5caC | 0.42 | ||
| Girard's P reagent | 5fC | 0.03 | 104 |
| 5caC | 0.75 | ||
| 5fC | 0.03 | 105 | |
| 5fU | 0.05 | ||
| 4-(2-(Trimethylammonio)ethoxy)benzenaminium halide (4-APC) | 5fC | 0.05 | 105 |
| 5fU | 0.09 | ||
| Hydrazino-s-triazine based glycan labelling reagents i-Pr2N | 5fC | 0.01 | 106 |
| 5caC | 0.025 | ||
| Hydrazino-s-triazine based glycan labelling reagents Me2N | 5fC | 0.05 | |
| Hydrazino-s-triazine based glycan labelling reagents Et2N | 5fC | 0.0125 | |
| Dansylhydrazine (DNSH) | 5hmC | 0.04 | 107 |
| 2-Bromoacetophenone | 5mC | 22.7 | 131 |
We and others previously utilized 2-bromo-1-(4-dimethylaminophenyl)-ethanone (BDAPE), 2-bromo-1-(4-diethylaminophenyl)-ethanone (BDEPE), and 4-(dimethylamino) benzoic anhydride to simultaneously label 5mC, 5hmC, 5fC, and 5caC in DNA,98–101 which led to a 313-fold increase of the detection sensitivities of DNA modifications. Similarly, some other chemical derivatization strategies were developed to enable the sensitive detection of DNA modifications, such as the glycosylation of 5hmC by T4 β-glucosyltransferase,102 derivatization of 5fC, 5caC and 5fU by Girard's reagents (Girard D, T, and P)103–105 and hydrazine reagents (Me2N, Et2N, and i-Pr2N),106 and derivatization of 5fC by dansylhydrazine (DNSH).107 In addition, 8-(diazomethyl)quinoline (8-DMQ) was used to derivatize the phosphate group of NTPs and modified NTPs, which resulted in the discovery of a series of new modifications occurring in NTPs.108
2.5. Capillary electrophoresis
The separation of analytes by capillary electrophoresis (CE) is mainly based on the equilibrium of charged analytes between the stationary phase and the mobile phase. Several detection techniques, including UV-absorbance, MS, and laser-induced fluorescence (LIF) detection, can be coupled with CE to determine the DNA modifications from clinical samples and plant tissues. CE-LIF and CE-MS generally can offer better detection sensitivity than CE-UV in detecting 5mC and 5hmC in genomic DNA. CE-MS usually provides better detection sensitivity than CE-UV, and 5mC and 5hmC could be easily detected using only subnanogram DNA samples.109 CE-LIF has been used to quantify 6mA, with the detection limit being as low as 1.4 amol.110 Our group developed a sensitive 5mC detection method based on capillary liquid chromatography using a hyper-cross-linked polymer monolithic column carrying phenyl and quaternary ammonium groups.111 The LOD of 5mC was 0.014 pmol, which is comparable to that obtained by MS analysis.
The CE method has been used to address many interesting questions, including the examination of DNA methylation in cancer cell lines,112 the investigation of the association between the DNA methylation and clinicopathological data from leukemia patients,113 and the evaluation of the in vivo activity of mammalian DNA methyltransferases.114 In the above-described studies, the typical sample amounts required were in the range of 0.1–10 μg of genomic DNA. Although the CE method provides higher separation efficiency than the LC method, the separation reproducibility can be affected by slight variations.
2.6. Thin layer chromatography
Thin layer chromatography (TLC) is based on the differences in net charge, polarity, and hydrophobicity between analytes.115 The genomic DNA is enzymatically hydrolyzed to nucleosides and then labeled with [32P]ATP by T4 polynucleotide kinase, followed by separation on cellulose TLC plates. The relative intensity of the spots can be determined using a phosphorimager. Many DNA modifications, such as 5hmC116,117 and 5caC,8 were initially discovered in the genomic DNA of mammalian cells by TLC and then further confirmed by HRMS.
Compared to one-dimensional (1D) chromatography, two-dimensional (2D) separation can effectively improve the separation capability. 2D-TLC has been widely utilized to detect various DNA modifications such as 5mC,118 base J,119 and OG.120 It is simple to identify and quantify 32P-labeled modified nucleotides down to the femtomole level. For the TLC-based detection of DNA modifications, there is no need for sophisticated instrumentation. However, the analytical procedure with radioactive labeling by TLC is relatively time-consuming.
2.7. Immunostaining-based detection
Immunostaining has been widely used to evaluate the global levels of various DNA modifications. Specific antibodies can recognize 5mC,121 5hmC,122 5fC,123 5caC,123 6mA,124 base J125 and OG126 for visualization. Although this method is simple, it has the disadvantages of potential low antibody specificity and bias towards DNA modifications. Immunostaining methods generally provide semi-quantitative information.
Enzyme-linked immunosorbent assay (ELISA) coupled with electrochemiluminescence (ECL) detection was used to detect 5mC127 and OG.128 The developed method employed an anti-5mC antibody labeled with acetylcholinesterase, which can convert acetylthiocholine (substrate) to thiocholine (product). The thiocholine was accumulated on a gold electrode surface, making it possible to observe bright and distinctive ECL by applying a potential to the gold electrode. The detection limit of 5mC was 0.18 pmol.
2.8. Fluorescence labeling-based detection
Fluorescence labeling is a well-established strategy in the detection of DNA modifications. High detection sensitivity can be achieved by selecting suitable fluorescent reagents.129,130 For example, 2-bromoacetophenone was used to selectively label cytosine moieties from the DNA of salmon testes and nucleic acids from plants, human blood, and earthworms, followed by the detection of these derivatives using LC with a spectrofluorimetric detector.131 The LOD of 5mC can reach 16.6 fmol. In general, the fluorescent moiety can be efficiently attached to aldehyde-containing modifications (such as 5fC and 5fU) by amine hydrazine or hydroxylamine through aldol-type condensation reactions,132–138 or by the direct condensation of aminobenzaldehyde with cyano reagents.139,140 In addition, by the chemical conversion of hydroxymethyl group to an aldehyde group, the quantitative detection of 5hmC in different mouse tissues has been achieved using a similar strategy.141 The linear correlation between the concentration of labeled modifications and the fluorescence intensity allows us to quantitatively determine the levels of DNA modifications. It should be noted that some developed methods have been successfully used to analyze the DNA modifications of biological samples;132,136,138,139,142 however, some methods were established using synthesized DNA carrying modifications.133–135,137,140
2.9. Electrochemical detection
Electrochemical biosensing strategies can offer fast and sensitive detection towards DNA modifications.143,144 Based on the decrease in charge density between the C·G base pairs caused by DNA methylation, a simple electrochemical strategy was reported for the continuous monitoring of the dynamic DNA methylation process.145 Although most of the studies can only detect the overall level of 5mC in DNA, the analysis of single DNA methylation sites also has been successfully accomplished.146
Signal amplification is a prevalent strategy to increase the detection sensitivity of electrochemical biosensors. The signal amplification technologies based on enzymatic amplification147 or redox cycles148 were reported to realize the sensitive quantification of 5mC and 5hmC in genomic DNA. Additionally, analytical approaches for signal amplification through carbon nanocomposite materials,149 new nano-microspheres,150 gold nanoparticles,151 and other novel nanomaterials152 were developed for the analysis of DNA modifications of various biological samples. Using these methods, the detection limit of 5hmC was as low as 9.06 fM,148 while the detection limit of OG was 1 pM.151
3. Methods for mapping DNA modifications
Methods for the overall detection of DNA modifications require the digestion or release of modifications from DNA before measurements, which will not provide information of the positions of modifications within the DNA sequence. Rapidly improving technologies, especially the next-generation DNA sequencing, revolutionize the study of genome-wide mapping of DNA modifications, allowing us to study the site-specific dynamic changes of epigenetic modifications.153 A variety of methods have been established for the genome-wide mapping of DNA modifications, which is essential and critical to reveal the biological functions of DNA modifications (Table 2).
Summary of methods for mapping DNA modifications.
| Modifications | Methods | Treatment | Resolution | Principles of sequencing | Ref. |
|---|---|---|---|---|---|
| 5mC, 5hmC | BS-seq | Bisulfite treatment | Single-base resolution | C, 5fC and 5caC undergo deamination and all of them are read as T, while 5mC and 5hmC are resistant to deamination and are read as C. | 166 |
| TAPS | Oxidation of 5mC, 5hmC and 5fC by TET proteins; reduction of 5caC by pyridine borane | TET oxidizes 5mC, 5hmC and 5fC to 5caC, then pyridine borane reduces 5caC to DHU, which is read as T in PCR and thereby realize the C-to-T conversion of 5mC and 5hmC. | 182 | ||
| EM-seq | Oxidation of 5mC by TET2 and glycosylation of 5hmC by β-GT; deamination of C by APOBEC3A | TET2 oxidation and β-GT glycosylation protect 5mC, 5hmC and 5fC from APOBEC3A deamination; only C is converted to U, which is read as T in sequencing. 5mC, 5hmC and 5fC are read as C. | 204 | ||
| 5mC | TAPSβ | Glycosylation of 5hmC by β-GT; TET oxidation and pyridine borane reduction of 5mC | Single-base resolution | β-GT glycosylation protects 5hmC from TET oxidation and pyridine borane reduction; only 5mC is converted to DHU, which is read as T in sequencing. | 182 |
| 5hmC | oxBS-seq | Oxidation of 5hmC by KRuO4 | Single-base resolution | KRuO4 oxidizes 5hmC to 5fC, which is read as T in BS-seq, while 5mC is read as C. | 167 |
| TAB-seq | Glycosylation of 5hmC by β-GT and oxidation of 5mC by TET proteins | Glycosylation of 5hmC to 5gmC by β-GT; TET oxidizes 5mC to 5fC and 5caC, both of which are read as T in BS-seq, while the original 5hmC is read as C. | 168 | ||
| hmC-CATCH | Labeling of 5fC by EtONH2; oxidation of 5hmC by K2RuO4 and labeling by 1,3-indandione (AI) | Protection of 5fC by EtONH2; K2RuO4 oxidizes 5hmC to 5fC followed by AI labeling, which induces a C-to-T transition in sequencing. | 178 | ||
| CAM-Seq | KRuO4 oxidation and azi-BP labeling of 5hmC | Protection of 5fC by hydroxylamine; KRuO4 oxidizes 5hmC to 5fC followed by azi-BP labeling, which induces C-to-T conversion in sequencing. | 180 | ||
| CAPS | Oxidation of 5hmC by KRuO4 and reduction by pyridine borane | KRuO4 oxidizes 5hmC to 5fC, then is reduced to DHU by pyridine borane. DHU is read as T in sequencing. | 182 | ||
| hmC-seq | Oxidation of 5hmC by peroxotungstate | Peroxotungstate converts 5hmC to trihydroxylated thymine (thT), leading to a C-to-T transition in polymerase extension. | 181 | ||
| AMD-seq, ACE-seq | Glycosylation of 5hmC by β-GT; deamination of C by APOBEC3A | β-GT glycosylation protects 5hmC from APOBEC3A deamination and 5gmC is read as C; C and 5mC are read as T. | 202 and 203 | ||
| hmC-seq | Glycosylation of 5hmC by β-GT and precipitation by JBP1 | Genome-wide | β-GT converts 5hmC to 5gmC that can be pulled down by J-binding protein 1 coupled to magnetic beads. | 195 | |
| nano-hmC-Seal | Glycosylation of 5hmC by β-GT | Labeling of 5hmC to 6-N3-β-glucosyl-5hmC by β-GT, then a biotin tag is installed onto the azido group for pull down using click chemistry. | 197 | ||
| 5fC | redBS-seq | Reduction of 5fC by NaBH4 | Single-base resolution | NaBH4 reduces 5fC to 5hmC, which is converted to CMS by bisulfite treatment. The 5fC site is identified by comparing the output of redBS-seq (where 5fC is read as C) with that of BS-seq (where 5fC is read as T). | 170 |
| fCAB-seq | Conversion of 5fC to oxime by EtONH2 | EtONH2 converts 5fC to oxime, which resists deamination by bisulfite treatment. The 5fC site is identified by comparing the output of fCAB-seq (where 5fC is read as C) with that of BS-seq (where 5fC is read as T). | 171 | ||
| fC-CET | Labeling of 5fC by 1,3-indandione (AI) | Labeling of 5fC by AI enables a subsequent C-to-T transition in PCR. | 191 | ||
| CLEVER-seq | Labeling of 5fC by malononitrile | Labeling of 5fC by malononitrile induces a C-to-T conversion in sequencing. | 192 | ||
| fC-seq | Labeling of 5fC by 2-(5-chlorobenzo[d] thiazol-2-yl)acetonitrile (CBAN) or azi-BP | CBAN or azi-BP reacts with 5fC to generate an intramolecular cyclization nucleobase, leading to a C-to-T conversion in polymerase extension. | 142 and 193 | ||
| fC-seq | Labeling of 5fC by O-(biotinylcarbazoylmethyl) hydroxylamine (ARP) | Genome-wide | Labeling of 5fC by ARP to form a biotinylated 5fC, which can be enriched by streptavidin-coated magnetic beads and then sequenced. | 160 | |
| 5caC | CAB-seq | Labeling of 5caC by EDC-catalyzed xylene-based primary amine | Single-base resolution | Labeling of 5caC with xylene-based primary amine, which protects 5caC from deamination. The labeled 5caC is read as C in BS-seq. | 172 |
| caMAB-seq | Reduction of 5fC by NaBH4; methylation of C by M.SssI enzyme | 5fC is reduced to 5hmC by NaBH4; methylation of C by M.SssI. 5caC is sequenced as T in BS-seq, whereas C, 5mC, 5hmC and 5fC are read as C. | 174 | ||
| 5fC, 5caC | MAB-seq | Methylation of C by M.SssI enzyme | Single-base resolution | Methylation of CpG by M.SssI protects unmodified C from bisulfite conversion to U. 5fC/5caC is read as T in sequencing. | 175 |
| 4mC | 4mC-TAB-seq | Oxidation of 5mC by TET proteins | Single-base resolution | Oxidation of 5mC to 5caC by TET proteins followed by bisulfite treatment; 5mC is read as thymine, while 4mC is read as cytosine in sequencing. | 177 |
| 5hmU | hmU-seq | Oxidation of 5hmU by KRuO4 | Single-base resolution | Oxidation of 5hmU to 5fU by KRuO4 induces a T-to-C base transition in polymerase extension. | 179 |
| hmU-seq | Glycosylation of 5hmU by base J glucosyltransferase (J-GT) | Genome-wide | Labeling of 5hmU with N3-glucose by J-GT followed by adding biotin tag and enrichment with streptavidin-coupled beads. | 199 | |
| 5fU | fU-seq | Labeling of 5fU by azi-BIAN | Genome-wide | Azi-BIAN labeling of 5fU enables pull down of 5fU-containing DNA fragments for sequencing. | 188 |
| OG | OG-seq | Oxidation of OG by K2IrBr6 | Genome-wide | K2IrBr6 oxidizes OG to a covalent adduct of a primary-amine-terminated biotin, allowing for enrichment and sequencing. | 189 |
| 6mA | 6mA-seq | Deamination of adenine by sodium nitrite (NaNO2) | Single-base resolution | NaNO2 deaminates unmethylated adenines to hypoxanthine bases, which are read as guanine by polymerase and reverse transcriptase. 6mA site resists deamination and is read as adenine. | 194 |
3.1. Immunoprecipitation enrichment-sequencing
A number of immunoprecipitation-based enrichment strategies combined with sequencing have been developed for mapping DNA modifications in genomes.49 These methods typically include the enrichment of DNA fragments that contain the modified base, sequencing of the enriched fragments, and aligning of the sequence reads to the reference genomes. The position of a modification in genomic DNA can be approximately mapped by fragmenting the DNA into short pieces, and the peak analysis could improve the mapping resolution for modifications.
The immunoprecipitation-mediated enrichment of DNA fragments using antibodies or specific affinity binding proteins has been demonstrated to be very useful for mapping DNA modifications. The immunoprecipitation of methylated DNA is commonly achieved by the isolation of methylated DNA fragments using antibodies for 5mC (MeDIP-seq)154 or the isolation of DNA fragments containing methylated CpG dinucleotides using methyl-CpG binding domain (MBD) proteins (MBD-seq).155,156 A secondary antibody captures the methylated DNA-antibody/protein complex (MethylCap-seq, MIRA-seq),157,158 which can then be further analyzed using PCR, array-based methods, or sequencing technologies. Similarly, affinity-based enrichment of 5hmC,159 5fC,160 6mA,161,162 base J,163 and OG,164 followed by sequencing, were also developed to map these modifications in genomes of various species.
Although immunoprecipitation-mediated sequencing is straightforward, it does not offer base-resolution information of DNA modifications. In addition, the results of independent DIP-seq studies often show considerable variations between the profiles of the identical genomes and between the profiles obtained by alternative methods, which might be attributed to the low specificity of antibodies and the intrinsic affinity of IgG for short unmodified DNA repeats.165
3.2. Chemical conversion-sequencing
Specific conversion of modified nucleosides or unmodified nucleosides by chemical treatments can lead to the subsequent characteristic sequencing events for modifications, which therefore can be employed for mapping modified nucleosides in DNA. The selective and efficient chemical conversion is the critical issue in this method.
3.2.1. Bisulfite sequencing
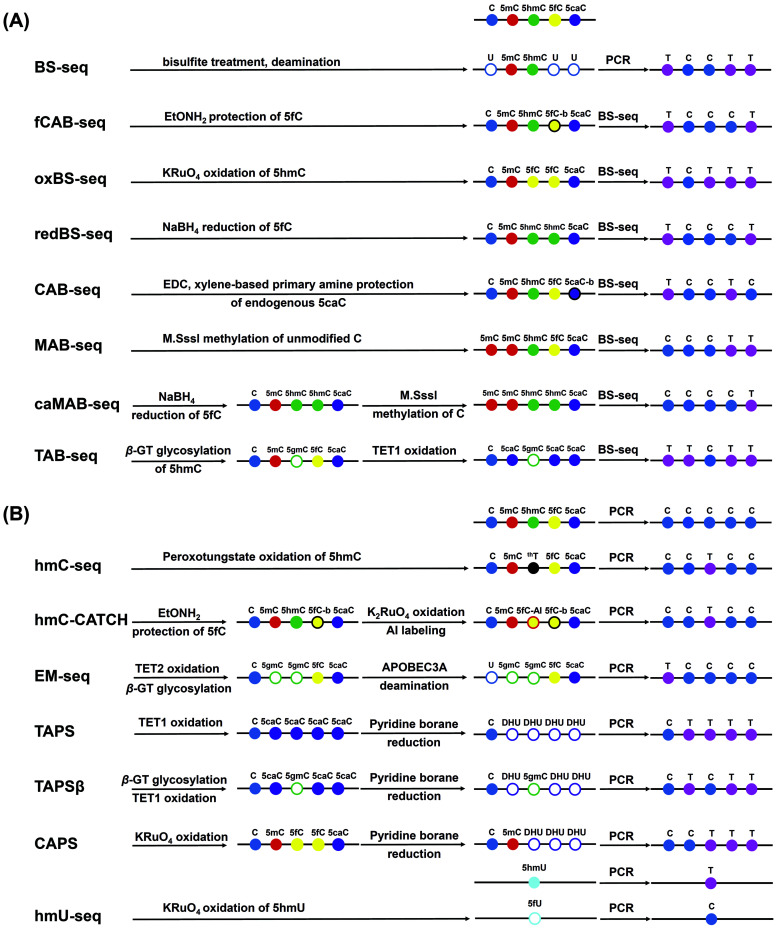
Bisulfite sequencing (BS-seq) is the gold standard for the genome-wide mapping of 5mC at single-base resolution.166 In BS-seq, cytosine, 5fC, and 5caC undergo deamination by bisulfite treatment and are read as thymines, while both 5mC and 5hmC are resistant to deamination by bisulfite treatment and are read as cytosines.167,168 Therefore, traditional bisulfite sequencing cannot distinguish 5fC or 5caC from cytosine, nor can it differentiate 5hmC from 5mC.
Some methods have been developed by combining chemical labeling and bisulfite treatment to achieve the genome-wide mapping of 5hmC, 5fC, and 5caC (Fig. 2A). Oxidative bisulfite sequencing (oxBS-seq)167 and TET-assisted bisulfite sequencing (TAB-Seq)168 methods were established to map 5hmC in genomic DNA. The oxBS-seq approach is based on the specific oxidation of 5hmC by KRuO4 to produce 5fC, which can be converted to uracil under bisulfite treatment.167 With the oxBS-seq approach, 5hmC is read as thymine, but 5mC is read as cytosine in DNA. In the TAB-seq approach, β-GT specifically transfers a glycosyl group to 5hmC to produce β-glucosyl-5-hydroxymethylcytosine (5gmC), which resists the oxidation by TET proteins.168 But TET proteins oxidize 5mC to 5fC and 5caC, both of which are read as thymines by bisulfite sequencing. The remaining cytosine signals come from the glycosylated 5hmC, offering a strategy for the single-base resolution mapping of 5hmC. A mirror bisulfite sequencing for 5hmC detection at a single CpG site with glycosylation by β-GT, methylation by M.SssI methylase and bisulfite conversion also achieved the site-specific analysis of 5hmC at the CpG sites of DNA.169
Fig. 2. Schematic illustration of the chemical conversion-sequencing methods for mapping DNA modifications. (A) Bisulfite sequencing. (B) Bisulfite-free sequencing.
Reduced bisulfite sequencing (redBS-seq)170 was developed to map 5fC based on the selective reduction of 5fC to 5hmC by sodium borohydride combined with bisulfite sequencing. 5fC sites could be detected by comparing the bisulfite-treated reduced and non-reduced DNA. In addition, the fCAB-seq approach171 used O-ethylhydroxylamine (EtONH2) to convert 5fC to oxime, which resists deamination by bisulfite treatment. Comparing the results of fCAB-seq (where 5fC is read as C) with those of BS-seq (where 5fC is read as T) enables the mapping of 5fC at single-base resolution. For 5caC mapping analysis, a chemical modification-assisted bisulfite sequencing (CAB-seq) method was established.172 The carboxyl group of 5caC reacts with the primary amine group, thereby protecting the labeled 5caC from deamination by bisulfite treatment. Based on CAB-Seq, a DNA immunoprecipitation-coupled CAB-Seq (DIP-CAB-Seq) was further developed and successfully used in the mapping of 5fC and 5caC in the genomes of mouse embryonic stem cells.173 In addition, the combination of bisulfite treatment with the use of M.SssI methyltransferase could achieve the base-resolution mapping of 5fC and 5caC in DNA (MAB-seq and caMAB-seq).174,175
In addition to the 5-substituted cytosine modifications, 4mC was also found to be present in the DNA of thermophilic bacteria and many bacterial mesophiles.176 Traditional bisulfite sequencing cannot distinguish 4mC from 5mC because both 4mC and 5mC resist deamination and will then be read as cytosines. A 4mC-Tet-assisted-bisulfite-sequencing (4mC-TAB-seq) approach was established for the genome-wide mapping of 4mC.177 This 4mC-TAB-seq approach can accurately identify the 4mC sites without interference from 5mC. By coupling the oxidation of all 5mC to 5caC mediated by excessive TET proteins under optimized bisulfite treatment conditions, 5caC is read as T. At the same time, 4mC is partially deaminated by bisulfite treatment, and about half of the 4mC sites are read as C. With this 4mC-TAB-seq approach, three 4mC-containing motifs were identified in the genomes of Caldicellulosiruptor kristjanssonii.
3.2.2. Bisulfite-free sequencing
The treatment condition by bisulfite is harsh, which could cause the degradation of the majority of input DNA. Thus, mild chemical treatment could provide an advantage in processing a small amount of DNA samples. In addition, bisulfite treatment converts abundant unmodified cytosines to thymines rather than non-canonical modifications, which reduces the sequence complexity of template DNA. Thus, BS-seq often involves low hybridization and mapping selectivity in sequencing. In this respect, bisulfite-free sequencing methods were developed for mapping DNA modifications (Fig. 2B).
The hmC-CATCH approach combines the EtONH2 protection of endogenous 5fC with the selective oxidation of 5hmC to 5fC using K2RuO4.178 1,3-Indandione (AI) was used for the selective labeling of the converted 5fC, which would lead to a C-to-T transition in the subsequent PCR amplification and sequencing.178 Similarly, the oxidation of 5hmU to form 5fU by KRuO4 was used for the mapping of 5hmU.179 In this approach, the chemical conversion of 5hmU to 5fU induced a T-to-C base transition in the subsequent polymerase extension, which was utilized to map 5hmU at single-base resolution. KRuO4 was also used to transform 5hmC to 5fC followed by azi-BP labeling. The C-to-T transition was induced because the labeled 5fC lost the exocyclic 4-amino group, enabling the mapping of 5hmC in DNA.180 Similarly, peroxotungstate can convert 5hmC to trihydroxylated thymine (thT), leading to a C-to-T transition during the polymerase extension and single-base resolution mapping of 5hmC.181
TET-assisted pyridine borane sequencing (TAPS)182,183 includes the TET-mediated oxidation of 5mC, 5hmC and 5fC to 5caC and the pyridine borane reduction of 5caC to dihydrouracil (DHU). DHU was read as thymine in PCR, thereby realizing the C-to-T conversion of 5mC and 5hmC, while unmodified cytosines still were read as C. Thus, 5mC and 5hmC can be differentiated from cytosines, but 5mC and 5hmC cannot be differentiated from each other by TAPS. In this respect, 5hmC was glycosylated using β-GT, and the formed 5gmC resisted the TET oxidation and pyridine borane reduction. Thus, 5hmC was read as C, while 5mC was read as T, realizing the differentiation of 5mC and 5hmC (TAPSβ assay).182
Iodine shows high selectivity in the cleavage of PT modifications in DNA.184 Thus, iodine-induced cleavage quantitative real-time PCR (IC-qPCR) has been developed to evaluate the frequency of PT modifications at a given site in bacterial DNA.185 The high sensitivity of the IC-qPCR method with a LOQ of 5 copies or 10 copies per reaction suggested that the IC-qPCR method was suitable for quantifying PT modifications at low frequencies. In addition, the PT-IC-seq method was based on the iodine-induced selective cleavage at the PT sites, and a high-throughput sequencing was developed to map the PT modifications in genomic DNA.186,187 In PT-IC-seq, iodine treatment could selectively induce DNA cleavage at the modified phosphodiester linkage, which produces the characteristic signatures. The PT sites can then be obtained from the cleavage sites that are examined by sequencing analysis. The mapping results for PT modifications indicated a remarkable target selection by the PT-modification proteins, and PT modifications could function in epigenetic control.
3.3. Chemical labeling-sequencing
3.3.1. Chemical labeling-assisted enrichment
In addition to the immunoprecipitation-mediated sequencing, chemical labeling-based strategies have been developed for the efficient enrichment of DNA fragments with modifications followed by sequencing. The critical point of chemical pull-down is the efficiency and selectivity of the chemical labeling reaction.
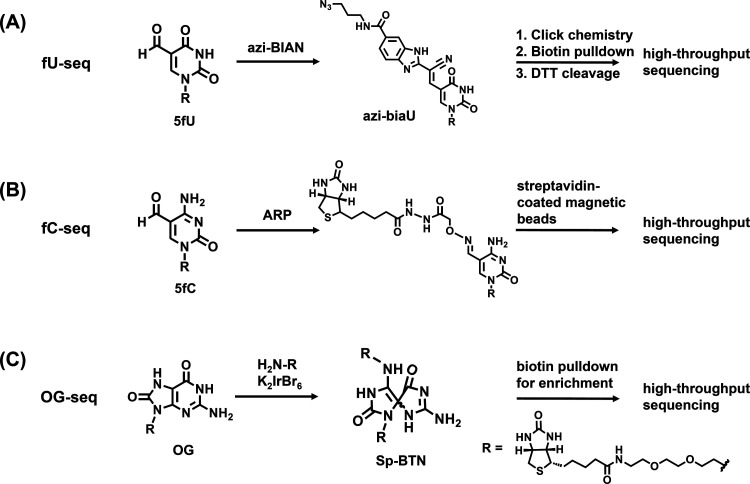
The reagent of (2-benzimidazolyl)-acetonitrile (azi-BIAN) was used to selectively label 5fU followed by the enrichment and genome-wide mapping of 5fU in human and mouse tissues (Fig. 3A).188 The mapping results indicated that most of the 5fU sites mainly exist in the intergenic regions and introns. Similarly, aldehyde-reactive reagents of O-(biotinylcarbazoylmethyl) hydroxylamine (ARP)160 and amine-terminal biotin189 could achieve the selective labeling of 5fC and OG in DNA, respectively, which can be further enriched by streptavidin (Fig. 3B and C). The ARP labeling-assisted enrichment and sequencing of 5fC showed that 5fC mainly occurred in the CpG islands (CGIs),160 supporting the role of 5fC in the maintenance of hypomethylation at CGIs in the ES cells. However, abasic sites as off-target aldehydes could also be labeled by ARP,190 which may lead to the inaccurate mapping of 5fC. As for the OG-seq approach,189 a mild one-electron oxidant (K2IrBr6) was used to selectively oxidize OG to form a covalent adduct of primary-amine terminated biotin (Fig. 3C). The resolution of OG-seq was 0.15 kb. The genome-wide mapping through OG-seq demonstrated that OG was mainly enriched in the promoter and UTR regulatory regions flanking protein-coding sequences.
Fig. 3. Reaction of the chemical labeling-assisted enrichment sequencing of fU-seq (A), fC-seq (B) and OG-seq (C) for mapping DNA modifications.
3.3.2. Chemical labeling-mediated base transition
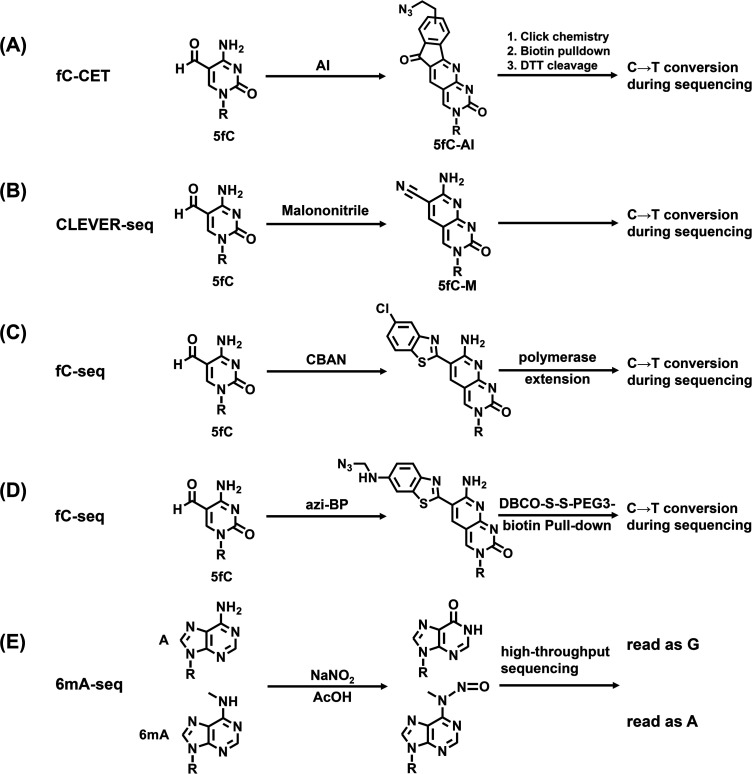
Chemical labeling of DNA modifications may affect their base pairing property, which can be utilized to distinguish DNA modifications from other nucleobases during sequencing. Yi's group developed an approach termed fC-CET sequencing for the genome-wide mapping of 5fC based on the 1,3-indandione (AI) selective labeling of 5fC (Fig. 4A).191 The AI-labeled 5fC enabled the subsequent C-to-T transition during PCR. However, AI is not an ideal reagent due to its poor solubility. In a recent approach of CLEVER-seq, malononitrile (M) was employed for 5fC labeling (Fig. 4B).192 Similar to 5fC-AI, 5fC-M enables a C-to-T conversion during sequencing. In this approach, malononitrile showed good solubility, high reactivity, and good biocompatibility. In addition, 2-(5-chlorobenzo[d]thiazol-2-yl)acetonitrile (CBAN)193 and its functional derivative of azi-BP142 were also demonstrated to be suitable reagents for 5fC labeling (Fig. 4C and D).
Fig. 4. Reaction of the chemical labeling-mediated base transition sequencing of fC-CET (A), CLEVER-seq (B), fC-seq (C-D) and 6mA-seq (E) for mapping DNA modifications.
A single-nucleotide resolution mapping analysis of 6mA in DNA and RNA has been recently conducted using nitrite labeling-mediated sequencing (Fig. 4E).194 In this study, treatment of DNA or RNA by sodium nitrite under acidic conditions led to the chemoselective deamination of unmethylated adenines, but not the 6mA. The deamination of adenines resulted in hypoxanthine bases, which were read as guanine during replication and reverse transcription, while 6mA resisted deamination and was read as adenine. The approach coupled with high-throughput sequencing enabled the identification of 6mA sites in DNA within various sequence contexts.
3.4. Enzyme-mediated labeling-sequencing
Enzyme-mediated labeling methods have been developed to achieve the efficient enrichment of DNA fragments that carry certain modifications. A typical example is the selective glycosylation of 5hmC by β-GT to produce 5gmC (Fig. 5A).195 In this respect, an engineered glucose moiety containing an azido group can be labeled to the hydroxyl group of 5hmC (Fig. 5B).196,197 The azido group on the glucose was then utilized to attach a dibenzocyclooctyne-PEG3-biotin through bioorthogonal chemistry for subsequent affinity enrichment and sequencing. Moreover, the combination of the glycosylation of 5hmC by β-GT, oxidation by sodium periodate, and biotinylation by an aldehyde-reactive probe (GLIB approach) enabled the enrichment and sequencing of 5hmC (Fig. 5C).198 In addition to β-GT, base J glucosyltransferase (J-GT) was also used to label and enrich 5hmU in genomic DNA (Fig. 5D).199 J-GT can selectively glycosylate 5hmU to produce glucosylhydroxymethyluracil (base J), which can be pulled down by an antibody against base J or J-binding protein 1. The enriched DNA was then subjected to sequencing analysis.
Fig. 5. Reaction of the enzyme-mediated labeling-sequencing of hmC-seq (A), hmC-seal (B), GLIB (C) and hmU-seq (D) for mapping DNA modifications.
Deciphering the biological impact of OG requires the single-base resolution sequencing of OG in genomes. Recently, the click-code-seq strategy has been established to enable the base-resolution mapping of OG by coupling the use of repair enzymes with a click DNA ligation reaction to insert a biocompatible locator code.200 In this approach, DNA was treated with the base excision repair proteins, formamidopyrimidine DNA glycosylase and apurinic/apyrimidinic endonuclease (APE1), to remove OG and yield a gap. The gap was filled with a synthetic O-3′-propargyl-modified nucleotide, which was labeled with a code sequence suitable for sequencing the location of the original OG site. With this approach, thousands of OG sites were discovered with distinct patterns related to transcription, chromatin architecture, and chemical oxidation potential.
3.5. Deamination-mediated sequencing
Some natural enzymes can selectively recognize specific nucleosides and convert them into other nucleosides, which can be utilized to map modifications in genomic DNA. For example, APOBEC3A (apolipoprotein B mRNA-editing catalytic polypeptide-like 3A) can efficiently deaminate cytosine, 5mC, and 5hmC, but shows no observable deamination activity toward glycosylated 5hmC.201 Using this unique property of APOBEC3A, we and others reported the APOBEC3A-mediated deamination sequencing (AMD-seq)202 and APOBEC-coupled epigenetic sequencing (ACE-seq)203 for the mapping of 5hmC in DNA at single-base resolution. In these approaches, the original C and 5mC in DNA were deaminated by APOBEC3A to form U and T, respectively, both of which were read as T during sequencing, while the glycosylated 5hmC was resistant to deamination and read as C during sequencing. Therefore, the remaining C in the sequence context came from the original 5hmC, which offered the single-base resolution analysis of 5hmC in DNA. These methods based on the unique property of DNA deaminase enzymes can effectively distinguish the modified state of cytosine, and enabled the mapping of 5hmC from 2 ng of input genomic DNA.203,204
3.6. Polymerase-mediated sequencing
We recently developed an approach for the single-base resolution mapping of OG in DNA using the differential coding properties of Tth and Bsu DNA polymerases.205 The anti and syn conformations of OG led to the dual coding property, i.e., the anti-conformation of OG engages in Watson–Crick base pairs with cytosine, while the syn-conformation of OG allows stable Hoogsteen base pairs with adenine.206 We found that Bsu DNA polymerase incorporated adenine opposite OG while Tth DNA polymerase incorporated cytosine opposite OG. The comparison of the primer extension products by Bsu and Tth polymerases followed by sequencing provided single-base resolution site information of OG in DNA. The analytical strategy was simple, and the analysis of sequencing data was relatively easy to perform.
3.7. Nuclease digestion-mediated sequencing
The nuclease digestion-mediated sequencing method is based on the fact that certain enzymes can specifically recognize specific DNA modifications. Therefore, by combining with various detection methods (such as gel electrophoresis, PCR, and sequencing), the resulting pattern can provide readouts of DNA modifications.
Some restriction enzymes are inhibited by 5mC in the sequence context CpG, so the cutting patterns by such enzymes can provide a readout of DNA methylation. The frequently used methylation-sensitive restriction enzymes for the DNA methylation studies are HpaII and SmaI, because they each have an isoschizomer (MspI for HpaII) or neoschizomer (XmaI for SmaI) that is not inhibited by CpG methylation.207 Similarly, the methods of DNA-modification-dependent restriction endonuclease AbaSI coupled with sequencing (Aba-seq208 and scAba-seq209) were developed to map 5hmC, and the assay of restriction endonuclease PvuRts1I coupled with sequencing (Pvu-Seal-seq) was created to map 5hmC and 5fC.210
DpnI-assisted N6-methyladenine sequencing (DA-6mA-seq) used DpnI to cleave methylated adenine sites (5′-G6mATC-3′) in duplex DNA.211 The cutting pattern was then employed to realize the base-resolution mapping of 6mA in sequencing. 6mA cross-linking exonuclease sequencing (6mACE-seq) utilized 6mA-specific antibodies cross-linked to protect the 6mA-DNA fragments from subsequent exonuclease treatment.212,213 In addition, the 6mA-CLIP-exo combines the immunoprecipitation enrichment, photo-crosslinking, and exonuclease digestion to map the 6mA sites.14 However, 6mA can only be detected at the sites recognized by these enzymes, and the 6mA site information may be lost if 6mA exists in other motifs.214
3.8. Nanopore sequencing
The third-generation sequencing technology of nanopore sequencing has become a promising approach in DNA sequencing.215–219 In nanopore sequencing, different nucleotides passing through nanopores generate different electric currents, which are measured and designated to the corresponding nucleotides or modified nucleotides. Nanopore sequencing technology has been employed to successfully distinguish 5mC from cytosine of human DNA,220 and 5mC from 5hmC in synthesized DNA strands,221 thus leading to the mapping of these modifications at single-base resolution. Besides, the 5hmC base was chemically labeled by thiolation in a synthesized single-stranded DNA under bisulfite treatment conditions and detected using nanopore sequencing.222 Nanopore-based sequencing was also successfully established to distinguish the five known C5-cytosine bases using synthesized DNA.223 In addition, the α-hemolysin (α-HL) nanopore was employed to map OG in ssDNA. In this work, OG was coupled with 1,12-dodecanediamine and then incubated with cucurbit[7]uril to form a host–guest complex-modified DNA hybrid. Translocation of this DNA hybrid generates current signatures reflective of the presence of OG in DNA.224
3.9. Single-molecule real-time sequencing
In addition to nanopore sequencing, single-molecule real-time (SMRT) sequencing is another promising third-generation sequencing technology.225 SMRT sequencing is performed by continuously observing the incorporation of a fluorescently labeled nucleotide with DNA polymerase. On the basis of the differences in the duration of nucleotide incorporation between the unmodified bases and the modified bases, SMRT sequencing can detect the positions and types of base modifications. SMRT sequencing has been applied to the direct detection of 5mC,226 6mA,227–229 5hmC,230 and PT.186 The detection of DNA modifications depends on the magnitude of the effect of the modification on polymerase kinetics. Because the methyl group of 5mC does not directly contribute to base pairing, 5mC shows a subtler impact on nucleotide incorporation than 6mA.231 Therefore, the detection of 5mC requires relatively high-coverage sequencing or oxidation of 5mC to 5caC using TET1 for signal enhancement.232
The emergence of third-generation sequencing technology provides complementary methods for mapping DNA modifications in genomes. Nevertheless, a recent report showed that this method usually overestimates the number of 6mA modifications measured in eukaryotic genomes.233 SMRT sequencing is also expensive, which impedes its extensive application to larger eukaryotic genomes.
4. Conclusions
The existing modifications in genomes broaden our views of the structures and functions of DNA. Elucidation of the biological roles of the modifications in genomes is critical for epigenetic-orientated biological sciences. We highlight the advances in the methods for the quantification and mapping of DNA modifications across prokaryotes and eukaryotes. The development of sensitive analytical methods and techniques in future studies will promote the discovery of new modifications and expand the list of natural modifications in genomes. The knowledge of DNA modifications is still growing, and deciphering the functions of DNA modifications remains an ongoing challenge. Developing appropriate and straightforward sequencing methods for mapping modifications in genomes and elucidating their dynamic changes will facilitate the study of their functions in human diseases, potentially leading to new strategies for therapies. Moreover, the determination of DNA modifications is also valuable in clinical diagnostics and forensics.
Although cells in living organisms have the same DNA sequence context, they can function differently. Single-cell epigenetic studies will provide insights into how heterogeneous DNA modifications may affect the transcriptional output and consequently their functions. The study of DNA modifications in individual cells will offer a new understanding of cell fate decisions, cell development, and disease progression. Mapping DNA modifications in a single cell is particularly valuable in embryos, which may uncover the mystery of epigenetic regulation in embryonic development. Current assays for single-cell DNA modifications frequently suffer from high false-discovery rates, low mapping rates and low detection sensitivity. There is still an urgent need for developing new methods to effectively decipher single-cell DNA modifications, especially for rare DNA modifications.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
The work is supported by the National Natural Science Foundation of China (22074110, 21635006, 21721005), and the Fundamental Research Funds for the Central Universities (2042021kf0212).
Biographies
Biography
Yi Dai .

Yi Dai is a master's student in the Department of Chemistry, Wuhan University, China. She received her BSc degree in Chemistry from the Beijing Normal University in China. Presently, she is working under the guidance of Prof. Bi-Feng Yuan, focusing on the development and applications of analytical methods for the study of nucleic acid modifications.
Biography
Bi-Feng Yuan .

Bi-Feng Yuan studied biochemistry and biophysics at Wuhan University in China, where he received his BSc and PhD degrees in 2001 and 2006, respectively. From 2006–2007 and 2007–2010, he worked as a postdoctoral researcher in the Department of Biological Sciences at the National University of Singapore and in the Department of Chemistry at the University of California Riverside, respectively. He joined Wuhan University in 2011 and is currently a professor of chemistry. His research focuses on the development and applications of new analytical techniques in the investigation of the occurrences, locations, and biological functions of nucleic acid modifications.
Biography
Yu-Qi Feng .
Yu-Qi Feng received his BSc and master's degrees from Lanzhou University in China in 1982 and 1985, respectively. From 1986 to 1991, he worked in Central China Normal University in China. He then studied chemistry at Chiba University in Japan, where he received his PhD degree in 1996. He joined Wuhan University in 1996 and became a full professor in 2000. His research focuses on sample pretreatment and LC/MS-based metabolomics.
References
- Zhao Y. Zuo X. Li Q. Chen F. Chen Y. R. Deng J. Han D. Hao C. Huang F. Huang Y. Ke G. Kuang H. Li F. Li J. Li M. Li N. Lin Z. Liu D. Liu J. Liu L. Liu X. Lu C. Luo F. Mao X. Sun J. Tang B. Wang F. Wang J. Wang L. Wang S. Wu L. Wu Z. S. Xia F. Xu C. Yang Y. Yuan B. F. Yuan Q. Zhang C. Zhu Z. Yang C. Zhang X. B. Yang H. Tan W. Fan C. Sci. China: Chem. 2021;64:171–203. doi: 10.1007/s11426-020-9864-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark B. McGouran J. F. Nat. Rev. Chem. 2018;2:332–348. [Google Scholar]
- Liu T. Ma C. J. Yuan B. F. Feng Y. Q. Sci. China: Chem. 2018;61:381–392. [Google Scholar]
- Greenberg M. V. C. Bourc'his D. Nat. Rev. Mol. Cell Biol. 2019;20:590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- Rausch C. Hastert F. D. Cardoso M. C. J. Mol. Biol. 2020;432:1731–1746. doi: 10.1016/j.jmb.2019.12.018. [DOI] [PubMed] [Google Scholar]
- Luo C. Hajkova P. Ecker J. R. Science. 2018;361:1336–1340. doi: 10.1126/science.aat6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Zhang Y. Nat. Rev. Genet. 2017;18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- He Y. F. Li B. Z. Li Z. Liu P. Wang Y. Tang Q. Ding J. Jia Y. Chen Z. Li L. Sun Y. Li X. Dai Q. Song C. X. Zhang K. He C. Xu G. L. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilyard M. K. Becker S. Balasubramanian S. Curr. Opin. Chem. Biol. 2020;57:1–7. doi: 10.1016/j.cbpa.2020.01.014. [DOI] [PubMed] [Google Scholar]
- Sood A. J. Viner C. Hoffman M. M. J. Cheminform. 2019;11:30. doi: 10.1186/s13321-019-0349-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. Yuan B. F. Feng Y. Q. Chem. Res. Toxicol. 2019;32:808–819. doi: 10.1021/acs.chemrestox.9b00042. [DOI] [PubMed] [Google Scholar]
- Heyn H. Esteller M. Cell. 2015;161:710–713. doi: 10.1016/j.cell.2015.04.021. [DOI] [PubMed] [Google Scholar]
- Luo G. Z. He C. Nat. Struct. Mol. Biol. 2017;24:503–506. doi: 10.1038/nsmb.3412. [DOI] [PubMed] [Google Scholar]
- Fu Y. Luo G. Z. Chen K. Deng X. Yu M. Han D. Hao Z. Liu J. Lu X. Dore L. C. Weng X. Ji Q. Mets L. He C. Cell. 2015;161:879–892. doi: 10.1016/j.cell.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer E. L. Blanco M. A. Gu L. Sendinc E. Liu J. Aristizabal-Corrales D. Hsu C. H. Aravind L. He C. Shi Y. Cell. 2015;161:868–878. doi: 10.1016/j.cell.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. Huang H. Liu D. Cheng Y. Liu X. Zhang W. Yin R. Zhang D. Zhang P. Liu J. Li C. Liu B. Luo Y. Zhu Y. Zhang N. He S. He C. Wang H. Chen D. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- Koziol M. J. Bradshaw C. R. Allen G. E. Costa A. S. Frezza C. Gurdon J. B. Nat. Struct. Mol. Biol. 2016;23:24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. Zhu Y. Luo G. Z. Wang X. Yue Y. Wang X. Zong X. Chen K. Yin H. Fu Y. Han D. Wang Y. Chen D. He C. Nat. Commun. 2016;7:13052. doi: 10.1038/ncomms13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. P. Wang T. Seetin M. G. Lai Y. Zhu S. Lin K. Liu Y. Byrum S. D. Mackintosh S. G. Zhong M. Tackett A. Wang G. Hon L. S. Fang G. Swenberg J. A. Xiao A. Z. Nature. 2016;532:329–333. doi: 10.1038/nature17640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C. L. Zhu S. He M. Chen D. Zhang Q. Chen Y. Yu G. Liu J. Xie S. Q. Luo F. Liang Z. Wang D. P. Bo X. C. Gu X. F. Wang K. Yan G. R. Mol. Cell. 2018;71:306–318. doi: 10.1016/j.molcel.2018.06.015. [DOI] [PubMed] [Google Scholar]
- Huang W. Xiong J. Yang Y. Liu S. M. Yuan B. F. Feng Y. Q. RSC Adv. 2015;5:64046–64054. [Google Scholar]
- Hao Z. Wu T. Cui X. Zhu P. Tan C. Dou X. Hsu K. W. Lin Y. T. Peng P. H. Zhang L. S. Gao Y. Hu L. Sun H. L. Zhu A. Liu J. Wu K. J. He C. Mol. Cell. 2020;78:382–395 e388. doi: 10.1016/j.molcel.2020.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musheev M. U. Baumgartner A. Krebs L. Niehrs C. Nat. Chem. Biol. 2020;16:630–634. doi: 10.1038/s41589-020-0504-2. [DOI] [PubMed] [Google Scholar]
- Liu X. Lai W. Li Y. Chen S. Liu B. Zhang N. Mo J. Lyu C. Zheng J. Du Y. R. Jiang G. Xu G. L. Wang H. Cell Res. 2021;31:94–97. doi: 10.1038/s41422-020-0317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F. Beraldi D. Hardisty R. E. McInroy G. R. van Delft P. Balasubramanian S. Genome Biol. 2017;18:23. doi: 10.1186/s13059-017-1150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen R. G. Simon M. Marmur J. J. Mol. Biol. 1962;5:248–250. doi: 10.1016/s0022-2836(62)80087-4. [DOI] [PubMed] [Google Scholar]
- Rae P. M. Science. 1976;194:1062–1064. doi: 10.1126/science.988637. [DOI] [PubMed] [Google Scholar]
- Pfaffeneder T. Spada F. Wagner M. Brandmayr C. Laube S. K. Eisen D. Truss M. Steinbacher J. Hackner B. Kotljarova O. Schuermann D. Michalakis S. Kosmatchev O. Schiesser S. Steigenberger B. Raddaoui N. Kashiwazaki G. Muller U. Spruijt C. G. Vermeulen M. Leonhardt H. Schar P. Muller M. Carell T. Nat. Chem. Biol. 2014;10:574–581. doi: 10.1038/nchembio.1532. [DOI] [PubMed] [Google Scholar]
- Borst P. Sabatini R. Annu. Rev. Microbiol. 2008;62:235–251. doi: 10.1146/annurev.micro.62.081307.162750. [DOI] [PubMed] [Google Scholar]
- Bullard W. Lopes da Rosa-Spiegler J. Liu S. Wang Y. Sabatini R. J. Biol. Chem. 2014;289:20273–20282. doi: 10.1074/jbc.M114.579821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J. H. Chen G. D. Hao F. Chen H. Fang Z. Chen F. F. Pang B. Yang Q. L. Wei X. Fan Q. Q. Xin C. Zhao J. Deng X. Wang B. A. Zhang X. J. Chu Y. Tang H. Yin H. Ma W. Chen L. Ding J. Weinhold E. Kohli R. M. Liu W. Zhu Z. J. Huang K. Tang H. Xu G. L. Nature. 2019;569:581–585. doi: 10.1038/s41586-019-1160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. M. Acc. Chem. Res. 2012;45:588–597. doi: 10.1021/ar2002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S. A. Gordenin D. A. Nat. Rev. Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B. F. Jiang Y. Wang Y. Chem. Res. Toxicol. 2010;23:11–19. doi: 10.1021/tx9004264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs A. Nussenzweig A. Cell. 2017;168:644–656. doi: 10.1016/j.cell.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B. F. Chem. Res. Toxicol. 2020;33:695–708. doi: 10.1021/acs.chemrestox.9b00372. [DOI] [PubMed] [Google Scholar]
- Fleming A. M. Burrows C. J. DNA Repair. 2017;56:75–83. doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L. Zhu B. Hao W. Zeng X. Vlahopoulos S. A. Hazra T. K. Hegde M. L. Radak Z. Bacsi A. Brasier A. R. Ba X. Boldogh I. J. Biol. Chem. 2016;291:25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming A. M. Ding Y. Burrows C. J. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X. Cao B. Aquino P. Chiu T. P. Chen C. Jiang S. Deng Z. Chen S. Rohs R. Wang L. Galagan J. E. Dedon P. C. Proc. Natl. Acad. Sci. U. S. A. 2020;117:14322–14330. doi: 10.1073/pnas.2002933117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T. Chen S. Wang L. Tang Y. Ryu J. Y. Jiang S. Wu X. Chen C. Luo J. Deng Z. Li Z. Lee S. Y. Chen S. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E2988–E2996. doi: 10.1073/pnas.1721916115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. Jiang S. Deng Z. Dedon P. C. Chen S. FEMS Microbiol. Rev. 2019;43:109–122. doi: 10.1093/femsre/fuy036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville J. J. Kellner S. M. Yuan Y. Hutinet G. Thiaville P. C. Jumpathong W. Mohapatra S. Brochier-Armanet C. Letarov A. V. Hillebrand R. Malik C. K. Rizzo C. J. Dedon P. C. de Crecy-Lagard V. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E1452–E1459. doi: 10.1073/pnas.1518570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutinet G. Kot W. Cui L. Hillebrand R. Balamkundu S. Gnanakalai S. Neelakandan R. Carstens A. B. Fa Lui C. Tremblay D. Jacobs-Sera D. Sassanfar M. Lee Y. J. Weigele P. Moineau S. Hatfull G. F. Dedon P. C. Hansen L. H. de Crecy-Lagard V. Nat. Commun. 2019;10:5442. doi: 10.1038/s41467-019-13384-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson M. A. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- Haws S. A. Leech C. M. Denu J. M. Trends Biochem. Sci. 2020;45:731–747. doi: 10.1016/j.tibs.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U. Rando O. J. Cell Metab. 2017;25:544–558. doi: 10.1016/j.cmet.2017.02.003. [DOI] [PubMed] [Google Scholar]
- Zhao L. Y. Song J. Liu Y. Song C. X. Yi C. Protein Cell. 2020;11:792–808. doi: 10.1007/s13238-020-00733-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H. He B. Yi C. ChemBioChem. 2019;20:1898–1905. doi: 10.1002/cbic.201900035. [DOI] [PubMed] [Google Scholar]
- Hofer A. Liu Z. J. Balasubramanian S. J. Am. Chem. Soc. 2019;141:6420–6429. doi: 10.1021/jacs.9b01915. [DOI] [PubMed] [Google Scholar]
- Armstrong K. M. Bermingham E. N. Bassett S. A. Treloar B. P. Roy N. C. Barnett M. P. Biotechnol. J. 2011;6:113–117. doi: 10.1002/biot.201000267. [DOI] [PubMed] [Google Scholar]
- Lai W. Y. Mo J. Z. Yin J. F. Lyu C. Wang H. L. TrAC, Trends Anal. Chem. 2019;110:173–182. [Google Scholar]
- Liu S. Wang Y. Chem. Soc. Rev. 2015;44:7829–7854. doi: 10.1039/c5cs00316d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X. J. Liu T. Ma C. J. Qi C. B. Tong Y. Zhao X. Yuan B. F. Feng Y. Q. Anal. Chem. 2019;91:10477–10483. doi: 10.1021/acs.analchem.9b01318. [DOI] [PubMed] [Google Scholar]
- Qi C. B. Jiang H. P. Xiong J. Yuan B. F. Feng Y. Q. Chin. Chem. Lett. 2019;30:553–557. [Google Scholar]
- Chen B. Yuan B. F. Feng Y. Q. Anal. Chem. 2019;91:743–756. doi: 10.1021/acs.analchem.8b04078. [DOI] [PubMed] [Google Scholar]
- Dai Y. Qi C. B. Feng Y. Cheng Q. Y. Liu F. L. Cheng M. Y. Yuan B. F. Feng Y. Q. Anal. Chem. 2021;93:6938–6946. doi: 10.1021/acs.analchem.0c04630. [DOI] [PubMed] [Google Scholar]
- Liu F. L. Ye T. T. Ding J. H. Yin X. M. Yang X. K. Huang W. H. Yuan B. F. Feng Y. Q. Anal. Chem. 2021;93:6848–6856. doi: 10.1021/acs.analchem.1c00915. [DOI] [PubMed] [Google Scholar]
- Li Q. Y. Yuan B. F. Feng Y. Q. Chem. Lett. 2018;47:1453–1459. [Google Scholar]
- Lan M. D. Yuan B. F. Feng Y. Q. Chin. Chem. Lett. 2019;30:1–6. [Google Scholar]
- Huang W. Qi C. B. Lv S. W. Xie M. Feng Y. Q. Huang W. H. Yuan B. F. Anal. Chem. 2016;88:1378–1384. doi: 10.1021/acs.analchem.5b03962. [DOI] [PubMed] [Google Scholar]
- Wang L. Chen S. Vergin K. L. Giovannoni S. J. Chan S. W. DeMott M. S. Taghizadeh K. Cordero O. X. Cutler M. Timberlake S. Alm E. J. Polz M. F. Pinhassi J. Deng Z. Dedon P. C. Proc. Natl. Acad. Sci. U. S. A. 2011;108:2963–2968. doi: 10.1073/pnas.1017261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. Tang Y. Dong X. Zheng Y. Y. Haruehanroengra P. Mao S. Lin Q. Sheng J. ACS Chem. Biol. 2020;15:1301–1305. doi: 10.1021/acschembio.0c00163. [DOI] [PubMed] [Google Scholar]
- Boulias K. Greer E. L. Methods Mol. Biol. 2021;2198:79–90. doi: 10.1007/978-1-0716-0876-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R. Mao S. Q. Zhao B. Chong Z. Yang Y. Zhao C. Zhang D. Huang H. Gao J. Li Z. Jiao Y. Li C. Liu S. Wu D. Gu W. Yang Y. G. Xu G. L. Wang H. J. Am. Chem. Soc. 2013;135:10396–10403. doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- Liu S. Wang J. Su Y. Guerrero C. Zeng Y. Mitra D. Brooks P. J. Fisher D. E. Song H. Wang Y. Nucleic Acids Res. 2013;41:6421–6429. doi: 10.1093/nar/gkt360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin R. Mo J. Lu M. Wang H. Anal. Chem. 2015;87:1846–1852. doi: 10.1021/ac5038895. [DOI] [PubMed] [Google Scholar]
- Wang J. Yuan B. F. Guerrero C. Bahde R. Gupta S. Wang Y. Anal. Chem. 2011;83:2201–2209. doi: 10.1021/ac103099s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q. Wu T. P. Gimple R. C. Li Z. Prager B. C. Wu Q. Yu Y. Wang P. Wang Y. Gorkin D. U. Zhang C. Dowiak A. V. Lin K. Zeng C. Sui Y. Kim L. J. Y. Miller T. E. Jiang L. Lee C. H. Huang Z. Fang X. Zhai K. Mack S. C. Sander M. Bao S. Kerstetter-Fogle A. E. Sloan A. E. Xiao A. Z. Rich J. N. Cell. 2018;175:1228–1243. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. Ye T. T. Ma C. J. Cheng Q. Y. Yuan B. F. Feng Y. Q. Nucleic Acids Res. 2019;47:1268–1277. doi: 10.1093/nar/gky1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner S. DeMott M. S. Cheng C. P. Russell B. S. Cao B. You D. Dedon P. C. Nat. Chem. Biol. 2017;13:888–894. doi: 10.1038/nchembio.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T. Hackner B. Truss M. Munzel M. Muller M. Deiml C. A. Hagemeier C. Carell T. Angew. Chem., Int. Ed. 2011;50:7008–7012. doi: 10.1002/anie.201103899. [DOI] [PubMed] [Google Scholar]
- Ito S. Shen L. Dai Q. Wu S. C. Collins L. B. Swenberg J. A. He C. Zhang Y. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. L. Shen F. Huang W. Qi J. H. Wang Y. Feng Y. Q. Liu S. M. Yuan B. F. Clin. Chem. 2013;59:824–832. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. Zhang J. Guo Z. Ma Q. Xu Z. Zhou Y. Xu Z. Li Z. Liu Y. Ye X. Li X. Yuan B. Ke Y. He C. Zhou L. Liu J. Ci W. Cell Res. 2016;26:103–118. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeze L. I. van der Reijden B. A. Jansen J. H. Biochim. Biophys. Acta. 2015;1855:144–154. doi: 10.1016/j.bbcan.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Ficz G. Gribben J. G. Genomics. 2014;104:352–357. doi: 10.1016/j.ygeno.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. F. Qi C. B. Yuan B. F. Feng Y. Q. Anal. Chim. Acta. 2019;1081:103–111. doi: 10.1016/j.aca.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Wilschefski S. C. Baxter M. R. Clin. Biochem. Rev. 2019;40:115–133. doi: 10.33176/AACB-19-00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel K. Landero Figueroa J. A. Zaina S. Lund G. Wrobel K. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2010;878:609–614. doi: 10.1016/j.jchromb.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Kim I. Y. Suh S. H. Lee I. K. Wolfe R. R. Exp. Mol. Med. 2016;48:e203. doi: 10.1038/emm.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. C., Labuschagne C. F., Vousden K. H. and Maddocks O. D. K., in Cancer Metabolism: Methods and Protocols, ed. M. Haznadar, Springer New York, New York, NY, 2019, pp. 55–67 [Google Scholar]
- Iwan K. Rahimoff R. Kirchner A. Spada F. Schröder A. S. Kosmatchev O. Ferizaj S. Steinbacher J. Parsa E. Müller M. Carell T. Nat. Chem. Biol. 2018;14:72–78. doi: 10.1038/nchembio.2531. [DOI] [PubMed] [Google Scholar]
- Liu B. Liu X. Lai W. Wang H. Anal. Chem. 2017;89:6202–6209. doi: 10.1021/acs.analchem.7b01152. [DOI] [PubMed] [Google Scholar]
- Feng Y. Xie N. B. Tao W. B. Ding J. H. You X. J. Ma C. J. Zhang X. Yi C. Zhou X. Yuan B. F. Feng Y. Q. CCS Chem. 2020;2:994–1008. [Google Scholar]
- Schiesser S. Hackner B. Pfaffeneder T. Muller M. Hagemeier C. Truss M. Carell T. Angew. Chem., Int. Ed. 2012;51:6516–6520. doi: 10.1002/anie.201202583. [DOI] [PubMed] [Google Scholar]
- Zoccali M. Tranchida P. Q. Mondello L. TrAC, Trends Anal. Chem. 2019;118:444–452. [Google Scholar]
- Rossella F. Polledri E. Bollati V. Baccarelli A. Fustinoni S. Rapid Commun. Mass Spectrom. 2009;23:2637–2646. doi: 10.1002/rcm.4166. [DOI] [PubMed] [Google Scholar]
- Tang Y. Gao X. D. Wang Y. Yuan B. F. Feng Y. Q. Anal. Chem. 2012;84:7249–7255. doi: 10.1021/ac301727c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podmore I. D. Griffiths H. R. Herbert K. E. Mistry N. Mistry P. Lunec J. Nature. 1998;392:559. doi: 10.1038/33308. [DOI] [PubMed] [Google Scholar]
- Qi B. L. Liu P. Wang Q. Y. Cai W. J. Yuan B. F. Feng Y. Q. TrAC, Trends Anal. Chem. 2014;59:121–132. [Google Scholar]
- Liu F. L. Qi C. B. Cheng Q. Y. Ding J. H. Yuan B. F. Feng Y. Q. Anal. Chem. 2020;92:2301–2309. doi: 10.1021/acs.analchem.9b05122. [DOI] [PubMed] [Google Scholar]
- Feng Y. Ma C. J. Ding J. H. Qi C. B. Xu X. J. Yuan B. F. Feng Y. Q. Anal. Chim. Acta. 2020;1098:56–65. doi: 10.1016/j.aca.2019.11.016. [DOI] [PubMed] [Google Scholar]
- Cheng Q. Y. Xiong J. Ma C. J. Dai Y. Ding J. H. Liu F. L. Yuan B. F. Feng Y. Q. Chem. Sci. 2020;11:1878–1891. doi: 10.1039/c9sc05094a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B. F. Zhu Q. F. Guo N. Zheng S. J. Wang Y. L. Wang J. Xu J. Liu S. J. He K. Hu T. Zheng Y. W. Xu F. Q. Feng Y. Q. Anal. Chem. 2018;90:3512–3520. doi: 10.1021/acs.analchem.7b05355. [DOI] [PubMed] [Google Scholar]
- Cheng Q. Y. Xiong J. Wang F. Yuan B. F. Feng Y. Q. Chin. Chem. Lett. 2018;29:115–118. [Google Scholar]
- Zeng H. Qi C. B. Liu T. Xiao H. M. Cheng Q. Y. Jiang H. P. Yuan B. F. Feng Y. Q. Anal. Chem. 2017;89:4153–4160. doi: 10.1021/acs.analchem.7b00052. [DOI] [PubMed] [Google Scholar]
- Tang Y. Zheng S. J. Qi C. B. Feng Y. Q. Yuan B. F. Anal. Chem. 2015;87:3445–3452. doi: 10.1021/ac504786r. [DOI] [PubMed] [Google Scholar]
- Huang W. Lan M. D. Qi C. B. Zheng S. J. Wei S. Z. Yuan B. F. Feng Y. Q. Chem. Sci. 2016;7:5495–5502. doi: 10.1039/c6sc01589a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. Li X. Zhang L. Liu D. Du W. Yin D. Lyu N. Zhao G. Guo C. Tang D. Oncotarget. 2017;8:91248–91257. doi: 10.18632/oncotarget.20093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J. Liu X. Cheng Q. Y. Xiao S. Xia L. X. Yuan B. F. Feng Y. Q. ACS Chem. Biol. 2017;12:1636–1643. doi: 10.1021/acschembio.7b00170. [DOI] [PubMed] [Google Scholar]
- Tang Y. Chu J.-M. Huang W. Xiong J. Xing X.-W. Zhou X. Feng Y.-Q. Yuan B.-F. Anal. Chem. 2013;85:6129–6135. doi: 10.1021/ac4010869. [DOI] [PubMed] [Google Scholar]
- Hong H. Wang Y. Anal. Chem. 2007;79:322–326. doi: 10.1021/ac061465w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y. Xiong J. Jiang H. P. Zheng S. J. Feng Y. Q. Yuan B. F. Anal. Chem. 2014;86:7764–7772. doi: 10.1021/ac5016886. [DOI] [PubMed] [Google Scholar]
- Jiang H. P. Liu T. Guo N. Yu L. Yuan B. F. Feng Y. Q. Anal. Chim. Acta. 2017;981:1–10. doi: 10.1016/j.aca.2017.06.009. [DOI] [PubMed] [Google Scholar]
- Yu Y. Yuan F. Zhang X. H. Zhao M. Z. Zhou Y. L. Zhang X. X. Anal. Chem. 2019;91:13047–13053. doi: 10.1021/acs.analchem.9b03227. [DOI] [PubMed] [Google Scholar]
- Zhang H. Y. Xiong J. Qi B. L. Feng Y. Q. Yuan B. F. Chem. Commun. 2016;52:737–740. doi: 10.1039/c5cc07354e. [DOI] [PubMed] [Google Scholar]
- Jiang H. P. Xiong J. Liu F. L. Ma C. J. Tang X. L. Yuan B. F. Feng Y. Q. Chem. Sci. 2018;9:4160–4167. doi: 10.1039/c7sc05472f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F. Zhang X. H. Nie J. Chen H. X. Zhou Y. L. Zhang X. X. Chem. Commun. 2016;52:2698–2700. doi: 10.1039/c5cc10155g. [DOI] [PubMed] [Google Scholar]
- Krais A. M. Cornelius M. G. Schmeiser H. H. Electrophoresis. 2010;31:3548–3551. doi: 10.1002/elps.201000357. [DOI] [PubMed] [Google Scholar]
- Chen M. L. Liu Y. L. Xing X. W. Zhou X. Feng Y. Q. Yuan B. F. Chem. – Eur. J. 2013;19:1035–1041. doi: 10.1002/chem.201203129. [DOI] [PubMed] [Google Scholar]
- Paz M. F. Fraga M. F. Avila S. Guo M. Pollan M. Herman J. G. Esteller M. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Lyko F. Stach D. Brenner A. Stilgenbauer S. Dohner H. Wirtz M. Wiessler M. Schmitz O. J. Electrophoresis. 2004;25:1530–1535. doi: 10.1002/elps.200305830. [DOI] [PubMed] [Google Scholar]
- Mund C. Musch T. Strodicke M. Assmann B. Li E. Lyko F. Biochem. J. 2004;378:763–768. doi: 10.1042/BJ20031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodi Z. Fray R. G. Methods Mol. Biol. 2017;1562:79–87. doi: 10.1007/978-1-4939-6807-7_6. [DOI] [PubMed] [Google Scholar]
- Tahiliani M. Koh K. P. Shen Y. Pastor W. A. Bandukwala H. Brudno Y. Agarwal S. Iyer L. M. Liu D. R. Aravind L. Rao A. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriaucionis S. Heintz N. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowher H. Leismann O. Jeltsch A. EMBO J. 2000;19:6918–6923. doi: 10.1093/emboj/19.24.6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooijes D. Chaves I. Kieft R. Dirks-Mulder A. Martin W. Borst P. Nucleic Acids Res. 2000;28:3017–3021. doi: 10.1093/nar/28.16.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maatouk I. Bouaïcha N. Plessis M. J. Périn F. Toxicol. Mech. Methods. 2006;16:313–322. doi: 10.1080/15376520600616909. [DOI] [PubMed] [Google Scholar]
- Thu K. L. Pikor L. A. Kennett J. Y. Alvarez C. E. Lam W. L. J. Cell. Physiol. 2010;222:522–531. doi: 10.1002/jcp.22009. [DOI] [PubMed] [Google Scholar]
- Inoue A. Zhang Y. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A. Shen L. Dai Q. He C. Zhang Y. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. Zhang M. Guo M. Discovery Med. 2020;29:85–90. [PubMed] [Google Scholar]
- van Leeuwen F. Taylor M. C. Mondragon A. Moreau H. Gibson W. Kieft R. Borst P. Proc. Natl. Acad. Sci. U. S. A. 1998;95:2366–2371. doi: 10.1073/pnas.95.5.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machella N. Regoli F. Santella R. M. Aquat. Toxicol. 2005;71:335–343. doi: 10.1016/j.aquatox.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Kurita R. Arai K. Nakamoto K. Kato D. Niwa O. Anal. Chem. 2012;84:1799–1803. doi: 10.1021/ac202692f. [DOI] [PubMed] [Google Scholar]
- Sun J. Lou X. D. Wang H. D. Sollazzo A. Harms-Ringdahl M. Skog S. He E. Haghdoost S. Int. J. Diabetes Dev. Countries. 2015;35:368–373. [Google Scholar]
- Hori Y. Otomura N. Nishida A. Nishiura M. Umeno M. Suetake I. Kikuchi K. J. Am. Chem. Soc. 2018;140:1686–1690. doi: 10.1021/jacs.7b09713. [DOI] [PubMed] [Google Scholar]
- Song C. X. Diao J. Brunger A. T. Quake S. R. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4338–4343. doi: 10.1073/pnas.1600223113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Torres A. Yanez Barrientos E. Wrobel K. Wrobel K. Anal. Chem. 2011;83:7999–8005. doi: 10.1021/ac2020799. [DOI] [PubMed] [Google Scholar]
- Guo P. Yan S. Hu J. Xing X. Wang C. Xu X. Qiu X. Ma W. Lu C. Weng X. Zhou X. Org. Lett. 2013;15:3266–3269. doi: 10.1021/ol401290d. [DOI] [PubMed] [Google Scholar]
- Hu J. Xing X. Xu X. Wu F. Guo P. Yan S. Xu Z. Xu J. Weng X. Zhou X. Chem. – Eur. J. 2013;19:5836–5840. doi: 10.1002/chem.201300082. [DOI] [PubMed] [Google Scholar]
- Xu L. Chen Y. C. Chong J. Fin A. McCoy L. S. Xu J. Zhang C. Wang D. Angew. Chem., Int. Ed. 2014;53:11223–11227. doi: 10.1002/anie.201406220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. Chen Y. C. Nakajima S. Chong J. Wang L. Lan L. Zhang C. Wang D. Chem. Sci. 2014;5:567–574. doi: 10.1039/C3SC51849C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Liu C. Yang W. Zou G. Zhang X. Wu F. Yu S. Luo X. Zhou X. Chem. Commun. 2018;54:1497–1500. doi: 10.1039/c7cc08715b. [DOI] [PubMed] [Google Scholar]
- Samanta B. Seikowski J. Hobartner C. Angew. Chem., Int. Ed. 2016;55:1912–1916. doi: 10.1002/anie.201508893. [DOI] [PubMed] [Google Scholar]
- Liu C. X. Chen Y. Q. Wang Y. F. Wu F. Zhang X. Yang W. Wang J. Q. Chen Y. He Z. Y. Zou G. R. Wang S. R. Zhou X. Nano Res. 2017;10:2449–2458. [Google Scholar]
- Liu C. Wang Y. Zhang X. Wu F. Yang W. Zou G. Yao Q. Wang J. Chen Y. Wang S. Zhou X. Chem. Sci. 2017;8:4505–4510. doi: 10.1039/c7sc00637c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q. Li K. Li L. L. Yu K. K. Zhang H. Shi L. Chen H. Yu X. Q. Anal. Chem. 2019;91:9366–9370. doi: 10.1021/acs.analchem.9b02499. [DOI] [PubMed] [Google Scholar]
- Shahal T. Green O. Hananel U. Michaeli Y. Shabat D. Ebenstein Y. Methods Appl. Fluoresc. 2016;4:044003. doi: 10.1088/2050-6120/4/4/044003. [DOI] [PubMed] [Google Scholar]
- Wang Y. Liu C. Zhang X. Yang W. Wu F. Zou G. Weng X. Zhou X. Chem. Sci. 2018;9:3723–3728. doi: 10.1039/c8sc00493e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y. Yang G. Chen H. Zhang H. Feng J. J. Cai C. Analyst. 2018;143:2051–2056. doi: 10.1039/c7an02049j. [DOI] [PubMed] [Google Scholar]
- Chen X. Huang J. Zhang S. Mo F. Su S. Li Y. Fang L. Deng J. Huang H. Luo Z. Zheng J. ACS Appl. Mater. Interfaces. 2019;11:3745–3752. doi: 10.1021/acsami.8b20144. [DOI] [PubMed] [Google Scholar]
- Zhang L. Xiao X. Xu Y. Chen D. Chen J. Ma Y. Dai Z. Zou X. Biosens. Bioelectron. 2018;100:184–191. doi: 10.1016/j.bios.2017.08.013. [DOI] [PubMed] [Google Scholar]
- Huang J. Zhang S. Mo F. Su S. Chen X. Li Y. Fang L. Huang H. Deng J. Liu H. Yang X. Zheng J. Biosens. Bioelectron. 2019;127:155–160. doi: 10.1016/j.bios.2018.12.022. [DOI] [PubMed] [Google Scholar]
- Beaujean N. Salvaing J. Hadi N. A. A. Pennings S. Methods Mol. Biol. 2018;1708:59–80. doi: 10.1007/978-1-4939-7481-8_4. [DOI] [PubMed] [Google Scholar]
- Cui L. Hu J. Wang M. Li C. C. Zhang C. Y. Anal. Chem. 2019;91:1232–1236. doi: 10.1021/acs.analchem.8b04663. [DOI] [PubMed] [Google Scholar]
- Meng Y. Bai W. Zhang Y. Sun H. Li Y. Talanta. 2020;210:120597. doi: 10.1016/j.talanta.2019.120597. [DOI] [PubMed] [Google Scholar]
- Chen H. Y. Wei J. R. Pan J. X. Zhang W. Dang F. Q. Zhang Z. Q. Zhang J. Biosens. Bioelectron. 2017;91:328–333. doi: 10.1016/j.bios.2016.12.039. [DOI] [PubMed] [Google Scholar]
- Fan J. Liu Y. Xu E. Zhang Y. Wei W. Yin L. Pu Y. Liu S. Anal. Chim. Acta. 2016;946:48–55. doi: 10.1016/j.aca.2016.10.022. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Li Y. Wei Y. Sun H. Wang H. Talanta. 2017;170:546–551. doi: 10.1016/j.talanta.2017.04.051. [DOI] [PubMed] [Google Scholar]
- Qi C. B. Ding J. H. Yuan B. F. Feng Y. Q. Chin. Chem. Lett. 2019;30:1618–1626. [Google Scholar]
- Weber M. Davies J. J. Wittig D. Oakeley E. J. Haase M. Lam W. L. Schubeler D. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Rauch T. Pfeifer G. P. Lab. Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- Serre D. Lee B. H. Ting A. H. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M. Kadam S. Xiong W. Rauch T. A. Jin S. G. Pfeifer G. P. Epigenomics. 2015;7:695–706. doi: 10.2217/epi.15.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O. Fernandez-Minan A. Tena J. J. de la Calle-Mustienes E. Gomez-Skarmeta J. L. Methods. 2013;62:207–215. doi: 10.1016/j.ymeth.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Thomson J. P. Meehan R. R. Methods Mol. Biol. 2018;1708:679–696. doi: 10.1007/978-1-4939-7481-8_35. [DOI] [PubMed] [Google Scholar]
- Raiber E. A. Beraldi D. Ficz G. Burgess H. E. Branco M. R. Murat P. Oxley D. Booth M. J. Reik W. Balasubramanian S. Genome Biol. 2012;13:R69. doi: 10.1186/gb-2012-13-8-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao B. Li Y. Wang Z. Chen L. Poidevin M. Zhang C. Lin L. Wang F. Bao H. Jiao B. Lim J. Cheng Y. Huang L. Phillips B. L. Xu T. Duan R. Moberg K. H. Wu H. Jin P. Mol. Cell. 2018;71:848–857 e846. doi: 10.1016/j.molcel.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Lai W. Zhang N. Wang H. Anal. Chem. 2018;90:5546–5551. doi: 10.1021/acs.analchem.8b01087. [DOI] [PubMed] [Google Scholar]
- Cliffe L. J. Siegel T. N. Marshall M. Cross G. A. Sabatini R. Nucleic Acids Res. 2010;38:3923–3935. doi: 10.1093/nar/gkq146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara M. Jiang L. Akatsuka S. Suyama M. Toyokuni S. DNA Res. 2014;21:603–612. doi: 10.1093/dnares/dsu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini A. Lagerwall C. Vikingsson S. Mjoseng H. K. Douvlataniotis K. Vogt H. Green H. Meehan R. R. Benson M. Nestor C. E. Nat. Methods. 2018;15:499–504. doi: 10.1038/s41592-018-0038-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin V. V. Kutueva L. I. Vanyushin B. F. Methods Mol. Biol. 2020;2138:297–312. doi: 10.1007/978-1-0716-0471-7_21. [DOI] [PubMed] [Google Scholar]
- Booth M. J. Branco M. R. Ficz G. Oxley D. Krueger F. Reik W. Balasubramanian S. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- Yu M. Hon G. C. Szulwach K. E. Song C. X. Zhang L. Kim A. Li X. Dai Q. Shen Y. Park B. Min J. H. Jin P. Ren B. He C. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D. Chung T. H. Sun X. Jia X. Y. Anal. Chem. 2018;90:13200–13206. doi: 10.1021/acs.analchem.8b02832. [DOI] [PubMed] [Google Scholar]
- Booth M. J. Marsico G. Bachman M. Beraldi D. Balasubramanian S. Nat. Chem. 2014;6:435–440. doi: 10.1038/nchem.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. X. Szulwach K. E. Dai Q. Fu Y. Mao S. Q. Lin L. Street C. Li Y. Poidevin M. Wu H. Gao J. Liu P. Li L. Xu G. L. Jin P. He C. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Song C. X. Szulwach K. Wang Z. Weidenbacher P. Jin P. He C. J. Am. Chem. Soc. 2013;135:9315–9317. doi: 10.1021/ja4044856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X. Han D. Zhao B. S. Song C. X. Zhang L. S. Dore L. C. He C. Cell Res. 2015;25:386–389. doi: 10.1038/cr.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. Wu X. Zhang Y. Nat. Protoc. 2016;11:1081–1100. doi: 10.1038/nprot.2016.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F. Incarnato D. Krepelova A. Parlato C. Oliviero S. Nat. Protoc. 2016;11:1191–1205. doi: 10.1038/nprot.2016.063. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. Wilson G. G. Kuo K. C. Gehrke C. W. J. Bacteriol. 1987;169:939–943. doi: 10.1128/jb.169.3.939-943.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M. Ji L. Neumann D. A. Chung D. H. Groom J. Westpheling J. He C. Schmitz R. J. Nucleic Acids Res. 2015;43:e148. doi: 10.1093/nar/gkv738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H. He B. Xia B. Bai D. Lu X. Cai J. Chen L. Zhou A. Zhu C. Meng H. Gao Y. Guo H. He C. Dai Q. Yi C. J. Am. Chem. Soc. 2018;140:13190–13194. doi: 10.1021/jacs.8b08297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki F. Martinez Cuesta S. Beraldi D. Mahtey A. Hardisty R. E. Carrington M. Balasubramanian S. Angew. Chem., Int. Ed. 2018;57:9694–9696. doi: 10.1002/anie.201804046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Zhang X. Wu F. Chen Z. Zhou X. Chem. Sci. 2019;10:447–452. doi: 10.1039/c8sc04272a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi G. Koyama K. Shiota H. Kamio A. Umeda T. Nagae G. Aburatani H. Okamoto A. J. Am. Chem. Soc. 2016;138:14178–14181. doi: 10.1021/jacs.6b06428. [DOI] [PubMed] [Google Scholar]
- Liu Y. Siejka-Zielinska P. Velikova G. Bi Y. Yuan F. Tomkova M. Bai C. Chen L. Schuster-Bockler B. Song C. X. Nat. Biotechnol. 2019;37:424–429. doi: 10.1038/s41587-019-0041-2. [DOI] [PubMed] [Google Scholar]
- Liu Y. Cheng J. Siejka-Zielinska P. Weldon C. Roberts H. Lopopolo M. Magri A. D'Arienzo V. Harris J. M. McKeating J. A. Song C. X. Genome Biol. 2020;21:54. doi: 10.1186/s13059-020-01969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gish G. Eckstein F. Science. 1988;240:1520–1522. doi: 10.1126/science.2453926. [DOI] [PubMed] [Google Scholar]
- Chen Y. Zheng T. Li J. Cui J. Deng Z. You D. Yang L. Sci. Rep. 2019;9:7485. doi: 10.1038/s41598-019-44011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B. Chen C. DeMott M. S. Cheng Q. Clark T. A. Xiong X. Zheng X. Butty V. Levine S. S. Yuan G. Boitano M. Luong K. Song Y. Zhou X. Deng Z. Turner S. W. Korlach J. You D. Wang L. Chen S. Dedon P. C. Nat. Commun. 2014;5:3951. doi: 10.1038/ncomms4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. Chen Y. Zheng T. Kong L. Zhu S. Sun Y. Deng Z. Yang L. You D. PLoS Genet. 2019;15:e1008026. doi: 10.1371/journal.pgen.1008026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Liu C. Wu F. Zhang X. Liu S. Chen Z. Zeng W. Yang W. Zhang X. Zhou Y. Weng X. Wu Z. Zhou X. iScience. 2018;9:423–432. doi: 10.1016/j.isci.2018.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y. Fleming A. M. Burrows C. J. J. Am. Chem. Soc. 2017;139:2569–2572. doi: 10.1021/jacs.6b12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide H. Akamatsu K. Kimura Y. Michiue K. Makino K. Asaeda A. Takamori Y. Kubo K. Biochemistry. 1993;32:8276–8283. doi: 10.1021/bi00083a031. [DOI] [PubMed] [Google Scholar]
- Xia B. Han D. Lu X. Sun Z. Zhou A. Yin Q. Zeng H. Liu M. Jiang X. Xie W. He C. Yi C. Nat. Methods. 2015;12:1047–1050. doi: 10.1038/nmeth.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. Gao Y. Guo H. Xia B. Song J. Wu X. Zeng H. Kee K. Tang F. Yi C. Cell Stem Cell. 2017;20:720–731 e725. doi: 10.1016/j.stem.2017.02.013. [DOI] [PubMed] [Google Scholar]
- Liu C. Wang Y. Yang W. Wu F. Zeng W. Chen Z. Huang J. Zou G. Zhang X. Wang S. Weng X. Wu Z. Zhou Y. Zhou X. Chem. Sci. 2017;8:7443–7447. doi: 10.1039/c7sc03685j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi-Amiri Y. Chung Kim Chung K. Hili R. Chem. Sci. 2021;12:606–612. doi: 10.1039/d0sc03509b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson A. B. Dahl J. A. Vagbo C. B. Tripathi P. Krokan H. E. Klungland A. Nucleic Acids Res. 2011;39:e55. doi: 10.1093/nar/gkr051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. X. Szulwach K. E. Fu Y. Dai Q. Yi C. Li X. Li Y. Chen C. H. Zhang W. Jian X. Wang J. Zhang L. Looney T. J. Zhang B. Godley L. A. Hicks L. M. Lahn B. T. Jin P. He C. Nat. Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. Lu X. Shih A. H. Nie J. You Q. Xu M. M. Melnick A. M. Levine R. L. He C. Mol. Cell. 2016;63:711–719. doi: 10.1016/j.molcel.2016.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A. Pape U. J. Huang Y. Henderson H. R. Lister R. Ko M. McLoughlin E. M. Brudno Y. Mahapatra S. Kapranov P. Tahiliani M. Daley G. Q. Liu X. S. Ecker J. R. Milos P. M. Agarwal S. Rao A. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard W. Kieft R. Sabatini R. Biol. Methods Protoc. 2017;2:bpw006. doi: 10.1093/biomethods/bpw006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. McKeague M. Sturla S. J. J. Am. Chem. Soc. 2018;140:9783–9787. doi: 10.1021/jacs.8b03715. [DOI] [PubMed] [Google Scholar]
- Schutsky E. K. Nabel C. S. Davis A. K. F. DeNizio J. E. Kohli R. M. Nucleic Acids Res. 2017;45:7655–7665. doi: 10.1093/nar/gkx345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. Y. Xie N. B. Xiong J. Yuan B. F. Feng Y. Q. Anal. Chem. 2018;90:14622–14628. doi: 10.1021/acs.analchem.8b04833. [DOI] [PubMed] [Google Scholar]
- Schutsky E. K. DeNizio J. E. Hu P. Liu M. Y. Nabel C. S. Fabyanic E. B. Hwang Y. Bushman F. D. Wu H. Kohli R. M. Nat. Biotechnol. 2018;36:1083–1090. doi: 10.1038/nbt.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z. Vaisvila R. Hussong L. M. Yan B. Baum C. Saleh L. Samaranayake M. Guan S. Dai N. Corrêa, Jr. I. R. Pradhan S. Davis T. B. Evans, Jr. T. C. Ettwiller L. M. Genome Res. 2021;31:291–300. doi: 10.1101/gr.265306.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F. Liu S. Li Q. Y. Yuan J. Li L. Wang Y. Yuan B. F. Feng Y. Q. Chem. Sci. 2019;10:4272–4281. doi: 10.1039/c8sc04946g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirouac K. N. Ling H. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3210–3215. doi: 10.1073/pnas.1013909108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajares M. J. Palanca-Ballester C. Urtasun R. Alemany-Cosme E. Lahoz A. Sandoval J. Methods. 2021;187:3–12. doi: 10.1016/j.ymeth.2020.06.021. [DOI] [PubMed] [Google Scholar]
- Sun Z. Terragni J. Borgaro J. G. Liu Y. Yu L. Guan S. Wang H. Sun D. Cheng X. Zhu Z. Pradhan S. Zheng Y. Cell Rep. 2013;3:567–576. doi: 10.1016/j.celrep.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooijman D. Dey S. S. Boisset J. C. Crosetto N. van Oudenaarden A. Nat. Biotechnol. 2016;34:852–856. doi: 10.1038/nbt.3598. [DOI] [PubMed] [Google Scholar]
- Sun Z. Dai N. Borgaro J. G. Quimby A. Sun D. Correa, Jr. I. R. Zheng Y. Zhu Z. Guan S. Mol. Cell. 2015;57:750–761. doi: 10.1016/j.molcel.2014.12.035. [DOI] [PubMed] [Google Scholar]
- Luo G. Z. Wang F. Weng X. Chen K. Hao Z. Yu M. Deng X. Liu J. He C. Nat. Commun. 2016;7:11301. doi: 10.1038/ncomms11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh W. S. S. Methods Mol. Biol. 2021;2198:369–377. doi: 10.1007/978-1-0716-0876-0_28. [DOI] [PubMed] [Google Scholar]
- Koh C. W. Q. Goh Y. T. Toh J. D. W. Neo S. P. Ng S. B. Gunaratne J. Gao Y. G. Quake S. R. Burkholder W. F. Goh W. S. S. Nucleic Acids Res. 2018;46:11659–11670. doi: 10.1093/nar/gky1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. Z. Blanco M. A. Greer E. L. He C. Shi Y. Nat. Rev. Mol. Cell Biol. 2015;16:705–710. doi: 10.1038/nrm4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasianowicz J. J. Brandin E. Branton D. Deamer D. W. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller A. Nivon L. Brandin E. Golovchenko J. Branton D. Proc. Natl. Acad. Sci. U. S. A. 2000;97:1079–1084. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D. W. Branton D. Acc. Chem. Res. 2002;35:817–825. doi: 10.1021/ar000138m. [DOI] [PubMed] [Google Scholar]
- Branton D. Deamer D. W. Marziali A. Bayley H. Benner S. A. Butler T. Di Ventra M. Garaj S. Hibbs A. Huang X. Jovanovich S. B. Krstic P. S. Lindsay S. Ling X. S. Mastrangelo C. H. Meller A. Oliver J. S. Pershin Y. V. Ramsey J. M. Riehn R. Soni G. V. Tabard-Cossa V. Wanunu M. Wiggin M. Schloss J. A. Nat. Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J. Balasubramanian S. Church G. M. Gilbert W. Rogers J. Schloss J. A. Waterston R. H. Nature. 2017;550:345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- Simpson J. T. Workman R. E. Zuzarte P. C. David M. Dursi L. J. Timp W. Nat. Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- Laszlo A. H. Derrington I. M. Brinkerhoff H. Langford K. W. Nova I. C. Samson J. M. Bartlett J. J. Pavlenok M. Gundlach J. H. Proc. Natl. Acad. Sci. U. S. A. 2013;110:18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. W. Gong L. Bayley H. Angew. Chem., Int. Ed. 2013;52:4350–4355. doi: 10.1002/anie.201300413. [DOI] [PubMed] [Google Scholar]
- Wescoe Z. L. Schreiber J. Akeson M. J. Am. Chem. Soc. 2014;136:16582–16587. doi: 10.1021/ja508527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. Li Y. Li T. Xie J. Chen C. Liu Q. Zhang S. Wu H. C. Anal. Chem. 2016;88:1073–1077. doi: 10.1021/acs.analchem.5b04102. [DOI] [PubMed] [Google Scholar]
- Ardui S. Ameur A. Vermeesch J. R. Hestand M. S. Nucleic Acids Res. 2018;46:2159–2168. doi: 10.1093/nar/gky066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg B. A. Webster D. R. Lee J. H. Travers K. J. Olivares E. C. Clark T. A. Korlach J. Turner S. W. Nat. Methods. 2010;7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z. Shen L. Cui X. Bao S. Geng Y. Yu G. Liang F. Xie S. Lu T. Gu X. Yu H. Dev. Cell. 2018;45:406–416. doi: 10.1016/j.devcel.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Zhou C. Wang C. Liu H. Zhou Q. Liu Q. Guo Y. Peng T. Song J. Zhang J. Chen L. Zhao Y. Zeng Z. Zhou D. X. Nat. Plants. 2018;4:554–563. doi: 10.1038/s41477-018-0214-x. [DOI] [PubMed] [Google Scholar]
- Fang G. Munera D. Friedman D. I. Mandlik A. Chao M. C. Banerjee O. Feng Z. Losic B. Mahajan M. C. Jabado O. J. Deikus G. Clark T. A. Luong K. Murray I. A. Davis B. M. Keren-Paz A. Chess A. Roberts R. J. Korlach J. Turner S. W. Kumar V. Waldor M. K. Schadt E. E. Nat. Biotechnol. 2012;30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C. X. Clark T. A. Lu X. Y. Kislyuk A. Dai Q. Turner S. W. He C. Korlach J. Nat. Methods. 2012;9:75–77. doi: 10.1038/nmeth.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T. A. Murray I. A. Morgan R. D. Kislyuk A. O. Spittle K. E. Boitano M. Fomenkov A. Roberts R. J. Korlach J. Nucleic Acids Res. 2012;40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T. A. Lu X. Luong K. Dai Q. Boitano M. Turner S. W. He C. Korlach J. BMC Biol. 2013;11:4. doi: 10.1186/1741-7007-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brown Z. K. Boulias K. Wang J. Wang S. Y. O'Brown N. M. Hao Z. Shibuya H. Fady P. E. Shi Y. He C. Megason S. G. Liu T. Greer E. L. BMC Genomics. 2019;20:445. doi: 10.1186/s12864-019-5754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]