Abstract
Mutations in ARID1A rank amongst the most common molecular aberrations in human cancer. However, oncogenic consequences of ARID1A mutation in human cells remain poorly defined due to lack of forward genetic models. Here, CRISPR/Cas9-mediated ARID1A knockout in primary TP53−/− human gastric organoids induced morphologic dysplasia, tumorigenicity and mucinous differentiation. Genetic Wnt/β-catenin activation rescued mucinous differentiation, but not hyperproliferation, suggesting alternative pathways of ARID1A KO-mediated transformation. ARID1A mutation induced transcriptional regulatory modules characteristic of MSI and EBV subtype human gastric cancer, including FOXM1-associated mitotic genes and BIRC5/survivin. Convergently, high-throughput compound screening indicated selective vulnerability of ARID1A-deficient organoids to inhibition of BIRC5/survivin, functionally implicating this pathway as an essential mediator of ARID1A KO-dependent early-stage gastric tumorigenesis. Overall, we define distinct pathways downstream of oncogenic ARID1A mutation, with non-essential Wnt-inhibited mucinous differentiation in parallel with essential transcriptional FOXM1/BIRC5-stimulated proliferation, illustrating the general utility of organoid-based forward genetic cancer analysis in human cells.
Keywords: ARID1A, Organoids, CRISPR/Cas9, FOXM1, BIRC5/survivin, WNT, Gastric Cancer
Introduction
Alterations in the epigenetic landscape are a hallmark of cancer(1). The epigenetic state defines the permissible transcriptome as chromatin topology determines responses to oncogenes and tumor suppressors. Thus, chromatin regulators play critical roles in tumorigenesis, and their mutation is now appreciated as a pervasive feature of malignancy. The mammalian SWI/SNF (mSWI/SNF, BAF) chromatin remodeling complex actively remodels chromatin in an ATP-dependent fashion and renders DNA accessible to transcription factors and other DNA binding proteins(2) to govern development, homeostasis and disease(3-5).
ARID1A, also designated BAF250a, encodes a multifunctional BAF complex subunit that targets BAF to AT-rich enhancer DNA sequences, regulates transcription and recruits topoisomerase II to chromatin(6,7). ARID1A mutations rank amongst the most common molecular aberrations in human cancer(8-11) and are frequent in multiple cancer types such as ovarian clear-cell carcinoma (~57%), endometrioid carcinoma (~30%), urothelial carcinoma (~26%), cholangiocarcinoma (~19%), pancreatic ductal adenocarcinoma (~8%), and colorectal carcinomas (~8%)(12). Mutations in ARID1A occur in ~31% of all gastric adenocarcinomas, particularly in microsatellite instability (MSI) and Epstein-Barr virus-associated (EBV) subtypes, but also in the chromosomal instability (CIN) subtype with lower frequency(13-15). ARID1A mutations dysregulate BAF complex-mediated chromatin remodeling since this subunit directly interfaces with DNA and recruits other transcriptional co-activators(16). ARID1A’s function as a global chromatin conformation regulator underlies the pleiotropic effects observed when this gene is disrupted, and renders the study of ARID1A’s role in oncogenesis especially challenging. Transgenic Arid1a knockout mouse models in embryo(17), ovarian(18), colon(19), small intestine(20), endometrium(21), pancreas(22-25), liver(26), and hematopoietic cells(27) have provided tremendous insight into ARID1A-associated tumorigenesis. However, despite these extensive mouse studies, forward genetic human models are crucially needed to elaborate mechanisms of ARID1A-dependent oncogenic transformation in a more clinically relevant context.
Organoid culture is a robust in vitro culture method that recapitulates many essential attributes of primary human tissue including 3-dimensional (3D) structure, multilineage differentiation, signaling nodes, histology, and pathology with high fidelity and thus represents an emerging approach to cancer biology(28). Bridging cell and tissue scales, organoids offer an attractive hybrid between transgenic mouse models and transformed 2D human cancer cell lines that enables an engineered “bottom up” approach to study temporal and sequential oncogenic events and permits the functional validation of oncogenic loci. Successful multi-hit oncogenic transformation of normal wild-type organoids to adenocarcinoma has been achieved by introducing simultaneous oncogenic mutations into tissues such as colon, stomach and pancreas(29-32).
Here, we utilize wild-type human gastric organoids to establish the first forward genetic human ARID1A-deficient oncogenic transformation model, using CRISPR/Cas9-engineered ARID1A depletion alongside mutation of TP53, a co-occurring tumor suppressor. These engineered ARID1A-deficient organoids mirror several clinicopathologic features of ARID1A-mutant gastric cancer. Coupled with a regulatory network-based analysis and high-throughput drug screening, we have leveraged this human organoid model to discover potential mechanisms underlying the role of ARID1A during oncogenic transformation of gastric epithelium.
Results
Establishment of clonal TP53 and TP53/ARID1A knockout human gastric organoid lines
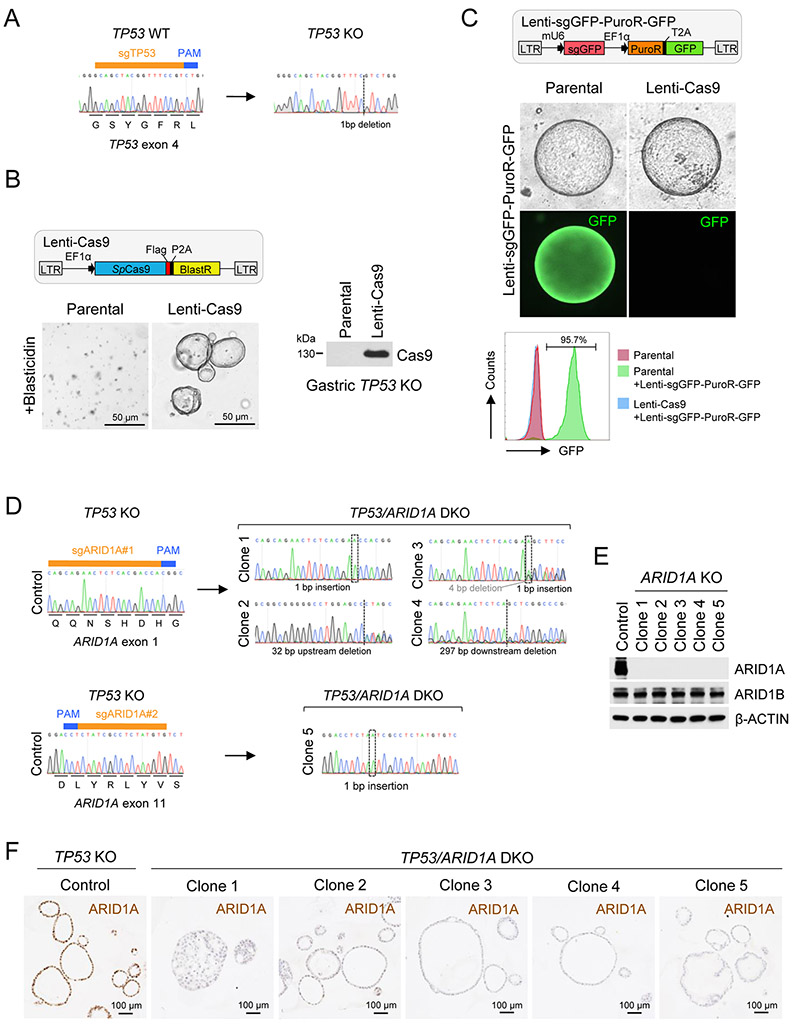
Arid1a is indispensable for stem-cell maintenance and self-renewal, as genetic deletion results in lethal compromise of gastrulation at E6.5, and knockout embryonic stem cells cannot be established(17,20,27,33). Consistent with these observations, using wild-type human gastric corpus organoids(34) from partial gastrectomy obesity surgeries, we could not expand and maintain ARID1A CRISPR/Cas9 KO derivatives in long-term culture. Four independent experiments were attempted, and a total of 12 clonal ARID1A CRISPR/Cas9 KO organoid lines were continuously tracked for at least two weeks. However, all of these ARID1A KO organoid lines eventually failed to grow, leading us to surmise that additional bypass mutation(s) could be needed. Thus, to establish an ARID1A-deficient human gastric cancer transformation model, we first disrupted TP53, the most frequently mutated locus (~49%) in gastric adenocarcinoma(14), by CRISPR/Cas9 into the same wild-type human gastric corpus organoids, followed by secondary CRISPR/Cas9 KO of ARID1A. Transient transfection of an all-in-one construct expressing both Cas9 and sgRNA targeting TP53 exon 4 followed by a recently developed nutlin-3 functional selection(30,31) yielded numerous organoid colonies, whereas no growth was seen in non-transfected cells. After clonal expansion, a nutilin-3-resistant organoid clone harboring a 1 bp cytosine deletion (327delC; TTCCG to TTCG) within TP53 exon 4 was chosen for further analysis (Fig. 1A).
Figure 1. Establishment of clonal TP53/ARID1A knockout human gastric organoid lines.
A, The TP53 indel created by CRISPR/Cas9 cleavage was determined by Sanger sequencing. B, Establishment of a stable Cas9-expressing engineered TP53 KO human gastric organoid line. After antibiotic (Blasticidin) selection, Cas9 expression was confirmed by immunoblot analysis. C, Highly efficient CRISPR/Cas9 cleavage in Cas9-expressing TP53 KO organoids. A lentiviral construct containing a GFP guide RNA targeting the GFP reporter in the same construct was delivered into control TP53 KO and the Cas9-expressing TP53 KO organoids. After antibiotic (Puromycin) selection, GFP-positive cells were quantified by flow cytometry. D, Five different TP53/ARID1A DKO clones were established. ARID1A indels were determined by Sanger sequencing. E, Immunoblot analysis of ARID1A and ARID1B expression. F, IHC staining of ARID1A in TP53 KO versus TP53/ARID1A DKO organoids.
Serial genome editing in primary human organoids to generate sequential oncogenic mutations has been largely restricted by limited absolute knockout efficiency and a paucity of available functional selection strategies(35). To overcome these limitations and introduce inactivating mutations in ARID1A in these newly generated TP53 KO gastric organoids, we utilized a two-vector, sequential lentiviral-based CRISPR/Cas9 system. First, TP53 KO organoids were transfected with a Cas9 construct conferring blasticidin resistance and constitutive Cas9 protein expression was verified (Fig. 1B). Cas9-expressing organoids did not exhibit growth defects, suggesting low Cas9 toxicity after blasticidin selection. To quantify the efficiency of CRISPR/Cas9 cleavage, we delivered a second lentivirus containing a sgRNA targeting the GFP reporter in the same construct and a puromycin resistance gene (Fig. 1C). In the parental TP53 KO organoids, nearly all cells showed GFP expression after puromycin selection. However, in Cas9-expressing TP53 KO organoids, over 95% of cells were GFP-negative as quantified by flow cytometry, indicating highly efficient CRISPR cleavage (Fig. 1C).
We next applied this dual lentiviral system to ARID1A genetic knockout in TP53-null organoids. Of note, CRISPR can be mutagenic by introducing random insertions or deletions (indels) during cleavage, resulting in heterogenous cell populations. To address this potential pitfall and more precisely characterize sequelae of ARID1A loss in gastric tumorigenesis, we established a spectrum of clonal TP53/ARID1A DKO organoid lines by sgRNA targeting of ARID1A exon 1 or exon 11 in a lentiviral vector with BFP reporter. After lentivirus sgRNA-BFP delivery of Cas9-TP53 KO organoids, single dissociated BFP positive cells were sorted into single wells of a 96-well plate, clonally expanded and ARID1A indels at sgRNA-targeted regions were confirmed by Sanger sequencing (Fig. 1D). The corresponding wild-type organoids possessed wild-type TP53 (Fig. 1A) and ARID1A (Fig. 1D) alleles. The loss of ARID1A expression, but not ARID1B, was further confirmed by Western blotting (Fig. 1E) and immunohistochemical (IHC) staining (Fig. 1F). A total of five TP53/ARID1A DKO organoids lines were chosen for this study. In parallel, an empty lentiviral sgRNA-BFP vector was transduced into the same Cas9-TP53 KO organoids, and represented the control. The TP53 KO organoids (control) and TP53/ARID1A DKO organoids (ARID1A KO) were established, grown, maintained and passaged using identical culture conditions throughout this study. We performed whole-genome sequencing of control TP53 KO, TP53/ARID1A DKO clone 3 (indels in ARID1A exon 1), and TP53/ARID1A DKO clone 5 (indels in ARID1A exon 11) at 3 months after ARID1A sgRNA delivery to outline the genetic background of these engineered organoids. The genome of the parental wild-type organoids was used as the reference. As expected, TP53 mutation induced a moderate degree of chromosomal instability in both TP53 KO and TP53/ARID1A DKO organoids (Supplementary Fig. 1A), and a few shared and clonal-specific nonsynonymous mutations were detected (Supplementary Fig. 1B). Importantly, no additional canonical TCGA gastric cancer driver mutations were identified in either TP53 KO or TP53/ARID1A DKO organoids, thus excluding promiscuous alteration of additional oncogenes or tumor suppressors (Supplementary Fig. 1B).
Loss of ARID1A promotes gastric malignancy
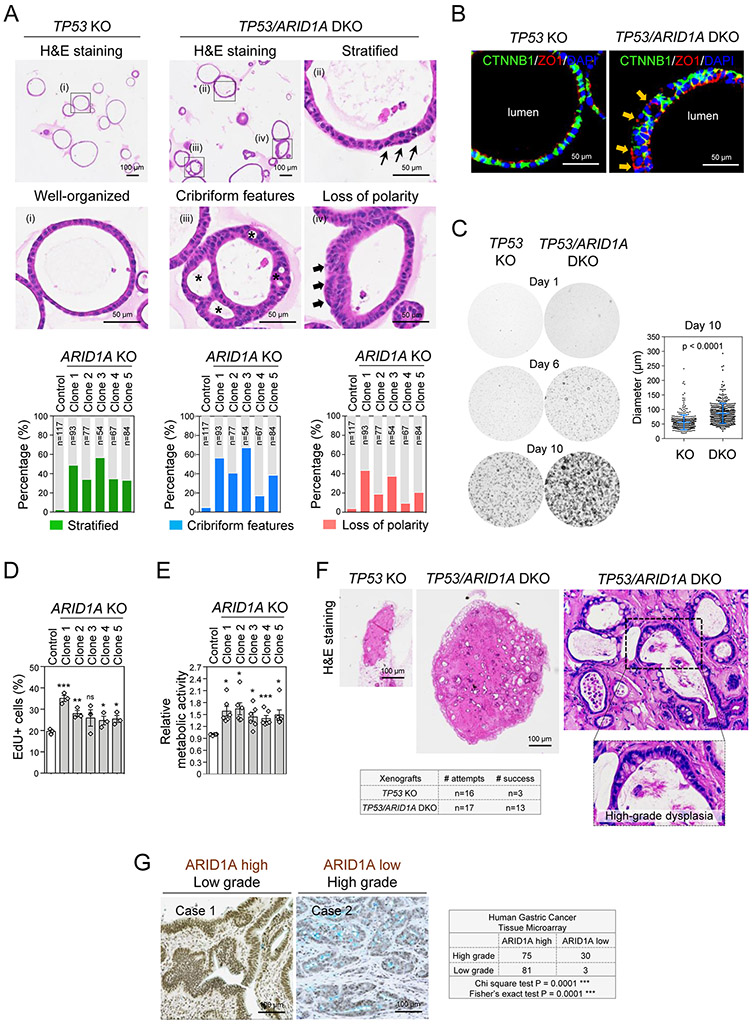
To elucidate consequences of ARID1A loss in gastric tumorigenesis, we initially examined histology of TP53 KO and TP53/ARID1A DKO organoids by hematoxylin and eosin (H&E) staining. TP53 KO organoids harboring control sgRNA predominantly grew as variably-sized acini composed of a single layer of polarized epithelium (Fig. 2A). Cytologically, the cells in TP53 KO organoids were well-organized with an apically oriented cytoplasm and basally placed nucleus, indicating preservation of apicobasal polarity. In contrast, all five TP53/ARID1A DKO organoid lines exhibited different degrees of architectural complexity and cytologic changes characteristic of high-grade dysplasia, including but not limited to cribriform growth, stratification, increased nuclear to cytoplasmic ratios, and nuclear pleomorphism with nuclear membrane irregularities (Fig. 2A). These features were rarely identified in the control TP53 KO organoids. The cribriform features of ARID1A KO organoids resulted in multi-cystic organoids containing several lumina (Fig. 2A), and epithelia were haphazardly arranged with loss of the distinctive apicobasal orientation evident on H&E stained histologic sections in TP53 KO organoids. The latter observations, along epithelial stratification, raise the possibility that cell intrinsic apicobasal polarity is disrupted in ARID1A-deficient organoids (Fig. 2A). Immunofluorescence staining of the apical-specific marker ZO1 further confirmed inappropriate basolateral ZO1 expression facing the extracellular matrix in a subset of TP53/ARID1A DKO organoid cells, suggesting disrupted apicobasal polarity (Fig. 2B). Additionally, TP53/ARID1A DKO organoids exhibited several high-grade dysplasia cytologic features, including nuclear pleomorphism, nuclear membrane irregularities and conspicuous nucleoli (Supplementary Fig. 2)(36,37). Functionally, TP53/ARID1A DKO organoid lines proliferated more rapidly than TP53 KO organoids, resulting in the larger size (Fig. 2C), as well as increased EdU-positive cells (Fig. 2D), revealing a growth advantage conferred by ARID1A loss. Consistent with these results, compared to TP53 KO organoids, TP53/ARID1A DKO organoids exhibited higher metabolic activity (Fig. 2E). Subcutaneous xenografts of TP53 KO organoids showed poor in vivo engraftment and diminutive outgrowth (n=16, 18.75% success rate); however, TP53/ARID1A DKO organoids engrafted at a significantly greater rate (n=17, 76.47% success rate) and formed larger masses (Fig. 2F). Of note, TP53/ARID1A DKO xenografts in vivo also reflected high-grade dysplasia (Fig. 2F). Taken together, these results suggested that ARID1A mutation morphologically and functionally enhances tumorigenesis in primary human gastric organoids. Additionally, our review of histopathology and immunohistochemical ARID1A expression in a gastric cancer tissue microarray of 197 patients from Stanford Hospital indicated a significant inverse association between ARID1A staining and tumor grade (Fig. 2G).
Figure 2. CRISPR KO of ARID1A promotes gastric malignancy.
A, TP53 KO (control) organoids were typically well-organized morphologically; however, TP53/ARID1A DKO organoids exhibited different degrees of architectural complexity. H&E staining. Quantitation revealed increased epithelial stratification (green bar), structural complexity (blue bar), and loss of polarity (red bar) in all five TP53/ARID1A DKO clones. B, Immunofluorescence staining of the apical-specific marker ZO1 (red) showed disruptions in apicobasal polarity in a subset of TP53/ARID1A DKO organoid cells. The arrow (orange) indicates loss of polarity with inappropriate basolateral ZO1 expression. Cell membrane was stained with CTNNB1 (green). Nuclei were stained with DAPI (blue). C, ARID1A-deficient organoids exhibit hyperproliferation. TP53 KO and TP53/ARID1A DKO organoids were grown from 20,000 single FACS-sorted BFP+ cells. Brightfield images were taken after cell sorting. Quantification of organoid size is shown (n=400 per group). D, Quantification of EdU-positive proliferating cells in TP53 KO and TP53/ARID1A DKO organoids from independent experiments (N=3) at day 6 after passage. E, Quantification of metabolic activity from independent experiments (N=6) was determined by Alamar blue assay at day 12 after passage. Relative metabolic activity was normalized to TP53 KO organoids (Control). Dots indicate independent experiments. The horizontal bar indicates mean. The error bar represents SEM. *P<0.05, ***P<0.005. ns, not significant. F, ARID1A-deficient organoids exhibited efficient in vivo tumor formation upon subcutaneous xenografting into NSG mice. TP53/ARID1A DKO xenografts formed larger tumors compare with TP53 KO xenografts. H&E staining. G, A significant negative correlation between ARID1A expression and tumor grade was identified in a human gastric cancer tissue microarray (total 197 patients). ARID1A expression was assessed by IHC.
Loss of ARID1A induces mucinous metaplasia
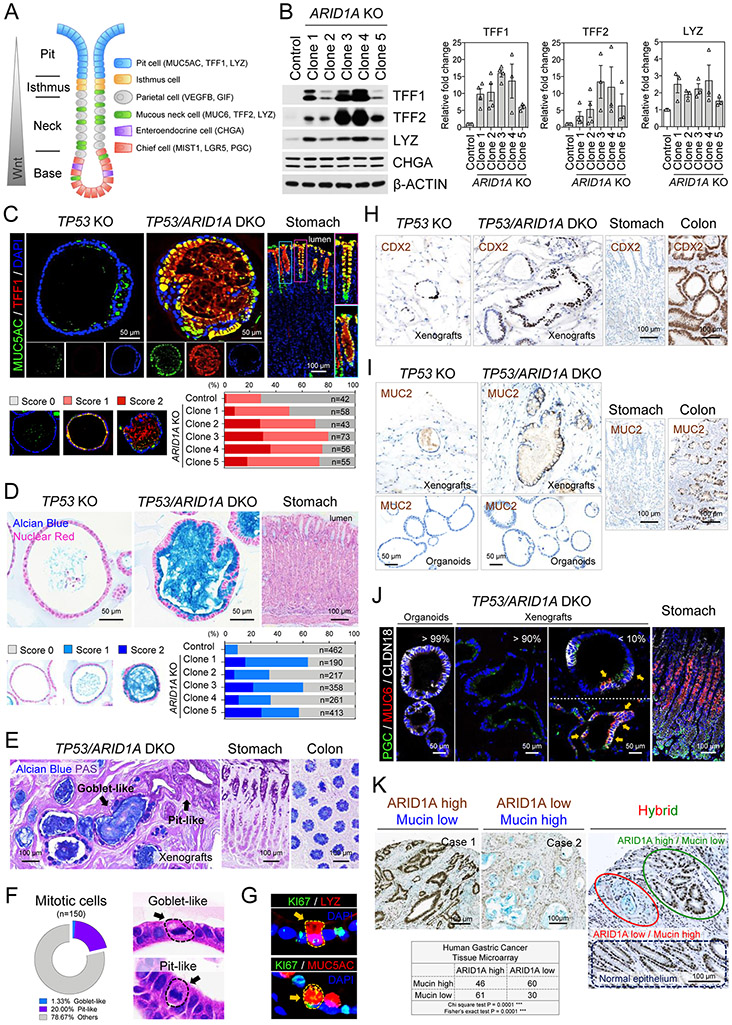
Precancerous transformation of gastric epithelial cells is incited by alterations in genes involved in lineage differentiation and stem cell activity(38). Human gastric homeostasis is maintained, in part, by a gradient of canonical Wnt/β-catenin activity generated from the gland base, where the chief cells reside, extending to mucin-producing populations such as neck (TFF2+, LYZ+) and upper gland pit cells (TFF1+, MUC5AC+)(39,40). To determine if ARID1A loss altered organoid differentiation, we assessed several lineage-specific markers (Fig. 3A). In comparison to TP53 KO organoids, TP53/ARID1A DKO organoids upregulated TFF1, TFF2 and LYZ but the enteroendocrine marker CHGA was unaltered (Fig. 3B). We further confirmed significantly up-regulation of additional pit cell markers, GKN1 and GKN2, in TP53/ARID1A DKO organoids (Supplementary Fig. 3A).
Figure 3. ARID1A knockout induces mucinous metaplasia.
A, Schematic illustration of gastric epithelium. Different cell lineages and specific lineage markers are indicated. B, Western blot of mucin-producing pit cell and mucous neck cell markers, TFF1, TFF2 and LYZ, reveals upregulation in TP53/ARID1A DKO organoids. Quantification of expression from independent experiments (N>3) was shown. Dots indicate independent experiments. C, Immunofluorescence staining of MUC5AC (green) and TFF1 (red) in engineered organoids and the donor primary gastric tissues. Nuclei were stained with DAPI (blue). Quantification of MUC5AC-positive organoids is shown. D, Mucin production in engineered organoids and donor primary gastric tissues detected by Alcian blue staining. Nuclei were counterstained by nuclear fast red. Quantification of Alcian blue-positive organoids indicate increased mucin in all five TP53/ARID1A DKO organoids lines. E, TP53/ARID1A DKO xenografts in a subcutaneously xenografted NSG mice retain their mucin-secreting phenotype in vivo. Alcian blue and PAS staining. Goblet-like (Alcian blue -positive) and pit-like (PAS positive) cells were indicated. F, Quantification of mitotic cells. Goblet-like and pit-like cells with mitotic figures were shown. H&E staining. G, Immunofluorescence staining of TP53/ARID1A DKO organoids showing LYZ-positive (red) or MUC5-positive (red) proliferating cells (KI67+, green). H, IHC staining of CDX2 in xenografts and the donor primary gastric tissues. Colon tissues were used as positive control. I, IHC staining of MUC2 in organoids, xenografts and the donor primary gastric tissues. Colon tissues were used as positive control. J, Immunofluorescence staining of CLDN18 (white), MUC6 (red) and PGC (green) in TP53/ARID1A DKO organoids, xenografts and the donor primary gastric tissues. Cells within SPEM features (MUC6 and PGC double positive) are marked by arrows. K, A significant negative correlation between ARID1A (brown) IHC expression and mucin (blue, Alcian blue) production was identified in a human gastric cancer tissue microarray (total 197 patients).
Next, we performed TFF1, MUC5AC, TFF2 and LYZ immunofluorescence staining of engineered organoids and the original cognate donor primary gastric tissues. In primary healthy tissues, TFF1 and MUC5AC were specifically expressed in the pit domain at gland tops (Fig. 3C). In addition, TFF2 specifically marked mucous neck cells (Supplementary Fig. 3B) and LYZ labeled pit cells, with additional scattered positivity in gland bases (Supplementary Fig. 3C). TP53 KO organoids expressed very low levels of TFF1 and only sporadically expressed MUC5AC, TFF2 and LYZ. In contrast, TP53/ARID1A DKO organoids profoundly induced TFF1, MUC5AC, TFF2, and LYZ, consistent with acquisition of a mucinous phenotype (Fig. 3C and Supplementary Fig. 3B-C). Chief cell mRNAs LGR5, MIST1, PGC, and CPB1 were down-regulated in TP53/ARID1A DKO organoids (Supplementary Fig. 3D).
Gastrointestinal cell fate decisions can increase mucin production in reaction to injury, a phenomenon termed mucous cell metaplasia(41,42). During metaplasia the epithelium is repopulated by cell lineages non-endemic to gastric tissues. Importantly, metaplastic transformation occurs in the earliest stages of progression of precancerous lesions to gastric cancer. We tested if ARID1A loss induced mucous cell metaplasia by Alcian blue staining, which marks acidic mucins in mucinous cancers but not normal stomach, and further does not stain pH-neutral mucins in healthy gastric epithelium (Fig. 3D). Accordingly, Alcian blue-positive cells were significantly increased in all five TP53/ARID1A DKO lines, versus TP53 KO organoids (Fig. 3D). In addition, in vivo xenografts from TP53/ARID1A DKO organoids retained the mucinous phenotype with Periodic Acid-Schiff (PAS)-positive gastric pit cell-like and Alcian blue-positive intestinal goblet cell-like dysplastic cells (Fig. 3E).
Interestingly, some Alcian blue and PAS double-positive mucin lakes were rimmed by Alcian blue-negative pit-like cells, suggesting an intermediate differentiation state between gastric-type and intestinal-type mucin-producing cells (Supplementary Fig. 3E). ARID1A deficient mucinous organoid cells were indeed proliferative, as 21.3% of mitotic cells exhibited mucinous histology, which could be subdivided into goblet-like (1.3%) and pit-like (20%) cells (Fig. 3F). Moreover, KI67-positive proliferating mucinous cells were identified (Fig. 3G). To further investigate the gastric versus intestinal mucinous state in organoid xenografts, we performed IHC staining of CDX2, an intestinal epithelium specific transcription factor. Both TP53 KO and TP53/ARID1A DKO xenografts exhibited clusters of CDX2-positive cells, indicating foci of intestinal metaplasia (IM) in vivo (Fig. 3H). Interestingly, intestinal goblet cell-like MUC2+ cells were exclusively identified in TP53/ARID1A DKO xenografts, but not in TP53/ARID1A DKO organoids (Fig. 3I), suggesting potential host tumor microenvironmental regulation of the IM phenotype. Accordingly, the gastric epithelium-specific tight junction protein CLDN18, was dramatically decreased in TP53/ARID1A DKO xenografts in vivo, versus TP53/ARID1A DKO organoids in vitro (Fig. 3J). Of note, a small proportion of CLDN18-positive TP53/ARID1A DKO xenografts (<10%) resembled spasmolytic polypeptide-expressing metaplasia (SPEM)(42), a metaplastic mucous cell lineage, by co-expressing the chief cell digestive enzyme PGC and mucous neck cell specific marker MUC6 (Fig. 3J).
We further confirmed these findings in gastric cancer patients by demonstrating a significant inverse correlation between ARID1A expression and mucin production by simultaneous ARID1A and Alcian blue staining of a 197-patient gastric cancer tissue microarray (Fig. 3K). Of note, in a few cases of heterogeneous ARID1A tumor expression, mucin was present in association with tumor areas having low, but not high ARID1A expression (Fig. 3K), again reiterating the mucous cell metaplasia associated with ARID1A loss.
Loss of ARID1A inhibits canonical Wnt/β-catenin activity
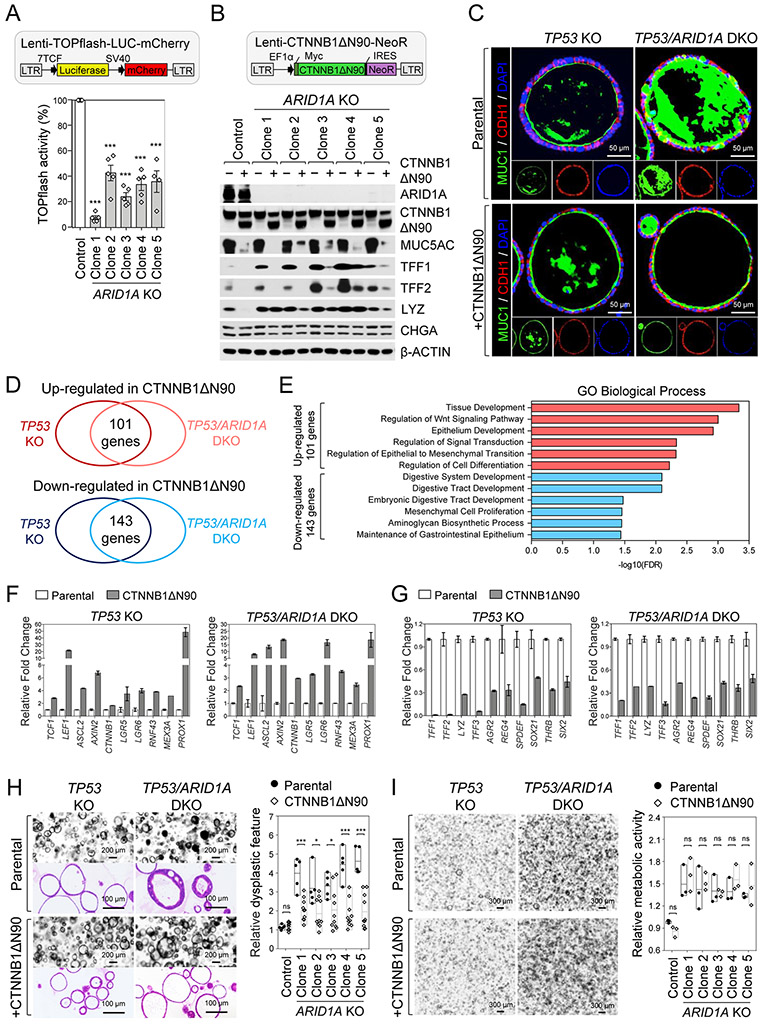
Wnt activity is inversely correlated with gastric mucinous differentiation since canonical Wnt signaling is lowest in the mucinous pit cell-containing regions occupying the apical-most domains of gastric glands(43) and withdrawal of Wnt and R-spondin from human gastric organoids directs cell fate from gland-type to mucin-expressing pit lineages(34). We thus hypothesized that organoid ARID1A KO induced the mucin-producing pit-like cell phenotype by impairing Wnt/β-catenin signaling (Fig. 3). This was directly tested by delivering a Wnt-activated TOPflash luciferase construct containing an mCherry reporter by lentiviral-based transduction into our engineered organoids. An equivalent number of mCherry-positive single cells were sorted from TP53 KO and TP53/ARID1A DKO organoids followed by quantification of luciferase activity. Consistent with this model, Wnt/β-catenin-induced reporter activity was significantly reduced in all five ARID1A-deficient lines (Fig. 4A) despite their increased proliferation (Fig. 2C-D).
Figure 4. Loss of ARID1A inhibits canonical Wnt/β-catenin activity.
A, Wnt/β-catenin-induced activity was decreased in TP53/ARID1A DKO organoids infected by lentivirus containing TOPflash Wnt reporter and mCherry followed by luciferase assay on 20,000 sorted mCherry-positive cells. Quantification of luciferase activity from independent experiments (N=5) is shown. Luciferase activity was normalized to TP53 KO organoids (Control). B, The mucin-producing phenotype was genetically rescued by lentiviral expression of an N-terminal truncated gain-of-function β-catenin mutant (CTNNB1ΔN90). After virus transduction and antibiotic (Neomycin) selection, protein expression in the engineered organoids was analyzed by Western blot as indicated. C, Immunofluorescence staining of apically-restricted transmembrane MUC1 (green) and membrane protein CDH1 (red) demonstrates that CTNNB1ΔN90 reduces mucin production and architectural complexity of TP53/ARID1A DKO organoids. D, Venn diagram indicates overlap of genes that are significantly increased (101 genes) or decreased (143 genes) at least 2-fold in organoids with CTNNB1ΔN90 alleles. E, Gene ontology analysis identified top key terms significantly associated with transcriptional profiles in CTNNB1ΔN90 organoids. F, Wnt/β-catenin target genes were upregulated in CTNNB1ΔN90 organoids. G, Gastric mucous cell and intestinal goblet cell markers were significantly downregulated in CTNNB1ΔN90 organoids. The expression of transcription factors SPDEF, SOX21, THRB, SIX2 was shown. H, Phenotypic changes induced by ARID1A loss were partially restored by lentivirus CTNNB1ΔN90. H&E staining and brightfield images. Relative stratification was quantified by counting the number of cells per length of perimeter of individual organoids. I, Constitutive Wnt signaling activation by CTNNB1ΔN90 did not rescue ARID1A KO-mediated proliferation. Single cells (20,000/40 μL Matrigel) from TP53 KO and TP53/ARID1A DKO organoids with and without lentivirus CTNNB1ΔN90 underwent Alamar blue quantification of cell viability at day 12. Relative cell viability was normalized to control TP53 KO organoids (Control). Three independent experiments (N=3) were performed. In A, H and I, dots indicate independent experiments, horizontal bars indicate mean and error bars represent SEM. *P<0.05, ***P<0.005. ns, not significant.
To determine if the mucinous metaplasia induced by ARID1A loss could be rescued by constitutively activated Wnt signaling, we transduced an N-terminal truncated gain-of-function β-catenin (CTNNB1ΔN90) lentivirus bearing neomycin resistance into TP53 KO and TP53/ARID1A DKO organoids, yielding CTNNB1ΔN90/TP53 KO and CTNNB1ΔN90/TP53/ARID1A DKO organoid lines. The gain-of-function β-catenin mutant strongly induced TOPflash reporter activity (Supplementary Fig. 4A) and extinguished the ectopic MUC5AC, TFF1 and TFF2 expression in TP53/ARID1A DKO organoids, while LYZ was relatively unaffected (Fig. 4B). Similarly, induction of MUC1, an apically-restricted, gastric cancer-associated transmembrane mucin(44), in TP53/ARID1A DKO organoids was profoundly reversed by CTNNB1ΔN90, reverting these organoids to a non-mucinous phenotype with re-establishment of apicobasal polarity indicated by uniformly apical MUC1 expression (Fig. 4C). To delineate the inhibitory effect of extracellular Wnt and R-Spondin on the mucin-producing phenotype, organoids were grown for 9 days in the fully supplemented culture medium (WENR) followed by withdrawal of Wnt and R-Spondin from the medium (EN) for an additional 5 days to induce mucous cell differentiation. The expression of TFF1 and TFF2, but not LYZ and CHGA were increased in both TP53 KO and TP53/ARID1A DKO organoids in the absence of Wnt and R-Spondin, suggesting withdrawal of Wnt stimulation is sufficient to induce mucinous differentiation (Supplementary Fig. 4B). Taken together, these results suggested that the mucin-producing phenotype of TP53/ARID1A DKO organoids results from inhibition of Wnt/β-catenin activity, indicating a redirection of gland- to pit-like cell fate determination.
To mechanistically investigate ARID1A mutation-repressed canonical Wnt/β-catenin signaling and mucous cell differentiation, we studied Wnt/β-catenin-regulated transcripts upon CTNNB1ΔN90 rescue of either TP53 KO or TP53/ARID1A DKO organoids. (Fig. 4D and Supplementary Table 1). Gene Ontology (GO) analysis of up-regulated genes showed enrichment of biological processes that are associated with Wnt activation, such as tissue development, regulation of Wnt signaling and epithelial to mesenchymal transition (EMT) (Fig. 4E). As expected, Wnt/β-catenin target genes such as TCF1, LEF1, ASCL2, AXIN2, CTNNB1, LGR5, LGR6 and RNF43 were induced along with MEX3A(45), and PROX1(46) which mark injury-inducible intestinal stem cells (Fig. 4F). In contrast, down-regulated gene GO terms implicated digestive tract development (Fig. 4E). Consistent with CTNNB1ΔN90 abrogation of the mucinous phenotype, markers of gastric pit cells (TFF1, LYZ), gastric mucous neck cells (TFF2, LYZ, AGR2), and intestinal goblet cells (TFF3, AGR2, REG4) were significantly decreased (Fig. 4F) alongside transcription factors SPDEF, SOX21, THRB and SIX2 (Fig. 4G). Notably, SPDEF is a master transcription factor regulating mucin-producing cell differentiation and maturation across many tissue types, such as gastric mucous neck cells(47) and intestinal goblet cells(48,49). On balance, these results suggested ARID1A- and Wnt-dependent control of mucous cell differentiation via SPDEF regulation.
In addition to the mucinous phenotype, CTNNB1ΔN90/TP53/ARID1A DKO organoids rescued many dysplastic features characteristic of TP53/ARID1A DKO organoids, with reduced epithelial stratification and architectural complexity (Fig. 4H). We next examined if the hyperproliferation phenotype of TP53/ARID1A DKO organoids (Fig. 2C) could be reverted by activated Wnt signaling. However, CTNNB1ΔN90 notably did not rescue the elevated cell proliferation of any of the five clonal TP53/ARID1A DKO organoid lines (Fig. 4I). These results thus dissociated the non-essential Wnt repression-dependent mucous metaplasia from alternative undefined yet essential mechanisms governing ARID1A loss-associated hyperproliferation.
ARID1A loss-associated gene regulatory modules recapitulates TCGA gastric cancers
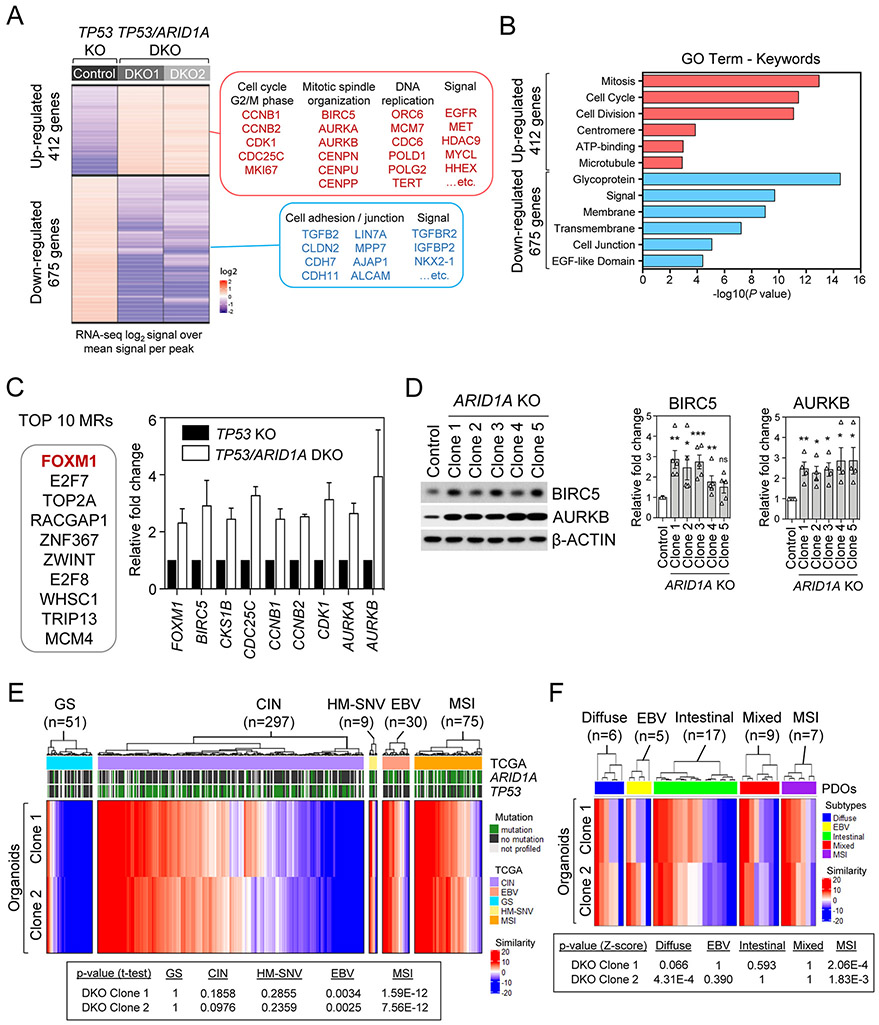
To identify the critical Wnt-independent biological processes governing the hyperproliferation associated with ARID1A loss, we investigated ARID1A-associated transcripts by bulk RNA-sequencing (RNA-seq) in the control TP53 KO and two of the TP53/ARID1A DKO organoid lines. Compared to TP53 KO organoids, the TP53/ARID1A DKO biological replicates contained 1,087 differentially expressed genes that were consistently up-regulated (472 genes) or down-regulated (675 genes) (Fig. 5A and Supplementary Table 2). GO enrichment analysis of up-regulated genes in ARID1A-deficient organoids indicated several key biological processes including regulation of mitotic cell cycle, cell division, chromatin segregation, and cytoskeletal organization (Fig. 5B and Supplementary Fig. 5). On the other hand, the top GO terms of the down-regulated genes in ARID1A-deficient cells included cell morphogenesis, nervous system development, cell differentiation, cell adhesion, cell migration, and negative regulation of cellular response to growth factor stimulus (Fig. 5B and Supplementary Fig. 5). These findings were in agreement with our conclusions that ARID1A loss altered cell proliferation and differentiation in TP53/ARID1A DKO organoids (Figs. 2, 3). Of note, the abnormal mitotic and chromatin segregation signatures suggested that ARID1A loss might be implicated in chromosome instability, consistent with our prior studies(7).
Figure 5. ARID1A loss-associated gene master regulatory modules identify a FOXM1/BIRC5 node and recapitulate TCGA MSI and EBV human gastric cancers.
A, Heatmap of significant differentially expressed genes with at least 2-fold change in each TP53/ARID1A DKO lines, compared with TP53 KO control line. A total of 412 up-regulated genes and 675 down-regulated genes were identified. Selected genes and signaling pathways are listed. B, Gene ontology analysis identified top key terms significantly associated with transcriptional profiles in TP53/ARID1A DKO organoids. C, Top 10 master regulators from ARACNe and VIPER prediction that were activated in TP53/ARID1A DKO organoids versus control TP53 KO are reported. Several FOXM1 targets, including BIRC5, CKS1B, CDC25C, CCNB1, CCNB2, CDK1, AURKA and AURKB were significantly upregulated in ARID1A-deficient cells. D, Western immunoblotting analysis demonstrated that FOXM1 targets, BIRC5 and AURKB, were upregulated in TP53/ARID1A DKO organoids. Quantification of BIRC5 and AURKB expression from independent experiments (N>3) was shown. Dots indicate independent experiments. The horizontal bar indicates mean. The error bar represents SEM. E, Comparison of master transcriptional regulators in ARID1A KO organoids to TCGA STAD gastric cancer patient cases indicated significant similarities between organoids and TCGA MSI and EBV subtypes. The p-value computed by t-test (one sample) with the alternative hypothesis of true mean of the similarity score is greater than zero. Red and blue colors indicate high and low similarity concurrence, respectively. F, Comparison of master transcriptional regulators in ARID1A-deficient organoids to gastric cancer patient-derived organoids (PDOs) indicated significant similarities between engineered TP53/ARID1A DKO organoids and MSI subtype PDOs.
To gain deeper insights into how ARID1A loss influences gene regulatory architecture, we performed master regulator (MR) analysis using the VIPER(50) algorithm to elucidate ARID1A-regulated gene hierarchies(51). Akin to highly multiplexed gene reporter assays, VIPER infers the activity of 2,782 regulator proteins based on expression of their positively regulated and repressed transcriptional targets. Transcriptional targets were identified by analyzing a set of 200 TCGA stomach adenocarcinoma (STAD) gene expression profiles(14) using the ARACNe(52) algorithm. VIPER analysis identified several MRs representing candidate effector proteins that were significantly associated with ARID1A loss in two independent TP53/ARID1A DKO organoid lines (Fig. 5C and Supplementary Table 3). FOXM1, a classical proliferation-associated transcription factor that is intimately involved in tumorigenesis(53), was listed as the top-ranked MR that was differentially enriched in both TP53/ARID1A DKO organoid biological replicates versus the control TP53 KO organoids. Consistent with this result, several FOXM1 targets, such as BIRC5, CKS1B, CDC25C, CCNB1, CCNB2, CDK1, AURKA and AURKB, were simultaneously upregulated in parallel with FOXM1 in ARID1A-deficient organoids (Fig. 5C and Supplementary Table 4). The upregulation of the FOXM1 targets BIRC5 and AURKB was further confirmed by Western blotting (Fig. 5D). Notably, the global MR profile of ARID1A-deficient organoids revealed strong overlap with MRs independently identified in TCGA STAD gastric cancers, with particularly significant similarity to STAD MSI (p<7.56E-12) and EBV (p<0.03) clusters where ARID1A mutations are highly enriched(14,15) but not GS (p>1), CIN (p>0.97), and HM-SNV (p>0.23) subtypes (Fig. 5E). In addition, compared to a gastric cancer patient-derived organoid (PDO) data set that was established in previous studies(54), the MR profile of ARID1A-deficient organoids again exhibited significant similarity to the MSI subtype PDOs (Fig. 5F).
ARID1A deletion confers therapeutic vulnerability to Survivin inhibition
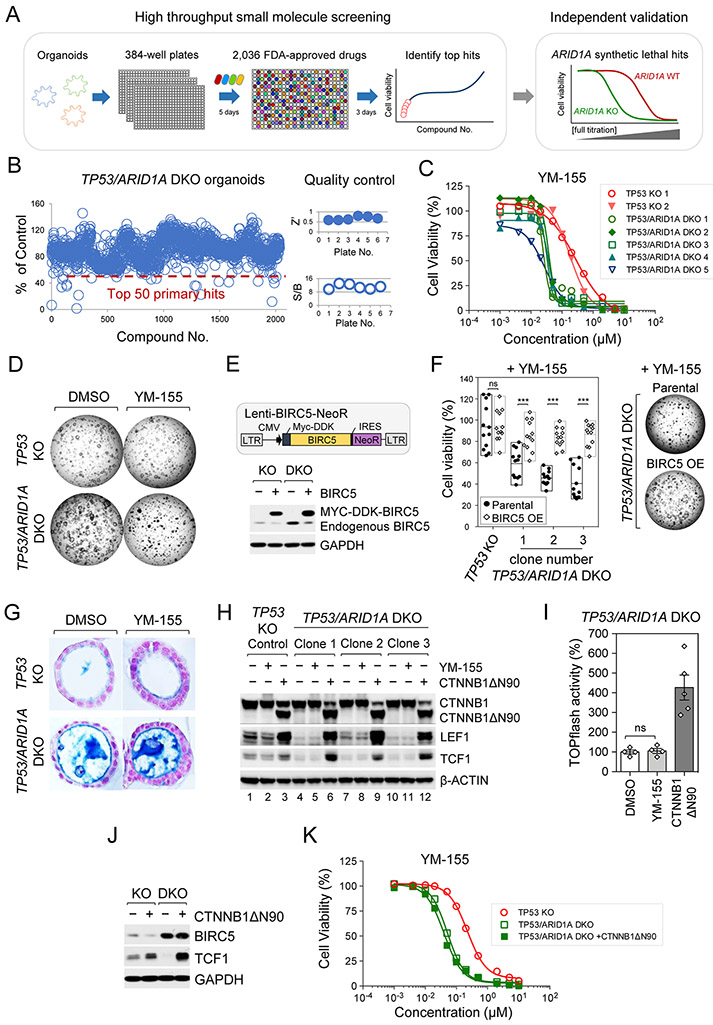
A potential advantage of the use of isogenic paired TP53 KO and TP53/ARID1A DKO organoids engineered from non-neoplastic gastric tissue is a reduced likelihood of simultaneous confounding co-occurring mutations that are common to transformed cancer cell lines, as confirmed by lack of driver alterations upon whole genome sequencing (Supplementary Fig. 1). Thus, the syngeneic and low background somatic mutational burden of these engineered organoids provided a unique opportunity to study ARID1A growth dependencies in a system having reduced interference from modifier loci. Thus, we tested ARID1A-specific growth dependencies by high-throughput small molecule screening of an FDA-approved and bioactive chemical library (2,036 compounds) in TP53/ARID1A DKO versus control TP53 organoid lines (Fig. 6A). TP53/ARID1A DKO organoids were dissociated into smaller clusters, re-plated into 384-well plates, cultured for 5 days followed by drug treatment and cell viability was quantified after 3 additional days (Fig. 6A). Notably, this screening system exhibited robust assay performance, with signal-to-background (S/B) ratio > 8 and Z’ > 0.5 (Fig. 6B). To discover compounds exhibiting selective synthetic lethality with ARID1A deficiency, we performed 12-point concentration counter-screening in the control TP53 KO versus two additional TP53/ARID1A DKO lines for the top 50 hits from the initial primary DKO organoid screening. Among these, several candidates such as YM-155, BMS-526924, HS-173 and Torin-2 selectively inhibited proliferation of TP53/ARID1A DKO versus TP53 KO organoids, whereas many hits such as AP-26113 showed no obvious differences (Supplementary Fig. 6A).
Figure 6. ARID1A deletion confers therapeutic vulnerability to BIRC5/survivin inhibition.
A, High-throughput small molecule and bioactive screening in engineered organoids. B, Histogram of high-throughput screening of an FDA-approved small molecule compound library (2,036 compounds) in TP53/ARID1A DKO organoids. Organoids were dissociated into smaller clusters, re-plated into 384-well plates, and cultured for 5 days before drug treatment. Cell viability was quantified 3 days after compound treatment. The signal-to-background (S/B) ratio and Z’ indicated robust assay performance. The top 50 primary hits are indicated below the dashed red line and were selected for counter screening. C, YM-155, a BIRC5/survivin inhibitor, exhibited ARID1A-specific synthetic lethality. Fully-titrated counter screening for YM-155 was performed in two TP53 KO lines versus five additional TP53/ARID1A DKO clones. D, Brightfield images after organoid treatment with YM-155 (IC50, 0.03 μM) for 3 days. YM-155 selectively inhibited growth of TP53/ARID1A DKO but not TP53 KO organoids. E, Establishment of stable BIRC5 over-expressing BIRC5/TP53 KO and BIRC5/TP53/ARID1A DKO organoid lines. After antibiotic (Neomycin) selection, BIRC5 expression was confirmed by immunoblot analysis. F, Constitutive expression of BIRC5 rescued the YM-155-associated sensitivity in TP53/ARID1A DKO organoids. Organoids were treated with YM-155 (IC50, 0.03 μM) for 3 days. Three independent experiments (N=3) were performed. G, YM-155 treatment did not alter mucin production in TP53/ARID1A DKO organoids. Alcian blue staining. Nuclei were counterstained by nuclear fast red. H, Western immunoblotting analysis indicated that a gain-of-function β-catenin mutant (CTNNB1ΔN90) was sufficient to induce Wnt/β-catenin targets, LEF1 and TCF1; however, YM-155 treatment did not affect Wnt/β-catenin activity. I, YM-155 IC50 treatment (0.03 μM) did not affect Wnt/β-catenin-induced TOPflash reporter activity. Quantification of luciferase activity from independent experiments (N=4) is shown. Luciferase activity was normalized to DMSO treatment. A gain-of-function β-catenin mutant (CTNNB1ΔN90) organoid line was used as the positive control. J, Lentiviral expression of CTNNB1ΔN90 did not rescue the BIRC5 expression, Western blot. K, Lentiviral expression of CTNNB1ΔN90 did not rescue the selective YM-155 sensitivity of ARID1A-deficient cells. Fully-titrated YM-155 treatment was performed in TP53 KO versus TP53/ARID1A DKO and TP53/ARID1A DKO plus CTNNB1ΔN90 organoid clones. Alamar blue, three independent experiments (N=3).
We then performed secondary counter-screening with repurchased compounds to repeat and further confirm enhanced sensitivity in TP53/ARID1A DKO organoids. While some variability in the magnitude of sensitivities were observed, the results of the secondary confirmatory assay were generally consistent with our primary screen, yielding 14 candidate compounds that selectively enhanced killing of ARID1A-mutant organoids (Supplementary Fig. 6B). Consistent with previous studies of ARID1A-mutated cancer cells, engineered ARID1A-deficient gastric organoids were selectively sensitive to histone deacetylase (HDAC) inhibitors(55,56) and PI3K/AKT inhibitors(57,58) (Supplementary Fig. 6B). Among these compounds, ARID1A-deficient gastric organoids were also sensitive to YM-155, a small molecule inhibitor of BIRC5/survivin(59), a member of the inhibitor of apoptosis (IAP) family, which inhibits caspase-mediated apoptosis(60) and controls mitotic spindle dynamics and chromosome segregation(61) (Supplementary Fig. 6A-B). We additionally confirmed the potent YM-155 repression of BIRC5 protein in TP53 KO and TP53/ARID1A DKO organoids (Supplementary Fig. 6C). Crucially, YM-155 exhibited selective lethality with ARID1A mutation consistently across all five TP53/ARID1A DKO lines (average IC50=0.03 μM) versus the two TP53 KO lines (average IC50=0.23 μM) (Fig. 6C-D).
We further evaluated the therapeutic effect of YM-155 in conventional 2D gastric cancer cell lines, as opposed to oncogene-engineered organoids. Six isogenic pairs of ARID1A wild-type and mutant cancer cell lines were generated by CRISPR/Cas9, and sensitivities to YM-155 were compared. In contrast to 3D engineered organoids, ARID1A KO 2D gastric cancer cell lines did not exhibit selective sensitivity to YM-155 (Supplementary Fig. 7). These results indicated that highly transformed gastric cancer cell lines are less dependent on BIRC5/survivin after ARID1A loss than our DKO organoids, which appear to harbor only TP53 and ARID1A oncogenic driver mutations and thus model early gastric cancer (Supplementary Fig. 1A-B).
Rescue and functional independence of ARID1A KO-regulated BIRC5/survivin and Wnt pathways
To test if constitutive expression of BIRC5 was sufficient to rescue YM-155-associated ARID1A synthetic lethality, we lentivirally overexpressed MYC-DDK-tagged BIRC5 in TP53 KO versus TP53/ARID1A DKO organoid lines (Fig. 6E). As expected, single TP53 KO control organoids exhibited YM-155 insensitivity at the IC50 of 0.03 μM for TP53/ARID1A DKO organoids, which was not altered by BIRC5 overexpression. However, the YM-155 hypersensitivity of multiple independent TP53/ARID1A DKO organoid lines was significantly rescued by BIRC5 overexpression, which additionally confirmed the specificity of YM-155 for BIRC5 (Fig. 6F). This unexpected convergence between the MR analysis, in which the mostly highly ranked hit was a ARID1A KO-induced FOXM1→BIRC5/survivin regulatory node with concurrent upregulation of FOXM1 mRNA and BIRC5 mRNA and protein (Fig. 5), and the small molecule screen, revealing selective sensitivity of ARID1A KO organoids to the BIRC5/survivin inhibitor YM-155 (Fig. 6), functionally implicated FOXM1→BIRC5/survivin as an essential pathway mediating hyperproliferation following ARID1A loss. Consistent with these findings in the organoids, BIRC5 expression was significantly higher in TCGA STAD patients harboring ARID1A mutations (Supplementary Table 5).
We then probed the functional independence of the ARID1A KO-induced, YM-155-senstive, FOXM1→BIRC5/survivin essential proliferation pathway, as distinct from the non-essential Wnt-regulated mucinous differentiation pathway. Importantly, YM-155 did not inhibit Wnt-dependent mucous metaplasia in ARID1A-deficient organoids (Fig. 6G). LEF1 and TCF1 are two Wnt/β-catenin targets that are robustly induced by the CTNNB1ΔN90 gain-of-function β-catenin mutant (Fig. 6H, lane 1 versus lane 3). As expected, LEF1 and TCF1 proteins were decreased in TP53/ARID1A DKO organoids having impaired Wnt signaling, versus control TP53 organoid lines (Fig. 6H, lane 1 versus lanes 4, 7 and 10). However, YM-155 did not revert the ARID1A KO-associated decrease in LEF1 or TCF1 protein (Fig. 6H, lanes 1 versus 2, 4 versus 5, 7 versus 8, 10 versus 11), indicating that YM-155 did not affect Wnt/β-catenin activity. Consistent with these observations, Wnt/β-catenin-induced TOPflash reporter activity was also not altered by YM-155 treatment (Fig. 6I). Conversely, CTNNB1ΔN90 Wnt pathway activation did not rescue the expression of BIRC5 (Fig. 6J), or the selective YM-155 proliferation sensitivity of ARID1A-deficient organoids (Fig. 6K) despite potently reversing the mucinous metaplasia phenotype (Fig. 4B-C). In total, these selective perturbation results confirmed the independent functionality of the ARID1A KO-induced Wnt/mucinous metaplasia versus FOXM1→BIRC5/survivin-mediated proliferation pathways.
Discussion
Primary human organoids have proven to be invaluable models of tumorigenesis(28). Organoids mimic oncogenic transformation on a collective tissue scale and accurately replicate the in vivo biology of their original native tissues. Coupled with contemporary experimental methods, organoid systems provide enormous experimental flexibility and capacity for studying molecular mechanisms of gene function in human cells. CRISPR/Cas9 gene editing of primary human organoids from various tissues including colon(30,31,62), stomach(32), pancreas(63,64) , breast(65) and liver(66) has contributed tremendous mechanistic insight into the functional basis of diverse oncogenic loci identified from large-scale next-generation sequencing studies of human cancers. Here, we leveraged primary human gastric organoids to establish the first forward genetic human ARID1A transformation model, whose multi-omic analysis revealed phenotypic and functional recapitulation of numerous features of ARID1A-mutated gastric cancer.
The inability to establish ARID1A KO organoids from wild-type human gastric organoids could originate in the anaphase bridge formation and G2/M cell cycle arrest upon loss of BAF subunits(7). TP53 deficiency, as in the current study, may bypass this arrest, allowing establishment of organoids mutated in both ARID1A and TP53. Although concomitant mutation of ARID1A and TP53 occurs sporadically (~4-13%) in human gastric cancers and ~30% of MSI gastric cancer(14,15,67), the engineered TP53/ARID1A DKO organoids nevertheless faithfully recapitulate numerous features of ARID1A-mutated gastric cancer; similar studies could be extended to model ARID1A loss in the context of other driver mutations.
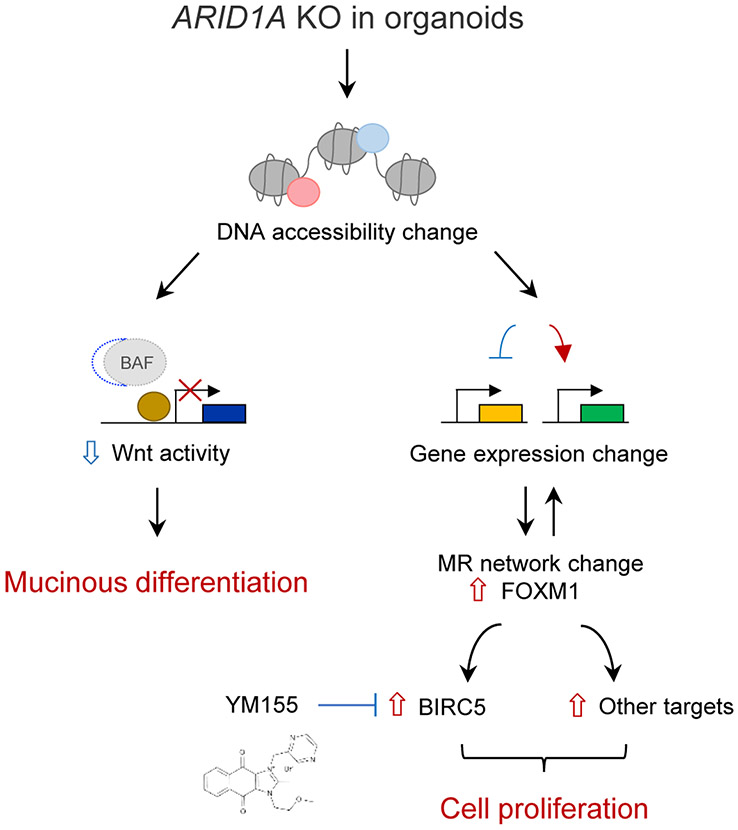
Notably, ARID1A KO elicits global transcriptional regulatory programs significantly reminiscent of MSI- and EBV-type gastric cancers, precisely those subtypes in which ARID1A mutation is most prevalent(14,15). Moreover, the absence of engineered MSI mutations in ARID1A-defieicient organoids suggests that ARID1A loss may be a major determinant of the overall transcriptional regulatory program of MSI stomach adenocarcinoma. Crucially, our multiscale analysis of ARID1A KO organoids, integrating transcriptional, small molecule and computational approaches, defines a bifurcated model of ARID1A-dependent oncogenic transformation where non-essential Wnt-regulated mucinous metaplasia is distinct from essential YM-155-sensitive, FOXM1→BIRC5-regulated proliferation (Fig. 7).
Figure 7. Model of ARID1A loss-mediated oncogenic transformation in early human gastric cancer.
ARID1A loss induces functionally independent transformation pathways during early gastric tumorigenesis in which non-essential Wnt-regulated mucinous differentiation operates in parallel with versus essential YM-155-sensitive FOXM1/BIRC5-regulated cell proliferation.
Mechanistically, ARID1A loss inhibits canonical Wnt/β-catenin activity leading to a redirection of gland- to pit-like cell fate determination. During homeostasis, gastric cell determination is maintained by a gradient of canonical Wnt/β-catenin activity that is established and most intense at the gland base, and extends up toward the mucin-producing pit cells in the upper gland where canonical Wnt/β-catenin activity is virtually absent(43). Emerging evidence suggests Wnt and R-Spondin agonists are critical microenvironmental cues for maintaining gastric stem cells(34,68,69). Consistent with this, ARID1A-deficient organoids displayed reduced canonical Wnt/β-catenin signaling, accompanied by a shift to pit-like mucin-producing lineage differentiation which was potently rescued by constitutive β-catenin activation. Of note, constitutive β-catenin activation significantly downregulated several gastric mucous cell and intestinal goblet cell genes, including SPDEF, encoding a transcription factor regulating epithelial goblet cell differentiation(47,48). Consistent with these results, previous studies identify SPDEF as a Wnt-responsive gene(48) that functions as a colorectal cancer tumor suppressor by regulating Wnt signaling(70,71). Together with the inverse relationship between ARID1A expression and mucinous differentiation in human gastric cancer microarrays, our findings confirm prior transgenic mouse studies where Arid1a loss promotes mucinous tumorigenesis in colon(19) and pancreas(22-24) but where a molecular mechanism was not established. In contrast, non-mucinous differentiation associated with Arid1a mutation occurs in ovarian and uterine tumors(18,21). Thus, lineage metaplasia may be a pervasive feature of ARID1A-deficient cancer, which our studies reveal can be driven by Wnt pathway dysregulation.
Surprisingly, despite robust Wnt-dependency of ARID1A loss-induced mucous metaplasia, this pathway did not regulate cell division, indicating non-essentiality. Instead, the unexpected convergence of our master regulator and small molecule selective lethal screens identified a YM-155-sensitive FOXM1→BIRC5 transcriptional node as a essential regulator of ARID1A KO-induced proliferation. The functional independence of the Wnt/mucin versus FOXM1→BIRC5/proliferation pathways is attested by the inability of β-catenin rescue to alter YM-155 sensitivity, while conversely YM-155 does not reverse Wnt-dependent mucinous differentiation or target expression.
Several studies have pursued discovery of targets exhibiting synthetic lethality with ARID1A deficiency in transformed cancer cell lines. Such ARID1A selective lethal compounds include inhibitors of EZH2 methyltransferase, a PRC2 core subunit that opposes BAF complex activity(72,73) and glutathione synthesis antagonists(74). Our results clearly indicate that ARID1A mutation confers selective sensitivity to BIRC5/survivin inhibition in engineered gastric organoids, reflecting early-stage gastric tumorigenesis. In contrast, multiple conventional 2D cancer cell lines did not exhibit selective sensitivity to BIRC5/survivin inhibition. Thus, BIRC5 dependency appears more stringent during earlier stages of ARID1A-deficient gastric oncogenesis, as in engineered organoids, while late-stage gastric cancers may possess redundant pro-survival mechanisms. We also cannot exclude confounding effects on drug sensitivity from 2D cancer cell line versus 3D engineered organoid culture, which can influence oncogenic phenotypes(75), or from genetic drift and resistance mechanisms in long-passaged cell lines. Thus, further work will be required to explore YM-155 efficacy in established gastric cancer and define the range of ARID1A-deficient malignancies for which YM-155 may be effective.
Mouse models have proven invaluable for study of molecular mechanisms underlying gastric metaplasia and its neoplastic progression. However murine models, while recapitulating early-stage gastric mucous cell hyperplasia and SPEM, are limited in modeling the later stages of carcinogenesis in humans, such as progression to IM, high-grade dysplasia, and infiltrating adenocarcinoma. Our engineered TP53/ARID1A DKO human organoids recapitulate high-grade dysplasia in vitro and acquired intestinal goblet cell features in vivo, the latter suggesting that stromal and/or inflammatory cells within the tumor microenvironment may promote development of late-stage gastric tumors. Interestingly, upon in vivo implantation, ~10% of TP53/ARID1A DKO organoid cells exhibit SPEM features, indicating that ARID1A loss could potentially predispose to SPEM, possibly in concert with environmental cues. Thus, engineered human tumor organoids together with in vivo xenotransplantation provide a valuable platform for studying previously inaccessible stages of human gastric cancer development. Future studies will be required to determine whether additional oncogenic drivers or microenvironmental cues facilitate evolution of ARID1A-deficient cells to metastatic adenocarcinoma.
Overall, our forward genetic study of engineered ARID1A-deficient human gastric organoids enabled a functional deconstruction of essential versus non-essential mechanisms of early ARID1A-dependent tumorigenesis. These analyses were greatly facilitated by the synthesis of genome-scale omics approaches, high-throughput small molecule screening and computational models, affording mechanistic insights into the genesis of ARID1A-deficient gastric cancer. Conceivably, analogous multimodal approaches to oncogene-engineered organoids may be further generalizable to additional cancer-associated loci and malignancies, yielding clinically relevant insights regarding cancer initiation and ultimately therapy.
Methods
Cell lines and maintenance.
L-WRN cells (ATCC, CRL-3276) that produced Wnt-3A/R-spondin/Noggin conditional media, and HEK293T cells were maintained in DMEM (Life Technologies, #11995-073) supplemented with 10% FBS. Gastric cancer cell lines were purchased from ATCC. SNU-16, AGS, NCI-N87 and MKN7 cells were maintained in RPMI1640 supplemented with 10% FBS. HGC27 cells were maintained in DMEM supplemented with 10% FBS. KATO-III cells were maintained in DMEM supplemented with 20% FBS. All cells were cultured at 37°C with 5% CO2. All cell lines have been tested for mycoplasma at least once every 6 months.
Organoid culture media.
The organoid culture media contained Advanced DMEM/F-12 (Thermo Fisher Scientific, #12634028) with 0.5% Penicillin/Streptomycin/Glutamine (Thermo Fisher Scientific, #10378016), 5% FBS, 1 mM HEPES (Thermo Fisher Scientific, #15630080), 1 mM N-Acetylcysteine (Sigma, A9165), 1X B-27 Supplement (Thermo Fisher Scientific, #12587001), 500 nM A83-01 (Tocris Bioscience, #2939), 1X GlutaMax Supplement (Thermo Fisher Scientific, #35050061), 10 μM SB-202190 (Biogems, #1523072), 10 mM Nicotinamide (Sigma, #N0636), 50 ng/mL EGF (PeproTech, AF-100-15), 100 μg/mL Normocin (InvivoGen, ant-nr-1), 10 mM Gastrin (Sigma, G9145), 200 ng/mL fibroblast growth factor (FGF) (Peprotech, #100-26), and 50% Wnt-3A/R-spondin/Noggin conditioned media.
Establishment of normal gastric organoid cultures.
Clinical samples used for gastric organoid establishment were obtained from gastric corpus of patients at Stanford University Hospital’s Tissue Procurement Shared Resource facility. Healthy gastric tissues were collected by surgical resection. Gastric organoids were established as previously reported(34). Briefly, surgical specimens were washed vigorously three times with sterile, cold phosphate-buffered saline (PBS) in a 15 mL conical tube, and then were dissected into smaller pieces in cold chelation buffer (5.6 mM Na2HPO4, 8.0 mM KH2PO4, 96.2 mM NaCl, 1.6 mM KCl, 43.4 mM Sucrose, 54.9 mM D-Sorbitol, 0.5 mM DTT) plus 10 mM EDTA. The tissues were incubated 4°C for 3-5 hours in a rocking chamber. After incubation, tissues were washed by fresh cold chelation buffer and vigorously shaken by hands. After shaking, the supernatant was checked for the presence of gastric crypts. This step was repeated 8-10 times and each wash produced supernatant containing gastric crypts that were examined under bright-field microscope. Finally, crypts collected from different fractions were combined and centrifuged at 600 g at 4°C for 5 minutes. Gastric crypts were resuspended in Matrigel (R&D systems, Basement Membrane Extract type 2) and plated in a 24-well plate. After Matrigel polymerization, organoid culture media was added to each well (described above) plus 10 μM Y-27632 (Peprotech, #1293823) and 3 μM CHIR-99021 (R&D Systems, #4223). After 3 days, the media was changed to organoid culture media without Y-27632 and CHIR-99021, and cultures were maintained in organoid culture media with routine media changes occurring every 3-4 days until subsequent passage. Fibroblast growth factor (FGF) was dispensable for engineered TP53 KO and TP53/ARID1A DKO organoids. Organoids were passaged to prevent overgrowth every 10-14 days. For passaging, organoids were washed by PBS and mechanically dissociated into smaller pieces by pipetting and resuspension in TrypLE™ (Invitrogen, #12604-012) at 37°C for 10-20 minutes. After incubation, fetal bovine serum (FBS) was added to quench TrypLE™ activity. Organoids were then centrifuged at 600 g for 5 minutes and washed once using organoid culture media before resuspension in matirgel and plating onto a new 24-well plate.
Guide RNA expression vector cloning.
The lentiviral sgRNA vectors were generously provided by Dr. Jonathan Weissman(76,77). The sgRNA vector was digested by BstXI (New England BioLabs, R0113) and BlpI (New England BioLabs, R0585) at 37°C for 6 hours. The linearized vectors were separated on a 1% agarose gel. Linearized vectors were cut and then purified by QIAquick Gel Extraction Kit (Qiagen, #28706). The lentiviral sgRNA expression vectors were cloned by inserting annealed sgRNA oligos into the linearized sgRNA vectors. The ligation of the linearized vectors and the annealed sgRNA oligos were completed by T4 DNA ligase (New England BioLabs, M0202) at 25°C for 2 hours. Ligation reactions were transformed into Stellar Competent E. coli Cells (TaKaRa, #636763) following the manufacturer’s instructions. Competent cells were plated on LB agar plates supplemented with 100 μg/mL carbenicillin and incubated at 37°C overnight. Colonies were randomly picked from each plate and inoculated into 4 mL LB supplemented with 100 μg/mL carbenicillin and then grown at 37°C for 14 hours. The lentiviral sgRNA expression vectors were purified by QIAprep Spin Miniprep Kit (Qiagen, #27106) for subsequent confirmation by Sanger-sequencing. The sgRNA sequences used in this study were listed in the key resources table.
Generation of clonal organoid lines.
Organoids were washed by PBS dissociated with TrypLE™ (Invitrogen, #12604-012) for 30 minutes at 37°C. Cell clumps were removed using 35 mm cell strainer (BD Falcon, #352235) and the flow-through was pelleted at 600 g at 4°C for 5 minutes. Cells pellets were resuspended in organoid culture media with 10 μM Y-27632 (Peprotech, #1293823). Single cells were sorted in single wells of a 96-well plate. The 96-well plate was pre-coated by 10 μL Matrigel (R&D systems, Basement Membrane Extract type 2) and covered by 100 μL organoid culture media. FACS Aria II (BD Biosciences) equipped with a 100 mm nozzle was used for cell sorting. Wells containing a single organoid 12-14 days after cell sorting were dissociated with TrypLE™ and replated for clonal expansion. The clonal lines were verified by Sanger-sequencing, immunoblot analysis, or immunostaining. For Sanger-sequencing, genomic DNA was isolated from organoids by using DNeasy blood and tissue kit (Qiagen, #69506). The targeted loci were amplified by PCR using Phusion High-Fidelity DNA Polymerase (New England BioLabs, M0530) and then sequenced directly. Primers for PCR amplification and Sanger-sequencing used in this study were listed in the key resources table.
Generation of lentivirus.
Lentiviral plasmids were co-transfected with viral packaging plasmid psPAX2 (Addgene, #12260) and viral envelope plasmid pCMV-VSV-G (Addgene, #8454) into 293T cells by Lipofectamine 2000 (Invitrogen, #11668-019) following the manufacturer’s instructions. Lentiviral supernatants were collected at 48 hours and 72 hours post-transfection and concentrated by PEG-it Virus Precipitation Solution (System Biosciences, LV825A-1). Precipitated lentiviral particles were pelleted at 1500 g at 4°C for 30 minutes and resuspended in organoid culture media containing 10 μM Y-27632 (Peprotech, #1293823). Lentiviral plasmids used in this study were listed in the key resources table.
Lentiviral transduction of organoids.
Organoids were washed by PBS and dissociated into smaller clusters with TrypLE™ (Invitrogen, #12604-012) for 15 minutes at 37°C. Organoids were resuspended into 500 μL transduction solution containing 10 μM Y-27632 (Peprotech, #1293823), 8 μg/mL polybrene (Sigma, #107689) and concentrated lentivirus in organoid culture media. Spinoculation of resuspended organoids was performed at 800 g for 1 hour at 32°C. After spinoculation, organoids were incubated for 12-14 hours at 37°C and then replated onto a new 24-well plate.
Immunoblotting.
Western blot analyses were performed using standard method. Briefly, the pellet was lysed in RIPA buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl at pH 7.5) with protease inhibitor cocktail (Roche, #04-693-124-001) and phosphatase inhibitor cocktail (Sigma, P5726). Protein concentration was measured by the BCA kit (Thermo Scientific, #23227). Cell lysates were separated by SDS Poly-acrylamide-gel-electrophoresis (Invitrogen, NP0323). PageRuler Plus Prestained Protein Ladder (Thermo Scientific, #26619) was used as molecular weight marker. Proteins were transferred to a PVDF membrane (Millipore, IPVH00010), blocked by 5% non-fat dry milk in 1X TBS buffer at pH 7.4 (Quality Biological, #351-086-151) with 0.05% Tween-20, and then probed with the indicated primary antibodies at 4°C overnight. Bound antibodies were visualized by chemiluminescence (Thermo Scientific, #34580) using a horseradish peroxidase-conjugated secondary antibody and exposure of AccuRay blue X-Ray films (E&K Scientific, EK5129). Antibodies used for immunoblotting were listed in the key resources table.
Immunohistochemistry and immunofluorescence staining.
Organoids were fixed with 2% paraformaldehyde (Electron Microscopy Sciences, #15714) in PBS for 30 minutes at room temperature, washed with PBS twice, embedded in HistoGel™ (Thermo Scientific, HG-4000-012), and then transferred to 70% ethanol for paraffin-embedding. Organoids were sectioned at 5-mm thickness. Paraffin-embedded sections were deparaffinized and rehydrated before staining. For immunohistochemistry staining, antigen retrieval was achieved in sodium citrate buffer (10 μM sodium citrate at pH 6.0). Slides were incubated in 3% H2O2 solution (Fisher Scientific, H325-100) in methanol at room temperature for 10 minutes to block endogenous peroxidase activity. After washing, slides were blocked in Avidin/Biotin Solution (Vector Laboratories, SP2001) at room temperature for 30 minutes and then in blocking buffer (5% normal goat or donkey serum in PBS) for 1 hour. After blocking, slides were incubated with primary antibody in blocking buffer at 4°C overnight. Slides were washed by PBST (PBS with 0.05% Tween-20) and incubated with secondary antibody at room temperature for 30 minutes. Slides were washed by PBST and ABC reagent was applied (Vector Laboratories, PK-6101). After washing with PBST, DAB staining was performed for signal detection (Vector Laboratories, SK-4100). The slides were counterstained with hematoxylin (Sigma, MHS16) for 2 minutes and rinsed with water for 1 minute. Subsequent treatment with 1% acid alcohol 3 times to differentiate nuclear detail was performed along with sequential treatment with 0.2% ammonia for bluing, each of these steps were followed by a water rinse for 1 minute. Following this, the slides were rehydrated and mounted using mounting solution (Thermo Scientific, #4112).
For immunofluorescence staining, deparaffinization and rehydration procedures were as described above. Slides were blocked in blocking buffer at room temperature for 1 hour. After blocking, slides were incubated with primary antibody in blocking buffer at 4°C overnight. Slides were washed by PBST and incubated with secondary antibody at room temperature for 30 minutes. After washes with PBST, slides were mounted by mounting solution with DAPI (Vector Laboratories, H-1500). Imaging was performed using fluorescence microscopy (Keyence, BZ-X700 series). For Alcian blue staining, slides were stained with Alcian blue (Thermo Fisher, #88043) following the manufacturer's instructions. Antibodies used for immunostaining were listed in the key resources table.
Real-time quantitative PCR.
Total RNA from organoids was isolated with the RNeasy kit (Qiagen, #74106). The on-column DNase digestion (Qiagen, #79254) was used to eliminate genomic DNA. A total of 0.5-1 μg RNA was used to synthesize complementary DNA using iScript™ Reverse Transcription Supermix (Bio-Rad, #1708841). Quantitative PCR was performed with Power SYBR™ Green PCR Master Mix (Thermo Scientific, #4368708). The primers used for quantitative PCR were listed in the key resources table.
Cell proliferation and viability assay.
Organoids were dissociated into smaller aggregates and single cells were sorted by FACS Aria II (BD Biosciences) as described above. A total of 20,000 cells were resuspended into 40μL Matrigel (R&D systems, Basement Membrane Extract type 2) and plated in a well of a 24-well plate. Over a period of 14 days, organoid growth was recorded daily by bright-field microscopy. YM-155 (Cayman Chemicals, #11490) was dissolved in DMSO. For 12-point full titration treatment of YM-155, a total of 5,000 cells were resuspended into 10 μL Matrigel and cultured in a well of a 96-well plate for 5 days before drug treatment. Cell viability was quantified 3 days after YM-155 treatment. For the cell viability assay, AlamarBlue™ Cell Viability Reagent (Invitrogen, DAL1100) in organoid culture media was added into the plate and incubated with organoids for 4 hours before being quantified using a Synergy H1 Hybrid Multi-mode Plate Reader (BioTek).
Luciferase assay.
A total of 20,000 TOPflash mCherry-positive single cells were sorted by FACS Aria II (BD Biosciences) as described above. Cells were washed by PBS and the pellet was lysed in Passive Lysis Buffer (Promega, E194A). Firefly luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, D1980).
sgRNA design.
Knockout sgRNA were designed using a combination of empirical data and on-target and off-target predictions. When available, empirical data from published CRISPR screens were used to pick the most active sgRNAs(78-80), otherwise the sgRNAs were designed as described previously(78). The sgRNA sequences used in this study are listed in the key resources table.
High-throughput compound screening.
Screening of the Emory Enriched Bioactive Library (EEBL), which includes 2,036 U.S. Food and Drug Administration (FDA) approved and bioactive compounds(81), was carried out using our miniaturized organoid culture platform in a 384-well format for HTS. Briefly, organoids grown in a 50 μl Matrigel droplet on a single well of a 24-well plate were harvested as described and re-suspended in ice-cold Matrigel (R&D systems, Basement Membrane Extract type 2) to form a cell/Matrigel mixture. 8 μL/well of the cells/Matrigel (~1,000 cells/well) mixture was dispensed onto a 384-well plate using a Multidrop Combi dispenser (Thermo Fisher Scientific). The plates were immediately centrifuged at 500 rpm for 1 min and incubated for 30 min at cell culture incubator to allow Matrigel solidification. 35 μL per well of organoid culture media was dispensed into the wells. The plates were sealed using gas permeable plate sealer (Breathe-Easy Sealing Film, Diversified Biotech, #BEM-1) and incubated for 5 days in cell culture incubator to allow organoid formation. Then, 0.1 μL of library compounds diluted in DMSO were added to each well using Pin-tool integrated with Beckman NX automated liquid handling system (Beckman Coulter, Danaher Corporation). The plates were centrifuged at 800 rpm for 5 min to ensure the uniform distribution of the compound into the wells. The final compound concentration was 4.6 μM and the final DMSO concentration was 0.2%. The plates were sealed with gas permeable plate sealer. After incubating with compound for 3 days, the viability of organoids was determined by CellTiter Blue reagent (Promega). Briefly, 5 μL of CellTiter Blue reagent was added to each well in 384-well plates using a MultiDrop Combi. After incubating at 37°C for 4 hours, the fluorescence intensity (FI), which is correlated with the number of viable cells, was measured using PHERAstar FSX multi-label plate reader (BMG LABTECH) with excitation at 540/20 nm and Emission at 590/20 nm.
Data analysis for high throughput drug screening.
Screening data were analyzed using CambridgeSoft Bioassay software. The performance of the organoids HTS viability assay in 384-well format was evaluated by Z’ factor and Signal-to-background (S/B) ratio and were calculated as the following equations:
Where SD DMSO and SD blank are the standard deviations, and FI DMSO control and FI blank are the corresponding average FI signal for the wells with DMSO control or blank with medium only without cells, respectively. A Z’ factor between 0.5 and 1.0 indicating that the assay is robust for HTS(82). The effect of compound on the growth of organoids was expressed as % of control and calculated based on per plate as the following equations:
The dose-response effect of selected hit compounds from HTS on the growth of organoids was analyzed using GraphPad Prism 7 (GraphPad Software, Inc.).
Master regulator analysis.
The context-specific regulatory network used in this analysis was reverse-engineered from a collection of 200 gene expression profiles from STAD patients in TCGA(14) using the ARACNe algorithm(52). Specifically, ARACNe was used to infer regulatory targets of 1,813 transcription factors—including genes annotated in Gene Ontology molecular function database (GO) as ‘transcription factor activity’, ‘DNA binding’, ‘transcription regulator activity’, or ‘regulation of transcription’ (GO:0003700, GO:0004677, GO:0030528, GO:0004677, GO: 0045449)—and a manually curated list of 969 transcriptional cofactors—including genes annotated as ‘transcription cofactor activity’, (GO:0003712, GO:0030528, GO:0045449). For each of these regulators, its protein activity was computed by VIPER(50) analysis of genes differentially expressed in TP53/ARID1A DKO compared to TP53 KO samples, using the STAD-specific ARACNe regulatory network. The list of regulators and of their inferred differential activity in TP53/ARID1A DKO samples were then compared to the VIPER inferred protein activity profiles of all TCGA-STES patient samples, using the ‘viperSimilarity’ method of the VIPER package. This method computes the similarity between two samples based on the conservation of their differentially active proteins. This is accomplished by performing a gene set enrichment analysis of statistically significantly differentially active proteins in one context (e.g., TP53/ARID1A DKO) to protein differentially active in the other context (e.g. STES patients) and vice versa, using the aREA algorithm, an analytic extension of GSEA(83). The similarity scores obtained from viperSimilarity method are the z-scores of enrichment analysis. TCGA-STES samples and their subtype annotations were obtained from literature(84). The context-specific regulatory network of the TCGA-STES samples was reverse-engineered using ARACNe algorithm, and protein activity profiles of all samples were computed by VIPER analysis of genes differentially expressed in each TCGA-STES sample compared to the average gene expression in all samples, using STES specific ARACNe regulatory network.
RNA-seq and data analysis.
For the RNA-seq, two technical duplicates were included for each sample. RNA-seq libraries were generated by using NEBNext Ultra II Directional RNA Library Prep Kit coupled with Poly(A) mRNA Magnetic Isolation Module and NEBNext multiplex oligos for Illumina (New England Biolabs). The deep sequencing was performed on the NextSeq 500 sequencing system (Illumina) with 75-cycle, paired-end sequencing. RNA-seq data were aligned to hg38 human genome assembly using kallisto (v 0.44.0) with default parameters. Differential gene expression analysis was performed using DESeq2(85). Change in gene expression between two conditions was defined as significant if ∣log2FC∣>0.5 and adjusted p-value <0.05. ComplexHeatmap was used to produce heat maps(86).
Somatic variant calling.
Short reads produced by WGS on the Illumina platform were aligned to hg38 using BWA (v0.7.17). Following GATK (v4.1.4.1) best practice workflow(87), the raw alignment files (BAMs) were then pre-processed through marking duplicated reads and base recalibration. SNV and INDEL calls were made using MuTect2 in GATK package. The calls were then filtered and annotated using FilterMutectCalls and Funcotator in GATK. Somatic copy number aberrations (SCNAs) were estimated using CNVkit (v0.9.6)(88).
Subcutaneous xenografts.
NOD-scid IL2Rgammanull (NSG) immunodeficient mice were obtained from the Jackson Laboratory (#005557). For xenograft studies, male adult NSG mice (~8-10 weeks old) were randomly divided into experimental groups. Mice were subcutaneously injected with organoids (1.5 x 106 cells in 150 μL 100% Matrigel per injection). Mice were sacrificed 3 months after inoculation of organoids. All mouse studies were approved by the Stanford Institutional Animal Care and Use Committee (IACUC).
Supplementary Material
Statement of significance.
We establish the first human forward genetic modeling of a commonly mutated tumor suppressor gene, ARID1A. Our study integrates diverse modalities including CRISPR/Cas9 genome editing, organoid culture, systems biology and small molecule screening to derive novel insights into early transformation mechanisms of ARID1A-deficient gastric cancers.
Acknowledgements
We thank generous support from NIH fellowship K00CA212433 (Y-H.L.), a Swedish Research Council International Postdoctoral Fellowship (K.K.), Stanford Bio-X Undergraduate Fellowship (W.D.S.), the NCI Cancer Target Discovery and Development (CTD2) Network (C.J.K., H.F., A.C. and C.N.C.), NIH U01CA217875 to H.F., NIH R01CA163915 to G.R.C and grants from the NIH (U01CA217851, U54CA224081, U01CA199241 and U19 AI116484), Emerson Collective and Ludwig Cancer Research to C.J.K.
Footnotes
Conflict of Interest Statement
The authors declare no potential conflicts of interest
Data availability.
The datasets generated in this study are available from the corresponding author on reasonable request. Raw and processed sequencing data were deposited into the Gene Expression Omnibus (GEO) under accession code GSE164179.
References
- 1.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357:eaal2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hodges C, Kirkland JG, Crabtree GR. The many roles of BAF (mSWI/SNF) and PBAF complexes in cancer. Cold Spring Harb Perspect Med. 2016;6:a026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mashtalir N, D’Avino AR, Michel BC, Luo J, Pan J, Otto JE, et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell. 2018;175:1272–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu JI, Lessard J, Crabtree GR. Understanding the Words of Chromatin Regulation. Cell. 2009;136:200–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pulice JL, Kadoch C. Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb Symp Quant Biol. 2016;81:53–60. [DOI] [PubMed] [Google Scholar]
- 7.Dykhuizen EC, Hargreaves DC, Miller EL, Cui K, Korshunov A, Kool M, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;497:624–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet. 2013;45:592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadoch C, Crabtree GR. Reversible disruption of mSWI/SNF (BAF) complexes by the SS18-SSX oncogenic fusion in synovial sarcoma. Cell. 2013;153:71–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson BG, Wang X, Shen X, McKenna ES, Lemieux ME, Cho YJ, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell. 2010;18:316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBride MJ, Pulice JL, Beird HC, Ingram DR, D’Avino AR, Shern JF, et al. The SS18-SSX Fusion Oncoprotein Hijacks BAF Complex Targeting and Function to Drive Synovial Sarcoma. Cancer Cell. 2018;33:1128–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu JN, Roberts CWM. ARID1A mutations in cancer: Another epigenetic tumor suppressor? Cancer Discov. 2013;3:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, Mcpherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. [DOI] [PubMed] [Google Scholar]
- 14.Bass AJ, Thorsson V, Shmulevich I, Reynolds SM, Miller M, Bernard B, et al. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 16.Chandler RL, Brennan J, Schisler JC, Serber D, Patterson C, Magnuson T. ARID1a-DNA Interactions Are Required for Promoter Occupancy by SWI/SNF. Mol Cell Biol. 2013;33:265–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler RL, Damrauer JS, Raab JR, Schisler JC, Wilkerson MD, Didion JP, et al. Coexistent ARID1A-PIK3CA mutations promote ovarian clear-cell tumorigenesis through pro-tumorigenic inflammatory cytokine signalling. Nat Commun. 2015;27:6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathur R, Alver BH, San Roman AK, Wilson BG, Wang X, Agoston AT, et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet. 2017;49:296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiramatsu Y, Fukuda A, Ogawa S, Goto N, Ikuta K, Tsuda M, et al. Arid1a is essential for intestinal stem cells through Sox9 regulation. Proc Natl Acad Sci U S A. 2019;116:1704–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilson MR, Reske JJ, Holladay J, Wilber GE, Rhodes M, Koeman J, et al. ARID1A and PI3-kinase pathway mutations in the endometrium drive epithelial transdifferentiation and collective invasion. Nat Commun. 2019;10:3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimura Y, Fukuda A, Ogawa S, Maruno T, Takada Y, Tsuda M, et al. ARID1A Maintains Differentiation of Pancreatic Ductal Cells and Inhibits Development of Pancreatic Ductal Adenocarcinoma in Mice. Gastroenterology. 2018;155:194–209. [DOI] [PubMed] [Google Scholar]
- 23.Wang W, Friedland SC, Guo B, O’Dell MR, Alexander WB, Whitney-Miller CL, et al. ARID1A, a SWI/SNF subunit, is critical to acinar cell homeostasis and regeneration and is a barrier to transformation and epithelial-mesenchymal transition in the pancreas. Gut. 2019;68:1245–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SC, Nassour I, Xiao S, Zhang S, Luo X, Lee J, et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut. 2019;68:1259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livshits G, Alonso-Curbelo D, Morris JP, Koche R, Saborowski M, Wilkinson JE, et al. Arid1a restrains Kras-dependent changes in acinar cell identity. Elife. 2018;7:e35216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun X, Wang SC, Wei Y, Luo X, Jia Y, Li L, et al. Arid1a Has Context-Dependent Oncogenic and Tumor Suppressor Functions in Liver Cancer. Cancer Cell. 2017;32:574–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han L, Madan V, Mayakonda A, Dakle P, Woon TW, Shyamsunder P, et al. Chromatin remodeling mediated by ARID1A is indispensable for normal hematopoiesis in mice. Leukemia. 2019;33:2291–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo Y-H, Karlsson K, Kuo CJ. Applications of organoids for cancer biology and precision medicine. Nat Cancer. 2020;1:761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Nadauld L, Ootani A, Corney DC, Pai RK, Gevaert O, et al. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat Med. 2014;20:769–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21:256–62. [DOI] [PubMed] [Google Scholar]
- 31.Drost J, Van Jaarsveld RH, Ponsioen B, Zimberlin C, Van Boxtel R, Buijs A, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521:43–7. [DOI] [PubMed] [Google Scholar]
- 32.Nanki K, Toshimitsu K, Takano A, Fujii M, Shimokawa M, Ohta Y, et al. Divergent Routes toward Wnt and R-spondin Niche Independency during Human Gastric Carcinogenesis. Cell. 2018;174:856–69. [DOI] [PubMed] [Google Scholar]
- 33.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartfeld S, Bayram T, Van De Wetering M, Huch M, Begthel H, Kujala P, et al. In vitro expansion of human gastric epithelial stem cells and their responses to bacterial infection. Gastroenterology. 2015;148:126–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujii M, Clevers H, Sato T. Modeling Human Digestive Diseases With CRISPR-Cas9–Modified Organoids. Gastroenterology. 2019;156:562–76. [DOI] [PubMed] [Google Scholar]
- 36.Uhler C, Shivashankar GV. Nuclear Mechanopathology and Cancer Diagnosis. Trends in Cancer. 2018;4:320–31. [DOI] [PubMed] [Google Scholar]
- 37.Webster M, Wikin KL, Cohen-Fix O. Sizing up the nucleus: Nuclear shape, size and nuclear-envelope assembly. J Cell Sci. 2009;122:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan P, Yeoh KG. Genetics and Molecular Pathogenesis of Gastric Adenocarcinoma. Gastroenterology. 2015;149:1153–62. [DOI] [PubMed] [Google Scholar]
- 39.Mills JC, Shivdasani RA. Gastric epithelial stem cells. Gastroenterology. 2011;140:412–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hayakawa Y, Fox JG, Wang TC. The Origins of Gastric Cancer From Gastric Stem Cells: Lessons From Mouse Models. CMGH. 2017;3:331–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giroux V, Rustgi AK. Metaplasia: Tissue injury adaptation and a precursor to the dysplasia-cancer sequence. Nat Rev Cancer. 2017;17:594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen CP, Mills JC, Goldenring JR. Murine Models of Gastric Corpus Preneoplasia. CMGH. 2017;3:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stange DE, Koo BK, Huch M, Sibbel G, Basak O, Lyubimova A, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang XT, Kong FB, Mai W, Li L, Pang LM. MUC1 immunohistochemical expression as a prognostic factor in gastric cancer: Meta-analysis. Dis Markers. 2016;9421571:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barriga FM, Montagni E, Mana M, Mendez-Lago M, Hernando-Momblona X, Sevillano M, et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;20:801–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan KS, Gevaert O, Zheng GXY, Anchang B, Probert CS, Larkin KA, et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell. 2017;21:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horst D, Gu X, Bhasin M, Yang Q, Verzi M, Lin D, et al. Requirement of the epithelium-specific Ets transcription factor Spdef for mucous gland cell function in the gastric antrum. J Biol Chem. 2010;285:35047–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gregorieff A, Stange DE, Kujala P, Begthel H, van den Born M, Korving J, et al. The Ets-Domain Transcription Factor Spdef Promotes Maturation of Goblet and Paneth Cells in the Intestinal Epithelium. Gastroenterology. 2009;137:1333–45. [DOI] [PubMed] [Google Scholar]
- 49.Noah TK, Kazanjian A, Whitsett J, Shroyer NF. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp Cell Res. 2010;316:452–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Hilda Ye B, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat Genet. 2016;48:838–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Califano A, Alvarez MJ. The recurrent architecture of tumour initiation, progression and drug sensitivity. Nat Rev Cancer. 2017;17:116–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–90. [DOI] [PubMed] [Google Scholar]
- 53.Wierstra I FOXM1 (Forkhead box M1) in tumorigenesis. Overexpression in human cancer, implication in tumorigenesis, oncogenic functions, tumor-suppressive properties, and target of anticancer therapy. Adv Cancer Res. 2013;119:191–419. [DOI] [PubMed] [Google Scholar]
- 54.Yan HHN, Siu HC, Law S, Ho SL, Yue SSK, Tsui WY, et al. A Comprehensive Human Gastric Cancer Organoid Biobank Captures Tumor Subtype Heterogeneity and Enables Therapeutic Screening. Cell Stem Cell. 2018;23:882–97. [DOI] [PubMed] [Google Scholar]
- 55.Bitler BG, Wu S, Park PH, Hai Y, Aird KM, Wang Y, et al. ARID1A-mutated ovarian cancers depend on HDAC6 activity. Nat Cell Biol. 2017;19:962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fukumoto T, Park PH, Wu S, Fatkhutdinov N, Karakashev S, Nacarelli T, et al. Repurposing Pan-HDAC Inhibitors for ARID1A-Mutated Ovarian Cancer. Cell Rep. 2018;22:3393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee D, Yu EJ, Ham IH, Hur H, Kim YS. AKT inhibition is an effective treatment strategy in ARID1A-deficient gastric cancer cells. Onco Targets Ther. 2017;10:4153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samartzis EP, Gutsche K, Dedes KJ, Fink D, Stucki M, Imesch P. Loss of ARID1A expression sensitizes cancer cells to PI3K- and AKT-inhibition. Oncotarget. 2014;5:5295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakahara T, Takeuchi M, Kinoyama I, Minematsu T, Shirasuna K, Matsuhisa A, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–21. [DOI] [PubMed] [Google Scholar]
- 60.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer. 2003;3:46–54. [DOI] [PubMed] [Google Scholar]
- 61.Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci. 2019;132:jcs223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drost J, Van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, et al. Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science. 2017;358:234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee J, Snyder ER, Liu Y, Gu X, Wang J, Flowers BM, et al. Reconstituting development of pancreatic intraepithelial neoplasia from primary human pancreas duct cells. Nat Commun. 2017;8:14686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Seino T, Kawasaki S, Shimokawa M, Tamagawa H, Toshimitsu K, Fujii M, et al. Human Pancreatic Tumor Organoids Reveal Loss of Stem Cell Niche Factor Dependence during Disease Progression. Cell Stem Cell. 2018;22:454–67. [DOI] [PubMed] [Google Scholar]
- 65.Dekkers JF, Whittle JR, Vaillant F, Chen H-R, Dawson C, Liu K, et al. Modeling Breast Cancer Using CRISPR-Cas9–Mediated Engineering of Human Breast Organoids. JNCI J Natl Cancer Inst. 2019;112:540–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, Tapia P, et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell. 2019;24:927–43. [DOI] [PubMed] [Google Scholar]
- 67.Chen K, Yang D, Li X, Sun B, Song F, Cao W, et al. Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc Natl Acad Sci U S A. 2015;112:1107–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sigal M, Logan CY, Kapalczynska M, Mollenkopf HJ, Berger H, Wiedenmann B, et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature. 2017;548:451–5. [DOI] [PubMed] [Google Scholar]
- 69.Barker N, Huch M, Kujala P, van de Wetering M, Snippert HJ, van Es JH, et al. Lgr5+ve Stem Cells Drive Self-Renewal in the Stomach and Build Long-Lived Gastric Units In Vitro. Cell Stem Cell. 2010;6:25–36. [DOI] [PubMed] [Google Scholar]
- 70.Lo YH, Noah TK, Chen MS, Zou W, Borras E, Vilar E, et al. SPDEF Induces Quiescence of Colorectal Cancer Cells by Changing the Transcriptional Targets of β-catenin. Gastroenterology. 2017;153:205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Noah TK, Lo YH, Price A, Chen G, King E, Washington MK, et al. SPDEF functions as a colorectal tumor suppressor by inhibiting β-catenin activity. Gastroenterology. 2013;144:1012–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim KH, Kim W, Howard TP, Vazquez F, Tsherniak A, Wu JN, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med. 2015;21:1491–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov A V., et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ogiwara H, Takahashi K, Sasaki M, Kuroda T, Yoshida H, Watanabe R, et al. Targeting the Vulnerability of Glutathione Metabolism in ARID1A-Deficient Cancers. Cancer Cell. 2019;35:177–90. [DOI] [PubMed] [Google Scholar]
- 75.Han K, Pierce SE, Li A, Spees K, Anderson GR, Seoane JA, et al. CRISPR screens in cancer spheroids identify 3D growth-specific vulnerabilities. Nature. 2020;580:136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, et al. Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife. 2016;5:e19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun. 2018;9:5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mo X, Tang C, Niu Q, Ma T, Du Y, Fu H. HTiP: High-Throughput Immunomodulator Phenotypic Screening Platform to Reveal IAP Antagonists as Anti-cancer Immune Enhancers. Cell Chem Biol. 2019;26:331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. [DOI] [PubMed] [Google Scholar]
- 83.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, et al. Comparative Molecular Analysis of Gastrointestinal Adenocarcinomas. Cancer Cell. 2018;33:721–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 87.Depristo MA, Banks E, Poplin R, Garimella K V., Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Talevich E, Shain AH, Botton T, Bastian BC. CNVkit: Genome-Wide Copy Number Detection and Visualization from Targeted DNA Sequencing. PLoS Comput Biol. 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated in this study are available from the corresponding author on reasonable request. Raw and processed sequencing data were deposited into the Gene Expression Omnibus (GEO) under accession code GSE164179.