Summary
Biomolecules may undergo liquid–liquid phase separation (LLPS) to spatiotemporally compartmentalize and regulate diverse biological processes. Because the number of tools to directly probe LLPS is limited (ie. FRAP, FRET, fluorescence microscopy, fluorescence anisotropy, circular dichroism, etc.), the physicochemical traits of phase-separated condensates remain largely elusive. Here, we introduce a light-switching dipyrene probe (Pyr-A) that forms monomers in either hydrophobic or viscous environments, and intramolecular excimers in aqueous solutions. By exploiting their distinct fluorescence emission spectra, we used fluorescent microscopic imaging to study phase-separated condensates formed by in vitro protein droplets and membraneless intracellular organelles (centrosomes). Ratiometric measurement of excimer and monomer fluorescence intensities showed that protein droplets became hydrophobic and viscous as their size increased. Moreover, centrosomes became hydrophobic and viscous during maturation. Our results show that Pyr-A is a valuable tool to characterize LLPS and enhance our understanding of phase separation underlying biological functions.
Subject areas: Biophysical Chemistry, Biological Sciences, Biochemistry, Biophysics, Biological Sciences Research Methodologies
Graphical abstract
Highlights
-
•
A dipyrene probe Pyr-A is able to monitor LLPS without complicated steps
-
•
Pyr-A switches excimers (E) to monomers (M) when environments became hydrophobic/viscous
-
•
Ratio-metric measurement (E/M) can evaluate physicochemical dynamics of LLPS
-
•
Phase-separated biomolecules became hydrophobic and viscous during increase in size
Biophysical chemistry; Biological sciences; Biochemistry; Biophysics; Biological sciences research methodologies
Introduction
Many subcellular compartments are formed by liquid–liquid phase separation (LLPS) of biomolecules. The resulting condensates, which are free of lipid bilayers and therefore called membraneless organelles, include large transcriptional apparatuses mediated by transcriptional coactivators (ie. Bromodomain protein, mediators), centrosomes, stress granules, nucleoli, processing bodies, Cajal bodies, nuclear speckles, and many others (Alberti et al., 2019; Elbaum-Garfinkle et al., 2015; Feric et al., 2016; Franzmann et al., 2018; Gomes and Shorter, 2019; Hernández-Vega et al., 2017; Brangwynne et al., 2009; Hyman et al., 2014; Langdon et al., 2018; Posey et al., 2018; Shin and Brangwynne, 2017; Shin et al., 2018; Wegmann et al., 2018; Wei et al., 2017; Zhang et al., 2019, 2020). In principle, phase-separated condensates can achieve a locally high concentration of biomolecules without global upregulation of protein synthesis. LLPS therefore provides a dynamic infrastructure for biological reactions (Alberti et al., 2019; Gomes and Shorter, 2019; Hyman et al., 2014). In order to understand the molecular determinants of intracellular LLPS, it is important to establish tools to investigate the microenvironment during growth and disassembly processes.
Several methods have been established to examine the properties of biomolecular condensates formed by LLPS (Burke et al., 2015; Conicella et al., 2016; Ganser and Myong, 2020; Mitrea et al., 2018; Murthy and Fawzi, 2020; Reichheld et al., 2017). Phase separation assays using bright-field imaging or fluorescence microscopy can measure the effect of various conditions on LLPS, measure droplet formation and dissolution kinetics, and test the miscibility of differentially labeled molecules over time. Fluorescence recovery after photo bleaching and particle tracking using microrheology probe the diffusion of molecules within droplets, whereas droplet fusion assays, right angle imaging, and surface wetting determine droplet surface tension (Mitrea et al., 2018; Ganser and Myong 2020; Murthy and Fawzi 2020). Additionally, NMR probes the chemical environment and secondary structure via chemical shifts, proximal interactions via nuclear overhauser effects and paramagnetic relaxation enhancement, and dynamics (Mitrea et al., 2018). However, these assays have many limitations. All fluorescent-based assays require conjugation of fluorescent materials to proteins of our interest. NMR assays require samples with high purity and concentration, and suffer limitations due to spectral overlap and signal broadening resulting from low sequence complexity, high molecular size, or increased viscosity. Moreover, data processing and interpretation yield unclear results. Thus, there are currently no suitable, convenient tools to monitor phase separation processes. Therefore, developing novel probes monitoring LLPS without complicated steps (e.g. protein purification, genetic engineering) will overcome methodological restrictions underlying current approaches.
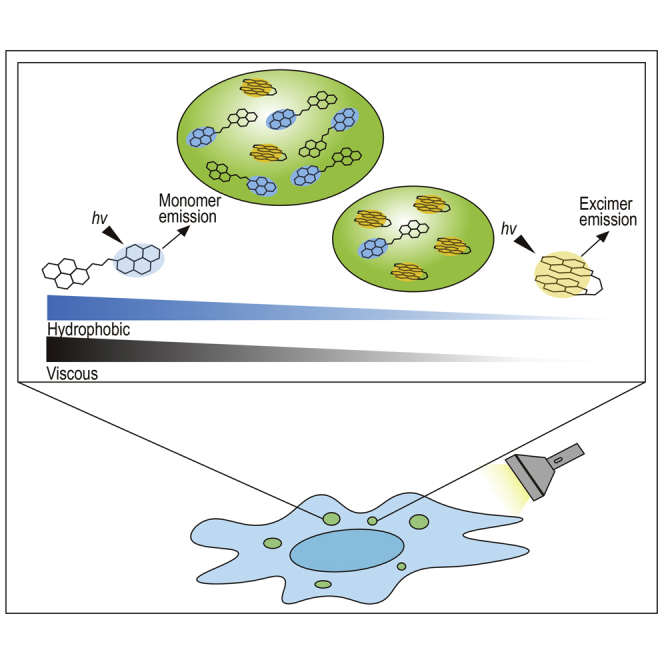
Pyrene is a polycyclic aromatic hydrocarbon consisting of four fused benzene rings and has dual fluorescence activity that is modulated by the solvent environment. Pyrenes can form excimers when an excited pyrene monomer interacts with another monomer in the ground state (Somerharju, 2002) in a concentration-dependent manner. Therefore, lipid-linked pyrenes are suitable for studying biophysical properties such as membrane fluidity (Gadella et al., 1990; Somerharju, 2002). For example, excimer formation by monopyrene lipids provides a measure of lateral lipid diffusion, while dipyrene species, when present at low concentrations, form only intramolecular excimers (Somerharju, 2002). Further, a conformational expansion of pyrene-labeled proteins during phase separation was successively monitored by evaluating intramolecular excimer emission (Majumdar et al., 2019). Recently, we used solvents with a wide range of polarities, including poly(dimethylsiloxane) and oligo(dimethylsiloxane), to evaluate aromatic donor–acceptor interactions between pyrene and pyromellitic diimide derivatives (Amemori et al., 2021). Because the photophysical properties of pyrene are sensitive to solvent polarity and viscosity (Somerharju, 2002), dipyrene analogues can be further used to evaluate physicochemical dynamics of LLPS (Figure 1A).
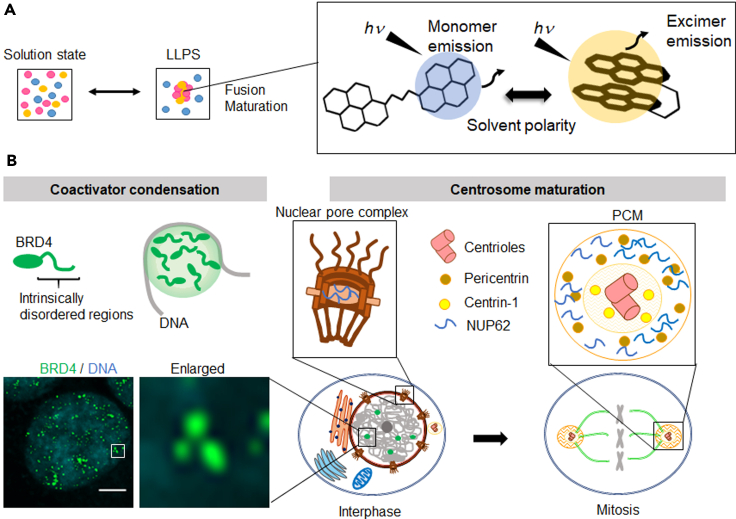
Figure 1.
Schematic illustration of quantitative measurement of phase-separated regions using a light-switching pyrene probe
(A) Evaluation of condensates in living cells using the fluorescent properties of the dual-pyrene probe pyr-A.
(B) Examples of phase-separated microenvironments, such as bromodomain-containing protein 4 (BRD4) coactivator condensates (left) and centrosomes (right).
In this study, we developed a novel dipyrene analogue, Pyr-A, and used it to probe biomolecule condensates formed by LLPS. These include in vitro droplets formed by intrinsically disordered regions (IDRs) of bromodomain-containing protein 4 (BRD4) (Caputo et al., 2021; Colino-Sanguino et al., 2019; Sabari et al., 2018), and centrosomes, which are dynamic and spherical membraneless intracellular organelles (Tiwary and Zheng, 2019) (Figure 1B). Centrosomes are surrounded by pericentriolar material (PCM) (Andersen et al., 2003). During the cell cycle, centrosomes replicate in the S phase, sequentially expand the PCM by assembling a set of proteins, organize the mitotic spindle to ensure equal distribution of duplicated genomes to two daughter cells (Figure 1B), and finally disassemble at the end of cell division (Tiwary and Zheng, 2019). Using fluorescence microcopy, the above centrosome dynamics can be monitored in living cells by imaging a fluorescently labeled centrosome component such as centrin-1 (Imai et al., 2010). Here, we show that Pyr-A can identify increased hydrophobicity and viscosity during increase in size of phase-separated molecular condensates.
Results
Synthesis and photophysical properties of Pyr-A
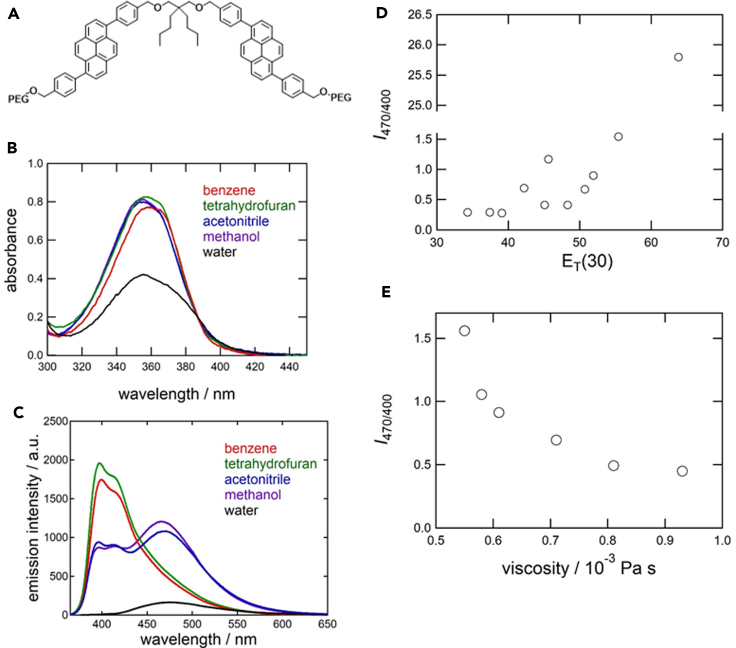
We prepared the dipyrene analogue Pyr-A (Figure 2A), with attached water-soluble poly(ethylene glycol) groups, from the substitution reaction between dipyrene derivative 4, with terminal hydroxyl groups, and poly(ethylene glycol) methyl ether tosylate (average Mn = 900 g mol−1) according to Scheme S1. The dipyrene derivative 4 was obtained from the Suzuki-Miyaura cross-coupling reaction between pyrene derivative 1 and diboronic acid derivative 3, the latter of which was synthesized from 2,2-dibutyl-1,3-propanediol in a three-step process (see Scheme S1 and Data S1–S7).
Figure 2.
In vitro spectroscopic properties of Pyr-A
(A) Structure of Pyr-A.
(B–E) (B) UV-Vis absorption and (C) fluorescence emission spectra of Pyr-A were acquired in various solvents. The fluorescence intensity ratio I470/400 of excimer emission (at 470 nm) and monomer emission (at 400 nm) was measured as a function of (D) the solvent polarity parameter ET(30) and (E) the viscosity of methanol/PEG 600 mixtures ([Pyr-A] = 1.0 μM; excitation wavelength, 358 nm). Viscosities of the solvent mixtures were calculated from the molar fraction of PEG 600, based on the relationship reported previously (Mousavi et al., 2020).
Absorption and fluorescence spectra of Pyr-A were measured in various solvents to elucidate its emission behavior as a function of microenvironment (Figure 2). An intense absorption band was observed in the range 320–400 nm (Figure 2B), which are similar to that observed for 1,6-diphenylpyrene derivatives (Imai et al., 2012) and red-shifted, intensified, and broadened (or vibrationally unstructured) compared with unmodified pyrenes due to the modification of the π-conjugation system of pyrene moieties by adjacent phenyl rings. An intense absorption band was observed in the range 320–400 nm (Figure 2B), similar to that observed for 1,6-diphenylpyrene derivatives. The molar absorption coefficients in nonaqueous solutions (ε = 7.6–8.2 × 104 cm−1 M−1 at the peak wavelength (Table S1)) were approximately twice as large as those of 1,6-diphenylpyrene (ε = 4 × 104 cm−1 M−1 in chloroform [Imai et al., 2012]), indicating that the spectroscopic properties of Pyr-A in the near-UV and visible regions are attributable to their two 1,6-diphenylpyrene chromophores. These absorption bands were markedly broadened and weakened in aqueous solution, observations that have previously been attributed to aggregation of ground-state pyrene chromophores (Winnik et al., 1987).
Fluorescence emission spectra of Pyr-A comprised two bands near 400 nm and 470 nm, whose intensities varied with solvent type (Figure 2C). The shorter- and longer-wavelength bands are similar to those of monomer and excimer emission of other pyrenes except that sharp emission peaks in the monomer band were smeared out by adjacent phenyl rings, as reported previously (Imai et al., 2012; Mizoshita and Inagaki, 2018). These assignments were supported by time-resolved fluorescence measurements (Figure S1A, Table 1); although the fluorescence lifetime was shorter than that of unmodified pyrenes due to the radiation decay rate enhanced by adjacent phenyl rings, the longer-wavelength emission decayed slowly (tens of nanoseconds), whereas the shorter-wavelength emission decayed rapidly (within several nanoseconds), consistent with the long-lived nature of pyrene excimers (Weltman et al., 1973). The excimer emission was independent of Pyr-A concentration, indicating that the excimer formation is intramolecular (Figure S1B). The emission spectrum in aqueous solution exhibited peculiar behavior, with only the excimer band emerging with a markedly lower intensity. The fluorescence quenching was also evidenced by a shorter fluorescence lifetime (Figure S1B), which likely results from intramolecular dimer formation of ground-state pyrene chromophores (Lemmetyinen et al., 1991), considering the modification of the absorption spectra. The resulting low-fluorescence quantum yield in aqueous solution (Table S1) indicates that Pyr-A is an insensitive probe for water-rich environments.
In nonaqueous solutions, the excimer emission of Pyr-A was enhanced in polar protic and aprotic solvents compared with less-polar solvents such as tetrahydrofuran and benzene. The excimer emission efficiency was evaluated using the ratio of monomer (400 nm) and excimer (470 nm) emission intensities, I470/400. When plotted as a function of the polarity parameter, ET(30), of pure solvents (Figure 2D) and binary mixtures of methanol and tetrahydrofuran (Figure S1C), the I470/400 value was observed to increase at higher solvent polarities. The excimer emission was more prominent in polar aprotic solvents with lower viscosities, such as acetonitrile (3.4 × 10−4 Pa∙s) and acetone (3.0 × 10−4 Pa∙s), than in more polar protic solvents with higher viscosities, such as 1-propanol (2.8 × 10−3 Pa∙s) and 1-octanol (7.4 × 10−3 Pa∙s). The influence of solvent viscosity was examined for binary mixtures of methanol and polyethylene glycol 600 (PEG 600), whose viscosities increase monotonically as a function of the molar fraction of PEG 600 (Mousavi et al., 2020), but whose ET(30) values remain almost unchanged (54.2–55.4). The value of I470/400 was found to decrease with increasing viscosity of the solvent mixtures (Figure 2E), indicating that the ratio of monomer and excimer fluorescence emissions of Pyr-A is sensitive to both the polarity and viscosity of its surrounding environment. Notably, excimer emission was dominant in more polar and less viscous environments, whereas monomer emission was dominant in less polar and more viscous environments, such as those formed during LLPS.
Fluorescence microscopic analysis of LLPS in vitro and in vivo
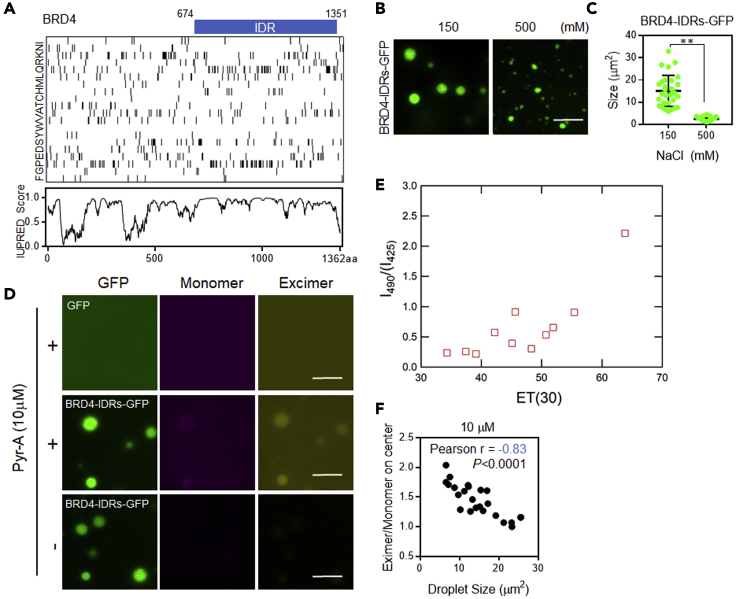
BRD4 is a coactivator involved in transcription regulation and its IDR forms droplets in vitro (Sabari et al., 2018). Therefore, we prepared a recombinant fusion protein comprising the IDR of BRD4 (BRD4-IDR) and monomeric enhanced green fluorescent protein (mEGFP), and analyzed BRD4-IDR–mEGFP protein droplets using fluorescent microscopic imaging (Figure 3A). Consistent with a previous report (Sabari et al., 2018), we first confirmed that the size distribution of BRD4-IDR–mEGFP droplets shifted to smaller values at high NaCl concentrations (Figures 3B and 3C). Next, we probed BRD4-IDR–mEGFP droplets using 10μM of Pyr-A and found that Pyr-A distributed inside microenvironments (Figure 3D). During these microscopic experiments, we detected monomer and excimer emission at 425 nm and 490 nm, respectively, due to the used optical filter characteristics, yielding the intensity ratios that show qualitatively similar solvent dependence to the case of I470/400 (Figure 3E). While a ratio metric measurement of the excimer/monomer fluorescence intensity (E/M) in buffer was around 2.2–2.3, the E/M values inside protein droplets showed an inverse relationship between E/M and droplet size (Figure 3F), suggesting that the protein droplets became hydrophobic and viscous during maturation. Importantly, both the size and the physicochemical traits of protein droplets were independent of Pyr-A concentrations (Figures S2A–S2D). In addition, the saturating concentrations of the protein droplets in the presence of Pyr-A was similar with that in the absence of Pyr-A (Figure S3). Collectively, Pyr-A itself rarely alter the phase behavior of the proteins by introducing some abnormal interactions.
Figure 3.
In vitro fluorescence microscopic analysis of protein droplets
(A) Amino acid composition of BRD4 (upper). The blue bar represents the previously identified intrinsically disordered regions (IDRs) of BRD4 (Sabari et al., 2018). Each row represents information pertaining to a single amino acid. Black bars represent the occurrence of the indicated amino acid at that position in BRD4. Analysis of the BRD4 protein sequence for IDRs used the IUPred2A (https://iupred2a.elte.hu) (Dosztányi, 2018) combined web interface (lower). The amino acid position in the linear sequence is plotted on the horizontal axis, and the disorder tendency (IUPRED Score) is displayed on the vertical axis.
(B) Representative images of droplets at different salt concentrations. Recombinant fusion protein comprising IDRs of BRD4 and membrane-anchored enhanced green fluorescent protein mEGFP (BRD4-IDR–mEGFP) was diluted in droplet formation buffer to a final concentration of 10 μM and the indicated NaCl concentration.
(C) Quantification of droplets in each salt concentration (n = 10, Mean ± SD). ∗∗p<0.01, comparisons between groups were made using an unpaired t test.
(D) Representative images of Pyr-A (10 μM) in droplets containing mEGFP (Upper) or BRD4-IDR–mEGFP (Middle). BRD4-IDRs-mEGFP in the absence of Pyr-A stands as negative control (Bottom).
(E) The fluorescence intensity ratio I490/425 of excimer emission (at 490 nm) and monomer emission (at 425 nm) as a function of the solvent polarity parameter Et(30). The respective intensities were integrated over wavelength regions covered by the used optical filters (397–453 nm for monomer and 470–510 nm for excimer).
(F) Ratiometric measurement of excimer/monomer fluorescence intensity (E/M). The E/M ratio inside droplets was quantified (n = 24).
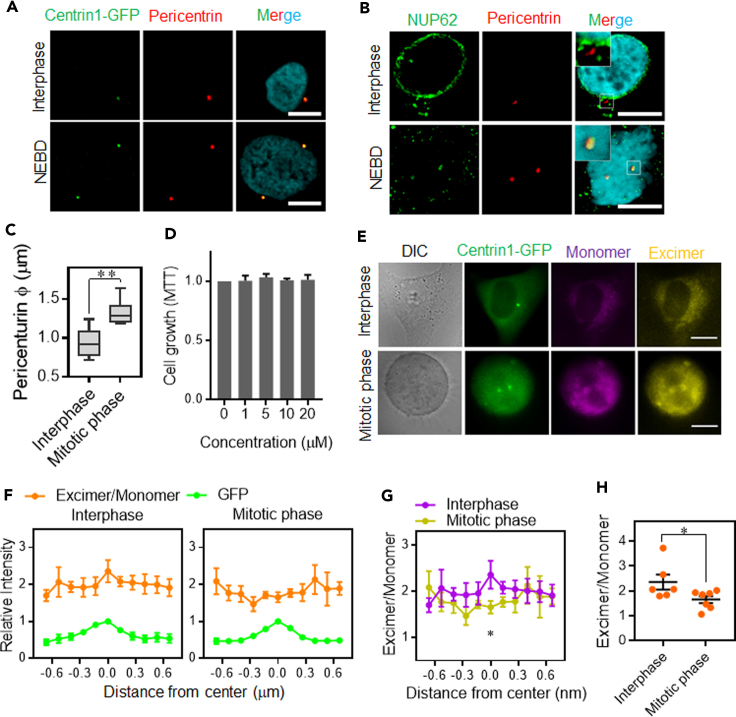
Additionally, we used Pyr-A to probe centrosomes inside cells. Centrosomes consist of a pair of centrioles embedded in PCM, whose constituents include centrin-1 and pericentrin (Figures 1B and 4A). After nuclear envelope breakdown, certain components of the nuclear pore complex, such as NUP62, localize to centrosomes (Figure 4B) and are involved in PCM layer maturation. Unsurprisingly, the size of centrosomes (measured from the spatial distribution of pericentrin fluorescence) in the mitotic phase was significantly larger than that in interphase (Figure 4C). To visualize centrosomes in live cells, we first established cells stably expressing the centrin1–GFP fusion protein (Figure S4). Next, we synchronized cells using thymidine blocking to enrich a mitotic population. Last, 10 μM of Pyr-A was applied before fluorescence microscopic imaging. Importantly, Pyr-A did not have any cytotoxic effects (Figure 4D).
Figure 4.
In vivo fluorescence microscopic analysis of centrosomes
(A and B) Confocal imaging of (A) centrosome markers centrin1 (green) and pericentrin (red) in HeLa cells expressing centrin1–GFP, and of (B) NUP62 (green) and pericentrin (red) in untransfected HeLa cells, acquired in the interphase and during nuclear envelope breakdown (NEBD) in the mitotic phase (scale bar, 10 μm).
(C) Quantification of centrosome foci in the interphase and mitotic phase (n = 10). Box plot element: medians with interquartile range and whiskers (min to max). ∗∗p<0.01, comparisons between groups were made using an unpaired t test.
(D) The effects of Pyr-A on cell viability, evaluated using the MTT assay (n = 3, mean ± SEM).
(E) Imaging of centrin1–GFP (green) and Pyr-A (monomer, purple; excimer, yellow) in HeLa cells expressing centrin1–GFP (scale bar, 20 μm).
(F–H) Line profiling of E/M in the centrosome region. (F) Intensity was normalized relative to the maximum GFP signal. E/M values of centrin1 in interphase and mitotic centrosomes were compared (G) along the lines and (H) at their center (interphase, n = 6; mitotic phase, n = 7; mean ± SEM). ∗p<0.05, comparisons between groups were made using an unpaired t test.
Both excimer and monomer emissions were observed inside cells, indicating that Pyr-A is cell permeable (Figure 4E). To profile E/M values in centrosomes, we measured E/M using line profile analyses (Figures 4F–H). The E/M value in the regions centered on centrin1–GFP signals was significantly lower in mitotic cells compared with interphase cells (Figures 4G and 4H). These results suggested that centrosomes in dividing cells grow by expanding the PCM and changing the microenvironment to be either more viscous or less polar.
Discussion
In this study, we developed a light-switching pyrene probe, Pyr-A, to monitor physicochemical properties of phase separation in centrosomes and purified protein. Importantly, monomer emission of Pyr-A was enhanced in hydrophobic and viscous solvents. The mean E/M value of Pyr-A reflects changes in the above physiochemical conditions, and fluorescent microscopic imaging of Pyr-A successfully monitored LLPS in in vitro protein droplets and centrosomes.
We demonstrated that the fluorescence emission of Pyr-A switched from excimer to monomer as the solvents became less polar and more viscous. This emission behavior is related to the favorable energetics and dynamics of the excimer formation. The intrinsically hydrophobic pyrene moieties favor intramolecular association in water-rich environments but behave as isolated monomers in hydrophobic environments. Excimer formation also requires a diffusion-controlled encounter between excited-state and ground-state pyrene moieties. Because the lifetime of the excited pyrene is limited, it is difficult to form excimers as solvent viscosities increase, resulting in predominant monomer emission.
Fluorescent proteins and chemical probes are valuable tools for studying dynamic processes within living cells. However, one rarely achieves a quantitative biophysical/physiochemical understanding of condensates. Here, we have revealed that the hydrophobicity and viscosity of target regions can be quantitatively probed by measuring the E/M ratio of Pyr-A. Although the current approach requires counterstaining with specific markers to spatially define the targets, further modification with functionalized nuclear localization signal peptides or antisense sequences specific for target RNAs will allow us to control the intracellular Pyr-A destination (Leung et al., 2018; Xu et al., 2010). These aspects will be addressed in the near future.
Centrosomes increase in size during mitosis by establishing a clouded PCM structure (Fu and Glover, 2012). Our results might reflect both the ability of the mitotic centrosomes to compartmentalize via LLPS and the integrity of the assembled PCM, which could be critical for forming two equally sized spindle poles during mitosis. Recent studies demonstrated that phosphorylation of the PCM by Polo-like kinase 1 is required for PCM expansion (Cabral et al., 2019), and also phosphatase activity of protein phosphatase 2A is involved in regulation of centrosome clustering in cancer cells (Antao et al., 2019). In addition, acetylation/deacetylation of RNA helicase DDX3X controls LLPS and stress granule assembly (Saito et al., 2019), and hyperacetylated proteins form amyloid-like intracellular aggregates (Olzscha et al., 2017). These findings underscore the role of post-translational modifications in LLPS growth and disassembly. Therefore, how these modifications affect the physicochemical dynamics of the microenvironment during LLPS needs to be addressed.
In conclusion, we have demonstrated that the bipyrene analogue Pyr-A can probe the polarity and viscosity of the microenvironment within phase-separated liquid droplets. Integrative analysis of these physicochemical dynamics and biological processes could provide important steps toward understanding the molecular determinants of phase separation.
Limitation of the study
In this study, E/M ratio data of current Pyr-A reflect both polarity and viscosity. To improve this convoluted evaluation, adjusting hydrophobic properties of pyrene will be required to specifically evaluate either polarity or viscosity. Nonetheless, we demonstrated that Pyr-A have great potential as chemical probe to evaluate phase-separated condensates without complicated steps. Combined approach with other analysis as such FRAP will enhance our understanding on LLPS, and we would like to address those issue in near future.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-b-actin rabbit polyclonal antibody | Cell Signaling Technology | Cat#4967; RRID:AB_330288 |
| anti-GFP mouse monoclonal antibody | FUJIFILM Wako Pure Chmicals | Cat#012-20461; RRID:AB_664697 |
| anti-Nup62 rat monoclonal antibody | Sigma-Aldrich | Cat#N1163; RRID:AB_1079511 |
| anti-Pericentrin rabbit polyclonal antibody | Abcam | Cat#ab4448; RRID:AB_304461 |
| Goat anti-Mouse Secondary Antibody Alexa Fluor 488 | Invitrogen | Cat#A11029; RRID:AB_138404 |
| Goat anti-Rabbit Secondary Antibody Rhodamine Red-X | Invitrogen | Cat#R6394; RRID:AB_2556551 |
| Goat anti-Rat Secondary Antibody Alexa Fluor 488 | Invitrogen | Cat#A11006; RRID:AB_141373 |
| Anti-mouse IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7076; RRID:AB_330924 |
| Anti-rabbit IgG, HRP-linked Antibody | Cell Signaling Technology | Cat#7074; RRID:AB_2099233 |
| Bacterial and virus strains | ||
| NEB 5-alpha Competent E. coli (subcloning Efficiency) | New England Biolabs | Cat#C2988J |
| Chemicals, peptides, and recombinant proteins | ||
| BamH1 | TOYOBO | Cat#BAH-111 |
| Kpn1 | New England Biolabs | Cat#R0142S |
| pEGFP-C1 | TaKaRa Clontech | Cat#6084-1 |
| LB broth (Lennox) | Sigma-Aldrich | Cat#L3022 |
| Kanamycin Sulfate | FUJIFILM Wako Pure Chmicals | Cat#117-00704 |
| PEG-Pyr | this study | N/A |
| FBS | Gibco | Cat#10270-106 |
| G418 Disulfate | Nacalai Tesque | Cat#16512-52 |
| Thymidine | FUJIFILM Wako Pure Chmicals | Cat#203-19423 |
| Lipofectamine™ 2000 Transfection Reagent | Invitrogen | Cat#11668019 |
| Opti-MEM™ | Gibco | Cat#31985-070 |
| Penicillin- Streptomycin-AnphotericinB suspension | FUJIFILM Wako Pure Chmicals | Cat#161-23181 |
| MTT 2-(4,5-dimethyl-2-thiazolyl)-3,5-diphenyl-2H-tetrazolium, monobromide | Cayman Chemical | Cat#21795 |
| DMF | Nacalai Tesque | Cat#13016-65 |
| Paraformaldehyde | Nacalai Tesque | Cat#26123-55 |
| BSA | FUJIFILM Wako Pure Chmicals | Cat#015-23295 |
| ProLong™ Diamond Antifade Mountant with DAPI | Invitrogen | Cat#P36962 |
| Protease inhibitor cocktail (EDTA free) (100x) | Nacalai Tesque | Cat#03969-21 |
| Immobilon Western Chemiluminescent HRP Substrate | Merck | Cat#WBKLS0500 |
| Immobilon-P PVDF | Merck | Cat#IPVH00010 |
| Tween20 Polyoxyethylene Sorbitan Monolaurate | FUJIFILM Wako Pure Chmicals | Cat#167-11515 |
| Pyr-A | This manuscript | N/A |
| Experimental models: Cell lines | ||
| HeLa | ATCC | N/A |
| Recombinant DNA | ||
| pEGFP-C1-Centrin1 | This study | N/A |
| Software and algorithms | ||
| Axio Vision 4.8 | Zeiss | N/A |
| FV10-ASW4.2 | Olympus | N/A |
| Graph Pad Prism 7 | Graph Pad | N/A |
| Multi Gauge V3.2 | FUJIFILM Wako Pure Chmicals | N/A |
| Other | ||
| 35mm glass bottom dish | MATSUNAMI | Cat#D11130H |
| flow view FV10i | Olympus | N/A |
| LAS-4000 image analyzer | FUJIFILM Wako Pure Chmicals | N/A |
| Multiskan GO | Thermo Fisher Scientific | N/A |
| Zeiss LSM5 EXCITER confocal microscope | Zeiss | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Masaharu Hazawa (masaharu.akj@gmail.com)
Materials availability
All unique reagents generated in this study are available from the lead contact with a completed Material Transfer Agreement.
Experimental model and subject details
Cell lines and primary cultures
The following cancer cell lines were used in this study.
1. HeLa cells (RRID: CVCL_0030)
Origin: Human endocervical adenocarcinoma cell line obtained from a 30-year-old female.
HeLa cells were obtained from ATCC and cultured in Dulbecco's Modified Eagle Medium (Invitrogen) with 10% fetal bovine serum and penicillin/streptomycin in a humidified atmosphere with 5% CO2 at 37°C. We did not authenticate this cell line in our laboratory.
2. SCC13 cells (RRID: CVCL_4029)
Origin: Human skin squamous cell carcinoma cell line obtained from a 56-year-old female.
SCC13 cells were kindly provided by Dr Timothy Chan (Memorial Sloan Kettering Cancer Center) and were cultured in Dulbecco's Modified Eagle Medium (Invitrogen) with 10% fetal bovine serum and penicillin/streptomycin in a humidified atmosphere with 5% CO2 at 37°C. The cell line was authenticated by short tandem repeat analysis with the Geneprint 10 System Kit (B9510, Promega, Madison, WI, USA) in 2014 described previously (Hazawa et al., 2017).
Method details
Synthesis of Pyr-A
Synthesis of 1
The mixture of 1,6-dibromopyrene (1.80 g, 5.0 mmol), 4-(hydroxymethyl)phenylbronic acid (0.76 g, 5.0 mmol), potassium carbonate (2 M in water, 13 mL) and toluene (80 mL) was deoxygenated by bubbling with nitrogen gas for 40 min. To the mixture was added Pd(PPh3)4 (208 mg, 0.18 mmol) was added to the mixture, and further deoxygenated by bubbling with nitrogen gas for 10 min, heated at 100°C under N2. While heating, deoxygenated ethanol (10 mL) was added to the mixture. After heating for 20 h, the resulting mixture was cooled to room temperature, poured into water, then extracted with mixture of CH2Cl2. The organic layer was washed with water, dried over anhydrous Na2SO4, then evaporated to dryness. The residue was purified by silica gel column chromatography (methanol/CH2Cl2=1/99) to obtain 1 as a pale yellow solid (1.05 g, 2.7 mmol, 54%).
Synthesis of 2
Under N2, 2,2,-dibutyl-1,3-propanediol (3.85 g, 20.5 mmol) in THF (70 mL) was slowly added to NaH (60% dispersion in oil, 2.45 g, 61.4 mmol) in THF (110 mL) at 0°C. After the mixture was stirred at 0°C for 1 hour, 4-bromobenzyl bromide (15.36 g, 61.4 mmol) in THF (15 mL) was slowly added at 0°C, and then warmed up to room temperature. Dry DMF (50 mL) was added to the mixture and stirred 1 week at room temperature. The resulting mixture was poured into ice water and then extracted with mixture of hexane and ethylacetate (3:1). The organic layer was washed with water, and dried over anhydrous Na2SO4, followed by evaporation to dryness. The residue was purified by silica gel column chromatography (CH2Cl2/Hexane=5/95) to obtain 2 as a white solid (6.81 g, 12.9 mmol, 63%).
Synthesis of 3
The mixture of 1 (1.05 g, 2.0 mmol), bis(pinacolato)diboron (1.12 g, 4.4 mmol), potassium acetate (1.30 g, 13.2 mmol) and dioxane (14 mL) was deoxygenated by bubbling with nitrogen gas for 40 min. [1,1′-bis(diphenylphosphino)ferrocene]dichloropalladium dichloromethane adduct (50 mg) was added to the mixture, and further deoxygenated by bubbling with nitrogen gas for 20 min, heated at 80°C under N2. Additional Pd catalyst (50 mg) and bis(pinacolato)diboron (67 mg) were added to the mixture. After heating for 30 h, the resulting mixture was cooled to room temperature, poured into water, and then extracted with a mixture of hexane and ethylacetate (4:1). The organic layer was washed with brine, and dried over anhydrous Na2SO4, followed by evaporation to dryness. The residue was purified by silica gel column chromatography (Ethylacetate/Hexane=5/95) to obtain 3 as a transparent oil (0.56 g, 0.9 mmol, 45%).
Synthesis of 4
The mixture of 3 (400 mg, 0.64 mmol), 1 (548 mg, 1.41 mmol), potassium carbonate (2 M in water, 2 mL), ethanol (2 mL) and toluene (4 mL) was deoxygenated by bubbling with nitrogen gas for 30 min. Pd(PPh3)4 (45 mg, 0.04 mmol) was added to the mixture and further deoxygenated by bubbling with nitrogen gas for 20 min, and refluxed under N2 for 15 h. The resulting mixture was cooled to room temperature and then filtrated. The residue was washed with water, methanol and CH2Cl2, and dried in vacuo to obtain compound 4 as a pale yellow solid (531 mg). This compound was used for the next reaction without further purification. HRMS(FAB) Calcd for C71H65O4 [(M+H)+]: m/z 981.4883, Found: m/z 981.4894.
Synthesis of Pyr-A
NaH (60% dispersion in oil, 20 mg, 0.29 mmol) was slowly added to the mixture of 4 (36 mg, 0.06 mmol), dry THF (0.5 mL) and dry DMF (0.6 mL) at 0°C under N2. After stirring for 2 h at 0°C, to the mixture was added poly(ethylene glycol) methyl ether tosylate (Mn = 900 gmol-1, 156 mg, 0.24 mmol) and warmed up to room temperature, and then stirred for 35 h. The resulting mixture was poured into saturated NH4Cl aq. and then extracted with CH2Cl2. The organic layer was washed with water, and dried over anhydrous Na2SO4, followed by evaporation to dryness. The residue was purified by silica gel column chromatography (methanol/CH2Cl2=2/98 to 5/95) and size exclusion chromatography to obtain Pyr-A as a pale yellow solid (48 mg, 0.019 mmol, 32%).
Instrumentation
1H and 13C NMR measurements were recorded on a Bruker Avance Neo 400 instrument (1H 400 MHz and 13C 100 MHz). Elemental analysis, FAB mass spectroscopy and EI mass spectroscopy were performed at the Research Institute for Instrumental Analysis, Advanced Science Research Center, Kanazawa University.
Cell proliferation assay
Cells were seeded into a 96-well plate at 3000 cells/well and cultured under the indicated concentrations of Pyr-A. Cell viability was assessed using the MTT (3-(4, 5-dimethylthiazol-2-yl) -2, 5-diphenyltetrazolium bromide) method (Hazawa et al., 2018, 2020). In brief, 10 μL of 12 mM MTT solution was added into each well followed by 3 h incubation. The reaction was stopped by adding 100 μL of STOP solution (2% acetic acid, 16% SDS, 42% DMF). Samples were mixed thoroughly, and absorbance was measured at 570 nm.
cDNA vector
The centrin1–EGFP vector was made by cloning cDNA from HeLa cells into a C1–EGFP vector using KpnI/BamHI restriction sites. IDRs of BRD4 (674-1351aa) was picked out from the pcDNA5-Flag-BRD4-WT vector (addgene # 90331) and ligated into the N1-EGFP vector using XhoI and BamhI (TOYOBO, BAH-111) with following primers: F1: 5’-CTCGAGGCCACCATG TGTTTGCGGAAGAAAAGG-3’; R1: 5’-GGATCCCGACTCTGGAAATTCATG-3’. Further, BRD4-IDRs-EGFP was amplified with F2: 5’-GAATTCATGTGTTTGCGGAAGAAAAGGAAACC-3’; R2: 5’-GCGGCCGCTTACTTGTACAGCTCGTC-3’, and ligated into the pET28a vector using EcoRI/NotI.
Transfection
DNA transfection was performed using Lipofectamine 2000 (Life Technologies). HeLa cells stably expressing centrin1–EGFP were selected following G418 treatment.
Protein purification
Constructed pET plasmid was transformed into Arctic Express cells and grown as follows. A fresh bacterial colony was inoculated into LB media containing kanamycin and grown 16 hours at 37°C. The LB solution containing cells were diluted 1:50 in 1 L LB solution and grown 3 hours at 30°C. IPTG was added to 1 mM and growth continued for 24 hours at 13°C. Cells were collected and stored frozen at 80°C.
Pellets from 1L LB were resuspended in 30ml buffer A (50 mM Tris HCl pH 7.5, 500 mM NaCl, 10 mM Imidazole) containing a protease inhibitor and disrupted using a French press. The supernatant obtained after lysate centrifugation was applied to His Trap HP 1ml (Cytiva) pre-equilibrated with buffer A. The column was washed with buffer A and eluted with buffer B (50 mM Tris HCl pH 7.5, 500 mM NaCl, 200 mM Imidazole) containing a protease inhibitor. Eluted protein was then concentrated by centrifuge filter unit (Amicon Ultra). BRD4-IDR-GFP was further purified by gel filtration chromatography. Glycerol was added to the purified protein to 10% and stored at -80°C until use.
Spectrophotomeric and spectrofluoromeric measurements
UV-visible absorption and fluorescence spectra of Pyr-A in various solvents were measured using a spectrophotometer (JASCO, V-750) and spectrofluorometer (JASCO, FP-8300), respectively. The temperature was maintained at 25±1°C during measurement. The concentration of Pyr-A was around 1.0 μM. Fluorescence quantum yield (Φf) was determined using quinine-sulfate (Φstd = 0.55) as a fluorescence reference standard. ET(30) values of solvent mixtures were evaluated from the wavelengths of absorption maxima of Betaine 30.
Time-resolved fluorescence measurements
Fluorescence decay profiles of Pyr-A were monitored using a high-speed photodiode detector following excitation by a pulsed laser. The excitation pulses at 355 nm (5 ns, 1.0 mJ) were provided by the third harmonic of a Nd:YAG laser (Amplitude, Minilite I). The fluorescence of monomer and excimer were selected by placing 400 nm and 470 nm bandpass filters (Thorlabs, FWHM 10 nm) respectively, in front of the detector, and recorded using a digital oscilloscope (Tektronix, TDS784D). The instrumental response function was measured to be 5 ns by monitoring the temporal profile of scattered light of the excitation pulses by pure methanol solvent. The fluorescence lifetimes of monomer (τM) and excimer (τE) were determined by fitting the data with a monoexponential decay function after deconvolution with the instrumental response function.
Immunocytochemistry and confocal microscopy
Anti-GFP (012-20461) mouse monoclonal antibody was obtained from Wako. Anti-pericentrin rabbit polyclonal antibody (ab4448) was acquired from Abcam. Anti-Nup62 rat monoclonal antibody (N1163) was obtained from Sigma-Aldrich. Secondary antibodies were acquired from Molecular Probes (Life Technologies). For immunofluorescence, synchronized HeLa cells were washed in phosphate-buffered saline (PBS, used at a 1× dilution) and fixed for 10 min in 4% paraformaldehyde at room temperature. Cells were then permeabilized with 0.3% Triton X-100 in PBS (1×) for 10 min at room temperature. For Nup62 staining, cells were washed with PBS (1×) and permeabilized with 0.3% Triton X-100 in PBS for 4 min before fixation, then fixed with 4% paraformaldehyde for 10 min. Coverslips were blocked with 4% bovine serum albumin in PBS (1×) for 30 min and incubated with primary antibodies for 2 h at room temperature and then secondary antibodies for 2 h at room temperature. Coverslips were mounted onto glass slides using Pro-Long Gold Antifade reagent (Life Technologies) and examined using a confocal microscope (FluoView® FV10i, Olympus, objective ×60/1.2). All confocal images were analyzed using FV10i software (version 4.1).
In vitro droplet assay
Recombinant mEGFP protein was obtained from ORIGENE (Sku#TP790050). Recombinant mEGFP fusion protein (BRD4aa674–1351; BRD4-IDR–mEGFP; see supplemental information) was concentrated to an appropriate level using Amicon Ultra centrifugal filters (50K MWCO, Millipore). Recombinant protein was diluted in to a final concentration of 10 μM in droplet buffer (50 mM Tris–HCl pH 7.5, 10% glycerol, 1 mM DTT, 10% PEG) containing the desired NaCl concentration, with or without 10 μM of Pyr-A, then immediately loaded on a glass-bottom dish (MATSUNAMI) and covered with a coverslip. Samples were imaged using a Zeiss LSM5 EXCITER microscope with a mercury lamp and a Plan-Apochromat 100×/1.4 oil objective. Monomer emission was examined using a customized fluorescent cube I (Ex365/DM390/Em425, Zeiss), while excimer emission was monitored using fluorescent cube II (Ex365/DM400/Em490, Zeiss). Axio Vision software (version 4.8) was used for image acquisition.
Live-cell fluorescence microscopic imaging
Cells were seeded onto a glass-bottom dish (MATSUNAMI) and incubated for 24 h to allow attachment. Then, cells were synchronized using thymidine block to enrich a mitotic population (Hashizume et al., 2013) and incubated in medium containing Pyr-A (10 μM). After 1 h treatment, cells were rinsed twice with PBS (1×) and fresh culture medium was added. Cells were then examined using a Zeiss LSM5 EXCITER microscope with a mercury lamp and a Plan-Apochromat 100×/1.4 oil objective. Axio Vision software (version 4.8) was used for acquisition. To identify the centrosome regions, approximately 30 image slices (0.2 mm thickness) were obtained, and the location with maximum centrin1–GFP signal was selected for line profile analysis.
Quantification and statistical analysis
Comparisons between groups were made using an unpaired t test. p < 0.05 was considered statistically significant. The Pearson correlation coefficient, r, was determined using GraphPad Prism.
Acknowledgments
This work was supported by Cross-Disciplinary Research Promotion Expenses (InFiniti, Kanazawa University; to M.H. and S.A.), the Center of Innovation Science and Technology based Radical Innovation and Entrepreneurship Program (COI stream, (JPMJCE1315)) aimed at the “Construction of next generation infrastructure using innovative materials: Realization of a safe and secure society that can coexist with the Earth for centuries” (to K.T.), the Ministry of Education, Culture, Sports, Science and Technology (MEXT) and the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers Grant Number 20K07568 (to M.H.), 18K14187 (to S.A.), 19K05521 and 19H04674 (to Y.N.), 21K19043 (to R.W.W.) and the Kobayashi International Scholarship Foundation (to R.W.W.), and the Shimadzu Science Foundation (to R.W.W.). We thank Edanz Group (https://en-author-services.edanz.com/ac) for editing the English text of a draft of this manuscript.
Author contributions
M.H., S.A., Y.N., and R.W.W. devised the research strategy. Y.I. and M.H. performed biological experiments and in vivo fluorescence imaging. S.A. synthesized the fluorescent probe. Y.N. characterized the in vitro fluorescent properties. Y.I. and A.K. constructed plasmid DNA and purified recombinant protein. M.H., S.A., and Y.N. wrote the initial draft of the manuscript, and H.N., M.M., K.T., and R.W.W. joined to finalize the manuscript.
Declaration of interests
Kanazawa University holds patent (2021–93,100) on the pyrene analogue to probe phase separated biomolecules.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102865.
Contributor Information
Masaharu Hazawa, Email: masaharu.akj@gmail.com.
Richard W. Wong, Email: rwong@staff.kanazawa-u.ac.jp.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. This study did not generate such unique code.
References
- Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemori S., Kikuchi K., Mizuno M. Poly(dimethylsiloxane) and oligo(dimethylsiloxane) solvent effects on aromatic donor-acceptor interactions. Chem. Commun. 2021;57:1141–1144. doi: 10.1039/d0cc06638a. [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Antao N.V., Marcet-Ortega M., Cifani P., Kentsis A., Foley E.A. A cancer-associated missense mutation in PP2A-Aα increases centrosome clustering during mitosis. IScience. 2019;19:74–82. doi: 10.1016/j.isci.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., Hyman A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- Burke K.A., Janke A.M., Rhine C.L., Fawzi N.L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral G., Laos T., Dumont J., Dammermann A. Differential requirements for centrioles in mitotic centrosome growth and maintenance. Dev. Cell. 2019;50:355–366.e6. doi: 10.1016/j.devcel.2019.06.004. [DOI] [PubMed] [Google Scholar]
- Caputo V.S., Trasanidis N., Xiao X., Robinson M.E., Katsarou A., Ponnusamy K., Prinjha R.K., Smithers N., Chaidos A., Auner H.W. Brd2/4 and Myc regulate alternative cell lineage programmes during early osteoclast differentiation in vitro. IScience. 2021;24:101989. doi: 10.1016/j.isci.2020.101989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colino-Sanguino Y., Cornett E.M., Moulder D., Smith G.C., Hrit J., Cordeiro-Spinetti E., Vaughan R.M., Krajewski K., Rothbart S.B., Clark S.J. A read/write mechanism connects p300 bromodomain function to H2A.Z. Acetylation. Iscience. 2019;21:773–788. doi: 10.1016/j.isci.2019.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conicella A.E., Zerze G.H., Mittal J., Fawzi N.L. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z. Prediction of protein disorder based on IUPred. Protein Sci. 2018;27:331–340. doi: 10.1002/pro.3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C.H., Eckmann C.R., Myong S., Brangwynne C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. U S A. 2015;112:7189–7194. doi: 10.1073/pnas.1504822112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M., Vaidya N., Harmon T.S., Mitrea D.M., Zhu L., Richardson T.M., Kriwacki R.W., Pappu R.V., Brangwynne C.P. Coexisting liquid phases underlie nucleolar subcompartments. Cell. 2016;165:1686–1697. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann T.M., Jahnel M., Pozniakovsky A., Mahamid J., Holehouse A.S., Nüske E., Richter D., Baumeister W., Grill S.W., Pappu R.V. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359:eaao5654. doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- Fu J., Glover D.M. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadella T.W.J., Moritz A., Westerman J., Wirtz K.W.A. Enzymatic synthesis of pyrene-labeled Polyphosphoinositides and their behavior in organic solvents and phosphatidylcholine bilayers. Biochemistry. 1990;29:3389–3395. doi: 10.1021/bi00465a035. [DOI] [PubMed] [Google Scholar]
- Ganser L.R., Myong S. Methods to study phase-separated condensates and the underlying molecular interactions. Trends Biochem. Sci. 2020;45:1004–1005. doi: 10.1016/j.tibs.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E., Shorter J. The molecular language of membraneless organelles. J. Biol. Chem. 2019;294:7115–7127. doi: 10.1074/jbc.TM118.001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume C., Moyori A., Kobayashi A., Yamakoshi N., Endo A., Wong R.W. Nucleoporin Nup62 maintains centrosome homeostasis. Cell Cycle. 2013;12:3804–3816. doi: 10.4161/cc.26671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M., Lin D.C., Handral H., Xu L., Chen Y., Jiang Y.Y., Mayakonda A., Ding L.W., Meng X., Sharma A. ZNF750 is a lineage-specific tumour suppressor in squamous cell carcinoma. Oncogene. 2017;36:2243–2254. doi: 10.1038/onc.2016.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M., Lin D., Kobayashi A., Jiang Y., Xu L., Dewi F.R.P., Mohamed M.S., Hartono, Nakada M., Meguro-Horike M. ROCK -dependent phosphorylation of NUP 62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018;19:73–88. doi: 10.15252/embr.201744523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M., Sakai K., Kobayashi A., Yoshino H., Iga Y., Iwashima Y., Lim K.S., Chih-Cheng Voon D., Jiang Y.Y., Horike S.ichi. Disease-specific alteration of karyopherin-α subtype establishes feed-forward oncogenic signaling in head and neck squamous cell carcinoma. Oncogene. 2020;39:2212–2223. doi: 10.1038/s41388-019-1137-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Vega A., Braun M., Scharrel L., Jahnel M., Wegmann S., Hyman B.T., Alberti S., Diez S., Hyman A.A. Local nucleation of microtubule bundles through tubulin concentration into a condensed tau phase. Cell Rep. 2017;20:2304–2312. doi: 10.1016/j.celrep.2017.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- Imai K., Sasaki T., Abe J., Nemoto N. Synthesis and properties of poly(tetramethyl-1,6-silpyrenylenesiloxane) derivative with phenyl groups on pyrenylene moiety. Polym. Bull. 2012;68:1589–1601. [Google Scholar]
- Imai J.H., Wang X., Shi S.H. Kaede-Centrin1 labeling of mother and daughter centrosomes in mammalian neocortical neural progenitors. Curr. Protoc. Stem Cell Biol. 2010:1–14. doi: 10.1002/9780470151808.sc05a05s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdon E.M., Qiu Y., Niaki A.G., Mclaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M. mRNA structure determines specificity of a polyQ-driven phase separation. Science. 2018;360:922–927. doi: 10.1126/science.aar7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmetyinen H., Ikonen M., Mikkola J. A kinetic study of monomer and excimer fluorescence of fyrene lecithin in Langmuir-Blodgett Films. Thin Solid Films. 1991;204:417–439. [Google Scholar]
- Leung H.M., Chan M.S., Liu L.S., Wong S.W., Lo T.W., Lau C.H., Tin C., Lo P.K. Dual-function, cationic, peptide-coated nanodiamond systems: facilitating nuclear-targeting delivery for enhanced gene therapy applications. ACS Sustain. Chem. Eng. 2018;6:9671–9681. [Google Scholar]
- Majumdar A., Dogra P., Maity S., Mukhopadhyay S. Liquid-liquid phase separation is driven by large-scale conformational unwinding and fluctuations of intrinsically disordered protein molecules. J. Phys. Chem. Lett. 2019;10:3929–3936. doi: 10.1021/acs.jpclett.9b01731. [DOI] [PubMed] [Google Scholar]
- Mitrea D.M., Chandra B., Ferrolino M.C., Gibbs E.B., Tolbert M., White M.R., Kriwacki R.W. Methods for physical characterization of phase-separated bodies and membrane-less organelles. J. Mol. Biol. 2018;430:4773–4805. doi: 10.1016/j.jmb.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoshita N., Inagaki S. Enhanced photoluminescence of mesostructured organosilica films with a high density of fluorescent chromophores. Macromol. Chem. Phys. 2018;219:1–7. [Google Scholar]
- Mousavi Z., Pirdashti M., Rostami A.A., Dragoi E.N. Thermophysical properties analysis of poly (ethylene glycol) 600 + methanol, ethanol, 1-propanol, and 2-propanol binary liquid mixtures. Int. J. Thermophys. 2020;41:1–26. [Google Scholar]
- Murthy A.C., Fawzi N.L. The (un)structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy. J. Biol. Chem. 2020;295:2375–2384. doi: 10.1074/jbc.REV119.009847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzscha H., Fedorov O., Kessler B.M., Knapp S., La Thangue N.B. CBP/p300 bromodomains regulate amyloid-like protein aggregation upon aberrant lysine acetylation. Cell Chem. Biol. 2017;24:9–23. doi: 10.1016/j.chembiol.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey A.E., Holehouse A.S., Pappu R.V. Elsevier Inc.; 2018. Phase Separation of Intrinsically Disordered Proteins. [DOI] [PubMed] [Google Scholar]
- Reichheld S.E., Muiznieks L.D., Keeley F.W., Sharpe S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl. Acad. Sci. U S A. 2017;114:E4408–E4415. doi: 10.1073/pnas.1701877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabari B.R., Dall’Agnese A., Boija A., Klein I.A., Coffey E.L., Shrinivas K., Abraham B.J., Hannett N.M., Zamudio A.V., Manteiga J.C. Coactivator condensation at super-enhancers links phase separation and gene control. Science. 2018;361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito M., Hess D., Eglinger J., Fritsch A.W., Kreysing M., Weinert B.T., Choudhary C., Matthias P. Acetylation of intrinsically disordered regions regulates phase separation. Nat. Chem. Biol. 2019;15:51–61. doi: 10.1038/s41589-018-0180-7. [DOI] [PubMed] [Google Scholar]
- Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;80:357. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- Shin Y., Chang Y.C., Lee D.S.W., Berry J., Sanders D.W., Ronceray P., Wingreen N.S., Haataja M., Brangwynne C.P. Liquid nuclear condensates mechanically sense and restructure the genome. Cell. 2018;175:1481–1491.e13. doi: 10.1016/j.cell.2018.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerharju P. Pyrene-labeled lipids as tools in membrane biophysics and cell biology. Chem. Phys. Lipids. 2002;116:57–74. doi: 10.1016/s0009-3084(02)00020-8. [DOI] [PubMed] [Google Scholar]
- Tiwary A.K., Zheng Y. Protein phase separation in mitosis. Curr. Opin. Cell Biol. 2019;60:92–98. doi: 10.1016/j.ceb.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann S., Eftekharzadeh B., Tepper K., Zoltowska K.M., Bennett R.E., Dujardin S., Laskowski P.R., MacKenzie D., Kamath T., Commins C. Tau protein liquid–liquid phase separation can initiate tau aggregation. EMBO J. 2018;37:1–21. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M.T., Elbaum-Garfinkle S., Holehouse A.S., Chen C.C.H., Feric M., Arnold C.B., Priestley R.D., Pappu R.V., Brangwynne C.P. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017;9:1118–1125. doi: 10.1038/nchem.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weltman J.K., Szaro R.P., Frackelton A.R., Dowben R.M., Bunting J.R., Cathou B.E. N-(3-pyrene)maleimide: a long lifetime fluorescent sulfhydryl reagent. J. Biol. Chem. 1973;248:3173–3177. [PubMed] [Google Scholar]
- Winnik F.M., Winnik M.A., Tazuke S. Synthesis and characterization of pyrene-labeled (Hydroxypropyl) cellulose and its fluorescence in solution. Macromolecules. 1987;20:38–44. [Google Scholar]
- Xu Y., Suzuki Y., Ito K., Komiyama M. Telomeric repeat-containing RNA structure in living cells. Proc. Natl. Acad. Sci. U S A. 2010;107:14579–14584. doi: 10.1073/pnas.1001177107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Bertulat B., Tencer A.H., Ren X., Wright G.M., Black J., Cardoso M.C., Kutateladze T.G. MORC3 forms nuclear condensates through phase separation. IScience. 2019;17:182–189. doi: 10.1016/j.isci.2019.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ma Z., Wang Y., Boyer J., Ni G., Cheng L., Su S., Zhang Z., Zhu Z., Qian J. Streptavidin promotes DNA binding and activation of cGAS to enhance innate immunity. IScience. 2020;23:101463. doi: 10.1016/j.isci.2020.101463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. This study did not generate such unique code.